Analysis Report: HEK293_TMG_2hB vs HEK293_DMSO_2hB
WANG Ziyi
Date: 19 5月 2022
Summary: HEK293_TMG_2hB vs HEK293_DMSO_2hB
1. Differentially Expressed Genes
Heatmap
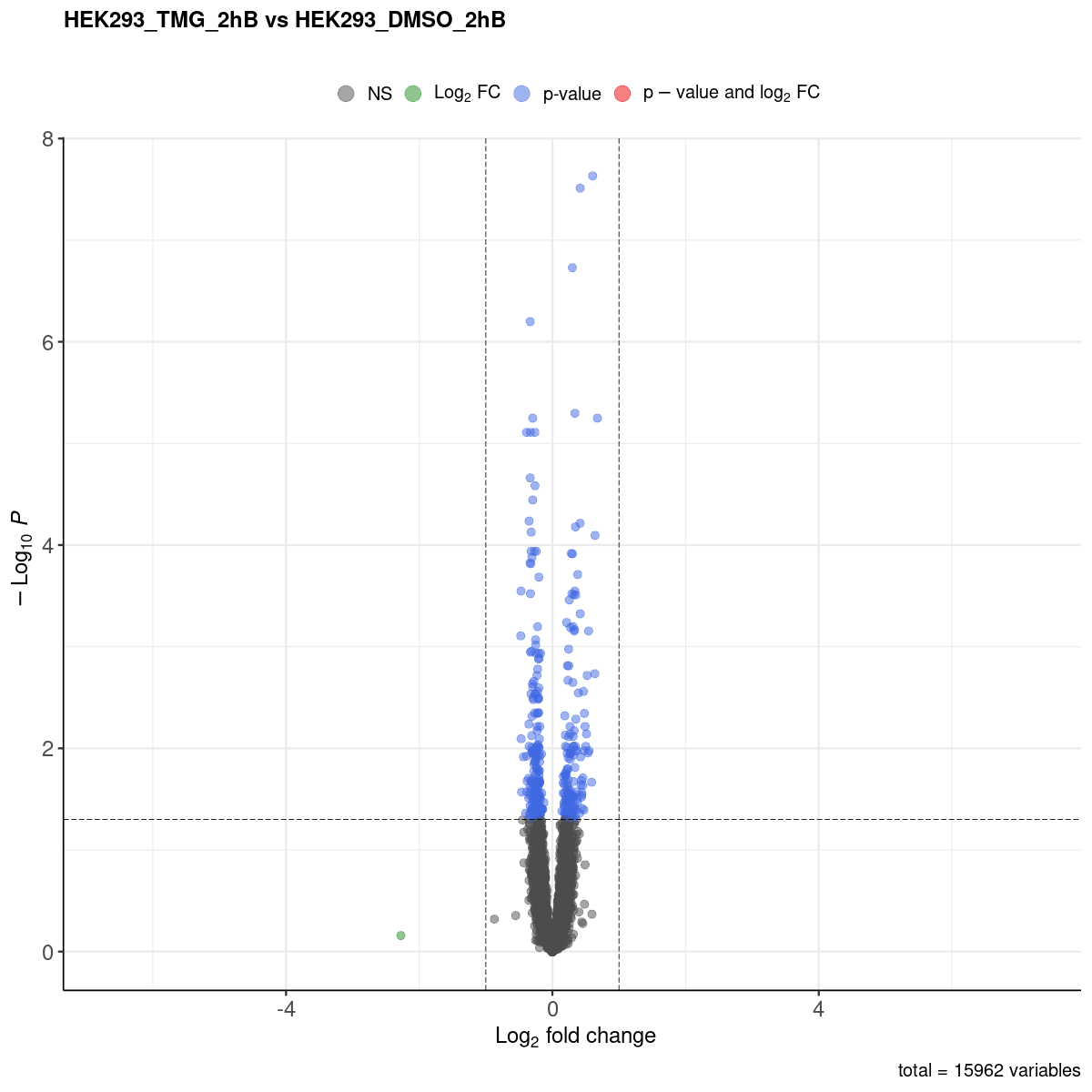
No significcant DEGs were detected under the threshold of P<0.05
Volcano plot
DEGs list
Spreadsheet
No significcant DEGs (P<0.05 and |log2FC|>1) or AS event (P<0.05 and dIF>0.05) were detected.
- Note:
- The spreadsheet above contains the genes with significantly changes in expression (P-value < 0.05 and |log2FC| > 1). Please Click HERE to check all mapped genes in a Microsoft .excel file.
- The comparisons made can be identified from the website name. “HEK293_TMG_2hB vs HEK293_DMSO_2hB” means that “HEK293_DMSO_2hB” is considered the ground state and “HEK293_TMG_2hB” the changed state. This also means that a positive log2FC value indicates that the gene expression is increased in “HEK293_TMG_2hB” compared to “HEK293_DMSO_2hB”.
- Click HERE to check all parameters of DESeq2 model for all detected genes of the current samples set in a Microsoft .excel file. For more detials of the DESeq2 model, please Click HERE.
Gene Biotype
## No significcant DEGs (P<0.05 and |log2FC|>1) or AS event (P<0.05 and dIF>0.05) were detected.AS in non-DEGs
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_TMG_2hB | SE_Intercept_HEK293_TMG_2hB | Intercept_HEK293_DMSO_2hB | SE_Intercept_HEK293_DMSO_2hB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000002016 | 5893 | RAD52 | protein_coding | P43351 | FUNCTION: Involved in double-stranded break repair. Plays a central role in genetic recombination and DNA repair by promoting the annealing of complementary single-stranded DNA and by stimulation of the RAD51 recombinase. {ECO:0000269|PubMed:12379650, ECO:0000269|PubMed:8702565}. | 3D-structure;Alternative splicing;DNA damage;DNA recombination;DNA repair;DNA-binding;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene shares similarity with Saccharomyces cerevisiae Rad52, a protein important for DNA double-strand break repair and homologous recombination. This gene product was shown to bind single-stranded DNA ends, and mediate the DNA-DNA interaction necessary for the annealing of complementary DNA strands. It was also found to interact with DNA recombination protein RAD51, which suggested its role in RAD51 related DNA recombination and repair. A pseudogene of this gene is present on chromosome 2. Alternative splicing results in multiple transcript variants. Additional alternatively spliced transcript variants of this gene have been described, but their full-length nature is not known. [provided by RefSeq, Jul 2014]. | hsa:5893; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; protein-DNA complex [GO:0032993]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; single-stranded DNA binding [GO:0003697]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to oxidative stress [GO:0034599]; DNA double-strand break processing involved in repair via single-strand annealing [GO:0010792]; DNA recombinase assembly [GO:0000730]; DNA recombination [GO:0006310]; double-strand break repair [GO:0006302]; double-strand break repair via homologous recombination [GO:0000724]; double-strand break repair via single-strand annealing [GO:0045002]; mitotic recombination [GO:0006312]; regulation of nucleotide-excision repair [GO:2000819] | 11456495_The ring-shaped quaternary structure of RAD52 and the formation of higher ordered complexes of rings appear to contribute to the extreme thermal stability of RAD52. 11691922_reduces double-strand break-induced homologous recombination with overexpression in mammalian cells 11809887_differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human tumor cell line 12023982_Observational study of gene-disease association. (HuGE Navigator) 12036913_Observational study of gene-disease association. (HuGE Navigator) 12139939_Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA 12191481_Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. 12370410_crystal structure of the single-strand annealing domain 12372413_RAD52 may play a role in transcription regulation and in targeting DNA damage on transcription active loci to recombinational repair 12376524_women with Ser346ter nonsense polymorphism of RAD52 not at increased risk of breast cancer in case-control study 12750383_coordinated WRN and RAD52 activities are involved in replication fork rescue after DNA damage 12883740_Observational study of gene-disease association. (HuGE Navigator) 14690434_Rad52 facilitates homologous recognition between single-stranded DNA and duplex-DNA through a process that involves unwinding or transient unpairing of the interacting duplex via a novel three-stranded intermediate that does not lead to strand exchange. 14734547_the ternary complex of hRad52 and XPF/ERCC1 is the active species that processes recombination intermediates generated during the repair of DNA double strand breaks and in homology-dependent gene targeting events 15205482_For both yeast Rad52 and HsRad52, the yield of strand-exchange reactions was proportional to the fractional A.T content of the DNA substrates, but both enzymes catalyzed exchange with substrates that contained up to at least 50% G.C 15205484_formation of a stoichiometric complex between HsRad52 and single-stranded DNA was found to be critical for strand exchange activity 15571718_analysis of residues important for DNA binding in the full-length human Rad52 protein 15670896_Observational study of gene-disease association. (HuGE Navigator) 15670896_RAD52 Y415X polymorphism is not associated with epithelial ovarian cancer in Australian women 15766559_purified hRad51 and hRad52 interact with each other as well as with Mini chromosome maintenance (MCM) proteins in HeLa cell extracts 15958648_Observational study of gene-disease association. (HuGE Navigator) 16018971_Interestingly, presence of hRad52 restores the ability of hRad51 binding to such DNA targets as well. 16367760_DNA-induced disassembly of higher-order forms of Rad51 and Rad52 proteins as steps that precede protein assembly during hRad51 presynapsis on DNA, in vitro. 16638864_Observational study of gene-disease association. (HuGE Navigator) 16956909_Observational study of gene-disease association. (HuGE Navigator) 17040915_Rad52 protein functions by binding to single-stranded DNA formed as intermediates of recombination rather than by binding to the unprocessed DNA double-strand break. 18086758_Observational study of gene-disease association. (HuGE Navigator) 18203022_germline mutations in RAD51, RAD51AP1, RAD51L1, RAD51L3, RAD52 and RAD54L are unlikely to be causal of an inherited predisposition to CLL. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18313388_Data show that DNA repair synthesis, catalyzed by human DNA polymerase eta (poleta) acting upon the priming strand of a D loop, leads to capture and annealing of the second end of a resected DSB in reactions mediated by RAD52 protein. 18449888_Observational study of gene-disease association. (HuGE Navigator) 18593704_Rad52 aligns two recombining DNA molecules within the first and second DNA binding sites to stimulate the homology search and strand invasion processes. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18990028_Observational study of gene-disease association. (HuGE Navigator) 19064565_Observational study of gene-disease association. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074292_model for hRad52-mediated DNA annealing where ssDNA release and dsDNA zippering are coordinated through successive rearrangement of overlapping nucleoprotein complexes 19092295_Observational study of gene-disease association. (HuGE Navigator) 19092295_Of the 21 loci screened, RAD52 2259 and RAD52 GLN221GLU may be of importance to disease process and may be associated with papillary thyroid cancer risk in Saudi Arabian population. 19124506_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19506022_Results indicate that RAD52 cooperates with OGG1 to repair oxidative DNA damage and enhances the cellular resistance to oxidative stress. 19530647_These data show that phosphorylated RPA promoted formation of a complex with monomeric Rad52 and caused the transfer of single stranded DNA from RPA to Rad52. 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19661089_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19799994_This study demonstrated a positive correlation between molecular beacon fluorescence intensity, RAD52 gene expression and both gamma ionising radiation and antineoplastic concentration in human TK6 cells. 19996105_aRPA interacted with both Rad52 and Rad51 and stimulated Rad51 strand exchange. 20081207_hRad52 stably binds and wraps both, protein free and replication protein A-coated ssDNA. 20150366_Observational study of gene-disease association. (HuGE Navigator) 20190268_the possibility of sumoylation playing an important role in the nuclear transport of RAD52 20453000_Observational study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21148102_Rad52 can respond to DNA double-strand breaks and replication stalling independently of BRCA2 21804533_RAD52(Y104pCMF) specifically targets and wraps ssDNA. Phosphorylation at Y104 enhances ssDNA annealing activity of RAD52 by attenuating dsDNA binding. 22001234_Silencing of the Rad52 gene in fractionated group of A549 cells made the cells radiosensitive. 22860035_The recruitment kinetics of Rad52 is slower than that of Mdc1, but exhibits the same dependence on LET 22964643_RAD52 is an alternative repair pathway of RAD51-mediated homologous recombination, and a target for therapy in cells deficient in the BRCA1-PALB2-BRCA2 repair pathway. 23188672_rs7963551 located at the hsa-let-7 binding site may alter expression of RAD52 and contribute to the development of breast cancer in Chinese women. 23209746_Both RAD52 variants and protein expression can predict platinum resistance, and RAD52 variants appeared to predict prognosis in cervical cancer patients. 23339595_Single nucleotide polymorphisms in RAD52 are associated with myelodysplastic syndromes. 23836560_RAD52 mutation is associated with leukemia. 24047694_Nuclear/chromatin PTEN mediates DNA damage repair through interacting with and modulating the activity of Rad52. 24204313_Our findings reveal a novel RAD52/MUS81-dependent mechanism that promotes cell viability and genome integrity in checkpoint-deficient cells, and disclose the involvement of MUS81 to multiple processes after replication stress 24415301_Our data did not show association between LIG4 and RAD52 SNPs and SLE, its clinical manifestations or ethnicity in the tested population. 24729511_This study found that only the RAD52 rs7963551 single nucleotide polymorphism was significantly associated with hepatitis B virus related - hepatocellular carcinoma risk. 25012956_RAD52 rs7963551 single nucleotide polymorphism was significantly associated with glioma risk. 25365323_We have demonstrated that Rad51 and Rad52 dependent homologous recombination is coupled to HSV-1 DNA replication. 25395584_BRG1-RAD52 complex mediates the replacement of RPA with RAD51 on single-stranded DNA (ssDNA) to initiate DNA strand invasion. Loss of BRG1 results in a failure of RAD51 loading onto ssDNA, abnormal homologous recombination repair 25793373_these results implicate increased RAD52 expression in both genetic susceptibility and tumorigenesis of upper aerodigestive tract and lung squamous cell carcinoma tumors. 26013599_Study discovered two cis-expression quantitative trait loci SNPs in the RAD52 gene that are associated with its expression and are also associated with lung squamous cell carcinomas (LUSC) risk. 26735576_RAD52 gene polymorphism is associated with colorectal cancer. 27362509_The C-terminal region of yRad52, but not of hRAD52, is involved in ssDNA annealing. This suggests that the second DNA binding site is required for the efficient ssDNA annealing by yRad52. We propose an updated model of Rad52-mediated ssDNA annealing. 27487923_Structure of the human DNA-repair protein RAD52 containing surface mutations has been reported. 27984745_The mitotic DNA synthesis is RAD52 dependent, and RAD52 is required for the timely recruitment of MUS81 and POLD3 to common fragile sites in early mitosis. 28176781_Rad52 competes with Ku for binding to S-region double-strand DNA breaks (DSB) free ends, where it facilitates a DSB synaptic process, which favours intra-S region recombination. 28329678_DNA-bound RAD52 is efficient at capturing ssDNA in trans. 28549257_Human RAD52-null cells retain a significant level of single-strand annealing (SSA) activity demonstrating perforce that additional SSA-like activities must exist in human cells. Moreover, the SSA activity associated with RAD52 is involved in, but not absolutely required for, most homology-directed repair (HDR) subpathways. Specifically, a deficiency in RAD52 impaired the repair of DNA DSBs. 28551686_Data suggest RAD52 binds tightly to RPA/ssDNA complex in presynaptic complex and inhibits RPA turnover; during presynaptic complex assembly, most of RPA and RAD52 is displaced from ssDNA, but some RAD52/RPA/ssDNA complexes persist as interspersed clusters surrounded by RAD51 filaments. (RAD52 = Rad52 DNA repair/recombination protein; RPA = replication protein A; ssDNA = single-stranded DNA; RAD51 = Rad51 recombinase) 28602639_Rad52 inverse strand exchange plays an important role in RNA-templated double strand break repair in vivo. 29024686_n a cohort of patients with typical symptoms of ischemic heart disease, a common single nucleotide polymorphism of the human RAD52 gene has a role in increased hazard of death, showing that it may influence aging 29145865_The mechanism by which RAD52 depletion causes synthetic lethality in BRCA1 mutant cancer cells depends on the 5' endonuclease EEPD1, which normally functions to cleave stressed replication forks to initiate HR repair. 29245274_our study demonstrated that RAD52 polymorphisms were associated with colorectal cancer in a Chinese Han cohort 29334356_Importantly, p53 defects or depletion unexpectedly allow mutagenic RAD52 and POLtheta; pathways to hijack stalled replication forks, which the authors find reflected in p53 defective breast-cancer patient COSMIC mutational signatures. These data uncover p53 as a keystone regulator of replication homeostasis within a DNA restart network. 29590107_inhibition of ataxia telangiectasia mutated (ATM) protein by siRNA or inhibitor treatment demonstrated that the acetylation of RAD52 at DSB sites is dependent on the ATM protein kinase activity, through the formation of RAD52, p300/CBP, SIRT2, and SIRT3 foci at DSB sites 29845285_Due to its similarity to RAD52, we hypothesized that RDM1 potentially repairs DNA doublestrand breaks arising through DNA replication. 30590106_RAD52 protein, just as BRCA2, interacts with pCHK1 checkpoint protein and helps maintain the checkpoint control in BRCA2 deficient cells during DNA damage response 30692206_RAD52 and SLX4 mediate distinct postreplicative DNA repair processes that maintain ALT telomere stability and cancer cell viability 30926821_RAD52 limits excessive remodelling of stalled replication forks, thus indirectly assisting RAD51 and BRCA2 in protecting forks from unscheduled degradation and preventing genome instability. 31015609_Study shows that ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. 31171703_lead to mitotic DNA synthesis (MiDAS) at telomeres mediated by RAD52 through its highly conserved N-terminal domain 31381562_RAD52 and POLQ are both synthetic lethal with loss of the BRCA1 and BRCA2 tumor suppressor genes. Furthermore, RAD52 and POLQ have been implicated in chromosomal break repair events that use flanking repeats to restore the chromosome. Combined disruption of RAD52 and POLQ causes at least additive hypersensitivity to cisplatin and a synthetic reduction in replication fork restart velocity. 31495919_Human RECQL4 represses the RAD52-mediated single-strand annealing pathway after ionizing radiation or cisplatin treatment. 31777915_ROS-induced telomeric R-loops promote repair of telomeric DSBs through CSB-RAD52-POLD3-mediated BIR, a previously unknown pathway protecting telomeres from ROS. ROS-induced telomeric SSBs may not only give rise to DSBs indirectly, but also promote DSB repair by inducing R-loops, revealing an unexpected interplay between distinct ROS-induced DNA lesions. 31799622_Our work introduces for the first time RAD52 as another interacting partner of DSS1 and shows that both proteins are important players in the SSA and BIR pathways of DSB repair. 31832684_We show pronounced suppression of gene-conversion with increasing DNA double-strand breaks load that is not due to RAD51 availability and which is delimited but not defined by 53BP1 and RAD52. 32143539_Aberrant Expression of RAD52, Its Prognostic Impact in Rectal Cancer and Association with Poor Survival of Patients. 32175645_The RAD52 S346X variant reduces risk of developing breast cancer in carriers of pathogenic germline BRCA2 mutations. 32401173_RAD52 variants influence NSCLC risk in the Chinese population in a high altitude area. 32945515_RAD52 aptamer regulates DNA damage repair and STAT3 in BRCA1/BRCA2deficient human acute myeloid leukemia. 33275133_The function of RAD52 N-terminal domain is essential for viability of BRCA-deficient cells. 33440161_RAD52 Adjusts Repair of Single-Strand Breaks via Reducing DNA-Damage-Promoted XRCC1/LIG3alpha Co-localization. 33536619_BRCA1 and RNAi factors promote repair mediated by small RNAs and PALB2-RAD52. 34789290_Nickel nanoparticle-induced cell transformation: involvement of DNA damage and DNA repair defect through HIF-1alpha/miR-210/Rad52 pathway. 35190531_Rad52 mediates class-switch DNA recombination to IgD. 35210412_SF3B4 promotes ovarian cancer progression by regulating alternative splicing of RAD52. | ENSMUSG00000030166 | Rad52 | 450.944667 | 0.9981595 | -0.002657715 | 0.14155305 | 3.541976e-04 | 9.849846e-01 | No | Yes | 443.851839 | 93.487108 | 454.919902 | 95.834259 | ||
| ENSG00000003402 | 8837 | CFLAR | protein_coding | O15519 | FUNCTION: Apoptosis regulator protein which may function as a crucial link between cell survival and cell death pathways in mammalian cells. Acts as an inhibitor of TNFRSF6 mediated apoptosis. A proteolytic fragment (p43) is likely retained in the death-inducing signaling complex (DISC) thereby blocking further recruitment and processing of caspase-8 at the complex. Full length and shorter isoforms have been shown either to induce apoptosis or to reduce TNFRSF-triggered apoptosis. Lacks enzymatic (caspase) activity. {ECO:0000269|PubMed:9880531}. | 3D-structure;Alternative splicing;Apoptosis;Host-virus interaction;Reference proteome;Repeat | The protein encoded by this gene is a regulator of apoptosis and is structurally similar to caspase-8. However, the encoded protein lacks caspase activity and appears to be itself cleaved into two peptides by caspase-8. Several transcript variants encoding different isoforms have been found for this gene, and partial evidence for several more variants exists. [provided by RefSeq, Feb 2011]. | hsa:8837; | CD95 death-inducing signaling complex [GO:0031265]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; death-inducing signaling complex [GO:0031264]; membrane raft [GO:0045121]; ripoptosome [GO:0097342]; cysteine-type endopeptidase activity involved in apoptotic process [GO:0097153]; cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:0097199]; cysteine-type endopeptidase activity involved in execution phase of apoptosis [GO:0097200]; death receptor binding [GO:0005123]; enzyme activator activity [GO:0008047]; protease binding [GO:0002020]; protein-containing complex binding [GO:0044877]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; cellular response to dexamethasone stimulus [GO:0071549]; cellular response to epidermal growth factor stimulus [GO:0071364]; cellular response to estradiol stimulus [GO:0071392]; cellular response to hypoxia [GO:0071456]; cellular response to insulin stimulus [GO:0032869]; cellular response to nitric oxide [GO:0071732]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cardiac muscle cell apoptotic process [GO:0010667]; negative regulation of cellular response to transforming growth factor beta stimulus [GO:1903845]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of extrinsic apoptotic signaling pathway via death domain receptors [GO:1902042]; negative regulation of hepatocyte apoptotic process [GO:1903944]; negative regulation of myoblast fusion [GO:1901740]; negative regulation of reactive oxygen species biosynthetic process [GO:1903427]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of extracellular matrix organization [GO:1903055]; positive regulation of glomerular mesangial cell proliferation [GO:0072126]; positive regulation of hepatocyte proliferation [GO:2000347]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of neuron projection development [GO:0010976]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; regulation of necroptotic process [GO:0060544]; regulation of skeletal muscle satellite cell proliferation [GO:0014842]; response to testosterone [GO:0033574]; skeletal muscle atrophy [GO:0014732]; skeletal muscle tissue development [GO:0007519]; skeletal muscle tissue regeneration [GO:0043403]; skeletal myofibril assembly [GO:0014866]; wound healing [GO:0042060] | 11830587_The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. 11877293_Higher levels of expression of FLIP were present in TRAIL-resistant multiple myeloma cells, and sensitivity to TRAIL was restored by lowered FLIP protein levels. 11940602_An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis 12031968_cFLIP may have an impact on the outcome of death receptor-triggered responses by directing the intracellular signals from beta-cell death to beta-cell survival 12060768_switches Fas-mediated glucose signaling in human pancreatic beta cells from apoptosis to cell replication switches Fas-mediated glucose signaling in human pancreatic beta cells from apoptosis to cell 12115181_FLICE-inhibitory protein expression in synovial fibroblasts and at sites of cartilage and bone erosion in rheumatoid arthritis. 12393527_results demonstrate a definite role for FLIP in the stem cell factor-induced protection of erythroid colony forming cells from IFNgamma-initiated apoptosis 12407100_These results provide new insights into the mechanisms of bile acid cytotoxicity and the proapoptotic effects of cFLIP phosphorylation in TRAIL signaling. 12432255_c-FLIP may play a critical role in regulating Fas-mediated apoptosis in prostate cancer cells 12477972_The consequences of FasL overexpression depend on the subcellular compartment and species in which FasL enforced expression is targeted and this is at least partially related to FLIP levels 12496285_c-FLIP expression is regulated by calcium/calmodulin-dependent protein kinase II and modulates Fas-mediated signaling in glioma cells 12496481_In response to doxorubicin, the level of FLIP decreased in all prostatic cell lines tested and correlated with the onset and magnitude of CASP8 and PARP cleavage in PC3 cells. 12496482_The metabolism of FLIP is essential to the death of prostatic cells in culture. 12552004_Adenovirus E1A inhibited TNF-alpha-dependent induction of c-FLIP(S) mRNA and stimulated ubiquitination- and proteasome-dependent degradation of c-FLIP(S) protein in Hela cells 12556488_Our results show that c-FLIP(L) and c-FLIP(S) potently control TRAIL responses, both by distinct regulatory features, and further imply that the differentiation state of malignant cells determines their sensitivity to death receptor signals. 12574377_Constitutive expression of the long form of human FLIP leads to an enhanced and prolonged inflammatory response in the central nervous system during experimental allergic encephalitis in DBA mice. 12592338_Constitutively active Akt1 protects HL60 leukemia cells from TRAIL-induced apoptosis through a mechanism involving NF-kappaB activation and up-regulation of this protein 12716387_Expression is increased in stomach cancer 12746452_cFLIP-L exerts its anti-apoptotic activity, in part, by inhibiting p38 MAPK activation, an additional anti-apoptotic effect for this protein. 12820373_cFLIP is an important determinant of susceptibility to death receptor-induced apoptosis in bladder carcinomas. 14562111_FLIP(L) and FLIP(S) are differentially regulated, and that the relative levels of both isoforms play a role in the regulation of apoptosis in myelodysplastic syndrome 14578361_In T cells, c-FLIP expression led to inhibition of IL-2 production, in contrast to the readily detectable c-FLIP-induced activation in Jurkat cells. 14637155_In this study, we show that c-FLIP(L) but not c-FLIP(S) physically binds to Daxx through interaction between C-terminal domain of c-FLIP(L) and Fas-binding domain of Daxx, an alternative Fas signaling adaptor. 14662022_Akt activity promotes human gastric cancer cell survival against TRAIL-induced apoptosis via upregulation of FLIP(S), and that the cytotoxic effect of TRAIL can be enhanced by modulating the Akt/FLIP(S) pathway in human gastric cancers. 15024054_Results demonstrate that FLIP(p43) processed by caspase 8 specifically interacts with TRAF2 and subsequently induces activation of the NF-kappaB signaling pathway. 15078899_the selective down-regulation of c-FLIP by small interfering RNA oligoribonucleotides was sufficient to sensitize Hodgkin/Reed-Sternberg cells to CD95 and tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis 15096587_down-regulation of cellular FLICE-inhibitory protein through the use of specific small inhibitory RNAs leads to reduced viability of the L428 and L591 HL-derived cell lines 15183989_The frequent expression and coexpression of Fas, FasL, and c-FLIP in urothelial carcinomas implicates c-FLIP as an inhibitor of the Fas-FasL-induced death pathway in these tumors. 15258564_FLIP plays a significant role in the regulation of apoptosis in human ovarian cancer cells and their sensitivity to cisplatin. 15273717_Regulation by AMP-activated protein kinase-related kinase 5 15297424_the inhibitory protein c-FLIP(L) is involved in resistance to CD95-mediated apoptosis in ovarian carcinoma cells with wild-type p53 15304499_When fusesd with Tat protein, prevesnts adverse apoptosis nd prolongs survival in tumor cells. 15334061_Malignant mesothelioma cells develop an intrinsic resistance to apoptosis induced by death receptors upregulating the expression of the antiapoptotic protein c-FLIP. 15354734_strong c-FLIP expression in nodular lymphocyte-predominant Hodgkin lymphoma was associated with transformation to diffuse large B-cell lymphoma; the majority of DLBCL cases tested were strongly c-FLIP-positive. 15459191_cFLIP(L) is not only a central antiapoptotic modulator of TRAIL-mediated apoptosis but also an inhibitor of TRAIL-induced NF-kappaB activation and subsequent proinflammatory target gene expression 15485835_c-FLIPL is recruited to death receptor 5 independent of Fas-associated protein with death domain (FADD) 15540114_Results indicate that some tumor cells are resistant to death receptor-mediated apoptosis by expressing cellular FLIP, and that histone deacetylase inhibitors sensitize such resistant tumor cells by directly downregulating cellular FLIP mRNA. 15557152_Constitutive overexpression of c-FLIP (long form) in T cells is sufficient to drive Th2 polarization of effector T cell responses and indicates that it might function as a key regulator of T helper (Th) cell differentiation. 15644494_DDB2 regulates TNF signaling-mediated apoptosis via cFLIP and contributes to acquired cross-resistance. 15653751_T cell blasts surviving activation-induced cell deathare memory CD44high cells with increased c-FLIP expression. 15686714_Heart graft rejection biopsies have elevated FLIP mRNA expression levels. 15701649_c-FLIPR, an isoform of c-FLIP, is a new regulator of death receptor-induced apoptosis. 15701651_CLARP inhibits caspase-8 induced, ASC-mediated NF-kappaB activation and IL-8 production 15722350_cFLIP/CFLAR is an essential NF-kappa B-dependent antiapoptotic gene in the tumor necrosis factor alpha-regulated pathway in epidermal keratinocytes 15731171_c-FLIP promoter was shown to contain multiple functional androgen response elements. 15760909_c-FLIP(L) functions primarily as an inhibitor of death receptor-mediated apoptosis through TRAIL and caspase-8 activation 15761846_Established and analyzed a transgenic mouse model that overexpresses human c-FLIPS (CFLAR) in thymocytes and peripheral T cells. Data suggests a specific in vivo function for c-FLIPS in the maintenance of restimulated T cells. 15815586_differential spatial and temporal regulation of cFLIP-alpha and cFLIP-delta expression that may influence the magnitude of cell death 15832422_c-FLIP is specially overexpressed in colon cancers and it might contribute to carcinogenesis of normal colonic mucosa. 15843523_A mechanism by which c-FLIP(long form)(CFLAR) influences effector T cell function in CFLAR transgenic mice is via its activation of caspase-8, which in turn cleaves CFLAR to allow receptor interacting protein (RIP)1 recruitment and NF-kappa B activation. 15864316_Our results suggest for the first time a critical role for FLIP in the regulation of apoptosis triggered by TRAIL in endometrial carcinoma cells. 15886205_The conformation-based predisposition of c-FLIP(S) to ubiquitin-mediated degradation introduces a novel concept to the regulation of the death-inducing signaling complex 15899875_FLIP protected mouse lung endothelial cells against hypoxia/reoxygenation by blocking both caspase 8/Bid and Bax/mitochondrial apoptotic pathways. 15917295_c-FLIP knock-down with a small interfering RNA significantly restores Fas-mediated apoptosis in infected cells 15956881_The majority of Langerhans cell histiocytosis cells express this protein. 16014121_c-FLIP might contribute to the carcinogenesis and aggressiveness of endometrial carcinoma and might be a useful prognostic factor in the tumor 16052233_findings show that E2F1 triggers apoptosis in lung adenocarcinoma cell lines by a mechanism involving the specific downregulation of the cellular FLICE-inhibitory protein short, leading to caspase-8 activation at the death-inducing signaling complex 16052516_results demonstrate a critical role for PKCdelta/NF-kappaB in the regulation of FLIP in human colon cancer cells 16077198_Cycloheximide sensitizes colorectal tumor cells to TNF-alpha induced apoptosis by reducing FLIP levels. 16211288_the PI 3-K/Akt signaling pathway may, in part, regulate Fas-mediated apoptosis in HL-60 cells through c-FLIP expression 16247474_c-FLIP inhibits apoptosis in response to chemotherapy in colorectal cancer cells 16298825_Expression of FLIP molecules may be, at least in part, an early prognostic indicator in the treatment of elderly acute myeloid leukemia patients. 16304056_overexpression of c-FLIP protects ALK+ ALCL cells from FAS-induced apoptosis and may contribute to ALCL pathogenesis. 16403915_c-FLIP confers Tax-mediated resistance toward CD95-mediated apoptosis. 16436054_Taken together, Tax inhibits Fas-mediated apoptosis by up-regulating c-FLIP expression in HTLV-I-infected cells, and NF-kappaB activity plays an essential role in the up-regulation of c-FLIP. 16441226_Review focuses on the role of c-FLIP as a tumour progression factor, with particular emphasis on contribution of c-FLIP to pathogenesis of Hodgkin's lymphoma. 16472594_Inhibition of Flip by antisense oligonucleotide reverted the resistance of CD LPLs to FAS-induced apoptosis. 16611896_results suggested that the TNF-alpha-induced apoptotic pathway is inhibited by a sustained c-FLIP expression associated with the expression of HCV core protein, which may play a role in HCV-mediated pathogenesis 16720717_Results suggest that, in addition to its proapoptotic function, par-4 acts as a novel transcription cofactor for androgen receptors to target c-FLIP gene expression. 16740746_The NFkappaB-mediated overexpression of cFLIP(long)represent alternative mechanisms for deregulating the extrinsic apoptotic pathway in LBCL subtypes defined by gene expression profiling. 16901543_Anti-apoptotic signaling of CD40 involves induction of c-FLIP proteins. 17056549_These data demonstrate specific activation of c-FLIP by HCMV IE2 and indicate a novel role for c-FLIP in the pathogenesis of HCMV retinitis. 17070520_Treatment with sodium arsenite resulted in upregulation of the endogenous TRAIL and downregulation of the cFLIP gene expression followed by cFLIP protein degradation and, finally, by acceleration of TRAIL-induced apoptosis 17106251_Doxorubicin-mediated downregulation of cFLIPS at the post-transcriptional level is sufficient to enhance TRAIL sensitivity in PC3 prostate carcinoma cells. 17272514_Enforced long-form cFLIP (cFLIP(L)) expression reduced release of cathepsin B from lysosomes and attenuated apoptosis. 17376892_induction of the short c-FLIP isoforms inhibits the onset of CD95-induced apoptosis in primary CD40-stimulated ALL cells despite high CD95 expression. 17440816_Downregulation of either FLIP or XIAP but not Bcl-2 restored sensitivity of Colo320 cells to Apo2L/TRAIL. 17441421_There was a negative relationship between the expression of FLIP and PTEN in laryngeal squamous cell carcinoma. 17450141_Observational study of gene-disease association. (HuGE Navigator) 17513603_c-FLIP plays a pivotal role in modulating drug-induced apoptosis in breast cancer cells 17559541_The expression of Fas, FasL and c-FLIP in colorectal carcinoma implicates c-FLIP as an inhibitor of the Fas-FasL-induced death pathway in these tumors. Moreover, c-FLIP conveys independent prognostic information in the presence of classic prognosticators. 17573774_cFLIP-L is prone to aggregate and impairs ubiquitin-proteasome system function, which could be involved in the pathological function of cFLIP-L expressed in certain cancer cells 17581950_FLIP(L) is essential for tumor necrosis factor (TNF)/tumor necrosis factor receptor I (TNFRI)-mediated neuroprotection after glucose deprivation in transgenic mice. 17638906_RIP and c-FLIP-mediated assembly of the death-inducing signaling complex in nonrafts is a critical upstream event in TRAIL resistance 17646662_enhanced proliferative nature of human leukemia cells is caused by elevated NF-kappaB and FLIP responses 17659339_bortezomib and TRAIL combination caused further down-regulation of cFLIP protein and increased apoptosis in CHL cells 17697742_VPA significantly increased sensitivity of leukemic cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and led to downregulation of c-FLIP (L) expression. 17726263_Caspase-8 and cFLIP are upregulated during colorectal carcinogenesis. 17762208_The mechanism by which antioxidant status alters FLIP levels following neonatal HI may be related to the ability to detoxify H2O2 produced following neonatal HI. 17912957_Human keratinocytes were transfected with either Flip, Faim, or Lifeguard (LFG). Our results suggest that heterotopic expression of antiapoptotic proteins can induce the resistance of keratinocytes to a major mechanism of rejection. 17922291_Overexpression of cFLIP(L) is a frequent event in head and neck squamous cell carcinoma 17932249_reconstitution of ATM kinase activity decreases FLIP protein levels and restores Fas sensitivity in Hodgkin lymphoma-derived cells 17982483_CK2 regulates endometrial carcinoma cell sensitivity to TRAIL and Fas by regulating FLIP levels. 17988665_IFN-alpha causes a transient upregulation of c-FLIP expression, at least through PKCalpha-mediated activation of NF-kappaB 18073330_(c-FLIP), which is dependent on the NF-kappaB pathway in normal muscle cells, is down-regulated in limb-girdle muscular dystrophy type 2A biopsies. 18084329_cFLIP represents an attractive therapeutic target for melanoma treatment, especially in combination with TRAIL receptor agonists. 18189268_Sanguinarine-mediated apoptosis is blocked by ectopic expression of Bcl-2 and cFLIPs 18243739_Study indicated that c-FLIP(L) might be a suppressor of Fas-mediated apoptosis in Fas antigen expressing colon carcinoma cells. 18257744_interaction of calmodulin cellular FLICE-like inhibitory protein 18264131_Since Gli2 silencing not only downregulates cFlip, but also Bcl-2, Gli2 could be a key target for a novel therapeutic approach in basal cell carcinoma. 18314443_cFLIP was overexpressed in CD4+/CD26- tumor cells with normal or enhanced Fas expression 18339897_TTP plasma-mediated apoptosis appears to involve IFNG-induced acceleration of c-FLIP degradation, sensitizing cells to TRAIL-mediated caspase-8 activation and cell death. 18391984_FOXO3a mediates the androgen-dependent regulation of FLIP and contributes to TRAIL-induced apoptosis of LNCaP cells. 18398344_c-Flip (CASP8 and FADD-like apoptosis regulator) is a key regulator of the cardiac response to ventricular pressure overload. 18414015_c-FLIPL promotes the motility of HeLa cells by activating FAK and ERK, and increasing MMP-9 expression. 18509086_preventing autocrine CD95L signaling by c-FLIP facilitates T-cell effector function and an efficient immune response 18593367_reversion of Fas resistance is mediated through CD40/CD40L ligation rather than IFN-gamma stimulation by inhibiting synthesis of c-FLIP 18603835_cFLIP is an essential pro-survival factor for granulosa cells, and it prevents granulosa cell apoptosis by inhibiting procaspase-8 activation. 18726983_Androgenic protection from TRAIL-induced apoptosis is via enhanced transcription of FLIP in prostate cancer cells. Loss of androgen-sensitivity in androgen-depletion-independent prostate cancer cells. Potential target for therapy of prostate cancer. 18767116_anti-apoptotic functions of cFLIPs may be attributed to inhibit oxaliplatin-induced apoptosis through the sustained XIAP protein level and Akt activation. 18820704_These results suggest that simultaneously targeting both FLIP and XIAP may prove useful in the treatment of cancers, particularly those in which the intrinsic mitochondrial apoptotic pathway has been compromised. 18823309_Observational study of gene-disease association. (HuGE Navigator) 18838202_Overexpression of FLIP reduced TRAIL and TNF-alpha-induced apoptosis in ML-1 cells. However, while FLIPL completely abrogated apoptosis, FLIPS allowed for BID cleavage and caspase-3 activation. 18840411_an apoptotic inhibitory complex comprised of DR5, FADD, caspase-8, and c-FLIP(L) exists in MCF-7 cells, and the absence of c-FLIP(L) from this complex induces DR5- and FADD-mediated caspase-8 activation in the death inducing signaling complex 19090833_Ubiquitination of cFLIP(L) inhibits the interaction between cFLIP(L) and Itch in T. cruzi-infected cells. 19109151_While dispensable for the development of bone marrow precursor B cells, cFLIP is necessary for B cell maturation in the periphery. 19115040_These data suggest caspase 8, but not cFLIP, activation induced by C5a leads to cell death if protein synthesis of antiapoptotic protein(s) is blocked. 19161534_FLIP expression was strong in most (103/107; 96.3%) of the prostate cancers, and only four cancers (3.7%) showed decreased immunoreactivities compared with the normal cells. 19177145_modulation of the CD95 signaling pathway by cFLIP in basal keratinocytes may explain the spatial localization of spongiosis in suprabasal epidermal layers, and provides new insights into the pathogenesis of spongiosis formation in eczematous dermatitis 19203346_the loss of Bcr-Abl in imatinib-resistant K562 cells led to the down-regulation of c-FLIP and subsequent increase of TRAIL sensitivity 19223508_Increase in FLIP is associated with prostate tumors. 19243385_inhibition of c-FLIP(L) expression might be a potential strategy for lung cancer therapy, especially for those lung cancers resistant to the agonistic antibody against death receptors 19249545_Pig islets expressing human c-FLIP(L) are significantly resistant to human cytotoxic lymphocyte killing and exhibit beneficial effects to prolong xenograft survival. 19282655_Over-expression of c-FLIP confers the resistance to TRAIL-induced apoptosis on gallbladder carcinoma. 19321593_Overexpression of c-FLIPL is associated with ovarian cancer. 19339247_TNFalpha facilitates the reduction of FLIP(L) protein, which is dependent on the phosphatidylinositol 3-kinase/Akt signaling. 19343040_S193 phosphorylation is mediated by PKC-aplha and PKC-beta and reveal a connection between c-FLIP phosphorylation and ubiquitylation, as S193 mutations differently affect c-FLIP ubiquitylation. 19363595_gp120 plays an important role, via involvement of c-FLIPL, in T-cell apoptotic cell death due to HIV-1 infection. 19372246_c-FLIP expression is induced in the abnormal alveolar epithelium of patients with idiopathic pulmonary fibrosis/usual interstitial pneumonia 19398149_the short isoform of c-FLIP and bcl-2 are key regulators in TRAIL-Myricetin mediated cell death in malignant glioma. 19409438_In summary, the short isoform of c-FLIP is a key regulator in TRAIL-Embelin-mediated apoptosis in malignant glioma. 19433309_These findings describe a novel function of c-FLIP-L involved in AP-1 activation and cell proliferation. 19439735_production of either c-FLIP(S) or c-FLIP(R) in humans is defined by a single nucleotide polymorphism in a 3' splice site of the c-FLIP gene (rs10190751A/G); increased lymphoma risk associated with the rs10190751 A genotype causing c-FLIP(R) expression. 19476635_Increased c-FLIP is associated with cervical intraepithelial neoplasia and cervical carcinoma. 19543235_FADD-Like Apoptosis Regulating Protein silencing sensitized non-small-cell lung cancer cells but not normal cells to chemotherapy in vitro, and silencing FLIP in vivo retarded non-small-cell lung cancer cells xenograft. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19604093_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19710364_The interaction between Mib1 and cFLIP decreases the association of caspase-8 with cFLIP, which activates caspase-8 and induces cell death. 19728335_IKKalpha, IKKbeta, RANK, Maspin, c-FLIP, Cip2 and cyclinD1 were found to show significant differences between hepatocellular tumor tissue and its corresponding adjacent tissue. 19734124_CD133-high cells may be resistant to TRAIL due to high expression of FLIP. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19798106_Data suggest that c-FLIP downregulation represents a mechanism by which diverse anticancer regimens can facilitate tumor cell execution by CD95/Fas through the direct pathway of caspase activation. 19802016_Results suggest that Akt confers resistance, in part, by modulating CDDP-induced, p53-dependent FLIP ubiquitination. 19816511_Results identify c-FLIP(L) as a susceptibility factor under the influence of epistatic modifiers for the development of autoimmunity. 19890350_Knockdown of c-FLIP by a enhanced basic apoptosis rates in cutaneous lymphoma cells and diminished the CD30-mediated suppression of apoptosis, thus proving the significance of c-FLIP in this context 19896469_These results indicate that AMPK activators facilitate the activation by TRAIL of an apoptotic cell death program through a mechanism independent of AMPK and dependent on the down-regulation of cFLIP levels. 19906927_our results demonstrate that apoptosis of HSV-1-infected iDCs requires both c-FLIP downregulation and diminished expression of viral LAT. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19949310_CDODO-Me-12 and CDODO-Me-11 downregulated the levels of anti-apoptosis protein c-FLIP in HL-60, U937 and K562 leukemic cell lines. 19956887_eupatolide could augment TRAIL-induced apoptosis in human breast cancer cells by down-regulating c-FLIP expression through the inhibition of AKT phosphorylation 20016063_role for nuclear cFLIP-L in the modulation of Wnt signaling 20087343_Egr-1 drives c-FLIP expression and the short splice variant of c-FLIP (c-FLIP(S)) specifically inhibits DR5 activation. 20218968_The role of c-FLIP is critical in the protection of erythroid-differentiated cells from apoptosis or in the determination of their sensitivity to TNF-mediated programmed cell death. 20224598_FLIP expression is transcriptionally regulated by hnRNP K and nucleolin, and may be a potential prognostic and therapeutic marker for nasopharyngeal carcinoma. 20227749_Data demonstrate that c-FLIP(L) exhibits multiple functions in ovarian cancer: first by concomitantly evading the natural immunity mediated by TRAIL-induced cell death, and second by augmenting cell motility and invasion in vivo. 20335528_Although deletion of cFLIP does not alter the primary development of B cells, adoptively transferred cFLIP-deficient follicular B cells do not effectively participate in the germinal center response. 20372864_Disclose a novel regulatory mechanism inHepG2 cells where down-regulation of c-FLIP by miR-512-3p contributed to taxol-induced apoptosis. 20449949_Data suggest that FLIP potentially extends the lifespan of synovial cells and thus contributes to the progression of joint destruction in juvenile idiopathic arthritis. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20561424_mRNA expressions of DLK1, c-FLIPL and c-FLIPS are abnormal in bone marrow mononuclear cells of myelodysplastic syndrome patients. 20568250_Observational study of gene-disease association. (HuGE Navigator) 20595005_PDCD4 plays an important role in mediating the sensitivity of gastric cancer cells to TRAIL-induced apoptosis through FLIP suppression. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20689807_Observational study of gene-disease association. (HuGE Navigator) 20696707_The function of c-FLIP(L) as a pro- or antiapoptotic protein in DR-mediated apoptosis is important for understanding the regulation of CD95-induced apoptosis, where subtle differences in c-FLIP concentrations determine life or death of the cells. 20802294_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20812013_CD30/CD95 crosstalk experiments revealed that CD30 ligation leads to NFkappaB-mediated cFLIP upregulation in cALCL cells, which in turn enhanced resistance to CD95-mediated apoptosis. 20876774_Both the long and the short isoform of the antiapoptotic protein c-FLIP are critical regulators of death receptor-induced apoptosis in pancreatic carcinoma cells and are suppressed by chemotherapeutics. 20882347_Triggering of death receptor apoptotic signaling by human papillomavirus 16 E2 protein in cervical cancer cell lines is mediated by interaction with c-FLIP. 20975036_These studies indicate that cFLIP protein is a crucial component of the signaling pathway involved in cardiac remodeling and heart failure. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21071136_review of molecular mechanisms that control c-FLIP expression and current research into inhibitors of the protein [review] 21107885_Itch/AIP4-independent proteasomal degradation of cFLIP induced by the histone deacetylase inhibitor SAHA sensitizes breast tumour cells to TRAIL-induced apoptosis. 21153369_regulation of cell death by c-FLIP phosphorylation 21307400_Reveal unique regulatory networks in acute myeloid leukemia whereby FLIP regulation of HO-1 provides cells with secondary anti-apoptotic protection against TNF induced death signals in these highly death-resistant cells. 21324892_Modeling reveals that dynamic regulation of c-FLIP levels determines cell-to-cell distribution of CD95-mediated apoptosis. 21403465_Silencing FLIP(L) modifies TNF-induced apoptotic protein expression. 21435442_KSR1 is overexpressed in endometrial carcinoma and regulates proliferation and TRAIL-induced apoptosis by modulating FLIP levels 21454681_Cellular FLICE-inhibitory protein (cFLIP) isoforms block CD95- and TRAIL death receptor-induced gene induction irrespective of processing of caspase-8 or cFLIP in the death-inducing signaling complex 21480219_BAY 11-7085 rapidly inhibited c-FLIP(L) expression in colon and pancreatic cancer cell lines during adhesion to mesothelial cells. 21525012_Epidermal growth factor receptor-mediated tissue transglutaminase overexpression couples acquired tumor necrosis factor-related apoptosis-inducing ligand resistance and migration through c-FLIP and MMP-9 proteins in lung cancer cells 21562857_increased expression of cFLIP protects cells from both interferon-alpha and death receptor mediated apoptosis 21638304_cFLIP expression showed no significant correlation to DLBCL subtypes (GCB or non-GCB) but was associated with a worse clinical outcome. 21691254_Data suggest that persistent Flice inhibitory protein-S expression is involved in the longevity of neonatal polymorphonuclear leukocytes. 21803845_PMLRARalpha binds to Fas and suppresses Fas-mediated apoptosis through recruiting c-FLIP in vivo. 21856935_our study reveals a novel link between NF-kappaB and PI3K/Akt and establishes c-FLIP as an important regulator of FasL-mediated cell death. 21868755_a novel mechanism through which the regulatory effects of c-FLIP on death receptor signaling in neoplastic cells are controlled by glycogen synthase kinase-3 21912376_results demonstrate that the H204 residue is responsible for c-FLIP(L) binding to CaM, which mediates the anti-apoptotic function of c-FLIP(L) 22027693_This study identifies SART1 as a previously unidentified regulator of c-FLIP and drug-induced activation of caspase-8. 22072062_Endoplasmic reticulum stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. 22126763_Kashin-Beck disease patients have significant increased FADD expression in the middle layer but decreased FLIP expression in the upper layer of the cartilage. 22190004_It was diminished expression of the c-FLIP(L) isoform in urothelial carcinoma tissues as well as in established carcinoma cell lines compared with normal urothelial tissues and cells, whereas c-FLIP(S) was unchanged. 22219201_an important role for TRAF7 in the activation of JNK following TNFalpha stimulation and involvement of this protein in regulating the turnover of c-FLIP 22288650_data suggest that Ro52/SSA is involved in death receptor-mediated apoptosis by regulating c-FLIP(L) 22345097_API-1 reduces c-FLIP and enhances TRAIL-induced apoptosis independent of its Akt-inhibitory activity 22393362_BCL-X(L) and BCL-2 but not FLIP(L) acts in synergy with MYC to drive AML development 22504646_Survival of activated and also of immature dendritic cells is regulated by BAK and shows that tumor necrosis factor (TNF) is protective only in the presence of FLIPL. 22683265_analysis of stoichiometry of the CD95 death-inducing signaling complex and demonstration of how procaspase-8, procaspase-10, and c-FLIP form DED chains at the DISC, enabling the formation of dimers and efficient activation of caspase-8 22753273_CHOP represses cFLIPL expression in Caki cells. 22781394_review of the structural and functional biology of c-FLIP with direct relevance to carcinogenesis [review] 22842544_TRAIL-induced apoptosis in human renal cancer cells by upregulation of DR5 as well as downregulation of c-FLIP and Bcl-2. 22948392_Suppression of HSP70 expression sensitizes NSCLC cell lines to TRAIL-induced apoptosis by upregulating DR4 and DR5 and downregulating c-FLIP-L expressions 23028678_the 'biomarker signature' of FLIP/Sp1/Sp3 combined with Gleason score predicted disease recurrence 23167276_Adult acute myeloid leukemia patients with higher-than-median mRNA expression of the long splice form(but not the short splice form) had significantly lower 3 year overall survival than those with low expression. 23230268_Apoptosis-related genes such as the caspase-8, FLIP, and DR5 genes specifically interfere with interferon-induced apoptosis. 23235765_Data show that alterations of the apoptosis-related protein FLIP is common in granulosa cell tumor (GCT), and suggest that expression of FLIP might play role in the pathogenesis of GCT, possibly by inhibiting apoptosis. 23247197_SIRT1 inhibition increases Ku70 acetylation, and the acetylated Ku70 with a decreased function mediates the induction of DR5 and the down-regulation of c-FLIP by up-regulating c-Myc/ATF4/CHOP pathway,promotes the TRAIL-induced apoptosis 23255321_c-FLIP may play an important role in the metastatic potential of osteosarcoma to the lung 23319802_findings provide the first evidence showing that mTORC2 stabilizes FLIP(S), hence connecting mTORC2 signaling to the regulation of death receptor-mediated apoptosis 23322903_Studies indicate that the anti-apoptotic protein c-FLIP is an important regulator of death receptor signaling, including TNFR1, Fas, DR4, and DR5. 23371318_ERK controls epithelial cell death receptor signalling and c-FLIP in ulcerative colitis 23483900_A novel mechanism of HBx-induced NF-kappaB activation in which ternary complex formation is involved among HBx, p22-FLIP and NEMO, is reported. 23518915_An increased expression of c-FLIP may be an important factor in the progression of cervical cancer 23519470_novel ROS-dependent post-translational modifications of the c-FLIP protein that regulate its stability, thus impacting sensitivity of cancer cells to TRAIL. 23615398_CHOP-mediated DR5 upregulation and proteasome-mediated downregulation of c-FLIP. 23696271_cFLIP upregulated the expression of viral re | ENSMUSG00000026031 | Cflar | 510.406702 | 1.1781073 | 0.236470992 | 0.13126949 | 3.242903e+00 | 7.173341e-02 | 3.612815e-01 | No | Yes | 518.200486 | 64.558158 | 435.277934 | 54.254795 | |
| ENSG00000004777 | 115703 | ARHGAP33 | protein_coding | O14559 | FUNCTION: May be involved in several stages of intracellular trafficking. Could play an important role in the regulation of glucose transport by insulin. May act as a downstream effector of RHOQ/TC10 in the regulation of insulin-stimulated glucose transport (By similarity). {ECO:0000250}. | Alternative splicing;GTPase activation;Methylation;Phosphoprotein;Protein transport;Reference proteome;SH3 domain;Transport | This gene encodes a member of the sorting nexin family. Members of this family contain a phox (PX) domain, which is a phosphoinositide binding domain, and are involved in intracellular trafficking. Alternative splice variants encoding different isoforms have been identified in this gene. [provided by RefSeq, Feb 2010]. | hsa:115703; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; GTPase activator activity [GO:0005096]; phosphatidylinositol binding [GO:0035091]; protein kinase binding [GO:0019901]; protein transport [GO:0015031]; small GTPase mediated signal transduction [GO:0007264] | 16777849_TCGAP interacts with Fyn and is phosphorylated by Fyn, with tyrosine-406 in the GTPase-activating protein (GAP) domain as a major Fyn-mediated phosphorylation site. 17664338_Results show that neurite outgrowth multiadaptor RhoGAP protein, NOMA-GAP, plays an essential role downstream of NGF in promoting neurite outgrowth and extension by recruiting SHP2 and activating Cdc42. 18660489_Observational study of gene-disease association. (HuGE Navigator) 26839058_ARHGAP33 is associated with brain phenotypes of patients with schizophrenia. | ENSMUSG00000036882 | Arhgap33 | 371.391373 | 0.9945381 | -0.007901523 | 0.23259609 | 1.201072e-03 | 9.723537e-01 | No | Yes | 390.703052 | 75.492577 | 398.597597 | 76.880327 | ||
| ENSG00000005801 | 7748 | ZNF195 | protein_coding | O14628 | FUNCTION: May be involved in transcriptional regulation. | Acetylation;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein belonging to the Krueppel C2H2-type zinc-finger protein family. These family members are transcription factors that are implicated in a variety of cellular processes. This gene is located near the centromeric border of chromosome 11p15.5, next to an imprinted domain that is associated with maternal-specific loss of heterozygosity in Wilms' tumors. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2012]. | hsa:7748; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; regulation of transcription, DNA-templated [GO:0006355] | 24817947_ZNF195 and SBF1 are potential biomarkers for gemcitabine sensitivity in head and neck squamous cell carcinoma cell lines. | 1016.599947 | 1.0351406 | 0.049826786 | 0.09480585 | 2.752527e-01 | 5.998298e-01 | 8.509616e-01 | No | Yes | 1082.849076 | 166.713895 | 1055.531841 | 162.555706 | |||
| ENSG00000005961 | 3674 | ITGA2B | protein_coding | P08514 | FUNCTION: Integrin alpha-IIb/beta-3 is a receptor for fibronectin, fibrinogen, plasminogen, prothrombin, thrombospondin and vitronectin. It recognizes the sequence R-G-D in a wide array of ligands. It recognizes the sequence H-H-L-G-G-G-A-K-Q-A-G-D-V in fibrinogen gamma chain. Following activation integrin alpha-IIb/beta-3 brings about platelet/platelet interaction through binding of soluble fibrinogen. This step leads to rapid platelet aggregation which physically plugs ruptured endothelial cell surface. | 3D-structure;Alternative splicing;Calcium;Cell adhesion;Cleavage on pair of basic residues;Direct protein sequencing;Disease variant;Disulfide bond;Glycoprotein;Integrin;Membrane;Metal-binding;Pyrrolidone carboxylic acid;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the integrin alpha chain family of proteins. The encoded preproprotein is proteolytically processed to generate light and heavy chains that associate through disulfide linkages to form a subunit of the alpha-IIb/beta-3 integrin cell adhesion receptor. This receptor plays a crucial role in the blood coagulation system, by mediating platelet aggregation. Mutations in this gene are associated with platelet-type bleeding disorders, which are characterized by a failure of platelet aggregation, including Glanzmann thrombasthenia. [provided by RefSeq, Jan 2016]. | hsa:3674; | blood microparticle [GO:0072562]; cell surface [GO:0009986]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; integrin complex [GO:0008305]; plasma membrane [GO:0005886]; platelet alpha granule membrane [GO:0031092]; extracellular matrix binding [GO:0050840]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; cell-matrix adhesion [GO:0007160]; integrin-mediated signaling pathway [GO:0007229] | 11204574_Observational study of gene-disease association. (HuGE Navigator) 11260063_Observational study of gene-disease association. (HuGE Navigator) 11260064_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11341496_Meta-analysis of gene-disease association. (HuGE Navigator) 11367863_Observational study of gene-disease association. (HuGE Navigator) 11467947_Reduction of disulfide bonds within the redox site of integrin alphaIIbbeta3 leads to transitions in the integrin's activation state, which is the switch from 'off' to 'on' that regulates its ligand binding affinity. 11575217_Observational study of gene-disease association. (HuGE Navigator) 11583324_Distinct domains of alphaIIbbeta3 support different aspects of outside-in signal transduction and platelet activation. 11698306_Observational study of gene-disease association. (HuGE Navigator) 11719362_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 11728949_Observational study of gene-disease association. (HuGE Navigator) 11776052_Observational study of gene-disease association. (HuGE Navigator) 11812775_Lateral clustering of platelet GP Ib-IX complexes leads to up-regulation of the adhesive function of integrin alpha IIbbeta 3 11858493_GPIIb-IIIa complexes are not in an activated conformational state after dissociation of abciximab unless there is an additional source of activation. 11918133_Observational study of gene-disease association. (HuGE Navigator) 11994301_Relationships between Rap1b, affinity modulation of integrin alpha IIbbeta 3, and the actin cytoskeleton (integrin alpha(IIb)beta(3)) 12023286_describes regions within CIB and alpha(IIb) that interact with one another 12031826_Factor XIII mediates adhesion of platelets to endothelial cells through alpha(v)beta(3) and glycoprotein IIb/IIIa integrins. 12042322_binding of Aup1 plays a crucial role in the alpha(IIb)beta(3) inside-out signaling 12049640_collagen receptor glycoprotein VI and alphaIIbbeta3 trigger distinct patterns of receptor signalling in platelets, leading to tyrosine phosphorylation of PLCgamma2 (integrin alphaiibbeta3) 12071877_Observational study of genotype prevalence. (HuGE Navigator) 12140290_Integrin alpha IIbbeta 3 has agonist-specific activation states and causes intrasubunit and intersubunit allosteric effects 12200372_the effects of either the Cys5Ala or Cys435Ala substitution of GPIIIa on the ability to bind GPIIb and the adhesive properties of the resulting GPIIb-IIIa complex 12297512_co-stimulation of G(12/13) and G(i) pathways is sufficient to activate GPIIb/IIIa in human platelets in a mechanism that involves intracellular calcium 12388784_three-dimensional structure at 20-A resolution of the unliganded; low-affinity state of the human platelet integrin alpha(IIb)beta(3) derived by electron cryomicroscopy and single particle image reconstruction 12393463_Identification of distal regulatory regions in the human alpha IIb gene locus necessary for consistent, high-level megakaryocyte expression 12399140_patients with schizophrenia have increased platelet expression of alpha IIb, IIIa which may contribute to their increased risk of cardiovascular illness 12408998_Observational study of gene-disease association. (HuGE Navigator) 12411794_Observational study of gene-disease association. (HuGE Navigator) 12426312_Ligand binding promotes the entropy-driven oligomerization of this protein 12446192_Observational study of gene-disease association. (HuGE Navigator) 12506038_A naturally occurring Tyr143His alpha IIb mutation abolishes alpha IIb beta 3 function for soluble ligands but retains its ability for mediating cell adhesion and clot retraction. The functional defect is likely caused by its allosteric effect. 12511588_Integrin alphaiibbeta3 plays a role in signal transduction that leads to a defect in leukocyte adhesion 12586134_Observational study of gene-disease association. (HuGE Navigator) 12586134_possible association between the GPIIb/IIIa PIA1/A2 polymorphism and the occurrence of cryptogenic stroke in young patients 12609844_data suggest that despite partial disruption of calf-1 or calf-2 domain by mutation in Glanzmann thrombasthenia, glycoprotein IIb/IIIa complex is formed but its transport from the endoplasmic reticulum is impaired 12637342_Review. A signal is initiated at the cytoplasmic tail to transform the extracellular domain of alphaIIbbeta3 into a functional receptor for fibrinogen or von Willebrand factor to support platelet aggregation & thrombus formation. 12668663_Intercellular calcium communication is primarily mediated by a signaling mechanism operating between this protein, integrin beta 3 and the adenosine diphosphate purinergic receptor P2Y12. 12719784_binding of thrombin to GPIbalpha induces fibrin binding to resting alphaIIbbeta3 leading to fibrin-dependent platelet aggregation and clot retraction, that can be selectively inhibited by alphaIIbbeta3 antagonists 12724616_Observational study of gene-disease association. (HuGE Navigator) 12730600_results suggest that homomeric associations involving transmembrane domains provide a driving force for integrin activation; results also suggest a structural basis for the coincidence of integrin activation and clustering 12736272_the extracellular Ig domain of IAP(CD47), when bound to thrombospondin, interacts with integrin alphaIIbbeta3 and can change alphaIIbbeta3 in a high affinity state without the requirement of intracellular signaling 12799374_identification of binding site in gamma C-domain of fibrinogen 12827240_Pathways dependent on the platelet alpha(2)beta(3) integrin physiology could be implicated in the pathogenesis of Type 2 diabetes. 12860973_Bidirectional signaling of ITGA2B requires the beta3 cytoplasmic domain. 12871362_Observational study of gene-disease association. (HuGE Navigator) 12871379_reduced availability of GPIIb-mRNA associated with G188A mutation is due to inefficient RNA splicing or utilization of alternative intronic donor sites that generate an in-frame STOP codon resulting in activation of nonsense-mediated mRNA decay, or both 12871468_high prevalence of Glanzmann thrombasthenia patients homozygous for the so-called French gypsy mutation (IVS15[ + 1]G-->A) in alphaIIb 12871600_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 12911597_Glycoprotein IIb/IIIa has a role in platelet-activating factor-induced platelet activation with protein kinase C activity 12938014_Observational study of gene-disease association. (HuGE Navigator) 14521607_the E749ATSTFTN756 region of the beta3-tail stabilizes the binding of soluble and surface-bound ligand to integrin alphaIIbbeta3 via a mechanism that involves the phosphorylation of FAK 14525764_proteins regulated by CBFA2 are required for inside-out signal transduction-dependent activation of GPIIb-IIIa 14597981_Substitution of conserved serine residues at position 1 in beta strand A of all seven repeats of alpha(IIb) similarly inhibited ligand binding to alpha(IIb)beta(3). 14671618_Observational study of gene-disease association. (HuGE Navigator) 14675395_Observational study of genetic testing. (HuGE Navigator) 15056669_analysis of the three-dimensional model of integrin alphaIIbbeta3 15121870_ERK promotes megakaryocyte differentiation by coordinate regulation of nuclear factors that synergize in GPIIb promoter regulation. 15131115_integrin alpha(IIb)beta3 adhesive function is regulated by platelet FXIII and calpain 15134555_This review discusses the major role played by glycoprotein IIb/IIIa (GPIIb/IIIa) in the regulation of platelet adhesion and aggregation during hemostasis. 15166241_alpha IIb beta 3 signaling in platelets is regulated by SHIP1 and Lyn kinase 15205468_studies reveal a previously unrecognized role for integrin alpha 2b whereby the alpha subunit cytoplasmic tail localizes the machinery for initiating and temporally maintaining the regulatory signaling activity of protein phosphatase 1 15219201_Phe171 plays an essential role in the interface between the beta-propeller domain of alpha(IIb) and the betaA domain of beta(3) 15226180_CD151 is essential for normal platelet function and that disruption of CD151 induced a moderate outside-in integrin alpha(IIb)beta(3) signaling defect 15227729_HPA-3 polymorphism was associated with MI in Korean individuals younger than 56 years of age, but other polymorphisms of GP 15227729_Observational study of gene-disease association. (HuGE Navigator) 15316595_Neither liver transplantation nor liver resection influences GPIIb/IIIa and P-selectin expression on circulating platelets. 15355503_Observational study of genetic testing. (HuGE Navigator) 15630502_Observational study of gene-disease association. (HuGE Navigator) 15634267_a novel missense mutation of Ile304 to Asn in addition to the missense mutation His280 to Pro in the integrin beta3 gene as a cause of the absence of platelet alphaIIbbeta3 in Glanzmann's thrombasthenia 15678278_There was no significant correlation between the incidence of autoimmune thrombocytopenia disorder and the HPA-3 gene polymorphism pattern; there is a greater variation in HPA-3 frequencies than in the HPA-1 gene. 15701653_integrin alphaIIbbeta3 has a role in mechanotransduction for shear activation of platelets 15701721_alphaIIb-mediated outside-in signaling resulting in TxA(2) production and granule secretion is negatively regulated by a sequence of residues in the membrane distal beta3 cytoplasmic domain sequence RKEFAKFEEER. 15730528_Observational study of genotype prevalence. (HuGE Navigator) 15748238_the novel mutation InsC, leading to a frameshift that affected the transmembrane domain and the cytoplasmic tail, formed a complex with beta3, but had not been transported to the Golgi apparatus 15842360_SHPS-1 negatively regulates platelet function via CD47, especially alpha(IIb)beta(3)-mediated outside-in signaling 15847651_Observational study of genetic testing. (HuGE Navigator) 15863506_TPO integrates G(i), but not G(q), stimulation, supports integrin alpha(IIb)beta(3) activation platelet aggregation independently of phospholipase C but requires PI3-kinase and Rap1B 15886807_a homozygous mutation (1619delC) in GPIIb may have a role in Glanzmann thrombasthenia 15890274_electron tomography analysis of the three-dimensional structure of integrin alphaIIbbeta3 in the active state 15917997_PI3-K and calcium oscillation are synergistically operated and form a positive-feedback regulation in integrin alpha(IIb)beta3-mediated outside-in signaling 15976180_reactive oxygen species produced in platelets significantly affected alphaIIbbeta3 integrin activation 15978110_platelet membrane integrins alpha IIb(beta)3 (HPA-1b/Pl) and alpha2(beta)1 (alpha807TT) polymorphisms may have a role in premature myocardial infarction 16051597_the engagement of alphaIIb beta3 by the C-terminal sequence of the fibrinogen gamma-chain initiates signals that suppress subsequent fibronectin assembly by spread platelets 16102042_GPVI and integrin alphaIIb beta3 have roles in signaling during platelet adhesion and platelet aggregation [review] 16115959_Data show that protein-tyrosine phosphatase (PTP)-1B is an essential positive regulator of the initiation of outside-in alphaIIbbeta3 integrin signaling in platelets. 16228296_PI3K and granular released ADP have important roles in coordinating the feedback regulations in integrin alpha(IIb)beta3-mediated platelet activation 16248996_Observational study of gene-disease association. (HuGE Navigator) 16322781_analysis of platelet integrin alphaIIbbeta3 structure and function (review) 16324093_Observational study of gene-disease association. (HuGE Navigator) 16357324_Outside-in signaling through integrin alpha2beta1 triggered inside-out activation of integrin alphaIIbbeta3 and promoted fibrinogen binding. (alpha2beta1 AND integrin alphaIIbbeta3 16359515_three beta-propeller mutations do not affect the production of pro-alpha(IIb), its ability to complex with beta3, or its stability, but do cause variable defects in transport of pro-alpha(IIb)beta3 complexes from the endoplasmic reticulum to the Golgi 16469512_Inhibition of alphaIIbbeta3 activation by NAD(P)H oxidase inhibitors and superoxide scavengers was independent of NO/cGMP signaling demonstrating a direct role of platelet NAD(P)H oxidase-generated ROS for integrin alphaIIbbeta3 activation. 16487966_Together, these data demonstrate a novel interaction between collagen XVII and alphaIIb integrin and also suggest a possibility to use tirofiban to inhibit the invasion and progression of alphaIIb expressing SCC tumors. 16573563_Observational study of genotype prevalence. (HuGE Navigator) 16582881_Increase in affinity of alphaIIb beta3-mediated cell adhesion through binding to calcium- and integrin-binding protein following agonist stimulation is impaired in patients because of mutations. 16636497_P-selectin expression, but not activated GPIIb/IIIa, is enhanced in ADP-activated platelets in the inflammatory stage of Takayasu's arteritis. SELP may play a significant role in the inflammatory and thrombotic responses associated with intractable TA 16716076_while the C-terminal region of the alpha(IIb) tail minimally influences alpha(IIb)beta(3) activation, the C-terminal region of the beta(3) tail is critically involved 16773503_Observational study of gene-disease association. (HuGE Navigator) 16822941_Results suggest that shear stress directly modulates alpha(IIb)beta(3) function and that the shear-induced signaling contributes to regulation of platelet aggregation by directing the release of constraining cytoskeletal elements from the beta(3)-tail. 16825200_Data report the structural characteristics of calcium- and integrin-binding protein 1 in solution and the mechanistic details of its interaction with a synthetic peptide derived from the alphaIIb integrin cytoplasmic domain. 16877710_interactions between alphaIIbbeta3 & ligands echistatin & fibrinogen's gamma-module; studies differentiate priming ligands, which bind to resting receptor & perturb its conformation, from regulated ligands, where binding-site remodeling must first occur 16879215_analysis of platelet integrin alphaIIbbeta3 mutations that may play a role in Glanzmann thrombasthenia 16879224_the KVGFFKR motif regulates integrin alphaIIbbeta3 activation state and cytoskeletal reorganization 16895913_Protein kinase C delta is positively regulated by platelet alpha IIb beta 3 outside-in signaling in platelet function. 16905953_Observational study of gene-disease association. (HuGE Navigator) 17032655_platelet alphaIIbbeta3 is activated by an exogenous peptide corresponding to the transmembrane domain of alphaIIb 17138951_Our results indicate that prothrombotic genetic factors may interact with smoking by modifying the stroke phenotype and affecting midterm survival. 17160992_Observational study of genotype prevalence. (HuGE Navigator) 17320454_Observational study of genotype prevalence. (HuGE Navigator) 17332246_the surface density of fibrinogen affects alpha II b beta 3-mediated platelet signaling, adhesion, and spreading 17337041_in patients with stable coronary artery disease, an increased platelet reactivity after exercise, is specifically associated with an increased expression of platelet GP IIb/IIIa receptor 17346829_Observational study of gene-disease association. (HuGE Navigator) 17414216_Discussion of recent developments in elucidating the mechanism of integrin activation, with particular focus on the platelet integrin alphaIIbbeta3, is provided in this review 17468108_integrins alphaLbeta2 and alphaIIbbeta3 are activated by mutation of a conserved asparagine in the I-like domain 17488698_3 new mutations were found in GPIIb:c.440C->G/p.Leu116Val, c.1772_1773insG/p.Asp560GlyfsX16 & c.2438C->A/p.His782Asn). c.1772_1773insG caused an early stop codon.p.His782Asn compromised transport of the pro-GPIIb/IIIa complex. 17495265_Observational study of gene-disease association. (HuGE Navigator) 17538005_LOX-1 is important for ADP-stimulated inside-out activation of platelet alpha(IIb)beta(3) and alpha(2)beta(1) integrins 17561290_Observational study of gene-disease association. (HuGE Navigator) 17615290_separation of transmembrane domains is required for integrin outside-in signal transduction 17630485_Observational study of gene-disease association. (HuGE Navigator) 17635696_Regions of alpha(IIb) and beta(3) cytoplasmic tails, together with membrane segments of the subunits, contact each other to form a complex which restrains the integrin in a resting state. It is unclasping of this complex that induces integrin activation. 17644514_Beta2-integrins and acquired glycoprotein IIb/IIIa (GPIIb/IIIa) receptors cooperate in NF-kappaB activation of human neutrophils 17714854_Structural rearrangements undergone by integrin alpha(IIb)beta(3) after platelet activation were tested. 17721619_proportion of TF-positive or both TF- and platelet antigen CD41a-positive leukocytes was increased markedly in pericardial blood obtained during CPB. 17728329_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17763154_effects of EDTA on GPIIb-IIIa dissociation and loss of adhesive functions are largely but not completely reve 17767046_Observational study of gene-disease association. (HuGE Navigator) 17872456_Transduction with shRNA sequence targeting integrin alphaIIb caused a significant reduction of integrin alphaIIb beta3 expression in platelets. It also inhibited alphaIIb beta3 activation. 17922435_Observational study of genotype prevalence. (HuGE Navigator) 17954176_Observational study of gene-disease association. (HuGE Navigator) 18041653_oxidant systems external to platelets did not activate GP IIb/IIIa receptors while increased intra-platelet iron was associated with appearance of cytosolic oxidizing species and with GP IIb/IIIa receptor activation 18057877_Observational study of gene-disease association. (HuGE Navigator) 18057877_Polymorphisms in human platelet alloantigen (HPA)-1 and HPA-3 (GPIIb/IIIa), HPA-2 (GPIb/IX), HPA-4 (GPIIIa) and HPA-5 (GPIa/IIa) were found to be associated with the symptoms and recurrence of ischemic stroke. 18064323_Novel ancestral mutation found in a cluster of Jordanian Glanzmann thrombasthenia patients disrupts a conserved Cys549-Cys558 bond which results in reduced production of constitutively active alpha IIb beta 3. 18088350_platelet activation and aggregation is regulated by MMP-2 that specifically interacts with integrin alpha(IIb)beta(3) 18174155_the beta3 membrane-proximal and -distal residues cooperatively regulate talin-mediated alpha IIb beta3 activation. 18201749_Tyr178 of beta3 is critical for alphaIIb maturation and macromolecular ligand binding to alphaIIbbeta3 18211801_tyrosine phosphorylation of GIT1 by Src kinases may regulate cytoskeletal reorganization downstream of alpha(IIb)beta(3) by bringing the Rac guanine nucleotide exchange factor betaPIX to the vicinity of the integrin 18237139_Thermodynamic data demonstrate that entropy is the dominant factor stabilizing lphaIIbbeta3:echistatin binding, while transition-state thermodynamic parameters indicate that the rate of complex formation is enthalpy-limited. 18316480_Arginine-methylated RUNX1 regulates CD41 gene expression. 18316775_The numbers of antibody-binding sites of CD41, CD41a, CD41b, and CD61 on platelets of aplastic anemia patients and in idiopathic thrombocytopenic purpura patients, the numbers of sites for CD41 and CD41a were less than in normal controls. 18317590_support an unsuspected role of alphaIIbbeta3/serotonin transporter(SERT) associations as well as alphaIIbbeta3 activation in control of SERT activity in vivo 18328539_analysis of important residues within the central turn motifs present in the cytoplasmic tails of integrin alphaIIb and alphaV subunits 18331836_RN181 associates with the platelet-specific integrin alphaIIbbeta3 in human platelets through the conserved alphaIIb cytoplasmic KVGFFKR motif. 18334487_Protein phosphatase 2Ac (alpha) can negatively regulate integrin alpha(IIb)beta(3) signaling by suppressing the ERK1/2 signaling pathway 18383324_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18396070_Show that the Pl(A1/A2) polymorphism of Gp IIb/IIIa primarily affects the aggregation of platelets to monocytes in males. 18399841_Observational study of genotype prevalence. (HuGE Navigator) 18405917_Data show no dramatic height change upon Mn(2+)-induced activation of membrane-bound integrins when compared with an inactive integrin control group. 18433460_CRP at levels found during episodes of inflammation directly binds to the activated form of alphaIIbbeta3 and inhibits platelet aggregation. 18502778_Observational study of gene-disease association. (HuGE Navigator) 18573917_Talin recruitment to alphaIIbbeta3 by RIAM mediates agonist-induced alphaIIbbeta3 activation, with implications for hemostasis and thrombosis. 18638089_Observational study of gene-disease association. (HuGE Navigator) 18641368_Ligand binding to platelet alphaIIbbeta3 induces integrin cytoplasmic domain-dependent phosphorylation of FcgammaRIIa, which then enlists selected components of the immunoreceptor signaling cascade to transmit amplification signals into the cell. 18646302_Observational study of gene-disease association. (HuGE Navigator) 18791937_The alphaIIb G236E mutation causes Glanzmann's thrombasthenia by impairing the association with beta3 during biogenesis of the receptor. 18796633_UV-C appears to activate alphaIIbbeta3 not by affecting intracellular signal transduction, but by reduction of disulfide bonds regulating integrin conformation. 18826388_Splenic macrophages that take up opsonized platelets via FcgammaRI are major APCs for cryptic GPIIb/IIIa peptides, and are central to the maintenance of anti-platelet autoantibody production in ITP patients 18976939_A novel Pro126His mutation in alphaIIb compromised transport of the pro-alphaIIbbeta3 complex from the endoplasmic reticulum to the Golgi, leading to intracellular retention 18979362_interaction of the membrane proximal sequence 989KGVFFKR995 of cytoplasmic domain of alpha(IIb) with the acidic terminal 1000LEEDDEEGE1008 motif may be an important structural factor in platelet signaling, leading to platelet activation and aggregation 18979363_alpha(IIb)beta(3) provides a constitutive presence of factor H on platelets; activation of platelets increases platelet-bound factor which perhaps involves other binding sites for factor H on platelets. 18989530_Glutathione regulates integrin alpha(IIb)beta(3)-mediated cell adhesion under flow conditions. 18990338_Observational study of gene-disease association. (HuGE Navigator) 19082798_Observational study of gene-disease association. (HuGE Navigator) 19082798_The changes in amino-acids expression induced by the two loci brought no significant influence on GP I b and GP II b- III a activities. 19117493_Ligation of alpha(IIb)beta(3) by immobilized ligands during platelet adhesion induces a transmembrane conformation change in the integrin, resulting in unclasping of the complex between the membrane-proximal parts of cytoplasmic tails. 19132198_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19170775_Glanzmann's thrombasthenia mutation c.1028T>C in exon 12 of GPIIb gene. RFLP followed by gel electrophoresis revealed that the mutation was heterozygous in all the family members. 19172520_Six missense mutations, four deletions and two splice junction mutations were detected in the GPIIb gene of Indian patients with Glanzmann's thrombasthenia. 19232685_Reduced GPIIb expression is reported in platelets hyposensitive to catecholamines when activated with TRAP. 19262211_Glycoprotein IIB/IIIA inhibitor to reduce postpercutaneous coronary intervention myonecrosis and improve coronary flow in diabetics: the 'OPTIMIZE-IT' pilot randomized study. 19279667_The structurally unique, highly conserved integrin alphaIIbbeta3 transmembrane complex rationalizes bi-directional signalling and represents the first structure of a heterodimeric TM receptor complex. 19286442_HPA-3 but not HPA-1 polymorphism may be one of the inherited risk factors associated with the susceptibility of hantavirus infection and the disease severity of HFRS. 19329429_Three heterozygous mutations were identified in the genes encoding platelet integrin receptor alphaIIbbeta3 in a patient with an ill defined platelet disorder: one in the beta3 gene (S527F) and two in the alphaIIb gene (R512W and L841M). 19527732_Multiple approaches converged on the same structure of the resting integrin's transmembrane heterodimer, and this conformation likely reflects the integrin's native structure. 19562259_Observational study of gene-disease association. (HuGE Navigator) 19570064_Observational study of genetic testing. (HuGE Navigator) 19691478_Observational study of gene-disease association. (HuGE Navigator) 19702628_HPA 3a/3b & 3b/3b genotypes were associated with need for hospitalization. Only HPA-3b/3b was associated with vaso-occlusive crisis frequency, type and medication. 19702628_Observational study of gene-disease association. (HuGE Navigator) 19729601_Observational study of gene-disease association. (HuGE Navigator) 19734576_alphaIIbA477P (A446P) mutation distinctly reduces the expression of alphaIIbbeta3 complex on the membrane 19765213_initial activation-independent platelet adhesion to VWF via GPIb is resistant to NO, however, NO inhibits GPIb-mediated activation of alpha(IIb)beta(3) and MLC leading to reduced platelet spreading and aggregation. 19778317_In a Croatian population, the HPA-3 allele frequencies are 3a-0.575, 3b-0.425. 19804783_The authors demonstrate that the F0 domain of kindlin-1 is required for the ability of kindlin-1 to support talin-induced alphaIIbbeta3 integrin activation and for the localization of kindlin-1 to focal adhesions. 19805198_while a point mutation in the clasp interface modestly activates alphaIIb beta3, additional mutations in the transmembrane interface have a synergistic effect, leading to extensive integrin activation 19863457_Results describe the role of the membrane distal region of human beta(3) integrin cytoplasmic tail in platelet aggregation, secretion, alpha(IIb)beta(3) activation and fibrinogen binding. 19884334_The characterization of PadA and its interaction with human platelets at the fibrinogen receptor GPIIbIIIa are reported. 19888429_GPIIb/IIIa and alphavbeta3 integrins are important mediators in the pathology of cervical cancer. 19903699_Oxidatively modified fibrinogen showed less binding activity than native fibrinogen to GpIIb/IIIa coated micro beads and human platelets whereas slightly higher binding capacity to ADP induced stimulated platelets 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948007_Observational study of genotype prevalence. (HuGE Navigator) 19996122_Data suggest a role for GPIIbIIIa integrin in caspase-3 activation induced by these platelet agonists. 20002543_These data provide a mechanism for enhancement of platelet activity by alpha(IIb)beta(3) inhibitors, but also reveal a potentially important signaling pathway operating from the integrin to GPVI signaling. 20020534_analysis of ITGA2B and ITGB3 mRNA splicing, expression, and structure-function relationship in Glanzmann thrombasthenia 20075254_study demonstrates not only a function of integrin alphaIIbbeta3 as a noncanonical Galpha13-coupled receptor but also a new concept of Galpha13-dependent dynamic regulation of RhoA 20149160_Observational study of genotype prevalence. (HuGE Navigator) 20153702_Exocytosis of platelet integrin alpha 2b was reduced in Cystic fibrosis patients 20158572_Data show that PLT adhesion to HIMEC was ICAM-1, FKN and integrin alpha(v)beta(3) dependent. 20230421_results presented here strongly suggest that MMP-2 interacts with alpha(IIb)beta(3) to regulate the shedding of CD40L exposed on the surfaces of activated human platelets 20308600_the interaction between Trp110 of alphaIIb and Arg261 of beta3 is critical for alphaIIbbeta3 integrity and outside-in signaling-related functions 20363746_analysis of the effect of limiting extension at the alphaIIb genu on ligand binding to integrin alphaIIbbeta3 20486118_Presented is an analysis of the nature of the platelet proteins identified as integrin alpha 2b interactors by LC-MS/MS. 20514618_The present study describes a patient with GT associated with a novel homozygous deletion (c.175delG) in exon 1 of ITGA2B. This deletion led to a reading frameshift and caused a severely truncated form of GPIIb. 20584077_structure and mechanism of action of seems to depend on discrete transitions and to be more tightly coupled to the local environment than previously thought 20615878_Cross-talk between serine/threonine protein phosphatase 2A and protein tyrosine phosphatase 1B regulates Src activation and adhesion of integrin alphaIIbbeta3 to fibrinogen. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20652946_IL-13 enhances the expression of GPIIb in tumor cell lines through a mechanism dependent on the transcription factor STAT6 that is able to bind the GPIIb gene promoter and upregulates the promoter activity. 20667040_Observational study of genotype prevalence. (HuGE Navigator) 20723174_A new low-frequency alloantigen results from the Ser472Asn substitution in alphaIIb. This polymorphism has a limited effect, if any, on the alphaIIbbeta3 complex functions. 20819594_The alphaIIbbeta3 deficiency of the proband was caused by two compound ITGA2B mutations, which were first reported in Chinese Glanzmann thrombasthenia patients. 20828133_two orthogonal events regulate integrin alphaIIbbeta3's interactions with fibrinogen 21029361_Glanzmann thrombasthenia causing mutation in ITGA2B highlights the importance of calcium binding domains in the beta-propeller for intracellular trafficking of alphaIIbbeta(3). 21071690_Streptococcus gordonii binding to the alpha(IIb)beta(3)/FcgammaRIIa integrin/ITAM signaling complex results in platelet activation that likely contributes to the thrombotic complications of infective endocarditis. 21156831_Data show that the alphaIIb cytoplasmic domain is largely disordered, but it interacts with and influences the conformation of the beta3 cytoplasmic domain. 21190668_These data provide important quantitative and qualitative characteristics of alphaIIbbeta3-fibrinogen binding and unbinding that underlie the dynamics of platelet adhesion and aggregation in blood flow. 21388953_analysis of Ca2+-CIB1 and Mg2+-CIB1 and their interactions with the platelet integrin alphaIIb cytoplasmic domain 21454453_identified a novel, conserved heterozygous ITGA2B R995W mutation in 4 unrelated families with congenital macrothrombocytopenia. The surface expression of platelet alphaIIbbeta3 was decreased to 50% to 70% of control. 21487445_all carriers of the French Gypsy mutation c.1544+1G>A at intron 15 descended from a common ancestor 300-400 years ago. 21529934_GPIIb/IIIa is the primary receptor set involved in platelet adhesion to adsorbed fibrinogen and serum albumin irrespective of their degree of adsorption-induced unfolding, while the GPIb-IX-V receptor complex plays an insignificant role. 21645497_Only the threonine 410 gets dephosphorylated, possibly in an integrin aIIbb3-dependent manner 21832081_Intact alphaIIbbeta3 integrin is extended after activation 21917754_We now look at the repertoire of ITGA2B and ITGB3 gene defects, reexamine the relationship between phenotype and genotype, and review integrin structure in the many variant forms. 21966982_designed mutations at interface between alphaIIb-subunit thigh domain and beta3-subunit I-EGF2 domain to determine effect of this interface during integrin inside-out activation and outside-in signaling 22015616_Direct intracoronary bolus injection resulted in a more pronounced local inhibition of platelet function and a higher degree of GP IIb/IIIa receptor occupancy as compared to standard intravenous bolus injection. 22015659_Platelet alphaIIbeta3 and alpha2beta1 levels were measured by flow cytometry in 320 acute coronary syndrome patients and 128 normal subjects and compared with MPV, platelet count, ITGA2 rs1126643, and ITGB3 rs5918 alleles. 22078565_we show that while wi | ENSMUSG00000034664 | Itga2b | 77.602483 | 0.8697626 | -0.201306381 | 0.33581804 | 3.622125e-01 | 5.472800e-01 | No | Yes | 71.961248 | 14.349846 | 88.024897 | 17.532955 | ||
| ENSG00000006740 | 9912 | ARHGAP44 | protein_coding | Q17R89 | FUNCTION: GTPase-activating protein (GAP) that stimulates the GTPase activity of Rho-type GTPases. Thereby, controls Rho-type GTPases cycling between their active GTP-bound and inactive GDP-bound states. Acts as a GAP at least for CDC42 and RAC1 (PubMed:11431473). In neurons, is involved in dendritic spine formation and synaptic plasticity in a specific RAC1-GAP activity (By similarity). Limits the initiation of exploratory dendritic filopodia. Recruited to actin-patches that seed filopodia, binds specifically to plasma membrane sections that are deformed inward by acto-myosin mediated contractile forces. Acts through GAP activity on RAC1 to reduce actin polymerization necessary for filopodia formation (By similarity). In association with SHANK3, promotes GRIA1 exocytosis from recycling endosomes and spine morphological changes associated to long-term potentiation (By similarity). {ECO:0000250|UniProtKB:F1LQX4, ECO:0000250|UniProtKB:Q5SSM3, ECO:0000269|PubMed:11431473}. | Alternative splicing;Cell junction;Cell projection;Endosome;Exocytosis;GTPase activation;Phosphoprotein;Reference proteome;Synapse | hsa:9912; | cytosol [GO:0005829]; dendrite [GO:0030425]; dendritic spine [GO:0043197]; glutamatergic synapse [GO:0098978]; leading edge membrane [GO:0031256]; postsynaptic density [GO:0014069]; presynaptic active zone [GO:0048786]; recycling endosome [GO:0055037]; GTPase activator activity [GO:0005096]; phospholipid binding [GO:0005543]; small GTPase binding [GO:0031267]; exocytosis [GO:0006887]; modification of dendritic spine [GO:0098886]; negative regulation of filopodium assembly [GO:0051490]; negative regulation of Rac protein signal transduction [GO:0035021]; neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0098887]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of dendritic spine morphogenesis [GO:0061001]; regulation of GTPase activity [GO:0043087]; regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0099152]; regulation of Rac protein signal transduction [GO:0035020]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | 11431473_Demonstrates that the RhoGAP domains of RICH-1 and RICH-2 can catalyze GTP hydrolysis of both Rac1 and Cdc42, but not of RhoA. 19273615_Knocking down expression of RICH 2 causes loss of the apical actin network and apical microvilli, an increase in actin bundles at the basal surface, and a reduction in cell height. (RICH 2 protein) 21107268_Besides the already known chromosome 6 associations, the analysis of low-frequency single nucleotide polymorphisms brought up a new association in the RICH2 gene for progression to AIDS. 28069446_RICH2 is implicated in viremic control of HIV-1 in black South African individuals. 28527113_Rho GTPase activating protein 44 (ARHGAP44) expression is lower in lung carcinoma compared with normal tissues. The GTP hydrolysis activity on cell division cycle 42 (Cdc42) and the expression level of ARHGAP44 are negatively correlated with cell migration and invasion. 31136984_Rho GTPase-activating protein RICH2 (RICH2) is downregulated in hepatocellular carcinoma (HCC). 33223223_CD317 mediates immunocytolysis resistance by RICH2/cytoskeleton-dependent membrane protection. | ENSMUSG00000033389 | Arhgap44 | 514.419435 | 1.0073315 | 0.010538577 | 0.12562522 | 6.971439e-03 | 9.334578e-01 | 9.753664e-01 | No | Yes | 494.437358 | 35.455107 | 489.941770 | 35.005953 | ||
| ENSG00000009950 | 51085 | MLXIPL | protein_coding | Q9NP71 | FUNCTION: Transcriptional repressor. Binds to the canonical and non-canonical E box sequences 5'-CACGTG-3' (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Williams-Beuren syndrome | This gene encodes a basic helix-loop-helix leucine zipper transcription factor of the Myc/Max/Mad superfamily. This protein forms a heterodimeric complex and binds and activates, in a glucose-dependent manner, carbohydrate response element (ChoRE) motifs in the promoters of triglyceride synthesis genes. The gene is deleted in Williams-Beuren syndrome, a multisystem developmental disorder caused by the deletion of contiguous genes at chromosome 7q11.23. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2015]. | hsa:51085; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; carbohydrate response element binding [GO:0035538]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; anatomical structure morphogenesis [GO:0009653]; energy homeostasis [GO:0097009]; fatty acid homeostasis [GO:0055089]; glucose homeostasis [GO:0042593]; glucose mediated signaling pathway [GO:0010255]; negative regulation of oxidative phosphorylation [GO:0090324]; negative regulation of peptidyl-serine phosphorylation [GO:0033137]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of fatty acid biosynthetic process [GO:0045723]; positive regulation of glycolytic process [GO:0045821]; positive regulation of lipid biosynthetic process [GO:0046889]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; triglyceride homeostasis [GO:0070328] | 16644671_This evolutionally conserved mechanism may play an essential role in glucose-responsive gene regulation. 18193043_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18193044_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18193046_Genome-wide association study of gene-disease association. (HuGE Navigator) 18193046_Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. 18591247_Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity.( 18606808_Phosphorylation of ChREBP was essential for its interaction with CRM1 for export to the cytosol, whereas nuclear import of ChREBP requires dephosphorylated ChREBP to interact with importin alpha. 18946681_Observational study of gene-disease association. (HuGE Navigator) 18946681_tested the hypothesis that the MLXIPL rs3812316 variant predicts plasma triglyceride (TG) levels. We found no difference between individuals with high TG and controls, and no association between the variant and plasma TG levels among the controls 18950580_The transcription factor ChRepsilonBP is a major mediator of glucose action on lipogenic genes & a key determinant of lipid synthesis in vitro. Review. 19148283_Observational study of gene-disease association. (HuGE Navigator) 19252981_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19252981_we were not able to find any statistically significant association between the single nucleotide polymorphisms in the FAS, ChREBP and SREPB-1 genes and an increased risk of breast cancer 19487539_Observational study of gene-disease association. (HuGE Navigator) 19571538_G771C polymorphism significantly related to coronary artery disease in Chinese patients 19680233_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19995986_suppression of ChREBP led to a p53-dependent reduction in tumor growth. These results demonstrate that ChREBP plays a key role both in redirecting glucose metabolism to anabolic pathways and suppressing p53 activity 20025850_a new nuclear export signal site ('NES1') of ChREBP was reported. 20158509_Observational study of gene-disease association. (HuGE Navigator) 20167577_Observational study of gene-disease association. (HuGE Navigator) 20570916_Observational study of gene-disease association. (HuGE Navigator) 20580033_Observational study of gene-disease association. (HuGE Navigator) 20679960_Observational study of gene-disease association. (HuGE Navigator) 20965718_Observational study of gene-disease association. (HuGE Navigator) 20972250_Observational study of gene-disease association. (HuGE Navigator) 21036147_These results suggest that the O-linked glycosylation of ChREBP itself or other proteins that regulate ChREBP is essential for the production of functional ChREBP. 21145868_in immortalized hepatocytes and in HepG2 hepatoma cells, only SREBP1c was able to induce adiponutrin/PNPLA3 expression, whereas ChREBP was unable to modulate its expression 21282101_ChREBP is a critical and direct mediator of glucose repression of PPARalpha gene expression in pancreatic beta-cells 21665952_an important mechanism by which importin-alpha and 14-3-3 control movement of ChREBP in and out of the nucleus in response to changes in glucose levels in liver 21726544_The rs3812316 and the haplotypes in ChREBP gene appeared to be related to high susceptibility to CAD 21811631_ChREBP may function as a transcriptional repressor as well as an activator. 21835137_Our study reports that PP2A activity is dispensable for ChREBP activation in response to glucose and that dephosphorylation on Ser-196 is not sufficient to promote ChREBP nuclear translocation in the absence of a rise in glucose metabolism. 21840420_The dramatic increase of ChREBP mRNA and protein levels during preadipocyte differentiation suggests a role in adipogenesis. 21938000_Multiple linear regression models based on 2373 individuals of Asian origin showed that the H allele of the MLXIPL gene was significantly associated with decreased concentrations of plasma triglycerides. 22338092_sorcin retains ChREBP in the cytosol at low glucose concentrations and may act as a Ca(2+) sensor for glucose-induced nuclear translocation and the activation of ChREBP-dependent genes. 22466288_ChREBP-beta expression in human adipose tissue predicts insulin sensitivity, indicating that it may be an effective target for treating diabetes. 22546860_ChREBP overexpression induced expression of stearoyl-CoA desaturase 1 (Scd1), the enzyme responsible for the conversion of saturated fatty acids (SFAs) into MUFAs 23209190_Data from obese adolescents with prediabetes/early type 2 diabetes suggest that expression of ChREBP-alpha/beta in abdominal subcutaneous adipose tissue is inversely related to hyperglycemia severity and positively correlated to insulin resistance. 23443556_de novo lipogenesis predicts metabolic health in humans in a tissue-specific manner and is likely regulated by glucose-dependent carbohydrate-responsive element-binding protein activation. 23530060_Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. 23597489_Data suggest that CHREBP is a central regulator of glycolysis/lipogenesis in liver and apoptosis/proliferation in specific cell types. [REVIEW] 23604004_Data suggest that the activity of CHREBP is regulated via various mechanisms and that CHREBP is involved in the modulation of glucose and lipid metabolism in liver, pancreatic beta-cells, and adipose tissue. [REVIEW] 23803610_FOXO1 inhibits beta cell TXNIP transcription and suggest that FOXO1 confers this inhibition by interfering with ChREBP DNA binding at target gene promoters. 24055811_FLII is a component of the ChREBP transcriptional complex and negatively regulates ChREBP function in cancer cells. 24366300_The ChREBP expression may be reflective of an aerobic metabolic phenotype that may conflict with hypoxia-induced signalling but provide a mechanism for growth at the oxygenated edge of the tumours. 24448738_The MLXIPL-rs3812316 was associated with lower baseline triglycerides and lower hypertriglyceridemia. 24449882_ChREBP plays a key role in reprogramming glucose and lipid metabolism in human cytomegalovirus infection. 24616092_The ChREBP mutant, W130A, did not exhibit HG-induced lipid accumulation and fibrotic proteins, suggesting that the Trp-130 residue in the MCR3 domain is important in the development of glomerulosclerosis. 24664750_High glucose-induced, ChREBP-mediated, and normoxic HIF-1alpha activation that may be partially responsible for neovascularization in both diabetic and age-related retinopathy. 24845031_demonstrates that Chrebp interacts with AR and regulates its transcriptional activity 24989072_Significant linkage disequilibria were noted among ZNF259, BUD13 and MLXIPL SNPs and serum lipid levels. 25111846_results demonstrate that AGEs-RAGE signaling enhances cancer cell proliferation in which AGEs-mediated ChREBP induction plays an important role. 25288136_Single-nucleotide polymorphisms alleles near MLXIPL that were associated with higher coffee consumption. 25573592_Polymorphisms in lipid level modifier MLXIPL, GCKR, GALNT2, CILP2, ANGPTL3 and TRIB1 genes are highly associated with plasma lipid level changes. 26124292_A major function of Mio in mitosis is to regulate the activation/deactivation of Plk1 and Aurora A. 26147751_Metformin down-regulates high-glucose-induced TXNIP transcription by inactivating ChREBP and FOXO1 in endothelial cells, partially through AMP-activated protein kinase activation 26177557_the single nucleotide polymorphism of MLXIPL is significantly associated with Non-alcoholic Fatty Liver Disease. 26384380_Data suggest that expression of ChREBPbeta isoform is up-regulated in pancreatic beta-cells in response to elevated levels of glucose (i.e., hyperglycemic conditions). 26526060_Diet-induced obesity increases basal expression of ChREBPbeta, which may increase the risk of developing hepatic steatosis, and fructose-induced activation is independent of gluconeogenesis. 26910886_Evaluation of the conservation of ChREBP and MondoA sequences demonstrate that MondoA is better conserved and potentially mediates more ancient function in glucose metabolism. 27029511_The results revealed the novel mechanism by which HNF-4alpha promoted ChREBP transcription in response to glucose, and also demonstrated that ChREBP-alpha and HNF-4alpha synergistically increased ChREBP-beta transcription. 27033449_High glucose-mediated induction of PDGF-C via ChREBP in mesangial cells contributes to the development of glomerular mesangial expansion in diabetes. 27281235_results indicated that the age and total cholesterol concentrations were independent influential factors of ChREBP methylation and DNMT1 variants could probably influence LDL-C to further modify ChREBP DNA methylation 27599772_p = 6.69 x 10(-9) ] on chr7 at the carbohydrate-responsive element-binding protein-encoding (MLXIPL) gene locus displayed significant protective characteristics, while another variant rs6982502 [0.76 (0.68-0.84); p = 5.31 x 10(-7) ] on chr8 showed similar but weaker properties. 27669460_these findings support a carbohydrate-mediated, ChREBP-driven mechanism that contributes to hepatic insulin resistance. 27854512_This cross-sectional study suggests that MLXIPL rs3812316 genotypes may be associated with Triglyceride levels. there were significantly different genotype distributions in two TG categories: (1) subjects with normal TG values had a significantly higher G allele frequency than those with elevated TG levels 27919710_ChREBP regulates gene transcription related to glucose and lipid metabolism. Findings from knockout mice and human subjects suggest that ChREBP helps to induce hepatic steatosis, dyslipidemia, and glucose intolerance. [review] 28027934_ChREBP role in non-alcoholic fatty liver disease.The involvement of ChREBP in FASN promoter histone modification. 28123933_ChREBP and FGF21 constitute a signaling axis likely conserved in humans that mediates an essential adaptive response to fructose ingestion that may participate in the pathogenesis of NAFLD and liver fibrosis. 28606928_A nutrient-sensitive mTOR/ChREBP regulated transcriptional network could be a novel target to improve beta cell survival and glucose homeostasis in diabetes. 28768172_ChREBP was initially studied as a master regulator of lipogenesis in liver and fat tissue, it is now clear that ChREBP functions as a central metabolic coordinator in a variety of cell types in response to environmental and hormonal signals, with wide implications in health and disease. 29153407_AKT2 drives de novo lipogenesis in adipocytes by stimulating ChREBPbeta transcriptional activity and that cold induces the AKT2-ChREBP pathway in brown adipose tissue to optimize fuel storage and thermogenesis. 29764859_Data (including data from studies using tissues/cells from transgenic mice) suggest that ChREBPalpha up-regulates expression and activity of NRF2, initiating mitochondrial biogenesis in beta-cells; induction of NRF2 is required for ChREBPalpha-mediated effects and for glucose-stimulated beta-cell proliferation. [NRF2 = nuclear factor (erythroid-derived 2)-like 2 protein] 29858861_The results of this population-based study provide evidence for a relationship between lipid regulatory gene polymorphisms including GCKR (rs780094), GCKR (rs1260333), FADS (rs174547), and MLXIPL (rs3812316) with dyslipidemia in an Iranian population. 30079502_we provide evidence that the rs1051943 A allele creates a functional miR-1322 binding site in ChREBP 3'-UTR and post-transcriptionally down-regulates its expression, possibly associated with levels of plasma lipids and glucose. 30420491_The expression levels of ChREBP and several cytokines (TNF-alpha, IL-1beta, and IL-6) were up-regulated in type 2 diabetes mellitus patients. 31227231_The HCF-1:ChREBP complex resides at lipogenic gene promoters, where HCF-1 regulates H3K4 trimethylation to prime recruitment of the Jumonji C domain-containing histone demethylase PHF2 for epigenetic activation of these promoters.. these findings define HCF-1's interaction with ChREBP as a previously unappreciated mechanism whereby glucose signals are both relayed to ChREBP and transmitted for epigenetic regulation 31407220_Expressions of Carbohydrate Response Element Binding Protein and Glucose Transporters in Liver Cancer and Clinical Significance. 31409643_our results indicate that SMURF2 reduces aerobic glycolysis and cell proliferation by promoting ChREBP ubiquitination and degradation via the proteasome pathway in colorectal cancer cells. We conclude that the SMURF2-ChREBP interaction might represent a potential target for managing colorectal cancer 31413120_Two cases were hemizygous for the rare T allele at rs12539160 in MLXIPL, previously associated with Autism Spectrum Disorder. 31782782_TXNIP induced by MondoA, rather than ChREBP, suppresses cervical cancer cell proliferation, migration and invasion. 32583421_Carbohydrate response element-binding protein regulates lipid metabolism via mTOR complex1 in diabetic nephropathy. 32776146_The structure of importin alpha and the nuclear localization peptide of ChREBP, and small compound inhibitors of ChREBP-importin alpha interactions. 33320482_Transcription factor ChREBP - the coordinator of carbohydrate and lipid metabolism', trans 'Czynnik transkrypcyjny ChREBP - koordynator metabolizmu weglowodanow i lipidow. 34270325_Sugar-Sweetened Beverage Consumption May Modify Associations Between Genetic Variants in the CHREBP (Carbohydrate Responsive Element Binding Protein) Locus and HDL-C (High-Density Lipoprotein Cholesterol) and Triglyceride Concentrations. 34461288_High glucose mediates the ChREBP/p300 transcriptional complex to activate proapoptotic genes Puma and BAX and contributes to intervertebral disc degeneration. 34600826_ChREBP deficiency alleviates apoptosis by inhibiting TXNIP/oxidative stress in diabetic nephropathy. 34769488_The Roles of Carbohydrate Response Element Binding Protein in the Relationship between Carbohydrate Intake and Diseases. 34784250_Thyroid hormone signaling promotes hepatic lipogenesis through the transcription factor ChREBP. 34948094_Transcription Factor ChREBP Mediates High Glucose-Evoked Increase in HIF-1alpha Content in Epithelial Cells of Renal Proximal Tubules. | ENSMUSG00000005373 | Mlxipl | 232.451683 | 1.1948078 | 0.256778548 | 0.17566357 | 2.127168e+00 | 1.447079e-01 | No | Yes | 227.379601 | 38.999002 | 196.793898 | 33.827414 | ||
| ENSG00000011332 | 8193 | DPF1 | protein_coding | Q92782 | FUNCTION: May have an important role in developing neurons by participating in regulation of cell survival, possibly as a neurospecific transcription factor. Belongs to the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). {ECO:0000250}. | Alternative splicing;Cytoplasm;Isopeptide bond;Metal-binding;Neurogenesis;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:8193; | cytoplasm [GO:0005737]; nBAF complex [GO:0071565]; histone binding [GO:0042393]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription coregulator activity [GO:0003712]; zinc ion binding [GO:0008270]; apoptotic process [GO:0006915]; negative regulation of transcription, DNA-templated [GO:0045892]; nervous system development [GO:0007399]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 34696437_BAF45b Is Required for Efficient Zika Virus Infection of HAP1 Cells. | ENSMUSG00000030584 | Dpf1 | 376.188501 | 0.9253808 | -0.111880945 | 0.15861976 | 5.083670e-01 | 4.758466e-01 | No | Yes | 366.978536 | 41.742923 | 397.910553 | 45.004226 | |||
| ENSG00000013441 | 1195 | CLK1 | protein_coding | P49759 | FUNCTION: Dual specificity kinase acting on both serine/threonine and tyrosine-containing substrates. Phosphorylates serine- and arginine-rich (SR) proteins of the spliceosomal complex and may be a constituent of a network of regulatory mechanisms that enable SR proteins to control RNA splicing. Phosphorylates: SRSF1, SRSF3 and PTPN1. Regulates the alternative splicing of tissue factor (F3) pre-mRNA in endothelial cells and adenovirus E1A pre-mRNA. {ECO:0000269|PubMed:10480872, ECO:0000269|PubMed:19168442}. | 3D-structure;ATP-binding;Alternative splicing;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Tyrosine-protein kinase | This gene encodes a member of the CDC2-like (or LAMMER) family of dual specificity protein kinases. In the nucleus, the encoded protein phosphorylates serine/arginine-rich proteins involved in pre-mRNA processing, releasing them into the nucleoplasm. The choice of splice sites during pre-mRNA processing may be regulated by the concentration of transacting factors, including serine/arginine rich proteins. Therefore, the encoded protein may play an indirect role in governing splice site selection. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jun 2009]. | hsa:1195; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ATP binding [GO:0005524]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; protein autophosphorylation [GO:0046777]; regulation of RNA splicing [GO:0043484] | 12773558_Clk/Sty is found in the nucleus of several different cell types and is a factor directly involved in splicing control 16223727_ASF/SF2 is phosphorylated by SRPK1 and Clk/Sty 18814951_five genes (TNFSF10/TRAIL, IL1RN, IFI27, GZMB, and CCR5) were upregulated and three genes (CLK1, TNFAIP3 and BTG1) were downregulated in at least three out of four subpopulations during acute GVHD. 19168442_Cdc2-like kinases and DNA topoisomerase I regulate alternative splicing of tissue factor in human endothelial cells. 21682887_CLK1 Increases While CLK2 Decreases HIV-1 Gene Expression. 23707382_The data establish a new view of SRSF1 protein regulation in which SRPK1 and CLK1 partition activities based on Ser-Pro versus Arg-Ser placement rather than on N- and C-terminal preferences along the RS domain. 24869919_findings suggest that CLK1-dependent hyperphosphorylation is the result of a general mechanism in which the N-terminus acts as a bridge connecting the kinase domain and the RS domain of the SR protein. 25529026_Data suggest that proline phosphorylation by CLK1/CDC-like kinase 1 (but not by SRPK1/serine/arginine-rich splicing factor kinase 1) regulates conformation and alternative splicing function of SFRS1 (serine/arginine-rich splicing factor 1). 25961505_Nuclear CLK-1 mediates a retrograde signalling pathway that is conserved from Caenorhabditis elegans to humans and is responsive to mitochondrial reactive oxygen species, thus acting as a barometer of oxidative metabolism. 26443864_removal of the N-terminus or dilution of CLK1 induces monomer formation and reverses this specificity. CLK1 self-association also occurs in the nucleus 27015110_These results thus reveal a large program of CLK1-regulated periodic alternative splicing intimately associated with cell cycle control. 27126587_Clk1, Clk2 and Clk4 prevent chromatin breakage by regulating the Aurora B-dependent abscission checkpoint. 27397683_SRPK1 interacts with an RS-like domain in the N terminus of CLK1 to facilitate the release of phosphorylated SR proteins, which then promotes efficient splice-site recognition and subsequent spliceosome assembly. 28232751_Global CLK-dependent exon recognition and conjoined gene formation revealed with a novel small molecule inhibitor has been described. 28581641_that mitochondrial Clk1 regulated chemoresistance in glioma cells through AMPK/mTOR/HIF-1alpha mediated glycolysis pathway 29335301_We now show that the ability of SRPK1 to mobilize SRSF1 from speckles to the nucleoplasm is dependent on active CLK1. Diffusion from speckles is promoted by the formation of an SRPK1-CLK1 complex that facilitates dissociation of SRSF1 from CLK1 and enhances the phosphorylation of several serine-proline dipeptides in this SR protein 29802995_Data suggest that CLK1 activity in both transformed and normal cells is carefully regulated via CLK1 alternative splicing through both exon 4 skipping and intron 4 retention. CLK1 autoregulates by favoring the expression of truncated isoforms, CLK1T1 and CLKT2 in environmental stress. 31064840_found that CLK1 amplifies its presence in the nucleus by forming a stable complex with the Ser-Arg protein substrate 31782550_LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. 32333232_Phosphoproteomic analysis identifies CLK1 as a novel therapeutic target in gastric cancer. 32359191_A conserved sequence motif bridges two protein kinases for enhanced phosphorylation and nuclear function of a splicing factor. 33811140_CLK1 reorganizes the splicing factor U1-70K for early spliceosomal protein assembly. | ENSMUSG00000026034 | Clk1 | 2153.766215 | 1.0552082 | 0.077527668 | 0.07262877 | 1.149322e+00 | 2.836911e-01 | 6.542635e-01 | No | Yes | 2003.980949 | 394.788816 | 1865.294486 | 367.504769 | |
| ENSG00000015153 | 10138 | YAF2 | protein_coding | Q8IY57 | FUNCTION: Binds to MYC and inhibits MYC-mediated transactivation. Also binds to MYCN and enhances MYCN-dependent transcriptional activation. Increases calpain 2-mediated proteolysis of YY1 in vitro. Component of the E2F6.com-1 complex, a repressive complex that methylates 'Lys-9' of histone H3, suggesting that it is involved in chromatin-remodeling. {ECO:0000269|PubMed:11593398, ECO:0000269|PubMed:12706874, ECO:0000269|PubMed:9016636}. | 3D-structure;Alternative splicing;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a zinc finger containing protein that functions in the regulation of transcription. This protein was identified as an interacting partner of transcriptional repressor protein Yy1, and also interacts with other transcriptional regulators, including Myc and Polycomb. This protein can promote proteolysis of Yy1. Multiple alternatively spliced transcript variants have been found. [provided by RefSeq, Feb 2016]. | hsa:10138; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; transcription corepressor activity [GO:0003714]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription, DNA-templated [GO:0006355] | 11953439_YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. 19240061_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25603536_These findings uncovered an apoptotic signaling cascade linking YAF2, PDCD5, and TP53 during genotoxic stress responses. 30672021_LINC00341 interacts with miR-141 to suppress its functional binding to the 3'-untranslated region of YAF2 messenger RNA whereby supporting chondrocyte survival and preventing osteoarthritis progression. 30915747_miR-34b inhibits the migration/invasion and promotes apoptosis of non-small-cell lung cancer cells by YAF2. 33784512_YAF2 exerts anti-apoptotic effect in human tumor cells in a FANK1- and phosphorylation-dependent manner. | ENSMUSG00000022634 | Yaf2 | 416.591347 | 0.8210153 | -0.284518901 | 0.13412948 | 4.450375e+00 | 3.489358e-02 | No | Yes | 409.297178 | 64.620038 | 477.921156 | 75.474845 | ||
| ENSG00000015413 | 1800 | DPEP1 | protein_coding | P16444 | FUNCTION: Hydrolyzes a wide range of dipeptides including the conversion of leukotriene D4 to leukotriene E4 (PubMed:2303490, PubMed:6334084, PubMed:31442408). Hydrolyzes cystinyl-bis-glycine (cys-bis-gly) formed during glutathione degradation (By similarity). Possesses also beta lactamase activity and hydrolytically inactivates beta-lactam antibiotics (PubMed:6334084). {ECO:0000250|UniProtKB:P31428, ECO:0000269|PubMed:2303490, ECO:0000269|PubMed:31442408, ECO:0000269|PubMed:6334084}.; FUNCTION: Independently of its dipeptidase activity, acts as an adhesion receptor for neutrophil recruitment from bloodstream into inflamed lungs and liver. {ECO:0000250|UniProtKB:P31428}. | 3D-structure;Cell membrane;Cell projection;Dipeptidase;Direct protein sequencing;Disulfide bond;GPI-anchor;Glycoprotein;Hydrolase;Lipid metabolism;Lipoprotein;Membrane;Metal-binding;Metalloprotease;Protease;Reference proteome;Signal;Zinc | The protein encoded by this gene is a kidney membrane enzyme involved in the metabolism of glutathione and other similar proteins by dipeptide hydrolysis. The encoded protein is known to regulate leukotriene activity by catalyzing the conversion of leukotriene D4 to leukotriene E4. This protein uses zinc as a cofactor and acts as a disulfide-linked homodimer. [provided by RefSeq, Dec 2020]. | hsa:1800; | anchored component of membrane [GO:0031225]; apical part of cell [GO:0045177]; apical plasma membrane [GO:0016324]; cell junction [GO:0030054]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; microvillus membrane [GO:0031528]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; beta-lactamase activity [GO:0008800]; cysteine-type endopeptidase inhibitor activity involved in apoptotic process [GO:0043027]; GPI anchor binding [GO:0034235]; metallodipeptidase activity [GO:0070573]; metalloexopeptidase activity [GO:0008235]; modified amino acid binding [GO:0072341]; zinc ion binding [GO:0008270]; antibiotic metabolic process [GO:0016999]; cellular lactam catabolic process [GO:0072340]; cellular response to calcium ion [GO:0071277]; cellular response to nitric oxide [GO:0071732]; cellular response to xenobiotic stimulus [GO:0071466]; glutathione catabolic process [GO:0006751]; glutathione metabolic process [GO:0006749]; homocysteine metabolic process [GO:0050667]; lipid metabolic process [GO:0006629]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell migration [GO:0030336]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154] | 12144777_Crystal structure of human renal dipeptidase involved in beta-lactam hydrolysis 15145522_DPEP1 has a role in colorectal carcinoma 20031578_CPS1, MUT, NOX4, and DPEP1 is associated with plasma homocysteine in healthy Women. 20031578_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20824289_DPEP1 is expressed in the early stages of colon carcinogenesis and affects cancer cell invasiveness. 21076463_we identified a novel immunohistochemical marker, dipeptidase 1, to distinguish primary mucinous ovarian cancers from ovarian metastasis of colorectal cancers. 22363658_DPEP1 plays a role in pancreatic cancer aggressiveness. 23839495_Dipeptidase 1 has been identified as an excellent marker of high-grade IEN and CRC, and may thus be applied for screening of early neoplastic lesions and for prognostic stratification. 26392408_In this study, we present an analysis of Neanderthal introgression at the dipeptidase 1 gene, DPEP1. 26824987_The results suggest that DPEP1 promotes cancer metastasis by regulating E-cadherin plasticity and that it might be a potential therapeutic target for preventing the progression of colon cancer. 31541079_DPEP1 is a direct target of miR-193a-5p and promotes hepatoblastoma progression by PI3K/Akt/mTOR pathway. 32068254_DPEP1 expression promotes proliferation and survival of leukaemia cells and correlates with relapse in adults with common B cell acute lymphoblastic leukaemia. 34291562_The relationship between common variants in the DPEP1 gene and the susceptibility and clinical severity of osteoarthritis. 34426578_A single genetic locus controls both expression of DPEP1/CHMP1A and kidney disease development via ferroptosis. | ENSMUSG00000019278 | Dpep1 | 20.255876 | 1.2468120 | 0.318243987 | 0.57107361 | 3.049801e-01 | 5.807770e-01 | No | Yes | 18.717992 | 5.666542 | 15.618103 | 4.639324 | ||
| ENSG00000032219 | 5926 | ARID4A | protein_coding | P29374 | FUNCTION: DNA-binding protein which modulates activity of several transcription factors including RB1 (retinoblastoma-associated protein) and AR (androgen receptor) (By similarity). May function as part of an mSin3A repressor complex (PubMed:14581478). Has no intrinsic transcriptional activity (By similarity). Plays a role in the regulation of epigenetic modifications at the PWS/AS imprinting center near the SNRPN promoter, where it might function as part of a complex with RB1 and ARID4B (By similarity). Involved in spermatogenesis, together with ARID4B, where it acts as a transcriptional coactivator for AR and enhances expression of genes required for sperm maturation. Regulates expression of the tight junction protein CLDN3 in the testis, which is important for integrity of the blood-testis barrier (By similarity). Plays a role in myeloid homeostasis where it regulates the histone methylation state of bone marrow cells and expression of various genes involved in hematopoiesis. May function as a leukemia suppressor (By similarity). {ECO:0000250|UniProtKB:F8VPQ2, ECO:0000269|PubMed:14581478}. | 3D-structure;Alternative splicing;Chromatin regulator;DNA-binding;Differentiation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a ubiquitously expressed nuclear protein. It binds directly, with several other proteins, to retinoblastoma protein (pRB) which regulates cell proliferation. pRB represses transcription by recruiting the encoded protein. This protein, in turn, serves as a bridging molecule to recruit HDACs and, in addition, provides a second HDAC-independent repression function. The encoded protein possesses transcriptional repression activity. Multiple alternatively spliced transcripts have been observed for this gene, although not all transcript variants have been fully described. [provided by RefSeq, Jul 2008]. | hsa:5926; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; transcription cis-regulatory region binding [GO:0000976]; erythrocyte development [GO:0048821]; establishment of Sertoli cell barrier [GO:0097368]; histone H3-K4 trimethylation [GO:0080182]; histone H3-K9 trimethylation [GO:0036124]; histone H4-K20 trimethylation [GO:0034773]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of gene expression by genetic imprinting [GO:0006349]; regulation of transcription by RNA polymerase II [GO:0006357]; spermatogenesis [GO:0007283] | 14581478_RBP1 and the mSin3 histone deacetylase complex bind to BRMS1 and repress transcription 16873370_there is an E2-dependent, ER isoform-specific transcriptional activation of the RBBP1 gene, which in part, is explained by the differential activity of ER AF1 and enhancer element binding 18211900_Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. 22247551_chromobarrel domain of RBBP1 is responsible for recognizing methylated histone tails in chromatin remodeling and epigenetic regulation 22732186_ARID4a (AT-rich interacting domain 4a, also known as RBP1) and CCL5 (C-C motif ligand 5) are targets for miR-302. 24379399_The RBBP1 Tudor domain binds both double- and single-stranded DNA with an affinity of 10-100 muM; no apparent DNA sequence specificity was detected. 29408527_Consistently, our ITC assays also showed that DNA does not significantly enhance the histone binding ability of the chromo barrel domain of RBBP1. 29797600_ARID4A and ARID4B may play the role as tumor suppressor gene in prostate cancer by inhibiting cell proliferation, migration, and invasion. Moreover, co-downregulation of ARID4A and ARID4B can predict poorer prognosis in prostate cancer, suggesting they might be the novel marker of prognosis and potential therapeutic targets for prostate cancer in human. 34506790_Structural Insight into Chromatin Recognition by Multiple Domains of the Tumor Suppressor RBBP1. | ENSMUSG00000048118 | Arid4a | 553.834945 | 1.4590774 | 0.545056460 | 0.11750003 | 2.157210e+01 | 3.407735e-06 | 7.008840e-04 | No | Yes | 650.260345 | 165.165988 | 431.268345 | 109.608239 | |
| ENSG00000034677 | 25897 | RNF19A | protein_coding | Q9NV58 | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from E2 ubiquitin-conjugating enzymes UBE2L3 and UBE2L6 in the form of a thioester and then directly transfers the ubiquitin to targeted substrates, such as SNCAIP or CASR. Specifically ubiquitinates pathogenic SOD1 variants, which leads to their proteasomal degradation and to neuronal protection. {ECO:0000269|PubMed:11237715, ECO:0000269|PubMed:12145308, ECO:0000269|PubMed:12750386, ECO:0000269|PubMed:15456787, ECO:0000269|PubMed:16513638}. | Alternative splicing;Cytoplasm;Cytoskeleton;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Transferase;Transmembrane;Transmembrane helix;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a member of the ring between ring fingers (RBR) protein family, and the encoded protein contains two RING-finger motifs and an in between RING fingers motif. This protein is an E3 ubiquitin ligase that is localized to Lewy bodies, and ubiquitylates synphilin-1, which is an interacting protein of alpha synuclein in neurons. The encoded protein may be involved in amyotrophic lateral sclerosis and Parkinson's disease. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:25897; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; glutamatergic synapse [GO:0098978]; hippocampal mossy fiber to CA3 synapse [GO:0098686]; integral component of membrane [GO:0016021]; postsynapse [GO:0098794]; ubiquitin ligase complex [GO:0000151]; metal ion binding [GO:0046872]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin protein ligase activity [GO:0061630]; microtubule cytoskeleton organization [GO:0000226]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; protein polyubiquitination [GO:0000209]; regulation of protein catabolic process at postsynapse, modulating synaptic transmission [GO:0099576]; ubiquitin-dependent protein catabolic process [GO:0006511] | 12145308_Dorfin has a role in ubiquitylating mutant SOD1 proteins and targeting them for proteasomal degradation 12750386_results suggest that synphilin-1 has an important role in the formation of aggregates and cytotoxicity in Parkinson disease and that Dorfin may be involved in the pathogenic process by ubiquitylation of synphilin-1 12875980_These results suggest that Dorfin plays a crucial role in the formation of ubiquitylated inclusions of alpha-synucleinopathy and amyotrophic lateral sclerosis. 15030390_Reducing the accumulation of mutant superoxide dismutase 1 [SOD1] in the mitochondria may be a new therapeutic strategy for mutant SOD1-associated familial amyotrophic lateral sclerosis, and Dorfin may play a significant role in this 15456787_Valosin-containing protein functionally regulates Dorfin through direct interaction 17157513_Dorfin-CHIP(L) rescued neuronal cells from mutant SOD1-associated toxicity and reduced the aggresome formation induced by mutant SOD1 more effectively than did Dorfin(WT). 18721867_Findings suggest that RNF19 has acquired a new promoter and alternative exons via continuous retrotransposition. 22493721_A more than 2-fold higher level of RNF19A mRNA in the blood of patients with prostate cancer than in healthy controls makes it an early diagnosis marker for this disease. 34184814_Ring finger protein 19A is overexpressed in non-small cell lung cancer and mediates p53 ubiquitin-degradation to promote cancer growth. 34789768_RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination. | ENSMUSG00000022280 | Rnf19a | 794.884872 | 1.3960210 | 0.481320634 | 0.11845729 | 1.677734e+01 | 4.203232e-05 | 4.533340e-03 | No | Yes | 933.069107 | 207.836374 | 683.682659 | 152.322481 |
| ENSG00000044090 | 9820 | CUL7 | protein_coding | Q14999 | FUNCTION: Core component of the 3M and Cul7-RING(FBXW8) complexes, which mediates the ubiquitination of target proteins. Core component of the 3M complex, a complex required to regulate microtubule dynamics and genome integrity. It is unclear how the 3M complex regulates microtubules, it could act by controlling the level of a microtubule stabilizer (PubMed:24793695). Interaction with CUL9 is required to inhibit CUL9 activity and ubiquitination of BIRC5 (PubMed:24793696). Core component of a Cul7-RING ubiquitin-protein ligase with FBXW8, which mediates ubiquitination and consequent degradation of target proteins such as GORASP1, IRS1 and MAP4K1/HPK1 (PubMed:21572988, PubMed:24362026). Ubiquitination of GORASP1 regulates Golgi morphogenesis and dendrite patterning in brain (PubMed:21572988). Mediates ubiquitination and degradation of IRS1 in a mTOR-dependent manner: the Cul7-RING(FBXW8) complex recognizes and binds IRS1 previously phosphorylated by S6 kinase (RPS6KB1 or RPS6KB2) (PubMed:18498745). The Cul7-RING(FBXW8) complex also mediates ubiquitination of MAP4K1/HPK1: recognizes and binds autophosphorylated MAP4K1/HPK1, leading to its degradation, thereby affecting cell proliferation and differentiation (PubMed:24362026). Acts as a regulator in trophoblast cell epithelial-mesenchymal transition and placental development (PubMed:20139075). Does not promote polyubiquitination and proteasomal degradation of p53/TP53 (PubMed:16547496, PubMed:17332328). While the Cul7-RING(FBXW8) and the 3M complexes are associated and involved in common processes, CUL7 and the Cul7-RING(FBXW8) complex may be have additional functions. {ECO:0000269|PubMed:16547496, ECO:0000269|PubMed:17332328, ECO:0000269|PubMed:18498745, ECO:0000269|PubMed:20139075, ECO:0000269|PubMed:21572988, ECO:0000269|PubMed:24362026, ECO:0000269|PubMed:24793695, ECO:0000269|PubMed:24793696}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Disease variant;Dwarfism;Golgi apparatus;Host-virus interaction;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a component of an E3 ubiquitin-protein ligase complex. The encoded protein interacts with TP53, CUL9, and FBXW8 proteins. Defects in this gene are a cause of 3M syndrome type 1 (3M1). Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2009]. | hsa:9820; | 3M complex [GO:1990393]; anaphase-promoting complex [GO:0005680]; centrosome [GO:0005813]; Cul7-RING ubiquitin ligase complex [GO:0031467]; cullin-RING ubiquitin ligase complex [GO:0031461]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; perinuclear region of cytoplasm [GO:0048471]; ubiquitin protein ligase binding [GO:0031625]; epithelial to mesenchymal transition [GO:0001837]; Golgi organization [GO:0007030]; microtubule cytoskeleton organization [GO:0000226]; mitotic cytokinesis [GO:0000281]; placenta development [GO:0001890]; positive regulation of dendrite morphogenesis [GO:0050775]; protein ubiquitination [GO:0016567]; proteolysis [GO:0006508]; regulation of mitotic nuclear division [GO:0007088]; ubiquitin-dependent protein catabolic process [GO:0006511]; vasculogenesis [GO:0001570] | 16142236_25 distinct mutations in the gene cullin 7 mappped to chromosome 6 were identified in 29 families with 3-M syndrome. 16547496_CUL7 functions to promote cell growth through, in part, antagonizing the function of p53 16875676_p53-binding domain of CUL7 contributes to the cytoplasmic localization of CUL7 17298945_CPH domain interaction surface of p53 resides in the tetramerization domain and is formed by residues contributed by at least two subunits 17332328_PARC and CUL7 subcomplexes exhibit E3 ubiquitin ligase activity in vitro. 17586686_In a proteomic screen for p53 interactors the cullin protein Cul7 efficiently associates with p53. 17675530_A novel homozygous 4582insT mutation in CUL7 resulted in a frameshift mutation & a premature stop codon at 1553 (Q1553X)in Yakuts with short stature syndromes. 17942889_CUL7 is a new oncogene that cooperates with Myc in transformation by blocking Myc-induced apoptosis in a p53-dependent manner. 19225462_in 33 novel cases of 3M syndrome, we identified deleterious CUL7 mutations in 23/33 patients, including 19 novel mutations & 1 paternal isodisomy of chromosome 6 encompassing a CUL7 mutation; findings also support genetic heterogeneity of this disease 20005570_CUL7 expression in placenta is up-regulated up to 10 times in intra-uterine growth restriction (IUGR) and up to 15 times in preeclampsia associated with IUGR; the CUL7 promoter is hypomethylated in IUGR. 21396581_CUL7 appears to be the major gene responsible for 3M syndrome accounting for 77.5% of cases while OBSL1 mutations accounts for 16.3%[review] 21737058_We propose that CUL7, OBSL1, and CCDC8 are members of a pathway controlling mammalian growth. 21946088_binding of Cul1-Rbx1 to Cul7-Rbx1 is mediated via heterodimerization of Fbxw8 with other F-box proteins which function to recruit substrates into the E3 ligase complex 22156540_discussion of roles of CUL7, OBSL1 (obscurin-like 1), and CCDC8 (coiled-coil domain containing protein 8) in growth and development using findings from patients with Miller-McKusick-Malvaux syndrome and Silver-Russell syndrome [REVIEW] 22524683_Dysregulation of Cul7 and Fbxw8 expression might affect trophoblast turnover in intrauterine growth restriction. 22942238_This study demonstrates specific genomic alterations in HCC/MS and points to CUL7 as a novel gene potentially involved in liver carcinogenesis associated with metabolic Syndrome, the amplification of which might influence cell proliferation. 23018678_Mutations in CUL7, OBSL1 and CCDC8 in 3-M syndrome lead to disordered growth factor signalling. 23029530_Growth factor-stimulated TBC1D3 ubiquitination and degradation are regulated by its interaction with CUL7-Fbw8. 23517720_Homozygous deletion in exon 18 of the CUL7 gene, which has not been previously described, could be responsible for the 3-M syndrome. 24362026_CUL7/Fbxw8 ubiquitin ligase-mediated HPK1 degradation revealed a direct link and novel role of CUL7/Fbxw8 ubiquitin ligase in the MAPK pathway, which plays a critical role in cell proliferation and differentiation. 24711643_CUL7, OBSL1 and CCDC8 modulate the alternative splicing of the INSR 24793695_The CUL7, OBSL1, and CCDC8 proteins form a 3M complex that functions in maintaining microtubule and genome integrity and normal development. 25003318_study provided evidence that Cullin7 functions as a novel oncogene in breast cancer and may be a potential therapeutic target for breast cancer management 25706399_our study provided evidence that Cullin7 functions as a novel oncogene in lung cancer and may be a potential therapeutic target for lung cancer management. 26488604_report an adult female with 3-M syndrome that was caused by novel compound heterozygous mutations (c.4023-1 G>A in splice acceptor site of exon 22 and c.4359_4363dupGGCTG in exon 23) in the CUL7 gene 26850509_We report a family with variable phenotypic features of 3-M syndrome and we describe the prenatal and postnatal growth pattern of two affected sisters with a novel homozygous CUL7 mutation (c.3173-1G>C), showing a pre- and post-natal growth deficiency and a normal cranial circumference. 26962950_Cullin7 may serve as an indicator of poor prognosis in patients with epithelial ovarian cancer. 27053346_Cullin7 promotes epithelial-mesenchymal transformation of cancer cells. 28739496_Hepatocellular carcinoma patients with positive expression for both Rabl3 and Cullin7 had a remarkably shorter survival time compared with patients with negative expression for both proteins. 29207184_Study shows that cullin 7 is highly expressed in breast cancer cells and suggests that positive expression is associated with the malignant phenotype and a predictor of poor prognosis. Cullin 7 is involved in cell proliferation and invasion by regulating the cell cycle and microtubule stability. 29207970_overexpression of Cullin7 plays an important role in the pathogenesis and progression of hepatocellular carcinoma and may be a valuable marker for hepatocellular carcinoma management. 29393450_CUL7 expression was associated with EC progression and poor prognosis. CUL7 may promote EMT via the ERKSNAI2 pathway in EC. 30807646_CUL7 prevents Caspase-8 activation by promoting Caspase-8 modification with non-degradative polyubiquitin chains at K215. CUL7 knockdown sensitized cancer cells to TRAIL-induced apoptosis in vitro and in nude mice. 30945686_Mutation in Cul7 gene is associated with 3M syndrome. 30980518_The mutational spectrum of CUL7, OBSL1, and investigation of genotype-phenotype correlation in 3M syndrome has been reported. 31898234_REVIEW: Cullin-RING E3 Ubiquitin Ligase 7 in Growth Control and Cancer 32141654_Identification of two CUL7 variants in two Chinese families with 3-M syndrome by whole-exome sequencing. 32252802_Cullin-7 (CUL7) is overexpressed in glioma cells and promotes tumorigenesis via NF-kappaB activation. 32278698_A novel mutation within intron 17 of the CUL7 gene results in appearance of premature termination codon. 33258289_A rare cause of syndromic short stature: 3M syndrome in three families. 34597859_Natural history of facial and skeletal features from neonatal period to adulthood in a 3M syndrome cohort with biallelic CUL7 or OBSL1 variants. | ENSMUSG00000038545 | Cul7 | 1269.526263 | 1.0795100 | 0.110376547 | 0.09862514 | 1.267321e+00 | 2.602698e-01 | 6.323698e-01 | No | Yes | 1108.589118 | 117.509342 | 1045.317597 | 110.675483 |
| ENSG00000046653 | 2824 | GPM6B | protein_coding | Q13491 | FUNCTION: May be involved in neural development. Involved in regulation of osteoblast function and bone formation. Involved in matrix vesicle release by osteoblasts; this function seems to involve maintenance of the actin cytoskeleton. May be involved in cellular trafficking of SERT and thereby in regulation of serotonin uptake. {ECO:0000269|PubMed:21638316}. | Alternative splicing;Cell membrane;Developmental protein;Differentiation;Glycoprotein;Membrane;Neurogenesis;Osteogenesis;Phosphoprotein;Protein transport;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a membrane glycoprotein that belongs to the proteolipid protein family. Proteolipid protein family members are expressed in most brain regions and are thought to be involved in cellular housekeeping functions such as membrane trafficking and cell-to-cell communication. This protein may also be involved in osteoblast differentiation. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are located on chromosomes Y and 22. [provided by RefSeq, Jan 2016]. | hsa:2824; | integral component of membrane [GO:0016021]; membrane raft [GO:0045121]; plasma membrane [GO:0005886]; extracellular matrix assembly [GO:0085029]; negative regulation of protein localization to cell surface [GO:2000009]; negative regulation of serotonin uptake [GO:0051612]; nervous system development [GO:0007399]; neuron projection development [GO:0031175]; ossification [GO:0001503]; positive regulation of bone mineralization [GO:0030501]; protein transport [GO:0015031]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of focal adhesion assembly [GO:0051893] | 15214007_it is unlikely that mutations in M6B have a role in Pelizaeus-Merzbacher-like syndrome, as examined in eight patients 16416265_Observational study of gene-disease association. (HuGE Navigator) 17918249_Observational study of gene-disease association. (HuGE Navigator) 18581270_Data suggest that M6B regulates serotonin uptake by affecting cellular trafficking of the serotonin transporter. 21638316_Microarray expression analysis of GPM6B-depleted osteogenic human mesenchymal stem cells revealed significant changes in genes involved in cytoskeleton organization and biogenesis. 23950870_Data show the Differences in high resolution melting analysis in promoters of tumor markers neuronal membrane glycoprotein M6-B, melanoma antigen family A12 and immunoglobulin superfamily Fc receptor indicated invasiveness of hepatocellular carcinoma. 24696529_Circulating levels of DR6 and Gpm6B correlate with breast cancer tumor grade. 24916915_Data indicate that neuronal membrane glycoproteins GPM6A and GPM6B may act as novel oncogenes in the development of lymphoid leukemia. 25113253_Increase in the expression levels of mRNA and protein for the Gpm6B is associated with various types of gynaecological malignancy. 30372567_Findings demonstrate that GPM6B plays a crucial role in SMC differentiation and regulates SMC differentiation through the activation of TGF-beta-Smad2/3 signaling via direct interactions with TbetaRI. 33294057_GPM6B Inhibit PCa Proliferation by Blocking Prostate Cancer Cell Serotonin Absorptive Capacity. | ENSMUSG00000031342 | Gpm6b | 1040.942741 | 0.9016546 | -0.149353292 | 0.09471354 | 2.475491e+00 | 1.156333e-01 | 4.545160e-01 | No | Yes | 1053.531751 | 163.244877 | 1175.218180 | 181.969316 | |
| ENSG00000048028 | 57646 | USP28 | protein_coding | Q96RU2 | FUNCTION: Deubiquitinase involved in DNA damage response checkpoint and MYC proto-oncogene stability. Involved in DNA damage induced apoptosis by specifically deubiquitinating proteins of the DNA damage pathway such as CLSPN. Also involved in G2 DNA damage checkpoint, by deubiquitinating CLSPN, and preventing its degradation by the anaphase promoting complex/cyclosome (APC/C). In contrast, it does not deubiquitinate PLK1. Specifically deubiquitinates MYC in the nucleoplasm, leading to prevent MYC degradation by the proteasome: acts by specifically interacting with isoform 1 of FBXW7 (FBW7alpha) in the nucleoplasm and counteracting ubiquitination of MYC by the SCF(FBW7) complex. In contrast, it does not interact with isoform 4 of FBXW7 (FBW7gamma) in the nucleolus, allowing MYC degradation and explaining the selective MYC degradation in the nucleolus. Deubiquitinates ZNF304, hence preventing ZNF304 degradation by the proteasome and leading to the activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) in a subset of colorectal cancers (CRC) cells (PubMed:24623306). {ECO:0000269|PubMed:16901786, ECO:0000269|PubMed:17558397, ECO:0000269|PubMed:17873522, ECO:0000269|PubMed:18662541, ECO:0000269|PubMed:24623306}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;Hydrolase;Isopeptide bond;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation;Ubl conjugation pathway | The protein encoded by this gene is a deubiquitinase involved in the DNA damage pathway and DNA damage-induced apoptosis. Overexpression of this gene is seen in several cancers. [provided by RefSeq, Oct 2016]. | hsa:57646; | cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; cysteine-type endopeptidase activity [GO:0004197]; thiol-dependent deubiquitinase [GO:0004843]; cell population proliferation [GO:0008283]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to UV [GO:0034644]; DNA damage checkpoint signaling [GO:0000077]; DNA repair [GO:0006281]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; protein deubiquitination [GO:0016579]; Ras protein signal transduction [GO:0007265]; regulation of protein stability [GO:0031647]; response to ionizing radiation [GO:0010212]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11597335_molecular cloning of USP28 & characterization of alternatively spliced products and tissue-specific isoforms 16901786_Using a human cell line that faithfully recapitulated the Chk2-p53-PUMA pathway, we show that USP28 is required to stabilize Chk2 and 53BP1 in response to DNA damage. 17558397_High expression levels of USP28 are found in colon and breast carcinomas, and stabilization of MYC by USP28 is essential for tumour-cell proliferation. 17873522_Usp28 dissociates from Fbw7alpha in response to UV irradiation, providing a mechanism how Fbw7-mediated degradation of Myc is enhanced upon DNA damage. 22144179_A new pathway that could be targeted at the level of GSK-3, Fbw7, or USP28 to influence HIF-1alpha-dependent processes like angiogenesis and metastasis. 23832602_USP28 gene expression is down regulated by oxidative stress through the mediation of reactive oxygen species 24075993_Study reveals a critical mechanism underlying the epigenetic regulation by USP28. 24347490_Usp28 expression was indentified as a independent predictors of survival (P = 0.001) and potentially valuable in prognostic evaluation of bladder cancer 24687851_USP28 is not a critical factor in double-strand break metabolism and is unlikely to be an attractive target for therapeutic intervention aimed at chemotherapy sensitization. 24960159_identified Usp28 as a c-MYC target gene highly expressed in colorectal cancers, which indicates that USP28 and c-MYC form a positive feedback loop that maintains high c-MYC protein levels in tumors 25359778_USP28 has a chain preference activity for Lys(11), Lys(48), and Lys(63) diubiquitin linkages 25437563_Dual regulation of Fbw7 activity by Usp28 is a safeguard mechanism for maintaining physiological levels of proto-oncogenic Fbw7 substrates, which is equivalently disrupted by loss or overexpression of Usp28. 25656529_Data indicte that deubiquitinating enzyme USP28 was targeted by microRNA miR-4295. 26209720_These results showed that USP28 is overexpressed in human glioblastomas and it contributes to glioma tumorigenicity. 26268556_findings provide a first insight into understanding how the enzymatic activity of Usp28 is regulated by its non-catalytic UBR and endogenous ligands. 27371829_The authors identified 53BP1 and USP28 as essential components acting upstream of p53, evoking p21-dependent cell cycle arrest in response not only to centrosome loss, but also to other distinct defects causing prolonged mitosis. 27432896_USP28-53BP1-p53-p21 signaling pathway is also required to arrest cell growth after a prolonged prometaphase. 27432897_analysis of centrinone resistance identified a 53BP1-USP28 module as critical for communicating mitotic challenges to the p53 circuit and TRIM37 as an enforcer of the singularity of centrosome assembly. 27546791_53BP1-USP28 cooperation is essential for normal p53-promoter element interactions and gene transactivation-associated events, yet dispensable for 53BP1-dependent DNA double-strand repair regulation. 29089421_USP28 controls activation of both the TP53 branch and the GATA4/NFkB branch that controls the senescence-associated secretory phenotype (SASP) 29545478_Lack of USP28 promotes a more malignant state of breast cancer cells, indicated by an epithelial-to-mesenchymal (EMT) transition, elevated proliferation, migration, and angiogenesis as well as a decreased adhesion. 29880484_USP28 enhances MAPK activity through the stabilization of RAF family members and is a key factor in BRAF inhibitor resistance. 30206969_Knockdown of USP28 enhanced the radiosensitivity of esophageal cancer cells by destabilizing c-Myc and enhancing the accumulation of HIF-1alpha. Therefore, USP28 may serve as a novel therapeutic target to overcome esophageal cancer radioresistance. 30543854_consequential impacts of USP28-mediated stabilization of LIN28A protein on enhancing cancer cell viability, migration and ultimately augmenting LIN28A-mediated tumor progression. Overall, these data suggest that a synergistic, combinatorial approach of targeting LIN28A with USP28 would contribute to effective cancer therapeutics. 30910399_The effect of USP28 on cell proliferation was mediated by regulating the expression of p53, p21 and p16(INK4a). 30926242_We confirm oligomeric states of USP25 and USP28 in cells and show that modulating oligomerization affects substrate stabilization in accordance with in vitro activity data 30926243_Our work led to the identification of significant differences between USP25 and USP28 and provided the molecular basis for the development of new and highly specific anti-cancer drugs 31604991_The MTH1 inhibitor TH588 is a microtubule-modulating agent that eliminates cancer cells by activating the mitotic surveillance pathway. 31938050_Deubiquitinase USP28 inhibits ubiquitin ligase KLHL2-mediated uridine-cytidine kinase 1 degradation and confers sensitivity to 5'-azacytidine-resistant human leukemia cells. 31982308_A non-canonical role of caspase-8 exploited by cancer cells to override the p53-dependent G2/M cell-cycle checkpoint through cleavage of USP28. 32053284_MicroRNA-216b suppresses the cell growth of hepatocellular carcinoma by inhibiting Ubiquitin-specific peptidase 28 expression. 32128997_Maintaining protein stability of Np63 via USP28 is required by squamous cancer cells. 32578360_USP28 and USP25 are downregulated by Vismodegib in vitro and in colorectal cancer cell lines. 33664871_Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. 34106567_USP28 promotes aerobic glycolysis of colorectal cancer by increasing stability of FOXC1. 34584067_USP28 facilitates pancreatic cancer progression through activation of Wnt/beta-catenin pathway via stabilising FOXM1. 34962618_Loss of USP28 and SPINT2 expression promotes cancer cell survival after whole genome doubling. | ENSMUSG00000032267 | Usp28 | 1562.965181 | 1.1860757 | 0.246196125 | 0.08674080 | 8.111123e+00 | 4.399447e-03 | 7.989468e-02 | No | Yes | 1770.858463 | 294.106144 | 1474.818774 | 244.982134 | |
| ENSG00000048707 | 55187 | VPS13D | protein_coding | Q5THJ4 | FUNCTION: Functions in promoting mitochondrial clearance by mitochondrial autophagy (mitophagy), also possibly by positively regulating mitochondrial fission (PubMed:29307555, PubMed:29604224). Mitophagy plays an important role in regulating cell health and mitochondrial size and homeostasis. {ECO:0000269|PubMed:29307555, ECO:0000269|PubMed:29604224}. | Acetylation;Alternative splicing;Disease variant;Neurodegeneration;Phosphoprotein;Reference proteome | This gene encodes a protein belonging to the vacuolar-protein-sorting-13 gene family. In yeast, vacuolar-protein-sorting-13 proteins are involved in trafficking of membrane proteins between the trans-Golgi network and the prevacuolar compartment. While several transcript variants may exist for this gene, the full-length natures of only two have been described to date. These two represent the major variants of this gene and encode distinct isoforms. [provided by RefSeq, Jul 2008]. | hsa:55187; | extracellular exosome [GO:0070062]; extrinsic component of membrane [GO:0019898]; mitochondrion organization [GO:0007005]; positive regulation of mitophagy [GO:1901526]; protein retention in Golgi apparatus [GO:0045053]; protein targeting to vacuole [GO:0006623] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20610895_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 25896417_genetic polymorphism is associated with il-6 production and mortality in patients with septic shock 29518281_findings underline the importance of the VPS13 complex in neurological diseases and a possible role in mitochondrial function 29604224_heterozygous mutations in VPS13D cause movement disorders along the ataxia-spasticity spectrum 31876103_VPS13D-related disorders presenting as a pure and complicated form of hereditary spastic paraplegia. 31943017_A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. 32020600_Genetic heterogeneity in Leigh syndrome: Highlighting treatable and novel genetic causes. 33623047_An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. 33758164_The lncRNA Snhg1-Vps13D vesicle trafficking system promotes memory CD8 T cell establishment via regulating the dual effects of IL-7 signaling. 33891012_VPS13D promotes peroxisome biogenesis. 33891013_VPS13D bridges the ER to mitochondria and peroxisomes via Miro. 34133214_VPS13D interacts with VCP/p97 and negatively regulates endoplasmic reticulum-mitochondria interactions. | ENSMUSG00000020220 | Vps13d | 1453.420124 | 1.1536712 | 0.206232140 | 0.10396943 | 3.998469e+00 | 4.554162e-02 | 2.872164e-01 | No | Yes | 1287.358871 | 152.698668 | 1107.987110 | 131.571712 | |
| ENSG00000049323 | 4052 | LTBP1 | protein_coding | Q14766 | FUNCTION: Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:2022183, PubMed:8617200, PubMed:8939931). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (PubMed:8617200, PubMed:8939931, PubMed:15184403). Outcompeted by LRRC32/GARP for binding to LAP regulatory chain of TGF-beta (PubMed:22278742). {ECO:0000269|PubMed:15184403, ECO:0000269|PubMed:2022183, ECO:0000269|PubMed:22278742, ECO:0000269|PubMed:8617200, ECO:0000269|PubMed:8939931}. | 3D-structure;Alternative splicing;Direct protein sequencing;Disulfide bond;EGF-like domain;Extracellular matrix;Glycoprotein;Growth factor binding;Hydroxylation;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal | The protein encoded by this gene belongs to the family of latent TGF-beta binding proteins (LTBPs). The secretion and activation of TGF-betas is regulated by their association with latency-associated proteins and with latent TGF-beta binding proteins. The product of this gene targets latent complexes of transforming growth factor beta to the extracellular matrix, where the latent cytokine is subsequently activated by several different mechanisms. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:4052; | collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; microfibril [GO:0001527]; protein-containing complex [GO:0032991]; calcium ion binding [GO:0005509]; microfibril binding [GO:0050436]; transforming growth factor beta binding [GO:0050431]; transforming growth factor beta-activated receptor activity [GO:0005024]; regulation of transforming growth factor beta activation [GO:1901388]; sequestering of TGFbeta in extracellular matrix [GO:0035583] | 12429738_Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein 14626352_The current study was carried out to understand hormonal regulation of LTBP-1 expression in normal and SV-40 virus transformed human lung fibroblasts 15564041_The LTBP1 protein expressed from a cDNA encoding only the TGF-beta-LAP-binding domain was incapable of supporting alphavbeta6-integrin-mediated activation. 16825507_Observational study of gene-disease association. (HuGE Navigator) 16825507_The single nucleotide polymorphisms -202G/C and +20A/C on the LTBP-1L promoter may affect the clinical outcome of ovarian cancer patients, probably via up-regulating protein expression. 17293099_LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 17580303_heparan sulfate proteoglycans may play a critical role in regulating TGF-beta availability by controlling the deposition of LTBP1 into the extracellular matrix in association with fibronectin 18602101_MT1-MMP-mediated proteolytic processing of ECM-bound LTBP-1 is a mechanism to release latent TGF-beta from the subendothelial matrix. 19258388_TGFB1 as an important candidate gene for further biological studies of IgA nephropathy and as a possible target for therapy. Our data also indicate a possibility of a gender effect in the genetic background of IgA nephropathy 19339270_Observational study of gene-disease association. (HuGE Navigator) 19431147_Data identify LTBP-1 as an important modulator of TGF-beta activation in glioma cells, which may contribute to the malignant phenotype. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19829035_codon 10 polymorphism in TGFB1 may have a significant influence on the development of PCa and BPH and that the T allele of the TGFB1 gene has a dominant effect on the development of PCa and BPH 19895311_TGF-beta(1) augmented Tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-induced production of IL-8 and MCP-1 by retinal pigment epithelial cells. 19897194_Observational study of gene-disease association. (HuGE Navigator) 19998449_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20446778_TGF-beta1 gene -509C/T polymorphism was associated with severe TID. The higher value in serum concentration of TGF-beta1 was also associated with severe TID and the T T genotype/T allele. 20587546_Observational study of gene-disease association. (HuGE Navigator) 20599762_extracellular HSP90beta is a negative regulator for the activation of latent TGF-beta1 modulating TGF-beta signaling in the extracellular domain. 20699357_large latent TGFbeta complex (LLC), which contains latent TGFbeta-binding protein 1 (LTBP-1), is directly dependent on the pericellular assembly of fibrillin microfibrils, which interact with fibronectin during higher-order fibrillogenesis. 21071877_Study indicates marked differences in the expression pattern of LTBP1 mRNA and protein in early lesions compared with advanced lesions. 21106222_A strong negative correlation between LTBP-1 and P-Smad2 immunoreactivity was found, implying that LTBP-1 is not likely to contribute directly to the increased levels of TGF-beta activity in malignant mesothelioma. 21161366_a spliced variant form of latent transforming growth factor beta binding protein 1, LTBPD41 is highly expressed in advanced atherosclerotic lesions 21843572_genetic polymorphism is associated with both lack of microfilariae and differential microfilarial loads in the blood of persons infected with lymphatic filariasis 21924590_results suggest that the CC genotype of the TGF-beta1 gene increases the risk to develop LOAD and is also associated with depressive symptoms in AD. 22015652_We have shown that TGF-beta1 expression in gastric tumor tissue with HER2/neu-negative status is of prognostic relevance in gastric cancer. 22169532_Force-induced latency-associated peptide conformational changes result in transforming growth factor (TGF)-beta1 release. Mechanical pulling opens the straitjacket structure that keeps TGF-beta1 latent. 22295116_susceptibility genes associated with alcohol drinking 22322529_Increased generation of reactive oxygen species (ROS) and enhanced expression of NADPH oxidases were found to be associated with the TGF-beta1-induced autophagy. 22410565_The expression of PAI-1 mRNA and TGF-beta1 mRNA was significantly higher in the cDNA samples obtained from the atopic cataracts than those obtained from the senile cataracts. 22414291_Elevated expression of either TGF-beta1 or TGF-beta2 in cells that represent the early stages in the development of human squamous cell carcinoma results in a more aggressive phenotype. 22632886_Data show that plasma TGF-beta1 was elevated in the circulation of patients with brain tumors and that significant decreases in TGF-beta1 levels were observed after the removal of benign and malignant tumors. 22820497_Dysregulated expression of TWIST1, TGF-beta1 and SMAD3 mRNA observed in osteoarthritis bone is reflected in the functionality of the osteoblast when these cells are cultured ex vivo. 23395279_The high incidence of intimal hyperplasia in patients with surgically repaired congenital heart disease (CHD) is correlated with TGF-beta1 expression and may contribute to the development of atherosclerotic coronary artery disease in CHD patients. 23442056_these results suggested that the C allele could increase TGF-beta secretion which suppresses antitumour immune responses and may affect the Oral squamous cell carcinoma risk. 23462118_High transforming Growth Factor beta1 expression is associated with myelofibrosis. 23696034_Lipocalin-2 negatively modulates the epithelial-to-mesenchymal transition in hepatocellular carcinoma through the epidermal growth factor (TGF-beta1)/Lcn2/Twist1 pathway. 23998914_The expression of TGF-beta1 is increased while the Smad7 expression is diminished in dendritic cell infiltration decrease in liver gastrointestinal cancer metastasis. 24164868_There is an increasing anti-cancer protection with increased numbers of alleles associated with higher levels of TGF-beta1 and IL-4. 24489852_The LTBP1 C-terminus adopts a flexible 'knotted rope' structure, which may facilitate cell matrix interactions. 25190493_LTBP-1 showed both an accumulation and a striking co-localization with Notch3-ECD deposits suggesting specific recruitment into aggregates in brain in CADASIL. 25359706_Significantly higher frequency of TT genotype of transforming growth factor-Beta 1 in asthma patients was found compared to controls. 25369932_LTBP-1 an extracellular matrix protein and key regulator of TGF-beta bioavailability is a novel HtrA1 target.Attenuation of TGF-beta signaling caused by a lack of HtrA1-mediated LTBP-1 processing as mechanism underlying CARASIL pathogenesis. 25374198_This review concludes that TGF-beta1 29T/C polymorphism does not play a role in breast cancer susceptibility in overall or ethnicity-specific manner. 25527259_The aim of this study is to evaluate whether specific inhibition of c-Abl by siRNA can influence the transforming growth factor-beta1 (TGF-beta1)-induced fibrotic responses. 25529149_Combination of EGF with TGF-beta1, but not EGF or TGF-beta1 alone, caused assembly of cells to a new two-dimensional structure, being characterized by dense aggregates connected by branches of few cells. 25605036_Dental pulp stem cells suppress the proliferation of lymphocytes via transforming growth factor-beta1 25759826_role in olive-pollen sensitization 25858550_LTBP-1 and LTBP-2 are involved in the keratinization of oral epithelium. 26884881_LTBP-1 may be a promising biomarker for distinguishing early stage hepatocellular carcinoma from the chronic hepatitis B or liver cirrhosis patients. 27297409_Eosinophils enhance Wnt-5a, TGF-beta1, fibronectin, and collagen gene expression in airway smooth muscle cells and promote proliferation of these cells in asthma. 27391803_All members of the tolloid family bind the N terminus of latent TGFb-binding protein-1, providing support for their role in TGFb signalling. 28331138_Expression levels of TGF-beta1 and BMP-2 appeared to be delayed in nonunion patients which could play an important role in developing an early marker of fracture union condition and facilitate improved patient's management. 28669633_LTBP1 anchors itself to FBN1 using two independent epitopes. 29257223_The present study demonstrated that in human osteoarthritis fibroblastlike synoviocytes, TGF-beta signals via p-Smad2 and p-Smad1/5/8. Furthermore, it was demonstrated that LTBP-1 may modulate the activity of TGF-beta in human osteoarthritis fibroblastlike synoviocytes. 29311025_Compared with the control, miR-340 was significantly lower in the serum of hepatocellular carcinoma patients (p<0.01). miR-340 was lower in HCC tissues than in adjacent; however, DcR3, TGF-beta1 and Smad2 were higher. 29361522_our results suggest that the ED-A domain enhances association of the latent TGF-b1 by promoting weak direct binding to LTBP-1 and by enhancing heparin-mediated protein interactions through HepII in fibronectin. 29365054_The important role of GP73 in regulating EMT and metastasis in HCC partly by targeting TGF-beta1/Smad2 signaling. 29504479_Differentiation is initially influenced by ECM rigidity, but is ultimately superseded by TGF-beta1. 29549584_It is a protein that promotes renal fibrosis in diabetic nephropathy. 29924680_these results show peripheral blood levels of TGF beta are significantly increased in colorectal patients 30447345_The relationship between TGF-beta1 and RELN and uncovered the important role of RELN in suppressing cell migration in HCC cells. 30486878_A negative correlation between miR-663a and TGF-beta1 expression was also confirmed from the clinical samples of HCC. 30575211_The phosphorylation of smad2 in Panc1/LRG was increased in comparison with parental Panc1 under TGF-beta1 stimulation. 31196201_MiR-27a-5p was expressed 11 times higher in OD-Exo, while miR-27a-5p promoted odontogenic differentiation of DPSCs and significantly upregulated TGFbeta1, TGFR1, p-Smad2/3, and Smad4 by downregulating the inhibitory molecule LTBP1. 31343988_Fibulin-1c regulates transforming growth factor-beta activation in pulmonary tissue fibrosis. 31628042_LTBP1 S-nitrosylation results in proteasome-dependent LTBP1 protein degradation. 32216815_LTBP1 promotes esophageal squamous cell carcinoma progression through epithelial-mesenchymal transition and cancer-associated fibroblasts transformation. 32245290_LTBP1 and Smad6 expression levels were evaluated by real-time PCR. 33059753_LTBP1 plays a potential bridge between depressive disorder and glioblastoma. 33991472_Bi-allelic premature truncating variants in LTBP1 cause cutis laxa syndrome. 34411563_POGLUT2 and POGLUT3 O-glucosylate multiple EGF repeats in fibrillin-1, -2, and LTBP1 and promote secretion of fibrillin-1. | ENSMUSG00000001870 | Ltbp1 | 1499.639357 | 0.8786213 | -0.186686614 | 0.08153273 | 5.196935e+00 | 2.262675e-02 | 1.989175e-01 | No | Yes | 1370.099556 | 106.135890 | 1580.775038 | 122.306993 | |
| ENSG00000051825 | 10198 | MPHOSPH9 | protein_coding | Q99550 | FUNCTION: Negatively regulates cilia formation by recruiting the CP110-CEP97 complex (a negative regulator of ciliogenesis) at the distal end of the mother centriole in ciliary cells (PubMed:30375385). At the beginning of cilia formation, MPHOSPH9 undergoes TTBK2-mediated phosphorylation and degradation via the ubiquitin-proteasome system and removes itself and the CP110-CEP97 complex from the distal end of the mother centriole, which subsequently promotes cilia formation (PubMed:30375385). {ECO:0000269|PubMed:30375385}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Golgi apparatus;Isopeptide bond;Membrane;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:10198; | centriole [GO:0005814]; centrosome [GO:0005813]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; membrane [GO:0016020]; negative regulation of cilium assembly [GO:1902018] | 19879194_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20546594_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24130709_The MPHOSPH9 polymorphism was associated with greater attack severity in Multiple sclerosis 30375385_MPP9 acts as a regulator of ciliogenesis by regulating the localization of CP110-CEP97 at the mother centriole. 33174370_M-Phase Phosphoprotein 9 upregulation associates with poor prognosis and activates mTOR signaling in gastric cancer. | ENSMUSG00000038126 | Mphosph9 | 1737.777390 | 1.0870217 | 0.120380793 | 0.11896804 | 1.026881e+00 | 3.108923e-01 | 6.770487e-01 | No | Yes | 1602.243754 | 300.465184 | 1469.847985 | 275.694267 | ||
| ENSG00000052795 | 57600 | FNIP2 | protein_coding | Q9P278 | FUNCTION: Binding partner of the GTPase-activating protein FLCN: involved in the cellular response to amino acid availability by regulating the mTORC1 signaling cascade controlling the MiT/TFE factors TFEB and TFE3 (PubMed:18663353, PubMed:31672913). In low-amino acid conditions, component of the lysosomal folliculin complex (LFC) on the membrane of lysosomes, which inhibits the GTPase-activating activity of FLCN, thereby inactivating mTORC1 and promoting nuclear translocation of TFEB and TFE3 (PubMed:31672913). Upon amino acid restimulation, disassembly of the LFC complex liberates the GTPase-activating activity of FLCN, leading to activation of mTORC1 and subsequent cytoplasmic retention of TFEB and TFE3 (PubMed:31672913). Together with FLCN, regulates autophagy: following phosphorylation by ULK1, interacts with GABARAP and promotes autophagy (PubMed:25126726). In addition to its role in mTORC1 signaling, also acts as a co-chaperone of HSP90AA1/Hsp90: inhibits the ATPase activity of HSP90AA1/Hsp90, leading to activate both kinase and non-kinase client proteins of HSP90AA1/Hsp90 (PubMed:18403135). Acts as a scaffold to load client protein FLCN onto HSP90AA1/Hsp90 (PubMed:18403135). Competes with the activating co-chaperone AHSA1 for binding to HSP90AA1, thereby providing a reciprocal regulatory mechanism for chaperoning of client proteins (PubMed:18403135). May play a role in the signal transduction pathway of apoptosis induced by O6-methylguanine-mispaired lesions (By similarity). {ECO:0000250|UniProtKB:Q80TD3, ECO:0000269|PubMed:18403135, ECO:0000269|PubMed:18663353, ECO:0000269|PubMed:25126726, ECO:0000269|PubMed:31672913}. | 3D-structure;Alternative splicing;Cytoplasm;DNA damage;Lysosome;Membrane;Phosphoprotein;Reference proteome | This gene encodes a protein that binds to the tumor suppressor folliculin and to AMP-activated protein kinase (AMPK), and may play a role cellular metabolism and nutrient sensing by regulating the AMPK-mechanistic target of rapamycin signaling pathway. The encoded protein may also be involved in regulating the O6-methylguanine-induced apoptosis signaling pathway. This gene has a closely related paralog that encodes a protein with similar binding activities. Both related proteins also associate with the molecular chaperone heat shock protein-90 (Hsp90) and negatively regulate its ATPase activity and facilitate its association with folliculin. [provided by RefSeq, Jul 2017]. | hsa:57600; | centriolar satellite [GO:0034451]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lysosomal membrane [GO:0005765]; ATPase inhibitor activity [GO:0042030]; chaperone binding [GO:0051087]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of protein-containing complex assembly [GO:0031334]; protein phosphorylation [GO:0006468]; regulation of protein phosphorylation [GO:0001932] | 18403135_The identification and characterization of a novel FNIP1 homolog FNIP2 that also interacts with FLCN and AMPK, is reported. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23201403_this study obtained evidence to show that stabilization of MAPO1 is regulated through its specific interaction with folliculin and AMPK. 28039480_SCFbeta-TRCP negatively regulates the FLCN complex by promoting FNIP2 degradation in Birt-Hogg-Dube syndrome-associated renal cancer. 33459596_Loss of FLCN-FNIP1/2 induces a non-canonical interferon response in human renal tubular epithelial cells. | ENSMUSG00000061175 | Fnip2 | 842.288225 | 0.9784509 | -0.031428590 | 0.11462434 | 7.533702e-02 | 7.837189e-01 | 9.301413e-01 | No | Yes | 852.985840 | 126.868309 | 868.706814 | 129.247835 | |
| ENSG00000054654 | 23224 | SYNE2 | protein_coding | Q8WXH0 | FUNCTION: Multi-isomeric modular protein which forms a linking network between organelles and the actin cytoskeleton to maintain the subcellular spatial organization. As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. Specifically, SYNE2 and SUN2 assemble in arrays of transmembrane actin-associated nuclear (TAN) lines which are bound to F-actin cables and couple the nucleus to retrograde actin flow during actin-dependent nuclear movement. May be involved in nucleus-centrosome attachment. During interkinetic nuclear migration (INM) at G2 phase and nuclear migration in neural progenitors its LINC complex association with SUN1/2 and probable association with cytoplasmic dynein-dynactin motor complexes functions to pull the nucleus toward the centrosome; SYNE1 and SYNE2 may act redundantly. During INM at G1 phase mediates respective LINC complex association with kinesin to push the nucleus away from the centrosome. Involved in nuclear migration in retinal photoreceptor progenitors. Required for centrosome migration to the apical cell surface during early ciliogenesis. {ECO:0000250|UniProtKB:Q6ZWQ0, ECO:0000269|PubMed:12118075, ECO:0000269|PubMed:18396275, ECO:0000269|PubMed:19596800, ECO:0000269|PubMed:20724637, ECO:0000269|PubMed:22945352}. | 3D-structure;Acetylation;Actin-binding;Alternative splicing;Cell junction;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Disulfide bond;Emery-Dreifuss muscular dystrophy;Membrane;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;Sarcoplasmic reticulum;Transmembrane;Transmembrane helix | The protein encoded by this gene is a nuclear outer membrane protein that binds cytoplasmic F-actin. This binding tethers the nucleus to the cytoskeleton and aids in the maintenance of the structural integrity of the nucleus. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:23224; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; filopodium membrane [GO:0031527]; focal adhesion [GO:0005925]; integral component of membrane [GO:0016021]; intermediate filament cytoskeleton [GO:0045111]; lamellipodium membrane [GO:0031258]; meiotic nuclear membrane microtubule tethering complex [GO:0034993]; mitochondrion [GO:0005739]; nuclear envelope [GO:0005635]; nuclear lumen [GO:0031981]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; sarcoplasmic reticulum [GO:0016529]; sarcoplasmic reticulum membrane [GO:0033017]; Z disc [GO:0030018]; actin binding [GO:0003779]; actin filament binding [GO:0051015]; cytoskeleton-nuclear membrane anchor activity [GO:0140444]; centrosome localization [GO:0051642]; nuclear migration [GO:0007097]; nuclear migration along microfilament [GO:0031022]; nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration [GO:0021817]; positive regulation of cell migration [GO:0030335]; regulation of cilium assembly [GO:1902017] | 15671068_Nesprin-2 binds lamin and emerin at the nuclear envelope in skeletal muscle. 15843432_Nesprin-2 has a scaffolding function at the nuclear membrane 16079285_The Nesprin-2 the conserved C-terminal amino acids PPPX is essential for the interaction with a C-terminal region in Sun1. 17462627_The characterisation of the residues both in emerin and in nesprin-1alpha and -2beta which are involved in their interaction is reported. 17761684_Screening for DNA variations in the genes encoding nesprin-1 (SYNE1) and nesprin-2 (SYNE2) in 190 probands with Emery Dreifuss muscular dystrophy identified four heterozygous missense mutations. 17881656_propose nesprin-2 giant as a structural reinforcer at the nuclear envelope in LMNA S143F progeria cells 18477613_Nesprin-2 is an important scaffold protein implicated in the maintenance of nuclear envelope architecture. 18827015_association with human nesprin-3 appeared to be stronger for torsinADeltaE than for torsinA. TorsinA also associated with the KASH domains of nesprin-1 and -2 19861416_Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. 20108321_Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20801886_Nesprin-2 interacts with {alpha}-catenin and regulates Wnt signaling at the nuclear envelope. 21820406_novel isoform, nesprin-2-epsilon, was found to be the major mRNA and protein product of the nesprin-2 gene. 22632968_Study presents crystal structures of the human SUN2-KASH1/2 complex, i.e. SUN2 complexed with the C-terminal 29 residues of human Nesprin-1 or -2 (the core of the LINC complex). 22768332_Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. 23707952_ectopic expression of BRAP2 inhibits nuclear localization of HMG20A and NuMA1, and prevents nuclear envelope accumulation of SYNE2. 23977161_Each mutation in LMNA has a distinct impact on the Nersprin-2 interaction that substantially explains how distinct mutations in widely expressed genes lead to the formation of phenotypically different diseases. 24080406_High Nesprin-2 expression is associated with colorectal cancer. 24718612_The significance of these shorter isoforms of nesprin, were evaluated. 24931616_nesprin-1 and nesprin-2 both regulate nuclear and cytoplasmic architecture. 25516977_nesprin-dependent recruitment of kinesin-1 to the nuclear envelope through the interaction of a conserved LEWD motif with kinesin light chain might be a general mechanism for cell-type-specific nuclear positioning during development. 26506308_We show that AMPH-1/BIN1 binds to nesprin and actin, as well as to the microtubule-binding protein CLIP170 in both species. We propose that BIN1 has a direct and evolutionarily conserved role in nuclear positioning, altered in myopathies. 27321956_these data identify N-terminal nesprin-2 variants as novel regulators of beta-catenin signaling. 27502069_variants of EGFR and SYNE2 play an important role in p21 regulation and are associated with the clinical outcome of HBV-related hepatocellular carcinoma in a TP53-indenpdent manner 27554857_The authors identified the nuclear envelope protein nesprin-2 as a binding partner for fascin in a range of cell types in vitro and in vivo. Nesprin-2 interacts with fascin through a direct, F-actin-independent interaction, and this binding is distinct and separable from a role for fascin within filopodia at the cell periphery. 29934494_Study shows that modulation of matrix pore size or of lamin A expression known to modulate nuclear stiffness directly impinges on levels of MT1-MMP-mediated pericellular collagenolysis by cancer cells. This response requires an intact connection between the nucleus and the centrosome via the linker of nucleoskeleton and cytoskeleton (LINC) complex protein nesprin-2 and dynein adaptor Lis1. 30054381_CRISPR/Cas9-mediated knockout of Syne-2 in cell culture led to an overexpression and mislocalization of Pcnt and to ciliogenesis defects. This suggests that the Pcnt-Syne-2 complex is important for ciliogenesis and outer segment formation during retinal development and plays a role in nuclear migration. 31144711_Results indicate that sperm associated antigen 4 (SPAG4L/SPAG4Lbeta) transcript isoform interacts with spectrin repeat containing nuclear envelope protein 2 (Nesprin2) in the meiotic process. 31578382_Nesprin-1-alpha2 associates with kinesin at myotube outer nuclear membranes, but is restricted to neuromuscular junction nuclei in adult muscle. 32184094_A novel SYNE2 mutation identified by whole exome sequencing in a Korean family with Emery-Dreifuss muscular dystrophy. 33472039_Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. 33686165_The SUN2-nesprin-2 LINC complex and KIF20A function in the Golgi dispersal. | ENSMUSG00000063450 | Syne2 | 881.763575 | 1.5978034 | 0.676089926 | 0.11567171 | 3.441928e+01 | 4.443002e-09 | 5.640604e-06 | No | Yes | 890.393437 | 146.674659 | 570.075696 | 93.995720 | |
| ENSG00000058056 | 8975 | USP13 | protein_coding | Q92995 | FUNCTION: Deubiquitinase that mediates deubiquitination of target proteins such as BECN1, MITF, SKP2 and USP10 and is involved in various processes such as autophagy and endoplasmic reticulum-associated degradation (ERAD). Component of a regulatory loop that controls autophagy and p53/TP53 levels: mediates deubiquitination of BECN1, a key regulator of autophagy, leading to stabilize the PIK3C3/VPS34-containing complexes. Also deubiquitinates USP10, an essential regulator of p53/TP53 stability. In turn, PIK3C3/VPS34-containing complexes regulate USP13 stability, suggesting the existence of a regulatory system by which PIK3C3/VPS34-containing complexes regulate p53/TP53 protein levels via USP10 and USP13. Recruited by nuclear UFD1 and mediates deubiquitination of SKP2, thereby regulating endoplasmic reticulum-associated degradation (ERAD). Also regulates ERAD through the deubiquitination of UBL4A a component of the BAG6/BAT3 complex. Mediates stabilization of SIAH2 independently of deubiquitinase activity: binds ubiquitinated SIAH2 and acts by impairing SIAH2 autoubiquitination. Has a weak deubiquitinase activity in vitro and preferentially cleaves 'Lys-63'-linked polyubiquitin chains. In contrast to USP5, it is not able to mediate unanchored polyubiquitin disassembly. Able to cleave ISG15 in vitro; however, additional experiments are required to confirm such data. {ECO:0000269|PubMed:17653289, ECO:0000269|PubMed:21571647, ECO:0000269|PubMed:21659512, ECO:0000269|PubMed:21811243, ECO:0000269|PubMed:21962518, ECO:0000269|PubMed:22216260, ECO:0000269|PubMed:24424410}. | 3D-structure;Alternative splicing;Autophagy;Hydrolase;Isopeptide bond;Metal-binding;Phosphoprotein;Protease;Reference proteome;Repeat;Thiol protease;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | hsa:8975; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; BAT3 complex binding [GO:1904288]; chaperone binding [GO:0051087]; cysteine-type endopeptidase activity [GO:0004197]; Lys48-specific deubiquitinase activity [GO:1990380]; proteasome binding [GO:0070628]; thiol-dependent deubiquitinase [GO:0004843]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-like protein ligase binding [GO:0044389]; zinc ion binding [GO:0008270]; autophagy [GO:0006914]; cell population proliferation [GO:0008283]; maintenance of unfolded protein involved in ERAD pathway [GO:1904378]; melanocyte differentiation [GO:0030318]; positive regulation of ERAD pathway [GO:1904294]; protein deubiquitination [GO:0016579]; protein K29-linked deubiquitination [GO:0035523]; protein K6-linked deubiquitination [GO:0044313]; protein K63-linked deubiquitination [GO:0070536]; protein stabilization [GO:0050821]; regulation of autophagy [GO:0010506]; regulation of transcription, DNA-templated [GO:0006355]; ubiquitin-dependent protein catabolic process [GO:0006511] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21659512_study identifies a new layer of Siah2 regulation mediated by USP13 binding to ubiquitinated Siah2 protein with a concomitant inhibitory effect on its activity under normoxia 21811243_Through deubiquitination, USP13 stabilizes and upregulates MITF protein levels. 21962518_USP10 mediates the deubiquitination of p53, regulating deubiquitination activity of USP10 and USP13 by Beclin1 provides a mechanism for Beclin1 to control the levels of p53. 23940278_to our knowledge, USP13 is the first deubiquitinating enzymes identified to modulate STAT1 and play a role in the antiviral activity of IFN against DEN-2 replication. 24270891_USP13 is a tumour-suppressing protein that functions through deubiquitylation and stabilization of PTEN. 24424410_The authors identify USP13 as a gp78-associated deubiquitinase that eliminates ubiquitin conjugates from Ubl4A to maintain the functionality of Bag6. 26453058_Down-regulation of USP13 mediates PTEN protein loss and fibroblast phenotypic change, and thereby plays a crucial role in idiopathic pulmonary fibrosis pathogenesis. 27869170_our results demonstrate that AR can promote melanoma metastasis via altering the miRNA-539-3p/USP13/MITF/AXL signal and targeting this newly identified signal with AR degradation enhancer ASC-J9 may help us to better suppress the melanoma metastasis. 27892457_These results reveal an important metabolism-centric role of USP13, which may lead to potential therapeutics targeting USP13 in ovarian cancers. 27923907_USP13 and FBXL14 play opposing roles in the regulation of glioma stem cells (GSCs) through reversible ubiquitination of c-Myc. 28498477_in the samples of 293T cell lines after the overexpression of USP13 and USP13 C345S, vinculin exhibited an increased expression, suggesting that it may be a candidate substrate of USP13. 28569838_USP13 is a regulator of DNA repair.USP13 regulates RAP80-BRCA1 complex foci formation.The role of USP13 in the neoplasm drug resistance. 29335437_Deubiquitinating enzyme 13 (USP13) regulates myeloid cell leukemia sequence 1 protein (MCL1) stability in lung and ovarian cancer cells. 29567855_As unanchored ubiquitin chains are preferred substrates for USP5, we suggest that USP5 regulates the assembly and disassembly of heat-induced stress granules by mediating the hydrolysis of unanchored ubiquitin chains while USP13 regulates stress granules through deubiquitylating protein-conjugated ubiquitin chains. 30329047_We found that USP13 independently regulates parkin and alpha-synuclein ubiquitination in models of alpha-synucleinopathies. USP13 shRNA knockdown increases alpha-synuclein ubiquitination and clearance, in a parkin-independent manner..These studies provide novel evidence of USP13 effects on parkin and alpha-synuclein metabolism and suggest that USP13 is a potential therapeutic target in the alpha-synucleinopathies 30986623_Downregulation of USP13 dramatically inhibited A549. 31200745_the present study demonstrates the tumor suppressor role of USP13 in BC and the potential regulatory loop network of NF-kB/USP13/PTEN. USP13 has been identified to be a target of miR-130b/301b and a downstream effector of NF-kB. 31594232_Ubiquitin Specific Protease 13 Regulates Tau Accumulation and Clearance in Models of Alzheimer's Disease. 32101753_Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. 32129945_Potent USP10/13 antagonist spautin-1 suppresses melanoma growth via ROS-mediated DNA damage and exhibits synergy with cisplatin. 32255563_Knockdown of ubiquitin-specific peptidase 13 inhibits cell growth of hepatocellular carcinoma by reducing c-Myc expression. 32735883_Molecular profiling of immune cell-enriched Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) interacting protein USP13. 33334891_USP13 interacts with cohesin and regulates its ubiquitination in human cells. 33592542_USP13 regulates the replication stress response by deubiquitinating TopBP1. 33627786_The deubiquitinase (DUB) USP13 promotes Mcl-1 stabilisation in cervical cancer. 33707416_SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. 34001947_Identification of targets of JS-K against HBV-positive human hepatocellular carcinoma HepG2.2.15 cells with iTRAQ proteomics. 34688658_Inhibition of ubiquitin-specific protease 13-mediated degradation of Raf1 kinase by Spautin-1 has opposing effects in naive and primed pluripotent stem cells. | ENSMUSG00000056900 | Usp13 | 1222.467320 | 1.1761498 | 0.234071842 | 0.13366046 | 3.156154e+00 | 7.564121e-02 | 3.682151e-01 | No | Yes | 1399.968222 | 215.433010 | 1150.443894 | 177.069918 | ||
| ENSG00000064651 | 6558 | SLC12A2 | protein_coding | P55011 | FUNCTION: Cation-chloride cotransporter which mediates the electroneutral transport of chloride, potassium and/or sodium ions across the membrane (PubMed:32294086). Plays a vital role in the regulation of ionic balance and cell volume. {ECO:0000269|PubMed:32081947, ECO:0000269|PubMed:32294086, ECO:0000269|PubMed:7629105}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Chloride;Deafness;Disease variant;Disulfide bond;Ion transport;Membrane;Mental retardation;Non-syndromic deafness;Phosphoprotein;Potassium;Potassium transport;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene mediates sodium and chloride transport and reabsorption. The encoded protein is a membrane protein and is important in maintaining proper ionic balance and cell volume. This protein is phosphorylated in response to DNA damage. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jan 2012]. | hsa:6558; | apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; cell body [GO:0044297]; cell body membrane [GO:0044298]; cell periphery [GO:0071944]; cell projection [GO:0042995]; cell projection membrane [GO:0031253]; cytoplasmic vesicle membrane [GO:0030659]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular vesicle [GO:1903561]; integral component of plasma membrane [GO:0005887]; lateral plasma membrane [GO:0016328]; membrane [GO:0016020]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; ammonium transmembrane transporter activity [GO:0008519]; cation:chloride symporter activity [GO:0015377]; chaperone binding [GO:0051087]; Hsp90 protein binding [GO:0051879]; metal ion transmembrane transporter activity [GO:0046873]; potassium ion transmembrane transporter activity [GO:0015079]; potassium:chloride symporter activity [GO:0015379]; protein kinase binding [GO:0019901]; sodium:potassium:chloride symporter activity [GO:0008511]; aging [GO:0007568]; ammonium transport [GO:0015696]; cell volume homeostasis [GO:0006884]; cellular chloride ion homeostasis [GO:0030644]; cellular potassium ion homeostasis [GO:0030007]; cellular response to chemokine [GO:1990869]; cellular response to potassium ion [GO:0035865]; cellular sodium ion homeostasis [GO:0006883]; chloride ion homeostasis [GO:0055064]; chloride transmembrane transport [GO:1902476]; gamma-aminobutyric acid signaling pathway [GO:0007214]; hyperosmotic response [GO:0006972]; inorganic anion import across plasma membrane [GO:0098658]; inorganic cation import across plasma membrane [GO:0098659]; ion transport [GO:0006811]; maintenance of blood-brain barrier [GO:0035633]; negative regulation of vascular wound healing [GO:0061044]; positive regulation of aspartate secretion [GO:1904450]; positive regulation of cell volume [GO:0045795]; potassium ion homeostasis [GO:0055075]; potassium ion import across plasma membrane [GO:1990573]; regulation of matrix metallopeptidase secretion [GO:1904464]; regulation of spontaneous synaptic transmission [GO:0150003]; sodium ion homeostasis [GO:0055078]; sodium ion import across plasma membrane [GO:0098719]; sodium ion transmembrane transport [GO:0035725]; T cell chemotaxis [GO:0010818]; transepithelial ammonium transport [GO:0070634]; transepithelial chloride transport [GO:0030321]; transport across blood-brain barrier [GO:0150104] | 11943682_intracellular Cl(-) (Cl) regulates the activity of protein kinase C (PKC)-delta and thus the activation of Na-K-Cl cotransport 12054469_NKCC1 was found in human HuH-7 hepatoma cells. The data suggest a role of NKCC1 in stellate cell transformation, hepatic volume regulation, and long-term adaption to dehydrating conditions. 12355171_most CFTR-positive ADPKD cysts also express NKCC1 suggests that transepithelial Cl(-) secretion in ADPKD involves molecular mechanisms similar to secretory epithelia. 12657561_all of the CCCs examined (NKCC1, NKCC2, KCC1, KCC3, and KCC4) can promote NH4(+) translocation, presumably through binding of the ion at the K(+) site 12740379_data conclusively link PASK with the phosphorylation and activation of NKCC1 14982922_the stimulation of either luminal or basolateral P2R increased NKCC1 activity, which was observed in the basolateral membrane, but not in the luminal membrane 15280386_the uncovered interacting domains are probably a major determinant of the NKCC1 conformational landscape 15347682_Data indicate that binding of hsp90 to the Na(+)-K(+)-Cl(-) cotransporter (NKCC1) may be required for sodium-potassium-chloride cotransport to occur at the cell surface. 15899883_role of the complex of serine/threonine protein kinases and a protein phosphatase is probably the maintenance of optimal phosphorylation of NKCC1 coincident with its physiological function in epithelial absorption and secretion 16222701_Taken together, the results of the present study indicate that the signal transduction protein, controlled by the Na+/K+/Cl- cotransporter, must be downstream of the PKC, and at/or upstream to MEK in the Ras/Raf/MEK/ERK cascade. 16227993_NKCC1 transporter facilitates seizures in the developing brain. 17478539_persistent NKCC1 activation by cAMP is constrained by a Ca(2+)-dependent cycle of co-transporter internalization, degradation and re-expression; this is a novel mechanism to limit intestinal fluid loss. 18032481_Ammonium was transported on NKCC1 in T84 cells nearly as well as potassium 18400987_Reveal a novel role for the EGFR in the chronic regulation of epithelial secretory capacity through upregulation of NKCC1 expression. 18550547_PKCdelta acts upstream of SPAK to increase activity of NKCC1 during hyperosmotic stress 19023125_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19523148_Observational study of gene-disease association. (HuGE Navigator) 19686239_membrane rafts render KCC2 inactive and NKCC1 active 19916249_NKCC1 mRNA is expressed predominately in small- and medium-diameter primary afferent neurons,21 and the NKCC1 population of sensory neurons includes a large proportion that express TrpV1 and thus are presumably capsaicin-sensitive nociceptorscu 20044742_decreased NKCC1 may contribute to the feature of the pathogenesis of salt-sensitive hypertension seen in African Americans. 20061948_NKCC1 modulates blood pressure through vascular and renal effects--REVIEW 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20442269_phosphorylation-induced activation of NKCC1 by osmotic shrinkage does not involve AMP-activated protein kinase and is likely to be due to STE20/SPS1-related proline/alanine-rich kinase activation 20471979_The result points at secretion as the main mechanism of cyst filling, and NKCC1 as the key candidate of fluid transport. 20732874_PKC{delta} and PKC{epsilon}-regulated NKCC1 surface expression plays an important role in the regulation of chloride secretion 20819947_The coupling between salt and water transport in NKCC1 represents a novel aspect of cellular water homeostasis where cells can change their volume independently of the direction of an osmotic gradient across the membrane. 20819979_In schizophrenia, increased expression levels of OXSR1 and WNK3 may shift the balance of chloride transport by NKCC1 and KCC2 and alter the nature of gamma-aminobutyric acid neurotransmission in the prefrontal cortex. 21613606_The Wnk3 protein isoforms have a similar effect on SLC12 cotransporters. NKCC1/2 and NCC were inhibited, even in hypertonicity, while KCCs were activated, even in isotonic conditions. 21847667_NKCC1 plays an important and yet partial role in RVI in C-20/A4 chondrocytes. 22178882_NKCC1 activation occurred through time-dependent increases in protein-protein interaction between ERK1/2 and NKCC1, which were proportional to EGF concentration 22437837_predictions of homology models of NKCC1 and demonstrate important roles for TM3 residues in ion translocation and loop diuretic inhibition. 22570591_NKCC1 deficiency increases the size of focal adhesions. 22643131_A significant association is found between single nucleotide polymophisms in SLC12A2 and CTXN3 and schizophrenia in a Thai population. 22989884_Data show that intracellular association between WNK1 and oxidative stress-responsive 1 (OSR1) is required for stimulation of OSR1 and Na(+), K(+), Cl(-)-Cotransporter NKCC1 and NKCC2 activities by osmotic stress. 23139219_Capsaicin inhibits chloride secretion in part by causing NKCC1 internalization, but by a mechanism that appears to be independent of TRPV1. 23224734_The rs10089 single nucleotide polymorphism was associated with increased susceptibility to noise-induced hearing loss. 23317544_These findings suggest that NKCC1 and AQP1 participate in meningioma biology and invasion 23515529_The results indicate a role for COMMD1 in the regulation of NKCC1 membrane expression and ubiquitination. 23894354_Reduced KCC2/NKCC1 ratio in the cerebrospinal fluid of Rett Syndrome patients suggests a disturbed process of GABAergic neuronal maturation and open up a new therapeutic perspective. 23921125_Altered hippocampal area function and coupling in DISC1 and SLC12A2 minor allele carriers. 24173102_The hormone aldosterone was found to upregulate NKCC1 by increasing protein stability. 24339991_Functional expression of human NKCC1 from a synthetic cassette-based cDNA: introduction of extracellular epitope tags and removal of cysteines. 24451383_NKCC activation involves movement of TM12 relative to TM10, which is likely tied to movement of the large C terminus 24555568_our data show a novel role for the WNK1/OSR1/NKCC1 pathway in glioma migration 24944475_Suggest that the expression of NKCC1 in esophageal squamous cell carcinoma may affect the G2/M checkpoint and may be related to the degree of histological differentiation of SCCs. 25218052_NKCC1 labeling was seen only in the basolateral membrane of the secretory coils. 25620102_The NKCC-1 is a Na(+)- dependent Cl(-) transporter that mediates the movement of Na(+), K(+), and Cl(-) ions across the plasma membrane and maintains cell volume and intracellular K(+) and Cl(-) homeostasis. 26559081_NKCC1 high expression predicted a bad clinical outcome for lung adenocarcinoma patients and EGFR-mutated subgroup. Therefore, NKCC1 may play a role in lung adenocarcinoma and novel therapeutic tactics could be developed by targeting NKCC1 protein. 26955005_Study describes a functional missense variant in SLC12A2 in human schizophrenia, and suggest that genetically encoded dysregulation of NKCC1 may be a risk factor for, or contribute to the pathogenesis of, schizophrenia 27170636_NKCC1 and KCCs are coordinately regulated by L-WNK1 isoforms. 28453575_We also discovered and replicated three genome-wide significant variants in previously unreported loci for RDW (SLC12A2 rs17764730, PSMB5 rs941718), and hematocrit (PROX1 rs3754140) and an upstream anti-sense long-noncoding RNA, LINC01184, as the likely causal variant 28574574_we introduce the molecular mechanism by which flavonoids, specifically quercetin, act through elevation of [Cl(-) ]c via activation of NKCC1 on important factors controlling various body and cellular functions, such as (1) antihypertensive actions controlling blood volume dependent on the amounts of renal Na(+) reabsorption via expression of the epithelial Na(+) channel 28631939_This new functional assay offered a robust working model for NKCC1 in determining reliable and concordant rank orders of the test compounds supporting its sensitivity and specificity. 28679472_NKCC1 not only controls cell volume and Cl- concentration, but it can also regulate the actin cytoskeleton through Cofilin 1. 30159893_NKCC1 activity promotes the epithelial-mesenchymal transition-like process in gliomas via RhoA and Rac1 signaling pathways. 30759289_Study significantly lower NKCC1 DNA methylation and significantly higher KCC2 DNA methylation levels in patients with juvenile myoclonic epilepsy (JME) compared with the healthy controls. This implies that NKCC1 expression can be higher and KCC2 expression can be reduced in affected people. 30840784_An increase in Na-K-2Cl cotransporter-1 transcripts was found in the caudate nucleus of Huntington's disease patients. 31241262_We found a significant relationship between TLE [temporal lobe epilepsy] and methylation on the NKCC1. The methylation of NKCC1 can be a mecha-nism of refractory temporal lobe epilepsy. 31441327_Does the circadian clock make RPE-mediated ion transport ''tick'' via SLC12A2 (NKCC1)? 31655271_Novel Human NKCC1 Mutations Cause Defects in Goblet Cell Mucus Secretion and Chronic Inflammation. 32039487_A mutation in the Na-K-2Cl cotransporter-1 leads to changes in cellular metabolism. 32081947_Report a cryo-EM structure of NKCC1 captured in a partially loaded, inward-open state. NKCC1 assembles into a dimer, with the first ten transmembrane (TM) helices harboring the transport core and TM11-TM12 helices lining the dimer interface. 32294086_study suggests that variants affecting exon 21 of the SLC12A2 transcript are responsible for hereditary hearing loss in humans 32333139_Expression of Cl(-) channels/transporters in nasal polyps. 32393472_Blockade of Cell Volume Regulatory Protein NKCC1 Increases TMZ-Induced Glioma Apoptosis and Reduces Astrogliosis. 32658972_SLC12A2 variants cause a neurodevelopmental disorder or cochleovestibular defect. 33500540_Clinical characterization and further confirmation of the autosomal recessive SLC12A2 disease. 33597714_The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2. 33662382_GWAS Identifies LINC01184/SLC12A2 as a Risk Locus for Skin and Soft Tissue Infections. 34213892_All-Atom Simulations Uncover the Molecular Terms of the NKCC1 Transport Mechanism. 34374074_PNPT1, MYO15A, PTPRQ, and SLC12A2-associated genetic and phenotypic heterogeneity among hearing impaired assortative mating families in Southern India. | ENSMUSG00000024597 | Slc12a2 | 1833.172412 | 1.0605560 | 0.084820849 | 0.07855901 | 1.161224e+00 | 2.812118e-01 | 6.518780e-01 | No | Yes | 2191.070925 | 376.426172 | 2045.229317 | 351.278837 | |
| ENSG00000064692 | 9627 | SNCAIP | protein_coding | Q9Y6H5 | FUNCTION: Isoform 2 inhibits the ubiquitin ligase activity of SIAH1 and inhibits proteasomal degradation of target proteins. Isoform 2 inhibits autoubiquitination and proteasomal degradation of SIAH1, and thereby increases cellular levels of SIAH. Isoform 2 modulates SNCA monoubiquitination by SIAH1. {ECO:0000269|PubMed:16595633, ECO:0000269|PubMed:19224863}. | 3D-structure;ANK repeat;Alternative splicing;Coiled coil;Cytoplasm;Neurodegeneration;Parkinson disease;Parkinsonism;Reference proteome;Repeat;Ubl conjugation | This gene encodes a protein containing several protein-protein interaction domains, including ankyrin-like repeats, a coiled-coil domain, and an ATP/GTP-binding motif. The encoded protein interacts with alpha-synuclein in neuronal tissue and may play a role in the formation of cytoplasmic inclusions and neurodegeneration. A mutation in this gene has been associated with Parkinson's disease. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2015]. | hsa:9627; | cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; cytosol [GO:0005829]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; presynaptic membrane [GO:0042734]; synaptic vesicle [GO:0008021]; identical protein binding [GO:0042802]; ubiquitin protein ligase binding [GO:0031625]; cell death [GO:0008219]; dopamine metabolic process [GO:0042417]; regulation of inclusion body assembly [GO:0090083]; regulation of neurotransmitter secretion [GO:0046928] | 11427316_Observational study of gene-disease association. (HuGE Navigator) 11784370_Observational study of gene-disease association. (HuGE Navigator) 11958831_The amino acid sequence of synphilin-1 shows extensive homology with its human counterpart, especially in regions containing ankyrin-like motifs and the coiled-coil domain. Expression of mouse synphilin-1 in tissues is similar to its human counterpart. 12750386_results suggest that synphilin-1 has an important role in the formation of aggregates and cytotoxicity in Parkinson disease and that Dorfin may be involved in the pathogenic process by ubiquitylation of synphilin-1 12761037_a causative role of the R621C mutation in the synphilin-1 gene in Parkinson's disease 12915459_Changes in synuclein expression presage neurodegeneration in a Drosophila model of Parkinson disease. 14506261_Siah-1 was found to abrogate the inhibitory effects of synphilin-1 on dopamine release 14627698_role of aggresomes in cell viability was addressed in the context of over-expressing alpha-synuclein and its interacting partner synphilin-1 14639662_Observational study of gene-disease association. (HuGE Navigator) 14645218_Casein kinase II (CKII) phosphorylates synphilin-1; beta subunit of this enzyme complex binds to synphilin-1. CKII-mediated phosphorylation of synphilin-1, rather than alpha-synuclein, modulates the aggregation into inclusion bodies. 15322916_role of synphilin-1 in synaptic function and protein degradation and in the molecular mechanisms leading to neurodegeneration in Parkinson disease 15728840_Parkin is a dual-function ubiquitin ligase. K63-linked ubiquitination of synphilin-1 by parkin may be involved in the formation of Lewy body inclusions associated with Parkinson disease. 15894486_We confirm that synphilin-1 and parkin are components of majority of Lewy Bodies in Parkinson's disease and that both proteins are susceptible to proteasomal degradation. 16174773_GSK3beta modulates synphilin-1 ubiquitylation and cellular inclusion formation by SIAH 16595633_Synphilin-1A may contribute to neuronal degeneration in alpha-synuclein mutations and provides insights into the role of inclusion bodies in neurodegenerative disorders. 16877356_These results suggest that NUB1 indeed targets synphilin-1 to the proteasome for its efficient degradation, which, because of the resultant reduction in synphilin-1, suppresses the formation of synphilin-1-positive inclusions. 17327361_a novel specific interaction of synphilin-1 with the regulatory proteasomal protein S6 ATPase (tbp7) in aggresome-like intracytoplasmic inclusions 17467279_These findings suggest that parkin and synphilin-1 isoform expression changes play a significant role in the pathogenesis of LB diseases. 17982729_review:Isoform Synphilin-1A inclusions recruit both alpha-synuclein and synphilin-1. Aggregation of synphilin-1 and synphilin-1A seems to be protective to cells 18292964_specific effects of C621 mutant synphilin-1 on gene expression that correlate with its role as a susceptibility factor in Parkinson's disease 18335262_All four alpha-synuclein isoforms were affected in dementia with LB (Lewy bodies), most parkin transcript variants in common LB disease, and all synphilin-1 isoforms in Parkinson disease. 18366718_Observational study of gene-disease association. (HuGE Navigator) 18366718_We found no evidence for association between genetic variability in synphilin-1 and Parkinson's disease 18635553_translocation to aggresomes required a special aggresome-targeting signal within the sequence of synphilin 1, an ankyrin-like repeat domain. 18782602_Synphilin-1 might be involved in motor function, and its accumulation in the central nervous system can cause motor impairments. 19224863_synphilin-1A has a novel role as a regulator of SIAH activity, modulating alpha-synuclein, and formation of Lewy body-like inclusions 19730898_Data show that periphilin displays an overlapping expression pattern with synphilin-1 in cellular and animal models and in Lewy bodies of Parkinson's disease (PD) patients, and support involvement of periphilin in PD. 19762560_alpha-synuclein-synphilin-1 interaction significantly promotes the formation of cytoplasmic alpha-synuclein inclusions, which may have implications for Lewy body formation in neural cells 19857556_expression of synphilin-1 shortens N1E-115 cell division doubling time, promotes neurite outgrowth, and protects against Rotenone-induced toxicity; synphilin-1 displays a neurotrophic effect in vitro, may play a neuroprotective role in Parkinson's disease 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20604806_Knockdown of Herp gene unexpectedly facilitated the degradation of synphilin-1, and improved cell viability during proteasomal inhibition. 20816781_Neuronal survival factor MEF2D is decreased in human and experimental Parkinson's disease, a decrease that is specifically associated with alpha-synuclein accumulation and aggregation. 20849899_Although serine-129 phosphorylation of alpha-synuclein facilitates tubulin polymerization promoting protein (TPPP)-mediated alpha-SYN oligomerization, this modification does not seem to play an inevitable role in the early step of alpha-SYN oligomer formation. 21103907_Synphilin-1 inhibits alpha-synuclein degradation by the proteasome. 21344240_Mutation screening of SNCAIP identifies novel sequence variants using a bioinformatic approach; further studies are necessary to determine their possible functional consequences in South African patients with Parkinson's disease. 22828940_Overexpression of SP1 in neurons, but not peripheral cells, increased the body weight of flies compared with that of non-transgenic controls. SP1 increased food intake but did not affect locomotor activity 24829096_Overexpression of human synphilin-1 in mice resulted in hyperphagia and obesity. 25545246_Synphilin-1 binds ATP, but not CTP. 26465922_Differential expression of synphilin-1 isoforms (and alpha-synuclein and parkin) was found in multiple system atrophy brains compared to control brain. 30316984_results indicated that synphilin-1 may play neuroprotective roles in Parkinson Disease pathogenesis by inhibiting ROS production and apoptosis. | ENSMUSG00000024534 | Sncaip | 25.523806 | 0.6749363 | -0.567176669 | 0.53533509 | 1.090898e+00 | 2.962723e-01 | No | Yes | 17.844440 | 6.119979 | 24.583483 | 8.346355 | ||
| ENSG00000065320 | 9423 | NTN1 | protein_coding | O95631 | FUNCTION: Netrins control guidance of CNS commissural axons and peripheral motor axons. Its association with either DCC or some UNC5 receptors will lead to axon attraction or repulsion, respectively. Binding to UNC5C might cause dissociation of UNC5C from polymerized TUBB3 in microtubules and thereby lead to increased microtubule dynamics and axon repulsion (PubMed:28483977). Involved in dorsal root ganglion axon projection towards the spinal cord (PubMed:28483977). It also serves as a survival factor via its association with its receptors which prevent the initiation of apoptosis. Involved in tumorigenesis by regulating apoptosis (PubMed:15343335). {ECO:0000269|PubMed:15343335, ECO:0000269|PubMed:28483977}. | 3D-structure;Apoptosis;Cytoplasm;Disease variant;Disulfide bond;Glycoprotein;Laminin EGF-like domain;Reference proteome;Repeat;Secreted;Signal | Netrin is included in a family of laminin-related secreted proteins. The function of this gene has not yet been defined; however, netrin is thought to be involved in axon guidance and cell migration during development. Mutations and loss of expression of netrin suggest that variation in netrin may be involved in cancer development. [provided by RefSeq, Jul 2008]. | hsa:9423; | basement membrane [GO:0005604]; cytoplasm [GO:0005737]; extracellular region [GO:0005576]; animal organ morphogenesis [GO:0009887]; anterior/posterior axon guidance [GO:0033564]; apoptotic process [GO:0006915]; Cdc42 protein signal transduction [GO:0032488]; cell-cell adhesion [GO:0098609]; chemorepulsion of axon [GO:0061643]; dendrite development [GO:0016358]; inner ear morphogenesis [GO:0042472]; mammary gland duct morphogenesis [GO:0060603]; motor neuron axon guidance [GO:0008045]; negative regulation of axon extension [GO:0030517]; neuron migration [GO:0001764]; nuclear migration [GO:0007097]; positive regulation of axon extension [GO:0045773]; positive regulation of cell motility [GO:2000147]; positive regulation of cell population proliferation [GO:0008284]; Ras protein signal transduction [GO:0007265]; regulation of cell migration [GO:0030334]; regulation of synapse assembly [GO:0051963]; substrate-dependent cell migration, cell extension [GO:0006930]; tissue development [GO:0009888] | 12810718_Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin 14602071_Data demonstrate that alpha6beta4 integrin mediates pancreatic epithelial cell adhesion to Netrin-1, whereas recruitment of alpha6beta4 and alpha3beta1 regulate the migration of putative pancreatic progenitors on Netrin-1. 15491747_DCC/netrin-1 signaling may commit cells to the transition of endometrial gland architecture or function from a proliferating to a secretory phase. 15573119_Binding of netrin-1 to its receptors inhibits tumour suppressor p53-dependent apoptosis (review) 15574733_Netrin binds through multiple domains to both DCC and Unc5c. 15811950_Raft localization of DCC is required for netrin-1-induced DCC-dependent ERK activation, and netrin-1-mediated axon outgrowth requires lipid raft integrity. 16158190_Review suggests possible roles of netrin-1 in nervous system development, neovascularisation, adhesion and tumorigenesis. 16181408_Ligand-mediated down-regulation of deleted in colorectal cancer might participate in loss of netrin-responsiveness in developing nervous system. 16203981_Endothelial expression of netrin-1 may inhibit basal cell migration into tissues and its down-regulation with the onset of sepsis/inflammation may facilitate leukocyte recruitment. 18253061_Both deleted in colorectal cancer (DCC) and neogenin become tyrosine phosphorylated in cortical neurons in response to netrin-1. 18353983_Netrin-1 expression observed in a large fraction of human metastatic breast tumors confers a selective advantage for tumor cell survival. 18400890_Although cAMP can alter response of axons to netrin-1, we conclude that netrin-1 does not alter cAMP levels in axons attracted by this cue and that soluble adenyl cyclase is not required for axon attraction to netrin-1. 18439993_Data show that Netrin-1 expressing cells inhibited angiogenic sprouting of unc5b expressing blood vessels, but had no pro-angiogenic activity at any stage of development examined. 18455953_In adults, decreased expression within the spinal cord injury lesion; likely an inhibitor of regenerating neural progenitors 18469807_PIKE-L acts as a downstream survival effector for netrin-1 through UNC5B in the nervous system 18653556_Netrin 1, through its receptor DCC, inhibits RhoA in embryonic spinal commissural neurons. 18692059_NF-kappaB activation that occurs in response to inflammation confers a selective advantage for tumor development through NF-kappaB-mediated netrin-1 up-regulation 18796601_netrin-1 is not only an attractive cue for developing commissural axons but also promotes their survival 18922894_the transcriptionally active TAp73alpha tumor suppressor is implicated in the apoptosis induced by netrin-1 in a p53-independent and DCC/ubiquitin-proteasome dependent manner. 19122655_HIF-1alpha-dependent induction of netrin-1 attenuates hypoxia-elicited inflammation at mucosal surfaces 19211441_High levels of netrin-1 found in 43 of the 92 NSCLC tumor samples. Interference with netrin-1 in human lung cancer cell lines was associated with UNC5H-mediated cell death in vitro and with tumor growth inhibition and/or regression in xenografted mice. 19349462_Netrin-1 up-regulation is a potential marker for poor prognosis in stage 4 neueroblastoma in infants. 19822088_Netrin-1 inhibited migration of synovial fibroblasts from patients with rheumatoid arthritis and osteoarthitis. 19826074_Netrin-1 protein functions might vary with its localization in the placenta and probably with time of gestation. 19940358_Study suggests that Netrin-1 promotes melanoma cell invasion and migration and therefore has an important role in the progression of malignant melanoma. 20007677_Netrin-1 is an early, predictive biomarker of acute kidney injury after cardiopulmnoary bypass. 20029409_Observational study of gene-disease association. (HuGE Navigator) 20036673_Netrin-1 has a role in cardioprotection 20075388_Pulmonary netrin-1 levels are repressed during acute lung injury. 20080097_Netrin-1 might be an important regulator of pancreatic tumor growth that functions in tumor and endothelial cells. 20305387_Data propose that induction of netrin-1 expression via NFkappaB in inflammatory bowel diseases patients could affect colorectal tumor promotion and progression 20609112_our findings might indicate also an important role for DCC and netrin-1 in human foetal central nervous system development 20620466_Urinary netrin-1 levels are increased in patients with various forms of acute kidney injury, and may therefore serve as a biomarker. 21193949_Netrin-1 may regulate the development of placental vessels and plays a key role in the pathogenesis of fetal growth restriction. 21303223_Plasma netrin-1 can be used as diagnostic biomarker of many human cancers. 21505994_Netrin-1 enhanced the viability, migration and tube formation of human placental microvascular endothelial cells. 21733374_Gene silencing of netrin-1 could inhibit viability, proliferation, migration, and tubal formation of HUVECs, and placental angiogenesis. 21740336_Properties and perspectives of uNGAL and Netrin-1 for their appropriate clinical utilization. 21789787_overexpression is predictive of ovarian malignancies 21980448_an autocrine function for netrin-1 and netrin-3 in U87 and U373 cells that slows migration 22231519_It was shown that netrin-1 was secreted by macrophages in human and mouse atheroma, where it inactivated macrophage migration out of atherosclerotic plaques. 22252496_Hypoxia-inducible factor 1 controls the expression of the uncoordinated-5-B receptor, but not of netrin-1, in first trimester human placenta 22332546_netrin-1 protein expression may be related to tumorigenesis and tumor progression of gastric cancer. 22588384_Netrin-1 and DCC are increased in diseased lumbar intervertebral discs and may play a role in the process of neurovascular growth. 22685302_knockdown of DSCAM inhibits netrin-induced tyrosine phosphorylation of UNC5C and Fyn as well as the interaction of UNC5C with Fyn. The double knockdown of both receptors abolishes the induction of Fyn tyrosine phosphorylation by netrin-1 22779713_Netrin 1 gene mutation found in patients diagnosed with superior semicircular canal dehiscence syndrome. 22871610_Data indicate that DeltaN-netrin-1 stimulated cell proliferation in vitro and tumor growth in vivo. 22889408_There was no relationship between the basal netrin-1 and decline in netrin-1 concentrations and the progression-free survival and overall survival in gastric cancer. 23195957_Netrin-1 promotes glioblastoma cell invasiveness and angiogenesis by multiple pathways including activation of RhoA, cathepsin B, and cAMP-response element-binding protein 23512105_In addition, eight genes classified as 'second tier' hits in the original study (PAX7, THADA, COL8A1/FILIP1L, DCAF4L2, GADD45G, NTN1, RBFOX3 and FOXE1) showed evidence of linkage and association in this replication sample. 23526078_Netrin-1 was upregulated in kidney carcinoma. 23538444_Netrin-1 protects hypoxia-induced mitochondrial apoptosis through HSP27 expression via DCC- and integrin alpha6beta4-dependent Akt, GSK-3beta, and HSF-1 in mesenchymal stem cells. 23549787_netrin-1 forms a complex with both Notch2 and Jagged1. 23666553_Higher netrin-1 and lower UNC5B expression in all prostate carcinoma cell lines indicated that netrin-1 and UNC5B could be used to predict metastasis. 24092381_Tubular atrophy and low netrin-1 gene expression are associated with delayed kidney allograft function. 24122613_Data indicate that dysregulation of expression of netrin-1 NTN1 in smooth muscle cell (SMC) and its chemorepulsive receptor UNC5B in macrophages are involved in the development of atherosclerosis and unstable plaques. 24174661_Netrin-1 increases the frequency and amplitude of excitatory postsynaptic p otentials (EPSPs) recorded from cortical pyramidal neurons. 24262644_promotes angiogenesis and accelerates atherosclerosis, protects the heart against ischemia-reperfusion injury, and reduces the infarct size. 24293316_The netrin-1 upregulation which appears to be p53-dependent is a survival mechanism as netrin-1 silencing by siRNA is associated with a potentiation of cancer cell death upon Doxorubicin treatment. 24338005_Studies indicate that elevating netrin-1 expression increased migration and invasion in colorectal cancer cells. 24442237_These results suggest that in hypoxic HCC cells, netrin-1 activates downstream signaling pathways to induce EMT activation with subsequent production of multiple inflammatory mediators which in turn promotes cancer invasion. 24528886_In bladder cancer cells, netrin-1 expression is increased compared to control cells. 24584118_Netrin-1 is highly expressed in obese but not lean adipose tissue, where it directs the retention of macrophages, promoting inflammation. NTN1 also promotes insulin resistance. 24647424_Data indicate both total and nuclear netrin-1 expression as prognostic factors in brain metastases patients in contrast to other prognostic markers in oncology such as patient age, number of brain metastases or Ki67 proliferation index. 24716747_Increased maternal netrin-4 levels in IUGR neonates may reflect in utero hypoxia, while the negative correlations between fetal netrin-1 and -4 levels may exert the dynamic balance between their angio- and anti-angiogenic properties. 24738865_Ntn-1 induces MMP-12-dependent E-cad degradation via the distinct activation of PKCalpha and FAK/Fyn, which is necessary to govern the activation of ERK, JNK, and NF-KB in promoting motility of umbilical cord blood-derived mesenchymal stem cells. 24778312_novel model for the spatial regulation of axon branching by Netrin-1, in which localized plasma membrane expansion occurs via TRIM9-dependent regulation of SNARE-mediated vesicle fusion. 24812271_Netrin-1 promotes medulloblastoma cell invasiveness and angiogenesis, and demonstrates elevated expression in tumor tissue and urine of patients with pediatric medulloblastoma. 24827071_Netrin-1 ameliorates myocardial infarction-induced myocardial injury. 25001177_netrin-1 promoted pancreatic cancer cell proliferation by upregulation of murine double minute 2 (Mdm2). 25022751_results of another in vitro assay, in which endothelial cells were co-cultured with human fibroblasts, however, showed that Mel2a-netrin1 CM inhibited tube formation through blocking elongation and coalescence of human umbilical vein endothelial cells 25154466_early urinary biomarker in the diagnosis of septic acute kidney injury 25289643_These studies demonstrate that semaphorin 3A and netrin-1 can be useful early diagnostic biomarkers of AKI after liver transplantation. 25353184_Careful phenotyping of patients combined with molecular and functional studies in zebrafish identifed netrin-1 as a potential shared genetic factor for cardiac and thyroid congenital defects. 25483983_These results identify Netrin-1 as a key regulator of osteoclast differentiation that may be a new target for bone therapies. 25569157_Investigation of the mechanisms that trigger local translation revealed a role for calcium-dependent retrograde netrin-1/DCC receptor signaling. 25903786_results demonstrate that netrin 1 is an important regulator of blood-brain barrier maintenance that protects the central nervous system against inflammatory conditions such as multiple sclerosis. 25950802_NTN1-DCC pathway contains targets of FDA-approved drugs and may offer promise for guiding applied clinical research on preventive and therapeutic interventions for AMD 26039999_Netrin-1 stimulates phosphatase 1A to dephosphorylate YAP, which leads to decreased ubiquitination and degradation, enhancing YAP accumulation and signaling 26393880_MUC4 has a role in promoting neural invasion mediated by the axon guidance factor Netrin-1 in pancreatic ductal adenocarcinoma 26573873_Akt-FoxO1 signaling pathway plays an essential role during liver regeneration 26749424_Enhanced detection of netrin-1-expressing CD14(low) cells in patients with systemic sclerosis-related interstitial lung disease was observed, and antibody-mediated netrin-1 neutralization attenuated detection of fibrocytes in all settings. 26859457_identify the V-2 domain of netrin-1 to be important for its interaction with the Ig1/Ig2 domains of UNC5H2 26944719_High Plasma netrin-1 levels are associated with peripheral artery disease in smokers. 27031829_Netrin-1 enhanced infectivity of hepatitis C virus particles and promoted viral entry by increasing the activation and decreasing the recycling of the epidermal growth factor receptor (EGFR), a protein that is dysregulated in hepatocellular carcinoma. 27060954_Netrin-1 reduced the Abeta1- 40-induced Abeta1-42 increase, increased sAbetaPPalpha, and reversed the Abeta-induced sAbetaPPalpha decrease in vitro 27067437_Data show that netrin-1 concentrations elevated in advanced non-small cell lung cancer compared to a healthy control group, and netrin-1 concentrations decreased with chemotherapy. 27087488_Data suggest that urinary netrin-1 may be a new biomarker for determining early tubular injury in obese children. 27400127_netrin-1 degradation products are capable of modulating vascular permeability in diabetic retinopathy 27422368_NETRIN1 signaling pathway is identified as a candidate pathway for major depressive disorder and should be explored in further large population studies. 27651158_cell proliferation and survival were decreased following targeted deletion of Netrin-1. Cell invasion was dramatically diminished in Netrin-1 knockdown Glioblastoma stem cells. Moreover, Netrin-1 knockdown Glioblastoma stem cells exhibited less proangiogenic activity. 27727089_Our study has shown an increase in netrin-1 expression that could be linked with macrophage infiltration and polarization in the epicardial adipose tissue of patients with coronary artery disease 27793362_upregulation of netrin-1 expression in different forms of cancer, and the increased expression of netrin-1 has been linked to its functions as a survival and invasion promoting factor. 27815019_that netrin-1 may function as a positive regulator of hypoxia-triggered malignant behavior in prostate cancer by activating the Yes-associated protein signaling 28069038_Results demonstrate that NTN1 is an important regulator of stemness and motility of glioblastoma cells. Furthermore, it deciphers a novel mechanism where NTN1 activates Notch signaling and subsequent stemness in invasive glioblastoma cells. 28241866_Netrin-1 acts as a non-canonical angiogenic factor produced by human Wharton's jelly mesenchymal stem cells. 28250059_Netrin-1 restores cell injury and impaired angiogenesis in vascular endothelial cells upon high glucose via PI3K/AKT-eNOS signaling. 28443497_we review the Netrin-1 mediated regulation of cancer, its potential use as a biomarker, and the targeting of the Netrin-1 pathway to treat cancers 28475012_Our observations revealed that SOX6 is a tumor suppressor in ovarian cancer cells, and SOX6 exerts an inhibitory effect on the proliferation, invasion, and tumor cell-induced angiogenesis of ovarian cancer cells, whereas nerin-1 plays an opposite role and its expression is inversely correlated with SOX6. 28489749_NTN1 rs9788972 is identified as a risk locus for nonsyndromic orofacial clefts susceptibility in a northern Chinese population; SNPs in PAX7 were not associated with any increased risk 28677863_Netrin-1 decreased in experimental autoimmune encephalomyelitis and in multiple sclerosis patients, mainly during relapse, suggesting an anti-inflammatory role of netrin-1 28704853_Conclusion RBP4 may be used as a predictive factor of diabetic nephropathy patients complicated with silent cerebral infarction (SCI) and is positively correlated with cognitive dysfunction. RBP4/Lp-PLA2/Netrin-1 pathway activation may be one of the occurrence mechanisms in diabetic nephropathy complicated with SCI. 28945198_Three mutations in exon 7 of NTN1 in 2 unrelated families and 1 sporadic case with isolated congenital mirror movements (CMM), a disorder characterized by involuntary movements of one hand that mirror intentional movements of the opposite hand. 29199452_increased concentrations of urinary SEMA-3A and urinary Netrin-1 are found in urine from children with severe hydronephrosis and that their concentrations are related to the degree of obstruction. 29202173_This study implicates DCC and NTN1 mutations in the pathophysiology of CHH consistent with the role of these two genes in the ontogeny of GnRH neurons in mice. 29234153_the current data suggest that netrin-1 can act as a pro-metastatic factor in NSCLC by enhancing cell invasion, migration, and VM via PI3K/AKT and ERK-mediated EMT process, thereby implicating netrin-1 as a novel promising therapeutic target against aggressive non-small cell lung cancer. 29277614_Such data indicate that cooperation between the UPR and UNC5A depletion as previously observed by ourselves in HCC patients samples may foster liver cancer development and growth. 29297981_Four genes-ACT1, PIN1, DNMT1 and NTN1-emerged as having roles in pathways that may influence Primary Biliary Cholangitis pathogenesis in British Columbia First Nations people. 29305865_Collectively, our results defined netrin-1 as a positive regulator of malignant tumor metastasis in GC by activating the YAP signaling, with potential implications for new approaches to GC therapy. 29328435_miR214 reduces chemoresistance by targeting netrin1 in bladder cancer cell lines. 30106432_High NTN1 expression is associated with gastric cancer. 30197049_Findings indicate that variation in the NETRIN1 signaling pathway may confer risk for major depressive disorder through effects on a number of white matter tracts. 30402937_The GxE interaction analysis revealed a significant interaction between maternal environmental contact with agrotoxics and rs16969816 (OR: 0.25, 95% CI: 0.08-0.74, p = .01), and pairwise interaction test with NTN1 rs1880646 yielded significant p values in the 1,000 permutation test for rs16969681, rs16969816, and rs16969862. 30479344_netrin-1 as a major signal that mediates the dynamic crosstalk between inflammation and chronic erosion of the extracellular matrix in Abdominal aortic aneurysm, is reported. 30506619_Our study suggested allele G at rs4791774 in NTN1 gene is risk of NSCLO, which could greatly increase the risk to have a baby with cleft. 30578605_rs4791774 exhibited a nominal association with missing of maxillary canine and premolar. 30719925_We found a significantly higher netrin-1 level in patients with CO poisoning. However, we were unable to find any significant difference in the patients with neurological involvement with respect to netrin-1 level, netrin-1 may present subclinically neurological effects. Therefore, we believe that netrin-1 cannot be used as a marker of CO poisoning severity. 30852966_Elevated serum netrin-1 levels were associated with improved prognosis at 3 months after ischemic stroke, suggesting that serum netrin-1 may be a potential prognostic biomarker for ischemic stroke. 30968155_Netrin1 mediated epithelialmesenchymal transition of A549 and PC9 cells in vitro, which may be associated with the PI3K/AKT pathway. This effect was not observed under normoxic conditions. Serum concentration of netrin1 was found to be significantly higher in nonsmall cell lung cancer (NSCLC) patients compared with healthy donors. Findings highlight a novel role for netrin1 in NSCLC development under hypoxia. 31047878_Serum netrin-1 might represent a potential biomarker for reflecting severity, inflammation and prognosis of human aneurysmal subarachnoid hemorrhage. 31088838_Netrin-1, a navigation cue during embryonic development, is upregulated in cancer-associated fibroblasts and regulates cancer cell stemness. 31162046_We further provide a combination of protein localisation and phenotypic evidence in chick, humans, mice and zebrafish that Netrin-1 has an evolutionarily conserved and essential requirement for f optic fissure closure (OFC), and is likely to have an important role in palate fusion. 31219796_High netrin-1 levels are associated with early diabetic kidney injury in type 1 diabetes. 31226147_Human TUBB3 mutations specifically perturb netrin-1/UNC5C-mediated repulsion. 31407237_The authors suggest that Netrin-1 enhances N-cadherin junctions to promote liver cancer cell collective migration in 3D cell culture and may subsequently increase liver cancer metastasis. 31769036_Netrin-1 and its receptor Unc5b may have essential roles in periodontal inflammation 31780810_The functional variant of NTN1 contributes to the risk of nonsyndromic cleft lip with or without cleft palate. 31806640_Netrin-1 and Its Receptor DCC Are Causally Implicated in Melanoma Progression. 31953174_Macrophage-derived netrin-1 is critical for neuroangiogenesis in endometriosis. 32151395_The identification and function of a Netrin-1 mutation in a pedigree with premature atherosclerosis. 32228196_Serum netrin-1 levels at presentation and delayed neurological sequelae in unintentional carbon monoxide poisoning. 32231305_Netrin-1 promotes naive pluripotency through Neo1 and Unc5b co-regulation of Wnt and MAPK signaling. 32267322_High Serum Netrin-1 and IL-1beta in Elderly Females with ACS: Worse Prognosis in 2-years Follow-up.', trans 'Niveis Elevados de Netrina-1 e IL-1beta em Mulheres Idosas com SCA: Pior Prognostico no Acompanhamento de Dois Anos. 32319367_is abnormal expression of Netrin-1 in serum of children with acute lymphoblastic leukemia 32417215_Serum netrin-1 concentrations are associated with clinical outcome in acute intracerebral hemorrhage. 32474463_Serum netrin-1 as a biomarker for colorectal cancer detection. 32912541_Decreased Serum Netrin-1 as a Predictor for Post-Stroke Depression in Chinese Patients with Acute Ischemic Stroke. 32917498_Decreased serum netrin-1 is associated with ischemic stroke: A case-control study. 32926844_IGF2BP1 silencing inhibits proliferation and induces apoptosis of high glucose-induced non-small cell lung cancer cells by regulating Netrin-1. 33095273_Growth cone repulsion to Netrin-1 depends on lipid raft microdomains enriched in UNC5 receptors. 33351190_Netrin-1 and its receptor DCC modulate survival and death of dopamine neurons and Parkinson's disease features. 33546947_Association between serum netrin-1 and prognosis of ischemic stroke: The role of lipid component levels. 33741358_Serum netrin-1 as a potential biomarker for functional outcome of traumatic brain injury. 34470826_Delta40p53 isoform up-regulates netrin-1/UNC5B expression and potentiates netrin-1 pro-oncogenic activity. 34585999_Netrin-1 expression and targeting in multiple myeloma. 35063900_The role of serum netrin-1 level in the detection of early-onset preeclampsia. | ENSMUSG00000020902 | Ntn1 | 1830.364656 | 0.8630300 | -0.212517377 | 0.07248100 | 8.573011e+00 | 3.411825e-03 | 6.873389e-02 | No | Yes | 1717.916191 | 189.570091 | 2018.693684 | 222.430708 | |
| ENSG00000065609 | 9892 | SNAP91 | protein_coding | O60641 | FUNCTION: Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. Binding of AP180 to clathrin triskelia induces their assembly into 60-70 nm coats (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Coated pit;Glycoprotein;Membrane;Methylation;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:9892; | clathrin-coated pit [GO:0005905]; clathrin-coated vesicle [GO:0030136]; extrinsic component of presynaptic endocytic zone membrane [GO:0098894]; synaptic vesicle [GO:0008021]; 1-phosphatidylinositol binding [GO:0005545]; clathrin heavy chain binding [GO:0032050]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; protein kinase binding [GO:0019901]; SNARE binding [GO:0000149]; clathrin coat assembly [GO:0048268]; clathrin-dependent endocytosis [GO:0072583]; protein transport [GO:0015031]; regulation of clathrin-dependent endocytosis [GO:2000369]; synaptic vesicle budding from presynaptic endocytic zone membrane [GO:0016185]; vesicle budding from membrane [GO:0006900] | 12493563_AP180 had an overall expression similar to synaptophysin, the immunoreactivity for the two proteins did not always co-localize. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19450545_RNA interference-mediated knockdown of AP180 reduces the generation of Abeta1-40 and Abeta1-42, whereas CALM knockdown has no effect on Abeta generation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20511563_Observational study of gene-disease association. (HuGE Navigator) 20847448_AP180 is significantly decreased in the hippocampus of Alzheimer's disease patients and in hippocampal neurons of transgenic mice. 21808019_unique mechanism of SNARE motif-dependent endocytic sorting and identify the ANTH domain proteins AP180 and CALM as cargo-specific adaptors for synaptobrevin endocytosis 22639918_AP180 and CALM are endocytic adaptors dedicated to the sorting of small soluble N-ethylmaleimide-sensitive-factor attachment protein receptors. (Review) 26204806_intrinsically disordered domains are highly potent drivers of membrane curvature 26412491_To sustain efficient neurotransmission and synaptic vesicle formation, AP180 and VAMP2 are required. 27574975_Study concludes that the novel clathrin interaction sites identified here in CALM and AP180 have a major role in how these proteins interface with clathrin. This work advances the case that AP180 and CALM are required to use a combination of standard clathrin N-terminal domain binding motifs and the sequence identified here for optimal binding and assembling clathrin. | ENSMUSG00000033419 | Snap91 | 95.722144 | 1.3200470 | 0.400589253 | 0.29990953 | 1.724083e+00 | 1.891684e-01 | No | Yes | 108.311385 | 30.691821 | 85.823360 | 24.283041 | |||
| ENSG00000066651 | 60487 | TRMT11 | protein_coding | Q7Z4G4 | FUNCTION: Catalytic subunit of an S-adenosyl-L-methionine-dependent tRNA methyltransferase complex that mediates the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs. {ECO:0000250}. | Acetylation;Alternative splicing;Methyltransferase;RNA-binding;Reference proteome;S-adenosyl-L-methionine;Transferase;tRNA processing;tRNA-binding | hsa:60487; | cytoplasm [GO:0005737]; methyltransferase activity [GO:0008168]; tRNA (guanine-N2-)-methyltransferase activity [GO:0004809]; tRNA binding [GO:0000049] | 22386179_SNPs associated with androgen deprivation failure in advanced prostate cancer | ENSMUSG00000019792 | Trmt11 | 602.191993 | 1.1212468 | 0.165103899 | 0.11566753 | 2.056320e+00 | 1.515758e-01 | 5.104276e-01 | No | Yes | 672.333772 | 130.730815 | 588.760638 | 114.586827 | ||
| ENSG00000067606 | 5590 | PRKCZ | protein_coding | Q05513 | FUNCTION: Calcium- and diacylglycerol-independent serine/threonine-protein kinase that functions in phosphatidylinositol 3-kinase (PI3K) pathway and mitogen-activated protein (MAP) kinase cascade, and is involved in NF-kappa-B activation, mitogenic signaling, cell proliferation, cell polarity, inflammatory response and maintenance of long-term potentiation (LTP). Upon lipopolysaccharide (LPS) treatment in macrophages, or following mitogenic stimuli, functions downstream of PI3K to activate MAP2K1/MEK1-MAPK1/ERK2 signaling cascade independently of RAF1 activation. Required for insulin-dependent activation of AKT3, but may function as an adapter rather than a direct activator. Upon insulin treatment may act as a downstream effector of PI3K and contribute to the activation of translocation of the glucose transporter SLC2A4/GLUT4 and subsequent glucose transport in adipocytes. In EGF-induced cells, binds and activates MAP2K5/MEK5-MAPK7/ERK5 independently of its kinase activity and can activate JUN promoter through MEF2C. Through binding with SQSTM1/p62, functions in interleukin-1 signaling and activation of NF-kappa-B with the specific adapters RIPK1 and TRAF6. Participates in TNF-dependent transactivation of NF-kappa-B by phosphorylating and activating IKBKB kinase, which in turn leads to the degradation of NF-kappa-B inhibitors. In migrating astrocytes, forms a cytoplasmic complex with PARD6A and is recruited by CDC42 to function in the establishment of cell polarity along with the microtubule motor and dynein. In association with FEZ1, stimulates neuronal differentiation in PC12 cells. In the inflammatory response, is required for the T-helper 2 (Th2) differentiation process, including interleukin production, efficient activation of JAK1 and the subsequent phosphorylation and nuclear translocation of STAT6. May be involved in development of allergic airway inflammation (asthma), a process dependent on Th2 immune response. In the NF-kappa-B-mediated inflammatory response, can relieve SETD6-dependent repression of NF-kappa-B target genes by phosphorylating the RELA subunit at 'Ser-311'. Phosphorylates VAMP2 in vitro (PubMed:17313651). {ECO:0000269|PubMed:11035106, ECO:0000269|PubMed:12162751, ECO:0000269|PubMed:15084291, ECO:0000269|PubMed:15324659, ECO:0000269|PubMed:17313651, ECO:0000269|PubMed:9447975}.; FUNCTION: [Isoform 2]: Involved in late synaptic long term potention phase in CA1 hippocampal cells and long term memory maintenance. {ECO:0000250|UniProtKB:Q02956}. | ATP-binding;Alternative promoter usage;Alternative splicing;Cell junction;Cytoplasm;Endosome;Inflammatory response;Kinase;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Zinc;Zinc-finger | Protein kinase C (PKC) zeta is a member of the PKC family of serine/threonine kinases which are involved in a variety of cellular processes such as proliferation, differentiation and secretion. Unlike the classical PKC isoenzymes which are calcium-dependent, PKC zeta exhibits a kinase activity which is independent of calcium and diacylglycerol but not of phosphatidylserine. Furthermore, it is insensitive to typical PKC inhibitors and cannot be activated by phorbol ester. Unlike the classical PKC isoenzymes, it has only a single zinc finger module. These structural and biochemical properties indicate that the zeta subspecies is related to, but distinct from other isoenzymes of PKC. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:5590; | apical cortex [GO:0045179]; apical plasma membrane [GO:0016324]; axon hillock [GO:0043203]; bicellular tight junction [GO:0005923]; cell junction [GO:0030054]; cell leading edge [GO:0031252]; cell-cell junction [GO:0005911]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome [GO:0005768]; extracellular exosome [GO:0070062]; glutamatergic synapse [GO:0098978]; membrane [GO:0016020]; membrane raft [GO:0045121]; microtubule organizing center [GO:0005815]; myelin sheath abaxonal region [GO:0035748]; nuclear envelope [GO:0005635]; nuclear matrix [GO:0016363]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; Schaffer collateral - CA1 synapse [GO:0098685]; stress fiber [GO:0001725]; vesicle [GO:0031982]; 14-3-3 protein binding [GO:0071889]; ATP binding [GO:0005524]; calcium-dependent protein kinase C activity [GO:0004698]; insulin receptor substrate binding [GO:0043560]; metal ion binding [GO:0046872]; phospholipase binding [GO:0043274]; potassium channel regulator activity [GO:0015459]; protein kinase activity [GO:0004672]; protein kinase binding [GO:0019901]; protein kinase C activity [GO:0004697]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein-containing complex binding [GO:0044877]; activation of phospholipase D activity [GO:0031584]; activation of protein kinase B activity [GO:0032148]; cell migration [GO:0016477]; cell surface receptor signaling pathway [GO:0007166]; cellular response to insulin stimulus [GO:0032869]; establishment of cell polarity [GO:0030010]; inflammatory response [GO:0006954]; intracellular signal transduction [GO:0035556]; long-term memory [GO:0007616]; long-term synaptic potentiation [GO:0060291]; membrane depolarization [GO:0051899]; membrane hyperpolarization [GO:0060081]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of apoptotic process [GO:0043066]; negative regulation of hydrolase activity [GO:0051346]; negative regulation of insulin receptor signaling pathway [GO:0046627]; negative regulation of peptidyl-tyrosine phosphorylation [GO:0050732]; negative regulation of protein-containing complex assembly [GO:0031333]; neuron projection extension [GO:1990138]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cell-matrix adhesion [GO:0001954]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of excitatory postsynaptic potential [GO:2000463]; positive regulation of insulin receptor signaling pathway [GO:0046628]; positive regulation of interleukin-10 production [GO:0032733]; positive regulation of interleukin-13 production [GO:0032736]; positive regulation of interleukin-4 production [GO:0032753]; positive regulation of interleukin-5 production [GO:0032754]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of protein transport [GO:0051222]; positive regulation of T-helper 2 cell cytokine production [GO:2000553]; positive regulation of T-helper 2 cell differentiation [GO:0045630]; protein kinase C signaling [GO:0070528]; protein localization to plasma membrane [GO:0072659]; protein phosphorylation [GO:0006468]; regulation of neurotransmitter receptor localization to postsynaptic specialization membrane [GO:0098696]; signal transduction [GO:0007165]; vesicle transport along microtubule [GO:0047496] | 11740573_Isoforms of protein kinase C and their distribution in human adrenal cortex and tumors 11765038_Investigation of the inhibitory effects of chelerythrine chloride on the translocation of the protein kinase C betaI, betaII, zeta in human neutrophils 11960776_PKC-zeta is required in EGF protection of microtubules and intestinal barrier integrity against oxidant injury. 12021260_role of PKCzeta in T cells through the control of NFAT function by modulating the activity of its transactivation domain 12056906_PKC zeta expressed in human neutrophils can phosphorylate p47phox and induce both its translocation and NADPH oxidase activation as well as the binding of p47phox to the cytosolic fragment of p22phox. 12093536_REVIEW:Interaction of protein kinase C isozymes with membranes containing anionic phospholipids utilizing fluorescent phorbol esters to probe the properties of the C1 domains 12105221_shows for the first time that topoisomerase II beta is a substrate for PKC zeta, and that PKC zeta may significantly influence topoisomerase II inhibitor-induced cytotoxicity by altering topoisomerase II beta activity through its kinase function 12223351_EGF protects against oxidative disruption of the intestinal barrier by stabilizing the cytoskeleton in large part through the activation of PKC-zeta and downregulation of iNOS 12242277_The adapter protein ZIP binds Grb14 and regulates its inhibitory action on insulin signaling by recruiting protein kinase Czeta. 12244101_catalytic domain of PKC zeta is intrinsically inactive and dependent on the transphosphorylation of the activation lo 12435813_the role of PKCzeta in the negative regulation of drug-induced SM-CER pathway. 12493764_Protein kinase C zeta-GATA-2 signaling regulates VCAM1 stimulation by thrombin in endothelial cells 12671055_hypoxia decreases Na,K-ATPase activity in alveolar epithelial cells by triggering its endocytosis through mitochondrial reactive oxygen species and PKC-zeta-mediated phosphorylation of the Na,K-ATPase alpha(1) subunit 12748064_PKC zeta has a role in the actin organization in ET-1-stimulated cells may play a role in myometrial contraction in pregnant women. 12783114_The novel varepsilon and eta and atypical zeta, but not the conventional alpha and beta and the novel delta PKCs, may be involved in the signaling pathways involved in thrombin-induced human platelet P-selectin expression 12791393_PKCzeta has a role in regulating cell survival in tumor cells 12882907_Defective aPKC activation contributes to skeletal muscle insulin resistance in glucose intolerance and type 2 diabetes 12900386_enteropathogenic E coli infection of intestinal epithelial cells activates several signaling pathways including PKC zeta and ERK that lead to NF-kappa B activation, thus ensuring the proinflammatory response. 12905622_Observational study of gene-disease association. (HuGE Navigator) 12905768_Observational study of gene-disease association. (HuGE Navigator) 12920244_disregulation of the function of atypical zeta PKC might be involved in the acquisition of an invasive and metastatic phenotype in pancreatic adenocarcinoma cells. 12970910_PRKCZ gene may be associated with type 2 diabetes in Han population in North China. The haplotypes at five SNP sites in this gene may be responsible for this association. 14570876_protein kinase C iota/lambda binds to Rab 2, which inhibits aPKCiota/lambda-dependent GADPH phosphorylation [Protein kinase C lambda] 14744756_PKCzeta is responsible for the activation of HIF-alpha by inhibiting the mRNA expression of FIH-1 thus promoting the transcription of hypoxia-inducible genes. 15069075_phosphorylation of Ser(318) by PKC-zeta might contribute to the inhibitory effect of prolonged hyperinsulinemia on IRS-1 function. 15081397_identification of neuronal protein KIBRA as a novel substrate 15159477_There was expression of protein kinase C zeta in abnormal muscle fibers. Protein kinase C isoforms may play a role in the pathogenesis of myofibrillar myopathy. 15172966_involvement of PKC zeta in thrombin-induced permeability changes in human umbilical vein endothelial cells 15210811_GTPase RhoA and atypical protein kinase Czeta are required for TLR2-mediated gene transcription. 15254234_PKCzeta activity is regulated by nucleotide exchange factor ECT2 15285019_Single nucleotide polymorphisms in protein kinase Cz (PKCZ) gene probably play a role in the susceptibility to type 2 diabetes by affecting the expression level of the relevant genes. 15313379_PKC-zeta, is critical to regulation of LPS-induced TLR4 lipid raft mobilization within macrophages. 15358551_PKCzeta could act as a modulator of nucleotide excision repair activity by regulating the expression of XPC/hHR23B heterodimer 15362041_The differential regulation of protein kinase C-zeta by enteropathogenic E. coli and enterohemorrhagic E. coli may in part explain the less profound effect of the latter on the barrier function of tight junctions. 15499829_We analyzed the dependence of the expression of some selected protein kinase C isoenzymes on the availability and/or action of androgens. 15544481_Disruption of the cellular environment through genetic mutation, disease, injury, or exposure to pro-oxidants, alcohol, or other insults can induce pathological PKC activation. 15604116_A role for PKCzeta in relaxin-mediated stimulation of cAMP was demonstrated. 15630457_stromal cell-derived factor 1 triggered PKC-zeta phosphorylation, translocation to the plasma membrane, and kinase activity 15808853_Results indicate that protein kinase C (PKC) zeta modulates hMutS alpha (MSH2, MSH6)stability and protein levels, and suggest a role for PKC zeta in genome stability by regulating mismatch repair activity. 15870274_Ajuba is a new cytosolic component of the IL-1 signaling pathway modulating IL-1-induced NF-kappaB activation by influencing the assembly and activity of the aPKC/p62/TRAF6 multiprotein signaling complex 15887250_PKCZ mediates opposing effects on Na(+)-K(+)-Exchanging ATPase activity in a breast neoplasm cell line. 15935276_data support the notion that PKCzeta is essential for LPS-induced NF-kB p65 subunit nuclear translocation in human myometrial cells 15956717_described a novel bifurcated pathway by which relaxin stimulates Gs alpha and 1-Phosphatidylinositol 3-Kinase /PKCzeta leading to increased cyclic AMP production 16011831_In this process p62-dependent Akt phosphorylation occurred via the release of Akt from PKCzeta by association of p62 and PKCzeta, which is known as a negative regulator of Akt activation. 16287866_phosphorylation and inhibition of caspase 9 by PKCzeta restrain the intrinsic apoptotic pathway during hyperosmotic stress 16407220_PKC zeta-dependent AMP-activated protein kinase (AMPK) activation may play an important role in regulating not only cellular energy metabolism but also signaling pathways that control cell growth, differentiation, and survival. 16611744_Myosin II-B resides in a complex with p21-activated kinase 1 (PAK1) and atypical protein kinase C (PKC) zeta (aPKCzeta) and the interaction between these proteins is EGF-dependent. 16644736_atypical PKCs are required for insulin-stimulated glucose transport in myocytes and adipocytes 16798739_PKCzeta is a necessary component of the IL-1 and TNF signaling pathways in chondrocytes that result in catabolic destruction of extracellular matrix proteins in osteoarthritic cartilage 16931574_Protein kinase Czeta/mammalian target of rapamycin/S6 kinase pathway plays an important role for the transition of androgen-dependent to androgen-independent prostate cancer cells. 16940160_Studies indicate that PKC-zeta inhibits colon cancer cell growth and enhances differentiation and apoptosis, while inhibiting the transformed phenotype of these cells. 16943418_These studies have revealed a novel mechanism of Trichostatin A action through derecruitment of a repressor from the Luteinizing hormone receptor gene promoter in a PI3K/PKCzeta-induced Sp1 phosphorylation-dependent manner. 17024099_the primary signaling pathway for LPC-dependent dendrite formation in human melanocytes involves the activation of PKCzeta and PKCzeta phosphorylation is Rac dependent 17313651_activated form phosphorylates VAMP2 in vitro 17346701_We propose that increased water influx through AQP9 is critically involved in the formation of membrane protrusions, and that AQP9-induced actin polymerization is augmented by activation of Cdc42 and PKC(zeta). 17369850_aPKCzeta cortical loading is associated with Lgl cytoplasmic release and tumor growth in Drosophila and human epithelia 17389234_In tumor cells, protein kinase C (PKC) zeta was activated to phosphorylate RhoGDI-1, which liberated RhoGTPases, leading to their activation. 17398070_MAP3K8 and PRKCZ cooperate in the regulation of the transcriptional activity of NFATC2 through the phosphorylation of its amino-terminal domain. 17525161_PKCzeta may function as a physiological Bax kinase to directly phosphorylate and interact with Bax, which leads to sequestration of Bax in cytoplasm and abrogation of the proapoptotic function of Bax 17541951_In osteoblasts of rheumatoid arthritis & osteoarthritis patients, PKC-zeta decreased when compared with normal cells. Treatment with IL-1beta & TNF-alpha significantly decreased PKC-zeta expression in post-traumatic osteoblasts. 17544492_first study to show that allergic disease is associated with altered expression of T-cell PKC isozymes in the neonatal period 17682059_Data show that in renal cell carcinoma, the Cut-like homeodomain protein is involved in FIH-1 transcriptional regulation and is controlled by a specific signaling event involving protein kinase C zeta. 17786270_Hemoglobin promptly and markedly modified the levels of expression of both calcium-dependent PKCalpha and calcium-independent PKCzeta. 17850789_These data suggest that PKCzeta activation plays a predominant role in regulation of PGE(2)-dependent melanocyte dendricity. 17850925_In peripheral T-cells from severe cases of Alzheimer's patients, Abeta(1-42) treatment stimulates two distinct PKC-delta- and PKC-zeta-producing T-cell subpopulations which are not noticeable in healthy adult and elderly subjects. 18050214_PKCzeta is involved in regulation of IL-1beta-induced NF-kappaB signaling in human osteoarthritis chondrocytes, which regulates downstream expression of ADAMTS-4 and NOS2 via NF-kappaB. 18067888_Data suggest that foreign body giant cell formation is supported by PKCbeta, PKCdelta, and PKCzeta in combined diacylglycerol-dependent (PKCbeta and PKCdelta) and -independent (PKCzeta) signaling pathways. 18094075_PKCzeta is the predominant isoform mediating stretch-induced COX-2 expression 18166155_PKC zeta, but not PKCalpha, positively regulates Casp-2S mRNA assembly triggered by topoisomerase inhibitors 18296444_Protein kinase C-epsilon regulates sphingosine 1-phosphate-mediated migration of human lung endothelial cells through activation of phospholipase D2, protein kinase C-zeta, and Rac1 18313384_Results show that protein kinase C zeta phosphorylates and specifically stabilizes SRC-3 in a selective ER-dependent manner. 18319301_Data identify PKCzeta as regulators of EC lumenogenesis in 3D collagen matrices. 18343365_Spisulosine inhibits the growth of the prostate tumor cells through intracellular ceramide accumulation and PKCzeta activation. 18354190_modulation of PKC may have therapeutic potential in the prevention of smoke-related lung injury 18385757_PKCzeta is an important transduction molecule downstream of TNF-alpha signaling and is associated with increased expression of CD1d that may enhance CD1d-natural killer T cell interactions in psoriasis lesions. 18571841_Atypical protein kinase C (PKC-iota/-zeta) phosphorylates IKKalphabeta in transformed non-malignant and malignant prostate cell survival. 18615853_Observational study of gene-disease association. (HuGE Navigator) 18615853_PRKCZ gene variants associated with the development of type 2 diabetes in this study needs to be investigated in a larger population to reveal any potential effects on metabolism. 18668687_FIC1 signals to FXR via PKC zeta. FIC1-related liver disease is likely related to downstream effects of FXR on bile acid homeostasis. 18760839_These results show a new function of HIV-1 Tat, its ability to regulate CXCR4 expression via PKCzeta. 18989463_role for host PKCsigma (PKCzeta) on the invasion of hepatocytes by sporozoites 19033536_PKC-zeta dependent stimulation of the human MCT1 promoter involves transcription factor AP2. 19061073_These results indicate that insulin induces dynamic associations between PKCzeta, 80K-H, and munc18c and that 80K-H may act as a key signaling link between PKCzeta and munc18c in live cells. 19086264_the role of protein kinase C zeta (PKCzeta) in interleukin (IL)-8-mediated activation of Mac-1 (CD11b/CD18) in human neutrophils 19164129_crucial implication of PKC-delta, PKC-zeta, ERK-1/2, and p38 in human blood eosinophil migration through extracellular matrix components 19187446_Protein kinase C zeta controls glioblastoma cell migration and invasion by regulating the cytoskeleton rearrangement. 19201988_PKCzeta is required for CSF-1-induced chemotaxis of macrophages. 19304661_There is a novel ceramide binding domain (C20zeta) in the carboxyl terminus of aPKC. 19339512_Enhancement of calcium transport in Caco-2 monolayer through PKCzeta-dependent Cav1.3-mediated transcellular and rectifying paracellular pathways by prolactin. 19380829_Atypical protein kinase C isozyme PKCzeta associates functionally with small GTPase RhoA and acts as a signaling component downstream of RhoA. 19460843_Serine proteases decrease intestinal epithelial ion permeability by activation of protein kinase Czeta. 19502590_Data show that cholesteryl ester formation was dependent on protein kinase zeta/ extracellular signal-related kinase 1/2 (PKCzeta/ERK1/2) activation. 19751727_These results strongly suggest that DGKalpha positively regulates TNF-alpha-dependent NF-kappaB activation via the PKCzeta-mediated Ser311 phosphorylation of p65/RelA. 19809379_Findings support the hypothesis that hepatocyte FIC1 enhances FXR signaling via a PKCzeta-dependent signaling pathway. 19812540_Patients who had an elevated anti-PKCzeta titer developed kidney allograft rejection, which was significantly more likely to result in graft loss. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20236250_These results suggest that beta-catenin play an inhibitory role for dendrite formation through the modulation of PKCzeta and PKCdelta. 20236512_RHOA and PRKCZ, and their downstream effectors, can represent important pharmacological targets that could potentially control the highly metastatic attitude of pancreatic ductal adenocarcinoma. 20407013_Protein kinase C(PKC)zeta is a novel mitogenic downstream mediator of epidermal growth factor (EGF) receptor, and a therapeutic target as well, in some carcinomas. 20416077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20417648_Inhibition of PKCzeta may be a novel drug target for thrombin-induced inflammatory hyperpermeability. 20463008_These data identify atypical PKC isozymes as STAT and ERK activators that mediate c-fos and collagenase expression. 20501645_PKC zeta is differentially expressed in ovarian carcinomas and is phosphorylated in response to apoptotic stimuli in primary ovarian carcinoma cells. 20538799_Results show that PKCzeta binds and phosphorylates ERK5, thereby decreasing eNOS protein stability and contributing to early events of atherosclerosis. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20679531_PKC zeta function in T helper cells is required for directional secretion of CD40 ligand (CD40L) and interferon (IFN)-gamma toward dendritic cells. 20844151_low levels of expression are associated with poorly differentiated tumours and a poor outcome in breast cancer patients 20941506_aPKC-zeta had moderate sensitivity as a marker of gastric dysplasia and additional studies are needed to establish its role in the diagnosis of dysplasia. 20949042_found that protein kinase C (PKC) zeta binds and phosphorylates LRRK2 both in vitro and in vivo 21081127_Results show that both PKC epsilon and PKC zeta isoforms are involved in the formation of ROS by C2-ceramide and the opening of the mPTP. 21390241_The prognostic impact of TGF-beta1, NF-kappaB p105, PKC-zeta, Par-6alpha, E-cadherin and vimentin in non-gastrointestinal stromal tumor soft tissue sarcomas, was investigated. 21454526_that upon DAMGO treatment, MOR activates PKCzeta through a PDK1-dependent signaling pathway to induce CCR5 phosphorylation and desensitization. 21549621_Data indicate that both tumor focality and Par3/Par6/atypical protein kinase C (APKC) expression were significantly associated with tumor recurrence. 21619587_Results indicate that phosphorylation of human DNMT1 by protein kinase C is isoform-specific and provides the first evidence of cooperation between PKCzeta and DNMT1 in the control of the DNA methylation patterns of the genome. 21624955_The PKCzeta activation by d-flow induces endothelial cell (EC) apoptosis by regulating p53. 21645497_human platelets express PKCzeta, and it may be constitutively phosphorylated at the activation loop threonine 410 and the turn motif threonine 560 under basal resting conditions, which are differentially dephosphorylated by outside-in signaling 21667320_Changes in PKC iota, PKC zeta, and non-muscle MyoIIA expression are likely to participate in pathogenesis of epithelial barrier function in response to local pro-inflammatory signals. 21780947_two key (hub) PPARgamma direct target genes, PRKCZ and PGK1, were experimentally validated to be repressed upon PPARgamma activation by its natural ligand, 15d-PGJ2 in three prostrate cancer cell lines 21849550_Inhibition of protein kinase M zeta results in a reduction of synaptic PSD-95 accumulation in developing visual cortex 21871968_The LNO(2) mediated signaling in lung type II epithelial cells occurs via a unique pathway involving PKCzeta. 21895402_High levels of protein kinase C zeta expression were associated with lymphatic metastasis in squamous cervical cancer. 22231931_Single nucleotide polymorphisms in protein kinase C zeta are associated with bipolar affective disorder. 22242160_HGF induced functional CXCR4 receptor expression in breast cancer cells. The effect of HGF was specifically mediated by PKCzeta activity. 22324796_Report role of PKC-zeta induction in bronchial inflammation and airyway hyperresponsiveness. 22390153_Western Blot data showed decreased expression (p < 0,05) of Munc18c and phospho-PKC Zeta in polycystic ovary-insulin resistant endometria (PCOSE-IR) with respect to the control. 22475628_Protein kinase C (PKC) zeta expression was significantly higher in normal than in cancerous tissues. Similarly, PKC zeta expression was down-regulated in four renal cancer cell lines compared to immortalized benign renal tubular cells. 22475757_SZ95 sebocytes express the conventional cPKCalpha; the novel nPKCdelta, epsilon, and eta; and the atypical aPKCzeta 22644296_A novel sequence was identified within the 3'-terminal domain of human PRKCZ. 22740332_It was concluded that protein kinase C zeta regulated protein kinase phosphorylation, which in turn regulated the proteolytic activity of phorbol dibutyrate-induced podosomes by influencing the recruitment of protein kinase C zeta and MMP9 to podosomes. 22750245_these findings suggest that PKC-zeta is involved in the phosphorylation of HMGB1, and the phosphorylation of specific serine residues in the nuclear localization signal regions is related to enhanced HMGB1 secretion in colon cancer cells. 22791907_Stat3 forms a multiprotein complex with Rac1 and PKC in an hypoxia-reoxygenation-dependent manner. 22812606_A proapoptotic role for protein kinase C zeta in the binding and phosphorylating Bcl10 at the nuclear envelope. 23004934_The findings suggest a potential role for the use of PKCzeta levels in cord blood T cells as a presymptomatic test to predict allergy risk in children. 23266528_Results indicate the importance of p62-associated PKCzeta in the overactive state of pagetic osteoclasts (OCs) and in the activation of NF-kappaB, particularly in the presence of the p62(P392L) mutation. 23349801_Data indicate that the phosphorylation of GSK-3beta was reduced by siRNA of PKCdelta, PKCepsilon, and PKCzeta. 23374352_Study reports that PKCzeta-deficient cells reprogram their metabolism for the utilization of glutamine instead of glucose through the serine biosynthetic cascade controlled by 3-phosphoglycerate dehydrogenase (PHGDH). 23868949_the PKC family genes may play a role in the pathogenesis of MS relapse through modulating the association between 25(OH)D and relapse. 24015205_STAT3 is an important downstream mediator of the pro-carcinogenic effects of PRKCZ in pancreatic cancer cells. 24447338_These data indicate for the first time that HIV-1 Gag phosphorylation on Ser487 is mediated by atypical PKC and that this kinase may regulate the incorporation of Vpr into HIV-1 virions and thereby supports virus infectivity. 24753582_Report up-regulation of MT1-MMP and atypical protein kinase C in hormone receptor-negative breast tumors in association with a higher risk of metastasis. Silencing of aPKC impaired delivery of MT1-MMP from storage compartments and inhibited invasion. 24786829_The results indicate that induction and activation of PKCzeta promote TNBC growth, invasion and metastasis. 24920238_Results indicate that PKCzeta regulates survivin expression levels and inhibits apoptosis in colon cancer cells. 24990612_PKCzeta and PKMzeta are overexpressed in TCF3-rearranged paediatric acute lymphoblastic leukaemia and may have a role in thiopurine sensitivity 25070947_Neuronal NF1/RAS regulation of cyclic AMP requires atypical PKC zeta activation, which is perturbed in neurofibromatosis type 1. 25075435_PRKCZ methylation is associated with sunlight exposure 25817572_PKCzeta inhibition prevented alternative cleavage and release of TROP2, suggesting that these events require endocytic uptake and exosomal release of the corresponding microvesicles. 25874946_Over-expression of PRKCZ results in gene and/or protein expression alterations of insulin-like growth factor 1 receptor (IGF1R) and integrin beta 3 (ITGB3) in SKOV3 and OVCAR3 cells. 26187466_Data show that aPKC scaffold protein p62 tethers Atypical protein kinase C (aPKC) in an active conformation. 26711256_The PKC-zeta - induced phosphorylation of GSK-3 beta stimulates GSK-3 beta activity. 26887939_data suggest that the interaction between this novel region in Galphaq and the effector PKCzeta is a key event in Galphaq signaling. 27040869_Study provides evidence for a novel PKC-zeta to p47phox interaction that is required for cell transformation from blebbishields and ROS production in cancer cells. 27143478_FRET-based translocation assays reveal that insulin promotes the association of both p62 and aPKC with the insulin-regulated scaffold IRS-1. 27490967_Drug discovery efforts have been hindered due to the non-availability of the protein structure and hence in the present study we attempted to build the open and closed models of the protein PKMzeta using homology modeling. 27516147_Here we provide the first evidence that PKC-zeta is a potential target for the treatment of COPD by selective small molecules 27911275_these results conclude that miR-25 targets PKCzeta and protects osteoblastic cells from Dex via activating AMPK signaling. 28159873_The data demonstrate that PKCzeta expression regulates the maturation of neonatal T-cells into specific functional phenotypes and that environmental influences may work via PKCzeta to regulate these phenotypes and disease susceptibility. 28366812_we found that Wnt3a treatment rapidly induces hyperphosphorylation and stabilization of Dvl2 and Dvl3. Our findings suggest a model of positive regulation of PKCzeta-mediated Dvl signaling activity, to produce a strong and sustained response to Wnt3a treatment by stabilizing Dvl protein levels. 28476658_Protein kinase C acts as a tumor suppressor.Cancer-associated mutations in protein kinase C are generally loss-of-function mutations.[review] 28515165_This study demonstrated that zinc upregulates PKCzeta by activating GPR39 to enhance the abundance of ZO-1, thereby improving epithelial integrity in S. typhimurium-infected Caco-2 cells. 28518146_PKCzeta was specifically involved in ACOT7 depletion-mediated cell cycle arrest as an upstream molecule of the p53-p21 signaling pathway in MCF7 human breast carcinoma and A549 human lung carcinoma cells. 28631559_Inhibition of protein kinase C zeta expression in prostate cancer cells promoted chemotaxis of peripheral macrophages and acquisition of M2 phenotypic features. These results were further supported by the finding that silencing of endogenous protein kinase C zeta promoted the expression of prostate cancer cell-derived interleukin-4 and interleukin-10 28652146_reduced expression of PKCzeta/Pard3/Pard6 contributes to non-small-cell lung cancer epithelial-mesenchymal transition, invasion, and chemoresistance. 28726782_Intestinal I/R induced the membrane translocation and phosphorylation of PKCzeta. Pretreatment with the PKCzeta activator phosphatidylcholine remarkably attenuated gut injury by suppressing apoptosis. H/R induced PKCzeta to combine with TRAF2, which was phosphorylated by PKCzeta at Ser(55), but not at Ser(11), under intestinal I/R or H/R conditions 28983601_PKCzeta promoted lung adenocarcinoma invasion and metastasis, and its expression was associated with MMP2 and MMP9 expression. 29048609_We also carried out PKC-iota and PKC-zeta directed siRNA treatments to prove the above observations. Immunoprecipitation data suggested an association between PKC-iota and vimentin and PKC-iota siRNA treatments confirmed that PKC-iota activates vimentin by phosphorylation. These results further suggested that PKC-iota is involved in signaling pathways which upregulate EMT and which can be effectively suppressed using A... 29408512_PKC-zeta may be responsible for the abnormal growth, proliferation, and migration of metastatic LOVO colon cancer cells via PKC-zeta/Rac1/Pak1/beta-Catenin pathway. 29520890_Defective PKCz control of interphase centrosome anchoring may underlie distinct categories of mitotic slippage that shape the development of low- or high-grade colorectal cancer phenotypes. 30221381_CCAT1 knockdown promoted M2 macrophage polarization by up-regulation of miR-148a and down-regulation of PKCzeta expression in prostate cancer cells. 30504065_Our findings demonstrate the mechanism of PKCzeta as a new phosphorylase of SIRT6 on maintaining tumor fatty acid beta-oxidation and define the new role of PKCzeta in lipid homeostasis. 30552022_Atypical protein kinase C zeta and protein kinase C lambda/iota expression are reduced in human serrated tumors. 30600259_Intracellular sphingosine 1-phosphate (S1P) directly bound to the purified kinase domain of atypical protein kinase C (aPKC) isozymes (aPKC) and relieved autoinhibitory constraints, thereby activating the kinase. 30940726_Indomethacin impairs mitochondrial dynamics by activating the PKCzeta-p38-DRP1 pathway and inducing apoptosis in gastric cancer and normal mucosal cells. 32330121_Mitochondrial nucleoid remodeling and biogenesis are regulated by the p53-p21(WAF1)-PKCzeta pathway in p16(INK4a)-silenced cells. 32580209_A polybasic domain in aPKC mediates Par6-dependent control of membrane targeting and kinase activity. 33444686_Bone metastatic breast cancer cells display downregulation of PKC-zeta with enhanced glutamine metabolism. 34063174_Cord Blood T Cells Expressing High and Low PKCzeta Levels Develop into Cells with a Propensity to Display Th1 and Th9 Cytokine Profiles, Respectively. 35017658_Methylation-mediated silencing of protein kinase C zeta induces apoptosis avoidance through ATM/CHK2 inactivation in dedifferentiated chondrosarcoma. 35124330_Human placenta and trophoblasts simultaneously express three isoforms of atypical protein kinase-c. | ENSMUSG00000029053 | Prkcz | 860.786267 | 0.8559362 | -0.224424809 | 0.09834690 | 5.205889e+00 | 2.251050e-02 | 1.981478e-01 | No | Yes | 778.165272 | 66.899523 | 906.332739 | 77.719732 | |
| ENSG00000067955 | 865 | CBFB | protein_coding | Q13951 | FUNCTION: Forms the heterodimeric complex core-binding factor (CBF) with RUNX family proteins (RUNX1, RUNX2, and RUNX3). RUNX members modulate the transcription of their target genes through recognizing the core consensus binding sequence 5'-TGTGGT-3', or very rarely, 5'-TGCGGT-3', within their regulatory regions via their runt domain, while CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX. The heterodimers bind to the core site of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, LCK, IL3 and GM-CSF promoters. CBF complexes repress ZBTB7B transcription factor during cytotoxic (CD8+) T cell development. They bind to RUNX-binding sequence within the ZBTB7B locus acting as transcriptional silencer and allowing for cytotoxic T cell differentiation. {ECO:0000250|UniProtKB:Q08024}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome | The protein encoded by this gene is the beta subunit of a heterodimeric core-binding transcription factor belonging to the PEBP2/CBF transcription factor family which master-regulates a host of genes specific to hematopoiesis (e.g., RUNX1) and osteogenesis (e.g., RUNX2). The beta subunit is a non-DNA binding regulatory subunit; it allosterically enhances DNA binding by alpha subunit as the complex binds to the core site of various enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers and GM-CSF promoters. Alternative splicing generates two mRNA variants, each encoding a distinct carboxyl terminus. In some cases, a pericentric inversion of chromosome 16 [inv(16)(p13q22)] produces a chimeric transcript consisting of the N terminus of core-binding factor beta in a fusion with the C-terminal portion of the smooth muscle myosin heavy chain 11. This chromosomal rearrangement is associated with acute myeloid leukemia of the M4Eo subtype. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:865; | core-binding factor complex [GO:0016513]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; sequence-specific DNA binding [GO:0043565]; transcription coactivator activity [GO:0003713]; cell maturation [GO:0048469]; definitive hemopoiesis [GO:0060216]; lymphocyte differentiation [GO:0030098]; myeloid cell differentiation [GO:0030099]; negative regulation of CD4-positive, alpha-beta T cell differentiation [GO:0043371]; negative regulation of transcription by RNA polymerase II [GO:0000122]; osteoblast differentiation [GO:0001649]; positive regulation of CD8-positive, alpha-beta T cell differentiation [GO:0043378]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein polyubiquitination [GO:0000209]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription by RNA polymerase II [GO:0006366] | 12239155_has a role in hematopoiesis; preturbations result from expression of the leukemogenic fusion gene Cbfb-MYH11 12495904_review: genetics of CBFB and RUNX1 and roles in hematopoiesis and leukemogenesis, with emphasis on human and knockout mice studies 15386419_Expression of CBFB is down regulated in a significant portion of gastric cancer cases; may be involved in gastric carcinogenesis 15585652_Plag1 and Plagl2 are novel leukemia oncogenes that act by expanding hematopoietic progenitors expressing CbF beta-SMMHC. 16502584_Detection of acute myeloid leukemic cells that are characterized by a CBFB-MYH11 gene fusion. 16504290_These observations suggest that when abdominal GS is diagnosed, an analysis of the CBFB/MYH11 fusion gene is necessary to make an appropriate decision regarding treatment options, even if no chromosomal abnormalities are found. 16767164_Agents interacting with the outer surface of the CBFbeta-SMMHC ACD that prevent multimerization may be effective as novel therapeutics in AML 17287858_Rare fusion transcripts were correlated with an atypical cytomorphology not primarily suggestive for the FAB subtype acute myelocytic leukemia. 17379770_Cbfb enhances osteogenic differentiation of mesenchymal stem cells by stabilizing Cbfa-1 proteins. 17571080_Examine consequences of expression of abnormal chimeric protein CBFbeta-MYH11 in acute myelomonocytic leukemia. 18695000_interaction with PEBP2-beta leads to the phosphorylation of RUNX1, which in turn induces p300 phosphorylation 19156145_high CBFB protein level was an independent predictor of survival in colorectal cancer 19179469_CBFbeta is essential for TEL-AML1's ability to promote self-renewal of B cell precursors in vitro. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19603346_For routine clinical practice, it may be meaningful to screen for C-KIT mutations in AML1/ETO-positive patients, as well as for FLT3(D835) mutations in CBF-AML. 20225274_Loss of DNA binding, but not nuclear localization or CBF-beta heterodimerization, causes RUNX2 haploinsufficiency in patients with the RUNX2(R131G) mutation. 20306249_The structural features of RUNX1/CBFbeta and their derivatives, their roles in transcriptional control, and their contributions to normal and malignant hematopoiesis are discussed. Review. 20306685_Studies show that FISH technic for the detection of CBF chromosomal aberrations was significantly higher than conventional cytogenetics. 20433876_The expression of Cbfbeta which were the key factors in osteogenic differentiation was also up-regulated. 20591170_conclude that CBFbeta is required for a subset of Runx2-target genes that are sufficient to maintain the invasive phenotype of the cells 20607802_Data collectively suggest that CBFbeta is required for malignant phenotype in prostate and ovarian cancer cells. 22190036_Vif and CBF-beta physically interact, and that the amino-terminal region of Vif is required for this interaction 22190037_CBF-beta is required for Vif-mediated degradation of APOBEC3G and therefore for preserving HIV-1 infectivity 22205746_Vif proteins of human and simian immunodeficiency viruses require cellular CBFbeta to degrade APOBEC3G. 22725134_These separation-of-function mutants demonstrate that HIV-1 Vif and the RUNX transcription factors interact with cellular CBFbeta on genetically distinct surfaces. 23098073_A comparison of heat capacity changes supports a model in which CBFbeta prestabilizes Vif((1-192)) relative to Vif((95-192)) 23152542_Our data indicate that the CBFbeta-SMMHC's C-terminus is essential to induce embryonic hematopoietic defects and leukemogenesis. 23160462_We conclude that non-type A CBFB-MYH11 fusion types associate with distinct clinical and genetic features, including lack of KIT mutations, and a unique gene-expression profile in acute myeloid leukemia 23175372_Authors revealed that different lengths and regions are required for CBFbeta to assist HIV-1 Vif or RUNX1. 23878140_This report of recurring FLT3 N676 mutations in core-binding factor (CBF) leukemias suggests a defined subgroup of CBF leukemias. 24002588_Transcriptional analysis revealed that upon fusion protein knockdown, a small subset of the CBFbeta-MYH11 target genes show increased expression, confirming a role in transcriptional repression 24390320_In the absence of CBFbeta, Vif does not bind Cul5, thus preventing the assembly of the E3 ligase complex. 24390335_CBF-beta is critical for the formation of the Vif-ElonginB/ElonginC-Cul5 core E3 ubiquitin ligase complex. 24402281_data reveal the structural basis for Vif hijacking of the CBF-beta and CUL5 E3 ligase complex, laying a foundation for rational design of novel anti-HIV drugs 24418540_Vif conserved residues E88/W89 are crucial for CBFbeta binding. 24522927_Authors propose that CBFbeta acts as a chaperone to stabilize HIV-1 Vif during and after synthesis and to facilitate interaction of Vif with cellular cofactors required for the efficient degradation of APOBEC3G. 24648201_Suggest that CBFbeta retention in the midbody during cytokinesis reflects a novel function that contributes to epigenetic control. 24651404_Our findings indicate that RUNX1 and CBF-beta cooperate in cells to modulate HIV-1 replication 25079347_CBFB contributes to the transcriptional regulation of ribosomal gene expression and provide further understanding of the epigenetic role of CBFB-SMMHC in proliferation and maintenance of the leukemic phenotype. 25122780_suggest that a different mechanism exists for the Vif-APOBEC interaction and that non-primates are not suitable animal models for exploring pharmacological interventions that disrupt Vif-CBF-beta interaction 25266220_we report a novel hypomethylation pattern, specific to CBFB-MYH11 fusion resulting from inv(16) rearrangement in acute myeloid leukemia the expression of which correlated with PBX3 differential methylation 25424878_These results provide important information on the assembly of the Vif-CUL5-E3 ubiquitin ligase and identify a new viV binding interface with CBF-beta at the C-terminus of HIV-1 Vif. 25582776_CBF-beta promoted steady-state levels of HIV-1 Vif by inhibiting the degradation of HIV-1 Vif through the proteasome pathway. 26163765_Our findings demonstrate that HSPCs exposed to non-cytotoxic levels of environmental chemicals and chemotherapeutic agents are prone to topoisomerase II-mediated DNA damage at the leukemia-associated genes MLL and CBFB. 26418744_Thus, an NGF/TrkA-MAPK-CBFbeta pathway converges with Islet1-Runx1 signaling to promote Runx1/CBFbeta holocomplex formation and nonpeptidergic nociceptor maturation. 27650511_the presented study demonstrates that CBFB-MYH11-based MRD status during the first 3 months after allo-HCT, but not KIT mutations, can be used to identify patients with a high risk of relapse. 27758855_Vif stabilization by CBFbeta is mainly caused by impairing MDM2-mediated degradation. 28196984_results suggest that CBFbeta-SMMHC has complex actions on human ribosome biogenesis at both the genomic and posttranscriptional level 28253536_The co-existence of BCR-ABL1 and CBFB rearrangement is associated with poor outcome and a clinical course similar to that of CML-BP, and unlike de novo AML with CBFB rearrangement, suggesting that high-intensity chemotherapy with TKI should be considered in these patients. 28299663_discussion of the role of CBFB in diseases caused by their mutations or deletions (review) 28302150_Mutational analysis of CBFbeta revealed that F68 and I55 residues are important and participate in a tripartite hydrophobic interaction with W5 of Vif to maintain a stable and functional Vif-CBFbeta complex. 28506695_Both c-kit receptor (KIT) D816V and KIT N822K mutations underwent autophosphorylation in the absence of growth factor in leukemia TF-1 cell line. 28516844_Moreover, using a CBF-beta loss-of-function mutant, the authors demonstrated that the interaction between CBF-beta and Vif was not sufficient for Vif assistance; a region including F68 in CBF-beta was also required for the stability and function of Vif. 29192243_our present study strongly suggests that an autonomous RUNX1-p53-CBFB regulatory triangle plays a vital role in the maintenance and the acquisition of chemo-resistance of AML cells, and potentially provides novel therapeutic targets for anti-leukemia strategy. 29386218_we identified a novel gastric cancer-associated lncRNA-LINC01234 and first uncover that LINC01234 is an oncogenic lncRNA that promotes cell proliferation and inhibits cell apoptosis through miR-204-5p-CBFB axis in human gastric cancer. 29523836_Type-III IFN-triggered and IL-10-induced CBFbeta are crucial factors for inhibiting Hepatitis B virus (HBV) replication, and the HBx-CBFbeta-HBsAg axis reveals a new molecular mechanism of interaction between HBV and its hosts. 29902450_This study investigated the role of circ-CBFB in chronic lymphocytic leukemia. The ID of circ-CBFB in circBase is hsa_circ_0000707, which locates at chromosome 16q22.1 and derived from the back-splicing of CBFB transcript. 29932212_All de novo and relapsed AMLs with CSF3R mutations were associated with genetic alterations in transcription factors, including RUNX1-RUNX1T1, CBFB-MYH11, double-mutated CCAAT/enhancer binding protein alpha (CEBPAdm), and NPM1 mutations; and core-binding factor gene abnormalities and CEBPAdm accounted for 90.5% (19 of 21 patients) 30851937_CBFB is essential for HIV-1 Vif to arrest host cells at G2/M phase. Vif-CBFB interaction is required for Vif-triggered cell cycle regulation. 31061501_Study found that CBFB binds to and enhances the translation of RUNX1 mRNA, which encodes the binding partner of CBFB. CBFB binds and regulates the translation of hundreds of mRNAs through hnRNPK and facilitate translation initiation by eIF4B. Data propose that breast cancer cells evade translation and transcription surveillance simultaneously through downregulating CBFB. 31610798_Transcriptional activation of CBFbeta by CDK11(p110) is necessary to promote osteosarcoma cell proliferation 31792451_Vif and CBFbeta form a platform to recruit A3F, revealing a direct A3F-recruiting role of CBFbeta beyond Vif stabilization. 31941780_Inhibition of Vif-Mediated Degradation of APOBEC3G through Competitive Binding of Core-Binding Factor Beta. 32005976_A role for CBFbeta in maintaining the metastatic phenotype of breast cancer cells. 32124467_Platelet CD34 expression in a patient with a partial deletion of transcription factor subunit CBFB. 32711101_The transcription factor CBFB mutations indicate an improved survival in HR+/HER2- breast cancer. 32893454_Multifaceted HIV-1 Vif interactions with human E3 ubiquitin ligase and APOBEC3s. 32926565_Core-binding factor acute myeloid leukemia with inv(16): Older age and high white blood cell count are risk factors for treatment failure. 33945523_CBFB cooperates with p53 to maintain TAp73 expression and suppress breast cancer. 34010414_Core-binding factor leukemia hijacks the T-cell-prone PU.1 antisense promoter. 34027799_Circular RNA_0062582 promotes osteogenic differentiation of human bone marrow mesenchymal stem cells via regulation of microRNA-145/CBFB axis. 34117074_Incidental identification of inv(16)(p13.1q22)/CBFB-MYH11 variant transcript in a patient with therapy-related acute myeloid leukemia by routine leukemia translocation panel screen: implications for diagnosis and therapy. 34739701_Non-age-related neoplastic loss of sex chromosome correlated with prolonged survival in real-world CBF-AML patients. 35128634_TET2 deficiency cooperates with CBFB-MYH11 to induce acute myeloid leukaemia and represents an early leukaemogenic event. 35184217_3'CBFB deletion in CBFB-rearranged acute myeloid leukemia retains morphological features associated with inv(16), but patients have higher risk of relapse and may require stem cell transplant. | ENSMUSG00000031885 | Cbfb | 2724.592254 | 1.0270976 | 0.038573242 | 0.06818096 | 3.175471e-01 | 5.730855e-01 | 8.417818e-01 | No | Yes | 2808.407281 | 579.495893 | 2679.964255 | 552.971690 | |
| ENSG00000068400 | 56850 | GRIPAP1 | protein_coding | Q4V328 | FUNCTION: Regulates the endosomal recycling back to the neuronal plasma membrane, possibly by connecting early and late recycling endosomal domains and promoting segregation of recycling endosomes from early endosomal membranes. Involved in the localization of recycling endosomes to dendritic spines, thereby playing a role in the maintenance of dendritic spine morphology. Required for the activity-induced AMPA receptor recycling to dendrite membranes and for long-term potentiation and synaptic plasticity (By similarity). {ECO:0000250|UniProtKB:Q9JHZ4}.; FUNCTION: [GRASP-1 C-terminal chain]: Functions as a scaffold protein to facilitate MAP3K1/MEKK1-mediated activation of the JNK1 kinase by phosphorylation, possibly by bringing MAP3K1/MEKK1 and JNK1 in close proximity. {ECO:0000269|PubMed:17761173}. | Acetylation;Alternative splicing;Cell junction;Cell projection;Coiled coil;Direct protein sequencing;Endosome;Membrane;Phosphoprotein;Protein transport;Reference proteome;Synapse;Transport | This gene encodes a guanine nucleotide exchange factor for the Ras family of small G proteins (RasGEF). The encoded protein interacts in a complex with glutamate receptor interacting protein 1 (GRIP1) and plays a role in the regulation of AMPA receptor function. [provided by RefSeq, Aug 2013]. | hsa:56850; | axon [GO:0030424]; blood microparticle [GO:0072562]; cytosol [GO:0005829]; dendrite [GO:0030425]; extrinsic component of postsynaptic early endosome membrane [GO:0098998]; glutamatergic synapse [GO:0098978]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; postsynaptic recycling endosome [GO:0098837]; recycling endosome membrane [GO:0055038]; identical protein binding [GO:0042802]; neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0098887]; regulation of modification of synaptic structure [GO:1905244]; regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0099152]; regulation of recycling endosome localization within postsynapse [GO:0099158] | 15897011_GRASP-1 was identified as an soluble autoantigen that has numerous coiled-coil domains throughout the protein. 17761173_Results suggest that GRASP-1 serves as a scaffold protein to facilitate MEKK-1 activation of JNK signaling in neurons. 28106924_We identified a new TFE3 fusion partner, GRIPAP1, in translocation renal cell carcinoma 28285821_results demonstrate a requirement for normal recycling endosome function in AMPAR-dependent synaptic function and neuronal connectivity in vivo, and suggest a potential role for GRASP1 in the pathophysiology of human cognitive disorders. | ENSMUSG00000031153 | Gripap1 | 1118.454555 | 1.1299629 | 0.176275407 | 0.08888824 | 3.962329e+00 | 4.652929e-02 | 2.898978e-01 | No | Yes | 1154.299857 | 100.917657 | 1012.077557 | 88.455811 | |
| ENSG00000068831 | 10235 | RASGRP2 | protein_coding | Q7LDG7 | FUNCTION: Functions as a calcium- and DAG-regulated nucleotide exchange factor specifically activating Rap through the exchange of bound GDP for GTP. May also activates other GTPases such as RRAS, RRAS2, NRAS, KRAS but not HRAS. Functions in aggregation of platelets and adhesion of T-lymphocytes and neutrophils probably through inside-out integrin activation. May function in the muscarinic acetylcholine receptor M1/CHRM1 signaling pathway. {ECO:0000269|PubMed:10918068, ECO:0000269|PubMed:14702343, ECO:0000269|PubMed:17576779, ECO:0000269|PubMed:17702895, ECO:0000269|PubMed:24958846, ECO:0000269|PubMed:27235135}. | 3D-structure;Alternative splicing;Calcium;Cell junction;Cell membrane;Cell projection;Cytoplasm;Disease variant;Guanine-nucleotide releasing factor;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Synapse;Synaptosome;Zinc;Zinc-finger | The protein encoded by this gene is a brain-enriched nucleotide exchanged factor that contains an N-terminal GEF domain, 2 tandem repeats of EF-hand calcium-binding motifs, and a C-terminal diacylglycerol/phorbol ester-binding domain. This protein can activate small GTPases, including RAS and RAP1/RAS3. The nucleotide exchange activity of this protein can be stimulated by calcium and diacylglycerol. Four alternatively spliced transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jan 2016]. | hsa:10235; | cytosol [GO:0005829]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; synapse [GO:0045202]; calcium ion binding [GO:0005509]; diacylglycerol binding [GO:0019992]; guanyl-nucleotide exchange factor activity [GO:0005085]; lipid binding [GO:0008289]; cellular response to calcium ion [GO:0071277]; positive regulation of GTPase activity [GO:0043547]; Ras protein signal transduction [GO:0007265]; regulation of cell growth [GO:0001558]; signal transduction [GO:0007165] | 12239348_CalDAG-GEFI plays a role in inside-out signaling to alphaIIbbeta3 20606303_analyzed the 5'-flanking region of rasgrp2 gene by a luciferase assay, which revealed that not only a promoter but also silencer regions were present upstream of D1E, suggesting rasgrp2 expression is controlled by a combination of promotion and repression 23530823_NIH3T3 cells were found nonpermissive to mtHSV but they became permissive following transformation with the Rasgrp2 gene. This effect was linked to the activation of the Ras-PKR signaling pathway. 23563504_RasGRP2 increases cell viability and cell-matrix adhesion through increased Ras expression and Rap1 activation, respectively, in endothelial cells. 23611601_phosphorylation of CalDAG-GEFI is a critical mechanism by which PKA controls Rap1b-dependent platelet aggregation 24958846_Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding. 27022025_RasGRP2 is exceptional in that its C1 domain has very weak binding affinity (Kd = 2890 +/- 240 nm for [(3)H]phorbol 12,13-dibutyrate. We have identified four amino acid residues responsible for this lack of sensitivity. Replacing Asn(7), Ser(8), Ala(19), and Ile(21) with the corresponding residues from RasGRP1/3 (Thr(7), Tyr(8), Gly(19), and Leu(21), respectively) conferred potent binding affinity (Kd = 1.47 +/- 0.03 nm). 27107697_These studies identify RasGRP2 as a novel substrate of ERK1/2 and define a negative-feedback loop that regulates the BRaf-MEK-ERK signaling cascade. This negative-feedback loop determines the amplitude and duration of active ERK1/2. 27235135_These patients are the first cases of a CalDAG-GEFI deficiency due to homozygous RASGRP2 mutations that are linked to defects in both leukocyte and platelet integrin activation. 28637664_Eleven cases with unexplained bleeding or platelet disorders harbored 11 different, previously unreported RASGRP2 variants that were biallelic and likely pathogenic. 28726538_we here describe a novel mutation in RASGRP2 that affects both expression and function of CalDAG-GEFI and that causes impaired platelet adhesive function and significant bleeding in humans. 29970392_results indicate that CD38 promotes RasGRP2/Rap1-mediated CLL cell adhesion and migration by increasing intracellular Ca2+ levels. 30076153_RASGRP2 was identified to be involved in the pathogenesis of rheumatoid arthritis by promoting adhesion, migration and IL-6 production from fibroblast-like synoviocytes 31724816_RASGRP2 mutation is associated with bleeding disorders. 32041177_RasGRP2 Structure, Function and Genetic Variants in Platelet Pathophysiology. 32609603_A novel missense variant in the RASGRP2 gene in patients with moderate to severe bleeding disorder. 33536515_RasGRP2 inhibits glyceraldehyde-derived toxic advanced glycation end-products from inducing permeability in vascular endothelial cells. 33711653_Mutations in RASGRP2 gene identified in patients misdiagnosed as Glanzmann thrombasthenia patients. 34681791_The Role of RASGRP2 in Vascular Endothelial Cells-A Mini Review. 34830306_CalDAG-GEFI Deficiency in a Family with Symptomatic Heterozygous and Homozygous Carriers of a Likely Pathogenic Variant in RASGRP2. 35122233_Novel RASGRP2 variants in platelet function defects: Indian study. | ENSMUSG00000032946 | Rasgrp2 | 21.520057 | 0.6363084 | -0.652202022 | 0.57236982 | 1.273563e+00 | 2.590993e-01 | No | Yes | 14.221629 | 4.342753 | 24.440641 | 7.408027 | ||
| ENSG00000071205 | 79658 | ARHGAP10 | protein_coding | A1A4S6 | FUNCTION: GTPase activator for the small GTPases RhoA and Cdc42 by converting them to an inactive GDP-bound state. Essential for PTKB2 regulation of cytoskeletal organization via Rho family GTPases. Inhibits PAK2 proteolytic fragment PAK-2p34 kinase activity and changes its localization from the nucleus to the perinuclear region. Stabilizes PAK-2p34 thereby increasing stimulation of cell death (By similarity). {ECO:0000250, ECO:0000269|PubMed:11432776}. | 3D-structure;Cell membrane;Cytoplasm;GTPase activation;Membrane;Phosphoprotein;Reference proteome;SH3 domain | hsa:79658; | cytosol [GO:0005829]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; cytoskeleton organization [GO:0007010]; negative regulation of apoptotic process [GO:0043066]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | 15471851_PS-GAP is a novel regulator of caspase-activated PAK-2 19911011_Observational study of gene-disease association. (HuGE Navigator) 20158304_In the Australian GWAS, one SNP achieved genomewide significance (p < 5 x 10(-8)) for ND (rs964170 in ARHGAP10 on chromosome 4, p = 4.43 x 10(-8)). 20332263_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27010858_ARHGAP10 may serve as a tumor suppressor through inactivating Cdc42, as well as inhibiting cell cycle, replication and BER pathways. 31445707_These findings suggest that CXCL12/CXCR4 promotes ovarian cancer cell invasion by suppressing ARHGAP10 expression, which is mediated by VEGF/VEGFR2 signaling. 31789419_miR3373p inhibits gastric tumor metastasis by targeting ARHGAP10. 32344433_GRAF2, WDR44, and MICAL1 mediate Rab8/10/11-dependent export of E-cadherin, MMP14, and CFTR DeltaF508. 32699248_ARHGAP10, which encodes Rho GTPase-activating protein 10, is a novel gene for schizophrenia risk. | ENSMUSG00000037148 | Arhgap10 | 377.323224 | 0.9582388 | -0.061542930 | 0.14014265 | 1.925613e-01 | 6.607933e-01 | No | Yes | 360.477531 | 41.691829 | 371.043247 | 42.820616 | |||
| ENSG00000071575 | 28951 | TRIB2 | protein_coding | Q92519 | FUNCTION: Interacts with MAPK kinases and regulates activation of MAP kinases. Does not display kinase activity (By similarity). {ECO:0000250|UniProtKB:Q28283, ECO:0000250|UniProtKB:Q96RU8}. | Cytoplasm;Cytoskeleton;Protein kinase inhibitor;Reference proteome | This gene encodes one of three members of the Tribbles family. The Tribbles members share a Trb domain, which is homologous to protein serine-threonine kinases, but lacks the active site lysine and probably lacks a catalytic function. The Tribbles proteins interact and modulate the activity of signal transduction pathways in a number of physiological and pathological processes. This Tribbles member induces apoptosis of cells mainly of the hematopoietic origin. It has been identified as a protein up-regulated by inflammatory stimuli in myeloid (THP-1) cells, and also as an oncogene that inactivates the transcription factor C/EBPalpha (CCAAT/enhancer-binding protein alpha) and causes acute myelogenous leukemia. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:28951; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; nucleus [GO:0005634]; mitogen-activated protein kinase kinase binding [GO:0031434]; protein kinase inhibitor activity [GO:0004860]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase regulator activity [GO:0055106]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of interleukin-10 production [GO:0032693]; negative regulation of protein kinase activity [GO:0006469]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; regulation of MAP kinase activity [GO:0043405] | 15950723_Anti-TRB2 antibody activities were detected in several autoimmune uveitis patients but not in control subjects, suggesting that TRB2 is a uveitis-associated candidate autoantigen. 17545167_TRB2-Mcl-1 axis plays an important role in survival factor withdrawal-induced apoptosis of TF-1 erythroleukemia cells. 18952906_Downregulation potentiates IL-8 production by human monocytes via enhanced activation of the ERKs and JNK/MAPK pathways 19656773_Data show that SNPs associated with TG in normolipidemic samples, including APOA5, TRIB1, TBL2, GCKR, GALNT2 and ANGPTL3 were significantly associated with HLP types 2B, 3, 4 and 5. 20005259_Trb-2 expression was reduced in acute myeloid leukaemia compared to healthy controls 20160349_Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. 20208562_Overexpression of TRIB2 is associated with malignant melanoma. 20468071_Observational study of gene-disease association. (HuGE Navigator) 20614846_studies have found increased autoantibodies against Tribbles homolog 2 in narcolepsy 20614847_A study reported an increased prevalence of autoantibodies against Tribbles homolog 2 (TRIB2) in patients with narcolepsy 20805362_our data demonstrate that Trib2 can bind both COP1 and C/EBP-alpha, leading to degradation of C/EBP-alpha. 20944657_Observational study of gene-disease association. (HuGE Navigator) 21399661_TRIB2 as a potential driver of lung tumorigenesis through a mechanism that involves downregulation of C/EBPalpha 22271508_Low Trib2 is associated with inflammatory bowel disease. 22589742_We have identified a SNP near the TRIB2 locus that is associated with pericardial fat but not with body mass index or visceral abdominal fat. 22775572_Our findings here support the ability of high TRIB2 expression to reveal a T cell profile in both T cell acute lymphoblastic leukaemias and acute myeloid leukaemias. 23071539_results illustrate that miR-511 and miR-1297 act as tumor suppressor genes, which could suppress A549 cell proliferation in vitro and in vivo by suppressing TRIB2 and further increasing C/EBPalpha expression 23108367_Positive natural selection of TRIB2, a novel gene that influences visceral fat accumulation, in East Asia. 23550039_We discuss the role of Tribs as central signaling mediators in different subtypes of acute leukemia and propose that inhibition of dysregulated Trib signaling may be therapeutically beneficial. 23850892_through regulating the expression of TRIB2 and its downstream factors, let-7c can effectively inhibit A549 cell proliferation in vitro and in vivo 24089522_impaired phosphorylation and ubiquitination by p70S6K and Smurf1 increase the protein stability of TRIB2 in liver cancer 24211582_this study reveals that the stability and ubiquitination of TRIB2 can be also controlled by Ubiquitin E3 ligase SCFb-TRCP. 24516045_we identify the contribution of dysregulated C/EBPalpha and E2F1 to elevated Trib2 expression and leukemic cell survival 25311538_TRIB2 negatively regulates Wnt activity through a reduction in protein stability of TCF4 and beta-Catenin 25583260_Data suggest that TRB2 exhibits metal-independent autophosphorylation activity, low affinity ATP binding/hydrolysis, and a conserved catalytic lysine residue in the active site (rather than the asparagine in many metal-dependent kinases). 25586991_The results suggest that TRIB2 is a meaningful biomarker reflecting diagnosis and progression of melanoma, as well as predicting clinical response to chemotherapy. 26202930_High TRIB2 reinforces the oncogenic transcriptional program controlled by the TAL1 complex in T-cell acute lymphoblastic leukemia. 26517922_Studies suggest that pseudo-kinase family of tribbles (TRIB) proteins TRIB1, TRIB2 and TRIB3 play roles in pathogenesis of rheumatoid arthritis (RA) and osteoarthritis. 26517925_Studies suggest that pseudo-kinase family of tribbles (TRIB) proteins TRIB1, TRIB2 and TRIB3 were involved in the pathogenesis of inflammation. 26517928_Studies suggest that tribbles homolog 2 (Drosophila) protein (TRIB2) as a meaningful biomarker to both diagnose and stage melanoma. 26517929_Studies indicate that the molecular interactions that take place between tribbles homolog 2 (Drosophila) protein (TRIB2) and factors involved in the ubiquitin proteasome system (UPS) are varied and have differential downstream effects. 26517930_Studies indicate that small molecules can reveal rate-limiting signalling outputs and functions of pseudo-kinase family of tribbles (TRIB) proteins TRIB1, TRIB2 and TRIB3 in cells and intact organisms, serving as guides for the development of new drugs. 26517933_Studies show that TRIB1 and TRIB2 are highly expressed in molecularly-defined sub-types of acute myeloid leukemia (AML). 26517934_Studies show a remarkable reduction in tribbles 2 protein (Trib2) expression during oocyte maturation whereas tribbles 1 protein (Trib1) and tribbles 3 protein (Trib3) expression was significantly increased during this process. 26996668_our data show that excess p30 cooperated with TRIB2 only in the presence of p42 to accelerate acute myeloid leukaemia (AML), and the direct interaction and degradation of C/EBPa p42 is required for TRIB2-mediated AML. 27515988_O-GlcNAcylation of TRIB2 might be critical for diabetes-associated liver cancer. 27563873_Data show that TRIB2-mediated degradation of CDC25C is associated with lysine-48-linked CDC25C polyubiquitination driven by the TRIB2 kinase-like domain. 28005074_miR-206 and miR-140, as tumor suppressors, induced lung adenocarcinoma cell death and inhibited cell proliferation by modifying oncogenic TRIB2 promoter activity through p-Smad3. 28212671_Studied effect of TRIB2 SNP on the expression of genes involved in adaptive thermogenesis using messenger RNAs prepared from adipose tissues of Japanese adults. Of the five thermogenic genes, DIO2, CIDEA, PPARGC1A, and PRDM16 showed significantly higher transcript levels in SAT of individuals with the AA genotype relative to those with the AT + TT genotype. 28276427_Data indicate a regulatory mechanism underlying drug resistance and suggest that tribbles homologue 2 (TRIB2) functions as a regulatory component of the PI3K network, activating AKT in cancer cells. 28670762_Tumors from resistant patients expressed the lowest levels of TRIB2. Downregulation of TRIB2 contributes to platin-resistance. 29170476_Trib2 is important for maintaining self-renewal in ES cells and for pluripotency induction during the reprogramming process. 29436678_Due to the downregulation of MDR1 and MRP1, the intracellular accumulation of ADM was increased in the transfected cells compared with that in the parental K562/ADM cells..Our research revealed that protein expression of the ERK signaling pathway was inhibited by downregulating TRIB2, indicating that the ERK pathway was involved in cell drug resistance and proliferation. 30031847_The expression profiles of TRIB1, TRIB2, and TRIB3 in human and murine hematopoietic stem, progenitor and mature cells, and in human leukemia datasets have been mapped. 30541550_TRIB2 suppresses cellular senescence through interaction with AP4 to down-regulate p21 expression. 31318172_Combined elevation of TRIB2 and MAP3K1 indicates poor prognosis and chemoresistance to temozolomide in glioblastoma. 31621188_Pseudokinases: a tribble-edged sword. 31954160_Knockdown of circ_0084043 suppressed the malignant development of melanoma presumably through modulating miR429/TRIB2 axis and inactivating Wnt/beta-catenin signaling pathway 32513531_LncRNA ZEB1-AS1 promotes pancreatic cancer progression by regulating miR-505-3p/TRIB2 axis. 33386706_Tribbles homolog 2 promotes hepatic fibrosis and hepatocarcinogenesis through phosphatase 1A-Mediated stabilization of yes-associated protein. 33860836_Upregulation of TRIB2 by Wnt/beta-catenin activation in BRAF(V600E) papillary thyroid carcinoma cells confers resistance to BRAF inhibitor vemurafenib. 34326276_TRIB2 Stimulates Cancer Stem-Like Properties through Activating the AKT-GSK3beta-beta-Catenin Signaling Axis. 34586732_A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. 34808454_Potential innate immunity-related markers of endometrial receptivity and recurrent implantation failure (RIF). | ENSMUSG00000020601 | Trib2 | 262.280887 | 1.1391442 | 0.187950414 | 0.19784615 | 8.903579e-01 | 3.453803e-01 | No | Yes | 256.635776 | 33.354077 | 228.072354 | 29.593527 | ||
| ENSG00000071889 | 60343 | FAM3A | protein_coding | P98173 | FUNCTION: May act as a defensin against invading fungal microorganisms. | Alternative splicing;Disulfide bond;Reference proteome;Secreted;Signal | This gene encodes a cytokine-like protein. The expression of this gene may be regulated by peroxisome proliferator-activated receptor gamma, and the encoded protein may be involved in the regulation of glucose and lipid metabolism. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2013]. | hsa:60343; | extracellular space [GO:0005615]; antifungal humoral response [GO:0019732]; antimicrobial humoral immune response mediated by antimicrobial peptide [GO:0061844]; negative regulation of antifungal innate immune response [GO:1905035]; negative regulation of gluconeogenesis [GO:0045721]; regulation of lipid biosynthetic process [GO:0046890] | 12160727_Expressed prominently in the vascular endothelium, particularly capillaries, also in islets of Langerhans. 18212107_In the present study, we report a putative human homologue of the antimicrobial Drosophila-derived drosomycin, designated drosomycin-like defensin (DLD), with specific antifungal activity. 23562554_Upregulation of FAM3A by PPARgamma activation is correlated with increased pAkt level in liver cells 24806753_FAM3A plays crucial roles in the regulation of glucose and lipid metabolism in the liver, where it activates the PI3K-Akt signaling pathway by way of a Ca(2+) /CaM-dependent mechanism 27688071_These results suggest that C/EBPbeta plays an important role in regulating FAM3A promoter activity and FAM3A inhibits adipocyte differentiation. 29532774_Fam3A may regulate high glucose-induced reactive oxygen species production in HUVECs via the p38 MAPK signaling pathway. 31000420_our data demonstrate that FAM3A positively regulates angiogenesis through activation of VEGFA transcription, suggesting that FAM3A may constitute a novel molecular therapeutic target for ischaemic vascular disease. 31910817_In this case prenatal ultrasound images were suggestive of a serious but non-lethal skeletal dysplasia. Due to the uncertain prognosis the parents were offered Whole Exome Sequencing (WES), which identified a specific gene mutation in the FAMIIIa gene | ENSMUSG00000031399 | Fam3a | 1771.289620 | 0.8410769 | -0.249690312 | 0.07798961 | 1.021177e+01 | 1.395474e-03 | 4.141466e-02 | No | Yes | 1589.869865 | 186.828682 | 1914.461773 | 224.633689 | |
| ENSG00000072195 | 10290 | SPEG | protein_coding | Q15772 | FUNCTION: Isoform 3 may have a role in regulating the growth and differentiation of arterial smooth muscle cells. | 3D-structure;ATP-binding;Alternative promoter usage;Alternative splicing;Differentiation;Disease variant;Disulfide bond;Immunoglobulin domain;Kinase;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | This gene encodes a protein with similarity to members of the myosin light chain kinase family. This protein family is required for myocyte cytoskeletal development. Along with the desmin gene, expression of this gene may be controlled by the desmin locus control region. Mutations in this gene are associated with centronuclear myopathy 5. [provided by RefSeq, Jun 2016]. | hsa:10290; | nucleus [GO:0005634]; ATP binding [GO:0005524]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; muscle cell differentiation [GO:0042692]; muscle organ development [GO:0007517]; negative regulation of cell population proliferation [GO:0008285] | 15784173_Genomic rearrangement on APEG1 in arteriosclerosis was studied. 16354304_the RGD motif might play a role not only in the adhesion of Aortic Preferentially Expressed Protein-1 and extracellular proteins but also in intracellular protein-protein interactions 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25087613_SPEG is present in cardiac muscle, where it plays a critical role; therefore, individuals with SPEG mutations additionally present with dilated cardiomyopathy. 30412272_Clinicians should consider evaluating a centronuclear myopathies patient for SPEG mutations even in the absence of centronuclear myopathies features 31625632_Novel SPEG variant cause centronuclear myopathy in China. 31790338_Striated muscle-specific serine/threonine-protein kinase beta segregates with high versus low responsiveness to endurance exercise training. 32683896_Loss of SPEG Inhibitory Phosphorylation of Ryanodine Receptor Type-2 Promotes Atrial Fibrillation. 32925938_A Novel Recessive Mutation in SPEG Causes Early Onset Dilated Cardiomyopathy. 33794647_Homozygous SPEG Mutation Is Associated With Isolated Dilated Cardiomyopathy. | ENSMUSG00000026207 | Speg | 226.119383 | 0.9588014 | -0.060696016 | 0.22687178 | 7.177724e-02 | 7.887665e-01 | No | Yes | 174.375854 | 29.447113 | 191.832032 | 32.283054 | ||
| ENSG00000073584 | 6605 | SMARCE1 | protein_coding | Q969G3 | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). Required for the coactivation of estrogen responsive promoters by SWI/SNF complexes and the SRC/p160 family of histone acetyltransferases (HATs). Also specifically interacts with the CoREST corepressor resulting in repression of neuronal specific gene promoters in non-neuronal cells. {ECO:0000250|UniProtKB:O54941, ECO:0000303|PubMed:12672490, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | 3D-structure;Alternative splicing;Chromatin regulator;Coiled coil;DNA-binding;Disease variant;Isopeptide bond;Mental retardation;Methylation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is part of the large ATP-dependent chromatin remodeling complex SWI/SNF, which is required for transcriptional activation of genes normally repressed by chromatin. The encoded protein, either alone or when in the SWI/SNF complex, can bind to 4-way junction DNA, which is thought to mimic the topology of DNA as it enters or exits the nucleosome. The protein contains a DNA-binding HMG domain, but disruption of this domain does not abolish the DNA-binding or nucleosome-displacement activities of the SWI/SNF complex. Unlike most of the SWI/SNF complex proteins, this protein has no yeast counterpart. [provided by RefSeq, Jul 2008]. | hsa:6605; | chromatin [GO:0000785]; nBAF complex [GO:0071565]; npBAF complex [GO:0071564]; nuclear chromosome [GO:0000228]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; SWI/SNF complex [GO:0016514]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; N-acetyltransferase activity [GO:0008080]; nuclear receptor binding [GO:0016922]; protein N-terminus binding [GO:0047485]; RNA binding [GO:0003723]; transcription coactivator activity [GO:0003713]; chromatin remodeling [GO:0006338]; negative regulation of transcription, DNA-templated [GO:0045892]; neurogenesis [GO:0022008]; nucleosome disassembly [GO:0006337]; regulation of transcription by RNA polymerase II [GO:0006357] | 16135788_BAF57-mediated cell death is associated with up-regulation of proapoptotic genes including the tumor suppressor familial cylindromatosis (CYLD), which is found to be a direct target of BAF57. 16199878_protein levels of BAF155/170 dictate the maximum cellular amount of BAF57 16769725_BAF57 is a critical regulator of estrogen receptor function in breast cancer cells 17669635_Ability of SMARCE 1 in modulating the replication efficiency of HBV. 20460533_Knockdown of BAF57 resulted in an accumulation of cells in the G(2)-M phase, inhibition of colony formation, and impaired growth in soft agar. Knockdown of BAF57 also caused transcriptional misregulation of various cell cycle-related gene. 20829358_Data show that the mechanism of BAF155-mediated stabilization of BAF57 involves blocking its ubiquitination by preventing interaction with TRIP12. 21465167_mutations in BAF57 could impinge on several oncogenic signaling pathways contributing to the origin and/or development of breast cancer. 22419023_BAF57 expression was significantly associated with the surgical stage, grade of the tumor, myometrial invasion, lympho-vascular space invasion and lymph node metastasis in 111 endometrial carcinomas. 23377182_Our findings identify multiple-spinal-meningioma disease as a new discrete entity and establish a key role for the SWI/SNF complex in the pathogenesis of both meningiomas and tumors with clear-cell histology. 23493350_Data indicate that BAF57 deregulation predisposes to metastasis. 24880093_Since both TTF1 and SMARCE1 are involved in chromatin remodeling, our results imply an epigenetic regulatory mechanism for T-cell recruitment that invites deciphering. 25081545_BAF complex gene SMARCE1 is mutated in Coffin-Siris syndrome patients. 25143307_these results demonstrate that loss of SMARCE1 is relevant to cranial as well as spinal meningiomas 25168959_Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SmarCE1 gene. 25611552_The results suggested that BAF57 is involved in ovarian cancer cell growth and sensitivity to anticancer agents, and that BAF57 may be a target for ovarian cancer therapy. 25656847_Addition of the EGFR inhibitor gefitinib restores the sensitivity of SMARCE1-knockdown cells to MET and ALK inhibitors in NSCLCs. Our findings link SMARCE1 to EGFR oncogenic signaling and suggest targeted treatment options for SMARCE1-deficient tumors. 26803492_a family with a pediatric CCM patient and an adult CCM patient and several asymptomatic relatives carrying a germline SMARCE1 mutation. 27149204_An exhaustive analysis of the BAF57 molecular and biochemical properties, cellular functions, loss-of-function phenotypes in living organisms and pathological manifestations in cases of human mutations. [review] 27264197_We report here three additional individuals with clinical features consistent with CSS and alterations in SMARCE1, one of which is novel. The probands all exhibited dysmorphic facial features, moderate developmental and cognitive delay, poor growth, and hypoplastic digital nails/phalanges, including digits not typically affected in the other genes associated with CSS. 27495308_SMARCE1 plays an essential role in breast cancer metastasis by protecting cells against anoikis through the HIF1A/PTK2 pathway. 27891692_SMARCE1 mutational hits, including novel SMARCE1 mutations, were found in six of eight tested patients with clear cell meningioma 28377514_High SMARCE1 expression is associated with eventual relapse and metastasis in breast cancer. 28474749_Study showed, for the first time, that SMARCE1 immunostaining is a highly sensitive biomarker for clear cell meningioma, useful as a routine diagnostic biomarker. 28620005_the malignancy risk in schwannomatosis is not well defined but may include an increased risk of malignant peripheral nerve sheath tumor in SMARCB1 Imaging protocols are also proposed for SMARCB1 and LZTR1 schwannomatosis and SMARCE1-related meningioma predisposition. 28716547_BAF57, BAF60a and SNF5 might act as novel pro-senescence factors in both normal and tumor human skin cells 28740345_miR-29a promotes hepatitis B virus (HBV) replication and expression through regulating SMARCE1 in HBV-infected HepG2.2.15 cells. 30548424_A de novo splicing mutation in SMARCE1 was identified in a patient with Angelman-like syndrome. 31611549_MicroRNA-802 induces hepatitis B virus replication and replication through regulating SMARCE1 expression in hepatocellular carcinoma. 32783402_Long noncoding RNA HOTTIP promotes the metastatic potential of ovarian cancer through the regulation of the miR-615-3p/SMARCE1 pathway. 33010889_Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA. 33185983_The importance of genetic counseling and screening for people with pathogenic SMARCE1 variants: A family study. 33319313_Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1. 33670012_BAF57/SMARCE1 Interacting with Splicing Factor SRSF1 Regulates Mechanical Stress-Induced Alternative Splicing of Cyclin D1. 34732413_BAF57 Is a Potential Determinant of Colorectal Cancer Malignancy. 35158202_Assembly and interaction of core subunits of BAF complexes and crystal study of the SMARCC1/SMARCE1 binary complex. | ENSMUSG00000037935 | Smarce1 | 4437.556050 | 1.0334693 | 0.047495542 | 0.06046124 | 6.151847e-01 | 4.328423e-01 | 7.666849e-01 | No | Yes | 4672.471791 | 669.565336 | 4498.641628 | 644.626607 | |
| ENSG00000075891 | 5076 | PAX2 | protein_coding | Q02962 | FUNCTION: Transcription factor that may have a role in kidney cell differentiation (PubMed:24676634). Has a critical role in the development of the urogenital tract, the eyes, and the CNS. {ECO:0000269|PubMed:24676634}. | Alternative splicing;DNA-binding;Developmental protein;Differentiation;Disease variant;Nucleus;Paired box;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | PAX2 encodes paired box gene 2, one of many human homologues of the Drosophila melanogaster gene prd. The central feature of this transcription factor gene family is the conserved DNA-binding paired box domain. PAX2 is believed to be a target of transcriptional supression by the tumor suppressor gene WT1. Mutations within PAX2 have been shown to result in optic nerve colobomas and renal hypoplasia. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Dec 2014]. | hsa:5076; | centriolar satellite [GO:0034451]; chromatin [GO:0000785]; lysosome [GO:0005764]; microtubule organizing center [GO:0005815]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; protein-DNA complex [GO:0032993]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; transcription factor binding [GO:0008134]; aging [GO:0007568]; anatomical structure development [GO:0048856]; axonogenesis [GO:0007409]; brain morphogenesis [GO:0048854]; branching involved in ureteric bud morphogenesis [GO:0001658]; camera-type eye development [GO:0043010]; cell fate determination [GO:0001709]; cellular response to epidermal growth factor stimulus [GO:0071364]; cellular response to glucose stimulus [GO:0071333]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to retinoic acid [GO:0071300]; cochlea development [GO:0090102]; cochlea morphogenesis [GO:0090103]; glial cell differentiation [GO:0010001]; inner ear morphogenesis [GO:0042472]; mesenchymal to epithelial transition [GO:0060231]; mesenchymal to epithelial transition involved in metanephros morphogenesis [GO:0003337]; mesodermal cell fate specification [GO:0007501]; mesonephros development [GO:0001823]; metanephric collecting duct development [GO:0072205]; metanephric distal convoluted tubule development [GO:0072221]; metanephric epithelium development [GO:0072207]; metanephric mesenchymal cell differentiation [GO:0072162]; metanephric mesenchyme development [GO:0072075]; metanephric nephron tubule formation [GO:0072289]; negative regulation of apoptotic process [GO:0043066]; negative regulation of apoptotic process involved in metanephric collecting duct development [GO:1900215]; negative regulation of apoptotic process involved in metanephric nephron tubule development [GO:1900218]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis [GO:0072305]; negative regulation of mesenchymal cell apoptotic process involved in metanephros development [GO:1900212]; negative regulation of programmed cell death [GO:0043069]; negative regulation of reactive oxygen species metabolic process [GO:2000378]; negative regulation of transcription, DNA-templated [GO:0045892]; nephric duct formation [GO:0072179]; neural tube closure [GO:0001843]; optic chiasma development [GO:0061360]; optic cup morphogenesis involved in camera-type eye development [GO:0002072]; optic nerve development [GO:0021554]; optic nerve morphogenesis [GO:0021631]; optic nerve structural organization [GO:0021633]; positive regulation of branching involved in ureteric bud morphogenesis [GO:0090190]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis [GO:0072108]; positive regulation of metanephric DCT cell differentiation [GO:2000594]; positive regulation of metanephric glomerulus development [GO:0072300]; positive regulation of optic nerve formation [GO:2000597]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; pronephric field specification [GO:0039003]; pronephros development [GO:0048793]; protein kinase B signaling [GO:0043491]; reactive oxygen species metabolic process [GO:0072593]; regulation of metanephric nephron tubule epithelial cell differentiation [GO:0072307]; regulation of metanephros size [GO:0035566]; regulation of transcription by RNA polymerase II [GO:0006357]; response to nutrient levels [GO:0031667]; retinal pigment epithelium development [GO:0003406]; stem cell differentiation [GO:0048863]; ureter development [GO:0072189]; ureter maturation [GO:0035799]; urogenital system development [GO:0001655]; vestibulocochlear nerve formation [GO:0021650]; visual perception [GO:0007601] | 11730657_The causal relationship between PAX2 gene mutations and renal-coloboma syndrome is further supported 11826030_The absence of PAX2 mutations has been identified in two families with histories of clinical overlap of Okihiro and acro-renal-ocular syndromes. 11940591_The HMG-I/Y-related protein p8 binds to p300 and Pax2 trans-activation domain-interacting protein to regulate the trans-activation activity of the Pax2A and Pax2B transcription factors on the glucagon gene promoter. 12141441_PAX2 has a role in urogenital tract development and disease [review] 12970747_The PAX2 gene was frequently expressed in a panel of 406 common primary tumor tissues and endogenous PAX gene expression is often required for the growth and survival of cancer cells 14561758_Pax2 protein regulates expression of secreted frizzled related protein 2 14566649_PAX2 mutation is associated with Optic nerve dysplasia and renal insufficiency of the renal-coloboma syndrome 14627715_expression of Pax2 by Kaposi's sarcoma cells correlated with an enhanced resistance against apoptotic signals and with the proinvasive phenotype 15502805_PAX-2 is a reliable marker for clear cell renal cell carcinomas of lower grades but not for higher grades. 15652857_A new PAX2 missense mutation, R71T, may cause macular abnormalities in addition to anomalies of the optic disk and the kidney. 15808183_Molecular genetic analysis of the PAX2 gene in combination with renal ultrasonography can help in making an earlier diagnosis of the disease. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16400326_PAX2 is a specific and sensitive immunohistochemical marker in identification and differential diagnosis of nephrogenic adenoma. 16436683_Over expression of Pax2 is associated with apoptosis resistance and angiogenesis favoring renal tumor growth 16509931_Observational study of gene-disease association. (HuGE Navigator) 16609680_Pax2 overexpression in renal cell carcinoma cells contributes to cisplatin resistance. 16735463_Powerful effects of PAX2 on renal branching morphogenesis and final nephron number may be mediated by activation of Naip which then suppresses apoptosis in ureter bud cells. 16814811_Results showed no direct involvement of PAX-8 genes in Wilms tumor pathogenesis. 16971658_Mutations in PAX2 is associated with renal hypodysplasia 17269592_Renal-coloboma syndrome: a single nucleotide deletion in the PAX2 gene at Exon 8 is associated with a highly variable phenotype 17513325_Observational study of gene-disease association. (HuGE Navigator) 17513325_Subtle renal hypoplasia in normal newborns may be partially due to a common variant of the PAX2 gene that reduces mRNA expression during kidney development 17529925_PAX2 was identified in ovarian papillary serous carcinoma cells derived from Mullerian epithelium or surrounding ovary. 18056486_Herpesvirus 8, Human infection may activate an embryonic angiogenic program in human microvascular endothelial cells by inducing the expression of PAX2 oncogene. 18379529_familial renal failure with ocular abnormality 18439754_PAX2 is a potential therapeutic gene target in renal cancer and suggest that adjunctive PAX2 knockdown may enhance the efficacy of other chemotherapeutic agents. 18609495_The association of optic nerve and renal malformations should lead to the suspicion of papillorenal syndrome with PAX2 mutation. 18685487_Conclude that PAX-2 is a useful marker for distinguishing metastatic clear cell renal carcinoma from its potential morphologic mimics. 18941400_potential diagnostic utility of Pax 2 in distinction of (i) oncocytoma from chromophobe RCC, (ii) clear cell RCC and papillary RCC from renal tumors with Xp11.2 translocation and (iii) high-grade clear cell RCC from urothelial carcinoma 19048125_WNT5A is regulated by PAX2 and may be involved in blastemal predominant Wilms tumorigenesis 19118900_PAX2 oncogene suppresses hBD1 expression in cancer and further implicate PAX2 as a novel therapeutic target for prostate cancer treatment. 19228645_PAX-2 seems to have a significant role in renal neogenesis and may represent a novel therapeutic target. 19401348_Study concluded that in ccRCC, PAX2 reactivation is driven by HIF-dependent mechanisms following pVHL loss. 19467152_A previously undescribed intron 9 and exon 10 containing splice variant of PAX2 in B-cell neoplasia and in solid tumors on mRNA and protein level, is described. 19517575_PAX2 may be a novel therapeutic target for the treatment of carcinomas such as prostate cancer via the down-regulation of its expression by targeting the AT1R signaling pathways. 19525924_a statistically significant difference in PAX2 mRNA expression among ovarian tumors of low malignant potential, low-grade, and high-grade carcinoma samples 19556301_Renal recovery after injury: the role of Pax-2. Review. 19851209_PAX 2 is a promising new, sensitive, and specific mullerian immunomarker for ovarian serous carcinomas (primary and metastatic) 19959718_Observational study of gene-disease association. (HuGE Navigator) 20061933_PAX2 distinguishes benign mesonephric and mullerian glandular lesions of the cervix from endocervical adenocarcinoma, including minimal deviation adenocarcinoma. 20358591_report on two fetuses with fetal renal hypodysplasia with PAX2 mutations; identified a de novo single G deletion of nucleotide 935 in exon 3 of the PAX2 resulting in a frameshift mutation 20413145_PAX-2 is expressed by immunohistochemistry in seminal vesicles and the ejaculatory ducts, but not in prostatic adenocarcinoma. 20631067_PAX2 and PTEN protein loss occurs independently and accumulates with increasing age in latent precancers of normal premenopausal endometrium. Loss of both characterizes the emergence of a premalignant lesion which carries forward to carcinoma. 20670131_PAX-2 is a sensitive and specific marker for metastatic renal cell carcinoma. 21108633_PAX2 mutations may be associated with isolated renal hypoplasia. 21263247_The PAX2(-), PAX8(-), and inhibin A(+) profile supports the diagnosis of hemangioblastoma with a sensitivity of 95%, specificity of 100%, and positive predictive value of 100%. 21380624_Mutations in HNF1Beta and PAX2 commonly cause syndromic urinary tract malformation. 21575608_these results suggest that PHD3 targets Pax2 for destruction. 21696512_The renal phenotypic feature of PRS is known to have a wide variation from only mild proteinuria without renal dysfunction to end-stage renal disease (ESRD) at school age to adulthood, even in patients with the same PAX2 gene mutations. 21730820_PAX2 is a sensitive marker of Wilm's tumor, but antibody shows weak-to-moderate-intensity nuclear staining of rhabdomyosarcoma and B-cell acute lymphoblastic lymphoma. 21836481_Diffuse expression of PAX2 and PAX8 in the cystic epithelium of mixed epithelial stromal tumor, angiomyolipoma with epithelial cysts, and primary renal synovial sarcoma supports renal tubular differentiation. 21876729_PAX2 can regulate ADAM10 expression, a metalloproteinase known to play important roles in melanoma metastasis. 21934480_PAX8 and PAX2 IHC may facilitate the diagnosis of primary epithelial neoplasms of the male genital tract. 21989345_PAX8 supersedes PAX2 as probably the best epithelial marker hitherto for primary or metastatic mullerian epithelial tumors. 22080059_We propose a unique co-variable in benign oviductal epithelium-the PAX2-null SCOUT-that reflects underlying dysregulation in genes linked to serous neoplasia. 22138676_This review outlines the current evidence supporting involvement of PAX2 in the pathologic processes involving the kidney.[review] 22169637_The high-grade serous carcinomas had a significantly higher expression level of p53 and Ki-67, while low-grade carcinomas had a significantly higher expression level of Pax2. 22213154_update of PAX2 mutations in renal coloboma syndrome; review of published cases and the collective diagnostic experience of 3 laboratories in the United States, France and New Zealand identified 55 unique mutations in 173 individuals from 86 families; a locus-specific database was created 22274645_These findings suggest that PAX2 may play a role in endometrial carcinogenesis. 22473392_in endometriosis patients, paired box protein Pax2 is down-regulated in the lesions compared with the eutopic tissue, possibly due to low epidermal growth factor (EGF) signalling 22495365_PAX-2, PAX-8, and hKIM-1 should be used cautiously in distinguishing renal cell carcinoma from nonseminomatous germ cell neoplasia and also adds to the growing list of nonrenal tumors that express these 3 markers. 22552444_PAX2 expression was detected in the majority of medulloblastoma samples and correlated with less differentiated histology 22660956_In both monozygotic twins with renal coloboma syndrome, a novel de novo mutation of PAX2 was detected, which leads to the substitution of a highly conserved cysteine (p.C52Y). 22683154_Mutations in PAX2 may not be ac ommon cause of Mullerian Duct Abnormalities. 22727345_No association was found between Pax-2 expression and disease persistance/progression in endometrial hyperplasia treated with progestins. 22807381_PAX8 and PAX2 are diagnostically useful adjuncts in confirming the diagnosis of renal cell carcinoma in cytology specimens. 22984579_cross-talk between p53 and Pax2 provides a transcriptional platform that promotes nephrogenesis, thus contributing to nephron endowment 23135283_PAX2 protein induces expression of cyclin D1 through activating AP-1 protein and promotes proliferation of colon cancer cells. 23137159_PAX2 gene is pivotal in kidney development and it is implicated in the pathogenesis of renal interstitial fibrosis (RIF) and glomerulosclerosis (GS). 23194047_investigation of expression of PAX2 and PAX8 in renal biopsy samples as diagnostic markers in the differential diagnosis of various types of primary and secondary renal cell carcinomas 23196794_PAX2 has little practical value in the diagnosis of peritoneal epithelioid mesotheliomas 23202787_PAX2 is effective in discriminating cervical adenocarcinoma in situ from benign endocervical glandular epithelium 23328975_Immunostains for PAX2 and PAX8 are useful in the detection of nephrogenic adenomas and particularly unveil those nephrogenic adenomas with flat pattern. 23503645_PAX2 and napsin A have high specificity but low sensitivity and only have limited value in the differential diagnosis of mesotheliomas and renal cell carcinomas 23539225_Report high rate of mutations in PAX2 genes known to be involved in monogenic syndromic Congenital anomalies of the kidney and urinary tract. 23686327_Cryptorchidism-associated papillorenal syndrome/PAX2 mutation has not been reported. 23756089_other genes and/or locus control regions regulating PAX2 may be involved in the pathogenesis of PAX2 mutation-negative cases of RCS. 23765687_Our data suggested that PAX2 hyper-expression promotes the development of the metastatic state in prostate cancer cells, presumably through upregulating the expression of cell membrane proteins. 23861881_miR-1915 and miR-1225-5p regulated the expression of important markers of renal progenitors, such as CD133 and PAX2, and important genes involved in the repair mechanisms of adult renal progenitor cells, such as TLR2. 23890189_Pax-2 expression might be used as an ancillary tool to discriminate chromophobe renal cell tumour from oncocytomas with overlapping morphological features. 24040464_Case Report: sporadic renal hemangioblastoma expressing PAX2. 24141260_cotransfection of Pax2 and Math1 strongly stimulates in situ hair cell regeneration in neomycin-damaged cochlear explants. 24334997_PAX2 and PAX8 are valuable diagnostic markers for metastatic mullerian carcinomas and renal cell carcinoma in effusion cytology 24344503_These data suggest that in difficult diagnostic cases both PAX2 and mesothelin immunohistochemical study may be useful in discriminating between PMRCC and primary pancreatic carcinoma. 24633556_study suggests an association between PAX2 gene polymorphisms and the development of vesicoureteral reflux, but not with ureteropelvic junction obstruction and multicystic dysplastic kidney 24676634_Exome sequencing in an index family with dominant FSGS revealed a nonconservative, disease-segregating variant in the PAX2 gene. Sequencing in probands of a familial FSGS cohort revealed seven rare and private heterozygous SNPs (4% of individuals). 24897005_PAX2 was strongly positive in all alveolar rhabdomyosarcomas and in two-thirds of the kidney clear cell sarcomas, and displayed variable expression in one-half of the embryonal rhabdomyosarcomas. 24992169_PAX2 and PAX8 are useful biomarker in the differential diagnosis of ovarian serous and mucinous tumors 25106525_Pax2 may play a role in kidney development by regulating the expression of TBX1. 25130537_study, for the first time, links PAX2-null with proliferating fetal and adult oviductal cells undergoing basal and ciliated differentiation and shows that this expression state is maintained in high-grade serous cancer precursors 25132193_PAX2 promotes cancer cell growth in androgen-independent prostate cancer by regulating androgen receptor gene expression through an epigenetic mechanism. 25180270_Novel PAX2 targets included multiple genes encoding proteins with predicted functions in the epididymis epithelium. 25279712_Immunohistochemical distinction of renal cell carcinoma from other carcinomas with clear-cell histomorphology: utility of CD10 and CA-125 in addition to PAX-2, PAX-8, RCCma, and adipophilin. 25385517_The 798C>T/909A>C PAX2 genotype did not increase the susceptibility to HenochSchonlein purpura (HSP), but it was likely to increase the susceptibility of kidney involvement in HSP patients. 25500505_data suggested the differential regulation of hDAO expression by two promoters whose activities may be modulated by the binding of PAX2 and PAX5 25613757_Results showed that PAX2 expression is significantly overexpressed in esophageal squamous cell carcinoma tissues and IL-5 is identified as PAX2's effector for metastasis. 25631048_PPM1B interacts with Groucho 4 and is localized to DNA in a Groucho-dependent manner, and phosphatase activity is required for transcriptional silencing. 25854853_Homozygosity for the risk alleles of RET and PAX2 was not seen in the late onset group in primary hyperoxaluria type 1 patients. 25912758_Report increased Pax2 expression in the ureter epithelium of children with vesicoureteric reflux. 26404914_a substantial minority of solitary fibrous tumors express nuclear PAX8 and PAX2 26571382_The results of this study identify several new mutations of PAX2, and sequence variants in four additional genes, including a novel potentially pathogenic mutation in KIF26B, which may play a role in the pathogenesis of Renal Coloboma Syndrome. 26797858_PAX2, PAX8, CDX2 immunostains was preformed to the TMA slides. 26823795_PAX2 and PAX5 are useful biomarker in the differential diagnosis of lung cancer 26972711_Vimentin, Nestin and WT1. Sox2 was expressed by the stem/progenitor cells of the ventricular zone, whereas the postmitotic neurons of the cortical plate were immunostained by PAX2 and NSE. 27324546_PAX2 gene silencing can significantly inhibit the process of epithelial-mesenchymal transition (EMT) of renal tubular cells in rats with advanced fibrosis. 27513319_we demonstrated that miR-497 is decreased in human ovarian cancer tissues and cells, and acts as a tumor suppressor. We also confirmed PAX2 is a target gene of miR-497 27722936_The polymorphism rs12266644 of PAX2 might be a risk factor for mullerian duct anomalies in Chinese Han females 27764784_PAX2 is involved in the carcinogenesis of endometrial cancer by stimulating cell growth and promoting cell motility; the overexpression of PAX2 in endometrial cancer is regulated by promoter hypermethylation and the transcription factor MZF1 27991925_Mutant p53 and PTEN loss negatively regulated PAX2 and PAX2 re-expression in high-grade serous ovarian cancer cells induced cell death. 28426529_Diagnostic Utility of Pax8, Pax2, and NGFR Immunohistochemical Expression in Pediatric Renal Tumors. 28674456_PAX2 is dispensable for mesenchymal-to-epithelial transition of nephron progenitors, but is required for morphological development of glomerular parietal epithelial cells, during nephron formation from human iPS cells in vitro. 28855599_The C (minor) allele decreased EPHA2 transcriptional activity relative to the T allele by reducing the binding affinity of PAX2 to the EPHA2 promoter region. 28918284_During the ensuing weeks, the PAX2/FOXA1 boundary progressively extended cranially such that by 21 weeks the entire vaginal epithelium was FOXA1-reactive and PAX2-negative. This observation supports Bulmer's proposal that human vaginal epithelium derives solely from urogenital sinus epithelium 29054766_This case unravels a previously unrecognized phenotype of camptodactyly due to a significant skeletal deformity of Papillorenal syndrome with a heterogeneous PAX2 mutation of hexanucleotide duplication. 29857823_Our data suggested that high expression of PAX2 could be associated with better survival in estrogen receptor positive tamoxifen-treated breast carcinoma patients. 30132866_Our results provide evidence that CP-31398 could inhibit EMT and promote apoptosis of cervical cancer cells to curb cervical cancer tumor growth by downregulating PAX2. 30400021_Decreased PAX2 expression is associated with clinical progression of endometrial intraepithelial neoplasia lesions. PAX2 expression was not prognostic in endometrial endometrioid carcinoma. 31001663_Study findings demonstrate high frequency of PAX2 mutations in familial form of steroid-resistant nephrotic syndrome and further expand the phenotypic spectrum of PAX2 heterozygous mutations to include autosomal dominant childhood-onset focal segmental glomerulosclerosis. 31922217_PAX2 promotes epithelial ovarian cancer progression involving fatty acid metabolic reprogramming. 31983104_Suppression of PAX2 inhibits the growth of the OVACAR-3 cells while PAX2 overexpression could avoid the growth inhibitory effects of miR-375 in OVACAR-3 cells. 32174259_Comparative modeling and structure based drug repurposing of PAX2 transcription factor for targeting acquired chemoresistance in pancreatic ductal adenocarcinoma. 32203253_Clinical and genetic variability of PAX2-related disorder in the Japanese population. 32279431_Our findings suggest that ASH2L participates in the promotion of endometrial cancer progression, if not totally at least partially, via upregulation of PAX2 transcription. 32381599_Pax2 and Pax8 Proteins Regulate Urea Transporters and Aquaporins to Control Urine Concentration in the Adult Kidney. 32776440_Whole genome sequence analysis identifies a PAX2 mutation to establish a correct diagnosis for a syndromic form of hyperuricemia. 32843722_The proapoptotic gene interferon regulatory factor-1 mediates the antiproliferative outcome of paired box 2 gene and tamoxifen. 33509057_PAX2 promoter methylation and AIB1 overexpression promote tamoxifen resistance in breast carcinoma patients. 33654185_Next-generation sequencing in patients with familial FSGS: first report of collagen gene mutations in Tunisian patients. 33828085_Silencing PTEN in the fallopian tube promotes enrichment of cancer stem cell-like function through loss of PAX2. 33907292_Identification of candidate PAX2-regulated genes implicated in human kidney development. 34545858_Reliable Identification of Endometrial Precancers Through Combined Pax2, beta-Catenin, and Pten Immunohistochemistry. 34696790_Phenotypic spectrum and genetics of PAX2-related disorder in the Chinese cohort. 34906559_Identification of of a PAX2 mutation from maternal mosaicism causes recurrent renal disorder in siblings. 35387670_PAX2, PAX8, and PR are correlated with ovarian seromucinous borderline tumor with endometriosis. | ENSMUSG00000004231 | Pax2 | 80.451421 | 0.9711471 | -0.042238219 | 0.32380912 | 1.736875e-02 | 8.951499e-01 | No | Yes | 78.140213 | 14.077487 | 78.050661 | 14.014281 | ||
| ENSG00000076662 | 3385 | ICAM3 | protein_coding | P32942 | FUNCTION: ICAM proteins are ligands for the leukocyte adhesion protein LFA-1 (integrin alpha-L/beta-2) (PubMed:1448173). ICAM3 is also a ligand for integrin alpha-D/beta-2. In association with integrin alpha-L/beta-2, contributes to apoptotic neutrophil phagocytosis by macrophages (PubMed:23775590). {ECO:0000269|PubMed:1448173, ECO:0000269|PubMed:23775590}. | 3D-structure;Cell adhesion;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phagocytosis;Phosphoprotein;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the intercellular adhesion molecule (ICAM) family. All ICAM proteins are type I transmembrane glycoproteins, contain 2-9 immunoglobulin-like C2-type domains, and bind to the leukocyte adhesion LFA-1 protein. This protein is constitutively and abundantly expressed by all leucocytes and may be the most important ligand for LFA-1 in the initiation of the immune response. It functions not only as an adhesion molecule, but also as a potent signalling molecule. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Feb 2016]. | hsa:3385; | extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; integrin binding [GO:0005178]; signaling receptor binding [GO:0005102]; cell adhesion [GO:0007155]; cell-cell adhesion [GO:0098609]; phagocytosis [GO:0006909] | 11784723_A novel serine-rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting 11799126_identification of DC-SIGN binding sites 12021323_interactions with DC-SIGN does not promote DC-SIGN mediated HIV-1 transmission 12571844_Expression of DC-SIGN and its ligand, ICAM-3, is found in substantial amounts only in RA synovium, suggesting that their interaction is implicated in the additional activation of synovial macrophages that leads to the production of EMMPRIN and MMP-1. 12600815_the expression of ICAM-1 might involve both p38 MAPK and NF-kappaB activities, whereas the regulation of CD11b, CD18, and ICAM-3 expressions might be mediated through p38 MAPK but not NF-kappaB. 12743567_ICAM-3 is highly expressed on the surface of human eosinophils and has a role in the downregulation of GM-CSF production. 14704632_Relationship of intercellular adhesion molecule-3 and hepatocyte growth factor with amyloidosis A in chronic renal-failure patients. There was a higher density of intercellular adhesion molecule-3-positive cells in the patients with amyloidosis A. 14726630_ICAM-3 is expressed on human bone marrow endothelial cells and controls endothelial integrity via reactive oxygen species-dependent signaling. 14970226_soluble DC-SIGN bound to gp120-Fc more than 100- and 50-fold better than ICAM-2-Fc and ICAM-3-Fc, respectively. Binding sites are described. 15163761_acts as a costimulating molecule to increase HIV-1 transcription and viral replication, a process allowing productive infection of quiescent CD4+ T lymphocytes. 15880373_Expression of ICAM-3 can be used as a valuable biomarker to predict the radiation resistance in cervical cancer that occurs during radiotherapy. 15958383_the hybrid domain of integrin alphaL beta2 has different requirements of affinity states for ICAM-1 and ICAM-3 binding 17145745_The results suggest that ICAM-3 assists in the interaction of granulocytes with DC-SIGN of dendritic cells. 17570115_Observational study of gene-disease association. (HuGE Navigator) 17570115_Patients with SARS homozygous for ICAM3 Gly143 showed significant association with higher lactate dehydrogenase levels and lower total white blood cell counts. 17591777_talin induced an intermediate affinity alphaLbeta2 that adhered constitutively to its ligand intercellular adhesion molecule ICAM-1 but not ICAM-3. 17913807_Here we demonstrate that leukocyte function-associated antigen 1 (LFA-1), intercellular adhesion molecule 1 (ICAM-1), and ICAM-3 are enriched at the VS and that inhibition of these interactions influences conjugate formation and reduces VS assembly. 18261116_This is the first case of CD20 positive mycosis fungoides involving a lymph node to be reported in the literature. 18354203_extended alpha(L)beta(2) with an open headpiece is required for ICAM-3 adhesion 19225705_Lower expression of ICAM-3 and higher expression of ICAM-1 suggest that AMs may be involved in the pathogenesis of scleroderma. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19801714_Observational study of gene-disease association. (HuGE Navigator) 19801714_no significant risk association was found for SARS infection for the ICAM-3 Asp143Gly SNP. 19898481_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19956847_ICAM-3 enhances the migratory and invasive potential of human non-small cell lung cancer cells by inducing MMP-2 and MMP-9 via Akt and CREB 20086017_CCR1 antagonist, BX471, did not significantly alter ICAM-3 expression in relapsing-remitting multiple sclerosis patients. 20331378_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21239057_Single nucleotide polymorphisms in ICAM3 gene is associated with lymphoma. 21381019_induction of morphological polarization in primary T lymphocytes and Jurkat cells enhances Kidins220/ARMS colocalization with ICAM-3 21712539_the cross-talk between neutrophils and NK cells is mediated by ICAM-3 and CD11d/CD18, respectively. 22117198_ICAM-3 may be an important adhesion molecule involved in chemotaxis to apoptotic human leukocytes. 22205703_the molecular basis of allergen-induced Th2 cell polarization 22396536_analysis of activated apoptotic cells induce dendritic cell maturation via engagement of Toll-like receptor 4 (TLR4), dendritic cell-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN), and beta2 integrins 22479382_Results indicate that the ICAM-3 gene promoter is negatively regulated by RUNX3. 23144795_Intercellular adhesion molecule (ICAM)-3 mRNA is upregulated in non-adherent endothelial forming cells. 23775590_ICAM3 acts as recognition receptors in the phagocytosis portals of macrophages for engulfment of apoptotic neutrophils. 24177012_this data clearly indicate that ICAM-3 promotes drug resistance via inhibition of apoptosis. 24474251_with larger patient groups and preferably detailed histopathological and clinical evaluations, are needed to explain the severity of ICAM-1, ICAM-2, and ICAM-3 molecules in Barrett's esophagus 27552332_Increased expression of PECAM-1, ICAM-3, and VCAM-1 in colonic biopsies from patients with inflammatory bowel disease (IBD) in clinical remission is associated with subsequent flares; this suggests that increases in the expression of these proteins may be early events that lead to flares in patients with IBD 29477378_we identify a potential CSC regulator and suggest a novel mechanism by which ICAM3 governs cancer cell stemness and inflammation. 29671117_Lewis-antigen-containing ICAM-2/3 on Jurkat leukemia cells interact with DC-SIGN to regulate DC functions. 29729315_exploration of the underlying mechanism demonstrated that ICAM3 not only binds to LFA-1 with its extracellular domain and structure protein ERM but also to lamellipodia with its intracellular domain which causes a tension that pulls cells apart (metastasis). 32013031_Changes in the Surface Expression of Intercellular Adhesion Molecule 3, the Induction of Apoptosis, and the Inhibition of Cell-Cycle Progression of Human Multidrug-Resistant Jurkat/A4 Cells Exposed to a Random Positioning Machine. 32371447_Vascular injury biomarkers and stroke risk: A population-based study. 33999358_Association of Circulating ICAM3 Concentrations with Severity and Short-term Outcomes of Acute Ischemic Stroke. | 56.572804 | 0.7705210 | -0.376093838 | 0.36220674 | 1.089304e+00 | 2.966255e-01 | No | Yes | 55.438253 | 9.357446 | 73.916320 | 11.433084 | ||||
| ENSG00000077238 | 3566 | IL4R | protein_coding | P24394 | FUNCTION: Receptor for both interleukin 4 and interleukin 13. Couples to the JAK1/2/3-STAT6 pathway. The IL4 response is involved in promoting Th2 differentiation. The IL4/IL13 responses are involved in regulating IgE production and, chemokine and mucus production at sites of allergic inflammation. In certain cell types, can signal through activation of insulin receptor substrates, IRS1/IRS2. {ECO:0000269|PubMed:8124718}.; FUNCTION: Soluble IL4R (sIL4R) inhibits IL4-mediated cell proliferation and IL5 up-regulation by T-cells. {ECO:0000269|PubMed:8124718}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Glycoprotein;Immunity;Membrane;Phosphoprotein;Receptor;Reference proteome;Secreted;Signal;Transmembrane;Transmembrane helix | This gene encodes the alpha chain of the interleukin-4 receptor, a type I transmembrane protein that can bind interleukin 4 and interleukin 13 to regulate IgE production. The encoded protein also can bind interleukin 4 to promote differentiation of Th2 cells. A soluble form of the encoded protein can be produced by proteolysis of the membrane-bound protein, and this soluble form can inhibit IL4-mediated cell proliferation and IL5 upregulation by T-cells. Allelic variations in this gene have been associated with atopy, a condition that can manifest itself as allergic rhinitis, sinusitus, asthma, or eczema. Polymorphisms in this gene are also associated with resistance to human immunodeficiency virus type-1 infection. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Apr 2012]. | hsa:3566; | centriolar satellite [GO:0034451]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; interleukin-4 receptor activity [GO:0004913]; defense response to protozoan [GO:0042832]; immune response [GO:0006955]; immunoglobulin mediated immune response [GO:0016064]; negative regulation of T-helper 1 cell differentiation [GO:0045626]; ovulation [GO:0030728]; positive regulation of chemokine production [GO:0032722]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of immunoglobulin production [GO:0002639]; positive regulation of macrophage activation [GO:0043032]; positive regulation of mast cell degranulation [GO:0043306]; positive regulation of myoblast fusion [GO:1901741]; positive regulation of T-helper 2 cell differentiation [GO:0045630]; production of molecular mediator involved in inflammatory response [GO:0002532]; regulation of cell population proliferation [GO:0042127]; response to estrogen [GO:0043627]; response to odorant [GO:1990834]; signal transduction [GO:0007165] | 11035134_Observational study of gene-disease association. (HuGE Navigator) 11164908_Observational study of gene-disease association. (HuGE Navigator) 11233912_Observational study of gene-disease association. (HuGE Navigator) 11354638_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11491529_Observational study of gene-disease association. (HuGE Navigator) 11528525_Observational study of gene-disease association. (HuGE Navigator) 11544427_Observational study of gene-disease association. (HuGE Navigator) 11678851_Observational study of gene-disease association. (HuGE Navigator) 11709756_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11709756_gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma 11714803_Tyrosine-phosphorylated IL4R peptides coprecipitate SH2-containing tyrosine phosphatase-1, SH2-containing tyrosine phosphatase-2, and SH2-containing inositol 5'-phosphatase, demonstrating a regulatory role for the ITIM motif in IL-4-induced proliferation. 11786020_high-affinity interaction of human IL-4 and the receptor alpha chain is constituted by two independent binding clusters 11801567_internalization of interleukin 4 receptor alpha increases cytotoxic effect of interleukin 4-receptor-targeted cytotoxin in cancer cells. 11811777_Characterization of IL-4 receptor components expressed on monocytes and monocyte-derived macrophages: variation associated with differential signaling by IL-4 11922633_Observational study of gene-disease association. (HuGE Navigator) 11922633_These results suggest TXA2 receptor polymorphism strongly interacts with IL-4R alpha polymorphism as a major determinant of high serum immunoglobulin E levels in atopic dermatitis. 11991671_Endogenous interferon-alpha production by differentiating human monocytes regulates expression and function of the IL-2/IL-4 receptor gamma chain 12020266_Observational study of gene-disease association. (HuGE Navigator) 12047364_a single gene effect of IL4Ralpha variants or any other gene on chromosome 16 could not be shown in a selected population of children with asthma 12047365_no evidence for linkage of the IL4R gene locus with sarcoidosis 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12100043_Observational study of gene-disease association. (HuGE Navigator) 12115161_Observational study of gene-disease association. (HuGE Navigator) 12133990_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12171893_Observational study of gene-disease association. (HuGE Navigator) 12401728_association of haplotype with type 1 diabetes 12420205_Observational study of gene-disease association. (HuGE Navigator) 12422339_Observational study of gene-disease association. (HuGE Navigator) 12442007_Observational study of gene-disease association. (HuGE Navigator) 12454749_up-regulated 5-fold in B cell chronic lymphocytic leukemia cells, probably leading to increased responsiveness to IL4 and resistance to apoptosis 12459556_role of Tyr-713 in the interleukin-4 receptor alpha in regulating dephosphorylation of Stat6 12508140_Observational study of gene-disease association. (HuGE Navigator) 12558814_Observational study of genotype prevalence. (HuGE Navigator) 12594065_Allele frequency of IL4RA polymorphism was associated with rapid decline of lung function in smokers. 12594065_Observational study of gene-disease association. (HuGE Navigator) 12743452_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12748907_IL4R is associated with diabetes mellitus, type 1 in Filipinos. 12748907_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12753568_Observational study of gene-disease association. (HuGE Navigator) 12871855_Th2 cytokines enhance TARC expression in human airway smooth muscle cells in IL-4Ralpha genotype-dependent fashion. 12897746_No genotypic effects on total serum IgE levels. 12897746_Observational study of gene-disease association. (HuGE Navigator) 12900808_Observational study of gene-disease association. (HuGE Navigator) 12940513_IL-4R alpha chain 576R/R genotypes confer genetic susceptibility to allergic asthma in Chinese. 12940513_Observational study of gene-disease association. (HuGE Navigator) 12973929_Observational study of gene-disease association. (HuGE Navigator) 14523823_Observational study of gene-disease association. (HuGE Navigator) 14523823_The prevalence of the mutant variant of the IL-4ra gene was lower in neonates with necrotizing enterocolitis (NEC) compared with those without NEC, suggesting that this mutation might protect against the development of NEC in VLBW infants. 14615367_Observational study of gene-disease association. (HuGE Navigator) 14745651_Observational study of gene-disease association. (HuGE Navigator) 14745651_functional variants within the IL4R gene predispose to hip osteoarthritis in Caucasian females 14984008_Observational study of gene-disease association. (HuGE Navigator) 14984008_investigated the frequency and genotypes of S503P and Q576R SNPs and their association with traits of allergic asthma in a Hawaii population 15005726_Observational study of gene-disease association. (HuGE Navigator) 15007345_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15007345_polymorphisms of IL1A (G/T at +4845) and IL4RA (T/C at +22446) show an epistatic effect on the risk of atopy 15007352_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15007352_variants in the IL4, IL13, and IL4RA genes play an important role in controlling specific IgE response in atopy 15021309_Observational study of gene-disease association. (HuGE Navigator) 15057902_Observational study of gene-disease association. (HuGE Navigator) 15122773_Observational study of gene-disease association. (HuGE Navigator) 15161635_Certain interstitial pneumonia patients can be modulated in a manner that is dependent on the IL-4 and IL-13 receptor subunit expression by these cells. 15170937_Observational study of gene-disease association. (HuGE Navigator) 15194285_Observational study of gene-disease association. (HuGE Navigator) 15298559_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15356556_V50R551 IL-4R alpha variant has enhanced function alone, but with Q110 IL-13 variant the 2 have a synergistic effect on IL-13-dependent gene induction. 15361128_Observational study of genotype prevalence. (HuGE Navigator) 15367225_Observational study of gene-disease association. (HuGE Navigator) 15479272_Observational study of gene-disease association. (HuGE Navigator) 15497451_Observational study of gene-disease association. (HuGE Navigator) 15497451_Polymorphisms of IL-4R previously associated with other immune mediated diseases, do not confer susceptibility to Graves' disease in white Caucasians in the United Kingdom. 15521376_differences in the potency of IL-13- and IL-4-mediated induction of eotaxin-3 might be explained by expression of types 1 and 2 IL-4 receptors in bronchial epithelium 15564773_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15566952_Observational study of gene-disease association. (HuGE Navigator) 15660293_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15712015_Observational study of gene-disease association. (HuGE Navigator) 15733644_Observational study of gene-disease association. (HuGE Navigator) 15737301_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15842262_Observational study of gene-disease association. (HuGE Navigator) 15969687_The IL-4RalphaQ576R polymorphism may involve in the development of penicillins allergy, and through modulating specific serum IgE levels. 15981567_Observational study of gene-disease association. (HuGE Navigator) 16024651_Observational study of gene-disease association. (HuGE Navigator) 16046318_Observational study of gene-disease association. (HuGE Navigator) 16100774_Observational study of genotype prevalence. (HuGE Navigator) 16184405_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16189667_IL$R alpha V50 homozygosity associates with slow progression and that exon 12 U-haplotypes might be associated with both susceptibility to infection via parenteral route and resistance to infection via sexual exposure. 16189667_Observational study of gene-disease association. (HuGE Navigator) 16226465_The soluble receptor balance (sIL-2R/sIL-4R) in patients with severe hemolytic uremic syndrome may shift, depending on the disease state of the patients 16264039_In conclusion, these results suggest that viral airway infection may enhance interleukin-4-induced eotaxin-3 production through upregulation of the interleukin-4 receptor in airway epithelial cells. 16280132_Observational study of gene-disease association. (HuGE Navigator) 16313303_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16313681_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16387595_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16403098_Observational study of gene-disease association. (HuGE Navigator) 16503977_Observational study of gene-disease association. (HuGE Navigator) 16538488_No association between type 1 diabetes and any SNP or haplotype was found by the transmission disequilibrium test. 16538488_Observational study of gene-disease association. (HuGE Navigator) 16551465_Observational study of gene-disease association. (HuGE Navigator) 16551465_This is the first demonstration of sex-specific association of the two foremost genes of the IL-4 signalling cascade with chronic inflammatory arthropathies. 16600026_Observational study of gene-disease association. (HuGE Navigator) 16613701_Observational study of gene-disease association. (HuGE Navigator) 16625214_Multifactor dimensionality reduction (MDR) test was applied to detect epistasis, and identified single-IL4R(Q576R)- and three-IL4R(Q576R), IL5RA(-80), CD14(-260)- locus association models that predict multiple sclerosis risk with 75-76% accuracy. 16625214_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16640778_A structure/function analysis of the IL-4 ligand-receptor interaction, using various mutations of both the ligand and the receptor, is reported. 16646030_Observational study of gene-disease association. (HuGE Navigator) 16681592_Observational study of gene-disease association. (HuGE Navigator) 16699452_Observational study of gene-disease association. (HuGE Navigator) 16750991_Observational study of gene-disease association. (HuGE Navigator) 16757160_Observational study of gene-disease association. (HuGE Navigator) 16842617_Observational study of gene-disease association. (HuGE Navigator) 16867043_Observational study of gene-disease association. (HuGE Navigator) 16914241_Observational study of gene-disease association. (HuGE Navigator) 16917945_Results show sequence variations as a possible way of altering alternative splicing selection of IL4R in vivo. 16938461_Observational study of gene-disease association. (HuGE Navigator) 16950281_Observational study of gene-disease association. (HuGE Navigator) 16961803_Observational study of gene-disease association. (HuGE Navigator) 17001290_Observational study of gene-disease association, gene-gene interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17006724_Homozygotes with the low activity allele of the A398G polymorphism in the IL4R gene had a modest increase in risk of atrophic gastritis (OR = 1.52, 95% CI: 1.05-2.21), compared with homozygotes of the high activity allele. 17006724_Observational study of gene-disease association. (HuGE Navigator) 17045041_Observational study of gene-disease association. (HuGE Navigator) 17045041_The Ile50Val polymorphism of IL-4R alpha gene is not associated with bronchial asthma in Han nationality patients. 17083349_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17083349_Variants in the IL4RA gene alone may not exert any major influence on susceptibility to asthma-related diseases in childhood, but in combination with other genes, such as IL9R, IL4RA may be an important gene for disease susceptibility 17091279_Observational study of gene-disease association. (HuGE Navigator) 17115186_Observational study of gene-disease association. (HuGE Navigator) 17121586_Observational study of gene-disease association. (HuGE Navigator) 17143971_Observational study of gene-disease association. (HuGE Navigator) 17143971_Our findings of higher frequency of IL4 and IL4RA genotypes and alleles with rheumatoid arthritis. 17152005_Observational study of gene-disease association. (HuGE Navigator) 17170387_Observational study of gene-disease association. (HuGE Navigator) 17170387_SNPs in IL4Ralpha, which are more common in African Americans, are associated with severe asthma exacerbations, lower lung function, and increased mast cell-related tissue inflammation 17179726_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17201240_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17209513_Observational study of gene-disease association. (HuGE Navigator) 17257312_Observational study of genotype prevalence. (HuGE Navigator) 17258448_Observational study of gene-disease association. (HuGE Navigator) 17284225_Observational study of gene-disease association. (HuGE Navigator) 17291854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17303794_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17315188_Observational study of gene-disease association. (HuGE Navigator) 17362266_Observational study of gene-disease association. (HuGE Navigator) 17420820_IL4+33 and IL4R*Q551 polymorphisms may have a promoting role of TH2 mediators in African MS descendants. IL4 and IL4R genes are susceptibility factors for Brazilian MS but may be able to modify ethnicity-dependent disease risk and penetrance. 17420820_Observational study of gene-disease association. (HuGE Navigator) 17444864_Observational study of gene-disease association. (HuGE Navigator) 17523428_Observational study of gene-disease association. (HuGE Navigator) 17536219_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17548690_IL4 and IL4R polymorphisms and haplotypes were neither significantly associated with IgE levels in controls nor associated with glioma status 17548690_Observational study of gene-disease association. (HuGE Navigator) 17586032_A meta-analysis of results from case-control studies strongly supports the conclusion that the R551 IL4R variant imparts a modest yet significant risk for atopic asthma 17586032_Meta-analysis of gene-disease association. (HuGE Navigator) 17600229_Observational study of gene-disease association. (HuGE Navigator) 17627763_Observational study of genotype prevalence. (HuGE Navigator) 17688234_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17688234_The IL-4R 75V variant was associated with increased risk of cervical tumors, cases homozygote for 75V had an odds ratio of 1.91 (1.27-2.86) with a tendency that the association was stronger in noncarriers of the DQB1*0602 allele. 17703412_Observational study of gene-disease association. (HuGE Navigator) 17714919_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17823973_IL4R gene polymorphisms may have roles in asthma in Chinese populations 17823973_Observational study of gene-disease association. (HuGE Navigator) 17889143_Observational study of gene-disease association. (HuGE Navigator) 17900677_analysis of polymorphisms of Ile50Val (rs.1805010), Ser478Pro (rs.1805015), and Gln551Arg (rs.1801275) in the IL4R gene between Japanese patients with Stevens-Johnson syndrome and Japanese healthy volunteers 17901851_IL-4R overexpression on newborns' monocytes and lymphocytes could be an early risk marker of allergy development. 17988330_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17988330_The IL-4R Ile50/Ile50 and IL-10R2 G520/G520 and G520/A520 genotypes were shown to determine the susceptibility to SLE(systemic lupus erythematosus)in a Chinese population 18006935_Observational study of gene-disease association. (HuGE Navigator) 18031948_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18031948_we studied the association of variants in IL-4 (C-589T, G3017T) and IL-4R alpha (Gln576Arg) with allergies and with risk of pancreatic cancer 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18179773_IL-4 and IL-4RA gene polymorphisms concur in selecting the H. pylori infecting strain, probably influencing the IL-4 signalling pathway 18179773_Observational study of gene-disease association. (HuGE Navigator) 18263811_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18263811_combined polymorphisms in the IL-13/IL-4R signaling pathway were associated with SJS/TEN with ocular surface complications. 18389618_Observational study of gene-disease association. (HuGE Navigator) 18425216_Observational study of gene-disease association. (HuGE Navigator) 18433792_Analyses of genotype distributions and allele frequencies of study subjects revealed that rs 180275 polymorphism in IL4R was associated with an increase in BMI in Korean population. 18433792_Observational study of gene-disease association. (HuGE Navigator) 18448485_Observational study of gene-disease association. (HuGE Navigator) 18538381_Observational study of gene-disease association. (HuGE Navigator) 18547691_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18576348_Observational study of gene-disease association. (HuGE Navigator) 18576348_our data indicate that the AA genotype of the IL4R 150V SNP is associated with joint erosions in psoriatic arthritis 18610831_Observational study of gene-disease association. (HuGE Navigator) 18610832_Observational study of gene-disease association. (HuGE Navigator) 18625055_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18625055_There is a lack of association or interactions between the IL-4, IL-4Ralpha and IL-13 genes, and rheumatoid arthritis. 18628242_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18632425_Observational study of genotype prevalence. (HuGE Navigator) 18633131_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18636124_Observational study of gene-disease association. (HuGE Navigator) 18674658_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18691306_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18715339_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18773331_Observational study of gene-disease association. (HuGE Navigator) 18774776_This study demonstrated upregulation of type 1 IL-4R by IFN-gamma, which resulted in enhanced IL-4-induced production of CCL26 from keratinocytes. 18781131_Observational study of gene-disease association. (HuGE Navigator) 18809513_The urine evaluation of the balance between IFN-gammaR1 and IL4R receptors might be informative for the immune states of HUS patients. 18813073_Observational study of gene-disease association. (HuGE Navigator) 18818748_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18849614_Observational study of gene-disease association. (HuGE Navigator) 18849614_Single nucleotide polymorphisms of IL4RA are not associated with olive pollen allergy or asthma, but the interaction between IL4RA I50V/Q551R was strongly associated with the asthma phenotype. 18927306_Observational study of gene-disease association. (HuGE Navigator) 18927306_Polymorphisms in the interleukin-4 receptor gene are associated with better survival in patients with glioblastoma 18931892_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18936436_Observational study of genotype prevalence. (HuGE Navigator) 18957266_data indicate inactivation of receptor-associated protein tyrosine phosphatase activity by cytokine-generated reactive oxygen species is a physiologic mechanism for amplification of IL-4 receptor activation revealing a role for ROS in cytokine crosstalk 18974840_Observational study of gene-disease association. (HuGE Navigator) 19011907_Observational study of gene-disease association. (HuGE Navigator) 19019335_Observational study of gene-disease association. (HuGE Navigator) 19028820_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19109239_IL-4 activates signaling pathways through type I IL-4Rs qualitatively differently from IL-13, which cooperate to induce optimal gene expression. 19117745_Observational study of gene-disease association. (HuGE Navigator) 19131662_Meta-analysis of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19263529_Observational study of gene-disease association. (HuGE Navigator) 19264973_Observational study of gene-disease association. (HuGE Navigator) 19279357_IL4RA (1902/G:G) polymorphism was found to be positively (susceptible) associated with chronic obstructive pulmonary disease in Macedonians. 19332120_Observational study of gene-disease association. (HuGE Navigator) 19382262_Observational study of gene-disease association. (HuGE Navigator) 19382262_The present study for the first time suggests an association of IL-4Ralpha I50 allele with increased likelihood of HIV-1 infection in North Indian population. 19408823_Observational study of genotype prevalence. (HuGE Navigator) 19414811_IL4Ralpha is up-regulated in both myeloid populations but its presence correlates with an immunosuppressive phenotype only when mononuclear cells, but not granulocytes, of tumor-bearing patients are considered. 19420105_Observational study of gene-disease association. (HuGE Navigator) 19421745_Observational study of gene-disease association. (HuGE Navigator) 19421745_The IL4Ralpha Q576 allele is related to penicillin allergy, and the IL-4Ralpha I75 allele is associated with the symptom of urticaria. The Q576/I75 haplotype may be related with an allergy to penicillin. 19447482_These results suggest that day care attendance is associated with serum IgE levels, and this effect is modified by CD14-550C/T and IL4R Ile50Val polymorphisms. 19470040_Observational study of gene-disease association. (HuGE Navigator) 19479237_Observational study of gene-disease association. (HuGE Navigator) 19505916_Observational study of gene-disease association. (HuGE Navigator) 19508433_Observational study of gene-disease association. (HuGE Navigator) 19515749_Observational study of gene-disease association. (HuGE Navigator) 19515749_These data support the role for host germline gene variations of immunologically important factors like the IL4R I75V gene variation to predict the survival in diffuse large B-cell lymphoma patients. 19527514_Observational study of gene-disease association. (HuGE Navigator) 19531027_Data show that IL-4 receptors are functionally competent in pancreatic beta-cells and that they signal via PI3K and JAK/STAT pathways. 19544559_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19548368_IL4Ralpha could be a candidate gene for assessing the risk of renal cell carcinoma. 19548368_Observational study of gene-disease association. (HuGE Navigator) 19555572_Observational study of gene-disease association. (HuGE Navigator) 19559392_Observational study of gene-disease association. (HuGE Navigator) 19573080_Observational study of gene-disease association. (HuGE Navigator) 19590686_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19594368_Observational study of genotype prevalence. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19657898_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19674346_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19683555_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19729601_Observational study of gene-disease association. (HuGE Navigator) 19739012_The urinary soluble IL-4 receptor levels did not correlate with the clinical severity of vesicoureteral reflux 19773279_Observational study of gene-disease association. (HuGE Navigator) 19773451_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19783684_association of Syk to IL-4R(alpha) is of biological significance and IL-4R(alpha) is a new candidate to be added to the few cytokine receptor components which associate with Syk. 19819209_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19860911_Observational study of gene-disease association. (HuGE Navigator) 19862939_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19863607_Observational study of gene-disease association. (HuGE Navigator) 19863607_The results suggest that the genetic polymorphisms of IL-4R and TGF-beta1 are associated with the risk of colorectal cancer in a Korean population. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19956098_38 single-nucleotide polymorphisms in IL4R were genotyped using the Sequenom iPLEX Gold MassARRAY technology in 2042 multiplex families from nine cohorts. 20002627_Observational study of gene-disease association. (HuGE Navigator) 20002627_Variations within the IL4R gene are associated with allergic diseases in children, preferably with eczema and disease phenotypes of ARC and asthma. 20040389_Data show that flavones inhibited the phosphorylation of STAT6, janus kinase 3, and IL-4Ralpha, whereas IL-4 signaling mediated through type II IL-4R was unaffected by flavones. 20072140_Observational study of gene-disease association. (HuGE Navigator) 20085599_Observational study of gene-disease association. (HuGE Navigator) 20085599_Significant associations of single nucleotide polymorphisms with wheeze in the past year were detected in only four genes (IL4R, TLR4, MS4A2, TLR9). Variants in IL4R and TLR4 were also related to allergen-specific IgE. 20128416_association between promoter polymorphisms and asthma in Iranian patients 20140262_Observational study of gene-disease association. (HuGE Navigator) 20163933_Observational study of gene-disease association. (HuGE Navigator) 20176658_Proliferation appears to be directly mediated via IL4Ralpha on the epithelial tumor cells 20213229_Observational study of gene-disease association. (HuGE Navigator) 20219689_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20298583_Observational study of gene-disease association. (HuGE Navigator) 20299965_Observational study of gene-disease association. (HuGE Navigator) 20378664_Observational study of gene-disease association. (HuGE Navigator) 20394509_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20416077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20417488_Observational study of gene-disease association. (HuGE Navigator) 20424473_Observational study of gene-disease association. (HuGE Navigator) 20444266_IL4R SNPs, rs1801275 and rs1805010, are associated with rheumatoid nodules in autoantibody-positive African-American rheumatoid arthritis patients with at least one HLA-DRB1 allele encoding the shared epitope. 20444266_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20452482_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20459687_Observational study of gene-disease association. (HuGE Navigator) 20484924_IL4R polymorphisms were associated with asthma in the asthma 20484924_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20503287_Observational study of gene-disease association. (HuGE Navigator) 20517665_Observational study of gene-disease association. (HuGE Navigator) 20536507_Observational study of gene-disease association. (HuGE Navigator) 20568250_Observational study of gene-disease association. (HuGE Navigator) 20603037_Observational study of gene-disease association. (HuGE Navigator) 20603050_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20654748_Observational study of gene-disease association. (HuGE Navigator) 20673868_Observational study of gene-disease association. (HuGE Navigator) 20703737_Observational study of gene-disease association. (HuGE Navigator) 20716621_Observational study of gene-disease association. (HuGE Navigator) 20729386_Data show differences in IL-4/IL-13 receptor subunit expression and responsiveness to IL-4 based on the extent of airway epithelial cell differentiation and suggest that these differences may have functional consequences in airway inflammation. 20811626_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21054877_Observational study of gene-disease association. (HuGE Navigator) 21061265_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21070662_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21071541_Observational study of gene-disease association. (HuGE Navigator) 21211370_IL-4RA Q576R polymorphisms impart a modest yet significant risk for asthma. 21251883_We found that levels of sIL-4R were strikingly reduced in all forms of TB, particularly meningeal. 21489615_Single nucleotide polymorphisms of CD14, IL-13 and IL-4 receptor are associated with hexamethylene diisocyanate asthma. 21497535_IL-4Ralpha polymorphisms were associated with susceptibility to rheumatoid arthritis and may be helpful in early detection of erosive rheumatoid arthritis. 21536546_IL-4R drives dedifferentiation, mitogenesis, and metastasis in rhabdomyosarcoma. 21729106_IL4RA polymorphism may make a modest but significant contribution to a person's genetic susceptibility to chagasic cardiomyopathy. 21733492_The Q551R IL-4RA polymorphism is a putative risk indicator for severe chronic periodontitis, but was not significant associated to AP. 21734400_Studies indicate that RAD50 and PTPRE of crude associations with asthma at a Bonferroni-corrected level of significance, while IL4R, CCL5 and TBXA2R of nominal significance. 21913997_did not identify any SNPs in the IL-4, IL-4R and IL-13Ralpha2 genes that were associated with atopic dermatitis 21918367_Interactions of the IL-4RA loci may play a role both conferring susceptibility and modulating severity of Atopic dermatitis (AD). 22045834_Data showed that polymorphisms in IL4, IL13, and their receptors do not play a role in SSc and do not influence the exp | ENSMUSG00000030748 | Il4ra | 335.078009 | 0.8908830 | -0.166692077 | 0.15450108 | 1.170885e+00 | 2.792195e-01 | No | Yes | 310.469020 | 36.345008 | 349.158088 | 40.550569 | ||
| ENSG00000079102 | 862 | RUNX1T1 | protein_coding | Q06455 | FUNCTION: Transcriptional corepressor which facilitates transcriptional repression via its association with DNA-binding transcription factors and recruitment of other corepressors and histone-modifying enzymes (PubMed:12559562, PubMed:15203199, PubMed:10688654). Can repress the expression of MMP7 in a ZBTB33-dependent manner (PubMed:23251453). Can repress transactivation mediated by TCF12 (PubMed:16803958). Acts as a negative regulator of adipogenesis (By similarity). The AML1-MTG8/ETO fusion protein frequently found in leukemic cells is involved in leukemogenesis and contributes to hematopoietic stem/progenitor cell self-renewal (PubMed:23812588). {ECO:0000250|UniProtKB:Q61909, ECO:0000269|PubMed:10688654, ECO:0000269|PubMed:10973986, ECO:0000269|PubMed:16803958, ECO:0000269|PubMed:23251453, ECO:0000269|PubMed:23812588, ECO:0000303|PubMed:12559562, ECO:0000303|PubMed:15203199}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a member of the myeloid translocation gene family which interact with DNA-bound transcription factors and recruit a range of corepressors to facilitate transcriptional repression. The t(8;21)(q22;q22) translocation is one of the most frequent karyotypic abnormalities in acute myeloid leukemia. The translocation produces a chimeric gene made up of the 5'-region of the runt-related transcription factor 1 gene fused to the 3'-region of this gene. The chimeric protein is thought to associate with the nuclear corepressor/histone deacetylase complex to block hematopoietic differentiation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2010]. | hsa:862; | nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor binding [GO:0140297]; metal ion binding [GO:0046872]; transcription corepressor activity [GO:0003714]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of transcription, DNA-templated [GO:0045892]; transcription, DNA-templated [GO:0006351] | 11869944_no conclusive evidence as yet that the AML1/ETO chimeric gene is sufficient per se to induce leukemia. 11983111_Analysis of nuclear distribution of the AML1/ETO protein and its homology domains led to identification of domains in ETO responsible for intranuclear transport and subnuclear distribution of AML1/ETO. 11986950_Potential involvement of the AML1-MTG8 fusion protein in the granulocytic maturation characteristic of the t(8;21) acute myelogenous leukemia cell 12393523_data demonstrate the capacity of AML1-ETO to promote the self-renewal of human hematopoietic cells and therefore support a causal role for t(8;21) translocations in leukemogenesis 12427969_2 independent subnuclear targeting signals in the N- and C-terminal regions of ETO direct ETO to the same binding sites occupied by AML1ETO. This provides a molecular basis for aberrant subnuclear targeting of AML1ETO, the defect in t(8;21)-related AML. 12557226_ETO rearrangements leading to the AML1-ETO fusion gene are frequently the result of small hidden interstitial insertions. 12773394_In t(8;21) leukaemic cells expressing the aberrant fusion protein AML1-ETO, we demonstrate that this protein is part of a transcription factor complex binding to extended sequences of the c-FMS intronic regulatory region rather than the promoter. 12874834_Data identify ETO as a partner for Gfi-1 and Gfi-1B, and suggest that Gfi-1 proteins repress transcription through recruitment of histone deacetylase-containing complexes. 14551142_ETO is a bona fide corepressor that links the transcriptional pathogenesis of acute leukemias and B-cell lymphomas and offers a compelling target for transcriptional therapy of hematologic malignancies. 14751048_RT-PCR for the detection of AML1/ETO in children with acute non-lymphoid leukemia (ANLL) was quick, convenient and sensitive, and could be regarded as a useful method for the diagnosis and prognosis of ANLL. 15203865_Present in acute myeloid leukemia of M2 subtype. 15295650_cloning, expression, purification and crystallization of NHR3 domain 15298716_These findings suggest a central role of RUNX1-CBFA2T1 in the maintenance of the leukaemia. 15333839_study identifes E proteins as AML1-ETO targets whose dysregulation may be important for t(8;21) leukemogenesis 15377655_N-CoR utilizes repression domains I and III for interaction and co-repression with ETO 15676213_DiffeRential expression of the ETO homologues suggests that they may have a potential role in hematopoietic differentiation. 15723339_Alternative splicing in the AML1-MTG8 fusion gene occurs in leukemia cell lines as well as in cells of leukemia paatients with a(8;21) translocation. 15735013_Results suggest a novel mechanism for gene silencing mediated by RUNX1/MTG8 and support the combination of HDAC and DNMT inhibitors as a novel therapeutic approach for t(8;21) AML. 15829516_AML1 and ETO are fused together at the t8;21 translocation breakpoint, resulting in the expression of a chimeric protein called AML1-ETO, which acts as a constitutive transcriptional repressor. 16502583_Translocations in acute myeloid leukemia (AML) is the t(8;21) is characterized by AML1-MTG8 (ETO) mutation. 16616331_Oligomerization is also required for AML1/ETO's interactions with ETO, MTGR1, and MTG16, but not with other corepressor molecules. 16652140_Suppression of AML1/MTG8 results in the increased expression of genes associated with myeloid differentiation, such as AZU1, BPI, CTSG, LYZ and RNASE2 as well as of antiproliferative genes such as IGFBP7, MS4A3 and SLA both in blasts and in cell lines 16741927_Conditional expression of AML1-ETO by the ecdysone-inducible system dramatically increases the expression of connexin 43 together with growth arrest at G1 phase in leukemic U937 cells. 16892037_AML1T1, an alternatively spliced isoform of the t(8;21) transcript, promotes leukemogenesis. 16990610_The leukemia-associated fusion protein AML1-ETO could aberrantly transactivate the EEN gene through an AML1 binding site. 17058450_As a result, we identified 14 unique proteins deregulated in AML1-ETO-carrying leukemic cells, including 3 up-regulated such as hairy and enhancer of split 5 (HES5) and 11 down-regulated such as MAT1 (menage a trois-1. 17244680_AML1/ETO participates in a protein complex with the RA receptor alpha (RARalpha) at RA regulatory regions on RARbeta2. 17284535_Loss of p21(WAF1) facilitates AML1-ETO-induced leukemogenesis. 17560011_Presence of AML1-ETO fusion protein increases the susceptibility of cells to chemical carcinogens, which favors the development of additional genetic alterations. 17625612_AML1-ETO and JAK2 may have a role in leukemogenesis, as shown by a myeloproliferative syndrome progressing to acute myeloid leukemia [case report] 17649722_Isoform AML1/ETO9a was correlated to acute myeloid leukemia-M2 subtype. 17875504_Our study suggests that KIT activating mutations in AML with t(8; 21) are associated with diminished CD 19 and positive CD56 expression on leukemic blasts and, thus, can be phenotypically distinguished from AML1-ETO leukemias 17989718_constitutively and overexpressed AML1-ETO protein was cleaved to four fragments of 70, 49, 40 and 25 kDa by activated caspase-3 during apoptosis induction by extrinsic mitochondrial and death receptor signaling pathways. 17996649_miR-223 is a direct transcriptional target of AML1/ETO 18039847_MTG8/ETO and Mtg16 (ETO2) associated with TCF4 18156164_These results demonstrate that scl is an important mediator of the ability of AML1-ETO to reprogram hematopoietic cell fate decisions, suggesting that scl may be an important contributor to AML1-ETO-associated leukemia. 18183572_VP-16 causes the ETO gene repositioning which allows AML1 and ETO genes to be localized in the same nuclear layer; data corroborate the so called 'breakage first' model of the origins of recurrent reciprocal translocation 18205948_The interaction between SIN3B and ETO required an intact amino-terminus of ETO and the NHR2 domain. 18258796_Knockdown of TLE1 or TLE4 levels increased the rate of cell division of the AML1-ETO-expressing Kasumi-1 cell line, whereas forced expression of either TLE1 or TLE4 caused apoptosis and cell death. 18332109_In functional assays, corepressor ETO, but not AML1/ETO, augments SHARP-mediated repression in an histone deacetylase-dependent manner. 18519037_AML1-ETO promotes leukemogenesis by blocking cell differentiation through inhibition of Sp1 transactivity. 18548094_RUNX1/RUNX1T1 siRNAs compromise the engraftment and/or self-renewal capacities of t(8;21) leukaemia-initiating cells. 18586123_ETO family member-mediated oligomerization and repression can be distinct events and that interaction between ETO family members and hSIN3B or N-CoR may not necessarily strengthen transcriptional repression. 18687517_survivin gene acts as a critical mediator of AML1/ETO-induced late oncogeneic events. 18952841_the crucial role of the NHR4 domain in determination of cellular fate during AML1-ETO-associated leukemogenesis. 19001502_novel role for the leukemia-related AML1-ETO protein in epigenetic control of cell growth through upregulation of ribosomal gene transcription mediated by RNA Pol I. 19043539_a major role for the functional interaction of AML1/ETO with AML1 and HEB in transcriptional regulation determined by the fusion protein. 19100510_Four copies of RUNX1T1 were found. 19179469_CBFbeta is essential for TEL-AML1's ability to promote self-renewal of B cell precursors in vitro. 19289505_Data show that E proteins contain another conserved ETO-interacting region, termed DES, and that differential associations with AD1 and DES allow ETO to repress transcription through both chromatin-dependent and chromatin-independent mechanisms. 19448675_Findings indicate a role for replication-independent pathways in RUNX and RUNX1-ETO senescence, and show that the context-specific oncogenic activity of RUNX1 fusion proteins is mirrored in their distinctive interactions with fail-safe responses. 19458628_AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) AML-M2. 19581587_CalpainB is required for AML1-ETO-induced blood cell disorders in Drosophila.[AML1-ETO] 20090777_NHR4 domain mutations of ETO are probably very infrequent in AML1-ETO positive myeloid leukemia cells. 20430957_Existence of an essential structural motif (hot spot) at the NHR2 dimer-tetramer interface, suitable for a molecular intervention in t(8;21) leukemias. 20487545_The critical role for an evolutionary conserved GATA binding site in transcriptional regulation of the ETO gene in cells of erythroid/megakaryocytic potential. 20688956_Expression of AML1-ETO caused an expansion of hematopoietic precursors in Drosophila, which expressed high levels of reactive oxygen species 20708017_ETO nervy homology region (NHR) 3 domain-PKA(RIIalpha) protein interaction does not appear to significantly contribute to AML1-ETO's ability to induce leukemia. 21200020_N-Ras(G12D) induces features of stepwise transformation in preleukemic umbilical cord blood cultures expressing the AML1-ETO fusion gene. 21245488_Elevated levels of AES mRNA and protein were confirmed in AML1/ETO-expressing leukemia cells, as well as in other acute myeloid leukemia specimens. 21499216_Data show that low RUNX1T1 expression was highly associated with hepatic metastases. 21540640_SMAD4 knockdown accelerated this re-silencing process, suggesting that normal TGF-beta signaling is essential for the maintenance of RunX1T1 expression 21571369_RUNX1T1 point mutation may be rare and passenger mutations in acute leukemias, lung and breast cancers 21764752_study found AML1-ETO is acetylated by the transcriptional coactivator p300 in leukemia cells isolated from t(8;21) AML patients, and this acetylation is essential for its self-renewal-promoting effects in cord blood CD34(+) cells and its leukemogenicity 22025082_Submicroscopic deletion of RUNX1T1 gene confirmed by high-resolution microarray in acute myeloid leukemia with RUNX1/RUNX1T1 rearrangement 22101339_The cooperative effect of the expression of mutated KIT and AML1-ETO oncogenes is crucial for selective toxic action of binase on malignant cells. 22201794_In this review, we detail the structural features, functions, and models used to study both RUNX1 and RUNX1-ETO in hematopoiesis over the past two decades 22343733_selective removal of RUNX1/ETO leads to a widespread reversal of epigenetic reprogramming and a genome-wide redistribution of RUNX1 binding, resulting in the inhibition of leukemic proliferation and self-renewal, and the induction of differentiation. 22498736_PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. 22995345_Study showed that in a widely used AML cell line, Kasumi-1, the chimeric AML1/ETO gene, is transcribed from the promoter P2 of AML1 gene, and this process does not depend on the formation of distant interactions between this promoter and the fused segment of ETO gene. 23223432_Our results indicated a feedback circuitry involving miR-193a and AML1/ETO/DNMTs/HDACs, cooperating with the PTEN/PI3K signaling pathway and contributing to leukemogenesis. 23426948_Importance of the degree of dysregulation by AML1-ETO in cellular transformation. 24402164_t(8;21)/RUNX1-RUNX1T1-positive AML shows a high frequency of additional genetic alterations. 24456692_The AML1/ETO target gene LAT2 interferes with differentiation of normal hematopoietic precursor cells. 24586690_Runx1t1 epigenetically regulates the proliferation and nitric oxide production of microglia. 24616160_Discordant MRD results were observed in 10/71 (14%) of the samples: in three samples from two patients who relapsed, RUNX1-RUNX1T1 was detectable only on DNA, while RUNX1-RUNX1T1 was detectable only on RNA in seven samples. 24727677_A reduction in KIT mRNA levels was also observed in AE-silenced cells, but silencing KIT expression reduced cell growth but did not induce apoptosis. 24783204_Our data shows that upregulated of the RUNX1-RUNX1T1 gene set maybe an important factor contributing to the etiology of ccRCC. 24897507_RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors are rescued by activating c-Kit mutations 24973361_identify a high-frequency mutation in t(8;21)/RUNX1-RUNX1T1 acute myekoid leukemia (AML) and identify the need for future studies to investigate the clinical and biological relevance of ASXL2 mutations in this unique subset of AML 24976338_RUNX1T1 gene may participate in t(8;21)(q22;q22)-dependent leukemic transformation due to its multiple interactions in cell regulatory network particularly through synergistic or antagonistic effects in relation to activity of RUNX1-RUNX1T1 fusion gene. 25082877_minimal residual disease determined by RUNX1/RUNX1T1 transcript levels could identify allogeneic hematopoietic stem cell transplantation t(8;21) (q22;q22) acute myeloid leukemia patients who are at high risk for relapse, together with c-KIT mutations 25101977_In t(8;21) acute myelogenous leukemia (AML), the acquisition of AML1/ETO is not sufficient, and the subsequent upregulation of AML1/ETO and the additional c-KIT mutant signaling are critical steps for transformation into leukemic stem cells. 25928846_A substantial fraction of AML1-ETO/p300 co-localization occurs near TSSs in promoter regions associated with transcriptionally active loci. 26320575_Exon splicing patterns in the human RUNX1-RUNX1T1 fusion gene present in acute myeloid leukemia patients have been decoded. 26546158_AML1/ETO transactivates gene expression through recruiting AP-1 to the AML1/ETO-AML1 complex. 26706127_Data suggest that biosynthesis and folding of leukemogenic fusion oncoprotein AML1-ETO/RUNX1-RUNX1T1 is facilitated by interaction with the chaperonin TRiC/CCT1/TCP1 and HSP70 (heat shock protein 70). 27252013_The specific association of ZBTB7A mutations with t(8;21) rearranged acute myeloid leukaemia points towards leukemogenic cooperativity between mutant ZBTB7A and the RUNX1/RUNX1T1 fusion protein has been reported. 27276256_Study demonstrated that TRiC's contribution to the activity of the DNA-binding domain (AML1-175) of AML1-ETO is consistent with its contribution to the activity of full-length AML1-ETO and is mediated through its direct association with the DNA-binding domain 27522676_The data suggest that the tumor suppressor activity of both RUNX1t1 and TFF1 are mechanistically connected to CEBPB and that cross-regulation between CEBPB-RUNX1t1-TFF1 plays an important role in gastric carcinogenesis. 27725192_the AML1-ETO fusion protein increases the expression of SIRT1, possibly by binding to the promoter region of SIRT1 to activate its transcription in t(8;21) AML. 27770540_A feedback circuitry involving miR-9-1 and RUNX1-RUNX1T1. 27798886_Our data revealed a novel role for RUNX1T1 as a tumor-suppressor gene in colorectal cancer through modulation of multiple cellular pathways affecting the cell cycle, DNA damage, DNA replication, estrogen signaling, and drug resistance 28092997_PKM2 as a novel target of RUNX1-ETO and is specifically downregulated in RUNX1-ETO positive AML patients, indicating that PKM2 level might have a diagnostic potential in RUNX1-ETO associated AML. 28166825_RUNX1-RUNX1T1 transcript levels were measured in bone marrow samples collected from 208 patients at scheduled time points after transplantation. Over 90% of the 175 patients who were in continuous complete remission had a >/=3-log reduction in RUNX1-RUNX1T1 transcript levels from the time of diagnosis at each time point after transplantation and a >/=4-log reduction at >/=12 months. 28322996_The role of RUNX1T1 in t(8;21) acute myeloid leukemia and miRNA expression regulation 28539478_Altogether, these results revealed an unexpected and important epigenetic mini-circuit of AML1-ETO/THAP10/miR-383 in t(8;21) acute myeloid leukaemia, in which epigenetic suppression of THAP10 predicts a poor clinical outcome and represents a novel therapeutic target. 28611288_Data indicate miR-29b-1 as a regulator of the AML1-ETO protein (RUNX1-RUNX1T1), and that miR-29b-1 expression in t(8;21)-carrying leukemic cell lines partially rescues the leukemic phenotype. 28640846_RUNX1T1 serves as a common angiogenic driver for vaculogenesis and functionality of endothelial lineage cells 28650479_Leukaemogenesis by AML1-ETO requires enhanced C/D box snoRNA/RNP formation. 29413154_At levels of condFDR < 0.01 and conjFDR < 0.05; we identified 5 ADHD-associated loci, 3 of these being shared between ADHD and EA. 29472719_identified most of these factors as RUNX1-RUNX1T1 targets, with Ras Homolog Family Member (RHOB) overexpression being the core of this network 29568097_Minimal residual disease (MRD) monitoring and mutational landscape in AML with RUNX1-RUNX1T1. 30050054_In an analysis of an independent validation cohort, KIAA0125 again showed a significant influence with respect to the impact of the RUNX1/RUNX1T1 fusion in mediation of acute myeloid leukemia. 30093631_As most transcriptional repressor proteins do not comprise tetramerization domains, our results provide a possible explanation as to the reason that RUNX1 is recurrently found translocated to ETO family members 30499136_lncRNA RUNX1-IT1 was down regulated both in colorectal cancer tissues and cell lines; besides, lncRNA RUNX1-IT1 could serve as a potential diagnostic biomarker and play a tumour-suppressive role owing to its good diagnostic efficacy and inhibition of colorectal cancer cell proliferation and migration 30569130_Study reports differential regulation of gene expression by RUNX1-RUNX1T1 oncoprotein and MAPK1 as determined by genome-wide expression analysis responsible for the phenotypic features of acute myeloid leukaemia with karyotypic discernible translocation (t)(8;21)(q22;q22). 30637949_found that the binding capacity of RUNX1-ETO9a, a truncated RUNX1-ETO isoform, to the c-KIT promoter was stronger than that of RUNX1-ETO, suggesting RUNX1-ETO9a as another valuable therapeutic target in t(8;21) AML 31119862_Targeting miR-193a-AML1-ETO-beta-catenin axis by melatonin suppresses the self-renewal of leukaemia stem cells in leukaemia with t (8;21) translocation. 31247675_Negative CD19 expression is associated with inferior relapse-free survival in children with RUNX1-RUNX1T1-positive acute myeloid leukaemia: results from the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 study. 31645082_Long non-coding RNA EPB41L4A-AS2 suppresses progression of ovarian cancer by sequestering microRNA-103a to upregulate transcription factor RUNX1T1. 31699991_Findings highlight the role of EZH1 in non-histone lysine methylation, indicating that cooperation between AML1-ETO and EZH1 and AML1-ETO site-specific lysine methylation promote AML1-ETO transcriptional repression in leukemia. 31725954_Morphological characteristics, cytogenetic profile, and outcome of RUNX1-RUNX1T1-positive acute myeloid leukemia: Experience of an Indian tertiary care center. 31839036_fusion transcript level after the first induction therapy in RUNX1-RUNX1T1-positive AML children is an independent factor influencing the long-term prognosis 31869378_AML1-ETO as a single oncogenic hit in a non-mutated background blocks granulocytic differentiation, deregulates the gene program via altering the acetylome of the differentiating granulocytic cells, and induces t(8;21) AML associated leukemic characteristics. 32024815_LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. 32071087_Histone deacetylase 3 preferentially binds and collaborates with the transcription factor RUNX1 to repress AML1-ETO-dependent transcription in t(8;21) AML. 32311890_In acute myeloid leukemia patients with positive RUNX1-RUNX1T1 receiving intensive consolidation therapy, the white blood cell counts at onset of leukemia, C-KIT mutations in exon 17, and FLT3-ITD gene mutations suggest poor prognosis 33067946_[Clinical Prognostic Factors Analysis of Initially Treated AML Children with t(8;21)/RUNX1-RUNX1T1()]. 33084222_Identification of RUNX1T1 as a potential epigenetic modifier in small-cell lung cancer. 33152396_RUNX1/ETO and mutant KIT both contribute to programming the transcriptional and chromatin landscape in t(8;21) acute myeloid leukemia. 33217477_The RUNX1/RUNX1T1 network: translating insights into therapeutic options. 33382982_Definition of a small core transcriptional circuit regulated by AML1-ETO. 33431365_Relationships between AML1-ETO and MLL-AF9 fusion gene expressions and hematological parameters in acute myeloid leukemia. 33483506_RUNX1/RUNX1T1 mediates alternative splicing and reorganises the transcriptional landscape in leukemia. 33524443_Optimized clinical application of minimal residual disease in acute myeloid leukemia with RUNX1-RUNX1T1. 33971010_Myeloid lncRNA LOUP mediates opposing regulatory effects of RUNX1 and RUNX1-ETO in t(8;21) AML. 34120331_Droplet digital polymerase chain reaction assay for the detection of the minor clone of KIT D816V in paediatric acute myeloid leukaemia especially showing RUNX1-RUNX1T1 transcripts. 34264479_RUNX1T1, a potential prognostic marker in breast cancer, is co-ordinately expressed with ERalpha, and regulated by estrogen receptor signalling in breast cancer cells. 34331016_AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms. 34369622_Leukemia associated RUNX1T1 gene reduced proliferation and invasiveness of glioblastoma cells. | ENSMUSG00000006586 | Runx1t1 | 97.871487 | 1.8544041 | 0.890955662 | 0.37617496 | 5.601965e+00 | 1.794035e-02 | No | Yes | 161.434683 | 45.378980 | 81.686041 | 23.006007 | ||
| ENSG00000080224 | 285220 | EPHA6 | protein_coding | Q9UF33 | FUNCTION: Receptor tyrosine kinase which binds promiscuously GPI-anchored ephrin-A family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling (By similarity). {ECO:0000250}. | ATP-binding;Alternative splicing;Glycoprotein;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | hsa:285220; | integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; transmembrane-ephrin receptor activity [GO:0005005]; axon guidance [GO:0007411]; positive regulation of kinase activity [GO:0033674]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 19850283_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20011078_During development of the retinal vasculature, migration of ligand-bearing astrocytes is slowed along Eph-A6 expression gradient through repellent Eph-A6 - ephrin-A1 and -A4 signaling. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20950786_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 25583493_Two predominant genes, ephrin type A receptor 6 (EPHA6) and folliculin (FLCN), with mutations exclusive to African American CRCs, are by genetic and biological criteria highly likely African American CRC driver genes. 26041887_EphA6 mRNA expression is higher in 112 CaP tumor samples compared with benign tissues from 58 benign prostate hyperplasia patients. Positive correlation was identified between EphA6 expression and vascular invasion, neural invasion, PSA level, and TNM staging in CaP cases. 27582484_Gene-based analysis identified EPHA6 as the gene most significantly associated with paclitaxel-induced neuropathy...This first study sequencing EPHA genes revealed that low-frequency variants in EPHA6, EPHA5, and EPHA8 contribute to the susceptibility to paclitaxel-induced neuropathy 29208002_The EPHA6 rs4857055 C > T SNP is a novel candidate gene for hypertension in the Korean population. 31483918_our data imply that EPHA6 expression is beneficial for glioblastoma multiforme inhibition, particularly in combination with activation of BMP-2 signaling.These results suggest that EPHA6 expression or protein levels could be used as biomarkers for identification of subsets of glioblastoma multiforme patients who might benefit from BMP treatment. | ENSMUSG00000055540 | Epha6 | 143.465454 | 1.0946179 | 0.130427384 | 0.25673908 | 2.594618e-01 | 6.104901e-01 | No | Yes | 173.505710 | 32.922644 | 160.515015 | 30.362167 | |||
| ENSG00000080503 | 6595 | SMARCA2 | protein_coding | P51531 | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Binds DNA non-specifically (PubMed:22952240, PubMed:26601204). Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). {ECO:0000250|UniProtKB:Q6DIC0, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | 3D-structure;ATP-binding;Acetylation;Activator;Alternative splicing;Bromodomain;DNA-binding;Disease variant;Helicase;Hydrolase;Hypotrichosis;Isopeptide bond;Mental retardation;Neurogenesis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Schizophrenia;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a member of the SWI/SNF family of proteins and is highly similar to the brahma protein of Drosophila. Members of this family have helicase and ATPase activities and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SNF/SWI, which is required for transcriptional activation of genes normally repressed by chromatin. Alternatively spliced transcript variants encoding different isoforms have been found for this gene, which contains a trinucleotide repeat (CAG) length polymorphism. [provided by RefSeq, Jan 2014]. | hsa:6595; | chromatin [GO:0000785]; intermediate filament cytoskeleton [GO:0045111]; intracellular membrane-bounded organelle [GO:0043231]; nBAF complex [GO:0071565]; npBAF complex [GO:0071564]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SWI/SNF complex [GO:0016514]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATP-dependent activity, acting on DNA [GO:0008094]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; helicase activity [GO:0004386]; histone binding [GO:0042393]; transcription cis-regulatory region binding [GO:0000976]; transcription coactivator activity [GO:0003713]; chromatin remodeling [GO:0006338]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; nervous system development [GO:0007399]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; spermatid development [GO:0007286] | 11719516_Brm could also drive expression of CD44; Brm can compensate for BRG-1 loss as pertains to RB sensitivity 11850427_Brm-containing SWI/SNF complex subfamily (trithorax-G) and a complex including YY1 and HDACs (Polycomb-G) counteract each other to maintain transcription of exogenously introduced genes 12493776_human adrenal carcinoma cells can spontaneously transition between two subtypes by switching expression of BRG1 and Brm at the post-transcriptional level 12566296_This report provides supportive evidence that BRG1 and BRM act as tumor suppressor proteins and implicates a role for their loss in the development of non-small cell lung cancers. 12620226_BRG1 and BRM complexes may direct distinct cellular processes by recruitment to specific promoters through protein-protein interactions that are unique to each ATPase. 14657023_Cell culture in the presence of HDAC inhibitors facilitates the isolation of clones overexpressing Brm. 15240517_BRM and BRG1 participate in two distinct chromosome remodeling complexes that are functionally complementary in non-small cell lung cancer 16341228_on genes regulated by SWI/SNF, Brm contributes to the crosstalk between transcription and RNA processing by decreasing RNAPII elongation rate and facilitating recruitment of the splicing machinery to variant exons with suboptimal splice sites 16749937_Observational study of gene-disease association. (HuGE Navigator) 16749937_family-based and case-control association study suggest that there is no association between the trinucleotide repeat polymorphism within SMARCA2 and schizophrenia 17075831_Aberrant expression of BRM genes is associated with disease development and progression in prostate cancers. 17546055_Loss of BRM through epigenetic silencing is associated with neoplasms 17938176_p53 activity is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes 18006815_Brm is required for villin expression, a definitive marker of intestinal metaplasia and differentiation 18042045_at the TERT gene locus in human tumour cells containing a functional SWI/SNF complex, Brm, and possibly BRG1, in concert with p54(nrb), would initiate efficient transcription and could be involved in the subsequent splicing of TERT transcripts 18082132_C/EBPbeta and GATAs may developmentally regulate the expression of brm by mutually exclusive binding. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18923443_Hotspot mutation of Brahma in non-melanoma skin cancer. 19144648_BRM and BRG1 SWI/SNF complexes have roles in differentiation 19183483_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19363039_A missense polymorphism (rs2296212)induced a lower nuclear localization of BRM was associated with low SMARCA2 expression in the postmortem prefrontal cortex of schizophrenia patients. 19371634_Cdx2 regulates intestinal villin expression through recruiting Brm-type SWI/SNF complex to the villin promoter. 19488910_Loss of heterozygosity at the 9p21-24 region and identification of BRM as a candidate tumor suppressor gene in head and neck squamous cell carcinoma. 19784067_Data suggest that heterogeneous SWI/SNF complexes composed of either the BRG1 or BRM subunit promote expression of distinct and overlapping MITF target genes. 20011120_show that SWI/SNF activity favors (from subunits Brg1/Brm) loading of HP1 proteins to chromatin both in vivo and in vitro 20333683_Knockdown of either BRM or BRG1 resulted in an inhibition of cell proliferation in monolayer cultures. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20457675_The SWI/SNF chromatin-remodelin complex asa key component of the genetic architecture of schizophrenia. 20460684_REQ functions as an efficient adaptor protein between the SWI/SNF complex and RelB/p52 and plays important roles in noncanonical NF-kappaB transcriptional activation and its associated oncogenic activity. 20719309_The methylation levels of CpG islands within Brahma increased during spermatogenesis and decreased during oogenesis 21070662_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21079652_The hBrm/Brg1 switch is an indicator of the responsiveness of a gene to heat-shock or IFNgamma stimulation and may represent an 'on-off switch' of gene expression in vivo. 21092585_The expression of BRG1 and BRM correlates with the development of prostatic cancer. 21189327_results reveal that miR-199a and Brm form a double-negative feedback loop through Egr1, leading to the generation of two distinct cell types during carcinogenesis. 21646426_multiple distinct transcriptional patterns of GR and Brm interdependence 22366787_sequenced the exomes of ten individuals with Nicolaides-Baraitser syndrome and identified heterozygous variants in SMARCA2 in eight of them. 22721696_SWI/SNF chromatin remodeling complex catalytic subunits Brg1 and Brm modulate cisplatin cytotoxicity by facilitating efficient repair of the cisplatin DNA lesions. 23088494_Reduced expression of BRM may contribute to the carcinogenesis of hepatocellular carcinoma. 23163725_Loss of BRG1 and BRM was frequent in E-cadherin-low, TTF-1-low, and vimentin-high cases. 23276717_SMARCA2 rs2296212 and rs4741651 variants were associated with oligodendroglioma risk. 23322154_BRM expression was lost in 25% of cell lines and 16% of tumors. 23359823_findings suggest that BRM promoter polymorphism (BRM-1321) could regulate BRM expression and may serve as a potential marker for genetic susceptibility to HCC 23524580_the mitogen-activated protein kinase pathway regulates both BRM acetylation and BRM silencing as MAP kinase pathway inhibitors both induced BRM as well as caused BRM deacetylation. 23770848_loss of BRM epigenetically induces C/EBPbeta transcription, which then directly induces alpha5 integrin transcription to promote the malignant behavior of mammary epithelial cells 23872584_High SMARCA2 expression is associated with lung cancer 23897427_Data indicate that transient knockdown of BRG1 or BRM reduces hypoxia induction of several known HIF1 and HIF2 target genes in Hep3B cells. 23963727_Data suggest that Brg1 and Brm integrate various proinflammatory cues into cell adhesion molecule transactivation in endothelial injury. 24421395_A SMARCA2-containing residual SWI/SNF complex underlies the oncogenic activity of SMARCA4 mutant cancers. 24471421_loss of BRM expression is a common feature among poorly differentiated tumours in clear cell renal cell carcinomas. We hypothesize that loss of BRM expression is involved in tumor de-differentiation in clear cell RCCs. 24519853_Findings suggest that the BRM promoter double insertion homozygotes may be associated with an increased risk of early-stage UADT cancers independent of smoking status and histology. 24520176_Depletion of BRM in BRG1-deficient cancer cells leads to a cell cycle arrest, induction of senescence, and increased levels of global H3K9me3. 24913006_these data show that the mechanism of BRM silencing contributes to the pathogenesis of Rhabdoid tumors 25026375_Over-expression of BRM in melanoma cells that harbor oncogenic BRAF promoted changes in cell cycle progression and apoptosis consistent with a tumor suppressive role. 25081545_BAF complex gene SMARCA2 is mutated in Coffin-Siris syndrome patients. 25169058_Phenotype and genotype in Nicolaides-Baraitser syndrome patients with SMARCA2 mutations 25496315_We report, for the first time, co-inactivation and frequent mutations of SMARCB1, SMARCA2 and PBRM1 in MRTs. 25673149_The miR-199a/Brm/EGR1 axis is a determinant of anchorage-independent growth in epithelial tumor cell lines 25808524_Knockout of BRG1 or BRM using CRISPR/Cas9 technology resulted in the loss of viability, consistent with a requirement for both enzymes in triple negative breast cancer cells 26356327_SMARCA4 loss, either alone or with SMARCA2, is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type, restoration of either SWI/SNF ATPase can inhibit the growth of SCCOHT cell lines 26551623_this study shows for the first time novel SMARCA4-deficient and SMARCA2-deficient variants in undifferentiated gastrointestinal tract carcinomas 26564006_our data suggest that concomitant loss of SMARCA2 and SMARCA4 is another hallmark of small cell carcinoma of the ovary, hypercalcemic type-a finding that offers new opportunities for therapeutic interventions. 27264538_We conclude that their features better resemble Coffin-Siris syndrome, rather than Nicolaides-Baraitser syndrome and that these features likely arise from SMARCA2 over-dosage. Pure 9p duplications (not caused by unbalanced translocations) are rare. Copy number analysis in patients with features that overlap with Coffin-Siris syndrome is recommended to further determine their genetic aspects. 27487558_Two functional promoter BRM polymorphisms were not associated with pancreatic adenocarcinoma risk, but are strongly associated with survival. 27665729_This de novo SMARCA2 missense mutation c.3721C>G, p.Gln1241Glu is the only reported mutation on exon 26 outside the ATPase domain of SMARCA2 to be associated with Nicolaides-Baraitser syndrome and adds to chromatin remodeling as a pathway for epileptogenesis. 27827316_Epigenetic regulatory molecules bind to two BRM promoter sequence variants but not to their wild-type sequences. These variants are associated with adverse overall and progression-free survival. Decreased BRM gene expression, seen with these variants, is also associated with worse overall survival 28038711_SMARCA4 and SMARCA2 deficiency is observed in 5.1% and 4.8% of non-small cell lung cancer 28070921_BRM gene mutation, chromosome 9 monosomy or BRM deletion and CpG methylation contribute collectively to the loss of BRM expression in poorly differentiated clear cell renal cell carcinoma 28232072_BRG1/BRM and c-MYC have an antagonistic relationship regulating the expression of cardiac conduction genes that maintain contractility, which is reminiscent of their antagonistic roles as a tumor suppressor and oncogene in cancer. 28292935_We also demonstrate that tazemetostat, a potent and selective EZH2 inhibitor currently in phase II clinical trials, induces potent antiproliferative and antitumor effects in SCCOHT cell lines and xenografts deficient in both SMARCA2 and SMARCA4. These results exemplify an additional class of rhabdoid-like tumors that are dependent on EZH2 activity for survival. 28296015_Although Genome-Wide Association studies have not been carried out in the field of alcohol-related hepatocellular carcinoma (HCC), common single nucleotide polymorphisms conferring a small increase in the risk of liver cancer risk have been identified. Specific patterns of gene mutations including CTNNB1, TERT, ARID1A and SMARCA2 exist in alcohol-related HCC. [review] 28427211_two promoter BRM germline variants were associated with worse outcome in our esophageal adenocarcinoma (EAC) patients. This significantly poorer outcome was independent of TNM classification at diagnosis or other clinic-demographic variables. 28571677_BRM-741 and BRM-1321 insertion polymorphisms are associated with susceptibility to cancer. 28602977_BRM could activate JAK2/STAT3 pathway to promote pancreatic cancer growth and chemoresistance. 28678310_Expression of BRM and MMP2 in the thoracic aortic aneurysm and aortic dissection is very high, indicating that BRM and MMP2 may play important roles in the occurrence and development of thoracic aortic aneurysm and aortic dissection. 28706277_association of the BRG1/hBRM bromodomain with nucleosomes plays a regulatory rather than targeting role 28892201_Two BRM promoter polymorphisms were strongly associated with hepatocellular carcinoma (HCC) prognosis but were not associated with increased HCC susceptibility. 29087303_PRC2-mediated repression of SMARCA2 predicts EZH2 inhibitor activity in SWI/SNF mutant tumors. 29273066_at genes where BRG1 and BRM antagonize one another we observe a nearly complete rescue of gene expression changes in the combined BRG/BRM double knockdown 29391527_High expression of SMARCA2 is associated with benign differentiated tumors. 29848589_Here, the authors show that C-terminally truncated forms of both SMARCA2 and SMARCA4, produced by caspase-mediated cleavage, accumulate in cells infected with different RNA or DNA viruses. The levels of truncated SMARCA2 or SMARCA4 strongly correlate with the degree of cell damage and death observed after virus infection. 29894502_Indel polymorphisms in the promoter region of the Brahma gene is associated with colorectal cancer. 30287812_The sensitivity of SWI/SNF-deficient cells to DNA damage induced by UV irradiation and cisplatin treatment depends on GTF2H1 levels. 30447346_Inactivation of SMARCA2 by promoter hypermethylation is associated with lung cancer development. 30478150_targeting of BRM in combination with radiotherapy is supposed to improve the therapeutic outcome of lung cancer patients harboring BRG1 mutations.The present study shows that the moderate radioresponsiveness of NSCLC cells with BRG1 mutations can be increased upon BRM depletion that is associated with a prolonged Rad51-foci prevalence at DNA DSBs. 30522882_In both mouse keratinocytes and HaCaT cells, Brm deficiency led to an increased cell population growth following ultraviolet radiation (UVR) exposure compared to cells with normal levels of Brm. The loss of Brm in keratinocytes exposed to UVR enables them to resume proliferation while harboring DNA photolesions, leading to an increased fixation of mutations and, consequently, increased carcinogenesis. 30722027_Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1-associated atypical neurofibromas. 30790683_Transcriptional regulation of miR-302a-3p by BRM potentiates pancreatic cancer cell metastasis by epigenetically suppressing SOCS5 expression and activating STAT3 signaling. 30946989_BRM suppresses the migration and invasion of breast cancer cells via epigenetically activating the transcription of Claudins. 31288860_SMARCA2 variants in Nicolaides-Baraitser syndrome 31355511_Never-smokers who carry BRM homozygous variants have an increased chance of developing MPM, which results in worse prognosis. In contrast, in ever-smokers, there may be a protective effect, with no difference in overall survival. Mechanisms for the interaction between BRM and smoking require further study. 31375262_results demonstrate that SMARCA2 mutations cause impaired differentiation through enhancer reprogramming via inappropriate targeting of SMARCA4. 31406271_SMARCA2-deficiency confers sensitivity to targeted inhibition of SMARCA4 in esophageal squamous cell carcinoma cell lines. 31751681_SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. 31906887_This work describes, for the first time, loss of one of the SWI/SNF ATPase subunit proteins in a large number of adenocarcinomas of the oesophagus. 32019955_AATF and SMARCA2 are associated with thyroid volume in Hashimoto's thyroiditis patients. 32073734_SWI/SNF chromatin remodeling complex and glucose metabolism are deregulated in advanced bladder cancer. 32312722_Review of the role of SMARCA2 and SMARCA4 mutations in various neoplasms. 32376693_The transcription factor GLI1 cooperates with the chromatin remodeler SMARCA2 to regulate chromatin accessibility at distal DNA regulatory elements. 32502208_BRM-SWI/SNF chromatin remodeling complex enables functional telomeres by promoting co-expression of TRF2 and TRF1. 32657847_Two cases of Nicolaides-Baraitser syndrome, one with a novel SMARCA2 variant. 32694869_De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. 32855269_Loss of SWI/SNF Chromatin Remodeling Alters NRF2 Signaling in Non-Small Cell Lung Carcinoma. 33027072_SMARCA4/SMARCA2-deficient Carcinoma of the Esophagus and Gastroesophageal Junction. 33481850_The clinicopathological significance of SWI/SNF alterations in gastric cancer is associated with the molecular subtypes. 33602783_SMARCA2 Is a Novel Interactor of NSD2 and Regulates Prometastatic PTP4A3 through Chromatin Remodeling in t(4;14) Multiple Myeloma. 34289068_The Bromodomains of the mammalian SWI/SNF (mSWI/SNF) ATPases Brahma (BRM) and Brahma Related Gene 1 (BRG1) promote chromatin interaction and are critical for skeletal muscle differentiation. 34518526_SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by restricting IP3R3-mediated Ca(2+) flux to mitochondria. 35289322_SMARCA2 deficiency in NSCLC: a clinicopathologic and immunohistochemical analysis of a large series from a single institution. | ENSMUSG00000024921 | Smarca2 | 961.252794 | 1.1598194 | 0.213900223 | 0.13515919 | 2.432308e+00 | 1.188578e-01 | 4.584177e-01 | No | Yes | 989.235574 | 122.390367 | 833.852730 | 103.286682 | |
| ENSG00000081019 | 54665 | RSBN1 | protein_coding | Q5VWQ0 | FUNCTION: Histone demethylase that specifically demethylates dimethylated 'Lys-20' of histone H4 (H4K20me2), thereby modulating chromosome architecture. {ECO:0000250|UniProtKB:Q80T69}. | Alternative splicing;Chromatin regulator;Dioxygenase;Iron;Isopeptide bond;Metal-binding;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:54665; | nucleus [GO:0005634]; dioxygenase activity [GO:0051213]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325] | 18305142_Observational study of gene-disease association. (HuGE Navigator) 19565500_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000044098 | Rsbn1 | 1033.563652 | 1.0032222 | 0.004641201 | 0.08927325 | 2.702254e-03 | 9.585421e-01 | 9.850507e-01 | No | Yes | 1156.896444 | 232.921758 | 1123.214289 | 226.134019 | ||
| ENSG00000081189 | 4208 | MEF2C | protein_coding | Q06413 | FUNCTION: Transcription activator which binds specifically to the MEF2 element present in the regulatory regions of many muscle-specific genes. Controls cardiac morphogenesis and myogenesis, and is also involved in vascular development. Enhances transcriptional activation mediated by SOX18. Plays an essential role in hippocampal-dependent learning and memory by suppressing the number of excitatory synapses and thus regulating basal and evoked synaptic transmission. Crucial for normal neuronal development, distribution, and electrical activity in the neocortex. Necessary for proper development of megakaryocytes and platelets and for bone marrow B-lymphopoiesis. Required for B-cell survival and proliferation in response to BCR stimulation, efficient IgG1 antibody responses to T-cell-dependent antigens and for normal induction of germinal center B-cells. May also be involved in neurogenesis and in the development of cortical architecture (By similarity). Isoforms that lack the repressor domain are more active than isoform 1. {ECO:0000250|UniProtKB:Q8CFN5, ECO:0000269|PubMed:11904443, ECO:0000269|PubMed:15340086, ECO:0000269|PubMed:15831463, ECO:0000269|PubMed:15834131, ECO:0000269|PubMed:9069290, ECO:0000269|PubMed:9384584}. | Acetylation;Activator;Alternative splicing;Apoptosis;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Disease variant;Epilepsy;Isopeptide bond;Mental retardation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This locus encodes a member of the MADS box transcription enhancer factor 2 (MEF2) family of proteins, which play a role in myogenesis. The encoded protein, MEF2 polypeptide C, has both trans-activating and DNA binding activities. This protein may play a role in maintaining the differentiated state of muscle cells. Mutations and deletions at this locus have been associated with severe cognitive disability, stereotypic movements, epilepsy, and cerebral malformation. Alternatively spliced transcript variants have been described. [provided by RefSeq, Jul 2010]. | hsa:4208; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; postsynapse [GO:0098794]; protein-containing complex [GO:0032991]; sarcoplasm [GO:0016528]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; histone deacetylase binding [GO:0042826]; minor groove of adenine-thymine-rich DNA binding [GO:0003680]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; apoptotic process [GO:0006915]; B cell homeostasis [GO:0001782]; B cell proliferation [GO:0042100]; B cell receptor signaling pathway [GO:0050853]; blood vessel development [GO:0001568]; blood vessel remodeling [GO:0001974]; cardiac ventricle formation [GO:0003211]; cell differentiation [GO:0030154]; cell morphogenesis involved in neuron differentiation [GO:0048667]; cellular response to calcium ion [GO:0071277]; cellular response to fluid shear stress [GO:0071498]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to parathyroid hormone stimulus [GO:0071374]; cellular response to transforming growth factor beta stimulus [GO:0071560]; cellular response to trichostatin A [GO:0035984]; cellular response to xenobiotic stimulus [GO:0071466]; chondrocyte differentiation [GO:0002062]; endochondral ossification [GO:0001958]; epithelial cell proliferation involved in renal tubule morphogenesis [GO:2001013]; excitatory postsynaptic potential [GO:0060079]; germinal center formation [GO:0002467]; glomerulus morphogenesis [GO:0072102]; heart development [GO:0007507]; heart looping [GO:0001947]; humoral immune response [GO:0006959]; learning or memory [GO:0007611]; MAPK cascade [GO:0000165]; melanocyte differentiation [GO:0030318]; muscle cell fate determination [GO:0007521]; muscle organ development [GO:0007517]; myotube differentiation [GO:0014902]; negative regulation of blood vessel endothelial cell migration [GO:0043537]; negative regulation of gene expression [GO:0010629]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of ossification [GO:0030279]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of vascular associated smooth muscle cell migration [GO:1904753]; negative regulation of vascular associated smooth muscle cell proliferation [GO:1904706]; negative regulation of vascular endothelial cell proliferation [GO:1905563]; nephron tubule epithelial cell differentiation [GO:0072160]; nervous system development [GO:0007399]; neural crest cell differentiation [GO:0014033]; neuron development [GO:0048666]; neuron differentiation [GO:0030182]; neuron migration [GO:0001764]; osteoblast differentiation [GO:0001649]; outflow tract morphogenesis [GO:0003151]; platelet formation [GO:0030220]; positive regulation of alkaline phosphatase activity [GO:0010694]; positive regulation of B cell proliferation [GO:0030890]; positive regulation of behavioral fear response [GO:2000987]; positive regulation of bone mineralization [GO:0030501]; positive regulation of cardiac muscle cell differentiation [GO:2000727]; positive regulation of cardiac muscle cell proliferation [GO:0060045]; positive regulation of gene expression [GO:0010628]; positive regulation of macrophage apoptotic process [GO:2000111]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of myoblast differentiation [GO:0045663]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of osteoblast differentiation [GO:0045669]; positive regulation of skeletal muscle cell differentiation [GO:2001016]; positive regulation of skeletal muscle tissue development [GO:0048643]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; primary heart field specification [GO:0003138]; regulation of AMPA receptor activity [GO:2000311]; regulation of dendritic spine development [GO:0060998]; regulation of germinal center formation [GO:0002634]; regulation of megakaryocyte differentiation [GO:0045652]; regulation of neuron apoptotic process [GO:0043523]; regulation of neurotransmitter secretion [GO:0046928]; regulation of NMDA receptor activity [GO:2000310]; regulation of synapse assembly [GO:0051963]; regulation of synaptic activity [GO:0060025]; regulation of synaptic plasticity [GO:0048167]; regulation of synaptic transmission, glutamatergic [GO:0051966]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; renal tubule morphogenesis [GO:0061333]; response to ischemia [GO:0002931]; secondary heart field specification [GO:0003139]; sinoatrial valve morphogenesis [GO:0003185]; skeletal muscle tissue development [GO:0007519]; smooth muscle cell differentiation [GO:0051145]; ventricular cardiac muscle cell differentiation [GO:0055012] | 11744164_MEF2C carboxy terminus (aa 387-473) deletion enhances transcriptional activation. AA 312-367 coupled with aa 1-86 activates transcription. Deletion of aa 312-350 decreases transcription. 11792813_The C-terminal region in MEF2C contains signals that are necessary to localize the histone deacetylase 4/MEF2 complex to the nucleus. 12061776_The DNA-binding TEA domain of transcription factor TEF-1 interacts with the MADS/MEF2 domain of MEF2C. 12135755_upregulation of expression by C-terminus of myogenin 12376544_The skeletal muscle-specific transcription cofactor vestigial-like 2 interacts with the C-terminal domain of MEF2C. 15084602_myogenin and myocyte enhancer factor-2 expression are triggered by membrane hyperpolarization during human myoblast differentiation 15831463_MEF2C is acetylated by p300 both in vitro and in vivo, which enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. 15834131_A conserved pattern of alternative splicing in vertebrate MEF2 (myocyte enhancer factor 2) genes generates an acidic activation domain in MEF2 proteins selectively in tissues where MEF2 target genes are highly expressed. (MEF2) 16469744_data show a dosage-dependent cardiomyopathic phenotype and a progressive reduction in ventricular performance associated with MEF2A or MEF2C overexpression 16478538_Phosphorylation of MEF2C at S396 aids sumoylation at K391, which recruits yet unidentified corepressors to inhibit transcription. Sumoylation motifs with a phosphorylated serine or an acidic residue at the +5 position might be more efficiently sumoylated. 16504037_Study demonstrates that human intestinal cell BCMO1 expression is dependent on the functional cooperation between peroxisome proliferator-activated receptor-gamma and myocyte enhancer factor 2 isoforms. 17611778_Overexpression of MEF2C is associated with hepatocellular carcinoma 17903302_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18067759_Results show the altered interaction between the different variants in the PGC-1alpha gene and MEF2C may attribute to the susceptibility to type 2 diabetes in the southern Chinese population. 18079734_In T-ALL cell lines, MEF2C is activated by NKX2-5 at both the RNA and protein levels. MEF2C consistently inhibits expression of NR4A1/NUR77, which regulates apoptosis via BCL2 transformation. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 19011954_Thus, ERK5 phosphorylation contributes to MEF2C activation and subsequent HASMC hypertrophy induced by Ang II 19065516_the association of the 482G/A polymorphism of the PGC-1alpha gene with type 2 diabetes and the quantitative and qualitative binding force changes between the PGC-1alpha domain mutant and MEF2C 19093215_MEF2 proteins are an important component in Galpha13-mediated angiogenesis. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19592390_These results strongly suggest that haploinsufficiency of MEF2C is responsible for severe mental retardation with stereotypic movements, seizures and/or cerebral malformations. 19720801_MEF2C is involved in the effect of insulin on skeletal muscle in type 2 diabetes. 19751190_Studies indicate that MAML1 functions as a coactivator for the tumor suppressor p53, MEF2C, beta-catenin and Notch signaling. 19828686_Muscle-growth geene MEF2C gene expression is enhanced by essential amino acid ingestion. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20333642_These deletions further support that haploinsufficiency of MEF2C is responsible for severe mental retardation, seizures, and hypotonia. 20412115_we present two additional patients with severe mental retardation, autism spectrum disorder and epilepsy, carrying a very small deletion encompassing the MEF2C gene. 20513142_Data indicate that MEF2C missense de novo mutations in severe mental retardation showed diminished MECP2 and CDKL5 expression. 20513142_Observational study of gene-disease association. (HuGE Navigator) 20610535_Binding sequences for the Mef2 family of transcription factors were enriched in the AR-bound regions, and data show that several Mef2c-dependent genes are direct targets of AR, suggesting a functional interaction between Mef2c and AR in skeletal muscle. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20709755_RNA-binding protein Muscleblind-like 3 (MBNL3) disrupts myocyte enhancer factor 2 (Mef2) {beta}-exon splicing 21060863_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21481790_NKX2-1, NKX2-2, and MEF2C define oncogenic pathways in T cell acute lymphoblastic leukemia (T-ALL). 21652706_knockdown of bone-specific transcription factors, Runx2 and osterix by shRNAi knockdown of Mef2c, suggests that Mef2c lies upstream of these two important factors in the cascade of gene expression in osteoblasts. 21849497_MEF2C is a transcriptional regulator of homeostasis in rod photoreceptor cells 21901155_Data report that forced expression of constitutively active MEF2C (MEF2CA) generates significantly greater numbers of neurons with dopaminergic properties in vitro. 22073279_suggest that PRL-3 functions downstream of the VEGF/MEF2C pathway in endothelial cells and may play an important role in tumor angiogenesis 22275376_a novel signaling cascade that links RhoA-mediated calcium sensitivity to MEF2-dependent myocardin expression in VSMCs through a mechanism involving p38 MAPK, PP1alpha, and CPI-17. 22363514_The present study investigated the effect of heart failure aetiology on Ca(+2) handling proteins and NFAT1, MEF2C and GATA4 (transcription factors) in the same cardiac tissue. 22449245_Mutations in MEF2C are probably a very rare cause of Rett syndrome. 22718505_15 bp-deletion and C-insertion in the 5'UTR region of MEF2C could affect hypertrophic cardiomyopathy, potentially by affecting expression of MEF2C. 22798246_genetic association study in population of 1,012 Han women in China: Data suggest that an SNP in MEF2C (rs1366594) is associated with bone mineral density of lumbar spine and hip joint in aging women. 22997707_Allele frequencies of three Alu insertions that are located in MEF2C (two of them) and TAX1BP1 genes significantly differ between cohorts of healthy donors and ALL(acute lymphoblastic leukemia) patients. 23001426_A targeted search for MEF2C mutations could be applied to patients with a severe intellectual deficiency associated with absence of language and hypotonia, strabismus, and epilepsy. 23174904_Validated miR-223 targets MEF2C and PTBP2 were significantly upregulated in chronic myeloid leukemia samples. 23226416_SREBP-1 regulate muscle protein synthesis through the downregulation of the expression of MYOD1, MYOG and MEF2C factors. 23468913_Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. 23572186_variants at MEF2C were associated with forearm bone mineral density (BMD), implicating this gene in the determination of BMD at forearm. [meta-analysis] 23776548_One variant, rs2194025 on chromosome 5q14 near the myocyte enhancer factor 2C MEF2C gene, was associated with retinal arteriolar caliber in meta-analysis. 23888946_We identified MEF2C as a novel transcription factor that regulates Nampt expression through specific interaction sites at the promoter; its regulatory role was highly dependent on epigenetic modulations, especially under hypoxia conditions 24008018_MEF2C alpha- variants are significantly expressed during neuronal cell differentiation, indicating a putative role of these variants in development. 24290359_Results identify redox-mediated protein posttranslational modifications, including S-nitrosylation and sulfonation of a critical cysteine residue in MEF2, as an early event contributing to neuronal damage in Parkinson's disease induced by mitochondrial toxins. 24337390_MEF2C binding in inflammatory pathways is associated with its role in bone density 24412363_MEF2 regulatory network is disrupted in myotonic dystrophy cardiac tissue leading to altered expression of a large number of miRNA and mRNA targets. 24988463_MEF2C/alpha-2-macroglobulin axis functions in endothelial cells as a negative feed-back mechanism that adapts sprouting activity to the oxygen concentration thus diminishing inappropriate and excess angiogenesis. 25328135_The overall effect of MEF2C in hepatocellular carcinoma progression regulation was dictated by its subcellular distribution. 25336633_MEF2 is the key cis-acting factor that regulates expression of a number of transcriptional targets involved in pulmonary vascular homeostasis, including microRNAs 424 and 503, connexins 37, and 40, and Kruppel Like Factors 2 and 4. 25352737_Single nucleotide polymorphisms in ALDOB, MAP3K1, and MEF2C are associated with cataract. 25404735_Alternative splicing of the alpha-exon of MEF2C regulates myogenesis. 25416133_Combinations that resulted in higher protein levels of Mef2c enhanced reprogramming efficiency of cardiac myocytes. 25691421_this is the first report of a Greek-Cypriot patient with a MEF2C-related phenotype highlighting the rich variability in phenotypic expression and the ethnic diversity associated with this condition. 25733682_MEF2C and MEF2D interact with the E3 ligase F-box protein SKP2, which mediates their subsequent degradation through the ubiquitin-proteasome system. 25789873_MEF2C regulates the expression of G2/M checkpoint genes (14-3-3gamma, Gadd45b and p21) and the sub-cellular localization of CYCLIN B1. 26172269_BCL2 inhibitors may be a therapeutic candidate in vivo for patients with ETP-ALL with high expression levels of MEF2C. 26184978_MiR-135b-5p and MiR-499a-3p Promote Cell Proliferation and Migration in Atherosclerosis by Directly Targeting MEF2C 26426104_The finding of a jugular pit in this patient facilitated the diagnosis, and he is, to our knowledge, the third case of jugular pit in association with 5q14.3 deletion incorporating the MEF2C locus. 26487643_Data show that high myocyte enhancer factor 2C (MEF2C) mRNA expression leads to overexpression of MEF2C protein, and these findings provide the rationale for therapeutic targeting of MEF2C transcriptional activation in acute myeloid leukemia. 26864752_We describe the prenatal identification of 5q14.3 duplication, including MEF2C, in a monochorionic twin pregnancy with corpus callosum anomalies, confirmed by autopsy. To the best of our knowledge, this cerebral finding has been observed for the first time in 5q14.3 duplication patients, possibly widening the neurological picture of this scarcely known syndrome. A pathogenetic role of MEF2C overexpression in brain develop 26900922_early B cell factor-1 (EBF1) was identified as a co-regulator of gene expression with MEF2C. 26921792_MEF2C mRNA level is up-regulated in both sporadic and SOD1 + ALS patients. 26923194_Study provides evidence that Mef2c cooperated with Sp1 to activate human AQP1 transcription by binding to its proximal promoter in human umbilical cord vein endothelial cells indicating that AQP1 is a direct target of Mef2c in regulating angiogenesis and vasculogenesis of endothelial cells. 27144530_Key role for miR-214 in modulation of MEF2C-MYOCD-LMOD1 signaling. 27255693_New MEF2C mutation in MEF2C haploinsufficiency syndrome 27268728_Long non-coding RNA uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. 27276684_MEF2C rs190982 polymorphism has a role in late-onset Alzheimer's disease in Han Chinese 27297623_a MEF2C and CEBPA correlation in CML disease progression 27337099_Data show that microRNA miR-27a was essential for the shift of mesenchymal stem cells (MSCs) from osteogenic differentiation to adipogenic differentiation in osteoporosis by targeting myocyte enhancer factor 2 c (Mef2c). 27479909_Single nucleotide polymorphism in MEF2C gene is associated with major depressive disorder. 27553283_Findings suggest that a single introduction of the three cardiomyogenic transcription factor (GATA4, cand TBX5)genes using polyethyleneimine (PEI)-based transfection is sufficient for transdifferentiation of adipose-derived stem cells (hADSCs) towards the cardiomyogenic lineage. 27616567_we identified novel associations in WLS , ARHGAP1 , and 5' of MEF2C ( P- values < 8x10 - 5 ; false discovery rate (FDR) q-values < 0.01) that were much more strongly associated with BMD compared to the GWAS SNPs. 27664809_Our analysis consistently identified significant sub-networks associated with the interacting transcription factors MEF2C and TWIST1, genes not previously associated with spontaneous preterm births , both of which regulate processes clearly relevant to birth timing. 27907007_The mRNA expressions of PPP3CB and MEF2C were significantly up-regulated, and CAMK1 and PPP3R1 were significantly down-regulated in mitral regurgitation(MR) patients compared to normal subjects. Moreover, MR patients had significantly increased mRNA levels of PPP3CB, MEF2C and PLCE1 compared to aortic valve disease patients 28017720_Mef2c is highly expressed in the retina where it modulates photoreceptor-specific gene expression 28456137_We report heterozygous MEF2C loss-of-function as a possible cause of question mark ear associated with intellectual deficiency. 28473437_Endothelial Mef2c regulates the endothelial actin cytoskeleton and inhibits smooth muscle cell migration into the intima. 28482719_MEF2C expression levels were significantly associated with or may even be predictive of the response to glucocorticoid treatment. 28799067_Overexpression of MEF2C decreased miR-448-induced VSMCs proliferation and migration. 28821601_Combined with automated 2D nano-scale chromatography, Accumulated ion monitoring achieved subattomolar limits of detection of endogenous proteins in complex biological proteomes. This allowed quantitation of absolute abundance of the human transcription factor MEF2C at approximately 100 molecules/cell, and determination of its phosphorylation stoichiometry from as little as 1 mug of extracts isolated from 10,000 human ... 28902616_This study indicates MEF2C as a new gene responsible for human dilated cardiomyopathy (DCM), which provides novel insight into the mechanism underpinning DCM, suggesting potential implications for development of innovative prophylactic and therapeutic strategies for DCM, the most prevalent form of primary myocardial disease. 29104469_the mutation significantly diminished the synergistic activation between MEF2C and GATA4, another cardiac core transcription factor that has been causally linked to Congenital heart disease (CHD). 29112298_MEF2C mRNA expression levels in AD subjects are significantly lower than those in control subjects and are correlated with disease severity. 29413154_At levels of condFDR < 0.01 and conjFDR < 0.05; we identified 5 ADHD-associated loci, 3 of these being shared between ADHD and EA. 29431698_High levels of phosphorylation of S222 of MEF2C are associated with primary chemotherapy resistance in an independent cohort of cytogenetically normal and Acute Myeloid Leukemia. 29468350_This study firstly associates MEF2C loss-of-function mutation with double outlet right ventricle in humans, which provides novel insight into the molecular pathogenesis of congenital heart diseases. 29526696_Targeting of LKB1 or SIK3 diminishes histone acetylation at MEF2C-bound enhancers and deprives leukemia cells of the output of this essential TF. 29678826_HuR upregulates MEF2C mRNA expression by protecting MEF2C mRNA from degradation, and consequently, the elevated MEF2C enhances SCN5A mRNA transcription. 29714661_The regulation mechanism of MIG6 and suggests potential implications for the therapeutic strategies of gefitinib resistance through inhibiting MEF2C in hepatic cancer cells. 29863696_Fourteen patients were studied including four patients with mutations in the MEF2C gene revealed by exome sequencing 29995456_MEF2 family, particularly MEF2C, is a key regulator underlying the transcriptional signature of bed rest and, hence, ultimately also skeletal muscle alterations induced by systemic unloading in humans. 30006604_The previously identified major depressive disorder risk variant rs10514299 in TMEM161B-MEF2C predicts neuronal correlates of reward processing in an alcohol dependence phenotype, possibly explaining part of the shared pathophysiology and comorbidity between the disorders. 30335237_MEF2C SNPs rs1366594, rs12521522 and rs11951031 were examined in women for northern Mexico. No significant association was found with bone mineral density, although some trends were seen. 30376817_Novel MEF2C point mutations in Chinese patients with Rett (-like) syndrome or non-syndromic intellectual disability have been reported. 30445463_our results disentangle a complex regulatory network governing neuronal MEF2C expression that involves multiple distal enhancers. In addition, the characterized neuronal enhancers pose as novel candidates to screen for mutations in neurodevelopmental disorders, such as Rett-like syndrome. 31094920_A novel MEF2C-SS18 gene fusion and unique histologic and immunophenotypic features characterize a heretofore undefined low-grade salivary adenocarcinoma for described as a microsecretory adenocarcinoma. 31140610_The results above showed that MEF2C was involved in the process of promoting the differentiation of stem cells into cardiac myocytes by miR-199a-3p inhibitors. 31254364_homeostasis and physiological function of AQP1 in endothelial health are maintained by the MEF2C and miR-133a-3p.1 regulatory circuit 31900516_A novel MEF2C mutation in lymphoid neoplasm diffuse large B-cell lymphoma promotes tumorigenesis by increasing c-JUN expression. 32003456_Identification of FOXH1 mutations in patients with sporadic conotruncal heart defect. 32017034_High expression of myocyte enhancer factor 2C predicts poor prognosis for adult acute myeloid leukaemia with normal karyotype. 32046534_MEF2C is associated with ADHD risk. 32186750_Methylationassociated silencing of miR638 promotes endometrial carcinoma progression by targeting MEF2C. 32410261_MicroRNA-23 suppresses osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting the MEF2C-mediated MAPK signaling pathway. 32975584_Genes influenced by MEF2C contribute to neurodevelopmental disease via gene expression changes that affect multiple types of cortical excitatory neurons. 33270893_Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression. 33303006_MiR-218 affects hypertrophic differentiation of human mesenchymal stromal cells during chondrogenesis via targeting RUNX2, MEF2C, and COL10A1. 33568691_MEF2C shapes the microtranscriptome during differentiation of skeletal muscles. 33831796_Electroclinical features of MEF2C haploinsufficiency-related epilepsy: A multicenter European study. 33984142_MEF2C silencing downregulates NF2 and E-cadherin and enhances Erastin-induced ferroptosis in meningioma. 33999292_Identification of a novel mutation in MEF2C gene in an atypical patient with frontotemporal lobar degeneration. 34022131_Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss-of-function mechanisms. 34184825_Comprehensive investigation of the phenotype of MEF2C-related disorders in human patients: A systematic review. 34570228_A dominant-negative SOX18 mutant disrupts multiple regulatory layers essential to transcription factor activity. 34731014_MEF2 is a key regulator of cognitive potential and confers resilience to neurodegeneration. 34779502_MicroRNA190b expression predicts a good prognosis and attenuates the malignant progression of pancreatic cancer by targeting MEF2C and TCF4. 34946961_Polymorphisms in Genes Involved in Osteoblast Differentiation and Function Are Associated with Anthropometric Phenotypes in Spanish Women. 34991657_Progress on the roles of MEF2C in neuropsychiatric diseases. 35130621_MEF2C ameliorates learning, memory, and molecular pathological changes in Alzheimer's disease in vivo and in vitro 35357565_MEF2C gene variations are associated with ADHD in the Chinese Han population: a case-control study. 35406681_Activin A Causes Muscle Atrophy through MEF2C-Dependent Impaired Myogenesis. | ENSMUSG00000005583 | Mef2c | 241.669568 | 1.1842196 | 0.243936621 | 0.21198059 | 1.322462e+00 | 2.501506e-01 | No | Yes | 295.400166 | 90.651089 | 245.840499 | 75.507479 | ||
| ENSG00000081307 | 79876 | UBA5 | protein_coding | Q9GZZ9 | FUNCTION: E1-like enzyme which specifically catalyzes the first step in ufmylation (PubMed:15071506, PubMed:18442052, PubMed:25219498, PubMed:20368332, PubMed:27653677, PubMed:26929408, PubMed:27545674, PubMed:30412706, PubMed:27545681). Activates UFM1 by first adenylating its C-terminal glycine residue with ATP, and thereafter linking this residue to the side chain of a cysteine residue in E1, yielding a UFM1-E1 thioester and free AMP (PubMed:20368332, PubMed:27653677, PubMed:26929408, PubMed:30412706). Activates UFM1 via a trans-binding mechanism, in which UFM1 interacts with distinct sites in both subunits of the UBA5 homodimer (PubMed:27653677). Trans-binding also promotes stabilization of the UBA5 homodimer, and enhances ATP-binding (PubMed:29295865). Transfer of UFM1 from UBA5 to the E2-like enzyme UFC1 also takes place using a trans mechanism (PubMed:27653677). Ufmylation is involved in reticulophagy (also called ER-phagy) induced in response to endoplasmic reticulum stress (PubMed:32160526). Ufmylation is essential for erythroid differentiation of both megakaryocytes and erythrocytes (By similarity). {ECO:0000250|UniProtKB:Q8VE47, ECO:0000269|PubMed:15071506, ECO:0000269|PubMed:18442052, ECO:0000269|PubMed:20368332, ECO:0000269|PubMed:25219498, ECO:0000269|PubMed:26929408, ECO:0000269|PubMed:27545674, ECO:0000269|PubMed:27545681, ECO:0000269|PubMed:27653677, ECO:0000269|PubMed:29295865, ECO:0000269|PubMed:30412706, ECO:0000269|PubMed:32160526}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Disease variant;Endoplasmic reticulum;Epilepsy;Golgi apparatus;Membrane;Mental retardation;Metal-binding;Neurodegeneration;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation pathway;Zinc | This gene encodes a member of the E1-like ubiquitin-activating enzyme family. This protein activates ubiquitin-fold modifier 1, a ubiquitin-like post-translational modifier protein, via the formation of a high-energy thioester bond. Alternative splicing results in multiple transcript variants. A pseudogene of this gene has been identified on chromosome 1. [provided by RefSeq, Feb 2016]. | hsa:79876; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; ATP binding [GO:0005524]; protein homodimerization activity [GO:0042803]; UFM1 activating enzyme activity [GO:0071566]; zinc ion binding [GO:0008270]; erythrocyte differentiation [GO:0030218]; megakaryocyte differentiation [GO:0030219]; neuromuscular process [GO:0050905]; protein K69-linked ufmylation [GO:1990592]; protein modification by small protein conjugation [GO:0032446]; protein ufmylation [GO:0071569]; regulation of intracellular estrogen receptor signaling pathway [GO:0033146]; response to endoplasmic reticulum stress [GO:0034976]; reticulophagy [GO:0061709] | 18442052_UBE1DC1 greatly activated SUMO2 in the nucleus or transferred activated-SUMO2 to nucleus after it conjugated SUMO2 in the cytoplasm. 20368332_Studies reveal structural features of UBA5 that further understanding of the enzyme reaction mechanism and provide insight into the evolution of ubiquitin activation. 24915089_Crystallization of Uba5 residues 57-363 was performed at 277 K using PEG 3350 as the precipitant, and crystals optimized by microseeding diffracted to 2.95 A resolution, with unit-cell parameters a=b=97.66, c=144.83 A, alpha=beta=90, gamma=120 degrees . 24966333_binding of ATP to Uba5 approximately Ufm1 thioester was required for efficient transfer of Ufm1 from Uba5 to Ufc1 via transthiolation. 25084390_Uba5 residues 364-404 were demonstrated to be necessary for the transthiolation of Ufm1 to Ufc1, and Uba5 381-404 was identified to be the minimal region for Ufc1 recognition 26872069_The study of UBA5-R246X revealed a dramatically decreased half-life and loss of UFM1 activation due to the absence of the catalytic cysteine Cys250. 26929408_study provides important structural and functional insights into the interaction between UBA5 and Ub-like modifiers, improving the understanding of the biology of the ufmylation pathway. 27545674_Biallelic Variants in UBA5 Link Dysfunctional UFM1 Ubiquitin-like Modifier Pathway to Severe Infantile-Onset Encephalopathy 27545681_clinical, biochemical, and experimental findings support our finding of UBA5 mutations as a pathophysiological cause for early-onset encephalopathies due to abnormal protein ufmylation 27653677_These findings explicitly elucidate the role of UBA5 dimerization in UFM1 activation. 28360427_UFM1 His 70 resembles UBA5 His336 and enters a negatively charged pocked on the other UFM1 molecule. 28965491_Heterozygous mutations in the UBA5 gene in two sisters with early-onset epileptic encephalopathy . 29286531_Likely pathogenic variants were identified in SOX5 gene, not previously associated with epilepsy, and UBA5, a recently associated with epilepsy gene 29295865_results make a connection between the binding of UFM1 to UBA5 and the latter's affinity to ATP, so we propose a novel mechanism for the regulation of ATP's binding to E1. 29663568_our results further demonstrate the importance of post-translational modifications such as the addition of an ubiquitin-fold modifier 1 (UFM1) to target proteins (ufmylation) for normal neuronal networks activity, and reveal that the dysfunction of the ubiquitous UBA5 protein is a cause of EME. 29902590_Hemizygous UBA5 missense mutation was identified in a patient with infantile-onset encephalopathy, acquired microcephaly, small cerebellum, movement disorder and severe neurodevelopmental delay. 30412706_Stimulates transfer of UFM1 from UBA5 to the E2, UFC1. 30990354_An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5. 32179706_A homozygous UBA5 pathogenic variant causes a fatal congenital neuropathy. 33853163_Homozygous UBA5 Variant Leads to Hypomyelination with Thalamic Involvement and Axonal Neuropathy. 34299007_A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1. 34508858_Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism. 34588452_Structural basis for UFM1 transfer from UBA5 to UFC1. | ENSMUSG00000032557 | Uba5 | 2651.572618 | 0.9904550 | -0.013836689 | 0.07054346 | 3.807964e-02 | 8.452833e-01 | 9.491576e-01 | No | Yes | 2747.456786 | 427.735427 | 2756.216212 | 429.066272 | |
| ENSG00000082146 | 55437 | STRADB | protein_coding | Q9C0K7 | FUNCTION: Pseudokinase which, in complex with CAB39/MO25 (CAB39/MO25alpha or CAB39L/MO25beta), binds to and activates STK11/LKB1. Adopts a closed conformation typical of active protein kinases and binds STK11/LKB1 as a pseudosubstrate, promoting conformational change of STK11/LKB1 in an active conformation (By similarity). {ECO:0000250, ECO:0000269|PubMed:14517248}. | ATP-binding;Alternative splicing;Cell cycle;Cytoplasm;Nucleotide-binding;Nucleus;Reference proteome | This gene encodes a protein that belongs to the serine/threonine protein kinase STE20 subfamily. One of the active site residues in the protein kinase domain of this protein is altered, and it is thus a pseudokinase. This protein is a component of a complex involved in the activation of serine/threonine kinase 11, a master kinase that regulates cell polarity and energy-generating metabolism. This complex regulates the relocation of this kinase from the nucleus to the cytoplasm, and it is essential for G1 cell cycle arrest mediated by this kinase. The protein encoded by this gene can also interact with the X chromosome-linked inhibitor of apoptosis protein, and this interaction enhances the anti-apoptotic activity of this protein via the JNK1 signal transduction pathway. Two pseudogenes, located on chromosomes 1 and 7, have been found for this gene. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2011]. | hsa:55437; | aggresome [GO:0016235]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; ATP binding [GO:0005524]; activation of protein kinase activity [GO:0032147]; cell cycle [GO:0007049]; cell morphogenesis [GO:0000902]; JNK cascade [GO:0007254]; negative regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001240]; protein export from nucleus [GO:0006611] | 12048196_a novel XIAP-interacting protein that acts as a co-factor enhancing XIAP-mediated activation of JNK1 and the caspase-independent protection of XIAP against apoptosis | ENSMUSG00000026027 | Stradb | 1685.803915 | 0.8831677 | -0.179240664 | 0.08398950 | 4.533103e+00 | 3.324529e-02 | 2.460430e-01 | No | Yes | 1660.181743 | 265.288148 | 1822.426530 | 291.169547 | |
| ENSG00000082458 | 1741 | DLG3 | protein_coding | Q92796 | FUNCTION: Required for learning most likely through its role in synaptic plasticity following NMDA receptor signaling. | 3D-structure;Acetylation;Alternative splicing;Mental retardation;Phosphoprotein;Reference proteome;Repeat;SH3 domain | This gene encodes a member of the membrane-associated guanylate kinase protein family. The encoded protein may play a role in clustering of NMDA receptors at excitatory synapses. It may also negatively regulate cell proliferation through interaction with the C-terminal region of the adenomatosis polyposis coli tumor suppressor protein. Mutations in this gene have been associated with X-linked cognitive disability. Alternatively spliced transcript variants have been described. [provided by RefSeq, Oct 2009]. | hsa:1741; | AMPA glutamate receptor complex [GO:0032281]; basolateral plasma membrane [GO:0016323]; bicellular tight junction [GO:0005923]; cell junction [GO:0030054]; cytosol [GO:0005829]; extracellular space [GO:0005615]; glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; kinase binding [GO:0019900]; phosphatase binding [GO:0019902]; ubiquitin protein ligase binding [GO:0031625]; cell-cell adhesion [GO:0098609]; establishment of planar polarity [GO:0001736]; establishment or maintenance of epithelial cell apical/basal polarity [GO:0045197]; negative regulation of cell population proliferation [GO:0008285]; positive regulation of protein tyrosine kinase activity [GO:0061098]; receptor clustering [GO:0043113]; receptor localization to synapse [GO:0097120]; regulation of postsynaptic membrane neurotransmitter receptor levels [GO:0099072] | 15185169_Loss may lead to altered synaptic plasticity and may explain the intellectual impairment observed in individuals with DLG3 mutations 19086053_Observational study of gene-disease association. (HuGE Navigator) 19167192_The results of this study suggested a putative role for DLG3/SAP102 in cortical hyperexcitability and epileptogenicity of malformations of cortical development. 19795139_Results identified a novel splice site mutation in the disc-large homolog 3 (DLG3) gene, encoding the synapse-associated protein 102 (SAP102) in one out of 300 families with moderate to severe non-syndromic mental retardation. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21252287_DLG3 was identified by genome-wide gene expression analyses as correlated with cellular sensitivity to cisplatin and carboplatin. DLG3 was also found to correlate with cellular sensitivity to platinating agents in NCI-60 cancer cell lines. 21369957_DLG3 did not associate with non-syndromic mental retardation in Chinese Han population; however, further studies are needed. 21384559_A total of six novel and 11 known single nucleotide polymorphisms were identified. Further studies are warranted to analyze the candidate genes at Xq11.1-q21.33. 22745750_Synapse associated protein 102 (SAP102) binds the C-terminal part of the scaffolding protein neurobeachin. 23103165_The PDZ-independent interaction between SAP102 and GluN2B mediates the synaptic clearance of GluN2B-containing NMDARs.(SAP102 protein) 24381070_The data of this study suggested that DLG3 is down-regulated in this cancer type. 24507884_This study identified DLG3 significantly associated loci with a biologically plausible role in schizophrenia. 24739954_miR-1246 might play a role in neurological pathogenesis of human enterovirus 71 by regulating DLG3 gene in infected cells. 25268382_Trans-homophilic interaction of CADM1 activates PI3K by forming a complex with MAGuK-family proteins MPP3 and Dlg. 25555912_These data shed new light on the role of SAP102 in the regulation of NMDAR trafficking. 27222290_Insertion of a guanine into the DLG3 5' UTR, 7 bp upstream of the start codon, down regulated DLG3 protein levels. This non-coding variant segregates with X-linked intellectual disability in a large family. 27222290_The dupG DLG3 variant segregated with non-syndromic X-linked intellectual disability in a large family and was predicted to disrupt folding of the mRNA. 27466188_Following the critical period NMDA receptor function was unaffected by loss of SAP102 but there was a reduction in the divergence of TC connectivity. These data suggest that changes in synaptic function early in development caused by mutations in SAP102 result in changes in network connectivity later in life. 28777483_This family broadens the mutational and phenotypical spectrum of DLG3-associated non-syndromic X-linked intellectual disability and demonstrates that heterozygous female mutation carriers can be as severely affected as males. 29282697_These data provide evidence for a novel mechanism in regulating SAP102 function and glutamate receptor trafficking. 31271664_High expression of DLG3 is associated with decreased survival from breast cancer. 32593652_Silence of lncRNA MIAT-mediated inhibition of DLG3 promoter methylation suppresses breast cancer progression via the Hippo signaling pathway. | ENSMUSG00000000881 | Dlg3 | 684.140977 | 0.9281825 | -0.107519557 | 0.11564555 | 8.621957e-01 | 3.531250e-01 | 7.078034e-01 | No | Yes | 750.810280 | 97.769720 | 790.626495 | 102.995962 | |
| ENSG00000082781 | 3693 | ITGB5 | protein_coding | P18084 | FUNCTION: Integrin alpha-V/beta-5 (ITGAV:ITGB5) is a receptor for fibronectin. It recognizes the sequence R-G-D in its ligand.; FUNCTION: (Microbial infection) Integrin ITGAV:ITGB5 acts as a receptor for adenovirus type C. {ECO:0000269|PubMed:20615244}. | Cell adhesion;Disulfide bond;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Integrin;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a beta subunit of integrin, which can combine with different alpha chains to form a variety of integrin heterodimers. Integrins are integral cell-surface receptors that participate in cell adhesion as well as cell-surface mediated signaling. The alphav beta5 integrin is involved in adhesion to vitronectin. [provided by RefSeq, Aug 2017]. | hsa:3693; | cell surface [GO:0009986]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; integrin alphav-beta5 complex [GO:0034684]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; integrin binding [GO:0005178]; signaling receptor activity [GO:0038023]; virus receptor activity [GO:0001618]; cell adhesion mediated by integrin [GO:0033627]; cell migration [GO:0016477]; cell-matrix adhesion [GO:0007160]; endodermal cell differentiation [GO:0035987]; epithelial cell-cell adhesion [GO:0090136]; integrin-mediated signaling pathway [GO:0007229]; stress fiber assembly [GO:0043149]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 11877043_Overexpression of the beta5 integrin in hematopoietic cells was associated with the inhibition of cell proliferation and apoptosis. 12270930_identification of a new alphavbeta5 integrin-interacting motif that is highly conserved in the fas-1 domains of many proteins suggesting that fas-1 domain-containing proteins may perform their biological functions by interacting with integrins 14741360_alphav subunit cleavage is essential for integrin function and has a considerable impact on integrin-dependent events, especially those leading to cell migration 15456946_Data suggest that modulating the expression of integrin subunits beta3/5 in human neurons may enhance adenoviral infectivity via the coxsackie-adenovirus receptor. 15866865_urokinase receptor-derived SRSRY peptide regulates cross-talk between fMLP and vitronectin receptors 15979906_different beta5 integrins: repeated-FNK (FNKFNK764-769) and single-FNK (FNK764-766) amino acid sequences in the cytoplasmic domain. 16005200_integrin alphavbeta6, alphavbeta3, alphavbeta5, alpha5beta1 and alpha9beta1 binding to osteopontin is controlled by specific structural motifs that are recognized by extracellular proteases 16385340_Expression of integrins alphavbeta1, alphavbeta3, and alphavbeta5 in cerebral arteriovenous malformations and cavernous malformations. (Integrins alphavbeta1, alphavbeta3, and alphavbeta5) 16614246_While tolerogenic DCs are not induced via alphavbeta5, coligation of CR3 and alphavbeta5 maintains the DC's tolerogenic profile. 16672769_Alphavbeta5 integrin may play a role in the adhesion and migration of VSMCs during the pathogenesis of atherosclerosis. 16675963_upregulated expression of alphavbeta5 contributes to autocrine TGF-beta signaling in localized scleroderma fibroblasts 17074516_Cell surface expression of alphavbeta5 resulted in an attenuation of alphavbeta3-mediated migration on vitronectin 17963729_Our studies suggest that the phagocytic function of beta5 integrin is regulated by an unconventional NPxY-talin-independent activation signal and argue for the existence of molecular switches in the beta5 cytoplasmic tail for adhesion and phagocytosis. 18162078_Real-time PCR showed that ETOH significantly altered the expression of genes involved in cell adhesion. There was an increase in the expression of alpha and beta Laminins 1, beta Integrins 3 and 5, Secreted phosphoprotein1 and Sarcoglycan epsilon. 18445685_data identify PAI1 as a novel regulator of fibronectin matrix assembly, and indicate that this regulation occurs through a previously undescribed crosstalk between the alphavbeta5 and alpha5beta1 integrins 18550570_Observational study of gene-disease association. (HuGE Navigator) 18590706_Oxidized LDL impairs angiogenic properties of endothelial progenitor cells at sub-apoptotic levels by downregulation of E-selectin and integrin alphavbeta5, both substantial mediators of EPC-endothelial cell 18648521_human umbilical vein endothelial cells adhere to immobilized CXCL4 through alphavbeta3 integrin, and also through other integrins, such as alphavbeta5 and alpha5beta1 18844213_urokinase-derived antagonists of alphavbeta5 integrin activation inhibit migration and invasion of carcinoma cells 19255147_The active site within the Sdc1 core protein was identified and a peptide inhibitor called synstatin (SSTN) that disrupts Sdc1's interaction with alpha(v)beta(3) and alpha(v)beta(5) integrins was derived. 19267182_Compound 4, having the DGR retro-sequence, is a low micromolar ligand for the alphavbeta5 integrin. 19502044_Observational study of gene-disease association. (HuGE Navigator) 19695571_Periostin mediates vascular smooth muscle cell migration through the integrins alphavbeta3 and alphavbeta5 and focal adhesion kinase (FAK) pathway 19811096_Beta1 integrins are required for adhesion and proliferation of induced pluripotent stem cells on Matrigel. On vitronectin, the integrin alphavbeta5 is required for initial attachment, but both alphavbeta5 and beta1 is required for proliferation. 19910644_The angiogenic effects of leptin are mediated by circulating angiogenic cells and involve src kinase dependent phosphorylation of integrin alphavbeta5. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20026907_Definitive endoderm highly express the integrins alphaV and beta5, which have the ability to bind to vitronectin. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20400979_Ovarian cancer ascites induces FAK and Akt activation in an alphavbeta5 integrin-dependent pathway, which confers protection from TRAIL-induced cell death and caspase activation. 20404485_The beta5-integrin adhesions contribute to the TGFbeta-induced EMT and the tumorigenic potential of carcinoma cells. 20431064_Src kinase-mediated activation of STAT3 and subsequent angiogenic gene expression mediate the effects of integrin alpha v beta 5 and may be exploited to enhance the paracrine activities of circulating angiogenic cells. 20507994_p21-activated kinase 4 phosphorylation of integrin beta5 Ser-759 and Ser-762 regulates cell migration 20615244_Integrin alphavbeta5 is a primary receptor for adenovirus. 20626753_Data show that knockdown of integrin alphav and erbB3 by small-interfering RNAs significantly inhibited cell migration induced by HRG. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20719960_PAK4 is activated by cell attachment to vitronectin as mediated by PAK4 binding partner integrin alphavbeta5. 21098231_beta5 integrin is identified as the most important partner of the alpha vitronectin-coupled integrin dimer in monocyte-derived macrophages, thus rendering these cells more susceptible to NF-kappaB-dependent HIV-1 infection. 21344378_CCN3 enhances the migration of chondrosarcoma cells by increasing MMP-13 expression through the alphavbeta3/alphavbeta5 integrin receptor, FAK, PI3K, Akt, p65, and NF-kappaB signal transduction pathway. 21978494_alphanubeta3 and alphanubeta5 have roles in photon-induced hypermigration of malignant glioma cells 22503669_Letter: loss of integrin beta5 in polyomavirus positive Merkel cell carcinoma may influence cell adhesion/migration. 22547695_Cysteine-rich protein Cyr61 activates interleukin (IL)-6 production via the alphavbeta5/Akt/NF-kappaB signaling pathway in rheumatoid arthritis. 22561002_High beta5-integrin protein expression is associated with aggressive behavior in gastric cancer. 22566688_knockdown of endogenous alphavbeta5 expression or treatment with a function-blocking alphavbeta5 antibody significantly decreased stabilin-2-mediated phagocytosis in the absence of soluble factors 22659470_Active MMP-2 regulates VEGF-A in melanoma cells on a transcriptional level via an integrin alphavbeta5/phosphoinositide-3-kinase-dependent pathway. 22692860_Data indicate the role of CYR61 and alpha(V)beta5 integrin as proteins that cooperate to mediate cancer cell migration. 22824793_Depletion of integrin beta5 in triple-negative breast carcinoma cells markedly reduced tumor take, growth and tumor angiogenesis, whereas reexpression of integrin beta5 rescued this phenotype; findings show a critical role for integrin beta5 in the tumorigenic potential of breast carcinoma cells 22879933_These studies implicate ASM as a mediator of apoptosis induced by inhibition of integrins alphavbeta3/alphavbeta5, and for the first time place c-Abl as an upstream regulator of ASM expression and activity. 23028753_Saccharomyces boulardii supernatant contains motogenic factors that enhance cell restitution through multiple pathways, including the dynamic fine regulation of alphavbeta5 integrin binding activity. 23227240_CTGF increased IL-6 production in osteoarthritic synovial fibroblasts via the alphavbeta5 integrin, ASK1, p38/JNK, and AP-1/NF-kappaB signaling pathways. 23261238_integrin alphavbeta5 might play an important role in invasion and metastases of laryngeal squamous cell carcinoma 23269786_study demonstrate that binding of integrin alphavbeta5 and alpha defensin 5 have opposite effects on the elastic response of adenovirus type 35, revealing a direct link between virus-host interactions and the mechanical properties of the capsid 23462327_Data suggest that the integrin alphaVbeta5 receptor is largely responsible for mediating the differentiation process in pancreatic cells. 23874483_BDNF enhances the migration of chondrosarcoma by increasing beta5 integrin expression through a signal transduction pathway that involves the TrkB receptor, PI3K, Akt, and NF-kappaB. 23962022_molecular model of integrin b5 structure was prepared and stereo chemical model quality was checked. Integrin b5 active sites were identified based on insilico analysis tools. 23984888_The top primary GWAS association of Airway hyperresponsiveness was in rs848788(P-value 7.2E-07) located within the ITGB5 genes. 24036928_Integrin-beta5 and zyxin mediate formation of ventral stress fibers in response to transforming growth factor beta. 24294359_Brain metastases ITGB5 expression exhibits considerable heterogeneity according to tumor origin. 24389192_findings suggest that Alpha(v)Beta5 integrins and their natural ligand VN are involved in P. fluorescens adherence and invasion in human epithelial cells 24455718_we demonstrated via antibody blocking experiments that aV b5 and a6 significantly promoted hESC attachment in 2%oxygen only, whereas blockage of CD44 inhibited cell attachment in 21% O2 alone 24639195_Integrins alphavbeta5 and alphavbeta3 both control myofibroblast differentiation by activating latent TGF-beta1 24899686_FAK activity regulates beta5 integrin expression and anchorage-independent cell growth in ovarian carcinoma. 24997346_Myo7a interacts with integrin beta5 and selectively promotes integrin alphavbeta5-mediated cell migration 25150423_Immunohistochemical analysis for alphavbeta3, alphavbeta5 and alphavbeta6 integrin subunits was performed and correlated with Ki67 and hypoxia-inducible factor (HIF)-1alpha indexes 25448675_FRMD5 regulates tumor cell motility via a dual pathway involving FRMD5 binding to integrin beta5 tail and to ROCK1 25472585_ITGA3, ITGA6, ITGB3,ITGB4 and ITGB5 are associated with GC susceptibility (rs2675), ITGA3, ITGA6, ITGB3, ITGB4 and ITGB5 are associated with gastric cancer susceptibility tumor stage and lymphatic metastasis in Chinese Han population 26244551_Data suggest that the binding, but not the internalization of photoreceptor outer segments (POS) confers protective effects on retinal pigment epithelium (RPE) cells through the alphavbeta5 integrin/focal adhesion kinase (FAK)/PGC-1alpha pathway. 26500056_Findings indicate integrin inhibition as a promising strategy to block both aryl hydrocarbon receptor (AhR) and transforming growth factor beta (TGF-beta)-controlled features of malignancy in glioblastoma. 26758421_High ITGB5 expression is associated with cervical cancer. 29386044_Our data reveal that the miR-185-ITGB5-beta-catenin pathway plays an important role in HCC tumorigenesis, and ITGB5 may be a promising specific target for HCC therapy. 31956038_Zika Virus Targets Glioblastoma Stem Cells through a SOX2-Integrin alphavbeta5 Axis. 33859325_Integrin alphavbeta5 heterodimer is a specific marker of human pancreatic beta cells. 34626585_Scavenger receptor MARCO contributes to macrophage phagocytosis and clearance of tumor cells. | ENSMUSG00000022817 | Itgb5 | 1078.529387 | 0.8015880 | -0.319067114 | 0.09015451 | 1.244655e+01 | 4.187652e-04 | 2.104295e-02 | No | Yes | 814.128144 | 113.339198 | 1079.342119 | 150.056234 | |
| ENSG00000083097 | 23033 | DOP1A | protein_coding | Q5JWR5 | FUNCTION: May be involved in protein traffic between late Golgi and early endosomes. {ECO:0000250|UniProtKB:Q03921}. | Golgi apparatus;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:23033; | cytosol [GO:0005829]; endosome [GO:0005768]; Golgi membrane [GO:0000139]; trans-Golgi network [GO:0005802]; Golgi to endosome transport [GO:0006895]; protein transport [GO:0015031] | 25424701_Assessment of REST-N50 and DOPEY1v2 may prove useful in diagnostic blood tests of breast cancer. REST-N50 shows a high potential as a blood biomarker for evaluating the effectiveness of therapy in the neoadjuvant setting. | ENSMUSG00000034973 | Dop1a | 795.837556 | 1.2361636 | 0.305869744 | 0.10137634 | 9.149279e+00 | 2.488161e-03 | 5.715660e-02 | No | Yes | 744.782493 | 127.102955 | 609.811295 | 104.131135 | ||
| ENSG00000084072 | 10450 | PPIE | protein_coding | Q9UNP9 | FUNCTION: Involved in pre-mRNA splicing as component of the spliceosome (PubMed:11991638, PubMed:28076346). Combines RNA-binding and PPIase activities (PubMed:8977107, PubMed:18258190, PubMed:20677832, PubMed:20460131). Binds mRNA and has a preference for single-stranded RNA molecules with poly-A and poly-U stretches, suggesting it binds to the poly(A)-region in the 3'-UTR of mRNA molecules (PubMed:8977107, PubMed:18258190, PubMed:20460131). Catalyzes the cis-trans isomerization of proline imidic peptide bonds in proteins (PubMed:8977107, PubMed:18258190, PubMed:20677832, PubMed:20541251). Inhibits KMT2A activity; this requires proline isomerase activity (PubMed:20677832, PubMed:20541251, PubMed:20460131). {ECO:0000269|PubMed:11991638, ECO:0000269|PubMed:18258190, ECO:0000269|PubMed:20460131, ECO:0000269|PubMed:20541251, ECO:0000269|PubMed:20677832, ECO:0000269|PubMed:28076346, ECO:0000269|PubMed:8977107}. | 3D-structure;Alternative splicing;Isomerase;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Rotamase;Spliceosome;mRNA processing;mRNA splicing | The protein encoded by this gene is a member of the peptidyl-prolyl cis-trans isomerase (PPIase) family. PPIases catalyze the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and accelerate the folding of proteins. This protein contains a highly conserved cyclophilin (CYP) domain as well as an RNA-binding domain. It was shown to possess PPIase and protein folding activities, and it also exhibits RNA-binding activity. Alternative splicing results in multiple transcript variants. A related pseudogene, which is also located on chromosome 1, has been identified. [provided by RefSeq, Aug 2010]. | hsa:10450; | catalytic step 2 spliceosome [GO:0071013]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; secretory granule lumen [GO:0034774]; U2-type catalytic step 2 spliceosome [GO:0071007]; cyclosporin A binding [GO:0016018]; mRNA binding [GO:0003729]; peptidyl-prolyl cis-trans isomerase activity [GO:0003755]; poly(A) binding [GO:0008143]; RNA binding [GO:0003723]; mRNA splicing, via spliceosome [GO:0000398]; positive regulation of viral genome replication [GO:0045070]; protein folding [GO:0006457]; protein peptidyl-prolyl isomerization [GO:0000413]; regulation of transcription, DNA-templated [GO:0006355] | 18258190_The results show that hCyP33 binds specifically to mRNA, namely poly(A)(+)RNA, and that binding stimulates the PPIase activity of hCyP33. 20460131_Data provide insight into the multiple functions of Cyp33 RRM and suggest a Cyp33-dependent mechanism for regulating the transcriptional activity of MLL. 20532202_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20541251_PPIase domain of CyP33 regulates the conformation of MLL1 through proline isomerization within the PHD3-Bromo linker, thereby disrupting the PHD3-Bromo interface and facilitating binding of the MLL1-PHD3 domain to the CyP33-RRM domain. 20677832_binding of H3K4me3 to PHD3 domain of MLL and binding of the CYP33 RRM domain to PHD3 are mutually inhibitory, implying that PHD3 is a molecular switch for the transition between activation and repression of target genes 21887220_CypE is a host restriction factor that inhibits the functions of nucleoprotein, as well as viral replication and transcription, by impairing the formation of the viral ribonucleoprotein complex. 30389953_Study of datasets analyzing the gene expression profiles and DNA methylation data of gestational Diabetes Mellitus (GDM) and confirmed through real time PCR in placenta tissue of three GDM samples and three normal samples identified Oas1, Ppie, Polr2g as possible pathogenic target genes of GDM by combining protein-protein interaction analysis. 33606679_Cyp33 binds AU-rich RNA motifs via an extended interface that competitively disrupts the gene repressive Cyp33-MLL1 interaction in vitro. | ENSMUSG00000028651 | Ppie | 1814.941939 | 1.0293439 | 0.041725043 | 0.07519598 | 3.067562e-01 | 5.796775e-01 | 8.438661e-01 | No | Yes | 1818.110588 | 241.318967 | 1787.322268 | 237.202955 | |
| ENSG00000087263 | 55239 | OGFOD1 | protein_coding | Q8N543 | FUNCTION: Prolyl 3-hydroxylase that catalyzes 3-hydroxylation of 'Pro-62' of small ribosomal subunit uS12 (RPS23), thereby regulating protein translation termination efficiency. Involved in stress granule formation. {ECO:0000269|PubMed:20154146, ECO:0000269|PubMed:24550447, ECO:0000269|PubMed:24550462}. | 3D-structure;Alternative splicing;Cytoplasm;Dioxygenase;Iron;Metal-binding;Nucleus;Oxidoreductase;Reference proteome;Vitamin C | hsa:55239; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; iron ion binding [GO:0005506]; L-ascorbic acid binding [GO:0031418]; peptidyl-proline 3-dioxygenase activity [GO:0031544]; peptidyl-proline dioxygenase activity [GO:0031543]; cell population proliferation [GO:0008283]; peptidyl-proline hydroxylation [GO:0019511]; protein hydroxylation [GO:0018126]; regulation of translational termination [GO:0006449]; stress granule assembly [GO:0034063] | 20154146_Data suggest that OGFOD1 plays important proapoptotic roles in the regulation of translation and HRI-mediated phosphorylation of eIF2alpha in cells subjected to arsenite-induced stress. 20579638_OGFOD1 plays an important role in ischemic cell survival and an OGFOD1 iron binding residue is required for ATPAF1 gene expression. 24550447_OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. 25909288_Propose that OGFOD1 is required for breast cancer cell proliferation and is associated with poor prognosis in breast cancer. 31112528_The ribosomal prolyl-hydroxylase OGFOD1 decreases during cardiac differentiation and modulates translation and splicing. 32002629_High OGFOD1 expression is associated with papillomavirus-infected laryngeal papillomas. | ENSMUSG00000033009 | Ogfod1 | 3017.206060 | 1.0460779 | 0.064990281 | 0.07707434 | 7.083658e-01 | 3.999869e-01 | 7.436376e-01 | No | Yes | 3737.683490 | 564.504894 | 3567.498778 | 538.810300 | ||
| ENSG00000088387 | 23348 | DOCK9 | protein_coding | Q9BZ29 | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Overexpression induces filopodia formation. {ECO:0000269|PubMed:12172552, ECO:0000269|PubMed:19745154}. | 3D-structure;Alternative splicing;Coiled coil;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;Repeat | hsa:23348; | cytosol [GO:0005829]; endomembrane system [GO:0012505]; membrane [GO:0016020]; cadherin binding [GO:0045296]; guanyl-nucleotide exchange factor activity [GO:0005085]; positive regulation of GTPase activity [GO:0043547]; small GTPase mediated signal transduction [GO:0007264] | 12172552_Sequence comparison combined with mutational analysis identified a new domain, which we named CZH2, that mediates direct interaction with Cdc42 17728666_DOCK9 contributes to both risk and increased illness severity in bipolar disorder. 17728666_Observational study of gene-disease association. (HuGE Navigator) 17935486_novel functions for the N-terminal region of zizimin1. 18056264_DOCK2 and DOCK9 specifically recognize Rac2 and Cdc42 through their switch 1 as well as beta2-beta3 regions and the mode of recognition via switch 1 appears to be conserved among diverse Rac-specific DHR-2 GEFs 18729074_interaction between Smad2/3 and the Cdc42 guanine nucleotide exchange factor, Zizimin1, in response to TGF-beta1 19745154_through structural analysis of DOCK9-Cdc42 complexes, we identify a nucleotide sensor within the alpha10 helix of the DHR2 domain that contributes to release of guanine diphosphate (GDP) and then to discharge of the activated GTP-bound Cdc42 19809089_Studies indicate that that many of the mechanistic principles of the exchange process are conserved in the DOCK9-catalyzed reaction. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 26641546_c.2262A>C substitution in DOCK9 leads to a splicing aberration. However, because the mutation effect was observed in vitro, a definitive relationship between DOCK9 and KTCN phenotype could not be established. 34741163_Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. | ENSMUSG00000025558 | Dock9 | 811.023322 | 0.9414811 | -0.086995999 | 0.10596551 | 6.676306e-01 | 4.138789e-01 | 7.538347e-01 | No | Yes | 758.216525 | 100.963238 | 820.963928 | 109.398137 | ||
| ENSG00000088888 | 57506 | MAVS | protein_coding | Q7Z434 | FUNCTION: Required for innate immune defense against viruses (PubMed:16125763, PubMed:16127453, PubMed:16153868, PubMed:16177806, PubMed:19631370, PubMed:20451243, PubMed:23087404, PubMed:20127681, PubMed:21170385, PubMed:24990078). Acts downstream of DHX33, DDX58/RIG-I and IFIH1/MDA5, which detect intracellular dsRNA produced during viral replication, to coordinate pathways leading to the activation of NF-kappa-B, IRF3 and IRF7, and to the subsequent induction of antiviral cytokines such as IFNB and RANTES (CCL5) (PubMed:16125763, PubMed:16127453, PubMed:16153868, PubMed:16177806, PubMed:19631370, PubMed:20451243, PubMed:23087404, PubMed:25636800, PubMed:20127681, PubMed:21170385, PubMed:20628368, PubMed:33110251). Peroxisomal and mitochondrial MAVS act sequentially to create an antiviral cellular state (PubMed:20451243). Upon viral infection, peroxisomal MAVS induces the rapid interferon-independent expression of defense factors that provide short-term protection, whereas mitochondrial MAVS activates an interferon-dependent signaling pathway with delayed kinetics, which amplifies and stabilizes the antiviral response (PubMed:20451243). May activate the same pathways following detection of extracellular dsRNA by TLR3 (PubMed:16153868). May protect cells from apoptosis (PubMed:16125763). {ECO:0000269|PubMed:16125763, ECO:0000269|PubMed:16127453, ECO:0000269|PubMed:16153868, ECO:0000269|PubMed:16177806, ECO:0000269|PubMed:19631370, ECO:0000269|PubMed:20127681, ECO:0000269|PubMed:20451243, ECO:0000269|PubMed:20628368, ECO:0000269|PubMed:21170385, ECO:0000269|PubMed:23087404, ECO:0000269|PubMed:24990078, ECO:0000269|PubMed:25636800, ECO:0000269|PubMed:33110251}. | 3D-structure;Alternative splicing;Antiviral defense;Host-virus interaction;Immunity;Innate immunity;Membrane;Methylation;Mitochondrion;Mitochondrion outer membrane;Peroxisome;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Ubl conjugation | This gene encodes an intermediary protein necessary in the virus-triggered beta interferon signaling pathways. It is required for activation of transcription factors which regulate expression of beta interferon and contributes to antiviral innate immunity. [provided by RefSeq, Jul 2020]. | hsa:57506; | integral component of membrane [GO:0016021]; mitochondrial membrane [GO:0031966]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; peroxisomal membrane [GO:0005778]; CARD domain binding [GO:0050700]; protein kinase binding [GO:0019901]; signaling adaptor activity [GO:0035591]; activation of innate immune response [GO:0002218]; cellular response to exogenous dsRNA [GO:0071360]; defense response to bacterium [GO:0042742]; defense response to virus [GO:0051607]; innate immune response [GO:0045087]; negative regulation of viral genome replication [GO:0045071]; positive regulation of chemokine (C-C motif) ligand 5 production [GO:0071651]; positive regulation of defense response to virus by host [GO:0002230]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of interferon-alpha production [GO:0032727]; positive regulation of interferon-beta production [GO:0032728]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of interleukin-8 production [GO:0032757]; positive regulation of IP-10 production [GO:0071660]; positive regulation of myeloid dendritic cell cytokine production [GO:0002735]; positive regulation of protein import into nucleus [GO:0042307]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of response to cytokine stimulus [GO:0060760]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of type I interferon-mediated signaling pathway [GO:0060340]; regulation of peroxisome organization [GO:1900063]; signal transduction [GO:0007165] | 16125763_Mitochondrial antiviral signaling (MAVS) mediates the activation of NF-kappaB and IRF 3 in response to viral infection, and transmembrane domain targets MAVS to the mitochondria, implicating a new role of mitochondria in innate immunity. 16153868_Data suggest that VISA is critically involved in both virus-triggered TLR3-independent and TLR3-mediated antiviral interferon signaling. 16177806_KIAA1271 (aka Cardif, MAVS, VISA, IPS-1) has been shown to be targeted by hepatitis c virus (HCV) NS3/4A protease. This cleavage interferes with the Rig-I mediated dsRNA sensing pathway and therefore with the activation of IRF-3. 16585524_HCV infection transiently induces RIG-I- and IPS-1-dependent IRF-3 activation 16707574_HCV blocks the dsRNA signaling by an NS3/4A-independent mechanism, in addition to the NS3/4A-dependent cleavage of MAVS/IPS-1. 16731946_HCV NS3-4A cleavage of human MAVS/IPS-1/VISA/Cardif/K1271 results in its dissociation from the mitochondrial membrane and disrupts signaling to the antiviral immune response 16858409_Cardif induces interferon (IFN)alpha through a direct and specific interaction with the TRAF domain of TRAF3, implicating Cardif as the link between cytoplasmic viral receptors and TRAF3. 16984921_IPS-1, IRF3, and IFNbeta have critical roles in Legionella infection of lung epithelium 17093192_GBV-B NS3/4A protease specifically cleaves VISA and dislodges it from mitochondria, thereby disrupting its function as a RIG-I adaptor and blocking downstream activation of both interferon regulatory factor 3 and nuclear factor kappa B. 17911629_IPS-1 is an essential component of the pathway relevant to polyinosinic-polycytidylic acid signaling of type I interferon in intestinal epithelial cells. 18207245_This study uncovers MAVS splicing variants of diverse biological function. 18307765_Models show a very different charge profile for the equivalent surfaces compared to IPS-1/MAVS/VISA/Cardif CARD. 18550535_RIG-1 - MAVS interacts with cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I - NF- kappa B-inducing kinase signaling pathway 18692023_The role of MAVS in apoptosis was investigated. 18692023_This paper describes the biochemical regulation and behavior of MAVS during apoptosis. 18756281_Inactivation of Trif and Cardif can also occur through cellular caspases activated by various pro-apoptotic signals. 18927075_PKR facilitates the host innate immune response and apoptosis in virus-infected cells by mediating IRF-3 activation through the mitochondrial IPS-1 signal transduction pathway 18977754_Upon Sendai virus (SeV) infection, TBK1s is induced in both human and mouse cells and binds to RIG-1, disrupting the interaction between RIG-I and VISA 18984593_IPS-1 requires TRAF6 and MEKK1 to activate NF-kappaB and mitogen-activated protein kinases that are critical for the optimal induction of type I interferons 19028691_PKR, in addition to IPS-1 and IRF3 but not TRIF, was required for maximal type I IFN-beta induction and the induction of apoptosis by both transfected PRNAs and polyinosinic-polycytidylic acid. 19193783_The activation of RIG-I/MDA-5 leads to the C-terminal transmembrane-dependent dimerization of the MAVS N-terminal caspase recruitment domain, thereby providing an interface for direct binding to and activation of the downstream effector TRAF3. 19351494_Data show that DeltaIPS-1 could decrease the secretory volume of IFN-beta in HEK293T cell and could not completely suppress the CPE of the cell infected by HSV-1. 19380491_recruitment of IKKepsilon to the mitochondria upon MAVS K500 ubiquitination plays a modulatory role in the cascade leading to NF-kappaB activation and expression of inflammatory and antiviral genes 19546225_Polo-like kinase 1 (PLK1) regulates interferon (IFN) induction by MAVS 19591957_Report shows that Rig-I, but not MAVS, is cleaved during cytomegalovirus infection. 19644511_show that interferon-beta promoter stimulator 1 (IPS-1) binds DAP3 and induces anoikis by caspase activation. 19690333_Data revealed that Mfn2 interacted with the carboxyl-terminal region of MAVS through a heptad repeat region, providing a structural perspective on the regulation of the mitochondrial antiviral response. 19701189_recognition of a viral ssRNA genome, Nod2 used the adaptor protein MAVS to activate IRF3. 19881509_PCBP2-AIP4 axis defines a new signaling cascade for MAVS degradation and 'fine tuning' of antiviral innate immunity. 19902255_Observational study of gene-disease association. (HuGE Navigator) 19914245_c-Abl modulates innate immune response through MAVS. 20032188_study reports MAVS protein level is reduced in Dengue virus-infected cells & that caspases 1 & 3 cleave MAVS at residue D429; MAVS is also a proapoptotic molecule that triggers disruption of the mitochondrial membrane potential & activation of caspases 20044805_the pre-activation status of the endogenous IFN system in the liver of patients with CHC is in part regulated by cleavage of MAVS. 20127681_DDX3 can bind viral RNA to join it in the IPS-1 complex. The 622-662 a.a DDX3 C-terminal region directly bound to the IPS-1 CARD-like domain. 20154210_Chlamydia pneumoniae infection of vascular endothelial cells activates mitochondrial antiviral signaling protein (MAVS) which stimulates IRF3- and IRF7-dependent signals controlling bacterial growth and modulating development of vascular lesions. 20538852_Influenza virus A PB2 protein interacts with MAVS and inhibits MAVS-mediated beta interferon expression. 20554965_Study suggests that HBV can target the RIG-I signaling by HBX-mediated MAVS downregulation, thereby attenuating the antiviral response of the innate immune system. 20661427_viral infection as well as transfection with 5'ppp-RNA resulted in the redistribution of IPS-1 to form speckle-like aggregates in cells. 20699220_These findings demonstrate that the viral polymerase plays an important role for regulating host anti-viral response through the binding to IPS-1 and inhibition of IFNbeta production. 20711230_indicate that Hepatitis B virus X protein inhibits signaling by components upstream but not downstream of VISA 20877624_Observational study of gene-disease association. (HuGE Navigator) 20962078_Chikungunya virus induces via IPS-1 accumulation of IRF-3 dependent antiviral gene mRNA. 21068253_The hepatitis B virus X protein-IPS-1 protein interaction was confirmed in plasmid-transfected HepG2 cells by reciprocal coimmunoprecipitation and Western blotting. 21127988_Polymorphisms in the VISA gene may be related to disease susceptibility and manifestations of systemic lupus erythematosus. 21130742_these results showed that conjunctival epithelial cells express RIG-I and MDA5, and IPS-1, an adaptor molecule common to RIG-I and MDA5, contributes to polyI:C-inducible cytokine production in conjunctival epithelial cells. 21170385_Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction 21187438_RIG-I or melanoma differentiation-associated gene (MDA)5 signaling through mitochondrial antiviral signaling protein MAVS is required for activation of interferon (IFN)-beta production by rotavirus-infected intestinal epithelial cells. 21200404_A functional C-terminal TRAF3-binding site in mitochondrial antiviral signaling protein participates in positive and negative regulation of the interferon antiviral response 21268286_In African-American systemic lupus erythematosus, patients, the C79F allele was associated with low type I interferon production and absence of anti-RNA-binding protein autoantibodies. 21285412_Data sugget that DeltaPsi(m) and MAVS are coupled at the same stage in the RLR antiviral signaling pathway. 21421666_NOD2 genotypes also influence the microbial composition in humans. 21782231_Viral infection induces the formation of very large MAVS aggregates, which potently activate IRF3 in the cytosol; results suggest that a prion-like conformational switch of MAVS activates and propagates the antiviral signaling cascade. 21813773_Mechanistically, the tetratrico-peptide repeat motif (E164/E165) of IFIT3 interacts with the N terminus (K38) of TBK1, thus bridging TBK1 to MAVS on the mitochondrion 21865020_The mitochondrial antiviral signaling (MAVS) adapter protein orchestrates the innate host response to RNA virus infections. (Review) 21957149_The ability of DHX9 in sensing both poly I:C and RNA virus is mediated by IPS-1 in myeloid dendritic cells. 22105485_Our findings unravel a critical role of PCBP1 in regulating MAVS for both fine-tuning the antiviral immunity and preventing inflammation 22131337_UXT-V1 represents a novel integral component of the MAVS signalosome on mitochondria, mediating the innate antiviral signal transduction. 22341464_The authors show that FAK interacts with MAVS at the mitochondrial membrane in a virus infection-dependent manner and potentiates MAVS-mediated signaling via a kinase-independent mechanism. 22383950_NS1 binds to MAVS and this binding inhibits the MAVS-RIG-I interaction required for IFN production 22623778_Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. 22626058_Study reports that RLR activation triggers MAVS ubiquitination on lysine 7 and 10 by the E3 ubiquitin ligase TRIM25 and marks it for proteasomal degradation concomitantly with downstream signaling. 22674996_Using a flow cytometry-based assay, the authors demonstrate that the PB1-F2 protein inhibits MAVS-mediated IFN synthesis by decreasing the mitochondrial membrane potential (MMP). 22792062_model proposed where dsDNA and dsRNA sensing induces the formation of membrane-bound compartments originating from the Golgi, which mediate the dynamic association of TRAF3 with MAVS leading to an optimal induction of innate immune responses 22870331_Canine hepacivirus NS3 serine protease can cleave the human adaptor proteins MAVS and TRIF. 22901541_Ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. 23028806_These results suggest that the comprehensive regulation of MAVS in response to foreign RNA may be essential to antiviral host defenses. 23087404_Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. 23246644_The influenza A virus PB2 MAVS-binding domain unexpectedly coincided with its PB1-binding domain, indicating an important dual functionality for this region of PB2. 23308256_IPS-1 oligomerization is essential for the formation of a multiprotein signaling complex. 23460740_rim44 functions as a positive regulator of the virus-triggered immune response by enhancing the stability of VISA. 23574001_The results establish that the mitochondrial location of IPS-1 is essential to its ability to induce apoptosis. 23582325_Findings reveal a previously undescribed mechanism by which NLRP3 localization to the mitochondria and inflammasome activation are regulated by the mitochondrial adaptor protein MAVS, and identify an N-terminal sequence in NLRP3 that supports its association with MAVS. 24048902_results suggest that MAVS facilitates the recruitment of NLRP3 to the mitochondria and may enhance its oligomerization and activation by bringing it in close proximity to mitochondrial reactive oxygen species. 24285545_data thus implicate two parallel pathways by which Gp78 regulates MAVS signaling 24390337_MDA5 and MAVS cleavages are both mediated by coxsackievirus B3 2A protease. 24391214_In virus-infected cells, a Bax/Bak-independent pathway involves dsRNA-induced innate immune signaling via mitochondrial antiviral signaling (MAVS) and caspase-8. 24529381_Study reports an example of alternative translation as a means of regulating innate immune signaling. MAVS, a regulator of antiviral innate immunity, is expressed from a bicistronic mRNA encoding a second protein, miniMAVS. 24569476_Elucidate the structural mechanism of MAVS polymerization, and explain how an alpha-helical domain uses distinct chemical interactions to form self-perpetuating filaments. 24613846_Collectively, these results demonstrated that eEF1Bgamma functions as a positive regulator of NF-small ka, CyrillicB signal by targeting MAVS for activation, which provides a new regulating mechanism of antiviral responses. 24623417_Overall, this study suggests a role for MAVS and RIG-I signaling during different stages of the Kaposi's sarcoma-associated herpesvirus life cycle. 24643253_MAVS protein is attenuated by rotavirus nonstructural protein 1. 24659800_plays an essential role in stress granule formation 24729608_that Smurf2 is an important nonredundant negative regulator of virus-triggered type I IFN signaling by targeting VISA for K48-linked ubiquitination and degradation. 24782566_Data indicate that DEAH-box RNA helicase DHX15/PRP43 stimulates the NF-kappaB and MAPK pathways downstream of virus-induced signaling adapter protein MAVS and contributes to MAVS-mediated cytokine production and apoptosis. 24889249_NS3 protease of GB virus C cleaved MAVS but not TRIF, and it inhibited interferon responses sufficiently to enhance growth of an interferon-sensitive virus. 25142606_The DLAIS motif in MAVS was found to be critical for MIB2 binding, the ligation of K63-linked ubiquitin to TANK-binding kinase 1, and phosphorylation-mediated IRF3/7 activation. 25288302_MAVS did not reveal significant single-SNP associations with multiple sclerosis risk. 25609814_Hepatitis C virus NS3-4A similarly diminished both human and mouse MAVS-dependent signaling in human and mouse cells and MAVS induces both type I and type III interferons, which together control the hepatitis C virus replication. 25636800_findings show MAVS and STING harbor 2 conserved serine and threonine clusters that are phosphorylated by IKK and/or TBK1 in response to stimulation; results reveal phosphorylation of innate adaptor proteins is an essential and conserved mechanism that selectively recruits IRF3 to activate the type I IFN pathway 25640825_Polymorphisms in IPS1 are independently associated with treatment response to PEG-IFN among Chinese HBeAg-positive CHB patients. 25833049_Notably, in Lymphocytic Choriomeningitis Virus-infected cells, RIG-I was dispensable for virus-induced apoptosis via MAVS. 25950488_IPS-1 induces anticancer activity through upregulation of pro-apoptotic gene TRAIL and downregulation of the anti-apoptotic genes BCL2, BIRC3 and PRKCE via IRF3 and IRF7 in type I interferon-dependent and -independent manners. 26179906_pVHL Negatively Regulates Antiviral Signaling by Targeting MAVS for Proteasomal Degradation 26183716_An autoinhibitory mechanism modulates MAVS activity in unstimulated cells and, on viral infection, individual regions of MAVS are released following MAVS filament formation to activate antiviral signalling cascades. 26221961_MAVS50, exposing a degenerate TRAF-binding motif within its N-terminus, effectively competed with full-length MAVS for recruiting TRAF2 and TRAF6 26223644_During Crimean-Congo hemorrhagic fever virus infection, RIG-I mediated a type I interferon response via MAVS. 26246171_MARCH5 binds MAVS only during viral stimulation when MAVS forms aggregates, and these interactions require the RING domain of MARCH5 and the CARD domain of MAVS. 26317833_Results show MAVS-transmembrane domain is shown to oligomerize in response to changes in the outer mitochondrial lipid membrane properties caused by treatment with mitochondrial reactive oxygen species inducers or by Sendai virus infection. 26385923_Transmembrane motif T6BM2-mediated TRAF6 binding is required for MAVS-related antiviral response. 26437794_These results demonstrate that poliovirus infection actively suppresses the host type I interferon response by blocking activation of IRF-3 and suggests that this is not mediated by cleavage of MDA-5 or IPS-1. 26512076_These results suggest that vIRF-1 is the first example of a viral protein to inhibit mitochondrial antiviral signaling through lipid raft-like microdomains. 26588843_indicate comparable activation of the IFN response by pex- and mito-mitochondrial antiviral-signaling protein in hepatocytes and efficient counteraction of both MAVS species by the HCV nonstructural protein 3 protease 26646717_Using bone marrow derived macrophages from knockout mice we demonstrate that hBD3 suppresses the polyI:C-induced TLR3 response mediated by TICAM1 (TRIF), while exacerbating the cytoplasmic response through MDA5 (IFIH1) and MAVS (IPS1/CARDIF). 26893477_deliberately targeting the evolutionarily conserved MDA-5-IPS-1 antiviral pathway in tumors can provoke parallel tumoricidal and immunostimulatory effects that bridge innate and adaptive immune responses for the therapeutic treatment of cancer 26906558_Pyruvate carboxylase activates the RIG-I-like receptor-mediated antiviral immune response by targeting the MAVS signalosome. 26954674_This study identified new functional alterations in antiviral signalling based on MAVS polymorphisms 26998762_Data show that the NS3 protein of dengue virus bound to 14-3-3 epsilon protein (14-3-3varepsilon) and prevented translocation of retinoic acid-inducible gene-I protein (RIG-I) to the adaptor MAVS protein and thereby blocked antiviral signaling. 27213432_Mechanistic studies showed that HACE1 exerts its inhibitory role on virus-induced signaling by disrupting the MAVS-TRAF3 complex. 27226371_Deficiency of MAVS in hematopoietic cells resulted in increased mortality and delayed West Nile Virus clearance from the brain. 27438769_this study shows that keratinocytes are an important source of IFN-beta and MAVS is critical to this function, and demonstrates how the epidermis triggers unwanted skin inflammation under disease conditions 27553710_Studied association of genetic variants of the MAVS, MITA and MFN2 genes with leprosy in Han Chinese from Southwest China; found no association between the variants and susceptibility to leprosy. 27593154_this study shows that MAVS silencing upregulates IFN-beta production via upregulation of NF-kappaB and IRF3 signaling 27605671_Can activate the RLR/MAVS pathway. 27629939_MCCC1 plays an essential role in virus-triggered, MAVS-mediated activation of NF-kappaB signaling. 27652379_this analysis did not indicate the association of the MAVS locus with susceptibility to Addison's disease and type 1 diabetes 27705941_our results demonstrate that miR-22 negatively regulates poly(I:C)-induced production of type I interferon and inflammatory cytokines via targeting MAVS. 27736772_Results indicate that TAX1BP1 functions as an adaptor molecule for Itch to target MAVS during RNA virus infection and thus restrict virus-induced apoptosis. 27899525_findings suggest that oxidative stress-induced MAVS oligomerization in SLE patients may contribute to the type I IFN signature that is characteristic of this syndrome. 27980081_DDX3 directly regulates TRAF3 ubiquitination and acts as a scaffold to co-ordinate assembly of signaling complexes downstream from MAVS. 28011935_TTLL12 as a negative regulator of RNA-virus-induced type I IFN expression by inhibiting the interaction of VISA with other proteins. 28024153_this study identified three single nucleotide polymorphisms within MAVS that showed significant differences in plasma HIV-1 viral load 28222744_Herpes simplex virus 1 blocks MAVS-Pex mediated early interferon-stimulated gene activation through VP16 to dampen the immediate early antiviral innate immunity signaling from peroxisomes. 28414768_GPATCH3 interacts with VISA and disrupts the assembly of virus-induced VISA signalosome therefore acts as a negative regulator of RLR-mediated innate antiviral immune responses. 28471483_This study demonstrates a novel pathway for elevated IFNbeta signaling in SLE that is not dependent on stimulation by immune complexes but rather is cell intrinsic and critically mediated by IFNbeta and MAVS. 28480979_The down regulation of TRIF, TLR3, and mitochondrial antiviral signaling protein (MAVS) expressions in chronic hepatitis C correlates with the disease severity and the outcome of hepatitis C virus infection 28566380_Therefore, Seneca Valley virus suppressed antiviral interferon production to escape host antiviral innate immune responses by cleaving host adaptor molecules MAVS, TRIF, and TANK by its 3C protease. 28594325_Taken together, these findings reveal an essential role of CypA in boosting RIG-I-mediated antiviral immune responses by controlling the ubiquitination of RIG-I and MAVS. 28607490_MAVS isoforms are truncated, which prevents its spontaneous aggregation in antiviral innate immune signalling 28928438_Zyxin promotes MAVS-mediated IFNB1 promoter activation. 28956771_Results show that during HCV infection, NLRX1 targets MAVS and induces its degradation. 28965816_findings reveal a negative feedback loop of RLR signaling generated by Tetherin-MARCH8-MAVS-NDP52 axis and provide insights into a better understanding of the crosstalk between selective autophagy and optimal deactivation of type I IFN signaling. 29097393_Using MAVS as a platform, NLRP11 degrades TRAF6 to attenuate the production of type I IFNs as well as virus-induced apoptosis. Our findings reveal the regulatory role of NLRP11 in antiviral immunity by disrupting MAVS signalosome. 29155878_In the late phase of RNA viral infection, iRhom2 mediates proteasome-dependent degradation of the E3 ubiquitin ligase MARCH5 and impairs mitochondria-associated degradation (MAD) of VISA. 29165031_BST2 recruits the E3 ubiquitin ligase MARCH8 to catalyze the K27-linked ubiquitination of MAVS for CALCOCO2-directed autophagic degradation. 29280086_results suggest that ASC, as a negative regulator of the MAVS-mediated innate immunity, may play an important role in host protection upon virus infection 29317664_Data show that human cytomegalovirus (HCMV; human betaherpesvirus 5) glycoprotein US9 inhibits the IFN-beta response by targeting the mitochondrial antiviral-signaling protein (MAVS) and stimulator of interferon genes (STING)-mediated signaling pathways. 29385716_The mitochondrial antiviral signaling adaptor protein (MAVS) oligomers and high MW aggregates coexist upon constitutively active retinoic acid-inducible gene I (RIG-I) ectopic expression and virus infection. Anchoring of MAVS to intracellular membranes is essential for an appropriate polymerization process allowing functional high MW aggregates to occur. 29743353_Low MAVS expression is associated with RNA virus infections. 29916539_these results demonstrated that HAUS8 may function as a positive regulator of RLRVISA dependent antiviral signaling by targeting the VISA complex, providing a novel regulatory mechanism of antiviral responses 30258449_Beclin-1 underwent K63-polyubiquitination upon RIG-I activation, and the ubiquitination decreased in TRAF6-deficient cells. This suggests that the RIG-I-MAVS-TRAF6 axis induced K63-linked polyubiquitination of Beclin-1, which has been implicated in triggering autophagy. 30390472_studies demonstrated that TARBP2 interacted with MAVS and targeted MAVS to abrogate MAVS-RIG-I and MAVS-TRAF3 association. 30391331_The identification of multiple regulatory elements in the MAVS 3'UTR offers new insights into the precise control of MAVS expression in innate immunity. 30460894_Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. 30463990_MAVS rs6052130 confer genetic susceptibility to cervical precancerous lesions. 30472208_These findings establish FAF1 as a modulator of MAVS and uncover mechanisms that regulate FAF1 to insure timely activation of antiviral defense. 30527812_RACK1 interacts with VISA to repress downstream signaling and downregulates virus-induced IFN-beta production in the RIG-I/VISA signaling pathway. 30878284_Apoptotic caspases control innate immunity and maintain immune homeostasis against viral infection, suppressing type I interferon production via the cleavage of cGAS, MAVS, and IRF3 30930359_Relatively weak inducible IFN-beta production in hepatitis b virus infected patients with IPS-1 rs7269320 SNP or wild-type may contribute to chronic virus infection. 30952814_findings thus provided novel insights into the regulatory cascade of the cellular antiviral response through YOD1-mediated K63-linked deubiquitination and aggregation of MAVS. 31235549_We identified human nucleus accumbens-associated 1 (NAC1), a member of the BTB/POZ family, as a bridge for MAVS and TBK1 that positively regulates the RIG-I-like receptors-mediated induction of type I IFN 31270229_MAVS degradation leads to the inhibition of the downstream IFN-beta pathway and therefore benefits virus proliferation. 31304625_Data propose a critical role for autophagy in mediating the degradation of MAVS aggregates, and identified RNF34 as a key regulator of mitochondrial turnover and the antiviral response by promoting the K27-/K29-linked ubiquitination of MAVS. 31324787_Nemo like kinase (NLK) interacts with and phosphorylates mitochondrial antiviral signaling protein (MAVS), and a peptide derived from MAVS promotes viral-induced interferon beta (IFN-beta) production and antagonizes viral replication. 31390091_Results found that the activity of MAVS-Region III depends on its multimerization state. TRAF3IP3 binds to multimerized and active MAVS-Region III. 31767975_MicroRNA-33/33* inhibit the activation of MAVS through AMPK in antiviral innate immunity. 31806367_Formation of SUMO3-conjugated chains of MAVS induced by poly(dA:dT), a ligand of RIG-I, enhances the aggregation of MAVS that drives the secretion of interferon-beta in human keratinocytes. 31806368_SNX5 inhibits RLR-mediated antiviral signaling by targeting RIG-I-VISA signalosome. 31829086_The RNA binding protein Quaking represses host interferon response by downregulating MAVS. 31881323_Dual targeting of RIG-I and MAVS by MARCH5 mitochondria ubiquitin ligase in innate immunity. 31915282_Thioredoxin 2 Negatively Regulates Innate Immunity to RNA Viruses by Disrupting the Assembly of the Virus-Induced Signaling Adaptor Complex. 31941397_Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. 31968249_The Mitochondrial Protein MAVS Stabilizes p53 to Suppress Tumorigenesis. 31996459_The Japanese Encephalitis Virus NS1' Protein Inhibits Type I IFN Production by Targeting MAVS. 32190169_The Reduced Oligomerization of MAVS Mediated by ROS Enhances the Cellular Radioresistance. 32301534_SIRT5 impairs aggregation and activation of the signaling adaptor MAVS through catalyzing lysine desuccinylation. 32386456_PB1-F2 protein of highly pathogenic influenza A (H7N9) virus selectively suppresses RNA-induced NLRP3 inflammasome activation through inhibition of MAVS-NLRP3 interaction. 32511263_Virus subtype-specific suppression of MAVS aggregation and activation by PB1-F2 protein of influenza A (H7N9) virus. 32708557_MAVS Genetic Variation Is Associated with Decreased HIV-1 Replication In Vitro and Reduced CD4(+) T Cell Infection in HIV-1-Infected Individuals. 32776819_Suppression of innate immune signaling molecule, MAVS, reduces radiation-induced bystander effect. 33110251_SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. 33139700_RNF115 plays dual roles in innate antiviral responses by catalyzing distinct ubiquitination of MAVS and MITA. 33177519_MAVS is energized by Mff which senses mitochondrial metabolism via AMPK for acute antiviral immunity. 33412226_Mitochondrial reactive zones in antiviral innate immunity. 33540654_The VP3 Protein of Bluetongue Virus Associates with the MAVS Complex and Interferes with the RIG-I-Signaling Pathway. 33582548_Mitochondrial DUT-M potentiates RLR-mediated antiviral signaling by enhancing VISA and TRAF2 association. 33593967_Human Herpesvirus 6B U26 Inhibits the Activation of the RLR/MAVS Signaling Pathway. 33603735_O-Linked N-Acetylglucosamine Modification of Mitochondrial Antiviral Signaling Protein Regulates Antiviral Signaling by Modulating Its Activity. 33628342_Next Generation Exome Sequencing of Pediatric Asthma Identifies Rare and Novel Variants in Candidate Genes. 34016972_USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. 34171297_Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. 34435375_A conventional immune regulator mitochondrial antiviral signaling protein blocks hepatic steatosis by maintaining mitochondrial homeostasis. 34452514_The Ebola Virus Interferon Antagonist VP24 Undergoes Active Nucleocytoplasmic Trafficking. | ENSMUSG00000037523 | Mavs | 2055.054552 | 0.8323649 | -0.264712031 | 0.09085522 | 8.526118e+00 | 3.500855e-03 | 6.973325e-02 | No | Yes | 1983.196147 | 202.935341 | 2413.262432 | 246.891788 | |
| ENSG00000089053 | 51433 | ANAPC5 | protein_coding | Q9UJX4 | FUNCTION: Component of the anaphase promoting complex/cyclosome (APC/C), a cell cycle-regulated E3 ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle. The APC/C complex acts by mediating ubiquitination and subsequent degradation of target proteins: it mainly mediates the formation of 'Lys-11'-linked polyubiquitin chains and, to a lower extent, the formation of 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains. {ECO:0000269|PubMed:18485873}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a tetratricopeptide repeat-containing component of the anaphase promoting complex/cyclosome (APC/C), a large E3 ubiquitin ligase that controls cell cycle progression by targeting a number of cell cycle regulators such as B-type cyclins for 26S proteasome-mediated degradation through ubiquitination. The encoded protein is required for the proper ubiquitination function of APC/C and for the interaction of APC/C with transcription coactivators. It also interacts with polyA binding protein and represses internal ribosome entry site-mediated translation. Multiple transcript variants encoding different isoforms have been found for this gene. These differences cause translation initiation at a downstream AUG and result in a shorter protein (isoform b), compared to isoform a. [provided by RefSeq, Nov 2008]. | hsa:51433; | anaphase-promoting complex [GO:0005680]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle [GO:0005819]; protein phosphatase binding [GO:0019903]; anaphase-promoting complex-dependent catabolic process [GO:0031145]; cell cycle [GO:0007049]; cell division [GO:0051301]; positive regulation of mitotic metaphase/anaphase transition [GO:0045842]; protein K11-linked ubiquitination [GO:0070979] | 15082755_Results show that Apc5 binds the poly(A) binding protein (PABP), which directly binds the internal ribosome entry site (IRES) element of platelet-derived growth factor 2 mRNA. 16319895_APC5 and APC7 suppress E1A-mediated transformation in a CBP/p300-dependent manner, indicating that these components of the APC/C may be targeted during cellular transformation 19826416_Studies indicate that APC/C(Cdh1) is required to maintain genomic stability. 20686030_Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. 28731177_coexpression of APC5 and Axin genes significantly downregulated Wnt signaling in human SW480 CRC cells and inhibited cell growth. | ENSMUSG00000029472 | Anapc5 | 7328.025179 | 1.0473304 | 0.066716635 | 0.05653639 | 1.379137e+00 | 2.402482e-01 | 6.122694e-01 | No | Yes | 7139.767472 | 1009.387506 | 6799.349452 | 961.160831 |
| ENSG00000089123 | 55617 | TASP1 | protein_coding | Q9H6P5 | FUNCTION: Protease responsible for KMT2A/MLL1 processing and activation (PubMed:14636557). It also activates KMT2D/MLL2 (By similarity). Through substrate activation, it controls the expression of HOXA genes, and the expression of key cell cycle regulators including CCNA1, CCNB1, CCNE1 and CDKN2A (By similarity) (PubMed:14636557). {ECO:0000250|UniProtKB:Q8R1G1, ECO:0000269|PubMed:14636557}. | 3D-structure;Alternative splicing;Autocatalytic cleavage;Direct protein sequencing;Disease variant;Hydrolase;Protease;Reference proteome;Threonine protease;Zymogen | This gene encodes an endopeptidase that cleaves specific substrates following aspartate residues. The encoded protein undergoes posttranslational autoproteolytic processing to generate alpha and beta subunits, which reassemble into the active alpha2-beta2 heterotetramer. It is required to cleave MLL, a protein required for the maintenance of HOX gene expression, and TFIIA, a basal transcription factor. Alternatively spliced transcript variants have been described, but their biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:55617; | cytoplasm [GO:0005737]; identical protein binding [GO:0042802]; threonine-type endopeptidase activity [GO:0004298]; positive regulation of transcription, DNA-templated [GO:0045893]; protein maturation [GO:0051604]; proteolysis [GO:0006508] | 14636557_purification and cloning of threonine aspartase 1 responsible for cleaving MLL; RNAi-mediated knockdown of Taspase1 results in the appearance of unprocessed MLL and the loss of proper HOX gene expression 16537915_Transfected taspase 1 enhances cleavage of TFIIA, and RNA interference knockdown of endogenous taspase 1 diminishes cleavage of TFIIA in vivo. 18395097_TASP1, EPS15R, and PRPF3 expression were significantly induced in HCCs of transgenic EGF2B mice as was P2 promoter-driven HNF4alpha 20516119_Taspase1 is overexpressed in primary human cancers, and deficiency of Taspase1 in cancer cells not only disrupts proliferation but also enhances apoptosis. 21084304_Cell-based analysis of structure-function activity of threonine aspartase 1. 21418451_Taspase1 appears to exploit the nuclear export activity of importin-alpha/nucleophosmin to gain transient access to the cytoplasm required to also cleave its cytoplasmic substrates. 22570686_inefficient heterodimerization appears to be the mechanism by which inactive Taspase1 variants fail to inhibit wild type Taspase1's activity in trans. 25996597_Our results provide first evidence that Taspase1 processing affects TFIIA regulation of TFIID and suggest that Taspase1 processing of TFIIA is required to establish INR-selective core promoter activity in the presence of NC2. 26137584_simultaneous expression of the leukemogenic AF4-MLL and dnTASP1 causes the disappearance of the leukemogenic oncoprotein, because the uncleaved AF4-MLL protein (328 kDa) is subject to proteasomal degradation. 26657154_Studies indicate that threonine Aspartase1 (Taspase1) is overexpressed in numerous liquid and solid malignancies and was characterized as a 'non-oncogene addiction' protease. 28992066_Taspase 1 processing promotes TFIIA's nuclear accumulation by evolutionary conserved nuclear export signal (NES, amino acids 21VINDVRDIFL30) masking, and modulates its transcriptional activity. 29097782_Proteolytic processing of TFIIA by Taspase1 was found to mask evolutionary conserved nuclear export signal, thereby promoting nuclear localization and transcriptional activation of TFIIA target genes, such as CDKN2A. 29633245_TASP1 is a novel disease-related gene that is associated with a disease phenotype overlapping with Wiedemann-Steiner syndrome as both are caused by defects in the same pathway. 32071545_TASP1 Promotes Gallbladder Cancer Cell Proliferation and Metastasis by Up-regulating FAM49B via PI3K/AKT Pathway. 33784495_Structural insights into the function of the catalytically active human Taspase1. 34012990_TASP1 Promotes Proliferation and Migration in Gastric Cancer via EMT and AKT/P-AKT Pathway. 34197029_Silencing of circTASP1 inhibits proliferation and induces apoptosis of acute myeloid leukaemia cells through modulating miR-515-5p/HMGA2 axis. | ENSMUSG00000039033 | Tasp1 | 290.419207 | 1.0881870 | 0.121926466 | 0.15757745 | 6.012421e-01 | 4.381045e-01 | No | Yes | 325.943493 | 43.321199 | 297.565861 | 39.617000 | ||
| ENSG00000089639 | 51291 | GMIP | protein_coding | Q9P107 | FUNCTION: Stimulates, in vitro and in vivo, the GTPase activity of RhoA. {ECO:0000269|PubMed:12093360}. | 3D-structure;Alternative splicing;Coiled coil;GTPase activation;Metal-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | This gene encodes a member of the ARHGAP family of Rho/Rac/Cdc42-like GTPase activating proteins. The encoded protein interacts with the Ras-related protein Gem through its N-terminal domain. Separately, it interacts with RhoA through a RhoGAP domain, and stimulates RhoA-dependent GTPase activity. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]. | hsa:51291; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; activation of GTPase activity [GO:0090630]; intracellular signal transduction [GO:0035556]; negative regulation of GTPase activity [GO:0034260]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12093360_signalling pathways controlled by two proteins of the Ras superfamily, RhoA and Gem, are linked via the action of the RhoGAP protein Gmip 16086184_Observational study of gene-disease association. (HuGE Navigator) 16086184_Variations in the GMIP gene can confer susceptibility to major depressive disorder. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20351714_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22438581_during exocytosis, actin depolymerization commences near the secretory organelle, not the plasma membrane, and secretory granules use a JFC1- and GMIP-dependent molecular mechanism to traverse cortical actin. 25173885_These findings demonstrate a new role of Gem/Gmip/RhoA signaling in cortical actin regulation during early mitotic stages | ENSMUSG00000036246 | Gmip | 129.456098 | 0.7327397 | -0.448627397 | 0.23312498 | 3.670480e+00 | 5.538424e-02 | No | Yes | 110.673173 | 17.156523 | 139.777750 | 21.501024 | ||
| ENSG00000089820 | 393 | ARHGAP4 | protein_coding | P98171 | FUNCTION: Inhibitory effect on stress fiber organization. May down-regulate Rho-like GTPase in hematopoietic cells. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;GTPase activation;Phosphoprotein;Reference proteome;SH3 domain | This gene encodes a member of the rhoGAP family of proteins which play a role in the regulation of small GTP-binding proteins belonging to the RAS superfamily. The protein encoded by the orthologous gene in rat is localized to the Golgi complex and can redistribute to microtubules. The rat protein stimulates the activity of some Rho GTPases in vitro. Genomic deletions of this gene and a neighboring gene have been found in patients with nephrogenic diabetes insipidus. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2009]. | hsa:393; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; growth cone [GO:0030426]; microtubule [GO:0005874]; GTPase activator activity [GO:0005096]; identical protein binding [GO:0042802]; cytoskeleton organization [GO:0007010]; negative regulation of axon extension [GO:0030517]; negative regulation of cell migration [GO:0030336]; negative regulation of fibroblast migration [GO:0010764]; regulation of small GTPase mediated signal transduction [GO:0051056]; Rho protein signal transduction [GO:0007266] | 11754100_A novel type of contiguous gene deletion of ARHGAP4 has been identified in unrelated Japanese kindreds with nephrogenic diabetes insipidus. 12736724_FNBP2, ARHGAP13, ARHGAP14 and ARHGAP4 constitute the FNBP2 family characterized by FCH, RhoGAP and SH3 domains. 18489790_ARHGAP4 may play some role in lymphocyte differentiation but partial loss of ARHGAP4 does not result in clinical immunodeficiency 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22009749_PRL-1 binding to p115 RhoGAP provides a coordinated mechanism underlying ERK1/2 and RhoA activation 22965914_X-linked nephrogenic diabetes insipidus (NDI) and intellectual disability in two dizygotic twin brothers was caused by a novel contiguous deletion of 17,905 bp of the entire AVPR2 gene and intron 7 of the ARHGAP4 gene. 24043878_ARHGAP4 rs2269368 was associated with risk of schizophrenia in a Han Chinese population. 26707211_The relation between ARHGAP4 mutation and Mental retardation(MR) clinical characteristic is needed to be illuminated with participation of more MR patients 30958531_ARHGAP4 regulates the cell migration and invasion of pancreatic cancer by the HDAC2/beta-catenin signaling pathway. 32378260_Comprehensive analysis on the whole Rho-GAP family reveals that ARHGAP4 suppresses EMT in epithelial cells under negative regulation by Septin9. 34524873_ARHGAP4-SEPT2-SEPT9 complex enables both up- and down-modulation of integrin-mediated focal adhesions, cell migration, and invasion. | ENSMUSG00000031389 | Arhgap4 | 96.296521 | 1.0704062 | 0.098158316 | 0.30281245 | 1.086893e-01 | 7.416413e-01 | No | Yes | 99.364001 | 26.322559 | 95.779893 | 25.329600 | ||
| ENSG00000090238 | 83719 | YPEL3 | protein_coding | P61236 | FUNCTION: Involved in proliferation and apoptosis in myeloid precursor cells. {ECO:0000250}. | Alternative splicing;Metal-binding;Nucleus;Reference proteome;Ubl conjugation;Zinc | hsa:83719; | nucleolus [GO:0005730]; metal ion binding [GO:0046872]; positive regulation of cellular senescence [GO:2000774] | 20388804_Findings point to YPEL3 being a novel tumor suppressor, which upon induction triggers a permanent growth arrest in human tumor and normal cells. 20508983_Observational study of gene-disease association. (HuGE Navigator) 21196260_In this review we show that transcriptionally active forms of p73 and p63, family members of p53, can transactivate the human YPEL3 gene[review] 21267786_YPEL3 is downregulated in colon tumors. 21671470_YPEL3 expression, which is induced by the removal of estrogen or treatment with tamoxifen triggers cellular senescence in MCF-7 cells while loss of YPEL3 increases the growth rate of MCF-7 cells. 27400785_Data show that YPEL3 is downregulated in nasopharyngeal carcinoma (NPC) cell lines and tissue samples and it suppresses NPC EMT and metastasis by suppressing the Wnt/beta-catenin signaling pathway. 29988027_Expression of the YPEL3 gene was upregulated in human colonic adenocarcinoma tissue. 32544203_yippee like 3 (ypel3) is a novel gene required for myelinating and perineurial glia development. 34675221_Steroid sulfatase deficiency causes cellular senescence and abnormal differentiation by inducing Yippee-like 3 expression in human keratinocytes. | ENSMUSG00000042675 | Ypel3 | 158.965717 | 0.9898911 | -0.014658343 | 0.22362797 | 4.312940e-03 | 9.476382e-01 | No | Yes | 158.321110 | 23.259748 | 162.978798 | 23.818004 | |||
| ENSG00000090621 | 8761 | PABPC4 | protein_coding | Q13310 | FUNCTION: Binds the poly(A) tail of mRNA. May be involved in cytoplasmic regulatory processes of mRNA metabolism. Can probably bind to cytoplasmic RNA sequences other than poly(A) in vivo (By similarity). {ECO:0000250}. | Alternative splicing;Cytoplasm;Direct protein sequencing;Isopeptide bond;Methylation;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation | Poly(A)-binding proteins (PABPs) bind to the poly(A) tail present at the 3-prime ends of most eukaryotic mRNAs. PABPC4 or IPABP (inducible PABP) was isolated as an activation-induced T-cell mRNA encoding a protein. Activation of T cells increased PABPC4 mRNA levels in T cells approximately 5-fold. PABPC4 contains 4 RNA-binding domains and proline-rich C terminus. PABPC4 is localized primarily to the cytoplasm. It is suggested that PABPC4 might be necessary for regulation of stability of labile mRNA species in activated T cells. PABPC4 was also identified as an antigen, APP1 (activated-platelet protein-1), expressed on thrombin-activated rabbit platelets. PABPC4 may also be involved in the regulation of protein translation in platelets and megakaryocytes or may participate in the binding or stabilization of polyadenylates in platelet dense granules. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. This protein has also been found to interact with coronavirus nucleocapsid proteins and is thought to inhibit coronavirus replication. [provided by RefSeq, Nov 2021]. | hsa:8761; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA 3'-UTR binding [GO:0003730]; poly(A) binding [GO:0008143]; poly(U) RNA binding [GO:0008266]; RNA binding [GO:0003723]; blood coagulation [GO:0007596]; myeloid cell development [GO:0061515]; regulation of mRNA stability [GO:0043488]; RNA catabolic process [GO:0006401]; RNA processing [GO:0006396]; translation [GO:0006412] | 21646427_RNA binding masks nuclear import signals within the cytoplasmic poly(A) binding protein RNA recognition motifs, thereby ensuring ef fi cient cytoplasmic retention of this protein in normal cells 21940797_Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export 22884093_PABPC4 is highly expressed in human colorectal cancer. 23300856_although function of PABPN1 may be compensated by nuclear translocation of PABP4 and perhaps by increase the cytoplasmic abundance of PABP5, these were not sufficient to prevent apoptosis of cells. PABPN1 may have an anti apoptotic role in mammalian cells 26005159_The PABPC4 rs4660293 SNP is associated with serum lipid levels. 33259899_A novel long non-coding RNA RP11-286H15.1 represses hepatocellular carcinoma progression by promoting ubiquitination of PABPC4. 33730015_Gene variants of coagulation related proteins that interact with SARS-CoV-2. 34612687_PABPC4 Broadly Inhibits Coronavirus Replication by Degrading Nucleocapsid Protein through Selective Autophagy. | ENSMUSG00000011257 | Pabpc4 | 8399.987021 | 1.0403235 | 0.057032259 | 0.04897925 | 1.362617e+00 | 2.430845e-01 | 6.155566e-01 | No | Yes | 8355.219545 | 645.372360 | 7980.995736 | 616.407164 | |
| ENSG00000091039 | 114882 | OSBPL8 | protein_coding | Q9BZF1 | FUNCTION: Lipid transporter involved in lipid countertransport between the endoplasmic reticulum and the plasma membrane: specifically exchanges phosphatidylserine with phosphatidylinositol 4-phosphate (PI4P), delivering phosphatidylserine to the plasma membrane in exchange for PI4P, which is degraded by the SAC1/SACM1L phosphatase in the endoplasmic reticulum. Binds phosphatidylserine and PI4P in a mutually exclusive manner (PubMed:26206935). Binds oxysterol, 25-hydroxycholesterol and cholesterol (PubMed:17428193, PubMed:17991739, PubMed:21698267). {ECO:0000269|PubMed:17428193, ECO:0000269|PubMed:17991739, ECO:0000269|PubMed:21698267, ECO:0000269|PubMed:26206935}. | 3D-structure;Acetylation;Alternative splicing;Endoplasmic reticulum;Lipid transport;Lipid-binding;Membrane;Nucleus;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of a family of proteins containing an N-terminal pleckstrin homology domain and a highly conserved C-terminal oxysterol-binding protein-like sterol-binding domain. It binds mutliple lipid-containing molecules, including phosphatidylserine, phosphatidylinositol 4-phosphate (PI4P) and oxysterol, and promotes their exchange between the endoplasmic reticulum and the plasma membrane. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:114882; | cortical endoplasmic reticulum [GO:0032541]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nuclear membrane [GO:0031965]; cholesterol binding [GO:0015485]; phosphatidylinositol-4-phosphate binding [GO:0070273]; phosphatidylserine binding [GO:0001786]; phosphatidylserine transfer activity [GO:0140343]; phospholipid transporter activity [GO:0005548]; sterol binding [GO:0032934]; sterol transporter activity [GO:0015248]; activation of protein kinase B activity [GO:0032148]; fat cell differentiation [GO:0045444]; negative regulation of cell migration [GO:0030336]; negative regulation of sequestering of triglyceride [GO:0010891]; phosphatidylserine acyl-chain remodeling [GO:0036150]; phospholipid transport [GO:0015914]; positive regulation of glucose import [GO:0046326]; positive regulation of insulin receptor signaling pathway [GO:0046628]; positive regulation of protein kinase B signaling [GO:0051897]; protein localization to nuclear pore [GO:0090204] | 17672918_occurrence of an unusual TG 3' splice site in intron 2 17991739_Data identify ORP8 as a negative regulator of ABCA1 expression and macrophage cholesterol efflux. ORP8 may, thus, modulate the development of atherosclerosis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21698267_results reveal that ORP8 has the capacity to modulate lipid homeostasis and SREBP activity, probably through an indirect mechanism, and provide clues of an entirely new mode of ORP action 23028956_ORP8 overexpression resulted in reduced expression of the aP2 mRNA, while down-regulation of adiponectin and aP2 was observed in ORP11 silenced cells. ORP8 overexpression or silencing of ORP11 markedly decreased cellular triglyceride storage. 24333576_Data indicate that miR-143 impairs insulin action via downregulation of oxysterol-binding protein-related protein 8 (ORP8). 25596532_role of ORP8 in Fas translocation to the plasma membrane and its down-regulation by miR-143 offer a putative mechanistic explanation for HCC resistance to apoptosis 26206935_ORP5 and ORP8 could mediate PI4P/phosphatidylserine (PS) countertransport between the endoplasmic reticulum (ER) and the plasma membrane (PM), thus delivering PI4P to the ER-localized PI4P phosphatase Sac1 for degradation and PS from the ER to the PM. 27113756_mammalian ORP5 and ORP8 proteins localize to ER-mitochondrial MCS, in addition to ER-PM contact sites. 27530118_the present study suggests that ORP8 may mediate the cytotoxicity of 25-hydroxycholesterol. 27983927_we found that overexpression of ORP8 significantly inhibits GC cell proliferation and implanted tumor growth in vivo. Induction of ER stress, inhibition of Wnt signaling, and apoptotic cell death are involved in ORP8-induced inhibition of GC cell proliferation. 28970484_ORP5/8 are endoplasmic reticulum (ER) membrane proteins implicated in lipid trafficking that localize to ER-plasma membrane (PM) contacts and maintain membrane homeostasis. Here the authors show that PtdIns(4,5)P 2 plays a critical role in the targeting and function of ORP5/8 at the PM. 29409900_Examination of a series of deletion constructs demonstrated that both the N-terminal polybasic region and the PH domain are required for proper targeting of the short splice variant ORP8S to the PM-ER contact site in Chinese hamster ovary cells. 29472386_ORP5/8 recruitment to the plasma membrane occurs through interactions with the N-terminal Pleckstrin homology domains and adjacent basic residues of ORP5/8 with both phosphatidylinositol 4-phosphate and Phosphatidylinositol 4,5-bisphosphate. 29748134_ORP5/8 regulate Ca(2+) signaling at specific membrane contact sites foci. 32323800_ORP8 induces apoptosis by releasing cytochrome c from mitochondria in nonsmall cell lung cancer. | ENSMUSG00000020189 | Osbpl8 | 2816.595274 | 1.0332735 | 0.047222150 | 0.06490117 | 5.307338e-01 | 4.662985e-01 | 7.856179e-01 | No | Yes | 3392.443190 | 622.903219 | 3268.955300 | 600.239226 | |
| ENSG00000091073 | 113878 | DTX2 | protein_coding | Q86UW9 | FUNCTION: Regulator of Notch signaling, a signaling pathway involved in cell-cell communications that regulates a broad spectrum of cell-fate determinations. Probably acts both as a positive and negative regulator of Notch, depending on the developmental and cell context. Mediates the antineural activity of Notch, possibly by inhibiting the transcriptional activation mediated by MATCH1. Functions as a ubiquitin ligase protein in vitro, suggesting that it may regulate the Notch pathway via some ubiquitin ligase activity. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Metal-binding;Methylation;Notch signaling pathway;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | DTX2 functions as an E3 ubiquitin ligase (Takeyama et al., 2003 [PubMed 12670957]).[supplied by OMIM, Nov 2009]. | hsa:113878; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; Notch signaling pathway [GO:0007219]; protein ubiquitination [GO:0016567] | 12670957_It is reported that BBAP and the human family of DTX proteins (DTX1, DTX2, and DTX3) function as E3 ligases based on their capacity for self-ubiquitination. 17286044_DTX2 is a gene encoding a WWE-RING-finger protein and involved in regulating heart development and heart functions. | ENSMUSG00000004947 | Dtx2 | 398.580254 | 0.8599030 | -0.217754250 | 0.13814284 | 2.475205e+00 | 1.156543e-01 | No | Yes | 351.862256 | 46.596653 | 406.740885 | 53.626138 | |
| ENSG00000092098 | 55072 | RNF31 | protein_coding | Q96EP0 | FUNCTION: E3 ubiquitin-protein ligase component of the LUBAC complex which conjugates linear ('Met-1'-linked) polyubiquitin chains to substrates and plays a key role in NF-kappa-B activation and regulation of inflammation (PubMed:17006537, PubMed:19136968, PubMed:20005846, PubMed:21455173, PubMed:21455180, PubMed:21455181, PubMed:22863777, PubMed:28481331, PubMed:28189684). LUBAC conjugates linear polyubiquitin to IKBKG and RIPK1 and is involved in activation of the canonical NF-kappa-B and the JNK signaling pathways (PubMed:17006537, PubMed:19136968, PubMed:20005846, PubMed:21455173, PubMed:21455180, PubMed:21455181, PubMed:22863777, PubMed:28189684). Linear ubiquitination mediated by the LUBAC complex interferes with TNF-induced cell death and thereby prevents inflammation (PubMed:21455173, PubMed:28189684). LUBAC is recruited to the TNF-R1 signaling complex (TNF-RSC) following polyubiquitination of TNF-RSC components by BIRC2 and/or BIRC3 and to conjugate linear polyubiquitin to IKBKG and possibly other components contributing to the stability of the complex (PubMed:20005846, PubMed:27458237). The LUBAC complex is also involved in innate immunity by conjugating linear polyubiquitin chains at the surface of bacteria invading the cytosol to form the ubiquitin coat surrounding bacteria (PubMed:28481331, PubMed:34012115). LUBAC is not able to initiate formation of the bacterial ubiquitin coat, and can only promote formation of linear polyubiquitins on pre-existing ubiquitin (PubMed:28481331). Recruited to the surface of bacteria by RNF213, which initiates the bacterial ubiquitin coat (PubMed:34012115). The bacterial ubiquitin coat acts as an 'eat-me' signal for xenophagy and promotes NF-kappa-B activation (PubMed:28481331, PubMed:34012115). Together with OTULIN, the LUBAC complex regulates the canonical Wnt signaling during angiogenesis (PubMed:23708998). RNF31 is required for linear ubiquitination of BCL10, thereby promoting TCR-induced NF-kappa-B activation (PubMed:27777308). Binds polyubiquitin of different linkage types (PubMed:23708998). {ECO:0000269|PubMed:17006537, ECO:0000269|PubMed:19136968, ECO:0000269|PubMed:20005846, ECO:0000269|PubMed:21455173, ECO:0000269|PubMed:21455180, ECO:0000269|PubMed:21455181, ECO:0000269|PubMed:22863777, ECO:0000269|PubMed:23708998, ECO:0000269|PubMed:27458237, ECO:0000269|PubMed:27777308, ECO:0000269|PubMed:28189684, ECO:0000269|PubMed:28481331, ECO:0000269|PubMed:34012115}. | 3D-structure;Alternative splicing;Cytoplasm;Isopeptide bond;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:17006537, ECO:0000269|PubMed:19136968, ECO:0000269|PubMed:20005846, ECO:0000269|PubMed:28481331}. | The protein encoded by this gene contains a RING finger, a motif present in a variety of functionally distinct proteins and known to be involved in protein-DNA and protein-protein interactions. The encoded protein is the E3 ubiquitin-protein ligase component of the linear ubiquitin chain assembly complex. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2015]. | hsa:55072; | CD40 receptor complex [GO:0035631]; cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; LUBAC complex [GO:0071797]; identical protein binding [GO:0042802]; K48-linked polyubiquitin modification-dependent protein binding [GO:0036435]; K63-linked polyubiquitin modification-dependent protein binding [GO:0070530]; linear polyubiquitin binding [GO:1990450]; metal ion binding [GO:0046872]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; CD40 signaling pathway [GO:0023035]; defense response to bacterium [GO:0042742]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of protein targeting to mitochondrion [GO:1903955]; positive regulation of xenophagy [GO:1904417]; protein linear polyubiquitination [GO:0097039]; protein polyubiquitination [GO:0000209]; T cell receptor signaling pathway [GO:0050852] | 15093743_ZIBRA was identified and its genomic organization, regulation, and expression studied in breast cancer cells. 19237537_RNF31 functions to stabilize DAX-1; RNF31 appears to be required for DAX-1 to repress transcription 20588308_Observational study of gene-disease association. (HuGE Navigator) 22427816_analysis of differentially expressed genes indicates systemic role for SF-1 in adrenal cortex affecting differentiation, proliferation and steroidogenesis and establishes RNF31 as an important regulator of adrenal steroidogenesis 22430200_The study highlights the versatility and specificity of protein-protein interactions involving Ub-like domain (UBL) of HOIL-1L/the Ub-associated domain (UBA) of HOIP and their cognate proteins. 22863777_The data showed how HOIP combines a general RING-IBR-RING ubiquitin ligase mechanism with unique, 'Linear ubiquitin chain Determining Domain'-dependent specificity for producing linear ubiquitin chains. 23986494_The HOIP component of the linear ubiquitin assembly complex catalyzes the formation of M1-polyubiquitin chains in response to interleukin-1. 24030825_target specificity toward NEMO is determined by multiple LUBAC components, whereas linear ubiquitin chain elongation is realized by a specific interplay between HOIP and ubiquitin. 24141947_crystal structure of the catalytic core of HOIP in its apo form and in complex with ubiquitin 24441041_RNF31, via stabilizing ESR1 levels, controls the transcription of estrogen-dependent genes linked to breast cancer cell proliferation. 24491438_Two rare germline polymorphisms affecting the LUBAC subunit RNF31 were identified and enriched among patients with activated B cell-like subtype of diffuse large B-cell lymphoma. 24726323_Phosphorylation of OTULIN prevents HOIP binding, whereas unphosphorylated OTULIN is part of the endogenous LUBAC complex. 24726327_HOIP binding to OTULIN is required for the recruitment of OTULIN to the TNF receptor complex. 26008899_human HOIP is essential for the assembly and function of LUBAC, which includes HOIL-1, and for various processes governing inflammation and immunity in both hematopoietic and nonhematopoietic cells 26038114_we identified BCL10 as a bona fide target of BCR-induced linear ubiquitylation and demonstrated an important role of the linear ubiquitin ligase HOIP in BCR-induced phosphorylation 26148235_RNF31 decreased p53 stability. RNF31 depletion caused cell cycle arrest and cisplatin-induced apoptosis in a p53-dependent manner. RNF31 associated with the p53/MDM2 complex, facilitating p53 polyubiquitination and degradation by stabilizing MDM2. 26231797_These findings collectively indicated an oncogene role of RNF31 in PCa progression which can be regulated by miR-503, suggesting that RNF31 could serve as a potential prognostic biomarker and therapeutic target for PCa. 26578682_This study describes a novel posttranslational regulation of the HOIP-containing linear-ubiquitin-chain assembly complex (LUBAC)-mediated linear ubiquitination that is critical for specifically directing TLR4-mediated NF-kappaB activation. 26789245_crystal structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation 27460922_This review highlights recent discoveries on RNF31 functions in nuclear factor modifications, breast cancer progression and possible therapeutic inhibitors targeting RNF31. [review] 27545878_SPATA2 has been described as a previously unrecognized factor in the linear ubiquitin chain assembly complex-dependent signaling pathways that serves as an adaptor between HOIP and CYLD, thereby enabling recruitment of CYLD to signaling complexes. 27591049_The data reveal SPATA2 as a high-affinity binding partner of CYLD and HOIP, and a regulatory component of linear ubiquitin chain assembly complex-mediated NF-kappaB signaling. 27669734_Study found that RNF31 is cleaved under apoptosis conditions through various stimulations; findings elucidate a novel regulatory loop between cell death and the survival signal 27810922_LUBAC components control TLR3-mediated innate immunity, thereby preventing development of immunodeficiency and autoinflammation. 28189684_These results indicate that caspase-mediated cleavage of HOIP divides critical functional regions of HOIP, and that this regulates linear (de)ubiquitination of substrates upon apoptosis. 28978479_The binding of SHARPIN or HOIL-1L facilitates the E2 loading of HOIP. 30389786_Human Treg cells that ectopically expressed RNF31 displayed stronger immune-suppressive capacity, suggesting that RNF31 positively regulates both FOXP3 stability and Treg cell function. 30936877_Second Case of HOIP Deficiency Expands Clinical Features and Defines Inflammatory Transcriptome Regulated by LUBAC. 32122970_Cross-regulation between LUBAC and caspase-1 modulates cell death and inflammation. 32403254_Linear Ubiquitin Code: Its Writer, Erasers, Decoders, Inhibitors, and Implications in Disorders. 33215740_Site-specific ubiquitination of the E3 ligase HOIP regulates apoptosis and immune signaling. 34416243_RNF31 mediated ubiquitination of A20 aggravates inflammation and hepatocyte apoptosis through the TLR4/MyD88/NF-kappaB signaling pathway. 34695570_Effects of removing a highly conserved disulfide bond in ubiquitin-associated domain of human HOIP on biochemical characteristics. | ENSMUSG00000047098 | Rnf31 | 925.083493 | 1.1261753 | 0.171431423 | 0.12708530 | 1.855673e+00 | 1.731250e-01 | 5.378004e-01 | No | Yes | 1015.737018 | 109.459146 | 901.599152 | 97.274911 |
| ENSG00000095777 | 53904 | MYO3A | protein_coding | Q8NEV4 | FUNCTION: Probable actin-based motor with a protein kinase activity. Probably plays a role in vision and hearing (PubMed:12032315). Required for normal cochlear hair bundle development and hearing. Plays an important role in the early steps of cochlear hair bundle morphogenesis. Influences the number and lengths of stereocilia to be produced and limits the growth of microvilli within the forming auditory hair bundles thereby contributing to the architecture of the hair bundle, including its staircase pattern. Involved in the elongation of actin in stereocilia tips by transporting the actin regulatory factor ESPN to the plus ends of actin filaments (By similarity). {ECO:0000250|UniProtKB:Q8K3H5, ECO:0000269|PubMed:12032315}. | 3D-structure;ATP-binding;Actin-binding;Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Deafness;Hearing;Kinase;Motor protein;Myosin;Non-syndromic deafness;Nucleotide-binding;Reference proteome;Repeat;Sensory transduction;Serine/threonine-protein kinase;Transferase;Vision | The protein encoded by this gene belongs to the myosin superfamily. Myosins are actin-dependent motor proteins and are categorized into conventional myosins (class II) and unconventional myosins (classes I and III through XV) based on their variable C-terminal cargo-binding domains. Class III myosins, such as this one, have a kinase domain N-terminal to the conserved N-terminal motor domains and are expressed in photoreceptors. The protein encoded by this gene plays an important role in hearing in humans. Three different recessive, loss of function mutations in the encoded protein have been shown to cause nonsyndromic progressive hearing loss. Expression of this gene is highly restricted, with the strongest expression in retina and cochlea. [provided by RefSeq, Jul 2008]. | hsa:53904; | cytoplasm [GO:0005737]; filamentous actin [GO:0031941]; filopodium [GO:0030175]; filopodium tip [GO:0032433]; myosin complex [GO:0016459]; stereocilium tip [GO:0032426]; actin binding [GO:0003779]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; microfilament motor activity [GO:0000146]; plus-end directed microfilament motor activity [GO:0060002]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cochlea morphogenesis [GO:0090103]; protein autophosphorylation [GO:0046777]; response to stimulus [GO:0050896]; sensory perception of sound [GO:0007605]; visual perception [GO:0007601] | 12032315_mutation in humans causes progressive nonsyndromic hearing loss DFNB30 12672820_class III myosin is an actin-based motor protein having a protein kinase activity 16385451_Observational study of gene-disease association. (HuGE Navigator) 17012748_myosin IIIA can spend a majority of its ATP hydrolysis cycling time on actin 17074769_the actomyosin-ADP state may be important for the ability of myosin III to function as a cellular transporter and actin cross-linker in the actin bundles of sensory cells 18229949_A model in which the activity and concentration of myosin IIIA localized to the tips of actin bundles mediates the morphology of the tips in sensory cells. 19229853_Observational study of gene-disease association. (HuGE Navigator) 19229853_behavioral inhibition-associated SNPs appear to be associated with differences in MYO3A- but not GAD2 lymphoblastoid-mRNA expression levels 20090771_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20610541_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20826793_Results suggest that Myo3A motor activity is regulated through a mechanism involving concentration-dependent autophosphorylation. 24214986_The differential regulation of the kinase and motor activities allows for MYO3A to precisely self-regulate its concentration in the actin bundle-based structures of cells. 25402663_Data suggest that, in enterocytes, MYO3A autophosphorylation of kinase domain at Thr184/Thr188 regulates kinase activity, translocation of MYO3A to tips of microvilli, and stability of actin cytoskeleton. 26785147_The structures of Myo3 in complex with Espin1 not only elucidate the mechanism of the binding, but also reveal a Myo3-induced release of Espin1 auto-inhibition mechanism. 26841241_Study reports an amino acid substitution in MYO3A motor-head domain disrupting its ATPase activity that can cause autosomal dominant progressive hereditary hearing loss. Also, these results uncovered a novel interaction between MYO3A and PCDH15 shedding new light on the function of myosin IIIA at stereocilia tips. 27063751_A homozygous mutation, MYO3A:c.1841C>T (p.S614F), was identified to be responsible for non-syndromic congenital deafness in two members of a Kazakh family in China. 27582493_MYO3A is more efficient than MYO3B at increasing formation and elongation of stable microvilli on the surface of cultured epithelial cells. 29880844_Characterization of a novel MYO3A missense mutation associated with a dominant form of late onset hearing loss. 32519820_A novel missense variant in MYO3A is associated with autosomal dominant high-frequency hearing loss in a German family. 33078831_Whole exome sequencing reveals pathogenic variants in MYO3A, MYO15A and COL9A3 and differential frequencies in ancestral alleles in hearing impairment genes among individuals from Cameroon. 33953343_Frequency and origin of the c.2090T>G p.(Leu697Trp) MYO3A variant associated with autosomal dominant hearing loss. 34788109_Deafness mutation in the MYO3A motor domain impairs actin protrusion elongation mechanism. | ENSMUSG00000025716 | Myo3a | 157.888932 | 1.1862233 | 0.246375558 | 0.25254316 | 9.501522e-01 | 3.296806e-01 | No | Yes | 169.366841 | 42.714035 | 136.734797 | 34.526774 | ||
| ENSG00000099999 | 200312 | RNF215 | protein_coding | Q9Y6U7 | Glycoprotein;Membrane;Metal-binding;Reference proteome;Transmembrane;Transmembrane helix;Zinc;Zinc-finger | hsa:200312; | endosome [GO:0005768]; Golgi transport complex [GO:0017119]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; trans-Golgi network [GO:0005802]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; Golgi to vacuole transport [GO:0006896]; protein targeting to vacuole [GO:0006623]; ubiquitin-dependent protein catabolic process [GO:0006511] | ENSMUSG00000003581 | Rnf215 | 668.190258 | 0.9329644 | -0.100106017 | 0.10645548 | 8.850562e-01 | 3.468206e-01 | 7.038277e-01 | No | Yes | 582.228801 | 56.598530 | 634.521954 | 61.444937 | ||||
| ENSG00000100068 | 91355 | LRP5L | transcribed_unprocessed_pseudogene | A4QPB2 | Alternative splicing;Reference proteome;Repeat | coreceptor activity [GO:0015026]; adipose tissue development [GO:0060612]; anterior/posterior pattern specification [GO:0009952]; bone marrow development [GO:0048539]; bone morphogenesis [GO:0060349]; bone remodeling [GO:0046849]; branching involved in mammary gland duct morphogenesis [GO:0060444]; cholesterol homeostasis [GO:0042632]; gastrulation with mouth forming second [GO:0001702]; osteoblast development [GO:0002076]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0061178]; retinal blood vessel morphogenesis [GO:0061304] | 32789677_The novel mutation P36R in LRP5L contributes to congenital membranous cataract via inhibition of laminin gamma1 and c-MAF. | 143.862113 | 1.2800890 | 0.356244073 | 0.22860666 | 2.467937e+00 | 1.161904e-01 | No | Yes | 148.659254 | 18.494215 | 116.404754 | 14.581293 | |||||||
| ENSG00000100083 | 26088 | GGA1 | protein_coding | Q9UJY5 | FUNCTION: Plays a role in protein sorting and trafficking between the trans-Golgi network (TGN) and endosomes. Mediates the ARF-dependent recruitment of clathrin to the TGN and binds ubiquitinated proteins and membrane cargo molecules with a cytosolic acidic cluster-dileucine (DXXLL) motif (PubMed:11301005, PubMed:15886016). Mediates export of the GPCR receptor ADRA2B to the cell surface (PubMed:27901063). Required for targeting PKD1:PKD2 complex from the trans-Golgi network to the cilium membrane (By similarity). Regulates retrograde transport of proteins such as phosphorylated form of BACE1 from endosomes to the trans-Golgi network (PubMed:15886016, PubMed:15615712). {ECO:0000250|UniProtKB:Q8R0H9, ECO:0000269|PubMed:11301005, ECO:0000269|PubMed:15615712, ECO:0000269|PubMed:15886016, ECO:0000269|PubMed:27901063}. | 3D-structure;Acetylation;Alternative splicing;Endosome;Golgi apparatus;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport;Ubl conjugation | This gene encodes a member of the Golgi-localized, gamma adaptin ear-containing, ARF-binding (GGA) protein family. Members of this family are ubiquitous coat proteins that regulate the trafficking of proteins between the trans-Golgi network and the lysosome. These proteins share an amino-terminal VHS domain which mediates sorting of the mannose 6-phosphate receptors at the trans-Golgi network. They also contain a carboxy-terminal region with homology to the ear domain of gamma-adaptins. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:26088; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; endosome membrane [GO:0010008]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; protein-containing complex [GO:0032991]; phosphatidylinositol binding [GO:0035091]; small GTPase binding [GO:0031267]; ubiquitin binding [GO:0043130]; Golgi to plasma membrane protein transport [GO:0043001]; intracellular protein transport [GO:0006886]; positive regulation of protein catabolic process [GO:0045732]; protein localization [GO:0008104]; protein localization to cell surface [GO:0034394]; protein localization to ciliary membrane [GO:1903441]; retrograde transport, endosome to Golgi [GO:0042147]; toxin transport [GO:1901998] | 11859376_X-ray structure of the GGA1 VHS domain alone, and in complex with the carboxy-terminal peptide of cation-independent mannose 6-phosphate receptor containing an ACLL sequence 12135764_endocytosis and intracellular transport of memapsin 2, mediated by its cytosolic domain, may involve the binding of GGA1 and GGA2 12668765_The 2.4-A crystal structure of the GAT domain of human GGA1 reveals a three-helix bundle, with a long N-terminal helical extension that is not conserved in GAT domains that do not bind ARF. 12679809_X-ray crystal structures of the human GGA1-GAT domain and the complex between ARF1-GTP and the N-terminal region of the GAT domain 12767220_Crystal structure of the human GGA1 GAT domain 14636058_A hydrophobic surface patch on the C-terminal three-helix bundle motif of the GGA1 GAT domain is directly involved in binding with a coiled-coil region of rabaptin-5. 14973137_GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region 15143060_Rabaptin-5, ubiquitin, and TSG101 bind to overlapping but distinct binding sites on the trihelical bundle subdomain of GGA-1 protein 15466887_serine phosphorylation of BACE is a physiologically relevant post-translational modification that regulates trafficking in the juxtanuclear compartment by interaction with GGA1 15615712_GGA proteins funstion with the phosphorylated ACDL in the memasin 2-recycling pathway from endosomes to trans Golgi on the way back to the cell surface. 15886016_Our data indicate that GGA proteins are not only involved in the sorting at the TGN but also mediate the retrograde transport of cargo proteins from endosomes to the TGN. 16407204_the trafficking of adiponectin through its secretory pathway is dependent on GGA-coated vesicles 17005855_GGA1 prevented APP beta-cleavage products from becoming substrates for gamma-secretase. Direct binding of GGA1 to BACE was not required for these effects, but the integrity of the GAT (GGA1 and TOM) domain of GGA1 was. 17151287_GGA1 alters the proteolytic processing of beta-amyloid precursor protein (APP) and the secretion of APPs and amyloid-beta, suggesting a role of GGA1 in Alzheimer's disease pathogenesis. 17494868_These results show that the dual roles of PI4P can promote specific GGA targeting and cargo recognition at the trans-Golgi network. 17506864_the interaction between the hinge region and the GAE domain underlies the autoregulation of GGA function in clathrin-mediated trafficking through competing with the accessory proteins and the AP-1 complex 17596511_p56 tightly cooperates with the GGAs in the sorting of cathepsin D to lysosomes, probably by enabling the movement of GGA-containing transport carriers. 19788741_GGA overexpression causes various sorting defects as measured by recycling of CD-MPR, internalization of transferrin receptor, and the subcellular localization of proteins like Tsg101, ubiquitin, and Hrs. 22621900_On basis of these results, propose that GGA1 facilitates LR11 endocytic traffic and that LR11 modulates A-beta levels by promoting amyloid-beta precursor protein traffic to the endocytic recycling compartment 24407285_These data indicate that clathrin is required for the function of AP-1- and GGA-coated carriers at the trans-Golgi network but may be dispensable for outward traffic en route to the plasma membrane. 24866237_These findings show that the AD-like phenotype of NPC model cells can be partly reverted by promoting a non-amyloidogenic processing of APP through the upregulation of GGA1 supporting its preventive role against AD 27901063_full length alpha2B-AR associated with GGA2 but not GGA1, its third intracellular loop was found to directly interact with both GGA1 and GGA2. More interestingly, further mapping of interaction domains showed that the GGA1 hinge region and the GGA2 GAE domain bound to multiple subdomains of the loop. 29142073_The adaptor, GGA1, and retromer are essential to mediate rapid trafficking of phosphorylated BACE1 to recycling endosomes. Therefore, post-translational phosphorylation of DISLL enhances the exit of BACE1 from early endosomes, a pathway mediated by GGA1 and retromer, which is important in regulating amyloid beta production. 33206455_Inactivation of the three GGA genes in HeLa cells partially compromises lysosomal enzyme sorting. | ENSMUSG00000033128 | Gga1 | 1894.380498 | 1.1322147 | 0.179147509 | 0.07484701 | 5.704509e+00 | 1.692139e-02 | 1.744302e-01 | No | Yes | 2057.435949 | 180.035936 | 1845.873119 | 161.501280 | |
| ENSG00000100206 | 11144 | DMC1 | protein_coding | Q14565 | FUNCTION: Participates in meiotic recombination, specifically in homologous strand assimilation, which is required for the resolution of meiotic double-strand breaks. {ECO:0000269|PubMed:21307306}. | 3D-structure;ATP-binding;Alternative splicing;Cell cycle;Chromosome;DNA-binding;Meiosis;Nucleotide-binding;Nucleus;Reference proteome | This gene encodes a member of the superfamily of recombinases (also called DNA strand-exchange proteins). Recombinases are important for repairing double-strand DNA breaks during mitosis and meiosis. This protein, which is evolutionarily conserved, is reported to be essential for meiotic homologous recombination and may thus play an important role in generating diversity of genetic information. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2013]. | hsa:11144; | chromosome [GO:0005694]; chromosome, telomeric region [GO:0000781]; condensed nuclear chromosome [GO:0000794]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP-dependent activity, acting on DNA [GO:0008094]; DNA binding [GO:0003677]; DNA strand exchange activity [GO:0000150]; double-stranded DNA binding [GO:0003690]; identical protein binding [GO:0042802]; single-stranded DNA binding [GO:0003697]; chromosome organization involved in meiotic cell cycle [GO:0070192]; DNA recombinase assembly [GO:0000730]; female gamete generation [GO:0007292]; homologous chromosome pairing at meiosis [GO:0007129]; male meiosis I [GO:0007141]; meiotic cell cycle [GO:0051321]; mitotic recombination [GO:0006312]; oocyte maturation [GO:0001556]; ovarian follicle development [GO:0001541]; reciprocal meiotic recombination [GO:0007131]; spermatid development [GO:0007286]; spermatogenesis [GO:0007283]; strand invasion [GO:0042148] | 14764457_p53 might be involved in homologous recombination and/or checkpoint function by directly binding to DMC1 protein to repress genomic instability in meiotic germ cells 15125839_The monomeric structure of the Dmc1 protein closely resembled those of the human and archaeal Rad51 proteins. We found another hydrogen bonding interaction at the polymer interface. 15164066_hDmc1-mediated DNA recombination initiates through the nucleation of hDmc1 onto ssDNA to form a helical nucleoprotein filament 15917243_N-terminal domain of DMC1 is required for the formation of the octamer, which may support the proper DNA binding activity of the DMC1 protein. 15917244_activation of hDmc1 is mediated through conformational changes induced by free Ca2+ ion binding to a protein site that is distinct from the Mg2+.ATP-binding center 17541404_BRCA2 binds the meiosis-specific recombinase DMC1 and define the primary DMC1 interaction site to a 26 amino-acid region 17639081_Hop2/Mnd1 greatly stimulates Dmc1 to promote synaptic complex formation on long duplex DNAs 18166824_Data show that DMC1 mutations may be one explanation for premature ovarian failure. 18166824_Observational study of gene-disease association. (HuGE Navigator) 18355319_The interaction of DMC1 and the homologue of the large subunit of CAF-1 is reported. 18535008_Results indicate that Rad51 and Dmc1 filaments are essentially identical with respect to several structural parameters, including persistence length, helical pitch, filament diameter, DNA base pairs per helical turn and helical handedness. 18566005_Biochemical analyses revealed that the human DMC1-M200V variant had reduced stability, and was moderately defective in catalyzing in vitro recombination reactions. 19004837_Reversibility, equilibration, and fidelity of strand exchange reaction between short oligonucleotides promoted by RecA protein from escherichia coli and human Rad51 and Dmc1 proteins. 19076215_DMC1-I37N polymorphism may be a source of improper meiotic recombination, causing meiotic defects in humans 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20378615_Observational study of gene-disease association. (HuGE Navigator) 20600108_the Dmc1 filament formed on single-stranded DNA has a mass per unit length expected from approximately 6.5 subunits per turn 21151113_D-loops in a human DMC1-driven reaction are substantially more resistant to dissociation by branch-migration proteins such as RAD54 than those formed by RAD51. 21307306_RAD51AP1 foci colocalize with a subset of DMC1 foci in spermatocytes. 21903585_RAD51-associated protein 1 (RAD51AP1) interacts with the meiotic recombinase DMC1 through a conserved motif. 22156371_The results suggested that PSF may function as an activator for the meiosis-specific recombinase DMC1. 23182424_conserved lysine in the Walker A motif of hDMC1 is critical for ATP binding. 23265958_results suggested mutations in the coding sequence of DMC1 are not associated with premature ovarian failure in Chinese women 23545642_truncated DMC1 octamers further stack with alternate polarity into a filament 26088134_In contrast to RAD51, stabilization of the presynaptic filament via ATP hydrolysis attenuation is insufficient for enhancement of the DMC1-catalyzed recombination reaction. 26709229_Dmc1 can reject divergent DNA sequences while bypassing a few mismatches in the DNA sequence. 26976601_BRCA2 protein stimulates DMC1-mediated DNA strand exchange between RPA-ssDNA complexes and duplex DNA, thus identifying BRCA2 as a mediator of DMC1 recombination function. 27052786_Data indicate that ahomologous pairing by DMC1 protein or RAD51 recombinase in chromatin containing nucleosome-depleted double-stranded DNA (dsDNA) regions. 29331980_To the best of our knowledge, this is the first report identifying DMC1 as the causative gene for human non-obstructive azoospermia and premature ovarian insufficiency. 30085085_In this study, we created a mouse model of a putative infertility allele, DMC1M200V. DMC1 encodes a RecA homolog essential for meiotic recombination and fertility in mice..we found that Dmc1M200V/M200V male and female mice are fully fertile and do not exhibit any gonadal abnormalities 33446654_Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures. 34515795_A pathogenic DMC1 frameshift mutation causes nonobstructive azoospermia but not primary ovarian insufficiency in humans. 34871438_Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination. | ENSMUSG00000022429 | Dmc1 | 465.471783 | 0.9783602 | -0.031562374 | 0.16190408 | 3.810201e-02 | 8.452384e-01 | 9.491576e-01 | No | Yes | 450.386879 | 92.336269 | 448.255297 | 91.890998 | |
| ENSG00000100362 | 5816 | PVALB | protein_coding | P20472 | FUNCTION: In muscle, parvalbumin is thought to be involved in relaxation after contraction. It binds two calcium ions. | 3D-structure;Acetylation;Calcium;Direct protein sequencing;Metal-binding;Muscle protein;Phosphoprotein;Reference proteome;Repeat | The protein encoded by this gene is a high affinity calcium ion-binding protein that is structurally and functionally similar to calmodulin and troponin C. The encoded protein is thought to be involved in muscle relaxation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2015]. | hsa:5816; | axon [GO:0030424]; cuticular plate [GO:0032437]; cytoplasm [GO:0005737]; neuronal cell body [GO:0043025]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; stereocilium [GO:0032420]; synapse [GO:0045202]; calcium ion binding [GO:0005509]; identical protein binding [GO:0042802]; protein-containing complex binding [GO:0044877]; cochlea development [GO:0090102]; excitatory chemical synaptic transmission [GO:0098976]; gene expression [GO:0010467]; inhibitory chemical synaptic transmission [GO:0098977] | 12867516_In subjects with schizophrenia a reduction in parvalbumin [PV] and GAD67 mRNA expression in prefrontal cortex neurons was noted 15059934_Slow relaxation caused by alpha-Tm mutants can be corrected by modifying calcium handling with parvalbumin. 15257133_In dopaminergic cells of the substantia nigra in Parkinson disease an increased parvalbumin content is detected reflecting a natural protective mechanism against putative increase of intracellular calcium caused by excitotoxic injury and oxidative stress. 17405923_demonstration of a reduced number of parvalbumin-immunoreactive projection neurons in the mammillary bodies in schizophrenia 17850980_There is no correlation between total neuronal loss and PV-ir neuronal loss in any of the hippocampal fields in epileptic patient. 18297277_The results of this study indicate a significant reduction in the number of PV interneurons within layer 2 of the multiple sclerosis primary motor cortex 18469313_sequencing of PVALB was performed in 132 cases of Gitelman's syndrome in whom only one or no (N = 79) mutant SLC12A3 allele was found 20093724_parvalbumin has a role in calcium handling in cardiac diastolic dysfunction [review] 20926528_PVALB is a novel diagnostic marker for Hurthle ademonas of the thyroid. 22272358_Data show calbindin (CB)- and tyrosine hydroxylase (TH)-cells were distributed in the three striatal territories, and the density of calretinin (CR) and parvalbumin (PV) interneurons were more abundant in the associative and sensorimotor striatum. 23254904_A subpopulation of projection neurons containing calcium-binding protein parvalbumin (PV) is identified in a precise mapping of the GABAergic cortical distribution. 23764361_This study demonistrated that loss of parvalbium immunoreactive axons in anterolateral columns of spinal cord in patient of lateral sclerosis spinal cords 23825418_Parvalbumin neurons decrease the drive of the input to the visual cortex in contrast sensitivity. 23891794_mRNA labeling at the single cell level shows a significant decrease in parvalbumin expression in Parkinson's (PD) cases; however, neuronal density of parvalbumin-positive neurons was not significantly different between PD patients and controls. 24217255_This study showed that the parvalbumin basket cell inputs in the dorsolateral prefrontal cortex were lower in patient with schizophrenia. 24628518_This study demonistrated that differentially expressed in PV neurons in subjects with schizophrenia, including genes associated with WNT (wingless-type), NOTCH, and PGE2 (prostaglandin E2) signaling. 24764033_TRPV1-, TRPV2-, P2X3-, and parvalbumin-immunoreactivity neurons in the human nodose ganglion innervate the pharynx and epiglottis through the pharyngeal branch and superior laryngeal nerve 26081613_Increased numbers of PVALB neurons and fiber labeling in focal cortical dysplasia compared to nondysplastic epileptic temporal neocortex and postmortem controls may be related to cortical malformation. 27173597_This study demonstrated that A reduction of PV-positive cells and PV-immunoreactivity was observed exclusively in FCD type I/III specimens compared with cryptogenic tissue from control patients with a poor postsurgical outcome. 27444795_excitatory synapse density is lower selectively on parvalbumin interneurons in schizophrenia and predicts the activity-dependent down-regulation of parvalbumin and GAD67 27458244_Our data support the hypothesis that the loss of PVALB plays a role in the pathogenesis of thyroid tumors. 28074478_The results of this study suggested that innervation from PV-containing thalamic nuclei extends across superficial and middle layers of the human dorsolateral prefrontal cortex. 28835159_These results demonstrate a specific association between elevated PVALB methylation and METH-induced psychosis. 28859333_This study showed that the density of parvalbumin was significantly diminished in the basolateral complex in patients with Parkinson Disease. 29688033_These results provide the first evidence that PVALB promoter methylation is abnormal in schizophrenia and suggest that this epigenetic finding may relate to the reduction of PV expression seen in the disease. 30545133_the Calreticulin deficiency-mediated increase in cell death was not prevented by calbindin-D28k or Parvalbumin. 31588185_PVALB promoter methylation in major depressive disorder and its correlation with major depressive disorder severity indicating a role for epigenetics in this psychiatric disorder. 31981822_Age-related changes in the number of cresyl-violet-stained, parvalbumin and NMDAR 2B expressing neurons in the human spiral ganglion. 32497062_GluN2D-mediated excitatory drive onto medial prefrontal cortical PV+ fast-spiking inhibitory interneurons. 32514083_Transcriptional and imaging-genetic association of cortical interneurons, brain function, and schizophrenia risk. 32524185_Parvalbumin immunohistochemical expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. 33386125_Propofol sedation-induced alterations in brain connectivity reflect parvalbumin interneurone distribution in human cerebral cortex. 34439824_Strontium Binding to alpha-Parvalbumin, a Canonical Calcium-Binding Protein of the ''EF-Hand'' Family. | ENSMUSG00000005716 | Pvalb | 16.837118 | 0.6084650 | -0.716753905 | 0.64019681 | 1.246688e+00 | 2.641861e-01 | No | Yes | 15.470057 | 3.987814 | 21.044984 | 5.162390 | ||
| ENSG00000100413 | 171568 | POLR3H | protein_coding | Q9Y535 | FUNCTION: DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts, such as Epstein-Barr virus-encoded RNAs (EBERs) induce type I interferon and NF- Kappa-B through the RIG-I pathway (By similarity). {ECO:0000250, ECO:0000269|PubMed:19609254, ECO:0000269|PubMed:19631370}. | 3D-structure;Alternative splicing;Antiviral defense;DNA-directed RNA polymerase;Immunity;Innate immunity;Nucleus;Reference proteome;Transcription | hsa:171568; | centrosome [GO:0005813]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; RNA polymerase III complex [GO:0005666]; DNA binding [GO:0003677]; DNA-directed 5'-3' RNA polymerase activity [GO:0003899]; defense response to virus [GO:0051607]; innate immune response [GO:0045087]; nucleobase-containing compound metabolic process [GO:0006139]; transcription by RNA polymerase III [GO:0006383]; transcription initiation from RNA polymerase III promoter [GO:0006384] | 19246067_The authors demonstrate the interaction of both RNA polymerase I and III with hepatitis delta virus RNA, both in vitro and in human cells. 20843307_Findings reveal that RNA polymerase III-dependent EBER expression through induction of cellular transcription factors and add to the repertoire of EBNA1's transcription-regulatory properties. 30830215_A pathogenic homozygous missense mutation (c.149A>G; p.Asp50Gly) in the POLR3H gene in two unrelated families with primary ovarian insufficiency was identified. | ENSMUSG00000022476 | Polr3h | 4571.893173 | 0.9671494 | -0.048189347 | 0.06850670 | 4.994744e-01 | 4.797312e-01 | 7.928877e-01 | No | Yes | 4257.243172 | 481.788566 | 4361.098980 | 493.443093 | ||
| ENSG00000100425 | 23774 | BRD1 | protein_coding | O95696 | FUNCTION: Scaffold subunit of various histone acetyltransferase (HAT) complexes, such as the MOZ/MORF and HBO1 complexes, that acts as a regulator of hematopoiesis (PubMed:16387653, PubMed:21753189, PubMed:21880731). Plays a key role in HBO1 complex by directing KAT7/HBO1 specificity towards histone H3 'Lys-14' acetylation (H3K14ac), thereby promoting erythroid differentiation (PubMed:21753189). {ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:21753189, ECO:0000269|PubMed:21880731}. | 3D-structure;Acetylation;Alternative splicing;Bromodomain;Chromatin regulator;Chromosome;Erythrocyte maturation;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a bromodomain-containing protein that localizes to the nucleus and can interact with DNA and histone tails. The encoded protein is a component of the MOZ/MORF acetyltransferase complex and can stimulate acetylation of histones H3 and H4, thereby potentially playing a role in gene activation. Variation in this gene is associated with schizophrenia and bipolar disorder in some study populations. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2017]. | hsa:23774; | chromosome [GO:0005694]; dendrite [GO:0030425]; histone H3-K14 acetyltransferase complex [GO:0036409]; MOZ/MORF histone acetyltransferase complex [GO:0070776]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; perikaryon [GO:0043204]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; erythrocyte maturation [GO:0043249]; histone H3 acetylation [GO:0043966]; histone H3-K14 acetylation [GO:0044154]; positive regulation of erythrocyte differentiation [GO:0045648]; response to electrical stimulus [GO:0051602]; response to immobilization stress [GO:0035902] | 19693800_BRD1 may have a role in schizophrenia and bipolar affective disorder 19693800_Observational study of gene-disease association. (HuGE Navigator) 19763615_These results suggest a general role of BRD1 in the cell and stress that the two BRD1 variants may play different roles in the etiology of psychiatric disease. 19908236_Observational study of gene-disease association. (HuGE Navigator) 19908236_The association of BRD1 with schizophrenia in a Japanese population, was analysed. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21880731_Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). 22820306_The PHD finger of BRPF2 can potentially bind DNA non-specifically with an evolutionarily conserved and positively charged surface. 27142060_Findings show that BRD1 primarily binds in close proximity to transcription start sites and regulates expression of numerous genes, many of which are involved with brain development and susceptibility to mental disorders. 28095495_Reduced BRD1 expression is associated with Schizophrenia. 28334966_Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2 have been presented. 28431065_TIMP-4, NT-proANP, NT-proBNP were strongest associated with PAF and AHRE. The discriminatory performance of CHADS2-VASc for PAF was increased by addition of selected biomarkers. 29453316_Results show that interaction of sulfatide with BRD1 induced acetylation and promoted integrin alpha V gene transcription in hepatocellular carcinoma cells. 29794561_There was no association between SNPs in the BRD1 and ZBED4 genes and schizophrenia in the Chinese population. 30042400_our data point to a cell type- and a stimulus-specific function of BRD1. Inhibiting BRD1 could have potential beneficial effects in rheumatoid arthritis via decreasing the proliferation of synovial fibroblasts. Anti-inflammatory effects were limited and only observed in monocyte-derived macrophages 33556623_Crystal structure of the BRPF2 PWWP domain in complex with DNA reveals a different binding mode than the HDGF family of PWWP domains. 34259319_HBO1 is a versatile histone acyltransferase critical for promoter histone acylations. 34482774_Expression Characteristics and Clinical Correlations of BRD1 in Colorectal Cancer Samples. | ENSMUSG00000022387 | Brd1 | 1596.452091 | 0.9280496 | -0.107726230 | 0.07921559 | 1.850716e+00 | 1.737002e-01 | 5.388301e-01 | No | Yes | 1483.966021 | 91.339055 | 1588.770354 | 97.604585 | |
| ENSG00000100614 | 5494 | PPM1A | protein_coding | P35813 | FUNCTION: Enzyme with a broad specificity. Negatively regulates TGF-beta signaling through dephosphorylating SMAD2 and SMAD3, resulting in their dissociation from SMAD4, nuclear export of the SMADs and termination of the TGF-beta-mediated signaling. Dephosphorylates PRKAA1 and PRKAA2. Plays an important role in the termination of TNF-alpha-mediated NF-kappa-B activation through dephosphorylating and inactivating IKBKB/IKKB. {ECO:0000269|PubMed:16751101, ECO:0000269|PubMed:18930133}. | 3D-structure;Alternative splicing;Cytoplasm;Hydrolase;Lipoprotein;Magnesium;Manganese;Membrane;Metal-binding;Myristate;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome | The protein encoded by this gene is a member of the PP2C family of Ser/Thr protein phosphatases. PP2C family members are known to be negative regulators of cell stress response pathways. This phosphatase dephosphorylates, and negatively regulates the activities of, MAP kinases and MAP kinase kinases. It has been shown to inhibit the activation of p38 and JNK kinase cascades induced by environmental stresses. This phosphatase can also dephosphorylate cyclin-dependent kinases, and thus may be involved in cell cycle control. Overexpression of this phosphatase is reported to activate the expression of the tumor suppressor gene TP53/p53, which leads to G2/M cell cycle arrest and apoptosis. Three alternatively spliced transcript variants encoding distinct isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:5494; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; calmodulin-dependent protein phosphatase activity [GO:0033192]; magnesium ion binding [GO:0000287]; manganese ion binding [GO:0030145]; protein serine phosphatase activity [GO:0106306]; protein serine/threonine phosphatase activity [GO:0004722]; protein threonine phosphatase activity [GO:0106307]; R-SMAD binding [GO:0070412]; cellular response to transforming growth factor beta stimulus [GO:0071560]; dephosphorylation [GO:0016311]; N-terminal protein myristoylation [GO:0006499]; negative regulation of BMP signaling pathway [GO:0030514]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of NIK/NF-kappaB signaling [GO:1901223]; negative regulation of SMAD protein complex assembly [GO:0010991]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; peptidyl-threonine dephosphorylation [GO:0035970]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of protein export from nucleus [GO:0046827]; positive regulation of transcription, DNA-templated [GO:0045893]; protein dephosphorylation [GO:0006470]; regulation of cell cycle [GO:0051726] | 16751101_This work demonstrates that PPM1A/PP2Calpha, through dephosphorylation of Smad2/3, plays a critical role in terminating TGFbeta signaling. 16931515_PPM1A plays an important role in controlling BMP signaling through catalyzing Smad1 dephosphorylation 17729405_Aberrant location of expression/staining intensity of PTEN, PPM1A and P-Smad2 in hepatocellular carcinoma may impact disease progression. 18482992_PTEN abrogates TGF-beta-induced Smad2/3 phosphorylation. This study establishes a novel role for nuclear PTEN in the stabilization of PPM1A. 18829461_Overexpression of PPM1A and the related PPM1B greatly reduced Cdk9 T-loop phosphorylation 18930133_Protein phosphatase 1A is essential to terminate I-kappa B Kinase-mediated NF-kappaappaB activation through binding to the activated form of I-kappa B Kinase and dephosphorylating I-kappa B Kinase at the conserved residues Ser177 and Ser181. 19404668_The present data indicate that PPM1A plays a critical role in the regulation of normal placentation by inhibiting trophoblast migration and invasion. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20970119_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22041443_High expression of LMP2 and low expression of PPM1A might play an important role in the motility and invasiveness of trophoblast cells and malignant transformation of hydatidiform mole. 22519956_Studies indicate that phosphatase PPM1G is a component of the spliceosome and binds to protein YB-1 to affect alternative splicing. 22727189_PPM1A inhibits HIV-1 infection and gene expression. PPM1A depletion in resting CD4+ T cells increases HIV-1 gene expression. 23560844_PPM1A negatively regulates ERK by directly dephosphorylating its pThr202 position early in epidermal growth factor stimulation. Additional kinetic studies reveal that key residues participate in phospho-ERK recognition by PPM1A. 23779087_a nuclear envelope-localized mechanism of inactivating TGF-beta signaling in which MAN1 competes with transcription factors for binding to Smad2 and Smad3 and facilitates their dephosphorylation by PPM1A. 23812431_PPM1A is a RelA phosphatase that regulates NF-kappaB activity and that PPM1A has tumor suppressor-like activity. 23906386_phosphatase activity toward phosphopeptide substrates by PP2Calpha and Wip1 requires the binding of a Mg(2)+ ion to the low-affinity site. 24901250_The TGF-beta/Smad signaling system decreases its activity through strong negative regulation. We provide evidence for a new negative feedback loop through PPM1A upregulation. 25026293_Loss of PPM1A is associated with the development of tumor invasion in bladder cancer patients. 25815785_findings demonstrate a novel regulatory circuit in which STING and TBK1 reciprocally regulate each other to enable efficient antiviral signaling activation, and PPM1A dephosphorylates STING and TBK1 27004401_These data suggest that PPM1A, which had previously been shown to play a role in the antiviral response to Herpes Simplex virus infection, also governs the antibacterial response of macrophages to bacteria, or at least to Mycobacterium tuberculosis infection 27121309_Present study suggests that HBx-induced degradation of PPM1a is a novel mechanism for over-activation of TGF-beta pathway in HCC development. 27195906_Report a tumor-suppressive function of PPM1A and an independent relationship to Smad4 in pancreatic ductal adenocarcinoma. 27301936_In a nested case control study of ischemic stroke, there was an epigenome-wide association for cg04985020 (PPM1A; P=1.78x10(-07)) with vascular recurrence in patients treated with aspirin. 27328942_establish PPM1A as a novel repressor of the SMAD3 pathway in renal fibrosis 28176854_Here the authors establish PPM1A as a checkpoint target used by Mycobacterium tuberculosis to suppress macrophage apoptosis. Overproduction of PPM1A suppressed apoptosis of Mycobacterium tuberculosis-infected macrophages by a mechanism that involves inactivation of the c-Jun N-terminal kinase (JNK). 28283039_Findings show that HCV infection and replication decreased PPM1A abundance, mediated by NS3, in hepatoma cells. Compared to normal liver tissues, the expression of PPM1A was significantly decreased in the HCC tumor tissues and adjacent non-tumor tissues through its regulation by NS3 which promotes its ubiquitination and proteasomal degradation. 28481111_hydrogen/deuterium exchange-mass spectrometry and molecular dynamics to characterize conformational changes in PP2Calpha between the active and inactive states 29343725_PPM1A as a negative threshold regulator of M1-type monocyte-to-macrophage differentiation. 29602904_Enzyme kinetics of PPM1Acat toward a phosphopeptide substrate supported a random-order, bi-substrate mechanism, with substantial interaction between the bound substrate and the labile metal ion 29898761_Our findings suggest that TRIM52 up-regulation promotes proliferation, migration and invasion of HCC cells through the ubiquitination of PPM1A. 30201805_CSIG enhanced the phosphatase activity of PPM1A and further inhibited TGF-beta signaling. 31686644_Hepatitis B Virus-Encoded MicroRNA (HBV-miR-3) Regulates Host Gene PPM1A Related to Hepatocellular Carcinoma. 31791585_Protein phosphatase magnesium-dependent 1A induces inflammation in rheumatoid arthritis. 31792727_Correlation of PPM1A Downregulation with CYP3A4 Repression in the Tumor Liver Tissue of Hepatocellular Carcinoma Patients. 31909517_Protein phosphatase Mg(2+) /Mn(2+) dependent-1A and PTEN deregulation in renal fibrosis: Novel mechanisms and co-dependency of expression. 32024237_PPM1A Controls Diabetic Gene Programming through Directly Dephosphorylating PPARgamma at Ser273. 32348176_TRIM59 inhibits PPM1A through ubiquitination and activates TGF-beta/Smad signaling to promote the invasion of ectopic endometrial stromal cells in endometriosis. 32878540_PPM1A suppresses the proliferation and invasiveness of RCC cells via Smad2/3 signaling inhibition. 33724144_MicroRNA-487a-3p inhibits the growth and invasiveness of oral squamous cell carcinoma by targeting PPM1A. 34336002_lncRNA TSPEAR-AS2, a Novel Prognostic Biomarker, Promotes Oral Squamous Cell Carcinoma Progression by Upregulating PPM1A via Sponging miR-487a-3p. 34637963_Role of active site arginine residues in substrate recognition by PPM1A. 34861123_A comprehensive overview of PPM1A: From structure to disease. | ENSMUSG00000021096 | Ppm1a | 1518.180014 | 1.1001620 | 0.137715972 | 0.09826172 | 1.930885e+00 | 1.646617e-01 | 5.268698e-01 | No | Yes | 1794.691105 | 276.245813 | 1663.563162 | 256.093509 | |
| ENSG00000100650 | 6430 | SRSF5 | protein_coding | Q13243 | FUNCTION: Plays a role in constitutive splicing and can modulate the selection of alternative splice sites. | Acetylation;Alternative splicing;Direct protein sequencing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;mRNA processing;mRNA splicing | The protein encoded by this gene is a member of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, which constitute part of the spliceosome. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. The RS domain is rich in serine and arginine residues and facilitates interaction between different SR splicing factors. In addition to being critical for mRNA splicing, the SR proteins have also been shown to be involved in mRNA export from the nucleus and in translation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:6430; | cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; mRNA binding [GO:0003729]; protein kinase B binding [GO:0043422]; RNA binding [GO:0003723]; cellular response to insulin stimulus [GO:0032869]; liver regeneration [GO:0097421]; mRNA processing [GO:0006397]; mRNA splice site selection [GO:0006376]; mRNA splicing, via spliceosome [GO:0000398]; positive regulation of RNA splicing [GO:0033120]; regulation of cell cycle [GO:0051726] | 12665590_role in c-H-ras alternative splicing regulation 15123677_SR proteins 9G8, SC35, ASF/SF2, and SRp40 have effects on the utilization of the A1 to A5 splicing sites of HIV-1 RNA 15163745_activates the ESE (exon splicing enhancer) which regulates HIV-1 rev, env, vpu, and nef gene expression 16103121_SRp40 regulates the switch in splicing from production of CREMtau(2)alpha to CREMalpha 16990281_SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 interact competitively at the HIV-1 Tat exon 2 splicing site 19843576_SRp40 antagonizes ASF/SF2 and SRp55 by competing for binding to certain sites in exon 5, thereby promoting TF exon 5 exclusion, an event unique to asTF biosynthesis. 20376328_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20427542_Here, the authors report that specific SR proteins, particularly SRp40 and SRp55, promote human immunodeficiency virus type 1 (HIV-1) Gag translation from unspliced (intron-containing) viral RNA. 22100336_We show that changes in alternative splicing of hnRNP A/B, affected by up regulation of SRSF5 (SRp40) or by treatment with C6-ceramide, occur within supraspliceosomes. 22205602_Relative levels of SRp20, SRp30c, and SRp40 in TM cells control differential expression of the two alternatively spliced isoforms of the GR and thereby regulate GC responsiveness. 25665148_Suggest that SRp40 might be associated with GRalpha transcripts in systemic lupus erythematosus patients. 26670336_Enhanced SRSF5 Protein Expression Reinforces Lamin A mRNA Production in HeLa Cells and Fibroblasts of Progeria Patients 27565915_the up-regulated expression of SRSF 5-7 proteins in LC with much more profound up-regulation in SCLC than in NSCLC and suggest that up-regulation of the SRSFs is related to SCLC proliferation. Moreover, we identified SRSF5 as a novel detection marker for SCLC and pleural metastatic cancer cells. 28536481_Cold induction of serine and arginine rich splicing factor 5 (SRSF5) is independent of cold-inducible RNA-binding protein (CIRP) and RNA-binding motif protein 3 (RBM3). 28592444_Posttranslational modification of SR proteins underlies the regulation of their mRNA export activities and distinguishes pluripotent from differentiated cells. 28854257_this study demonstrates that HRS acts as a key component of TLR7 signaling to orchestrate immune and inflammatory responses during EV71 infection 28867611_we show that ErbB3 interacts with the ESCRT-0 subunit Hrs both in the presence and absence of heregulin. This indicates an ESCRT-mediated sorting of ErbB3 to late endosomes and lysosomes, and in line with this we show that impaired ESCRT function leads to an endosomal accumulation of ErbB3. 29857020_Study found that SRSF5 is a novel target of SRSF3. SRSF5 is overexpressed in oral squamous cell carcinoma (OSCC) and functions as an oncogene. Its downregulation in OSCC cell lines slows cell growth, cycle progression, and tumor growth. The expression of SRSF5 seems to controlled by an autoregulation mechanism. 29891722_In this study, we uncover an alternative role for the ESCRT-0 component hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) in promoting the constitutive recycling of transmembrane proteins. We find that endosomal localization of the actin nucleating factor Wiscott-Aldrich syndrome protein and SCAR homologue (WASH) requires HRS, which occupies adjacent endosomal subdomains 29942010_upon glucose intake, the splicing factor SRSF5 is specifically induced through Tip60-mediated acetylation on K125, which antagonizes Smurf1-mediated ubiquitylation. SRSF5 promotes the alternative splicing of CCAR1 to produce CCAR1S proteins, which promote tumor growth by enhancing glucose consumption and acetyl-CoA production. 31106485_Antitumor activity of SR splicing-factor 5 knockdown by downregulating pyruvate kinase M2 in non-small cell lung cancer cells. 32400287_Genes involved in glucocorticoid receptor signalling affect susceptibility to mood disorders. | ENSMUSG00000021134 | Srsf5 | 6834.341587 | 0.9665687 | -0.049055860 | 0.05216928 | 8.819899e-01 | 3.476572e-01 | 7.041485e-01 | No | Yes | 6671.930789 | 1252.257777 | 6957.760298 | 1305.793880 | |
| ENSG00000100906 | 4792 | NFKBIA | protein_coding | P25963 | FUNCTION: Inhibits the activity of dimeric NF-kappa-B/REL complexes by trapping REL dimers in the cytoplasm through masking of their nuclear localization signals. On cellular stimulation by immune and proinflammatory responses, becomes phosphorylated promoting ubiquitination and degradation, enabling the dimeric RELA to translocate to the nucleus and activate transcription. {ECO:0000269|PubMed:7479976}. | 3D-structure;ANK repeat;Cytoplasm;Disease variant;Ectodermal dysplasia;Host-virus interaction;Hydroxylation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | This gene encodes a member of the NF-kappa-B inhibitor family, which contain multiple ankrin repeat domains. The encoded protein interacts with REL dimers to inhibit NF-kappa-B/REL complexes which are involved in inflammatory responses. The encoded protein moves between the cytoplasm and the nucleus via a nuclear localization signal and CRM1-mediated nuclear export. Mutations in this gene have been found in ectodermal dysplasia anhidrotic with T-cell immunodeficiency autosomal dominant disease. [provided by RefSeq, Aug 2011]. | hsa:4792; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; I-kappaB/NF-kappaB complex [GO:0033256]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; NF-kappaB binding [GO:0051059]; nuclear localization sequence binding [GO:0008139]; transcription factor binding [GO:0008134]; ubiquitin protein ligase binding [GO:0031625]; apoptotic process [GO:0006915]; cellular response to cold [GO:0070417]; cytoplasmic sequestering of NF-kappaB [GO:0007253]; cytoplasmic sequestering of transcription factor [GO:0042994]; I-kappaB kinase/NF-kappaB signaling [GO:0007249]; lipopolysaccharide-mediated signaling pathway [GO:0031663]; negative regulation of DNA binding [GO:0043392]; negative regulation of lipid storage [GO:0010888]; negative regulation of macrophage derived foam cell differentiation [GO:0010745]; negative regulation of myeloid cell differentiation [GO:0045638]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of Notch signaling pathway [GO:0045746]; nucleotide-binding oligomerization domain containing 1 signaling pathway [GO:0070427]; nucleotide-binding oligomerization domain containing 2 signaling pathway [GO:0070431]; positive regulation of cellular protein metabolic process [GO:0032270]; positive regulation of cholesterol efflux [GO:0010875]; positive regulation of inflammatory response [GO:0050729]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein import into nucleus [GO:0006606]; regulation of cell population proliferation [GO:0042127]; regulation of NIK/NF-kappaB signaling [GO:1901222]; response to exogenous dsRNA [GO:0043330]; response to muramyl dipeptide [GO:0032495]; response to muscle stretch [GO:0035994]; toll-like receptor 4 signaling pathway [GO:0034142]; tumor necrosis factor-mediated signaling pathway [GO:0033209] | 11799106_IkappaBalpha x p53 complex plays an important role in responses involving growth regulation, apoptosis, and hypoxic stress 11913973_A novel in vitro assay for deubiquitination of I kappa B alpha 12028809_NF-kappa Beta activation in BAL cells may play in important role in initiation and progression of silica-induced pulmonary inflammation, cellular damage, and fibrosis 12084717_results demonstrate that SLPI prevents LPS-induced NF-kappaB activation by inhibiting degradation of IkappaBalpha without affecting the LPS-induced phosphorylation and ubiquitination of IkappaBalpha 12086926_Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C and Ikappab-alpha 12167702_The protein IkappaBalpha is a novel substrate of recombinant c-Abl in HEK cells. c-Abl-mediated phosphorylation at tyrosine 305 is associated with an increase of the IkappaBalpha protein stability. 12193729_A membrane-transducing mutant of I kappa B alpha has been generated that efficiently enters cells, associates with NF-kappa B p65 subunit, and inhibits NF-kappa B-mediated transcription and binding to its consensus sequence. 12244195_Increased nuclear accumulation of I kappa B alpha in neutrophils is associated with the inhibition of NF-kappa B activity and the induction of apoptosis in these cells. 12297126_NFkB, I-kB and I-kB kinase are present in platelets; upon platelet activation, the NF-kappaB/I-kappaBalpha complex is dissociated by phosphorylation of I-kB and proteolysis. 12377412_polymorphism associated with an increased risk of multiple myeloma 12388747_These data suggest that the subcellular location of I(kappa)B(alpha) is a critical determinant in ionizing radiation-induced nuclear factor-kappaB activation. 12406554_Observational study of gene-disease association. (HuGE Navigator) 12419806_IkappaBalpha attenuates NF-kappaBeta transcriptional activity which is an important process that restores the latent state in post-induced cells 12433922_found in mitochondria; regulates mitochondrial gene expression 12518988_Our data suggest that the serine and tyrosine phosphorylation of IkappaB-alpha may play a role in determining the radiosensitivity of malignant glioma cells. 12589049_IkappaBalpha and p65 have roles in regulating the cytoplasmic shuttling of nuclear corepressors 12620896_data suggest that NO may play a major role in the regulation of IkappaBalpha levels in aortic endothelial cells and that the application of low shear flow increases NF-kappaB activity by attenuating NO generation and thus IkappaBalpha levels. 12651903_Data show that increased nuclear factor-kappaB (NF-kB) activity in the amnion during labor is associated with an increase in the expression of NF-kBp65 and of the NF-kB binding proteins IkBa, IkBb-1 and IkBb-2. 12704657_Induced stabilization of IkappaBalpha can facilitate its re-synthesis and prevent sequential degradation. 12711606_H2O2 induces NF-kappaB activation, not through serine phosphorylation or degradation of IkappaBalpha, but through Syk-mediated tyrosine phosphorylation of IkappaBalpha 12748192_results indicate that protein kinase CKII may control IkappaBalpha and p27Kip1 degradation and thereby G1/S phase transition through the phosphorylation of threonine 10 within CKBBP1 protein 12897149_IKBA is degraded by HSP27 through the 26s proteasome 12921778_Okadaic acid induces degradation of the nuclear IKBA in neutrophils. 12944982_Inhibitor kappa B-alpha promoter polymorphisms is associated with pulmonary sarcoidosis 12972430_IkappaBalpha regulates the transcriptional activity of homeodomain-containing proteins positively through cytoplasmic sequestration of HDAC1 and HDAC3 13680285_single nucleotide polymorphisms in the 3'-UTR were significantly increased only in Crohn's disease patients without a variation in the CARD15 gene. 14576361_IkappaBalpha was markedly degraded at 1 h, and NF-kappaB-DNA-binding activity markedly increased 2 h after beta(2) integrin aggregation 14623898_Down-regulation of PTEN by NIK/NF-kappaB results in activation of the PI3K/Akt pathway. This effect is associated with a lack of an inhibitor of kappaB (IkappaB)-alpha autoregulatory loop. 14961554_Observational study of gene-disease association. (HuGE Navigator) 15068390_PCR measure of I kappa B-alpha mRNA levels is a rapid, sensitive, and powerful method to quantify the transcriptional power of NF-kappa B. 15073126_nuclear factor-kappa B and I kappa B alpha proteins have roles in development of prostatic adenocarcinomas 15173174_I kappa B-NF kappa B participates on ERK2-mediated survival mechanisms 15184376_IkappaBalpha is inhibited by ultraviolet rays and activates NFkappaB 15202778_Calpain plays an important role in IkB alpha degradation, a crucial event in T cell activation. 15330761_results indicate that the PKC pathway leading to SOD2 induction proceeds at least in part through NF-kappaB and that inhibition of IkappaBalpha synthesis might serve as a potential pharmacological approach to up-regulate MnSOD 15337789_IkappaBalpha mutation can result in different clinical syndromes within one family 15389633_demonstrated that TRAIL mediates the recruitment of PI-3K/Akt and NF-B/IB pathways in leukaemic cells, namely Jurkat T cells 15536134_IkappaBalpha is recruited to the promoter regions of the Notch-target gene, hes1 15613239_IkappaB-alphaS32/36A, a proteolysis-resistant inhibitor of NF-kappaB, potently inhibits the growth of HIV-1 and SIVmac239 in cell cultures. 15671028_Results indicate that polymyxin B induces a partial maturation of human dendritic cells through increased adhesion and activation of the IkappaB-alpha/NF-kappaB pathway, and that increased ERK1/2 activation inhibits maturation. 15811852_gene transactivation by the transcription factor NF-kappaB is subject to the regulation of a dynamic balance between the coactivators and corepressors in the IkappaB alpha promoter 15858823_Observational study of gene-disease association. (HuGE Navigator) 15979856_IkappaB-alpha is involved in the proliferation of human Burkitt lymphoma Daudi cells, possibly through the MAP kinase pathway. 16046415_Galpha13-induced VASP phosphorylation that involves activation of RhoA and MEKK1, phosphorylation and degradation of IkappaB, release of PKA catalytic subunit from the complex with IkappaB and NF-kappaB, and subsequent phosphorylation of VASP 16105840_p65 phosphorylated on serine 536 is not associated with or regulated by IkappaBalpha, but it has a distinct set of target genes and may represent a noncanonical NF-kappaB pathway that is independent of IkappaBalpha regulation 16258173_IkappaB kinase and IkappaBalpha have NF-kappaB-dependent as well as NF-kappaB-independent pathways of HAS1 activation 16407234_Herpes simplex virus disrupts NF-kappaB regulation by blocking its recruitment on the IkappaBalpha promoter 16426569_ST2 negatively regulates LPS-induced IL-6 production via the inhibition of IkappaB degradation in THP-1 cells 16540234_Observational study of gene-disease association. (HuGE Navigator) 16540234_Significant differences in the frequency of particular polymorphisms across the NFKBIA gene were noted between patients and controls, and analysis may lead to associations with disease progression and survival and thus more personalized therapy. 16737960_TLR8-mediated MEKK3-dependent IKKgamma phosphorylation might play an important role in the activation of IKK complex, leading to IkappaBalpha phosphorylation 16756995_Surface plasmon resonance (SPR) data showed that the IkappaBalpha and NF-kappaB associate rapidly but dissociate very slowly, leading to an extremely stable complex. 16919536_glucose intake induces an immediate increase in intranuclear NF-kappaB binding, a fall in IkappaBalpha, an increase in IKKalpha, IKKbeta, IKK activity, and messenger RNA expression of TNF-alpha in MNCs in healthy subjects. 16931600_Results indicate that 14-3-3 proteins facilitate the nuclear export of IkappaBalpha-p65 complexes and are required for the appropriate regulation of NFkappaB signaling. 16955245_heat shock increases IkappaBalpha gene expression primarily by increasing Ikappa Balpha mRNA stability and this effect is partially dependent on p38 MAP kinase. 17003112_Human HIF asparaginyl hydroxylase, factor inhibiting HIF (FIH), also efficiently hydroxylates specific asparaginyl (Asn)-residues within proteins of the IkappaB family. 17062574_DSCR1 attenuates NF-kappaB-mediated transcriptional activation by stabilizing its inhibitory protein, IkappaBalpha 17115186_Observational study of gene-disease association. (HuGE Navigator) 17148610_IkappaBalpha conformational flexibility and regions of IkappaBalpha folding upon binding to NF-kappaB are important attributes for its regulation of NF-kappaB transcriptional activity 17284228_Observational study of gene-disease association. (HuGE Navigator) 17284228_The IkappaBalpha-826 T and -550 A alleles are associated with susceptibility to rheumatoid arthritis . 17307728_Immature intestinal epithelial cells have increased IKKbeta expression & phosphorylation compared with adult IEC. 17316064_I-kappa B alpha, the inhibitory subunit of NF-kappa B (a transcription factor activated by oxidative stress), was upregulated following wounding. 17318773_Observational study of gene-disease association. (HuGE Navigator) 17318773_Polymorphisms in NFKBIA gene associated with type 1 diabetes. 17333217_Observational study of gene-disease association. (HuGE Navigator) 17351338_IkappaBalpha acts as a sensor of viral infection. 17354114_Chinese individuals >or=50 years of age carrying the AG genotype of NFKBIA may be at an increased risk of developing colorectal cancer, and the GG genotype of NFKBIA may be considered as a prognostic factor for Swedish patients. 17354114_Observational study of gene-disease association. (HuGE Navigator) 17463416_NFKBIA polymorphisms associate with susceptibility to pneumococcal disease. 17463416_Observational study of gene-disease association. (HuGE Navigator) 17492467_Observational study of gene-disease association. (HuGE Navigator) 17492467_Polymorphisms in NF-kappaB1 and NF-kappaBIalpha genes is associated with melanoma 17593226_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17607549_Hyaluronan of intrinsic molecular weight suppresses LPS-stimulated production of proinflammatory cytokines via ICAM-1 through down-regulation of NF-kappaB and IkappaB 17681786_Berberine suppressed Il-1beta/TNF-alpha production in lung inflammation models. Suppression was dependent on inhibition of IkappaB-alpha phosphorylation and degradation. 17701919_NFKB and NFKBI polymorphisms have roles in susceptibility of tumour and other diseases [review] 17703412_Observational study of gene-disease association. (HuGE Navigator) 17910475_Tyrosine nitration of IKBA reveals a novel mechanism for NF-kappaB activation. 17942396_IkappaB-alpha negatively regulates the HIV-1 expression and replication in an NF-kappaB-independent manner by directly binding to Tat 17991436_reduction of phosphorylation leads to nuclear retention of p65, which might be partly responsible for altered transcriptional behavior of p65 serine mutants 18071880_IkappaBalpha -826 T nucleotide promoter polymorphism may be a risk factor for the development of systemic lupus erythematosus in Taiwanese. 18071880_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18199400_There was a negative correlation between the expression of I Kappa B-alpha mRNA level and acute physiology in multiple organ dysfunction syndrome. 18215193_cytoplasmic pI kappaB-alpha expression in non-small cell lung cancer that independently predict overall survival have been identified. 18260825_The canonical IKK2/IkappaBalpha pathway of NF-kappaB activation mediates the up-regulation of RGS4 expression in response to IL-1beta. 18356846_NF-kappaB, IkappaB-alpha, IkappaB-beta mRNA decreased significantly after weight loss. 18434448_Observational study of gene-disease association. (HuGE Navigator) 18474597_H(2)O(2) prolongs NF-kappaB activation in co-stimulated cells by suppressing the negative regulatory functions of Cezanne and IkappaBalpha. 18511071_These data demonstrate clearly that the coupled folding and binding of IkappaBalpha is critical for its precise control of NF-kappaB transcriptional activity. 18515365_Interleukin (IL) 1beta induction of IL-6 is mediated by a novel phosphatidylinositol 3-kinase-dependent AKT/IkappaB kinase alpha pathway targeting activator protein-1 18555796_Cys(189) of IkappaB alpha is a target for S-glutathionylation. 18600435_promoter polymorphisms is associated with susceptibility to primary Sjogren's syndrome in Taiwan 18606063_Transfection of IkappaBalpha can inhibit NF-kappaB activity, thus inhibit cell invasion of A549, which may be through the down-regulation of MMP-2 and MMP-9 expressions. 18636124_Observational study of gene-disease association. (HuGE Navigator) 18636537_Blocking NF-kappaB with IKKbeta- or RelA siRNA substantially sensitized Adriamycin-induced cytotoxicity 18660489_Observational study of gene-disease association. (HuGE Navigator) 18716880_NFKBIA 3'UTR GG genotype associated with an increased risk for extensive colitis in Hungarian inflammatory bowel disease patients 18716880_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18818748_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 18981184_tumor suppressor protein SMAR1 can modulate NF-kappaB transactivation and inhibit tumorigenesis by regulating NF-kappaB target genes 19019440_c-mip inhibits the degradation of I-kappaBalpha and impedes the dissociation of the NF-kappaB/I-kappaBalpha complexes. 19046417_These data suggest a mechanism for maintaining NF-kappaB activity in human T cells through the binding of the Caspase-3-generated carboxy-terminal fragment of p65/RelA to IkappaBalpha in order to protect wild-type p65/RelA from IkappaBalpha inhibition. 19187648_The findings indicate that IkappaBalphaM inhibits the activation of NF-kappaB. It significantly up-regulates TIMP-2 expression in human malignant glioma cells and down-regulates the expression of MMP-9. 19213954_overexpression of Nur77 significantly increased IkappaBalpha promoter activity via directly binding to a Nur77 response element in the IkappaBalpha promoter 19223558_Observational study of gene-disease association. (HuGE Navigator) 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19296948_Compared with normal endometrium, progesterone receptor isoform B (PR-B) and IkappaBalpha immunoreactivity were statistically significantly reduced in ectopic as well as eutopic endometrium from women with adenomyosis. 19297320_Adenosine signaling mediates SUMO-1 modification of IkappaBalpha during hypoxia and reoxygenation. 19363678_IkBalpha promoter polymorphisms are related to susceptibility to ankylosing spondylitis in Taiwan. 19363678_Observational study of gene-disease association. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19456861_Data suggest that IkappaB alpha, owing to its interactions with NF-kappaB and HIF-1alpha, may play a pivotal role in the crosstalk between the molecular events that underlie inflammatory and hypoxic responses. 19500386_NFKBIA and NFKBIB are not likely to harbor ovarian cancer risk alleles. 19500386_Observational study of gene-disease association. (HuGE Navigator) 19507254_Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. 19522780_Constitutive inactivation of the NF-kappaB pathway in IkappaBalpha-double negative transgenic mice attenuates the non-permissive properties of reactive astrocytes, leading to an increased ability of neurons to sustain injury and regenerate their axons. 19527514_Observational study of gene-disease association. (HuGE Navigator) 19543524_20-Hydroxycholecalciferol, product of vitamin D3 hydroxylation by P450scc, decreases NF-kappaB activity by increasing IkappaB alpha levels in human keratinocytes 19554506_The expression of NFKBIA gene of NF-kappaB pathway in multiple myeloma using bone marrow aspirates obtained at diagnosis is reported. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19591507_The nuclear magnetic resonance results and residual dipolar coupling experiments presented here strongly indicate that this fragment of IkappaappaBalpha (67-206) represents the well-folded part of the IkappaappaBalpha ankyrin repeat domain. 19648161_NFKBIA low mutation frequency plays a minor role in the NF-kappaB activity of nodular lymphocyte-predominant Hodgkin's lymphoma (NLPHL), suggesting different mechanisms of NF-kappaB activation in NLPHL compared to classical Hodgkin's lymphoma (cHL). 19673747_Observational study of gene-disease association. (HuGE Navigator) 19733238_Results indicate that regulation of the IkappaBalpha level is one of the underlying mechanisms by which C/EBP-beta controls NF-kappaappaB-associated signalling. 19758175_Data show that actively invading CD4(+) cells in PM and sIBM contained both p65 and p50, whereas I-kappaB alpha was absent. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19773451_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19797428_NFKBIA -826T and -881AG were associated with the risk of hepatocellular carcinoma in the subjects infected with HBV genotype C 19798070_Subjects with at least one A allele for NFKBIA rs1951276 had approximately 29% lower insulin sensitivity compared to individuals homozygous for the G allele... 19816602_Expression and activity of MMP-2 was enhanced by the IkappaBalpha siRNA in human ciliary muscle cells through the activation of the NF-kappaB signaling pathway. 19886988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19891769_Aurora-A, via activating Akt, stimulated nuclear factor-kappaB signaling pathway to promote cancer cell survival. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19941056_Observational study of genotype prevalence. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20068069_Elevated levels of sCLU promote prostate cancer cell survival by facilitating degradation of COMMD1 and I-kappaB, thereby activating the canonical NF-kappaB pathway. 20080849_Case report. this patient's IkappaBalpha mutation caused GH and IGF-l resistance which, in turn, contributed to his growth failure. 20132559_I kappaB alpha rs2233408 T heterozygotes were associated with reduced risk of gastric cancer, especially for the development of certain subtypes of gastric cancer in Chinese population. 20164548_IKBA -826T allele, IKBA -881A -826T -550A -519C -297C haplotype, and IKBA -881A -826T -550A -519T -297C haplotype may be related to susceptibility to Behcet's disease. IKBA -826T/T is associated with skin lesions in patients with Behcet's disease. 20164548_Observational study of gene-disease association. (HuGE Navigator) 20193848_NFKBIA mRNA was strongly expressed in H-RS cells from Hodgkin lymphoma sections, and little was detected in the reactive surrounding lymphocytes. 20304615_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20304615_The allele frequency and genotype distribution of NFKBIA gene polymorphism did not differ between myocardial infarction and control group. 20335171_Proteasome inhibitor PS-341 (bortezomib) induces calpain-dependent IkappaB(alpha) degradation. 20379146_Observational study and genome-wide association study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20385620_Data show that RSK2 is activated by treatment with tumor necrosis factor-alpha (TNF-alpha) and directly phosphorylates IkappaBalpha at Ser-32, leading to IkappaBalpha degradation. 20388715_Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation 20448286_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20495844_Meta-analysis of gene-disease association. (HuGE Navigator) 20495844_This meta-analysis demonstrates that autoimmune and inflammatory diseases are associated with NFKBIA gene -826C/T polymorphism, but not with 2758A/G, -881A/G, and -279C/T. 20503287_Observational study of gene-disease association. (HuGE Navigator) 20546338_Total and phosphorylated IkappaBalpha protein expression might serve as markers for NF-kappaB activation in human ovarian carcinoma. 20563630_Observational study of gene-disease association. (HuGE Navigator) 20563630_promoter polymorphism is associated with susceptibility to rheumatoid arthritis in Taiwain 20568250_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20652762_alpha-mangostin also strongly inhibited PMA-induced degradation of inhibitor of kappaBalpha (IkappaBalpha) 20659425_TGase 2-mu-calpain system is significant in the NF-kappaB pathway via I-kappaBalpha polymerization and subsequent degradation. 20674643_Observational study of gene-disease association. (HuGE Navigator) 20674643_gene polymorphisms is associated with the development of asthma in Korea 20696864_Although p65 recruitment to TNF-alpha, IL-1ss, and IL-6 promoters is inhibited by nuclear IkappaBkappaalpha, recruitment to interleukin (IL)-8 promoter is not repressed. 20716621_Observational study of gene-disease association. (HuGE Navigator) 20797629_Nuclear IKKbeta acts as an adaptor protein for IkappaBalpha degradation in UV-induced NF-kappaB activation. NF-kappaB activated by the nuclear IKKbeta adaptor protein suppresses anti-apoptotic gene expression and promotes UV-induced cell death. 20811626_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20836841_Polymorphism in NFkB was associated with colorectal cancer. 20890592_Acrolein causes the decrease in nitric oxide production as an I-kappaBalpha-independent downregulator of NF-kappaB activity in human malignant keratinocytes. 20953189_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20953190_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21175304_Deletion of NFKBIA has an effect that is similar to the effect of EGFR amplification in the pathogenesis of glioblastoma and is associated with comparatively short survival. 21220295_Studies indicate that even at high concentrations of DNA, IkappaBalpha remains associated with NF-kappaB. 21228035_Data show that IKBalpha, NFKB2, and TRAF3 gene polymorphisms play a role in the development of multiple myeloma and in the response to bortezomib therapy. 21263074_alpha-1-antitrypsin blocks nuclear translocation of NF-kappaB p50/p65 despite an unexpected elevation in associated phosphorylated and ubiquitinated IkappaBalpha. 21376060_rs17103265 deletion homozygote is a genetic risk factor for gastric cancer, especially for certain subtypes in a southern Chinese population 21454528_CCL5 signaling induces GLI2 through a PI3K-AKT-IkappaBalpha-p65 pathway and requires GLI2 transcriptional activity to modulate IL-6 expression and Ig secretion in vitro and in vivo 21606193_Data show that IkappaB kinase beta (IKKbeta) can be activated in the nucleus by Nkx3.2 in the absence of exogenous IKK-activating signals, allowing constitutive nuclear degradation of IkappaB-alpha. 21778941_deletion of NFKBIA and amplification of EGFR show a pattern of mutual exclusivity in glioblastoma multiforme[review] 22022389_These results suggest the existence of a reservoir of monoubiquitylated IkappaBalpha resistant to TNFalpha-induced proteolysis. 22078572_A novel mutation in the IKBA gene was discovered in an infant with autosomal-dominant anhidroticectodermal dysplasia with immune deficiency syndrome 22156201_in glioma cell lines that microRNA-30e* (miR-30e*) directly targets the IkappaBalpha 3'-UTR and suppresses IkappaBalpha expression 22184009_MIB1 negatively regulates TNFalpha- and IL1beta- induced NF-kappaB target gene activation 22302838_The endothelium plays important roles in obesity- and age-related disorders through intracellular NF-kappaB signaling, thereby ultimately affecting life span. 22393418_bortezomib-induced autophagy confers relative DLBCL cell drug resistance by eliminating I-kappaBalpha. 22509384_Our results suggest that NFKB1 -94 ATTG2, NFKBIA -826 T, and -881 G alleles are associated with oral carcinogenesis 22935973_Our findings clearly demonstrate changes in the levels of IkappaBalpha in Sjogren's syndrome monocytes, suggesting that the attenuated expression of IkappaBalpha could contribute to the deregulation of NF-kappaB pathways in Sjogren's syndrome pathogenesis. 23046941_The expressions of NF-kappaBp65 and IkappaBalpha are negatively correlated in gestational trophoblastic tumor tissues. 23054188_Pokemon activates the expression of both p65 and IkappaBalpha by sequence-specific binding to their promoters and plays a dual role in regulating NF-kappaB signaling. 23069812_Epithelial-mesenchymal transformation, along with acquisition of a tumor stem cell-like phenotype mediated by IKKbeta/IkappaBalpha/RelA signal pathway via Snail, contributes to a low concentration of arsenite-induced tumorigenesis. 23250755_TOPK directly interacted with and phosphorylated IkappaBalpha at Ser-32, leading to p65 nuclear translocation and NF-kappaB activation. 23274114_YLTA mutations stabilize the I-kappa-B-alpha to proteasomal degradation. 23285182_miR-126 may play roles in UC inflammatory activity by down-regulating the expression of IKBA, an important inhibitor of NF-kappaB signaling pathway. 23322360_a common susceptible mechanism of inflammation in lung induced by genetic variants in NFkappaB1 (-94del>ins ATTG) or IkappaBalpha (2758G>A) to predict risk of COPD or lung cancer. 23377923_the analysis of IkappaBalpha expression at salivary gland epithelial cell level could be a potential new hallmark of Sjogren's syndrome progression. 23422506_The findings from this human cell study in vitro indicate that both relatively high single-dose and chronic opioid exposure may induce the structural changes in the developing human brain and the adult brain by altering the expression of neuronal differentiation- and neurite outgrowth-related genes IkappaBa and TrkB in vivo. 23439505_the silencing of NFKBIA may play an important role in the carcinogenesis of adenocarcinomas independent of EGFR/K-ras mutations or EML4-ALK fusion. 23453579_Authors discover that HIV-1 infection elevates the phosphorylation of IkappaBalpha and p100, and that this increase is greatly reduced when a Vpr-negative HIV-1 is used for infection. 23457512_NFKB1 -94 Ins promoter polymorphism increased the risk of hepatocellular carcinoma (HCC), and may be applied as a predictive factor for the clinical stage and tumor size in female HCC patients. 23487427_Immunologically relevant genetic variation in the promoter of NFKBIA is differentially susceptibile to severe bronchiolitis following infection with respiratory syncytial virus, airway hyperresponsiveness, and severe bronchopulmonary dysplasia. 23555990_These data show that I-kappa-B-alpha can be used as a prognostic marker and target for therapy in Activated B-cell lymphoma, one of the three subtypes of Diffuse Large B-cell Lymphoma. 23563313_CCDC22 participates in NF-kappaB activation and its deficiency leads to decreased IkappaB turnover 23697845_Data indicate that following bortezomib treatment there was accumulation of IkappaB-alpha (IkappaBalpha) without affecting its phosphorylation status at an early time point. 23996241_No statistically significant CRC risk association was found for the NFKBIA polymorphisms. 24085292_long-term incubation with PIs activates NF-kappaB, which is mediated by IkappaBalpha degradation via the lysosome in an IKK-dependent and IKK-independent manner. 24248593_Data indicate that the p97-UFD1L-NPL4 protein complex specifically associates with ubiquitinated IkappaBalpha via the interactions between p97 and the SCF(beta-TRCP) ubiquitin ligase. 24295474_NF-kB expression was downregulated and its cytoplasmatic inhibitor IKBalpha was increased in CTLA4-Ig treated macrophages. 24324738_Association of common polymorphisms in TNFA, NFkB1 and NFKBIA with risk and prognosis of esophageal squamous cell carcinoma in northern Indian population, was investigated. 24330732_data indicate that NFKBIA deletions are present but not frequent in Glioblastoma multiforme (GBM); the deletions become frequent in GBM neurospheres (NS), an event that seems to be affected by the presence of EGF in the culture medium 24361600_The results of this study suggested that oligodendroglial IkappaBalpha expression and NF-kappaB are activated early in the course of MSA and their balance contributes to the decision of cellular demise. 24367071_during IkappaBalpha-mediated dissociation of NF-kappaB from DNA, a ternary complex forms, and rapidly rearranges to release DNA 24368589_This study reveals that polymorphisms in the IkB-alpha promoter (-881 A/G, -826 C/T) are strongly associated with the susceptibility of Iranian Multiple Sclerosis patients 24457201_IkappaBalpha sequesters not only p65 but also RPS3 in the cytoplasm. 24463357_miR-196a can directly interact with IkappaBalpha 3'-UTR to suppress IkappaBalpha expression and subsequently promote activation of NF-kappaB 24504166_MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. 24578542_No association was observed between NFKBIA variants and risk of liver cancer. 25112903_data suggest that the NFKBIA 126 G/A polymorphism might be potentially helpful to identify liver transplant recipients with an increased susceptibility to develop recurrent acute rejections 25215581_The single nucleotide polymorphism rs1957106 CT and TT genotypes were found to be associated with lower NFKBIA protein levels and a poor prognosis of pateints with glioblastoma. 25326706_NFKBIA-rs2233419AG was associated with a significantly increased risk of developing recurrent wheezing. 25352423_Study demonstrates an association between functional polymorphisms of IkappaBalpha rs696 and smoking with the risk of defective spermatogenesis, suggesting some interaction between the NF-kappaB signalling pathway and smoking-related ROS in human spermatogenesis. 25361605_IkappaBetaalpha inhibits apoptosis at the outer mitochondrial membrane independently of NF-kappaB retention. 25367031_the wild genotypes of both two single nucleotide polymorphisms (ins/ins and AA genotypes) and ins/ins/AA combined NFKB1 rs28362491 and NFKBIA rs696 genotype are strongly associated with enhanced risk of Behcet's disease in a Turkish population. 25374080_Expression of IkappaBalpha in human bladder cancer cells is negatively correlated with epithelial-mesenchymal transition and tumor invasion in vitro. 25577468_Data suggest that activity of IKBalpha can be regulated by dietary factors; dietary supplementation with luteolin inhibits vascular endothelial inflammation by suppressing IKBalpha/NFkappaB signaling. 25930675_Data indicate the involvement of the p38 MAPK/NF-kappaB/IkappaBalpha pathway in Newcastle disease virus (NDV) infection and subsequent induction of apoptosis in clear cell renal cell carcinoma (ccRCC) cells. 25937534_SM22alpha is a phosphorylation-regulated suppressor of IKK-IkappaBalpha-NF-kappaB signaling casca | ENSMUSG00000021025 | Nfkbia | 767.887686 | 1.1412466 | 0.190610549 | 0.10032164 | 3.601733e+00 | 5.771938e-02 | 3.215338e-01 | No | Yes | 805.441571 | 74.025822 | 715.019792 | 65.780371 | |
| ENSG00000101966 | 331 | XIAP | protein_coding | P98170 | FUNCTION: Multi-functional protein which regulates not only caspases and apoptosis, but also modulates inflammatory signaling and immunity, copper homeostasis, mitogenic kinase signaling, cell proliferation, as well as cell invasion and metastasis. Acts as a direct caspase inhibitor. Directly bind to the active site pocket of CASP3 and CASP7 and obstructs substrate entry. Inactivates CASP9 by keeping it in a monomeric, inactive state. Acts as an E3 ubiquitin-protein ligase regulating NF-kappa-B signaling and the target proteins for its E3 ubiquitin-protein ligase activity include: RIPK1, CASP3, CASP7, CASP8, CASP9, MAP3K2/MEKK2, DIABLO/SMAC, AIFM1, CCS and BIRC5/survivin. Ubiquitinion of CCS leads to enhancement of its chaperone activity toward its physiologic target, SOD1, rather than proteasomal degradation. Ubiquitinion of MAP3K2/MEKK2 and AIFM1 does not lead to proteasomal degradation. Plays a role in copper homeostasis by ubiquitinating COMMD1 and promoting its proteasomal degradation. Can also function as E3 ubiquitin-protein ligase of the NEDD8 conjugation pathway, targeting effector caspases for neddylation and inactivation. Ubiquitinates and therefore mediates the proteosomal degradation of BCL2 in response to apoptosis (PubMed:29020630). Regulates the BMP signaling pathway and the SMAD and MAP3K7/TAK1 dependent pathways leading to NF-kappa-B and JNK activation. Acts as an important regulator of innate immune signaling via regulation of Nodlike receptors (NLRs). Protects cells from spontaneous formation of the ripoptosome, a large multi-protein complex that has the capability to kill cancer cells in a caspase-dependent and caspase-independent manner. Suppresses ripoptosome formation by ubiquitinating RIPK1 and CASP8. Acts as a positive regulator of Wnt signaling and ubiquitinates TLE1, TLE2, TLE3, TLE4 and AES. Ubiquitination of TLE3 results in inhibition of its interaction with TCF7L2/TCF4 thereby allowing efficient recruitment and binding of the transcriptional coactivator beta-catenin to TCF7L2/TCF4 that is required to initiate a Wnt-specific transcriptional program. {ECO:0000269|PubMed:11447297, ECO:0000269|PubMed:12121969, ECO:0000269|PubMed:14685266, ECO:0000269|PubMed:17560374, ECO:0000269|PubMed:17967870, ECO:0000269|PubMed:19473982, ECO:0000269|PubMed:20154138, ECO:0000269|PubMed:21145488, ECO:0000269|PubMed:22103349, ECO:0000269|PubMed:22304967, ECO:0000269|PubMed:29020630, ECO:0000269|PubMed:9230442}. | 3D-structure;Apoptosis;Cytoplasm;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Protease inhibitor;Reference proteome;Repeat;S-nitrosylation;Thiol protease inhibitor;Transferase;Ubl conjugation;Ubl conjugation pathway;Wnt signaling pathway;Zinc;Zinc-finger | This gene encodes a protein that belongs to a family of apoptotic suppressor proteins. Members of this family share a conserved motif termed, baculovirus IAP repeat, which is necessary for their anti-apoptotic function. This protein functions through binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2 and inhibits apoptosis induced by menadione, a potent inducer of free radicals, and interleukin 1-beta converting enzyme. This protein also inhibits at least two members of the caspase family of cell-death proteases, caspase-3 and caspase-7. Mutations in this gene are the cause of X-linked lymphoproliferative syndrome. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 2 and 11.[provided by RefSeq, Feb 2011]. | hsa:331; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle microtubule [GO:0005876]; cysteine-type endopeptidase inhibitor activity involved in apoptotic process [GO:0043027]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; protein serine/threonine kinase binding [GO:0120283]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to DNA damage stimulus [GO:0006974]; copper ion homeostasis [GO:0055070]; inhibition of cysteine-type endopeptidase activity [GO:0097340]; inhibition of cysteine-type endopeptidase activity involved in apoptotic process [GO:1990001]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; neuron apoptotic process [GO:0051402]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of JNK cascade [GO:0046330]; positive regulation of protein linear polyubiquitination [GO:1902530]; positive regulation of protein ubiquitination [GO:0031398]; regulation of apoptotic process [GO:0042981]; regulation of BMP signaling pathway [GO:0030510]; regulation of cell cycle [GO:0051726]; regulation of cell population proliferation [GO:0042127]; regulation of inflammatory response [GO:0050727]; regulation of innate immune response [GO:0045088]; regulation of nucleotide-binding oligomerization domain containing signaling pathway [GO:0070424]; Wnt signaling pathway [GO:0016055] | 11597143_genomic organization reported; a novel transcript homologous to BIRC4 and expressed solely in the testis was identified 11927604_The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3- and caspase 9-interacting sites. 11972398_caspase 3-mediated focal adhesion kinase processing in human ovarian cancer cells: possible regulation by X-linked inhibitor of apoptosis protein 12048196_ILPIP, a novel XIAP-interacting protein acts as a co-factor enhancing XIAP-mediated activation of JNK1 and the caspase-independent protection of XIAP against apoptosis 12121969_XIAP has a role in degrading smac and protecting cells from mitochondial damage 12121983_calpain-mediated XIAP degradation contributes to initiation of apoptosis in normal neutrophils and dysfunction of this regulatory pathway can lead to pathological neutrophil accumulation. 12218061_Smac-penetratin fusion peptide crossed the cellular membrane, bound XIAP and cIAP1, displaced caspase-3 from cytoplasmic aggregates, and enhanced drug-induced caspase action in situ 12243753_results indicate that IAPs alone are not the main factor responsible for the resistance of non-small-cell lung cancer cells to treatment 12388702_Reovirus-induced apoptosis involves reduction of cellular XIAP protein levels 12482981_nuclear ribonucleoproteins C1 and C2 are part of the RNP complex that forms on XIAP IRES, the cellular levels of hnRNPC1 and -C2 parallel the activity of XIAP IRES and the overexpression of hnRNPC1-C2 specifically enhanced translation of XIAP IRES 12592339_findings indicate that the levels of XIAP and Bcl-X(L) are regulated by distinct pathways during monocytic differentiation, and that upregulation of these proteins contributes to the increased longevity of cells in the monocytic lineage 12624662_the overexpression of XIAP inhibits taxol-induced apoptosis through the decrease of caspase-3 activity and inhibition of the processing of pro-caspase-3. 12691733_RING domain was essentially required for the proteasomal association of XIAP and for its ubiquitination. 12725530_Expression of XIAP and downregulation of Fas-L are linked to chemoresistance in ovarian carcinoma cells and may represent one of the potential antiapoptotic mechanisms involved during this process. 12747801_identification of ubiquitination sites 12835328_identification as substrate for mitochondrial serine protease Omi/HtrA2 12851723_although cIAP-1, cIAP-2 and XIAP transcripts were highly upregulated, their expression of endogenous proteins were not increased in HUVECs stimulated with LPS 12855663_XIAP protein has a role in preventing apoptosis in human lung cancer cells 12970762_cytokines regulate the expression of XIAP in leukemic cell lines and primary AML blasts 14512414_role in relief of caspase inhibition mediated by Smac 14523016_XIAP is regulated by Smac3, a Smac/DIABLO splicing variant 14532997_XIAP is a principal inhibitor of apoptosis overexpressed in human hepatocellular carcinoma and that XIAP may be a potential target for gene therapy of human HCCs 14570909_Grim promoted XIAP ubiquitination and degradation. 14759516_XIAP interacts with CHEK1 during mitosis. 15029247_Data suggest that ARTS induces apoptosis by antagonizing IAPs, including XIAP. 15037009_no difference was observed in XIAP expression between young and aged subjects. 15044484_XIAP up-regulation requires nuclear factor kappa b and has a role in Cyr61-induced resistance to apoptosis in breast cancer cells 15173080_x-linked inhibitor of apoptosis protein and survivin have roles in progression of childhood de novo acute myeloid leukemia 15201285_role of antagonizing XIAP in induction of caspase-dependent cell death in concert with authocatalytic processing of HtrA2/Omi 15207275_First evidence is provided that increased XIAP levels protect the neonatal brain of transgenic XIAP-overexpressing mice against hypoxia-ischemia. 15282301_These data demonstrate that full-length X-linked inhibitor of apoptosis (XIAP) inhibits caspase activation required for mitochondrial amplification of death receptor signals. 15292176_ERK suppresses stress-induced apoptosis downstream of mitochondrial alterations by maintaining XIAP levels and oxidants block this effect through activation of p38 and protein phosphatases 15297970_XIAP-mediated inhibition of apoptosis has a role during progression of clear-cell clear cell renal carcinomas.XIAP expression is a new independent prognostic marker in this tumor type. 15337764_Data indicate that phenylurea-based XIAP antagonists block interaction of downstream effector caspases with XIAP, thus inducing apoptosis of tumor cell lines through a caspase-dependent, Bcl-2/Bax-independent mechanism. 15359644_In drug-induced apoptosis XIAP & its BIR3-RING cleavage product redistribute into large nuclear inclusions, implying a new unknown function of XIAP and its BIR3-RING fragment. 15531913_Treatment with rottlerin downregulated the protein levels of survivin and X-chromosome-linked IAP (XIAP), two major caspase inhibitors. 15570290_The XIAP expression was significantly lower in patients with favorable than intermediate or poor cytogenetics (n = 74; P < 0.05). 15580265_XIAP binds directly to the active-site pockets of effector caspases 15677500_Genetically modified human islets expressing XIAP are resistant to the negative effects of immunosuppressive drugs on insulin secretion and cell viability. 15749826_XIAP functions as ubiquitin ligase toward mature caspase-9 and Smac to inhibit apoptosis. 16142363_IAP-2, XIAP, and survivin may make an important contribution to the resistance to the apoptotic effect of cisplatin in prostate cancer 16211302_downregulation of the XIAP or survivin enhances cell death by TRAIL and increases sensitivity against some chemotherapeutic agents in hepatocellular carcinoma cells 16278380_XIAP expression in colorectal cancer is regulated by hepatocyte growth factor/C-met pathway via Akt signaling 16343440_The ratio of XAF1(A) and XAF1C mRNA expression differs amongst the cell lines tested, suggesting differential mRNA stabilities and/or the existence of tissue- or cell type-specific splicing regulation. 16344307_These data identify an important function of XIAP in cardiomyocytes and point to a striking similarity in the regulation of apoptosis in postmitotic cells. 16394139_XIAP has a pivotal role in apoptosis deregulation in cancer [review] 16543147_These data provide an unsuspected link between copper homeostasis and the regulation of cell death through XIAP and may contribute to the pathophysiology of copper toxicosis disorders. 16603637_ALK7-induced apoptosis is at least in part through two Smad-dependent pathways, Bax/Bcl-2 and Xiap. 16732928_Increased XIAP protein level contributes to the drug resistance in HL-60 cells induced by adhesion to fibronectin. 16799641_Detachment-induced upregulation of XIAP and cIAP2 delays anoikis of intestinal epithelial cells. 16868249_down-regulation of XIAP by RNA interference markedly enhanced the susceptibility of HL cells for CTL-mediated cytotoxicity 16869888_We show here that alpha-fetoprotein cancels XIAP-mediated inhibition of endogenous active caspases in cytosolic lysates of tumor cells, as well as XIAP-induced blockage of active recombinant caspase 3 in a reconstituted cell-free system. 16932741_A sharp XIAP concentration threshold separates conditions of efficient apoptosis execution and inhibition 16932741_high levels of XIAP control caspase activation and substrate cleavage, and may promote apoptosis resistance and sublethal caspase activation in vivo 16964381_These studies identify XIAP as a new substrate of Raf-1. 16972754_data suggest that the sensitization to radiation results from NBS1-siRNA-mediated suppression of DNA repair and/ or X-ray-induced cell survival signaling pathways through NFKB and XIAP. 16983704_Differential expression of IAPs in B-cell lymphomas suggests differences in pathogenesis that may have implications for novel treatment strategies targeting IAPs. 17008917_XLX, a Xenopus laevis inhibitor of apoptosis (IAP) family member, exhibits characteristics typical of an IAP, such as caspase inhibition and autoubiquitylation.XLX is phosphorylated during meiosis by protein kinases of the MAPK and MPF pathways. 17016456_Reviewing current knowledge of the caspase-inhibitory potential of the human IAPs shows that XIAP is probably the only bona fide caspase inhibitor. 17035597_Deguelin has great potential for chemosensitization of XIAP and could represent a new therapeutic agent for treatment of breast cancer. 17050666_These results implicate elevated XIAP levels caused by high basal NF-kappaB activity in TRAIL resistance and suggest that therapeutic strategies involving TRAIL can be abetted by inhibition of NF-kappaB and/or XIAP only in tumor cells. 17069460_In particular, the stability of cIAP-2 is modulated by the presence of X-linked IAP and their interaction is stabilized in infected cells. 17080092_by identifying an XLP immunodeficiency that is caused by mutations in XIAP, we show that XIAP is a potent regulator of lymphocyte homeostasis in vivo 17094439_XIAP is overexpressed in pancreatic cancer and contributes to chemoresistance 17144666_Definition of the consensus sequences of the motifs that interact with the three BIR domains in an unbiased manner. 17179183_The ability of the wild-type XIAP BIR3 domain as well as its Trp323Ser variant in inhibition of human caspase-9, binding to AVPFVASLPN (SMAC-peptide), SMAC protein, and mature caspase-9 was investigated. 17287399_cytoplasmic hnRNP A1 is a negative regulator of XIAP internal ribosome entry site (IRES)-dependent translation, indicating a novel function for the cytoplasmic form of this protein. 17291493_Endogenously expressed XIAP bound active forms of both caspase-9 and caspase-3. However, downregulation of XIAP were unable to induce caspase-9 activity, indicating that it is not a major determinant in blocking caspase-9 in NSCLC cells. 17331366_In cancerization of oral mucosa, XIAP protein could play an important antiapoptotic role by overexpression, while XAF1 protein does not appear to antagonize effectively the role of XIAP. 17332680_XIAP regulates IL-6 transcription via NF-kappaB in cooperation with AP1 and C/EBP-beta. 17339366_Overexpression of X-linked inhibitor of apoptosis proteins is associated with pancreatic cancers 17350081_XIAP is upregulated in mesothelioma effusions and peritoneal mesotheliomas, suggesting a prosurvival role in malignant mesothelioma cells. 17437405_Results demonstrate that cleavage by caspase 3 does not activate caspase 9, but enhances apoptosis by alleviating XIAP inhibition of the apical caspase. 17440816_Downregulation of either FLIP or XIAP but not Bcl-2 restored sensitivity of Colo320 cells to Apo2L/TRAIL. 17450518_Preservation of XIAP proteins represents a key mechanism by which PGI2 protects endothelial cells from oxidant-induced apoptosis. 17471152_An increased expression ratio of XIAP to XAF1 in combination with a disturbed expression of the XAF1 splice variants could be shown in gastric adenocarcinomas. 17534699_XIAP and Survivin are not involved in the cytoprotective effect of HGF against antineoplastic agents. 17579071_Early Kupffer cell survival, even in the presence of tumor necrosis factor (TNF)-alpha, during Pseudomonas aeruginosa strain PA103 infection is mediated by stabilization of XIAP. 17603079_Inhibitor of apoptosis proteins, nuclear factor-kappa, Smac/DIABLO and apoptosis inducing factor were increased in colon cancer cells. 17611394_XIAP is highly expressed in esophageal cancer and its downregulation by RNAi sensitizes esophageal carcinoma cell lines to chemotherapeutics 17613533_XAF1 mediates Survivin down-regulation through a complex containing XIAP, supporting dual roles for XAF1 in apoptosis 17626072_Bortezomib inhibited expression of cIAP-1, cIAP-2, and XIAP, which are regulated by NF-kappaB and function as inhibitors of apoptosis. 17630106_XIAP was highly specific for papillary thyroid carcinomas among the thyroid neoplasms thus it may be a useful marker for differential diagnosis. 17698078_Data report the crystal structure of XIAP and suggest that dimerization may be important for its action in TGF-beta and BMP signaling and the action of cIAP1 and cIAP2 in TNF receptor signaling. 17721914_adhesion-dependent activation of the PI3K/Akt/XIAP pathway may be one of the factors involved in the CAM-DR of U937 cells. 17724022_XIAP homotrimerizes via its C-terminal Ring domain, making its inhibitory activity toward caspase-3 more susceptible to Smac. 17936246_Studies using reporter constructs and NF-kappaB Rel-A deficient mouse embryonic fibroblasts showed that NF-kappaB signaling is required for the induction of Sod2 by XIAP. XIAP also reduced oxidative stress in the PC6.3 cells. 17947468_XIAP is expressed at higher levels in prostate cancers compared with matched normal tissues and may have a role in reducing risk of tumor recurrence 17951200_Strong XIAP immunostaining differentiates malignant from benign and hyperplastic mesothelial cell populations. 17967870_identify apoptosis-inducing factor as a new XIAP binding partner and indicate a role for XIAP in regulating cellular reactive oxygen species 17993464_Data show levels of XIAP are significantly elevated in the COMP-expressing cells and down-regulation XIAP protein levels by small interfering RNAs blocks the ability of COMP to enhance survival. 18022123_findings suggest that the potent apoptotic inhibitor XIAP may be a biomarker in head and neck SCCs. 18024305_The pathological grade of the colorectal cancer tumors was associated with the expression level of XIAP. 18041764_Ectopic expression of Par-4 decreases levels of XIAP protein in thapsigargin-treated cells, caused in part by XIAP protein instability & caspase activation. XIAP instability & Akt inactivation are important in cellular pathways affected by Par-4. 18068114_These results indicate that Nulp1 plays a role in cell death control and may influence tumor growth. 18068526_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18068526_These results suggest that XIAP polymorphisms do not significantly affect susceptibility to lung cancer in Koreans. 18071906_Altogether these results highlight an intertwined role for specific Akt isoforms and XIAP in chemoresistance of uterine cancer cells. 18187663_the role of triptolide as a sensitizer to TRAIL-induced apoptosis in part by independent modulation of XIAP expression and p53 signaling. 18251743_XIAP elevation may be correlated with increasing melanoma thickness and tumor progression. 18309651_XIAP is expressed higher in laryngeal squamous carcinomas than in normal laryngeal tissues, and is associated with tumor clinicopathological characteristics. 18415656_Anti-myeloma effect of homoharringtonine with concomitant targeting of the myeloma-promoting molecules, Mcl-1, XIAP, and beta-catenin 18432259_XIAP expression appears to be useful for distinguishing atypical adenomatous hyperplasia from adenomatous hyperplasia of the lung 18438971_Smac/DIABLO interacts in a similar way with both XIAP and survivin through its amino terminal residues. 18467439_XIAP plays a putative in vivo role in counteracting TNFalpha-induced apoptosis in endometrial tumor cells. 18520160_No defects were observed in the coding sequence of XIAP in a cohort of 30 hypogammaglobulinemic patients; X-linked lymphoproliferative syndrome caused by XIAP deficiency may be a very rare differential diagnosis in male patients with CVID. 18520160_Observational study of gene-disease association. (HuGE Navigator) 18521960_study provides evidence that during T cell proliferation the intracellular caspase inhibitor X-linked inhibitor-of-apoptosis protein (XIAP) interacts with caspases-3/-7, thereby blocking their full activation, substrate cleavage, and cell death 18560353_A mitochondrial block and expression of XIAP lead to resistance to TRAIL-induced apoptosis during progression to metastasis of a colon carcinoma. 18566024_XIAP and HIAP-1 in myelin lesions were co-localized with microglia and T cells in multiple sclerosis 18590777_confirmed a protective role of XIAP upstream of mitochondrial permeabilisation during TRAIL-induced apoptosis 18645029_The effect of XIAP inhibition and PPARgamma activation in colon cancer, was explored. 18647593_These findings suggest that calpain inhibition delays neutrophil apoptosis via cyclic AMP-independent activation of PKA and PKA-mediated stabilization of Mcl-1 and XIAP. 18666224_Important molecule in controlling hepatocellular carcinoma metastasis. XIAP can be a molecular target subject to intervention to reduce metastasis and recurrence. 18703998_XIAP expression postischemic injury is delayed and may continue for several days. Potentiation of XIAP expression may be neuroprotective in the developing brain. 18704457_Down-regulation of XIAP in HCT116 cells with or without troglitazone treatment was associated with changes of gene expression that favor increased tendency of apoptosis, decreased cell proliferation, and angiogenesis potential. 18755525_XIAP over-expression in transgenic mice reduces age-related hearing loss and hair cell death in the cochlea. 18761086_Through interaction with MEKK2, XIAP functions in an ubiquitin ligase dependent manner to evoke a second wave of NF-kappaB activation, resulting in the modulation of NF-kappaB target gene expression. 18767116_anti-apoptotic functions of cFLIPs may be attributed to inhibit oxaliplatin-induced apoptosis through the sustained XIAP protein level and Akt activation. 18795889_COMMD1 mutant which was unable to bind to XIAP demonstrated a complete loss of basal ubiquitination 18807090_High expression of XIAP and low expression of XAF1 is associated with gastric adenocarcinomas. 18820704_These results suggest that simultaneously targeting both FLIP and XIAP may prove useful in the treatment of cancers, particularly those in which the intrinsic mitochondrial apoptotic pathway has been compromised. 18829553_A combination of XIAP inhibition plus TRAIL is a promising strategy to overcome apoptosis resistance of pancreatic cancer. 18831009_The purpose of this study was to test the feasibility of immunocytochemically detecting markers that may be affected by therapy or are predictive of therapeutic responsiveness, including phosphohistone H1, XIAP and p63. 18851976_analysis of 4-substituted azabicyclo[5.3.0]alkane smac mimetics and their binding to the BIR3 domain of XIAP 19001278_XIAP knockdown using RNA interference enhanced drug sensitivity and decreased tumor formation in NOD/SCID mice 19011619_These data show an unexpected role of XIAP and cellular-inhibitor of apoptosis proteins in the turnover of C-RAF protein, thereby modulating the MAPK signalling pathway and cell migration. 19171073_inhibition of apoptosis in rheumatoid arthritis synovium was observed downstream of caspase-3 and may involve the caspase-3 inhibitors, survivin and xIAP. 19223549_cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. 19273858_Nitrosative stress contributes to Parkinson's disease pathogenesis through the impairment of prosurvival proteins such as parkin and XIAP through different mechanisms. 19288545_A rapid flow cytometric screening test for X-linked lymphoproliferative disease due to XIAP deficiency. 19289587_EMA is a better marker than XIAP or GLUT-1 for the diagnosis of malignant mesothelioma 19355825_XIAP-positive staining was more prevalent in PTCs than in the benign thyroid disorders. Although BRAF(V600E) and XIAP expression are commonly seen in papillary thyroid carcinoma, their presence together seems unrelated 19369629_XIAP expression in thymoma suggests a possible role in the pathogenesis of thymoma and may be helpful in differentiating thymic hyperplasia from thymoma. 19393243_Data report the X-ray crystal structure of XIAP-BIR3 domain in complex with a two-headed compound (compound 3) with improved efficacy relative to its monomeric form. 19397802_Data determined an imbalance in XIAP/XAF1 mRNA expression levels correlated to overall patient survival, and that high XIAP immunoreactivity was a poor prognostic factor. 19398375_Results show that XLP due to BIRC4 mutation is not associated with decreased populations of iNKT cells, and that XIAP is likely not a requirement for iNKT cell development. 19411066_Upregulation of XIAP in MDM2-overexpressing cancer cells in response to irradiation resulted in resistance of these cells to radiation-induced apoptosis 19473982_XIAP acts as an E3 ubiquitin ligase for PTEN and promotes Akt activity by regulating PTEN content and compartmentalization. 19506533_Data show that combination therapy resulted in robust apoptotic induction with a concomitant survivin and XIAP reduction in the MIA PaCa-2 cells with little effect in the PANC-1 cells. 19531477_xIAP has a role in mediating oncogenic signaling by TGF-beta and NF-kappaB in breast cancer cells 19562673_NF-kappaB and its downstream target XIAP were essential for the growth and drug resistance of small avascular tumor. 19649722_The results indicate that serum activates Akt and ERK pathways earlier than XIAP expression. 19654940_Multivariate analyses revealed that XIAP overexpression was an independent unfavorable prognostic factor for relapse-free survival 19667203_Data suggest a role for XIAP in regulating innate immune responses by interacting with NOD1 and NOD2 through interaction with RIP2. 19670614_Overexpression of XIAP might cause delayed apoptosis of Hep-2 cells after gamma radiation. 19698783_The RING domain of XIAP (and probably cIAP-1 and cIAP-2) associates with GSK3, GSK3 acts upstream of the apoptosome to promote intrinsic apoptosis, and the association between XIAP and GSK3 may block the pro-apoptotic function of GSK3. 19723899_Observational study of gene-disease association. (HuGE Navigator) 19726736_Selective inhibition of AKT renders normally resistant first-trimester trophoblast cells sensitive to FAS-mediated apoptosis by regulating XIAP expression. 19738422_Data show that the N-t-boc-Daidzein induced apoptosis is characterized by caspase activation, XIAP and AKT degradation. 19758744_XIAP gene RNAi inhibited proliferation of ovarian carcinoma cells and caused cells to be sensitive to cisplatin through the reduction of its mRNA and protein. XIAP is an ovarian cancer-related gene and potential target for therapeutic anti-cancer drugs. 19763917_Arsenic trioxide suppresses transcription of XIAP mRNA in NB-4 cells. 19782107_the inhibition of apoptosis by BMP2 and GDF5 does not depend on more complex signal transduction pathways such as smad and MAPK signaling but on direct stabilization of XIAP by BMPR2. 19787196_Downregulation of XIAP and activation of capase-3 play an important role in mediating the PPAR-gamma-dependent cell death. 19854829_Data demonstrate that XIAP antagonists in combination with Fas ligand (FasL) or the death receptor 5 (DR5) agonist antibody synergistically stimulate death in cancer cells and inhibit tumor growth. 19859091_Observational study of genetic testing. (HuGE Navigator) 19875445_Membrane-associated XIAP induces mitochondrial outer membrane permeabilization leading to cytochrome c and Smac release, which is dependent on Bax and Bak. 19877056_Observational study of gene-disease association. (HuGE Navigator) 19877056_The 423Q polymorphism of the X-linked inhibitor of apoptosis gene (XIAP) is a predisposing factor for idiopathic periodic fever development, possibly through its influence on monocyte function. 19885569_the overexpression of survivin in the majority of NSCLCs together with the abundant or upregulated expression of HBXIP and XIAP suggest that tumours are endowed with resistance against a variety of apoptosis-inducing conditions. 19897582_p53 activation enhances XIAP inhibition-induced cell death by promoting mitochondrial release of second mitochondria-derived activator of caspases (SMAC) and by inducing the expression of caspase-6. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19946707_NHT did not influence XIAP expression. We speculate that the inhibition of XIAP expression may reinforce the apoptotic effect of NHT and improve the prognosis in patients with prostate cancer. 19949310_CDODO-Me-12 and CDODO-Me-11 downregulated the levels of anti-apoptosis protein XIAP in HL-60, U937 and K562 leukemic cell lines. 20067634_XIAP has a more central role in regulating death receptor mediated apoptosis than it does the intrinsic pathway mediated cell death 20154138_Results describe the identification of the copper chaperone for superoxide dismutase as a mediator of copper delivery to XIAP in cells. 20171186_HAX-1 suppresses the polyubiquitination of XIAP; formation of the HAX-1-XIAP complex inhibits apoptosis by enhancing the stability of XIAP against proteosomal degradation. 20213810_XIAP, an inhibitor of activated caspase 3, was significantly up-regulated(approximately 3-fold) at the protein and mRNA levels in ADAM15-transfected chondrocytes upon camptothecin treatment 20381828_Kaplan-Meier survival analysis revealed a pattern of X-linked inhibitor of apoptosis protein expression with impaired overall and disease-free survival in patients with invasive ductal breast cancer. 20395960_The inhibition of XIAP activity proved to be crucial for full effector caspase activity and clonogenic execution. 20406824_survivin cleavage by Granzyme M triggers degradation of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex to free caspase activity leading to cytolysis of target tumor cells 20406946_XIAP is required for survival of cells with acquired resistance to GW583340. 20431038_XIAP inhibits multiple aspects of protein degradation in skeletal muscle during chronic kidney disease. 20484174_Proapoptotic response to benzyl isothiocyanate is mediated by suppression of X-linked inhibitor of apoptosis protein expression. 20515940_Rac1 knockdown by RNAi interference confirmed the specificity of NSC23766 and requirement for Rac1 in the regulation of cyclin D1, survivin, and XIAP in breast cancer cells. 20517649_Suggest a role of BIRC4/XIAP variants as modifier gene of Wilson's disease. 20582956_expression of XIAP is highly increased by post-transcriptional regulation in C-ALL and is associated with poor in vivo glucocorticoid response and outcome in T-cell ALL . 20625944_embelin results in human leukemia cells apoptosis through caspase-dependent mechanisms involving down-regulation of XIAP. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20632385_Data suggest that XIAP expression is a poor prognostic factor in diffuse large B-cell lymphoma (DLBCL) and the XIAP-AKT relationship should be explored further as a potential therapeutic target in DLBCL. 20670888_that SNO-caspase transnitrosylates (transfers its NO group) to XIAP, forming SNO-XIAP, and thus promotes cell injury and death. 20676365_Results indicate that downregulation of XIAP by hUCBSC treatment induces apoptosis, which led to the death of the glioma cells and xenograft cells. 20677802_ex vivo transduction of islets with adenoviral vector encoding XIAP can protect them from inflammatory cytokines and improve their viability and function 20682709_The function of XIAP seems to be conducive to the process of malignant transformation and/or progression. 20712893_XIAP gene expression and function is positively regulated by exposure to the three TGF-beta isoforms in a Smad-dependent manner, similar to constitutive XIAP gene expression which depends on autocrine TGF-beta/Smad signalling 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20856198_Data show that show here the XIAP depletion to essentially require initial caspase-mediated cleavage. 20862799_The expression of XIAP in epithelial ovarian cancer was significantly lower than that in benign ovarian tumor and normal ovarian tissue. 20938744_XIAP antisense oligonucleotide (AEG35156) achieves target knockdown and induces apoptosis preferentially in CD34+38- cells in a phase 1/2 study of patients with relapsed/refractory AML. 20959405_Observational study of gene-disease association. (HuGE Navigator) 20979872_The expressions of PED/PEA-15 and XIAP are elevated in hepatocellular carcinoma as compared with adjacent tissues and normal tissues. 20980180_Results describe molecular docking and dynamics simulations of the binding of XIAP-BIR3 with three groups of Smac mimetics. 21102524_XIAP belongs to the HuR-regulated RNA operon of antiapoptotic genes, which, along with Bcl-2, Mcl-1 and ProTalpha, contributes to the regulation of cell survival. 21119115_Mutations in the gene XIAP is associated with X-linked lymphoproliferative syndrome type 2. 21317880_Co-expression of caspase-3 and XIAP identified high-grade ovarian serous carcinomas with different prognosis. 21325798_S100P has slightly lower sensitivity and higher specificity of pancreatic ductal adenocarcinoma than XIAP in endoscopic ultrasound-guided fine needle aspirations specimens. 21354220_The dimerization of Smac is critical for the XIAP-mediated retention of Smac at or inside the mitochondria. 21364655_The physiological or elevated XIAP levels can affect the kinetics or the extent of Smac release. 21367892_Thus, sendai virus infection-mediated apoptosis is temporally regulated by the prevention of XIAP degradation by PI3K. 21387310_our experiments have identified XIAP as a novel substrate of HtrA1 and the degradation of XIAP by HtrA1 contributes to cell response to chemotherapy. 21402697_XIAP was found to negatively regulate RhoGDI SUMOylation, which might affect its activity in controlling cell motility 21442307_Inhibition of Bcl-2 and XIAP has a non-linear effect on sensitization towards apoptosis in leukemia and lymphoma cells. 21518480_There was a significant increase of XIAP mRNA expression in accelerated and blastic phase of CML, compared with the patients in chronic phase. 21535957_Bortezomib can inhibit the proliferation of K562 cells, and induce apoptosis by down-regulating the expression of XIAP. 21543760_Human X-linked variable immunodeficiency caused by a hypomorphic mutation in XIAP in association with a rare polymorphism in CD40LG. 21628460_TRIM32 sensitizes TNFalpha-induced apoptosis by antagonizing XIAP 21674762_Seven novel mutations in PRF1, UNC13D, and XIAP were identified in Chinese EBV-HLH patients. Only a fraction of Chinese children with EBV-HLH have genetic defects in PRF1, UNC13D, and XIAP. 21712378_Resveratrol induces p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated Bax protein oligomerization on mitochondria to initiate cytochrome c release and caspase activation. 21869827_Translocation of ARTS initiates a first wave of caspase activation leading to the subsequent release of additional mitochondrial factors, including cytochrome C and SMAC/Diablo. 21908615_Syk drives EBV+ B cell lymphoma survival through PI3K/Akt activation, which prevents the HtrA2-dependent loss of XIAP. Syk, Akt, and XIAP antagonists may present potential new therapeutic strategies for PTLD 21935275_Lateral neck lymph node metastases were more frequent in patients negative for XIAP expression 22039248_Overexpression of human XIAP protects ARPE-19 cells against H(2)O(2)-induced oxidative cell death by acting downstream on the apoptotic pathway. 22072751_IKKepsilon regulated Sendai virus-induced apoptosis via the phosphorylation-dependent turnover of XIAP. 22083305_The induction of apoptosis was also accompanied by the downregulation of PI3K/Akt and the inhibitor of apoptosis protein (IAP) family proteins. 22103349_Data indicate that XIAP blocks cell death inducing activity of apoptosis inducing factor (AIF) (not NADH oxidation) through nondegradative ubiquitination; specific residue of AIF (K255) in DNA-binding site appears to be involved in this interaction. 22194841_These results provide the first structural information of XIAP-UBA and map its interaction with mono-ubiquitin, Lys48-linked and linear-linked diubiquitins. 22273571_Human XIAP may be partially associated with the pathogenesis of Parkinson's disease and dementia with Lewy bodies. 22320973_The levels of cellular apoptosis-associated proteins such as Smac/DIABLO, Cyto C, and the activated fragment of caspase-3 increased in pancreatic cancer cells, but the expression | ENSMUSG00000025860 | Xiap | 1643.629496 | 0.9951505 | -0.007013326 | 0.08492783 | 6.911484e-03 | 9.337439e-01 | 9.754215e-01 | No | Yes | 1528.881063 | 338.219166 | 1494.739105 | 330.681264 | |
| ENSG00000102710 | 55578 | SUPT20H | protein_coding | Q8NEM7 | FUNCTION: Required for MAP kinase p38 (MAPK11, MAPK12, MAPK13 and/or MAPK14) activation during gastrulation. Required for down-regulation of E-cadherin during gastrulation by regulating E-cadherin protein level downstream from NCK-interacting kinase (NIK) and independently of the regulation of transcription by FGF signaling and Snail (By similarity). Required for starvation-induced ATG9A trafficking during autophagy. {ECO:0000250, ECO:0000269|PubMed:19893488}. | Alternative splicing;Autophagy;Developmental protein;Gastrulation;Nucleus;Phosphoprotein;Reference proteome | hsa:55578; | SAGA complex [GO:0000124]; SAGA-type complex [GO:0070461]; transcription coregulator activity [GO:0003712]; autophagy [GO:0006914]; gastrulation [GO:0007369]; monoubiquitinated histone deubiquitination [GO:0035521]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; regulation of transcription by RNA polymerase II [GO:0006357] | 11340631_mRNA is down-regulated in human prostate cancer, also described as C13 12070015_Novel transcription factors identified in human CD34 antigen-positive hematopoietic stem cells. 15978328_C13orf19 mRNA is down-regulated in matched prostate tissues compared to prostate carcinoma tissues 16685401_C13orf19 is expressed independently of the androgen. No interaction between C13orf19 and p38MAPK was identified. 19114550_hSPT20 and other hSAGA subunits, together with RNA polymerase II, are specifically recruited to genes induced by endoplasmic reticulum (ER) stress. 19893488_Manipulation of p38IP and p38alpha alters mAtg9 localization, suggesting p38alpha regulates, through p38IP, the starvation-induced mAtg9 trafficking to forming autophagosomes. 24220028_p38IP inhibits ubiquitination-induced GCN5 degradation and therefore promotes alpha-tubulin acetylation, facilitating spindle formation and G2/M progression. 27531877_provide the first report that p38-p38IP is required for the Snail-induced E-cadherin down-regulation and cell invasion in HNSCC 31210371_Our results indicate that FAM48A is a kind of sensor that is required for compensatory autophagy induction upon proteasome impairment. | ENSMUSG00000027751 | Supt20 | 1144.770900 | 1.0110510 | 0.015855826 | 0.09639866 | 2.733997e-02 | 8.686701e-01 | 9.566155e-01 | No | Yes | 1188.209952 | 218.081995 | 1177.740072 | 216.153547 | ||
| ENSG00000102786 | 26512 | INTS6 | protein_coding | Q9UL03 | FUNCTION: Component of the Integrator (INT) complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267). May have a tumor suppressor role; an ectopic expression suppressing tumor cell growth (PubMed:15254679, PubMed:16239144). {ECO:0000269|PubMed:15254679, ECO:0000269|PubMed:16239144, ECO:0000269|PubMed:23904267, ECO:0000305|PubMed:16239144}. | 3D-structure;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. The protein encoded by this gene is a DEAD box protein that is part of a complex that interacts with the C-terminus of RNA polymerase II and is involved in 3' end processing of snRNAs. In addition, this gene is a candidate tumor suppressor and is located in the critical region of loss of heterozygosity (LOH). Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2015]. | hsa:26512; | actin cytoskeleton [GO:0015629]; integrator complex [GO:0032039]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transmembrane signaling receptor activity [GO:0004888]; snRNA 3'-end processing [GO:0034472]; snRNA processing [GO:0016180] | 11939413_Molecular characterization of the tumor suppressor gene in lung carcinoma cells 12527901_Somatic mutations were identified in three patients (3/56, 5%), and one novel polymorphism was identified in 3% of ESCC patients (4/136) and 3% of healthy individuals (6/232). We conclude that DICE1 mutations occur in ESCC but are infrequent. 15254679_DICE1 has a growth-suppressing activity and interferes with anchorage-independent growth of IGF-IR transformed tumor cells dependent upon IGF-I signaling 16271964_Not mutated in human cancer cell lines which demonstratse 13q14 deletions. 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 23280823_Findings suggest that EBV encoded miR-BART3* miRNA targets DICE1 tumor suppressor to promote cellular growth and transformation in nasopharyngeal cancer(NPC). 23986477_In response to the DNA damage response, INTS3-hSSB1-INTS6 complex relocates to the DNA damage sites. 25660097_DICE1 appears to be involved in prostate cancer progression rather than in the initiation of prostate cancer. 25686840_Findings demonstrate INTS6P1 and INTS6 exert the tumor suppressive roles through competing for oncomiR-17-5p. 28899352_the results of our study show that down-regulated INTS6 expression is associated with a poorer prognosis in hepatocellular carcinoma (HCC) patients. This newly identified INTS6/WIF-1 axis indicates the molecular mechanism of HCC and may represent a therapeutic target in HCC patients. 29895194_Downregulation of Wnt/beta-catenin signaling was observed during the activation period, and the impairment of beta-catenin degradation reversed the tumor suppressor effects of INTS6. 34004147_The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. 34508742_INTS6 promotes colorectal cancer progression by activating of AKT and ERK signaling. | ENSMUSG00000035161 | Ints6 | 1090.127035 | 1.2569733 | 0.329954037 | 0.10886611 | 9.242455e+00 | 2.364678e-03 | 5.580712e-02 | No | Yes | 1099.143000 | 225.725636 | 861.626906 | 176.964227 | |
| ENSG00000102981 | 50855 | PARD6A | protein_coding | Q9NPB6 | FUNCTION: Adapter protein involved in asymmetrical cell division and cell polarization processes. Probably involved in the formation of epithelial tight junctions. Association with PARD3 may prevent the interaction of PARD3 with F11R/JAM1, thereby preventing tight junction assembly. The PARD6-PARD3 complex links GTP-bound Rho small GTPases to atypical protein kinase C proteins (PubMed:10873802). Regulates centrosome organization and function. Essential for the centrosomal recruitment of key proteins that control centrosomal microtubule organization (PubMed:20719959). {ECO:0000269|PubMed:10873802, ECO:0000269|PubMed:20719959}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Membrane;Phosphoprotein;Reference proteome;Tight junction | This gene is a member of the PAR6 family and encodes a protein with a PSD95/Discs-large/ZO1 (PDZ) domain and a semi-Cdc42/Rac interactive binding (CRIB) domain. This cell membrane protein is involved in asymmetrical cell division and cell polarization processes as a member of a multi-protein complex. The protein also has a role in the epithelial-to-mesenchymal transition (EMT) that characterizes the invasive phenotype associated with metastatic carcinomas. Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008]. | hsa:50855; | apical plasma membrane [GO:0016324]; bicellular tight junction [GO:0005923]; cell cortex [GO:0005938]; centriolar satellite [GO:0034451]; centrosome [GO:0005813]; cytosol [GO:0005829]; nucleus [GO:0005634]; PAR polarity complex [GO:0120157]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; GTP-dependent protein binding [GO:0030742]; protein kinase C binding [GO:0005080]; small GTPase binding [GO:0031267]; transcription factor binding [GO:0008134]; cell division [GO:0051301]; cell-cell junction maintenance [GO:0045217]; centrosome cycle [GO:0007098]; establishment or maintenance of cell polarity [GO:0007163]; establishment or maintenance of epithelial cell apical/basal polarity [GO:0045197]; negative regulation of protein phosphorylation [GO:0001933]; positive regulation of protein localization to centrosome [GO:1904781]; positive regulation of protein secretion [GO:0050714]; regulation of cellular localization [GO:0060341]; viral process [GO:0016032] | 14976222_Partitioning-defective protein 6 associated constitutively with endogenous aPKCs. 15590654_crystal structure of the complex of PKCiota and Par6alpha PB1 domains to a resolution of 1.5 A 15744531_A rare A/G polymorphism in the promoter of the Par6alpha gene is associated with reduced fasting glycaemia, increased glucose tolerance and reduced serum nonesterified fatty Acids concentrations. 15744531_Observational study of gene-disease association. (HuGE Navigator) 15782111_findings demonstrated that G-protein-activated phospholipase C-beta interacts with cell polarity proteins Par3 and Par6 to form protein complexes and to mediate downstream signal transduction 17050699_Studies in this review confirm that signaling by Par6alpha controls the migration of immature granule neurons down the Bergmann glial fibers into the internal granule cell layer in which they establish synaptic connections. 17057644_We provide evidence for the existence of a distinct PAR protein complex in endothelial cells. Both PAR-3 and PAR-6 associate directly with the adherens junction protein vascular endothelial cadherin (VE-cadherin). 17335965_Par6alpha-mediated inhibition of insulin-dependent glycogen synthesis in C2C12 cells depends on the direct interaction of Par6alpha with aPKC and on aPKC-mediated T34 phosphorylation of Akt1. 17420281_Par6 was characterized as a dual-location protein. 17586613_Shear stress-induced directed migratory polarity is modulated by exogenous growth factors and dependent on Par6 activity and shear stress direction in endothelial cells 17976838_In vivo binding studies identified a novel mechanism of Par6 interaction, suggesting that the cell polarity machinery may serve to spatially restrict Rit signaling. 18562307_Neph1-Nephrin proteins bind the Par3-Par6-atypical protein kinase C (aPKC) complex to regulate podocyte cell polarity 18922891_in addition to regulating cell polarization processes, Par6 is an inducer of cell proliferation in breast epithelial cells. 19617897_Ect2 and PKCiota are genetically and functionally linked in NSCLC, acting to coordinately drive tumor cell proliferation and invasion through formation of an oncogenic PKCiota-Par6alpha-Ect2 complex. 19667198_the TGFbeta-Par6 polarity pathway has a role in breast cancer progression 20719959_Par6alpha controls centrosome organization through its association with the dynactin subunit p150(Glued). 21170030_Data show that DDR1 coordinates the Par3/Par6 cell-polarity complex through its carboxy terminus, binding PDZ domains in Par3 and Par6. 21549621_Data indicate that both tumor focality and Par3/Par6/atypical protein kinase C (APKC) expression were significantly associated with tumor recurrence. 22339630_Pak6 is a binding partner and a outatuve effector protein for the atypical rho GTPase cdc42 homologous protein. 23249950_Atypical protein kinase C kinase activity, as well as an association with PAR6, were found to be important for PAR6 phosphorylation. 23341197_Par6 negatively regulates trophoblast fusion via its roles on tight junctions and cytoskeleton dynamics and provide novel insight into the contribution of this polarity marker in altered trophoblast cell fusion typical of preeclampsia. 23439680_Morg1 facilitates Par6-aPKC binding to Crb3 for definition of apical identity of epithelial cells. 23762244_BDNF can regulate formation of functional synapses by increasing the expression of the RhoA inhibitors, Par6C and Rnd3. 25756394_TGFbeta induced Par6 phosphorylation on Ser345 and its recruitment to the leading edge of the membrane ruffle in migrating PC-3U cells, where it colocalised with aPKCzeta. The p-Par6-aPKCzeta complex is important for cell migration and invasion. 26050620_Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition 27624926_We first demonstrate that Hook2 is essential for the polarized Golgi re-orientation towards the migration front. Depletion of Hook2 results in a decrease of PAR6alpha at the centrosome during cell migration, while overexpression of Hook2 in cells induced the formation of aggresomes with the recruitment of PAR6alpha, aPKC and PAR3 28590507_Roles of partitioning-defective protein 6 (Par6) and its complexes in the proliferation, migration and invasion of cancer cells 29696344_Studies indicate that the PAR3-PAR6-aPKC complex are important for the establishment of neuronal polarity [Review]. 29992488_In T84 cells, overexpression of Par-6 causes intestinal barrier dysfunction. Lipopolysaccharide (LPS)-induced intestinal epithelial barrier dysfunction and increase in Par-6 expression was prevented by AhR activation. 30408122_In Par6A knockout cells, we find that active Cdc42 is more mobile at the apical membrane compared to control cells and that wild type Cdc42 is more diffusely localized throughout the cell, indicating that Par6A is required to restrict Cdc42 signaling. 31914615_The pathological analysis confirmed the correlation between Par6 expression and the prognosis in human glioma tissues, suggesting the regulation of Par6 expression regulates glioma tumorigenesis and progression. 32580209_A polybasic domain in aPKC mediates Par6-dependent control of membrane targeting and kinase activity. 35379775_Partitioning defective 6 homolog alpha (PARD6A) promotes epithelial-mesenchymal transition via integrin beta1-ILK-SNAIL1 pathway in ovarian cancer. | ENSMUSG00000005699 | Pard6a | 32.830816 | 0.6873961 | -0.540786397 | 0.46787878 | 1.311773e+00 | 2.520738e-01 | No | Yes | 28.092193 | 6.591789 | 39.742934 | 8.921546 | ||
| ENSG00000102984 | 55565 | ZNF821 | protein_coding | O75541 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;Coiled coil;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a protein with two C2H2 zinc finger motifs and a score-and-three (23)-amino acid peptide repeat (STPR) domain. The STPR domain of the encoded protein binds to double stranded DNA and may also contain a nuclear localization signal, suggesting that this protein interacts with chromosomal DNA. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:55565; | nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000031728 | Zfp821 | 191.769635 | 0.9428481 | -0.084902791 | 0.20353891 | 1.699661e-01 | 6.801419e-01 | No | Yes | 189.184828 | 18.423247 | 202.953884 | 19.672399 | ||
| ENSG00000103021 | 29070 | CCDC113 | protein_coding | Q9H0I3 | FUNCTION: Component of centriolar satellites contributing to primary cilium formation. {ECO:0000269|PubMed:25074808}. | Alternative splicing;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Reference proteome | hsa:29070; | axoneme [GO:0005930]; centriolar satellite [GO:0034451]; ciliary basal body [GO:0036064]; protein-containing complex [GO:0032991]; cilium assembly [GO:0060271] | Mouse_homologues 17971504_Ccdc113 is most abundantly expressed in tissues rich in highly ciliated cells, such as olfactory sensory neurons, and is predicted to be important to cilia. | ENSMUSG00000036598 | Ccdc113 | 370.870181 | 0.8836224 | -0.178498038 | 0.15105320 | 1.397156e+00 | 2.372003e-01 | No | Yes | 421.804575 | 50.981505 | 465.481475 | 56.211362 | |||
| ENSG00000103227 | 64788 | LMF1 | protein_coding | Q96S06 | FUNCTION: Involved in the maturation of specific proteins in the endoplasmic reticulum. Required for maturation and transport of active lipoprotein lipase (LPL) through the secretory pathway. Each LMF1 molecule chaperones 50 or more molecules of LPL. {ECO:0000250|UniProtKB:Q3U3R4, ECO:0000269|PubMed:24909692}. | Alternative splicing;Chaperone;Endoplasmic reticulum;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:64788; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; protein maturation [GO:0051604]; triglyceride metabolic process [GO:0006641] | 19471043_Results show that cotransfection of LPL with wild-type Lmf1 restores its ability to support normal lipase maturation. 19820022_Second novel pathogenic mutation in LMF1 gene in a patient with severe hypertriglyceridemia. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24909692_Thus we provide evidence for the critical role of the N-terminus of LMF1 for the maturation of LPL and relevant ratio of chaperone to substrate. 25817768_Our results suggest that LMF1 mutations are involved in a substantial proportion of cases with severe primary hypertriglyceridemia, putting together the moderate-aggressive effect of rare mutations with polymorphisms classically associated with this disease. 28534127_Triglyceride-raising variant alleles of the LMF1 encoding lipase maturation factor 1, associated with clinical Cardiovascular endpoints. 29910226_Compound heterozygous mutation of LMF1 gene is associated with Hypertriglyceridemia. 30037590_rare genetic variants of LMF1 gene identified in severe hypertriglyceridemia 30068531_LMF1 is needed for secretion of some ER client proteins that require reduction of non-native disulfides during their folding. 30885219_A heterozygous LMF1 nonsense variant was found in a hypertriglyceridemia-acute pancreatitis patient with severe obesity and heavy smoking, highlighting an important interplay between genetic and lifestyle factors in the etiology of hypertriglyceridemia. 34062067_Assessment of Zinc- alpha2 glycoprotein (ZAG) and Lipase Maturation Factor 1 (LMF1) concentration in children with chronic kidney disease. | ENSMUSG00000002279 | Lmf1 | 457.524846 | 0.7718179 | -0.373667578 | 0.12721796 | 8.627131e+00 | 3.311928e-03 | No | Yes | 358.166309 | 42.122940 | 469.167216 | 54.799224 | |||
| ENSG00000103326 | 6650 | CAPN15 | protein_coding | O75808 | Alternative splicing;Hydrolase;Metal-binding;Phosphoprotein;Protease;Reference proteome;Repeat;Thiol protease;Zinc;Zinc-finger | This gene encodes a protein containing zinc-finger-like repeats and a calpain-like protease domain. The encoded protein may function as a transcription factor, RNA-binding protein, or in protein-protein interactions during visual system development. [provided by RefSeq, Jul 2008]. | hsa:6650; | cytoplasm [GO:0005737]; calcium-dependent cysteine-type endopeptidase activity [GO:0004198]; metal ion binding [GO:0046872]; proteolysis [GO:0006508] | 32885237_Biallelic variants in the small optic lobe calpain CAPN15 are associated with congenital eye anomalies, deafness and other neurodevelopmental deficits. 33410501_Biallelic deletion in a minimal CAPN15 intron in siblings with a recognizable syndrome of congenital malformations and developmental delay. | ENSMUSG00000037326 | Capn15 | 2393.789882 | 1.0415226 | 0.058694076 | 0.07156883 | 6.764128e-01 | 4.108247e-01 | 7.522795e-01 | No | Yes | 2160.865619 | 264.672385 | 2093.339864 | 256.333581 | ||
| ENSG00000104133 | 80208 | SPG11 | protein_coding | Q96JI7 | FUNCTION: May play a role in neurite plasticity by maintaining cytoskeleton stability and regulating synaptic vesicle transport. {ECO:0000269|PubMed:24794856}. | Alternative splicing;Amyotrophic lateral sclerosis;Cell projection;Charcot-Marie-Tooth disease;Cytoplasm;Hereditary spastic paraplegia;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a potential transmembrane protein that is phosphorylated upon DNA damage. Defects in this gene are a cause of spastic paraplegia type 11 (SPG11). Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2009]. | hsa:80208; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; dendrite [GO:0030425]; lysosomal membrane [GO:0005765]; nucleolus [GO:0005730]; plasma membrane [GO:0005886]; synapse [GO:0045202]; axo-dendritic transport [GO:0008088]; axon extension [GO:0048675]; chemical synaptic transmission [GO:0007268]; phagosome-lysosome fusion involved in apoptotic cell clearance [GO:0090389]; synaptic vesicle transport [GO:0048489]; walking behavior [GO:0090659] | 17322883_mutations in the SPG11 gene causes spastic paraplegia with thin corpus callosum 18067136_Frameshift, nonsense mutations, and splice mutations in SPG11. Mutations are major cause of autosomal recessive hereditary spastic paraplegia with thin corpus callosum associated with severe motor and cognitive impairment. 18079167_The study reveals the high frequency of SPG11 mutations in patients with HSP, a TCC and cognitive impairment, including in isolated patients, and extends the associated phenotype. 18332254_Autosomal recessive HSP-TCC is a frequent subtype of complicated HSP in Tunisia and is clinically and genetically heterogeneous. SPG11 and SPG15 are the major loci for this entity. 18337587_Mutations on KIAA1840 are frequent in complex autosomal recessive hereditary spastic paraplegia, but they are an infrequent cause of sporadic complex hereditary spastic paraplegia. 18361476_Mutations of the SPG11 gene encoding the spatacsin protein have been identified as a major cause of hereditary spastic paraplegia. 18408091_Genetic and phenotypic data on five patients from two Taiwanese/Chinese families with ARHSP-TCC. 18439221_SPG11 mutations should be suspected in patients with isolated or recessive HSP, thin corpus callosum and mental retardation. 18663179_Loss-of-function SPG11 mutations are the major cause of autosomal recessive hereditary spastic paraparesis with thin corpus callosum in Southern Europe, even in apparently sporadic cases. 18717728_5 new spatacsin mutations were found in complex autosomal recessive hereditary spastic paraplegia:p.C133LfsX154, p.Q1875X, p.K2386QfsX2393,c.2834 + 1G > T & c.6754 + 4insTG. 18835492_This study widens the mutation spectrum of the SPG11 gene and the mutations in the SPG11 gene are also the major causative gene for HSP-TCC in the Chinese Hans. 19040626_Abnormal MRI signal in the region of the forceps minor of the corpus callosum is a characteristic early imaging finding of HSP-TCC with SPG11 mutations. 19084844_ZFYVE26 is the second gene responsible for spastic paraplegia with thinning of the corpus callosum in the Italian population 19087158_This study confirms heterogeneity amongst Italian families with hereditary spastic paraplegia/thin corpus callosum and reports a new mutation predicted to affect splicing in the spatacsin gene. 19105190_While expanding the spectrum of mutations in SPG11, this larger series also corroborated the notion that even within apparently homogeneous population a molecular diagnosis cannot be achieved without full gene sequencing. 19194956_Degeneration of the central retina is a common and previously unrecognized feature in SPG11 related disease. 19196735_Findings expand the mutation spectrum of SPG11 and suggest that SPG11 mutations may occur more frequently in familial than sporadic forms of cHSP without TCC. 19224311_Evidence that parkinsonism may initiate SPG11-linked HSP TCC and that SPG11 may cause juvenile parkinsonism. 19513778_we report three novel and one known heterozygous compound SPG11 mutations in patients with hereditary spastic paraplegia with thin corpus callosum; these are the first cases of genetically confirmed SPG11 mutations in the Korean population. 19917823_phenotype and mutation frequency compared with SPG15 in complicated hereditary spastic paraplegia 20110243_Up to 12 sequence alterations in the spatacsin gene have been identified in unrelated pedigrees with autosomal recessive juvenile amyotrophic lateral sclerosis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20390432_analysis of SPG11 mutations in Asian kindreds and disruption of spatacsin function in the zebrafish 20669327_We identified genetic deficits in spatacsin that were associated with Levodopa responsive parkinsonism with pyramidal signs. 20971220_Data support the importance of SPG11 as a frequent cause for ARHSP-TCC, and expands the clinical SPG11 spectrum. 21035867_Retinal changes, an integral part of SPG11 mutations in this series of patients, are only observed once the paraplegia has become apparent. 21440262_SPG11, the most frequent gene associated with hereditary spastic paraplegia with thin corpus callosum (HSP-TCC) encodes spatacsin, demonstrating the extensive genetic heterogeneity of this condition. 21545838_Spatacsin was strongly expressed in cortical and spinal motor neurons and in embryos. It partially co-localized with multiple organelles, particularly with protein-trafficking vesicles, endoplasmic reticulum and microtubules. 22154821_This study confirmed that SPG11 as a genetic cause of juvenile amyotrophic lateral sclerosis and indicate that SPG11 mutations could be associated with 2 different clinical phenotypes within the same family. 22237444_The analysis shows that the high number of repeated elements in SPG11 together with the presence of recombination hotspots and the high intrinsic instability of the 15q locus all contribute toward making this genomic region more prone to large gene rearrangements. 22696581_mutations result in white and grey matter abnormalities 23121729_This study widens the spectrum of mutations in SPG11 23221952_There was a characteristic gradation in the reduction of microstructural integrity among fiber types and within the CC in patients with the SPG11 mutation 23438842_A novel homozygous nonsense mutation in exon 15 of the SPG11 gene (c.2678G>A; p.W893X) found in two Spanish siblings with a complicated forms of hereditary spastic paraplegia. 23825025_We propose AP-5, SPG15, SPG11 form a coat-like complex, with AP-5 involved in protein sorting, SPG15 facilitating docking of the coat onto membranes by interacting with PI3P via its FYVE domain, and SPG11 (possibly together with SPG15) forming a scaffold. 24085347_SPG11 mutations were identified in autosomal recessive juvenile Amyotrophic lateral sclerosis. 24090761_This study identified novel compound heterozygous mutations in the SPG11 gene of the patients as follows: a nonsense mutation c.6856C>T (p.R2286X) in exon 38 and a deletion mutation c.2863delG (p.Glu955Lysfs*8) in exon 16. 24112408_widespread accumulation of spatacsin observed in pathologic alpha-synuclein-containing inclusions suggests that spatacsin may be involved in the pathogenesis of alpha-synucleinopathies 24315199_We have identified an Hereditary spastic paraplegia patient who inherited the c.5121_5122insAG mutation from his mother and the c.6859C>T mutation from his father 24794856_Study provides evidence that SPG11 is implicated in axonal maintenance and cargo trafficking. 25365221_spastizin and spatacsin were essential components for the initiation of lysosomal tubulation. Together, these results link dysfunction of the autophagy/lysosomal biogenesis machinery to neurodegeneration. 25769290_SPG11 mutation has been identified in a Turkish familial hypobetalipoproteinemia family with hereditary spastic paraplegia. 26003865_novel compound heterozygous mutations in SPG11 are associated with HSP and lower motor neuron involvement, mild cerebellar signs and dysgenesis of the corpus callosum 26374131_SPG11 and CYP7B1 were the most common cause of autosomal recessive hereditary spastic paraplegia in Greece. A novel variant in SPG11, which led to disease with later onset was identified in the Greek population. 26556829_SPG11 is the causative gene of a wide spectrum of clinical features, including autosomal recessive axonal Charcot-Marie-Tooth disease. 1 26671123_a novel homozygous mutation in the splice site donor of intron 30 (c.5866+1G>A) in consanguineous Japanese SPG11 siblings showing late-onset spastic paraplegia (whole-exome sequencing) 27071356_Screening of large cohorts of hereditary spastic paraplegia (HSP) patients identified 83 alleles with 'small' mutations and 13 alleles with large genomic rearrangements. These findings widen the spectra of mutations and mutational mechanisms in SPG11, underscore the pivotal role played by Alu elements, and are of high diagnostic relevance for a wide spectrum of clinical phenotypes including the most frequent form of HSP. 27217339_SPG11 defects were found to be by far the commonest cause of complex hereditary spastic paraplegia in the UK, accounting for 30.9% of cases. 27256065_We identified two novel compound heterozygous mutations in SPG11 in 2 affected individuals with autosomal recessive hereditary spastic paraplegia (ARHSP) with thin corpus callosum 27544499_This study identified novel compound heterozygous mutations in SPG11 in a complex hereditary spastic paraplegia family with thin corpus callosum and severe axonal sensory-motor polyneuropathy as a late manifestation of the disease. 27900367_These hereditary spastic paraplegia patients are compound heterozygous for variants in the SPG11 gene, including the paternally inherited c.6856C>T (p.Arg2286 *) variant and the novel maternally inherited c.2316+5G>A splice-donor region variant. 28681766_Compound heterozygous mutations of the SPG11 gene were identified in the index patient and her younger brother, while the parents were carriers 28933964_SPG11 was suspected to be the most common subtype of autosomal recessive hereditary spastic paraplegia in China, whereas SPG15, SPG5 or SPG7 are rare. The core symptoms of Chinese SPG11 patients showed no difference when compared to SPG11 in western countries, and clinical heterogeneity also existed in our SPG11 patients. 29732542_increased levels of homocarnosine do not seem to be a biomarker for SPG11 in our patients 29946510_SPG11-related hereditary spastic paraplegia is characterized by selective neuronal vulnerability, in which a precocious and widespread white matter involvement is later followed by a restricted but clearly progressive grey matter degeneration. 30081747_ZFYVE26 and SPG11 are differently involved in autophagy and endocytosis. 31281085_Whereas, a previously reported variant c.5769delT (p.Ser1923Argfs*28) in the SPG11 gene was identified in family B manifesting clinical features of SPG11 in 3 affected individuals 31900114_Investigated spastic paraplegia type 11 (SPG11) mutations in Chinese patients with autosomal recessive hereditary spastic paraplegia. 32007496_Two types of recessive hereditary spastic paraplegia in Roma patients in compound heterozygous state; no ethnically prevalent variant found. 32007754_A novel variant in the spatacsin gene causing SPG11 in a Malian family. 32040826_Identification of a Mutation in SPG11 in an Iranian Patient with Spastic Paraplegia and Ears of the Lynx Sign. 32383541_Description of combined ARHSP/JALS phenotype in some patients with SPG11 mutations. 32593884_Hereditary spastic paraplegia type 11 (SPG11) is associated with obesity and hypothalamic damage. 32671691_Phenotypic and genotypic features of patients diagnosed with ALS in the city of Sakarya, Turkey. 33581793_Hereditary spastic paraplegia type 11: Clinicogenetic lessons from 339 patients. 34031922_Transactivation response DNA-binding protein of 43 kDa proteinopathy and lysosomal abnormalities in spastic paraplegia type 11. 34140036_Circ-SPG11 knockdown hampers IL-1beta-induced osteoarthritis progression via targeting miR-337-3p/ADAMTS5. | ENSMUSG00000033396 | Spg11 | 987.460847 | 1.2275873 | 0.295825636 | 0.09405398 | 9.974348e+00 | 1.587362e-03 | 4.423712e-02 | No | Yes | 700.129536 | 82.120137 | 573.024977 | 67.217727 | |
| ENSG00000104228 | 23087 | TRIM35 | protein_coding | Q9UPQ4 | FUNCTION: E3 ubiquitin-protein ligase that participates in multiple biological processes including cell death, glucose metabolism, and in particular, the innate immune response. Mediates 'Lys-63'-linked polyubiquitination of TRAF3 thereby promoting type I interferon production via DDX58/RIG-I signaling pathway (PubMed:32562145). Can also catalyze 'Lys-48'-linked polyubiquitination and proteasomal degradation of viral proteins such as influenza virus PB2 (PubMed:32562145). Acts as a negative feedback regulator of TLR7- and TLR9-triggered signaling. Mechanistically, promotes the 'Lys-48'-linked ubiquitination of IRF7 and induces its degradation via a proteasome-dependent pathway (PubMed:25907537). Reduces FGFR1-dependent tyrosine phosphorylation of PKM, inhibiting PKM-dependent lactate production, glucose metabolism, and cell growth (PubMed:25263439). {ECO:0000269|PubMed:25263439, ECO:0000269|PubMed:25907537, ECO:0000269|PubMed:32562145}. | Acetylation;Alternative splicing;Apoptosis;Coiled coil;Cytoplasm;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The function of this protein has not been identified. [provided by RefSeq, Jul 2008]. | hsa:23087; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; apoptotic process [GO:0006915]; innate immune response [GO:0045087]; negative regulation of mitotic cell cycle [GO:0045930]; positive regulation of apoptotic process [GO:0043065]; protein ubiquitination [GO:0016567]; suppression of viral release by host [GO:0044790] | 14662771_gene located on chromosome 8p21, a region implicated in leukemias and solid tumors; enforced expression inhibited cell growth, clonogenicity, and tumorigenicity; conceivable HLS5 is one of the tumor suppressor genes thought to reside at the 8p21 locus 25263439_TRIM35 regulates the Warburg effect and tumorigenicity through interaction with PKM2 in hepatocellular carcinoma. 25576919_Data suggest that pyruvate kinase isoform M2 (PKM2)/tripartite motif-containing 35 (TRIM35) expression could be a biomarker for the prognosis of hepatocellular carcinoma (HCC) and target for cancer therapy. 28394882_Authors subsequently investigated whether miR-4417 and TRIM35 regulate HCC cell proliferation and apoptosis through PKM2 Y105 phosphorylation, and the results supported our speculation that miR-4417 targets TRIM35 and regulates the Y105 phosphorylation of PKM2 to promote hepatocarcinogenesis. 32293015_Tripartite motif containing 35 contributes to the proliferation, migration, and invasion of lung cancer cells in vitro and in vivo. 34124276_Suppression of DLBCL Progression by the E3 Ligase Trim35 Is Mediated by CLOCK Degradation and NK Cell Infiltration. | ENSMUSG00000022043 | Trim35 | 609.299062 | 0.8868607 | -0.173220509 | 0.11124511 | 2.428924e+00 | 1.191146e-01 | 4.588196e-01 | No | Yes | 576.063802 | 43.845916 | 651.215878 | 49.519975 |
| ENSG00000104450 | 6674 | SPAG1 | protein_coding | Q07617 | FUNCTION: May play a role in the cytoplasmic assembly of the ciliary dynein arms (By similarity). May play a role in fertilization. Binds GTP and has GTPase activity. {ECO:0000250, ECO:0000269|PubMed:11517287, ECO:0000269|PubMed:1299558}. | 3D-structure;Alternative splicing;Ciliopathy;Cytoplasm;Fertilization;GTP-binding;Hydrolase;Kartagener syndrome;Nucleotide-binding;Phosphoprotein;Primary ciliary dyskinesia;Reference proteome;Repeat;TPR repeat | The correlation of anti-sperm antibodies with cases of unexplained infertility implicates a role for these antibodies in blocking fertilization. Improved diagnosis and treatment of immunologic infertility, as well as identification of proteins for targeted contraception, are dependent on the identification and characterization of relevant sperm antigens. The protein expressed by this gene is recognized by anti-sperm agglutinating antibodies from an infertile woman. Furthermore, immunization of female rats with the recombinant human protein reduced fertility. This protein localizes to the plasma membrane of germ cells in the testis and to the post-acrosomal plasma membrane of mature spermatozoa. Recombinant polypeptide binds GTP and exhibits GTPase activity. Thus, this protein may regulate GTP signal transduction pathways involved in spermatogenesis and fertilization. Two transcript variants of this gene encode the same protein. [provided by RefSeq, Jul 2008]. | hsa:6674; | chaperone complex [GO:0101031]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dynein axonemal particle [GO:0120293]; GTP binding [GO:0005525]; hydrolase activity [GO:0016787]; axonemal dynein complex assembly [GO:0070286]; single fertilization [GO:0007338] | 12846798_The h-Sp-1 molecule is expressed in sperm and testes and plays a role in fertilization. 16368546_HSD-3.8 (SPAG1), interacts with G-protein beta 1 subunit and activates extracellular signal-regulated kinases 1 and 2 16983343_Immunocytochemical analysis demonstrated colocalization of SPAG1 with microtubules, and their association was confirmed by co-immunoprecipitation; subsequent motility assays further substantiated a potential role of SPAG1 in cancer cell motility 24055112_SPAG1 probably plays a role in the cytoplasmic assembly. 31118266_Demonstrated that a SPAG1 sub-fragment, containing a putative P-loop motif, cannot efficiently bind and hydrolyze GTP in vitro Our data challenge the interpretation of SPAG1 possessing GTPase activity. We propose instead that SPAG1 regulates nucleotide hydrolysis activity of the HSP and RUVBL1/2 partners. 32619574_Bioinformatics analysis of the genes involved in the extension of prostate cancer to adjacent lymph nodes by supervised and unsupervised machine learning methods: The role of SPAG1 and PLEKHF2. 33739091_Optimizing the First TPR Domain of the Human SPAG1 Protein Provides Insight into the HSP70 and HSP90 Binding Properties. 35178554_The role of SPAG1 in the assembly of axonemal dyneins in human airway epithelia. 35227274_Comprehensive analysis of SPAG1 expression as a prognostic and predictive biomarker in acute myeloid leukemia by integrative bioinformatics and clinical validation. | ENSMUSG00000037617 | Spag1 | 289.675468 | 0.8464064 | -0.240577620 | 0.16924690 | 1.969840e+00 | 1.604648e-01 | No | Yes | 228.043462 | 48.939230 | 252.923698 | 54.229737 | ||
| ENSG00000104763 | 427 | ASAH1 | protein_coding | Q13510 | FUNCTION: Lysosomal ceramidase that hydrolyzes sphingolipid ceramides into sphingosine and free fatty acids at acidic pH (PubMed:10610716, PubMed:7744740, PubMed:15655246, PubMed:11451951). Ceramides, sphingosine, and its phosphorylated form sphingosine-1-phosphate are bioactive lipids that mediate cellular signaling pathways regulating several biological processes including cell proliferation, apoptosis and differentiation (PubMed:10610716). Has a higher catalytic efficiency towards C12-ceramides versus other ceramides (PubMed:7744740, PubMed:15655246). Also catalyzes the reverse reaction allowing the synthesis of ceramides from fatty acids and sphingosine (PubMed:12764132, PubMed:12815059). For the reverse synthetic reaction, the natural sphingosine D-erythro isomer is more efficiently utilized as a substrate compared to D-erythro-dihydrosphingosine and D-erythro-phytosphingosine, while the fatty acids with chain lengths of 12 or 14 carbons are the most efficiently used (PubMed:12764132). Has also an N-acylethanolamine hydrolase activity (PubMed:15655246). By regulating the levels of ceramides, sphingosine and sphingosine-1-phosphate in the epidermis, mediates the calcium-induced differentiation of epidermal keratinocytes (PubMed:17713573). Also indirectly regulates tumor necrosis factor/TNF-induced apoptosis (By similarity). By regulating the intracellular balance between ceramides and sphingosine, in adrenocortical cells, probably also acts as a regulator of steroidogenesis (PubMed:22261821). {ECO:0000250|UniProtKB:Q9WV54, ECO:0000269|PubMed:10610716, ECO:0000269|PubMed:11451951, ECO:0000269|PubMed:12764132, ECO:0000269|PubMed:12815059, ECO:0000269|PubMed:15655246, ECO:0000269|PubMed:17713573, ECO:0000269|PubMed:22261821, ECO:0000269|PubMed:7744740, ECO:0000303|PubMed:10610716}.; FUNCTION: [Isoform 2]: May directly regulate steroidogenesis by binding the nuclear receptor NR5A1 and negatively regulating its transcriptional activity. {ECO:0000305|PubMed:22927646}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Disease variant;Disulfide bond;Epilepsy;Glycoprotein;Hydrolase;Lipid metabolism;Lysosome;Neurodegeneration;Nucleus;Reference proteome;Secreted;Signal;Sphingolipid metabolism | PATHWAY: Lipid metabolism; sphingolipid metabolism. {ECO:0000269|PubMed:10610716, ECO:0000269|PubMed:7744740}. | This gene encodes a member of the acid ceramidase family of proteins. Alternative splicing results in multiple transcript variants, at least one of which encodes a preproprotein that is proteolytically processed. Processing of this preproprotein generates alpha and beta subunits that heterodimerize to form the mature lysosomal enzyme, which catalyzes the degradation of ceramide into sphingosine and free fatty acid. This enzyme is overexpressed in multiple human cancers and may play a role in cancer progression. Mutations in this gene are associated with the lysosomal storage disorder, Farber lipogranulomatosis, and a neuromuscular disorder, spinal muscular atrophy with progressive myoclonic epilepsy. [provided by RefSeq, Oct 2015]. | hsa:427; | extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; ficolin-1-rich granule lumen [GO:1904813]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; nucleus [GO:0005634]; tertiary granule lumen [GO:1904724]; ceramidase activity [GO:0102121]; fatty acid amide hydrolase activity [GO:0017064]; hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds [GO:0016810]; hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides [GO:0016811]; N-acylsphingosine amidohydrolase activity [GO:0017040]; nuclear receptor binding [GO:0016922]; transcription corepressor activity [GO:0003714]; cellular response to tumor necrosis factor [GO:0071356]; ceramide biosynthetic process [GO:0046513]; ceramide catabolic process [GO:0046514]; fatty acid metabolic process [GO:0006631]; keratinocyte differentiation [GO:0030216]; negative regulation of nucleic acid-templated transcription [GO:1903507]; regulation of programmed necrotic cell death [GO:0062098]; regulation of steroid biosynthetic process [GO:0050810]; sphingosine biosynthetic process [GO:0046512] | 12764132_acid ceramidase has a central role in sphingolipid metabolism 12815059_biochemistry of acid ceramidase reaction with acid sphingomyelinase 15088070_p53-dependent expression of acid ceramidase and blockage of A-SMase activation play pivotal roles in protection from gamma-radiation of cells with endogenous functional p53. 16500425_These results provide the first characterization of the Acid Ceremidase promoter from any species and demonstrate that Kruppel-like factor 6 (KLF6) is one transcription factor involved in the regulation of AC gene expression. 17713573_upregulation of haCER1 and AC mediates the Ca2+(o)-induced growth arrest and differentiation of keratinocytes by generating sphingosine and its phosphate 17881906_AC is a critical regulator of prostate cancer progression by affecting not only tumor cell proliferation and migration but also responses to drug therapy 18245333_Positive selection is possibly operating on ASAH1 in the modern human population. 18477771_acid ceramidase was constitutively overexpressed in leukemic LGLs and its inhibition induced apoptosis of leukemic LGLs 19298866_These data identify the ACTH/cAMP signaling pathway and CREB as transcriptional regulators of the ASAH1 gene in the human adrenal cortex. 19905902_high ASAH1 expression correlates with grading & estrogen receptor (ER) status in breast cancer; high ASAH1 expression was associated with larger tumor size; a better prognosis of patients with higher ASAH1 expression in ER-positive subgroup was detected 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20520628_Both acidic ceramidase (aCDase) and neutral ceramidase (nCDase) activities declined after low- and high-UVB, but returned to normal only in low-UVB cells 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20871013_Acid ceramidase (aCDase) is responsible for ceramide degradation within mammalian cells. An inherited deficiency of aCDase activity results in Farber disease. 21335555_cells deficient in acid ceramidase (aCDase) also exhibited defects in CCL5 induction, whereas cells deficient in sphingosine kinase-1 and -2 exhibited higher levels of CCL5. 21375364_This study supports that the ASAH1 gene may be a potential candidate gene for schizophrenia in Han Chinese subjects. 21493710_a mechanism through which genistein promotes sphingolipid metabolism and support a role for ASAH1 in breast cancer cell growth. 21504271_the level of AC did not correlate with the sensitivity of HNSCC cells to Fas-induced apoptosis. 21700700_down-regulation of aCDase alone or in combination with DTIC may represent a useful tool in the treatment of metastatic melanoma. 21846728_Identification of cystatin SA as a novel inhibitor of acid ceramidase. 21893389_A report of two siblings with Farber disease who carry a novel V97G ASAH1 mutation with the parents and a sister being asymptomatic carriers. 22261821_ASAH1 as a pivotal regulator of steroidogenic capacity in the human adrenal cortex. 22322590_Acid ceramidase, through sphingosine 1-phosphate, promotes an invasive phenotype in prostate cancer by causing overexpression and secretion of cathepsin B through activation and nuclear expression of Ets1. 22515519_ASAH1 inhibition synergistically sensitizes lung cancer cells resistant to the antiproliferative effect of choline kinase alpha inhibitors. 22703880_Our results reveal a wide phenotypic spectrum associated with ASAH1 mutations in spinal muscular atrophy associated with progressive myoclonal epilepsy. 22927646_results demonstrate that ASAH1 is a novel coregulatory protein that represses SF-1 function by directly binding to the receptor on SF-1 target gene promoters and identify a key role for nuclear lipid metabolism in regulating gene transcription 23423838_These observations confirm ASAH1 as a therapeutic target in advanced and chemoresistant forms of prostate cancer 23518908_Acid ceramidase is a prognostic factor in epithelial ovarian cancer 23707712_This work unravels for the first time the mutations underlying the neonatal form of Farber disease and represents the first report of a large deletion identified in the ASAH1 gene. 23777806_Depletion of both Akt2 and ASAH1 is much more potent than depleting each alone at inhibiting neoplastic cell viability/proliferation and invasion. 24091326_Immunohistochemical analysis of human prostate cancer tissues revealed higher levels of ASAH1 after radiotherapy failure. 24098536_Acid ceramidase promotes nuclear export of PTEN through sphingosine 1-phosphate mediated Akt signaling. 24355074_novel ASAH1 mutations affecting polypyrimidine tract deletion, and exon skipping and resulting in Farber lipogranulomatosis 25131496_high ASAH1 expression is generally associated with an improved prognosis in invasive breast cancer independent of adjuvant treatment and could also be valuable as prognostic factor for pre-invasive DCIS. 25888580_Data suggest up-regulation of ASAH1 activity by androgen in androgen-sensitive prostate cancer cells (not other cancer cells) is due to prolonged stability of ASAH1 by androgen-stimulated induction of USP2 (ubiquitin specific peptidase 2) expression. 26553872_AC-controlled sphingolipid metabolism may play an important role in the control of melanoma proliferation 26687835_Genetic or pharmacological acid ceramidase inhibition promotes cisplatin cytotoxicity in head and neck tumor cells. 26945816_Our findings indicate that hypomorphic mutations in ASAH1 may result in an osteoarticular phenotype with a juvenile phase resembling rheumatoid arthritis that evolves to osteolysis as the final stage in the absence of neurologic signs. This observation delineates a novel type of recessively inherited peripheral osteolysis and illustrates the long-term skeletal manifestations of acid ceramidase deficiency (Farber's disease 27026573_This study describes for the first time the association between ASAH1 variants and an adult SMA phenotype with no myoclonic epilepsy nor death in early age, thus expanding the phenotypic spectrum of ASAH1-related SMA. 27411168_We report an atypical presentation of Farber disease with her pathology and associated genetic defect. This case expands the phenotypic spectrum of Farber disease to include novel mutations of ASAH1, which pose a diagnostic challenge. 27650050_The present report describes a 9-year-old girl with novel clinical phenotype of a patient with polyarticular arthritis followed by symptoms of SMA due to acid ceramidase deficiency. Whole exome sequencing identified compound heterozygous pathogenic mutation in the N-acylsphingosine amidohydrolase 1 gene. 27825124_Acid ceramidase plays a critical role in acute myeloid leukemia cell survival via regulation of both sphingolipid levels and Mcl-1. 28251733_we describe an individual with a sporadic atypical spinal muscular atrophy, in whom clinical DNA sequencing reported one pathogenic ASAH1 mutation .Transcriptome sequencing on patient leukocytes identified a highly significant and atypical ASAH1 isoform not explained by c.458A>G(p<10(-16) ). 28785021_Acid ceramidase ablation blocks cell cycle progression and accelerates senescence. Importantly, ASAH1-null cells also lose the ability to form cancer-initiating cells and to undergo self-renewal, which is suggestive of a key role for acid ceramidase in maintaining malignancy and self-renewal of invasive melanoma cells. 28905881_ASAH1 variant Leu401Pro co-segregates with keloid phenotype in a large Yoruba family. 29259036_results demonstrate a tissue-specific role for AC in regulating neutrophilic inflammation and cytokine production. 29278706_Collectively, our data show the novel discovery of anti-inflammatory and anti-apoptotic effects of acid ceramidase in host cells exposed to periodontal bacteria, and the attenuation of the expression of host-protective acid ceramidase in periodontal lesions. 29320752_The expression of ASAH1 was associated significantly with lymph nodes metastasis, indicating that ASAH1 may serve as a biomarker to predict patients' lymph nodes status in breast cancers. 29518574_AC may be implicated in the response of rectal cancer to Neoadjuvant chemoradiotherapy. We propose its further assessment as a novel potential biomarker and therapeutic target. 29692406_Autocleavage of ASAH1 triggers a conformational change exposing a hydrophobic channel leading to the active site. 30254208_We also identified ASAH1 as a new target of MITF, thereby involving MITF in the regulation of sphingolipid metabolism. Together, our findings provide new cues to the mechanisms underlying the phenotypic plasticity of melanoma cells and identify new anti-metastatic targets. 30413652_induction of ASAH1 could rescue retinal cells from oxidative stress by hydrolyzing excess Cers 30815900_Mutational analyses revealed new mutations in the ASAH1 gene and a broad diversity of phenotypes without a genotype/phenotype correlation in Farber disease patients. 30988134_The data suggest that endoreplication occurs independent of ASAH1 while neosis is ASAH1-dependent in both prostate and lung cancer cells. 31505435_Pediatric ependymoma: GNAO1, ASAH1, IMMT and IPO7 protein expression and 5-year prognosis correlation. 32111095_Acid Ceramidase Depletion Impairs Neuronal Survival and Induces Morphological Defects in Neurites Associated with Altered Gene Transcription and Sphingolipid Content. 32449975_ASAH1 pathogenic variants associated with acid ceramidase deficiency: Farber disease and spinal muscular atrophy with progressive myoclonic epilepsy. 32569620_The interaction of ASAH1 and NGF gene involving in neurotrophin signaling pathway contributes to schizophrenia susceptibility and psychopathology. 32627310_Spinal muscular atrophy and Farber disease due to ASAH1 variants: A case report. 32706452_Farber disease in a patient from China. 32875576_ASAH1-related disorders: Description of 15 novel pediatric patients and expansion of the clinical phenotype. 33334013_Targeting Acid Ceramidase to Improve the Radiosensitivity of Rectal Cancer. 33766731_N-acylsphingosine amidohydrolase 1 promotes melanoma growth and metastasis by suppressing peroxisome biogenesis-induced ROS production. 34102611_Acid ceramidase promotes senescent cell survival. | ENSMUSG00000031591 | Asah1 | 1494.036352 | 0.9455522 | -0.080771052 | 0.07956413 | 1.033713e+00 | 3.092881e-01 | 6.750603e-01 | No | Yes | 1296.088172 | 173.391099 | 1376.572658 | 184.057077 |
| ENSG00000104870 | 2217 | FCGRT | protein_coding | P55899 | FUNCTION: Cell surface receptor that transfers passive humoral immunity from the mother to the newborn. Binds to the Fc region of monomeric immunoglobulin gamma and mediates its selective uptake from milk (PubMed:7964511, PubMed:10933786). IgG in the milk is bound at the apical surface of the intestinal epithelium. The resultant FcRn-IgG complexes are transcytosed across the intestinal epithelium and IgG is released from FcRn into blood or tissue fluids. Throughout life, contributes to effective humoral immunity by recycling IgG and extending its half-life in the circulation. Mechanistically, monomeric IgG binding to FcRn in acidic endosomes of endothelial and hematopoietic cells recycles IgG to the cell surface where it is released into the circulation (PubMed:10998088). In addition of IgG, regulates homeostasis of the other most abundant circulating protein albumin/ALB (PubMed:24469444, PubMed:28330995). {ECO:0000250|UniProtKB:P13599, ECO:0000269|PubMed:10933786, ECO:0000269|PubMed:10998088, ECO:0000269|PubMed:24469444, ECO:0000269|PubMed:28330995, ECO:0000269|PubMed:7964511}.; FUNCTION: (Microbial infection) Acts as an uncoating receptor for a panel of echoviruses including Echovirus 5, 6, 7, 9, 11, 13, 25 and 29. {ECO:0000269|PubMed:30808762, ECO:0000269|PubMed:31104841}. | 3D-structure;Cell membrane;Direct protein sequencing;Disulfide bond;Endosome;Glycoprotein;IgG-binding protein;Immunoglobulin domain;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a receptor that binds the Fc region of monomeric immunoglobulin G. The encoded protein transfers immunoglobulin G antibodies from mother to fetus across the placenta. This protein also binds immunoglobulin G to protect the antibody from degradation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2009]. | hsa:2217; | endosome membrane [GO:0010008]; external side of plasma membrane [GO:0009897]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; beta-2-microglobulin binding [GO:0030881]; IgG binding [GO:0019864]; IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor [GO:0002416] | 11717196_Human FcRn binds selectively to human, rabbit and guinea pig IgG but not significantly to rat, bovine, sheep or mouse IgG (except for weak binding to mouse IgG2b). 12006623_Assembly of the FcRn alpha-chain with beta(2)microglobulin is important for both transport of FcRn from the ER to the cell surface and efficient pH-dependent IgG binding. 12023961_Functional reconstitution in Madin-Darby canine kidney cells requires co-expressed human beta 2-microglobulin 12144784_although a secreted soluble form of human FcRn does not dimerize, the membrane-anchored receptor can form both non-covalent and covalent dimers; dimerization of human FcRn occurs in the absence of its ligand, IgG 12538789_Expression of FcRn is demonstrated along the human fetal intestine and in a human nonmalignant fetal intestinal epithelial cell line (H4), which by location indicates that FcRn could play a role in the uptake and transport of IgG in the human fetus. 12972260_data show that it is possible to confer binding of mouse immunoglobulin G on human FcRn by mutagenesis of selected residues; observations are of direct relevance to understanding the molecular nature of the human FcRn-IgG interaction 14764666_Analysis of the dynamics and properties of trafficking of FcRn in live microvascular endothelial cells shows that the primary site at which FcRn sorts IgGs for either salvage or lysosomal degradation is the sorting endosome. 14767057_strong cell surface polarity displayed by hFcRn results from dominant basolateral sorting by motifs in the cytoplasmic tail that nonetheless allows for a cycle of bidirectional transcytosis 15258288_FCRN is involved in IgG exocytosis. 15644205_residues encompassing and extending away from the interaction site on the alpha2 helix of FcRn play a significant and most likely indirect role in FcRn-IgG interactions 15654966_expression of a functional FcRn in normal human epidermal keratinocytes. 15689494_FcRn leaves sorting endosomes in Rab4(+)Rab11(+) or Rab11(+) compartments 16229888_These transport and localization data are in accordance with efficient hFcRn-mediated apical IgG recycling and basolateral directed IgG transcytosis in placental trophoblasts. 16549777_FcRn binds IgG and albumin, salvages both from a degradative fate, and maintains their physiologic concentrations 16805790_a variable number of tandem repeats promoter polymorphism influences the expression of the FcRn receptor, leading to different IgG-binding capacities 16849638_FcRn fulfills a major role in IgG-mediated phagocytosis 17046328_FcRn-mediated recycling is a major contributor to the high endogenous concentrations of IgG and serum albumin, two important plasma proteins. 17048273_Our results show clear evidence that the conserved H166 is a key player in the FcRn-albumin interaction. 17384151_Elucidation of intracellular recycling pathways leading to exocytosis of FCRN was facilitated by using multifocal plane micoscopy. 17674040_Neonatal Fc receptor (FcRn) was expressed in various cells of the human skin (including keratinocytes, melanocytes, and histiocytes). 17703228_This review summarizes FcRn biology. 17709515_These data provide the first evidence that NF-kappaB signaling via intronic sequences regulates FcRn expression and function. 18003977_These results suggest a novel mechanism for regulation of IgG transport by calmodulin-dependent sorting of FcRn and its cargo away from a degradative pathway and into a bidirectional transcytotic route. 18566411_JAK/STAT-1 signaling pathway was necessary and sufficient to mediate the down-regulation of FcRn gene expression by IFN-gamma 18599440_A previously undescribed role for FcRn in mediating the presentation of antigens by dendritic cells when antigens are present as a complex with antibody, is shown. 18637944_The impact of two free cysteine residues (C48 and C251) of the FcRn heavy chain on the overall structure and function of soluble human FcRn is explored. 18684948_Intracellular trafficking of FcRn is regulated by its intrinsic sorting information and/or an interaction with major histocompatibility (MHC) class II invariant chain (Ii). 18843053_Results indicate that FcRn-dependent internalization of IgG may be important not only in cells taking up IgG from an extracellular acidic space, but also in endothelial cells participating in homeostatic regulation of circulating IgG levels. 19164298_N-glycans in FcRn contribute significantly to the steady-state membrane distribution and direction of IgG transport in polarized epithelia. 19362735_The role of hFcRn in IgG transport and trafficking in syncytiotrophoblasts cultures in vitro. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19451275_a single structurally and functionally heterogeneous recycling endosome compartment that traffics FcRn to both cell surfaces while discriminating between recycling and transcytosis pathways polarized in their direction of transport 19462839_It plays a role in intracellular IGG transfer and immune surveillance. (review) 19772792_Observational study of gene-disease association. (HuGE Navigator) 19772792_VNTR polymorphisms within the FCGRT promoter are not associated with LN in the Chinese population. 20006594_A recombinant truncated HSA variant, HSA(Bartin), does not interact with FcRn, which gives a molecular explanation for the low serum levels. 20018855_No binding of albumin was observed at physiological pH to neonatal Fc receptor. At acidic pH, a 100-fold difference in binding affinity was observed. 20083659_Affinities to FcRn of clinically used therapeutic proteins are closely correlated with the serum half-lives reported from clinical studies, and suggest an important role of FcRn in regulating the serum half-lives of the therapeutic proteins. 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20452034_Observational study of gene-disease association. (HuGE Navigator) 20452034_promoter polymorphism is not associted with the rate of maternal-fetal IgG transfer 20592032_the x-ray crystal structure of a representative monomeric peptide in complex with human FcRn 20627700_influence of FcRn expression on disease phenotype and the catabolism of therapeutically administered intravenous immunoglobulins in 28 patients with common variable immunodeficiency 20930418_Thirty-three genetic variations of FCGRT, including 17 novel ones, were found. 21256596_Methionine (Met) oxidation can result in a significant reduction of the serum circulation half-life and the magnitude of the change correlates well with the extent of Met oxidation and changes in FcRn binding affinities. 21368166_These studies demonstrate that FcRn-mediated transport is a mechanism by which IgG can act locally in the female genital tract in immune surveillance and in host defense against sexually transmitted diseases. 21690327_FcRn transgene blockade is a primary contributing factor toward reduction in arthritis severity; engineering of antibody Fc regions to generate potent FcRn blockers holds promise for the therapy of antibody-mediated autoimmunity in experimental arthritis. 22215085_Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor 22453095_This mAb panel provides a powerful resource for probing the biology of human FcRn and for the evaluation of therapeutic FcRn blockade strategies. 22570488_Serum half-life of IgG is controlled by the neonatal Fc receptor (FcRn) that interacts with the IgG Fc region and may be increased or decreased as a function of altered FcRn binding. 23220220_Human FcRn was visualized in epithelial cells of Tg276 mice, but low serum hIgG levels were obtained 23286945_genetic polymorphism is associated with the efficiency of Ig replacement therapy in common variable immunodeficiency 23384837_Data indicate that Fc-neonatal Fc receptor (FcRn) interaction is pH dependent. 23741050_Only the unbound receptor or FcRn bound to monomeric IgG is sorted into recycling tubules emerging from early endosomes. 23751752_The FCGRT promoter VNTR may influence mAbs' distribution in the body. CNV of FCGRT cannot be used as a relevant pharmacogenetic marker because of its low frequency. 23765230_Analytical FcRn chromatography allows differentiation of IgG samples and variants by peak pattern and retention time profile. 23917469_Studies indicate that high levels of endogenous IgG can compete with the monoclonal antibodies (mAbs) for binding to the neonatal Fc receptor (FcRn). 24057047_Molecular dynamic simulations of human FcRn-Fc binding structures proposed that the protein-protein binding interface is composed of three subsites. 24072267_In intestine, there was an increasing proximal-distal gradient of mucosal FcRn mRNA and protein expression. 24278022_The Neonatal Fc receptor (FcRn) enhances human immunodeficiency virus type 1 (HIV-1) transcytosis across epithelial cells. 24469444_analysis of neonatal Fc receptor-based recycling mechanisms through identification of the human Fc interaction with human FcRn 24550358_FcRn has the potential to interact with IgG-Fc domains in the ciliary epithelium and retinal and choroidal vasculature, which might affect the half-life and distribution of intravitreally injected Fc-carrying molecules. 24652290_Extending serum half-life of albumin by engineering neonatal Fc receptor (FcRn) binding. 24764301_A cluster of conserved tryptophan residues of FcRn is required for binding to albumin and anti-FcRn albumin blocking antibodies. 24802048_Characterization and screening of IgG binding to the neonatal Fc receptor. 25030041_These results suggest that hFcRn Tgm are a valuable and useful tool for pharmacokinetic screening of mAbs and Fc-fusion proteins in the preclinical stage. 25344603_domain I and III of albumin required for optimal pH-dependent binding to the neonatal Fc receptor 25652137_These data indicate that human FcRn facilitates the transepithelial transport of IgE in the form of IgG anti-IgE/IgE ICs. 25658443_The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. 25823782_none of the three functional polymorphisms in FcgammaR genes explored here, the FCGR3A F158V and FCGR2B I232T nsSNPs and the VNTR in FCGRT, showed an association with the response to TNFi in patients with rheumatoid arthritis 26252948_analysis of binding motifs in the hFCGRT promoter that interact with their corresponding (Sp1, Sp2, Sp3, c-Fos, c-Jun, YY1, and C/EBPbeta or C/EBPdelta) transcription factors (TFs) suggests their involvement in regulation of human FCGRT gene expression 26254986_FcRn binding activity of a large set of Fc-fusion samples after thermal stress, was investigated. 26260795_Critical Role of the Neonatal Fc Receptor (FcRn) in the Pathogenic Action of Antimitochondrial Autoantibodies Synergizing with Anti-desmoglein Autoantibodies in Pemphigus Vulgaris. 26337808_Data indicate improved pharmacokinetics through enhanced neonatal Fc receptor (FcRn) interactions were apparent for a complementarity-determining region (CDR) charge-patch normalized monoclonal antibody (mAb) which was affected by non-specific clearance. 26634928_the localization of FcRn alpha-chain in fixed nasal tissue, was studied. 27016466_This review summarizes the main findings on Fc Receptor neonatal biology, function and distribution throughout different tissues. 27469170_The results suggest that regardless of hyperglycemia degree, it decreases FcRn expression in placenta and blood cells and compromises the production and transfer of antibodies from maternal blood to newborns. 27836572_FcRn is involved in transport of aflibercept through REC in vitro. 27974681_we have demonstrated that loss of FcRn expression promotes tumor cell growth and proliferation. Our data support a model in which FcRn-mediated recycling of albumin reduces amino acid availability to fuel metabolic pathways. 28081504_this study shows that FcRn may be associated with the transport and metabolism of IgG in thyrocytes and that transport is independent of IgG type, and that FcRn may be involved in Hashimoto's thyroiditis pathogenesis 28637874_Data suggest that, unlike albumin with low FcRn-binding affinity, albumin with high FcRn-binding affinity (due to genetic variation/genetic engineering) is directed less to lysosomes and more to endosomes, suggestive of FcRn-directed albumin salvage from lysosomal degradation. (FcRn = neonatal Fc receptor) 28760885_these findings establish a novel mechanism of humoral protection in the eye involving FcRn and may facilitate vaccine and therapeutic development for other ocular surface diseases 28991504_Domains distal from the Fc could contribute to FcRn binding of IgG and alter antibody pharmacokinetics. 28991911_An arginine-to-histidine replacement at residue 435 in the binding domain of IgG3 to FcRn increases the transplacental transfer and half-life of malaria-specific IgG3 in young infants and is associated with reduced risk of clinical malaria during infancy. 29302759_Regulation of the Human Fc-Neonatal Receptor alpha-Chain Gene FCGRT by MicroRNA-3181. 29523681_As expected, recombinant factor IX (without albumin fusion) and an FcRn interaction-defective albumin variant localized to the lysosomal compartments of both FcRn-expressing and nonexpressing cells. These results indicate that FcRn-mediated recycling via the albumin moiety is a mechanism for the half-life extension of rIX-FP observed in clinical studies. 29540212_the presence of human serum albumin (HSA) or immunoglobulin G (IgG), which are both protected from intracellular degradation by the interaction with the neonatal Fc receptor (FcRn), results in a decreased amount of internalized fibrinogen. 29745444_this study shows that the IgG transfer from maternal serum to the fetus is positively correlated with FcRn expression in placental tissue throughout gestati 29782488_small molecule binds into a conserved cavity of the heterodimeric, extracellular domain composed of an alpha-chain and beta2-microglobulin 29908223_Down-regulation of FcRn is speculated to play a protective role in Hashimoto's thyroiditis pathogenesis by mainly reducing IgG1 transport in thyrocytes. 30326650_the measurement of FcRn protein may be preferred to FcRn mRNA for quantitative applications. Significant differences were found in FcRn expression in transgenic mice, Swiss Webster mice, and human tissues, which may have implications for the use of mouse models in the assessment of monoclonal antibody disposition, efficacy, and safety. 30808762_We show that loss of expression of FcRn or its binding partner beta 2 microglobulin (b2M) renders cells resistant to infection by a panel of echoviruses at the stage of virus attachment, and that a blocking antibody to b2M inhibits echovirus infection in cell lines and in primary human intestinal epithelial cells 30893823_This is the first time that FcRn expression and mAb transcytosis has been shown in a model of human nasal respiratory epithelium in vitro. 30955709_Placental FcRn expression in endothelial cells and macrophages is analogous to the expression pattern in other organs. FcRn expression suggests an involvement of FcRn in IgG transcytosis and/or participation in recycling/salvaging of maternal IgG present in the fetal circulation. FcRn expression in placental macrophages may account for recycling of monomeric IgG and/or processing and presentation of immune complexes. 31089170_Reduced FcRn-mediated transcytosis of IgG2 due to a missing Glycine in its lower hinge. 31209240_Contribution of DNA methylation to the expression of FCGRT in human liver and myocardium. 31289263_Study shows that human cytomegalovirus US11 inhibits the assembly of FcRn with beta2m and retains FcRn in the endoplasmic reticulum (ER), consequently blocking FcRn trafficking to the endosome. US11 recruits the ubiquitin enzymes Derlin-1, TMEM129 and UbE2J2 to engage FcRn, consequently initiating the dislocation of FcRn from the ER to the cytosol and facilitating its degradation. 32461366_FcRn, but not FcgammaRs, drives maternal-fetal transplacental transport of human IgG antibodies. 32658257_FcRn is a CD32a coreceptor that determines susceptibility to IgG immune complex-driven autoimmunity. 33025844_Functional humanization of immunoglobulin heavy constant gamma 1 Fc domain human FCGRT transgenic mice. 33077645_V Region of IgG Controls the Molecular Properties of the Binding Site for Neonatal Fc Receptor. 33360096_Demonstration of fibrinogen-FcRn binding at acidic pH by means of Fluorescence Correlation Spectroscopy. 34634960_Neonatal Fc receptor expression in lymphoid and myeloid cells in systemic lupus erythematosus. 34704415_Research progress on neonatal Fc receptor and its application. 34753797_An Expanded Genome-Wide Association Study of Fructosamine Levels Identifies RCN3 as a Replicating Locus and Implicates FCGRT as the Effector Transcript. 34996950_Human IgE does not bind to human FcRn. | ENSMUSG00000003420 | Fcgrt | 491.059612 | 0.8486239 | -0.236802755 | 0.14183594 | 2.808419e+00 | 9.377074e-02 | 4.103332e-01 | No | Yes | 454.261741 | 59.975968 | 544.419536 | 71.547668 | |
| ENSG00000105072 | 84167 | C19orf44 | protein_coding | Q9H6X5 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:84167; | ENSMUSG00000052794 | 1700030K09Rik | 370.514070 | 1.0053484 | 0.007695558 | 0.14830706 | 2.691010e-03 | 9.586283e-01 | No | Yes | 349.074379 | 32.058919 | 346.203989 | 31.779751 | ||||||
| ENSG00000105088 | 93145 | OLFM2 | protein_coding | O95897 | FUNCTION: Involved in transforming growth factor beta (TGF-beta)-induced smooth muscle differentiation. TGF-beta induces expression and translocation of OLFM2 to the nucleus where it binds to SRF, causing its dissociation from the transcriptional repressor HEY2/HERP1 and facilitating binding of SRF to target genes (PubMed:25298399). Plays a role in AMPAR complex organization (By similarity). Is a regulator of vascular smooth-muscle cell (SMC) phenotypic switching, that acts by promoting RUNX2 and inhibiting MYOCD binding to SRF. SMC phenotypic switching is the process through which vascular SMCs undergo transition between a quiescent contractile phenotype and a proliferative synthetic phenotype in response to pathological stimuli. SMC phenotypic plasticity is essential for vascular development and remodeling (By similarity). {ECO:0000250|UniProtKB:Q568Y7, ECO:0000250|UniProtKB:Q8BM13, ECO:0000269|PubMed:25298399}. | Cell junction;Coiled coil;Cytoplasm;Disulfide bond;Glycoprotein;Membrane;Nucleus;Reference proteome;Secreted;Signal;Synapse | hsa:93145; | AMPA glutamate receptor complex [GO:0032281]; cytoplasm [GO:0005737]; extracellular region [GO:0005576]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; positive regulation of smooth muscle cell differentiation [GO:0051152]; protein secretion [GO:0009306]; regulation of vascular associated smooth muscle cell dedifferentiation [GO:1905174] | 17122126_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17122126_The Arg144Gln mutation in OLFM2 is a possible disease-causing mutation in Japanese patients with OAG. Common polymorphisms in OLFM2 and OPTN may interactively contribute to the development of OAG, indicating a polygenic etiology. 25298399_Olfm2 physically interacts with serum response factor (SRF) without affecting the SRF-myocardin interaction. 27844144_Our study indicates that OLFM2 is likely to be important in mammalian eye development and disease and should be considered as a gene for human ocular anomalies. 29553861_plasma OLFM2 is a potential biomarker for restenosis and may be a novel target for the treatment of restenosis. | ENSMUSG00000032172 | Olfm2 | 258.291918 | 0.7606797 | -0.394638998 | 0.18088738 | 4.675294e+00 | 3.059946e-02 | No | Yes | 215.667072 | 27.623350 | 278.957972 | 35.271867 | |||
| ENSG00000105202 | 2091 | FBL | protein_coding | P22087 | FUNCTION: S-adenosyl-L-methionine-dependent methyltransferase that has the ability to methylate both RNAs and proteins (PubMed:24352239, PubMed:30540930, PubMed:32017898). Involved in pre-rRNA processing by catalyzing the site-specific 2'-hydroxyl methylation of ribose moieties in pre-ribosomal RNA (PubMed:30540930). Site specificity is provided by a guide RNA that base pairs with the substrate (By similarity). Methylation occurs at a characteristic distance from the sequence involved in base pairing with the guide RNA (By similarity). Probably catalyzes 2'-O-methylation of U6 snRNAs in box C/D RNP complexes (PubMed:32017898). U6 snRNA 2'-O-methylation is required for mRNA splicing fidelity (PubMed:32017898). Also acts as a protein methyltransferase by mediating methylation of 'Gln-105' of histone H2A (H2AQ104me), a modification that impairs binding of the FACT complex and is specifically present at 35S ribosomal DNA locus (PubMed:24352239, PubMed:30540930). {ECO:0000250|UniProtKB:P15646, ECO:0000269|PubMed:24352239, ECO:0000269|PubMed:30540930, ECO:0000269|PubMed:32017898}. | 3D-structure;Acetylation;Isopeptide bond;Methylation;Methyltransferase;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;S-adenosyl-L-methionine;Transferase;Ubl conjugation;rRNA processing | This gene product is a component of a nucleolar small nuclear ribonucleoprotein (snRNP) particle thought to participate in the first step in processing preribosomal RNA. It is associated with the U3, U8, and U13 small nuclear RNAs and is located in the dense fibrillar component (DFC) of the nucleolus. The encoded protein contains an N-terminal repetitive domain that is rich in glycine and arginine residues, like fibrillarins in other species. Its central region resembles an RNA-binding domain and contains an RNP consensus sequence. Antisera from approximately 8% of humans with the autoimmune disease scleroderma recognize fibrillarin. [provided by RefSeq, Jul 2008]. | hsa:2091; | box C/D RNP complex [GO:0031428]; Cajal body [GO:0015030]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; granular component [GO:0001652]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; small-subunit processome [GO:0032040]; ATPase binding [GO:0051117]; histone-glutamine methyltransferase activity [GO:1990259]; RNA binding [GO:0003723]; rRNA methyltransferase activity [GO:0008649]; TFIID-class transcription factor complex binding [GO:0001094]; box C/D RNA 3'-end processing [GO:0000494]; histone glutamine methylation [GO:1990258]; osteoblast differentiation [GO:0001649]; rRNA methylation [GO:0031167]; rRNA processing [GO:0006364]; snoRNA localization [GO:0048254] | 17461797_Increased levels of exogenous BRAG2 in nucleoli result in redistribution of FBL to the nucleolar periphery. 17603021_Our data suggest that fibrillarin would play a critical role in the maintenance of nuclear shape and cellular growth. 18292223_Evidence that BIG1 and nucleolin, but not fibrillarin, can be present with p62 at the nuclear envelope confirms the presence of BIG1 and nucleolin in dynamic molecular complexes that change in composition while moving through nuclei 19331828_Data demonstrate that fibrillarin and Nop56 directly interact in vivo, and that this interaction is indispensable for the association of both proteins with the box C/D snoRNPs. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21536856_p32 is a new rRNA maturation factor involved in the remodeling from pre-90S particles to pre-40S and pre-60S particles that requires the exchange of FBL for Nop52. 22909121_NS1 protein of the human H3N2 virus interacts primarily via the C-terminal NLS2/NoLS and to a minor extent via the N-terminal NLS1 with the main nucleolar proteins, nucleolin, B23 and fibrillarin. 24029231_FBL overexpression contributes to tumorigenesis and is associated with poor survival in patients with breast cancer. 27682166_Nucleolar Methyltransferase Fibrillarin: Evolution of Structure and Functions. 30190478_Fibrillarin acts as a central node in a regulatory network engaged in imparting immunity against bacterial pathogens. 30540930_SIRT7-dependent deacetylation impacts nucleolar activity by an FBL-driven circuitry that mediates cell-cycle-dependent fluctuation of ribosomal DNA transcription. 32011831_This is the first report on the clinical aspect of ribosome biogenesis in pediatric BCP-ALL [B-cell precursor acute lymphoblastic leukemia ], and it shows that overexpression of CMYC and C/D box nucleoproteins FBL and NOP56 is an antecedent event in patients who subsequently relapse. 32384686_Fibrillarin Ribonuclease Activity is Dependent on the GAR Domain and Modulated by Phospholipids. 33522570_Phase transition of fibrillarin LC domain regulates localization and protein interaction of fibrillarin. 33795875_A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation. | ENSMUSG00000046865 | Fbl | 12832.957957 | 1.0203104 | 0.029008148 | 0.04841119 | 3.600152e-01 | 5.484978e-01 | 8.304938e-01 | No | Yes | 12997.886873 | 1589.416638 | 12834.404879 | 1569.259928 | |
| ENSG00000105221 | 208 | AKT2 | protein_coding | P31751 | FUNCTION: AKT2 is one of 3 closely related serine/threonine-protein kinases (AKT1, AKT2 and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth and angiogenesis. This is mediated through serine and/or threonine phosphorylation of a range of downstream substrates. Over 100 substrate candidates have been reported so far, but for most of them, no isoform specificity has been reported. AKT is responsible of the regulation of glucose uptake by mediating insulin-induced translocation of the SLC2A4/GLUT4 glucose transporter to the cell surface. Phosphorylation of PTPN1 at 'Ser-50' negatively modulates its phosphatase activity preventing dephosphorylation of the insulin receptor and the attenuation of insulin signaling. Phosphorylation of TBC1D4 triggers the binding of this effector to inhibitory 14-3-3 proteins, which is required for insulin-stimulated glucose transport. AKT regulates also the storage of glucose in the form of glycogen by phosphorylating GSK3A at 'Ser-21' and GSK3B at 'Ser-9', resulting in inhibition of its kinase activity. Phosphorylation of GSK3 isoforms by AKT is also thought to be one mechanism by which cell proliferation is driven. AKT regulates also cell survival via the phosphorylation of MAP3K5 (apoptosis signal-related kinase). Phosphorylation of 'Ser-83' decreases MAP3K5 kinase activity stimulated by oxidative stress and thereby prevents apoptosis. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 at 'Ser-939' and 'Thr-1462', thereby activating mTORC1 signaling and leading to both phosphorylation of 4E-BP1 and in activation of RPS6KB1. AKT is involved in the phosphorylation of members of the FOXO factors (Forkhead family of transcription factors), leading to binding of 14-3-3 proteins and cytoplasmic localization. In particular, FOXO1 is phosphorylated at 'Thr-24', 'Ser-256' and 'Ser-319'. FOXO3 and FOXO4 are phosphorylated on equivalent sites. AKT has an important role in the regulation of NF-kappa-B-dependent gene transcription and positively regulates the activity of CREB1 (cyclic AMP (cAMP)-response element binding protein). The phosphorylation of CREB1 induces the binding of accessory proteins that are necessary for the transcription of pro-survival genes such as BCL2 and MCL1. AKT phosphorylates 'Ser-454' on ATP citrate lyase (ACLY), thereby potentially regulating ACLY activity and fatty acid synthesis. Activates the 3B isoform of cyclic nucleotide phosphodiesterase (PDE3B) via phosphorylation of 'Ser-273', resulting in reduced cyclic AMP levels and inhibition of lipolysis. Phosphorylates PIKFYVE on 'Ser-318', which results in increased PI(3)P-5 activity. The Rho GTPase-activating protein DLC1 is another substrate and its phosphorylation is implicated in the regulation cell proliferation and cell growth. AKT plays a role as key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development and synapse formation. Signals downstream of phosphatidylinositol 3-kinase (PI(3)K) to mediate the effects of various growth factors such as platelet-derived growth factor (PDGF), epidermal growth factor (EGF), insulin and insulin-like growth factor I (IGF-I). AKT mediates the antiapoptotic effects of IGF-I. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. May be involved in the regulation of the placental development.; FUNCTION: One of the few specific substrates of AKT2 identified recently is PITX2. Phosphorylation of PITX2 impairs its association with the CCND1 mRNA-stabilizing complex thus shortening the half-life of CCND1. AKT2 seems also to be the principal isoform responsible of the regulation of glucose uptake. Phosphorylates C2CD5 on 'Ser-197' during insulin-stimulated adipocytes. AKT2 is also specifically involved in skeletal muscle differentiation, one of its substrates in this process being ANKRD2. Down-regulation by RNA interference reduces the expression of the phosphorylated form of BAD, resulting in the induction of caspase-dependent apoptosis. Phosphorylates CLK2 on 'Thr-343'. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Apoptosis;Carbohydrate metabolism;Cell membrane;Cytoplasm;Developmental protein;Diabetes mellitus;Disease variant;Disulfide bond;Endosome;Glucose metabolism;Glycogen biosynthesis;Glycogen metabolism;Glycoprotein;Kinase;Manganese;Membrane;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Serine/threonine-protein kinase;Sugar transport;Transferase;Translation regulation;Transport;Ubl conjugation | This gene is a putative oncogene encoding a protein belonging to a subfamily of serine/threonine kinases containing SH2-like (Src homology 2-like) domains, which is involved in signaling pathways. The gene serves as an oncogene in the tumorigenesis of cancer cells For example, its overexpression contributes to the malignant phenotype of a subset of human ductal pancreatic cancers. The encoded protein is a general protein kinase capable of phophorylating several known proteins, and has also been implicated in insulin signaling. [provided by RefSeq, Nov 2019]. | hsa:208; | cell cortex [GO:0005938]; cytosol [GO:0005829]; early endosome [GO:0005769]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; ruffle membrane [GO:0032587]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; activation of GTPase activity [GO:0090630]; carbohydrate transport [GO:0008643]; cellular protein modification process [GO:0006464]; cellular response to high light intensity [GO:0071486]; cellular response to insulin stimulus [GO:0032869]; fat cell differentiation [GO:0045444]; glucose metabolic process [GO:0006006]; glycogen biosynthetic process [GO:0005978]; insulin receptor signaling pathway [GO:0008286]; intracellular protein transmembrane transport [GO:0065002]; intracellular signal transduction [GO:0035556]; mammary gland epithelial cell differentiation [GO:0060644]; negative regulation of apoptotic process [GO:0043066]; negative regulation of long-chain fatty acid import across plasma membrane [GO:0010748]; peptidyl-serine phosphorylation [GO:0018105]; peripheral nervous system myelin maintenance [GO:0032287]; positive regulation of cell migration [GO:0030335]; positive regulation of cell motility [GO:2000147]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of fatty acid beta-oxidation [GO:0032000]; positive regulation of glucose import [GO:0046326]; positive regulation of glucose metabolic process [GO:0010907]; positive regulation of glycogen biosynthetic process [GO:0045725]; positive regulation of mitochondrial membrane potential [GO:0010918]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of vesicle fusion [GO:0031340]; protein localization to plasma membrane [GO:0072659]; regulation of cell cycle [GO:0051726]; regulation of cell migration [GO:0030334]; regulation of translation [GO:0006417]; retinal rod cell apoptotic process [GO:0097473]; signal transduction [GO:0007165] | 12114503_In renal tubular epithelial cell, Akt phosphorylation is up-regulated in an effort to minimize cell death. 12480711_Activation of AKT consequent to binding of albumin by CLL cells blocks chlorambucil- and radiation-induced apoptosis. 12482965_a new mechanism for androgen-mediated prostate cancer cell survival and establish FKHR as nuclear target for both AKT-dependent and -independent survival signals in prostate cancer cells. 12517337_Results describe the structure of an inactive and unliganded Akt2 kinase domain, and several features that distinguish it from other kinases. 12517798_data indicate that AKT2 mediates PI3-K-dependent effects on adhesion, motility, invasion, and metastasis in vivo 12545160_These results indicate that PI3k/Akt pathway is involved in the signaling cascade of glioma cells required to induce cell migration. 12663464_results show that a defect in the ability of insulin to activate Akt-2 and -3 may explain the impaired insulin-stimulated glucose transport in insulin resistance 12733712_both p70S6K and Akt are activated in the majority of human papillary cancer cells. 12808085_Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation 14504284_Akt2 phosphorylates MLK3, which results in the disassembly of the JNK complex bound to POSH and down-regulation of the JNK signaling pathway 14612499_Akt2 may be an important regulator of both Xiap and p53 contents in ovarian cancer after CDDP challenge. 14637151_Our data suggest that Akt is an endogenous inhibitor during TRAIL-mediated synovial cell apoptotic pathway. 14654898_Findings suggest that Akt2 is a novel independent predictor for the development and progression of hepatocellular carcinoma . 14699494_Lipid rafts(LR) may play important role in determining function of PI 3-K/Akt2 signaling, including stimulation of intestinal Na absorption. LR-associated Akt2 may be involved in enterocyte differentiation. 14735903_data suggest that upstream perturbations of the PI3K/AKT pathway contribute to frequent activation of AKT2 in pancreatic cancer, which may contribute to the pathogenesis of this highly aggressive form of human malignancy 15010337_Thus a physiological concentration of insulin stimulated Akt-1 & Akt-2 phosphorylation in human skeletal muscle in the absence of hyperglycemia, but Akt-2 expression is impaired in muscle of obese patients with atypical diabetes with severe hyperglycemia. 15102693_activated Akt and Akt2 have roles in progression of pancreatic ductal adenocarcinoma 15111130_These results suggest that cross-talk between the PKB and caspase-8 pathways may regulate the balance between cell survival and cell death in ECV304. 15166380_a mutation in AKT2 in a family showing autosomal dominant inheritance of severe insulin resistance and diabetes mellitus is described; the mutant kinase in cultured cells disrupted insulin signaling and inhibited the function of coexpressed, wild-type AKT 15531580_Akt2 is a critical kinase that regulates ezrin phosphorylation and activation 15557754_AKT2 plays an important role in glioma cell motility and invasion 15890450_Kinetic analysis of GST-AKT2 demonstrates that phosphorylation of Thr309 in the activation loop of the kinase is largely responsible for observed reduction in Km & for a subsequent 150-fold increase in the catalytic efficiency (k(cat)/Km) of the enzyme. 15987444_in breast cancer patients, Akt activation is associated with tumour proliferation and poor prognosis, particularly in the subset of patients with ErbB2-overexpressing tumours 16365168_In this study, we provide evidence for isoform-specific positive and negative roles for Akt1 and -2 in regulating growth factor-stimulated phenotypes in breast epithelial cells 16402276_results suggest that AKT pathway may play an important role in the development and progression of gliomas 16982699_These data show that specific interaction of the Akt2 isoform with p21 is key to its negative effect on normal cell cycle progression. 17012749_Akt1 and Akt2 have opposing roles in Rac/Pak signaling and cell migration 17276404_Akt1 induces CREB phosphorylation at Ser-133 and CREB target gene expression. 17327441_Observational study of gene-disease association. (HuGE Navigator) 17327441_Sequencing of the entire coding region and splice junctions of AKT2 and the relationship of genetic variations in the gene with multiple metabolic diseases is reported. 17332325_Twist as a positive transcriptional regulator of AKT2 expression; Twist-AKT2 signaling is involved in promoting invasive ability and survival of breast cancer cells. 17372934_Changes in tissue pressure during inflammation may regulate macrophage phagocytosis by activation of PI-3K, which activates Akt2, mTOR, and p70S6K. 17482291_the PI3K/AKT/mTOR signaling pathway is involved in regulation of SphK1, with AKT2 playing a key role in PDGF-induced SphK1 expression 17576055_Observational study of gene-disease association. (HuGE Navigator) 17576055_Two variants in 5' regulatory region of Akt2 gene are associated and may modulate susceptibility to insulin resistance and related metabolic abnormalities. 17804734_AKT2 up-regulation is characteristic of skin squamous cell carcinoma and coincident AKT2 activation through serine phosphorylation correlates with malignancy. 17825284_Pressure did not stimulate translocation of AKT2 to the plasma membrane. 17895832_PKB[alpha] and/or PKB[gamma] and not PKB[beta] alone are involved in gemcitabine resistance mechanisms. 17908691_Akt2 is required for rapamycin-induced vascular smooth muscle cell differentiation, whereas Akt1 appears to oppose contractile protein expression. 17914025_Akt1 and Akt2 mediate GPIb-IX signaling via the cGMP-dependent signaling pathway. 18048359_a key regulatory role of the Akt/mTOR pathway in the generation of the effects of As(2)O(3) 18281467_Functional convergence of Twist and AKT2 underscores the importance of this signaling pathway in tumor development and progression. 18316591_Dlx5 can act as an oncogene by cooperating with Akt2 to promote lymphomagenesis 18353613_Results suggest that Akt2 directly mediates EGF-induced chemotactic signaling pathways through PKCzeta and its expression is critical during the extravasation of circulating cancer cells. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18768676_Observational study of gene-disease association. (HuGE Navigator) 18768676_Polymorphisms in two components of the insulin signaling pathway, AKT2 and GSK3B, are associated with polycystic ovary syndrome. 18842333_Akt2 down-regulation sensitizes ovarian cancer cells to paclitaxel-induced apoptosis, and inhibits survivin expression 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074768_an important role of PKC-delta and PI3K/Akt1,2 pathways in activating mTOR as an endogenous modulator to ensure a tight regulation of NF-kappaB signaling of ICAM-1 expression in endothelial cells. 19075230_the consequence of PTEN loss and Akt2 overexpression function synergistically to promote metastasis 19079138_mtDNA depletion prevents detachment-induced apoptosis (anoikis) and promotes migratory capabilities onto basement membrane proteins through upregulation of p85 and p110 phosphatidylinositol 3-kinase (PI3K) subunits, which results in Akt2 activation 19110052_Akt2 up-regulates beta1 integrins & promotes adhesion & invasion of breast cancer cells in vitro & also metastasis in vivo. It opposes Akt1. Review. 19164214_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19164214_genetic variations in akt2 were associated with survival in esophageal cancer 19197940_Akt2 plays an essential role in both CSF-1- and chemokine-induced chemotaxis of macrophages. 19261608_Bcr-Abl represses the expression of PHLPP1 and PHLPP2 and continuously activates Akt1, -2, and -3 via phosphorylation on Ser-473, resulting in the proliferation of CML cells 19435822_Findings indicate that AP-1 has an important function in pancreatic cancer cells and provide evidence for a previously unknown Akt-mediated mechanism of c-Jun activation. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19771908_The positive rate of AKT2 protein expression in laryngeal squamous cell carcinoma was significantly higher than that in peri-cancer tissue and normal laryngeal epithelium, and was correlated with tumor site, lymph node metastasis and clinical stage. 19797172_mTOR/Akt2 is required for optimal PPARgamma activation. Patients who receive SESs during concomitant RSG treatment may be at risk for delayed stent healing. 19825827_induction of EMT is controlled by microRNAs whose abundance depends on the balance between Akt1 and Akt2 rather than on the overall activity of Akt 19843246_Taken together, these results showed that, in addition to the MAPK pathway previously reported being a downstream target of stimulated Grm1, AKT2 is another downstream target in Grm1 mediated melanocyte transformation. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19933843_These findings provide further evidence of the importance of mediators of the growth factor signaling in ER regulation, introducing the Akt2/FoxO3a axis as a pursuable target in therapy for ER-positive breast cancer. 20018949_Akt2 was located in the mitochondria of human cell lines. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20056908_Plasmin triggers chemotaxis of monocyte-derived dendritic cells through an Akt2-dependent pathway and promotes a T-helper type-1 response. 20059950_By using yeast two-hybrid systems with full-length human Akt2 as bait, we found a stable interaction between the tetratricopeptide (TPR) repeat domain 3 (TTC3) and Akt; finding could underlie important clinical manifestations of Down syndrome. 20102399_In breast cancer AKT3 amplifications and AKT1 and AKT2 deletions were seen by FISH 20109457_Results represent the first indication that Akt isoforms regulate intermediate filament protein expression and support the hypothesis that IFs are involved in PI3K/Akt pathway. 20116920_Akt2 may play a critical role in the development of gliomas and present a potential therapeutic target for malignant gliomas 20133737_Data show that AKT1 and AKT2 appeared to regulate growth through FOXO proteins, but not through either GSK3beta or mTOR, and in contrast, inactivation of PDPK1 affected GSK3beta and mTOR activation. 20167810_Our data suggest that Akt2 and Akt3 play an important role in the viability of human malignant glioma cells. 20186503_Observational study of gene-disease association. (HuGE Navigator) 20186503_We evaluated the presence of mutations in PIK3CA, AKT1, AKT2, AKT3, PTEN, and PDPK1 genes in 83 papillary thyroid carcinomas 20354455_AKT2 mutation coexisted with EGFR and PIK3CA mutations was associated with lung cancer. 20354455_Observational study of gene-disease association. (HuGE Navigator) 20398329_Studies indicate that three different isoforms Akt1, Akt2, and Akt3 have distinct expression patterns and functions. 20409325_MYCN contributes to tumorigenesis, in part, by repressing miR-184, leading to increased levels of AKT2, a direct target of miR-184 20447721_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20514445_AKT2 expression is associated with more advanced and especially aggressive gliomas and critical for cell survival and invasion. 20602615_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20687898_SGK3 is not required for insulin-induced PFK-2 activation and that this effect is likely mediated by PKBalpha. 20688159_The expression levels and activities of exogenous Akt1 and Akt2 are almost identical, Akt2 exerted a greater inhibitory effect on both proliferation and cell migration. 20691427_Observational study of gene-disease association. (HuGE Navigator) 20848774_The positive expression of p-AKT2 in lung adenocarcinoma was higher than that in lung squamous carcinomas. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21050850_Akt signaling regulates the stability of palladin 21092549_Expressions of Akt1, Akt2 and PI3K were decreased in ovarian epithelial cancer cells transduced with PTEN. 21179235_Advanced glycation end products of BSA can induce endothelial-to-mesenchymal transition of cultured human and monkey endothelial cells and AKT2 may play a role in this process. 21182834_Data suggest that Akt2/B23 functions as an oncogenic unit to drive tumorigenesis of A549 lung cancer cells. 21297943_Akt2 augments cell proliferation by facilitating cell cycle progression through the upregulation of the cell cycle engine, and protects a cell from pathological autophagy by modulating mitochondrial homeostasis. 21432781_analysis of the roles of Akt1 and Akt2 in cancer [review] 21507933_loss of Akt1 or Akt2 decreased proliferation of Pten wild-type astrocytes, whereas combined loss of multiple isoforms was needed to inhibit proliferation of Pten-null astrocytes 21518566_The Akt2 gene is not a major cause of diabetes in a non-obese Chinese Han population characterized by insulin resistance. 21590431_Transcriptional activation of the Akt2 pathway indicates that it is involved in lumbar disc degeneration. 21618512_Data suggest a regulation pathway of B-cell-lymphoma-2 (Bcl-2) by cisplatin, via the activation of PKC and Akt2, which has impact on resistance to cisplatin-induced apoptosis in endometrial cancer cells. 21636708_High AKT2 is associated with glioma progression. 21680733_Angiopoietin-2, an angiogenic regulator, promotes initial growth and survival of breast cancer metastases to the lung through the integrin-linked kinase (ILK)-AKT1,2-B cell lymphoma 2 (Bcl-2) pathway. 21722267_AKT1 and AKT2 both contribute to cell survival, albeit via different mechanisms; the effects on cell growth and migration are predominantly regulated by AKT1 21932427_This study revealed a novel mechanism of Akt2 regulation by ErbB2 in prostate cancer cells. 21979934_3 unrelated children with unexplained, recurrent and severe fasting hypoglycemia and asymmetrical growth were found to carry the same de novo mutation, p.Glu17Lys, in AKT2, in 2 cases as heterozygotes and in 1 case in mosaic form 21979951_Akt2 kinase suppresses glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-mediated apoptosis in ovarian cancer cells via phosphorylating GAPDH at threonine 237 and decreasing its nuclear translocation 22031698_analysis of resistance of Akt kinase and PP2A to dephosphorylation through ATP-dependent conformational plasticity 22107784_The prognostic impact of Akt (Akt1) phosphorylated at threonine308 and serine473, Akt2, Akt3, PI3K and PTEN, alone and in coexpression with ER and PgR in non-gastrointestinal stromal tumor soft tissue sarcomas. 22158034_Unexpected roles of Akt2 in transcriptional control and phosphorylation of Thr45 in histone H3 as a new epigenetic mark related to Snail1 and Akt2 action. 22261254_Akt2 is a negative regulator of NFAT activation through its ability to inhibit calcium mobilization from the endoplasmic reticulum. 22480544_study concludes that Akt2 is indispensable for the regulation of preadipocyte and adipocyte number, whereas Akt1 and Akt2 are equally important for the regulation of insulin-stimulated metabolic pathways in adipocytes 22556379_Although there is an association of PI3 kinase signaling and chemoresistance in advanced ovarian cancer, there is no clear evidence that this is driven specifically by AKT2 mutations. 22748472_amplification of AKT1 and/or AKT2 and high-level polysomy were found in 16% of total lung carcinoma cases, and this defined subset was characterized by the overexpression/activation of Akt, reciprocal to EGFR aberrations. 22778840_In transfected HEK2 cells, basal activity of Akt2 regulates DAT cell surface expression. 22809628_these data define an inhibitory role for both AKT1 and AKT2 in prostate cancer migration and invasion and highlight the cell type-specific actions of AKT kinases in the regulation of cell motility. 23018889_An Akt2/PKBbeta-targeted siRNA inhibited the 8-Cl-cAMP- and AICAR-mediated phosphorylation of AMPK and p38 MAPK. 23092922_CD40L-provoked signaling results in the production of several cytokines. Among these, IL-6 expression is mediated through Akt and NF-kappaB pathways. 23250987_In cultured human aortic vascular smooth muscle cells, AKT2 inhibited the expression of MMP-9 and stimulated the expression of TIMP-1 by preventing the binding of transcription factor forkhead box protein O1 to the MMP-9 and TIMP-1 promoters. 23305873_Data indicate that the prognostic value of Akt2 increases with higher oestrogen receptor (ER) expression, motivating further mechanistic studies on the role of Akt2 in ER+ breast cancer. 23321478_Data suggest that although COOH-terminal dephosphorylation is likely necessary for glycogen synthase (GS) activation, Akt2-dependent NH2-terminal dephosphorylation is the site for 'fine-tuning' insulin-mediated GS activation in skeletal muscle. 23444369_Insulin signaling via Akt2 switches plakophilin 1 function from stabilizing cell adhesion to promoting cell proliferation. 23464484_In a manner that depended on the level of phosphorylated AKT1/2 protein. 23468863_Akt2 regulates metastatic potential in neuroblastoma. 23567263_AKT1 and AKT2 have non-redundant roles in the regulation of prostate cancer cell proliferation and migration. 23777806_Depletion of both Akt2 and ASAH1 is much more potent than depleting each alone at inhibiting neoplastic cell viability/proliferation and invasion. 23823123_the direct interaction of AKT2 and EF2 was found to be dynamically regulated in embryonic rat cardiomyocytes 23900341_Findings suggest an important role of Gli1 as a tumor suppressor in neuroblastoma, and offer a mechanism by which AKT2 regulates the subcellular localization, and in turn, inhibits the tumor-suppressive function of Gli1 in neuroblastoma. 23929892_detected and measured all three AKT isoforms 1, 2 and 3 to enable the study of the multiple and variable roles that these isoforms play in AKT breast tumorigenesis 24030155_overexpression of 4EBP1, p70S6K, Akt1 or Akt2 could promote the Coxsackievirus B3-induced apoptosis. 24039187_AKT2 is closely linked to Type II diabetes and the implications of various types of mutations are discussed 24056770_results disclose a new function of Akt2 and identify a potential therapeutic target for preserving glomerular function in CKD 24123662_Akt2 downregulation favors limbal keratinocyte stem cell maintenance as a result of a gain of FOXO functions. 24244431_Knockdown of RPS7 resulted in increased expression of P85alpha, P110alpha, and AKT2. 24247267_Experimental validation indicate that AKT1, AKT2 and AKT3 proteins may all be novel unfavorable prognostic factors for patients with hepatocellular carcinoma. 24321521_in a defined subset of bone and soft tissue tumors, including benign tumors, Akt was frequently overexpressed and activated, and AKT1/2 copy number was increased 24337067_Results show that miR-302b inhibits hepatocellular carcinoma cell proliferation and growth in vitro and in vivo by targeting AKT2. 24565443_AA induces Akt2 activation and invasion in MDA-MB-231 cells. Akt2 activation requires the activity of Src, EGFR, and PIK3, whereas migration and invasion require Akt, PI3K, EGFR and metalloproteinases activity. 24642468_AKT2 inhibition reduced heterotypic aggregation of neutrophils and platelets isolated from sickle cell disease patients, suggesting that AKT2 is important for neutrophil recruitment and neutrophil-platelet interactions. 24656454_AKT2 may play an important role in the development of meningioma. High AKT2 labeling index indicates higher grade of meningioma, and therefore AKT2 may be a useful molecular marker for predicting the prognosis of meningioma. 24699302_We examined the response of 80 samples of primary cells from acute myeloid leukemia patients to selective inhibitors of the phosphatidylinositol 3 kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) axis. 24769357_CK2-dependent phosphorylation is therefore a crucial event which, discriminating between Akt1 and Akt2, can account for different substrate specificities. 24819169_SchA-FAK-p85 complex subsequently selectively recruited and activated Akt2, not Akt1 24838891_The specific role of AKT2 in tumor maintenance provides a rationale for the development of isoform-specific inhibitors for patients with PTEN-deficient cancers. 24946858_Findings suggest that adding chloroquine to EGF receptor (EGFR) and AKT2 inhibition has the potential to improve tumor responses in EGFR M+ non-small cell lung cancer (NSCLC). 24970808_Findings indicate that miR-137 is a valuable biomarker for hepatocellular carcinoma (HCC) prognosis and the forkhead box D3 (FoxD3)/miR-137/AKT2 regulatory network plays an important role in HCC progression. 25025572_Role of the guanine nucleotide exchange factor in Akt2-mediated plasma membrane translocation of GLUT4 in insulin-stimulated skeletal muscle. 25059120_Knockdown of Akt2 using siRNAs or the PI3K inhibitor Ly294002 inhibited TGF-beta1-induced phosphorylation of GSK3beta and expression of Snail1 25134663_the biological effect of Akt2 in colorectal cancer cells 25162660_AKT isoform-specific knock down combined with quantitative phosphoproteomics provided a powerful strategy to unravel AKT isoform-specific signaling 25173755_Mutations in genes such as AKT2, CCNA1, MAP3K4, and TGFBR1, were associated significantly with Epstein-Barr-positive gastric tumors, compared with EBV-negative tumors. 25233414_Data suggest that three isoforms of the serine/threonine protein kinase Akt (Akt1, Akt2, Akt3) regulate cell survival, cell growth, cell proliferation, and cell metabolism in breast cancer cells. [REVIEW] 25246356_The individual contribution of each Akt isoform in p120 RasGAP fragment N-mediated cell protection against Fas ligand induced cell death, was investigated. 25253241_Akt1 and Akt2 are involved in albumin endocytosis, and phosphorylation of Dab2 by Akt induces albumin endocytosis in proximal tubule epithelial cells. 25263462_miR-29b plays an important role in TGF-beta1-mediated epithelial-mesenchymal transition in ARPE-19 cells by targeting Akt2. 25285168_MiR-194 deregulation contributes to colorectal carcinogenesis via reducing the expression AKT2. 25288334_let-7b/g inhibited AKT2 expression by directly binding to its 3'UTR, reduced p-AKT (S473) activation and suppressed expression of the downstream effector pS6. 25304263_Our study uncovers a novel molecular mechanism by which AEG-1 augments glioma progression and offers a rationale to block AEG-1-Akt2 signaling function as a novel GBM treatment. 25428377_findings suggested that AKT2 may be one of the targets of miR29s in gastric cancer 25733895_The results indicate that BCAM-AKT2 expression is a new mechanism of AKT2 kinase activation in high-grade serous ovarian carcinoma. 25771729_PIK3CA and AKT2 mutations occurred at low frequency in gastric cancer. 25856297_miR-615-5p inhibits pancreatic cancer cell proliferation, migration, and invasion by targeting AKT2 25894377_we assayed expression of myosin II.. this assay is a powerful predictor of the use of ZEB2/Akt2 as a marker for tumor progression in serous ovarian cancers. 25936945_QSAR based docking studies identify marine algal callophycin A as inhibitor of protein kinase B beta. 25951903_Up-regulation of AKT2 was associated with chemoresistance in renal cell carcinoma. 26102366_miR-137 which is frequently down-regulated in gastric cancer is potentially involved in gastric cancer tumorigenesis and metastasis by regulating AKT2 related signal pathways 26158514_miR-612 directly suppressed AKT2, which in turn inhibited the downstream epithelial-mesenchymal transition-related signaling pathway. 26229955_the correlation between RLN2 and p-AKT or RLN2 and p-ERK1/2 expression was investigated. 26234648_Akt2 role in the human lung cancer cell proliferation, growth, motility, invasion and endothelial cell tube formation 26254095_miR-302b inhibits SMMC-7721 cell invasion and metastasis by targeting AKT2 26265781_Combined PI3K/Akt and Hsp90 targeting synergistically suppresses essential functions of alloreactive T cells and increases Tregs. 26318486_AKT1 but not AKT2 protects islet cells from apoptosis and drives proliferation. 26475619_High constitutive Akt2 activity in U937 promonocytes: effective reduction of Akt2 phosphorylation by the histamine H2-receptor and the beta2-adrenergic receptor 26512921_High AKT2 expression is associated with ovarian cancer. 26711268_Active, phosphorylated Akt2 translocates to the nucleus in Notch-expressing cells, resulting in GSK-3beta inactivation in this compartment. 26803515_High AKT2 expression is associated with gallbladder cancer. 26828791_No association has been found between AKT2 polymorphisms and oesophageal squamous cell carcinoma risk. 26855332_Akt1 and Akt2 activated both SREBP-1 and SREBP-2, whereas Akt3 upregulated SREBP-1 to enhance hepatitis C virus translation. 26953242_Studies provide evidence that AKT2 counteracts oxidative-stress-induced apoptosis and is required for alpha-beta thymocyte survival and differentiation. Also, it plays a critical role in antagonizing cardiomyocyte apoptosis. [review] 26971877_controls endothelial Jagged1 expression and, thereby, Notch signalling regulating VSMC maintenance. 27067543_TGF-beta signaling through Akt2 induces phosphorylation of heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) at serine-43 (p-hnRNP E1), driving epithelial to mesenchymal transition and metastasis. 27105349_Data indicate that proto-oncogene protein c-akt (Akt) phosphorylates distinct sites than SRPK1 protein within the arginine-serine (RS) domain of Lamin B Receptor (LBR). 27189341_AKT2 can regulate miR-200a in a histology- or stage-specific manner and this regulation is independent of subsequent involvement of miR-200a in epithelial-mesenchymal transition. 27545875_IRF5 and IRF5 disease-risk variants increase glycolysis and human m1 macrophage polarization by regulating proximal signaling and Akt2 activation. 27663592_Analysis of genomic data from TCGA demonstrated coamplification of CCNE1 and AKT2 Overexpression of Cyclin E1 and AKT isoforms, in addition to mutant TP53, imparted malignant characteristics in untransformed fallopian tube secretory cells, the dominant site of origin of high-grade serous ovarian cancer 27871477_The expression levels of AKT2 and CDC25C showed lower expression in neural tube defects. And the percentage of methylated region of AKT2 promoter were increased in neural tube defects. 27878243_This was further verified in gene expression analyses, showing downregulation of genes involved in glucose metabolism. Additionally, both AKT1 KO and AKT2 KO demonstrated an impaired fatty acid metabolism. However, genes were upregulated in the Wnt and cell proliferation pathways, which could oppose this effect. AKT inhibition should therefore be combined with other effectors to attain the best effect. 27878305_miR-148a functions as tumor suppressor in Renal cell carcinoma by targeting AKT2. 27906180_This study demonstrates novel regulatory circuits involving miR-148a-3p/ERBB3/AKT2/c-myc and DNMT1 that controls bladder cancer progression, which may be useful in the development of more effective therapies against bladder cancer. 28026019_Both in our animal model and in human Age-related macular degeneration (AMD), the AKT2-NF-kappaB-LCN-2 signalling axis is involved in activating the inflammatory response. 28036396_Quinoline-type inhibitors bind in the Akt2 PH domain. 28068324_Data identify MTSS1 as a new Akt2-regulated gene, and point to suppression of MTSS1 as a key step in the metastasis-promoting effects of Akt2 in CRC cells. 28129626_MiR-650 could inhibit the proliferation, migration and invasion of rheumatoid arthritis synovial fibroblasts through targeted regulation of AKT2 expression. 28271235_High expressions of AKT1 and AKT2 through possible relation with androgen may cause granulosa cells dysfunction in the +HA PCOS patients. 28287129_Data show while overexpression of AKT serine/threonine kinase 1 (AKT1) promoted local tumor growth, downregulation of AKT1 or overexpression of AKT serine/threonine kinase 2 (AKT2) promoted peritumoral invasion and lung metastasis. 28341696_identified a novel association between the coding variant (p.Pro50Thr) in AKT2 and fasting plasma insulin (FI), a gene in which rare fully penetrant mutations are causal for monogenic glycemic disorders. Carriers of the FI-increasing allele had increased 2-h insulin values, decreased insulin sensitivity, and increased risk of type 2 diabetes. 28404925_miR-143-3p acts as a novel tumor suppressive miRNA by regulating gastric tumor growth, migration and invasion through directly targeting AKT2 gene 28440469_Our findings suggest that Akt2 might be associated with the resistance to anti-EGFR therapies, especially the use of erlotinib against PC, and that this resistance can be overcome by combined treatment with a PI3K inhibitor. Akt2 expression could become a predictive biomarker for erlotinib resistance in PC. 28455433_Results indicate that AKT2 modulates pulmonary fibrosis through inducing TGF-beta1 and IL-13 production by macrophages, and inhibition of AKT2 may be a potential strategy for treating Idiopathic pulmonary fibrosis. 28456993_Studied action of linoleic acid (LA) on cell migration and neoplasm invasiveness of breast cancer cells. Findings show Akt2 activation requires EGFR and PI3K activity, whereas migration and invasion are dependent on FFAR4, EGFR and PI3K/Akt activity. 28534950_miR296 is downregulated in tissue from patients with pancreatic cancer and pancreatic carcinoma cell lines. These findings suggested that it may function as a tumor suppressor via inhibiting the growth, migration and invasion of pancreatic cancer cells. AKT2 was validated as a direct target of miR296 in pancreatic cancer cells. 28541532_The p.Glu17Lys mutation of AKT2 confers low-level constitutive activity upon the kinase and produces hypoglycemia with suppressed fatty acid release from adipose tissue, but not fatty liver, hypertriglyceridemia, or elevated hepatic de novo lipogenesis. 28557977_This review summarises and discusses the consequences of genetic deletions of Akt isoforms in adult mice and their implications for cancer therapy. Whereas combined Akt1 and Akt2 rapidly induced mortality, hepatic Akt inhibition induced liver injury that promotes hepatocellular carcinoma. 28586057_miR2965p/AKT2 axis serves important roles in Hepatocellular carcinoma carcinogenesis and progression. 28650848_Recent studies reveal that AKT2-NOX2 signaling has critical roles in Ca mobilization, ROS generation, degranulation, and control of the ligand-binding function of cell surface molecules, thereby promoting heterotypic cell-cell interactions in thromboinflammation. 28689659_Akt2, Erk2, and IKK1/2 phosphorylate Bcl3, converting Bcl3 into a transcriptional coregulator by facilitating its recruitment to DNA. 28837154_Findings demonstrate that DSBs trigger pro-survival autophagy in an ATM- and p53-dependent manner, which is curtailed by AKT2 signaling. 28931550_Report frequency of genetic variation in Akt2 and discuss link to type 2 diabetes. 29025710_AKT1 and AKT2 isoforms have opposing roles in smooth muscle cell proliferation, migration, differentiation, and rapamycin response in vitro and in vascular injury in vivo. 29075783_Authors present the first evidence that miR-608 behaves as a tumour suppressor in A549 and SK-LU-1 cells through the regulation of AKT2. 29141982_genetic association studies in population of men in Finland: Data suggest that a partial loss-of-function variant in AKT2 (p.Pro50Thr) is associated with type 2 diabetes in the population studied; this AKT2 variant is associated with reduced insulin-mediated glucose uptake in multiple insulin-sensitive tissues. 29153407_AKT2 driv | ENSMUSG00000004056 | Akt2 | 2918.949585 | 0.9881127 | -0.017252473 | 0.06764584 | 6.462094e-02 | 7.993360e-01 | 9.347379e-01 | No | Yes | 3281.549064 | 372.962326 | 3314.869755 | 376.666107 | |
| ENSG00000105245 | 9253 | NUMBL | protein_coding | Q9Y6R0 | FUNCTION: Plays a role in the process of neurogenesis. Required throughout embryonic neurogenesis to maintain neural progenitor cells, also called radial glial cells (RGCs), by allowing their daughter cells to choose progenitor over neuronal cell fate. Not required for the proliferation of neural progenitor cells before the onset of embryonic neurogenesis. Also required postnatally in the subventricular zone (SVZ) neurogenesis by regulating SVZ neuroblasts survival and ependymal wall integrity. Negative regulator of NF-kappa-B signaling pathway. The inhibition of NF-kappa-B activation is mediated at least in part, by preventing MAP3K7IP2 to interact with polyubiquitin chains of TRAF6 and RIPK1 and by stimulating the 'Lys-48'-linked polyubiquitination and degradation of TRAF6 in cortical neurons. {ECO:0000269|PubMed:18299187, ECO:0000269|PubMed:20079715}. | 3D-structure;Cytoplasm;Developmental protein;Neurogenesis;Phosphoprotein;Reference proteome;Ubl conjugation pathway | hsa:9253; | cytoplasm [GO:0005737]; adherens junction organization [GO:0034332]; axonogenesis [GO:0007409]; cytokine-mediated signaling pathway [GO:0019221]; lateral ventricle development [GO:0021670]; nervous system development [GO:0007399]; neuroblast division in subventricular zone [GO:0021849]; positive regulation of neurogenesis [GO:0050769]; protein metabolic process [GO:0019538] | 16899352_Observational study of gene-disease association. (HuGE Navigator) 18940473_Both gene sequence alterations and amplifications of LNX1 and Numbl are present in a subset of human gliomas. 18940473_Observational study of gene-disease association. (HuGE Navigator) 20079715_NUMBL interacts with TRAF6 and promotes the degradation of TRAF6 in vivo, leading to the inhibition of NF-kappaB signaling pathway. 22593207_data suggest that Numbl regulates glioma cell migration and invasion by abrogating TRAF5-induced activation of NF-KappaB 23440423_Numbl-Klf4 signaling is critical to maintain multiple nodes of metastatic progression, including persistence of cancer-initiating cells. 23681800_Numbl might be involved in the inhibition of growth, proliferation, and invasion of 95-D lung cancer cells. 26069237_Numb/Numbl control VEGF receptor endocytosis, signaling, and recycling in endothelial cells, which promotes the angiogenic growth of blood vessels. 27383182_These findings highlight the importance of Numb and Numbl in the control of myoepithelial cell fate determination, epithelial identity, and lactogenesis 27521217_let-7c inhibits Notch and progression markers but up-regulates Numbl in pancreatic cancer treated with quercetin 27613838_NumbL can act as an independent tumor suppressor inhibiting the Notch pathway and regulating the cancer stem cell pool 28067668_investigations revealed that NUMB and NUMBL interacted with small GTPase Rab7 to transition ERBB2 from early to late endosome for degradation. | ENSMUSG00000063160 | Numbl | 870.699960 | 0.8791078 | -0.185888008 | 0.11389646 | 2.671047e+00 | 1.021888e-01 | 4.267032e-01 | No | Yes | 825.080948 | 86.903081 | 929.022457 | 97.748596 | ||
| ENSG00000105339 | 22898 | DENND3 | protein_coding | A2RUS2 | FUNCTION: Guanine nucleotide exchange factor (GEF) activating RAB12. Promotes the exchange of GDP to GTP, converting inactive GDP-bound RAB12 into its active GTP-bound form (PubMed:20937701). Regulates autophagy in response to starvation through RAB12 activation. Starvation leads to ULK1/2-dependent phosphorylation of Ser-472 and Ser-490, which in turn allows recruitment of 14-3-3 adapter proteins and leads to up-regulation of GEF activity towards RAB12 (By similarity). Also plays a role in protein transport from recycling endosomes to lysosomes, regulating, for instance, the degradation of the transferrin receptor and of the amino acid transporter PAT4 (PubMed:20937701). Starvation also induces phosphorylation at Tyr-858, which leads to up-regulated GEF activity and initiates autophagy (By similarity). {ECO:0000250|UniProtKB:A2RT67, ECO:0000269|PubMed:20937701}. | Alternative splicing;Cytoplasm;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:22898; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; guanyl-nucleotide exchange factor activity [GO:0005085]; cellular protein catabolic process [GO:0044257]; endosome to lysosome transport [GO:0008333]; regulation of Rab protein signal transduction [GO:0032483] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25925668_a novel signaling pathway identified whereby starvation-induced activation of ULK leads to phosphorylation of endogenous DENND3, with subsequent activation of Rab12 and initiation of membrane trafficking events required for autophagy 28249939_DENND3 is the exchange factor for the small GTPase Rab12 regulated through an intramolecular interaction 29352104_show that DENND3 binds actin through a surface of positively charged residues on the PHenn domain of Ran12 | ENSMUSG00000036661 | Dennd3 | 343.057343 | 0.9871167 | -0.018707485 | 0.16529891 | 1.300186e-02 | 9.092174e-01 | No | Yes | 330.871370 | 43.176274 | 336.174525 | 43.783230 | |||
| ENSG00000105443 | 9266 | CYTH2 | protein_coding | Q99418 | FUNCTION: Acts as a guanine-nucleotide exchange factor (GEF). Promotes guanine-nucleotide exchange on ARF1, ARF3 and ARF6. Activates ARF factors through replacement of GDP with GTP (By similarity). The cell membrane form, in association with ARL4 proteins, recruits ARF6 to the plasma membrane (PubMed:17398095). Involved in neurite growth (By similarity). {ECO:0000250|UniProtKB:P63034, ECO:0000269|PubMed:17398095}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Guanine-nucleotide releasing factor;Lipid-binding;Membrane;Reference proteome;Tight junction | The protein encoded by this gene is a member of the PSCD family. Members of this family have identical structural organization that consists of an N-terminal coiled-coil motif, a central Sec7 domain, and a C-terminal pleckstrin homology (PH) domain. The coiled-coil motif is involved in homodimerization, the Sec7 domain contains guanine-nucleotide exchange protein (GEP) activity, and the PH domain interacts with phospholipids and is responsible for association of PSCDs with membranes. Members of this family appear to mediate the regulation of protein sorting and membrane trafficking. The encoded protein exhibits GEP activity in vitro with ARF1, ARF3, and ARF6 and is 83% homologous to CYTH1. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008]. | hsa:9266; | adherens junction [GO:0005912]; bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi membrane [GO:0000139]; growth cone [GO:0030426]; membrane [GO:0016020]; plasma membrane [GO:0005886]; guanyl-nucleotide exchange factor activity [GO:0005085]; inositol 1,4,5 trisphosphate binding [GO:0070679]; lipid binding [GO:0008289]; actin cytoskeleton organization [GO:0030036]; endocytosis [GO:0006897]; regulation of ARF protein signal transduction [GO:0032012] | 12641750_The overexpression of ARNO, another mammalian GEF, produces extensive neuritogenesis in Aplysia neurons 12920129_cytohesin 2 binds to IPCEF1, which modifies its activity 15277685_the N-terminal coiled-coil and parts of the Sec7 domain of cytohesin-2 are required for serum-mediated transcriptional activation in nonimmune cells 16027149_Endogenous levels of ARNO/cytohesin-2 present in HEK293 cells are sufficient and necessary for sustained activation of the MAP kinase signaling pathway 16484220_The transport and regulation of ARNO in polartized epithelial cells, and its interactions with ARF6 in endocytosis are reported. 17398095_Study shows that three related Arf-like GTPases Arl4a, Arl4c, and Arl4d, are able to recruit ARNO and other cytohesins to the plasma membrane by binding to their PH domains irrespective of whether they are in the diglycine or triglycine form. 17623778_data suggest cells ruffle upon CaSR (calcium sensing receptor)stimulation via a mechanism that involves translocation of beta-arrestin 1 pre-assembled with the CaSR or ARNO (Arf nucleotide binding site opener) 20016009_protein-protein interaction mediated by ARNO coiled-coil domain required for ARNO induced motility; coiled-coil domain promotes assembly of multiprotein complex containing ARNO and Dock180; assembly of complex requires coiled-coil domain, GRASP and IPCEF 20153292_Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO. 20525696_cytohesin-2, through a previously unexplored complex formation with paxillin, regulates preadipocyte migration; paxillin plays a previously unknown role as a scaffold protein of Arf guanine-nucleotide exchange factor 21118813_Arno behaves as a bistable switch, having an absolute requirement for activation by an Arf protein but, once triggered, becoming highly active through the positive feedback effect of Arf1-GTP. 21307348_Data that aldolase forms a complex with ARNO/Arf6 and the V-ATPase and that it may contribute to remodeling of the actin cytoskeleton. 22002459_role for endothelial ARNO in VEGF-dependent initiation of angiogenesis by regulation of VEGFR-2 internalization in endothelial cells, resulting in the activation of the Akt pathway, vessel permeability, and ultimately endothelial proliferation 22341462_ARNO in turn triggered WAVE regulatory complex recruitment and activation, which was dramatically enhanced when ARNO cooperated with Salmonella SopE. 22454518_The PH domains of cytohesin 2/ARNO and cytohesin 3/GRP1 are responsible for the differential effects of these proteins on cell adhesion to fibronectin. 22659138_Expression of CYTH2 mutant deficient of the EFh2 domain in cells also inhibits Arf6 activation and neurite extension. 23095975_Arf6/ ARNO signaling mediates phospholipase-D, ERK1/2 and cofilin activation in pancreatic beta-cells. 23255605_Kinetics of interaction between ADP-ribosylation factor-1 (Arf1) and the Sec7 domain of Arno guanine nucleotide exchange factor, modulation by allosteric factors, and the uncompetitive inhibitor brefeldin A 23288846_The N termini of a-subunit isoforms are involved in signaling between vacuolar H+-ATPase (V-ATPase) and cytohesin-2 23545718_There is an association between cytohesin-2 expression and overall survival and disease-free survival in patients with hepatocellular carcinoma. 24083777_Phosphorylation of Ser392 of ARNO stabilized the C-terminal alpha-helix via formation of salt bridges between phospho-Ser392 and Arg390, Lys395, and Lys396. 24581425_Cytohesin-2 constitutively suppresses platelet dense granule secretion and aggregation by keeping ARF6 in a GTP-bound state 24618737_inhibiting cytohesins or ARNO as cytoplasmic activators of EGFR and IGF-I in colorectal cancer resulted in anti-proliferation, reduced invasion, decreased migration, and suppressed growth in vivo and in vitro. 25736891_The authors show that Shigella flexneri IpgD phosphatase activity is required for recruitment of the ARF6 guanine nucleotide exchange factor (GEF) ARF nucleotide binding site opener (ARNO) to bacterial entry sites. 26378252_Data establish a role for cytohesin-2/ARNO as a regulator of R-Ras and integrin recycling and suggest that ARF-regulated trafficking of R-Ras is required for R-Ras-dependent effects on spreading and adhesion formation. 27203102_Authors' experiments argue against ARNO being a robust modifier of EGFR catalytic activity. 28007915_ARNO-ARF1 regulates formation of podosomes by inhibition of RhoA/myosin-II and promotion of actin core assembly. 30355448_INAVA-CUPID exhibits dual functions, coordinated directly by ARNO, that bridge epithelial barrier function with extracellular signals and inflammation. 31780432_Molecular Architecture of a Network of Potential Intracellular EGFR Modulators: ARNO, CaM, Phospholipids, and the Juxtamembrane Segment. | ENSMUSG00000003269 | Cyth2 | 1639.517180 | 0.8990718 | -0.153491800 | 0.07399679 | 4.286933e+00 | 3.840639e-02 | 2.643316e-01 | No | Yes | 1527.350840 | 165.153877 | 1723.450116 | 186.385082 | |
| ENSG00000105483 | 22900 | CARD8 | protein_coding | Q9Y2G2 | FUNCTION: Inflammasome sensor, which mediates inflammasome activation in response to various pathogen-associated signals, leading to subsequent pyroptosis of CD4(+) T-cells and macrophages (PubMed:11821383, PubMed:11408476, PubMed:15030775, PubMed:32840892, PubMed:32051255, PubMed:33542150, PubMed:34019797). Inflammasomes are supramolecular complexes that assemble in the cytosol in response to pathogens and other damage-associated signals and play critical roles in innate immunity and inflammation (PubMed:11821383, PubMed:11408476, PubMed:15030775). Acts as a recognition receptor (PRR): recognizes specific pathogens and other damage-associated signals, such as HIV-1 protease activity or Val-boroPro inhibitor, and mediates CARD8 inflammasome activation (PubMed:32840892, PubMed:33542150). In response to pathogen-associated signals, the N-terminal part of CARD8 is degraded by the proteasome, releasing the cleaved C-terminal part of the protein (Caspase recruitment domain-containing protein 8, C-terminus), which polymerizes to initiate the formation of the inflammasome complex: the CARD8 inflammasome directly recruits pro-caspase-1 (proCASP1) independently of PYCARD/ASC and promotes caspase-1 (CASP1) activation, which subsequently cleaves and activates inflammatory cytokines IL1B and IL18 and gasdermin-D (GSDMD), leading to pyroptosis (PubMed:33053349, PubMed:32840892, PubMed:32051255, PubMed:33542150). Ability to sense HIV-1 protease activity leads to the clearance of latent HIV-1 in patient CD4(+) T-cells after viral reactivation; in contrast, HIV-1 can evade CARD8-sensing when its protease remains inactive in infected cells prior to viral budding (PubMed:33542150). Also acts as a negative regulator of the NLRP3 inflammasome (PubMed:24517500). May also act as an inhibitor of NF-kappa-B activation (PubMed:11551959, PubMed:12067710). {ECO:0000269|PubMed:11408476, ECO:0000269|PubMed:11551959, ECO:0000269|PubMed:11821383, ECO:0000269|PubMed:12067710, ECO:0000269|PubMed:15030775, ECO:0000269|PubMed:24517500, ECO:0000269|PubMed:32051255, ECO:0000269|PubMed:32840892, ECO:0000269|PubMed:33053349, ECO:0000269|PubMed:33542150, ECO:0000269|PubMed:34019797}.; FUNCTION: [Caspase recruitment domain-containing protein 8]: Constitutes the precursor of the CARD8 inflammasome, which mediates autoproteolytic processing within the FIIND domain to generate the N-terminal and C-terminal parts, which are associated non-covalently in absence of pathogens and other damage-associated signals. {ECO:0000269|PubMed:22087307}.; FUNCTION: [Caspase recruitment domain-containing protein 8, N-terminus]: Regulatory part that prevents formation of the CARD8 inflammasome: in absence of pathogens and other damage-associated signals, interacts with the C-terminal part of CARD8 (Caspase recruitment domain-containing protein 8, C-terminus), preventing activation of the CARD8 inflammasome (PubMed:33542150). In response to pathogen-associated signals, this part is ubiquitinated by the N-end rule pathway and degraded by the proteasome, releasing the cleaved C-terminal part of the protein, which polymerizes and forms the CARD8 inflammasome (Probable) (PubMed:32558991). {ECO:0000269|PubMed:33542150, ECO:0000303|PubMed:32558991, ECO:0000305|PubMed:33053349}.; FUNCTION: [Caspase recruitment domain-containing protein 8, C-terminus]: Constitutes the active part of the CARD8 inflammasome (PubMed:32840892, PubMed:34019797). In absence of pathogens and other damage-associated signals, interacts with the N-terminal part of CARD8 (Caspase recruitment domain-containing protein 8, N-terminus), preventing activation of the CARD8 inflammasome (PubMed:33542150). In response to pathogen-associated signals, the N-terminal part of CARD8 is degraded by the proteasome, releasing this form, which polymerizes to form the CARD8 inflammasome complex: the CARD8 inflammasome complex then directly recruits pro-caspase-1 (proCASP1) and promotes caspase-1 (CASP1) activation, leading to gasdermin-D (GSDMD) cleavage and subsequent pyroptosis (PubMed:32840892, PubMed:33542150). {ECO:0000269|PubMed:32840892, ECO:0000269|PubMed:33542150, ECO:0000269|PubMed:34019797}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Disease variant;Host-virus interaction;Hydrolase;Immunity;Inflammasome;Inflammatory response;Innate immunity;Necrosis;Nucleus;Protease;Reference proteome;Ubl conjugation | The protein encoded by this gene belongs to the caspase recruitment domain (CARD)-containing family of proteins, which are involved in pathways leading to activation of caspases or nuclear factor kappa-B (NFKB). This protein may be a component of the inflammasome, a protein complex that plays a role in the activation of proinflammatory caspases. It is thought that this protein acts as an adaptor molecule that negatively regulates NFKB activation, CASP1-dependent IL1B secretion, and apoptosis. Polymorphisms in this gene may be associated with a susceptibility to rheumatoid arthritis. Alternatively spliced transcript variants have been described for this gene. [provided by RefSeq, May 2010]. | hsa:22900; | CARD8 inflammasome complex [GO:0140634]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; inflammasome complex [GO:0061702]; NLRP3 inflammasome complex [GO:0072559]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; CARD domain binding [GO:0050700]; cysteine-type endopeptidase activator activity [GO:0140608]; cysteine-type endopeptidase activator activity involved in apoptotic process [GO:0008656]; endopeptidase activity [GO:0004175]; NACHT domain binding [GO:0032089]; pattern recognition receptor activity [GO:0038187]; peptidase activity [GO:0008233]; protein homodimerization activity [GO:0042803]; protein self-association [GO:0043621]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; antiviral innate immune response [GO:0140374]; CARD8 inflammasome complex assembly [GO:0140633]; defense response to virus [GO:0051607]; inflammatory response [GO:0006954]; inhibition of cysteine-type endopeptidase activity [GO:0097340]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of interleukin-1 beta production [GO:0032691]; negative regulation of lipopolysaccharide-mediated signaling pathway [GO:0031665]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of NLRP3 inflammasome complex assembly [GO:1900226]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; positive regulation of interleukin-1 beta production [GO:0032731]; protein homooligomerization [GO:0051260]; pyroptosis [GO:0070269]; regulation of apoptotic process [GO:0042981]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; self proteolysis [GO:0097264] | 11821383_CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. (card-8 protein) 11956601_Expression and characterization of NDPP1 12067710_TUCAN/CARDINAL and DRAL participate in a common pathway for modulation of NF-kappaB activation. 16796750_TUCAN does not play a role in inhibition of procaspase-9 and in determining the sensitivity to cisplatin in Non Small Cells Lung Cancer. 17030188_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17030188_association between a likely functional polymorphism in TUCAN and Crohn disease 17484912_no significant association between the risk allele 'A' at Cys10Stop and risk for Crohn's disease or ulcerative colitis was detected in patients of German and Norwegian descent 17595233_Observational study of gene-disease association. (HuGE Navigator) 17878386_deleterious polymorphism of CARD8 may help predict the severity of rheumatoid arthritis 18092344_Analyzing 3 independent European IBD cohorts, we found no evidence that the C10X variant in CARD8 confers susceptibility for CD. 18092344_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18212821_isoforms of CARD8 differ in their N-termini, resulting in diverse predicted molecular weights (47, 48, 51, 54 and 60 kDa) and multiple outcomes for the variant including Cys10Stop, Cys34Stop, Phe52Ile and Phe102Ile 18263599_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18311798_Observational study of genotype prevalence. (HuGE Navigator) 18311798_The NALP3 and TUCAN single-nucleotide polymorphisms may explain the increased IL-1beta levels and inflammatory symptoms observed, but further studies are needed to reveal a functional relationship. 18841008_Observational study of gene-disease association. (HuGE Navigator) 18841008_Women, but not men, carrying the CARD8 AA genotype (truncated protein) had a 2.39-fold higher risk of developing Alzheimer's disease than subjects with the CARD8 TT genotype (full-length protein) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19252766_Gene-gene interaction between CARD8 and interleukin-6 reduces Alzheimer's disease risk. 19252766_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19319132_CARD8 and NALP3 genes combined polymorphism has a role of developing Crohn disease in men. 19319132_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19443463_Carriage of CARD8-X is associated with a worse disease course in early rheumatoid arthritis. 19664744_Observational study of gene-disease association. (HuGE Navigator) 19843337_Observational study of gene-disease association. (HuGE Navigator) 19843337_Variation in the innate immunity genes CARD4, CARD8 and CARD15 is unlikely to play a major role in the susceptibility to CRC in the German population. 19940360_Study shows an essential role for apoptosis signal-regulating kinase 1 (ASK1), together with both c-jun-N-terminal kinase (JNK) and p38 pathways, and caspase-8 in Fas-induced apoptosis. 20182451_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20182451_propose that CARD8-NALP3 genotype combinations protect against gut inflammation by preventing the NALP3 inflammasome from producing excessive interleukin-1be 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20385562_Data show that CARD8 represents a novel molecular switch involved in the endogenous regulation of NOD2-dependent inflammatory processes in epithelial cells. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 21248762_CARD8 variants might have roles in the pathogenesis of Crohn's disease and ulcerative colitis in Koreans 21308686_Caspase8 polymorphism IVS12-19 G>A but not CASP8 -652 6N del polymorphism may modulate risk of esophageal squamous cell carcinoma and its survival outcome in northern Indian population. 21621776_analysis of interaction of the inflammasome genes CARD8 and NLRP3 in abdominal aortic aneurysms 22087307_results identify a function for the FIIND and show that CARD8 and NLRP1 are ZU5-UPA domain-containing autoproteolytic proteins, thus suggesting a novel mechanism for regulating innate immune responses 22128899_Variations in the CARD8 and NLRP3 genes are not associated with rheumatoid arthritis in the French as well general Tunisian population. 22227487_Twelve single nucleotide polymorphisms within NLRP1, NLRP3, NLRC4, CARD8, CASP1, and IL1B genes were analyzed in 150 HIV-1-infected Brazilian subjects. 23053059_we performed a genetic association study in patients with pneumococcal meningitis and found that single-nucleotide polymorphisms in the inflammasome genes CARD8 and NLRP1 are associated with poor disease outcome. 23088220_Results suggest that the CARD8 rs2043211 gene variant does not in susceptibility to rheumatoid arthritis (RA) or in the development of cardiovascular disease in patients with RA. 23506543_Mutation in the CARD8, a component of inflammasome, is associated with lower levels of antibodies directed to mannans and glucans at least in Crohn's disease patients. 23507658_The results of this study support the novel association between CARD8 gene and HIV+tuberculosis coinfection, demonstrating that inflammasome genetics could influence HIV-1 infection and the development of opportunistic infection. 23547871_In a Swedish population, the minor allele of CARD8-C10X is associated with a decreased risk of AS, but not with levels of faecal calprotectin or disease phenotype. 23563199_Genetic variation in the inflammasome affects atopic dermatitis susceptibility. 23611467_Data indicate that CARD8 (caspase recruitment domain 8) mRNA was highly expressed in atherosclerotic plaques, and the minor allele was associated with lower expression of CARD8 in the plaques, suggesting that CARD8 may promote inflammation. 23695559_The first crystal structure of the CARD8 caspase-recruitment domain is reported. It adopts a six-helix bundle fold with a unique conformation of the alpha6 helix that is described for the first time. 24098386_levels of inflammasome-produced cytokines as a measure of inflammasome activation in healthy individuals carrying Q705K polymorphism in the NLRP3 gene combined with C10X in the CARD8 gene 24385277_ANRIL may increase the risk of ischemic stroke through regulation of the CARD8 pathway 24517500_we show that CARD8 plays a role as a negative regulator of NLRP3 inflammasome through its binding with NLRP3 25564880_genetic polymorphism is associated with susceptibility to Crohn's disease under the dominant model and homozygote contrast in the European population; meta-analysis 25790751_Patients carrying genotype TT of CARD8 rs2043211 polymorphism had higher triglycerides levels compared to those carrying the AA genotype. 25895569_The polymorphism of rs2043211 in CARD8 may be a relevant host susceptibility factor for the development of preeclampsia in the Chinese Han population. 25921775_CARD8 might not play a role in the pathogenesis of Tourette syndrome in Chinese Han population 26095808_Data indicate 3 variants in 3 novel genes myc target 1 protein (MYCT1), caspase recruitment domain family member 8 (CARD8) and zinc finger protein 543 (ZNF543), associated with familial IgA nephropathy (IgAN). 26283210_A novel association between CARD8 and increased risk of surgical recurrence in Crohn's disease was observed and CARD8 could be a new marker for risk stratification and prevention of recurrent surgery. 26462562_There is evidence for association of gout with functional variants in CARD8, IL1B and CD14. 26462578_In patients with ileal, stenotic or fistulizing Crohn's disease, the mutant-type CARD8 rs2043211 polymorphism may generate a potentially protective effect. (Meta-analysis) 27110561_The NLRP3 rs10754558 gene polymorphism was significantly associated with the occurrence of CAD, while the CARD8 rs2043211 gene polymorphism was not involved. 27550484_The CARD8 rs2043211 genetic variants was not implicated in the development of gout in the male Korean population. However, we found that in a pair-wise comparison of the CA/TT P2X7R and CARD8 genotype combination was shown to have an increased trend for the risk of gout. 27810076_High CARD8 expression is associated with malignant melanoma. 28135700_The over expression of CARD8 due to higher promoter activity of the TT genotype may result in a dramatically inflammatory immune response and then increase the susceptibility to Arteriosclerosis Obliterans. 28137891_these results identify a new CARD8 variant associated with periodic fever with aphthous stomatitis, pharyngitis, and cervical adenitis, and further suggest that disruption of the interaction between CARD8 and NLRP3 can regulate autoinflammation in patients 28185410_Review/Meta-analysis: CARD8 p.C10X SNP were not associated with the susceptibility to rheumatoid arthritis. 29097263_this study found that the AT genotype of CARD8 (rs2043211) was significantly higher compared to TT genotype in high and intermediate risk chronic myeloid leukemia patients 29230505_Our results showed that rs10754558 NLRP3 and rs2043211 CARD8 polymorphisms are associated with rheumatoid arthritis development (p value = 0.044, OR = 1.77, statistical power = 0.999) and severity measured by Health Assessment Questionnaire (HAQ) (p value = 0.03), respectively. 29408806_inflammasome activation studies revealed that intact but not mutated CARD8 prevented NLRP3 deubiquitination and serine dephosphorylation. Crohn's disease (CD) due to a CARD8 mutation was not effectively treated by anti-TNF-alpha, but did respond to IL-1beta inhibitors. Thus, patients with anti-TNF-alpha-resistant CD may respond to this treatment option. 30088494_The study showed that CARD8 rs2043211 polymorphism is associated with cardiovascular diseases (systematic review and meta-analysis). 30211233_this study showed that the CARD8-C10X (rs2043211) AT genotype contributed to the susceptibility of multiple myeloma 30816317_Polymorphisms in CARD8 and NLRP3 are associated with extrapulmonary TB and poor clinical outcome in active TB in Ethiopia. 31529732_Patients with combined wild type of both NLRP3 rs35829419 and CARD8 rs2043211 had significantly smaller odds for early implant loosening (odds ratio 0.33, p = 0.02). 32080163_Relationship of CARD8 Gene Polymorphisms with Susceptibility to Ankylosing Spondylitis: A Case-Control Study. 32104685_A New Risk Polymorphism rs10403848 of CARD8 Significantly Associated with Psoriasis Vulgaris in Northeastern China. 32396255_Association of CARD8 Activating Polymorphism With Bone Erosion in Cholesteatoma Patients. 32613553_SNP rs2043211 (p.C10X) in CARD8 Is Associated with Large-Artery Atherosclerosis Stroke in a Chinese Population. 32689633_Combined polymorphisms in genes encoding the inflammasome components NLRP3 and CARD8 confer risk of ischemic stroke in men. 32840892_CARD8 inflammasome activation triggers pyroptosis in human T cells. 33053349_Activation of the CARD8 Inflammasome Requires a Disordered Region. 33067783_CARD8 polymorphism rs2043211 protects against noise-induced hearing loss by causing the dysfunction of CARD8 protein. 33154409_Expression of CARD8 in human atherosclerosis and its regulation of inflammatory proteins in human endothelial cells. 33164551_Genetic variation in CARD8, a gene coding for an NLRP3 inflammasome-associated protein, alters the genetic risk for diabetic nephropathy in the context of type 2 diabetes mellitus. 33420028_Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8. 33420033_Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. 33542150_CARD8 is an inflammasome sensor for HIV-1 protease activity. 34019797_Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. 34587665_Associations of NLRP3 and CARD8 gene polymorphisms with alcohol dependence and commonly related psychiatric disorders: a preliminary study. | 829.538739 | 1.1142117 | 0.156023434 | 0.11012152 | 2.008441e+00 | 1.564260e-01 | 5.148104e-01 | No | Yes | 845.399766 | 126.608086 | 738.831373 | 110.791271 | |||
| ENSG00000105676 | 93436 | ARMC6 | protein_coding | Q6NXE6 | Alternative splicing;Methylation;Phosphoprotein;Reference proteome;Repeat | The function of this gene's protein product has not been determined. A related protein in mouse suggests that this protein has a conserved function. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2010]. | hsa:93436; | cytosol [GO:0005829]; hematopoietic progenitor cell differentiation [GO:0002244] | ENSMUSG00000002343 | Armc6 | 2020.561732 | 0.9501802 | -0.073727027 | 0.07624040 | 9.243255e-01 | 3.363418e-01 | 6.956518e-01 | No | Yes | 2020.346591 | 300.904511 | 2160.459559 | 321.576240 | |||
| ENSG00000105738 | 23094 | SIPA1L3 | protein_coding | O60292 | FUNCTION: Plays a critical role in epithelial cell morphogenesis, polarity, adhesion and cytoskeletal organization in the lens (PubMed:26231217). {ECO:0000269|PubMed:26231217}. | Acetylation;Cataract;Cell membrane;Chromosomal rearrangement;Coiled coil;GTPase activation;Membrane;Phosphoprotein;Reference proteome | This gene belongs to the signal induced proliferation associated 1 family of genes, which encode GTPase-activating proteins specific for the GTP-binding protein Rap1. Rap1 has been implicated in regulation of cell adhesion, cell polarity, and organization of the cytoskeleton. Like other members of the family, the protein encoded by this gene contains RapGAP and PDZ domains. In addition, this protein contains a C-terminal leucine zipper domain. This gene is proposed to function in epithelial cell morphogenesis and establishment or maintenance of polarity. Consistently, expression of the protein in cell culture showed localization to cell-cell borders in apical regions, and downregulation of the gene in 3D Caco2 cell culture resulted in abnormal cell polarity and morphogenesis. Allelic variants of this gene have been associated with congenital cataracts in humans. [provided by RefSeq, Feb 2016]. | hsa:23094; | apical part of cell [GO:0045177]; apical plasma membrane [GO:0016324]; cytoplasm [GO:0005737]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; stress fiber [GO:0001725]; tricellular tight junction [GO:0061689]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; cytoskeleton organization [GO:0007010]; epithelial cell morphogenesis [GO:0003382]; establishment of epithelial cell polarity [GO:0090162]; eye development [GO:0001654]; hematopoietic progenitor cell differentiation [GO:0002244]; regulation of small GTPase mediated signal transduction [GO:0051056] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25804400_Whole-exome sequencing identified a homozygous nonsense variant in SIPA1L3 in two sisters with congenital cataract. 26231217_Abnormalities of SIPA1L3 contribute to lens and eye defects, and we identify a critical role for SIPA1L3 in epithelial cell morphogenesis, polarity, adhesion and cytoskeletal organization. 30924596_smoking cessation benefited survival of LUAD patients with low methylation at cg02268510SIPA 1L3 . The results have implications for not only smoking cessation after diagnosis, but also possible methylation-specific drug targeting. | ENSMUSG00000030583 | Sipa1l3 | 943.193733 | 0.7976178 | -0.326230534 | 0.09460162 | 1.183525e+01 | 5.811977e-04 | 2.557270e-02 | No | Yes | 790.996890 | 97.994855 | 979.171222 | 121.250726 | |
| ENSG00000105866 | 6671 | SP4 | protein_coding | Q02446 | FUNCTION: Binds to GT and GC boxes promoters elements. Probable transcriptional activator. | Activator;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene is a transcription factor that can bind to the GC promoter region of a variety of genes, including those of the photoreceptor signal transduction system. The encoded protein binds to the same sites in promoter CpG islands as does the transcription factor SP1, although its expression is much more restricted compared to that of SP1. This gene may be involved in bipolar disorder and schizophrenia. [provided by RefSeq, May 2016]. | hsa:6671; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; regulation of transcription by RNA polymerase II [GO:0006357] | 11943774_Sp4-mediated differential activation of the beta-PDE transcription defines the first specific Sp4 target gene reported 15345676_DIM-C-pPhCF3 induced p21 expression through a novel mechanism that involves PPARgamma interactions with both Sp1 and Sp4 proteins bound to the proximal GC-rich region of the p21 promoter. 15788387_has an intimate part in transcriptional regulation of the lh receptor gene. 16574784_Sp3 and Sp4 cooperatively interact with ERalpha to activate VEGFR2 17356515_Observational study of genotype prevalence. (HuGE Navigator) 17356515_While digenic disease with the SP4 Asn306Ser and the GNB1 intronic variant alleles has not been established, neither has it been ruled out. This leaves open the possibility of a cooperative involvement of SP4 and GNB1 in the normal function of the retina. 18593936_Curcumin also decreased bladder tumor growth in athymic nude mice bearing KU7 cells as xenografts and this was accompanied by decreased Sp1, Sp3, and Sp4 protein levels in tumors. 19401786_Observational study of gene-disease association. (HuGE Navigator) 19401786_Out of ten SNPs selected from the SP4 genomic locus, four displayed significant association with bipolar disorder in European Caucasian families. 19656107_Association of A80807T polymorphism of the transcriptional factor Sp4 gene with PT ICA was established. 19656107_Observational study of gene-disease association. (HuGE Navigator) 19934275_Data show that Sp4 was constitutively bound to the GC-box in the proximal region of the AS promoter regardless of arginine availability in all three cell lines. 20125088_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20298200_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20487506_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20538607_Results of Sp1, Sp3, and Sp4 knockdown by RNA interference demonstrate that both p50 and p65 are Sp-regulated genes. 20634195_Sp4 hypomorphic mice could therefore serve as a genetic model to investigate impaired NMDA functions resulting from loss-of-function mutations of human SP4 gene in schizophrenia and/or other psychiatric disorders. 21919647_Ascorbic acid decreased RKO and SW480 colon cancer cell proliferation and induced apoptosis and necrosis, accompanied by downregulation of Sp1, Sp3, and Sp4 proteins. 22017217_This study demonistrated that The transcription factor SP4 is reduced in postmortem cerebellum of bipolar disorder subjects. 22614877_Significantly increased SP4 relative expression levels are observed in patients with Alzheimer's disease, compared with controls. 23540600_This study showed that SP1, SP3 and SP4 protein levels inversely correlated with negative symptoms in the cerebellum. 23941741_This study shows reduced SP4 protein levels in first-episode psychosis in lymphocytes, suggesting that these transcription factors are potential peripheral biomarkers of psychotic spectrum disorders in the early stages. 25175639_The results of this study suggest that SP4 and SP1 upregulation may be part of the mechanisms deregulated downstream of glutamate signalling pathways in schizophrenia 25915526_Results found that SP4 S770 phosphorylation was significantly increased in lymphocytes in first-episode psychosis compared to controls. 26049820_Results indicate that SP4 S770 phosphorylation is increased in the cerebellum in bipolar disorder that committed suicide and in severe schizophrenia, and may be part of a degradation signal that controls Sp4 abundance in cerebellar granule neurons 26431879_O-GlcNAc modification of Sp3 and Sp4, but not Sp2 transcription factors negatively regulates their transcriptional activities. 26450579_Common risk factors in the SP4 gene are associated with schizophrenia, although not with MDD, in the Han Chinese population. 26967243_Sp1, Sp3 and Sp4 are non-oncogene addiction (NOA) genes and are attractive drug targets for individual and combined cancer chemotherapies. 28081371_IN TMD patients, a novel locus at genome-wide level of significance (rs73460075, OR = 0.56, P = 3.8 x 10(-8)) in the intron of the dystrophin gene DMD (X chromosome), and a suggestive locus on chromosome 7 (rs73271865, P = 2.9 x 10(-7)) upstream of the Sp4 Transcription Factor ( SP4) gene were identified in the discovery cohort, but neither of these was replicated. | ENSMUSG00000025323 | Sp4 | 609.237065 | 1.0366548 | 0.051935497 | 0.11617156 | 2.020021e-01 | 6.531096e-01 | 8.755649e-01 | No | Yes | 694.544053 | 136.788194 | 656.143607 | 129.387034 | |
| ENSG00000106078 | 23242 | COBL | protein_coding | O75128 | FUNCTION: Plays an important role in the reorganization of the actin cytoskeleton. Regulates neuron morphogenesis and increases branching of axons and dendrites. Regulates dendrite branching in Purkinje cells (By similarity). Binds to and sequesters actin monomers (G actin). Nucleates actin polymerization by assembling three actin monomers in cross-filament orientation and thereby promotes growth of actin filaments at the barbed end. Can also mediate actin depolymerization at barbed ends and severing of actin filaments. Promotes formation of cell ruffles. {ECO:0000250, ECO:0000269|PubMed:21816349}. | 3D-structure;Actin-binding;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Membrane;Phosphoprotein;Reference proteome;Repeat | This gene encodes a protein that contains WH2 domains (WASP, Wiskott-Aldrich syndrome protein, homology domain-2) that interact with actin. The encoded actin regulator protein is required for growth and assembly of brush border microvilli that play a role in maintaining intestinal homeostasis. A similar protein in mouse functions in midbrain neural tube closure. A pseudogene of this gene is located on chromosome X. [provided by RefSeq, Oct 2016]. | hsa:23242; | actin filament [GO:0005884]; axon [GO:0030424]; axonal growth cone [GO:0044295]; cell cortex [GO:0005938]; dendrite [GO:0030425]; dendritic growth cone [GO:0044294]; membrane [GO:0016020]; neuronal cell body [GO:0043025]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; terminal web [GO:1990357]; actin monomer binding [GO:0003785]; actin filament network formation [GO:0051639]; actin filament polymerization [GO:0030041]; collateral sprouting in absence of injury [GO:0048669]; digestive tract development [GO:0048565]; embryonic axis specification [GO:0000578]; floor plate development [GO:0033504]; liver development [GO:0001889]; neural tube closure [GO:0001843]; notochord development [GO:0030903]; positive regulation of dendrite development [GO:1900006]; positive regulation of ruffle assembly [GO:1900029]; somite specification [GO:0001757] | 19058789_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21816349_Cordon-Bleu works as a dynamizer of actin assembly by combining many properties of profilin with weak filament nucleating and powerful filament severing activities and sequestration of ADP-actin, which generate oscillatory polymerization kinetics. 24415668_Filament severing displayed the greatest stringency and was abolished in all mutated forms of Cobl-KAB. 26334624_Ca2+/Calmodulin modulates Cobl's actin binding properties and furthermore promotes Cobl's previously identified interactions with the membrane-shaping F-BAR protein syndapin I, which accumulated with Cobl at nascent dendritic protrusion sites. 27419633_Data show that six patients had large interstitial deletions starting within intronic regions of COBL at diagnosis, which is ~611 Kb downstream of IKZF1, suggesting that COBL is a hotspot for IKZF1 deletion (DeltaIKZF1). 27770636_Two single-nucleotide polymorphisms at novel loci, rs112404845 (P = 3.8 x 10-8), upstream of COBL, and rs16961023 (P = 4.6 x 10-8), downstream of SLC10A2, obtained genome-wide significant evidence of association with the posterior liability of late-onset Alzheimer's disease in African Americans. | ENSMUSG00000020173 | Cobl | 525.386712 | 0.9831875 | -0.024461529 | 0.12998800 | 3.576163e-02 | 8.500086e-01 | 9.507016e-01 | No | Yes | 533.365449 | 59.624341 | 543.469234 | 60.729127 | |
| ENSG00000106133 | 260294 | NSUN5P2 | transcribed_unprocessed_pseudogene | This locus represents a transcribed pseudogene of a nearby locus on chromosome 7, which encodes a putative methyltransferase. There is also a third closely related pseudogene locus in this region. There is extensive alternative splicing at this locus. [provided by RefSeq, Jul 2013]. | 811.322634 | 1.0687915 | 0.095980487 | 0.12120387 | 6.118426e-01 | 4.340949e-01 | 7.672775e-01 | No | Yes | 774.752642 | 103.216644 | 758.316878 | 100.987722 | |||||||||
| ENSG00000106477 | 95681 | CEP41 | protein_coding | Q9BYV8 | FUNCTION: Required during ciliogenesis for tubulin glutamylation in cilium. Probably acts by participating in the transport of TTLL6, a tubulin polyglutamylase, between the basal body and the cilium. {ECO:0000269|PubMed:22246503}. | Alternative splicing;Autism;Autism spectrum disorder;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Joubert syndrome;Methylation;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a centrosomal and microtubule-binding protein which is predicted to have two coiled-coil domains and a rhodanese domain. In human retinal pigment epithelial cells the protein localized to centrioles and cilia. Mutations in this gene have been associated with Joubert Syndrome 15; an autosomal recessive ciliopathy and neurological disorder. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2012]. | hsa:95681; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytosol [GO:0005829]; membrane [GO:0016020]; cilium assembly [GO:0060271]; protein polyglutamylation [GO:0018095]; protein transport [GO:0015031] | 21438139_Three rare potentially pathogenic variants were identified in the TSGA14 gene, which encodes a centrosomal protein. 22246503_The data identified CEP41 mutations as a cause of Joubert syndrome and implicated tubulin post-translational modification in the pathogenesis of human ciliary dysfunction. 22456293_In cortices, the MEST promoter was hemimethylated, as expected for a differentially methylated imprinting control region, whereas the COPG2 and TSGA14 promoters were completely demethylated, typical for transcriptionally active non-imprinted genes. 30664616_Missense variants in one gene, CEP41, associated significantly with Autism Spectrum Disorder (ASD). Homozygous gene-disrupting variants in CEP41 were initially found to be responsible for recessive Joubert syndrome. 31885126_CEP41-mediated ciliary tubulin glutamylation drives angiogenesis through AURKA-dependent deciliation. | ENSMUSG00000029790 | Cep41 | 731.165663 | 0.9570098 | -0.063394355 | 0.13807341 | 2.103185e-01 | 6.465179e-01 | 8.721045e-01 | No | Yes | 788.674180 | 107.991656 | 840.627362 | 115.031659 | |
| ENSG00000106948 | 80709 | AKNA | protein_coding | Q7Z591 | FUNCTION: Centrosomal protein that plays a key role in cell delamination by regulating microtubule organization (By similarity). Required for the delamination and retention of neural stem cells from the subventricular zone during neurogenesis (By similarity). Also regulates the epithelial-to-mesenchymal transition in other epithelial cells (By similarity). Acts by increasing centrosomal microtubule nucleation and recruiting nucleation factors and minus-end stabilizers, thereby destabilizing microtubules at the adherens junctions and mediating constriction of the apical endfoot (By similarity). In addition, may also act as a transcription factor that specifically activates the expression of the CD40 receptor and its ligand CD40L/CD154, two cell surface molecules on lymphocytes that are critical for antigen-dependent-B-cell development (PubMed:11268217). Binds to A/T-rich promoters (PubMed:11268217). It is unclear how it can both act as a microtubule organizer and as a transcription factor; additional evidences are required to reconcile these two apparently contradictory functions (Probable). {ECO:0000250|UniProtKB:Q80VW7, ECO:0000269|PubMed:11268217, ECO:0000305}. | Activator;Alternative splicing;Cytoplasm;Cytoskeleton;DNA-binding;Microtubule;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | hsa:80709; | centriole [GO:0005814]; centrosome [GO:0005813]; cytosol [GO:0005829]; fibrillar center [GO:0001650]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; microtubule [GO:0005874]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; delamination [GO:0060232]; epithelial to mesenchymal transition [GO:0001837]; neuroblast delamination [GO:0060234]; neuroblast division in subventricular zone [GO:0021849]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of inflammatory response [GO:0050727] | 15869410_AKNA expresses multiple transcripts and protein isoforms as a result of alternative promoter usage, splicing, and polyadenylation. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20507260_Data indicate that AKNA appears to be an important genetic factor associated with the risk cervical cancer. 20507260_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21297967_significantly decreased expression of CD18 and AKNA suggests a role for both proteins in the pathogenesis of Vogt-Koyanagi-Harada syndrome 25373726_Results support the hypothesis that akna is a cervical cancer susceptibility genetic factor and suggest that akna transcriptional regulation has a role in the disease; identified an association between high akna expression levels and cervical cancer and squamous intraepithelial lesion, but its direction differs in each disease stage. 28484714_identified HIF1A Pro582Ser T allele and C/T genotype as well as AKNA -1372C>A polymorphism A/A genotype as genetic factors associated with Primary Sjogren's Syndrome. 29368274_Authors suggest that regulatory and coding polymorphisms of the inflammatory modulator gene AKNA can influence the development of KOA. 29618620_We studied the expression profile of the 60 genes located at that genomic region. POLE3 and AKNA were the only two genes deregulated in resistant tumors harboring the 9q32-q33.1 gain 30787442_Epithelial-like neural stem cells divide in the ventricular zone at the ventricles of the embryonic brain, self-renew and generate basal progenitors that delaminate and settle in the subventricular zone in enlarged brain regions; the length of time that cells stay in the subventricular zone is essential for controlling further amplification and fate determination; interphase AKNA has a key role in this process 32367404_An exome-first approach to aid in the diagnosis of primary ciliary dyskinesia. 32460535_Evaluation of Common Variants in the AKNA Gene and Susceptibility to Knee Osteoarthritis Among the Han Chinese. 32462010_AKNA Is a Potential Prognostic Biomarker in Gastric Cancer and Function as a Tumor Suppressor by Modulating EMT-Related Pathways. 34680889_A Homozygous AKNA Frameshift Variant Is Associated with Microcephaly in a Pakistani Family. | ENSMUSG00000039158 | Akna | 122.273955 | 1.1047099 | 0.143667616 | 0.25005752 | 3.313817e-01 | 5.648466e-01 | No | Yes | 120.661500 | 15.218823 | 106.008041 | 13.158763 | |||
| ENSG00000107521 | 3257 | HPS1 | protein_coding | Q92902 | FUNCTION: Component of the BLOC-3 complex, a complex that acts as a guanine exchange factor (GEF) for RAB32 and RAB38, promotes the exchange of GDP to GTP, converting them from an inactive GDP-bound form into an active GTP-bound form. The BLOC-3 complex plays an important role in the control of melanin production and melanosome biogenesis and promotes the membrane localization of RAB32 and RAB38 (PubMed:23084991). {ECO:0000269|PubMed:23084991}. | Albinism;Alternative splicing;Disease variant;Guanine-nucleotide releasing factor;Hermansky-Pudlak syndrome;Reference proteome;Repeat;Sensory transduction;Vision | This gene encodes a protein that may play a role in organelle biogenesis associated with melanosomes, platelet dense granules, and lysosomes. The encoded protein is a component of three different protein complexes termed biogenesis of lysosome-related organelles complex (BLOC)-3, BLOC4, and BLOC5. Mutations in this gene are associated with Hermansky-Pudlak syndrome type 1. Alternative splicing results in multiple transcript variants. A pseudogene related to this gene is located on chromosome 22. [provided by RefSeq, Aug 2015]. | hsa:3257; | BLOC-3 complex [GO:0031085]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; lysosome [GO:0005764]; guanyl-nucleotide exchange factor activity [GO:0005085]; protein dimerization activity [GO:0046983]; lysosome organization [GO:0007040]; melanosome assembly [GO:1903232]; response to stimulus [GO:0050896]; vesicle-mediated transport [GO:0016192]; visual perception [GO:0007601] | 12125811_Description of mutations in HPS genes that cause Hermansky-Pudlak syndrome (review) 12442288_Four novel mutations were discovered and the diagnosis of HPS-1, available only on molecular grounds, has important prognostic and treatment implications. 12663659_identification as a component of two complexes, BLOC-3 and BLOC-4, involved in the biogenesis of lysosome-related organelles 12756248_observations demonstrate that the Hermansky-Pudlak syndrome 1(HPS1) and HPS4 proteins are components of a cytosolic complex that is involved in the biogenesis of lysosomal-related organelles 12847290_Biogenesis of lysosome-related organelles complex 3 (BLOC-3): a complex containing the Hermansky-Pudlak syndrome (HPS) proteins HPS1 and HPS4. 16020891_Mutations in this gene are associated with Hermansky-Pudlak syndrome type 1. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16417222_Observational study of genotype prevalence. (HuGE Navigator) 16431308_Observational study of genotype prevalence. (HuGE Navigator) 17975119_Observational study of gene-disease association. (HuGE Navigator) 18463683_Observational study of gene-disease association. (HuGE Navigator) 19023099_Observational study of gene-disease association. (HuGE Navigator) 19320733_Observational study of gene-disease association. (HuGE Navigator) 19665357_The first case report of a Chinese Hermansky-Pudlak syndrome patient with a novel mutation on HPS1 gene. 19865097_Observational study of gene-disease association. (HuGE Navigator) 20048159_Data show that recombinant HPS1-HPS4 produced in insect cells can be efficiently isolated as a 1:1 heterodimer, and might function as a Rab9 effector in the biogenesis of lysosome-related organelles. 20514622_Three different mutations in the HPS1 gene were found in the two families. 20662851_a previously unreported missense mutation (G313S) at the 3' splice junction of exon 10 of Hermansky-Pudlak syndrome 1 protein resulted in activation of a cryptic intronic splice site causing an aberrantly spliced HPS1 mRNA 21833017_Seven mutations (six previously unreported) were described in the HPS1, HPS4, and HPS5 genes among Hermansky-Pudlak Syndrome patients of Mexican, Uruguayan, Honduran, Cuban, Venezuelan, and Salvadoran ancestries. 23084991_BLOC-3 is a Rab32 and Rab38 guanine nucleotide exchange factor, with a specific function in the biogenesis of lysosome-related organelles. Silencing of the BLOC-3 subunits Hps1 and Hps4 results in the mislocalization of Rab32 and Rab38. 27593200_HPS1 mutation is associated with high hypopigmentation in Hermansky-Pudlak syndrome. 33224134_HPS1 Regulates the Maturation of Large Dense Core Vesicles and Lysozyme Secretion in Paneth Cells. 33878481_Hermansky-Pudlak syndrome: Five Chinese patients with novel variants in HPS1 and HPS6. 34736469_Modeling of lung phenotype of Hermansky-Pudlak syndrome type I using patient-specific iPSCs. 35328057_Delineating Novel and Known Pathogenic Variants in TYR, OCA2 and HPS-1 Genes in Eight Oculocutaneous Albinism (OCA) Pakistani Families. | ENSMUSG00000025188 | Hps1 | 1692.820429 | 0.9388297 | -0.091064669 | 0.07642501 | 1.425924e+00 | 2.324308e-01 | 6.045845e-01 | No | Yes | 1777.501652 | 169.225410 | 1891.525849 | 179.887865 | |
| ENSG00000107560 | 22841 | RAB11FIP2 | protein_coding | Q7L804 | FUNCTION: A Rab11 effector binding preferentially phosphatidylinositol 3,4,5-trisphosphate (PtdInsP3) and phosphatidic acid (PA) and acting in the regulation of the transport of vesicles from the endosomal recycling compartment (ERC) to the plasma membrane. Involved in insulin granule exocytosis. Also involved in receptor-mediated endocytosis and membrane trafficking of recycling endosomes, probably originating from clathrin-coated vesicles. Required in a complex with MYO5B and RAB11 for the transport of NPC1L1 to the plasma membrane. Also acts as a regulator of cell polarity. Plays an essential role in phagocytosis through a mechanism involving TICAM2, RAC1 and CDC42 Rho GTPases for controlling actin-dynamics. {ECO:0000269|PubMed:12364336, ECO:0000269|PubMed:15304524, ECO:0000269|PubMed:16251358, ECO:0000269|PubMed:16775013, ECO:0000269|PubMed:19542231, ECO:0000269|PubMed:30883606}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Endosome;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | hsa:22841; | cell projection [GO:0042995]; cytoplasmic vesicle membrane [GO:0030659]; endosome [GO:0005768]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; phagocytic cup [GO:0001891]; recycling endosome membrane [GO:0055038]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; protein kinase binding [GO:0019901]; small GTPase binding [GO:0031267]; establishment of cell polarity [GO:0030010]; insulin secretion involved in cellular response to glucose stimulus [GO:0035773]; phagocytosis [GO:0006909]; positive regulation of GTPase activity [GO:0043547]; regulated exocytosis [GO:0045055]; TRAM-dependent toll-like receptor 4 signaling pathway [GO:0035669] | 11994279_Rab11-FIP2 functions downstream of Rab11 in endosomal trafficking. 16775013_Phosphorylation of Rab11-FIP2 on serine 227 by MARK2 regulates an alternative pathway modulating the establishment of epithelial polarity 17626244_Rab11-FIP2 regulates trafficking at multiple points within the apical recycling system of polarized cells 18621683_the FIP2 C2 mutant caused a failure at the final budding step in the RSV virus life cycle 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22420646_Rab11A/myosin Vb/Rab11-FIP2 complex frames two late recycling steps of langerin from the ERC to the plasma membrane 23006599_The recruitment of FIP2 is specific since other members of the Rab11-Family of Interacting Proteins do not associate with the chlamydial inclusions. 24372966_Rab11-FIP2 binding to MYO5B is essential for vesicular trafficking in Hela and MDCK cells. 26502090_our findings reveal a novel mechanism underlying the role of Rab11-FIP2 in gastric cancer dissemination 26792722_Our data suggested a potential role of Rab11-FIP2 in tumor progression and provided novel insights into the mechanism of how Rab11-FIP2 positively regulated cell migration and invasion in CRC cells. 28073833_miR-142-3p, a microRNA enriched in exosomes during acute cellular rejection, is transferred to endothelial cells and compromises endothelial barrier function via down-regulation of RAB11FIP2. 28228550_an interaction of Eps15 and Rab11-FIP2 at the appropriate time and location in polarizing cells is necessary for proper establishment of epithelial polarity. 30006518_miR-192/215-Rab11-FIP2 axis appears to represent a new molecular mechanism underlying gastric cancer progression. 30471866_Mechanistic investigations showed that Rab11-FIP2 interacted with the glycolytic kinase PGK1 and promoted its ubiquitination in NSCLC cells, leading to inactivation of the oncogenic AKT/mTOR signaling pathway. 30659120_The integral function of the endocytic recycling compartment is regulated by RFFL-mediated ubiquitylation of Rab11 effectors. 31452257_Knockdown Rab11-FIP2 inhibits migration and invasion of nasopharyngeal carcinoma via suppressing Rho GTPase signaling. 32744247_Crystal structure of the Rab-binding domain of Rab11 family-interacting protein 2. | ENSMUSG00000040022 | Rab11fip2 | 1262.795196 | 1.0608079 | 0.085163469 | 0.08541865 | 1.000524e+00 | 3.171837e-01 | 6.825237e-01 | No | Yes | 1581.801461 | 346.796826 | 1466.032400 | 321.469864 | ||
| ENSG00000107643 | 5599 | MAPK8 | protein_coding | P45983 | FUNCTION: Serine/threonine-protein kinase involved in various processes such as cell proliferation, differentiation, migration, transformation and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK8/JNK1. In turn, MAPK8/JNK1 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN, JDP2 and ATF2 and thus regulates AP-1 transcriptional activity (PubMed:18307971). Phosphorylates the replication licensing factor CDT1, inhibiting the interaction between CDT1 and the histone H4 acetylase HBO1 to replication origins (PubMed:21856198). Loss of this interaction abrogates the acetylation required for replication initiation. Promotes stressed cell apoptosis by phosphorylating key regulatory factors including p53/TP53 and Yes-associates protein YAP1 (PubMed:21364637). In T-cells, MAPK8 and MAPK9 are required for polarized differentiation of T-helper cells into Th1 cells. Contributes to the survival of erythroid cells by phosphorylating the antagonist of cell death BAD upon EPO stimulation (PubMed:21095239). Mediates starvation-induced BCL2 phosphorylation, BCL2 dissociation from BECN1, and thus activation of autophagy (PubMed:18570871). Phosphorylates STMN2 and hence regulates microtubule dynamics, controlling neurite elongation in cortical neurons. In the developing brain, through its cytoplasmic activity on STMN2, negatively regulates the rate of exit from multipolar stage and of radial migration from the ventricular zone. Phosphorylates several other substrates including heat shock factor protein 4 (HSF4), the deacetylase SIRT1, ELK1, or the E3 ligase ITCH (PubMed:20027304, PubMed:17296730, PubMed:16581800). Phosphorylates the CLOCK-ARNTL/BMAL1 heterodimer and plays a role in the regulation of the circadian clock (PubMed:22441692). Phosphorylates the heat shock transcription factor HSF1, suppressing HSF1-induced transcriptional activity (PubMed:10747973). Phosphorylates POU5F1, which results in the inhibition of POU5F1's transcriptional activity and enhances its proteosomal degradation (By similarity). Phosphorylates JUND and this phosphorylation is inhibited in the presence of MEN1 (PubMed:22327296). In neurons, phosphorylates SYT4 which captures neuronal dense core vesicles at synapses (By similarity). Phosphorylates EIF4ENIF1/4-ET in response to oxidative stress, promoting P-body assembly (PubMed:22966201). {ECO:0000250|UniProtKB:P49185, ECO:0000250|UniProtKB:Q91Y86, ECO:0000269|PubMed:10747973, ECO:0000269|PubMed:16581800, ECO:0000269|PubMed:17296730, ECO:0000269|PubMed:18307971, ECO:0000269|PubMed:18570871, ECO:0000269|PubMed:20027304, ECO:0000269|PubMed:21095239, ECO:0000269|PubMed:21364637, ECO:0000269|PubMed:21856198, ECO:0000269|PubMed:22327296, ECO:0000269|PubMed:22441692, ECO:0000269|PubMed:22966201}.; FUNCTION: JNK1 isoforms display different binding patterns: beta-1 preferentially binds to c-Jun, whereas alpha-1, alpha-2, and beta-2 have a similar low level of binding to both c-Jun or ATF2. However, there is no correlation between binding and phosphorylation, which is achieved at about the same efficiency by all isoforms. | 3D-structure;ATP-binding;Alternative splicing;Biological rhythms;Cell junction;Cytoplasm;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;S-nitrosylation;Serine/threonine-protein kinase;Synapse;Transferase | The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various cell stimuli, and targets specific transcription factors, and thus mediates immediate-early gene expression in response to cell stimuli. The activation of this kinase by tumor-necrosis factor alpha (TNF-alpha) is found to be required for TNF-alpha induced apoptosis. This kinase is also involved in UV radiation induced apoptosis, which is thought to be related to cytochrom c-mediated cell death pathway. Studies of the mouse counterpart of this gene suggested that this kinase play a key role in T cell proliferation, apoptosis and differentiation. Several alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Apr 2016]. | hsa:5599; | axon [GO:0030424]; basal dendrite [GO:0097441]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; ATP binding [GO:0005524]; enzyme binding [GO:0019899]; histone deacetylase binding [GO:0042826]; histone deacetylase regulator activity [GO:0035033]; JUN kinase activity [GO:0004705]; MAP kinase activity [GO:0004707]; protein phosphatase binding [GO:0019903]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine kinase binding [GO:0120283]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cellular response to amino acid starvation [GO:0034198]; cellular response to cadmium ion [GO:0071276]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to mechanical stimulus [GO:0071260]; cellular response to reactive oxygen species [GO:0034614]; cellular senescence [GO:0090398]; Fc-epsilon receptor signaling pathway [GO:0038095]; intracellular signal transduction [GO:0035556]; JNK cascade [GO:0007254]; JUN phosphorylation [GO:0007258]; negative regulation of apoptotic process [GO:0043066]; negative regulation of protein binding [GO:0032091]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cell killing [GO:0031343]; positive regulation of cyclase activity [GO:0031281]; positive regulation of deacetylase activity [GO:0090045]; positive regulation of gene expression [GO:0010628]; positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway [GO:1900740]; positive regulation of protein metabolic process [GO:0051247]; protein phosphorylation [GO:0006468]; regulation of circadian rhythm [GO:0042752]; regulation of DNA replication origin binding [GO:1902595]; regulation of DNA-binding transcription factor activity [GO:0051090]; regulation of macroautophagy [GO:0016241]; regulation of protein localization [GO:0032880]; response to mechanical stimulus [GO:0009612]; response to oxidative stress [GO:0006979]; response to UV [GO:0009411]; rhythmic process [GO:0048511]; stress-activated MAPK cascade [GO:0051403] | 11912216_Polycystin-1 activation of c-Jun N-terminal kinase and AP-1 is mediated by heterotrimeric G proteins 11931768_Results show that the N-CoR-HDAC3 complex inhibits JNK activation through the associated GPS2 subunit and thus could potentially provide an alternative mechanism for hormone-mediated antagonism of AP-1 function. 11971973_These data strongly suggest that in TNF-induced apoptosis, Hsp72 specifically interferes with the Bid-dependent apoptotic pathway via inhibition of JNK. 12058026_novel role for the I kappa B kinase complex-associated protein (IKAP) in the regulation of activation of the mammalian stress response via the c-Jun N-terminal kinase (JNK)-signaling pathway 12058028_role in stabilizing p21(Cip1) by phosphorylation 12079429_the NOx-induced cell proliferation via activation of JNK1 might contribute to lung tissue damage caused by NOx 12135322_Galpha13 can induce ppET-1 gene expression through a JNK-mediated pathway. 12140754_Elevated JNK activation contributes to the pathogenesis of human brain tumors 12143039_Jun kinase modulates tumor necrosis factor-dependent apoptosis in liver cells 12148599_Unimpaired activation of c-Jun NH2-terminal kinase (JNK) 1 upon CD40 stimulation in B cells of patients with X-linked agammaglobulinemia. 12206715_description of the signaling of JNK and p38 MAPK in apoptosis after stimulation by antioxidants 12296995_TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. 12354774_JNK has isoform-selective gene regulation and distinct JNK isoforms have a role in specific cellular responses 12359245_Relationship of Mcl-1 isoforms, ratio p21WAF1/cyclin A and this protein phosphorylation to apoptosis in human breast carcinomas. 12413764_Psoriatic epidermis shows selective activation of ERK and JNK, which might be related to hyperproliferation and abnormal differentiation of psoriatic epidermis. 12421945_JNK activation is predominantly involved in the induction of CD44 expression in monocytic cells via lipopolysaccharide-mediated signaling. 12478662_JNK-1 and p38 play a role in apoptosis induced by capsaicin in H-ras-transformed tumor cells 12514174_phosphorylation of JNK1 and WOX1 is necessary for their physical interaction and functional antagonism 12538493_JNK is required for growth of prostate carcinoma cells in vitro and in vivo 12592382_Western blot demonstrated that phosphorylation of JNK was induced only by TPA during 30 min to 1 h. 12646240_Data suggest that epidermal growth factor (EGF) stimulated c-Jun N-terminal kinase phosphorylation of c-Jun is uncoupled from protein kinase D suppression in cancer cells. 12707267_JNK1 has a role in the synergistic effect of TRAIL combined with DNA damage by mediating signals independent of p53 leading to apoptosis 12810082_Results suggest that tissue or plasma fibronectin may modulate the intestinal epithelial response to repetitive deformation through inhibted activation of p38 and jun kinases. 12847227_c-Jun N-terminal kinase plays a negative role in the production of IL-12 from human macrophages stimulated by lipopolysaccharide. 12859962_Data show that inhibition of arachidonate 5-lipoxygenase induces rapid activation of c-Jun N-terminal kinase (JNK) in human prostate cancer cells which is prevented by the 5-lipoxygenase metabolite, 5(S)-HETE. 12878610_Axin utilizes distinct regions for competitive MEKK1 and MEKK4 binding and JNK activation. 12902351_adipose cytokines and JNK are key mediators between obesity and hormone-resistant prostate cancer 12917434_Threonine 668 within the Amyloid beta protein precursor intracellular domain is indeed phosphorylated by JNK1; although JIP-1 can facilitate this phosphorylation, it is not required for this process. 14500675_Findings strongly suggest that the JNK/AP-1 transcription factor signaling pathway has little or no impact on the generation of inflammatory mediators in neutrophils. 14514687_late but not early JNK1 activation is associated with the induction of apoptosis 14532003_Results show that tumor necrosis factor alpha-mediated caspase 8 cleavage and apoptosis require a sequential pathway involving c-Jun N-terminal kinase, Bid, and Smac/DIABLO. 14557276_c-Jun N-terminal kinase activation in T cell receptor signaling is mediated by SH3 domain-containing adaptor HIP-55 14561739_JNK inhibition with SP600125 also blocked binding of Sp1 to the DR5/TRAIL-R2 promoter. 14637155_Fas-induced cell death and JNK activation are sensitive to Fas stimulation in cell lines carrying undetectable level of c-FLIP(L). 14688370_Stress-activated protein kinase 1 is involved in the control of monocyte chemoattractant protein-1-induced migration of MonoMac6 cells. 14699155_JNK activation is important for lipopolysacchairde-induced MCP-1 expression but not for TNF-alpha or IL-8 expression 14701702_data support an essential role for JNK signaling in the induction of growth inhibition and apoptosis by As(2)O(3) 14724588_activation of JNK is important for the induction of apoptosis following stresses that function at different cell cycle phases, and that basal JNK activity is necessary to promote proliferation and maintain diploidy in breast cancer cells 14729602_Calcium signaling in ovarian surface epithelial cells not only induces telomerase activity via JNK but also activates Pyk2. 14766760_JNK regulates the expression of HIPK3 in prostate cancer cells, which leads to increased resistance to Fas receptor-mediated apoptosis by reducing the interaction between FADD and caspase-8 14981905_Inducible expression of RbAp46 activated the c-Jun N-terminal kinase (JNK) signaling pathway and triggered apoptosis in Saos-2 cells xenografted into nude mice. 15013949_JNK signaling regulates the phosphorylation state of several kinases in skeletal muscle. JNK activation is unlikely to be the major mechanism by which contractile activity increases glycogen synthase activity in skeletal muscle. 15238629_The c-Jun-N-terminal kinase(JNK)cascade mediates the stimulatory effect Angiotensin II has on the proximal renin promoter in humans. 15456887_Data show that endogenous germinal center kinase is activated by agonists that require TRAF6 for c-Jun N-terminal kinase activation. 15474087_SAPKgamma/JNK1 and SAPKalpha/JNK2 may be important mediators of stress-induced responses in early implanting conceptuses that could mediate embryo loss. 15516492_Antiproliferative and prodifferentiation effects of BMP4 were Smad1 dependent with JNK also contributing to differentiation. 15527495_crucial role of JNK signalling pathway in N. meningitidis invasion in human brain microvascular endothelial cells 15528994_Cooperation of CD99 engagement with suboptimal TCR/CD3 signals resulted in enhanced CD4+ T cell proliferation, elevated expression of CD25 and GM1, increased apoptosis, augmented activation of JNK, and increased AP-1 activation 15542843_activation of p38 MAPK and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and TNFR1/TNFa expression 15569856_The ACE-inhibitor mediated activation of the c-Jun N-terminal kinase (JNK)/c-Jun pathway, results in an enhanced endothelial ACE expression 15629131_While JNK1 is a downstream effector of the Tumor Necrosis Factor (TNF) signaling, Zfra regulation of the TNF cytotoxic function is likely due to its interaction, in part, with JNK1 15637062_Caspase 3-cleaved SH3 domain of HIP-55 is likely involved in PRAM-1-mediated JNK activation upon arsenic trioxide-induced differentiation of NB4 cells. 15655348_ERK1/2 and JNK1/2 signaling is stimulated by radiation and can promote cell cycle progression in human colon cancer cells 15657352_Alpha-tocopheryl sulfsatse showed increased levels in prostate tumor cells. 15665513_TNF-alpha causes a net up-regulation of MUC2 gene expression in cultured colon cancer cells because NF-kappaB transcriptional activation of this gene is able to counter-balance the suppressive effects of the JNK pathway 15696159_JNK phosphorylates 14-3-3 proteins, which regulate nuclear targeting of c-Abl in the apoptotic response to DNA damage 15755722_PKD is a critical mediator in H2O2- but not TNF-induced ASK1-JNK signaling 15769735_raft-associated acid sphingomyelinase and JNK activation and translocation are induced by UV-C light on a nuclear signal 15778501_p53 participates in a feedback mechanism with JNK to regulate the apoptotic process and is oppositely regulated by JNK1 and JNK2. 15860507_Some green tea catechins induce pro-MMP-7 production via O2- production and the activation of JNK1/2. 15890690_JNK may act via c-Myc and Egr-1, which were shown to be important for B-lymphoma survival and growth. 15981086_rF1-induced JNK MAPK activity was correlated to the functional activation of macrophages by demonstrating the inhibition of NO, TNF-alpha production and microtubule polymerization 16086581_results suggest that activated JNK can, in turn, activate not only jun but also raf that, in turn, activates MEK that can then cross-activate JNK in a positive feedback loop 16105650_We concluded that JNK pathway might play an important role in mediating cisplatin-induced apoptosis in A2780 cells, and the duration of JNK activation might be critical in determining whether cells survive or undergo apoptosis. 16166642_JAMP is a membrane-anchored regulator of the duration of JNK1 activity in response to diverse stress stimuli 16176806_The activation of JNK1 is required for the triptolide-induced inhibition of tumor proliferation. 16243842_Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages 16260419_protein kinase Cdelta and JNK have roles in Ifn-alpha induced expression of phospholipid scramblase 1 through STAT1 16260609_c-Jun N-terminal kinase signaling is regulated by a stabilization-based feed-forward 16282329_Foxo1 is involved in the nucleocytoplasmic translocation of PDX-1 by oxidative stress and the JNK pathway 16283431_Results describe the opposite effect of ERK1/2 and JNK on p53-independent p21WAF1/CIP1 activation involved in the arsenic trioxide-induced human epidermoid carcinoma A431 cellular cytotoxicity. 16307741_gemcitabine-induced apoptosis in human non-small cell lung cancer H1299 cells requires activation of the JNK1 signaling pathway 16321971_in gastric epithelial cells, H. pylori up-regulates MMP-1 in a type IV secretion system-dependent manner via JNK and ERK1/2 16328781_Results suggest that VEGF induced by hyperbaric oxygen is through c-Jun/AP-1 activation, and through simultaneous activation of ERK and JNK pathways. 16339571_TNF-alpha induced PTX3 expression in human lung cell lines and primary epithelial cells; knockdown of either JNK1 or JNK2 with small interfering RNA also significantly reduced the regulated PTX3 expression 16381010_results indicate that the aberrant p-JNK1/2 expression and the co-expressed p-JNK1/2 and p-p38 in breast tissues may play a role in the carcinogenesis of breast infiltrating ductal carcinoma 16407310_Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1 16412424_Data report that cepharanthine induces apoptosis in HuH-7 cells through activation of JNK1/2 and the downregulation of Akt. 16434970_Both activation of JNK and inhibition of Akt play a role in translocation of Nur77 from the nucleus to the cytoplasm. 16465391_We investigated whether Jun-N-terminal kinase activation is increased in inflammatory bowel disease and analyzed the effects of SP600125, which decreases inflammatory cytokine synthesis by inhibiting the phosphorylation of this kinase. 16569638_matrix metalloproteinase-1 expression is regulated by JNK through Ets and AP-1 promoter motifs 16648634_Inhibition of JNK in epidermal keratinocytes is sufficient to initiate their differentiation program and suggest that augmenting JNK activity could be used to delay cornification and enhance wound healing. 16687404_JNK-mediated feedback phosphorylation of MLK3 regulates its activation and deactivation states by cycling between Triton-soluble and Triton-insoluble forms 16699726_ERK1/2, JNK1/2 and p38 mapk pathways are all required for B[a]P-induced G1/S transition 16760468_JNK is a critical component downstream of PI 3-kinase that may be involved in PDGF-stimulated chemotaxis presumably by modulating the integrity of focal adhesions by phosphorylating its components 16794185_Paclitaxel increases endothelial TF expression via its stabilizing effect on microtubules and selective activation of JNK 16802349_JNK (c-Jun N-terminal kinase) function might be modulated by targeting MKK-7 to suppress cytokine-mediated fibroblast-like synoviocytes (FLS) activation while leaving other stress responses intact. 16814421_Results show that activation of c-Jun N-terminal protein kinase, ERKs 1 and 2, and p38 MAP kinase is critical for Hs683 glioma cell migration induced by GDNF. 16815888_PP1-JNK pathway plays a role in H(2)O(2)-induced Sp1 phosphorylation in lung epithelial cells 16824735_JNK1 is associated with UV signal transduction in human epidermis and SCCA1 is a suppressor of this process. 16895791_TNF-alpha down-regulates human Cu/Zn superoxide dismutase 1 promoter via JNK/AP-1 signaling pathway 16912864_Leptin stimulates proliferation and inhibits apoptosis in colon cancer cells. This effect involves JAK2, PI3 kinase and JNK and activation of the oncogenic transcription factors signal transducer and activator of transcription 16927023_This study is the first to demonstrate that H2O2 induces a Rac1/JNK1/p38 signaling cascade, and that JNK and p38 activation is important for H2O2-induced apoptosis as well as apoptosis-inducing factor/Bax translocation of retinal pigment epithelial cells. 16972261_Data show that pharmacologic inhibitors of extracellular signal-regulated kinase (ERKs) and c-Jun NH(2)-terminal kinase (JNK) decrease glutathione content and sensitize human promonocytic leukemia cells to arsenic trioxide-induced apoptosis. 16983342_active JNK1 inhibits ubiquitination of C/EBPalpha possibly by phosphorylating in its DBD 17008315_STAT3 activation by G alpha(s) distinctively requires protein kinase A, JNK, and phosphatidylinositol 3-kinase 17054907_Collectively, our results suggest that the inhibition of the interaction between JNK and c-Jun may be an integral part of the mechanism underlying the negative regulation of the JNK signaling pathway by NO. 17074809_JNK1 and JNK2 differentially regulate TBP through Elk-1, controlling c-Jun expression and cell proliferation 17079291_Overall, these results demonstrate the importance of the JNK pathway for varicella-zoster virus replication. 17158878_PSK1-alpha is a bifunctional kinase that associates with microtubules, and JNK- and caspase-mediated removal of its C-terminal microtubule-binding domain permits nuclear translocation of the N-terminal region of PSK1-alpha and its induction of apoptosis 17178870_JNK1 is activated in response to collagen I, which increases tumorigenesis by up-regulating N-cadherin expression and by increasing motility. 17255354_Oxidative stress response regulates DKK1 expression through the JNK signaling cascade in multiple myeloma plasma cells. 17296730_Human Rev7 (hRev7)/MAD2B/MAD2L2 is an interaction partner for Elk-1 and hRev7 acts to promote Elk-1 phosphorylation by the c-Jun N-terminal protein kinase (JNK) MAP kinases. 17303384_These results suggest that in human tracheal smooth muscle cells, activation of p42/p44 MAPK, p38, and JNK pathways, at least in part, mediated through NF-kappaB, is essential for lipopolysaccharide-induced VCAM-1 gene expression. 17317777_Increased expression of stress-activated kinases and IKK and their phosphorylated forms in omental fat occurs in obesity. 17453826_The regulation of three major mitogen-activated protein kinases phosphorylation, ERKp44/p42, p38, and JNK, was determined. The influence of specific mitogen-activated protein kinase inhibitors on IL-1 beta protein levels during beta-endorphin stimulation. 17478078_Proinvasive activity of BMP7 through SMAD4/src-independent and ERK/Rac/JNK-dependent signaling pathways in colon cancer cells is reported. 17481915_These results indicate that binding of the alpha3beta1 integrin results in a suppression in the activation of the IL-1 induced intracellular signaling pathway from JNK to AP-1. 17496921_In this review, the interplay between NF-kappa B and JNK1 provides a paradigm that shows how crosstalk between different signaling pathways decides the function of the cell signaling circuitry. 17541429_Gemin5 functions as a scaffold protein for the ASK1-JNK1 signaling module and thereby potentiates ASK1-mediated signaling events. 17545598_Findings showed that TOPK positively modulated UVB-induced JNK1 activity and played a pivotal role in JNK1-mediated cell transformation induced by H-Ras. 17568996_IGF-1R and PDGFR co-inhibition caused an increased cell death in two human glioma cell lines and induced the radiosensitization of the JNK1 expressing cell line. 17584736_GCK is required for JNK and, unexpectedly, p38 activation by three bacterial PAMPs, lipopolysaccharide, peptidoglycan, and flagellin 17603935_TNF-alpha induced reactive oxygen species formation is mediated by JNK1, which regulates ferritin degradation and thus the level of highly reactive iron. 17620321_These results support a significant role for ALK1 as a negative regulator of endothelial cell migration and suggest the implication of JNK and ERK as mediators of this effect. 17626013_JNK and p38 mitogen-activated protein kinases were activated by TAK1. 17640761_This study reveals a novel pathway of gene regulation by alcohol which involves the activation of JNK and the consequent mRNA stabilization. 17652454_These results indicate that Ask1 oxidation is required at a step subsequent to activation for signaling downstream of Ask1 after H(2)O(2) treatment. 17690186_Data suggest that JNK activation and decreased expression of MKP-1 may play important roles in progression of urothelial carcinoma. 17693927_Studies suggest that the target of regulation by PP2A includes upstream kinases in the JNK MAPK pathway. 17699782_Findings suggest that the JNK/PTEN and NF-kappaB/PTEN pathways play a critical role in normal intestinal homeostasis and colon carcinogenesis. 17702750_long duration KOR antagonists disrupt KOR signaling by activating JNK 17703233_findings establish a major role for DAPk and its specific interaction with PKD in regulating the JNK signaling network under oxidative stress. 17704768_Results demonstrate that Fbl10 is a key regulator of c-Jun function. 17719653_Costimulation by anti-CD3 and anti-CD28 antibodies could activate JNK, p38 MAPK and NF-kappaB. The upregulation of IL-25 receptors were differentially regulated by intracellular JNK, p38 MAPK and NF-kappaB. 17785464_Study provides evidence during rolling and adhesion of platelets to vWF that platelet GPIb-vWF interaction triggers alphaIIbbeta3 activation in a JNK1-dependent manner; this was confirmed with a Glanzmann thrombastenic patient lacking alphaIIbbeta3. 17883418_Streptococcus intermedius histon-like DNA binding protein (Si-HLP) stimulation induced the activation of cell signal transduction pathways, extracellular signal-regulated kinase 1/2 (ERK1/2) and c-Jun N-terminal kinase (JNK). 17904874_Our data show that basic JNK activity plays an important role in the progression of the cell cycle at G2/M cell phase. 17908987_JNK1/2 activity is commonly increased in head and neck squamous cell carcinoma 17913539_These results collectively indicated that Chlamydophilal antigens induce foam cell formation mainly via Toll-like receptor 2 and c-Jun NH2 terminal kinase. 17933493_The induction of IFIT4 transcription by IFN-alpha depends upon sequential activation of PKCdelta, JNK and STAT1, and the influence of PKCdelta or JNK on IFN-alpha-mediated induction of IFIT4 is dependent upon the phosphorylation of STAT1 at Ser-727. 17942603_IFNalpha-induced apoptosis requires activation of ERK1/2, PKCdelta, and JNK downstream of PI3K and mTOR, and it can occur in a nucleus-independent manner, thus demonstrating that IFNalpha induces apoptosis in the absence of de novo transcription. 17967471_Critical role in cell transformation induced by EBV LMP1. 17982228_crude extract of D. farinae induces ICAM-1 expression in EoL-1 cells through signaling pathways involving both NF-kappaB and JNK 18025271_Cyclin G2 expression is modulated by HER2 signaling through multiple pathways including phosphoinositide 3-kinase, c-jun NH(2)-terminal kinase, and mTOR signaling. 18036196_Observational study of gene-disease association. (HuGE Navigator) 18036196_The G/G genotype of MAPK8 SNP-1066 did not affect T2DM susceptibility despite specific binding of AP2alpha. 18055217_We conclude that endogenous SOCS3 inhibits AP-1 activity through blocking of JNK phosphorylation. 18082745_Nonalcoholic steatohepatitis is specifically associated with (1) failure to generate sXBP-1 protein and (2) activation of JNK. 18086557_Activation of protease-activated receptors but not stimulation with lipopolysaccharides leads to ERK1/2 and JNK-mediated production of IL-8. 18087676_These results clearly indicate that CCDC134 is a novel member of the secretory family and down-regulates the Raf-1/MEK/ERK and JNK/ SAPK pathways. 18094581_The majority of acute myeloid leukemia cases did not show any levels of Mitogen-Activated Protein Kinases activation except for two cases, which were associated with an extremely high white blood cell count, chromosomal aberration. 18164704_Our data first suggest that JNK participates in bFGF-mediated surface cadherin downregulation. Loss of surface cadherins may affect the cell-cell interaction between endothelial cells and facilitate angiogenesis. 18181766_ERK and JNK are involved in PMA-mediated MD-2 gene expression during HL-60 cell differentiation. 18199680_JNK1 activation is necessary to phosphorylate Sp1 and to shield Sp1 from the ubiquitin-dependent degradation pathway during mitosis in tumor cell lines. 18212053_c-Jun amino-terminal kinase (JNK) was important for neurite outgrowth stimulated by both Wnt-3a and Dkk1. 18218857_corneal inflammation is significantly impaired in JNK1 knockout mice compared with control mice, and in mice treated with the JNK inhibitor compared with vehicle control. 18219313_These results identify NLRX1 as a NLR that contributes to the link between reactive oxygen species generation at the mitochondria and innate immune responses. 18249102_Our results suggest that clivorine has direct toxicity on HEK293 cells, and phosphorylated JNK may play some role in counteracting the toxicity of clivorine on HEK293 cells. 18253836_activator protein-1 (AP-1) was activated through phosphorylation of cJun and cFos, induced by JNK and p38, respectively. 18256527_Study shows that basal c-Jun N-terminal kinases (JNKs) are required for mitotic histone H3-S10 phosphorylation in human primary fibroblast IMR90 cells 18276794_Glutamate cysteine ligase iz induced by hydroxynonenal through the c-Jun N-terminal kinase (JNK) pathway in respiratory epithelium. 18286207_the activity of JIP1-JNK complexes is downregulated by VRK2 in response to interleukin-1beta 18288129_both constitutive and sIgM-induced phosphorylation of p38 and JNK is inhibited by LAIR-1 through an ITIM-dependent signal 18292600_PP2A and AIP1 cooperatively induce activation of ASK1-JNK signaling and vascular endothelial cell apoptosis. 18293403_Preventing the TAK1/JNK1 signaling cascade in astrocytes might provide a fruitful strategy for treating intractable neuropathic pain. 18297686_These results identify JNK, and not NFkappaB, as a critical mediator of TNF-alpha repressory effect on connexin 43 gene expression. 18316600_ERK and JNK MAPK/Elk-1/Egr-1 signal cascade is required for p53-independent transcriptional activation of p21(Waf1/Cip1) in response to curcumin in U-87MG human glioblastoma cells 18316603_c-Jun translocates B23 and ARF from the nucleolus after JNK activation by means of protein interactions 18325654_Oxidative stress-activated JNK signaling pathway is involved in METH-induced cell death. 18337589_Phosphorylation aids GR sumoylation and that cross talk of JNK and SUMO pathways fine tune GR transcriptional activity. 18344085_The protein expression rates of p-JNK and P-glycoprotein in gastric cancer were significantly higher than those in normal gastric tissue. 18348163_Data show that JNK modulates the effect of caspases and NF-kappaB in the TNF-alpha-induced down-regulation of Na+/K+ATPase in HepG2 cells. 18356158_H. pylori mediates CagA-independent signaling that promotes cell motility through the beta1 integrin-JNK pathway 18373696_Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in cervical cancer. 18401423_Although STAT1 phosphorylation required JNK and p38MAPK activation, only JNK activation was essential for IRF1 promoter activation by Tie2-R849W. 18405916_These data demonstrate the role of Cx43 in the proliferation and migration of human saphenous vein smooth muscle cells and angiotensin II-induced Cx43 expression via mitogen-activated protein kinases (MAPK)-AP-1 signaling pathway. 18429822_Study demonstrates that the serine/threonine kinase PKN1 plays a critical role in regulating constitutive IKK/JNK activity in unstimulated cells and report on the molecular mechanism. 18439101_These findings suggest that JNK1 and JNK2 are involved in TNF-alpha-induced neutrophil apoptosis and GM-CSF-mediated antiapoptotic effect on neutrophils, respectively. 18457359_is the key enzyme mediating melanogenesis in B16F10 cell. 18495129_inhibition of JNK and p38 activation interrupts CD40 induced endothelial cell activation and apoptosis 18506470_JNK is involved in regulation of proinflammatory mediators of endometrium 18524773_JNK1 is a critical transcriptional target of FoxM1 that contributes to FoxM1-regulated cell cycle progression, tumor cell migration, invasiveness, and anchorage-independent growth 18540881_Although JNK activation may be a primary inducing factor, further phosphorylation of tau is required for neuronal death and NFT formation in neurodegenerative diseases, including those characterized by tauopathy. 18541008_JNK, and in particular the JNK1 isoform, support the growth of melanoma cells, by controlling either cell cycle progression or apoptosis depending on the cellular context. 18547751_Nanosilver acts through ROS and JNK to induce apoptosis via the mitochondrial pathway. 18570871_JNK1 mediates starvation-induced Bcl-2 phosphorylation, Bcl-2 dissociation from Beclin 1, and autophagy activation 18573678_Data show that pokeweed antiviral protein (PAP) does not inhibit protein translation, but induces the activation of c-Jun NH2-terminal kinase (JNK), which was specific to rRNA depurination. 18594007_c-Jun NH2-terminal protein kinase activation caused by tubulin depolymerization and DNA damage has a crucial role in moscatilin-induced apoptosis in human colorectal cancer cells 18603327_Data show that M. bovis BCG-induced human beta-defensin mRNA expression in A549 cells is regulated at least in part through activation of signaling proteins of PKC, JNK and PI3K. 18620777_WWOX induces apoptosis and inhibits human hepatocellular carcinoma cell growth through a mechanism enhanced by JNK inhibition. 18636174_A functional analysis of JNK1 and M-RIP with RNA interference reveals a critical role for this cascade in the invasive behavior of cancer cells. 18651223_These findings suggest JNK to have an important pro-apoptotic function following ultraviolet rays B irradiation in human melanocytes, by acting upstream of lysosomal membrane permeabilization and Bim phosphorylation. 18663379_Shikonin-induced oxidative injury operates at a proximal point in apoptotic signaling cascades, and subsequently activates the stress-related JNK pathway, triggers mitochondrial dysfunction, cytochrome c release, and caspase activation. 18667537_necrosis factor-alpha-elicited stimulation of gamma-secretase is mediated by c-Jun N-terminal kinase-dependent phosphorylation of presenilin and nicastrin 18681908_SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes 18682391_Histamine-induced Egr-1 expression is dependent on the activation of the H1 receptor, and rapidly and transiently activates PKCdelta, ERK1/2, p38 kinase, and JNK prior to Egr-1 induction. 18703151_These results demonstrate that ERKs and JNKs are responsible for the decrease of cyclin D1 and CDK4 expression levels in human embryonic lung fibroblasts induced by silica. 18713649_These results suggest that phosphorylation of paxillin on Ser 178 by JNK is required for the association of paxillin with FAK, and subsequent tyrosine phosphorylation of paxillin. 18713996_JNK is differentially regulated by MKK4 and MKK7 depending on the stimulus. 18718914_15d-PGJ(2) induces vascular endothelial cell apoptosis through the signaling of JNK and p38 MAPK-mediated p53 activation both in vitro and in vivo 18723442_whether MAP kinase phosphatase (MKP)-1, a negative regulator of p38 and JNK, mediates the antiinflammatory effects of shear stress. 18757369_a link between RhoA, JNK, c-Jun, and MMP2 activity that is functionally involved in the reduction in osteosarcoma cell invasion by the statin. This suggests a novel strategy targeting RhoA-JNK-c-Jun signaling to reduce osteosarcoma cell tumorigenesis. 18769111_A speculative model for understanding the interrelationship between autophagy and apoptosis regulated by JNK1-mediated Bcl-2 phosphorylation was proposed. 18782768_pneumolysin selectively induced expression of MKP1 via a TLR4-dependent MyD88-TRAF6-ERK pathway, which inhibited the PAK4-JNK signaling pathway,leading to up-regulation of MUC5AC mucin production 18815275_These results establish a novel function of filamin B as a molecular scaffold in the JNK signaling pathway for type I IFN-induced apoptosis. 18818208_analysis of JNK-mediated phosphorylation of paxillin in adhesion assembly and tension-induced cell death by the adenovirus death factor E4orf4 18845538_a novel function for Parkin in modulating the expression of Eg5 through the Hsp70-JNK-c-Jun signaling pathway. 18922473_These results indicate that TGF-beta activates JNK and p38 through a mechanism similar to that operating in the interleukin-1beta/Toll-like receptor pathway. 18936517_JNK may play an important role in posttranscriptional control of LDL receptor expression, thus constituting a novel mechanism to enhance plasma LDL clearance by liver cells. 18950845_Observational study of gene-disease association. (HuGE Navigator) 18978303_dynasore may stimulate PAI-1 protein expression and enhance TGF-beta(1) activity through activation of JNK-mediated signaling in human pleural mesothelial cells 18979912_The changes of phospho-JNK expression after skin burned might correlate with wound healing. 18982452_Data show that an increase in JNK activation in the presence of NFkappaB inhibition significantly increased the expression of IGFBP6. 18989785_Results show that Docetaxel-induced apoptosis is mediated by induction of ER stress, through activation of JNK and downstream targets of JNK. 18996088_These results suggest that JNK, but not caspase 8, involves in Fas-mediated CH11-induced autophagy in HeLa cells, and this autophagy plays a protective role in CH11-induced cell death. 19033664_a mechanistic link between JNK activity and liver cell proliferation via p21 and c-Myc and suggest JNK1 targeting can be considered as a new therapeutic approach for HCC treatment. 19036714_Upon UVB-induced stress in keratinocytes, ROCK1 was activated, bound to JIP-3, and activated the JNK pathway 19037093_AMPK controls the molecular mechanism underlying the differential biological functions of JNK, providing a novel explanation for the antiapoptotic role of LKB1. 19041150_JNK1 plays important roles in the development of human HCC partially through the epigenetic mechanisms. 19052872_c-Abl and p53 are important for execution of the cell death program initiated in A2E-laden RPE cells exposed to blue light, while JNK might play an anti-apoptotic role 19056926_JNK1 stimulated and mediated the effects of IFN and TNF-alpha on XAF1 expression through transcriptional regulation by induction of IRF-1. 19060920_activation of JNK pathway can mediate Beclin 1 expression, which plays a key role in autophagic cell death in cancer cel | ENSMUSG00000021936 | Mapk8 | 1334.147843 | 1.0652048 | 0.091130899 | 0.08685205 | 1.101051e+00 | 2.940355e-01 | 6.626063e-01 | No | Yes | 1788.526473 | 350.875197 | 1705.926214 | 334.711514 | |
| ENSG00000107738 | 64115 | VSIR | protein_coding | Q9H7M9 | FUNCTION: Immunoregulatory receptor which inhibits the T-cell response (PubMed:24691993). May promote differentiation of embryonic stem cells, by inhibiting BMP4 signaling (By similarity). May stimulate MMP14-mediated MMP2 activation (PubMed:20666777). {ECO:0000250|UniProtKB:Q9D659, ECO:0000269|PubMed:20666777, ECO:0000269|PubMed:24691993}. | 3D-structure;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:64115; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; endopeptidase activator activity [GO:0061133]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; negative regulation of CD4-positive, alpha-beta T cell proliferation [GO:2000562]; negative regulation of CD8-positive, alpha-beta T cell proliferation [GO:2000565]; negative regulation of interferon-gamma production [GO:0032689]; negative regulation of interleukin-10 production [GO:0032693]; negative regulation of interleukin-17 production [GO:0032700]; negative regulation of tumor necrosis factor production [GO:0032720]; positive regulation of cell migration [GO:0030335]; positive regulation of collagen catabolic process [GO:0120158]; positive regulation of endopeptidase activity [GO:0010950]; positive regulation of gene expression [GO:0010628]; positive regulation of regulatory T cell differentiation [GO:0045591]; regulation of immune response [GO:0050776]; zymogen activation [GO:0031638] | 20666777_Results suggest that GI24 contributes to tumor-invasive growth in the collagen matrix by augmenting cell surface MT1-MMP. 25279955_overexpression of the newly described co-stimulatory molecule, PD1 homologue (PD-1H) in human monocyte/macrophages is sufficient to induce spontaneous secretion of multiple cytokines. 26228159_p53-induced expression of DD1alpha prevents persistence of cell corpses and ensures efficient generation of precise immune responses in mice. 28236118_Taken together, the results indicated that the VISTA high and CD8 low group, as an immunosuppressive subgroup, might be associated with a poor prognosis in primary OSCC. These findings indicated that VISTA might be a potential immunotherapeutic target in OSCC treatment. 28258694_this review describes the functions of VISTA in the context of cancer immunotherapy 28776578_This study is the first to describe the expression of VISTA-expressing lymphocytes in melanoma samples and in the context of acquired resistance to immune checkpoint inhibitors 29203588_The immunomodulatory role of VISTA in human NSCLC. 29216931_VISTA expression supports immune-complex inflammation in collagen antibody-induced arthritis and VISTA is expressed in human synovium 29720116_VISTA protein expression in hepatocellular carcinoma showed cell specific and displayed different prognosis 29737375_High VISTA expression is associated with primary cutaneous melanoma. 29771768_Data indicate that VISTA is predominantly expressed and up-regulated in the high-density infiltrated immune cells but minimal in pancreatic cancerous cells. 30106102_EGFR may be involved in immune evasion, possibly through regulation of B7H5 expression in nonsmall cell lung carcinoma. 30128738_High VISTA expression is associated with colorectal carcinoma. 30220083_this study shows that VSIG-3 as a ligand of VISTA inhibits human T-cell function 30306644_This study is the first to demonstrate that in the CNS, VISTA is expressed by microglia, and that VISTA is differentially expressed in CNS pathologies. 30382166_The data suggest that VISTA is a novel immunosuppressive factor within the tumour microenvironment, as well as a new target for cancer immunotherapy. 30635425_VISTA may be a relevant immunotherapy target for effective treatment of patients with pancreatic cancer. 31088847_Hypoxia-Induced VISTA Promotes the Suppressive Function of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. 31363159_Expression of V-set immunoregulatory receptor in malignant mesothelioma. 31484064_Structure and Functional Binding Epitope of V-domain Ig Suppressor of T Cell Activation. 31537897_V-domain Ig-containing suppressor of T-cell activation (VISTA), a potentially targetable immune checkpoint molecule, is highly expressed in epithelioid malignant pleural mesothelioma. 31645726_findings identify a mechanism by which VISTA may engender resistance to anti-tumour immune responses, as well as an unexpectedly determinative role for pH in immune co-receptor engagement 31781843_VISTA was detected in 51.4% of all samples and 46.6% of PD-L1-negative samples in patients with high-grade serous ovarian cancer. Furthermore, VISTA expression was associated with pathologic type and PD-L1 expression. Moreover, VISTA expression in tumor cells, but not in immune cells, was associated with prolonged progression-free and overall survival in patients with high-grade serous ovarian cancer. 31883303_Overexpression of B7H5/CD28H is associated with worse survival in human gastric cancer. 31901178_VISTA protein (B7-H5) is a ligand for transmembrane and immunoglobulin domain containing 2 protein (CD28H) and is widely expressed in tumor cells. B7-H5 expression is closely related to the prognosis of the tumor [Review]. 31940493_FOXD3 Regulates VISTA Expression in Melanoma. 31949051_VISTA is therefore a distinctive negative checkpoint regulator of naive T cells that is critical for steady-state maintenance of quiescence and peripheral tolerance. 32060343_Prognostic value of VISTA in solid tumours: a systematic review and meta-analysis. 32117584_Expression of TIM3/VISTA checkpoints and the CD68 macrophage-associated marker correlates with anti-PD1/PDL1 resistance: implications of immunogram heterogeneity. 32205423_Galectin-9 and VISTA Expression Define Terminally Exhausted T Cells in HIV-1 Infection. 32266446_Low VISTA expression is associated with breast cancer. 32600443_VISTA: an immune regulatory protein checking tumor and immune cells in cancer immunotherapy. 32873829_The prognostic significance of VISTA and CD33-positive myeloid cells in cutaneous melanoma and their relationship with PD-1 expression. 33086339_High VISTA Expression Correlates With a Favorable Prognosis in Patients With Colorectal Cancer. 33178206_VISTA Re-programs Macrophage Biology Through the Combined Regulation of Tolerance and Anti-inflammatory Pathways. 33250890_The Expression Pattern and Clinical Significance of the Immune Checkpoint Regulator VISTA in Human Breast Cancer. 33329552_Ligand-Receptor Interactions of Galectin-9 and VISTA Suppress Human T Lymphocyte Cytotoxic Activity. 33396515_LAG-3, TIM-3 and VISTA Expression on Tumor-Infiltrating Lymphocytes in Oropharyngeal Squamous Cell Carcinoma-Potential Biomarkers for Targeted Therapy Concepts. 33770210_Expression of VISTA on tumor-infiltrating immune cells correlated with short intravesical recurrence in non-muscle-invasive bladder cancer. 33889438_VISTA and PD-L1 synergistically predict poor prognosis in patients with extranodal natural killer/T-cell lymphoma. 34106206_VISTA is an activating receptor in human monocytes. 34465822_Superpixel image segmentation of VISTA expression in colorectal cancer and its relationship to the tumoral microenvironment. 34660778_Soluble B7-H5 Is a Novel Diagnostic, Severity, and Prognosis Marker in Acute Pancreatitis. 34728682_The immune checkpoint VISTA exhibits high expression levels in human gliomas and associates with a poor prognosis. 34752423_Kidney VISTA prevents IFN-gamma/IL-9 axis-mediated tubulointerstitial fibrosis after acute glomerular injury. 35056382_VISTA, PDL-L1, and BRAF-A Review of New and Old Markers in the Prognosis of Melanoma. 35187955_Different clinical significance of novel B7 family checkpoints VISTA and HHLA2 in human lung adenocarcinoma. | ENSMUSG00000020101 | Vsir | 11.615169 | 1.3807592 | 0.465461778 | 0.76533107 | 3.791881e-01 | 5.380381e-01 | No | Yes | 16.537650 | 9.344426 | 11.239634 | 6.247478 | |||
| ENSG00000107819 | 81855 | SFXN3 | protein_coding | Q9BWM7 | FUNCTION: Mitochondrial serine transporter that mediates transport of serine into mitochondria, an important step of the one-carbon metabolism pathway (PubMed:30442778). Mitochondrial serine is converted to glycine and formate, which then exits to the cytosol where it is used to generate the charged folates that serve as one-carbon donors (PubMed:30442778). {ECO:0000269|PubMed:30442778}. | Acetylation;Amino-acid transport;Membrane;Mitochondrion;One-carbon metabolism;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:81855; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrion [GO:0005739]; serine transmembrane transporter activity [GO:0022889]; transmembrane transporter activity [GO:0022857]; mitochondrial transmembrane transport [GO:1990542]; one-carbon metabolic process [GO:0006730]; serine import into mitochondrion [GO:0140300] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000025212 | Sfxn3 | 125.148115 | 1.4087510 | 0.494416600 | 0.26186556 | 3.654457e+00 | 5.591937e-02 | No | Yes | 135.770790 | 18.167461 | 93.491448 | 12.663552 | |||
| ENSG00000107890 | 22852 | ANKRD26 | protein_coding | Q9UPS8 | FUNCTION: Acts as a regulator of adipogenesis. Involved in the regulation of the feeding behavior. {ECO:0000250|UniProtKB:Q811D2}. | ANK repeat;Alternative splicing;Coiled coil;Phosphoprotein;Reference proteome;Repeat | This gene encodes a protein containing N-terminal ankyrin repeats which function in protein-protein interactions. Mutations in this gene are associated with autosomal dominant thrombocytopenia-2. Pseudogenes of this gene are found on chromosome 7, 10, 13 and 16. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:22852; | centrosome [GO:0005813]; negative regulation of fat cell differentiation [GO:0045599] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 21211618_mutations in the 5' UTR of ANKRD26 are implicated in thrombocytopenia 2. 21467542_The ANKRD26-related thrombocytopenia has to be taken into consideration in the differential diagnosis of isolated thrombocytopenias. 23223974_Ubiquitin/proteasome-rich particulate cytoplasmic structures are a characteristic feature of ANKRD26-related thrombocytopenia platelets and megakaryocytes. 23869080_the missense mutations may paly a role in the pathogenesis of Autosomal-dominant nonsyndromic thrombocytopenia-2 24030261_Studies indicate that ANKRD26-RT is an insidious form of inherited thrombocytopenias that exposes patients to a low risk of bleeding but predisposes them to hematologic myeloid malignancies. 24430186_ANKRD26 regulatory region mutations induce MAPK hyperactivation in familial thrombocytopenia 24628296_The study supports the association of ANKRD26 mutations with thrombocytopenia 2 and a predisposition to myeloid malignancies. 25902755_thrombocytopenia with 5'UTR ANKRD26 gene mutation must be considered in case of a constitutional isolated thrombocytopenia, with a low bleeding tendency, associated with autosomal dominant transmission and normal platelet volume. 26175287_WASP, RUNX1, and ANKRD26 genes are important for normal TPO signaling and the network underlying thrombopoiesis. 27108925_Molecular analysis identified a mutation located in the promoter of the ankyrin repeat domain 26 (ANKRD26) gene, c.-127A>T in normocytic thrombocytopenia. 27123948_The findings of lifelong thrombocytopenia with mild/absent bleeding, family history of thrombocytopenia with normal platelet size and myeloid neoplasms should raise the suspicion of ANKRD26 mutated thrombocytopenia. 28100250_investigation of one patient with the c.3G>A showed that mutation was associated with strong ANKRD26 overexpression in vivo, which is the proposed mechanism for predisposition to AML in THC2 patients 28277066_in a cohort of patients with suspected familial thrombocytopenia, the c.-140C>G mutation seems to be the most frequent ANKRD26 mutation. 28976612_Two cases with mutant ANKRD26 highlight that patients with thrombocytopenia 2 are at risk of being misdiagnosed with myelodysplastic syndrome and receiving undue myelosuppressive treatments. Because dysmegakaryopoiesis is a feature also of other forms of inherited thrombocytopenia, a genetic disorder must always be considered when a patient presents with isolated thrombocytopenia and dysmegakaryopoiesis. 29545013_The overall purpose of this review is to point out that important progresses have been made in understanding the pathogenesis of ANKRD26-Related Thrombocytopenia and MYH9-Related Diseases and new therapeutic approaches have been proposed and tested. 30747248_A novel nucleotide substitution in the 5' untranslated region of ANKRD26 gene is associated with inherited thrombocytopenia. 31425920_ANKRD26-RET is a novel rearrangement of the RET gene, associated with RET expression in thyroid tissue 31607198_Successful management of a pregnant woman with severe ANKRD26-related thrombocytopenia and anti-HPA-5b alloimmunization. 31801613_Study demonstrates that downregulation of ANKRD26 gene caused by promoter hypermethylation at specific CpGs represents a common abnormality in obese patients. These changes correlate to the pro-inflammatory profile and the cardio-metabolic risk factors of obese individuals, suggesting they mark the adverse health outcome occurring in some of these patients. 32659145_Relation between mutations in the 5' UTR of ANKRD26 gene and inherited thrombocytopenia with predisposition to myeloid malignancies. An Egyptian study. 32944898_A novel RUNX1 mutation with ANKRD26 dysregulation is related to thrombocytopenia in a sporadic form of myelodysplastic syndrome. 33857290_Familial thrombocytopenia due to a complex structural variant resulting in a WAC-ANKRD26 fusion transcript. | 620.766116 | 1.0899774 | 0.124298173 | 0.11549295 | 1.166785e+00 | 2.800628e-01 | 6.500986e-01 | No | Yes | 517.705020 | 108.309550 | 467.436505 | 97.856220 | |||
| ENSG00000108448 | 147166 | TRIM16L | transcribed_unprocessed_pseudogene | Q309B1 | 3D-structure;Alternative splicing;Cytoplasm;Reference proteome | hsa:147166; | cytosol [GO:0005829]; plasma membrane [GO:0005886] | 275.308043 | 0.9012401 | -0.150016663 | 0.16173122 | 8.652356e-01 | 3.522777e-01 | No | Yes | 272.529604 | 34.502331 | 306.202084 | 38.619010 | |||||||
| ENSG00000108509 | 23125 | CAMTA2 | protein_coding | O94983 | FUNCTION: Transcription activator. May act as tumor suppressor. {ECO:0000269|PubMed:11925432}. | ANK repeat;Activator;Alternative splicing;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a member of the calmodulin-binding transcription activator protein family. Members of this family share a common domain structure that consists of a transcription activation domain, a DNA-binding domain, and a calmodulin-binding domain. The encoded protein may be a transcriptional coactivator of genes involved in cardiac growth. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Jan 2010]. | hsa:23125; | chromatin [GO:0000785]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; histone deacetylase binding [GO:0042826]; sequence-specific DNA binding [GO:0043565]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cardiac muscle hypertrophy in response to stress [GO:0014898]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 29110692_Mutation of CAMTA2 resulting in post-transcriptional inhibition of its own gene activity likely underlies a novel syndromic tremulous dystonia. | ENSMUSG00000040712 | Camta2 | 411.649702 | 1.2104516 | 0.275545371 | 0.50327655 | 3.133897e-01 | 5.756075e-01 | No | Yes | 540.072335 | 92.292825 | 458.812511 | 79.719678 | ||
| ENSG00000109255 | 10874 | NMU | protein_coding | P48645 | FUNCTION: [Neuromedin-U-25]: Ligand for receptors NMUR1 and NMUR2 (By similarity). Stimulates muscle contractions of specific regions of the gastrointestinal tract. In humans, NmU stimulates contractions of the ileum and urinary bladder. {ECO:0000250|UniProtKB:P12760}.; FUNCTION: [Neuromedin precursor-related peptide 33]: Does not function as a ligand for either NMUR1 or NMUR2. Indirectly induces prolactin release although its potency is much lower than that of neuromedin precursor-related peptide 36. {ECO:0000250|UniProtKB:P12760}.; FUNCTION: [Neuromedin precursor-related peptide 36]: Does not function as a ligand for either NMUR1 or NMUR2. Indirectly induces prolactin release from lactotroph cells in the pituitary gland, probably via the hypothalamic dopaminergic system. {ECO:0000250|UniProtKB:P12760}. | Amidation;Cleavage on pair of basic residues;Neuropeptide;Oxidation;Reference proteome;Secreted;Signal | This gene encodes a member of the neuromedin family of neuropeptides. The encoded protein is a precursor that is proteolytically processed to generate a biologically active neuropeptide that plays a role in pain, stress, immune-mediated inflammatory diseases and feeding regulation. Increased expression of this gene was observed in renal, pancreatic and lung cancers. Alternative splicing results in multiple transcript variants encoding different isoforms. Some of these isoforms may undergo similar processing to generate the mature peptide. [provided by RefSeq, Jul 2015]. | hsa:10874; | extracellular region [GO:0005576]; terminal bouton [GO:0043195]; neuromedin U receptor binding [GO:0042922]; signaling receptor binding [GO:0005102]; type 1 neuromedin U receptor binding [GO:0031839]; type 2 neuromedin U receptor binding [GO:0031840]; energy homeostasis [GO:0097009]; G protein-coupled receptor signaling pathway [GO:0007186]; neuropeptide signaling pathway [GO:0007218]; positive regulation of smooth muscle contraction [GO:0045987]; positive regulation of synaptic transmission [GO:0050806]; regulation of feeding behavior [GO:0060259]; regulation of grooming behavior [GO:2000821]; temperature homeostasis [GO:0001659] | 14623274_These results suggest that NMU plays a role in feeding behavior and catabolic functions via corticotropin-releasing hormone. 15187020_NmU expression is related to Myb and that the NmU/NMU1R axis constitutes a previously unknown growth-promoting autocrine loop in myeloid leukemia cells 15331768_irreversible binding of NmU to its receptors 16878152_Overexpression of neuromedin U is associated with bladder tumor formation, lung metastasis and cancer cachexia 16984985_Amino acid variants in NMU associate with overweight and obesity, suggesting that NMU is involved in energy regulation in humans. 16984985_Observational study of gene-disease association. (HuGE Navigator) 17018595_NMU & its cancer-specific receptors, as well as its target genes, are frequently overexpressed in clinical samples of lung cancer and in cell lines, and that those gene products play indispensable roles in the growth and progression of lung cancer cells. 19118941_NmU may be involved in the HGF-c-Met paracrine loop regulating cell migration, invasiveness and dissemination of pancreatic ductal adenocarcinoma. 19519756_[review] Taken together with its vascular actions, NMU may be a functional link between energy balance and the cardiovascular system and may provide a future target for therapies directed against the disorders that comprise metabolic syndrome. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21791076_Inactivation of the von Hippel-Lindau tumour suppressor gene induces Neuromedin U expression in renal cancer cells. 23936460_a role for NMU gene through interaction with ADRB2 gene in bone strength regulation 24876102_our results defined NmU as a candidate drug response biomarker for HER2-overexpressing cancers and as a candidate therapeutic target to limit metastatic progression and improve the efficacy of HER-targeted drugs. 25871004_polymorphisms in the gene may be useful in identifying women at risk for osteoporosis. 27279246_Overexpression of Nmu may be involved in the process of regional metastasis of HNSCC, and may serve as a novel and valuable biomarker for predicting regional metastasis in patients with HNSCC. 28235053_This study shows an association between a NMU haplotype and anthropometric indices, mainly linked to fat mass, which appears to be age- and sex-specific in children. 28340506_Results indicate a mechanism of action for neuromedin U (NmU) in HER2-overexpressing breast cancer that enhances resistance to HER2-targeted drugs through conferring cancer stem cell (CSC) characteristics and expansion of the CSC phenotype. 28423716_NMU might contribute to progression of NMUR2-positive breast cancer 29107108_The anti-tumorigenic effect of HAND2-AS1 in endometrioid endometrial carcinoma was mediated by down-regulating neuromedin U expression. 30096454_Results identified NMU as highly expressed in non-small cell lung cancer (NSCLC) tissues and cell lines. Furthermore, integrative bioinformatics analyses demonstrate that NMU may confer the alectinib resistance in NSCLC via multiple mechanisms. 31326439_Association between variants of neuromedin U gene and taste thresholds and food preferences in European children: Results from the IDEFICS study. 32013887_The prognostic value of neuromedin U in patients with hepatocellular carcinoma. | ENSMUSG00000029236 | Nmu | 153.478472 | 0.9838326 | -0.023515163 | 0.22481173 | 1.117350e-02 | 9.158166e-01 | No | Yes | 145.674390 | 22.777015 | 149.138312 | 23.248593 | ||
| ENSG00000109534 | 54433 | GAR1 | protein_coding | Q9NY12 | FUNCTION: Required for ribosome biogenesis and telomere maintenance. Part of the H/ACA small nucleolar ribonucleoprotein (H/ACA snoRNP) complex, which catalyzes pseudouridylation of rRNA. This involves the isomerization of uridine such that the ribose is subsequently attached to C5, instead of the normal N1. Each rRNA can contain up to 100 pseudouridine ('psi') residues, which may serve to stabilize the conformation of rRNAs. May also be required for correct processing or intranuclear trafficking of TERC, the RNA component of the telomerase reverse transcriptase (TERT) holoenzyme. {ECO:0000269|PubMed:10757788, ECO:0000269|PubMed:15044956}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein;Ribosome biogenesis;Ubl conjugation;rRNA processing | This gene is a member of the H/ACA snoRNPs (small nucleolar ribonucleoproteins) gene family. snoRNPs are involved in various aspects of rRNA processing and modification and have been classified into two families: C/D and H/ACA. The H/ACA snoRNPs also include the DKC1, NOLA2 and NOLA3 proteins. These four H/ACA snoRNP proteins localize to the dense fibrillar components of nucleoli and to coiled (Cajal) bodies in the nucleus. Both 18S rRNA production and rRNA pseudouridylation are impaired if any one of the four proteins is depleted. These four H/ACA snoRNP proteins are also components of the telomerase complex. The encoded protein of this gene contains two glycine- and arginine-rich domains and is related to Saccharomyces cerevisiae Gar1p. Two splice variants have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:54433; | box H/ACA scaRNP complex [GO:0072589]; box H/ACA snoRNP complex [GO:0031429]; box H/ACA telomerase RNP complex [GO:0090661]; chromosome, telomeric region [GO:0000781]; fibrillar center [GO:0001650]; nucleoplasm [GO:0005654]; telomerase holoenzyme complex [GO:0005697]; box H/ACA snoRNA binding [GO:0034513]; RNA binding [GO:0003723]; telomerase RNA binding [GO:0070034]; snoRNA guided rRNA pseudouridine synthesis [GO:0000454]; telomere maintenance via telomerase [GO:0007004] | 17903301_Genome-wide association study of gene-disease association. (HuGE Navigator) 18989882_Observational study of gene-disease association. (HuGE Navigator) 18989882_heterozygous point mutations in NOLA1 gene are not responsible for aplastic anemia in our patients at least acting via telomere 19917616_The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. 20332099_Observational study of gene-disease association. (HuGE Navigator) 23707062_The deregulated expression and function of H/ACA snoRNPs may underlie specific pathological features of human disease. 33526451_SUMOylation- and GAR1-Dependent Regulation of Dyskerin Nuclear and Subnuclear Localization. | ENSMUSG00000028010 | Gar1 | 2201.089788 | 1.0161628 | 0.023131486 | 0.06819121 | 1.156889e-01 | 7.337581e-01 | 9.123299e-01 | No | Yes | 2283.547410 | 203.815960 | 2229.566866 | 198.965333 | |
| ENSG00000109572 | 1182 | CLCN3 | protein_coding | P51790 | FUNCTION: [Isoform 1]: Strongly outwardly rectifying, electrogenic H(+)/Cl(-)exchanger which mediates the exchange of chloride ions against protons (By similarity). The CLC channel family contains both chloride channels and proton-coupled anion transporters that exchange chloride or another anion for protons (PubMed:29845874). The presence of conserved gating glutamate residues is typical for family members that function as antiporters (PubMed:29845874). {ECO:0000250|UniProtKB:P51791, ECO:0000303|PubMed:29845874}.; FUNCTION: [Isoform 2]: Strongly outwardly rectifying, electrogenic H(+)/Cl(-)exchanger which mediates the exchange of chloride ions against protons. {ECO:0000269|PubMed:11967229}. | ATP-binding;Alternative splicing;Antiport;CBS domain;Cell membrane;Cell projection;Chloride;Endosome;Glycoprotein;Golgi apparatus;Ion transport;Lysosome;Membrane;Nucleotide-binding;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the voltage-gated chloride channel (ClC) family. The encoded protein is present in all cell types and localized in plasma membranes and in intracellular vesicles. It is a multi-pass membrane protein which contains a ClC domain and two additional C-terminal CBS (cystathionine beta-synthase) domains. The ClC domain catalyzes the selective flow of Cl- ions across cell membranes, and the CBS domain may have a regulatory function. This protein plays a role in both acidification and transmitter loading of GABAergic synaptic vesicles, and in smooth muscle cell activation and neointima formation. This protein is required for lysophosphatidic acid (LPA)-activated Cl- current activity and fibroblast-to-myofibroblast differentiation. The protein activity is regulated by Ca(2+)/calmodulin-dependent protein kinase II (CaMKII) in glioma cells. Multiple alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Aug 2011]. | hsa:1182; | apical plasma membrane [GO:0016324]; axon terminus [GO:0043679]; cell surface [GO:0009986]; cytoplasmic vesicle [GO:0031410]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; endosome membrane [GO:0010008]; external side of plasma membrane [GO:0009897]; GABA-ergic synapse [GO:0098982]; glutamatergic synapse [GO:0098978]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; inhibitory synapse [GO:0060077]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; integral component of postsynaptic membrane [GO:0099055]; integral component of synaptic vesicle membrane [GO:0030285]; intracellular membrane-bounded organelle [GO:0043231]; late endosome [GO:0005770]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; recycling endosome [GO:0055037]; ruffle membrane [GO:0032587]; secretory granule [GO:0030141]; specific granule [GO:0042581]; synaptic vesicle [GO:0008021]; vesicle membrane [GO:0012506]; antiporter activity [GO:0015297]; ATP binding [GO:0005524]; chloride channel activity [GO:0005254]; PDZ domain binding [GO:0030165]; solute:proton antiporter activity [GO:0015299]; voltage-gated chloride channel activity [GO:0005247]; volume-sensitive chloride channel activity [GO:0072320]; adult locomotory behavior [GO:0008344]; chloride transmembrane transport [GO:1902476]; endosomal lumen acidification [GO:0048388]; negative regulation of cell volume [GO:0045794]; phagocytosis, engulfment [GO:0006911]; photoreceptor cell maintenance [GO:0045494]; positive regulation of reactive oxygen species biosynthetic process [GO:1903428]; regulation of pH [GO:0006885]; synaptic transmission, GABAergic [GO:0051932]; synaptic transmission, glutamatergic [GO:0035249]; synaptic vesicle lumen acidification [GO:0097401] | 12183454_ClC-3 is an important molecular component underlying VSOACs and the RVD process in HeLa cells 12471024_results show that the ClC-3B PDZ-binding isoform resides in the Golgi where it co-localizes with a small amount of CFTR (cystic fibrosis transmembrane conductance regulator) 12842831_results suggest a fundamental role of endogenous ClC-3 in the swelling-activated Cl- channels function and cell volume regulation in human gastric epithelial cells 12843258_ClC-2 and ClC-3 channels are specifically upregulated in glioma membranes and endow glioma cells with an enhanced ability to transport Chloride 15073168_ClC-3 and ZnT3 reside in a common vesicle population where they functionally interact to determine vesicle luminal composition. 15596438_volume-regulated ClC-3 Cl(-) channels play important role in the regulation of [Cl(-)](i) and cell proliferation of vascular smooth muscle cells 16033995_Clcn3 was considered the most likely candidate of Cl- channel involved in volume regulation of human sperm. 16522634_ClC-3 is required for normal neutrophil oxidative function, phagocytosis, and transendothelial migration 17360969_This study demonstrates that superoxide flux across the endothelial cell plasma membrane occurs through chloride channel-3 channels and induces intracellular calcium release, which activates mitochondrial superoxide generation. 17869465_The relative density of CLC-3 mRNA was 0.22+/-0.09 and 0.12+/-0.05 in HNECs treated with 3 and 0.9% saline, respectively. 17882904_CLC-3 is upregulated in ethmoid mucosa and may affect the development of chronic rhinosinusitis without nasal polyps. 17908687_inhibition of the NADPH oxidase or ClC-3 in otherwise unstimulated cells elicited a phenotype similar to that seen after endotoxin priming, suggesting that basal oxidant production helps to maintain cellular quiescence. 17976378_oxidation induces surface expression of ClC-3 and activation of a ClC-3-dependent anion permeability in K562 cells 17977943_ClC-3 overexpression induced a pH-sensitive conductance in HEK293T cells that is biophysically similar to ClC-4 and ClC-5. 18077605_These data confirm that ClC-3 is important in VRAC function and cell volume regulation, is associated with the I(Cl,LPA) current activity, and participates in the fibroblast-to-myofibroblast transition. 18986326_An essential role of sClC-3 in native volume-sensitive outwardly rectifying anion channels function in mouse atrial myocytes. 20139089_CaMKII is a molecular link translating intracellular calcium changes, which are intrinsically associated with glioma migration, to changes in ClC-3 conductance required for cell movement 20945353_ClC-3 is involved in the activation of volume-activated chloride currents but not of stretch-activated chloride currents in hepatocellular carcinoma cells. 21115901_High ClC-3 is associated with extensive migration and invasion in glioma. 21792908_ClC-3 may be a main component of background chloride channels which can be activated under isotonic conditions by autocrine/paracrine ATP through purinergic receptor pathways; the background current is involved in maintenance of basal cell volume. 22049206_study demonstrates that premitotic condensation involves the activation of ClC-3 by Ca(2+)/calmodulin-dependent protein kinase II in glioma cells 22108225_ClC-3 protein may be considered as a potential tumor marker and therapeutic target for human nasopharyngeal carcinoma. 22371056_results reveal a cell cycle-dependent change of the subcellular distribution of ClC-3 and strongly suggest that ClC-3 has nucleocytoplasmic shuttling dynamics that may play key regulatory roles during different stages of the cell cycle in tumor cells. 22496242_ClC-3 is a candidate of the channel proteins that mediate or regulate the acid-activated chloride current in nasopharyngeal carcinoma cells. 23006728_Data indicate in umbilical vein endothelial cells transfected with ClC-3 siRNA showed activation of NF-kappaB. 23270726_our data suggest that the ClC-3 chloride channel is an important target of cyclin D1. Cyclin D1 may regulate the functional activities of the chloride channel via CDK4 and CDK6, and/or the expression of the chloride channel. 23345219_K(Ca)3.1 and ClC-3 are expressed in tissue samples obtained from patients diagnosed with grade IV gliomas. Both K(Ca)3.1 and ClC-3 colocalize to the invading processes of glioma cells 23408563_Suggest that ClC-3 suppression causes the inhibition of Akt and autophagy, which can enhance the therapeutic benefit of cisplatin in U251 cells. 23873092_ClC-3 deficiency inhibited Ang II-induced EPC apoptosis via suppressing ROS generation derived from NADPH oxidase. 24284495_swelling-activated Cl currents and CLC-3 play a role in pulmonary artery smooth muscle cell proliferation, but CLC-3 channels do not underlie swelling in these cells 25421907_Authors summarize the function of CLC-3 in cancer and discuss the mechanisms by which CLC-3 contributes to proliferation, apoptosis and drug resistance in cancer cells. [Review] 25514499_CLC3 is required in the activation and migration of human blood eosinophils. 25537517_Data indicate that cytoplasmic chloride channel-3 (ClC-3) plays an active and key role in tumor metastasis and may be a valuable prognostic biomarker and a therapeutic target to prevent tumor spread. 25973047_these results demonstrated that ClC-3 is involved in the proliferation and migration of osteosarcoma cell 26965430_ClC-3 promotes endometriotic cell migration and invasion. 27064645_Study provided novel and compelling evidence for the functional role of the unique CLC-3, which are significantly upregulated during ischemia, in the protection of the heart under stress 27451945_Transfection of cells with ClC-3 siRNA decreased the expression of cyclin D1, cyclin dependent kinase 4 and 6, and increased the expression of cyclin dependent kinase inhibitors (CDKIs), p21 and p27. Pretreatments of cells with p21 and p27 siRNAs depleted the inhibitory effects of ClC-3 siRNA on the expression of CDK4 and CDK6, but not on that of cyclin D1 28419445_ClC-3 is a potential target of 17beta-estradiol and is modulated by the ERalpha in breast cancer cells. 28972156_Unique oligomerization properties of ClC-4 permit regulated targeting of ClC-4 to various endosomal compartment systems via expression of different ClC-3 splice variants. 29749557_we demonstrated that ClC-3 can arrest the cell cycle at the G1 phase to inhibit cell proliferation, suggesting that ClC-3 has the potential to be a novel target for hepatocellular carcinoma (HCC) therapy and potentially improve the prognosis of HCC patients 29787012_CLC-3 may get involved in proliferation, invasion, and migration of ovarian cancer cells and thus may be a useful therapeutic target. 29945325_Findings suggest chloride channel 3 protein (ClC-3) a therapeutic target in multiple myeloma (MM). 30044661_ittle, if any, active H(+) transport, supported by ATP, occurred. Major transporters include cariporide-sensitive NHE1 in basolateral membranes and ClC3 and ClC5 in apical osteoblast membranes. The mineralization inhibitor levamisole reduced bone formation and expression of alkaline phosphatase, NHE1, and ClC5. 30217218_The expression and function of CLC-3 were promoted after XRCC5 overexpression, and the promotion effects were reversed by the CLC-3 knockdown. The mechanistic study revealed that knockdown of XRCC5 suppressed the binding of XRCC5 to the CLC-3 promoter and subsequent promoter activity, thus regulating CLC-3 expression at the transcriptional level by interacting with PARP1. 30230544_that ClC-3 expression in cancer cells induces mediating multidrug resistance by upregulating NF-kappaB-signaling-dependent P-glycoprotein expression involving another new mechanism for ClC-3 in the development of drug resistance of cancers 30678806_up-regulation of CIC-3 markedly correlated with tumor size and overall prognosis 30905432_ClC-3 participates in the processes of SCC cell migration and invasion and regulates MMP-9 expression via the PI3K/Akt/mTOR signaling pathway. 30992317_Starvation-induced autophagy is up-regulated via ROS-mediated ClC-3 chloride channel activation in the nasopharyngeal carcinoma cell line. 31281178_the expression of ClC-3 chloride channel in chondrosarcoma was examined. 32772229_ClC-3 induction protects against cerebral ischemia/reperfusion injury through promoting Beclin1/Vps34-mediated autophagy. 33069359_Suppression of CLC-3 reduces the proliferation, invasion and migration of colorectal cancer through Wnt/beta-catenin signaling pathway. 33093458_ClC-3/SGK1 regulatory axis enhances the olaparib-induced antitumor effect in human stomach adenocarcinoma. 33383561_Swelling-activated ClC-3 activity regulates prostaglandin E2 release in human OUMS-27 chondrocytes. 34186028_Unique variants in CLCN3, encoding an endosomal anion/proton exchanger, underlie a spectrum of neurodevelopmental disorders. | ENSMUSG00000004319 | Clcn3 | 3648.821246 | 0.8719294 | -0.197716785 | 0.06513213 | 9.213089e+00 | 2.402908e-03 | 5.622043e-02 | No | Yes | 4122.618762 | 781.919411 | 4734.968920 | 897.923071 | |
| ENSG00000110080 | 6484 | ST3GAL4 | protein_coding | Q11206 | FUNCTION: A beta-galactoside alpha2-3 sialyltransferase involved in terminal sialylation of glycoproteins and glycolipids (PubMed:8288606, PubMed:8611500). Catalyzes the transfer of sialic acid (N-acetyl-neuraminic acid; Neu5Ac) from the nucleotide sugar donor CMP-Neu5Ac onto acceptor Galbeta-(1->3)-GalNAc- and Galbeta-(1->4)-GlcNAc-terminated glycoconjugates through an alpha2-3 linkage (PubMed:8288606, PubMed:8611500). Plays a major role in hemostasis. Responsible for sialylation of plasma VWF/von Willebrand factor, preventing its recognition by asialoglycoprotein receptors (ASGPR) and subsequent clearance. Regulates ASGPR-mediated clearance of platelets (By similarity). Participates in the biosynthesis of the sialyl Lewis X epitopes, both on O- and N-glycans, which are recognized by SELE/E-selectin, SELP/P-selectin and SELL/L-selectin. Essential for selectin-mediated rolling and adhesion of leukocytes during extravasation (PubMed:25498912). Contributes to adhesion and transendothelial migration of neutrophils likely through terminal sialylation of CXCR2 (By similarity). In glycosphingolipid biosynthesis, sialylates GM1 and GA1 gangliosides to form GD1a and GM1b, respectively (PubMed:8288606). Metabolizes brain c-series ganglioside GT1c forming GQ1c (By similarity). Synthesizes ganglioside LM1 (IV3Neu5Ac-nLc4Cer), a major structural component of peripheral nerve myelin (PubMed:8611500). {ECO:0000250|UniProtKB:P61131, ECO:0000250|UniProtKB:Q91Y74, ECO:0000269|PubMed:25498912, ECO:0000269|PubMed:8288606, ECO:0000269|PubMed:8611500}. | Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Lipid metabolism;Membrane;Reference proteome;Secreted;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000305|PubMed:8288606}.; PATHWAY: Glycolipid biosynthesis. {ECO:0000305|PubMed:8288606}. | This gene encodes a member of the glycosyltransferase 29 family, a group of enzymes involved in protein glycosylation. The encoded protein is targeted to Golgi membranes but may be proteolytically processed and secreted. The gene product may also be involved in the increased expression of sialyl Lewis X antigen seen in inflammatory responses. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:6484; | extracellular region [GO:0005576]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; beta-galactoside (CMP) alpha-2,3-sialyltransferase activity [GO:0003836]; monosialoganglioside sialyltransferase activity [GO:0047288]; N-acetyllactosaminide alpha-2,3-sialyltransferase activity [GO:0008118]; neolactotetraosylceramide alpha-2,3-sialyltransferase activity [GO:0004513]; sialyltransferase activity [GO:0008373]; cognition [GO:0050890]; glycolipid biosynthetic process [GO:0009247]; glycoprotein biosynthetic process [GO:0009101]; keratan sulfate biosynthetic process [GO:0018146]; lipid glycosylation [GO:0030259]; O-glycan processing [GO:0016266]; oligosaccharide biosynthetic process [GO:0009312]; positive regulation of blood coagulation [GO:0030194]; positive regulation of leukocyte tethering or rolling [GO:1903238]; protein glycosylation [GO:0006486]; protein sialylation [GO:1990743]; sialylation [GO:0097503]; viral protein processing [GO:0019082] | 12375029_down-regulation of ST3Gal IV mRNA may be one of the factors associated with the malignant progression of human renal cell carcinoma. 12565846_Transcriptional regulation of human Galbeta1,3GalNAc/Galbeta1, 4GlcNAc alpha2,3-sialyltransferase (hST3Gal IV) gene in testis and ovary cell lines. 17054948_IL-1 beta-induced sLeX expression on HuH-7 cells was suppressed by transfection of gene-specific small interference RNAs against FUT VI and ST3Gal IV but not against FUT IV and ST3Gal III. 19520807_Thrombocytopenia in mice deficient in the St3gal4 sialyltransferase gene (St3Gal-IV(-/-) mice) is caused by the recognition of terminal galactose residues exposed on the platelet surface in the absence of sialylation. 19781661_Expression of ST3Gal IV in several gastrointestinal cell lines is correlated with the expression of sialyl Lewis x at the cell surface. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20383541_These data suggest that the ST3GAL4 gene is responsible for biosynthesis of the viral receptor and may play a crucial role in infection of H5N1 avian influenza virus in humans. 21140242_These results indicated that gastric cancer tissues expressed high levels of alpha 2,3-linked sialic acid residues, ST3Gal IV, and ST6Gal I. 22691873_TNF increases the expression of alpha2,3-sialyltransferase gene ST3GAL4 23726834_the role of ST3Gal IV in the acquisition of adhesive, migratory and metastatic capabilities and, secondly, in analyzing the expression of ST3Gal III and ST3Gal IV in pancreatic adenocarcinoma tissues, was investigated. 23799130_The expression of ST3GAL4 leads to SLe(x) antigen expression in gastric cancer cells which in turn induces an increased invasive phenotype through the activation of c-Met 24606438_The polymorphism analysis showed that at SNP rs10893506, genotypes CC and CT of the ST3GAL4 B3 promoter were associated with the presence of premalignant lesions and cervical cancer. 25086711_There may be a racial/ethnic-specific association, and/or sex-specific association between the ST3GAL4 rs11220462 SNP and serum lipid parameters in some ethnic groups. 25498912_ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes 26258617_Expression levels of sialyltransferases ST3GAL1 and ST3GAL4 were upregulated in the HRMECs after high-glucose stimulation. 27584569_study illustrates the power of next-generation sequencing in the discovery of new genetic variants and a significant ethnic diversity in the ST3GAL4 gene 27821620_TNF-responsive element in an intronic region of the ST3GAL4 gene, whose TNF-dependent activity is repressed by ERK/p38 and MSK1/2 inhibitors. This TNF-responsive element contains potential binding sites for ETS1 and ATF2 transcription factors related to TNF signaling. 28512058_ST3GAL IV affects apoptotic signal, cell proliferation and the effectiveness of imatinib treatment in chronic myeloid leukemia cells. 29667779_MiR-193a-3p and miR-224 promoted the cell proliferation and decreased apoptosis by targeting alpha-2,3-sialyltransferase IV in renal cell carcinoma. 29749491_The V1 transcript of the ST3GAL4 demonstrated significant decreased expression in premalignant and malignant cervical tissues. 31914669_a cross-restoration of each of the three genes in ST3GAL6 KO cells showed that overexpression of ST3GAL6 sufficiently rescued the total alpha2,3-sialylation levels, cell morphology, and alpha2,3-sialylation of EGFR, whereas the alpha2,3-sialylation levels of beta1 were greatly enhanced by an overexpression of ST3GAL4 32295993_Identification and characterization of the V3 promoter of the ST3GAL4 gene. 33080315_MiR-193b modulates osteoarthritis progression through targeting ST3GAL4 via sialylation of CD44 and NF-small ka, CyrillicB pathway. 33524390_Rab11-mediated post-Golgi transport of the sialyltransferase ST3GAL4 suggests a new mechanism for regulating glycosylation. | ENSMUSG00000032038 | St3gal4 | 587.592090 | 0.8991207 | -0.153413236 | 0.11829632 | 1.659754e+00 | 1.976365e-01 | 5.671309e-01 | No | Yes | 548.892662 | 58.434473 | 622.678052 | 65.988870 |
| ENSG00000110318 | 57562 | CEP126 | protein_coding | Q9P2H0 | FUNCTION: Participates in cytokinesis (PubMed:19799413). Necessary for microtubules and mitotic spindle organization (PubMed:24867236). Involved in primary cilium formation (PubMed:24867236). {ECO:0000269|PubMed:19799413, ECO:0000269|PubMed:24867236}. | Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Reference proteome | hsa:57562; | centrosome [GO:0005813]; ciliary base [GO:0097546]; cytoplasm [GO:0005737]; midbody [GO:0030496]; cilium assembly [GO:0060271]; cytoplasmic microtubule organization [GO:0031122]; mitotic spindle organization [GO:0007052]; non-motile cilium assembly [GO:1905515] | 22264561_KIAA1377 and C5orf42 gene synergistically play a role as susceptibility genes for monomelic amyotrophy. 24867236_CEP126 is a regulator of microtubule organisation at the centrosome that acts through modulation of the transport of pericentriolar satellites, and consequently, of the organisation of cell structure 30958603_It was exhibited that KIAA1377 was able to promote the proliferation and motility of both KYSE-150 and HeLa cells, which can be reverted by re-expression of let-7b-5p. The luciferase reporter assay verified that let-7b-5p can diametrically target KIAA1377. Collectively, our data demonstrated that let-7b-5p can directly but negatively regulate KIAA1377 in SCC cell lines, Ecal109, and HeLa cells. | ENSMUSG00000040729 | Cep126 | 87.989481 | 1.0336168 | 0.047701390 | 0.28783232 | 2.796561e-02 | 8.671897e-01 | No | Yes | 90.355633 | 21.731698 | 82.292109 | 19.823794 | |||
| ENSG00000110330 | 329 | BIRC2 | protein_coding | Q13490 | FUNCTION: Multi-functional protein which regulates not only caspases and apoptosis, but also modulates inflammatory signaling and immunity, mitogenic kinase signaling, and cell proliferation, as well as cell invasion and metastasis. Acts as an E3 ubiquitin-protein ligase regulating NF-kappa-B signaling and regulates both canonical and non-canonical NF-kappa-B signaling by acting in opposite directions: acts as a positive regulator of the canonical pathway and suppresses constitutive activation of non-canonical NF-kappa-B signaling. The target proteins for its E3 ubiquitin-protein ligase activity include: RIPK1, RIPK2, RIPK3, RIPK4, CASP3, CASP7, CASP8, TRAF2, DIABLO/SMAC, MAP3K14/NIK, MAP3K5/ASK1, IKBKG/NEMO, IKBKE and MXD1/MAD1. Can also function as an E3 ubiquitin-protein ligase of the NEDD8 conjugation pathway, targeting effector caspases for neddylation and inactivation. Acts as an important regulator of innate immune signaling via regulation of Toll-like receptors (TLRs), Nodlike receptors (NLRs) and RIG-I like receptors (RLRs), collectively referred to as pattern recognition receptors (PRRs). Protects cells from spontaneous formation of the ripoptosome, a large multi-protein complex that has the capability to kill cancer cells in a caspase-dependent and caspase-independent manner. Suppresses ripoptosome formation by ubiquitinating RIPK1 and CASP8. Can stimulate the transcriptional activity of E2F1. Plays a role in the modulation of the cell cycle. {ECO:0000269|PubMed:15665297, ECO:0000269|PubMed:18082613, ECO:0000269|PubMed:21145488, ECO:0000269|PubMed:21653699, ECO:0000269|PubMed:21931591, ECO:0000269|PubMed:23453969}. | 3D-structure;Activator;Alternative splicing;Apoptosis;Cytoplasm;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | The protein encoded by this gene is a member of a family of proteins that inhibits apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2, probably by interfering with activation of ICE-like proteases. This encoded protein inhibits apoptosis induced by serum deprivation and menadione, a potent inducer of free radicals. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2012]. | hsa:329; | CD40 receptor complex [GO:0035631]; cytoplasm [GO:0005737]; cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; membrane raft [GO:0045121]; nucleus [GO:0005634]; XY body [GO:0001741]; chaperone binding [GO:0051087]; cysteine-type endopeptidase inhibitor activity involved in apoptotic process [GO:0043027]; FBXO family protein binding [GO:0098770]; identical protein binding [GO:0042802]; protein N-terminus binding [GO:0047485]; protein-containing complex binding [GO:0044877]; transcription coactivator activity [GO:0003713]; transferase activity [GO:0016740]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; cell surface receptor signaling pathway [GO:0007166]; I-kappaB kinase/NF-kappaB signaling [GO:0007249]; necroptotic process [GO:0070266]; negative regulation of apoptotic process [GO:0043066]; negative regulation of necroptotic process [GO:0060546]; negative regulation of ripoptosome assembly involved in necroptotic process [GO:1902443]; NIK/NF-kappaB signaling [GO:0038061]; placenta development [GO:0001890]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of protein K48-linked ubiquitination [GO:1902524]; positive regulation of protein K63-linked ubiquitination [GO:1902523]; positive regulation of protein monoubiquitination [GO:1902527]; positive regulation of protein ubiquitination [GO:0031398]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein polyubiquitination [GO:0000209]; regulation of apoptotic process [GO:0042981]; regulation of cell cycle [GO:0051726]; regulation of cell differentiation [GO:0045595]; regulation of cell population proliferation [GO:0042127]; regulation of cysteine-type endopeptidase activity [GO:2000116]; regulation of inflammatory response [GO:0050727]; regulation of innate immune response [GO:0045088]; regulation of necroptotic process [GO:0060544]; regulation of NIK/NF-kappaB signaling [GO:1901222]; regulation of nucleotide-binding oligomerization domain containing signaling pathway [GO:0070424]; regulation of reactive oxygen species metabolic process [GO:2000377]; regulation of RIG-I signaling pathway [GO:0039535]; regulation of toll-like receptor signaling pathway [GO:0034121]; response to cAMP [GO:0051591]; response to ethanol [GO:0045471]; response to hypoxia [GO:0001666]; tumor necrosis factor-mediated signaling pathway [GO:0033209] | 12208731_Expression of cIAP1, a target for 11q22 amplification, correlates with resistance of cervical cancers to radiotherapy. 12209092_CD40 engagement enhances eosinophil survival through induction of cellular inhibitor of apoptosis protein 2 expression: implications for allergic inflammation 12218061_Smac-penetratin fusion peptide crossed the cellular membrane, bound XIAP and cIAP1, displaced caspase-3 from cytoplasmic aggregates, and enhanced drug-induced caspase action in situ 12243753_These results indicate that IAPs alone are not the main factor responsible for the resistance of non-small-cell lung cancer cells to treatment. 12388702_Reovirus-induced apoptosis involves reduction of cellular IAP1 protein levels 12525502_Cellular inhibitors of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. 12571250_TRAF2, TRAF3, cIAP1, Smac, and lymphotoxin beta receptor associate and are involved in apoptosis 12603340_c-IAP1 is an important intracellular modulator of Fas- as well as TNF-alpha death signalling pathways in human vascular smooth muscle cells. 12651874_cIAP1 and cIAP2 are potential oncogenes and are overexpressed in multiple lung cancers with or without higher copy numbers 12851723_although cIAP-1, cIAP-2 and XIAP transcripts were highly upregulated, their expression of endogenous proteins were not increased in HUVECs stimulated with LPS 12888921_cIAP-1 has a role in regulating cell survival in endometrial cancer cells 14708638_Relative risk of death was lower for cytoplasmic c-IAP1, cytoplasmic c-IAP2, and nuclear c-IAP2 expression. It was higher for nuclear c-IAP1 expression. 14960576_levels of c-IAP1 and c-IAP2 are regulated by Smac/DIABLO through the ubiquitin/proteasome pathway 14960583_HIAP2 is translationally induced during endoplasmic reticulum stress 15183896_X-linked XIAP is present in Chronic lymphocytic leukemia cells and is up-regulated in conditions where apoptosis is prevented. 15187025_c-IAP1 cellular location has a role in regulating cell differentiation 15318034_No association of the TAP2 gene with schizophrenia in the Korean population. 15359644_CIAP1 is downregulated and/or cleaved in a dose-dependent manner upon by anti-cancer drugs. cIAP-1's mitochondrial localization & liberation indicate a profoundly different function of this protein despite its similar modular structure to XIAP. 15501771_expression in urethral epithelium upregulated by Neisseria gonorrhoeae PorB IB and upregulation dependent on NF-kappaB activation 15665297_role for overexpressed cIAP1 in genetic instability, possibly by interfering with mitotic functions 15911110_nuclear cIAP-1 expression appears to be a useful marker for predicting poor prognosis in head and neck squamous cell carcinoma (HNSCC), and may play roles in HNSCCs through the signaling pathway mediated by Smac/DIABLO and caspase-3 16080516_There is endogenous cLAP1 expression in MKN45 cells, which may be a factor in the presumed anti-apoptotic system in these human gastric cancer cells. 16180223_IAP1 protects neural progenitor cells against TRAIL-induced apoptosis and suppresses caspase-3 activation. 16282325_a single cIAP can direct its E3 ligase activity toward different substrates and can alter the cellular functions of different protein targets, including TRAF2 and SMAC, in accordance with differences in the specificity of individual BIR domains 16339151_cIAP1 and cIAP2 bind but do not inhibit caspases 16510124_Since F-box proteins are specificity determining subunits of SCF ubiquitin protein ligases, our results suggest that Fbxo7 can mediate the ubiquitination of cIAP1 by SCF ubiquitin protein ligase and in the regulation of cIAP1 function. 16701639_Results indicate a novel mechanism by which HIAP2 can regulate ER-initiated apoptosis by modulating the activity of caspase-2. 16775178_High cIAP membranous expression is associated with epithelial ovarian cancer 16929535_IAP family proteins may the prognosis of multiple myeloma patients in association with chemotherapy-induced overexpression of MDR1 or LRP. 16983704_Differential expression of IAPs in B-cell lymphomas suggests differences in pathogenesis that may have implications for novel treatment strategies targeting IAPs. 17220297_TNFR2 signaling induces selective c-IAP1-dependent ASK1 ubiquitination and terminates mitogen-activated protein kinase signaling 17626072_Bortezomib inhibited expression of cIAP-1, cIAP-2, and XIAP, which are regulated by NF-kappaB and function as inhibitors of apoptosis. 17822677_Importantly, our findings suggest that a paradox exists whereby Nrf2 activity is beneficial in non-malignant cells but in cancer cells it may provide a selective advantage for clonal expansion. 17993464_Data show that Cartilage oligomeric matrix protein protects cells against death by elevating cIAP1 proteins. 18230607_ME-BS directly interacts with the BIR3 domain of cIAP1, promotes auto-ubiquitylation dependent on its RING domain, and facilitates proteasomal degradation of cIAP1. 18239673_Results suggest that HSP90 beta prevents auto-ubiquitination and degradation of its client protein c-IAP1, whose depletion would be sufficient to inhibit cell differentiation. 18434593_the RING domain of cIAP1 mediates the degradation of RING-bearing inhibitor of apoptosis proteins by distinct pathways 18566024_HIAP-1 and HIAP-2 mRNA levels were elevated in resting T cells while NAIP mRNA was increased in whole blood in multiple sclerosis 18570872_cIAP1 and cIAP2 promote cancer cell survival by functioning as E3 ubiquitin ligases that maintain constitutive ubiquitination of the RIP1 adaptor protein. 18621506_Nuclear, cytoplasmic and concurrent cIAP-1 immunoreactions were significantly correlated with lymph node metastasis in tongue squamous cell carcinomas. 18621737_c-IAP1 and c-IAP2 are required for TNFalpha-stimulated RIP1 ubiquitination and NF-kappaB activation. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18827186_once in the cytoplasm, cIAP1 is involved in the degradation of the adaptor protein tumor necrosis factor receptor-associated factor 2 (TRAF2) by the proteosomal machinery 18846111_Inhibitor of Apoptosis Proteins 1 is an important member of the inhibitor of apoptosis family of proteins and is involved in the regulation of the NF-kappa B-signalling pathway downstream of the Tumor Necrosis Factor receptor. 18931663_The ubiquitin-associated domain is essential for the oncogenic potential of cIAP1. 19029953_PERK activity inhibits the ER stress-induced apoptotic program through the induction of cellular inhibitor of apoptosis (cIAP1 and cIAP2) proteins 19154529_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19223549_cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. 19336370_Observational study of gene-disease association. (HuGE Navigator) 19397698_Immortalized cell lines from human gingival mucosal keratinocytes with epithelial mesenchymal transition (EMT) expressed significantly elevated levels of cIAP-1, Bclx and p27(kip) higher than non-EMT cells. 19416853_ARIA knockdown significantly increased inhibitor of apoptosis cIAP-1 and cIAP-2 protein expression 19567200_Results demonstrated that the inhibition of spontaneous B cell apoptosis by CpG DNA was correlated to up-regulation of Bcl-xL, IAP and down-regulation of p53 and caspase 3 mediated through PI3K/AKT signaling. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19720604_Flt-1 expression influences apoptotic susceptibility of vascular smooth muscle cells through the NF-kappaB/IAP-1 pathway. 19893574_The data presented are consistent with a model in which translation of cIAP1 is governed, at least in part, by NF45, a novel cellular IRES trans-acting factor. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20078866_IAPs could be involved in prostate disorder (BPH, PIN and PC) development since might be provoke inhibition of apoptosis and subsequently cell proliferation 20097753_virus-triggered ubiquitination of TRAF3 and TRAF6 by cIAP1 and cIAP2 is essential for type I IFN induction and cellular antiviral response 20447407_Results describe the complex between baculoviral IAP repeat (BIR) 1 of cIAP1/2 and the coiled-coil region of TRAF2. 20458734_These data suggest a novel role for BIRC2 in lipopolysaccharide-induced autophagy in vascular endothelial cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20667824_cIAP1 cooperatively interacts with oligomerized processed caspase-9 in the apoptosome and blocks procaspase-3 activation. 20932475_A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. 20951133_TRAIL-mediated apoptosis proceeds through caspase 8-dependent degradation of cIAP-1. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21113135_The authors demonstrate that c-IAP1 and UbcH5 family promote K11-linked polyubiquitination of receptor-interacting protein 1 (RIP1) in vitro and in vivo. 21331077_Data show that inhibitor-induced rapid degradation of cIAPs requires binding to TRAF2, and reveal an unexpected difference between cIAP1 and cIAP2. 21393245_Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization 21414315_Data show that NIPER-4 induces cIAP1-mediated ubiquitylation of CRABP-II, resulting in the proteasomal degradation. 21549626_The structure of the caspase recruitment domain (CARD) of cIAP1 is required for cIAP1 autoregulation. 21653699_one of the functions of cIAP1 when localized in the nucleus is to regulate E2F1 transcriptional activity. 21795072_Data show that inhibitor of apoptosis protein 1 (cIAP1) and cellular inhibitor of apoptosis protein (cIAP2) expression was significantly increased in bladder cancer compared with normal bladder urothelium. 21817100_cIAP1/2 are important regulators of inflammatory processes in endothelial cells. 21849505_The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. 21931591_cIAP1 and cIAP2 are direct E3 ubiquitin ligases for all four RIP proteins 22403404_resistance to Vpr-induced apoptosis is specifically mediated by cIAP1/2 genes independent of Bcl-xL and Mcl-1, which play a key role in maintaining cell viability. 22434933_c-IAP1 and c-IAP2 were required for canonical activation of NF-B and MAPK by members of the TNFR family. 22591684_Src, Akt, NF-kappaB, BCL-2 and c-IAP1 may be involved in an anti-apoptotic effect in patients with BCR-ABL positive and BCR-ABL negative acute lymphoblastic leukemia. 22733500_BIRC2 is amplified in a subset of squamous cell carcinoma of the uterine cervix. 22815481_cIAP1 plays a critical role in beta-cell survival under endoplasmic reticulum stress: by promoting ubiquitination and degradation of C/EBP homologous protein (CHOP) 22927431_IAP inhibitors and lexatumumab synergistically trigger apoptosis in a RIP1-dependent but TNFalpha-independent manner in RMS cells 23085658_c-IAP1 acts on both secretion of PCSK9 and its lysosomal localization. The novel pathway described here will open new avenues for exploring novel disease treatments. 23109427_a molecular mechanism for cIAP1's regulation in the NOD2 signaling pathway 23250032_BIRC2 and BIRC3 act as a molecular brake to rein in activation of the JNK signalling pathway and to fine-tune NF-kappaB and JNK signalling to ensure transcriptional responses are appropriately matched to extracellular inputs. 23269702_Data suggest that inhibitor of apoptosis (IAP) antagonist-based cancer treatment may be compromised by osteoporosis and enhanced skeletal metastasis, which may be prevented by antiresorptive agents. 23275033_The cIAP1 expression was decreased by siRNA silencing of Foxa2, but increased by Foxa2-expressing vectors. 23524849_These results suggest that OTUB1 regulates NF-kappaB and MAPK signalling pathways and TNF-dependent cell death by modulating c-IAP1 stability. 23605047_The 3'untranslated region of BIRC2 contains a cis-acting element that decreases stability of the message but increase protein expression 24008361_palmitate did not decrease cIAP-1 and cIAP-2 mRNA expression in the cells 24083782_Data indicate that a potent BIR2-selective inhibitor with in vivo pharmacokinetic properties which potentiates apoptotic signaling. 24093940_Data indicate the discovery of benzodiazepinone 36, a selective XIAP BIR2 inhibitors. 24194568_we note that the compounds that sensitize cancer cells to TRAIL are the most efficacious in binding to X-linked IAP, and in inducing cellular-IAP (cIAP)-1 and cIAP-2 degradation 24276241_we show that cIAP1 regulates TNF-induced actin cytoskeleton reorganization through a cdc42-dependentpathway 24371121_Data indicate that silencing of IAPs, IAP1 IAP2 and XIAP, reduced the TRAP-evoked RhoA activation. 24382102_higher expression in the ovarian endometriomas 24422988_Identify xIAP and cIAP1 as molecular targets of ceramide and show ceramide analog LCL85 is an effective sensitizer in overcoming resistance of metastatic colon and breast cancers to apoptosis induction to suppress metastasis in vivo. 24425875_cisplatin significantly triggered the proteasomal degradation of cellular inhibitor of apoptosis protein 1 and 2 (c-IAP1 and c-IAP2), and X-linked inhibitor of apoptosis (XIAP) in a ROS-dependent manner 24559922_Data suggest IAP1 is involved in pancreatic beta cell survival during endoplasmic reticulum stress; switching between IAP1 and transcription factor CHOP in unfolded protein response allows cells to survive or kills cells through apoptosis. [REVIEW] 24577083_Selenite caused CYLD upregulation via LEF1 and cIAP downregulation, both of which contribute to the degradation of ubiquitin chains on RIP1 and subsequent caspase-8 activation and colorectal tumor cell apoptosis. 24633224_cIAPs constitutively downregulate PACS-2 by polyubiquitination and proteasomal degradation, thereby restraining TRAIL-induced killing of liver cancer cells 24704827_Results identify IGF2BP1 as a critical translational regulator of cIAP1-mediated apoptotic resistance in rhabdomyosarcoma. 24841289_XIAP, cIAP1, and cIAP2, members of inhibitor of apoptosis (IAP) proteins, are critical regulators of cell death and survival and are attractive targets for new cancer therapy 24975362_Ubiquitin-dependent regulation of MEKK2/3-MEK5-ERK5 signaling module by XIAP and cIAP1.cIAP1 role in the physical and functional disassembly of ERK5-MAPK module and human muscle cell differentiation. 25046208_CIAP1 and cIAP2 represent novel therapeutic targets for the prevention of spontaneous preterm birth. 25079338_ARC is regulated via BIRC2/MAP3K14 signalling and its overexpression in AML or MSCs can function as a resistant factor to birinapant-induced leukaemia cell death. 25113061_cIAP1 and cIAP2 expression is increased in placenta from women with pre-existing maternal obesity and in response to treatment with pro-inflammatory cytokines 25139236_study demonstrate EndoG interacts with cIAP1; results indicate IAPs interact and ubiquitinate EndoG via K63-mediated isopeptide linkages without affecting EndoG levels and EndoG-mediated cell death, suggesting EndoG ubiquitination by IAPs may serve as a regulatory signal independent of proteasomal degradation 25273171_these findings suggest that GDC-0152 results in human leukemia apoptosis through caspase-dependent mechanisms involving down-regulation of IAPs and inhibition of PI3K/Akt signaling. 25383668_Although motions within each interface of the 'closed' monomer are insufficient to activate cIAP1, they enable associations with catalytic partners and activation factors. 25469840_cIAP1 undergoes a dramatic conformational change during activation that is now shown to be due to the dynamic and metastable nature of the closed form of the enzyme. 25549803_Data indicate that Smac/DIABLO showed an inverse correlation with inhibitor of apoptosis proteins XIAP, cIAP-1 and cIAP-2. 25635055_Survivin affected the stability of cIAP1 through binding, contributing to cell sensitivity to antineoplastic drug YM155. 25669656_The E3 ubiquitin ligase activity of XIAP and cIAP1 activates NFkappaB signalling, leading to the direct binding of p65 to the promoter of Beclin 1 and to its transcriptional activation and induction of autophagy. 25911380_Data suggest that the seventh zinc finger motif of DNA-binding protein A20 plays important role in NFkappaB-mediated apoptosis induced by tumor necrosis factor-alpha; A20 appears to bind and thus down-regulate inhibitor of apoptosis proteins cIAP1/2. 26011589_Data show that ectopic expression of interferon regulatory factor 1 (IRF-1) reduces NF-kappa B activity and suppresses TNF receptor-associated factor 2 (TRAF2) and inhibitor of apoptosis 1 protein (cIAP1) expression in breast cancer cells. 26168135_cIAP1 is associated with tumor progression in human ovarian cancer. 26314849_AZD5582 draws Mcl-1 down-regulation for induction of apoptosis through targeting of cIAP1 and XIAP in human pancreatic cancer 26666816_Overexpression of cIAP1 is associated with glioma. 26733177_High expression of cIAP1 is associated with triple-negative breast cancer. 27014915_High cIAP1 expression is associated with colorectal cancer. 27070702_cIAP1 and cIAP2 mediate BCL10 ubiquitination essential for BCR-dependent NF-kB activity in the ABC subtype of DLBCL. cIAP1/2 attach K63-linked polyubiquitin chains on themselves and on BCL10, recruiting IKK and LUBAC essential for IKK activation. 27197231_High cIAP1 expression is associated with Lung Tumorigenesis. 27608596_The baseline tumor necrosis factor tumor (TNFalpha) expression correlates with the sensitivity to Inhibitors of apoptosis proteins (IAPs) antagonist T-3256336. 27637083_we conclude that DMBT1 by binding with CRNDE and c-IAP1 associated with PI3K-AKT pathway is crucial for GBC carcinogenesis, and targeting this pathway may be pivotal in the treatment of GBC. 27693792_cIAP1-induced mitophagy led to dysfunctional mitochondria that resulted in abrogation of mitochondrial oxygen consumption rate along with the decrease in ATP levels. The ubiquitination of cIAP1 was found to be the critical regulator of mitophagy. 28515292_These data suggest that molluscum contagiosum virus MC159 competitively binds to NEMO to prevent cIAP1-induced NEMO polyubiquitination. 28542143_These results reveal an additional level of regulation of the stability and the activity of E2F1 by a non-degradative K63-poly-ubiquitination and uncover a novel function for the E3-ubiquitin ligase cIAP1. 28852129_Our results provide the first evidence for a mediator function of cIAP1 and collaborative activity of cIAP1 and CHIP, suggesting that maintaining balanced levels of these E3 ligases might be beneficial for normal cell growth. 28871172_the cytoplasmic retinoic acid receptor gamma (RARgamma) controls receptor-interacting protein kinase 1 (RIP1)-initiated cell death when cellular inhibitor of apoptosis (cIAP) activity is blocked. 29021293_We further identified this underlying mechanism also involved a PPARgamma-induced ANXA1-dependent autoubiquitination of cIAP1, the direct E3 ligase of RIP1, shifting cIAP1 toward proteosomal degradation..our study provides first insight for the suitability of using drug-induced expression of ANXA1 as a new player in RIP1-induced death machinery in triple-negative breast cancer 29518103_NAIP expression is most abundant in M2 macrophages, while cIAP1 and cIAP2 show an inverse pattern of expression in polarized cells, cIAP2 is preferentially expressed in M1-macrophages and cIAP1 in M2-macrophages. IAP antagonist treatment of resting M0 macrophages preceding polarization stimulation, induced upregulation of NAIP in M2 and downregulation of cIAP1 in M1 and M2 but an induction of cIAP2 in M1 macrophages. 29674627_cIAP1 regulates the EGFR/Snai2 axis in triple-negative breast cancer cells 30181285_study provides an interesting example using chemical biological approaches for determining distinct biological consequences from inhibiting vs. activating an E3 ubiquitin ligase and suggests a potential broad therapeutic strategy for targeting c-MYC in cancer treatment by pharmacologically modulating cIAP1 E3 ligase activity. 30352681_Pfn2 levels, regulated by cIAP1, affected intracellular levels of reactive oxygen species. 30359437_BIRC2 is a target gene for the E2F1 Transcription factor and is required for chromatin binding. 30421089_High cIAP1 expression is correlated with Head and Neck Cancer. 30523153_results disclose the mechanism by which cIAP1 RING dimer activates UbcH5B approximately Ub and indicate that noncovalent Ubiquitin (Ub) binding further stabilizes the cIAP1-UbcH5B approximately Ub complex in the active conformation to stimulate Ub transfer 30902881_cIAP1 is an oncological protein abundant in gallbladder cancer tissues, which enhances proliferation and immigration and blocks TNF-alpha from apoptosis through NF-kappaB pathway in vitro. 31217499_PAR-4 overcomes chemo-resistance in breast cancer cells by antagonizing cIAP1. 31266830_Dual Antagonist of cIAP/XIAP ASTX660 Sensitizes HPV(-) and HPV(+) Head and Neck Cancers to TNFalpha, TRAIL, and Radiation Therapy. 31922244_SMAC mimetic birinapant inhibits hepatocellular carcinoma growth by activating the cIAP1/TRAF3 signaling pathway. 31957840_MiR-5195-3p inhibits the proliferation of glioma cells by targeting BIRC2. 32020226_Tumor necrosis factorrelated apoptosisinducing ligand as a therapeutic option in urothelial cancer cells with acquired resistance against firstline chemotherapy. 32028675_A20 Promotes Ripoptosome Formation and TNF-Induced Apoptosis via cIAPs Regulation and NIK Stabilization in Keratinocytes. 32053868_Future Therapeutic Directions for Smac-Mimetics. 32322338_Increased Expression of BIRC2, BIRC3, and BIRC5 from the IAP Family in Mesenchymal Stem Cells of the Umbilical Cord Wharton's Jelly (WJSC) in Younger Women Giving Birth Naturally. 32553630_Stabilization of C-terminal binding protein 2 by cellular inhibitor of apoptosis protein 1 via BIR domains without E3 ligase activity. 32846130_BIRC2 Expression Impairs Anti-Cancer Immunity and Immunotherapy Efficacy. 33660894_Development of a protein signature to enable clinical positioning of IAP inhibitors in colorectal cancer. 34389694_Clinical Positioning of the IAP Antagonist Tolinapant (ASTX660) in Colorectal Cancer. 34681681_Inhibition of BIRC2 Sensitizes alpha7-HPV-Related Cervical Squamous Cell Carcinoma to Chemotherapy. 34997070_BIRC2-BIRC3 amplification: a potentially druggable feature of a subset of head and neck cancers in patients with Fanconi anemia. 35204822_Cytoplasmic and Nuclear Functions of cIAP1. | ENSMUSG00000057367 | Birc2 | 1228.360790 | 1.0935288 | 0.128991258 | 0.10258893 | 1.559504e+00 | 2.117379e-01 | 5.833886e-01 | No | Yes | 1048.897320 | 220.577955 | 946.036390 | 198.959686 | |
| ENSG00000111052 | 8825 | LIN7A | protein_coding | O14910 | FUNCTION: Plays a role in establishing and maintaining the asymmetric distribution of channels and receptors at the plasma membrane of polarized cells. Forms membrane-associated multiprotein complexes that may regulate delivery and recycling of proteins to the correct membrane domains. The tripartite complex composed of LIN7 (LIN7A, LIN7B or LIN7C), CASK and APBA1 associates with the motor protein KIF17 to transport vesicles containing N-methyl-D-aspartate (NMDA) receptor subunit NR2B along microtubules (By similarity). This complex may have the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Ensures the proper localization of GRIN2B (subunit 2B of the NMDA receptor) to neuronal postsynaptic density and may function in localizing synaptic vesicles at synapses where it is recruited by beta-catenin and cadherin. Required to localize Kir2 channels, GABA transporter (SLC6A12) and EGFR/ERBB1, ERBB2, ERBB3 and ERBB4 to the basolateral membrane of epithelial cells. {ECO:0000250|UniProtKB:Q8JZS0, ECO:0000269|PubMed:12967566}. | Cell junction;Cell membrane;Exocytosis;Membrane;Postsynaptic cell membrane;Protein transport;Reference proteome;Synapse;Tight junction;Transport | The protein encoded by this gene is involved in generating and maintaining the asymmetric distribution of channels and receptors at the cell membrane. The encoded protein also is required for the localization of some specific channels and can be part of a protein complex that couples synaptic vesicle exocytosis to cell adhesion in the brain. [provided by RefSeq, May 2016]. | hsa:8825; | basolateral plasma membrane [GO:0016323]; bicellular tight junction [GO:0005923]; cell-cell junction [GO:0005911]; extracellular exosome [GO:0070062]; MPP7-DLG1-LIN7 complex [GO:0097025]; plasma membrane [GO:0005886]; postsynaptic density membrane [GO:0098839]; presynapse [GO:0098793]; synapse [GO:0045202]; L27 domain binding [GO:0097016]; exocytosis [GO:0006887]; inner ear development [GO:0048839]; maintenance of epithelial cell apical/basal polarity [GO:0045199]; neurotransmitter secretion [GO:0007269]; protein localization to basolateral plasma membrane [GO:1903361]; protein transport [GO:0015031]; protein-containing complex assembly [GO:0065003]; synaptic vesicle transport [GO:0048489] | 12110687_coordinated folding and association of the LIN-2, -7 domain 17237226_LIN7A is a PDZ protein that interacts with human papillomavirus-16 E6 and forms a tripartite complex with MPP7 and DLG1, regulating the stability and localization of DLG1 to cell junctions. 18286632_Allelic and haplotype association was found between both BDNF and adult ADHD, and LIN-7 and adult ADHD. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24658322_LIN7A depletion disrupts cerebral cortex development, contributing to intellectual disability in 12q21-deletion syndrome. 26887652_This study therefore shows that LIN7A has a crucial role in the polarity abnormalities associated with breast carcinogenesis. 27320196_Overexpression of LIN7 or IRSp53 did not prevent the formation of hyperfused mitochondria in cells coexpressing the Drp1 K38A mutant, thus suggesting that LIN7-IRSp53 complex requires functional Drp1 to regulate mitochondrial morphology. 29749382_miR-501-3p suppresses metastasis and progression of hepatocellular carcinoma through targeting LIN7A. | ENSMUSG00000019906 | Lin7a | 251.492856 | 1.3408231 | 0.423118950 | 0.23846832 | 2.931514e+00 | 8.686601e-02 | No | Yes | 207.360033 | 40.288991 | 158.679623 | 30.856443 | ||
| ENSG00000111203 | 55846 | ITFG2 | protein_coding | Q969R8 | FUNCTION: As part of the KICSTOR complex functions in the amino acid-sensing branch of the TORC1 signaling pathway. Recruits, in an amino acid-independent manner, the GATOR1 complex to the lysosomal membranes and allows its interaction with GATOR2 and the RAG GTPases. Functions upstream of the RAG GTPases and is required to negatively regulate mTORC1 signaling in absence of amino acids. In absence of the KICSTOR complex mTORC1 is constitutively localized to the lysosome and activated. The KICSTOR complex is also probably involved in the regulation of mTORC1 by glucose. {ECO:0000269|PubMed:28199306}. | Alternative splicing;Lysosome;Membrane;Phosphoprotein;Reference proteome;Repeat | hsa:55846; | cytosol [GO:0005829]; KICSTOR complex [GO:0140007]; lysosomal membrane [GO:0005765]; nucleoplasm [GO:0005654]; cellular response to amino acid starvation [GO:0034198]; cellular response to glucose starvation [GO:0042149]; germinal center B cell differentiation [GO:0002314]; negative regulation of TORC1 signaling [GO:1904262]; regulation of TOR signaling [GO:0032006] | 28199306_identification of a protein complex (KICSTOR) that is composed of four proteins, KPTN, ITFG2, C12orf66 and SZT2, and that is required for amino acid or glucose deprivation to inhibit mTORC1 in cultured human cells | ENSMUSG00000001518 | Itfg2 | 446.972146 | 1.3321247 | 0.413729169 | 0.12930959 | 1.023025e+01 | 1.381559e-03 | No | Yes | 477.287256 | 42.938008 | 363.977703 | 32.928731 | |||
| ENSG00000111653 | 51147 | ING4 | protein_coding | Q9UNL4 | FUNCTION: Component of HBO1 complexes, which specifically mediate acetylation of histone H3 at 'Lys-14' (H3K14ac), and have reduced activity toward histone H4 (PubMed:16387653). Through chromatin acetylation it may function in DNA replication (PubMed:16387653). May inhibit tumor progression by modulating the transcriptional output of signaling pathways which regulate cell proliferation (PubMed:15251430, PubMed:15528276). Can suppress brain tumor angiogenesis through transcriptional repression of RELA/NFKB3 target genes when complexed with RELA (PubMed:15029197). May also specifically suppress loss of contact inhibition elicited by activated oncogenes such as MYC (PubMed:15029197). Represses hypoxia inducible factor's (HIF) activity by interacting with HIF prolyl hydroxylase 2 (EGLN1) (PubMed:15897452). Can enhance apoptosis induced by serum starvation in mammary epithelial cell line HC11 (By similarity). {ECO:0000250|UniProtKB:Q8C0D7, ECO:0000269|PubMed:15029197, ECO:0000269|PubMed:15251430, ECO:0000269|PubMed:15528276, ECO:0000269|PubMed:15897452, ECO:0000269|PubMed:16387653}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cell cycle;Chromatin regulator;Citrullination;Coiled coil;Metal-binding;Nucleus;Reference proteome;Tumor suppressor;Zinc;Zinc-finger | This gene encodes a tumor suppressor protein that contains a PHD-finger, which is a common motif in proteins involved in chromatin remodeling. This protein can bind TP53 and EP300/p300, a component of the histone acetyl transferase complex, suggesting its involvement in the TP53-dependent regulatory pathway. Multiple alternatively spliced transcript variants have been observed that encode distinct proteins. [provided by RefSeq, Jul 2008]. | hsa:51147; | cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; intermediate filament cytoskeleton [GO:0045111]; MOZ/MORF histone acetyltransferase complex [GO:0070776]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; methylated histone binding [GO:0035064]; transcription coactivator activity [GO:0003713]; apoptotic process [GO:0006915]; cell cycle [GO:0007049]; chromatin organization [GO:0006325]; DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator [GO:0006978]; DNA replication [GO:0006260]; histone H3 acetylation [GO:0043966]; histone H4-K12 acetylation [GO:0043983]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; histone modification [GO:0016570]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of growth [GO:0045926]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of apoptotic process [GO:0043065]; positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator [GO:1902164]; protein acetylation [GO:0006473]; regulation of cell cycle [GO:0051726]; regulation of cell cycle G2/M phase transition [GO:1902749]; regulation of cell growth [GO:0001558]; regulation of DNA biosynthetic process [GO:2000278]; regulation of transcription, DNA-templated [GO:0006355] | 12750254_p29ING4 and p28ING5 may be significant modulators of p53 function. 15029197_In mice, xenografts of human glioblastoma U87MG, which has decreased expression of ING4, grow significantly faster and have higher vascular volume fractions than control tumours 15251430_ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells 15528276_detected deletion of the ING4 locus in 10-20% of human breast cancer cell lines and primary breast tumors, supporting the possibility that ING4 might be a tumor suppressor gene 15882981_NLS domain of ING4 is essential for the binding of ING4 to p53 and the function of ING4 associated with p53 15897452_ING4 represses activation of the hypoxia inducible factor. 15935570_Frequent deletion and decreased mRNA expression of ING4 suggested it as a class two tumor suppressor gene and may play an important role in head and neck cancer. 16096374_mediates the ability of HIF to activate transcription of its downstream target genes [review] 16973615_Splice variants of ING4 are associated with cell growth and motility of cancer cells 17325660_data suggest that alternative splicing could modulate the activity of ING4 tumor suppressor protein 17517644_provide evidence for the role of ING4 in mediating the effect of low K intake on renal outer medullary K channel activity by stimulation of p38 and ERK mitogen-activated protein kinase 17848618_ING4 exerts an inhibitory effect on the production of proangiogenic molecules and consequently on MM-induced angiogenesis 18399550_may play an inhibitory role in lung adenocarcinoma by up-regulation or down-regulation of cell proliferation-regulating proteins such as p27, cyclinD1, SKP2, and Cox2 by means of inactivation of Wnt-1/beta-catenin signaling 18775696_Data suggest that the two wobble-splicing events at the exon 4-5 boundary influence subnuclear localization and degradation of ING4. 18779315_ING4 may specifically regulate the activity of NF-kappaB molecules that are bound to target gene promoters. 18789575_HCC risk associated with heavy alcohol intake and current smoking differed by this polymorphism among CLD patients. IL-1B -31T/C polymorphism may modify HCC risk in relation to alcohol intake or smoking 19034511_Findings suggest that exogenous ING4 can enhance A549 apoptosis via regulating the expression of Bcl-2 family proteins and the activation of mitochondrial apoptotic pathway. 19250543_Report down-regulation of the inhibitor of growth family member 4 (ING4) in different forms of pulmonary fibrosis. 19430401_ING4 has a potential role in growth suppression and apoptosis enhancement of melanoma through the activation of the mitochondrial-induced apoptotic pathway and the hindrance of the cell cycle. The deregulation of ING4 might be involved in melanomagenesis. 19571607_ING4 has a potential role on the growth suppression and apoptosis enhancement in gliomas U87MG via the activation of mitochondrial-induced apoptotic pathway and the hindrance of the cell cycle progression. 19775294_our data suggest an essential role for ING-4 in human astrocytoma development and progression possibly through regulation of the NF-kappaB-dependent expression of genes involved in tumor invasion 20053357_results show that ING4 is a bivalent reader of the chromatin H3K4me3 modification and suggest a mechanism for enhanced targeting of the HBO1 complex to specific chromatin sites 20381459_over-expression of miR-650 in gastric cancer may promote proliferation and growth of cancer cells, at least partially through directly targeting ING4. 20501848_A dominant mutant allele of the ING4 tumor suppressor found in human cancer cells exacerbates MYC-initiated mouse mammary tumorigenesis. 20705953_Mutations in ING4 is associated with cancer. 20707719_Loss of ING4 is associated with breast carcinoma. 20716169_Demonstrated decreased ING4 mRNA and expression in 100% (50/50) lung tumour tissues. Furthermore, ING4 expression was lower in grade III than in grades I-II tumours. Reduced ING4 mRNA correlated with lymph node metastasis. 21056991_ING4 is induced by BRMS1 and that it inhibits melanoma angiogenesis by suppressing NF-kappaB activity and IL-6 expression. 21177815_EBNA3C negatively regulate p53-mediated functions by interacting with ING4 and ING5. 21310648_results suggest that the decreases in nuclear ING4 may play important roles in tumorigenesis, progression and tumor differentiation in head and neck squamous cell carcinoma. 21315418_These results sustain the view that ING4 is a tumor suppressor in breast cancer and suggest that ING4 deletion may contribute to the pathogenesis of HER2-positive breast cancer. 21454715_Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53 21626442_Downregulated expression of inhibitor of growth 4 is associated with colorectal cancers. 21971889_In this review, the different properties of ING4 are discussed, and its activities are correlated with different aspects of cell physiology. [Review] 22078444_Mechanism of ING4 mediated inhibition of the proliferation and migration of human glioma cell line U251. 22228137_Data suggest that ING4 may be a promising target for the treatment for ovarian cancer. 22334692_crystal structure of the ING4 N-terminal domain 22436625_Inhibitor of growth 4 may represent an important biomarker for assessing the severity of breast cancer 22767438_Data suggested that miR-650 is correlated with the pathogenesis of hepatocellular carcinoma (HCC) and is involved in the HCC tumorigenesis process by inhibiting the expression of ING4. 23055189_Loss of ING4 expression is associated with lymphatic metastasis in colon cancer. 23056468_ING4 negatively regulates NF-kappaB in breast cancer 23181555_Report up-regulation of ING4 expression in sarcoid granulomas. 23504291_The low expression level of ING4 protein was correlated with high-risk gastrointestinal stromal tumors. 23603392_ING4 may regulate c-MYC translation by its association with AUF1. 23604125_These findings support a critical role for ING4 expression in normal cells in the non-cell-autonomous regulation of tumor growth. 23624912_ING4 acts as an E3 ubiquitin ligase to induce ubiquitination of p65 and degradation, which is critical to terminate NFkappaB activation. 23967213_The ING4 Binding with p53 and Induced p53 Acetylation were Attenuated by Human Papillomavirus 16 E6. 23969950_these findings suggest that ING4 may be a feasible modulator for the MDR phenotype of gastric carcinoma cells 24057236_ING4 level elevation mediated proliferation and invasion inhibition may be tightly associated with the suppression of NF-kappaB signaling pathway. 24130172_KAI1 overexpression increases ING4 expression in melanoma. 24157826_JWA has an important role in ING4-regulated melanoma angiogenesis, and ING4/JWA/ILK are promising prognostic markers and may be used as anti-angiogenic therapeutic targets for melanoma. 24762396_loss of ING4, either directly or indirectly through loss of Pten, promotes Myc-driven prostate oncogenesis. 25490312_work suggests that ING4 can suppress osteosarcoma progression through signaling pathways such as mitochondria pathway and NF-kappaB signaling pathway and ING4 gene therapy is a promising approach to treating osteosarcoma. 25571952_The enhanced antitumor activity generated by Ad.RGD-ING4-PTEN was closely associated with activation of the intrinsic and extrinsic apoptotic pathways and additive inhibition of tumor angiogenesis both in vitro and in vivo. 25790869_Data suggest a close connection between aberrant ING4 expression and the carcinogenesis of human bladder cells. 25792601_SCF(JFK) as a bona fide E3 ligase for ING4 and unraveled the JFK-ING4-NF-kappaB axis as an important player in the development and progression of breast cancer 25968091_This review summarizes the recent published literature that investigates the role of ING4 in regulating tumorigenesis and progression, and explores its potential for cancer treatment. [review] 26278569_MiR-761 directly targeted ING4 and TIMP2. 26544625_Data show that Ras protein regulates inhibitor of growth protein 4 (ING4)-thymine-DNA glycosylase (TDG)-Fas protein axis to promote apoptosis resistance in pancreatic cancer. 26803518_ING4 can facilitate cancer cell sensitivity to chemotherapy and radiotherapy. Although ING4 loss is observed for many types of cancers, increasing evidences show that ING4 can be used for gene therapy. In this review, the recent progress of ING4 regulating tumorigenesis is discussed 26936485_ING4 inhibits CRC invasion and metastasis probably via a switch from mesenchymal marker N-cadherin to epithelial marker E-cadherin through downregulation of Snail1 epithelial-mesenchymal transition (EMT)-inducing transcription factor (EMT-TF). 27381846_Inhibitor of growth 4 upregulation plus radiotherapy induced synergistic tumor suppression in SPC-A1 xenografts implanted in athymic nude mice. Thus, the restoration of inhibitor of growth 4 function might provide a potential strategy for non-small cell lung cancer radiosensitization. 27421660_results indicate that the combination of ING4 and PTEN may provide an effective therapeutic strategy for HCC 27471108_Low ING4 expression is associated with malignant phenotype and temozolomide chemoresistance in glioblastomas. 27527891_ING4 directly binds the Miz1 promoter and is required to induce Miz1 mRNA and protein expression during luminal cell differentiation. 27806345_Results found that ING4 expression was significantly reduced in CRC tissues and associated with increased lymph node metastasis, advanced TNM stage and poor overall survival. Also, ING4 suppressed CRC angiogenesis by inhibition of Sp1 expression and transcriptional activity through destabilization and ubiquitin degradation and down-regulation of Sp1 downstream pro-angiogenic factors MMP-2 and COX-2. 27926782_ING4 binds double-stranded DNA through its central region with micromolar affinity. 28050784_The oncogenic role of miR-330 in Hepatocellular Carcinoma Cells is linked to downregulation of ING4. 29207034_These results demonstrated that overexpression of ING4 can induce the apoptosis of melanoma cells and CD3+ T cells through signaling pathways such as the Fas/FasL pathway, and that ING4 gene therapy for melanoma treatment is a novel approach. 29489009_Both CELSR2 and ING4 display increased cytoplasmic staining in breast cancer cells compared to benign epithelium, suggesting a possible role of both genes in the pathogenesis of human mammary neoplasia. 30390334_Low ING4 expression is associated with cell proliferation and migration of renal cell carcinoma . 30403588_Splicing type of ING4 affects the translocation of ING4 proteins into the nucleus. 30643005_We also present the current understanding concerning the role of ING4 in the development of neoplastic and non-neoplastic diseases. These studies offer inspiration for pursuing novel therapeutics for various cancers. 31239073_These findings suggest that ING4 may inhibit hypoxia-induced EMT via decreasing HIF-1alpha and snail in HK2 cells, indicating the potential of ING4 as a therapeutic target for renal fibrosis. 31754246_Inhibitor of Growth 4 (ING4) is a positive regulator of rRNA synthesis. 31773467_differential expression profile revealed the potential role of ING4 in prostate cancer pathogenesis possibly through modulation of key genes, pathways and biological networks that are central drivers of the disease. 32008173_Found antibodies binding to p29ING4. 32033221_Macromolecular Crowding Increases the Affinity of the PHD of ING4 for the Histone H3K4me3 Mark. 33334698_ING4 Expression Landscape and Association With Clinicopathologic Characteristics in Breast Cancer. 33778834_[Expression levels of MMP-14, ING4 and HIF-1alpha in 60 patients with oral squamous cell carcinoma and their clinical significance]. 34787253_ING3 and ING4 immunoexpression and their relation to the development of benign odontogenic lesions. | ENSMUSG00000030330 | Ing4 | 337.381057 | 1.1023936 | 0.140639433 | 0.16492557 | 7.107155e-01 | 3.992064e-01 | No | Yes | 328.039606 | 34.877985 | 311.566516 | 33.082009 | ||
| ENSG00000112234 | 26235 | FBXL4 | protein_coding | Q9UKA2 | Cytoplasm;Disease variant;Leucine-rich repeat;Methylation;Mitochondrion;Nucleus;Primary mitochondrial disease;Reference proteome;Repeat;Ubl conjugation pathway | This gene encodes a member of the F-box protein family, which are characterized by an approximately 40 amino acid motif, the F-box. F-box proteins constitute one subunit of modular E3 ubiquitin ligase complexes, called SCF complexes, which function in phosphorylation-dependent ubiquitination. The F-box domain mediates protein-protein interactions and binds directly to S-phase kinase-associated protein 1. In addition to an F-box domain, the encoded protein contains at least 9 tandem leucine-rich repeats. The ubiquitin ligase complex containing the encoded protein may function in cell-cycle control by regulating levels of lysine-specific demethylase 4A. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:26235; | cytosol [GO:0005829]; mitochondrial intermembrane space [GO:0005758]; nuclear speck [GO:0016607]; SCF ubiquitin ligase complex [GO:0019005]; ubiquitin ligase complex [GO:0000151]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146]; ubiquitin-dependent protein catabolic process [GO:0006511] | 21757720_SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein 23993193_Mutations in FBXL4 are disease causing and establish FBXL4 as a mitochondrial protein with a possible role in maintaining mtDNA integrity and stability. 23993194_These data strongly support a role for FBXL4 in controlling bioenergetic homeostasis and mtDNA maintenance. 25868664_A clinical pattern of early-onset encephalopathy, persistent lactic acidosis, profound muscular hypotonia and typical facial dysmorphism should prompt initiation of molecular genetic analysis of FBXL4. 27182039_On clinical indication of mitochondrial encephalomyopathy, sequencing of FBXL4 should be performed, even when the activity levels of the MRC enzymes are normal 27743463_Overall, FBXL4 defects account for at least 0.7% (6 out of 808) of subjects suspected to have a mitochondrial disorder, and as high as 14.3% (4 out of 28) in young children with congenital lactic acidosis and clinical features of mitochondrial disease. Including FBLX4 in the mitochondrial diseases panel should be particularly important for patients with congenital lactic acidosis 28698647_Study found a common genomic copy number loss at 6q16.1-16.2, containing the FBXL4 gene in prostate cancer bone metastases. Loss of FBXL4 was also detected in primary tumors and it was highly associated with prognostic factors including high Gleason score, PSA, as well as poor patient survival, suggesting that FBXL4 loss contributes to prostate cancer progression. 28940506_Biallelic pathogenic variants in FBXL4 are associated with an encephalopathic mtDNA maintenance defect syndrome that is a multisystem disease. 31442532_FBXL4 protein promotes mitochondrial fusion. The C584R FBXL4 variant cannot promote mitochondrial fusion. 32525278_FBXL4 deficiency increases mitochondrial removal by autophagy. 32559514_Novel homozygous mutation in the FBXL4 gene is associated with mitochondria DNA depletion syndrome-13. | ENSMUSG00000040410 | Fbxl4 | 913.345880 | 0.9280545 | -0.107718610 | 0.09278864 | 1.350735e+00 | 2.451497e-01 | 6.178694e-01 | No | Yes | 844.949015 | 115.305466 | 892.021561 | 121.701318 | ||
| ENSG00000112511 | 5252 | PHF1 | protein_coding | O43189 | FUNCTION: Polycomb group (PcG) that specifically binds histone H3 trimethylated at 'Lys-36' (H3K36me3) and recruits the PRC2 complex. Involved in DNA damage response and is recruited at double-strand breaks (DSBs). Acts by binding to H3K36me3, a mark for transcriptional activation, and recruiting the PRC2 complex: it is however unclear whether recruitment of the PRC2 complex to H3K36me3 leads to enhance or inhibit H3K27me3 methylation mediated by the PRC2 complex. According to some reports, PRC2 recruitment by PHF1 promotes H3K27me3 and subsequent gene silencing by inducing spreading of PRC2 and H3K27me3 into H3K36me3 loci (PubMed:18285464 and PubMed:23273982). According to another report, PHF1 recruits the PRC2 complex at double-strand breaks (DSBs) and inhibits the activity of PRC2 (PubMed:23142980). Regulates p53/TP53 stability and prolonges its turnover: may act by specifically binding to a methylated from of p53/TP53. {ECO:0000269|PubMed:18086877, ECO:0000269|PubMed:18285464, ECO:0000269|PubMed:18385154, ECO:0000269|PubMed:23142980, ECO:0000269|PubMed:23150668, ECO:0000269|PubMed:23273982}. | 3D-structure;Alternative splicing;Chromatin regulator;Chromosomal rearrangement;Cytoplasm;Cytoskeleton;DNA damage;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a Polycomb group protein. The protein is a component of a histone H3 lysine-27 (H3K27)-specific methyltransferase complex, and functions in transcriptional repression of homeotic genes. The protein is also recruited to double-strand breaks, and reduced protein levels results in X-ray sensitivity and increased homologous recombination. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2009]. | hsa:5252; | cytoplasm [GO:0005737]; microtubule organizing center [GO:0005815]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone methyltransferase binding [GO:1990226]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; methylated histone binding [GO:0035064]; transcription corepressor binding [GO:0001222]; cellular response to DNA damage stimulus [GO:0006974]; chromatin organization [GO:0006325]; negative regulation of histone H3-K27 methylation [GO:0061086]; positive regulation of histone H3-K27 methylation [GO:0061087]; regulation of transcription, DNA-templated [GO:0006355] | 18086877_The role of PHF1 in H3K27 methylation and Hox gene silencing is reported. 18285464_PHF1 modulates the activity of Ezh2 in favor of the repressive H3K27me3 mark 18385154_PHF1 interacts physically with Ku70/Ku80, suggesting that PHF1 promotes nonhomologous end-joining processes. 18722875_predicted 684-amino acid JAZF1/PHF1 chimeric protein retained one zinc finger domain from JAZF1 and the two zinc finger domains from PHF1, and its oncogenic mechanism should be similar to that of the JAZF1/SUZ12 protein 19204726_Observational study of gene-disease association. (HuGE Navigator) 19851445_Observational study of gene-disease association. (HuGE Navigator) 22796436_The PHF1 gene, previously shown to be the 3'-partner of fusion genes in endometrial stromal tumors, is also recurrently involved in the pathogenesis of ossifying fibromyxoid tumors. 22918161_Morphological features, immunoprofile and fluorescence in situ hybridization rearrangements of JAZF1 and PHF1 genes were correlated with tumor category and outcome in endometrial sarcomas 23104054_findings show that the interaction of Phf19 with H3K36me2 and H3K36me3 is essential for PRC2 complex activity and for proper regulation of gene repression in embryonic stem cells 23142980_findings suggest that PHF1 can mediate deposition of the repressive H3K27me3 mark and acts as a cofactor in early DNA-damage response 23150668_Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis 23211293_All endometrial stromal sarcomas showing sex cords had PHF1 genetic rearrangement, suggesting that such rearrangements may induce sex cord differentiation. 23228662_The histone H3K36me3 binding by the Tudor domains of PHF1, PHF19 and likely MTF2 provide another recruitment and regulatory mechanism for the PRC2 complex. 23879974_PHF1b may be a molecular transducer of GABA A receptor function and thus GABA-mediated neurotransmission in the central nervous system. 23887158_Underscore the likely importance of PHF1 rearrangements in the pathogenesis of ossifying fibromyxoid tumors of soft parts. 23954330_full-length PHF1 in HEK293 cells co-localizes with histone K27me3, but not with K36me3, and this co-localization depends on the trimethyllysine binding pocket indicating that K27me3 is an in vivo target for the PHF1 Tudor domain 24285434_ZC3H7B-BCOR and MEAF6-PHF1 fusions occurred predominantly in S100 protein-negative and malignant OFMT. 24285434_ZC3H7B-BCOR and MEAF6-PHF1 fusions occurred predominantly in S100 protein-negative and malignant ossifying fibromyxoid tumors. 24530230_present two more ESS with MEAF6/PHF1 detected by transcriptome sequencing (case 1) and RT-PCR (case 2), proving that this fusion is recurrent in ESS 25923537_the F61L/S86F mutant of MTF2 Tudor-PHD1 was able to bind to H3K36me3 as strong as the PHF1 Tudor bound to this PTM . We concluded that the hydrophobic patch plays an essential role in binding of these Tudors to methylated chromatin 26494712_PCL1 binds to and stabilizes p53 to induce cellular quiescence 28082396_the increase of DNA accessibility within the H3K36me3-containing nucleosome, instigated by the Tudor domain of PHF1 binding to H3K36me3, is dramatically enhanced by the PHF1 N-terminal domain. 29846670_PHF1 promotes cell proliferation, invasion, and tumorigenesis in vivo and in vitro and its expression is markedly upregulated in a variety of human cancers 31932680_PHF1 fusions cause distinct gene expression and chromatin accessibility profiles in ossifying fibromyxoid tumors and mesenchymal cells. 32237188_A novel MBTD1-PHF1 gene fusion in endometrial stromal sarcoma: A case report and literature review. 33179613_Primary malignant ossifying fibromyxoid tumour of the bone. A clinicopathologic and molecular report of two cases. 34560087_A PHF1-TFE3 fusion atypical ossifying fibromyxoid tumor with prominent collagenous rosettes: Case report with a brief review. | ENSMUSG00000024193 | Phf1 | 150.600565 | 1.2818647 | 0.358243971 | 0.21348203 | 2.812698e+00 | 9.352099e-02 | No | Yes | 158.168926 | 23.100207 | 126.211721 | 18.714566 | ||
| ENSG00000113369 | 57561 | ARRDC3 | protein_coding | Q96B67 | FUNCTION: Adapter protein that plays a role in regulating cell-surface expression of adrenergic receptors and probably also other G protein-coupled receptors (PubMed:20559325, PubMed:21982743, PubMed:23208550). Plays a role in NEDD4-mediated ubiquitination and endocytosis af activated ADRB2 and subsequent ADRB2 degradation (PubMed:20559325, PubMed:23208550). May recruit NEDD4 to ADRB2 (PubMed:20559325). Alternatively, may function as adapter protein that does not play a major role in recruiting NEDD4 to ADRB2, but rather plays a role in a targeting ADRB2 to endosomes (PubMed:23208550). {ECO:0000269|PubMed:20559325, ECO:0000269|PubMed:23208550}. | 3D-structure;Cell membrane;Cytoplasm;Endosome;Lysosome;Membrane;Reference proteome;Repeat | This gene encodes a member of the arrestin family of proteins, which regulate G protein-mediated signaling. The encoded protein is thought to act as a regulator of breast cancer growth and progression by binding to a phosphorylated form of integrin beta4, a tumor-related antigen, targeting the integrin for internalization and degradation. [provided by RefSeq, Jul 2016]. | hsa:57561; | cytoplasm [GO:0005737]; early endosome [GO:0005769]; endosome [GO:0005768]; lysosome [GO:0005764]; plasma membrane [GO:0005886]; beta-3 adrenergic receptor binding [GO:0031699]; fat pad development [GO:0060613]; negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway [GO:0071878]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of heat generation [GO:0031651]; negative regulation of locomotion involved in locomotory behavior [GO:0090327]; positive regulation of ubiquitin-protein transferase activity [GO:0051443]; protein transport [GO:0015031]; skin development [GO:0043588]; temperature homeostasis [GO:0001659] | 16269462_[Thioredoxin-binding protein-2-like inducible membrane protein, TLIMP] TLIMP, a novel VD3- or PPARgamma ligand-inducible membrane-associated protein, plays a regulatory role in cell proliferation and PPARgamma activation. 19605364_Txnip regulates cellular metabolism independent of its binding to thioredoxin and the arrestin domains are crucial structural elements in metabolic functions of alpha-arrestin proteins 20306291_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20559325_Data show that ARRDC3 interacts with NEDD4 through two conserved PPXY motifs and recruits NEDD4 to the activated beta2-adrenergic receptor. 20603614_ARRDC3 directly binds to a phosphorylated form of ITGbeta4 leading to its internalization, ubiquitination and ultimate degradation, and suppress breast cancer progression. 21652578_single-nucleotide polymorphisms in ARRDC3, FLT1, and SKAP1 were significant predictors for survival androgen-deprivation therapy in prostate cancer patients. 21982743_The obesity risk locus contains a single gene, arrestin domain-containing 3 (ARRDC3), which interacts directly with beta-adrenergic receptors and is gender-specific. 23208550_Beta 2 adrenergic receptor endocytosis requires beta-arrestin2 but not ARRDC3. 24379409_High affinity binding of full-length ARRDC3 and Nedd4 is driven by the avid interaction of both PPXY motifs with either the WW2-WW3 or WW3-WW4 combinations. 25148870_Promoter hypermethylation is an important mechanism of the transcriptional inactivation of ARRDC3 in invasive ductal breast carcinoma. 25220262_The ARRDC3 is one of six known human alpha-arrestins, and has been implicated in the downregulation of the beta2-adrenergic receptor (beta2AR). 26490116_We found that ARRDC3 is required for ALIX ubiquitination induced by activation of PAR1 27109471_For the in vivo experiment, miR-182-5p overexpression also promoted the growth and progression of prostate cancer tumors. In this regard, we suggest that miR-182-5p may be a key androgen receptor-regulated factor that contributes to the development and metastasis of Chinese prostate cancers and may be a potential target for the early diagnosis and therapeutic studies of prostate cancer. 27226565_ARRDC3 functions as a switch to modulate the endosomal residence time and subsequent intracellular signaling of the beta2AR 28782483_correlation analysis indicated that the expression of ARRDC3 was negatively correlated with ITGbeta4 in clinical prostate cancer (PCa) tissues and cell lines. Our data revealed that ARRDC3 can serve as a tumor suppressor to inhibit PCa progression and an independent marker to predict the risk of biochemical recurrence and metastasis after radical resection of PCa. 29348172_a critical link between the tumor suppressor ARRDC3 and regulation of GPCR trafficking and signaling in breast cancer 29364502_ARRDC3 suppresses colorectal cancer progression through destabilizing the oncoprotein YAP 30412241_ARRDC3 is involved in the infectious entry of human papillomavirus into the cell 31295851_Authors identified miR-200b as a major target gene of ARRDC3. miR-200b played an essential role in mediating ARRDC3 dependent reversal of EMT phenotypes and chemo-resistance to DNA damaging agents in TNBC cells. Expression of miR-200b also increased the expression of ARRDC3 as well in TNBC cells, suggesting a positive feedback loop between these two molecules. 31341404_Study observed that the depletion of ARRDC3 in human hepatocytes resulted in the downregulation of inflammasome pathway-associated genes and the enhancement of apoptosis of hepatic stellate cells treated with their conditioned media. Results demonstrated ARRDC3 may play a role in the development of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. 31665660_Upregulated ARRDC3 limits trophoblast cell invasion and tube formation and is associated with preeclampsia. 32634127_Arrestin domain containing 3 promotes Helicobacter pylori-associated gastritis by regulating protease-activated receptor 1. 33722977_alpha-Arrestin ARRDC3 tumor suppressor function is linked to GPCR-induced TAZ activation and breast cancer metastasis. 34415232_MicroRNA-624-mediated ARRDC3/YAP/HIF1alpha axis enhances esophageal squamous cell carcinoma cell resistance to cisplatin and paclitaxel. 34748117_ARRDC3 polymorphisms may affect the risk of glioma in Chinese Han. | ENSMUSG00000074794 | Arrdc3 | 313.305562 | 1.1979304 | 0.260544047 | 0.20132182 | 1.601279e+00 | 2.057220e-01 | No | Yes | 359.482591 | 88.002462 | 310.239632 | 76.065442 | ||
| ENSG00000113532 | 7903 | ST8SIA4 | protein_coding | Q92187 | FUNCTION: Catalyzes the polycondensation of alpha-2,8-linked sialic acid required for the synthesis of polysialic acid (PSA), which is present on the embryonic neural cell adhesion molecule (N-CAM), necessary for plasticity of neural cells. | 3D-structure;Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | The protein encoded by this gene catalyzes the polycondensation of alpha-2,8-linked sialic acid required for the synthesis of polysialic acid, a modulator of the adhesive properties of neural cell adhesion molecule (NCAM1). The encoded protein, which is a member of glycosyltransferase family 29, is a type II membrane protein that may be present in the Golgi apparatus. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:7903; | Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity [GO:0003828]; sialic acid binding [GO:0033691]; cellular protein modification process [GO:0006464]; ganglioside biosynthetic process [GO:0001574]; N-glycan processing [GO:0006491]; nervous system development [GO:0007399]; oligosaccharide metabolic process [GO:0009311]; protein glycosylation [GO:0006486] | 12138100_SIAT8D has a role in neural development and sialic acid synthesis on NCAM 12227654_Pancreatitis risk was highest in individuals with abnormalities in pancreatic duct(CFTR) and acini (PST1) indicating that PST1 is a modifier gene for CFTR-related idiopathic chronic pancreatitis. 15710344_The upregulation of ST8SIA4 and the donwregulation of ST8SIA2 by valproic acid in HUVEC and tumor cell lines are reported. 16027151_polysialyltransferase ST8Sia IV/PST recognizes specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule 16229822_Observational study of gene-disease association. (HuGE Navigator) 18384787_PSA-NCAM and ST8Sia-II/IV Expression Is Increased in Pancreatic Carcinomas 19336400_Amino acid substitutions in conserved sequences are critical for the protein-specific polysialylation of NCAM. 19578876_Observational study of gene-disease association. (HuGE Navigator) 19725832_polysialylated NCAM persistence, up-regulated polysialyltransferase-1 mRNA and previously uncovered defective myelin-associated glycoprotein may be early pathogenetic events in adult-onset autosomal-dominant leukodystrophy 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22184126_The ST8SiaIV/PST polybasic region plays a critical role in substrate recognition. 23801331_ST8SiaIV synthesized polySia selectively on a NRP2 glycoform that was characterized by the presence of sialylated core 1 and core 2 O-glycans. 24531716_this study indicated that sialylation involved in the development of MDR of AML cells probably through ST3GAL5 or ST8SIA4 regulating the activity of PI3K/Akt signaling and the expression of P-gp and MRP1. 25855199_ST8SIA4 gene is involved in the development of multidrug resistance via PI3K/Akt pathway in chronic myeloid leukemia. 26884342_Sequence Requirements for Neuropilin-2 Recognition by ST8SiaIV and Polysialylation of Its O-Glycans. 27074314_This is the first report to demonstrate a role for a glycosyltransferase in human pluripotent stem cell lineage specification. 27527856_Data show that miR-181c was inversely correlated with the levels of ST8SIA4 expression in chronic myelocytic leukemia (CML) cell lines and samples. 28032858_changes in the glycosylation patterns and sialylation levels may be useful markers of the progression of breast cancer, as well as miR-26a/26b may be widely involved in the regulation of sialylation machinery by targeting ST8SIA4. 28233978_The polybasic region of the polysialyltransferase ST8Sia-IV binds directly to NCAM. 28427206_our results demonstrate that miR-146a and miR-146b promote proliferation, migration and invasion of FTC via inhibition of ST8SIA4. 28810663_Different properties of polysialic acids synthesized by the polysialyltransferases ST8SIA2 and ST8SIA4 have been described. | ENSMUSG00000040710 | St8sia4 | 44.395668 | 1.2068382 | 0.271232311 | 0.41951144 | 4.177431e-01 | 5.180653e-01 | No | Yes | 53.714703 | 15.917819 | 45.909522 | 13.476716 | |
| ENSG00000113645 | 23286 | WWC1 | protein_coding | Q8IX03 | FUNCTION: Probable regulator of the Hippo/SWH (Sav/Wts/Hpo) signaling pathway, a signaling pathway that plays a pivotal role in tumor suppression by restricting proliferation and promoting apoptosis. Along with NF2 can synergistically induce the phosphorylation of LATS1 and LATS2 and can probably function in the regulation of the Hippo/SWH (Sav/Wts/Hpo) signaling pathway. Acts as a transcriptional coactivator of ESR1 which plays an essential role in DYNLL1-mediated ESR1 transactivation. Regulates collagen-stimulated activation of the ERK/MAPK cascade. Modulates directional migration of podocytes. Acts as a substrate for PRKCZ. Plays a role in cognition and memory performance. {ECO:0000269|PubMed:15081397, ECO:0000269|PubMed:16684779, ECO:0000269|PubMed:18190796, ECO:0000269|PubMed:18596123, ECO:0000269|PubMed:18672031, ECO:0000269|PubMed:20159598, ECO:0000269|PubMed:23778582}. | 3D-structure;Activator;Alternative splicing;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Membrane;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a cytoplasmic phosphoprotein that interacts with PRKC-zeta and dynein light chain-1. Alleles of this gene have been found that enhance memory in some individuals. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2010]. | hsa:23286; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; protein-containing complex [GO:0032991]; ruffle membrane [GO:0032587]; kinase binding [GO:0019900]; molecular adaptor activity [GO:0060090]; transcription coactivator activity [GO:0003713]; cell migration [GO:0016477]; establishment of cell polarity [GO:0030010]; negative regulation of hippo signaling [GO:0035331]; negative regulation of organ growth [GO:0046621]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of MAPK cascade [GO:0043410]; regulation of hippo signaling [GO:0035330]; regulation of intracellular transport [GO:0032386]; regulation of transcription, DNA-templated [GO:0006355] | 12559952_Data describe the isolation of a cDNA coding for a novel protein, KIBRA, possessing two amino-terminal WW domains, an internal C2-like domain and a carboxy-terminal glutamic acid-rich stretch 15081397_identification as novel substrate for protein kinase C zeta and regulation of cellular function by same enzyme 16684779_the DLC1-KIBRA interaction is essential for ER transactivation in breast cancer cells 17053149_Genome-wide association study of gene-disease association. (HuGE Navigator) 17053149_gene expression studies showed that KIBRA was expressed in memory-related brain structures; KIBRA allele-dependent differences in hippocampal activations during memory retrieval were detected 17353070_Observational study of gene-disease association. (HuGE Navigator) 17353070_The impact of KIBRA on memory is most likely of high relevance in elderly subjects as it is in young. 17707552_Observational study of gene-disease association. (HuGE Navigator) 17707552_The current study reveals that KIBRA (rs17070145) T allele (CT and TT genotypes) is associated with an increased risk (OR 2.89; p=0.03) for very-late-onset (after the age of 86 years) AD. 18190796_KIBRA may play a role in how the reproductive state influences the mammary epithelial cell to respond to changing cell-context information, such as experienced during the tissue remodeling events of mammary gland development. 18194457_KIBRA T-->C polymorphism contributes to modulate episodic memory amongst community-dwelling older adults free of dementia 18194457_Observational study of gene-disease association. (HuGE Navigator) 18205171_No association is found between Kibra and memory performance in multiple memory tasks. 18205171_Observational study of gene-disease association. (HuGE Navigator) 18378080_Observational study of gene-disease association. (HuGE Navigator) 18378080_the KIBRA genotype could affect memory performance in a different way in those that complain of memory deficits compared to those that do not. 18789830_Observational study of gene-disease association. (HuGE Navigator) 18789830_This study findings suggest that KIBRA is associated with both individual variation in normal episodic memory and predisposition to late-onset Alzheimer's disease. 19058786_Observational study of gene-disease association. (HuGE Navigator) 19397951_Results suggest a role for the T-->C substitution in intron 9 of KIBRA in a component of episodic memory involved in long-term storage in an aged population. 19606085_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19606085_study concludes that variation in KIBRA influences cognitive flexibility in a population-specific way, and that current smoking status moderates this effect 19804789_This study showed that the KIBRA and CLSTN2 genes interactively modulate episodic memory performance. 20125193_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20150879_Even if T/C polymorphism of the KIBRA gene induces memory disturbances, they may be unspecific and unselective for recurrent depressive disorder. 20150879_Observational study of gene-disease association. (HuGE Navigator) 20185150_Observational study of gene-disease association. (HuGE Navigator) 20185150_This study replicates the association between the KIBRA gene and episodic memory and suggests a possibly differential effect of the polymorphism in psychotic and non-psychotic individuals. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20509760_KIBRA is highly expressed in the brain and kidneys, and is reported to be involved in synaptic plasticity. Therefore, we first tried to replicate the association between the SNP and memory performance in a Japanese subjects. 20509760_Observational study of gene-disease association. (HuGE Navigator) 20691427_Observational study of gene-disease association. (HuGE Navigator) 20881395_Observational study of gene-disease association. (HuGE Navigator) 20881395_Results show a strong association between the KIBRA gene and episodic memory impairment in Alzheimer's disease (AD), but show no influence on AD in our Japanese cohort. KIBRA might have an effect similar to cognitive reserve. 21173572_KIBRA methylation occurs frequently in B-cell acute lymphocytic leukemia but not in epithelial cancers and is linked to specific genetic event in B-ALL. 21185624_The results of this study suggested a modest role for KIBRA as a cognition and AD risk gene, and also highlight the multifactorial complexity of its genetic associations. 21204039_a pooled GWAS study undertaken to find novel genes associated with episodic memory performance; a genomic locus for KIBRA was found to be associated with memory performance in three cognitively normal cohorts from Switzerland and the USA 21233212_KIBRA associates with and activates Lats (large tumor suppressor) 1 and 2 kinases by stimulating their phosphorylation on the hydrophobic motif 21346737_Potential associations of the common KIBRA rs17070450 genotype are comprehensively tested with cognitive functions in a medically well-characterized cohort of community-dwelling elderly individuals. 21497093_The results strongly support the notion that KIBRA regulates epithelial cell polarity by suppressing apical exocytosis through direct inhibition of aPKC kinase activity in the PAR3-aPKC-PAR6 complex. 21643791_No increased risk of any type of late development, and cognitive impairment was associated with KIBRA (rs17070145) 21878642_KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. 21943600_These results indicate that KIBRA regulates higher brain function by regulating AMPAR trafficking and synaptic plasticity. 21976506_Enhanced memory performance in KIBRA T allele carriers is linked to elevated hippocampal functioning, rather than to neural compensation in noncarriers. 22129841_KIBRA genotype, as well as the number of intake drug abuse problems and a younger age, were associated with an increased risk of relapse 22430796_analysis of clinical-pathological parameters showed that KIBRA methylation was associated with unfavorable biological prognostic parameters, including unmutated IGHV genes (p = 0.007) and high CD38 expression (p < 0.05). 22614006_Decreased expression of KIBRA is associated with breast cancer. 22784093_identified KIBRA Ser(542) and Ser(931) as main phosphorylation sites for CDK1 both in vitro and in vivo 22794909_The results suggest that the KIBRA rs17070145 T-allele effects are specific to episodic memory and support the hypothesis that associations between rs17070145 variation and memory are disparate between healthy and impaired populations. 22904328_the KIBRA-Aurora-Lats2 protein complexes form a novel axis that regulates precise mitosis. 23065961_findings indicate that SNP rs17070145 located within KIBRA explains 0.5 percent of the variance for episodic memory tasks and 0.1 percent of the variance for working memory tasks in samples of primarily Caucasian background 23163744_High expression of KIBRA is correlated with lymphatic (P = 0.046) and venous invasion (P = 0.039). 23266749_findings suggest that the polymorphism in the KIBRA gene affects matter volume and the function of the default-mode network and executive control network 23582269_study reveals an association between two WWC1 SNPs and the likelihood of PTSD development. 23733450_KIBRA/WWC1 rs17070145 polymorphism has effect on spatial memory in humans and age differences in the reliance on landmark and boundary-related spatial information. 23778582_exonic missense variants in the C2 domain modify lipid binding and cognitive performance 23926262_This study demonistrated that KIBRA T-allele (rs17070145) had a larger hippocampal volume relative to noncarriers. The structural differences observed were specific to the CA fields and DG regions of the hippocampus. 24072042_Data indicate transcription factor TCF7L2 as a regulator of KIBRA gene expression. 24190487_KIBRA might associate with younger Alzheimer's disease patients (=74 years) in a Northern Han Chinese population. 24269383_KIBRA knockdown impaired cell migration and proliferation in breast cancer cells. 24290728_Results show a dynamic relationship between WWC1 rs17070145 polymorphism and increasing age on neuronal activity in the hippocampal region and cognitive performance 24642126_KIBRA is involved in human tumorigenesis and other physiological processes including cell polarity, membrane/vesicular trafficking, mitosis and cell migration. 25023289_overexpression of Kibra rescues the increased cell migration and aberrant three-dimensional morphogenesis induced by knockdown of PTPN14, and this rescue is mediated through the activation of the upstream LATS1 kinase 25080189_The results of this study supports the view that effects of KIBRA and CLSTN2 polymorphisms genetic polymorphisms on cognitive functioning may be most easily disclosed at suboptimal levels of cognitive ability, such as in old-age depression. 25146696_the study demonstrates an association between the KIBRA gene and episodic memory (immediate free recall) and suggests a differential effect of this genetic variant in early-onset schizophrenia and healthy siblings 25656173_In brief, we showed that individuals with MD[major depression] that were WWC1 rs17070145TT carriers reported more severe 'concentration difficulties' than C-carriers. 25800888_KIBRA rs17070145 affects specific phenotypes (high alcohol consumption, APOE synergism) of Japanese patients with Alzheimer disease in an age-dependent manner. Right inferior frontal gyrus hyperperfusion was seen in the C allele carrier group. 26156558_Study observed a trend for a better memory performance and significantly higher volumes and better microstructural integrity in the subfields comprising the CA2/3 and CA4/DG of the hippocampus in KIBRA T-allele carriers compared with non-T-carriers 26405221_WWC1 variants are involved in AMPA receptor trafficking and neuronal plasticity, which are associated with cognitive impairment and schizophrenia susceptibility. 26415670_Study did not find support for an association of KIBRA either alone or in combination with CLSTN2 with memory performance or hippocampal volume, nor did variation in these genes influence longitudinal memory decline or hippocampal atrophy in older adults 26768155_decreased expression of the KIBRA gene on both mRNA and protein levels in depression, is reported. 26929199_Phosphorylation-dependent regulation of the DNA damage response of Kibra protein in cancer cells 27041503_study provides a mechanistic link between aberrant tau acetylation, KIBRA deficiency, and dementia in Alzheimer's disease 27060629_No significant differences in allelic or gene frequency distribution of KIBRA SNP rs17070145 were observed between schizophrenic and healthy control groups. 27123786_Ability to navigate in the wilderness benefits from episodic memory (KIBRA). 27220053_show that KIBRA is overexpressed in human prostate tumors. Our studies uncover unexpected results and identify KIBRA as a tumor promoter in prostate cancer 27620974_that APOE and KIBRA have region-dependent additive and epistatic interactions on brain connectivity in healthy young adults 28079891_we report for the first time that CRB3 acts as an upstream regulator of the Hippo pathway to regulate contact inhibition by recruiting other Hippo molecules, such as Kibra and/or FRMD6, in mammary epithelial cells. 28380666_our results suggest that the effect of the WWC1 rs17070145 polymorphism on memory performance and Alzheimer's disease might be due to a differential regulation of the MAPK signaling, a key pathway involved in memory and learning processes 28472652_Study identifies four likely Tourette disorder risk genes with multiple de novo damaging variants in unrelated probands: WWC1 (WW and C2 domain containing 1), CELSR3 (Cadherin EGF LAG seven-pass G-type receptor 3), NIPBL (Nipped-B-like), and FN1 (fibronectin 1). 28859866_the overall data in the present meta-analysis revealed a significant association of KIBRA polymorphism rs17070145 and risk of Alzheimer disease. [meta-analysis] 28982981_Data (including data from studies in knockout mice) suggest that KIBRA plays important role in regulating HPO activity, YAP signaling, and actin cytoskeletal dynamics in podocytes; expression of KIBRA and YAP plus phosphorylation of YAP are up-regulated in glomeruli of patients with focal segmental glomerulosclerosis. (KIBRA = kidney/brain protein-KIBRA; HPO = hepatopoietin protein; YAP = Yes associated protein-1) 28990091_the study indicated that loss of WWC1 may contribute to podocyte apoptosis by inducing nuclear relocation of dendrin protein, which provided novel insight into the molecular events in podocyte apoptosis. 29032191_In younger adults, no effects of KIBRA were found 29046731_Data show that promoter methylation is a major mechanism involved in KIBRA expression regulation. KIBRA expression levels are reduced in clear cell renal cell carcinoma (ccRCC) and alterations in the balance of KIBRA/SP1 binding by promoter methylation may be involved in the onset and/or progression of ccRCC. 29391469_KIBRA rs17070145 is associated with accelerated cognitive decline in APOE epsilon4-positive cognitively normal adults with high Abeta-amyloid burden. 29562176_KIBRA functions co-operatively with the protein tyrosine phosphatase PTPN14 to trigger mechanotransduction-regulated signals that inhibit the nuclear localization of oncogenic transcriptional co-activators YAP/TAZ. Our results argue that the selective advantage produced by 5q loss involves reduced dosage of KIBRA, promoting oncogenic functioning of YAP/TAZ in TNBC. 29724824_These results indicate that KIBRA C2 domain association with membranes is calcium-independent and involves distinctive C2 domain-membrane relative orientations. 29793439_Overall low-KIBRA expression has an independent effect on the recurrence-free survival (RFS) and predicts the RFS outcome of luminal breast cancer patients who received endocrine therapy and breast cancer patients who received chemotherapy. 30121333_theses results reveal crosstalk between H3K27me3 inhibition catalyzed by EZH2 and CpG island methylation mediated by DNMT1 within the wwc1 promoter, which synergistically silence wwc1 gene expression in triple negative breast cancer. 30134813_Our findings suggest that the T-allele of KIBRA rs17070145 supports recollection based episodic memory retrieval and contributes to memory accuracy in a gender dependent manner. 30201328_Epistasis between variants of WWC1 and TLN2 in association to Alzheimer's. 30372890_MiR-21 suppressed the Hippo signaling pathway and promoted the progression of lung adenocarcinoma through targeting KIBRA. 30729420_KIBRA polymorphism modulates gray matter volume to influence cognitive ability in the elderly. 30953258_Our results confirm a role of the KIBRA T allele on progression of cognitive decline 31112471_The Influence of Hippocampal Dopamine D2 Receptors on Episodic Memory Is Modulated by BDNF and KIBRA Polymorphisms. 31560173_SOX2 antagonizes WWC1 to drive YAP1 activation in esophageal squamous cell carcinoma. 31597702_We show that recombinant human KIBRA WW2 domain is primarily disordered, binds SYNPO with relatively weak affinity and remains largely disordered in the complex. 32057830_Interaction of COMT and KIBRA modulates the association between hippocampal structure and episodic memory performance in healthy young adults. 33901448_KIBRA connects Hippo signaling and cancer. 34441043_Low KIBRA Expression Is Associated with Poor Prognosis in Patients with Triple-Negative Breast Cancer. | ENSMUSG00000018849 | Wwc1 | 1209.877661 | 0.9365894 | -0.094511359 | 0.09925730 | 9.157630e-01 | 3.385899e-01 | 6.976909e-01 | No | Yes | 1254.934799 | 88.969342 | 1354.325018 | 95.884267 | |
| ENSG00000113739 | 8614 | STC2 | protein_coding | O76061 | FUNCTION: Has an anti-hypocalcemic action on calcium and phosphate homeostasis. | Disulfide bond;Glycoprotein;Hormone;Phosphoprotein;Reference proteome;Secreted;Signal | This gene encodes a secreted, homodimeric glycoprotein that is expressed in a wide variety of tissues and may have autocrine or paracrine functions. The encoded protein has 10 of its 15 cysteine residues conserved among stanniocalcin family members and is phosphorylated by casein kinase 2 exclusively on its serine residues. Its C-terminus contains a cluster of histidine residues which may interact with metal ions. The protein may play a role in the regulation of renal and intestinal calcium and phosphate transport, cell metabolism, or cellular calcium/phosphate homeostasis. Constitutive overexpression of human stanniocalcin 2 in mice resulted in pre- and postnatal growth restriction, reduced bone and skeletal muscle growth, and organomegaly. Expression of this gene is induced by estrogen and altered in some breast cancers. [provided by RefSeq, Jul 2008]. | hsa:8614; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum lumen [GO:0005788]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; perinuclear region of cytoplasm [GO:0048471]; enzyme binding [GO:0019899]; heme binding [GO:0020037]; hormone activity [GO:0005179]; protein homodimerization activity [GO:0042803]; cellular calcium ion homeostasis [GO:0006874]; cellular response to hypoxia [GO:0071456]; decidualization [GO:0046697]; embryo implantation [GO:0007566]; endoplasmic reticulum unfolded protein response [GO:0030968]; negative regulation of gene expression [GO:0010629]; negative regulation of multicellular organism growth [GO:0040015]; regulation of hormone biosynthetic process [GO:0046885]; regulation of store-operated calcium entry [GO:2001256]; response to oxidative stress [GO:0006979]; response to peptide hormone [GO:0043434]; response to vitamin D [GO:0033280] | 15367391_STC-2 can act as a potent growth inhibitor and reduce intramembranous and endochondral bone development and skeletal muscle growth, implying that these tissues are specific physiological targets of stanniocalcins 15486227_Recombinant human and fish STC2 proteins were generated and found to be N-glycosylated homodimers. STC2 is a functional homodimeric glycoprotein, and thecal cell-derived STC2 could play a paracrine role during follicular development. 17545519_NTN4, TRA1, and STC2 have roles in progression of breast cancer 17909264_The induction of GDF15 and STC2 is likely specific to MK-4, vitamin K2 analog. 18355956_elevated expression of STC-1 or STC-2 act as survival factors also for breast cancer cells and thereby contribute to tumor dormancy. 18394600_STC2 was aberrantly hypermethylated in human cancer cells. 18492817_Stanniocalcin 2 expression is regulated by hormone signalling and negatively affects breast cancer cell viability in vitro. 19298603_stanniocalcin 2 overexpression in a prostate cancer cell line promoted prostate cancer cell growth, indicating its oncogenic property 19415750_Increased STC2 gene expression is associated with colorectal cancer. 19582875_Stanniocalcin 2 promotes invasion and is associated with metastatic stages in neuroblastoma. 19786016_STC2 is a HIF-1 target gene and is involved in the regulation of cell proliferation. 20174869_Human stanniocalcin-1 or -2 expressed in mice reduces bone size and severely inhibits cranial intramembranous bone growth. 20422456_High expression level of Stanniocalcin 2 is associated with gastric cancer. 20424473_Observational study of gene-disease association. (HuGE Navigator) 20619259_STC2 is a positive regulator in tumor progression at hypoxia. 20734150_High STC2 is associated with lymph node metastases in squamous cell esophageal carcinoma. 22503972_These findings point to three novel functions of STC2, and suggest that STC2 interacts with HO1 to form a eukaryotic 'stressosome' involved in the degradation of heme. 23187001_STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration. 23548070_High STC2 expression levels are associated with disease recurrence in patients with gastric cancer. 23664860_Stanniocalcin-1 and -2 promote angiogenic sprouting in HUVECs via VEGF/VEGFR2 and angiopoietin signaling pathways. 23906305_High STC2 expression is associated with lymph node metastasis in squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. 24100594_Our results indicate that STC2 could be a useful molecular blood marker for predicting tumor progression by monitoring CTCs in patients with gastric cancer. 24375080_stanniocalcin 2 (STC2) was the most highly upregulated gene in anti-VEGF antibody-treated tumors 24503185_Interleukin-1-induced changes in the glioblastoma secretome suggest its role in tumor progression. 24606961_STC2 overexpression correlates to poor prognosis for nasopharyngeal carcinomas. 24729417_High STC2 expression is associated with glioma. 24743310_Circulating STC1 and STC2 mRNA are potentially useful blood markers for LSCC 25234931_The aim of this study was to evaluate the clinical value of measuring expression levels of STC2 in colorectal cancer (CRC) patients. 25463045_These findings suggest that STC2 can be a potential lung cancer biomarker and plays a positive role in lung cancer metastasis and progression. 25674244_Stanniocalcin 2 may contribute to tumor development and radioresistance in cervical cancer. 25771305_This study utilized ER+ IBC to identify a metagene including ABAT and STC2 as predictive biomarkers for endocrine therapy resistance. 25830567_STC2 may inhibit epithelial-mesenchymal transition at least partially through the PKC/Claudin-1-mediated signaling in human breast cancer cells. 26165228_The results showed that the expression of HMGA2 and STC2 was positively correlated. Furthermore, STC2 expression was significantly associated with tumor grade and histotype. 26361149_STC2 has a role in promoting cell proliferation and cisplatin resistance in cervical cancer 26424558_Up-regulation of CDK2 and CDK4 and down-regulation of cell cycle inhibitors p16 and p21 were observed after the delivery of STC2. Furthermore, STC2 transduction activated pAKT and pERK 1/2 signal pathways. 26874357_Data suggest that stanniocalcin 1 and 2 (STC1, STC2) participate in inhibition of proteolytic activity of pregnancy-associated plasma protein-A (PAPP-A) during folliculogenesis. 26983002_STC2 is involved in regulating PAPP-A activity during the development of atherosclerosis 27346255_Our results demonstrated the contrasting effects of STC1 and STC2-derived peptides on human macrophage foam cell formation associated with ACAT1 expression and on HASMC migration. 27662663_STC2 promotes colorectal cancer tumorigenesis and epithelial-mesenchymal transition (EMT) progression through activating ERK/MEK and PI3K/AKT signaling pathways. 27863406_Results show that STC2 promotes head and neck squamous cell carcinoma cell proliferation, tumor growth, and metastasis through the PI3K/AKT/Snail pathway. 27878259_These findings indicated that STC2 may promote osteoblast differentiation and mineralization by regulating ERK activation 27939696_Mus81 knockdown suppresses proliferation and survival of HCC cells likely by downregulating STC2 expression, implicating Mus81 as a therapeutic target for HCC. 28887636_MiR-184 was confirmed to directly target STC2 in glioblastoma cells 28990324_STC2, is a possible early marker during development of diabetes mellitus. 29712555_In STEMI patients, Stanniocalcin-2 and IGFBP-4 emerged as independent predictors of all-cause death and readmission due to HF. The Stanniocalcin-2/PAPP-A/IGFBP-4 axis exhibits a significant role in STEMI risk stratification. 29748538_these findings newly identify STC2 as the first stanniocalcin responsible for mediating the immunomodulatory effects of MSCs on allogeneic T cells and STC2 contribute to MSC-based treatment for ACD mainly via reducing the CD8(+) Tc1 cells. 29956502_In conclusion, STC2 expression is abundant in male breast cancer where it is an independent prognostic factor for disease-free survival. 30661222_high-expression of STC2 is a negative prognostic factor, associated with increased risk of recurrence in endometrioid endometrial cancer 30962272_The high expression of STC2 mRNA and protein expression in hepatocellular carcinoma (HCC) may be associated with the occurrence, development, and prognosis of HCC. STC2 may also be possible to help developing new therapeutic strategies for HCC. 31075234_LINC00460 and STC2 were highly expressed while miR-206 was poorly expressed in head and neck squamous cell carcinoma . Besides, miR-206 was found to bind to both LINC00460 and STC2. 31100207_Study shows for the first time an opposite effect of hSTC-1 and hSTC-2 on glyceride-glycerol generation from glucose in epididymal white adipose tissue (eWAT) of fed rats and provides evidence for a direct role of hSTC-1 and hSTC-2 in the regulation of lipid and glucose metabolism in eWAT of rats. 31103608_Total IGFBP 5, PAPPA, PAPPA2 and Stanniocalcin-2. 31173256_The results demonstrated that STC2 participates in the development and progression of colorectal cancer (CRC) by promoting CRC cell proliferation, survival and migration and activating the Wnt/betacatenin signaling pathway. 31236000_Study found that STC2 is overexpressed in colon adenocarcinoma tissues and positively correlated with poor prognosis. The binding site of the transcription factor Sp1 is widely located in the promoter region of STC2. Knocking down the expression of Sp1 significantly inhibited the transcription activity of STC2. 31845230_Results showed that the expression level of STC2 gene in breast cancer tissues was significantly higher than in normal breast tissues. The expression of STC2 gene was correlated with lymph node metastasis, distant metastasis, TNM stage and histological grade. 31982135_STC2 modulates ERK1/2 signaling to suppress adipogenic differentiation of human bone marrow mesenchymal stem cells. 32296395_New Insights Into Physiological and Pathophysiological Functions of Stanniocalcin 2. 32445747_Stanniocalcin 2 contributes to aggressiveness and is a prognostic marker for oral squamous cell carcinoma. 32900244_Antioncogenic Effect of MicroRNA-206 on Neck Squamous Cell Carcinoma Through Inhibition of Proliferation and Promotion of Apoptosis and Autophagy. 33234031_BIRC7 and STC2 Expression Are Associated With Tumorigenesis and Poor Outcome in Extrahepatic Cholangiocarcinoma. 33774276_The novel prognostic risk factor STC2 can regulate the occurrence and progression of osteosarcoma via the glycolytic pathway. 33797657_Stanniocalcin-2 promotes cell EMT and glycolysis via activating ITGB2/FAK/SOX6 signaling pathway in nasopharyngeal carcinoma. 33813422_Clinical Significance of Stanniocalcin2 mRNA Expression in Patients With Colorectal Cancer. 34099278_Prognostic Correlation of Glycolysis-Related Gene Signature in Patients with Laryngeal Cancer. 34612765_The significance of Stanniocalcin 2 in malignancies and mechanisms. 34881630_Blocking stanniocalcin 2 reduces sunitinib resistance in clear cell renal cell carcinoma. 35157929_Stanniocalcin2 inhibits the epithelial-mesenchymal transition and invasion of trophoblasts via activation of autophagy under high-glucose conditions. | ENSMUSG00000020303 | Stc2 | 2000.340507 | 0.8218436 | -0.283064242 | 0.07515963 | 1.415167e+01 | 1.686470e-04 | 1.136886e-02 | No | Yes | 2033.829614 | 211.923233 | 2480.860425 | 258.275537 | |
| ENSG00000113790 | 1962 | EHHADH | protein_coding | Q08426 | FUNCTION: Peroxisomal trifunctional enzyme possessing 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and delta 3, delta 2-enoyl-CoA isomerase activities. Catalyzes two of the four reactions of the long chain fatty acids peroxisomal beta-oxidation pathway (By similarity). Can also use branched-chain fatty acids such as 2-methyl-2E-butenoyl-CoA as a substrate, which is hydrated into (2S,3S)-3-hydroxy-2-methylbutanoyl-CoA (By similarity). Optimal isomerase for 2,5 double bonds into 3,5 form isomerization in a range of enoyl-CoA species (Probable). Also able to isomerize both 3-cis and 3-trans double bonds into the 2-trans form in a range of enoyl-CoA species (By similarity). With HSD17B4, catalyzes the hydration of trans-2-enoyl-CoA and the dehydrogenation of 3-hydroxyacyl-CoA, but with opposite chiral specificity (PubMed:15060085). Regulates the amount of medium-chain dicarboxylic fatty acids which are essential regulators of all fatty acid oxidation pathways (By similarity). Also involved in the degradation of long-chain dicarboxylic acids through peroxisomal beta-oxidation (PubMed:15060085). {ECO:0000250|UniProtKB:P07896, ECO:0000250|UniProtKB:Q9DBM2, ECO:0000269|PubMed:15060085, ECO:0000305|PubMed:15060085}. | Acetylation;Alternative splicing;Disease variant;Fatty acid metabolism;Isomerase;Lipid metabolism;Lyase;Multifunctional enzyme;NAD;Oxidoreductase;Peroxisome;Phosphoprotein;Reference proteome | PATHWAY: Lipid metabolism; fatty acid beta-oxidation. {ECO:0000269|PubMed:15060085}. | The protein encoded by this gene is a bifunctional enzyme and is one of the four enzymes of the peroxisomal beta-oxidation pathway. The N-terminal region of the encoded protein contains enoyl-CoA hydratase activity while the C-terminal region contains 3-hydroxyacyl-CoA dehydrogenase activity. Defects in this gene are a cause of peroxisomal disorders such as Zellweger syndrome. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2009]. | hsa:1962; | cytosol [GO:0005829]; peroxisomal matrix [GO:0005782]; peroxisome [GO:0005777]; 3-hydroxyacyl-CoA dehydrogenase activity [GO:0003857]; dodecenoyl-CoA delta-isomerase activity [GO:0004165]; enoyl-CoA hydratase activity [GO:0004300]; enzyme binding [GO:0019899]; intramolecular oxidoreductase activity, transposing C=C bonds [GO:0016863]; long-chain-3-hydroxyacyl-CoA dehydrogenase activity [GO:0016509]; long-chain-enoyl-CoA hydratase activity [GO:0016508]; NAD+ binding [GO:0070403]; fatty acid beta-oxidation [GO:0006635]; fatty acid beta-oxidation using acyl-CoA oxidase [GO:0033540] | 18660489_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 24401050_Mistargeting of peroxisomal EHHADH disrupts mitochondrial metabolism and leads to renal Fanconi's syndrome; this indicates a central role of mitochondria in proximal tubular function. 33430801_EHHADH contributes to cisplatin resistance through regulation by tumor-suppressive microRNAs in bladder cancer. 33997049_Increased EHHADH Expression Predicting Poor Survival of Osteosarcoma by Integrating Weighted Gene Coexpression Network Analysis and Experimental Validation. | ENSMUSG00000022853 | Ehhadh | 254.499726 | 0.9809277 | -0.027781220 | 0.18469174 | 2.312380e-02 | 8.791356e-01 | No | Yes | 292.865896 | 48.966322 | 289.626496 | 48.411432 | |
| ENSG00000113838 | 55171 | TBCCD1 | protein_coding | Q9NVR7 | FUNCTION: Plays a role in the regulation of centrosome and Golgi apparatus positioning, with consequences on cell shape and cell migration. {ECO:0000269|PubMed:20168327}. | Alternative splicing;Cytoplasm;Cytoskeleton;Reference proteome | hsa:55171; | cytoplasm [GO:0005737]; spindle pole centrosome [GO:0031616]; cell morphogenesis [GO:0000902]; maintenance of centrosome location [GO:0051661]; maintenance of Golgi location [GO:0051684]; regulation of cell migration [GO:0030334]; regulation of cell shape [GO:0008360] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000004462 | Tbccd1 | 509.804153 | 1.0506147 | 0.071233676 | 0.12340574 | 3.323723e-01 | 5.642655e-01 | 8.381040e-01 | No | Yes | 552.977513 | 104.513987 | 542.442590 | 102.486286 | ||
| ENSG00000113851 | 51185 | CRBN | protein_coding | Q96SW2 | FUNCTION: Substrate recognition component of a DCX (DDB1-CUL4-X-box) E3 protein ligase complex that mediates the ubiquitination and subsequent proteasomal degradation of target proteins, such as MEIS2 or ILF2 (PubMed:33009960). Normal degradation of key regulatory proteins is required for normal limb outgrowth and expression of the fibroblast growth factor FGF8 (PubMed:20223979, PubMed:24328678, PubMed:25043012, PubMed:25108355). Maintains presynaptic glutamate release and consequently cognitive functions, such as memory and learning, by negatively regulating large-conductance calcium-activated potassium (BK) channels in excitatory neurons (PubMed:18414909, PubMed:29530986). Likely to function by regulating the assembly and neuronal surface expression of BK channels via its interaction with KCNT1 (PubMed:18414909). May also be involved in regulating anxiety-like behaviors via a BK channel-independent mechanism (By similarity). {ECO:0000250|UniProtKB:Q8C7D2, ECO:0000269|PubMed:18414909, ECO:0000269|PubMed:20223979, ECO:0000269|PubMed:24328678, ECO:0000269|PubMed:25043012, ECO:0000269|PubMed:25108355, ECO:0000269|PubMed:29530986, ECO:0000305}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Membrane;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway;Zinc | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:20223979, ECO:0000305|PubMed:25108355}. | This gene encodes a protein related to the Lon protease protein family. In rodents and other mammals this gene product is found in the cytoplasm localized with a calcium channel membrane protein, and is thought to play a role in brain development. Mutations in this gene are associated with autosomal recessive nonsyndromic cognitive disability. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2010]. | hsa:51185; | Cul4A-RING E3 ubiquitin ligase complex [GO:0031464]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; metal ion binding [GO:0046872]; transmembrane transporter binding [GO:0044325]; locomotory exploration behavior [GO:0035641]; negative regulation of large conductance calcium-activated potassium channel activity [GO:1902607]; negative regulation of protein-containing complex assembly [GO:0031333]; positive regulation of protein-containing complex assembly [GO:0031334]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein ubiquitination [GO:0016567] | 15557513_A nonsense mutation causing a premature stop codon in CRBN was found in a large kindred with mild mental retardation. ATP-dependent degradation of proteins may play a role in memory and learning. 17380424_This may suggest new functions of CRBN in cell nucleolus besides its mitochondria protease activity in cytoplasm. 18414909_Nonsense mutation (R419X) CRBN disturbs the development of adult brain BK(Ca) isoforms. These changes are predicted to result in BK(Ca) channels with a higher intracellular Ca(2+) sensitivity, faster activation, and slower deactivation kinetics. 20131966_The results suggest that mutCRBN may cause ARNSID by disrupting the developmental regulation of BK(Ca) in brain regions that are critical for memory and learning. 21232561_CRBN directly interacts with the alpha1 subunit of AMP-activated protein kinase (AMPK alpha1) and inhibits the activation of AMPK activation. 21860026_CRBN is an essential requirement for immunoregulatory drugs activity and a possible biomarker for the clinical assessment of antimyeloma efficacy. 22698399_Data show that in a cereblon-dependent fashion, lenalidomide downregulates IRF4 and SPIB, transcription factors that together prevent IFNbeta production. 23026050_These findings suggest that CRBN may modulate proteasome activity by directly interacting with the beta7 subunit. 23233657_We conclude that CRBN expression may be associated with the clinical efficacy of thalidomide. 23434730_We investigated the role of CRBN in response to lenalidomide in myelodysplastic syndrome infants without chromosome 5 deletion. 23983124_Findings suggest an approach for the treatment of mental retardation associated with cereblon nonsense mutation. 23992230_CRBN and IRF4 have been shown to be associated with response to lenalidomide in patients, these findings do not translate back to HMCLs, which could be attributable to factors present in the bone marrow microenvironment. 24118365_CRBN expression may provide a biomarker to predict response to Immunomodulatory drugs in patients with MM and its high expression can serve as a marker of good prognosis. 24129344_Data indicate that low cereblon (CRBN) expression can predict resistance to immunomodulatory drug (IMiD monotherapy and is a predictive biomarker for survival outcomes. 24166296_In the three intrinsically IMiD-resistant cell lines that clearly express detectable levels of cereblon, the absence of CRBN and DDB1 mutations suggest that potential cereblon-independent mechanisms of resistance exist 24328678_Presents a molecular model in which drug binding to cereblon results in the interaction of Ikaros and Aiolos to CRL4(CRBN) , leading to their ubiquitination, subsequent proteasomal degradation and T cell activation. 24687382_CRBN expression in bone marrow myeloma cells is associated with superior treatment response to lenalidomide in refractory myeloma and thalidomide/dexamethasone in new patients. CRBN is a crucial factor for the anti-MM effect of immunomodulators. 24925210_Data suggest that cereblon (CRBN) expression in peripheral blood chronic lymphocytic leukemia (CLL) cells is independent of prognostic factors and may be considered a potential biomarker. 25043012_structures of the DDB1-CRBN complex bound to thalidomide, lenalidomide and pomalidomide 25108355_The study presents the crystal structure of human CRBN bound to DDB1 and the drug lenalidomide. 25284710_High levels of full-length CRBN mRNA in lower risk 5q deletion patients are necessary for the efficacy of lenalidomide. 25569776_Studied the protein cereblon , which has been recently indentified as a primary target of thalidomide and its C-terminal part as responsible for binding thalidomide within a domain carrying several invariant cysteine and tryptophan residues. 25858704_A copy number gain of CRBN gene might be responsible for developmental delay/intellectual disability. 26021757_Our data are consistent with the idea that the CUL4A/B-DDB1-CRBN complex catalyses the polyubiquitination and thus controls the degradation of CLC-1 channels. 26024445_Structural dynamics of the cereblon ligand binding domain. 26119939_The anti-PEL effects of IMiDs involved cereblon-dependent suppression of IRF4 and rapid degradation of IKZF1, but not IKZF3. Small hairpin RNA-mediated knockdown of MYC enhanced the cytotoxicity of IMiDs 26186254_dual assay demonstrated superior Cereblon IHC measurement in MM samples compared with the single IHC assay using a published commercial rabbit polyclonal Cereblon antibody and could be used to explore the potential utility of Cereblon as a biomarker in the clinic 26188093_Although abnormal CRBN function may be associated with intellectual disability disease onset, its cellular mechanism is still unclear. Here, we examine the role of CRBN in aggresome formation and cytoprotection. 26990986_GS is acetylated at lysines 11 and 14, yielding a degron that is necessary and sufficient for binding and ubiquitylation by CRL4(CRBN) and degradation by the proteasome. 27142104_These results support the notion that the cereblon binding partner AGO2 plays an important role in regulating MM cell growth and survival and AGO2 could be considered as a novel drug target for overcoming IMiD resistance in MM cells. 27329811_results suggest that CRBN binds to Ikaros via its N-terminal region and regulates transcriptional activities of Ikaros and its downstream target, enkephalin 27365142_45.2 of multiple myeloma patients were CRBN(+). Among patients treated with thalidomide-based regimens, CRBN(+) patients showed a better treatment response than did CRBN-negative patients. There was no significant difference in survival outcomes between thalidomide- and bortezomib-based regimens in CRBN(+) patients. 27417535_mitochondrially expressed CRBN exhibited protease activity, and was induced by oxidative stress. 27458004_Compared with newly diagnosed multiple myeloma, an increased prevalence of mutations in the Ras pathway genes KRAS, NRAS, and/or BRAF (72%), as well as TP53 (26%), CRBN (12%), and CRBN pathway genes (10%) was observed. 27460676_overview of the current understanding of mechanism of action of Immunomodulatory drugs via CRBN and prospects for the development of new drugs that degrade protein of interest. 27468689_CRBN negatively regulates TLR4 signaling via attenuation of TRAF6 and TAB2 ubiquitination. 27601648_we identify Rabex-5 as a Immunomodulatory drugs (IMiDs) target molecule that functions to restrain TLR activated auto-immune promoting pathways. We propose that release of Rabex-5 from complex with Cereblon enables the suppression of immune responses, contributing to the antiinflammatory properties of IMiDs. 27618360_CRBN expression is of prognostic value in MM patients ineligible for ASCT treated with thalidomide as an immunomodulatory drug. With low expression indicating a possible suboptimal treatment outcome, measurement of CRBN expression might serve as additional prognostic tool in the personalized treatment approach. 27751757_We sequenced CRBN-thalidomide binding region in 38 thalidomide embryopathy individuals and 136 Brazilians without congenital anomalies, and performed in silico analyses. Eight variants were identified, seven intronic and one in 3'UTR. 28017969_High CRBN expression is associated with multiple myeloma. 28083618_IRF4 and CRBN polymorphisms affect risk and response to treatment in multiple myeloma 28143899_These findings thus contribute to a growing list of ID disorders caused by CRBN mutations, broaden the spectrum of phenotypes attributable to Autosomal-recessive non-syndromic intellectual disability (ARNS-ID)and provide new insight into genotype-phenotype correlations between CRBN mutations and the aetiology of ARNS-ID. 28848067_Plasmablast differentiation, whether induced by BAFF or CD40L, is prevented by modulation of CRBN activity with CC-220, which induces degradation of the B cell differentiation factors Aiolos and Ikaros. Downregulation of these B cell differentiation processes by CC-220 modulation of CRBN provides a new therapeutic approach to treating B cell-mediated pathology in SLE. 29272390_Cereblon suppresses the formation of pathogenic protein aggregates in a p62-dependent manner. 29449372_insight paves the way for studies of CRBN-dependent proteasome-targeting molecules in nonprimate models and provides a new understanding of CRBN's substrate-recruiting function 30190590_SALL4 has a role mediating teratogenicity as a thalidomide-dependent cereblon substrate 30234487_the ubiquitin-conjugating enzymes UBE2G1 and UBE2D3 cooperatively promote the K48-linked polyubiquitination of CRL4(CRBN) neomorphic substrates 30458989_This work suggests that the binding domain in CRBN is a critical factor which influences the regulation of immunomodulatory drugs on the ubiquitination and stability of these CRBN-interacting partners. 30683842_CRBN could protect cells from DNA damage-induced apoptosis, which provides a novel understanding of physiological roles of CRBN in p53-mediated apoptosis. 31115923_Multivariate analysis showed that patients with the CRBN CC genotype (rs1672753) had more than a 14-fold higher risk of peripheral polyneuropathy compared to patients with other variants of the investigated SNP [odds ratio (OR) = 14.29]. 31619706_Cereblon gene variants and clinical outcome in multiple myeloma patients treated with lenalidomide. 31620128_CRBN Is a Negative Regulator of Bactericidal Activity and Autophagy Activation Through Inhibiting the Ubiquitination of ECSIT and BECN1. 31746254_Cereblon (CRBN) gene polymorphisms predict clinical response and progression-free survival in relapsed/refractory multiple myeloma patients treated with lenalidomide: a pharmacogenetic study from the IMMEnSE consortium. 31865141_CRBN knockdown mitigates lipopolysaccharide-induced acute lung injury by suppression of oxidative stress and endoplasmic reticulum (ER) stress associated NF-kappaB signaling. 31873151_Disordered region of cereblon is required for efficient degradation by proteolysis-targeting chimera. 31964914_CRL4-Cereblon complex in Thalidomide Embryopathy: a translational investigation. 31983437_Cereblon-mediated degradation of the amyloid precursor protein via the ubiquitin-proteasome pathway. 32061138_decreased expression of IKZF1 and increased expression of KPNA2 compared to that of CRBN mRNA predicts poor outcomes of Len and dexamethasone therapy 32071327_Thalidomide Inhibits Human iPSC Mesendoderm Differentiation by Modulating CRBN-dependent Degradation of SALL4. 32132601_Genome-wide screening reveals a role for subcellular localization of CRBN in the anti-myeloma activity of pomalidomide. 32200025_Targeting the E3 ubiquitin ligases DCAF15 and cereblon for cancer therapy. 32219443_Aromatase is a novel neosubstrate of cereblon responsible for immunomodulatory drug-induced thrombocytopenia. 32251415_Thalidomide-dependent degradation of the embryonic transcription factor SALL4 by the CRL4(CRBN) E3 ubiquitin ligase is a plausible major driver of thalidomide teratogenicity. The structure of the second zinc finger of SALL4 in complex with pomalidomide, cereblon and DDB1 reveals the molecular details of recruitment. 32333926_Ubiquitin-dependent proteasomal degradation of AMPK gamma subunit by Cereblon inhibits AMPK activity. 32466489_CUL4-DDB1-CRBN E3 Ubiquitin Ligase Regulates Proteostasis of ClC-2 Chloride Channels: Implication for Aldosteronism and Leukodystrophy. 32677118_Role of cereblon in angiogenesis and in mediating the antiangiogenic activity of immunomodulatory drugs. 32901087_L-Arginine prevents cereblon-mediated ubiquitination of glucokinase and stimulates glucose-6-phosphate production in pancreatic beta-cells. 33009960_Cereblon Promotes the Ubiquitination and Proteasomal Degradation of Interleukin Enhancer-Binding Factor 2. 33206504_Cereblon Modulators Target ZBTB16 and Its Oncogenic Fusion Partners for Degradation via Distinct Structural Degrons. 33443552_Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. 33830389_Circ_0114428 Regulates Sepsis-Induced Kidney Injury by Targeting the miR-495-3p/CRBN Axis. 33972400_Cereblon Regulates the Proteotoxicity of Tau by Tuning the Chaperone Activity of DNAJA1. 34115836_Cereblon enhancer methylation and IMiD resistance in multiple myeloma. 34373585_Mutations in CRBN and other cereblon pathway genes are infrequently associated with acquired resistance to immunomodulatory drugs. 34392531_Cereblon regulates NK cell cytotoxicity and migration via Rac1 activation. 34489457_The E3 ubiquitin ligase component, Cereblon, is an evolutionarily conserved regulator of Wnt signaling. 34764413_PLZF and its fusion proteins are pomalidomide-dependent CRBN neosubstrates. 35013300_A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues. | ENSMUSG00000005362 | Crbn | 1509.447745 | 1.0541734 | 0.076112233 | 0.09486697 | 6.342391e-01 | 4.258046e-01 | 7.615324e-01 | No | Yes | 1541.822128 | 276.503511 | 1432.610461 | 256.943178 |
| ENSG00000114331 | 23527 | ACAP2 | protein_coding | Q15057 | FUNCTION: GTPase-activating protein (GAP) for ADP ribosylation factor 6 (ARF6). {ECO:0000269|PubMed:11062263}. | 3D-structure;ANK repeat;Coiled coil;Endosome;GTPase activation;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:23527; | endosome membrane [GO:0010008]; membrane [GO:0016020]; ruffle [GO:0001726]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; actin filament-based process [GO:0030029]; cellular response to nerve growth factor stimulus [GO:1990090]; endocytic recycling [GO:0032456] | 11050434_Centaurin beta2/KIAA0041, is a member of the centaurin family of ADP-ribosylation factor directed GTPase-activating proteins. Binding of phosphoinositides to a pleckstrin homology domain may regulate subcellular localisation and activity. 18003747_ACAP2 has robust, constitutive Arf6 GAP activity in vivo, with little activity toward Arf1. 25853217_ACAP2 is a functional homolog of C. elegans CNT-1 and its inactivation or downregulation in human cells may contribute to cancer development 28566286_Data suggest that Rab35, interacting with TBC1D10A, functions in vascular endothelial cells as a negative regulator of histamine-evoked, Ca2+-dependent Weibel-Palade body exocytosis, most likely acting through the downstream effectors ACAP2 and Arf6. (Rab35 = rab GTP-binding protein 53; TBC1D10A = TBC1 domain family member 10A; ACAP2 = centaurin beta2; Arf6 = ADP-ribosylation factor 6) 30212824_The down-regulation of circRNA-ACAP2 promoted miR-21-5p expression, which further suppressed the transcription and translation of Tiam1. 30905672_analysis of structural basis for effector recognition in Rab35/ACAP2 and Rab35/RUSC2 complexes 31863774_Elevated expression of circACAP2 in breast cancer tissues leads to malignant phenotype upon cancerous cells. CircACAP2-miR-29a/b-3p-COL5A1 axis leads to breast cancer tumorigenesis 34085707_Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. | ENSMUSG00000049076 | Acap2 | 1326.873123 | 1.0137758 | 0.019738649 | 0.07995699 | 6.095738e-02 | 8.049891e-01 | 9.361892e-01 | No | Yes | 1364.800157 | 259.179246 | 1335.024495 | 253.538652 | ||
| ENSG00000115274 | 83444 | INO80B | protein_coding | Q9C086 | FUNCTION: Induces growth and cell cycle arrests at the G1 phase of the cell cycle. {ECO:0000269|PubMed:15556297}.; FUNCTION: Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair. {ECO:0000269|PubMed:15556297}. | 3D-structure;Coiled coil;DNA damage;DNA recombination;DNA repair;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a subunit of an ATP-dependent chromatin remodeling complex, INO80, which plays a role in DNA and nucleosome-activated ATPase activity and ATP-dependent nucleosome sliding. Readthrough transcription of this gene into the neighboring downstream gene, which encodes WW domain-binding protein 1, generates a non-coding transcript. [provided by RefSeq, Feb 2011]. | hsa:83444; | Ino80 complex [GO:0031011]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; metal ion binding [GO:0046872]; chromatin remodeling [GO:0006338]; DNA recombination [GO:0006310]; DNA repair [GO:0006281]; nucleosome mobilization [GO:0042766]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of cell cycle [GO:0051726]; regulation of chromosome organization [GO:0033044]; regulation of DNA replication [GO:0006275]; regulation of DNA strand elongation [GO:0060382] | 25143393_pathway-guided genome-wide meta-analysis and mutational analysis identified INO80B gene is a susceptibility gene for pre-eclampsia. | ENSMUSG00000030034 | Ino80b | 604.632909 | 1.0453546 | 0.063992380 | 0.11317932 | 3.204276e-01 | 5.713508e-01 | 8.412588e-01 | No | Yes | 647.820959 | 70.860700 | 631.143477 | 68.927296 | |
| ENSG00000115392 | 55120 | FANCL | protein_coding | Q9NW38 | FUNCTION: Ubiquitin ligase protein that mediates monoubiquitination of FANCD2 in the presence of UBE2T, a key step in the DNA damage pathway (PubMed:12973351, PubMed:16916645, PubMed:17938197, PubMed:19111657, PubMed:24389026). Also mediates monoubiquitination of FANCI (PubMed:19589784). May stimulate the ubiquitin release from UBE2W. May be required for proper primordial germ cell proliferation in the embryonic stage, whereas it is probably not needed for spermatogonial proliferation after birth. {ECO:0000269|PubMed:12973351, ECO:0000269|PubMed:16916645, ECO:0000269|PubMed:17938197, ECO:0000269|PubMed:19111657, ECO:0000269|PubMed:19589784, ECO:0000269|PubMed:24389026}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;DNA damage;DNA repair;Fanconi anemia;Metal-binding;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a ubiquitin ligase that is a member of the Fanconi anemia complementation group (FANC). Members of this group are related by their assembly into a common nuclear protein complex rather than by sequence similarity. This gene encodes the protein for complementation group L that mediates monoubiquitination of FANCD2 as well as FANCI. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2018]. | hsa:55120; | cytoplasm [GO:0005737]; Fanconi anaemia nuclear complex [GO:0043240]; intracellular membrane-bounded organelle [GO:0043231]; nuclear body [GO:0016604]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; gamete generation [GO:0007276]; interstrand cross-link repair [GO:0036297]; protein monoubiquitination [GO:0006513]; regulation of cell population proliferation [GO:0042127] | 12417526_Deficiency of Fancl (also called Pog) is the cause of gcd mouse, which has a reduced number of primordial germ cells during the embryonic stage. 12606378_FANCL is necessary for primordial germ cell proliferation during the embryonic stage but not necessary for spermatogonia proliferation in adulthood. Thus, mouse FancL-/- males are infertile at 7 to 12 weeks but gain fertility thereafter. 12973351_data suggest that PHF9 has a crucial role in the Fanconi anemia pathway as the likely catalytic subunit required for monoubiquitination of FANCD2 16474167_FANCL, via its WD40 region, binds the FA complex and, via its PHD, recruits an as-yet-unidentified E2 for mono-ubiquitination of FANCD2 17106252_Abnormal FANCL expression is the cause leading to a defective Ranconi anemia-BRCA pathway, conferring sensitivity of a lung cancer cell line to mitomycin C> 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18607065_the first report to describe hypermethylation of FANCC in leukemia 19589784_Upon the occurrence of DNA damage, FANCI becomes monoubiquitinated on Lys-523 by the UBE2T-FANCL pair. 19690177_Observational study of gene-disease association. (HuGE Navigator) 19737859_Observational study of gene-disease association. (HuGE Navigator) 19737859_results rule out a major role of FANCL in familial breast cancer susceptibility 20407210_expression of a novel splice variant of FA complementation group L (FANCL), named FAVL, can impair the FA pathway in non-FA tumor cells and act as a tumor promoting factor 21297076_FANCL is associated with acute lung injury in mice 21543111_genetic diversity in FANCA, FANCC and FANCL does not support an association of these genes with cervical cancer susceptibility in the Swedish population. 21697891_FA DNA repair genes, FANCD2, FANCL, and FANCC, are transcriptionally upregulated differently in melanoma compared with non-melanoma skin cancer 22653977_Suppression of FANCL expression in normal CD34(+) stem and progenitor cells results in fewer beta-catenin active cells and inhibits expansion of multilineage progenitors. 22828653_FAVL elevation can increase the tumorigenic potential of bladder cancer cells, including the invasive potential that confers the development of advanced bladder cancer. 23783032_a signal transduction pathway involved in self-renewal and survival of hematopoietic stem cells also functions to stabilize FANCL and suggesting that FANCL participates directly in support of stem cell function. 25754594_Loss-of-Function FANCL Mutations Associate with Severe Fanconi Anemia Overlapping the VACTERL Association 26385482_Using small interfering RNA (siRNA), knockdown of FANCF, FANCL, or FANCD2 inhibited function of the FA/BRCA pathway in A549, A549/DDP and SK-MES-1 cells, and potentiated sensitivity of the three cells to cisplatin. 28419882_A novel homozygous mutation c.822_823insCTTTCAGG (p.Asp275LeufsX13) in the FANCL gene identified in a Chinese patient with Fanconi anemia. 31513304_A founder variant in the South Asian population leads to a high prevalence of FANCL Fanconi anemia cases in India. 32048394_FANCL gene mutations in premature ovarian insufficiency. 32420600_Characterization of FANCL variants observed in patient cancer cells. 33394227_Severe telomere shortening in Fanconi anemia complementation group L. | ENSMUSG00000004018 | Fancl | 975.556047 | 0.9776734 | -0.032575539 | 0.10261392 | 9.953571e-02 | 7.523875e-01 | 9.209939e-01 | No | Yes | 970.466800 | 163.845942 | 987.430113 | 166.688103 |
| ENSG00000115507 | 5013 | OTX1 | protein_coding | P32242 | FUNCTION: Probably plays a role in the development of the brain and the sense organs. Can bind to the BCD target sequence (BTS): 5'-TCTAATCCC-3'. | DNA-binding;Developmental protein;Homeobox;Isopeptide bond;Nucleus;Reference proteome;Ubl conjugation | This gene encodes a member of the bicoid sub-family of homeodomain-containing transcription factors. The encoded protein acts as a transcription factor and may play a role in brain and sensory organ development. A similar protein in mouse is required for proper brain and sensory organ development and can cause epilepsy. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2015]. | hsa:5013; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; diencephalon morphogenesis [GO:0048852]; inner ear morphogenesis [GO:0042472]; metencephalon development [GO:0022037]; midbrain development [GO:0030901]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 11901358_Fine mapping of the chromosome 2p12-16 dyslexia susceptibility locus candidate gene 18849347_OTR1, OTX2 and CRX act as positive modulators of the BEST1 promoter in the retinal pigment epithelium. 19018235_Observational study of gene-disease association. (HuGE Navigator) 19414065_In the human fetal eye, OTX1 expression was confined to anterior retina. 19893048_This study identifies OTX1 as a molecular marker for high-grade germinal center derived Non-Hodgkin Lymphoma 20354145_The early expression of OTX1 in proliferative cell layers of the human fetal brain supports the concept that this homeobox gene is important in neuronal cell development and differentiation. 21478910_established that the p53 protein directly induces OTX1 expression by acting on its promoter 21750575_XPO1 and OXT1 may contribute to ASD in 2p15-p16.1 deletion cases and non-deletion cases of ASD mapping to this chromosome region. 24388989_overexpression of OTX1 results in accumulation of colorectal cancer (CRC) cell proliferation and invasion in vitro and tumor growth in vivo, whereas ablation of OTX1 expression inhibits the proliferative and invasive capability of CRC cell lines 25203062_Genitalia defects in these patients could result from the effect of OTX1. 28348423_OTX1 and OTX2 genes might have a role in the pathogenesis of different types of sinonasal neoplasms. 28599093_We speculate that ACTR2 and MEIS1 might respectively play a role in the pathogenesis of the observed deafness and cardiomyopathy...the patient carrying a 2p14p15 deletion including OTX1 had normal kidneys and genitalia, thus confirming that OTX1 haploinsufficiency is not invariably associated with genitourinary defects. 30066897_High OTX1 expression is associated with gastric cancer. 30797919_The OTX1 gene was expressed in 52% of 60 Brazilian medulloblastoma patients. Expression varied with age (higher in adults), location (predominantly by hemisphere), and histological type (desmoplastic). 32325080_Long non-coding RNA HNF1A-AS1 upregulates OTX1 to enhance angiogenesis in colon cancer via the binding of transcription factor PBX3. 32419651_miR-3196 acts as a Tumor Suppressor and Predicts Survival Outcomes in Patients With Gastric Cancer. 32549762_MicroRNA-4516 suppresses pancreatic cancer development via negatively regulating orthodenticle homeobox 1. 32838760_OTX1 exerts an oncogenic role and is negatively regulated by miR129-5p in laryngeal squamous cell carcinoma. 33015792_OTX1 is a novel regulator of proliferation, migration, invasion and apoptosis in lung adenocarcinoma. 33336731_Long noncoding RNA MAFG-AS1 facilitates the progression of hepatocellular carcinoma via targeting miR-3196/OTX1 axis. 34306176_miR-223-5p Suppresses OTX1 to Mediate Malignant Progression of Lung Squamous Cell Carcinoma Cells. 34559577_Orthodenticle homeobox OTX1 is a potential prognostic biomarker for bladder cancer. 35303522_Overexpression of OTX1 promotes tumorigenesis in patients with esophageal squamous cell carcinoma. | ENSMUSG00000005917 | Otx1 | 100.976318 | 1.0946280 | 0.130440613 | 0.26807399 | 2.414156e-01 | 6.231855e-01 | No | Yes | 98.890166 | 15.485903 | 89.484134 | 14.072379 | ||
| ENSG00000115966 | 1386 | ATF2 | protein_coding | P15336 | FUNCTION: Transcriptional activator which regulates the transcription of various genes, including those involved in anti-apoptosis, cell growth, and DNA damage response. Dependent on its binding partner, binds to CRE (cAMP response element) consensus sequences (5'-TGACGTCA-3') or to AP-1 (activator protein 1) consensus sequences (5'-TGACTCA-3'). In the nucleus, contributes to global transcription and the DNA damage response, in addition to specific transcriptional activities that are related to cell development, proliferation and death. In the cytoplasm, interacts with and perturbs HK1- and VDAC1-containing complexes at the mitochondrial outer membrane, thereby impairing mitochondrial membrane potential, inducing mitochondrial leakage and promoting cell death. The phosphorylated form (mediated by ATM) plays a role in the DNA damage response and is involved in the ionizing radiation (IR)-induced S phase checkpoint control and in the recruitment of the MRN complex into the IR-induced foci (IRIF). Exhibits histone acetyltransferase (HAT) activity which specifically acetylates histones H2B and H4 in vitro (PubMed:10821277). In concert with CUL3 and RBX1, promotes the degradation of KAT5 thereby attenuating its ability to acetylate and activate ATM. Can elicit oncogenic or tumor suppressor activities depending on the tissue or cell type. {ECO:0000269|PubMed:10821277, ECO:0000269|PubMed:15916964, ECO:0000269|PubMed:18397884, ECO:0000269|PubMed:22304920}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cytoplasm;DNA damage;DNA-binding;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a transcription factor that is a member of the leucine zipper family of DNA binding proteins. The encoded protein has been identified as a moonlighting protein based on its ability to perform mechanistically distinct functions This protein binds to the cAMP-responsive element (CRE), an octameric palindrome. It forms a homodimer or a heterodimer with c-Jun and stimulates CRE-dependent transcription. This protein is also a histone acetyltransferase (HAT) that specifically acetylates histones H2B and H4 in vitro; thus it may represent a class of sequence-specific factors that activate transcription by direct effects on chromatin components. The encoded protein may also be involved in cell's DNA damage response independent of its role in transcriptional regulation. Several alternatively spliced transcript variants have been found for this gene [provided by RefSeq, Jan 2014]. | hsa:1386; | CCAAT-binding factor complex [GO:0016602]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; H4 histone acetyltransferase complex [GO:1902562]; mitochondrial outer membrane [GO:0005741]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II transcription regulator complex [GO:0090575]; site of double-strand break [GO:0035861]; cAMP response element binding [GO:0035497]; cAMP response element binding protein binding [GO:0008140]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; H2B histone acetyltransferase activity [GO:0044013]; H4 histone acetyltransferase activity [GO:0010485]; histone acetyltransferase activity [GO:0004402]; identical protein binding [GO:0042802]; leucine zipper domain binding [GO:0043522]; metal ion binding [GO:0046872]; promoter-specific chromatin binding [GO:1990841]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; protein kinase binding [GO:0019901]; protein-containing complex binding [GO:0044877]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; abducens nucleus development [GO:0021742]; adipose tissue development [GO:0060612]; amelogenesis [GO:0097186]; apoptotic process involved in development [GO:1902742]; BMP signaling pathway [GO:0030509]; brainstem development [GO:0003360]; cellular lipid metabolic process [GO:0044255]; cellular response to anisomycin [GO:0072740]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to leucine starvation [GO:1990253]; cellular response to oxidative stress [GO:0034599]; cellular response to virus [GO:0098586]; detection of cell density [GO:0060245]; facial nucleus development [GO:0021754]; growth plate cartilage chondrocyte differentiation [GO:0003418]; growth plate cartilage chondrocyte proliferation [GO:0003419]; hematopoietic progenitor cell differentiation [GO:0002244]; hepatocyte apoptotic process [GO:0097284]; histone H2B acetylation [GO:0043969]; histone H4 acetylation [GO:0043967]; hypoglossal nucleus development [GO:0021743]; in utero embryonic development [GO:0001701]; intrinsic apoptotic signaling pathway in response to hypoxia [GO:1990144]; JNK cascade [GO:0007254]; liver development [GO:0001889]; mitotic intra-S DNA damage checkpoint signaling [GO:0031573]; motor neuron apoptotic process [GO:0097049]; mRNA transcription by RNA polymerase II [GO:0042789]; negative regulation of angiogenesis [GO:0016525]; negative regulation of epithelial cell proliferation [GO:0050680]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neurofilament cytoskeleton organization [GO:0060052]; NK T cell differentiation [GO:0001865]; outflow tract morphogenesis [GO:0003151]; p38MAPK cascade [GO:0038066]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of cardiac muscle myoblast proliferation [GO:0110024]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of gene expression [GO:0010628]; positive regulation of mitochondrial membrane permeability involved in apoptotic process [GO:1902110]; positive regulation of neuron apoptotic process [GO:0043525]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus [GO:1901522]; positive regulation of transforming growth factor beta2 production [GO:0032915]; protein import into nucleus [GO:0006606]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; response to organic cyclic compound [GO:0014070]; response to osmotic stress [GO:0006970]; response to water deprivation [GO:0009414]; vacuole organization [GO:0007033]; white fat cell differentiation [GO:0050872] | 11836564_Infrequent mutations of the activating transcription factor-2 gene in human lung cancer, neuroblastoma and breast cancer 12110590_activation by growth factors via phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38 12270648_Observational study of gene-disease association. (HuGE Navigator) 12592382_Phosphorylation of one of the downstream transcriptional factors of MAPK cascade, ATF2, was 3.2- and 2.0-fold induced by TPA and Saikosaponin a, respectively. 12663670_JNK-dependent phosphorylation of ATF2 plays an important role in the drug resistance phenotype likely by mediating enhanced DNA repair by a p53-independent mechanism. 12833146_ATF-2 and ATF3 seem to play an important role in the protective response of human cells to ionizing radiation 14630918_ATF4 and ATF2 have roles in regulating CHOP expression 14678960_In melanoma strong cytoplasmic ATF2 expression was associated with primary specimens rather than metastases and with better survival. Strong nuclear ATF2 expression was associated with metastatic specimens and with poor survival. 14734562_differentiation-dependent expression and phosphorylation of ATF2 protein physically and functionally interacts with C/EBPalpha and coativator ASC-2 and synergizes to induce target gene transcription during granulocytic differentiation 14988408_ATF2 and HO-1 are regulated and induced by biliverdin reductase 15691875_genes affected by ATF2 and ATF2-sm appear to belong to discrete groups 15878807_ATF2 expression in the neuron of normal human brain. But downregulation in the Neurodegenerative Disease( Alzheimer disease, Huntington disease and Parkinson disease). 15916964_Data demonstrate that the protein kinase ATM phosphorylates ATF2 on serines 490 and 498 following ionizing radiation (IR). 16049073_activity of the AP-1 components c-Jun, ATF2, and c-Fos is altered in renal cystic tissue of patients with autosomal dominant polycystic kidney disease 16418168_Resistin induces PTEN expression by activating stress signaling p38 pathway, which may activate target transcription factor ATF-2, which in turn induces PTEN expression 16869889_These studies establish p38 MAP kinase-mediated activation of ATF-2 as a significant mechanism in amylin-evoked beta-cell death, which may serve as a target for pharmaceutical intervention and effective suppression of beta-cell failure in type-2 diabetes. 16896160_gamma-gene induction by butyrate and trichostatin A involves ATF-2 and CREB1 activation via p38 MAPK signaling. 17054722_Sin1 may contribute to ATF-2 signaling specificity by acting as a nuclear scaffold. 17082618_Binding of ATF2 to the CD1A promoter in human monocytes suggests a role for ATF/cAMP response element binding protein family members in regulation of CD1A expression. 17244683_Shear stress and KLF2 inhibit nuclear activity of ATF2, providing a potential mechanism by which endothelial cells exposed to laminar flow are protected from basal proinflammatory, atherogenic gene expression. 17258390_This study demonstrates that ATF2 mediates the TGF-beta-induced MMP-2 transcriptional activation, elucidating a molecular mechanism for the malignant progression of human breast epithelial cells exerted by TGF-beta. 17337306_ATF-2 may be a key regulator of the human insulin promoter possibly stimulating activity in response to extracellular signals. 17626013_ATF2, but not CREB, was a target for the TAK1-JNK pathway, and p38 negatively regulated TAK1 activity through TAB1 phosphorylation. 17681939_The transcription factor ATF2, which is phosphorylated and activated by JNK, is a critical mediator for inducible expression of DUSP1 and DUSP10 in this signaling pathway. 17869487_ATF2/STAT3 signaling pathways are activated and may play a role in development of eccrine porocarcinoma and eccrine poroma. 18077426_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18348191_review of signaling pathways that activate ATF-2, as well as its downstream targets [review] 18397884_analysis of regulation of ATM activation by ATF2-dependent control of TIP60 stability and activity 18547788_Overexpression of phosphorylated-ATF2 and STAT3 in cutaneous squamous cell carcinoma, Bowen's disease and basal cell carcinoma. 18671972_IRF2-BP1 is a JDP2-binding protein enhancing the polyubiquitination of JDP2 and represses ATF2-mediated transcriptional activation from a CRE-containing promoter. 18677098_This review summarizes the current understanding of ATF2 regulation and function. 18700251_overexpression of p-ATF2, p-STAT3 and possibly p53, but not p63 or p73, may contribute to the tumorigenesis of cutaneous vascular tumors. 19082758_p53 and ATF-2 partly mediate the overexpression of COX-2 in H(2)O (2)-induced premature senescence of human fibroblasts. 19176525_the phosphorylation of ATF-2 at Ser-121 plays a key role in the c-Jun-mediated activation of transcription that occurs in response to TPA. 19184334_Data show that individuals who are homozygous for the short AP-2 beta allele display higher depression scores when psychosocial adversity is taken into account. 19278424_P-ATF2 and p-STAT3 are concordantly overexpressed in extramammary Paget's disease and their expressions may possibly be associated with the tumor stage. 19331149_Breast cancer patients with high ATF2 expression as detected by WB had a significantly shorter overall survival. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19487697_Phospholipase D1 acts as an important regulator in house dust mite allergen Der f 2-induced expression and production of IL-13 through activation of activating transcription factor-2 activation in human bronchial epithelial cells. 19712049_show that Activating transcription factor 2 induction enhances hypoxia-inducible factor 1alpha protein stability via direct protein interaction. 19822663_Data reveal a new signaling pathway activated by amino acid starvation leading to ATF2 phosphorylation and subsequently positively affecting the transcription of amino acid-regulated genes. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19923798_JNK/ATF2 pathway is involved in iodinated contrast media-induced apoptosis 19944700_Stability and DNA-binding ability of the bZIP dimers formed by the ATF-2 and c-Jun. 20051382_rapid phosphorylation of p38 mitogen-activated protein kinase and activating transcription factor 2 also takes place in IM-9 lymphoblastoid cells, which naturally express GPR55, after stimulation with LPI 20116378_JNK-ATF-2 inhibits thrombomodulin (TM) expression by recruiting histone deacetylase4 (HDAC4) and forming a transcriptional repression complex in the TM promoter. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20507983_Growth factor-independent signaling pathways converge in the formation of an active c-Jun.AFT2 dimer, which induces the expression of the anti-apoptotic factor Bcl-X(L) that mediates a pro-survival response. 20581861_Overexpression of ATF2 resulted in significant increase in ATF3 promoter activity, and electrophoretic mobility shift assay identified this region as a core sequence to which ATF2 binds 20619956_calorimetric studies of atf-2 and c-jun transcription factors 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20675274_Results suggest that Fra-2 protein may be more effective than ATF-2 protein in cyst formation originated from epithelial cells of dental follicles. 21098032_hyperosmotic stress can activate the Plk3 signaling pathway that subsequently regulates the AP-1 complex by directly phosphorylating ATF-2 independent from the effects of JNK and p38 activation. 21203491_MITF is downregulated by ATF2 in the skin of Atf2-/- mice, in primary human melanocytes, and in melanoma cell lines. 21278380_ATF2 interacts with beta-cell-enriched transcription factors, MafA, Pdx1, and beta2, and activates insulin gene transcription. 21384452_Data suggest that competition between GSTpi and active JNK for the substrate ATF2 may be responsible for the inhibition of JNK catalysis by GSTpi. 21858082_Data show that ATF7-4 is an important cytoplasmic negative regulator of ATF7 and ATF2 transcription factors. 21901137_Data concluded that IR-induced up-regulation of ATF2 was coordinately enhanced by suppression of miR-26b in lung cancer cells, which may enhance the effect of IR in the MAPK signaling pathway. 21990224_Our data suggest regulatory roles for ATF2 in TNF-related mechanisms of Head and Neck Squamous Cell Carcinoma. Its perturbation and nuclear activation are associated with significant effects on survival and cytokine production. 22275354_ATF2 subcellular localization is probably modulated by multiple mechanisms 22304920_The ability of ATF2 to reach the mitochondria is determined by PKCepsilon, which directs ATF2 nuclear localization. Genotoxic stress attenuates PKCepsilon effect on ATF2; enables ATF2 nuclear export and localization at the mitochondria. 22351776_we report the kinetic mechanism for JNK1beta1 with transcription factors ATF2 and c-Jun along with interaction kinetics for these substrates. 22685333_Phosphorylation of ATF2 by PKCepsilon is the master switch that controls its subcellular localization and function. 22843696_ATF2-Jun heterodimers bind IFNb in both orientations alone and in association with IRF3 and HMGI 23416976_results therefore suggest that c-MYC induces stress-mediated activation of ATF2 and ATF7 and that these transcription factors regulate apoptosis in response to oncogenic transformation of B cells 23589174_Data indicate that small molecules that block the oncogenic addiction to PKCepsilon signaling by promoting ATF2 nuclear export, resulting in mitochondrial membrane leakage and melanoma cell death. 23591579_Increasing of ATF2 expression is mediated via oxidative stress induced by arsenic in SV-HUC-1 cells, and MAPK pathways are involved. 23656735_these studies show that the IL-1beta-induced increase in intestinal tight junction permeability was regulated by p38 kinase activation of ATF-2 and by ATF-2 regulation of MLCK gene activity 23800081_ATF2 knockdown revealed ATF2-triggered p21(WAF1) protein expression, suggesting p21(WAF1) transactivation through ATF2. 23866832_The expression of ATF2 in chondrocytes is involved in apoptosis in Kashin-Beck disease. 23966864_we establish that ATF2 family members physically and functionally interact with TCF1/LEF1 factors to promote target gene expression and hematopoietic tumor cell growth 24223142_There is synergism between developmental stage-specific recruitments of the ATF2 protein complex and expression of gamma-globin during erythropoiesis. 24289970_Cytoplasmic ATF2 expression was less frequently seen than nuclear expression in malignant mesenchymal tumors. Benign mesenchymal tumors mostly showed much lower nuclear and cytoplasmic ATF2 expression. 24338393_An association between ATF2 polymorphisms and heavy alcohol consumption is only weakly supported. 24780446_Data show that salvianolic acid B protects endothelial progenitor cells against oxidative stress by modulating Akt/mTOR/4EBP1, p38 MAPK/ATF2, and ERK1/2 signaling pathways. 24966171_ATF-2 knockdown blocked VEGF-A-stimulated VCAM-1 expression and endothelial-leukocyte interactions. ATF-2 was also required for other endothelial cell outputs, such as cell migration and tubulogenesis. 24968707_Study identified a potential target of miR-451, ATF2, and revealed a novel role of miR-451 in the inhibition of the migratory ability of hepatoma cell lines. 24982422_zymosan-induced il23a mRNA expression is best explained through coordinated kappaB- and ATF2-dependent transcription; and (iii) il23a expression relies on complementary phosphorylation of ATF2 on Thr-69 and Thr-71 dependent on PKC and MAPK activities. 25041846_study revealed that, autocrine soluble factors regulate dual but differential role of ATF-2 as a transcription factor or DNA repair protein, which collectively culminate in radioresistance of A549 cells. 25096061_in human HCC tissues, SPTBN1 expression correlated negatively with expression levels of STAT3, ATF3, and CREB2; SMAD3 expression correlated negatively with STAT3 expression 25141981_While expression of ATF-2 is not associated with outcome. 25258251_Study demonstrates the role of miR-622 in suppressing glioma invasion and migration mediated by ATF2, and shows that miR-622 expression inversely correlates with ATF2 in glioma patients 25417007_More terminally differentiated human odontoblasts was ATF-2 positive, as compared to pulpal fibroblasts at various stages of differentiation: ATF-2 is more associated with cell survival rather than cell proliferation. 25456131_suppression of tumorigenesis by JNK requires ATF2. 26153406_Neisseria meningitidis caused a high level of E-selectin expression elicited by the activity of phosphorylated ATF2 transcription factor on the E-selectin promoter. 26462148_Results reveal that mitochondrial ATF2 is associated with the induction of apoptosis and BRAF inhibitor resistance through Bim activation. 26645581_increased expression of the gene encoding PKCepsilon and abundance of phosphorylated, transcriptionally active ATF2 were observed in advanced-stage melanomas and correlated with decreased FUK expression 26729199_the variant alleles of TSG101 rs2292179 and ATF2 rs3845744 were associated with a reduced risk of breast cancer, particularly for subjects with BMI <24 (kg/m(2)) and postmenopausal women, respectively 26747248_CARMA1- and MyD88-dependent activation of Jun/ATF-type AP-1 complexes is a hallmark of ABC diffuse large B-cell lymphomas. 26935060_miR-204 may act as a tumor suppressor by directly targeting ATF2 in non-small cell lung cancer 27094031_these observations suggest that CD99 is involved in the regulation of CD1a transcription and expression by increasing ATF-2. 27210757_These findings point to an oncogenic function for ATF2 in melanoma development that appears to be independent of its transcriptional activity. 27377902_Results show that ATF2 is highly expressed in renal cell carcinoma (RCC) tissues and promotes RCC cell proliferation, migration and invasion. The study suggests that ATF2 exerts an oncogenic role in RCC. 27582466_We further demonstrated the suppressive function of lncRNA#32 in hepatitis B virus and hepatitis C virus infection. lncRNA#32 bound to activating transcription factor 2 (ATF2) and regulated ISG expression. Our results reveal a role for lncRNA#32 in host antiviral responses. 27588402_ATF2 regulated by miR-204 might also play an important role in the regulation of malignant behavior of glioblastoma. 27708346_activation of JNK was found to be dependent on muscarinic acid receptor induced Ca(2+)/CAMKII as well as ROS. JNK dependent phosphorylation of ATF2/c-Jun transcription factors resulted in TGF-beta transcription and its signaling. 27821620_TNF induces the binding of ATF2 to the TNF-responsive element. 28212892_This review provides an overview of the currently known upstream regulators and downstream targets of ATF2. [review] 28429654_Our study found that miR-451 regulates the drug resistance of renal cell carcinoma by targeting ATF-2 28618963_Noxin facilitated the expression of Cyclin D1 and Cyclin E1 through activating P38-activating transcription factor 2 signaling pathway, thus enhanced cell growth of breast cancer 28904175_this study demonstrates that CPEB2 alternative splicing is a major regulator of key cellular pathways linked to anoikis resistance and metastasis. 28916425_p38alpha and ATF2 expression play a crucial role in the malignant phenotypes of ovarian tumor cells and are a markers of poor prognosis in patients with ovarian serous adenocarcinomas. 29382922_ARHGEF39 promotes tumor growth and invasion by activating the Rac1-P38-ATF2 signaling pathway, as well as increasing the expression of Cyclin A2, Cyclin D1, and MMP2 in NSCLC cells. 29453334_beta-catenin-independent regulation of Wnt target genes by RoR2 and ATF2/ATF4 in colon cancer cells 29850528_deregulation of the miR-144-5p/ATF2 axis plays an important role in non-small-cell lung cancer cell radiosensitivity. 29885262_Demonstration of an interaction between PCSK9 and ATF-2, which reduces ATF-2/c-Jun dimerization and ATF-2/c-Jun binding to the IFNbeta enhancer. This novel function of PCSK9 should have important implications in optimizing the clinical use of PCSK9 inhibitors. 30106255_Analgesic effect of fentanyl in the cold pressor-induced pain test was significantly associated with rs7583431 SNP in the ATF2 region. 30229828_In non-small cell lung cancer cells and tissues, ATF2 was up-regulated and MiR-451a was down-regulated. 30367508_inhibition-promoted ATF-2 expression was responsible for epithelium to mesenchyme transition and invasion of pancreatic cancer cells, while the inhibition of ATF-2 confers to gemcitabine sensitivity in human pancreatic cancer cells in vitro. 31204513_miR-204 could inhibit proliferation and autophagy and induce apoptosis of cervical cancer cells by targeting ATF2 31760171_ATF-2 and Tpl2 regulation of endothelial cell cycle progression and apoptosis. 32222864_Mechano growth factor pretreatment yield mechanical stimuli induced cell stress responses in ligament fibroblasts of osteoarthritis via activating ATF-2. 32476275_Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells. 32587775_KIF18B promotes tumor progression in osteosarcoma by activating beta-catenin. 32710569_miRNA profiling of biliary intraepithelial neoplasia reveals stepwise tumorigenesis in distal cholangiocarcinoma via the miR-451a/ATF2 axis. 32789554_Sprouty2 Inhibits Migration and Invasion of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis by Down-regulating ATF2 Expression and Phosphorylation. 32993798_Aberration of the modulatory functions of intronic microRNA hsa-miR-933 on its host gene ATF2 results in type II diabetes mellitus and neurodegenerative disease development. 33188182_Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38. 33198803_Activating transcription factor-2 (ATF2) is a key determinant of resistance to endocrine treatment in an in vitro model of breast cancer. 33631165_Protective effect of SIRT6 on cholesterol crystal-induced endothelial dysfunction via regulating ACE2 expression. 33724979_Micro-RNA-451 Reduces Proliferation of B-CPAP Human Papillary Thyroid Cancer Cells by Downregulating Expression of Activating Transcription Factor 2. 34195276_Long Noncoding RNA RP11-357H14.17 Plays an Oncogene Role in Gastric Cancer by Activating ATF2 Signaling and Enhancing Treg Cells. 34273954_MARCH6 promotes hepatocellular carcinoma development through up-regulation of ATF2. 34498716_Long noncoding RNA ZSCAN16AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA451a and consequently increasing ATF2 expression. 34983301_Protein disulfide-isomerase A3 knockdown attenuates oxidized low-density lipoprotein-induced oxidative stress, inflammation and endothelial dysfunction in human umbilical vein endothelial cells by downregulating activating transcription factor 2. | ENSMUSG00000027104 | Atf2 | 1472.241791 | 1.1230008 | 0.167359018 | 0.07791648 | 4.598352e+00 | 3.200271e-02 | 2.400611e-01 | No | Yes | 1734.931742 | 248.207431 | 1522.887619 | 217.905123 | |
| ENSG00000116138 | 23341 | DNAJC16 | protein_coding | Q9Y2G8 | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:23341; | integral component of membrane [GO:0016021] | ENSMUSG00000040697 | Dnajc16 | 817.069983 | 1.0081572 | 0.011720587 | 0.13617547 | 7.479839e-03 | 9.310800e-01 | 9.746090e-01 | No | Yes | 897.672330 | 158.097465 | 885.417428 | 155.876721 | ||||
| ENSG00000116539 | 55870 | ASH1L | protein_coding | Q9NR48 | FUNCTION: Histone methyltransferase specifically trimethylating 'Lys-36' of histone H3 forming H3K36me3 (PubMed:21239497). Also monomethylates 'Lys-9' of histone H3 (H3K9me1) in vitro (By similarity). The physiological significance of the H3K9me1 activity is unclear (By similarity). {ECO:0000250|UniProtKB:Q99MY8, ECO:0000269|PubMed:21239497}. | 3D-structure;Acetylation;Activator;Alternative splicing;Bromodomain;Cell junction;Chromatin regulator;Chromosome;Disease variant;Isopeptide bond;Mental retardation;Metal-binding;Methylation;Methyltransferase;Nucleus;Phosphoprotein;Reference proteome;Repeat;S-adenosyl-L-methionine;Tight junction;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the trithorax group of transcriptional activators. The protein contains four AT hooks, a SET domain, a PHD-finger motif, and a bromodomain. It is localized to many small speckles in the nucleus, and also to cell-cell tight junctions. [provided by RefSeq, Jul 2008]. | hsa:55870; | bicellular tight junction [GO:0005923]; chromosome [GO:0005694]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone methyltransferase activity (H3-K36 specific) [GO:0046975]; histone methyltransferase activity (H3-K4 specific) [GO:0042800]; histone methyltransferase activity (H3-K9 specific) [GO:0046974]; histone-lysine N-methyltransferase activity [GO:0018024]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; decidualization [GO:0046697]; flagellated sperm motility [GO:0030317]; histone H3-K36 dimethylation [GO:0097676]; negative regulation of acute inflammatory response [GO:0002674]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of MAPK cascade [GO:0043409]; positive regulation of transcription by RNA polymerase II [GO:0045944]; post-embryonic development [GO:0009791]; single fertilization [GO:0007338]; skeletal system development [GO:0001501]; tarsal gland development [GO:1903699]; uterine gland development [GO:1903709]; uterus morphogenesis [GO:0061038] | 17923682_These data suggest that ASH1L occupies most, if not all, active genes and methylates histone H3 in a nonredundant fashion at a subset of genes. 21239497_human ASH1L specifically methylates histone H3 Lys-36. 22140534_ASH1 and MLL1 synergize in activation of Hox genes. 22541069_Long Non-coding RNA, DBE-T recruits the Trithorax group protein Ash1L to the FSHD locus, driving histone H3 lysine 36 dimethylation, chromatin remodeling, and 4q35 gene transcription. 22696682_induction of Cdk5 activity is a novel mechanism through which hASH1 may regulate migration in lung carcinogenesis 24019522_all of the H3K36-specific methyltransferases, including ASH1L, HYPB, NSD1, and NSD2 were inhibited by ubH2A, whereas the other histone methyltransferases, including PRC2, G9a, and Pr-Set7 were not affected by ubH2A. 24244179_Our results uncover a novel regulatory cascade orchestrated by Ash1l with RAR and provide insights into mechanisms underlying the establishment of the transcriptional activation that counteracts Polycomb silencing 25238203_These data demonstrate that miR-142-3p downregulation has a role in thyroid tumorigenesis, by regulating ASH1L and MLL1. 26002201_Both ASH1L and SETD2 are H3K36 specific methyltransferases but only SETD2 can trimethylate this mark. 26292256_Structural features were identified in ASH1L related to its' function and enzymatic activity. 27154821_Epigenetic regulators play vital roles in cancer pathogenesis and represent a new frontier in therapeutic targeting. Our studies provide basic mechanistic insight into the role of H3K36me2 in transcription activation and MLL leukemia pathogenesis and implicate ASH1L histone methyltransferase as a promising target for novel molecular therapy. 27434206_Our data suggest a role for ASH1L, a methyl transferase protein and member of the trithorax (Trx) family, in regulation of the HBB gene and as a potential modifier of beta-thalassaemia severity. 28041684_suppresses matrix metalloproteinase through mitogen-activated protein kinase signaling pathway in pulpitis 28394464_There has been increasing evidence that heterozygous mutation of ASH1L is associated with ID and autism spectrum disorders. We suggest that ASH1L abnormalities may cause a novel MCA/ID syndrome. 29109511_DDB2 promotes cyclobutane pyrimidine dimer excision by recruiting the histone methyltransferase ASH1L, which methylates lysine 4 of histone H3. In turn, methylated H3 facilitates the docking of the XPC complex to nucleosomal histone octamers. 29724719_Study shows that mutations associated with acute myelogenous leukemia (AML) result both in loss of miR-142-3p function and in decreased miR-142-5p expression. During hematopoietic maturation, loss of Mir142 increased ASH1L protein expression and consequently resulted in the aberrant maintenance of Hoxa gene expression in myeloid-committed hematopoietic progenitors. 29753921_A novel loss of function mutation in ASH1L was identified in a patient with an emergent neurodevelopmental syndrome. 30827841_ASH1L activation by MRG15 represents a delicate regulatory mechanism for how a cofactor activates an SET domain HMTase by releasing autoinhibition 30827843_Ash1L stimulates H3K36 methyltransferase activity through Mrg15 binding 31673123_Mutations in ASH1L confer susceptibility to Tourette syndrome. 32398261_Novel role of ASH1L histone methyltransferase in anaplastic thyroid carcinoma. 34373061_ASH1L mutation caused seizures and intellectual disability in twin sisters. 34782621_Deficiency of autism risk factor ASH1L in prefrontal cortex induces epigenetic aberrations and seizures. 35147301_Exploration of INSM1 and hASH1 as additional markers in lung cytology samples of high-grade neuroendocrine carcinoma with indeterminate neuroendocrine differentiation. 35307981_ASH1L may contribute to the risk of Tourette syndrome: Combination of family-based analysis and case-control study. | ENSMUSG00000028053 | Ash1l | 2133.347720 | 1.3728140 | 0.457136172 | 0.33440306 | 1.906266e+00 | 1.673787e-01 | 5.294338e-01 | No | Yes | 1876.831483 | 362.967485 | 1350.480517 | 261.212705 | |
| ENSG00000116668 | 54823 | SWT1 | protein_coding | Q5T5J6 | Reference proteome | hsa:54823; | nucleus [GO:0005634] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 20200978_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000052748 | Swt1 | 162.237617 | 1.0644167 | 0.090063066 | 0.22270182 | 1.594721e-01 | 6.896430e-01 | No | Yes | 169.612494 | 32.708745 | 153.185546 | 29.624833 | ||||
| ENSG00000116698 | 9887 | SMG7 | protein_coding | Q92540 | FUNCTION: Plays a role in nonsense-mediated mRNA decay. Recruits UPF1 to cytoplasmic mRNA decay bodies. Together with SMG5 is thought to provide a link to the mRNA degradation machinery involving exonucleolytic pathways, and to serve as an adapter for UPF1 to protein phosphatase 2A (PP2A), thereby triggering UPF1 dephosphorylation. {ECO:0000269|PubMed:15546618, ECO:0000269|PubMed:15721257}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Nonsense-mediated mRNA decay;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat | This gene encodes a protein that is essential for nonsense-mediated mRNA decay (NMD); a process whereby transcripts with premature termination codons are targeted for rapid degradation by a mRNA decay complex. The mRNA decay complex consists, in part, of this protein along with proteins SMG5 and UPF1. The N-terminal domain of this protein is thought to mediate its association with SMG5 or UPF1 while the C-terminal domain interacts with the mRNA decay complex. This protein may therefore couple changes in UPF1 phosphorylation state to the degradation of NMD-candidate transcripts. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Aug 2011]. | hsa:9887; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; intermediate filament cytoskeleton [GO:0045111]; nucleus [GO:0005634]; telomerase holoenzyme complex [GO:0005697]; protein phosphatase 2A binding [GO:0051721]; telomerase RNA binding [GO:0070034]; telomeric DNA binding [GO:0042162]; mRNA export from nucleus [GO:0006406]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; regulation of dephosphorylation [GO:0035303] | 14636577_Data show that phosphorylated hUPF1, the human ortholog of UPF1/SMG-2, forms a complex with human orthologs of the Caenorhabditis elegans proteins SMG-5 and SMG-7. 20237496_Observational study of gene-disease association. (HuGE Navigator) 25013172_The study demonstrates that SMG5-SMG7 and SMG6 exhibit different and non-overlapping modes of UPF1 recognition, thus pointing at distinguished roles in integrating the complex nonsense-mediated mRNA decay interaction network. 25211080_Depletion of nonsense-mediated mRNA decay pathway components Upf1, Smg5, and Smg7 led to increased levels of viral proteins and and virus release. 26783109_We showed SMG7 mRNA levels in peripheral blood mononuclear cells correlated inversely with antinuclear antibody titres of patients with systemic lupus erythematosus. The inverse relationship between levels of the risk allele-associated SMG7 mRNAs and antinuclear antibody suggested the novel contribution of mRNA surveillance pathway to systemic lupus erythematosus pathogenesis. 27771886_in gastric and colorectal cancers, study found SMG7 somatic mutations, which consisted of frameshift mutations by a deletion (c.2454delA (p. Met849fsx1) and a duplication (c.2545dupA (p. Met849Asnfsx10)) within the repeat 27864472_Transcriptome-wide identification of nonsense-mediated mRNA decay-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. 28461625_Nonsense-mediated mRNA decay (NMD) substrates with PTCs undergo constitutive SMG6-dependent endocleavage, rather than SMG7-dependent exonucleolytic decay. In contrast, the turnover of NMD substrates containing upstream ORFs and long 3' UTRs involves both SMG6- and SMG7-dependent endo- and exonucleolytic decay, respectively; extent to which SMG6 and SMG7 degrade NMD substrates is determined by the mRNA architecture. 31511540_Characterization of SMG7 14-3-3-like domain reveals phosphoserine binding-independent regulation of p53 and UPF1. 32602581_Nonsense-mediated decay factor SMG7 sensitizes cells to TNFalpha-induced apoptosis via CYLD tumor suppressor and the noncoding oncogene Pvt1. 33820915_Critical role of SMG7 in activation of the ATR-CHK1 axis in response to genotoxic stress. 34172724_SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. | ENSMUSG00000042772 | Smg7 | 3895.159918 | 1.1264817 | 0.171823933 | 0.06987405 | 6.130412e+00 | 1.328759e-02 | 1.525227e-01 | No | Yes | 4346.672814 | 375.591639 | 3829.254446 | 330.909414 | |
| ENSG00000117593 | 55157 | DARS2 | protein_coding | Q6PI48 | 3D-structure;ATP-binding;Acetylation;Aminoacyl-tRNA synthetase;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;Reference proteome;Transit peptide | The protein encoded by this gene belongs to the class-II aminoacyl-tRNA synthetase family. It is a mitochondrial enzyme that specifically aminoacylates aspartyl-tRNA. Mutations in this gene are associated with leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation (LBSL). [provided by RefSeq, Nov 2009]. | hsa:55157; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; aspartate-tRNA ligase activity [GO:0004815]; aspartate-tRNA(Asn) ligase activity [GO:0050560]; ATP binding [GO:0005524]; protein homodimerization activity [GO:0042803]; tRNA binding [GO:0000049]; aspartyl-tRNA aminoacylation [GO:0006422]; mitochondrial asparaginyl-tRNA aminoacylation [GO:0070145]; tRNA aminoacylation [GO:0043039] | 15779907_The gene for mitochondrial aspartyl-tRNA synthetase is described and the initial characterization of the enzyme is reported. Genes for the remaining missing human synthetases have also been found with the exception of glutaminyl-tRNA synthetase. 17384640_Mutations in DARS2, which encodes mitochondrial aspartyl-tRNA synthetase, in affected individuals with leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. 19209188_Meta-analysis of gene-disease association. (HuGE Navigator) 19592391_DARS2 mutations cause childhood-to-adolescence onset leucoencephalopathy, but they do not seem to be associated with multiple sclerosis. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20878420_This report describes two novel heterozygote composite mutations in the DARS2 gene 21749991_Case demonstrates that DARS2 mutation homozygosity is not lethal, as suggested earlier, but compatible with a rather benign disease course. 21792730_We describe two new cases of Leukoencephalopathy with brainstem and spinal cord involvement and elevated lactate with a novel pathogenic mutation in the DARS2 gene 21815884_A novel homozygous mutation of DARS2 may cause a severe LBSL (Leukoencephalopathy with brain stem and spinal cord involvement with lactate elevation) variant. 22023289_Leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation is associated with cell-type-dependent splicing of mtAspRS mRNA. 23216004_Pathogenic mutations causing LBSL affect mitochondrial aspartyl-tRNA synthetase in diverse way 23275545_A comparison of biophysical properties of human mitochondrial aspartyl-tRNA synthetase, HsaDRS2, with them to those of a bacterial (E. coli) homolog, EcoDRS. 23652419_Cognitive impairment seems to be common among patients with leukoencephalopathy with brainstem and spinal cord involvement and elevated lactate and DARS2 mutations 24566671_60 different DARS2 mutations were identified in 78 patients with leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation, 13 of which have not been reported before 25527264_This study identified DARS2-associated leukoencephalopathy with hypomyelination with brainstem and spinal cord involvement and leg spasticity. 26620921_Mutations with mild effects on solubility occur in patients as allelic combinations whereas those with strong effects on solubility or on aminoacylation are necessarily associated with a partially functional allele. 29052520_Results show that DARS2 is strongly upregulated in hepatocellular carcinoma (HCC) and is associated with HCC progression. DARS2 promotes HCC tumorigenesis by accelerating cell cycle progression and attenuating cell apoptosis. 30352563_Whole exome sequencing of the proband identified two compound heterozygous variants (NM_018122.4:c.1762C > G and c.563G > A) in DARS2 32766765_DARS2 is indispensable for Purkinje cell survival and protects against cerebellar ataxia. 33972171_Phenotypic diversity of brain MRI patterns in mitochondrial aminoacyl-tRNA synthetase mutations. | ENSMUSG00000026709 | Dars2 | 4423.251606 | 1.0080534 | 0.011572128 | 0.05402994 | 4.593375e-02 | 8.302963e-01 | 9.452372e-01 | No | Yes | 4646.498030 | 877.304981 | 4589.423339 | 866.423756 | ||
| ENSG00000117602 | 11123 | RCAN3 | protein_coding | Q9UKA8 | FUNCTION: Inhibits calcineurin-dependent transcriptional responses by binding to the catalytic domain of calcineurin A. Could play a role during central nervous system development (By similarity). {ECO:0000250}. | Alternative splicing;Reference proteome | hsa:11123; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; calcium-dependent protein serine/threonine phosphatase regulator activity [GO:0008597]; phosphatase binding [GO:0019902]; RNA binding [GO:0003723]; troponin I binding [GO:0031013]; anatomical structure morphogenesis [GO:0009653]; calcium-mediated signaling [GO:0019722] | 16516408_The yeast cotransformation and GST fusion protein assay demonstrated the interaction between this new DSCR1L2 variant and the human cardiac troponin I. 17270291_human RCAN3, encoded by the RCAN3 (also known as DSCR1L2) gene, interacts physically and functionally with calcineurin 18022329_The interaction between TNNI3 and the new R variants of RCAN3, RCAN3-2,3,5 and RCAN3-2,4,5, is reported. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19819266_our data have elucidated the molecular and cellular mechanism whereby PGF(2alpha) regulates CXCL8 expression via the FP receptor in endometrial adenocarcinomas and have highlighted RCAN1-4 as a negative regulator of CXCL8 expression 21042787_Data shows RCAN3 inhibits HUVEC proliferation both basally and under VEGF or phorbol 12-myristate 13-acetate-stimulated conditions, however it does not modulate gene expression of the chosen inflammatory genes. 21961037_Studies indicate that RCAN3 is composed of a multigene system that includes at least 21 alternative spliced isoforms. 25916653_The tumor suppressor effect of RCAN3 in breast cancer depends on the interaction with calcineurin. 32345470_Functional implications of miR-145/RCAN3 axis in the progression of cervical cancer. | ENSMUSG00000059713 | Rcan3 | 648.936345 | 1.2429521 | 0.313770696 | 0.12577351 | 6.243575e+00 | 1.246446e-02 | 1.473739e-01 | No | Yes | 700.379973 | 103.950665 | 552.164710 | 81.954558 | ||
| ENSG00000118420 | 90025 | UBE3D | protein_coding | Q7Z6J8 | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from specific E2 ubiquitin-conjugating enzymes, and transfers it to substrates, generally promoting their degradation by the proteasome. {ECO:0000269|PubMed:15749827}. | Acetylation;Cytoplasm;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | hsa:90025; | cytosol [GO:0005829]; nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; cyclin binding [GO:0030332]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-like protein conjugating enzyme binding [GO:0044390]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein autoubiquitination [GO:0051865]; protein monoubiquitination [GO:0006513]; protein polyubiquitination [GO:0000209] | 15749827_The carboxyl terminal half of H10BH was able to bind cyclin B and ubiquitinylate cyclin B in vitro. 25872646_A novel missense SNV, rs7739323, is strongly associated with age-related macular degeneration in an East Asian population. 26962152_With the exception of associations of BEGAIN with severe and UBE3D with moderate chronic periodontitis (CP) , no other loci were associated with CP in ARIC or aggressive periodontitis in the German sample. | ENSMUSG00000032415 | Ube2cbp | 379.484869 | 1.0646246 | 0.090344749 | 0.13928045 | 4.232077e-01 | 5.153408e-01 | No | Yes | 401.979024 | 68.683299 | 380.648381 | 65.084475 | ||
| ENSG00000118816 | 10983 | CCNI | protein_coding | Q14094 | Alternative splicing;Cyclin;Membrane;Nucleus;Reference proteome | The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle. Cyclins function as regulators of CDK kinases. Different cyclins exhibit distinct expression and degradation patterns which contribute to the temporal coordination of each mitotic event. This cyclin shows the highest similarity with cyclin G. The transcript of this gene was found to be expressed constantly during cell cycle progression. [provided by RefSeq, Jan 2017]. | hsa:10983; | cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; mitotic cell cycle phase transition [GO:0044772]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079]; spermatogenesis [GO:0007283] | 17507299_The increased levels of cyclin I and GDI2 found to be associated with pancreatic carcinoma were further confirmed by Western blot analyses in an independent series of serum samples and/or pancreatic juice samples 23907122_Knockdown of Cyclin I induced cell cycle arrest at S/G2/M phases. 26698249_A cyclin I-Cdk5 complex forms a critical antiapoptotic factor in the process of generating cisplatin resistance in cervical cancer. | ENSMUSG00000063015 | Ccni | 6661.806166 | 1.0468477 | 0.066051633 | 0.05213956 | 1.600046e+00 | 2.058967e-01 | 5.768662e-01 | No | Yes | 7384.818871 | 913.081757 | 7011.533944 | 866.839219 | ||
| ENSG00000118873 | 25782 | RAB3GAP2 | protein_coding | Q9H2M9 | FUNCTION: Regulatory subunit of a GTPase activating protein that has specificity for Rab3 subfamily (RAB3A, RAB3B, RAB3C and RAB3D). Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Rab3 GTPase-activating complex specifically converts active Rab3-GTP to the inactive form Rab3-GDP. Required for normal eye and brain development. May participate in neurodevelopmental processes such as proliferation, migration and differentiation before synapse formation, and non-synaptic vesicular release of neurotransmitters. {ECO:0000269|PubMed:9733780}. | Alternative splicing;Cataract;Cytoplasm;Disease variant;GTPase activation;Mental retardation;Phosphoprotein;Reference proteome | The protein encoded by this gene belongs to the RAB3 protein family, members of which are involved in regulated exocytosis of neurotransmitters and hormones. This protein forms the Rab3 GTPase-activating complex with RAB3GAP1, where it constitutes the regulatory subunit, whereas the latter functions as the catalytic subunit. This gene has the highest level of expression in the brain, consistent with it having a key role in neurodevelopment. Mutations in this gene are associated with Martsolf syndrome.[provided by RefSeq, Oct 2009]. | hsa:25782; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; enzyme activator activity [GO:0008047]; enzyme regulator activity [GO:0030234]; GTPase activator activity [GO:0005096]; small GTPase binding [GO:0031267]; establishment of protein localization to endoplasmic reticulum membrane [GO:0097051]; intracellular protein transport [GO:0006886]; positive regulation of autophagosome assembly [GO:2000786]; positive regulation of endoplasmic reticulum tubular network organization [GO:1903373]; positive regulation of protein lipidation [GO:1903061]; regulation of GTPase activity [GO:0043087] | 18485483_Data show that at chemical synapses, Rab3 performs specific functions in synpatic vesicle exocytosis. 18849981_KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator RAB3GAP2 occurs in a GTP-dependent manner through MADD. 20967465_functionally severe RAB3GAP2 mutations cause Warburg Micro syndrome while hypomorphic RAB3GAP2 mutations can result in the milder Martsolf phenotype 23176487_Micro syndrome has been associated with causative mutations in three disease genes: RAB3GAP1, RAB3GAP2 and RAB18. Martsolf syndrome has been associated with a mutation in RAB3GAP2. [Review] 23420520_One-hundred and forty-four Micro and nine Martsolf families were investigated, identifying mutations in RAB3GAP1 in 41% of cases, mutations in RAB3GAP2 in 7% of cases, and mutations in RAB18 in 5% of cases 24891604_Rab18 and a Rab18 GEF complex of Rab3GAP1 and Rab3GAP2 have roles in the endoplasmic reticulum structure 25495476_RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy 28342870_RAB18 modulates macroautophagy and proteostasis, and is dependent on activity of RAB3GAP1 and RAB3GAP2. 28575017_show that FOXC1 regulates the expression of RAB3GAP1, RAB3GAP2 and SNAP25 29419336_Mutation in RAB3GAP2 gene is associated with Warburg micro syndrome and Martsolf syndrome. 32248620_The Warburg Micro Syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I. 32376645_Hypogonadotropic hypogonadism due to variants in RAB3GAP2: expanding the phenotypic and genotypic spectrum of Martsolf syndrome. 32740904_Micro and Martsolf syndromes in 34 new patients: Refining the phenotypic spectrum and further molecular insights. | ENSMUSG00000039318 | Rab3gap2 | 1922.589020 | 1.0639832 | 0.089475371 | 0.07215097 | 1.536990e+00 | 2.150664e-01 | 5.870413e-01 | No | Yes | 2052.026263 | 403.662965 | 1956.435887 | 384.873361 | |
| ENSG00000118961 | 60526 | LDAH | protein_coding | Q9H6V9 | FUNCTION: Probable serine lipid hydrolase associated with lipid droplets. Appears to lack cholesterol esterase activity. Appears to lack triglyceride lipase activity. Highly expressed in macrophage-rich areas in atherosclerotic lesions, suggesting that it could promote cholesterol ester turnover in macrophages. {ECO:0000250|UniProtKB:Q8BVA5}. | Alternative splicing;Endoplasmic reticulum;Hydrolase;Lipid degradation;Lipid droplet;Lipid metabolism;Reference proteome | hsa:60526; | endoplasmic reticulum [GO:0005783]; lipid droplet [GO:0005811]; lipase activity [GO:0016298]; lipid catabolic process [GO:0016042]; lipid storage [GO:0019915] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20676098_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22662242_Our results provide further support for association of the C2orf43, FOXP4, GPRC6A and RFX6 genes with prostate cancer in Eastern Asian populations 24357060_The data identify lipid droplet-associated serine hydrolase (LDAH) a candidate target to promote reverse cholesterol transport from atherosclerotic lesions. 30169630_We then amassed convergent genomic evidence showing population level associations between LDAHexpression and occurrence of prostate cancer . We further identified loss of LDAH in both tissues and cell lines derived from human prostate cancer, and generated Ldah-/- mouse model, which recapitulated many of the clinical findings in DGAP056. | ENSMUSG00000037669 | Ldah | 382.221195 | 1.0252550 | 0.035982743 | 0.14238088 | 6.458218e-02 | 7.993949e-01 | No | Yes | 477.837473 | 109.010357 | 471.625576 | 107.567034 | |||
| ENSG00000119397 | 11064 | CNTRL | protein_coding | Q7Z7A1 | FUNCTION: Involved in cell cycle progression and cytokinesis. During the late steps of cytokinesis, anchors exocyst and SNARE complexes at the midbody, thereby allowing secretory vesicle-mediated abscission. {ECO:0000269|PubMed:12732615, ECO:0000269|PubMed:16213214}. | Alternative splicing;Cell cycle;Cell division;Chromosomal rearrangement;Coiled coil;Cytoplasm;Cytoskeleton;Leucine-rich repeat;Phosphoprotein;Reference proteome;Repeat | This gene encodes a centrosomal protein required for the centrosome to function as a microtubule organizing center. The gene product is also associated with centrosome maturation. One version of stem cell myeloproliferative disorder is the result of a reciprocal translocation between chromosomes 8 and 9, with the breakpoint associated with fibroblast growth factor receptor 1 and centrosomal protein 1. [provided by RefSeq, Jul 2008]. | hsa:11064; | centriolar subdistal appendage [GO:0120103]; centrosome [GO:0005813]; cytosol [GO:0005829]; Flemming body [GO:0090543]; meiotic spindle pole [GO:0090619]; membrane [GO:0016020]; microtubule organizing center [GO:0005815]; mitotic spindle pole [GO:0097431]; perinuclear region of cytoplasm [GO:0048471]; cytoskeletal protein binding [GO:0008092]; aorta development [GO:0035904]; cell cycle [GO:0007049]; cell division [GO:0051301]; coronary vasculature development [GO:0060976]; kidney development [GO:0001822]; regulation of cytoskeleton organization [GO:0051493]; ventricular septum development [GO:0003281] | 11956314_CEP110 is essential for the reformation of specific aspects of the interphase centrosome architecture following mitosis as well as being required for the centrosome to function as a MTOC. 16213214_We propose that centriolin anchors protein complexes required for vesicle targeting and fusion and integrates membrane-vesicle fusion with abscission. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20508983_Observational study of gene-disease association. (HuGE Navigator) 23777766_CNTRL and FGFR1 have roles in myeloid and lymphoid malignancies in both human and mouse models 31250523_In this study, we present two new cases of CNTRL-FGFR1 fusion | ENSMUSG00000057110 | Cntrl | 457.962098 | 1.2834467 | 0.360023419 | 0.15393160 | 5.569874e+00 | 1.827207e-02 | No | Yes | 439.925302 | 112.689390 | 337.517288 | 86.533134 | ||
| ENSG00000119537 | 2531 | KDSR | protein_coding | Q06136 | FUNCTION: Catalyzes the reduction of 3-ketodihydrosphingosine (KDS) to dihydrosphingosine (DHS). {ECO:0000269|PubMed:28575652}. | Alternative splicing;Chromosomal rearrangement;Disease variant;Endoplasmic reticulum;Lipid metabolism;Membrane;NADP;Oxidoreductase;Palmoplantar keratoderma;Proto-oncogene;Reference proteome;Signal;Sphingolipid metabolism;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; sphingolipid metabolism. | The protein encoded by this gene catalyzes the reduction of 3-ketodihydrosphingosine to dihydrosphingosine. The putative active site residues of the encoded protein are found on the cytosolic side of the endoplasmic reticulum membrane. A chromosomal rearrangement involving this gene is a cause of follicular lymphoma, also known as type II chronic lymphatic leukemia. The mutation of a conserved residue in the bovine ortholog causes spinal muscular atrophy. [provided by RefSeq, Jul 2008]. | hsa:2531; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; 3-dehydrosphinganine reductase activity [GO:0047560]; 3-keto-sphinganine metabolic process [GO:0006666]; sphingolipid biosynthetic process [GO:0030148] | 15328338_FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane 16711541_Observational study of gene-disease association. (HuGE Navigator) 17420465_Describes an Ala-175 to Thr mutation in the bovine ortholog that causes spinal muscular atrophy. This residue is conserved in several species, including human. 18395445_Data show that mutations in FVT1 do not contribute significantly to the cause of motor neuron diseases in the human population. 19019774_FVT1 is significantly underexpressed by germinal center-type diffuse large B-cell lymphoma compared with non-germinal center-type DLBCL, follicular lymphoma, & normal tonsil control samples. Increased expression of FVT1 correlated with decreased survival. 19773279_Observational study of gene-disease association. (HuGE Navigator) 20855565_Observational study of gene-disease association. (HuGE Navigator) 28575652_mutations in KDSR (3-ketodihydrosphingosine reductase), encoding an enzyme in the ceramide synthesis pathway, lead to a previously undescribed recessive Mendelian disorder in the progressive symmetric erythrokeratoderma spectrum. 30467204_Depletion of kdsr causes thrombocytopenia due to impaired proplatelet formation and sphingolipid dysregulation. 34686882_Formation of keto-type ceramides in palmoplantar keratoderma based on biallelic KDSR mutations in patients. | ENSMUSG00000009905 | Kdsr | 1453.218881 | 1.0096491 | 0.013853954 | 0.08078520 | 2.950879e-02 | 8.636095e-01 | 9.548533e-01 | No | Yes | 1360.971496 | 109.907083 | 1343.995285 | 108.449657 |
| ENSG00000119599 | 26094 | DCAF4 | protein_coding | Q8WV16 | FUNCTION: May function as a substrate receptor for CUL4-DDB1 E3 ubiquitin-protein ligase complex. {ECO:0000269|PubMed:16949367, ECO:0000269|PubMed:16964240}. | 3D-structure;Alternative splicing;Reference proteome;Repeat;Ubl conjugation pathway;WD repeat | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a WD repeat-containing protein that interacts with the Cul4-Ddb1 E3 ligase macromolecular complex. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2009]. | hsa:26094; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; nucleoplasm [GO:0005654]; protein ubiquitination [GO:0016567] | 22366774_The minor allele of WDR21A may induce a good response to inhaled corticosteroids possibly through competition with the Gbeta(1) proteins for binding to GRs 25624462_Genome-wide association (GWA) meta-analysis and de novo genotyping of 20 022 individuals revealed a novel association (p=6.4x10(-10)) between LTL and rs2535913, which lies within DCAF4 28383684_our results suggest that rs12587742 is associated with an increased lung cancer risk, possibly by up-regulating mRNA expression and decreasing methylation status of DCAF4. | ENSMUSG00000021222 | Dcaf4 | 540.873422 | 1.2055838 | 0.269731952 | 0.12603535 | 4.517387e+00 | 3.355204e-02 | 2.470469e-01 | No | Yes | 579.906639 | 41.888496 | 486.649276 | 35.301312 |
| ENSG00000119638 | 91754 | NEK9 | protein_coding | Q8TD19 | FUNCTION: Pleiotropic regulator of mitotic progression, participating in the control of spindle dynamics and chromosome separation. Phosphorylates different histones, myelin basic protein, beta-casein, and BICD2. Phosphorylates histone H3 on serine and threonine residues and beta-casein on serine residues. Important for G1/S transition and S phase progression. Phosphorylates NEK6 and NEK7 and stimulates their activity by releasing the autoinhibitory functions of Tyr-108 and Tyr-97 respectively. {ECO:0000269|PubMed:12840024, ECO:0000269|PubMed:14660563, ECO:0000269|PubMed:19941817}. | 3D-structure;ATP-binding;Acetylation;Cell cycle;Cell division;Coiled coil;Cytoplasm;Direct protein sequencing;Disease variant;Kinase;Magnesium;Metal-binding;Mitosis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the NimA (never in mitosis A) family of serine/threonine protein kinases. The encoded protein is activated in mitosis and, in turn, activates other family members during mitosis. This protein also mediates cellular processes that are essential for interphase progression. [provided by RefSeq, Jul 2016]. | hsa:91754; | cytosol [GO:0005829]; nucleus [GO:0005634]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein kinase binding [GO:0019901]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell cycle [GO:0007049]; cell division [GO:0051301] | 12101123_binds the Ran GTPase and regulates mitotic progression 12840024_Activated in mitosis, and activates nek6 and nek7 kinase. 14660563_mediates certain cellular processes, which are ultimately essential for interphase progression 17443675_The disruption of a nuclear function of NEK9 by adenovirus E1A-associated cellular proteins is reported. 19941817_The activity of Nek6 and Nek7, but not the tyrosine mutant, is increased by interaction with the Nek9 noncatalytic C-terminal domain, suggesting a mechanism in which the tyrosine is released from its autoinhibitory position. 20508983_Observational study of gene-disease association. (HuGE Navigator) 21454704_DYNLL/LC8 protein controls signal transduction through the Nek9/Nek6 signaling module by regulating Nek6 binding to Nek9. 21642957_Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. 21735226_The interaction between the human NimA-like protein kinase Nek9 and the Helicobacter HcpC has been validated by ELISA and surface plasmon resonance. 22818914_Nek9 phosphorylates NEDD1 on Ser377 driving its recruitment and thereby that of gamma-tubulin to the centrosome in mitotic cells. 23482567_Structural analysis of LC8 with both Nek9 peptides, together with different biophysical experiments, explains the observed diminished binding affinity of Nek9 to LC8 upon phosphorylation on Ser(944) within the Nek9 sequence 23665325_NEK9 inhibition represents a novel anti-cancer strategy by induction of mitotic catastrophe via impairment of spindle dynamics, cytokinesis and mitotic checkpoint control. 25131192_The findings demonstrate that a novel NEK9 network regulates the growth of cancer cells lacking functional p53. 26522158_The C-terminal domain of Nek9 activates Nek7 through promoting back-to-back dimerization. 26676776_Overall, these results highlight the complexity of virus-host interactions and identify a new role for the cellular protein Nek9 during human adenovirus infection, suggesting a role for Nek9 in regulating p53 target gene expression. 26908619_Recessive NEK9 mutation is associated with lethal skeletal dysplasia. 26956052_High expression level of NEK9 is associated with recurrence in glioblastoma. 27153399_Somatic Mutations in NEK9 Cause Nevus Comedonicus. 28630147_Signaling cascade of the NIMA-related kinases (Neks) Nek6, Nek7, and Nek9 is required for the localization and function of two kinesins essential for cytokinesis, Mklp2 and Kif14 to properly coordinate cytokinesis. 29276125_Eg5 localization and centrosome separation in prophase depend on the nuclear microtubule-associated protein TPX2, a pool of which localizes to the centrosomes before nuclear envelope breakdown. This localization involves the kinase Nek9, which phosphorylates TPX2 nuclear localization signal preventing its interaction with importin and nuclear import. 29472518_This study supports a role for NEK9 and MAP2K4 in mediating buparlisib resistance and demonstrates the value of unbiased omic analyses in uncovering resistance mechanisms to targeted therapy in Triple-Negative Breast Cancers. 31857374_NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. 32098687_Decreased Nek9 expression correlates with aggressive behaviour and predicts unfavourable prognosis in breast cancer. 32184261_EML4-ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7. 33500736_NEK9, a novel effector of IL-6/STAT3, regulates metastasis of gastric cancer by targeting ARHGEF2 phosphorylation. 33664869_The short isoform of PRLR suppresses the pentose phosphate pathway and nucleotide synthesis through the NEK9-Hippo axis in pancreatic cancer. 33819539_Three novel pathogenic NEK9 variants in patients with nevus comedonicus: A case series. | ENSMUSG00000034290 | Nek9 | 4676.977702 | 0.9773317 | -0.033079776 | 0.05888862 | 3.182013e-01 | 5.726906e-01 | 8.415819e-01 | No | Yes | 4610.522874 | 332.812584 | 4678.052055 | 337.654008 | |
| ENSG00000119685 | 23093 | TTLL5 | protein_coding | Q6EMB2 | FUNCTION: Polyglutamylase which modifies tubulin, generating polyglutamate side chains on the gamma-carboxyl group of specific glutamate residues within the C-terminal tail of tubulin. Preferentially mediates ATP-dependent initiation step of the polyglutamylation reaction over the elongation step. Preferentially modifies the alpha-tubulin tail over a beta-tail (By similarity). Required for CCSAP localization to both polyglutamylated spindle and cilia microtubules (PubMed:22493317). Increases the effects of transcriptional coactivator NCOA2/TIF2 in glucocorticoid receptor-mediated repression and induction and in androgen receptor-mediated induction (PubMed:17116691). {ECO:0000250|UniProtKB:Q8CHB8, ECO:0000269|PubMed:17116691, ECO:0000269|PubMed:22493317}. | ATP-binding;Alternative splicing;Cell projection;Cilium;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Disease variant;Ligase;Magnesium;Metal-binding;Microtubule;Nucleotide-binding;Nucleus;Reference proteome;Transcription | This gene encodes a member of the tubulin tyrosine ligase like protein family. This protein interacts with two glucocorticoid receptor coactivators, transcriptional intermediary factor 2 and steroid receptor coactivator 1. This protein may function as a coregulator of glucocorticoid receptor mediated gene induction and repression. This protein may also function as an alpha tubulin polyglutamylase.[provided by RefSeq, Feb 2010]. | hsa:23093; | centrosome [GO:0005813]; cilium [GO:0005929]; cytosol [GO:0005829]; microtubule [GO:0005874]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; tubulin binding [GO:0015631]; tubulin-glutamic acid ligase activity [GO:0070740]; microtubule cytoskeleton organization [GO:0000226]; protein polyglutamylation [GO:0018095]; retina development in camera-type eye [GO:0060041] | 17116691_STAMP is an important new, downstream component of GR action in both gene activation and gene repression. 20374646_This study indicates that a physiological function of STAMP in several settings is to modify cell growth rates in a manner that can be independent of steroid hormones. 24791901_this study has performed exome sequencing in 28 individuals with a similar disease phenotype and subsequently used a casecontrol approach to identify mutations in TTLL5 as a cause of recessive retinal dystrophy. 28173158_5 homozygous variants [p.(Asp594fs), p.(Gln117*), p.(Met712fs), p.(Ile756Phe) and p.(Glu543Lys)] in TTLL5, in 8 patients from 6 families were identified. 2 male patients carrying truncating TTLL5 variants also displayed a reduction in sperm motility and infertility, whereas those carrying missense changes were fertile. TTLL has multiple viable isoforms, being highly expressed in retina, testis and spermatozoon flagellum. 28356705_in a study of 3 family members from 2 generations, identified in a previously misdiagnosed incomplete congenital stationary night blindness (icCSNB) case a splice-site mutation in intron 3 of TTLL5 (c.182-3_182-1delinsAA); reinvestigation of the clinical data corrected the diagnosis to cone dystrophy 30517872_Binding of CSAP to TTLL5 promotes relocalization of TTLL5 toward microtubules. 34203883_Novel TTLL5 Variants Associated with Cone-Rod Dystrophy and Early-Onset Severe Retinal Dystrophy. 35365235_Expanding the phenotype of TTLL5-associated retinal dystrophy: a case series. | 640.873408 | 0.9210469 | -0.118653535 | 0.18234002 | 4.218927e-01 | 5.159941e-01 | 8.122384e-01 | No | Yes | 689.658990 | 107.463919 | 756.191467 | 117.881568 | |||
| ENSG00000119812 | 25940 | FAM98A | protein_coding | Q8NCA5 | FUNCTION: Positively stimulates PRMT1-induced protein arginine methylation (PubMed:28040436). Involved in skeletal homeostasis (By similarity). Positively regulates lysosome peripheral distribution and ruffled border formation in osteoclasts (By similarity). Promotes colorectal cancer cell malignancy (PubMed:28040436). {ECO:0000250|UniProtKB:Q3TJZ6, ECO:0000269|PubMed:28040436}. | Reference proteome | hsa:25940; | tRNA-splicing ligase complex [GO:0072669]; protein methyltransferase activity [GO:0008276]; RNA binding [GO:0003723]; lysosome localization [GO:0032418]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of gene expression [GO:0010628]; positive regulation of ruffle assembly [GO:1900029]; protein methylation [GO:0006479] | 26503212_FAM98A is expressed in numerous ovarian cancer cell lines and is important for the malignant characteristics of ovarian cancer cells. 28040436_the two structural homologs FAM98A and FAM98B included in a novel complex with DDX1 and C14orf166 are required for PRMT1 expression in colorectal cancer cell lines 31114934_Mechanistically, it was demonstrated that miR-142-3p directly targeted FAM98A, and modulated its expression. 34783205_Circular RNA intraflagellar transport 80 facilitates endometrial cancer progression through modulating miR-545-3p/FAM98A signaling. | ENSMUSG00000002017 | Fam98a | 3226.901365 | 1.0124800 | 0.017893449 | 0.05880726 | 9.281355e-02 | 7.606303e-01 | 9.240023e-01 | No | Yes | 3518.574508 | 602.012690 | 3471.042464 | 593.842020 | ||
| ENSG00000119899 | 26503 | SLC17A5 | protein_coding | Q9NRA2 | FUNCTION: Transports glucuronic acid and free sialic acid out of the lysosome after it is cleaved from sialoglycoconjugates undergoing degradation, this is required for normal CNS myelination. Mediates aspartate and glutamate membrane potential-dependent uptake into synaptic vesicles and synaptic-like microvesicles. Also functions as an electrogenic 2NO(3)(-)/H(+) cotransporter in the plasma membrane of salivary gland acinar cells, mediating the physiological nitrate efflux, 25% of the circulating nitrate ions is typically removed and secreted in saliva. {ECO:0000269|PubMed:10581036, ECO:0000269|PubMed:11751519, ECO:0000269|PubMed:15510212, ECO:0000269|PubMed:21781115, ECO:0000269|PubMed:22778404}. | Alternative splicing;Amino-acid transport;Cell junction;Cell membrane;Cytoplasmic vesicle;Disease variant;Glycoprotein;Lysosome;Membrane;Phosphoprotein;Reference proteome;Symport;Synapse;Transmembrane;Transmembrane helix;Transport | This gene encodes a membrane transporter that exports free sialic acids that have been cleaved off of cell surface lipids and proteins from lysosomes. Mutations in this gene cause sialic acid storage diseases, including infantile sialic acid storage disorder and and Salla disease, an adult form. [provided by RefSeq, Jul 2008]. | hsa:26503; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; membrane [GO:0016020]; plasma membrane [GO:0005886]; synaptic vesicle membrane [GO:0030672]; carbohydrate:proton symporter activity [GO:0005351]; sialic acid transmembrane transporter activity [GO:0015136]; sialic acid:proton symporter activity [GO:0015538]; transmembrane transporter activity [GO:0022857]; amino acid transport [GO:0006865]; anion transport [GO:0006820]; ion transport [GO:0006811]; response to bacterium [GO:0009617]; sialic acid transport [GO:0015739] | 12359136_expression, localization, and targeting of the wild-type sialin, as well as two mutant polypeptides in sialic acid storage disorders 15006695_In primary neuronal cultures sialin was not targeted into lysosomes but rather revealed a punctate staining along the neuronal processes and was also seen in the plasma membrane. 15172005_Molecular studies showed that all four affected individuals were homozygous for the same novel 983G > A mutation in exon 8 of the SLC17A5 gene, replacing glycine with glutamic acid at position 328 of the sialin protein 15510212_Two missense mutations and one small, in-frame deletion in sialin are associated with ISSD abolished transport, the mutation causing Salla disease (R39C) slowed down, but did not stop the transport cycle. 15516337_there is a direct correlation between sialin function and the disease state of sialic acid storage disorders 16170568_a SLC17A5 p.K136E mutation may have a role in a case of Italian severe Salla disease 17933575_study assessed the effect of missense mutations in the sialin gene (G328E and G409E) and found complete loss of measurable transport activity with both and impaired trafficking of the G409E protein 18399798_The lysosomal localization of human sialin was not or only partially affected by pathogenic missense mutations; in contrast, all pathogenic mutations abolished transport of sialic acid. 18695252_sialin possesses dual physiological functions and acts as a vesicular aspartate/glutamate transporter 19557856_Mutations in the SLC17A5 gene must be considered in two siblings with hypomyelination, even in the absence of sialuria. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20424173_analysis of crucial residues and substrate-induced conformational changes in SLC17 transporter sialin 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21781115_Human SLC17A5 carrying mutations that causes both phenotypes of Salla disease and mutations that cause infantile sialic acid storage disease showed no transport activity 22334707_the substrate-binding site of sialin (SLC17A5) 22778404_These data demonstrate that sialin mediates nitrate influx into salivary gland and other cell types. 27872510_Elevated levels of AST are Associated with Cardiovascular disease. 28187749_study describes a novel pathogenic variant in SLC17A5, namely an intronic transposal insertion, in a patient with mild biochemical and clinical phenotypes. The presence of a small fraction of normal transcript may explain the mild phenotype. This case illustrates the importance of including lysosomal sialic acid storage disease in the differential diagnosis of developmental delay with postnatal onset and hypomyelination. | ENSMUSG00000049624 | Slc17a5 | 773.802417 | 0.8979706 | -0.155259847 | 0.10513778 | 2.161557e+00 | 1.415013e-01 | 4.952037e-01 | No | Yes | 741.425619 | 139.387764 | 815.866626 | 153.289078 | |
| ENSG00000120159 | 79886 | CAAP1 | protein_coding | Q9H8G2 | FUNCTION: Anti-apoptotic protein that modulates a caspase-10 dependent mitochondrial caspase-3/9 feedback amplification loop. {ECO:0000269|PubMed:21980415}. | Alternative splicing;Apoptosis;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:79886; | apoptotic process [GO:0006915]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:2001268] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21980415_The data suggest that C9orf82 functions as an anti-apoptotic protein that modulates a caspase-10 dependent mitochondrial caspase-3/9 feedback amplification loop. It is designated as Conserved Anti-Apoptotic Protein (CAAP). 32315812_MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. 33336752_LncRNA LncOGD-1006 alleviates OGD-induced ischemic brain injury regulating apoptosis through miR-184-5p/CAAP1 axis. | ENSMUSG00000028578 | Caap1 | 984.340175 | 1.0696890 | 0.097191468 | 0.09515750 | 1.038837e+00 | 3.080921e-01 | 6.746459e-01 | No | Yes | 1097.246338 | 186.285524 | 1014.296932 | 172.201723 | ||
| ENSG00000120697 | 29880 | ALG5 | protein_coding | Q9Y673 | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Glycosyltransferase;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene encodes a member of the glycosyltransferase 2 family. The encoded protein participates in glucosylation of the oligomannose core in N-linked glycosylation of proteins. The addition of glucose residues to the oligomannose core is necessary to ensure substrate recognition, and therefore, effectual transfer of the oligomannose core to the nascent glycoproteins. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2008]. | hsa:29880; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; dolichyl-phosphate beta-glucosyltransferase activity [GO:0004581]; oligosaccharyl transferase activity [GO:0004576]; determination of left/right symmetry [GO:0007368]; protein glycosylation [GO:0006486]; protein N-linked glycosylation [GO:0006487]; protein N-linked glycosylation via asparagine [GO:0018279] | ENSMUSG00000036632 | Alg5 | 2054.743074 | 0.9086681 | -0.138174701 | 0.07496149 | 3.356780e+00 | 6.692880e-02 | 3.477781e-01 | No | Yes | 1993.763748 | 297.108172 | 2186.450017 | 325.661738 | ||
| ENSG00000120868 | 317 | APAF1 | protein_coding | O14727 | FUNCTION: Oligomeric Apaf-1 mediates the cytochrome c-dependent autocatalytic activation of pro-caspase-9 (Apaf-3), leading to the activation of caspase-3 and apoptosis. This activation requires ATP. Isoform 6 is less effective in inducing apoptosis. {ECO:0000269|PubMed:10393175, ECO:0000269|PubMed:12804598}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Calcium;Cytoplasm;Direct protein sequencing;Nucleotide-binding;Reference proteome;Repeat;WD repeat | This gene encodes a cytoplasmic protein that initiates apoptosis. This protein contains several copies of the WD-40 domain, a caspase recruitment domain (CARD), and an ATPase domain (NB-ARC). Upon binding cytochrome c and dATP, this protein forms an oligomeric apoptosome. The apoptosome binds and cleaves caspase 9 preproprotein, releasing its mature, activated form. Activated caspase 9 stimulates the subsequent caspase cascade that commits the cell to apoptosis. Alternative splicing results in several transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:317; | apoptosome [GO:0043293]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; secretory granule lumen [GO:0034774]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; cysteine-type endopeptidase activator activity involved in apoptotic process [GO:0008656]; heat shock protein binding [GO:0031072]; identical protein binding [GO:0042802]; nucleotide binding [GO:0000166]; activation of cysteine-type endopeptidase activity [GO:0097202]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c [GO:0008635]; aging [GO:0007568]; apoptotic process [GO:0006915]; cardiac muscle cell apoptotic process [GO:0010659]; cell differentiation [GO:0030154]; cellular response to transforming growth factor beta stimulus [GO:0071560]; forebrain development [GO:0030900]; intrinsic apoptotic signaling pathway [GO:0097193]; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress [GO:0070059]; kidney development [GO:0001822]; nervous system development [GO:0007399]; neural tube closure [GO:0001843]; neuron apoptotic process [GO:0051402]; positive regulation of apoptotic process [GO:0043065]; positive regulation of apoptotic signaling pathway [GO:2001235]; regulation of apoptotic DNA fragmentation [GO:1902510]; regulation of apoptotic process [GO:0042981]; response to G1 DNA damage checkpoint signaling [GO:0072432]; response to hypoxia [GO:0001666]; response to nutrient [GO:0007584] | 11864614_Apaf1 WD40 repeat domains form propellar-like structures that are important for procaspase 9 binding to the CARD domain. 12021264_lack of role in caspase-9 activation in Sendai virus-infected cells 12149244_APAF-1 is under transcriptional regulation of E2F-1 and initiates a caspase cascade 12615903_Data show that the trace amount of cytochrome c present in neutrophils is both necessary and sufficient for apoptotic protease-activating factor 1 (Apaf-1)-dependent caspase activation in these cells. 12637514_function of Apaf-1 is not only to oligomerize procaspase-9 but also to maintain the interaction of the caspase-9 protease domain after processing 12804598_alternatively spliced APAF-1-ALT is a molecule that is deficient and impeded for mediating apoptosis and may contribute to the resistance to DNA damage-induced treatment observed in LNCaP 12963020_Bcl-XL directly binds to the ATPase domain of Apaf-1. 14566819_AMF regulates expression of Apaf-1 and caspase-9 genes via a complex signaling pathway and indirectly regulates formation of the apoptosome. 14747474_the cytochrome c-Apaf-1-procaspase-9 complex functions in the caspase amplification rather than in its initiation 14993223_the functional apoptosome complex in apoptotic cells consists primarily of Apaf-1 and processed caspase-9 15009102_Loss of APAF-1 expression can be considered an indicator of malignant transformation in melanoma. 15026369_Allelic imbalance of 12q22-23 associated with APAF-1 locus correlates with poor disease outcome in cutaneous melanoma 15033720_Apaf-1 proteolytic degradation does not significantly abrogate either the apoptotic morphology or the cleavage of canonical targets. 15305193_Apaf-1 expression is significantly reduced in human melanoma and Apaf-1 may serve as a therapeutic target in melanoma. 15378005_results suggest that APAF-1 gene haploinsufficiency caused by AI increases with tumor progression, and relates to hepatic metastasis 15590702_Limiting Apaf-1 activity may alleviate both pathological protein aggregation and neuronal cell death in HD. 15649154_there is an inverse correlation between Apaf-1 expression and pathologic stage of melanoma 15692060_noncytosolic localization of Apaf-1 may constitute a novel mechanism of chemoresistance in B lymphoma 15703181_Protein kinase A regulates caspase-9 activation by Apaf-1 downstream of cytochrome c 15832174_Apaf-1 expression in 15 melanoma cell lines 15832175_Apaf-1 levels vary in melanoma 15863166_Loss of Apaf-1 expression may represent a marker of aggressive tumor behavior since it correlates significantly with the occurrence of lymph node metastasis in cervical cancer. 15972851_Methylation silencing is a mechanism of the inactivation of APAF1 in acute leukemia. 16046141_Using different cell lines of neuronal origin and modulating the expression of both mutant SOD1s and Apaf1, we show that the removal of Apaf1 prevents cells death. 16098052_No coorelation between apaf-1 expression in melanoma and malignancy 16231040_The depression-associated alleles thus have a common phenotype that is distinct from that of non-associated variants 16232302_Apaf-1 gene structure and function and its role in apoptotic machinery. Involvement in melanoma progression and chemoresistance, as well as clinico-pathological relevance of these findings in treatment of this deadly disease. Review. 16331630_poor prognosis was observed in patients with loss of APAF-1 expression and additional p53 mutation; thus, loss of APAF-1 may become relevant when additional core apoptosis signaling components are disrupted 16420245_These results suggest that Apaf-1 expression may become a prognostic marker for progress of human cutaneous melanoma and further support the notion that loss of Apaf-1 may be an important contributory factor in the development of the disease. 16595687_progression of UV-induced apoptosis requires IRES-mediated translation of Apaf-1 to ensure continuous levels of Apaf-1 despite an overall suppression of protein synthesis 16951219_The promoter hypermethylation of APAF-1 is a marker of aggressive renal cell carcinoma and provides independent prognostic information on disease outcome. 17133271_Methylation of APAF-1 is associated with bladder and kidney cancer 17348858_Data support a model in which Apaf-1 is necessary for the cleavage or activation of all procaspases and the promotion of mitochondrial apoptotic events induced by genotoxic drugs. 17361096_mRNA elevation of apaf1 during blast crisis indicates an involvement in chronic myelogenous leukemia disease progression. 17534194_Observational study of gene-disease association. (HuGE Navigator) 17541304_APAF-1 methylation is related to transcriptional activity of EZH2 expression in early-stage tumor disease of the bladder. 17603079_Fas receptor and Apaf-1 were down-regulated in stage III colorectal cancer cell line. 17876870_The expression of Apaf-1 gene is low in gastric cancer tissues. Methylation of Apaf-1 gene promoter and LOH in domain of 12q22-23 are the main reasons for the expression and altered expression of Apaf-1 gene. 17882496_Reduced expression of Apaf-1 is associated with colorectal adenocarcinoma 17885667_These findings suggest that the efflux of K(+) is prerequisite not only for the formation of the apoptosome but also for the downstream apoptotic signal-transduction pathways. 18042457_These data point to a role for Apaf-1 as a bona fide tumor suppressor. 18093951_Apaf-1 has a role in regulating cytochrome c induction of apoptosis in brain tumors but not in normal neural tissues 18373966_Frameshift mutation at mononucleotide repeat in Apaf-1 is rare in gastric carcinomas. 18439902_PHAPI, CAS, and Hsp70 function together to accelerate nucleotide exchange on Apaf-1 and prevent inactive Apaf-1/cytochrome c aggregation. 18587251_The methylation profile observed demonstrates a substantial role of the APAF-1 gene in urogenital tumorigenesis. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18719738_it has been shown that only the isoforms with the extra WD-40 repeat region activate procaspase-9, it suggests that low procaspase-9 activation may also be involved in the deregulation of apoptosis and chemotherapy resistance in acute myeloid leukemia. 18834886_Frequent HDAC2 mutations are found in MSI tumors and HDAC2 plays a major role in mediating apoptotic response to HDAC inhibitors through direct regulation of APAF1. 19058789_Observational study of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19141860_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19238172_The oxidative modification of caspase-9 by reactive oxygen species can mediate its interaction with apoptotic protease-activating factor 1, and can promote its formation of disulfide-linked complexes. 19455599_results show that the APAF1 variants associated with risk for MDD in the Utah pedigrees are very rare in Northern European and European-American populations 19603830_Data report here the caspase recruitment domain (CARD) of recombinant human apoptotic protease activating factor-1 (Apaf-1) can be induced to undergo amyloid-like fibrillation. 19625176_Observational study of gene-disease association. (HuGE Navigator) 19662370_Expression of Apaf-1 was up-regulated in MCF-7 cells treated with 5-Aza-CdR. 19675677_A drug targeting Apaf-1 allows protection from apoptosis as well as regeneration in the course of inflammation-induced tissue injury 19692168_Observational study of gene-disease association. (HuGE Navigator) 19801675_analysis of a model of the transition of APAF-1 from inactive monomer to caspase-activating apoptosome 19809088_Studies suggest that the apoptosome contains a 1:1 Apaf-1:caspase-9 stoichiometry. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20216471_IL-24 has a role in sensitizing melanoma cells to erlotinib through modulation of the Apaf-1 and Akt signaling pathways 20345447_APAF-1 expression was significantly higher in low-risk, compared to high-risk myelodysplastic syndrome, according to IPSS (P < 0.0001), FAB (P = 0.0265), and cytogenetic risk (P = 0.0134). 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20959405_Observational study of gene-disease association. (HuGE Navigator) 20977544_higher expression of APAF-1 is related to an undifferentiated state in the testicular germ cell tumor (TGCT) pathway. 21104989_Hypermethylation of APAF1 gene is associated with neuroblastoma. 21297999_in the absence of nucleotide such as ATP, direct association of procaspase-9 with Apaf-1 leads to defective molecular timer, and thus, inhibits apoptosome-mediated caspase activation 21528061_Results show a significant inverse correlation between GLUT1 expression and Apaf-1 expression in colorectal adenocarcinomas, and patients with GLUT1 expression demonstrate poor overall survival. 21659556_These results suggest that, in addition to the classical cytochrome c/Apaf-1-dependent pathway of caspase-9 activation, staurosporine can induce caspase-9 activation and apoptosis independently of the apoptosome. 21922274_APAF-1, DAPK-1 and SPARC may have a role in progression and higher tumor stage of renal cell carcinoma in North Indian population 22101335_Curcumin treatment of cancer cells led to Apaf-1 upregulation both at the protein and mRNA levels. Deficiency of Apaf-1 inhibits caspase activation and cell death. 22246185_Collectively, these results identify a novel locus of apoptosomal regulation wherein MAPK signalling promotes Rsk-catalysed Apaf-1 phosphorylation and consequent binding of 14-3-3varepsilon, resulting in decreased cellular responsiveness to cytochrome c. 22335196_melatonin inhibits cell proliferation and induces apoptosis in MDA-MB-361 breast cancer cells in vitro by simultaneously suppressing the COX-2/PGE2, p300/NF-kappaB, and PI3K/Akt/signaling and activating the Apaf-1/caspase-dependent apoptotic pathway. 22996741_Both the silencing of miR-155 and the overexpression Apaf-1 greatly increased the sensitivity of A549 cells to cisplatin. 23085065_The 5' untranslated region of Apaf-1 mRNA directs translation under apoptosis conditions via a 5' end-dependent scanning mechanism. 23207240_Data indicate that the formation of cytochrome c-Apaf-1 apoptosome and the presence of Smac are absolutely required for PSAP-induced apoptosis. 23326121_Apoptosis of HGC-27 cells induced by oridonin may be associated with differential expression of Apaf-1, caspase-3 and cytochrome c, which are highly dependent upon the mitochondrial pathway 23521171_Apaf-1 conformational changes during apoptosome assembly. 23822711_Amiloride modulates glioblastoma multiforme cell radiosensitivity involving the Akt phosphorylation and the alternative splicing of APAF1. 24038028_Using gene reporter assays, we show that promoter variations in 11 intrinsic apoptosis genes, including ADPRT, APAF1, BCL2, BAD, BID, MCL1, BIRC4, BCL2L1, ENDOG, YWHAB, and YWHAQ, influence promoter activity in an allele-specific manner. 24142530_APAF-1 plays an important role in iron-induced erythroid apoptosis increase in myelodysplastic syndrome 24416260_a role for Apaf1 at the mitochondria 24424093_The inhibitory effect of Ab42 on the apoptotic pathway is associated with its interaction with procaspase-9 and consequent inhibition of Apaf-1 apoptosome assembly 25330150_results provide evidence that Apaf-1 pharmacological inhibition has therapeutic potential for the treatment of apoptosis-related diseases. 25599959_Our results indicate that the APAF1, BAX, and FLASH genes not only harbor frameshift mutations but also demonstrate mutational ITH, which together might play a role in the tumorigenesis of CRC with MSI-H by affecting the apoptosis of cancer cells. 25701323_This study suggested that miR-23a, acting as an oncogenic regulator by directly targeting APAF1 in pancreatic cancer, is a useful potential biomarker in diagnosis and treatment of pancreatic cancer. 25895636_The auto-assembly of truncated Apaf1 molecules disappeared after several hours without any enzyme activation. 26014357_cytochrome c and the WD domains of Apaf-1 interact, with bifurcated salt bridges involved in apoptosome assembly 26179084_Different selenium concentrations had varying effects on BAK1 and APAF1 levels. APAF1 may play an important role in the pathogenesis of KBD 26183023_NF-kappaB response element, located close to the p53RE#1, mediates APAF1 transcriptional repression by affecting interaction between KAISO and p53 26290316_Inhibition of Caspase-9 restricted, while Apaf-1 promoted, Chlamydia pneumoniae infection in HEp-2, HeLa, and mouse epithelial fibroblast (MEF) cells. 26740177_These results suggest that CED-4 forms a complex with ced-3 mRNA and delivers it to ribosomes for translation. 26743285_the markers Ets-1 and APAF-1 relative to p53, Ki-67 and PTEN expression in colon adenomas/polyps, were investigated. 26851285_HDAC inhibitors can induce p53 acetylation at lysine 120, which in turn enhances mitochondrion-mediated apoptosis through transcriptional up-regulation of Apaf-1. 27021436_miR21 is transferred from cancer-associated adipocytes (CAAs) and fibroblasts (CAFs) to the cancer cells, where it suppresses ovarian cancer apoptosis and confers chemoresistance by binding to its direct novel target, APAF1. 27032384_Inhibition of TRIAP1 in RPMI8226 cells increased the percentage of apoptotic cells, accompanied by increased expression of APAF1 and Caspase 9, and Caspase 9 and Caspase 3/7 activity. 27141571_Renal APAF1 expression is increased in diabetes and diabetic nephropathy. 27480415_Loss of APAF-1 expression is associated with early recurrence in stage I-III colorectal cancer, suggesting that APAF-1 may have clinical value as a predictive marker of early recurrence. 27863378_Primary cells derived from patients with diffuse large B cell lymphomas show membrane raft sequestration of the apoptosome adaptor protein, Apaf-1, which may mediate drug resistance. 28143931_Results indicate that the apoptotic protease-activating factor 1 (Apaf-1) apoptosome activates caspase-9 in part through sequestration of the inhibitory caspase recruitment domains (CARDs) domain. 28423356_Knockdown of microRNA-27a increased the expression level of Apaf-1, enhancing the formation of Apaf-1-caspase-9 complex and subsequently promoting the TRAIL-induced apoptosis in colorectal cancer stem cells. Findings suggested that knockdown of microRNA-27a in colorectal cancer stem cells by the specific antioligonucleotides was potential to reverse the chemoresistance to TRAIL. 28429233_A significant correlation between changes in the levels of expression and methylation was detected for the three apoptosis-regulatory genes (APAF1, DAPK1, and BCL2). The results suggest that methylation play an important role in the regulation of the apoptosis system genes in breast cancer. 28453462_Authors found significant up-regulation of miR-221 and significant down-regulation of Apaf-1 expression in LSCC tissues compared to normal nearby laryngeal tissues. Significant associations between up-regulated miR-221 and down-regulated Apaf-1 expressions and clinical stage and lymph node (LN) metastasis were found. 28627586_we sought to investigate mechanisms mediated by Hsp70 acetylation in relation to apoptotic and autophagic programmed cell death. Upon stress-induced apoptosis, Hsp70 acetylation inhibits apoptotic cell death, mediated by Hsp70 association with apoptotic protease-activating factor (Apaf)-1 and apoptosis-inducing factor (AIF), key modulators of caspase-dependent and -independent apoptotic pathways, respectively 28895780_miR-300 regulates the cellular sensitivity to ionizing radiation through targeting p53 and apaf1 in lung cancer cells. 28982084_APAF-1 was found to be reduced in non-small-cell lung cancer tissue samples along with high expression of miR-484. 29352212_Results strongly suggest that rare mutations in apoptosis-related genes including CASP9, APAF1, and CASP3 contribute to etiology of neural tube defects in Han Chinese population. 29358613_APAF1 mutations cause recurrent folate-resistant neural tube defects. 29395479_Results suggest that low levels of Apaf-1 as an adaptor protein might be considered as a possible regulatory barrier by which differentiating cells control cell death upon rise in ROS production and cytochrome c release from mitochondria. 29410086_interactions between endogenous Apaf-1 and DeltaApaf-1 is stronger than its interaction with native exogenous Apaf-1 as indicated by dominant negative effect of DeltaApaf-1 on caspase-3 processing 29511177_Cytochrome c and 14-3-3epsilon interaction blocked 14-3-3epsilon-mediated Apaf-1 inhibition. 30066861_Study suggests a potential underlying molecular mechanism of apoptosis inhibition via APAF1 downregulation in human neuroblastoma BE(2)C cells with miRNA36133p overexpression. 30072015_Down-regulation of Apaf-1 protein and the overexpression of Cyclin D1 and AQP-5 proteins possibly contribute to an aggressive serous ovarian carcinoma with a high risk of recurrence and poor response to the first-line chemotherapy. 31094091_the Apaf-1-mediated suppression of HepG2 cell malignancy is achieved by inhibiting the wnt/beta-catenin pathway. 31131537_Silencing of APAF1 reduced the sensitivity of bladder cancer cells to cisplatin chemotherapy. Furthermore, Circular RNA Cdr1as could directly sponge miR-1270 and abolish its effect on APAF1. 31255735_Results show that demonstrates that APAF1 expression is regulated by CBX8 in esophageal squamous cell carcinoma cells. 31329620_These results show that ubiquitinated Apaf-1 may activate caspase-9 under conditions of proteasome impairment. 31498791_A novel mechanism has been revealed that EV71 utilizes for virus release via a 3C protease-hnRNPA1-apaf1-caspase-3-apoptosis axis. 32190895_Hypoxia-induced up-regulation of miR-27a promotes paclitaxel resistance in ovarian cancer. 32461338_The Lumiptosome, an engineered luminescent form of the apoptosome can report cell death by using the same Apaf-1 dependent pathway. 32653692_Long Noncoding RNA Hypoxia-Inducible Factor-1 Alpha-Antisense RNA 1 Regulates Vascular Smooth Muscle Cells to Promote the Development of Thoracic Aortic Aneurysm by Modulating Apoptotic Protease-Activating Factor 1 and Targeting let-7g. 33132160_Loss of WD2 subdomain of Apaf-1 forms an apoptosome structure which blocks activation of caspase-3 and caspase-9. 33177505_EMC6 regulates acinar apoptosis via APAF1 in acute and chronic pancreatitis. 33308446_Apaf-1 Pyroptosome Senses Mitochondrial Permeability Transition. 33386492_Intracellular leucine-rich alpha-2-glycoprotein-1 competes with Apaf-1 for binding cytochrome c in protecting MCF-7 breast cancer cells from apoptosis. 34298080_Contribution of Apaf-1 to the pathogenesis of cancer and neurodegenerative diseases. 34507064_Apaf1 nanoLuc biosensors identified lentinan as a potent synergizer of cisplatin in targeting hepatocellular carcinoma cells. 34898374_Exosomal microRNA rectangle93 rectangle3p secreted by bone marrow mesenchymal stem cells downregulates apoptotic peptidase activating factor 1 to promote wound healing. 35030371_Apoptotic protease activating factor-1 gene and MicroRNA-484: A possible interplay in relapsing remitting multiple sclerosis. | ENSMUSG00000019979 | Apaf1 | 1065.653592 | 1.0919145 | 0.126859922 | 0.08870029 | 2.042326e+00 | 1.529755e-01 | 5.116928e-01 | No | Yes | 916.196949 | 157.686463 | 833.738945 | 143.491356 | |
| ENSG00000121413 | 65982 | ZSCAN18 | protein_coding | Q8TBC5 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:65982; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 24948044_A high negative correlation between promoter DNA methylation and gene expression was observed for CDO1, ZNF331 and ZSCAN18 in gastrointestinal tumors. | ENSMUSG00000070822 | Zscan18 | 257.259101 | 1.0629318 | 0.088048970 | 0.17907345 | 2.409193e-01 | 6.235430e-01 | No | Yes | 251.438403 | 25.968978 | 233.053958 | 23.996311 | |||
| ENSG00000121753 | 576 | ADGRB2 | protein_coding | O60241 | FUNCTION: Orphan G-protein coupled receptor involved in cell adhesion and probably in cell-cell interactions. Activates NFAT-signaling pathway, a transcription factor, via the G-protein GNAZ (PubMed:20367554, PubMed:28891236). Involved in angiogenesis inhibition (By similarity). {ECO:0000250|UniProtKB:Q8CGM1, ECO:0000269|PubMed:20367554, ECO:0000269|PubMed:28891236}. | Alternative splicing;Cell membrane;Direct protein sequencing;Disease variant;Disulfide bond;G-protein coupled receptor;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transducer;Transmembrane;Transmembrane helix | This gene encodes a a seven-span transmembrane protein that is thought to be a member of the secretin receptor family. The encoded protein is a brain-specific inhibitor of angiogenesis. The mature peptide may be further cleaved into additional products (PMID:20367554). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2014]. | hsa:576; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; G protein-coupled receptor activity [GO:0004930]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189]; calcineurin-NFAT signaling cascade [GO:0033173]; cell surface receptor signaling pathway [GO:0007166]; G protein-coupled receptor signaling pathway [GO:0007186]; negative regulation of angiogenesis [GO:0016525]; peripheral nervous system development [GO:0007422] | 12218411_involvement of BAI2 in the ischemia-induced brain angiogenesis 20367554_BAI2 is a functional GPCR regulated by proteolytic processing and activates the NFAT pathway 20677014_Observational study of gene-disease association. (HuGE Navigator) 21787750_This is the first study reporting BAI2 as an interaction partner of GIP. 28891236_These studies provide new insights into the signaling capabilities of the adhesion GPCR BAI2/ADGRB2 and shed light on how an apparent gain-of-function mutation to the receptor's C-terminus may lead to human disease. | ENSMUSG00000028782 | Adgrb2 | 763.795127 | 0.9443095 | -0.082668297 | 0.12005566 | 4.769996e-01 | 4.897845e-01 | 7.971392e-01 | No | Yes | 679.574925 | 72.175539 | 714.511462 | 75.697802 | |
| ENSG00000122390 | 79903 | NAA60 | protein_coding | Q9H7X0 | FUNCTION: N-alpha-acetyltransferase that specifically mediates the acetylation of N-terminal residues of the transmembrane proteins, with a strong preference for N-termini facing the cytosol (PubMed:25732826). Displays N-terminal acetyltransferase activity towards a range of N-terminal sequences including those starting with Met-Lys, Met-Val, Met-Ala and Met-Met (PubMed:21750686, PubMed:25732826, PubMed:27550639, PubMed:27320834). Required for normal chromosomal segregation during anaphase (PubMed:21750686). May also show histone acetyltransferase activity; such results are however unclear in vivo and would require additional experimental evidences (PubMed:21981917). {ECO:0000269|PubMed:21750686, ECO:0000269|PubMed:25732826, ECO:0000269|PubMed:27320834, ECO:0000269|PubMed:27550639, ECO:0000305|PubMed:21981917}. | 3D-structure;Acetylation;Acyltransferase;Alternative promoter usage;Alternative splicing;Chromatin regulator;Chromosome partition;Golgi apparatus;Membrane;Reference proteome;Transferase | This gene encodes an enzyme that localizes to the Golgi apparatus, where it transfers an acetyl group to the N-terminus of free proteins. This enzyme acts on histones, and its activity is important for chromatin assembly and chromosome integrity. Alternative splicing and the use of alternative promoters results in multiple transcript variants. The upstream promoter is located in a differentially methylated region (DMR) and undergoes imprinting; transcript variants originating from this position are expressed from the maternal allele. [provided by RefSeq, Nov 2015]. | hsa:79903; | Golgi membrane [GO:0000139]; H4 histone acetyltransferase activity [GO:0010485]; histone acetyltransferase activity [GO:0004402]; peptide alpha-N-acetyltransferase activity [GO:0004596]; protein homodimerization activity [GO:0042803]; cell population proliferation [GO:0008283]; chromosome segregation [GO:0007059]; histone H3 acetylation [GO:0043966]; histone H4 acetylation [GO:0043967]; N-terminal peptidyl-methionine acetylation [GO:0017196]; N-terminal protein amino acid acetylation [GO:0006474]; nucleosome assembly [GO:0006334] | 21750686_Naa60 is an N-terminal acetyltransferase defined as NatF. Human and Drosophila melanogaster Naa60 acetylate Met-Lys- and other Met-starting N-termini. In Drosophila cells, NAA60 knockdown induced chromosomal segregation defects. 21981917_HAT4 is an important player in the organization and function of the genome and may contribute to the diversity and complexity of higher eukaryotic organisms [HAT4] 25732826_Naa60 is necessary for maintenance of the Golgi ribbon through its Nt-acetylation of substrate protein(s) that is/are involved in Golgi ribbon structural and/or functional organization. 26164078_Knockdown of the N-terminal acetyltransferase Naa60 compromises Golgi ribbon integrity. 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that NAA60 (NAT15) is a putative expression variability marker for the 3-day, 8-cell embryo stage. 27320834_The first crystal structure of the N-terminal acetyltransferase Naa60 reveals two elongated loops important for substrate-specific binding not found in other NAT structures. 27550639_The hNaa60 protein contains an amphipathic helix following its GNAT domain that may contribute to Golgi localization of hNaa60, and the beta7-beta8 hairpin adopted different conformations in the hNaa60(1-242) and hNaa60(1-199) crystal structures. 28196861_analysis of the mode of cytosolic Naa60 anchoring to the Golgi apparatus, most likely occurring post-translationally and specifically facilitating post-translational N-terminal acetylation of many transmembrane proteins | ENSMUSG00000005982 | Naa60 | 1125.439333 | 0.9799987 | -0.029148259 | 0.10870098 | 7.335002e-02 | 7.865202e-01 | 9.308155e-01 | No | Yes | 1223.928188 | 139.522370 | 1242.954397 | 141.620724 | |
| ENSG00000123124 | 11059 | WWP1 | protein_coding | Q9H0M0 | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Ubiquitinates ERBB4 isoforms JM-A CYT-1 and JM-B CYT-1, KLF2, KLF5 and TP63 and promotes their proteasomal degradation. Ubiquitinates RNF11 without targeting it for degradation. Ubiquitinates and promotes degradation of TGFBR1; the ubiquitination is enhanced by SMAD7. Ubiquitinates SMAD6 and SMAD7. Ubiquitinates and promotes degradation of SMAD2 in response to TGF-beta signaling, which requires interaction with TGIF. {ECO:0000269|PubMed:12535537, ECO:0000269|PubMed:15221015, ECO:0000269|PubMed:15359284}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Host-virus interaction;Membrane;Nucleus;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | WW domain-containing proteins are found in all eukaryotes and play an important role in the regulation of a wide variety of cellular functions such as protein degradation, transcription, and RNA splicing. This gene encodes a protein which contains 4 tandem WW domains and a HECT (homologous to the E6-associated protein carboxyl terminus) domain. The encoded protein belongs to a family of NEDD4-like proteins, which are E3 ubiquitin-ligase molecules and regulate key trafficking decisions, including targeting of proteins to proteosomes or lysosomes. Alternative splicing of this gene generates at least 6 transcript variants; however, the full length nature of these transcripts has not been defined. [provided by RefSeq, Jul 2008]. | hsa:11059; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ubiquitin ligase complex [GO:0000151]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; central nervous system development [GO:0007417]; ion transmembrane transport [GO:0034220]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of protein catabolic process [GO:0045732]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; signal transduction [GO:0007165]; viral entry into host cell [GO:0046718] | 12450395_First demonstration of ubiquitin-protein ligase WWP1 from human lung cDNA library recruited by penton base proteins of human adenovirus serotypes Ad2 and Ad3 in vitro and in vivo. 12450395_the interaction of nonenveloped viruses with ubiquitin-protein ligases of host cells. 12535537_Data show that the crystal structure of the HECT domain of the human ubiquitin ligase WWP1/AIP5 maintains a two-lobed structure like the HECT domain of the human ubiquitin ligase E6AP. 15221015_WWP1 negatively regulates TGF-beta signaling in cooperation with Smad7. 16223724_KLF5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells 16785210_Full-length expressed WWP1 could interact in vitro with the cytoplasmic domain of human Notch1, which also regulate the nuclear localization of WWP1. 16924229_these findings identify the first instance of a ubiquitin ligase that causes stabilization of p53 while inactivating its transcriptional activities. 17016436_WWP1 overexpression is a common mechanism involved in the inactivation of TGFbeta function in human cancer. 17330240_genomic aberrations of WWP1 may contribute to the pathogenesis of breast cancer 17609263_the interaction between Gag and WWP1 is required for functions other than Gag ubiquitination 18724389_WWP1 may promote cell proliferation and survival partially through suppressing RNF11-mediated ErbB2 and EGFR downregulation in human cancer cells. 18806757_WWP1 may have a context-dependent role in regulating cell survival through targeting different p63 proteins for degradation. 19035836_analysis of the interaction between ubiquitin ligase WWP1 and Nogo-A 19047365_WWP1 ubiquitinated and caused the degradation of HER4 but not of EGFR, HER2, or HER3. 19267401_Overexpression of WWP1 is associated with the estrogen receptor and insulin-like growth factor receptor 1 in breast carcinoma 19307600_Experiments suggest functions for WWP1 and SPG20 in the regulation of lipid droplet turnover and potential pathological mechanisms in Troyer syndrome. 19561640_WWP1 and its family members suppress the ErbB4 expression and function in breast cancer 19580544_SPG20 interacts with AIP4 and AIP5. 20951678_WWP1 regulates DeltaNp63 transcriptional activity, acting thus as a potential regulator of the proliferation and survival of epithelial-derived cells. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21191027_The authors show that WWP1 changes the ubiquitination status of ARRDC1, suggesting that the arrestins may provide a platform for ubiquitination in PPXY-dependent budding. 21480222_WWP1 depletion-induced apoptosis was rescued by the overexpression of the wild-type WWP1 but not the E3 ligase inactive WWP1-C890A mutant in MCF7 cells. 21795702_WWP1 markedly inhibited the replicative senescence induced by p27(Kip1) by promoting p27(Kip1) degradation. 21996799_WWP1 mutations are not a common cause of human dystroglycanopathy. 22045023_TAZ promotes breast cell growth partially through protecting KLF5 from WWP1-mediated degradation and enhancing KLF5's activities. 22051607_WWP1 may function as the E3 ligase for several PY motif-containing proteins, such as Smad2, KLF5, p63, ErbB4/HER4, RUNX2, JunB, RNF11, SPG20, and Gag, as well as several non-PY motif containing proteins, such as TbetaR1, Smad4, KLF2, and EPS15. Review. 22266093_WWP1 negatively regulates cell migration to CXCL12 by limiting CXCR4 degradation to promote breast cancer metastasis to bone. 23293026_WWP1 down-regulates AMPKalpha2 under high glucose culture conditions via the ubiquitin-proteasome pathway 23573293_LATS1 is critical in mediating WWP1-induced increased cell proliferation in breast cancer cells. 23849376_results reveal that WWP1 might play an oncogenic role in oral cancer cells. 24792179_Knocking down WWP1 promoted cleaved caspase3 protein and p53 expression in hepatocellular carcinoma cells, and caspase3 inhibition could prevent cell apoptosis induced by the knockdown of WWP1. 25051198_Results suggest that elevated transcription and expression levels of ubiquitin ligase E3s WWP1, Smurf1 and Smurf2 genes may be the mechanisms of occurrence, development and metastasis of prostate cancer. 25071155_Data show that WWP1, which specifically ubiquitinates and degrades DeltaNp73 heterodimerizes with another E3 ligase, WWP2. 25293520_Most notably, WWP1 downregulation both inactivated PTEN-Akt signaling pathway in MKN-45 and AGS cells. our findings suggest that WWP1 acts as an oncogenic factor and should be considered as a novel interfering molecular target for gastric carcinoma 25755729_miR-21 overexpression or WWP1 knockdown in endothelial progenitor cells significantly activates the TGFbeta signaling pathway and inhibits cell proliferation. 25814387_PTPN14, a Pez mammalian homolog, is degraded by overexpressed Su(dx) or Su(dx) homologue WWP1 in mammalian cells. 26152726_The cancer-driven alteration of WWP1 culminates in excessive TbetaRI degradation and attenuated TGFbeta1 cytostatic signaling, a consequence that could conceivably confer tumorigenic properties to WWP1. 26506518_Overexpression of WWP1 promotes tumorigenesis in patients with hepatocellular carcinoma. 27070713_Study shows that WWP1 was upregulated in prostate cancer (PCa) clinical specimens and contributed to cancer cell invasion, indicating that this target of Mir-452 functioned as an oncogene. 28426804_Knock-down of WWP1 abrogates DNA damage-induced down-regulation of DeltaNp63alphaand partially rescues cell apoptosis 28431583_Results show that WWP1 is frequently upregulated in gastric cancer (GC) tissues and cells, and that its 3'-UTR is a putative target of miR-584 which suppresses its protein expression by mRNA degradation. 28461335_results demonstrate that WWP1 catalyzes the formation of Ub chains through a sequential addition mechanism, in which Ub monomers are transferred in a successive fashion to the substrate, and that ubiquitination by WWP1 requires the presence of a low-affinity, noncovalent Ub-binding site within the HECT domain. 28475870_Describe an autoinhibitory mechanism for WWP1 ubiquitin ligase involving a linker-HECT domain interaction. This intramolecular interaction traps the HECT enzyme in its inactive state and can be relieved by linker phosphorylation. 28768865_study describes a physical and functional interaction between Ebola virus VP40 (eVP40) and WWP1, a host E3 ubiquitin ligase that ubiquitinates VP40 and regulates virus-like particles egress. 29209041_Provide molecular insights into the anti-cancer potential of WWP1 inhibition. 29635000_Study identified beta-dystroglycan as a substrate of WWP1 and found that the muscular dystrophy-causing mutation of WWP1 renders the enzyme hyperactive by relieving autoinhibition. 29888632_WWP1 expression was significantly associated with histological grade, invasion depth and lymph node metastasis in patients with cutaneous squamous cell carcinoma (CSCC). 30978403_WWP1 overexpression decreased miR-30a-5p expression and inhibited glioma cell malignant behaviors via inhibiting NF-kappaB p65. 31348766_RT-qPCR and Western blot showed that WWP1 was positively regulated by SNHG12 and negatively regulated by miR-129-5p at the mRNA level and protein level. Overexpression of WWP1 significantly increased proliferation and invasion of laryngeal cancer cells. 32459922_n this study involving patients with disorders resulting in a predisposition to the development of multiple malignant neoplasms without PTEN germline mutations, we confirmed the function of WWP1 as a cancer-susceptibility gene through direct aberrant regulation of the PTEN-PI3K signaling axis. 33113605_[Abnormal expression of WWP1 in chronic lymphocytic leukemia and its clinical significance]. 33198785_MicroRNA-15b shuttled by bone marrow mesenchymal stem cell-derived extracellular vesicles binds to WWP1 and promotes osteogenic differentiation. 33470109_Ubiquitin Ligase Activities of WWP1 Germline Variants K740N and N745S. 33516665_BAP1 antagonizes WWP1-mediated transcription factor KLF5 ubiquitination and inhibits autophagy to promote melanoma progression. 33648498_CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. 34035464_miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/beta-catenin signaling pathway. 34139860_Targeting E3 Ubiquitin Ligase WWP1 Prevents Cardiac Hypertrophy Through Destabilizing DVL2 via Inhibition of K27-Linked Ubiquitination. 34404733_AMOTL2 mono-ubiquitination by WWP1 promotes contact inhibition by facilitating LATS activation. 34404764_WWP1 targeting MUC1 for ubiquitin-mediated lysosomal degradation to suppress carcinogenesis. 34559578_Ubiquitin ligase Wwp1 gene deletion attenuates diastolic dysfunction in pressure-overload hypertrophy. 34907909_WWP1 inactivation enhances efficacy of PI3K inhibitors while suppressing their toxicities in breast cancer models. 35154481_alpha-Catulin promotes cancer stemness by antagonizing WWP1-mediated KLF5 degradation in lung cancer. | ENSMUSG00000041058 | Wwp1 | 1105.501624 | 1.0158929 | 0.022748327 | 0.10681388 | 4.516929e-02 | 8.316931e-01 | 9.452725e-01 | No | Yes | 1236.204576 | 254.990255 | 1230.188841 | 253.771532 |
| ENSG00000124596 | 221443 | OARD1 | protein_coding | Q9Y530 | FUNCTION: ADP-ribose glycohydrolase that hydrolyzes ADP-ribose and acts on different substrates, such as proteins ADP-ribosylated on glutamate and O-acetyl-ADP-D-ribose (PubMed:23481255, PubMed:23474714, PubMed:21849506). Specifically acts as a glutamate mono-ADP-ribosylhydrolase by mediating the removal of mono-ADP-ribose attached to glutamate residues on proteins (PubMed:23481255, PubMed:23474714). Does not act on poly-ADP-ribosylated proteins: the poly-ADP-ribose chain of poly-ADP-ribosylated glutamate residues must by hydrolyzed into mono-ADP-ribosylated glutamate by PARG to become a substrate for OARD1 (PubMed:23481255). Deacetylates O-acetyl-ADP ribose, a signaling molecule generated by the deacetylation of acetylated lysine residues in histones and other proteins (PubMed:21849506). Catalyzes the deacylation of O-acetyl-ADP-ribose, O-propionyl-ADP-ribose and O-butyryl-ADP-ribose, yielding ADP-ribose plus acetate, propionate and butyrate, respectively (PubMed:21849506). {ECO:0000269|PubMed:21849506, ECO:0000269|PubMed:23474714, ECO:0000269|PubMed:23481255}. | 3D-structure;Acetylation;Chromosome;Disease variant;Hydrolase;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a deacylase that can convert O-acetyl-ADP-ribose to ADP-ribose and acetate, O-propionyl-ADP-ribose to ADP-ribose and propionate, and O-butyryl-ADP-ribose to ADP-ribose and butyrate. The ADP-ribose product is able to inhibit these reactions through a competitive feedback loop. [provided by RefSeq, Jul 2016]. | hsa:221443; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; site of DNA damage [GO:0090734]; ADP-ribosylglutamate hydrolase activity [GO:0140293]; O-acetyl-ADP-ribose deacetylase activity [GO:0061463]; purine nucleoside binding [GO:0001883]; cellular response to DNA damage stimulus [GO:0006974]; peptidyl-glutamate ADP-deribosylation [GO:0140291]; protein de-ADP-ribosylation [GO:0051725]; purine nucleoside metabolic process [GO:0042278] | 21849506_Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. 23481255_C6orf130 enzymatic activity has a role in the turnover and recycling of protein ADP-ribosylation in neurodegenerative disease. 26091342_Studies indicate that poly (ADP-ribose) glycohydrolase (PARG) and terminal ADP-ribose glycohydrolase 1 (TARG1) are key enzymes in poly(ADP-ribose) polymerases (PARPs)-mediated ADP-ribosylation. 32427867_Comparative analysis of MACROD1, MACROD2 and TARG1 expression, localisation and interactome. 34508355_TARG1 protects against toxic DNA ADP-ribosylation. | ENSMUSG00000040771 | Oard1 | 395.775550 | 1.0100878 | 0.014480687 | 0.14340324 | 1.019238e-02 | 9.195843e-01 | No | Yes | 370.206368 | 46.742953 | 364.896445 | 46.070154 | ||
| ENSG00000124615 | 4337 | MOCS1 | protein_coding | Q9NZB8 | FUNCTION: Isoform MOCS1A and isoform MOCS1B probably form a complex that catalyzes the conversion of 5'-GTP to cyclic pyranopterin monophosphate (cPMP). MOCS1A catalyzes the cyclization of GTP to (8S)-3',8-cyclo-7,8-dihydroguanosine 5'-triphosphate and MOCS1B catalyzes the subsequent conversion of (8S)-3',8-cyclo-7,8-dihydroguanosine 5'-triphosphate to cPMP. {ECO:0000269|PubMed:11891227}. | 4Fe-4S;Acetylation;Alternative splicing;Disease variant;GTP-binding;Iron;Iron-sulfur;Lyase;Metal-binding;Molybdenum cofactor biosynthesis;Nucleotide-binding;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine | PATHWAY: Cofactor biosynthesis; molybdopterin biosynthesis. | Molybdenum cofactor biosynthesis is a conserved pathway leading to the biological activation of molybdenum. The protein encoded by this gene is involved in this pathway. This gene was originally thought to produce a bicistronic mRNA with the potential to produce two proteins (MOCS1A and MOCS1B) from adjacent open reading frames. However, only the first open reading frame (MOCS1A) has been found to encode a protein from the putative bicistronic mRNA, whereas additional splice variants are likely to produce a fusion between the two open reading frames. This gene is defective in patients with molybdenum cofactor deficiency, type A. A related pseudogene has been identified on chromosome 16. [provided by RefSeq, Nov 2017]. | hsa:4337; | cytosol [GO:0005829]; molybdopterin synthase complex [GO:0019008]; nucleus [GO:0005634]; 4 iron, 4 sulfur cluster binding [GO:0051539]; cyclic pyranopterin monophosphate synthase activity [GO:0061799]; GTP 3',8'-cyclase activity [GO:0061798]; GTP binding [GO:0005525]; metal ion binding [GO:0046872]; Mo-molybdopterin cofactor biosynthetic process [GO:0006777]; molybdopterin cofactor biosynthetic process [GO:0032324] | 12754701_Review: A total of 32 different disease-causing mutations, including several common to more than one family, have been identified in molybdenum cofactor-deficient patients and their relatives 15180982_MOCS1A is an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis 17065069_genetic and protein structural analysis of MOCS1 in molybdenum cofactor deficiency [case report] 29368224_Case Report: in a mild case of molybdenum cofactor deficiency detected two proteins: a truncated MOCS1AB protein and a 22.4 kDa protein representing MOCS1B, demonstrating an unusual mechanism of translation re-initiation in the MOCS1 transcript, which results in trace amounts of functional MOCS1B protein being sufficient to partially protect the patient from the most severe symptoms. 31477743_ETHE1 and MOCS1 deficiencies: Disruption of mitochondrial bioenergetics, dynamics, redox homeostasis and endoplasmic reticulum-mitochondria crosstalk in patient fibroblasts. 31996372_Alternative splicing of the bicistronic gene molybdenum cofactor synthesis 1 (MOCS1) uncovers a novel mitochondrial protein maturation mechanism. | ENSMUSG00000064120 | Mocs1 | 470.682214 | 0.9222731 | -0.116734009 | 0.12586293 | 8.617088e-01 | 3.532609e-01 | 7.078034e-01 | No | Yes | 445.273036 | 53.610739 | 478.397148 | 57.483625 |
| ENSG00000124782 | 6239 | RREB1 | protein_coding | Q92766 | FUNCTION: Transcription factor that binds specifically to the RAS-responsive elements (RRE) of gene promoters (PubMed:9305772, PubMed:15067362, PubMed:8816445, PubMed:10390538, PubMed:17550981). Represses the angiotensinogen gene (PubMed:15067362). Negatively regulates the transcriptional activity of AR (PubMed:17550981). Potentiates the transcriptional activity of NEUROD1 (PubMed:12482979). Promotes brown adipocyte differentiation (By similarity). May be involved in Ras/Raf-mediated cell differentiation by enhancing calcitonin expression (PubMed:8816445). {ECO:0000250|UniProtKB:Q3UH06, ECO:0000269|PubMed:10390538, ECO:0000269|PubMed:12482979, ECO:0000269|PubMed:15067362, ECO:0000269|PubMed:17550981, ECO:0000269|PubMed:8816445, ECO:0000269|PubMed:9305772}. | Activator;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a zinc finger transcription factor that binds to RAS-responsive elements (RREs) of gene promoters. It has been shown that the calcitonin gene promoter contains an RRE and that the encoded protein binds there and increases expression of calcitonin, which may be involved in Ras/Raf-mediated cell differentiation. Multiple transcript variants encoding several different isoforms have been found for this gene. [provided by RefSeq, Dec 2009]. | hsa:6239; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; nuclear body [GO:0016604]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; multicellular organism development [GO:0007275]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of brown fat cell differentiation [GO:0090336]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of lamellipodium morphogenesis [GO:2000394]; positive regulation of mammary gland epithelial cell proliferation [GO:0033601]; positive regulation of substrate adhesion-dependent cell spreading [GO:1900026]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of wound healing, spreading of epidermal cells [GO:1903691]; Ras protein signal transduction [GO:0007265]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; transcription by RNA polymerase II [GO:0006366] | 12700664_The p16 promoter can be downregulated by transfected human RREB, in a Ras- or Mek-dependent manner, and that the BALB/c promoter is more sensitive than DBA/2 to regulation by RREB, a ras-responsive transcriptional element with zinc-finger binding motifs. 15067362_Finb functions as a sequence-specific transcriptional repressor of the hANG gene 18394891_essential to reduce cell-cell adhesion when epithelial cells within an interconnected group undergo dynamic changes in cell shape 19558368_Findings provide evidence that RREB-1 participates in modulating p53 transcription in response to DNA damage. 19802870_Ras pathway and activation of RREB-1 are involved in hZIP1 down-regulation and may play a role in the decrease of the transporter expression in prostate cancer. 19890057_This demonstration is the first of a repressor factor of HLA-G transcriptional activity taking part in HLA-G repression by epigenetic mechanisms. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20884846_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 21159816_KRAS and RREB1 are targets of miR-143/miR-145, revealing a feed-forward mechanism that potentiates Ras signaling 21360563_RREB-1 overexpression results in down-regulation of hZIP1 and contributes to the loss of hZIP1 expression and zinc in prostate cancer. 21613827_Data show that the combination of concurrent zinc, ZIP3, and RREB-1 changes represent early events in the development of adenocarcinoma. 21703425_RREB1 transcription factor splice variants are associated with urologic cancer. 21792086_Data indicate the upregulation of RREB1, PDE6B, and CD209 suggests that these proteins might play important roles in the differentiation of primitive gut tube cells from embryonic stem cells (hESCs) and in primitive gut tube development into pancreas. 22427155_These results support a concept that downregulation of RREB-1 causes downregulation of ZIP3, which results in decreased zinc in pancreatic premalignant and carcinoma cells 22751122_RREB1 is overexpressed in colorectal adenocarcinoma tumors. RREB1 repressed miR-143/145 modulates KRAS signaling. 24348900_Data show the transcriptional activation of the cholecystokinin gene by DJ-1 through interaction of DJ-1 with RREB1 and the effect of DJ-1 on the cholecystokinin level. 24418439_Thus HNT and c are functional homologs at the level of DNA binding, transcriptional regulation and developmental control 25027322_RREB1 is a novel candidate gene for type 2 diabetes associated end-stage kidney disease. 25050557_The pathway of RREB1/ZIP3/Zinc and its downregulation during oncogenesis exist to prevent the accumulation of cytotoxic levels of zinc during the development and progression of the malignant cells in pancreatic adenocarcinoma. 26608785_Histone modifier genes (JMJD1C, RREB1, MINA, KDM7A) alter conotruncal heart phenotypes in 22q11.2 deletion syndrome. 27185405_RREB1 and CCND1 gains are common in nail apparatus melanoma 28341660_RREB1 cooperates with noncoding RNA linc-ADAMTS5 to inhibit ADAMTS5 expression, thereby affecting degeneration of the extracellular matrix (ECM) of the intervertebral disc. 29099273_Of the 465,447 CpG sites analyzed, 12 showed differential methylation (false discovery rate <0.15), including markers within genes associated with monogenic diabetes (HNF4A) or obesity (RREB1). The overall methylation at HNF4A showed inverse correlations with mRNA expression levels, though non significant 29912715_Ectomesenchymal chondromyxoid tumors are characterized by an RREB1-MKL2 fusion gene. 30355676_Functional data indicate that modulating Ras-responsive element-binding protein 1 (RREB1) expression in human DLBCL cell lines in vitro alters KRAS expression, signaling, and proliferation; thus, suggesting that this proto-oncogene is a common mechanism of RAS/MAPK hyperactivation in human DLBCL. 30814490_Data show that RREB1-induced upregulation of AGAP2-AS1 regulates cell proliferation and migration in PC partly through suppressing ANKRD1 and ANGPTL4 by recruiting EZH2. 30982491_Findings demonstrated that RREB1 blocks granulocytic differentiation of myeloid leukemia cells by inhibiting the expression of miR-145 and downstream targets of the RAS signal pathway. 31915377_identification of RAS-responsive element binding protein 1 (RREB1), a RAS transcriptional effector, as a key partner of TGF-beta-activated SMAD transcription factors in epithelial-to-mesenchymal transitions 32210733_Transcription Factor RREB1: from Target Genes towards Biological Functions. 32938917_Haploinsufficiency of RREB1 causes a Noonan-like RASopathy via epigenetic reprogramming of RAS-MAPK pathway genes. 34666611_Knockdown of RREB1 inhibits cell proliferation via enhanced p16 expression in gastric cancer. | ENSMUSG00000039087 | Rreb1 | 1108.599935 | 1.1591955 | 0.213123892 | 0.10143212 | 4.462279e+00 | 3.465124e-02 | 2.511419e-01 | No | Yes | 1235.774233 | 122.030825 | 1074.959401 | 106.132565 | |
| ENSG00000125122 | 26231 | LRRC29 | transcribed_unitary_pseudogene | Q8WV35 | FUNCTION: Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation. | Leucine-rich repeat;Reference proteome;Repeat;Ubl conjugation pathway | This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbls class and, in addition to an F-box, contains 9 tandem leucine-rich repeats. Two transcript variants encoding the same protein have been found for this gene. Other variants may occur, but their full-length natures have not been characterized. [provided by RefSeq, Jul 2008]. | 64.759691 | 0.8797155 | -0.184890985 | 0.32923744 | 3.154755e-01 | 5.743395e-01 | No | Yes | 52.382332 | 9.948703 | 60.093702 | 11.272375 | |||||||
| ENSG00000125247 | 84899 | TMTC4 | protein_coding | Q5T4D3 | FUNCTION: Transfers mannosyl residues to the hydroxyl group of serine or threonine residues. The 4 members of the TMTC family are O-mannosyl-transferases dedicated primarily to the cadherin superfamily, each member seems to have a distinct role in decorating the cadherin domains with O-linked mannose glycans at specific regions. Also acts as O-mannosyl-transferase on other proteins such as PDIA3. {ECO:0000269|PubMed:28973932}. | Alternative splicing;Endoplasmic reticulum;Membrane;Reference proteome;Repeat;TPR repeat;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:28973932}. | This gene encodes a transmembrane protein that belongs to family of proteins containing an N-terminal transmembrane domain and a C-terminal tetratricopeptide repeat (TPR) domain. TPR domains mediate protein-protein interactions in various cellular processes, such as synaptic vesicle fusion, protein folding, and protein translocation. A pseudogene of this gene has been defined on chromosome 5. [provided by RefSeq, Apr 2017]. | hsa:84899; | endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; ATPase binding [GO:0051117]; dolichyl-phosphate-mannose-protein mannosyltransferase activity [GO:0004169]; mannosyltransferase activity [GO:0000030]; endoplasmic reticulum unfolded protein response [GO:0030968]; outer hair cell apoptotic process [GO:1905584]; positive regulation of endoplasmic reticulum calcium ion concentration [GO:0032470]; protein O-linked mannosylation [GO:0035269]; sensory perception of sound [GO:0007605] | 33436046_Conserved sequence motifs in human TMTC1, TMTC2, TMTC3, and TMTC4, new O-mannosyltransferases from the GT-C/PMT clan, are rationalized as ligand binding sites. 33925440_Transmembrane and Tetratricopeptide Repeat Containing 4 Is a Novel Diagnostic Marker for Prostate Cancer with High Specificity and Sensitivity. | ENSMUSG00000041594 | Tmtc4 | 680.261074 | 0.9511213 | -0.072298816 | 0.11009169 | 4.318953e-01 | 5.110604e-01 | 8.115359e-01 | No | Yes | 559.060216 | 50.966996 | 593.548344 | 53.996354 |
| ENSG00000125319 | 78995 | HROB | protein_coding | Q8N3J3 | FUNCTION: DNA-binding protein involved in homologous recombination that acts by recruiting the MCM8-MCM9 helicase complex to sites of DNA damage to promote DNA repair synthesis. {ECO:0000269|PubMed:31467087}. | Alternative splicing;Chromosome;DNA damage;DNA recombination;DNA repair;DNA synthesis;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome | hsa:78995; | nucleus [GO:0005634]; site of DNA damage [GO:0090734]; single-stranded DNA binding [GO:0003697]; cellular response to DNA damage stimulus [GO:0006974]; DNA synthesis involved in DNA repair [GO:0000731]; female gamete generation [GO:0007292]; interstrand cross-link repair [GO:0036297]; male gamete generation [GO:0048232]; recombinational repair [GO:0000725] | 19079262_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 30370607_The C17orf53 SNP rs227584 is associated with human bone density and bone fractures risk. 31467087_C17orf53/HROB is an OB-fold-containing factor involved in HR that acts by recruiting the MCM8-MCM9 helicase to sites of DNA damage to promote DNA synthesis 32528060_MCM8IP activates the MCM8-9 helicase to promote DNA synthesis and homologous recombination upon DNA damage. 32853826_C17orf53 is identified as a novel gene involved in inter-strand crosslink repair. 34707299_Meiotic genes in premature ovarian insufficiency: variants in HROB and REC8 as likely genetic causes. | ENSMUSG00000034773 | Hrob | 464.324859 | 1.0780030 | 0.108361203 | 0.13248102 | 6.706843e-01 | 4.128131e-01 | 7.535081e-01 | No | Yes | 454.373995 | 47.605549 | 431.841928 | 45.236670 | ||
| ENSG00000125637 | 23550 | PSD4 | protein_coding | Q8NDX1 | FUNCTION: Guanine nucleotide exchange factor for ARF6 and ARL14/ARF7. Through ARL14 activation, controls the movement of MHC class II-containing vesicles along the actin cytoskeleton in dendritic cells. Involved in membrane recycling. Interacts with several phosphatidylinositol phosphate species, including phosphatidylinositol 3,4-bisphosphate, phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 4,5-bisphosphate. {ECO:0000269|PubMed:12082148, ECO:0000269|PubMed:21458045}. | Alternative splicing;Cell membrane;Cell projection;Coiled coil;Guanine-nucleotide releasing factor;Lipid-binding;Membrane;Phosphoprotein;Reference proteome | hsa:23550; | membrane [GO:0016020]; ruffle membrane [GO:0032587]; guanyl-nucleotide exchange factor activity [GO:0005085]; phospholipid binding [GO:0005543]; regulation of ARF protein signal transduction [GO:0032012] | 11050434_One of the cytohesin family of guanine nucleotide exchange factors, Tic may act on the ARF family of GTP-binding proteins. A PH domain with similar sequence and relative position to those in other cytohesins may allow association with phosphoinositides. 25115298_results identify EFA6B as a novel antagonist in breast cancer and they point to its regulatory and signaling pathways as rational therapeutic targets in aggressive forms of this disease 33850160_EFA6B regulates a stop signal for collective invasion in breast cancer. 33932127_TNF-alpha/NF-kappaB signaling epigenetically represses PSD4 transcription to promote alcohol-related hepatocellular carcinoma progression. 35140331_DDR1 promotes hepatocellular carcinoma metastasis through recruiting PSD4 to ARF6. | ENSMUSG00000026979 | Psd4 | 87.099006 | 1.2746944 | 0.350151431 | 0.27857538 | 1.589650e+00 | 2.073761e-01 | No | Yes | 90.633828 | 18.238363 | 75.525419 | 15.126239 | |||
| ENSG00000125703 | 84938 | ATG4C | protein_coding | Q96DT6 | FUNCTION: Cysteine protease that plays a key role in autophagy by mediating both proteolytic activation and delipidation of ATG8 family proteins (PubMed:21177865, PubMed:29458288, PubMed:30661429). The protease activity is required for proteolytic activation of ATG8 family proteins: cleaves the C-terminal amino acid of ATG8 proteins MAP1LC3 and GABARAPL2, to reveal a C-terminal glycine (PubMed:21177865). Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy (By similarity). In addition to the protease activity, also mediates delipidation of ATG8 family proteins (PubMed:29458288, PubMed:33909989). Catalyzes delipidation of PE-conjugated forms of ATG8 proteins during macroautophagy (PubMed:29458288, PubMed:33909989). Compared to ATG4B, the major protein for proteolytic activation of ATG8 proteins, shows weaker ability to cleave the C-terminal amino acid of ATG8 proteins, while it displays stronger delipidation activity (PubMed:29458288). In contrast to other members of the family, weakly or not involved in phagophore growth during mitophagy (PubMed:33773106). {ECO:0000250|UniProtKB:Q9Y4P1, ECO:0000269|PubMed:21177865, ECO:0000269|PubMed:29458288, ECO:0000269|PubMed:30661429, ECO:0000269|PubMed:33773106, ECO:0000269|PubMed:33909989}. | Acetylation;Autophagy;Cytoplasm;Hydrolase;Phosphoprotein;Protease;Protein transport;Reference proteome;Thiol protease;Transport;Ubl conjugation pathway | Autophagy is the process by which endogenous proteins and damaged organelles are destroyed intracellularly. Autophagy is postulated to be essential for cell homeostasis and cell remodeling during differentiation, metamorphosis, non-apoptotic cell death, and aging. Reduced levels of autophagy have been described in some malignant tumors, and a role for autophagy in controlling the unregulated cell growth linked to cancer has been proposed. This gene encodes a member of the autophagin protein family. The encoded protein is also designated as a member of the C-54 family of cysteine proteases. Alternate transcriptional splice variants, encoding the same protein, have been characterized. [provided by RefSeq, Jul 2008]. | hsa:84938; | cytoplasm [GO:0005737]; cysteine-type endopeptidase activity [GO:0004197]; cysteine-type peptidase activity [GO:0008234]; autophagy [GO:0006914]; protein delipidation [GO:0051697]; protein transport [GO:0015031]; proteolysis [GO:0006508] | 22248718_miR-376b controls autophagy by directly regulating intracellular levels of two key autophagy proteins, ATG4C and BECN1. 27742532_association between SNPs and Kashin-Beck disease 28423511_We demonstrate that, in breast cancer cells, ATM and ATG4C are essential drivers of mammosphere formation, suggesting that their targeting may improve current approaches to eradicate breast cancer cells with a stem-like phenotype. 30661429_Human HAP1 and HeLa cells lacking ATG4B exhibit a severe but incomplete defect in LC3/GABARAP processing and autophagy. By further genetic depletion of ATG4 isoforms using CRISPR-Cas9 and siRNA we uncover that ATG4A, ATG4C and ATGD all contribute to residual priming activity, which is sufficient to enable lipidation of endogenous GABARAPL1 on autophagic structures. 31291988_our results suggested that increased ATG4C expression was associated with worse prognosis in glioma patients. Knockdown of ATG4C suppressed glioma progression by inducing cell cycle arrest and promoting apoptosis of glioma cells possibly through increasing ROS production. 32401768_CAV1 and ATG4C serve as useful prognostic biomarkers and candidate therapeutic targets in EOC 33773106_ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. 34619495_MicroRNA-142-3p inhibits autophagy and promotes intracellular survival of Mycobacterium tuberculosis by targeting ATG16L1 and ATG4c. 34699312_Epigallocatechin-3-gallate protects cardiomyocytes from hypoxia-reoxygenation damage via raising autophagy related 4C expression. | ENSMUSG00000028550 | Atg4c | 1277.352067 | 0.8655586 | -0.208296665 | 0.09398749 | 4.898482e+00 | 2.688031e-02 | 2.195101e-01 | No | Yes | 1283.248485 | 300.990947 | 1491.485048 | 349.759010 | |
| ENSG00000125740 | 2354 | FOSB | protein_coding | P53539 | FUNCTION: Heterodimerizes with proteins of the JUN family to form an AP-1 transcription factor complex, thereby enhancing their DNA binding activity to gene promoters containing an AP-1 consensus sequence 5'-TGA[GC]TCA-3' and enhancing their transcriptional activity (PubMed:12618758, PubMed:28981703). As part of the AP-1 complex, facilitates enhancer selection together with cell-type-specific transcription factors by collaboratively binding to nucleosomal enhancers and recruiting the SWI/SNF (BAF) chromatin remodeling complex to establish accessible chromatin (By similarity). Together with JUN, plays a role in activation-induced cell death of T cells by binding to the AP-1 promoter site of FASLG/CD95L, and inducing its transcription in response to activation of the TCR/CD3 signaling pathway (PubMed:12618758). Exhibits transactivation activity in vitro (By similarity). Involved in the display of nurturing behavior towards newborns (By similarity). May play a role in neurogenesis in the hippocampus and in learning and memory-related tasks by regulating the expression of various genes involved in neurogenesis, depression and epilepsy (By similarity). Implicated in behavioral responses related to morphine reward and spatial memory (By similarity). {ECO:0000250|UniProtKB:P13346, ECO:0000269|PubMed:12618758, ECO:0000269|PubMed:28981703}.; FUNCTION: [Isoform 11]: Exhibits lower transactivation activity than isoform 1 in vitro (By similarity). The heterodimer with JUN does not display any transcriptional activity, and may thereby act as an transcriptional inhibitor (By similarity). May be involved in the regulation of neurogenesis in the hippocampus (By similarity). May play a role in synaptic modifications in nucleus accumbens medium spiny neurons and thereby play a role in adaptive and pathological reward-dependent learning, including maladaptive responses involved in drug addiction (By similarity). Seems to be more stably expressed with a half-life of ~9.5 hours in cell culture as compared to 1.5 hours half-life of isoform 1 (By similarity). {ECO:0000250|UniProtKB:P13346}. | 3D-structure;Alternative splicing;DNA-binding;Disulfide bond;Nucleus;Phosphoprotein;Reference proteome | The Fos gene family consists of 4 members: FOS, FOSB, FOSL1, and FOSL2. These genes encode leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. As such, the FOS proteins have been implicated as regulators of cell proliferation, differentiation, and transformation. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:2354; | chromatin [GO:0000785]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription factor binding [GO:0008134]; cellular response to calcium ion [GO:0071277]; cellular response to hormone stimulus [GO:0032870]; female pregnancy [GO:0007565]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357]; response to cAMP [GO:0051591]; response to corticosterone [GO:0051412]; response to mechanical stimulus [GO:0009612]; response to morphine [GO:0043278]; response to progesterone [GO:0032570]; response to xenobiotic stimulus [GO:0009410]; transcription by RNA polymerase II [GO:0006366] | 12371906_demonstrated that a functional AP-1 site mediates MMP-2 transcription in cardiac cells through the binding of distinctive Fra1-JunB and FosB-JunB heterodimers. The synthesis of MMP-2 is considered to be independent of the AP-1 transcriptional complex 12618758_AP-1 (c-Jun & FosB) binds to a site in the 5' untranslated region of the CD95L gene. Transdominant negative Jun mutants reduce CD95L promoter activity. FosB dimerized with c-Jun has an important role in TCR/CD3-mediated activation-induced cell death. 14741347_VEGF and PlGF induced expression of both full-length FosB mRNA and an alternatively spliced variant. 15926923_In hepatoma-associated anorexia-cachexia FpsB was induced in several brain areas of thes forebrain. 17495958_Coordinated down- and up-regulation of the various AP-1 subunits in the course of epidermal wound healing is important for its undisturbed progress, putatively by influencing inflammation and cell-cell communication. 17601350_Observational study of gene-disease association. (HuGE Navigator) 18207134_Observational study of gene-disease association. (HuGE Navigator) 18490653_activation of protein kinase A elicits an immediate response through induction of genes such as ID2 and FosB, followed by sustained secretion of bone-related cytokines such as BMP-2, IGF-1, and IL-11 18491952_DeltaFosB was able to trigger partial Pref-1-mediated de-differentiation of adipocytes, which also retained their adipocytic cell phenotype. 18802113_An IFN-gamma-mediated homeostatic loop limits the potential for tissue damage associated with inflammation and identifies transcriptional factor AP-1 that regulates matrix metalloproteinase expression in myeloid cells in inflammatory settings. 18977241_Observational study of gene-disease association. (HuGE Navigator) 19133651_MicroRNA-101, which is aberrantly expressed in hepatocellular carcinoma, could repress the expression of the FOS oncogene. 19381435_FosB is induced in PMA treated K562 cells in a sustained manner and forms an active AP-1 protein-DNA complex. Down-regulation of FosB with specific shRNAs inhibited the induction of CD41, a specific cell surface marker of megakaryocytes. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19795327_Results suggest that in addition to clinically prognostic factors, FOS-B expression has a debatable impact on patient survival. 20571966_RGS16 and FosB are underexpressed in pancreatic cancer with lymph node metastasis and associated with reduced survival 20813842_transcription factor FosB/activating protein-1 (AP-1) activation is a prominent downstream signal of the extracellular nucleotide receptor P2RX7 in monocytic and osteoblastic cells 20826776_Findings indicate that neurobehavioral stress leads to FosB-driven increases in IL8, which is associated with increased tumor growth and metastases. 21543584_AP-1 protein induction during monopoiesis favors C/EBP: AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. 21616539_Induction of a DeltaFOSB mediated transcriptional pattern in the prefrontal cortex is opposite to the down-regulation observed in the nucleus accumbens in patients with major depressive disorder 23614275_Report Fos-B expression in skin keratinocytes/fibroblasts and keloid fibroblasts exposed to genistein. 23810250_Data indicate a significant correlation in miR-181b, FOS and miR-21 expression glioma tissues. 23933656_This study describing the expres-sion pattern of FosB/ FosB immunoreactivity inpost mortem basal ganglia sections from Parkinson disease patients. 24374978_Pseudomyogenic haemangioendothelioma consistently displays a SERPINE1-FOSB fusion gene, resulting from a translocation between chromosomes 7 and 19. 25043949_The abnormalities were screened by FISH in 44 epithelioid hemangioma (EH) from different locations with seven additional EH revealing FOSB gene rearrangements, all except one being fused to ZFP36. 25446562_studies of FosB are providing new insight into the molecular basis of depression and antidepressant action. 26608367_our findings show that DeltaFosB increases the expression of MMP-9 and exhibits a significantly high survival and proliferation in MCF-7 cells. 26949019_These results suggest that SETDB1- mediated FosB expression is a common molecular phenomenon, and might be a novel pathway responsible for the increase in cell proliferation that frequently occurs during anticancer drug therapy. 27248170_FOSB overexpression results in TNBC cell death, whereas inhibition of calcium signaling eliminates FOSB induction and blocks TP4-induced TNBC cell death 27494187_FosB expression in the prefrontal cortex and hippocampus of the cocaine addicted and depression patients 27515856_Diffuse and strong FOSB expression was specific for pseudomyogenic hemangioendothelioma in the current series and FOSB immunohistochemistry is an effective tool for differentiating between PHE and its histological mimics. 27889568_Our results using a number of approaches, such as promoter reporter assay, FosB knock down and Chip assay, suggest that the expression of miR-22 is regulated transcriptionally by FosB. 28009608_FOSB is a highly sensitive and diagnostically useful marker for pseudomyogenic hemangioendothelioma. 28387432_results shows the involvement of Spry2 in regulation of FosB and Runx2 genes, MAPK signaling and proliferation of mesenchymal stem cells. 28653602_miR-144-3p plays an important role in pancreatic cancer cell proliferation, migration, and invasion by targeting FOSB 28724635_The NFATc3 first induced the expression of its interaction partner FosB before forming the heterodimeric NFATc3-FosB transcription factor complex, which bound the proximal AP-1 site in the TF gene promoter and activated TF expression. 28981703_the mechanism underlying redox-regulation of AP-1 Fos/Jun transcription factors and provide structural insight for therapeutic interventions targeting AP-1 proteins. 29527734_FOSB immunohistochemistry is sensitive in the diagnosis of angiolymphoid hyperplasia with eosinophilia, and allows differentiation from its histological mimics 29858576_Findings iindicate a human bone tumour defined by mutations of FOS and FOSB. 30256258_Recurrent ACTB-FOSB fusions are found in pseudomyogenic hemangioendothelioma. 30459475_Recurrent ACTB-FOSB fusion occurs in pseudomyogenic hemangioendothelioma. 31262712_We identified increased level of Fos-B mRNA, the binding target of FUS, in FUS-mutant MNs. While Fos-B reduction using si-RNA or an inhibitor ameliorated the observed aberrant axon branching, Fos-B overexpression resulted in aberrant axon branching even in vivo. The commonality of those phenotypes was further confirmed with other amyotrophic lateral sclerosis (ALS) causative mutation than FUS 31735020_Poly(ADP-ribose) Polymerases (PARP) cleavage and positive annexin V level are increased during piperlongumine (PL) treatment with FosB overexpression in MCF7 breast cancer cells, whereas PARP cleavage and positive annexin V level were decreased during PL treatment with siFosB transfection, implying that FosB is a pro-apoptotic protein for induction of cell death in PL-treated MCF7 breast cancer cells. 31894310_Association of FOSB exon 4 unmethylation with poor prognosis in patients with latestage nonsmall cell lung cancer. 32482537_Clinicopathologic study of 6 cases of epithelioid osteoblastoma of the jaws with immunoexpression analysis of FOS and FOSB. 32819575_MicroRNA-27a-3p directly targets FosB to regulate cell proliferation, apoptosis, and inflammation responses in immunoglobulin a nephropathy. 32891613_FosB recruits KAT5 to potentiate the growth and metastasis of papillary thyroid cancer in a DPP4-dependent manner. 34153682_Knockout of FosB gene changes drug sensitivity and invasion activity via the regulation of Bcl-2, E-cadherin, beta-catenin, and vimentin expression. | ENSMUSG00000003545 | Fosb | 93.167027 | 0.9414393 | -0.087059969 | 0.28096023 | 9.633647e-02 | 7.562711e-01 | No | Yes | 86.903349 | 14.701702 | 94.071908 | 15.781967 | ||
| ENSG00000125772 | 56261 | GPCPD1 | protein_coding | Q9NPB8 | FUNCTION: May be involved in the negative regulation of skeletal muscle differentiation, independently of its glycerophosphocholine phosphodiesterase activity. {ECO:0000250}. | 3D-structure;Cytoplasm;Hydrolase;Phosphoprotein;Reference proteome | hsa:56261; | cytosol [GO:0005829]; glycerophosphocholine phosphodiesterase activity [GO:0047389]; starch binding [GO:2001070]; glycerophospholipid catabolic process [GO:0046475]; skeletal muscle tissue development [GO:0007519] | 22343285_common genetic variants in GPCPD1 are associated with scaling of visual cortical surface area in humans 22570503_We have identified EDI3, a key enzyme controlling GPC and choline metabolism; Because inhibition of it corrects the GPC/PC ratio and decreases the migration capacity of tumor cells, it represents a possible target for therapeutic intervention. 25482527_EDI3 links choline metabolism to integrin expression, cell adhesion and spreading 34626691_Circular RNA circSNX6 promotes sunitinib resistance in renal cell carcinoma through the miR-1184/GPCPD1/ lysophosphatidic acid axis. | ENSMUSG00000027346 | Gpcpd1 | 569.865644 | 0.9898933 | -0.014655065 | 0.11488626 | 1.622255e-02 | 8.986493e-01 | 9.638194e-01 | No | Yes | 625.906897 | 147.843838 | 635.380403 | 150.100290 | ||
| ENSG00000125844 | 6238 | RRBP1 | protein_coding | Q9P2E9 | FUNCTION: Acts as a ribosome receptor and mediates interaction between the ribosome and the endoplasmic reticulum membrane. {ECO:0000250}. | Acetylation;Alternative splicing;Endoplasmic reticulum;Isopeptide bond;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Translocation;Transmembrane;Transmembrane helix;Transport;Ubl conjugation | This gene encodes a ribosome-binding protein of the endoplasmic reticulum (ER) membrane. Studies suggest that this gene plays a role in ER proliferation, secretory pathways and secretory cell differentiation, and mediation of ER-microtubule interactions. Alternative splicing has been observed and protein isoforms are characterized by regions of N-terminal decapeptide and C-terminal heptad repeats. Splicing of the tandem repeats results in variations in ribosome-binding affinity and secretory function. The full-length nature of variants which differ in repeat length has not been determined. Pseudogenes of this gene have been identified on chromosomes 3 and 7, and RRBP1 has been excluded as a candidate gene in the cause of Alagille syndrome, the result of a mutation in a nearby gene on chromosome 20p12. [provided by RefSeq, Apr 2012]. | hsa:6238; | endoplasmic reticulum [GO:0005783]; integral component of endoplasmic reticulum membrane [GO:0030176]; membrane [GO:0016020]; ribosome [GO:0005840]; RNA binding [GO:0003723]; signaling receptor activity [GO:0038023]; osteoblast differentiation [GO:0001649]; protein transport [GO:0015031]; translation [GO:0006412] | 15184079_Results identify the ribosome receptor, p180, as a binding partner of the kinesin heavy chain isoform KIF5B. 17634287_These data suggest that p180 mediates interactions between the endoplasmic reticulum and microtubules mainly through the novel microtubule-binding and -bundling domain MTB-1. 18083570_over-expression of p180R failed to increase secretory pathway proteins calnexin, SEC61beta, and calreticulin, or ribosome biogenesis 19037105_results obtained demonstrate that p180 is both necessary & sufficient to induce a secretory phenotype in mammalian cells; findings support a central role for p180 in the terminal differentiation of secretory cells & tissues 19425502_the discovery overexpression of GPD1 and RRBP1 proteins and lack of expression for HNRNPH1 and SERPINB6 proteins which are new candidate biomarkers of colon cancer. 19932094_A novel function of p180-abundant endoplasmic reticulum on the trans-Golgi network expansion, both of which are highly developed in various professional secretory cells. 20647306_p180 plays crucial roles in enhancing collagen biosynthesis at the entry site of the secretory compartments by a novel mechanism that mainly involves facilitating ribosome association on the ER. 21111237_p180 is concentrated in ER sheets by interacting with polysomes and may play a role in ER morphology. 22679391_p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. 23318453_RRBP1 could alleviate ER stress and help cancer cell survive 24019514_Data indicate that p180 is required for the efficient targeting of placental alkaline phosphatase (ALPP) mRNA to the endoplasmic reticulum (ER). 26196185_High RRBP1 expression facilitates colorectal cancer progression and predicts an unfavourable post-operative prognosis. 30684972_RRBP1 expression was an independent prognostic factor for overall survival (OS) and disease-free survival (DFS) in patients with endometrial carcinoma (EC) (both P < 0.05). 31285390_Expression of RRBP1 in epithelial ovarian cancer and its clinical significance. 32289433_we found that E2F1 could bind to the RRBP1 promoter and promote the transcription of RRBP1, and the E2F1/RRBP1 axis played an important role in the process of high glucose promoting the proliferation, migration and invasion of HepG2 cells (hepatocellular carcinoma) 32536673_Correlation between reticulum ribosome-binding protein 1 (RRBP1) overexpression and prognosis in cervical squamous cell carcinoma. 33762722_RRBP1 rewires cisplatin resistance in oral squamous cell carcinoma by regulating Hippo pathway. 34445467_Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma. | ENSMUSG00000027422 | Rrbp1 | 5532.507980 | 0.9418084 | -0.086494436 | 0.05817920 | 2.218358e+00 | 1.363780e-01 | 4.872215e-01 | No | Yes | 4988.817329 | 396.864602 | 5289.748258 | 420.427246 | |
| ENSG00000125968 | 3397 | ID1 | protein_coding | P41134 | FUNCTION: Transcriptional regulator (lacking a basic DNA binding domain) which negatively regulates the basic helix-loop-helix (bHLH) transcription factors by forming heterodimers and inhibiting their DNA binding and transcriptional activity. Implicated in regulating a variety of cellular processes, including cellular growth, senescence, differentiation, apoptosis, angiogenesis, and neoplastic transformation. Inhibits skeletal muscle and cardiac myocyte differentiation. Regulates the circadian clock by repressing the transcriptional activator activity of the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). {ECO:0000250}. | Alternative splicing;Biological rhythms;Cytoplasm;Developmental protein;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation | The protein encoded by this gene is a helix-loop-helix (HLH) protein that can form heterodimers with members of the basic HLH family of transcription factors. The encoded protein has no DNA binding activity and therefore can inhibit the DNA binding and transcriptional activation ability of basic HLH proteins with which it interacts. This protein may play a role in cell growth, senescence, and differentiation. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:3397; | centrosome [GO:0005813]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; proteasome binding [GO:0070628]; protein C-terminus binding [GO:0008022]; protein dimerization activity [GO:0046983]; protein N-terminus binding [GO:0047485]; transcription regulator inhibitor activity [GO:0140416]; angiogenesis [GO:0001525]; blood vessel endothelial cell migration [GO:0043534]; blood vessel morphogenesis [GO:0048514]; brain development [GO:0007420]; cell differentiation [GO:0030154]; cell-abiotic substrate adhesion [GO:0036164]; cellular response to dopamine [GO:1903351]; cellular response to epidermal growth factor stimulus [GO:0071364]; cellular response to nerve growth factor stimulus [GO:1990090]; cellular response to peptide [GO:1901653]; circadian regulation of gene expression [GO:0032922]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of dendrite morphogenesis [GO:0050774]; negative regulation of DNA binding [GO:0043392]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of endothelial cell differentiation [GO:0045602]; negative regulation of protein binding [GO:0032091]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription by transcription factor localization [GO:0010621]; negative regulation of transcription, DNA-templated [GO:0045892]; neuron differentiation [GO:0030182]; positive regulation of actin filament bundle assembly [GO:0032233]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of gene expression [GO:0010628]; regulation of vasculature development [GO:1901342]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 11729207_Id1, a dominant negative inhibitor of basic helix-loop-helix proteins, is a direct target gene for BMP 11896613_Id-1 expression is linked with the loss of NF-1/Rb/HDAC-1 transcription repressor complex in metastatic breast cancer 12016143_role in stimulating serum-independent prostate cancer cell proliferation through inactivation of p16(INK4a)/pRB pathway 12020803_TGF beta 1 may be one of the upstream regulators of Id-1 12203366_immunohistochemical studies on NPC samples showed that expression of Id-1 was present in NPC cells but absent in normal tissues. This study demonstrates that Id-1 plays an important role in cell proliferation in NPC cells 12296825_Data suggest that signalling of BMP-2 to stimulate the expression of Id1 would be transduced by BMPR-IA and mediated by Smad1 and Smad4. 12576450_Level of protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumors. 12719589_expression in human endothelial cells induced by Kaposi sarcoma-associated herpesvirus LANA protein, expression in Kaposi sarcoma tumor cells and in vivo 12787042_expression may relate to the metastatic behaviour in human oral squamous cell carcinoma 12823438_dysregulated Id-1 may not only contribute to delaying the senescence program in keratinocytes, it may also contribute to the escape of the relatively undifferentiated tumor cells in BCC from immune surveillance. 12881706_Our results strongly suggest that Id-1 may be one of the upstream regulators of NF-kappaB and activation of NF-kappaB signalling pathway may be essential for Id-1 induced cell proliferation through protection against apoptosis. 12947323_Significantly overexpressed in papillary thyroid cancer. Primarily localized to cytoplasm of thyroid follicular cells. Activation of the mitogen intracellular protein kinase A and protein kinase C signaling pathways up-regulated Id-1 mRNA expression. 12949053_over-expression of Id-1 induces cell proliferation in HCC through inactivation of p16INK4a/retinoblastoma pathway 14688027_Expression of Id-1 was able to reduce androgen-stimulated growth and S phase fraction of the cell cycle in prostatic cancer cells. 14690332_Id1 protein is involved in the process of differentiation of keratinocytes seeen in normal skin and Id1 pathway is activated in psoriasis. 14742319_Id-1 may be an upstream regulator of the Raf/MEK signalling pathway, which plays an essential role in protection against taxol-induced apoptosis 14755252_ID1 may contribute to oncogenesis not only by inhibiting transcriptional activity of basic helix-loop-helix transcription factors and abrogate differentiation but also by subverting centrosome duplication. 15026801_ID-1 expression significantly associated with intratumoral microvessel density in pancreatic cancer 15041724_Id-1 and Id-2 proteins control prostate cancer cell phenotypes and could serve as molecular markers of aggressive human prostate cancer. 15064751_Id1 induction by LMP1 depends on its NF-kappaB activation domain at the COOH-terminal region, CTAR1 and CTAR2. This may facilitate clonal expansion of premalignant nasopharyngeal epithelial cells infected with EBV & promote malignant transformation. 15138269_silencing Id1 expression in young cells by RNA interference induced an increased p16(INK4a) level and premature cellular senescence 15163661_MyoD modulates the rate of Id1 degradation and suggest a dynamic interplay of these factors 15322112_induction of Id1 not only blocks transcriptional activity but also induces myogenin degradation by blocking formation of myogenin-E47 protein complexes. 15370294_Id1 can be up-regulated and p53 down-regulated by a statin in endothelial cells. Regulation of these proteins in endothelial cells may account for the proangiogenic effect of statins. 15489884_Id1 has cell cycle regulatory functions that are similar to those of c-Myc. 15494533_Overexpression of Id1 enhances expression of ICAM-1 and E-selectin, and induces angiogenic processes such as transmigration, matrix metalloproteinase-2 and -9 expression, and tube formation in cultured vascular endothelial cells. 15575081_Idl protein may play an important role in the process of gastric carcinogenesis, and high-level Id I expression may be related to the malignant potential of tumor cells 15579766_Id1 is overexpressed in hyperplastic and neoplastic thyroid tissue and directly regulates the growth of thyroid cancer cells of follicular cell origin, but is not a marker of aggressive phenotype in differentiated thyroid cancer. 15645115_Id1, 2 and 3 might play a role in the early stages of hepatocarcinogenesis, but not in the development of advanced carcinoma, and might consequently be related to HCC dedifferentiation 15694377_Thus, we speculate that the regulation of cell growth mediated by CASK may be involved in Id1. 15701714_constitutive expression of Id1 inhibits eosinophil development, whereas in contrast neutrophil differentiation was modestly enhanced 15744343_These results indicate ID1 and ID2 are important retinoic acid responsive genes in acute promyelocytic leukemia [APL], and suggest that inhibition of specific bHLH transcription factor complexes may play a role in the therapeutic effect of ATRA in APL 15877825_In mature human B cells, BMP-6 inhibited cell growth, and rapidly induced an upregulation of Id1. 15905202_Downregulation of Id-1 may be a novel target to inhibit the growth of metastatic cancers through the suppression of angiogenesis. 16002046_Furthermore, by using DCs derived from Id1(-/-) mice that are defective in Flt-1 signaling, we demonstrated that the inhibitory function of VEGF on DC function is most likely mediated by Flt-1. 16007194_degradation is modulated by E12 and E47 16029118_Id1 gene expression may be associated with thyroid cancer cell growth and differentiation. 16115231_Expression of Id1 in HM was significantly higher than that in normal placenta. 16123120_Id-1 protein may be regulated by TNFalpha through the ubiquitin/proteasome degradation pathway and the stability of the Id-1 protein appears to correlate with the sensitivity of TNFalpha-induced apoptosis 16189525_study supports a significant role of the ID1 protein in melanoma progression and patient prognosis; inverse relation between ID1 and TSP-1 expression support an important role of ID1 in regulation of this complex multitarget protein. 16271072_Id1, 3 double-knockdown significantly impaired the ability of gastric cancer cells to form peritoneal metastasis 16287090_results indicate that increased Id-1 expression in prostate cancer cells may play a protective role against apoptosis, and downregulation of Id-1 may be a potential target to increase sensitivity of taxol-induced apoptosis in prostate cancer cells 16469432_Id-1 overexpression is associated with breast tumor angiogenesis 16506209_Study provides evidence for the first time that Id-1 plays a role in both proliferation and survival of esophageal cancer cells. 16525633_Up-regulation of Id-1 was associated with increased EGFR expression, clinical staging and the invasion ability of bladder cancer cells, and inactivation of Id-1 may be a potential therapeutic target to inhibit the invasion by bladder cancer cells. 16552541_The IGFBP3, hRas, JunB, Egr-1, Id1 and MIDA1 genes were up-regulated in psoriatic involved skin compared with uninvolved skin. 16682435_ID proteins (ID1, ID2, ID3 and ID4) were significantly increased in Mecp2-deficient Rett syndrome brain, ID genes are ideal targets for MeCP2 regulation of neuronal maturation that may explain the molecular pathogenesis of Rett syndrome 16894355_Interferon-alpha downregulates ID1 ID in cultured human vascular endothelial cells 17072841_Expression of ID1 decreased significantly in chondrocytes while the opposite was seen in mesenchymal stem cells. 17102133_expression of ATF-3 in hypoxic cells represses Id-1 and prevents their loss 17145808_Id-1 is a novel angiogenic factor for HCC metastasis and down-regulation of Id-1 may be a novel target to inhibit HCC metastasis through suppression of angiogenesis. 17202424_Id1 transcriptional inhibitor is a crucial player in mediating cell dedifferentiation of renal tubular epithelium and suggest that epithelial-to-mesenchymal transition is a multistep process. 17426247_In conclusion, we provide the molecular mechanism of the cross talk between HIF-1alpha and Id-1, which may play a critical role in tumor angiogenesis. 17537403_PLZF upregulates apoptosis-inducer TP53INP1, ID1, and ID3 genes, and downregulates the apoptosis-inhibitor TERT gene 17565736_Id1 expression is primarily regulated at the transcriptional level in radial growth phase melanomas and expect that therapies that target Id1 gene expression may be useful in the treatment of Id-associated malignancies. 17599389_significantly enhanced expression of Id-1 has been detected in malignant prostate cell lines and in primary carcinomas; the increase in Id-1 expression was significantly associated with the increasing Gleason score of carcinomas 17855368_caveolin-1 is a novel Id-1 binding partner that mediates the function of Id-1 in promoting prostate cancer progression through activation of the Akt pathway leading to cancer cell invasion and resistance to anticancer drug-induced apoptosis 17891176_Dysregulates centrosome homeostasis at least in part by interfering with S5A/Rpn10 activities at the centrosome. 17916352_a novel tumorigenic role of Id-1 through reorganization of actin cytoskeleton and disassembly of cell-cell adhesion in response to TGF-beta1 in human prostate epithelial cells 18000500_The Id-1 may promote distant metastasis in oesophageal squamous cell carcinoma(ESCC), and both Id-1 may be used for prognostication for ESCC patients. 18072288_These data demonstrate that BMP-6 promotes migration and invasion of prostate cancer cells, potentially through activation of Id-1 and MMP activation. 18158619_Id-1 regulates Bcl-2 and Bax expression through p53 and NF-kappaB in MCF-7 breast cancer cells 18202790_Id-1 overexpression plays an important role in colorectal cancer progression. 18240291_Id1 may contribute to the malignant conversion of primary human melanocytes through extension of cellular lifespan. 18353975_The association of Id1 and Cdh1 is dependent on the canonical destruction box motif of Id1, the increased binding of which may compete with the interaction between Cdh1 and Aurora A, leading to stabilization of Aurora A in Id1-overexpressing cells. 18372912_a novel function of Id-1 in promoting chromosomal instability through modification of APC/C activity during mitosis and provide a novel molecular mechanism accounted for the function of Id-1 as an oncogene. 18413773_Id1 may contribute to early breast cancer by promoting excessive proliferation through cyclin D1. 18436795_The integration of signals at the level of Id gene expression may contribute to the pathogenesis of familial pulmonary arterial hypertension. 18542061_Id1 immortalizes hematopoietic progenitors in vitro and promotes a myeloproliferative disease in vivo. 18556654_ID1 is an effector of the p53-dependent DNA damage response pathway. 18559972_Id1 is an important target of constitutively activated tyrosine kinases and may be a therapeutic target for leukemias associated with oncogenic tyrosine kinases. 18583319_Id1 inhibited DNA binding by Dril1, and the two proteins co-localized in vitro and in vivo, providing a potential mechanism for suppression of fibrosis by Id1 through inhibition of the profibrotic function of Dril1. 18648363_study reports that the expression of the E6 onco-protein of the high-risk human papillomaviruses is correlated with Id-1 overexpression in the majority of invasive breast cancer tissue samples studied 18674781_These results demonstrated a novel function of Id-1 in regulating HBX protein stability through interaction with the proteasome. 18752043_Inhibitor of DNA binding-1 overexpression is associated with prostate cancer. 18922905_both ATF3 and Smad were crucially and synergistically involved in down-regulation of Id-1, which regulated JNK phosphorylation in REIC/Dkk-3-induced apoptosis. 19002177_In uterine cervical cancer, ID-1 might work on tumour advancement through angiogenic activity. ID-1 expression correlated with microvessel counts and with histoscore. 19014499_Id-1, took part in development and progression of colorectal carcinomas and that Id-1 was associated with regulations of EGFR and VEGF. 19031736_The expression of ID1 gene was up-regulated in ATRA-induced NB4 cells and APL cells from two patients and was independent of other protein synthesis. 19079342_Id-1 is a novel PTEN inhibitor that could activate the Akt pathway and its downstream effectors, the Wnt/TCF pathway and p27(Kip1) phosphorylation 19079362_Id1 can be strongly up-regulated by Transforming Growth Factor beta1 in the human mammary gland epithelial cell line MCF10A. 19201527_Id-1 expression in androgen-dependent prostate cancer was negatively regulated by androgen in a receptor-dependent way. 19217708_ERbeta1 inhibits breast cancer cell growth through binding with Id1 and upregulating p21 gene expression. 19288008_up-regulation of Id-1 in bladder cancer cells lead to increased cell viability in response to epirubicin by its improved anti-apoptotic role. 19330020_Targeting Id dimerization may therefore be effective for triggering differentiation and restraining neuroblastoma cell tumorigenicity. 19423615_Neuroblastoma cell adaptation to hypoxia involves the modulation of HIF1alpha and VEGF expression, nuclear translocation of ID1 and ID2 transcription factors, and activation of a gene expression program consistent with pro-metastatic events. 19438642_These data suggest that high-risk human papillomaviruses can enhance the progression of human cervical cancer through Id-1 regulation. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19491902_High ID1 expression is associated with lymph node metastasis and peritoneal dissemination in gastric cancer. 19515385_role in the development of peritoneal metastasis of pancreatic cancer 19551863_Id-1 overexpression is associated with esophageal cancer. 19567783_Examined regulatory mechanisms of p16 expression. Exogenously expressed Id1 suppressed up-regulation of p16 by Hepatocyte growth factor and the antiproliferative effect of HGF. Knockdown of Id1 sig. enhanced activity of the p16 promoter with ERK. 19571317_ID1 promotes expansion and survival of primary erythroid cells and is a target of JAK2V617F-STAT5 signaling 19643984_High Id1 expression is associated with acute myeloid leukemia. 19644139_data suggest that targeting the ID1 and C/EBPalpha transcriptional regulators may be of benefit in the design of novel therapies for low-risk myelodysplasia. 19710505_These results implicate Id1 and TSP1 as downstream mediators of Epac/Rap1, inhibitory regulators of the angiogenic process 19766362_may have important roles in the development of salivary adenoid cystic carcinoma 19843640_Abl interactor 1 regulates Src-Id1-matrix metalloproteinase 9 axis and is required for invadopodia formation, extracellular matrix degradation and tumor growth of human breast cancer cells 19906510_Positive Id-1 expression is associated with high malignancy/poor prognosis. Protein expression status may help assess tumor malignancy and patient prognosis. 19926394_Anti-proliferative and chemosensitization effect of gamma-tocotrienol on breast cancer cells cells may be mediated through downregulation of Id1 protein. 19951400_KLF17 has a role in transcriptional regulation of metastasis by Id1 [review] 20003244_ID-1 increased in ovarian cancer cells during tumor progression 20010941_In prostate cancer cells, Id-1 has the ability to modulate bone cell differentiation favouring metastatic bone disease. 20070914_shows that not only ectopic expression in tissue culture but endogenous levels of ID1 modulate centrosome numbers. 20080245_The level of Id-1 protein expression was associated with the malignant potential of gastric tumors 20186495_Id1 promotes cancer/tumor morphology, cell cycle and epithelial to mesenchymal transition by influencing AP1, tnf, tgfbeta, PdgfBB and estradiol pathways. 20191379_Id1/3 peptide aptamer could represent a nontoxic exogenous agent that can significantly provoke antiproliferative and apoptotic effects in breast cancer cells, which are associated with deregulated expression of Id1 and Id3. 20388787_ID1 enhances docetaxel cytotoxicity in prostate cancer cells through inhibition of p21 20484992_Id1 activation may need to occur at discrete stages in cooperation with additional gene dysregulation to repress and induce epigenetic silencing of tumor suppressor genes during melanoma progression. 20515335_Supratentorial PNETs expressed significantly higher levels of SOX2, NOTCH1, ID1, and ASCL-1 transcripts. 20522807_Prostacyclin analogues inhibited smooth muscle cell proliferation and prevented progression of pulmonary hypertension while enhancing Smad1/5 phosphorylation and Id1 gene expression. 20574154_Id-1 expression is at least partially regulated by HBx and may serve as a potential prognostic marker for HBV-related HCC. 20697353_Id1 may contribute to tumor development through Polycomb group -mediated epigenetic regulation. 20709421_ID1 was expressed in the nucleus in 70% of squamous cell carcinomas and 50% of non-squamous cell carcinomas and in vascular endothelium of non-tumor tissue 20863724_in human liver, not only HAMP, but also SMAD7 and Id1 mRNA significantly correlate with the extent of hepatic iron burden. However, this correlation is lost in patients with HFE-HC, but maintained in subjects with non-hemochromatotic iron overload. 20881502_Silencing of the Id1 gene has an in-vivo preventive effect against the development of prostate cancer in a mouse model. 20887552_Id1 contributed to hyperproliferation of keratinocytes via enhancement of cell cycle progression, removal of cell cycle inhibition, and simultaneously increased keratin production. 21106423_Id-1 proteins were overexpressed in human mammary invasive ductal carcinoma and were increased from benign to premalignant and malignant lesions. 21106425_significant role of Id-1 protein in the gastric tumorigenesis 21467165_MUC18 is involved in cell signaling regulating the expression of Id-1 and ATF-3, thus contributing to melanoma metastasis 21486943_Study uniquely identifies ID1 and ID3 as negative regulators of the hPSC-hematopoietic transition from a hemogenic to a committed hematopoietic fate, and demonstrates that this is conserved between hESCs and hiPSCs. 21506108_Induction of Id-1 by FGF-2 involves activity of EGR-1 and sensitizes neuroblastoma cells to cell death. 21536374_Nude mice study further confirmed an increased tumor growth in Id1-overexpressing cells and a decreased tumor growth in Id1-knockdowned cells. In conclusion, inactivation of Id1 may provide a novel strategy for treatment of lung cancer patients 21606196_Data show that ID1 is induced by nicotinic acetylcholine receptor (nAChR) and epidermal growth factor receptor (EGFR) signaling in a panel of NSCLC cell lines and primary cells from the lung. 21630092_These results demonstrate that Id1-induced beta1-integrin expression in endothelial cells and the function of Id1 in cell migration and tubulogenesis are dependent on p53. 21670698_Inhibitor of differentiation 1 expression is correlated to human papillomavirus infection in cervical carcinoma. 21688151_ID-1 is implicated in the pathogenesis and progression of gastric cancer. 21701587_Study revealed that Id1 interacts with LMP1 by binding to the CTAR1 domain of LMP1. N-terminal region of Id1 is required for the interaction with LMP1. 21732357_Results suggest that IL-3 plays an inhibitory role in osteoclast differentiation by regulating c-Fos and Id1, and also exerts anti-bone erosion effects. 21896993_Id1 could be considered as a prognostic predictor for stage III esophageal squamous cell carcinoma patients. 21921026_Bone morphogenetic proteins and ID1 exerted an anti-apoptotic effect in mesangial cells by inhibition of USF2 transcriptional activity. 21921784_Malignant rhabdoid tumors express many stem cell-associated transcription factors, which may be regulated by the expression of EZH2 and the Id family of proteins. 21955753_These results indicate that increased migration of MDA-MB-231 cells following cyclin D1 silencing can be mediated by Id1 and is linked to an increase in EMT markers. 21993163_Older muscle contained significantly more transcript for Forkhead Box O 1 (FoxO1, p=0.001), Inhibitor of DNA binding 1 (ID1, p=0.009), and Inhibitor of DNA Binding 3 (ID3, p=0.043) than young muscle. 22076920_Silencing of Id-1 protein inhibited the growth of A375 cells. 22139627_Acheron regulates vascular endothelial proliferation and angiogenesis together with Id1 during wound healing. 22226665_Results suggest that inhibitor of differentiation or DNA binding -1 (Id-1) may play roles in tumor progression and epithelial-to-mesenchymal transition (EMT) activation in bladder cancer. 22278018_Id1 shuttles between nucleus and cytoplasm, and promotes peritubular inflammation and tubular epithelial dedifferentiation, suggesting that these two events are intrinsically coupled during renal fibrogenesis. 22301282_combined analysis of Id-1 and NF-kappaB/p65 can be useful for identifying patients at risk for unfavorable clinical outcomes. Id-1 or/and NF-kappaB/p65 enhanced tumor cell migration, which is associated with the secretion of MMP-9 22393458_Notch activation provides Id1-expressing tumor cells with selective advantages in growth and survival 22480390_High level of ID-1 expression suggested poor prognosis for patients with esophageal squamous cell carcinoma. 22580608_using Bcl-3 knockdown in prostate cancer cells, identified the inhibitor of DNA-binding (Id) family of helix-loop-helix proteins as potential Bcl-3-regulated genes 22592405_It was shown that the ID1-p65 interaction modulates activation of the NF-kappa B signaling pathway. By affecting p65 nuclear translocation, ID1 was essential in regulating p65 recruitment to the promoter of cellular inhibitor of apoptosis protein 2. 22684559_These results indicate that Id1 may down-regulate the ability of PC3 cells to form osteolytic lesions in vivo and the signal pathway needs to be further investigated. 22698403_Data show that regulation of p21 by ID1 and ID3 is a central mechanism preventing the accumulation of excess DNA damage and subsequent functional exhaustion of cancer-initiating cells (C-ICs). 22781717_High expression level of ID1 gene was mostly seen in acute myeloid leukemia patients with adverse cytogenetics and older age (age >/= 60 years), and might be associated with poor prognosis. 22819717_shRNA knockdown of ID1 attenuates androgen-stimulated hepatocellular carcinoma cell migration and invasion. 23060149_These findings suggest an essential role of Id1 and Id3 in TGFbeta1 effects on proliferation and migration in prostate cancer cells. 23243024_Our results suggest that Id-1 regulates multiple tumor-promoting pathways in glioblastoma 23308043_Results suggests a definite role for Id1 in mediating nicotine-induced proliferation and invasion of pancreatic cancer cells. 23311395_Id1 and Id3 co-expression seems associated with a poor clinical outcome in patients with locally advanced NSCLC treated with definitive chemoradiotherapy 23342268_increased Id1 and Id3 expression attenuates all three cyclin-dependent kinase inhibitors (CDKN2B, -1A, and -1B) resulting in a more aggressive PCa phenotype. 23377983_these results suggest that ID1 may be a new prognostic marker for Glioblastoma multiforme 23555842_E47 interacts with Id1 in E47 overexpressing MDCK cells that underwent a full epithelial-mesenchymal transition as well as in mesenchymal breast carcinoma and melanoma cell lines 23558671_inhibitor of differentiation-1 is expressed during fetal embryogenesis and in different mouse and human cancer types 23645773_Higher ID1 expression was associated with advanced breast cancer. 23714001_Id1 can enhance EPC angiogenesis in ovarian cancer, which is mainly mediated by the PI3K/Akt and NF-kappaB/MMP-2 signaling pathways 23723304_This study revealed that there was a positive correlation between Id-1 expression and the expression of p-Akt, p-GSK3beta and p-HSF1 in oral squamous cell carcinoma. 23771884_Id proteins, and particularly Id1 and Id3, are critical downstream effectors of BMP signaling in pulmonary artery smooth muscle cell. 23872946_Ectopic expression of ID proteins partially rescues the senescence-like phenotype induced by loss of DPY30. 23900621_EGCG induces apoptosis and inhibits proliferation of poorly differentiated AGS gastric cancer cells, and Id1 may be one of the target genes regulated by EGCG in cancer inhibition. 24129125_Higher Id1 mRNA expression levels might predict a higher hazard ratio for progression and a shorter disease-free survival in prostate cancer. 24133588_Id-1 is considered to be a candidate for new therapeutic target and a prognostic factor in non-small cell lung cancer. 24295493_The results demonstrate that Id-1B decreases the malignancy of lung and prostate cancer cells and counteracts the protumorigenic role of the classical form of Id-1. 24332369_Id1 induces mesenchymal-to-epithelial transition and pulmonary metastatic colonization by antagonizing Twist1 activity. 24378760_ID1 is a synthetic sick/lethal gene that interacts with the R175H TP53 mutant. 24403496_High ID-1 expression is associated with high microvessel density in ovarian cancer. 24434151_Investigated the effect of Id1 on TGF-beta-induced collagen expression in dermal fibroblasts. When Id1-b isoform was overexpressed, TGF-beta-induced collagen expression was markedly inhibited. 24480377_ID1 expression in pancreatic adenocarcinoma was associated with angiogenesis as measured by microvessel density. 24572994_Results show that Id1 and NF-kappaB regulate the expression of CD133 and BMI-1 in an additive or synergistic manner in oral squamous cell carcinoma. 24599583_Inhibitor of differentiation 1 is a prognostic marker for multicentric Castleman's disease. 24599933_Data indicate that the inhibitor of DNA binding 1 (Id1)-IGF-II-IGF-IR-AKT signaling cascade plays an important role in esophageal cancer progression. 24620998_We conclude that Id1 is potently angiogenic and can be up-regulated in endothelial progenitor cells 24628854_Id-1 cooperates with HPV E6E7 proteins, which involves inactivation of p53 and pRb in host cells, to promote cervical carcinogenesis. 24662327_Id-1, a protein repressed by miR-29b, facilitates TGFbeta1-induced EMT in human ovarian cancer cells and represents a promising therapeutic target for treating ovarian cancer. 24695670_Id1 as an important modulator of molecular events during DPSC commitment and differentiation, which should be considered in dental research on tissue engineering. 24804700_Studies demostrated the regulatory role of ID1 in the proliferation and metastasis of colorectal cancer. 24861919_Id-1 and TGF-beta1 played important roles in the progression of gastric cancer, in which Id-1 might act as a downstream mediator of TGF-beta1 signaling through a regulatory mechanism involving N-cadherin and beta-catenin. 24948111_Our results suggest that Id1 promotes breast cancer metastasis by the suppression of S100A9 expression 24970809_Data indicate that grap2 and cyclin D1 interacting protein (GCIP) and inhibitor of of DNA binding/differentiation 1 (Id1) are inversely expressed in non-small cell lung cancer (NSCLC) cell lines and specimens. 25010525_Smad1 as a novel binding protein of KSHV latency-associated nuclear antigen (LANA). LANA interacted with and sustained BMP-activated p-Smad1 in the nucleus and enhanced its loading on the Id promoters. 25028095_Overexpression of ID1 in two different cell lines induced STMN3 and GSPT1 at the transcriptional level, while depletion of ID1 reduced their expression. 25031707_This study elucidated the potential mechanism underlying Id1 participation in the progression of prostate cancer 25323535_LIF has a role in negatively regulating tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers 25344919_NSCLC cells with high Id1 protein expression were vulnerable to the treatment of paclitaxel and cisplatin 25449776_High Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. 25496992_berberine's anti-proliferative and anti-invasive activities could be partially rescued by Id-1 overexpression in HCC models, revealing a novel anti-cancer/anti-invasive mechanism of berberine via Id-1 suppression. 25514034_ERbeta1 inhibits the migration and invasion of breast cancer cells and upregulated E-cadherin expression in a Id1-dependent manner. 25549282_Downregulation of ID1 by gene silencing can lead to acceleration of TGF-beta1-induced hESC differentiation into ECs and inhibition of proliferation and migration of ECs. 25623217_Overexpression of ID1 is associated with colorectal neoplasia. 25778840_Inhibitor of differentiation 4 (ID4) acts as an inhibitor of ID-1, -2 and -3 and promotes basic helix loop helix (bHLH) E47 DNA binding and transcriptional activity. 25924227_Advanced melanoma patients, relative to healthy controls, express much higher levels of ID1 in myeloid peripheral blood cells. 25938540_these findings provide in vivo genetic evidence of Id1 functions as an oncogene in breast cancer 26072160_Four more genes (BMP4, BMPR1B, SMAD1 and SMAD4) of the ID1 pathway were investigated and only one (BMPR1B) shows the same down regulation 26191235_ID1 overexpression may be associated with higher risk karyotype classification and act as an independent risk factor in young non-M3 acute myeloid leukemia patients. 26475334_Our findings define an intricate E2F1-dependent mechanism by which Id1 increases thymidylate synthase and IGF2 expressions to promote cancer chemoresistance. The Id1-E2F1-IGF2 regulatory axis has important implications for cancer prognosis and treatment. 26577912_Peritoneal VEGF-A expression is regulated by TGF-beta1 through an ID1 pathway in women with endometriosis 26797271_Therefore, we identified a tumorigenic role of Id-1 in OS and suggested a potential therapeutic target for OS patients. 26858249_Simultaneous high expression of ID1 and c-Jun or c-Fos was correlated with poor survival in esophageal squamous cell carcinoma patients. 27044543_Results show that ID1, ID3 and IGJ genes are highly expressed in adult B-ALL and correlate with poor prognosis in Hispanic patients. 27087608_Reducing ID1 gene expression reduces metastatic spread of salivary gland neoplasms. 27466488_ID1 down-regulation induced parallel changes in the IGF and AKT pathways. The crosstalk of these pathways may enhance malignant phenotypes in salivary gland cancer. 27477274_a loss of CULLIN3 represents a common signaling node for controlling the activity of intracellular WNT and SHH signaling pathways mediated by ID1 27546618_High ID1 expression is associated with breast cancer. 27610466_MVD was determined by immunohistochemistry, and the expressions of mRNA and protein of inhibitor of differentiation-1 (ID1) and vascular endothelial growth factor (VEGF) were detected in gastric cancer. 27633352_High ID-1 expression is associated with colorectal carcinoma. 27978873_ID1 expression was positively related to drug resistance of EGFR-TKI in non-small cell lung cancer. 28510612_ID1 expression impacts the sensitivity of colon cancer cells to 5-FU and may be considered as a potential predictive marker in colorectal carcinoma treatment. 28549790_Id1 enables lung cancer liver colonization by activating an epithelial mesenchymal transformation program in tumor cells and establishing the pre-metastatic niche. 29039489_ID1 expression was associated with the proliferation, invasion and migration of SACC cells. The observed inhibition of SACC cell growth, invasion and migration following knockdown of ID1 expression in the present study, may have been due to restoration of the balance between oncogenic and tumor-suppressive effects resulting from changes in the expression of downstream genes or associated proteins. 29079782_CaMKII can directly phosphorylate Beclin 1 at Ser90 to promote K63-linked ubiquitination of Beclin 1 and activation of autophagy; it also promotes K63-linked ubiquitination of inhibitor of differentiation 1/2 (Id-1/2) by catalyzing phosphorylation of Id proteins and recruiting TRAF-6 29159 | ENSMUSG00000042745 | Id1 | 389.390575 | 1.0737064 | 0.102599524 | 0.14435481 | 5.033397e-01 | 4.780363e-01 | No | Yes | 405.607224 | 52.707175 | 377.334971 | 49.115064 | ||
| ENSG00000126216 | 10426 | TUBGCP3 | protein_coding | Q96CW5 | FUNCTION: Gamma-tubulin complex is necessary for microtubule nucleation at the centrosome. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Cytoskeleton;Microtubule;Phosphoprotein;Reference proteome | hsa:10426; | centriole [GO:0005814]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; equatorial microtubule organizing center [GO:0000923]; gamma-tubulin complex [GO:0000930]; gamma-tubulin small complex [GO:0008275]; membrane [GO:0016020]; polar microtubule [GO:0005827]; spindle [GO:0005819]; gamma-tubulin binding [GO:0043015]; structural constituent of cytoskeleton [GO:0005200]; structural molecule activity [GO:0005198]; cytoplasmic microtubule organization [GO:0031122]; meiotic cell cycle [GO:0051321]; microtubule nucleation [GO:0007020]; mitotic cell cycle [GO:0000278]; single fertilization [GO:0007338]; spindle assembly [GO:0051225] | 19299467_Stability of the small gamma-tubulin complex (gamma tubulin/GCP2/GCP3) requires HCA66, a protein of the centrosome and the nucleolus. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20858683_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 23886939_Authors show genetically that GCP3/Spc98 function is fully conserved with Alp6 across species but that functional differences exist between GCP2/Spc97 and Alp4. 26079448_This study demonstrated that Overexpression and Nucleolar Localization gcp3 in glioblastoma. 28851027_NMR secondary structure and interactions of recombinant human MOZART1 protein with its binding partner, GCP3, has been presented. | ENSMUSG00000000759 | Tubgcp3 | 942.963935 | 0.9851858 | -0.021532217 | 0.09104939 | 5.586576e-02 | 8.131538e-01 | 9.383525e-01 | No | Yes | 920.199953 | 99.695745 | 929.384374 | 100.687401 | ||
| ENSG00000126249 | 84306 | PDCD2L | protein_coding | Q9BRP1 | FUNCTION: Over-expression suppresses AP1, CREB, NFAT, and NF-kB transcriptional activation, and delays cell cycle progression at S phase. {ECO:0000269|PubMed:17393540}. | Acetylation;Cell cycle;Phosphoprotein;Reference proteome | hsa:84306; | cytoplasm [GO:0005737]; membrane [GO:0016020]; cell cycle [GO:0007049] | 16311922_To study the role of PDCD2_C domain in apoptosis, the cDNAs of two isoforms of PDCD2 and MGC13096 were cloned. PDCD2 (NM_002598) was over expressed when endothelial cells treated with leukotriene D4 or natural killer cells were activated by IL-2. 17393540_MGC13096 over-expression restrained proliferation of HEK293T cells. DNA/flow cytometry analysis showed that the over-expression of MGC13096 severely delays cell cycle progression at S phase. 18486760_Overexpression of PDCD2-like gene attenutates TNF-alpha release in Daudi cells 27697862_Findings uncover the existence of an extra-ribosomal complex consisting of PDCD2L, RPS2, and PRMT3 and support a role for PDCD2L in the late maturation of 40S ribosomal subunits. | ENSMUSG00000002635 | Pdcd2l | 633.493601 | 0.9588119 | -0.060680336 | 0.11092720 | 2.993698e-01 | 5.842778e-01 | 8.466668e-01 | No | Yes | 601.501263 | 49.309260 | 629.571189 | 51.396118 | ||
| ENSG00000126705 | 27245 | AHDC1 | protein_coding | Q5TGY3 | Acetylation;DNA-binding;Isopeptide bond;Mental retardation;Methylation;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | This gene encodes a protein containing two AT-hooks, which likely function in DNA binding. Mutations in this gene were found in individuals with Xia-Gibbs syndrome. [provided by RefSeq, Jun 2014]. | hsa:27245; | DNA binding [GO:0003677] | 24791903_this study hasidentified AHDC1 de novo truncating mutations that most likely cause syndromic expressive language delay, hypotonia, and sleep apnea. 30615951_Microdeletion and microduplication of 1p36.11p35.3 involving AHDC1 contribute to neurodevelopmental disorder. 30729726_De novo heterozygous variants in AHDC1 gene were identified in two patients with partial growth hormone deficiency. 31390932_we unveiled that LINC01133 may function as a ceRNA for miR-4784 to advance AHDC1 expression, intensifying CC cell malignant phenotypes and EMT process, which may demonstrate the implied value of LINC01133 as a therapeutic target for CC patients. 31737670_Three rare mutations of AHDC1 in patients with OSA in Chinese Hanindividuals. 33644933_Phenotypic and protein localization heterogeneity associated with AHDC1 pathogenic protein-truncating alleles in Xia-Gibbs syndrome. 34229113_Phenotypic heterogeneity and mosaicism in Xia-Gibbs syndrome: Five Danish patients with novel variants in AHDC1. | ENSMUSG00000037692 | Ahdc1 | 477.435692 | 1.0174388 | 0.024942001 | 0.13731202 | 3.300011e-02 | 8.558501e-01 | 9.523387e-01 | No | Yes | 589.412506 | 85.282389 | 563.375937 | 81.347077 | ||
| ENSG00000127824 | 7277 | TUBA4A | protein_coding | P68366 | FUNCTION: Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain. | Acetylation;Alternative splicing;Amyotrophic lateral sclerosis;Cytoplasm;Cytoskeleton;Direct protein sequencing;GTP-binding;Methylation;Microtubule;Neurodegeneration;Nitration;Nucleotide-binding;Phosphoprotein;Reference proteome | Microtubules of the eukaryotic cytoskeleton perform essential and diverse functions and are composed of a heterodimer of alpha and beta tubulin. The genes encoding these microtubule constituents are part of the tubulin superfamily, which is composed of six distinct families. Genes from the alpha, beta and gamma tubulin families are found in all eukaryotes. The alpha and beta tubulins represent the major components of microtubules, while gamma tubulin plays a critical role in the nucleation of microtubule assembly. There are multiple alpha and beta tubulin genes and they are highly conserved among and between species. This gene encodes an alpha tubulin that is a highly conserved homolog of a rat testis-specific alpha tubulin. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jun 2013]. | hsa:7277; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; protein kinase binding [GO:0019901]; structural constituent of cytoskeleton [GO:0005200]; microtubule cytoskeleton organization [GO:0000226]; mitotic cell cycle [GO:0000278] | 12054644_increased levels in paclitaxel-resistant breast cancer cells 15934946_Some protein-protein interactions in rafts were disrupted in 3-nitrotyrosine-containing proteins, e.g. caveolin-1 was dissociated from a complex with flotillin-1 and alpha-tubulin. 16039987_Our results suggest that MIZIP might play an important role in mammalian cells by associating with tubulin and thus might provide a link between MCHR1 and tubulin functions. 16257120_These results provide the first characterisation of endogenously nitrated tubulin from human tumour samples. 20546612_Observational study of gene-disease association. (HuGE Navigator) 20668689_Subunits of alphaB crystallin that exchange dynamically with the alphaB crystallin complex can interact with tubulin subunits to regulate the equilibrium between tubulin and microtubules. 20686780_The results suggested that cytoskeletal alterations in the amygdala determined by tubulin seem to be involved in the pathophysiology of drug addiction 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 24268707_TUBA4A is significantly increased in spermatazoa from men with azoospermia. 25374358_TUBA4A mutants destabilize the microtubule network, diminishing its repolymerization capability 25893256_This study mutational screening revealed the presence of four novel heterozygous variants of TUBA4A mutation in four sporadic amyotrophic lateral sclerosis. 26103986_Data show that overexpression of the 14-3-3sigma isoform resulted in a disruption of the tubulin cytoskeleton mediated by binding Tau protein. 26465396_In conclusion, our current results did not find an association between TUBA4A and ALS in Chinese patients, and further studies will be required. 26675813_TUBA4A mutations are not associated with FTD. 27447976_alphaTubulin expression might be an independent prognostic factor for pancreatic cancer after surgical resection 28069311_Its mutations were seen in patients with ALS and frontotemporal dementia. 28478440_The elevated selenium species levels in the TUBA4A patient may have a genetic etiology and/or represent a pathogenic pathway through which this mutation favors disease onset. 29540513_The frequency of TUBA4A mutations suggests that TUBA4A is a rare cause of Amotrophic Lateral Sclerosis in Chinese patients. 30030593_Data indicate a microRNA-1825/TBCB/TUBA4A pathway as a putative pathogenic cascade in both familial ALS (fALS) and both sporadic (sALS). 30760556_Mice carrying a missense mutation in the Tuba4a gene, and a patient with a monoallelic double missense mutation in the TUBA4A gene displayed macrothrombocytopenia and structural abnormalities in the marginal band of platelets. | ENSMUSG00000026202 | Tuba4a | 100.279802 | 0.6521548 | -0.616713540 | 0.26445088 | 5.380164e+00 | 2.036697e-02 | No | Yes | 78.620066 | 12.422567 | 122.531209 | 19.125175 | ||
| ENSG00000127995 | 64921 | CASD1 | protein_coding | Q96PB1 | FUNCTION: O-acetyltransferase that catalyzes 9-O-acetylation of sialic acids (PubMed:20947662, PubMed:26169044). Sialic acids are sugars at the reducing end of glycoproteins and glycolipids, and are involved in various processes such as cell-cell interactions, host-pathogen recognition (PubMed:20947662, PubMed:26169044). {ECO:0000269|PubMed:26169044, ECO:0000305|PubMed:20947662}. | Acyltransferase;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Transferase;Transmembrane;Transmembrane helix | hsa:64921; | integral component of Golgi membrane [GO:0030173]; N-acetylneuraminate 7-O(or 9-O)-acetyltransferase activity [GO:0047186]; carbohydrate metabolic process [GO:0005975] | 20947662_human Cas1 protein is directly involved in O-acetylation of alpha2-8-linked sialic acids 26169044_CASD1 is a sialate O-acetyltransferase and serves as key enzyme in the biosynthesis of 9-O-acetylated sialoglycans. | ENSMUSG00000015189 | Casd1 | 473.107563 | 1.1348343 | 0.182481713 | 0.12504272 | 2.127831e+00 | 1.446453e-01 | 4.991157e-01 | No | Yes | 463.139410 | 114.788119 | 398.529274 | 98.730540 | ||
| ENSG00000128536 | 222256 | CDHR3 | protein_coding | Q6ZTQ4 | FUNCTION: Cadherins are calcium-dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types.; FUNCTION: (Microbial infection) Acts as a receptor for rhinovirus C. {ECO:0000269|PubMed:25848009}. | 3D-structure;Alternative splicing;Asthma;Calcium;Cell adhesion;Cell membrane;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Membrane;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | hsa:222256; | adherens junction [GO:0005912]; catenin complex [GO:0016342]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; calcium ion binding [GO:0005509]; virus receptor activity [GO:0001618]; adherens junction organization [GO:0034332]; calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules [GO:0016339]; cell morphogenesis [GO:0000902]; cell-cell adhesion via plasma-membrane adhesion molecules [GO:0098742]; cell-cell junction assembly [GO:0007043]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156] | 24241537_the role of CDHR3 variants in the development of asthma with severe exacerbations based on genome-wide association study in Denmark 25848009_the asthma susceptibility gene product CDHR3 mediates RV-C entry into host cells, and suggest that rs6967330 mutation could be a risk factor for RV-C wheezing illnesses. 26270739_CDHR3 is a susceptibility locus for early childhood Asthma with severe exacerbations. 27923563_we have determined that carriers of rs6967330 are at significantly increased risk for CRS. 28318885_Our study supports the concept that the CDHR3 variant is an important susceptibility factor for severe adult asthma in individuals who develop the disease in early life. 28782631_In summary, the CDHR3 asthma-risk variant at rs6967330 was not associated with severe RSV bronchiolitis. 29121479_A genome-wide association study (GWAS) focusing on childhood asthma with recurrent severe exacerbations revealed a nonsynonymous SNP (rs6967330) in the CDHR3 (cadherin-related family member 3) gene that was associated specifically with this phenotype. 30659817_A single nucleotide polymorphism (rs6967330) in CDHR3 increases cell surface expression of this protein and, as a result, also promotes Human rhinovirus-C infections and illnesses. 30916989_Polymorphisms in CDHR3 rs6967330 locus (G-->A) previously associated with childhood asthma were related to differences in CDHR3 expression and epithelial cell function. The rs6967330 A allele was associated with higher overall protein expression and RV-C binding and replication compared with the rs6967330 G allele. The rs6967330 A allele was associated with earlier ciliogenesis and higher FOXJ1 expression. 30930175_The rs6967330 SNP of CDHR3 confers risk of severe childhood asthma exacerbations, likely through increasing HRV-C infection levels and protein surface localization. 31610042_Cadherin-related family member 3 gene impacts childhood asthma in Chinese children. 31841129_Genetic susceptibility to severe childhood asthma and rhinovirus-C maintained by balancing selection in humans for 150 000 years. 32056219_Cadherin-related family member 3 upregulates the effector functions of eosinophils. 32615023_Genetic impact of CDHR3 on the adult onset of asthma and COPD. 33213402_Polymorphisms in the airway epithelium related genes CDHR3 and EMSY are associated with asthma susceptibility. 34322716_The role of CDHR3 in susceptibility to otitis media. | ENSMUSG00000035860 | Cdhr3 | 68.430685 | 1.4992476 | 0.584238706 | 0.32111701 | 3.358167e+00 | 6.687246e-02 | No | Yes | 86.256413 | 20.079606 | 60.773765 | 14.136456 | |||
| ENSG00000128652 | 3232 | HOXD3 | protein_coding | P31249 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene belongs to the homeobox family of genes. The homeobox genes encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, located on different chromosomes, consisting of 9 to 11 genes arranged in tandem. This gene is one of several homeobox HOXD genes located at 2q31-2q37 chromosome regions. Deletions that removed the entire HOXD gene cluster or 5' end of this cluster have been associated with severe limb and genital abnormalities. The protein encoded by this gene may play a role in the regulation of cell adhesion processes. [provided by RefSeq, Jul 2008]. | hsa:3232; | aggresome [GO:0016235]; chromatin [GO:0000785]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; cartilage development [GO:0051216]; cell-matrix adhesion [GO:0007160]; embryonic skeletal system morphogenesis [GO:0048704]; Notch signaling pathway [GO:0007219]; positive regulation of gene expression [GO:0010628]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; thyroid gland development [GO:0030878]; transcription, DNA-templated [GO:0006351] | 12405287_transduction of antisense DNA into human melanoma cells results in decreased invasive and motile activities 14610084_Hox D3 coordinately regulates the expression of integrin alpha5beta1 and integrin alphavbeta3 during angiogenesis in vivo. 14633614_HoxD3 may provide a means to directly improve collagen deposition, angiogenesis and closure in poorly healing diabetic wounds. 15545924_HOXD3 may play an important role in regulating cerebral angiogenesis, and that gene transfer of HOXD3 may provide a novel and potent means to stimulate angiogenesis 17126050_The karyotype of our patient suggests another possible locus of the Duane syndrome, and the mapped genes around the deleted region, 1q42.13-43, contain possible candidate genes such as a homeobox gene. 17785556_Study identified hypermethylation and gene inactivation of HOXA4 and HOXA5 was frequently observed (26-79%) in all types of leukemias studied. 19283074_Further validation of candidate genes on a separate cohort of low and high grade prostate cancers by quantitative MethyLight analysis has allowed us to confirm DNA hypermethylation of HOXD3 and BMP7... 19453261_Observational study of gene-disease association. (HuGE Navigator) 19938081_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20212450_HOXD3 methylation distinguishes low-grade prostate cancers from intermediate and high-grade ones. 21207416_quantitative increase in promoter methylation levels of HOXD3 is associated with prostate cancer progression 22935821_High expression of HOXD3 correlates with invasive breast cancer. 24127533_describe familial cases of TH in two generations (proband and his father), in addition to other two sporadic cases. We have found polymorphisms in the HOXB3, HOXD3, and a new synonymous variant, and PITX2 genes 24718283_These results validate the association between promoter hypermethylation of ABHD9 and HOXD3 and prostate cancer recurrence 24847526_HOXD3 promoter hypermethylation is correlated with clinicopathologic features in prostate cancer. This correlation is more common in older, higher risk patients. 27244890_EGR1 is a key player in the transcriptional control of miR-203a, and that miR-203a acts as an anti-oncogene to suppress HCC tumorigenesis by targeting HOXD3 through EGFR-related cell signaling pathways. 27499213_the HOXD3 gene promotes colorectal cancer cell growth and plays a pivotal role in the development and survival of malignant human colorectal cancer cells. 29402992_The findings indicate that miR-203a inhibits hepatocellular carcinoma cell invasion, metastasis, and angiogenesis by negatively targeting HOXD3 and suppressing cell signaling through the VEGFR pathway. 30823921_Nuclear lncRNA HOXD-AS1 suppresses colorectal carcinoma growth and metastasis via inhibiting HOXD3-induced ITGB3 transcriptional activation and MAPK/AKT signaling. 32557953_HOXD3 was negatively regulated by YY1 recruiting HDAC1 to suppress progression of hepatocellular carcinoma cells via ITGA2 pathway. 34928233_HOXD3 Up-regulating KDM5C Promotes Malignant Progression of Diffuse Large B-Cell Lymphoma by Decreasing p53 Expression | ENSMUSG00000079277 | Hoxd3 | 53.835115 | 1.7406330 | 0.799612046 | 0.38334672 | 4.271062e+00 | 3.876669e-02 | No | Yes | 57.188543 | 18.602204 | 33.028921 | 10.979164 | ||
| ENSG00000128805 | 58504 | ARHGAP22 | protein_coding | Q7Z5H3 | FUNCTION: Rho GTPase-activating protein involved in the signal transduction pathway that regulates endothelial cell capillary tube formation during angiogenesis. Acts as a GTPase activator for the RAC1 by converting it to an inactive GDP-bound state. Inhibits RAC1-dependent lamellipodia formation. May also play a role in transcription regulation via its interaction with VEZF1, by regulating activity of the endothelin-1 (EDN1) promoter (By similarity). {ECO:0000250}. | Alternative splicing;Angiogenesis;Coiled coil;Cytoplasm;Developmental protein;Differentiation;GTPase activation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene encodes a member of the GTPase activating protein family which activates a GTPase belonging to the RAS superfamily of small GTP-binding proteins. The encoded protein is insulin-responsive, is dependent on the kinase Akt and requires the Akt-dependent 14-3-3 binding protein which binds sequentially to two serine residues. The result of these interactions is regulation of cell motility. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:58504; | cytosol [GO:0005829]; focal adhesion [GO:0005925]; glutamatergic synapse [GO:0098978]; nucleus [GO:0005634]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; angiogenesis [GO:0001525]; cell differentiation [GO:0030154]; regulation of postsynapse organization [GO:0099175]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21310492_We identified a genetic association for susceptibility to retinopathy in 5 novel chromosomal regions and PLXDC2 and ARHGAP22, the latter 2 of which are genes implicated in endothelial cell angiogenesis and increased capillary permeability. 21926414_The interaction of the IL1RAPL1 family of proteins with PTPdelta and RhoGAP2 reveals a pathophysiological mechanism of cognitive impairment associated with a novel type of trans-synaptic signaling. 21969604_Identification of RhoGAP22 as an Akt-dependent regulator of cell motility in response to insulin 22952583_a weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode 28544509_There is a significant association between single nucleotide polymorphism in the ARHGAP22 gene and diabetic retinopathy risk in a Han Chinese population 31602702_DNA hypermethylation is associated with invasive phenotype of malignant melanoma. 31976761_rs10491034 of gene ARHGAP22 is associated with diabetic retinopathy incidence and severity among Chinese Hui population. | ENSMUSG00000063506 | Arhgap22 | 464.823032 | 0.9430087 | -0.084657033 | 0.12469094 | 4.601870e-01 | 4.975366e-01 | 8.025053e-01 | No | Yes | 469.170800 | 52.333375 | 494.091504 | 54.926325 | |
| ENSG00000129173 | 79733 | E2F8 | protein_coding | A0AVK6 | FUNCTION: Atypical E2F transcription factor that participates in various processes such as angiogenesis and polyploidization of specialized cells. Mainly acts as a transcription repressor that binds DNA independently of DP proteins and specifically recognizes the E2 recognition site 5'-TTTC[CG]CGC-3'. Directly represses transcription of classical E2F transcription factors such as E2F1: component of a feedback loop in S phase by repressing the expression of E2F1, thereby preventing p53/TP53-dependent apoptosis. Plays a key role in polyploidization of cells in placenta and liver by regulating the endocycle, probably by repressing genes promoting cytokinesis and antagonizing action of classical E2F proteins (E2F1, E2F2 and/or E2F3). Required for placental development by promoting polyploidization of trophoblast giant cells. Acts as a promoter of sprouting angiogenesis, possibly by acting as a transcription activator: associates with HIF1A, recognizes and binds the VEGFA promoter, which is different from canonical E2 recognition site, and activates expression of the VEGFA gene. {ECO:0000269|PubMed:15897886, ECO:0000269|PubMed:16179649, ECO:0000269|PubMed:18202719, ECO:0000269|PubMed:22903062}. | 3D-structure;Activator;Cell cycle;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a member of a family of transcription factors which regulate the expression of genes required for progression through the cell cycle. The encoded protein regulates progression from G1 to S phase by ensuring the nucleus divides at the proper time. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Jan 2012]. | hsa:79733; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; RNA polymerase II transcription regulator complex [GO:0090575]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity [GO:0001217]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; cell cycle comprising mitosis without cytokinesis [GO:0033301]; cell population proliferation [GO:0008283]; chorionic trophoblast cell differentiation [GO:0060718]; hepatocyte differentiation [GO:0070365]; negative regulation of cytokinesis [GO:0032466]; negative regulation of transcription by RNA polymerase II [GO:0000122]; placenta development [GO:0001890]; positive regulation of DNA endoreduplication [GO:0032877]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; sprouting angiogenesis [GO:0002040]; trophoblast giant cell differentiation [GO:0060707] | 15897886_The similarities between E2F-7 and E2F-8 define a new subgroup of the E2F family, and further imply that E2F-7 and E2F-8 may act through overlapping mechanisms in mediating cell cycle control. 16179649_E2F8 may have an important role in repressing the expression of E2F-target genes in the S-phase of the cell cycle. 18202719_E2F7 and E2F8 act upstream of E2F1, and influence the ability of cells to undergo a DNA-damage response. 20068156_Analyses indicated that E2F8 could bind to regulatory elements of cyclin D1, regulating its transcription and promoting accumulation of S-phase cells. Findings suggest that E2F8 contributes to the oncogenic potential of HCC 23064264_The results identify E2F8 as a repressor and E2F1 as an activator of a transcriptional network controlling polyploidization in mammalian cells. 25892397_transcription factor E2F8 is involved in the polyploidization during mouse and human decidualization 26089541_E2F8 is overexpressed in Lung Cancer and is required for the growth of LC cells. E2F8 knockdown reduced the expression of UHRF1. These findings implicate E2F8 as a novel therapeutic target for LC treatment. 26992224_High E2F8 expression is associated with breast cancer. 27454291_E2F8-mediated transcriptional repression is a critical tumor suppressor mechanism during postnatal liver development 27683099_Results showed that E2F8 is up-regulated in metastatic prostate cancer and associated with poor prognosis. These results indicate that E2F8 is a crucial transcription regulator controlling cell cycle and survival in prostate cancer cells. 28270228_Study provides evidence that E2F8 functions as a proliferation-related oncogene in papillary thyroid cancer progression. Moreover, miR-144 appears to be a tumor suppressor through direct inhibition of E2F8. 28605876_E2F8 can shorten cisplatin induced G2/M arrest by promoting MASTL mediated mitotic progression in ER+ breast cancer cells, conferring drug resistance. 29615147_miR-223-5p suppressed NSCLC progression through targeting E2F8. 30144184_MiR-1258 may function as a suppressive factor by negatively controlling E2F8 30951518_Authors detected the expression of hsa_circ_0002468, miR-561, and E2F8 by using quantitative real-time polymerase chain reaction (qRT-PCR) analyses. 31471336_These results demonstrate that E2F8 modulates the growth of human colon cancer cells through promoting the NF-kappaB pathway. 31733123_NONO post-transcriptionally regulates the expression of cell proliferation-related genes by binding to their mRNAs, as exemplified by S-phase-associated kinase 2 and E2F transcription factor 8. 31929759_E2F8 regulates the proliferation and invasion through epithelial-mesenchymal transition in cervical cancer. 31990034_Carcinogenesis effects of E2F transcription factor 8 (E2F8) in hepatocellular carcinoma outcomes: an integrated bioinformatic report. 31995441_Cell cycle oscillators underlying orderly proteolysis of E2F8. 32703494_E2F transcription factor 8 promotes proliferation and radioresistance in glioblastoma. 32823614_E2F8 Induces Cell Proliferation and Invasion through the Epithelial-Mesenchymal Transition and Notch Signaling Pathways in Ovarian Cancer. | ENSMUSG00000046179 | E2f8 | 507.375815 | 1.1047492 | 0.143718873 | 0.13228080 | 1.190648e+00 | 2.751990e-01 | 6.450728e-01 | No | Yes | 522.850993 | 98.425474 | 467.976547 | 88.173603 | |
| ENSG00000130159 | 51295 | ECSIT | protein_coding | Q9BQ95 | FUNCTION: Adapter protein of the Toll-like and IL-1 receptor signaling pathway that is involved in the activation of NF-kappa-B via MAP3K1. Promotes proteolytic activation of MAP3K1. Involved in the BMP signaling pathway. Required for normal embryonic development (By similarity). {ECO:0000250}.; FUNCTION: As part of the MCIA complex, involved in the assembly of the mitochondrial complex I. {ECO:0000269|PubMed:32320651}. | Alternative splicing;Cytoplasm;Immunity;Innate immunity;Mitochondrion;Nucleus;Reference proteome;Transit peptide | hsa:51295; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; innate immune response [GO:0045087]; regulation of oxidoreductase activity [GO:0051341]; regulation of protein complex stability [GO:0061635] | 17344420_Study shows a mitochondrial function for Ecsit in the assembly of mitochondrial complex I, an N-terminal targeting signal directs Ecsit to mitochondria, where it interacts with assembly chaperone NDUFAF1. 20877624_Observational study of gene-disease association. (HuGE Navigator) 22588174_TRIM59 interacts with ECSIT as an adaptor protein required for the TLR-mediated transduction pathway. 25228397_mediates virus-triggered type I IFN induction by bridging RIG-I and MDA5 to the VISA complex 25355951_Ubiquitination of ECSIT might have a role in the regulation of NF-kappaB activity in TLR4 signaling. 25371197_Data show that the ECSIT (evolutionarily conserved signaling intermediate in Toll pathways) complex, including MEKK7 (TAK1) and TNF receptor-associated factor 6 (TRAF6), plays a role in Toll-like receptor 4 -mediated signals to activate NF-kappa B. 25449573_Authors conclude that ECSIT appears to be a novel Hepatitis B virus X protein-interacting signal molecule and their interaction is mechanistically important in IL-1beta induction of NF-kappaB activation. 29291352_We added thalidomide to the conventional dexamethasone-containing therapy regimen for two patients with HPS who expressed ECSIT-V140A, and we observed reversal of their HPS and disease-free survival for longer than 3 years. These findings provide mechanistic insights and a potential therapeutic strategy for extranodal natural killer/T cell lymphoma-associated Hemophagocytic syndrome. 31620128_CRBN Is a Negative Regulator of Bactericidal Activity and Autophagy Activation Through Inhibiting the Ubiquitination of ECSIT and BECN1. 33054138_No association between ECSIT germline mutations and hemophagocytic lymphohistiocytosis in natural killer/T-cell lymphoma. 34032637_ECSIT is a critical limiting factor for cardiac function. | ENSMUSG00000066839 | Ecsit | 1062.119555 | 0.9801726 | -0.028892220 | 0.09395982 | 9.467712e-02 | 7.583133e-01 | 9.233919e-01 | No | Yes | 1049.790524 | 171.603693 | 1088.040739 | 177.777601 | ||
| ENSG00000130477 | 23025 | UNC13A | protein_coding | Q9UPW8 | FUNCTION: Plays a role in vesicle maturation during exocytosis as a target of the diacylglycerol second messenger pathway. Involved in neurotransmitter release by acting in synaptic vesicle priming prior to vesicle fusion and participates in the activity-dependent refilling of readily releasable vesicle pool (RRP). Essential for synaptic vesicle maturation in most excitatory/glutamatergic but not inhibitory/GABA-mediated synapses. Facilitates neuronal dense core vesicles fusion as well as controls the location and efficiency of their synaptic release (By similarity). Also involved in secretory granule priming in insulin secretion. Plays a role in dendrite formation by melanocytes (PubMed:23999003). {ECO:0000250|UniProtKB:Q4KUS2, ECO:0000250|UniProtKB:Q62768, ECO:0000269|PubMed:23999003}. | Calcium;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Differentiation;Exocytosis;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Synapse;Zinc;Zinc-finger | This gene encodes a member of the UNC13 family. UNC13 proteins bind to phorbol esters and diacylglycerol and play important roles in neurotransmitter release at synapses. Single nucleotide polymorphisms in this gene may be associated with sporadic amyotrophic lateral sclerosis. [provided by RefSeq, Feb 2012]. | hsa:23025; | neuromuscular junction [GO:0031594]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; presynaptic active zone [GO:0048786]; presynaptic active zone cytoplasmic component [GO:0098831]; presynaptic membrane [GO:0042734]; synaptic vesicle membrane [GO:0030672]; terminal bouton [GO:0043195]; calcium ion binding [GO:0005509]; calmodulin binding [GO:0005516]; diacylglycerol binding [GO:0019992]; phospholipid binding [GO:0005543]; syntaxin-1 binding [GO:0017075]; cell differentiation [GO:0030154]; dense core granule priming [GO:0061789]; intracellular signal transduction [GO:0035556]; neuromuscular junction development [GO:0007528]; neuronal dense core vesicle exocytosis [GO:0099011]; neurotransmitter secretion [GO:0007269]; positive regulation of dendrite extension [GO:1903861]; presynaptic dense core vesicle exocytosis [GO:0099525]; regulation of synaptic transmission, glutamatergic [GO:0051966]; synaptic transmission, glutamatergic [GO:0035249]; synaptic vesicle docking [GO:0016081]; synaptic vesicle maturation [GO:0016188]; synaptic vesicle priming [GO:0016082] | 19734901_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19734901_rs12608932 Single Nucleotide Polymorphism is located at 19p13.3 and maps to a haplotype block within the boundaries of UNC13A, which regulates the release of neurotransmitters such as glutamate at neuromuscular synapses. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20385924_Observational study of gene-disease association. (HuGE Navigator) 20385924_Results do not provide evidence of an association between a variant in the UNC13A gene and susceptibility to sporadic Amyotrophic lateral sclerosis in a French homogeneous population. 22118904_Our results further corroborate the role of UNC13A in amyotrophic lateral sclerosis pathogenesis. 22248876_Here, we report that, like Rab3A, RIM and Munc13 are present in human sperm and that they play a functional role in acrosomal exocytosis before the acrosomal calcium efflux 22921269_This study demonistrated that UNC13A influences survival in Italian amyotrophic lateral sclerosis patients. 23801330_CAPS1 binds to the full-length of cytoplasmic syntaxin-1 with preference to its 'open' conformation, whereas Munc13-1 binds to the first 80 N-terminal residues of syntaxin-1. 23830992_Munc13-1, on account of its role in both insulin and neurotransmitter exocytosis and through its binding properties, may be an important factor contributing to the development or progression of diabetic neuropathy. 24931836_UNC13A provides a novel link between amyotrophic lateral sclerosis and frontotemporal dementia and identifies changes in neurotransmitter release and synaptic function as a converging mechanism in the pathogenesis of ALS and FTD-TDP. 26162714_UNC13A rs12608932 is a risk factor for ALS and a modifying factor for survival and disease progression rate in a Spanish cohort. 27584932_This study demonstrated that the population specific rare variants of UNC13A may modulate survival in ALS in United kingdom. 28192369_Synaptic UNC13A protein variant causes increased neurotransmission and dyskinetic movement disorder 30368160_Its polymorphism contributes to frontotemporal disease in sporadic amyotrophic lateral sclerosis. 31201598_rs12608932(C) is associated with increased ALS susceptibility, especially in Caucasian and European subjects, and that the CC genotype of rs12608932 is associated with reduced ALS patient survival. 32627229_The Distinct Traits of the UNC13A Polymorphism in Amyotrophic Lateral Sclerosis. 33818064_Probing the Diacylglycerol Binding Site of Presynaptic Munc13-1. | ENSMUSG00000034799 | Unc13a | 515.764337 | 0.9266919 | -0.109838315 | 0.12658676 | 7.612100e-01 | 3.829501e-01 | 7.305076e-01 | No | Yes | 497.284751 | 68.450273 | 531.026306 | 72.981951 | |
| ENSG00000130529 | 54795 | TRPM4 | protein_coding | Q8TD43 | FUNCTION: Calcium-activated non selective (CAN) cation channel that mediates membrane depolarization (PubMed:12015988, PubMed:29211723, PubMed:30528822). While it is activated by increase in intracellular Ca(2+), it is impermeable to it (PubMed:12015988). Mediates transport of monovalent cations (Na(+) > K(+) > Cs(+) > Li(+)), leading to depolarize the membrane. It thereby plays a central role in cadiomyocytes, neurons from entorhinal cortex, dorsal root and vomeronasal neurons, endocrine pancreas cells, kidney epithelial cells, cochlea hair cells etc. Participates in T-cell activation by modulating Ca(2+) oscillations after T lymphocyte activation, which is required for NFAT-dependent IL2 production. Involved in myogenic constriction of cerebral arteries. Controls insulin secretion in pancreatic beta-cells. May also be involved in pacemaking or could cause irregular electrical activity under conditions of Ca(2+) overload. Affects T-helper 1 (Th1) and T-helper 2 (Th2) cell motility and cytokine production through differential regulation of calcium signaling and NFATC1 localization. Enhances cell proliferation through up-regulation of the beta-catenin signaling pathway. Plays a role in keratinocyte differentiation (PubMed:30528822). {ECO:0000269|PubMed:11535825, ECO:0000269|PubMed:12015988, ECO:0000269|PubMed:12799367, ECO:0000269|PubMed:14758478, ECO:0000269|PubMed:15121803, ECO:0000269|PubMed:15331675, ECO:0000269|PubMed:15472118, ECO:0000269|PubMed:15550671, ECO:0000269|PubMed:15590641, ECO:0000269|PubMed:15845551, ECO:0000269|PubMed:16186107, ECO:0000269|PubMed:16407466, ECO:0000269|PubMed:16424899, ECO:0000269|PubMed:16806463, ECO:0000269|PubMed:20625999, ECO:0000269|PubMed:20656926, ECO:0000269|PubMed:29211723, ECO:0000269|PubMed:30528822}. | 3D-structure;ATP-binding;Adaptive immunity;Alternative splicing;Calcium;Calmodulin-binding;Cell membrane;Coiled coil;Disease variant;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Immunity;Ion channel;Ion transport;Membrane;Metal-binding;Nucleotide-binding;Palmoplantar keratoderma;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport;Ubl conjugation | The protein encoded by this gene is a calcium-activated nonselective ion channel that mediates transport of monovalent cations across membranes, thereby depolarizing the membrane. The activity of the encoded protein increases with increasing intracellular calcium concentration, but this channel does not transport calcium. [provided by RefSeq, Mar 2016]. | hsa:54795; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; sodium channel complex [GO:0034706]; spanning component of membrane [GO:0089717]; spanning component of plasma membrane [GO:0044214]; ATP binding [GO:0005524]; calcium activated cation channel activity [GO:0005227]; calcium ion binding [GO:0005509]; calmodulin binding [GO:0005516]; cation channel activity [GO:0005261]; identical protein binding [GO:0042802]; ligand-gated calcium channel activity [GO:0099604]; sodium channel activity [GO:0005272]; adaptive immune response [GO:0002250]; calcium ion transmembrane transport [GO:0070588]; calcium-mediated signaling [GO:0019722]; cation transmembrane transport [GO:0098655]; cellular response to ATP [GO:0071318]; dendritic cell chemotaxis [GO:0002407]; inorganic cation transmembrane transport [GO:0098662]; membrane depolarization during AV node cell action potential [GO:0086045]; membrane depolarization during bundle of His cell action potential [GO:0086048]; membrane depolarization during Purkinje myocyte cell action potential [GO:0086047]; negative regulation of bone mineralization [GO:0030502]; negative regulation of osteoblast differentiation [GO:0045668]; positive regulation of adipose tissue development [GO:1904179]; positive regulation of atrial cardiac muscle cell action potential [GO:1903949]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of fat cell differentiation [GO:0045600]; positive regulation of heart rate [GO:0010460]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774]; positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization [GO:1904199]; positive regulation of vasoconstriction [GO:0045907]; protein homotetramerization [GO:0051289]; protein sumoylation [GO:0016925]; regulation of heart rate by cardiac conduction [GO:0086091]; regulation of T cell cytokine production [GO:0002724]; regulation of ventricular cardiac muscle cell action potential [GO:0098911]; sodium ion import across plasma membrane [GO:0098719]; vasoconstriction [GO:0042310] | 12799367_Voltage dependence is not due to block by divalent cations or to voltage-dependent binding of intracellular Ca2+ to an activator site, indicating that TRPM4 is a transient receptor potential channel with an intrinsic voltage-sensing mechanism. 15550671_TRPM4-mediated depolarization modulates Ca2+ oscillations, with downstream effects on cytokine production in T lymphocytes 15590641_the Ca(2+) sensitivity of TRPM4 is regulated by ATP, PKC-dependent phosphorylation, and calmodulin binding at the C terminus. 15845551_analysis of selectivity filter of the cation channel TRPM4 16186107_hydrolysis of PI(4,5)P(2) underlies desensitization of TRPM4, and PI(4,5)P(2) is a general regulator for the gating of TRPM ion channels 16424899_PIP2 is a strong positive modulator of TRPM4 and implicate the C-terminal PH domain in PIP2 action. 17217063_evidence indicates a role as a regulator of membrane potential, and thus the driving force for Ca2+ entry from the extracellular medium--{REVIEW} 18262493_This study is believed to provide the first clear evidence that TRPM4b interacts physically with TRPC3. 19063936_Depolarizing currents generated by TRPM4 are an important component in the control of intracellular Ca(2+) signals necessary for insulin secretion and perhaps glucagon from alpha-cells. 19726882_In 3 branches of a large South African Afrikaner pedigree with an autosomal-dominant form of progressive familial heart block type I, identified the mutation c.19G-->A, which attenuated deSUMOylation of the TRPM4 channel. 20562447_TRPM4 gene is a causative gene in isolated cardiac conduction disease with mutations resulting in a gain of function and TRPM4 channel being highly expressed in cardiac Purkinje fibers. 20625999_These results identify TRPM4 as an important, unanticipated regulator of the beta-catenin pathway, where aberrant signaling is frequently associated with cancer. 20884614_Cys(1093) residue of TRPM4 is crucial for the H(2)O(2)-mediated loss of desensitization. 21887725_In eight probands with atrioventricular block or right bundle branch block-five familial cases and three sporadic cases-a total of six novel and two published TRPM4 mutations were identified. 23116477_TRPM4 possesses the biophysical properties and upstream cellular signaling and regulatory pathways that establish it as a major physiological player in smooth muscle membrane depolarization. 23160238_TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. 23382873_Because of its effect on the resting membrane potential, reduction or increase of TRPM4 channel function may both reduce the availability of sodium channel and thus lead to Brugada syndrome . 23796873_No role was found for TRPM4 in the peripheral blood compartment of multiple sclerosis patients. 24333049_Used the Xenopus laevis oocyte expression system for expression, purification and extraction of functional human TRPM4 protein. Investigated the supra-molecular assembly of TRPM4. 24518820_TRPM4 protein is a ROS-modulated non-selective cationic channel that performs several cell functions, including regulating intracellular Ca(2+) overload and Ca(2+) oscillation 24866019_Contraction of cerebral artery smooth muscle cells requires the integration of pressure-sensing signaling pathways and depolarization through the activation of TRPM4. 25001294_TRPM4 is an important regulator of Ca2+ signals generated by histamine in hASCs and is required for adipogenesis. 25047048_These results showed that the cell surface expression of TRPM4 channels is mediated by 14-3-3gamma binding. 25231975_Casein kinase 1 phosphorylates S839 and is responsible for the basolateral localization of TRPM4. 25909699_TRPM4 acts to maintain endothelial features and its loss promotes fibrotic conversion via TGF-beta production. 26071843_new insight into the ligand binding domains of the TRPM4 channel 26110647_we demonstrate that the inhibition of TRPM4 activity alters cellular contractility in vivo, affecting cutaneous wound healing. 26172285_these data suggest an important role for the Sur1-Trpm4 channel in the pathophysiology of postischemic cell death 26496025_Identify TRPM4 as a regulator of store operated calcium entry in prostate tumor cells, and demonstrate a role for TRPM4 in cancer cell migration. 26571400_The PKC-dependent effect of GLP-1 on membrane potential and electrical activity was mediated by activation of Na(+)-permeable TRPM4 and TRPM5 channels by mobilization of intracellular Ca(2+) from thapsigargin-sensitive Ca(2+) stores 26590985_TRPM4 protein expression is widely expressed in benign and cancerous prostate tissue 26791488_TRPM4 channels regulate human detrusor smooth muscle excitability and contractility and are critical determinants of human urinary bladder function 26820365_A large pedigree diagnosed with progressive familial heart block type I was linked to a mutation of the TRPM4 ion channel. 27207958_The loss-of-function variants A432T/G582S found in 2 unrelated patients with atrioventricular block are most likely caused by misfolding-dependent altered trafficking. 28248435_TRPM4 protein expression is up-regulated in diffuse large B cell lymphoma 28315637_Study supports the view that TRPM4 variants could be responsible for about 2% of LQT syndrome cases. The impact of these variants results in electrophysical disturbances. 28494446_We identified a double heterozygosity for pathogenic mutations in SCN5A and TRPM4 in a Brugada syndrome patient. 28614631_This study presents further evidence to show that TRPM4 regulates beta-catenin signaling and enhances the proliferation of prostate cancer cell lines, through a calcium-dependent regulation of Akt1 and GSK-3beta activity. 29211723_electron cryo-microscopy structure of the most widespread CAN channel, human TRPM4, bound to the agonist Ca(2+) and the modulator decavanadate 29217581_study presents 2 structures of TRPM4 embedded in lipid nanodiscs at ~3-angstrom resolution, as determined by single-particle cryo-electron microscopy; these structures, with and without calcium bound, reveal a general architecture for this major subfamily of TRP channels and a well-defined calcium-binding site within the intracellular side of the S1-S4 domain 29240297_Data suggest that TRPM4 exhibits binding sites for calmodulin (CaM) and S100 calcium-binding protein A1 (S100A1) located in very distal part of TRPM4 N-terminus. 29463718_The electron cryomicroscopy structure of human transient receptor potential melastatin subfamily member 4 (TRPM4) in a closed, Na(+)-bound, apo state at pH 7.5 to an overall resolution of 3.7 A is reported. 30021168_Rational mutagenesis of TRPM4 at position 432 revealed that the bulkiness of the amino acid was key to TRPM4(A432T)'s aberrant gating. 30142439_Compound heterozygous TRPM4 null mutations were identified in a patient with Brugada type 1 electrocardiogram. 30160201_Sulfonylurea Receptor 1, Transient Receptor Potential Cation Channel Subfamily M Member 4, and KIR6.2:Role in Hemorrhagic Progression of Contusion. 30343491_To evaluate whether TRPM4 overexpression was related to the spreading capability of tumors, TRPM4 was knocked down by using shRNAs in PC3 prostate cancer cells and the effect on cellular migration and invasion was analyzed. PC3 cells with reduced levels of TRPM4 (shTRPM4) display a decrease of the migration/invasion capability. 30391667_Here, we report the functional effects of previously uncharacterized variants of uncertain significance (VUS) that we have found while performing a 'genetic autopsy' in individuals who have suffered sudden unexpected death. We have identified thirteen uncommon missense VUS in TRPM4 by testing 95 targeted genes implicated in channelopathy and cardiomyopathy in 330 cases 30482841_Differentiated normal bronchial epithelial (NHBE) cells and tracheal cells from patients with cystic fibrosis (CFT1-LC3) expressed only TRPM4 and all three isoforms of NCXs. Blocking the activity of TRPM4 or NCX proteins abrogated MUC5AC secretion from NHBE and CFT1-LC3 cells. 30484364_Downstream TRPM4 Polymorphisms Are Associated with Intracranial Hypertension and Statistically Interact with ABCC8 Polymorphisms in a Prospective Cohort of Severe Traumatic Brain Injury. 30789900_Trpm4-mediated interneurons that express calcium (Ca2+)-activated nonselective cationic current is indispensable for motor output but not the rhythmogenic core mechanism of the breathing central pattern generator. 31087102_Molecular and genetic insights into progressive cardiac conduction disease. 31358308_Progressive symmetric erythrokeratodermia: Activating mutations of TRPM4 31441200_TRPM4 plays a versatile role in cancer cell proliferation, cell cycle, and invasion. 32147520_TRPM4 Modulates Right Ventricular Remodeling Under Pressure Load Accompanied With Decreased Expression Level. 32301552_KCTD5, a novel TRPM4-regulatory protein required for cell migration as a new predictor for breast cancer prognosis. 32320859_Aberrant TRPM4 expression in MLL-rearranged acute myeloid leukemia and its blockade induces cell cycle arrest via AKT/GLI1/Cyclin D1 pathway. 32484822_TRPM4 is overexpressed in breast cancer associated with estrogen response and epithelial-mesenchymal transition gene sets. 32681584_Domain zipping and unzipping modulates TRPM4's properties in human cardiac conduction disease. 33047172_TRPM4 non-selective cation channel in human atrial fibroblast growth. 33058873_Small Molecular Inhibitors Block TRPM4 Currents in Prostate Cancer Cells, with Limited Impact on Cancer Hallmark Functions. 33562811_TRPM4 in Cancer-A New Potential Drug Target. 33959666_Whole-Exome Sequencing Identifies a Novel TRPM4 Mutation in a Chinese Family with Atrioventricular Block. 34309670_Genetic Variants Associated With Intraparenchymal Hemorrhage Progression After Traumatic Brain Injury. 34445219_Theoretical Investigation of the Mechanism by which A Gain-of-Function Mutation of the TRPM4 Channel Causes Conduction Block. | ENSMUSG00000038260 | Trpm4 | 377.972537 | 0.9336623 | -0.099027290 | 0.15175820 | 4.307631e-01 | 5.116147e-01 | No | Yes | 350.203048 | 49.076596 | 380.152888 | 53.181723 | ||
| ENSG00000130540 | 25830 | SULT4A1 | protein_coding | Q9BR01 | FUNCTION: Atypical sulfotransferase family member with very low affinity for 3'-phospho-5'-adenylyl sulfate (PAPS) and very low catalytic activity towards L-triiodothyronine, thyroxine, estrone, p-nitrophenol, 2-naphthylamine, and 2-beta-naphthol. May have a role in the metabolism of drugs and neurotransmitters in the CNS. {ECO:0000269|PubMed:17425406}. | 3D-structure;Alternative splicing;Cytoplasm;Lipid metabolism;Phosphoprotein;Reference proteome;Steroid metabolism;Transferase | This gene encodes a member of the sulfotransferase family. The encoded protein is a brain-specific sulfotransferase believed to be involved in the metabolism of neurotransmitters. Polymorphisms in this gene may be associated with susceptibility to schizophrenia. [provided by RefSeq, Jul 2008]. | hsa:25830; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; identical protein binding [GO:0042802]; sulfotransferase activity [GO:0008146]; dendrite arborization [GO:0140059]; steroid metabolic process [GO:0008202]; sulfation [GO:0051923] | 14623933_the first immunohistochemical localization of SULT4A1 in human brain 16152568_Candidate gene for schizophrenia susceptibility. 18823757_Observational study of gene-disease association. (HuGE Navigator) 18823757_These results provide the first evidence of how genetic variation in Sult4A1 may be related to clinical symptoms and cognitive function in schizophrenia 19125109_Observational study of gene-disease association. (HuGE Navigator) 19125109_The lack of polymorphisms in the coding region of the SULT4A1 gene is highly unusual and, along with its high conservation between species, suggests that SULT4A1 may have an important function in vivo. 19343046_Observational study of gene-disease association. (HuGE Navigator) 19439498_Cytosolic SULT4A1 interacts with PIN1. 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21521020_determination of SULT4A1-1 haplotype status might be useful for identifying patients who show an enhanced response to long-term olanzapine treatment. 24956247_This study provides a second replication of superior olanzapine response in SULT4A1-1-positive subjects compared with SULT4A1-1-negative subjects. 24988429_These results show that SULT4A1 is widely expressed in human tissues, but mostly as a splice variant that produces a rapidly degraded protein. Dimerization protects the protein from degradation. 25340730_Across three psychiatric disorders (n=2815 patients), we observed no consistent association between SULT4A1-1 status and atypical antipsychotic effect 30606728_Study using SH-SY5Y cells, human induced pluripotent stem cells, and mouse embryonic tissue revealed time-dependent changes in SULT4A1 protein and MBNL/CELF protein during differentiation supported their role in correctly splicing the SULT4A1 mRNA. Furthermore, ectopic expression of each factor produced efficient splicing in the minigene assay as well as correct splicing of the endogenous SULT4A1 mRNA. 31266751_Study in SULT4A1 knockout mice demonstrates that depletion of SULT4A1 induces oxidative stress in neurons and expression of SULT4A1 in human SH-SY5Y cells protects against oxidative-stress-induced mitochondrial dysfunction and cell death. Results suggest that SULT4A1 may have a crucial protective function against mitochondrial dysfunction and oxidative stress. 32152050_Interaction of the Brain-Selective Sulfotransferase SULT4A1 with Other Cytosolic Sulfotransferases: Effects on Protein Expression and Function. | ENSMUSG00000018865 | Sult4a1 | 217.469215 | 1.1878028 | 0.248295362 | 0.18357107 | 1.828751e+00 | 1.762751e-01 | No | Yes | 248.180926 | 34.637900 | 210.298952 | 29.470198 | ||
| ENSG00000130584 | 140685 | ZBTB46 | protein_coding | Q86UZ6 | FUNCTION: Functions as a transcriptional repressor for PRDM1. {ECO:0000250}. | Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:140685; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; transcription cis-regulatory region binding [GO:0000976]; negative regulation of dendritic cell differentiation [GO:2001199]; negative regulation of granulocyte differentiation [GO:0030853]; negative regulation of macrophage differentiation [GO:0045650]; negative regulation of monocyte differentiation [GO:0045656]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of dendritic cell differentiation [GO:2001200]; regulation of transcription by RNA polymerase II [GO:0006357] | 23739915_A single nucleotide polymorphisn in ZBTB46, rs6062314, is associated with an increased genetic risk for multiple sclerosis. 28692046_Authors report a novel tumor promoter, ZBTB46, which is negatively regulated by AR signaling via microRNA (miR)-1-mediated downregulation. ZBTB46 is associated with malignant prostate cancer and is essential for metastasis. 29743654_ZBTB46 is able to clearly define the dendritic cell identity of many previously unclassified histiocytic disease subtypes. All examined cases of Langerhans cell histiocytosis and histiocytic sarcoma expressed ZBTB46, while all cases of blastic plasmacytoid dendritic cell neoplasm, chronic myelomonocytic leukemia, juvenile xanthogranuloma, Rosai-Dorfman disease, and Erdheim-Chester disease failed to express ZBTB46. 29884909_In vitro results using endothelial cells (ECs) showed that cell confluence and laminar shear stress, both known physiological conditions promoting EC quiescence, led to upregulation of ZBTB46 expression. 30312731_ZBTB46 is inversely correlated with SPDEF and is increased in higher tumor grades and small-cell NE prostate cancer (SCNC) patients, which are positively associated with PTGS1. Findings suggest that the induction of ZBTB46 results in increased PTGS1 expression, which is associated with NEPC progression and linked to the dysregulation of the AR-SPDEF pathway. 33398073_Nerve growth factor interacts with CHRM4 and promotes neuroendocrine differentiation of prostate cancer and castration resistance. 33723576_Single nucleotide variations in ZBTB46 are associated with post-thrombolytic parenchymal haematoma. 34815524_PCK1 regulates neuroendocrine differentiation in a positive feedback loop of LIF/ZBTB46 signalling in castration-resistant prostate cancer. | ENSMUSG00000027583 | Zbtb46 | 219.667177 | 0.9370277 | -0.093836338 | 0.19235756 | 2.358307e-01 | 6.272337e-01 | No | Yes | 207.337511 | 34.169164 | 213.846945 | 35.139998 | |||
| ENSG00000130751 | 4861 | NPAS1 | protein_coding | Q99742 | FUNCTION: May control regulatory pathways relevant to schizophrenia and to psychotic illness. May play a role in late central nervous system development by modulating EPO expression in response to cellular oxygen level (By similarity). Forms a heterodimer that binds core DNA sequence 5'-TACGTG-3' within the hypoxia response element (HRE) leading to transcriptional repression on its target gene TH (By similarity). {ECO:0000250, ECO:0000250|UniProtKB:P97459}. | Alternative splicing;DNA-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a member of the basic helix-loop-helix (bHLH)-PAS family of transcription factors. Studies of a related mouse gene suggest that it functions in neurons. The exact function of this gene is unclear, but it may play protective or modulatory roles during late embryogenesis and postnatal development. [provided by RefSeq, Jul 2008]. | hsa:4861; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; protein heterodimerization activity [GO:0046982]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; central nervous system development [GO:0007417]; maternal behavior [GO:0042711]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357]; startle response [GO:0001964] | 27782878_Here the authors examined the crystal structures of multi-domain NPAS1-ARNT and NPAS3-ARNT-DNA complexes, discovering each to contain four putative ligand-binding pockets. 28499489_Study provides a clear, unbiased view of the full spectrum of genes regulated by NPAS1 and NPAS3 and shows that these transcription factors are master regulators of neuropsychiatric function. These findings expose the molecular pathophysiology of NPAS1/3 mutations and provide a striking example of the shared, combinatorial nature of molecular pathways that underlie diagnostically distinct neuropsychiatric conditions. | ENSMUSG00000001988 | Npas1 | 443.186702 | 1.0283868 | 0.040383060 | 0.13111287 | 9.554256e-02 | 7.572458e-01 | No | Yes | 433.481111 | 65.225160 | 421.805440 | 63.436198 | ||
| ENSG00000130816 | 1786 | DNMT1 | protein_coding | P26358 | FUNCTION: Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells (PubMed:24623306). Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs) (PubMed:24623306). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing (PubMed:24623306). Promotes tumor growth (PubMed:24623306). {ECO:0000269|PubMed:16357870, ECO:0000269|PubMed:18413740, ECO:0000269|PubMed:18754681, ECO:0000269|PubMed:24623306}. | 3D-structure;Acetylation;Activator;Alternative splicing;Chromatin regulator;DNA-binding;Deafness;Disease variant;Isopeptide bond;Metal-binding;Methylation;Methyltransferase;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes an enzyme that transfers methyl groups to cytosine nucleotides of genomic DNA. This protein is the major enzyme responsible for maintaining methylation patterns following DNA replication and shows a preference for hemi-methylated DNA. Methylation of DNA is an important component of mammalian epigenetic gene regulation. Aberrant methylation patterns are found in human tumors and associated with developmental abnormalities. Variation in this gene has been associated with cerebellar ataxia, deafness, and narcolepsy, and neuropathy, hereditary sensory, type IE. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:1786; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; replication fork [GO:0005657]; DNA (cytosine-5-)-methyltransferase activity [GO:0003886]; DNA binding [GO:0003677]; DNA-methyltransferase activity [GO:0009008]; methyl-CpG binding [GO:0008327]; promoter-specific chromatin binding [GO:1990841]; RNA binding [GO:0003723]; zinc ion binding [GO:0008270]; cellular response to amino acid stimulus [GO:0071230]; cellular response to bisphenol A [GO:1903926]; DNA methylation [GO:0006306]; DNA methylation involved in embryo development [GO:0043045]; DNA methylation-dependent heterochromatin assembly [GO:0006346]; maintenance of DNA methylation [GO:0010216]; negative regulation of gene expression [GO:0010629]; negative regulation of histone H3-K9 methylation [GO:0051573]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of vascular associated smooth muscle cell apoptotic process [GO:1905460]; negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching [GO:1905931]; positive regulation of DNA methylation-dependent heterochromatin assembly [GO:0090309]; positive regulation of gene expression [GO:0010628]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of vascular associated smooth muscle cell proliferation [GO:1904707]; Ras protein signal transduction [GO:0007265] | 11834837_recruited to the RARbeta2 promoter by the PML-RAR fusion protein 11932749_DNMT1 and DNMT3b cooperate to silence genes in human cancer cells 12145218_the human de novo enzymes hDNMT3a and hDNMT3b form complexes with the major maintenance enzyme hDNMT1. 12496760_DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. 12530095_DNMT1 gene copy number does not influence susceptibility to development of malignant lymphoproliferative disease. 12538344_Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway 12548018_DNA methylation is tightly coupled to replication through physical interaction of DNMT1 and core components of the replication machinery. 12576480_reduction in DNMT1 triggers intra-S-phase arrest of DNA replication proposed to protect the genome from extensive DNA demethylation. 12594811_Decrease of DNA methyltransferase 1 expression relative to cell proliferation in transitional cell carcinoma. 12637155_mutational inactivation of the DNMT1 gene that potentially causes a genome-wide alteration of DNA methylation status may be a rare event during human carcinogenesis 12738984_Over-expressed in squamous cell carcinoma of the mouth. 12789259_We investigated occupancy of ER-alpha promoter by pRb2/p130-E2F4/5-HDAC1-SUV39 H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 complexes, and provided a link between pRb2/p130 and chromatin-modifying enzymes in the regulation of ER-alpha transcription 12804601_structural homology model for human DNA methyltransferase 1 14559786_DNMT1 plays a key role in methylation maintenance, DNMT3b may act as an accessory to support the function in ovarian cancer cells. 14577574_gene expression as a likely mechansism in underlying causes of changes in DNA methylation in aging and tumorigenesis 14583449_Activation of p53 reduces binding and relieves transcriptional repression of the Dnmt1gene, whereas loss of p53, a frequent, early event in tumorigenesis, may significantly contribute to aberrant genomic methylation. 14684836_hypothesis that the increase of DNA-methyltransferase 1 expression in telencephalic GABAergic interneurons of schizophrenia patients causes a promoter hypermethylation of reelin and GAD(67) and perhaps of other genes expressed in these interneurons 15087453_DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation 15220328_DNMT1 activity contributes to the preservation of the correct organization of large heterochromatic regions 15289832_inhibition of DNA methylation by DNMT1 by an antisense oligodeoxynucleotide influences cell morphology and adhesion 15340041_Expression levels of DNMT1 in tumor cells may affect the effectiveness of doxorubicin in chemotherapy. 15375672_Observational study of gene-disease association. (HuGE Navigator) 15526354_The average mRNA level of Dnmt1 gene from cancerous tissue was higher and that of mbd2 gene from cancerous tissue was lower than that from non-cancerous tissue 15657147_DNMT1- and p53-mediated methylation of the survivin promoter, suggesting cooperation between p53 and DNMT1 in gene silencing. 15735013_Results suggest a novel mechanism for gene silencing mediated by RUNX1/MTG8 and support the combination of HDAC and DNMT inhibitors as a novel therapeutic approach for t(8;21) AML. 15755728_DNMT1 protein levels are elevated in breast cancer tissues and in MCF-7 breast cancer cells relative to normal human mammary epithelial cells without a concomitant increase in DNMT1 mRNA or proliferative fraction. 15762053_Increased DNMT1 gene expression appears to be in lymphomas and maybe associated with oncogenesis in this group of tumors. 15799776_Unlike Dnmt1, pre-existing cytosine methylation at CpG sites or non-CpG sites does not stimulate Dnmt3a activity in vitro and in vivo. 15870198_STAT3 may transform cells by inducing epigenetic silencing of SHP-1 in cooperation with DNMT1 and histone deacetylase 1 15956212_Direct role of Dnmt1 in the restoration of epigenetic information during DNA repair. 16053511_These data suggest that increased DNMT1 protein expression participates in multistage pancreatic carcinogenesis from the precancerous stage to malignant progression of ductal carcinomas and may be a biological predictor of poor prognosis. 16227093_necessary for both class switching and somatic hypermutation in B cells 16314526_DNMT1 is necessary for proper PcG body assembly independent of DNMT-associated histone deacetylase activity. 16322686_Breast cancer cells have a prominent loss of DNA methylation accompanied by altered expression of maintenance DNA methyltransferase DNMT1. 16423997_data suggests a potential role for DNMT1 in the initiation of promoter CpG island hypermethylation in human cancer cells 16424002_human cancer cells may differ in their reliance on DNMT1 for maintaining DNA methylation 16497664_down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells 16537562_DNMT1 protein overexpression may be responsible for aberrant DNA methylation during multistage carcinogenesis of the pancreas 16769694_In human cells maintenance of XIST methylation is controlled differently than global genomic methylation and in the absence of both DNMT1 and DNMT3B. 16801630_Inhibitors of DNMT1 may have clinical relevance for immune modulation by augmentation of cytokine effects and/or expression of tumor-associated antigens. 16861352_STAT3 binds in vitro to 2 STAT3 SIE/GAS-binding sites identified in promoter 1 and enhancer 1 of the DNMT1 gene 16897079_suppression of DNMT-1 might be related to the pathogenesis of atopic dermatitis, especially in whom serum IgE level is high 16960727_study demonstrated that expression of DNMT1 is clearly regulated in both impaired spermatogenesis and development of embryonal carcinoma, while HDAC1 expression is not regulated during aberrant germ cell differentiation 16963560_Data suggest that DNMT1 might be essential for maintenance of DNA methylation, proliferation, and survival of cancer cells. 16998846_Sex-specific time windows for concomitant upregulation of DNMT1 are associated with prenatal remethylation of the human male and female germ line. 17015478_depletion of DNMT1 with either antisense or small interfering RNA (siRNA) specific to DNMT1 activates a cascade of genotoxic stress checkpoint proteins 17030625_AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability 17053888_The increased DNMT1 expression was more frequent in adenocarcinoma vs. squamous cell carcinoma (42% vs 19%). Smokers with ~65 packyears showed 4.17 times higher risk of increased DNMT1 expression compared to those who smoked 45 packyears in adenocarcinoma 17067458_DNMT1 expression was correlated to the methylation of RASSF1A, tumor grade and stage, which implied that DNMT1 may contribute to the carcinogenesis and development of adenoid cystic carcinoma. 17081533_The genes DNMT1, DNMT3A, and DNMT3B were over-expressed in the ectopic endometrium as compared with normal control subjects or the eutopic endometrium of women with endometriosis. 17085482_Direct cooperation between DNMT1 and G9a provides a mechanism of coordinated DNA and H3K9 methylation during cell division. 17093909_These findings suggested that DNMT1 was associated with the malignant phenotype, and dysregulation of DNMT1 expression was present in tumor cells of colorectal cancer. 17178861_LMP1 induces formation of a transcriptional repression complex, composed of DNMT1 and histone deacetylase, which locates on E-cadherin gene promoter. 17312023_DNMT1 is essential for the maintenance of DNA methylation patterns in human cells. 17322882_DNMT1 is required for faithfully maintaining DNA methylation patterns in human cancer cells and is essential for their proliferation and survival. 17470536_Direct interactions between HP1 and DNMT1 mediate silencing of euchromatic genes. 17492476_LAT, ZAP-70, and DNMT1 protein levels in CD4(+) T cells can be associated with systemic lupus erythematosus. 17532557_DNMT1 plays a key role in maintenance of methylation, and DNMT3B may act as an accessory DNA methyltransferase to epigenetically silence CXCL12 expression in MCF-7 and AsPC1 cells 17538945_Progressive up-regulation of the gene encoding DNMT1 was found in the colorectal adenoma-carcinoma sequence. 17657744_SUV39H1 is significantly associated with DNMT1, but not with euchromatic promoter methylation in colorectal cancer 17673620_data suggest that UHRF1 may help recruit DNMT1 to hemimethylated DNA to facilitate faithful maintenance of DNA methylation 17698033_mahanine can reverse an epigenetically silenced gene, RASSF1A in prostate cancer cells by inhibiting DNMT activity that in turn down-regulates a key cell cycle regulator, cyclin D1 17716861_These results suggested that PKB enhanced DNMT1 stability and maintained DNA methylation and chromatin structure, which might contribute to cancer cell growth. 17893234_DNA methyltransferase (DNMT) 1 over-expression is not a secondary result of increased cell proliferative activity but is significantly correlated with the CpG island methylator phenotype. 17931718_establish a link between HESX1 and DNMT1 and suggest a novel mechanism for the repressing properties of HESX1 17934516_The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. 17965595_DNMT1 expression is increased in schizophrenia (SZ) and bipolar (BP) disorder brains. [REVIEW] 17991895_reveals novel functions for DNMT1 as a component of the cellular response to DNA damage, which may help optimize patient responses to this agent in the future 18038118_mutant p53 loses its ability to suppress DNMT1 expression, and thus enhances methylation levels of the p16 ( ink4A ) promoter and subsequently down-regulates p16(ink4A )protein. 18049164_In hepatocellular carcinoma, DNMT1 is necessary to maintain the methylation of CpG islands in certain tumor-related genes. 18198215_DNMT-1 has a direct suppressive role in 15-LOX-1 transcriptional silencing that is independent of 15-LOX-1 promoter DNA methylation 18202356_Parental Dnmt1 is a modifier of transmission of alleles at an unlinked chromosomal region and perhaps has a role in the genesis of transmission ratio distortion. 18204201_The subcellular localization of DNMT1 can be altered by the addition of IL-6, and this process is greatly enhanced by phosphorylation of the DNMT1 nuclear localization signal (NLS) by PKB/AKT. 18252747_Study shows that the maintenance DNA methyltransferase Dnmt1, which preserves the patterns of CpG methylation, plays a key role in CAG repeat instability in human cells and in the male and female mouse germlines. 18253830_DNMT1 was over expressed in gastric neoplasms. 18404674_DNMT1 protein overexpression might contribute to aberrant DNA hypermethylation of specific tumor suppressor genes in endometrial cancers. 18413740_Chromatin immunoprecipitation analysis of the BAG-1 promoter in DNMT1-overexpressing or DNMT3B-overexpressing colon cells show a permissive chromatin status assoscsiated with DNA binding of BORIS. 18414412_DNMT1 may play an important role in modulating NSCLC patient survival and thus be useful for identifying NSCLC patients who would benefit most from aggressive therapy. 18499700_Observational study of gene-disease association. (HuGE Navigator) 18499700_The association between sequence variants of DNMT1 and 3B and mutagen sensitivity induced by BPDE supports the involvement of these DNMTs in protecting the cell from DNA damage. 18505931_histone deacetylase inhibition promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells 18563322_DNMT1 and DNMT3b will increase their biological effects and have a synergistic effect on suppressing the growth of cholangiocarcinoma 18567946_The HIV-1 responsive element resides in the 5' most 420 bp of the -1634 to +71 DNMT1 promoter; positioning of this truncated promoter proximal to a hybrid SV40-DNMT1 reporter results in HIV-1-dependent regulation 18680430_study shows endogenous DNMT1 & CFP1 interact in vivo; CFP1 interaction with Setd1A or Setd1B not required for its interaction with DNMT1; result indicates CFP1 intersects cytosine methylation machinery independently of its association with Setd1 complexes 18754681_The CXXC domain in the amino terminus region of DNMT1 cooperates with the catalytic domain for DNA methyltransferase activity. 18829110_elevated DNMT1 expression may in particular contribute to ineffective erythropoiesis in MDS. 18931722_DNMT1 and HLA-DRalpha are prognostic factors for hepatocelluar carcinoma. 19016755_Interaction between DNMT1 and DNA replication reactions in the SV40 in vitro replication system is reported. 19019634_The ratio of MBD-2/DNMT-1 might be valuable in explanation of hypomethylation and evaluation of clinical activity of systemic luopus erythematosus. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124506_Observational study of gene-disease association. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19173286_mapped the dimerization domain to the targeting sequence TS that is located in the center of the N-terminal domain (amino acids 310-629) 19211935_miR-29b down-regulates DNMT1 indirectly by targeting Sp1. 19236003_Incorporation of 6-thioguanine into the CpG site affects the methylation of the cytosine residue by both DNMT1 and HpaII. The effect on cytosine methylation is dependent on the position of 6-thioguanine with respect to the cytosine to be methylated. 19246518_Observational study of gene-disease association. (HuGE Navigator) 19275888_p21(WAF1)-p300-DNMT1 pathway may play a pivotal role to ensure regulated DNMT1 expression and DNA methylation in mammalian cell division. 19279403_Panobinostat treatment depletes DNMT1 levels and enhances decitabine mediated repression of JunB and loss of survival of human acute leukemia cells. 19282482_signaling through SET7 represents a means of DNMT1 enzyme turnover 19292979_DNMT1 levels in tumor cells may determine the effectiveness of doxorubicin in chemotherapy. 19339266_EBV latent membrane protein 2A (LMP2A) up-regulated DNMT1, leading to an increase in methylation of the PTEN promoter in stomach cancer. 19386473_In patient with schizophrenia the DNA-methyltransferase 1 was up regulation in telencephalic GABAergic neurons. 19394279_Data showed that the CD70, perforin and KIR2DL4 promoters are demethylated in CD4(+)CD28(-) T cells, and that DNA methyltransferase 1 (Dnmt1) and Dnmt3a levels are decreased in this subset. 19399408_Silencing DNMT3b and DNMT1 could inhibit the cell growth and promote apoptosis of bladder carcinoma cells. 19399937_DNA methyltransferase 1 (DNMT1) expression was increased in hepatocellular carcinoma compared to non-neoplastic liver tissues and the incidence of DNMT1 immunoreactivity in HCCs correlated significantly with poor tumor differentiation. 19424621_Higher expression of DNMT1 and HDAC1 correlated with advanced stages of the disease and reflect the malignancy of pancreatic carcinoma. 19450230_SUMOylation significantly enhances the methylase activity of DNMT1 both in vitro and in chromatin. 19465937_the role of promoter region DNA hypermethylation as a mediator of gene silencing in glioblastoma multiforme cell line using RNAi directed against DNMT1 and DNMT3b was investigated. 19468253_Our results were not able to demonstrate a clear correlation between DNMT1 and DNMT3a immunoexpression and salivary gland neoplasms development. 19531770_Increase of DNMT1 expression is associated with multiple myeloma and plasma cell leukemias. 19539327_genetic and phenotypic consequences of silencing DNMT1 in PC3 cells are markedly different from those in colon and gastric cancers, indicating that DNMT1 preferentially targets certain gene promoters 19763880_The expression of DNMT1, DNMT2, DNMT3A and DNMT3B in pediatric acute lymphoblastic leukemia patients, was investigated. 19798569_Our data suggest there is no apparent association of common DNMT-1 and DNMT-3B polymorphisms with the risk of breast cancer in Chinese women. 19896490_Observational study of gene-disease association. (HuGE Navigator) 19932585_Our results indicate that DNMT1 plays the main role in maintenance of methylation of CXCR4 promoter, while DNMT3B may function as an accessory DNA methyltransferase to modulate CXCR4 expression in AsPC1 cells. 19936946_Observational study of gene-disease association. (HuGE Navigator) 19957555_Downregulation of DNMT1 can inhibit cervical cancer cell proliferation and induce cell apoptosis. 20044957_The DNMT 1 is the key enzyme responsible for DNA methylation, which often occurs in CpG islands located near the regulatory regions of genes and affects transcription of specific genes. Two intron polymorphisms of DNMT1 might affect HBV clearance. 20071334_DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta 20071580_Data suggest that KSHV miRNA targets multiple pathways to maintain KHSV latency, including repression of the viral protein Rta and a cellular factor, Rbl2, and increased cellular DNMT, in regulating global epigenetic reprogramming. 20081831_DNMT1 is essential for epidermal progenitor cell function 20093774_Carcinogne-induced DNMT1 accumulation and subsequent hypermethylation of the promoter of tumor suppressor genes may lead to tumorigenesis and poor prognosis. 20147412_Hepatitis B virus-induced overexpression of DNMTs leads to viral DNA methylation and decreased viral gene expression and also leads to methylation of host CpG islands. 20192566_No association between DNMT1 and colorectal cancer in Iranian patients. 20192608_No association between DNMT1 polymorphisms and gastric cancer. 20192608_Observational study of gene-disease association. (HuGE Navigator) 20228804_Dnmt1 and Dnmt3a transgenes are required for synaptic plasticity, learning and memory through their overlapping roles in maintaining DNA methylation and modulating neuronal gene expression in adult central nervous system neurons. 20335008_Our results suggest that one potential mechanism by which varenicline may decrease cigarette smoking in schizophrenia is by decreasing DNMT1 mRNA. 20354000_Our results indicate that DNMT-mediated gene silencing may play a role in inflammation-associated colon tumorigenesis. 20381114_expression of DNA methyltransferase 1, 3a, and 3b showed significantly higher levels in stage IV tumors than in stage I or II tumors 20398055_DNMT1 and DNMT3b silencing sensitizes human hepatoma cells to TRAIL-mediated apoptosis via up-regulation of TRAIL-R2/DR5 and caspase-8. 20428781_thymine DNA glycosylase was not up-regulated in DNMT1 nor 3B knockdown cancer cells 20570896_Data provided compelling evidence that deregulation of DNMT1 is associated with gain of transcriptional activation of Sp1 and/or loss of repression of p53. 20588031_Increased expression of DNMT1 in non-neoplastic epithelium may precede or be a relatively early event in ulcerative colitis-associated tumorigenesis. 20592467_interaction between hNaa10p and DNMT1 was required for E-cadherin silencing through promoter CpG methylation, and E-cadherin repression contributed to the oncogenic effects of hNaa10p. 20593030_Observational study of gene-disease association. (HuGE Navigator) 20613874_results reveal that the disruption of Dnmt1/PCNA/UHRF1 interactions acts as an oncogenic event and that one of its signatures (i.e. the low level of mMTase activity) is a molecular biomarker associated with a poor prognosis in GBM patients 20620135_Regulation of DNMT1 and DNMT3A by HBx promoted hypermethylation of p16(INK4A) promoter region 20840813_The expression of DNMT1 was significantly higher in lung cancer tissues than in corresponding normal lung tissues. 20875141_In MCF7 cells, ANT2 knockdown enhanced the expression and activity of DNA methyltransferase 1 (DNMT1). 20920981_Observational study of gene-disease association. (HuGE Navigator) 20920981_The current work shows that polymorphisms of the DNMT1 gene in exons may affect the individual intraductal carcinoma risk in women of the northeast of China. 20937307_The anti-TNFalpha biological agents do not seem to affect DNA methylation and mRNA expressions of DNMT1 and MBD2 in RA 20940144_our studies provide compelling additional evidence for DNMT1 acting as a regulator of genome integrity and as an early responder to DNA DSBs. 20951977_lower levels of DNMT1 may be related with smoking habit. significantly higher mean percentage of DNMT1 immunoreactivity in non-smokers 20970125_The mRNA of DNMT1 (DNA-methyltransferase 1) is expressed in the endometrium across the menstrual cycle. 20980350_Depletion of DNMT1 did not induce cellular invasion in MCF-7 and ZR-75-1 non-invasive breast cancer cell lines. 21042757_HPV-16 E6 may act through p53/ DNMT1 to regulate the development of cervical cancer. 21045206_a previously unknown mode of regulation of DNMT1 protein stability through the coordinated action of an array of DNMT1-associated proteins. 21078759_This study demonstrated that patients with SLE had a significantly lower level of DNA methylation than the controls, and that expression of both DNMT1 and MBD2 mRNA was significantly increased in the SLE patients compared with controls. 21151116_Modifications to DNMT1 mediated by AKT1 and SET7, affect cellular DNMT1 levels. 21163962_structure of DNMT1 composed of CXXC, tandem bromo-adjacent homology (BAH1/2), and methyltransferase domains bound to DNA-containing unmethylated CpG sites; studies identify an autoinhibitory mechanism of DNA methylation 21229291_we report significant overexpression of DNMT1 and DNMT3B in glioma 21266713_Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. 21268065_Findings suggest that, by balancing Dnmt1 ubiquitination, Usp7 and Uhrf1 fine tune Dnmt1 stability. 21296890_association of Dnmt1 and alpha-synuclein might mediate aberrant subcellular localization of Dnmt1 21311766_Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. 21316665_Expression level of DNMT1 is significantly higher in secretory-phase endometrium compared with proliferative endometrium and menstrual endometrium. 21321201_Mitochondrial DNMT1 appears to be responsible for mtDNA cytosine methylation. 21347439_DNA methylation recovery was mediated by the major human DNA methyltransferase, DNMT1 21353307_Results implied that IL-6 expression was regulated by promoter demethylation induced by down-regulation of DNMT activity. 21389349_the naked DNA- and polynucleosome-binding activities of Dnmt1 are inhibited by the RFTS domain, which functions by virtue of binding the catalytic domain to the exclusion of DNA 21395176_Increased expression of DNMT1 may initiate the oncogenesis of laryngeal squamous cell carcinoma. Smoking may induce expression. 21458988_Low DNMT1 expression defines a subgroup of GC patients with better outcomes following platinum/5FU-based neoadjuvant chemotherapy. In vitro data support a functional relationship between DNMT1 and cisplatin sensitivity. 21459093_Study provides evidence for nuclear receptor mediated regulation of Dnmt1 expression through ERRgamma and SHP crosstalk. 21478913_These findings show an important role for p53 in the progression of serous borderline ovarian tumors to an invasive carcinoma, and suggest that downregulation of E-cadherin by DNMT1-mediated promoter methylation contributes to this process. 21523767_DNMT1 could be a significant clinical predictor for stage and treatment response of bladder cancer. 21532572_Here we show that mutations in DNMT1 cause both central and peripheral neurodegeneration in one form of hereditary sensory and autonomic neuropathy with dementia and hearing loss. 21539677_The proportion of cells that express DNMT1 at the mRNA and protein levels in esophageal squamous cell carcinoma clinical samples is generally higher than in paired non-cancerous tissues. 21565170_these results suggest that dysregulation of cell cycle via CDKs could induce abnormal phosphorylation of DNMT1 and lead to DNA hypermethylation often observed in cancer cells. 21565830_findings describe a new mechanism for the regulation of DNMT1 and aberrant DNA hypermethylation in colorectal cancer 21573703_DNA methylation of the BNIP3 promoter was mediated by DNMT1 via the MEK pathway 21592522_Data show that the levels of DNMT1 mRNA were significantly decreased in a depressive but not in a remissive state of MDD and BPD. 21619587_Results indicate that phosphorylation of human DNMT1 by protein kinase C is isoform-specific and provides the first evidence of cooperation between PKCzeta and DNMT1 in the control of the DNA methylation patterns of the genome. 21636528_These results offer an explanation for how and why unmethylated microsatellite repeats can be destabilized in cells with decreased DNMT1 levels and uncover a novel and important role for PARP in this process. 21735817_DNMT1 as a key enzyme in maintaing of proper methylation pattern is a attractive target in anti-tumor therapy. 21756783_PARP1 could regulate DNA methylation by inhibiting the enzyme activity of DNMT1 in bronchial epithelial cells exprosed to B(a)P. 21757290_Data show that Core inhibited p16 expression by inducing promoter hypermethylation via up-regulation of DNMT1 and DNMT3b. 21791605_Data show that trichostatin A treatment reduces global DNA methylation and the DNMT1 protein level and alters DNMT1 nuclear dynamics and interactions with chromatin. 21826395_Dnmt1 was principally expressed in the nucleus & cytoplasm of NeuN-positive neurons, but not in GFAP-positive astrocytes. Levels were significantly increased in patients with temporal lobe epilepsy. 21887463_Significantly higher levels of CXCR4, DNMT3A, DNMT3B and DNMT1 transcript (p=0.0058, 0.0163, 0.0003 and <0.0001, respectively) levels in cancer tissue as compared to normal samples. 21947282_SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities 21962230_MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma 21966451_Results suggest that DNMT1 operates either as a functional intermediary or in cooperation with E2F1 inhibiting AR gene expression in a methylation independent manner. 21999220_DNMT1 silencing is associated with malignant phenotype and methylated tumor suppressor gene expression in cervical cancer 22048249_analysis of DNMT1, expression showed peaks of mRNA transcripts in primary spermatocytes and in mature ejaculated spermatozoa, with DNMT1 transcript level being the most abundant in all cell stages. I 22072770_Elevated expression of DNMT1 or DNMT3B alone is not required to maintain restricted latency. 22110720_DNMT1 and DNMT3a are regulated by GLI1 in pancreatic cancer, and DNMT1 is its direct target gene 22167392_DNMT1 was over-expressed in primary tumors and cell lines, while knockdown of DNMT1 using siRNA could decrease methylation level of miR-148a promoter and restore its expression 22219193_Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. 22236544_Increased expression of DNMT1 was observed in oral lichen planus compared to the control group 22264301_DNMT1 hypermethylation is a late part of glioma progression. 22295098_reduced microRNA-152 can lose an inhibitory effect on DNA methyltransferase, which leads to hypermethylation of the ERalpha gene and a decrease of ERalpha level 22301400_Increased DNMT1 gene expression is associated with carcinogenesis in pancreatic ductal adenocarcinoma. 22317856_supported the idea that NNK-induced DNMT1 expression may result from protein stabilization. Increased DNMT1 protein expression may play a critical role in the malignant progression of larynx. 22328086_Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. 22349384_Decreased expression levels of DNMT1, DNMT3a, and DNMT3b in the ectopic endometrium and eutopic endometrium may play a role in patients with abnormal epigenetics which may lead to endometriosis. 22362755_In human hepatocellular carcinoma, increased DNMT1 expression is negatively correlated with SHP levels. 22403725_Wnt7a is lost by methylation in a subset of tumors and that this methylation is maintained by DNMT1 22455563_MeCP2, DNMT1 and H4Ac expression levels did not correlate with any of tested clinicopathological parameters in breast invasive ductal carcinoma tissues 22520950_data suggest that overexpression of DNMT1, DNMT3a, and DNMT3b might represent a critical event responsible for the epigenetic inactivation of multiple tumor suppressor genes, leading to the development of aggressive forms of sporadic breast cancer 22547080_Pregnancy and the DNMT1a genotype independently influence the arsenic methylation phenotype 22739025_induction of DNMT1 expression in the chronically inflamed colon may release IL-6 signaling towards signal transducer and activator of transcription 3 from inhibition through SOCS3 increasing the propensity to malignant transformation. 22768205_The data from this study provided the first evidence for differential expression of DNMTs proteins in ovarian cancer tissues and their associations with clinicopathological and survival data in sporadic ovarian cancer patients. 22795133_Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells 22879518_MT-ND6 transcriptional regulation dependent on the enhanced expression of the mitochondrial-targeted isoform of the DNA (cytosine-5) methyl transferase 1 (DNMT1). 22894906_A review of the various regulatory mechanisms and interactions of DNMT1. 22898998_increased Dnmt1 expression in DDX20-deficient cells hypermethylated the promoters of metallothionein genes, resulting in decreased metallothionein expression leading to enhanced NF-kappaB activity 22942708_DNMT1, DNMT2 and DNMT3A may play important roles in gastric cancer carcinogenesis. 22975348_these data demonstrated for the first time that ERalpha could upregulate DNMT1 expression by directly binding to the DNMT1 promoter region in MFC-7(ER )/PTX cells. 23039890_The DNMT1+32204GG genotype correlates with DNA hypomethylation. 23041765_Ciprofloxacin has antifibrotic actions in Systemic sclerosis dermal and lung fibroblasts via the downregulation of Dnmt1, the upregulation of Fli1. 23049933_Results suggested that SNPs of DNMT1 could be used as genotypic markers for predicting genetic susceptibilities to H.pylori infection and risks in gastric atrophy. 23064049_results indicate DNMT1 is essential for maintenance of colon cancer stem-like cells/cancer-initiating cells and short-term suppression of DNMT1 might be sufficient to disrupt cancer stem-like cells/cancer-initiating cells 23072722_Data suggest that LSD1 (lysine-specific demethylase 1) is critical in the regulation of cell proliferation, but not an absolute requirement for the stabilization of either p53 or DNMT1(DNA methyltransferase 1). 23079992_Two SNPs, rs16999593 in DNMT1 and rs2424908 in DNMT3B, are significantly associated with breast cancer risk. 23125218_Upregulation of DNMT1 is associated with neoplastic transformation. 23127209_Abnormal expression levels of DNMT1 and methyl-CpG binding domain protein 2 demethylase mRNA may be important causes of the global hypomethylation observed in CD4+T cells in systemic lupus erythematosus. 23242655_mutant p53 protein binds to the promoter of ESR1 through direct interaction with HDAC1 and indirect interaction with DNMT1, MeCP2 proteins in the ER-negative MDA-MB-468 cells. 23249948_HBP1 represses the DNMT1 gene through binding a high-affinity site in the DNMT1 promoter. 23254386_Higher levels of expression of DNMT1 in prostate cancer cells is associated with a more aggressive phenotype. 23318422_were involved in ovarian cancer cisplatin resistance in vitro and in vivo by targeting DNMT1 directly. 23364257_Both expression level and activity of DNMT1 were inversely correlated with the expression level of BNIP3 in colon carcinoma cells after treatment with chemotherapeutic agents and radiation 23365052_DNMT1 mutations in patients presenting with familial frontotemporal dementia or primary memory decline may also have sensory neuropathy and hearing loss, or phenotype for narcolepsy. 23393137_mammalian DNMT1, DN | ENSMUSG00000004099 | Dnmt1 | 9143.907196 | 1.1399094 | 0.188919197 | 0.05266666 | 1.294143e+01 | 3.213892e-04 | 1.776479e-02 | No | Yes | 9585.120722 | 709.593093 | 8433.372380 | 624.384983 | |
| ENSG00000130940 | 54897 | CASZ1 | protein_coding | Q86V15 | FUNCTION: Transcriptional activator (PubMed:23639441, PubMed:27693370). Involved in vascular assembly and morphogenesis through direct transcriptional regulation of EGFL7 (PubMed:23639441). {ECO:0000269|PubMed:23639441, ECO:0000269|PubMed:27693370}. | Alternative splicing;DNA-binding;Developmental protein;Disease variant;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a zinc finger transcription factor. The encoded protein may function as a tumor suppressor, and single nucleotide polymorphisms in this gene are associated with blood pressure variation. Alternative splicing results in multiple transcript variants that encode different protein isoforms. [provided by RefSeq, Jul 2012]. | hsa:54897; | chromatin [GO:0000785]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of neuron differentiation [GO:0045664] | 16631614_CASZ1 is expressed in a number of human tumors and localizes to a chromosomal region frequently lost in tumors of neuroectodermal origin. 19430479_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 20479155_Observational study of gene-disease association. (HuGE Navigator) 21252912_CASZ1 is a critical modulator of neural cell development, and that somatically acquired disruption of normal CASZ1 expression contributes to the malignant phenotype of human Neuroblastoma. 21490919_The study indicates that although their mechanisms of regulation may be distinct, both CASZ1b and CASZ1a have largely redundant but critical roles in suppressing tumor cell growth. 22068036_determined that the tumor suppressors CLU, NGFR, and RUNX3 were also directly repressed by EZH2 like CASZ1 in NB cells 22262398_Papillomavirus DNA integration is associated with loss of CASZ1 gene and thus cervical carcinogenesis. 22331471_This study identifies key domains needed for CASZ1b to regulate gene transcription; a link between loss of CASZ1b transcriptional activity and attenuation of CASZ1b-mediated inhibition of neuroblastoma growth and tumorigenicity 23892435_CASZ1 inhibits cell cycle progression in neuroblastoma by restoring retinoblastoma protein activity. 26296975_CASZ1b binds to chromatin and recruits NuRD complexes to orchestrate epigenetic-mediated transcriptional programs 27270431_These findings provide insight into mechanisms by which CASZ1 regulates transcription, and suggests that regulation of CASZ1 subcellular localization may impact its function in normal development and pathologic conditions such as NB tumorigenesis. 27693370_the current study firstly identifies CASZ1 as a new gene predisposing to congenital heart disease in humans 28099117_The current study reveals CASZ1 as a new gene responsible for human dilated cardiomyopathy (DCM), which provides novel mechanistic insight and potential therapeutic target for CASZ1-associated DCM, implying potential implications in improved prophylactic and therapeutic strategies for DCM, the most common type of primary myocardial disease. 28945738_Collectively, our proteomic, biochemical, genetic, and structural studies suggest that the physical interaction between TBX20 and CASZ1 is required for cardiac homeostasis, and further, that reduction or loss of this critical interaction leads to dilated cardiomyopathy (DCM) 29467767_these data identify Casz1 as a new Th plasticity regulator having important clinical implications for autoimmune inflammation and mucosal immunity. 29660117_only rs11121615 (CASZ1)reached a nominal significance level of P < .05. Results of original GWAS and replication studies were combined by a meta-analysis, and polymorphisms listed above as well as rs111434909 (ANGPT1) and rs4463578 passed a genome-wide significant threshold. 31570750_A variant of the castor zinc finger 1 (CASZ1) gene is differentially associated with the clinical classification of chronic venous disease. 31642367_Epigenome-Wide Association Study for All-Cause Mortality in a Cardiovascular Cohort Identifies Differential Methylation in Castor Zinc Finger 1 (CASZ1). 32060262_CASZ1 up-regulates MYOD signature genes and induces skeletal muscle differentiation in normal myoblasts and Embryonal rhabdomyosarcoma. 33205602_Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. | 203.966507 | 0.9833458 | -0.024229246 | 0.18996876 | 1.630776e-02 | 8.983849e-01 | No | Yes | 201.977884 | 25.631988 | 212.332417 | 26.800641 | ||||
| ENSG00000131089 | 23229 | ARHGEF9 | protein_coding | O43307 | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for CDC42. Promotes formation of GPHN clusters (By similarity). {ECO:0000250|UniProtKB:Q9QX73, ECO:0000269|PubMed:10559246}. | 3D-structure;Alternative splicing;Cell junction;Cytoplasm;Disease variant;Epilepsy;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome;SH3 domain;Synapse | The protein encoded by this gene is a Rho-like GTPase that switches between the active (GTP-bound) state and inactive (GDP-bound) state to regulate CDC42 and other genes. This brain-specific protein also acts as an adaptor protein for the recruitment of gephyrin and together these proteins facilitate receceptor recruitement in GABAnergic and glycinergic synapses. Defects in this gene are the cause of startle disease with epilepsy (STHEE), also known as hyperekplexia with epilepsy, as well as several other types of cognitive disability. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2017]. | hsa:23229; | cytosol [GO:0005829]; postsynaptic density [GO:0014069]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of small GTPase mediated signal transduction [GO:0051056] | 11727829_Here we identified residues critical for interaction with gephyrin in the linker region between the SH3 and the DH domains of collybistin. 15215304_translocates gephyrin to submembrane microaggregates; collybistin mutation (G55A)is found in exon 2 of the ARHGEF9 gene in a patient with clinical symptoms of both hyperekplexia and epilepsy 18208356_Results show that hPEM-2 is a target protein of Smurf1. 19911011_Observational study of gene-disease association. (HuGE Navigator) 20622020_Study propose that the collybistin-gephyrin complex has an intimate role in the clustering of GABA(A)Rs containing the alpha2 subunit. 21633362_Data indicate that ARHGEF9 is likely to be responsible for syndromic X-linked mental retardation associated with epilepsy. 21807943_major regulator of GABAergic postsynaptic gephyrin clustering 22033413_These results reveal that G(s) and G(q) signalings regulate hPEM-2 functions through PKA and c-Src in Neuro-2a neuroblastoma cells, respectively. 22778260_Phosphorylation of gephyrin in hippocampal neurons by cyclin-dependent kinase CDK5 at Ser-270 is dependent on collybistin. 24297911_The enhancement of Cb-induced gephyrin clustering by GTP-TC10 does not depend on the guanine nucleotide exchange activity of Cb but involves an interaction that resembles reported interactions of other small GTPases with their effectors 25678704_Impairment of the membrane lipid binding activity of Collybistin R290H and a consequent defect in inhibitory synapse maturation represent a likely molecular pathomechanism of epilepsy and mental retardation in humans. 25898924_Collybistin forms a complex with mTOR and eIF3 and by sequestering these proteins downregulates mTORC1 signaling and protein synthesis potentially contributing to intellectual disability and autism. 27238888_Autism spectrum disorder patient with the smallest inactivating deletion in the collybistin gene. 29130122_Study identified a novel mutation in ARHGEF9, c.868C > T/p.R290C, which co-segregated with epileptic encephalopathy, and validated its association with epileptic encephalopathy. Further analysis revealed that all ARHGEF9 mutations were associated with intellectual disability. 31283007_Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. 31942680_Clinical and Molecular Characterization of Three Novel ARHGEF9 Mutations in Patients with Developmental Delay and Epilepsy. 32939676_De novo ARHGEF9 missense variants associated with neurodevelopmental disorder in females: expanding the genotypic and phenotypic spectrum of ARHGEF9 disease in females. | ENSMUSG00000025656 | Arhgef9 | 655.012582 | 1.0920510 | 0.127040186 | 0.11883953 | 1.155642e+00 | 2.823713e-01 | 6.530574e-01 | No | Yes | 682.773556 | 78.631770 | 629.431474 | 72.492082 | |
| ENSG00000131400 | 9476 | NAPSA | protein_coding | O96009 | FUNCTION: May be involved in processing of pneumocyte surfactant precursors. | Aspartyl protease;Direct protein sequencing;Disulfide bond;Glycoprotein;Hydrolase;Protease;Reference proteome;Secreted;Signal;Zymogen | This gene encodes a member of the peptidase A1 family of aspartic proteases. The encoded preproprotein is proteolytically processed to generate an activation peptide and the mature protease. The activation peptides of aspartic proteinases function as inhibitors of the protease active site. These peptide segments, or pro-parts, are deemed important for correct folding, targeting, and control of the activation of aspartic proteinase zymogens. The encoded protease may play a role in the proteolytic processing of pulmonary surfactant protein B in the lung and may function in protein catabolism in the renal proximal tubules. This gene has been described as a marker for lung adenocarcinoma and renal cell carcinoma. [provided by RefSeq, Feb 2016]. | hsa:9476; | alveolar lamellar body [GO:0097208]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; lysosome [GO:0005764]; multivesicular body lumen [GO:0097486]; aspartic-type endopeptidase activity [GO:0004190]; endopeptidase activity [GO:0004175]; peptidase activity [GO:0008233]; membrane protein proteolysis [GO:0033619]; proteolysis [GO:0006508]; surfactant homeostasis [GO:0043129] | 12151090_Pronapsin A gene expression in normal and malignant human lung and mononuclear blood cells 12698189_Napsin is a promising marker for the diagnosis of primary lung adenocarcinoma. 13129928_role in the N- and C-terminal processing of pro-surfactant protein-B in type-II pneumocytes 18195689_NAPSA suppresses tumor growth independent of its catalytic activity. 18216060_A general defect in napsin A or cathepsin H expression or activity was not the specific cause for abnormal surfactant accumulation in juvenile pulmonary alveolar proteinosis 19740516_The combined use of napsin A and thyroid transcription factor-1 results in improved sensitivity and specificity for identifying pulmonary adenocarcinoma in primary lung tumors and in a metastatic setting. 20830690_Although TTF-1 had a higher sensitivity, napsin-A was useful as a surrogate marker when encountering a poorly differentiated lung adenocarcinoma. 21164283_Immunohistochemical staining using a combination of TTF-1, napsin A, p63, and CK5/6 allows subclassification of poorly differentiated non-small cell carcinomas on small lung biopsies in most cases. 21464700_in combination with thyroid transcription factor-1-positive immunostaining helps in differentiating primary lung adenocarcinoma from metastatic carcinoma in the lung 21717589_Data suggest that the panels of Napsin A, TTF-1, and p63 were effective in identifying histologic type in NSCLC, and that a combination of either Napsin A and p63 or TTF-1 and p63 should be chosen, depending on morphology. 21838611_Diagnostic utility of PAX8, TTF-1 and napsin A for discriminating metastatic carcinoma from primary adenocarcinoma of the lung. 22156835_napsin A is a useful marker that can assist in the diagnosis of both lung adenocarcinomas and renal cell carcinomas[review] 22198009_None of the 90 squamous cell carcinomas of the lung exhibited napsin A positivity in the neoplastic cells; however, strong napsin A reactivity was observed in hyperplastic type II pneumocytes and in intra-alveolar macrophages entrapped within the tumor. 22288963_Nap-A was more specific than thyroid transcription factor 1 for primary lung adenocarcinoma versus all tumors, excluding kidney, independent of tumor type 22418245_absence of napsin A was an independent prognostic factor for reduced survival time in lung adenocarcinoma 22495379_a useful immunohistochemical marker for differentiation of lung squamous cell carcinoma and adenocarcinoma from other subtypes 22914608_Napsin-A seems to be a useful marker in classifying primary pulmonary neoplasm as adenocarcinomas. 23194051_investigation of expression of NAPSA (a potential diagnostic marker) in lungs/respiratory mucosa of subjects with pulmonary sclerosing hemangioma 23333608_Napsin B was duplicated from napsin A during the early stages of primate evolution, and the subsequent loss of napsin B function during primate evolution reflected ongoing human-specific napsin B pseudogenization 23355200_Mucin-producing neoplasms of the lung infrequently express napsin A, suggesting that immunohistochemical assessment of napsin A may have limited diagnostic usefulness for distinguishing primary and metastatic mucinous adenocarcinomas involving the lung. 23503645_PAX2 and napsin A have high specificity but low sensitivity and only have limited value in the differential diagnosis of mesotheliomas and renal cell carcinomas 23681073_A minority of anaplastic and poorly differentiated micropapillary pattern thyroid carcinomas are napsin A positive. 23899066_24 cases each of pulmonary and esophageal adenocarcinoma were stained with TTF-1, napsin A, CDX2, 34betaE12, N-cadherin, and IMP3 in an attempt to find an optimal panel for differentiation. IMP3, CDX2, and N-cadherin are superior to either TTF-1 or napsin A. 24145649_Napsin A is a sensitive and specific biomarker of the clear cell histotype in endometrial carcinomas and accordingly may have diagnostic utility in their histotyping. 24191930_Napsin A as a marker of clear cell ovarian carcinoma. 24331839_TTF-1 is more sensitive than napsin for detection of lung sarcomatoid carcinoma, and no cases were positive for napsin but negative for TTF-1 24479710_These data suggest that napsin A may be a useful marker for identifying metastatic adenocarcinomas of pulmonary origin 24721826_napsin A is another sensitive and specific marker for distinguishing ovarian clear cell tumors (especially adenocarcinomas) from other ovarian tumors 25389337_Our study showed that napsin A is an extremely sensitive (100%) marker of ovarian clear cell carcinomas 25521803_Data indicate that polyclonal but not monoclonal napsin A antibody has a virtually universal nonspecific labeling in mucinous adenocarcinomas of various sites. 25551297_In diagnosis of ovarian clear cell carcinoma, Napsin A is specific but of intermediate sensitivity. 25889632_Napsin A is expressed in a broad spectrum of renal neoplasms. 25971546_Napsin A is frequently expressed in ovarian and endometrial clear cell carcinomas. 25982999_Low expression levels of NAPSA is associated with lung adenocarcinoma. 26339401_Combining HNF-1beta and napsin A may distinguish clear cell carcinoma from high-grade serous carcinoma, endometrioid adenocarcinoma and metastatic Krukenberg tumors. 26400099_napsin A is aberrantly expressed in a subset of lymphomas 26447895_it may be useful to combine NAPA and TTF-1 for increased sensitivity in lung cancer diagnostics. There is no substantial difference between monoclonal and polyclonal p40 and between different NAPA clones, whereas there is a difference between the TTF-1 clones 8G7G3/1 and SPT24 26469326_CK7, TTF-1 and napsin A are predominantly expressed in primary lung adenocarcinoma patients, with CDX-2 being inconsistently expressed. 26696549_The immunocytochemical expressions of napsin A and p40 in imprint cytology seem to be of great utility for the accurate histological differentiation of lung cancers. 26710975_Napsin-A, and Desmocollin-3 were sensitive and specific markers for the diagnosis of AC and SCC, respectively. Both markers allowed classification of 54/60 cases into either AC or SCC. 26808134_To the best of our knowledge, expression of monoclonal Napsin A in lymphomas has never been reported. ALK-DLBCL should be considered in the differential diagnosis when evaluating a Napsin A-positive tumor of poorly differentiated morphology and of unknown primary 26842346_It is important for pathologists to be aware that breast carcinomas with apocrine features can express napsin A. 26945446_Although napsin A is infrequently expressed in endometrial carcinomas, positive results of napsin A immunostaining in endometrial neoplasms might support the diagnosis of clear cell carcinoma when the pathologic differential diagnosis includes other histologic subtypes 27045515_Immunohistochemical detection of thyroid transcription factor 1, Napsin A, and P40 fragment of TP63 can be used in the subclassification of non-small cell lung carcinomas. 27412420_High Napsin A expression is associated with adenocarcinoma in non-small cell lung carcinoma. 29845258_These data indicated that napsin A expression may inhibit TGFbetalinduced EMT and was negatively associated with EMTmediated erlotinib resistance, suggesting that napsin A expression may improve the sensitivity of lung cancer cells to EGFRTKI through the inhibition of EMT. 29901522_Napsin A is of value in resolving diagnostic confusion between gastric-type adenocarcinoma (GAS) and clear cell carcinoma (CCC), whereas HNF1beta lacks specificity and its use in this setting is discouraged. 29956857_Raspberry bodies and hyaline globules with positive napsin A immunoexpression are useful features in diagnosing clear cell carcinoma. 31130231_Data indicate that lung napsin A aspartic peptidase (napsin A) in many mammals is a heterogeneous enzyme. 31361605_Napsin-A and AMACR are Superior to HNF-1beta in Distinguishing Between Mesonephric Carcinomas and Clear Cell Carcinomas of the Gynecologic Tract. 31953311_Surfactant Expression Defines an Inflamed Subtype of Lung Adenocarcinoma Brain Metastases that Correlates with Prolonged Survival. 32270298_Immunohistochemical expression of Napsin A in normal human fetal lungs at different gestational ages and in acquired and congenital pathological pulmonary conditions. 32561332_Napsin A is a highly sensitive marker for nephrogenic adenoma: an immunohistochemical study with a specificity test in genitourinary tumors. 33247694_Role of Napsin A and Survivin Immunohistochemical Expression in Bronchogenic Adenocarcinoma. 33876717_A study on the immunohistochemical expression of napsin A in oral squamous cell carcinomas, intraepithelial neoplasia, and normal oral mucosa. | ENSMUSG00000002204 | Napsa | 33.993379 | 0.9288295 | -0.106514345 | 0.46027597 | 5.498030e-02 | 8.146133e-01 | No | Yes | 30.953756 | 7.221817 | 33.079145 | 7.617706 | ||
| ENSG00000131626 | 8500 | PPFIA1 | protein_coding | Q13136 | FUNCTION: May regulate the disassembly of focal adhesions. May localize receptor-like tyrosine phosphatases type 2A at specific sites on the plasma membrane, possibly regulating their interaction with the extracellular environment and their association with substrates. {ECO:0000269|PubMed:7796809}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;Phosphoprotein;Reference proteome;Repeat | The protein encoded by this gene is a member of the LAR protein-tyrosine phosphatase-interacting protein (liprin) family. Liprins interact with members of LAR family of transmembrane protein tyrosine phosphatases, which are known to be important for axon guidance and mammary gland development. This protein binds to the intracellular membrane-distal phosphatase domain of tyrosine phosphatase LAR, and appears to localize LAR to cell focal adhesions. This interaction may regulate the disassembly of focal adhesion and thus help orchestrate cell-matrix interactions. Alternatively spliced transcript variants encoding distinct isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:8500; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite [GO:0030425]; focal adhesion [GO:0005925]; presynaptic active zone [GO:0048786]; protein-containing complex [GO:0032991]; cell-matrix adhesion [GO:0007160]; negative regulation of protein localization to plasma membrane [GO:1903077]; negative regulation of stress fiber assembly [GO:0051497]; signal transduction [GO:0007165]; synapse organization [GO:0050808] | 12923177_the interaction between ERC2 and liprin-alpha may be involved in the presynaptic localization of liprin-alpha and the molecular organization of presynaptic active zones 16313174_liprin binds to ATP-agarose, challenged by free ATP, not by free GTP. Liprin LH region mutations inhibit liprin phosphorylation stabilized association of liprin with ATP-agarose. Suggests that liprin autophosphorylation regulates its association with LAR. 17419996_regulated degradation of liprinalpha1 is important for proper LAR receptor distribution, and could provide a mechanism for localized control of dendrite and synapse morphogenesis by activity and CaMKII. 18196592_Amplification and overexpression of PPFIA1, a putative 11q13 invasion suppressor gene, is associated with head and neck squamous cell carcinoma 19690048_Liprin is an essential regulator of cell motility that contributes to the effectiveness of cell-edge protrusion. 19787783_Amplification of both PPFIA1 and CCND1 were significantly associated with high-grade breast cancer phenotype but were unrelated to tumor stage or nodal stage 20096687_Data show that increased levels of liprin-alpha1 affected the localization of inactive, low-affinity integrins, while increasing the average size of beta1 integrin-positive focal adhesions. 21157931_These results suggest that an increased expression of Liprin-alpha1 in the brain may be associated with human Intractable epilepsy . 22072677_The results of this study finding suggested that a model by which the self-assembly of SYD-2/Liprin-alpha proteins mediated by the coiled-coil LH1 domain is one of the key steps to the accumulation of presynaptic components at nascent synaptic junctions 22295056_PPFIA1 as a potential functional candidate acute lung injury risk gene 23453270_PPFIA1 was frequently co-amplified with the Cyclin D1 gene in oral carcinomas and could present a biomarker as well as a novel target for specific gene therapy. 24982445_Liprin-alpha1, ERC1a and LL5 also define new highly polarized and dynamic cytoplasmic structures uniquely localized near the protruding cell edge 25492966_results suggested that PPFIA1 functioned with PP2A to promote the dephosphorylation of Kif7, triggering Kif7 localization to the tips of primary cilia and promoting Gli transcriptional activity. 26663347_The human tumor cell line MDA-MB-231 expresses liprin-alpha 1 and is able to promote the formation of metastasis in mice. 27075696_These results indicate that liprin-alpha1 localizes to different adhesion and cytoskeletal structures to regulate vimentin intermediate filament network, thereby altering the invasion and growth properties of the cancer cells. 27876801_This study shows that PPFIA1 is required for FN polymerization-dependent vascular morphogenesis, both in vitro and in the developing zebrafish embryo. 28720060_Based on these findings, we infer that high PPFIA1 expression might be an independent prognostic indicator of increased metastatic relapse risk in patients with estrogen receptor+/N- breast cancer, but not in estrogen receptor+/N+ or estrogen receptor- patients. 29348417_The results indicate that liprin-alpha1, LL5 and ERC1 define a novel dynamic membrane-less compartment that regulates matrix degradation by affecting invadosome motility. 30005669_liprin-alpha1 as a novel regulator of CD82 30805892_We found that PPFIA1 and ALG3 were distinctively overexpressed at the mRNA level in HNSCC tissues compared with normal tissues, they had a significant co-occurrence relationship. Patients without both PPFIA1 and ALG3 mRNA expression alterations had better overall survival and disease/progression-free survival compared with patients with both PPFIA1 and ALG3 alterations. 33215388_Quantifying Polarized Extracellular Matrix Secretion in Cultured Endothelial Cells. 33761347_Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. | ENSMUSG00000037519 | Ppfia1 | 3155.081785 | 1.0301549 | 0.042861342 | 0.07934913 | 2.967031e-01 | 5.859567e-01 | 8.477345e-01 | No | Yes | 3298.252888 | 441.284439 | 3182.405384 | 425.777036 | |
| ENSG00000131944 | 91442 | FAAP24 | protein_coding | Q9BTP7 | FUNCTION: Plays a role in DNA repair through recruitment of the FA core complex to damaged DNA. Regulates FANCD2 monoubiquitination upon DNA damage. Induces chromosomal instability as well as hypersensitivity to DNA cross-linking agents, when repressed. Targets FANCM/FAAP24 complex to the DNA, preferentially to single strand DNA. {ECO:0000269|PubMed:17289582}. | 3D-structure;DNA damage;DNA repair;DNA-binding;Nucleus;Reference proteome | FAAP24 is a component of the Fanconi anemia (FA) core complex (see MIM 227650), which plays a crucial role in DNA damage response (Ciccia et al., 2007 [PubMed 17289582]).[supplied by OMIM, Mar 2008]. | hsa:91442; | Fanconi anaemia nuclear complex [GO:0043240]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; interstrand cross-link repair [GO:0036297] | 17289582_FAAP24 targets FANCM to structures that mimic intermediates formed during the replication/repair of damaged DNA. 18174376_FANCM is an anchor required for recruitment of the FA core complex to chromatin, and the FANCM/FAAP24 interaction is essential for this chromatin-loading activity 18206976_FAAP24 is dispensable for DNA binding and branch migration activity of FANCM. 18995830_DNA damage recognition and remodeling activities of FANCM and FAAP24 cooperate to promote efficient activation of DNA damage checkpoints in Fanconi anemia. 20670894_FANCM/FAAP24 plays a role in ICL-induced checkpoint activation through regulating RPA recruiment at ICL-stalled replication forks. 23333308_FANCM and FAAP24 play multiple, while not fully epistatic, roles in maintaining genomic integrity. 23932590_Crystal structure of the FANCM-FAAP24 complex. 23999858_These results demonstrate dual roles of FAAP24 in DNA damage response against crosslinking lesions, one through the formation of FANCM/FAAP24 heterodimer and the other via its ssDNA-binding activity required in optimized checkpoint activation. 24003026_Results show that the first HhH motif of FAAP24 is a potential binding site for DNA, which plays a critical role in targeting FANCM-FAAP24 to chromatin. 27473539_This is the first report of an FAAP24 loss of function mutation found in human patients with EBV-associated lymphoproliferation. | ENSMUSG00000030493 | Faap24 | 246.170337 | 0.7318326 | -0.450414361 | 0.21052948 | 4.609538e+00 | 3.179460e-02 | No | Yes | 223.720986 | 28.733972 | 309.191750 | 39.454199 | ||
| ENSG00000131966 | 55860 | ACTR10 | protein_coding | Q9NZ32 | Cytoplasm;Cytoskeleton;Reference proteome | hsa:55860; | axon cytoplasm [GO:1904115]; azurophil granule lumen [GO:0035578]; cytosol [GO:0005829]; dynactin complex [GO:0005869]; extracellular region [GO:0005576]; ficolin-1-rich granule lumen [GO:1904813]; microtubule-based movement [GO:0007018]; retrograde axonal transport of mitochondrion [GO:0098958] | ENSMUSG00000021076 | Actr10 | 1596.992192 | 0.8765998 | -0.190009798 | 0.08724576 | 4.679242e+00 | 3.052921e-02 | 2.351919e-01 | No | Yes | 1465.244410 | 258.235022 | 1676.703146 | 295.404586 | ||||
| ENSG00000132016 | 79173 | BRME1 | protein_coding | Q0VDD7 | FUNCTION: Meiotic recombination factor component of recombination bridges involved in meiotic double-strand break repair. Modulates the localization of recombinases DMC1:RAD51 to meiotic double-strand break (DSB) sites through the interaction with and stabilization of the BRCA2:HSF2BP complex during meiotic recombination. Indispensable for the DSB repair, homologous synapsis, and crossover formation that are needed for progression past metaphase I, is essential for spermatogenesis and male fertility. {ECO:0000250|UniProtKB:Q6DIA7}. | Alternative splicing;Chromosome;Meiosis;Phosphoprotein;Reference proteome | hsa:79173; | chromosome [GO:0005694]; double-strand break repair involved in meiotic recombination [GO:1990918]; female meiosis I [GO:0007144]; spermatogenesis [GO:0007283] | Mouse_homologues 32345962_BRCA2 binds to the C-terminus of MEILB2, resulting in the formation of the BRCA2-MEILB2-BRME1 ternary complex. 32845237_A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. 33250349_The novel male meiosis recombination regulator coordinates the progression of meiosis prophase I. | ENSMUSG00000008129 | Brme1 | 406.590392 | 0.9928135 | -0.010405377 | 0.13404775 | 6.035433e-03 | 9.380762e-01 | No | Yes | 336.260526 | 31.371471 | 342.058709 | 31.715753 | |||
| ENSG00000132128 | 10489 | LRRC41 | protein_coding | Q15345 | FUNCTION: Probable substrate recognition component of an ECS (Elongin BC-CUL2/5-SOCS-box protein) E3 ubiquitin ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. {ECO:0000269|PubMed:15601820}. | Alternative splicing;Leucine-rich repeat;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | hsa:10489; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; protein ubiquitination [GO:0016567] | Mouse_homologues 22709582_The authors identified MUF1 as the first substrate for RhoBTB-Cul3 ubiquitin ligase complexes. | ENSMUSG00000028703 | Lrrc41 | 3490.439883 | 0.9768778 | -0.033750041 | 0.06367442 | 2.794505e-01 | 5.970616e-01 | 8.504829e-01 | No | Yes | 3477.977550 | 432.820524 | 3574.295010 | 444.695480 | |
| ENSG00000132323 | 80895 | ILKAP | protein_coding | Q9H0C8 | FUNCTION: Protein phosphatase that may play a role in regulation of cell cycle progression via dephosphorylation of its substrates whose appropriate phosphorylation states might be crucial for cell proliferation. Selectively associates with integrin linked kinase (ILK), to modulate cell adhesion and growth factor signaling. Inhibits the ILK-GSK3B signaling axis and may play an important role in inhibiting oncogenic transformation. {ECO:0000269|PubMed:14990992}. | Acetylation;Cytoplasm;Hydrolase;Magnesium;Manganese;Metal-binding;Phosphoprotein;Protein phosphatase;Reference proteome | The protein encoded by this gene is a protein serine/threonine phosphatase of the PP2C family. This protein can interact with integrin-linked kinase (ILK/ILK1), a regulator of integrin mediated signaling, and regulate the kinase activity of ILK. Through the interaction with ILK, this protein may selectively affect the signaling process of ILK-mediated glycogen synthase kinase 3 beta (GSK3beta), and thus participate in Wnt signaling pathway. [provided by RefSeq, Jul 2008]. | hsa:80895; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; protein serine phosphatase activity [GO:0106306]; protein threonine phosphatase activity [GO:0106307]; protein dephosphorylation [GO:0006470] | 21782789_these results suggested that palladin played a specific role in modulating the subcellular localization of the cytoplasmic ILKAP and promoting the ILKAP-induced apoptosis. 23329845_ILKAP is a nuclear protein that regulates cell survival and apoptosis through the regulation of RSK2 signaling. 24743186_Data show involvement of ANXA5 and ILKAP in susceptibility to malignant melanoma 25872452_The expression of ILKAP was considerably lower in eutopic and ectopic endometrial samples from women with endometriosis than in control endometrium. Expression varied depending on the phase of the menstrual cycle. 26460618_our data suggest an essential role of PINCH1, ILK and ILKAP for the radioresistance of p53-wildtype glioblastoma multiforme cells 29742494_ILKAP physically interacted with HIF-1A and induced its dephosphorylation. Both the HIF-1A-p53 interaction and apoptosis relied on ILKAP. | ENSMUSG00000026309 | Ilkap | 1641.447014 | 1.0382666 | 0.054176924 | 0.07719361 | 4.892431e-01 | 4.842651e-01 | 7.956130e-01 | No | Yes | 1679.391257 | 131.312534 | 1627.661729 | 127.194354 | |
| ENSG00000132386 | 5176 | SERPINF1 | protein_coding | P36955 | FUNCTION: Neurotrophic protein; induces extensive neuronal differentiation in retinoblastoma cells. Potent inhibitor of angiogenesis. As it does not undergo the S (stressed) to R (relaxed) conformational transition characteristic of active serpins, it exhibits no serine protease inhibitory activity. {ECO:0000269|PubMed:7592790, ECO:0000269|PubMed:8226833}. | 3D-structure;Direct protein sequencing;Dwarfism;Glycoprotein;Osteogenesis imperfecta;Phosphoprotein;Pyrrolidone carboxylic acid;Reference proteome;Secreted;Signal | This gene encodes a member of the serpin family that does not display the serine protease inhibitory activity shown by many of the other serpin proteins. The encoded protein is secreted and strongly inhibits angiogenesis. In addition, this protein is a neurotrophic factor involved in neuronal differentiation in retinoblastoma cells. Mutations in this gene were found in individuals with osteogenesis imperfecta, type VI. [provided by RefSeq, Aug 2016]. | hsa:5176; | axon hillock [GO:0043203]; basement membrane [GO:0005604]; collagen-containing extracellular matrix [GO:0062023]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; melanosome [GO:0042470]; perinuclear region of cytoplasm [GO:0048471]; serine-type endopeptidase inhibitor activity [GO:0004867]; aging [GO:0007568]; cellular response to cobalt ion [GO:0071279]; cellular response to dexamethasone stimulus [GO:0071549]; cellular response to glucose stimulus [GO:0071333]; cellular response to retinoic acid [GO:0071300]; kidney development [GO:0001822]; negative regulation of angiogenesis [GO:0016525]; negative regulation of endopeptidase activity [GO:0010951]; negative regulation of endothelial cell migration [GO:0010596]; negative regulation of epithelial cell proliferation involved in prostate gland development [GO:0060770]; negative regulation of gene expression [GO:0010629]; negative regulation of inflammatory response [GO:0050728]; negative regulation of neuron death [GO:1901215]; ovulation cycle [GO:0042698]; positive regulation of neurogenesis [GO:0050769]; positive regulation of neuron projection development [GO:0010976]; response to acidic pH [GO:0010447]; response to arsenic-containing substance [GO:0046685]; response to peptide [GO:1901652]; retina development in camera-type eye [GO:0060041]; short-term memory [GO:0007614] | 12200129_role of PEDF in protecting retinal pericytes from oxidant injury 12237317_Results describe the first collagen-binding site described for a serpin, pigment epithelium-derived factor (PEDF), which is distinct from its heparin-binding region, neurotrophic active site, and its serpin exposed loop. 12599204_EPC-1 may play a role in the entry of early passage fibroblasts into a G(0) state or the maintenance of such a state once reached 12603315_reactive center loop, specifically Gly376 and Leu377, is involved in the interaction of PEDF with components of the quality control system in the endoplasmic reticulum, thus ensuring its efficient secretion 12670505_VEGF secreted by retinal pigment epithelial cells upregulates pigment epithelium-derived factor expression via VEGFR-1 in an autocrine manner. 12687338_pigment epithelium-derived factor content in the aqueous humor of diabetic patients strongly predicts who among them will develop progression of retinopathy 12711260_PEDF may block the angiogenic effects of leptin through its anti-oxidative properties 12737624_Present in plasma at approx. 100 nM (5 microg/ml) or twice the level required to inhibit aberrant blood-vessel growth in the eye 12827055_Pterygia exhibit significantly lower PEDF but higher VEGF levels than those in normal corneas and conjunctivae. The decreased PEDF level in pterygia may play a role in the formation and progression of pterygia. 12837042_Observational study of gene-disease association. (HuGE Navigator) 12860293_human oral squamous cell carcinoma cells produce VEGF, which in turn regulates PEDF production, and this balance may be contributing to neovascularization in tumors. 12878936_Gene transfer of pigment epithelium-derived factor suppresses tumor vascularization and growth, while prolonging survival in syngeneic murine models of thoracic malignancies. 12920663_level of the natural ocular anti-angiogenic agent pigment epithelium-derived factor (PEDF) is inversely associated with proliferative retinopathy 14991838_Malignant peripheral nerve sheath tumors (MPNST) Schwann cells lose the ability to downregulate PEDF upon axonal contact. 15096582_effectively abates vascular endothelial growth factor-induced vascular permeability 15140209_potential role of the age-associated decline in EPC-1 expression in tissue remodeling and in the development of skin diseases with excessive angiogenesis 15150108_PEDF expression suggests a more favorable prognosis than in patients with ductal pancreatic adenocarcinomas. 15239109_Role in angiogenesis of hepatocellular carcinoma(HCC). Reduction of serum PEDF concentration associated with development of chronic liver diseases may contribute to progression of HCC. Gene therapy using PEDF may be efficient treatment for HCC. 15374885_novel role of extracellular phosphorylation is shown here to completely change the nature of PEDF from a neutrophic to an antiangiogenic factor. 15377265_PEDF-6 affects different second messenger biosynthesis systems in HL-60 cells. and inhibits phosphatidylinositol-specific phospholipase C stimulated by aluminum tetrafluoride anions 15713745_PEDF downregulates Fyn through Fes, resulting in inhibition of FGF-2-induced capillary morphogenesis of endothelial cells 15846509_PEDF inhibits Ang-II-induced EC activation by suppressing NADPH-oxidase-mediated ROS generation and that PEDF may play a protective role in the development 15856012_PEDF gene contains a response element specific for p63 and p73 in its promoter region 15994443_PEDF may play a significant role in determining the balance of angiogenesis/ antiangiogenesis during atherogenesis 16102727_In tumor xenografts, the overexpression of wild-type PEDF significantly suppressed tumor growth, whereas a mutant of the collagen I-binding site of PEDF (Col-mut PEDF) did not inhibit tumor growth 16196102_In painless neuropathies, there is decreased level of PEDF in cerebrospinl fluid, compared with patients with painful neuropathies. 16322471_Phosphorylation induces variable effects of PEDF, and therefore contributes to the complexity of PEDF action, shich might be used to generate effective antiangiogenic or neurotrophic drugs. 16409998_REVIEW: this paper describes the unbalanced expressions of VEGF and PEDF as a cause of CNV 16596284_Results show that low levels of PEDF in lung tumor tissues was associated with a significantly shorter survival. 16707486_Central mechanism for PEDF inhibition of the AGE signaling to vascular permeability is by suppression of NADPH oxidase-mediated reactive oxygen species generation and subsequent VEGF expression. 16740777_These observations collectively support the hypothesis that a lack of PEDF expression is a potent factor for the enhancement of tumor growth and angiogenesis in breast cancer. 16777976_study is the first to demonstrate a role of pigment epithelium-derived factor (PEDF) in ovarian surface epithelium biology and ovarian cancer and suggests that the loss of PEDF may be of relevance in carcinogenesis 16797605_PEDF may play a protective role against early diabetic retinopathy by attenuating the deleterious effect of AGE 16896539_The analysis of the promoter region of the EPC-1/PEDF gene in this paper suggests the age- and cell cycle-dependent expression of specific transcriptional factor(s). 17188371_Increase in serum PEDF during ulcerative colitis, especially in severe forms of disease suggests its involvement in ulcerative colitis pathogenesis. 17202143_heparin induces a conformational change in the vicinity of Lys(178) 17213275_High levels of PEDF in the plasma may be related to the progression of diabetic retinopathy. 17479108_PEDF-overexpressing tumors exhibited reduced intratumoral angiogenesis, and PEDF may be a new and promising approach for the treatment of osteosarcoma. 17525281_These results demonstrate that PEDF could inhibit neointimal formation via suppression of NADPH oxidase-mediated reactive oxygen species generation. 17593873_hepatic PEDF levels may be elevated to counteract the effects of oxidative stress 17604022_The correlation between PEDF and VEGF striatal levels in PD patients suggests that concerted neurotrophic functions of these factors or structural changes in blood vessel walls play an important role in the pathophysiology of PD. 17651710_PEDF induces human umbilical vein endothelial cells apoptosis through the sequential induction of PPARgamma and p53 overexpression. 17658465_These results demonstrate that the PEDF gene, in cooperation with the VEGF gene, may contribute to the development of diabetic retinopathy. 17870167_PEDF induces macrophage apoptosis and necrosis through the signaling of PPARgamma. 18084848_Results demonstrate that the aqueous humour level of asymmetric dimethylarginine is correlated with pigment epithelium-derived factor in humans and suggest that both ADMA and PEDF may be elevated in response to inflammation in uveitis. 18180304_GPR39 protects from cell death by increasing secretion of pigment epithelium-derived growth factor 18191271_There is a novel role for PEDF in hepatic triglyceride homeostasis through binding to ATGL. Both localize to adiposomes in hepatocytes. 18226801_Observational study of gene-disease association. (HuGE Navigator) 18226801_Our data suggest that the PEDF Met72Thr T allele may be a risk factor for wet AMD in the Taiwan Chinese population. PEDF may play a role in the pathogenesis of wet AMD. 18312852_These results give the first direct demonstration that PEDF might represent a target for PARP inhibition treatment and the effects of PEDF on endothelial cells growth are context dependent. 18455830_PEDF levels are significantly higher in type 1 diabetic patients with retinopathy compared to the patients without it. 18523656_The levels of plasma PEDF increases with advances in both diabetic retinopathy and nephropathy. 18625531_serum level of PEDF was one of the independent determinants of resting heart rate in Japanese outpatients 18676622_In moderate hypoxia, PEDF is downregulated such that the VEGF-to-PEDF ratio increases (and angiogenesis is facilitated). 18677713_Serum levels of PEDF were positively associated with metabolic components and TNF-alpha in Japanese patients with type 2 diabetes. 18704312_HIF-1alpha-specific shRNA can effectively silence the HIF-1alpha gene, and consequently down-regulate VEGF and up-regulate PEDF expression in retinal pigment epithelial cells under hypoxic conditions. 18715664_Data show that serum PEDF levels (mean (S.D.)) were increased in 96 Type 2 diabetic vs. 54 non-diabetic subjects. 18787046_Plasma SERPINF1 level is strongly associated with body adiposity, especially with the visceral fat depot in the non-diabetic general population. 18787502_Observational study of gene-disease association. (HuGE Navigator) 18792873_PEDF could improve the Advanced glycation end products-elicited insulin resistance in Hep3B cells by inhibiting JNK- and IkappaB kinase-dependent serine phosphorylation of IRS-1 via suppression of Rac-1 activation. 18805795_The HA-binding activity of PEDF may contribute to deposition in the extracellular matrix and to its reported antitumor/antimetastatic effects. 18845835_An apoptosis-inducing factor (AIF)-related pathway is an essential target of PEDF-mediated neuroprotection. 19091957_Cytosolic phospholipase A2-alpha is an early apoptotic activator in PEDF-induced endothelial cell apoptosis. 19180572_review summarizes the current knowledge on the biochemical properties of PEDF and its receptors and the therapeutic potential of PEDF 19182495_study found a difference in protein levels of expression of VEGF and PEDF between diabetic and non-diabetic epiretinal membranes 19223990_Observational study of gene-disease association. (HuGE Navigator) 19223990_data suggest that none of the investigated PEDF polymorphisms is likely a major risk factor for exudative age-related macular degeneration in a white European population 19224861_Results suggest that the laminin receptor is a pigment epithelium-derived factor (PEDF)receptor that mediates PEDF angiogenesis inhibition. 19247457_Transduced PEDF reduces retinal ganglion cell(RGC) loss and vision decline in DBA/2J mice, possibly via reduction of TNF and IL-18, and downregulation of GFAP. Novel approach to prevention of glaucomatous RGC death. 19253105_The reduction in PEDF expression levels may be, in part, responsible for tumor malignancy in VEGF-low ovarian tumors 19298519_Data suggest a role for PEDF in the regulation of angiogenesis in the heart and propose PEDF as a possible therapeutic target in heart disease. 19342598_Late outgrowth endothelial cells derived from Wharton jelly in human umbilical cord reduce neointimal formation after vascular injury: involvement of pigment epithelium-derived factor. 19503741_Genotyped the Met72Thr variant (rs1136287) and report a lack of association between the PEDF Met72Thr variant and either neovascular age-related macular degeneration (AMD) or polypoidal choroidal vasculopathy (PCV) in a Japanese population. 19503741_Observational study of gene-disease association. (HuGE Navigator) 19513563_Showed that reduced PEDF levels in lung cancer tissues significantly correlated with lymph node metastasis and an overall poor prognosis in the lung cancer patients. 19583952_The pigment epithelium-derived factor is a negative regulator of insulin action in obesity. 19608741_NCoR1 expression is required to maintain IEC in a proliferative state, and PEDF is a novel transcriptional target for NCoR1 repressive action 19635565_Serum PEDF levels may be elevated in response to circulating advanced glycation end products(AGEs) as a counter-system against the AGE-elicited tissue damage. 19637042_Decreased PEDF expression contributes to tumor progression, possibly through increased tumor cell proliferation and increased angiogenesis. 19661223_Lentivirus-mediated gene transfer of PEDF decreased the growth of ocular melanoma and its hepatic micrometastasis in a mouse ocular melanoma model. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948828_PEDF may exert anti-apoptotic effects through inhibition of lysosomal degradation of Bcl-xL in camptothecin-treated HepG2 cells. 20087951_Data suggests serum pigment epithelium derived factor (PEDF) is an independent marker of metabolic syndrome in a Caucasian population. 20104255_Decreased expression of PEDF in primary human lens epithelial cells resulted in a decrease in the expression of vimentin and the increase of alphaB-crystallin expression, two proteins critical for maintaining lens clarity. 20132989_Gene variants in both CFH and HTRA1 contribute significantly to the AMD (age-related macular degeneration) phenotype in a Japanese population. 20141354_Our results suggest lack of association of PRKCB1 gene promoter polymorphisms and moderate protective association of PEDF gene polymorphism with diabetic retinopathy in the south Indian population. 20228934_polarity is an important determinant of the level of PEDF and VEGF secretion in pigment epithelial cells 20229034_PEDF expression is a potentially useful prognostic marker for breast cancer survival. 20237999_The results suggest a molecular pathway by which PEDF ligand/receptor interactions on the cell surface could generate a cellular signal. 20381502_Our present study suggests that substitution of PEDF proteins may be a promising strategy for the treatment of diabetic nephropathy. 20412062_Size-exclusion ultrafiltration assays showed that recombinant human PEDF formed a complex with recombinant yeast F(1)-ATP synthase. 20504225_55% lower plasma level of PEDF in colorectal cancer patients than in a healthy control group. 20504225_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20631025_PEDF concentrations decrease significantly after weight loss in association with blood pressure. PEDF seems to be involved in human adipocyte biology. 20664971_PEDF gene loaded in PLGA nanoparticles could effectively inhibit the growth of mouse colonic carcinoma. 20678803_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20685859_Plasma PEDF was significantly associated with the presence of the metabolic syndrome and predicted the development of the metabolic syndrome in Chinese men. 20709803_Reduced pigment epithelium-derived factor is associated with liver cirrhosis and fibrosis. 20857208_These results indicate that PEDF may play an inhibitory role on growth and migration of HaCaT cells through dephosphorylation of ERK1/2. 21058962_Three-step serum proteome analysis reveals that excessive alcohol drinking increases the PEDF level 21067572_Observational study of gene-disease association. (HuGE Navigator) 21122834_Significantly lower levels of peritoneal fluid PEDF in patients with endometriosis compared with patients without endometriosis, suggest that peritoneal fluid PEDF plays a role in the pathogenesis of this disorder. 21152082_Studies indicate that preferred codon usage is advantageous to translational efficiency of biologically active PEDF in E coli. 21174599_Detection of SNPs in the PEDF gene was not found to be significantly associated with exudative age-related macular degeneration. 21209034_The serum PEDF level is elevated in women with polycystic ovary syndrome and is associated with insulin resistance 21211310_The expression of PEDF and the incidence of prostate cancer have a negative correlation. 21236558_Data indicate that bone marrow stem cells play an important role in regulating neovascularization, and that the ratio of VEGF and PEDF may be an indicator of the pro- or anti-angiogenic activities of BMSCs. 21275514_VEGF and PEDF may independently influence retinal vascular permeability in CRVO patients with macular edema. 21281801_Overexpression of PEDF inhibits retinal inflammation and neovascularization. 21292512_Expressional change of PEDF and TNF-alpha is in relation to angiogenesis of bladder tumor 21323605_The combined assessment of pigment epithelium-derived factor (PEDF), Haptoglobin and Tau protein levels, using Iterative Marginal Optimization, improved the differential diagnosis of Alzheimer's Disease and other dementias. 21330424_PEDF may be a useful biomarker for assessing the effects of angiotensin II type 1 receptor blockers independent of a reduction in blood pressure. 21336718_PEDF expression had a negative correlation with IL-8 in papillary urothelial neoplasm of low malignant potential. 21353196_These data provide genetic evidence for SERPINF1 involvement in human bone homeostasis. 21570865_Data show that PEDF inhibits the production of IL8 in human hormone-refractory prostate cancer cells, through PEDF receptor/phospholipase A2 regulation of NFkappaB and PPARgamma, and delays the growth of these cells in vitro. 21652703_Pigment epithelium-derived factor (PEDF) shares binding sites in collagen with heparin/heparan sulfate proteoglycans. 21673604_PEDF modulated diverse categories of genes known to be involved in angiogenesis and migration in human melanoma cells. 21697716_may be involved in cross talk between adipose tissue and skeletal muscle [review] 21777264_Data suggest that plasma PEDF concentration is significantly associated with BP, and incident hypertension. 21826736_Loss of pigment epithelium-derived factor function constitutes a novel mechanism for osteogenesis imperfecta and shows its involvement in bone. mineralization. 21846721_a novel mechanism for the tumoricidal activity of PEDF, which involves tumor cell killing via PPARgamma-mediated TRAIL induction in macrophages. 21851923_We found that ficolin-3 levels were elevated in the vitreous fluid of patients with proliferative diabetic retinopathy (PDR); and may be used as a new therapeutic target for treatment of PDR. 21947986_PEDF may play a role in renal dysfunction in coronary artery disease patients. 22028839_a novel nuclear localization signal and mechanism for serpinF1 nuclear import 22029535_No evidence was found to support a role for the Met72Thr variant in susceptibility to either PCV or nAMD in a Han Chinese cohort. 22051848_Lower levels of serum PEDF were found in women with endometriosis than in those without endometriosis. 22113968_the patients are homozygous for a frameshift mutation in exon 4 of the SERPINF1 gene, which leads to lack of the transcription/translation product, likely a key factor in bone deposition and remodeling 22115973_MITF acts through PEDF to inhibit RPE cell migration and to play a significant role in regulating RPE cellular function. 22120651_elevated sera levels in psoriasis patients 22215693_findings establish PEDF as both a metastatic suppressor and a neuroprotectant in the brain, highlighting its role as a double agent in limiting brain metastasis and its local consequences 22218393_Results indicate that adeno-associated virus-mediated PEDF gene expression may offer an active approach to inhibit Lewis lung carcinoma growth. 22234980_In patients with pancreatic ductal adenocarcinoma, both tissue and serum levels of PEDF were decreased, stromal TIP47 expression was higher and the tissue VEGF to PEDF ratio was increased. 22315956_PEDF processing is associated with coronary plaque rupture. 22457728_findings show that hypoxic conditions encountered during primary melanoma growth downregulate antiangiogenic and antimetastasic PEDF by a posttranslational mechanism involving degradation by autophagy 22457810_functional common genetic variant in the gene locus encoding PEDF contributes to overall body adiposity, obesity-related insulin resistance, and circulating leptin levels 22547925_Show that long-term survival and neurotrophic potential of hRPE cells can be enhanced by FDA-approved plastic-based microcarriers. Growth of hRPE cells on microcarriers led to sustained levels of PEDF and VEGF-A in conditioned media for two months. 22570532_This paper highlights the current understanding of probable mechanism of how PEDF blocks deterioration of diabetic retinopathy through its antioxidative properties and application prospects of PEDF as a novel therapeutic target in diabetic retinopathy. 22669302_results suggest that lack of circulating PEDF is a common characteristic of osteogenesis imperfecta type VI, regardless of the nature of the underlying SERPINF1 mutation 22701300_The differential effects exhibited by StVOrth-2 and StVOrth-3 in this orthotopic model of osteosarcoma may be related to the functional epitopes on the PEDF glycoprotein that they represent. 22792709_PEDF (44-77) suppressed proliferation of endothelial cells by 53% and inhibited endothelial cell tube formation at the concentration of 1 nM. 22819570_Data suggest that down-regulation of PEDF expression and increased microvascular density in ovarian endometriotic lesions play roles in pathogenesis of ovarian endometriosis; lesions were compared to endometrium from patients and control subjects. 22827961_Serum levels of PEDF are independently associated with fasting apoB48 levels in carotid atherosclerosis. 22837306_Data suggest that patients exhibit elevated circulating PEDF in obesity, hypertension, dyslipidemia, and insulin resistance compared with subjects with normal glucose tolerance. Plasma PEDF might be useful biomarker for atherosclerosis. 22884488_The present study reveals that serum levels of PEDF are independently associated with P-III-P levels, suggesting that PEDF level is a novel biomarker of liver fibrosis in patients with nonalcoholic fatty liver disease. 22915824_Adenoviral E4 region stimulated expression and secretion of PEDF by human renal epithelial cells that acted as a survival factor for glomerulus-derived endothelial cells. 22917444_these data validate that MSCs-PEDF can migrate and deliver PEDF to target glioma cells, which may be a novel and promising therapeutic approach for refractory brain tumour. 22930782_Plasma PEDF is elevated in overweight youth and is positively associated with insulin resistance. These findings suggest that PEDF may play a role in the development of cardiometabolic dysfunction in youth. 23038613_PEDF acts directly on monocytes/macrophages by inducing their migration and differentiation into M1-type cells in prostate. 23055574_Increased levels of PEDF may be a response to counterbalance the activity of angiogenic and fibrogenic factors in proliferative diabetic retinopathy (PDR), proliferative vitreoretinopathy (PVR). 23075882_the dynamic expression of PEDF that inversely portrays VEGF expression may imply its putative role as a physiological negative regulator of follicular angiogenesis. 23123582_Adenovirus mediated PEDF is potent in retarding the growth and invasiveness of endometrial cancer cells. 23151593_PEDF silencing confers resistance to tamoxifen in breast cancer cells and its stable expression sensitizes resistant cells to endocrine therapy 23218576_Circulating levels of PEDF were independently associated with asymmetrical dimethylarginine levels. 23318725_Liver, but not adipose tissue, might be the source of increased circulating PEDF linked to insulin resistance. 23346798_Dry form of age related retinal degeneration was associated with lower plasma PEDF levels. A strong positive correlation between VEGF and PEDF concentrations was seen in patients with wet form and in patients with bilateral disease. 23359450_PEDF suppression of oxidative stress delays cellular senescence and allows greater expansion of mesenchymal stem cells. 23393224_Our data showed that the elevated AZGP1 and decreased PEDF and PRDX2 expressions in CRC serum and tissues were correlated with liver metastases. 23547472_Only full length PEDF was shown to form amyloid like fibril structures, but not the truncated form. Accumulation of fibrils results in fibroblasts destruction and might be the cause of changes in structure of Tenon's capsule observed in myopia 23547730_Serum PEDF levels are positively associated with coronary artery disease in a Chinese population. 23553951_PEDF stimulated limbal stem cell proliferation. 23569025_PEDF may be involved in the pathogenesis of condyloma acuminatum. 23600479_Data suggest that serum PEDF (pigment epithelium-derived factor) is positively associated with insulin resistance and negatively associated with HDL level in prediabetes (impaired fasting glucose), diabetes type 2, and control subjects. 23722394_We demonstrated a marginal association of the PEDF SNP, rs12603825, with myopic CNV in extremely myopic patients 23818523_PEDF-R is required for the survival and antiapoptotic effects of PEDF on retina cells. 23844817_This review discusses the role of PEDF in cardiometabolic disorders and the utility of measuring its levels. 23924722_Here, PEDF was purified from human plasma by use of a dermatan sulfate affinity column, and then hydroxyapatite, gel filtration and ion exchange columns 23939834_PEDF activated ERK and AKT signaling pathways in MSCs to induce expression of osteoblastic-related genes. These data suggest that PEDF is involved in MSCs osteoblastic differentiation. 24078214_Loss of PEDF expression is associated with malignant peripheral nerve sheath tumours. 24105071_serum PEDF levels are closely associated with hs-CRP in women with PCOS. PEDF may play a role in the development of chronic inflammation in PCOS 24161393_the present study suggests that atheroprotective effects of PEDF might be partly ascribed to its caveolin-1-interacting properties. 24255038_iPS-RPE possesses the machinery to process retinoids for support of visual pigment regeneration. Inhibition of all-trans retinyl ester accumulation by NEM confirms LRAT is active in iPS-RPE. 24342618_PEDF causes anti-angiogenic, anti-inflammatory and anti-thrombogenic reactions in myeloma cells through the interaction with LR. 24361362_PEDF is emerging as an ovarian factor, which has unexpected reactive oxygen species (ROS)-augmenting activities in the human ovary; it may be involved in ovarian ROS homeostasis and may contribute to oxidative stress 24698153_PEDF may play a pivotal role in skin homoeostasis and the response of keratinocytes to injury or inflammatory insults. 24760990_Data (including data from transgenic mice expressing human PEDF) suggest white adipose tissue exhibits near-normal angiogenesis/growth (adiposity)/lipid metabolism/insulin sensitivity in response to mild PEDF overexpression (as seen in obesity). 24766673_TFAP2B overexpression contributes to tumor growth and a poor prognosis of human lung adenocarcinoma through modulation of ERK and VEGF/PEDF signaling. 24769282_The expression levels of PEDF are inversely correlated to that of vascular endothelial growth factor during the menstrual cycle. 24810046_Suggest that PEDF/low-dose chemotherapy may represent a new therapeutic alternative for castration-refractory prostate cancer. 25030625_these results identify PEDF as a novel transcriptional target of MITF and support a relevant functional role for the MITF-PEDF axis in the biology of melanoma. 25064413_PEDF represents a novel intrinsic antioxidant of granulosa cells. 25127091_Heterozygous SERPINF1 mutation carriers had no detectable abnormalities in fat and bone, despite decreased PEDF expression. 25159325_Data suggest that healthy adult stem cells exposed to vitreous/aqueous humors of subjects with proliferative diabetic retinopathy results in increased expression of serpin F1, CXCL4 (platelet factor 4), and endothelin-1 (aqueous only). 25166721_Elevated PEDF levels may represent a compensatory change in type 2 diabetic patients with renal disease and appear to be a useful marker for evaluating the progression of diabetic nephropathies. 25172543_study supports the possible influence of sICAM-1 and PEDF on the pathophysiology of retinal neovascularization in SCD patients. 25284724_PEDF inhibited breast cancer cell migration and invasion by down-regulating fibronectin 25293868_plasma PEDF similar in type 2 diabetes mellitus and obese groups of children 25363869_pigment epithelium derived factor may regulate Sost expression by osteocytes leading to enhanced osteoblastic differentiation and increased matrix mineralization 25447045_MITF-regulated PEDF has a role in melanoma progression 25450390_PEDF is anti-angiogenic and is associated with the growth inhibition of hemangioma-derived endothelial cells. 25462587_The review discuss the evolving role of PEDF as a novel metabolic regulatory protein that plays a causal role in insulin resistance probably through proposed mechanism as inflammation, lipolytic free fatty acid mobilization or mitochondrial dysfunction. 25562624_PEDF inhibits retinal microvascular dysfunction induced by 12/15-lipoxygenase-derived eicosanoids. 25678686_PEDF peptide derivative may be an innovative strategy for tissue engineering and repair therapy in partial LSC deficiency diseases. 25700221_Data suggest that pigment epithelium-derived factor (PEDF) and adipose triglyceride lipase (ATGL) may serve as therapeutic targets for managing vascular hyperpermeability in sepsis. 25705004_Serum PEDF levels were one of the independent correlates of circulating DPP-4 levels in cardiovascular disease patients. 25820866_the results suggest that PEDF is not a major susceptibility gene for age-related macular degeneration (AMD) and polypoidal choroidal vasculopathy (PCV) in the overall population. 25821324_Retinal pigment epithelial cells exposed to adiponectin showed decreased expression of VEGF mRNA, protein whereas PEDF protein is unaltered and PEDF mRNA was increased. 25844034_Elevated PEDF levels may be involved in promoting the development of COPD by performing proinflammatory functions. 25868797_MC3T3-E1 osteoblasts stably overexpressing SERPINF1 with the p.Ala91_Ser93dup mutation had decreased collagen type I deposition and mineralization 25881671_Studies indicate that role of pigment epithelium-derived factor (PEDF) in diabetic and hypoxia-induced angiogenesis, and the pathways mediating PEDF's effects under these conditions. 25948043_PEDF binds VEGFR-1 and VEGFR-2 in vascular endothelial cells. 25992628_PEDF sustained glioma stem cell self-renewal by Notch1 cleavage. 26304116_region composed of positions 98-114 of PEDF contains critical residues for PEDF-R interaction that mediates survival effects, the findings reveal distinct small PEDF fragments with neurotrophic effects on photoreceptors 26308290_Data suggest differences in regulation of expression of PEDF (up-regulation) vs. VEGF (down-regulation) in granulosa cells explain reduced risk of ovarian hyperstimulation syndrome due to ovulation induction using GnRH/GNRHR agonists rather than hCG. 26333415_Thus PEDF could be involved in the establishment of the avascular nature of seminiferous tubules and after puberty androgens may further reinforce this feature. 26427478_Studies indicate that pigment epithelium-derived factor (PEDF) is a natural protein of the retina . 26450919_We demonstrate that rPEDF may serve as a useful intervention to alleviate the risk of tamoxifen-induced endometrial pathologies 26612427_hCG-induced PEDF downregulation and VEGF upregulation are mediated by similar signaling cascades emphasizes the delicate regulation of ovarian angiogenesis. 26693895_We confirmed that expression of SERPINF1 in the liver restored the serum level of PEDF. We also demonstrated that PEDF secreted from the liver was biologically active by showing the expected metabolic effects of increased adiposity and impaired glucose tolerance in Serpinf1(-/-) mice. 26697494_We showed that transplantation of pigment epithelial cells overexpressing PEDF can restore a permissive subretinal environment for RPE and photoreceptor maintenance, while inhibiting choroidal blood vessel growth. 26700654_Study demonstrated the inhibitory effect of PEDF on insulin-dependent molecular mechanisms of glucose homeostasis, and suggests that PEDF could be a specific target in the management of metabolic disorders. 26746675_Study discuss the anti-tumor activities of PEDF and focus on its dual role as an inhibitor (e.g., angiogenesis) and as an inducer of various vital biological processes that lead to the therapeutic effect via different mechanisms of action. [review] 26815784_we report on two apparently unrelated children with OI type VI who had the same unusual homozygous variant in intron 6 of SERPINF1 26921338_results demonstrate that PEDF maintains tumor-suppressive functions in fibroblasts to prevent CAF conversion and illustrat | ENSMUSG00000000753 | Serpinf1 | 726.003038 | 0.8126476 | -0.299298147 | 0.13162017 | 5.023454e+00 | 2.500624e-02 | 2.098600e-01 | No | Yes | 663.794301 | 75.006304 | 834.807313 | 93.887452 | |
| ENSG00000132561 | 4147 | MATN2 | protein_coding | O00339 | FUNCTION: Involved in matrix assembly. {ECO:0000250}. | Alternative splicing;Coiled coil;Disulfide bond;EGF-like domain;Glycoprotein;Reference proteome;Repeat;Secreted;Signal | This gene encodes a member of the von Willebrand factor A domain containing protein family. This family of proteins is thought to be involved in the formation of filamentous networks in the extracellular matrices of various tissues. This protein contains five von Willebrand factor A domains. The specific function of this gene has not yet been determined. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:4147; | collagen-containing extracellular matrix [GO:0062023]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; calcium ion binding [GO:0005509]; extracellular matrix structural constituent [GO:0005201] | 11852232_matrilin-2, a filament-forming protein widely distributed in extracellular matrices. 12164922_matrilin-2 is expressed in normal skin by keratinocytes and fibroblasts and may thus contribute to cutaneous homeostasis. 12180907_study of interactions by which matrilin-2 can be integrated into extracellular filamentous networks 16401863_These results suggest that matrilin-2 may be a specific and clinically useful biomarker for discriminating between indolent and clinically aggressive pilocytic astrocytoma. 18328806_DeltaNp63/BMP-7 signaling pathway modulates wound healing process through the regulation of matrilin-2. 18386166_data indicate matrilin-2 is a novel basement membrane component in the liver, synthesized during sinusoidal 'capillarization' in cirrhosis & in hepatocellular carcinoma 19730683_Observational study of gene-disease association. (HuGE Navigator) 19834535_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26271809_Matrilin-2 induces post-burn inflammatory responses as an endogenous danger signal, partly through a TLR4-mediated mechanism 27105914_Taken together, our results showed that high-glucose-induced Matrilin-2 expression that was mediated by the TGF-beta1/Smad3 signaling pathway might play a role in Diabetic nephropathy (DN) pathogenesis and our finding provided a potential diagnostic and/or therapeutic target for DN. 27923659_Data show that the mRNA and protein levels of matrilin-2 were increased after irradiation treatment in both mouse lung tissue and human pulmonary alveolar epithelial cells (HPAEpiC). 28533472_findings showed that YopK binds to the cell surface-exposed endogenous MATN2 and that purified YopK protein strongly inhibits the bacterial adherence to HeLa cells 30408806_miR-202-5p could target MATN2 to induce M2 polarization involved in allergic rhinitis. 31493245_MiR-202-5p/MATN2 are associated with regulatory T-cells differentiation and function in allergic rhinitis. 32046115_Mechanistic Roles of Matrilin-2 and Klotho in Modulating the Inflammatory Activity of Human Aortic Valve Cells. 33633437_Extracellular matrix changes in corneal opacification vary depending on etiology. | ENSMUSG00000022324 | Matn2 | 53.117778 | 1.5103032 | 0.594838191 | 0.35509362 | 2.804401e+00 | 9.400593e-02 | No | Yes | 73.662944 | 21.722920 | 49.842899 | 14.621139 | ||
| ENSG00000132950 | 9205 | ZMYM5 | protein_coding | Q9UJ78 | FUNCTION: Functions as a transcriptional regulator. {ECO:0000269|PubMed:17126306}. | 3D-structure;Alternative splicing;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:9205; | nucleus [GO:0005634]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122] | ENSMUSG00000040123 | Zmym5 | 222.358303 | 1.1289470 | 0.174977741 | 0.17902738 | 9.633658e-01 | 3.263403e-01 | No | Yes | 227.260045 | 53.389535 | 195.089857 | 45.896118 | ||||
| ENSG00000132964 | 1024 | CDK8 | protein_coding | P49336 | FUNCTION: Component of the Mediator complex, a coactivator involved in regulated gene transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional pre-initiation complex with RNA polymerase II and the general transcription factors. Phosphorylates the CTD (C-terminal domain) of the large subunit of RNA polymerase II (RNAp II), which may inhibit the formation of a transcription initiation complex. Phosphorylates CCNH leading to down-regulation of the TFIIH complex and transcriptional repression. Recruited through interaction with MAML1 to hyperphosphorylate the intracellular domain of NOTCH, leading to its degradation. {ECO:0000269|PubMed:10993082, ECO:0000269|PubMed:15546612, ECO:0000269|PubMed:30905399}. | 3D-structure;ATP-binding;Activator;Alternative splicing;Autism spectrum disorder;Disease variant;Kinase;Mental retardation;Nucleotide-binding;Nucleus;Reference proteome;Repressor;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase | This gene encodes a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are known to be important regulators of cell cycle progression. This kinase and its regulatory subunit, cyclin C, are components of the Mediator transcriptional regulatory complex, involved in both transcriptional activation and repression by phosphorylation of the carboxy-terminal domain of the largest subunit of RNA polymerase II. This kinase regulates transcription by targeting the cyclin-dependent kinase 7 subunits of the general transcription initiation factor IIH, thus providing a link between the Mediator complex and the basal transcription machinery. Multiple pseudogenes of this gene have been identified. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2016]. | hsa:1024; | mediator complex [GO:0016592]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; ATP binding [GO:0005524]; cyclin-dependent protein serine/threonine kinase activity [GO:0004693]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA polymerase II CTD heptapeptide repeat kinase activity [GO:0008353]; protein phosphorylation [GO:0006468] | 17212659_CDK8 play positive roles in transcriptional activation. 17612495_RNA interference experiments demonstrate that CDK8 functions as a coactivator within the p53 transcriptional program. 18418385_Within T/G-Mediator, cdk8 phosphorylates serine-10 on histone H3, which in turn stimulates H3K14 acetylation by GCN5L within the complex. 18794899_by retaining RB1 and amplifying CDK8, colorectal tumour cells select conditions that collectively suppress E2F1 and enhance the activity of beta-catenin 18794900_CDK8 kinase activity was necessary for beta-catenin-driven transformation and for expression of several beta-catenin transcriptional targets 19047373_Med12--but not Med13--is essential for activating the CDK8 kinase. 19240132_Med12 and Med13 are critical for human CDK8 subcomplex -dependent repression, whereas CDK8 kinase activity is not 19790197_Overexpression of CDK8 is associated with beta-catenin activation and colon cancer. 20083228_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20098423_The authors show, using a human tumor cell line, that CDK8 is a positive regulator of genes within the serum response network, including several members of the activator protein 1 and early growth response family of oncogenic transcription factors. 20408451_plays a role in mechanisms of transcriptional regulation upon protein phosphorylation. (review) 20514474_CDK8 may identify a subset of colon cancer patients with poor prognosis. 21067790_The Walleye dermal sarcoma virus cyclin functions as a structural ortholog of cyclin C in spite of its limited amino acid sequence identity with C cyclins or with any known cyclins and activates Cdk8 and Cdk3. 21179167_significant inverse correlation between mH2A and CDK8 expression levels exists in melanoma patient samples 21344156_beta-catenin expression was suppressed by CDK8 interference in the gastric adenocarcinoma cell lines. 21806996_2.2-A crystal structure of CDK8/CycC in complex with sorafenib; the CDK8 activation loop appears not to be phosphorylated. Based on the structure, we discuss an alternate mode of CDK8 activation to the general CDK activation by T-loop phosphorylation 22117896_Microarrays identified target genes for each CDK, and we selected six genes: two target genes of CDK8, two target genes of CDK19 and two genes that were targets for both. 22345154_we identify a role for the CDK8 oncogene in regulating tumor differentiation and stem cell pluripotency 22379099_Retroviral cyclin enhances cyclin-dependent kinase-8 activity. 22869755_Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities 22945643_The phosphorylation of S375 by CDK8 regulates E2F1 ability to repress transcription of beta-catenin/TCF-dependent genes, as well as activation of E2F1-dependent genes. 23352233_CDK8 is a key regulator of STAT1 and antiviral responses. 23454913_results show a reverse correlation between CDK8 levels and several key features of the endometrial cancer cells, including cell proliferation, migration and invasion as well as tumor formation in vivo 23563140_Authors found that interaction of the CDK8 kinase module with Mediator's middle module interferes with C-terminal domain-dependent RNAPII binding to a previously unknown middle-module CTD-binding site and with the holoenzyme formation process. 23630251_analysis of the structure-kinetic relationship of the cyclin-dependent kinase 8 (CDK8)/cyclin C (CycC) complex 23746844_Study reports that HIF1A employs a specific variant of the Mediator complex to stimulate RNAPII elongation; the Mediator-associated kinase CDK8, but not the paralog CDK19, is required for induction of many HIF1A target genes. 23749998_CDK8 and CDK19, individually interact with PRMT5 and WDR77, and their interactions with PRMT5 cause transcriptional repression of C/EBPbeta target genes. 24009531_Here, loss of CDK8 suppressed the reduced mRNA expression and RNAPII occupancy levels of CTD truncation mutants. 24139904_CDK8 regulates the transcription factors turnover, C-terminal domain phosphorylation, and regulates the activator or repressor functions. (Review) 24439911_Cdk8 is required for stress-induced Mdv1-Dnm1 interaction and mitochondrial fragmentation. 24754906_Cancer-mediated CDK8 point mutations (D173A and D189N) change the binding pattern of cdk8 to its partner, CycC. 24840924_A novel transcriptional repression mechanism of hMED18 mediated by hCDK8 and further a novel positive role of free CDK/cyclin module in transcriptional activation is described. 25400821_Suggest that miR-107 plays a key role in cisplatin resistance by targeting the CDK8 protein in non small cell lung cancer cells. 25818643_The Skp2-mH2A1-CDK8 axis has a critical role in breast cancer development via dysregulation of the G2/M transition, polyploidy, cell growth dysregulation, and loss of tumor suppression. 26002960_Data suggest that, during neurogenesis, Mediator complex cyclin-dependent kinases (CDK8, CDK19) interact directly with PRC2 (polycomb repressive complex 2) subunit EZH2 (enhancer of zeste homolog 2), as well as SUZ12 (suppressor of zeste 12 homolog). 26006748_Current state of the art confirms that further development of CDK8 inhibitors will translate into targeted therapies in oncology 26042770_our results suggest that mTORC1 activation in NAFLD and insulin resistance results in down-regulation of the CDK8-CycC complex and elevation of lipogenic protein expression. 26182352_Data suggest that MED13, MED12, CDK8 and cyclin C (CycC) comprise a four-subunit 'kinase' module of the Mediator complex that functions as a major ingress of oncogenic and developmental signaling/gene expression in humans. [REVIEW] 26286725_miR-101 is a potent tumor repressor that directly represses CDK8 expression 26349978_our data provided evidence that CDK-8 played a significant role in colon cancer hepatic metastasis by regulating the Wnt/beta-catenin signal pathway 26416749_Mediator-associated kinases CDK8 and CDK19 restrain increased activation of key super-enhancer-associated genes in acute myeloid leukaemia (AML) cells 26452386_the expression of CDK8 and its interactive genes has a profound impact on the response to adjuvant therapy in breast cancer in accordance with the role of CDK8 in chemotherapy-induced tumor-supporting paracrine activities--{REVIEW} 26496706_Collectively, our results implicate Ste12 and Tec1 as general and important contributors to the Cdk8, RNAPII-CTD regulatory circuitry as it relates to the maintenance of genome integrity 27678455_Moreover, it identified CDK19 and CDK8 to be specifically overexpressed during prostate cancer progression, highlighting their potential as novel therapeutic targets in advanced prostate cancer. 28147342_These results identify CDK8 as a novel downstream mediator of estrogen receptors (ER) and suggest the utility of CDK8 inhibitors for ER-positive breast cancer therapy 28151579_On stimulation of TLR9, CDK8/19 positively regulates inflammatory gene transcription in cooperation with NF-kappaB and C/EBPbeta. 28220308_Our data suggest for the first time that CDK8 appears to contribute to the malignant mechanism of LSCC and may represent a significant prognostic marker for LSCC patients. 28805801_identification of the Mediator-associated kinase CDK8, and its paralog CDK19, as negative regulators of IL-10 production during innate immune activation 28855340_Genes coregulated by CDK8/19 and NFkappaB include IL8, CXCL1, and CXCL2. 29440396_These findings confirm that UF-linked mutations in MED12 disrupt composite Mediator-associated kinase activity and identify CDK8/19 as prospective therapeutic targets in uterine fibroids. 29856990_Compared with adjacent normal tissues, pancreatic cancer tissues showed upregulation of CDK8 expression, which was inversely correlated with T grade, liver metastasis, size, lymph node metastasis and poor survival. CDK8 overexpression promoted angiogenesis in pancreatic cancer via activation of the CDK8-beta-catenin-KLF2 signaling axis. 29967145_these observations suggest that Zyxin promotes colon cancer tumorigenesis in a mitotic-phosphorylation-dependent manner and through CDK8-mediated YAP activation. 30585107_The CDK8 may enable metabolic and transcriptional reprogramming through enhancers and chromatin looping. 30905399_missense mutations in CDK8 cause a developmental disorder that has phenotypic similarity to syndromes associated with mutations in other subunits of the Mediator kinase module 30967300_The present results obtained forcefully proved that miR-152-3p exhibited an antineoplastic activity via targeting CDK8 and might be served as a potential therapeutic target for the treatment of hepatocellular carcinoma 31044597_siRNA knockdown of CDK8 or CDK19 had no effect on HIV transcription. This result was confirmed using CDK8 or CDK19 inhibitors, Cortistatin A and Senexin A. these results indicate that CDK8 or CDK19 are not required for HIV transcription. 31087707_The microRNA-141-3p/ CDK8 pathway regulates the chemosensitivity of breast cancer cells to trastuzumab. 31628323_Cyclin dependent kinase 8 (CDK8) is essential for survival of BCR-ABL1p185+ leukemic cells. 31945608_MiR-297 alleviates LPS-induced A549 cell and mice lung injury via targeting cyclin dependent kinase 8. 31988137_The N-terminal portion of MED12 wraps around CDK8, whereby it positions an 'activation helix' close to the T-loop of CDK8 for its activation. 32183180_Suppression of a Subset of Interferon-Induced Genes by Human Papillomavirus Type 16 E7 via a Cyclin Dependent Kinase 8-Dependent Mechanism. 32407143_LncRNA HCP5 Regulates Pancreatic Cancer Progression by miR-140-5p/CDK8 Axis. 32560467_Cyclin-Dependent Kinases 8 and 19 Regulate Host Cell Metabolism during Dengue Virus Serotype 2 Infection. 32705162_Knockdown of SNHG16 suppresses the proliferation and induces the apoptosis of leukemia cells via miR193a5p/CDK8. 32945439_miR592 acts as an oncogene and promotes medullary thyroid cancer tumorigenesis by targeting cyclindependent kinase 8. 33067521_Pathogenesis of CDK8-associated disorder: two patients with novel CDK8 variants and in vitro and in vivo functional analyses of the variants. 33291686_Pharmacological Inhibition of CDK8 in Triple-Negative Breast Cancer Cell Line MDA-MB-468 Increases E2F1 Protein, Induces Phosphorylation of STAT3 and Apoptosis. 33357429_CDK8 Fine-Tunes IL-6 Transcriptional Activities by Limiting STAT3 Resident Time at the Gene Loci. 33577031_Long non-coding RNA HEIH modulates CDK8 expression by inhibiting miR-193a-5p to accelerate nasopharyngeal carcinoma progression. 33605551_De novo filament formation by human hair keratins K85 and K35 follows a filament development pattern distinct from cytokeratin filament networks. 33655711_LINC01224 accelerates malignant transformation via MiR-193a-5p/CDK8 axis in gastric cancer. 33727660_CDK8 maintains stemness and tumorigenicity of glioma stem cells by regulating the c-MYC pathway. 33818788_CircFAT1 facilitates cervical cancer malignant progression by regulating ERK1/2 and p38 MAPK pathway through miR-409-3p/CDK8 axis. 34056708_MED12 interacts with the heat-shock transcription factor HSF1 and recruits CDK8 to promote the heat-shock response in mammalian cells. 34329458_Loss of Cyclin C or CDK8 provides ATR inhibitor resistance by suppressing transcription-associated replication stress. 35181333_Inhibition of Cyclin-Dependent Kinase 8/Cyclin-Dependent Kinase 19 Suppresses Its Pro-Oncogenic Effects in Prostate Cancer. | ENSMUSG00000029635 | Cdk8 | 956.229060 | 1.0876768 | 0.121249948 | 0.09953837 | 1.472824e+00 | 2.249007e-01 | 5.989913e-01 | No | Yes | 941.337312 | 176.525686 | 869.927029 | 163.113362 | |
| ENSG00000133226 | 10250 | SRRM1 | protein_coding | Q8IYB3 | FUNCTION: Part of pre- and post-splicing multiprotein mRNP complexes. Involved in numerous pre-mRNA processing events. Promotes constitutive and exonic splicing enhancer (ESE)-dependent splicing activation by bridging together sequence-specific (SR family proteins, SFRS4, SFRS5 and TRA2B/SFRS10) and basal snRNP (SNRP70 and SNRPA1) factors of the spliceosome. Stimulates mRNA 3'-end cleavage independently of the formation of an exon junction complex. Binds both pre-mRNA and spliced mRNA 20-25 nt upstream of exon-exon junctions. Binds RNA and DNA with low sequence specificity and has similar preference for either double- or single-stranded nucleic acid substrates. {ECO:0000269|PubMed:10339552, ECO:0000269|PubMed:10668804, ECO:0000269|PubMed:11739730, ECO:0000269|PubMed:12600940, ECO:0000269|PubMed:12944400, ECO:0000269|PubMed:9531537}. | 3D-structure;Acetylation;Alternative splicing;Citrullination;DNA-binding;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Spliceosome;Ubl conjugation;mRNA processing;mRNA splicing | hsa:10250; | catalytic step 2 spliceosome [GO:0071013]; cytosol [GO:0005829]; nuclear matrix [GO:0016363]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; RNA binding [GO:0003723]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380]; RNA splicing, via transesterification reactions [GO:0000375] | 11991360_1H, 13C, and 15N resonance assignments and secondary structure of the PWI domain from SRm160 was determined using reduced dimensionality NMR. 12581738_Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins and plays a role in spliceosome assembly 12624182_two contiguous sequences that independently target SRm160 to nuclear matrix sites at splicing speckled domains: amino acids 300-350 and 351-688 15024032_Data show that SRm160, a splicing coactivator and component of the exon junction complex (EJC) involved in RNA export, has an adenosine triphosphate (ATP)-dependent mobility. 16159877_found that the majority of proteins identified in SRm160-containing complexes are associated with pre-mRNA processing. Interestingly, SRm160 is also associated with factors involved in chromatin regulation and sister chromatid cohesion 16354706_SRm160, a splicing coactivator, regulates CD44 alternative splicing in a Ras-dependent manner. 30590765_We discovered that two factors, SRRM1 and SF3B1, affect not only cis-SAGe chimeras, but also other types of chimeric RNAs in a genome-wide fashion. | ENSMUSG00000028809 | Srrm1 | 6138.321612 | 1.1016433 | 0.139657141 | 0.06098531 | 5.190031e+00 | 2.271681e-02 | 1.992960e-01 | No | Yes | 6833.162184 | 572.815399 | 6142.590041 | 515.013115 | ||
| ENSG00000133316 | 54663 | WDR74 | protein_coding | Q6RFH5 | FUNCTION: Regulatory protein of the MTREX-exosome complex involved in the synthesis of the 60S ribosomal subunit (PubMed:26456651). Participates in an early cleavage of the pre-rRNA processing pathway in cooperation with NVL (PubMed:29107693). Required for blastocyst formation, is necessary for RNA transcription, processing and/or stability during preimplantation development (By similarity). {ECO:0000250|UniProtKB:Q8VCG3, ECO:0000269|PubMed:26456651, ECO:0000269|PubMed:29107693}. | Alternative splicing;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:54663; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; preribosome, large subunit precursor [GO:0030687]; blastocyst formation [GO:0001825]; ribosomal large subunit biogenesis [GO:0042273]; RNA metabolic process [GO:0016070]; rRNA processing [GO:0006364] | 26456651_results suggest that WDR74 is a novel regulatory protein of the MTR4-exsosome complex whose interaction is regulated by NVL2 and is involved in ribosome biogenesis 29107693_knockdown of WDR74 leads to significant defects in the pre-rRNA cleavage within the internal transcribed spacer 1, occurring in an early stage of the processing pathway. When the dissociation of WDR74 from the MTR4-containing exonuclease complex was impaired upon expression of mutant NVL2, the same processing defect, with partial migration of WDR74 from the nucleolus towards the nucleoplasm, was observed. 30594465_Through direct interactions with Smad proteins, WDR74 enhances TGF-beta-mediated phosphorylation and nuclear accumulation of Smad2 and Smad3. 31838084_WDR74 induces nuclear beta-catenin accumulation and activates Wnt-responsive genes to promote lung cancer growth and metastasis. 32005977_WDR74 modulates melanoma tumorigenesis and metastasis through the RPL5-MDM2-p53 pathway. | ENSMUSG00000042729 | Wdr74 | 1522.365974 | 0.8684190 | -0.203536736 | 0.12911340 | 2.369222e+00 | 1.237487e-01 | 4.677066e-01 | No | Yes | 1386.154792 | 175.395616 | 1673.347008 | 211.668248 | ||
| ENSG00000133794 | 406 | ARNTL | protein_coding | O00327 | FUNCTION: Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. ARNTL/BMAL1 positively regulates myogenesis and negatively regulates adipogenesis via the transcriptional control of the genes of the canonical Wnt signaling pathway. Plays a role in normal pancreatic beta-cell function; regulates glucose-stimulated insulin secretion via the regulation of antioxidant genes NFE2L2/NRF2 and its targets SESN2, PRDX3, CCLC and CCLM. Negatively regulates the mTORC1 signaling pathway; regulates the expression of MTOR and DEPTOR. Controls diurnal oscillations of Ly6C inflammatory monocytes; rhythmic recruitment of the PRC2 complex imparts diurnal variation to chemokine expression that is necessary to sustain Ly6C monocyte rhythms. Regulates the expression of HSD3B2, STAR, PTGS2, CYP11A1, CYP19A1 and LHCGR in the ovary and also the genes involved in hair growth. Plays an important role in adult hippocampal neurogenesis by regulating the timely entry of neural stem/progenitor cells (NSPCs) into the cell cycle and the number of cell divisions that take place prior to cell-cycle exit. Regulates the circadian expression of CIART and KLF11. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The NPAS2-ARNTL/BMAL1 heterodimer positively regulates the expression of MAOA, F7 and LDHA and modulates the circadian rhythm of daytime contrast sensitivity by regulating the rhythmic expression of adenylate cyclase type 1 (ADCY1) in the retina. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence (PubMed:23229515). CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3' (PubMed:23229515). The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3' (PubMed:23229515). Essential for the rhythmic interaction of CLOCK with ASS1 and plays a critical role in positively regulating CLOCK-mediated acetylation of ASS1 (PubMed:28985504). Plays a role in protecting against lethal sepsis by limiting the expression of immune checkpoint protein CD274 in macrophages in a PKM2-dependent manner (By similarity). Regulates the diurnal rhythms of skeletal muscle metabolism via transcriptional activation of genes promoting triglyceride synthesis (DGAT2) and metabolic efficiency (COQ10B) (By similarity). {ECO:0000250|UniProtKB:Q9WTL8, ECO:0000269|PubMed:11441146, ECO:0000269|PubMed:12738229, ECO:0000269|PubMed:18587630, ECO:0000269|PubMed:23785138, ECO:0000269|PubMed:23955654, ECO:0000269|PubMed:24005054, ECO:0000269|PubMed:28985504}.; FUNCTION: (Microbial infection) Regulates SARS coronavirus-2/SARS-CoV-2 entry and replication in lung epithelial cells probably through the post-transcriptional regulation of ACE2 and interferon-stimulated gene expression. {ECO:0000269|PubMed:34545347}. | 3D-structure;Acetylation;Activator;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a basic helix-loop-helix protein that forms a heterodimer with CLOCK. This heterodimer binds E-box enhancer elements upstream of Period (PER1, PER2, PER3) and Cryptochrome (CRY1, CRY2) genes and activates transcription of these genes. PER and CRY proteins heterodimerize and repress their own transcription by interacting in a feedback loop with CLOCK/ARNTL complexes. Defects in this gene have been linked to infertility, problems with gluconeogenesis and lipogenesis, and altered sleep patterns. The protein regulates interferon-stimulated gene expression and is an important factor in viral infection, including COVID-19. [provided by RefSeq, Oct 2021]. | hsa:406; | aryl hydrocarbon receptor complex [GO:0034751]; chromatin [GO:0000785]; chromatoid body [GO:0033391]; CLOCK-BMAL transcription complex [GO:1990513]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; transcription regulator complex [GO:0005667]; aryl hydrocarbon receptor binding [GO:0017162]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; E-box binding [GO:0070888]; Hsp90 protein binding [GO:0051879]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; circadian regulation of gene expression [GO:0032922]; circadian rhythm [GO:0007623]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of glucocorticoid receptor signaling pathway [GO:2000323]; negative regulation of TOR signaling [GO:0032007]; negative regulation of transcription, DNA-templated [GO:0045892]; oxidative stress-induced premature senescence [GO:0090403]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of circadian rhythm [GO:0042753]; positive regulation of protein acetylation [GO:1901985]; positive regulation of skeletal muscle cell differentiation [GO:2001016]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of cell cycle [GO:0051726]; regulation of cellular senescence [GO:2000772]; regulation of hair cycle [GO:0042634]; regulation of insulin secretion [GO:0050796]; regulation of neurogenesis [GO:0050767]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; regulation of type B pancreatic cell development [GO:2000074]; response to redox state [GO:0051775]; spermatogenesis [GO:0007283] | 11875063_The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. 12897057_BMAL1 and CLOCK have roles in circadian system control 14750904_Transcripts of BMAL1 underwent circadian oscillation. 16474406_a molecular genetic screen in mammalian cells to identify mutants of the circadian transcriptional activators CLOCK and BMAL1. 16507006_Observational study of gene-disease association. (HuGE Navigator) 16528748_Observational study of gene-disease association. (HuGE Navigator) 16628007_the CLOCK(NPAS2)/BMAL1 complex is post-translationally regulated by cry1 and cry2 17274950_We have examined the circadian expression of clock genes in human leukocytes and found that Bmal1 mRNA showed weak rhythm. 17457720_variations associated with seasonal affective disorder 17728404_Observational study of gene-disease association. (HuGE Navigator) 17728404_rat models to human provides evidence of a causative role of Bmal1 variants in pathological components of the metabolic syndrome 17994337_CLOCK/BMAL1-mediated activation of PER1 by AP1 and E-Box elements is distinct from peripheral transcriptional modulation via cAMP-induced CREB and C/EBP. 18228528_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18411297_DEC1, along with DEC2, plays a role in the finer regulation and robustness of the molecular clock CLOCK/BMAL1 18430226_The regulators of clock-controlled transcription PER2, CRY1 and CRY2 differ in their capacity to interact with each single component of the BMAL1-CLOCK heterodimer and, in the case of BMAL1, also in their interaction sites. 18517031_Individual PER3 rhythms correlated significantly with sleep-wake timing and the timing of melatonin and cortisol, but those of PER2 and BMAL1 did not reach significance. 18587630_The transcription of human nocturnin gene displayed circadian oscillations in Huh7 cells (a human hepatoma cell line) and was regulated by CLOCK/BMAL1 heterodimer via the E-box of nocturnin promoter. 18663240_Several new polymorphisms in ARNTL gene are reported. 19277210_The shRNA barcode screening technique identified ARNTL as being involved in p53 regulation. cells having suppressed ARNTL are unable to arrest upon p53 activation associated with an inability to activate the p53 target gene p21(CIP1). 19296127_expression of BMAL1 is significantly associated with lymph node metastasis and poor prognosis in breast cancer 19328558_Observational study of gene-disease association. (HuGE Navigator) 19470168_Observational study of gene-disease association. (HuGE Navigator) 19693801_Observational study of gene-disease association. (HuGE Navigator) 19839995_Observational study of gene-disease association. (HuGE Navigator) 19861541_Epigenetic inactivation of the circadian clock gene BMAL1 is associated with hematologic malignancies. 19861640_BMAL1 mRNA levels were correlated only with age (beta = -.50, p < .001). 19912323_results suggest that a peripheral molecular clock, as reflected in the dampened expression of the clock genes BMAL1 in total leukocytes, is altered in Parkinson's disease patients 19913121_Observational study of gene-disease association. (HuGE Navigator) 19934327_Observational study of gene-disease association. (HuGE Navigator) 20072116_Observational study of gene-disease association. (HuGE Navigator) 20174623_Observational study of gene-disease association. (HuGE Navigator) 20180986_Observational study of gene-disease association. (HuGE Navigator) 20222832_There was no circadian rhythm of bmal1(brain and muscle ARNT-like 1) and cry1(cryptochrome 1) in PBMC of preterm neonates 20368993_ARNTL and NPAS2 SNPs were associated with reproduction and with seasonal variation. 20368993_Observational study of gene-disease association. (HuGE Navigator) 20446921_In this salivary gland cell line, there is a rhythm in the core oscillator components BMAL1 and REV-ERBalpha, an indication that circadian-based transcriptional regulation can be modelled in this peripheral cell type. 20554694_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20861012_ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression 21363938_Data indicate that fetal adrenal gland showed circadian expression of Bmal-1 and Per-2 as well as of Mt1 and Egr-1. 21380491_Overexpression of the Bmal1 gene and reduced expression of the Per1 gene may thus be useful predictors of liver metastasis. 21411511_Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. 21799909_rhythmic modulation of circulating microRNAs targeting Bmal1 21966465_Data conclude that BMAL1 is a crucial factor in the regulation of energy homeostasis, and disorders of the functions of BMAL1 lead to the development of metabolic syndrome. 21990077_Examines the effect of acute sleep deprivation under light conditions on the expression of hPer2 and hBmal1. 40-hour acute sleep deprivation under light conditions may affect the expression of hPer2 in PBMCs. 22105622_This study demonistraed that BMAL1 exhibited rhythmic expression in the control group, but not the Adult attention-deficit hyperactivity disorder group. 22217103_treatment with isoprenaline or dexamethasone induces circadian expression of hPer1, hPer2, hPer3, and hBMAL1 22510946_findings indicate that BMAL1 has a critical role in MPM and could serve as an attractive therapeutic target for MPM 22990020_Data indicate that SNPs in SERPINE1 and ARNTL and an nucleotide polymorphisms (SNPs) associated with the expression of MUC3 were robustly associated with circulating levels of PAI-1. 23003921_DNA methylation levels at different CpG sites of CLOCK, BMAL1 and PER2 genes were analyzed in sixty normal-weight, overweight and obese women following a 16-week weight reduction program. 23033538_Data indicate that Clock-Bmal1 regulates the response to glucocorticoids in peripheral tissues through acetylation of the glucocorticoid receptor (GR), possibly antagonizing the biologic actions of diurnally fluctuating circulating cortisol. 23129285_Expression of the CLOCK, BMAL1, and PER1 circadian genes in human oral mucosa cells as dependent on CLOCK gene polymorphic variants. 23206673_The data suggest that the impairment of the BMAL1 clock gene expression is closely associated with GDM susceptibility. 23229515_The phospho-mimicking S78E mutant of BMAL1 efficiently blocks DNA binding, which provides a molecular rationale for the possibility of rhythmic binding of CLOCK-BMAL1 during circadian cycle. 23323702_These data document for the first time that the expression of BMAL1, PER3, PPARD and CRY2 genes is altered in gestational diabetes compared to normal pregnant women. deranged expression of clock genes may play a pathogenic role in GDM. 23337503_this study demonstrated that BMAL1 was modified with an O-linked beta-N-acetylglucosamine (O-GlcNAc), which stabilized BMAL1 and enhanced its transcriptional activity. 23358160_The SNPs most strongly associated in the single-marker analysis of the combined Danish samples were rs4757144 in ARNTL (P=3.78 x 10-6) and rs8057927 in CDH13. 23395176_O-GlcNAc transferase (OGT) promotes expression of BMAL1/CLOCK target genes and affects circadian oscillation of clock genes in vitro and in vivo. 23449886_Variants in ARNTL have been associated with seasonality and seasonal affective disorder, phenotypes that could reflect circadian rhythm disruption. 23546644_Studies indicate that in the cytoplasm, PER3 protein heterodimerizes with PER1, PER2, CRY1, and CRY2 proteins and enters into the nucleus, resulting in repression of CLOCK-BMAL1-mediated transcription. 23563360_Bmal1 is a tumor suppressor, capable of suppressing cancer cell growth and invasiveness. 23584858_Data suggest that clock genes Per1, Cry1, Clock, and Bmal1 and their protein products may be directly involved in the daytime-dependent regulation and adaptation of hormone synthesis. 23606611_there is significant daily variation in PER2, PER3, and ARNTL1 expression with earlier timing of expression in women than in men 24005054_Knockdown of either BMAL1 or Period1 in human anagen hair follicles significantly prolonged anagen. 24103761_These results suggest that DNA methylation of the BMAL1 gene is critical for interfering with circadian rhythms. 24277452_The progression-free survival of patients with high Bmal1 expression is significantly longer than that of patients with low Bmal1 expression. 24418196_The rhythm of Bmal1 mRNA in human plaque-derived vascular smooth muscle cells is altered. 24636202_The results of this study suggest that the ARNTL gene may be associated with the lithium prophylactic response in bipolar illness. 25070164_There is not a significant difference in the expression of CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension regardless of patient genotype. 25175925_These results suggested that ARNTL may be a tumor suppressor and is epigenetically silenced in ovarian cancer. 25182847_Data indicate that the inner nuclear membrane protein MAN1 directly binds the transcription activator BMAL1 promoter and enhances its transcription. 25203321_FAL1 associates with the epigenetic repressor BMI1 and regulates its stability in order to modulate the transcription of a number of genes including CDKN1A. 25310406_PER1 and BMAL1 operate as cell-autonomous modulators of human pigmentation and may be targeted for future therapeutic strategies 25480789_temporal signals of fasting and refeeding hormones regulate the transcription of Bmal1, a key transcription activator of molecular clock, in the liver 25741730_Bmal1-dependent oscillators of arginine vasopressin neurons modulate the coupling of the suprachiasmatic nucleus. 25799324_rs2290036-C variant of ARNTL was over-represented in psychosis patients, and the variants rs934945-G and rs10462023-G of PER2 were associated with a more severe psychotic disorder 26134245_CLOCK, ARNTL, and NPAS2 gene polymorphisms may have a role in seasonal variations in mood and behavior 26164627_these findings suggest that sumoylation plays a critical role in the spatiotemporal co-activation of CLOCK-BMAL1 by CBP for immediate-early Per induction and the resetting of the circadian clock. 26168277_In men undergoing acute total sleep deprivation, BMAL1 gene expression was decreased in skeletal muscle compared with controls. 26247999_possible circadian rhythm in full-term placental expression 26283580_a 4-locus CSNK1E haplotype encompassing the rs1534891 SNP (Z-score=2.685, permuted p=0.0076) and a 3-locus haplotype in ARNTL (Z-score=3.269, permuted p=0.0011) showed a significant association with Bipolar Disorder 26370682_found that overexpression of both Clock and Bmal1 suppressed cell growth 26507264_ARNTL and PER1 were associated with PD. 26562283_Data suggest that cryptochromes mediate periodic binding of Ck2b (casein kinase 2beta) to Bmal1 (aryl hydrocarbon receptor nuclear translocator-like protein) and thus inhibit Bmal1-Ser90 phosphorylation by Ck2a (casein kinase 2alpha). [SYNOPSIS] 26657859_define a regulatory mechanism that links chondrocyte BMAL1 to the maintenance and repair of cartilage 26683776_Bmal1 could directly bind to the p53 gene promoter and thereby transcriptionally activate the downstream tumor suppressor pathway in a p53-dependent manner in pancreatic tumors. 26753996_Our results indicate that activation of TGF-beta1 promotes the transcriptional induction of BMAL1. 26782499_The BMAL1 rs2278749 T/C was associated with Alzheimer disease (AD) risk, and T carriers in BMAL1 rs2278749 T/C showed a higher risk of AD than did non-carriers. 26850841_when overexpressed, c-MYC is able to repress Per1 transactivation by BMAL1/CLOCK via targeting selective E-box sequences. Importantly, upon serum stimulation, MYC was detected in BMAL1 protein complexes 26873744_research describes an association between changes in the methylation of the BMAL1 gene with the intervention and the effects of a weight loss intervention on blood lipids levels 26915801_decreased expression of Bmal1 is correlated with tumor progression and poor prognosis in pancreatic ductal adenocarcinoma, with potential to be used as a biomarker for diagnosis and prognosis 27060253_Synchronized cells exhibit an autonomous ultradian mitochondrial respiratory activity which is abrogated by silencing the master clock gene BMAL1. 27117143_The level of BMAL1 expression in granulosa cells the polycystic ovary syndrome (PCOS) group was lower than that of the group without PCOS. We also analyzed estrogen synthesis and aromatase expression in KGN cell lines. Both were downregulated after BMAL1 and SIRT1 knock-down and, conversely, upregulated after overexpression treatments of these two genes in KGN cells. 27253997_Bmal1 is a key clock gene to involve in cartilage homeostasis mediated through sirt1. 27285754_PI3K-PTEN upregulated-mTORC1 and mTORC2 complex plays a critical role in controlling BMAL1, establishing a connection between PI3K signaling and the regulation of circadian rhythm, ultimately resulting in deregulated BMAL1 in tumor cells with disrupted PI3K signaling 27373683_TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1 27739341_Methylation at cg05733463 of ARNTL was significantly higher in bipolar disorder (BD) than in controls. Thus, the study suggests that altered epigenetic regulation of ARNTL might provide a mechanistic basis for better understanding circadian rhythms and mood swings in BD. 27821487_our results identified Bmal1 as a novel tumor suppressor gene that elevates the sensitivity of cancer cells to paclitaxel, with potential implications as a chronotherapy timing biomarker in tongue squamous cell carcinoma 27883893_Study found rhythmic methylation of BMAL1 was altered in Alzheimer's disease brains and fibroblasts and correlated with transcription cycles. Results indicate that cycles of DNA methylation contribute to the regulation of BMAL1 rhythms in the brain. 27884645_NR1D1 and BMAL1 mRNA and protein levels were significantly reduced in OA compared to normal cartilage. In cultured human chondrocytes, a clear circadian rhythmicity was observed for NR1D1 and BMAL1. 27913791_this study shows that BMAL1 can regulate cellular innate immunity against specific RNA viruses 28487473_These results suggest that the circadian clock system can be recovered through BMAL1 expression induced by aza-dC within a day. 28543681_M. tuberculosis infection caused enhanced MMP-1, -9, and miR-223 expression, with inhibited BMAL1 expression. MiR-223 modulated BMAL1 expression via the direct binding of BMAL1 3'-UTR. 28708003_ARNTL rs7107287 was associated with a cyclothymic temperament, depressive and stress symptoms 29172799_lost BMAL1 and Ki-67 overexpression are associated with poor overall survival of nasopharyngeal carcinoma patients 29217191_Determined a novel role of TNF-alpha in inducing Bmal1 via dual calcium dependent pathways; Roralpha was up-regulated in the presence of Ca(2+) influx and Rev-erbalpha was down-regulated in the absence of that. 29276151_BMAL1 Deficiency Contributes to Mandibular Dysplasia by Upregulating MMP3. 29324865_results of this study suggest that genetic variability in the ARNTL and CLOCK genes might be associated with risk for multiple sclerosis 29396463_YY1 transcriptionally activated miR-135b and formed a 'miR-135b-BMAL1-YY1' loop, which holds significant predictive and prognostic value for patients with pancreatic cancer. 29508277_It identified BMAL1 as a cAMP-responsive coactivator of HDAC5 to regulate hepatic gluconeogenesis 29871923_This work identifies UBE2O as a critical regulator in the ubiquitin-proteasome system, which modulates BMAL1 transcriptional activity and circadian function by promoting BMAL1 ubiquitination and degradation under normal physiological conditions. 29935055_Ubiquitin E3 ligase TRAF2 reduces BMAL1 protein level.BMAL1 interacts with the zinc finger domain in TRAF2.TRAF2 promotes BMAL1 ubiquitination and degradation.TRAF2 attenuates the BMAL1 transcriptional activity. 29943823_the messenger RNA expression of brain and muscle ARNT-like 1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK) genes is altered by low doses (5 mJ/cm(2) ) of ultraviolet B in the immortalized HaCat human keratinocyte cell line. 30005017_The basal level of HIV transcription on antiretroviral therapy can vary significantly and is modulated by the circadian regulator BMAL-1, amongst other factors. 30195196_A two-way ANOVA revealed significant differences between the clock genes profiles of controls and RBD patients for hPer1 (P = 0.0324), hPer2 (P = 0.0039), hBmal1 (P = 0.0160), and hNr1d1 (P = 0.0046). 30287810_our present research illustrates a novel insight into the circadian clock proteins, CLOCK and BMAL1, which can promote the proliferation, migration, and invasion of cancer cells by affecting the formation of F-actin. 30316844_study revealed that TP, as a Bmal1-enhancing natural compound, alleviated redox imbalance via strengthening Keap1/Nrf2 antioxidant defense pathway and ameliorating mitochondrial dysfunction in a Bmal1-dependent manner 30375470_we found that knockout of BMAL1 gene in two mammalian cells affects the apoptotic response and invasion properties differently in respect to the contribution to carcinogenesis. 30502053_Study found that H. pylori up-regulated the expression of LIN28A which directly bound to BMAL1 promoter, activating the transcription of BMAL1 and increases its expression. BMAL1 in turn promoted transcription of TNF-alpha by directly binding to the E-box elements on its promoter to increase its secretion. 30523262_these results suggest a role for hBMAL1a protein isoform as a negative regulator of the mammalian molecular clock 30621723_GSEA assay found that ARNTL was associated with cell cycle and ectopic ARNTL overexpression could induce G2-M phase arrest. 30843665_The study indicates that SNPs in BMAL1 genes might contribute to the clinical outcome of gastric cancer through their impact on gene expression. 30943793_PER2 showed relatively stable oscillations compared with BMAL1. 30985884_Impaired decidualization caused by downregulation of circadian clock gene BMAL1 contributes to human recurrent miscarriagedagger. 31099187_PIWIL1 suppresses circadian rhythms through GSK3beta-induced phosphorylation and degradation of CLOCK and BMAL1 in cancer cells. 31244920_Bmal1 coordinates temporal expressions of DBP (a MRP2 activator), REV-ERBalpha (an E4BP4 repressor) and E4BP4 (a MRP2 repressor), generating diurnal MRP2 expression. 31252075_Study showed that abnormal expression levels of Per2, Clock, and Bmal1 were detected in patients with traumatic brain injury-related sleep disorders. 31300350_We identified BMAL1 as a potential marker and functional player in resistance to clinically-in-use VEGFA-neutralizing antibody (bevacizumab) in preclinical, clinical and cellular mechanistic studies of colorectal cancer. 31355331_our findings reveal a new pathway, initiated by the autophagic removal of ARNTL, that facilitates ferroptosis induction. 31580742_rs3816360 and rs2290035 Polymorphisms in BMAL1 Genes are associated with Risk of Lung Cancer. 31612734_Association of insulin resistance with polymorphic variants of Clock and Bmal1 genes: A case-control study. 31919052_Circadian Regulator CLOCK Recruits Immune-Suppressive Microglia into the GBM Tumor Microenvironment. 32041778_The E3 ubiquitin ligase STUB1 attenuates cell senescence by promoting the ubiquitination and degradation of the core circadian regulator BMAL1. 32225100_MYC-Associated Factor MAX is a Regulator of the Circadian Clock. 32231148_BMAL1 Suppresses Proliferation, Migration, and Invasion of U87MG Cells by Downregulating Cyclin B1, Phospho-AKT, and Metalloproteinase-9. 32284354_BMAL1 coordinates energy metabolism and differentiation of pluripotent stem cells. 32316196_Extracellular Acidosis Promotes Metastatic Potency via Decrease of the BMAL1 Circadian Clock Gene in Breast Cancer. 32319143_ARNT-dependent CCR8 reprogrammed LDH isoform expression correlates with poor clinical outcomes of prostate cancer. 32506468_Circadian genes polymorphisms, night work and prostate cancer risk: Findings from the EPICAP study. 32522976_The circadian clock gene Bmal1 facilitates cisplatin-induced renal injury and hepatization. 32784474_Clock Protein Bmal1 and Nrf2 Cooperatively Control Aging or Oxidative Response and Redox Homeostasis by Regulating Rhythmic Expression of Prdx6. 32858102_ARID1A-dependent permissive chromatin accessibility licenses estrogen-receptor signaling to regulate circadian rhythms genes in endometrial cancer. 32925059_Epigenetic Regulation of BMAL1 with Sleep Disturbances and Alzheimer's Disease. 33106415_The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. 33111200_Transcriptome analysis of the circadian clock gene BMAL1 deletion with opposite carcinogenic effects. 33114015_Elevated CLOCK and BMAL1 Contribute to the Impairment of Aerobic Glycolysis from Astrocytes in Alzheimer's Disease. 33374803_Proinflammatory Cytokines Perturb Mouse and Human Pancreatic Islet Circadian Rhythmicity and Induce Uncoordinated beta-Cell Clock Gene Expression via Nitric Oxide, Lysine Deacetylases, and Immunoproteasomal Activity. 33788000_Alterations in Rev-ERBalpha/BMAL1 ratio and glycated hemoglobin in rotating shift workers: the EuRhythDia study. 33791812_Clock gene Bmal1 controls diurnal rhythms in expression and activity of intestinal carboxylesterase 1. 33792447_The circadian clock gene Bmal1: Role in COVID-19 and periodontitis. 33799903_Genetic Association of Hepatitis C-Related Mixed Cryoglobulinemia: A 10-Year Prospective Study of Asians Treated with Antivirals. 33888702_Circadian clock dysfunction in human omental fat links obesity to metabolic inflammation. 34065633_BMAL1 Knockdown Leans Epithelial-Mesenchymal Balance toward Epithelial Properties and Decreases the Chemoresistance of Colon Carcinoma Cells. 34124933_Rewiring of Lactate-Interleukin-1beta Autoregulatory Loop with Clock-Bmal1: a Feed-Forward Circuit in Glioma. 34160901_Circadian clock protein BMAL1 regulates melanogenesis through MITF in melanoma cells. 34261484_Decreased expression of the clock gene Bmal1 is involved in the pathogenesis of temporal lobe epilepsy. 34481895_IL-1beta induces changes in expression of core circadian clock components PER2 and BMAL1 in primary human chondrocytes through the NMDA receptor/CREB and NF-kappaB signalling pathways. 34515147_Downregulation of Arntl mRNA Expression in Women with Hypertension: A Case-Control Study. 34542664_L-Theanine inhibits melanoma cell growth and migration via regulating expression of the clock gene BMAL1. 34727291_BMAL1 induces colorectal cancer metastasis by stimulating exosome secretion. 34807912_NF-kappaB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1. 34815366_BMAL1 may be involved in angiogenesis and peritumoral cerebral edema of human glioma by regulating VEGF and ANG2. 35112498_Mutational scanning identified amino acids of the CLOCK exon 19-domain essential for circadian rhythms. 35263812_Effects of BMAL1 on dentinogenic differentiation of dental pulp stem cells via PI3K/Akt/mTOR pathway. | ENSMUSG00000055116 | Arntl | 102.546282 | 1.2362135 | 0.305927978 | 0.30228719 | 1.025053e+00 | 3.113235e-01 | No | Yes | 127.040258 | 27.914920 | 108.199927 | 23.720838 | ||
| ENSG00000134138 | 4212 | MEIS2 | protein_coding | O14770 | FUNCTION: Involved in transcriptional regulation. Binds to HOX or PBX proteins to form dimers, or to a DNA-bound dimer of PBX and HOX proteins and thought to have a role in stabilization of the homeoprotein-DNA complex. Isoform 3 is required for the activity of a PDX1:PBX1b:MEIS2b complex in pancreatic acinar cells involved in the transcriptional activation of the ELA1 enhancer; the complex binds to the enhancer B element and cooperates with the transcription factor 1 complex (PTF1) bound to the enhancer A element; MEIS2 is not involved in complex DNA-binding. Probably in complex with PBX1, is involved in transcriptional regulation by KLF4. Isoform 3 and isoform 4 can bind to a EPHA8 promoter sequence containing the DNA motif 5'-CGGTCA-3'; in cooperation with a PBX protein (such as PBX2) is proposed to be involved in the transcriptional activation of EPHA8 in the developing midbrain. May be involved in regulation of myeloid differentiation. Can bind to the DNA sequence 5'-TGACAG-3'in the activator ACT sequence of the D(1A) dopamine receptor (DRD1) promoter and activate DRD1 transcription; isoform 5 cannot activate DRD1 transcription. {ECO:0000269|PubMed:10764806, ECO:0000269|PubMed:11279116, ECO:0000269|PubMed:21746878}. | 3D-structure;Activator;Alternative splicing;Cytoplasm;DNA-binding;Developmental protein;Disease variant;Homeobox;Mental retardation;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene encodes a homeobox protein belonging to the TALE ('three amino acid loop extension') family of homeodomain-containing proteins. TALE homeobox proteins are highly conserved transcription regulators, and several members have been shown to be essential contributors to developmental programs. Multiple transcript variants encoding distinct isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:4212; | chromatin [GO:0000785]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription factor binding [GO:0008134]; animal organ morphogenesis [GO:0009887]; brain development [GO:0007420]; embryonic pattern specification [GO:0009880]; eye development [GO:0001654]; negative regulation of myeloid cell differentiation [GO:0045638]; negative regulation of transcription by RNA polymerase II [GO:0000122]; pancreas development [GO:0031016]; positive regulation of cardiac muscle myoblast proliferation [GO:0110024]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of mitotic cell cycle [GO:0045931]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; response to growth factor [GO:0070848]; response to mechanical stimulus [GO:0009612]; visual learning [GO:0008542] | 18408019_Analysis of an Affymetrix data set in the public domain showed high expression of MEIS1 in human endometrium 18973687_co-expression of PBX1 and MEIS1/2 in granulosa cells in normal human ovaries suggested that MEIS1/2 might control PBX1 sublocalization, as seen in other systems 19559479_co-operation between TLX1 and MEIS proteins may have a significant role in T-cell leukemogenesis. 19584346_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20195266_Clinical trial and genome-wide association study of gene-disease association. (HuGE Navigator) 20195266_The Single Nucleotide Polymorphism in Meis homeobox 2 (MEIS2) mediated the effects of risperidone on hip circumference (q=0.004). 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20523026_Data demonstrate by in situ hybridization and immunohistochemistry that the two homeobox genes Pax6 and MEIS2 are expressed during early fetal brain development in humans. 20553494_This work suggests that the transcriptional activity of all members of the Meis/Prep Hth protein family is subject to autoinhibition by their Hth domains, and that the Meis3.2 splice variant encodes a protein that bypasses this autoinhibitory effect. 21368052_Transgenic mice lacking microRNAs miR-9-2 and miR-9-3 exhibit multiple defects of telencephalic structures which may be brought about by dysregulation of Foxg1, Nr2e1, Gsh2, and Meis2 expression. 21746878_Klf4 recruits a complex of Meis and Pbx proteins to DNA, resulting in Meis2 transcriptional activation domain-dependent activation of a subset of Klf4 target genes. 23541832_The data suggest existence of a complex regulatory pathway in the trabecular meshwork part of which includes interactions between FOXC1, miR-204, MEIS2, and ITGbeta1. 24678003_Our results show that MEIS2 is a gene needed for palate closure. In syndromic cases of cleft palate, MEIS2 should be considered among the candidate genes, for example, in cases without 22q11.2 deletions. 25210800_FOXM1 is a direct target gene of MEIS2 and is required for MEIS2 to upregulate mitotic genes. 25712757_This is the first report showing a de novo small intragenic mutation in MEIS2 and further confirms the important role of this gene in normal development. 26512644_Meis2 as the target of miR-134 in the regulation of cardiomyocyte progenitor cell proliferation. 27225850_we have identified a novel nonsense MEIS2 mutation in a female patient with cleft palate, cardiac septal defect, severe ID, developmental delay and feeding difficulty with gastro-esophageal reflux. Our findings strongly suggest neurocristopathy be included in the clinical features associated with MEIS2 alterations. 27346355_High expression of MEIS2 impairs repressive DNA binding of AML1-ETO, inducing increased expression of genes such as the druggable proto-oncogene YES1. 29337667_Mechanistic analyses integrate this unrecognized anti-atrial function of ISL1 with known and newly identified atrial inducers. In this revised view, ISL1 is antagonized by retinoic acid signaling via a novel player, MEIS2. 29382709_Moderate elevation of MEIS2A expression reduced proliferation of MYCN-amplified human neuroblastoma cells, induced neuronal differentiation and impaired the ability of these cells to form tumors in mice 29716922_These data implicate a functional role for MEIS proteins in regulating cancer progression, and support a hypothesis whereby tumor expression of MEIS1 and MEIS2 expression confers a more indolent prostate cancer phenotype, with a decreased propensity for metastatic progression 30055086_these are the first described de novo missense variants in MEIS2, expanding the known mutation spectrum of the newly recognized human disorder caused by aberrations in this gene. 30291340_Loss-of-function MEIS2 mutations were identified in nine patients with palatal defects, congenital heart defects, and intellectual disability. 30526668_TAL1 acts as a downstream gene mediating the function of MEIS2 during early hematopoiesis. 30594396_The present study indicated that Meis2 repress the osteoblastic transdifferentiation of aortic valve interstitial cells through the Notch1/Twist1 signaling pathway. 30742945_Study shows that MEIS2 expression is regulated in bladder neoplasm via alternative splicing by PTBP1. 30859572_MEIS2 might be involved in the Wnt/beta-catenin pathway. 31115559_Study demonstrated that MEIS2 acted as a promoter of metastasis in colorectal cancer (CRC). Knockdown of MEIS2 significantly suppressed CRC migration, invasion and EMT. MEIS2 was associated with a shorter overall survival time for patients with CRC. These results suggest that MEIS2 may serve as a novel biomarker for CRC. 31315484_Inhibition of Senescence-Associated Genes Rb1 and Meis2 in Adult Cardiomyocytes Results in Cell Cycle Reentry and Cardiac Repair Post-Myocardial Infarction. 31623651_MEISC/D promote hepatocellular carcinoma development via Wnt/beta-catenin and Hippo/YAP signaling pathways 31640805_Results strongly indicate that aberrant DNA hypermethylation is associated with epigenetic silencing of MEIS2 transcriptional expression in prostate cancer (PC) and validate MEIS2 as a potential prognostic biomarker for PC. 33091211_MEIS2 sequence variant in a child with intellectual disability and cardiac defects: Expansion of the phenotypic spectrum and documentation of low-level mosaicism in an unaffected parent. 33427397_Intellectual disability associated with craniofacial dysmorphism, cleft palate, and congenital heart defect due to a de novo MEIS2 mutation: A clinical longitudinal study. 33864110_Expression of ISL1 and its partners in prostate cancer progression and neuroendocrine differentiation. 33975520_Meis homeobox 2 (MEIS2) inhibits the proliferation and promotes apoptosis of thyroid cancer cell and through the NF-kappaB signaling pathway. | ENSMUSG00000027210 | Meis2 | 395.212122 | 0.9717374 | -0.041361617 | 0.14111922 | 8.572424e-02 | 7.696849e-01 | No | Yes | 457.227411 | 52.691165 | 469.265186 | 54.087501 | ||
| ENSG00000134278 | 56907 | SPIRE1 | protein_coding | Q08AE8 | FUNCTION: Acts as an actin nucleation factor, remains associated with the slow-growing pointed end of the new filament (PubMed:11747823, PubMed:21620703). Involved in intracellular vesicle transport along actin fibers, providing a novel link between actin cytoskeleton dynamics and intracellular transport (PubMed:11747823). Required for asymmetric spindle positioning and asymmetric cell division during meiosis (PubMed:21620703). Required for normal formation of the cleavage furrow and for polar body extrusion during female germ cell meiosis (PubMed:21620703). Also acts in the nucleus: together with FMN2, promotes assembly of nuclear actin filaments in response to DNA damage in order to facilitate movement of chromatin and repair factors after DNA damage (PubMed:26287480). {ECO:0000269|PubMed:11747823, ECO:0000269|PubMed:21620703, ECO:0000269|PubMed:26287480}. | 3D-structure;Acetylation;Actin-binding;Alternative splicing;Cell membrane;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | Spire proteins, such as SPIRE1, are highly conserved between species. They belong to the family of Wiskott-Aldrich homology region-2 (WH2) proteins, which are involved in actin organization (Kerkhoff et al., 2001 [PubMed 11747823]).[supplied by OMIM, Mar 2008]. | hsa:56907; | cell cortex [GO:0005938]; cytoplasmic vesicle membrane [GO:0030659]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; integral component of mitochondrial outer membrane [GO:0031307]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; actin binding [GO:0003779]; actin cytoskeleton organization [GO:0030036]; actin filament network formation [GO:0051639]; actin filament polymerization [GO:0030041]; actin nucleation [GO:0045010]; cleavage furrow formation [GO:0036089]; establishment of meiotic spindle localization [GO:0051295]; formin-nucleated actin cable assembly [GO:0070649]; Golgi vesicle transport [GO:0048193]; intracellular transport [GO:0046907]; polar body extrusion after meiotic divisions [GO:0040038]; positive regulation of double-strand break repair [GO:2000781]; positive regulation of mitochondrial fission [GO:0090141]; protein transport [GO:0015031]; vesicle-mediated transport [GO:0016192] | 18042452_The multifunctional character of the WH2 domains allows Spire to sequester four G-actin subunits binding cooperatively in a tight SA(4) complex and to nucleate, sever, and cap filaments at their barbed ends. 19605360_both mammalian Spir proteins, Spir-1 and Spir-2, interact with mammalian Fmn subgroup proteins formin-1 and formin-2 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21705804_analysis of the molecular basis of the Spir1/formin-2 interaction 24213528_Spire-1 is specifically recruited at invadosomes and is part of a multi-molecular complex containing Src kinase, the formin mDia1 and actin. 24586110_Spire recruits Fmn2 and facilitates its association with actin filaments barbed ends. 26287480_This DNA damage-induced nuclear actin assembly requires two biologically and physically linked nucleation factors: Formin-2 and Spire-1/Spire-2. 26305500_The authors propose Spire1C isoform cooperates with INF2 to regulate actin assembly at endoplasmic reticulum-mitochondrial contacts. 26668326_Data suggest formin homology domain (FH2) of Fmn2 binds actin at filament barbed end as weak capper and plays a role in displacing WASP homology domain 2 (WH2) domains of Spire-1 from actin; competitive binding of Fmn2 vs Spire-1 aids actin assembly. 27627128_interferes with bacterial binding in Salmonella typhimurium host cell invasion 34863993_Lnc-SMaRT Translational Regulation of Spire1, A New Player in Muscle Differentiation. 35084586_Spire1 and Myosin Vc promote Ca(2+)-evoked externalization of von Willebrand factor in endothelial cells. | ENSMUSG00000024533 | Spire1 | 1220.588662 | 0.7396576 | -0.435070516 | 0.11733762 | 1.390869e+01 | 1.919091e-04 | 1.212317e-02 | No | Yes | 1296.709604 | 233.365887 | 1768.708146 | 318.344286 | |
| ENSG00000134897 | 54841 | BIVM | protein_coding | Q86UB2 | Mouse_homologues NA; + ;NA | Alternative splicing;Cytoplasm;Nucleus;Reference proteome | Mouse_homologues NA; + ;NA | hsa:54841; | cytoplasm [GO:0005737]; extracellular space [GO:0005615]; nucleus [GO:0005634] | 19773279_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000026048+ENSMUSG00000041684 | Ercc5+Bivm | 786.945578 | 1.1924146 | 0.253885915 | 0.10354742 | 6.001103e+00 | 1.429694e-02 | 1.578759e-01 | No | Yes | 819.226124 | 128.019462 | 693.505157 | 108.429129 | |
| ENSG00000135119 | 84900 | RNFT2 | protein_coding | Q96EX2 | Alternative splicing;Membrane;Metal-binding;Reference proteome;Transmembrane;Transmembrane helix;Zinc;Zinc-finger | hsa:84900; | integral component of membrane [GO:0016021]; metal ion binding [GO:0046872]; ubiquitin protein ligase activity [GO:0061630]; positive regulation of ERAD pathway [GO:1904294] | 31990690_The RNFT2/IL-3Ralpha axis regulates IL-3 signaling and innate immunity. 33517265_Tissue RNFT2 Expression Levels Are Associated With Peritoneal Recurrence and Poor Prognosis in Gastric Cancer. | ENSMUSG00000032850 | Rnft2 | 120.577869 | 0.8096460 | -0.304636784 | 0.25017538 | 1.468360e+00 | 2.256046e-01 | No | Yes | 113.098507 | 16.675905 | 143.693339 | 20.846043 | ||||
| ENSG00000135362 | 79899 | PRR5L | protein_coding | Q6MZQ0 | FUNCTION: Associates with the mTORC2 complex that regulates cellular processes including survival and organization of the cytoskeleton (PubMed:17461779). Regulates the activity of the mTORC2 complex in a substrate-specific manner preventing for instance the specific phosphorylation of PKCs and thereby controlling cell migration (PubMed:22609986). Plays a role in the stimulation of ZFP36-mediated mRNA decay of several ZFP36-associated mRNAs, such as TNF-alpha and GM-CSF, in response to stress (PubMed:21964062). Required for ZFP36 localization to cytoplasmic stress granule (SG) and P-body (PB) in response to stress (PubMed:21964062). {ECO:0000269|PubMed:17461779, ECO:0000269|PubMed:21964062, ECO:0000269|PubMed:22609986}. | Alternative splicing;Phosphoprotein;Reference proteome;Signal transduction inhibitor;Ubl conjugation | hsa:79899; | TORC2 complex [GO:0031932]; ubiquitin protein ligase binding [GO:0031625]; cellular response to oxidative stress [GO:0034599]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of signal transduction [GO:0009968]; positive regulation of intracellular protein transport [GO:0090316]; positive regulation of mRNA catabolic process [GO:0061014]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein phosphorylation [GO:0001934]; regulation of fibroblast migration [GO:0010762]; TORC2 signaling [GO:0038203] | 17461779_It was demonstrated that immunoprecipitation of Protor-1 or Protor-2 results in the co-immunoprecipitation of other mTORC2 subunits, but not Raptor, a specific component of mTORC1. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21964062_Protor-2 associates with tristetraprolin (TTP) to accelerate TTP-mediated mRNA turnover and functionally links the control of TTP-regulated mRNA stability to mTORC2 activity | ENSMUSG00000032841 | Prr5l | 39.691119 | 1.5222117 | 0.606169048 | 0.47105034 | 1.555916e+00 | 2.122642e-01 | No | Yes | 49.779839 | 12.384470 | 36.280667 | 9.138867 | |||
| ENSG00000135452 | 6302 | TSPAN31 | protein_coding | Q12999 | Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. This encoded protein is thought to be involved in growth-related cellular processes. This gene is associated with tumorigenesis and osteosarcoma. [provided by RefSeq, Jul 2008]. | hsa:6302; | integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; positive regulation of cell population proliferation [GO:0008284] | 15024701_CDK4, MDM2, SAS and GLI genes are amplified in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 32207169_TSPAN31 suppresses cell proliferation in human cervical cancer through down-regulation of its antisense pairing with CDK4. | ENSMUSG00000006736 | Tspan31 | 655.706826 | 0.8265409 | -0.274841909 | 0.11991838 | 5.161203e+00 | 2.309690e-02 | 2.004454e-01 | No | Yes | 613.234382 | 94.097474 | 744.355998 | 113.973677 | ||
| ENSG00000135740 | 6553 | SLC9A5 | protein_coding | Q14940 | FUNCTION: Involved in pH regulation to eliminate acids generated by active metabolism or to counter adverse environmental conditions. Major proton extruding system driven by the inward sodium ion chemical gradient. Plays an important role in signal transduction (By similarity). {ECO:0000250}. | Antiport;Glycoprotein;Ion transport;Membrane;Phosphoprotein;Reference proteome;Sodium;Sodium transport;Transmembrane;Transmembrane helix;Transport | hsa:6553; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; potassium:proton antiporter activity [GO:0015386]; sodium:proton antiporter activity [GO:0015385]; ion transport [GO:0006811]; potassium ion transmembrane transport [GO:0071805]; regulation of intracellular pH [GO:0051453]; sodium ion import across plasma membrane [GO:0098719] | 12205089_observations demonstrate that NHE5 is localized to the recycling endosomal pathway and is dynamically regulated by phosphatidylinositol 3'-kinase and by the state of F-actin assembly 15699339_beta-Arrestins bind and decrease cell-surface abundance of the Na+/H+ exchanger NHE5 isoform. 16920332_RACK1 activates NHE5 both by integrin-dependent and independent pathways, which may coordinate cellular ion homeostasis during cell-matrix adhesion. 19276089_SCAMP2 regulates NHE5 transit through recycling endosomes and promotes its surface targeting in an Arf6-dependent manner. 21296876_discrete elements of an elaborate sorting signal in NHE5 contribute to beta-arrestin2 binding and trafficking along the recycling endosomal pathway. 21551074_overexpression of NHE5 inhibits spine growth in response to neuronal activity 24936055_These data reveal a unique role for AMPK and NHE5 in regulating the pH homeostasis of hippocampal neurons during metabolic stress. 31595389_High NHE5 expression is associated with glioma. | ENSMUSG00000014786 | Slc9a5 | 176.298158 | 0.9556931 | -0.065380623 | 0.20543877 | 1.023313e-01 | 7.490497e-01 | No | Yes | 160.025448 | 24.496068 | 171.701837 | 26.209046 | |||
| ENSG00000135924 | 3300 | DNAJB2 | protein_coding | P25686 | FUNCTION: Functions as a co-chaperone, regulating the substrate binding and activating the ATPase activity of chaperones of the HSP70/heat shock protein 70 family (PubMed:7957263, PubMed:22219199). In parallel, also contributes to the ubiquitin-dependent proteasomal degradation of misfolded proteins (PubMed:15936278, PubMed:21625540). Thereby, may regulate the aggregation and promote the functional recovery of misfolded proteins like HTT, MC4R, PRKN, RHO and SOD1 and be crucial for many biological processes (PubMed:12754272, PubMed:20889486, PubMed:21719532, PubMed:22396390, PubMed:24023695). Isoform 1 which is localized to the endoplasmic reticulum membranes may specifically function in ER-associated protein degradation of misfolded proteins (PubMed:15936278). {ECO:0000269|PubMed:12754272, ECO:0000269|PubMed:15936278, ECO:0000269|PubMed:20889486, ECO:0000269|PubMed:21625540, ECO:0000269|PubMed:21719532, ECO:0000269|PubMed:22219199, ECO:0000269|PubMed:22396390, ECO:0000269|PubMed:24023695, ECO:0000269|PubMed:7957263}. | 3D-structure;Acetylation;Alternative splicing;Chaperone;Cytoplasm;Endoplasmic reticulum;Lipoprotein;Membrane;Methylation;Neurodegeneration;Nucleus;Phosphoprotein;Prenylation;Reference proteome;Repeat;Ubl conjugation | This gene is almost exclusively expressed in the brain, mainly in the neuronal layers. It encodes a protein that shows sequence similarity to bacterial DnaJ protein and the yeast homologs. In bacteria, this protein is implicated in protein folding and protein complex dissociation. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jul 2011]. | hsa:3300; | cytoplasm [GO:0005737]; cytoplasmic side of endoplasmic reticulum membrane [GO:0098554]; cytosol [GO:0005829]; extrinsic component of endoplasmic reticulum membrane [GO:0042406]; inclusion body [GO:0016234]; intrinsic component of endoplasmic reticulum membrane [GO:0031227]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; ATPase activator activity [GO:0001671]; chaperone binding [GO:0051087]; Hsp70 protein binding [GO:0030544]; polyubiquitin modification-dependent protein binding [GO:0031593]; proteasome binding [GO:0070628]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-dependent protein binding [GO:0140036]; unfolded protein binding [GO:0051082]; chaperone-mediated protein folding [GO:0061077]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of inclusion body assembly [GO:0090084]; negative regulation of protein binding [GO:0032091]; negative regulation of protein deubiquitination [GO:0090086]; neuron cellular homeostasis [GO:0070050]; positive regulation of ATPase activity [GO:0032781]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; positive regulation of protein ubiquitination [GO:0031398]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein refolding [GO:0042026]; regulation of chaperone-mediated protein folding [GO:1903644]; regulation of protein localization [GO:0032880]; regulation of protein ubiquitination [GO:0031396]; response to unfolded protein [GO:0006986]; ubiquitin-dependent ERAD pathway [GO:0030433] | 12754272_data provide evidence that cytoplasmic chaperones HSJ1a and HSJ1b when targeted to the endoplasmic reticulum can influence the folding and processing of rhodopsin 15936278_HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. 16604191_Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase 17601350_Observational study of gene-disease association. (HuGE Navigator) 18321953_Damaging exercise induced the expression of capZalpha, MCIP1, CARP1, DNAJB2, c-myc, and junD, each of which are likely involved in skeletal muscle growth, remodeling, and stress management. 18977241_Observational study of gene-disease association. (HuGE Navigator) 20395441_Data show that DNAJB2 is expressed in skeletal muscle at the neuromuscular junction of normal fibers, in the cytoplasm and membrane of regenerating fibers, and in protein aggregates and vacuoles in protein aggregate myopathies. 22522442_a mutation causing a loss-of-function of HSJ1 is linked to a pure lower motor neuron disease, strongly suggesting that HSJ1 also plays an important and specific role in motor neurons. 24023695_HSJ1a acts on mutant SOD1 through a combination of chaperone, co-chaperone and pro-ubiquitylation activity. 25274842_The results of this study confirm that HSJ1 mutations are a rare but detectable cause of autosomal recessive dHMN and CMT2. 27449489_Study describes the identi fi cation of the fi rst deletion reported at the DNAJB2 locus, further expanding its phenotypic and genotypic spectrums as well as its disease-associated mechanisms with spinal muscular atrophy and parkinsonism. 28031292_Our results disclose a novel interplay between ubiquitin- and phosphorylation-dependent signalling, and represent the first report of a regulatory mechanism for UIM-dependent function. They also suggest that CK2 inhibitors could release the full neuroprotective potential of HSJ1, and deserve future interest as therapeutic strategies for neurodegenerative disease. 31682009_DNAJB2 expression in healthy human palatal mucosa is strongly negatively correlated with serum cotinine levels | ENSMUSG00000026203 | Dnajb2 | 657.757501 | 0.9458328 | -0.080342858 | 0.11183262 | 5.107550e-01 | 4.748122e-01 | 7.890341e-01 | No | Yes | 660.082916 | 70.569660 | 682.046124 | 72.660194 | |
| ENSG00000136536 | 64844 | MARCHF7 | protein_coding | Q9H992 | FUNCTION: E3 ubiquitin-protein ligase which may specifically enhance the E2 activity of HIP2. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfer the ubiquitin to targeted substrates (PubMed:16868077). May be involved in T-cell proliferation by regulating LIF secretion (By similarity). May play a role in lysosome homeostasis (PubMed:31270356). {ECO:0000250|UniProtKB:Q9WV66, ECO:0000269|PubMed:16868077, ECO:0000269|PubMed:31270356}. | Acetylation;Alternative splicing;Metal-binding;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | MARCH7 is a member of the MARCH family of membrane-bound E3 ubiquitin ligases (EC 6.3.2.19). MARCH proteins add ubiquitin (see MIM 191339) to target lysines in substrate proteins, thereby signaling their vesicular transport between membrane compartments (Bartee et al., 2004 [PubMed 14722266]).[supplied by OMIM, Mar 2010]. | hsa:64844; | cytosol [GO:0005829]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; MDM2/MDM4 family protein binding [GO:0097371]; transferase activity [GO:0016740]; ubiquitin binding [GO:0043130]; ubiquitin conjugating enzyme binding [GO:0031624]; zinc ion binding [GO:0008270]; negative regulation of DNA damage response, signal transduction by p53 class mediator [GO:0043518]; negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:1902166]; negative regulation of proteasomal protein catabolic process [GO:1901799]; negative regulation of protein autoubiquitination [GO:1905524]; negative regulation of T cell proliferation [GO:0042130]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of protein polyubiquitination [GO:1902916]; protein autoubiquitination [GO:0051865]; protein monoubiquitination [GO:0006513]; protein stabilization [GO:0050821]; regulation of tolerance induction [GO:0002643] | 17161353_Data suggest that clinical auto-graft versus host disease inversely correlates with axotrophin transcript expression. 18410486_MARCH7 can be stabilized by both USP9X and USP7, which deubiquitylate MARCH7 in the cytosol and nucleus, respectively 19901269_Axotrophin/MARCH-7 has an expression profile that is distinct from that of other MARCH family members. 27302477_USP7 and MARCH7 are involved in the progression of Epithelial ovarian cancer. 29295817_The findings uncover a novel mechanism for the regulation of Mdm2 and reveal MARCH7 as an important regulator of the Mdm2-p53 pathway. 29794480_MARCH7 may function as a competing endogenous RNA (ceRNA) to regulate the expression of ATG7 by competing with miR-200a. MARCH7 silencing inhibited autophagy invasion and metastasis of SKOV3 cells both in vitro and in vivo. 31006800_Data show that miR-27b-3p inhibited ubiquitin E3 ligase membrane-associated RING-CH-type finger 7 (MARCH7) in endometrial cancer (EC) cells and MARCH7 promoted cell invasion through snail transcription factors (Snail)-mediated pathway. 33729124_Identification of novel interactions between host and non-structural protein 2C of foot-and-mouth disease virus. | ENSMUSG00000026977 | Marchf7 | 2570.004996 | 1.0639383 | 0.089414427 | 0.06282407 | 2.031022e+00 | 1.541170e-01 | 5.121718e-01 | No | Yes | 3362.253567 | 557.101003 | 3117.856141 | 516.687082 |
| ENSG00000136603 | 6498 | SKIL | protein_coding | P12757 | FUNCTION: May have regulatory role in cell division or differentiation in response to extracellular signals. | 3D-structure;Alternative splicing;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is a component of the SMAD pathway, which regulates cell growth and differentiation through transforming growth factor-beta (TGFB). In the absence of ligand, the encoded protein binds to the promoter region of TGFB-responsive genes and recruits a nuclear repressor complex. TGFB signaling causes SMAD3 to enter the nucleus and degrade this protein, allowing these genes to be activated. Four transcript variants encoding three different isoforms have been found for this gene. [provided by RefSeq, Oct 2011]. | hsa:6498; | acrosomal vesicle [GO:0001669]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; protein-containing complex [GO:0032991]; transcription regulator complex [GO:0005667]; chromatin binding [GO:0003682]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; protein domain specific binding [GO:0019904]; protein-containing complex binding [GO:0044877]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; SMAD binding [GO:0046332]; blastocyst formation [GO:0001825]; lens fiber cell differentiation [GO:0070306]; lymphocyte homeostasis [GO:0002260]; negative regulation of BMP signaling pathway [GO:0030514]; negative regulation of cell differentiation [GO:0045596]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; positive regulation of axonogenesis [GO:0050772]; positive regulation of extrinsic apoptotic signaling pathway via death domain receptors [GO:1902043]; positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage [GO:1902231]; regulation of cell cycle [GO:0051726]; response to antibiotic [GO:0046677]; response to cytokine [GO:0034097]; response to growth factor [GO:0070848]; skeletal muscle tissue development [GO:0007519]; spermatogenesis [GO:0007283]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 12764135_the ability of Ski and SnoN to repress the growth inhibitory function of the Smad proteins is required for their transforming activity. 15677458_SnoN acts as a positive mediator of TGF-beta-induced transcription and cell cycle arrest in lung epithelial cells 15809735_Sno expression was identified as an important mechanism to shut off antiproliferative TGF-beta signaling in malignant melanoma 16109768_mechanism of regulation of TGF-beta signaling via differential subcellular localization of SnoN 16314499_A novel role of SnoN in the transforming activity of TGF-beta in fibroblasts was demonstrated. 16442497_SnoN also seems to regulate negatively the TGF-beta-responsive SMAD, mothers against DPP homolog 7 gene by binding and repressing its promoter in a similar way to Ski 16966324_SnoN is directly regulated by sumoylation leading to the enhancement of the ability of SnoN to repress transcription in a promoter-specific manner 17062133_snoN protein has both oncogenic and tumor suppressive properties in colorectal tumorigenesis. 17074815_SnoN plays both pro-tumorigenic and antitumorigenic roles at different stages of mammalian malignant progression 17469184_These results indicate that impaired competition with p300 is the possible cause of dysfunction of c-Ski/SnoN in scleroderma fibroblasts and that this might contribute to maintenance of the autocrine TGFbeta loop in this disease. 17510063_Arkadia induces degradation of SnoN and c-Ski in addition to Smad7. 17591695_Results show that Arkadia specifically activates transcription via Smad3/Smad4 binding sites by inducing degradation of the transcriptional repressor SnoN. 17625116_CREB activation, in concert with Sp1, constitutes a molecular switch that confers the cell type-specific induction of SnoN in response to HGF stimulation 18261624_Ski and SnoN proteins are overexpressed in Barrett's esophagus 18612694_SnoN overexpression is associated with depth of invasion and recurrence in patients with esophageal squamous cell carcinoma. 18782659_c-Ski and SnoN, mediators in TGF-beta resistance, might be implicated in melanoma growth and progression. 19096149_SnoN and Ski were overexpressed both in adenomas with severe dysplasia and colorectal carcinomas. 19189315_Data show that dominant-negative transforming growth factor beta type II receptor decreases matrix metalloproteinase 2 in hepatic stellate cells, and upregulates SKI-like oncogene, which antagonizes TGF-beta signaling. 19383336_results implicate SnoN levels in multiple roles during ovarian carcinogenesis: promoting cellular proliferation in ovarian cancer cells and as a positive mediator of cell cycle arrest and senescence in non-transformed ovarian epithelial cells 19538364_SnoN is involved in differentiation in normal skin and benign and nonmetastatic skin tumors, but plays a proto-oncogenic role in undifferentiated squamous cell carcinoma 19889106_Regulation of TGF-beta-co-repressor (SnoN) is greatly affected suggesting that SnoN as a cardinal player in cholestasis-induced fibrogenesis. 19898560_Inhibition of Smad signaling may be achieved at the transcriptional level through c-Ski/receptor-Smad/co-mediator Smad4 interactions--REVIEW 20093492_BMP-7 prevents TGF-beta-mediated loss of the transcriptional repressor SnoN and hence specifically limits Smad3 DNA binding. 20457602_The endogenous SnoN plays a role in regulating ADAM12 expression in response to TGFbeta1. 20460516_SnoN elevation is associated with mammary gland branching morphogenesis, postlactational involution, and mammary tumorigenesis. 20957027_The flexibility in the putative protein binding groove enables SnoN to recognize multiple interaction partners. 22227247_SnoN level promotes ERalpha signaling and possibly breast cancer progression. 22674574_the SNON-SMAD4 complex negatively regulated basal SKIL gene expression through binding the promoter and recruiting histone deacetylases 22710172_SnoN may have broad functions in the embryonic development and tissue morphogenesis [Review] 22710173_analysis of SnoN signaling in proliferating cells and postmitotic neurons [review] 22767605_SnoN mediates a negative feedback mechanism evoked by TGF-beta to inhibit BMP signaling and, subsequently, hypertrophic maturation of chondrocytes. 23154181_These results support our observation that cancer tissues have lower expression levels of SnoN, miR-720, and miR-1274A compared to adjacent normal tissues from esophageal squamous cell carcinoma patients. 23154981_SNON predominantly associates with SMAD2 at the promoters of primitive streak (PS) and early DE marker genes 23178716_Data suggest that SKIL expression is modulated by antineoplastic agents and may be involved in drug resistance in ovarian carcinoma; up-regulation of SKIL expression by arsenic trioxide and reduction of apoptosis involves activation of PI3K pathway. 23418461_These data strongly suggest that SnoN can function as a tumor suppressor at early stages of tumorigenesis in human cancer tissues. 23446947_SnoNspecific siRNA is capable of effectively inhibiting the expression of SnoN in human HepG2 cells, and the downregulation of SnoN expression induces growth inhibition and apoptosis 23621864_Phospholipid Scramblase 1, an interferon-regulated gene located at 3q23, is regulated by SnoN/SkiL in ovarian cancer cells. 23764425_these studies identify TLOC1 and SKIL as driver genes at 3q26 and more broadly suggest that cooperating genes may be coamplified in other regions with somatic copy number gain. 23832742_High SnoN expression is associated with metastasis in breast cancer. 24637302_The results indicate that protein ubiquitination promotes megakaryopoiesis via degrading SnoN, an inhibitor of CD61 expression, strengths the roles of ubiquitination in cellular differentiation. 25059663_Data indicate that tripartite motif containing 33 protein TIF1gamma promotes sumoylation of SKI-like proto-oncogene protein SnoN1 and regulates epithelial-mesenchymal transition (EMT). 25464936_Whole exome sequencing of the blood of the patient and both parents revealed a de novo germline SKIL mutation in the child that was not present in either parent 25749039_SKIL knockdown led to growth arrest in PC-3 and LNCaP cell line models of prostate cancer, and its overexpression led to increased invasiveness in RWPE-1 cells. 25907906_RNAi-mediated downregulation of SnoN effectively inhibited proliferation and enhanced apoptosis of pancreatic cells. 26743567_the findings of this study demonstrate that the downregulation of SnoN expression in hRPTECs under high-glucose conditions is mediated by the increased expression of Smurf2 through the TGF-b1/Smad signaling pathway. 27237790_SnoN interacts with multiple components of the Hippo pathway to inhibit the binding of Lats2 to TAZ and the subsequent phosphorylation of TAZ, leading to TAZ stabilization. 27430247_suggest that SnoN suppresses TGF-betainduced epithelial-mesenchymal transition and invasion of bladder cancer cells in a TIF1gammadependent manner 28115165_signal transducer and activator of transcription (Stat)3 represses Smad3 in synergy with the potent negative regulators of TGF-beta signaling, c-Ski and SnoN, whereby renders gefitinib-sensitive HCC827 cells resistant 28397834_Our results show that SnoN, and SMAD heteromers can form a joint structural core for the binding of other transcription modulators. 28707079_It is a critical negative regulator of TGF-beta1/Smad signal pathway, involving in tubule epithelial-mesenchymal transition (EMT), extracellular matrix (ECM) accumulation, and tubulointerstitial fibrosis. 29734252_Ski, SnoN, and Smad4 seem to play a role in oral squamous cell carcinoma oncogenesis, and it is likely that Ski and SnoN functions may take place independent of Smad4. 31471872_SKIL role in the tumorigenesis and metastasis of colorectal cancer.SNHG14 promotes the tumorigenesis and metastasis of colorectal cancer through miR-32-5p/ski-oncogene-like SKIL axis.SKIL is a downstream target gene of miR-32-5p. 31836508_Our study verified that miR-130a-3p facilitates the TGF-b1/Smad pathway in renal tubular epithelial cells and may participate in renal fibrosis by targeting SnoN, which could be a possible strategy for renal fibrosis treatment. 32770107_PIAS1 and TIF1gamma collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition. 33268765_SKIL facilitates tumorigenesis and immune escape of NSCLC via upregulating TAZ/autophagy axis. 35136033_ATAD3B and SKIL polymorphisms associated with antipsychotic-induced QTc interval change in patients with schizophrenia: a genome-wide association study. | ENSMUSG00000027660 | Skil | 773.700984 | 0.9238595 | -0.114254666 | 0.10970255 | 1.076276e+00 | 2.995322e-01 | 6.671021e-01 | No | Yes | 911.520210 | 158.713391 | 980.423388 | 170.781919 | |
| ENSG00000136826 | 9314 | KLF4 | protein_coding | O43474 | FUNCTION: Transcription factor; can act both as activator and as repressor. Binds the 5'-CACCC-3' core sequence. Binds to the promoter region of its own gene and can activate its own transcription. Regulates the expression of key transcription factors during embryonic development. Plays an important role in maintaining embryonic stem cells, and in preventing their differentiation. Required for establishing the barrier function of the skin and for postnatal maturation and maintenance of the ocular surface. Involved in the differentiation of epithelial cells and may also function in skeletal and kidney development. Contributes to the down-regulation of p53/TP53 transcription. {ECO:0000269|PubMed:17308127, ECO:0000269|PubMed:20071344}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein that belongs to the Kruppel family of transcription factors. The encoded zinc finger protein is required for normal development of the barrier function of skin. The encoded protein is thought to control the G1-to-S transition of the cell cycle following DNA damage by mediating the tumor suppressor gene p53. Mice lacking this gene have a normal appearance but lose weight rapidly, and die shortly after birth due to fluid evaporation resulting from compromised epidermal barrier function. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Sep 2015]. | hsa:9314; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; euchromatin [GO:0000791]; nucleoplasm [GO:0005654]; transcription regulator complex [GO:0005667]; beta-catenin binding [GO:0008013]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; promoter-specific chromatin binding [GO:1990841]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II sequence-specific DNA-binding transcription factor recruiting activity [GO:0001010]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; zinc ion binding [GO:0008270]; canonical Wnt signaling pathway [GO:0060070]; cellular response to growth factor stimulus [GO:0071363]; cellular response to laminar fluid shear stress [GO:0071499]; cellular response to leukemia inhibitory factor [GO:1990830]; defense response to tumor cell [GO:0002357]; epidermal cell differentiation [GO:0009913]; epidermis morphogenesis [GO:0048730]; fat cell differentiation [GO:0045444]; mesodermal cell fate determination [GO:0007500]; negative regulation of angiogenesis [GO:0016525]; negative regulation of cell migration involved in sprouting angiogenesis [GO:0090051]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of chemokine (C-X-C motif) ligand 2 production [GO:2000342]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; negative regulation of gene expression [GO:0010629]; negative regulation of heterotypic cell-cell adhesion [GO:0034115]; negative regulation of inflammatory response [GO:0050728]; negative regulation of interleukin-8 production [GO:0032717]; negative regulation of leukocyte adhesion to arterial endothelial cell [GO:1904998]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of response to cytokine stimulus [GO:0060761]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of cellular protein metabolic process [GO:0032270]; positive regulation of core promoter binding [GO:1904798]; positive regulation of gene expression [GO:0010628]; positive regulation of hemoglobin biosynthetic process [GO:0046985]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of protein metabolic process [GO:0051247]; positive regulation of sprouting angiogenesis [GO:1903672]; positive regulation of telomerase activity [GO:0051973]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; post-embryonic camera-type eye development [GO:0031077]; post-embryonic hemopoiesis [GO:0035166]; regulation of axon regeneration [GO:0048679]; regulation of blastocyst development [GO:0120222]; regulation of cell differentiation [GO:0045595]; regulation of transcription by RNA polymerase II [GO:0006357]; response to retinoic acid [GO:0032526]; stem cell population maintenance [GO:0019827]; transcription, DNA-templated [GO:0006351] | 8702718_This is the first paper that describes the identification and characterization of KLF4 (also called GKLF) and that KLF4 is inhibitor of cell growth. 9218496_This paper describes that the nuclear localization signals within KLF4 define a closely related subfamily of Kruppel-like factors including KLF1, KLF2 and KLF4. 9428642_This study examines the expression of KLF4 during embryonic and postnatal development and in intestinal tumorigenesis. 9443972_This paper reports a consensus DNA binding sequence for KLF4 using an unbiased screen. 9651398_This study reports the first identified target gene for KLF4 was CYP1A1, and the mechanism by which KLF4 regulates CYP1A1 expression. 10556311_This study characterized the gene and promoter structures of the mouse KLF4 gene. 10666450_This paper examines the structure-function relationship of KLF4 in activating target gene expression and in suppressing growth. 10749849_This paper describes that KLF4 is transcriptionally activated by the p53 tumor suppressor after DNA damage and results in the transcriptional activation of the p21 cell cycle inhibitor gene. 11521200_This paper reports that KLF4 is regulated by the APC tumor suppressor in colorectal cancer cells through CDX2. 12087155_This paper describes the opposite effect of KLF4 and KLF5 on transcription of the KLF4 gene. 12297499_induction of GKLF mRNA and protein expression by interferon-gamma treatment was associated with reduction of ornithine decarboxylase (ODC) gene expression and enzyme activity in colon cancer HT-29 cells 12427745_KLF4 is an essential mediator of p53 in controlling G(1)/S progression of the cell cycle following DNA damage 12439907_down-regulation of gut-enriched Kruppel-like factor in esophageal squamous cancer 12581631_role in cell cycle regulation and epithelial differentiation 12776194_Over-expression of KLF4 in human colon cancer cells reduces tumorigenecity. 12853980_Regulation of A33 antigen expression by GKLF. 12901861_inactivation of KLF4 is one of the frequent steps towards bladder carcinogenesis 12919939_intestinal alkaline transactivation by Kruppel-like factor-4 is likely mediated through a critical region located within the proximal IAP promoter region 14627709_KLF4 is necessary for preventing the entry into mitosis following DNA damage 14627709_role of KLF4 in maintaining the integrity of the G2/M checkpoint following DNA damage 14670968_KLF4 can act to repress histidine decarboxylase gene expression by Sp1-dependent and -independent mechanisms 14724568_KLF4 is a tumor suppressor in colorectal cancer. 15031282_KLF4 is a novel regulator of u-PAR expression that drives the synthesis of u-PAR in the luminal surface epithelial cells of the colon 15051827_transactivation of Kruppel-like factor 4(KLF4) by butyrate appears to be mediated through interaction with a Sp1 transcription factor-binding domain on the promoter 15102675_KLF4 has a role in the aggressive phenotype of early-stage infiltrating ductal carcinoma 15561714_a longer isoform of gut-enriched Kruppel-like factor 4 (GKLF) we term GKLFa interacts with the CD11d promoter 15674344_KLF4 can function in the nucleus to induce squamous epithelial dysplasia. 15740636_A review article that summarizes the mechanisms by which KLF4 and KLF5 regulate cell proliferation. 15805274_Promoter hypermethylation and hemizygous deletion contributed to the down-regulation of KLF4 expression and the induction of apoptosis contributed to the antitumor activity of KLF4; alteration of KLF4 expression plays a role in gastric cancer development. 15806166_KLF4 is both necessary and sufficient in preventing centrosome amplification following gamma-radiation-induced DNA damage. 15937668_Observational study of gene-disease association. (HuGE Navigator) 16507986_the cross talk of KLF4 and beta-catenin plays a critical role in homeostasis of the normal intestine as well as in tumorigenesis of colorectal cancers. 16632465_cGMP-dependent protein kinase expression is regulated by Rho and Kruppel-like transcription factor-4 17016435_KLF4 may be an important determinant of cell fate following gamma-radiation-induced DNA damage. 17017123_KLF4 exerts a global inhibitory effect on macromolecular biosynthesis that is beyond its established role as a cell cycle inhibitor. 17135299_KLF-4 and AP-2 is regulating the activity of the hSMVT promoter in the intestine and provide direct in vivo confirmation of hSMVT promoter activity. 17339326_Kruppel-like factor 4 as a novel regulator of endothelial activation in response to pro-inflammatory stimuli. 17508399_A review article that summarizes the biological and pathobiological functions in the intestinal epithelium. 17614846_Genetic and epigenetic alterations of the KLF4 gene might play a minor role in gastric carcinogenesis. 17671182_This study reports that haploinsufficiency of Klf4 gene in transgenic mice with targeted deletion of one of the Klf4 alleles promotes intestinal tumorigenesis when crossbred with the ApcMin mice. 17688680_Observational study of gene-disease association. (HuGE Navigator) 17688680_analysis of SNPs, located in the KLF2, KLF4 and KLF5 gene did not show an association with Type 2 diabetes in this French population 17762869_KLF4 is a critical regulator in the transcriptional network controlling monocyte differentiation. 17908689_KLF4 might function as an activator or repressor of transcription depending on whether it interacts with co-activators such as p300 and CREB-binding protein or co-repressors such as HDAC3. 17932114_Data show that human testis strongly expresses KLF4 and they were localized to nuclei of round spermatids during normal spermatogenesis stages II-IV. 18056793_Transient transfection of Kruppel-like factor 4 suppressed LDLR, steroidogenic acute regulatory protein, and CYP11A 18157115_Using ectopic expression of Oct4, Sox2, Klf4 and Myc, we have derived iPS cells from fetal, neonatal and adult human primary cells 18264726_Loss of KLF4 protein expression might contribute to assessing prognosis in colorectal cancer with lymph node metastasis 18376139_KLF4 expression is associated with human skin SCC progression and metastases 18390749_The inflammation-selective effects of loss-of-KLF4 and the gain-of-KLF4-induced monocytic differentiation in HL60 cells identify KLF4 as a key regulator of monocytic differentiation 18977346_Reduced levels of KLF4 tumor suppressor activity in colon tumors may be driven by elevated beta-catenin/Tcf signaling. 19074836_Notch signaling suppresses KLF4 expression in intestinal tumors and colorectal cancer cells 19197145_KLF4 pathway is a potential effector of retinoid signaling in tumor prevention. Of several oncogenes that independently induce malignant transformation of RK3E epithelial cells, only KLF4 showed substantial sensitivity to retinoids. 19327128_Results indicate that KLF4 positively regulates human ghrelin expression via binding to a KLF-responsive region in the promoter. 19361279_Luciferase reporter gene assays revealed that survivin promoter activity is repressed upon overexpression of KLF4 in EC9706 cells. 19409607_Increased miR-145 expression inhibits human embryonic stem cells self-renewal, represses expression of pluripotency genes, and induces lineage-restricted differentiation; loss of miR-145 impairs differentiation and elevates OCT4, SOX2, and KLF4. 19458492_Studies show that the expression of genes belonging to the OCT3/SOX2/NANOG/KLF4 core circuitry that acts at the highest level in regulating stem cell biology. 19495417_N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b 19503094_Studies discovered a novel molecular network between p53, KLF4 and ERalpha. 19505997_The finding suggests that KLF4 may be an important factor for the maintenance of the developmental and the tumorigenic potential of IGCNU as well as for the malignancy of seminoma. 19522013_Data showed that both KLF4 and PBX1 mRNA and protein expression were downregulated during hESC differentiation. Overexpression of KLF4 and PBX1 upregulated NANOG promoter activity and NANOG protein expression. 19541842_inhibition of miR-25 in cytokine-stimulated ASM cells up-regulates KLF4 expression via a post-transcriptional mechanism. 19544095_KLF4 binds to the p57(Kip2) promoter and transcriptionally upregulates its expression, which in turn inhibits the stress activated protein kinase cascade and c-Jun phosphorylation. 19549847_Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. 19696409_Induced pluripotent stem cells programmed with OCT4/SOX2/KLF4 and without c-MYC yield proficient cardiogenesis for functional heart chimerism. 19717984_Results suggest that KLF4 could contribute to breast tumor progression by activating synthesis of Notch1 and by promoting signaling through a non-canonical Notch1 pathway. 19737957_The expression of KLF4 resulted in marked inhibition of cell growth and clonogenic formation. The tumor-suppressive effect was associated with in up-regulating p21 and down-regulating cyclin D1, leading to cell cycle arrest at the G(1)-S checkpoint. 19763260_human neural stem cells are capable of reprogramming into a pluripotent state by forced expression of Oct3/4 and Klf4 19779097_KLF4 is potentially a reliable marker of HNSCC, and myrAkt transgenic mice are valuable tools for preclinical research of HNSCC. 19816951_show that Klf4 interacts directly with Oct4 and Sox2 when expressed at levels sufficient to induce induced pluripotent stem cells. 19858207_Kruppel-like factor 4 is a novel mediator of Kallistatin in inhibiting endothelial inflammation via increased endothelial nitric-oxide synthase expression. 19968965_these data identify a significant degree of mechanistic and functional conservation between KLF2 and KLF4, and importantly, provide further insights into the complex regulatory networks governing endothelial vasoprotection. 20030945_Expression of KLF-4 increases under IL-1beta and TNF-alpha stimulation, and may participate in regulation of endothelial activity. 20043075_down-regulated KLF4, CHGA, GPX3, SST and LIPF, together with up-regulated SERPINH1, THY1 and INHBA is an 8-gene signature for gastric cancer 20071344_Klf4 functions upstream of Nanog in ES cell self-renewal and in preventing ES cell differentiation 20072650_Data show that that KLF4 is inactivated by either genetic or epigenetic mechanisms in a large subset of medulloblastomas and that it likely functions as a tumor suppressor gene in the pathogenesis of medulloblastoma. 20075075_overexpression of miR-10b in KYSE140 and KYSE450 cells led to a reduction of endogenous KLF4 protein, whereas silencing of miR-10b in EC9706 cells caused up-regulation of KLF4 protein in esophageal cancer 20215880_Reduced KLF4 is associated with lung cancer cell invasion by suppression of SPARC expression 20356845_Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression 20385121_KLF4 is identified as a novel transcription factor that controls NAG-1 promoter activity in human and mouse colorectal cancers. And PPARgamma agonists up-regulate KLF4 expression in receptor-dependent manner. 20442780_the two factors Klf4 and Sp3 exert an overlapping repressor function through their binding to the Notch1 promoter. 20479568_In terms of human reproductive tissue, KLF4 may be a factor concerning cell cycle, directly responsive to progesterone receptor activation. 20519630_Loss of KLF4 is associated with B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. 20551324_KLF4 expression largely reproduces the protective phenotype in endothelial cellsdecreased angiogenic, migratory, and inflammatory potential 20629177_hTERT is one of the major targets of KLF4 in cancer and stem cells to maintain long-term proliferation potential. 20699379_KLF4 expression is significantly downregulated in colon cancer, and loss of KLF4 is an independent predictor of survival and recurrence 20718868_Data indicate up-regulation of KLF4 expression(by high-density lipoproteins, especially HDL3) causes up-regulation of expression of SR-BI (scavenger receptor class B type I); KLF4 binds to promoter for SR-BI. 20727893_a splice variant of KLF4 (KLF4alpha) is up-regulated in aggressive pancreatic cancer cells and human pancreatic tumor tissues 20849362_These results provide new insights toward understanding the molecular basis of mesenchymal stromal cells stemness maintenance and underline the ability of KLF4 to maintain cells in an undifferentiated state. 20972454_tumor-predisposing p53 mutations hijack p63 to a different location on the promoter, turning it into an activator of KLF4. 21075859_Dopamine acting through its D(2) receptor, inhibits IGF-I-induced proliferation of gastric adenocarcinoma cells by up-regulating KLF4, a negative regulator of the cell cycle through down regulation of IGF-IR and AKT phosphorylation. 21125297_Longitudinal cell formation in the small intestine was regulated by the colocalization of Hath1 and Klf4 that converted Paneth cell differentiation into goblet cell differentiation. 21132436_reduced expression in skin cancer lesions 21159640_Results suggest that KLF4 functions as an inhibitor of tumor cell growth and migration in prostate cancer and decreased expression has prognostic value for predicting prostate cancer metastasis. 21166929_MEK5 activation by laminar shear stress inhibits inflammatory responses in microvascular endothelial cells, in part through ERK5-dependent induction of KLF4 21177849_KLF4 is profoundly degraded in response to TGF-beta signaling 21219537_Low KLF4 is associated with head and neck squamous cell carcinoma. 21224073_Loss of KLF4 expression is closely related to the genomic loss, and its restoration inhibits cancer cell proliferation, suggesting a key suppressor role in pancreatic tumorigenesis. 21242971_study provides evidence that KLF4 has a potent oncogenic role in mammary tumorigenesis likely by maintaining stem cell-like features and by promoting cell migration and invasion 21312053_results suggest that PAF receptor plays a pivotal role in POVPC-induced migration of human BMSCs through PAF receptor-mediated expression of KLF4 21412023_These results suggest a relationship between D9S105 deletion and downregulation of KLF4 gene expression as an early event in pancreatic ductal adenocarcinoma progression. 21502426_KLF4 is another marker of monocytic differentiation in monocytic leukemias 21518959_Results identify a KLF4-miR-206 feedback pathway that oppositely affects protein translation in normal cells and cancer cells. 21531817_Restored expression of KLF4, a putative tumor suppressor, downregulated IFITM3 expression in colon cancer cells in vitro. 21539536_Our data supports a model of antagonistic interaction of KLF4/CREBBP trans-factors in HBG regulation. 21586797_KLF4 plays a role in maintenance of high glycolytic metabolism by transcriptional activation of the PFKP gene in breast cancer cells. 21652709_KLF4-siRNA inhibits PKCdelta-dependent p21(Cip1) promoter activity. PKCdelta increases KLF4 expression leading to enhanced KLF4 interaction with the GC-rich elements in the p21(Cip1) promoter to activate transcription 21670502_These data suggest that KLF4 is a novel regulator of macrophage polarization. 21673106_down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. 21674249_Loss of nuclear expression of KLF4 is associated with in situ and invasive breast carcinomas. 21689550_Findings suggest that KLF4 is able to promote odontoblastic differentiation of and inhibit proliferation of human dental pulp cells. 21746878_Klf4 recruits a complex of Meis and Pbx proteins to DNA, resulting in Meis2 transcriptional activation domain-dependent activation of a subset of Klf4 target genes. 21833590_Data sugget that KLF4 regulates the cellular sensitivity to cisplatin in hepatocarcinoma stem-like cells and hepatocarcinoma cells by elevating intracellular glutathione levels. 22170051_Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4: implications for colon cancer. 22170594_KLF4 may function as a tumor suppressor in cervical carcinoma by inhibiting cell growth and tumor formation. 22209344_Patients with oral cancer with negative nuclear Kruppel-like factor 4 expression in tumor cells had poor prognoses and a 2.5-fold higher death risk. 22267480_miR-92a coregulates KLF4 and KLF2 expression in arterial endothelium and contributes to phenotype heterogeneity associated with regional atherosusceptibility and protection in vivo. 22282354_TGF-beta1 downregulates AT1 receptor expression via PKC-delta-mediated Sp1 dissociation from KLF4 and Smad-mediated PPAR-gamma association with KLF4. 22364861_ZNF750 directly links a tissue-specifying factor, p63, to an effector of terminal differentiation, KLF4. 22384261_KLF4 affects intestinal epithelial cell morphology by regulating proliferation, differentiation and polarity of the cells 22389506_suppression of pVHL in response to estrogen signaling results in elevation of KLF4, which mediates estrogen-induced mitogenic effect 22407433_KLF4 inhibits beta-catenin expression and regulates the beta-catenin-mediated biological behaviors of gastric cancer cells. 22430140_The present study demonstrated a key regulatory role of KLF4 in the endothelial ASS1 expression and NO production in response to laminar shear stress. 22491752_data suggest that KLF4 inhibits cell proliferation, migration and adhesion and that loss of KLF4 promotes skin tumorigenesis 22528804_KLF4 has a suppressive effect on the proliferation and metastasis of the breast cancer. 22539002_Data suggest that there is a partial functional redundancy between Sox2 and Klf4 in the disruption of cellular homeostasis and activation of regulatory networks that define pluripotency. 22592916_KLF4 translation level is associated with the differentiation stage of different leukemias and is independent of other parameters of risk stratification. 22653776_KLF4 is induced in a bacterial DNA-TLR9-Src-dependent manner and regulates IL-10 expression 22677193_KLF4 binds to the promoter of VDR to regulate its expression altering signal transduction and contributing to the progression of hepatocellular carcinoma. 22751119_MiR-29 expression is suppressed by progestins in breast cancer cells. 22766303_Both Sox9 and KLF4 interact with beta-catenin in an immunoprecipitation assay and reduce its binding to TCF4. 22835519_LIF combined with VEGF can maintain the preliminary, progenitor phenotype of EPCs and alleviate cell differentiation by upregulating KLF4, which may provide new insights into transcriptional regulation in endothelial progenitor cells 22842522_Our findings establish the impact of Oct1, KLF4 and c-Myc on cancer bioenergetics and evidence a link between oncosecretomics and cellular bioenergetics profile. 22854048_We conclude that KLF4 upregulation by HGF represents a novel mechanism mediating HGF-induced cell scattering and perhaps other associated events such as cell migration and invasion 22937066_Kruppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. 23007394_the HIC-5- and KLF4-dependent mechanism transactivates p21(Cip1) in response to anchorage loss 23045286_KLF4 contributes to the favorable disease outcome by directly mediating the growth and lineage determination of neuroblastoma cells. 23089465_High Klf4 expression in the cytoplasm is associated with prostate cancer. 23097599_LMP3 induced the successful osteogenic differentiation of AFSC by inducing the expression of osteogenic markers and osteospecific transcription factors. 23118962_the cooperative binding of KLF4 and p53 to DNA exemplifies a regulatory mechanism that contributes to p53 target selectivity 23160196_KLF4 differentially regulated pertinent endothelial targets via competition for the coactivator p300. 23202735_CDX2-driven leukemogenesis involves KLF4 repression and deregulated PPARgamma signaling. 23249717_Data indicat that the SPARC overexpression may play an important role in the initiation and development of non-small cell lung cancer (NSCLC), whereas KLF4 inhibits this process. 23275339_up-regulation of KLF4 upon PPARgamma activation is mediated through the PPRE in the KLF4 promoter 23348505_Meningioma mutations in TRAF7 commonly occurred with a recurrent mutation (K409Q) in KLF4, a transcription factor known for its role in inducing pluripotency, or with AKT1, a mutation known to activate the PI3K pathway. 23376074_these results suggest that E-cadherin expression in cancer cells is controlled by a balance between ZEB2 and KLF4 expression levels. 23384942_Results suggest that miR-7 and KLF4 may serve as biomarkers or therapeutic targets for brain metastasis of breast cancer. 23395444_Data show that ACTL6a prevents SWI/SNF complex binding to promoters of KLF4 and other differentiation genes and that SWI/SNF catalytic subunits are required for full induction of KLF4 targets. 23404370_The findings of this study suggested an essential contribution of combined KLF4 K409Q and TRAF7 mutations in the genesis of secretory meningioma and demonstrate a role for TRAF7 alterations in other non-NF2 meningiomas. 23418515_Data suggest that KLF4 can be a potential therapeutic target for treating colon cancer. 23440423_Numbl-Klf4 signaling is critical to maintain multiple nodes of metastatic progression, including persistence of cancer-initiating cells. 23515309_the importance KLF4 sumoylation in regulating pluripotency and adipocyte differentiation. 23585530_Kruppel-like factor 4 plays a role in controlling cell cycle and resistance to alkylating agents in multiple myeloma cells. 23599428_KLF4 is part of a multiprotein complex that interacts that the hINV promoter distal regulatory region to drive differentiation-dependent hINV gene expression in epidermis. 23696417_MiR-10a restores the differentiation capability of aged hMSCs through repression of KLF4. 23722653_KLF4 is a putative tumor suppressor gene epigenetically silenced in renal cell cancers by promoter CpG methylation and that it has prognostic value for renal cell progression. 23737434_KLF4 modulates maintenance of myeloid-derived suppressor cells (MDSCs) in bone marrow by inducing GM-CSF production via CXCL5 and regulates recruitment of MDSCs into the primary tumors through the CXCL5/CXCR2 axis 23758499_Patients with nasopharyngeal carcinoma (NPC) showing lower cytoplasmic KLF4 expression had a significantly shorter overall survival time than those with high NPC KLF4 expression. 23861801_Attenuation of kruppel-like factor 4 facilitates carcinogenesis by inducing g1/s phase arrest in clear cell renal cell carcinoma. 23867820_microRNA-15a has a key role in the KLF4 suppressions of proliferation and angiogenesis in endothelial and vascular smooth muscle cells 23884216_downregulated expression in human monocyte in patients with coronary atherosclerosis 23921950_Here we report that the combination of p63, a master regulator of epidermal development and differentiation, and KLF4, a regulator of epidermal differentiation, is sufficient to convert dermal fibroblasts to a keratinocyte phenotype. 23926105_Co-transfection of KLF-4 and HDAC1,2 expression plasmids in breast cancer cells results in synergistic repression of VEGF expression and inhibition of angiogenic potential 24018236_KLF4 may function as a tumor suppressor gene in urothelial cancer since down-regulation of KLF4 by promoter hypermethylation would promote cancer progression. 24030402_Human aortic aneurysms demonstrate significantly higher transgenic KLF4 expression that is localized to smooth muscle cells. 24060351_ChIP analysis established that PDGF-BB-induced repression of Myocd gene expression is most likely regulated by enhanced binding of Klf4 and Klf5 to a lesser extent, to the PRR of PrmM 24065485_loss of KLF4 expression correlates with diffuse-type gastric cancer and immunoreactivity to Fas, and are inversely linked with p53 nuclear accumulation. 24067139_The present findings revealed that KLF4 is overexpressed in Burkitt pediatric lymphoma and is a potential biomarker for inferior overall survival. 24105022_KLF4 may play an anti-oncogenic role in gastric tumorigenesis. 24114958_Radiation exposure of the tissues around brain tumour during radiation therapy causes KLF4 overexpression in astrocytes, which induces more DSB and reduces SSB. 24156273_Endothelial KLF4 regulates the transcription of genes involved in key pathways implicated in pulmonary arterial hypertension. 24281002_Both miR-29a high expression and KLF4 mRNA low expression were associated with metastasis. 24318462_our results indicated that KLF4 is a key transcriptional regulator of BTB function by regulating expressions of tight junction related proteins, which would draw growing attention to KLF4 as a potential target for glioma therapy. 24388984_analysis of Klf4 ubiquitination sites using several Klf4 deletion fragments and bioinformatics predictions showed that the lysine sites which are signaling for Klf4 protein degradation lie in its N-terminal domain (aa 1-296) 24415058_Our data suggest that the effects of miR-34a on apoptosis occur due to the downregulation of KLF4 24551169_KLF4 gene is inactivated by methylation-induced silencing mechanisms in a large subset of cervical carcinomas. 24573354_KLF4 and HOXD10 were identified as direct targets of miR-10b in bladder cancer cells 24626089_Results argue that KLF5 acts as a fundamental core regulator of intestinal oncogenesis at the stem-cell level, and they suggest KLF5 targeting as a rational strategy to eradicate stem-like cells in colorectal cancer. 24755985_Hemodynamics influence endothelial KLF4 expression through DNA methyltransferase enrichment/myocyte enhancer factor-2 inhibition mechanisms of KLF4 promoter CpG methylation with regional consequences for atherosusceptibility. 24763828_NS suppression in the AGS cell line caused some morphological alterations, a cell cycle arrest at G1 phase, and an upregulation of iPS genes: Nanog, Sox2, and Klf4. 24789055_The acetylated histone H3 located more at KLF4 promoter region. 24812666_KLF4 overexpression in cultured podocytes increased expression of nephrin and other epithelial markers and reduced mesenchymal gene expression. 24897024_High cytoplasmic expression of KLF4 was associated with better disease-specific survival and was an independently favorable prognostic factor in hepatocellular carcinoma. 24928509_study implicates the A2bAR as a regulator of adipocyte differentiation and the A2bAR-KLF4 axis as a potentially significant modulator of adipose biology. 24931170_WISP2 has a role in regulating tumor cell susceptibility through EMT by inducing the TGF-beta signaling pathway, KLF-4 expression and miR-7 inhibition. 24947925_KLF4 underexpression is correlated with disease stage and pancreatic cancer progression. 24978920_p53, KLF4, and p21 showed altered expression patterns in pulmonary neuroendocrine neoplasms. Lack of KLF4 and p21 expression was associated with accumulation of aggressive features in typical carcinoids. 24993134_Silencing of KLF4 expression resulted in the abolishment of LPA-stimulated migration and proliferation of PC-3M-luc-C6 cells. 25037230_This study identified a novel miR-2909-KLF4 molecular axis able to differentiate between the pathogeneses of pediatric B- and T-cell ALLs, and which may represent a new diagnostic/prognostic marker. 25060774_Low KLF4 level is associated with colorectal cancer. 25100730_KLF4 is a regulator of cardiac hypertrophy by modulating the expression and the activity of myocardin. 25109409_Inhibition of KLF4 by NOTCH1 regulates proliferation and differentiation of bladder cancer cells. 25137052_KLF4 functions as a tumor suppressor gene in ovarian cancer cells by inhibiting TGFB-induced epithelial mesenchymal transition. 25146389_Twist1-Jagged1/KLF4 axis is essential both for transdifferentiation of tumour cells into endothelial cells and for chemoresistance acquisition. 25175483_Enhanced expression of ABCG2 and KLF4 in the recurrent PCa tissues postulates the suggestion that enrichment for cells with stem cell characteristics in these tissues might be playing a critical role for chemoresistance and recurrence of cancer. 25181544_miR-7 acts as an oncomiR in the epithelial cellular context, where through the negative regulation of KLF4-dependent signaling pathways, miR-7 promotes cellular transformation and tumor growth. 25218589_miR-152 inhibited the cell proliferation, migration, invasion, and promoted apoptosis of glioblastoma stem cells by directly targeting KLF4. 25341045_HDAC1 and Klf4 are potential new molecular markers and targets for clinical diagnosis, prognosis, and treatment of myeloid leukemia. 25400747_KLF4 might act as a tumor suppressor in esophageal squamous cell carcinoma and the expression status of KLF4 could be considered as a prognosis predictor. 25531393_Overexpression of KLF4 inhibited virus-induced activation of ISRE and IFN-b promoter in cells, while knockdown of KLF4 potentiated viral infection-triggered induction of IFNB1 and downstream genes and attenuated viral replication. 25644173_These results suggest that KLF4 as a major transcription factor that suppresses the expression of T-cell associated genes, thus inhibiting T-ALL progression. 25652467_In this review, we focus on the functions, roles, and regulatory networks of these five KLFs in HCC, summarize key pathways, and propose areas for further investigation 25756955_KLF4 acts as a counter-regulatory transcription factor in pneumococci-related proinflammatory activation of lung epithelial cells. 25772473_KLF4 N-terminal variance modulates induced reprogramming to pluripotency 25789974_These results identify KLF4 and KLF5 as cooperating protumorigenic factors and critical participants in resistance to lapatinib, furthering the rationale for combining anti-MCL1/BCL-XL inhibitors with conventional HER2-targeted therapies. 25791170_induction of a miR-200c-SUMOylated KLF4 feedback loop is a significant aspect of the PDGF-BB proliferative response in VSMCs and that targeting Ubc9 represents a novel approach for the prevention of restenosis. 25851906_Decreased KLF4 was associated with Esophageal Squamous Cell Carcinoma. 25860962_upregulated Siat7A expression, which was paralleled by the increased Klf4 in the ischemic myocardium, contributed to cardiomyocyte apoptosis following myocardial infarction 25879941_Nuclear factor I-C regulates E-cadherin via control of KLF4 in breast cancer 25892014_The hypermethylation of KLF4 directly mediated by Dnmt1 contributes to the progression of EMT in renal epithelial cells. 25907774_expression of OCT4, SOX2, KLF4 and MYC (OSKM) induced pluripotent stem cell differentiation 25954999_Results showed that KLF4 plays an important role in inhibiting the aggressiveness of hepatocellular carcinoma cells via upregulation of TIMP-1 and TIMP-2. 25985364_Our findings indicate that the contribution of SMCs to atherosclerotic plaques has been greatly underestimated, and that KLF4-dependent transitions in SMC phenotype are critical in lesion pathogenesis 26046571_In the atherosclerosis mouse model they found that loss of Klf4 exclusively in SMCs resulted in a 50% reduction in atherosclerosis 26050649_demonstrated that BAALC blocks ERK-mediated monocytic differentiation of acute myeloid leukemia cells by trapping Kruppel-like factor 4 (KLF4) in the cytoplasm and inhibiting its function in the nucleus 26075898_KLF4 promotes angiogenesis by transcriptionally activating vascular endothelial growth factor expression, thus activating the vascular endothelial growth factor signaling pathway in human retinal microvascular endothelial cells. 26087183_Authors demonstrate that F-box only protein 22 (FBXO22) interacts with and thereby destabilizes KLF4 via polyubiquitination. As a result, FBXO22 could promote HCC cells proliferation. 26097551_down-regulation of KLF4 and up-regulation of KLF5 may stimulate oral carcinoma progression through the dedifferentiation of carcinoma cells. 26108068_Angiotensin II can modulate epigenetic regulation in podocytes and renin-angiotensin blockade inhibits these actions in part via KLF4 in proteinuric kidney diseases. 26109433_Data indicte that Kruppel-like factor 4 | ENSMUSG00000003032 | Klf4 | 115.626149 | 1.0374379 | 0.053025031 | 0.24464769 | 4.709554e-02 | 8.281966e-01 | No | Yes | 114.827883 | 18.787332 | 110.505671 | 18.072246 | ||
| ENSG00000136828 | 9649 | RALGPS1 | protein_coding | Q5JS13 | FUNCTION: Guanine nucleotide exchange factor (GEF) for the small GTPase RALA. May be involved in cytoskeletal organization (By similarity). Guanine nucleotide exchange factor for. {ECO:0000250, ECO:0000269|PubMed:10747847, ECO:0000269|PubMed:10889189}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Reference proteome | hsa:9649; | cytoplasm [GO:0005737]; plasma membrane [GO:0005886]; guanyl-nucleotide exchange factor activity [GO:0005085]; intracellular signal transduction [GO:0035556]; regulation of Ral protein signal transduction [GO:0032485]; small GTPase mediated signal transduction [GO:0007264] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21494904_The guanine-nucleotide exchange factor (GEF) RalGPS1a activates small GTPase Ral proteins such as RalA and RalB by stimulating the exchange of Ral bound GDP to GTP, thus regulating various downstream cellular processes. 34745385_Ral GEF with the PH Domain and SH3 Binding Motif 1 Regulated by Splicing Factor Junction Plakoglobin and Pyrimidine Metabolism Are Prognostic in Uterine Carcinosarcoma. | ENSMUSG00000038831 | Ralgps1 | 316.351869 | 1.0734682 | 0.102279516 | 0.19601201 | 2.690587e-01 | 6.039639e-01 | No | Yes | 297.662234 | 54.211009 | 272.933778 | 49.713464 | |||
| ENSG00000136997 | 4609 | MYC | protein_coding | P01106 | FUNCTION: Transcription factor that binds DNA in a non-specific manner, yet also specifically recognizes the core sequence 5'-CAC[GA]TG-3' (PubMed:24940000, PubMed:25956029). Activates the transcription of growth-related genes (PubMed:24940000, PubMed:25956029). Binds to the VEGFA promoter, promoting VEGFA production and subsequent sprouting angiogenesis (PubMed:24940000, PubMed:25956029). Regulator of somatic reprogramming, controls self-renewal of embryonic stem cells (By similarity). Functions with TAF6L to activate target gene expression through RNA polymerase II pause release (By similarity). Positively regulates transcription of HNRNPA1, HNRNPA2 and PTBP1 which in turn regulate splicing of pyruvate kinase PKM by binding repressively to sequences flanking PKM exon 9, inhibiting exon 9 inclusion and resulting in exon 10 inclusion and production of the PKM M2 isoform (PubMed:20010808). {ECO:0000250|UniProtKB:P01108, ECO:0000269|PubMed:20010808, ECO:0000269|PubMed:24940000, ECO:0000269|PubMed:25956029}. | 3D-structure;Acetylation;Activator;Alternative splicing;Chromosomal rearrangement;DNA-binding;Glycoprotein;Isopeptide bond;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | Mouse_homologues NA; + ;NA | This gene is a proto-oncogene and encodes a nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation. The encoded protein forms a heterodimer with the related transcription factor MAX. This complex binds to the E box DNA consensus sequence and regulates the transcription of specific target genes. Amplification of this gene is frequently observed in numerous human cancers. Translocations involving this gene are associated with Burkitt lymphoma and multiple myeloma in human patients. There is evidence to show that translation initiates both from an upstream, in-frame non-AUG (CUG) and a downstream AUG start site, resulting in the production of two isoforms with distinct N-termini. [provided by RefSeq, Aug 2017]. | hsa:4609; | chromatin [GO:0000785]; mitochondrion [GO:0005739]; Myc-Max complex [GO:0071943]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; RNA polymerase II transcription repressor complex [GO:0090571]; core promoter sequence-specific DNA binding [GO:0001046]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; E-box binding [GO:0070888]; protein dimerization activity [GO:0046983]; protein-containing complex binding [GO:0044877]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; transcription coregulator binding [GO:0001221]; amino acid transport [GO:0006865]; branching involved in ureteric bud morphogenesis [GO:0001658]; cell population proliferation [GO:0008283]; cellular iron ion homeostasis [GO:0006879]; cellular response to angiotensin [GO:1904385]; cellular response to arsenite(3-) [GO:1903841]; cellular response to carbohydrate stimulus [GO:0071322]; cellular response to cycloheximide [GO:0071409]; cellular response to dimethyl sulfoxide [GO:1904620]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to endothelin [GO:1990859]; cellular response to epidermal growth factor stimulus [GO:0071364]; cellular response to estrogen stimulus [GO:0071391]; cellular response to fibroblast growth factor stimulus [GO:0044344]; cellular response to growth hormone stimulus [GO:0071378]; cellular response to hydrostatic pressure [GO:0071464]; cellular response to hypoxia [GO:0071456]; cellular response to insulin stimulus [GO:0032869]; cellular response to interferon-gamma [GO:0071346]; cellular response to interleukin-1 [GO:0071347]; cellular response to lectin [GO:1990858]; cellular response to phorbol 13-acetate 12-myristate [GO:1904628]; cellular response to platelet-derived growth factor stimulus [GO:0036120]; cellular response to prolactin [GO:1990646]; cellular response to putrescine [GO:1904586]; cellular response to retinoic acid [GO:0071300]; cellular response to testosterone stimulus [GO:0071394]; cellular response to UV [GO:0034644]; cellular response to xenobiotic stimulus [GO:0071466]; chromatin remodeling [GO:0006338]; chromosome organization [GO:0051276]; DNA-templated transcription, initiation [GO:0006352]; energy reserve metabolic process [GO:0006112]; ERK1 and ERK2 cascade [GO:0070371]; fibroblast apoptotic process [GO:0044346]; G0 to G1 transition [GO:0045023]; G1/S transition of mitotic cell cycle [GO:0000082]; glucose metabolic process [GO:0006006]; hypothalamus development [GO:0021854]; in utero embryonic development [GO:0001701]; inner mitochondrial membrane organization [GO:0007007]; lactic acid secretion [GO:0046722]; liver regeneration [GO:0097421]; MAPK cascade [GO:0000165]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell division [GO:0051782]; negative regulation of fibroblast proliferation [GO:0048147]; negative regulation of gene expression [GO:0010629]; negative regulation of glucose import [GO:0046325]; negative regulation of monocyte differentiation [GO:0045656]; negative regulation of stress-activated MAPK cascade [GO:0032873]; negative regulation of transcription by RNA polymerase II [GO:0000122]; ovarian follicle development [GO:0001541]; ovulation [GO:0030728]; oxygen transport [GO:0015671]; positive regulation of ATP biosynthetic process [GO:2001171]; positive regulation of cell cycle [GO:0045787]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; positive regulation of DNA binding [GO:0043388]; positive regulation of DNA biosynthetic process [GO:2000573]; positive regulation of DNA methylation [GO:1905643]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of gene expression [GO:0010628]; positive regulation of glial cell proliferation [GO:0060252]; positive regulation of glycolytic process [GO:0045821]; positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator [GO:1902255]; positive regulation of mesenchymal cell proliferation [GO:0002053]; positive regulation of metanephric cap mesenchymal cell proliferation [GO:0090096]; positive regulation of mitochondrial membrane potential [GO:0010918]; positive regulation of oxidative phosphorylation [GO:1903862]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of response to DNA damage stimulus [GO:2001022]; positive regulation of smooth muscle cell migration [GO:0014911]; positive regulation of smooth muscle cell proliferation [GO:0048661]; positive regulation of telomerase activity [GO:0051973]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein-DNA complex disassembly [GO:0032986]; pyruvate transport [GO:0006848]; re-entry into mitotic cell cycle [GO:0000320]; regulation of cell cycle process [GO:0010564]; regulation of gene expression [GO:0010468]; regulation of mitotic cell cycle [GO:0007346]; regulation of somatic stem cell population maintenance [GO:1904672]; regulation of telomere maintenance [GO:0032204]; regulation of transcription by RNA polymerase II [GO:0006357]; response to estradiol [GO:0032355]; response to ethanol [GO:0045471]; response to gamma radiation [GO:0010332]; response to growth factor [GO:0070848]; response to human chorionic gonadotropin [GO:0044752]; response to wounding [GO:0009611]; response to xenobiotic stimulus [GO:0009410]; transcription by RNA polymerase II [GO:0006366] | 3277717_Alternative translation initiation from an upstream, in-frame non-AUG (CUG) and a downstream AUG start site results in the production of two isoforms with distinct N-termini. 11182042_Observational study of gene-disease association. (HuGE Navigator) 11443860_Review: EBV regulates c-MYC, apoptosis, and tumorigenicity in Burkitt's lymphoma 11777933_We have located a region in the c-myc promoter that is required for the complete activation by the immunoglobulin heavy chain gene enhancer 11795828_In Situ studies demonstrate enhanced mRNA expression of the proto-oncogene c-myc in stenotic venous bypass grafts. 11804592_c-Myc physically interacts with Smad2 and Smad3 involved in TGF-beta signaling.c-Myc promotes cell growth and cancer development partly by inhibiting the growth inhibitory functions of Smads 11817538_Uteroglobin promoter-targeted c-MYC expression in transgenic mice cause hyperplasia of Clara cells and malignant transformation of T-lymphoblasts and tubular epithelial cells 11840336_initiates illegitimate recombination of ribonucleotide reductase 2 gene 11848444_Acute hyperglycaemia induces an up-regulation of seven major genes, four of which were not previously reported in the literature. Northern blot analyses, performed on these 4 genes, confirm macroarrays results for alphav, beta4, c-myc, and MUC18. 11848471_Activation of c-MYC and c-MYB proto-oncogenes is associated with decreased apoptosis in tumor colon progression. 11850082_amplification in acute nonlymphocytic leukemia 11861398_Repression of alpha-fetoprotein gene expression under hypoxic conditions in human hepatoma cells: characterization of a negative hypoxia response element that mediates opposite effects of hypoxia inducible factor-1 and c-Myc. 11877042_The cytoplasmic domain of gp130 is involve the induction of c-myc expression and the cell proliferation in Meg-01 cell. 11877298_Retinoic acid-induced cell cycle arrest of human myeloid cell lines is associated with sequential down-regulation of c-Myc and cyclin E. 11909963_Scatter factor/hepatocyte growth factor stimulation of glioblastoma cell cycle progression through G(1) is c-Myc dependent and independent of p27 suppression, Cdk2 activation, or E2F1-dependent transcription. 11916966_complex with Nmi and BRCA1 inhibits c-Myc-induced human telomerase reverse transcriptase gene promoter activity in breast cancer 11956070_c-myc-induced apoptosis in polycystic kidney disease is independent of FasL/Fas interaction. 11960382_The N-myc and c-myc downstream pathways include the chromosome 17q genes nm23-H1 and nm23-H2 11968011_estrogen and Myc negatively regulate EphA2 expression in mammary epithelial cells 12027803_A novel form of the RelA nuclear factor kappaB subunit is induced by and forms a complex with this proto-oncogene protein 12031912_overexpressed in acute myeloid leukemia while translocations associated with this gene are absent 12032779_The proto-oncogene c-myc in hematopoietic development and leukemogenesis 12049739_c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function 12072203_Overexpression of c-myc mRNA was found in an Achilles tendon clear cell sarcoma and may have a role in its malignant progression. 12077335_TRRAP binding and the recruitment of histone H3 and H4 acetyltransferase activities are required for the transactivation of a silent TERT gene in exponentially growing human fibroblasts by c-Myc or N-Myc protein. 12082260_Effects of fluid shear stress on expression of proto-oncogenes c-fos and c-myc in cultured human umbilical vein endothelial cells. 12130502_the in vitro effects of iron on the proliferation of a primary, human synovial fibroblast cell line and the involvement of c-myc in this process as a model for c-myc proto-oncogene expression in hemophilic synovitis 12149248_role of c-Myc increasing susceptibility to tumor necrosis factor mediated apoptosis 12177005_Myc activates transcription by stimulating elongation and that P-TEFb is a key mediator of this process 12209953_dysregulated beta-catenin may cause a transcriptional upregulation of the c-myc gene; the c-Myc protein expression appears to be further regulated by posttranscriptional mechanism(s) during neoplastic progression in colorectal adenocarcinomas 12213716_C-Myc may be a downstream target of anaplastic lymphoma kinase (ALK)signaling and its expression a defining characteristic of ALK-positive anaplastic large cell lymphomas 12226097_c-myc induces programmed cell death in a process requiring glutathione in human tumor cells 12237776_Amplifications of c-myc and CCND1 are associated with detrusor-muscle-invasive transitional cell carcinoma 12356725_results reveal a novel cytoskeletal function for Myc and indicate the feasibility of quantitative whole-proteome analysis in mammalian cells 12360279_This review describes the role of MYC in tumor progression 12362975_c-myc and c-erbB2 amplification in breast cancer 12379776_Marked intratumoral heterogeneity of this and cyclinD1 but not of c-erbB2 amplification in breast cancer 12394763_Beta-catenin mutations correlate with over expression of C-myc and cyclin D1 Genes in bladder cancer. 12420224_CD19+ cells from transgenic mice with a lamba-humanMYC construct driven by B-cell elements overexpressed both C-MYC and protein kinase A-Cbeta. 12435808_c-Myc promotes cell survival under stressful conditions. 12452058_the mechanism of apoptosis induced by c-myc gene in oral squamous cell 12490316_Reduction in the c-myc protein was correlated with neck metastasis in nasopharyngeal carcinoma. 12509468_c-myc translation is regulated by hnRNP C via internal ribosomal entry site binding 12529648_c-Myc binds to TFIIIB, a pol III-specific general transcription factor, and directly activates pol III transcription 12538578_examination of functionality of basic domains compared with Mad 12545156_repression of differentiation-induced p21 expression through Miz-1 may be an important mechanism by which Myc blocks differentiation 12562237_Expression of c-Myc in primary central nervous system diffuse large B-cell lymphoma may be a prognostic marker for poor overall survival. 12637327_results show that Ser727/Tyr701-phosphorylated Stat1 plays a key role as a prerequisite for the ATRA-induced down-regulation of c-Myc; cyclins A, B, D2, D3, and E; and simultaneous up-regulation of p27Kip1, associated with arrest in the G0/G1 phase 12637527_results indicate that Nijmegen breakage syndrome 1 gene (NBS1) is a direct transcriptional target of c-Myc and links the function of c-Myc to the regulation of DNA double-strand break repair pathway 12646176_c-myc binds to the hTERT promoter and is involved in the pathway for regulation of cellular immortalization through BRCA1 12673205_A detailed structure-function analysis of the Myc N-terminal domain in deletion & point mutants studied their ability to induce cell cycle progression, apoptosis & transformation as well as repress & activate endogenous target genes. 12688321_Up-regulation of c-Myc protein could disturb the signaling pathway of ceramide & sphingosine, endogenous modulators mediating apoptosis. c-Myc protein is a mediator of cytoprotective effect of PKC pathway in HL-60 cells. 12721301_Myc down-regulation might directly mediate the growth-inhibitory properties of 3'-5' RNA exonuclease 12729735_inverse correlation between TMEFF2 and c-Myc expression 12748187_findings suggest that Myc-mediated functions can be negatively regulated by KRAB box containing zinc finger protein (Krim-1) 12776177_MYC recruits the TIP60 histone acetyltransferase complex to chromatin 12783888_c-myc gene is regulated by nitric oxide via inactivating NF-kappa B complex 12808131_c-Myc together with its heterodimeric partner, Max, occupy >15% of gene promoters tested in Burkitt lymphoma cells. Overexpressed c-Myc plays a role in global transcriptional regulation in some cancer cells and functions in malignant transformation. 12818373_the PI3K/p70 S6K/c-Myc cascade plays an important role in neutrophilic proliferation in HL-60 cells. 12821782_Induction of hTERT by Myc/E6 was independent of Myc phosphorylation at Thr-58/Ser-62 within the transactivation domain. E6 associates with Myc complexes & activates a Myc-responsive gene, hTERT. 12824159_Inhibition of this oncoprotein limits the growth of human melanoma cells by inducing cellular crisis 12829833_expression of MYC oncogene is reduced at the mRNA level and MYC protein has an increased half-life in Adenovirus infected Hela cells resulting in constant steady state levels. 12842909_results demonstrate that MYC directly stimulates transcription of the Werner syndrome gene,WRN; propose that WRN up-regulation by MYC may promote MYC-driven tumorigenesis by preventing cellular senescence 12855588_hematopoietic growth factors induce cell cycle progression via internal ribosome entry site-mediated translation of c-myc though the PI3K pathway in human factor-dependent leukemic cells 12909717_overexpression of MYC disrupts the repair of double-strand DNA breaks, resulting in a several-magnitude increase in chromosomal breaks and translocations 12970749_the mutated version of the c-myc IRES that is prevalent in patients with multiple myeloma bound hnRNPK more efficiently in vitro and was stimulated by hnRNPK to a greater extent in vivo. 14532990_p53 and c-Myc expression may have a role in regulation of telomerase activity in ovarian tumours 14576301_c-Myc regulates cell growth and proliferation by the coordinated induction of cdk activity and rRNA processing 14625288_contribution of JNK to the regulation of c-Myc protein stability under normal growth conditions 14645543_identified an evolutionarily conserved boundary at which the c-myc transcription unit is separated from the flanking condensed chromatin enriched in lysine 9-methylated histone H3 14645547_PLZF expression maintains a cell in a quiescent state by repressing c-myc expression and preventing cell cycle progression. 14663479_we review Myc-induced pathways that contribute to the apoptotic response. 14663583_These data also suggest that location of intragenic PTEN mutations and their coexistence with the CMYC amplification may play a crucial part in the development of various subtypes of endometrial carcinoma. 14672406_Data suggest that cyclin dependent kinase 4 is involved in the development of tobacco-mediated oral carcinogenesis, and that c-myc expression is absent in normal and high in later stages of oral cancer development. 14697225_c-myc may have a role in the development of Burkitt's lymphoma, through an RNA-mediated gene silencing pathway 14724288_c-Myc may control the activity of multiple signal pathways involved in cellular transformation by induction of HSP90A 14744757_Aurora-A induces telomerase activity and hTERT by up-regulation of c-Myc 14760071_MYC has a role in tumor progression in BRCA1-associated breast cancers 14769798_c-Myc expression is regulated by cytosolic phospholipase A2 in a process that involves B-myb 14991929_c-Myc does not play a major role in gene regulation of the catalytic subunit of telomerase (hTERT) in human hepatocellular carcinoma. 15067010_c-Myc can support self-renewal of HSCs as a downstream mediator of Notch and HOXB4. 15071503_Myc is an integral part of a novel HIF-1alpha pathway, which regulates a distinct group of Myc target genes in response to hypoxia. 15077166_Levels of both c-myc and beta-catenin increased in Cyr61 stably transfected H520 cells. 15083194_Gene amplification of c-myc may play key role in regulating expression of its mRNA and protein in high-grade breast cancers 15108364_c-Myc can specifically recognize the HIV-1 initiator element surrounding the start site of transcription and linker scanning mutagenesis experiments confirmed a loss of c-Myc-mediated repression in the absence of this region 15126105_High nuclear expression of c-myc is correlated with locally advanced colorectal cancer 15160911_C-Myc over-expression was significantly associated with high sVEGF and normal sFlt-1 level in DLBCL patients, suggesting a complex interrelationship between c-Myc oncogene expression and angiogenic regulators 15190416_expression of c-Myc protein is increased not only in uterine cervix cancer but also in the premalignant lesions 15191563_Translocations involving the MYC locus were detected in six cases of B-cell lymphoma, five of them derived from a MYC/IGH juxtaposition and one from a translocation involving a non-IG gene partner. 15192039_c-myc transactivates rat and human adrenomedullin genes and accelerates the degradation rate of adrenomedullin mRNA. 15198123_c-myc mRNA was detected in 18 of 59 cases, and was associated with shorter patient survival times on both univariate and multivariate analyses. The presence of c-myc mRNA was also significantly associated with tumor anaplasia. 15199070_cMyc is a target of ARF tumor suppressor 15199147_Myc binds well to conserved canonical E boxes, but not nonconserved E boxes. Results show Myc is an important regulator of glycolytic genes, suggesting that MYC plays a role in a switch to glycolytic metabolism during cell proliferation or tumorigenesis. 15243561_c-myc oncogene has not shown to be a prognostic factor for laryngeal squamous cell carcinoma of the supraglottic larynx. 15282543_c-MYC activated transcription from the upstream binding factor promoter 15286700_A prolactin signalling cascade in W53 cells involves Src kinases that mediate stimulation of c-Myc expression. 15287031_There is no obvious correlation between breakpoint locations within the immunoglobulin H locus and the amount of MYC mRNA. 15355849_polyamine-induced nuclear c-Myc interacts with Max, binds to the specific DNA sequence, and plays an important role in stimulation of normal intestinal epithelial cell proliferation. 15361884_p53-independent checkpoint to prevent c-Myc-mediated tumorigenesis 15457447_Inhibition of c-Myc (and c-Raf-1) significantly reduced the growth and invasiveness of rheumatoid arthritis synovial fibroblasts in the SCID mouse model. 15459488_There was a significant positive correlation between the levels of c-myc mRNA expression and the occurrence of apoptosis in 59 hepatocellular carcinomas. 15468060_novel iron-dependent cell cycle regulatory mechanism involving modulation of translocated c-myc gene expression 15474507_c-myc downregulation and release from the endogenous p21WAF1/CIP1 promoter contributes to transcriptional activation of the p21WAF1/CIP1 in HeLa cells 15516975_Myc is essential for the regulation of a large number of growth-related genes in B cells, and cannot be replaced by other serum-induced factors. 15522869_CREB binding protein is essential for keeping c-myc in a repressed state in G(1) and thereby preventing inappropriate entry of cells into S phase. 15528212_c-Myc antagonized the induction of p21Cip1 mediated by oncogenic H-, K-, and N-Ras and by constitutively activated Raf and ERK2 15542830_chromosomal c-myc replicator activity can be altered by transcription factors that induce transcription 15580293_Myc stimulates VEGF production by a rapamycin- and LY294002-sensitive pathway. 15595642_LMP1 activates telomerase via c-myc 15601838_c-myc expression is regulated by TFIIH using an expanded proximal promoter 15629428_the increased affinity for the duplex state due to mutation in the nuclease hypersensitive element could play a functional role in the aberrant regulation of c-myc 15645079_c-myc and mad1 can regulate the hTERT transcript in a different manner in hTERT positive cells, but not in normal cells 15663936_the entire N and C-terminal regions of c-Myc transactivation domain act in concert to achieve high specificity and affinity to two structurally and functionally orthogonal target proteins 15688026_Tyrosine residues become phosphorylated following receptor engagement and, as such, form two Grb2 binding sites, which have been proposed to be differentially coupled to Myc-dependent survival. 15697230_This paper reports the first solution structure of a G-quadruplex found to form in the promoter region of an oncogene (c-MYC). 15716988_frequency and chromosomal features of this der(8)t(8;14;18) in a series of acute leukaemias and malignant lymphomas 15723053_c-Myc coordinates the activity of all three nuclear RNA polymerases, and thereby plays a key role in regulating ribosome biogenesis and cell growth 15723054_stimulation of rRNA synthesis by c-Myc is a key pathway driving cell growth and tumorigenesis 15739117_Stimulation of islet expression by 30 mmol/l glucose results from activation of a distinct, probably oxidative-stress-dependent signalling pathway. 15743499_c-Myc has a pivotal function in the development of breast cancer; data show that decreasing the c-Myc protein level in MCF-7 cells by RNAi could significantly inhibit tumor growth both in vitro and in vivo 15756435_c-MYC amplification is an early event in breast cancer progression, while ZNF217 and Her2/neu amplification may play a role in the later stage of tumor development 15763593_expression of ESRA, bcl-2 and c-myc gene expression in fibroadenomas and adjacent normal breast is related to nodule size, hormonal and reproductive features 15769738_observed, subsequent to knocking down CRD-BP/IMP1, decreased c-myc expression, increased IGF-II mRNA levels, and reduced cell proliferation rates 15800668_p16(INK4A) reconstitution in p16(INK4A)-deficient T-ALL cells induced cell cycle arrest in the presence of cyclin E and c-Myc expression, uncoupled growth from cell cycle progression and caused a sequential process of growth, differentiation and apoptosis 15839202_Data show a coordinating inhibition of the proliferation of MCF-7 by enhancing expression of p53,p21 and decreasing expression of c-myc 15856024_elevated levels of Myc counteract p53 activity in human tumor cells.This mechanism could contribute to explain the c-Myc deregulation frequently found in cancer 15878876_chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z 15929079_Observational study of gene-disease association. (HuGE Navigator) 15937962_the c-MYC gene is sufficient to induce carcinogenesis for prostate cancer 15944709_c-Myc activates expression of a cluster of six miRNAs on human chromosome 13 15965094_results are consistent with the possibility that IL-5 inhibits apoptosis in JYTF-1 cells via the upregulation of c-myc expression 15972952_The presence of activated beta-catenin and c-myc in the epidermis of chronic wounds may serve as a molecular marker of impaired healing 15986448_conclude that alterations of the CMYC gene, including copy number gains and amplifications, are linked to genetically unstable bladder cancers that are characterized by a high histologic grade and/or invasive growth 15988031_A pivotal role for Myc in regulating mitochondrial biogenesis was shown. 15991278_Observational study of gene-disease association. (HuGE Navigator) 15992821_tumor-specific isoforms of Bin1 are precluded from interaction with c-Myc through an intramolecular polyproline-SH3 domain interaction. Furthermore, c-Myc/Bin1 interaction can be inhibited by phosphorylation of c-Myc at Ser62. 16007143_c-Myc overexpression showed an upregulation of beta4 promoter activity 16085756_Kinetic analysis of the interconversion between the duplex and the quadruplex forms of the human c-myc promoter. 16094360_mutant MYC proteins, by selectively disabling a p53-independent pathway, enable tumour cells to evade p53 action during lymphomagenesis 16107691_p53 represses c-myc transcription through a mechanism that involves histone deacetylation 16107734_17-beta-estradiol promotes survival signals in breast cancer cells through mammalian target of rapamycin-dependent increase in Myc expression 16116477_MYC family genes might affect oncogenesis through distinct sets of targets, in particular implicating the importance of transcriptional repression 16126174_p300 can acetylate DNA-bound Myc:Max complexes and that acetylated Myc:Max heterodimers efficiently interact with Miz-1 16139224_The MYC protein phosphorylation and turnover are thus linked to cell cycle exit in primary mouse CGNP cultures and the developing cerebellum. 16167342_replicative senescence-specific factors may block c-Myc inhibition of Miz-1 activation of hMad4 expression, and the continual presence of hMad4 protein may transcriptionally repress selected c-Myc target genes 16169462_Increased wild-type MYC expression occurs frequently in human cancers, except in Burkitt's lymphoma. 16172399_c-MYC binds to the genomic MTA1 locus and recruits transcriptional coactivators, which is essential for the transformation potential of c-MYC. 16173081_transcription activity is repressed by various MM-1 isoforms 16174239_expression of cyclin D1 and c-Myc in epithelial ovarian cancer reaffirms the notion that they are crucial components in the pathway of tumorigenesis 16201965_data demonstrate that the sphingosine-recycling pathway for the generation of endogenous long-chain ceramide in response to exogenous C6-cer is regulated by ROS, and plays an important biological role in controlling c-Myc function 16205115_Myc induces nuclear encoded mitochondrial gene expression and mitochondrial biogenesis, thereby directly linking Myc's transcriptional properties to mitochondrial ROS production, promotion of genomic oxidative damage, and genomic instability. 16210249_These results identify glycogen synthase kinase 3beta and FBW7 as potential cancer therapeutic targets and MYC as a critical substrate in the GSK3beta survival-signaling pathway. 16260605_c-Myc isoforms differentially regulate cell growth and apoptosis in a species-specific manner 16269333_Site-specific ubiquitination regulates the switch between an activating and a repressive state of the Myc protein, and they suggest a strategy to interfere with Myc function in vivo. 16287840_dual roles for p300-CBP-associated factor in Myc regulation: as a Myc coactivator that stabilizes Myc and as an inducer of Myc instability via direct Myc acetylation 16293596_decreased internal ribosome entry site (IRES)-dependent Myc mRNA translation accounts for the phenotypic changes induced by inhibition of the BCR/ABL-ERK-dependent HNRPK translation-regulatory function. 16295419_C-myc expression shows a positive association with increasing grade of breast carcinoma. 16308474_We aimed to investigate the expression pattern of p53, Bcl-2 and C-Myc in colorectal cancer(CRC)tissues obtained from Egyptian colorectal cancer patients divided in two different groups, associated with and without Schistosoma mansoni. 16316993_up-regulation of mitochondrial CLIC4, together with a reduction in Bcl-2 and Bcl-xL, sensitizes Myc-expressing cells to the proapoptotic action of Bax. 16328057_Down-regulation of MYC is not necessary to abolish malignant phenotypes by induction of terminal monocyte-macrophage differentiation in leukaemic cells carrying t(9;11)(p22;q23). 16352593_Miz-1 is essential for Myc-mediated apoptosis 16367922_interferon-gamma, which induces HLA-DR antigens on the cell surface, also suppresses c-myc expression in situ, and is a possible non-immunological mechanism involved in the better long-term survival of colorectal cancer patients 16376880_Data show that hepatitis B virus X protein blocks the ubiquitination of c-myc through a direct interaction with the F box region of Skp2 and destabilization of the SCF(Skp2) complex. 16378632_dominant-negative forms of c-Myc block transformation by activated Notch1, E6 and E7, suggesting that c-Myc is required for HPV16-mediated transformation 16410719_study shows that p14ARF directly associates with Myc and relocates Myc from the nucleoplasm to the nucleolus, in addition, p14ARF down regulates Myc activated transcription 16410805_overexpression of c-myc protects melanoma cells from IFN-beta-mediated growth inhibition 16423995_PEG10 is a direct target of c-MYC; findings link cancer genetics and epigenetics by showing that a classic proto-oncogene, MYC, acts directly upstream of a proliferation-positive imprinted gene, PEG10 16423996_c-myc seems to play a causal role in inducing anaplasia in medulloblastoma; it is proposed that c-myc dysregulation is involved in the progression of these malignant embryonal neoplasms 16432227_Myc overexpression causes DNA damage in vivo and the ATM-dependent response to this damage is critical for p53 activation, apoptosis, and the suppression of tumor development 16466700_Modifications in iron metabolism, resulting from the strong basal expression of c-Myc, and amplified by iron addition, could lead to a disruption in homeostasis and consequently to growth arrest in Burkitt's lymphoma. 16478988_abnormal expression of REST/NRSF and MYC in undifferentiated neural stem/progenitor cells causes cerebellum-specific tumors. Furthermore, these results suggest that such a mechanism plays a role in the formation of human medulloblastoma. 16490593_Positive C-MYC staining is detected mainly in the cytoplasm of esophageal cancer cells. 16494045_Inhibition of hTERT expression by RNAi could increase the expression of C-myc protein in Hep-2 cells. 16508012_These findings provide a molecular basis for increased TFRC1 expression in human tumors, illuminate the role of TFRC1 in the c-Myc target gene network 16537449_p16(INK4a) expression was regulated by the Polycomb group repressor Bmi-1, which is shown as a direct transcriptional target of c-Myc. 16537801_Doubling the hMYC level by breeding homozygous transgenic animals switched the phenotype from primarily monocytic tumors to exclusively T-cell tumors. Results imply that MYC level affects the spontaneous acquisition of synergistic oncogenic mutations. 16541139_In both Richter's transformation and prolymphocytic transformation, high-levels of AID mRNA expression and high-frequency mutations of c-MYC genes were detected. 16543245_there is an alternative mechanism for the hypoxic induction of VEGF in colon cancer that does not depend upon HIF-1alpha but instead requires the activation of PI3K/Rho/ROCK and c-Myc 16552729_Results showed that certain regulation involved in c-myc, c-fos, and c-jun was present in the apoptosis, and the c-Myc dependent-on and Jun N-terminal kinase (JNK) pathway also play roles. 16554306_Bcl2, in addition to its survival function, may also suppress DNA repair in a novel mechanism involving c-Myc and APE1, which may lead to an accumulation of DNA damage in living cells, genetic instability, and tumorigenesis 16572399_Taken together, these findings suggest that Bax and caspase activation, together with PKCdelta signaling are involved in c-Myc-dependent etoposide-induced apoptosis. 16582589_Myc and E2F1 engage the ATM signaling pathway to activate p53 and induce apoptosis [review] 16596619_These data suggest that an increase in c-Myc phosphorylation in response to prolonged ERK phosphorylation negatively auto-regulates c-Myc gene expression, leading to the suppression of its target gene expression and cell cycle block. 16628215_Results describe the roles of the FarUpStream Element (FUSE), FUSE Binding Protein (FBP), FBP Interacting Repressor (FIR), and TFIIH in the regulation of c-myc expression. 16631470_APL-like case lacking t(15;17) and the retinoic acid receptor alpha (RARA) breakpoint and also has the deletion MYC of 8q24 associated with the occurrence of MYC amplification on double-minute chromosomes (dmin). 16675879_Results show that the CT-element is not especially susceptible to the formation of breakpoints leading to chromosomal translocations in Burkitt's lymphoma. 16682952_MYC activation, triggered by the insertion of human papillomavirus DNA sequences, can be an important genetic event in cervical oncogenesis. 16690525_in diffuse large B-cell lymphoma, molecular alterations in ice, bcl-2, c-myc and p53 are present in hematopoietic cells from bone marrow as well as in primitive hematopoietic progenitors 16706751_p21Cip1 is one of the direct mediators of induced c-Myc following increased polyamines and that p21Cip1 repression by c-Myc is implicated in stimulation of normal intestinal epithelial cell proliferation 16724113_This study provides the first evidence for regulation of global chromatin structure by an oncoprotein and may explain the broad effects of Myc on cell behavior and tumorigenesis. 16785237_TGFbeta suppresses TERT by Smad3 interactions with c-Myc and the hTERT gene 16788099_c-Myc is only required for pre-TCR-induced proliferation but is dispensable for developmental progression from the DN to the DP stage 16797278_Having an S allele in the L-myc gene may increase the risk of renal failure. 16797278_Observational study of gene-disease association. (HuGE Navigator) 16809765_proteins implicated in replication initiation are selectively and differentially bound across the c-myc replicator, dependent on discrete structural elements in DNA or chromatin 16857742_Ras and c-Myc play important roles in the up-regulation of nucleophosmin/B23 during proliferation of cells associated with a high degree of malignancy, thus outlining a signaling cascade involving these factors in the cancer cells. 16874304_Together, these results demonstrate that ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway is an important mechanism of oncogenic c-myc activation in pancreatic cancer. 16940161_Ketogenesis is an undesirable metabolic characteristic of the proliferating cell, which is down-regulated through c-Myc-mediated repression of the key metabolic gene HMGCS2. 16964280_Functional promoter analyses revealed that both the Myc-binding site cluster and the C/EBPalpha-binding site are essential for strong transcriptional activation, and that Myc and C/EBPalpha synergistically activate the WS5 promoter 16984727_c-myc and VEGFA may be involved in the regulation of angiogenesis in esophageal adenocarcinoma 16996503_provide a mechanism for elevated Myc expression in hormone-dependent and hormone-independent breast cancer 17001014_Epstein-Barr virus (EBV) acts as an antiapoptotic in Burkitt lymphoma (BL) by selecting among three transcriptional programs, all of which, unlike the full virus growth-transforming program, remain compatible with high c-myc expression. 17028906_Several genes in the MYCN amplicon, including the DEAD box polypeptide 1 (DDX1) gene, and neuroblastoma-amplified gene (NAG gene), have been found to be frequently co-amplified with MYCN in NB. 17046748_Nuclear arrangement of c-myc genes and transcripts was conserved during cell differentiation and, therefore, independent of the level of differentiation-specific c-myc gene expression. 17050536_NDRG2 expression is repressed by Myc via Miz-1-dependent interaction with the NDRG2 core promoter 17070983_Fluorescent protocol can successfully be applied to diagnostic needle biopsies to identify relative 8q gain in prostate carcinomas and that patients with a MYC/CEP18 ratio > or = 1.5 present a significantly higher risk of dying from the disease. 17082179_repression of BCL2 transcription is the single essential consequence of targeting the MIZ-1 pathway during apoptosis induction, which explains a copperative interaction between c-MYC and BCL2 17082780_Max as a novel co-activator of C/EBPalpha functions, thereby suggesting a possible link between C/EBPalpha and Myc-Max-Mad network. 17093053_Results provide a snapshot of genome-wide, unbiased characterization of direct Myc binding targets in a model of human B lymphoid tumor using ChIP coupled with pair-end ditag sequencing analysis (ChIP-PET). 17114293_identify c-MYC as an essential mediator of NOTCH1 signaling and integrate NOTCH1 activation with oncogenic signaling pathways upstream of c-MYC 17141659_FOXM1c transactivates the human c-myc P1 and P2 promoters synergistically with Sp1, a transcription factor known to bind and transactivate these two promoters 17145812_The c-Myc oncogene expression was very low in normal and cancer tissues but highly increased in papillomatosis. 17146437_E-cadherin repression is necessary for c-Myc-induced cell transformation. 17146439_establish PML-mediated destabilization of Myc and the derepression of cell cycle inhibitor genes as an important regulatory mechanism that allows cell differentiation 17151361_Our data suggest that Mel-18 regulates Bmi-1 expression during senescence via down-regulation of c-Myc. 17158641_MYC amplification is not associated with 'basal-like' phenotype and proved to be an independent prognostic factor for breast cancer patients treated with anthracycline-based chemotherapy. 17159920_c-MYC functions are cellular-context-dependent and | ENSMUSG00000049086+ENSMUSG00000022346 | Bmyc+Myc | 4008.597713 | 0.9075870 | -0.139892112 | 0.05884130 | 5.623621e+00 | 1.772000e-02 | 1.779690e-01 | No | Yes | 4092.136081 | 318.553082 | 4460.103971 | 346.941128 |
| ENSG00000137038 | 90871 | DMAC1 | protein_coding | Q96GE9 | FUNCTION: Required for the assembly of the mitochondrial NADH:ubiquinone oxidoreductase complex (complex I). Involved in the assembly of the distal region of complex I. {ECO:0000269|PubMed:27626371}. | Alternative splicing;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:90871; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 19240061_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000028398 | Dmac1 | 1067.686827 | 0.9361462 | -0.095194232 | 0.09246376 | 1.048196e+00 | 3.059228e-01 | 6.728389e-01 | No | Yes | 989.189825 | 95.456328 | 1048.829224 | 100.985304 | ||
| ENSG00000137177 | 63971 | KIF13A | protein_coding | Q9H1H9 | FUNCTION: Plus end-directed microtubule-dependent motor protein involved in intracellular transport and regulating various processes such as mannose-6-phosphate receptor (M6PR) transport to the plasma membrane, endosomal sorting during melanosome biogenesis and cytokinesis. Mediates the transport of M6PR-containing vesicles from trans-Golgi network to the plasma membrane via direct interaction with the AP-1 complex. During melanosome maturation, required for delivering melanogenic enzymes from recycling endosomes to nascent melanosomes by creating peripheral recycling endosomal subdomains in melanocytes. Also required for the abcission step in cytokinesis: mediates translocation of ZFYVE26, and possibly TTC19, to the midbody during cytokinesis. {ECO:0000269|PubMed:19841138, ECO:0000269|PubMed:20208530}. | ATP-binding;Alternative splicing;Cell cycle;Cell division;Coiled coil;Cytoplasm;Cytoskeleton;Endosome;Golgi apparatus;Membrane;Microtubule;Motor protein;Nucleotide-binding;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a member of the kinesin family of microtubule-based motor proteins that function in the positioning of endosomes. This family member can direct mannose-6-phosphate receptor-containing vesicles from the trans-Golgi network to the plasma membrane, and it is necessary for the steady-state distribution of late endosomes/lysosomes. It is also required for the translocation of FYVE-CENT and TTC19 from the centrosome to the midbody during cytokinesis, and it plays a role in melanosome maturation. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Aug 2011]. | hsa:63971; | centrosome [GO:0005813]; endosome membrane [GO:0010008]; Golgi membrane [GO:0000139]; kinesin complex [GO:0005871]; microtubule [GO:0005874]; midbody [GO:0030496]; trans-Golgi network membrane [GO:0032588]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; microtubule binding [GO:0008017]; microtubule motor activity [GO:0003777]; cell cycle [GO:0007049]; cell division [GO:0051301]; endosome to lysosome transport [GO:0008333]; Golgi to plasma membrane protein transport [GO:0043001]; intracellular protein transport [GO:0006886]; melanosome organization [GO:0032438]; microtubule-based movement [GO:0007018]; plus-end-directed vesicle transport along microtubule [GO:0072383]; regulation of cytokinesis [GO:0032465]; vesicle cargo loading [GO:0035459] | 11106728_Functionally characterizes the homologous mouse protein. 19328558_Observational study of gene-disease association. (HuGE Navigator) 19841138_show that the clathrin adaptor AP-1 and the kinesin motor KIF13A together create peripheral recycling endosomal subdomains in melanocytes required for cargo delivery to maturing melanosomes. 20208530_PtdIns(3)P production is essential for proper cytokinesis. PtdIns(3)P-binding centrosomal protein FYVE-CENT and TTC19 control cytokinesis through their translocation from the centrosome to the midbody mediated by the kinesin protein KIF13A. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21873978_the kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization 24462287_KIF13A interacts and cooperates with RAB11 to generate endosomal tubules. 24859087_kjh kiiou ouiy 8yt 8iy 7i9y 29061883_that KIF13A plays an important role in the transport of influenza A viral ribonucleoproteins 29980677_the crystal structure of a catalytically active kinesin-13 monomer (Kif2A) in complex with two bent alphabeta-tubulin heterodimers in a head-to-tail array, providing a view of these interactions, is reported. 30049714_KIF13A-mediated endosomal trafficking modulates RhoB plasma membrane localization to control membrane blebbing and blebby amoeboid migration. 31469403_Identification of novel splicing patterns and differential gene expression in RE+/FECD- samples provides new insights and more relevant gene targets that may be protective against FECD disease in vulnerable patients with TCF4 CTG TNR expansions. 33526817_Postzygotic inactivating mutation of KIF13A located at chromosome 6p22.3 in a patient with a novel mosaic neuroectodermal syndrome. 35122963_Coordination of two kinesin superfamily motor proteins, KIF3A and KIF13A, is essential for pericellular matrix degradation by membrane-type 1 matrix metalloproteinase (MT1-MMP) in cancer cells. | ENSMUSG00000021375 | Kif13a | 984.445035 | 1.1807263 | 0.239674600 | 0.10985450 | 4.836251e+00 | 2.786741e-02 | 2.238505e-01 | No | Yes | 1056.837918 | 136.620859 | 892.562110 | 115.456999 | |
| ENSG00000137221 | 93643 | TJAP1 | protein_coding | Q5JTD0 | FUNCTION: Plays a role in regulating the structure of the Golgi apparatus. {ECO:0000250|UniProtKB:Q9DCD5}. | Acetylation;Alternative splicing;Cell junction;Cell membrane;Coiled coil;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Tight junction | This gene encodes a tight junction-associated protein. Incorporation of the encoded protein into tight junctions occurs at a late stage of formation of the junctions. The encoded protein localizes to the Golgi and may function in vesicle trafficking. Alternatively spliced transcript variants have been described. A related pseudogene exists on the X chromosome. [provided by RefSeq, Mar 2009]. | hsa:93643; | bicellular tight junction [GO:0005923]; Golgi apparatus [GO:0005794]; plasma membrane [GO:0005886]; trans-Golgi network [GO:0005802]; Golgi organization [GO:0007030] | ENSMUSG00000012296 | Tjap1 | 1611.578398 | 1.0023229 | 0.003347292 | 0.08121385 | 1.690679e-03 | 9.672019e-01 | 9.884466e-01 | No | Yes | 1651.750176 | 145.133993 | 1677.370404 | 147.292554 | ||
| ENSG00000137411 | 57176 | VARS2 | protein_coding | Q5ST30 | ATP-binding;Alternative splicing;Aminoacyl-tRNA synthetase;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Primary mitochondrial disease;Protein biosynthesis;Reference proteome;Transit peptide | This gene encodes a mitochondrial aminoacyl-tRNA synthetase, which catalyzes the attachment of valine to tRNA(Val) for mitochondrial translation. Mutations in this gene cause combined oxidative phosphorylation deficiency-20, and are also associated with early-onset mitochondrial encephalopathies. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Aug 2014]. | hsa:57176; | cytosol [GO:0005829]; mitochondrion [GO:0005739]; aminoacyl-tRNA editing activity [GO:0002161]; ATP binding [GO:0005524]; valine-tRNA ligase activity [GO:0004832]; valyl-tRNA aminoacylation [GO:0006438] | 18400783_Data indicate that variations in the levels of VARS2L between tissue types and patients could underlie the difference in clinical presentation between individuals homoplasmic for the 1624C>T MTTV (equivalent to mt-tRNAVal C25U) mutation. 19851445_Observational study of gene-disease association. (HuGE Navigator) 20503108_Observational study of gene-disease association. (HuGE Navigator) 20503108_VARS2 V552V may be considered as a prognostic factor for survival in patients with early breast cancer. 20877624_Observational study of gene-disease association. (HuGE Navigator) 25404243_VARS2 polymorphism significantly associated with chronic hepatitis B in Korean population. 29137650_VARS2 mutation underlies a novel autosomal recessive syndrome with epilepsy, mental retardation, short stature, growth hormone deficiency, and hypogonadism. 29314548_this gene should be considered in early-onset mitochondrial encephalomyopathies or encephalocardiomyopathies. 30458719_The novel compound heterozygous pathogenic VARS2 mutations c.643 C > T (p. His215Tyr) and c.1354 A > G (p. Met452Val) has been reported in a female infant. These heterozygous mutations were carried individually by the proband's parents and elder sister. 31064326_report two additional cases of two non-related young girls from Sardinia, born from non-consanguineous and healthy parents, carrying the aforesaid homozygous VARS2 variant. At onset both the patients presented with worsening psychomotor delay, muscle hypotonia and brisk tendon reflexes 31529142_Study shows A combination of two novel VARS2 variants causes a mitochondrial disorder associated with failure to thrive and pulmonary hypertension | ENSMUSG00000038838 | Vars2 | 1997.663542 | 0.9522646 | -0.070565596 | 0.06905918 | 1.047381e+00 | 3.061109e-01 | 6.728389e-01 | No | Yes | 1922.900672 | 193.726377 | 2028.833489 | 204.314562 | ||
| ENSG00000137434 | 347744 | C6orf52 | protein_coding | Q5T4I8 | Alternative splicing;Reference proteome | hsa:347744; | 51.830160 | 0.9166719 | -0.125522671 | 0.38935804 | 1.030465e-01 | 7.482039e-01 | No | Yes | 48.035718 | 9.479454 | 50.132364 | 9.743814 | ||||||||
| ENSG00000138074 | 8884 | SLC5A6 | protein_coding | Q9Y289 | FUNCTION: Sodium-dependent multivitamin transporter that transports pantothenate, biotin and lipoate (PubMed:10329687, PubMed:15561972, PubMed:21570947, PubMed:20980265, PubMed:22927035, PubMed:22015582, PubMed:25971966, PubMed:25809983, PubMed:28052864, PubMed:27904971, PubMed:31754459). Required for biotin and pantothenate uptake in the instestine (By similarity). Plays a role in the maintenance of intestinal mucosa integrity, by providing the gut mucosa with biotin (By similarity). May play a role in the transport of biotin and pantothenate into the brain across the blood-brain barrier (PubMed:25809983). May also be involved in the sodium-dependent transport of iodide ions (PubMed:20980265). {ECO:0000250|UniProtKB:Q5U4D8, ECO:0000269|PubMed:10329687, ECO:0000269|PubMed:15561972, ECO:0000269|PubMed:20980265, ECO:0000269|PubMed:21570947, ECO:0000269|PubMed:22015582, ECO:0000269|PubMed:22927035, ECO:0000269|PubMed:25809983, ECO:0000269|PubMed:25971966, ECO:0000269|PubMed:27904971, ECO:0000269|PubMed:28052864, ECO:0000269|PubMed:31754459}. | Biotin;Cell membrane;Disease variant;Glycoprotein;Ion transport;Membrane;Neurodegeneration;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | hsa:8884; | apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; brush border membrane [GO:0031526]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; plasma membrane [GO:0005886]; vesicle membrane [GO:0012506]; biotin transmembrane transporter activity [GO:0015225]; pantothenate transmembrane transporter activity [GO:0015233]; sodium-dependent multivitamin transmembrane transporter activity [GO:0008523]; symporter activity [GO:0015293]; vitamin transmembrane transporter activity [GO:0090482]; biotin import across plasma membrane [GO:1905135]; biotin transport [GO:0015878]; iodide transmembrane transport [GO:1904200]; pantothenate transmembrane transport [GO:0015887]; sodium ion transport [GO:0006814]; transport across blood-brain barrier [GO:0150104] | 12646417_in intestinal and liver epithelial cells, SMVT is the major (if not the only) biotin uptake system that operates. 15561972_biotin uptake by human renal epithelial cells occurs via the hSMVT system and that the process is regulated by intracellular PKC- and Ca(2+)/calmodulin-mediated pathways. 16749865_A sodium-dependent multivitamin transporter, SMVT, responsible for biotin uptake and transport, was identified and functionally characterized in MDCK-MDR1 cells. 16959947_Human intestinal biotin uptake is adaptively regulated but is not mediated via changes in hSMVT RNA stability. 17135299_KLF-4 and AP-2 is regulating the activity of the hSMVT promoter in the intestine and provide direct in vivo confirmation of hSMVT promoter activity. 17904341_findings from this study are consistent with the theory that HCS senses biotin, and that biotin regulates its own cellular uptake by participating in HCS-dependent chromatin remodeling events at the SMVT promoter 1 locus in Jurkat cells. 19211916_Conclude that the COOH tail of hSMVT contains several determinants important for polarized targeting and biotin transport in epithelial cells. 19898482_Observational study of gene-disease association. (HuGE Navigator) 20962270_These results show important role for His(1)(1) and His(2) residues in hSMVT function, which is most probably mediated via an effect on level of hSMVT expression at the cell membrane. 20980265_hSMVT may play an important role in the homeostasis of I(-) in the body 21183659_PDZD11 is an interacting partner with hSMVT in intestinal epithelial cells and that this interaction affects hSMVT function and cell biology. 21570947_Human SMVT protein is glycosylated, and that glycosylation is important for its function. The study also shows a role for the putative PKC-phosphorylation site Thr(286) of hSMVT in the PKC-mediated regulation of biotin uptake. 22015582_Cys(294) is essential for the function of the human sodium-dependent multivitamin transporter. 22304562_PCR analysis had confirmed the existence of FR-alpha, SMVT, and B ((0, +)) in Y-79 and ARPE-19 cells. 22732670_This study for the first time confirms the molecular expression of SMVT and demonstrates that SMVT, responsible for biotin uptake, is functionally active in PC-3 prostate cancer cells 22927035_This study shows for the first time the functional and molecular presence of SMVT in immortalized human corneal epithelial (HCEC) and retinal pigment epithelial (D407) cells 23142496_these studies demonstrated for the first time the functional and molecular expression of sodium dependent multivitamin transporter (SMVT) in human derived breast cancer (T47D) cells 25809983_SLC5A6 is responsible for the supplies of biotin and pantothenic acid into the brain across the blood brain barrier. 25971966_SMVT-mediated transport is highly specific for R-(+)-alpha-lipoic acid. 25999427_S. typhimurium infection inhibits intestinal biotin uptake by SLC5A6, and that the inhibition is mediated via the action of proinflammatory cytokines. 28052864_This study shows for the first time that LPS inhibits colonic biotin uptake via decreasing membrane expression of its transporter and that these effects likely involve a CK2-mediated pathway. 31894266_Integrated profiling identifies SLC5A6 and MFAP2 as novel diagnostic and prognostic biomarkers in gastric cancer patients. | ENSMUSG00000006641 | Slc5a6 | 3806.635513 | 1.0357435 | 0.050666714 | 0.07075402 | 5.182211e-01 | 4.716015e-01 | 7.876947e-01 | No | Yes | 4082.379975 | 394.990459 | 4003.574357 | 387.189826 | ||
| ENSG00000138080 | 11117 | EMILIN1 | protein_coding | Q9Y6C2 | FUNCTION: May be responsible for anchoring smooth muscle cells to elastic fibers, and may be involved not only in the formation of the elastic fiber, but also in the processes that regulate vessel assembly. Has cell adhesive capacity. | 3D-structure;Alternative splicing;Cell adhesion;Coiled coil;Collagen;Disulfide bond;Extracellular matrix;Glycoprotein;Reference proteome;Secreted;Signal | This gene encodes an extracellular matrix glycoprotein that is characterized by an N-terminal microfibril interface domain, a coiled-coiled alpha-helical domain, a collagenous domain and a C-terminal globular C1q domain. The encoded protein associates with elastic fibers at the interface between elastin and microfibrils and may play a role in the development of elastic tissues including large blood vessels, dermis, heart and lung. [provided by RefSeq, Sep 2009]. | hsa:11117; | collagen trimer [GO:0005581]; collagen-containing extracellular matrix [GO:0062023]; EMILIN complex [GO:1990971]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; integrin alpha4-beta1 complex [GO:0034668]; extracellular matrix constituent conferring elasticity [GO:0030023]; identical protein binding [GO:0042802]; integrin binding involved in cell-matrix adhesion [GO:0098640]; aortic valve morphogenesis [GO:0003180]; cell adhesion [GO:0007155]; cell adhesion mediated by integrin [GO:0033627]; cell migration [GO:0016477]; cell-matrix adhesion [GO:0007160]; elastic fiber assembly [GO:0048251]; negative regulation of angiogenesis [GO:0016525]; negative regulation of cell activation [GO:0050866]; negative regulation of collagen biosynthetic process [GO:0032966]; negative regulation of collagen fibril organization [GO:1904027]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of gene expression [GO:0010629]; negative regulation of macrophage migration [GO:1905522]; negative regulation of pathway-restricted SMAD protein phosphorylation [GO:0060394]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; negative regulation of vascular endothelial growth factor receptor signaling pathway [GO:0030948]; positive regulation of angiogenesis [GO:0045766]; positive regulation of cell-substrate adhesion [GO:0010811]; positive regulation of defense response to bacterium [GO:1900426]; positive regulation of extracellular matrix assembly [GO:1901203]; positive regulation of gene expression [GO:0010628]; regulation of blood pressure [GO:0008217]; regulation of cell population proliferation [GO:0042127] | 12456677_beta1 Integrin-dependent cell adhesion to this protein is mediated by its gC1q domain 15017143_NMR assignments for the C-terminal globular domain of EMILIN-1 17988845_EMILIN1 interacts with anthrax protective antigen and inhibits toxin action in vitro. EMILIN1 may be a potential target and/or a protein useful for countermeasures against B. anthracis toxin lethality. 18411305_EMILIN1 is a novel local regulator of lymphangiogenesis 18463100_EMILIN1 gC1q-alpha4beta1 represents a unique ligand/receptor system, with a requirement for a 3-fold arrangement of the interaction site. 18564921_Observational study of gene-disease association. (HuGE Navigator) 19922630_Observational study of gene-disease association. (HuGE Navigator) 19922630_Our findings don't support positive association of Emilin1 gene with EH, but the interaction of age and genotype variation of rs3754734 and rs2011616 might increase the risk to hypertension 20186130_Observational study of gene-disease association. (HuGE Navigator) 20186130_rs2289360, rs2011616, and rs2304682 in the human EMILIN1 gene, as well as the haplotype constructed using rs2536512, rs2011616, and rs17881426, are useful genetic markers of essential hypertension in Japanese men. 20701466_EMILIN-1 may regulate the formation of oxytalan fibers and play a role in their homeostasis. 21753788_All three SNPs in introns 1 and 5 (rs2289360, rs2011616 and rs7424556) of EMILIN1 were in strong pair-wise linkage disequilibrium and were significantly associated with hypertension. 22639547_There were significant associations of rs2011616 and rs2304682 polymorphisms in the EMILIN1 gene with hypertension among Japanese. 22814752_Emilin-1 produced by vascular smooth muscle cells acts as a main regulator of resting blood pressure levels by controlling the myogenic response in resistance arteries through TGF-beta. 24513040_The present findings highlight the peculiar activity of PMN elastase in disabling EMILIN1 suppressor function. 25445627_Data suggested mechanisms for homo- and hetero-typic EMILINs multimers formation: EMILIN1 or EMILIN2 alone can form trimers and multimers in the absence of each other or they can co-polymerize in a head-to-tail fashion to form hetero-typic multimers. 26462740_These findings collectively suggest that EMILIN1 may represent a new disease gene associated with an autosomal-dominant connective tissue disorder. 27090767_This study is the first to identify EMILIN-1 and ILK as prospective markers of islet regenerative function in human mesenchymal stem cells. 29037761_Study discloses a novel mechanism of interaction occurring between the trimeric gC1q domain of EMILIN1 and the alpha4beta1 integrin and determines that the three E933 residues (one from each monomer) are all required for ligand binding. Furthermore, R904 was identified as a synergistic residue for cell adhesion. 30354220_Report reduced EMILIN-1 and enhanced myogenic tone, dependent on increased TGF-beta-EGFR signaling, in resistance arteries from hypertensive patients. 30408617_Data provide evidence that the novel 'regulatory structural' role of EMILIN-1 in the lymphangiogenic process is played by the integrin binding site within its gC1q domain. 31242895_EMILIN1 induces anti-tumor effects by up-regulating TSPAN9 expression in gastric cancer. Hence, membrane proteins TSPAN9 and EMILIN1 may represent novel therapeutic targets for the treatment of gastric cancer. 34783040_Elastin MIcrofibriL INterfacer1 (EMILIN-1) is an alternative prosurvival VLA-4 ligand in chronic lymphocytic leukemia. | ENSMUSG00000029163 | Emilin1 | 43.251895 | 0.9566736 | -0.063901302 | 0.42877435 | 2.109412e-02 | 8.845229e-01 | No | Yes | 37.035160 | 7.236845 | 41.140061 | 7.724892 | ||
| ENSG00000138111 | 79847 | MFSD13A | protein_coding | Q14CX5 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:79847; | integral component of membrane [GO:0016021] | 31237042_The allele G of rs2001389 weakened the binding activity with CTCF, and it was related to the lower expression of a putative antioncogene MFSD13A whose knockdown promoted proliferation of PC cells. By integrating analysis on multiomics data, association studies and functional assays, we proposed that the common variant rs2001389 and the gene MFSD13A might be genetic modifiers of PC tumorigenesis. 31615651_Using a homology model of TMEM180, we experimentally determined that the protein has 12 transmembrane domains, and that its N-terminal and C-termini are exposed extracellularly. Moreover, we found that the putative cation-binding site of TMEM180 is conserved among orthologs, and that its position is similar to that of melibiose transporter MelB. 33768244_Integrative Analyses Followed by Functional Characterization Reveal TMEM180 as a Schizophrenia Risk Gene. | ENSMUSG00000025227 | Mfsd13a | 690.082720 | 1.1428623 | 0.192651580 | 0.10869550 | 3.162440e+00 | 7.535050e-02 | 3.682151e-01 | No | Yes | 716.524021 | 72.960364 | 639.307618 | 64.990342 | |||
| ENSG00000138593 | 9728 | SECISBP2L | protein_coding | Q93073 | FUNCTION: Binds SECIS (Sec insertion sequence) elements present on selenocysteine (Sec) protein mRNAs, but does not promote Sec incorporation into selenoproteins in vitro. | Alternative splicing;Phosphoprotein;Reference proteome | hsa:9728; | ribonucleoprotein complex [GO:1990904]; mRNA 3'-UTR binding [GO:0003730]; ribonucleoprotein complex binding [GO:0043021]; RNA binding [GO:0003723]; selenocysteine insertion sequence binding [GO:0035368]; selenocysteine incorporation [GO:0001514] | 21566536_SLAN, also known as KIAA0256. The novel protein suppressed in lung cancer down-regulated in lung cancer tissues retards cell proliferation and inhibits the oncokinase Aurora-A. 22530054_SECISBP2L (SBP2L) interacts with all known human SECIS RNAs in vitro and selenoprotein mRNAs co-immunoprecipitate with endogenous SBP2L, suggesting a role in regulating selenoprotein expression. 31314582_SLAN mediates the Aurora-A-triggered cytokinesis bypass and SLAN plays dual roles in that process depending on its phosphorylation status. | ENSMUSG00000035093 | Secisbp2l | 903.394606 | 1.0286988 | 0.040820590 | 0.11097366 | 1.325008e-01 | 7.158530e-01 | 9.073014e-01 | No | Yes | 930.572040 | 191.615934 | 885.137457 | 182.314571 | ||
| ENSG00000138658 | 55345 | ZGRF1 | protein_coding | Q86YA3 | Alternative splicing;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Zinc;Zinc-finger | The encoded protein contains GRF zinc finger (zf-GRF) and transmembrane domains. GRF zinc fingers are found in a number of DNA-binding proteins. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Apr 2017]. | hsa:55345; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; 5'-flap endonuclease activity [GO:0017108]; helicase activity [GO:0004386]; RNA binding [GO:0003723]; zinc ion binding [GO:0008270]; replication fork reversal [GO:0071932] | 20332099_Observational study of gene-disease association. (HuGE Navigator) 26599207_Two novel loci IDH1 and ZGRF1 are associated with adiposity. [meta-analysis] 32640219_The ZGRF1 Helicase Promotes Recombinational Repair of Replication-Blocking DNA Damage in Human Cells. | 1003.916995 | 1.2924335 | 0.370090095 | 0.10917048 | 1.170294e+01 | 6.240146e-04 | 2.679022e-02 | No | Yes | 884.340619 | 192.119319 | 687.529290 | 149.440602 | ||||
| ENSG00000138670 | 153020 | RASGEF1B | protein_coding | Q0VAM2 | FUNCTION: Guanine nucleotide exchange factor (GEF) with specificity for RAP2A, it doesn't seems to activate other Ras family proteins (in vitro). {ECO:0000269|PubMed:19645719, ECO:0000269|PubMed:23894443}. | Alternative splicing;Endosome;Guanine-nucleotide releasing factor;Reference proteome | hsa:153020; | early endosome [GO:0005769]; late endosome [GO:0005770]; midbody [GO:0030496]; guanyl-nucleotide exchange factor activity [GO:0005085]; small GTPase mediated signal transduction [GO:0007264] | 19645719_Mutation of the Ser39 in Rap1 changed the specificity and allowed the nucleotide exchange of Rap1(S39F) to be stimulated by RasGEF1B 22303795_As an addition to PRKG2 and RASGEFIB genes, we propose to include BMP3 gene as the principal determinant of the observed common phenotype. 27362560_We show that knockdown of the expression of mcircRasGEF1B reduces LPS-induced ICAM-1 expression. Additionally, we demonstrate that mcircRasGEF1B regulates the stability of mature ICAM-1 mRNAs. 31044621_study provides evidence for two novel candidate genes, SPG7 and RASGEF1B, associating with white coat effect | ENSMUSG00000089809 | Rasgef1b | 250.214026 | 1.1320136 | 0.178891314 | 0.17971877 | 9.788327e-01 | 3.224872e-01 | No | Yes | 286.709140 | 61.454257 | 252.079340 | 54.037538 | |||
| ENSG00000138756 | 55589 | BMP2K | protein_coding | Q9NSY1 | FUNCTION: May be involved in osteoblast differentiation. {ECO:0000250|UniProtKB:Q91Z96}. | 3D-structure;ATP-binding;Alternative splicing;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene is the human homolog of mouse BMP-2-inducible kinase. Bone morphogenic proteins (BMPs) play a key role in skeletal development and patterning. Expression of the mouse gene is increased during BMP-2 induced differentiation and the gene product is a putative serine/threonine protein kinase containing a nuclear localization signal. Therefore, the protein encoded by this human homolog is thought to be a protein kinase with a putative regulatory role in attenuating the program of osteoblast differentiation. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:55589; | cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; AP-2 adaptor complex binding [GO:0035612]; ATP binding [GO:0005524]; phosphatase regulator activity [GO:0019208]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; positive regulation of Notch signaling pathway [GO:0045747]; protein phosphorylation [GO:0006468]; regulation of bone mineralization [GO:0030500]; regulation of clathrin-dependent endocytosis [GO:2000369] | 15254968_PGE2 produced by COX-2 increased BMP-2 expression via binding the EP4 receptor. 16753015_The association of the BMP-2 gene polymorphisms Ser37Ala and Arg190Ser with osteoporosis in 6353 men and women from the Rotterdam Study was studied. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19927351_BMP2K gene 1379 G/A variant is strongly correlated with high myopia and may contribute to a genetic risk factor for high degrees of myopic pathogenesis. 19927351_Observational study of gene-disease association. (HuGE Navigator) 26853940_Results present the first structures of AAK1 and BIKE which reveal that all members of the Numb-associated kinase family share unusual activation segment architecture. 32792513_Regulation of BMP2K in AP2M1-mediated EGFR internalization during the development of gallbladder cancer. 32795391_Splicing variation of BMP2K balances abundance of COPII assemblies and autophagic degradation in erythroid cells. | ENSMUSG00000034663 | Bmp2k | 886.397410 | 1.0438677 | 0.061938816 | 0.10909558 | 3.276659e-01 | 5.670367e-01 | 8.402215e-01 | No | Yes | 868.866708 | 148.052359 | 835.731543 | 142.453020 | |
| ENSG00000138796 | 3033 | HADH | protein_coding | Q16836 | FUNCTION: Mitochondrial fatty acid beta-oxidation enzyme that catalyzes the third step of the beta-oxidation cycle for medium and short-chain 3-hydroxy fatty acyl-CoAs (C4 to C10) (PubMed:10231530, PubMed:11489939, PubMed:16725361). Plays a role in the control of insulin secretion by inhibiting the activation of glutamate dehydrogenase 1 (GLUD1), an enzyme that has an important role in regulating amino acid-induced insulin secretion (By similarity). {ECO:0000250|UniProtKB:Q61425, ECO:0000269|PubMed:10231530, ECO:0000269|PubMed:11489939, ECO:0000269|PubMed:16725361}. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Fatty acid metabolism;Hydroxylation;Lipid metabolism;Mitochondrion;NAD;Oxidoreductase;Reference proteome;Transferase;Transit peptide | PATHWAY: Lipid metabolism; fatty acid beta-oxidation. {ECO:0000269|PubMed:11489939, ECO:0000269|PubMed:16725361}. | This gene is a member of the 3-hydroxyacyl-CoA dehydrogenase gene family. The encoded protein functions in the mitochondrial matrix to catalyze the oxidation of straight-chain 3-hydroxyacyl-CoAs as part of the beta-oxidation pathway. Its enzymatic activity is highest with medium-chain-length fatty acids. Mutations in this gene cause one form of familial hyperinsulinemic hypoglycemia. The human genome contains a related pseudogene of this gene on chromosome 15. [provided by RefSeq, May 2010]. | hsa:3033; | cytoplasm [GO:0005737]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; 3-hydroxyacyl-CoA dehydrogenase activity [GO:0003857]; identical protein binding [GO:0042802]; NAD+ binding [GO:0070403]; transferase activity [GO:0016740]; fatty acid beta-oxidation [GO:0006635]; negative regulation of insulin secretion [GO:0046676]; positive regulation of cold-induced thermogenesis [GO:0120162]; regulation of insulin secretion [GO:0050796]; response to activity [GO:0014823]; response to insulin [GO:0032868]; response to xenobiotic stimulus [GO:0009410] | 9185222_Patients with the G1528C mutation of 3-hyroxyacyl-CoA dehydrogenase exhibit hepatomegaly and steatosis of the liver, as well as accumulation of fat in the myocardium, renal tubules, and skeletal muscle 11451959_To investigate its function in this catalytic dyad, Glu(170) was replaced with glutamine (E170Q), and the mutant enzyme was characterized. Substrate and cofactor binding were unaffected by the mutation; E170Q exhibited diminished catalytic activity 14693719_SCHAD deficiency can result in persistent hyperinsulinemic hypoglycemia of infancy 17065362_Observational study of gene-disease association. (HuGE Navigator) 17065362_Unlikely that variation in HADHSC plays a major role in the pathogenesis of type 2 diabetes in the examined cohorts. 19318379_This case indicates that mutations of the HADH gene should be sought in hyperinsulinemic patients in whom diffuse form of hyperinsulinemic hypoglycemia. 20332099_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 20931292_Congenital hyperinsulinism due to mutations in HNF4A and HADH. 21252247_We recommend that HADH sequence analysis is considered in all patients with diazoxide-responsive hyperinsulinemic hypoglycemia when recessive inheritance is suspected 21347589_Clinical, biochemical and molecular findings of four new Caucasian patients with HADH deficiency. 22583614_Loss of function mutations in 3-Hydroxyacyl-CoA Dehydrogenase (HADH) cause leucine sensitive hyperinsulinaemic hypoglycaemia. 23273570_Next-generation sequencing reveals deep intronic cryptic ABCC8 and HADH splicing founder mutations causing hyperinsulinism by pseudoexon activation. 26268944_in a cohort of hyperinsulinemic hypoglycemia patients from Isfahan, Iran, 78% were noted to have disease-causing mutations: 48% had HADH mutations and 26% had ABCC8 mutations. 26316438_We present clinical and laboratory findings together with the long-term clinical course of a case with a deep intronic HADH splicing mutation (c.636+471G>T) causing neonatal-onset hyperinsulinemic hypoglycemia with mild progression 26361074_Paretic muscle in hemiparetic stroke survivors had lower HAD concentration. 27181376_The most frequently seen mutations in Turkish patients with congenital hyperinsulinism (CHI) were ATP binding cassette subfamily C member 8 (ABCC8) gene, followed by 3-hydroxyacyl CoA dehydrogenase (HADH) and kcnj11 channel (KCNJ11) genes. 32876354_Functional evaluation of 16 SCHAD missense variants: Only amino acid substitutions causing congenital hyperinsulinism of infancy lead to loss-of-function phenotypes in vitro. 33881965_Differentially expressed genes PCCA, ECHS1, and HADH are potential prognostic biomarkers for gastric cancer. 34736508_Genetic pathogenesis, diagnosis, and treatment of short-chain 3-hydroxyacyl-coenzyme A dehydrogenase hyperinsulinism. | ENSMUSG00000027984 | Hadh | 3043.964168 | 0.9316781 | -0.102096565 | 0.07934133 | 1.624334e+00 | 2.024885e-01 | 5.730467e-01 | No | Yes | 3041.949097 | 437.693420 | 3278.017690 | 471.601761 |
| ENSG00000139117 | 144402 | CPNE8 | protein_coding | Q86YQ8 | FUNCTION: Probable calcium-dependent phospholipid-binding protein that may play a role in calcium-mediated intracellular processes. {ECO:0000250|UniProtKB:Q99829}. | Alternative splicing;Calcium;Metal-binding;Phosphoprotein;Reference proteome;Repeat | Calcium-dependent membrane-binding proteins may regulate molecular events at the interface of the cell membrane and cytoplasm. This gene is one of several genes that encode a calcium-dependent protein containing two N-terminal type II C2 domains and an integrin A domain-like sequence in the C-terminus. [provided by RefSeq, Jul 2008]. | hsa:144402; | extracellular exosome [GO:0070062]; plasma membrane [GO:0005886]; calcium-dependent phospholipid binding [GO:0005544]; metal ion binding [GO:0046872]; cellular response to calcium ion [GO:0071277] | Mouse_homologues 12670487_characterization of the human ortholog, CPNE8. 19795140_Results identify Cpne8, a member of the copine family, as the most promising candidate gene for prion disease incubation time. | ENSMUSG00000052560 | Cpne8 | 485.856196 | 1.1722296 | 0.229255114 | 0.15257329 | 2.241078e+00 | 1.343872e-01 | 4.825501e-01 | No | Yes | 584.262722 | 142.787427 | 499.937167 | 122.209015 | |
| ENSG00000139354 | 283431 | GAS2L3 | protein_coding | Q86XJ1 | FUNCTION: Cytoskeletal linker protein. May promote and stabilize the formation of the actin and microtubule network. {ECO:0000269|PubMed:21561867}. | Actin-binding;Cytoplasm;Cytoskeleton;Microtubule;Phosphoprotein;Reference proteome | hsa:283431; | actin cytoskeleton [GO:0015629]; actin filament [GO:0005884]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; actin binding [GO:0003779]; actin filament binding [GO:0051015]; cytoskeletal anchor activity [GO:0008093]; microtubule binding [GO:0008017]; actin crosslink formation [GO:0051764]; actin cytoskeleton organization [GO:0030036]; microtubule cytoskeleton organization [GO:0000226] | 21561867_tubulin acetylation drives GAS2-like 3 localization to MTs and may provide functional insights into the role of GAS2-like 3. 22344256_GAS2L3, a target gene of the DREAM complex, is required for cytokinesis and genomic stability. 23469016_show that the Gas2l3 protein is targeted for ubiquitin-mediated proteolysis by the APC/C(Cdh1) complex, but not by the APC/C(Cdc20) complex, and is phosphorylated by Cdk1 in mitosis 24571573_show that the GAR domain of GAS2L3 is required for localization of GAS2L3 to the constriction zone | ENSMUSG00000074802 | Gas2l3 | 510.006496 | 1.1415276 | 0.190965735 | 0.12791282 | 2.210527e+00 | 1.370718e-01 | 4.886786e-01 | No | Yes | 561.311325 | 115.418227 | 496.333457 | 102.154556 | ||
| ENSG00000139437 | 84260 | TCHP | protein_coding | Q9BT92 | FUNCTION: Tumor suppressor which has the ability to inhibit cell growth and be pro-apoptotic during cell stress. Inhibits cell growth in bladder and prostate cancer cells by a down-regulation of HSPB1 by inhibiting its phosphorylation. May act as a 'capping' or 'branching' protein for keratin filaments in the cell periphery. May regulate K8/K18 filament and desmosome organization mainly at the apical or peripheral regions of simple epithelial cells (PubMed:15731013, PubMed:18931701). Is a negative regulator of ciliogenesis (PubMed:25270598). {ECO:0000269|PubMed:15731013, ECO:0000269|PubMed:18931701, ECO:0000269|PubMed:25270598}. | Apoptosis;Cell junction;Cell membrane;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Isopeptide bond;Membrane;Mitochondrion;Reference proteome;Tumor suppressor;Ubl conjugation | hsa:84260; | apical cortex [GO:0045179]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; desmosome [GO:0030057]; keratin filament [GO:0045095]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; apoptotic process [GO:0006915]; cell projection organization [GO:0030030]; negative regulation of cell growth [GO:0030308]; negative regulation of cilium assembly [GO:1902018] | 15731013_trichoplein is a keratin 8/18-binding protein that may be involved in the organization of the apical network of keratin filaments and desmosomes in simple epithelial cells 18931701_MITOSTATIN was found within a 3.2-kb transcript for an approximately 62 kDa mitochondrial protein with tumor suppressor activity. It inhibits cell growth, is proapoptotic and downregulates Hsp27. 20930847_Trichoplein/mitostatin is a new regulator of mitochondria-endoplasmic reticulum juxtaposition. 21325031_Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. 24403067_These findings underscore the complexity of PGC-1alpha-mediated mitochondrial homeostasis and establish mitostatin as a key regulator of tumor cell mitophagy and angiostasis. 26880200_Ndel1 acts as a novel upstream regulator of the trichoplein-Aurora A pathway to inhibit primary cilia assembly. 29080840_Soluble matrix-derived cues being transduced downstream of receptor engagement converge upon a newly-discovered nexus of autophagic machinery consisting of Peg3 for endothelial cell autophagy and mitostatin for tumor cell mitophagy. | 608.173265 | 1.1612290 | 0.215652497 | 0.11495148 | 3.492533e+00 | 6.164618e-02 | 3.339904e-01 | No | Yes | 638.390808 | 58.708738 | 569.984623 | 52.423015 | ||||
| ENSG00000139697 | 55206 | SBNO1 | protein_coding | A3KN83 | Acetylation;Alternative splicing;Coiled coil;Phosphoprotein;Reference proteome | hsa:55206; | nucleus [GO:0005634]; chromatin DNA binding [GO:0031490]; histone binding [GO:0042393]; regulation of transcription, DNA-templated [GO:0006355] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22449796_Enhanced expression of Cdx2 by stimulation with bile acids may induce intestinal differentiation of esophageal columnar cells by interaction with the Notch signaling pathway. | ENSMUSG00000038095 | Sbno1 | 1773.872979 | 1.0861205 | 0.119184186 | 0.08191502 | 2.138018e+00 | 1.436874e-01 | 4.979882e-01 | No | Yes | 2335.823598 | 389.229773 | 2125.457504 | 354.249963 | |||
| ENSG00000140006 | 112840 | WDR89 | protein_coding | Q96FK6 | Reference proteome;Repeat;WD repeat | hsa:112840; | ENSMUSG00000045690 | Wdr89 | 746.467913 | 0.9696487 | -0.044465995 | 0.11218626 | 1.588189e-01 | 6.902463e-01 | 8.922935e-01 | No | Yes | 848.416619 | 189.391859 | 860.972765 | 192.159667 | |||||
| ENSG00000140022 | 85439 | STON2 | protein_coding | Q8WXE9 | FUNCTION: Adapter protein involved in endocytic machinery. Involved in the synaptic vesicle recycling. May facilitate clathrin-coated vesicle uncoating. {ECO:0000269|PubMed:11381094, ECO:0000269|PubMed:11454741, ECO:0000269|PubMed:21102408}. | 3D-structure;Alternative splicing;Cell junction;Cytoplasm;Endocytosis;Membrane;Phosphoprotein;Reference proteome;Repeat;Synapse;Synaptosome;Ubl conjugation | This gene encodes a protein which is a membrane protein involved in regulating endocytotic complexes. The protein product is described as one of the clathrin-associated sorting proteins, adaptor molecules which ensure specific proteins are internalized. The encoded protein has also been shown to participate in synaptic vesicle recycling through interaction with synaptotagmin 1 required for neurotransmission. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2012]. | hsa:85439; | clathrin-coated vesicle [GO:0030136]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; membrane [GO:0016020]; neuron projection [GO:0043005]; nucleolus [GO:0005730]; synaptic vesicle [GO:0008021]; clathrin adaptor activity [GO:0035615]; regulation of endocytosis [GO:0030100]; synaptic vesicle endocytosis [GO:0048488]; vesicle-mediated transport [GO:0016192] | 16459302_The ability of stonin 2 to facilitate endocytosis of synaptotagmin is dependent on its association with AP-2, an intact mu-homology domain, and functional AP-2 heterotetramers. 18166656_Our data identify the molecular determinants for recognition of synaptotagmin by stonin 2 or its Caenorhabditis elegans orthologue UNC-41B. 21407139_STON2 may be a susceptibility gene for Chinese Han patients with schizophrenia. 23785397_The present study demonstrated that the functional variant of the STON2 gene could alter cortical surface area on the right inferior temporal and contribute to the pathogenesis of schizophrenia. 28758939_STON2 plays an important role in the progression and prognosis of ovarian carcinoma, especially in platinum resistance, intraperitoneal metastasis, and recurrence. STON2 can be a novel antitumor drug target and biomarker which predicts an unfavorable prognosis for EOC patients. 30325582_this study revealed that miR-199b-5p functions as antioncogene miRNA in papillary thyroid carcinoma cells and that the miR-199b-5p/STON2 axis might be a potential treatment option for papillary thyroid carcinoma 30518424_The functional significance of this STON2-DNMT1/MUC1 pathway is supported by the observation that STON2 overexpression suppresses MUC1-induced sphere formation of OCSCs. 32575107_Narrowing down the Common Cytogenetic Deletion 14q to a 5.6-Mb Critical Region in 1p/19q Codeletion Oligodendroglioma-Relapsed Patients Points to Two Potential Relapse-Related Genes: SEL1L and STON2. 32737415_Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. | ENSMUSG00000020961 | Ston2 | 481.611790 | 1.1264056 | 0.171726453 | 0.14735676 | 1.386717e+00 | 2.389604e-01 | 6.114264e-01 | No | Yes | 515.357709 | 66.386258 | 454.900104 | 58.709412 | |
| ENSG00000140044 | 122953 | JDP2 | protein_coding | Q8WYK2 | FUNCTION: Component of the AP-1 transcription factor that represses transactivation mediated by the Jun family of proteins. Involved in a variety of transcriptional responses associated with AP-1 such as UV-induced apoptosis, cell differentiation, tumorigenesis and antitumogeneris. Can also function as a repressor by recruiting histone deacetylase 3/HDAC3 to the promoter region of JUN. May control transcription via direct regulation of the modification of histones and the assembly of chromatin. {ECO:0000269|PubMed:12707301, ECO:0000269|PubMed:12903123, ECO:0000269|PubMed:16026868, ECO:0000269|PubMed:16518400}. | Alternative splicing;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:122953; | chromatin [GO:0000785]; nucleus [GO:0005634]; cAMP response element binding [GO:0035497]; chromatin binding [GO:0003682]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; leucine zipper domain binding [GO:0043522]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of histone deacetylation [GO:0031065]; regulation of transcription by RNA polymerase II [GO:0006357] | 14627710_c-Jun dimerization protein 2 inhibits cell transformation and has a role as a tumor suppressor gene 16026868_JDP2 is a cellular survival protein whose presence is necessary for normal cellular function 18396163_JDP2 acts as a repressor and could be functionally associated with HDAC3 to inhibit CHOP transcription 18671972_IRF2-BP1 is a JDP2-binding protein enhancing the polyubiquitination of JDP2 and represses ATF2-mediated transcriptional activation from a CRE-containing promoter. 19553667_A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. 20452405_3 SNPs (2 intronic: rs741846 & rs175646; & 1 in the untranslated region: rs8215) & their genotype distribution showed significant association in the Japanese & Korean but not Dutch intracranial aneurysm patients. 20452405_Observational study of gene-disease association. (HuGE Navigator) 20677166_JDP2 expression was downregulated in pancreatic carcinoma & this correlated with metastasis & decreased post-surgery survival. 20950777_The molecular mechanisms that underlie the action of JDP2 in cellular aging and replicative senescence by mediating the dissociation of polycomb repressive complexes from the p16(Ink4a)/Arf locus are discussed. 21525011_JDP2 is crucial to triggering reactivation from latency to lytic replication 22989952_the recruitment of multiple HDAC members to JDP2 and ATF3 is part of their transcription repression mechanism. 24120378_Preeclamptic plasma induces transcription modifications involving the AP-1 transcriptional regulator JDP2 in endothelial cells. 24232097_Results suggest that JDP2 is an integral component of the Nrf2-MafK complex and that it modulates antioxidant and detoxification programs by acting via the ARE. 28315425_In hepatocellular carcinoma, high expression of JDP2 is significantly correlated with smaller tumor size, early stage HCC and better survival. 29941549_These studies establish JDP2 as a novel oncogene in high-risk T cell acute lymphoblastic leukemia. 30721335_JDP2 was downregulated in myelodysplastic syndrome. Its expression was inversely related to disease aggressiveness and AML transformation. JDP2 suppression is a direct result of reduced PU.1. PU.1 and JDP2 expression correlate and are concurrently reduced with the extent of differentiation arrest and aggression/prognosis in MDS/AML. It was upregulated upon azacytidine treatment. 30877624_JDP2 is downregulated in HCC tissues and cells, and overexpressed JDP2 facilitated HCC cell invasion and EMT. 32150333_Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. 33108704_JDP2 is directly regulated by ATF4 and modulates TRAIL sensitivity by suppressing the ATF4-DR5 axis. | ENSMUSG00000034271 | Jdp2 | 74.539656 | 1.1863395 | 0.246516915 | 0.36464272 | 4.763190e-01 | 4.900944e-01 | No | Yes | 112.014532 | 26.134146 | 90.178541 | 20.762401 | |||
| ENSG00000140905 | 2653 | GCSH | protein_coding | P23434 | FUNCTION: The glycine cleavage system catalyzes the degradation of glycine. The H protein (GCSH) shuttles the methylamine group of glycine from the P protein (GLDC) to the T protein (GCST). {ECO:0000269|PubMed:1671321}. | Lipoyl;Mitochondrion;Reference proteome;Transit peptide | Degradation of glycine is brought about by the glycine cleavage system, which is composed of four mitochondrial protein components: P protein (a pyridoxal phosphate-dependent glycine decarboxylase), H protein (a lipoic acid-containing protein), T protein (a tetrahydrofolate-requiring enzyme), and L protein (a lipoamide dehydrogenase). The protein encoded by this gene is the H protein, which transfers the methylamine group of glycine from the P protein to the T protein. Defects in this gene are a cause of nonketotic hyperglycinemia (NKH). Two transcript variants, one protein-coding and the other probably not protein-coding,have been found for this gene. Also, several transcribed and non-transcribed pseudogenes of this gene exist throughout the genome.[provided by RefSeq, Jan 2010]. | hsa:2653; | cytoplasm [GO:0005737]; glycine cleavage complex [GO:0005960]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; aminomethyltransferase activity [GO:0004047]; glycine catabolic process [GO:0006546]; glycine decarboxylation via glycine cleavage system [GO:0019464]; protein lipoylation [GO:0009249] | 12402263_Heterozygous GCSH gene mutation in transient neonatal hyperglycinemia. 16450403_Observational study of genotype prevalence. (HuGE Navigator) 19299230_Genetic analysis showed a non-previously described mutation affecting a consensus splice site (IVS2-1G > C 3) in the AMT gene encoding the T protein of the glycine cleavage system. 19844255_Observational study of gene-disease association. (HuGE Navigator) 21539457_There is no detectable glycine cleavage enzyme activity in human skin fibroblasts. 25231368_Data indicate no mutation was found in glycine cleavage system protein-H (GCSH) and suggest that mutations in both glycine decarboxylase (GLDC) and aminomethyltransferase (AMT) are the main cause of glycine encephalopathy in Malaysian population. 30337557_GCSH protein is overexpressed in both, breast cancer tissue and breast cancer cell lines. A shorter (391 bp) transcript variant (Tv*) was amplified with an increased expression in healthy breast cells and a decreased expression in breast cancer samples. A GCSH-equilibrium at the transcript level is likely conceivable for optimal glycine degradation. 33890291_Biallelic start loss variant, c.1A > G in GCSH is associated with variant nonketotic hyperglycinemia. | ENSMUSG00000034424 | Gcsh | 1994.998672 | 1.0530986 | 0.074640523 | 0.07722273 | 9.326309e-01 | 3.341802e-01 | 6.940038e-01 | No | Yes | 2180.826585 | 372.311238 | 2064.922133 | 352.518080 | |
| ENSG00000141013 | 2622 | GAS8 | protein_coding | O95995 | FUNCTION: Component of the nexin-dynein regulatory complex (N-DRC), a key regulator of ciliary/flagellar motility which maintains the alignment and integrity of the distal axoneme and regulates microtubule sliding in motile axonemes. Plays an important role in the assembly of the N-DRC linker (By similarity). Plays dual roles at both the primary (or non-motile) cilia to regulate hedgehog signaling and in motile cilia to coordinate cilia movement. Required for proper motile cilia functioning (PubMed:26387594, PubMed:27120127, PubMed:27472056). Positively regulates ciliary smoothened (SMO)-dependent Hedgehog (Hh) signaling pathway by facilitating the trafficking of SMO into the cilium and the stimulation of SMO activity in a GRK2-dependent manner (By similarity). {ECO:0000250|UniProtKB:Q60779, ECO:0000250|UniProtKB:Q7XJ96, ECO:0000269|PubMed:26387594, ECO:0000269|PubMed:27120127, ECO:0000269|PubMed:27472056}. | Alternative splicing;Cell projection;Ciliopathy;Cilium;Coiled coil;Cytoplasm;Cytoskeleton;Flagellum;Golgi apparatus;Microtubule;Primary ciliary dyskinesia;Reference proteome | This gene includes 11 exons spanning 25 kb and maps to a region of chromosome 16 that is sometimes deleted in breast and prostrate cancer. The second intron contains an apparently intronless gene, C16orf3, that is transcribed in the opposite orientation. This gene is a putative tumor suppressor gene. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2013]. | hsa:2622; | axoneme [GO:0005930]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytosol [GO:0005829]; extracellular region [GO:0005576]; Golgi apparatus [GO:0005794]; microtubule [GO:0005874]; motile cilium [GO:0031514]; plasma membrane [GO:0005886]; sperm flagellum [GO:0036126]; microtubule binding [GO:0008017]; small GTPase binding [GO:0031267]; axoneme assembly [GO:0035082]; brain development [GO:0007420]; cellular protein localization [GO:0034613]; cilium movement involved in cell motility [GO:0060294]; determination of left/right symmetry [GO:0007368]; epithelial cilium movement involved in extracellular fluid movement [GO:0003351]; flagellated sperm motility [GO:0030317]; negative regulation of cell population proliferation [GO:0008285]; positive regulation of protein localization to cilium [GO:1903566]; positive regulation of smoothened signaling pathway [GO:0045880]; regulation of microtubule binding [GO:1904526] | 19995461_A two-hybrid screen for proteins that interact with NS1 from influenza A yielded growth arrest-specific protein 8. Gas8 associated with NS1 in vitro and in vivo. 26387594_Loss-of-Function GAS8 Mutations Cause Primary Ciliary Dyskinesia and Disrupt the Nexin-Dynein Regulatory Complex. 27120127_Study identifies bi-allelic loss-of-function mutations in GAS8 as a cause for primary ciliary dyskinesia, and unveils the key importance of the protein encoded by this gene in N-DRC integrity and in the proper alignment of axonemal microtubules in humans. 27472056_Here, we generate the first mouse with a Gas8 mutation and show that it causes severe Primary Ciliary Dyskinesia (PCD)phenotypes; however, there were no overt Hh pathway phenotypes. In addition, we identified two human patients with missense variants in Gas8 29327301_In papillary thyroid carcinoma (PTC) cell lines, GAS8-AS1 inhibited proliferation, activated autophagy, and increased ATG5 expression. Downregulation of ATG5 reversed GAS8-AS1-mediated activation of autophagy leading to cell death, revealing a novel mechanism of the GAS8-AS1-ATG5 axis in PTC cell lines. 30228180_Expression of GAS8-AS1 or GAS8 is significantly decreased in hepatocellular carcinoma tissues and is associated with a poor prognosis among hepatocellular carcinoma patients. 31003831_The current study shows significance of GAS8 and GAS8-AS1 in the pathogenesis of MS and the putative role of GAS8-AS1 as a diagnostic biomarker in a subset of patients. 32165089_Expression analysis of growth arrest specific 8 and its anti-sense in breast cancer tissues. 33942556_LncRNA GAS8-AS1 downregulates lncRNA NEAT1 to inhibit glioblastoma cell proliferation. | ENSMUSG00000040220 | Gas8 | 510.086259 | 1.0522174 | 0.073432850 | 0.13179206 | 3.125330e-01 | 5.761300e-01 | 8.426151e-01 | No | Yes | 518.374881 | 59.944456 | 490.275732 | 56.682501 | |
| ENSG00000141086 | 1506 | CTRL | protein_coding | P40313 | Disulfide bond;Glycoprotein;Hydrolase;Protease;Reference proteome;Serine protease;Signal;Zymogen | This gene encodes a serine-type endopeptidase with chymotrypsin- and elastase-2-like activities. The gene encoding this zymogen is expressed specifically in the pancreas and likely functions as a digestive enzyme. [provided by RefSeq, Sep 2016]. | hsa:1506; | extracellular space [GO:0005615]; serine-type endopeptidase activity [GO:0004252]; serine-type peptidase activity [GO:0008236]; protein catabolic process [GO:0030163]; proteolysis [GO:0006508] | 25578495_We found CT-like activity to be an independent predictor of high-risk PCa, and as such, it may be a good candidate as a biomarker for high-risk PCa detection and stratification. | ENSMUSG00000031896 | Ctrl | 86.161204 | 0.6793153 | -0.557846824 | 0.29415790 | 3.505009e+00 | 6.118350e-02 | No | Yes | 70.112950 | 23.355797 | 109.986163 | 36.543916 | |||
| ENSG00000141391 | 10650 | PRELID3A | protein_coding | Q96N28 | FUNCTION: In vitro, the TRIAP1:PRELID3A complex mediates the transfer of phosphatidic acid (PA) between liposomes and probably functions as a PA transporter across the mitochondrion intermembrane space. Phosphatidic acid import is required for cardiolipin (CL) synthesis in the mitochondrial inner membrane. {ECO:0000305|PubMed:26071602}. | 3D-structure;Alternative splicing;Lipid transport;Mitochondrion;Reference proteome;Transport | hsa:10650; | mitochondrial intermembrane space [GO:0005758]; phosphatidic acid transfer activity [GO:1990050]; phospholipid transport [GO:0015914] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 26071602_crystal structures of free TRIAP1 and TRIAP1-SLMO1 complex reveal how the PRELI domain is chaperoned during import into the intermembrane mitochondrial space; structural resemblance of PRELI-like domain of SLMO1 with that of mammalian phoshatidylinositol transfer proteins suggest they share similar lipid transfer mechanisms | ENSMUSG00000024530 | Prelid3a | 130.374986 | 0.9393635 | -0.090244600 | 0.23286395 | 1.502630e-01 | 6.982842e-01 | No | Yes | 132.672593 | 16.928175 | 145.807084 | 18.569346 | |||
| ENSG00000141577 | 22994 | CEP131 | protein_coding | Q9UPN4 | FUNCTION: Component of centriolar satellites contributing to the building of a complex and dynamic network required to regulate cilia/flagellum formation (PubMed:17954613, PubMed:24185901). In proliferating cells, MIB1-mediated ubiquitination induces its sequestration within centriolar satellites, precluding untimely cilia formation initiation (PubMed:24121310). In contrast, during normal and ultraviolet or heat shock cellular stress-induced ciliogenesis, its non-ubiquitinated form is rapidly displaced from centriolar satellites and recruited to centrosome/basal bodies in a microtubule- and p38 MAPK-dependent manner (PubMed:24121310, PubMed:26616734). Acts also as a negative regulator of BBSome ciliary trafficking (PubMed:24550735). Plays a role in sperm flagellar formation; may be involved in the regulation of intraflagellar transport (IFT) and/or intramanchette (IMT) trafficking, which are important for axoneme extension and/or cargo delivery to the nascent sperm tail (By similarity). Required for optimal cell proliferation and cell cycle progression; may play a role in the regulation of genome stability in non-ciliogenic cells (PubMed:22797915, PubMed:26297806). Involved in centriole duplication (By similarity). Required for CEP152, WDR62 and CEP63 centrosomal localization and promotes the centrosomal localization of CDK2 (PubMed:26297806). Essential for maintaining proper centriolar satellite integrity (PubMed:30804208). {ECO:0000250|UniProtKB:Q62036, ECO:0000269|PubMed:17954613, ECO:0000269|PubMed:22797915, ECO:0000269|PubMed:24121310, ECO:0000269|PubMed:24185901, ECO:0000269|PubMed:24550735, ECO:0000269|PubMed:26297806, ECO:0000269|PubMed:26616734, ECO:0000269|PubMed:30804208}. | Alternative splicing;Cell cycle;Cell projection;Cilium biogenesis/degradation;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Developmental protein;Differentiation;Phosphoprotein;Reference proteome;Spermatogenesis;Transport;Ubl conjugation | hsa:22994; | acrosomal vesicle [GO:0001669]; centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; ciliary transition zone [GO:0035869]; cytosol [GO:0005829]; intercellular bridge [GO:0045171]; intracellular membrane-bounded organelle [GO:0043231]; manchette [GO:0002177]; microtubule cytoskeleton [GO:0015630]; sperm head-tail coupling apparatus [GO:0120212]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; cell cycle [GO:0007049]; cilium assembly [GO:0060271]; intraciliary transport involved in cilium assembly [GO:0035735]; intramanchette transport [GO:1990953]; manchette assembly [GO:1905198]; non-motile cilium assembly [GO:1905515]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of intracellular protein transport [GO:0090316]; protein localization to centrosome [GO:0071539]; regulation of centrosome duplication [GO:0010824]; sperm axoneme assembly [GO:0007288] | 22797915_These data therefore highlight the importance of human Cep131 for maintaining genomic integrity 23137637_Analysis of the data suggested that SP1 is a pivotal transcription factor for the regulation of CEP131 expression, consequently leading the control of centrosome functions. 26616734_CEP131 is the key regulatory target of MK2 and 14-3-3 in centriolar satellite remodeling. 28694105_CEP131 serves as a potential prognostic biomarker in HCC 30538195_RNA of AZI1 functions as the 'initiator' RNA to induce TMPRSS2-ERG fusion. 30804208_PLK4 phosphorylates CEP131 at Ser-78 to maintain centriolar satellite integrity. 31358734_These findings demonstrate that Cep131 is a novel substrate of Plk4, and that phosphorylation or dysregulated Cep131 overexpression promotes Plk4 stabilization and therefore centrosome amplification, establishing a perspective in understanding a relationship between centrosome amplification and cancer development. | ENSMUSG00000039781 | Cep131 | 723.681350 | 0.9582991 | -0.061452051 | 0.11097036 | 3.094204e-01 | 5.780361e-01 | 8.434415e-01 | No | Yes | 666.076325 | 88.926427 | 693.045591 | 92.465157 | ||
| ENSG00000141646 | 4089 | SMAD4 | protein_coding | Q13485 | FUNCTION: In muscle physiology, plays a central role in the balance between atrophy and hypertrophy. When recruited by MSTN, promotes atrophy response via phosphorylated SMAD2/4. MSTN decrease causes SMAD4 release and subsequent recruitment by the BMP pathway to promote hypertrophy via phosphorylated SMAD1/5/8. Acts synergistically with SMAD1 and YY1 in bone morphogenetic protein (BMP)-mediated cardiac-specific gene expression. Binds to SMAD binding elements (SBEs) (5'-GTCT/AGAC-3') within BMP response element (BMPRE) of cardiac activating regions (By similarity). Common SMAD (co-SMAD) is the coactivator and mediator of signal transduction by TGF-beta (transforming growth factor). Component of the heterotrimeric SMAD2/SMAD3-SMAD4 complex that forms in the nucleus and is required for the TGF-mediated signaling (PubMed:25514493). Promotes binding of the SMAD2/SMAD4/FAST-1 complex to DNA and provides an activation function required for SMAD1 or SMAD2 to stimulate transcription. Component of the multimeric SMAD3/SMAD4/JUN/FOS complex which forms at the AP1 promoter site; required for synergistic transcriptional activity in response to TGF-beta. May act as a tumor suppressor. Positively regulates PDPK1 kinase activity by stimulating its dissociation from the 14-3-3 protein YWHAQ which acts as a negative regulator. {ECO:0000250, ECO:0000269|PubMed:17327236, ECO:0000269|PubMed:25514493, ECO:0000269|PubMed:9389648}. | 3D-structure;Acetylation;Cytoplasm;DNA-binding;Disease variant;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation;Zinc | This gene encodes a member of the Smad family of signal transduction proteins. Smad proteins are phosphorylated and activated by transmembrane serine-threonine receptor kinases in response to transforming growth factor (TGF)-beta signaling. The product of this gene forms homomeric complexes and heteromeric complexes with other activated Smad proteins, which then accumulate in the nucleus and regulate the transcription of target genes. This protein binds to DNA and recognizes an 8-bp palindromic sequence (GTCTAGAC) called the Smad-binding element (SBE). The protein acts as a tumor suppressor and inhibits epithelial cell proliferation. It may also have an inhibitory effect on tumors by reducing angiogenesis and increasng blood vessel hyperpermeability. The encoded protein is a crucial component of the bone morphogenetic protein signaling pathway. The Smad proteins are subject to complex regulation by post-translational modifications. Mutations or deletions in this gene have been shown to result in pancreatic cancer, juvenile polyposis syndrome, and hereditary hemorrhagic telangiectasia syndrome. [provided by RefSeq, Aug 2017]. | hsa:4089; | activin responsive factor complex [GO:0032444]; centrosome [GO:0005813]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; heteromeric SMAD protein complex [GO:0071144]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SMAD protein complex [GO:0071141]; transcription regulator complex [GO:0005667]; chromatin binding [GO:0003682]; collagen binding [GO:0005518]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; filamin binding [GO:0031005]; I-SMAD binding [GO:0070411]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; protein homodimerization activity [GO:0042803]; R-SMAD binding [GO:0070412]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sulfate binding [GO:0043199]; transcription cis-regulatory region binding [GO:0000976]; transcription coactivator binding [GO:0001223]; adrenal gland development [GO:0030325]; aging [GO:0007568]; anatomical structure morphogenesis [GO:0009653]; atrioventricular canal development [GO:0036302]; atrioventricular valve formation [GO:0003190]; axon guidance [GO:0007411]; BMP signaling pathway [GO:0030509]; brainstem development [GO:0003360]; branching involved in ureteric bud morphogenesis [GO:0001658]; cardiac conduction system development [GO:0003161]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; cellular iron ion homeostasis [GO:0006879]; cellular response to BMP stimulus [GO:0071773]; cellular response to glucose stimulus [GO:0071333]; developmental growth [GO:0048589]; embryonic digit morphogenesis [GO:0042733]; endocardial cell differentiation [GO:0060956]; endothelial cell activation [GO:0042118]; epithelial to mesenchymal transition involved in endocardial cushion formation [GO:0003198]; ERK1 and ERK2 cascade [GO:0070371]; female gonad morphogenesis [GO:0061040]; formation of anatomical boundary [GO:0048859]; gastrulation with mouth forming second [GO:0001702]; in utero embryonic development [GO:0001701]; interleukin-6-mediated signaling pathway [GO:0070102]; intracellular signal transduction [GO:0035556]; left ventricular cardiac muscle tissue morphogenesis [GO:0003220]; mesendoderm development [GO:0048382]; metanephric mesenchyme morphogenesis [GO:0072133]; negative regulation of cardiac muscle hypertrophy [GO:0010614]; negative regulation of cardiac myofibril assembly [GO:1905305]; negative regulation of cell death [GO:0060548]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of protein catabolic process [GO:0042177]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; nephrogenic mesenchyme morphogenesis [GO:0072134]; neural crest cell differentiation [GO:0014033]; neuron fate commitment [GO:0048663]; osteoblast differentiation [GO:0001649]; outflow tract septum morphogenesis [GO:0003148]; ovarian follicle development [GO:0001541]; positive regulation of BMP signaling pathway [GO:0030513]; positive regulation of cardiac muscle cell apoptotic process [GO:0010666]; positive regulation of cell proliferation involved in heart valve morphogenesis [GO:0003251]; positive regulation of epithelial to mesenchymal transition [GO:0010718]; positive regulation of follicle-stimulating hormone secretion [GO:0046881]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of histone H3-K9 acetylation [GO:2000617]; positive regulation of luteinizing hormone secretion [GO:0033686]; positive regulation of pathway-restricted SMAD protein phosphorylation [GO:0010862]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of SMAD protein signal transduction [GO:0060391]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus [GO:1901522]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of transforming growth factor beta receptor signaling pathway [GO:0030511]; regulation of binding [GO:0051098]; regulation of hair follicle development [GO:0051797]; regulation of transforming growth factor beta receptor signaling pathway [GO:0017015]; regulation of transforming growth factor beta2 production [GO:0032909]; response to hypoxia [GO:0001666]; response to transforming growth factor beta [GO:0071559]; sebaceous gland development [GO:0048733]; secondary palate development [GO:0062009]; seminiferous tubule development [GO:0072520]; single fertilization [GO:0007338]; SMAD protein complex assembly [GO:0007183]; SMAD protein signal transduction [GO:0060395]; somite rostral/caudal axis specification [GO:0032525]; spermatogenesis [GO:0007283]; transcription, DNA-templated [GO:0006351]; transforming growth factor beta receptor signaling pathway [GO:0007179]; uterus development [GO:0060065]; ventricular septum morphogenesis [GO:0060412]; wound healing [GO:0042060] | 9741623_structure of DNA-binding domain 10871368_DNA-binding protein 11783110_Alterations in tumor-suppressor gene DPC4 may play an important role during the tumorigenesis of pancreatic cancer. 11836524_A point mutation in Smad4 abolished binding to SMIF. 11866987_smad4 may play an important role in the regulation of TGFbeta inducible gene expression and subsequent growth inhibition. 11920286_Common deletion of SMAD4 in juvenile polyposis is a mutational hotspot. 12097320_TLV-1 tax represses Smad-mediated TGF-beta signaling. 12136244_Observational study of gene-disease association. (HuGE Navigator) 12191474_Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. 12202987_Observational study of genotype prevalence. (HuGE Navigator) 12209716_In this study, the expression of Smad4 protein appeared to be correlated with the depth of invasion of esophageal SCC 12226080_interactions between AR, Smad3, and Smad4 may result in the differential regulation of the AR transactivation, which further strengthens their roles in the prostate cancer progression 12414627_Restoration of transforming growth factor Beta signaling by functional expression of smad4 induces anoikis. 12417513_Observational study of gene-disease association. (HuGE Navigator) 12419246_determined the crystal structure of a complex between a conserved Smad4 binding fragment of Ski and the MH2 domain of Smad4 at 2.85 A resolution 12429655_Smad4/DPC4 has a role in TGF-beta-mediated inhibition of cell proliferation in vitro and in vivo 12531695_Phenotypic and functional changes associated with TGF-beta1-induced fibroblast terminal differentiation are differentially regulated by Smad2, Smad3, and Smad4. 12548549_the re-expression of the Smad4 gene by either method partially restored TGF-beta responsiveness in FaDu cells with respect to both growth inhibition and expression of p21WAF1/CIP1 and p15INK4B 12569386_Smad4 point mutations are prevalent in pancreatic carcinoma, they are infrequent in early stages (I-III) of colorectal cancer. 12576474_Interaction domains of Smad4 and ERalpha are mapped and shown to be essential for transcriptional repression of ERalpha by Smad4. 12618756_Smad4 is not required for nuclear translocation of Smad2 and Smad3, but is needed for activation of at least certain transcriptional responses. 12631740_Results suggest that the transcriptional cross talk between the TGFbeta-regulated Smads 3 and 4 and hepatocyte nuclear factor-4 is mediated by specific functional domains in the two types of transcription factors. 12700666_Human HCC transfectants express a mutant Smad2(3S-A). Serine residues of SSXS motif were changed to alanine. They have impaired Smad2 signaling. Forced expression of Smad2(3S-A induced TGFB secretion & resistance to TGFB-induced growth inhibition. 12720172_results suggest carcinogenesis in the biliary tract epithelium in anomalous pancreaticobiliary ductal union (APBDU) is accompanied by multistep mutational events; inactivation of DPC-4 gene accumulates late in progression of biliary tract adenocarcinoma 12740389_Smad4 is sumoylated and has a role in the regulation of TGF-beta signaling through Smads 12758167_Smad-4 had no effect on the basal activity of the MCP-1 promoter, but showed the ability to decrease both Smad-3 and Tat-induced transcription of the MCP promoter in human astrocytic cells 12794086_oligo-ubiquitination positively regulates Smad4 function, whereas poly-ubiquitination primarily occurs in unstable cancer mutants and leads to protein degradation 12802277_duration of TGF-beta-Smad signaling is a critical determinant of the specificity of the TGF-beta response. 12813045_SUMO-1 modification serves to protect Smad4 from ubiquitin-dependent degradation and consequently enhances the growth inhibitory and transcriptional responses of Smad4. 12894231_A G/A transition at 31 bp upstream-nontranslated regions of exon 8 of Smad 4 was found in cervical cancer cells, highlighting an important role for Smad 4 in human cervical tumors. 12917407_Smad4 interaction with CAN/Nup214, and nuclear import requires structural elements present only in the full-length Smad4; Smad3 and Smad4 have different susceptibility to inhibition of import by cytoplasmic retention factor SARA 12952364_Smad4 was expressed in all thyroid cell lines and controls analyzed, differently from other classes of tumors where Smad4 expression was deleted. 14514699_Sumoylation of Smad4 mainly occurs at lysine 159 and facilitates Smad-dependent transcriptional activation; PIAS-mediated sumoylation of Smad4 is regulated by the p38 MAP kinase pathway 14525983_DACH1 bound to endogenous NCoR and Smad4 in cultured cells; Smad4 was required for DACH1 repression of TGF-beta induction of Smad signaling 14555988_Smad2, Smad3 and Smad4 contribute to the regulation of TGF-beta responses to varying extents, and exhibit distinct roles in different cell types 14607700_Inactivation of DPC4 gene late in neoplastic progression of pancreatic carcinoma. Variation of DPC4 gene activation in biliary tract carcinoma. Common bile duct carcinoma and pancreatic carcinoma have similar molecular alternations. 14630914_SMAD3 and SMAD4 activate gadd45beta through its third intron to facilitate G2 progression following TGFbeta treatment 14639103_DPC4/Smad4 inactivation by mutation or deletion appears to be very rare in pancreatic endocrine tumors. 14647410_Mutations in SMAD4 abrogate its function in transducing the signaling of TGF-beta, which plays an important role in various stages of cancer formation. 14647445_DPC4 mutations in appendiceal adenocarcinomas suggests involvement of DPC4 and nearby genes on chromosome 18q (DCC and/or JV-18) in the pathogenesis of appendiceal adenocarcinomas 14669329_DPC4 is involved in the development of pancreatic carcinoma and is a late event in pancreatic carcinogenesis. 14671321_DLX1 is expressed in hematopoietic cells in a lineage-dependent manner and that DLX1 interacts with Smad4 through its homeodomain 14701756_CHIP can interact with the Smad1/Smad4 proteins and block BMP signal transduction through the ubiquitin-mediated degradation of Smad proteins. 14715079_Two missense mutations in the C-terminal domain of Smad4, D351H (Asp351-->His) & D537Y (Asp537-->Tyr), from colorectal cancer cells cannot interact with either TGF-beta-induced phosphorylated Smad2 or Smad3. 14727154_Observational study of gene-disease association. (HuGE Navigator) 14766211_SMAD4 has a role in regulation of large-scale chromatin unfolding 14988407_Smad4 protein stability is regulated by ubiquitin ligase SCF beta-TrCP1 15014009_Homozygous deletion, followed by inactivating nonsense or frameshift mutations, is the predominant form of MADH4 inactivation in pancreatic cancers. 15033661_Smad4 showed most significant prognostic differences in stage I gastric cancer patients. 15042598_decreased nuclear Smad4 expression associated with progression to prostate cancer; loss of BMP2 and Smad4 related to progression to a more aggressive phenotype 15063137_Inactivation of the DPC4 gene contributes to the genesis of colorectal carcinoma through allelic loss 15069531_Thirty-eight colon carcinomas were analyzed by immunohistochemistry for cell adhesion molecules (E-cadherin, beta-catenin, CD44), cell cycle regulatory proteins (cyclin D1, p27, p21), mismatch repair proteins (hMLH1, hMSH2), cyclooxygenase-2 and DPC4 15107966_exon 2 (belonging to the MH1 domain) and exons 8, 10, 11 (belonging to the MH2 domain) are not altered in renal cell carcinoma 15157044_a significant role of impaired SMAD4 function in the pathogenesis of small intestinal adenocarcinomas 15166010_The most transcriptionally active splice variants of Smad4 are made in macrophages (but not SMCs) of fibrofatty lesions and are upregulated after cell differentiation from monocytes. Cyclin inhibitors are induced by Smads. Fibrous plaque SMCs make Smad4. 15173084_Dpc4 may have a role in invasiveness of intraductal carcinoma 15235019_Observational study of genotype prevalence. (HuGE Navigator) 15240101_Our findings suggest that BAMBI transcription is regulated by TGF-beta signaling through direct binding of SMAD3 and SMAD4 to the BAMBI promoter. 15280432_Regulated cytoplasmic and nuclear retention may play a role in determining the distribution of Smads between the cytoplasm and the nucleus in both uninduced cells and upon TGF-beta induction. 15367885_DPC4 mutations were found in 40% of pancreatic adenocarcinoma cell lines and 58% of primary tumors. They were mostly deletions in exons 8-11, and 1 frameshift in exon 9. 15531914_Squamous cell cervical cancer showed loss of Smad4 protein expression or reduced expression. 15592526_gene expression regulation by TGF beta under Smad4 knockdown 15637079_Daxx suppresses Smad4-mediated transcriptional activity by direct interaction with the sumoylated Smad4 and has a role in regulating transforming growth factor beta signaling 15736060_expression of Smad4 was significantly lower in diffuse-type gastric adenocarcinoma than intestinal-type gastric adenocarcinomas 15799969_TGF-beta signaling suppresses nuclear export of Smad4 by chromosome region maintenance 1 and targets Smad4 into the nucleus; mutations in Smad4 that affect its interaction with Smad2 or Smad3 impair nuclear accumulation of Smad4 in response to TGF-beta 15814640_The level of expression of SMAD4 was found to be a more sensitive marker than 18q21 allelic imbalance and SMAD4 mutations, which were of no prognostic significance for colorectal cancer patients. 15817471_Smad4 is targeted for degradation by multiple ubiquitin ligases that can simultaneously act on R-Smads and signaling receptors 15846069_DPC4 inactivation was found in 75% of patients examined for lethal metastatic pancreatic neoplasms. 15855639_Cancer cell lines harboring Smad4 point mutations exhibited rapid Smad4 protein degradation due to the effect of SCF(beta-TrCP1). 15867212_Results suggest that loss of Smad4 expression may be involved in HPV16-induced carcinogenesis of head and neck squamous cell carcinomas. 15881652_Tsc-22 binds to and modulate the transcriptional activity of Smad3 and Smad4 15886208_demonstrate that Smad4 induces apoptosis by regulating Bim splicing as an initial intrinsic signal in ERalpha-positive cells 15940269_Smad4 is both frequently mutated and deregulated by aberrant splicing in thyroid tumours and these alterations may contribute as an early event to thyroid tumorigenesis. 16082587_SMAD4 gene alteration was found in patients, diagnosed with multiploid colorectal carcinomas. 16135802_There are two populations of TGF-beta target genes that are distinguished by their dependency on Smad4. 16146757_Smad4, but not Smad2, mediates TGF-beta1-induced MMP-2 expression in invasive extravillous trophoblasts 16172383_Functional evidence was provided for a switch of the Smad4 pathway, from tumor-suppressor to prometastatic, in the development of breast cancer bone metastasis. 16223572_deletion constructs of the promoter and mutational deletion of specific transcription factor binding sites indicated that Smad3/4 and AP-1 binding sites mediated the TGF-beta1 response on LTBP-3 16288847_Smad4 plays the role in the transcriptional activation of NF-kappaB. 16436638_Observational study of genotype prevalence. (HuGE Navigator) 16478646_A nonsense mutation of the SMAD4 gene in exon 5 codon 245 CAG (glut) -->TAG (stop) was found in neck squamous cell carcinoma cell line CAL27. 16613914_Hereditary haemorrhagic telangiectasia and juvenile polyposis in patients with SMAD4 mutations. 16627986_Biallelic inactivation of SMAD4 through homozygous deletion in breast cancer cell lines and invasive ductal carcinomas.( 16754688_One mechanism for positive regulation of TLR2 induction involves functional cooperation between the TGF-betaR-Smad3/4 pathway and NF-kappaB pathway. Another involves (MKP-1)-dependent inhibition of p38 MAPK, a known negative regulator for TLR2 induction 16953227_These data define the expression control of an essential BM component as a novel function for the tumor suppressor Smad4. 17016646_shRNA interference suppresses endogenous Smad4 gene expression and subsequently modulates cell growth and apoptosis. 17023741_aberration of the Transforming Growth factor-beta pathway, as indicated by a reduction or absence of Smad4 expression, promotes carcinogenesis of oral squamous cell carcinoma 17043799_identified a total of 47 protein species with a Smad4-dependent expression 17053951_mRNA expressed in human granulosa-luteal cells at oocyte retrieval. 17132729_Cooperates with lymphoid enhancer-binding factor 1 to activate c-myc expression. 17151782_DPC4 regulates MMP9 and may inhibit the proliferation of colon cancer cell by restraining growth and inducing apoptosis 17167985_Loss of SMAD4 expression was significantly more frequent in poorly differentiated carcinoma and signet-ring cell carcinoma of the colorectum 17200344_SMAD4 loss in gastric carcinomas is due to several mechanisms, including LOH, hypermethylation, and proteasome degradation 17301079_Smad4 mediates down-regulation of E-cadherin induced by TGF-beta in PANC-1 cells, at least in part, through Snail and Slug induction. 17353364_Transgenic Smad4 has a role in the maintenance of hematopoietic stem cell self-renewal and reconstituting capacity. 17390050_Our results indicate that absence of Smad4 expression correlated significantly with liver metastases regardless of the time of their occurrence and represents a promising new biomarker to predict liver metastasis in colorectal cancer patients. 17436386_Mutations of K-ras and Dpc4 genes can accumulate already in non-malignant, inflammatory pancreatic tissue, suggesting its applicability in monitoring of further destruction of pancreatic tissue and progression into malignancy. 17469085_Correlation of the expression of S100A8 and S100A9 revealed that the microenvironments of tumours which lacked expression of Smad4, had significantly reduced numbers of S100A8-immunoreactive (p = 0.023) but not S100A9-immunoreactive (p = 0.21) cells. 17476473_examined the possible deterioration in the pathway in human squamous cancer cell lines, focusing on intracellular localization of S100C/A11 and its functional partners Smad3 and Smad4 17478078_Proinvasive activity of BMP7 through SMAD4/src-independent and ERK/Rac/JNK-dependent signaling pathways in colon cancer cells is reported. 17591695_Results show that Arkadia specifically activates transcription via Smad3/Smad4 binding sites by inducing degradation of the transcriptional repressor SnoN. 17643425_Smad4 siRNA treatment completely abolished TGFb-induced early gene upregulation, indicating the absence of the rapid activation of Smad signaling. 17659731_combined with screening of K-ras mutations and allelic losses of tumor suppressors p16 and DPC4 represents a very sensitive approach in screening for pancreatic malignancy. 17847004_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17854080_study suggests that SMAD4 is an important marker for confirming a diagnosis of pancreatic adenocarcinoma as a primary tumor, as well as when it presents as a metastatic tumor on small fine-needle aspirate samples 17873119_5 nonsense, 6 frameshift and 4 missense mutations (2 new) were associated with juvenile polyposis syndrome. 17873119_Observational study of gene-disease association. (HuGE Navigator) 17875924_Mutant p53 attenuates TGF-beta1 signaling. This was exhibited by a reduction in SMAD2/3 phosphorylation and an inhibition of both the formation of SMAD2/SMAD4 complexes and the translocation of SMAD4 to the cell nucleus. 17994767_functional characterization of a tumorigenic mutation in Smad4(E330A); findings show this mutant & a Smad3 mutant (Smad3 E239A) failed to activate transcription in response to TGFbeta stimulation because of defects in oligomerization 17997817_Smad4 has a role in extracellular matrix composition in cervical cancer 18178612_Large genomic deletions of SMAD4, BMPR1A and PTEN are a common cause of JPS. 18213629_We failed to find evidence of genomic deletions or amplifications affecting the Smad4 locus on chromosome 18 in advanced prostate cancer. 18310076_loss of Smad4 contributes to aberrant RON expression and cross-talk of Smad4-independent TGF-beta signaling and the RON pathway promotes an invasive phenotype 18310088_vidence for a cross talk between Smad4 and the Wnt/beta-catenin pathway in pancreatic carcinoma cells. 18321803_Data show that the expression of key transcription factors, phosphorylated Smad1 protein, and the nuclear accumulation of Smad1 and Smad4 are inhibited by Ubc9 silencing. 18413775_Observational study of gene-disease association. (HuGE Navigator) 18425078_Smad4 is a target molecule for functional inactivation in cervical cancer. 18471510_In MSI CRC this is associated predominantly with impaired BMPR2 expression and in MSS CRC with impaired SMAD4 expression. 18505344_DPC$ might be involved in preventing the tumor metastasis by inhibiting tumor angiogenesis. 18519565_TGF-beta-induced and basal state spontaneous nuclear import of Smad4 require importin 7 and 8. 18519681_loss of Smad4, leading to aberrant activation of STAT3, contributes to the switch of TGFbeta from a tumor-suppressive to a tumor-promoting pathway in pancreatic cancer 18568018_Data demonstrate that in response to TGFbeta stimulation the transcriptional regulator TAZ binds heteromeric Smad2/3-4 complexes and is recruited to TGFbeta response elements. 18620728_Deleted in pancreatic carcinoma locus 4 might be an important biomarker for malignant transformation and be involved in inducing apoptosis by modulating Bcl-2/Bax balance. 18664273_Smad4 mediates transcriptional regulation through three mechanisms: Smad4 binding to a functional SBE site in the LAMA3 promoter, Smad4 binding to AP1 (and Sp1) sites via interaction with AP1 family, and Smad4 impact on transcription of AP1 factors 18823382_Observational study of gene-disease association. (HuGE Navigator) 18823382_The overall prevalence of SMAD4 and BMPR1A point mutations and deletions in JPS was 45% in the largest series of patients to date 18949401_TGF-beta1-induced cell growth inhibition by up-regulating p16 expression and cellular apoptosis by activating caspase 3 was Smad4-dependent 18985820_Inhibition of pancreatic carcinoma cell growth in vitro by DPC4 gene transfection. 19064568_Observational study of gene-disease association. (HuGE Navigator) 19064568_The inactivation of SMAD4 is similar in familial pancreatic adenocarcinomas as in sporadic pancreatic adenocarcinomas. 19144825_The association between v-ErbA and Smad4 is essential for the dysregulation of TGF-beta signaling. 19183329_There was no association between SMAD4(MAD homolog 4) protein expression, SMAD4 copy number, family history, Microsatellite instability status, tumour stage or grade in patients with early onset colorectal cancer 19211612_Observational study of gene-disease association. (HuGE Navigator) 19247629_Observational study of gene-disease association. (HuGE Navigator) 19266212_Both solitary and SMAD4 mutated JP case showed extensive co-localization of the alterations described above: decreased expression of SMAD4, CDX2 and partially also MUC2 and de novo expression of MUC5AC. 19270646_The majority of the cases of Adenosquamous carcinoma of the pancreas had loss of Dpc4 protein 19270816_mutational analysis of the SMAD4 gene in Korean patients with hereditary hemorrhagic telangiectasia. 19273710_Observational study of gene-disease association. (HuGE Navigator) 19284991_The data suggested that restoration of Smad4 in Smad4-deficient cells may provide a potential therapeutic strategy for intervention of colon cancer migration and metastasis. 19321257_Smad4 is dispensable for enhanced invasiveness of human colorectal cancer cells due to BMP-4 overexpression. 19341727_Results suggest a mechanism by which a balance between Smad4 and Smad7 in human gastric cancer is critical for differentiation, metastasis, and apoptosis of tumor cells. 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19463221_The prevalence of germline mutations of BMPR1A and SMAD4 are about 20% each in the patient with JPS. 19527492_Observational study of gene-disease association. (HuGE Navigator) 19538865_Low expression of Smad4 and TbetaRII may promote metastasis of oral squamous cell carcinoma. 19548527_Observational study of gene-disease association. (HuGE Navigator) 19558215_Study shows that Sp1 alone or the combination of Smad2 and Smad4 activated the alpha(1)(I) collagen promoter in transfected LX-2 stellate cells. 19584151_Data show that SMAD4 gene inactivation is associated with poorer prognosis in patients with surgically resected adenocarcinoma of the pancreas. 19624886_Smad4 expression was related with tumor differentiation and Lauren classification of gastric cancer. 19626374_Strong Smad4 expression is associated with hepatocellular carcinoma. 19686287_Smad4 was found in the regions relatively distant from the transcription start sites, while Smad2/3 binding regions were more often present near the transcription start sites. 19688145_Study data demonstrates downregulated SMAD4 expression in psoriatic skin. 19690946_threonine 9 (Thr9) and Serine 138 (Ser138) within the N-terminal Mad homology1 (MH1) domain of Smad4 could be phosphorylated by NLK 19730683_Observational study of gene-disease association. (HuGE Navigator) 19752858_Linoleic acid induces PAI-1 expression in breast cancer cells through SMAD4. 19834456_4E-BP1 gene appears critical for TGFbeta/Smad4-mediated inhibition of cell proliferation. 19841536_connection between Smad4 and the Fanc/Brca pathway and highlight the impact of epithelial Smad4 loss on inflammation 19856310_a predictive marker of pancreatic ductal adenocarcinoma cell permissiveness for oncolytic infection with parvovirus H-1PV 19913121_Observational study of gene-disease association. (HuGE Navigator) 19916025_Mutations and/or alterations in expression of TGF-beta receptors and loss of Smad 4 are frequent in human ovarian cancers. 19996292_BMP9 acts as a proliferative factor for immortalized ovarian surface epithelial cells and ovarian cancer cell lines, signaling predominantly through an ALK2/Smad1/Smad4 pathway, the major BMP9 receptor in endothelial cells. 20012971_Expression of TGF-beta1 and its downstream effectors Smad4 and Smad7 was assessed & only Smad4 was found to have possible predictive value for esophageal squamous cell carcinoma in patients receiving neoadjuvant chemoradiotherapy. 20101697_analysis of SMAD4 mutations with juvenile polyposis (JP) and JP-HHT syndrome 20118412_The ectopic expression of miR-224 can enhance TGF-beta1-induced GC proliferation through targeting Smad4. 20165854_The expression of TGF-beta1, its receptor TGFbetaRII, and signaling proteins Smad4 and Smad7 was observed in the majority of colorectal cancer specimens. 20200332_Observational study of gene-disease association. (HuGE Navigator) 20235236_SMAD4 is not required for maintenance of the undifferentiated state of human embryonic stem cells, but rather to stabilize that state. 20237276_BMP2 is both negatively and positively regulated by Smad4 and YY1 bound to a promoter region in cerebral cortex neurons. 20307265_Smad4-reconstituted colon carcinoma cells respond to TNFalpha by increased laminin-332 expression; coincubation with TGFbeta and TNFalpha leads to synergistic induction and to the secretion of large amounts of the heterotrimer. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20350217_We assessed the immunohistochemical expression profiles of Smad2, P-Smad2, Smad4, and p21/WAF1 proteins in 34 cases of osteosarcoma 20404275_SMAD4 regulates inositol-1,4,5-trisphosphate 5-phosphatase (SHIP1) and interleukin-1 receptor-associated kinase 3 (IRAK-M) expression during lipopolysaccharide-induced development of endotoxin tolerance. 20473902_Our study indicates for the first time, that oncogenic ras and loss of Smad signaling cooperate to upregulate EGFR and erbB2, which plays a role in promoting invasion. 20565773_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20565773_Observed a significant association of SMAD4 gene aberrations with KRAS mutant status suggesting the involvement of at least two molecules in the advanced tumor grade in colorectal cancers in a Kashmiri population. 20577838_RETINOIC ACID UPREGULATES MIR146A AND DOWNREGULATES SMAD4 IN APL CELLS 20581473_Study identified driver mutations in three known pancreatic cancer driver genes P53, SMAD4 and CDKN2A. 20622003_Smad4 loss alters the tumor's interaction with stromal myeloid cells and the tumor cells' response to the stromal chemokine, S100A8. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20682711_different routes to neoplasia in Juvenile polyposis syndrome caused by germline SMAD4 mutation seem to be operative 20682989_Results provide further evidence for a role of SMAD4 as a regulator of invasion, a process of prime importance in carcinogenesis but hitherto poorly understood in molecular terms. 20685810_Reduction of SMAD4 may play a significant role in thyroid carcinogenesis. 20734064_Observational study of gene-disease association. (HuGE Navigator) 20734429_cooperative regulation of estrogen signaling by FHL2 and Smad4 in breast cancer cells, and might provide a new regulation mechanism underlying breast cancer development and progression 20797318_Smad4 suppresses human ovarian cancer cell metastasis potential through its effect on the expressions of PAI-1, E-cadherin and VEGF. 20862427_New Smad4 dependent and independent TGF-beta responses in colon carcinoma cells, were identified. 20885978_TGFbeta1 down-regulated expression of prolactin and IGFBP-1 in endometrial stromal cells in SMAD4-dependent and SMAD4-independent manners. 21036691_Observational study of gene-disease association. (HuGE Navigator) 21036691_SMAD4 gene promoter haplotype -462T(14)/-4T(10) was found in 85% of pancreatic cancer tissues, but it was not present in colorectal cancer tissues 21068203_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21095583_PARP-1 dissociates Smad complexes from DNA by ADP-ribosylating Smad3 and Smad4, which attenuates Smad-specific gene responses and TGF-beta-induced epithelial-mesenchymal transition. 21105199_These results suggest that genetic variants in the SMAD4 gene play a protective role in gastric cancer in a Chinese population. 21112326_Findings identify miR-483-3p as a potent regulator of DPC4/Smad4, which may provide a novel therapeutic strategy for the treatment of DPC4/Smad4-driven pancreatic cancer. 21126430_Individuals with the SMAD4 (rs10502913) AA genotype had an increased risk of coal work's pneumoconiosis. 21245094_Results define a molecular mechanism that explains how loss of the tumor suppressor Smad4 promotes colorectal cancer progression 21259057_the aim of this study was to investigate the prognosis and clinicopathologic roles of beta-catenin, Wnt1, Smad4, Hoxa9, and Bmi-1 in esophageal squamous cell carcinoma 21263249_Loss of Dpc4 expression provides the most useful immunohistochemical evidence for establishing the pancreaticobiliary tract as the most likely source of these metastatic mucinous carcinomas in the ovary. 21289291_Data indicate that hypoxia represses hepcidin expression through inhibition of BMP/SMAD4 signaling. 21289624_This model-informed progression analysis, together with genetic, functional and translational studies, establishes SMAD4 as a key regulator of prostate cancer progression in mice and humans. 21294585_proposed two possible model structures of Hoxa9 and Smad4 complex. 21330551_show that Gata4 and Smad4 cooperatively activated the Id2 promoter, that human GATA4 mutations abrogated this activity, and that Id2 deficiency in mice could cause atrioventricular septal defects 21352803_these findings identify miR-421 as a potent regulator of DPC4/Smad4 in pancreas cancer. 21412070_Juvenile polyps with a SMAD4 germline mutation were predominantly type B, whereas type A was more common among juvenile polyps with a BMPR1A germline mutation. 21421563_The predicted transcription factor binding site profiles for each of the four SMAD4 promoters shared few transcription factors in common, but were conserved across several species. 21465659_report of an individual whose family history was positive for aortopathy, mitral valve dysfunction, and juvenile polyposis syndrome; mutation analysis of SMAD4 implicates this gene for these phenotypes in this family 21492476_Data suggest a regulatory circuitry involving Smad4 dependent up-regulation of KRT23 (directly or indirectly) which in turn modulates the interaction between KRT23 and 14-3-3epsilon leading to a cytoplasmic sequestration of 14-3-3epsilon. 21540640_SMAD4 knockdown accelerated this re-silencing process, suggesting that normal TGF-beta signaling is essential for the maintenance of RunX1T1 expression 21597466_TIF1gamma binds to and represses the plasminogen activator inhibitor 1 promoter, demonstrating a direct role of TIF1gamma in TGF-beta-dependent gene expression 21705453_Data indicate that Cal27 cells stably transfected with Smad4 showed similar resveratrol effects as parental Cal27, suggegsting that a lack of resveratrol effect in Det562 cells was independent of Smad4 status in these cells. 21709185_Data shows that Smad4(Dpc4) immunostaining correlated with the pattern of disease progression in pancreatic adenocarcinoma. 21726607_Smad4 overexpression promotes apoptosis in tongue squamous carcinoma cells, and decreases TGF-beta1-mediated cell migration and metastasis. 21782795_The present finding that NF1/Smad4 repressor complexes are formed through TGF-beta signaling pathways suggests a new, but much broader, role for these complexes in the initiation or maintenance of the growth-inhibited state. 21791112_SMAD4 may play an inhibitive role during the development of glioma and may be a potential prognosis predictor of glioma. 21835029_in breast cancer cases SMAD4 was significantly over-expressed 21898662_A putative splice site mutation in SMAD4 resulted in moderate transcript loss due to compromised splicing efficiency in PAH, pulmonary arterial hypertension. 21945631_Results show that BMP4-induced changes in OvCa cell morphology and motility are Smad-dependent with shRNA targeting Smads 1, 4, and 5. 21964812_novel findings that high SMAD4 expression predicts a better prognosis suggests that SMAD4 immunohistochemistry could constitute a prognostic marker in combination with CIMP and MSI screening status 21968601_It was shown that miR-146a functions as a novel negative regulator to modulate myofibroblast transdifferentiation during TGF-beta1 induction by targeting SMAD4. 22002709_SMAD4, occurring in 12% of colorectal adenocarcinomas, correlated with the presence of lymph node metastases. 22020746_miR-146a can directly target SMAD4, and suggest that miR-146a may play a role in the development of gastric cancer by modulating cell proliferation and apoptosis. 22024061_Smad4 may help to identify a subset of colorectal cancer patients with early recurrence after curative therapy. 22028478_Data show two miR-130a binding sites were identified in the 3'-untranslated region of the Smad4 mRNA. 22109972_Smad4 C324Y mutation plays an important role in thyroid carcinogenesis and can be considered as a new prognostic and therapeutic target for thyroid cancer 22115830_Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. 22130069_Smad4-expressed cancer tissues not only show the elevated expression of PAK1, but also support our hypothesis that Smad4 induces PUMA-mediated cell death through PAK1 suppression. 22158539_We identified three distinct heterozygous missense SMAD4 mutations affecting the codon for Ile500 in 11 individuals with Myhre syndrome. 22209340_Compared with conventional carcinomas, serrated adenocarcinomas showed significantly reduced SMAD4 22243968_The present report identified a previously unrecognized class of mutations in the SMAD4 gene with profound impact on development and growth. 22266936_Genetic alt | ENSMUSG00000024515 | Smad4 | 1847.124090 | 0.9903967 | -0.013921648 | 0.08252609 | 2.806133e-02 | 8.669647e-01 | 9.556819e-01 | No | Yes | 2811.145068 | 494.497894 | 2816.874471 | 495.531959 | |
| ENSG00000141664 | 54877 | ZCCHC2 | protein_coding | Q9C0B9 | Alternative splicing;Metal-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | hsa:54877; | cytoplasm [GO:0005737]; nucleic acid binding [GO:0003676]; phosphatidylinositol binding [GO:0035091]; zinc ion binding [GO:0008270] | ENSMUSG00000038866 | Zcchc2 | 678.558982 | 1.2278444 | 0.296127775 | 0.12052761 | 6.113436e+00 | 1.341580e-02 | 1.531487e-01 | No | Yes | 704.034353 | 93.254464 | 573.962534 | 76.098560 | ||||
| ENSG00000142632 | 128272 | ARHGEF19 | protein_coding | Q8IW93 | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for RhoA GTPase. {ECO:0000250}. | Alternative splicing;GTPase activation;Guanine-nucleotide releasing factor;Reference proteome;SH3 domain | Guanine nucleotide exchange factors (GEFs) such as ARHGEF19 accelerate the GTPase activity of Rho GTPases (see RHOA, MIM 165390).[supplied by OMIM, Dec 2008]. | hsa:128272; | cytosol [GO:0005829]; guanyl-nucleotide exchange factor activity [GO:0005085]; activation of GTPase activity [GO:0090630]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of small GTPase mediated signal transduction [GO:0051056]; Wnt signaling pathway, planar cell polarity pathway [GO:0060071]; wound healing [GO:0042060] | 24405610_overcomes the suppressive influence of miR-503 in hepatocellular carcinoma cells 29164615_These findings suggest that ARHGEF19 upregulation, due to the low expression of miR-29 in non-small cell lung cancer tissues, may play a crucial role in non-small cell lung cancer tumorigenesis by activating MAPK signaling 32993957_ARHGEF19 regulates MAPK/ERK signaling and promotes the progression of small cell lung cancer. | ENSMUSG00000028919 | Arhgef19 | 46.539596 | 0.9173485 | -0.124458191 | 0.39437503 | 9.642169e-02 | 7.561668e-01 | No | Yes | 41.317263 | 7.706440 | 44.775269 | 8.343145 | ||
| ENSG00000142700 | 63950 | DMRTA2 | protein_coding | Q96SC8 | FUNCTION: May be involved in sexual development. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Zinc | hsa:63950; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; cerebral cortex regionalization [GO:0021796]; dopaminergic neuron differentiation [GO:0071542]; neuron fate specification [GO:0048665]; positive regulation of neuroblast proliferation [GO:0002052]; regulation of transcription by RNA polymerase II [GO:0006357]; sex differentiation [GO:0007548]; skeletal muscle cell differentiation [GO:0035914] | 25082981_This study reveals, for the first time, the requirement of DMRTA2 for normal human female embryonic germ cell development. 26757254_Data suggest that loss of function of doublesex and mab-3-related transcription factor a2 (DMRTA2) leads to disorder of cortical development. | ENSMUSG00000047143 | Dmrta2 | 39.469464 | 0.8179759 | -0.289869682 | 0.41986030 | 4.803892e-01 | 4.882461e-01 | No | Yes | 35.134032 | 6.650130 | 44.408412 | 8.072500 | |||
| ENSG00000143319 | 81875 | ISG20L2 | protein_coding | Q9H9L3 | FUNCTION: 3'-> 5'-exoribonuclease involved in ribosome biogenesis in the processing of the 12S pre-rRNA. Displays a strong specificity for a 3'-end containing a free hydroxyl group. {ECO:0000269|PubMed:18065403}. | Exonuclease;Hydrolase;Nuclease;Nucleus;Reference proteome;Ribosome biogenesis | This gene encodes a 3'-5' exoribonuclease that may be involved in the processing of the 12S pre-rRNA. Pseudogenes have been identified on chromosomes 6 and 11. [provided by RefSeq, Dec 2014]. | hsa:81875; | nucleolus [GO:0005730]; nucleus [GO:0005634]; 3'-5'-exoribonuclease activity [GO:0000175]; exonuclease activity [GO:0004527]; RNA binding [GO:0003723]; ribosome biogenesis [GO:0042254] | 18065403_ISG20L2 is a 3' to 5' exoribonuclease involved in ribosome biogenesis at the level of 5.8 S rRNA maturation, more specifically in the processing of the 12 S precursor rRNA. 30394818_miR-139-3p target genes ISG20L2, RAD54B, KIAA0101, and PIGS were significantly negatively correlated with miR-139-3p expression in patients with hepatocellular carcinoma. | ENSMUSG00000048039 | Isg20l2 | 4006.528477 | 1.1362919 | 0.184333546 | 0.05427261 | 1.153836e+01 | 6.817461e-04 | 2.757704e-02 | No | Yes | 4552.470546 | 381.972183 | 3977.886069 | 333.789579 | |
| ENSG00000143498 | 9015 | TAF1A | protein_coding | Q15573 | FUNCTION: Component of the transcription factor SL1/TIF-IB complex, which is involved in the assembly of the PIC (pre-initiation complex) during RNA polymerase I-dependent transcription. The rate of PIC formation probably is primarily dependent on the rate of association of SL1/TIF-IB with the rDNA promoter. SL1/TIF-IB is involved in stabilization of nucleolar transcription factor 1/UBTF on rDNA. Formation of SL1/TIF-IB excludes the association of TBP with TFIID subunits. {ECO:0000269|PubMed:15970593, ECO:0000269|PubMed:7801123}. | Alternative splicing;DNA-binding;Direct protein sequencing;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene encodes a subunit of the RNA polymerase I complex, Selectivity Factor I (SLI). The encoded protein is a TATA box-binding protein-associated factor that plays a role in the assembly of the RNA polymerase I preinitiation complex. Alternate splicing results in multiple transcript variants encoding multiple isoforms.[provided by RefSeq, Jan 2011]. | hsa:9015; | microtubule cytoskeleton [GO:0015630]; nucleoplasm [GO:0005654]; RNA polymerase I transcription regulator complex [GO:0000120]; DNA binding [GO:0003677]; transcription by RNA polymerase I [GO:0006360]; transcription by RNA polymerase II [GO:0006366] | 15113842_This study identifies the first nuclear import sequence within the TBP-Associated Factor subunits of Selectivity Factor 1. 19246067_The authors demonstrate the interaction of both RNA polymerase I and III with hepatitis delta virus RNA, both in vitro and in human cells. 32432789_TNFAIP8 regulates cisplatin resistance through TAF-Ialpha and promotes malignant progression of esophageal cancer. | ENSMUSG00000072258 | Taf1a | 396.928494 | 1.0980387 | 0.134928913 | 0.14150210 | 9.049537e-01 | 3.414569e-01 | No | Yes | 444.398435 | 63.768032 | 401.988475 | 57.722588 | ||
| ENSG00000143502 | 55061 | SUSD4 | protein_coding | Q5VX71 | FUNCTION: Acts as complement inhibitor by disrupting the formation of the classical C3 convertase. Isoform 3 inhibits the classical complement pathway, while membrane-bound isoform 1 inhibits deposition of C3b via both the classical and alternative complement pathways. {ECO:0000269|PubMed:23482636}. | Alternative splicing;Complement pathway;Disulfide bond;Glycoprotein;Immunity;Innate immunity;Membrane;Reference proteome;Repeat;Secreted;Signal;Sushi;Transmembrane;Transmembrane helix | hsa:55061; | extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; complement activation, classical pathway [GO:0006958]; innate immune response [GO:0045087]; negative regulation of complement activation, alternative pathway [GO:0045957]; negative regulation of complement activation, classical pathway [GO:0045959]; regulation of complement activation [GO:0030449] | 20237496_Observational study of gene-disease association. (HuGE Navigator) 20348246_analysis of cloning, expression, refolding, and tissue distribution of Sushi domain-containing protein 4 23482636_Membrane-bound isoform SUSD4a inhibits the classical and alternative complement pathways when expressed on the surface of Chinese hamster cells but not when expressed as a soluble, truncated protein. 26480818_SUSD4 expression in both breast cancer cells and T cells infiltrating the tumor-associated stroma is useful to predict better prognosis of breast cancer patients. | ENSMUSG00000038576 | Susd4 | 53.476399 | 0.8525313 | -0.230175335 | 0.43032079 | 2.857710e-01 | 5.929434e-01 | No | Yes | 41.307279 | 10.699445 | 48.900497 | 12.623786 | |||
| ENSG00000144426 | 65065 | NBEAL1 | protein_coding | Q6ZS30 | Alternative splicing;Reference proteome;Repeat;WD repeat | hsa:65065; | cytosol [GO:0005829]; membrane [GO:0016020]; protein kinase binding [GO:0019901]; protein localization [GO:0008104] | 15193433_isolation of human neurobeachin-like 1 (NBEAL1); highly expressed in the brain, kidney, prostate, and testis, and in biopsies of different grade glioma [NBEAL1] | ENSMUSG00000073664 | Nbeal1 | 278.740130 | 0.9152821 | -0.127711592 | 0.16551112 | 5.879963e-01 | 4.431950e-01 | No | Yes | 154.231547 | 40.209353 | 173.717771 | 45.344694 | ||||
| ENSG00000144451 | 79582 | SPAG16 | protein_coding | Q8N0X2 | FUNCTION: Necessary for sperm flagellar function. Plays a role in motile ciliogenesis. May help to recruit STK36 to the cilium or apical surface of the cell to initiate subsequent steps of construction of the central pair apparatus of motile cilia (By similarity). {ECO:0000250|UniProtKB:Q8K450}. | Alternative splicing;Cell projection;Cilium;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Flagellum;Phosphoprotein;Reference proteome;Repeat;WD repeat | Cilia and flagella are comprised of a microtubular backbone, the axoneme, which is organized by the basal body and surrounded by plasma membrane. SPAG16 encodes 2 major proteins that associate with the axoneme of sperm tail and the nucleus of postmeiotic germ cells, respectively (Zhang et al., 2007 [PubMed 17699735]).[supplied by OMIM, Jul 2008]. | hsa:79582; | axonemal central apparatus [GO:1990716]; axoneme [GO:0005930]; extracellular region [GO:0005576]; sperm flagellum [GO:0036126]; axoneme assembly [GO:0035082]; cerebrospinal fluid circulation [GO:0090660]; cilium assembly [GO:0060271]; mucociliary clearance [GO:0120197]; sperm axoneme assembly [GO:0007288] | 12391165_Data describe the cloning of human and mouse Pf20 proteins that interact with Spag6 protein. 17699735_Heterozygous mutation that affects both SPAG16L and SPAG16S does not cause male infertility in man, but is associated with reduced stability of the interacting proteins of the central apparatus. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20463177_Observational study of gene-disease association. (HuGE Navigator) 22963137_Analysis of SPAG16 regions encoding conserved WD repeats revealed no evidence for association of mutations or genetic variation with sperm motility in infertile males. 23956247_SPAG16 influences MMP-3 regulation and protects against joint destruction in autoantibody-positive rheumatoid arthritis. 25086173_SPAG16 is a novel autoantibody target in a subgroup of MS patients and in combination with other diagnostic criteria, elevated levels of anti-SPAG16 Abs could be used as a biomarker for diagnosis. 28137312_Short-SOX5 regulates transcription of human SPAG16L gene via directly binding to the promoter of SPAG16L. 34445527_Identification and Classification of Novel Genetic Variants: En Route to the Diagnosis of Primary Ciliary Dyskinesia. | ENSMUSG00000053153 | Spag16 | 432.273531 | 0.9722402 | -0.040615255 | 0.13270963 | 9.320242e-02 | 7.601447e-01 | No | Yes | 446.481006 | 71.990440 | 443.807459 | 71.566221 | ||
| ENSG00000144524 | 64708 | COPS7B | protein_coding | Q9H9Q2 | FUNCTION: Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of SCF-type E3 ligase complexes, leading to decrease the Ubl ligase activity of SCF-type complexes such as SCF, CSA or DDB2. The complex is also involved in phosphorylation of p53/TP53, JUN, I-kappa-B-alpha/NFKBIA, ITPK1 and IRF8/ICSBP, possibly via its association with CK2 and PKD kinases. CSN-dependent phosphorylation of TP53 and JUN promotes and protects degradation by the Ubl system, respectively. {ECO:0000269|PubMed:11285227, ECO:0000269|PubMed:11337588, ECO:0000269|PubMed:12628923, ECO:0000269|PubMed:12732143}. | 3D-structure;Acetylation;Alternative splicing;Coiled coil;Cytoplasm;Host-virus interaction;Nucleus;Phosphoprotein;Reference proteome;Signalosome | hsa:64708; | COP9 signalosome [GO:0008180]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; COP9 signalosome assembly [GO:0010387]; protein deneddylation [GO:0000338] | 33503427_CSN7B defines a variant COP9 signalosome complex with distinct function in DNA damage response. | ENSMUSG00000026240 | Cops7b | 2268.810740 | 1.0619745 | 0.086749190 | 0.06740782 | 1.656115e+00 | 1.981286e-01 | 5.674383e-01 | No | Yes | 2417.238036 | 150.568345 | 2258.942095 | 140.607724 | ||
| ENSG00000145016 | 9711 | RUBCN | protein_coding | Q92622 | FUNCTION: Inhibits PIK3C3 activity; under basal conditions negatively regulates PI3K complex II (PI3KC3-C2) function in autophagy. Negatively regulates endosome maturation and degradative endocytic trafficking and impairs autophagosome maturation process. Can sequester UVRAG from association with a class C Vps complex (possibly the HOPS complex) and negatively regulates Rab7 activation (PubMed:20974968, PubMed:21062745). {ECO:0000269|PubMed:20974968, ECO:0000269|PubMed:21062745}.; FUNCTION: Involved in regulation of pathogen-specific host defense of activated macrophages. Following bacterial infection promotes NADH oxidase activity by association with CYBA thereby affecting TLR2 signaling and probably other TLR-NOX pathways. Stabilizes the CYBA:CYBB NADPH oxidase heterodimer, increases its association with TLR2 and its phagosome trafficking to induce antimicrobial burst of ROS and production of inflammatory cytokines (PubMed:22423966). Following fungal or viral infection (implicating CLEC7A (dectin-1)-mediated myeloid cell activation or DDX58/RIG-I-dependent sensing of RNA viruses) negatively regulates pro-inflammatory cytokine production by association with CARD9 and sequestering it from signaling complexes (PubMed:22423967). {ECO:0000269|PubMed:22423966, ECO:0000269|PubMed:22423967}. | 3D-structure;Alternative splicing;Autophagy;Endocytosis;Endosome;Immunity;Lysosome;Neurodegeneration;Phagocytosis;Phosphoprotein;Reference proteome | The protein encoded by this gene is a negative regulator of autophagy and endocytic trafficking and controls endosome maturation. This protein contains two conserved domains, an N-terminal RUN domain and a C-terminal DUF4206 domain. The RUN domain is involved in Ras-like GTPase signaling, and the DUF4206 domain contains a diacylglycerol (DAG) binding-like motif. Mutation in this gene results in deletion of the DAG binding-like motif and causes a recessive ataxia. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Apr 2014]. | hsa:9711; | cytosol [GO:0005829]; early endosome [GO:0005769]; intracellular membrane-bounded organelle [GO:0043231]; late endosome [GO:0005770]; lysosome [GO:0005764]; nucleoplasm [GO:0005654]; phosphatidylinositol phosphate binding [GO:1901981]; autophagy [GO:0006914]; endocytosis [GO:0006897]; immune system process [GO:0002376]; multivesicular body sorting pathway [GO:0071985]; negative regulation of autophagosome maturation [GO:1901097]; negative regulation of autophagy [GO:0010507]; negative regulation of endocytosis [GO:0045806]; negative regulation of phosphatidylinositol 3-kinase activity [GO:0043553]; phagocytosis [GO:0006909] | 19270696_Two Beclin 1 associated proteins, Atg14L and Rubicon, were identified. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20826435_we report the identification of a family with three children affected with a new form of recessive ataxia, which we suggest naming 'Salih ataxia', and of a frameshift mutation of KIAA0226 (rundataxin) that segregates with the disease 20943950_Rubicon and PLEKHM1 specifically and directly interact with Rab7 via their RH domain; this interaction is critical for their function; show Rubicon but not PLEKHM1 uniquely regulates membrane trafficking via simultaneously binding both Rab7 and PI3-kinase 20974968_Rubicon serves as a previously unknown Rab7 effector to ensure the proper progression of the endocytic pathway. 21062745_a critical role of the Rubicon RUN domain in PI3KC3 and autophagy regulation 22423966_Rubicon may thus be pivotal to generating an optimal intracellular immune response against microbial infection. 22423967_Rubicon differentially targets signaling complexes, depending on environmental stimuli, and may function to coordinate various immune responses against microbial infection. 22527681_New DNA sequencing technologies are enabling us to investigate the whole or large targeted proportions of the genome in a rapid, affordable, and comprehensive way. Exome and targeted sequencing rundataxin genes causing ataxia. 23728897_This study demonistrated that KIAA0226 mutation impairs Rubicon endosomal localization 25807108_HCV, by differentially inducing the expression of Rubicon and UVRAG, temporally regulated the autophagic flux to enhance its replication. 28392573_Rubicon thus functions as an important negative regulator of the innate immune response, enhances viral replication and may play a role in viral immune evasion. 29215797_Rubicon (Rubcn) was identified as a negative regulator of canonical autophagy and endosomal trafficking [Review]. 31752345_Rubicon as a novel substrate of HUNK. 32165681_Identification of highly potent and selective inhibitor, TIPTP, of the p22phox-Rubicon axis as a therapeutic agent for rheumatoid arthritis. 32632011_Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex. 32943718_Hepatitis C virus enhances Rubicon expression, leading to autophagy inhibition and intracellular innate immune activation. 34932879_Epigenome-wide three-way interaction study identifies a complex pattern between TRIM27, KIAA0226, and smoking associated with overall survival of early-stage NSCLC. | ENSMUSG00000035629 | Rubcn | 634.783366 | 1.0691197 | 0.096423394 | 0.12414826 | 6.010265e-01 | 4.381866e-01 | 7.709670e-01 | No | Yes | 701.491801 | 74.667276 | 630.254689 | 67.096109 | |
| ENSG00000145868 | 81545 | FBXO38 | protein_coding | Q6PIJ6 | FUNCTION: Substrate recognition component of a SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of PDCD1/PD-1, thereby regulating T-cells-mediated immunity (PubMed:30487606). Required for anti-tumor activity of T-cells by promoting the degradation of PDCD1/PD-1; the PDCD1-mediated inhibitory pathway being exploited by tumors to attenuate anti-tumor immunity and facilitate tumor survival (PubMed:30487606). May indirectly stimulate the activity of transcription factor KLF7, a regulator of neuronal differentiation, without promoting KLF7 ubiquitination (By similarity). {ECO:0000250|UniProtKB:Q8BMI0, ECO:0000269|PubMed:30487606}. | Adaptive immunity;Alternative splicing;Cytoplasm;Disease variant;Immunity;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:30487606}. | This gene encodes a large protein that contains an F-box domain and may participate in protein ubiquitination. The encoded protein is a transcriptional co-activator of Krueppel-like factor 7 (Klf7). A heterozygous mutation in this gene was found in individuals with autosomal dominant distal hereditary motor neuronopathy type IID. There is a pseudogene for this gene on chromosome 4. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2013]. | hsa:81545; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; SCF ubiquitin ligase complex [GO:0019005]; adaptive immune response [GO:0002250]; positive regulation of neuron projection development [GO:0010976]; positive regulation of T cell mediated immune response to tumor cell [GO:0002842]; protein K48-linked ubiquitination [GO:0070936]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146] | 14729953_This publication identifies the F-box protein 38 mouse ortholog and characterizes its interaction with KLF7 during regulation of cell cycle progression. 20549515_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 26643602_Our findings provide direct evidence for the association of FBXO38 and AP3B2 with severe chronic periodontitis in the Han Chinese population. 30487606_data indicate that FBXO38 regulates PD-1 expression and highlight an alternative method to block the PD-1 pathway 30804394_USP7 Regulates Cytokinesis through FBXO38 and KIF20B. 31269066_Study found that a COPD associated alternative splicing variant of previously unknown function may contribute to the inclusion of a new exon in the FBXO38 gene. | ENSMUSG00000042211 | Fbxo38 | 844.756036 | 1.3421366 | 0.424531470 | 0.11327721 | 1.388855e+01 | 1.939765e-04 | 1.216687e-02 | No | Yes | 1004.788628 | 193.438927 | 757.010480 | 145.699137 |
| ENSG00000145907 | 10146 | G3BP1 | protein_coding | Q13283 | FUNCTION: ATP- and magnesium-dependent helicase that plays an essential role in innate immunity (PubMed:30510222). Participates in the DNA-triggered cGAS/STING pathway by promoting the DNA binding and activation of CGAS. Enhances also DDX58-induced type I interferon production probably by helping DDX58 at sensing pathogenic RNA (PubMed:30804210). In addition, plays an essential role in stress granule formation (PubMed:12642610, PubMed:20180778, PubMed:23279204). Unwinds preferentially partial DNA and RNA duplexes having a 17 bp annealed portion and either a hanging 3' tail or hanging tails at both 5'- and 3'-ends (PubMed:9889278). Unwinds DNA/DNA, RNA/DNA, and RNA/RNA substrates with comparable efficiency (PubMed:9889278). Acts unidirectionally by moving in the 5' to 3' direction along the bound single-stranded DNA (PubMed:9889278). Phosphorylation-dependent sequence-specific endoribonuclease in vitro (PubMed:11604510). Cleaves exclusively between cytosine and adenine and cleaves MYC mRNA preferentially at the 3'-UTR (PubMed:11604510). {ECO:0000269|PubMed:11604510, ECO:0000269|PubMed:12642610, ECO:0000269|PubMed:20180778, ECO:0000269|PubMed:23279204, ECO:0000269|PubMed:30510222, ECO:0000269|PubMed:30804210, ECO:0000269|PubMed:9889278}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;DNA-binding;Direct protein sequencing;Endonuclease;Helicase;Host-virus interaction;Hydrolase;Immunity;Innate immunity;Isopeptide bond;Methylation;Nuclease;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transport;Ubl conjugation | This gene encodes one of the DNA-unwinding enzymes which prefers partially unwound 3'-tailed substrates and can also unwind partial RNA/DNA and RNA/RNA duplexes in an ATP-dependent fashion. This enzyme is a member of the heterogeneous nuclear RNA-binding proteins and is also an element of the Ras signal transduction pathway. It binds specifically to the Ras-GTPase-activating protein by associating with its SH3 domain. Several alternatively spliced transcript variants of this gene have been described, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Jul 2008]. | hsa:10146; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; nucleus [GO:0005634]; perikaryon [GO:0043204]; ribonucleoprotein complex [GO:1990904]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; DNA/RNA helicase activity [GO:0033677]; endonuclease activity [GO:0004519]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; defense response to virus [GO:0051607]; innate immune response [GO:0045087]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; positive regulation of stress granule assembly [GO:0062029]; positive regulation of type I interferon production [GO:0032481]; Ras protein signal transduction [GO:0007265]; stress granule assembly [GO:0034063] | 15471883_involvement of cellular protein G3BP in transcription of intermediate stage genes may regulate the transition between early and late phases of vaccinia virus replication 15602692_G3BPs are scaffolding proteins linking signal transduction to RNA metabolism (review) 16996479_Hepatitis C virus viral gene and proteins may regulate the presence of host cellular proteins in detergent resistant membrane 17210633_Caprin-1/G3BP-1 complex is likely to regulate the transport and translation of mRNAs of proteins involved with synaptic plasticity in neurons 17253181_The expressions of G3BP and OPN proteins have a close relationship with lymphoid metastasis and survival in esophageal squamous carcinoma patients. 17297477_Both G3BP1 and G3BP2 isoforms may act as negative regulators of tumor suppressor protein p53. 17696235_The expression of G3BP and RhoC protein is closely related to the lymph node metastasis and survival in esophageal squamous carcinoma (ESC) patients. G3BP and RhoC proteins can be considered as predictors of prognosis in ESC patients. 20180778_Results illustrated a role for MK-STYX in regulating the ability of G3BP1 to integrate changes in growth-factor stimulation and environmental stress with the regulation of protein synthesis. 20424128_these results strongly indicate that (-)-epigallocatechin gallate suppresses lung tumorigenesis through its binding with G3BP1 20643132_The kinetics of assembly of stress granules(SGs) in living cells demonstrated that Tudor-SN co-localizes with G3BP and is recruited to the same SGs in response to different stress stimuli. 20663914_Molecular and functional studies indicate that the interaction of G3BP1 with beta-F1 mRNA inhibits its translation at the initiation level, supporting a role for G3BP1 in the glycolytic switch that occurs in cancer. 21206022_The nuclear transport factor 2-like (NTF2-like) domain of human G3BP1 was subcloned, overexpressed in Escherichia coli and purified. 21257637_TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. 21266361_CD24 may play a role in the inhibition of cell invasion and metastasis, and that intracellular CD24 inhibits invasiveness and metastasis through its influence on the posttranscriptional regulation of BART mRNA levels via G3BP RNase activity. 21304914_interaction between IncA and G3BP1 of Hep-2 cells infected with Chlamydophila psittaci reduces c-Myc concentration 21665939_overexpression of the amino (N)-terminal region of G3BP, including the binding region for BART mRNA, dominant-negatively inhibits formation of the complex between endogenous G3BP and BART mRNA, and increases the expression of BART. 22205990_arguments against G3BP1 being a genuine RasGAP-binding partner 22703643_In this report, we demonstrate that a novel peptide GAP161 blocked the functions of G3BP and markedly suppressed HCT116 cell growth through the induction of apoptosis 22767504_These findings establish a novel function for Poly(ADP-ribose) in the formation of G3BP-induced stress granules upon genotoxic stress. 22833567_Data indicate that assembly of large RasGAP SH3-binding protein (G3BP)-induced stress granules precedes phosphorylation of eukaryotic initiation factor 2alpha (eIF2alpha). 23087212_Data show that the nsP3/G3BP interaction also blocks stress granules (SGs) induced by other stresses than virus infection. 23163895_MK-STYX inhibits stress granule formation independently of G3BP-1 phosphorylation at Ser149. 23279204_both G3BP1 and G3BP2 play a role in the formation of SGs in various human cells and thereby recovery from these cellular stresses. 24157923_Data revealed that knockdown of G3BP inhibited the migration and invasion of human lung carcinoma cells through the inhibition of Src, FAK, ERK and NF-kappaB and decreased levels of MMP-2, MMP-9 and uPA. 24321297_G3BP1 regulation of cell proliferation in breast cancer cells, may occur via a regulatory effect on PMP22 expression. 24324649_Binding motifs specificity has been determined for human G3BP1 NTF2-like domain. 24992036_G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. 24998844_These findings disclose a novel mechanism of resveratrol-induced p53 activation and resveratrol-induced apoptosis by direct targeting of G3BP1. 25229650_G3BP1 has a role in modulating stress granule assembly during HIV-1 infection 25520508_Authors show that the PXXP domain within G3BP1 is essential for the recruitment of PKR to stress granules, for eIF2alpha phosphorylation driven by PKR, and for nucleating stress granules of normal composition. 25578447_eQTLs acting across multiple tissues are significant carriers of inherited risk for CAD. FLYWCH1, PSORSIC3, and G3BP1 are novel master regulatory genes in CAD that may be suitable targets. 25653451_Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. 25658430_ICP8 binding to G3BP also inhibits SG formation, which is a novel function of HSV ICP8. 25784705_The G3BP1-Caprin1-PKR complex represents a new mode of PKR activation and is important for antiviral activity of G3BP1 and PKR during infection with mengovirus. 25800057_these findings demonstrate a critical role for YB-1 in stress granule formation through translational activation of G3BP1, and highlight novel functions for stress granules in tumor progression. 25809930_Our data define G3BP1 as a novel independent prognostic factor that is correlated with gastric cancer progression. 25847539_G3BP1 is essential for normal stress granule-processing body interactions and stress granule function. 25962958_Our findings identified a novel function of G3BP1 in the progression of breast cancer via activation of the epithelial-to-mesenchymal transition 26350772_G3BP1 granules were assembled independently of TIA-1 and had a negative impact on Dengue virus replication. 26432022_Host G3BP1 captures HIV-1 RNA transcripts and thereby restricts mRNA translation, viral protein production and virus particle formation. 27022092_G3BP mediates the condensation of stress granules by shifting between two different states that are controlled by the phosphorylation of S149 and by binding to Caprin1 or USP10. 27303792_Based on insights from the structures and existing biochemical data, the existence of an evolutionarily conserved ribonucleoprotein (RNP) complex consisting of Caprin-1, FMRP and G3BP1 is proposed. 27383630_Results show the crystal structure of the NTF2-like domain of G3BP-1 in complex with nsP3 protein revealing a poly-complex of G3BP-1 dimers interconnected through the FGDF motifs in nsP3. Although in vitro and in vivo binding studies revealed a hierarchical interaction of the two FGDF motifs with G3BP-1, viral growth curves clearly demonstrated that two intact FGDF motifs are required for efficient viral replication. 27601476_G3BP1 is differentially methylated on specific arginine residues by protein arginine methyltransferase (PRMT) 1 and PRMT5 in its RGG domain. 27920254_These data support a role for casein kinase 2 in regulation of protein synthesis by downregulating stress granule formation through G3BP1. 28011284_The data suggested that JNK-enhanced Tudor-SN phosphorylation promotes the interaction between Tudor-SN and G3BP and facilitates the efficient recruitment of Tudor-SN into stress granules under conditions of sodium arsenite-induced oxidative stress. 28523344_Activated glucocorticoid receptor induced phosphorylation of v-AKT Murine Thymoma Viral Oncogene Homologue (AKT) kinase, which in turn phosphorylated and promoted nuclear translocation of G3BP1. The nuclear G3BP1 bound to the G3BP1 consensus sequence located on primary miR-15b~16-2 and miR-23a~27a~24-2 to inhibit their maturation. 28755480_G3BP1 interacts directly with the foot-and-mouth disease virus internal ribosome entry site and negatively regulates translation. 28972166_JMJD6 is a novel Stress Granule component that interacts with G3BP1 complexes, and its expression reduces G3BP1 monomethylation and asymmetric dimethylation at three Arg residues. 29463567_Ras GTPase-activating protein SH3-domain-binding protein (G3BP1) shows alternating binding in nanocores and anomalous diffusion in the liquid phase of stress granules. 29588351_Study demonstrated that the cytosolic lncRNA P53RRA is downregulated in cancers and functions as a tumor suppressor by inhibiting cancer progression. P53RRA bound G3BP1 and cytosolic P53RRA-G3BP1 interaction displaced p53 from a G3BP1 complex, resulting in greater p53 retention in the nucleus, which led to cell-cycle arrest, apoptosis, and ferroptosis. 29626090_High G3BP1expression is associated with arteriosclerosis. 29717134_G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. 30006004_The disruption of stress granules (SGs) during the late stage of Enterovirus 71 (EV71) infection is caused by viral protease 3C-mediated cleavage of G3BP stress granule assembly factor 1 (G3BP1). Over-expression of G3BP1-SGs negatively impacts viral replication at the cytopathic effect (CPE), protein, RNA, and viral titer levels. 30510222_this paper shows that G3BP1 promotes DNA binding and activation of cyclic GMP-AMP synthase 30804210_data reveal G3BP1 as a critical component of RIG-I signaling and possibly acting as a co-sensor to promote RIG-I recognition of pathogenic RNA. 30989663_results here elucidate that G3BP1-depletion suppresses proliferation, migration, and invasion capabilities of esophageal cancer cells via the inactivation of Wnt/beta-catenin and PI3K/AKT signaling pathways 31199850_These data confirm that G3BP is a ribosomal binding protein and reveal that alphaviral nsP3 uses G3BP to concentrate viral replication complexes and to recruit the translation initiation machinery, promoting the efficient translation of viral mRNAs. 31481087_YBX1 interacts with G3BP1 to promote metastasis of RCC by activating the YBX1/G3BP1-SPP1-NF-kappaB signaling axis. 31481451_Replacement of endogenous G3BP1 with the K376Q mutant or the RNA binding-deficient F380L/F382L mutant interfered with SG formation. Significant G3BP1 K376 acetylation was detected during SG resolution, and K376-acetylated G3BP1 was seen outside SGs. G3BP1 acetylation is regulated by histone deacetylase 6 (HDAC6) and CBP/p300. 31501480_G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. 31560169_Elevated expression of G3BP1 associates with YB1 and p-AKT and predicts poor prognosis in nonsmall cell lung cancer patients after surgical resection. 31827077_G3BP1 inhibits RNA virus replication by positively regulating RIG-I-mediated cellular antiviral response. 31941779_Typical Stress Granule Proteins Interact with the 3' Untranslated Region of Enterovirus D68 To Inhibit Viral Replication. 31941782_Sensitivity of Alphaviruses to G3BP Deletion Correlates with Efficiency of Replicase Polyprotein Processing. 31956030_UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1. 32302571_G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. 32302572_RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. 32406909_G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. 32663095_Depletion of lncRNA MALAT1 inhibited sunitinib resistance through regulating miR-362-3p-mediated G3BP1 in renal cell carcinoma. 32692974_Translational Repression of G3BP in Cancer and Germ Cells Suppresses Stress Granules and Enhances Stress Tolerance. 32989225_G3BP1 interacts with YWHAZ to regulate chemoresistance and predict adjuvant chemotherapy benefit in gastric cancer. 33000280_Overexpression of G3BP1 facilitates the progression of colon cancer by activating betacatenin signaling. 33020468_G3BP1 controls the senescence-associated secretome and its impact on cancer progression. 33131924_Role of Chikungunya nsP3 in Regulating G3BP1 Activity, Stress Granule Formation and Drug Efficacy. 33199441_RNA partitioning into stress granules is based on the summation of multiple interactions. 33497611_G3BPs tether the TSC complex to lysosomes and suppress mTORC1 signaling. 33536604_G3BP1 promotes human breast cancer cell proliferation through coordinating with GSK-3beta and stabilizing beta-catenin. 33547245_Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. 33726799_LncRNA SPOCD1-AS from ovarian cancer extracellular vesicles remodels mesothelial cells to promote peritoneal metastasis via interacting with G3BP1. 33941659_G3BP1 Inhibition Alleviates Intracellular Nucleic Acid-Induced Autoimmune Responses. 33945510_Huntington's disease mice and human brain tissue exhibit increased G3BP1 granules and TDP43 mislocalization. 33987877_G3BP1 may serve as a potential biomarker of proliferation, apoptosis, and prognosis in oral squamous cell carcinoma. 34287041_Orf Virus ORF120 Protein Positively Regulates the NF-kappaB Pathway by Interacting with G3BP1. 34432484_Resveratrol and related stilbene derivatives induce stress granules with distinct clearance kinetics. 34517025_Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation. 34614161_G3BP1 binds to guanine quadruplexes in mRNAs to modulate their stabilities. 34739333_Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. 34795264_G3BP1 inhibits Cul3(SPOP) to amplify AR signaling and promote prostate cancer. 34967276_Loss of Ras GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) inhibits the progression of ovarian cancer in coordination with ubiquitin-specific protease 10 (USP10). 35075101_SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. 35085371_TDRD3 is an antiviral restriction factor that promotes IFN signaling with G3BP1. 35100495_Small nucleolar RNA SNORA71A promotes epithelial-mesenchymal transition by maintaining ROCK2 mRNA stability in breast cancer. 35176431_The roles of G3BP1 in human diseases (review). | ENSMUSG00000018583 | G3bp1 | 12247.061521 | 1.0431396 | 0.060932205 | 0.04999455 | 1.474974e+00 | 2.245626e-01 | 5.987432e-01 | No | Yes | 13656.265060 | 2920.300290 | 13049.878485 | 2790.483635 | |
| ENSG00000146648 | 1956 | EGFR | protein_coding | P00533 | FUNCTION: Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses (PubMed:2790960, PubMed:10805725, PubMed:27153536). Known ligands include EGF, TGFA/TGF-alpha, AREG, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF (PubMed:2790960, PubMed:7679104, PubMed:8144591, PubMed:9419975, PubMed:15611079, PubMed:12297049, PubMed:27153536, PubMed:20837704, PubMed:17909029). Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules (PubMed:27153536). May also activate the NF-kappa-B signaling cascade (PubMed:11116146). Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling (PubMed:11602604). Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin (PubMed:11483589). Positively regulates cell migration via interaction with CCDC88A/GIV which retains EGFR at the cell membrane following ligand stimulation, promoting EGFR signaling which triggers cell migration (PubMed:20462955). Plays a role in enhancing learning and memory performance (By similarity). {ECO:0000250|UniProtKB:Q01279, ECO:0000269|PubMed:10805725, ECO:0000269|PubMed:11116146, ECO:0000269|PubMed:11483589, ECO:0000269|PubMed:11602604, ECO:0000269|PubMed:12297049, ECO:0000269|PubMed:12297050, ECO:0000269|PubMed:12620237, ECO:0000269|PubMed:12873986, ECO:0000269|PubMed:15374980, ECO:0000269|PubMed:15590694, ECO:0000269|PubMed:15611079, ECO:0000269|PubMed:17115032, ECO:0000269|PubMed:17909029, ECO:0000269|PubMed:19560417, ECO:0000269|PubMed:20462955, ECO:0000269|PubMed:20837704, ECO:0000269|PubMed:21258366, ECO:0000269|PubMed:27153536, ECO:0000269|PubMed:2790960, ECO:0000269|PubMed:7679104, ECO:0000269|PubMed:8144591, ECO:0000269|PubMed:9419975}.; FUNCTION: Isoform 2 may act as an antagonist of EGF action.; FUNCTION: (Microbial infection) Acts as a receptor for hepatitis C virus (HCV) in hepatocytes and facilitates its cell entry. Mediates HCV entry by promoting the formation of the CD81-CLDN1 receptor complexes that are essential for HCV entry and by enhancing membrane fusion of cells expressing HCV envelope glycoproteins. {ECO:0000269|PubMed:21516087}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Developmental protein;Direct protein sequencing;Disease variant;Disulfide bond;Endoplasmic reticulum;Endosome;Glycoprotein;Golgi apparatus;Host cell receptor for virus entry;Host-virus interaction;Hydroxylation;Isopeptide bond;Kinase;Lipoprotein;Membrane;Methylation;Nucleotide-binding;Nucleus;Palmitate;Phosphoprotein;Proto-oncogene;Receptor;Reference proteome;Repeat;Secreted;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a transmembrane glycoprotein that is a member of the protein kinase superfamily. This protein is a receptor for members of the epidermal growth factor family. EGFR is a cell surface protein that binds to epidermal growth factor, thus inducing receptor dimerization and tyrosine autophosphorylation leading to cell proliferation. Mutations in this gene are associated with lung cancer. EGFR is a component of the cytokine storm which contributes to a severe form of Coronavirus Disease 2019 (COVID-19) resulting from infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). [provided by RefSeq, Jul 2020]. | hsa:1956; | basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; cell junction [GO:0030054]; cell surface [GO:0009986]; clathrin-coated endocytic vesicle membrane [GO:0030669]; cytoplasm [GO:0005737]; early endosome membrane [GO:0031901]; endoplasmic reticulum membrane [GO:0005789]; endosome [GO:0005768]; endosome membrane [GO:0010008]; extracellular space [GO:0005615]; focal adhesion [GO:0005925]; Golgi membrane [GO:0000139]; integral component of plasma membrane [GO:0005887]; intracellular vesicle [GO:0097708]; membrane [GO:0016020]; membrane raft [GO:0045121]; multivesicular body, internal vesicle lumen [GO:0097489]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; receptor complex [GO:0043235]; ruffle membrane [GO:0032587]; Shc-EGFR complex [GO:0070435]; actin filament binding [GO:0051015]; ATP binding [GO:0005524]; ATPase binding [GO:0051117]; cadherin binding [GO:0045296]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; enzyme binding [GO:0019899]; epidermal growth factor binding [GO:0048408]; epidermal growth factor-activated receptor activity [GO:0005006]; identical protein binding [GO:0042802]; kinase binding [GO:0019900]; MAP kinase kinase kinase activity [GO:0004709]; protein phosphatase binding [GO:0019903]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; transmembrane signaling receptor activity [GO:0004888]; ubiquitin protein ligase binding [GO:0031625]; virus receptor activity [GO:0001618]; activation of phospholipase A2 activity by calcium-mediated signaling [GO:0043006]; activation of phospholipase C activity [GO:0007202]; cell differentiation [GO:0030154]; cell morphogenesis [GO:0000902]; cell surface receptor signaling pathway [GO:0007166]; cell-cell adhesion [GO:0098609]; cellular response to amino acid stimulus [GO:0071230]; cellular response to cadmium ion [GO:0071276]; cellular response to epidermal growth factor stimulus [GO:0071364]; cellular response to estradiol stimulus [GO:0071392]; cellular response to reactive oxygen species [GO:0034614]; cerebral cortex cell migration [GO:0021795]; digestive tract morphogenesis [GO:0048546]; embryonic placenta development [GO:0001892]; epidermal growth factor receptor signaling pathway [GO:0007173]; eyelid development in camera-type eye [GO:0061029]; hair follicle development [GO:0001942]; learning or memory [GO:0007611]; morphogenesis of an epithelial fold [GO:0060571]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cardiocyte differentiation [GO:1905208]; negative regulation of epidermal growth factor receptor signaling pathway [GO:0042059]; negative regulation of protein catabolic process [GO:0042177]; ossification [GO:0001503]; peptidyl-tyrosine autophosphorylation [GO:0038083]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of cell growth [GO:0030307]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0045737]; positive regulation of DNA repair [GO:0045739]; positive regulation of DNA replication [GO:0045740]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of nitric oxide mediated signal transduction [GO:0010750]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of phosphorylation [GO:0042327]; positive regulation of production of miRNAs involved in gene silencing by miRNA [GO:1903800]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of protein kinase C activity [GO:1900020]; positive regulation of protein localization to plasma membrane [GO:1903078]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein autophosphorylation [GO:0046777]; protein insertion into membrane [GO:0051205]; regulation of ERK1 and ERK2 cascade [GO:0070372]; regulation of JNK cascade [GO:0046328]; regulation of nitric-oxide synthase activity [GO:0050999]; regulation of peptidyl-tyrosine phosphorylation [GO:0050730]; regulation of phosphatidylinositol 3-kinase signaling [GO:0014066]; response to UV-A [GO:0070141]; salivary gland morphogenesis [GO:0007435]; signal transduction [GO:0007165]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11467954_A truncated form of the hEGFR ectodomain comprising residues 1-501, unlike the full-length hEGFR ectodomain (residues 1-621), binds hEGF and hTGF-alpha with high affinity and is a competitive inhibitor of EGF-stimulated mitogenesis. 11504770_Observational study of gene-disease association. (HuGE Navigator) 11531336_Chemical/biological model for EGFR activation. 11533659_EGFR has been detected in the nucleus and might function as a transcription factor to activate gene transcription. 11751923_The data presented here demonstrate that, in contrast to activation by the cytokine, growth hormone (GH), the activation of STAT5b by the growth factor, epidermal growth factor (EGF), requires overexpression of the EGF receptor (EGFR). 11788593_effects of receptor-selective retinoid ligands on EGFR-associated signal transduction 11796728_Interaction of the extracellular domain of the epidermal growth factor receptor with gangliosides 11814623_this study, 11 analogs of a fragment of the B-loop of EGF-related peptides from several species were synthesized to study binding to A431 human epidermoid carcinoma using both 125I-EGF and [3'4'-3H-Tyr(22,29), Abu(20,31)]EGF(20-31)-NH(2). 11831486_CsA affects EGF-r metabolism in gingival keratinocytes resulting in an increased number of cell surface receptors 11853560_Calmodulin binds to the EGFR 11875501_data demonstrate that prostaglandin E2 transactivates EGFR and triggers mitogenic signaling in gastric epithelial and colon cancer cells as well as in rat gastric mucosa in vivo 11886870_sequestration in non-caveolar lipid rafts inhibits lipid binding 11894095_Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors 11897506_estrogen transactivates the epidermal growth factor receptor (EGFR) to MAP K signaling axis via GPR30;implications for breast cancer biology 11912208_Identification of epidermal growth factor receptor as a target of Cdc25A protein phosphatase 11916499_contig map for the EGFR region and markers positioned on its associated physical map to the analysis of 7p11.2 amplifications in a series of glioblastomas. 11935304_EGFR is often strongly expressed and is a potential therapeutic target in patients with malignant thymic tumors 11953893_Antisense epidermal growth factor receptor RNA transfection in human glioblastoma cells down-regulates telomerase activity and telomere length. 11966576_epidermal growth factor receptor activity on fertilization capacity of testicular harvested spermatozoa 11968000_results suggest a potential mechanism by which maintenance of low levels of EGFR expression and subsequent EGFR upregulation may be attributed to the loss of transcriptional repression of EGFR gene expression in hormone-dependent breast cancer cells 11983694_Src-dependent phosphorylation of the EGFR at Tyr-845 is required for EGFR transactivation and zinc-induced Ras activation 11992543_Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-kappaB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. 11994282_blocking of ubiquitination by inhibiting Src family kinases 12006662_novel mechanism of EGFR internalization that does not require ligand binding, receptor kinase activity, or ubiquitylation and does not direct the receptor into a degradative pathway 12018405_Dephosphorylation of the EGFR and the consequent suppression of EGFR signalling. Review. 12023273_second cysteine-rich region contains targeting information for caveolae/rafts 12099696_Helicobacter pylori-stimulated EGF receptor transactivation requires metalloprotease cleavage of HB-EGF 12105206_determination of decorin binding site 12127695_Expression of this molecule and its correlation with prognostic markers in patients with head and neck tumors 12133497_Detection of serum epidermal growth factor receptor in the diagnosis of proliferation of pituitary adenomas 12134064_These data demonstrate a distinct radiation response profile at the transcriptional level that is dependent on enhanced EGFR/Ras/MAPK signaling. 12134089_These results indicate that epidermal growth factor (EGF) receptors can form a ligand-independent inactive dimer and that receptor dimerization and activation are mechanistically distinct and separable events. 12135609_ErbB1 and ErbB2 employ different mechanisms of plasma membrane targeting during keratinocyte differentiation; cytoskeletal association may facilitate the coupling of activated ErbB1 and ERK. 12147707_Rac activation upon cell-cell contact formation is dependent on signaling from here 12152785_Differential EGFR patterns by interphase cytogenetics in malignant peripheral nerve sheath tumor and morphologically similar spindle cell neoplasms. 12161422_Results show that the juxtamembrane region of the epidermal growth factor receptor is necessary for accurate polarized expression of the native molecule. 12167618_The results of this study indicate that dual inhibition of focal adhesion kinase (FAK) and epidermal growth factor receptor (EGFR) signaling pathways can cooperatively enhance apoptosis in breast cancers. 12180964_Stimulation of cells with EGF rapidly leads to phosphorylation of Hrs, raising the question whether the EGF receptor tyrosine kinase phosphorylates Hrs directly. Several downstream kinases, rather than the active receptor kinase are responsible. 12181310_These results demonstrate that 1,25(OH)(2)D(3) alters EGFR membrane trafficking and down-regulates EGFR growth signaling. 12192610_EGF receptor expression was elevated in the prefrontal cortex in schizophrenic 12218189_Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. 12223352_findings suggest an increase in functional TGF-alpha and activation of the EGFr in response to IFN-gamma 12234920_Data show that human sprouty2 potentiates epidermal growth factor receptor signalling by specifically intercepting c-Cbl-mediated effects on receptor down-regulation. 12243760_Data indicate that phospholipase A2 downregulates the EGF receptor-mediated intracellular signal transduction that may be mediated by arachidonic acid and/or ceramide. 12297049_Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. 12297050_Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains 12354760_caveolin-1/EGFR association and is critical for the EGF-induced tyrosine phosphorylation of caveolin-1 that is associated with its inhibition of EGFR activation. 12381737_determination of whether G protein-coupled receptor kinase-2 can phosphorylate and desensitize epidermal growth factor receptor 12388817_Human cytomegalovirus infection inhibits epidermal growth factor (EGF) signalling by targeting EGF receptors 12397069_ligand-independent activation of the epidermal growth factor receptor is triggered by cholesterol depletion from the plasma membrane 12399475_Endocytosis is regulated by Grb2-mediated recruitment of the Rab5 GTPase-activating protein RN-tre. 12419588_Cell proliferation, nuclear ploidy, and EGFr and HER2/neu tyrosine kinase oncoproteins in infiltrating ductal breast carcinoma. 12429632_epidermal growth factor receptor may have a role in disease relapse and progression to androgen-independence in human prostate cancer 12429742_mediation of synergism with c-src by STAT5b 12435727_mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo 12444032_hydrogen peroxide increases cPLA(2) activity through its phosphorylation utilizing an epithelial growth factor/Ras/extracellular signal-regulated kinase and p38 pathway. 12446727_EGF receptor is activated in a pathway that requires ARF4 to induce phospholipase D2 12456372_Mutual interaction of EGFR with c-Src is required for many EGFR-mediated cellular functions including proliferation, migration, survival and EGFR endocytosis, as discussed in this review. 12479108_EGFR, DI, and the diploid are valuable targets for judging metastasis and recurrence of gastric cancer before and after operation. 12487410_Expression of the invasive phenotype in breast epithelial cells requires increased EGF receptor signaling, involving both PI 3-kinase and Erk 1,2 activities, which leads to enhanced secretion of MMP-9 and transcription of invasion-related genes. 12508124_Levels of this receptor are lowered in cryptorchism. 12517767_targeting in cancer, involved in apoptosis (REVIEW) 12517803_activation regulates mutation and epidermal growth factor receptor activation regulates vascular endothelial growth factor mRNA expression in human glioblastoma cells by transactivating the proximal VEGF promoter. 12527890_TPA-induced AP-1 activation requires EGFR protein, but not EGFR tyrosine kinase and EGFR autophosphorylation at tyrosine(1173), whereas both EGFR tyrosine kinase and EGFR autophosphorylation at Y(1173) play a critical role in EGF-induced AP-1 activation. 12532415_SKMG-3 cells produced high levels of EGFR protein. Signals from overexpressed EGFRs contribute to the constitutive phosphorylation of Erk, but these signals may not be required for the constitutive activation of PI3K or AKT1. 12534349_role in mediating thromboxane A2-dependent -signal-regulated kinase activation 12556561_the mechanism of attenuated ERK signaling in EGFR-overexpressing cells is a sequestration of ERKs at the cell membrane in EGFR-containing complexes. 12568494_Review. Mucin transcription in response to both gram-positive bacteria and tobacco smoke is mediated through activation of the epidermal growth factor receptor (EGFR). 12579331_Expression of EGFR seems to play an important role in metastasis, especially liver metastasis and recurrence of pancreatic cancer. 12582944_in glioblastoma multiforme patients a complex relationship exists between epidermal growth factor receptor expression and age 12586780_results demonstrate that tamoxifen resistant MCF-7 cell growth is mediated by the autocrine release and action of an epidermal growth factor receptor-specific ligand inducing preferential epidermal growth factor receptor/c-erbB2 dimerization 12603863_Inhibition of erbB receptor family members protects HaCaT keratinocytes from ultraviolet-B-induced apoptosis. This inhibition was specific for the erbB receptor family and specific for ultraviolet-B-induced apoptosis. 12604776_EGFR binds to c-src and has a role in oncogenesis 12606946_Exogenous OPN increased EGFR mRNA expression, as well as EGFR kinase activity. Inhibition of EGFR significantly impaired the cell migration response to OPN. 12607604_Comparative study in the expression of p53, EGFR, TGF-alpha, and cyclin D1 in verrucous carcinoma, verrucous hyperplasia, and squamous cell carcinoma of head and neck region. 12615082_Five autophosphorylation sites in the extra-kinase C-terminal domain of EGFR are not required for the ability of EGFR to induce morphological differentiation of PC12 cells. 12631599_Observational study of genotype prevalence. (HuGE Navigator) 12642595_data indicate that growth hormone, by activating extracellular signal related kinases, can modulate epidermal growth factor-induced epidermal growth factor receptor trafficking and signaling 12643788_Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 & ERCC1 in DU145 & LNCaP prostate carcinoma through MAPK signaling indicating a capacity of the EGFR-ERK signaling to modulate DNA repair in cancer cells. 12653106_EGFR microsatellite polymorphism is associated with autosomal dominant polycystic kidney disease. 12653106_Observational study of gene-disease association. (HuGE Navigator) 12654182_Expression of EGF and EGFR is involved in the gallbladder carcinogenesis, and is related to high activity of cell proliferation. 12657642_propose a role of Ent-1 in the trafficking of EGFR to down-regulate intestinal mitogenic signals, highlighting the mechanisms of cell growth arrest associated with enterocytic differentiation 12664617_Human colon carcinoma cells that overexpress cyclooxygenase exhibit growth stimulation and induction of this protein. 12673202_Overexpression of an introduced EGFR, under an E1A-insensitive heterologous promoter, blocked E1A induction of apoptosis in SCC cells. E1A-mediated EGFR downregulation appears to be the cause not consequence of E1A-induced apoptosis in these cells. 12681285_Data show that by using RNA interference , the expression of endogenous erbB1 can be specifically and extensively suppressed in A431 human epidermoid carcinoma cells. 12683217_The study found that over-expression of EGFR occurred more often in cases of cervical cancer (50%) compared to breast cancer cases (36%), while in breast cancer EGFR expression correlated significantly with metastasis of the lymph nodes. 12686539_EGFR/HER2 heterodimers traffic as single entities; levels of HER2 in normal cells are barely at the threshold necessary to drive efficient heterodimerization 12694196_Nitric oxide and nitric oxide donors induce EGF receptor phosphorylations, in A431 tumor cells. 12704666_deltaEGFR may contribute to glioblastoma development 12708474_EGFRvIII expressed in human tumors is phosphorylated and hence activated; sustained activation of EGFRvIII is implicated in the pathogenesis of non-small cell lung cancers 12708492_ZO-1 bound to EGFR irrespective of the phosphorylation status of EGFR. EGFR associated ZO-1 was highly tyrosine-phosphorylated only in primary colorectal cancers but was dephosphorylated in the liver-metastasized cancers. 12719950_Observational study of gene-disease association. (HuGE Navigator) 12722480_The expression of this protein was not different in the clone cell variants or the A431 parental line. 12733059_Results suggest that epidermal growth factor receptor and protein kinase C activation are involved in 12- O-tetradecanoylphorbol-13-acetate-induced cell signaling for modulation of cadherin-dependent cell-cell adhesion and cell shape in Caco-2 cells. 12734385_signaling intensity determines intracellular protein interactions, ubiquitination, and internalization 12743604_Loss of PTEN/MMAC1/TEP in tumor cells expression this protein counteracts the antitumor action of EGFR tyrosine kinase inhibitors. 12746449_carboxyl-terminal mutation of the epidermal growth factor receptor alters tyrosine kinase activity and substrate specificity 12746839_androgens promote an increase in the activity of the epidermal growth factor (EGF)-network by increasing ErbB1 levels, and this activity of is essential for androgen-induced proliferation and survival of the prostate cancer LNCaP cell line. 12754251_findings establish Cbl protein as the major endogenous ubiquitin ligase responsible for epidermal growth factor receptor degradation 12754350_EGFR gene expression is identified in recurrent glioblastoma multiforme 12757445_Seasonal allergic rhinitis subject showed significant elevation in EGFR expression, consistent with the observation of mucus hypersecretion in allergic rhinitis. 12768436_Observational study of genotype prevalence. (HuGE Navigator) 12782602_Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. 12794748_HEGFR was expressed in the subventricular zone embryologically 12795333_Epidermal growth factor receptor negatively regulates intracellular kinase activation in the absence of ligand 12814937_Characterization and expression of novel 60-kDa and 110-kDa EGFR isoforms in human placenta. 12819035_Data show that stimulation of epidermal growth factor receptors differentially regulates chemokine expression in keratinocytes. 12824187_PGE2 regulates cell migration via the EGFR, and affects cell division and neoplasm invasiveneess. 12825853_Data support that EGFR and HER-2/neu play an important role in cell cycle control in ductal carcinoma in situ. 12828935_The results suggest that activation and nuclear localization of EGFR may be needed for induction of NOS-2 in response to elevated intraocular pressure in glaucomatous optic neuropathy. 12829707_Ligand-induced EGFR degradation is preceded by proteasome-dependent EGFR de-ubiquitination. 12839682_Bcl-2 and Bax, being nuclear matrix associated proteins, are probably involved in the EGFR-cDNA induced malignant conversion of glioblastoma cells by introducing EGFR cDNA into the tumor cells. 12844146_Data show that detachment-induced expression of Bim requires a lack of beta(1)-integrin engagement, downregulation of EGF receptor (EGFR) expression and inhibition of Erk signalling. 12871937_PLSCR1, through its interaction with Shc, promotes Src kinase activation through the EGF receptor. 12879076_EGFR is a necessary component for HCMV-triggered signalling and viral entry 12897150_endosomal epidermal growth factor receptor stimulates cell growth 12900408_EGFR has a role in beta2 tyrosine phosphorylation of AP-2 by interacting at receptor 974YRAL and di-leucine motifs 12925580_Results demonstrate that mice humanised for epidermal growth factor receptor (EGFR) display tissue-specific hypomorphic phenotypes and describe a novel function for EGFR in bone development. 12930839_EGF receptor down-regulation by UVA may play an important role in the execution of the cell suicide program by attenuating its anti-apoptotic function 12939263_EGFR is an aldosterone-induced protein and is involved in the manifold (patho)biological actions of aldosterone 12953099_EGFR overexpression is frequent in NSCLC, is most prominent in SCC, and correlates with increased gene copy number per cell. High gene copy numbers per cell showed a trend toward poor prognosis. 12954170_identification of ligand-induced site-specific phosphorylation of epidermal growth factor receptor 12955084_depending on their localization, oxytocin receptors transactivate EGFR and activate ERK1/2 using different signalling intermediates. The final outcome is a different temporal pattern of EGFR and ERK1/2 phosphorylation 13679441_These results indicate that the EGFR signaling pathway is involved in urothelial regeneration. 13679857_Mitogenic effects of gastrin-releasing peptide in head and neck squamous cells are mediated by activation of EGFR. 14506149_ERBB1 is overexpressed and may play a role in high-grade diffusely infiltrative pediatric brain stem glioma 14506242_Infection of primary cells with adenoviruses carrying the relevant point mutations confirmed the crucial role of putative YXX Phi and dileucine (LL) transport motifs within Ad2 10.4-14.5 for down-regulation of Fas, TRAIL-R1, TRAIL-R2, and EGFR. 14507652_Epidermal growth factor receptor (a potential therapeutic target) and SALL2 stained most cases of synovial sarcoma; staining was significantly less common among other tested sarcomas. 14512423_ganglioside GM3 inhibits integrin-induced, ligand-independent epidermal growth factor receptor phosphorylation (cross-talk) through suppression of Src family kinase and phosphatidylinositol 3-kinase signaling 14520461_Up-regulation of EGFR is associated with renal cell carcinoma 14530278_cholesterol depletion with cyclodextrin induced an increase in both basal and EGF-stimulated EGF receptor phosphorylation, at specific tyrosine sites, that was associated with an increase in the intrinsic kinase activity of the EGF receptor kinase. 14551192_the EGF receptor is abundantly expressed in epithelioid vascular smooth muscle cells and the activation of this receptor results in cell cycle arrest through activation of the mitogen-activated protein kinase pathway 14557654_lysosomal degradation by adenovirus E3 RIDalpha protein dependent on specific domains 14607699_findings suggest that the coexpression of c-erbB-2 oncogene protein, epidermal growth factor receptor, and TGF-beta1 in pancreatic ductal adenocarcinoma is related to the histopathological grades and clinical stages of tumors 14632199_heregulin/EGFR system as a possible important physiologic growth regulatory system in melanocytes in which multiple deregulations may occur during progression toward melanoma, all resulting in, or indicating, growth factor independence. 14647423_demonstration that metalloproteinase-mediated transactivation of the EGFR is a key mechanism of the cellular signalling network that promotes MAPK activation as well as tumour cell migration and invasion 14660604_transactivation of the EGFr is required for the full expression of cAMP-dependent Cl- secretory responses 14670955_data suggest that agonist-induced binding of Src kinase to the Src homology 3 binding sites in the P2Y(2) purinergic receptor facilitates Src activation and allows Src to efficiently phosphorylate the epidermal growth factor receptor 14681060_E-cadherin and epidermal growth factor receptor (EGFR) are associated in mammary epithelial cells and that E-cadherin engagement in these cells induces transient activation of EGFR, as previously seen in keratinocytes. 14688027_The Id-1-induced androgen-independent prostate cancer cell growth was correlated with up-regulation of EGF-R. 14690686_results support a structural model for oligomerization of EGF receptors in which dimers are positioned head-to-head with respect to the ligand-binding site 14701753_SHP-2/Gab1 association is critical for linking EGFR to NF-kappaB transcriptional activity via the PI3-kinase/Akt signaling axis in glioblastoma cells 14702346_sorting of endocytosed epidermal growth factor receptor into the degradation pathway requires both its kinase activity and actin-binding domain 14704150_involvement urokinase plasminogen activator secretion and cell motility in breast cancer cells 14711810_EGFR and ErbB-2 have roles in ligand-dependent apoptosis that could be a natural mechanism to protect tissues from unrestricted proliferation 14718570_the adhesion-dependent activation of EGF receptor signaling is promoted by L1-type CAMs in vitro and in vivo 14729599_the relationship between the egfr polymorphism and breast cancer risk 14734462_Overexpression of EGFR is associated with recurrent non-small cell lung cancer 14764825_PGF(2 alpha)-FP receptor may promote endometrial tumorigenesis via phospholipase C-mediated phosphorylation of EGF receptor and MAPK signaling pathways. 14960328_CaM co-immunoprecipitates with EGF-activated and non-activated receptors 14963038_EGFR and c-Src-mediated Stat-3 activation is facilitated by Pyk2 14977086_A significantly higher frequency of EGFR expression occurred in PC than in AC glottic cancer. 14977838_EGFR and COX-2 cooperate to promote cervical neoplasm progression 14978035_a novel and important role for metalloprotease activation and EGFR transmodulation in mediating the cellular response to TNF 14988406_Sustained hyaluronan depolymerization is expected to cause tissue kallikrein activation, EGF release, and EGFR signaling. 15010475_Trypsin exerts robust trophic action on colon cancer cells and underline the critical role of EGF-R transactivation. 15026342_Cell surface expression of EGFR is associated with osteosarcoma pathogenesis 15031710_whole gene amplifications of egfr are rare in invasive breast cancer and explain protein overexpression in only about 12.5% of invasive breast cancer cases 15039424_epidermal growth factor receptor and CD95 activation are triggered by Src family kinase Yes 15042583_The EGFR pathway may be a specific, signal transduction pathway that regulates reactive astrocytes to form cavernous spaces in the glial scars following CNS injury and in the compressed optic nerve in glaucomatous optic nerve neuropathy. 15054105_PPARgamma and EGFR signalling have roles in urothelial terminal differentiation 15057284_EGFR regulation by E-cadherin was associated with complex formation between EGFR and E-cadherin that depended on the extracellular domain of E-cadherin but was independent of beta-catenin binding or p120-catenin binding 15063762_Observational study of gene-disease association. (HuGE Navigator) 15082004_Intensive staining of EGF-R is associated with invasive tumours of bladder 15082764_Data show that epidermal growth factor receptor signaling results in phosphorylation of CUG-BP1, and leads to increased binding of CUG-BP1 to CCAAT/enhancer binding protein beta (C/EBP beta) mRNA and elevated expression of the C/EBPbeta LIP isoform. 15100232_misfolding of the LDLR epidermal growth factor-AB pair results from low density lipoprotein receptor familial hypercholesterolemia mutations 15118073_A subgroup of patients with non-small-cell lung cancer have specific mutations in the EGFR gene, which correlate with clinical responsiveness to the tyrosine kinase inhibitor gefitinib. 15118125_somatic mutations of EGFR were found in 15 of 58 non-small cell lung cancer tumors from Japan and 1 of 61 from the United States; EGFR mutations may predict sensitivity to gefitinib 15123705_Results provide an explanation for cell surface receptor cross-talk involving the Met receptor and link G protein-coupled receptors and the epidermal growth factor receptor to the oncogenic potential of Met signaling in human carcinoma cells. 15134305_P. 445:'The EGF receptor (EGFR), a glycoprotein of 170 kDA, has also been intensily studied and finally located on chromosome 17, region p13l13-q22.' 15143334_Overexpression of EGFR was observed in only a small fraction of colorectal carcinomas, but were frequently accompanied by gene amplification. 15166244_the epidermal growth factor receptor has a role in activating ERK with extracellular oxidation by taurine chloramine 15166495_Endothelial cell vessel assembly requires EGFR signaling transduction pathways. 15182358_Data demonstrate that at least four different sets of endogenously expressed gangliosides, including GD3, did not have a significant effect on epidermal growth factor receptor distribution in the plasma membrane. 15185528_Statistically significant relation existed between the ovarian cancer metastatic potential and EGFR expression level. 15192046_The overall time-dependent activation of EGFR autophosphorylation was identical in cells treated with 1 nm BTC or 1.5 nm EGF. 15205458_EGFR inhibition promotes desmosome assembly in oral squamous cell carcinoma cells, resulting in increased cell-cell adhesion 15211117_The cytoplasmic overexpression of EGFr plays a significant role in the progression of pancreatic ductal adenocarcinoma, especially in the invasion and acquisition of aggressive clinical behavior. 15213840_EGFR, PYK2, Yes, and SHP-2 are involved in transduction of the TF/FVIIa signal possibly via transactivation of the EGF receptor. 15215236_epidermal growth factor receptor is dephosphorylated at endomembranes after ligand-mediated endocytosis 15219825_transactivation through CCR3 is a critical pathway that elicits mitogen-activated protein kinase activation and cytokine production in bronchial epithelial cells 15219850_trans-activation by calcium-sensing receptor 15221011_The tumor-specific mutation of epidermal growth factor receptor promotes cells survival and dimerization with the wild-type EGFR. 15226397_EGF receptor traffic is disrupted by farnesyltransferase inhibitors through modulation of the RhoB GTPase 15233293_c-erbB-2 and EGF-R are overexpressed in breast neoplasms and have an inverse association with Estrogen Receptor expression 15245434_Human leukocyte elastase induces keratinocyte proliferation by proteolytic activation of an EGFR signaling cascade involving TGF-alpha. 15248827_betacellulin may play a role as a local growth factor in promoting the differentiated villous trophoblastic function via ErbB-1 in early placentas and in contributing to placental growth through EVT cell function via ErbB-4 in term placentas. 15253134_The epidermal growth factor receptor (EGFR) is one of signalling pathways activated during premalignant proliferative changes in the airway epithelium. 15253384_an interrelationship is now known to exist between the IGF and EGF receptors [review] 15254267_amphiregulin- and ErbB1-dependent mechanism by which autocrine ERK activation is maintained in normal keratinocytes 15254682_EGFR, c-erbB-2, VEGF and MMP-2 and MMP-9 play an important role in tumor growth, invasion and metastasis in squamous cell carcinoma of the head and neck 15269346_In each of these gliomas, the founding molecule was generated by a simple event that circularizes a chromosome fragment overlapping the epidermal growth factor receptor gene. 15271882_novel mechanism by which IGF-I induces ERK activation in a manner that is dependent on the basal level of EGFR-TK activity, but is independent of receptor transactivation. 15282549_Upregulation of leucine-rich repeats and immunoglobulin-like domains 1 is followed by enhanced ubiquitylation and degradation of EGFR 15284455_mutant EGFRs selectively transduce survival signals on which nonsmall cell lung cancers become dependent 15298855_Carbon black causes oxidative stress-mediated proliferation of airway epithelium, involving activation of EGF-R. 15300588_IL-1beta-dependent prolonged EGFR transactivation involves multiple pathways, including an IL-8-dependent pathway. 15302576_EGFR signaling involves reinforcing altered gene expression of uPAR,thus further inducing cell motility. 15335267_In patients with breast cancer, most CNS metastatic tumor deposits showed expression for either EGFR or HER-2/neu, and less often for both. 15337524_Syk acts a negative control element of EGFR signalling. 15337756_Data show that activation of ADAM-17 results in discrete cellular responses, while G protein-coupled receptor agonists promote activation of the Ras/MAPK pathway and cell proliferation via the epidermal growth factor receptor. 15342520_The timing of lethality caused by homozygosity for a null allele of the epidermal growth factor receptor in mice is strongly dependent on genetic background. 15358139_Data report that antagonism of the type 1 insulin-like growth factor receptor in combination with inhibitors of the epidermal growth factor receptor synergistically sensitizes human malignant glioma cells to CD95L-induced apoptosis. 15364923_Results describe the role of epidermal growth factor receptor regulation in antiapoptosis, cell migration, and cell proliferation. 15366372_Egfr Wa5 is a novel ENU-induced antimorphic allele caused by a kinase-dead receptor acting as a dominant negative 15383630_data explain how thrombin exerts robust trophic action on colon cancer cells and underline the critical role of EGFR transactivation 15389641_GqPCR-induced FAK activation is mediated by via a pathway involving transactivation of the EGFr and alterations in the actin cytoskeleton. 15447984_serum epidermal growth factor receptor and HER2 have roles in response of advanced non-small cell lung cancer to chemotherapy 15469987_a novel regulatory role for Galphas in EGF receptor degradation and provide mechanistic insights into the function of Galphas in endocytic sorting 15469991_NHERF stabilizes EGFR at the cell surface and slows the rate of endocytosis without affecting recycling 15485674_EGF signalling amplification is induced by dynamic clustering of EGFR 15494003_analysis of conformations of the epidermal growth factor receptor and antibody binding [review] 15519654_cholangiocarcinoma cells exhibit sustained EGFR activation due to defective receptor internalization 15522239_Data demonstrate that the alpha-hemolysin elevates the activity of receptor-like protein tyrosine phosphatase sigma (rPTPsigma). 15524283_regulation of LMP1 on the nuclear translocation of EGFR is critical for the process of nasopharyngeal carcinoma 15540509_For genotype of EGFR gene Bsr I polymorphism, there was statistically significant differences between systemic lupus erythematosus and controls. In addition, there was significant association between the two groups in allelic frequency of the T allele. 15540509_Observational study of gene-disease association. (HuGE Navigator) 15541730_Our results suggest a novel role for the juxtamembrane domain (JM) of EGFR in mediating intracellular dimerization and thus receptor kinase activation and function. 15542601_role for EGFR activity in the lifespan and inflammatory potential of RSV-infected epithelial cells 15545271_protein kinase B/Akt phosphorylation is stimulated by mechanical stretch in epidermal cells via angiotensin II type 1 receptor and epidermal growth factor receptor 15556605_S1P transactivates c-Met and EGFR in gastric cancer cells. 15556944_Gene 33 is a physiologi | ENSMUSG00000020122 | Egfr | 463.806457 | 0.8754245 | -0.191945344 | 0.14939304 | 1.685914e+00 | 1.941404e-01 | 5.634622e-01 | No | Yes | 444.066402 | 56.762734 | 509.012355 | 64.939414 | |
| ENSG00000147082 | 85417 | CCNB3 | protein_coding | Q8WWL7 | FUNCTION: Cyclins are positive regulatory subunits of the cyclin-dependent kinases (CDKs), and thereby play an essential role in the control of the cell cycle, notably via their destruction during cell division. Its tissue specificity suggest that it may be required during early meiotic prophase I. {ECO:0000269|PubMed:12185076}. | Alternative splicing;Cell cycle;Cell division;Cyclin;Meiosis;Nucleus;Reference proteome;Ubl conjugation | The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance through the cell cycle. Cyclins function as positive regulators of cyclin-dependent kinases (CDKs), and thereby play an essential role in the control of the cell cycle. Different cyclins exhibit distinct expression and degradation patterns, which contribute to the temporal coordination of each mitotic event. Studies of similar genes in chicken and drosophila suggest that this cyclin may associate with CDC2 and CDK2 kinases, and may be required for proper spindle reorganization and restoration of the interphase nucleus. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Oct 2011]. | hsa:85417; | centrosome [GO:0005813]; cyclin-dependent protein kinase holoenzyme complex [GO:0000307]; cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; cyclin-dependent protein serine/threonine kinase regulator activity [GO:0016538]; protein kinase binding [GO:0019901]; cell division [GO:0051301]; meiotic cell cycle [GO:0051321]; mitotic cell cycle phase transition [GO:0044772]; regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0000079]; regulation of G2/M transition of mitotic cell cycle [GO:0010389] | 12185076_cloning of a full length cDNA; is expressed in developing male germ cells and is restricted to testis in mature tissues 21404461_Increased expression of cyclin B3 maybe an important marker for identifying endoderm cells differentiated from human embryonic stem cells. 22387997_A new fusion was observed between BCOR (encoding the BCL6 co-repressor) and CCNB3 (encoding the testis-specific cyclin B3) on the X chromosome. 24805859_Study provides a detailed description of the histologic spectrum, immunohistochemical features, and clinical characteristic of BCOR-CCNB3 sarcoma justifying distinction from Ewing sarcoma with its typical EWS/FUS-ETS translocations. 25360585_This study adds to recent reports on the clinicopathologic spectrum of BCOR-CCNB3 fusion-positive sarcomas, a newly emerging entity within the undifferentiated unclassified sarcoma category 26037154_CCNB3 immunostaining was useful for differentiating BCOR-CCNB3 from a group of 'Ewing-like sarcomas' and may contribute to the evaluation of treatment strategies for bone sarcomas. 28817404_Case Reports: renal sarcomas with BCOR-CCNB3 gene fusion showing histological overlap with BCOR-related clear cell sarcoma of the kidney. 28877060_Immunohistochemistry for either CCNB3 or BCOR is not completely sensitive and specific in undifferentiated sarcoma with BCOR-CCNB3 fusion. 30064235_Detection of fusion transcripts BCOR-CCNB3 in the bone marrow suggests that undifferentiated sarcoma patients with positive findings are at high risk of tumor progression. 31876361_report two patients with clear cell sarcoma of the kidney showing BCOR-CCNB3 (where CCNB3 is cyclin B3) fusion 32840647_Imaging features and clinical course of undifferentiated round cell sarcomas with CIC-DUX4 and BCOR-CCNB3 translocations. 32938693_Biallelic variant in cyclin B3 is associated with failure of maternal meiosis II and recurrent digynic triploidy. | ENSMUSG00000051592 | Ccnb3 | 59.822918 | 1.5417060 | 0.624527670 | 0.35860387 | 3.050756e+00 | 8.069959e-02 | No | Yes | 83.481311 | 24.384275 | 53.110462 | 15.522841 | ||
| ENSG00000147180 | 7552 | ZNF711 | protein_coding | Q9Y462 | FUNCTION: Transcription regulator required for brain development. Probably acts as a transcription factor that binds to the promoter of target genes and recruits PHF8 histone demethylase, leading to activate expression of genes involved in neuron development, such as KDM5C. {ECO:0000269|PubMed:20346720}. | Alternative splicing;DNA-binding;Isopeptide bond;Mental retardation;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a zinc finger protein of unknown function. It bears similarity to a zinc finger protein which acts as a transcriptional activator. This gene lies in a region of the X chromosome which has been associated with cognitive disability. [provided by RefSeq, Jul 2008]. | hsa:7552; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of gene expression [GO:0010468]; regulation of transcription by RNA polymerase II [GO:0006357] | 20346720_A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation is reported. 21384559_A total of six novel and 11 known single nucleotide polymorphisms were identified. Further studies are warranted to analyze the candidate genes at Xq11.1-q21.33. 26188516_The KCTD5/cullin3 complex stabilizes ZNF711 transcription factor. 27993705_Two large four-generation families have been described with a total of 11 males affected with intellectual disability (ID) caused by mutations in ZNF711, all present with mild to moderate ID and poor speech accompanied by autistic features and mild facial dysmorphisms, suggesting that ZNF711mutations cause non-syndromic ID. 32406922_Characterization of the ZFX family of transcription factors that bind downstream of the start site of CpG island promoters. 34356104_Analysis of a Set of KDM5C Regulatory Genes Mutated in Neurodevelopmental Disorders Identifies Temporal Coexpression Brain Signatures. 34521054_ZNF711 down-regulation promotes CISPLATIN resistance in epithelial ovarian cancer via interacting with JHDM2A and suppressing SLC31A1 expression. 34992252_Clinical findings and a DNA methylation signature in kindreds with alterations in ZNF711. | ENSMUSG00000025529 | Zfp711 | 1110.731791 | 1.0610486 | 0.085490761 | 0.08785650 | 9.425144e-01 | 3.316320e-01 | 6.932742e-01 | No | Yes | 1114.336597 | 220.013784 | 1047.975477 | 206.920250 | |
| ENSG00000147234 | 84443 | FRMPD3 | protein_coding | Q5JV73 | Reference proteome | This gene encodes a protein that contains a PDZ (post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1) domain at the N-terminus followed by a FERM domain. The encoded protein is involved in signal transduction. The PDZ domain is thought to function in protein-protein interactions, mainly by binding to specific C-terminal peptides of other proteins. The FERM domain is found in proteins that are localized to the plasma membrane and are associated with the cytoskeleton. [provided by RefSeq, May 2017]. | hsa:84443; | cytoskeleton [GO:0005856]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; tertiary granule membrane [GO:0070821] | ENSMUSG00000042425 | Frmpd3 | 66.806839 | 1.0457525 | 0.064541471 | 0.34582626 | 3.597472e-02 | 8.495677e-01 | No | Yes | 63.637360 | 11.323495 | 61.473821 | 10.910547 | ||||
| ENSG00000147421 | 79618 | HMBOX1 | protein_coding | Q6NT76 | FUNCTION: Binds directly to 5'-TTAGGG-3' repeats in telomeric DNA (PubMed:23813958, PubMed:23685356). Associates with the telomerase complex at sites of active telomere processing and positively regulates telomere elongation (PubMed:23685356). Important for TERT binding to chromatin, indicating a role in recruitment of the telomerase complex to telomeres (By similarity). Also plays a role in the alternative lengthening of telomeres (ALT) pathway in telomerase-negative cells where it promotes formation and/or maintenance of ALT-associated promyelocytic leukemia bodies (APBs) (PubMed:23813958). Enhances formation of telomere C-circles in ALT cells, suggesting a possible role in telomere recombination (PubMed:23813958). Might also be involved in the DNA damage response at telomeres (PubMed:23813958). {ECO:0000250|UniProtKB:Q8BJA3, ECO:0000269|PubMed:23685356, ECO:0000269|PubMed:23813958}. | 3D-structure;Alternative splicing;Chromosome;Cytoplasm;DNA-binding;Homeobox;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Telomere;Transcription;Transcription regulation;Ubl conjugation | hsa:79618; | Cajal body [GO:0015030]; centrosome [GO:0005813]; chromatin [GO:0000785]; chromosome, telomeric region [GO:0000781]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; double-stranded telomeric DNA binding [GO:0003691]; identical protein binding [GO:0042802]; protein-containing complex binding [GO:0044877]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; telomeric DNA binding [GO:0042162]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of chromatin binding [GO:0035563]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomere maintenance via telomerase [GO:0032212]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of telomerase activity [GO:0051972] | 16825764_Hmbox1 is widely expressed in pancreas and the expression of this gene can also be detected in pallium, hippocampus and hypothalamus 19728927_decreased expression in hepatic carcinoma 19757162_These findings suggest a distinct role of HMBOX1b, and the control of mRNA splicing might be involved in homeobox genes regulation. 20182437_higher HMGB1(homeobox containing 1) levels in Tracheal aspirates are associated with the development of bronchopulmonary dysplasia or death in ventilated premature infants. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21706044_HMBOX1 negatively regulates the expression of NKG2D and the activation of the NKG2D/DAP10 signaling pathway in NK cells. 21839858_HMBOX1 may function as a negative regulator of IFN-gamma in NK cells. 22320217_our results revealed a novel regulatory mechanism: miR-30c-1(*) promoted NK cell cytotoxicity against hepatoma cells by targeting HMBOX1. 23685356_HOT1 supports telomerase-dependent telomere elongation. 23813958_a homeobox-containing protein known as HMBOX1 can directly bind telomeric double-stranded DNA and associate with PML nuclear bodies. 26456220_HMBOX1 regulates intracellular free zinc level by interacting with MT2A to inhibit apoptosis and promote autophagy in vascular endothelial cells. 28628186_Our data suggest a possible role of HMBOX1 in regulating radiosensitivity in cervical cancer cells. 28731165_HMBOX1 may be a potential diagnostic marker in glioma. 29709478_HMBOX1 played important role in the high-grade serous ovarian cancer through regulation of proliferation and apoptosis. 30015890_Low HMBOX1 expression is associated with liver cancer progression. 30032072_HMBOX1 in hepatocytes acts as a key immunosuppressive factor for inflammation 30257388_this study demonstrates for the first time that c-Fos/miR-18a/HMBOX1 axis plays a critical role in the progression of gliomas 30358079_TDD activates HMBOX1. 31005794_The association of HMBOX1 and CD133 with gastric neoplasm cell proliferation and migration are reported; the use of the levels of these proteins as prognostic markers are discussed. 33388421_Cancer-secreted exosomal miR-1468-5p promotes tumor immune escape via the immunosuppressive reprogramming of lymphatic vessels. | ENSMUSG00000021972 | Hmbox1 | 101.698644 | 1.2726123 | 0.347792937 | 0.28114551 | 1.566286e+00 | 2.107472e-01 | No | Yes | 96.946630 | 16.811022 | 77.074168 | 13.554172 | |||
| ENSG00000147576 | 137872 | ADHFE1 | protein_coding | Q8IWW8 | FUNCTION: Catalyzes the cofactor-independent reversible oxidation of gamma-hydroxybutyrate (GHB) to succinic semialdehyde (SSA) coupled to reduction of 2-ketoglutarate (2-KG) to D-2-hydroxyglutarate (D-2-HG). D,L-3-hydroxyisobutyrate and L-3-hydroxybutyrate (L-3-OHB) are also substrates for HOT with 10-fold lower activities. {ECO:0000269|PubMed:16435184}. | Acetylation;Alternative splicing;Lipid metabolism;Mitochondrion;Oxidoreductase;Phosphoprotein;Reference proteome;Transit peptide | The ADHFE1 gene encodes hydroxyacid-oxoacid transhydrogenase (EC 1.1.99.24), which is responsible for the oxidation of 4-hydroxybutyrate in mammalian tissues (Kardon et al., 2006 [PubMed 16616524]).[supplied by OMIM, Mar 2008]. | hsa:137872; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; alcohol dehydrogenase (NAD+) activity [GO:0004022]; hydroxyacid-oxoacid transhydrogenase activity [GO:0047988]; metal ion binding [GO:0046872]; 2-oxoglutarate metabolic process [GO:0006103]; glutamate catabolic process via 2-oxoglutarate [GO:0006539]; lipid metabolic process [GO:0006629] | 12592711_cloned and characterized an iron-activated alcohol dehydrogenase gene, Fe-containing alcohol dehydrogenase 1 (ADHFe1) 19013439_show that the ADHFe1 gene is related to bacterial GHB dehydrogenases and has a conserved NAD-binding site 20877624_Observational study of gene-disease association. (HuGE Navigator) 23517143_ADHFE1 has an important role of differentiation in CRC, as well as normal colorectal mucosa and embryonic developmental processes. 24886599_These results demonstrate that the promoter hypermethylation of ADHFE1 is frequently present in CRC and alcohol induces methylation-mediated down expression of ADHFE1 and proliferation of CRC cells. 29202474_data support the hypothesis that ADHFE1 and MYC signaling contribute to D-2HG accumulation in breast tumors and show that D-2HG is an oncogenic metabolite and potential driver of disease progression. 32317010_Genome-wide DNA methylation profiles of low- and high-grade adenoma reveals potential biomarkers for early detection of colorectal carcinoma. | ENSMUSG00000025911 | Adhfe1 | 18.435250 | 1.8126229 | 0.858078806 | 0.64889473 | 1.797152e+00 | 1.800571e-01 | No | Yes | 23.333227 | 8.082147 | 13.189882 | 4.638186 | ||
| ENSG00000147854 | 115426 | UHRF2 | protein_coding | Q96PU4 | FUNCTION: E3 ubiquitin ligase that plays important roles in DNA methylation, histone modifications, cell cycle and DNA repair (PubMed:15178429, PubMed:29506131, PubMed:27743347, PubMed:23404503). Acts as a specific reader for 5-hydroxymethylcytosine (5hmC) and thereby recruits various substrates to these sites to ubiquitinate them (PubMed:27129234, PubMed:24813944). This activity also allows the maintenance of 5mC levels at specific genomic loci and regulates neuron-related gene expression (By similarity). Participates in cell cycle regulation by ubiquitinating cyclins CCND1 and CCNE1 and thereby inducing G1 arrest (PubMed:15178429, PubMed:15361834, PubMed:21952639). Ubiquitinates also PCNP leading to its degradation by the proteasome (PubMed:14741369, PubMed:12176013). Plays an active role in DNA damage repair by ubiquitinating p21/CDKN1A leading to its proteosomal degradation (PubMed:29923055). Promotes also DNA repair by acting as an interstrand cross-links (ICLs) sensor. Mechanistically, cooperates with UHRF1 to ensure recruitment of FANCD2 to ICLs, leading to FANCD2 monoubiquitination and subsequent activation (PubMed:30335751). Contributes to UV-induced DNA damage response by physically interacting with ATR in response to irradiation, thereby promoting ATR activation (PubMed:33848395). {ECO:0000250|UniProtKB:Q7TMI3, ECO:0000269|PubMed:12176013, ECO:0000269|PubMed:14741369, ECO:0000269|PubMed:15178429, ECO:0000269|PubMed:15361834, ECO:0000269|PubMed:21952639, ECO:0000269|PubMed:23404503, ECO:0000269|PubMed:24813944, ECO:0000269|PubMed:27129234, ECO:0000269|PubMed:27743347, ECO:0000269|PubMed:29506131, ECO:0000269|PubMed:29923055, ECO:0000269|PubMed:30335751, ECO:0000269|PubMed:33848395}. | 3D-structure;Alternative splicing;Cell cycle;Chromosome;DNA-binding;Disulfide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a nuclear protein which is involved in cell-cycle regulation. The encoded protein is a ubiquitin-ligase capable of ubiquinating PCNP (PEST-containing nuclear protein), and together they may play a role in tumorigenesis. The encoded protein contains an NIRF_N domain, a PHD finger, a set- and ring-associated (SRA) domain, and a RING finger domain and several of these domains have been shown to be essential for the regulation of cell proliferation. This protein may also have a role in intranuclear degradation of polyglutamine aggregates. Alternative splicing results in multiple transcript variants some of which are non-protein coding. [provided by RefSeq, Feb 2012]. | hsa:115426; | heterochromatin [GO:0000792]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; DNA binding [GO:0003677]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; SUMO transferase activity [GO:0019789]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; cell cycle [GO:0007049]; cell differentiation [GO:0030154]; maintenance of DNA methylation [GO:0010216]; protein autoubiquitination [GO:0051865]; protein ubiquitination [GO:0016567]; regulation of cell cycle [GO:0051726] | 12176013_NIRF is involved in cell-cycle regulation 14741369_NIRF has ubiquitination activity, the hallmark of a ubiquitin ligase. PCNP was readily ubiquitinated in 293 and COS-7 cells, and NIRF ubiquitinated PCNP in vitro as well as in vivo. 14741369_NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP (PEST-containing nuclear protein). 15178429_Overexpression of NIRF induces cell cycle arrest at G1 phase. NIRF binds to the cyclin E-Cdk2 complex in vivo, and is phosphorylated by Cdk2 in vitro. 15178429_may participate in the G1/S transition regulation 19218238_UHRF-2 is an essential molecule for nuclear pQ degradation as a component of nuclear PQC machinery in mammalian cells 19818775_Growth-inhibitory effect of let-7a on the A549 cells in vitro and in vivo may be explained in part by le-7a-induced suppression of NIRF and elevation of p21(WAF1). 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21598301_the cooperative interplay of Uhrf2 domains may contribute to a tighter epigenetic control of gene expression in differentiated cells 21952639_NIRF is immediately adjacent to the single nucleotide polymorphism rs719725, which is reportedly associated with the risk of colorectal cancer. 22018986_NIRF is frequently upregulated in colorectal cancer and its oncogenicity can be suppressed by let-7a microRNA. 22072112_In the present study, ubiquitin ligase, NIRF, binds to hepatitis B virus core protein and leads to the proteasome-mediated degradation of hepatitis B virus core protein in vivo. 22673569_NIRF may contribute to the coupling between the cell cycle network and the epigenetic landscape 23244124_Low UHRF2 mRNA expression is associated with malignant glioma. 23404503_UHRF2, a ubiquitin E3 ligase, acts as a small ubiquitin-like modifier E3 ligase for zinc finger protein 131 23537643_UHRF2, the homolog of UHRF1, interacts with N-methylpurine DNA glycosylase (MPG) in cancer cells. 23833190_UHRF2 is a transcriptional target of E2F, that it directly interacts with E2F1. 24573556_UHRF2 may contribute to the progression of colon carcinogenesis and function as a novel prognostic indicator after curative operation 24813944_A hydrogen bond between the hydroxyl group of 5hmC and UHRF2-SRA is critical for their preferential binding. 27114453_Results show that UHRF2 down-regulates a number of epithelial-mesenchymal transition markers and up-regulates the expression of transcription factors. The study also suggests that UHRF2 play a role in tumor metastasis. 27129234_study suggests that ZNF618 is a key protein that regulates UHRF2 function as a specific 5hmC reader in vivo. 27738314_UHRF2 expression is reduced in some leukemia cell lines, this correlates with promoter hypermethylation, and similar UHRF2 methylation profiles are seen in primary human leukemia samples. Thus, UHRF2 and 5hmC are widely present in differentiated human tissues, and UHRF2 protein is poorly expressed or mislocalized in diverse human cancers. 27743347_TIP60 acted downstream of UHRF2 to regulate H3K9ac and H3K14ac expression. 28004105_The overexpression of UHRF2 increased the expression of H3K9ac in L02 normal cells (P<0.01), but decreased the expression of histone H3 lysine 9 acetylation in HepG2 cancer cells. 28951215_Here, we report the complex structure of PCNA and the peptide ((784)NEILQTLLDLFFPGYSK(800)) derived from UHRF2 that contains a PIP box. Structural analysis combined with mutagenesis experiments provide the molecular basis for the recognition of UHRF2 by PCNA via PIP-box. 29506131_Uncoupling of UHRF2 from the DNA methylation maintenance program is linked to differences in the molecular readout of chromatin signatures that connect UHRF1 to ubiquitination of histone H3. 29909415_UHRF2 may be a negative regulator of EMT and a novel prognostic biomarker for ESCC. 29923055_Immunoprecipitation and immunofluorescence staining reveled that UHRF2 combined with p21 in the nucleus. In addition, UHRF2 degraded p21 through ubiquitination and shortened the half-life of p21. 32372448_UHRF2 promotes intestinal tumorigenesis through stabilization of TCF4 mediated Wnt/beta-catenin signaling. 33848395_UV-induced activation of ATR is mediated by UHRF2. 33876395_HBx promotes hepatocarcinogenesis by enhancing phosphorylation and blocking ubiquitinylation of UHRF2. 34111398_UHRF2 commissions the completion of DNA demethylation through allosteric activation by 5hmC and K33-linked ubiquitination of XRCC1. 34162562_IGF2BP1/UHRF2 Axis Mediated by miR-98-5p to Promote the Proliferation of and Inhibit the Apoptosis of Esophageal Squamous Cell Carcinoma. 34400880_UHRF2 promotes Hepatocellular Carcinoma Progression by Upregulating ErbB3/Ras/Raf Signaling Pathway. 34826027_MiR-196a promotes the proliferation and migration of esophageal cancer via the UHRF2/TET2 axis. | ENSMUSG00000024817 | Uhrf2 | 1131.677898 | 1.2006345 | 0.263796985 | 0.08970780 | 8.605631e+00 | 3.351253e-03 | 6.782262e-02 | No | Yes | 1027.657955 | 170.040192 | 859.046307 | 142.202617 |
| ENSG00000148019 | 84131 | CEP78 | protein_coding | Q5JTW2 | FUNCTION: May be required for efficient PLK4 centrosomal localization and PLK4-induced overduplication of centrioles (PubMed:27246242). May play a role in cilium biogenesis (PubMed:27588451). {ECO:0000269|PubMed:27246242, ECO:0000269|PubMed:27588451}. | Alternative splicing;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Coiled coil;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Deafness;Phosphoprotein;Reference proteome | This gene encodes a centrosomal protein that is both required for the regulation of centrosome-related events during the cell cycle, and required for ciliogenesis. The encoded protein has an N-terminal leucine-rich repeat (LRR) domain with six consecutive LRR repeats, and a C-terminal coiled-coil domain. It interacts with the N-terminal catalytic domain of polo-like kinase 4 (PLK4) and colocalizes with PLK4 to the distal end of the centriole. Naturally occurring mutations in this gene cause defects in primary cilia that result in retinal degeneration and sensorineural hearing loss which are associated with cone-rod degeneration disease as well as Usher syndrome. Low expression of this gene is associated with poor prognosis of colorectal cancer patients. [provided by RefSeq, Mar 2017]. | hsa:84131; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytosol [GO:0005829]; cilium organization [GO:0044782] | 27246242_the interaction between Cep78 and the N-terminal catalytic domain of Plk4 is a new and important element in the centrosome overduplication process. 27357513_CEP78 functions as a tumor suppressor in colorectal cancer and low CEP78 expression leads to shorter survival in colorectal cancer patients. 27588451_data strongly suggest that mutations in CEP78 cause a previously undescribed clinical entity of a ciliary nature characterized by blindness and deafness but clearly distinct from Usher syndrome, a condition for which visual impairment is due to retinitis pigmentosa 27588452_truncating mutations in CEP78 result in a phenotype involving both the visual and auditory systems but different from typical Usher syndrome 27627988_Our results provide evidence that CEP78 is a novel disease-causing gene for Usher syndrome, demonstrating an additional link between ciliopathy and Usher protein network in photoreceptor cells and inner ear hair cells. 28242748_we identify Cep78 as a new player that regulates centrosome homeostasis by inhibiting the final step of the enzymatic reaction catalyzed by EDD-DYRK2-DDB1(Vpr)(BP). 30884127_Low CEP78 expression is associated with differentiated thyroid carcinoma. 31999394_Functional characterization of the first missense variant in CEP78, a founder allele associated with cone-rod dystrophy, hearing loss, and reduced male fertility. 33119552_Centrosome Protein 78 Is Overexpressed in Muscle-Invasive Bladder Cancer and Is Associated with Tumor Molecular Subtypes and Mutation Signatures. 34223797_Expanding the clinical phenotype in patients with disease causing variants associated with atypical Usher syndrome. 34259627_CEP78 functions downstream of CEP350 to control biogenesis of primary cilia by negatively regulating CP110 levels. | ENSMUSG00000041491 | Cep78 | 2411.553672 | 1.0552666 | 0.077607520 | 0.07469268 | 1.092742e+00 | 2.958645e-01 | 6.639497e-01 | No | Yes | 2258.588851 | 356.074907 | 2120.543585 | 334.337241 | |
| ENSG00000148411 | 138151 | NACC2 | protein_coding | Q96BF6 | FUNCTION: Functions as a transcriptional repressor through its association with the NuRD complex. Recruits the NuRD complex to the promoter of MDM2, leading to the repression of MDM2 transcription and subsequent stability of p53/TP53. {ECO:0000269|PubMed:22926524}. | Isopeptide bond;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:138151; | chromatin [GO:0000785]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; histone deacetylase binding [GO:0042826]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter [GO:1900477]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage [GO:1902231]; protein homooligomerization [GO:0051260]; protein-containing complex localization [GO:0031503]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22926524_RBB is a novel transcriptional repressor and an important regulator of p53 pathway. | ENSMUSG00000026932 | Nacc2 | 847.487747 | 0.9256346 | -0.111485371 | 0.09713099 | 1.323896e+00 | 2.498940e-01 | 6.229393e-01 | No | Yes | 785.502588 | 99.151515 | 833.546391 | 105.177186 | ||
| ENSG00000149054 | 7762 | ZNF215 | protein_coding | Q9UL58 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene is imprinted in a tissue-specific manner with preferential expression in the testis, and encodes a zinc finger protein that belongs to a family of zinc finger transcription factors. The encoded protein contains an N-terminal SRE-ZBP, Ctfin51, AW-1, and Number 18 (SCAN) domain, a kruppel-associated box A (KRABA) domain, and four C-terminal zinc finger domains. This gene is located within one of three regions on chromosome 11p15 associated with Beckwith-Wiedemann syndrome, called Beckwith-Wiedemann syndrome chromosome region-2 (BWSCR2), and is thought to play a role in the etiology of this disease. [provided by RefSeq, Aug 2017]. | hsa:7762; | nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 34091126_Loss of ZNF215 imprinting is associated with poor five-year survival in patients with cytogenetically abnormal-acute myeloid leukemia. | 363.934500 | 0.9497187 | -0.074427763 | 0.14942389 | 2.447625e-01 | 6.207874e-01 | No | Yes | 345.480410 | 60.222926 | 367.291335 | 63.958667 | ||||
| ENSG00000149289 | 85463 | ZC3H12C | protein_coding | Q9C0D7 | FUNCTION: May function as RNase and regulate the levels of target RNA species. {ECO:0000305}. | Alternative splicing;Endonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | hsa:85463; | cytoplasmic ribonucleoprotein granule [GO:0036464]; nucleus [GO:0005634]; endoribonuclease activity [GO:0004521]; metal ion binding [GO:0046872]; mRNA binding [GO:0003729]; RNA phosphodiester bond hydrolysis, endonucleolytic [GO:0090502] | 18178554_MCPIP1, 2, 3, and 4, encoded by four genes, Zc3h12a, Zc3h12b, Zc3h12c, and Zc3h12d, respectively, regulates macrophage activation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23360436_Thus Zc3h12c is an endogenous inhibitor of TNFalpha-induced inflammatory signalling in HUVECs and might be a therapeutic target in vascular inflammatory diseases. 25391383_The Gene-based analyses revealed four significant associations in the WT1, ZC3H12C, DLGAP2, and GPR1 genes at p < 0.05. in this study. 34215755_The RNase MCPIP3 promotes skin inflammation by orchestrating myeloid cytokine response. | ENSMUSG00000035164 | Zc3h12c | 368.012583 | 0.6467437 | -0.628733919 | 0.14107873 | 1.977101e+01 | 8.729668e-06 | No | Yes | 325.069815 | 58.968392 | 500.592377 | 90.648898 | |||
| ENSG00000149308 | 4863 | NPAT | protein_coding | Q14207 | FUNCTION: Required for progression through the G1 and S phases of the cell cycle and for S phase entry. Activates transcription of the histone H2A, histone H2B, histone H3 and histone H4 genes in conjunction with MIZF. Also positively regulates the ATM, MIZF and PRKDC promoters. Transcriptional activation may be accomplished at least in part by the recruitment of the NuA4 histone acetyltransferase (HAT) complex to target gene promoters. {ECO:0000269|PubMed:10995386, ECO:0000269|PubMed:10995387, ECO:0000269|PubMed:12665581, ECO:0000269|PubMed:12724424, ECO:0000269|PubMed:14585971, ECO:0000269|PubMed:14612403, ECO:0000269|PubMed:15555599, ECO:0000269|PubMed:15988025, ECO:0000269|PubMed:16131487, ECO:0000269|PubMed:17163457, ECO:0000269|PubMed:17826007, ECO:0000269|PubMed:17967892, ECO:0000269|PubMed:17974976, ECO:0000269|PubMed:9472014}. | Acetylation;Activator;Cell cycle;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | hsa:4863; | Cajal body [GO:0015030]; cytoplasm [GO:0005737]; Gemini of coiled bodies [GO:0097504]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein C-terminus binding [GO:0008022]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; transcription corepressor activity [GO:0003714]; in utero embryonic development [GO:0001701]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of gene expression [GO:0010468]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083] | 12473189_Results characterize the functional signals required for NPAT localization to the cell nucleus. 12665581_NPAT links E2F to the activation of S-phase-specific histone gene transcription. 12724424_the ability of p220 to promote S phase is independent of its ability to promote histone H4 transcription and p220 may link cyclin E/Cdk2 to multiple independent downstream functions 12887926_OCA-S, the multicomponent Oct-1 coactivator, intercts with NPAT. 14612403_p220 is an essential downstream component of the cyclin E/Cdk2 signaling pathway and functions to coordinate multiple elements of the G1/S transition. 17449237_NPAT, together with CUL5 and PPP2R1B, is implicated in the deregulation of the cell-cycle and apoptosis regulators and in the pathogenesis of B-CLL. 17967892_NPAT recruits the TRRAP-Tip60 complex to histone gene promoters to coordinate the transcriptional activation of multiple histone genes during the G(1)/S-phase transition 17974976_HiNF-P/P220NPAT regulates expression of nonhistone targets that influence competency for cell cycle progression. 18677100_Only the number of FLASH/NPAT histone gene locus bodies correlates with ploidy and only these organelles appear to be regulated during the cell cycle. 19170105_Results suggest that cyclin-dependent kinase inhibitors selectively control stimulation of the histone H4 gene promoter by the p220(NPAT)/HiNF-P complex. 19277982_The subnuclear organization of histone gene regulatory proteins and 3' end processing factors (NPAT/LSM10) of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19890848_Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20190802_NPAT is essential for histone mRNA 3' end processing and recruits CDK9 to replication-dependent histone genes. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21050672_Genetic variants of NPAT-ATM and AURKA are associated with an individual early adverse reaction in the gastrointestinal tract of patients with cervical cancer treated with pelvic radiation therapy 21050672_Observational study of gene-disease association. (HuGE Navigator) 21562039_NPAT is thus far the first gene implicated in nodular lymphocyte predominant Hodgkin lymphoma predisposition. 25339177_The conserved C-terminal domain shared by FLASH, YARP, and Mute recognizes the C-terminal sequence of NPAT orthologues, thus acting as a signal targeting proteins to histone locus bodies. 26429916_Cpn10 has a role in the spatial regulation of NPAT signaling 32722282_Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. 34197485_The genetic association of the transcription factor NPAT with glycemic response to metformin involves regulation of fuel selection. | ENSMUSG00000033054 | Npat | 1206.009372 | 1.0855972 | 0.118488863 | 0.08998608 | 1.742302e+00 | 1.868475e-01 | 5.552868e-01 | No | Yes | 1154.713542 | 248.125474 | 1053.027763 | 226.330181 | ||
| ENSG00000149761 | 84304 | NUDT22 | protein_coding | Q9BRQ3 | FUNCTION: Hydrolyzes UDP-glucose to glucose 1-phosphate and UMP and UDP-galactose to galactose 1-phosphate and UMP. Preferred substrate is UDP-glucose. {ECO:0000269|PubMed:29413322}. | 3D-structure;Alternative splicing;Hydrolase;Magnesium;Metal-binding;Reference proteome | hsa:84304; | nucleoplasm [GO:0005654]; GDP-mannose hydrolase activity [GO:0052751]; metal ion binding [GO:0046872]; UDP-sugar diphosphatase activity [GO:0008768] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000037349 | Nudt22 | 335.154871 | 0.9557617 | -0.065277095 | 0.15294635 | 1.818767e-01 | 6.697653e-01 | No | Yes | 347.826060 | 48.535572 | 361.242292 | 50.244765 | |||
| ENSG00000150556 | 130576 | LYPD6B | protein_coding | Q8NI32 | FUNCTION: Believed to act as a modulator of nicotinic acetylcholine receptors (nAChRs) activity. In vitro acts on nAChRs in a subtype- and stoichiometry-dependent manner. Modulates specifically alpha-3(3):beta-4(2) nAChRs by enhancing the sensitivity to ACh, decreasing ACh-induced maximal current response and increasing the rate of desensitization to ACh; has no effect on alpha-7 homomeric nAChRs; modulates alpha-3(2):alpha-5:beta-4(2) nAChRs in the context of CHRNA5/alpha-5 variant Asn-398 but not its wild-type sequence. {ECO:0000269|PubMed:26586467}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Signal | hsa:130576; | anchored component of membrane [GO:0031225]; extracellular region [GO:0005576]; plasma membrane [GO:0005886]; acetylcholine receptor regulator activity [GO:0030548] | 18360792_LYPD7 was especially highly expressed in testis, lung, stomach, and prostate. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 33019770_Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors. | ENSMUSG00000026765 | Lypd6b | 51.102131 | 0.7186935 | -0.476551468 | 0.37097628 | 1.661395e+00 | 1.974150e-01 | No | Yes | 47.586385 | 8.598285 | 67.330468 | 11.779418 | |||
| ENSG00000151136 | 121551 | BTBD11 | protein_coding | A6QL63 | ANK repeat;Alternative splicing;Membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix | hsa:121551; | integral component of membrane [GO:0016021]; protein heterodimerization activity [GO:0046982]; SMAD protein signal transduction [GO:0060395] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000020042 | Btbd11 | 363.705575 | 1.0590437 | 0.082762161 | 0.17322617 | 2.321875e-01 | 6.299065e-01 | No | Yes | 329.108131 | 36.766022 | 309.078722 | 34.451618 | ||||
| ENSG00000151150 | 288 | ANK3 | protein_coding | Q12955 | FUNCTION: In skeletal muscle, required for costamere localization of DMD and betaDAG1 (By similarity). Membrane-cytoskeleton linker. May participate in the maintenance/targeting of ion channels and cell adhesion molecules at the nodes of Ranvier and axonal initial segments. Regulates KCNA1 channel activity in function of dietary Mg(2+) levels, and thereby contributes to the regulation of renal Mg(2+) reabsorption (PubMed:23903368). {ECO:0000250, ECO:0000269|PubMed:17974005}.; FUNCTION: [Isoform 5]: May be part of a Golgi-specific membrane cytoskeleton in association with beta-spectrin. {ECO:0000305|PubMed:17974005}. | 3D-structure;ANK repeat;Alternative splicing;Autism spectrum disorder;Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Golgi apparatus;Lysosome;Membrane;Mental retardation;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Repeat;Synapse | Ankyrins are a family of proteins that are believed to link the integral membrane proteins to the underlying spectrin-actin cytoskeleton and play key roles in activities such as cell motility, activation, proliferation, contact, and the maintenance of specialized membrane domains. Multiple isoforms of ankyrin with different affinities for various target proteins are expressed in a tissue-specific, developmentally regulated manner. Most ankyrins are typically composed of three structural domains: an amino-terminal domain containing multiple ankyrin repeats; a central region with a highly conserved spectrin binding domain; and a carboxy-terminal regulatory domain which is the least conserved and subject to variation. Ankyrin 3 is an immunologically distinct gene product from ankyrins 1 and 2, and was originally found at the axonal initial segment and nodes of Ranvier of neurons in the central and peripheral nervous systems. Multiple transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Feb 2011]. | hsa:288; | axon initial segment [GO:0043194]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; cell surface [GO:0009986]; costamere [GO:0043034]; cytosol [GO:0005829]; dendrite [GO:0030425]; endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; intercalated disc [GO:0014704]; lateral plasma membrane [GO:0016328]; lysosome [GO:0005764]; neuromuscular junction [GO:0031594]; neuron projection [GO:0043005]; node of Ranvier [GO:0033268]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; sarcolemma [GO:0042383]; sarcoplasmic reticulum [GO:0016529]; spectrin-associated cytoskeleton [GO:0014731]; T-tubule [GO:0030315]; Z disc [GO:0030018]; cadherin binding [GO:0045296]; cytoskeletal anchor activity [GO:0008093]; cytoskeletal protein binding [GO:0008092]; protein-macromolecule adaptor activity [GO:0030674]; spectrin binding [GO:0030507]; structural constituent of cytoskeleton [GO:0005200]; transmembrane transporter binding [GO:0044325]; axonogenesis [GO:0007409]; cellular response to magnesium ion [GO:0071286]; establishment of protein localization [GO:0045184]; Golgi to plasma membrane protein transport [GO:0043001]; magnesium ion homeostasis [GO:0010960]; maintenance of protein location in plasma membrane [GO:0072660]; membrane assembly [GO:0071709]; mitotic cytokinesis [GO:0000281]; negative regulation of delayed rectifier potassium channel activity [GO:1902260]; neuromuscular junction development [GO:0007528]; neuronal action potential [GO:0019228]; plasma membrane organization [GO:0007009]; positive regulation of cation channel activity [GO:2001259]; positive regulation of cell communication by electrical coupling [GO:0010650]; positive regulation of gene expression [GO:0010628]; positive regulation of homotypic cell-cell adhesion [GO:0034112]; positive regulation of membrane depolarization during cardiac muscle cell action potential [GO:1900827]; positive regulation of membrane potential [GO:0045838]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of sodium ion transmembrane transporter activity [GO:2000651]; positive regulation of sodium ion transport [GO:0010765]; protein localization to axon [GO:0099612]; protein localization to plasma membrane [GO:0072659]; regulation of potassium ion transport [GO:0043266]; signal transduction [GO:0007165] | 11781319_The ankyrin-B C-terminal domain determines activity of ankyrin-B/G chimeras 11950515_Ankyrin G, a key protein of membrane remodeling after axonal injury, colocalizes with voltage gated sodium channels in human neuroma. 12507143_A propensity to overexpress ankyrin G after peripheral nerve trauma may turn out to be a factor in the development of painful neuromas and neuropathic pain. 14757759_ankyrin-G plays a pleiotropic role in assembly of lateral membranes of bronchial epithelial cells 15931389_Altered expression is associated with therapy failure and death in patients with multiple types of cancer. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16775201_ankyrin-G regulates neuronal excitability not only through clustering Nav channels but also by directly modifying their channel gating. 17033732_Ankyrin G may have a role in Hutchinson-Gilford progeria syndrome 17074766_ankyrin-G and beta(2)-spectrin are functional partners in biogenesis of the lateral membrane of epithelial cells 17373700_Observational study of gene-disease association. (HuGE Navigator) 17620337_E-cadherin requires both ankyrin-G and beta-2-spectrin for its cellular localization in early embryos as well as cultured epithelial cells. 18163421_Observational study of gene-disease association. (HuGE Navigator) 18163421_Significant association with late-onset Alzheimer's disease for 4 SNPs: rs1881747 near DKK1, rs2279420 in ANK3, rs2306402 in CTNNA3, and rs5030882 in CXXC6 in 1,160 cases and 1,389 controls. 18635543_Phosphorylation and ankyrin-G binding of the C-terminal domain regulate targeting and function of the ammonium transporter RhBG 18711365_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18711365_To identify susceptibility loci for bipolar disorder, we tested 1.8 million variants in 4,387 cases and 6,209 controls and identified a region of strong association (rs10994336, P = 9.1 x 10(-9)) in ANK3 (ankyrin G). 18768923_Ankyrin facilitates intracellular trafficking of alpha1-Na+-K+-ATPase in polarized cells. 19088739_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19088739_This study strongly support ANK3 as a bipolar disorder susceptibility gene and suggest true allelic heterogeneity. 20185149_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20185149_The association of ANK3 with schizophrenia is intriguing in light of recent associations of ANK3 with bipolar disorder, thereby supporting the hypothesis of an overlap in genetic susceptibility between these psychopathological entities. 20351715_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20386566_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20636642_there is genetic variation local to ANK3 gene affecting its expression, but that this variation is not responsible for increasing risk of bipolar disorder. 20877300_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21037240_Observational study of gene-disease association. (HuGE Navigator) 21304963_results suggest that allelic variation in ANK3 impacts cognitive processes associated with signal detection and this mechanism may relate to risk for Bipolar Disorder 21395576_we did not find evidence for association between the bipolar disorder risk polymorphisms rs10994336 in the ANK3 gene and rs1006737 in the CACNA1C gene in migraine 21438140_These findings supported the association between ANK3 and bipolar disorder, and also suggested the genomic region around rs1938526 as a common risk locus across ethnicities. 21676128_This study demonistreated that ANK3 genotype was associated with proneness to anhedonia. 21702894_results support a specific genetic contribution of ANK3 to bipolar disorder though failed to replicate findings for schizophrenia. 21767209_association of SNPs rs10994336 and rs9804190 with bipolar disorders and psychosis subphenotype 21893642_The ANK3 rs9804190 C allele increases the risk for schizophrenia by affecting ANK3 expression levels 21972176_These results further support that ANK3 is a susceptibility gene specific to bipolar disorder and that more than one risk locus is involved. 22180603_loss of AnkG expression may prevent the arrival of Cx43 to its final destination. 22328486_DNA sequencing revealed a novel low frequency (0.007) ANK3 SNP (ss469104599) which causes a non-conservative amino acid change at position 794 in the shorter isoforms of the ankyrin G protein. 22498896_Individuals carrying the bipolar disorder risk T-allele of ANK3 showed significantly reduced sensitivity in target detection, increased errors of commission, and atypical response latency variability. 22647524_The findings of this study do not support a strong genetic link between bipolar disorder and major depressive disorder for ANK3 genes. 22688190_data established a role for ankG in the human adaptive immune response against resident brain proteins, and they show that ankG immunization reduces brain beta-amyloid and its related neuropathology 22850628_These findings suggest a brain-specific cis-regulatory transcriptional effect of ANK3 that may be relevant to BD pathophysiology. 22865819_An association between ANK3 mutations and autism spectrum disorder susceptibility. 22986903_show novel expression of genes near regions of significantly associated SNPS, including TMEM26 and FOXA1 in airway epithelium and lung parenchyma, and ANK3 in alveolar macrophages in COPD 23109352_study concludes that ANK3 gene has a major influence on susceptibility to schizophrenia across populations 23129772_Cysteine 70 of ankyrin-G is S-palmitoylated and is required for function of ankyrin-G in membrane domain assembly. 23390136_inactivating mutations in the Ankyrin 3 (ANK3) gene in patients with severe cognitive deficits. 23715300_haplotype associated with bipolar disorder in Latino populations 23796624_A role of ANK3 in risk of stress-related and externalizing disorders, beyond its previous associations with bipolar disorder and schizophrenia. 24016415_ANK3 risk allele rs1938526 appears to be associated with general cognitive impairment and widespread cortical thinning in patients with first-episode psychosis 24108394_ANK3 SNP associated with brain connectivity changes in bipolar disorder. 24361380_results indicated that genetic variation within ANK3 may exert gene-specific modulating effects onworking memory deficits in schizophrenia. 24655771_These resultsindicatethatariskvariantwithin ANK3 may have an impact on neurocog- nitive function,suggesting a mechanism by which ANK3 confers risk for bipolar disorder. 24716743_This study demonistrated that ANK3 Bipolar disorder-associated variant rs139972937, responsible for an asparagine to serine. change (p = 0.042). 24809399_Here, we show that ANK3 gene expression in blood is significantly increased in bipolar disorder and schizophrenia compared with healthy controls. 24914473_Study showed a significant association between LMAN2L and risk of both bipolar disorder and schizophrenia 25307106_Data suggest that death domain of ankyrin G (ANK3-DD; located near C-terminus) exhibits C-terminal tail that curves back so that aromatic ring of a phenylalanine residue anchors flexible tail onto core domain; ANK3-DD exists as monomer in solution. 25311363_ANK3 rs10761482 showed a significant association with bipolar disorder 25616663_Kidney AE1 actually associates with epithelial ankyrin-G and renal ammonium transporter RhBG, which also binds ankyrin-G. 25711502_ANK3 bipolar-risk polymorphisms are associated with hyperactivation in the ventral anterior cingulate cortex in bipolar disorder. 25998125_Phosphorylation of KCNQ2 and KCNQ3 anchor domains by protein kinase CK2 augments binding to AnkG. 26055424_Whole-exome sequencing in ASD patients from each family identified a second rare inherited genetic variant, affecting either the ANK3 genes encoding NLGN4X interacting proteins expressed in inhibitory or in excitatory synapses. 26227746_we investigated the association of CACNA1C and ANK3 with SZ using meta-analytic techniques. 26574545_ankyrin-G associates with and inhibits the endocytosis of VE-cadherin cis dimers. 26682468_This study identified a SNP in ANK3 with a strong protective effect for Bipolar Disorder and Schizophrenia. 27177275_we found significant associations between rs10994336 and the N170 during the facial affect processing across two independent samples of healthy adults. In both samples, the risk allele carriers (TT/TC) consistently showed reduced N170. Our result showed clear evidence that rs10994336 may impact on the early neural processes (as reflected in N170) of facial affect processing. 27240527_Decreases in UF FA were observed among BD subjects carrying the ANK3 rs9804190 risk allele (T), compared to CC and T-carrier control subjects and BD CC homozygotes. 27488254_The ankyrin 3 genotype may be associated with pathogenesis of age-related neurodegeneration, and, in part, of bipolar disorder. 27534968_ANK3 expression is associated with androgen receptor stability, invasiveness, and lethal outcome in prostate cancer patients. 27811378_an association between ANK3 rs10994336, rs10994338, rs4948418 and rs958852 and schizophrenia risk in a northern Chinese Han population. 27956739_Thus ANK3's important association with human bipolar susceptibility may arise from imbalance between AnkG function in interneurons and principal cells and resultant excessive circuit sensitivity and output. 28079488_The haplotype analysis results suggest that ANK3 variants rs1938526 and rs10994336 may confer susceptibility for BD in the Korean population. Association analysis revealed a probable genetic difference between Korean and Caucasian populations in the degree of ANK3 involvement in BD pathogenesis. 28648753_The combination of tract-based spatial statistics (TBSS) with genotyping can be powerful to unveil the role of white matter in bipolar disorder, in conjunction with risk genes, ANK3 and ZNF804A. 28687526_Nonsense mutation in ANK3 was identified in a patient with speech impairment, intellectual disability and autistic features. 29068871_A significant association was found between bipolar disorder and rs10994336 (OR=1.18; 95% confidence interval: 1.06-1.31; P=0.0027) as well as rs1938526 (OR=1.16; 95% confidence interval: 1.06-1.28; P=0.0016) in ANK3. 29109170_Polymorphism in ANK3 gene is associated with Fetal Alcohol Spectrum Disorders. 30297702_Study shows that an elevated expression of a minor isoform of ANK3 is a risk factor for bipolar disorder. 30504823_Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP. 30941650_This study indicates that ANK3 is related to AR signalling pathway and is associated with BC prognosis. 31372216_this genome-wide scan of cleft lip triads identifies parent-of-origin interaction effects between ANK3 and maternal smoking, and between ARHGEF10 and alcohol consumption 32651551_Overrepresentation of genetic variation in the AnkyrinG interactome is related to a range of neurodevelopmental disorders. 32911427_Age-related atrophy of cortical thickness and genetic effect of ANK3 gene in first episode MDD patients. 33410423_Human ankyrins and their contribution to disease biology: An update. 33556457_Axonal TAU Sorting Requires the C-terminus of TAU but is Independent of ANKG and TRIM46 Enrichment at the AIS. 33729739_A linkage and exome study of multiplex families with bipolar disorder implicates rare coding variants of ANK3 and additional rare alleles at 10q11-q21. 34218362_ANK3 related neurodevelopmental disorders: expanding the spectrum of heterozygous loss-of-function variants. 35091539_Mapping the expression of an ANK3 isoform associated with bipolar disorder in the human brain. | ENSMUSG00000069601 | Ank3 | 690.310270 | 1.2215880 | 0.288757737 | 0.12969560 | 4.850960e+00 | 2.763075e-02 | 2.229620e-01 | No | Yes | 738.564771 | 176.505975 | 608.693431 | 145.479321 | |
| ENSG00000151422 | 2241 | FER | protein_coding | P16591 | FUNCTION: Tyrosine-protein kinase that acts downstream of cell surface receptors for growth factors and plays a role in the regulation of the actin cytoskeleton, microtubule assembly, lamellipodia formation, cell adhesion, cell migration and chemotaxis. Acts downstream of EGFR, KIT, PDGFRA and PDGFRB. Acts downstream of EGFR to promote activation of NF-kappa-B and cell proliferation. May play a role in the regulation of the mitotic cell cycle. Plays a role in the insulin receptor signaling pathway and in activation of phosphatidylinositol 3-kinase. Acts downstream of the activated FCER1 receptor and plays a role in FCER1 (high affinity immunoglobulin epsilon receptor)-mediated signaling in mast cells. Plays a role in the regulation of mast cell degranulation. Plays a role in leukocyte recruitment and diapedesis in response to bacterial lipopolysaccharide (LPS). Plays a role in synapse organization, trafficking of synaptic vesicles, the generation of excitatory postsynaptic currents and neuron-neuron synaptic transmission. Plays a role in neuronal cell death after brain damage. Phosphorylates CTTN, CTNND1, PTK2/FAK1, GAB1, PECAM1 and PTPN11. May phosphorylate JUP and PTPN1. Can phosphorylate STAT3, but the biological relevance of this depends on cell type and stimulus. {ECO:0000269|PubMed:12972546, ECO:0000269|PubMed:14517306, ECO:0000269|PubMed:19147545, ECO:0000269|PubMed:19339212, ECO:0000269|PubMed:19738202, ECO:0000269|PubMed:20111072, ECO:0000269|PubMed:21518868, ECO:0000269|PubMed:22223638, ECO:0000269|PubMed:7623846, ECO:0000269|PubMed:9722593}. | 3D-structure;ATP-binding;Alternative promoter usage;Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Kinase;Lipid-binding;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;Transferase;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a member of the FPS/FES family of non-transmembrane receptor tyrosine kinases. It regulates cell-cell adhesion and mediates signaling from the cell surface to the cytoskeleton via growth factor receptors. Alternative splicing results in multiple transcript variants. A related pseudogene has been identified on chromosome X. [provided by RefSeq, Apr 2015]. | hsa:2241; | cell cortex [GO:0005938]; cell junction [GO:0030054]; cell projection [GO:0042995]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; nucleus [GO:0005634]; actin binding [GO:0003779]; ATP binding [GO:0005524]; cadherin binding [GO:0045296]; epidermal growth factor receptor binding [GO:0005154]; gamma-catenin binding [GO:0045295]; lipid binding [GO:0008289]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase binding [GO:0019901]; protein phosphatase 1 binding [GO:0008157]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; signaling receptor binding [GO:0005102]; small GTPase binding [GO:0031267]; actin cytoskeleton reorganization [GO:0031532]; cell adhesion [GO:0007155]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; cell-cell adhesion mediated by cadherin [GO:0044331]; cellular response to insulin stimulus [GO:0032869]; cellular response to macrophage colony-stimulating factor stimulus [GO:0036006]; cellular response to reactive oxygen species [GO:0034614]; chemotaxis [GO:0006935]; cytokine-mediated signaling pathway [GO:0019221]; diapedesis [GO:0050904]; extracellular matrix-cell signaling [GO:0035426]; Fc-epsilon receptor signaling pathway [GO:0038095]; innate immune response [GO:0045087]; insulin receptor signaling pathway via phosphatidylinositol 3-kinase [GO:0038028]; interleukin-6-mediated signaling pathway [GO:0070102]; intracellular signal transduction [GO:0035556]; Kit signaling pathway [GO:0038109]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of mast cell activation involved in immune response [GO:0033007]; peptidyl-tyrosine phosphorylation [GO:0018108]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of actin filament polymerization [GO:0030838]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of epidermal growth factor receptor signaling pathway [GO:0042058]; regulation of fibroblast migration [GO:0010762]; regulation of lamellipodium assembly [GO:0010591]; regulation of mast cell degranulation [GO:0043304]; regulation of protein phosphorylation [GO:0001932]; response to lipopolysaccharide [GO:0032496]; response to platelet-derived growth factor [GO:0036119]; substrate adhesion-dependent cell spreading [GO:0034446]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; tyrosine phosphorylation of STAT protein [GO:0007260] | 11994747_Closing in on the biological functions of Fps/Fes and Fer. A review. 12871378_Fps/Fes and Fer are expressed in human and mouse platelets, and are activated following stimulation with collagen and collagen-related peptide (CRP), suggesting a role in GPVI receptor signaling 16732323_Fer is a regulator of cell-cycle progression in malignant cells and a potential target for cancer intervention. 18985748_FerT coexist in the acroplaxome with phosphorylated cortactin, a regulator of F-actin dynamics[Fer testis ] 19147545_Fer tyrosine kinase level correlates with the development of prostate cancer and aggressiveness of prostate cancer cell lines 19339212_Results suggest that Fer may allow a bypass of focal adhesion kinase-related cell anchorage dependency for intracellular signal transduction in hepatocytes. 19738202_Overexpression of Fer enhanced lamellipodia formation and cell migration in a manner dependent on PLD activity and the PA-FX interaction. 19835603_FER plays a role in the invasion and metastasis of hepatocellular carcinoma cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21122136_tyrosine phosphorylation of RhoGDIalpha by Fer as a mechanism to regulate binding of RhoGDIalpha to Rac 21518868_Overexpression of FER from a cDNA confers quinacrine resistance to several different types of cancer cell lines. 21700879_The A allele of SNP rs10447248 in the FER locus was nominally associated with lower MMW (P = 0.016) but was not associated with LMW (P > 0.05) adiponectin levels 22223638_Transcription of the ferT gene in CC cells was found to be driven by an intronic promoter residing in intron 10 of the fer gene and to be regulated by the Brother of the Regulator of Imprinted Sites (BORIS) transcription factor. 22238358_Fer, a non-receptor-type tyrosine kinase, plays a critical role in synthesis of the laminin-binding glycans on alpha-DG. 23445469_Fer expression correlates with renal cell carcinoma cell proliferation both in vitro and in vivo, and with tumor progression and survival 23699534_data show that Fer kinase is elevated in non-small cell lung cancer tumors and is important for cellular invasion and metastasis 23873028_FER kinase promotes breast cancer metastasis by regulating alpha6- and beta1-integrin-dependent cell adhesion and anoikis resistance. 23906537_Fer contributes to aberrant androgen receptor signaling via pSTAT3 cross-talk during castrate-resistant prostate cancer progression. 23931849_Detection of PJA2-FER fusion mRNA is correlated with poor postoperative survival periods in non-small cell lung cancer. 25533491_FER encodes a cytosolic non-receptor tyrosine kinase that influences neutrophil chemotaxis and endothelial permeability. 25867068_Fer serves as a crucial mediator and amplifier of Src-induced tumor progression. 28245430_Many human tumor types and cancer cell lines express the MAN2A1-FER fusion, which increases proliferation and invasiveness of cancer cell lines and has liver oncogenic activity in mice. 28851893_The FER rs4957796 TT genotype remained a significant covariate for the 90-day mortality risk in the multivariate analysis (hazard ratio, 4.62; 95% CI, 1.58-13.50; p = 0.0050). In conclusion, FER rs4957796 might act as a prognostic variable for survival in patients with severe ARDS due to pneumonia. 29038547_Data show that FER tyrosine kinase (Fer/FerT) activity was disrupted by E260, which selectively evokes metabolic stress in cancer cells by imposing mitochondrial dysfunction and deformation. 29099290_FER mediated HGF-independent regulation of HGFR/MET activates RAC1-PAK1 pathway to potentiate metastasis in ovarian cancer. 29208465_We propose a model for the regulation of Fer based on an intramolecular interaction and the curvature-dependent membrane binding mediated by its intrinsically disordered region. 29540831_FER enhances IGF-1R expression, phosphorylation, and signaling to promote cooperative growth and adhesion signaling that may facilitate cancer progression. 29907877_FER overexpression improves survival through STAT activation enhancing innate immunity and accelerating bacterial clearance in the lung. 29920310_Targeting of both FER and MET may be an effective strategy for therapeutic intervention in ovarian cancer. 31746983_This study supports the critical role of FER and FES tyrosine kinase fusions in the pathogenesis of follicular T-cell lymphoma and provides additional evidence that these can drive follicular T-cell lymphoma in the absence of RHOA mutations. 33411917_Phosphorylation of PKCdelta by FER tips the balance from EGFR degradation to recycling. 33430475_Fer and FerT Govern Mitochondrial Susceptibility to Metformin and Hypoxic Stress in Colon and Lung Carcinoma Cells. 33806191_Loss of Fer Jeopardizes Metabolic Plasticity and Mitochondrial Homeostasis in Lung and Breast Carcinoma Cells. | ENSMUSG00000000127 | Fer | 663.659769 | 1.1837546 | 0.243370092 | 0.11623842 | 4.329133e+00 | 3.746532e-02 | 2.615632e-01 | No | Yes | 853.899642 | 209.573526 | 702.704048 | 172.551030 | |
| ENSG00000151502 | 112936 | VPS26B | protein_coding | Q4G0F5 | FUNCTION: Acts as component of the retromer cargo-selective complex (CSC). The CSC is believed to be the core functional component of retromer or respective retromer complex variants acting to prevent missorting of selected transmembrane cargo proteins into the lysosomal degradation pathway. The recruitment of the CSC to the endosomal membrane involves RAB7A and SNX3. The SNX-BAR retromer mediates retrograde transport of cargo proteins from endosomes to the trans-Golgi network (TGN) and is involved in endosome-to-plasma membrane transport for cargo protein recycling. The SNX3-retromer mediates the retrograde transport of WLS distinct from the SNX-BAR retromer pathway. The SNX27-retromer is believed to be involved in endosome-to-plasma membrane trafficking and recycling of a broad spectrum of cargo proteins. The CSC seems to act as recruitment hub for other proteins, such as the WASH complex and TBC1D5. May be involved in retrograde transport of SORT1 but not of IGF2R. Acts redundantly with VSP26A in SNX-27 mediated endocytic recycling of SLC2A1/GLUT1 (By similarity). {ECO:0000250|UniProtKB:O75436, ECO:0000250|UniProtKB:Q8C0E2}. | Cytoplasm;Endosome;Membrane;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:112936; | cytosol [GO:0005829]; early endosome [GO:0005769]; endosome [GO:0005768]; late endosome [GO:0005770]; phagocytic vesicle [GO:0045335]; retromer complex [GO:0030904]; retromer, cargo-selective complex [GO:0030906]; cellular response to interferon-gamma [GO:0071346]; intracellular protein transport [GO:0006886]; regulation of macroautophagy [GO:0016241]; retrograde transport, endosome to Golgi [GO:0042147] | 21920005_Colocalization of Vps26 paralogues with different endosomally located Rab proteins shows prolonged association of Vps26B-retromer with maturing endosomes relative to Vps26A-retromer. 26113136_Authors propose that PAR-2 plasma membrane repopulation is regulated by Vps26B-retromer, describing a potential novel role for this complex. | ENSMUSG00000031988 | Vps26b | 2091.782211 | 1.1405441 | 0.189722200 | 0.07030171 | 7.325920e+00 | 6.796717e-03 | 1.036382e-01 | No | Yes | 2123.370004 | 127.081517 | 1878.233123 | 112.375583 | ||
| ENSG00000151690 | 54842 | MFSD6 | protein_coding | Q6ZSS7 | Mouse_homologues FUNCTION: MHC class I receptor. Binds only to H-2 class I histocompatibility antigen, K-D alpha chain (H-2K(D)). {ECO:0000269|PubMed:17010536}. | Acetylation;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:54842; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; antigen processing and presentation of exogenous peptide antigen via MHC class I [GO:0042590] | 19401682_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20978832_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000041439 | Mfsd6 | 86.221355 | 2.1592559 | 1.110534207 | 0.29321654 | 1.434549e+01 | 1.521436e-04 | No | Yes | 131.160956 | 32.128281 | 61.480231 | 15.108205 | |||
| ENSG00000151746 | 636 | BICD1 | protein_coding | Q96G01 | FUNCTION: Regulates coat complex coatomer protein I (COPI)-independent Golgi-endoplasmic reticulum transport by recruiting the dynein-dynactin motor complex. | Alternative splicing;Coiled coil;Golgi apparatus;Host-virus interaction;Reference proteome | This gene encodes an adaptor protein that belongs to the bicaudal D family of dynein cargo adaptors. The encoded protein acts as an intracellular cargo transport cofactor that regulates the microtubule-based loading of cargo onto the dynein motor complex. It also controls dynein motor activity and coordination. It has a domain architecture consisting of coiled-coil domains at the N- and C-termini that are highly conserved in other family members. Naturally occurring mutations in this gene are associated with short telomere length and emphysema. [provided by RefSeq, Aug 2017]. | hsa:636; | centrosome [GO:0005813]; cytoplasmic vesicle [GO:0031410]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; membrane [GO:0016020]; perinuclear region of cytoplasm [GO:0048471]; secretory vesicle [GO:0099503]; trans-Golgi network [GO:0005802]; cytoskeletal anchor activity [GO:0008093]; dynactin binding [GO:0034452]; dynein complex binding [GO:0070840]; dynein intermediate chain binding [GO:0045505]; protein kinase binding [GO:0019901]; proteinase activated receptor binding [GO:0031871]; small GTPase binding [GO:0031267]; structural constituent of cytoskeleton [GO:0005200]; anatomical structure morphogenesis [GO:0009653]; intracellular mRNA localization [GO:0008298]; microtubule anchoring at microtubule organizing center [GO:0072393]; minus-end-directed organelle transport along microtubule [GO:0072385]; negative regulation of phospholipase C activity [GO:1900275]; negative regulation of phospholipase C-activating G protein-coupled receptor signaling pathway [GO:1900737]; positive regulation of protein localization to centrosome [GO:1904781]; positive regulation of receptor-mediated endocytosis [GO:0048260]; protein localization to organelle [GO:0033365]; regulation of microtubule cytoskeleton organization [GO:0070507]; regulation of proteinase activated receptor activity [GO:1900276]; RNA processing [GO:0006396]; stress granule assembly [GO:0034063]; viral process [GO:0016032] | 16320833_Various processes in which BicD is involved during Drosophilian development (review) 17101644_findings show BICD1 localized to Chlamydia trachomatis inclusions in a biovar-specific manner and that EGFP-BICD1 is recruited to the inclusion in a microtubule- and Golgi apparatus-independent but chlamydial gene expression-dependent mechanism 17139249_These results imply that GSK-3beta may function in transporting centrosomal proteins to the centrosome by stabilizing the BICD1 and dynein complex, resulting in the regulation of a focused microtubule organization. 17707369_The brain-specific Rab6B via Bicaudal-D1 is linked to the dynein/dynactin complex, suggesting a regulatory role for Rab6B in the retrograde transport of cargo in neuronal cells. 18487243_BICD1 plays a similar role in telomere length homeostasis in humans. 20080650_Observational study of gene-disease association. (HuGE Navigator) 20164183_the protease-activated receptor-1 interactor, Bicaudal D1, regulates G protein signaling and internalization 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20686608_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20709820_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20709820_Variants in BICD1 are associated with length of telomeres, which suggests that a mechanism linked to accelerated aging may be involved in the pathogenesis of emphysema. 25792135_Graft BICD1 polymorphisms apart from the association with telomere length, affect early kidney function after transplantation 27496426_Kidney transplant recipients' polymorphisms of genes associated with telomere length, BICD1 and chromosome 18, but not hTERT, affect kidney allograft early and long-term function after transplantation. 28215293_Data suggest that BICD1 and BICD2 are highly expressed in the nervous system during development and are important in neuronal homeostasis. [REVIEW] 28410362_This study showed that rs2735940 hTERT CX-TT donor-recipient genotype pair was associated with almost five times higher odds (OR=4.82; 95% CI: 1.32-18; p=0.016) of delayed graft function (DGF), and that rs2735940 hTERT, rs2630578 BICD1, and rs7235755 chromosome 18 polymorphisms combined pairs were not associated with acute rejection (AR). 30464225_BICD1 mediates HIF1alpha nuclear translocation in mesenchymal stem cells during hypoxia adaptation. 31541015_The disrupting Bicd1/Fignl1 interaction induced motor axon pathfinding defects characteristic of Fignl1 gain or loss of function, respectively. 31846791_BICD1 genes may contribute to the decrease in forced vital capacity levels by interacting with PM10 exposure 32088084_BICD1 functions as a prognostic biomarker and promotes hepatocellular carcinoma progression. | ENSMUSG00000003452 | Bicd1 | 297.204614 | 1.0625505 | 0.087531466 | 0.18989048 | 2.145274e-01 | 6.432418e-01 | No | Yes | 217.348837 | 38.913953 | 195.811585 | 35.050825 | ||
| ENSG00000151892 | 2674 | GFRA1 | protein_coding | P56159 | FUNCTION: Receptor for GDNF. Mediates the GDNF-induced autophosphorylation and activation of the RET receptor (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Endosome;GPI-anchor;Glycoprotein;Golgi apparatus;Lipoprotein;Membrane;Receptor;Reference proteome;Repeat;Signal | This gene encodes a member of the glial cell line-derived neurotrophic factor receptor (GDNFR) family of proteins. The encoded preproprotein is proteolytically processed to generate the mature receptor. Glial cell line-derived neurotrophic factor (GDNF) and neurturin (NTN) are two structurally related, potent neurotrophic factors that play key roles in the control of neuron survival and differentiation. This receptor is a glycosylphosphatidylinositol (GPI)-linked cell surface receptor for both GDNF and NTN, and mediates activation of the RET tyrosine kinase receptor. This gene is a candidate gene for Hirschsprung disease. Alternative splicing results in multiple transcript variants, at least one of which encodes a preproprotein that is proteolytically processed. [provided by RefSeq, Jan 2016]. | hsa:2674; | anchored component of membrane [GO:0031225]; axon [GO:0030424]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; extrinsic component of membrane [GO:0019898]; Golgi apparatus [GO:0005794]; multivesicular body [GO:0005771]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; plasma membrane protein complex [GO:0098797]; receptor complex [GO:0043235]; glial cell-derived neurotrophic factor receptor activity [GO:0016167]; integrin binding [GO:0005178]; neurotrophin receptor activity [GO:0005030]; signaling receptor activity [GO:0038023]; signaling receptor binding [GO:0005102]; aging [GO:0007568]; cell migration [GO:0016477]; cell surface receptor signaling pathway [GO:0007166]; kidney development [GO:0001822]; male gonad development [GO:0008584]; nervous system development [GO:0007399]; neuron projection development [GO:0031175]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731] | 12065680_The expression of GFRA1 in normal infants and normoganglionic colon of patients with Hirschsprung's disease was restricted to receptor tyrosine kinase(RET)-negative glial cells and RET-positive neurons of the ganglionic plexus. 12490080_GFRA1-193C > G and 537T > C could be in linkage disequilibrium with other loci responsible for medullary thyroid cancer 12490080_Observational study of gene-disease association. (HuGE Navigator) 12624147_Observational study of gene-disease association. (HuGE Navigator) 14514671_analysis of binding surface for the GDNF-GFR alpha 1 16175604_Observational study of gene-disease association. (HuGE Navigator) 16385451_Observational study of gene-disease association. (HuGE Navigator) 16551639_Gas1 is related to the GDNF alpha receptors and regulates Ret signaling 16644101_Loss of dopaminergic neurons in the substantia nigra may induce changes in the expression of GDNF but not its receptor snd Parkinson disease. 16813162_Observational study of gene-disease association. (HuGE Navigator) 17298301_The role of heparin and heparan sulfate in GDNF signalling remains unclear, but the present study indicates that it does not occur in the first step of the pathway, namely GDNF-GFRalpha1 engagement. 17825269_GFRalpha-1 were observed within sensory and motor nuclei of cranial nerves, dorsal column nuclei, olivary nuclear complex, reticular formation, pontine nuclei, locus caeruleus, raphe nuclei, substantia nigra, and quadrigeminal plate. 18089803_GDNF can act as an important component of the inflammatory response in breast cancers and its effects aare mediated by GFR alpha 1 receptors. 18353777_direct receptor-receptor interactions are not required for high affinity GDNF binding to NCAM but play an important role in the regulation of NCAM-mediated cell adhesion by GFRalpha1 18394855_GDNF is a key component to preserve several cell populations in the nervous system and also participates in the survival and differentiation of peripheral neurons. 18845535_analysis of how GDNF.GFR alpha 1 can mediate cell adhesion and how heparin might inhibit GDNF signaling through RET 19019765_38 cases of germ cell tumors: 26 cases contained immature teratoma, of which 24 had immature neuroepithelium and showed strong membrane staining for GFRalpha-1. staining for GFRalpha-1 in immature neuroepithelium may facilitate its identification. 19188437_Ape1 is a novel physiological regulator of GDNF responsiveness, and Ape1-induced GFRalpha1 expression may play important roles in pancreatic cancer progression and neuronal cell survival. 19282698_Observational study of gene-disease association. (HuGE Navigator) 19896648_GFR-alpha1 mRNA transcripts were detected in oocytes and GCs from all samples from fetuses, girls and adult women. 20116071_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20116071_This study found nominally-significant evidence for interactions between GFRA1, 2 and 3 associated with schizophrenia and clozapine response, consistent with the locations of these three genes within linkage regions for schizophrenia. 20347960_Here we report that human nigral dopaminergic neurons express GFRalpha1 and RET receptors at all ages. There was no reduction in the number of neurons expressing these receptors as a function of age. 20350599_Results identify persephin, a GDNF family member, as a novel ligand for GFRalpha1/RET receptor complex. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20424473_Observational study of gene-disease association. (HuGE Navigator) 20669561_MEN2 arises from activating missense mutations in RET, causing autodimerization under certain conditions. The position of missense mutations influences medullary thyroid carcinoma aggressiveness. Review. 21133924_Post-synaptic transgenic GFRalpha1 has profound effects on the development of dopamine neurons, resulting in a 40% increase in the adult number. 22729463_Mutations in GFRA1 gene is associated with urinary tract malformations. 23351331_The expression of GFRalpha1 and/or GFRalpha3, especially when combined with ARTN expression, may be useful predictors of disease progression and outcome in specific subtypes of mammary carcinoma. 23881409_The study shows co-localization of RET with GFRA1 and GFRA2 in myenteric ganglia of the adult human colon. 24139947_In the cochlea, GFRalpha-1 was identified mainly in the cell bodies of the spiral neurons. In the organ of Corti, GFRalpha-1 was demonstrated in the Deiters' cells, Hensen cells, inner pillar cells, and weakly in inner hair cells but not in the outer hair cells. 24778213_These findings collectively demonstrate that GFRalpha1 released by nerves enhances perineural invasion through GDNF-RET signaling and that GFRalpha1 expression by cancer cells enhances but is not required for it. 25009298_Methylation changes of GFRA1, SRF, and ZNF382 may be a potential biomarker set for prediction of gastric carcinoma metastasis. 25253858_GFRalpha1 levels in neurons from autopsied AD brains are significantly decreased. 26400951_This study demonstrate, using a knock-in mouse model in which GFRalpha1 is no longer located in lipid rafts, that the developmental functions of GDNF in the periphery require the translocation of the GDNF receptor complex into lipid rafts. 26904955_Hox proteins coordinate motor neuron differentiation and connectivity programs through Ret/Gfra genes. 27346872_that the consequences of this is that GFRalpha-1-mediated signalling is altered during the ageing process 27533506_RET c.1296A may be a common susceptibility allele for nephron underdosing-related diseases. The 5'-UTR and intronic variants near exon 5 of GFRA1 are not associated with nephron endowment. 27566576_the methylation status of CpG sites in GFRA1 and GSTM2 may have a role and could be used as potential biomarkers for the screening of rectal cancer 27754745_GFRA1 regulates AMPK-dependent autophagy by promoting SRC phosphorylation independent of proto-oncogene RET kinase 28952330_identified a novel susceptibility locus (rs3781545in GFRA1) with suggestive significance for migraine risk in Han Chinese 29037220_circGFRA1 may function as a competing endogenous RNA (ceRNA) to regulate GFRA1 expression through sponging miR-34a to exert regulatory functions in TNBC. 30040982_Data suggest that GDNF family receptor alpha 1 protein (GFRA1) is a protein target of ST3 beta-galactoside alpha-23-sialyltransferase 1 (ST3GAL1). 31642440_Low expression of EZH2 in the colon tissue of children with Hirschsprung's disease may be one of the causes of inadequate expression of GFRalpha1 and onset of Hirschsprung's disease 31988584_GFRA1 expression is frequently reactivated by DNA demethylation in colon cancer (CC) tissues and is significantly associated with a poor prognosis in patients with CC, especially those with metastatic CC. GFRA1 can promote the proliferation/growth of CC cells, probably by the activation of AKT and ERK pathways. 32841458_LncRNA LINC00210 regulated radiosensitivity of osteosarcoma cells via miR-342-3p/GFRA1 axis. 33020172_Biallelic Pathogenic GFRA1 Variants Cause Autosomal Recessive Bilateral Renal Agenesis. 33175846_Hypomethylation of GDNF family receptor alpha 1 promotes epithelial-mesenchymal transition and predicts metastasis of colorectal cancer. 33233403_Generation of an Oncolytic Herpes Simplex Viral Vector Completely Retargeted to the GDNF Receptor GFRalpha1 for Specific Infection of Breast Cancer Cells. 34391932_Deficiency of GFRalpha1 promotes hepatocellular carcinoma progression but enhances oxaliplatin-mediated anti-tumor efficacy. 34668628_CircGFRA1 facilitates the malignant progression of HER-2-positive breast cancer via acting as a sponge of miR-1228 and enhancing AIFM2 expression. | ENSMUSG00000025089 | Gfra1 | 239.723181 | 1.0227418 | 0.032442019 | 0.17364526 | 3.506459e-02 | 8.514604e-01 | No | Yes | 255.413742 | 35.146125 | 253.975283 | 34.854035 | ||
| ENSG00000152672 | 165530 | CLEC4F | protein_coding | Q8N1N0 | FUNCTION: Receptor with an affinity for galactose and fucose. Could be involved in endocytosis (By similarity). {ECO:0000250}. | Alternative splicing;Endocytosis;Glycoprotein;Lectin;Membrane;Receptor;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | hsa:165530; | external side of plasma membrane [GO:0009897]; integral component of membrane [GO:0016021]; carbohydrate binding [GO:0030246]; endocytosis [GO:0006897] | Mouse_homologues 23762286_CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in alpha-galactosylceramide presentation in liver. 30590594_The CLEC4f protein is expressed in spleen in non-human primates and the close evolutionary relationship of the CLEC4f protein to langerin (CD207) suggest that it may function in the immune system, possibly as a pathogen receptor. 33993195_Kupffer cell receptor CLEC4F is important for the destruction of desialylated platelets in mice. | ENSMUSG00000014542 | Clec4f | 9.560743 | 1.1545515 | 0.207332474 | 0.92659012 | 4.429183e-02 | 8.333116e-01 | No | Yes | 9.532643 | 4.022385 | 8.279209 | 3.591408 | |||
| ENSG00000153208 | 10461 | MERTK | protein_coding | Q12866 | FUNCTION: Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to several ligands including LGALS3, TUB, TULP1 or GAS6. Regulates many physiological processes including cell survival, migration, differentiation, and phagocytosis of apoptotic cells (efferocytosis). Ligand binding at the cell surface induces autophosphorylation of MERTK on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with GRB2 or PLCG2 and induces phosphorylation of MAPK1, MAPK2, FAK/PTK2 or RAC1. MERTK signaling plays a role in various processes such as macrophage clearance of apoptotic cells, platelet aggregation, cytoskeleton reorganization and engulfment (PubMed:32640697). Functions in the retinal pigment epithelium (RPE) as a regulator of rod outer segments fragments phagocytosis. Plays also an important role in inhibition of Toll-like receptors (TLRs)-mediated innate immune response by activating STAT1, which selectively induces production of suppressors of cytokine signaling SOCS1 and SOCS3. {ECO:0000269|PubMed:17005688, ECO:0000269|PubMed:32640697}. | 3D-structure;ATP-binding;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Receptor;Reference proteome;Repeat;Retinitis pigmentosa;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | This gene is a member of the MER/AXL/TYRO3 receptor kinase family and encodes a transmembrane protein with two fibronectin type-III domains, two Ig-like C2-type (immunoglobulin-like) domains, and one tyrosine kinase domain. Mutations in this gene have been associated with disruption of the retinal pigment epithelium (RPE) phagocytosis pathway and onset of autosomal recessive retinitis pigmentosa (RP). [provided by RefSeq, Jul 2008]. | hsa:10461; | cytoplasm [GO:0005737]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; photoreceptor outer segment [GO:0001750]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; rhabdomere [GO:0016028]; ATP binding [GO:0005524]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; cell migration [GO:0016477]; cell surface receptor signaling pathway [GO:0007166]; cell-cell signaling [GO:0007267]; natural killer cell differentiation [GO:0001779]; negative regulation of cytokine production [GO:0001818]; negative regulation of leukocyte apoptotic process [GO:2000107]; negative regulation of lymphocyte activation [GO:0051250]; nervous system development [GO:0007399]; neutrophil clearance [GO:0097350]; phagocytosis [GO:0006909]; platelet activation [GO:0030168]; positive regulation of kinase activity [GO:0033674]; positive regulation of phagocytosis [GO:0050766]; protein kinase B signaling [GO:0043491]; protein phosphorylation [GO:0006468]; retina development in camera-type eye [GO:0060041]; secretion by cell [GO:0032940]; spermatogenesis [GO:0007283]; substrate adhesion-dependent cell spreading [GO:0034446]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; vagina development [GO:0060068] | 11727200_retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively 12768229_Gas6 receptors were not upregulated in any of the allograft groups, except for the Axl receptor, which increased only in acute tubular necrosis. 15111602_The present study reports the identification of R844C, the first putative pathogenic MERTK missense mutation that results in severe retinal degeneration with childhood onset 15130911_mer, presumably through activation by its ligand Gas6, participates in regulation of platelet function in vitro and platelet-dependent thrombosis in vivo. 15733062_Axl, Sky and Mer are Gas6 receptors that enhance platelet activation and regulate thrombotic responses 16675557_Transforming Mer signals may contribute to T-cell leukemogenesis, and abnormal Mer expression may be a novel therapeutic target in pediatric acute lymhpocytic leukemia therapy. 16710167_Mutations in the MERTK gene are relatively rare in Japanese patients with autosomal recessive retinitis pigmentosa. 16837475_Observational study of gene-disease association. (HuGE Navigator) 17005688_Here we show the involvement of members of the Tyro3 receptor tyrosine kinase family-Axl, Dtk, and Mer-in cell entry of filoviruses. 17047157_cleavage results in production of a soluble Mer protein that prevented Gas6-mediated stimulation of membrane-bound Mer. Mer inhibition led to defective macrophage-mediated engulfment of apoptotic cells and decreased platelet aggregation 17626743_Mer is highly expressed on Jurkat cells, and could inhibit cell apoptosis via Bcl-2 signaling pathway. 18039660_Tyr-867 in Mer receptor tyrosine kinase allows for dissociation of multiple signaling pathways for phagocytosis of apoptotic cells and down-modulation of lipopolysaccharide-inducible NF-kappaB transcriptional activation 18174230_Observational study of gene-disease association. (HuGE Navigator) 18246816_Overexpressed Mer tyrosine kinase receptor can inhibit the migration and angiogenesis of HMEC-1 cells through VEGF-C/VEGFR-2 signal pathway. 18250462_MerTK macrophage receptor is an essential component required for serum-stimulated phagocytosis of apoptotic cells. 18587056_MERTK, a cell surface receptor that recognizes apoptotic cells, is expressed on human alveolar macrophages (AMs), and its expression is up-regulated in AMs of cigarette smokers. 18620092_Possible strategies for targeted inhibition of the TAM family in the treatment of human cancer. 18815424_IVS16+1G>T disrupts the splice donor site causing exon 16 skipping. Absence of exon 16 causes a frameshift and the introduction of a premature termination codon into exon 17 creating an altered mRNA with a seriously affected tyrosine kinase domain. 18922854_human protein S can inhibit the expression and activity of SR-A through Mer RTK in macrophages, suggesting that human protein S is a modulator for macrophage functions in uptaking of modified lipoproteins 19403518_The splice site mutation c.2189+1G>T in MERTK causes rod-cone dystrophy with a distinct macular phenotype. 19541935_there was a negative correlation between Gas6 and soluble Axl and Mer in established multiple sclerosis lesions. In addition, increased levels of soluble Axl and Mer were associated with increased levels of mature ADAM17, mature ADAM10, and Furin 19922767_data suggest that growth arrest-specific-6 (GAS6)/MER receptor tyrosine kinase axis regulates homing and survival of the E2A/PBX1-positive B-cell precursor acute lymphoblastic leukemia in the bone marrow niche 20130272_Homozygosity mapping and mutation analysis in the distant family member affected by RP revealed a homozygous mutation in MERTK, but no CEP290 mutations. 20300561_The phenotype associated with these identified MERTK mutations is of a childhood onset rod-cone dystrophy with early macular atrophy. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20538656_results strongly suggest that the nonsense mutation in MERTK, leading to premature termination of the protein, is responsible for retinitis pigmentosa phenotype in the affected individuals of the Pakistani family. 20546121_Gas6 and Tyro3, Axl and Mer (TAM) receptors have roles in human and murine platelet activation and thrombus stabilization 20591486_Observational study of genetic testing. (HuGE Navigator) 20637106_correlation of decreased protein S levels with lupus disease activity is consistent with a role for the TAM receptors in scavenging apoptotic cells and controlling inflammation 20654219_The lower activity of PKC and the higher expression of MERTK are very important for sustaining the phagocytic process of ROS by human retinal pigment epithelial cells. 20664904_Findings indicate genetic association between MERTK and TYRO3 allelic variants and carotid atherosclerosis. 20664904_Observational study of gene-disease association. (HuGE Navigator) 20801516_Observational study of genetic testing. (HuGE Navigator) 20959443_PMN-Ect interacted with the macrophages by activation of the MerTK pathway responsible for down-modulation of the proinflammatory signals generated by ZymA. 20978472_Tubby and Tulp1 are bridging molecules with their N-terminal region as MERTK-binding domain and C-terminal region as phagocytosis prey-binding domain. 21347448_MERTK gene is a novel risk gene for multiple sclerosis susceptibility 21384080_overexpressed in atherosclerotic carotid plaques 21496228_plasma concentrations of sMer and sTyro3 were significantly increased in patients with active systemic lupus erythematosis and Rheumatoid arthritis 21542987_Gene expression profile is changed by the dysfunction of Mer during retinal pigment epithelium phagocytosis.( 21677792_A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. 21792939_This study identified galectin-3 as a new MerTK ligand by an advanced dual functional cloning strategy. 22180149_MERTK mutations lead to severe retinitis pigmentosa with discrete dot-like autofluorescent deposits at early stages, which are a hallmark of this MERTK-specific dystrophy. 22363695_The present study explores Mer behavior following prolonged exposure to Gas6. 22469987_Mer receptor tyrosine kinase promotes invasion and survival in glioblastoma multiforme. 22841784_The single nucleotide polymorphisms rs4374383 and rs9380516 were linked to the functionally related genes MERTK and TULP1, which encode factors involved in phagocytosis of apoptotic cells by macrophages. 22890323_These results indicate that Mer and Axl have complementary and overlapping roles in Non-small cell lung cancer 22942426_MerTK expression levels adapt to changing immunologic environment, being suppressed in M1 and M2a macrophages and in dendritic cells. 22997156_Oxidized of polyunsaturated fatty acids contained in the tissue close to necrotic core of human carotid plaques are strong inducers of Adam17, which in turn may cleave the extracellular domain of Mertk giving rise to sMer. 23065156_The tyrosine kinase receptor MER is activated by PROS and mediates its inhibitory effect on VEGF-A-induced EC proliferation. 23390493_MERTK signaling in the retinal pigment epithelium involves a cohort of SH2-domain proteins with the potential to regulate both cytoskeletal rearrangement and membrane movement. 23474756_data suggest a role for Mer in acute myeloid leukemogenesis and indicate that targeted inhibition of Mer may be an effective therapeutic strategy in pediatric and adult AML 23585477_MERTK expression correlates with disease progression in metastatic melanomas, primary melanomas, and nevi 23617806_MERTK has a role in regulating melanoma cell migration and survival and differentially regulates cell behavior relative to AXL 23662598_[review] Receptor tyrosine kinases Tyro-3, Axl and Mer, collectively designated as TAM, are involved in the clearance of apoptotic cells. 23835724_MerTK expression in circulating innate immune cells is increased in patients with septic shock in comparison with healthy volunteers and trauma patients and its persistent overexpression after septic shock is associated with adverse outcome. 24741600_Both mMer and sMer levels significantly increased in SLE and positively correlated with disease activity and severity. The upregulation of MerTK expression may serve as a biomarker of the disease activity and severity of SLE. 24939420_The MER receptor pathway promotes wound repair in macrophages and epithelial cell growth. 25074926_These studies demonstrate that, despite their similarity, TYRO3, AXL, and MER are likely to perform distinct functions in both immunoregulation and the recognition and removal of apoptotic cells 25074939_These data collectively identify MERTK as a significant link between cancer progression and efferocytosis, and a potentially unrealized tumor-promoting event when MERTK is overexpressed in epithelial cells. 25102945_Inhibition of the Gas6 receptor Mer or therapeutic targeting of Gas6 by warfarin is a promising strategy for the treatment of multiple myeloma. 25428221_Mer expression correlates with CNS positivity upon initial diagnosis in t(1;19)-positive pediatric acute lymphoblastic leukemia patients. 25450174_The key role of the MERTK could be demonstrated in HMDM engulfing dying cells using gene silencing as well as blocking antibodies. Similar pathways were found upregulated in living ARPE-19 engulfing anoikic ARPE-19 cells. 25479139_Patients with ACLF have increased numbers of immunoregulatory monocytes and macrophages that express MERTK and suppress the innate immune response to microbes. The number of these cells correlates with disease severity and the inflammatory response. 25624460_MERTK on DCs controls T cell activation and expansion through the competition for PROS1 interaction with MERTK in the T cells. MERTK is a potent suppressor of T cell response. 25695599_results identify Mer as a receptor uniquely capable of both tethering ACs to the macrophage surface and driving their subsequent internalization. 25762638_UNC1666 is a novel potent small molecule tyrosine kinase inhibitor that decreases oncogenic signaling and myeloblast survival by dual Mer/Flt3 inhibition. 25826078_Mer enhances malignant phenotype and pharmacological inhibition of Mer overcomes resistance of non-small cell lung cancer to EGFR-targeted agents. 25878564_Significantly increased levels of sMer, sTyro3 and sAxl may be important factors contributing to the deficit in phagocytosis ability in systemic lupus erythematosus . 25881761_The mRNA expression levels of Tyro-3, Axl were decreased in pSS patients. When considering the plasma level, increased levels of soluble Mer was observed with statistically significant difference. 26263531_Upon differentiation of these iPSC towards RPE, patient-specific RPE cells exhibited defective phagocytosis, a characteristic phenotype of MERTK deficiency observed in human patients and animal models 26316303_Utilizing an ex vivo co-cultivation approach to model key cellular and molecular events found in vivo during infarction, cardiomyocyte phagocytosis was found to be inefficient, in part due to myocyte-induced shedding of macrophage MERTK 26427420_Studies indicate that c-Mer receptor tyrosine kinase MERTK mutations cause retinal degenerations. 26427450_Data indicate that AAV2-VMD2-c-mer proto-oncogene protein (hMERTK) provided up to 6.5 months photoreceptor rescue in the RCS rat, and also had a major protective effect in Mertk-null mice. 26427488_Data show that activated AMP-activated protein kinase (AMPK) limits retinal pigment epithelial cells (RPE) phagocytic activity by abolishing retinal photoreceptor cell outer segment (POS)-induced activation of c-mer proto-oncogene tyrosine kinase (MerTK). 26596542_The rs4374383 AA genotype, associated with lower intrahepatic expression of MERTK, is protective against F2-F4 fibrosis in patients with non-alcoholic fatty liver disease (NAFLD). 26768676_STK 11 testing can confirm those at risk of Peutz-Jeghers syndrome, who require lifelong surveillance, and possibly release those with a simple dermatosis, such as Laugier-Hunziker syndrome, from invasive and thus potentially harmful surveillance. 26819373_Combined Mertk (and Mfge8) deficiency in macrophages blunted VEGFA release from infarcted hearts. 26962228_The current study demonstrates the contribution of the TAM receptor MerTK to the phagocytosis of myelin by human adult microglia and monocyte-derived macrophages. 26990204_One of the associated variants was also found to be linked with increased expression of MERTK in monocytes and higher expression of MERTK was associated with either increased or decreased risk of developing MS, dependent upon HLA-DRB1*15:01 status. 27028863_these data suggest that endogenous GAS6 and Mer receptor signaling contribute to the establishment of prostate cancer stem cells in the bone marrow microenvironment 27081701_MERTK is frequently overexpressed in head and neck squamous cell carcinoma and plays an important role in tumor cell motility. 27122965_We report a novel missense mutation (c.3G>A, p.0?) in the MERTK gene that causes severe vision impairment in a patient. 27486820_Study identified the Gas6/TAM receptor pathway with Tyro3 and Mer as novel targets in colorectal cancer. 27649555_The broad-spectrum activity mediated by UNC2025 in leukemia patient samples and xenograft models, alone or in combination with cytotoxic chemotherapy, supports continued development of MERTK inhibitors for treatment of leukemia 27753136_Knockdown of MERTK by shRNA in prostate cancer cells induced a decreased ratio of P-Erk1/2 to P-p38, increased expression of p27, NR2F1, SOX2, and NANOG, induced higher levels of histone H3K9me3 and H3K27me3, and induced a G1/G0 arrest, all of which are associated with dormancy. 27801848_In this paper, we review the biology of the Gas6/Tyro3, Axl, and MerTK(collectively named TAM system)and the current evidence supporting its potential role in the pathogenesis of multiple sclerosis . 27827458_Study describes a novel cellular pathway involved in diabetic efferocytosis, wherein diabetes-induced decrease in miR-126 expression results in upregulation of ADAM9 expression that in-turn leads proteolytic cleavage of MerTK and formation of inactive soluble Mer. Decrease in MerTK phosphorylation leads to reduced downstream cytoskeletal signaling required for engulfment and thus decreases efferocytosis. 28067670_evidence that proteolytic cleavage of the macrophage efferocytosis receptor c-Mer tyrosine kinase (MerTK) reduces efferocytosis and promotes plaque necrosis and defective resolution. 28127639_The expression of MerTK and AxlTK varied according to the deposition of immunoglobulin and complements on glomeruli. Both MerTK and AxlTK expressions were increased on glomeruli and varied according to pathological classifications. 28184013_Phosphatidylserine mediated hyperactivation of Mertk.MERTK promotes epithelial cell efferocytosis in a tyrosine kinase-dependent manner.MERTK role in AKT-dependent drug resistance. 28251492_Small molecule and antibody inhibitors of AXL and MER have recently been described, and some of these have already entered clinical trials. The optimal design of treatment strategies to maximize the clinical benefit of these AXL and MER targeting agents are discussed in relation to the different cancer types and the types of resistance encountered. 28324114_A 48 bp insertion sequence was buried within the breakpoint; 18 bps shared homology to MIR4435-2HG and LINC00152, and 30 bp mapped to MERTK. The deletion cosegregated with arRP in the family. 28334911_MERTK G > A variant affects liver disease, nutrient oxidation and glucose metabolism in NAFLD. 28462455_Sequence analysis revealed that the proband was a compound heterozygote with two independent mutations in MERTK, a novel nonsense mutation (c.2179C > T) and a previously reported missense variant (c.2530C > T). The proband's affected brother also had both mutations 28668213_Patients with macroalbuminuria diabetes had higher circulating levels of sMer and more urinary soluble Tyro3 and sMer than normoalbuminuric diabetics. Increased clearance of sTyro3 and sMer was associated with loss of tubular Tyro3 and Mer expression in diabetic nephropathy tissue. During in vitro diabetes, human kidney cells had down-regulation of Tyro3 and Mer mRNA and increased shedding of sTyro3 and sMer. 28851810_Monocyte-induced MerTK cleavage on proreparative MHCII(LO) cardiac macrophages is a novel contributor to myocardial ischemic reperfusion injury. 28916522_this study shows that viral infection sensitizes fetal membranes by MERTK Inhibition 29299721_The present study provides a meaningfully negative result demonstrating that rare variants in MERTK are not associated with AMD. The study also demonstrates the role of large sample size genetic studies utilizing whole-genome sequencing as a powerful tool that can resolve clinically relevant questions regarding the genetic basis of ophthalmic disease. 29359540_We observed that the frequency for the wild-type haplotype was higher in the control group, compared to that in the group of patients with COPD, in the subgroup analysis of current smokers, although the difference was not statistically significant 29437494_The targeted NGS strategy employed provides an efficient tool for RP pathogenic gene detection. This study identified a new autosomal recessive mutation in the RP-related gene MERTK, which expands the spectrum of RP disease-causing mutations 29553850_MerTK mediates STAT3-KRAS/SRC-signaling axis for glioma stem cell maintenance 29659094_Mutations in MERTK have been associated with severe autosomal recessive retinal dystrophies in the RCS rat and in humans. We present here a comprehensive review of all reported MERTK disease causing variants with the associated phenotype 30091033_cooperation between CD14 and MerTK may foster the clearance of apoptotic neutrophils by human monocytes/macrophages 30093568_MERTK expression was increased in cell lines and patient-derived xenografts treated with AXL inhibitors. 30254055_activation of MerTK in human macrophages led to ERK-mediated expression of the gene encoding SERCA2, which decreased the cytosolic Ca(2+) concentration and suppressed the activity of calcium/calmodulin-dependent protein kinase II 30416333_Patients with retinitis pigmentosa (RP) in this study were carriers of two novel allelic mutations in the MERTK gene, a missense variant in exon 17 and an approximate 91 kb genomic deletion. Mapping of the deletion breakpoints allowed molecular testing of a cohort of patients with RP with allele-specific PCR. 30541554_Axl and Tyro3, but not Mertk, have an important role in platelet activation and thrombus formation 30619243_data do not support a particular role for TAM receptors or for activated CD11b in the association of platelet-derived EVs with monocytes and granulocytes in the circulation. 30765874_Mertk expression was not significantly higher in non-muscle invasive bladder cancers (MIBCs) and MIBCs, compared to normal urothelium. Loss-of-function experiments in vitro and in a mouse xenograft model showed that Mertk depletion had only a minor impact on cell viability. 30790467_Three novel loss-of-function mutations in MERTK gene were identified in Chinese patients with retinitis pigmentosa. 30851773_MERTK has a role in retinal pigment epithelium as a regulator of rod outer segments' phagocytosis. Due to c.1647T > G substitution, the stop codon (p.Tyr549Ter) appears early in the transcript. 30944303_Authors report that SAV1, a Hippo signaling component, inhibits Akt, a function independent of its role in Hippo signaling. Binding to a proline-tyrosine motif in the Akt-PH domain, SAV1 suppresses Akt activation by blocking Akt's movement to plasma membrane. Authors further identify cancer-associated SAV1 mutations with impaired ability to bind Akt, leading to Akt hyperactivation. 31221805_Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. 31230815_we identify an unexpected role of TAM kinases (Tyro3, Axl, and Mer)as promoters of necroptosis, a pro-inflammatory necrotic cell death. Pharmacologic or genetic targeting of TAM kinases results in a potent inhibition of necroptotic death in various cellular models. We identify phosphorylation of MLKL Tyr376 as a direct point of input from TAM kinases into the necroptosis signaling 31451482_TAM Family Receptor Kinase Inhibition Reverses MDSC-Mediated Suppression and Augments Anti-PD-1 Therapy in Melanoma. 31583026_Gas6/MERTK signaling components are upregulated in liver fibrosis. (Review) 31655933_Increased sMer, but not sAxl, sTyro3, and Gas6 relate with active disease in juvenile systemic lupus erythematosus. 31805065_These results suggest that deletion of MERTK in human pulmonary microvascular endothelial cells in vitro and in all cells in vivo aggravates the inflammatory response. 31839486_Macrophage MerTK Promotes Liver Fibrosis in Nonalcoholic Steatohepatitis. 32051695_New Insights into the Role of Tyro3, Axl, and Mer Receptors in Rheumatoid Arthritis. 32160519_MERTK-Dependent Ensheathment of Photoreceptor Outer Segments by Human Pluripotent Stem Cell-Derived Retinal Pigment Epithelium. 32454903_Role of Gas6 and TAM Receptors in the Identification of Cardiopulmonary Involvement in Systemic Sclerosis and Scleroderma Spectrum Disorders. 32640697_Mertk Interacts with Tim-4 to Enhance Tim-4-Mediated Efferocytosis. 32973744_Potential Oncogenic Effect of the MERTK-Dependent Apoptotic-Cell Clearance Pathway in Starry-Sky B-Cell Lymphoma. 32976546_Pathogenic variants of AIPL1, MERTK, GUCY2D, and FOXE3 in Pakistani families with clinically heterogeneous eye diseases. 33101282_Expression of TAM-R in Human Immune Cells and Unique Regulatory Function of MerTK in IL-10 Production by Tolerogenic DC. 33119085_A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding. 33234241_Recent advancements in role of TAM receptors on efferocytosis, viral infection, autoimmunity, and tissue repair. 33234243_TAM receptors and their ligand-mediated activation: Role in atherosclerosis. 33234244_Post-translational modifications of the ligands: Requirement for TAM receptor activation. 33234245_Immunological role of TAM receptors in the cancer microenvironment. 33353011_Bi-Allelic Pathogenic Variations in MERTK Including Deletions Are Associated with an Early Onset Progressive Form of Retinitis Pigmentosa. 33512463_Mer tyrosine kinase as a possible link between resolution of inflammation and tissue fibrosis in IgG4-related disease. 33532004_Role of the Gas6/TAM System as a Disease Marker and Potential Drug Target. 33571503_MerTK activity is not necessary for the proliferation of glioblastoma stem cells. 33609509_Deterioration of phagocytosis in induced pluripotent stem cell-derived retinal pigment epithelial cells established from patients with retinitis pigmentosa carrying Mer tyrosine kinase mutations. 33761885_Differential polarization and the expression of efferocytosis receptor MerTK on M1 and M2 macrophages isolated from coronary artery disease patients. 33859405_Microglia use TAM receptors to detect and engulf amyloid beta plaques. 34035216_Ligand-dependent kinase activity of MERTK drives efferocytosis in human iPSC-derived macrophages. 34207717_MERTK-Mediated LC3-Associated Phagocytosis (LAP) of Apoptotic Substrates in Blood-Separated Tissues: Retina, Testis, Ovarian Follicles. 34289798_MERTK retinopathy: biomarkers assessing vision loss. 34415994_MERTK on mononuclear phagocytes regulates T cell antigen recognition at autoimmune and tumor sites. 35036076_MerTK-mediated efferocytosis promotes immune tolerance and tumor progression in osteosarcoma through enhancing M2 polarization and PD-L1 expression. 35163068_High Glucose Impairs Expression and Activation of MerTK in ARPE-19 Cells. | ENSMUSG00000014361 | Mertk | 563.231915 | 0.9261019 | -0.110757124 | 0.15907578 | 4.644317e-01 | 4.955601e-01 | 8.010378e-01 | No | Yes | 592.751644 | 114.491116 | 628.141085 | 121.162131 | |
| ENSG00000153531 | 113622 | ADPRHL1 | protein_coding | Q8NDY3 | FUNCTION: Required for myofibril assembly and outgrowth of the cardiac chambers in the developing heart (By similarity). Appears to be catalytically inactive, showing no activity against O-acetyl-ADP-ribose (By similarity). {ECO:0000250|UniProtKB:Q6AZR2, ECO:0000250|UniProtKB:Q8BGK2}. | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome | ADP-ribosylation is a reversible posttranslational modification used to regulate protein function. ADP-ribosyltransferases (see ART1; MIM 601625) transfer ADP-ribose from NAD+ to the target protein, and ADP-ribosylhydrolases, such as ADPRHL1, reverse the reaction (Glowacki et al., 2002 [PubMed 12070318]).[supplied by OMIM, Mar 2008]. | hsa:113622; | sarcomere [GO:0030017]; ADP-ribosylarginine hydrolase activity [GO:0003875]; magnesium ion binding [GO:0000287]; cardiac chamber ballooning [GO:0003242]; cardiac myofibril assembly [GO:0055003]; protein de-ADP-ribosylation [GO:0051725] | 27217161_Hybrid human-Xenopus Adprhl1 protein constructs were produced by utilizing restriction sites from one species and engineering the same site into the second species by PCR. Hearts containing human ADPRHL1 developed normally. Engineered human-Xenopus hybrid proteins cause extensive branching of myofibrils with sarcomere division occurring at the actin-Z-disc boundary. 34528284_Non-genetic and genetic rewiring underlie adaptation to hypomorphic alleles of an essential gene. | ENSMUSG00000031448 | Adprhl1 | 141.483831 | 0.7343014 | -0.445555833 | 0.22664232 | 3.783310e+00 | 5.176617e-02 | No | Yes | 124.726408 | 20.740315 | 169.258585 | 27.982380 | ||
| ENSG00000154548 | 135295 | SRSF12 | protein_coding | Q8WXF0 | FUNCTION: Splicing factor that seems to antagonize SR proteins in pre-mRNA splicing regulation. {ECO:0000269|PubMed:11684676}. | Nucleus;RNA-binding;Reference proteome;mRNA processing;mRNA splicing | hsa:135295; | cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; RS domain binding [GO:0050733]; unfolded protein binding [GO:0051082]; mRNA 5'-splice site recognition [GO:0000395]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of mRNA splicing, via spliceosome [GO:0048025]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; spliceosomal tri-snRNP complex assembly [GO:0000244] | ENSMUSG00000054679 | Srsf12 | 344.948024 | 0.9817669 | -0.026547606 | 0.14425231 | 3.402960e-02 | 8.536439e-01 | No | Yes | 353.511001 | 50.408157 | 360.977398 | 51.517596 | ||||
| ENSG00000155100 | 51633 | OTUD6B | protein_coding | Q8N6M0 | FUNCTION: [Isoform 1]: Deubiquitinating enzyme that may play a role in the ubiquitin-dependent regulation of protein synthesis, downstream of mTORC1 (PubMed:21267069, PubMed:27864334). May associate with the protein synthesis initiation complex and modify its ubiquitination to repress translation (PubMed:27864334). May also repress DNA synthesis and modify different cellular targets thereby regulating cell growth and proliferation (PubMed:27864334). May also play a role in proteasome assembly and function (PubMed:28343629). {ECO:0000269|PubMed:21267069, ECO:0000269|PubMed:27864334, ECO:0000269|PubMed:28343629}.; FUNCTION: [Isoform 2]: Stimulates protein synthesis. Influences the expression of CCND1/cyclin D1 by promoting its translation and regulates MYC/c-Myc protein stability. {ECO:0000269|PubMed:27864334}. | Acetylation;Alternative splicing;Disease variant;Epilepsy;Hydrolase;Mental retardation;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | This gene encodes a member of the ovarian tumor domain (OTU)-containing subfamily of deubiquitinating enzymes. Deubiquitinating enzymes are primarily involved in removing ubiquitin from proteins targeted for degradation. This protein may function as a negative regulator of the cell cycle in B cells. [provided by RefSeq, Nov 2013]. | hsa:51633; | thiol-dependent deubiquitinase [GO:0004843]; cell population proliferation [GO:0008283]; negative regulation of translation [GO:0017148]; positive regulation of translation [GO:0045727]; proteasome assembly [GO:0043248]; protein deubiquitination [GO:0016579] | 21267069_down-regulation of Otud-6b expression after prolonged cytokine stimulation may be required for cell proliferation in B lymphocytes 27864334_The global OTUD6B expression level does not change significantly between nonneoplastic and malignant tissues, suggesting that modifications of splicing factors during the process of transformation are responsible for this isoform switch. 28343629_OTUD6B encodes a deubiquitinating enzyme; study reports biallelic pathogenic variants in OTUD6B in 12 individuals from 6 families with intellectual disability syndrome associated with seizures & dysmorphic features; other features include developmental delay, microcephaly, absent speech, hypotonia, growth retardation, feeding difficulties, structural brain abnormalities, malformations of heart & musculoskeleton. 31156645_OTUD6B-AS1 Might Be a Novel Regulator of Apoptosis in Systemic Sclerosis. 32323143_DUB-independent regulation of pVHL by OTUD6B suppresses hepatocellular carcinoma. 34680978_Compound Heterozygote of Point Mutation and Chromosomal Microdeletion Involving OTUD6B Coinciding with ZMIZ1 Variant in Syndromic Intellectual Disability. 35110537_Deubiquitylase OTUD6B stabilizes the mutated pVHL and suppresses cell migration in clear cell renal cell carcinoma. | ENSMUSG00000040550 | Otud6b | 1006.227543 | 1.1203472 | 0.163945862 | 0.09260279 | 3.154789e+00 | 7.570448e-02 | 3.682151e-01 | No | Yes | 1053.700130 | 214.966836 | 932.831233 | 190.380074 | |
| ENSG00000155380 | 6566 | SLC16A1 | protein_coding | P53985 | FUNCTION: Proton-coupled monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate. Depending on the tissue and on cicumstances, mediates the import or export of lactic acid and ketone bodies. Required for normal nutrient assimilation, increase of white adipose tissue and body weight gain when on a high-fat diet. Plays a role in cellular responses to a high-fat diet by modulating the cellular levels of lactate and pyruvate, small molecules that contribute to the regulation of central metabolic pathways and insulin secretion, with concomitant effects on plasma insulin levels and blood glucose homeostasis. {ECO:0000269|PubMed:17701893}. | 3D-structure;Alternative splicing;Cell membrane;Disease variant;Membrane;Phosphoprotein;Reference proteome;Symport;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is a proton-linked monocarboxylate transporter that catalyzes the movement of many monocarboxylates, such as lactate and pyruvate, across the plasma membrane. Mutations in this gene are associated with erythrocyte lactate transporter defect. Alternatively spliced transcript variants have been found for this gene.[provided by RefSeq, Oct 2009]. | hsa:6566; | apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; cell junction [GO:0030054]; centrosome [GO:0005813]; extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; lateral plasma membrane [GO:0016328]; membrane [GO:0016020]; plasma membrane [GO:0005886]; synapse [GO:0045202]; carboxylic acid transmembrane transporter activity [GO:0046943]; identical protein binding [GO:0042802]; lactate transmembrane transporter activity [GO:0015129]; mevalonate transmembrane transporter activity [GO:0015130]; monocarboxylic acid transmembrane transporter activity [GO:0008028]; organic cyclic compound binding [GO:0097159]; symporter activity [GO:0015293]; behavioral response to nutrient [GO:0051780]; carboxylic acid transmembrane transport [GO:1905039]; cellular response to organic cyclic compound [GO:0071407]; centrosome cycle [GO:0007098]; glucose homeostasis [GO:0042593]; lipid metabolic process [GO:0006629]; mevalonate transport [GO:0015728]; monocarboxylic acid transport [GO:0015718]; plasma membrane lactate transport [GO:0035879]; regulation of insulin secretion [GO:0050796]; response to food [GO:0032094]; transport across blood-brain barrier [GO:0150104] | 11882670_substrate-induced regulation of human colonic MCT1. The basis of this regulation is a butyrate-induced increase in mRNA abundance, resulting from the dual control of gene transcription and stability of the transcript 11944921_the structural organization of the human MCT1 gene and 5'-flanking region which contains potential binding sites for a variety of transcription factors with known association with butyrate's action in the colon 11953883_Carcinoma samples displaying reduced levels of MCT1 were found to express the high affinity glucose transporter, GLUT1, suggesting that there is a switch from butyrate to glucose as an energy source in colonic epithelia during transition to malignancy 12479094_MCT1 and NHE1 genes play important regulation roles in proliferation and growth of tumor cells, probably by affecting pHi. 12759536_MCT1 is involved in cellular transportation of butyrate, which induces cellular differentiation. Down-regulation is suggested to be involved in human breast cancers. 12949353_MCT mediates biotin uptake in human lymphoid cells. 14724187_MCT1 content in skeletal muscle in Type 2 diabetes is lower compared with healthy men. Strength training increases MCT1 content in healthy men and in Type 2, thus normalizing the content in Type 2. 15135232_studies demonstrate that the opposing plasma membranes of human syncytiotrophoblast are polarized with respect to both monocarboxylate transporter MCT1 and MCT4 activity and expression 15765403_These results show the importance of MCT1 to the ability of butyrate to induce cell-cycle arrest and differentiation, and suggest fundamental differences in the mechanisms by which butyrate modulates specific aspects of cell function. 15804185_MCT1 is functionally active and is the only MCT isoform involved in the apical uptake of monocarboxylates by retinal pigmented epithelial ARPE-19 cells 15901598_Distinct MCT isoforms may be involved in short-chain fatty acid transport across the apical or basolateral membranes in polarized colonic epithelial cells. 16136905_MCT1 gene plays an important role in pHi regulation, lactate transport and cell growth in tumor cells. 16150873_studies demonstrate inhibition of MCT1-mediated butyrate uptake in Caco-2 cells in response to enteropathogenic Escherichia coli infection 16370372_The inhibition of MCT1 during T lymphocyte activation results in selective and profound inhibition of the extremely rapid phase of T cell division essential for an effective immune response. 16403470_Monocarboxylate transporter MCT1 is strongly expressed by glial cells often associated with blood vessels that were identified as astrocytes. (Monocarboxylate transporter MCT1) 16408234_this study shows conflicting adaptations in MCT1 and MCT4 protein and mRNA levels following training, which may indicate post-transcriptional regulation of MCT expression in human muscle. 16441976_demonstrated greater activity of the RBC monocarboxylate cotransporter MCT-1, lower RBC deformability and impaired hematological indices in sickle cell trait (SCT) carriers compared to control subjects 17016429_Targeting the Map kinase signal transduction cascademay provide a potential therapeutic approach in lymphomas and related malignancies that exhibit high levels of the MCT-1 protein. 17082373_A single bout of high-intensity exercise decreased both MCT relative abundance (MCT1 and MCT4) in membrane preparations. 17182800_DL-2-Hydroxy-(4-methylthio)butanoic acid is transported into the colonic cancer cell line cell membrane by a transport mechanissm involving MCT1. 17701893_These studies show that promoter-activating mutations in exercise-induced hyperinsulinism induce SLC16A1 expression in beta cells, permitting pyruvate uptake and pyruvate-stimulated insulin release despite ensuing hypoglycemia. 18079261_In obesity, MCT1 expression appears linked both to changes in oxidative parameters and to changes in visceral adipose tissue content. 18188595_Expression of MCT1 and MCT4 showed a significant gain in plasma membranes of colorectal neoplasms. 18539591_the effective movement of H(+) into the bulk cytosol is increased by CAII, thus slowing the dissipation of the H(+) gradient across the cell membrane, which drives MCT1 activity 18832090_Report effects of high-intensity training on muscle MCT1/4 and postexercise recovery of muscle lactate and hydrogen ions in women. 19033536_PKC-zeta dependent stimulation of the human MCT1 promoter involves transcription factor AP2. 19033663_MCT1 was found to be expressed by an array of primary human tumors, we suggest that MCT1 inhibition has clinical antitumor potential. 19427019_miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. 19850519_carriers of the A1470T polymorphism in the MTC1 (monocarboxylate transporter 1) gene exhibit a worse lactate transport capability 19876643_Data suggest that hypoxia increases lactate release from adipocytes and modulates MCT expression in a type-specific manner, with MCT1 and MCT4, but not MCT2 expression, being hypoxia-inducible transcription factor-1 (HIF-1) dependent. 19881260_Observational study of genotype prevalence. (HuGE Navigator) 19881260_the -363-855T>C is a novel mutation. The 1282G>A (Val(428)Ile) is a novel SNP and was found as heterozygotic in 4 subjects. The 1470T>A (Asp(490)Glu) was found to be a common polymorphism in this study. 19898482_Observational study of gene-disease association. (HuGE Navigator) 19905008_close correlation between the [Ca2+](in) level and pH(in), and NHEs were involved with the MCT mediated uptake process 19913121_Observational study of gene-disease association. (HuGE Navigator) 20035863_Monocarboxylic acid transporter 1 was expressed in the human corneal epithelium at mRNA and protein levels. 20454640_MCT1, MCT2, and MCT4 protein expression in breast, colon, lung, and ovary neoplasms, as well as CD147 and CD44, were analysed. 20501436_Increased MCT1 association with CD147 at the apical membrane in response to somatostatin (SST) is p38 MAP kinase dependent and underlies the stimulatory effects of SST on butyrate uptake in human intestinal epithelial cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20636790_results provide evidence for a prognostic value of MCT1 in breast carcinoma 20877624_Observational study of gene-disease association. (HuGE Navigator) 21081165_Loss of MCT1 on brain microvessels is mechanistically involved in the pathophysiology of drug-resistant temporal lobe epilepsy; re-expression of MCT1 may represent a novel therapeutic approach for this disease. 21306479_We found that the expression of the monocarboxylate transporters MCT1 and MCT4, but not MCT5, in human lung cancer cell lines was significantly correlated with invasiveness 21646425_miR-29a, miR-29b selectively target MCT1 3'UTR ; the miR-29 isoforms are highly expressed in islets and contribute to silencing Mct1 in beta cells; miR-29 isoforms contribute to beta-cell-specific silencing of the MCT1 transporter and may affect insulin release 21787388_Data show that a significant increase of MCT2 and MCT4 expression in the cytoplasm of tumour cells and a significant decrease in both MCT1 and CD147 expression in prostate tumour cells was observed when compared to normal tissue. 21870331_both MCT1 and CD147, but not MCT4, were associated with GLUT1 and CAIX expression in a large series of invasive breast carcinoma samples 21930917_CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. 21987487_Reduced SLC16A1 is associated with impaired butyrate oxidation in ulcerative colitis. 22184616_our findings identify MCT1 as a target for p53 repression and they suggest that MCT1 elevation in p53-deficient tumors allows them to adapt to metabolic needs by facilitating lactate export or import depending on the glucose availability. 22281667_Co-expression of monocarboxylate transporter 1 (MCT1) and its chaperone (CD147) is associated with low survival in patients with gastrointestinal stromal tumors. 22428047_The lactate transporter monocarboxylate transporter 1 (MCT1) is the main regulator of HIF-1 activation by lactate in endothelial cells. 22516692_The data suggested that MCT1 polymorphism influenced lactate transport across sarcolemma in males but not in females. 23073708_Data suggest that MCT1 is expressed in insulin-secreting cells; overexpression of MCT1 insulin-secreting cells (1 patient versus 3 organ donors) can possibly lead to exercise-induced hyperinsulinaemic hypoglycemia. [CASE REPORT] 23187830_Combined application of GLUT-1, MCT-1, and MCT-4 immunohistochemistry might be useful in differentiating malignant pleural mesothelioma from reactive mesothelial hyperplasia. 23574725_MCT1 and MCT4 biomarkers were employed to determine the metabolic state of proliferative cancer cells. 23628675_MCT1 gene A1470T polymorphism is associated with endurance athlete status and blood lactate level after intensive exercise. 24012639_MCT1 inhibition and or gene knockdown has antitumor potential associated with the nuclear factor kappa B pathway. 24013424_MCT1 is an efficient lactate exporter in MM cells and plays a role in tumor growth within an acidic microenvironment. 24166504_Study identifies the mitochondrion as a glucose sensor promoting tumor cell migration, MCT1 is also revealed as a transducer of this response, providing a new rationale for the use of MCT1 inhibitors in cancer. 24277449_Small cell lung cancer patients with tumors expressing MCT1 and lacking in MCT4 are most likely to respond to the antineoplastic agent AZD3965. 24338019_Coexpression of CAIV with MCT1 and MCT4 resulted in a significant increase in MCT transport activity. 24485392_The findings indicated that theMCT1 TT genotype was overrepresented in sprint/power athletes compared to both endurance athletes and non-athlete controls. 24486553_Both L- and D-CysNO also inhibited cellular pyruvate uptake and caused S-nitrosation of thiol groups on monocarboxylate transporter 1, a proton-linked pyruvate transporter 24708341_T-allele frequency was significantly higher among swimmers compared with runners. 24797797_These results indicate there are no additional benefits of IHT when compared to similar normoxic training. Hence, the addition of the hypoxic stimulus on anaerobic performance or MCT expression after a three-week training period is ineffective. 24858043_Study demonstrated that PTEN loss and MCT-1 induction synergistically promoted the neoplastic multinucleation via the Src/p190B signaling activation. 25068893_Data indicate that pteridine showed substantial anticancer activity in monocarboxylate transporter 1 (MCT1)-expressing Raji lymphoma cells. 25225794_We provide novel information of MCT1 as a candidate marker for prognostic stratification in NSCLC. 25241983_We demonstrate, for the first time, that 3 different pHi regulators responsible for acid extruding, i.e. NHE and NBC, and MCT, are functionally co-existed in cultured radial artery smooth muscle cells. 25263481_The possible cooperative role of CD147 and MCT1 in determining cisplatin resistance. 25371203_H(+)-coupled 5-oxoproline transport is mediated solely by SLC16A1 in astrocytes 25390740_exome sequencing in a patient with ketoacidosis and identified a homozygous frameshift mutation in the gene SLC16A1. Genetic analysis in 96 patients suspected of having ketolytic defects yielded seven additional inactivating mutations in MCT1 25447800_MCT1 was up-regulated by exposure to DL-2-hydroxy-(4-methylthio)butanoic acid(HMTBA). Moreover, total monolayer MCT1 immunoreactivity increased 1.8-fold in HMTBA-supplemented cultures, this effect mainly being localised at the apical membrane. 25456395_Overexpression of MCT1, MCT4, and CD147 predicts tumor progression in clear cell renal cell carcinoma. 25492048_SNPs in MCT1 and MCT2 genes may affect clinical outcomes and can be used to predict the response to adjuvant chemotherapy in CRC patients who received surgical treatment 25578492_Our findings suggest that SNPs in MCT1 and MCT2 genes may affect clinical outcomes and can be used to predict the response to adjuvant chemotherapy in NSCLC patients who received surgical treatment once validated in future study. 25656974_Data suggest that expression of MCT1 in intestinal mucosa can be altered by diet; here, expression of MCT1 is down-regulated in Caco-2 Cells by products of fermentation of dietary proteins by intestinal microbes. 25755717_CD147 interacts with MCT1 to regulate tumor cell glycolysis, resulting in the progression of thyroid carcinoma. 25957999_The first IF method has been developed and optimised for detection of MCT 1 and MCT4 in cancer patient circulating tumour cells 26024713_interaction of rmEMMPRINex with U937 cells leads to inhibition of MCT1 membrane expression, intracellular activation of procaspase-9, followed by DNA fragmentation and apoptosis. 26045776_Down regulation of MCT1 promote the sensitivity to cisplatin in ovarian cancer cells. This effect appeared to be mediated by antagonizing Fas. 26340466_The aim of this study was to investigate the association between the MCT1 A1470T polymorphism and fat-free mass in young Italian elite soccer players. The MCT1 T allele is associated with the percentage of fat-free mass in young elite male soccer players. 26539827_MCT1 and GLUT1 may be potential prognostic markers in adenocarcinoma. 26563366_MCT1 expression associates with the SCC type and metastatic behavior of AC, whereas MCT4 expression concomitantly increases from in situ SCC to invasive SCC and is significantly associated with the AC type. Consistently, FOXM1 expression is statistically associated with MCT1 positivity in SCC, whereas the expression of FOXO3a, a FOXM1 functional antagonist, is linked to MCT1 negativity 26595136_The most important finding from the present study was that wild-type allele carriers, 2917(1414)T and IVS3-17C, have been associated with higher blood lactate levels at submaximal intensities of exercise for the first time. 26615136_MCT1 is highly expressed in anaplastic thyroid cancer compared to non-cancerous thyroid tissue and papillary thyroid cancer. 26765963_Data suggest that inhibition of mnocarboxylate transporters MCT1 and MCT4 may have clinical relevance in pancreatic ductal adenocarcinoma (PDAC). 26779534_MCT1 expression was not clearly associated with overall or disease-free survival. MCT4 and CD147 expression correlate with worse prognosis across many cancer types. These results warrant further investigation of these associations. 26854723_MCT1 may be acting as an uptake transporter and MCT4 as an efflux system across the basolateral membrane for ferulic acid, and that this process is stimulated by butyric acid. 26876179_MCT1 inhibition impairs proliferation of glycolytic breast cancer cells co-expressing MCT1 and MCT4 via disruption of pyruvate rather than lactate export. 26944480_The reversible H(+)/lactate(-) symporter MCT1 cotransports lactate and proton, leading to the net extrusion of lactic acid in glycolytic tumors. A model of its role in pH control in tumor cells is described. Review. 27026015_MCT1 affects the plasma lactate decrease . 27105345_MCT1 and MCT4 expression levels were associated with worse prognosis and shorter overall survival. 27127175_MCT1 expression, independent of transporter activity, is required for growth factor-induced tumor cell motility. 27144334_our finding that the expression of MCT1 and MCT4 is reduced in mutant IDH1 gliomas highlights the unusual metabolic reprogramming that occurs in mutant IDH1 tumors and has important implications for our understanding of these tumors and their treatment 27240355_Results found that MCT contributes to endothelial cell growth and tube formation via up-regulation of angiopoietin-1 expression suggesting that MCT plays an important role in pancreatic cancer angiogenesis and tumor growth via activating the angiopoietin-1 pathway. 27331625_Hypoxia-induced MCT1 supports glioblastoma glycolytic phenotype, being responsible for lactate efflux and an important mediator of cell survival and aggressiveness 27373212_The targeting of MCT1 and PFKFB3 regulated cell proliferation. 27634412_This study investigated MCT1 protein abundance in various human intestinal tissues. 27813046_AA genotype of the MCT1 T1470A polymorphism is over-represented in wrestlers compared with controls and is associated with lower blood lactate concentrations after 30-s Wingate Anaerobic test and during intermittent sprint tests in Japanese wrestlers 27957817_Monocarboxylate transporters (MCTs) are expressed in normal and cancer cells and are involved in cell metabolism and survival. 28099149_Increased miR-210 and concomitant decreased ISCU RNA levels were found in ~40% of tumors and this was significantly associated with HIF-1alpha and CAIX, but not MCT1 or MCT4, over-expression. 28235486_Data show that metastasis-associated in colon cancer-1 (MACC1) and monocarboxylate transporter 1 (MCT1) are highly expressed in gastric cancer indicating poor prognosis. 28260082_After indirect co-culture, OP was increased in the BxPc-3 and Panc-1 cells; correspondingly, succinate dehydrogenase, FH and MCT expression were increased. After the MCT1-specific inhibitor removed 'tumor-stromal' metabolic coupling, the migration and invasion abilities of the pancreatic cancer cells were decreased. 28559188_The structures and functions of hMCT1 and hMCT4 transporters. 28762551_Data suggest that targeting monocarboxylate transporter 1 (MCT1) in both tumor cells and brain endothelial cells (EC) may be a promising therapeutic strategy for the treatment of Glioblastoma (GBM). 28827372_Silencing or genetic deletion of MCT1 in vivo inhibited migration, invasion, and spontaneous metastasis. 28846107_Metabolism-dependent clonal growth of HCT15 colorectal cancer cells was induced by Nrf2-dependent activation of MCT1-driven lactate exchange. 28923861_MCT1 inhibitor AZD3965 increased MCT4-dependent accumulation of intracellular lactate, inhibiting monocarboxylate influx and efflux. 29066459_Using in vitro models, we demonstrate that tumor-excreted branched-chain amino acid (BCKA)s can be taken up and re-aminated to BCAAs by tumor-associated macrophages. Our data further suggest that the anti-proliferative effects of MCT1 knockdown observed by others might be related to the blocked excretion of BCKAs. 29248132_TOMM20 and MCT1 were highly expressed in diffuse large B-cell lymphoma lymphocytes, indicating an OXPHOS phenotype, whereas non-neoplastic lymphocytes in the control samples did not express these markers. 29248133_TOMM20, MCT1, and MCT4 expression was significantly different in Hodgkin and Reed Sternberg (HRS) cells. HRS have high expression of MCT1, while tumor associated macrophages have absent MCT1 expression. Tumor-infiltrating lymphocytes have absent MCT1 expression. Reactive lymph nodes in contrast to cHL tumors had low TOMM20, MCT1, and MCT4 expression in lymphocytes and macrophages. 29351758_Activation of autophagy can promote metastasis and glycolysis in HCC cells, and autophagy induces MCT1 expression by activating Wnt/beta-catenin signaling. 29481555_Study demonstrated that the high mRNA level of both MCT1 and GLUT1 correlated with poor prognosis, high- Fuhrman grade clear-cell renal cell carcinoma and metabolic reprogramming. 29572438_7ACC2 is an inhibitor of mitochondrial pyruvate transport; the blockade of pyruvate import into mitochondria prevents extracellular lactate uptake as efficiently as a MCT1 inhibitor 29657088_High MCT-1 expression is associated with clear cell renal cell carcinoma. 29775610_These results demonstrate that Monocarboxylate transporters tend to play a role in the aggressive breast cancer subtypes through the dynamic interaction between breast cancer cells and adipocytes. 29809145_Study shows that proton-driven lactate flux is enhanced by the intracellular carbonic anhydrase CAII, which is colocalized with the monocarboxylate transporter MCT1 in MCF-7 breast cancer cells; the results suggest that CAII features a moiety that exclusively mediates proton exchange with the MCT to facilitate transport activity. 29882205_MCT1 might act as a new regulator to improve invasion and migration of NPC cells and be correlated with activating the PI3K/Akt pathway. 29985759_Sub-Saharan African groups show extremely high values of the T allele of 1470T > A polymorphism. The TT genotype preeminence in African groups could explain the better predisposition to sprint/power performances of African athletes. Caucasian and Asian populations show variable proportions of TT and AA genotypes allowing inter-individual differences in lactate transport. 30115973_Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer 30290372_the MCT1 A allele is associated with forward soccer player status 30655616_MCT1 expression was significantly increased in HPV-negative tumours, and inhibition suppressed tumour cell invasion, colony formation and promoted radiosensitivity. HPV-positive and negative head and neck squamous cell carcinoma (HNSCC) have different metabolic profiles which may have potential therapeutic applications. 30720131_MCT1 mRNA expression in esophageal squamous cell carcinoma (ESCC) was analyzed in The Cancer Genome Atlas database and in ESCC cells. MCT1 expression was found to correlate with neoplasm stage. Survival analysis showed that patients in a highMCT1 group had a lower overall and progressionfree survival. Downregulation of MCT1 suppressed proliferation and survival of ESCC cells in vitro. 31101938_we were interested in the molecular mechanism responsible for the difference in substrate specificity between hMCT1 and hMCT4. Therefore, we generated 3D structure models of hMCT1 and hMCT4 to identify amino acid residues involved in the substrate specificity of these transporters. We found that the substrate specificity of hMCT1 was regulated by residues involved in turnover number (M69) and substrate affinity 31209810_Lactate promoted Wnt activity and increased the expression of CD133 cancer stem cell organoids. Silencing MCT1, the prominent path for lactate uptake in human tumor with siRNA significantly impaired organoid forming capacity of oral squamous cell carcinoma cells. 31395464_Monocarboxylate transporters in cancer. 31404019_Monocarboxylate transporter 1 (MCT1) expression is associated with better prognosis and reduced nodal metastasis in pancreatic cancer. 31564440_Hyperpolarized MRI of Human Prostate Cancer Reveals Increased Lactate with Tumor Grade Driven by Monocarboxylate Transporter 1. 31593685_K38 plays an essential role in hMCT1 transport activity 31605138_Neural crest-derived tumor neuroblastoma and melanoma share 1p13.2 as susceptibility locus that shows a long-range interaction with the SLC16A1 gene. 31678436_The concentration gradients (Api - BL) of glucose and lactate correlated with the gene expression of respective SLC2A1 and SLC16A1 transporters. 31723238_CAIX forms a transport metabolon with monocarboxylate transporters in human breast cancer cells. 31727576_Relationships between plasma lactate, plasma alanine, genetic variations in lactate transporters and type 2 diabetes in the Japanese population. 31729681_CD147 augmented monocarboxylate transporter-1/4 expression through modulation of the Akt-FoxO3-NF-kappaB pathway promotes cholangiocarcinoma migration and invasion. 31853067_MCT1 inhibition had little effect on the growth of primary subcutaneous tumours 32111097_Monocarboxylate Transporter 1 (MCT1) in Liver Pathology. 32123312_Targeting metabolic activity in high-risk neuroblastoma through Monocarboxylate Transporter 1 (MCT1) inhibition. 32703414_Identification of the HT-29 cell line as a model for investigating MCT1 transporters in sigmoid colon adenocarcinoma. 32748357_SLC16A1-AS1 enhances radiosensitivity and represses cell proliferation and invasion by regulating the miR-301b-3p/CHD5 axis in hepatocellular carcinoma. 32761447_Delayed Myelination Pattern and an Abnormal Thyroid Profile Caused by a Novel Mutation in the SLC16A2 Gene. 32819565_Identification of the essential extracellular aspartic acids conserved in human monocarboxylate transporters 1, 2, and 4. 32839325_Hyperpolarized [1-(13)C]pyruvate-to-[1-(13)C]lactate conversion is rate-limited by monocarboxylate transporter-1 in the plasma membrane. 32863950_LncRNA-SLC16A1-AS1 induces metabolic reprogramming during Bladder Cancer progression as target and co-activator of E2F1. 32872409_Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. 32887638_Tissue expression of lactate transporters (MCT1 and MCT4) and prognosis of malignant pleural mesothelioma (brief report). 33212210_Immunoreactivity of receptor and transporters for lactate located in astrocytes and epithelial cells of choroid plexus of human brain. 33559958_Monocarboxylate transporter 1 promotes proliferation and invasion of renal cancer cells by mediating acetate transport. 33633176_Monocarboxylate transporter-1 (MCT1) protein expression in head and neck cancer affects clinical outcome. 33641091_Overexpression of Cell-Surface Marker SLC16A1 Shortened Survival in Human High-Grade Gliomas. 34022218_Excess exogenous pyruvate inhibits lactate dehydrogenase activity in live cells in an MCT1-dependent manner. 34077729_Therapy-induced DNA methylation inactivates MCT1 and renders tumor cells vulnerable to MCT4 inhibition. 34480633_Influence of the MCT1-T1470A polymorphism (rs1049434) on repeated sprint ability and blood lactate accumulation in elite football players: a pilot study. 34619707_Coexpression of MCT1 and MCT4 in ALK-positive Anaplastic Large Cell Lymphoma: Diagnostic and Therapeutic Implications. 34769160_miR-31-NUMB Cascade Modulates Monocarboxylate Transporters to Increase Oncogenicity and Lactate Production of Oral Carcinoma Cells. 35023320_A Novel Metabolic Reprogramming Strategy for the Treatment of Diabetes-Associated Breast Cancer. | ENSMUSG00000032902 | Slc16a1 | 18784.329743 | 0.9329642 | -0.100106301 | 0.04966455 | 4.037732e+00 | 4.449357e-02 | 2.849587e-01 | No | Yes | 18147.808232 | 3598.614026 | 19399.759293 | 3846.511224 | |
| ENSG00000155666 | 79831 | KDM8 | protein_coding | Q8N371 | FUNCTION: Bifunctional enzyme that acts both as an endopeptidase and 2-oxoglutarate-dependent monooxygenase (PubMed:28847961, PubMed:29459673, PubMed:28982940, PubMed:29563586). Endopeptidase that cleaves histones N-terminal tails at the carboxyl side of methylated arginine or lysine residues, to generate 'tailless nucleosomes', which may trigger transcription elongation (PubMed:28847961, PubMed:29459673, PubMed:28982940). Preferentially recognizes and cleaves monomethylated and dimethylated arginine residues of histones H2, H3 and H4. After initial cleavage, continues to digest histones tails via its aminopeptidase activity (PubMed:28847961, PubMed:29459673). Upon DNA damage, cleaves the N-terminal tail of histone H3 at monomethylated lysine residues, preferably at monomethylated 'Lys-9' (H3K9me1). The histone variant H3F3A is the major target for cleavage (PubMed:28982940). Additionnally, acts as Fe(2+) and 2-oxoglutarate-dependent monooxygenase, catalyzing (R)-stereospecific hydroxylation at C-3 of 'Arg-137' of RPS6 and 'Arg-141' of RCCD1, but the biological significance of this activity remains to be established (PubMed:29563586). Regulates mitosis through different mechanisms: Plays a role in transcriptional repression of satellite repeats, possibly by regulating H3K36 methylation levels in centromeric regions together with RCCD1. Possibly together with RCCD1, is involved in proper mitotic spindle organization and chromosome segregation (PubMed:24981860). Negatively regulates cell cycle repressor CDKN1A/p21, which controls G1/S phase transition (PubMed:24740926). Required for G2/M phase cell cycle progression. Regulates expression of CCNA1/cyclin-A1, leading to cancer cell proliferation (PubMed:20457893). Also, plays a role in regulating alpha-tubulin acetylation and cytoskeletal microtubule stability involved in epithelial to mesenchymal transition (PubMed:28455245). Regulates the circadian gene expression in the liver (By similarity). Represses the transcriptional activator activity of the CLOCK-ARNTL/BMAL1 heterodimer in a catalytically-independent manner (PubMed:30500822). Negatively regulates the protein stability and function of CRY1; required for AMPK-FBXL3-induced CRY1 degradation (PubMed:30500822). {ECO:0000250|UniProtKB:Q9CXT6, ECO:0000269|PubMed:20457893, ECO:0000269|PubMed:24740926, ECO:0000269|PubMed:24981860, ECO:0000269|PubMed:28455245, ECO:0000269|PubMed:28847961, ECO:0000269|PubMed:28982940, ECO:0000269|PubMed:29459673, ECO:0000269|PubMed:29563586, ECO:0000269|PubMed:30500822}. | 3D-structure;Alternative splicing;Aminopeptidase;Biological rhythms;Cell cycle;Chromatin regulator;Chromosome;Dioxygenase;Hydrolase;Iron;Metal-binding;Nucleus;Oxidoreductase;Protease;Reference proteome;Transcription;Transcription regulation | This gene likely encodes a histone lysine demethylase. Studies of a similar protein in mouse indicate a potential role for this protein as a tumor suppressor. Alternatively spliced transcript variants have been described.[provided by RefSeq, Feb 2009]. | hsa:79831; | chromosome [GO:0005694]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 2-oxoglutarate-dependent dioxygenase activity [GO:0016706]; aminopeptidase activity [GO:0004177]; chromatin binding [GO:0003682]; endopeptidase activity [GO:0004175]; histone H3-methyl-lysine-36 demethylase activity [GO:0051864]; metal ion binding [GO:0046872]; methylated histone binding [GO:0035064]; peptidyl-arginine 3-dioxygenase activity [GO:0106157]; chromatin organization [GO:0006325]; circadian regulation of gene expression [GO:0032922]; G2/M transition of mitotic cell cycle [GO:0000086]; histone H3-K36 demethylation [GO:0070544]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription, DNA-templated [GO:0045893]; protein destabilization [GO:0031648] | 16858412_Identifies five novel candidate tumor suppressor genes in mouse, including Pou2f2, Hivep3, Jmjd5, Fbxl10 and a protein similar to human N4BP3. 20457893_show that JMJD5 (now renamed KDM8), demethylates H3K36me2 and is required for cell cycle progression. 21115819_Data show that both Arabidopsis jmjd5 mutant seedlings and mammalian cell cultures deficient for the human ortholog of this gene have similar fast-running circadian oscillations compared with WT. 22375008_JMJD5 is a post-translational co-repressor for NFATc1 that attenuates osteoclastogenesis. 22851697_The study reports high-resolution crystal structures of the human JMJD5 catalytic domain in complex with the substrate 2-oxoglutarate (2-OG) and the inhibitor N-oxalylglycine (NOG). 23948433_These results reveal that the N-terminal domain is essential for the nuclear localization of JMJD5 and its normal enzymatic function towards substrates in the nucleus 24100311_Comparison of the structure of JMJD5 with that of FIH, a well characterized protein hydroxylase, reveals that human JMJD5 might function as a protein hydroxylase. 24344305_JMJD5 has a role in regulating PKM2 nuclear translocation and reprogramming HIF-1alpha-mediated glucose metabolism 24740926_we provide genetic and biochemical evidence that the JMJD5/CDKN1A (p21) axis is essential to maintaining the short G1 phase which is critical for pluripotency in human embryonic stem cells 24981860_RCCD1 and KDM8 form a histone demethylase complex. 26025680_results reveal that JMJD5 is a novel binding partner of p53 and it functions as a positive modulator of cell cycle and cell proliferation mainly through the repression of p53 pathway. 26261525_Suggest JMJD5 as a potential oncogene in colon carcinogenesis. 26710852_Data suggest that JMJD5 partially accumulates on mitotic spindles during mitosis; depletion of JMJD5 results in significant mitotic arrest, spindle assembly defects, and sustained activation of spindle assembly checkpoint. 26760772_JMJD5 is a tumor suppressor gene in HCC pathogenesis, and the epigenetic silencing of JMJD5 promotes HCC cell proliferation by directly down-regulating CDKN1A transcription. 26792738_These results suggest that direct interaction of JMJD5 with HBx facilitates hepatitis B virus replication through the hydroxylase activity of JMJD5. 27715397_JMJD5 depletion increases the susceptibility of cancer cells to microtubule-destabilizing agents. 28847961_The protease activities of JMJD5 and JMJD7 represent a mechanism for removal of histone tails bearing methylated arginine residues and define a potential mechanism of transcription regulation. 28982940_Here, we report that JMJD5, a Jumonji C (JmjC) domain-containing protein, is a Cathepsin L-type protease that mediates histone H3 N-tail proteolytic cleavage under stress conditions that cause a DNA damage response. 29459673_Recognition between JMJD5/JMJD7 and histone substrates is specific, which is reflected by the binding data between enzymes and substrates. High structural similarity between JMJD5 and JMJD7 is reflected by the shared common substrates and high binding affinity. 29563586_JMJD5 catalyzes stereoselective C-3 hydroxylation of arginine residues in sequences from human RCCD1 and ribosomal protein S6. 30072740_KDM8/JMJD5exhibits a novel property as a dual coactivator of AR and PKM2 and as such, it is a potent inducer of castration and therapy resistance.Its oncogenic properties in prostate cancer come from its direct interaction with AR to affect androgen response and with PKM2 to regulate tumor metabolism. 30551455_JMJD5 expression is increased in oral squamous cell carcinoma tissue samples and tumor cell lines. 32678829_Downregulation of Jumonji-C domain-containing protein 5 inhibits proliferation by silibinin in the oral cancer PDTX model. 32747552_JMJD5 couples with CDK9 to release the paused RNA polymerase II. 35176452_Jumonji-C domain-containing protein 5 suppresses proliferation and aerobic glycolysis in pancreatic cancer cells in a c-Myc-dependent manner. | ENSMUSG00000030752 | Kdm8 | 262.319966 | 0.9508165 | -0.072761109 | 0.16438180 | 1.960237e-01 | 6.579497e-01 | No | Yes | 232.427771 | 28.814221 | 249.713881 | 30.835058 | ||
| ENSG00000156096 | 7363 | UGT2B4 | protein_coding | P06133 | FUNCTION: UDP-glucuronosyltransferase (UGT) that catalyzes phase II biotransformation reactions in which lipophilic substrates are conjugated with glucuronic acid to increase the metabolite's water solubility, thereby facilitating excretion into either the urine or bile (PubMed:18719240, PubMed:23288867). Essential for the elimination and detoxification of drugs, xenobiotics and endogenous compounds (PubMed:18719240, PubMed:23288867). Catalyzes the glucuronidation of the endogenous estrogen hormones such as estradiol and estriol (PubMed:18719240, PubMed:23288867). {ECO:0000269|PubMed:18719240, ECO:0000269|PubMed:23288867}. | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Glycosyltransferase;Lipid metabolism;Membrane;Reference proteome;Signal;Transferase;Transmembrane;Transmembrane helix | hsa:7363; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; glucuronosyltransferase activity [GO:0015020]; cellular glucuronidation [GO:0052695]; estrogen metabolic process [GO:0008210] | 12806625_Farnesoid X receptor (FXR) induces the UGT2B4 enzyme in hepatocytes; this study identifies UGT2B4 as a novel FXR target gene. 12810707_UGT2B4 expression is regulated by PPARalpha 15318931_Observational study of gene-disease association. (HuGE Navigator) 15319348_Observational study of genotype prevalence. (HuGE Navigator) 15319348_P.1053: '...UGT2B4...is...fairly common in Caucasians...and Africans...'but...still rare in ...Japanese...' P.1054: '...genotyping UGT2B4...in Japanese...would be useful for studies on... association between haplotypes and pharmacokinetic...parameters.' 16513443_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16849011_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17263731_Results strongly indicated that the presence of an aromatic residue at position 33 is important for the activity and specificity of UGTB4. 18161889_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19898482_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20139797_Extensive splicing of transcripts of the bile acid-conjugating enzyme UGT2B4 modulates glucuronidation. 20142249_Observational study of gene-disease association. (HuGE Navigator) 20214802_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 21660508_The variation pattern upstream UGT2B4 is highly unusual and may be the result of balancing selection. 22092298_Methadone inhibits CYP2D6 and UGT2B7/2B4 in vivo 22367021_The UGT1A8 and UGT2B4 genotypes associated with decreased predicted enzyme activities, were significantly associated with an increased risk of esophageal squamous cell carcinoma. 25948711_These data indicate that knocking down PAPSS increases UGT2B4 transcription and mRNA stability as a compensatory response to the loss of SULT2A1 activity 27282283_UGT2B4 previously implicated in the risk of breast cancer is associated with menarche timing in Ukrainian females. 29138287_Clopidogrel carboxylic acid is metabolized mainly by UGT2B7 and UGT2B4 in the liver and by UGT2B17 in the small intestinal wall. 32170986_Absence of significant association between UGT2B4 genetic variants and the susceptibility to anti-tuberculosis drug-induced liver injury in a Western Chinese population. | 10.556306 | 1.0904236 | 0.124888671 | 0.82113144 | 2.119769e-02 | 8.842417e-01 | No | Yes | 10.576519 | 5.751463 | 8.694355 | 4.738801 | |||||
| ENSG00000156170 | 137682 | NDUFAF6 | protein_coding | Q330K2 | FUNCTION: Involved in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I) at early stages. May play a role in the biogenesis of complex I subunit MT-ND1. {ECO:0000269|PubMed:18614015, ECO:0000269|PubMed:22019594}. | Alternative splicing;Cytoplasm;Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Nucleus;Primary mitochondrial disease;Reference proteome;Transit peptide | This gene encodes a protein that localizes to mitochondria and contains a predicted phytoene synthase domain. The encoded protein plays an important role in the assembly of complex I (NADH-ubiquinone oxidoreductase) of the mitochondrial respiratory chain through regulation of subunit ND1 biogenesis. Mutations in this gene are associated with complex I enzymatic deficiency. [provided by RefSeq, Nov 2011]. | hsa:137682; | cytoplasm [GO:0005737]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; transferase activity [GO:0016740]; biosynthetic process [GO:0009058]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 22019594_C8orf38 is a crucial factor required for the translation and/or integration of ND1 into an early-stage assembly intermediate 23509070_In a forward genetic screen to identify genes that cause neurodegeneration, we identified sicily, the Drosophila melanogaster homologue of human C8ORF38, the loss of which causes Leigh syndrome. 27466185_Affected kidney and lung showed specific loss of the mitochondria-located NDUFAF6 isoform and ultrastructural characteristics of mitochondrial dysfunction. Accordingly, affected tissues had defects in mitochondrial respiration and complex I biogenesis that were corrected with NDUFAF6 cDNA transfection. Our results demonstrate that the Acadian variant of Fanconi Syndrome results from mitochondrial respiratory chain complex 27623250_This paper confirms NDUFAF6 as a genuine morbid gene and proposes the coupling of exome sequencing with mRNA analysis as a method useful for enhancing the exome sequencing detection rate when the simple application of classical inheritance models fails. 28476317_NDUFAF6 encodes a complex I assembly factor and mutations result in complex I deficiency, Leigh syndrome or Acadian variant Fanconi syndrome. Human NDUFAF6 is a mitochondria-targeted 333-amino acid protein belonging to the family of squalene and phytoene synthases. 30642748_NDUFAF6-related Leigh syndrome is a relevant cause of childhood onset dystonia and isolated bilateral striatal necrosis [review] 35237031_Genetic Effects of NDUFAF6 rs6982393 and APOE on Alzheimer's Disease in Chinese Rural Elderly: A Cross-Sectional Population-Based Study. | ENSMUSG00000050323 | Ndufaf6 | 665.730512 | 1.1672429 | 0.223104827 | 0.10677043 | 4.369046e+00 | 3.659747e-02 | 2.587274e-01 | No | Yes | 721.378040 | 97.797968 | 621.035083 | 84.213221 | |
| ENSG00000156218 | 57188 | ADAMTSL3 | protein_coding | P82987 | Alternative splicing;Disulfide bond;Extracellular matrix;Glycoprotein;Reference proteome;Repeat;Secreted;Signal | hsa:57188; | extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; intracellular membrane-bounded organelle [GO:0043231]; extracellular matrix organization [GO:0030198] | 14667842_Highest expression of ADAMTSL3 was found in liver, kidney, heart and skeletal muscle. 17597111_ADAMTSL3 is expressed in numerous tissues, suggesting a broader regulatory role than in colorectal epithelium alone, and that colorectal cancer has both structural mutations as well as decreased expression of ADAMTSL3. 18391952_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19266077_Observational study of gene-disease association. (HuGE Navigator) 20397748_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20546612_Observational study of gene-disease association. (HuGE Navigator) 21239144_The results of this study suggested that the ADAMTSL3 as a candidate gene for schizophrenia. 24793337_Compared with the George-Abraham' study [2012], ADAMTSL3 might be more related to the cardiac disorders in tetrasomy 15q patients 29162515_A137T in ADAMTSL3 may be a susceptibility mutation for diabetes. 32266537_knocking out either the ADAMTSL3 or PTEN genes promoted either the proliferation or metastasis of hepatocellular carcinoma cells, respectively | ENSMUSG00000070469 | Adamtsl3 | 32.171034 | 1.3915232 | 0.476664973 | 0.51814295 | 8.496615e-01 | 3.566481e-01 | No | Yes | 37.488135 | 13.370991 | 27.421028 | 9.713191 | ||||
| ENSG00000156313 | 6103 | RPGR | protein_coding | Q92834 | FUNCTION: Could be a guanine-nucleotide releasing factor. Plays a role in ciliogenesis. Probably regulates cilia formation by regulating actin stress filaments and cell contractility. Plays an important role in photoreceptor integrity. May play a critical role in spermatogenesis and in intraflagellar transport processes (By similarity). May be involved in microtubule organization and regulation of transport in primary cilia. {ECO:0000250, ECO:0000269|PubMed:21933838}. | 3D-structure;Alternative splicing;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Deafness;Disease variant;Flagellum;Golgi apparatus;Guanine-nucleotide releasing factor;Lipoprotein;Methylation;Phosphoprotein;Prenylation;Reference proteome;Repeat;Retinitis pigmentosa;Sensory transduction;Vision | This gene encodes a protein with a series of six RCC1-like domains (RLDs), characteristic of the highly conserved guanine nucleotide exchange factors. The encoded protein is found in the Golgi body and interacts with RPGRIP1. This protein localizes to the outer segment of rod photoreceptors and is essential for their viability. Mutations in this gene have been associated with X-linked retinitis pigmentosa (XLRP). Multiple alternatively spliced transcript variants that encode different isoforms of this gene have been reported, but the full-length natures of only some have been determined. [provided by RefSeq, Dec 2008]. | hsa:6103; | centrosome [GO:0005813]; ciliary basal body [GO:0036064]; Golgi apparatus [GO:0005794]; photoreceptor outer segment [GO:0001750]; sperm flagellum [GO:0036126]; guanyl-nucleotide exchange factor activity [GO:0005085]; RNA binding [GO:0003723]; cilium assembly [GO:0060271]; intracellular protein transport [GO:0006886]; intraciliary transport [GO:0042073]; response to stimulus [GO:0050896]; visual perception [GO:0007601] | 11754050_Insertional/deletional mutations observed in the three families with X-linked retinitis pigmentosa are all different and new, and are predicted to lead to a frameshift, resulting in a truncated protein. 11857109_X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15 11875055_A mutation in this gene causes X-linked cone dystrophy. This type of hereditary retinal degeneration is distinct from retinitis pigmentosa. 11992260_A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. 12140192_RPGR and RPGRIP isoforms are distributed and co-localized at restricted foci throughout the outer segments of human and bovine, but not mice rod photoreceptors. 12160730_Identification of an RPGR mutation in atrophic macular degeneration expands the phenotypic range from retinitis pigmentosa. 12657579_RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15. 14516808_Different RPGR mutations lead to distinct RP (retinitis pigmentosa) phenotypes, with a highly variable inter- and intrafamilial phenotypic spectrum of disease 14564670_Among patients with RPGR mutations, those with ORF15 mutations had, on average, a significantly larger visual field area and a borderline larger ERG amplitude than did patients with RPGR mutations in exons 1-14 14564670_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 14566651_Mutations in the RPGR gene is associated with X-linked retinitis pigmentosa 15364249_Sequencing revealed skipping of exon 2 in the mutated transcript, leading to in-frame deletion of 42 amino acids affecting the critical RCC1-like domain. 15772089_RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. 15914600_Two families were identified with nonsense mutations, and clinical evaluation revealed them both to have a similar phenotype. 16043481_RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. 16052169_This 30 kb deletion contains the exons coding for the RCC1-like domain of RPGR. It is the first report of a macrodeletion that spans the entire RCC1-like domain of RPGR in X-linked retinitis pigmentosa patients 16086276_Finding expands the spectrum of RPGR mutations causing X-linked dominant retinitis pigmentosa in Chinese family and is useful for further genetic consultation and genetic diagnosis. 16273303_Observational study of gene-disease association. (HuGE Navigator) 16273303_the mutational risk in the RPGR gene appears not to be altered by the haplotype background 16387007_This report expands the clinical heterogeneity spectrum caused by RPGR mutations and our knowledge concerning the molecular pathologic condition that pertains to Coats'-like RP. 16917484_We describe a novel PRPF31 mutation and present the first case of a homozygous mutation in the RPGR gene in a female individual. 16935610_A defect in trafficking of opsins to outer segments exists in a carrier with the RPGR Gly436Asp mutation. 16936086_In this cohort of XLRP families, as has happened in previous studies, RP3 also seems to be the most prevalent form of XLRP, and, based on the results, the authors propose a four-step protocol for molecular diagnosis of XLRP families. 16969763_26 new mutations in RP2 and RPGR patients with X-linked Retinitis Pigmentosa. 16969763_Observational study of genotype prevalence. (HuGE Navigator) 17093403_Three ORF15 mutations and one RP2 mutation in five Japanese retinitis pigmentosa families. 17195164_All disease-causing mutations occur in one or more RPGR isoforms containing the carboxyl-terminal exon open reading frame 15 (ORF15), which are widely expressed but show their highest expression in the connecting cilia of rod and cone photoreceptors. 17249551_the ORF15 and RCC1-like domain of RPGR play a crucial role in the human retina 17325176_Observational study of gene-disease association. (HuGE Navigator) 17325176_Patients with X-linked retinitis pigmentosa due to RPGR mutations lose visual acuity and visual field more rapidly than do patients with dominant retinitis pigmentosa due to RHO mutations. 17405150_The nucleotide substitution affects regulated alternative splicing of the novel RPGR isoform and suggests a tight adjustment of splicing as a prerequisite for proper function of photoreceptors. 17480003_These results indicate that an additional gene (or genes), linked to RPGR, modulate disease expression in severely affected carriers. 17724181_The proportion of RP2-mediated XLRP in the Danish population is higher and the proportion of RPGR-ORF15 is lower than reported in other studies. 17893654_Associated with retinitis pigmentosa in all hemizygous males and four of five heterozygous female carriers in Chinese family. These results revealed broader xlRP genotypic and phenotypic spectrum of RPGR mutations. 17898302_Inner retinal laminar abnormalities in RPGR-XLRP are likely to reflect a neuronal-glial retinal remodeling response to photoreceptor loss and are detectable relatively early in the disease course. 17923551_We identified a novel mutation in the 3' end of a highly repetitive region of exon open reading frame 15 (ORF15) and documented the detailed phenotypes of the patients with XLRP with the mutation. 18188948_Retinal phenotype of an X-linked pseudo-Usher syndrome in association with the G173R mutation in the RPGR gene is reported. 18332319_An identical mutation in RPGR-ORF15 manifested distinct clinical phenotypes in individuals of the same family. 19218993_This novel mutation in RPGRcauses X-Linked RP with complete penetrance in males and females and affected females are highly myopic but retain better visual function than affected males. 19429592_Four different RPGR ORF15 mutations were found in four probands and all mutations in the ORF15 exon resulted in premature truncation of the RPGR protein. 19815619_RPGR is involved in cilia-dependent cascades during development in zebrafish. 19834030_These findings show that splicing of RPGR is precisely regulated in a tissue-dependent fashion and suggest that mutations in RPGR frequently interfere with the expression of alternative transcript isoforms. 20021257_Our results expand the frequency and spectrum of mutations at RPGR and RP2 as well as their associated clinical phenotypes in Chinese patients. 20064120_The pedigree we have investigated here represents the first Czech family with an identified molecular genetic cause of retinitis pigmentosa 20090203_Studies highlight the recent developments in understanding the mechanism of cilia-dependent photoreceptor degeneration due to mutations in RPGR and PGR-interacting proteins in severe genetic diseases. 20238008_Recent advances on understanding the role of RPGR in photoreceptor protein trafficking, are summarized. 20574030_Differentially expressed genes were identified in mutant retinas. At 7 and 16 weeks, a combination of nonclassic anti- and proapoptosis genes and mitochondria-related genes appear to be involved in photoreceptor degeneration. 20591486_Observational study of genetic testing. (HuGE Navigator) 20631154_RPGR modulates intracellular localization and function of RAB8A. 20801516_Observational study of genetic testing. (HuGE Navigator) 20806050_The novel mutation in RPGR ORF15 causes a serious retinitis pigmentosa phenotype in males and noretinitis pigmentosa phenotype in female carriers. 21174525_Mutations in the RPGR gene lead to X-linked Retinitis pigmentosa (XLRP), one of the most severe and early onset forms of RP. Gene therapy is considered a potential therapeutic option and is currently under investigation. 21227725_a novel deletion mutation in the retinitis pigmentosa GTPase regulator gene, gORF15+556delA, in a Han Chinese family with retinitis pigmentosa 21683121_a micro-deletion through prenatal genetic diagnosis and another novel nonsense mutation in RPGR-ORF15. 21857984_Data show that the minor allele (N) of I393N in IQCB1 and the common allele (R) of R744Q in RPGRIP1L were associated with severe disease in XlRP with RPGR mutations. 21866333_Expression of RPGR mutations in this particular region appears to be relatively homogeneous and predisposed to cones. 21914266_This novel mutation in RPGR ORF15 causes serious retinitis pigmentosa phenotype in males and no RP phenotype in female carriers. 21933838_The function of RPGR was analysed by RNA interference-mediated translational suppression. 22183348_Genetic variation of RPGRIP1L and IQCB1 may affect severity in RPGR mutation X-linked retinitis pigmentosa. 22577079_the human RPGR proximal promoter region in which a 3-kb fragment contained sufficient regulatory elements to control RPGR expression in mouse retina and other tissues. 22888088_Mutations in RPGR were found in two patients and relatives with primary ciliary dyskinesia and retinitis pigmentosa. Reduced ciliary orientation and coordination of ciliary bundles suggest RPGR may play a role in respiratory cilia orientation. 23150612_Based on our findings, we suggest that RPGR should be considered as a first tier gene for screening isolated males with retinal degeneration. 23372056_Mutations in RPGR are one of the most common causes of all forms of retinitis pigmentosa. 23559067_RPGR is acting as a scaffold protein recruiting cargo-loaded PDE6D and Arl3 to release lipidated cargo into cilia. 23681342_While visual acuity and electroretinography phenotypes are concordant in only some patients carrying identical mutations, assessment of phenotypes revealed stronger phenotypic conservation. 24428633_Although carriers of XLRP are usually asymptomatic or have a mild disease of late onset, the proband presented here exhibited an early-onset, aggressive form of the disease. 24454928_RPGR mutations associated with X-linked retinitis pigmentosa. 24489377_A novel RPGR gene was found in a retinal dystrophy patient in a family with Stargardt disease. 24555744_Profound visual loss occurred by the second decade of life with progression to near no light perception by age 60 in this kindred of X-linked RP associated with the RPGR genotype. 24664734_We will summarize the functional characterization of RPGR and highlight recent studies in animal models, which will not only shed light on the disease mechanisms in X linked retinitis pigmentosa but will also provide therapeutic strategies for treatment. 25301933_X-linked retinitis pigmentosa caused by mutations in the RPGR gene is a severe and early onset form of retinal degeneration. [review] 25352739_Severe retinal degeneration is found in a Czech family women with a c.2543del mutation in ORF15 of the RPGR gene. 25556114_The edge of the ellipsoid zone in each patient with X-linked retinitis pigmentosa indicates a transition zone between relatively healthy and relatively degenerate retina. 25569437_Coverage-based analysis indicated that the RPGR open reading frame (ORF)15 was located in an uncovered or low-depth region. Through additional screening of ORF15, we identified pathogenic mutations in 14% (7/50) of patients. 26431479_The regulator of chromosome condensation 1-like domain of RPGR was conserved in vertebrates and invertebrates, but RPGR(ORF15) was unique to vertebrates. 26936822_Together with a physical interaction between RPGR and the C-terminal domain of CEP290, our data suggest that RPGR and CEP290 genetically interact and highlight the involvement of hypomorphic alleles of genes as potential modifiers of heterogeneous retinal ciliopathies. 27323122_Three mutations were identified in the ORF15 exon of RPGR. No RP2 mutations were found among the examined families. Mutation screening of RP patients is essential to understand the mechanism behind this disease and develop treatments 27798110_We also correlated the features observed in patients with those of three Rpgr-mutant (Rpgr-ko, Rd9, and Rpgr-cko) mice. In patients, there was pronounced macular disease 27911705_Studies indicate taht the majority of patients with X-linked RP have mutations in the retinitis pigmentosa GTPase regulator (RPGR) or retinitis pigmentosa 2 protein (RP2) genes. 27995965_A novel frame-shift mutation in exon ORF15 of RPGR gene attributes of this heterozygosity suggest that gain-of-function mechanism could give rise to pathologic myopia via a degenerative cell-cell remodeling of the retinal structures. 28172980_RPGR interacts with PDE6delta and INPP5E. PDE6delta binds selectively to the C-terminus of RPGR and that this interaction is critical for RPGR's localization to cilia. INPP5E associates with the N-terminus of RPGR and trafficking of INPP5E to cilia is dependent upon the ciliary localization of RPGR. 28294154_four frameshift mutations including three novel mutations of c.1059 + 1 G > T, c.2002dupC and c.2236_2237del CT, as well as a previously reported mutation of c.2899delG were detected in the RPGR gene in the other four families. Our study further expands the mutation spectrum of RP2 and RPGR, and will be helpful for further study molecular pathogenesis of X-linked retinitis pigmentosa. 28814713_In induced pluripotent stem cells and mouse knockouts that RPGR mediates actin dynamics in photoreceptors via the actin-severing protein, gelsolin. 29135076_this is the first report of a molecular genetic diagnosis of XLRP in a patient with Turner syndrome. The X-linked RP in this woman may not be related to Turner syndrome, but may be a manifestation of the lack of a normal paternal X chromosome with in tact but mutate d RPGR 29528978_Based on best-corrected visual acuity survival probabilities, the intervention window for gene therapy for RPGR-associated retinal dystrophies is relatively broad in patients with RP. RPGR-ORF15 mutations were associated with COD/CORD and with a more severe phenotype in RP. 29721948_A novel RPGR mutation (p. Lys857Glu fs221X) was found in all but one affected members of a consanguineous Pakistani family with retinal degeneration. 29721984_RPGR mutations are associated with the majority of X-linked forms of retinal degenerations. The deregulation of intracellular pathways involving RPGR may underlie the pathogenesis of severe retinal degeneration. Review. 29847648_In general, good interocular symmetry exists; however, there was both variation between subjects and with the use of various metrics. Our findings will guide patient selection and design of RPGR treatment trials, and provide clinicians with specific prognostic information to offer patients affected by this condition. 30021045_A novel c.1059_1059+2delGGT, p.(?) variant in RPGR was identified as hemizygous in the affected boy and heterozygous in his mother. This case study expands the genotypic spectrum of RPGR variants associated with systemic manifestations. 30193314_This paper documents the first clinically validated next-generation sequencing (NGS) method for reliable, high-throughput sequencing of RPGR ORF15. 30208424_Measurement of rod sensitivity under dark-adapted conditions averaged across a small region showed the greatest potential for detectability of progression in the shortest period. 30289068_Co-Existence of Novel PDE6A Mutations and A Recurrent RPGR Mutation: A Potential Explanation for Phenotypic Diversity in Female RPGR Mutation Carriers. 30312579_Ellipzoid zone (EZ) metrics are sensitive structural biomarkers for measuring residual extent and progression in RPGR-associated retinopathy. 30313097_A Chinese family with Retinitis pigmentosa, 5 males with night blindness and decreased vision, and 8 females with different severities of myopia. Targeted exome capture sequencing in 2 males revealed a novel variant in the RPGR gene. The mutation cosegregated with the disease phenotype in the family. 31033374_A complete family history allowed determination of the inheritance pattern providing genetic counseling for patients and their families. The geno-phenotypic attributes of this heterozygosity suggest a correlation between retinitis pigmentosa and pathologic myopia. 31652454_A novel missense variant c.G644A (p.G215E) of the RPGR gene in a Chinese family causes X-linked retinitis pigmentosa. 31775781_We found two novel nonsense mutations of RPGR in two Chinese families with X-linked retinitis pigmentosa and analysed the association between genotypes and phenotypes. 31953110_X-Chromosome Inactivation Is a Biomarker of Clinical Severity in Female Carriers of RPGR-Associated X-Linked Retinitis Pigmentosa. 32012938_RPGR-Associated Dystrophies: Clinical, Genetic, and Histopathological Features. 32702353_Detailed comparison of phenotype between male patients carrying variants in exons 1-14 and ORF15 of RPGR. 32788070_A novel missense mutation of RPGR identified from retinitis pigmentosa affects splicing of the ORF15 region and causes loss of transcript heterogeneity. 32839555_A novel mutation of the RPGR gene in a Chinese X-linked retinitis pigmentosa family and possible involvement of X-chromosome inactivation. 32970112_Association of a Novel Intronic Variant in RPGR With Hypomorphic Phenotype of X-Linked Retinitis Pigmentosa. 33355362_RPGR isoform imbalance causes ciliary defects due to exon ORF15 mutations in X-linked retinitis pigmentosa (XLRP). 33446141_X-linked dominant RPGR gene mutation in a familial Coats angiomatosis. 33467000_X-Linked Retinitis Pigmentosa Caused by Non-Canonical Splice Site Variants in RPGR. 33620278_Multimodal imaging of an RPGR carrier female. 33805381_Cone Dystrophy Associated with a Novel Variant in the Terminal Codon of the RPGR-ORF15. 33808286_The Major Ciliary Isoforms of RPGR Build Different Interaction Complexes with INPP5E and RPGRIP1L. 34390733_Homozygous females for a X-linked RPGR-ORF15 mutation in an Iranian family with retinitis pigmentosa. | ENSMUSG00000031174 | Rpgr | 432.482574 | 1.1475107 | 0.198507634 | 0.17747225 | 1.206314e+00 | 2.720634e-01 | No | Yes | 426.253111 | 84.369795 | 377.118233 | 74.702185 | ||
| ENSG00000156453 | 5097 | PCDH1 | protein_coding | Q08174 | FUNCTION: May be involved in cell-cell interaction processes and in cell adhesion. | 3D-structure;Alternative splicing;Calcium;Cell adhesion;Cell junction;Cell membrane;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene belongs to the protocadherin subfamily within the cadherin superfamily. The encoded protein is a membrane protein found at cell-cell boundaries. It is involved in neural cell adhesion, suggesting a possible role in neuronal development. The protein includes an extracelllular region, containing 7 cadherin-like domains, a transmembrane region and a C-terminal cytoplasmic region. Cells expressing the protein showed cell aggregation activity. Alternative splicing occurs in this gene. [provided by RefSeq, Jul 2008]. | hsa:5097; | cell junction [GO:0030054]; cell-cell junction [GO:0005911]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; calcium ion binding [GO:0005509]; cell adhesion [GO:0007155]; cell-cell signaling [GO:0007267]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; nervous system development [GO:0007399] | 19729670_PCDH1 is a novel gene for BHR in adults and children. Its identification as a BHR susceptibility gene may suggest that a structural defect in the integrity of the airway epithelium and contributes to the development of BHR. 21929597_PCDH1 gene variant IVS3-116 associates with eczema in two independent birth cohorts in Netherlands. 21982948_PCDH1 transcripts display remarkable variability in expression of conserved intracellular signaling domains; enhanced PCDH1 expression levels strongly correlate with differentiation of bronchial epithelial cells. 23050600_genetic polymorphism is associated with subphenotypes of asthma in German children 23988763_Common variations in PCDH1 increase the risk of developing both transient early asthma and atopic dermatitis in early childhood 26209277_PCDH1 and SMAD3 act in a single pathway in asthma susceptibility that affects sensitivity of the airway epithelium to TGF-beta. 26227965_PCDH1 is important for airway function as a physical barrier, and its dysfunction is involved in the pathogenesis of allergic airway inflammation. 27701444_Results show that PCDH1 is localized to the cell membrane of bronchial epithelial cells basolateral to the adherens junction. Its expression is not reduced nor delocalized in asthma even though PCDH1 contributes to homotypic adhesion, epithelial barrier formation and repair. 30464266_identification of the human asthma-associated gene protocadherin-1 as an essential determinant of entry and infection in pulmonary endothelial cells by two hantaviruses that cause hantavirus pulmonary syndrome, Andes virus and Sin Nombre virus; the surface glycoproteins of ANDV and SNV directly recognize the outermost extracellular repeat domain of PCDH1-a member of the cadherin superfamily--to exploit PCDH1 for entry | ENSMUSG00000051375 | Pcdh1 | 205.530969 | 0.8071326 | -0.309122438 | 0.21126148 | 2.132750e+00 | 1.441818e-01 | No | Yes | 184.187933 | 28.375118 | 230.113373 | 35.187724 | ||
| ENSG00000157036 | 9941 | EXOG | protein_coding | Q9Y2C4 | FUNCTION: Endo/exonuclease with nicking activity towards supercoiled DNA, a preference for single-stranded DNA and 5'-3' exonuclease activity. {ECO:0000269|PubMed:18187503}. | 3D-structure;Alternative splicing;Endonuclease;Hydrolase;Membrane;Metal-binding;Mitochondrion;Mitochondrion inner membrane;Nuclease;Reference proteome;Transit peptide | This gene encodes an endo/exonuclease with 5'-3' exonuclease activity. The encoded enzyme catalyzes the hydrolysis of ester linkages at the 5' end of a nucleic acid chain. This enzyme is localized to the mitochondria and may play a role in programmed cell death. Alternatively spliced transcript variants have been described. A pseudogene exists on chromosome 18. [provided by RefSeq, Feb 2009]. | hsa:9941; | mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; 5'-3' exonuclease activity [GO:0008409]; endonuclease activity [GO:0004519]; endoribonuclease activity [GO:0004521]; metal ion binding [GO:0046872]; nucleic acid binding [GO:0003676]; single-stranded DNA endodeoxyribonuclease activity [GO:0000014]; apoptotic DNA fragmentation [GO:0006309] | 17415550_ENDOGL1 is a candidate disease-susceptibility gene for type 2 diabetes in a Japanese population 17415550_Observational study of gene-disease association. (HuGE Navigator) 18187503_Human endonuclease G-like-1 is a dimeric mitochondrial enzyme that displays 5'-3' exonuclease activity and further differs from EndoG in substrate specificity 20877624_Observational study of gene-disease association. (HuGE Navigator) 21768646_EXOG depletion induces persistent SSBs in the mtDNA, enhances ROS levels, and causes apoptosis in normal cells but not in mt genome-deficient rho0 cells. 23986585_These data indicate that HSV-1 UL12.5 deploys cellular proteins, including ENDOG and EXOG, to destroy mtDNA and contribute to a growing body of literature highlighting roles for ENDOG and EXOG in mtDNA maintenance. 28466855_Data suggest a role of exo/endonuclease G nuclease (hEXOG) in pathway for mitochondrial DNA repair. 30949702_ExoG has a role as a unique 5'-exonuclease to mediate the flap-independent RNA primer removal process during mtDNA replication to maintain mitochondrial genome integrity | ENSMUSG00000042787 | Exog | 897.786007 | 1.0900905 | 0.124447908 | 0.09518734 | 1.713282e+00 | 1.905601e-01 | 5.607597e-01 | No | Yes | 1004.806559 | 170.243345 | 910.286410 | 154.285115 | |
| ENSG00000157259 | 57798 | GATAD1 | protein_coding | Q8WUU5 | FUNCTION: Component of some chromatin complex recruited to chromatin sites methylated 'Lys-4' of histone H3 (H3K4me), with a preference for trimethylated form (H3K4me3). {ECO:0000269|PubMed:20850016}. | Cardiomyopathy;Disease variant;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene contains a zinc finger at the N-terminus, and is thought to bind to a histone modification site that regulates gene expression. Mutations in this gene have been associated with autosomal recessive dilated cardiomyopathy. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jun 2012]. | hsa:57798; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; sequence-specific DNA binding [GO:0043565]; zinc ion binding [GO:0008270]; chromatin organization [GO:0006325]; chromatin remodeling [GO:0006338]; regulation of transcription, DNA-templated [GO:0006355] | 20546612_Observational study of gene-disease association. (HuGE Navigator) 21965549_GATAD1 mutation is associated with autosomal recessive dilated cardiomyopathy. 24462704_GATAD1 expression was significantly decreased in the third-trimester placentas associated with preeclampsia than those associated with normal pregnancy. 29266303_GATAD1 plays a pivotal oncogenic role in hepatocellular carcinoma by directly inducing PRL3 transcription to activate the Akt signaling pathway. GATAD1 may serve as an independent poor prognostic factor for hepatocellular carcinoma patients | ENSMUSG00000007415 | Gatad1 | 1689.643852 | 1.0749942 | 0.104328915 | 0.07575349 | 1.886971e+00 | 1.695440e-01 | 5.328525e-01 | No | Yes | 1751.372154 | 248.903671 | 1618.071132 | 229.948193 | |
| ENSG00000157350 | 6483 | ST3GAL2 | protein_coding | Q16842 | FUNCTION: A beta-galactoside alpha2-3 sialyltransferase primarily involved in terminal sialylation of ganglio and globo series glycolipids (PubMed:8920913, PubMed:9266697). Catalyzes the transfer of sialic acid (N-acetyl-neuraminic acid; Neu5Ac) from the nucleotide sugar donor CMP-Neu5Ac onto acceptor Galbeta-(1->3)-GalNAc-terminated glycoconjugates through an alpha2-3 linkage (PubMed:8920913, PubMed:9266697, PubMed:25916169). Sialylates GM1/GM1a, GA1/asialo-GM1 and GD1b gangliosides to form GD1a, GM1b and GT1b, respectively (PubMed:8920913, PubMed:9266697). Together with ST3GAL3, primarily responsible for biosynthesis of brain GD1a and GT1b that function as ligands for myelin-associated glycoprotein MAG on axons, regulating MAG expression and axonal myelin stability and regeneration (By similarity). Via GT1b regulates TLR2 signaling in spinal cord microglia in response to nerve injury (By similarity). Responsible for the sialylation of the pluripotent stem cell- and cancer stem cell-associated antigen SSEA3, forming SSEA4 (PubMed:12716912). Sialylates with low efficiency asialofetuin, presumably onto O-glycosidically linked Galbeta-(1->3)-GalNAc-O-Ser (PubMed:9266697, PubMed:25916169). {ECO:0000250|UniProtKB:Q11204, ECO:0000269|PubMed:12716912, ECO:0000269|PubMed:25916169, ECO:0000269|PubMed:8920913, ECO:0000269|PubMed:9266697}. | Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Lipid metabolism;Membrane;Reference proteome;Secreted;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000305|PubMed:9266697}.; PATHWAY: Glycolipid biosynthesis. {ECO:0000305|PubMed:9266697}. | The protein encoded by this gene is a type II membrane protein that catalyzes the transfer of sialic acid from CMP-sialic acid to galactose-containing substrates. The encoded protein is normally found in the Golgi but can be proteolytically processed to a soluble form. This protein, which is a member of glycosyltransferase family 29, can use the same acceptor substrates as does sialyltransferase 4A. [provided by RefSeq, Jul 2008]. | hsa:6483; | extracellular region [GO:0005576]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; beta-galactoside (CMP) alpha-2,3-sialyltransferase activity [GO:0003836]; monosialoganglioside sialyltransferase activity [GO:0047288]; protein homodimerization activity [GO:0042803]; sialyltransferase activity [GO:0008373]; ganglioside biosynthetic process via lactosylceramide [GO:0010706]; globoside biosynthetic process via lactosylceramide [GO:0010707]; glycolipid biosynthetic process [GO:0009247]; glycoprotein biosynthetic process [GO:0009101]; keratan sulfate biosynthetic process [GO:0018146]; lipid glycosylation [GO:0030259]; O-glycan processing [GO:0016266]; oligosaccharide biosynthetic process [GO:0009312]; protein glycosylation [GO:0006486]; protein sialylation [GO:1990743]; sialylation [GO:0097503]; viral protein processing [GO:0019082] | 12504121_Genomic structure, expression, and transcriptional regulation of ST3GALII. 12716912_ST3Gal II is a MSGb5 (stage-specific embryonic antigen-4) synthase and its increased expression level is closely related to renal carcinogenesis 25916169_This suggests that the C-terminal domain of ST3Gal-II depends on N-glycosylation to attain an optimum conformation for proper exit from the endoplasmic reticulum 28698248_Data suggest that ganglioside glycosyltransferases ST3GAL5, ST8SIA1, and B4GALNT1 are S-acylated at conserved cysteine residues located close to cytoplasmic border of their transmembrane domains; ST3Gal-II is acylated at conserved cysteine residue in N-terminal cytoplasmic tail; for B4GALNT1 and ST3Gal-II, dimer formation controls their S-acylation status. | ENSMUSG00000031749 | St3gal2 | 841.132433 | 0.8579397 | -0.221051898 | 0.10575510 | 4.359778e+00 | 3.679710e-02 | 2.588970e-01 | No | Yes | 939.784346 | 127.899254 | 1074.001512 | 145.881923 |
| ENSG00000157450 | 54778 | RNF111 | protein_coding | Q6ZNA4 | FUNCTION: E3 ubiquitin-protein ligase (PubMed:26656854). Required for mesoderm patterning during embryonic development (By similarity). Acts as an enhancer of the transcriptional responses of the SMAD2/SMAD3 effectors, which are activated downstream of BMP (PubMed:14657019, PubMed:16601693). Acts by mediating ubiquitination and degradation of SMAD inhibitors such as SMAD7, inducing their proteasomal degradation and thereby enhancing the transcriptional activity of TGF-beta and BMP (PubMed:14657019, PubMed:16601693). In addition to enhance transcription of SMAD2/SMAD3 effectors, also regulates their turnover by mediating their ubiquitination and subsequent degradation, coupling their activation with degradation, thereby ensuring that only effectors 'in use' are degraded (By similarity). Activates SMAD3/SMAD4-dependent transcription by triggering signal-induced degradation of SNON isoform of SKIL (PubMed:17591695). Associates with UBE2D2 as an E2 enzyme (PubMed:22411132). Specifically binds polysumoylated chains via SUMO interaction motifs (SIMs) and mediates ubiquitination of sumoylated substrates (PubMed:23751493). Catalyzes 'Lys-63'-linked ubiquitination of sumoylated XPC in response to UV irradiation, promoting nucleotide excision repair (PubMed:23751493). Mediates ubiquitination and degradation of sumoylated PML (By similarity). The regulation of the BMP-SMAD signaling is however independent of sumoylation and is not dependent of SUMO interaction motifs (SIMs) (By similarity). {ECO:0000250|UniProtKB:Q99ML9, ECO:0000269|PubMed:14657019, ECO:0000269|PubMed:16601693, ECO:0000269|PubMed:17591695, ECO:0000269|PubMed:22411132, ECO:0000269|PubMed:23751493, ECO:0000269|PubMed:26656854}. | 3D-structure;Alternative splicing;Cytoplasm;DNA damage;DNA repair;Developmental protein;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:14657019, ECO:0000269|PubMed:16601693, ECO:0000269|PubMed:17591695, ECO:0000269|PubMed:22411132, ECO:0000269|PubMed:23751493, ECO:0000269|PubMed:26656854}. | The protein encoded by this gene is a nuclear RING-domain containing E3 ubiquitin ligase. This protein interacts with the transforming growth factor (TGF) -beta/NODAL signaling pathway by promoting the ubiquitination and proteosomal degradation of negative regulators, like SMAD proteins, and thereby enhances TGF-beta target-gene transcription. As a modulator of the nodal signaling cascade, this gene plays a critical role in the induction of mesoderm during embryonic development. Alternative splicing of this gene results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2012]. | hsa:54778; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; protein-containing complex [GO:0032991]; metal ion binding [GO:0046872]; SMAD binding [GO:0046332]; SUMO polymer binding [GO:0032184]; ubiquitin protein ligase activity [GO:0061630]; global genome nucleotide-excision repair [GO:0070911]; pattern specification process [GO:0007389]; positive regulation of protein ubiquitination [GO:0031398]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of transforming growth factor beta receptor signaling pathway [GO:0030511]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; ubiquitin-dependent protein catabolic process [GO:0006511]; ubiquitin-dependent SMAD protein catabolic process [GO:0030579] | 11298452_Functional analysis of the mouse counterpart 11298453_Functional analysis of the mouse counterpart 14657019_Silencing of the Arkadia gene resulted in repression of transcriptional activities induced by TGF-beta and BMP, and accumulation of the Smad7 protein. 17510063_Arkadia induces degradation of SnoN and c-Ski in addition to Smad7. 17591695_Results show that Arkadia specifically activates transcription via Smad3/Smad4 binding sites by inducing degradation of the transcriptional repressor SnoN. 18059455_Arkadia stimulates renal tubular epithelial to mesenchymal cell transition through degradation of Smad7. 19898560_Arkadia protein intensifies TGF-beta-induced effects by marking c-Ski and inhibitory Smad7 for destruction--REVIEW 19959502_findings show that Arkadia regulates the levels of expression of c-Ski protein in cell-type-dependent fashion, and exhibits a tumour suppressor function by inhibiting tumour cell growth 20965945_Arkadia complexes with clathrin adaptor AP2 and regulates EGF signalling. 21795712_RB1CC1 thus appears to play a unique role as a modulator of TGF-beta signaling by restricting substrate specificity of Arkadia. 21998011_Arkadia enhances transforming growth factor-beta signaling responses and supports its tumor-suppressing properties in colorectal cancer. 22411132_Determined the nuclear magnetic resonance solution structure of Arkadia's RING-H2 domain and revealed a (beta)betabetaalpha fold, fully consistent with the expected 'cross-brace' mode of Zn(II)-ligation. 23212909_FHL2 increases the stability of the TGF-beta pathway positive regulator Arkadia by inhibiting its ubiquitination and cooperates with Arkadia to activate TGF-beta signaling. 23246292_We established that ARX polyA alterations damage the regulation of KDM5C expression. 23394999_RNF111-dependent neddylation activates DNA damage-induced ubiquitination 23467611_findings indicate that Arkadia is not critical for regulating tumor growth per se, but is required for the early stages of cancer cell colonization at the sites of metastasis 23530056_Arkadia is involved in arsenic-induced degradation of polysumoylated PML protein. 23751493_Our findings establish RNF111 as a new STUbL that directly links nonproteolytic ubiquitylation and SUMOylation in the DNA damage response. 24912682_Arkadia can both promote and inhibit gene expression, indicating that Arkadia's activity in transcriptional control may depend on the epigenetic context, defined by Polycomb repressive complexes and DNA methylation. 26151477_These data suggest that RNF111, together with the CRL4(DDB2) ubiquitin ligase complex, is responsible for sequential XPC ubiquitylation, which regulates the recruitment and release of XPC and is crucial for efficient progression of the NER reaction. 26238425_Results suggest that -459CpG methylation in Sp1-binding site of RNF111 promoter transcriptionally decreases RNF111 expression, which inhibits TGF-beta/Smad signaling associated invasion in NSCLC cells. 26522722_Results indicate a tumor-suppressive role of the ring finger protein 111 (Arkadia)-epithelial splicing regulatory protein 2 (ESRP2) axis in clear-cell renal cell carcinoma (ccRCC). 26656854_Study reports that the monomeric RING domains from the human E3 ligases Arkadia and Ark2C bind directly to free ubiquitin with an affinity comparable to that of other dedicated ubiquitin-binding domains. 28647409_These data show that the position occupied by W972 within wild-type Arkadia is critical for the function of its RING domain and that it depends on the nature of the residue at this position. 29286088_rkadia may regulate the pathogenesis and progression of hepatic fibrosis. 29597191_Arkadia/RNF111 is able to lys-48 ubiquitination of Nrf2 protects Nrf2 from degradation thereby allowing Nrf2-dependent gene transcription. 31417085_Arkadia specifically selects substrates carrying SUMO1-capped SUMO2/3 hybrid conjugates and targets them for proteasomal degradation. 34740826_Quantitative Ubiquitylome Analysis Reveals the Specificity of RNF111/Arkadia E3 Ubiquitin Ligase for its Degradative Substrates SKI and SKIL/SnoN in TGF-beta Signaling Pathway. | ENSMUSG00000032217 | Rnf111 | 1007.636648 | 1.1077632 | 0.147649522 | 0.09328872 | 2.498276e+00 | 1.139710e-01 | 4.507868e-01 | No | Yes | 998.743153 | 236.043531 | 905.817944 | 214.078357 |
| ENSG00000157869 | 9364 | RAB28 | protein_coding | P51157 | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Cell projection;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;GTP-binding;Lipoprotein;Membrane;Methylation;Nucleotide-binding;Phosphoprotein;Prenylation;Reference proteome | This gene encodes a member of the Rab subfamily of Ras-related small GTPases. The encoded protein may be involved in regulating intracellular trafficking. Alternative splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 9 and X. [provided by RefSeq, Apr 2009]. | hsa:9364; | ciliary basal body [GO:0036064]; ciliary rootlet [GO:0035253]; cytoplasm [GO:0005737]; endomembrane system [GO:0012505]; plasma membrane [GO:0005886]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; intracellular protein transport [GO:0006886]; toxin transport [GO:1901998] | 19026641_Crystal structures of Rab28 in the active (GppNHp-bound) and inactive (GDP-3'P-bound) forms at 1.5 and 1.1A resolution were reported. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23746546_Autosomal-recessive cone-rod dystrophy is associated with RAB28 mutations. 25356532_Deleterious mutations in RAB28 result in a classic CRD phenotype and are an infrequent cause of CRD in the Spanish population. 28388261_In summary, we identified a novel rare 'likely pathogenic' variant--RAB28 c.68C>T--in a Korean patient with cone-rod dystrophy. 32084271_In conclusion, RAB28 variants are a rare cause of cone-rod dystrophy in various ethnic groups. These variants cause a comparable ophthalmological phenotype and furthermore could also be a cause of postaxial polydactyly. 33396523_Expanding the Clinical and Genetic Spectrum of RAB28-Related Cone-Rod Dystrophy: Pathogenicity of Novel Variants in Italian Families. | ENSMUSG00000029128 | Rab28 | 530.276997 | 1.1250828 | 0.170031143 | 0.11962861 | 2.020344e+00 | 1.552040e-01 | 5.135144e-01 | No | Yes | 523.465091 | 104.961729 | 461.822036 | 92.656805 | ||
| ENSG00000158019 | 9577 | BABAM2 | protein_coding | Q9NXR7 | FUNCTION: Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX (PubMed:17525341, PubMed:19261746, PubMed:19261749, PubMed:19261748). In the BRCA1-A complex, it acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the BRCA1-BARD1 heterodimer (PubMed:21282113, PubMed:19261748). Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates (PubMed:19214193, PubMed:24075985, PubMed:25283148, PubMed:26195665). Within the BRISC complex, acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity (PubMed:21282113). The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression (PubMed:24075985). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985). May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway. May regulate TNF-alpha signaling through its interactions with TNFRSF1A; however these effects may be indirect (PubMed:15465831). {ECO:0000269|PubMed:14636569, ECO:0000269|PubMed:19261748, ECO:0000269|PubMed:19261749, ECO:0000269|PubMed:24075985, ECO:0000269|PubMed:26195665, ECO:0000305|PubMed:15465831}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cell cycle;Cell division;Chromatin regulator;Cytoplasm;DNA damage;DNA repair;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation pathway | This gene encodes an anti-apoptotic, death receptor-associated protein that interacts with tumor necrosis factor-receptor-1. The encoded protein acts as an adapter in several protein complexes, including the BRCA1-A complex and the BRISC complex. The BRCA1-A complex possesses ubiquitinase activity and targets sites of double strand DNA breaks, while the BRISC complex exhibits deubiquitinase activity and is involved in mitotic spindle assembly. This gene is upregulated in several types of cancer. [provided by RefSeq, Jun 2016]. | hsa:9577; | BRCA1-A complex [GO:0070531]; BRISC complex [GO:0070552]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear ubiquitin ligase complex [GO:0000152]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; peroxisome targeting sequence binding [GO:0000268]; polyubiquitin modification-dependent protein binding [GO:0031593]; tumor necrosis factor receptor binding [GO:0005164]; apoptotic process [GO:0006915]; cell division [GO:0051301]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to ionizing radiation [GO:0071479]; chromatin organization [GO:0006325]; double-strand break repair [GO:0006302]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; negative regulation of apoptotic process [GO:0043066]; positive regulation of DNA repair [GO:0045739]; protein K63-linked deubiquitination [GO:0070536]; response to ionizing radiation [GO:0010212]; signal transduction [GO:0007165] | 15465831_BRE mediates antiapoptosis by inhibiting the mitochondrial apoptotic machinery 15582573_the enhanced tumor growth is more likely due to the antiapoptotic activity of BRE than any direct effect of the protein on cell proliferation 17704801_Antiapoptotic in vivo; Bre levels are regulated post-transcriptionally in the liver, which is not observed in human hepatocellular carcinoma (HCC) and non-HCC cell lines. 18756325_results implied that BRE plays a significant role in mediating antiapoptotic and proliferative responses in esophageal carcinoma cells 19757177_A novel stress-responsive gene called BRE which interacts with TNF-receptor-1 and blocks the apoptotic effect of TNF-alpha, was identified. 20035718_These results show that BRE over-expression can indeed promote growth, though not initiation, of liver tumors. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20861917_overexpression of the BRE gene is predominantly found in MLL-rearranged AML with t(9;11)(p22;q23). 21282113_NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes 21937695_High BRE expression defines a novel subtype of adult acute myeloid leukemia characterized by a favorable prognosis. 22555662_High BRE and high EVI1 expression are mutually exclusive in MLL-AF9-positive acute myeloid leukemia patients. 27733185_Results show that BRE expression is regulated by HOTTIP LncRNA. Its over-expression promotes cell proliferation and cell cycle progression inhibiting apoptosis of glioma cells. 28871137_C-terminal BRE might be an important contributor to this program because in a case with relapsed AML, we observed an ins(11;2) fusing CHORDC1 to BRE at the region where intragenic transcription starts in KMT2A-rearranged and KAT6A-CREBBP AML. 29416040_show that BRE facilitates deubiquitylation of CDC25A by recruiting ubiquitin-specific-processing protease 7 (USP7) in the presence of DNA damage 30287910_The BRE rs7572644 and NRG3 rs1649942 genetic variants were validated in an independent cohort of EOC Portuguese patients. | ENSMUSG00000052139 | Babam2 | 1790.025928 | 0.9772965 | -0.033131812 | 0.07273838 | 2.067017e-01 | 6.493650e-01 | 8.733317e-01 | No | Yes | 1837.336057 | 169.655765 | 1861.229974 | 171.759368 | |
| ENSG00000158290 | 8450 | CUL4B | protein_coding | Q13620 | FUNCTION: Core component of multiple cullin-RING-based E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:14578910, PubMed:16322693, PubMed:16678110, PubMed:18593899, PubMed:29779948, PubMed:30166453, PubMed:33854232, PubMed:33854239, PubMed:22118460). The functional specificity of the E3 ubiquitin-protein ligase complex depends on the variable substrate recognition subunit (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:29779948, PubMed:22118460). CUL4B may act within the complex as a scaffold protein, contributing to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:22118460). Plays a role as part of the E3 ubiquitin-protein ligase complex in polyubiquitination of CDT1, histone H2A, histone H3 and histone H4 in response to radiation-induced DNA damage (PubMed:14578910, PubMed:16678110, PubMed:18593899). Targeted to UV damaged chromatin by DDB2 and may be important for DNA repair and DNA replication (PubMed:16678110). A number of DCX complexes (containing either TRPC4AP or DCAF12 as substrate-recognition component) are part of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation (PubMed:29779948). The DCX(AMBRA1) complex is a master regulator of the transition from G1 to S cell phase by mediating ubiquitination of phosphorylated cyclin-D (CCND1, CCND2 and CCND3) (PubMed:33854232, PubMed:33854239). The DCX(AMBRA1) complex also acts as a regulator of Cul5-RING (CRL5) E3 ubiquitin-protein ligase complexes by mediating ubiquitination and degradation of Elongin-C (ELOC) component of CRL5 complexes (PubMed:30166453). Required for ubiquitination of cyclin E (CCNE1 or CCNE2), and consequently, normal G1 cell cycle progression (PubMed:16322693, PubMed:19801544). Regulates the mammalian target-of-rapamycin (mTOR) pathway involved in control of cell growth, size and metabolism (PubMed:18235224). Specific CUL4B regulation of the mTORC1-mediated pathway is dependent upon 26S proteasome function and requires interaction between CUL4B and MLST8 (PubMed:18235224). With CUL4A, contributes to ribosome biogenesis (PubMed:26711351). {ECO:0000269|PubMed:14578910, ECO:0000269|PubMed:16322693, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:18235224, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:19801544, ECO:0000269|PubMed:22118460, ECO:0000269|PubMed:26711351, ECO:0000269|PubMed:29779948, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854239}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;DNA damage;DNA repair;Disease variant;Dwarfism;Isopeptide bond;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:10230407, ECO:0000269|PubMed:16678110, ECO:0000269|PubMed:18593899, ECO:0000269|PubMed:29779948, ECO:0000269|PubMed:30166453, ECO:0000269|PubMed:33854232, ECO:0000269|PubMed:33854239}. | This gene is a member of the cullin family. The encoded protein forms a complex that functions as an E3 ubiquitin ligase and catalyzes the polyubiquitination of specific protein substrates in the cell. The protein interacts with a ring finger protein, and is required for the proteolysis of several regulators of DNA replication including chromatin licensing and DNA replication factor 1 and cyclin E. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:8450; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; Cul4B-RING E3 ubiquitin ligase complex [GO:0031465]; cullin-RING ubiquitin ligase complex [GO:0031461]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleoplasm [GO:0005654]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; cellular response to DNA damage stimulus [GO:0006974]; G1/S transition of mitotic cell cycle [GO:0000082]; histone H2A monoubiquitination [GO:0035518]; neuron projection development [GO:0031175]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of protein catabolic process [GO:0045732]; proteasomal protein catabolic process [GO:0010498]; protein ubiquitination [GO:0016567]; ribosome biogenesis [GO:0042254]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146]; UV-damage excision repair [GO:0070914] | 16322693_human CUL4B and cyclin E proteins interact with each other and the CUL4B complexes can polyubiquitinate the CUL4B-associated cyclin E 16407252_Cul4B, PCNA, and DDB1 are involved in the degradation of Cdt1 after ultraviolet radiation 17236139_The relatively high frequency of CUL4B mutations in this series indicates that it is one of the most commonly mutated genes underlying XLMR and suggests that its introduction into clinical diagnostics should be a high priority. 17273978_Mutation in CUL4B causes X-linked mental retardation 17392787_a fat-soluble ligand-dependent ubiquitin ligase complex in human cell lines, in which dioxin receptor (AhR) is integrated as a component of a novel cullin 4B ubiquitin ligase complex, CUL4B(AhR) 18235224_CUL4-DDB1 ubiquitin ligase interacts with Raptor and regulates the mTORC1-mediated signaling pathway through ubiquitin-dependent proteolysis. 18593899_DDB1-CUL4B(DDB2) E3 ligase may have a distinctive function in modifying the chromatin structure at the site of UV lesions to promote efficient NER. 19056892_CUL4A and CUL4B are therefore components of a conserved Wnt-induced proteasome targeting (WIPT) complex that regulates p27(KIP1) levels and cell cycle progression in mammalian cells. 19295130_Cells depleted of Dda1 spontaneously accumulated double-stranded DNA breaks in a similar way to Cul4A-, Cul4B- or Wdr23-depleted cells, indicating that Dda1 interacts physically and functionally with cullin-RING E3 ligases complexes. 19801544_Data show that that RNA interference of CUL4B led to an inhibition of cell proliferation and a prolonged S phase, due to the overaccumulation of cyclin E. 19818632_Studies indicate that CUL4 uses a large beta-propeller protein, DDB1, as a linker to interact with a subset of WD40 proteins. 20002452_The CUL4B gene is associated with X-linked mental retardation syndrome. 20005570_CUL4B is over-expressed in placenta in intra-uterine growth restriction. 20064923_the interplay between CUL4A and CUL4B in pathogenesis of CUL4B-deficiency in humans 20932471_This study identifies CRL4-Cdt2 ubiquitin ligase to promote the ubiquitin-dependent proteolysis of the histone H4 methyltransferase Set8 during S-phase of the cell cycle and after UV-irradiation in a reaction that is dependent on PCNA. 20951943_Increased PRMT5 activity mediates key events associated with cyclin D1-dependent neoplastic growth, including CUL4 repression, CDT1 overexpression, and DNA rereplication. 21352845_the unexpected association of defective CUL4B with syndromal X-linked mental retardation in humans 21795677_Cullin 4B protein ubiquitin ligase targets peroxiredoxin III for degradation. 21816345_CUL4B targets WDR5 for ubiquitylation and degradation in the nucleus. 22992378_The data suggest that unneddylated Cul4B isoforms specifically inhibits beta-catenin degradation during mitosis. 23238014_Cullin4B-Ring E3 ligase complex (CRL4B) is physically associated with PRC2. CRL4B possesses an intrinsic transcription repressive activity by promoting H2AK119 monoubiquitination. CUL4B promotes cancer cell proliferation, invasion, and tumorigenesis in vitro and in vivo. 23348097_Our results suggest that XLID CUL4B mutants are defective in promoting TSC2 degradation and positively regulating mTOR signaling in neocortical neurons 23357576_Studies indicate Jun activation domain-binding protein Jab1 as a substrate for CUL4B E3 ligase. 23479742_the up-regulation of CDK2 by CUL4B is achieved via the repression of miR-372 and miR-373, which target CDK2. 23649548_Investigated CUL4B expression pattern in patients with colon cancer; immunohistochemistry and PCR study showed that high CUL4B expression was significantly associated with colon cancer progression and pathogenesis. 24292684_CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing 24452595_these observations establish an important negative regulatory role of CUL4B on p53 stability. 24719410_HIV-1 Vpr can trigger G2 cell cycle arrest in the absence of either CUL4A or CUL4B. 24898194_The intellectual disability phenotype is caused by aberrant splicing and removal of intron 7 from CUL4B gene primary transcript. 25189186_Results demonstrated that CUL4B promotes cell proliferation and inhibits the apoptosis of osteosarcoma cells. 25385192_Data show that CUL4B variants are associated with a wide range of cerebral malformations and suggest an important role in brain through its interaction with WDR62, a protein in which variants were identified in patients with cerebral malformations. 25430888_CUL4B can up-regulate Wnt/beta-catenin signalling in human HCC through transcriptionally repressing Wnt antagonists and thus contributes to the malignancy of HCC. 25464270_results established a critical role of CUL4B in negatively regulating the p53-ROS positive feedback loop that drives cellular senescence 25542213_Results show that CUL4A- and CUL4B-mediated polyubiquitination of gamma-tubulin for its degradation. 25970626_FBXO44-mediated degradation of RGS2 protein uniquely depends on a Cul4B/DDB1 complex. 26021757_Our data are consistent with the idea that the CUL4A/B-DDB1-CRBN complex catalyses the polyubiquitination and thus controls the degradation of CLC-1 channels. 26617747_these results showed that knockdown of CUL4B inhibit proliferation and promotes apoptosis of colorectal cancer cells through suppressing the Wnt/beta-catenin signaling pathway 27656838_these results suggest that knockdown of CUL4B inhibited the proliferation and invasion through suppressing the Wnt/beta-catenin signaling pathway in NSCLC cells. Therefore, CUL4B may represent a novel therapeutic target for the treatment of NSCLC. 27899484_CUL4B protein levels in human subcutaneous adipose tissue is negatively correlated with body mass index. 27974468_findings revealed that CUL4A and CUL4B are differentially associated with etiologic factors for pulmonary malignancies and are independent prognostic markers for the survival of distinct lung cancer subtypes 28164432_This study found that microRNA-194 (miR-194) and CUL4B protein were inversely correlated in cancer specimens and demonstrated that miR-194 could downregulate CUL4B by directly targeting its 3'-UTR. 28225217_CUL4B regulates protein turnover and homeostasis in response to dopamine stimulation. 28816568_lack of any significant mutations in patients with azoospermia due to Sertoli-cell-only syndrome 28886238_The CUL4B interacts with WD-40 proteins through the adaptor protein DNA damage-binding protein 1 (DDB1) to target substrates for ubiquitylation. 29106389_High CUL4B expression promotes gastric cancer invasion and metastasis. 30229816_In osteosarcoma tissues, expression of CUL4B is higher and expression of miRNA-708 is lower. 30483755_Taken together, the findings of this study suggest that the miR381/miR489mediated expression of CUL4B modulates the proliferation and invasion of GC cells via the Wnt/betacatenin pathway, which indicates that the miR381/miR489CUL4B axis is critical in the control of GC tumourigenesis. 30609075_Authors demonstrate that CUL4B upregulates the expression of C-MYC at post-transcriptional level through epigenetic silencing of miR-33b-5p. CUL4B-induced oncogenic activity in PCa by targeting C-MYC is repressed by miR-33b-5p. 30612524_Enhanced CUL4B expression was discovered in diffuse large B-cell lymphoma tissues and cells. CUL4B promoted the growth of diffuse large B-cell lymphoma cells both in vitro and in vivo, which was by autophagy mediated by JNK signaling. 30883036_Results demonstrate a key contribution of CUL4B overexpression in the malignant behavior of head and neck squamous cell carcinoma (HNSCC) cells, at least in part through the stimulation of angiogenesis and the activation of the Wnt/beta-catenin signaling pathway. 30898011_An autophagy-related (ATG) protein that plays a critical role in autophagosome biogenesis, is a direct substrate of CUL4-RING ubiquitin ligases (CRL4s). 30945295_miR-194-5p, was significantly downregulated in intervertebral disc degeneration samples and could bind to the three prime untranslated regions (3'-UTRs) of both CUL4A and CUL4B, thereby downregulating their expression. 31111526_CUL4B regulates cancer stem-like traits of prostate cancer cells by targeting BMI1 via miR200b/c, which might give novel insight into how CUL4B promotes prostate cancer progression through regulating cancer stem-like traits. 31329620_These results show that ubiquitinated Apaf-1 may activate caspase-9 under conditions of proteasome impairment. 31407591_circZFR exhibited a carcinogenic role by sponging miR-101-3p and regulating CUL4B expression in NSCLC 31448526_NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. 31729179_Alu-mediated Xq24 deletion encompassing CUL4B, LAMP2, ATP1B4, TMEM255A, and ZBTB33 genes causes Danon disease in a female patient. 32275162_Downregulation of lncRNA ZEB1-AS1 Represses Cell Proliferation, Migration, and Invasion Through Mediating PI3K/AKT/mTOR Signaling by miR-342-3p/CUL4B Axis in Prostate Cancer. 32466489_CUL4-DDB1-CRBN E3 Ubiquitin Ligase Regulates Proteostasis of ClC-2 Chloride Channels: Implication for Aldosteronism and Leukodystrophy. 32587774_CUL4 E3 ligase regulates the proliferation and apoptosis of lung squamous cell carcinoma and small cell lung carcinoma. 32622365_Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. 33022894_Cullin 4B regulates cell survival and apoptosis in clear cell renal cell carcinoma as a target of microRNA-217. 33227394_CUL4B promotes aggressive phenotypes of renal cell carcinoma via upregulating c-Met expression. 33506897_Effect and mechanism of miR-217 on drug resistance, invasion and metastasis of ovarian cancer cells through a regulatory axis of CUL4B gene silencing/inhibited Wnt/beta-catenin signaling pathway activation. 33638154_CUL4B renders breast cancer cells tamoxifen-resistant via miR-32-5p/ER-alpha36 axis. 34002487_LncRNA SNHG12 regulates the miR-101-3p/CUL4B axis to mediate the proliferation, migration and invasion of non-small cell lung cancer. 34011786_Cul4b Promotes Progression of Malignant Cutaneous Melanoma Patients by Regulating CDKN2A. 34026424_CUL4B Promotes Breast Carcinogenesis by Coordinating with Transcriptional Repressor Complexes in Response to Hypoxia Signaling Pathway. 34119472_CUL4(high) Lung Adenocarcinomas Are Dependent on the CUL4-p21 Ubiquitin Signaling for Proliferation and Survival. 34153655_Cullin-4B promotes cell proliferation and invasion through inactivation of p53 signaling pathway in colorectal cancer. 34338146_Inhibition of MicroRNA miR-101-3p on prostate cancer progression by regulating Cullin 4B (CUL4B) and PI3K/AKT/mTOR signaling pathways. | ENSMUSG00000031095 | Cul4b | 4461.752111 | 1.0897887 | 0.124048474 | 0.05966409 | 4.302239e+00 | 3.806223e-02 | 2.631245e-01 | No | Yes | 4799.493476 | 865.977615 | 4359.467656 | 786.563546 |
| ENSG00000158691 | 9753 | ZSCAN12 | protein_coding | O43309 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Mouse_homologues mmu:22758; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 35039565_Methylation statuses of NCOR2, PARK2, and ZSCAN12 signify densities of tumor-infiltrating lymphocytes in gastric carcinoma. | ENSMUSG00000036721 | Zscan12 | 850.271567 | 1.1631495 | 0.218036500 | 0.09606584 | 5.152664e+00 | 2.321074e-02 | 2.007408e-01 | No | Yes | 1062.119928 | 225.255858 | 896.062336 | 190.108726 | ||
| ENSG00000158813 | 1896 | EDA | protein_coding | Q92838 | FUNCTION: Cytokine which is involved in epithelial-mesenchymal signaling during morphogenesis of ectodermal organs. Functions as a ligand activating the DEATH-domain containing receptors EDAR and EDA2R (PubMed:8696334, PubMed:11039935, PubMed:27144394, PubMed:34582123). May also play a role in cell adhesion (By similarity). {ECO:0000250|UniProtKB:O54693, ECO:0000269|PubMed:11039935, ECO:0000269|PubMed:27144394, ECO:0000269|PubMed:34582123, ECO:0000269|PubMed:8696334}.; FUNCTION: [Isoform 1]: Binds only to the receptor EDAR, while isoform 3 binds exclusively to the receptor EDA2R. {ECO:0000269|PubMed:11039935, ECO:0000269|PubMed:27144394}.; FUNCTION: [Isoform 3]: Binds only to the receptor EDA2R. {ECO:0000269|PubMed:11039935, ECO:0000269|PubMed:27144394}. | 3D-structure;Alternative splicing;Cell membrane;Cleavage on pair of basic residues;Collagen;Developmental protein;Differentiation;Disease variant;Ectodermal dysplasia;Glycoprotein;Membrane;Reference proteome;Secreted;Signal-anchor;Transmembrane;Transmembrane helix | The protein encoded by this gene is a type II membrane protein that can be cleaved by furin to produce a secreted form. The encoded protein, which belongs to the tumor necrosis factor family, acts as a homotrimer and may be involved in cell-cell signaling during the development of ectodermal organs. Defects in this gene are a cause of ectodermal dysplasia, anhidrotic, which is also known as X-linked hypohidrotic ectodermal dysplasia. Several transcript variants encoding many different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:1896; | apical part of cell [GO:0045177]; collagen trimer [GO:0005581]; cytoskeleton [GO:0005856]; endoplasmic reticulum membrane [GO:0005789]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; lipid droplet [GO:0005811]; membrane [GO:0016020]; plasma membrane [GO:0005886]; death receptor agonist activity [GO:0038177]; death receptor binding [GO:0005123]; receptor ligand activity [GO:0048018]; signaling receptor binding [GO:0005102]; tumor necrosis factor receptor binding [GO:0005164]; animal organ development [GO:0048513]; cell differentiation [GO:0030154]; cell-matrix adhesion [GO:0007160]; cytokine-mediated signaling pathway [GO:0019221]; gene expression [GO:0010467]; hair follicle placode formation [GO:0060789]; immune response [GO:0006955]; odontogenesis of dentin-containing tooth [GO:0042475]; pigmentation [GO:0043473]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of gene expression [GO:0010628]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; regulation of NIK/NF-kappaB signaling [GO:1901222]; salivary gland cavitation [GO:0060662]; trachea gland development [GO:0061153] | 12673367_The structure of the EDA1 gene in a patient with anhidrotic ectodermal dysplasia 12682853_Identified ED1 mutations including one novel mutation in the 5' splice site following exon 8 (IVS8+5delG) and another missense mutation (A959G; Y320C), which has been reported previously. 12920369_The ED1 gene was identified as a responsive gene for X-LINKED HYPOHIDROTIC ECTODERMAL DYSPLASIA. 12930312_identified ED1 mutations including three novel mutations by sequencing genomic DNAs from eight unrelated Japanese X-linked hypohidrotic ectodermal dysplasia families 15663448_point mutation (G1149A) in exon 8 changes codon 291 from glycine to arginine causing X-linked hypohidrotic ectodermal dysplasia 16423472_isoforms of EDA-A5 and A5',activated NF-kappaB through receptors EDAR and XEDAR 16752854_EDA signaling has its biological significance in inducing development and morphogenesis of sweat glands and in maintaining physiological function of skin. 17102627_EDA signaling has a role in skin appendage development [review] 17256800_An amino acid substitution in ectodysplasin A is associated with X-linked dominant incisor hypodontia. 17478381_Report of a novel insertion mutation in EDA1 gene in a Pakistani family with x-linked hypohidrotic ectodermal dysplasia. 17970812_study reports the molecular analyses of four patients from India with hypohidrotic ectodermal dysplasia, three who harbour novel mutations, two in the EDA gene and one in the EDAR gene 18076698_Three novel missense mutations of the EDA gene have been identified. This provides evidence for an unequal homologous recombination between two LINE-1 elements as the molecular mechanism in the pathogensis of LXHED. 18427821_a novel deletion mutation is described in a Chinese family with X-linked hypohidrotic ectodermal dysplasia 18545687_results indicate that these novel missense mutations in EDA are associated with the isolated tooth agenesis and provide preliminary explanation for the abnormal clinical phenotype at a molecular structural level 18633626_In TGF-beta1 incubated CLPF mRNA amount of FN and isoforms ED-A and ED-B was slightly increased. IFN-gamma only decreased FN in CLPF, TNF significantly reduced FN-mRNA by 40%, FN ED-A mRNA by 25%, and ED-B mRNA by 50%. 18657636_congenital absence of maxillary and mandibular central incisors, lateral incisors and canines, with the high possibility of persistence of maxillary and mandibular first permanent molars as a pattern of tooth agenesis, suggests an EDA mutation 18666859_Screening of EDA1 gene in X-linked anhiderotic ectodermal dysplasia using DHPLC: identification of 14 novel mutations in Italian patients is reported. 18688569_Missense mutation in the EDA gene is associated with X-linked recessive isolated hypodontia 18702659_the c.952delG mutation of the EDA1 gene is likely to be the disease-causing mutation for XLHED in this family. 18821982_data confirm that mutations could cause both X-linked hipohydrotic ectodermal dysplasia and isolated hypodontia and provide evidence that EDA is a strong candidate gene for tooth genesis 19127222_Case Reports:Two novel mutations in the ED1 gene in Japanese families with X-linked hypohidrotic ectodermal dysplasia. 19278982_genetic defect could result in non-syndromic oligodontia in affected males 19438931_results expand the allelic series for mutations underlying hypohidrotic ectodermal dysplasia 19504606_We present a new mutation in the EDA gene which causes selective tooth agenesis and demonstrates the phenotype variation that can be encountered in the ectodermal dysplasia syndrome (HED) with the highest prevalence worldwide. 19551394_Recurrent mutations in functionally-related EDA and EDAR genes underlie X-linked isolated hypodontia and autosomal recessive hypohidrotic ectodermal dysplasia. 19590514_Observational study of gene-disease association. (HuGE Navigator) 19623212_An in vitro functional analysis was performed of six selective tooth agenesis-causing EDA mutations (one novel and five known) that are located in the C-terminal tumor necrosis factor homology domain of the protein. 19657145_The collagen domain activates EDA1 by multimerization, whereas the proteoglycan-binding domain may restrict the distribution of endogeneous EDA1 in vivo. 19816326_EDA has been identified as a nonsyndromic tooth agenesis gene, X-linked. 20195514_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20236127_Data show that 25 different mutations on EDA and EDAR genes were detected in HED patients. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20486090_analysis of Missense mutation of the EDA gene in a Jordanian family with X-linked hypohidrotic ectodermal dysplasia [case report] 20501644_A crucial role of the EDA-A2/ectodysplasin A2 (XEDAR) interaction is revealed in the p53-signaling pathway. 20628232_Direct sequencing of the EDA-A1 gene in affected individuals of the 3 families revealed the same missense mutation. Microsatellite marker analysis showed a shared haplotype among the affected members of both families, suggesting a common founder mutation. 20979233_EDA1 gene was the most common hypohidrotic/anhidrotic ectodermal dysplasia disease-causing gene 21091672_This study further confirms the differential effect of the mutations in EDA gene that define the pathogenic basis of X-linked recessive isolated hypodontia. 21357618_Systematic mapping of EDA mutations together with the analysis of objective clinical data may help to distinguish functionally crucial mutations from those allowing residual activity of the gene product. 21457804_there exists a correlation between the phenotypes and genotypes of XLHED and NSH subjects harboring EDA mutations 21724072_association in dental crowding in the Hong Kong Chinese population 21916884_The finding that EDAR370A attenuates hypohidrotic ectodermal dysplasia symptoms provides the first in vivo evidence that allele is a more potent signalling molecule than EDAR370V. 22004506_we report a novel mutation of the EDA gene identified in a Korean family with X-linked hypohidrotic ectodermal dysplasia. 22008666_Direct DNA sequencing of the whole coding region of EDA revealed a novel missense mutation, p.Leu354Pro in a patient affected with XLHED. 22835214_The mutation described resulted in a deletion of the highly conserved TNF homology sequence responsible for binding to EDA1R. 22875504_Various mutations of ED1 gene were detected. 23293949_Case of hypohidrotic ectodermal dysplasia caused by a large deletion mutation in the EDA gene. In a Japanese boy, 3 .months old 23603338_identified heterozygous nonsense mutation c.874G>T (p.Glu292X) in TNF homology domain of EDA in all affected females. phenotype variability in heterozygous female carriers may occur due to differential pattern of X-chromosome inactivation 23625373_identified novel missense mutation (c.779 T>G) in Nonsyndromic Hypodontia. mutation results in Ile260Ser substitution in the TNF homology domain. alteration may induce conformational change in hydrophobic center of TNF homology domain. 23626789_most of these Chinese XLHED carriers' have hypermethylated EDA promoter. 23635427_novel one-nucleotide deletion mutation (c.855delG) of EDA in exon 8 which caused premature termination of the polypeptide at amino acid 307 was confirmed 23744312_Identification of a novel c.822 G>T mutation of EDA gene in a Chinese family with X-linked hypohidrotic ectodermal dysplasia. 23926003_The deletion and missense mutation in ED1 gene is associated with X-linked hypohidrotic ectodermal dysplasia families. 24312213_WNT10A and EDA digenic mutations could result in oligodontia and syndromic tooth agenesis in the Chinese population. Moreover, our results will greatly expand the genotypic spectrum of tooth agenesis. 24503206_hemizygous nonsense mutation c.739C>T (p.Q247X) in exon 4 associated with x-linked hypohidrotic ectodermal dysplasia 24554542_involvement of PAX9, EDA, SPRY2, SPRY4, and WNT10A as risk factors for MLIA. uncovered 3 strong synergistic interactions between MLIA liability and MSX1-TGFA, AXIN2-TGFA, and SPRY2-SPRY4 gene pairs. 1st evidence of sprouty genes in MLIA susceptibility. 24985548_dentified a novel deletion mutation located in exon 1 which if expressed would produce a highly truncated protein in a Chinese Han family with X-linked hypohidrotic ectodermal dysplasia 25203534_novel non-synonymous mutation in ectodysplasin A (EDA) associated with non-syndromic X-linked dominant congenital tooth agenesis 25296636_novel nonsense mutation in Chinese family 25438642_hemizygous frame-shift mutation c. 731delG (p.R244Qfs*36) underlies hypohidrotic ectodermal dysplasia in a Japanese family 25626993_We found a novel missense mutation in exon 1 of the EDA1 gene in a putative Mayan family from Mexico with XL-HED. 25846883_A novel missense mutation in the EDA gene in a Chinese family with X-linked hypohidrotic ectodermal dysplasia. 26411740_we identified a novel and three reported EDA missense mutations in four of six patients with X-linked hypohidrotic ectodermal dysplasia. Missense mutations and the mutations affecting the tumor necrosis factor homology domain were correlated with fewer missing teeth. 26634545_Our findings indicate that a novel mutation (c.878T>G) of EDA is associated with XLHED and adds to the repertoire of EDA mutations. 26659383_EDA-A2 and its receptor XEDAR are overexpressed in epithelial cells of salivary glands in Sjogren's syndrome patients, in comparison with healthy individuals. The EDA-A2/XEDAR system in these cells is involved in the induction of apoptosis via CASP3 activation. 26753551_Based on a computerized protein structure analysis, we suggest that the change p.Arg289His in EDA impairs protein stabilization and thus might possibly be involved in the development of oligodontia concomitant with a mild ED phenotype. 27054699_EDA is an important candidate gene for two developmental diseases sharing the common feature of the congenital lack of teeth. In addition, these results can support the hypothesis that X-linked HED and EDA-related NTA are the same disease with different degrees of severity. 28045201_The new EDA variants expand the mutational pro fi le of hypohidrotic ectodermal dysplasia, and in two patients, a de novo mutation was identi fi ed. 28052341_rs132630321 (c.1013C>T) in EDA gene was found in non-syndromic X-linked oligodontia male patients. Heterozygous female carriers for this EDA mutation may present a highly variable and milder dental phenotype. Replacement of T338 native (hydrophilic) by an M338 mutated residue (hydrophobic) was predicted to alter the conformation of the 337-342 loop leading to a potential impact on EDA-A1 receptor binding properties. 28498389_Case Report: EDA mutation causing hypohidrotic ectodermal dysplasia with hyperplasia of the sebaceous glands in a Chinese patient. 28655773_Data suggest that EDA is highly expressed in meibomian glands and is detectible in human tears but not serum; EDA protein is secreted from meibomian glands and promotes corneal epithelial cell proliferation through regulation of EGFR signaling pathway. (EGFR = epidermal growth factor receptor) 28782908_EDA gene expression contributes to the maintenance of epithelial barrier function. 28813629_This is the first analysis of the role of Eda in the root, showing a direct role for this pathway during postnatal mouse development, and it suggests that changes in proliferation and angle of HERS may underlie taurodontism in a range of syndromes. 29444360_In conclusion, we propose that mutation c.648_683del36 and c.925-2A>G can indeed cause the HED, by destroying the structure and interaction properties of EDA protein. 29676859_A novel functional skipping-splicing EDA mutation was considered to be the cause of HED in the two pedigrees reported here. 30117778_Mutations were identified in all seven families, including four previously reported missense mutations (p.M1T, p.R156C, p.G299S, and p.A349T) and three novel mutations; missense mutation (p.Q358 L), indel (P228Tfs*52), as well as a large deletion. 30125999_Study analyzed two Japanese families with hypohidrotic ectodermal dysplasia and identified two novel mutations in the EDA gene. 30417976_One novel mutation (c.441_442insACTCT) and three reported mutations (c.252delT, c.463C>T, and c.1013C>T) in EDA were identified in families with ectodermal dysplasia. The novel EDA mutation was co-segregated with phenotype. 30526585_The EDA mutations of c.878 T > G, c.663-697del and c.587-615del may be responsible for the pathogenesis of autosomal recessive hypohidrotic ectodermal dysplasia in their pedigrees. 31129666_9 EDA mutations (3 reported previously, 6 novel) were found in 10 Korean X-linked hypohidrotic ectodermal dysplasia patients from 9 families. 2 of the novel mutations caused frameshift in protein synthesis (p.Pro229Profs*51, p.Ala238Leufs*42). 1 was a nonsense mutation (p.Gly73*). The remaining 3 novel missense variants (p.Thr378Lys, p.Val322Ala, p.Val251Met) were predicted to affect protein function. 31241787_Study of a two-generation X-linked hypohidrotic ectodermal dysplasia (HED) family found a novel frameshift mutation (NM_001399.4: c.381_382delinsG, p.Q128Rfs*9) in EDA gene. Proband had typical clinical features of HED and the mother had identical but milder features. Interestingly, some phenotypes of the mother appeared asymmetrically between the right and left side of the body that were not reported in previous studies. 31376704_A novel missense mutation p.S305R of EDA gene causes X-linked hypohidrotic ectodermal dysplasia in a Chinese family 31526774_ectodysplasin A aggravates steatosis by striking balance between lipid deposition and elimination; it was a potential biomarker of nonalcoholic fatty liver disease 31652981_The missense variant (c.1133C>T p.Thr378Met) has been mapped on the TNF-like domain of EDA and is predicted to change its functional nature. The binding of EDA variants to EDAR alters the recruitment of its adaptor protein, EDARADD, which is crucial in activating the NF-kappaB signaling pathway. 31796081_study conducted to date in the Spanish population affected by ED. The EDA, EDAR, EDARADD and WNT10A genes constitute the molecular basis in 70.8% of patients with a 74.6% yield in Hypohidrotic ectodermal dysplasia and 44.4% in non-syndromic tooth agenesis. 32736705_Ectodysplasin-A2 induces dickkopf 1 expression in human balding dermal papilla cells overexpressing the ectodysplasin A2 receptor. 33205897_Missense mutations in EDA and EDAR genes cause dominant syndromic tooth agenesis. 33746906_Ectodysplasin A Is Increased in Non-Alcoholic Fatty Liver Disease, But Is Not Associated With Type 2 Diabetes. 34573371_Gene Mutations of the Three Ectodysplasin Pathway Key Players (EDA, EDAR, and EDARADD) Account for More than 60% of Egyptian Ectodermal Dysplasia: A Report of Seven Novel Mutations. 34582123_Two novel ectodysplasin A gene mutations and prenatal diagnosis of X-linked hypohidrotic ectodermal dysplasia. 34817077_Understanding the impact of missense mutations on the structure and function of the EDA gene in X-linked hypohidrotic ectodermal dysplasia: A bioinformatics approach. 34863015_Ectodysplasin pathogenic variants affecting the furin-cleavage site and unusual clinical features define X-linked hypohidrotic ectodermal dysplasia in India. | ENSMUSG00000059327 | Eda | 603.611197 | 0.9992874 | -0.001028463 | 0.11177411 | 8.449437e-05 | 9.926659e-01 | 9.974203e-01 | No | Yes | 573.623958 | 51.976439 | 582.559372 | 52.699048 | |
| ENSG00000158985 | 56990 | CDC42SE2 | protein_coding | Q9NRR3 | FUNCTION: Probably involved in the organization of the actin cytoskeleton by acting downstream of CDC42, inducing actin filament assembly. Alters CDC42-induced cell shape changes. In activated T-cells, may play a role in CDC42-mediated F-actin accumulation at the immunological synapse. May play a role in early contractile events in phagocytosis in macrophages. {ECO:0000269|PubMed:10816584, ECO:0000269|PubMed:15840583}. | Cell membrane;Cell projection;Cell shape;Cytoplasm;Cytoskeleton;Lipoprotein;Membrane;Palmitate;Phagocytosis;Phosphoprotein;Reference proteome | hsa:56990; | cell projection [GO:0042995]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; phagocytic cup [GO:0001891]; plasma membrane [GO:0005886]; signaling adaptor activity [GO:0035591]; small GTPase binding [GO:0031267]; phagocytosis [GO:0006909]; regulation of cell shape [GO:0008360]; regulation of Rho protein signal transduction [GO:0035023]; regulation of signal transduction [GO:0009966] | 15840583_Recruitment of SPEC2 within Jurkat T cells to the antigen-presenting cell interface occurred following incubation with staphylococcal enterotoxin E superantigen-loaded B cells and colocalized there with F-actin and Cdc42. T cell receptor 17030554_Observational study of gene-disease association. (HuGE Navigator) 17030554_haplotypes underlying the SPEC2/PDZ-GEF2/acyl-CoA synthetase long-chain family member 6 region are associated with schizophrenia 18804346_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 23833064_Cdc42 is an important regulator of corneal epithelial wound repair. To promote healing, Cdc42 may interact with receptor tyrosine kinase-activated signaling cascades that participate in cell migration and cell-cycle progression. | ENSMUSG00000052298 | Cdc42se2 | 948.665336 | 1.0208766 | 0.029808487 | 0.09248059 | 1.036085e-01 | 7.475415e-01 | 9.191987e-01 | No | Yes | 982.776707 | 215.293560 | 961.385870 | 210.587137 | ||
| ENSG00000159761 | 388284 | C16orf86 | protein_coding | Q6ZW13 | Reference proteome | hsa:388284; | ENSMUSG00000013158 | 4933405L10Rik | 13.674185 | 1.0273154 | 0.038879137 | 0.72420465 | 2.676465e-03 | 9.587402e-01 | No | Yes | 11.982976 | 3.535175 | 11.840423 | 3.517989 | ||||||
| ENSG00000160282 | 10841 | FTCD | protein_coding | O95954 | FUNCTION: Folate-dependent enzyme, that displays both transferase and deaminase activity. Serves to channel one-carbon units from formiminoglutamate to the folate pool. {ECO:0000269|PubMed:12815595}.; FUNCTION: Binds and promotes bundling of vimentin filaments originating from the Golgi. {ECO:0000250|UniProtKB:O88618}. | Alternative splicing;Cytoplasm;Cytoskeleton;Disease variant;Folate-binding;Golgi apparatus;Histidine metabolism;Lyase;Multifunctional enzyme;Phosphoprotein;Reference proteome;Transferase | PATHWAY: Amino-acid degradation; L-histidine degradation into L-glutamate; L-glutamate from N-formimidoyl-L-glutamate (transferase route): step 1/1. {ECO:0000269|PubMed:12815595}. | The protein encoded by this gene is a bifunctional enzyme that channels 1-carbon units from formiminoglutamate, a metabolite of the histidine degradation pathway, to the folate pool. Mutations in this gene are associated with glutamate formiminotransferase deficiency. Alternatively spliced transcript variants have been found for this gene.[provided by RefSeq, Dec 2009]. | hsa:10841; | centriole [GO:0005814]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; plasma membrane [GO:0005886]; smooth endoplasmic reticulum membrane [GO:0030868]; folic acid binding [GO:0005542]; formimidoyltetrahydrofolate cyclodeaminase activity [GO:0030412]; glutamate formimidoyltransferase activity [GO:0030409]; microtubule binding [GO:0008017]; cytoskeleton organization [GO:0007010]; folic acid-containing compound metabolic process [GO:0006760]; histidine catabolic process to glutamate and formamide [GO:0019556]; histidine catabolic process to glutamate and formate [GO:0019557]; tetrahydrofolate interconversion [GO:0035999] | 12815595_Disease-causing mutations have been identified in the FTCD gene in three patients with the putative autosomal recessive disorder glutamate formiminotransferase deficiency. 19048631_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19204726_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20209057_Scyl1 interacts with 58K/formiminotransferase cyclodeaminase (FTCD) and golgin p115, and is required for the maintenance of Golgi morphology 20494980_The FTCD promoter is activated by serum depletion according to promoter reporter assays in HEK 293 cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 24686083_FTCD is a positive regulator of the hypoxia-HIF signaling pathway and has an important role in cell proliferation, metabolism and migration in HepG2 cells 29178637_The formiminotransferase-cyclodeaminase allelic spectrum comprised of 12 distinct variants including 5 missense alterations, an in-frame deletion, two frameshift variants and four nonsense variants with the remaining alterations predicted to affect mRNA processing/stability. 29927301_The rs914246 variant, but not the rs914245 variant, of the FTCD gene modulated accuracy in the task for younger, but not older, people under high working memory 30784016_The Diagnostic Value of Arginase-1, FTCD, and MOC-31 Expression in Early Detection of Hepatocellular Carcinoma (HCC) and in Differentiation Between HCC and Metastatic Adenocarcinoma to the Liver. 30893314_the minor allele (A) of rs61735836 (p.Val101Met) in exon 3 of FTCD was associated with increased urinary Inorganic arsenics % (P = 8x10-13), increased mono-methylated arsenic % (P = 2x10-16) and decreased di-methylated arsenic % (P = 6x10-23). 33555040_p97 and p47 function in membrane tethering in cooperation with FTCD during mitotic Golgi reassembly. | ENSMUSG00000001155 | Ftcd | 12.622848 | 0.7958868 | -0.329364775 | 0.84609349 | 1.573767e-01 | 6.915833e-01 | No | Yes | 10.075488 | 5.135859 | 12.216207 | 6.307821 | |
| ENSG00000160360 | 26086 | GPSM1 | protein_coding | Q86YR5 | FUNCTION: Guanine nucleotide dissociation inhibitor (GDI) which functions as a receptor-independent activator of heterotrimeric G-protein signaling. Keeps G(i/o) alpha subunit in its GDP-bound form thus uncoupling heterotrimeric G-proteins signaling from G protein-coupled receptors. Controls spindle orientation and asymmetric cell fate of cerebral cortical progenitors. May also be involved in macroautophagy in intestinal cells. May play a role in drug addiction. {ECO:0000269|PubMed:11024022, ECO:0000269|PubMed:12642577}. | Alternative splicing;Cell membrane;Cytoplasm;Developmental protein;Differentiation;Endoplasmic reticulum;Golgi apparatus;Membrane;Methylation;Neurogenesis;Phosphoprotein;Reference proteome;Repeat;TPR repeat | G-protein signaling modulators (GPSMs) play diverse functional roles through their interaction with G-protein subunits. This gene encodes a receptor-independent activator of G protein signaling, which is one of several factors that influence the basal activity of G-protein signaling systems. The protein contains seven tetratricopeptide repeats in its N-terminal half and four G-protein regulatory (GPR) motifs in its C-terminal half. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2011]. | hsa:26086; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; Golgi membrane [GO:0000139]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; GDP-dissociation inhibitor activity [GO:0005092]; cell differentiation [GO:0030154]; negative regulation of GTPase activity [GO:0034260]; negative regulation of guanyl-nucleotide exchange factor activity [GO:1905098]; nervous system development [GO:0007399]; positive regulation of macroautophagy [GO:0016239] | 10969064_The GPR motif in this protein and other proteins is actually associated with activity as a GDI - guanine nucleotide dissociation inhibitor and not GTPase activator as initially suggested. 12642577_AGS3 is involved in an early event during the autophagic pathway probably prior to the formation of the autophagosome. 12925752_cortical localization in mitotic cell culture systems and requirement for normal cell cycle progression 17991770_data support a model wherein AGS3 modulates the protein trafficking along the TGN/plasma membrane/endosome loop 18566450_The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G-proteins. 20065032_These data present AGS3, G-proteins, and mInsc as candidate proteins involved in regulating cellular stress associated with protein-processing pathologies. 20126274_unlike wild-type AGS3, over-expression of an AGS3 mutant lacking this modification fails to enhance macroautophagic activity. These observations imply that AGS3 phosphorylation may participate in the modulation of macroautophagy 20237496_Observational study of gene-disease association. (HuGE Navigator) 20716524_AGS3 receptor coupling to both Galphabetagamma and GPR-Galpha(i) offer additional flexibility for systems to respond and adapt to challenges and orchestrate complex behaviors 21209316_These results provide mechanistic insights into how reversible modulation of Galpha(i3) activity by AGS3 and GIV maintains the delicate equilibrium between promotion and inhibition of autophagy. 23945395_Data identified three new loci for type 2 diabetes with genome-wide significance: MIR129-LEP, GPSM1 and SLC16A13. 24307516_Our data indicate that decreased expression of AGS3 is correlated with reduced levels of p-CREB in the apoptotic model. The negative role of AGS3 in cell apoptosis was further confirmed by knocking down AGS3. 24510965_these results indicate that GRK6 complexes with AGS3-Galphai2 to regulate CXCR2-mediated leukocyte functions at different levels, including downstream effector activation, receptor trafficking, and expression at the cell membrane. 25480567_Results confirmed that Ric-8A can directly bind to AGS3S but failed to facilitate Galpha(i)-induced suppression of adenylyl cyclase, suggesting that it may not serve as a guanine exchange factor for AGS3/Galpha(i/o)-GDP complex in a cellular environment. 25740824_High GPSM1 gene methylation is associated with gastric tumor aggressiveness. 25812748_Activator of G-protein Signaling 3 is an important regulator of esophageal squamous cell carcinoma proliferation 26987813_Data support the notion that the Galpha, but not Gbetagamma, arm of the Gi/o signalling is involved in TRPC4 activation and unveil new roles for RGS and AGS3 in fine-tuning TRPC4 activities. 27270970_The G-protein Regulatory motif of AGS3 is critical for regulating MUC1/Muc1 expression and cytokine production in the inflammatory microenvironment. 31215992_the data indicate that the effect of AGS3 in prostate cancer development and progression is probably mediated via a MAPK/AR-dependent pathway. 31959871_Genome-wide meta-analysis associates GPSM1 with type 2 diabetes, a plausible gene involved in skeletal muscle function. 33172018_AGS3 and Galphai3 Are Concomitantly Upregulated as Part of the Spindle Orientation Complex during Differentiation of Human Neural Progenitor Cells. 33220708_Depletion of GPSM1 enhances ovarian granulosa cell apoptosis via cAMP-PKA-CREB pathway in vitro. 34257610_Knockdown of GPSM1 Inhibits the Proliferation and Promotes the Apoptosis of B-Cell Acute Lymphoblastic Leukemia Cells by Suppressing the ADCY6-RAPGEF3-JNK Signaling Pathway. | ENSMUSG00000026930 | Gpsm1 | 1156.648913 | 0.9275790 | -0.108457961 | 0.08557181 | 1.606552e+00 | 2.049771e-01 | 5.758100e-01 | No | Yes | 1014.852395 | 110.413500 | 1095.404827 | 119.011006 | |
| ENSG00000160602 | 284086 | NEK8 | protein_coding | Q86SG6 | FUNCTION: Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia (By similarity). Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway. {ECO:0000250, ECO:0000269|PubMed:23418306}. | ATP-binding;Cell projection;Ciliopathy;Cilium;Cytoplasm;Cytoskeleton;Disease variant;Kinase;Magnesium;Metal-binding;Nephronophthisis;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the serine/threionine protein kinase family related to NIMA (never in mitosis, gene A) of Aspergillus nidulans. The encoded protein may play a role in cell cycle progression from G2 to M phase. Mutations in the related mouse gene are associated with a disease phenotype that closely parallels the juvenile autosomal recessive form of polycystic kidney disease in humans. [provided by RefSeq, Jul 2008]. | hsa:284086; | centrosome [GO:0005813]; ciliary base [GO:0097546]; ciliary inversin compartment [GO:0097543]; cilium [GO:0005929]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; animal organ morphogenesis [GO:0009887]; chromosome segregation [GO:0007059]; determination of left/right symmetry [GO:0007368]; heart development [GO:0007507]; regulation of hippo signaling [GO:0035330] | 15019993_Data demonstrate for the first time that Nek8 is a novel tumor associated gene, and shares considerable sequence homology with the Nek family of protein kinases and may be involved in G(2)/M progression. 15872312_characterization of the proteome in mice that have a double point mutation in the related gene. 18199800_mutations cause nephronophthisis; mutant forms show defects in ciliary localization 21068128_Observational study of gene-disease association. (HuGE Navigator) 22106379_study finds that induction of ciliogenesis upon cell cycle exit is accompanied by both activation and proteasomal degradation of Nek8, and that activation is dependent upon phosphorylation within the catalytic domain 23026745_NPHP9 promotes signalling through the transcriptional co-activator TAZ. 23418306_NEK8 is essential for organ development and that the complete loss of NEK8 perturbs multiple signalling pathways resulting in a severe early embryonic phenotype. 23793029_ANKS6 as a new NPHP family member that assembles a distinct module of nephronophthisis-associated proteins, encompassing NEK8, INVS and NPHP3. 23973373_Mutation in NEK8 is associated with renal ciliopathies 25451921_NEK8 may be a new target gene of HIFs; pVHL can down-regulate NEK8 via HIFs to maintain the primary cilia structure in human renal cancer cells 26697755_The mutations: c.2069_2070insC variant (p.Ter693LeufsTer86), and a c.1043C>T variant (p.Thr348Met) in RCC1 domain of NEK8 in two brothers with cardiac, renal, and hepatic anomalies 26967905_our study demonstrates that NEK8 human mutations cause major organ developmental defects due to altered ciliogenesis and cell differentiation/proliferation through deregulation of the Hippo pathway 27892797_NEK8 plays a critical role in replication fork stability through its regulation of the DNA repair and replication fork protection protein RAD51. 31633649_Homozygous NEK8 Mutations in Siblings With Neonatal Cholestasis Progressing to End-stage Liver, Renal, and Cardiac Disease. | ENSMUSG00000017405 | Nek8 | 81.764608 | 0.7735429 | -0.370446714 | 0.32779313 | 1.306268e+00 | 2.530715e-01 | No | Yes | 57.593284 | 9.684672 | 73.473716 | 12.319390 | ||
| ENSG00000160766 | 2630 | GBAP1 | transcribed_unprocessed_pseudogene | 28983119_Using skin-derived induced pluripotent stem cells of PD patients with GBA mutations and controls, we observed a significant GBA up-regulation during dopaminergic differentiation, paralleled by down-regulation of miR-22-3p. Our results describe the first microRNA controlling GBA and suggest that the GBAP1 non-coding RNA functions as a GBA ceRNA 30951202_Pseudogene GBAP1 contributes to the development and progression of gastric cancer by sequestering the miR-212-3p from binding to GBA. 31670439_Parkinson's Disease: Glucocerebrosidase 1 Mutation Severity Is Associated with CSF Alpha-Synuclein Profiles. | 159.413189 | 0.9108356 | -0.134737481 | 0.23656753 | 3.102299e-01 | 5.775391e-01 | No | Yes | 122.353499 | 20.278577 | 135.423269 | 22.362012 | ||||||||||
| ENSG00000161048 | 222236 | NAPEPLD | protein_coding | Q6IQ20 | FUNCTION: D-type phospholipase that hydrolyzes N-acyl-phosphatidylethanolamines (NAPEs) to produce bioactive N-acylethanolamines/fatty acid ethanolamides (NAEs/FAEs) and phosphatidic acid (PubMed:14634025, PubMed:16527816, PubMed:27571266, PubMed:25684574). Cleaves the terminal phosphodiester bond of diacyl- and alkenylacyl-NAPEs, primarily playing a role in the generation of long-chain saturated and monounsaturated NAEs in the brain (By similarity). May control NAPE homeostasis in dopaminergic neuron membranes and regulate neuron survival, partly through RAC1 activation (By similarity). As a regulator of lipid metabolism in the adipose tissue, mediates the crosstalk between adipocytes, gut microbiota and immune cells to control body temperature and weight. In particular, regulates energy homeostasis by promoting cold-induced brown or beige adipocyte differentiation program to generate heat from fatty acids and glucose. Has limited D-type phospholipase activity toward N-acyl lyso-NAPEs (By similarity). {ECO:0000250|UniProtKB:Q8BH82, ECO:0000269|PubMed:14634025, ECO:0000269|PubMed:16527816, ECO:0000269|PubMed:25684574, ECO:0000269|PubMed:27571266}. | 3D-structure;Acetylation;Endosome;Golgi apparatus;Hydrolase;Lipid degradation;Lipid metabolism;Membrane;Metal-binding;Nucleus;Phospholipid degradation;Phospholipid metabolism;Reference proteome;Zinc | NAPEPLD is a phospholipase D type enzyme that catalyzes the release of N-acylethanolamine (NAE) from N-acyl-phosphatidylethanolamine (NAPE) in the second step of the biosynthesis of N-acylethanolamine (Okamoto et al., 2004 [PubMed 14634025]).[supplied by OMIM, Oct 2008]. | hsa:222236; | cytoplasm [GO:0005737]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; membrane-bounded organelle [GO:0043227]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; photoreceptor outer segment membrane [GO:0042622]; bile acid binding [GO:0032052]; identical protein binding [GO:0042802]; N-acetylphosphatidylethanolamine-hydrolysing phospholipase activity [GO:0102200]; N-acylphosphatidylethanolamine-specific phospholipase D activity [GO:0070290]; zinc ion binding [GO:0008270]; host-mediated regulation of intestinal microbiota composition [GO:0048874]; N-acylethanolamine metabolic process [GO:0070291]; N-acylphosphatidylethanolamine metabolic process [GO:0070292]; phospholipid catabolic process [GO:0009395]; positive regulation of brown fat cell differentiation [GO:0090336]; positive regulation of inflammatory response [GO:0050729]; retinoid metabolic process [GO:0001523]; temperature homeostasis [GO:0001659] | 19715685_The human NAPE-PLD was activated by phosphatidylethanolamine and inhibited by the beta-lactamase substrate nitrocefin. 20369362_During the menstrual cycle, NAPE-PLD immunoreactivity was down-regulated in the secretory epithelial gland compared to the proliferative epithelial gland and unaffected in the stroma. 20885390_Observational study of gene-disease association. (HuGE Navigator) 20885390_a common haplotype in NAPEPLD, an enzyme involved in endocannabinoid synthesis, was protective against obesity. 21251115_The results of this study suggested that NAPEPLD was alterated in multip;e sclerosis. 22827915_NAPE-PLD expression increase steadily after infancy, peaking in adulthood. 23122699_The differential expression of NAPE-PLD and FAAH suggests that Anandamide could play an important role in the pathophysiology of preeclampsia. 24018423_a major physiological role of NAPE-PLD 24126189_our results do not support a clear role of FAAH, CNR1 and NAPE-PLD in BD and lithium response. 25684574_NAPE-PLD forms homodimers partly separated by an internal channel and uniquely adapted to associate with phospholipids. 29652995_The AC genotype and C allele of NAPE-PLD rs12540583 locus are risk factors for schizophrenia. 30399323_Transcriptome analysis revealed that deletion of NAPE-PLD activates a transcriptional program for nutrient transportation, including lipids and lipoproteins, and inactivates cell-cycle or mitosis-related genes in Caco-2 cells. | ENSMUSG00000044968 | Napepld | 208.216548 | 1.3015773 | 0.380260994 | 0.24865794 | 2.266439e+00 | 1.322033e-01 | No | Yes | 235.671930 | 63.726767 | 172.691040 | 46.732743 | ||
| ENSG00000161551 | 84765 | ZNF577 | protein_coding | Q9BSK1 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:84765; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 21424380_Single nucleotide polymorphisms in ZNF577 gene is associated with breast cancer. 32390948_ZNF577 Methylation Levels in Leukocytes From Women With Breast Cancer Is Modulated by Adiposity, Menopausal State, and the Mediterranean Diet. | 119.980799 | 1.1189425 | 0.162135883 | 0.26026466 | 3.866311e-01 | 5.340756e-01 | No | Yes | 102.862939 | 26.126423 | 94.801823 | 24.037555 | |||||
| ENSG00000162062 | 80178 | TEDC2 | protein_coding | Q7L2K0 | FUNCTION: Acts as a positive regulator of ciliary hedgehog signaling. Required for centriole stability. {ECO:0000250|UniProtKB:Q6GQV0}. | Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:80178; | centriole [GO:0005814]; cilium [GO:0005929]; cytoplasm [GO:0005737]; positive regulation of smoothened signaling pathway [GO:0045880] | ENSMUSG00000024118 | Tedc2 | 968.231932 | 1.0186290 | 0.026628668 | 0.09568053 | 7.764170e-02 | 7.805191e-01 | 9.298207e-01 | No | Yes | 1002.553081 | 118.623529 | 991.178250 | 117.212829 | |||
| ENSG00000162073 | 124222 | PAQR4 | protein_coding | Q8N4S7 | Alternative splicing;Membrane;Receptor;Reference proteome;Transmembrane;Transmembrane helix | hsa:124222; | integral component of membrane [GO:0016021]; signaling receptor activity [GO:0038023] | 29228296_Data indicate that PAQR4 has a tumorigenic effect on human breast cancers, and such effect is associated with a modulatory activity of PAQR4 on protein degradation of CDK4 30322804_PAQR4 (Progestin and AdipoQ Receptor 4) expression is closely associated with progression of many cancers and microRNA (miRNA) processing. 32206121_PAQR4 promotes chemoresistance in non-small cell lung cancer through inhibiting Nrf2 protein degradation. 32291319_Golgi-Localized PAQR4 Mediates Antiapoptotic Ceramidase Activity in Breast Cancer. 34718369_PAQR4 promotes the development of hepatocellular carcinoma by activating PI3K/AKT pathway. 35240821_Overexpressed PAQR4 predicts poor overall survival and construction of a prognostic nomogram based on PAQR family for hepatocellular carcinoma. | ENSMUSG00000023909 | Paqr4 | 1785.394613 | 0.9197026 | -0.120760750 | 0.07308820 | 2.726951e+00 | 9.866749e-02 | 4.191236e-01 | No | Yes | 1611.034935 | 186.932132 | 1783.401089 | 206.705559 | |||
| ENSG00000162241 | 283130 | SLC25A45 | protein_coding | Q8N413 | Alternative splicing;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | SLC25A45 belongs to the SLC25 family of mitochondrial carrier proteins (Haitina et al., 2006 [PubMed 16949250]).[supplied by OMIM, Mar 2008]. | hsa:283130; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; acyl carnitine transmembrane transporter activity [GO:0015227]; acyl carnitine transport [GO:0006844]; amino acid transport [GO:0006865] | 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. | ENSMUSG00000024818 | Slc25a45 | 64.699846 | 0.8928390 | -0.163528052 | 0.33016127 | 2.390644e-01 | 6.248826e-01 | No | Yes | 65.979793 | 11.044915 | 69.298785 | 11.487129 | |||
| ENSG00000162522 | 57648 | KIAA1522 | protein_coding | Q9P206 | Alternative splicing;Lipoprotein;Methylation;Myristate;Phosphoprotein;Reference proteome | hsa:57648; | cell differentiation [GO:0030154] | 27098511_high expression of KIAA1522 can be used as an independent biomarker for predication of poor survival and platinum-resistance of NSCLC patients, and aberrant KIAA1522 might be a new target for the therapy of the disease. 30177391_miR-125b-5p functions as a tumor suppressor and regulates breast cancer cell progression through targeting KIAA1522 32854746_KIAA1522 potentiates TNFalpha-NFkappaB signaling to antagonize platinum-based chemotherapy in lung adenocarcinoma. | ENSMUSG00000050390 | C77080 | 1461.419790 | 1.0788639 | 0.109512876 | 0.08854304 | 1.542025e+00 | 2.143166e-01 | 5.864815e-01 | No | Yes | 1358.296794 | 132.698051 | 1254.037452 | 122.392296 | |||
| ENSG00000162600 | 115209 | OMA1 | protein_coding | Q96E52 | FUNCTION: Metalloprotease that is part of the quality control system in the inner membrane of mitochondria (PubMed:20038677, PubMed:25605331, PubMed:32132706, PubMed:32132707). Activated in response to various mitochondrial stress, leading to the proteolytic cleavage of target proteins, such as OPA1, UQCC3 and DELE1 (PubMed:20038677, PubMed:25275009, PubMed:32132706, PubMed:32132707). Following stress conditions that induce loss of mitochondrial membrane potential, mediates cleavage of OPA1 at S1 position, leading to OPA1 inactivation and negative regulation of mitochondrial fusion (PubMed:20038677, PubMed:25275009). Also acts as a regulator of apoptosis: upon BAK and BAX aggregation, mediates cleavage of OPA1, leading to the remodeling of mitochondrial cristae and allowing the release of cytochrome c from mitochondrial cristae (PubMed:25275009). In depolarized mitochondria, may also act as a backup protease for PINK1 by mediating PINK1 cleavage and promoting its subsequent degradation by the proteasome (PubMed:30733118). May also cleave UQCC3 in response to mitochondrial depolarization (PubMed:25605331). Also acts as an activator of the integrated stress response (ISR): in response to mitochondrial stress, mediates cleavage of DELE1 to generate the processed form of DELE1 (S-DELE1), which translocates to the cytosol and activates EIF2AK1/HRI to trigger the ISR (PubMed:32132706, PubMed:32132707). Its role in mitochondrial quality control is essential for regulating lipid metabolism as well as to maintain body temperature and energy expenditure under cold-stress conditions (By similarity). Binds cardiolipin, possibly regulating its protein turnover (By similarity). Required for the stability of the respiratory supercomplexes (By similarity). {ECO:0000250|UniProtKB:Q9D8H7, ECO:0000269|PubMed:20038677, ECO:0000269|PubMed:25275009, ECO:0000269|PubMed:25605331, ECO:0000269|PubMed:30733118, ECO:0000269|PubMed:32132706, ECO:0000269|PubMed:32132707}. | Alternative splicing;Autocatalytic cleavage;Disulfide bond;Hydrolase;Lipid-binding;Membrane;Metal-binding;Metalloprotease;Mitochondrion;Mitochondrion inner membrane;Protease;Reference proteome;Transit peptide;Transmembrane;Transmembrane helix;Zinc;Zymogen | hsa:115209; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; mitochondrial inner membrane [GO:0005743]; mitochondrial membrane [GO:0031966]; lipid binding [GO:0008289]; metal ion binding [GO:0046872]; metalloendopeptidase activity [GO:0004222]; cristae formation [GO:0042407]; diet induced thermogenesis [GO:0002024]; energy homeostasis [GO:0097009]; glucose metabolic process [GO:0006006]; HRI-mediated signaling [GO:0140468]; integrated stress response signaling [GO:0140467]; lipid metabolic process [GO:0006629]; mitochondrial protein processing [GO:0034982]; mitochondrial respiratory chain complex assembly [GO:0033108]; negative regulation of mitochondrial fusion [GO:0010637]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cold-induced thermogenesis [GO:0120162]; protein autoprocessing [GO:0016540]; protein quality control for misfolded or incompletely synthesized proteins [GO:0006515]; proteolysis involved in cellular protein catabolic process [GO:0051603]; regulation of apoptotic process [GO:0042981]; regulation of cristae formation [GO:1903850]; zymogen activation [GO:0031638] | 17615298_These results provide evidence for different substrate specificities of m-AAA proteases composed of different subunits and reveal a striking evolutionary switch of proteases involved in the proteolytic processing of dynamin-like GTPases in mitochondria. 20038677_OMA1 controls OPA1 cleavage and function. 20877624_Observational study of gene-disease association. (HuGE Navigator) 24634514_Cleavage of the inner membrane fusion factor L-OPA1 is prevented due to the failure to activate the inner membrane protease OMA1 in mitochondria that have a collapsed membrane potential. 24719224_OMA1 is cleaved to a short form (S-OMA1) by itself upon mitochondrial membrane depolarization; S-OMA1 is degraded quickly but could be stabilized by CCCP treatment or Prohibitin knockdown in cells. 25112877_These findings demonstrate that (a) p53 and Oma1 mediate L-Opa1 processing, (b) mitochondrial fragmentation is involved in CDDP-induced apoptosis in OVCA and CECA cells, and (c) dysregulated mitochondrial dynamics 25275009_The mitochondrial metalloprotease OMA1 was activated in a Bax- and Bak-dependent fashion. 26923599_differential stress-induced degradation of YME1L and OMA1 as a mechanism for sensitively adapting mitochondrial inner membrane protease activity and function in response to distinct types of cellular insults. 29545505_we identify AFG3L2 [matrix (m)-AAA complex] as the major protease mediating this event, which acts by maturing the 60 kDa pre-pro-OMA1 to the 40 kDa pro-OMA1 form by severing the N-terminal portion without recognizing a specific consensus sequence. 29748581_leptin targeting the GSK3/OMA1/OPA1 signaling pathway can optimize hMSCs therapy for cardiovascular diseases such 30714136_Studies present evidence that OMA1 activation promotes cell death and its inhibition is protective against it. OMA1 activation on gene expression and protein levels is regulated by many tumor proteins. [review] 30733118_Mitochondrial membrane potential loss-dependent PINK1 import arrest does not result solely from Tim23 inactivation but also through an actively regulated 'tug of war' between Tom7 and OMA1. 31611601_Depletion of mitochondrial protease OMA1 alters proliferative properties and promotes metastatic growth of breast cancer cells. 31729644_Overexpression of ROMO1 and OMA1 are Potentially Biomarkers and Predict Unfavorable Prognosis in Gastric Cancer. 32132707_mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway; the OMA1-DELE1-HRI pathway represents a potential therapeutic target that could enable fine-tuning of the integrated stress response for beneficial outcomes in diseases that involve mitochondrial dysfunction 32236861_A Common Missense Variant in OMA1 Associated with the Prognosis of Heart Failure. 33130089_OMA1-An integral membrane protease? 33200421_Mitochondrial Safeguard: a stress response that offsets extreme fusion and protects respiratory function via flickering-induced Oma1 activation. 34400172_Tau phosphorylation and OPA1 proteolysis are unrelated events: Implications for Alzheimer's Disease. 34414449_Circular RNA OMA1 regulates the progression of breast cancer via modulation of the miR1276/SIRT4 axis. 35163244_The Mitochondrial PHB2/OMA1/DELE1 Pathway Cooperates with Endoplasmic Reticulum Stress to Facilitate the Response to Chemotherapeutics in Ovarian Cancer. | ENSMUSG00000035069 | Oma1 | 810.819889 | 0.9923088 | -0.011138880 | 0.09754774 | 1.303636e-02 | 9.090976e-01 | 9.672991e-01 | No | Yes | 814.805494 | 149.040334 | 825.657141 | 150.993502 | ||
| ENSG00000162631 | 22854 | NTNG1 | protein_coding | Q9Y2I2 | FUNCTION: Involved in controlling patterning and neuronal circuit formation at the laminar, cellular, subcellular and synaptic levels. Promotes neurite outgrowth of both axons and dendrites. {ECO:0000269|PubMed:21946559}. | 3D-structure;Alternative splicing;Cell membrane;Developmental protein;Differentiation;Direct protein sequencing;Disulfide bond;GPI-anchor;Glycoprotein;Laminin EGF-like domain;Lipoprotein;Membrane;Neurogenesis;Reference proteome;Repeat;Signal | This gene encodes a preproprotein that is processed into a secreted protein containing eukaroytic growth factor (EGF)-like domains. This protein acts to guide axon growth during neuronal development. Polymorphisms in this gene may be associated with schizophrenia. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Aug 2015]. | hsa:22854; | anchored component of plasma membrane [GO:0046658]; anchored component of presynaptic active zone membrane [GO:0099029]; extracellular region [GO:0005576]; glutamatergic synapse [GO:0098978]; laminin complex [GO:0043256]; plasma membrane [GO:0005886]; Schaffer collateral - CA1 synapse [GO:0098685]; cell adhesion molecule binding [GO:0050839]; cell-cell adhesion mediator activity [GO:0098632]; animal organ morphogenesis [GO:0009887]; axonogenesis [GO:0007409]; basement membrane assembly [GO:0070831]; cell migration [GO:0016477]; modulation of chemical synaptic transmission [GO:0050804]; regulation of neuron migration [GO:2001222]; regulation of neuron projection arborization [GO:0150011]; regulation of neuron projection development [GO:0010975]; substrate adhesion-dependent cell spreading [GO:0034446]; synaptic membrane adhesion [GO:0099560]; tissue development [GO:0009888] | 14595443_Netrin-G1 is an important part of the NGL-1 receptor and functions to promote the outgrowth of dorsal thalamic axons. 15508520_Observational study of gene-disease association. (HuGE Navigator) 15508520_findings suggest that netrin G1 or a nearby gene may contribute to overall genetic risk for schizophrenia 15705354_Specific haplotypes encompassing alternatively spliced exons of NTNG1 were associated with schizophrenia, and concordantly, messenger ribonucleic acid isoform expression was significantly different between schizophrenic and control brains. 15870826_Sequence analysis of the cloned junction fragment indicated that on chromosome 1 the predominantly brain-expressed Netrin G1 (NTNG1) gene is disrupted, whereas on chromosome 7 there was no indication for a truncated gene 15901489_NTNG1 may use alternative splicing to diversify its function in a developmentally and tissue-specific manner. 16502428_Mutations in the NTNG1 gene appear to be a rare cause of Rett syndrome but NTNG1 function demands further investigation in relation to the central nervous system pathophysiology of the disorder. 17507910_The data of this stusty implicate NTNG1 in the pathophysiology of schizophrenia and bipolar disorder, but do not support the hypothesis that altered mRNA expression is the mechanism by which genetic variation of NTNG1 may confer disease susceptibility. 17903671_Netrin G1 is not involved in atypical Rett syndrome or in unexplained encephalopathy with epilepsy, but in specific forms to be delineated better in the future. 18384956_Observational study of gene-disease association. (HuGE Navigator) 18628988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20029409_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21079607_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22227940_Genotype and allele frequencies of single nucleotide polymorphisms on NTNG1 are significantly associated with schizophrenia. 23769687_Our finding nominates the minor G allele of the NTNG1 rs628117 single nucleotide polymorphism as a risk factor for ischemic stroke at least in Armenian population. 23986473_Interaction between the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses. 25325217_examined the hypothesis that NTNG1 allelic variation contributes to the risk for schizophrenia 28074533_Significant genetic family-based associations were detected between NTNG1 polymorphisms and cocaine dependence. NTNG1 expression in BA10, BA46 and the cerebellum, however, were not significantly associated with any allele or haplotype of this gene. 32503821_A 117-base pair SARS-CoV-2 orf1b sequence matched a sequence in the human genome with 94.6% identity. The sequence was in chromosome 1p within an intronic region of the netrin G1 (NTNG1) gene, implicated in schizophrenia. 34073619_A Genetic Study of Cerebral Atherosclerosis Reveals Novel Associations with NTNG1 and CNOT3. | ENSMUSG00000059857 | Ntng1 | 25.427661 | 0.5483695 | -0.866779692 | 0.53270053 | 2.645783e+00 | 1.038250e-01 | No | Yes | 18.537254 | 6.611897 | 35.179552 | 12.464001 | ||
| ENSG00000162849 | 55083 | KIF26B | protein_coding | Q2KJY2 | FUNCTION: Essential for embryonic kidney development. Plays an important role in the compact adhesion between mesenchymal cells adjacent to the ureteric buds, possibly by interacting with MYH10. This could lead to the establishment of the basolateral integrity of the mesenchyme and the polarized expression of ITGA8, which maintains the GDNF expression required for further ureteric bud attraction. Although it seems to lack ATPase activity it is constitutively associated with microtubules (By similarity). {ECO:0000250}. | ATP-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Developmental protein;Disease variant;Microtubule;Motor protein;Nucleotide-binding;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene is an intracellular motor protein thought to transport organelles along microtubules. The encoded protein is required for kidney development. Elevated levels of this protein have been found in some breast and colorectal cancers. [provided by RefSeq, Mar 2017]. | hsa:55083; | cytoplasm [GO:0005737]; kinesin complex [GO:0005871]; microtubule [GO:0005874]; ATP binding [GO:0005524]; microtubule binding [GO:0008017]; microtubule motor activity [GO:0003777]; establishment of cell polarity [GO:0030010]; positive regulation of cell-cell adhesion [GO:0022409]; ureteric bud invasion [GO:0072092] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23585914_High expression of KIF26B in breast cancer associates with poor prognosis. 25652119_KIF26B plays an important role in colorectal carcinogenesis and functions as a novel prognostic indicator and a potential therapeutic target for CRC. 28581513_KIF26B, a novel oncogene regulated by miR-372, promotes proliferation and metastasis through the VEGF pathway in gastric cancer 29880787_Upregulation of KIF26B enhanced proliferation and migration of ovarian cancer cells in vitro 30151950_KIF26B may play a critical role in the brain development and, when mutated, cause pontocerebellar hypoplasia with arthrogryposis. 30248545_KIF26B promoted the development and progression of breast cancer and might act as a potential therapeutic target for treating breast cancer. 31077021_Our results did prove a statistical association of both rs2228557 and rs12407427 genotypes (TT and CT + CC) and allele (T) with KTCN susceptibility in Iranian population. 34817735_ELK1-induced up-regulation of KIF26B promotes cell cycle progression in breast cancer. | ENSMUSG00000026494 | Kif26b | 309.156305 | 0.8673047 | -0.205389124 | 0.17742648 | 1.372168e+00 | 2.414398e-01 | No | Yes | 303.356674 | 41.465074 | 346.782308 | 47.422530 | ||
| ENSG00000162885 | 148789 | B3GALNT2 | protein_coding | Q8NCR0 | FUNCTION: Beta-1,3-N-acetylgalactosaminyltransferase that synthesizes a unique carbohydrate structure, GalNAc-beta-1-3GlcNAc, on N- and O-glycans. Has no galactose nor galactosaminyl transferase activity toward any acceptor substrate. Involved in alpha-dystroglycan (DAG1) glycosylation: acts coordinately with GTDC2/POMGnT2 to synthesize a GalNAc-beta3-GlcNAc-beta-terminus at the 4-position of protein O-mannose in the biosynthesis of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan, which is required for binding laminin G-like domain-containing extracellular proteins with high affinity. {ECO:0000269|PubMed:14724282, ECO:0000269|PubMed:23453667, ECO:0000269|PubMed:23929950}. | Alternative splicing;Congenital muscular dystrophy;Disease variant;Dystroglycanopathy;Endoplasmic reticulum;Glycoprotein;Glycosyltransferase;Golgi apparatus;Lissencephaly;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene encodes a member of the glycosyltransferase 31 family. The encoded protein synthesizes GalNAc:beta-1,3GlcNAc, a novel carbohydrate structure, on N- and O-glycans. Alternatively spliced transcript variants that encode different isoforms have been described. [provided by RefSeq, Mar 2013]. | hsa:148789; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; acetylgalactosaminyltransferase activity [GO:0008376]; glycosyltransferase activity [GO:0016757]; UDP-glycosyltransferase activity [GO:0008194]; protein glycosylation [GO:0006486]; protein O-linked glycosylation [GO:0006493] | 14724282_Although the GalNAcbeta1-3GlcNAcbeta1-R structure has not been reported in humans or other mammals, we have discovered a novel human glycosyltransferase producing this structure on N- and O-glycans. 23453667_Results demonstrate a role for B3GALNT2 in the glycosylation of alpha-DG and show that B3GALNT2 mutations can cause dystroglycanopathy with muscle and brain involvement. 24084573_B3GALNT2 is a gene associated with congenital muscular dystrophy with brain malformations. 24285400_B3GALNT2 overexpression is associated with breast cancer. 29273094_Mutations in B3GALNT2 give rise to a novel muscular dystrophy-dystroglycanopathies syndrome presentation, characterized by intellectual disability and seizure, but without any apparent muscular involvement. 29618368_B3GALNT2 reduced expression of some metabolic enzymes and thus downregulated levels of secreted acetoacetate. This relieved the activity of MIF and enhanced macrophage recruitment to promote tumor growth. 30898876_B3GALNT2 primarily transferred LDN to intracellular glycoproteins, thereby clearly delineating proteins that carry type-I or type-II LacdiNAcs 33290285_Novel mutations in B3GALNT2 gene causing alpha-dystroglycanopathy in Chinese patients. 35338537_Prenatal diagnosis of Walker-Warburg syndrome due to compound mutations in the B3GALNT2 gene. | ENSMUSG00000039242 | B3galnt2 | 2304.086614 | 1.0266606 | 0.037959365 | 0.06939952 | 3.015002e-01 | 5.829435e-01 | 8.463418e-01 | No | Yes | 2309.728942 | 331.661780 | 2237.357745 | 321.212230 |
| ENSG00000163075 | 200373 | CFAP221 | protein_coding | Q4G0U5 | FUNCTION: May play a role in cilium morphogenesis. {ECO:0000250|UniProtKB:A9Q751}. | Alternative splicing;Calmodulin-binding;Cell projection;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Reference proteome | hsa:200373; | axoneme [GO:0005930]; cilium [GO:0005929]; extracellular region [GO:0005576]; sperm flagellum [GO:0036126]; calmodulin binding [GO:0005516]; cerebrospinal fluid circulation [GO:0090660]; cilium assembly [GO:0060271]; motile cilium assembly [GO:0044458]; mucociliary clearance [GO:0120197] | 18039845_plays an important role in ciliary and flagellar biogenesis and motility sperm flagella and the cilia of respiratory epithelial cells and brain ependymal cells in both mice and humans 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 31636325_These results show that genetic variants in CFAP221 cause Pprimary ciliary dyskinesia (PCD) and that CFAP221 should be considered a candidate gene in cases where PCD is suspected but cilia structure and beat frequency appear normal. | ENSMUSG00000036962 | Cfap221 | 40.595100 | 1.1560383 | 0.209189140 | 0.41707577 | 2.440444e-01 | 6.213002e-01 | No | Yes | 45.908455 | 15.771806 | 38.435701 | 13.176714 | |||
| ENSG00000163374 | 55249 | YY1AP1 | protein_coding | Q9H869 | FUNCTION: Associates with the INO80 chromatin remodeling complex, which is responsible for transcriptional regulation, DNA repair, and replication (PubMed:27939641). Enhances transcription activation by YY1 (PubMed:14744866). Plays a role in cell cycle regulation (PubMed:17541814, PubMed:27939641). {ECO:0000269|PubMed:14744866, ECO:0000269|PubMed:17541814, ECO:0000269|PubMed:27939641}. | Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | The encoded gene product presumably interacts with YY1 protein; however, its exact function is not known. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:55249; | cytoplasm [GO:0005737]; fibrillar center [GO:0001650]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription coregulator activity [GO:0003712]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; regulation of cell cycle [GO:0051726]; regulation of transcription, DNA-templated [GO:0006355] | 11856496_human liver cancer associated gene 14744866_YY1AP is a novel co-activator of YY1 17541814_HCCA2 may play a novel role in cell cycle regulation. 25597408_YY1AP1 may serve as a key molecular target for EpCAM(+) AFP(+) HCC subtype 27939641_Loss-of-Function Mutations in YY1AP1 Lead to Grange Syndrome and a Fibromuscular Dysplasia-Like Vascular Disease. 28332498_Data indicate WW-binding protein 2 (WBP2) as an important co-factor of YY1 associated protein 1 (YAP) that enhances YAP/TEAD-mediated gene transcription. 29330849_Study proposed that the overexpression of YAP and TAZ around the human molluscum contagiosum (MC) virus infected skin lesions supports the hypothesis that the Hippo signaling pathway plays a key role in the development of MC. It is also conceivable that MCV contributes to the development of an infectious environment by increasing the expression of YAP/TAZ and subsequently inhibiting TBK1. 30556293_This is the first report of biallelic YY1AP1 variants in noncoding regions and just the second family with multiple affected siblings. 31270375_Hemorrhagic stroke and renovascular hypertension with Grange syndrome arising from a novel pathogenic variant in YY1AP1. 31481532_The results suggest that YAP/TAZ may be modulating cell volume in combination with cytoskeletal tension during cell cycle progression. 31989743_Prostaglandin E2 and its receptor EP2 trigger signaling that contributes to YAP-mediated cell competition. 33155207_CUL4A promotes proliferation and inhibits apoptosis of colon cancer cells via regulating Hippo pathway. 33971976_Whole genome sequencing reveals a frameshift mutation and a large deletion in YY1AP1 in a girl with a panvascular artery disease. 34808502_KAT6A is associated with sorafenib resistance and contributes to progression of hepatocellular carcinoma by targeting YAP. 35121738_ROCK1 mechano-signaling dependency of human malignancies driven by TEAD/YAP activation. | ENSMUSG00000054199 | Gon4l | 2183.892843 | 1.0442924 | 0.062525686 | 0.06616958 | 8.909918e-01 | 3.452087e-01 | 7.031012e-01 | No | Yes | 2258.240604 | 148.809508 | 2172.066597 | 143.158946 | |
| ENSG00000163376 | 84541 | KBTBD8 | protein_coding | Q8NFY9 | FUNCTION: Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin ligase complex that acts as a regulator of neural crest specification (PubMed:26399832). The BCR(KBTBD8) complex acts by mediating monoubiquitination of NOLC1 and TCOF1: monoubiquitination promotes the formation of a NOLC1-TCOF1 complex that acts as a platform to connect RNA polymerase I with enzymes responsible for ribosomal processing and modification, leading to remodel the translational program of differentiating cells in favor of neural crest specification (PubMed:26399832). {ECO:0000269|PubMed:26399832}. | Alternative splicing;Cytoplasm;Cytoskeleton;Golgi apparatus;Kelch repeat;Reference proteome;Repeat;Translation regulation;Ubl conjugation pathway | hsa:84541; | Cul3-RING ubiquitin ligase complex [GO:0031463]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; spindle [GO:0005819]; neural crest cell development [GO:0014032]; neural crest formation [GO:0014029]; protein monoubiquitination [GO:0006513]; regulation of translation [GO:0006417] | 19893584_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 29999490_The authors found that CUL3 complexed with KBTBD8 monoubiquitylates its essential targets only after these have been phosphorylated in multiple motifs by CK2, a kinase whose levels gradually increase during embryogenesis. 33109073_Downregulation of the ubiquitin ligase KBTBD8 prevented epithelial ovarian cancer progression. | ENSMUSG00000030031 | Kbtbd8 | 760.742342 | 1.0607638 | 0.085103453 | 0.11699655 | 5.268699e-01 | 4.679258e-01 | 7.857976e-01 | No | Yes | 820.591540 | 178.966473 | 762.078875 | 166.139286 | ||
| ENSG00000163482 | 27148 | STK36 | protein_coding | Q9NRP7 | FUNCTION: Serine/threonine protein kinase which plays an important role in the sonic hedgehog (Shh) pathway by regulating the activity of GLI transcription factors (PubMed:10806483). Controls the activity of the transcriptional regulators GLI1, GLI2 and GLI3 by opposing the effect of SUFU and promoting their nuclear localization (PubMed:10806483). GLI2 requires an additional function of STK36 to become transcriptionally active, but the enzyme does not need to possess an active kinase catalytic site for this to occur (PubMed:10806483). Required for postnatal development, possibly by regulating the homeostasis of cerebral spinal fluid or ciliary function (By similarity). Essential for construction of the central pair apparatus of motile cilia. {ECO:0000250|UniProtKB:Q69ZM6, ECO:0000269|PubMed:10806483}. | ATP-binding;Alternative splicing;Cilium biogenesis/degradation;Cytoplasm;Developmental protein;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the serine/threonine kinase family of enzymes. This family member is similar to a Drosophila protein that plays a key role in the Hedgehog signaling pathway. This human protein is a positive regulator of the GLI zinc-finger transcription factors. Knockout studies of the homologous mouse gene suggest that defects in this human gene may lead to congenital hydrocephalus, possibly due to a functional defect in motile cilia. Because Hedgehog signaling is frequently activated in certain kinds of gastrointestinal cancers, it has been suggested that this gene is a target for the treatment of these cancers. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Aug 2011]. | hsa:27148; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular region [GO:0005576]; nucleus [GO:0005634]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transcription corepressor binding [GO:0001222]; brain development [GO:0007420]; cilium assembly [GO:0060271]; epithelial cilium movement involved in extracellular fluid movement [GO:0003351]; positive regulation of hh target transcription factor activity [GO:0007228]; positive regulation of smoothened signaling pathway [GO:0045880]; post-embryonic development [GO:0009791]; protein phosphorylation [GO:0006468]; regulation of DNA-binding transcription factor activity [GO:0051090]; smoothened signaling pathway [GO:0007224] | 15268766_The FU gene and its genomic structure was identified. 25278022_Fu ubiquitination and cleavage is one of the key elements connecting the MID1-PP2A protein complex with GLI3 activity control 28543983_Data identified homozygous loss-of-function mutations in STK36 in one PCD-affected individual with situs solitus. Transmission electron microscopy analysis demonstrates that STK36 is required for cilia orientation in human respiratory epithelial cells, with a probable localization of STK36 between the RS and CP. STK36 screening can now be included for this rare and difficult to diagnose PCD subgroup. | ENSMUSG00000033276 | Stk36 | 966.219502 | 1.1251624 | 0.170133213 | 0.11692064 | 2.128462e+00 | 1.445858e-01 | 4.991087e-01 | No | Yes | 1007.379424 | 157.710778 | 894.736553 | 140.098111 | |
| ENSG00000163686 | 57406 | ABHD6 | protein_coding | Q9BV23 | FUNCTION: Lipase that preferentially hydrolysis medium-chain saturated monoacylglycerols including 2-arachidonoylglycerol (PubMed:22969151). Through 2-arachidonoylglycerol degradation may regulate endocannabinoid signaling pathways (By similarity). Also has a lysophosphatidyl lipase activity with a preference for lysophosphatidylglycerol among other lysophospholipids (By similarity). Also able to degrade bis(monoacylglycero)phosphate (BMP) and constitutes the major enzyme for BMP catabolism (PubMed:26491015). BMP, also known as lysobisphosphatidic acid, is enriched in late endosomes and lysosomes and plays a key role in the formation of intraluminal vesicles and in lipid sorting (PubMed:26491015). {ECO:0000250|UniProtKB:Q8R2Y0, ECO:0000269|PubMed:22969151, ECO:0000269|PubMed:26491015}. | 3D-structure;Endosome;Hydrolase;Lipid metabolism;Lysosome;Membrane;Mitochondrion;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | hsa:57406; | AMPA glutamate receptor complex [GO:0032281]; GABA-ergic synapse [GO:0098982]; glutamatergic synapse [GO:0098978]; integral component of membrane [GO:0016021]; integral component of postsynaptic membrane [GO:0099055]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; mitochondrial membrane [GO:0031966]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; acylglycerol lipase activity [GO:0047372]; phospholipase activity [GO:0004620]; acylglycerol catabolic process [GO:0046464]; arachidonic acid metabolic process [GO:0019369]; long-term synaptic depression [GO:0060292]; lysobisphosphatidic acid metabolic process [GO:2001311]; monoacylglycerol catabolic process [GO:0052651]; negative regulation of cell migration [GO:0030336]; negative regulation of cold-induced thermogenesis [GO:0120163]; phospholipid catabolic process [GO:0009395]; positive regulation of lipid biosynthetic process [GO:0046889]; regulation of endocannabinoid signaling pathway [GO:2000124] | 18360779_report the tissue distribution, subcellular location and differential distribution among cancer cell lines of Abhd6, one unannotated member of this group 19793082_High expression of ABHD6 in Ewing family tumors (EFT) in comparison to normal tissues and other tumors suggests that ABHD6 might be an interesting new diagnostic or therapeutic target for EFT. 22827915_ABHD6 increases from neonatal age. 22969151_Data show that the three hydrolases are genuine MAG lipases; medium-chain saturated MAGs were the best substrates for hABHD6 and hMAGL, whereas hABHD12 preferred the 1 (3)- and 2-isomers of arachidonoylglycerol. 24534757_Results confirm the genetic association of the locus 3p14.3 with systemic lupus erythematosus in Europeans and point to the ABHD6 and not PXK, as the major susceptibility gene in the region. 26491015_data suggest that ABHD6 controls BMP catabolism and is therefore part of the late endosomal/lysosomal lipid-sorting machinery 27114538_The hydrolase activity of ABHD6 was not required for the effects of ABHD6 on AMPAR function in either neurons or transfected HEK293T cells. Thus, these findings reveal a novel and unexpected mechanism governing AMPAR trafficking at synapses through ABHD6. 28880480_Data suggest that ABHD6 plays important role in regulation of signaling via monoacylglycerols (MAGs) in both central and peripheral tissues; alterations in MAG signaling are involved in type 2 diabetes, obesity, and metabolic syndrome. [REVIEW] 30728209_We discovered a regulatory role of ABHD6 in human plasmacytoid dendritic cells (pDCs) through modulating the local abundance of its substrate, the endocannabinoid 2-arachidonyl glycerol (2-AG), and elucidated a hitherto unknown cannabinoid receptor 2-mediated regulatory role of 2-AG on IFN-alpha induction by pDCs. 30894461_It demonstrate that ABHD6 affect Bis(monoacylglycerol)phosphate (BMP) metabolism in mice and humans. 32143183_Enhanced monoacylglycerol lipolysis by ABHD6 promotes NSCLC pathogenesis. | ENSMUSG00000025277 | Abhd6 | 485.109657 | 0.9059254 | -0.142535770 | 0.12569444 | 1.273815e+00 | 2.590523e-01 | 6.312439e-01 | No | Yes | 449.824882 | 46.560504 | 505.851890 | 52.082967 | ||
| ENSG00000163702 | 84818 | IL17RC | protein_coding | Q8NAC3 | FUNCTION: Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity (By similarity). Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity. Receptor for IL17A and IL17F homodimers as part of a heterodimeric complex with IL17RA (PubMed:16785495). Receptor for the heterodimer formed by IL17A and IL17B as part of a heterodimeric complex with IL17RA (PubMed:18684971). Has also been shown to be the cognate receptor for IL17F and to bind IL17A with high affinity without the need for IL17RA (PubMed:17911633). Upon binding of IL17F homodimer triggers downstream activation of TRAF6 and NF-kappa-B signaling pathway (PubMed:16785495, PubMed:32187518). Induces transcriptional activation of IL33, a potent cytokine that stimulates group 2 innate lymphoid cells and adaptive T-helper 2 cells involved in pulmonary allergic response to fungi (By similarity). Promotes sympathetic innervation of peripheral organs by coordinating the communication between gamma-delta T cells and parenchymal cells. Stimulates sympathetic innervation of thermogenic adipose tissue by driving TGFB1 expression (By similarity). Binding of IL17A-IL17F to IL17RA-IL17RC heterodimeric receptor complex triggers homotypic interaction of IL17RA and IL17RC chains with TRAF3IP2 adapter through SEFIR domains. This leads to downstream TRAF6-mediated activation of NF-kappa-B and MAPkinase pathways ultimately resulting in transcriptional activation of cytokines, chemokines, antimicrobial peptides and matrix metalloproteinases, with potential strong immune inflammation (PubMed:18684971, PubMed:17911633). Primarily induces neutrophil activation and recruitment at infection and inflammatory sites (By similarity). Stimulates the production of antimicrobial beta-defensins DEFB1, DEFB103A, and DEFB104A by mucosal epithelial cells, limiting the entry of microbes through the epithelial barriers (By similarity). {ECO:0000250|UniProtKB:Q8K4C2, ECO:0000269|PubMed:16785495, ECO:0000269|PubMed:17911633, ECO:0000269|PubMed:18684971, ECO:0000269|PubMed:32187518}.; FUNCTION: [Isoform 5]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 6]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 7]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 8]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}. | 3D-structure;Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;Glycoprotein;Inflammatory response;Membrane;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | This gene encodes a single-pass type I membrane protein that shares similarity with the interleukin-17 receptor (IL-17RA). Unlike IL-17RA, which is predominantly expressed in hemopoietic cells, and binds with high affinity to only IL-17A, this protein is expressed in nonhemopoietic tissues, and binds both IL-17A and IL-17F with similar affinities. The proinflammatory cytokines, IL-17A and IL-17F, have been implicated in the progression of inflammatory and autoimmune diseases. Multiple alternatively spliced transcript variants encoding different isoforms have been detected for this gene, and it has been proposed that soluble, secreted proteins lacking transmembrane and intracellular domains may function as extracellular antagonists to cytokine signaling. [provided by RefSeq, Feb 2011]. | hsa:84818; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; interleukin-17 receptor activity [GO:0030368]; inflammatory response [GO:0006954] | 16688746_IL-17RL exists as multiple isoforms due to extensive alternative splicing. Changes in RNA IL-17RL splicing occur in advanced cancers. 16785495_The biologic activity of IL-17 is dependent on a complex composed of receptors IL-17RA and IL-17RC. 17911633_IL-17RC functions as a receptor for both IL-17A and IL-17F; a soluble version of this protein should be an effective antagonist of IL-17A and IL-17F mediated inflammatory diseases. 18097068_IL-17A-induced IL-6, IL-8, and CCL20 secretion was dependent on both IL-17RA and IL-17RC, which are overexpressed in RA patients. 18684971_Interleukin-17F is inhibited by the IL-17RC receptor, a combination of soluble IL-17RA/IL-17RC receptors is required for inhibition of the IL-17F/IL-17A activity. 20173024_IL-17 and its receptor IL-17RC are involved in rheumatoid arthritis synovial fluid-mediated chemotaxis in human lung microvascular endothelial cell culture. 22744455_Overall, our study found a significant association of IL-17RC gene polymorphisms with AIS in a Chinese Han population, indicating IL-17RC gene may be as a susceptibility gene for AIS. 22898922_Data show that IL-17RA, IL-17RC, IL-22R1, ERK1/2 MAPK and NF-kappaB pathways are involved in Th17 cytokine-induced proliferation. 22999050_IL-17RC predisposes to the development of adolescent idiopathic scoliosis in a Chinese Han population. 24885153_results suggested a potential involvement of IL-17RC+CD8+ T cells in pathogenesis of ocular sarcoidosis 25918342_human IL-17RC is essential for mucocutaneous immunity to C. albicans but is otherwise largely redundant. 26731132_methylation of IL17RC could play as a marker in CNV and degeneration of RPE cells in vitro. 27155366_Interleukin 17A (IL17a) and interleukin-23 (IL-23) - dependent interleukin-17 receptor C (IL-17RC) are expressed by sputum and neutrophils in deltaF508-CFTR protein (F508del) cystic fibrosis patients. 29584788_IL-17RC rs708567 polymorphism in A/A genotype, G/G genotype, and G/a genotype did not seem to influence RA susceptibility in Tunisian population. 29695654_Genetic Variants on IL-17 are associated with development of atherosclerotic diseases. 29764467_Five SNPs in the IL17RC (and COL6A1) genes were found to be associated with susceptibility to ossification of the posterior longitudinal ligament in Han Chinese patients. 30024651_Genetic study revealed no association between IL-17RC, and SNPs and acute kidney transplant graft rejection. Nevertheless, a significant improvement of graft survival was found in kidney transplant recipients carrying the IL-17RC*G/G, and *G/A genotypes. 31291973_An increase in IL17RC gene expression levels in peripheral blood samples was found in ossification of the posterior longitudinal ligament patients. 31481525_Interleukin-17 receptor C gene polymorphism reduces treatment effect and promotes poor prognosis of ischemic stroke. 32187518_The crystal structure of the interleukin 17 receptor C (IL-17RC):interleukin 17F (IL-17F) complex provides a structural basis for IL-17F signaling through IL-17RC. 35167487_IL-17 Receptor C Signaling Controls CD4(+) TH17 Immune Responses and Tissue Injury in Immune-Mediated Kidney Diseases. | ENSMUSG00000030281 | Il17rc | 203.269961 | 0.9973197 | -0.003872025 | 0.20463425 | 3.511991e-04 | 9.850483e-01 | No | Yes | 168.663043 | 26.048977 | 172.306178 | 26.467662 | ||
| ENSG00000163872 | 55689 | YEATS2 | protein_coding | Q9ULM3 | FUNCTION: Chromatin reader component of the ATAC complex, a complex with histone acetyltransferase activity on histones H3 and H4 (PubMed:18838386, PubMed:19103755, PubMed:27103431). YEATS2 specifically recognizes and binds histone H3 crotonylated at 'Lys-27' (H3K27cr) (PubMed:27103431). Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors (PubMed:27103431). {ECO:0000269|PubMed:18838386, ECO:0000269|PubMed:19103755, ECO:0000269|PubMed:27103431}. | 3D-structure;Coiled coil;Epilepsy;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Summary: The protein encoded by this gene is a scaffolding subunit of the ATAC complex, which is a complex with acetyltransferase activity on histones H3 and H4. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, May 2017]. | hsa:55689; | ATAC complex [GO:0140672]; mitotic spindle [GO:0072686]; histone binding [GO:0042393]; modification-dependent protein binding [GO:0140030]; TBP-class protein binding [GO:0017025]; histone H3 acetylation [GO:0043966]; histone H3-K14 acetylation [GO:0044154]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of cell cycle [GO:0051726]; regulation of cell division [GO:0051302]; regulation of embryonic development [GO:0045995]; regulation of histone deacetylation [GO:0031063]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; regulation of tubulin deacetylation [GO:0090043] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 27103431_YEATS2 is a selective histone crotonylation reader. 29057918_YEATS2 gene is highly amplified in human non-small cell lung cancer (NSCLC) and is required for cancer cell growth and survival 31539032_our findings suggest that benign adult familial myoclonic epilepsy type 4 is caused by the insertions of the intronic TTTCA repeats in YEATS2 32749678_YEATS2 is a target of HIF1alpha and promotes pancreatic cancer cell proliferation and migration. 34587874_YEATS domain-containing 2 (YEATS2), targeted by microRNA miR-378a-5p, regulates growth and metastasis in head and neck squamous cell carcinoma. | ENSMUSG00000041215 | Yeats2 | 1704.816277 | 1.1871181 | 0.247463438 | 0.08241161 | 9.111571e+00 | 2.539976e-03 | 5.789574e-02 | No | Yes | 1696.688294 | 138.265027 | 1443.392912 | 117.637887 | |
| ENSG00000164050 | 5364 | PLXNB1 | protein_coding | O43157 | FUNCTION: Receptor for SEMA4D (PubMed:19843518, PubMed:20877282, PubMed:21912513). Plays a role in GABAergic synapse development (By similarity). Mediates SEMA4A- and SEMA4D-dependent inhibitory synapse development (By similarity). Plays a role in RHOA activation and subsequent changes of the actin cytoskeleton (PubMed:12196628, PubMed:15210733). Plays a role in axon guidance, invasive growth and cell migration (PubMed:12198496). {ECO:0000250|UniProtKB:Q8CJH3, ECO:0000269|PubMed:12196628, ECO:0000269|PubMed:12198496, ECO:0000269|PubMed:15210733, ECO:0000269|PubMed:19843518, ECO:0000269|PubMed:20877282, ECO:0000269|PubMed:21912513}. | 3D-structure;Alternative splicing;Cell membrane;Coiled coil;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | hsa:5364; | extracellular region [GO:0005576]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; semaphorin receptor complex [GO:0002116]; GTPase activating protein binding [GO:0032794]; GTPase activator activity [GO:0005096]; semaphorin receptor activity [GO:0017154]; semaphorin receptor binding [GO:0030215]; transmembrane signaling receptor activity [GO:0004888]; cell migration [GO:0016477]; inhibitory synapse assembly [GO:1904862]; intracellular signal transduction [GO:0035556]; negative regulation of cell adhesion [GO:0007162]; negative regulation of osteoblast proliferation [GO:0033689]; neuron projection morphogenesis [GO:0048812]; ossification involved in bone maturation [GO:0043931]; positive regulation of axonogenesis [GO:0050772]; positive regulation of GTPase activity [GO:0043547]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; regulation of cell migration [GO:0030334]; regulation of cell shape [GO:0008360]; regulation of cytoskeleton organization [GO:0051493]; regulation of GTPase activity [GO:0043087]; semaphorin-plexin signaling pathway [GO:0071526]; semaphorin-plexin signaling pathway involved in axon guidance [GO:1902287]; semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis [GO:1900220] | 11937491_The plexin-B1/Rac interaction inhibits PAK activation and enhances Sema4D ligand binding 12196628_LARG plays a critical role in plexin-B1 signaling to stimulate Rho activation and cytoskeletal reorganization. 12220504_Interaction of plexin-B1 with PDZ domain-containing Rho guanine nucleotide exchange factors 12406905_Plexin-B1 is an easily accessible receptor for CD100 within the immune system. The crosstalk operated by the CD100/Plexin-B1 interaction is not malignancy related but reproduces a mechanism used by normal CD5+ B cells. 12533544_cleavage by proprotein convertases is a novel regulatory step for semaphorin receptors localized at the cell surface. 15184888_In some human neoplastic lines, PLXB1 is overexpressed, constitutively tyrosine phosphorylated, and associated with Scatter Factor Receptors. 15210733_ErbB-2-mediated phosphorylation of plexin-B1 is critically involved in Sema4D-induced RhoA activation. 15632204_demonstrate that Sema4D is angiogenic in vitro and in vivo and that this effect is mediated by its high-affinity receptor, Plexin B1, and that biologic effects elicited by Plexin B1 require coupling and activation of the Met tyrosine kinase 15929008_the minimal Rac1 GTPase binding domain of plexin-B1 has a ubiquitin fold, as shown by NMR 16055703_Semaphorin 4D/plexin-B1 induces endothelial cell migration through the activation of PYK2, Src, and the phosphatidylinositol 3-kinase-Akt pathway 17383649_Plexin-B1 contributes to trophoblast-endometrium interactions, most likely by enhancing adhesion properties. 17855350_plexin-B1 promotes endothelial cell motility through RhoA and ROK by regulating the integrin-dependent signaling networks that result in the activation of PI3K and Akt 17916560_Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain 18024597_13 somatic missense mutations in the cytoplasmic domain of the Plexin-B1 gene hinder Rac & R-Ras binding & R-RasGAP activity, & increase cell motility, invasion, adhesion, & lamellipodia extension. 18025083_a novel mechanism by which plexin-mediated signaling can be regulated and explains how Sema4D can exert different biological activities through the differential association of its receptor with ErbB-2 and Met. 18275816_NMR solution structure of the Rho GTPase binding domain; study suggests that the oncogenic behavior of the mutants can be rationalized with reference to the structure of the RBD of plexin-B1 18279812_Plexin B1 protein is absent in more than 80% of renal cell carcinomas. when we have induced plexin B1 expression with an expression vector in the renal adenocarcinoma cell line ACHN, a marked reduction in proliferation rate was produced. 18321527_Mapping of the Rac1 GTPase surface that contacts the Rho GTPase binding domain of plexin-B1 by NMR confirms the plexin domain as a GTPase effector protein and regions neighboring the GTPase switch I and II regions are also involved in the interaction. 18417270_Plexin B1 expression is reduced in the group of 'uncoupled' stem cell-like breast cancer tumors 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19444311_Plexin-B1 is a dual functional GTPase activating protein for R-Ras and M-Ras, remodelling axon and dendrite morphology, respectively. 19483722_B-Raf/MKK/ERK provides a permissive environment for melanoma genesis by modulating plexin B1. 19805522_show here that activation of plexin-B1 by Semaphorin 4D and its subsequent tyrosine phosphorylation creates docking sites for the SH2 domains of phospholipase Cgamma. 19843518_the monomeric intracellular plexin-B1 binds R-Ras but not H-Ras. These findings suggest that the monomeric form of the intracellular region is primed for GAP activity and extend a model for plexin activation. 19940359_Data show that Plex-B1 assumes a predictive value for unfavourable outcome when co-expressed with Met. 20040080_In the endometrium, both Glycodelin and Plexin-B1 are exhibiting a cyclic pattern suggesting a possible steroid regulation and a role in endometrial receptivity 20164843_Plexin B1 may function as a tumor promoter in melanomas not driven by c-Met activation. 20610402_Sema4D/Plexin-B1 promotes the dephosphorylation and activation of PTEN through the R-Ras GAP activity, inducing growth cone collapse. 20877282_crystal structures of cognate complexes of the semaphorin-binding regions of plexins B1 and A2 with semaphorin ectodomains (human PLXNB1(1-2)-SEMA4D(ecto) and murine PlxnA2(1-4)-Sema6A(ecto)), plus unliganded structures of PlxnA2(1-4) and Sema6A(ecto) 21029396_Plexin B1 abrogates integrin-dependent migration and activation of pp125(FAK). 21059203_Plexin-B1 expression correlates with malignant phenotypes of serous ovarian tumors, probably via phosphorylation of AKT at Ser473 21216304_plexin-B1, a target of miR-214, may function as an oncogene in human cervical cancer HeLa cells 21812859_The binding of Sema4D to plexinB1 induced small GTPase Ras homolog gene family, member A activation and resulted in the phosphorylation of MAPK and Akt. 21912513_Two crystal structures of the human Plexin-B1 cytoplasmic region in complex with a constitutively active RhoGTPase, Rac1, are reported. 22378040_ErbB-2 overexpression in human breast & ovarian cancer cell lines leads to phosphorylation & activation of Plexin-B1. This was required for ErbB-2-dependent activation of RhoA & RhoC & promoted invasive behavior. 22404908_PlexinB1 mutations block plexinB1-mediated signalling pathways that inhibit cell motility. 23603360_Data indicate that Rnd1 efficiently displaces Rac1 from its complex with Plexin-B1 but not vice versa. 23775445_Activation of endogenous plexin-B1 enhances cell migration and tumor invasiveness in prostate cancer cells. 25982277_results show that Sema4D/plexin-B1 signaling promotes the translocation of androgen receptor to the nucleus and thereby enhances AR transcriptional activity 26035216_Results show that decreased expression of Sema4D, plexin-B1 and -B2 was associated with local recurrence and poor prognosis of breast neoplasm. 26051877_Plexin-B1 induces cutaneous squamous cell carcinoma cell proliferation, migration, and invasion by interacting with Sema4D. Plexin-B1 might serve as a useful biomarker and/or as a novel therapeutic target for cSCC. 26275342_Blocking of CD100, plexin B1 and/or B2 in adhesion experiments have shown that both CD100 and plexins act as adhesion molecules involved in monocyte-endothelial cell binding. 26341082_Dysregulation of the vascular endothelial growth factor and semaphorin ligand-receptor families in prostate cancer metastasis 26944058_Plexin-B1 mediates RhoA/integrin alphavbeta3 involved in the PI3K/Akt pathway and SRPK1 to influence the growth of glioma cell, angiogenesis, and motility in vitro and in vivo. 27456345_The positive expression of both Sema4D and PlexinB1 was found to be an independent risk factor for a worse survival in colorectal cancer. 28004109_Plexin B1 expression was regulated by TMPRSS2-ERG fusion gene in prostate cancer. 28739743_Loss of plexin B1 expression might play a pivotal role in enhancing the metastatic potential of breast cancer cells. 29040270_Analysis of the interaction of Plexin-B1 and Plexin-B2 with Rnd family proteins shows lack of binding specificity. 29939944_Decreased expression of sema4D and Plexin-B1 may be responsible for the deficiency in Met signalling and the development of preeclampsia. 30762724_Plexin-B1 level showed a significant positive correlation both with overall survival and disease free survival of Caucasian breast cancer patients 30937968_Sema4D/PlexinB1 promotes endothelial differentiation of dental pulp stem cells via activation of AKT and ERK1/2 signaling. 31861264_PlexinB1 Promotes Nuclear Translocation of the Glucocorticoid Receptor. 32601713_Plexin-Bs enhance their GAP activity with a novel activation switch loop generating a cooperative enzyme. 34854398_Sema4D/Plexin-B1 promotes the progression of osteosarcoma cells by activating Pyk2-PI3K-AKT pathway. 35170806_PLXNB1 mutations in the etiology of idiopathic hypogonadotropic hypogonadism. | ENSMUSG00000053646 | Plxnb1 | 295.885948 | 1.0838499 | 0.116165026 | 0.18525673 | 3.867959e-01 | 5.339884e-01 | No | Yes | 259.300867 | 47.609990 | 247.030761 | 45.325840 | |||
| ENSG00000164073 | 256471 | MFSD8 | protein_coding | Q8NHS3 | FUNCTION: May be a carrier that transport small solutes by using chemiosmotic ion gradients. {ECO:0000305}. | Alternative splicing;Disease variant;Glycoprotein;Lysosome;Membrane;Neurodegeneration;Neuronal ceroid lipofuscinosis;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a ubiquitous integral membrane protein that contains a transporter domain and a major facilitator superfamily (MFS) domain. Other members of the major facilitator superfamily transport small solutes through chemiosmotic ion gradients. The substrate transported by this protein is unknown. The protein likely localizes to lysosomal membranes. Mutations in this gene are correlated with a variant form of late infantile-onset neuronal ceroid lipofuscinoses (vLINCL). [provided by RefSeq, Oct 2008]. | hsa:256471; | integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; transmembrane transporter activity [GO:0022857]; autophagosome maturation [GO:0097352]; lysosome organization [GO:0007040]; neuron development [GO:0048666]; regulation of autophagy [GO:0010506]; regulation of lysosomal protein catabolic process [GO:1905165]; TORC1 signaling [GO:0038202] | 17564970_MFSD8 gene is involved in late-infantile-onset neuronal ceroid lipofuscinose;it was mapped to chromosome 4q28.1-q28.2. 18850119_Results describe a novel mutation in the MFSD8 gene, responsible for neuronal ceroid lipofuscinoses, in a consanguineous Egyptian family 19177532_Study contributes to a better molecular characterization of Italian NCL cases, and will facilitate medical genetic counseling in such families. 19201763_CLN7/MFSD8 defects are not restricted to the Turkish population, as initially anticipated, but are a relatively common cause of NCL in different populations. 19277732_Data show that neuronal ceroid lipofuscinosis in a Saudi family is due to a homozygous novel mutation in the most recently described NCL gene (MFSD8). 20826447_Expression and lysosomal targeting of CLN7 are reported. 24423645_This study showed that Gene disruption of Mfsd8 provides animal model for CLN7 disease. 25227500_In this study, we identified variants in MFSD8 as a novel cause of nonsyndromic autosomal recessive macular dystrophy with central cone involvement. 25270050_A mutation in MFSD8, c.472G>A (p.Gly158Ser), segregates with the disease phenotype in variant late infantile neuronal ceroid lipofuscinosis. 25439737_MFSD8 genetic testing should also be considered in patients with Rett like phenotype at onset and negative MECP2 mutation 28586915_This study highlights a hierarchy of MFSD8 variant severity, predicting three consequences of mutation: (1) nonsyndromic localized maculopathy, (2) nonsyndromic widespread retinopathy, or (3) syndromic neurological disease. 29514215_Quantification revealed that the amounts of 12 different soluble lysosomal proteins were significantly reduced in Cln7 ko MEFs compared with wild-type controls. One of the most significantly depleted lysosomal proteins was Cln5 protein that underlies another distinct neuronal ceroid lipofuscinosis disorder 30144815_We identified a novel homozygous mutation in MFSD8 gene. 30382371_that MFSD8-associated lysosomal dysfunction may contribute to frontotemporal lobar degeneration pathology 31006324_Here and for the first time, we reported on two previously variant late-infantile neuronal ceroid lipofuscinoses-associated variants in MFSD8 but in association with a form of cone-rod dystrophy known as non-syndromic macular dystrophy with central cone involvement. 33226711_Mutation analysis of MFSD8 in an amyotrophic lateral sclerosis cohort from mainland China. 35087090_Aberrant upregulation of the glycolytic enzyme PFKFB3 in CLN7 neuronal ceroid lipofuscinosis. 35216386_A Novel, Apparently Silent Variant in MFSD8 Causes Neuronal Ceroid Lipofuscinosis with Marked Intrafamilial Variability. 35457110_Contribution of Whole-Genome Sequencing and Transcript Analysis to Decipher Retinal Diseases Associated with MFSD8 Variants. | ENSMUSG00000025759 | Mfsd8 | 1049.698379 | 0.9649831 | -0.051424397 | 0.08900530 | 3.326251e-01 | 5.641174e-01 | 8.381040e-01 | No | Yes | 943.080733 | 184.389757 | 980.150619 | 191.550467 | |
| ENSG00000164076 | 79012 | CAMKV | protein_coding | Q8NCB2 | FUNCTION: Does not appear to have detectable kinase activity. | Alternative splicing;Calmodulin-binding;Cell membrane;Cytoplasmic vesicle;Membrane;Phosphoprotein;Reference proteome | hsa:79012; | cytoplasmic vesicle membrane [GO:0030659]; glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; peptidyl-serine phosphorylation [GO:0018105]; regulation of modification of postsynaptic structure [GO:0099159] | 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) | ENSMUSG00000032936 | Camkv | 349.064107 | 1.0061875 | 0.008899133 | 0.14335313 | 3.874394e-03 | 9.503680e-01 | No | Yes | 318.018927 | 40.027710 | 326.918788 | 41.072125 | |||
| ENSG00000164078 | 4486 | MST1R | protein_coding | Q04912 | FUNCTION: Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to MST1 ligand. Regulates many physiological processes including cell survival, migration and differentiation. Ligand binding at the cell surface induces autophosphorylation of RON on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with the PI3-kinase subunit PIK3R1, PLCG1 or the adapter GAB1. Recruitment of these downstream effectors by RON leads to the activation of several signaling cascades including the RAS-ERK, PI3 kinase-AKT, or PLCgamma-PKC. RON signaling activates the wound healing response by promoting epithelial cell migration, proliferation as well as survival at the wound site. Plays also a role in the innate immune response by regulating the migration and phagocytic activity of macrophages. Alternatively, RON can also promote signals such as cell migration and proliferation in response to growth factors other than MST1 ligand. {ECO:0000269|PubMed:18836480, ECO:0000269|PubMed:7939629, ECO:0000269|PubMed:9764835}. | 3D-structure;ATP-binding;Alternative splicing;Cleavage on pair of basic residues;Disease variant;Disulfide bond;Glycoprotein;Immunity;Innate immunity;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | This gene encodes a cell surface receptor for macrophage-stimulating protein (MSP) with tyrosine kinase activity. The mature form of this protein is a heterodimer of disulfide-linked alpha and beta subunits, generated by proteolytic cleavage of a single-chain precursor. The beta subunit undergoes tyrosine phosphorylation upon stimulation by MSP. This protein is expressed on the ciliated epithelia of the mucociliary transport apparatus of the lung, and together with MSP, thought to be involved in host defense. Alternative splicing generates multiple transcript variants encoding different isoforms that may undergo similar proteolytic processing. [provided by RefSeq, Jan 2016]. | hsa:4486; | basal plasma membrane [GO:0009925]; cell surface [GO:0009986]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; stress fiber [GO:0001725]; vacuole [GO:0005773]; ATP binding [GO:0005524]; enzyme binding [GO:0019899]; macrophage colony-stimulating factor receptor activity [GO:0005011]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; cell migration [GO:0016477]; defense response [GO:0006952]; innate immune response [GO:0045087]; nervous system development [GO:0007399]; phagocytosis [GO:0006909]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of protein kinase B signaling [GO:0051897]; response to virus [GO:0009615]; signal transduction [GO:0007165]; single fertilization [GO:0007338]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 12214279_overexpression of human wild-type RON causes the formation of lung tumors with unique biological characteristics in vivo 12419829_Transgenic mice in which surfactant protein C promoter was used to express human wild-type RON in distal lung epithelial cells developed multiple RON-expressing lung adenomas with distinct morphology & growth pattern which progressed to adenocarcinoma. 12527888_These data suggest that RON expression is altered in certain primary colon cancers. Abnormal accumulation of RON variants may play a role in the progression of certain colorectal cancers in vivo 12676986_HYAL2 receptor protein is associated with the RON receptor tyrosine kinase, rendering it functionally silent. 12766581_immunostaining and Western blot showed Ron in normal kidney and in all oncocytomas but never in renal cell carcinomas or in the renal carcinoma cell line Caki-1; Ron was expressed in phosphorylated, i.e., active, form 12802274_Ron tyrosine kinase receptor desensitization mediated by c-Cbl and its binding partner Grb2. 12915129_These data suggest that coexpression of the MET and RON receptors confer a selective advantage to ovarian cancer cells and might promote ovarian cancer progression. 12919677_Data show that macrophage stimulating protein (MSP) and its receptor (Ron) induce phosphorylation of both Ron and alpha6beta4 integrin, and result in activation of alpha3beta1 integrin. 14597639_the ron-sema domain can suppress the growth of macrophage-stimulating protein-responsive cells in culture 15001985_persistent RON expression and activation cause the loss of epithelial phenotypes 15184888_RON receptor can interact with each of the three members of the class B Plexins and control tumor invasive growth. 15378025_altered expression of RON in colon cancer cells is required to maintain tumorigenic phenotypes 15557181_Data suggest that RON initiates signaling pathways that negatively regulate HIV-1 transcription in monocytes/macrophages, and that HIV-1 suppresses RON function by decreasing protein levels in the brain to assure efficient replication. 15764806_Receptor RON and uts ligand macrophage stimulating protein are involved in the pathophysiology of endometriosis. 15788670_co-expression of RON and MET is associated with aggressive phenotype in node-negative breast cancer patients 15929040_The proto-oncogene, c-Cbl, which modulates ubiquitylation of RON, was increased in glia in both multiple sclerosis brains and experimental autoimmune encephalomyelitis spinal cords 16166096_altered exon usage in the juxtamembrane domain of mouse and human RON regulates receptor activity and signaling specificity 16166746_Mst1r tyrosine kinase-domain deficient-mice have decreased survival times, alterations in cytokine and nitric oxide regulation, and an earlier onset of pulmonary pathology compared with control mice during acute lung injury 17311308_RON activation induced molecular and cellular alterations consistent with epithelial-mesenchymal (EMT) transition. 17395888_These data suggest that high molecular form hyaluronan is broken down by reactive oxygen species to form low-molecular-weight fragments that signal via RHAMM and RON to stimulate beat frequency. 17456594_overexpression of MSP, MT-SP1, and MST1R was a strong independent indicator of both metastasis and death in human breast cancer 17588532_This study reveals for the first time that RON alone is sufficient to induce complete and stabilized EMT in MDCK cells. 17611409_RONdelta170 is a naturally occurring variant with dominant negative activities and has potential for inhibiting RON-mediated tumorigenic activities in colorectal cancer cells. 17616662_RON receptor signaling may contribute to pancreatic carcinogenesis 18019691_high expression of Ron in the colorectal mucosa of ulcerative colitis patients suggests that this receptor might play roles in the pathophysiological background of this disease 18073207_Translocation of c-Met to the nucleus depends upon the adaptor protein Gab1 and importin beta1, and formation of Ca(2+) signals in turn depends upon this translocation 18165235_a clear link between Sp1-dependent RON tyrosine kinase expression and invasion of breast carcinoma cells. 18200509_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18200509_the linkage disequilibrium with these functional MST1R variants implicate this gene as having a possible role in Crohn Disease pathogenesis. 18204077_RON activation differentially regulates claudin expression in epithelial cells 18209063_RON represses HIV transcription at multiple transcriptional check points including initiation, elongation and chromatin organization. 18221954_We found that RON was over expressed in bladder cancer but RON over expression was not associated with overall survival or other clinical parameters 18310076_loss of Smad4 contributes to aberrant RON expression and cross-talk of Smad4-independent TGF-beta signaling and the RON pathway promotes an invasive phenotype 18593918_The regulation of human RON gene expression by nuclear factor-kappaB; a potential therapeutic role for curcumin in blocking RON tyrosine kinase-mediated invasion of carcinoma cells. 18606710_study demonstrates that HIV-1 Tat targets the receptor tyrosine kinase recepteur d'origine nantais (RON), which negatively regulates inflammation & HIV transcription, for proteosome degradation 18620091_Ron is a critical factor in tumorigenesis. 18836480_RON has 2 alternative SRC-kinase-mediated modes of signaling that can contribute to oncogenic behavior in normal breast epithelial cells: an MSP-dependent increase in cell proliferation & migration and MSP-independent increased cell survival. 18950514_The entire C-terminus is required for RON or RON160-mediated intracellular signaling events leading to various cellular activities. 19040718_RON expression is significant in gastric carcinoma tissue and corresponding paraneoplastic tissue, but is not expressed in normal gastric mucosa. RON expression in gastric carcinoma tissue was not related to patient survival rates. 19224914_Results demonstrate that Ron plays an essential role in maintaining malignant phenotypes of colon cancer cells through regulating mutant PI3K activity. 19242504_RON is involved in mesangial cell proliferation under both physiological and pathological conditions 19307182_RON is a novel molecular target of HIF-1alpha. 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19519771_The expression of several oncogenic RON splice variants in malignant gliomas suggests that these could represent candidate targets for treatment with agents inhibiting RON activity 19657358_Observational study of gene-disease association. (HuGE Navigator) 19838218_Data show that the knockdown of Ron in PC-3 or DU145 cells results in a significant decrease in angiogenic chemokine production and is associated with a decreased activation of NF-kappaB. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19956854_analysis of how a defined extracellular region of the RON receptor tyrosine kinase promote RON-mediated motile and invasive phenotypes in epithelial cells 20026054_RON expression in MCF-10A breast epithelial cells lead to an alteration of cell-surface hyaluronan compared to the parental cells. 20103639_Findings indicate that RON signaling mediates cell survival and in vivo resistance to gemcitabine in pancreatic cancer. 20358644_RON signaling results in MAPK-mediated VEGF secretion by pancreatic cancer cells and promotion of microtubule formation. 20428780_RONDelta85 is an antagonist to the MSP-RON pathway, which has potential for regulating RON/RON160-mediated tumorigenic activities. 20434834_Ron overexpression on a Chk2*1100 deletion background accelerates the development of mammary tumors. 20498137_Homodimerization of RON, a receptor tyrosine kinase, usually occurs in cells stimulated by a ligand and leads to the downstream activation of signaling pathways in bladder cancer cells. 20596668_TGase-4 expression is intrinsically linked to the activation of RON in prostate cancer cells and that this autoactivation of RON contributes to the increased cell motility in TGase-4 expressing cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20664977_RON-targeted siRNA could be therapeutically efficacious by inducing cell apoptosis through the modulation of the NF-kappaB and Bcl-2 family in gastric cancer cells. 20689759_A novel connection between the Ron receptor tyrosine kinase and an important mechanism of tamoxifen resistance in breast cancer. 20800603_Observational study of gene-disease association. (HuGE Navigator) 21081472_results suggest that MSP induces uPAR expression via MAPK, AP-1 and NF-kappaB signaling pathways 21114864_Alterations in the first IPT domain in extracellular region differentially regulate RON mediated tumorigenic activities. 21212418_Our findings establish that oncogene addiction to MET involves Ron transactivation 21543897_Role of RON in gastroesophageal cancer and its role in cooperative signaling with MET in gastroesophageal tissues and cell lines 21659546_The impact of RON on the proliferation of transformed B cells suggests that RON may be a novel therapeutic target for EBV-associated lymphoproliferative diseases. 21723047_The results suggest that RON signaling seems to play at least some role in the pathogenesis of Merkel cell carcinoma 21799005_Nrf2 inducer SFN stabilized Nrf2 and inhibited RON expression in carcinoma cells from various tumor types. 21874262_macrophage stimulating 1 receptor is associated with invasive and oncogenic phenotypes such as tumor cell migration, invasion, resistance to apoptosis and cell cycle arrest 21875933_These findings suggest that the MSP/RON signaling pathway may be regulated by hepsin in tissue homeostasis and in disease pathologies, such as in cancer and immune disorders. 21901254_Results indicate that knockdown of RON inhibits AP-1 activity and induces apoptosis and cell cycle arrest through the modulation of Akt/FoxO signaling in human colorectal cancer cells. 22086736_results suggest cytoplasmic phosphorylated RON is a potential marker for poor prognosis in Esophageal squamous cell carcinomapatients. 22095683_tumor promoter induces RON expression via Egr-1, which, in turn, stimulates cell invasiveness in AGS cells 22515290_Findings suggest the involvement of RON expression in the development of breast cancer, and that an autocrine/paracrine loop of RON seems to affect tumor invasiveness. 22585712_High RON expression is associated with small and non-small cell lung cancers. 22613539_Elevated RON expression may contribute to the occurrence, progression and metastasis of non-small cell lung cancer(NSCLC), inferring that it could be useful as a new prognostic indicator for patients with NSCLC. 22848655_the structure of RON Sema-PSI domains at 1.85 A resolution 22902361_High RON expression is associated with acute myeloid leukemia. 22958871_RON expression is not associated with prognosis or therapeutic responsiveness in resected pancreatic cancer. 22974584_These results suggest an important role of RON in the tumorigenesis and metastasis of nasopharyngeal carcinoma. 22975341_A new splice variant of RON suggests a novel role for the RON receptor in the progression of metastasis in colorectal cancer. 22993024_study presents two novel splicing variants of RON in the partial splicing events that involve exons 5 and 6; the difference of these two isoforms is the inclusion or skipping of intron 6 23235762_Knockdown of RON inhibited invasive growth & the activation of oncogenic signaling pathways including Akt, MAPK and beta-catenin. RON up-regulation was associated with tumor size, lymphatic metastasis, invasiveness, tumor stage and poor survival. 23294341_RON may influence carcinogenesis in a number of systemic malignancies. 23483216_the function of RON in tumors may be multidimensional, not just as a tumor suppressor or oncogene. 23542172_findings identify the pathway of Ron-c-Abl-PCNA as a mechanism of oncogene-induced cell proliferation, with potentially important implications for development of combination therapy of breast cancer 23558571_Suggest that MET and MST1R are independent prognostic factors in classical classical Hodgkin's lymphoma, and may allow the identification of a subgroup of cHL patients who require more intensive therapy. 23597200_Data suggest that oncogenic RON160 is frequently expressed in primary invasive ductal, lobular, and lymph node-involved breast cancer tissues; RON160 overexpression is predominantly observed in invasive ductal and lymph node-involved cases. 23745832_The MET(-) RON(-) phenotype retained its prognostic impact after subgroup analysis. 23799848_Ron synergises with EGFR to confer certain adverse features in head and neck squamous cell carcinomas. 23811285_Demonstrate of the important role of RON in mediating lapatinib resistance in breast cancer cells. 24155930_intron retention in the RON gene was investigated. 24189591_Through double-base and single-base substitution analysis of the 2-nt RNA, this study demonstrated that the GA, CC, UG and AC dinucleotides on exon 11, in addition to the wild-type AG sequence, function as enhancers for exon 11 inclusion of the Ron pre-mRNA. 24287326_RON expression was dominantly observed in laryngeal SCC tissues relative to adjacent normal mucosa. Knockdown of RON resulted in significantly reduced cell invasion and cell migration in human laryngeal SCC cells. 24388747_RON promotes breast cancer metastasis through altered DNA methylation. 24518495_Stusies indicate that RON receptor tyrosine kinase is overexpressed and activated in pancreatic tumor specimens. 24628993_Data shows that MET and RON are regulated by PAX8 in non-small cell lung cancer. 24903148_Results pointed to a novel function for RON as a transcriptional regulator that promotes the survival of cancer cells subjected to hypoxia. 24954505_Studies demonstrate that DEK overexpression, due in part to Ron receptor activation, drives breast cancer progression through the induction of Wnt/beta-catenin signaling. 25193665_These results support the hypothesis that the alpha-chain of MSPalphabeta mediates RON dimerization. 25220236_SRSF2 promotes exon 11 inclusion of Ron proto-oncogene through targeting exon 11. 25512530_approach identified 18 kinase and kinase-related genes whose overexpression can substitute for EGFR in EGFR-dependent PC9 cells, and these genes include seven of nine Src family kinase genes, FGFR1, FGFR2, ITK, NTRK1, NTRK2, MOS, MST1R, and RAF1. 25874493_the prognostic significance of MET is limited in early stage disease. MET+/RON+ patients had higher overall recurrence rates than those with the other expression patterns 25997828_Data demonstrated upregulated RON isoform expression and significant changes in splicing factor expression in primary ovarian cancer suggesting a regulatory interplay of splicing factor and RON alternative splicing pattern in ovarian cancer. 26152593_Activation of RON as an alternate mechanism in the development of CRPC. 26189249_RON expression in endometrial adenocarcinoma was significantly higher than that in normal endometrial tissues. 26477314_Findings indicate a role of the RON tyrosine kinase receptor in pancreatic cancer and suggest RON as a potential therapeutic target. 26721331_Aberrant glycosylation of the RON receptor was shown as an alternative mechanism of oncogenic activation. 26772202_Results show that under hypoxia, nuclear RON activates non-homologous end joining DNA repair by interacting with Ku70 and DNA-PKcs and confers chemoresistance. 26775595_Study identified four novel uniquely spliced RON transcripts in small cell lung carcinoma (SCLC) and Non-SCLC cell lines. 26872377_High RON expression is associated with castration resistant prostate cancer. 26930004_hnRNP A1 directly binds to the 5' untranslated region of the RON mRNA and activates its translation through G-quadruplex RNA secondary structures 26951679_the MST1R variant c.G917A:p.R306H is highly associated with nasopharyngeal carcinoma (odds ratio of 9.0). 27323855_RON isoforms may comprise half of total RON transcript in human pancreatic cancer and their expression is regulated at least in part by promoter hypermethylation. 27533251_Forkhead box C1 protein (FOXC1) promotes melanoma cell function by regulating macrophage stimulating 1 receptor (MST1R) and activating MST1R/PI3K/AKT pathway. 27919987_complete loss of one or both MET and RON, as well as their overexpression, is a poor prognostic factor in patients with extrahepatic cholangiocarcinoma, probably due to the high rate of lymph-node metastasis 28075465_High expression of RON is associated with drug resistance in bladder cancer. 28100548_rare variants in MST1R in pNTMPEX/scoliosis patients could contribute to Lady Windermere syndrome by decreasing airway ciliary function and reducing IFN-gamma production in response to pulmonary nontuberculous mycobacterial disease 28123075_RON has a role in cancer-induced bone destruction and osteoporosis 28388571_RONDelta165E2 variant promoted tumor progression while activating the PI3K/AKT pathway via PTEN phosphorylation in colorectal carcinoma. 28440432_These findings indicate that RON and c-Met facilitate metastasis through ERK1/2 signaling and that targeting RON and c-Met with foretinib may be an attractive therapeutic option for suppressing Prostate cancer metastasis 30120239_the cis-regulatory landscape that determines alternative splicing of exon 11 in the proto-oncogene MST1R (RON), was examined. 30121008_this study demonstrates the functional significance of RON during prostate cancer progression and provides a strong rationale for targeting RON signaling in prostate cancer as a means to limit resistance to androgen deprivation therapy. 30223007_Study conclude that the splice variants of RON lacking exon 11 and exons 11-13 were detected in several lung cancer cell lines. Novel variant formed by skipping exons 11-13, the sequence of which code for transmembrane region, is predicted to code for a truncated isoform that may be secreted out. 30859654_Results suggest that high levels of RON may play a role in predisposing African Americans men to develop aggressive prostate cancer. 30967626_study demonstrates the functional significance of Mst1r during pancreatic cancer initiation and progression 31085796_These findings establish a link between the receptor tyrosine kinase RON and beta-catenin and provide insight into the mechanism by which they contribute to gastric cancer progression. 31254927_Comparative characterization of the HGF/Met and MSP/Ron systems in primary pancreatic adenocarcinoma. 31268163_High expression of RON is associated with epithelial mesenchymal transition through regulating MAPK signaling pathway in oral squamous cell carcinoma. 31792339_OPN promotes the aggressiveness of non-small-cell lung cancer cells through the activation of the RON tyrosine kinase. 32324988_RON and MET Co-overexpression Are Significant Pathological Characteristics of Poor Survival and Therapeutic Targets of Tyrosine Kinase Inhibitors in Triple-Negative Breast Cancer. 32342233_MST1R (RON) expression is a novel prognostic biomarker for metastatic progression in breast cancer patients. 32399910_Short-form RON (sf-RON) enhances glucose metabolism to promote cell proliferation via activating beta-catenin/SIX1 signaling pathway in gastric cancer. 32669614_The HNRNPA2B1-MST1R-Akt axis contributes to epithelial-to-mesenchymal transition in head and neck cancer. 32962738_MET and RON receptor tyrosine kinases in colorectal adenocarcinoma: molecular features as drug targets and antibody-drug conjugates for therapy. 33257837_RON signalling promotes therapeutic resistance in ESR1 mutant breast cancer. 33410267_Recepteur d'origine nantais contributes to the development of endometriosis via promoting epithelial-mesenchymal transition of a endometrial epithelial cells. 33508385_Tumor cell intrinsic RON signaling suppresses innate immune responses in breast cancer through inhibition of IRAK4 signaling. 33673346_Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer. 34347914_A potential signaling axis between RON kinase receptor and hypoxia-inducible factor-1 alpha in pancreatic cancer. 34821550_EGFR transactivates RON to drive oncogenic crosstalk. | ENSMUSG00000032584 | Mst1r | 19.156122 | 0.9086400 | -0.138219242 | 0.62167691 | 4.663732e-02 | 8.290215e-01 | No | Yes | 11.236057 | 5.972421 | 14.616884 | 7.754484 | ||
| ENSG00000164171 | 3673 | ITGA2 | protein_coding | P17301 | FUNCTION: Integrin alpha-2/beta-1 is a receptor for laminin, collagen, collagen C-propeptides, fibronectin and E-cadherin. It recognizes the proline-hydroxylated sequence G-F-P-G-E-R in collagen. It is responsible for adhesion of platelets and other cells to collagens, modulation of collagen and collagenase gene expression, force generation and organization of newly synthesized extracellular matrix.; FUNCTION: (Microbial infection) Integrin ITGA2:ITGB1 acts as a receptor for Human rotavirus A. {ECO:0000269|PubMed:12941907}.; FUNCTION: (Microbial infection) Integrin ITGA2:ITGB1 acts as a receptor for Human echoviruses 1 and 8. {ECO:0000269|PubMed:8411387}. | 3D-structure;Calcium;Cell adhesion;Direct protein sequencing;Disulfide bond;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Integrin;Magnesium;Membrane;Metal-binding;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes the alpha subunit of a transmembrane receptor for collagens and related proteins. The encoded protein forms a heterodimer with a beta subunit and mediates the adhesion of platelets and other cell types to the extracellular matrix. Loss of the encoded protein is associated with bleeding disorder platelet-type 9. Antibodies against this protein are found in several immune disorders, including neonatal alloimmune thrombocytopenia. This gene is located adjacent to a related alpha subunit gene. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2012]. | hsa:3673; | axon terminus [GO:0043679]; basal part of cell [GO:0045178]; cell surface [GO:0009986]; external side of plasma membrane [GO:0009897]; focal adhesion [GO:0005925]; integrin alpha2-beta1 complex [GO:0034666]; integrin complex [GO:0008305]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; amyloid-beta binding [GO:0001540]; collagen binding [GO:0005518]; collagen binding involved in cell-matrix adhesion [GO:0098639]; collagen receptor activity [GO:0038064]; heparan sulfate proteoglycan binding [GO:0043395]; integrin binding [GO:0005178]; laminin binding [GO:0043236]; metal ion binding [GO:0046872]; protein-containing complex binding [GO:0044877]; virus receptor activity [GO:0001618]; animal organ morphogenesis [GO:0009887]; blood coagulation [GO:0007596]; cell adhesion [GO:0007155]; cell adhesion mediated by integrin [GO:0033627]; cell population proliferation [GO:0008283]; cell-matrix adhesion [GO:0007160]; cell-substrate adhesion [GO:0031589]; cellular response to estradiol stimulus [GO:0071392]; cellular response to mechanical stimulus [GO:0071260]; collagen-activated signaling pathway [GO:0038065]; detection of mechanical stimulus involved in sensory perception of pain [GO:0050966]; establishment of protein localization [GO:0045184]; female pregnancy [GO:0007565]; focal adhesion assembly [GO:0048041]; hepatocyte differentiation [GO:0070365]; hypotonic response [GO:0006971]; integrin-mediated signaling pathway [GO:0007229]; mammary gland development [GO:0030879]; mesodermal cell differentiation [GO:0048333]; positive regulation of alkaline phosphatase activity [GO:0010694]; positive regulation of cell adhesion [GO:0045785]; positive regulation of cell projection organization [GO:0031346]; positive regulation of collagen binding [GO:0033343]; positive regulation of collagen biosynthetic process [GO:0032967]; positive regulation of DNA binding [GO:0043388]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of inflammatory response [GO:0050729]; positive regulation of leukocyte migration [GO:0002687]; positive regulation of phagocytosis, engulfment [GO:0060100]; positive regulation of positive chemotaxis [GO:0050927]; positive regulation of smooth muscle cell migration [GO:0014911]; positive regulation of smooth muscle cell proliferation [GO:0048661]; positive regulation of smooth muscle contraction [GO:0045987]; positive regulation of translation [GO:0045727]; positive regulation of transmission of nerve impulse [GO:0051971]; response to amine [GO:0014075]; response to hypoxia [GO:0001666]; response to L-ascorbic acid [GO:0033591]; response to muscle activity [GO:0014850]; response to parathyroid hormone [GO:0071107]; response to xenobiotic stimulus [GO:0009410]; skin morphogenesis [GO:0043589]; substrate-dependent cell migration [GO:0006929] | 11238113_Observational study of genotype prevalence. (HuGE Navigator) 11246537_Observational study of gene-disease association. (HuGE Navigator) 11323022_Observational study of gene-disease association. (HuGE Navigator) 11395045_Observational study of gene-disease association. (HuGE Navigator) 11472360_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11568114_Observational study of gene-disease association. (HuGE Navigator) 11698306_Observational study of gene-disease association. (HuGE Navigator) 11728949_Observational study of gene-disease association. (HuGE Navigator) 11775028_Bromodeoxyuridine induces integrin expression at transcriptional (alpha2 subunit) and post-transcriptional (beta1 subunit) levels, and alters the adhesive properties of two human lung tumour cell lines. 11776052_Observational study of gene-disease association. (HuGE Navigator) 11809527_Reduction in integrin alpha 2 is related to changes seen during immortalization and malignant progression 11812069_Observational study of gene-disease association. (HuGE Navigator) 11835340_Observational study of genotype prevalence. (HuGE Navigator) 11877061_The C807T polymorphism of integrin alpha2 gene in Suzhou Han population was different from that observed in other populations. 11958806_Trimucytin, a collagen-like snake venom protein, activates platelets independent of I-domain within alpha2 subunit of alpha2beta1 integrin. 11978651_Observational study of gene-disease association. (HuGE Navigator) 12038776_Observational study of gene-disease association. (HuGE Navigator) 12070018_the rGPIa/IIa-collagen interaction dominates the adhesion of rGPIa/IIa-Ib alpha-liposomes to the collagen surface at low shear rates; the rGPIa/IIa-collagen and rGPIb alpha-VWF interactions synergistically support liposome adhesion at high shear rates. 12071877_Observational study of genotype prevalence. (HuGE Navigator) 12073410_Observational study of gene-disease association. (HuGE Navigator) 12082590_Observational study of gene-disease association. (HuGE Navigator) 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12181354_Data report the identification of signaling pathways required for suppression of integrin alpha2beta1 function by c-erbB2. 12208476_Observational study of gene-disease association. (HuGE Navigator) 12372459_initial adhesion of endometrial cells to mesothelium is not mediated by alpha(2)beta(1)integrin 12392763_findings demonstrate that E-cadherin can interact with alpha2beta1 and suggest that heterotypic interactions between E-cadherin and integrins may be more common than originally thought 12412731_Observational study of gene-disease association. (HuGE Navigator) 12412731_role of the GP Ia C807T/G873A polymorphism as a risk factor for thrombosis in Behcet disease 12486862_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12499711_Observational study of gene-disease association. (HuGE Navigator) 12529399_R-Ras promotes focal adhesion formation by signaling to FAK and p130(Cas) through a novel mechanism that differs from but synergizes with the alpha2beta1 integrin. 12540964_genetic variation of platelet glycoprotein Ia may play a particularly important role during the advanced stages of diabetic retinopathy 12544734_twins developed ischaemic strokes and were shown to be homozygous for the alpha2 807T allele 12615788_Observational study of gene-disease association. (HuGE Navigator) 12657625_linkage with extracellular signal-regulated kinase is functionally linked in highly malignant autocrine transforming growth factor-alpha-driven colon cancer cells 12681287_Data show that NADPH oxidase activation production of reactive oxygen species are involved in the increase of alpha2beta1-integrin plasma membrane expression in Caco-2 cells stimulated with type IV collagen. 12690916_role of polymorphisms in cardiovascular thrombotic disease [review] 12717361_signaling through both constitutively expressed alpha2 integrin and Matrigel-induced alpha3 integrin expression is required to acquire a differentiated phenotype in Caco-2 cells. 12724616_Observational study of gene-disease association. (HuGE Navigator) 12788934_capillary morphogenesis requires endothelial alpha2beta1 integrin engagement of a single type I collagen integrin-binding site, possibly signaling via p38 MAPK and focal adhesion disassembly/FAK inactivation. 12791669_adhesion to the alpha2beta1-specific peptide was disulfide-exchange dependent and protein disulfide isomerase (PDI) mediated in platelets 12871362_results of a case-control study involving 180 stroke patients and 172 controls do not support a role for the integrin alpha2 C807T and GPVI Q317L polymorphisms in the development of first-ever ischemic stroke 12871600_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 12915563_Monkey rotavirus binding to alpha2beta1 integrin requires the alpha2 I domain and is facilitated by the homologous beta1 subunit. 12928694_Observational study of gene-disease association. (HuGE Navigator) 13679375_role in inhibiting Fas-mediated apoptosis in T-lymphocytes 14556196_Observational study of gene-disease association. (HuGE Navigator) 14556196_There were significant differences in the distribution of T and C alleles between myocardial infarction and control groups. 14563646_human platelet deposition on collagen depends on the concerted interplay of several receptors: GPIb in synergy with alpha(2)beta(1) mediating primary adhesion, reinforced by activation through GPVI, which further regulates the thrombus formation. 14671618_Observational study of gene-disease association. (HuGE Navigator) 14679206_results suggest that matrix metalloproteinase-1 can stimulate dephosphorylation of Akt protein and neuronal death through a non-proteolytic mechanism that involves changes in integrin alpha2beta1 signaling 14687991_Since the platelet alpha2 C807T gene polymorphism is associated with alpha2beta1 receptor density on the platelet surface, the level of alpha2beta1 on platelets may be an additional factor affecting Glanzmann's thromboasthenia clinical expression. 14701832_virus attachment to alpha(2) integrin on the cell surface was found to result in integrin clustering, which can give rise to signaling and facilitate the initiation of the viral entry 14746139_Observational study of genotype prevalence. (HuGE Navigator) 15104219_Observational study of gene-disease association. (HuGE Navigator) 15132990_VEGF-A induced alpha1 & alpha2 integrins, promoting lymphatic endothelial tube formation & haptotactic migration. Lineage-specific integrin receptor expression contribute to the distinct dynamics of wound-associated angiogenesis & lymphangiogenesis. 15205592_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15227729_Observational study of gene-disease association. (HuGE Navigator) 15240572_alpha2beta1 integrin has a role in endorepellin-induced endothelial cell disassembly of actin cytoskeleton and focal adhesions 15265786_alpha2beta1 integrin and GPVI regulate stress fiber formation in megakarocytes, the primary actin structures needed for cell contraction 15292257_alpha2beta1 integrin and CD44/CSPG receptor binding on human melanoma cell activation has been evaluated herein using triple-helical peptide ligands incorporating collagen peptides. 15304053_ILK regulates alpha 2 beta 1 in HEL cells, is activated in platelets and associates with beta 1-integrins 15350465_Observational study of gene-disease association. (HuGE Navigator) 15355503_Observational study of genetic testing. (HuGE Navigator) 15355504_Observational study of gene-disease association. (HuGE Navigator) 15514009_the interaction of platelet GP Ib with VWF mediates the activation of alpha2beta1, increasing its affinity for collagen 15522237_Data demonstrate that type I collagen synergistically enhances platelet-derived growth factor (PDGF)-induced smooth muscle cell proliferation through Src-dependent crosstalk between the alpha2beta1 integrin and the PDGF receptor beta. 15546585_Observational study of gene-disease association. (HuGE Navigator) 15630502_Observational study of gene-disease association. (HuGE Navigator) 15630502_alpha2807TT genotype of the platelet membrane integrin alpha2beta1 is associated with premature onset of early fetal loss. It appears that this risk factor does not induce the pathomechanism, but modulates the course of fetal loss 15647274_Data show that the recombinant Scl protein p176 promotes adhesion and spreading of human lung fibroblast cells through an alpha2beta1 integrin-mediated interaction. 15699160_Alpha2beta1 integrin is required for up-regulation of IL-1 beta-dependent airway smooth muscle secretory responses by fibronectin; while alpha2beta1 is also an important transducer for type I collagen. 15730528_Observational study of genotype prevalence. (HuGE Navigator) 15777792_Binds to vimentin, but this association is lost after prolonged adhesion of endothelial cells to collagen. 15847651_Observational study of genetic testing. (HuGE Navigator) 15892865_Observational study of gene-disease association. (HuGE Navigator) 15947241_Observational study of gene-disease association. (HuGE Navigator) 15947241_individual effect of each polymorphism located either in the coding or promoter sequence of the alpha2 gene may act in combination to modulate variations in platelets alpha2beta1 receptor density 15978109_Observational study of genotype prevalence. (HuGE Navigator) 15978109_analysis of platelet glycoprotein I(b)alpha and integrin alpha2beta1 polymorphisms in diverse populations 15978110_platelet membrane integrins alpha IIb(beta)3 (HPA-1b/Pl) and alpha2(beta)1 (alpha807TT) polymorphisms may have a role in premature myocardial infarction 16043429_analysis of binding between collagen type III and integrins alpha1beta1 and alpha2beta1 16055706_the signaling network involving Smad-dependent TGFbeta, PKCdelta, and integrin alpha2beta1/alpha3beta1, regulates cell spreading, motility, and invasion of the SNU16mAd gastric carcinoma cell variant 16113793_alpha2beta1 and signaling via autocrine mediators facilitate and amplify the GPVI procoagulant activity of fibrillar and non-fibrillar collagens 16140647_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16157382_Observational study of gene-disease association. (HuGE Navigator) 16214444_Observational study of gene-disease association. (HuGE Navigator) 16304451_Observational study of gene-disease association. (HuGE Navigator) 16317580_Integrin alpha-2 gene polymorphisms is associated with breast cancer 16317580_Observational study of gene-disease association. (HuGE Navigator) 16357324_Outside-in signaling through integrin alpha2beta1 triggered inside-out activation of integrin alphaIIbbeta3 and promoted fibrinogen binding. (alpha2beta1 AND integrin alphaIIbbeta3 16380674_Observational study of gene-disease association. (HuGE Navigator) 16380674_Our results did not show the causative relationship between the existence of platelets GP Ia mutations and venous system thrombosis in the women in labor. 16390868_Results suggest that loss of E-cadherin function is linked to regulation of cell-cell and cell-matrix adhesion, based in part on cell surface expression of alpha2, alpha3 and beta1 integrins. 16421008_The Platelet surface integrin, alpha 2-collagen interaction is an early step associated with platelet adhesion and activation and plays an important role in arterial thrombosis. 16513317_Common polymorphism (C807/T807), affecting the GPIa gene expression, in the development of oral cancer. 16513317_Observational study of gene-disease association. (HuGE Navigator) 16573563_Observational study of genotype prevalence. (HuGE Navigator) 16622460_malignant phenotype of pancreatic cancer on type I collagen is mediated specifically by the alpha2beta1 integrin 16697311_Genetic polymorphism C807T of platelet glycoprotein Ia increases the risk for premature myocardial infarction. It is also an independent predictor for the release of sCD40L during acute MI and persists 1 year after event. 16697311_Observational study of gene-disease association. (HuGE Navigator) 16732726_Acquisition of multiple antitumor drugs was accompanied by a drastically reduced expression of alpha2beta1 in the adenocarcinoma cells. 16820192_Meta-analysis of gene-disease association. (HuGE Navigator) 16820192_This meta-analysis does not support an association between the C807T polymorphism of ITAG2 gene and stroke. 16828471_The ectopic expression of TM4SF5 in Cos7 cells reduced integrin signaling under serum-containing conditions, but increased integrin signaling upon serum-free replating on substrates. 16875034_Observational study of genotype prevalence. (HuGE Navigator) 16882656_extracellular matrix fragments produced by apoptotic EC initiate a state of resistance to apoptosis in fibroblasts via an alpha2beta1 integrin/SFK (Src and Fyn)/phosphatidylinositol 3-kinase (PI3K)-dependent pathway 16905953_Observational study of gene-disease association. (HuGE Navigator) 16908762_Cell attachment was significantly inhibited by function-blocking anti-alpha2 (56%) and -beta1 (98%) integrin antibodies. 16972245_Results show that alpha2beta1 integrin signalling enhances cyclooxygenase-2 expression in intestinal epithelial cells. 17003923_Observational study of genotype prevalence. (HuGE Navigator) 17023078_Meta-analysis of gene-disease association. (HuGE Navigator) 17023078_Our findings support the view that C807T polymorphism of the GPla gene is not a significant risk factor for CAD, either alone or in combination with other major cardiovascular risk factors. 17036337_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17070428_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17127487_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17157856_C807T/G873A polymorphisms, but not T837C, are associated with higher platelet reactivity and residual platelet activity after treatment with anti-platelet agents 17157856_Observational study of gene-disease association. (HuGE Navigator) 17160992_Observational study of genotype prevalence. (HuGE Navigator) 17164499_Observational study of gene-disease association. (HuGE Navigator) 17164499_glycoprotein Ia 807C/T and 873G/A dimorphisms were not shown as risk factors for VTE 17179151_activated EGF receptor transiently modulates integrin alpha2 cell surface expression and stimulates integrin alpha2 trafficking via caveolae/raft-mediated endocytosis 17184645_Clinical trial of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17196570_Observational study of gene-disease association. (HuGE Navigator) 17264949_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17312461_pancretic cancer cells attach to 3D type I collagen scaffolds in an alpha2beta1-specific manner 17331499_Adhesion of breast cancer cells through the VLA integrins alpha2beta1 and alpha5beta1 was significantly reduced by an apoptosis-inducing natural triterpenoid, dehydrothyrsiferol (DT), when studied on low amounts of extracellular matrix. 17346829_Observational study of gene-disease association. (HuGE Navigator) 17408410_both the gpIb-VWF interaction and the integrin alpha(2)beta(1)-collagen interaction contribute to platelet adhesion under high shear stress; integrin alpha(II)beta(1) makes a greater contribution to adhesion to type I collagen because less VWF is bound 17495265_study indicates that the GPIa 807 C/T polymorphism does not represent a risk factor for Buerger's disease itself, but could be associated with premature onset of this disorder in predisposed individuals 17498594_findings suggest an important role of integrin alpha2beta1, alpha3beta1, and alpha5beta1 in the architectural characteristics of ameloblastomas and adenomatoid odentogenic tumor 17534386_Observational study of gene-disease association. (HuGE Navigator) 17534386_genetic variability within ITGA2 may confer risk for ischemic stroke independent of conventional risk factors. 17538005_LOX-1 is important for ADP-stimulated inside-out activation of platelet alpha(IIb)beta(3) and alpha(2)beta(1) integrins 17598123_There is no indication that the presence of 807C/T polymorphism is present in platelet glycoprotein Ia gene and a risk factor for retinal vein occlusion. 17630485_Observational study of gene-disease association. (HuGE Navigator) 17638891_CD44+alpha2beta1+ cell population is enriched in tumor-initiatin prostate cancer cells 17669516_thrombopoietin-induced in vitro differentiation of primary human cord blood mononuclear cells into megakaryocytes, we observed rapid, progressive CpG methylation of ITGA1, but not PELO or ITGA2. 17728329_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17890945_Observational study of gene-disease association. (HuGE Navigator) 17890945_Our results do not support an independent association between the 807C/T polymorphism and stroke of undetermined etiology. 18026855_The result suggested that cytoskeleton was a possible mechanical sensor to centrifugal stimuli, and the cytoskeleton regulation to centrifugal loading was in an extracellular matrix-dependent and integrin-mediated manner. 18057877_Polymorphisms in human platelet alloantigen (HPA)-1 and HPA-3 (GPIIb/IIIa), HPA-2 (GPIb/IX), HPA-4 (GPIIIa) and HPA-5 (GPIa/IIa) were found to be associated with the symptoms and recurrence of ischemic stroke. 18231737_Observational study of gene-disease association. (HuGE Navigator) 18231737_Platelet membrane glycoprotein Ia variations may have an important impact on aspirin resistance. 18362184_Initiation of the signal requires two collagen receptors, alpha2beta1 integrin and discoidin domain receptor (DDR). Each receptor propagates signals through separate pathways that converge to up-regulate N-cadherin. 18413316_induction of decorin expression in angiogenic, as opposed to quiescent, endothelial cells promotes a motile phenotype in an interstitial collagen I-rich environment by both signaling through IGF-IR and influencing alpha2beta1 integrin activity 18417478_collagen and a jararhagin-derived disintegrin peptide competitively bind to the integrin alpha2-I domain 18448666_alpha2beta1 integrin clustering defines its own entry pathway that is Pak1 dependent but clathrin and caveolin independent and that is able to sort cargo to caveosomes 18491034_The data suggest that phosphoenolpyruvate -dependent decrease of collagen biosynthesis in cultured human skin fibroblasts may undergo through depression of alpha(2)beta(1) integrin and IGF-IR signaling. 18502778_Observational study of gene-disease association. (HuGE Navigator) 18513389_Observational study of gene-disease association. (HuGE Navigator) 18554474_Integrin alpha2beta1 may promote migration and invasion of neuroblastoma cells. 18587047_These results indicate that NSP4 interaction with integrin alpha1 and alpha2 is an important component of enterotoxin function and rotavirus pathogenesis, further distinguishing this viral virulence factor from other microbial enterotoxins. 18608122_807C/T polymorphism of glycoprotein Ia (GPIa) gene were associated with higher risk of ischaemic stroke in atrial fibrillation 18608122_Observational study of gene-disease association. (HuGE Navigator) 18613064_Generated high throughput platform to examine interactions of type I collagen receptor alpha(2)beta(1) and fibronectin receptor alpha(5)beta(1) with peptide ligands to evaluate the effects of integrin cross-talk on adhesive responses. 18638089_Observational study of gene-disease association. (HuGE Navigator) 18787945_SEMA3A suppression of tumor cell migration is dependent on alpha2beta1, the expression of which is stimulated in breast tumor cells by an autocrine SEMA3A pathway. 18806884_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18820259_analysis of distinct roles of beta1 metal ion-dependent adhesion site (MIDAS), adjacent to MIDAS (ADMIDAS), and ligand-associated metal-binding site (LIMBS) cation-binding sites in ligand recognition by integrin alpha2beta1 18826391_high-affinity binding for alpha2beta1 can control the overall platelet-adhesive activity of native collagens. 18830231_Observational study of gene-disease association. (HuGE Navigator) 18836731_ITGA2 807C>T polymorphism may be associated with reduced colorectal cancer risk. 18836731_Observational study of gene-disease association. (HuGE Navigator) 18936436_Observational study of genotype prevalence. (HuGE Navigator) 18974842_Observational study of gene-disease association. (HuGE Navigator) 18983487_NO targets activation-dependent adhesion mediated by alpha(2)beta(1), possibly by reducing bioavailability of platelet-derived ADP, but has no effect on activation-independent adhesion mediated by GPVI 18990704_analysis of a prokaryotic Scl1 collagen sequence motif that mediates binding to human collagen receptors, integrins alpha2beta1 and alpha11beta1 19008959_Meta-analysis of gene-disease association. (HuGE Navigator) 19082798_Observational study of gene-disease association. (HuGE Navigator) 19110485_Observational study of gene-disease association. (HuGE Navigator) 19131662_Meta-analysis of gene-disease association. (HuGE Navigator) 19132198_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19202951_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19204726_Observational study of gene-disease association. (HuGE Navigator) 19235843_The increase in human platelet antigen, HPA-5b allelic frequency in Hepatitis C infection may indicate a possible association between HCV infection and HPAs. 19263529_Observational study of gene-disease association. (HuGE Navigator) 19336370_Observational study of gene-disease association. (HuGE Navigator) 19350519_Observational study of gene-disease association. (HuGE Navigator) 19388931_Observational study of gene-disease association. (HuGE Navigator) 19395705_alpha2beta1 integrin acquired core3 O-glycans in cells expressing core3 synthase with decreased maturation of beta1 integrin, leading to decreased levels of the alpha2beta1 integrin complex 19411307_alpha(2)-, alpha(3)-, and beta(1)-integrins and E-cadherin expression in normal human lung and bronchopulmonary sequestration and congenital cystic adenomatoid malformation were evaluated using Western blot and immunohistochemistry. 19420105_Observational study of gene-disease association. (HuGE Navigator) 19422573_Increased expression of integrin alpha2 and the increased response to TGF-beta1 of hereditary gingival fibromatosis (HGF) fibroblasts may be related to the excessive collagen deposition in HGF patients. 19451223_Nox1 knockdown led to a loss of directional migration which takes place through a RhoA-dependent alpha2/alpha3 integrin switch. 19479237_Observational study of gene-disease association. (HuGE Navigator) 19500323_nucleotide substitution at position 2235 induces a Q716H amino acid change in the GPIa mature protein that causes Fetal/neonatal alloimmune thrombocytopenia. 19530321_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19549780_alpha2-integrin expression and Rho kinase activity are regulated by protein kinase signaling cascades upon activation of alpha7 nicotinic receptor 19559392_Observational study of gene-disease association. (HuGE Navigator) 19570064_Observational study of genetic testing. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19587357_Meta-analysis of gene-disease association. (HuGE Navigator) 19622836_A GPVI-independent signaling role of alpha2beta1 in response to collagen stimulation. 19635510_Altogether these data suggest that integrins alpha2 and alpha v beta 3 do not play a major role in the rotavirus entry process. 19700757_a central role for alpha2beta1 integrin in experimental and developmental angiogenesis 19702628_Observational study of gene-disease association. (HuGE Navigator) 19724904_AsialoGM1 and integrin alpha2beta1 mediate prostate cancer progression. 19729601_Observational study of gene-disease association. (HuGE Navigator) 19740098_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19740098_genetic polymorphisms in ITGA2 and P2RY1 combine with plasma VWF:Ag levels to modulate baseline platelet reactivity in response to collagen plus EPI, while genetic differences in P2RY1 and GP1BA significantly effect platelet responses to collagen plus ADP 19778317_In a Croatian population, the HPA-5 allele frequencies are 5a-0.895 and 5b-0.105. 19789264_TM4SF5 in hepatocytes negatively regulates integrin alpha2 function via an interaction between the extracellular loop 2 of TM4SF5 and integrin alpha2 during cell spreading on and migration through collagen I environment 19789387_SHP-1 coprecipitates with integrin alpha2. A novel functional interaction between the integrin alpha2 subunit and SHP-1 is described. 19827952_TGF-beta-mediated up-regulation of the expression of the integrin subunits alpha(2) and alpha(6) is mainly mediated in MSC by Smad2. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19927126_in its entry strategy human echovirus 1 seems to rely on the activation of signalling pathways that are dependent on alpha2beta1 clustering, but do not require the conformational regulation of the receptor 19948007_Observational study of genotype prevalence. (HuGE Navigator) 19995941_Collagen type I inhibits the secretion of IL-8 by human neutrophils in a selective manner and this effect is mediated by the interaction of collagen with integrin alpha2beta1. 20063990_Data suggest that integrin alpha(2)beta(1), glycoprotein Ib and vWf interactions with collagen I and III contribute to platelet adhesion under high shear flow. 20076847_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20076847_the 807T allele of GP Ia and the PlA2 allele of GP IIIa, and specially its combination confer an additional risk for development of carotid atherosclerosis and arterial thrombosis in type 2 diabetes 20149160_Observational study of genotype prevalence. (HuGE Navigator) 20178602_Prostaglandin E2 enhances the migration of chondrosarcoma cells by increasing alpha2beta1 integrin expression through the EP1/PLC/PKCalpha/c-Src/NF-kappaB signal transduction pathway. 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20204402_Observational study of gene-disease association. (HuGE Navigator) 20336352_Observational study of gene-disease association. (HuGE Navigator) 20336352_Polymorphisms in the alpha2 gene of integrin alpha2beta1 in patients with von Willebrand disease could significantly impact platelet function. 20351310_Measurement of thrombus height after specific receptor blockade or use of altered proportions of peptides indicates a signaling rather than adhesive role for glycoprotein VI, and primarily adhesive roles for both alpha(2)beta(1) and the VWF axis 20436081_Androgens increased INT alpha1 and alpha2 subunits in tubuloepithelial cells and in healthy labial salivary glands. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20485444_Observational study of gene-disease association. (HuGE Navigator) 20485444_Single-nucleotide polymorphism in ITGA2 is associated with coronary atherosclerosis. 20532885_Observational study of gene-disease association. (HuGE Navigator) 20536507_Observational study of gene-disease association. (HuGE Navigator) 20598296_The syndecan- and alpha2beta1 integrin-binding peptides synergistically affect cells and accelerate cell adhesion. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20667040_Observational study of genotype prevalence. (HuGE Navigator) 20691446_Observational study of gene-disease association. (HuGE Navigator) 20806289_alpha2beta1 integrin may be a major regulatory molecule in Th17 cell functions. 20939067_The extracellular matrix rigidity affects the osteogenic outcome of mesenchymal stem cells through mechanotransduction events that are mediated by alpha(2)-integrin. 21034162_Observational study of gene-disease association. (HuGE Navigator) 21126803_Domain V of perlecan, a known alpha2 integrin ligand, inhibits brain amyloid-beta neurotoxicity in an alpha2 integrin-dependent manner. 21134100_Results suggest a novel mechanism for pathogen entry into host cells as well as a new function for C1q- alpha2beta1 integrin interactions. 21135504_alpha2beta1 integrin functionally inhibits breast tumor metastasis, and alpha2 expression may serve as an important biomarker of metastatic potential and patient survival 21193198_Polymorphisms in IL5RA, LPL, ITGA2 and NOS3 genes were independently associated with ischemic stroke in Chinese diabetic population. 21262375_molecular forces of the alpha2beta1 integrin-collagen interaction 21370991_Expression of laminin beta1 and integrin alpha2 is elevated in the anterior temporal neocortex tissue from patients with intractable epilepsy. 21474814_in endothelial cells, ligation of alpha2beta1 and alpha6beta1 integrins induces the Notch pathway, and we disclose a novel role of basement membrane proteins in the processes controlling tip vs stalk cell selection 21508388_Single nucleotide polymorphisms of integrin alpha-2 gene is not associated with colorectal cancer. 21558389_PHLDA1 expression marks the putative epithelial stem cells, downregulates ITGA2 and ITGA6, and contributes to intestinal tumorigenesis 21596751_endorepellin requires both the alpha2beta1 integrin and VEGFR2 for its angiostatic activity 21632096_Findings do not support the influence of the IRGA2807T allele in the development of retinopathy in type 2 diabetes. 21647271_Integrin alpha2beta1 might play a more crucial role in maintaining the mechanical creep properties of the collagen matrix than does integrin alpha1beta1. 21652699_data suggest that extracellular membrane-bound CAIV, but not cytosolic CAII, augments transport activity of MCT2 in a non-catalytic manner, possibly by facilitating a proton pathway other than His-88 21658756_Blocking the alpha(2)beta(1) receptor on human mesenchymal stem cells also resulted in a reduction of cell adhesion on both types of collagen-like peptide surfaces. 21672359_Rapid initiation of collagen-induced platelet aggregation may be associated with the platelet membrane GPIa T807 allele, which may be important in unstable angina pectoris pathogenesis. 21734795_Data suggest that the ITGA2 gene C807T polymorphism may be associated with an increased risk of gastric cancer, differentiation and invasion of gastric cancer. 21743959_CDKN2A, GATA3, CREBBP, ITGA2, NBL1 and TGM4 were down-regulated in the prostate carcinoma glands compared to the corresponding normal glands 21765051_Single-nucleotide polymorphisms in ITAG2 is associated with chronic kidney disease in Type 2 diabetes. 21787362_Under convertase inhibition, alpha2beta1 engagement led to enhanced phosphorylation of both FAK (focal adhesion kinase) and MAPK (mitogen-activated protein kinase). 21796158_The disturbance of alpha2beta1 mediated interactions to collagen I results in a tremendous reduction of hMSC numbers owing to mitochondrial leakage accompanied by Bcl-2-associated X protein upregulation. 21850018_Analysis of the data led to the elucidation of a new molecular mechanism by which SPARC promotes cathepsin B-mediated melanoma invasiveness using collagen I and alpha2beta1 integrins as mediators. 22015659_Platelet alphaIIbeta3 and alpha2beta1 levels were measured by flow cytometry in 320 acute coronary syndrome patients and 128 normal subjects and compared with MPV, platelet count, ITGA2 rs1126643, and ITGB3 rs5918 alleles. 22049795_Inflammation can up-regulate the expressions of | ENSMUSG00000015533 | Itga2 | 513.769868 | 0.9920703 | -0.011485801 | 0.12722079 | 8.245364e-03 | 9.276483e-01 | 9.737949e-01 | No | Yes | 524.162725 | 122.973363 | 531.046439 | 124.545021 | |
| ENSG00000164305 | 836 | CASP3 | protein_coding | P42574 | FUNCTION: Involved in the activation cascade of caspases responsible for apoptosis execution (PubMed:7596430). At the onset of apoptosis it proteolytically cleaves poly(ADP-ribose) polymerase (PARP) at a '216-Asp-|-Gly-217' bond (PubMed:7774019). Cleaves and activates sterol regulatory element binding proteins (SREBPs) between the basic helix-loop-helix leucine zipper domain and the membrane attachment domain. Cleaves and activates caspase-6, -7 and -9 (PubMed:7596430). Involved in the cleavage of huntingtin (PubMed:8696339). Triggers cell adhesion in sympathetic neurons through RET cleavage (PubMed:21357690). Cleaves and inhibits serine/threonine-protein kinase AKT1 in response to oxidative stress (PubMed:23152800). Acts as an inhibitor of type I interferon production during virus-induced apoptosis by mediating cleavage of antiviral proteins CGAS, IRF3 and MAVS, thereby preventing cytokine overproduction (PubMed:30878284). Cleaves XRCC4 and phospholipid scramblase proteins XKR4, XKR8 and XKR9, leading to promote phosphatidylserine exposure on apoptotic cell surface (PubMed:23845944, PubMed:33725486). {ECO:0000269|PubMed:21357690, ECO:0000269|PubMed:23152800, ECO:0000269|PubMed:23845944, ECO:0000269|PubMed:30878284, ECO:0000269|PubMed:33725486, ECO:0000269|PubMed:7596430, ECO:0000269|PubMed:7774019, ECO:0000269|PubMed:8696339}. | 3D-structure;Acetylation;Apoptosis;Cytoplasm;Direct protein sequencing;Hydrolase;Phosphoprotein;Protease;Reference proteome;S-nitrosylation;Thiol protease;Zymogen | The protein encoded by this gene is a cysteine-aspartic acid protease that plays a central role in the execution-phase of cell apoptosis. The encoded protein cleaves and inactivates poly(ADP-ribose) polymerase while it cleaves and activates sterol regulatory element binding proteins as well as caspases 6, 7, and 9. This protein itself is processed by caspases 8, 9, and 10. It is the predominant caspase involved in the cleavage of amyloid-beta 4A precursor protein, which is associated with neuronal death in Alzheimer's disease. [provided by RefSeq, Aug 2017]. | hsa:836; | caspase complex [GO:0008303]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; death-inducing signaling complex [GO:0031264]; membrane raft [GO:0045121]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; aspartic-type endopeptidase activity [GO:0004190]; cyclin-dependent protein serine/threonine kinase inhibitor activity [GO:0004861]; cysteine-type endopeptidase activity [GO:0004197]; cysteine-type endopeptidase activity involved in apoptotic process [GO:0097153]; cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:0097199]; cysteine-type endopeptidase activity involved in execution phase of apoptosis [GO:0097200]; death receptor binding [GO:0005123]; peptidase activity [GO:0008233]; phospholipase A2 activator activity [GO:0016005]; protease binding [GO:0002020]; protein-containing complex binding [GO:0044877]; anterior neural tube closure [GO:0061713]; apoptotic process [GO:0006915]; apoptotic signaling pathway [GO:0097190]; axonal fasciculation [GO:0007413]; B cell homeostasis [GO:0001782]; cell fate commitment [GO:0045165]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to staurosporine [GO:0072734]; erythrocyte differentiation [GO:0030218]; execution phase of apoptosis [GO:0097194]; glial cell apoptotic process [GO:0034349]; heart development [GO:0007507]; hippocampus development [GO:0021766]; intrinsic apoptotic signaling pathway in response to osmotic stress [GO:0008627]; keratinocyte differentiation [GO:0030216]; learning or memory [GO:0007611]; leukocyte apoptotic process [GO:0071887]; luteolysis [GO:0001554]; negative regulation of activated T cell proliferation [GO:0046007]; negative regulation of apoptotic process [GO:0043066]; negative regulation of B cell proliferation [GO:0030889]; neuron apoptotic process [GO:0051402]; neuron differentiation [GO:0030182]; neurotrophin TRK receptor signaling pathway [GO:0048011]; platelet formation [GO:0030220]; positive regulation of amyloid-beta formation [GO:1902004]; positive regulation of apoptotic DNA fragmentation [GO:1902512]; positive regulation of apoptotic process [GO:0043065]; positive regulation of neuron apoptotic process [GO:0043525]; protein processing [GO:0016485]; proteolysis [GO:0006508]; regulation of macroautophagy [GO:0016241]; regulation of protein stability [GO:0031647]; response to amino acid [GO:0043200]; response to antibiotic [GO:0046677]; response to cobalt ion [GO:0032025]; response to estradiol [GO:0032355]; response to glucocorticoid [GO:0051384]; response to glucose [GO:0009749]; response to hydrogen peroxide [GO:0042542]; response to hypoxia [GO:0001666]; response to lipopolysaccharide [GO:0032496]; response to nicotine [GO:0035094]; response to tumor necrosis factor [GO:0034612]; response to UV [GO:0009411]; response to X-ray [GO:0010165]; response to xenobiotic stimulus [GO:0009410]; sensory perception of sound [GO:0007605]; striated muscle cell differentiation [GO:0051146]; T cell homeostasis [GO:0043029]; wound healing [GO:0042060] | 11437602_A soluble proform of caspase 3 containing a fortuitous mutation (W206R) is inactive when incubated with recombinant human caspase 8 and therefore can serve as a useful reagent to test the efficacy of caspase 8 inhibitors. 11787859_Enzyme activation of caspase-3 was observed in apoptosis of K562 human cell line. 11866986_caspase-3 may participate in the regulation mechanism of lymphoma cel apoptosis. 11960384_Activation of caspase-3 and cleavage of Rb are associated with p16-mediated apoptosis in human non-small cell lung cancer cells. 11972398_caspase 3-mediated focal adhesion kinase processing in human ovarian cancer cells: possible regulation by X-linked inhibitor of apoptosis protein 11981455_investigation of molecular mechanism of MPO-mediated apoptosis and caspase-3 activation 11989976_These data suggest that D4-GDI of Rho family GTPase may be regulated during apoptosis through the caspase-3 mediated cleavage of the GDI protein. 11992386_Clinical significance of caspase-3 expression in pathologic-stage I, nonsmall-cell lung cancer 12004072_translation of some IRES-containing mRNAs is regulated by proteolytic cleavage of PTB during apoptosis 12032677_results suggest that IGF-1/PI-3 kinase inhibited C2-ceramide-induced apoptosis due to relieving oxidative damage, which resulted from the inhibition of catalase by activated caspase-3 12036886_Expression levels of apoptosis-related proteins caspase 3, Bcl-2, and PI9 predict clinical outcome in anaplastic large cell lymphoma. 12044963_Identification of high caspase-3 mRNA expression as a unique signature profile for extremely old individuals 12055227_The significant expression of caspase-3 that occurs in monocytes during serum-deprived induction of apoptosis can be down-regulated to baseline levels by addition of platelets. 12070005_High numbers of active caspase 3-positive Reed-Sternberg cells in pretreatment biopsy specimens of patients with Hodgkin disease predict favorable clinical outcome. 12070657_Caspase-cleaved amyloid precursor protein and activated caspase-3 are co-localized in the granules of granulovacuolar degeneration in Alzheimer's disease and Down's syndrome brain. 12080079_Selective inhibition of dipeptidyl peptidase I, not caspases, prevents the partial processing of procaspase-3 in CD3-activated human CD8(+) T lymphocytes 12145703_Pro-CASP3 moved to the the mitochondria of U937 cells during TPA-induced differentiation. 12151338_caspase 3 cleaves CDC6 during apoptosis, which prevents wounded cell from replicating and facilitates death 12169388_Both oleandrin and radiation share a caspase-3 dependent mechanism of apoptosis in the PC-3 human prostate cacncer cell line. 12181128_Hypoxia-induced cleavage of caspase-3 and DFF45/ICAD in human failed cardiomyocytes. 12210761_Role of nuclear PKC delta in mediating caspase-3-upregulation in Jurkat T leukemic cells exposed to ionizing radiation 12228224_Data show that endoplasmic reticulum stress induced by thapsigargin not only activated the apoptosis effector caspase-3 but also caused a large and prolonged increase in the activity of glycogen synthase kinase-3beta. 12297281_caspase 3-independent function of Bak in the TNF-alpha-induced apoptotic pathway 12390838_The apoptosis resistance of mdr cells is not related to the abnormality of CPP32 but the upstream of caspase, the fact of which indicates promising prospect of the research on reversion of mdr cells using CPP32 as target. 12393901_caspase 3 activation and apoptosis are blocked by LIGHT protein in hepatocytes 12397210_A significant positive correlation between in-situ active caspase-3 in the sperm midpiece and DNA fragmentation was observed in the low motility fractions of patients. 12444137_IFN-gamma-mediated caspase-3 activation and C. burnetii killing depend on the expression of membrane TNF. 12483536_determination of levels of caspase-3 expression in breast tumor samples and to determine whether alterations in its expression can affect their ability to undergo apoptosis 12511568_ceramide increases oxidative damage by inhibition of ROS scavenging ability through caspase-3-dependent proteolysis of catalase 12515825_caspase 3 has a role in damaging mitochondrial function and generating reactive oxygen species after activation by cytochrome c 12566444_Presentation of nitric oxide regulates monocyte survival through effects on this enzyme and caspase-9 activation. 12576296_induction of ceramide accumulation by various triggers of ceramide generation triggered the activation of caspase-3. 12576443_mRNA and protein expression of this enzyme are examined in breast cancer to determine level of apoptosis. 12579342_CASP3 induced by H202 was completely blocked by Z-VAD-fmk. 12581734_caspase-3 is essential for efficient induction of apoptosis by staurosporine, but not for mitochondrial steps that occur earlier in the pathway 12598529_one or more distinct cellular mechanisms regulate Bid cleavage by caspases 8 and 3 in situ. 12605885_caspase-3 was upregulated in a region-specific manner with marked activation in the selectively vulnerable hippocampal after cerebral ischemia 12606589_Data report that human oocytes and fragmenting preimplantation embryos possess transcripts encoding Harakiri and caspase-3. 12611892_caspase-3-mediated proteolysis of FAK, an anti-apoptotic protein, is regulated by hsp72 12621124_in neutrophils, functional expression of caspase-3 in neutrophils may be regulated during ontogeny. 12643601_We conclude that the changes in the level of caspase-3 and survivin play an important role in the transformation from normal gastric mucosa to gastric cancer. 12677451_caspase-3 as a key effector of neuronal apoptosis in pneumococcal meningitis. 12686427_activated by coxsackievirus B3 (CVB3) infections and occurred in cells expressing full length CAR 12700630_the caspase 3 apoptosis program is not required for anti-inflammatory clearance by human macrophages 12700660_Caspase-3 mediates a mitochondrial amplification loop that is required for the optimal release of cytochrome c, mitochondrial permeability shift transition, & cell death during apoptosis induced by treatment with the microtubule-damaging agent paclitaxel. 12707329_Caspase 3-mediated cell death is central to the biological control of antigen-independent expansion of recent thymic emigrants from human cord blood. 12788227_CASP3 is activated in the oxidative stress-induced apoptosis in tendon fibroblasts in vitro. 12788938_existence of a regulatory mechanism of protein stability and PTEN-protein interactions during apoptosis, executed by caspase-3 in a PTEN phosphorylation-regulated manner. 12792782_caspase 3 has a role in cell death through apoptosis induced by ursolic acid 12804035_MCF-7 tumor cells are deficient in CASP3 and instead have TNF-alpha-induced apoptosis 12809603_Apoptotic cells secrete chemotactic factors that stimulate the attraction of monocytic cells and primary macrophages. The activation of caspase-3 in the apoptotic cell was found to be required for the release of these chemotactic factors. 12824190_In Kennedy syndrome, phosphorylation of the polyglutamine-expanded form of androgen receptor regulates its cleavage by caspase-3 and enhances cell death. 12833566_The crystal structure of the binding site of caspase-3 reveals critical side chain movements in a hydrophobic pocket. The positions of these side chains may have implications for the directed design of inhibitors of caspase-3 or caspase-7. 12854132_Chemical substances derived from the primary foci and metastatic microenvironment can inhibit the growth of metastatic cells by enhancing Caspase-3 expression and diminishing FasL expression in gastric cancer. 12867600_Data suggest that increased intrathecal release of Fas, but not FasL or caspase 3, in the cerebrospinal fluid of infants with hydrocephalus may serve as an indicator of brain injury from progressive ventricular dilatation. 12874324_Caspase 1 involvement in monocyte lysis induced by Actinobacillus actinomycetemcomitans leukotoxin. 12897143_Caspase 3-mediated inactivation of rac GTPases promotes drug-induced apoptosis in human lymphoma cells 14570914_Caspase 3 has a role in proteolysis of the N-terminal cytoplasmic domain of the human erythroid anion exchanger 1 (band 3) 14572611_Inhibition of the Src-family tyrosine kinases activity by PP2 and of caspase-3 by Z-DEVD-FMK reverses apoptosis 14612448_the cascade of pro-apoptotic events leading to Bax, mitochondria, and caspase-3 activation are regulated by calpastatin and calpain-1 14623896_apoptosis was preceded by proteolytic cleavage of caspases 2, 3, and 7, and wild type STAT1 also induced cleavage of caspase 7 14657946_Doxorubicin-induced cell death of ALK-positive anaplastic large-cell lymphoma (ALCL) cells involves CASP3 activation. CASP3 activation correlates with ALK expression in ALCL tumors. 14674699_caspase-3 activation, mitochondrial respiratory function and cytochrome c release have roles in induction of apoptosis by ceramides 14678329_activation of caspase-3 by the Dot/Icm virulence system of Legionella pneumophila is essential for halting biogenesis of the L. pneumophila-containing phagosome through the endosomal/lysosomal pathway 14684629_Caspase-3 is activated in oxidized low-density lipoprotein (ox-LDL) induces apoptosis in endothelial cells. 14710359_IGF-I activates specific apoptotic pathways (Caspase-3 activation, Annexin-V binding and DNA degradation in an osteosarcoma cell line. 14764700_In Jurkat cells caspase-3 is a component of the death-inducing signaling complex that colocalizes in lipid rafts with caspase-8, where caspase-3 activity is required for complete caspase-8 activation following Fas cross-linking. 14960581_Evidence indicates that casp3 activated by ricin acts on BAT3 at the caspase cleavage site, DEQD(1001) to release a C-terminal fragment designated CTF-131, which induces phosphatidylserine exposure, cell rounding, and chromatin condensation as ricin does 14970175_p38-MAPK can directly phosphorylate and inhibit the activities of caspase-8 and caspase-3 and thereby hinder neutrophil apoptosis, and, in so doing, regulate the inflammatory response. 14971566_IFN-gamma-activated monocytes may induce reactive oxygen metabolites in retinal pigment epithelial(RPE) cells through cell-to-cell contact and promote HRPE cell apoptosis via caspase-3 activation. 14976035_activated in normal erythropoiesis and in erythroblasts in culture 14982947_IL-15 does not increase IL-1alpha or IL-1beta production but induces IL-1Ra release, increases myeloid cell differentiation factor-1 stability, decreases the activity of caspase-3 and caspase-8, resulting in an inhibition of vimentin cleavage 14991812_Caspase-3 activation in human disease can play a prominent role in localized cellular degenerative processes without causing nuclear or cell death. 15014070_Mcl-1L degradation by either GrB or caspase-3 interferes with Bim sequestration by Mcl-1L 15015772_Synthetic activation of inducible caspase-3, but not of caspase-8, resulted in apoptosis in glioma cell lines. 15033720_Dispensable for the execution of apoptosis in a metastatic melanoma cell line. 15033815_glc-oxLDL increases TUNEL positivity and caspase-3 activation of human coronary smooth muscle cells 15041704_Transfected caspase-3 is involved in the reduction of Akt level, and its involvement is mediated through caspase-9 activation. 15098015_Caspase-3 is frequently overexpressed in hepatocellular carcinomas and is associated with high serum levels of alpha-fetoprotein. 15153940_mice overexpressing human caspase 3 are essentially normal, however, they have increased susceptibility to degenerative insults 15156398_Overexpression of antiapoptotic proteins Bcl-2 and Bcl-X(L) and down-regulation of caspase-3 activity may be associated with cisplatin resistance in human ovarian cancer. 15187159_granule pools of Zn may be distinct from those regulating activation of procaspase-3 and NF-kappaB. 15205454_caspase-8 and -3 have roles in human mast cell apoptosis induction by Pseudomonas aeruginosa exotoxin A 15220451_required for Parvovirus B19 virus-induced apoptosis in primary hepatocytes and hepatocellular carcinoma cell line HepG2 15241180_results suggest that matrix metalloproteinase-9 and matrix metalloproteinase-2 contribute to caspase-mediated brain endothelial cell death after hypoxia-reoxygenation by disrupting cell-matrix interactions and homeostatic integrin signaling 15246206_Neutrophils isolated from cirrhosis patients exhibited a decreased viability and a marked accelerated apoptosis and significantly higher caspase-3 activity. 15254227_BAP31 and caspase 3 are cleaved in a process involving capsase 8 in the mitochondrial membrane during apoptosis 15280469_induced and utilized by human Astrovirus Yuc8 to promote processing of the capsid precursor and dissemination of the viral particles in CAC0-2 cells. 15375594_hypoxia caused epithelial cell death induced by caspase-3-like activity-dependent apoptosis 15473998_These data suggest that increased proneness to caspase activation in lymphocytes could reflect an ongoing systemic response in neurodegenerative disease with pathogenetic implications. 15505416_CASP3 was expressed in the same tissues as its murine counterpart. 15564512_required for Sars virus 7a protein induction of apoptosis in cell lines from different organs 15569669_granzyme B targets a highly restricted range of substrates and orchestrates cellular demolition largely through activation of caspase-3 15569672_the early and temporary activation of PP2A in neutrophils impaired not only the p38 MAPK-mediated inhibition of caspase 3 but also restored the activity to caspase 3 that had already been phosphorylated and thereby inactivated 15569692_results suggest that active caspase-3 is translocated in association with a substrate-like protein(s) from the cytoplasm into the nucleus during progression through apoptosis 15599395_caspase-3 is crucial for the differentiation of bone marrow stromal stem cells by influencing TGF-beta/Smad2 pathway and cell cycle progression 15617521_results suggest that Anaplasma phagocytophilum inhibits human neutrophil apoptosis via transcriptional upregulation of bfl-1 and inhibition of mitochondria-mediated activation of caspase 3 15637055_removal of N-terminal domains of Bid by caspase-8 and Mcl-1 by caspase-3 enables the maximal mitochondrial perturbation that potentiates TRAIL-induced apoptosis 15638357_Expression correlates with intensity of apoptosis in colorectal adenocarcinoma. 15657349_Gossypol induced complete cytochrome c release from mitochondria amd increased caspases-3 and -9 activity in large cell lymphoma cells. 15657356_The increased ratio of Bax/Bcl-2 proteins after Epigallocatechin-3-gallate may lead to activation of caxpaxe-3, leading to apoptosis. 15670787_Taken together, our data suggested that the JNK/c-Jun signaling cascade plays a crucial role in Cd-induced neuronal cell apoptosis and provides a molecular linkage between oxidative stress and neuronal apoptosis. 15705385_Caspase-3 dependent apoptosis occurs in human granulosa cells and activates when follicles begin to leave the resting pool. 15716280_PKCdelta-dependent phosphorylation of caspase-3 is involved in the regulation of monocyte life span 15748158_Caspase 3 activation associated with cellular prion is closely related to its ability to undergo endocytosis. This is, to our knowledge, the first direct description of an endocytosis-dependent PrP(c)-associated function. 15748808_Stx1 and LPS trigger DNA fragmentation and caspase-3 activation, as evidenced by the cleavage of poly(ADP-ribose) polymerase (PARP). 15803189_Absence of active caspase 3-positive nasopharyngeal carcinoma predicts rapid fatal outcome. 15843892_procaspase-2S-mediated anti-apoptotic effects are associated with inhibition of the processing and activation of procaspase-3 in VP-16-treated cells 15848173_calcium could favour a necrotic mechanism by inducing the generation of a form of caspase 3 insensitive to mitochondrial activation 15869582_caspase 3 exists in microparticles from endothelial cells and may be associated with caspase 3 activation unrelated to apoptosis 15935070_The antiapoptotic property of Hualpha-Syn in neuronal cell lines is associated with the attenuation of caspase-3 activity without affecting the caspase-9 activity or the levels of cleaved, active caspase-3. 15996160_Caspase 3 plays a important role in apoptotic process of mesothelioma cells. 16009347_These findings provide biological evidence showing that (+)-alpha-tocopherol can amplify the apoptotic response by up-regulating the expression of pro-caspase-3. 16080782_The characteristics of the fragment of hPMCA4b produced by caspase-3 are reported. 16083554_Data show that Caspase-3 was involved in the apoptosis of glomeruli and renal tubuli of lupus nephritis. 16121124_cleaved caspase-3 positive cells was found in temporal lobe epilepsy sections but not controls; nuclear localization of apoptosis-inducing factor was limited and restricted to cells that were negative for cleaved caspase-3 16179347_involvement of Fas/caspase 8/caspase 3-dependent signaling in an enucleated cell leading to PS externalization, a central feature of erythrophagocytosis and erythrocyte biology 16203739_the prodomain acts as an intramolecular chaperone during assembly of the (pro)caspase-3 subunits and increases the efficiency of formation of the native conformation 16213496_Caspase 3 activation is a prominent feature in periodontitis-associated tissue injury. 16242324_caspase-3, -8, and -9 activity is enhanced by safrole oxidase in lung cancer cells 16286477_desmoglein 1 is a novel caspase-3 target that regulates apoptosis in keratinocytes 16300929_Thrombin is able to induce activation of caspases 3 and 9 in human platelets and significantly increases the amount in the cytoskeleton of the active forms of both caspases and the procaspases 3 and 9. 16320828_Apoptosis marker activated caspase-3 was analysed by immunohistochemistry, during the production of collagen type II, the adhesion and signal transduction receptor beta1-integrin. 16337360_These results provide evidence for a novel type 1 IFN-mediated pathway that regulates apoptosis of T cells through a mitochondrial-dependent and caspase-dependent and independent pathway. 16382148_tTGase is a new type of caspase 3 inhibitor in THG-mediated apoptosis. 16455648_NFATc2 activity is regulated by caspase-3 16463650_Caspase-3 was expressed increasingly and activated in HCMV-infected cells. 16498457_These results demonstrate that SSRP1 degradation during apoptosis is a two-step process coupling caspase cleavage and ubiquitin-dependent proteolysis. 16505307_Blood values increased in untreated Parkinson disease patientes. 16516911_Asymmetric dimethylarginine induces apoptosis of endothelial cell via elevation of intracellular oxidant production, which involves p38 MAPK/caspase-3-dependent signaling pathway. 16526059_170-kDa P-gp has been reported to counteract apoptosis, its cleavage may be a mechanism aimed at blocking an important cell survival component 16529748_CAD is downregulated at the mRNA and protein level during the erythroid differentiation in TF-1 cells. 16530181_These results suggest that caspases, including caspase 3, can act as substrates for non-caspase proteases in cells primed for necrosis induction. 16530191_Protein-protein interaction of NMTs revealed that m-calpain interacts with NMT1 while caspase-3 interacts with NMT2. 16567804_caspase-3 cleavage of EAAT2 is one mechanism responsible for the impairment of glutamate uptake in mutant SOD1-linked amyotrophic lateral sclerosis 16651613_SUMOylation and activation of ataxia-telangiectasia-mutated protein, PKCdelta, caspase-3, and nuclear factor kappaB signaling pathways modulate salivary adaptive responses to stress in cells exposed to either 1% O(2) or DFO. 16689860_caspase-3 cleavage in the spinous layer of the epidermis is a pathologic event contributing to spongiosis formation in acute atopic dermatitis 16716256_caspase-3 and caspase-9 activation has critical roles in hypoxia/reoxygenation induced apoptosis 16781734_Crystal structures were determined of caspase-3 complexes with the substrate analogs at resolutions of 1.7 A to 2.3 A. 16787777_The atomic resolution (1.06 Angstroms) crystal structure of the caspase-3 reveals the structural basis for substrate selectivity in the S4 pocket. 16791842_Thus, early activation of caspase-3 seems to be a non-apoptotic event required for cellular function. 16802364_AECA subsets in the sera of patients with systemic sclerosis (SSc) and diffuse SSc induce patterns of human dermal endothelial cells gene expression in the setting of apoptosis with increased caspase 3 activity and reexpression of fibrillin 1. 16808908_Expression level of intracellular activated caspase-3 in peripheral T cell subsets increases in patients with active systemic lupus erythematosus (SLE), compared to patients with inactive SLE and healthy controls. 16835231_Data show that decorin protein core inhibits tumor xenograft growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activation. 16882668_Caspase-3-dependent activation of calcium-independent phospholipase A2 enhances cell migration in non-apoptotic ovarian cancer cells 16884727_HDL prevents endothelial progenitor cell (EPC)apoptosis through inhibition of caspase-3 activity suggesting a possible mechanism for its positive effects on circulating EPC numbers 16894574_These data imply that confluent cells undergo spontaneous cell death mediated by cathepsin B; Lipopolysaccharides may accelerate this caspase-independent cell death through release of mitochondrial contents and reactive oxygen species. 16931214_Activated caspase-3-positive cells were present in glomeruli of 88.2% of lupus nephritis cases, observed in the glomerular tuft and cellular and fibrocellular crescents 16935258_peroxynitrite-mediated inhibition of human caspase-3 is impaired by CO2 16948902_Fas-FasL signal transduction pathway plays an important role in the induction of T cell apoptosis. 16949642_Active caspase 3-positive Reed-Sternberg cells were detected in 47 of 70 cases. 16955788_Results indicate that caspase-3 mRNA antisense oligodeoxynucleotides prevent HL-60 cells from apoptosis induced by gamma-radiation,and reduce expression of caspase-3 and its mRNA. 16977583_CASP3 was overexpressed in 40.8% of the 210 invasive ductal breast carcinomas studied 16983089_ROCK-1-dependent caspase-3 activation was coupled with the activation of PTEN and the subsequent inhibition of protein kinase B (Akt) activity, all of which was attenuated by siRNA directed against ROCK-1 expression. 17013756_This study demonstrates FHOD1 is cleaved by caspase-3 at the SVPD(616) site during apoptosis and the C-terminal FHOD1 cleavage product has the ability to inhibit RNA polymerase I transcription. 17013758_These data indicate that, in addition to its function as an effector caspase, caspase-3 plays an important role in maximizing the activation of apical caspases and crosstalk between the two major apoptotic pathways. 17023557_Collectively, these data suggest that cathepsin D activation of caspase 3/7 may be required for inducing one of the death pathways elicited by E. coli. 17038954_Immunoreactivities indicating single-stranded DNA and cleaved caspase-3 were higher in moyamoya disease than in controls and were located in smooth muscle cells of media. 17065568_Cell death in the penumbra of subacute infarcts is partially caspase-3 independent and may be attributed to nitric oxide. 17071630_Observational study of gene-disease association. (HuGE Navigator) 17071630_Single nucleotide polymorphismsin caspase 3 is associated with non-Hodgkin lymphoma 17080552_Observations suggest that PB-induced apoptosis occurs through a caspase-dependent pathway and detains the cell cycle in the G2/M phase. 17114647_Taken together, our results indicate that IL-4 inhibits caspase activity during the initial stages of human Th2 cell differentiation by regulating expression of several key players in the Fas-induced pathway. 17118559_Interaction of Ramos cells with immobilized alpha2,6-linked sialic acid suppresses cas[ase-3 activity and inhibits Igm-induced apoptosis. 17121821_p53-dependent staurosporine-induced caspase-3 activation is affected by the C-terminal products of cellular prion protein processing, C1 and C2 17123522_expression of FasL is upregulated in testes of patients with Sertoli cell-only syndrome & maturation arrest, suggesting that it may be associated with apoptotic elimination or altered maturation of Fas-expressing germ cells by activation of caspase-3. 17124493_These data demonstrate that huntingtin inhibits caspase-3 activity. 17130234_PTP-PEST actively contributes to the cellular apoptotic response and reveal the importance of caspases as regulators of PTPs in apoptosis. 17141888_The role of high gammaGT activity in HepG2 cells can be connected with production of reactive oxygen species and with S-thiolation with Cys and Cys-Gly that can influence activity of caspase 3. 17143787_Results discuss caspase-3 immunohistochemical expression as a marker of apoptosis, increased grade and early recurrence in intracranial meningiomas. 17167422_in erythroid precursors undergoing terminal differentiation, Hsp70 prevents active caspase-3 from cleaving GATA-1 and inducing apoptosis 17172814_Human tumor xenograft in nude mice is suppressed significantly by the treatment with a hTERT/re-Caspase-3 system. 17198755_Adult stem cell injections into blastocysts is influenced by apoptosis as shown by high caspase-3 activity. 17217622_Pro-caspase 3 appears to be a substrate of SAG/ROC-(beta-TCRP) E3 ubiquitin ligase, which protects cells from apoptosis by reducing the basal level of pro-caspase-3. 17219053_Data show that caspase-3 antisense oligodeoxynucleotides inhibit apoptosis in gamma-irradiated human leukemia HL-60 cells.( 17235653_The expression of Caspase 3 in peripheral blood mononuclear cells was significantly increased in SLE patients. 17245643_While polyploid, mature megakaryocytes from healthy subjects or myelodysplastic syndromes patients manifested caspase-3 activation during terminal differentiation, freshly isolated, immature MK from MDS died without caspase-3 activation 17272816_In this review, reversible glutathiolation of procaspase-3 by glutaredoxin provides further mechanistic insight into the role of reactive oxygen species in TNF-alpha-induced apoptosis. 17301822_Caspase-3 and the p38alpha MAP kinase were activated during TIMP-1-induced UT-7 cells erythroid differentiation. 17321792_Caspase 3 was found to be activated by elaidic acid and palmitic acid. 17324936_calcineurin B potentiates the activation of procaspase-3 by accelerating its proteolytic maturation 17364171_Comparing SIDS to non-SIDS cases, increased active caspase-3 expression was restricted to four nuclei in the caudal pons and two nuclei in the rostral medulla. 17379327_caspases-3 and caspase-9 play novel roles in transcription by regulating polycomb protein function through direct cleaving of Ring1B. 17412564_Cystatin A suppresses UVB-induced apoptosis of keratinocytes by the inhibition of caspase 3 activation. 17413988_In this test set, cyclooxygenase 2 and Caspase 3 seem to be immunohistochemical markers with prognostic significance for vulva cancer. 17430778_An 8.94-fold and 6.73-fold increase in expression of caspase-3mRNA in intracranial aneurysm and AAA, respectively, were obtained relative to the normal vessels. 17436592_A 3-fold increase in caspase-3 activity was observed in cells treated with TAC-101 colon cancer cells in comparison to the control cells. 17437405_Results demonstrate that cleavage by caspase 3 does not activate caspase 9, but enhances apoptosis by alleviating XIAP inhibition of the apical caspase. 17462862_NF-kappa-B inhibition enhances CASP3 degradation of Akt1 and apoptosis in response to campogthecin. 17495975_EXEL-0862 induced apoptotic death in EOL-1 cells and imatinib-resistant T674I FIP1L1-PDGFR-alpha-expressing cells, and resulted in significant downregulation of the antiapoptotic protein Mcl-1 through a caspase-3 dependent mechanism. 17498346_The activity of Caspase-3 increased significantly in MMT-induced apoptosis in PC-3M cells. 17505517_Survivin-DeltaEx3 plays a key role in the inhibition of caspase-3 activity. 17525639_CASP3 expression was higher in the helicobacter pylori infection (HP+) group, HP+ with intestinal metaplasia, and HP+ with dysplasia groups. 17534194_Observational study of gene-disease association. (HuGE Navigator) 17544405_Akt1 is cleaved in vitro at the caspase-3 consensus site DQDD(456). 17559062_Caspase 3 regulated the biochemical, morphological, and functional changes in cells undergoing staurosporine-induced apoptosis. Full-length Dsg2 was processed to a 70-kDa fragment which was released into the cytosol. 17597071_caspase-3 prodomain binding to heat shock protein 27 regulates monocyte apoptosis by inhibiting caspase-3 proteolytic activation 17606719_cleaved caspase-3 is an activated form of caspase-3 that has a role in preventing progression of gliomas 17606900_A specific transnitrosation reaction between procaspase-3 and thioredoxin-1 (Trx) occurs in cultured human T cells and prevents apoptosis. 17616526_RbAp48-mediated transformation of HPV16 is probably because of the regulation by RbAp48 of tumor suppressors retinoblastoma and p53, apoptosis-related enzymes caspase-3 and caspase-8, E6, E7, cyclin D1 (CCND1), and c-MYC. 17645689_demonstrate the cellular mechanisms of neuronal cell degeneration induced via c-Jun-N-terminal kinases and caspase-dependent signaling 17689858_caspase-3 is a key regulator of apoptosis in response to combined genistein and TRAIL in human gastric adenocarcinoma AGS cells through the activation of DR5 and mitochondrial dysfunction 17698513_Thimerosal caused Ca2+-independent apoptosis of gastric cancer cells via phosphorylation of p38 MAPK and caspase-3 activation. 17721932_S100A6 might be involved in the processing of apoptosis by modulating the transcriptional regulation of caspase-3. 17724022_XIAP homotrimerizes via its C-terminal Ring domain, making its inhibitory activity toward caspase-3 more susceptible to Smac. 17805550_Report caspase-3 activity, response to chemotherapy and clinical outcome in patients with colon cancer. 17880920_These findings suggest that caspase-7 facilitates the execution of apoptosis through down-regulation of the 26S proteasome, which regulates the turnover of proteins involved in the apoptotic process. 17908973_Caspase-3 may have a role in local recurrence in rectal cancer 17936584_Gastrins activate members of Rho family G proteins which in turn regulate different proteins of the Bcl-2 family leading to changes in caspase 3 activity. 17950632_Results describe the role of carbohydrate moiety of bleomycin-A2 in caspase-3 activation and internucleosomal chromatin fragmentation in apoptosis of laryngeal carcinoma cells. 17978580_These data indicate that cytosolic PDI is a substrate of caspase-3 and -7, and that it has an anti-apoptotic action. 17981569_bcl2 and Caspase-3 have roles in Shikonin analogue (SA) 93/637 induced apoptosis of human U937 cells 17983802_Two novel homozygously deleted genes in hepatocellular carcinomas are caspase 3 and CHES1. 17989718_constitutively and overexpressed AML1-ETO protein was cleaved to four fragments of 70, 49, 40 and 25 kDa by activated caspase-3 during apoptosis induction by extrinsic mitochondrial and death receptor signaling pathways. 18000616_release of caspase 3-containing microparticles may contribute to endothelial cell survival 18047842_apoE expression modulates capase-3 activity, but this has no significant impact on sensitivity to apoptosis and only a moderate impact on basal cholesterol efflux. 18054197_Enteropathogenic Escherichia coli induced apoptosis in T84 intestinal epithelial cells via induction of caspases-3, -8, and -9. 18055116_caspase-3 could be involved in recruitment of ischemic brain tissue being a marker of infarct growth. 18058802_Observational study of gene-disease association. (HuGE Navigator) 18058802_Single nucleotide polymorphisms in the Caspase-3 gene is associated with lung cancer 18060501_For the first time our data demonstrate that CSN-mediated deneddylation can be regulated by active Casp3 and that the CSN executes a specific function during the apoptotic process. 18083346_HSP105 appears to chaperone the | ENSMUSG00000031628 | Casp3 | 1074.218807 | 0.9278862 | -0.107980143 | 0.09656060 | 1.234223e+00 | 2.665874e-01 | 6.396362e-01 | No | Yes | 1113.988011 | 230.483801 | 1181.135725 | 244.307813 | |
| ENSG00000164318 | 133584 | EGFLAM | protein_coding | Q63HQ2 | FUNCTION: Involved in both the retinal photoreceptor ribbon synapse formation and physiological functions of visual perception. Necessary for proper bipolar dendritic tip apposition to the photoreceptor ribbon synapse. Promotes matrix assembly and cell adhesiveness (By similarity). {ECO:0000250}. | Alternative splicing;Cell junction;Cell projection;Direct protein sequencing;Disulfide bond;EGF-like domain;Extracellular matrix;Glycoprotein;Reference proteome;Repeat;Secreted;Signal;Synapse | hsa:133584; | basement membrane [GO:0005604]; cell projection [GO:0042995]; interstitial matrix [GO:0005614]; presynaptic active zone [GO:0048786]; synaptic cleft [GO:0043083]; calcium ion binding [GO:0005509]; glycosaminoglycan binding [GO:0005539]; animal organ morphogenesis [GO:0009887]; extracellular matrix organization [GO:0030198]; peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan [GO:0019800]; positive regulation of cell-substrate adhesion [GO:0010811]; tissue development [GO:0009888] | 19727342_Strong candidate gene for macular dystrophy, MCDR3 (human 5p15.33-p13.1). Conclusion is based on a massive expression data set for mouse (103 strains) and joint analysis of RetNet database. 28339009_rs1465567 of EGFLAM and rs113710653 of SPATC1L may be susceptibility loci for true aortic aneurysm and rs143881017 of RNASE13 may be such a locus for dissecting aortic aneurysm in Japanese individuals. 30829611_this is the first report to describe that EGFLAM promoted the proliferation, migration and invasion of GBM cells via the activation of PI3K/AKT pathway, and predicted poor prognosis in GBM patients. | ENSMUSG00000042961 | Egflam | 75.060413 | 0.8918105 | -0.165190913 | 0.30947566 | 2.780147e-01 | 5.980054e-01 | No | Yes | 59.249723 | 10.658218 | 67.022036 | 11.924507 | |||
| ENSG00000164463 | 153222 | CREBRF | protein_coding | Q8IUR6 | FUNCTION: Acts as a negative regulator of the endoplasmic reticulum stress response or unfolded protein response (UPR). Represses the transcriptional activity of CREB3 during the UPR. Recruits CREB3 into nuclear foci. {ECO:0000269|PubMed:18391022}. | Alternative splicing;Nucleus;Reference proteome;Repressor;Stress response;Transcription;Transcription regulation;Transport;Unfolded protein response | hsa:153222; | cytoplasm [GO:0005737]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of endoplasmic reticulum unfolded protein response [GO:1900102]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of intracellular transport [GO:0032388]; positive regulation of protein catabolic process [GO:0045732]; positive regulation of protein transport [GO:0051222]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; response to endoplasmic reticulum stress [GO:0034976]; response to unfolded protein [GO:0006986] | 27278737_Findings provide new insight into the molecular mechanisms underlying hypoxia-induced glioblastoma cell autophagy and indicate that the hypoxia/CREBRF/CREB3/ATG5 pathway plays a central role in malignant glioma progression. 27455349_Missense Mutation in CREBRF gene is associated with obesity. 28405013_The rs373863828-A allele was not found in both AN-speaking and non-AN-speaking Melanesians living in Papua New Guinea. Our results suggest that rs373863828-A of CREBRF, a promising thrifty variant, arose in recent ancestors of AN-speaking Polynesians. 28928463_Studied CREBRF genetic variants in association to height and weight in young children; found that the rs373863828 variant was significantly associated with growth at 4 years of age; also found the variants to be prevalent in other Pacific populations, including Maori. 29721634_The presence of each additional CREBRF rs373863828 A allele is associated with increased BMI yet reduced odds of type 2 diabetes in adults of Maori and Pacific (Polynesian) ancestry living in Aotearoa/New Zealand. 29729692_CREBRF promotes the proliferation of human gastric cancer cells via the AKT signaling pathway. 29877158_The rs373863828-A allele (a missense variant of the CREBRF gene) may not directly affect the level of serum HDL-cholesterol independent of BMI. 29958463_We found that CREBRF and TRIM2 mRNA were significantly upregulated by rHDL, particularly in response to its phospholipid component 1-palmitoyl-2-linoleoyl-phosphatidylcholine, however, protein expression was not significantly altered. 31280340_CREBRF G allele (rs12513649) and A allele (rs373863828)are associated with higher BMI and lower risk of diabetes in Pacific Islander (Guam and Saipan) populations. 31543511_The Maori and Pacific specific CREBRF variant and adult height. 32190945_A missense variant in CREBRF is associated with taller stature in Samoans. 32491157_Population-specific reference panels are crucial for genetic analyses: an example of the CREBRF locus in Native Hawaiians. 32654027_The Pacific-specific CREBRF rs373863828 allele protects against gestational diabetes mellitus in Maori and Pacific women with obesity. 32884101_A missense variant in CREBRF, rs373863828, is associated with fat-free mass, not fat mass in Samoan infants. 35144939_CREBRF missense variant rs373863828 has both direct and indirect effects on type 2 diabetes and fasting glucose in Polynesian peoples living in Samoa and Aotearoa New Zealand. 35218947_The minor allele of the CREBRF rs373863828 p.R457Q coding variant is associated with reduced levels of myostatin in males: Implications for body composition. | ENSMUSG00000048249 | Crebrf | 259.757361 | 1.2913226 | 0.368849513 | 0.16909785 | 4.743973e+00 | 2.940110e-02 | No | Yes | 334.153455 | 84.279679 | 246.952293 | 62.325774 | |||
| ENSG00000164818 | 54919 | DNAAF5 | protein_coding | Q86Y56 | FUNCTION: Cytoplasmic protein involved in the delivery of the dynein machinery to the motile cilium. It is required for the assembly of the axonemal dynein inner and outer arms, two structures attached to the peripheral outer doublet A microtubule of the axoneme, that play a crucial role in cilium motility. {ECO:0000269|PubMed:23040496, ECO:0000269|PubMed:25232951}. | Acetylation;Alternative splicing;Ciliopathy;Cilium biogenesis/degradation;Cytoplasm;Direct protein sequencing;Disease variant;Kartagener syndrome;Primary ciliary dyskinesia;Reference proteome;Repeat | The protein encoded by this gene is essential for the preassembly or stability of axonemal dynein arms, and is found only in organisms with motile cilia and flagella. Mutations in this gene are associated with primary ciliary dyskinesia-18, a disorder characterized by abnormalities of motile cilia. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Feb 2013]. | hsa:54919; | cytoplasm [GO:0005737]; dynein axonemal particle [GO:0120293]; dynein intermediate chain binding [GO:0045505]; cilium movement [GO:0003341]; inner dynein arm assembly [GO:0036159]; outer dynein arm assembly [GO:0036158] | 23040496_Identification of HEATR2 contributes to the growing number of genes associated with PCD identified in both individuals and model organisms and shows that exome sequencing in family studies facilitates the discovery of novel disease-causing gene mutations. | ENSMUSG00000025857 | Dnaaf5 | 2794.907072 | 0.9744830 | -0.037291100 | 0.09232439 | 1.653599e-01 | 6.842690e-01 | 8.894290e-01 | No | Yes | 2348.445194 | 179.963516 | 2404.313939 | 184.183410 | |
| ENSG00000164828 | 23353 | SUN1 | protein_coding | O94901 | FUNCTION: As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex involved in the connection between the nuclear lamina and the cytoskeleton (PubMed:18039933, PubMed:18396275). The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning (By similarity). Required for interkinetic nuclear migration (INM) and essential for nucleokinesis and centrosome-nucleus coupling during radial neuronal migration in the cerebral cortex and during glial migration (By similarity). Involved in telomere attachment to nuclear envelope in the prophase of meiosis implicating a SUN1/2:KASH5 LINC complex in which SUN1 and SUN2 seem to act at least partial redundantly (By similarity). Required for gametogenesis and involved in selective gene expression of coding and non-coding RNAs needed for gametogenesis (By similarity). Helps to define the distribution of nuclear pore complexes (NPCs) (By similarity). Required for efficient localization of SYNE4 in the nuclear envelope (By similarity). May be involved in nuclear remodeling during sperm head formation in spermatogenenis (By similarity). May play a role in DNA repair by suppressing non-homologous end joining repair to facilitate the repair of DNA cross-links (PubMed:24375709). {ECO:0000250|UniProtKB:Q9D666, ECO:0000269|PubMed:18039933, ECO:0000269|PubMed:18396275, ECO:0000269|PubMed:24375709}. | 3D-structure;Alternative splicing;Coiled coil;Differentiation;Disulfide bond;Isopeptide bond;Meiosis;Membrane;Nucleus;Phosphoprotein;Reference proteome;Signal-anchor;Spermatogenesis;Transmembrane;Transmembrane helix;Ubl conjugation | This gene is a member of the unc-84 homolog family and encodes a nuclear envelope protein with an Unc84 (SUN) domain. The protein is involved in nuclear anchorage and migration. Alternatively spliced transcript variants have been described. [provided by RefSeq, Jan 2019]. | hsa:23353; | cytoplasm [GO:0005737]; integral component of nuclear inner membrane [GO:0005639]; intracellular membrane-bounded organelle [GO:0043231]; meiotic nuclear membrane microtubule tethering complex [GO:0034993]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; cytoskeleton-nuclear membrane anchor activity [GO:0140444]; lamin binding [GO:0005521]; protein-membrane adaptor activity [GO:0043495]; centrosome localization [GO:0051642]; homologous chromosome pairing at meiosis [GO:0007129]; meiotic attachment of telomere to nuclear envelope [GO:0070197]; nuclear envelope organization [GO:0006998]; nuclear matrix anchoring at nuclear membrane [GO:0090292]; nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration [GO:0021817]; spermatogenesis [GO:0007283] | 10375507_Describes cloning and function of C. elegans unc-84 and cloning of human orthologs. 11593002_KIAA0810 is a novel 100-kDa transmembrane protein with similarity to Caenorhabditis elegans Unc-84A and resides in the inner nuclear membrane. It is likely to interact with the nuclear lamina. 12036294_Isolation of a cDNA encoding a mouse homolog of the human SUN1 (UNC84A) gene. 16079285_The Sun1 itself does not require functional A-type lamins for its localisation at the inner nuclear membrane in mammalian cells. 16445915_localization and anchoring of UNC84A is not dependent on the lamin proteins, in contrast to what had been observed for C. elegans UNC-84 17132086_SUN1 and SUN2 may form a physical interaction between the nuclear envelope and the centrosome 17631499_chromosome de-condensation needs the function of an inner nuclear membrane (INM) protein hsSUN1 and a membrane-associated histone acetyltransferase (HAT), hALP. 17724119_Results suggest that Sun1 represents an important determinant of nuclear pore complex distribution across the nuclear surface. 19240061_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 19933576_perturbations in lamin A-SUN1 and SUN2 protein interactions may underlie the opposing effects of EDMD and HGPS mutations on nuclear and cellular mechanics 20108321_Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21311568_These results demonstrate that the interplay between SUN1 and farnesylated prelamin A contributes to nuclear positioning in human myofibers and may be implicated in pathogenetic mechanisms. 21727197_POM121 and Sun1 interact transiently to promote early steps of interphase nuclear pore complexes assembly 22541428_Reduction of SUN1 overaccumulation in LMNA mutant fibroblasts and in cells derived from HGPS patients corrected nuclear defects and cellular senescence. 24375709_Results highlight the interactions at the nuclear envelope where mutations in the EMD and TMPO gene in combination with mutations in SUN1 have an impact on several components of the network. 24522183_Farnesylation of progerin enhances its interaction with SUN1 and reduces SUN1 mobility. 24662567_Codepletion of SUN1/2 slows cell proliferation and results in an accumulation of morphologically defective and disoriented mitotic spindles. 25057012_The Caenorhabditis elegans lamin, LMN-1, is required for nuclear migration and interacts with the nucleoplasmic domain of the SUN protein UNC-84. 25210889_An important role for SUN1 and SUN2 in muscle disease pathogenesis. 25482198_these data support a model whereby mitotic phosphorylation of SUN1 disrupts interactions with nucleoplasmic binding partners, promoting disassembly of the nuclear lamina and, potentially, its chromatin interactions 26356418_Nuclear envelope associated endosome-mediated transfer depends on the nuclear envelope proteins SUN1 and SUN2, as well as the Sec61 translocon complex. 26476453_SUN1 plays a role in hnRNP-involved mRNA export. 26962703_wndchrm revealed a consistent negative correlation between SUN1 expression and the size of nucleoli in human breast cancer tissues. 28747499_SUN1/SUN2 may function redundantly in early HIV-1 infection steps and therefore influence HIV-1 replication and pathogenesis. 28831067_Results provide evidence that SUN1 is involved in mRNA export and this function is regulated by phosphorylation of serine 113 in the N-terminal domain. 29408528_Thus, AID is a conserved functional domain in SUN proteins and this work provides the structural evidence to support the conversation of the AID-mediated autoinhibition of SUN proteins. 29539404_Mechanisms of SUN1 Oligomerization in the Nuclear Envelope 29643244_Low SUN1 expression is associated with HIV infections. 29813079_While Sun1 and Sun2 in HeLa cells are each able to bind KASH-domains, Sun1 is more efficiently incorporated into LINC complexes under normal growth conditions. Furthermore, the balance of Sun1 and Sun2 incorporated into LINC complexes is cell type-specific and is correlated with SRF/Mkl1-dependent gene expression. 30808750_Progerin overexpression increased levels of SUN1, which couples the nucleus to microtubules through nesprin-2G and dynein, and microtubule association with the nucleus. Reducing microtubule-nuclear connections through SUN1 depletion or dynein inhibition rescued the polarity defects. 31935926_SUN1/2 Are Essential for RhoA/ROCK-Regulated Actomyosin Activity in Isolated Vascular Smooth Muscle Cells. 32931086_The SUN1 splicing variants SUN1_888 and SUN1_916 differentially regulate nucleolar structure. 33686165_The SUN2-nesprin-2 LINC complex and KIF20A function in the Golgi dispersal. 34039995_The SUN1-SPDYA interaction plays an essential role in meiosis prophase I. 34580332_Nuclear restriction of HIV-1 infection by SUN1. | ENSMUSG00000036817 | Sun1 | 4210.869816 | 1.0220985 | 0.031534207 | 0.06489306 | 2.331489e-01 | 6.291988e-01 | 8.630015e-01 | No | Yes | 3597.846970 | 450.219082 | 3615.282728 | 452.276445 | |
| ENSG00000164877 | 79778 | MICALL2 | protein_coding | Q8IY33 | FUNCTION: Effector of small Rab GTPases which is involved in junctional complexes assembly through the regulation of cell adhesion molecules transport to the plasma membrane and actin cytoskeleton reorganization. Regulates the endocytic recycling of occludins, claudins and E-cadherin to the plasma membrane and may thereby regulate the establishment of tight junctions and adherens junctions. In parallel, may regulate actin cytoskeleton reorganization directly through interaction with F-actin or indirectly through actinins and filamins. Most probably involved in the processes of epithelial cell differentiation, cell spreading and neurite outgrowth (By similarity). {ECO:0000250}. | Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Endosome;LIM domain;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Tight junction;Zinc | hsa:79778; | bicellular tight junction [GO:0005923]; cell-cell junction [GO:0005911]; cytosol [GO:0005829]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; recycling endosome [GO:0055037]; stress fiber [GO:0001725]; actin filament binding [GO:0051015]; actinin binding [GO:0042805]; filamin binding [GO:0031005]; metal ion binding [GO:0046872]; small GTPase binding [GO:0031267]; actin cytoskeleton reorganization [GO:0031532]; actin filament polymerization [GO:0030041]; bicellular tight junction assembly [GO:0070830]; endocytic recycling [GO:0032456]; neuron projection development [GO:0031175]; positive regulation of protein targeting to mitochondrion [GO:1903955]; substrate adhesion-dependent cell spreading [GO:0034446] | 17891173_Involved in epithelial cell scattering. 25864591_Data indicate that MICAL-L2 may be an important regulator of epithelial-mesenchymal transition (EMT) in ovarian cancer cells. 28416812_We identified one new significant locus at 7p22.3 for the Stroop word interference time (rs11514810, P=3.42E-09 for discovery, P=0.01176 for replication and combined P=5.249E-09). Regulatory feature analysis and expression quantitative trait loci (eQTL) data showed that this locus contributes to MICALL2 expression in the human brain. 31034158_Up-regulation of MICAL-L2 is associated with gastric cancer cell migration. 33976349_JRAB/MICAL-L2 undergoes liquid-liquid phase separation to form tubular recycling endosomes. | ENSMUSG00000036718 | Micall2 | 168.916732 | 1.2108167 | 0.275980506 | 0.23901401 | 1.368794e+00 | 2.420191e-01 | No | Yes | 152.947564 | 28.980210 | 127.255955 | 24.172036 | |||
| ENSG00000164933 | 81034 | SLC25A32 | protein_coding | Q9H2D1 | FUNCTION: Transports folate across the inner membranes of mitochondria (PubMed:15140890, PubMed:29666258). Can also transport FAD across the mitochondrial inner membrane (PubMed:16165386). {ECO:0000269|PubMed:15140890, ECO:0000269|PubMed:16165386, ECO:0000269|PubMed:29666258}. | Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | hsa:81034; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; FAD transmembrane transporter activity [GO:0015230]; folic acid transmembrane transporter activity [GO:0008517]; folate import into mitochondrion [GO:1904947]; folic acid metabolic process [GO:0046655]; folic acid transport [GO:0015884]; mitochondrial FAD transmembrane transport [GO:1990548] | Mouse_homologues 29666258_These data demonstrate that the loss of functional Slc25a32 results in cranial neural tube defects (NTDs) in mice and has also been observed in a human NTD patient. | ENSMUSG00000022299 | Slc25a32 | 1574.917212 | 0.9727554 | -0.039851057 | 0.07524674 | 2.809571e-01 | 5.960746e-01 | 8.504829e-01 | No | Yes | 1677.263778 | 302.325165 | 1687.853332 | 304.178107 | ||
| ENSG00000165521 | 161436 | EML5 | protein_coding | Q05BV3 | FUNCTION: May modify the assembly dynamics of microtubules, such that microtubules are slightly longer, but more dynamic. {ECO:0000250}. | Alternative splicing;Cytoplasm;Cytoskeleton;Microtubule;Reference proteome;Repeat;WD repeat | hsa:161436; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; microtubule [GO:0005874]; catalytic activity [GO:0003824]; microtubule binding [GO:0008017] | ENSMUSG00000051166 | Eml5 | 422.819121 | 0.9632967 | -0.053947940 | 0.14721731 | 1.354955e-01 | 7.128008e-01 | No | Yes | 427.140598 | 91.744118 | 438.977604 | 94.346357 | ||||
| ENSG00000165731 | 5979 | RET | protein_coding | P07949 | FUNCTION: Receptor tyrosine-protein kinase involved in numerous cellular mechanisms including cell proliferation, neuronal navigation, cell migration, and cell differentiation upon binding with glial cell derived neurotrophic factor family ligands. Phosphorylates PTK2/FAK1. Regulates both cell death/survival balance and positional information. Required for the molecular mechanisms orchestration during intestine organogenesis; involved in the development of enteric nervous system and renal organogenesis during embryonic life, and promotes the formation of Peyer's patch-like structures, a major component of the gut-associated lymphoid tissue. Modulates cell adhesion via its cleavage by caspase in sympathetic neurons and mediates cell migration in an integrin (e.g. ITGB1 and ITGB3)-dependent manner. Involved in the development of the neural crest. Active in the absence of ligand, triggering apoptosis through a mechanism that requires receptor intracellular caspase cleavage. Acts as a dependence receptor; in the presence of the ligand GDNF in somatotrophs (within pituitary), promotes survival and down regulates growth hormone (GH) production, but triggers apoptosis in absence of GDNF. Regulates nociceptor survival and size. Triggers the differentiation of rapidly adapting (RA) mechanoreceptors. Mediator of several diseases such as neuroendocrine cancers; these diseases are characterized by aberrant integrins-regulated cell migration. Mediates, through interaction with GDF15-receptor GFRAL, GDF15-induced cell-signaling in the brainstem which induces inhibition of food-intake. Activates MAPK- and AKT-signaling pathways (PubMed:28846097, PubMed:28953886, PubMed:28846099). Isoform 1 in complex with GFRAL induces higher activation of MAPK-signaling pathway than isoform 2 in complex with GFRAL (PubMed:28846099). {ECO:0000269|PubMed:20064382, ECO:0000269|PubMed:20616503, ECO:0000269|PubMed:20702524, ECO:0000269|PubMed:21357690, ECO:0000269|PubMed:21454698, ECO:0000269|PubMed:28846097, ECO:0000269|PubMed:28846099, ECO:0000269|PubMed:28953886}. | 3D-structure;ATP-binding;Alternative splicing;Cell adhesion;Cell membrane;Chromosomal rearrangement;Disease variant;Disulfide bond;Endosome;Glycoprotein;Hirschsprung disease;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Reference proteome;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | This gene encodes a transmembrane receptor and member of the tyrosine protein kinase family of proteins. Binding of ligands such as GDNF (glial cell-line derived neurotrophic factor) and other related proteins to the encoded receptor stimulates receptor dimerization and activation of downstream signaling pathways that play a role in cell differentiation, growth, migration and survival. The encoded receptor is important in development of the nervous system, and the development of organs and tissues derived from the neural crest. This proto-oncogene can undergo oncogenic activation through both cytogenetic rearrangement and activating point mutations. Mutations in this gene are associated with Hirschsprung disease and central hypoventilation syndrome and have been identified in patients with renal agenesis. [provided by RefSeq, Sep 2017]. | hsa:5979; | axon [GO:0030424]; dendrite [GO:0030425]; early endosome [GO:0005769]; endosome membrane [GO:0010008]; integral component of plasma membrane [GO:0005887]; membrane raft [GO:0045121]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; plasma membrane protein complex [GO:0098797]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; signaling receptor activity [GO:0038023]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; axon guidance [GO:0007411]; cellular response to retinoic acid [GO:0071300]; embryonic epithelial tube formation [GO:0001838]; enteric nervous system development [GO:0048484]; glial cell-derived neurotrophic factor receptor signaling pathway [GO:0035860]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; innervation [GO:0060384]; lymphocyte migration into lymphoid organs [GO:0097021]; MAPK cascade [GO:0000165]; membrane protein proteolysis [GO:0033619]; neural crest cell migration [GO:0001755]; neuron cell-cell adhesion [GO:0007158]; neuron maturation [GO:0042551]; Peyer's patch morphogenesis [GO:0061146]; positive regulation of cell adhesion mediated by integrin [GO:0033630]; positive regulation of cell migration [GO:0030335]; positive regulation of cell size [GO:0045793]; positive regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001241]; positive regulation of gene expression [GO:0010628]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of metanephric glomerulus development [GO:0072300]; positive regulation of neuron maturation [GO:0014042]; positive regulation of neuron projection development [GO:0010976]; positive regulation of peptidyl-serine phosphorylation of STAT protein [GO:0033141]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of transcription, DNA-templated [GO:0045893]; posterior midgut development [GO:0007497]; protein phosphorylation [GO:0006468]; regulation of axonogenesis [GO:0050770]; regulation of cell adhesion [GO:0030155]; response to pain [GO:0048265]; response to xenobiotic stimulus [GO:0009410]; retina development in camera-type eye [GO:0060041]; signal transduction [GO:0007165]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; ureter maturation [GO:0035799]; ureteric bud development [GO:0001657] | 10850414_The presence of coiled-coil domains in the ktn1/ret fusion protein (PTC8) suggests ligand-independent dimerization and thus constitutive activation of the ret tyrosine kinase domain. 11238493_Observational study of gene-disease association. (HuGE Navigator) 11589684_Observational study of gene-disease association. (HuGE Navigator) 11692159_Three new somatic cell missense mutations of the RET proto-oncogene associated with sporadic medullary thyroid carcinoma (MTC). 11746981_A germline RET mutation at codon 603 in exon 10 is associated with both medullary and nonmedullary thyroid cancer in a kindred. 11803116_Differentiation of cardiac ganglionic cells is affected, after RETINOIC ACID treatment, by the down-regulation of c-Ret. 11839664_RET oligonucleotide microarray for the detection of RET mutations in multiple endocrine neoplasia type 2 syndromes 11883863_The finding of a somatic deletion in RET exon 15 clarified the sporadic nature of a medullary thyroid carcinoma suspected to be familial. A 12 bp deletion within the catalytic domain of the protooncogene RET. 11886862_role in regulating rac activity and lamellipodia formation 11900218_early detection of RET proto-oncogene mutation is crucial for prevention of thyroidectomy in multiple endocrine neoplasia type 2 children 11927965_if the association between Hashimoto's thyroiditis and thyroid cancer exists, its molecular basis is different from RET/PTC rearrangement 11932300_Familial medullary thyroid carcinoma: clinical variability and low aggressiveness associated with RET mutation at codon 804. 11935126_family in which the MEN 2A and the HSCR phenotypes are associated with a single point mutation in exon 10 of the RET proto-oncogene. polymorphic sequence variants of the RET proto-oncogene. 11949835_germline mutation of the RET proto-oncogene in members of Slovak families with multiple endocrine neoplasia 2 11950855_Observational study of gene-disease association. (HuGE Navigator) 11953745_Segregation at three loci explains familial and population risk in Hirschsprung disease. We show oligogenic inheritance of S-HSCR, the 3p21 and 19q12 loci as RET-dependent modifiers, and a parent-of-origin effect at RET. 11953748_Dissecting Hirschsprung disease. RET is the main gene conferring susceptibility. 11955539_Observational study of gene-disease association. (HuGE Navigator) 11955539_These observations lend support to the idea that both RET alleles have a role in pathogenesis of Hirschsprung's disease, in a dose-dependent fashion. We also showed that the c135G/A polymorphism modifies the phenotype. 11973622_Hirschsprung associated GDNF mutations do not prevent RET activation 11979448_the G691S and S904S variants of RET may somehow play a role on the age of onset of MEN 2A 12000816_Observational study of genotype prevalence. (HuGE Navigator) 12056817_Activation of RET tyrosine kinase regulates interleukin-8 production by multiple signaling pathways 12057919_RET activation closely parallels the morphological changes, that it is restricted to those areas of the tumor with the cytological alterations and that it is detectable in both mono- and polyclonal tumors 12085189_RET codon 691 polymorphism is associated with radiation induced tumors with a C-cell hyperplasia of thyroid tumors 12114746_RET/PTC rearrangement in thyroid tumors. Review 12161537_RET expression in papillary thyroid cancer from patients irradiated in childhood for benign conditions. 12182057_Analysis of mutation of protooncogene RET are presented in patients with thyroid medullary carcinoma 12182058_analysis of RET somatic mutations supports the differentiation between sporadic and inherited medullary thyroid carcinoma 12187076_5'-End RET splicing: absence of variants in normal tissues and intron retention in pheochromocytomas. 12193298_Possible pathogenesis of papillary thyroid carcinoma caused by exon 13 and 14 RET mutations that affect the intracellular domain of ret proto-oncogene protein. 12214285_a protective role of this low-penetrant haplotype in the pathogenesis of HSCR and demonstrate a possible functional effect linked to RET messenger RNA expression. 12355085_genetic interaction between mutations in RET and EDNRB is an underlying mechanism for Hirschsprung disease 12439935_relationship between RET oncogene and Chinese patients with Hirschsprung's disease 12466368_A novel Val648Ile substitution in RET protooncogene observed in a Cys634Arg multiple endocrine neoplasia type 2A kindred presenting with an adrenocorticotropin-producing pheochromocytoma. 12474140_A founding locus within the RET proto-oncogene may account for a large proportion of apparently sporadic Hirschsprung disease and a subset of cases of sporadic medullary thyroid carcinoma 12490841_Patients with RET codon 790/791 mutations seemed to have a less aggressive clinical course compared with patients with classic multiple endocrine neoplasia type 2A syndrome. 12519890_Amplification and overexpression of mutant RET in multiple endocrine neoplasia type 2-associated medullary thyroid carcinoma. RET germline mutation in codon 634. Tandem duplication.Genomic chromosome 10 abnormalities increase mutant RET mRNA. 12608895_RET proto-oncogene is often stimulated in follicular cell-derived thyroid tumors, not only in papillary carcinoma but also in follicular tumors (follicular adenomas and follicular carcinoma), and may contribute to tumorigenesis of these tumors. 12632375_Not only RET mutations but also RET polymorphic variants may contribute to the occurrence of total intestinal aganglionosis. 12637586_RET/PTC associates with STAT3 and activates it by the specific phosphorylation of the tyrosine 705 residue. STAT3 activation by the RET/PTC tyrosine kinase is one of the critical signaling pathways for the regulation of specific genes. 12670889_High prevalence of BRAF mutations in thyroid cancer is genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. 12720173_in RET mutation carriers in Hirschsprung's disease, the gut caliber change was almost identical to the histologic transition in cases of short segment aganglionosis, whereas these were markedly dissociated in cases exhibiting extensive aganglionosis 12720532_RET rearrangements may not play any distinctive role in driving histotype development and cancer progression in papillary thyroid carcinomas. 12767512_A deletion of the chromosomal region including the RET proto-oncogene is involved in the pathogenesis of SCLC 12788868_Specific nucleotide and amino acid exchanges at codon 634 might have a direct impact on tumor aggressiveness in MEN 2A. 12841548_Letter discussing RET mutations in distinguishing between sporadic and familial medullary thyroid carcinoma. 12872262_Four novel intronic mutations that have a strong association with the HSCR phenotype were identified in Hirschsprung disease patients 12872262_Observational study of gene-disease association. (HuGE Navigator) 12881714_The BRAF(V599E) mutation appears to be an alternative event to RET/PTC rearrangement rather than to RAS mutations, which are rare in PTC. BRAF(V599E) may represent an alternative pathway to oncogenic MAPK activation in PTCs without RET/PTC activation. 12884527_mutations of the RET protooncogene were analyzed in Russian patients with inherited or sporadic medullary thyroid carcinoma.The most common mutation affected codon 918 to cause substitution of methionine with threonine and accounted for 31.6% alleles. 12939698_findings support the notion that both RET alleles are involved in the pathogenesis of a subgroup of Hirschsprung disease patients in a dose-dependent fashion 12959980_Shp2 activity required for RetM918T-induced Akt activation. Shp2 downstream mediator of mutated receptors RetC634Y and RetM918T. Shp2 acts as limiting factor in Ret-associated endocrine tumors, in neoplastic syndromes multiple endocrine neoplasia. 14508694_Observational study of gene-disease association. (HuGE Navigator) 14555929_Observational study of gene-disease association. (HuGE Navigator) 14557473_there is a low-penetrance pheochromocytoma susceptibility locus in a region upstream of the putative loci for Hirschprung disease and apparently sporadic thyroid carcinoma. 14586073_Point mutation in exon 14 at codon 804 of the RET proto-oncogene locus in a case of lymph node metastases of medullary and papillary thyroid carcinoma. 14600022_Loss-of-function germline mutations of the RET proto-oncogene are reported in familial and sporadic cases of Hirschsprung disease (HSCR) with a variable frequency 14602786_new missense point mutation in exon 8 of the RET gene (1597G-->T) corresponding to a Gly(533)Cys substitution in the cysteine-rich domain of RET protein 14711813_Mass spectrometric analysis revealed that RET Tyr(806), Tyr(809), Tyr(900), Tyr(905), Tyr(981), Tyr(1062), Tyr(1090), and Tyr(1096) were autophosphorylation sites. 14739491_association of high-level Ret proto-oncogene protein expression with neuronal morphology suggests that the variable overexpression of Ret in pheochromocytomas might in part be an epiphenomenon, reflecting the known phenotypic plasticity of these tumors 14761598_Ret expression is significantly higher in thyroid papillary carcinoma than benign thyroid tissue; and this characteristic can have important diagnostic value. 14766744_Ret tyrosine 981 constitutes the major binding site of the Src homology 2 domain of Src and therefore the primary residue responsible for Src activation upon Ret engagement 14981541_Expression of the mitogenic and invasive phenotype of RET/PTC-transformed thyroid cells is stimulated by osteopontin. 15044950_Data report the crystal structure of GFRalpha1 domain 3, and the effects of specific mutations on GDNF binding and RET phosphorylation. 15129804_Observational study of genetic testing. (HuGE Navigator) 15138456_Observational study of gene-disease association. (HuGE Navigator) 15142370_association of RET IVS1-126G>T variant with sporadic medullary thyroid cancer in a cohort of 104 patients 15225646_Persephin/GFRalpha4 is unable to recruit RET protein into lipid rafts. 15231654_RET expression leads to increased HSF1 activation, which correlates with increased expression of stress response genes. RET may be directly responsible for expression of stress response proteins and the initiation of stress response. 15240649_Observational study of gene-disease association. (HuGE Navigator) 15240649_With this study we excluded influence of the G691S polymorphism on RET mRNA expression, development of somatic RET mutation, the linkage with germline RET mutation, younger onset of medullary thyroid carcinoma, and clinical outcome of the disease. 15240857_Observational study of gene-disease association. (HuGE Navigator) 15271413_Observational study of gene-disease association. (HuGE Navigator) 15273715_mutated in papillary thyroid cancer. 15277225_RET point mutants for follicular thyroid cells may account for the occurrence of papillary thyroid carcinoma in patients affected by familial medullary thyroid carcinoma 15286081_Dok-6 binds to the phosphorylated Ret Tyr(1062) residue resulting in phosphorylation of tyrosine residue(s) located within the unique C terminus of Dok-6 predominantly through a Src-dependent mechanism 15297606_RET/PTC expression phosphorylates the Y701 residue of STAT1, a type II interferon (IFN)-responsive protein. 15316058_Selective disruption of oncogenic RET signaling in medullary thyroid carcinoma in vitro and in vivo is associated with loss of the neoplastic phenotype of medullary thyroid carcinoma. 15320968_the RET proto-oncogene mutation Y791F, characterized by a low penetrance, occurs comparatively frequently among patients with normal serum calcitonin concentrations 15331579_mechanisms leading to RET oncogenic conversion 15350625_Observational study of gene-disease association. (HuGE Navigator) 15351743_RET requires coupling of Gab1 to phosphatidylinositol 3-kinase for function in human tumor cells 15355438_Germ-Line Mutation in RET proto-oncogene is associated with Multiple Endocrine Neoplasia 15469971_All Ret dominant-negative/+ mice died by 1 month of age and had distal intestinal aganglionosis reminiscent of Hirschsprung disease (HSCR) in humans 15491993_critical role of the immunoglobulin domain in regulation of the localization of human PTPmu in bovine cell lines 15502856_Papillary carcinomas with high RET/PTC1 expression showed an association trend for large tumor size. 15523405_The Cys630 RET genotype may have a more vigorous transforming activity than currently thought and can cause medullary thyroid carcinoma in RET gene carriers within the first year of life. 15548547_Observational study of gene-disease association. (HuGE Navigator) 15583857_RET has roles in neoplastic transformation [review] 15592804_Observational study of gene-disease association. (HuGE Navigator) 15632018_Expression of a human Ret proto-oncogene with the MEN 2B mutation does not cause any features of MEN 2B in mice. 15633231_Mutations of RET proto-oncogene may play an important role in the pathogenesis of Chinese patients with Hirschsprung disease. 15643606_A RET haplotype (A-C-A) composed of alleles at three SNPs is associated with reduced RET gene expression in Hirschsprung patients. 15657578_RET signals through focal adhesion kinase in medullary thyroid cancer cells. 15677445_These findings establish a mechanism for the differential down-regulation of RET9 and RET51 signaling that could explain the apparently paradoxical activities of these two RET isoforms. 15716612_RET/PTC and CK19 have roles in progression of papillary thyroid carcinoma 15741265_RET gene mutation may explain the wide clinical variability associated with germline mutations at codon 804 in medullary thyroid carcinoma/multiple endocrine neoplasia type 2A patients. 15753666_Single nucleotide polymorphisms in the RET oncogene may play a role in sporadic papillary thyroid carcinoma. 15759212_Of these 86 variations, 8 proved to be in regions highly conserved among different vertebrates and within putative transcription factor binding sites. We therefore considered these as candidate disease-associated variants. 15785245_Koreans showed increased RET gene expression in papillary thyroid carcinoma. 15829955_a common non-coding RET variant within a conserved enhancer-like sequence in intron 1 is significantly associated with Hirschsprung disease susceptibility 15834508_Single nucleotide polymorphisms of RET is associated with Hirschsprung disease 15841388_Observational study of gene-disease association. (HuGE Navigator) 15844786_medullary thyroid carcinoma manifested in new RET mutation and RET polymorphism. 15933516_Observational study of gene-disease association. (HuGE Navigator) 15940252_in the presence of RET oncoproteins, both RAI and GAB 1 are tyrosine-phosphorylated, and the stoichiometry of this interaction remarkably increases 15953945_Observational study of gene-disease association. (HuGE Navigator) 15956201_Substantial discrimination between predicted functional classes of RET mutations and disease severity even for a multigenic disease such as Hirschsprung disease. 15988377_Mutations in medullary thyroid cancer and in multiple endocrine neoplasia 2. 15994200_RET/PTC is able to phosphorylate the Y315 residue of PKB, an event that results in maximal activation of PKB for RET/PTC-induced thyroid tumorigenesis. 16007166_determination of mutation specific gene expression profiles in papillary thyroid carcinoma 16053382_C630R mirrors C634R in penetrance and in early age of onset of medullary thyroid carcinoma 16118333_Observational study of gene-disease association. (HuGE Navigator) 16127999_PHOX2A, but not PHOX2B, seems to act directly on the c-RET promoter 16144862_The histone acetylation level was evaluated by the chromatin immunoprecipitation method applied to cells displaying different degrees of endogenous RET expression. 16153436_direct interaction between RET and a broad range of effector molecules that may contribute to tumor pathogenesis 16181547_Observational study of gene-disease association. (HuGE Navigator) 16203990_The RET/PTC1 oncogene activates a proinflammatory program, provide a direct link between a transforming human oncogene, inflammation, and malignant behavior. 16227613_Y1062 is a critical regulator of Ret9 signaling and suggest that Ret51-specific motifs are likely to inhibit the activity of this isoform 16230779_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16230779_significant association of the S691 allele with medullary thyroid carcinoma 16269442_RET enhancer modulates expression in the enteric nervous system consistent with its proposed role in Hirschsprung disease 16314641_Germline mutation on the RET gene was present in patients with pheochromocytoma or functional paraganglioma. 16357163_Single nucleotide polymorphism in ret is associated with the aggressive growth of pancreatic cancers 16384843_The newly identified RET/N777S germline mutation is a low-penetrant cause of medullary thyroid carcinoma. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16388093_possibility that lower-penetrance RET mutations may contribute to the list of causes of familial pheochromocytomas 16419493_new missense point mutation in exon 5 in familial medullary thyroid carcinoma 16441254_Observational study of gene-disease association. (HuGE Navigator) 16441254_Two Hirschsprung disease-associated haplotypes derive from a single founding locus, extending up to intron 19 and successively rearranged in correspondence with a high recombination rate region located between the proximal and distal end of the RET gene. 16448984_gene expression impairment seems to be at the basis of the association of HSCR disease with several RET polymorphisms, allowing us to define a predisposing haplotype spanning from the promoter to exon 2. 16452504_These results reveal novel roles of key RET-dependent signaling pathways in embryonic kidney development and provide murine models and new insights into the molecular basis for CAKUT. 16469774_Characterization of the R833C substitution suggests that this tyrosine kinase mutation confers a weak activating potential upon RET but introduces an intracellular cysteine which activate RET. 16483615_frequency of RET rearrangements in papillary thyroid carcinoma for the Polish population 16484222_Erk8 has a role as a novel effector of RET/PTC3 and, therefore, RET biological functions 16525712_Mutational screening of RET revealed 9 different mutations, present in 26 of the 114 MEN 2 Spanish patients. 16534860_Observational study of gene-disease association. (HuGE Navigator) 16551639_Gas1 is related to the GDNF alpha receptors and regulates Ret signaling 16555159_Observational study of gene-disease association. (HuGE Navigator) 16556802_evaluation of noncoding sequences at the zebrafish ret locus conserved among teleosts, and at the human RET locus, conserved among mammals 16569669_Mutagenesis analysis revealed that Tyr981 within the intracellular domain of RET was crucial for the interaction with SH2-Bbeta. Morphological evidence showed that SH2-Bbeta and RET colocalized in mesencephalic neurons. 16596053_Medullary thyroid carcinoma as part of multiple endocrine neoplasia type 2 in a family with a mutation in RET proto oncogene. 16628270_Observational study of genetic testing. (HuGE Navigator) 16646689_Observational study of gene-disease association. (HuGE Navigator) 16732321_strong propensity to self-association in the RET-transmembrane underlies - and may be required for - dimer formation and oncogenic activation of juxtamembrane cysteine mutants of RET 16767674_Observational study of genotype prevalence. (HuGE Navigator) 16767674_Some patients with apparently sporadic pheochromacytoma were carrier of mutations in RET proto-oncogene. 16773224_In the human digestive and reproductive systems, a subset of epithelial cells exhibited GFRalpha3- and RET-like staining, suggesting co-localization. 16813162_Observational study of gene-disease association. (HuGE Navigator) 16816022_Analysis of the RET gene revealed neither linkage nor mutations in Hirschsprung's disease mapping. 16818057_Observational study of gene-disease association. (HuGE Navigator) 16847065_Cell type-specific functions involve a competitive recruitment of different phosphotyrosine binding adaptor molecules by RET that activate selective signaling pathways. 16849523_We show that RA-induced differentiation is mediated by a positive autocrine loop that sustains Ret downstream signaling and depends on glial cell-derived neurotrophic factor expression and release. 16868135_Timing and extent of prophylactic thyroidectomy can be modified by individual RET mutation 16877807_Observational study of gene-disease association. (HuGE Navigator) 16945332_the RET finger protein has a role in estrogen receptor-mediated transcription in tumor cells 16946010_Ret mutations in thyroid tumorigenesis. 16954442_First molecular studies on a complex germline RET mutation lying in the juxtamembrane region of the receptor are reported in medullary thyroid carcinoma 16979782_RET dysfunction has a crucial role and discusses RET as a potential therapeutic target. 16986122_A variant located in the 3' untranslated region of the RET gene, which slows down mRNA decay in patients with Hirschsprung disease. 17009072_Observational study of genotype prevalence. (HuGE Navigator) 17049487_These results suggest that RFP is a mediator connecting several MBD proteins and allowing the formation of a more potent transcriptional repressor complex. 17065770_Identification of a heterozygous germ line missense mutation at codon 634 of exon 11 in the RET gene that causes a cysteine to arginine amino acid substitution in a MEN2A patient. 17102080_Observational study of genotype prevalence. (HuGE Navigator) 17108762_Observational study of gene-disease association. (HuGE Navigator) 17138574_The molecular basis for HPT has been further elucidated by teh detection of inactivating germline mutations in the CaSR gene in familial hypocalciuric hypercalcemia syndrome and in the RET genes in the familial forms of HPT. 17185892_Observational study of gene-disease association. (HuGE Navigator) 17185892_results indicate a possible association between the presence of lymph node involvement at the time of diagnosis (extent of disease) of medullary thyroid carcinoma and L769L or S836S polymorphism. 17209045_tumor samples from FMTC patients showed strong nuclear staining of phosphorylated ERK1/2 and Ser(727) STAT3; FMTC-RET mutants activate a Ras/ERK1/2/STAT3 Ser(727) pathway, which plays an important role in cell mitogenicity and transformation. 17227125_Copy gain of PDGFB occurs in a subset of tumors showing no evidence of mutated BRAF or rearranged ret, suggesting that copy gain of PDGFB may underlie the increased expression of platelet-derived growth factor described recently in the literature. 17270245_study demonstrated RET amplification in all 3 cases of radiation-associated thyroid cancers(papillary thyroid cancer (PTC) & anaplastic thyroid cancer(ATC) but not in sporadic well-differentiated PTC; RET amplification was observed in all 6 cases of ATCs 17270543_Children of families with RET cysteine mutations may develop early metastatic medullary carcinoma of the thyroid gland. 17274802_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17316110_Observational study of genotype prevalence. (HuGE Navigator) 17384213_the absence of RET alterations in all cases of C-cell hyperplasia 17388787_our results suggest that SPRY2 regulates GDNF-dependent proliferation and differentiation of TGW neuroblastoma cells mediated by RET tyrosine kinase. 17397038_Observational study of gene-disease association. (HuGE Navigator) 17397038_These data highlight the pivotal role of the RET gene in both isolated and syndromic Hirschsprung disease. 17431108_RET, a receptor tyrosine kinase involved with differentiation, was consistently up-regulated throughout the time course of retinoic acid treatment in the majority of neuroblastic tumor cell lines. 17440194_study demonstrates that the interaction between RET and PHOX2B polymorphisms has a substantial impact on risk of Hirschsprung's disease 17464312_prevalence of BRAF mutation and RET/PTC were determined in diffuse sclerosing variant of papillary thyroid carcinoma; RET/PTC1 and RET/PTC3 were found in <50% of the cases investigated 17471236_overexpression of SH2B1beta, by enhancing phosphorylation/activation of RET transducers, potentiates the cellular differentiation and the neoplastic transformation thereby induced, and counteracts the action of RET inhibitors. 17490619_These findings provide evidence for a novel cooperative interaction between VEGFR2 and RET that mediates VEGF-A functions in ureteric bud cells. 17527003_primary hyperparathyroidism in RET 630 mutations might be associated with lower penetrance of primary hyperparthyoidism and pheochromocytoma 17554617_five RET sequence variants were detected in Hirschprung disease patients, including G15165A in exon 11, G20692A in exon 15, A18919G, T18888G, and a frameshift mutation 18974insG in exon 13 17573899_RET proto-oncogene mutations were restricted to codon 634 and 918 in Chinese families with MEN2A and MEN2B. 17576593_A heterozygous M918T mutation of the RET proto-oncogene was found in MEN 2B patients. 17599050_oncogenic precursor of RET(MEN 2B) is phosphorylated, interacts with adapter proteins and induces downstream signalling from the ER. 17605401_Observational study of genotype prevalence. (HuGE Navigator) 17605401_a change in the spectrum of mutations detected in the RET proto-oncogene in patients with hereditary MTC from the 'classical' mutation at codon 634 in exon 11 (level 2) to more cases with mutations in the exons 13-15 (level 1) and less aggressive disease 17610518_Patients harboring Y791F mutation may be considered at risk in an age earlier than 20 years and speculate that L769L variant of RET may be related to the early age of onset. 17623957_Observational study of genotype prevalence. (HuGE Navigator) 17623957_The reserachers found RET mutations common in an Iranian population, and these mutations are associated with the development of medullary thyroid carcinoma. 17639057_A fusion product between exon 11 of HOOK3 and exon 12 of RET gene was identified by 5'RACE, and the presence of chimeric HOOK3-RET protein of 88 kDa was detected by western blot analysis with an anti-RET antibody. 17639058_Observational study of genotype prevalence. (HuGE Navigator) 17664273_inhibition of RET is not impaired by mutation of the Val(804) gatekeeper residue, so MEN2 tumors may be less susceptible to acquired Sorafenib resistance 17727338_BRAF(V600E) mutation detected on fine-needle aspiration biopsy specimens, more than RET/PTC rearrangements, is highly specific for papillary thyroid carcinoma. 17786355_No RET rearrangement is associated with the papillary thyroid carcinomas 17825269_In the medulla oblongata, labekke nerve fibers, punctate structures, abd cell bodies occured in the caudate and interpolar parts of the spinal trigeminal nucleus. 17895320_We unexpectedly discovered a germline RET mutation in 35 of 481 (7.3%) apparently sporadic medullary thyroid carcinoma patients 17900235_expression of RET, nuclearRAS, and ERK proteins is greatly enhanced in papillary thyroid carcinoma cells and Hashimoto's thyroiditis oxyphil cells. 17934909_review summarizes how the different domains of the RET protein contribute to its normal function and how mutations in these domains affect the function of the receptor[Review] 17952863_This report reviews the RET involvement in the etiology of multiple endocrine neoplasia (MEN) 2 and medullary thyroid cancer (MTC) and updates the therapeutic approach in preclinical and clinical studies. 17954023_The frequency of RET rearrangement in Chinese patients is low and age related. RET/PTC-1 and RET/PTC-3 are associated with different clinical pathological characteristics but not with lymph node involvement. 17954268_A break-apart fluorescence in situ hybridization (FISH) assay should be able to detect translocations involving the RET gene in a papillary thyroid carcinoma cell line. 18058472_occurrence and the type of germline mutations in the RET gene in patients with medullary thyroid carcinoma. 18062802_Potential of DNA-based screening of all relevant RET exons. 18063059_The penetrance of pheochromocytoma varies between multiple endocrine neoplasia 2A RET codon mutations 18073307_Observational study of gene-disease association. (HuGE Navigator) 18073307_the presence of a somatic RET mutation correlates with a worse outcome of MTC patients, not only for the highest probability to have persistence of the disease, but also for a lower survival rate in a long-term follow up. 18090939_Child with hirschsprung disease and Wilms tumor showed mutations in the RET gene. 18189271_an important function for FRS2 is to concentrate RET in membrane foci, leading to an engagement of specific signaling complexes localized in these membrane domains 18222320_GDNF, NTN, GFRalpha-1, GFRalpha-2, and c-Ret proteins are differentially expressed in the different stages of hair follicle cycle. GFRalpha-mediated signaling involves c-Ret and may play a role in human HF biology. 18226854_frequency of the occurrence of BRAF mutation and/or RET/PTC in H4-PTEN positive tumors was extremely high (75%) in papillary thyroid carcinoma 18248648_There is a significant difference in MTC development with less extensive C-cell disease, higher cure rate and more frequent additional endocrinopathies in carriers of RET codon 791 mutations compared with carriers of codons 790 and 804 mutations. 18248681_Observational study of genotype prevalence. (HuGE Navigator) 18252215_Renal aplasia in humans is associated with RET mutations. 18258924_Observational study of genetic testing. (HuGE Navigator) 18273880_Observational study of genotype prevalence. (HuGE Navigator) 18273880_study demonstrates that the Ret Gly691Ser mutation is associated with primary vesicoureteral reflux and may be one of the genetic causes of this condition in the French-Canadian population in Quebec 18282654_A statistically significant correlation between the presence of RET somatic mutations with more advanced pathological neoplasm stages was observed in sporadic medullary thyroid carcinomas. 18284634_Observational study of gene-disease association. (HuGE Navigator) 18284634_RET genotypes including two intronic RET variants were associated with the risk of developing sporadic medullary thyroid cancer and to more aggressive behaviour. 18299477_uncommon coexistence of a germline mutation in two suppressor genes, RET and CDKN2A in multiple endocrine neoplasia 2a 18316595_oncogenic RET and loss of p18 cooperate in the multistep tumorigenesis of medullary thyroid carcinoma 18316596_a beta-catenin-RET kinase pathway is a critical contributor to the development and metastasis of thyroid carcinoma 18322301_Clinical and in vitro findings indicate that the transmembrane RET S649L mutation is associated with late-onset non-aggressive disease. 18331611_Observational study of gene-disease association. (HuGE Navigator) 18331611_The RET G691S variant could modulate the age of onset of sporadic medullary thyroid carcinoma as demonstrated previously for familial tumours. 18353552_These results suggest that engagement of different adaptor proteins by Ret results in very different downstream signaling and functions within neurons and that Dok recruitment leads to a rapid receptor relocation and formation of microspikes. 18355321_Differential membrane compartmentalization of RET by PTB-adaptor engagement is reported. 18365214_Oncogenic RET mutations may, however, vary between specific population groups. RET analysis in MEN has revolutionized the management of children of MEN2 and allowed surgical prediction and prophylaxis to take place. 18393128_No significan correlation between mutations in this protein and papillary thyroid cancer in Turkey. 18394855_Data show that the increased phosphorylation of tyrosine 1062 of the Ret receptor tyrosine kinase was induced by GDNF. GDNF signal through a molecular complex and the Ret receptor tyrosine kinase. 18402529_The RET proto-oncogene has become the target for molecularly designed drug therapy. Tyrosine kinase inhibitors targeting activated RET are currently in clinical trials for the treatment of patients | ENSMUSG00000030110 | Ret | 182.600170 | 0.7914935 | -0.337350677 | 0.20709246 | 2.656963e+00 | 1.030974e-01 | No | Yes | 140.061940 | 24.644968 | 181.618416 | 31.905391 | ||
| ENSG00000166169 | 27343 | POLL | protein_coding | Q9UGP5 | FUNCTION: DNA polymerase that functions in several pathways of DNA repair (PubMed:11457865, PubMed:19806195, PubMed:20693240, PubMed:30250067). Involved in base excision repair (BER) responsible for repair of lesions that give rise to abasic (AP) sites in DNA (PubMed:11457865, PubMed:19806195). Also contributes to DNA double-strand break repair by non-homologous end joining and homologous recombination (PubMed:19806195, PubMed:20693240, PubMed:30250067). Has both template-dependent and template-independent (terminal transferase) DNA polymerase activities (PubMed:10982892, PubMed:10887191, PubMed:12809503, PubMed:14627824, PubMed:15537631, PubMed:19806195). Has also a 5'-deoxyribose-5-phosphate lyase (dRP lyase) activity (PubMed:11457865, PubMed:19806195). {ECO:0000269|PubMed:10887191, ECO:0000269|PubMed:10982892, ECO:0000269|PubMed:11457865, ECO:0000269|PubMed:12809503, ECO:0000269|PubMed:14627824, ECO:0000269|PubMed:15537631, ECO:0000269|PubMed:19806195, ECO:0000269|PubMed:20693240, ECO:0000269|PubMed:30250067}. | 3D-structure;Alternative splicing;Chromosome;DNA damage;DNA repair;DNA replication;DNA synthesis;DNA-binding;DNA-directed DNA polymerase;Lyase;Manganese;Metal-binding;Nucleotidyltransferase;Nucleus;Reference proteome;Transferase | This gene encodes a DNA polymerase. DNA polymerases catalyze DNA-template-directed extension of the 3'-end of a DNA strand. This particular polymerase, which is a member of the X family of DNA polymerases, likely plays a role in non-homologous end joining and other DNA repair processes. Alternatively spliced transcript variants have been described. [provided by RefSeq, Mar 2010]. | hsa:27343; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; 5'-deoxyribose-5-phosphate lyase activity [GO:0051575]; DNA binding [GO:0003677]; DNA-directed DNA polymerase activity [GO:0003887]; metal ion binding [GO:0046872]; base-excision repair, gap-filling [GO:0006287]; DNA biosynthetic process [GO:0071897]; DNA replication [GO:0006260]; double-strand break repair via homologous recombination [GO:0000724]; double-strand break repair via nonhomologous end joining [GO:0006303]; nucleotide-excision repair [GO:0006289]; somatic hypermutation of immunoglobulin genes [GO:0016446] | 11821417_role in DNA repair 11974915_role in dna replication and DNA repair 12368291_complex between PCNA and pol lambda may play an important role in the bypass of abasic sites in human cells 12683997_DNA polymerase lambda has an intrinsic terminal deoxyribonucleotidyl transferase activity that preferentially adds pyrimidines onto 3'OH ends of DNA oligonucleotides and elongates an RNA primer hybridized to a DNA template. 12829698_mammalian Pol lambda has a role in non-homologous end-joining 14561766_Polymerase lambda is the primary gap-filling polymerase for accurate nonhomologous end joining 14627824_pol lambda Phe506Arg/Gly mutants possess very low polymerase and terminal transferase activities as well as greatly reduced abilities for processive DNA synthesis 15157109_fills short-patched DNA gaps in base excision repair pathways and participates in mammalian nonhomologous end-joining pathways to repair double-stranded DNA breaks 15202001_Results link p53 status with POLkappa expression and suggest that loss of p53 function may in part contribute to the observed POLkappa upregulation in human lung cancers. 15350147_A molecular mechanism is suggested for the observed high in vivo rate of frameshift generation by pol lambda and the remarkable ability of pol lambda to promote microhomology pairing between two DNA strands. 15358682_A helix-hairpin-helix domain of DNA polymerase lambda is important for primer binding and/or for proliferating cell nuclear antigen interaction. 15537631_determined that Fyn phosphorylated MAP-2c on tyrosine 67 15608652_crystal structures of Pol lambda representing three steps in filling a single-nucleotide gap 15665310_Human DNA polymerase kappa, an error-prone enzyme that is up-regulated in lung cancers, induces DNA breaks and stimulates DNA exchanges as well as aneuploidy. 15979954_Results suggest that Pol lambda plays a role in the short-patch base excision repair rather than contributes to the long-patch base excision repair pathway. 16174846_DNA polymerase lambda is phosphorylated in vitro by several cyclin-dependent kinase/cyclin complexes, including Cdk2/cyclin A, in its proline-serine-rich domain. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16522650_DNA polymerase lambda has the ability to create base pair mismatches and human replication protein A can suppress this intrinsic in vitro mutator phenotype. 16675458_DNA polymerase fidelity is controlled not by an accessory protein or a proofreading exonuclease domain but by an internal regulatory domain 16807316_DNA polymerase lambda is unable to differentiate between matched and mismatched termini during the DNA binding step, thus accounting for the relatively high efficiency of mismatch extension. 17005572_Kinetic studies on human DNA polymerase lambda reveal roles of a downstream strand and the 5'-terminal moieties 17321545_The erroneous nucleotide incorporations catalyzed by DNA polymerases lambda and beta as well as the subsequent ligation catalyzed by a DNA ligase during base excision repair are a threat to genomic integrity. 17653665_cloning, expression and tissue distribution in normal liver and hepatoma 17666409_DNA pol lambda active site residue tyrosine 505 determined the nucleotide selectivity opposite 1,2-dihydro-2-oxoadenine during error-free bypass of the 1,2-dihydro-2-oxoadenine lesion. 17686665_capacity of Pol lambda either to insert opposite 8oxoG or to extend correct base pairs containing such a damage could be beneficial for its role in NHEJ 18688254_Pol lambda is stabilized during cell cycle progression in late S and G2 phases, most likely allowing Pol lambda to correctly conduct repair of damaged DNA during and after S phase. 18692600_analysis of human DNA polymerase lambda with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies 19060005_RAGE is under genetic selection in the non-African populations. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19104052_replication protein A and proliferating cell nuclear antigen act as molecular switches to activate the DNA pol lambda- dependent highly efficient and faithful repair of A:8-oxo-G mismatches in human cells and to repress DNA pol beta activity. 19120024_dATP was a preferential substrate for DNA polymerase beta and dGMP was the only substrate for DNA polymerase lambda. 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19467241_These experiments suggest that polymerase lambda does not undergo major conformational changes during the catalysis in the solution phase. 19701199_when bound to a 2-nucleotide gap, Pol lambda scrunches the template strand and binds the additional uncopied template base in an extrahelical position within a binding pocket that comprises three conserved amino acids. 19806195_a natural mutator variant of human DNA polymerase lambda promotes chromosomal instability by compromising NHEJ 19820168_observe specific recruitment of MUTYH, DNA pol lambda, proliferating cell nuclear antigen (PCNA), flap endonuclease 1 (FEN1) and DNA ligases I and III from human cell extracts to A:8-oxo-G DNA, but not to undamaged DNA. 19900463_DNA polymerase lambda uses a novel sugar selection mechanism to discriminate against ribonucleotides, whereby the ribose 2'-hydroxyl group was excluded mostly by a backbone segment and slightly by the side chain of Y505. 20164241_Both Pol lambda- and (Pol kappa)-positive staining were associated with shorter survival in glioma patients. 20348107_analysis of the interaction between DNA Polymerase lambda and anticancer nucleoside analogs 20423048_DNA polymerase lambda can bypass a thymine glycol lesion on the template strand of gapped DNA substrates. 20435673_The results demonstrate that loop 1 is not essential for catalytic activity, but it is important for the fidelity of DNA synthesis and the accuracy of non-homologous end joining. 20693240_study found expression of PollambdaR438W sensitizes cells to camptothecin by affecting the homologous recombination pathway, whereas overexpression of pollambdaWT did not impact cell survival; this effect depends entirely on its DNA polymerase activity 20805875_both pol lambda and pol beta interact with the upstream DNA glycosylases for repair of alkylated and oxidized DNA bases 20851705_The fidelity of Pol lambda was maintained predominantly by a single residue, R517, which has minor groove interactions with the DNA template. 20975951_Studies indicate that codon-based models of gene evolution yielded statistical support for the recurrent positive selection of five NHEJ genes during primate evolution: XRCC4, NBS1, Artemis, POLlambda, and CtIP. 21486570_Studies indicate that pol lambda undergoes posttranslational modifications during the cell cycle that regulate its stability and possibly its subcellular localization. 21757740_In vitro gap-directed translesion DNA synthesis of an abasic site involving human DNA polymerases epsilon, lambda, and beta. 22317757_Pollambda may play a specialized role in the process of repair of these kinds of lesions 22373917_Results reveal that DNA pol lambda and DNA ligase I are sufficient to promote efficient microhomology-mediated end-joining repair of broken DNA ends in vitro. 22584622_A structural study shows how a ribonucleotide can be accommodated in the DNA polymerase lambda active site. 23118481_The results provides evidence that DNA pol lambda is required for cell cycle progression and is functionally connected to the S phase DNA damage response machinery in cancer cells. 23330920_Inactivation of polymerase (DNA directed) lambda lyase activity by 5'-(2-phosphoryl-1,4-dioxobutane prevents the enzyme from conducting polymerization following preincubation of the protein and DNA. 23935073_A specific N-terminal extension of the 8 kDa domain of DNA polymerase lambda is important for the non-homologous end joining function. 25015085_Structural basis for the binding and incorporation of nucleotide analogs with L-stereochemistry by human DNA polymerase lambda. 25460917_DNA polymerase lamda catalyzes lesion bypass across benzo[a]pyrene-derived DNA adducts. 25687118_Fen1 significantly stimulated trinucleotide repeats expansion by Pol beta, but not by the related enzyme Pol lambda. 25741586_pol lambda is responsible for a significant fraction of Fapy.dG-induced G --> T mutations. 26917111_Pol beta, to a greater extent than Pol lambda can incorporate rNMPs opposite normal bases or 8-oxo-G, and with a different fidelity. Further, the incorporation of rNMPs opposite 8-oxo-G delays repair by DNA glycosylases. 27481934_The authors demonstrate that Pol lambda has a flexible active site that can tolerate 8-oxo-dG in either the anti- or syn-conformation. Importantly, we show that discrimination against the pro-mutagenic syn-conformation occurs at the extension step and identify the residue responsible for this selectivity. 27621267_Data suggest that individuals who carry the rs3730477 POLL germline variant have an increased risk of estrogen-associated breast cancer. 27992186_Bond formation and cleavage reactions catalyzed by base excision repair DNA polymerases beta and lambda has been described. 28109743_T204 was identified as a main target for ATM/DNA-PKcs phosphorylation on human POLL, and this phosphorylation may facilitate the repair of a subset of IR-induced DSBs and the efficient POLL-mediated gap-filling during NHEJ. POLL phosphorylation might favor POLL interaction with the DNA-PK complex at DSBs. 28841305_When mutated or deregulated, DNA polymerase lambda can also be a source of genetic instability. Its multiple roles in DNA damage tolerance and its ability in promoting tumor progression make it also a possible target for novel anticancer approaches. [review] 30250067_PAXX, XLF and XRCC4 synergise in the efficient DNA double-strand breaks recruitment, substrate recognition and stimulation of Pol lambda enzymatic activity during nonhomologous end joining DNA repair. 33673424_A Role for Human DNA Polymerase lambda in Alternative Lengthening of Telomeres. | ENSMUSG00000025218 | Poll | 786.256232 | 0.9783231 | -0.031617058 | 0.11074479 | 8.109918e-02 | 7.758133e-01 | 9.288513e-01 | No | Yes | 829.688696 | 86.499361 | 860.080038 | 89.497924 | |
| ENSG00000166250 | 79827 | CLMP | protein_coding | Q9H6B4 | FUNCTION: May be involved in the cell-cell adhesion. May play a role in adipocyte differentiation and development of obesity. Is required for normal small intestine development. {ECO:0000269|PubMed:14573622, ECO:0000269|PubMed:15563274, ECO:0000269|PubMed:22155368}. | Cell junction;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Reference proteome;Repeat;Signal;Tight junction;Transmembrane;Transmembrane helix | This gene encodes a type I transmembrane protein that is localized to junctional complexes between endothelial and epithelial cells and may have a role in cell-cell adhesion. Expression of this gene in white adipose tissue is implicated in adipocyte maturation and development of obesity. This gene is also essential for normal intestinal development and mutations in the gene are associated with congenital short bowel syndrome. [provided by RefSeq, Aug 2015]. | hsa:79827; | bicellular tight junction [GO:0005923]; cell surface [GO:0009986]; cytoplasmic microtubule [GO:0005881]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; digestive tract development [GO:0048565] | 12851705_ASAM, IGSF11, CXADR and ESAM are type I transmembrane proteins and members of the same IGSF superfamily. 14573622_CLMP is a novel cell-cell adhesion molecule and a new component of epithelial tight junctions. 15563274_We identified ACAM (adipocyte adhesion molecule), a novel homologue of the CTX (cortical thymocyte marker in Xenopus) gene family, which may be the critical adhesion molecule in adipocyte differentiation and development of obesity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22155368_Loss-of-function mutations in CLMP cause congenital short bowel syndrome in human beings, likely by interfering with tight-junction formation, which disrupts intestinal development. 22718816_The PDZ1 and PDZ3 domains of MAGI-1 regulate the eight-exon isoform of the CXADR-like membrane protein. 22992863_Coxsackievirus and adenovirus receptor gene expression is induced in esophageal cancer cells by the HDAC inhibitor trichostatin A. 23460781_The key processes involved in intestinal epithelial development appear to be unaffected by wild type-CLMP or mutant-CLMP. 27352967_Novel inherited variants in CLMP were identified in three congenital short bowel syndrome patients derived from two unrelated families. 27527752_Inhibition of CFTR or histone deacetylase (HDAC) enhanced CAR expression while CFTR overexpression or restoration of the diminished HDAC activity in cystic fibrosis cells reduced CAR expression. 27720179_We describe a newborn presenting CSBS intestinal malrotation and chronic intestinal pseudo-obstruction syndrome (CIPS), compound heterozygous for two previously unreported heterozygous mutations in Coxsackie and adenovirus receptor-like membrane protein (CLMP) gene. We identified two heterozygous mutations in CLMP, one in intron 1 (c.28+1G>C) from the father, the other on exon 4 (c502C>T, p.R168X) from the mother. | ENSMUSG00000032024 | Clmp | 28.244700 | 0.3931386 | -1.346890077 | 0.50732929 | 7.033382e+00 | 8.000411e-03 | No | Yes | 20.952296 | 7.819645 | 52.924290 | 19.547823 | ||
| ENSG00000166321 | 25961 | NUDT13 | protein_coding | Q86X67 | FUNCTION: NAD(P)H pyrophosphatase that hydrolyzes NADH into NMNH and AMP, and NADPH into NMNH and 2',5'-ADP. Has a marked preference for the reduced pyridine nucleotides. Does not show activity toward NAD-capped RNAs; the NAD-cap is an atypical cap present at the 5'-end of some RNAs. {ECO:0000250|UniProtKB:Q8JZU0}. | Alternative splicing;Hydrolase;Magnesium;Manganese;Metal-binding;Mitochondrion;NAD;NADP;Reference proteome;Transit peptide | hsa:25961; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; metal ion binding [GO:0046872]; NAD+ diphosphatase activity [GO:0000210]; NADH pyrophosphatase activity [GO:0035529]; pyrophosphatase activity [GO:0016462]; NADH metabolic process [GO:0006734]; NADP catabolic process [GO:0006742]; nucleobase-containing small molecule interconversion [GO:0015949] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000021809 | Nudt13 | 71.532862 | 1.2419550 | 0.312612958 | 0.34453964 | 8.311577e-01 | 3.619380e-01 | No | Yes | 78.825112 | 22.390220 | 61.413238 | 17.530433 | |||
| ENSG00000166762 | 117155 | CATSPER2 | protein_coding | Q96P56 | FUNCTION: Voltage-gated calcium channel that plays a central role in calcium-dependent physiological responses essential for successful fertilization, such as sperm hyperactivation, acrosome reaction and chemotaxis towards the oocyte. {ECO:0000269|PubMed:21412338, ECO:0000269|PubMed:21412339}. | Alternative splicing;Calcium;Calcium channel;Calcium transport;Cell membrane;Cell projection;Cilium;Developmental protein;Differentiation;Flagellum;Ion channel;Ion transport;Membrane;Reference proteome;Spermatogenesis;Transmembrane;Transmembrane helix;Transport;Voltage-gated channel | This gene encodes a member of a family of cation channel proteins that localize to the flagellum of spermatozoa. Defects at this locus causes male infertility. Alternatively spliced transcript variants have been observed at this locus. Readthrough transcription originates upstream of this locus in diphosphoinositol pentakisphosphate kinase 1 pseudogene 1 and is represented by GeneID:110006325. Related pseudogenes are found next to this locus on chromosome 15 and on chromosome 5. [provided by RefSeq, Mar 2017]. | hsa:117155; | CatSper complex [GO:0036128]; motile cilium [GO:0031514]; plasma membrane [GO:0005886]; calcium activated cation channel activity [GO:0005227]; calcium channel activity [GO:0005262]; voltage-gated ion channel activity [GO:0005244]; fertilization [GO:0009566]; flagellated sperm motility [GO:0030317]; regulation of ion transmembrane transport [GO:0034765]; sperm capacitation [GO:0048240] | 16740636_CatSper1 and CatSper2 can associate with and modulate the function of the Ca(v)3.3 channel, which might be important in the regulation of sperm function. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23519396_Levels of Tektin 2 and CatSper 2 proteins are positively associated with sperm motility parameters. Measurements of Tektin 2 levels can be correlated with the clinical outcome of ICSI. 30629171_disruption of CATSPER2 current is a significant factor causing idiopathic male infertility; the specific copy number variation disrupts one gene copy in the region 43894500-43950000 in chromosome 15 | ENSMUSG00000033486 | Catsper2 | 46.680768 | 0.5721913 | -0.805430607 | 0.39028833 | 4.278953e+00 | 3.858711e-02 | No | Yes | 34.264376 | 12.016516 | 53.224974 | 18.747705 | ||
| ENSG00000167525 | 147011 | PROCA1 | protein_coding | Q8NCQ7 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:147011; | cyclin binding [GO:0030332]; phospholipase A2 activity [GO:0004623]; arachidonic acid secretion [GO:0050482]; phospholipid metabolic process [GO:0006644] | ENSMUSG00000044122 | Proca1 | 34.423415 | 0.8672879 | -0.205417178 | 0.46803564 | 1.844037e-01 | 6.676158e-01 | No | Yes | 31.889384 | 7.023377 | 39.042937 | 8.410193 | |||||
| ENSG00000167703 | 124935 | SLC43A2 | protein_coding | Q8N370 | FUNCTION: Sodium-, chloride-, and pH-independent high affinity transport of large neutral amino acids. {ECO:0000269|PubMed:15659399}. | Alternative splicing;Amino-acid transport;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the L-amino acid transporter-3 or SLC43 family of transporters. The encoded protein mediates sodium-, chloride-, and pH-independent transport of L-isomers of neutral amino acids, including leucine, phenylalanine, valine and methionine. This protein may contribute to the transfer of amino acids across the placental membrane to the fetus. [provided by RefSeq, Mar 2016]. | hsa:124935; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; amino acid transmembrane transporter activity [GO:0015171]; L-amino acid transmembrane transporter activity [GO:0015179]; neutral amino acid transmembrane transporter activity [GO:0015175]; amino acid transport [GO:0006865]; negative regulation of amino acid transport [GO:0051956]; negative regulation of leucine import [GO:0060358]; neutral amino acid transport [GO:0015804] | 15659399_In situ hybridization experiments show that LAT4 mRNA is restricted to the epithelial cells of the distal tubule and the collecting duct in the kidney. In the intestine, LAT4 is mainly present in the cells of the crypt. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 32879489_Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. 33077272_Plasma citrulline correlates with basolateral amino acid transporter LAT4 expression in human small intestine. | ENSMUSG00000038178 | Slc43a2 | 559.655806 | 0.7902816 | -0.339561223 | 0.11978404 | 8.038618e+00 | 4.579047e-03 | 8.148308e-02 | No | Yes | 480.941424 | 64.423388 | 612.097279 | 81.596302 | |
| ENSG00000167861 | 283987 | HID1 | protein_coding | Q8IV36 | FUNCTION: May play an important role in the development of cancers in a broad range of tissues. {ECO:0000269|PubMed:11281419}. | Alternative splicing;Cytoplasm;Golgi apparatus;Lipoprotein;Membrane;Myristate;Phosphoprotein;Reference proteome | hsa:283987; | cytoplasm [GO:0005737]; cytoplasmic microtubule [GO:0005881]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extrinsic component of Golgi membrane [GO:0090498]; Golgi apparatus [GO:0005794]; Golgi medial cisterna [GO:0005797]; Golgi trans cisterna [GO:0000138]; membrane [GO:0016020] | 21337012_mammalian HID-1 localized to the medial- and trans- Golgi apparatus as well as the cytosol. 29074564_Authors propose that HID-1 influences early steps in LDCV formation by controlling dense core formation at the TGN. 33999436_Mutations in HID1 Cause Syndromic Infantile Encephalopathy and Hypopituitarism. | ENSMUSG00000034586 | Hid1 | 352.246121 | 0.8275651 | -0.273055220 | 0.14576305 | 3.502889e+00 | 6.126189e-02 | No | Yes | 288.532239 | 35.092341 | 356.509167 | 43.073324 | |||
| ENSG00000167972 | 21 | ABCA3 | protein_coding | Q99758 | FUNCTION: Catalyzes the ATP-dependent transport of phospholipids such as phosphatidylcholine and phosphoglycerol from the cytoplasm into the lumen side of lamellar bodies, in turn participates in the lamellar bodies biogenesis and homeostasis of pulmonary surfactant (PubMed:16959783, PubMed:17574245, PubMed:28887056, PubMed:31473345, PubMed:27177387). Transports preferentially phosphatidylcholine containing short acyl chains (PubMed:27177387). In addition plays a role as an efflux transporter of miltefosine across macrophage membranes and free cholesterol (FC) through intralumenal vesicles by removing FC from the cell as a component of surfactant and protects cells from free cholesterol toxicity (PubMed:26903515, PubMed:25817392, PubMed:27177387). {ECO:0000269|PubMed:16959783, ECO:0000269|PubMed:17574245, ECO:0000269|PubMed:25817392, ECO:0000269|PubMed:26903515, ECO:0000269|PubMed:27177387, ECO:0000269|PubMed:28887056, ECO:0000269|PubMed:31473345}. | ATP-binding;Alternative splicing;Cytoplasmic vesicle;Disease variant;Disulfide bond;Endosome;Glycoprotein;Lipid transport;Lysosome;Membrane;Nucleotide-binding;Reference proteome;Repeat;Translocase;Transmembrane;Transmembrane helix;Transport | The membrane-associated protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intracellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, White). This protein is a member of the ABC1 subfamily. Members of the ABC1 subfamily comprise the only major ABC subfamily found exclusively in multicellular eukaryotes. The full transporter encoded by this gene may be involved in development of resistance to xenobiotics and engulfment during programmed cell death. [provided by RefSeq, Jul 2008]. | hsa:21; | alveolar lamellar body [GO:0097208]; alveolar lamellar body membrane [GO:0097233]; cytoplasmic vesicle membrane [GO:0030659]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; lamellar body [GO:0042599]; lamellar body membrane [GO:0097232]; late endosome [GO:0005770]; lysosomal membrane [GO:0005765]; multivesicular body membrane [GO:0032585]; plasma membrane [GO:0005886]; ABC-type xenobiotic transporter activity [GO:0008559]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATPase-coupled transmembrane transporter activity [GO:0042626]; lipid transporter activity [GO:0005319]; phosphatidylcholine flippase activity [GO:0140345]; phosphatidylcholine transfer activity [GO:0120019]; cellular protein metabolic process [GO:0044267]; lipid transport [GO:0006869]; lung development [GO:0030324]; organelle assembly [GO:0070925]; phosphatidylcholine metabolic process [GO:0046470]; phosphatidylglycerol metabolic process [GO:0046471]; phospholipid homeostasis [GO:0055091]; phospholipid transport [GO:0015914]; positive regulation of cholesterol efflux [GO:0010875]; positive regulation of phospholipid efflux [GO:1902995]; positive regulation of phospholipid transport [GO:2001140]; positive regulation of protein homooligomerization [GO:0032464]; regulation of lipid biosynthetic process [GO:0046890]; regulation of phosphatidylcholine metabolic process [GO:0150172]; response to glucocorticoid [GO:0051384]; response to xenobiotic stimulus [GO:0009410]; surfactant homeostasis [GO:0043129]; xenobiotic export [GO:0046618]; xenobiotic transmembrane transport [GO:0006855]; xenobiotic transport [GO:0042908] | 11940594_Identification of LBM180, a lamellar body limiting membrane protein of alveolar type II cells, as the ABC transporter protein ABCA3 15465012_Results suggest that ABCA3 shows ATPase activity, which is induced by lipids, and may be involved in the biogenesis of lamellar body-like structures. 15976379_Observational study of gene-disease association. (HuGE Navigator) 16415354_ABCA3 is required for lysosomal loading of phosphatidylcholine and conversion of lysosomes to lamellar body-like structures 16857811_ABCA3 has a role in drug resistance in childhood acute myeloid leukemia 16959783_ABCA3 mutation is associated with fatal surfactant deficiency 16968533_ABCA17P & ABCA3 form a complex of overlapping genes at their 5' ends. Non-coding & protein-coding ABC A-transporter RNAs are expressed. This is the 1st demonstration of expression of a pseudogene & its parent from a common overlapping human DNA region. 17429902_ABCA3 mutations were the basis for intractable respiratory distress syndrome in three Norwegian term infants 17574245_ABCA3 mediates ATP-dependent choline-phospholipids uptake into intracellular vesicles. 17597647_Finding of heterozygosity for ABCA3 mutations in severely affected infants with SFTPC I73T, and independent inheritance from disease-free parents supports that ABCA3 acts as a modifier gene for the phenotype associated with an SFTPC mutation. 17660803_In infants with a desquamative interstitial pneumonitis pattern, surfactant or ABCA3 mutations should be evaluated. 18024538_Dense abnormalities of lamellar bodies, characteristic of ABCA3 mutations, were seen by electron microscopy in all adequate specimens. 18246475_Observational study of gene-disease association. (HuGE Navigator) 18317237_ABCA3 mutations impart increased genetic risk for newborn respiratory distress syndrome. 18317237_Observational study of genotype prevalence. (HuGE Navigator) 18463677_found ABC transporter A3 to be expressed consistently in acute myeloid leukemia samples. Greater expression of ABCA3 is associated with unfavorable treatment outcome, and in vitro, elevated expression induces resistance to a broad spectrum of cytostatics 18603241_This newborn infant had a heterozygote mutation of ABCA3. 19220077_Mutations in the ABCA3 gene are associated with both fatal respiratory distress in the neonatal period and interstitial lung disease in older infants, children, and adults. 19252731_A case of newborn respiratory distress syndrome associated with ABCA3 transporter deficiency and a ABCA3 mutation is described. 19343046_Observational study of gene-disease association. (HuGE Navigator) 19861431_The identification of one copy of this novel mutation in a premature infant with chronic respiratory insufficiency suggests that ABCA3 haploinsufficiency together with lung prematurity may result in more severe, or more prolonged, respiratory failure 19902402_Decreased ABCA3 expression is associated with breast cancer. 20304423_The segregation of ABCA3 alleles, absence of ABCA3 immunostaining, lung pathology, and ultrastructural findings support genetic ABCA3 deficiency as the cause of lung disease. 20371530_Subclinical fibrotic changes may be present in family members of patients with SFTPC mutation-associated interstitial lung disease and suggest ABCA3 variants could affect disease pathogenesis. 20656946_Observational study of gene-disease association. (HuGE Navigator) 20863830_Data show that the ABCA3 N-terminus is proteolytically removed inside acidic LAMP3-positive vesicles MVB/LB. 21165348_Several genetic abnormalities have been associated with familial pulmonary fibrosis. The present study examined the genes coding for surfactant protein-C, ATPbinding cassette protein A3 and telomerase, and found no abnormalities. 21214890_Suggest that expression of partially or completely endoplasmic reticulum localized ABCA3 mutant proteins can increase the apoptotic cell death of the affected cells. 21526180_SALL4 was able to bind to the promoter region of ABCA3 and activate its expression while regulating the expression of ABCG2 indirectly. 21586796_A novel conserved targeting motif found in ABCA transporters mediates trafficking to early post-Golgi compartments. 21873242_Lymphoma exosomes shield target cells from antibody attack and that exosome biogenesis is modulated by the lysosome-related organelle-associated ATP-binding cassette (ABC) transporter A3 (ABCA3). 22068586_identification of new ABCA3 mutations in patients with life-threatening neonatal respiratory distress and/or pediatric interstitial lung disease (ILD); two mutations associated with ILD acted via different pathophysiological mechanisms despite similar clinical phenotypes 22145626_the ABCA3 missense mutation E292V had no remarkable effect on pulmonary outcome in VLBW infants. Present results do not rule out the possibility that E292V phenotype is associated with minor difference in the morbidity. 22337229_An intronic ABCA3 mutation is responsible for a fatal respiratory disease in newborns. 22434821_Data suggest the impairment of epithelial function as a mechanism by which ABCA3 mutations cause interstitial lung disease (ILD). 22455634_Genetic variants within ABCA3 may be the genetic cause of or a contributor to some unexplained refractory neonatal respiratory distress syndrome. 22800827_There is an association between a synonymous cSNP rs323043 and the development of neonatal respiratory distress syndrome. 22866751_partially reduced ABCA3 activity due to E292V is not a major risk factor for reduced lung function and COPD in the general population 23166334_Although ABCA3 mutations are individually rare, they are collectively common among European- and African-descent individuals in the general population. 23432194_Tyrosine kinase inhibitor exposure facilitates a protective loop of SALL4 and ABCA3 cooperation in persistent leukaemic cells. 23443156_Identification of novel compound heterozygous mutations in the coding exons of ABCA3 in two brothers with interstitial lung disease. 23846195_Report of ABCA3 mutations in a family with one child exhibiting interstitial lung disease. 24142515_cotranslational N-linked glycosylation at N124 and N140 is critical for ABCA3 stability, and its disruption results in protein destabilization and proteasomal degradation 24145140_Our findings indicate a considerable and direct relationship between mRNA expression levels of ABCA2, ABCA3, MDR1, and MRP1 genes and positive minimal residual disease (MRD) measured after one year of treatment. 24269975_A compound heterozygote for both novel mutations in the ABCA3 gene. 24420869_Two siblings are described who were homozygous for a 5,983 bp deletion in ABCA3 including exons 2-5 as well as the start AUG codon and a putative Golgi exit signal motif. 24628317_A novel missense mutation in ABCA3 was found to cause fatal congenital surfactant deficiency in two siblings. 24633979_Therapeutic strategies for chronic interstitial lung disease have been used successfully in cases of a mild clinical course in juvenile patients with ABCA3 gene mutation. In our patient with homozygous ABCA3 gene mutation,they were not effective. 24657120_ABCA3 protein, human genetic variants does not increase the risk of neonatal respiratory distress syndrome. 24730976_A large kindred is identified with a novel ABCA3 mutation causing pulmonary fibrosis. 24871971_Genotype-phenotype correlations exist for homozygous or compound heterozygous mutations in ABCA3 causing neonatal respiratory failure or childhood interstitial lung disease. 25056761_SLCO1B3 699GG and 344TT genotypes are associated with non-response to IM, while ABCA3 4548-91 CC/CA genotypes are related to poor CMR in CML patients treated with standard-dose imatinib. 25073622_analyzed the coding exons for rs117603931 (p.R288K) in exon 8 and rs149989682 (p.E292V) in exon 9 of ABCA3 in 224 Caucasian preterm infants ranging in GA from 29 to 36 weeks with respiratory distress syndrome; 10 mutations were found (6 at rs117603931 in exon 8 and 4 at rs149989682 in exon 9, combined allele frequency of 2.5% and all but one were present as heterozygotes 25406294_We identified a cataract-microcornea syndrome (cmcc) associated gene, ABCA3, which had heterozygous missense mutations in two autosomal dominant CCMC families. Another four heterozygous mutations, 2 missense and 2 splice site mutations were identified. 25817392_Accumulation of free cholesterol as a result of a loss of ABCA3 export function represents a novel pathomechanism in ABCA3-induced Diffuse parenchymal lung disease. 26295388_Data discussed the structural features of ABCA3 and how the use of bioinformatics tools could help researchers to obtain a reliable structural model to locate relevant mutations and make genotype/phenotype correlations of affected patients. [review] 26508177_The clinical features of ABCA3 mutations, including onset, severity or clinical course are very heterogeneous. In the two siblings we had a lightly discordant course that could be explained by exposure to different environmental stresses or variable penetrance. 26517903_Studies indicate that ATP-binding cassette (ABC), subfamily A, member 3 (ABCA3) is developmentally regulated. 26522252_The TGGAG haplotype may be a risk factor for Respiratory Distress Syndrome in preterm infants in this Chinese population. The haplotype TGGAG was significantly more frequent in RDS infants than in non-RDS infants. 26547207_Rare mutations in surfactant-associated genes contribute to neonatal respiratory distress syndrome. We resequenced all exons of the ATP-binding cassette member A3 (ABCA3) and we found three ABCA3 mutations in the Han [minor allele frequency (MAF)=0.003] and 7 in the Zhuang (MAF=0.011) cohorts. The contribution of these rare ABCA3 mutations to disease burden in the south China population is still unknown. 26903515_These results provide evidence of ABCA3 as an MLF efflux transporter in human macrophages and support its role in the direct antileishmanial effect of this alkylphosphocholine drug. 27352740_transporter oligomerization is crucial for ABCA3 function. 27374344_two ABCA3 mutations (p.R288K and p.R1474W) identified among term and late-preterm infants with respiratory distress syndrome, were characterized. 27516224_1153 patients with diffuse parenchymal lung disease (DPLD) were registered in the KLR. The DNA of 242 of these patients was sequenced for ABCA3 mutations. 69 patients had at least one variation in the ABCA3 gene. Of 40 patients with two disease-causing ABCA3 mutations, 22 patients were homozygous and 18 heterozygous. 28241820_Understanding the relationship between cholesterol and inflammation in the lung, and the role that ABC transporters play in this may illuminate new pathways to target for the treatment of inflammatory lung diseases 28468577_Genes ABCC7, A3, A8, A12, and C8 prevailed among the most upregulated or downregulated ones. In conclusion, the results supported our theory about general adenosine triphosphate-binding cassette gene expression profiles and their importance for cancer on clinical as well as research levels. 28642621_Our results showed that the allele frequency of p.G1205R, but not p.L39V, was significantly higher in ILD patients than in healthy controls. However, no additional subject carrying the variant p.S828F or p.V968M was detected in the cohort analysis. These results indicate that the heterozygous ABCA3 gene variants may contribute to susceptibility to diseases in the Chinese population. 28887056_a mutation (E292V) located in the first cytoplasmic loop of ABCA3 did not significantly affect lipid transport, but rather resulted in smaller vesicles. In addition to these findings, the assay used in this work for analysing the PC-lipid transport into ABCA3 positive vesicles will be useful to screen for compounds susceptible to restore function in mutated ABCA3 protein. 29255193_Heterozygous previously described rare or novel variants in surfactant proteins genes ABCA3, SFTPB and SFTPC were identified in 24 newborn infants with particularly severe respiratory distress syndrome. Ultrastructural analysis of lung tissue of one infant showed features suggesting ABCA3 dysfunction. 29325094_The aim of this study was to prove that disease causing misfolding ABCA3 mutations can be corrected in vitro and to investigate available options for correction. 29505158_we classified cellular consequences of missense ABCA3 sequence variations leading to pulmonary disease of variable severity. The corresponding molecular pathomechanisms of such ABCA3 variants may specifically be addressed by targeted treatments. 29569581_Mutations in SFTPC, NKX2.1, and FOXF1 were identified among Japanese infants and children with childhood interstitial lung disease, whereas ABCA3 mutations were rare. 31210424_Potentiation of ABCA3 lipid transport function by ivacaftor and genistein. 31331098_Mutations in adenosine triphosphate-binding cassette transporter A3 (ABCA3) (OMIM: 601615) gene constitute the most frequent genetic cause of severe neonatal respiratory distress syndrome (RDS) and interstitial lung disease (ILD) in children. 31473345_The assay presented in this work probes the choline containing lipid transport function of ABCA3. Visualization and quantification of fluorescence intensity of the labeled lipids inside ABCA3+ vesicles at equilibrium can specifically assess the transport function of ABCA3. 32196812_Functional characterization of four ATP-binding cassette transporter A3 gene (ABCA3) variants. 32238781_ABCA3 mutations in adult pulmonary fibrosis patients: a case series and review of literature. 32532878_Neonatal respiratory failure due to novel compound heterozygous mutations in the ABCA3 lipid transporter. 32692933_Functional Genomics of ABCA3 Variants. 33818848_ABCA3 deficiency dramatically improved by azithromycin administration. 34101541_Heterogeneity in Human Induced Pluripotent Stem Cell-derived Alveolar Epithelial Type II Cells Revealed with ABCA3/SFTPC Reporters. 34186035_Familial Pulmonary Fibrosis: Genetic Features and Clinical Implications. 34245068_A homozygous variant in ABCA3 is associated with severe respiratory distress and early neonatal death. 34449154_Whole exome sequencing identifies a novel variant in ABCA3 in an individual with fatal congenital surfactant protein deficiency. 34597626_ATP-binding cassette transporters mediate differential biosynthesis of glycosphingolipid species. 34638622_Structure-Based Understanding of ABCA3 Variants. 34715861_A novel synonymous ABCA3 variant identified in a Chinese family with lethal neonatal respiratory failure. 35170262_Biologic characterization of ABCA3 variants in lung tissue from infants and children with ABCA3 deficiency. 35394827_Cryo-EM structures of the human surfactant lipid transporter ABCA3. | ENSMUSG00000024130 | Abca3 | 2799.241808 | 0.8809119 | -0.182930381 | 0.07841151 | 5.489680e+00 | 1.912904e-02 | 1.840371e-01 | No | Yes | 2441.418371 | 257.748157 | 2790.697536 | 294.353242 | |
| ENSG00000168010 | 89849 | ATG16L2 | protein_coding | Q8NAA4 | FUNCTION: May play a role in regulating epithelial homeostasis in an ATG16L1-dependent manner. {ECO:0000250|UniProtKB:Q6KAU8}. | Alternative splicing;Coiled coil;Cytoplasm;Protein transport;Reference proteome;Repeat;Transport;WD repeat | hsa:89849; | Atg12-Atg5-Atg16 complex [GO:0034274]; autophagosome membrane [GO:0000421]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; phagophore assembly site membrane [GO:0034045]; autophagosome assembly [GO:0000045]; negative stranded viral RNA replication [GO:0039689]; protein lipidation [GO:0006497]; protein transport [GO:0015031] | 23850713_Three novel risk loci including ATG16L2 were discovered by genome-wide association study in the Korean Crohn's disease population, of which two showed patterns of association in the International Inflammatory Bowel Disease Genetics Consortium dataset. 24406150_Atg16L2 may play an important role in autophagy of T cells and serve as a potential biomarker to predict clinical relapse of Multiple sclerosis 26663301_A novel SNP-systemic lupus erythematosus association was identified between FCHSD2 and P2RY2, peaking at rs11235667 on a 33-kb haplotype upstream of ATG16L2. 27611316_ATG16L2 is a susceptibility gene for Crohn's disease in the Chinese population. The rs11235604 single nucleotide polymorphism is remarkably associated with downregulation of the expression of ATG16L2. 29454863_Patients with the ATG16L2 rs10898880 CC variant genotype had a better LRFS, PFS, and OS (adjusted hazard ratio = 0.59, 0.64, and 0.64; 95% confidence interval: 0.45-0.79, 0.48-0.84, and 0.48-0.86; p = 0.0004, 0.002, and 0.003, respectively), but a greater risk for development of severe RP (adjusted hazard ratio = 1.80, 95% confidence interval: 1.04-3.12, p = 0.037) than did patients with AA/AC genotypes. 31692259_Results found ATG16L2 rs10898880 was significantly associated with the occurrence of grade 3-4 oral mucositis and myelosuppression in nasopharyngeal carcinoma (NPC) patients treated with radiotherapy. 33113339_Identification of a novel differentially methylated region adjacent to ATG16L2 in lung cancer cells using methyl-CpG binding domain protein-enriched genome sequencing. | ENSMUSG00000047767 | Atg16l2 | 204.245260 | 0.9592403 | -0.060035790 | 0.18595559 | 1.041035e-01 | 7.469597e-01 | No | Yes | 176.845006 | 24.341876 | 188.702943 | 25.909044 | |||
| ENSG00000168026 | 199223 | TTC21A | protein_coding | Q8NDW8 | FUNCTION: Intraflagellar transport (IFT)-associated protein required for spermatogenesis (PubMed:30929735). Required for sperm flagellar formation and intraflagellar transport (PubMed:30929735). {ECO:0000269|PubMed:30929735}. | Alternative splicing;Differentiation;Disease variant;Reference proteome;Repeat;Spermatogenesis;TPR repeat | hsa:199223; | cilium [GO:0005929]; intraciliary transport particle A [GO:0030991]; flagellated sperm motility [GO:0030317]; intraciliary retrograde transport [GO:0035721]; protein localization to cilium [GO:0061512]; spermatid development [GO:0007286] | 30929735_bi-allelic mutations in TTC21A can induce asthenoteratospermia with defects of the sperm flagella and head-tail conjunction 31812070_Increased TTC21A expression in lung adenocarcinoma tissues is significantly correlated with pathological stage, tumor status and lymph nodes. Up-regulated TTC21A expression, negative results of pathological stage and distant metastasis are independent prognostic factors for good prognosis. | ENSMUSG00000032514 | Ttc21a | 77.351782 | 0.8951690 | -0.159768079 | 0.34860936 | 2.103825e-01 | 6.464678e-01 | No | Yes | 64.638982 | 20.109121 | 76.153784 | 23.654086 | |||
| ENSG00000168461 | 11031 | RAB31 | protein_coding | Q13636 | FUNCTION: The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Rabs cycle between an inactive GDP-bound form and an active GTP-bound form that is able to recruit to membranes different set of downstream effectors directly responsible for vesicle formation, movement, tethering and fusion. Required for the integrity and for normal function of the Golgi apparatus and the trans-Golgi network. Plays a role in insulin-stimulated translocation of GLUT4 to the cell membrane. Plays a role in M6PR transport from the trans-Golgi network to endosomes. Plays a role in the internalization of EGFR from the cell membrane into endosomes. Plays a role in the maturation of phagosomes that engulf pathogens, such as S.aureus and M.tuberculosis. {ECO:0000269|PubMed:17189207, ECO:0000269|PubMed:17678623, ECO:0000269|PubMed:19725050, ECO:0000269|PubMed:21255211, ECO:0000269|PubMed:21586568}. | 3D-structure;Cytoplasmic vesicle;Endosome;GTP-binding;Golgi apparatus;Lipoprotein;Membrane;Nucleotide-binding;Phosphoprotein;Prenylation;Reference proteome | Small GTP-binding proteins of the RAB family, such as RAB31, play essential roles in vesicle and granule targeting (Bao et al., 2002 [PubMed 11784320]).[supplied by OMIM, Jul 2009]. | hsa:11031; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; early phagosome membrane [GO:0036186]; endomembrane system [GO:0012505]; phagocytic cup [GO:0001891]; phagocytic vesicle [GO:0045335]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; trans-Golgi network membrane [GO:0032588]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; cellular response to insulin stimulus [GO:0032869]; Golgi to plasma membrane protein transport [GO:0043001]; intracellular protein transport [GO:0006886]; phagosome maturation [GO:0090382]; positive regulation of phagocytosis, engulfment [GO:0060100]; receptor internalization [GO:0031623]; regulated exocytosis [GO:0045055] | 17678623_Thus, Rab22B may have a role in anterograde exit from the trans-Golgi network . 17952591_uPAR-del4/5 and rab31 mRNA represent independent prognostic markers in breast cancer and may be components of different, but possibly associated, tumor-relevant signaling pathways. 19725050_Rab22B possibly plays role in regulating epidermal growth factor receptor (EGFR) and mannose 6-phosphate receptor (M6PR) transport into late endosomes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21586568_RIN3 specifically acts as a GEF for Rab31. 21848504_Neither UPARwt, nor UPAR-del4/5 nor RAB31 mRNA expression levels were found to be prognostic markers in advanced ovarian cancer. 22792175_MUC1-C and Rab31 function in an autoinductive loop that contributes to overexpression of MUC1-C in breast cancer cells. 22920728_Overexpression of rab31 in breast cancer cells leads to a switch from an invasive to a proliferative phenotype as indicated by an increased cell proliferation, reduced adhesion and invasion in vitro, and a reduced capacity to form lung metastases in vivo. 24644286_Data indicate that early endosome antigen 1 (EEA1) is important for the small GTPase Rab31-mediated enhancement of ligand-bound EGF receptor (EGFR) endocytic trafficking. 26044564_this study provides the evidence of Rab31 overexpression in HCC, and Rab31 is potentially used as a novel biomarker of poor prognosis in patients with HCC. PI3K/AKT/Bcl-2/BAX axis was involved in Rab31-promoting HCC progression. 26245486_the role of Rab31 in cancer progression, is reported. 30120935_The impaired EGFR signaling and its effects on epidermal differentiation were also observed in familial AI patients and Ncstn(DeltaKC) mice. Thus, our study showed that miR-30a-3p/RAB31/EGFR signaling pathway may play a key role in the pathogenesis of familial AI with NCSTN mutations. 30182384_Findings identified a breast cancer (BC) susceptibility SNP, rs6506689 G>T in RAB31. Functional analysis of this germline mutation suggested that the rs6506689-T allele may increase FOXA1-binding affinity and up-regulates the expression of RAB31, ultimately promoting the development of BC. 32707053_Upregulation of Rab31 is associated with poor prognosis and promotes colorectal carcinoma proliferation via the mTOR/p70S6K/Cyclin D1 signalling pathway. 32958903_RAB31 marks and controls an ESCRT-independent exosome pathway. 33469675_High expression of Rab31 confers a poor prognosis and enhances cell proliferation and invasion in oral squamous cell carcinoma. 34296983_Shigella escapes lysosomal degradation through inactivation of Rab31 by IpaH4.5. 34906074_Rab31-dependent regulation of transforming growth factor ss expression in breast cancer cells. 35072529_LncRNA HOXA10-AS Activated by E2F1 Facilitates Proliferation and Migration of Nasopharyngeal Carcinoma Cells Through Sponging miR-582-3p to Upregulate RAB31. | ENSMUSG00000056515 | Rab31 | 1314.816331 | 0.9349383 | -0.097056921 | 0.08723637 | 1.231238e+00 | 2.671664e-01 | 6.399534e-01 | No | Yes | 1356.177154 | 255.357266 | 1441.094481 | 271.340041 | |
| ENSG00000168502 | 23255 | MTCL1 | protein_coding | Q9Y4B5 | FUNCTION: Microtubule-associated factor involved in the late phase of epithelial polarization and microtubule dynamics regulation. Plays a role in the development and maintenance of non-centrosomal microtubule bundles at the lateral membrane in polarized epithelial cells. {ECO:0000269|PubMed:23902687}. | Alternative splicing;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Membrane;Phosphoprotein;Reference proteome | hsa:23255; | apical plasma membrane [GO:0016324]; apicolateral plasma membrane [GO:0016327]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular space [GO:0005615]; lateral plasma membrane [GO:0016328]; microtubule bundle [GO:0097427]; midbody [GO:0030496]; spindle pole [GO:0000922]; microtubule binding [GO:0008017]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; establishment or maintenance of epithelial cell apical/basal polarity [GO:0045197]; microtubule bundle formation [GO:0001578]; positive regulation of protein targeting to membrane [GO:0090314]; regulation of autophagy [GO:0010506] | 18649358_Observational study of gene-disease association. (HuGE Navigator) 25366663_MTCL1 cooperates with CLASPs and AKAP450/CG-NAP in the formation of the Golgi-derived microtubules 27521566_results suggest that the PPP2R5E phosphatase may contribute to microtubule organization by stabilizing MTCL1. 28787032_microtubule-regulating activity of microtubule crosslinking factor 1 30548255_We propose MTCL1 as a candidate gene for autosomal recessive cerebellar ataxia in humans. In addition, our study confirms the high diagnostic yield of NGS in early-onset cerebellar ataxias, with at least 50% detection rate in our ataxia cohort. 33587225_SOGA1 and SOGA2/MTCL1 are CLASP-interacting proteins required for faithful chromosome segregation in human cells. | ENSMUSG00000052105 | Mtcl1 | 470.662077 | 1.1343289 | 0.181838955 | 0.14467405 | 1.607877e+00 | 2.047905e-01 | 5.757778e-01 | No | Yes | 415.056115 | 50.193226 | 369.239725 | 44.733565 | ||
| ENSG00000168734 | 11142 | PKIG | protein_coding | Q9Y2B9 | FUNCTION: Extremely potent competitive inhibitor of cAMP-dependent protein kinase activity, this protein interacts with the catalytic subunit of the enzyme after the cAMP-induced dissociation of its regulatory chains. {ECO:0000250}. | Protein kinase inhibitor;Reference proteome | This gene encodes a member of the protein kinase inhibitor family. Studies of a similar protein in mice suggest that this protein acts as a potent competitive cAMP-dependent protein kinase inhibitor, and is a predominant form of inhibitor in various tissues. The encoded protein may be involved in osteogenesis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:11142; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; cAMP-dependent protein kinase inhibitor activity [GO:0004862]; negative regulation of cAMP-dependent protein kinase activity [GO:2000480]; negative regulation of protein import into nucleus [GO:0042308]; negative regulation of transcription by RNA polymerase II [GO:0000122] | 9218452_Functional studies of the mouse homolog 11742798_Functional studies of the mouse homolog 16870489_These results indicate that the downregulation of PKIgamma may be prerequisite for the PKA activation during the osteoblastic differentiation of precursor cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23963683_Results show that endogenous levels of Pkig reciprocally regulate osteoblast and adipocyte differentiation and that this reciprocal regulation is mediated in part by LIF. | ENSMUSG00000035268 | Pkig | 357.594280 | 0.9493509 | -0.074986633 | 0.14569785 | 2.622793e-01 | 6.085585e-01 | No | Yes | 336.575319 | 34.150039 | 353.021215 | 35.690859 | ||
| ENSG00000168970 | 8681 | JMJD7-PLA2G4B | protein_coding | P0C869 | FUNCTION: Calcium-dependent phospholipase A1 and A2 and lysophospholipase that may play a role in membrane phospholipid remodeling. {ECO:0000269|PubMed:10085124, ECO:0000269|PubMed:10358058, ECO:0000269|PubMed:16617059}.; FUNCTION: [Isoform 3]: Calcium-dependent phospholipase A2 and lysophospholipase. Cleaves the ester bond of the fatty acyl group attached to the sn-2 position of phosphatidylethanolamines, producing lysophospholipids that may be used in deacylation-reacylation cycles. Hydrolyzes lysophosphatidylcholines with low efficiency but is inefficient toward phosphatidylcholines. {ECO:0000269|PubMed:16617059}.; FUNCTION: [Isoform 5]: Calcium-dependent phospholipase A1 and A2 and lysophospholipase. Cleaves the ester bond of the fatty acyl group attached to the sn-1 or sn-2 position of diacyl phospholipids (phospholipase A1 and A2 activity, respectively), producing lysophospholipids that may be used in deacylation-reacylation cycles. Can further hydrolyze lysophospholipids enabling complete deacylation. Has no activity toward alkylacyl phospholipids. {ECO:0000269|PubMed:10085124, ECO:0000269|PubMed:10358058, ECO:0000269|PubMed:16617059}. | Alternative splicing;Calcium;Cytoplasm;Endosome;Hydrolase;Lipid degradation;Lipid metabolism;Membrane;Metal-binding;Mitochondrion;Reference proteome | Mouse_homologues NA; + ;NA | This locus represents naturally-occurring readthrough transcription between the neighboring jumonji domain containing 7 (JMJD7) and phospholipase A2, group IVB (cytosolic) (PLA2G4B) genes. Readthrough transcripts encode fusion proteins that share amino acid sequence with each individual gene product, including a partial JmjC domain and downstream C2 and phospholipase A2 domains. Alternatively spliced transcript variants have been observed. [provided by RefSeq, Oct 2013]. | hsa:100137049;hsa:8681; | cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extracellular region [GO:0005576]; mitochondrial inner membrane [GO:0005743]; calcium ion binding [GO:0005509]; calcium-dependent phospholipase A2 activity [GO:0047498]; calcium-dependent phospholipid binding [GO:0005544]; lysophospholipase activity [GO:0004622]; phosphatidyl phospholipase B activity [GO:0102545]; phospholipase A1 activity [GO:0008970]; phospholipase A2 activity [GO:0004623]; arachidonic acid metabolic process [GO:0019369]; calcium-mediated signaling [GO:0019722]; glycerophospholipid catabolic process [GO:0046475]; inflammatory response [GO:0006954]; parturition [GO:0007567]; phosphatidylcholine acyl-chain remodeling [GO:0036151]; phosphatidylethanolamine acyl-chain remodeling [GO:0036152]; phosphatidylglycerol acyl-chain remodeling [GO:0036148] | 11741884_circulating human neutrophils express groups V and X sPLA(2) (GV and GX sPLA(2)) mRNA and contain GV and GX sPLA(2) proteins, whereas GIB, GIIA, GIID, GIIE, GIIF, GIII, and GXII sPLA(2)s are undetectable 15274049_Observational study of gene-disease association. (HuGE Navigator) 15999343_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000098488+ENSMUSG00000033852 | Pla2g4b+Gm28042 | 238.856971 | 1.3375124 | 0.419552214 | 0.19196968 | 4.804319e+00 | 2.838849e-02 | No | Yes | 247.005215 | 34.109260 | 188.933463 | 26.324544 | |
| ENSG00000169062 | 65110 | UPF3A | protein_coding | Q9H1J1 | FUNCTION: Involved in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons by associating with the nuclear exon junction complex (EJC) and serving as link between the EJC core and NMD machinery. Recruits UPF2 at the cytoplasmic side of the nuclear envelope and the subsequent formation of an UPF1-UPF2-UPF3 surveillance complex (including UPF1 bound to release factors at the stalled ribosome) is believed to activate NMD. However, UPF3A is shown to be only marginally active in NMD as compared to UPF3B. Binds spliced mRNA upstream of exon-exon junctions. In vitro, weakly stimulates translation. {ECO:0000269|PubMed:11163187, ECO:0000269|PubMed:16601204}. | 3D-structure;Alternative splicing;Cytoplasm;Nonsense-mediated mRNA decay;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transport;mRNA transport | hsa:65110; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; exon-exon junction complex [GO:0035145]; intracellular membrane-bounded organelle [GO:0043231]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA binding [GO:0003723]; structural constituent of nuclear pore [GO:0017056]; telomeric DNA binding [GO:0042162]; mRNA transport [GO:0051028]; nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:0000184]; positive regulation of translation [GO:0045727] | Mouse_homologues 27040500_Analysis revealed that UPF3A is a broadly acting nonsense-mediated RNA decay inhibitor. This discovery implies that UPF3A and UPF3B do not primarily work in a complementary or redundant manner as previously supposed; instead, they oppose each other, allowing this paralog pair to serve as a molecular rheostat to modulate the level of gene expression during development. | ENSMUSG00000038398 | Upf3a | 591.886117 | 1.2273033 | 0.295491800 | 0.11306489 | 6.862776e+00 | 8.800972e-03 | 1.216184e-01 | No | Yes | 565.283862 | 72.083871 | 459.021903 | 58.666043 | ||
| ENSG00000169249 | 8233 | ZRSR2 | protein_coding | Q15696 | FUNCTION: Pre-mRNA-binding protein required for splicing of both U2- and U12-type introns. Selectively interacts with the 3'-splice site of U2- and U12-type pre-mRNAs and promotes different steps in U2 and U12 intron splicing. Recruited to U12 pre-mRNAs in an ATP-dependent manner and is required for assembly of the prespliceosome, a precursor to other spliceosomal complexes. For U2-type introns, it is selectively and specifically required for the second step of splicing. {ECO:0000269|PubMed:21041408, ECO:0000269|PubMed:9237760}. | Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein;Spliceosome;Ubl conjugation;Zinc;Zinc-finger;mRNA processing;mRNA splicing | This gene encodes an essential splicing factor. The encoded protein associates with the U2 auxiliary factor heterodimer, which is required for the recognition of a functional 3' splice site in pre-mRNA splicing, and may play a role in network interactions during spliceosome assembly. [provided by RefSeq, Jul 2008]. | hsa:8233; | nucleoplasm [GO:0005654]; spliceosomal complex [GO:0005681]; U12-type spliceosomal complex [GO:0005689]; U2AF complex [GO:0089701]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; pre-mRNA 3'-splice site binding [GO:0030628]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380]; spliceosomal complex assembly [GO:0000245] | 19204726_Observational study of gene-disease association. (HuGE Navigator) 21041408_Data show that through recognition of a common splicing element, Urp facilitates distinct steps of U2- and U12-type intron splicing. 22389253_In univariate analysis, mutated SRSF2 predicted shorter overall survival and more frequent acute myeloid leukemia progression compared with wild-type SRSF2, whereas mutated U2AF1, ZRSR2 had no impact on patient outcome. 25586593_ZRSR2 has a role in RNA splicing; dysregulated splicing of U12-type introns is a characteristic feature of ZRSR2 mutations in myelodysplastic syndrome 25964599_The mutational status of the SRSF2, U2AF1 and ZRSR2 did not affect the response rate or survival in MDS patients who had received first-line decitabine treatment. 28942350_We conclude that the common clinical features of patients with an isolated mutation of ZRSR2 are a macrocytic anemia without leukopenia, thrombocytopenia or an increase in marrow blast percentage 31124956_This meta-analysis indicates a positive effect of SF3B1 and an adverse prognostic effect of SRSF2, U2AF1, and ZRSR2 mutations in patients with myelodysplastic syndrome. 31361176_ZRSR2 mutation in a child with refractory macrocytic anemia and Down Syndrome. 33568749_ZRSR2 overexpression is a frequent and early event in castration-resistant prostate cancer development. | ENSMUSG00000044068 | Zrsr1 | 333.779019 | 1.2757614 | 0.351358521 | 0.15586813 | 5.011632e+00 | 2.517756e-02 | No | Yes | 373.901552 | 40.764639 | 299.410021 | 32.845728 | ||
| ENSG00000169570 | 285605 | DTWD2 | protein_coding | Q8NBA8 | FUNCTION: Catalyzes the formation of 3-(3-amino-3-carboxypropyl)uridine (acp3U) at position 20a in the D-loop of several cytoplasmic tRNAs (acp3U(20a)) (PubMed:31804502). Also has a weak activity to form acp3U at position 20 in the D-loop of tRNAs (acp3U(20)) (PubMed:31804502). {ECO:0000269|PubMed:31804502}. | Acetylation;Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase;tRNA processing | hsa:285605; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; tRNA-uridine aminocarboxypropyltransferase activity [GO:0016432]; tRNA modification [GO:0006400] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 23953852_The rs17144687 near DTWD2 associated with MaxDirnks. | ENSMUSG00000024505 | Dtwd2 | 496.357966 | 1.0870382 | 0.120402691 | 0.14012126 | 7.301054e-01 | 3.928490e-01 | 7.378278e-01 | No | Yes | 567.666249 | 141.867084 | 495.479679 | 123.846749 | ||
| ENSG00000170166 | 3233 | HOXD4 | protein_coding | P09016 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene belongs to the homeobox family of genes. The homeobox genes encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, located on different chromosomes, consisting of 9 to 11 genes arranged in tandem. This gene is one of several homeobox HOXD genes located at 2q31-2q37 chromosome regions. Deletions that removed the entire HOXD gene cluster or 5' end of this cluster have been associated with severe limb and genital abnormalities. The protein encoded by this gene may play a role in determining positional values in developing limb buds. Alternatively spliced variants have been described but their full length nature has not been determined. [provided by RefSeq, Jul 2008]. | hsa:3233; | cell junction [GO:0030054]; chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; embryonic organ development [GO:0048568]; embryonic skeletal system morphogenesis [GO:0048704]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 15776434_Observational study of gene-disease association. (HuGE Navigator) 19232136_microRNA-10a (miR-10a) targets a homologous DNA region in the promoter region of the hoxd4 gene and represses its expression at the transcriptional level 19453261_Observational study of gene-disease association. (HuGE Navigator) 19938081_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20686522_Observational study of gene-disease association. (HuGE Navigator) 20686522_The results suggest that genetic polymorphisms in HOXD4 may not be a major contributor to the observed variability in peak BMD in the lumbar spine and the hip in Chinese men and women 26545093_identified heterozygous HOXD4 loss-of-function mutations in three subjects with spinal extradural arachnoid cyst (SEDAC) | ENSMUSG00000101174 | Hoxd4 | 39.647217 | 1.4229563 | 0.508891341 | 0.42780989 | 1.407733e+00 | 2.354332e-01 | No | Yes | 43.425151 | 12.039998 | 29.715681 | 8.429352 | ||
| ENSG00000170248 | 10015 | PDCD6IP | protein_coding | Q8WUM4 | FUNCTION: Multifunctional protein involved in endocytosis, multivesicular body biogenesis, membrane repair, cytokinesis, apoptosis and maintenance of tight junction integrity. Class E VPS protein involved in concentration and sorting of cargo proteins of the multivesicular body (MVB) for incorporation into intralumenal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome. Binds to the phospholipid lysobisphosphatidic acid (LBPA) which is abundant in MVBs internal membranes. The MVB pathway requires the sequential function of ESCRT-O, -I,-II and -III complexes (PubMed:14739459). The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis (PubMed:17853893, PubMed:17556548). Adapter for a subset of ESCRT-III proteins, such as CHMP4, to function at distinct membranes. Required for completion of cytokinesis (PubMed:17853893, PubMed:17556548, PubMed:18641129). May play a role in the regulation of both apoptosis and cell proliferation. Regulates exosome biogenesis in concert with SDC1/4 and SDCBP (PubMed:22660413). By interacting with F-actin, PARD3 and TJP1 secures the proper assembly and positioning of actomyosin-tight junction complex at the apical sides of adjacent epithelial cells that defines a spatial membrane domain essential for the maintenance of epithelial cell polarity and barrier (By similarity). {ECO:0000250|UniProtKB:Q9WU78, ECO:0000269|PubMed:14739459, ECO:0000269|PubMed:17556548, ECO:0000269|PubMed:17853893, ECO:0000269|PubMed:18641129, ECO:0000269|PubMed:22660413}.; FUNCTION: (Microbial infection) Involved in HIV-1 virus budding. Can replace TSG101 it its role of supporting HIV-1 release; this function requires the interaction with CHMP4B. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as enveloped virus budding (HIV-1 and other lentiviruses). {ECO:0000269|PubMed:14505569, ECO:0000269|PubMed:14505570, ECO:0000269|PubMed:14519844, ECO:0000269|PubMed:17556548, ECO:0000269|PubMed:18641129}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Cell cycle;Cell division;Cell junction;Cytoplasm;Cytoskeleton;Direct protein sequencing;Host-virus interaction;Methylation;Phosphoprotein;Protein transport;Reference proteome;Secreted;Tight junction;Transport | This gene encodes a protein that functions within the ESCRT pathway in the abscission stage of cytokinesis, in intralumenal endosomal vesicle formation, and in enveloped virus budding. Studies using mouse cells have shown that overexpression of this protein can block apoptosis. In addition, the product of this gene binds to the product of the PDCD6 gene, a protein required for apoptosis, in a calcium-dependent manner. This gene product also binds to endophilins, proteins that regulate membrane shape during endocytosis. Overexpression of this gene product and endophilins results in cytoplasmic vacuolization, which may be partly responsible for the protection against cell death. Several alternatively spliced transcript variants encoding different isoforms have been found for this gene. Related pseudogenes have been identified on chromosome 15. [provided by RefSeq, Jan 2012]. | hsa:10015; | actomyosin [GO:0042641]; bicellular tight junction [GO:0005923]; cytosol [GO:0005829]; endoplasmic reticulum exit site [GO:0070971]; extracellular exosome [GO:0070062]; extracellular vesicle [GO:1903561]; Flemming body [GO:0090543]; focal adhesion [GO:0005925]; immunological synapse [GO:0001772]; intracellular membrane-bounded organelle [GO:0043231]; melanosome [GO:0042470]; membrane [GO:0016020]; microtubule organizing center [GO:0005815]; calcium-dependent protein binding [GO:0048306]; protein homodimerization activity [GO:0042803]; proteinase activated receptor binding [GO:0031871]; actomyosin contractile ring assembly [GO:0000915]; apoptotic process [GO:0006915]; bicellular tight junction assembly [GO:0070830]; maintenance of epithelial cell apical/basal polarity [GO:0045199]; midbody abscission [GO:0061952]; mitotic cytokinesis [GO:0000281]; multivesicular body assembly [GO:0036258]; positive regulation of exosomal secretion [GO:1903543]; positive regulation of extracellular exosome assembly [GO:1903553]; protein homooligomerization [GO:0051260]; protein transport [GO:0015031]; regulation of centrosome duplication [GO:0010824]; regulation of extracellular exosome assembly [GO:1903551]; regulation of membrane permeability [GO:0090559]; ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway [GO:0090611]; viral budding [GO:0046755]; viral budding via host ESCRT complex [GO:0039702] | 12360406_p95 has roles in regulating cell adhesion and morphology. 12588984_AIP1/Alix interacts with the apoptosis-linked protein ALG-2 and recognizes recognize the protein-protein binding motif YPXL/I, where Tyr, Pro, and Leu/Ile are crucial for its interactive properties 12771190_adaptor protein SETA (CIN85) and its binding partner AIP1 are involved with the cytoskeleton and in the regulation of cell adhesion 12860994_CHMP4b and Alix participate in formation of multivesicular bodies by cooperating with SKD1 14739459_role in control of the formation of multivesicular liposomes induced by lysobisphosphatidic acid (LBPA); regulated the organization of LBPA-containing endosomes in vivo 14999017_The region corresponding to amino acid residues 794 to 827 in the carboxy-terminal proline-rich region of Alix is sufficient to confer the ability to interact directly with ALG2. This region includes four-tandem PxY repeats. 15195070_results suggest that insulin-like growth factor I receptor-induced paraptosis is mediated by MAPKs, and inhibited by AIP-1/Alix 15456872_Alix negatively regulates the endocytic complex containing Cbl and SETA/CIN85, and thereby antagonizes receptor endocytosis. 15557335_Alix is negatively regulated by Src, via direct interaction and phosphorylation. 15849434_Partial colocalization of FLAG-tagged Rab GTPase-activating protein-like protein and green fluorescent protein (GFP)-fused Alix was observed at cell edges 15914539_Alix functions in the actin-dependent intracellular positioning of endosomes 16764724_The HIV-1 protein Nef contributes to the release of viral particles from infected cells through its interaction with AIP1. 17014699_Four proteins (TSG101,Hrs,Aip1/Alix, and Vps4B) of the ESCRT (endosomal sorting complex required for transport) machinery were localized in T cells and macrophages by quantitative electron microscopy. 17082185_Alix inhibits down-regulation of PDGFRbeta by modulating the interaction between c-Cbl and the receptor, thereby affecting the ubiquitination of the receptor 17158451_Alix fragment 364-716 is a potent, Alix binding site-specific inhibitor of HIV-1 assembly and release. 17166905_A YLDL sequence within the M protein showed L-domain activity, and its specific interaction with the N terminus of Alix/AIP1(1-211) was important for the budding of virus-like particles (VLPs) of M protein. 17250865_We also identified a host protein, AlP1/Alix, involved in apoptosis and efficient budding of several enveloped viruses as an interacting partner of the V and NP proteins. 17277784_Overexpression of the V domain inhibits HIV-1 release from cells. This inhibition of release is reversed by mutations that block binding of the Alix V domain to p6. 17350572_Crystal structure; ALIX serves as a flexible, extended scaffold that connects retroviral Gag proteins to ESCRT-III and other cellular-budding machinery 17389591_TNF-induced TRAF2-RIP1-AIP1-ASK1 complex formation and for the activation of ASK1-JNK/p38 apoptotic signaling. 17428861_ALIX can have a dramatic effect on HIV-1 release by binding at the CHMP4B site; the ability to use ALIX may allow HIV-1 to replicate in cells that express only low levels of Tsg101 17556548_study shows that two proteins involved in HIV-1 budding-Tsg101, a subunit of the endosomal sorting complex required for transport I (ESCRT-I), & Alix, an ESCRT-associated protein-were recruited to the midbody during cytokinesis by interaction with Cep55 17601348_Here, we report that dominant negative forms of Vps4A, Vps4B, and AIP1 inhibit HTLV-1 budding. 17673164_These results eliminate the possibility that the two transcript variants encode different isoforms of Alix protein and suggest that alternative polyadenylation is one of the mechanisms controlling Alix protein expression. 17853893_that ALIX and TSG101/ESCRT-I also bind a series of proteins involved in cytokinesis, including CEP55, CD2AP, ROCK1, and IQGAP1. 18032513_ALIX also binds to the nucleocapsid (NC) domain of HIV-1 Gag and that ALIX and its isolated Bro1 domain can be specifically packaged into viral particles via NC. 18380665_ALG-2-interacting protein 1 up-regulates dopamine D1 and D3 receptor expression and is important for their stability and protein trafficking. 18476810_Study demonstrate that formation or exposure of the p6(Gag)/p9(Gag) docking site in Alix is a regulated event and that Alix association with the membrane may play a positive role in this process. 18477395_C-terminal proline-rich domain of ALIX allows the access of its binding site to p6. 18511562_The Bro1 domain of ALIX binds specifically to C-terminal residues of the human CHMP4 proteins (CHMP4A-C). 18641129_the Cep55/Alix/ESCRT-III pathway has a role in cytokinesis and HIV-1 release 18644787_Alix down-expression decreases both LBPA levels and the lumenal vesicle content of late endosomes. Cellular cholesterol levels are also decreased, presumably because the storage capacity of endosomes is affected and thus cholesterol clearance accelerated 18684393_These results strongly suggest that the capacity of several mutants of Alix to block both caspase-dependent and independent cell death does not relate to their capacity to modulate autophagy. 18936101_Alix and ALG-2 are new actors of the TNF-R1 pathway 18940611_A Ca(2+)/EF3-driven arginine switch mechanism for ALG-2 binding to Alix. 18948538_crystal structure of the ESCRT and ALIX-binding region (EABR) of CEP55 bound to an ALIX peptide at a resolution of 2.0 angstroms; structure shows that EABR forms an aberrant dimeric parallel coiled coil 19016654_The native state of Alix does not have a functional Bro1 domain. 19393081_Results identified ALIX as an ubiquitination substrate of POSH and indicate that POSH and ALIX cooperate to facilitate efficient virus release. 19520058_Ca2+-loaded ALG-2 bridges Alix and TSG101 as an adaptor protein. 19523902_A crescent-shaped Alix dimer targets ESCRT-III CHMB4 filaments. 19596386_The authors determined that the p6-Alix interaction plays an important role in HIV-1 replication. 19692479_Data suggest that HSV-1 production is independent of ALIX and TSG101 expression. 19706535_Data show that galectin-3 is an inhibitory regulator of T-cell activation and functions intracellularly by promoting TCR down-regulation, possibly through modulating Alix's function at the IS. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20519395_Together these data support a model in which Alix recruits Nedd4-1 to facilitate HIV-1 release mediated through the LYPX(n)L/Alix budding pathway via a mechanism that involves Alix ubiquitination. 20605035_The results indicate YLDL motif in M protein is essential for efficient budding in the context of virus infection and suggest involvement of Alix/AIP1 in Sendai virus budding. 20669987_Examined changes in subcellular proteomes of different cellular compartments of human endothelial cells upon DENV2 infection. Double immunofluorescence staining revealed colocalization of Alix with late endosomal lysobisphosphatidic acid (LBPA). 20670214_Identification and biophysical assessment of the molecular recognition mechanisms between the human haemopoietic cell kinase Src homology domain 3 and ALG-2-interacting protein X 20691033_inability of the two-residue shorter ALG-2 isoform to bind Alix 20800603_Observational study of gene-disease association. (HuGE Navigator) 20929444_studies on intramolecular interactions:the relieving of specific, autoinhibitory interactions within ALIX regulates binding with ESCRT proteins or viral proteins and is critical for ALIX to participate in retroviral budding 20962096_Crystal structures revealed that anchoring tyrosines and nearby hydrophobic residues contact the ALIX V domain, revealing how SIV gag proteins employ a diverse family of late-domain sequences to bind ALIX and promote virus budding. 21129143_Data suggest that the boomerang-shaped Bro1 domain of Alix appears to escort hepatitis B virus naked capsids without ESCRT. 21248028_The authors demonstrate that ALIX/AIP1, an ESCRT-associated host protein, is required for the incorporation of the nucleoprotein of Mopeia virus, a close relative of Lassa virus, into Z-induced virus-like particles (VLPs). 21528537_HIV-1 infection affects the expression of host factors TSG101 and Alix 21715492_Mutations designed to destabilize the closed conformation of the V domain opened the V domain, increased ALIX membrane association, and enhanced HIV-1 budding. 21889351_Mutation of residues within the Phe105 loop of the Bro1 domain compromise Alix function in HIV-1 release. 22162750_structural analysis of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding 22641034_Structural recognition mechanisms between human Src homology domain 3 (SH3) and ALG-2-interacting protein X (Alix 22660413_Identify key role for syndecan-syntenin-ALIX in membrane transport and signalling processes. 22771033_At the midbody, BRCA2 influences the recruitment of endosomal sorting complex required for transport (ESCRT)-associated proteins, Alix and Tsg101, and formation of CEP55-Alix and CEP55-Tsg101 complexes during abscission. 22833563_Data indicate that AP-3 facilitates PAR1 interaction with ALIX. 22969426_BFRF1 recruits the ESCRT components to modulate nuclear envelope for the nuclear egress of Epstein Barr virus. 23201121_study reports that the V domain of ALIX binds directly and selectively to K63-linked polyubiquitin chains, exhibiting a strong preference for chains composed of more than three ubiquitins 23527201_Alix serves as an adaptor that allows human parainfluenza virus type 1 to interact with the host cell ESCRT system. 23664863_Lysobisphosphatidic acid recruits ALIX onto late endosomes via the calcium-bound Bro1 domain, triggering a conformational change in ALIX to mediate the delivery of viral nucleocapsids to the cytosol during infection. 23777424_Common genetic variations in PDCD6IP may influence hepatocellular carcinoma risk, possibly through promoter activity-mediated regulation. 23924735_The results of in vitro binding assays using purified recombinant proteins indicated that ALG-2 functions as a Ca(2)-dependent adaptor protein that bridges ALIX and ESCRT-I to form a ternary complex 24287454_ALIX regulates these mammalian cell-specific cytokinesis, exosome release, and virus budding. [Review] 24637612_Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. 24712823_Alix protein plays a critical role in the maintenance of the barrier function of T84 monolayers 24834918_ALIX is recruited to the neck of the assembling HIV-1 virion and is mostly recycled after virion release. 24870593_Results suggest that programmed cell death 6 interacting protein (PDCD6IP) insertion/deletion polymorphism was potentially related to non-small cell lung cancer (NSCLC) susceptibility in Chinese Han population. 25118280_HIV-1 Nef interacts with Alix in late endosomes, and this is required for efficient lysosomal targeting of CD4. 25451933_Aip1 has a role in actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring 25502766_The serum lever of Alzheimer's disease were decrease and the expression of ALIX strongly correlated with the Mini-Mental State Examination scores of the AD patients 25510652_Alix is critically involved in multivesicular body sorting of membrane receptors in mammalian cells. 25534348_Lack of ALG-2, ALIX or Vps4B each prevents shedding, and repair of the injured cell membrane 25732677_our findings identify heparanase as a modulator of the syndecan-syntenin-ALIX pathway, fostering endosomal membrane budding and the biogenesis of exosomes by trimming the heparan sulfate chains on syndecans 26063962_Findings indicate that the PDCD6IP 15bp insertion/deletion polymorphism decreases the risk of breast neoplasm in an Iranian population. 26139244_Our data reveal that AIP1, by inhibiting VEGFR2-dependent signaling in tumor niche, suppresses tumor EMT switch, tumor angiogenesis, and tumor premetastatic niche formation to limit tumor growth and metastasis. 26490116_We found that ARRDC3 is required for ALIX ubiquitination induced by activation of PAR1 26859355_phosphorylation of the intramolecular interaction site in the PRD is one of the major mechanisms that activates the ESCRT function of ALIX 26866605_homologous domain of human Bro1 domain-containing proteins, Alix and Brox, binds CHMP4B but not STAM2, despite their high structural similarity 26929449_Accordingly, ALIX depletion leads to furrow regression in cells with chromosome bridges, a phenotype associated with abscission checkpoint signaling failure. 26935291_These findings indicate that Alix binds to Ago2 and miRNAs, suggesting that it plays a key role in miRNA enrichment during extracellular vesicles biogenesis. 26962944_The authors find that HIV-1 nucleocapsid mimics the PDZ domains of syntenin, a membrane-binding adaptor involved in cell-to-cell contact/communication, to capture the Bro1 domain of ALIX, which is an ESCRTs recruiting cellular adaptor. 26980041_Alix plays an important role in the proliferation of glioma cells and overexpression in gliomas predicts poor survival. 27150162_Revealed the transition of diffuse ALIX protein signals into a multivesicular body-like pattern during adenoma-carcinoma sequence in colorectal neoplasms. 27244115_Alix acts in concert with endophilin A to promote clathrin-independent endocytosis of cholera toxin and to regulate cell migration. 27301021_ALIX regulates P2Y1 degradation. 27578500_our results identify the CD63-syntenin-1-ALIX complex as a key regulatory component in post-endocytic HPV trafficking. 27909058_farnesylation of K-Ras was required for its packaging within extracellular nanovesicles, yet expressing a K-Ras farnesylation mutant did not decrease the number of nanovesicles or the amount of Alix protein released per cell. 29891975_In summary, these data reveal the functional role of PYRE motif insertion towards the cooperative mechanism of ALIX/Nedd4-1 in HIV-1 release in the absence of PTAP/Tsg101 pathway. 30021161_ALIX is a regulator of both EGFR activity and PD-L1 surface presentation in basal-like breast cancer (BLBC) cells. 30599162_The results showed that ALIX both interacted and co-localized with Dengue virus NS3 protein and that upregulation of ALIX resulted in a significantly increased viral titer, while either siRNA or CRISPR-Cas9 mediated down regulation of ALIX significantly reduced viral production. 30760577_data suggest that PYxE insertion in Gag restores the ability of Gag to bind ALIX and correlates with enhanced viral fitness in the absence or presence of LPV and TAF. The high prevalence and increased replication fitness of the HIV-1C virus with PYxE insertion indicates the clinical importance of these viral variants. 30944935_ALIX knockout and overexpression regulate protein profile in induced pluripotent stem cells-derived exosome. 31138766_analysis of how flotillin-mediated endocytosis and ALIX-syntenin-1-mediated exocytosis protect the cell membrane from damage caused by necroptosis 31159502_Alix serves as a co-factor for the interaction between the E3-ubiquitin ligase NEDD4-1 and the ABC transporter targets, ABCG1 and ABCG4. 31172941_These results, for the first time, indicate that CCL2 mediates ALIX mobilization from F-actin and enhances Gag-p6 mediated HIV-1 release and fitness. 32049272_ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. 32213612_Alix-Mediated Rescue of Feline Immunodeficiency Virus Budding Differs from That Observed with Human Immunodeficiency Virus. 32286682_PDCD6IP, encoding a regulator of the ESCRT complex, is mutated in microcephaly. 32321914_Direct interactions between ALIX, syntenin and syndecan-4 are essential for proper enrichment of the ESCRT-III machinery at the cytokinetic abscission site. 32731849_Alix and Syntenin-1 direct amyloid precursor protein trafficking into extracellular vesicles. 32917811_Proline-rich domain of human ALIX contains multiple TSG101-UEV interaction sites and forms phosphorylation-mediated reversible amyloids. 33058236_Occludin, caveolin-1, and Alix form a multi-protein complex and regulate HIV-1 infection of brain pericytes. 33139753_ESCRT-III controls nuclear envelope deformation induced by progerin. 34287046_Structural Insight into the Interaction of Sendai Virus C Protein with Alix To Stimulate Viral Budding. 34688656_Mechanistic roles of tyrosine phosphorylation in reversible amyloids, autoinhibition, and endosomal membrane association of ALIX. 34851141_Roles of ESCRT Proteins ALIX and CHMP4A and Their Interplay with Interferon-Stimulated Gene 15 during Tick-Borne Flavivirus Infection. | 7744.671079 | 1.0718592 | 0.100115406 | 0.04977173 | 4.023199e+00 | 4.487852e-02 | 2.855436e-01 | No | Yes | 8380.780420 | 1844.620845 | 7812.948608 | 1719.535668 | |||
| ENSG00000170364 | 6419 | SETMAR | protein_coding | Q53H47 | FUNCTION: Protein derived from the fusion of a methylase with the transposase of an Hsmar1 transposon that plays a role in DNA double-strand break repair, stalled replication fork restart and DNA integration. DNA-binding protein, it is indirectly recruited to sites of DNA damage through protein-protein interactions. Has also kept a sequence-specific DNA-binding activity recognizing the 19-mer core of the 5'-terminal inverted repeats (TIRs) of the Hsmar1 element and displays a DNA nicking and end joining activity (PubMed:16332963, PubMed:16672366, PubMed:17877369, PubMed:17403897, PubMed:18263876, PubMed:22231448, PubMed:24573677, PubMed:20521842). In parallel, has a histone methyltransferase activity and methylates 'Lys-4' and 'Lys-36' of histone H3. Specifically mediates dimethylation of H3 'Lys-36' at sites of DNA double-strand break and may recruit proteins required for efficient DSB repair through non-homologous end-joining (PubMed:16332963, PubMed:21187428, PubMed:22231448). Also regulates replication fork processing, promoting replication fork restart and regulating DNA decatenation through stimulation of the topoisomerase activity of TOP2A (PubMed:18790802, PubMed:20457750). {ECO:0000269|PubMed:16332963, ECO:0000269|PubMed:16672366, ECO:0000269|PubMed:17403897, ECO:0000269|PubMed:17877369, ECO:0000269|PubMed:18790802, ECO:0000269|PubMed:20457750, ECO:0000269|PubMed:20521842, ECO:0000269|PubMed:21187428, ECO:0000269|PubMed:22231448, ECO:0000269|PubMed:24573677, ECO:0000303|PubMed:18263876}. | 3D-structure;Alternative splicing;Chromatin regulator;Chromosome;DNA damage;DNA repair;DNA-binding;Endonuclease;Hydrolase;Magnesium;Metal-binding;Methylation;Methyltransferase;Multifunctional enzyme;Nuclease;Nucleus;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase;Zinc | This gene encodes a fusion protein that contains an N-terminal histone-lysine N-methyltransferase domain and a C-terminal mariner transposase domain. The encoded protein binds DNA and functions in DNA repair activities including non-homologous end joining and double strand break repair. The SET domain portion of this protein specifically methylates histone H3 lysines 4 and 36. This gene exists as a fusion gene only in anthropoid primates, other organisms lack mariner transposase domain. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jan 2013]. | hsa:6419; | nucleolus [GO:0005730]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; DNA binding [GO:0003677]; DNA topoisomerase binding [GO:0044547]; double-stranded DNA binding [GO:0003690]; endonuclease activity [GO:0004519]; histone methyltransferase activity (H3-K36 specific) [GO:0046975]; histone methyltransferase activity (H3-K4 specific) [GO:0042800]; protein homodimerization activity [GO:0042803]; single-stranded DNA binding [GO:0003697]; single-stranded DNA endodeoxyribonuclease activity [GO:0000014]; zinc ion binding [GO:0008270]; cell population proliferation [GO:0008283]; chromatin organization [GO:0006325]; DNA catabolic process, endonucleolytic [GO:0000737]; DNA double-strand break processing [GO:0000729]; DNA integration [GO:0015074]; double-strand break repair via nonhomologous end joining [GO:0006303]; histone H3-K36 dimethylation [GO:0097676]; histone H3-K36 methylation [GO:0010452]; histone H3-K4 methylation [GO:0051568]; mitotic DNA integrity checkpoint signaling [GO:0044774]; negative regulation of chromosome organization [GO:2001251]; nucleic acid phosphodiester bond hydrolysis [GO:0090305]; positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity [GO:2000373]; positive regulation of double-strand break repair via nonhomologous end joining [GO:2001034]; replication fork processing [GO:0031297] | 16332963_Metnase is a nonhomologous end-joining repair protein that regulates genomic integration of exogenous DNA and establishes a relationship among histone modification, DNA repair, and integration. 16989604_These data suggest that vectors based on the Himar1 transposable element, in conjunction with the hyperactive mutant transposase C9, may be suitable vectors for gene therapy applications. 17130240_SETMAR is unlikely to catalyze transposition in the human genome, although the nicking activity may have a role in the DNA repair phenotype. 17403897_The activities of the SETMAR protein on transposon ends are described. 17877369_Results suggest that Metnase's DNA cleavage activity, unlike those of other eukaryotic transposases, is not coupled to its sequence-specific DNA binding. 18263876_hPso4 is necessary to bring Metnase to the DSB sites for its function(s) in DNA repair 18790802_Metnase physically interacts and co-localizes with Topoisomerase IIalpha, the key chromosome decatenating enzyme. 19458360_myeloid leukemia cells fail to arrest at the mitotic decatenation checkpoint, and their progression through this checkpoint is regulated by the DNA repair component Metnase (also termed SETMAR) 20416268_hPso4, once it forms a complex with Metnase, negatively regulates Metnase's TIR binding activity 20457750_results establish Metnase as a key factor that promotes restart of stalled replication forks, and implicate Metnase in the repair of collapsed forks. 20800603_Observational study of gene-disease association. (HuGE Navigator) 21124928_Data show that DBN1, SETMAR and HIG2 are direct transcriptional targets of the SOX11 protein. 21187428_DNA repair protein Metnase (also SETMAR), which has a SET histone methylase domain, localized to an induced DSB and directly mediated the formation of H3K36me2 near the induced DSB 21491884_a role for Metnase's endonuclease activity in promoting the joining of noncompatible ends 22231448_phosphorylation of Metnase S495 differentiates between these two functions, enhancing DSB repair and repressing replication fork restart. 24573677_a single mutation DDN(610) --> DDD(610), which restores the ancestral catalytic site, results in loss of function in Metnase 24607956_found known and novel SETMAR splice variants to be significantly increased in acute myeloid leukemia 24655462_293 T transfected with Metnase revealed a large number of rescued plasmids. 25333365_Metnase may possess an important role in DNA repair, topoisomerase II function, and the maintenance of stemness during colon cancer development. 25795785_methylation of snRNP70 by SETMAR regulates constitutive and/or alternative splicing 26437079_The SET domain is needed for the 5' end of ss-overhang cleavage with fork and non-fork DNA without affecting the Metnase-DNA interaction. This domain has a positive role in restart of replication fork and the 5' end of ss-overhang cleavage. 27974460_These results suggest that Metnase enhances Exo1-mediated exonuclease activity on the lagging strand DNA by facilitating Exo1 loading onto a single strand gap at the stalled replication fork. 28038463_Various SETMAR proteins can be synthesized in human glioblastoma that may each have specific biophysical and/or biochemical properties and characteristics. 30329085_ur data is consistent with a model in which SETMAR is part of an anthropoid primate-specific regulatory network centered on the subset of genes containing a transposon end. 31238295_The roles of the human SETMAR protein in illegitimate DNA recombination and non-homologous end joining repair were studied. Contrary to previous reports, it was found that wild type SETMAR had little to no effect on the rate of cell division, DNA integration into the genome or non-homologous end joining. 33621919_Mutation and expression alterations of histone methylation-related NSD2, KDM2B and SETMAR genes in colon cancers. 33812898_Two repeated motifs enriched within some enhancers and origins of replication are bound by SETMAR isoforms in human colon cells. | 453.743914 | 0.9582561 | -0.061516829 | 0.13844408 | 1.953276e-01 | 6.585190e-01 | No | Yes | 451.829194 | 96.077137 | 473.359210 | 100.580815 | ||||
| ENSG00000170634 | 98 | ACYP2 | protein_coding | P14621 | FUNCTION: Its physiological role is not yet clear. | Acetylation;Direct protein sequencing;Hydrolase;Phosphoprotein;Reference proteome | Acylphosphatase can hydrolyze the phosphoenzyme intermediate of different membrane pumps, particularly the Ca2+/Mg2+-ATPase from sarcoplasmic reticulum of skeletal muscle. Two isoenzymes have been isolated, called muscle acylphosphatase and erythrocyte acylphosphatase on the basis of their tissue localization. This gene encodes the muscle-type isoform (MT). An increase of the MT isoform is associated with muscle differentiation. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2016]. | hsa:98; | acylphosphatase activity [GO:0003998]; identical protein binding [GO:0042802]; phosphate-containing compound metabolic process [GO:0006796] | 10542090_Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. 11799398_effects of 40 single point mutations on the conversion of the denatured form of the alpha/beta protein acylphosphatase (AcP) into insoluble aggregates 15670608_studies of aggregates formed from human muscle acylphosphatase and disaggregation suggests that amyloid formation occurs in discrete steps whose reversibility is increasingly difficult, and dependent on the size of the aggregates 16084386_Most of the conserved glycine residues in this protein could have been maintained during evolution because of their ability to inhibit amyloid aggregation. 18649358_Observational study of gene-disease association. (HuGE Navigator) 18809411_extensive mutational analysis of aggregation and disaggregation of amyloid-like protofibrils of muscle acylphosphatase 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 22522822_The study used a stopped-flow device coupled to turbidometry detection to monitor the rapid conversion of human muscle acylphosphatase into oligomers with varying heparan sulfate and protein concentrations. 23754389_Enhanced stability is observed upon modifications of a loop region in the enzyme acylphosphatase and is achieved despite significant enthalpy losses. 25665007_The ACYP2 risk variant strongly predisposed these patients to precipitous hearing loss and was related to ototoxicity severity. 25858589_These findings suggest a minor role of the single nucleotide polymorphisms of ACYP2 investigated as genetic determinants of chronic oxaliplatin-induced peripheral neurotoxicity. 26928270_Results shown that the previously reported association of an ACYP2 variant with cisplatin-induced hearing loss in pediatric brain tumor patients could be replicated in an independent cohort of patients with osteosarcoma. Therefor, the ACYP2 variant should be considered a predictor of cisplatin-induced hearing loss in patients with osteosarcoma or other solid tumors 27552709_Data show that the acylphosphatase 2 (ACYP2) gene polymorphism significantly decreased the risk of high altitude pulmonary edema (HAPE). 27686078_the genetic polymorphisms of ACYP2 and TSPYL6 are associated with increased risk of developing ischemic stroke 27894080_Findings indicate significant associations between single nucleotide oolymorphism (SNPs) in the acylphosphatase 2 (ACYP2) gene and breast cancer (BC) risk in a Han Chinese population. 27974682_three SNPs in ACYP2 (rs1682111, rs11896604 and rs843720) associate with lung cancer in the Chinese Han population. 28039478_Genetic polymorphisms in the telomere length-related gene ACYP2 are associated with the risk of colorectal cancer in a Chinese Han population. 28353602_Rs6713088, rs843752, and rs17045754 associated with high-altitude pulmonary edema risk 28415712_Our results indicate that ACYP2 polymorphisms may influence the GC risk and may serve as a new precursory biomarker in the northwest Chinese Han population. 28424424_ACYP2 gene may be associated with an increased risk of esophageal carcinoma in Chinese Han populations. Future studies to address the biological function of this polymorphism in the development of esophageal carcinoma are warranted. 28445188_Results found an association between the ACYP2 rs1872328 polymorphism and cisplatin-induced ototoxicity. 29033240_Minor allele of rs1682111 and rs10439478 within acylphosphatase 2 gene and its interaction were associated with increased breast cancer in Chinese Han women. 29358504_This study has provided the first evidence for the role of ACYP2 rs1872328 in cisplatin-induced ototoxicity in patients with testicular cancer. 30305294_Traditional genome-wide association studies have identified single-nucleotide polymorphisms in ACYP2 and WFS1 associated with cisplatin-induced hearing loss. 31070019_rs6713088, rs843711 and rs11896604 of ACY2 gene were correlated with an increased risk of Gastrointestinal cancer. 31124313_ACYP2 rs1682111 was associated with the risk of cancer. ACYP2 gene high expression was found to be associated with better overall survial for all liver patients. 31140742_rs1682111 variant was significantly associated with a decreased laryngeal squamous cell carcinoma (LSCC)susceptibility. Polymorphisms of rs10439478, rs11125529, rs12615793, rs843711, rs11896604, rs17045754 were significantly associated with an increased LSCC risk (p < 0.05). The results of haplotype analysis indicated that 'TTCTCG' and 'TTCTAA' in block 1 and 'TG' in block 2 showed risk factor for development of LSCC. 31291640_findings suggested that polymorphisms (rs843720 and rs12615793) of ACYP2 may be pivotal in the development of IgAN 31487124_ACYP2 polymorphisms are associated with the risk of renal cell cancer. 32517717_ACYP2 contributes to malignant progression of glioma through promoting Ca(2+) efflux and subsequently activating c-Myc and STAT3 signals. 32937184_The influence of TERC, TERT and ACYP2 genes polymorphisms on plasma telomerase concentration, telomeres length and T2DM. 33921254_Telomere Length and Male Fertility. | ENSMUSG00000060923 | Acyp2 | 140.702388 | 1.1326441 | 0.179694631 | 0.22333437 | 6.545495e-01 | 4.184908e-01 | No | Yes | 146.046247 | 25.999111 | 128.749465 | 22.962282 | ||
| ENSG00000171084 | 100125556 | FAM86JP | transcribed_unprocessed_pseudogene | 75.459362 | 0.9434956 | -0.083912229 | 0.29983024 | 7.836362e-02 | 7.795274e-01 | No | Yes | 73.191058 | 10.183605 | 78.349230 | 10.784209 | |||||||||||
| ENSG00000171163 | 55657 | ZNF692 | protein_coding | Q9BU19 | FUNCTION: May act as an transcriptional repressor for PCK1 gene expression, in turn may participate in the hepatic gluconeogenesis regulation through the activated AMPK signaling pathway. {ECO:0000269|PubMed:17097062, ECO:0000269|PubMed:21910974}. | 3D-structure;Alternative splicing;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:55657; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of gluconeogenesis [GO:0006111]; regulation of transcription by RNA polymerase II [GO:0006357] | 17097062_cloning and characterization of a novel zinc finger transcription factor referred to as AREBP. AREBP is phosphorylated at Ser(470) by AMPK. 28669730_The results show that ZNF692 is expressed in LUAD tissues compared to adjacent normal tissues, and hyper-expression of ZNF692 in LUAD is an independent risk factor for worse overall survival in LUAD patients (HR: 8.800, 95%CI: 1.082-71.560, P = 0.042) by Tissue Microarray stain assay (TMA). 30466806_ZNF692 promotes CC cells proliferation and invasion through suppressing p27(kip1) transcription by directly binding its promoter region. 30816443_Results found that ZNF692 was upregulated in colon adenocarcinoma (COAD) tissues and cells and that high ZNF692 expression was significantly correlated with lymph node metastasis, distant metastasis and tumor stage in COAD patients. Furthermore, ZNF692 promoted COAD cell proliferation, migration and invasion via the PI3K/AKT pathway, suggesting that ZNF692 may serve as a novel oncogene for COAD. | ENSMUSG00000037243 | Zfp692 | 497.741815 | 1.1087601 | 0.148947286 | 0.12675391 | 1.374217e+00 | 2.410887e-01 | 6.132266e-01 | No | Yes | 503.661003 | 93.119682 | 463.900450 | 85.790245 | ||
| ENSG00000171307 | 84287 | ZDHHC16 | protein_coding | Q969W1 | FUNCTION: Palmitoyl acyltransferase that mediates palmitoylation of proteins such as PLN and ZDHHC6 (PubMed:28826475). Required during embryonic heart development and cardiac function, possibly by mediating palmitoylation of PLN, thereby affecting PLN phosphorylation and homooligomerization (By similarity). Also required for eye development (By similarity). Palmitoylates ZDHHC6, affecting the quaternary assembly of ZDHHC6, its localization, stability and function (PubMed:28826475). May play a role in DNA damage response (By similarity). May be involved in apoptosis regulation (By similarity). Involved in the proliferation of neural stem cells by regulating the FGF/ERK pathway (By similarity). {ECO:0000250|UniProtKB:B8A4F0, ECO:0000250|UniProtKB:Q9ESG8, ECO:0000269|PubMed:28826475}. | Acyltransferase;Alternative splicing;Apoptosis;DNA damage;Endoplasmic reticulum;Membrane;Reference proteome;Transferase;Transmembrane;Transmembrane helix | hsa:84287; | endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; palmitoyltransferase activity [GO:0016409]; protein-cysteine S-palmitoyltransferase activity [GO:0019706]; apoptotic process [GO:0006915]; cellular response to DNA damage stimulus [GO:0006974]; eye development [GO:0001654]; heart development [GO:0007507]; protein palmitoylation [GO:0018345]; telencephalon development [GO:0021537] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 17123647_APH2 contains a zf-DHHC domain (148-210aa), which is involved in protein-protein or protein-DNA interaction. 28826475_human ZDHHC6, which modifies key proteins of the endoplasmic reticulum, is controlled by an upstream palmitoyltransferase, ZDHHC16, revealing the first palmitoylation cascade. | ENSMUSG00000025157 | Zdhhc16 | 1775.201628 | 0.9318115 | -0.101889979 | 0.07225416 | 1.987123e+00 | 1.586420e-01 | 5.192561e-01 | No | Yes | 1732.839153 | 139.801434 | 1871.888492 | 150.877004 | ||
| ENSG00000171551 | 9427 | ECEL1 | protein_coding | O95672 | FUNCTION: May contribute to the degradation of peptide hormones and be involved in the inactivation of neuronal peptides. | Alternative splicing;Disease variant;Disulfide bond;Glycoprotein;Hydrolase;Membrane;Metal-binding;Metalloprotease;Protease;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix;Zinc | This gene encodes a member of the M13 family of endopeptidases. Members of this family are zinc-containing type II integral-membrane proteins that are important regulators of neuropeptide and peptide hormone activity. Mutations in this gene are associated with autosomal recessive distal arthrogryposis, type 5D. This gene has multiple pseudogenes on chromosome 2. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Mar 2014]. | hsa:9427; | integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; metalloendopeptidase activity [GO:0004222]; metallopeptidase activity [GO:0008237]; neuropeptide signaling pathway [GO:0007218]; protein processing [GO:0016485]; respiratory system process [GO:0003016] | 18192274_Sp1 recruits ATF3, c-Jun, and STAT3 to obtain the requisite synergistic effect in neuronal injury through DINE neuronal injury-inducible gene 23236030_We described a new and homogenous phenotype of DA associated with ECEL1 that resulted in symptoms involving rather the peripheral than the central nervous system and suggesting a developmental dysfunction 23261301_Mutations in ECEL1 cause distal arthrogryposis type 5D. 23808592_A novel missense c.1819G>A mutation (G607S) in the ECEL1 gene has been identified in a consanguineous pedigree of Turkish origin presenting with congenital contracture syndromes. 23829171_Three novel ECEL1 mutations have been identified in consanguineous pedigrees of Saudi Arabian origin presenting with distal arthrogryposis type 5D. 25173900_Our clinical findings are consistent with recessive ECEL1 mutations causing variably penetrant orbital dysinnervation phenotypes (ptosis and/or complex strabismus with abnormal synkinesis) 25708584_Mutation of a conserved residue in ECEL1 is linked with fetal arthrogryposis multiplex congenita. 29663639_A novel ECEL1 homozygote mutation in the patient with distal arthrogryposis multiplex congenita type 5D to include pretibial vertical skin creases. 33491998_Distal arthrogryposis type 5D in a South Indian family caused by novel deletion in ECEL1 gene. | ENSMUSG00000026247 | Ecel1 | 154.186964 | 0.6974725 | -0.519791766 | 0.21357275 | 5.902730e+00 | 1.511744e-02 | No | Yes | 113.871226 | 20.639391 | 166.257190 | 29.886065 | ||
| ENSG00000171827 | 148268 | ZNF570 | protein_coding | Q96NI8 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:148268; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000037001 | Zfp39 | 155.829786 | 0.9807083 | -0.028104045 | 0.21954112 | 1.628516e-02 | 8.984550e-01 | No | Yes | 141.060693 | 22.238860 | 141.856430 | 22.423226 | ||||
| ENSG00000172046 | 10869 | USP19 | protein_coding | O94966 | FUNCTION: Deubiquitinating enzyme that regulates the degradation of various proteins. Deubiquitinates and prevents proteasomal degradation of RNF123 which in turn stimulates CDKN1B ubiquitin-dependent degradation thereby playing a role in cell proliferation. Involved in decreased protein synthesis in atrophying skeletal muscle. Modulates transcription of major myofibrillar proteins. Also involved in turnover of endoplasmic-reticulum-associated degradation (ERAD) substrates. Regulates the stability of BIRC2/c-IAP1 and BIRC3/c-IAP2 by preventing their ubiquitination. Required for cells to mount an appropriate response to hypoxia and rescues HIF1A from degradation in a non-catalytic manner. Plays an important role in 17 beta-estradiol (E2)-inhibited myogenesis. Decreases the levels of ubiquitinated proteins during skeletal muscle formation and acts to repress myogenesis. Exhibits a preference towards 'Lys-63'-linked ubiquitin chains. {ECO:0000269|PubMed:19465887, ECO:0000269|PubMed:21849505, ECO:0000269|PubMed:22128162, ECO:0000269|PubMed:22689415}. | 3D-structure;Alternative splicing;Endoplasmic reticulum;Hydrolase;Membrane;Metal-binding;Phosphoprotein;Protease;Reference proteome;Repeat;Thiol protease;Transmembrane;Transmembrane helix;Ubl conjugation pathway;Zinc;Zinc-finger | Protein ubiquitination controls many intracellular processes, including cell cycle progression, transcriptional activation, and signal transduction. This dynamic process, involving ubiquitin conjugating enzymes and deubiquitinating enzymes, adds and removes ubiquitin. Deubiquitinating enzymes are cysteine proteases that specifically cleave ubiquitin from ubiquitin-conjugated protein substrates. This protein is a ubiquitin protein ligase and plays a role in muscle wasting. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2017]. | hsa:10869; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; Hsp90 protein binding [GO:0051879]; Lys48-specific deubiquitinase activity [GO:1990380]; metal ion binding [GO:0046872]; thiol-dependent deubiquitinase [GO:0004843]; ubiquitin protein ligase binding [GO:0031625]; negative regulation of proteasomal protein catabolic process [GO:1901799]; negative regulation of skeletal muscle tissue development [GO:0048642]; positive regulation of cell cycle process [GO:0090068]; protein deubiquitination [GO:0016579]; protein stabilization [GO:0050821]; regulation of cellular response to hypoxia [GO:1900037]; regulation of ERAD pathway [GO:1904292]; regulation of protein stability [GO:0031647]; response to endoplasmic reticulum stress [GO:0034976]; ubiquitin-dependent ERAD pathway [GO:0030433] | 19465887_USP19 is the first example of a membrane-anchored deubiquitinating enzymes involved in the turnover of endoplasmic-reticulum-associated degradation substrates. 21264218_ability of USP19 to regulate cell proliferation and p27(Kip1) levels; complete loss of USP19 function on cell growth may arise as a result of oncogenic transformation of cells. 21849505_The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. 22128162_ubiquitin-specific protease-19 (USP19) interacts with components of the hypoxia pathway including HIF-1alpha and rescues it from degradation independent of its catalytic activity 22810585_experimentally verified the targets heterogeneous nuclear ribonucleoprotein U, phosphatidylinositol-3-OH kinase, the WNK (with-no-lysine) kinase family and USP19 (ubiquitin-specific peptidase 19) as vulnerable nodes in the host cellular defence system against viruses 23500468_USP19 interacts with the ubiquitin ligases SIAH1 and SIAH2, which promote USP19 ubiquitylation and degradation by the proteasome. 24356957_These results clarify the role of USP19 in ERAD and suggest a novel DUB regulation that involves chaperone association and membrane integration 26048142_Expression of USP19 correlates with that of MuRF1 and MAFbx/atrogin-1 in skeletal muscles 26808260_In conjunction with HSP90, the cytoplasmic USP19 may play a key role in triage decision for the disease-related polyQ-expanded substrates, suggesting a function of USP19 in quality control of misfolded proteins by regulating their protein levels 26988033_USP19 positively regulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. 27129179_The regulation of CORO2A through the deubiquitinating activity of USP19 affected the transcriptional repression activity of the retinoic acid receptor (RAR), suggesting that USP19 may be involved in the regulation of RAR-mediated adipogenesis. 27517492_USP19 is a key factor in modulating DNA damage repair by targeting HDAC1/2 K63-linked ubiquitination, cells with deletion or decreased expression of USP19 might cause genome instability and even contribute to tumorigenesis. 27827840_HRD1 is a novel substrate for USP19. USP19 negatively regulates the ubiquitination of HRD1 and prevents it from undergoing proteasomal degradation. 28391724_findings establish USP19 is a negative regulator of the cellular type I IFN antiviral response to enterovirus 71 (EV71) infection; through the decrease of K63-linked polyubiquitination of TRAF3, USP19 suppresses cellular type I IFN signaling, supporting EV71 propagation in host cells 29093475_HSP90 interacts with Htt-N90 on the N-terminal amphipathic alpha-helix, and then recruits USP19 to modulate the protein level and aggregation of Htt-N90. 29901692_USP19 modulates GR levels and in so doing may modulate both insulin and glucocorticoid signaling, two critical pathways that control protein turnover in muscle and overall glucose homeostasis. 30386869_USP19 mRNA expression in human adipose tissue was positively correlated with the expression of important adipocyte genes in abdominal fat depots, but not subcutaneous fat depots. 30700749_USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth. 30716094_BAP1 is highly positively correlated with RBM15B and USP19 expression in invasive breast carcinoma, UM, and colon adenocarcinoma 32236633_Ubiquitin specific peptidase 19 is a prognostic biomarker and affect the proliferation and migration of clear cell renal cell carcinoma. 33978709_USP19 promotes hypoxia-induced mitochondrial division via FUNDC1 at ER-mitochondria contact sites. | ENSMUSG00000006676 | Usp19 | 3247.232875 | 1.0575207 | 0.080685861 | 0.07139974 | 1.285906e+00 | 2.568038e-01 | 6.294719e-01 | No | Yes | 3224.601765 | 338.640203 | 3063.548264 | 321.676184 | |
| ENSG00000172059 | 8462 | KLF11 | protein_coding | O14901 | FUNCTION: Transcription factor (PubMed:9748269, PubMed:10207080). Activates the epsilon- and gamma-globin gene promoters and, to a much lower degree, the beta-globin gene and represses promoters containing SP1-like binding inhibiting cell growth (PubMed:9748269, PubMed:10207080, PubMed:16131492). Represses transcription of SMAD7 which enhances TGF-beta signaling (By similarity). Induces apoptosis (By similarity). {ECO:0000250|UniProtKB:Q8K1S5, ECO:0000269|PubMed:10207080, ECO:0000269|PubMed:16131492}. | Activator;Alternative splicing;Apoptosis;DNA-binding;Diabetes mellitus;Disease variant;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene is a zinc finger transcription factor that binds to SP1-like sequences in epsilon- and gamma-globin gene promoters. This binding inhibits cell growth and causes apoptosis. Defects in this gene are a cause of maturity-onset diabetes of the young type 7 (MODY7). Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Apr 2010]. | hsa:8462; | chromatin [GO:0000785]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; apoptotic process [GO:0006915]; cellular response to peptide [GO:1901653]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of apoptotic process [GO:0043065]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083] | 15024015_TIEG2 is an activator of Monoamine oxidase B gene expression. 15300592_novel mechanism in TGF-beta-regulated gene expression: KLF11 potentiates Smad-signaling activity in normal epithelial cells through termination of the negative feedback loop imposed by Smad7. 15531587_in the presence of cholesterol, KLF11 acts as a dominant repressor of the caveolin-1 gene 15774581_KLF11 plays a role in the regulation of pancreatic beta cell physiology, and its variants may contribute to the development of diabetes. 15774581_Observational study of gene-disease association. (HuGE Navigator) 17114344_These results suggest a central role for KLF11 in TGFbeta-induced c-myc repression and antiproliferation. 17130512_KLF11 A347S and T220M mutations do not contribute to increased risk of diabetes in European-derived populations. 17130512_Observational study of gene-disease association. (HuGE Navigator) 17252542_results may help to increase the understanding of Tieg3-mediated transcriptional control and to characterize this TGF-beta-induced Sp1/Klf-like transcription factor 17308981_TIEG2 decreases the levels of the anti-apoptotic protein Bcl-X(L) and inhibits transcription driven by the Bcl-X(L) promoter. 17479246_Kruppel-like factor 11 inhibits human proinsulin promoter activity in pancreatic beta cells 18199129_Observational study of gene-disease association. (HuGE Navigator) 18505768_KLF11 may interfere with glucose homeostasis in a Danish general population and that STAT3-mediated up-regulation of KLF11 transcription was impaired by the -1659G>C variant. 18505768_Observational study of gene-disease association. (HuGE Navigator) 18593768_KLF11 is unlikely to play a major role in the etiology of T2 diabetes among this Native American population. 18593768_Observational study of gene-disease association. (HuGE Navigator) 19122346_19 polymorphisms, 6 of which are novel, were identified, but none of them showed association with type 2 diabetes. Functional analyses of these variants showed reduced effects on transcriptional activities of insulin, catalase1, & the Smad7 gene. 19122346_Observational study of gene-disease association. (HuGE Navigator) 19124506_Observational study of gene-disease association. (HuGE Navigator) 19843526_MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. 20002157_epigenetic inactivation and subsequent transcriptional repression of the KLF11 gene is quite frequent in MDS. 20124487_KLF11 is an integrator of progesterone receptor signaling and proliferation in uterine leiomyoma cells. 20154088_Kruppel-like factor 11 has a role in the regulation of prostaglandin E2 biosynthesis 20709022_KLF11, but not KLF15, was essential for UCP1 expression during brown adipocyte differentiation of muBM3.1. 21592955_an important role for KLF11 in the regulation of INS transcription via the novel c.-331 KLF site. 22318730_A PXVXL HP1-interacting domain identified at position 487-491 of KLF11 mediates the binding of HP1alpha and KLF11 in vitro and in cultured cells. 22375010_Here we report the characterization of two antagonistic, chromatin-mediated mechanisms by which KLF11 regulated the transcription of the dopamine D2 receptor. 22628545_KLF11 is an MAO A regulator and is produced in response to neuronal stress, which transcriptionally activates MAO A. 22801105_KLF11 has a role in fine-tuning insulin transcription in certain cellular situations rather than representing a major transcriptional activator or repressor of the insulin gene. 22930747_Data indicate that median methylation levels of BCAN, HOXD1, KCTD8, KLF11, NXPH1, POU4F1, SIM1, and TCF7L1 were >/=30% higher than in normal samples, representing potential biomarkers for tumor diagnosis. 23042817_Kruppel-like factor-11, a transcription factor involved in diabetes mellitus, suppresses endothelial cell activation via the nuclear factor-kappaB signaling pathway. 23555910_Data identify a novel pathogenic role for KLF11 in preventing de novo disease-associated fibrosis in endometriosis. 23589285_The A347S mutation disrupted KLF11-mediated increases in basal insulin levels and promoter activity and blunted glucose-stimulated insulin secretion. 23915421_Levels of KLF11 are significantly increased in the postmortem prefrontal cortex of subjects with alcohol dependence compared to controls. 24060634_KLF111 represses glycodelin-A levels and influences its role in the ovarian endometrium. 24069400_KLF11 is a critical transcription factor in the function of mesenchymal cells and, specifically, as a negative regulator of collagen secretion. 24885560_KLF11 regulates distinct gene networks. 25076120_KLF11 repressed most endometrial CYP enzymes 25502632_Results suggest a role for KLF11 in upregulating MAO-A in depressive disorder and chronic social stress, suggesting that inhibition of the pathways regulated by this transcription factor may aid in the therapeutics of neuropsychiatric illnesses 25504365_identified an evolutionarily conserved metabolic regulator, Kruppel-like factor 11 (KLF11), as a novel browning transcription factor in human adipocytes that is required for rosiglitazone-induced browning. 25931269_The data suggest a relationship between promoter DNA methylation and KLF11 gene expression in ovarian cancer tumorigenesis. 26935598_In endometrial-stromal fibroblasts, KLF11 recruited SIN3A/HDAC (histone deacetylase), resulting in COL1A1-promoter deacetylation and repression 28938437_Profibrotic gene expression was activated in a primary human peritoneal cell line in response to KLF11 short hairpin RNA and medroxyprogesterone acetate but not estradiol. 29923255_microRNA-30d mediated breast cancer invasion, migration, and epithelial-mesenchymal transition by targeting KLF11 and activating STAT3 pathway. 30509092_KLF11 has a critical role in regulating gastric cancer migration and invasion by increasing Twist1 expression 30602303_In human aortic smooth muscle cells, small interfering RNA-mediated knockdown of KLF11 increased tissue factor expression. 31124255_Specific variants of KLF11 (MODY7) with a dominant-negative effect underlie early childhood-onset type 1B diabetes with incomplete penetrance. This study documents a novel monogenic mutation associated with diabetes in children. 32451988_Prognostic Role of Kruppel-Like Factors 5, 9, and 11 in Endometrial Endometrioid Cancer. 32524199_DNA hypomethylation of the Kruppel-like factor 11 (KLF11) gene promoter: a putative biomarker of depression comorbidity in panic disorder and of non-anxious depression? 32633337_KLF11 protects chondrocytes via inhibiting p38 MAPK signaling pathway. 33507881_KLF11 protects against abdominal aortic aneurysm through inhibition of endothelial cell dysfunction. 33571129_Genome-wide CRISPR-Cas9 screen identified KLF11 as a druggable suppressor for sarcoma cancer stem cells. 33604390_Clinical and Functional Characteristics of a Novel KLF11 Cys354Phe Variant Involved in Maturity-Onset Diabetes of the Young. 35108381_Evaluation of Evidence for Pathogenicity Demonstrates That BLK, KLF11, and PAX4 Should Not Be Included in Diagnostic Testing for MODY. | ENSMUSG00000020653 | Klf11 | 712.157113 | 1.1341570 | 0.181620333 | 0.10757833 | 2.848994e+00 | 9.143115e-02 | 4.063403e-01 | No | Yes | 746.492273 | 79.169902 | 651.216115 | 69.248867 | |
| ENSG00000172671 | 93550 | ZFAND4 | protein_coding | Q86XD8 | Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:93550; | zinc ion binding [GO:0008270] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 25682742_In gastric cancer cells, ANUBL1 was upregulated and promoted proliferation of SGC-7901 cells. Over-expression decreased miR-182 expression. ANUBL1 expression was in turn directly downregulated by miR-182 in a negative feedback loop. 29074611_Suggest that ZFAND4 is a useful marker for predicting metastasis and poor prognosis in patients with oral squamous cell carcinoma. | ENSMUSG00000042213 | Zfand4 | 307.233041 | 1.0177112 | 0.025328248 | 0.16600012 | 2.358167e-02 | 8.779541e-01 | No | Yes | 284.137412 | 42.494916 | 280.403193 | 41.978259 | ||||
| ENSG00000172731 | 55222 | LRRC20 | protein_coding | Q8TCA0 | Alternative splicing;Leucine-rich repeat;Phosphoprotein;Reference proteome;Repeat | hsa:55222; | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20659327_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) | ENSMUSG00000037151 | Lrrc20 | 498.521357 | 0.7250411 | -0.463865296 | 0.13458347 | 1.168045e+01 | 6.316010e-04 | 2.695726e-02 | No | Yes | 456.771533 | 65.048595 | 633.010253 | 89.905135 | ||||
| ENSG00000173064 | 283450 | HECTD4 | protein_coding | F8VU57 | Proteomics identification;Reference proteome | Mouse_homologues mmu:269700; | Mouse_homologues glucose homeostasis [GO:0042593]; glucose metabolic process [GO:0006006] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23575436_Newly implicated variants (MYL2, C12orf51 and OAS1) were found to be significantly associated with 1-h plasma glucose as predisposing risk factors for type 2 diabetes. 25661349_There was no significant association between C12orf51 rs11066280 and survival of gastric cancer. 26675016_SNPs in the HECTD4 gene is associated with decreased thoracic-to-hip circumference ratio in male Koreans. 28212632_This exome-wide association study indicated that C12orf51 rs11066280, MYL2 rs12229654, and ALDH2 rs671 polymorphisms are linked to blood Pb levels in the Korean population. 34828409_A Genome-Wide Association Study of a Korean Population Identifies Genetic Susceptibility to Hypertension Based on Sex-Specific Differences. | ENSMUSG00000042744 | Hectd4 | 1671.805442 | 1.0050915 | 0.007326792 | 0.07919308 | 8.637849e-03 | 9.259512e-01 | 9.734792e-01 | No | Yes | 1356.089856 | 162.218216 | 1339.817532 | 160.403854 | |||
| ENSG00000173230 | 2804 | GOLGB1 | protein_coding | Q14789 | FUNCTION: May participate in forming intercisternal cross-bridges of the Golgi complex. | Acetylation;Alternative splicing;Coiled coil;Disulfide bond;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:2804; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; Golgi stack [GO:0005795]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; RNA binding [GO:0003723]; sequence-specific DNA binding [GO:0043565]; chondrocyte proliferation [GO:0035988]; Golgi organization [GO:0007030]; protein localization to pericentriolar material [GO:1905793]; regulation of transcription, DNA-templated [GO:0006355] | 12429822_Data report the characterization of a mammalian coiled-coil protein, CASP, a Golgi protein that shares with giantin a conserved histidine in its transmembrane domain. 17475246_Giantin binds to Rab6A and Rab1 proteins. 23422753_unbiased whole-genome search for genetic modifiers of stroke risk in sickle cell anemia was performed; mutation in GOLGB1 (Y1212C) and mutation in ENPP1 (K173Q) were confirmed as having significant associations with a decreased risk for stroke 23555793_the spatial organization of the Golgi ribbon is mediated by giantin, which also plays a role in cargo transport and sugar modifications 24046448_Partial depletion of giantin or of WDR34 leads to an increase in cilia length consistent with the concept that giantin acts through dynein-2. 25086069_Results show that ST3Gal1 uses GM130-GRASP65 and giantin, whereas C2GnT-L uses only giantin for Golgi targeting and defective giantin dimerization in PC-3 and DU145 prostate cancer cells causes fragmentation of the Golgi and prevents its targeting. 26664786_Single nucleotide polymorphisms (SNPs) rs1035798 in RAGE gene, rs2073617 and rs2073618 in TNFRSF11B, and rs3732410 in Golgb1 will be investigated on whether there is an association with hemorrhagic stroke (HS) in Chinese population. 28782625_In situ proximity ligation assays of Golgi localization of alpha-mannosidase IA at giantin versus GM130-GRASP65 site, and absence or presence of N-glycans terminated with alpha3-mannose on trans-Golgi glycosyltransferases may be useful for distinguishing indolent from aggressive prostate cancer cells. 29430628_common polymorphisms of GOLGB1 are not associated with nonsyndromic cleft palate susceptibility in the Brazilian population 30453527_Ethanol-induced Golgi disassembly was associated with de-dimerization of the largest Golgi matrix protein giantin, along with impaired transport of selected hepatic proteins. | ENSMUSG00000034243 | Golgb1 | 2429.819181 | 1.2046371 | 0.268598643 | 0.10341390 | 6.816520e+00 | 9.031837e-03 | 1.236495e-01 | No | Yes | 1783.404380 | 271.837355 | 1476.564581 | 225.093765 | ||
| ENSG00000173473 | 6599 | SMARCC1 | protein_coding | Q92922 | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. May stimulate the ATPase activity of the catalytic subunit of the complex (PubMed:10078207, PubMed:29374058). Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). {ECO:0000250|UniProtKB:P97496, ECO:0000269|PubMed:10078207, ECO:0000269|PubMed:11018012, ECO:0000269|PubMed:29374058, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | 3D-structure;Acetylation;Chromatin regulator;Coiled coil;Cytoplasm;Direct protein sequencing;Isopeptide bond;Methylation;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a member of the SWI/SNF family of proteins, whose members display helicase and ATPase activities and which are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SNF/SWI and contains a predicted leucine zipper motif typical of many transcription factors. [provided by RefSeq, Jul 2008]. | hsa:6599; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nBAF complex [GO:0071565]; npBAF complex [GO:0071564]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; SWI/SNF complex [GO:0016514]; XY body [GO:0001741]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; protein N-terminus binding [GO:0047485]; transcription coactivator activity [GO:0003713]; animal organ morphogenesis [GO:0009887]; chromatin remodeling [GO:0006338]; insulin receptor signaling pathway [GO:0008286]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; nervous system development [GO:0007399]; nucleosome disassembly [GO:0006337]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357] | 16199878_protein levels of BAF155/170 dictate the maximum cellular amount of BAF57 16568092_BAF155 and potentially INI1 are substrates for Akt phosphorylation 17513758_Constitutive expression of SRG3 inhibits positive selection processes in T-cell receptor transgenic mice. 18581278_An increased expression of SMARCC1 protein was found in prostate cancer positively correlated with tumour dedifferentiation, progression, metastasis and time to recurrence. 19156145_patients with tumours displaying high levels of CBFB and SMARCC1 proteins had a significantly better overall survival rate than patients with low levels 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20829358_Data show that the mechanism of BAF155-mediated stabilization of BAF57 involves blocking its ubiquitination by preventing interaction with TRIP12. 22139574_loss of BAF155 expression represents another mechanism for inactivation of SWI/SNF complex activity in the development in human cancer. 23799850_miR-320c regulates the resistance of pancreatic cancer cells to gemcitabine through SMARCC1. 23996527_Results show the secondary structure of SWIRM domain of BAF155 consists of five alpha-helices forming a typical histone fold for DNA interactions. 24365151_Wwp2 acts as a ubiquitin ligase of SRG3. 24434208_we identify BAF155 as a substrate for arginine methyltransferase CARM1. 26257059_NKX6.1 directly enhances the mRNA level of E-cadherin by recruiting BAF155 coactivator and represses that of vimentin and N-cadherin by recruiting RBBP7 (retinoblastoma binding protein 7) corepressor. 27190130_our data showed that Swi3 strongly affects haem/oxygen-dependent activation of respiration gene promoters whereas Swi2 affects only the basal, haem-independent activities of these promoters. using computational analysis and RNAi knockdown, we showed that the mammalian Swi3 BAF155 and BAF170 regulate respiration in HeLa cells. 28438634_Here, the authors first confirmed that SWIRM domain of BAF155 is responsible for its interaction with BAF47 and then narrowed down the SWIRM-binding region in BAF47 to the Repeat 1 (RPT1) domain. 30144500_Study found SMARCC1 as a direct target of miR-202-5p and promoted the growth and metastasis of colorectal carcinoma (CRC) cells. Furthermore, SMARCC1 could reverse the inhibitory effect of miR-202-5p on growth and metastasis of CRC cells. 31533543_BAF155 plays important roles in ubiquitin-independent degradation of hepatitis B virus X protein 32244797_A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction. 33931740_Multiple interactions of the oncoprotein transcription factor MYC with the SWI/SNF chromatin remodeler. 33953332_SWI/SNF subunit BAF155 N-terminus structure informs the impact of cancer-associated mutations and reveals a potential drug binding site. 35158202_Assembly and interaction of core subunits of BAF complexes and crystal study of the SMARCC1/SMARCE1 binary complex. | ENSMUSG00000032481 | Smarcc1 | 13170.845491 | 1.0841939 | 0.116622791 | 0.04720409 | 6.129216e+00 | 1.329658e-02 | 1.525227e-01 | No | Yes | 15008.361050 | 2239.989097 | 13628.923843 | 2034.040070 | |
| ENSG00000173846 | 1263 | PLK3 | protein_coding | Q9H4B4 | FUNCTION: Serine/threonine-protein kinase involved in cell cycle regulation, response to stress and Golgi disassembly. Polo-like kinases act by binding and phosphorylating proteins are that already phosphorylated on a specific motif recognized by the POLO box domains. Phosphorylates ATF2, BCL2L1, CDC25A, CDC25C, CHEK2, HIF1A, JUN, p53/TP53, p73/TP73, PTEN, TOP2A and VRK1. Involved in cell cycle regulation: required for entry into S phase and cytokinesis. Phosphorylates BCL2L1, leading to regulate the G2 checkpoint and progression to cytokinesis during mitosis. Plays a key role in response to stress: rapidly activated upon stress stimulation, such as ionizing radiation, reactive oxygen species (ROS), hyperosmotic stress, UV irradiation and hypoxia. Involved in DNA damage response and G1/S transition checkpoint by phosphorylating CDC25A, p53/TP53 and p73/TP73. Phosphorylates p53/TP53 in response to reactive oxygen species (ROS), thereby promoting p53/TP53-mediated apoptosis. Phosphorylates CHEK2 in response to DNA damage, promoting the G2/M transition checkpoint. Phosphorylates the transcription factor p73/TP73 in response to DNA damage, leading to inhibit p73/TP73-mediated transcriptional activation and pro-apoptotic functions. Phosphorylates HIF1A and JUN is response to hypoxia. Phosphorylates ATF2 following hyperosmotic stress in corneal epithelium. Also involved in Golgi disassembly during the cell cycle: part of a MEK1/MAP2K1-dependent pathway that induces Golgi fragmentation during mitosis by mediating phosphorylation of VRK1. May participate in endomitotic cell cycle, a form of mitosis in which both karyokinesis and cytokinesis are interrupted and is a hallmark of megakaryocyte differentiation, via its interaction with CIB1. {ECO:0000269|PubMed:10557092, ECO:0000269|PubMed:11156373, ECO:0000269|PubMed:11447225, ECO:0000269|PubMed:11551930, ECO:0000269|PubMed:11971976, ECO:0000269|PubMed:12242661, ECO:0000269|PubMed:14968113, ECO:0000269|PubMed:14980500, ECO:0000269|PubMed:15021912, ECO:0000269|PubMed:16478733, ECO:0000269|PubMed:16481012, ECO:0000269|PubMed:17264206, ECO:0000269|PubMed:17804415, ECO:0000269|PubMed:18062778, ECO:0000269|PubMed:18650425, ECO:0000269|PubMed:19103756, ECO:0000269|PubMed:19490146, ECO:0000269|PubMed:20889502, ECO:0000269|PubMed:20940307, ECO:0000269|PubMed:20951827, ECO:0000269|PubMed:21098032, ECO:0000269|PubMed:21264284, ECO:0000269|PubMed:21376736, ECO:0000269|PubMed:21840391, ECO:0000269|PubMed:9353331}. | 3D-structure;ATP-binding;Apoptosis;Cell cycle;Cytoplasm;Cytoskeleton;DNA damage;Golgi apparatus;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene is a member of the highly conserved polo-like kinase family of serine/threonine kinases. Members of this family are characterized by an amino-terminal kinase domain and a carboxy-terminal bipartite polo box domain that functions as a substrate-binding motif and a cellular localization signal. Polo-like kinases are important regulators of cell cycle progression. This gene has also been implicated in stress responses and double-strand break repair. In human cell lines, this protein is reported to associate with centrosomes in a microtubule-dependent manner, and during mitosis, the protein becomes localized to the mitotic apparatus. Expression of a kinase-defective mutant results in abnormal cell morphology caused by changes in microtubule dynamics and mitotic arrest followed by apoptosis. [provided by RefSeq, Sep 2015]. | hsa:1263; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; dendrite [GO:0030425]; Golgi stack [GO:0005795]; neuronal cell body [GO:0043025]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spindle pole [GO:0000922]; ATP binding [GO:0005524]; p53 binding [GO:0002039]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; apoptotic process [GO:0006915]; cellular response to DNA damage stimulus [GO:0006974]; cytoplasmic microtubule organization [GO:0031122]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; endomitotic cell cycle [GO:0007113]; G1/S transition of mitotic cell cycle [GO:0000082]; G2/M transition of mitotic cell cycle [GO:0000086]; Golgi disassembly [GO:0090166]; mitotic cell cycle [GO:0000278]; mitotic G1/S transition checkpoint signaling [GO:0044819]; negative regulation of apoptotic process [GO:0043066]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of chaperone-mediated autophagy [GO:1904716]; positive regulation of intracellular protein transport [GO:0090316]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia [GO:2000777]; protein kinase B signaling [GO:0043491]; protein phosphorylation [GO:0006468]; regulation of cell division [GO:0051302]; regulation of cytokinesis [GO:0032465]; regulation of signal transduction by p53 class mediator [GO:1901796]; response to osmotic stress [GO:0006970]; response to radiation [GO:0009314]; response to reactive oxygen species [GO:0000302] | Mouse_homologues 15107614_Data demonstrate that FGF-inducible kinase (Fnk) expression levels in transfected cells can be regulated by nuclear-cytoplasmic trafficking, ubiquitination, and proteosome-dependent degradation. 17264206_Plk3 localizes to the nucleolus and is involved in regulation of the G1/S phase transition. 18519666_Ectopic expression of the Plk3-kinase domain (Plk3-KD), but not its Polo-box domain or a Plk3-KD mutant, suppressed the nuclear accumulation of HIF-1 alpha induced by nickel or cobalt ions 19188452_The specificity of TTP for promoting the degradation of Plk3 was demonstrated by the unaltered decay of Plk3 mRNA in cell 20889502_Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1alpha 20940307_Plk3 as a new player in the regulation of the PI3K/PDK1/Akt signaling axis by phosphorylation and stabilization of PTEN. 21264284_Calcium- and integrin-binding protein 1 is involved in regulating endomitosis, perhaps through its interaction with Plk3 26949938_Polo-Like Kinase 3 Appears Dispensable for Normal Retinal Development Despite Robust Embryonic Expression 33514736_The oncogenicity of tumor-derived mutant p53 is enhanced by the recruitment of PLK3. | ENSMUSG00000028680 | Plk3 | 58.336121 | 1.2060758 | 0.270320537 | 0.34736763 | 6.084841e-01 | 4.353591e-01 | No | Yes | 57.198857 | 11.920863 | 46.545460 | 9.694383 | ||
| ENSG00000173960 | 165324 | UBXN2A | protein_coding | P68543 | Alternative splicing;Reference proteome | hsa:165324; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; nucleus [GO:0005634]; acetylcholine receptor binding [GO:0033130]; ubiquitin binding [GO:0043130]; autophagosome assembly [GO:0000045]; cellular response to leukemia inhibitory factor [GO:1990830]; Golgi organization [GO:0007030]; membrane fusion [GO:0061025]; nuclear membrane reassembly [GO:0031468]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of protein catabolic process [GO:0042176]; regulation of protein ubiquitination [GO:0031396] | 22454508_Ubx2 and Ubxd8 regulates lipid droplet homeostasis. 24625977_Suggest UBXN2A can reconstitute inactive p53-dependent apoptotic pathways in colonic neoplasms. 26188124_veratridine enhances transactivation of UBXN2A, resulting in upregulation of UBXN2A in the cytoplasm, where UBXN2A binds and inhibits the oncoprotein mortalin-2 26634371_UBXN2A binds to mortalin's binding pocket within the substrate-binding domain of mortalin. UBXN2A increases stability of p53 protein targeted by the mortalin-CHIP E3 ubiquitin ligase. 30107089_The existence of a multiprotein complex containing UBXN2A, CHIP, and mot-2 suggests a synergistic tumor suppressor activity of UBXN2A and CHIP in mot-2-enriched tumors. | ENSMUSG00000020634 | Ubxn2a | 1503.551209 | 0.9893817 | -0.015400881 | 0.07901497 | 3.814717e-02 | 8.451479e-01 | 9.491576e-01 | No | Yes | 1367.602967 | 217.480769 | 1358.669976 | 216.005984 | |||
| ENSG00000174137 | 152877 | FAM53A | protein_coding | Q6NSI3 | FUNCTION: May play an important role in neural development; the dorsomedial roof of the third ventricle. {ECO:0000250|UniProtKB:Q5ZKN5}. | Nucleus;Phosphoprotein;Reference proteome | hsa:152877; | nucleus [GO:0005634]; protein import into nucleus [GO:0006606] | 28179588_Expression levels of TP53BP2, FBXO28, and FAM53A genes were associated with patient survival specifically in ER-positive, TP53-mutated tumors. | ENSMUSG00000037339 | Fam53a | 51.481263 | 1.2024906 | 0.266025578 | 0.36972401 | 5.168390e-01 | 4.721932e-01 | No | Yes | 50.136011 | 9.308370 | 43.640871 | 8.103337 | |||
| ENSG00000174405 | 3981 | LIG4 | protein_coding | P49917 | FUNCTION: DNA ligase involved in DNA non-homologous end joining (NHEJ); required for double-strand break (DSB) repair and V(D)J recombination (PubMed:8798671, PubMed:9242410, PubMed:9809069, PubMed:12517771, PubMed:17290226). Catalyzes the NHEJ ligation step of the broken DNA during DSB repair by resealing the DNA breaks after the gap filling is completed (PubMed:9242410, PubMed:9809069, PubMed:12517771, PubMed:17290226). Joins single-strand breaks in a double-stranded polydeoxynucleotide in an ATP-dependent reaction (PubMed:9242410, PubMed:9809069, PubMed:12517771, PubMed:17290226). LIG4 is mechanistically flexible: it can ligate nicks as well as compatible DNA overhangs alone, while in the presence of XRCC4, it can ligate ends with 2-nucleotides (nt) microhomology and 1-nt gaps (PubMed:17290226). Forms a subcomplex with XRCC4; the LIG4-XRCC4 subcomplex is responsible for the NHEJ ligation step and XRCC4 enhances the joining activity of LIG4 (PubMed:9242410, PubMed:9809069). Binding of the LIG4-XRCC4 complex to DNA ends is dependent on the assembly of the DNA-dependent protein kinase complex DNA-PK to these DNA ends (PubMed:10854421). LIG4 regulates nuclear localization of XRCC4 (PubMed:24984242). {ECO:0000269|PubMed:10854421, ECO:0000269|PubMed:12517771, ECO:0000269|PubMed:17290226, ECO:0000269|PubMed:24984242, ECO:0000269|PubMed:8798671, ECO:0000269|PubMed:9242410, ECO:0000269|PubMed:9809069}. | 3D-structure;ATP-binding;Cell cycle;Cell division;DNA damage;DNA recombination;DNA repair;DNA replication;Direct protein sequencing;Disease variant;Ligase;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Reference proteome;Repeat;SCID | The protein encoded by this gene is a DNA ligase that joins single-strand breaks in a double-stranded polydeoxynucleotide in an ATP-dependent reaction. This protein is essential for V(D)J recombination and DNA double-strand break (DSB) repair through nonhomologous end joining (NHEJ). This protein forms a complex with the X-ray repair cross complementing protein 4 (XRCC4), and further interacts with the DNA-dependent protein kinase (DNA-PK). Both XRCC4 and DNA-PK are known to be required for NHEJ. The crystal structure of the complex formed by this protein and XRCC4 has been resolved. Defects in this gene are the cause of LIG4 syndrome. Alternatively spliced transcript variants encoding the same protein have been observed. [provided by RefSeq, Jul 2008]. | hsa:3981; | chromosome, telomeric region [GO:0000781]; condensed chromosome [GO:0000793]; cytoplasmic ribonucleoprotein granule [GO:0036464]; DNA ligase IV complex [GO:0032807]; DNA-dependent protein kinase-DNA ligase 4 complex [GO:0005958]; nonhomologous end joining complex [GO:0070419]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; DNA binding [GO:0003677]; DNA ligase (ATP) activity [GO:0003910]; DNA ligase activity [GO:0003909]; ligase activity [GO:0016874]; metal ion binding [GO:0046872]; protein C-terminus binding [GO:0008022]; cell cycle [GO:0007049]; cell division [GO:0051301]; cell population proliferation [GO:0008283]; cellular response to ionizing radiation [GO:0071479]; cellular response to lithium ion [GO:0071285]; central nervous system development [GO:0007417]; chromosome organization [GO:0051276]; DNA biosynthetic process [GO:0071897]; DNA ligation [GO:0006266]; DNA ligation involved in DNA recombination [GO:0051102]; DNA ligation involved in DNA repair [GO:0051103]; DNA replication [GO:0006260]; double-strand break repair [GO:0006302]; double-strand break repair via classical nonhomologous end joining [GO:0097680]; double-strand break repair via nonhomologous end joining [GO:0006303]; establishment of integrated proviral latency [GO:0075713]; immunoglobulin V(D)J recombination [GO:0033152]; in utero embryonic development [GO:0001701]; isotype switching [GO:0045190]; negative regulation of neuron apoptotic process [GO:0043524]; neuron apoptotic process [GO:0051402]; nucleotide-excision repair, DNA gap filling [GO:0006297]; positive regulation of chromosome organization [GO:2001252]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of neurogenesis [GO:0050769]; pro-B cell differentiation [GO:0002328]; response to gamma radiation [GO:0010332]; response to X-ray [GO:0010165]; single strand break repair [GO:0000012]; somatic stem cell population maintenance [GO:0035019]; T cell differentiation in thymus [GO:0033077]; T cell receptor V(D)J recombination [GO:0033153]; V(D)J recombination [GO:0033151] | 11702069_crystal structure of an Xrcc4-DNA ligase IV complex 11779494_mutations identified in patients exhibiting developmental delay and immunodeficiency 12036913_Observational study of gene-disease association. (HuGE Navigator) 12077346_Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. 12471202_Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination 12547193_Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment 12589063_DNA ligase IV has a role in binding with a subunit of the condensin complex 12750264_Observational study of gene-disease association. (HuGE Navigator) 14578164_Observational study of gene-disease association. (HuGE Navigator) 15126335_Observational study of gene-disease association. (HuGE Navigator) 15194694_stability is regulated by multiple factors, including interaction with XRCC4, phosphorylation status, and possibly Lig4 conformation 15256476_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15333585_Examination of recombinant mutant DNA ligase IV shows that the severity of LIG4 syndrome correlates with the level of residual ligase activity. 15509577_interactions between BLM and DNA ligase IV play a role in DNA repair in human cells 15520013_DNAPK was physically required for the mobilization of the XRCC4-ligase IV complex, and for stable recruitment of XRCC4; phosphorylation of either H2AX or XRCC4 was unnecessary for DNAPK or XRCC4-ligase IV recruitment 15609317_Observational study of gene-disease association. (HuGE Navigator) 15958648_Observational study of gene-disease association. (HuGE Navigator) 16357942_LIG4 mutations can result in either a developmental defect with immunological abnormalities or a severe combined immunodeficiency picture with normal development. 16407418_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16412978_We confirmed that DNA ligase IV is unstable in the absence of XRCC4, with a half-life of approximately 30-90 min. 16485136_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16571728_Cernunnos physically interacts with the XRCC4 x DNA-ligase IV complex 16638864_Observational study of gene-disease association. (HuGE Navigator) 16735143_residues 470-476 function as part of a molecular pincer that maintains the DNA in a conformation that is required for ligation 16857995_Observational study of gene-disease association. (HuGE Navigator) 17018785_Observational study of gene-disease association. (HuGE Navigator) 17241822_DNA-PK kinase activity results in disassembly of the Ku/DNA ligase IV/Xrcc4 complex 17290226_XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. 17492771_Results describe the activity requirements for DNA ligases III and IV in the pathways of non-homologous DNA end joining. 17541392_DNA ligase IV gene polymorphisms might have a role in acute lymphoblastic leukemia in children 17963495_DNA Ligase IV/XRCC4 recruitment by DNA-PK to DNA double-strand breaks prevents the formation of long ssDNA ends at double-strand breaks during the S phase. 18165945_Haplotype analysis revealed an association of glioma risk with genetic variants in LIG4 block 1 (global P=0.011), and XRCC4 blocks 2 and 4 (both global P<0.0001). 18165945_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18440984_DNA ligase IV is not required in microhomology-mediated end joining but facilitates Ku-dependent DNA nonhomologous end-joining. 18508926_DNA damage left unrepaired by DNA-ligase IV or DNA-PK might be the initiator for caspase activation by doxorubicin in cancer cells 18579371_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18830263_Observational study of gene-disease association. (HuGE Navigator) 18845326_Omenn syndrome is associated with mutations in DNA ligase IV. 18952251_In cells infected with E1B 55k-deficient adenovirus, ligase IV could not be found in XRCC4-containing complexes and was observed in a novel ligase IV/E4 34k/Cul5/Elongin BC complex. 18990028_Observational study of gene-disease association. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19092295_Observational study of gene-disease association. (HuGE Navigator) 19116388_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124506_Observational study of gene-disease association. (HuGE Navigator) 19127255_Observational study and meta-analysis of gene-disease association and gene-environment interaction. (HuGE Navigator) 19142223_DNA-PK regulates the dysbindin-1 isoforms A and B by phosphorylation in nucleus 19147782_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19251090_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19332554_The high-resolution crystal structure of human XRCC4 bound to the carboxy-terminal tandem BRCT repeat of DNA ligase IV, is reported. 19408343_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19418549_study reports 2 siblings with LIG4 syndrome; mutational analysis of the family revealed a novel sequence variant; homozygous trinucleotide deletion, 1762delAAG, was detected in the brother, resulting in deletion of a lysine at amino acid position 588 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19573080_Observational study of gene-disease association. (HuGE Navigator) 19604268_Observational study of gene-disease association. (HuGE Navigator) 19638463_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19661089_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19727724_role of NHEJ enzyme Ligase IV in the pathogenesis of MDS 19773279_Observational study of gene-disease association. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19837014_provide a first sight of the structural organization of the Lig4-Xrcc4 complex, which suggests that the BRCT domains could provide the link of the ligase to Xrcc4 while permitting some movements of the catalytic domains of Lig4. 19859091_Observational study of genetic testing. (HuGE Navigator) 20150366_Observational study of gene-disease association. (HuGE Navigator) 20368557_Polymorphisms in the LIG4, BTBD2, HMGA2, and RTEL1 genes, which are involved in the double-strand break repair pathway, are associated with glioblastoma multiforme survival. 20400235_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20400235_in the Chinese population, LIG4 Ile658Val has only a slight impact on the risk of developing cervical carcinoma 20453000_Observational study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20496270_Observational study of gene-disease association. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20568250_Observational study of gene-disease association. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20731661_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20811692_Observational study of gene-disease association. (HuGE Navigator) 20811692_these results suggest that some variants in XRCC4, LIG4 and Ku80 genes can contribute to thyroid cancer susceptibility 20813000_Meta-analysis of gene-disease association. (HuGE Navigator) 21717429_The current results indicated that NHEJ genetic polymorphisms, particularly LIG4 rs1805388, may modulate the risk of RP in patients with NSCLC who receive definitive radio(chemo)therapy 21974800_polymorphisms in LIG4 do not contribute to cancer risk in a population of Lynch syndrome patients with colorectal cancer 21982441_XRCC4 modulates the dynamic interaction of the Ligase IV/XRCC4 complex with the NHEJ machinery at double-stranded DNA breaks 22534089_The alpha2 helix in the Lig4 BRCT-1 domain is required for adenovirus-mediated degradation of Lig4. 22658747_The flexibility of the DNA ligase IV catalytic region is limited in a manner that affects the formation of the LigIV/XRCC4/XLF-Cernunnos complex. 22994770_No association between LIG4 gene polymorphisms (rs1805386 T>C, rs1805389 C>T, rs1805388 C>T and rs2232641 A>G) and breast cancer risk. 23219551_Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. 23275564_DNA Ligase IV is differentially required for certain chromosome fusion events induced by telomere dysfunction. 23630330_genetic association studies in population in China: Data suggest that SNPs in LIG4 (rs1805388, exon 2 54C>T, Thr9Ile) and RAG1 (recombination activating gene 1, rs2227973, A>G, K820R) are associated with male infertility. 23663450_These results indicate for the first time that LIG4 rs1805388 and X-ray Repair Cross Complementing-4 (XRCC4) rs1805377, alone or in combination, are associated with a risk of gliomas. 23794378_Characterization of the protein interaction domains that modulate the XRCC4/Ligase IV interaction. 23967291_DNA ligase IV and Artemis act cooperatively to promote nonhomologous end-joining 23994631_The chromatin binding of XRCC4 was dependent on the presence of LIG4. 24123394_study reports the identification of biallelic truncating LIG4 mutations in 11 patients with microcephalic primordial dwarfism presenting with restricted prenatal growth and extreme postnatal global growth failure 24282031_Loss of LIG4 is associated with colorectal cancer. 24415301_Our data did not show association between LIG4 and RAD52 SNPs and SLE, its clinical manifestations or ethnicity in the tested population. 24530422_DNA ligase IV expression of K562/DNR was also suppressed significantly with Sp1 family protein inhibition 24722796_LIG4 Thr9Ile polymorphism is associated with treatment response in advanced non-small cell lung cancer. 24837021_Ku-independent, LIGIV-dependent repair pathway exists in human somatic cells. 24984242_DNA Ligase IV is required for efficient localization of XRCC4 and XLF into cell nucleus. 25201414_Results show that in cells deficient for Lig4, chromosomal translocations junctions had significantly longer deletions and more microhomologies. 25314918_despite several limitations, this meta-analysis suggested that LIG4 T9I genetic variant is associated with a decreased risk of cancer among Caucasians, however, the rs1805386 gene polymorphism is not a risk factor of cancer. 25605214_High LIG4 expression is associated with less radiosensitivity of nasopharyngeal cancer. 25811031_The genetic polymorphisms in LIG4 rs1805388 and HSPB1 rs2868371 were not obviously correlated with the risk of radiation pneumonitis and radiation-induced lung injury of lung cancer. 25973104_our results suggest that LIG4 rs10131 polymorphism in the DNA repair pathways plays an important role in the risk of glioma in a Chinese population. 26134445_Our study identifies LIG4 as a predictor of an increased risk for early biochemical recurrence in prostate cancer 26201248_Our data showed that Cd altered the phosphorylation of DNA-PKcs, and reduced the expression of both XRCC4 and Ligase IV in irradiated cells. 26762768_Five novel mutations in LIG4 and a potential hotspot mutation (c.833G > T; p.R278L) in the Chinese population with LIG4 deficiency syndrome were identified. 26974709_The rs1805388 in LIG4 was associated with increased radioresistance. 27009971_LIG4 is highly upregulated in human colorectal cancer cells. 27497341_found that the rs228593, rs2267437 and rs1805388 functional polymorphisms probably alter the level of expression of the ATM, XRCC6 and LIG4 genes, respectively, being important in the maintenance of genomic instability in MDS 27508978_This study shown that both ligase IV and XRCC4 may act in concert to modulate the development of glioma. 27612988_marked overlap of dyskeratosis congenita with four other genetic syndromes, confounding accurate diagnosis and subsequent management. Patients with clinical features of dyskeratosis congenita need to have genetic analysis of USB1, LIG4 and GRHL2 in addition to the classical dyskeratosis congenita genes and telomere length measurements. 28453785_In a recombinant PNKP-XRCC4-LigIV complex, both the PNKP FHA and catalytic domains contact the XRCC4 coiled-coil and LigIV BRCT repeats. Multipoint contacts between PNKP and XRCC4-LigIV regulate PNKP recruitment and activity within NHEJ. 28695890_Cells doubly deficient in Pol theta; and Lig4 exhibit 100% gene-targeting efficiency because of virtually no random integration events. 28696258_Data suggest that stimulation of Artemis nuclease/DCLRE1C activity by XRCC4-DNA ligase IV hetero-complex and efficiency of blunt-end ligation are determined by structural configurations at the DNA ends. (XRCC4 = X-ray repair cross complementing 4) 28930678_Cellular NHEJ of diverse ends thus identifies the steps necessary for repair through LIG4-mediated sensing of differences in end structure and consequent dynamic remodeling of aligned ends. 28976792_We demonstrated an association between six previously published single nucleotide polymorphisms (rs15869 [ BRCA2], rs1805389 [ LIG4], rs8079544 [ TP53], rs25489 [ XRCC1], rs1673041 [ POLD1], and rs11615 [ ERCC1]) and subsequent CNS tumors in survivors of childhood cancer treated by radiation therapy. 28991497_study demonstrated that there was an association between DNA ligase 4 Thr9Ile polymorphism and male infertility and suggests CT genotype as a risk factor for male infertility 28992182_An increased frequency of binucleated lymphocytes with micronuclei was increased in carriers of the T/T genotype of the LIG4 (rs1805388) gene compared to miners harbouring the C/T genotype. 29676720_Our data show that DNA ligase IV can act outside of the nucleus to allow repair of dsDNA breaks in poxvirus genomes. 29980672_These studies provide a scaffold for defining impacts of LigIV catalytic core mutations and deficiencies in human LIG4 syndrome. 30038324_Study indicated that miR-1246 suppressed LIG4 expression by directly binding to its 3'-UTR inducing genomic DNA damage and reduced cell proliferation. 30496552_Disrupting the NAD+ recognition site in the BRCT domain(BRCA1 C-terminal domain) impairs non-homologous end joining (NHEJ) in cell. Taken together, our study reveals that in addition to ATP, Ligase IV may use NAD+ as an alternative adenylation donor for NHEJ repair and maintaining genomic stability. 30617623_The phenotypical spectrum of LIG4 deficiency. 31184023_The genetic polymorphism of the effects of human granulocytic anaplasmosis was revealed, and the most significant cytogenetical damage was found in the patients carrying the Ile/Ile genotype of the LIG4 Thr9Ile gene. 31243170_Our research revealed XRCC6 C1310G and LIG4 T9I polymorphisms are associated with diminished risk of colorectal cancer. However, to confirm obtained results, a further investigations should be carried out. 31330087_Complex genetic interactions between DNA polymerase beta and the NHEJ ligase. 31630206_Altered DNA ligase activity in human disease. 31667665_Downregulation of LIG4 was consistently associated with de novo myelodysplastic syndrome . 32534991_Ligase IV syndrome can present with microcephaly and radial ray anomalies similar to Fanconi anaemia plus fatal kidney malformations. 32879366_Inhibition of non-homologous end joining of gamma ray-induced DNA double-strand breaks by cAMP signaling in lung cancer cells. 33087274_Human DNA ligases in replication and repair. 33264406_NAD+ is not utilized as a co-factor for DNA ligation by human DNA ligase IV. 33586762_Hypomorphic mutations in human DNA ligase IV lead to compromised DNA binding efficiency, hydrophobicity and thermal stability. 34352203_Cryo-EM of NHEJ supercomplexes provides insights into DNA repair. 34614178_USP39 promotes non-homologous end-joining repair by poly(ADP-ribose)-induced liquid demixing. 34630384_Characterization of a Cohort of Patients With LIG4 Deficiency Reveals the Founder Effect of p.R278L, Unique to the Chinese Population. | ENSMUSG00000049717 | Lig4 | 204.051700 | 0.9668386 | -0.048652980 | 0.20542973 | 5.654262e-02 | 8.120464e-01 | No | Yes | 205.868219 | 44.954835 | 210.301396 | 45.927480 | ||
| ENSG00000174442 | 55055 | ZWILCH | protein_coding | Q9H900 | FUNCTION: Essential component of the mitotic checkpoint, which prevents cells from prematurely exiting mitosis. Required for the assembly of the dynein-dynactin and MAD1-MAD2 complexes onto kinetochores. Its function related to the spindle assembly machinery is proposed to depend on its association in the mitotic RZZ complex (PubMed:15824131). {ECO:0000269|PubMed:15824131}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Kinetochore;Mitosis;Phosphoprotein;Reference proteome | hsa:55055; | cytosol [GO:0005829]; kinetochore [GO:0000776]; RZZ complex [GO:1990423]; cell division [GO:0051301]; mitotic spindle assembly checkpoint signaling [GO:0007094]; protein localization to kinetochore [GO:0034501] | 12686595_Other name: HZwilch. Homologue of Drosophila Zwilch (CG18729). Localizes to the kinetochore during prometaphase and metaphase of HeLa cells. Complexed with human ROD and human ZW10, similar to situation in Drosophila. 19008095_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000032400 | Zwilch | 2233.923656 | 1.1000456 | 0.137563320 | 0.07273538 | 3.541594e+00 | 5.984791e-02 | 3.285505e-01 | No | Yes | 2580.738222 | 388.892275 | 2344.362264 | 353.202086 | ||
| ENSG00000174943 | 253980 | KCTD13 | protein_coding | Q8WZ19 | FUNCTION: Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex required for synaptic transmission (PubMed:19782033). The BCR(KCTD13) E3 ubiquitin ligase complex mediates the ubiquitination of RHOA, leading to its degradation by the proteasome (PubMed:19782033) Degradation of RHOA regulates the actin cytoskeleton and promotes synaptic transmission (By similarity). {ECO:0000250|UniProtKB:Q8BGV7, ECO:0000269|PubMed:19782033}. | 3D-structure;Autism;Autism spectrum disorder;Disease variant;Nucleus;Reference proteome;Schizophrenia;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:19782033}. | hsa:253980; | Cul3-RING ubiquitin ligase complex [GO:0031463]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; identical protein binding [GO:0042802]; protein domain specific binding [GO:0019904]; small GTPase binding [GO:0031267]; cell migration [GO:0016477]; negative regulation of Rho protein signal transduction [GO:0035024]; positive regulation of DNA replication [GO:0045740]; positive regulation of synaptic transmission [GO:0050806]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein homooligomerization [GO:0051260]; protein ubiquitination [GO:0016567]; stress fiber assembly [GO:0043149] | 16239304_PDIP1 mRNA was expressed in 3T3-L1 adipocytes and THP-1 macrophages 22596160_data suggest that KCTD13 is a major driver for the neurodevelopmental phenotypes associated with the 16p11.2 copy number variants (CNV), and reinforce the idea that one or a small number of transcripts within a CNV can underpin clinical phenotypes 27668412_Present study explores the role of KCTD13 in the development of SCZ and to provide a more complete picture of the allelic architecture at this risk locus. 30518945_Copy number variations in KCTD13 gene is associated with proximal syndromes with intellectual disability. 31402430_CRISPR/Cas9-mediated Knockout of the Neuropsychiatric Risk Gene KCTD13 Causes Developmental Deficits in Human Cortical Neurons Derived from Induced Pluripotent Stem Cells. | ENSMUSG00000030685 | Kctd13 | 571.041505 | 1.0158786 | 0.022728023 | 0.12033330 | 3.588447e-02 | 8.497543e-01 | 9.507016e-01 | No | Yes | 506.791214 | 44.864155 | 505.456492 | 44.631776 | |
| ENSG00000175029 | 1488 | CTBP2 | protein_coding | P56545 | FUNCTION: Corepressor targeting diverse transcription regulators. Functions in brown adipose tissue (BAT) differentiation (By similarity). {ECO:0000250}.; FUNCTION: Isoform 2 probably acts as a scaffold for specialized synapses. | 3D-structure;Alternative splicing;Cell junction;Differentiation;Direct protein sequencing;Host-virus interaction;Methylation;NAD;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repressor;Synapse;Transcription;Transcription regulation | This gene produces alternative transcripts encoding two distinct proteins. One protein is a transcriptional repressor, while the other isoform is a major component of specialized synapses known as synaptic ribbons. Both proteins contain a NAD+ binding domain similar to NAD+-dependent 2-hydroxyacid dehydrogenases. A portion of the 3' untranslated region was used to map this gene to chromosome 21q21.3; however, it was noted that similar loci elsewhere in the genome are likely. Blast analysis shows that this gene is present on chromosome 10. Several transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Feb 2014]. | hsa:1488; | GABA-ergic synapse [GO:0098982]; glutamatergic synapse [GO:0098978]; nucleus [GO:0005634]; photoreceptor ribbon synapse [GO:0098684]; presynaptic active zone cytoplasmic component [GO:0098831]; presynaptic cytosol [GO:0099523]; transcription repressor complex [GO:0017053]; chromatin binding [GO:0003682]; identical protein binding [GO:0042802]; NAD binding [GO:0051287]; oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [GO:0016616]; protein kinase binding [GO:0019901]; protein-containing complex binding [GO:0044877]; retinoic acid receptor binding [GO:0042974]; structural constituent of presynaptic active zone [GO:0098882]; transcription coactivator activity [GO:0003713]; transcription corepressor activity [GO:0003714]; transcription corepressor binding [GO:0001222]; cellular response to leukemia inhibitory factor [GO:1990830]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of chromatin binding [GO:0035563]; positive regulation of retinoic acid receptor signaling pathway [GO:0048386]; positive regulation of transcription by RNA polymerase II [GO:0045944]; synaptic vesicle docking [GO:0016081]; viral genome replication [GO:0019079]; white fat cell differentiation [GO:0050872] | 16356938_roles of acetylation in regulating subcellular localization and transcriptional activity of CtBP2. 16385451_Observational study of gene-disease association. (HuGE Navigator) 17023432_analysis of the CtBP2 corepressor complex induced by E1A and modulation of E1A transcriptional activity by CtBP2 17546044_E1A may gain access to cellular promoters through conservved sequence-dependent interaction with CtBP2. 18184656_PKA 1) induces metabolic changes in the adrenal cortex and 2) phosphorylates CtBP1 and 2 proteins, particularly CtBP1 at T144, resulting in CtBP protein partnering and ACTH-dependent CYP17 transcription 18264096_Genome-wide association study of gene-disease association. (HuGE Navigator) 18794092_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19318432_Observational study of gene-disease association. (HuGE Navigator) 19366831_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19423541_Observational study of gene-disease association. (HuGE Navigator) 19506021_Data show that loss of CtBP1/2 expression suppresses cell proliferation through a combination of apoptosis, reduction in cell cycle progression, and aberrations in transit through mitosis. 19549807_Observational study of gene-disease association. (HuGE Navigator) 19668232_Study shows that transcription corepressor CtBP2 directly binds acinus, which is regulated by nerve growth factor (NGF), inhibiting its stimulatory effect on cyclin A1, but not cyclin A2, expression in leukemia. 19754958_CtBP2 monomer interacts with a major CtBP-dependent repressor ZEB and HDAC and that the interaction of the two factors with the CtBP2 monomer was mutually exclusive. 19798104_Data suggest that ARF antagonism of CtBP repression of Bik and other BH3-only genes may have a critical role in ARF-induced p53-independent apoptosis and tumor suppression. 19866473_Observational study of gene-disease association. (HuGE Navigator) 19900942_Observational study of gene-disease association. (HuGE Navigator) 19902474_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20450899_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20460480_Observational study of gene-disease association. (HuGE Navigator) 20523059_CtBP2 selectively down-regulates Th2 cytokines, therefore it is a potential target for the treatment of allergic diseases. 20564319_Meta-analysis of gene-disease association. (HuGE Navigator) 20690139_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20717903_Observational study of gene-disease association. (HuGE Navigator) 20878950_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20964627_CtBP2 proteins are ubiquitously expressed in all lines and tumour samples. 21057548_we demonstrate that it is the interaction of CtBPs with transcriptional regulators and/or chromatin-modifying enzymes in the cell nucleus, rather than their role in Golgi fission, which is critical for the maintenance of mitotic fidelity. 21071540_Observational study of gene-disease association. (HuGE Navigator) 21315774_This study demonstrates that ataxin-1 occupies the promoter region of E-cadherin in vivo and that ataxin-1 activates the promoter in a CtBP2-mediated transcriptional regulation manner. 21334379_Doubly transgenic zebrafish exhibit a startle response and typical swimming behavior, indicating that there is no gross disruption of either hearing or vestibular function useful in the study of ribbon synapse development of the hair cell. 21665989_CtBP1 and CtBP2 promote the oligomerization of truncated APC through binding to the 15 amino acid repeats of truncated APC. 22945647_We propose that CtBP2 is an ovarian cancer oncogene that regulates gene expression program by modulating HDAC activity. 23255392_CtBP2 might contribute to the progression of esophageal squamous cell carcinoma through a negative transcriptional regulation of p16(INK4A). 23393140_Interaction with cyclin H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal binding protein 2 (CtBP2) and promotes cancer cell migration 23410750_these data demonstrate that CHIP regulates the steady-state level of CtBP2 as an E3 ubiquitin ligase and determines the expression levels of CtBP2 target genes. 23730208_BRCA1 expression is epigenetically repressed in sporadic ovarian cancer cells by overexpression of C-terminal binding protein 2.CtBP2 is an ovarian cancer oncogene. 23775127_CTBP2 is a transcriptional cofactor for RXR-alpha/RAR-alpha. 23853115_E2F7 recruits the co-repressor C-terminal-binding protein (CtBP) and that CtBP2 is essential for E2F7 to repress E2F1 transcription. 24012420_CtBP2 directly targeted stem cell core genes resulting in increased cancer cell stemness and increasing metastatic and tumorigenic potential. 24332637_High CTBP2 expression is associated with prostate cancer. 24522810_Overexpression of CtBP2 is associated with breast carcinoma. 24657618_Crystal structures of human CtBP1 and CtBP2 in complex with 4-Methylthio 2-oxobutyric acid and NAD. 24835310_CtBP2 is over-expressed in prostate cancer.CtBP2 promotes prostate cancer cell proliferation through c-Myc signaling. 25228652_results show how CtBP2 contributes to prostate cancer progression by modulating AR and oncogenic signaling 25686837_Enhanced CtBP2expression promoted HCC xenograft growth and induced EMT. 25820824_Our results indicated that CCNH/CDK7-CtBP2 axis may augment ESCC cell migration, and targeting the interaction of both may provide a novel therapeutic target of esophageal squamous cell carcinoma . 25895816_Releasing the key adipogenic regulator C/EBPalpha from CtBP2 binding. 26871283_Pinin and CtBP1 and CtBP2 are oncotargets that closely interact with each other to regulate transcription and pre-mRNA alternative splicing and promote cell adhesion and other epithelial characteristics of ovarian cancer cells. 27699603_Overall, our findings reveal that CtBP1/2 is essential to promote to human glioma cell growth through maintaining the DNA stability regulated by the MRN/ATR/Chk1/CDK2/HIF-1alpha signaling pathway. 28040410_This Gene-based tests suggest evidence of association with related genes, ZEB2, RND3, MCTP1, CTBP2, and beta EEG 28111233_CtBP2 ameliorated palmitic acid-induced insulin resistance via ROS-dependent JNK pathway. 28179207_CtBP2 may be a potential target to suppress tumorigenesis in neuroblastoma 28404932_this study shows that CtBP2 overexpression promotes tumor cell proliferation and invasion in gastric cancer and is associated with poor prognosis 28412731_these findings provide insight into the role CtBP2 plays in promoting proliferation and migration in breast cancer by the inhibition of p16INK4A. 28414304_CtBP2 is a druggable transforming oncoprotein critical for the evolution of neoplasia driven by Apc mutation. 28677795_High CTBP2 expression is associated with prostate cancer. 28826173_Results indicated that CTBP2 was direct target of miR-338-5p in glioma cells. CTBP2 silencing can rescued the phenotype changes induced by miR-338-5p inhibitor on cell proliferation and invasion in glioma. 29658564_the present study indicated that CtBP2 reduced the susceptibility of ECA109 cells to cisplatin by regulating the expression of apoptosis-related proteins, suggesting that it may be a promising therapeutic target in esophageal squamous cell carcinoma in the future. 29700119_Here, multi-angle light scattering (MALS) data established the NAD(+)- and NADH-dependent assembly of CtBP1 and CtBP2 into tetramers. 30151388_HBV may be involved in the occurrence and development of hepatocellular carcinoma by upregulating CtBP2 expression. 30334447_CtBP2 plays a crucial role in non-small cell lung cancer progression and cis-diamminedichloroplatinum sensitivity 30442980_CtBP promotes metastasis of breast cancer through repressing cholesterol and activating TGF-beta signaling 30585266_The results represent a conserved mechanism, by which CtBP2 serves as a NADH-dependent repressor of the p300-Runx2 transcriptional complex and thus affects bone formation. 31866012_Transcriptional co-repressor CtBP2 orchestrates epithelial-mesenchymal transition through a novel transcriptional holocomplex with OCT1. 32553630_Stabilization of C-terminal binding protein 2 by cellular inhibitor of apoptosis protein 1 via BIR domains without E3 ligase activity. 32804017_Low expression of CTBP2 and CASP8AP2 predicts risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a retrospective cohort study. 32971103_CtBP2 interacts with ZBTB18 to promote malignancy of glioblastoma. 32993576_Establishment of multifactor predictive models for the occurrence and progression of cervical intraepithelial neoplasia. 33264605_Cryo-EM structure of CtBP2 confirms tetrameric architecture. 33524397_NAD(H) phosphates mediate tetramer assembly of human C-terminal binding protein (CtBP). 33531645_microRNA-133a exerts tumor suppressive role in oral squamous cell carcinoma through the Notch signaling pathway via downregulation of CTBP2. 33580202_Low CtBP2 expression is associated with a stem cell-like signature and adverse clinical outcome in childhood B-cell lymphoblastic leukemia. 33972635_Endoplasmic reticulum stress regulates the intestinal stem cell state through CtBP2. 34052660_CtBP2 confers protection against oxidative stress through interactions with NRF1 and NRF2. 34728642_The transcriptional corepressor CtBP2 serves as a metabolite sensor orchestrating hepatic glucose and lipid homeostasis. 34781990_Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. 34878149_CtBP2 interacts with TGIF to promote the progression of esophageal squamous cell cancer through the Wnt/betacatenin pathway. 34997967_NADH/NAD(+) binding and linked tetrameric assembly of the oncogenic transcription factors CtBP1 and CtBP2. 35113002_Knockdown of receptor interacting protein 140 (RIP140) alleviated lipopolysaccharide-induced inflammation, apoptosis and permeability in pulmonary microvascular endothelial cells by regulating C-terminal binding protein 2 (CTBP2). | ENSMUSG00000030970 | Ctbp2 | 3651.071603 | 0.9746203 | -0.037087875 | 0.05650249 | 4.307847e-01 | 5.116042e-01 | 8.115359e-01 | No | Yes | 3645.506571 | 367.992803 | 3712.685448 | 374.686633 | |
| ENSG00000175066 | 256356 | GK5 | protein_coding | Q6ZS86 | ATP-binding;Alternative splicing;Glycerol metabolism;Kinase;Nucleotide-binding;Reference proteome;Transferase | PATHWAY: Polyol metabolism; glycerol degradation via glycerol kinase pathway; sn-glycerol 3-phosphate from glycerol: step 1/1. | hsa:256356; | mitochondrion [GO:0005739]; ATP binding [GO:0005524]; glycerol kinase activity [GO:0004370]; phosphotransferase activity, alcohol group as acceptor [GO:0016773]; glycerol catabolic process [GO:0019563]; glycerol metabolic process [GO:0006071]; glycerol-3-phosphate biosynthetic process [GO:0046167]; phosphorylation [GO:0016310]; triglyceride metabolic process [GO:0006641] | 25936394_miR-135b may function as an oncogene by inhibiting GK5 in glioblastoma. | ENSMUSG00000041440 | Gk5 | 849.719995 | 1.0717181 | 0.099925522 | 0.10769773 | 8.662044e-01 | 3.520082e-01 | 7.070545e-01 | No | Yes | 727.151592 | 195.098580 | 667.176545 | 179.049314 | ||
| ENSG00000175414 | 285598 | ARL10 | protein_coding | Q8N8L6 | GTP-binding;Nucleotide-binding;Reference proteome | hsa:285598; | GTP binding [GO:0005525]; GTPase activity [GO:0003924] | ENSMUSG00000025870 | Arl10 | 2893.257289 | 0.9756641 | -0.035543516 | 0.07471393 | 2.249030e-01 | 6.353292e-01 | 8.669299e-01 | No | Yes | 2462.644421 | 316.428751 | 2526.684562 | 324.644680 | ||||
| ENSG00000175595 | 2072 | ERCC4 | protein_coding | Q92889 | FUNCTION: Catalytic component of a structure-specific DNA repair endonuclease responsible for the 5-prime incision during DNA repair, and which is essential for nucleotide excision repair (NER) and interstrand cross-link (ICL) repair. {ECO:0000269|PubMed:10413517, ECO:0000269|PubMed:11790111, ECO:0000269|PubMed:19596235, ECO:0000269|PubMed:24027083, ECO:0000269|PubMed:32034146, ECO:0000269|PubMed:8797827}. | 3D-structure;Acetylation;Alternative splicing;Chromosome;Cockayne syndrome;DNA damage;DNA repair;DNA-binding;Disease variant;Dwarfism;Endonuclease;Fanconi anemia;Hydrolase;Isopeptide bond;Magnesium;Nuclease;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Xeroderma pigmentosum | The protein encoded by this gene forms a complex with ERCC1 and is involved in the 5' incision made during nucleotide excision repair. This complex is a structure specific DNA repair endonuclease that interacts with EME1. Defects in this gene are a cause of xeroderma pigmentosum complementation group F (XP-F), or xeroderma pigmentosum VI (XP6).[provided by RefSeq, Mar 2009]. | hsa:2072; | chromosome, telomeric region [GO:0000781]; ERCC4-ERCC1 complex [GO:0070522]; nucleoplasm [GO:0005654]; nucleotide-excision repair complex [GO:0000109]; nucleotide-excision repair factor 1 complex [GO:0000110]; nucleus [GO:0005634]; damaged DNA binding [GO:0003684]; endodeoxyribonuclease activity [GO:0004520]; identical protein binding [GO:0042802]; promoter-specific chromatin binding [GO:1990841]; protein C-terminus binding [GO:0008022]; protein N-terminus binding [GO:0047485]; single-stranded DNA binding [GO:0003697]; single-stranded DNA endodeoxyribonuclease activity [GO:0000014]; TFIID-class transcription factor complex binding [GO:0001094]; cellular response to UV [GO:0034644]; DNA repair [GO:0006281]; double-strand break repair via homologous recombination [GO:0000724]; double-strand break repair via nonhomologous end joining [GO:0006303]; negative regulation of double-stranded telomeric DNA binding [GO:1905768]; negative regulation of protection from non-homologous end joining at telomere [GO:1905765]; negative regulation of telomere maintenance [GO:0032205]; negative regulation of telomere maintenance via telomere lengthening [GO:1904357]; nucleotide-excision repair [GO:0006289]; nucleotide-excision repair involved in interstrand cross-link repair [GO:1901255]; nucleotide-excision repair, DNA incision [GO:0033683]; nucleotide-excision repair, DNA incision, 3'-to lesion [GO:0006295]; nucleotide-excision repair, DNA incision, 5'-to lesion [GO:0006296]; regulation of autophagy [GO:0010506]; resolution of meiotic recombination intermediates [GO:0000712]; response to UV [GO:0009411]; telomere maintenance [GO:0000723]; telomeric DNA-containing double minutes formation [GO:0061819]; UV protection [GO:0009650] | 11895912_Observational study of gene-disease association. (HuGE Navigator) 14625810_Observational study of genotype prevalence. (HuGE Navigator) 14652281_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14690602_Data reveal an unanticipated involvement of the ERCC1/XPF NER endonuclease in the regulation of telomere integrity. 14728600_XPF protein has important roles in psoralen ICL-mediated DNA repair and mutagenesis. 14734547_the ternary complex of hRad52 and XPF/ERCC1 is the active species that processes recombination intermediates generated during the repair of DNA double strand breaks and in homology-dependent gene targeting events 15095299_XPA, ERCC1 and XPF DNA repair protein expression is reduced in testis neoplasms 15184880_We show that RAD1 is an essential gene for sustained cell proliferation and that loss of Rad1 causes destabilization of Rad9 and Hus1 and consequently disintegration of the sliding-clamp complex. 15849729_Observational study of gene-disease association. (HuGE Navigator) 15886521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15886521_The combined effect of ERCC2 Asp(312)Asn and ERCC4 Ser(835)Ser genotypes might be associated with breast cancer risk in Korean women 15932882_The XPF binding sites of ERCC1 were located in helices H1 and H3 and in the C-terminal region, similar to the involved surface of XPF. 15936543_Observational study of gene-disease association. (HuGE Navigator) 16034668_A nuclear magnetic resonance examination of the DNA binding site. 16076955_XPF-ERCC1 recognizes a branched DNA substrate by binding the two ssDNA arms with the two HhH2 domains of XPF and ERCC1 and by binding the 5'-ssDNA arm with the central domain of ERCC1. 16195237_Observational study of gene-disease association. (HuGE Navigator) 16258177_Observational study of gene-disease association. (HuGE Navigator) 16284373_Observational study of gene-disease association. (HuGE Navigator) 16284380_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16338413_The ERCC1 domain folds properly only in the presence of the XPF domain, which implies a role for XPF as a scaffold for the folding of ERCC1. 16393248_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16399771_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16492920_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16537713_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16609022_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16638864_Observational study of gene-disease association. (HuGE Navigator) 16678501_XPF is required to form gamma-H2AX and likely double strand breaks in response to interstrand crosslinks in human cells 16806697_Observational study of gene-disease association. (HuGE Navigator) 16806697_results suggest that the polymorphic variants of these NER genes do not contribute to the risk of developing AD. 16823510_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17018596_A SNP (rs744154) in intron 1 was associated with recessive protection from breast cancer after adjustment for multiple testing in stage 2. It is in the first intron, in a region that is highly conserved across species, and could be causal. 17018596_Observational study of gene-disease association. (HuGE Navigator) 17055345_unanticipated nuclease-independent function of XPF in TRF2-mediated telomere shortening 17078101_Observational study of gene-disease association. (HuGE Navigator) 17210993_Observational study of gene-disease association. (HuGE Navigator) 17299578_Meta-analysis of gene-disease association. (HuGE Navigator) 17313739_Observational study of gene-disease association. (HuGE Navigator) 17438655_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17575242_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17682675_Observational study of gene-disease association. (HuGE Navigator) 17682675_The results suggest that the Arg415Gln polymorphism in ERCC4/XPF may not be linked with appearance and development of breast cancer. 17685459_Observational study of gene-disease association. (HuGE Navigator) 17912758_The XPF HhH homodimer has a larger interaction interface, aromatic stacking interactions, and additional hydrogen bond contacts as compared to the XPF/ERCC1 HhH complex, which accounts for its higher stability 17945097_Observational study of gene-disease association. (HuGE Navigator) 17945097_The gene polymorphism at ERCC4 gene had no effects on the DNA damage of lymphocytes in coke oven workers. 18006494_These results implicate the XPF-ERCC1 complex in initiating interstrand cross-links (ICL) repair by unhooking the ICL, which simultaneously induces a double strand break at a stalled fork. 18020456_Data show that incision deficiency correlates with reduced levels of DNA repair synthesis in Fanconi anemia cells and is not due to reduced levels of XPF. 18026184_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18068852_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18068852_the polymorphism rs3136038 on the promotor region of ERCC4 may contribute to the etiology of lung cancer. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18396111_To determine whether a human flap endonuclease could recognize and process this potential intermediate, the genetic requirement for the ERCC1/XPF heterodimer during LINE-1 retrotransposition was characterized 18544627_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18551366_Observational study of gene-disease association. (HuGE Navigator) 18551366_Single nucleotide polymorphism in XPF is associated with breast cancer. 18676680_Observational study of gene-disease association. (HuGE Navigator) 18701435_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18709642_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18767034_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18812185_XPF-ERCC1 controls TRF2 and telomere length maintenance through two distinctive mechanisms, with the former requiring its nuclease activity 18830263_Observational study of gene-disease association. (HuGE Navigator) 18838045_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18990748_Meta-analysis of gene-disease association. (HuGE Navigator) 19029193_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19116388_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19124519_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19124519_Variant ERCC4 genotypes are statistically significantly associated with benign breast disease, wspecially in women with a family history of breast cancer. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19258314_a model for the mechanism of ICL repair in mammalian cells that implicates the DNA glycosylase activity of NEIL1 downstream of XPF/ERCC1 and translesion DNA synthesis repair steps. 19297315_The smoking status was also predictive of both RPA.. and XPF levels.. after adjusting for age, sex ... 19339270_Observational study of gene-disease association. (HuGE Navigator) 19423537_Meta-analysis of gene-disease association. (HuGE Navigator) 19423537_Our data provide persuasive evidence against an overall association between invasive breast cancer risk and ERCC4 rs744154, TNF rs361525, CASP10 rs13010627, PGR rs1042838, and BID rs8190315 genotypes among women of European descent. 19434073_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19902366_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19920816_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19920816_the role of a SNP in intron 1 of the ERCC4 gene (rs744154), previously reported to be associated with a reduced risk of breast cancer in the general population, as a breast cancer risk modifier in BRCA1 and BRCA2 mutation carriers. 19956886_ERCC1 and XPF are upregulated during testicular germ cell tumor progression 20061190_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20062074_Observational study of gene-disease association. (HuGE Navigator) 20062074_XPF promoter -357A>C polymorphism may regulate the expression of XPF and thereby contribute to susceptibility to and prognosis of bladder cancer. 20150366_Observational study of gene-disease association. (HuGE Navigator) 20183911_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20199546_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20221251_This demonstrates that at least part of the DNA repair defect and symptoms associated with mutations in XPF are due to mislocalization of XPF-ERCC1 into the cytoplasm of cells. 20372803_squamous cell carcinoma metastases of the head and neck show increased levels of nucleotide excision repair protein XPF in vivo that correlate with increased chemoresistance ex vivo 20429839_Observational study of gene-disease association. (HuGE Navigator) 20429839_Studies indicate that a marginally statistically significant association was found for XRCC1 codon 399, XPD Asp312Asn and XRCC1 codon 194 variants and head and neck cancer. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20518486_sites of interaction of FANCG with ERCC1, which is different from the region of ERCC1 that binds to XPF 20522537_Observational study of gene-disease association. (HuGE Navigator) 20601096_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20731661_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20846399_Data indicate for the first time that the exceptional sensitivity of TTC and, therefore, very likely the curability of TGCT rests on their limited ICL repair due to low level of expression of ERCC1-XPF. 20864414_Observational study of gene-disease association. (HuGE Navigator) 20957144_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21277872_The association of MPM with DNA repair genes support the hypothesis that an increased susceptibility to DNA damage may favour asbestos carcinogenicity. 21423097_results indicated that Arg399Gln polymorphism of XRCC1 gene and Arg415Gln polymorphism of ERCC4 gene may not be associated with smoking- and drinking-related larynx cancer in Polish population 21424776_XPF Arg415Gln may be a low-penetrant risk factor in the Caucasian ethnicity for developing breast cancer. 21672525_These results indicated that persistent HBV infection might trigger NER impairment in part through upregulation of miR-192, which suppressed the levels of ERCC3 and ERCC4. 21737503_Data show that high XPF expression correlated with early time to progression both by univariate and multivariate analysis. 21839691_This work reveals a new role for ERCC1 distinct from its known function in DNA repair, which may be independent of XPF. 21896658_collaboration between hSNM1A and XPF-ERCC1 is necessary to initiate ICL repair in replicating human cells 22212909_ERCC1-XPF dependent DNA repair is critical for protecting prostate epithelial from transformation to cancer 22353549_these results suggest that the interaction between XPF and Eg5 plays a role in mitosis and DNA repair and offer new insights into the pathogenesis of XP-F and XFE. 22457069_XPF-ERCC1 has a preference for cleaving the phosphodiester bond positioned on the 3'-side of a T or a U, which is flanked by an upstream T or U suggesting that a T/U pocket may exist within the catalytic domain. 22547097_Multiple DNA binding domains mediate the function of the ERCC1-XPF protein in nucleotide excision repair 22609620_There is no evidence that G1244A and T2505 single nucleotide polymorphisms in XPF affect expression of ERCC1. 22768293_This meta-analysis suggests a lack of statistical evidence for the association between the four XPF SNPs and overall risk of cancers. 22771116_XPG endonuclease promotes DNA breaks and DNA demethylation at promoters allowing the recruitment of CTCF and gene looping, which is further stabilized by XPF. 22848636_two SNPs (rs2276466 and rs3136038) in ERCC4 may be functional and contribute to SCCHN susceptibility 23385459_Frameshift mutation XP11BE not only divests the XPF-interaction motif impairing DNA repair, but also reduces XPB solubility, leading to a lower intracellular level of transcription factor TFIIH and deficient transcription. 23415627_The ERCC4 tagSNPs, rs6498486 and rs254942, may play protective roles in gastric carcinogenesis, especially in the development of atrophic gastritis. 23435956_individuals whose expression of XRCC1, ERCC4, ERCC2, and ERCC1 are reduced may be at a higher risk of developing squamous intraepithelial lesion which eventually leads to invasive cervical carcinoma. 23537993_a significantly decreased risk of gastric cancer associated with the ERCC4 rs744154 GC/CC genotypes in a Chinese population 23580445_The compound interacts with the domain of XPF responsible for interaction with ERCC1. 23623386_Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. 23623389_Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. 23679285_Polymorphisms in rs180067, rs1799801, rs2276466 and rs744154 in XPF is associated with gastric cancer. 23909490_The present study shows that the ERCC1 rs11615 and XPF rs6498486 polymorphisms are associated with breast cancer risk in a Chinese population. 23982883_Overexpression of XPF decreased drug sensitivity in malignant melanoma. 23991957_The rs1800067 G and rs2276466 G allele frequencies in Xeroderma pigmentosum complementation group f polymorphisms influence risk of glioma. 24004570_ERCC4 rs1800124 and MBD4 rs10342 non-synonymous single nucleotide polymorphism variants were associated with DNA repair capacity. 24027083_the frequency of Spanish individuals heterozygous for pathogenic mutations in the ERCC4 gene is approximately 0.3%, and it does not differ between familial breast/ovarian cancer patients and healthy controls. 24318989_ERCC1 (rs11615, rs3212986C>A, and rs2298881) and ERCC4 (rs226466C>G, rs2276465, and rs6498486) were selected and genotyped 24412486_The C2169A nonsense mutation in XPF protein is closely associated with gastric carcinogenesis in the Chinese population.The XPF mutation is largely monoallelic indicating the haplo-insufficiency of XPF. 24465539_The contribution of ERCC4/FANCQ coding mutations to hereditary breast cancer in Central and Eastern Europe is likely to be small. 24709955_genetic variants in XPF might contribute to the susceptibility to ESCC 24861646_no significant associations between XPF polymorphisms in rs2276466 or rs6498486 and risk of colorectal cancer in a Chinese population 24938470_Our study suggests that the rs1800067 genetic variant of XPF functions in the development of glioma. 25019640_Low XPF expression in head and neck squamous cell carcinoma patients is associated with better response to induction chemoradiotherapy, while high XPF expression correlates with a worse response. 25292041_ERCC4 rs1800067 polymorphism is not associated with cancer risk. 25342505_This meta-analysis suggests that the 3 common XPF polymorphisms rs744154, rs6498486, and rs1799801 are not associated with gastric cancer risk. 25535740_XPF and XPC expression may be a potential predictive factor for bladder cancer, and smoking can not only influence the recurrence of bladder cancer as a single factor but also aggravate the results of the XPF defect and XPC defect. 25538220_these results establish USP45 as a new regulator of XPF-ERCC1 crucial for efficient DNA repair 25730007_we found no association between the rs11615 of the ERCC1 and rs2276466 of the XPF polymorphisms and the risk of colorectal cancer 25812040_Polymorphisms of ERCC4 gene are associated with HPV-positive cervical cancer. 25879486_Mus81-deficient cells fail to recover from exposure to low doses of replication inhibitors and cell viability is dependent on the XPF endonuclease. 26025908_ERCC1-XPF participates in DNA repair of the Top1-DNA damage complex. ERCC1-XPF and RPA form a DNA/protein complex on the nick DNA substrate. 26045829_Genotypes of ERCC1 (rs11615, rs3212986 and rs2298881) and XPF (rs2276465 and rs6498486) were performed by Polymerase Chain Reaction Restriction Fragment Length Polymorphism (PCR-RFLP) assay 26074087_ERCC4 is located on human chromosome 16p13.12 and consists of 11 exons spanning about 28.2 kb, the broad tissue expression pattern of ERCC4 is similar to that of ERCC1; ERCC1 and ERCC4 genes encode the two subunits of the ERCC1-XPF nuclease. This enzyme plays an important role in repair of DNA damage and in maintaining genomic stability. [Review] 26085086_F231L mutation results in only a small disturbance of the ERCC1-XPF interface, where, in contrast to Phe(231), Leu(231) lacks interactions stabilizing the ERCC1-XPF complex 26146099_Based on these results, we conclude that the XPF gene polymorphism Ser835Ser may be associated with a decreased risk of colorectal cancer. 26411687_Helicobacter Pylori introduces double-stranded DNA breaks by the nucleotide excision repair endonucleases XPF and XPG, which, together with RelA, are recruited to chromatin in a highly coordinated, type IV secretion system-dependent manner. 26427666_inherited abnormalities in DNA repair pathway related to XPF 30028C and TP53 Arg72Pro polymorphisms act as prognostic factors for progression free survival and overall survival of cutaneous melanoma patients. 26453996_SLX4 (FANCP) and XPF (FANCQ) proteins interact with each other and play a vital role in the Fanconi anemia (FA) DNA repair pathway. 26888738_C allele of the 30028T/C polymorphism significantly increased the risk of ischemic stroke. 27460091_Polymorphisms in XPF gene is associated with gastrointestinal stromal tumours. 27542841_silenced XPF significantly increased the sensitivity and survival following treatment with cisplatin in xenograft mice bearing renal cell tumor. 28028171_Based on structural models, NMR titrations, DNA-binding studies, site-directed mutagenesis, charge distribution, and sequence conservation, we propose that the HhH domain of ERCC1 binds to dsDNA upstream of the damage, and XPF binds to the non-damaged strand within a repair bubble 28130555_ERCC1 was not detectable in the nucleus of the XPF knockout cells indicating the necessity of a functional XPF/ERCC1 heterodimer to allow ERCC1 to enter the nucleus. 28607004_Strikingly, the addition of the single-stranded DNA (ssDNA)-binding replication protein A (RPA) selectively restores XPF-ERCC1 endonuclease activity on this structure. The 5'-3' exonuclease SNM1A can load from the XPF-ERCC1-RPA-induced incisions and digest past the crosslink to quantitatively complete the unhooking reaction. 29403087_Our results confirm that biallelic ERCC4 mutations cause a cerebellar ataxia-dominant phenotype with mild cutaneous symptoms, possibly accounting for a high proportion of the genetic causes of ARCA in Japan, where XP-F is prevalent. 29544698_common genetic variations in ERCC1/XPF genes predispose to neuroblastoma risk, which needs to be further validated by ongoing efforts. 29739952_XPF and MUS81 merit further validation in prospective clinical trials as biomarkers that may predict clinical response of esophageal adenocarcinoma (EAC) to oxaliplatin-based chemotherapy. 29741112_For 11985 A>G polymorphism, lung cancer subjects treated with irinotecan cisplatin/carboplatin regimen having heterozygous genotype (AG) was associated with high mortality risk (p = 0.0001). 673 C>T polymorphism was associated with increased lung cancer risk. 30059501_Study provides evidence that Replication forks can break quickly in S-phase upon DNA replication stress induction by an endonucleolytic mechanism independent of MUS81. Two nucleases ARTEMIS and XPF-ERCC1 are responsible for this Rapid-Replication Fork Breakage (RRFB) which takes place during S and G2 phases of the cell cycle. 30165384_we investigated how specific XPF mutations found in patients affected with xeroderma pigmentosum(XP) , XP combined with Cockayne syndrome or Fanconi anemia impair the activity of the ERCC1-XPF complex in response to DNA damage induction by UV irradiation. We show that XPF with an XP mutation is inefficiently recruited into the NER machinery but retains repair activity. 30527102_XPF rs1799801 was associated with severe preeclampsia and early-onset preeclampsia in Chinese Han Women. 30658521_Functional studies in human XPF-KO isogenic cells demonstrate that a defined cellular phenotype cannot be easily correlated to each pathogenic XPF mutation. Substituted positions along XPF sequences are not predictive of cellular phenotype nor reflect a particular disease. 31040199_Impact of XPF rs2276466 polymorphism on cancer susceptibility: a meta-analysis. 31226203_The results indicate that core nucleotide excision repair (NER) components XPA and XPF are equally required and that both global genome (GG-NER) and transcription coupled (TC-NER) subpathways contribute to the repair. 31392348_Our results confirm its dependence on XPA in NER and furthermore show that its engagement in ICLR is dependent on FANCD2. Interestingly, we find that two ICLR-defective XPF mutants (R689S and S786F) are less well recruited to ICLs. These studies highlight the differential mechanisms that regulate ERCC1-XPF activity in DNA repair 31493872_DCAF7 is a novel regulator of ERCC1-XPF protein level and cellular nucleotide excision repair activity. 31495888_The binding of SLX4IP to both SLX4 and XPF-ERCC1 not only is vital for maintaining the stability of SLX4IP protein, but also promotes the interaction between SLX4 and XPF-ERCC1, especially after DNA damage. Collectively, these results demonstrate a new regulatory role for SLX4IP in maintaining an efficient SLX4-XPF-ERCC1 complex in interstrand crosslinks (ICLs) repair. 31568607_ERCC4 rs6498486 polymorphism is associated with colorectal cancer risk. 31862643_results suggest a potential involvement of APE-1, XRCC-1 and XPF proteins in the pathogenesis of benign epithelial odontogenic lesions, especially in those with more aggressive biological behavior 32034146_TIP60 directly acetylates XPF at Lysine 911 following ultraviolet irradiation and that this acetylation is required for XPF-ERCC1 complex assembly and subsequent activation. 32683874_Association of XPG rs17655G>C and XPF rs1799801T>C Polymorphisms with Susceptibility to Cutaneous Malignant Melanoma: Evidence from a Case-Control Study, Systematic Review and Meta-Analysis', trans 'Vztah mezi polymorfismem XPG rs17655G>C a XPF rs1799801T>C a nachylnosti k malignimu melanomu kůže: důkazy ze studie připadů a kontrol, systematicky přehled a metaanalyza. 32739873_Tonicity-responsive enhancer-binding protein promotes stemness of liver cancer and cisplatin resistance. 33067872_Distribution and susceptibility of ERCC1/XPF gene polymorphisms in Han and Uygur women with breast cancer in Xinjiang, China. 33202356_DNA base excision repair and nucleotide excision repair proteins in malignant salivary gland tumors. 33347546_SLX4-XPF mediates DNA damage responses to replication stress induced by DNA-protein interactions. 33827099_Contribution of XPD and XPF Polymorphisms to Susceptibility of Non-Small Cell Lung Cancer in High-Altitude Areas. 33931939_The ZEB2-dependent EMT transcriptional programme drives therapy resistance by activating nucleotide excision repair genes ERCC1 and ERCC4 in colorectal cancer. 34416547_XPF -673C>T variation is associated with the susceptibility to breast cancer. 34633654_F-circEA1 regulates cell proliferation and apoptosis through ALK downstream signaling pathway in non-small cell lung cancer. 34837148_The link of ERCC2 rs13181 and ERCC4 rs2276466 polymorphisms with breast cancer in the Bangladeshi population. | ENSMUSG00000022545 | Ercc4 | 972.200669 | 1.1518311 | 0.203929168 | 0.09638789 | 4.458432e+00 | 3.472937e-02 | 2.511419e-01 | No | Yes | 916.158983 | 149.437930 | 785.558622 | 128.106659 | |
| ENSG00000175611 | 100128782 | LINC00476 | lncRNA | 33370431_LINC00476 Suppresses the Progression of Non-Small Cell Lung Cancer by Inducing the Ubiquitination of SETDB1. | 129.378753 | 0.8413564 | -0.249211098 | 0.23177993 | 1.157256e+00 | 2.820354e-01 | No | Yes | 119.227567 | 14.789404 | 139.798458 | 17.037737 | ||||||||||
| ENSG00000175826 | 23399 | CTDNEP1 | protein_coding | O95476 | FUNCTION: Serine/threonine protein phosphatase forming with CNEP1R1 an active phosphatase complex that dephosphorylates and may activate LPIN1 and LPIN2. LPIN1 and LPIN2 are phosphatidate phosphatases that catalyze the conversion of phosphatidic acid to diacylglycerol and control the metabolism of fatty acids at different levels. May indirectly modulate the lipid composition of nuclear and/or endoplasmic reticulum membranes and be required for proper nuclear membrane morphology and/or dynamics. May also indirectly regulate the production of lipid droplets and triacylglycerol. May antagonize BMP signaling. {ECO:0000269|PubMed:17420445, ECO:0000269|PubMed:22134922}. | Endoplasmic reticulum;Hydrolase;Membrane;Nucleus;Protein phosphatase;Reference proteome;Transmembrane;Transmembrane helix | hsa:23399; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; lipid droplet [GO:0005811]; Nem1-Spo7 phosphatase complex [GO:0071595]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; phosphoprotein phosphatase activity [GO:0004721]; protein serine phosphatase activity [GO:0106306]; protein serine/threonine phosphatase activity [GO:0004722]; protein threonine phosphatase activity [GO:0106307]; gamete generation [GO:0007276]; mesoderm development [GO:0007498]; mitotic nuclear membrane disassembly [GO:0007077]; nuclear envelope organization [GO:0006998]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of triglyceride biosynthetic process [GO:0010867]; protein dephosphorylation [GO:0006470]; protein localization to nucleus [GO:0034504] | Mouse_homologues 23360989_Dullard keeps BMP signalling at an appropriate level, which is required for nephron maintenance in the postnatal period. 23469192_Dullard may play a role in the fine-tuning of WNT signalling activity by modulating the expression of ligands/antagonists and the availability of Dvl2 protein during specification of the germ cell lineage. 25155999_Dullard is involved in suppression of TGF-beta signaling during endochondral ossification. 29521016_Dullard deficiency results in hemorrhagic ovarian cysts and infertility. 32105214_Dullard-mediated Smad1/5/8 inhibition controls mouse cardiac neural crest cells condensation and outflow tract septation. | ENSMUSG00000018559 | Ctdnep1 | 3115.074092 | 1.0009506 | 0.001370824 | 0.06085319 | 5.078132e-04 | 9.820214e-01 | 9.935391e-01 | No | Yes | 3144.134721 | 325.718883 | 3143.301030 | 325.461319 | ||
| ENSG00000175970 | 84747 | UNC119B | protein_coding | A6NIH7 | FUNCTION: Myristoyl-binding protein that acts as a cargo adapter: specifically binds the myristoyl moiety of a subset of N-terminally myristoylated proteins and is required for their localization. Binds myristoylated NPHP3 and plays a key role in localization of NPHP3 to the primary cilium membrane. Does not bind all myristoylated proteins. Probably plays a role in trafficking proteins in photoreceptor cells. {ECO:0000269|PubMed:22085962}. | 3D-structure;Acetylation;Cell projection;Cilium;Cilium biogenesis/degradation;Lipid-binding;Protein transport;Reference proteome;Transport | hsa:84747; | ciliary transition zone [GO:0035869]; cilium [GO:0005929]; cytosol [GO:0005829]; lipid binding [GO:0008289]; cilium assembly [GO:0060271]; lipoprotein transport [GO:0042953]; nervous system development [GO:0007399] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000046562 | Unc119b | 3130.030363 | 1.1685813 | 0.224758122 | 0.06441355 | 1.214445e+01 | 4.923413e-04 | 2.276847e-02 | No | Yes | 2777.092621 | 301.128412 | 2323.309959 | 251.935640 | ||
| ENSG00000176371 | 54993 | ZSCAN2 | protein_coding | Q7Z7L9 | FUNCTION: May be involved in transcriptional regulation during the post-meiotic stages of spermatogenesis. {ECO:0000250}. | Alternative splicing;DNA-binding;Developmental protein;Differentiation;Metal-binding;Nucleus;Reference proteome;Repeat;Spermatogenesis;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene contains several copies of zinc finger motif, which is commonly found in transcriptional regulatory proteins. Studies in mice show that this gene is expressed during embryonic development, and specifically in the testis in adult mice, suggesting that it may play a role in regulating genes in germ cells. Alternative splicing of this gene results in several transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:54993; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; cell differentiation [GO:0030154]; regulation of transcription by RNA polymerase II [GO:0006357]; spermatogenesis [GO:0007283] | 1937051_PMID:1937051 pertains to studies in mouse. | ENSMUSG00000038797 | Zscan2 | 312.200305 | 0.9210931 | -0.118581107 | 0.17943444 | 4.257513e-01 | 5.140811e-01 | No | Yes | 294.178584 | 40.333013 | 323.973484 | 44.374942 | ||
| ENSG00000176473 | 79446 | WDR25 | protein_coding | Q64LD2 | Alternative splicing;Reference proteome;Repeat;WD repeat | This gene encodes a protein containing 7 WD repeats. WD repeats are approximately 30 to 40-amino acid domains containing several conserved residues, typically having a Tryptophan-Aspartic acid dipeptide (WD) at the C-terminal end. WD domains are involved in protein-protein interactions in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Apr 2017]. | hsa:79446; | ENSMUSG00000040877 | Wdr25 | 262.332463 | 0.9215821 | -0.117815366 | 0.18803814 | 3.835002e-01 | 5.357359e-01 | No | Yes | 265.115795 | 35.794296 | 281.335535 | 37.859343 | |||||
| ENSG00000177150 | 125228 | FAM210A | protein_coding | Q96ND0 | FUNCTION: May play a role in the structure and strength of both muscle and bone. {ECO:0000250|UniProtKB:Q8BGY7}. | Coiled coil;Cytoplasm;Membrane;Mitochondrion;Reference proteome;Transmembrane;Transmembrane helix | hsa:125228; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 29618611_Genetic variation near FAM210A, a gene of previously unknown function, was strongly associated with both appendicular and whole body lean mass, as well as bone mineral density. | ENSMUSG00000038121 | Fam210a | 610.777873 | 1.0315699 | 0.044841527 | 0.12341700 | 1.318681e-01 | 7.165029e-01 | 9.073039e-01 | No | Yes | 699.801379 | 109.830146 | 656.581376 | 103.019300 | ||
| ENSG00000177335 | 286122 | LINC02904 | lncRNA | 27.225550 | 0.9125442 | -0.132033623 | 0.49998560 | 6.850639e-02 | 7.935240e-01 | No | Yes | 22.920299 | 5.447863 | 26.225988 | 6.346415 | |||||||||||
| ENSG00000177406 | 100049716 | NINJ2-AS1 | lncRNA | 27.657680 | 1.1734857 | 0.230800313 | 0.50275620 | 2.164611e-01 | 6.417497e-01 | No | Yes | 30.323860 | 7.708947 | 25.923973 | 6.563148 | |||||||||||
| ENSG00000179134 | 55095 | SAMD4B | protein_coding | Q5PRF9 | FUNCTION: Has transcriptional repressor activity. Overexpression inhibits the transcriptional activities of AP-1, p53/TP53 and CDKN1A. {ECO:0000269|PubMed:20510020}. | Cytoplasm;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation | hsa:55095; | cytosol [GO:0005829]; nucleus [GO:0005634]; P-body [GO:0000932]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; translation repressor activity [GO:0030371]; nuclear-transcribed mRNA poly(A) tail shortening [GO:0000289] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20510020_Results suggest that SAMD4B is a widely expressed gene involved in AP-1-, p53-and p21-mediated transcriptional signaling activity. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 34109425_miR451 suppresses the malignant characteristics of colorectal cancer via targeting SAMD4B. | ENSMUSG00000109336 | Samd4b | 4566.134128 | 0.9294860 | -0.105494926 | 0.05450125 | 3.738291e+00 | 5.317881e-02 | 3.108483e-01 | No | Yes | 4029.531286 | 455.510936 | 4396.746529 | 496.917250 | ||
| ENSG00000179364 | 23241 | PACS2 | protein_coding | Q86VP3 | FUNCTION: Multifunctional sorting protein that controls the endoplasmic reticulum (ER)-mitochondria communication, including the apposition of mitochondria with the ER and ER homeostasis. In addition, in response to apoptotic inducer, translocates BIB to mitochondria, which initiates a sequence of events including the formation of mitochondrial truncated BID, the release of cytochrome c, the activation of caspase-3 thereby causing cell death. May also be involved in ion channel trafficking, directing acidic cluster-containing ion channels to distinct subcellular compartments. {ECO:0000269|PubMed:15692563, ECO:0000269|PubMed:15692567}. | Alternative splicing;Apoptosis;Disease variant;Endoplasmic reticulum;Epilepsy;Host-virus interaction;Mitochondrion;Phosphoprotein;Reference proteome | hsa:23241; | endoplasmic reticulum [GO:0005783]; mitochondrion [GO:0005739]; transmembrane transporter binding [GO:0044325]; apoptotic process [GO:0006915]; autophagosome assembly [GO:0000045]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; mitochondrion-endoplasmic reticulum membrane tethering [GO:1990456]; protein localization to phagophore assembly site [GO:0034497]; protein localization to plasma membrane [GO:0072659] | 15692563_subcellular localization and function of polycystin-2 are directed by phosphofurin acidic cluster sorting protein (PACS)-1 and PACS-2 15692567_PACS-2 as a novel sorting protein that links the endoplasmic reticulum (ER)-mitochondria axis to ER homeostasis 18296443_PACS-2 is required for Nef action and sorting of itinerant membrane cargo in the TGN/endosomal system 18417615_the phosphorylation state of the calnexin cytosolic domain and its interaction with PACS-2 sort the chaperone between domains of the ER and the plasma membrane 19481529_Results identify PACS-2 as an essential TRAIL effector, and show that Akt cooperates with 14-3-3 to regulate the homeostatic and apoptotic properties of PACS-2 that mediate TRAIL action. 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 22496420_The sites on Nef and the PACS proteins required for their interaction, are identified. 22645134_Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein-induced lysosomal translocation of proapoptotic effectors is mediated by phosphofurin acidic cluster sorting protein-2. 24633224_cIAPs constitutively downregulate PACS-2 by polyubiquitination and proteasomal degradation, thereby restraining TRAIL-induced killing of liver cancer cells 28476937_Protein adaptation and the expanding roles of the PACS1 and PACS2 proteins in tissue homeostasis and disease has been summarized. (Review) 28867141_Missense Mutation in PACS2 gene is associated with Neurodevelopmental Disorders. 29656858_These findings support the causality of this recurrent de novo PACS2 heterozygous missense in Developmental and epileptic encephalopathies with facial dysmorphim and cerebellar dysgenesis. 30415949_Insulin acutely regulates SIRT1 activity by triggering recruitment of PACS-2 and DBC1 to the SIRT1 N terminal region creating a regulatory hub. 30684285_These findings confirmed our previous research and expanded the mutational spectrum of WEE2, making it a potential genetic diagnostic marker for those suffering from human fertilization failure. 31242668_the role of the multifunctional Mitochondria-associated endoplasmic reticulum membrane protein phosphofurin acidic cluster sorting protein 2 (PACS-2) in regulating Vascular Smooth Muscle Cell survival following a challenge by atherogenic lipids. 33243487_Clinical variations of epileptic syndrome associated with PACS2 variant. 33369122_Coloboma may be a shared feature in a spectrum of disorders caused by mutations in the WDR37-PACS1-PACS2 axis. 34405643_[Early infantile epileptic encephalopathy caused by PACS2 gene variation: three cases report and literature review]. | ENSMUSG00000021143 | Pacs2 | 1497.380438 | 1.0038989 | 0.005614034 | 0.09371954 | 3.644365e-03 | 9.518621e-01 | 9.830432e-01 | No | Yes | 1436.183885 | 165.304921 | 1431.702979 | 164.763616 | ||
| ENSG00000179950 | 22827 | PUF60 | protein_coding | Q9UHX1 | FUNCTION: DNA- and RNA-binding protein, involved in several nuclear processes such as pre-mRNA splicing, apoptosis and transcription regulation. In association with FUBP1 regulates MYC transcription at the P2 promoter through the core-TFIIH basal transcription factor. Acts as a transcriptional repressor through the core-TFIIH basal transcription factor. Represses FUBP1-induced transcriptional activation but not basal transcription. Decreases ERCC3 helicase activity. Does not repress TFIIH-mediated transcription in xeroderma pigmentosum complementation group B (XPB) cells. Is also involved in pre-mRNA splicing. Promotes splicing of an intron with weak 3'-splice site and pyrimidine tract in a cooperative manner with U2AF2. Involved in apoptosis induction when overexpressed in HeLa cells. Isoform 6 failed to repress MYC transcription and inhibited FIR-induced apoptosis in colorectal cancer. Isoform 6 may contribute to tumor progression by enabling increased MYC expression and greater resistance to apoptosis in tumors than in normal cells. Modulates alternative splicing of several mRNAs. Binds to relaxed DNA of active promoter regions. Binds to the pyrimidine tract and 3'-splice site regions of pre-mRNA; binding is enhanced in presence of U2AF2. Binds to Y5 RNA in association with RO60. Binds to poly(U) RNA. {ECO:0000269|PubMed:10606266, ECO:0000269|PubMed:10882074, ECO:0000269|PubMed:11239393, ECO:0000269|PubMed:16452196, ECO:0000269|PubMed:16628215, ECO:0000269|PubMed:17579712}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;DNA-binding;Direct protein sequencing;Disease variant;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Ribonucleoprotein;Transcription;Transcription regulation;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a nucleic acid-binding protein that plays a role in a variety of nuclear processes, including pre-mRNA splicing and transcriptional regulation. The encoded protein forms a complex with the far upstream DNA element (FUSE) and FUSE-binding protein at the myelocytomatosis oncogene (MYC) promoter. This complex represses MYC transcription through the core-TFIIH basal transcription factor. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2012]. | hsa:22827; | cell junction [GO:0030054]; nucleoplasm [GO:0005654]; cadherin binding [GO:0045296]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; RNA binding [GO:0003723]; alternative mRNA splicing, via spliceosome [GO:0000380]; apoptotic process [GO:0006915]; mRNA splice site selection [GO:0006376]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381] | 16628215_Results describe the roles of the FarUpStream Element (FUSE), FUSE Binding Protein (FBP), FBP Interacting Repressor (FIR), and TFIIH in the regulation of c-myc expression. 17579712_study identified PUF60 as a factor that promotes splicing of an intron with a weak 3' splice-site; demonstrated that PUF60 can functionally substitute for U2AF(65)in vitro, but splicing is strongly stimulated by the presence of both proteins 18974054_Puf60-UHM binds to ULM sequences in the splicing factors SF1, U2AF65, and SF3b155. 20420426_FIR is monomeric in solution but dimerizes upon DNA binding; DNA-induced dimerization is mediated by FIR's RNA recognition motif. 22496461_Data indicate that altered FIR and c-myc pre-mRNA splicing, in addition to c-Myc expression by augmented FIR/FIRDeltaexon2-SAP155 complex, potentially contribute to colorectal cancer development. 23113893_Circulating FIR variant mRNA in the peripheral blood of cancer patients were significantly overexpressed compared to that in healthy volunteers. 23594796_The interaction between SAP155 and FIR/FIRDeltaexon2 not only integrates cell-cycle progression and c-Myc transcription by modifying P27 and P89 expression. 24140112_Haploinsufficiency of each of SCRIB or PUF60 contribute uniquely to specific endophenotypes (e.g., coloboma, heart defects), and binary interaction potentially exacerbates other aspects of the clinical pathology of individuals with 8q24.3 deletion. 24811221_High FBP-interacting repressor expression is associated with hepatocellular carcinoma. 24824848_Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. 26177862_Concomitant over expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer cells. 26497854_Mutations in PUF60 gene is associated with idiopathic hypereosinophilic syndrome. 27756887_PUF60 auto-antibodies are detected in the sera of early-stage colon cancer patients and level decreases after surgery. 27804958_These results confirm that PUF60 is a major driver for the developmental, craniofacial, skeletal and cardiac phenotypes associated with the 8q24.3 microdeletion 28074499_3 Verheij syndrome patients with de novo pathogenic variants in PUF60 are described. Mutations in gene PUF60 were analyzed. 28327570_Heterozygote loss-of-function variants in PUF60 cause a phenotype comprising growth/developmental delay and craniofacial, cardiac, renal, ocular and spinal anomalies. 28471317_The present report describes a de novo missense mutation in PUF60, detected in a boy with multiple congenital anomalies. 28990276_Authors report the identification of 25 DNVs out of which five were classified as pathogenic or likely pathogenic. A two base pair deletion was identified in the PUF60 gene. Result adds to the growing evidence that PUF60 is responsible for the majority of the symptoms reported for carriers of a microdeletion across this region. 28993636_PUF60 is involved in: 1) up-regulation of core promoter activity through its interaction with transcription factor TCF7L2, 2) promotion of 3.5 kb RNA degradation and 3) suppression of 3.5 kb RNA splicing. 29541951_Anti-PUF60 antibodies were nonspecific for myositis, since they could be detected in other rheumatic diseases. 29788428_PUF60-activated exons uncover altered 3' splice-site selection by germline missense mutations in a single RNA recognition motif. 33061812_Poly(U) binding splicing factor 60 promotes renal cell carcinoma growth by transcriptionally upregulating telomerase reverse transcriptase. 33253191_Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60. | ENSMUSG00000002524 | Puf60 | 6164.405968 | 0.9633363 | -0.053888577 | 0.05718553 | 8.808155e-01 | 3.479784e-01 | 7.043220e-01 | No | Yes | 6080.168624 | 836.776059 | 6427.065324 | 884.335389 | |
| ENSG00000180479 | 51276 | ZNF571 | protein_coding | Q7Z3V5 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:51276; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 109.497016 | 1.4015831 | 0.487057329 | 0.25273756 | 3.713537e+00 | 5.397290e-02 | No | Yes | 118.712880 | 26.986426 | 82.319141 | 18.663347 | ||||||
| ENSG00000180628 | 84333 | PCGF5 | protein_coding | Q86SE9 | FUNCTION: Component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones; it mediates monoubiquitination of histone H2A 'Lys-119', rendering chromatin heritably changed in its expressibility (PubMed:26151332). Within the PRC1-like complex, regulates RNF2 ubiquitin ligase activity (PubMed:26151332). Plays a redundant role with PCGF3 as part of a PRC1-like complex that mediates monoubiquitination of histone H2A 'Lys-119' on the X chromosome and is required for normal silencing of one copy of the X chromosome in XX females (By similarity). {ECO:0000250|UniProtKB:Q3UK78, ECO:0000269|PubMed:26151332}. | 3D-structure;Alternative splicing;Metal-binding;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:84333; | centrosome [GO:0005813]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PcG protein complex [GO:0031519]; PRC1 complex [GO:0035102]; X chromosome [GO:0000805]; metal ion binding [GO:0046872]; histone H2A-K119 monoubiquitination [GO:0036353]; inactivation of X chromosome by genetic imprinting [GO:0060819]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385451_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000024805 | Pcgf5 | 1517.739106 | 1.0577316 | 0.080973531 | 0.07772683 | 1.087281e+00 | 2.970744e-01 | 6.650707e-01 | No | Yes | 909.163976 | 80.674405 | 846.463054 | 75.154627 | ||
| ENSG00000181192 | 55526 | DHTKD1 | protein_coding | Q96HY7 | FUNCTION: The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components: 2-oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3) (By similarity). {ECO:0000250}. | 3D-structure;Charcot-Marie-Tooth disease;Disease variant;Glycolysis;Mitochondrion;Neurodegeneration;Neuropathy;Oxidoreductase;Reference proteome;Thiamine pyrophosphate;Transit peptide | This gene encodes a component of a mitochondrial 2-oxoglutarate-dehydrogenase-complex-like protein involved in the degradation pathways of several amino acids, including lysine. Mutations in this gene are associated with 2-aminoadipic 2-oxoadipic aciduria and Charcot-Marie-Tooth Disease Type 2Q. [provided by RefSeq, May 2013]. | hsa:55526; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; oxoglutarate dehydrogenase (succinyl-transferring) activity [GO:0004591]; thiamine pyrophosphate binding [GO:0030976]; generation of precursor metabolites and energy [GO:0006091]; glycolytic process [GO:0006096]; hematopoietic progenitor cell differentiation [GO:0002244]; tricarboxylic acid cycle [GO:0006099] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 23141293_DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. 23141294_A nonsense mutation in DHTKD1 causes Charcot-Marie-Tooth disease type 2 in a large Chinese pedigree. 24076469_DHTKD1 contributes to mitochondrial biogenesis and function maintenance. 25860818_DHTKD1 encodes the E1 subunit of the alpha-ketoadipic acid dehydrogenase complex 32024885_Synthetic analogues of 2-oxo acids discriminate metabolic contribution of the 2-oxoglutarate and 2-oxoadipate dehydrogenases in mammalian cells and tissues. 32160276_DHTKD1 and OGDH display substrate overlap in cultured cells and form a hybrid 2-oxo acid dehydrogenase complex in vivo. 32303640_Structure-function analyses of the G729R 2-oxoadipate dehydrogenase genetic variant associated with a disorder of l-lysine metabolism. 32633484_Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex. 34484123_Knock-Out of DHTKD1 Alters Mitochondrial Respiration and Function, and May Represent a Novel Pathway in Cardiometabolic Disease Risk. 35052424_Heterozygous DHTKD1 Variants in Two European Cohorts of Amyotrophic Lateral Sclerosis Patients. | ENSMUSG00000025815 | Dhtkd1 | 2384.565128 | 1.0446193 | 0.062977272 | 0.06643247 | 9.035623e-01 | 3.418283e-01 | 6.997985e-01 | No | Yes | 2519.295561 | 227.879488 | 2379.453476 | 215.295363 | |
| ENSG00000182185 | 5890 | RAD51B | protein_coding | O15315 | FUNCTION: Involved in the homologous recombination repair (HRR) pathway of double-stranded DNA breaks arising during DNA replication or induced by DNA-damaging agents. May promote the assembly of presynaptic RAD51 nucleoprotein filaments. Binds single-stranded DNA and double-stranded DNA and has DNA-dependent ATPase activity. Part of the RAD21 paralog protein complex BCDX2 which acts in the BRCA1-BRCA2-dependent HR pathway. Upon DNA damage, BCDX2 acts downstream of BRCA2 recruitment and upstream of RAD51 recruitment. BCDX2 binds predominantly to the intersection of the four duplex arms of the Holliday junction and to junction of replication forks. The BCDX2 complex was originally reported to bind single-stranded DNA, single-stranded gaps in duplex DNA and specifically to nicks in duplex DNA. The BCDX2 subcomplex RAD51B:RAD51C exhibits single-stranded DNA-dependent ATPase activity suggesting an involvement in early stages of the HR pathway. {ECO:0000269|PubMed:11751635, ECO:0000269|PubMed:11751636, ECO:0000269|PubMed:11842113, ECO:0000269|PubMed:12441335, ECO:0000269|PubMed:23108668, ECO:0000269|PubMed:23149936}. | ATP-binding;Alternative splicing;Chromosomal rearrangement;DNA damage;DNA recombination;DNA repair;DNA-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a member of the RAD51 protein family. RAD51 family members are evolutionarily conserved proteins essential for DNA repair by homologous recombination. This protein has been shown to form a stable heterodimer with the family member RAD51C, which further interacts with the other family members, such as RAD51, XRCC2, and XRCC3. Overexpression of this gene was found to cause cell cycle G1 delay and cell apoptosis, which suggested a role of this protein in sensing DNA damage. Rearrangements between this locus and high mobility group AT-hook 2 (HMGA2, GeneID 8091) have been observed in uterine leiomyomata. [provided by RefSeq, Mar 2016]. | hsa:5890; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Rad51B-Rad51C-Rad51D-XRCC2 complex [GO:0033063]; replication fork [GO:0005657]; ATP binding [GO:0005524]; ATP-dependent activity, acting on DNA [GO:0008094]; DNA binding [GO:0003677]; double-stranded DNA binding [GO:0003690]; single-stranded DNA binding [GO:0003697]; blastocyst growth [GO:0001832]; DNA recombination [GO:0006310]; DNA repair [GO:0006281]; double-strand break repair via homologous recombination [GO:0000724]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of G2/M transition of mitotic cell cycle [GO:0010971]; reciprocal meiotic recombination [GO:0007131]; somite development [GO:0061053] | 11744692_This work describes the in vitro and in vivo identification of the RAD51B/RAD51C heterocomplex 11978964_involved in the frequently occurring t(6;14) (p21;q23-->q24) in pulmonary chondroid hamartomas 12427746_Rad51B and Rad51C function through interactions with the human Rad51 recombinase and play a crucial role in the homologous recombinational repair pathway 12441335_Rad51B protein may have a specific function in Holliday junction processing in the homologous recombinational repair pathway in humans 14704354_a fragment of Rad51B containing amino acid residues 1-75 interacts with the C-terminus and linker of Rad51C, residues 79-376, and this region of Rad51C also interacts with mRad51D and Xrcc3 15701685_motif in the N-terminus of Rad51B serves as an NLS that allows Rad51B to localize to the nucleus independent of Rad51C or BRCA2 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19329439_EVL protein is a novel recombination factor that may be required for repairing specific DNA lesions, and that may cause tumor malignancy by its inappropriate expression. 19330030_A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). 19330030_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19657362_BCR/ABL fragments were used for identifying the sites of BCR/ABL interaction with RAD51B 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20095854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20195514_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20237344_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20610542_polymorphisms in and haplotypes of the RAD51L1 gene, which is involved in the double-strand break repair pathway, modulate gamma-radiation-induced mutagen sensitivity. 21368091_findings support the notion that DNA repair genes, in particular RAD51L1, play a role in nasopharyngeal carcinoma etiology and development 21533530_our results suggest that RAD51L1 is unlikely to represent a high-penetrance breast cancer susceptibility gene. 21852249_rs11249433 at 1p.11.2, and two highly correlated single-nucleotide polymorphisms rs999737 and rs10483813 (r(2)= 0.98) at 14q24.1 (RAD51L1), for up to 46 036 invasive breast cancer cases and 46 930 controls from 39 studies, were genotyped. 22017238_Single Nucleotide Polymorphisms in RAD51L1 gene is associated with glioblastoma. 22454379_Single nucleotide polymorphism in RAD51L1 is associated with breast cancer. 23001122_SNP in RAD51B at 14q24.1 was significantly associated with male breast cancer risk 23108668_Study observed centrosome defects in the absence of XRCC3. While RAD51B and RAD51C act early in homologous recombination, XRCC3 functions jointly with GEN1 later in the pathway at the stage of Holliday junction resolution. 23810717_The HsRAD51B-HsRAD51C complex plays a role in stabilizing the HsRAD51 nucleoprotein filament during the presynaptic phase of homologous recombination. 24022229_This study provides robust evidence for an association of rheumatoid arthritis susceptibility with genes involved in B cell differentiation (BACH2) and DNA repair (RAD51B). 24139550_It confirms that RAD51 paralog mutations confer breast and ovarian cancer predisposition and are rare events. 24498017_Data indicate that complement factor H (CFH) R1210C and common variants in COL8A1 and RAD51B plus six genes contribute predictive information for advanced macular degeneration (AMD) beyond macular and behavioral phenotypes. 24526414_the risk of developing AMD exhibits dose dependency as well as an epistatic combined effect in rs17105278 T>C and rs4902566 C>T carriers and that the elevated risk for rs17105278 T>C carriers may be due to decreased transcription of RAD51B. 25255808_Relative excess risk of breast cancer due to interaction between RAD51L1 single-nucleotide polymorphism and BMI. 25600502_a novel germ line RAD51B nonsense mutation, and reduced expression of RAD51B in melanoma cells indicating inactivation of RAD51B 26261251_Mutations in epithelial ovarian cancer cases were more frequent in RAD51C (14 occurrences, 0.41%) and RAD51D (12 occurrences, 0.35%) than in RAD51B (two occurrences, 0.06%). 26339569_The aim of the present study was to evaluate the relationship between prostate cancer risk and the presence of single nucleotide polymorphisms in the genes involved in Homologous recombination repair, RAD51, RAD51B, XRCC2 and XRCC3. 27149063_common variation is significantly associated with familial breast cancer risk 27334422_our study suggested that miRNA-binding site genetic variants of RAD51B may modify the susceptibility to cervical cancer, which is important to identify individuals with differential risk for this malignancy and to improve the effectiveness of preventive intervention. 27651161_over-expression of RAD51B promoted cell proliferation, aneuploidy, and drug resistance, while RAD51B knockdown led to G1 arrest and sensitized cells to 5-fluorouracil 27683114_hypermethylation of homologous recombination DNA repair genes including RAD51B and XRCC3 is associated with an inflamed phenotype in squamous cell cancers of the head and neck, lung and cervix. 28361912_We successfully identified a common variant, rs911263, as being significantly associated with the disease status . In addition, this SNP was shown to be related to erosion, a clinical assessment of disease severity in RA (P = 2.89 x 10(-5), OR = 0.52). These findings shed light on the role of RAD51B in the onset and severity of Rheumatoid arthritis (RA). 31584931_Here we provide three coherent sets of isogenic mutants, both in transformed and non-transformed human cells. Importantly, using these mutant lines, we report the unanticipated result that RAD51B has a less crucial role in homologous recombination than the other four paralogs, and find that all RAD51 paralogs are critically important for early functions during homologous recombination 32669601_Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. 33622874_TBX15 rs98422, DNM3 rs1011731, RAD51B rs8017304, and rs2588809 Gene Polymorphisms and Associations With Pituitary Adenoma. 34060991_Serum Levels of ARMS2, COL8A1, RAD51B, and VEGF and their Correlations in Age-related Macular Degeneration. | ENSMUSG00000059060 | Rad51b | 341.716505 | 0.8424886 | -0.247270908 | 0.15108118 | 2.647346e+00 | 1.037230e-01 | No | Yes | 308.000906 | 52.775246 | 373.096783 | 63.866891 | ||
| ENSG00000183018 | 124976 | SPNS2 | protein_coding | Q8IVW8 | FUNCTION: Acts a a crucial lysosphingolipid sphingosine 1-phosphate (S1P) transporter involved in S1P secretion and function (PubMed:19074308, PubMed:23180825). S1P is a bioactive signaling molecule that regulates many physiological processes important for the development and for the immune system. Regulates levels of S1P and the S1P gradient that exists between the high circulating concentrations of S1P and low tissue levels that control lymphocyte trafficking (PubMed:19074308, PubMed:23180825). {ECO:0000269|PubMed:19074308, ECO:0000269|PubMed:23180825}. | Cell membrane;Deafness;Disease variant;Endosome;Lipid transport;Membrane;Non-syndromic deafness;Reference proteome;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is a transporter of sphingosine 1-phosphate, a secreted lipid that is important in cardiovascular, immunological, and neural development. Defects in this gene are a cause of early onset progressive hearing loss. [provided by RefSeq, Jul 2016]. | hsa:124976; | endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; sphingolipid transporter activity [GO:0046624]; transmembrane transporter activity [GO:0022857]; B cell homeostasis [GO:0001782]; bone development [GO:0060348]; lipid transport [GO:0006869]; lymph node development [GO:0048535]; lymphocyte migration [GO:0072676]; regulation of eye pigmentation [GO:0048073]; regulation of humoral immune response [GO:0002920]; sphingolipid biosynthetic process [GO:0030148]; sphingosine-1-phosphate receptor signaling pathway [GO:0003376]; T cell homeostasis [GO:0043029] | 21084291_The sphingosine 1-phosphate transporter, SPNS2, functions as a transporter of the phosphorylated form of the immunomodulating agent FTY720. 23180825_Spns2 is an S1P transporter in vivo that plays a role in regulation not only of blood S1P but also lymph node and lymph S1P levels and consequently influences lymphocyte trafficking and lymphatic vessel network organization. 25330231_Spns2 plays key roles in regulating the cellular functions in non-small cell lung cancer (NSCLC) cells 25356849_mammals. These findings indicate that Spns2 is required for normal maintenance of the endocochlear potential and hence for normal auditory function, and support a role for S1P signalling in hearing. 27343196_A novel role for Spns2 and S1P1&2 in the activation of p47(phox) and production of reactive oxygen species involved in hyperoxia-mediated lung injury. 27562371_Butyrate and bioactive proteolytic form of Wnt-5a regulate colonic epithelial proliferation and spatial development 29112690_In both macrophage and epithelial cell types, Spns2 was also found localized to cytoplasm and the nucleus, in line with a predicted bipartile Nuclear Localization Signal at the position aa282 of the human Spns2 sequence 29772789_our data demonstrate that Spns2 and S1P have a crucial effect on proximal tubular epithelial cells by regulating an inflammatory process and a subsequent fibrotic reaction, and also by influencing other tubular transporters that are involved in the pathophysiology of inflammatory and fibrotic kidney diseases. 30196234_Spinster 2 (SPNS2) was shown to act as a mediator of intracellular sphingosine-1-phosphate (S1P) release and play an important role in the regulation of S1P [Review]. 31206801_SPNS2 promotes the malignancy of colorectal cancer cells via regulating Akt and ERK pathway. 32253496_A possible contribution of the SPNS2 variation to POI was not strictly ruled out, but various data presented in the text including reported association of variations in related gene ALOX12 with menopause-age and role of ALOX12B in atretic bovine follicle formation argue in favor of ALOX12B. It is, therefore, concluded that the mutation in ALOX12B is the likely cause of POI in the pedigree. 33673355_Prognostic Significance of Cytoplasmic SPNS2 Expression in Patients with Oral Squamous Cell Carcinoma. 34671931_Long non-coding RNA HOXA-AS3 facilitates the malignancy in colorectal cancer by miR-4319/SPNS2 axis. | ENSMUSG00000040447 | Spns2 | 43.260166 | 1.3017420 | 0.380443523 | 0.41189600 | 8.615129e-01 | 3.533157e-01 | No | Yes | 28.020667 | 6.038756 | 21.253556 | 4.589559 | ||
| ENSG00000183250 | 84536 | LINC01547 | lncRNA | 397.189552 | 0.7690368 | -0.378875439 | 0.15144020 | 6.272186e+00 | 1.226477e-02 | No | Yes | 335.734774 | 47.090116 | 437.512384 | 61.025495 | |||||||||||
| ENSG00000183506 | 375133 | PI4KAP2 | transcribed_unitary_pseudogene | 874.445419 | 1.0739510 | 0.102928161 | 0.10458521 | 9.791831e-01 | 3.224006e-01 | 6.856013e-01 | No | Yes | 873.019483 | 120.053945 | 829.142229 | 114.182253 | ||||||||||
| ENSG00000183828 | 256281 | NUDT14 | protein_coding | O95848 | FUNCTION: Hydrolyzes UDP-glucose to glucose 1-phosphate and UMP and ADP-ribose to ribose 5-phosphate and AMP. The physiological substrate is probably UDP-glucose. Poor activity on other substrates such as ADP-glucose, CDP-glucose, GDP-glucose and GDP-mannose. | 3D-structure;Cytoplasm;Hydrolase;Magnesium;Reference proteome | The protein encoded by this gene is a member of the Nudix hydrolase family. Nudix hydrolases eliminate potentially toxic nucleotide metabolites from the cell and regulate the concentrations and availability of many different nucleotide substrates, cofactors, and signaling molecules. This enzyme contains a Nudix hydrolase domain and is a UDPG pyrophosphatase that hydrolyzes UDPG to produce glucose 1-phosphate and UMP. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2016]. | hsa:256281; | cytosol [GO:0005829]; ADP-ribose diphosphatase activity [GO:0047631]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; UDP-sugar diphosphatase activity [GO:0008768]; nucleoside phosphate metabolic process [GO:0006753]; protein N-linked glycosylation via asparagine [GO:0018279]; ribose phosphate metabolic process [GO:0019693] | 26781650_the interaction between the RL13 protein and NUDT14 protein may be involved in human cytomegalovirus DNA replication. | ENSMUSG00000002804 | Nudt14 | 383.308146 | 0.8643643 | -0.210288641 | 0.15828903 | 1.752928e+00 | 1.855090e-01 | No | Yes | 336.200245 | 49.141845 | 396.840709 | 57.755905 | ||
| ENSG00000183856 | 128239 | IQGAP3 | protein_coding | Q86VI3 | 3D-structure;Calmodulin-binding;Phosphoprotein;Reference proteome;Repeat | hsa:128239; | cell-cell junction [GO:0005911]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lateral plasma membrane [GO:0016328]; actin filament binding [GO:0051015]; calmodulin binding [GO:0005516]; GTPase activator activity [GO:0005096]; myosin VI light chain binding [GO:0070856]; small GTPase binding [GO:0031267]; cellular response to organic substance [GO:0071310]; ERK1 and ERK2 cascade [GO:0070371]; G1/S transition of mitotic cell cycle [GO:0000082]; negative regulation of gene expression [GO:0010629]; positive regulation of gene expression [GO:0010628]; positive regulation of mammary gland epithelial cell proliferation [GO:0033601]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of protein phosphorylation [GO:0001934]; Ras protein signal transduction [GO:0007265]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of cell size [GO:0008361]; regulation of GTPase activity [GO:0043087] | 20800603_Observational study of gene-disease association. (HuGE Navigator) 21299499_The first IQ-motifs from IQGAP2 and IQGAP3 form transient interactions with calmodulin in the absence of calcium. 24849319_IQGAP3 may contribute to the pathogenesis of lung cancer by modulating EGFR-ERK signaling 25229330_Mammalian IQGAP proteins may play a role in cytokinesis by regulating the localization of key cytokinesis regulatory proteins to the contractile apparatus during mitosis. 25722290_IQGAP1, IQGAP2, and IQGAP3 have diverse roles in vertebrate physiology, operating in the kidney, nervous system, cardiovascular system, pancreas, and lung. (Review) 28281966_IQGAP3 may be a potential therapeutic target in human breast cancer. 28810875_IQGAP3 functions as an important regulator of metastasis and epithelial-to-mesenchymal transition by constitutively activating the TGF-beta signaling pathway in Hepatocellular carcinoma. 29073199_IQGAP2 expression correlated positively with survivability, on the contrary, IQGAP3 expression levels correlated inversely with survivability 29131081_These results suggest that IQGAP3 plays an important role in gastric cancer stem cells (CSCs). The location of IQGAP3 on the cell membrane makes it a potential therapeutic target for gastric cancer (GC). 30446454_Both IQGAP3/BMP4 and IQGAP3/FAM107A ratios in ucfDNA were significantly higher in patients with BC than in those with hematuria. 31088707_Data found that the IQGAP3/BMP4 ratio in urinary cell-free DNA (ucfDNA) was significantly associated with recurrence-free survival (RFS) and progression-free survival (PFS) in non-muscle-invasive bladder cancer (NMIBC) patients suggesting that the IQGAP3/BMP4 ratio in ucfDNA may be an important independent factor for NMIBC prognosis. 31544570_IQGAP3 was upregulated in colorectal cancer at the tissue level and cellular level. Based on immunohistochemistry results of the tissue microarrays, authors demonstrated that higher expression of IQGAP3 was associated with higher tumor node metastasis stage (P = 0.005), higher incidence of lymph node metastasis (P = 0.004), and shorter overall survival (P = 0.022). 31605603_IQGAP3 may be a potential target gene for Kaempferol in the treatment of breast cancer. 32824461_Enhancement of Migration and Invasion of Gastric Cancer Cells by IQGAP3. 32896617_IQGAP3 interacts with Rad17 to recruit the Mre11-Rad50-Nbs1 complex and contributes to radioresistance in lung cancer. 34183451_IQGAP3, a YAP Target, Is Required for Proper Cell-Cycle Progression and Genome Stability. | ENSMUSG00000028068 | Iqgap3 | 1489.995880 | 1.0791227 | 0.109858871 | 0.08507485 | 1.670575e+00 | 1.961815e-01 | 5.661236e-01 | No | Yes | 1485.464257 | 192.648390 | 1388.953485 | 180.058798 | |||
| ENSG00000184381 | 8398 | PLA2G6 | protein_coding | O60733 | FUNCTION: Calcium-independent phospholipase involved in phospholipid remodeling with implications in cellular membrane homeostasis, mitochondrial integrity and signal transduction. Hydrolyzes the ester bond of the fatty acyl group attached at sn-1 or sn-2 position of phospholipids (phospholipase A1 and A2 activity respectively), producing lysophospholipids that are used in deacylation-reacylation cycles (PubMed:9417066, PubMed:10092647, PubMed:10336645, PubMed:20886109). Hydrolyzes both saturated and unsaturated long fatty acyl chains in various glycerophospholipid classes such as phosphatidylcholines, phosphatidylethanolamines and phosphatidates, with a preference for hydrolysis at sn-2 position (PubMed:10092647, PubMed:10336645, PubMed:20886109). Can further hydrolyze lysophospholipids carrying saturated fatty acyl chains (lysophospholipase activity) (PubMed:20886109). Upon oxidative stress, contributes to remodeling of mitochondrial phospholipids in pancreatic beta cells, in a repair mechanism to reduce oxidized lipid content (PubMed:23533611). Preferentially hydrolyzes oxidized polyunsaturated fatty acyl chains from cardiolipins, yielding monolysocardiolipins that can be reacylated with unoxidized fatty acyls to regenerate native cardiolipin species (By similarity). Hydrolyzes oxidized glycerophosphoethanolamines present in pancreatic islets, releasing oxidized polyunsaturated fatty acids such as hydroxyeicosatetraenoates (HETEs) (By similarity). Has thioesterase activity toward fatty-acyl CoA releasing CoA-SH known to facilitate fatty acid transport and beta-oxidation in mitochondria particularly in skeletal muscle (PubMed:20886109). Plays a role in regulation of membrane dynamics and homeostasis. Selectively hydrolyzes sn-2 arachidonoyl group in plasmalogen phospholipids, structural components of lipid rafts and myelin (By similarity). Regulates F-actin polymerization at the pseudopods, which is required for both speed and directionality of MCP1/CCL2-induced monocyte chemotaxis (PubMed:18208975). Targets membrane phospholipids to produce potent lipid signaling messengers. Generates lysophosphatidate (LPA, 1-acyl-glycerol-3-phosphate), which acts via G-protein receptors in various cell types (By similarity). Has phospholipase A2 activity toward platelet-activating factor (PAF, 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine), likely playing a role in inactivation of this potent proinflammatory signaling lipid (By similarity). In response to glucose, amplifies calcium influx in pancreatic beta cells to promote INS secretion (By similarity). {ECO:0000250|UniProtKB:A0A3L7I2I8, ECO:0000250|UniProtKB:P97570, ECO:0000250|UniProtKB:P97819, ECO:0000269|PubMed:10092647, ECO:0000269|PubMed:10336645, ECO:0000269|PubMed:18208975, ECO:0000269|PubMed:20886109, ECO:0000269|PubMed:23533611, ECO:0000269|PubMed:9417066}.; FUNCTION: [Isoform Ankyrin-iPLA2-1]: Lacks the catalytic domain and may act as a negative regulator of the catalytically active isoforms. {ECO:0000269|PubMed:9417066}.; FUNCTION: [Isoform Ankyrin-iPLA2-2]: Lacks the catalytic domain and may act as a negative regulator of the catalytically active isoforms. {ECO:0000269|PubMed:9417066}. | ANK repeat;Alternative splicing;Calmodulin-binding;Cell membrane;Cell projection;Chemotaxis;Cytoplasm;Disease variant;Dystonia;Hydrolase;Lipid metabolism;Membrane;Mitochondrion;Neurodegeneration;Parkinson disease;Parkinsonism;Pharmaceutical;Phospholipid metabolism;Reference proteome;Repeat;Transmembrane;Transmembrane helix | The protein encoded by this gene is an A2 phospholipase, a class of enzyme that catalyzes the release of fatty acids from phospholipids. The encoded protein may play a role in phospholipid remodelling, arachidonic acid release, leukotriene and prostaglandin synthesis, fas-mediated apoptosis, and transmembrane ion flux in glucose-stimulated B-cells. Several transcript variants encoding multiple isoforms have been described, but the full-length nature of only three of them have been determined to date. [provided by RefSeq, Dec 2010]. | hsa:8398; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; pseudopodium [GO:0031143]; 1-alkyl-2-acetylglycerophosphocholine esterase activity [GO:0003847]; ATP-dependent protein binding [GO:0043008]; calcium-independent phospholipase A2 activity [GO:0047499]; calmodulin binding [GO:0005516]; hydrolase activity [GO:0016787]; identical protein binding [GO:0042802]; lysophospholipase activity [GO:0004622]; myristoyl-CoA hydrolase activity [GO:0102991]; palmitoyl-CoA hydrolase activity [GO:0016290]; phosphatidyl phospholipase B activity [GO:0102545]; phospholipase A2 activity [GO:0004623]; protein kinase binding [GO:0019901]; serine hydrolase activity [GO:0017171]; antibacterial humoral response [GO:0019731]; cardiolipin acyl-chain remodeling [GO:0035965]; cardiolipin biosynthetic process [GO:0032049]; chemotaxis [GO:0006935]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; maternal process involved in female pregnancy [GO:0060135]; memory [GO:0007613]; negative regulation of synaptic transmission, glutamatergic [GO:0051967]; phosphatidic acid metabolic process [GO:0046473]; phosphatidylcholine catabolic process [GO:0034638]; phosphatidylethanolamine catabolic process [GO:0046338]; platelet activating factor metabolic process [GO:0046469]; positive regulation of arachidonic acid secretion [GO:0090238]; positive regulation of ceramide biosynthetic process [GO:2000304]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of exocytosis [GO:0045921]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774]; positive regulation of protein kinase C signaling [GO:0090037]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of release of cytochrome c from mitochondria [GO:0090200]; regulation of store-operated calcium channel activity [GO:1901339]; response to endoplasmic reticulum stress [GO:0034976]; urinary bladder smooth muscle contraction [GO:0014832]; vasodilation [GO:0042311] | 12208880_activation during apoptosis promotes the exposure of membrane lysophosphatidylcholine leading to binding by natural immunoglobulin M antibodies and complement activation 12423354_stimulation of three isoforms of PLA2 by thapsigargin liberates free AA that, in turn, induces capacitative calcium influx in human T-cells 14749286_Arachidonic acid produced by iPLA(2)beta-catalyzed hydrolysis of their substrates induces release of Ca(2+) from ER stores, an event thought to participate in glucose-stimulated insulin secretion. 15052324_The GVIB iPLA2 is widely expressed in human tissues but is enriched in heart, placenta, and skeletal muscle. 15249229_Here we show that the C-terminal region of human iPLA(2)gamma is responsible for the enzymatic activity. 15252038_iPLA2 may be dispensable for the apoptotic process to occur 15318030_Observational study of gene-disease association. (HuGE Navigator) 15364929_iPLA2epsilon (adiponutrin), iPLA2zeta (TTS-2.2), and iPLA2eta (GS2) are three novel TAG lipases/acylglycerol transacylases that likely participate in TAG hydrolysis and the acyl-CoA independent transacylation of acylglycerols 15385540_truncated iPLA(2) proteins associate with active iPLA(2) and down-regulate its activity during G(1) 15573142_Detailed characterization of group VIA phospholipase A2 beta suggests that the pancreatic islet beta-cells express multiple isoforms of iPLA2beta; the hypothesis in this review is that these isozymes participate in different cellular functions. 16585943_This study reviews the evidence and discusses the potential roles of phospholipase A2 Group 6A for schizophrenia with particular emphasis on published association studies. 16783378_mapped a locus for infantile neuroaxonal dystrophy (INAD) and neurodegeneration with brain iron accumulation (NBIA) to chromosome 22q12-q13 and identified mutations in PLA2G6, encoding a group VI phospholipase A2, in NBIA, INAD and Karak syndrome 16943248_Increase in iPLA(2) and accumulation of membrane phospholipid-derived metabolites in HCAEC exposed to hypoxia or thrombin have important implications in inflammation and arrhythmogenesis in atherosclerosis/thrombosis and myocardial ischemia. 16966332_iPLA2-VIA is a novel regulator of endothelial cell S phase progression, cell cycle residence, and angiogenesis 17003039_The role of calcium influx factor and PLA2G6 in the activation of CRAC channels and calcium entry in rat tumor cell lines is reported. 17033970_The disease gene was mapped to a 1.17-Mb locus on chromosome 22q13.1. 17082190_Transient receptor potential subfamily M member 8 (TRPM8) channel is stimulated by the Ca2+-independent phospholipase A2 (iPLA2) signaling pathway with its end products, lysophospholipids, acting as its endogenous ligands. 17188740_human coronary artery endothelial cells exposed to thrombin or tryptase stimulation demonstrated an increase in iPLA2 activity and arachidonic acid release 17254819_Cerebellar atrophy without cerebellar cortex hyperintensity in infantile neuroaxonal dystrophy (INAD) due to PLA2G6 mutation. 17275398_Secretion and activity of sPLA(2) were found to be similar in granulocyte-like PLB cells expressing or lacking cPLA(2)alpha, indicating that they are not under cPLA(2)alpha regulation. 17459165_oxytocin stimulation of uterine PGF2alpha production is mediated, at least in part, by up-regulation of PLA2G6 expression and activity 18208975_iPLA(2)beta and cPLA(2)alpha regulate monocyte migration from different intracellular locations, with iPLA(2)beta acting as a critical regulator of the cellular compass. 18562188_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18570303_PLA2G6 mutations are associated with infantile neuroaxonal dystrophy and have been reported previously to cause early cerebellar signs, and the syndrome was classified as neurodegeneration with brain iron accumulation (type 2). 18676680_Observational study of gene-disease association. (HuGE Navigator) 18775417_This review discusses the role of iPLA2 in cell growth with special emphasis placed on its role in cell signaling; the putative lipid signals involved are also discussed. 18790994_Tryptase stimulation of human small airway epithelial cells increased membrane-associated, calcium-independent phospholipase A(2)gamma (iPLA(2)gamma) activity, resulting in increased arachidonic acid and PGE(2) release. 18799783_Observational study of gene-disease association. (HuGE Navigator) 18799783_PLA2G6 mutations are associated with nearly all cases of classic infantile neuroaxonal dystrophy. 18826942_H2O2-mediated hyperoxidation of Prdx6 induces cell cycle arrest at the G2/M transition through up-regulation of iPLA2 activity. 19059366_iPLA2 activity is responsible for membrane phospholipid hydrolysis in response to tryptase or thrombin stimulation in pulmonary vascular endothelial cells 19087156_Different and even identical PLA2G6 mutations may cause neurodegenerative diseases with heterogeneous clinical manifestations, including dystonia-parkinsonism. 19138334_The nine novel mutations identified in this study suggest the uniqueness of the PLA2G6 mutation spectrum in Chinese patients 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19225567_Chromofungin or Catestatin , penetrate into PMNs, inducing extracellular calcium entry by a CaM-regulated iPLA2 pathway. 19556238_The region with the greatest change upon lipid binding in phospholipase A2 group VI was region 708-730. 19578365_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20053941_iPLA2 expression is increased in neutrophils from people with diabetes and mediates superoxide generation, presenting an alternate pathway independent of protein kinase C and phosphatidic acid phosphohydrolase-1 hydrolase signaling. 20171194_These data demonstrate the novel findings that iPLA2 inhibition activates p38 by inducing reactive species, and further suggest that this signaling kinase is involved in p53 activation, cell cycle arrest and cytostasis. 20186954_Observational study of gene-disease association. (HuGE Navigator) 20186954_Our data suggest that PLA2G6 mutations are unlikely to be an important cause of the common garden variety of Parkinson's disease patients with dystonia or a positive family history 20219570_Results characterize a pathway leading to NOX2 activation in which iPLA(2)-regulated p38 MAPK activity is a key regulator of S100A8/A9 translocation via S100A9 phosphorylation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20464283_Observational study of gene-disease association. (HuGE Navigator) 20584031_Compound heterozygosity for a large intragenic deletion and a nonsense mutation was found in one patient with infantile neuroaxonal dystrophy while the other is carrying two novel splice-site mutations 20619503_This report further defines the clinical features and neuropathology of PLA2G6 related childhood and adult onset dystonia-parkinsonism . 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20629144_Genetically determined NBIA cases from the Indian subcontinent suggest presence of unusual phenotypes of PANK2 and novel mutations 20647408_Observational study of gene-disease association. (HuGE Navigator) 20669327_We identified genetic deficits in PLA2G6 that were associated with Levodopa responsive parkinsonism with pyramidal signs. 20813170_This study demonistrated that pLA2G6 has a dynamic expression pattern both in terms of the location of expression and the differentiation state of expressing cells. 20881058_The A isoform of PLA2G6 was shown to play a role in controlling the architecture of the endoplasmic reticulum-golgi intermediate compartment proteins in a way that impacts transport. 20886109_Catalytic function of PLA2G6 impaired by mutations is associated with infantile neuroaxonal dystrophy. 20938027_Observational study of gene-disease association. (HuGE Navigator) 20938027_The results of this study suggested that PLA2G6 mutations should be considered in patients with early-onset l-dopa-responsive parkinsonism and dementia with frontotemporal lobar atrophy. 21191104_These results indicate that cardiac endothelial cell PAF production is dependent on iPLA(2)beta activation and that both iPLA(2)beta and iPLA(2)gamma may be involved in PGI(2) release. 21368765_PLA2G6 mutations are unlikely to be the major causes or risk factors of Parkinson's disease at least in Asian populations 21482170_data indicate that PLA2G6 mutation may not play a major role in general frontotemporal type of dementia 21700586_tHIS STUDY DEMONISTRATED THAT the PLA2G6 gene allocated PARK14 locus and is associated with autosomal recessive early-onset parkinsonism. 21812034_A possible involvement of calcium-independent group VI phospholipase A2 (iPLA2-VI) in the pathogenesis of Parkinson's disease has been proposed. 22213678_PLA2G6 mutations are associated with PARK14-linked young-onset parkinsonism and sporadic Parkinson's disease 22218592_acts as an inhibitory modulator of NKCC2 activity in thick ascending limb 22406380_Our result indicated that PLA2G6 mutations might not be a main cause of Chinese sporadic early-onset parkinsonism. 22459563_The results of this study suggested that PLA2G6 is not a susceptibility gene for parkinson disease in our population. 22549787_These data confirm the role of iPLA(2)beta as an essential mediator of endogenous store operated calcium entry. 22680611_The present study confirms the involvement of iPLA(2)-VIA in efficient retinal pigment epithelium phagocytosis of photoreceptor outer segments. 22903185_Neuronal phospholipid deacylation is essential for axonal and synaptic integrity through the action of iPLA2 and NTE. (Review) 23007400_membrane composition and the presence of nucleotides play key roles in recruiting and modulating GVIA-iPLA(2) activity in cells 23043102_Orai1 and PLA2g6 are involved in adhesion formation, whereas STIM1 participates in both adhesion formation and disassembly. 23074238_Findings reveal for the first time expression of iPLA(2)beta protein in human islet beta-cells and that induction of iPLA(2)beta during endoplasmic reticulum stress contributes to human islet beta-cell apoptosis. 23182313_identified four rare PLA2G6 mutations in 250 PD patients in Chinese population with Parkinson's disease 23196729_Mutations in PLA2G6 is often associated with rapidly progressive parkinsonism and with additional features including pyramidal signs, cognitive decline and loss of sustained Levodopa responsiveness. 23277130_The association with bipolar disorder of the iPLA2beta (PLA2G6) its genetic interaction with type 2 transient receptor potential channel gene TRPM2, was examined. 23587695_This study demonistrated that PLA2g8 expression was significantly decreased in patients treated with antipsychotic drug. 24130795_the phenotype of neurodegeneration associated with PLA2G6 mutations 24512906_our findings indicate that PRDX6 promotes lung tumor growth via increased glutathione peroxidase and iPLA2 activities 24522175_clinical findings may be helpful in distinguishing PLA2G6-related neurodegeneration from the other major cause of NBIA, recessive PANK2 mutations. 24745848_Novel PLA2G6 mutations were identified in all patients with Phospholipase A2 associated neurodegeneration. 24791136_The significance of calcium-independent phospholipase A, group VIA (iPLA2-VIA), in retinal pigment epithelial cell survival, was investigated. 24858037_The loss of PS2 could have a critical role in lung tumor development through the upregulation of iPLA2 activity by reducing gamma-secretase. 25004092_IL-1beta and IFNgamma induces mSREBP-1 and iPLA2beta expression and induce beta-cell apoptosis. 25207958_genetic association studies in a population of Han Chinese: Data suggest that SNPs in PLA2G6 (rs132984; rs2284060) are associated with type 2 diabetes and hypertriglyceridemia in the population studied. [Meta-Analysis included] 25348461_A homozygous novel mutation at position c.2277-1G>C in PLA2G6 gene presumed to give rise to altered splicing, was detected, thus confirming the diagnosis of infantile Neuroaxonal Dystrophy (INAD). 25482049_Stimulation of adrenoreceptors causes increased iPLA2 expression via MAP kinase/ERK 1/2. 25668476_Mutations in PANK2 and CoASY lead, respectively, to PKAN and CoPAN forms of Neurodegeneration with brain iron accumulation . Mutations in PLA2G6 lead to PLAN. Mutations in C19orf12 lead to MPAN 26001724_our findings demonstrate that loss of normal PLA2G6 gene activity leads to lipid peroxidation, mitochondrial dysfunction and subsequent mitochondrial membrane abnormalities. 26160611_Results demonstrated no significant impact of PLA2G6 and PLA2G4C gene polymorphisms on attenuated niacin skin flushing in schizophrenia patients. 26446356_Three catalytically active cPLA2, iPLA2, and sPLA2 are expressed in different areas within the human spermatozoon cell body. Spermatozoa with a significant low motility showed strong differences both in terms of total specific activity and of different intracellular distribution, compared with normal spermatozoa. Phospholipases could be potential biomarkers of asthenozoospermia. 26525102_genetic association study in Quebec City population: Data suggest total plasma n-6 fatty acid phospholipid levels and C-reactive protein are modulated by SNPs in PLA2G4A and PLA2G6 alone or in combination with fish oil dietary supplementation. 26668131_Mutations in PLA2G6 altered Golgi morphology, O-linked glycosylation and sialylation of protein in patients with neurodegeneration 26755131_Analysis of the cells from idiopathic Parkinson's disease patients reveals a significant deficiency in store-operated PLA2g6-dependent Ca(2+) signalling 27030050_This study demonstrated that elevated expression of alphaSyn/PalphaSyn in mitochondria appears to be the early response to PLA2G6-deficiency in neurons. 27196560_PLA2G6 mutations in Indian patients with infantile neuroaxonal dystrophy and atypical late-onset neuroaxonal dystrophy 27196560_PLA2G6 were identified in patients with a spectrum of neurodegenerative conditions, such as infantile neuroaxonal dystrophy, atypical late-onset neuroaxonal dystrophy and dystonia parkinsonism complex in Indian families 27268037_Finding suggest the broadness of the clinical spectrum of group VI phospholipases A2 (PLA2G6)-related neurodegeneration. 27317427_These results strongly suggest that PNPLA9, -6 and -4 play a key role in GPL turnover and homeostasis in human cells. A hypothetical model suggesting how these enzymes could recognize the relative concentration of the different GPLs is proposed 27513994_This study identifies a novel PLA2G6 mutation that is the possible genetic cause of FCMTE in this Chinese family. 27709683_This exome sequencing in a family and identified compound-heterozygous PLA2G6 mutations in 2 affected sisters. 27942883_Study performed direct sequencing and investigated copy number variations (CNVs) of this gene in 109 Japanese patients with parkinsonism. Results suggest that CNV in PLA2G6 is rare in parkinsonism, at least in the Japanese population, in contrast to the reports of its frequency in neurodegeneration associated with brain iron accumulation. 28091863_performed a longitudinal brain volumetry study in a couple of bicorial twins with PLA2G6-positive infantile neuronal axonal dystrophy 28150298_PLA2G6-associated neurodegeneration was caused by paternal isodisomy of the chromosome 22 (carrying c.680C>T mutation) following in vitro fertilization 28213071_the association of PLA2G6 with the pathogenesis of idiopathic PD, in addition to PARK14. 28295203_PLA2G6 gene mutations in 3 families, are reported. 28651698_We found no significant influence of the PLA2G6 and PLA2G4C polymorphisms on mean age at first hospital admission (P > 0.05) and that the investigated polymorphisms significantly influenced the clinical psychopathology only in male patients. The PLA2G4C polymorphism accounted for approximately 12% of negative symptom severity. 28821231_A novel missense mutation in PLA2G6 gene (c.3G > T:p.M1I) in one and half-year-old boy with muscle weakness and neurodevelopmental regression (speech, motor and cognition). 29325618_This study showed that PANK2 genes account for disease of patients diagnosed with an Neurodegeneration with brain iron accumulation disorder. 29342349_A lipidomics-based LC/MS assay was used to define the specificity of cPLA2, iPLA2, and sPLA2 toward a variety of phospholipids. A unique hydrophobic binding site for the cleaved fatty acid dominates each enzyme's specificity rather than its catalytic residues and polar headgroup binding site. 29454663_PLA2G6 mutations are associated with a continuous clinical spectrum from phospholipase A2-associated neurodegeneration to hereditary spastic paraplegia. 29472584_The catalytic domains of iPLA2beta form a tight dimer and surrounded by ankyrin repeat domains that adopt an outwardly flared orientation, poised to interact with membrane proteins. 29739362_A novel variant (NM_003560.2 c.1427 + 2 T > C) acting on a splice donor site and predicted to lead to skipping of exon 10 was found in PLA2G6 in two siblings diagnosed with infantile neuroaxonal dystrophy. This mutation seems to be pathogenic and was found in a homozygous state in the two patients and homozygous reference or heterozygous in five healthy family members. 29753029_PLA2G6 inherited genotype may influence melanoma BRAF/NRAS subtype development. 29909971_This study shows a hereto unknown role of PLA2G6/PARK14 in sphingolipid homeostasis and endolysosomal function. The suppression of PLA2G6 expression causes a reduction in Vps26 and Vps35 and leads to an elevation of ceramides and other sphingolipid intermediates in neuroblastoma cell line cells. 30232368_Two missense variants, p.R53C and p.T319M in PLA2G6 gene may contribute to Parkinson's disease susceptibility in Chinese population. 30302010_PLA2G6 mutation is associated with neurodegeneration presenting as a complicated form of hereditary spastic paraplegia. 30707893_Impaired iPLA2beta activity affects iron uptake and storage without iron accumulation: An in vitro study excluding decreased iPLA2beta activity as the cause of iron deposition in PLAN. 30772976_report a PLA2G6 compound complicated mutation in an atypical neuroaxonal dystrophy Chinese family, that is the p. A80T and p.D331Y mutation 31196701_A novel compound heterozygous mutation of the PLA2G6 gene, c.1648delC and c.991G>T, is associated with adult onset ataxia. 31277247_Proportion of calcium-independent (i)PLA2beta-containing mucosal mast cells (MCs) and the expression intensity of sPLA2-IIA was increased in Crohn's disease (CD). In vitro study showed that (i)PLA2beta is involved in the secretion of secretory phospholipases (sPLA2) from human MC line HMC-1. These results suggest that iPLA2beta-mediated release of sPLA2 from intestinal MCs may contribute to CD pathophysiology. 31492433_Study demonstrated significant, though weak, effects of the PLA2G6 polymorphism on the risk of nicotine dependence, as well as a significant, though weak, influence of the PLA2G6-smoking interaction on age of schizophrenia onset, with both effects manifested in a gender-specific fashion. 31493991_Genetic testing of the PLA2G6 confirmed presence of compound heterozygous novel mutations in Malaysian siblings with infantile neuroaxonal dystrophy 1 31506141_immunohistochemistry showed a reduction in the protein expression of PLAG6 in the muscular tissue of this child 31689548_Infantile onset progressive cerebellar atrophy and anterior horn cell Degeneration-A novel phenotype associated with mutations in the PLA2G6 gene. 31922589_The compound variants of c.668C>A (p.Pro223Gln) and c.2266C>T (p.Gln756Ter) of the PLA2G6 gene probably underlies infantile neuroaxonal dystrophy in the child 32771225_PLA2G6 variants associated with the number of affected alleles in Parkinson's disease in Japan. 33080873_Metabolic Effects of Selective Deletion of Group VIA Phospholipase A2 from Macrophages or Pancreatic Islet Beta-Cells. 33087576_PLA2G6 guards placental trophoblasts against ferroptotic injury. 33279242_Association of rare heterozygous PLA2G6 variants with the risk of Parkinson's disease. 33576074_Whole genome sequencing reveals biallelic PLA2G6 mutations in siblings with cerebellar atrophy and cap myopathy. 33766980_Typical MRI features of PLA2G6 mutation-related phospholipase-associated neurodegeneration (PLAN)/infantile neuroaxonal dystrophy (INAD). 34131139_iPLA2beta-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4. 34207793_iPLA2beta Contributes to ER Stress-Induced Apoptosis during Myocardial Ischemia/Reperfusion Injury. 34622992_Dissecting the Phenotype and Genotype of PLA2G6-Related Parkinsonism. 35092705_Novel insertion mutation in the PLA2G6 gene in an Iranian family with infantile neuroaxonal dystrophy. 35247231_Genome sequencing reveals novel noncoding variants in PLA2G6 and LMNB1 causing progressive neurologic disease. | ENSMUSG00000042632 | Pla2g6 | 235.868557 | 1.0752321 | 0.104648180 | 0.18417640 | 3.202801e-01 | 5.714393e-01 | No | Yes | 209.914764 | 34.820652 | 205.670758 | 34.124361 | ||
| ENSG00000184402 | 26039 | SS18L1 | protein_coding | O75177 | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | Activator;Alternative splicing;Calcium;Centromere;Chromatin regulator;Chromosome;Kinetochore;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | This gene encodes a calcium-responsive transactivator which is an essential subunit of a neuron-specific chromatin-remodeling complex. The structure of this gene is similar to that of the SS18 gene. Mutations in this gene are involved in amyotrophic lateral sclerosis (ALS). Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Sep 2014]. | hsa:26039; | cytosol [GO:0005829]; kinetochore [GO:0000776]; nBAF complex [GO:0071565]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription coactivator activity [GO:0003713]; chromatin organization [GO:0006325]; dendrite development [GO:0016358]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893] | 12696068_A synovial sarcoma of classic morphology contained a novel t(20;X) SS18L1(strong homology to SS18 on Ch20)/SSX1 fusion transcript in which nucleotide 1216 (exon 10) of SS18L1 was fused in-frame with nucleotide 422 (exon 6) of SSX1. 15866867_analysis of the CREST domain that inhibits dendritic growth in cultured neurons 16484776_SS18 and SS18L1 genes map within co-linear DNA segments that may have evolved through a relatively recent genomic duplication event. 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 23708140_This study demonistrated that CREST mutations inhibited activity-dependent neurite outgrowth in primary neurons, and CREST associated with the ALS protein FUS. 24360741_Its mutation causes amyotrophic lateral sclerosis. 25888396_calcium-responsive transactivator and certain other amyotrophic lateral sclerosis-linked proteins share several features implicated in amyotrophic lateral sclerosis pathogenesis 33398438_Protein expression pattern of calcium-responsive transactivator in early postnatal and adult testes. | ENSMUSG00000039086 | Ss18l1 | 518.204499 | 0.9371720 | -0.093614178 | 0.13396548 | 4.813284e-01 | 4.878212e-01 | 7.961416e-01 | No | Yes | 483.918113 | 94.359880 | 523.204989 | 102.030374 | |
| ENSG00000184949 | 646851 | FAM227A | protein_coding | F5H4B4 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:646851; | ENSMUSG00000042564 | Fam227a | 211.316963 | 1.1290497 | 0.175108979 | 0.19575341 | 8.091124e-01 | 3.683828e-01 | No | Yes | 206.903779 | 63.259961 | 185.347922 | 56.779099 | ||||||
| ENSG00000185158 | 114659 | LRRC37B | protein_coding | Q96QE4 | Mouse_homologues NA; + ;NA | Alternative splicing;Coiled coil;Glycoprotein;Leucine-rich repeat;Membrane;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA | hsa:114659; | integral component of membrane [GO:0016021] | 19291764_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000034239+ENSMUSG00000078632 | Gm884+Lrrc37a | 127.883469 | 0.9723519 | -0.040449494 | 0.23185753 | 3.063232e-02 | 8.610632e-01 | No | Yes | 120.106354 | 27.588895 | 127.206672 | 29.282082 | ||
| ENSG00000185596 | WASH3P | transcribed_unprocessed_pseudogene | 433.143259 | 1.0153172 | 0.021930490 | 0.14846980 | 2.147576e-02 | 8.834903e-01 | No | Yes | 432.184476 | 55.416755 | 425.125891 | 54.536977 | ||||||||||||
| ENSG00000185813 | 5833 | PCYT2 | protein_coding | Q99447 | FUNCTION: Ethanolamine-phosphate cytidylyltransferase that catalyzes the second step in the synthesis of phosphatidylethanolamine (PE) from ethanolamine via the CDP-ethanolamine pathway (PubMed:9083101, PubMed:31637422). Phosphatidylethanolamine is a dominant inner-leaflet phospholipid in cell membranes, where it plays a role in membrane function by structurally stabilizing membrane-anchored proteins, and participates in important cellular processes such as cell division, cell fusion, blood coagulation, and apoptosis (PubMed:9083101). {ECO:0000269|PubMed:31637422, ECO:0000269|PubMed:9083101, ECO:0000303|PubMed:9083101}. | 3D-structure;Alternative splicing;Disease variant;Hereditary spastic paraplegia;Lipid biosynthesis;Lipid metabolism;Neurodegeneration;Nucleotidyltransferase;Phospholipid biosynthesis;Phospholipid metabolism;Phosphoprotein;Reference proteome;Transferase | PATHWAY: Phospholipid metabolism; phosphatidylethanolamine biosynthesis; phosphatidylethanolamine from ethanolamine: step 2/3. {ECO:0000269|PubMed:31637422, ECO:0000269|PubMed:9083101}. | This gene encodes an enzyme that catalyzes the formation of CDP-ethanolamine from CTP and phosphoethanolamine in the Kennedy pathway of phospholipid synthesis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:5833; | endoplasmic reticulum membrane [GO:0005789]; ethanolamine-phosphate cytidylyltransferase activity [GO:0004306]; phosphatidylethanolamine biosynthetic process [GO:0006646]; phospholipid biosynthetic process [GO:0008654] | 16023412_The Pcyt2 promoter is driven by a functional CAAT box (-90/-73) and by negative (-385/-255) and positive regulatory elements (-255/-153) in the upstream regions. 18583706_EGR1 is an important transcriptional stimulator of the human PCYT2 and that conditions that modify EGR1 also affect the function of ECT and consequently PE synthesis 22339418_Pcyt2 expression is responsive to tumor nutritional micro-environment, up-regulated in response to metabolic stress under conditions of serum deprivation. 24519946_differences in phosphorylation between Pcyt2 isoforms 24802409_These results suggest that the N-terminal CT domain of hECT contributes to its catalytic reaction, but C-terminal CT domain does not. 31577958_Phosphoethanolamine Accumulation Protects Cancer Cells under Glutamine Starvation through Downregulation of PCYT2. 31637422_our data establish PCYT2 as a disease gene for a new complex hereditary spastic paraplegia and confirm that etherlipid homeostasis is important for the development and function of the brain | ENSMUSG00000025137 | Pcyt2 | 1147.627614 | 1.0358191 | 0.050772019 | 0.09161832 | 3.071537e-01 | 5.794321e-01 | 8.438661e-01 | No | Yes | 1229.974865 | 148.962767 | 1198.865229 | 145.225084 |
| ENSG00000186104 | 120227 | CYP2R1 | protein_coding | Q6VVX0 | FUNCTION: A cytochrome P450 monooxygenase involved in activation of vitamin D precursors. Catalyzes hydroxylation at C-25 of both forms of vitamin D, vitamin D(2) and D(3) (calciol) (PubMed:12867411, PubMed:15465040, PubMed:18511070). Can metabolize vitamin D analogs/prodrugs 1alpha-hydroxyvitamin D(2) (doxercalciferol) and 1alpha-hydroxyvitamin D(3) (alfacalcidol) forming 25-hydroxy derivatives (PubMed:15465040, PubMed:18511070). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase) (PubMed:12867411, PubMed:15465040, PubMed:18511070). {ECO:0000269|PubMed:12867411, ECO:0000269|PubMed:15465040, ECO:0000269|PubMed:18511070}. | 3D-structure;Disease variant;Endoplasmic reticulum;Heme;Iron;Lipid metabolism;Membrane;Metal-binding;Microsome;Monooxygenase;Oxidoreductase;Reference proteome;Signal | PATHWAY: Hormone biosynthesis; vitamin D biosynthesis. {ECO:0000305|PubMed:15465040, ECO:0000305|PubMed:18511070}. | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This enzyme is a microsomal vitamin D hydroxylase that converts vitamin D into the active ligand for the vitamin D receptor. A mutation in this gene has been associated with selective 25-hydroxyvitamin D deficiency. [provided by RefSeq, Jul 2008]. | hsa:120227; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; intracellular membrane-bounded organelle [GO:0043231]; D3 vitamins binding [GO:1902271]; heme binding [GO:0020037]; iron ion binding [GO:0005506]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen [GO:0016712]; protein homodimerization activity [GO:0042803]; steroid hydroxylase activity [GO:0008395]; vitamin D3 25-hydroxylase activity [GO:0030343]; calcitriol biosynthetic process from calciol [GO:0036378]; organic acid metabolic process [GO:0006082]; vitamin D metabolic process [GO:0042359]; vitamin metabolic process [GO:0006766]; xenobiotic metabolic process [GO:0006805] | 12867411_CYP2R1 is a strong candidate for the microsomal vitamin D 25-hydroxylase. 15465040_Results suggest that CYP2R1 plays a physiologically important role in the vitamin D 25-hydroxylation in humans. 16600026_Observational study of gene-disease association. (HuGE Navigator) 17223345_CYP27B1 gene could play a functional role in the pathogenesis of type 1 diabetes through modulation of its mRNA expression and influence serum levels of 1,25(OH)(2)D(3) via the -1260 C/A polymorphism 17607662_Novel association of CYP2R1 polymorphisms in patients with type 1 diabetes and with circulating levels of vitamin D. 17607662_Observational study of gene-disease association. (HuGE Navigator) 18476984_Observational study of gene-disease association. (HuGE Navigator) 18511070_The structure reveals the secosteroid binding mode in an extended active site and allows rationalization of the molecular basis of the inherited rickets associated with CYP2R1. 19783860_No significant differences were observed in genotype or allele frequencies between case and control groups for VDR, CYP27B1 or CYP2R1 SNPs 19783860_Observational study of gene-disease association. (HuGE Navigator) 19852851_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20007432_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20418485_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 20541252_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 20619365_Studies indicate that vitamin D synthesis, a two-step process, starts with a 25-hydroxylation primarily by CYP2R1 and a subsequent 1alpha-hydroxylation via CYP27B1. 20654748_Observational study of gene-disease association. (HuGE Navigator) 20809279_Observational study of gene-disease association. (HuGE Navigator) 20809279_The results suggest that the CYP2R1 and GC genes may contribute to the variation of serum 25(OH)D levels in healthy populations. 21270327_We found a lower gene and protein expression of CYP2R1 in samples with hypospermatogenesis and Sertoli-cell-only syndrome. 21441443_Study confirms that variation in CYP2R1 is associated with predisposition to autoimmune disease type 1 diabetes. 22576297_Our study is the first to confirm the association of variants in DHCR7 and CYP2R1 with 25(OH)-vitamin D levels in patients with chronic liver diseases. 22583563_genetic association study in population in Singapore: Data suggest that diet and SNPs in CYP2R1, CYP3A4 (cytochrome P450, subfamily 3A, polypeptide 4), and GC/VDBP (group-specific component) interact to contribute to vitamin D deficiency. 22801813_Our study found that variant genotypes of single nucleotide polymorphisms in the CYP2R1 gene, rs10766197, rs12794714, rs10741657, rs2060793 and rs1562902, were all significantly associated with plasma 25-hydroxyvitamin D levels. 22855339_CYP2R1 is a major vitamin D 25-hydroxylase that plays a fundamental role in activation of this essential vitamin. 23047203_In vitro and in vivo studies revealed that CYP2R1 expression in Leydig cells appeared to be hCG dependent. 23063903_Finding suggest that GG genotype of CYP2R1 polymorphism and/or CC genotype of CYP27B1 polymorphism increased the risk of developing of type 1 diabetes in Egyptian children. 23190755_genetic association studies in postmenopausal Caucasian women in US: Data suggest that 4 SNPs in CYP2R1 are associated with 25-hydroxyvitamin D in blood (i.e., vitamin D deficiency); vitamin D intake/seasonal sunlight exposure modify these effects. 23377224_Genetic variation in CYP2R1 associated with lower serum 25(OH)D has a decreased risk of aggressive prostate cancer. 23734184_Data indicate that the same nucleotide polymorphisms (SNPs) genotypes of CYP2R1, GC and DHCR7 that are associated with reduced 25(OH)D3 serum levels were found to be associated with hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC). 23793229_CYP2R1 rs10741657 A>G polymorphism is associated with colon cancer recurrence. 23826131_SNPs in the CYP24A1, CYP2R1, calcium sensing receptor (CASR), vitamin D binding protein (GC), retinoid X receptor-alpha (RXRA) and megalin (LRP2) genes were significantly associated with pancreas cancer risk. 24073854_The GAA haplotype of the CYP2R1 gene (P = 0.026) is associated with susceptibility to rickets. 24128439_Low baseline DNA methylation levels in the promoter region of CYP2R1 is associated with low vitamin D response. 24245571_Results show that upregulated gene expression of CYP2R1 may lead to misbalance of vitamin D metabolites and may contribute to the pathogenesis of RCC 24497297_Data indicate that 1,25(OH)2D3 treatment time-dependently increased Cytochrome P450 2R1 (CYP2R1) expression in oral squamous cell carcinomas. 24587115_common polymorphisms in GC and CYP2R1 are associated with serum 25(OH)D concentrations in the Caucasian population and that certain haplotypes may predispose to lower 25(OH)D concentrations in late summer in Denmark. 24732451_Studied whether abnormal endometrial expression of CYP27A1 and/or CYP2R1 may impair VDR-antiproliferative properties in endometrial carcinoma. 24974252_In this mendelian randomisation study, we generated an allele score (25[OH]D synthesis score) based on variants of genes that affect 25(OH)D synthesis or substrate availability (CYP2R1 and DHCR7) 25003556_The CYP2R1 rs10741657 polymorphism is a novel genetic marker for coronary artery disease. 25046415_genetic differences in the VDR gene may be involved in the development of AITD and the activity of GD, whereas the genetic differences in the GC and CYP2R1 genes may be involved with the intractability of GD. 25070320_The increase in [25(OH)D] attributable to vitamin D3 supplementation may vary according to common genetic differences in vitamin D 25-hydroxylase (CYP2R1), 24-hydroxylase (CYP24A1), and the vitamin D receptor (VDR) genes. 25079458_CYP2R1 variants had no significant association with serum 25-OHD3 levels among postmenopausal women of the Han ethnic group in Beijing. 25208829_Data indicate that genome-wide significant associations were found both at age 6 and 14 with single nucleotide polymorphisms (SNPs) on chromosome 11p15 in phosphodiesterase 3B, cGMP-inhibited protein PDE3B/cytochrome P-450 CYP2R1 genes. 25405862_Significant associations were found between the GC (rs2282679 and rs7041), CYP2R1 (rs10741657) single nucleotide polymorphisms and the active form of Vitamin D, 25(OH)D. 25527766_genetic association studies in populations in Denmark: Data suggest that an SNP in CYP2R1 (rs10741657) is associated with response to either UVB treatment or vitamin D3 fortification of bread and milk in prevention of vitamin D deficiency in winter. 25730676_Found a significant upregulation of the CYP2R1 gene in human brain pericytes challenged with tumor necrosis factor-alpha and interferon-gamma. Results suggest the existence of an autocrine/paracrine vitamin D system in the neurovascular unit. 25845986_The 25-hydroxylases CYP2R1 and CYP27A1 catalyze vitamin D to its circulating form 25-hydroxyvitamin D. 25942481_CYP2R1 alleles have dosage-dependent effects on vitamin D homeostasis. CYP2R1 mutations cause a novel form of genetic vitamin D deficiency with semidominant inheritance. 26038244_Allelic variations in CYP2R1 and GC affect vitamin D levels, but variant alleles on VDR and DHCR7 were not correlated with vitamin D deficiency. 26073892_A significant association was found between decreased ASD risk and child CYP2R1 AA-genotype. 26149120_Polymorphisms in CYP2R1-rs10766197 and DHCR7/NADSYN1-rs12785878 are associated with vitamin D deficiency in Uygur and Kazak ethnic populations 26177022_The VDR and CYP2R1 variants may be involved in genetic interactions in the pathogenesis of persistent allergic rhinitis. 26383826_The aim of this study was to investigate the association of three polymorphisms in the GC gene (rs7041 and rs4588) and CYP2R1 gene (rs10741657) on 25-(OH) VD serum concentration among Jordanians. 26661839_rs11023374 in CYP2R1 was significantly associated with serum 25(OH)D3. 26911666_Vitamin D related (VDR rs2228570 and CYP2R1 rs10741657) and IL28B rs12979860 genes polymorphisms accurately assure sustained viral response in naive CHC G4 patients treated with low cost standard therapy. 27160686_In a Pakistani population, no statistically significant associations between SNPs in VDR, DBP, and CYP2R1 and tuberculosis was demonstrated. 27314545_CYP3A4*22 and combined CYP3A genotypes are unlikely to provide additional information beyond CYP3A5 genotype. 27473187_CYP2R1 polymorphisms are important modulators of circulating 25-hydroxyvitamin D levels in elderly females with vitamin insufficiency. 27473561_This article concludes that mutations in CYP2R1 are responsible for an atypical form of vitamin D-deficiency rickets, which has been classified as vitamin D dependent rickets type 1B (VDDR1B, MIM 600081). [review] 27716192_our study for the first time reports a potentially causative role of CYP2R1 mutation in Vogt-koyanagi-harada disease. 27977320_Expression of CYP2R1 was in spermatozoa from healthy controls compared with infertile men, however the percentage of spermatozoa expressing CYP2R1 was not significantly higher. 28008453_genetic association study in population in north India: Data suggest (1) GT allele of VDBP SNP rs7041, (2) VDBP allelic combination (GC1F/1F: T allele rs4588; C allele rs7041), and (3) GA allele of CYP2R1 SNP rs2060793 are associated with vitamin D deficiency in women with PCOS (polycystic ovarian syndrome) in population studied. (VDBP = vitamin D-binding protein; CYP2R1 = cytochrome P450 family 2 subfamily R member 1) 28673024_We investigated the association between genetic polymorphisms in VDR, cytochrome P450 (CYP2R1, CYP24A1, and the CYP3A family) with plasma concentrations of vitamin D metabolites (25-hydroxyvitamin D3 (25(OH)D3) and proportion 24,25-dihydroxyvitamin D3 (24,25(OH)2D3)) among individuals of sub-Saharan African and European ancestry. Only CYP3A43 and VDR polymorphisms were associated with proportion 24,25(OH)2D. 28757204_The authors describe a low-frequency CYP2R1 coding variant that exerts the largest effect upon 25-hydroxyvitamin D levels identified to date in the general European population and implicates vitamin D in the etiology of multiple sclerosis. 28834557_The results of this study suggested that a role of CYP2R1 rs10766197 in both risk and progression of Muscle sclerosis with sex-related differences. 29502202_rs12794714 SNP is associated with asthma risk in a Tunisian population. 29528271_No significant differences were found between ischemic stroke patients and controls in terms of CYP24A1 rs927650 and CYP2R1 rs10741657 genotype frequencies. Polymorphic allele frequencies of CYP24A1 rs927650 and CYP2R1 rs10741657 were 0.414 and 0.660 in stroke patients, respectively. 29752008_Changes in serum vitamin D and metabolic profile following high dose supplementation with vitamin D have been shown to be associated with CYP2R1 polymorphism. 29804528_Genetic variants of CYP2R1 are key determinants of serum 25OHD levels and are highly associated with myocardial infarction risk. The three studied SNPs were associated with significantly different total 25OHD levels and their genotype distributions differed significantly between MI patients and controls where the high risk genotypes were AG/AA for rs2060793, AG/GG for rs1993116 and AG/AA for rs10766197. 30019995_There were nine genetic variants identified in CYP2R1 genes in three Chinese populations: Han, Tibetan and Uighur. 30081191_Results also showed for the first that polymorphisms of CYP2R1 gene (rs1993116 and rs10766197) were significantly associated with type 2 diabetes mellitus in a Chinese rural population. 30120973_Published articles provide evidence supporting a major role for the rs10741657 polymorphism of the CYP2R1 gene in determining 25(OH)D levels and the presence of vitamin D deficiency. [review] 30192652_Our results suggest that CYP2R1 polymorphisms are associated with a reduced risk of hypertension independent of the vitamin D level in the Han Chinese population. 30475961_A genetic risk score (GRS) was calculated based on 3 genetic variants [i.e., 7-dehydrocholesterol reductase (DHCR7) rs12785878, cytochrome P450 2R1 (CYP2R1) rs10741657 and group-specific component globulin (GC) rs2282679] related to circulating vitamin D levels. We found a significant interaction between dietary fat intake and vitamin D GRS on 2-y changes in whole-body bone mineral density. 30576350_Genotyping was performed for eight SNPs covering the CYP2R1 gene in 2868 men. Subjects were followed up concerning incidence of fracture during five years. There was a significant genetic association with circulating levels of 25(OH)D (4.6-18.5% difference in mean values between SNP alleles), but there were no correlations with levels of calcium, phosphate, PTH or FGF23 for any genetic variant. 30624776_rs10741657 variant of the CYP2R1 gene modulates the response to high-dose of vitamin D supplementation. 31206955_CYP2R1 rs12794714 and rs10766196 polymorphisms were associated with a higher risk of type 1 diabetes. Thus, polymorphisms in vitamin D metabolism may contribute to susceptibility to type 1 diabetes in Korean children. 31313056_The rs10766197 variant, near the CYP2R1 gene locus, significantly modified the efficacy of high-dose vitamin D3 supplementation for its effects on improving cognitive abilities. 31331440_rs1279414 and rs10766197 polymorphisms associated with serum vitamin D levels 31520221_Results indicated that genetic variations in the CYP2R1 gene (rs12794714, rs10741657, rs1562902, and rs10766197) were associated with susceptibility to HCV infection within a high-risk Chinese population, and the combined effect of risk alleles in the four SNPs contributed to a significantly elevated risk of HCV infection in a locus-dosage manner. 31625015_Genetic Polymorphisms in the Vitamin D Pathway and Non-small Cell Lung Cancer Survival. 32012190_rs10741657 was not associated with risk of colorectal cancer per se. rs10741657 may influence the association between vitamin D intake and risk of colorectal cancer. 32115644_Differential Frequency of CYP2R1 Variants Across Populations Reveals Pathway Selection for Vitamin D Homeostasis. 32344004_CYP2R1 and CYP27A1 genes: An in silico approach to identify the deleterious mutations, impact on structure and their differential expression in disease conditions. 32517587_Genetic Variation in Cytochrome P450 2R1 and Vitamin D Binding Protein Genes are associated with Vitamin D Deficiency in Adolescents. 32534577_Genetic variants of VDR and CYP2R1 affect BMI independently of serum vitamin D concentrations. 32764491_Vitamin D Pathway Genetic Variation and Type 1 Diabetes: A Case-Control Association Study. 32915988_Association of Vitamin D Pathway Gene CYP27B1 and CYP2R1 Polymorphisms with Autoimmune Endocrine Disorders: A Meta-Analysis. 32971523_Evaluation of the Associations of GC and CYP2R1 Genes and Gene-Obesity Interactions with Type 2 Diabetes Risk in a Chinese Rural Population. 33058307_Dysregulation of vitamin D synthesis pathway genes in colorectal cancer: A case-control study. 33109689_Low vitamin D and risk of bacterial pneumonias: Mendelian randomisation studies in two population-based cohorts. 33625596_SNP rs12794714 of CYP2R1 is associated with serum vitamin D levels and recurrent spontaneous abortion (RSA): a case-control study. 33626316_The CYP2R1 Enzyme: Structure, Function, Enzymatic Properties and Genetic Polymorphism. 33715104_Two novel CYP2R1 mutations in a family with vitamin D-dependent rickets type 1b. 33961971_Vitamin D pathway gene polymorphisms, vitamin D level, and cytokines in children with type 1 diabetes. 34093899_Association of Polymorphisms in Vitamin D-Metabolizing Enzymes DHCR7 and CYP2R1 with Cancer Susceptibility: A Systematic Review and Meta-Analysis. 34797893_Genetic variations of CYP2R1 (rs10741657) in Bangladeshi adults with low serum 25(OH)D level-A pilot study. 34906413_The influence of CYP2R1 polymorphisms and gene-obesity interaction with hypertension risk in a Chinese rural population. 34944511_New Variants of the Cytochrome P450 2R1 (CYP2R1) Gene in Individuals with Severe Vitamin D-Activating Enzyme 25(OH)D Deficiency. 34977256_Polymorphisms CYP2R1 rs10766197 and CYP27B1 rs10877012 in Multiple Sclerosis: A Case-Control Study. 35057442_Haplotypes in the GC, CYP2R1 and CYP24A1 Genes and Biomarkers of Bone Mineral Metabolism in Older Adults. | ENSMUSG00000030670 | Cyp2r1 | 304.266773 | 0.8268547 | -0.274294303 | 0.16278001 | 2.823125e+00 | 9.291532e-02 | No | Yes | 269.150046 | 47.556964 | 325.309102 | 57.243080 | |
| ENSG00000186174 | 283149 | BCL9L | protein_coding | Q86UU0 | FUNCTION: Transcriptional regulator that acts as an activator. Promotes beta-catenin transcriptional activity. Plays a role in tumorigenesis. Enhances the neoplastic transforming activity of CTNNB1 (By similarity). {ECO:0000250}. | 3D-structure;Acetylation;Activator;Alternative splicing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | hsa:283149; | beta-catenin-TCF complex [GO:1990907]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; beta-catenin binding [GO:0008013]; transcription coactivator activity [GO:0003713]; canonical Wnt signaling pathway [GO:0060070]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; positive regulation of epithelial to mesenchymal transition [GO:0010718]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of cell morphogenesis [GO:0022604] | 17052462_crystallographic analysis of how beta-catenin, BCL9, BCL9-2 and Tcf4 interact 20637214_Pygo2 PHD is the only known PHD finger that is capable of interacting simultaneously with two functional ligands, B9L and BCL9. 21703997_BCL9-2 promotes early phases of intestinal tumor progression in humans and in transgenic mice. BCL9-2 increases the expression of a subset of canonical Wnt target genes but also regulates genes that are required for early stages of tumor progression. 22109522_Data show that beta-catenin/BCL9-Like (BCL9L)/T-cell factor 4 (TCF4) signalling directly targets the GCM1/syncytin pathway and thereby regulates the fusion of human choriocarcinoma cells. 25149534_BCL9-2 induces ER positive breast cancers in vivo, regulates ER expression by a novel ss-catenin independent mechanism in breast cancer cells. 25678599_The inhibition of the transcriptional activity of BCL9-2 by WWOX and HDAC3 constitutes a new molecular mechanism and provides new insight for a broad range of cancers. 27713160_we identify BCL9L as a novel regulator of TGF-beta-induced EMT in pancreatic cancer. 28073006_BCL9L dysfunction contributes to aneuploidy tolerance in both TP53-WT and mutant cells by reducing basal caspase-2 levels and preventing cleavage of MDM2 and BID. 30698996_Dual-luciferase reporter confirmed that BCL9L is the target gene of both miR-22 and miR-214. 33767438_BCL9/BCL9L promotes tumorigenicity through immune-dependent and independent mechanisms in triple negative breast cancer. 34319913_Type I collagen promotes tumor progression of integrin beta1 positive gastric cancer through a BCL9L/beta-catenin signaling pathway. 34545187_The interactions of Bcl9/Bcl9L with beta-catenin and Pygopus promote breast cancer growth, invasion, and metastasis. | ENSMUSG00000063382 | Bcl9l | 395.442072 | 1.0415575 | 0.058742442 | 0.17381543 | 1.165065e-01 | 7.328548e-01 | No | Yes | 414.192945 | 62.143551 | 403.286732 | 60.445064 | |||
| ENSG00000186564 | 2306 | FOXD2 | protein_coding | O60548 | FUNCTION: Probable transcription factor involved in embryogenesis and somatogenesis. {ECO:0000250}. | DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene belongs to the forkhead family of transcription factors which is characterized by a distinct forkhead domain. The specific function of this gene has not yet been determined. [provided by RefSeq, Jul 2008]. | hsa:2306; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; anatomical structure morphogenesis [GO:0009653]; cell differentiation [GO:0030154]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 12621056_FOXD2 is a physiological regulator which increases cAMP sensitivity and sets the threshold for cAMP-mediated negative modulation of T cell activation 22306510_identified transcription factors FOXD2 and GABP that bind to specific elements in the promoter of the RhitH gene 32945354_Long noncoding RNA FOXD2AS1 regulates the tumorigenesis and progression of breast cancer via the S100 calcium binding protein A1/Hippo signaling pathway. 34043149_The potential roles of lncRNAs DUXAP8, LINC00963, and FOXD2-AS1 in luminal breast cancer based on expression analysis and bioinformatic approaches. | ENSMUSG00000055210 | Foxd2 | 120.111866 | 1.3674730 | 0.451512343 | 0.24711930 | 3.288990e+00 | 6.974592e-02 | No | Yes | 123.916045 | 18.789970 | 88.582494 | 13.444432 | ||
| ENSG00000186575 | 4771 | NF2 | protein_coding | P35240 | FUNCTION: Probable regulator of the Hippo/SWH (Sav/Wts/Hpo) signaling pathway, a signaling pathway that plays a pivotal role in tumor suppression by restricting proliferation and promoting apoptosis. Along with WWC1 can synergistically induce the phosphorylation of LATS1 and LATS2 and can probably function in the regulation of the Hippo/SWH (Sav/Wts/Hpo) signaling pathway. May act as a membrane stabilizing protein. May inhibit PI3 kinase by binding to AGAP2 and impairing its stimulating activity. Suppresses cell proliferation and tumorigenesis by inhibiting the CUL4A-RBX1-DDB1-VprBP/DCAF1 E3 ubiquitin-protein ligase complex. {ECO:0000269|PubMed:20159598, ECO:0000269|PubMed:20178741, ECO:0000269|PubMed:21167305}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Deafness;Disease variant;Membrane;Nucleus;Phosphoprotein;Reference proteome;Tumor suppressor;Ubl conjugation | This gene encodes a protein that is similar to some members of the ERM (ezrin, radixin, moesin) family of proteins that are thought to link cytoskeletal components with proteins in the cell membrane. This gene product has been shown to interact with cell-surface proteins, proteins involved in cytoskeletal dynamics and proteins involved in regulating ion transport. This gene is expressed at high levels during embryonic development; in adults, significant expression is found in Schwann cells, meningeal cells, lens and nerve. Mutations in this gene are associated with neurofibromatosis type II which is characterized by nervous system and skin tumors and ocular abnormalities. Two predominant isoforms and a number of minor isoforms are produced by alternatively spliced transcripts. [provided by RefSeq, Jul 2008]. | hsa:4771; | adherens junction [GO:0005912]; apical part of cell [GO:0045177]; cell body [GO:0044297]; cleavage furrow [GO:0032154]; cortical actin cytoskeleton [GO:0030864]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; early endosome [GO:0005769]; filopodium membrane [GO:0031527]; lamellipodium [GO:0030027]; membrane [GO:0016020]; neuron projection [GO:0043005]; nucleolus [GO:0005730]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; actin binding [GO:0003779]; integrin binding [GO:0005178]; actin cytoskeleton organization [GO:0030036]; cell-cell junction organization [GO:0045216]; ectoderm development [GO:0007398]; hippocampus development [GO:0021766]; lens fiber cell differentiation [GO:0070306]; mesoderm formation [GO:0001707]; negative regulation of cell migration [GO:0030336]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of cell-cell adhesion [GO:0022408]; negative regulation of cell-matrix adhesion [GO:0001953]; negative regulation of MAPK cascade [GO:0043409]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of receptor signaling pathway via JAK-STAT [GO:0046426]; negative regulation of tyrosine phosphorylation of STAT protein [GO:0042532]; odontogenesis of dentin-containing tooth [GO:0042475]; positive regulation of cell differentiation [GO:0045597]; positive regulation of stress fiber assembly [GO:0051496]; regulation of apoptotic process [GO:0042981]; regulation of cell cycle [GO:0051726]; regulation of gliogenesis [GO:0014013]; regulation of hippo signaling [GO:0035330]; regulation of neural precursor cell proliferation [GO:2000177]; regulation of protein localization to nucleus [GO:1900180]; regulation of protein stability [GO:0031647]; regulation of stem cell proliferation [GO:0072091]; Schwann cell proliferation [GO:0014010] | 11836375_A constitutional de novo interstitial deletion of 8 Mb on chromosome 22q12.1-12.3 encompassing the neurofibromatosis type 2 (NF2) locus in a dysmorphic girl with severe malformations. 11839955_Nonsense NF2 mutations and NF2 allele losses were found in all tumors in children of sporadic neurofibromatosis 2 patients in whom no NF2-mutations were found by screening their blood-DNA. 12191989_The motor protein kinesin-1 links neurofibromin and this protein in a common cellular pathway of neurofibromatosis 12444101_The binding of merlin to its partner, hepatocyte growth factor-regulated tyrosine kinase substrate, may facilitate its ability to function as a tumor suppressor. 12444102_Schwannomin inhibits Stat3 activation in schwannoma cells. 12471027_isolation and characterization of an aggresome determinant in this tumor suppressor gene 12478663_NF2 has a role in the development of multiple meningioma 12665675_Small NF2 mutations were identified in six out of seventeen meningiomas and schwannomas. Large deletions occurred in six meningiomas. These mutations could cause a truncated NF2 protein or loss of a large protein domain. 12684666_functional loss of the NF2 protein may be involved in the formation of a subset of human malignant mesothelioma 12896975_merlin is the first neuronal binding partner for PKA-RIbeta and may have a novel function in connecting neuronal cytoskeleton to PKA signaling 13679444_The lack of NF2 alterations supports the hypothesis that GI schwannomas represent a morphologically and genetically distinct group of peripheral nerve sheath tumors that are different from conventional schwannomas. 14566860_Review discusses NF2 involvement in cell motility, cell proliferation, and Rac signaling, and its implication in the development of sporadic schwannomas and meningiomas. 14612918_NF2 has a role in inhibiting schwannoma cell proliferation through promoting PDGFR degradation 14724586_the first direct demonstration that the S518D merlin mutation, which mimics merlin phosphorylation, impairs not only merlin growth and motility suppression but also leads to a novel phenotype previously ascribed to ERM proteins 14981079_both the merlin N and C termini are phosphorylated by PKA; phosphorylation of serine 518 promotes heterodimerization between merlin and ezrin 15123692_interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity; results provide the first clue to functional interaction between TRBP and merlin and suggest a novel mechanism for the tumor suppressor function of merlin 15190457_Observational study of gene-disease association. (HuGE Navigator) 15378014_S518 phosphorylation modulates ability of merlin to function as a tumor suppressor 15467741_role for merlin in receptor-mediated signaling at the cell surface 15580288_In glioma and osteosarcoma cells, endogenous merlin was targeted to the nucleus in a cell cycle-specific manner. 15609345_Aberrant NF2 hypermethylation is associated with the development of meningiomas 15635074_Mosaic NF2 may be the cause of about 8% of multiple meningiomas in sporadic adult cases. 15699051_NF2 tumor suppressor Merlin and the ERM proteins interact with N-WASP and regulate its actin polymerization function 15743831_Neurofibromin 2 (merlin) exerts antiproliferative effect via repression of PAK-induced cyclin D1 expression. 15797715_The widespread expression of merlin in brain and its association with protein kinase A suggest a role for merlin in brain biology. 15824172_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15980114_Involvement of the neurofibromatosis 2 tumour suppressor gene, NF2, on chromosome 22q in the high incidence of benign meningioma in the elderly. 16007223_Merlin can function as a tumor suppressor by inhibiting the RalGDS-mediated oncogenic signals. 16497727_Schwannomin inhibits tumorigenesis through direct interaction with the eukaryotic initiation factor subunit c (eIF3c) 16532029_Localization of HEI10 is dependent on merlin expression level. 16537381_Merlin and MLK3 can interact in situ and merlin can disrupt the interactions between B-Raf and Raf-1 or those between MLK3 and either B-Raf or Raf-1. 16612978_transitional and fibroblastic meningiomas harbor significantly more NF2 mutations than meningothelial meningiomas, indicating molecular subsets of these tumors 16652148_Merlin negatively regulates focal adhesion kinase, a major component of pathways that control cell motility and invasiveness. 16786152_Mutational analysis by denaturing HPLC showed that mutations in the NF2 gene play an important role in the development of sporadic meningiomas. 16824698_Functional duality of merlin may represent a paradigm in proteome complexity and is important in investigating multifactorial diseases such as cancer. 16885985_CPI-17 siRNA decreased the level of merlin phosphorylation and consequently Ras and ERK activity in human tumor cell lines 17007372_Merlin was expressed in vestibular schwannoma tissue. There were no correlations between merlin expression percentage and the age, gender, tumor diameter and clinical stage. 17210637_NF2-associated GTP binding protein is a tumor suppressor that regulates and requires merlin to suppress cell proliferation 17222329_There is a higher frequency of biallelic NF2 inactivation in fibroblastic (52%) versus meningothelial (18%) tumors, presence of macro-mutations on 22q also shows marked differences between fibroblastic (86%) and meningothelial (39%) meningioma subtypes. 17470137_Observational study of genotype prevalence. (HuGE Navigator) 17478763_The NF2 gene plays a role in the tumorigenesis of pediatric meningiomas and chromogenic in situ hybridization detects NF2 gene deletion in formalin-fixed, paraffin-embedded tissues. 17509660_Schwannomas and meningiomas, and to a lesser degree, ependymomas, express a high incidence of NF2 gene deletion, which supports the hypothesis that NF2 gene plays an important role in their tumorigenesis. 17566081_merlin plays a key role in the regulation of the Schwann cell microtubule cytoskeleton 17655741_We observed merlin haploinsufficiency in peripheral nerves of two different patients with NF2-related polyneuropathy. 17868749_study confirms the NF2 mutation in HEI-193 cells causes a splicing defect in the NF2 transcript and identified the protein product as merlin isoform 3 with attenuated growth suppressive activity 17989580_Merlin is inversely correlated with cyclin D1 in regard to subcellular localization. merlin compensates for the malignant effect of S518 phosphorylation by limiting the loss of contact inhibition of growth and controlling proliferation. 18033041_Observational study of gene-disease association. (HuGE Navigator) 18033041_patients with nonsense or frameshift mutations of NF2 were more frequently affected with meningiomas and spinal tumours, in addition to VIII nerve schwannomas 18071304_A role for merlin in mediating PKA-induced changes of the actin cytoskeleton. 18072270_The latter observation suggests that a four-hit mechanism involving the SMARCB1 and NF2 genes may be implicated in schwannomatosis-related tumorigenesis. 18173316_Patients with unilateral NF2-related vestibular schwannoma have a high risk of developing a contralateral tumor; and transmission risks are reduced for offspring, particularly in mosaic patients. 18285426_In all affected individuals with SMARCB1 mutations and available tumour tissue, bi-allelic somatic inactivation of the NF2 gene, was detected. 18332868_VprBP depletion abolished the in vivo interaction of Merlin and Roc1-Cullin4A-DDB1, which resulted in Merlin stabilization and inhibited ERK and Rac activation 18361411_Merlin is inactivated in DU145 prostate cancer cells by PAK-mediated constitutive phosphorylation, identifying a novel mechanism of merlin inactivation in neoplastic cells. 18445079_Data show the altered adhesive structures and their relation to RhoGTPase activation in merlin-deficient Schwannoma. 18632626_merlin is a potent inhibitor of high-grade human glioma 18670066_Mutations in the NF2 gene is associated with neurofibromatosis type 2 18725387_this is the first report of negative regulatory signaling from Merlin to YAP1 in mammalian cells and may shed light on mechanisms of malignant pleural mesothelioma development 18766994_Bilateral combined hamartomas of the retina and retinal pigment epithelium (RPE) in a young child should alert the clinician to the possibility of neurofibromatosis type 2. 18808065_Expression of c-Myc, neurofibromatosis Type 2, somatostatin receptor 2 and erb-B2 in human meningiomas: relation to grades or histotypes. 18822692_No alterations in NF2 gene expression in the quantitative analyses of the 5 gastrointestinal stromal tumors. 18835652_the disruption of NF2 signalling is essential for the development of human mesothelioma. 18923024_Schwannomin is necessary for the correct organization and regulation of axoglial and glio-glial contacts in transgenic mice. 18953429_results represent the first demonstration that merlin regulates cell growth in meningioma cells by suppressing YAP 19029950_Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas 19141242_The affinity of Merlin to CD44 was increased after phosphorylation at S518 in vestibular schwannoma. 19142715_involvement of phosphorylated merlin in tumorigenesis of sporadic vestibular Schwannomas. 19144871_p55-NF2 protein interaction may play a functional role in the regulation of apico-basal polarity and tumor suppression pathways in non-erythroid cells. 19328320_Mutations may be present in familial and sporadic schwannomaatosis. 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19451225_Data show that NF2 patient tumors and Nf2-deficient mouse embryonic fibroblasts demonstrate elevated mTORC1 signaling. 19451229_Data reveal that merlin-negative mesotheliomas display unregulated mTORC1 signaling and are sensitive to rapamycin, thus providing a preclinical rationale for prospective, biomarker-driven clinical studies of mTORC1 inhibitors in these tumors. 19687056_preimplantation genetic diagnosis in female carrier of de novo mutations in NF2 gene; informative markers flanking the NF2 gene were used 19778867_The actin binding site in the carboxy terminal is absent in merlin and in its closed conformation the indirect actin binding site in the FERM domain is also not available for the interaction of other proteins with it. 19924781_Features of patients with large intragenic deletions and individuals with mutations affecting single or multiple nucleotides of the NF2 gene are relatively similar in neurofibromatosis 2. 19968670_Observational study of gene-disease association. (HuGE Navigator) 20054297_NF2 mutations were found in non-VHL mutated clear cell renal cell carcinoma 20178741_Study concludes that Merlin suppresses tumorigenesis by translocating to the nucleus to inhibit CRL4(DCAF1). 20195187_Merlin depletion results in deregulation of ErbB receptor signaling, promotes a dedifferentiated state, and increases Schwann cell proliferation, suggesting critical steps toward schwannoma tumorigenesis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20445339_Mutational analysis of the NF2 gene in Korean patients with neurofibromatosis type 2. 20491622_Studies indicate that recent advances have improved basic understanding about merlin function and the major signaling pathways regulated by merlin and provided the foundation for future research in clinic cancers. 20553997_identified 8 at-risk haplotypes and 5 mutations in the NF2 gene of 39 patients with neurofibromatosis 2-associated tumors. 20600642_Silencing of merlin gene expression in vestibular schwannomas results in down-regulation of p21 (waf1). 20682713_Meningiomas displayed different patterns of genetic alterations during progression according to their NF2 status 20831745_a significant proportion of sporadic vestibular schwannomas (>40%) have unmethylated wild-type NF2 genes 20890305_These results establish merlin as a potent inhibitor of mixed lineage kinase 3, extracellular signal-regulated kinase, and c-jun N-terminal kinase activation in cancer. 20930055_A four-hit mechanism of tumour suppressor gene inactivation, involving SMARCB1 and NF2, might be operative in familial multiple meningiomas associated meningiomas. 21154335_the first report of familial neurofibromatosis type II with a splicing mutation of IVS3+ 3A to C of the NF2 gene 21167305_merlin mediates PICT-1-induced growth inhibition by translocating to the nucleolus and binding PICT-1 21182951_Results demonstrate that Schwannomin plays an essential role in inducing and/or maintaining the SC's spindle shape, and by stabilizing the bipolar morphology, Sch promotes the alignment of SCs with axons and ultimately influences myelin segment length. 21278391_Mutations within the NF2 gene in Neurofibromatosis type 2 patients are associated with the risk of developing meningiomas 21383154_Missense mutations in the NF2 gene result in the quantitative loss of merlin protein and minimally affect protein intrinsic function. 21481793_Depletion of Angiomotin in Nf2(-/-) Schwann cells attenuates the Ras-MAPK signaling pathway, impedes cellular proliferation in vitro and tumorigenesis in vivo 21563229_The effects of splicing mutations in NF2 are often complex and that information theory based analysis is helpful in elucidating the consequences of these mutations. 21743150_Merlin protein might contribute to the initiation of metastasis of non-small cell lung carcinoma. 21750658_Sequential phosphorylation of merlin C- and N-terminus by different oncogenic kinases targets merlin for degradation and thus downregulates its activity. 21965655_a novel mechanism for the loss of Merlin protein in breast cancer, and have developed a discriminatory model using Merlin and OPN expression in breast tumor tissues. 21971708_gastrointestinal stromal tumor cases exhibiting expression of phosphorylated Thr567 in the ezrin protein were associated with immunoactivities of KIT and merlin expression 22012890_The study reports the crystal structure of the human merlin head domain when crystallized in the presence of its tail domain. 22081132_Our data indicate that somatic mutation of NF2 gene is not prevalent in common human cancers, and its mutation somatically occurs in a minor fraction of HCC, lung cancer and acute leukaemia. 22247700_The mechanism by which merlin loss leads to increased Wnt/beta-catenin in NF2 was investigated. 22249120_NF2 gene inactivation by promoter hypermethylation is a rare or very uncommon mechanism of NF2 gene inactivation in sporadic VS. 22281088_We report the first description of a pigmented choroidal schwannoma in PTEN hamartoma tumor syndrome (PHTS). This rare tumor showed a unique combination of reduction of PTEN and absence of NF2 expression. 22295085_The molecular genetic changes in sporadic VS identified here included mutations and allelic loss, but no aberrant hypermethylation of the NF2 gene was detected 22325036_The C-terminal domains are important for merlin's morphogenic properties and growth inhibiting function. 22355270_alternative splicing and frequent codeletion of CHEK2 and NF2 contribute to the genomic instability and associated development of aggressive biologic behavior in meningiomas. 22431917_in addition to the tumor-suppressing activity of merlin, it also functions to maintain physiological angiogenesis in the nervous system by regulating antiangiogenic factors such as SEMA3F 22525268_our data suggest loss of merlin results in the Rac-dependent decrease of anterograde trafficking of exocytic vesicles, representing a possible mechanism controlling the concentration of growth factor receptors at the cell surface. 22911524_The frequency of NF2 allelic losses seen in Croatian patients is broadly similar to that reported in other populations and thus confirms the existing hypothesis regarding the tumorigenesis of schwannomas, and contributes to schwannoma genetic profile. 22912849_Merlin negatively regulates human melanoma growth, and that loss of merlin, or impaired merlin function, results in an opposite effect. 22989157_Data indicate that the truncating mutations in NF2 protein are associated with more severe phenotypes in neurofibromatosis 2 (NF2). 23036697_LOH of the NF2 gene was observed in an early stage of WDPMP, thus indicating that LOH of the NF2 gene is an early molecular alteration, and NF2 loss is a molecular mechanism associated not only with malignant pleural mesothelioma, but also with WDPMP. 23188051_this is the first NF2 case caused by a deep intronic mutation in which an in vitro antisense therapeutic approximation has been tested. These results open the possibility of using this approach in vivo for this type of mutation causing NF2. 23249734_Merlin's role in restricting cortical Ezrin may contribute to tumorigenesis by disrupting cell polarity, spindle orientation, and, potentially, genome stability 23308224_There is a novel role for NF2 as a suppressor of JC virus T-antigen-induced cell cycle regulation. 23413263_Loss of the SOX10 protein, which is vital for normal Schwann cell development, is also key to the pathology of Merlin-null schwannoma tumors. 23455610_A merlin-isoform 2-dependent complex is identified that promotes neurofilament heavy chain phosphorylation. 23468835_Ezrin and moesin are required for efficient T cell adhesion and homing to lymphoid organs. 23558725_Intracranial meningiomas are common in NF2. They are associated with poor prognosis factors 23666797_This reviews genetic properties of NF2 gene, molecular characteristics of merlin, summarizes mutational spectra and explains merlin's multifunctional roles regarding its involvement in neurofibromatosis associated tumorigenesis. 23792589_Structural variants unique to the malignant cell line inactivated: the neurofibromin2 (NF2) gene, a known tumor suppressor. 23921927_NF2 mutations may play a role in the pathophysiology of hearing loss as well as in the pattern of growth of vestibular schwannomas 24030433_Data indicate that neurofibromin 2 (NF2) somatic mutations in 34.5% of sporadic vestibular schwannomas, but no significant difference in the mutation detection rates between cystic vestibular schwannoma and vestibular schwannoma. 24166499_Findings indicate that merlin is sumoylated and that this post-translational modification is essential for tumor suppression. 24171707_A clear association was found among sporadic meningiomas cases between mutation of the NF2 gene and monosomy 22. 24249803_Use of a common set of endpoints should improve the quality of NF2 clinical trials and will foster comparison among studies for hearing loss and facial weakness. 24260485_CD43 promotes cells transformation by preventing merlin-mediated contact inhibition of growth. 24282279_characterization of NF2 shows it binds to and stabilizes microtubules through attenuation of tubulin turnover by lowering both rates of microtubule polymerization and depolymerization as well as by reducing the frequency of microtubule catastrophes. 24309211_Axonal merlin directly regulates Schwann cell behavior in patients with neurofibromatosis type 2. 24323642_Data indicate that neurofibromin 2 (NF2)/Merlin may serve as a potential target in the management of colorectal cancer. 24356468_integrin-linked kinase (ILK) plays a critical role in the suppression of the Hippo pathway via phospho-inhibition of MYPT1-PP1, leading to inactivation of Merlin 24357459_A deletion causing NF2 exon 9 skipping is associated with familial autosomal dominant intramedullary ependymoma. 24595234_The NF2 L64P allele is temperature sensitive. 24619252_results led us to suggest that high frequency of NF2 mutations may play a critical role in early tumorigenesis of young vestibular schwannomas 24706749_Data indicate that DCAF1 protein folds into a beta-hairpin structure and binds to the F3 lobe of neurofibromin 2 (Merlin) FERM domain. 24726726_NF2/Merlin tumor suppressor function 24728215_Study shows that overexpression of wild-type merlin inhibits cell proliferation, migration and adhesion. 24848258_Suggest that merlin deficiency predicts mesothelioma sensitivity to treatment with FAK inhibitors. 25026211_NF2 loss-driven derepressed CRL4(DCAF1) promotes activation of YAP by inhibiting hippo pathwat kinases Lats1 and 2 in the nucleus. 25043298_the phosphorylation of S518-Merlin in glioblastoma promotes oncogenic properties that are not only the result of inactivation of the tumor suppressor role of Merlin. 25217104_The p53/mouse double minute 2 homolog complex deregulation in merlin-deficient tumours. 25275700_These results suggest a novel tumor suppressor function of merlin in melanoma cells: the inhibition of the proto-oncogenic NHE1 activity, possibly including its downstream signaling pathways. 25452392_Findings suggest that the majority of NF2-associated vestibular schwannomas are polyclonal, such that the tumor mass represents a collision of multiple, distinct tumor clones. 25488749_Integrative analysis of mutations and somatic copy-number alterations revealed frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in pleural mesothelioma. 25549701_identified potential driver mutations in NF2 (neurofibromatosis type 2) and MN1 (meningioma 1). 25706233_Merlin coordinates collective migration of epithelial cells by acting as a mechanochemical transducer. 25783601_Our findings demonstrated that merlin exerts inhibitory effects on TNF-alpha-induced EMT by regulating hyaluronan endocytosis and the TAK1-p38MAPK signaling pathway. 25798586_Mutation in NF2 gene is associated with malignant peritoneal mesothelioma. 25823924_NF2 (frequently deleted in MPM) inhibited Snail-mediated p53 suppression and was stabilized by RKIP. 25965831_Stusies indicate that monosomy 22, which is often associated with mutations of the neurofibromin 2 (NF2) gene, has emerged as the most frequent alteration of meningiomas. 26045165_angiomotin and Merlin respectively interface cortical actin filaments and core kinases in Hippo signaling 26073919_showed that 77% of NF2 identified variants were detected by coding exons sequencing 26275417_The mortality of patients with NF2 diagnosed in more recent decades was lower than that of patients diagnosed earlier. 26359368_NF2/merlin inactivation augments mutant RAS signaling by promoting YAP/TEAD-driven transcription of oncogenic and wild-type RAS, resulting in greater MAPK output and increased sensitivity to MEK inhibitors. 26418719_Loss of Nf2 and Cdkn2a/b have synergistic effects with PDGF-B overexpression promoting meningioma malignant transformation. 26443326_(Delta2-4)Merlin variant disrupts the normal function of Merlin and promotes hepatocellular carcinoma metastasis. 26483206_findings demonstrated that Merlin critically regulated pancreatic cancer pathogenesis by suppressing FOXM1/beta-catenin signaling 26493618_homozygous deletions in CDKN2A and hemizygous loss of NF2 as detected by fluorescence in situ hybridization would confer a poor clinical outcome and may guide future treatment decisions for patients with peritoneal mesothelioma. 26549023_Data show that neurofibromin 2 (Merlin) suppresses proliferation and adhesion, at least partly, through inhibiting kinase suppressor of Ras 1 (KSR1) and DCAF1 protein. 26549232_Together our results uncover miRNAs as yet another negative mechanism controlling Merlin tumor suppressor functions. 26806348_AMOTL1 Promotes Breast Cancer Progression and Is Antagonized by Merlin 26840621_IL-1beta Induces NF2 Promoter Methylation in Meningioma/Leptomeningeal Cells. 26895810_An independent set of Sarcomatoid Renal Cell Carcinoma demonstrated mutations in NF2. NF2 mutations were mutually exclusive with TP53 but not with VHL mutations. 26923924_we demonstrate that NF2 negatively controls the invasiveness of Glioblastoma multiforme through YAP-dependent induction of CYR61/CCN1 and miR-296-3p. 27128293_Data suggest that, at least using the commercial antibodies used in this study, immunohistochemical staining for NF2 (neurofibromin 2), LATS2 (large tumor suppressor kinase 2), and YAP/TAZ (nuclear translocation of complex of Yes-associated protein [YAP] with transcriptional coactivator with PDZ-binding motif [TAZ]) is not helpful for differential diagnosis of mesothelioma versus a benign proliferation. 27285107_sustained activation of Wnt/beta-catenin signaling due to abrogation of Merlin-mediated inhibition of LRP6 phosphorylation may be a cause of Neurofibromatosis type II disease. 27289045_The authors proposed that NF2 behaves as a protein sensing tissue damage and aromatase-driven local estrogen formation, eventually leading to regulation of stem cells differentiation and tissue repair by liver cancer cells. (Review) 27378628_These findings uncover the significance of Merlin protein expression and Survivin labeling index as prognosticators for poor clinical outcome in two independent Malignant pleural mesothelioma cohorts. 27818180_co-deletion of Rac1 with Nf2 blocks tumor initiation but paradoxically exacerbates hepatomegaly induced by Nf2 loss, which can be suppressed either by treatment with pro-oxidants or by co-deletion of Yap. 27871951_NF2 localizes in nucleus when Ser518 is not phosphorylated, while phosphorylated form is present in cytoplasm and plasma membrane. Data suggest that binding of NF2 to TIMAP and EBP50 is critical in nuclear localization of NF2. (NF2 = neurofibromin 2; TIMAP = TGF-beta-inhibited membrane-associated protein; EBP50 = Ezrin-Radixin-Mosein binding phosphoprotein 50) 28003305_Genetic data coupled with transcriptomic data allowed the identification of a new malignant pleural mesothelioma (MPM)molecular subgroup, C2(LN), characterized by a co-occurring mutation in the LATS2 and NF2 genes in the same MPM. MPM patients of this subgroup presented a poor prognosis. Coinactivation of LATS2 and NF2 leads to loss of cell contact inhibition between MPM cells 28112165_Merlin loss increased oxidative stress causing aberrant activation of Hedgehog signaling in in vitro. 28396363_The acquired sensitivity to erlotinib supports the known crosstalk between MET and the HER family of receptors. For the first time, we show inactivation of NF2 during acquisition of resistance to MET-TKI that may explain the refractoriness to erlotinib in these cells. 28422417_Molecular analyses for NF2 mutations in blood of the irradiated individuals failed to detect disease-causing mutations 28429859_Four of the five had a mutation in the NF2 gene. Three had a family history of NF2; one of these patients also had a family history of intracranial aneurysm with NF2 28473293_Moesin and merlin regulate urokinase receptor-dependent endothelial cell migration, adhesion and angiogenesis 28474103_Study demonstrated a high frequency of structural variants, including novel truncating fusions of NF2, and an HRR-independent evolution of AC3 signature in low-dose radiation-induced meningiomas. 28692055_We suggest that PrP(C) and its interactor, LR/37/67 kDa, could be potential therapeutic targets for schwannomas and other Merlin-deficient tumours. 28710469_a correlation between the NF2 status and the growth patterns of sporadic vestibular schwannomas, is reported. 28729415_Low merlin expression is associated with meningioma and schwannoma. 28764788_Methylation of NF2 and DNMT1 was markedly increased, and miR-152-3p was downregulated in GBM tissues and glioma cells. Both knockdown of DNMT1 and overexpression miR-152-3p showed that demethylation activated the expression of NF2. 28775147_This study demonstrates that simultaneous inhibition of c-Met and Src signaling in MD-MSCs triggers apoptosis and reveals vulnerable pathways that could be exploited to develop NF2 therapies. 28775249_In this study, the authors perform an exome, methylation and RNA-seq analysis of 31 cases of radiation-induced meningioma and show NF2 rearrangement, an observation previously unreported in the sporadic tumors. 28849072_These results indicate that Merlin/YAP/cMyc/mTOR signaling axis promotes human cholangiocarcinoma (CCA) cell proliferation by overriding contact inhibition. We propose that overriding cMycmediated contact inhibition is implicated in the development of CCA. 28919412_Collectively, we provide for the first time in vivo evidence that the function of Merlin, as a tumor suppressor is independent of its conformational change. 28981922_The occurrance and evolvement of sporadic intraspinal Schwannomas have a close relationship with mutations of the NF2 gene. 29130106_Study identified missense NF2 mutations in 1.9% hepatocellular carcinoma (HCC) and 5.3% intrahepatic cholangiocarcinoma (ICC). Allele frequency of NF2 IVS4-39 A/A was significantly higher in HCCs. Also, NF2/Merlin showed a dual role as a tumorigenic gene and tumor-suppressor gene; Merlin was expressed at higher levels in HCC tumors ; while the rate of Merlin upregulation was lower in poorly differentiated ICCs. 29230670_Data indicate that mutations affecting SMARCB1 play a role in the development or progression of a small subset of spinal schwannomas and that biallelic inactivation of SMARCB1 may cooperate with deficiency of NF2 function in schwannoma tumorigenesis. 29587439_Study summarizes the current knowledge of molecular events triggered by NF2/merlin inactivation, which lead to the development of mesothelioma and other cancers. Genetic alterations in NF2 that abrogate merlin's functional activity are found in about 40% of malignant mesothelioma (MM), indicating the importance of NF2 inactivation in MM development and progression. [review] 29599333_The genetic alterations observed in the NF2 gene indicated that spinal schwannomas are associated with genetic alterations also found in other schwannomas and type 2 Neurofibromatosis, which reinforces the etiological role of this gene. 29626191_lipid binding results in the open conformation of neurofibromin 2 and lipid binding is necessary for inhibiting cell proliferation 29637450_NF2 promoter gene mutations occurred in medulloblastoma (MB) patients. The NF2 mRNA expression was higher in the controls than in patients; however NF2 protein expression was significantly higher in patients than in the controls. NF2 protein was mainly expressed in the nucleus in MB patients, while the NF2 protein was mainly expressed in the cytoplasm in the controls. 29893810_This is the first report of Merlin functioning as a molecular restraint on cellular metabolism in breast cancer. 29897904_Cell viability results showed that three agents (GSK2126458, Panobinostat, CUDC-907) had the greatest activity across schwannoma and meningioma cell systems, but merlin status did not significantly influence response 30135214_reduction or deletion of TbetaR2 or NF2 induces the TbetaR1-mediated oncogenic pathway, and therefore inhibition of the unbalanced TGFbeta signaling is a putative strategy for NF2-related cancers 30335132_we identify nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappaB)-inducing kinase (NIK) as a potential drug target driving NF-kappaB signaling and Merlin-deficient schwannoma genesis 30417500_Results suggest that fibroblast growth factor receptor 2 (FGFR2) expression might be negatively regulated by neurofibromin 2 (NF2) signaling in the mesothelial cells. 30816526_Study in human colorectal carcinoma and lung adenocarcinoma cells revealed downregulation of NF2 by miR92a3p via its wildtype 3'UTR, but not NF23'UTR with mutated miR92a3p MRE. miR92a3p overexpression in cancer cells promoted migration, proliferation and resistance to apoptosis, as well as altered Factin organization compared with controls. 31015291_identified a previously unreported Merlin-binding protein, apoptosis-stimulated p53 protein 2 (ASPP2, also called Tp53bp2), that bound to closed-conformation Merlin predominately through the FERM domain. 31018575_Merlin regulates contact inhibition and is an integral part of cell-cell junctions, while ERM proteins, ezrin, radixin and moesin, assist in the formation and maintenance of specialized plasma membrane structures and membrane vesicle structures. [review] 31231129_Hemizygous loss of NF2 detected by fluorescence in situ hybridization is useful for the diagnosis of malignant pleural mesothelioma. 31273341_Study has identified a very high probable mosaicism rate in de novo NF2, probably makin | ENSMUSG00000009073 | Nf2 | 1794.081141 | 1.0155132 | 0.022209035 | 0.08250642 | 7.242794e-02 | 7.878340e-01 | 9.311111e-01 | No | Yes | 1903.446506 | 192.448475 | 1893.076324 | 191.259442 | |
| ENSG00000186854 | 129293 | TRABD2A | protein_coding | Q86V40 | FUNCTION: Metalloprotease that acts as a negative regulator of the Wnt signaling pathway by mediating the cleavage of the 8 N-terminal residues of a subset of Wnt proteins. Following cleavage, Wnt proteins become oxidized and form large disulfide-bond oligomers, leading to their inactivation. Able to cleave WNT3A, WNT5, but not WNT11. Required for head formation. {ECO:0000269|PubMed:22726442}. | Alternative splicing;Cell membrane;Glycoprotein;Hydrolase;Membrane;Metal-binding;Metalloprotease;Protease;Reference proteome;Signal;Transmembrane;Transmembrane helix;Wnt signaling pathway | hsa:129293; | integral component of organelle membrane [GO:0031301]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; endopeptidase activity [GO:0004175]; metal ion binding [GO:0046872]; metalloendopeptidase activity [GO:0004222]; Wnt-protein binding [GO:0017147]; head development [GO:0060322]; negative regulation of Wnt signaling pathway [GO:0030178]; positive regulation of protein oxidation [GO:1904808]; positive regulation of protein-containing complex assembly [GO:0031334]; proteolysis [GO:0006508]; Wnt signaling pathway [GO:0016055] | 22726442_Tiki1 antagonizes Wnt function in embryos and human cells via a TIKI homology domain that is conserved from bacteria to mammals and acts likely as a protease to cleave eight amino-terminal residues of a Wnt protein, resulting in oxidized Wnt oligomers that exhibit normal secretion but minimized receptor-binding capability. 31282173_Metalloprotease TRABD2A Restriction of HIV-1 Production in Monocyte-Derived Dendritic Cells. 31801864_Inhibition of Metalloprotease TRABD2A Facilitates the Study of HIV-1 Replication in Resting CD4(+) T Cells. | 22.829346 | 0.6209885 | -0.687361538 | 0.55540394 | 1.518220e+00 | 2.178890e-01 | No | Yes | 15.573290 | 5.317675 | 24.271026 | 8.272800 | |||||
| ENSG00000186866 | 23275 | POFUT2 | protein_coding | Q9Y2G5 | FUNCTION: Catalyzes the reaction that attaches fucose through an O-glycosidic linkage to a conserved serine or threonine residue in the consensus sequence C1-X(2,3)-S/T-C2-X(2)-G of thrombospondin type 1 repeats where C1 and C2 are the first and second cysteines, respectively. O-fucosylates members of several protein families including the ADAMTS family, the thrombosporin (TSP) and spondin families. The O-fucosylation of TSRs is also required for restricting epithelial to mesenchymal transition (EMT), maintaining the correct patterning of mesoderm and localization of the definite endoderm (By similarity). Required for the proper secretion of ADAMTS family members such as ADAMSL1 and ADAMST13. {ECO:0000250, ECO:0000269|PubMed:11067851, ECO:0000269|PubMed:16464858, ECO:0000269|PubMed:17395588, ECO:0000269|PubMed:17395589, ECO:0000269|PubMed:22588082}. | 3D-structure;Alternative splicing;Carbohydrate metabolism;Disulfide bond;Endoplasmic reticulum;Fucose metabolism;Glycoprotein;Glycosyltransferase;Golgi apparatus;Reference proteome;Signal;Transferase | PATHWAY: Protein modification; protein glycosylation. | Fucose is typically found as a terminal modification of branched chain glycoconjugates, but it also exists in direct O-linkage to serine or threonine residues within cystine knot motifs in epidermal growth factor (EGF; MIM 131530)-like repeats or thrombospondin (THBS; see MIM 188060) type-1 repeats. POFUT2 is an O-fucosyltransferase that use THBS type-1 repeats as substrates (Luo et al., 2006 [PubMed 16464857]).[supplied by OMIM, Mar 2008]. | hsa:23275; | endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; peptide-O-fucosyltransferase activity [GO:0046922]; fucose metabolic process [GO:0006004]; mesoderm formation [GO:0001707]; positive regulation of protein folding [GO:1903334]; protein O-linked fucosylation [GO:0036066]; regulation of epithelial to mesenchymal transition [GO:0010717]; regulation of gene expression [GO:0010468]; regulation of secretion [GO:0051046] | 20549515_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22588082_Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. 25544610_POFUT2 and B3GLCT mediate a noncanonical endoplasmic reticulum quality-control mechanism that recognizes folded thrombospondin type 1 repeats and stabilizes them by glycosylation. 27773385_This study demonstrated that the alteration of POFUT2 expression in the superior temporal gyrus of elderly patients with schizophrenia. | ENSMUSG00000020260 | Pofut2 | 1338.412711 | 0.9008666 | -0.150614629 | 0.09597895 | 2.448311e+00 | 1.176514e-01 | 4.577691e-01 | No | Yes | 1311.541383 | 106.692324 | 1445.162474 | 117.331234 |
| ENSG00000188283 | 163087 | ZNF383 | protein_coding | Q8NA42 | FUNCTION: May function as a transcriptional repressor, suppressing transcriptional activities mediated by MAPK signaling pathways. {ECO:0000269|PubMed:15964543}. | Cytoplasm;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene is a KRAB-related zinc finger protein that inhibits the transcription of some MAPK signaling pathway genes. The repressor activity resides in the KRAB domain of the encoded protein. [provided by RefSeq, Sep 2016]. | hsa:163087; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity [GO:0003700]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 15964543_Overexpression of ZNF383 in cells inhibits the transcriptional activities of AP-1 and SRE, suggesting that ZNF383 may act as a negative regulator in MAPK-mediated signaling pathways. | ENSMUSG00000099689 | Zfp383 | 252.570638 | 1.0133873 | 0.019185650 | 0.17438381 | 1.212350e-02 | 9.123247e-01 | No | Yes | 219.371533 | 49.572022 | 214.578443 | 48.561513 | ||
| ENSG00000188338 | 10991 | SLC38A3 | protein_coding | Q99624 | FUNCTION: Sodium-dependent amino acid/proton antiporter. Mediates electrogenic cotransport of glutamine and sodium ions in exchange for protons. Also recognizes histidine, asparagine and alanine. May mediate amino acid transport in either direction under physiological conditions. May play a role in nitrogen metabolism and synaptic transmission. {ECO:0000250|UniProtKB:Q9JHZ9, ECO:0000269|PubMed:10823827}. | Amino-acid transport;Antiport;Cell membrane;Disulfide bond;Glycoprotein;Ion transport;Membrane;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | hsa:10991; | apical plasma membrane [GO:0016324]; basolateral plasma membrane [GO:0016323]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; amino acid transmembrane transporter activity [GO:0015171]; antiporter activity [GO:0015297]; L-alanine transmembrane transporter activity [GO:0015180]; L-asparagine transmembrane transporter activity [GO:0015182]; L-glutamine transmembrane transporter activity [GO:0015186]; L-histidine transmembrane transporter activity [GO:0005290]; symporter activity [GO:0015293]; amino acid transmembrane transport [GO:0003333]; amino acid transport [GO:0006865]; asparagine transport [GO:0006867]; brain development [GO:0007420]; cellular response to potassium ion starvation [GO:0051365]; female pregnancy [GO:0007565]; glutamine transport [GO:0006868]; histidine transport [GO:0015817]; L-alanine transport [GO:0015808]; positive regulation of glutamine transport [GO:2000487]; positive regulation of transcription from RNA polymerase II promoter in response to acidic pH [GO:0061402]; sodium ion transport [GO:0006814]; transport across blood-brain barrier [GO:0150104] | 12788082_SN1 is a target for the ubiquitin ligase Nedd4-2, which is inactivated by the serum and glucocorticoid inducible kinase SGK1, its isoform SGK3, and protein kinase B. 15094455_SNAT3 mRNA showed a 3-5 times stronger expression in gliomas than in metastases or control tissue, and was virtually absent from glioma cultures. Native glioblastoma immunostained positively with anti-SNAT3 antibody. 16432833_Transcription of the SLC38A3 gene was impaired in all 5 RCC cell lines analyzed. Our data indicate this gene as a putative tumour suppressor gene. 19892400_SNAT3 is expressed in the placenta in the early stage of pregnancy. 24854847_The regulation of SNAT3 gene expression by extracellular pH involves post-transcriptional and transcriptional mechanisms, the latter being distinct from the mechanisms that control the tissue-specific expression of the gene. 28202352_SLC38A3 activated PDK1/AKT signaling and promoted metastasis of non-small cell lung cancer cells through regulating glutamine and histidine transport. | ENSMUSG00000010064 | Slc38a3 | 277.260504 | 0.9130687 | -0.131204734 | 0.18403623 | 5.134828e-01 | 4.736351e-01 | No | Yes | 254.258040 | 35.942901 | 286.060426 | 40.196362 | |||
| ENSG00000188372 | 7784 | ZP3 | protein_coding | P21754 | FUNCTION: Component of the zona pellucida, an extracellular matrix surrounding oocytes which mediates sperm binding, induction of the acrosome reaction and prevents post-fertilization polyspermy. The zona pellucida is composed of 3 to 4 glycoproteins, ZP1, ZP2, ZP3, and ZP4. ZP3 is essential for sperm binding and zona matrix formation. | Alternative splicing;Cell membrane;Cleavage on pair of basic residues;Disease variant;Disulfide bond;Extracellular matrix;Fertilization;Glycoprotein;Membrane;Pyrrolidone carboxylic acid;Receptor;Reference proteome;Secreted;Signal;Transmembrane;Transmembrane helix | The zona pellucida is an extracellular matrix that surrounds the oocyte and early embryo. It is composed primarily of three or four glycoproteins with various functions during fertilization and preimplantation development. The protein encoded by this gene is a structural component of the zona pellucida and functions in primary binding and induction of the sperm acrosome reaction. The nascent protein contains a N-terminal signal peptide sequence, a conserved ZP domain, a C-terminal consensus furin cleavage site, and a transmembrane domain. It is hypothesized that furin cleavage results in release of the mature protein from the plasma membrane for subsequent incorporation into the zona pellucida matrix. However, the requirement for furin cleavage in this process remains controversial based on mouse studies. A variation in the last exon of this gene has previously served as the basis for an additional ZP3 locus; however, sequence and literature review reveals that there is only one full-length ZP3 locus in the human genome. Another locus encoding a bipartite transcript designated POMZP3 contains a duplication of the last four exons of ZP3, including the above described variation, and maps closely to this gene. [provided by RefSeq, Jul 2008]. | hsa:7784; | collagen-containing extracellular matrix [GO:0062023]; egg coat [GO:0035805]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; acrosin binding [GO:0032190]; carbohydrate binding [GO:0030246]; extracellular matrix structural constituent [GO:0005201]; identical protein binding [GO:0042802]; receptor ligand activity [GO:0048018]; structural constituent of egg coat [GO:0035804]; binding of sperm to zona pellucida [GO:0007339]; blastocyst formation [GO:0001825]; egg coat formation [GO:0035803]; humoral immune response mediated by circulating immunoglobulin [GO:0002455]; negative regulation of binding of sperm to zona pellucida [GO:2000360]; negative regulation of transcription, DNA-templated [GO:0045892]; oocyte development [GO:0048599]; positive regulation of acrosomal vesicle exocytosis [GO:2000368]; positive regulation of acrosome reaction [GO:2000344]; positive regulation of antral ovarian follicle growth [GO:2000388]; positive regulation of humoral immune response [GO:0002922]; positive regulation of inflammatory response [GO:0050729]; positive regulation of interferon-gamma production [GO:0032729]; positive regulation of interleukin-4 production [GO:0032753]; positive regulation of leukocyte migration [GO:0002687]; positive regulation of ovarian follicle development [GO:2000386]; positive regulation of T cell proliferation [GO:0042102]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of type IV hypersensitivity [GO:0001809] | 14673092_Eggs expressing glycoprotein huZP3, derived from transgenic mice, bind murine but not human sperm, implying that huZP3 acquires the same O-glycans as native mZP3. 15379548_human and mouse ZP3 proteins are quite similar, and alternative explanations of taxon-specific sperm binding warrant exploration 15950651_Binding sites for recombinant zona pellucida C (ZPC) glycoprotein are located both at the N- and C-terminus of proacrosin 16407501_Exposure of sperm to ZP proteins promoted acrosomal exocytosis and changed motility patterns. 17192598_Peptides rhuZP3a22 approximately 176 and rhuZP3b177 approximately 348 have a role similar to human ZP3. The mechanism of the response to the peptides involves influx of calcium, the G protein pathway, and a T-type calcium channel. 18033806_ZP proteins were detected in both the oocyte and the granulosa cells as early as the primordial follicle stage in the human. The detection of ZP proteins in the quiescent primordial follicle suggests that these proteins have been present since oogenesis. 18502569_may have a role in the development of primordial follicle before zona pellucida formation 18667750_induces acrosome reactions which are protein kinase-C, protein tyrosine kinase, T-type Ca2+ channels, and extracellular Ca2+ dependent 19004505_a significant decrease in acrosomal exocytosis mediated by both recombinant human ZP3 (p<0.005) and ZP4 (p<0.005) was observed in presence of the immune sera 19246320_The functional activity of human ZP3 resides in its C-terminal domain. 20237496_Observational study of gene-disease association. (HuGE Navigator) 22889493_observed sequence variations in exons of ZP3 gene in women with infertility of unknown origin who exhibit abnormal zona pellucida; sperm-ovum interactions appear relatively normal in these patients [CASE REPORTS] 24334245_Epididymal CRISP1 mediates sperm-zona pellucida binding through its interaction with ZP3. 28204536_The oocyte ZP3 expression was the main predictor of the fertilization capacity. 28646452_Mutations in ZP2 and ZP3 have dosage effects which can cause female infertility in humans. 28886344_Missense Mutation in ZP3 gene is associated with Empty Follicle Syndrome and Female Infertility. 30341457_The zona pellucida-3 (ZP3) protein plays a pivotal role in oocyte and gamete development. 30810869_Mutations in ZP1, ZP2, and ZP3 might affect the corresponding protein expression, secretion, and interaction, thus providing a mechanistic explanation for the phenotypes for female infertility. 32569527_Zona Pellucida Proteins, Fibrils, and Matrix. 32573113_Heterozygous mutations in ZP1 and ZP3 cause formation disorder of ZP and female infertility in human. 33140178_A novel mutation in ZP3 causes empty follicle syndrome and abnormal zona pellucida formation. 33604805_A novel homozygous variant in ZP2 causes abnormal zona pellucida formation and female infertility. 34736966_Novel expression of zona pellucida 3 protein in normal testis; potential functional implications. 34816529_A novel gene mutation in ZP3 loop region identified in patients with empty follicle syndrome. | ENSMUSG00000004948 | Zp3 | 79.541925 | 0.9299924 | -0.104709163 | 0.29107717 | 1.298994e-01 | 7.185363e-01 | No | Yes | 72.197796 | 13.547374 | 79.302744 | 14.690413 | ||
| ENSG00000188859 | 149297 | FAM78B | protein_coding | Q5VT40 | Reference proteome | hsa:149297; | 19064610_Observational study of gene-disease association. (HuGE Navigator) 19536175_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000060568 | Fam78b | 50.845589 | 0.8537097 | -0.228182486 | 0.39278550 | 3.492037e-01 | 5.545643e-01 | No | Yes | 48.564658 | 11.221297 | 57.763457 | 13.283887 | |||||
| ENSG00000189050 | 51136 | RNFT1 | protein_coding | Q5M7Z0 | FUNCTION: E3 ubiquitin-protein ligase that acts in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway, which targets misfolded proteins that accumulate in the endoplasmic reticulum (ER) for ubiquitination and subsequent proteasome-mediated degradation. Protects cells from ER stress-induced apoptosis. {ECO:0000269|PubMed:27485036}. | Alternative splicing;Endoplasmic reticulum;Membrane;Metal-binding;Reference proteome;Transferase;Transmembrane;Transmembrane helix;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:27485036}. | hsa:51136; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; metal ion binding [GO:0046872]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; positive regulation of ERAD pathway [GO:1904294]; protein autoubiquitination [GO:0051865] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000020521 | Rnft1 | 549.031931 | 1.0839059 | 0.116239509 | 0.11975415 | 9.348319e-01 | 3.336105e-01 | 6.938123e-01 | No | Yes | 565.304322 | 106.275473 | 527.518896 | 99.136172 | |
| ENSG00000196118 | 90835 | CFAP119 | protein_coding | A1A4V9 | Alternative splicing;Cell projection;Cilium;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Flagellum;Phosphoprotein;Reference proteome | hsa:90835; | acrosomal vesicle [GO:0001669]; cytoplasm [GO:0005737]; sperm principal piece [GO:0097228] | ENSMUSG00000057176 | Ccdc189 | 239.524959 | 0.9158314 | -0.126846091 | 0.17682469 | 5.059112e-01 | 4.769143e-01 | No | Yes | 204.755675 | 22.266628 | 231.506082 | 24.997266 | |||||
| ENSG00000196220 | 9901 | SRGAP3 | protein_coding | O43295 | FUNCTION: GTPase-activating protein for RAC1 and perhaps Cdc42, but not for RhoA small GTPase. May attenuate RAC1 signaling in neurons. {ECO:0000269|PubMed:12195014, ECO:0000269|PubMed:12447388}. | Alternative splicing;Chromosomal rearrangement;Coiled coil;GTPase activation;Phosphoprotein;Reference proteome;SH3 domain | hsa:9901; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; GTPase activator activity [GO:0005096]; negative regulation of cell migration [GO:0030336]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | 12195014_putative role in severe mental retardation 12736724_FNBP2, ARHGAP13, ARHGAP14 and ARHGAP4 constitute the FNBP2 family characterized by FCH, RhoGAP and SH3 domains. 16730001_Data suggest that MEGAP negatively regulates cell migration by perturbing the actin and microtubule cytoskeleton and by hindering the formation of focal complexes. 19433673_We found no association between SRGAP3/MEGAP haploinsufficiency and mental retardation. 19760623_Current evidence suggests that SRGAP3 is the major determinant of mental retardation in distal 3p deletions. 20035503_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23108406_conclude that srGAP3 has tumor suppressor-like activity in HMECs, likely through its activity as a negative regulator of Rac1 24300292_deletion of SRGAP3 provides the most convincing explanation for our patient's phenotype, and our observations lend further weight to a causative role of SRGAP3 haploinsufficiency in mental retardation. 24561795_Nuclear-localized srGAP3 interacts with Brg1. This interaction is mediated by the C-terminal of srGAP3 and the ATPase motif of Brg1. 25819436_A single PXXP motif in the C-terminal region of srGAP3 mediates binding to multiple SH3 domains. | ENSMUSG00000030257 | Srgap3 | 130.197311 | 2.1158155 | 1.081213826 | 0.27282387 | 1.567733e+01 | 7.511936e-05 | No | Yes | 153.709157 | 31.427157 | 77.995593 | 16.089772 | |||
| ENSG00000196338 | 54413 | NLGN3 | protein_coding | Q9NZ94 | FUNCTION: Cell surface protein involved in cell-cell-interactions via its interactions with neurexin family members. Plays a role in synapse function and synaptic signal transmission, and may mediate its effects by clustering other synaptic proteins. May promote the initial formation of synapses, but is not essential for this. May also play a role in glia-glia or glia-neuron interactions in the developing peripheral nervous system (By similarity). {ECO:0000250, ECO:0000269|PubMed:15620359}. | Alternative splicing;Asperger syndrome;Autism;Autism spectrum disorder;Cell adhesion;Cell junction;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Signal;Synapse;Transmembrane;Transmembrane helix | This gene encodes a member of a family of neuronal cell surface proteins. Members of this family may act as splice site-specific ligands for beta-neurexins and may be involved in the formation and remodeling of central nervous system synapses. Mutations in this gene may be associated with autism and Asperger syndrome. Multiple transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Oct 2009]. | hsa:54413; | asymmetric, glutamatergic, excitatory synapse [GO:0098985]; cell surface [GO:0009986]; endocytic vesicle [GO:0030139]; excitatory synapse [GO:0060076]; integral component of plasma membrane [GO:0005887]; integral component of postsynaptic membrane [GO:0099055]; integral component of postsynaptic specialization membrane [GO:0099060]; plasma membrane [GO:0005886]; presynapse [GO:0098793]; spanning component of membrane [GO:0089717]; symmetric, GABA-ergic, inhibitory synapse [GO:0098983]; synapse [GO:0045202]; cell adhesion molecule binding [GO:0050839]; neurexin family protein binding [GO:0042043]; scaffold protein binding [GO:0097110]; signaling receptor activity [GO:0038023]; adult behavior [GO:0030534]; axon extension [GO:0048675]; chemical synaptic transmission [GO:0007268]; inhibitory postsynaptic potential [GO:0060080]; learning [GO:0007612]; modulation of chemical synaptic transmission [GO:0050804]; neuron cell-cell adhesion [GO:0007158]; positive regulation of AMPA receptor activity [GO:2000969]; positive regulation of excitatory postsynaptic potential [GO:2000463]; positive regulation of synapse assembly [GO:0051965]; positive regulation of synaptic transmission, glutamatergic [GO:0051968]; postsynaptic membrane assembly [GO:0097104]; presynapse assembly [GO:0099054]; presynaptic membrane assembly [GO:0097105]; receptor-mediated endocytosis [GO:0006898]; regulation of respiratory gaseous exchange by nervous system process [GO:0002087]; rhythmic synaptic transmission [GO:0060024]; social behavior [GO:0035176]; synapse assembly [GO:0007416]; synapse organization [GO:0050808]; synaptic vesicle endocytosis [GO:0048488]; vocalization behavior [GO:0071625] | 12669065_report mutations in two X-linked genes encoding neuroligins NLGN3 and NLGN4 in siblings with autism-spectrum disorders 15274046_Observational study of genotype prevalence. (HuGE Navigator) 15389766_Observational study of genotype prevalence. (HuGE Navigator) 15622415_No structural variants were found in the NLGN3 gene when 96 unrelated patients with autism, 24 ADHD and 24 bipolar disorder patients were analyzed. 16077734_Neuroligin mutations most probably represent rare causes of autism; it is unlikely that the allelic variants in any of these genes would be major risk factors for autism. 16508939_Data indicate that coding mutations in neuroligin 3 are very rarely associated to autism spectrum disorders. 16648374_Splice variants of the NLGN3 gene are associated with autism. 17292328_Syntrophin-gamma2 (SNTG2) is a de novo binding partner of neuroligin 3, which correlates with autism-related mutations. 18189281_Observational study of gene-disease association. (HuGE Navigator) 18189281_no evidence for an involvement of NLGN3 and NLGN4X genetic variants with autism spectrum disorder on high functioning level 18628683_Observational study of gene-disease association. (HuGE Navigator) 19058789_Observational study of gene-disease association. (HuGE Navigator) 19406211_these data support the hypothesis that the autism-associated NL3 mutation affects information processing in neuronal networks by altering network architecture and synchrony 19645625_Observational study of gene-disease association. (HuGE Navigator) 20227402_further characterization of the R451C mutation in NLGN3;role in protein folding 21569590_study provides initial evidence that a common variant in NLGN3 gene may play a role in the etiology of ASDs among affected males in Chinese Han population. 22671294_Data from studies using cross-linking reagents suggest that neuroligins form dimers, including homodimers and, most notably, neuroligin 1/3 heteromers; autism-associated neuroligin mutant (neuroligin 3 R471C) forms heterodimers with neuroligin 1. 23431752_Neuroligins are adhesion proteins that bind to beta-neurexin to form functional synapses. 23468870_Lack of association between NLGN3, NLGN4, SHANK2 and SHANK3 gene variants and autism spectrum disorder in a Chinese population. 23851596_The present study explores, for the first time, the contribution of NLGN3 and NLGN4X genetic variants in Greek patients with autistic disorder. 24570023_Our data provided a further evidence for the involvement of NLGN3 and NLGN4X gene in the pathogenesis of autism in Chinese population. 25913192_The synaptic protein neuroligin-3 (NLGN3) was identified as the leading candidate mitogen, and soluble NLGN3 was sufficient and necessary to promote robust high-grade glioma cell proliferation. 27782075_No statistically significant haplotypes have been found associated to autism in the NLGN3 after logistic regression and permutation analysis. 27805570_e found that NLGN3 function at inhibitory synapses in rat CA1 depends on the presence of NLGN2 and identified a domain in the extracellular region that accounted for this functional difference between NLGN2 and 3 specifically at inhibitory synapses. 28948087_Our data suggest that these four previously described neuroligin mutations are not primary risk factors for autism. 28959975_high-grade gliomas growth depends on microenvironmental NLGN3, identify signalling cascades downstream of NLGN3 binding in glioma, and determine a therapeutically targetable mechanism of secretion 29503438_Wnt/beta-catenin signaling targets the trasncription of the autism-associated Neuroligin 3 gene. 29622757_the effects of rs1421589 within NRXN1, rs4844285 and rs11795613 within NLGN3, as well as rs5961397 within NLGX4X on Hirschsprung's disease phenotypes were also statistically significant. 29792861_NLGN3 protects retinal pigment epithelium (RPE) cells and retinal ganglion cells (RGCs) from H2O2. 31119867_We report gut symptoms in patients with the autism-associated R451C mutation encoding the neuroligin-3 protein. We show that many of the genes implicated in autism are expressed in mouse gut. The neuroligin-3 R451C mutation alters the enteric nervous system, causes gastrointestinal dysfunction, and disrupts gut microbe populations in mice. 31150649_NLGN3 promoted neuroblastoma cell proliferation and growth through activating PI3K/AKT pathway and providing a new target for neuroblastoma therapy 31184401_Our report of two missense variants affecting the normal localization of NLGN3 in a total of five affected individuals reinforces the involvement of the NLGN3 gene in a neurodevelopmental disorder characterized by ID and ASD. 31705895_Evidence for a Contribution of the Nlgn3/Cyfip1/Fmr1 Pathway in the Pathophysiology of Autism Spectrum Disorders. 34373757_Neuronal-driven glioma growth requires Galphai1 and Galphai3. | ENSMUSG00000031302 | Nlgn3 | 96.227724 | 0.9161608 | -0.126327283 | 0.26580370 | 2.244528e-01 | 6.356678e-01 | No | Yes | 81.511261 | 13.041267 | 86.691437 | 13.831920 | ||
| ENSG00000196498 | 9612 | NCOR2 | protein_coding | Q9Y618 | FUNCTION: Transcriptional corepressor (PubMed:20812024). Mediates the transcriptional repression activity of some nuclear receptors by promoting chromatin condensation, thus preventing access of the basal transcription. Isoform 1 and isoform 4 have different affinities for different nuclear receptors. Involved in the regulation BCL6-dependent of the germinal center (GC) reactions, mainly through the control of the GC B-cells proliferation and survival. Recruited by ZBTB7A to the androgen response elements/ARE on target genes, negatively regulates androgen receptor signaling and androgen-induced cell proliferation (PubMed:20812024). {ECO:0000269|PubMed:18212045, ECO:0000269|PubMed:20812024, ECO:0000269|PubMed:23911289}. | 3D-structure;Acetylation;Alternative splicing;Coiled coil;DNA-binding;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a nuclear receptor co-repressor that mediates transcriptional silencing of certain target genes. The encoded protein is a member of a family of thyroid hormone- and retinoic acid receptor-associated co-repressors. This protein acts as part of a multisubunit complex which includes histone deacetylases to modify chromatin structure that prevents basal transcriptional activity of target genes. Aberrant expression of this gene is associated with certain cancers. Alternate splicing results in multiple transcript variants encoding different isoforms.[provided by RefSeq, Apr 2011]. | hsa:9612; | chromatin [GO:0000785]; membrane [GO:0016020]; nuclear body [GO:0016604]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; glucocorticoid receptor binding [GO:0035259]; histone deacetylase binding [GO:0042826]; Notch binding [GO:0005112]; protein N-terminus binding [GO:0047485]; protein-containing complex binding [GO:0044877]; retinoid X receptor binding [GO:0046965]; transcription corepressor activity [GO:0003714]; estrous cycle [GO:0044849]; lactation [GO:0007595]; negative regulation of androgen receptor signaling pathway [GO:0060766]; negative regulation of production of miRNAs involved in gene silencing by miRNA [GO:1903799]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of cellular ketone metabolic process [GO:0010565]; regulation of transcription by RNA polymerase II [GO:0006357]; response to estradiol [GO:0032355]; response to organonitrogen compound [GO:0010243] | 11929748_the effect of STAT5b-RARalpha on the activity of myeloid transcription factors including STAT3, and STAT5 as well as its molecular interactions with the nuclear receptor corepressor, SMRT, and nuclear receptor coactivator, TRAM-1. 11929749_Sstable binding of the Stat5-RARalpha fusion protein to corepressor SMRT is accompanied by an impaired response to differentiation signals in hematopoietic cells 11972046_Interactions that determine the assembly of a retinoid X receptor/corepressor complex 12139968_The silencing mediator of retinoic acid and thyroid hormone receptors can interact with the aryl hydrocarbon (Ah) receptor but fails to repress Ah receptor-dependent gene expression. 12388540_SMRT has a role as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element 12441355_a significant role of SMRT in modulating androgen receptor transcriptional activity 12554770_Differing transcriptional properties appear to reflect the differing abilities of the three RAR isotypes to interact with the SMRT corepressor protein. 12771131_SMRT and DAX-1 repress agonist-dependent activity of both androgen and progesterone receptors 12840002_SANT motif interprets the histone code and promotes histone deacetylation 14690607_Data describe a 17 residue fragment from SMRT that binds to the BCL6 BTB domain, and report the crystal structure of the complex to 2.2 angstroms. 14715875_differential recruitment of steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors by estrogen receptor-alpha and beta in breast cancer may be central to the response of the tumor to endocrine treatment 15300237_Elevated SMRT levels are common in prostate cancer cells, resulting in suppression of target genes associated with antiproliferative action. 15319284_SMRTbeta expression may influence the binding and transcriptional capacities of nuclear receptors in tumor cells (SMRTbeta) 15632172_differential mRNA splicing of SMRT serves to customize corepressor function in different cells, allowing the transcriptional properties of nuclear receptors to be adapted to different contexts 15635693_No significant allelic/genotypic association between any of the five mutations in SMRT/N-CoR2 and bipolar phenotype and the CAG repeat did not demonstrate allelic instability. 15635693_Observational study of gene-disease association. (HuGE Navigator) 15713534_the subnuclear positioning of SMRT is influenced by the ligand-bound ERalpha, and this activity is dependent on the ratio of the co-expressed ERalpha and SMRT 15802375_N-CoR and SMRT play an active role in preventing tamoxifen from stimulating proliferation in breast cancer cells through repression of a subset of target genes involved in ERalpha function and cell proliferation 15843525_TRAC-1 (T cell RING protein identified in activation screen)is the first E3 ubiquitin ligase that serves a positive regulatory role in T cell activation. 16373395_SMRT and N-CoR corepressors are involved in transcriptional regulation by both agonist- and antagonist-bound AR and regulate the magnitude of hormone response, at least in part, by competing with coactivators. 16453284_Observational study of gene-disease association. (HuGE Navigator) 16480812_The aryl hydrocarbon receptor activates the retinoic acid receptoralpha through SMRT antagonism 16712523_This study shows that SMRT/NCoR-HDAC3 complex is a cofactor of CNOT2-mediated repression and suggest that transcriptional regulation by the Ccr4-Not complex involves regulation of chromatin modification. 17383980_expression of the silencing mediator of retinoid and thyroid receptor (SMRT) & histone deacetylase4 (HDAC4) enhances formation of Bach2 foci in nuclear matrix. SMRT mediates HDAC4 binding to Bach2, & HDAC4 facilitates retention of Bach2 in the foci. 17902051_NCOR2/SMRT is associated with poor patient outcome in breast cancer 17991421_These results would shed new insights for the molecular mechanisms of PML-RARalpha-associated leukemogenesis. 18052923_both SMRT and N-CoR are limited in cells and knocking down either of them results in co-repressor-free TR and consequently de-repression of TR target genes 18362166_Vitamin D receptor (VDR) with the retinoid X receptor (RXR) recruits NCoR and SMRT strictly in a VDR agonist-dependent manner. 18487509_CD40 signaling rapidly disrupts the ability of BCL6 to recruit the SMRT corepressor complex by excluding it from the nucleus, leading to histone acetylation, RNA polymerase II processivity, and activation of BCL6 target genes 18546531_Tamoxifen treatment of breast cancer cells reduced the expression of ER-alpha and increased the expression of SMRT. 18632669_SMRT and NCoR have important roles in the regulation of beta-catenin-TCF4-mediated gene transcription 18660489_Observational study of gene-disease association. (HuGE Navigator) 18838553_SMRT protein stability is regulated by Pin1 and Cdk2. 18852122_These findings show that AR antagonists can enhance corepressor recruitment by stabilizing a distinct antagonist conformation of the AR coactivator/corepressor binding site. 18950845_Observational study of gene-disease association. (HuGE Navigator) 19098224_Results indicate that the SMRT corepressor is directly involved in the vitamin D receptor-mediated repression in vivo via an ID1-specific interaction with the VDR. 19183483_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19223455_Activated MEK signaling cascade inhibits functional recruitment of corepressor SMRT to cyproterone acetate-bound AR in prostate cancer cells. 19417136_SMRT function restoration induces JAG2 down-regulation as well as multipe myeloma apoptosis. 19432593_SMRT decreased with oestradiol treatment in human skeletal muscle cells 19730683_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 19904269_Both NCOR2 and CITED2 mRNA levels were associated with MFS, that is, tumour aggressiveness, independently of traditional prognostic factors. 19934400_Observational study of gene-disease association. (HuGE Navigator) 20003447_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20392877_These data link the SMRT corepressor directly with SRC family coactivators in positive regulation of ERalpha-dependent gene expression. 20555024_Aberrant expression and modification of SMRT might be involved in the pathogenesis of tumoral cortisol resistance. 20965228_Data report that SMRT interacts with itself to form a protein dimer, and that Erk2, a mitogen-activated protein (MAP) kinase, disrupts this SMRT self-dimerization in vitro and in vivo. 21083371_This study independently verifies the influence of NCOR2 and IDH1 on HIV transmission, and its findings suggest that variation in these genes affects susceptibility to HIV infection in exposed individuals. 21131350_Aberrant corepressor interactions implicated in PML-RAR(alpha) and PLZF-RAR(alpha) leukemogenesis reflect an altered recruitment and release of specific NCoR and SMRT splice variants. 21143702_We immunohistochemically analyzed the expression of NCOR1 and 2 as well as HDAC1, 2, and 3 on a tissue microarray comprising tumor samples from 283 astrocytic gliomas 21240272_The crystal structure of the tetrameric oligomerization domain of TBL1, which interacts with both SMRT and GPS2, and the NMR structure of the interface complex between GPS2 and SMRT. 21518914_data strongly support a model in which EBNA2 association with NCoR-deficient RBPJ enhances transcription and EBNALP dismisses NCoR and RBPJ repressive complexes from enhancers 22185585_An equilibrium dissociation constant obtained for SMRT in the presence and absence of rifampicin indicates that the ligand does not enhance the affinity of the pregnane X receptor or its corepressor. 22230954_the structure of a complex between an HDAC and a co-repressor, namely, human HDAC3 with the deacetylase activation domain (DAD) from the human SMRT co-repressor (also known as NCOR2) 22292036_miR-16 targets SMRT and modulates NF-kappaB-regulated transactivation of the IL-8 gene. 22337871_Regulated HDAC3 degradation serves as a buffering mechanism to protect independent formation of N-CoR and SMRT corepressor complexes. 22576662_Thyroid hormone receptors induce TRAIL expression, and TRAIL thus synthesized acts in concert with simultaneously synthesized Bcl-xL to promote metastasis 22695118_altered recruitment and loss of corepressors SMRT/NCoR may provide a mechanism that changes the response of AR function to ligands and contributes to the progression of the advanced stages of human prostate cancer. 22944139_Corepressor molecules NCoR and SMRT are present at 1,25(OH)2D3 activated gene enhancers 23015261_There was no association between SMRT expression and overall survival for patients, regardless of whether they received tamoxifen 23055525_SMRT and GR act in a consistent manner with steroid hormones 23117886_Our findings highlight a novel splice variant of NCOR2 as a candidate biomarker in breast cancer that not only predicts tamoxifen response but may be targeted to overcome tamoxifen resistance 23221346_expression of the transcriptional corepressor complex subunits GPS2 and SMRT was significantly reduced in obese adipose tissue, inversely correlated to inflammatory status 23225884_Both MEK1 and SMRT bind to the c-Fos promoter and regulate its transcription; SMRT knockdown results in early-phase stimulation followed by a late-phase inhibition of T cell activation. 23562850_SMRT may be recruited in the SXR-cofactor complex even in the presence of ligand; SMRTmay be involved not only in SXR-mediated suppression without ligand, but also in ligand-activated transcription to suppress the overactivation of transcription 23690919_SMRT is responsible for basal repression of Wip1,a phosphatase that de-phosphorylates and inactivates Chk2, thus affecting a feedback loop responsible for licensing the correct timing of Chk2 activation and the proper execution of the DNA repair process. 23861398_a model in which TNFalpha-induced beta-TrCP1 accumulation promotes SMRT degradation and the subsequent induction of proinflammatory gene expression. 24268649_Phosphorylation of the CK2 site on SMRT significantly increased affinity for SHARP. 24449765_model where p53 binding to the SMRT deacetylase activation domain (DAD) limits HDAC3 interaction with this coregulator, thereby facilitating SMRT coactivation of p53-dependent gene expression 24971610_SMRT enhances cell growth of estrogen receptor-alpha-positive breast cancer cells by promotion of cell cycle progression and inhibition of apoptosis. 25821969_Significant methylation changes in the SLC23A2 and NCOR2 regulatory regions. 25953768_SNPs in Notch pathway genes may be predictors of cutaneous melanoma disease-specific survival. 26055705_A repeated peptide motif present in both SMRT and NCoR is sufficient to mediate specific interaction, with micromolar affinity, with all the Class IIa Histone Deacetylases (HDACs 4, 5, 7, and 9). 26161557_Molecular basis for the specific interactions between HDAC4 and the SMRT corepressor. 26836265_this study identifies NCOR2 as a new gene for FVC, indicating the importance of further research into the role of vitamin A intake/supplementation and its interactions with related genes in the regulation of FVC. 27239967_IL-6-mediated AR antagonism induced by cypermethrin is related to repress the recruitment of co-regulators SRC-1 and SMRT to the AR in a ligand-independent manner 28396297_The mechanism of IL-6-induced AR activation is mediated through enhancing AR-SRC-1 interaction and inhibiting AR-SMRT interaction. 29262348_Cellular Differentiation of Human Monocytes Is Regulated by Time-Dependent Interleukin-4 Signaling and the Transcriptional Regulator NCOR2. 29420220_These findings demonstrate that BQ323636.1(Splice Variant to NCOR2) can be a reliable biomarker to predict tamoxifen resistance in patients with ER-positive breast cancer 29470550_The pregnane X receptor (PXR) and the nuclear receptor corepressor 2 (NCoR2) modulate cell growth in head and neck squamous cell carcinoma 29764865_Direct epigenetic repression of the transcription-repressive regulators NCOR2 and ZBTB7A by the histone reader protein HP1gamma leads to activation of protumorigenic genes in lung adenocarcinoma. 30157580_this report tried to address the molecular basis for the direct interaction between CSL and SMRT. 30321390_SMRT binding does not activate the cryptic deacetylase activity of HDAC4; class IIa HDACs and the SMRT-HDAC3 complex are coordinated during gene regulation 30680848_MIR22HG inhibited HCC progression in part through the miR-10a-5p/NCOR2 signaling. 31208445_Data suggest that the nuclear receptor co-repressor 1/2 protein NCoR-1/NCoR-2 paralogs have been subject to a mix of shared and distinct selective pressures, resulting in the pattern of divergent and convergent alternative-splicing observed in extant species. 32940102_Circular RNA circ-NCOR2 accelerates papillary thyroid cancer progression by sponging miR-516a-5p to upregulate metastasis-associated protein 2 expression. 33109580_Thyroid Hormones, Silencing Mediator for Retinoid and Thyroid Receptors and Prognosis in Primary Breast Cancer. 33742141_Recurrent novel HMGA2-NCOR2 fusions characterize a subset of keratin-positive giant cell-rich soft tissue tumors. 34709749_KPNA1 regulates nuclear import of NCOR2 splice variant BQ323636.1 to confer tamoxifen resistance in breast cancer. 34864816_Low NCOR2 levels in multiple myeloma patients drive multidrug resistance via MYC upregulation. 34910907_Reduced NCOR2 expression accelerates androgen deprivation therapy failure in prostate cancer. 35039565_Methylation statuses of NCOR2, PARK2, and ZSCAN12 signify densities of tumor-infiltrating lymphocytes in gastric carcinoma. 35181586_Recurrent Fusion of the Genes for High-mobility Group AT-hook 2 (HMGA2) and Nuclear Receptor Co-repressor 2 (NCOR2) in Osteoclastic Giant Cell-rich Tumors of Bone. | ENSMUSG00000029478 | Ncor2 | 2370.184005 | 0.8374449 | -0.255933856 | 0.07301771 | 1.223747e+01 | 4.683932e-04 | 2.203441e-02 | No | Yes | 2160.846186 | 297.429233 | 2524.280681 | 347.405084 | |
| ENSG00000196511 | 27010 | TPK1 | protein_coding | Q9H3S4 | FUNCTION: Catalyzes the phosphorylation of thiamine to thiamine pyrophosphate. Can also catalyze the phosphorylation of pyrithiamine to pyrithiamine pyrophosphate. {ECO:0000269|PubMed:11342111}. | 3D-structure;ATP-binding;Alternative splicing;Disease variant;Kinase;Nucleotide-binding;Reference proteome;Transferase | PATHWAY: Cofactor biosynthesis; thiamine diphosphate biosynthesis; thiamine diphosphate from thiamine: step 1/1. | The protein encoded by this gene functions as a homodimer and catalyzes the conversion of thiamine to thiamine pyrophosphate, a cofactor for some enzymes of the glycolytic and energy production pathways. Defects in this gene are a cause of thiamine metabolism dysfunction syndrome-5. [provided by RefSeq, Apr 2017]. | hsa:27010; | cytosol [GO:0005829]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; kinase activity [GO:0016301]; thiamine binding [GO:0030975]; thiamine diphosphokinase activity [GO:0004788]; thiamine diphosphate biosynthetic process [GO:0009229]; thiamine metabolic process [GO:0006772] | 10567383_This is the first report on the primary sequence for mammalian thiamin pyrophosphokinase, a protein that catalyzes the pyrophosphorylation of thiamin with adenosine 5'-triphosphate to form thiamin pyrophosphate. 16262001_findings indicate the importance of the Sp1 transcription factor cis-element in regulating the human thiamin pyrophosphokinase gene (hTPK1) gene expression 17295612_Genomic variations in either the fetal or maternal TPK1 gene could contribute to variability of birth weight in normal humans. 17295612_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22152682_Mutation analysis of TPK1 uncovered three missense, one splice-site, and one frameshift mutation resulting in decreased TPK protein levels 23642734_These findings demonstrate that the genes involved in dictating thiamine homeostasis, such as SLC19A2, SLC25A19 and TPK-1, were significantly up-regulated in clinical tissues and breast cancer cell lines. 30483896_any disease mutants are directly or indirectly involved in thiamine binding. Thus, our study provided a novel rationale for thiamine supplementation, so far the major therapeutic intervention in TPK deficiency. 31964553_TPK1 as a predictive marker for the anti-tumour effects of simvastatin in gastric cancer. 32361878_Whole Exome Sequencing Identifies a Novel Mutation of TPK1 in a Chinese Family with Recurrent Ataxia. 33031988_Movement disorders associated with thiamine pyrophosphokinase deficiency: Intrafamilial variability in the phenotype. 34244791_The conserved Tpk1 regulates non-homologous end joining double-strand break repair by phosphorylation of Nej1, a homolog of the human XLF. | ENSMUSG00000029735 | Tpk1 | 35.601993 | 0.9679840 | -0.046944884 | 0.48328528 | 9.602965e-03 | 9.219365e-01 | No | Yes | 31.819089 | 9.351898 | 31.268687 | 9.232922 | |
| ENSG00000196628 | 6925 | TCF4 | protein_coding | P15884 | FUNCTION: Transcription factor that binds to the immunoglobulin enhancer Mu-E5/KE5-motif. Involved in the initiation of neuronal differentiation. Activates transcription by binding to the E box (5'-CANNTG-3'). Binds to the E-box present in the somatostatin receptor 2 initiator element (SSTR2-INR) to activate transcription (By similarity). Preferentially binds to either 5'-ACANNTGT-3' or 5'-CCANNTGG-3'. {ECO:0000250}. | 3D-structure;Activator;Alternative splicing;Corneal dystrophy;DNA-binding;Differentiation;Disease variant;Epilepsy;Mental retardation;Neurogenesis;Nucleus;Phosphoprotein;Primary microcephaly;Reference proteome;Transcription;Transcription regulation | This gene encodes transcription factor 4, a basic helix-loop-helix transcription factor. The encoded protein recognizes an Ephrussi-box ('E-box') binding site ('CANNTG') - a motif first identified in immunoglobulin enhancers. This gene is broadly expressed, and may play an important role in nervous system development. Defects in this gene are a cause of Pitt-Hopkins syndrome. In addition, an intronic CTG repeat normally numbering 10-37 repeat units can expand to >50 repeat units and cause Fuchs endothelial corneal dystrophy. Multiple alternatively spliced transcript variants that encode different proteins have been described. [provided by RefSeq, Jul 2016]. | hsa:6925; | beta-catenin-TCF complex [GO:1990907]; beta-catenin-TCF7L2 complex [GO:0070369]; chromatin [GO:0000785]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; beta-catenin binding [GO:0008013]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; E-box binding [GO:0070888]; identical protein binding [GO:0042802]; protein C-terminus binding [GO:0008022]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; TFIIB-class transcription factor binding [GO:0001093]; cell differentiation [GO:0030154]; nervous system development [GO:0007399]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein-DNA complex assembly [GO:0065004]; regulation of transcription by RNA polymerase II [GO:0006357] | 15351717_dHAND/E-protein (E2A, ME2, and ALF1) heterodimers have distinct DNA binding specificities 16126178_Competitive RT-PCR-based promoter activity assay showed that over-expression of ITF2B protein inhibited the expression of IL-2Ralpha gene in Jurkat cells in an NRE-dependent manner 17478476_haploinsufficiency of TCF4 causes PHS and suggest that D. rerio is a valuable model to study the molecular pathogenesis of Pitt-Hopkins syndrome and the role of TCF4 in brain development 18222743_Interstitial deletion involving TCF4 is associated with severe developmental delay and multiple abnormalities. 18371301_Protein sequence alignment of the closely related bHLH transcription factors ITF-2B, HeLa E box protein (HITF4), and the E2A proteins E12 and E47 revealed the presence of a highly conserved protein domain. 18627065_Gene disruption of TCF4 is associated with mental retardation but not always associated with Pitt-Hopkins syndrome. 18635522_based on results, it is proposed that the observed frequent epigenetic-mediated TCF4 silencing plays a role in tumor formation and progression 18781613_Nine novel deletion mutations in TCF4 in Pitt-Hopkins Syndrome are described. 18792017_findings suggest that the concurrent action of Spi-B and E2-2 controls the development of progenitor cells into the plasmacytoid dendritic cell lineage 18854153_These results identify E2-2 as a specific transcriptional regulator of the plasmacytoid dendritic cells lineage in mice and humans and reveal a key function of E proteins in the innate immune system. 19235238_Studies delineate an underdiagnosed mental retardation syndrome, highlighting TCF4 function during development and facilitating diagnosis within the first year of life. 19394332_ITF-2B is a tumor suppressor that has an important function at the adenoma to carcinoma transition. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19571808_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19635457_This data support the notion that the ITF2 gene on chromosome 18q is a tumor suppressor gene. 19938247_a genotype-phenotype correlation of increased seizure activity with TCF4 missense mutations was proposed in patients with Pitt-Hopkins syndrome. 20205897_The diagnosis of Pitt-Hopkins syndrome, an underdiagnosed cause of mental retardation, was based on clinical and genetic findings. Searching for TCF4 mutations is highly recommended when others overlapping syndromes was excluded. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20421335_These data suggest that TCF4, NRXN1, and CNTNAP2 may participate in a biological pathway that is altered in patients with schizophrenia and other neuropsychiatric disorders. 20585880_Inactivation of TCF4 by promoter methylation is associated with the early stage of gastric carcinoma progression. 20603605_Data suggest that that ITF2 is one of the CXCR4 targets, which is involved in CXCR4-dependent tumor growth and invasion of breast cancer cells. 20673877_Observational study of gene-disease association. (HuGE Navigator) 20673877_This study confirmed that common risk factors in the major histocompatibility complex region and TCF4 gene are associated with schizophrenia in Han Chinese. 20825314_Genetic variation in TCF4 contributes to the development of Fuch's corneal dystrophy. 20825314_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21037240_Observational study of gene-disease association. (HuGE Navigator) 21228604_VDM function is influenced by the TCF4 gene in schizophrenia patients. However, the elevated risk for schizophrenia is not conferred by TCF4-mediated VDM impairment. 21245398_The authors report the first independent replication of rs613872 conferring risk of late-onset Fuchs endothelial dystrophy 21533127_study presents evidence to support the role of the intronic TCF4 single nucleotide polymorphism rs613872 in late-onset FECD through both association and linkage studies 21543597_Sensorimotor gating is modulated by TCF4 genotype, indicating an influential role of TCF4 gene variations in the development of early information-processing deficits in schizophrenia. 21659310_Polymorphisms within TCF4, a gene which has been implicated in Fuch's corneal dystrophy susceptibility among Europeans, was also found to be strongly associated in Chinese 21671075_TCF4 (+/+) individuals were only moderately developmentally delayed while TCF4 (+/-) individuals failed to reach developmental milestones beyond those typically acquired by 12 months of age. 21722264_these findings suggest that FAM96B acts as a regulator of E2-2 through the control of its protein expression. 21789225_Usage of numerous 5' exons of the human TCF4 gene potentially yields in TCF4 protein isoforms with 18 different N-termini. 21791550_Common variants at TCF4 show genome-wide significant association with schizophrenia. 21812098_Association between schizophrenia and TCF4 is not mediated by a relatively common non-synonymous variant, or by a variant that alters mRNA expression as measured in adult human brain. 22045651_No obvious difference was observed between patients harboring truncating, missense mutations, or deletions, further supporting TCF4 haploinsufficiency as the molecular mechanism underlying Pitt-Hopkins syndrome. 22146553_We were unable to detect an association between TCF4 rs613872 genotype and the variation in corneal endothelial cell density or variation in cell morphology in a healthy young adult population. 22234156_variation in TCF4, genes are associated with Fuchs' endothelial dystrophy in Caucasian Australian cases 22383159_report on the screening of the TCF4 and MEF2C genes in a cohort of 81 classical, atypical, and incomplete atypical Rett syndrome (RTT) patients; results suggest that these genes are not commonly associated with RTT 22451930_Finding suggests that sensory gating is modulated by an interaction of TCF4 genotype with smoking, and both factors may play a role in early information processing deficits also in schizophrenia. 22460224_different Pitt-Hopkins syndrome-associated mutations impair the functions of TCF4 by diverse mechanisms and to a varying extent, possibly contributing to the phenotypic variability of Pitt-Hopkins syndrome patients. 22777675_TCF4 may modulate the expression of NRXN1 and CNTNAP2 thereby defining a regulatory network in Pitt-Hopkins syndrome. 22821403_GPR35 shows associations in both ulcerative colitis (UC) and primary sclerosing cholangitis (PSC), whereas TCF4 represents a PSC risk locus not associated with UC. Both loci may represent previously unexplored aspects of PSC pathogenesis. 22832956_Common TCF4 variants are involved in psychosis pathology, probably related to abnormal neurodevelopment. 22998502_We have shown that SNP rs613872 in the TCF4 gene is highly associated with Fuchs endothelial corneal dystrophy. 23110055_Association of FECD grade with TCF4 was highly significant (OR= 6.01 at rs613872; p = 4.8x10(-25)), and remained significant when adjusted for changes in central corneal thickness. 23129290_The schizophrenia-associated single nucleotide polymorphisms are in linkage disequilibrium with common variants within putative DNA regulatory elements, suggesting that regulation of expression may underlie association with schizophrenia. 23165966_A mosaic TCF4 point mutation can result in a significantly milder phenotype. 23185296_The TGC trinucleotide repeat expansion in TCF4 is strongly associated with Fuchs endothelial corneal dystrophy. 23249814_A single nucleotide polymorphism in TCF4 gene is associated with cognitive function in schizophrenic patients and healthy controls. 23671559_Regulation of TCF4-mediated BCL2 gene expression by BMI1 is universal. 23758498_Our results showed that there was no significant association between any of five reported SNPs of TCF4 and PTPRG genes and the occurrence of Fuchs' endothelial dystrophy; only rs7640737 in PTPRG showed an increased risk for corneal dystrophy. 24027308_mRNA isoforms of TCF4 is associated with Epstein-Barr virus lytic trans-activator BZLF1. 24058414_Knockdown of human TCF4 affects multiple signaling pathways involved in cell differentiation and survival in addition to a subset of clinically important mental retardation genes. 24094747_That AGBL1 interacts biochemically with the FCD-associated protein TCF4. 24210665_There is little genetic variation in non-coding region of TCF4 in schizophrenic patients. 24255041_That was the first independent replication of the expanded CTG 18.1 allele conferring significant risk for FECD (>30-fold increase). 24275585_The rs9960767 (C) of TCF4 appears to be associated with neurocognitive deficits in the Reasoning/Problem-Solving cognitive domain, in patients with episode psychosis. 24339136_results suggest that a schizophrenia risk variant in TCF4 is associated with neurophysiologic traits thought to index attention and working memory abnormalities in psychotic disorders. 24413739_Among the few schizophrenia-risk genes that have been consistently replicated is TCF4. This review integrates the animal and the human perspective. [review] 24594265_TCF4 is an important regulator of neurodevelopment and epithelial-mesenchymal transition. 24846398_In human colorectal cancer cell lines and tissue samples, ITF2 appears to prevent activation of the beta-catenin-TCF4 complex and transcription of its gene targets. 24862328_These findings indicate that beta-catenin/TCF-4 is an important pathway for restricted HIV-1 replication in monocytes and plays a significant role in potentiating HIV-1 replication as monocytes differentiate into macrophages. 25150259_High TCF4 expression is associated with acute myeloid leukemia progression. 25168903_Complete sequencing of the TCF4 genomic region revealed no single causative variant for Fuchs' endothelial corneal dystrophy (FECD). The intronic trinucleotide repeat expansion within TCF4 continues to be more strongly associated with FECD 25217366_This study showed that no effect of schizophrenia risk genes TGF4 on macroscopic brain structure. 25298419_Transethnic replication of the association between the CTG18.1 repeat expansion in the TCF4 gene and FECD suggests it is a common, causal variant shared in Eurasian populations conferring significant risk for the development of FECD. 25299301_TCF4 rs613872 is strongly associated with FCD in Caucasians. 25342617_TCF4 poses a major contributor to FECD manifestation globally, with a significant association of rs17089887 and CTG18.1 allele in the Indian population. 26010163_No significant association (P < 0.05) was found between TCF4 rare sequence variants and schizophrenia 26087656_This meta-analysis suggested a genetic association between four TCF4 polymorphisms (rs613872, rs2286812, rs17595731, and rs9954153) and the risk of Fuchs' endothelial dystrophy. 26200491_A monoallelic expansion of CTG18.1 contributes to increased disease severity and is causal at (CTG)n>103, whereas a biallelic expansion is sufficient to be causal for FCD at (CTG)n>40. 26218914_These findings show for the first time in a Japanese population the association of the TNR expansion in TCF4 with FECD. 26280645_expanded TGC allele with more than 50 TGC repeats in intron 2 and the described risk allele G of the polymorphism rs613872 in intron 3 of the TCF4 gene appear as an association to FECD. The chance to be affected by FECD is up to 30 times higher. With molecular genetics also donors with clinically unknown FECD may be detected. 26343600_Results suggested that the TCF4 rs2958182 variant may play an important role in the susceptibility to schizophrenia 26401622_The TCF4 triplet repeat expansion resulted in a more severe form of Fuchs endothelial corneal dystrophy , with clinical and surgical therapeutic implications. 26451375_The data suggest that changes in the transcript level containing constitutive TCF4 exon encoding the amino-terminal part of the protein seem not to contribute to disease pathogenesis. 26622166_ZEB1 mutations and TCF4 rs613872 changes are associated with late onset Fuchs endothelial corneal dystrophy in patients from Northern India. 26884349_Studied promoter methylation of ITF2 and APC and associated microsatellite instability in two case-case studies nested in colorectal cancer. 27103199_We found that the genotype 'AG' of rs9320010 and 'GA' of rs7235757 decreased SCZ risk. In the genetic model analysis, we also observed that the allele 'A' of rs9320010 and 'G' of rs7235757 were inversely related with the risk of SCZ in the dominant model. Our study indicated that rs9320010, rs7235757, and rs1452787 were prominently associated with SCZ. 27121161_Analysis of SLC4A11, ZEB1, LOXHD1, COL8A2 and TCF4 gene sequences in a multi-generational family with late-onset Fuchs corneal dystrophy found no evidence for found polymorophisms causing the disease in this specific pedigree. 27132474_We suggest that screening for mutations in TCF4 could be considered in the investigation of non-syndromic intellectual disability. 27179618_Although validation in additional patients is required, the findings suggest that the dysmorphic features and severe intellectual disability characteristic of PTHS are partially rescued by overexpression of those short TCF4 transcripts encoding a nuclear localization signal, a transcription activation domain, and the basic helix-loop-helix domain. 27305091_Among the TCF4 variants, rs12966547 and rs8766 were significantly associated with earlier schizophrenia age of onset. 27374225_Results show that MLL-AF9 reduces Id2 and increases E2-2 expression to drive and sustain leukemia stem cell potential in MLL-rearranged acute myeloid leukemia (AML). Low expression of Id2 or of an Id2 gene signature is associated with poor prognosis in not only MLL-rearranged but also t(8;21) AML patients. 27440233_Altered DNA methylation in TCF4 involves in the etiology of Bipolar disorder and Major Depressive disorder . 27485769_Examination of X-ray structures of the closely related TCF3 and USF1 bound to DNA suggests TCF3 can undergo a conformational shift to preferentially bind to 5hmC while the USF1 basic region is bulkier and rigid precluding a conformation shift to bind 5hmC. These results greatly expand the regulatory DNA sequence landscape bound by TCF4. 27542264_TCF4 knockdown promoted HepG-2 cell differentiation and inhibited tumor formation, and TCF4 could be the potential downstream target for clopidogrel therapy 27689884_Expression of TCF4 in a neural progenitor cell line derived from the developing human cerebral cortex was reduced using RNA interference. Genes that were differentially expressed following TCF4 knockdown were highly enriched for involvement in the cell cycle. There was a nonsignificant trend for genetic association between the differentially expressed gene set and schizophrenia. 27755191_We demonstrate that expansion of the CTG18.1 trinucleotide repeat in TCF4 is associated with a higher risk of corneal transplantation at a younger age, assessed for the first time in a multiracial population sample with Fuchs corneal dystrophy from the United States. 27846392_Results identify TCF4 as a crucial transcriptional regulator required for maintenance of blastic plasmacytoid dendritic cell neoplasm. 28051067_Hippo pathway transcription factor TEAD4 directly associates with the Wnt pathway transcription factor TCF4 via their DNA-binding domains, forming a complex on target genes. VGLL4 binds to this TEAD4-TCF4 complex to inhibit transactivation of both TCF4 and TEAD4. 28118661_Corneal endothelium from FECD patients harbors a unique signature of mis-splicing events due to CTG TNR expansion in the TCF4 gene, consistent with the hypothesis that RNA toxicity contributes to the pathogenesis of FECD. 28341444_Expression of TCF4 at the mRNA and protein level may be significant in the etiology of recurrent depressive disorder and it is not dependent on sex and age. 28574827_Data show that silencing of immunoglobulin transcription factor 2 (ITF-2) by siRNA significantly enhanced susceptibility to the MEK inhibitor selumetinib (AZD6244) in resistant cells. 28608272_The CTG18.1 repeat expansion may reduce gene expression of TCF4 and ZEB1, suggesting that a mechanism triggering a loss of function may contribute to FECD. 28631899_Array-comparative genomic hybridization confirmed a de novo paternal deletion of the 15q11.2q13 region and exome sequencing identified a second mutational event in both girls, which was a novel variant c.145+1G>A affecting a TCF4 canonical splicing site inherited from the mosaic mother 28790108_we found that CypA binds beta-catenin and is recruited to Wnt target gene promoters. By increasing the interaction between beta-catenin and TCF4, CypA enhances transcriptional activity. Our results demonstrate that CypA enhances GIC stemness, self-renewal, and radioresistance through Wnt/beta-catenin signaling 28807867_A frameshift-causing partial TCF4 gene deletion was identified in an adult patient with mild ID and nonspecific facial dysmorphisms but without the typical features of Pitt-Hopkins syndrome. A nonsense variant within exon 8 was identified in a child presenting with a severe phenotype largely mimicking PTHS. 28832669_repeat expansion showed stronger association than the most significantly associated SNP, rs613872, in TCF4, with the disease in the Australian cohort 28886202_Our work suggests that DM1 patients are at risk for Fuchs' endothelial corneal dystrophy (FECD). DMPK mutations contribute to the genetic burden of FECD but are uncommon. We establish a connection between two repeat expansion disorders converging upon RNA-MBNL1 foci and FECD. 28921696_In conclusion, this study provides genetic and some preliminary functional evidence to support the view that the TCF4 (NM_001243232) p.Pro29Thr mutation causes familial SAK. 29033371_The formation of the beta-catenin/TCF4 complex was disrupted by HI-B1 due to the direct interaction of HI-B1 with beta-catenin. Colon cancer patient-derived xenograft (PDX) studies showed that a tumor with higher levels of beta-catenin expression was more sensitive to HI-B1 treatment, compared to a tumor with lower expression levels of beta-catenin 29044056_rs613872, rs17595731, and CTG repeat expansions in intronic region of TCF4 are associated with increased risk of sporadic late-onset FECD in the Indian cohort studied 29105523_Our results showed that the mRNA level of TCF4 may be associated with schizophrenia, its psychopathology, IQ and cognitive impairments in an Iranian group of patients with schizophrenia. 29196769_Black patients with Fuchs' dystrophy were less likely than white patients to demonstrate CTG18.1 allele expansion. 29228394_TCF4 binding sites are found in a large number of neuronal genes that include many genetic risk factors for common neurodevelopmental disorders. 29666142_these results from tumor xenograft modeling depict a link between altered TCF4 expression and breast cancer chemoresistance. 29677349_Expanded CTG.CAG repeats in the context of the third intron of TCF4 are transcribed and translated via non-ATG initiation, providing evidence for RAN translation in corneal endothelium of patients with Fuchs' endothelial corneal dystrophy (FECD). 29695756_these results highlight TCF4 as a frequent cause of moderate to profound intellectual disabilities (ID) and broaden the clinical spectrum associated to TCF4 mutations to nonspecific ID. 29901121_Long noncoding RNA AFAP1-AS1 enhances cell proliferation and invasion in osteosarcoma through regulating miR-4695-5p/TCF4-beta-catenin signaling. 29905862_Thirteen genes within the 108 loci had both a TCF4 binding site +/-10kb and were differentially expressed in siRNA knockdown experiments of TCF4, suggesting direct TCF4 regulation. These findings confirm TCF4 as an important regulator of neural genes and point toward functional interactions with potential relevance for schizophrenia (SCZ). 29966009_Gene expression in the corneal endothelium of Fuchs endothelial corneal dystrophy patients with and without expansion of a trinucleotide repeat in TCF4 30098193_We observed instability of the TCF4 triplet repeat expansion in nearly a third of parent-child transmissions. Large mutant CTG18.1 alleles are prone to contraction, whereas intermediate mutant alleles tend to expand when unstably transmitted. Intergenerational instability of TCF4 repeat expansion has implications on FECD disease inheritance. 30288643_The rs8766 in TCF4 was selected for genotyping. rs8766 allele and genotype frequencies were not significantly different between case and control groups and a significant association cannot be suggested for the selected SNP for schizophrenia. 30315825_in cell fractions with erythroid lineage potential, TCF4 is expressed less in MDS patients than in healthy controls. This correlates with the low overall Hb levels seen in MDS patients compared with healthy individuals and is consistent with the positive impact of TCF4 on erythroid development while not having impact on white colonies. 30450687_Clinical and molecular characterization of carriers of TCF4 mosaic deletions and/or mutations contributes to our under-standing of the pathogenic mechanisms leading to Pitt Hopkins syndrome. 30527807_a serine determining the transcriptional activity of TCF-4 in lung carcinoma cells, was identified. 30593567_In contrast to HEB and E2A, which facilitate AML1-ETO-mediated leukemogenesis, E2-2 compromises the function of the AML1-ETO-containing transcription factor complex and negatively regulates leukemogenesis. 30682148_We found CTG repeat length to be a useful adjunct indicator which may be used to counsel a Fuchs' Endothelial Corneal Dystrophy (FECD) patient regarding his or her risk of significant clinical progression and keratoplasty over the next 10 years. There was more rapid clinical progression of FECD amongst patients who harbor an expanded TCF4 CTG18.1 allele (i.e. L40 group), during the first 5 years. 30705426_Findings provide new insight into the specific roles of Tcf4 molecular pathway in neocortical development and their relevance in the pathogenesis of neuropsychiatric diseases. 30733599_in this study we demonstrate a custom application of an amplification-free long-read sequencing method to specifically study the TCF4 repeat element at a nucleotide level 30771755_rs1273263 is a potential regulator of TCF4 expression, and might be associated with schizophrenia. 30811544_Our findings showed that TCF4 mRNA is upregulated in the corneal endothelium of patients with Fuchs' endothelial corneal dystrophy. 30973406_Our large German cohort demonstrated a significant association between the risk allele G in rs613872 and FECD, irrespective of TNR expansion, although this risk allele was more frequent in Fuchs endothelial corneal dystrophy cases with TNR expansion than without. 31028223_We confirmed that rs613872 in the TCF4 gene is strongly and statistically associated with late-onset FECD in a Greek population. 31081034_Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine has been described. 31209209_Study identify TCF4 as a key regulator of neural stem cells (NSCs). TCF4 interacts with Mediator, colocalizes with Mediator at super enhancers and regulates neurogenic transcription factor genes with super enhancers and broad H3K4me3 domains. Data suggest that high binding-affinity for Mediator is an important organizing feature in the transcriptional network that determines NSC identity. 31261288_Coexpression of TCF4 and CD123 is a highly reliable feature of blastic plasmacytoid dendritic cell neoplasms. 31276570_This first examination of the sequence structure of CAG repeats in CTG18.1 suggests that factors other than interruptions to the repeat structure account for the absence of disease in some elderly patients with repeat expansions in the TCF4 gene. 31288376_we observed that the beta-catenin/TCF4 complex mediates NRF3 expression by binding directly to the WRE site. 31535015_TCF4 may serve as a master regulator of a gene network dysregulated in schizophrenia at early stages of neurodevelopment. 31540772_these findings suggest that TINCR knockdown inhibits TCF4 by regulating miR-137 expression in colorectal cancer cells 31554942_Trinucleotide repeat expansion in the transcription factor 4 (TCF4) gene in Thai patients with Fuchs endothelial corneal dystrophy. 31666615_The subcellular localization of bHLH transcription factor TCF4 is mediated by multiple nuclear localization and nuclear export signals. 31678554_Knockdown of MFI2-AS1 increased cell viability but suppressed apoptosis, inflammatory response and extracellular matrix degradation in LPS-treated chondrocytes by increasing miR-130a-3p and decreasing TCF4, indicating a novel target for the treatment of osteoarthritis 31933004_TCF4 promotes colorectal cancer drug resistance and stemness via regulating ZEB1/ZEB2 expression. 32023836_Regulation of Transcription Factor E2-2 in Human Plasmacytoid Dendritic Cells by Monocyte-Derived TNFalpha. 32280673_Association between a TCF4 Polymorphism and Susceptibility to Schizophrenia. 32319632_Association between KCNQ2, TCF4 and RGS18 polymorphisms and silent brain infarction based on wholeexome sequencing. 32355234_Genetic variant rs613872 in transcription factor 4 (TCF4) is not associated with primary open-angle glaucoma. 32463444_Analyzing pre-symptomatic tissue to gain insights into the molecular and mechanistic origins of late-onset degenerative trinucleotide repeat disease. 32735996_TCF4-mediated Fuchs endothelial corneal dystrophy: Insights into a common trinucleotide repeat-associated disease. 32802179_Super-enhancer-driven AJUBA is activated by TCF4 and involved in epithelial-mesenchymal transition in the progression of Hepatocellular Carcinoma. 33054062_Transcription factor 4 (TCF4) expression predicts clinical outcome in RUNX1 mutated and translocated acute myeloid leukemia. 33069932_Various haploinsufficiency mechanisms in Pitt-Hopkins syndrome. 33116252_The Fuchs corneal dystrophy-associated CTG repeat expansion in the TCF4 gene affects transcription from its alternative promoters. 33122081_Causal links between major depressive disorder and insomnia: A Mendelian randomisation study. 33174523_Long Noncoding RNA PTPRG Antisense RNA 1 Reduces Radiosensitivity of Nonsmall Cell Lung Cancer Cells Via Regulating MiR-200c-3p/TCF4. 33414364_Transcription factor 4 and its association with psychiatric disorders. 33462220_Survival control of oligodendrocyte progenitor cells requires the transcription factor 4 during olfactory bulb development. 33782268_Relationship of Body Mass Index With Fuchs Endothelial Corneal Dystrophy Severity and TCF4 CTG18.1 Trinucleotide Repeat Expansion. 34021255_RBM24 exacerbates bladder cancer progression by forming a Runx1t1/TCF4/miR-625-5p feedback loop. 34134113_Molecular and Cellular Function of Transcription Factor 4 in Pitt-Hopkins Syndrome. 34240166_Increased miR-6875-5p inhibits plasmacytoid dendritic cell differentiation via the STAT3/E2-2 pathway in recurrent spontaneous abortion. 34518368_Isoform-Specific Reduction of the Basic Helix-Loop-Helix Transcription Factor TCF4 Levels in Huntington's Disease. 34519126_Epilepsy, electroclinical features, and long-term outcomes in Pitt-Hopkins syndrome due to pathogenic variants in the TCF4 gene. 34686186_Microarray profile analysis identifies ETS1 as potential biomarker regulated by miR-23b and modulates TCF4 in gastric cancer. 34748727_Functional consequences of TCF4 missense substitutions associated with Pitt-Hopkins syndrome, mild intellectual disability, and schizophrenia. 34779502_MicroRNA190b expression predicts a good prognosis and attenuates the malignant progression of pancreatic cancer by targeting MEF2C and TCF4. 34855896_Comparison of TCF4 repeat expansion length in corneal endothelium and leukocytes of patients with Fuchs endothelial corneal dystrophy. 34946954_Lower Fractions of TCF4 Transcripts Spanning over the CTG18.1 Trinucleotide Repeat in Human Corneal Endothelium. 35189377_Disruption and deletion of the proximal part of TCF4 are associated with mild intellectual disability: About three new patients. 35322736_Circ_0005526 contributes to interleukin-1beta-induced chondrocyte injury in osteoarthritis via upregulating transcription factor 4 by interacting with miR-142-5p. | ENSMUSG00000053477 | Tcf4 | 789.681474 | 1.0294220 | 0.041834505 | 0.11780144 | 1.280418e-01 | 7.204711e-01 | 9.087537e-01 | No | Yes | 1089.943456 | 243.787336 | 1006.462111 | 225.164606 | |
| ENSG00000196670 | 643836 | ZFP62 | protein_coding | Q8NB50 | FUNCTION: May play a role in differentiating skeletal muscle. {ECO:0000250}. | Alternative splicing;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:643836; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000046311 | Zfp62 | 1085.226808 | 1.0750334 | 0.104381459 | 0.10567767 | 9.924426e-01 | 3.191461e-01 | 6.835974e-01 | No | Yes | 1164.073632 | 197.034321 | 1072.045329 | 181.436130 | ||
| ENSG00000196696 | 283970 | PDXDC2P-NPIPB14P | lncRNA | This locus represents naturally-occurring readthrough transcription between two pseudogenes, PDXDC2P (pyridoxal dependent decarboxylase domain containing 2, pseudogene) and NPIPB14P (nuclear pore complex interacting protein family, member B14, pseudogene). The individual pseudogene loci are not curated as transcribed regions. Readthrough transcripts likely do not encode functional proteins. [provided by RefSeq, Feb 2017]. | 211.514884 | 1.3102172 | 0.389805981 | 0.21691137 | 3.238131e+00 | 7.194266e-02 | No | Yes | 210.697106 | 50.615998 | 171.683847 | 41.317371 | ||||||||||
| ENSG00000196839 | 100 | ADA | protein_coding | P00813 | FUNCTION: Catalyzes the hydrolytic deamination of adenosine and 2-deoxyadenosine (PubMed:8452534, PubMed:16670267). Plays an important role in purine metabolism and in adenosine homeostasis. Modulates signaling by extracellular adenosine, and so contributes indirectly to cellular signaling events. Acts as a positive regulator of T-cell coactivation, by binding DPP4 (PubMed:20959412). Its interaction with DPP4 regulates lymphocyte-epithelial cell adhesion (PubMed:11772392). Enhances dendritic cell immunogenicity by affecting dendritic cell costimulatory molecule expression and cytokines and chemokines secretion (By similarity). Enhances CD4+ T-cell differentiation and proliferation (PubMed:20959412). Acts as a positive modulator of adenosine receptors ADORA1 and ADORA2A, by enhancing their ligand affinity via conformational change (PubMed:23193172). Stimulates plasminogen activation (PubMed:15016824). Plays a role in male fertility (PubMed:21919946, PubMed:26166670). Plays a protective role in early postimplantation embryonic development (By similarity). {ECO:0000250|UniProtKB:P03958, ECO:0000250|UniProtKB:P56658, ECO:0000269|PubMed:11772392, ECO:0000269|PubMed:15016824, ECO:0000269|PubMed:16670267, ECO:0000269|PubMed:20959412, ECO:0000269|PubMed:21919946, ECO:0000269|PubMed:23193172, ECO:0000269|PubMed:26166670, ECO:0000269|PubMed:8452534}. | 3D-structure;Acetylation;Cell adhesion;Cell junction;Cell membrane;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Hereditary hemolytic anemia;Hydrolase;Lysosome;Membrane;Metal-binding;Nucleotide metabolism;Reference proteome;SCID;Zinc | This gene encodes an enzyme that catalyzes the hydrolysis of adenosine to inosine in the purine catabolic pathway. Various mutations have been described for this gene and have been linked to human diseases related to impaired immune function such as severe combined immunodeficiency disease (SCID) which is the result of a deficiency in the ADA enzyme. In ADA-deficient individuals there is a marked depletion of T, B, and NK lymphocytes, and consequently, a lack of both humoral and cellular immunity. Conversely, elevated levels of this enzyme are associated with congenital hemolytic anemia. [provided by RefSeq, Sep 2019]. | hsa:100; | cell junction [GO:0030054]; cell surface [GO:0009986]; cytoplasmic vesicle lumen [GO:0060205]; cytosol [GO:0005829]; external side of plasma membrane [GO:0009897]; lysosome [GO:0005764]; membrane [GO:0016020]; plasma membrane [GO:0005886]; 2'-deoxyadenosine deaminase activity [GO:0046936]; adenosine deaminase activity [GO:0004000]; zinc ion binding [GO:0008270]; adenosine catabolic process [GO:0006154]; allantoin metabolic process [GO:0000255]; AMP catabolic process [GO:0006196]; AMP salvage [GO:0044209]; cell adhesion [GO:0007155]; dAMP catabolic process [GO:0046059]; dATP catabolic process [GO:0046061]; deoxyadenosine catabolic process [GO:0006157]; embryonic digestive tract development [GO:0048566]; germinal center B cell differentiation [GO:0002314]; GMP salvage [GO:0032263]; hypoxanthine salvage [GO:0043103]; inosine biosynthetic process [GO:0046103]; liver development [GO:0001889]; lung alveolus development [GO:0048286]; negative regulation of adenosine receptor signaling pathway [GO:0060169]; negative regulation of inflammatory response [GO:0050728]; negative regulation of leukocyte migration [GO:0002686]; negative regulation of mature B cell apoptotic process [GO:0002906]; negative regulation of mucus secretion [GO:0070256]; negative regulation of penile erection [GO:0060407]; negative regulation of thymocyte apoptotic process [GO:0070244]; Peyer's patch development [GO:0048541]; placenta development [GO:0001890]; positive regulation of alpha-beta T cell differentiation [GO:0046638]; positive regulation of B cell proliferation [GO:0030890]; positive regulation of calcium-mediated signaling [GO:0050850]; positive regulation of germinal center formation [GO:0002636]; positive regulation of heart rate [GO:0010460]; positive regulation of smooth muscle contraction [GO:0045987]; positive regulation of T cell differentiation in thymus [GO:0033089]; positive regulation of T cell receptor signaling pathway [GO:0050862]; purine nucleotide salvage [GO:0032261]; purine-containing compound salvage [GO:0043101]; regulation of cell-cell adhesion mediated by integrin [GO:0033632]; response to hypoxia [GO:0001666]; T cell activation [GO:0042110]; trophectodermal cell differentiation [GO:0001829]; xanthine biosynthetic process [GO:0046111] | 11121182_Observational study of gene-disease association. (HuGE Navigator) 11354825_Observational study of gene-disease association. (HuGE Navigator) 11534018_Observational study of gene-disease association. (HuGE Navigator) 11901152_Clustered charged amino acids of human adenosine deaminase comprise a functional epitope for binding the adenosine deaminase complexing protein CD26/dipeptidyl peptidase IV 12104097_Relationship between plasma malondialdehyde levels and adenosine deaminase activities in preeclampsia. 12113294_serum adenosine deaminase-2 (but not adenosine deaminase-1) and cytidine deaminase levels were significantly higher in systemic lupus erythematosus patients 12381379_In humans, ADA1 was mainly purified concomitant with ADA-binding protein and dipeptidyl peptidase IV, was not adsorbed in adenosine-Sepharose, but was adsorbed in IgG anti-ADA1-Sepharose column; properties compared with chicken ADA1 12712614_The activity of this enzyme was studied in tissues, erythrocytes, and blood plasma of patients with peptic ulcer both in its uncomplicated course and in the development of complications. 12774669_The activity of ADA was determined in lymphocytes, esoinophils and blood serum in patients with bronchial asthma. 12809673_ADA was analyzed by linkage disequilibrium and no correlation was found between hot spots of equal and unequal homologous recombination. 12935677_Observational study of genotype prevalence. (HuGE Navigator) 14726805_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, genetic testing, and healthcare-related. (HuGE Navigator) 15016824_Adenosine deaminase binding is diminished by mutation of ADA residues known to interact with CD26. 15063762_Observational study of gene-disease association. (HuGE Navigator) 15257174_Observational study of gene-disease association. (HuGE Navigator) 15257174_adenosine-related gene variants do not appear to alter susceptibility to the disease in this group of essential hypertensives 15281007_adenosine deaminase contributes to the clinical manifestations of type 2 diabetes and probably also have a marginal influence on susceptibility to the disease 15632314_The adenosine deaminase (ADA) gene was highly up-regulated in Ara-C-resistant cells, while equilibrative nucleoside transporter 1 (ENT1) and several cell-cycle-related genes were down-regulated. 15761857_Observational study of gene-disease association. (HuGE Navigator) 15983379_ADA colocalizing with adenosine receptors on dendritic cells interact with CD26 expressed on lymphocytes. 16133068_Adenosine deaminase activity was abnormal in active nephrotic syndrome, and lymphocyte ADA demonstrated change both in active as well as remission stage of the disease. 16224193_Observational study of gene-disease association. (HuGE Navigator) 16410722_We tested whether p63 is implicated in transcriptional events related to sustaining cell proliferation by transactivation of antiapoptotic and cell survival target genes such as Adenosine Deaminase (ADA), an important gene involved in cell proliferation. 16670267_During acute hypoxia associated with vascular leakage and excessive inflammation, ADA inhibition may serve as therapeutic strategy. 16724628_Observational study of gene-disease association. (HuGE Navigator) 16754522_Observational study of gene-disease association. (HuGE Navigator) 16886895_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16970880_Findings suggest a possible role for a low-activity genotype of adenosine deaminase in the pathogenesis of mild mental retardation. No significant differences were found when comparing the group with moderate or severe mental retardation and controls. 16970880_Observational study of gene-disease association. (HuGE Navigator) 17181544_the ADA c7C/T mutation was estimated to be approximately 7,100 years old 17243920_Increased adenosine deaminase is associated with hydatidiform mole 17287605_ADA*2 allele may decrease genetic susceptibility to coronary artery disease. 17287605_Observational study of gene-disease association. (HuGE Navigator) 17340203_G22A polymorphism plays a minimal role in susceptibility to autism in North American families 17397971_The measurement of adenosine deaminase activity in ascites represents a diagnostic advance in tuberculous peritonitis among end-stage renal failure patients. 17536804_the transition-state structures of PfADA, HsADA, and bovine ADA (BtADA) solved using competitive kinetic isotope effects (KIE) and density functional calculations 17696452_This study shows that adenosine elimination on human airway epithelia is mediated by ADA1, CNT2, and CNT3, which constitute important regulators of adenosine-mediated inflammation. 18218852_CD4(+) T cells from ADA-severe combined immunodeficiency patients have severely compromised T cell receptor/CD28-driven proliferation and cytokine production, both at the transcriptional and protein levels. 18222177_Results describe the activities of ectonucleotide pyrophosphatase/phosphodiesterase and adenosine deaminase in patients with uterine cervix neoplasia. 18302529_the primary mechanism in men with ischemic stroke might involve the reduction of ADA1 activity 18357489_Suggest that adenosine deaminase can be used to distinguish between tuberculous and malignant pleural effusions. 18560234_Negative association between coronary artery disease and ADA*2 allele is only present in males 18562134_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18597230_the increased level of ADA activity in the subararachnoid hemorrhage patients with the poor clinical and consciousness level may have resulted from the ischemic cerebral insult 18673094_Elevated serum adenosine deaminase in patients with hyperemesis gravidarum may relate to high levels of E2 and progesterone. 18680557_Human ADA, apart from reducing the adenosine concentration and thus preventing A(1)R desensitization, binds to A(1)R behaving as an allosteric effector that markedly enhances agonist affinity and increases receptor functionality. 18768081_Observational study of gene-disease association. (HuGE Navigator) 18768081_Zygotes with low adenosine deaminase locus 1 activity and low F activity may experience the most favourable intrauterine conditions for a balanced development of the feto-placental unit. 18794722_Observational study of gene-disease association. (HuGE Navigator) 18794722_heterozygosity for the 22G>A variant of ADA, although reducing catalytic activity, does not enhance forearm reactive hyperaemia 19019667_Findings suggest that adenosine deaminase might play a crucial role in the development of aspirin-intolerant-asthma by mediation of A1 and A2A receptors. 19026999_Results hypothesized that altered ADA activity may be associated with altered immunity. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19172437_Serum adenosine deaminase activity was significantly higher in eclamptic pregnant women when compared with healthy pregnant women and non-pregnant women. 19237091_ADA may have a role in the cytokine network of the inflammatory cascade of familial Mediterranean fever; elevated ADA levels may be a part of the activated Th1 response in the disease 19470168_Observational study of gene-disease association. (HuGE Navigator) 19521708_serum catalytic concentration elevated in pregnancy, no difference between gestational diabetes and normal pregnancy 19628957_Decreased paraoxonase activity as well as increased adenosine deaminase and xanthine oxidase activities and nitrite levels indicate that oxidative stress is increased and purine metabolism is altered in pseudoexfoliation syndrome. 19633200_ADA-deficient SCID is associated with a specific microenvironment and bone phenotype characterized by RANKL/OPG imbalance and osteoblast insufficiency. 19668260_Addition of ADA to the co-cultures resulted in enhanced CD4(+) and CD8(+) T-cell proliferation and robust ADA-induced increase in cytokine production. 19703146_Adenosine deaminase increases TNF-alpha production in amniotic fluid 19789510_A significant interaction between ACP1 and ADA1 concerning susceptibility to type 1 diabetes, was revealed. 19845893_Observational study of gene-disease association. (HuGE Navigator) 19901882_Occult tuberculous pleurisy is significantly common in patients with pleural effusion ADA levels of 50 IU/L or less and who may otherwise be diagnosed with nonspecific pleurisy. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20147632_The CD8positive CD28positive T lymphocytes that express ADA have significantly greater telomerase activity than those that do not express ADA; however, ADA is progressively lost as cultures progress to senescence. 20174870_Data show that that adenosine deaminase influences human life-span in a sex and age specific way. 20174870_Observational study of gene-disease association. (HuGE Navigator) 20180986_Observational study of gene-disease association. (HuGE Navigator) 20306731_mean serum ADA level in the non-pregnant women was higher than that in the normal pregnant women. Amongst the pregnant women, mean serum ADA in the hypertensive and pre-eclamptic women was significantly higher than that in the normal pregnant group 20347569_ADA activity was positively correlated with lactate dehydrogenase and protein in patients with HIV positive and it was negatively correlated with glucose levels 20392501_Observational study of gene-disease association. (HuGE Navigator) 20412337_novel missense mutation in pedigree of Chinese family with dyschromatosis symmetrica hereditaria 20414589_Observational study of gene-disease association. (HuGE Navigator) 20414589_results suggest that the G/A genotype associated with low adenosine deaminase activity and, supposingly, with higher adenosine levels is less frequent among schizophrenic patients. 20453107_Human adenosine deaminase 2 role in innate immunity and adaptive immunity 20480728_From genetic association studies of SNP in children in Italy, a specific haplotype for ADA is associated with diabetes mellitus type I in girls; the association/susceptibility appears to be reversed in boys. 20480728_Observational study of gene-disease association. (HuGE Navigator) 20522203_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20548128_decrease of ADA activity may play an important role in the formation of pulmonary injury in COPD patients. 20554694_Observational study of gene-disease association. (HuGE Navigator) 20581655_Observational study of gene-disease association. (HuGE Navigator) 20581655_association with coronary artery disease is present in nondiabetic subjects only and dependent on male gender 20590444_Observational study of gene-disease association. (HuGE Navigator) 20590444_Polymorphic sites of ADA might influence cell-mediated anti-tumor immune responses controlling adenosine level and extraenzymatic protein functions. 20615890_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20651391_adenosine deaminase is involved in the regulatory system of chronic atrophic gastritis and gastric cancer risk. 20805743_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20836741_elevated in infants with bronchopulmonary dysplasia 20872567_Elevated adenosine deaminase levels is associated with celiac disease. 20943049_Observational study of gene-disease association. (HuGE Navigator) 20943049_Our data indicate a border line effect of ADA gene polymorphism on susceptibility to heumatoid arthritis that need to be confirmed in other clinical settings 20959412_ADA is a relevant modulator of CD4+ T cell differentiation, even in cells from immunologically compromised individuals. 21198750_effects of soluble human CD26 on human T-cell proliferation are mechanistically independent of both the enzyme activity and the adenosine deaminase-binding capability of soluble CD26 21269755_investigation of plasma activity of total ADA, ADA1, and ADA2 in menses, follicular, and luteal phases of menstrual cycle: significant correlation between monocyte number and ADA activities 21350874_ADA and its isoenzymes analysis in serum of patients could be used as a useful and non-invasive diagnostic tool in evaluation of systemic lupus erythematosus active phase and the disease severity. 21359089_SNP rs73598374 role in sleep modulated by caffeine 21365880_ADA levels rise in some infections & in hypertensive disorders. This may represent a compensatory mechanism resulting from increased adenosine levels & the release of hormones & inflammatory mediators stimulated by hypoxia. Review. 21461854_The new disease activity index that applies serum ADA may help in predicting disease activity in rheumatoid arthritis. 21496384_ADA1 may have a modulatory role in the implantation and duration of the pregnancy. 21593678_This study demonstrated elevated levels of total adenosine deaminase and adenosine deaminase 2 isozyme in patients with active disease. As the patient improves and becomes clinically stable these levels decrease, approaching normal values. 21640091_Serum adenosine deaminase was significantly increased in HIV and HIV-HBV co-infections 21725047_Myeloid dysplasia and bone marrow hypocellularity is associated with adenosine deaminase-deficient severe combined immune deficiency 21734253_This study demonstrated that genetic reduction of ADA activity elevates sleep pressure and plays a key role in sleep and waking quality in humans. 21856036_[review] Recent studies clarify the interactions of dipeptidyl peptidase IV (DPPIV) with adenosine deaminase (ADA) and the transcription transactivator Tat of human immunodeficiency virus (HIV) type-1. 21860532_Results evaluate the utility of the determination of adenosine deaminase (ADA) level in pleural fluid for the differential diagnosis between tuberculous pleural effusion and malignant pleural effusion. 21865054_report that a combination of functional SNPs within ADA and TNF-alpha genes can influence life-expectancy in a gender-specific manner and that males and females follow different pathways to attain longevity 21919946_alteration in ADA activity can lead to reduced adenosine levels, which may be involved in disturbing the fertility process 22062893_Mononuclear cell adenosine deaminase decreases rapidly after angioplasty, indicating that it is an early marker for reperfusion. 22086524_The results suggest that the adenosine deaminase *2 allele is associated with a low risk for recurrent spontaneous abortions, but this association is dependent on older age. 22104430_cutoff value of 9.5 U/L in CSF is a useful aid for the differential diagnosis of tuberculous meningitis and non-tuberculous meningitis 22184407_Data show that the alterations in the CD39/CD73 adenosinergic machinery and loss of function in ADA-deficient Tregs provide insights into a predisposition to autoimmunity and the underlying mechanisms causing defective peripheral tolerance in ADA-SCID. 22257696_ADA1 was found to be the ADA isoenzyme present in platelets of healthy people 22614344_results indicate that the G22A polymorphism of ADA in isolation or together with HLA-B*27 is not associated with ankylosing spondylitis 22719194_Ascitic fluid ADA measurement is helpful in the differential diagnosis of tuberculous peritonitis and peritoneal carcinomatosis. 22942500_Studies report that polymorphisms in the adenosine deaminase (ADA) gene explains phenotypic variance in responses to total sleep deprivation. 22952909_The present study verified the association between the ADA G22A polymorphism and changes in sleep electroencephalogram spectral power. 23066321_analysis of adenosine deaminase activity in tuberculous peritonitis among patients with underlying liver cirrhosis 23096558_Serum ADA could play an important role in adult-onset Still's disease and may be an important biomarker for its diagnosis. 23160984_Affordable parameters such as serum ADA and IgG correlated significantly with immune activation levels and markers of disease progression in untreated HIV-infection. 23193172_ADA open form, but not the closed one, that is responsible for the functional interaction with AR and AAR. 23240012_Taken together, these findings suggest that ADA would promote enhanced and correctly polarized T-cell responses in strategies targeting asymptomatic HIV-infected individuals. 23433854_ADA activity in serum and platelets is decreased in prostate cancer patients. 23768730_The determination of CSF and serum ADA activity is a simple and reliable test for differentiating tuberculous meningitis from other types of meningitis 23897810_Soluble ecto-5'-nucleotidase (5'-NT), alkaline phosphatase, and adenosine deaminase (ADA1) activities in neonatal blood favor elevated extracellular adenosine. 23959645_In Class 3 people the combination of high ACP f-isozyme concentration and the ADA*2 allele, lowers the rate of glycolysis that may reduce the amount of metabolic calories and activates Sirtuin genes that protect cells against age-related diseases. 24257613_ADA acts as a natural antagonist for DPP4-mediated entry of the Middle East respiratory syndrome coronavirus. 24305782_Serum ADA activity in patients with chronic tonsillitis was significantly higher than in healthy controls. 24453844_A strong correlation between ADA and fasting plasma glucose which suggests an association between ADA and nonobese type 2 diabetes mellitus subjects. 24640520_Association between ADA, genetic polymorphism (ADA1, ADA2 and ADA6) and coronary artery disease was studied.Differend pattern observed between males and females. 24682206_Data indicate that the well-being was worse in the adenosine deaminase (ADA) G/A genotype under sleep loss and n-back working memory performance appeared to be specifically susceptible to sleep-wake manipulation in this genotype. 24762755_A history of tuberculosis and hypertension were more common in the low-adenosine deaminase group than the high-adenosine deaminase group. 24896148_ADA rs73598374-A carriers showed lower telomerase activity and shorter leukocyte telomere length. 24965595_The frequency of the ADA2 *2 allele in Sardinia (Italy) is higher in Oristano than in Nuoro resulting in a higher frequency of the ADA1 *1/ADA2 *2 haplotype in Oristano. This suggests a selective advantage of this haplotype in a malarial environment. 24997584_This study suggests that serum ADA could be used as a biochemical marker in cutaneous anthrax. 25125338_ADA polymorphism is associated with type 1 diabetes mellitus. 25170811_No relationship between ADA rs452159 polymorphism and susceptibility to chronic heart failure was found in Chinese population. 25293959_Genetic association replicative and exploratory studies identify SNPs in ADA and MTR highly associated with isolated Neural tube defects (NTD)and SNP in ARID1A and ALDH1A2 associated with NTDs in whites and African Americans respectively. 25299872_We conclude that ADA is an effective test for the diagnosis of pleural tuberculosis; ADA values varied with age in our study population 25437848_Data provide evidence for a more distinct circadian modulation of nap sleep in G/A- compared to G/G-allele ADA carriers. They further indicate that REM sleep, being under strong circadian control, boosts working memory performance according to genotype in a time-of-day dependent manner. 25527815_increase in the salivary ADA and DPP-IV activities as well as in the lipid peroxidation could be related of the regulation to various aspects of adipose tissue function and inflammatory obesity 25647479_the diagnostic utility of pfADA in a low mTB incidence area 25689690_diagnostic value of serum adenosine deaminase (ADA) activity as a useful tool to differentiate HIV mono- and co-infection, was investigated. 25764155_Serum ADA levels were increased in secondary hemophagocytic lymphohistiocytosis suggesting a partial role of activated T-cell response in the disease pathophysiology. 25963491_Multiplex PCR and thoracoscopy are useful investigations in the diagnostic work-up of pleural effusions complicating CKD while the sensitivity and/or specificity of ADA and 65 kDa gene PCR is poor 26166670_activity of ADA isoenzymes and distribution of ADA1 G22 A genotypes were different among fertile and infertile men and more likely the GA genotype, which had lower ADA1 activity and was higher in fertile men is a protective factor against infertility. 26216523_ADA6, PTPN22 and ACP1 are involved in immune reactions: since endometriosis has an autoimmune component. 26261621_Pleural ADA activity is an accurate test for the diagnosis of tuberculous pleural effusions. 26310829_In autologous cocultures of T cells with HIV-1-pulsed dendritic cells, adenosine deaminase decreased HIV-1-induced CD4(+)CD25(hi) FOXP3(+) cells and enhanced the HIV-1-specific CD4(+) responder T cells. 26329539_ADA and ADK are involved in glioma progression; increased ADA and ADK levels in peritumoral tissues may be associated with epilepsy in glioma patients. 26376800_variable mutations are identified as a cause of immunodeficiency in a single Italian center 26659614_Mitochondrial Damage and Apoptosis Induced by Adenosine Deaminase Inhibition and Deoxyadenosine in Human Neuroblastoma Cell Lines 26707067_We conclude that pleural ADA activity could be integrated in the diagnostic procedures of pTB in low to medium tuberculosis prevalence settings. 26794633_High level of ADA is associated with obesity. 26918693_Genetic variability within the ADA gene may influence adenosine concentration and in turn the immune response by lymphocytes in solid tumors. 26980102_association between the occurrence of polycystic ovary syndrome with altered ADA activity and genetic distribution of G22A and A4223C polymorphisms of the ADA1 gene 26994767_In this study, we compared the activity of human adenosine deaminase (hADA) expressed in transgenic seeds of three different plant species: pea (Pisum sativum L.), Nicotiana benthamiana L. and tarwi (Lupinus mutabilis Sweet). All three species were transformed with the same expression vector containing the hADA gene driven by the seed-specific promoter LegA2 with an apoplast targeting pinII signal peptide. 27017482_Patients with tuberculous pleurisy who had high ADA values had a higher probability of manifesting pulmonary tuberculosis. High ADA values may help predict contagious pleuroparenchymal tuberculosis. The most common pulmonary involvement of tuberculous pleurisy showed a centrilobular nodular pattern. 27044834_This study aimed to investigate the activities of purinergic system ecto-enzymes present on the platelet surface as well as CD39 and CD73 expressions on platelets of sickle cell anemia treated patients. 27186641_our analysis shows that, mononuclear cells secretes ADA in response to leishmanial infection and the elevated serum ADA activity is associated with VL and PKDL, and serum ADA activity can be used as prognostic marker to monitor the course of the treatment. 27221867_Increase in serum ADA levels was observed in Oral Squamous Cell Carcinoma. 27273565_Platelet aggregation and adenosine deaminase (ADA) activity were evaluated in pregnant women living with some disease conditions including hypertension, diabetes mellitus and human immunodeficiency virus infection. 27557561_Simvastatin reversed IL-13-suppressed adenosine deaminase activity, leading to the down-regulation of adenosine signaling and inhibition osteopontin expression through the direct inhibition of IL-13-activated STAT6 pathway in COPD. 27663683_Adenosine deaminases ADA1 and ADA2 (ADAs) decrease the level of adenosine by converting it to inosine, which serves as a negative feedback mechanism. These results suggest the existence of a new mechanism, where the activation and survival of immune cells is regulated through the activities of ADA2 or ADA1 anchored to the cell surface. 28074903_The authors found significant neurological and cognitive alterations in untreated Adenosine Deaminase deficiency-severe combined immunodeficiency patients. These included motor dysfunction, EEG alterations, sensorineural hypoacusia, white matter and ventricular alterations in MRI as well as a low mental development index or IQ. 28748310_this study characterizes the long-term outcome of ADA-deficient patients, who survived 5 or more years after therapy 29061410_ADA levels were found to be higher among patients, and revealed a possible link between evening rise and severity of auditory hallucinations as well as morning rise and severity of avolition-apathy in patients with schizophrenia. 29194839_this study shows that ADA is biomarker candidate for endometriosis 29797475_Studied levels of mucin 16 (CA125), adenosine deaminase and midkine as tumor markers in nonsmall cell lung cancer-associated malignant pleural effusion. 30017738_Results provide evidence that ADA SNPs rs12190871 and rs121908723 are risk factors for contracting tuberculous pericarditis. 30394149_ADA levels were found to be increased in polycystic ovary syndrome patients compared to controls. 30444912_Alanine scanning results showed that His17, Gly184, Asp295, and Asp296 exerted the greatest effects on protein energy, suggesting that they played crucial roles in binding to inhibitors. 30641086_Inhibition of eADA blocked endothelial activation suggesting a crucial role of this enzyme in the control of vascular inflammation. This supports the concept of eADA targeted vascular protection therapy 30778076_Adenosine deaminase-1 is an immune regulator of the follicular helper T cell.Adenosine deaminase-1 interaction with CD26 is impaired during HIV infection. 30896318_Increased serum ADA activity as well as its association with obesity in Polycystic Ovary Syndrome (PCOS), while there was no change in serum DPP-4 activity in women with PCOS. 31233715_GLT would confer protection against renal injury by protecting against lipid accumulation and glutathione depletion, at least in part, through suppression of ADA/XO/UA pathway. 31379209_sequential detection of ADA screening and T-SPOT assay was found to be an accurate and rapid method for identifying TB pleurisy from pleural effusion 31709646_Clinical value of combined detection of reactive oxygen species modulator 1 and adenosine deaminase in pleural effusion in the identification of NSCLC associated malignant pleural effusion. 31953681_Exploring the binding modes of cordycepin to human adenosine deaminase 1 (ADA1) compared to adenosine and 2'-deoxyadenosine. 32245326_A Novel Non-frameshift ADA Deletion Detected by Whole Exome Sequencing in an Iranian Family with Severe Combined Immunodeficiency. 32438744_Adenosine Deaminase as a Biomarker of Tenofovir Mediated Inflammation in Naive HIV Patients. 32468861_A significant difference existed in pleural effusion ADA levels between malignant pleural mesothelioma (MPM) and benign disease patients. Pleural effusion ADA levels were significantly higher in MPM patients. 33007809_A Single Nucleotide ADA Genetic Variant Is Associated to Central Inflammation and Clinical Presentation in MS: Implications for Cladribine Treatment. 34510320_ADA gene haplotype is associated with coronary-in-stent-restenosis. 34981765_Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation. | ENSMUSG00000017697 | Ada | 448.080925 | 1.0177761 | 0.025420267 | 0.13126001 | 3.738833e-02 | 8.466766e-01 | No | Yes | 440.120087 | 40.647680 | 435.570621 | 40.142592 | ||
| ENSG00000197461 | 5154 | PDGFA | protein_coding | P04085 | FUNCTION: Growth factor that plays an essential role in the regulation of embryonic development, cell proliferation, cell migration, survival and chemotaxis. Potent mitogen for cells of mesenchymal origin. Required for normal lung alveolar septum formation during embryogenesis, normal development of the gastrointestinal tract, normal development of Leydig cells and spermatogenesis. Required for normal oligodendrocyte development and normal myelination in the spinal cord and cerebellum. Plays an important role in wound healing. Signaling is modulated by the formation of heterodimers with PDGFB (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;Cleavage on pair of basic residues;Developmental protein;Disulfide bond;Glycoprotein;Growth factor;Mitogen;Reference proteome;Secreted;Signal | This gene encodes a member of the protein family comprised of both platelet-derived growth factors (PDGF) and vascular endothelial growth factors (VEGF). The encoded preproprotein is proteolytically processed to generate platelet-derived growth factor subunit A, which can homodimerize, or alternatively, heterodimerize with the related platelet-derived growth factor subunit B. These proteins bind and activate PDGF receptor tyrosine kinases, which play a role in a wide range of developmental processes. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2015]. | hsa:5154; | cell surface [GO:0009986]; endoplasmic reticulum lumen [GO:0005788]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; Golgi lumen [GO:0005796]; Golgi membrane [GO:0000139]; microvillus [GO:0005902]; platelet alpha granule lumen [GO:0031093]; platelet-derived growth factor complex [GO:1990265]; collagen binding [GO:0005518]; growth factor activity [GO:0008083]; growth factor receptor binding [GO:0070851]; identical protein binding [GO:0042802]; platelet-derived growth factor binding [GO:0048407]; platelet-derived growth factor receptor binding [GO:0005161]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; actin cytoskeleton organization [GO:0030036]; angiogenesis [GO:0001525]; animal organ morphogenesis [GO:0009887]; cell activation [GO:0001775]; cell projection assembly [GO:0030031]; cell-cell signaling [GO:0007267]; cellular response to transforming growth factor beta stimulus [GO:0071560]; embryonic lung development [GO:1990401]; hair follicle development [GO:0001942]; inner ear development [GO:0048839]; lung alveolus development [GO:0048286]; male gonad development [GO:0008584]; negative chemotaxis [GO:0050919]; negative regulation of phosphatidylinositol biosynthetic process [GO:0010512]; negative regulation of platelet activation [GO:0010544]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of cell division [GO:0051781]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of mesenchymal cell proliferation [GO:0002053]; positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway [GO:0035793]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein autophosphorylation [GO:0031954]; positive regulation of protein kinase B signaling [GO:0051897]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling [GO:0060683]; regulation of peptidyl-tyrosine phosphorylation [GO:0050730]; regulation of smooth muscle cell migration [GO:0014910]; response to estradiol [GO:0032355]; response to hypoxia [GO:0001666]; response to inorganic substance [GO:0010035]; response to retinoic acid [GO:0032526]; response to wounding [GO:0009611]; response to xenobiotic stimulus [GO:0009410]; skin development [GO:0043588]; wound healing [GO:0042060] | 11678848_Observational study of gene-disease association. (HuGE Navigator) 12411321_involvement of Furin gene regulation in the maturation of PDGF-AB in differentiating megakaryoblastic cells 12576295_PDGF and similar cytokines may be important factors in airway remodeling by redistribution of smooth muscle cells during inflammation and that urokinase may be important in potentiating the response. 12615918_The preferential formation of disulfide-bonded heterodimers from an equimolar mixture of unfolded A- and B-chains is thus a kinetically controlled process 12645527_PDGF has a role in inducing beta-gamma-secretase-mediated cleavage of Alzheimer's amyloid precursor protein through a Src-Rac-dependent pathway 12670444_PDGF up-regulates the expression of transcription factors NF-E2, GATA-1 and c-Fos in megakaryocytic cell lines 12850832_a time-dependent release of TGFB and PDGF-AB occurred at neutral and alkaline pH, but not pH5, 15 min and 12h after platelet activation, suggesting relevance to wound healing 12972619_PDGF1 potently activates AP-1 ands causes cellular neoplastic transformation. 14573617_PDGF-A chain gene expression, under the control of KLF5, is cooperatively activated by the NF-kappaB p50 subunit and a pathophysiological stimulus. 14633628_Glucose causes a late increase in PDGF-dependent TGF-beta 1 translation by enhancing cellular sensitivity to PDGF. 14705808_cultured follicular keratinocytes synthesize both PDGF-A and PDGF-B, whereas, dermal papilla cells only express PDGF-A. 14708943_PDGF-A is a novel mitogenic target of 1,25-(OH)2D3 whose expression is induced via binding of hormone-activated VDR to a response element located far upstream of the transcription start site 14997209_PDGFA autocrine inhibition is associated with malignant gliomas 15631865_The PDGF expression, as a marker of angiogenesis, was evaluated in myomas obtained after surgery. 15829977_These results identify a novel mechanism of transcriptional enhancement involving ligand-independent activity of the VDR/RXR heterodimer and MAPK signaling pathways 15890262_thrombospondin-1 is not necessary for proliferation but is permissive for vascular smooth muscle cell responses to platelet-derived growth factor 16760468_JNK is a critical component downstream of PI 3-kinase that may be involved in PDGF-stimulated chemotaxis presumably by modulating the integrity of focal adhesions by phosphorylating its components 16850112_36% of the examined primary central nervous system lymphoma specimens showed expression of PDGF-A. Tumours expressing survivin occasionally co-expressed PDGF-A. 17087110_During the wound healing of nasal mucosa, the levels of PDGF, TNF-alpha, and hyaluronic acid are different at each postoperative stage. 17578349_expression level might be decreased during the secretory phase in the eutopic endometrium of women with advanced stage endometriosis 17852407_FGF and PDGF have roles in cell proliferation and migration in osteoblastic cells 17933214_Tretinoin inducibility of PDGFA is mediated by 5'-distal DNA motifs that overlap with basal enhancer and vitamin D response elements. 17944929_Increased expression of PDGF/Ralphabeta may play an important role in the mechanism of growth of...paediatric fibromatous lesions 17958740_thrombin may play an important role in the proliferation of A172 cells by inducing PDGF-AB secretion and that thrombin's action is mediated by its proteolytic activity 17984069_Ligand-mediated stabilization of G-quadruplex structures within the PDGF-A nuclease hypersensitive element can silence PDGF-A expression. 18084257_PDGFRs and the PDGF-A chain are frequently expressed in ovarian clear-cell adenocarcinomas. 18174235_tumor cell-secreted platelet-derived growth factor (PDGF) and phosphoinositide 3-kinase (PI3K) activation have roles in resistance of fibroblasts against oxidative damage 18284546_regularly expressed in variable levels in ameloblastomas 18326546_The proteins for PDGF-A were detected in oocytes, and in granulosa cells (GC) of 50% of the follicles from women/girls. 18332228_identified activin-mediated transforming growth factor (TGF)-beta signaling, platelet-derived growth factor (PDGF) signaling and fibroblast growth factor (FGF) signaling as the key pathways involved in MSC differentiation 18592007_Measuring PDGFA, bFGF, and HIF1a expression may contribute to a better understanding of the prognosis of patients with pancreatic cancer. 18808740_Results describe significant differences between controls and patients with renal cell carcinoma both pre-operatively and post-operatively in angiogenin, PDGF and MCP-1 serum levels. 18814141_OPG production by osteoblastic cells was stimulated by platelet-derived growth factor (PDGF) in two human osteosarcoma cell lines (MG63, Saos-2), a mouse pre-osteoblastic cell line (MC3T3-E1) and human bone marrow stromal cells (hMSC). 18992915_We showed that Prx I, PDGF-A, and PDGFR-alpha follow the same expression pattern during carcinoma ex pleomorphic adenoma progression. 19141566_A novel PDGF response in human preadipocytes that involves the pro-inflammatory kinase IKKbeta and demonstrate that it is required for the inhibition of adipogenesis. 19493486_SiO(2) may affect the expression of PDGF and synthesis of collagen through alveolar macrophage mediation and participate in the formation of lung fibrosis. 19559929_Immunohistochemistry for PDGF was found to be useful in differentiating various grades of oligodendroglioma, and therefore, it may be involved in tumor cell proliferation and malignant transformation. 19707201_concurrent expression of PDGFA and PDGFRA in different subtypes of gliomas, reinforce the recognised significance of this signalling pathway in gliomas. 19956642_analysis of how an enediynyl peptide inhibitor of furin blocks processing of proPDGF-A, B and proVEGF-C 20036812_Studies indicate communication between tumor cells and their microenvironment is through polypeptide growth factors EGF, FGF, PDGF and receptors for these growth factors. 20036815_Studies indicate that the imbalance of pro-angiogenic and anti-angiogenic factors VEGFA, Notch, Dll4, PDGF and angiopoietin-1 promotes tumor angiogenesis. 20083221_data suggest that advanced glycation end product(AGE)-modification of PDGF contributes to reduced wound healing in diabetic patients 20202860_Three of four angiogenic factors, VEGF, b-FGF and IGF-I, are higher concentration in human milk which collected from preterm mothers than those of terms. 20220555_PDGF-alpha diminished human adipose-derived stromal cell adipogenesis. 20224347_adipose tissue-derived stem cells have the capacity to differentiate into capillary structures and platelet-derived growth factor alpha plays a critical role in this process. 20485444_Observational study of gene-disease association. (HuGE Navigator) 20534510_analysis of the conserved, hydrophobic association mode of PDGF-A/propeptide complex 20730440_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20822908_arteriovenous malformation had higher levels of PDGF-A expression than cerebral cavernous malformation 21187969_the serum-induced increase in cell surface GLUT4 levels is not due to inhibition of its internalization and is not mediated by insulin, PDGF, IGF-1, or HGF 21559523_TMEFF2 can function to regulate PDGF signaling; it is hypermethylated and downregulated in glioma and several other cancers 21645144_PDGF and PDGFR-alpha may be involved in the pathophysiology of the human thymus. 21769672_Studies suggest that various isoforms of PDGF bind with differential affinities to two related tyrosine kinase receptors, denoted the PDGF alpha- and beta-receptors. 21771201_Our study was the first to show that CSF PDGF-AA is related to disease duration in relapsing-remitting multiple sclerosis. 21906875_BRAF mutations were found in 3 of 87 gastrointestinal stromal tumors. 21919032_Significant positive correlation was seen between serum PDGF-AB & microvascular density in multiple myeloma, & between pre- & post-treatment patients. PDGF-AB plays a role in a complex cytokine network inducing bone marrow neovascularization. 21931722_analysis of the cell of origin of NF1- and PDGF-driven glioblastomas 21993555_Melanoma cell-derived platelet-derived growth factor (PDGF)-AA and PDGF-CC were identified as major mediators that signal through PDGFR-alpha and thus induce HAS2-mediated HA synthesis in fibroblasts 22174424_Report increased PDGF-A, PDGF-B and FGF-2 expression in recurrence of salivary gland pleomorphic adenoma. 22266848_MUC1-induced invasion and proliferation occurs via increased exogenous production of PDGFA in pancreatic ductal carcinoma. 22393407_analysis of astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment 22588000_Somatostatin and somatostatin analogues reduce PDGF-induced endometrial cell proliferation and motility 22617683_The kidney takes part in the elimination of IL-6 and PDGF from systemic circulation. The kidney does not take part in the elimination of TGF-beta. 22704695_Findings suggest a role for brain derived neurotropic factor (BDNF) and platelet derived growth factors PDGF-AA in the patho-physiological mechanism of cerebrovascular disease in sickle cell anemia (SCA). 22733151_Results suggest that PDGFA may be used for cholangiocarcinoma prognosis and/or as diagnostic candidate marker. 22949635_Pdgf signaling potentiates Wnt2-Wnt7b signaling to promote high levels of Wnt activity in mesenchymal progenitors that is required for proper development of endoderm-derived organs, such as the lung 22964636_our studies establish that loss of SMAD1/5 leads to upregulation of PDGFA in ovarian granulosa cells 23025904_show that VEGF was down-regulated while platelet-derived growth factor-A (PDGF-A) was up-regulated when IFN-alpha treatment was re-initiated. 23536282_Platelet-derived growth factor-A and vascular endothelial growth factor-C might induce intimal proliferation in pulmonary arteries and contribute to the development of pulmonary tumor thrombotic microangiopathy. 23620752_The mRNA expressions of the proangiogenic growth factors VEGF, PDGF, bFGF and their receptors (VEGFR1, VEGFR2, PDGFRA, PDGFRB, FGFR1, FGFR2) were measured and compared in gastric ulcers of cirrhotic patients. 23944365_review elucidates the role of tumor stroma interactions, the roles of PDGF receptor signaling in cancer-associated fibroblasts via alteration of stromal matrix composition and the mitogenic effects of cancer-derived PDGFs. 24063997_Low concentration of PDGF-AB showed synergism with IFN-alpha in IFN-beta and -gamma induction. 24378341_PDGF positively modulates the TGF-beta-induced osteogenic differentiation of hMSCs through synergistic crosstalk between MEK- and PI3K/Akt-mediated signaling 24454980_The suppressive effects of PDGF-BB on Ca(2+) overload in neurons were more potent than those of PDGF-AA. 24480986_demonstrated that PDGFRA activation supported tumor cell proliferation 24995287_both VEGF and PDGF were significantly downregulated after completing a full CMP rush IgE desensitization. 25085844_AVP stimulates myofibroblast proliferation and induces PDGFA secretion, implying that PDGFA mediates local myofibroblast proliferation by an autocrine feedback loop and regulates epithelial proliferation and permeability by a paracrine mechanism. 25092176_PDGFA is a SOX11 direct target gene upregulated in MCL cells whose inhibition impaired SOX11-enhanced in vitro angiogenic effects on endothelial cells 25140996_Patients with Graves ophthalmopathy had significantly higher serum levels of PDGF-AA than controls. Immunosuppressive therapy removed differences. 25379550_VEGF, PDGF, and TGF-beta1 concentrations in platelets may be associated with prognosis of breast cancer. 25429785_Proprotein convertase 5/6 cleaves PDGFA in the human endometrium in preparation for embryo implantation. 25686533_Silencing the Col4-alpha1 gene or disrupting integrin engagement by blocking the antibody reduced the expression of platelet-derived growth factor A (PDGF-A), a potent chemotactic factor for fibroblasts. 25767870_MIC-1 and PDGF-A expression is elevated in both prostate tumors and structurally intact adjacent tissues. 25832656_our findings showed how FoxM1 activates expression of PDGF-A and STAT3 in a pathway required to maintain the self-renewal and tumorigenicity of glioma stem-like cells. 25869208_Demonstrate a novel positive regulatory feedback loop between FoxM1 and the PDGF-A/AKT signaling pathway, which contributes to breast cancer cell growth and tumorigenesis. 26088859_PDGF and TGFbeta1 regulate acute respiratory distress syndrome-associated lung fibrosis through distinct signaling pathway-mediated activation of fibrosis-related proteins. 26420039_Human uPAR activation and its association with beta1-integrin are required for PDGF-AB-induced migration. 26535645_The development and severity of age-related cataracts was related to the secretion and expression of PDGF-alpha. 26708839_Normalization of bone marrow microenvironment is paralleled by decreased expression of TIMP and PDGFA. 26769046_PDGF-AA impairs endothelium-dependent vasodilation and PDGF-AA mediates BMP4-induced adverse effect on endothelial cell function through SMAD1/5- and SMAD4-dependent mechanisms. 26841646_There was evidence for the involvement of PDGF, IGF-1, and EGF in primary pulmonary hypoplasia. PDGF deficiency plays a certain role in secondary pulmonary hypoplasia 27002148_in primary cultures of glioma stemlike cells that EGR1 contributes to stemness marker expression and proliferation by orchestrating a PDGFA-dependent growth-stimulatory loop. 27010479_These findings implicate DNA hypomethylation as a specific factor in mediating overexpression of genes associated with Biliary Atresia (BA) and identify PDGFA as a new candidate in BA pathogenesis. 27044757_Insulin treatment caused sustained Akt activity, whereas EGF or PDGF-AA promoted transient signaling; PDGF-BB produced sustained responses at higher concentrations.Transient responses to EGF were caused by negative feedback at the receptor level, as a second treatment yielded minimal responses, whereas parallel exposure to IGF-I caused full Akt activation 27110716_Nox4 and Duox1/Duox2 mediate redox activation of mesenchymal cell migration by PDGF. 27521890_these findings suggest that IGFBP-3 suppresses transcription of EGR1 and its target genes bFGF and PDGF through inhibiting IGF-1-dependent ERK and AKT activation. 27588483_this study uncovers a novel mechanism of the Nrf2/PDGFA regulatory loop that is crucial for AKT-dependent hepatocellular carcinoma progression 27666726_downregulated Rab5a led to slowed cell growth, decreased numbers of migrated cells, decreased numbers of cells at the G0G1 phase and a higher apoptosis rate. However, PDGF significantly rescued these phenomena caused by siRNA against Rab5a 27783304_In contrast to platelet-derived growth factor, all urokinase isoforms induced secretion of MMP-9 by mesenchymal stromal cells. 27931212_Tumor cell induced mesenchymal stromal cell chemotaxis appears to be mediated through paracrine secretion of PDGF-AA. 27941651_TLR-stimulated pancreatic cancer cells were specifically investigated for activated signaling pathways of VEGF/PDGF and anti-apoptotic Bcl-xL expression as well as tumor cell growth. 28000902_Based on a luciferase reporter assay, plateletderived growth factorA (PDGFA) was identified as a direct target gene of miR375. Additionally, overexpression of PDGFA significantly reversed the effect of miR375 on cell migration and invasion in oral squamous cell carcinoma cell. 28267575_There are four platelet-derived growth factor (PDGF) genes (PDGFA, PDGFB, PDGFC and PDGFD) that reside on chromosomes 7, 22, 4 and 11. 28275303_PDGFAA in tumor drainage and HER2 in peripheral vein blood may have roles in synchronous liver metastasis of colorectal cancer 28323386_The highest PDGF AA levels were found in mothers of fetuses with hypoplastic left heart syndrome . These findings may be useful in screening for congenital heart defects (CCHDs) and offer insight into their association with nuchal translucency 28828208_PDGFA was down-regulated in the malignant Middle Cerebral Artery Infarction. 28986588_These results establish novel insight into DUSP28 and PDGF-A related autonomous signaling pathway in pancreatic cancer. 29728363_Hypomethylation at a CpG site in PDGFA (encoding platelet-derived growth factor alpha) and PDGFA overexpression are both associated with increased T2 diabetes risk, hyperinsulinemia, increased insulin resistance, and increased steatohepatitis risk. 30340644_High PDGF expression is associated with cardiac fibrosis. 30890563_a reducing environment alters signaling through the PDGF-associated MAPK/Akt pathways, inducing chronic dephosphorylation of ERK1/2 at Thr202/Tyr204 and phosphorylation of Akt at Ser473 in a growth factor-independent manner. 30924403_Data suggest that a panel of mitogenic (PDGF), biochemical (albumin) and demographical (age) parameters may improve liver-fibrosis staging with a high degree of accuracy in those with a hepatitis C virus infection. 30929921_UTP14a promotes angiogenesis through upregulating transcription and secretion of PDGFA in colorectal cancer. 31129275_Results found PDGFA as a critical downstream target gene of FOXE1. A positive correlation is found between low PDGFA expression and clinical features of early stage papillary thyroid cancer. 31149049_The experiments disclosed positive regulation of NUPR1 expression by thyroid hormone T3/thyroid receptor through direct binding to the -2066 to -1910 region of the NUPR1 promoter. NUPR1 induced transcription of PDGFA through direct binding to the corresponding promoter region, mediating an angiogenetic effect. 31300060_This is the first report showing the activation of PDGFA expression by CEBPD through IL-1beta treatment and a novel CEBPD function in maintaining the self-renewal feature of glioma stem-like cells 31482267_High PDGFA expression is associated with glioblastoma. 31964549_Platelet-derived growth factor-AA-inducible epiregulin promotes elongation of human hair shafts by enhancing proliferation and differentiation of follicular keratinocytes. 32662506_PDGFA gene rs9690350 polymorphism increases biliary atresia risk in Chinese children. 32711436_The Role of Vitamin D, Platelet-Derived Growth Factor and Insulin-Like Growth Factor 1 in the Progression of Thyroid Diseases. 33760132_HaCaTconditioned medium supplemented with the small molecule inhibitors SB431542 and CHIR99021 and the growth factor PDGFAA prevents the dedifferentiation of dermal papilla cells in vitro. 33892919_Platelet-Derived Growth Factor Predicts Vulnerable Plaque in Patients with Non-ST Elevation Acute Coronary Syndrome. 34011067_High expression of PDGFA predicts poor prognosis of esophageal squamous cell carcinoma. 34885714_Lyophilised Platelet-Rich Fibrin: Physical and Biological Characterisation. 35105853_EPHA2 mediates PDGFA activity and functions together with PDGFRA as prognostic marker and therapeutic target in glioblastoma. 35241778_PDGF regulates guanylate cyclase expression and cGMP signaling in vascular smooth muscle. | ENSMUSG00000025856 | Pdgfa | 159.531605 | 0.8313373 | -0.266494175 | 0.21476883 | 1.515545e+00 | 2.182950e-01 | No | Yes | 141.612838 | 19.611702 | 167.413740 | 23.107856 | ||
| ENSG00000197472 | 57116 | ZNF695 | protein_coding | Q8IW36 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;Metal-binding;Nucleus;Reference proteome;Repeat;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA; + ;NA; + ;NA; + ;NA | hsa:57116; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 25273507_ZNF695 methylation was associated with the response to definitive chemoradiotherapy in esophageal squamous cell carcinoma. | ENSMUSG00000098905+ENSMUSG00000114923+ENSMUSG00000055341+ENSMUSG00000057842+ENSMUSG00000048280 | Zfp953+Gm49345+Zfp457+Zfp595+Zfp738 | 203.793184 | 1.1560640 | 0.209221279 | 0.18967710 | 1.215424e+00 | 2.702606e-01 | No | Yes | 203.626188 | 22.528057 | 178.624048 | 19.836141 | ||
| ENSG00000197535 | 4644 | MYO5A | protein_coding | Q9Y4I1 | FUNCTION: Processive actin-based motor that can move in large steps approximating the 36-nm pseudo-repeat of the actin filament. Involved in melanosome transport. Also mediates the transport of vesicles to the plasma membrane. May also be required for some polarization process involved in dendrite formation. {ECO:0000269|PubMed:10448864}. | 3D-structure;ATP-binding;Acetylation;Actin-binding;Alternative splicing;Calmodulin-binding;Coiled coil;Motor protein;Myosin;Nucleotide-binding;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | This gene is one of three myosin V heavy-chain genes, belonging to the myosin gene superfamily. Myosin V is a class of actin-based motor proteins involved in cytoplasmic vesicle transport and anchorage, spindle-pole alignment and mRNA translocation. The protein encoded by this gene is abundant in melanocytes and nerve cells. Mutations in this gene cause Griscelli syndrome type-1 (GS1), Griscelli syndrome type-3 (GS3) and neuroectodermal melanolysosomal disease, or Elejalde disease. Multiple alternatively spliced transcript variants encoding different isoforms have been reported, but the full-length nature of some variants has not been determined. [provided by RefSeq, Dec 2008]. | hsa:4644; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; filopodium tip [GO:0032433]; growth cone [GO:0030426]; insulin-responsive compartment [GO:0032593]; melanosome [GO:0042470]; membrane [GO:0016020]; myosin complex [GO:0016459]; neuron projection [GO:0043005]; ruffle [GO:0001726]; actin filament binding [GO:0051015]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; microfilament motor activity [GO:0000146]; RNA binding [GO:0003723]; small GTPase binding [GO:0031267]; actin filament organization [GO:0007015]; actin filament-based movement [GO:0030048]; cellular response to insulin stimulus [GO:0032869]; melanosome transport [GO:0032402]; post-Golgi vesicle-mediated transport [GO:0006892]; protein localization to plasma membrane [GO:0072659]; protein transport [GO:0015031]; vesicle transport along actin filament [GO:0030050]; vesicle-mediated transport [GO:0016192] | 11980908_role of function in melanosome transport 12148598_MYO5A mutations in Griscelli disease 12603861_Interactions of human Myosin Va isoforms in human melanocytes are tightly regulated by the tail domain. Interaction with rab27a and melanophilin. Myosin Va medial tail domain provides the globular tail domain with organelle-interacting specificity. 12897212_Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). 15772161_the endosome-associated protein hrs is a subunit of a protein complex containing actinin-4, BERP, and myosin V that is necessary for efficient TfR recycling but not for EGFR degradation 15788565_MYO5A transports dense core secretory vesicles in pancreatic MIN6 beta-cells. 17029413_exon B and its associated dynein light chain have a significant effect on the structure of parts of the coiled-coil tail domains and such a way could influence the regulation and cargo-binding function of myosin Va 17487986_present a kinetic model for the walking of myosin V on actin 17891151_Data suggest that myosin-V makes two brownian 90 degrees rotations per 36-nm step as it processively walks on actin filaments in a hand-over-hand fashion. 17898234_MyoVa directly mediates stable attachment of secretory granules at the plasma membrane. 18401430_Specific knockdown of MyoVa exon F isoforms resulted in transport inhibition of melanosomes to peripheral subcortical actin network in dendrite tips; perinuclear aggregation of melanosomes. May become innovative drug to treat hyperpigmentation. 18478159_During primate spermiogenesis, dynein, myosin Va, MyRIP and Rab27b that compose microtubule-based and actin-based vesicle transport systems are actually present in the manchette and might possibly be involved in intramanchette transport. 18676680_Observational study of gene-disease association. (HuGE Navigator) 19008234_related Rab protein, Rab10, can interact with myosin Va, myosin Vb, and myosin Vc 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19320733_Observational study of gene-disease association. (HuGE Navigator) 19521958_data demonstrate an essential role of myosin Va in cancer cell migration and metastasis, and suggest a novel target for Snail in its regulation of cancer progression 19590514_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19808891_Myosin-Va has a role in restraining Na(+)/K(+)-ATPase-containing vesicles within intracellular pools. 20203291_MARCKS and related chaperones bind to unconventional myosin V isoforms in airway epithelial cells 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20631136_Myo5a is activated in cells during HSV-1 infection to help transport virion- and glycoprotein-laden vesicles from the TGN, through the cortical actin, to the plasma membrane. 21245139_Myosin Va is required for P body but not stress granule formation. 21349835_Myo5a and Rab3A are direct binding partners and interact on synaptic vesicles and the Myo5a/Rab3A complex is involved in transport of neuronal vesicles 21740491_A Rab27a/MyRIP/myosin Va complex is involved in linking von-Willebrand factor (Vwf) to the peripheral actin cytoskeleton of endothelial cells to allow full maturation and prevent premature secretion of vWF. 22437832_Calmodulin bound to the first IQ motif is responsible for calcium-dependent regulation of myosin 5a. 23176491_Myosin Va plays a role in the transport and turnover of mRNA. [Review] 23652798_myosin-Va promotes adhesion dynamics, anchorage-independent survival, migration, and invasion in vitro 24006491_Data indicate that myosin Va interacted with multiple new Rab subfamilies including Rab6, Rab14 and Rab39B. 24097982_the cargo-binding domain (CBD) structures of the three human MyoV paralogs (Va, Vb, and Vc), revealing subtle structural changes that drive functional differentiation and a novel redox mechanism controlling the CBD dimerization process 24248336_several crystal structures of the myosin Va or the myosin Vb globular tail domain that gives insights into how the motor is linked to the recycling membrane compartments via Rab11 or the melanophilin adaptor that binds to Rab27a. 24339992_Structural insights into the globular tails of the human type v myosins Myo5a, Myo5b, And Myo5c. 25308080_These findings reveal a new fast-acting energy conservation strategy halting growth by immobilizing myosin V in a newly described state on selectively stabilized actin cables. 27129208_the inhibited Myo5a is equilibrated between the folded state, in which the Mlph-binding site is buried, and the preactivated state, in which the Mlph-binding site is exposed, and that Mlph is able to bind to the Myo5a in preactivated state and activates its motor function. 27477320_ETV6-NTRK3, MYO5A-NTRK3 and MYH9-NTRK3 fusions are identified in Spitz tumours and demonstrated that NTRK3 fusions constitutively activate the mitogen-activated protein kinase, phosphoinositide 3-kinase and phospholipase Cgamma1 pathways in melanocytes. 27939378_the essential melanocyte-specific transcription factor MITF regulates expression of the MYO5A gene, which encodes the molecular motor myosin-Va. 28193897_Mechanochemical cycle of myosin-V has been reported. 28266547_our studies revealed RPGRIP1L as a novel MyoVa-binding protein - the first to be demonstrated to interact with MyoVa at the centrosome - and uncover an unprecedented link between MyoVa and ciliogenesis, providing new perspectives for studies aiming to better understand why defects in MyoVa cause neurological disorders in Griscelli syndrome patients. 28939769_Data suggest that membrane tethering mediated by endosomal RAB11A is drastically and selectively stimulated by its cognate Rab effectors, class V myosins (MYO5A and MYO5B), in a GTP-dependent manner. (RAB11A = ras-related GTPase Rab-11A; MYO5 = myosin class V) 29298889_human cytomegalovirus capsids associate with nuclear myosin Va and F-actin and that antagonism of myosin Va impairs capsid localization toward the nuclear rim and nuclear egress. 29335527_Depletion of myosin-Va significantly inhibits the attachment of preciliary vesicles to the distal appendages of the mother centriole and decreases cilia assembly. Myosin-Va functions upstream of EHD1- and Rab11-mediated ciliary vesicle formation. 29670293_MYO5A, having rare amino acid mutations p.R849Q and p.V1601G, was involved in the biological network of known MODY genes. 29898384_demonstrate that high expression of myosin 5a may be an independent prognostic factor in patients with Esophageal squamous cell carcinoma 30733278_Spermine synthase (SMS) localized together with myosin Va (MyoVa) in cytoplasmic vesicles of breast cancer MCF-7 and neuroblastoma SH-SY5Y cell lines, known to produce exosomes, supporting a role for MyoVa in SMS expression and targeting. 32496561_LUZP1 and the tumor suppressor EPLIN modulate actin stability to restrict primary cilia formation. 32735728_Identification of three predictors of gastric cancer progression and prognosis. 33727250_LRRK2-phosphorylated Rab10 sequesters Myosin Va with RILPL2 during ciliogenesis blockade. 34680875_MYO5A Frameshift Variant in a Miniature Dachshund with Coat Color Dilution and Neurological Defects Resembling Human Griscelli Syndrome Type 1. 34935414_Modeling myosin Va liposome transport through actin filament networks reveals a percolation threshold that modulates transport properties. | ENSMUSG00000034593 | Myo5a | 1093.715304 | 1.0527631 | 0.074180814 | 0.11640812 | 4.153331e-01 | 5.192750e-01 | 8.138399e-01 | No | Yes | 1068.039326 | 203.086766 | 1000.166366 | 190.264723 | |
| ENSG00000197563 | 23556 | PIGN | protein_coding | O95427 | FUNCTION: Ethanolamine phosphate transferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers ethanolamine phosphate to the first alpha-1,4-linked mannose of the glycosylphosphatidylinositol precursor of GPI-anchor (By similarity). May act as suppressor of replication stress and chromosome missegregation. {ECO:0000250, ECO:0000269|PubMed:23446422}. | Disease variant;Endoplasmic reticulum;Epilepsy;GPI-anchor biosynthesis;Glycoprotein;Membrane;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Glycolipid biosynthesis; glycosylphosphatidylinositol-anchor biosynthesis. | This gene encodes a protein that is involved in glycosylphosphatidylinositol (GPI)-anchor biosynthesis. The GPI-anchor is a glycolipid found on many blood cells and serves to anchor proteins to the cell surface. This protein is expressed in the endoplasmic reticulum and transfers phosphoethanolamine (EtNP) to the first mannose of the GPI anchor. Two alternatively spliced variants, which encode an identical isoform, have been reported. [provided by RefSeq, Jul 2008]. | hsa:23556; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; mannose-ethanolamine phosphotransferase activity [GO:0051377]; GPI anchor biosynthetic process [GO:0006506]; preassembly of GPI anchor in ER membrane [GO:0016254] | 19690890_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21493957_Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. 22876578_PIGN encodes phosphatidylinositol-glycan biosynthesis class N protein. 24253414_Findings confirm that developmental delay, hypotonia, and epilepsy combined with congenital anomalies are common phenotypes of PIGN mutations and add progressive cerebellar atrophy to this clinical spectrum. 26364997_The mutated PIGN caused a significant decrease of the overall glycosylphoshatidylinositol-anchored proteins and CD24 expression which is sufficient to cause severe phenotypic expression. 26394714_PIGN mutation is associated with multiple congenital anomalies hypotonia seizures syndrome related epilepsy. 27038415_Study reports compound heterozygous mutations in PIGN in two siblings with Fryns syndrome, and a homozygous mutation in an unrelated affected individual. However, two further individuals with Fryns syndrome did not carry mutations in this gene, suggesting genetic heterogeneity in this syndrome. 27980068_PIGN-1/PIGN is required for quality control in Caenorhabditis elegans and in mammalian cells. 28187452_PIGN is a novel biomarker of CIN and leukemic transformation/progression in a subgroup of patients with MDS or AML-MRC. 28327575_Disease associated mutation L311W reduces enzymatic activity rather than affecting protein levels. 29330547_Loss of function PIGN alleles causes Fryns syndrome. Founder effect for PIGN intragenic deletion is observed in La Reunion and other Indian Ocean islands. 34051595_Novel insights into the clinico-radiological spectrum of phenotypes associated to PIGN mutations. 34561473_PIGN spatiotemporally regulates the spindle assembly checkpoint proteins in leukemia transformation and progression. | ENSMUSG00000056536 | Pign | 1288.910783 | 0.8716304 | -0.198211663 | 0.09045509 | 4.736534e+00 | 2.952850e-02 | 2.306979e-01 | No | Yes | 1189.397119 | 242.568314 | 1359.593277 | 277.155174 |
| ENSG00000197565 | 1288 | COL4A6 | protein_coding | Q14031 | FUNCTION: Type IV collagen is the major structural component of glomerular basement membranes (GBM), forming a 'chicken-wire' meshwork together with laminins, proteoglycans and entactin/nidogen. | Alternative splicing;Basement membrane;Cell adhesion;Chromosomal rearrangement;Collagen;Deafness;Disease variant;Disulfide bond;Extracellular matrix;Glycoprotein;Hydroxylation;Non-syndromic deafness;Reference proteome;Repeat;Secreted;Signal | This gene encodes one of the six subunits of type IV collagen, the major structural component of basement membranes. Like the other members of the type IV collagen gene family, this gene is organized in a head-to-head conformation with another type IV collagen gene, alpha 5 type IV collagen, so that the gene pair shares a common promoter. Deletions in the alpha 5 gene that extend into the alpha 6 gene result in diffuse leiomyomatosis accompanying the X-linked Alport syndrome caused by the deletion in the alpha 5 gene. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2013]. | hsa:1288; | collagen type IV trimer [GO:0005587]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular matrix structural constituent [GO:0005201]; extracellular matrix structural constituent conferring tensile strength [GO:0030020]; cell adhesion [GO:0007155]; cellular response to amino acid stimulus [GO:0071230]; collagen-activated tyrosine kinase receptor signaling pathway [GO:0038063]; extracellular matrix organization [GO:0030198] | 14592452_We provide a first indication that highly specialized patterns characteristic of COL4A5-COL4A6 expression in vivo arise from effects of distributed cis-acting regulatory elements on a bidirectional proximal promoter, itself transcriptionally competent. 15211113_Collagen chains alpha5(IV) and alpha6(IV) were frequently absent in basement membrane from pancreatic adenocarcinoma, and their absence might be related to the invasion of cancer cells. 15679046_Enhanced expression of collagen type IV is associated with liver metastases from gastrointestinal tumours 16507901_The expression of the alpha5(IV)/alpha6(IV) chains was down-regulated in colorectal cancer, and the loss of expression of the alpha5(IV)/alpha6(IV) chains was associated with the hypermethylation of their promoter region. 17955302_This study showed that in some cases of esophageal squamous cell carcinoma the alpha5/alpha6 (IV) chains are expressed linearly around the cancer cell nests and the alpha5/alpha6 (IV) chain expression is one prognostic factor of ESCC. 18074349_Type IV collagen alpha6 chain-derived noncollagenous domain 1 (alpha6NC1) is an endogenous inhibitor of angiogenesis and tumor growth. 18474427_Meningiomas increase in size through increased production of extracellular matrix; furthermore, the proliferation of cells typically associated with neoplasia requires considerable interaction with the extracellular matrix. 18782525_The expression of collagen type IV was significantly decreased in colorectal cancer tissues. 19422682_Results suggest that the down-regulation of alpha 6(IV) mRNA coincides with the acquisition of invasive growth properties, whereas alpha1(IV) and alpha1(VII) mRNAs were up-regulated already in dysplastic tissue. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20359090_The expression of Col I, Col IV and Fn was closely related to tumor invasion, the regional lymph node metastasis and other pathological features in laryngeal squamous cell carcinoma. 20452482_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20673868_Observational study of gene-disease association. (HuGE Navigator) 20951201_expression of collagen type IV alpha6 chain in the smooth muscle BM of the gastrointestinal tract is restricted to the esophagus in humans 21380622_In this paper we improve the definition of the COL4A5/COL4A6 deletions in three Alport syndrome with diffuse leiomyomatosis. 23551189_The expression of collagen type IV and its alpha chains (alpha1-6) was investigated in different endothelial cell culture systems in vitro qualitatively and quantitatively. 23621580_We showed that TPM2, CLU, and COL4A6 mRNA levels are downregulated in prostate cancer. 23714752_COL4A6 is associated with X-linked nonsyndromic hearing loss. 26179878_New deletion in COL4A6/COL4A5 related to diffuse esophageal leiomyomatosis associated with Alport syndrome in a Chinese family. 26811494_Mutation in COL4A6 gene is associated with gastric cancer peritoneal carcinomatosis. 27377778_although alpha5 and alpha6 (IV) chains are induced in the glomerular basement membrane in autosomal recessive Alport syndrome, their induction does not seem to play a major compensatory role 28275241_deletion breakpoints in five Alport syndrome-diffuse leiomyomatosis patients and show a contiguous COL4A6/COL4A5 deletion in each case, were characterized. 29422532_These findings indicate that alpha112:alpha556 (IV) network, which is the only network that includes the alpha6(IV) chain, is one regulator of KRT10 expression in keratinization of oral mucosal epithelium. 33840813_Confirmation of COL4A6 variants in X-linked nonsyndromic hearing loss and its clinical implications. | ENSMUSG00000031273 | Col4a6 | 451.333319 | 0.9500694 | -0.073895215 | 0.12713033 | 3.366595e-01 | 5.617638e-01 | No | Yes | 441.287389 | 42.329613 | 465.923772 | 44.573480 | ||
| ENSG00000197587 | 127343 | DMBX1 | protein_coding | Q8NFW5 | FUNCTION: Functions as a transcriptional repressor. May repress OTX2-mediated transactivation by forming a heterodimer with OTX2 on the P3C (5'-TAATCCGATTA-3') sequence. Required for brain development (By similarity). {ECO:0000250}. | Alternative splicing;DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a member of the bicoid sub-family of homeodomain-containing transcription factors. The encoded protein acts as a transcription factor and may play a role in brain and sensory organ development. Two transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:127343; | chromatin [GO:0000785]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; adult feeding behavior [GO:0008343]; adult locomotory behavior [GO:0008344]; brain development [GO:0007420]; central nervous system development [GO:0007417]; developmental growth [GO:0048589]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357] | 17990594_no indications were found for an association between the MBX gene and microphthalmia with congenital cataract in humans 27980695_Results found that CpG sites of C1orf106, DMBX1, and SIK3 mediate the genetic risk of psoriasis in Chinese Han population. 30928384_Authors found that the function of DMBX1 was dependent on p21 (CDKN1A), a key regulator of G1/S cell cycle progression. Co-IP assay revealed that DMBX1 directly bound to another homeobox transcription factor, OTX2. ChIP and luciferase reporter assay confirmed that OTX2 directly interacted with the promoter region of p21 to enhance its transcription, and DMBX1 repressed OTX2-mediated transcription of p21. | ENSMUSG00000028707 | Dmbx1 | 131.909551 | 0.9315927 | -0.102228762 | 0.24509841 | 1.766935e-01 | 6.742304e-01 | No | Yes | 134.861093 | 18.874304 | 142.155813 | 19.780412 | ||
| ENSG00000198105 | 57209 | ZNF248 | protein_coding | Q8NDW4 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:57209; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 27238579_Smoking and obesity are risk factors for LOA. ZNF248 confers increased susceptibility to LOA in African Americans. | ENSMUSG00000030145 | Zfp248 | 236.244950 | 1.1532762 | 0.205738039 | 0.18224908 | 1.261190e+00 | 2.614260e-01 | No | Yes | 273.305440 | 41.606931 | 234.449377 | 35.634389 | |||
| ENSG00000198237 | GUSBP13 | unprocessed_pseudogene | 41.701930 | 1.7211229 | 0.783350132 | 0.42293549 | 3.322358e+00 | 6.834376e-02 | No | Yes | 39.514173 | 21.715408 | 24.537952 | 13.665073 | ||||||||||||
| ENSG00000198276 | 54963 | UCKL1 | protein_coding | Q9NWZ5 | FUNCTION: May contribute to UTP accumulation needed for blast transformation and proliferation. {ECO:0000269|PubMed:12199906}. | ATP-binding;Alternative splicing;Cytoplasm;Host-virus interaction;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation | PATHWAY: Pyrimidine metabolism; CTP biosynthesis via salvage pathway; CTP from cytidine: step 1/3.; PATHWAY: Pyrimidine metabolism; UMP biosynthesis via salvage pathway; UMP from uridine: step 1/1. | The protein encoded by this gene is a uridine kinase. Uridine kinases catalyze the phosphorylation of uridine to uridine monophosphate. This protein has been shown to bind to Epstein-Barr nuclear antigen 3 as well as natural killer lytic-associated molecule. Ubiquitination of this protein is enhanced by the presence of natural killer lytic-associated molecule. In addition, protein levels decrease in the presence of natural killer lytic-associated molecule, suggesting that association with natural killer lytic-associated molecule results in ubiquitination and subsequent degradation of this protein. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2014]. | hsa:54963; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; ATP binding [GO:0005524]; uridine kinase activity [GO:0004849]; CTP salvage [GO:0044211]; pyrimidine nucleoside salvage [GO:0043097]; UMP salvage [GO:0044206] | 19653100_These results indicate a role for UCKL-1 in tumor cell survival. 30344298_The presence of IL28B rs8099917 TT and rs12979860 CC SNPs, but not the intensity of UCKL-1 expression, is strongly associated with increased chances of Hepatocellular Carcinoma development in hepatitis C virus -positive cirrhotic patients. | ENSMUSG00000089917 | Uckl1 | 1045.639781 | 1.1797227 | 0.238447805 | 0.09136217 | 6.793947e+00 | 9.146747e-03 | 1.246438e-01 | No | Yes | 1105.174612 | 116.781424 | 938.088132 | 99.072295 |
| ENSG00000198324 | 144717 | PHETA1 | protein_coding | Q8N4B1 | FUNCTION: Plays a role in endocytic trafficking. Required for receptor recycling from endosomes, both to the trans-Golgi network and the plasma membrane. {ECO:0000269|PubMed:21233288}. | 3D-structure;Alternative splicing;Cytoplasmic vesicle;Endosome;Golgi apparatus;Phosphoprotein;Reference proteome | This gene encodes a protein that localizes to the endosome and interacts with the enzyme, inositol polyphosphate 5-phosphatase OCRL-1. Alternate splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:144717; | clathrin-coated vesicle [GO:0030136]; cytosol [GO:0005829]; early endosome [GO:0005769]; recycling endosome [GO:0055037]; trans-Golgi network [GO:0005802]; protein homodimerization activity [GO:0042803]; endosome organization [GO:0007032]; receptor recycling [GO:0001881]; retrograde transport, endosome to Golgi [GO:0042147] | 20133602_Two closely related endocytic proteins, Ses1 and Ses2, which interact with OCRL, were identified. The interaction is mediated by a short amino acid motif similar to that used by the rab-5 effector APPL1. 21233288_Two novel OCRL1-binding proteins, termed inositol polyphosphate phosphatase interacting protein of 27 kDa (IPIP27)A and B (also known as Ses1 and 2), that also bind the related 5-phosphatase Inpp5b, were identified. 32152089_Deficiency in the endocytic adaptor proteins PHETA1/2 impairs renal and craniofacial development. | ENSMUSG00000044134 | Pheta1 | 489.164766 | 0.8825403 | -0.180265987 | 0.12682096 | 2.028874e+00 | 1.543349e-01 | 5.121718e-01 | No | Yes | 460.184413 | 63.210197 | 514.383054 | 70.379895 | |
| ENSG00000198496 | 10230 | NBR2 | lncRNA | This gene was identified by its close proximity on chromosome 17 to tumor suppressor gene BRCA1. Experimental evidence indicates that the two genes share a bi-directional promoter. Transcription for either gene is controlled individually by distinct transcriptional repressor factors. A short (112 amino acid) open reading frame is observed which includes a region derived from a LINE1 element. A strong Kozak signal is not observed for the putative ORF and the stop codon is more than 55 nucleotides upstream of the last splice site for the transcript, suggesting that the transcript is subject to nonsense-mediated decay. Therefore, this gene does not appear to encode a protein. Glucose starvation induces the expression of this gene and the long non-coding RNA transcribed by it functions with AMP-activated protein kinase in mediating the energy stress response. [provided by RefSeq, Aug 2016]. | 26999735_Data indicate that long non-coding RNAs (lncRNAs) NBR2 (neighbour of BRCA1 gene 2) interacts with AMP-activated kinase (AMPK) and promotes AMPK kinase activity during energy stress. 27792451_Data indicate that long non-coding RNAs (lncRNAs) NBR2 (neighbour of BRCA1 gene 2) modulates cancer cell sensitivity to phenformin through NBR2 regulation of GLUT1 expression at transcriptional level. 27792451_NBR2-GLUT1 axis may serve as an adaptive response in cancer cells to survive in response to phenformin treatment. 28249151_High NBR2 expression is associated with breast cancer. 28586153_data suggested that miR-19a negatively controlled the autophagy of hepatocytes attenuated in D-GalN/LPS-stimulated hepatocytes via regulating NBR2 and AMPK/PPARalpha signaling. 31599420_LncRNA NBR2 inhibits EMT progression by regulating Notch1 pathway in NSCLC. 31888414_This study revealed a novel mechanism by which long noncoding RNA NBR2 mediates curcumin suppression of colorectal cancer proliferation by activating adenosine monophosphate-activated protein kinase and inactivating the mTOR signaling pathway. 33378941_LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. 33651649_LncRNA NBR2 aggravates hepatoblastoma cell malignancy and promotes cell proliferation under glucose starvation through the miR-22/TCF7 axis. 34076984_NBR2 promotes the proliferation of glioma cells via inhibiting p15 expression. 34506209_Long non-coding RNA NBR2 suppresses the progress of colorectal cancer in vitro and in vivo by regulating the polarization of TAM. | 87.921531 | 1.1840042 | 0.243674249 | 0.28951705 | 6.953092e-01 | 4.043644e-01 | No | Yes | 90.296044 | 19.155987 | 76.391117 | 16.258093 | |||||||||
| ENSG00000198598 | 4326 | MMP17 | protein_coding | Q9ULZ9 | FUNCTION: Endopeptidase that degrades various components of the extracellular matrix, such as fibrin. May be involved in the activation of membrane-bound precursors of growth factors or inflammatory mediators, such as tumor necrosis factor-alpha. May also be involved in tumoral process. Cleaves pro-TNF-alpha at the '74-Ala-|-Gln-75' site. Not obvious if able to proteolytically activate progelatinase A. Does not hydrolyze collagen types I, II, III, IV and V, gelatin, fibronectin, laminin, decorin nor alpha1-antitrypsin. | Alternative splicing;Calcium;Cell membrane;Cleavage on pair of basic residues;Disulfide bond;Extracellular matrix;GPI-anchor;Glycoprotein;Hydrolase;Lipoprotein;Membrane;Metal-binding;Metalloprotease;Protease;Reference proteome;Repeat;Secreted;Signal;Zinc;Zymogen | This gene encodes a member of the peptidase M10 family and membrane-type subfamily of matrix metalloproteinases (MMPs). Proteins in this family are involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes, such as arthritis and metastasis. Members of this subfamily contain a transmembrane domain suggesting that these proteins are expressed at the cell surface rather than secreted. The encoded preproprotein is proteolytically processed to generate the mature protease. This protein is unique among the membrane-type matrix metalloproteinases in that it is anchored to the cell membrane via a glycosylphosphatidylinositol (GPI) anchor. Elevated expression of the encoded protein has been observed in osteoarthritis and multiple human cancers. [provided by RefSeq, Jan 2016]. | hsa:4326; | anchored component of membrane [GO:0031225]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; plasma membrane [GO:0005886]; enzyme activator activity [GO:0008047]; metalloaminopeptidase activity [GO:0070006]; metalloendopeptidase activity [GO:0004222]; zinc ion binding [GO:0008270]; collagen catabolic process [GO:0030574]; drinking behavior [GO:0042756]; extracellular matrix organization [GO:0030198]; kidney development [GO:0001822]; proteolysis [GO:0006508] | 12661033_down-regulation of most MT-MMPs is typical for prostate carcinoma; seems to occur mainly in epithelial cells 12962706_Data show that eosinophils constitutively express membrane type-4 matrix metalloproteinase (MT4-MMP), which is increased upon stimulation with tumor necrosis factor-alpha. 14701864_MT4-MMP and the proteoglycan form of syndecan-1 have roles in ADAMTS-4 activation on the cell surface 16686598_MT1-MT4-MMP chimaeras do not undergo normal trafficking and are not correctly processed to their fully active forms and, as a consequence, they are unable to activate pro-MMP-2 at the cell surface. 19426156_MT4-MMP promotes lung metastasis by disturbing the tumour vessel integrity and thereby facilitating tumour cell intravasation 20019845_Studies suggest a model of hypoxia induced metastasis through expression of HIF-1alpha, and SLUG regulation of MT4-MMP transcription. 20452482_Observational study of gene-disease association. (HuGE Navigator) 20587546_Observational study of gene-disease association. (HuGE Navigator) 20673868_Observational study of gene-disease association. (HuGE Navigator) 21828052_The data presented here provide a new insight into the characteristics of MT4-MMP and highlight the common and distinct properties of the glycosylphosphatidylinositol-anchored membrane type-matrix metalloproteinases. 22262494_It identifies MT4-MMP as a key intrinsic tumor cell determinant that contributes to the elaboration of a permissive microenvironment for metastatic dissemination. 22674854_CAV1 when found in lipid rafts does not allow the metalloproteinase MT4-MMP to localize into the lipid rafts, affecting its expression in the cell and probably its activity which is translated into the metastasis-associated activities of these cells. 25320013_A functional link between MT4-MMP and the growth factor receptor EGFR. 25963716_Screening of patients with inherited thoracic aortic aneurysms and dissections identified a missense mutation (R373H) in the MMP17 gene that prevented the expression of the protease in human transfected cells. 26663028_The MT4-MMP is internalized by the clathrin-independent carriers/GPI-enriched early endosomal compartments pathway, a mechanism that differs from that responsible for the internalization of other membrane-type MMP members. 28196064_Low MT4-MMP expression is associated with erlotinib resistance in breast cancer. 28531887_Three forms of MT4-MMP with molecular masses of 45 kDa, 58 kDa and 69 kDa were detected. Further, we demonstrate that the 58 kDa form is the mature protein in the cell membrane, while the 69 kDa form is its precursor found in intracellular compartments. 29500407_MT4-MMP targeting may constitute a novel strategy to boost patrolling monocyte activity in early inflammation. 30792164_In breast cancer cells, the overexpression of MT4-MMP modulates the expression of microRNAs involved in several biological processes associated with tumor formation and progression and with clinical relevance. 31813546_MT4-MMP promotes invadopodia formation and cell motility in FaDu head and neck cancer cells. | ENSMUSG00000029436 | Mmp17 | 135.604732 | 0.7669802 | -0.382738668 | 0.22787410 | 2.821171e+00 | 9.302853e-02 | No | Yes | 118.788884 | 20.478074 | 156.253007 | 26.690842 | ||
| ENSG00000198646 | 23054 | NCOA6 | protein_coding | Q14686 | FUNCTION: Nuclear receptor coactivator that directly binds nuclear receptors and stimulates the transcriptional activities in a hormone-dependent fashion. Coactivates expression in an agonist- and AF2-dependent manner. Involved in the coactivation of different nuclear receptors, such as for steroids (GR and ERs), retinoids (RARs and RXRs), thyroid hormone (TRs), vitamin D3 (VDR) and prostanoids (PPARs). Probably functions as a general coactivator, rather than just a nuclear receptor coactivator. May also be involved in the coactivation of the NF-kappa-B pathway. May coactivate expression via a remodeling of chromatin and its interaction with histone acetyltransferase proteins. | Acetylation;Activator;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a transcriptional coactivator that can interact with nuclear hormone receptors to enhance their transcriptional activator functions. This protein has been shown to be involved in the hormone-dependent coactivation of several receptors, including prostanoid, retinoid, vitamin D3, thyroid hormone, and steroid receptors. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jun 2011]. | hsa:23054; | cytosol [GO:0005829]; histone methyltransferase complex [GO:0035097]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; chromatin binding [GO:0003682]; enzyme binding [GO:0019899]; estrogen receptor binding [GO:0030331]; nuclear receptor coactivator activity [GO:0030374]; retinoid X receptor binding [GO:0046965]; thyroid hormone receptor binding [GO:0046966]; transcription coactivator activity [GO:0003713]; brain development [GO:0007420]; cellular response to DNA damage stimulus [GO:0006974]; DNA-templated transcription, initiation [GO:0006352]; heart development [GO:0007507]; myeloid cell differentiation [GO:0030099]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; response to hormone [GO:0009725] | 12482968_we show that ASC-2 belongs to a steady-state complex of approximately 2 MDa (ASC-2 complex [ASCOM]) in HeLa nuclei. ASCOM contains retinoblastoma-binding protein RBQ-3, alpha/beta-tubulins, and trithorax group proteins ALR-1, ALR-2, HALR, and ASH2. 12519782_The activation of DNA-PK in the absence of DNA ends by the coactivator TRBP suggests a novel mechanism of coactivator-stimulated DNA-PK phosphorylation in transcriptional regulation. 12519782_interacts with and stimulates its associated DNA-dependent protein kinase 12724417_ASC-2 is a physiologically important transcriptional coactivator of LXRs and demonstrate its pivotal role in the liver lipid metabolism 14645241_ASC-2 appears to contain at least three distinct nuclear receptor interaction domains; also, Rb and ASC-2 have roles in androgen receptor transactivation 14734562_ASC-2 has a role in inducing target gene transcription during granulocytic differentiation through binding to ATF2 14960326_results suggest that ASC-2 is a novel coactivator for HSF1 and heat shock stress may contribute to the strong active transcription complex through sequential recruitment of HSF1 and ASC-2. 15764585_ASC-2 is a bona fide coactivator of the xenobiotic nuclear receptor CAR and mediate the specific xenobiotic response by CAR in vivo. 18031289_The retinoid x receptor does not interact with the NR box-1 of ASC-2, but functions as an allosteric activator of LXR binding to NR box-2 of ASC-2. 18263591_interaction between RAP250, Smad2, and Smad3 constitutes an important bridging mechanism linking LXR and TGF-beta signaling pathways. 18462265_AIB3 contributes to the maintenance of beta-cell function in nondiabetic children and regulates gene expression in INS-1 cells. 18462265_Observational study of gene-disease association. (HuGE Navigator) 18552123_The coactivator NCOA6 mediates the mechanism of the synergistic activation of the CYP2C9 gene by CAR and HNF4alpha. 18660489_Observational study of gene-disease association. (HuGE Navigator) 19329434_Promoter analysis showed that PRIP acted through serum-responsive factor to regulate FOS gene expression. 19556342_ASCOM-MLL3 and ASCOM-MLL4 play redundant but essential roles in FXR transactivation via their histone 3 lysine 4 trimethylation activity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20721975_Observational study of gene-disease association. (HuGE Navigator) 21292004_NCOA6 is responsible for the synergistic activation of CYP2C9 by HNF4alpha and pregnane X receptor and NCOA6 differentially regulates CYP2C9 and CYP3A4 gene expression though both the genes are regulated by the same nuclear receptors. 21552418_diverse physiological function of NCOA6 may be mediated by multiple isoforms expressed in different tissues and localized in different subcellular compartments 31744881_NCOA6 associates with the GREB1 promoter and enhancer in an E2-independent manner and that NCOA6 knockout reduces chromatin looping, enhancer-promoter interactions, and basal GREB1 expression in the absence of estradiol. 32790097_Nuclear receptor coactivator 6 promotes HTR-8/SVneo cell invasion and migration by activating NF-kappaB-mediated MMP9 transcription. 33339270_Application of WES Towards Molecular Investigation of Congenital Cataracts: Identification of Novel Alleles and Genes in a Hospital-Based Cohort of South India. 35384116_Nuclear receptor coactivator-6 is essential for the morphological change of human uterine stromal cell decidualization via regulating actin fiber reorganization. | ENSMUSG00000038369 | Ncoa6 | 2034.494650 | 1.0480595 | 0.067720649 | 0.07130318 | 9.017144e-01 | 3.423224e-01 | 7.004858e-01 | No | Yes | 1965.329171 | 240.811849 | 1883.371723 | 230.811645 | |
| ENSG00000198791 | 29883 | CNOT7 | protein_coding | Q9UIV1 | FUNCTION: Has 3'-5' poly(A) exoribonuclease activity for synthetic poly(A) RNA substrate. Its function seems to be partially redundant with that of CNOT8. Catalytic component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. During miRNA-mediated repression the complex seems also to act as translational repressor during translational initiation. Additional complex functions may be a consequence of its influence on mRNA expression. Associates with members of the BTG family such as TOB1 and BTG2 and is required for their anti-proliferative activity. {ECO:0000269|PubMed:19605561, ECO:0000269|PubMed:20065043, ECO:0000269|PubMed:20634287, ECO:0000269|PubMed:23236473}. | 3D-structure;Alternative splicing;Cytoplasm;Exonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease;Nucleus;RNA-binding;RNA-mediated gene silencing;Reference proteome;Repressor;Transcription;Transcription regulation;Translation regulation | The protein encoded by this gene binds to an anti-proliferative protein, B-cell translocation protein 1, which negatively regulates cell proliferation. Binding of the two proteins, which is driven by phosphorylation of the anti-proliferative protein, causes signaling events in cell division that lead to changes in cell proliferation associated with cell-cell contact. The encoded protein downregulates the innate immune response and therefore provides a therapeutic target for enhancing its antimicrobial activity against foreign agents. Alternative splicing of this gene results in multiple transcript variants. Related pseudogenes have been identified on chromosomes 1 and X. [provided by RefSeq, Apr 2016]. | hsa:29883; | CCR4-NOT complex [GO:0030014]; CCR4-NOT core complex [GO:0030015]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; nuclear body [GO:0016604]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; P-body [GO:0000932]; 3'-5'-exoribonuclease activity [GO:0000175]; DNA-binding transcription factor binding [GO:0140297]; exoribonuclease activity [GO:0004532]; metal ion binding [GO:0046872]; poly(A)-specific ribonuclease activity [GO:0004535]; RNA binding [GO:0003723]; transcription corepressor activity [GO:0003714]; deadenylation-dependent decapping of nuclear-transcribed mRNA [GO:0000290]; defense response to virus [GO:0051607]; exonucleolytic catabolism of deadenylated mRNA [GO:0043928]; gene silencing by miRNA [GO:0035195]; gene silencing by RNA [GO:0031047]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of gene expression [GO:0010629]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of type I interferon-mediated signaling pathway [GO:0060339]; nuclear-transcribed mRNA poly(A) tail shortening [GO:0000289]; P-body assembly [GO:0033962]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of mRNA catabolic process [GO:0061014]; positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900153]; positive regulation of nuclear-transcribed mRNA poly(A) tail shortening [GO:0060213]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of viral genome replication [GO:0045070]; regulation of tyrosine phosphorylation of STAT protein [GO:0042509] | 12845644_CNOT7 is not the target tumor suppressor gene in the 8p22-23.1 colorectal cancer suppressor region. 17264152_CAF1 is a new regulator of PRMT1-dependent arginine methylation. 18084094_antiproliferative region of human Tob (residues 1-138) and intact hCaf1 were co-expressed in Escherichia coli, purified and successfully cocrystallized 19558367_Data show that Ccr4-Not function in RNA splicing and nuclear export, and that CNOT7 binds strongly with CNOT6. 19605561_Data show that efficient cell proliferation requires both CNOT7 and CNOT8, although combined knockdown of both subunits further reduces cell proliferation indicating partial redundancy between these proteins. 23236473_The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. 23386060_CNOT7 is an antimicrobial protein and a regulator of the innate immune response. 25038453_CNOT7/hCAF1 is involved in ICAM-1 and IL-8 regulation by TTP in HPMEC. 26471122_MEX3C associates with the cytoplasmic deadenylation complexes and ubiquitinates CNOT7. 28591869_Findings suggest a preferential involvement of CNOT7 variant 2 (CNOT7v2) in nuclear processes, such as arginine methylation and alternative splicing, rather than mRNA turnover 30984545_Active 1-hydroxy-xanthines inhibit both isolated Caf1 (CNOT7) enzyme and human Caf1-containing complexes that also contain the second nuclease subunit Ccr4 (CNOT6L) to a similar extent, indicating that the active site of the Caf1 nuclease subunit does not undergo substantial conformational change when bound to other Ccr4-Not subunits. 31180491_Data show that incorporation of ATP-dependent RNA helicase eIF4A-2 (eIF4A2) into the CCR4-NOT complex inhibits CCR4-NOT transcription complex subunit 7 (CNOT7) deadenylation activity. 32160402_CNOT7 depletion reverses natural killer cell resistance by modulating the tumor immune microenvironment of hepatocellular carcinoma. 33021411_Frequent loss of BTG1 activity and impaired interactions with the Caf1 subunit of the Ccr4-Not deadenylase in non-Hodgkin lymphoma. 33412213_CNOT7 modulates biological functions of ovarian cancer cells via AKT signaling pathway. 33757599_Coping with brain amyloid: genetic heterogeneity and cognitive resilience to Alzheimer's pathophysiology. 34038562_Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex. | ENSMUSG00000031601 | Cnot7 | 3788.394908 | 0.9691648 | -0.045186105 | 0.05563439 | 6.575383e-01 | 4.174304e-01 | 7.552689e-01 | No | Yes | 3485.594997 | 618.618401 | 3538.091648 | 627.920763 | |
| ENSG00000198825 | 22876 | INPP5F | protein_coding | Q9Y2H2 | FUNCTION: Inositol 4-phosphatase which mainly acts on phosphatidylinositol 4-phosphate. May be functionally linked to OCRL, which converts phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol, for a sequential dephosphorylation of phosphatidylinositol 4,5-bisphosphate at the 5 and 4 position of inositol, thus playing an important role in the endocytic recycling (PubMed:25869669). Regulator of TF:TFRC and integrins recycling pathway, is also involved in cell migration mechanisms (PubMed:25869669). Modulates AKT/GSK3B pathway by decreasing AKT and GSK3B phosphorylation (PubMed:17322895). Negatively regulates STAT3 signaling pathway through inhibition of STAT3 phosphorylation and translocation to the nucleus (PubMed:25476455). Functionally important modulator of cardiac myocyte size and of the cardiac response to stress (By similarity). May play a role as negative regulator of axon regeneration after central nervous system injuries (By similarity). {ECO:0000250|UniProtKB:Q8CDA1, ECO:0000269|PubMed:17322895, ECO:0000269|PubMed:25476455, ECO:0000269|PubMed:25869669}. | 3D-structure;Alternative splicing;Coated pit;Endosome;Hydrolase;Membrane;Phosphoprotein;Reference proteome | The protein encoded by this gene is an inositol 1,4,5-trisphosphate (InsP3) 5-phosphatase and contains a Sac domain. The activity of this protein is specific for phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2011]. | hsa:22876; | axon [GO:0030424]; clathrin-coated endocytic vesicle [GO:0045334]; clathrin-coated pit [GO:0005905]; dendrite [GO:0030425]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; intracellular membrane-bounded organelle [GO:0043231]; neuronal cell body [GO:0043025]; recycling endosome [GO:0055037]; inositol monophosphate 1-phosphatase activity [GO:0008934]; inositol monophosphate 3-phosphatase activity [GO:0052832]; inositol monophosphate 4-phosphatase activity [GO:0052833]; phosphatidylinositol phosphate 4-phosphatase activity [GO:0034596]; phosphatidylinositol phosphate 5-phosphatase activity [GO:0034595]; phosphatidylinositol-4-phosphate phosphatase activity [GO:0043812]; protein homodimerization activity [GO:0042803]; adult locomotory behavior [GO:0008344]; cardiac muscle hypertrophy in response to stress [GO:0014898]; clathrin-dependent endocytosis [GO:0072583]; negative regulation of axon regeneration [GO:0048681]; negative regulation of peptidyl-serine phosphorylation [GO:0033137]; negative regulation of tyrosine phosphorylation of STAT protein [GO:0042532]; phosphatidylinositol biosynthetic process [GO:0006661]; phosphatidylinositol catabolic process [GO:0031161]; phosphatidylinositol dephosphorylation [GO:0046856]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of receptor recycling [GO:0001921]; regulation of cell motility [GO:2000145]; regulation of endocytic recycling [GO:2001135]; regulation of protein kinase B signaling [GO:0051896] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 19875726_Inpp5f is a polyphosphoinositide phosphatase that regulates cardiac hypertrophic responsiveness. 25476455_These findings suggest that INPP5F is a potential tumor suppressor in gliomas via inhibition of STAT3 pathway, and that deregulation of INPP5F may lead to contribution to gliomagenesis. 25869669_Sac2 colocalizes with early endosomal markers and is recruited to transferrin-containing vesicles during endocytic recycling. 26430724_we identified inositol polyphosphate-5-phosphatase F (INPP5F) as a prognostic factor for progression-free survival in Chronic lymphocytic leukemia 32693431_Imprinting aberrations of SNRPN, ZAC1 and INPP5F genes involved in the pathogenesis of congenital heart disease with extracardiac malformations. | ENSMUSG00000042105 | Inpp5f | 583.947629 | 1.0255805 | 0.036440791 | 0.11721304 | 9.737071e-02 | 7.550080e-01 | 9.224051e-01 | No | Yes | 463.572577 | 73.025323 | 450.532789 | 71.045107 | |
| ENSG00000203667 | 116228 | COX20 | protein_coding | Q5RI15 | FUNCTION: Essential for the assembly of the mitochondrial respiratory chain complex IV (CIV), also known as cytochrome c oxidase (PubMed:23125284). Acts as a chaperone in the early steps of cytochrome c oxidase subunit II (MT-CO2/COX2) maturation, stabilizing the newly synthesized protein and presenting it to metallochaperones SCO1/2 which in turn facilitates the incorporation of the mature MT-CO2/COX2 into the assembling CIV holoenzyme (PubMed:24403053). {ECO:0000269|PubMed:23125284, ECO:0000269|PubMed:24403053}. | Acetylation;Alternative splicing;Disease variant;Membrane;Mitochondrion;Mitochondrion inner membrane;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a protein that plays a role in the assembly of cytochrome C oxidase, an important component of the respiratory pathway. It contains two transmembrane helices and localizes to the mitochondrial membrane. Mutations in this gene can cause mitochondrial complex IV deficiency, which results in ataxia and muscle hypotonia. There are multiple pseudogenes for this gene. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2015]. | hsa:116228; | integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; mitochondrial cytochrome c oxidase assembly [GO:0033617] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 22678713_These results suggest that HNRNPU, FAM36A, and NCRNA00201 are not major genes for microcephaly and corpus callosum abnormalities but are good candidates for intellectual disability (ID) and seizures. 23125284_The function of the human gene FAM36A/COX20 in complex IV assembly and role of the gene in complex IV deficiency. 24202787_This study deministrated that phenotypic spectrum of mutation in COX20 to a recessively inherited, early-onset dystonia-ataxia syndrome that is characterized by reduced complex IV activity. 24403053_COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. 29154948_data shows that by unbalancing the amount of TMEM177, newly synthesized COX2 accumulates in a COX20-associated state. 30656193_Study reports on four subjects with features that include childhood hypotonia, areflexia, ataxia, dysarthria, dystonia, and sensory neuropathy. Exome sequencing in all four subjects identified the same novel COX20 variants. One variant affected the splice donor site of intron-one (c.41A>G), while the other variant (c.157+3G>C) affected the splice donor site of intron-two. 31079202_COX20 mutation are associated with autosomal recessive axonal neuropathy and static encephalopathy. 33751098_Bi-allelic loss of function variants in COX20 gene cause autosomal recessive sensory neuronopathy. | ENSMUSG00000026500 | Cox20 | 741.717659 | 1.0394308 | 0.055793663 | 0.11093038 | 2.533002e-01 | 6.147608e-01 | 8.570215e-01 | No | Yes | 752.327786 | 121.454483 | 720.952055 | 116.403500 | |
| ENSG00000204406 | 55777 | MBD5 | protein_coding | Q9P267 | FUNCTION: Binds to heterochromatin. Does not interact with either methylated or unmethylated DNA (in vitro). | Alternative splicing;Chromosome;Mental retardation;Nucleus;Reference proteome | This gene encodes a member of the methyl-CpG-binding domain (MBD) family. The MBD consists of about 70 residues and is the minimal region required for a methyl-CpG-binding protein binding specifically to methylated DNA. In addition to the MBD domain, this protein contains a PWWP domain (Pro-Trp-Trp-Pro motif), which consists of 100-150 amino acids and is found in numerous proteins that are involved in cell division, growth and differentiation. Mutations in this gene cause an autosomal dominant type of cognitive disability. The encoded protein interacts with the polycomb repressive complex PR-DUB which catalyzes the deubiquitination of a lysine residue of histone 2A. Haploinsufficiency of this gene is associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Alternatively spliced transcript variants have been found, but their full-length nature is not determined. [provided by RefSeq, Jul 2017]. | hsa:55777; | chromocenter [GO:0010369]; chromosome [GO:0005694]; extracellular exosome [GO:0070062]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; glucose homeostasis [GO:0042593]; nervous system development [GO:0007399]; positive regulation of growth hormone receptor signaling pathway [GO:0060399]; regulation of behavior [GO:0050795]; regulation of multicellular organism growth [GO:0040014] | 19904302_Haploinsufficiency of MBD5 is associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures.( 20700456_MBD5 and MBD6 are unlikely to be methyl-binding proteins, yet they may contribute to the formation or function of heterochromatin. 21271666_2q23 de novo microdeletion involving the MBD5 gene in a patient with developmental delay, postnatal microcephaly and distinct facial features. 21981781_MBD5 is a single causal locus as shown by 2q23.1 microdeletion syndrome with roles in intellectual disability, epilepsy, and autism spectrum disorder 23055267_MBD5 was tied to neurodevelopmental disorders following the identification of microdeletions on chromosome 2q22-2q23. 23422940_Identified de novo intragenic deletions of MBD5 in three patients. 23587880_study demonstrates that haploinsufficiency of MBD5 causes diverse phenotypes, yields insight into the spectrum of resulting neurodevelopmental and behavioral psychopathology and provides clinical context for interpretation of MBD5 structural variations. 23632792_The features associated with a deletion, mutation or duplication of MBD5 and the gene expression changes observed support MBD5 as a dosage-sensitive gene critical for normal development. 24634419_We studied and showed that both MBD5 and MBD6 interact with the mammalian PR-DUB Polycomb protein complex in a mutually exclusive manner, and that the MBD of MBD5 and MBD6 is both necessary and sufficient to mediate this interaction. 24885232_A genomic copy number variant analysis implicates the MBD5 and HNRNPU genes in Chinese children with infantile spasms and expands the clinical spectrum of 2q23.1 deletion. 25271084_Circadian rhythm gene expression altered by haploinsufficiency of MBD5. 25853262_Results show that when MBD5 and RAI1 are haploinsufficient, they perturb several common pathways that are linked to neuronal and behavioral development. 25966365_Reduced MBD5 dosage leads to mRNA and microRNA expression patterns and DNA methylation patterns more characteristic of differentiating than proliferating neural stem cells. This balance change may underlie neurodevelopmental disorders. 28295210_Based on segregation analysis of a patient (case 296491) with an intragenic deletion of the MBD5 gene, we classified deletions of exons 3 to 4 of the 5' untranslated region (UTR) of this gene as probably benign. The deletion of exon 3 was shared with the father and paternal uncle, both unaffected 28807762_A novel frameshift mutation c.254_255delGA (p.Arg85Asnfs*6) in the MBD5 gene was identified in a family with intellectual disability and epilepsy. 33427406_MBD5-related intellectual disability in a Vietnamese child. 33510365_Long-read whole-genome sequencing identified a partial MBD5 deletion in an exome-negative patient with neurodevelopmental disorder. 34459404_Early-Onset Dementia Associated with a Heterozygous, Nonsense, and de novo Variant in the MBD5 Gene. 35385942_[Clinical phenotypes and genetic features of epilepsy children with MBD5 gene variants]. | ENSMUSG00000036792 | Mbd5 | 426.769040 | 0.8249234 | -0.277667910 | 0.13869877 | 4.039174e+00 | 4.445558e-02 | No | Yes | 466.357619 | 79.786417 | 593.447040 | 101.575178 | ||
| ENSG00000204599 | 56658 | TRIM39 | protein_coding | Q9HCM9 | FUNCTION: [Isoform 1]: E3 ubiquitin-protein ligase (PubMed:22529100). May facilitate apoptosis by inhibiting APC/C-Cdh1-mediated poly-ubiquitination and subsequent proteasome-mediated degradation of the pro-apoptotic protein MOAP1 (PubMed:19100260, PubMed:22529100). Regulates the G1/S transition of the cell cycle and DNA damage-induced G2 arrest by stabilizing CDKN1A/p21 (PubMed:23213251). Positively regulates CDKN1A/p21 stability by competing with DTL for CDKN1A/p21 binding, therefore disrupting DCX(DTL) E3 ubiquitin ligase complex-mediated CDKN1A/p21 ubiquitination and degradation (PubMed:23213251). {ECO:0000269|PubMed:19100260, ECO:0000269|PubMed:22529100, ECO:0000269|PubMed:23213251}.; FUNCTION: [Isoform 2]: Regulates the G1/S transition of the cell cycle and DNA damage-induced G2 arrest by stabilizing CDKN1A/p21 (PubMed:23213251). Positively regulates CDKN1A/p21 stability by competing with DTL for CDKN1A/p21 binding, therefore disrupting DCX(DTL) E3 ubiquitin ligase complex-mediated CDKN1A/p21 ubiquitination and degradation (PubMed:23213251). Negatively regulates the canonical NF-kappa-B signaling pathway via stabilization of CACTIN in an ubiquitination-independent manner (PubMed:26363554). {ECO:0000269|PubMed:23213251, ECO:0000269|PubMed:26363554}. | 3D-structure;Alternative splicing;Apoptosis;Cell cycle;Coiled coil;Cytoplasm;Metal-binding;Mitochondrion;Nucleus;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The function of this protein has not been identified. This gene lies within the major histocompatibility complex class I region on chromosome 6. Alternate splicing results in two transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:56658; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; apoptotic process [GO:0006915]; innate immune response [GO:0045087]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; positive regulation of apoptotic signaling pathway [GO:2001235]; protein stabilization [GO:0050821]; protein ubiquitination [GO:0016567]; regulation of cell cycle G1/S phase transition [GO:1902806]; regulation of gene expression [GO:0010468] | 19100260_Data suggest that TRIM39 can promote apoptosis signalling through stabilization of MOAP-1. 19851445_Observational study of gene-disease association. (HuGE Navigator) 20875797_these findings suggest that RNF39 and TRIM39 are involved in the etiology of Behcet's disease. 23213251_TRIM39 has a role in regulating cell cycle progression and the balance between cytostasis and apoptosis after DNA damage via stabilizing p21 23213260_analysis of ubiquitylation of p53 by the APC/C inhibitor Trim39 23707810_TRIM39R, but not TRIM39B, regulates type I interferon response 26363554_TRIM39 negatively regulates the NFkappaB signaling pathway possibly via stabilization of cactin. 33311639_Nasopharyngeal carcinoma MHC region deep sequencing identifies HLA and novel non-HLA TRIM31 and TRIM39 loci. 33846303_TRIM39 deficiency inhibits tumor progression and autophagic flux in colorectal cancer via suppressing the activity of Rab7. 35174936_TRIM39 is a poor prognostic factor for patients with estrogen receptor-positive breast cancer and promotes cell cycle progression. | ENSMUSG00000045409 | Trim39 | 632.498794 | 1.1030517 | 0.141500408 | 0.12613796 | 1.281423e+00 | 2.576346e-01 | 6.294719e-01 | No | Yes | 662.508064 | 49.294332 | 589.591226 | 43.927941 |
| ENSG00000204710 | 387778 | SPDYC | protein_coding | Q5MJ68 | FUNCTION: Promotes progression through the cell cycle via binding and activation of CDK1 and CDK2. Involved in the spindle-assembly checkpoint. Required for recruitment of MAD2L1, BUBR1 and BUB1 to kinetochores. Required for the correct localization of the active form of Aurora B in prometaphase. {ECO:0000269|PubMed:15611625, ECO:0000269|PubMed:20605920}. | Cell cycle;Cytoplasm;Reference proteome | hsa:387778; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; protein kinase binding [GO:0019901]; cell cycle [GO:0007049] | 18802405_CDK-Speedy/Ringo C complexes positively regulate cell cycle progression during the late S and G(2) phases of the cell cycle. 20605920_Results indicate a role for RINGO C in the mitotic checkpoint, which might be mediated by defective recruitment of SAC components and deregulation of the activity of Aurora kinase B. | 168.670559 | 0.8948213 | -0.160328505 | 0.23195419 | 4.573290e-01 | 4.988749e-01 | No | Yes | 140.585032 | 28.187635 | 161.645873 | 32.283180 | |||||
| ENSG00000205593 | 414918 | DENND6B | protein_coding | Q8NEG7 | FUNCTION: Guanine nucleotide exchange factor (GEF) for RAB14. Also has some, lesser GEF activity towards RAB35. {ECO:0000269|PubMed:22595670}. | Cytoplasm;Endosome;Guanine-nucleotide releasing factor;Reference proteome | hsa:414918; | cytosol [GO:0005829]; recycling endosome [GO:0055037]; guanyl-nucleotide exchange factor activity [GO:0005085] | ENSMUSG00000015377 | Dennd6b | 90.401105 | 1.1768505 | 0.234931103 | 0.27785520 | 7.187514e-01 | 3.965538e-01 | No | Yes | 96.175794 | 16.410016 | 82.044754 | 14.069170 | ||||
| ENSG00000205609 | 728689 | EIF3CL | protein_coding | B5ME19 | FUNCTION: Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis. The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre-initiation complex (43S PIC). The eIF-3 complex stimulates mRNA recruitment to the 43S PIC and scanning of the mRNA for AUG recognition. The eIF-3 complex is also required for disassembly and recycling of post-termination ribosomal complexes and subsequently prevents premature joining of the 40S and 60S ribosomal subunits prior to initiation. The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation, including cell cycling, differentiation and apoptosis, and uses different modes of RNA stem-loop binding to exert either translational activation or repression. {ECO:0000250|UniProtKB:Q99613}. | Acetylation;Cytoplasm;Initiation factor;Phosphoprotein;Protein biosynthesis;Reference proteome | The protein encoded by this gene is a core subunit of the eukaryotic translation initiation factor 3 (eIF3) complex. The encoded protein is nearly identical to another protein, eIF3c, from a related gene. The eIF3 complex binds the 40S ribosome and mRNAs to enable translation initiation. Several transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Dec 2015]. | hsa:728689; | eukaryotic 43S preinitiation complex [GO:0016282]; eukaryotic 48S preinitiation complex [GO:0033290]; eukaryotic translation initiation factor 3 complex [GO:0005852]; translation initiation factor activity [GO:0003743]; translation initiation factor binding [GO:0031369]; formation of cytoplasmic translation initiation complex [GO:0001732]; translational initiation [GO:0006413] | ENSMUSG00000030738 | Eif3c | 142.958460 | 0.9480594 | -0.076950649 | 0.26508116 | 8.244574e-02 | 7.740100e-01 | No | Yes | 172.713444 | 29.010193 | 188.653796 | 31.521196 | |||
| ENSG00000205643 | 150383 | CDPF1 | protein_coding | Q6NVV7 | Reference proteome | hsa:150383; | ENSMUSG00000064284 | Cdpf1 | 198.628619 | 0.7532573 | -0.408785351 | 0.19669759 | 4.259583e+00 | 3.902950e-02 | No | Yes | 168.901378 | 24.269861 | 221.346314 | 31.580957 | ||||||
| ENSG00000205771 | 440278 | CATSPER2P1 | transcribed_unprocessed_pseudogene | Catsper genes belong to a family of putative cation channels that are specific to spermatozoa and localize to the flagellum. This gene is part of a tandem repeat on chromosome 15q15; this copy of the gene is thought to be a pseudogene. [provided by RefSeq, Oct 2008]. | 30.872476 | 0.8565520 | -0.223387194 | 0.50744742 | 1.907434e-01 | 6.622986e-01 | No | Yes | 35.889223 | 8.708950 | 41.867204 | 9.993452 | ||||||||||
| ENSG00000206530 | 55779 | CFAP44 | protein_coding | Q96MT7 | FUNCTION: Flagellar protein involved in sperm flagellum axoneme organization and function. {ECO:0000250|UniProtKB:E9Q5M6}. | Alternative splicing;Cell projection;Cilium;Coiled coil;Cytoplasm;Cytoskeleton;Flagellum;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:55779; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; motile cilium [GO:0031514]; peptidase activity [GO:0008233]; cilium assembly [GO:0060271]; cilium-dependent cell motility [GO:0060285]; microtubule cytoskeleton organization [GO:0000226]; sperm axoneme assembly [GO:0007288] | 25602530_Results show that the rs13064411 polymorphism in the WDR52 gene was associated with a modified effect of statin therapy on serum LDL and total cholesterol levels. 28552195_biallelic mutations in either CFAP43 or CFAP44 can cause sperm flagellar abnormalities and impair sperm motility. 29277146_Study demonstrated that biallelic mutations in CFAP44 and CFAP43 cause multiple morphological abnormalities of the sperm flagella (MMAF). These results provide researchers with a new insight to understand the genetic etiology of MMAF and to identify new loci for genetic counselling of MMAF. 29449551_demonstrate that CFAP43 and CFAP44 have a similar structure with a unique axonemal localization and are necessary to produce functional flagella in species ranging from Trypanosoma to human 30904354_important cause of multiple morphological abnormalities of the sperm flagellum in the Chinese population | ENSMUSG00000071550 | Cfap44 | 199.335558 | 1.6788610 | 0.747482767 | 0.26656726 | 7.975925e+00 | 4.740354e-03 | No | Yes | 238.554696 | 65.314424 | 148.230397 | 40.768163 | |||
| ENSG00000213722 | 23564 | DDAH2 | protein_coding | O95865 | FUNCTION: Hydrolyzes N(G),N(G)-dimethyl-L-arginine (ADMA) and N(G)-monomethyl-L-arginine (MMA) which act as inhibitors of NOS. Has therefore a role in the regulation of nitric oxide generation. {ECO:0000269|PubMed:10493931}. | Cytoplasm;Direct protein sequencing;Hydrolase;Mitochondrion;Reference proteome | This gene encodes a dimethylarginine dimethylaminohydrolase. The encoded enzyme functions in nitric oxide generation by regulating the cellular concentrations of methylarginines, which in turn inhibit nitric oxide synthase activity. The protein may be localized to the mitochondria. Alternative splicing resulting in multiple transcript variants. [provided by RefSeq, Dec 2014]. | hsa:23564; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; mitochondrion [GO:0005739]; amino acid binding [GO:0016597]; catalytic activity [GO:0003824]; dimethylargininase activity [GO:0016403]; arginine catabolic process [GO:0006527]; arginine metabolic process [GO:0006525]; citrulline metabolic process [GO:0000052]; negative regulation of apoptotic process [GO:0043066]; nitric oxide biosynthetic process [GO:0006809]; nitric oxide mediated signal transduction [GO:0007263]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; regulation of nitric-oxide synthase activity [GO:0050999] | 12370443_In cultured endothelial cells, heterologously expressed human DDAH II was S-nitrosylated after cytokine induced expression of the inducible NOS isoforms. 14550280_DDAH2 expression in endothelial cells is altered by genetic variation in a basal promoter element 16920729_messenger RNA and protein were demonstrated in first trimester placental tissue, primary extravillous trophoblasts and extravillous trophoblast-derived cell lines. 16928504_Observational study of gene-disease association. (HuGE Navigator) 17002794_Allelic variation for a polymorphism in the DDAH II gene may influence asymmetrical dimethyl arginine concentrations, hence the severity of organ failure in septic shock patients. 17002794_Observational study of gene-disease association. (HuGE Navigator) 18251679_The purpose of this study was to investigate whether there is any association between preeclampsia and eNOS, DDAH, and VEGF gene polymorphisms.Polymorphisms in eNOS, DDAH, and VEGF gene do not seem to be risk factors for preeclampsia. 18342305_Lysophosphatidylcholine (LPC) impairs DDAH/ADMA/NOS/NO pathway, and DDAH2 gene transfer could improve the LPC-elicited impairments in endothelial cells. 19143821_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19250061_Observational study of gene-disease association. (HuGE Navigator) 19250061_results suggest that the DDAH2 common variant may play a protective role in the development of Intracerebral Hemorrhage, implicating that the DDAH2/ADMA pathway may act as a critical regulator of cerebral small-vessel disorders 19570459_Low expression of DDAH-2 in placenta and increased serum asymmetric dimethylarginine level might confer susceptibility to preeclampsia. 19666123_Observational study of gene-disease association. (HuGE Navigator) 19666123_Single nucleotide polymorphisms in the DDAH2 gene are associated with blood pressure levels, prevalence of hypertension, and left ventricular mass and function in the general population. 19822957_DDAH2 mRNA expression is inversely associated with some cardiovascular risk-related features. 19851445_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20010544_Observational study of gene-disease association. (HuGE Navigator) 20209122_DDAH1 and DDAH2 polymorphisms are strongly and additively associated with serum ADMA concentrations in individuals with type 2 diabetes 20209122_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20936901_Observational study of gene-disease association. (HuGE Navigator) 21677405_a possible association between A allele / AA genotype for DDAH2 SNP1 (-1151 C/A, rs805304) and G allele / GG genotype for SNP2 (-449 C/G, rs805305) with CVD in male 35-50 year-old Egyptian patients was observed. 21898353_DDAH-2 could play an important role in IL-1beta-induced NO production and in osteoarthritis pathogenesis. 22428028_The -449G single nucleotide polymorphism within the DDAH2 gene was associated with both decreased plasma asymmetric dimethylarginine and an increased likelihood of presenting with 'cold' shock in pediatric sepsis. 22558392_A functional polymorphism of the DDAH2 gene may confer increased risk for type 2 diabetes by affecting insulin sensitivity 22579530_Found that SNP rs2272592 in DDAH2 is associated with type 2 diabetes but SNP rs805304 in DDAH2 is not. DDAH2 SNP rs2272592 AG+GG genotypes are associated with genetic susceptibility to type 2 diabetes in Korean population. 22923027_No association was observed between the DDAH2 polymorphisms at rs805305 and rs2272592 and coronary heart disease. 23129820_DDAH2-1151 A/C polymorphism is associated with Chronic renal impairment in type 2 diabetes. 23171931_Suppression of DDAH2 expression is a culprit for homocysteine-induced impairments of DDAH/ADMA/NOS/NO pathway in endothelial cells. 23430976_Glucose-stimulated insulin secretion is increased in Ddah2-transgenic pancreatic islets by 33%, compared to its levels in wild-type mice. 23770786_There is a significant difference in distribution of DDAH2 gene polymorphism among hemodialysis patients compared to healthy individuals. 24125425_The rs9267551 functional variant of the DDAH2 gene is associated with chronic kidney disease with carriers of the C allele having a lower risk of renal dysfunction independently from several confounders 24134589_Enhancing pulmonary DDAH II activity attenuates LPS-mediated lung leak in acute lung injury. 24155659_Our approaches revealed signature candidates of differentially hypermethylated genes of DDAH2 and DUSP1 which can be further developed as potential biomarkers for OSCC as diagnostic, prognostic and therapeutic targets in the future. 24928011_-476 to -469 of the DDAH2 promoter was a NF-kB responsive element and is important for the transactivation of DDAH2. 24934151_Homocysteine disrupts EPCs function via inducing the hypermethylation of DDAH2 promoter, suggesting a key role of epigenetic mechanism in the progression of atherosclerosis 25152204_DDAH-1 is a specific molecular target for portal pressure reduction, through actions on ADMA-mediated regulation of eNOS activity. 25194333_increased ADMA levels in rheumatoid arthritis do not appear to relate to DDAH genetic polymorphisms 25236572_Our results suggest that the rs805304 C allele of the DDAH gene was associated with decreased risk of myocardial infarction and decreased risk of obesity. 25701782_The percentage of senescent endothelial progenitor cells increased while the expression of DDAH2 decreased concomitantly with an increase in the plasma levels of asymmetric dimethylarginine in patients with type 2 diabetes mellitus. 26082478_Inhibiting the expression of DDAH1, but not DDAH2, resulted in a significant increase in the sensitivity of the EVT cell line SGHPL-4 to tumour necrosis factor related apoptosis inducing ligand (TRAIL) induced apoptosis 26515557_expression of DDAH2 is associated with invasiveness of lung adenocarcinoma via tumor angiogenesis 26786611_The main finding from this study was that it demonstrated that the C-allele of rs3087894 in DDAH1 is a risk factor for hypertension in the Kazakh group but a protective factor in the Uygur group. In addition, we did not find any genotype of DDAH1 and DDAH2 associated with hypertension in the Han group. 28150318_Exogenous human DDAH2 gene promotes differentiation of rabbit bone marrow-derived endothelial progenitor cells into mature endothelial cells. 28590543_genetic variations in the DDAH2 gene may influence the ADMA concentration and erythropoietin resistance in MHD patients 30538005_The presence of the rs805305 SNP in the DDAH2 gene was determined using genomic DNA extraction from buffy coat and whole blood samples by LGC group (Hertfordshire, UK). 31409409_DDAH2 rs9267551 polymorphism is significantly associated with myocardial infarction in type 2 diabetes mellitus patients of European ancestry. 34283907_Polymorphism (-499C/G) in DDAH2 promoter may act as a protective factor for metabolic syndrome: A case-control study in Azar-Cohort population. 34787071_Interference of KLF9 relieved the development of gestational diabetes mellitus by upregulating DDAH2. | ENSMUSG00000007039 | Ddah2 | 403.018171 | 0.9396128 | -0.089861797 | 0.13489613 | 4.424326e-01 | 5.059507e-01 | No | Yes | 384.546629 | 61.421141 | 411.594874 | 65.609612 | ||
| ENSG00000213918 | 1773 | DNASE1 | protein_coding | P24855 | FUNCTION: Serum endocuclease secreted into body fluids by a wide variety of exocrine and endocrine organs (PubMed:2251263, PubMed:11241278, PubMed:2277032). Expressed by non-hematopoietic tissues and preferentially cleaves protein-free DNA (By similarity). Among other functions, seems to be involved in cell death by apoptosis (PubMed:11241278). Binds specifically to G-actin and blocks actin polymerization (By similarity). Together with DNASE1L3, plays a key role in degrading neutrophil extracellular traps (NETs) (By similarity). NETs are mainly composed of DNA fibers and are released by neutrophils to bind pathogens during inflammation (By similarity). Degradation of intravascular NETs by DNASE1 and DNASE1L3 is required to prevent formation of clots that obstruct blood vessels and cause organ damage following inflammation (By similarity). {ECO:0000250|UniProtKB:P00639, ECO:0000250|UniProtKB:P21704, ECO:0000250|UniProtKB:P49183, ECO:0000269|PubMed:11241278, ECO:0000269|PubMed:2251263, ECO:0000269|PubMed:2277032}. | 3D-structure;Actin-binding;Alternative splicing;Apoptosis;Calcium;Cytoplasmic vesicle;Direct protein sequencing;Disulfide bond;Endonuclease;Glycoprotein;Hydrolase;Nuclease;Nucleus;Pharmaceutical;Reference proteome;Secreted;Signal;Systemic lupus erythematosus | This gene encodes a member of the DNase family. This protein is stored in the zymogen granules of the nuclear envelope and functions by cleaving DNA in an endonucleolytic manner. At least six autosomal codominant alleles have been characterized, DNASE1*1 through DNASE1*6, and the sequence of DNASE1*2 represented in this record. Mutations in this gene have been associated with systemic lupus erythematosus (SLE), an autoimmune disease. A recombinant form of this protein is used to treat the one of the symptoms of cystic fibrosis by hydrolyzing the extracellular DNA in sputum and reducing its viscosity. Alternate transcriptional splice variants of this gene have been observed but have not been thoroughly characterized. [provided by RefSeq, Jul 2008]. | hsa:1773; | extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; nuclear envelope [GO:0005635]; nucleus [GO:0005634]; zymogen granule [GO:0042588]; actin binding [GO:0003779]; deoxyribonuclease I activity [GO:0004530]; DNA binding [GO:0003677]; apoptotic process [GO:0006915]; DNA catabolic process [GO:0006308]; DNA catabolic process, endonucleolytic [GO:0000737]; neutrophil activation involved in immune response [GO:0002283]; regulation of acute inflammatory response [GO:0002673]; regulation of neutrophil mediated cytotoxicity [GO:0070948] | 11332641_Observational study of genotype prevalence. (HuGE Navigator) 12005024_results show that the chief cells of the stomach produce DNase I 12708782_Deficiencies in this gene may contribute to SLE; gene therapy might be possible (REVIEW) 14613299_The A/T mutation of the DNASE1 gene is absent in both Tunisian systemic lupus erythematosus and in controls. 14688237_The studies of immunological properties and the tissue-distribution patterns of DNase I indicated that the canine enzyme is more closely related to the human DNase I than to other mammalian DNases I 15188364_serum Dnase1 in cooperation with the plasminogen system guarantees a fast and effective breakdown of chromatin during necrosis by the combined cleavage of DNA as well as of DNA binding proteins 15333586_Observational study of gene-disease association. (HuGE Navigator) 15363449_Observational study of genotype prevalence. (HuGE Navigator) 16352456_Results describe chromatin disposal during necrosis and the involvement of Deoxyribonuclease 1 in this process with respect to its possible role in the prevention of anti-nuclear auto-immunity. 16382368_Observational study of genotype prevalence. (HuGE Navigator) 16449364_Observational study of gene-disease association. (HuGE Navigator) 16771825_human DNASE1 expression is regulated through the use of alternative promoter and alternative splicing 16877481_Observational study of gene-disease association. (HuGE Navigator) 17032129_Observational study of genotype prevalence. (HuGE Navigator) 17320453_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17360785_Observational study of gene-disease association. (HuGE Navigator) 17405189_Observational study of genotype prevalence. (HuGE Navigator) 17405189_This study is the first to reveal an extremely high frequency of DNASE1*1 among African populations. Caucasians and Asians had a lower frequency of DNASE1*1 than the African groups. 17588132_Compared to other ethnic populations, Han Chinese had its own unique DNase I gene distribution characteristics 17588132_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 18174230_Observational study of gene-disease association. (HuGE Navigator) 18311594_Han Chinese myocardial infarction (MI) patients, but deoxyribonuclease I gene polymorphisms are not associated with susceptibility to MI in Han Chinese 18311594_Observational study of gene-disease association. (HuGE Navigator) 19022625_In patients with autoimmune thyroid disease (AITD), a novel mutation (1218G>A, exon 5) and multiple polymorphisms were identified in the DNASE1 gene. 19022625_Observational study of gene-disease association. (HuGE Navigator) 19055475_DNASE1 polymorphism appears to affect the specific activity, heat sensitivity, and optimum pH of the DNase I enzyme. 19055475_Observational study of genotype prevalence. (HuGE Navigator) 19181929_human recombinant DNase I up-regulates cell surface Fas expression and induces increased susceptibility of T cells to Fas-mediated apoptosis 19318394_DNase1 (deoxyribonuclease I) activity was significantly lower in patients with Systemic lupus erythematosus than in controls. 19360410_Observational study of gene-disease association. (HuGE Navigator) 19360410_Show no particular variant in exon2 sequence of DNASE1 gene among Tunisian patients affected with systemic lupus erythematosus, rheumatoid arthritis and Sjogren syndrome and healthy subjects. 19362700_deficiency is linked to systemic lupus erythematosus 19844716_Low DNase1 activity is associated to the active phase of type III or IV lupus erythromatosus nephropathy. 19863681_A significant association of HumDN1 Variable Number of Tandem Repeats polymorphism in DNASE1 gene with systemic lupus erythematosus, is reported. 19863681_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20417303_The objectives of this study were to clarify genetic and biochemical aspects of 12 non-synonymous SNPs in the human gene (DNASE1), potentially giving rise to an alteration in the in vivo DNase I activity levels. 20439745_show that serum endonuclease DNase1 is essential for disassembly of neutrophil extracellular traps. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 20856893_Reduction in renal Dnase1 expression and activity is limited to mice and SLE patients with signs of membranoproliferative nephritis, and may be a critical event in the development of severe forms of lupus nephritis. 21233855_Modified DNase1 can efficiently eliminate apoptosis-resistant cancer cells through apoptosis. 21235399_The genetic aspects of DNASE1 with regard to all the SNPs in wide-ranging ethnic groups could be first demonstrated. Further, there was no correlation of all the polymorphic SNPs other than nonsynonymous ones with serum DNase I activity levels. 21282512_In thymocyte-selected CD4 T cells (T-CD4 T cells) DNase I hypersensitive sites at the 3'-enhancer region of interleukin-4 play an essential role during the induction of IL-4 expression. 22094313_POU1F1 binds to multiple sites at deoxyribonuclease I hypersensitive site II. 22479529_silencing of renal DNaseI gene expression initiates a cascade of inflammatory signals including activation of Toll like receptors and Clec4e, leading to progression of both murine and human lupus nephritis 23183758_DNase I activity in systemic lupus erythematosus patients was lower than in healthy controls 23215638_The novel structure of rhDNase I presented herein reveals the precise location and coordination sphere of the Mg2+ ion bound at subsite IVb along with the orientation of key side chains in the active site. 23225239_a downregulation of the DNASE1 mRNA expression in patients with autoimmune thyroid disease. This might result in degrading less DNA from dying cells, thereby promoting the development of thyroid autoimmunity. 23273922_Early mesangial nephritis initiates a cascade of inflammatory signals that lead to up-regulation of Trap1 and a consequent down-regulation of renal DNaseI by transcriptional interference. 23963431_DNAse I Q222R polymorphism is a potential genetic risk factor for systemic lupus erythematosus in South Indian Tamils. In addition, the mutant allele confers a significant risk for lupus nephritis. 24206041_Single nucleotide polymorphisms in the DNASE1 is associated with autoimmune diseases. 24564745_Our results indicate that increased DNASE1 expression is common to both SLE and RA, while DNase1 reduction activity is specific to SLE. 24676545_In conclusion, elevated DNase I in diabetes may be related to pancreatic injury and could be one of the causes that induce diabetes. 24819173_Characterization of the nonsynonymous single-nucleotide polymorphism variants of human DNASE1. 25771153_HumDN1 polymorphisms were examined in blood of 11 worldwide populations by polymerase chain reaction. 15 genotypes were found leading to the conclusion that HumDN1 variable no. tandem repeats are ethnic group specific. 26065428_TNF-alpha amplifies DNaseI expression in renal tubular cells while IL-1beta promotes nuclear DNaseI translocation in inactive form in lupus nephritis. 26393465_Val66Met in the BDNF gene and two SNPs, Fokl and Apal, in the VDR gene may potentially be associated with DED. Additionally, the association between DED and Val66Met may vary by depression status. 26547219_in Iranians the 3/6 genotype frequency was significantly higher in systemic lupus erythematosus patients than in healthy controls which implies the possible role of this genotype in SLE susceptibility; the 3/4 and 4/6 genotype frequencies were remarkably high in the control group which indicated the protective role of these genotypes against the disease 27082840_DNase I activity was observed to be increased in type 2 diabetes, and high glucose combined with increased DNase I is suggested to aggravate beta-cell apoptosis. 27116004_Single nucleotide polymorphisms producing a loss-of-function variant of the enzymes in DNASE1, DNASE1L3, and DNASE2, possibly serving as a genetic risk factor for autoimmune diseases, were confirmed. 27606473_Although no significant association between Q222R polymorphism in DNAse I gene and the risk of systemic lupus erythematosus was found, the presence of the A allele was associated with an increased risk for the development of nephropathy 28263100_Serum DNase1-activity is significantly decreased in ALD patients, indicating its potential implication in their pathogenesis. Furthermore, DNase1-activity could be used as a new surrogate biomarker for predicting response to AIH treatment. 28321907_Genetic variants in DNase I hypersensitive sites play an important role in carcinogenesis. two novel single nucleotide polymorphisms rs12309362 (odds ratio = 0.64, P = 5.61 x 10(-6) ) and rs9970827 (odds ratio = 0.73, P = 7.23 x 10(-6) ) significantly associated with decreased risk of HCC. 28935869_Cigarette smoke significantly increases the expression of neutrophil and macrophage extracellular traps with coexpression of the pathogenic proteases, neutrophil elastase and matrix metalloproteinases 9 and 12. This response to cigarette smoke was significantly reduced by the addition of DNase 1, which also significantly decreased macrophage numbers and lung proteolysis. 30521874_EndoG is an endonuclease with the unique ability to inactivate another endonuclease, DNase I, and to modulate the development of apoptosis. 30936483_Human deoxyribonuclease I (DNaseI) is overexpressed and encapsidated in hepatitis B virus (HBV) particles in vitro in hypoxic environments and in vivo in cirrhotic patient livers as well as in the serum of infected patients. DNase I induces genome-free hepatitis B virions and is incorporated into hepatitis B viral capsids. A hypoxic environment enhances DNASE1 and restricts HBV. 31541133_Evaluation of the functional effects of genetic variantsmissense and nonsense SNPs, indels and copy number variationsin the gene encoding human deoxyribonuclease I potentially implicated in autoimmunity. 32813937_AAV-mediated gene transfer of DNase I in the liver of mice with colorectal cancer reduces liver metastasis and restores local innate and adaptive immune response. 33891165_Deoxyribonuclease 1 Q222R single nucleotide polymorphism and long-term mortality after acute myocardial infarction. 34006390_The Nexus of cfDNA and Nuclease Biology. | ENSMUSG00000005980 | Dnase1 | 266.900434 | 1.2373409 | 0.307242991 | 0.16434333 | 3.513257e+00 | 6.087967e-02 | No | Yes | 257.768416 | 27.403088 | 211.032635 | 22.513350 | ||
| ENSG00000214226 | 339210 | C17orf67 | protein_coding | Q0P5P2 | Reference proteome;Secreted;Signal | hsa:339210; | extracellular region [GO:0005576] | 19266077_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000072553 | Gm525 | 49.889568 | 0.9937170 | -0.009092996 | 0.39527655 | 5.057680e-04 | 9.820577e-01 | No | Yes | 45.047368 | 10.745147 | 46.973490 | 11.143644 | ||||
| ENSG00000214439 | FAM185BP | transcribed_unprocessed_pseudogene | 41.048132 | 0.9933280 | -0.009657878 | 0.42074009 | 5.257383e-04 | 9.817069e-01 | No | Yes | 40.367619 | 10.242557 | 37.928374 | 9.646633 | ||||||||||||
| ENSG00000214765 | 641977 | SEPTIN7P2 | transcribed_unprocessed_pseudogene | Septins, such as SEPT13, are conserved GTP-binding proteins that function as dynamic, regulatable scaffolds for the recruitment of other proteins. They are involved in membrane dynamics, vesicle trafficking, apoptosis, and cytoskeleton remodeling, as well as infection, neurodegeneration, and neoplasia (Hall et al., 2005 [PubMed 15915442]).[supplied by OMIM, Jul 2008]. | 518.997777 | 1.1438994 | 0.193960176 | 0.12476508 | 2.443738e+00 | 1.179947e-01 | 4.580972e-01 | No | Yes | 579.864834 | 128.826566 | 510.839548 | 113.539021 | |||||||||
| ENSG00000214783 | 84820 | POLR2J4 | lncRNA | 146.134375 | 1.3843632 | 0.469222468 | 0.23202242 | 3.997879e+00 | 4.555755e-02 | No | Yes | 124.411119 | 19.362522 | 96.998619 | 15.205207 | |||||||||||
| ENSG00000215041 | 84461 | NEURL4 | protein_coding | Q96JN8 | FUNCTION: Promotes CCP110 ubiquitination and proteasome-dependent degradation. By counteracting accumulation of CP110, maintains normal centriolar homeostasis and preventing formation of ectopic microtubular organizing centers. {ECO:0000269|PubMed:22261722, ECO:0000269|PubMed:22441691}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Ubl conjugation pathway | The protein encoded by this gene is predicted and it includes two isoforms resulting from two alternatively spliced transcript variants. [provided by RefSeq, Jul 2008]. | hsa:84461; | centriole [GO:0005814]; cytoplasm [GO:0005737]; ubiquitin protein ligase activity [GO:0061630] | 22261722_the NEURL4-HERC2 complex participates in the ubiquitin-dependent regulation of centrosome architecture 22441691_Neurl4 counteracts accumulation of CP110, thereby maintaining normal centriolar homeostasis and preventing formation of ectopic microtubular organizing centres 28385950_The data support a model in which the daughter centriole promotes ciliogenesis through Neurl-4-dependent regulation of CP110 levels at the mother centriole. 35157000_Neuralized-like protein 4 (NEURL4) mediates ADP-ribosylation of mitochondrial proteins. | ENSMUSG00000047284 | Neurl4 | 2352.510670 | 1.1065380 | 0.146052961 | 0.10146152 | 2.120927e+00 | 1.452985e-01 | 5.002025e-01 | No | Yes | 2461.846375 | 342.992737 | 2224.926750 | 310.006817 | |
| ENSG00000215252 | 440270 | GOLGA8B | protein_coding | A8MQT2 | FUNCTION: May be involved in maintaining Golgi structure. {ECO:0000250}. | Alternative splicing;Coiled coil;Golgi apparatus;Membrane;Reference proteome | hsa:440270; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; Golgi cis cisterna [GO:0000137]; Golgi cisterna membrane [GO:0032580]; Golgi organization [GO:0007030]; spindle assembly [GO:0051225] | 745.208450 | 0.8526597 | -0.229958056 | 0.16906197 | 1.843067e+00 | 1.745919e-01 | 5.400585e-01 | No | Yes | 518.476377 | 149.838049 | 614.028059 | 177.426817 | |||||
| ENSG00000221926 | 10626 | TRIM16 | protein_coding | O95361 | FUNCTION: E3 ubiquitin ligase that plays an essential role in the organization of autophagic response and ubiquitination upon lysosomal and phagosomal damages. Plays a role in the stress-induced biogenesis and degradation of protein aggresomes by regulating the p62-KEAP1-NRF2 signaling and particularly by modulating the ubiquitination levels and thus stability of NRF2. Acts as a scaffold protein and facilitates autophagic degradation of protein aggregates by interacting with p62/SQSTM, ATG16L1 and LC3B/MAP1LC3B. In turn, protects the cell against oxidative stress-induced cell death as a consequence of endomembrane damage. {ECO:0000269|PubMed:22629402, ECO:0000269|PubMed:27693506, ECO:0000269|PubMed:30143514}. | Alternative splicing;Coiled coil;Cytoplasm;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | The protein encoded by this gene is a tripartite motif (TRIM) family member that contains two B box domains and a coiled-coiled region that are characteristic of the B box zinc finger protein family. While it lacks a RING domain found in other TRIM proteins, the encoded protein can homodimerize or heterodimerize with other TRIM proteins and has E3 ubiquitin ligase activity. This gene is also a tumor suppressor and is involved in secretory autophagy. [provided by RefSeq, Jan 2017]. | hsa:10626; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; PML body [GO:0016605]; DNA binding [GO:0003677]; interleukin-1 binding [GO:0019966]; NACHT domain binding [GO:0032089]; transferase activity [GO:0016740]; zinc ion binding [GO:0008270]; histone H3 acetylation [GO:0043966]; histone H4 acetylation [GO:0043967]; positive regulation of interleukin-1 beta production [GO:0032731]; positive regulation of keratinocyte differentiation [GO:0045618]; positive regulation of retinoic acid receptor signaling pathway [GO:0048386]; positive regulation of transcription, DNA-templated [GO:0045893]; response to growth hormone [GO:0060416]; response to organophosphorus [GO:0046683]; response to retinoic acid [GO:0032526] | 11919186_role in regulating keratinocyte differentiation 16575408_These results provide evidence for a role of EBBP in innate immunity by enhancing the alternative secretion pathway of IL-1beta. 16636064_EBBP increased betaRARE-transactivating function through its coiled-coil domain 19147277_The estrogen-responsive B box protein (EBBP) restores retinoid sensitivity in retinoid-resistant cancer cells via effects on histone acetylation. 20729920_TRIM16 acts as a tumour suppressor, affecting neuritic differentiation, cell migration and replication through interactions with cytoplasmic vimentin and nuclear E2F1 in neuroblastoma cells. 22629402_via its unique structure, TRIM16 possesses both heterodimerization function with other TRIM proteins and also has E3 ubiquitin ligase activity. 23404198_TRIM16 can promote apoptosis by directly modulating caspase-2 activity in cancer cells. 23422002_data suggest that TRIM16 acts as a novel regulator of both neuroblastoma G 1/S progression and cell differentiation 25333256_Chromatin immunoprecipitation assays revealed TRIM16 directly bound the IFNbeta1 gene promoter. Low level TRIM16 expression in 91 melanoma patient samples, strongly correlated with lymph node metastasis, and, predicted poor patient prognosis. 25843803_results suggest that TRIM16 is a potential pharmacologic target for the treatment of NSCLC and promotion TRIM16 expression might represent a novel strategy to NSCLC metastasis 25866896_Expression of SDMGC and TRIM16 was upregulated in the distant metastasis tissues 26718507_TRIM16 directly regulated the degradation of Gli1 protein via the ubiquitinproteasome pathway. TRIM16 expression was low in breast cancer. It negatively correlated with metastasis and suppressed stem-cell properties in breast cancer cells. 26892350_Results found that the expression of TRIM16 was significantly downregulated in hepatocellular carcinoma cell (HCC) lesions and define TRIM16 as an inhibitor of epithelial-mesenchymal transition and metastasis in HCC. 26902425_Data suggest that TRIM16 and TDP43 are both good prognosis indicators; data shows that TRIM16 inhibits cancer cell viability by a novel mechanism involving interaction and stabilisation of TDP43 with consequent effects on E2F1 and pRb proteins. 27693506_The cooperation between TRIM16 and Galectin-3 in targeting and activation of selective autophagy protects cells from lysosomal damage and Mycobacterium tuberculosis invasion. 27737724_TRIM16 inhibits the migration and invasion via suppressing the Sonic hedgehog signaling pathway in ovarian cancer cells. 27748839_TRIM16 expression is decreased in prostate cancer tissues and overexpression of TRIM16 inhibits cell migration, invasion and the EMT process in vitro in prostate cancer through the transcription factor Snail. 29295721_High TRIM16 expression is associated with drug resistance in Non-Small Cell Lung Cancer. 30143514_TRIM16 acts as a scaffold protein and, by interacting with p62, ULK1, ATG16L1, and LC3B, facilitates autophagic degradation of protein aggregates. 30488195_Papillary thyroid carcinoma was characterized by high expression of ESR2 and AR, which was associated with expression and content of nuclear factors Brn-3A and TRIM16. 31521826_Galectin-3 and TRIM16 coregulate osteogenic differentiation of human bone marrow-derived mesenchymal stem cells at least partly via enhancing autophagy. 31812473_The results indicate that the RASSF6-TRIM16 axis is a key effector in ESCC progression and that RASSF6 serves as a potential target for the treatment of ESCC. 33046716_AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent Vimentin degradation. 33391467_A thermosensitive, reactive oxygen species-responsive, MR409-encapsulated hydrogel ameliorates disc degeneration in rats by inhibiting the secretory autophagy pathway. 34135057_Impaired TRIM16-Mediated Lysophagy in Chronic Obstructive Pulmonary Disease Pathogenesis. 34265287_TRIM16 overexpression inhibits the metastasis of colorectal cancer through mediating Snail degradation. 34452557_Prognostic Significance of Tripartite Motif Containing 16 Expression in Patients with Gastric Cancer. 35230201_CircPTK2 inhibits cell cisplatin (CDDP) resistance by targeting miR-942/TRIM16 axis in non-small cell lung cancer (NSCLC). | ENSMUSG00000047821 | Trim16 | 602.627117 | 0.9979391 | -0.002976281 | 0.13519777 | 4.848824e-04 | 9.824320e-01 | 9.935391e-01 | No | Yes | 578.058171 | 55.473447 | 585.376920 | 56.285462 | |
| ENSG00000222011 | 222234 | FAM185A | protein_coding | Q8N0U4 | Alternative splicing;Reference proteome | hsa:222234; | ENSMUSG00000047221 | Fam185a | 195.199018 | 1.1193824 | 0.162702951 | 0.19684949 | 6.830825e-01 | 4.085273e-01 | No | Yes | 205.788588 | 38.105071 | 177.418535 | 32.884401 | ||||||
| ENSG00000222020 | 101928111 | HDAC4-AS1 | lncRNA | 10.015766 | 1.1511936 | 0.203130489 | 0.81412236 | 6.341963e-02 | 8.011705e-01 | No | Yes | 12.276699 | 4.505683 | 10.549539 | 3.871847 | |||||||||||
| ENSG00000223573 | 257000 | TINCR | protein_coding | A0A2R8Y7D0 | Reference proteome | 163.607971 | 0.8644198 | -0.210195963 | 0.20959115 | 1.011759e+00 | 3.144819e-01 | No | Yes | 143.718532 | 22.335311 | 168.028468 | 26.105005 | |||||||||
| ENSG00000223960 | 101927027 | CHROMR | lncRNA | 247.322704 | 1.1554794 | 0.208491553 | 0.19130338 | 1.202961e+00 | 2.727307e-01 | No | Yes | 271.143319 | 50.469904 | 229.195533 | 42.716911 | |||||||||||
| ENSG00000225096 | 101927293 | lncRNA | 97.855248 | 0.8994766 | -0.152842318 | 0.26656104 | 3.316227e-01 | 5.647051e-01 | No | Yes | 91.063740 | 12.058494 | 103.624674 | 13.801966 | ||||||||||||
| ENSG00000226210 | 100288778 | WASH8P | unprocessed_pseudogene | 343.176885 | 1.0750391 | 0.104389168 | 0.15413455 | 4.598801e-01 | 4.976800e-01 | No | Yes | 373.569422 | 40.276107 | 350.565528 | 37.879032 | |||||||||||
| ENSG00000226686 | 101927667 | LINC01535 | lncRNA | 31273925_Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. 32329845_LINC01535 promotes proliferation and inhibits apoptosis in esophageal squamous cell cancer by activating the JAK/STAT3 pathway. 33174047_lncRNA LINC01535 upregulates BMP2 expression levels to promote osteogenic differentiation via sponging miR36195p. | 22.521931 | 0.7411338 | -0.432194119 | 0.55672712 | 5.898202e-01 | 4.424887e-01 | No | Yes | 18.646550 | 3.522427 | 24.629263 | 4.528521 | ||||||||||
| ENSG00000226864 | 100130887 | ATE1-AS1 | transcribed_unitary_pseudogene | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | 28.512702 | 0.9508658 | -0.072686419 | 0.48895614 | 2.252548e-02 | 8.806976e-01 | No | Yes | 29.979607 | 9.924660 | 32.439324 | 10.656788 | ||||||||||
| ENSG00000227372 | 57212 | TP73-AS1 | transcribed_unitary_pseudogene | 20477830_PDAM knockdown induces cisplatin resistance in glioma cells harboring wild-type p53. The finding suggests that PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes (BCL2 and BCL2L1). 20477830_The study suggested that PDAM might function as a non-protein-coding RNA.And PDAM deregulation play a role in oligodendroglial tumors development, PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes. 22388545_Report expression of TP73 isoforms in human cell lines. 26410378_KIAA0495 methylation was detected in none of both primary myeloma samples at diagnosis (N = 61) and at relapse/progression (N = 16). KIAA0495 methylation appeared unimportant in the pathogenesis or progression of myeloma. 26799587_High expression level of long non-coding RNA TP73-AS1 is associated with esophageal squamous cell carcinoma. 28379612_Study showed that TP73-AS1 was specifically upregulated in brain glioma tissues and cell lines, and was associated with poorer prognosis in patients with glioma. TP73-AS1 knock down suppressed human brain glioma cell proliferation and invasion in vitro. TP73-AS1 seems to act as an oncogenic lncRNA promoting brain glioma proliferation and invasion. 28639399_Study showed that TP73-AS1 was specifically upregulated in BC tissues and breast cancer (BC) cell lines and was correlated to a poor prognosis. Its knocking down suppressed BC cell proliferation in vitro through regulation of TFAM. Also, data revealed that TP73-AS1 could regulate miR-200a through direct targeting. 28857253_Data show a direct binding between TP73-AS1, miR-200a and ZEB1. TP73-AS1 promoted ZEB1 expression via competing with its 3'UTR for miR-200a binding. ZEB1 binds the promoter region of TP73-AS1 activating its expression. TP73-AS1 and ZEB1 expression was increased, whereas miR-200a expression was low in breast cancer (BC). These regulating loop TP73-AS1/miR-200a/ZEB1 in BC promotes cancer cell invasion and migration. 29412778_TP73-AS1 inhibited the brain glioma growth and metastasis as a competing endogenous RNA (ceRNA) through miR-124-dependent iASPP regulation. 29474580_miR-941 sponge region of nine high-affinity miR-941 binding sites clustering within 1 kb 29625110_lncRNA TP73-AS1 may serve as a tumor suppressor participated in bladder cancer progression, which provided a promising therapy strategy for patients with bladder cancer. 29750302_TP73-AS1 may be an oncogenic lncRNA that promotes the proliferation of ovarian cancer cells 29803931_An explicit oncogenic role of TP73-AS1 in the NSCLC tumorigenesis, suggesting a TP73-AS1-miR-449a-EZH2 axis and providing new insight for NSCLC tumorigenesis. 29864904_TP73-AS1 could function as a sponge of miR-142 to positively regulate Rac1 in osteosarcoma cells. 29904939_Study found that TP73-AS1 was upregulated in the ovarian cancer tissues and ovarian cancer cells, and correlated with poor prognosis. it promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. 29966969_TP73-AS1 was upregulated in cholangiocarcinoma, facilitating migration and invasive potential of tumor cells. 30010111_this study demonstrated that TP73-AS1 regulated Colorectal cancer (CRC)progression by acting as a competitive endogenous RNA to sponge miR-194 to modulate the expression of TGFa 30016766_TP73-AS1 knockdown activated PI3K/Akt/mTOR signaling pathway, while overexpression of TP73-AS1 induced inhibition of PI3K/Akt/mTOR pathway and these effects could be partly abolished by overexpression of KISS1. 30118890_Data showed that TP73-AS1 was enhanced in gastric cancer (GC) tissues and cells, and tightly associated with tumor size, TNM stage, and overall survival. Decreased TP73-AS1 could restrain cell growth and promoted apoptosis partly by regulating Bcl-2/caspase-3 pathway. Importantly, TP73-AS1 could promote xenograft growth in vivo. 30193732_Twist-related protein 1 (TWIST1) was a target of miR-490-3p and participated in long non-coding RNA TP73 antisense RNA 1 (TP73-AS1)/miR-490-3p-modulated MDA-MB-231cell vasculogenic mimicry (VM) formation. 30279010_TP73-AS1 was upregulated in gastric cancer tissues and cell lines. Furthermore, TP73-AS1 exerted oncogenic role in gastric cancer through promoting cell growth and metastasis. In addition, TP73-AS1 was certified as a ceRNA by regulating miR-194-5p/SDAD1 axis. 30472379_we showed that TP73AS1 inhibits CRC cell growth by functioning as a ceRNA (competing endogenous RNAs) to regulate PTEN levels. Our findings provide new insights into the underlying molecular mechanisms of TP73AS1-mediated CRC. 30541897_LncRNA TP73-AS1 promoted the progression of lung adenocarcinoma via PI3K/AKT pathway. 30643007_LncRNA TP73 antisense RNA 1T (TP73-AS1) positively regulates 3-hydroxybutyrate dehydrogenase type 2 (BDH2) by regulating miR-141. 30867410_results reveal that high TP73-AS1 predicts poor prognosis in primary glioblastoma multiform cohorts and that this lncRNA promotes tumor aggressiveness and temozolomide resistance in glioblastoma cancer stem cells. 31015368_LncRNA TP73-AS1 down-regulates miR-139-3p to promote retinoblastoma cell proliferation. 31081944_The lncRNA TP73-AS1 is a prognostic marker. 31129247_Results found that P73-AS1 was upregulated in non-small cell lung cancer (NSCLC) tissues and predicted poor survival. TP73-AS1 is a potential upstream positive regulator of miRNA to promote NSCLC cell migration and invasion. 31210297_LncRNA TP73-AS1 promotes malignant progression of hepatoma by regulating microRNA-103. 31232491_LncRNA TP73-AS1 is a novel regulator in cervical cancer via miR-329-3p/ARF1 axis. 31432138_the findings suggested that upregulation of TP73AS1 promoted cervical cancer progression by promoting CCND2 via the suppression of miR607 expression. 31549851_Association of TP73-AS1 gene polymorphisms with the risk and survival of gastric cancer in a Chinese Han Population 31652459_Role of long non-coding RNA TP73-AS1 in cancer. 31663517_LncRNA TP73-AS1 promoted proliferation of cervical cancer cell lines by targeting miR-329-3p to regulate the expression of the SMAD2 gene. A regulatory network was formed between lncRNA TP73-AS1, miR-329-3p, and SMAD2. 31678571_role of TP73-AS1 lncRNA in human cancers 31901156_Knockdown of long noncoding RNA TP73-AS1 suppresses the malignant progression of breast cancer cells in vitro through targeting miRNA-125a-3p/metadherin axis. 31978409_Long non-coding RNA TP73-AS1 in cancers. 32412786_lncRNA TP73-AS1 Regulates miR-21/PTEN Axis to Affect Cell Proliferation in Acute Myeloid Leukemia. 32818670_LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-beta1 signaling. 32901838_Long noncoding RNA TP73AS1 accelerates the progression and cisplatin resistance of nonsmall cell lung cancer by upregulating the expression of TRIM29 via competitively targeting microRNA34a5p. 33400379_LncRNA TP73-AS1 regulates miR-495 expression to promote migration and invasion of nasopharyngeal carcinoma cells through junctional adhesion molecule A. 33589576_Long non-coding RNA TP73-AS1 is a potential immune related prognostic biomarker for glioma. 33683827_LncRNA TP73-AS1 enhances the malignant properties of pancreatic ductal adenocarcinoma by increasing MMP14 expression through miRNA -200a sponging. 34007135_Long non-coding RNA TP73-AS1 promotes pancreatic cancer growth and metastasis through miRNA-128-3p/GOLM1 axis. 34103010_LncRNA TP73-AS1 promotes oxidized low-density lipoprotein-induced apoptosis of endothelial cells in atherosclerosis by targeting the miR-654-3p/AKT3 axis. 34115613_TP73-AS1 is induced by YY1 during TMZ treatment and highly expressed in the aging brain. | 1882.204967 | 1.0064523 | 0.009278862 | 0.08162183 | 1.285596e-02 | 9.097260e-01 | 9.675554e-01 | No | Yes | 1747.507048 | 143.821411 | 1715.598834 | 141.149754 | |||||||||
| ENSG00000227885 | 124901333 | lncRNA | 25.520302 | 0.8685592 | -0.203303844 | 0.53949024 | 1.333436e-01 | 7.149901e-01 | No | Yes | 21.181524 | 5.766377 | 23.202001 | 6.442597 | ||||||||||||
| ENSG00000228409 | 643253 | CCT6P1 | transcribed_unprocessed_pseudogene | 358.412320 | 1.1244613 | 0.169234067 | 0.14944086 | 1.294594e+00 | 2.552031e-01 | No | Yes | 363.749137 | 54.695496 | 319.065952 | 48.083895 | |||||||||||
| ENSG00000229267 | 101928103 | SNHG31 | lncRNA | 14.674321 | 0.9076431 | -0.139803004 | 0.72195655 | 3.417172e-02 | 8.533420e-01 | No | Yes | 9.943892 | 5.593597 | 11.843312 | 6.650872 | |||||||||||
| ENSG00000230590 | 100302692 | FTX | lncRNA | This gene is located upstream of XIST, within the X-inactivation center (XIC). It produces a spliced long non-coding RNA that is thought to positively regulate the expression of XIST, which is essential for the initiation and spread of X-inactivation. [provided by RefSeq, May 2015]. | 26992218_our study demonstrates that the novel pathway lncRNA Ftx/miR-545/RIG-I promotes hepatocellular carcinoma development 27065331_these findings suggest that lnc-FTX may act as a tumor suppressor in hepatocellular carcinoma (HCC) through physically binding miR-374a and MCM2. It may also be one of the reasons for HCC gender disparity and may potentially contribute to HCC treatment 29845188_Study showed that lncRNA Ftx was upregulated in human hepatocellular carcinoma (HCC) tissues and cell lines and associated with aggressive clinicopathological features. lncRNA Ftx overexpression promoted the proliferation, invasion and migration of HCC cells, whereas its knockdown resulted in the opposite effects. lncRNA Ftx seems to be a promoter of the Warburg effect and tumor progression, partly via the PPARgamma pa... 29925853_Data demonstrated an oncogenic role of lncRNA FTX in CRC tumorigenesis and progression via interaction with miR-215 and vimentin. 31709769_Quantitative Proteomics Analysis Revealed the Potential Role of lncRNA Ftx in Promoting Gastric Cancer Progression. 31920141_LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. 31957854_Long non-coding RNA FTX alleviates hypoxia/reoxygenation-induced cardiomyocyte injury via miR-410-3p/Fmr1 axis. 32176463_LncRNA FTX activates FOXA2 expression to inhibit non-small-cell lung cancer proliferation and metastasis. 32271421_Long non-coding RNA FTX promotes gastric cancer progression by targeting miR-215. 32476208_Long noncoding RNA FTX is associated with prognosis of glioma patients. 32660465_Down-regulation of FTX promotes the differentiation of osteoclasts in osteoporosis through the Notch1 signaling pathway by targeting miR-137. 33121404_The Function of LncRNA FTX in Several Common Cancers. 33389283_LncRNA FTX Promotes Colorectal Cancer Cells Migration and Invasion by miRNA-590-5p/RBPJ Axis. 33398336_Long non-coding RNA FTX predicts a poor prognosis of human cancers: a meta-analysis. 33736615_Silencing of FTX suppresses pancreatic cancer cell proliferation and invasion by upregulating miR-513b-5p. 34720057_Long non-coding RNA (lncRNA) five prime to Xist (FTX) promotes retinoblastoma progression by regulating the microRNA-320a/with-no-lysine kinases 1 (WNK1) axis. 35027040_Prognostic significance of long non-coding RNA five prime to XIST in various cancers. | 292.504052 | 0.9419022 | -0.086350900 | 0.15985229 | 2.877460e-01 | 5.916686e-01 | No | Yes | 227.166154 | 50.765653 | 243.064777 | 54.340514 | |||||||||
| ENSG00000231365 | 101929147 | WARS2-AS1 | lncRNA | 1118.497524 | 1.0709661 | 0.098912866 | 0.08685602 | 1.303791e+00 | 2.535219e-01 | 6.252140e-01 | No | Yes | 1088.257030 | 145.971327 | 1015.679609 | 136.315257 | ||||||||||
| ENSG00000232931 | LINC00342 | lncRNA | 293.282442 | 1.2629201 | 0.336763423 | 0.17163938 | 3.915797e+00 | 4.783432e-02 | No | Yes | 305.983943 | 97.649040 | 236.658809 | 75.608019 | ||||||||||||
| ENSG00000233611 | GBX2-AS1 | lncRNA | 86.273910 | 1.0816960 | 0.113295071 | 0.30465620 | 1.398383e-01 | 7.084419e-01 | No | Yes | 90.384274 | 17.129922 | 85.672566 | 16.117529 | ||||||||||||
| ENSG00000234028 | 101928403 | EIF2AK3-DT | lncRNA | 21.447124 | 0.5814566 | -0.782256500 | 0.58831052 | 1.774063e+00 | 1.828800e-01 | No | Yes | 14.460374 | 4.142486 | 21.859975 | 5.977036 | |||||||||||
| ENSG00000234350 | 101926913 | ERICH2-DT | lncRNA | 11.843711 | 1.5851024 | 0.664576027 | 0.77640632 | 7.524428e-01 | 3.857039e-01 | No | Yes | 18.593163 | 7.960401 | 10.349462 | 4.410099 | |||||||||||
| ENSG00000235257 | 101928153 | ITGA9-AS1 | lncRNA | 84.895724 | 0.9624642 | -0.055195241 | 0.28921665 | 3.640174e-02 | 8.486882e-01 | No | Yes | 77.524471 | 12.669090 | 79.176320 | 12.812884 | |||||||||||
| ENSG00000235944 | ZNF815P | transcribed_unprocessed_pseudogene | 90.530479 | 1.2209832 | 0.288043314 | 0.28249107 | 1.024704e+00 | 3.114059e-01 | No | Yes | 92.064865 | 13.396414 | 79.217067 | 11.552403 | ||||||||||||
| ENSG00000236753 | 100506881 | MKLN1-AS | lncRNA | 33000222_Long noncoding RNA MKLN1AS aggravates hepatocellular carcinoma progression by functioning as a molecular sponge for miR6543p, thereby promoting hepatomaderived growth factor expression. 33878313_Long non-coding RNA muskelin 1 antisense RNA (MKLN1-AS) is a potential diagnostic and prognostic biomarker and therapeutic target for hepatocellular carcinoma. 34983308_ETS Proto-Oncogene 1-activated muskelin 1 antisense RNA drives the malignant progression of hepatocellular carcinoma by targeting miR-22-3p to upregulate ETS Proto-Oncogene 1. | 109.449062 | 0.9629164 | -0.054517528 | 0.25252390 | 4.707127e-02 | 8.282402e-01 | No | Yes | 119.072880 | 15.368940 | 122.206435 | 15.891772 | ||||||||||
| ENSG00000236869 | 112840933 | ZKSCAN7-AS1 | lncRNA | 26.633560 | 1.2461765 | 0.317508401 | 0.51098029 | 3.856822e-01 | 5.345778e-01 | No | Yes | 31.736688 | 8.976271 | 24.776519 | 7.050925 | |||||||||||
| ENSG00000237489 | 387723 | C10orf143 | protein_coding | A0A1B0GUT2 | Reference proteome | Mouse_homologues mmu:77252; | ENSMUSG00000040139 | 9430038I01Rik | 152.639712 | 1.0377125 | 0.053406739 | 0.21968873 | 5.945425e-02 | 8.073606e-01 | No | Yes | 154.759458 | 16.963059 | 154.059459 | 16.879618 | ||||||
| ENSG00000237945 | 100506334 | LINC00649 | lncRNA | 32547537_Lnc-ITSN1-2, Derived From RNA Sequencing, Correlates With Increased Disease Risk, Activity and Promotes CD4(+) T Cell Activation, Proliferation and Th1/Th17 Cell Differentiation by Serving as a ceRNA for IL-23R via Sponging miR-125a in Inflammatory Bowel Disease. 32883226_LINC00649 underexpression is an adverse prognostic marker in acute myeloid leukemia. 33789312_LINC00649 Facilitates the Cellular Process of Bladder Cancer Cells via Signaling Axis miR-16-5p/JARID2. 33975517_Long non-coding RNA LINC00649 regulates YES-associated protein 1 (YAP1)/Hippo pathway to accelerate gastric cancer (GC) progression via sequestering miR-16-5p. 35088877_Long noncoding RNA LINC00649 functions as a microRNA4325p sponge to facilitate tumourigenesis in colorectal cancer by upregulating HDGF. 35188449_LncRNA LINC00649 promotes the growth and metastasis of triple-negative breast cancer by maintaining the stability of HIF-1alpha through the NF90/NF45 complex. | 37.771248 | 0.8198848 | -0.286506922 | 0.43093764 | 4.436652e-01 | 5.053587e-01 | No | Yes | 35.099886 | 10.306675 | 42.835024 | 12.583450 | ||||||||||
| ENSG00000241316 | 101927111 | SUCLG2-DT | lncRNA | 82.436850 | 0.8918381 | -0.165146292 | 0.31045649 | 2.883171e-01 | 5.913010e-01 | No | Yes | 74.847156 | 21.769886 | 82.373470 | 23.906769 | |||||||||||
| ENSG00000241852 | 541565 | C8orf58 | protein_coding | Q8NAV2 | Alternative splicing;Reference proteome | hsa:541565; | ENSMUSG00000044551 | 9930012K11Rik | 124.726418 | 1.2537325 | 0.326229599 | 0.25518818 | 1.644798e+00 | 1.996685e-01 | No | Yes | 134.410272 | 17.147860 | 107.343258 | 13.776198 | ||||||
| ENSG00000243667 | 116143 | DNAAF10 | protein_coding | Q96MX6 | FUNCTION: Key assembly factor specifically required for the stability of axonemal dynein heavy chains in cytoplasm. {ECO:0000250|UniProtKB:A8J3F6}. | 3D-structure;Alternative splicing;Apoptosis;Cytoplasm;Reference proteome;Repeat;WD repeat | This gene encodes a protein with two WD40 repeat domains thought to be involved in an apoptosis via activation of caspase-3. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2012]. | hsa:116143; | dynein axonemal particle [GO:0120293]; RPAP3/R2TP/prefoldin-like complex [GO:1990062]; ubiquitin binding [GO:0043130]; apoptotic process [GO:0006915]; axonemal dynein complex assembly [GO:0070286] | 16487927_Monad could be involved in apoptosis induced by TNF-alpha 16487927_Monad may function as a novel modulator of apoptosis pathway. 19450687_Is part of an RNA polymerase II-associated complex with possible chaperone activity. 23844004_Exosome-bound WD repeat protein Monad inhibits breast cancer cell invasion by degrading amphiregulin mRNA. | ENSMUSG00000078970 | Dnaaf10 | 572.837218 | 1.0137154 | 0.019652667 | 0.12124446 | 2.623091e-02 | 8.713377e-01 | 9.569981e-01 | No | Yes | 628.286840 | 90.753260 | 617.768759 | 89.199015 | |
| ENSG00000243701 | 344595 | DUBR | lncRNA | 33758355_DUBR suppresses migration and invasion of human lung adenocarcinoma cells via ZBTB11-mediated inhibition of oxidative phosphorylation. | 146.385464 | 1.0120694 | 0.017308225 | 0.26077833 | 4.302223e-03 | 9.477032e-01 | No | Yes | 151.432681 | 32.608954 | 153.789581 | 33.092535 | ||||||||||
| ENSG00000243708 | 100137049 | PLA2G4B | protein_coding | P0C869 | FUNCTION: Calcium-dependent phospholipase A1 and A2 and lysophospholipase that may play a role in membrane phospholipid remodeling. {ECO:0000269|PubMed:10085124, ECO:0000269|PubMed:10358058, ECO:0000269|PubMed:16617059}.; FUNCTION: [Isoform 3]: Calcium-dependent phospholipase A2 and lysophospholipase. Cleaves the ester bond of the fatty acyl group attached to the sn-2 position of phosphatidylethanolamines, producing lysophospholipids that may be used in deacylation-reacylation cycles. Hydrolyzes lysophosphatidylcholines with low efficiency but is inefficient toward phosphatidylcholines. {ECO:0000269|PubMed:16617059}.; FUNCTION: [Isoform 5]: Calcium-dependent phospholipase A1 and A2 and lysophospholipase. Cleaves the ester bond of the fatty acyl group attached to the sn-1 or sn-2 position of diacyl phospholipids (phospholipase A1 and A2 activity, respectively), producing lysophospholipids that may be used in deacylation-reacylation cycles. Can further hydrolyze lysophospholipids enabling complete deacylation. Has no activity toward alkylacyl phospholipids. {ECO:0000269|PubMed:10085124, ECO:0000269|PubMed:10358058, ECO:0000269|PubMed:16617059}. | Alternative splicing;Calcium;Cytoplasm;Endosome;Hydrolase;Lipid degradation;Lipid metabolism;Membrane;Metal-binding;Mitochondrion;Reference proteome | Mouse_homologues NA; + ;NA | This gene encodes a member of the cytosolic phospholipase A2 protein family. Phospholipase A2 enzymes hydrolyze the sn-2 bond of phospholipids, releasing lysophospholipids and fatty acids. This enzyme may be associated with mitochondria and early endosomes. Most tissues also express read-through transcripts from the upstream gene into this gene, some of which may encode fusion proteins combining the N-terminus of the upstream gene including its JmjC domain with the almost complete coding region of this gene, including the C2 and cytoplasmic phospholipase A2 domains. [provided by RefSeq, Jul 2008]. | hsa:100137049;hsa:8681; | cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extracellular region [GO:0005576]; mitochondrial inner membrane [GO:0005743]; calcium ion binding [GO:0005509]; calcium-dependent phospholipase A2 activity [GO:0047498]; calcium-dependent phospholipid binding [GO:0005544]; lysophospholipase activity [GO:0004622]; phosphatidyl phospholipase B activity [GO:0102545]; phospholipase A1 activity [GO:0008970]; phospholipase A2 activity [GO:0004623]; arachidonic acid metabolic process [GO:0019369]; calcium-mediated signaling [GO:0019722]; glycerophospholipid catabolic process [GO:0046475]; inflammatory response [GO:0006954]; parturition [GO:0007567]; phosphatidylcholine acyl-chain remodeling [GO:0036151]; phosphatidylethanolamine acyl-chain remodeling [GO:0036152]; phosphatidylglycerol acyl-chain remodeling [GO:0036148] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20167866_TLR2 cooperates with MyD88, PI3K, and Rac1 in lipoteichoic acid-induced cPLA2/COX-2-dependent airway inflammatory responses 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 28030848_JMJD7-PLA2G4B may serve as an important therapeutic target and prognostic marker for head and neck squamous cell carcinoma development and progression. | ENSMUSG00000098488+ENSMUSG00000033852 | Pla2g4b+Gm28042 | 52.957726 | 1.9646729 | 0.974289142 | 0.37085883 | 6.794770e+00 | 9.142532e-03 | No | Yes | 67.419118 | 26.651599 | 34.864592 | 13.661790 | |
| ENSG00000243970 | 728448 | PPIEL | transcribed_unprocessed_pseudogene | This transcribed pseudogene is related to PPIE (Gene ID: 10450). Expression of this pseudogene may be downregulated in non-small cell lung cancer (NSCLC). Differential DNA methylation of this locus may be associated with intellectual disability and bipolar disorder in human patients. [provided by RefSeq, Sep 2016]. | 218.464851 | 1.0334727 | 0.047500256 | 0.19763186 | 5.718323e-02 | 8.110047e-01 | No | Yes | 188.667803 | 30.745243 | 189.796971 | 30.991116 | ||||||||||
| ENSG00000244257 | PKD1P1 | unprocessed_pseudogene | 1612.168548 | 1.1066554 | 0.146206038 | 0.08785516 | 2.732913e+00 | 9.829983e-02 | 4.188218e-01 | No | Yes | 1513.456729 | 275.272026 | 1384.638949 | 251.857443 | |||||||||||
| ENSG00000244405 | 2119 | ETV5 | protein_coding | P41161 | FUNCTION: Binds to DNA sequences containing the consensus nucleotide core sequence 5'-GGAA.-3'. {ECO:0000269|PubMed:8152800}. | 3D-structure;Alternative splicing;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:2119; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; cell differentiation [GO:0030154]; cellular response to oxidative stress [GO:0034599]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 12387735_functional role for the ERMs (ezrin/radixin/moesin) as adaptor molecules in the interactions of adhesion receptors and intracellular tyrosine kinases 15534105_ERM has a role in progression of breast cancer 15620692_The erm gene expression is regulated by the conventional PKC (cPKC) pathway. 15652352_CD44-mediated hyaluronan binding in myeloid cells is regulated by phosphorylated ERM and the actin cytoskeleton. 15857832_ERM is subject to SUMO modification and this post-translational modification causes inhibition of transcription-enhancing activity 16613858_Erm is involved in SP-C regulation, which results from an interaction with TTF-1 16615918_analysis of NHERF recognition by ERM proteins 17489097_Testing various shorter fragments of ERM as bait indicated that the region essential for binding CHD3/ZFH is within the amino acid region 96-349, which contains the central inhibitory DNA-binding domain (CIDD) of ERM 17638886_ERM/ETV5 up-regulation activates MMP-2 and thus has a role during myometrial infiltration in endometrial cancer 18172298_A TMPRSS2:ETV5 gene fusion was identified in prostate cancer. 19692490_Observational study of gene-disease association. (HuGE Navigator) 19746409_Observational study of gene-disease association. (HuGE Navigator) 19812171_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19851340_Observational study of gene-disease association. (HuGE Navigator) 20215397_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20520848_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20647002_The flexible nature of transactivation domain may be instrumental for ERM recognition and binding to diverse molecular partners. 20712903_Observational study of gene-disease association. (HuGE Navigator) 20724581_Observational study of gene-disease association. (HuGE Navigator) 20816195_Observational study of gene-disease association. (HuGE Navigator) 20970160_results indicate concurrent mechanisms in expression of MMP-2 and -9, RUNX1/AML1 and ETV5/ERM, and several basement membrane components, which likely associate with the invasive stage of endometrioid endometrial and ovarian endometrioid carcinoma. 21520040_Upregulation of ETV5 induced the expression of cell adhesion molecules and enhanced cell survival in a spheroid model. 21689625_Pea3 and Erm, but not Er81, play an important role in the progression of esophageal squamous cell carcinoma 22266854_we propose ETV5-transcriptional regulation of the EMT process through a crosstalk with the tumor surrounding microenvironment, as a principal event initiating EC invasion. 22425584_Review evidence for a role of ETV1, 4 and 5 as oncoproteins and describe modes of their action. 22589409_we found that the downregulation of ETV5 reduced the expression of the oncogenic transcription factor FOXM1. 22771031_The homozygous +48845 G>T (TT allele) variant confers a higher risk for male infertility associated with nonobstructive azoospermia and Sertoli cell only syndrome in Australian men. 22968857_This data suggests that ETV5 has a significant role in regulating MMP2 expression and therefore matrix resorption in human chondrosarcoma, and thus may be a targetable upstream effector of the metastatic cascade in this cancer. 24756106_there is a higher expression of ERM/ETV5 in early stages of endometrioid endometrial carcinomas 25172658_Snail regulates the motility and invasiveness of oral cancer cells via RhoA/Cdc42/p-ERM pathway. 25203299_developed a novel RNA in situ hybridization-based assay for the in situ detection of ETV1, ETV4, and ETV5 in formalin-fixed paraffin-embedded tissues from prostate needle biopsies, prostatectomy, and metastatic PCa specimens using RNA probes 25801911_Our results reveal a novel ERM-based spatial mechanism that is coopted by DLBCL cells to sustain tumor cell growth and survival. 25924802_Increase of ETV5 protein expression is associated with endometrial cancer invasion. 26289026_ERM proteins contribute toward accelerated CD44 shedding by MT1-MMP through ERM protein-mediated interactions between their cytoplasmic tails. 26729407_Resistin, a fat-derived secretory factor, promotes metastasis of MDA-MB-231 human breast cancer cells through ERM activation 27276064_MiR-200b inhibition by transfection in MCF-7 markedly decreased miR-200b level, elevated ERM expression, and enhanced cell migration and invasion. MiR-200b overexpression in MDA-MB-231 obviously increased miR-200b level, reduced ERM expression, and weakened cell migration and invasion. 27783944_four oncogenic ETS (ERG, ETV1, ETV4, and ETV5), and no other ETS, interact with the Ewing's sarcoma breakpoint protein, EWS. 28161714_Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETV1, ETV4 and ETV5. 28408625_the prostate cancer-related oncogenic E26 transformation-specific (ETS) transcription factors, ETV1, ETV4, and ETV5, were required for TAZ gene transcription in PC3 prostate cancer cells 28645872_Novel ETV5 SNP rs7647305 is associated with childhood hypertension adjusted by obesity. 29729315_exploration of the underlying mechanism demonstrated that ICAM3 not only binds to LFA-1 with its extracellular domain and structure protein ERM but also to lamellipodia with its intracellular domain which causes a tension that pulls cells apart (metastasis). 29787563_High Etv5 expression is associated with glioma. 30650178_Study suggested that the transcription factor ETV5 could stimulate CRC malignancy and promote CRC angiogenesis by directly targeting PDGF-BB. 30952872_ETV5 links the FGFR3 and Hippo signalling pathways in bladder cancer. 31452441_ETS Factor ETV5 Activates the Mutant Telomerase Reverse Transcriptase Promoter in Thyroid Cancer. 31937834_The ETS transcription factor ETV5 is a target of activated ALK in neuroblastoma contributing to increased tumour aggressiveness. 32325133_ETV5 enhanced follicular thyroid cancer cell proliferation, migration, and epithelial-mesenchymal transition through the PIK3CA signaling pathway 32518154_ETS variant transcription factor 5 and c-Myc cooperate in derepressing the human telomerase gene promoter via composite ETS/E-box motifs. 33931578_The role of E26 transformation-specific variant transcription factor 5 in colorectal cancer cell proliferation and cell cycle progression. 34716308_Chromatin accessibility analysis identifies the transcription factor ETV5 as a suppressor of adipose tissue macrophage activation in obesity. 34736492_Biological and prognostic value of ETV5 in high-grade serous ovarian cancer. 34753799_Human Islet MicroRNA-200c Is Elevated in Type 2 Diabetes and Targets the Transcription Factor ETV5 to Reduce Insulin Secretion. | ENSMUSG00000013089 | Etv5 | 21.165816 | 0.4559167 | -1.133157951 | 0.56437141 | 3.913089e+00 | 4.791146e-02 | No | Yes | 9.024496 | 2.873171 | 20.779318 | 6.502471 | |||
| ENSG00000244625 | MIATNB | lncRNA | 57.625553 | 0.9146278 | -0.128743345 | 0.35237124 | 1.347857e-01 | 7.135208e-01 | No | Yes | 52.954318 | 9.857339 | 57.102526 | 10.817158 | ||||||||||||
| ENSG00000244754 | 10443 | N4BP2L2 | protein_coding | Q92802 | Alternative splicing;Coiled coil;Reference proteome | hsa:10443; | extracellular exosome [GO:0070062]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; enzyme binding [GO:0019899]; transcription corepressor activity [GO:0003714]; blastocyst development [GO:0001824]; negative regulation of hematopoietic stem cell differentiation [GO:1902037]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of hematopoietic stem cell proliferation [GO:1902035] | 19506020_Data suggest that expression of PFAAP5 allows neutrophil elastase to potentiate the repression of Gfi1 target genes. | ENSMUSG00000029655 | N4bp2l2 | 1647.413872 | 1.1760198 | 0.233912358 | 0.07513042 | 9.721774e+00 | 1.820977e-03 | 4.814199e-02 | No | Yes | 1844.027807 | 376.306934 | 1560.667247 | 318.573881 | |||
| ENSG00000245680 | 92285 | ZNF585B | protein_coding | Q52M93 | FUNCTION: May be involved in transcriptional regulation. | Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:92285; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 225.417051 | 0.8633341 | -0.212009130 | 0.20068601 | 1.125356e+00 | 2.887681e-01 | No | Yes | 218.349867 | 39.348211 | 252.667425 | 45.482132 | ||||||
| ENSG00000245849 | 100505648 | RAD51-AS1 | lncRNA | 26230935_results identify TPIP as a novel E2F1 co-activator, suggest a similar role for other TPTEs, and indicate that the TODRA lncRNA affects RAD51 dysregulation and RAD51-dependent DSB repair in malignancy 28667302_Overexpression of RAD51-AS1 promoted EOC cell proliferation, while silencing of RAD51-AS1 inhibited EOC cell proliferation, delayed cell cycle progression and promoted apoptosis in vitro and in vivo. 29749376_Corylin increases the sensitivity of hepatocellular carcinoma cells to chemotherapy through long noncoding RNA RAD51-AS1-mediated inhibition of DNA repair. 33314669_LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed proliferation, invasion and glycolysis of colorectal cancer. | 116.984218 | 1.3133689 | 0.393272198 | 0.27802941 | 2.057789e+00 | 1.514298e-01 | No | Yes | 122.411441 | 25.823546 | 90.829459 | 19.286184 | ||||||||||
| ENSG00000246596 | transcribed_unprocessed_pseudogene | 205.418805 | 1.0220389 | 0.031450102 | 0.19169219 | 2.656676e-02 | 8.705238e-01 | No | Yes | 177.837854 | 46.631304 | 176.702788 | 46.294486 | |||||||||||||
| ENSG00000249898 | MCPH1-AS1 | lncRNA | 30.116359 | 0.8881108 | -0.171188476 | 0.48647177 | 1.183210e-01 | 7.308627e-01 | No | Yes | 21.714780 | 6.709476 | 25.743984 | 7.873947 | ||||||||||||
| ENSG00000250802 | 728723 | ZBED3-AS1 | lncRNA | 33712067_Lnc13728 facilitates human mesenchymal stem cell adipogenic differentiation via positive regulation of ZBED3 and downregulation of the WNT/beta-catenin pathway. | 145.017375 | 1.2191250 | 0.285846042 | 0.21903834 | 1.694766e+00 | 1.929739e-01 | No | Yes | 153.535792 | 30.368239 | 124.513426 | 24.628799 | ||||||||||
| ENSG00000251369 | 162972 | ZNF550 | protein_coding | Q7Z398 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:162972; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 255.470730 | 1.2135159 | 0.279193018 | 0.18467863 | 2.335177e+00 | 1.264806e-01 | No | Yes | 287.873239 | 52.154038 | 229.979369 | 41.786059 | ||||||
| ENSG00000251474 | 132241 | RPL32P3 | transcribed_unprocessed_pseudogene | 448.196029 | 1.1982155 | 0.260887393 | 0.12909002 | 4.110670e+00 | 4.261347e-02 | No | Yes | 522.237453 | 89.260062 | 438.307061 | 74.993788 | |||||||||||
| ENSG00000253771 | TPTE2P1 | transcribed_unprocessed_pseudogene | 18.666132 | 0.8005134 | -0.321002529 | 0.60611176 | 2.819036e-01 | 5.954562e-01 | No | Yes | 15.574677 | 4.471090 | 19.155799 | 5.342790 | ||||||||||||
| ENSG00000254413 | 386593 | CHKB-CPT1B | protein_coding | H3BT56 | Reference proteome | The genes CHKB and CPT1B are adjacent on chromosome 22 and read-through transcripts are expressed that include exons from both loci. The read-through transcripts are candidates for nonsense-mediated mRNA decay (NMD) and are unlikely to express proteins. [provided by RefSeq, Jun 2009]. | cytoplasm [GO:0005737]; choline kinase activity [GO:0004103]; ethanolamine kinase activity [GO:0004305]; CDP-choline pathway [GO:0006657]; phosphatidylcholine biosynthetic process [GO:0006656]; phosphatidylethanolamine biosynthetic process [GO:0006646] | 586.511332 | 1.1630760 | 0.217945322 | 0.14447357 | 2.284548e+00 | 1.306682e-01 | 4.786931e-01 | No | Yes | 567.534159 | 89.719418 | 510.608409 | 80.799077 | ||||||
| ENSG00000254469 | 100133315 | XNDC1N | protein_coding | Q6ZNB5 | Alternative splicing;Reference proteome | Mouse_homologues mmu:102443350; | chromosome [GO:0005694]; nucleolus [GO:0005730]; damaged DNA binding [GO:0003684]; single strand break repair [GO:0000012] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000099481 | Xndc1 | 105.345204 | 0.8064060 | -0.310421766 | 0.26907965 | 1.333949e+00 | 2.481039e-01 | No | Yes | 102.051376 | 18.963189 | 123.746875 | 22.980174 | ||||
| ENSG00000254860 | 493900 | TMEM9B-AS1 | lncRNA | 134.359156 | 1.1418591 | 0.191384657 | 0.23669812 | 6.412964e-01 | 4.232417e-01 | No | Yes | 116.479784 | 23.204493 | 104.145524 | 20.746272 | |||||||||||
| ENSG00000255508 | 64852 | protein_coding | H3BRB1 | Mouse_homologues FUNCTION: Poly(A) polymerase that creates the 3'-poly(A) tail of specific pre-mRNAs. Localizes to nuclear speckles together with PIP5K1A and mediates polyadenylation of a select set of mRNAs, such as HMOX1. In addition to polyadenylation, it is also required for the 3'-end cleavage of pre-mRNAs: binds to the 3'UTR of targeted pre-mRNAs and promotes the recruitment and assembly of the CPSF complex on the 3'UTR of pre-mRNAs. In addition to adenylyltransferase activity, also has uridylyltransferase activity. However, the ATP ratio is higher than UTP in cells, suggesting that it functions primarily as a poly(A) polymerase. Acts as a specific terminal uridylyltransferase for U6 snRNA in vitro: responsible for a controlled elongation reaction that results in the restoration of the four 3'-terminal UMP-residues found in newly transcribed U6 snRNA. Not involved in replication-dependent histone mRNA degradation. {ECO:0000250|UniProtKB:Q9H6E5}. | Coiled coil;RNA-binding;Reference proteome;Transferase | This gene encodes a nucleotidyl transferase that functions as both a terminal uridylyltransferase and a nuclear poly(A) polymerase. The encoded enzyme specifically adds and removes nucleotides from the 3' end of small nuclear RNAs and select mRNAs and may function in controlling gene expression and cell proliferation.[provided by RefSeq, Apr 2009]. | Mouse_homologues mmu:70044; | intracellular membrane-bounded organelle [GO:0043231]; polynucleotide adenylyltransferase activity [GO:0004652]; RNA binding [GO:0003723] | 18288197_PIPKIalpha co-localizes at nuclear speckles and interacts with a newly identified non-canonical poly(A) polymerase, Star-PAP; the activity of Star-PAP can be specifically regulated by PtdIns4,5P2 18305108_These data indicate that CKIalpha, PIPKIalpha, and Star-PAP function together to modulate the production of specific Star-PAP messages. 21102410_The data support a model where Star-PAP binds to the pre-mRNA, recruits the CPSF complex to the 3'-end of pre-mRNA and then defines cleavage by CPSF 73 and subsequent polyadenylation of its target mRNAs. 22244330_PIPKIalpha, PI4,5P(2), and PKCdelta regulate Star-PAP control of BIK expression and induction of apoptosis. 23306079_Star-PAP and its regulatory molecules form a signaling nexus at the 3'-end of target mRNAs to control the expression of select group of genes including the ones involved in stress responses. (Review) 23416977_Star-PAP controls E6 mRNA polyadenylation and expression and modulates wild-type p53 levels. 23874977_The human TUT1 nucleotidyl transferase is a global regulator of microRNA abundance. 25142229_Nucleotidyl transferase TUT1 inhibits lipogenesis in osteosarcoma cells through regulation of microRNA-24 and microRNA-29a. 26496945_Star-PAP recognises a unique nucleotide motif on its target mRNA.CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding poly A polymerase. 28151486_Star-PAP possesses tumor-suppressing activity. 28589955_Crystallographic and biochemical studies of TUT1 revealed the molecular mechanism underlying the specific oligo-uridylylation of the 3'-end of U6 snRNA by TUT1. 28911096_Star-PAP-specific polyadenylation sites usage regulates the expression of the eukaryotic translation initiation factor EIF4A1, the tumor suppressor gene PTEN and the long non-coding RNA NEAT1. 29032201_KLHL7 is a novel regulator of the nucleolus associated with TUT1 ubiquitination, and pathogenic KLHL7 mutants may provide valuable information to elucidate a mechanism of retinitis pigmentosa etiology. 34576144_Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. | ENSMUSG00000071645 | Tut1 | 337.609588 | 0.9674410 | -0.047754392 | 0.18699689 | 6.546318e-02 | 7.980607e-01 | No | Yes | 307.341486 | 52.314766 | 317.867455 | 54.103168 | |||
| ENSG00000256591 | 54949 | protein_coding | F5GXW4 | Chaperone;Reference proteome | 141.408789 | 1.1182798 | 0.161281262 | 0.30793359 | 2.826617e-01 | 5.949620e-01 | No | Yes | 157.109551 | 39.640373 | 145.677159 | 36.751714 | ||||||||||
| ENSG00000256683 | 59348 | ZNF350 | protein_coding | Q9GZX5 | FUNCTION: Transcriptional repressor. Binds to a specific sequence, 5'-GGGxxxCAGxxxTTT-3', within GADD45 intron 3. {ECO:0000269|PubMed:11090615}. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:59348; | nuclear body [GO:0016604]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding [GO:0001162]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12872252_ZBRK1 mutation found in BRCA1 and BRCA2 mutation negative probands with breast or ovarian cancer. 14517299_ZBRK1 is a novel target for DNA damage-induced degradation 14660588_ZBRK1 zinc fingers have dual roles in sequence-specific DNA-binding and BRCA1-dependent transcriptional repression 15113441_Observational study of gene-disease association. (HuGE Navigator) 15496401_the CTRD is a novel protein interaction surface responsible for directing homotypic and heterotypic interactions necessary for ZBRK1-directed transcriptional repression 15596820_interacts with BBLF2/3 to provide a tethering point on oriLyt for the EBV replication complex 16485136_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16997916_KRAB-induced transcriptional repression is robust and active over a variety of genomic contexts that include at least the wide range of sites targeted by lentiviral integration 17557904_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17764113_Genetic variants and haplotypes of the ZBRK1 gene is associated with breast and ovarian cancer 17764113_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19279087_KRAB/KAP1 regulation is fully functional within the context of episomal DNA 19484476_BRCA1 and ZNF350 may jointly contribute to individuals' susceptibility of breast cancer in Chinese women. 19484476_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19996286_Our findings suggest that ZBRK1 acts to inhibit metastasis of cervical carcinoma, perhaps by modulating MMP9 expression. 20007691_Data found that HMGA2, along with a dozen of other genes, was co-repressed by ZBRK1, BRCA1, and CtIP. 20150366_Observational study of gene-disease association. (HuGE Navigator) 20221260_KRAB/KAP1 recruitment induces long-range repression through the spread of heterochromatin. 20306497_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20713352_RB.E2F1 complex plays a critical role in ZBRK1 transcriptional repression, and loss of this repression may contribute to cellular sensitivity of DNA damage, ultimately leading to carcinogenesis. 20926453_KRAB-containing zinc-finger transcriptional regulator, ZBRK1, an interaction partner of the SCA2 gene product ataxin-2. 21791101_high levels of gene activity in the genomic environment and the pre-deposition of repressive histone marks within a gene increase its susceptibility to KRAB/KAP1-mediated repression 22975076_ZBRK1 negatively regulates the HIV-1 LTR 23151675_promoter assays in two breast cancer cell lines identified two haplotypes (H11 and H12) stimulating significantly the expression of ZNF350 transcript compared with the common haplotype H8 23991171_results indicate that a loss of ZBRK1 contributes to the increased expression of KAP1, potentiating its role to enhance metastasis and invasion 24924633_The secondary structure of the ZBRK1-DNA complex is found to be significantly altered from the standard B-DNA conformation. 25107531_MAGE proteins bind to KAP1, a gene repressor and ubiquitin E3 ligase which also binds KRAB domain containing zinc finger transcription factors (KZNFs), and MAGE expression may affect KZNF mediated gene regulation. 25749518_ZBRK1 suppresses renal cancer progression perhaps by regulating VHL expression through formation of a complex with VHL and p300 in renal cancer 26658611_villin directly interacts with a transcriptional corepressor and ligand of the Slug promoter, ZBRK1. 27586871_Results indicated that SNP rs2278414 at ZNF350 may in fl uence an individual's susceptibility to age-related cataract (ARC) by affecting the binding af fi nity of miR-21-3p and miR-150-5p and regulating expression levels of the mRNAs, resulting in different levels of cellular DNA breaks, thus contributing to ARC. 29653063_Results indicate ZNF350 as an important gene mammary oncogenesis. 30061100_These data indicate that the KAP1-KZNF pathway contributes to genome stability and innate immune control in adult human cells. 30714292_BRCA1 forms a co-repressor complex with ZBRK1 that coordinately represses GOT2 expression via a ZBRK1 recognition element in the promoter of GOT2. 31289231_The work identifies the interaction interfaces in the KAP1 tripartite motif responsible for self-association and KRAB binding and establishes their role in retrotransposon silencing. 32470015_PFKP is transcriptionally repressed by BRCA1/ZBRK1 and predicts prognosis in breast cancer. 33407485_Correlation between ZBRK1/ZNF350 gene polymorphism and breast cancer. 34685484_Role of the Transcriptional Repressor Zinc Finger with KRAB and SCAN Domains 3 (ZKSCAN3) in Retinal Pigment Epithelial Cells. | 122.051539 | 0.7913437 | -0.337623634 | 0.23622471 | 2.029188e+00 | 1.543031e-01 | No | Yes | 98.753318 | 23.537262 | 122.337072 | 29.183079 | |||||
| ENSG00000258325 | 283440 | ITFG2-AS1 | lncRNA | 30.882106 | 1.0991761 | 0.136422516 | 0.47412074 | 8.072311e-02 | 7.763199e-01 | No | Yes | 37.981794 | 8.521776 | 35.479139 | 7.888273 | |||||||||||
| ENSG00000260027 | 3217 | HOXB7 | protein_coding | P09629 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene is a member of the Antp homeobox family and encodes a protein with a homeobox DNA-binding domain. It is included in a cluster of homeobox B genes located on chromosome 17. The encoded nuclear protein functions as a sequence-specific transcription factor that is involved in cell proliferation and differentiation. Increased expression of this gene is associated with some cases of melanoma and ovarian carcinoma. [provided by RefSeq, Jul 2008]. | hsa:3217; | chromatin [GO:0000785]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; embryonic organ development [GO:0048568]; embryonic skeletal system morphogenesis [GO:0048704]; myeloid cell differentiation [GO:0030099]; positive regulation of branching involved in ureteric bud morphogenesis [GO:0090190]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12697323_Identification of transcription factors that regulate HOXB7 expression. 16597639_HOXB7 is expressed from primordial and early primary stage follicles through to germinal vesicle (GV) oocytes. 16673309_Homeobox B7 (HoxB7) mRNA is overexpressed in biliary cancer cell lines and possibly involved in the pathogenesis of bile duct cancer. 17018609_HOXB7 promotes tumor invasion through activation of Ras/Rho pathway by up-regulating bFGF. HOXB7 overexpression causes epithelial-mesenchymal transition in epithelial cells, accompanied by aggressive properties of tumorigenicity, migration, & invasion. 17274802_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17514648_thalidomide inhibits the RA-induced HOXB7 expression in glioblastoma cells 19878270_Differential expression of HOXB7 gene in multiple myeloma and extramedullary multiple myeloma patients 19956843_HOXB7 may contribute to oral carcinogenesis by increasing tumor cell proliferation 20480203_Strongly reduced expression of the microRNA miR-196a in melanoma cells compared to healthy melanocytes leads to enhanced HOX-B7 mRNA and protein levels, which subsequently raise Ets-1 activity by inducing basic fibroblast growth factor (bFGF). 21183939_confirmed that HOXB7 overexpression by multiple myeloma (MM) cells stimulated tumor growth, increased MM-associated angiogenesis and the expression of pro-angiogenic genes by microarray analysis 21474578_Here, we showed that PI3K/AKT and MAPK pathways were activated because p-ERK, p-AKT and p-GSK3-beta were upregulated by HOXB7. 21690342_Up-regulation of EGFR occurs through direct binding of HOXB7 to the EGFR promoter, enhancing transcriptional activity. 21953534_Data indicate that HOXB7 and related HOX genes are upregulated in plasma cells of specific subsets of multiple myeloma patients lacking major immunoglobulin heavy chain locus translocations, irrespective of hyperdiploidy or other cytogenetic lesions. 22239410_The current study demonstrates that the expression of HOXB7 is an independent prognostic marker for oral squamous cell carcinoma. 22553335_the absence of HoxB7 leads to inefficient viral progeny production, as HAdV5 gene expression is highly regulated by HoxB7-mediated activation of various adenoviral promoters. 22603795_HOXB5, HOXB6, and HOXB7 are activated in Barrett esophagus, and the midcluster HOXB gene signature in BE most resembled the colon rather than other GI epithelia. 22844406_a novel mechanism whereby polyADP-ribosylation regulates transcriptional activities of HOX proteins such as HOXB7 and HOXA7 22879911_Measuring IGF2BP3, HOXB7 and NEK2 mRNA levels by RT-PCR in addition to cytology has the potential to improve detection of malignant biliary disorders from brush cytology specimens. 22911672_HOXB7 promotes LAC progression by enhancing proliferation and metastasis. 22914903_HOXB7 is frequently overexpressed in pancreatic adenocarcinoma, specifically promotes invasive phenotype, and is associated with lymph node metastasis and worse survival outcome. 23400877_findings suggest the disruption of the HOXB7/PBX2 complexes, miR-221&222 inhibition or even better their combination, as innovative therapeutic approaches 23861821_MicroRNA-196b regulates the HOXB7-VEGF axis in cervical cancer. 24088503_HOXB7 mRNA is overexpressed in pancreatic ductal adenocarcinomas and its knockdown induces cell cycle arrest and apoptosis. 24641834_HOXB7 was over-expressed and miR-337 was minimally expressed in pancreatic ductal adenocarcinoma 24853421_Results suggest that p53-regulated TUG1 is a growth regulator, which acts in part through control of HOXB7 in non-small cell lung cancer. 25183455_MiR-337 targets HOXB7 and effects significant suppression of pancreatic ductal adenocarcinoma (PDAC) cell proliferation and invasion. 25428821_Results demonstrate HOXB7 as a master factor driving progenitors behavior lifetime, providing a better understanding of bone senescence and leading to an optimization of MSC performance. 25542862_our results suggest that HOXB7 promotes tumor progression in a cell-autonomous and non-cell-autonomous manner through activation of the TGFbeta signaling pathway. 25565354_Myelomeningocele is significantly associated with HOXB7 hypomethylation. 26014856_Study reports >1,500 chromatin binding sites in the genome of a breast cancer cell line overexpressing HOXB7 and identifies some potential direct targets that are located nearby. 26076456_HOXB7 could promote cancer cell proliferation and might be an independent prognostic factor for patients with esophageal squamous cell carcinoma. 26135503_HOXB7 protein expression level is downregulated following siRNA transfection and downregulation of HOXB7 gene expression effectively inhibits MCF-7 cell proliferation. 26180042_HOXB7 Is an ERalpha Cofactor in the Activation of HER2 and Multiple ER Target Genes Leading to Endocrine Resistance 26307396_HOXB7 is generally overexpressed in gastric cancer, is associated with patient clinical characteristics, and specifically promotes gastric cancer cell malignant biological properties 26403398_HoxB7 is associated with tumor metastasis in patients with lung adenocarcinoma and HoxB7 may be implicated in promoting the development of lung adenocarcinoma through activation of the TGF-beta/SMAD3 signaling. 26902420_Results point out the complex interplay between the DSB DNA repair activity and the homeoproteins HoxB7 and Cdx2 in colon cancer cells. 26968988_The study suggests that HOXB7 has an oncogenic role in gastric cancer, which might be related to the modulation of Akt/PTEN activity to induce cell migration/invasion and anti-apoptotic effects. 26991799_These findings suggest that hoxb7 and hoxb9 proteins might play a role in salivary gland tumourigenesis, but in contrast to the reported for other human cancers, they were not significant prognostic determinants in the sample studied. 27272787_HOXB7 expression was significantly associated with larger tumor size and higher rate of biliary invasion in hepatocellular carcinoma. 27834359_the overexpression of HOXB7 in HSC-4 and KB/VCR cells reduces the sensitivity of the cells to chemo-radiotherapy-induced apoptosis and promotes oral cancer cell migration and invasion via upregulation of TGFbeta2. 27855613_Suggest that simultaneous targeting of key regulatory miRNA miR-222 and HOXB7 may be a useful strategy for prevention of colorectal cancer metastasis. 27901487_our results showed that increased expression of HOXB7 might play an important role in promoting GC proliferation, migration and invasion by inducing both AKT and MAPK pathways, thus resulting in progression of, and poor prognosis in GC patients. 27983923_downregulation of HOXB7 inhibited proliferation, invasion, and tumorigenesis, partly through suppressing the PI3K/Akt signaling pathway in osteosarcoma cells. 28454092_HOXB7 promotes HCC cell proliferation, migration, and invasion through the bFGF-induced MAPK/ERK pathway activation. It might be a novel prognostic factor in HCC and a promising therapeutic target for tumor metastasis and recurrence. 28482289_MiR-376c-3p suppresses the fission, proliferation, migration and invasion and induces cell apoptosis of oral squamous cancer cells via targeting HOXB7. 28533224_miR-196b-5p inhibition led to significantly increased colorectal cancer cell migration/invasion and metastases formation in mice, whereas ectopic overexpression showed the opposite phenotype. Molecular profiling and target confirmation identified an interaction between miR-196b-5p and HOXB7 and GALNT5, which in turn regulated colorectal cancer cell migration 28646927_Overexpression of HOXB7 was significantly correlated with poor prognosis of HCC. HOXB7 up-regulated c-Myc and Slug expression via the AKT pathway to promote the acquisition of stem-like properties and facilitate epithelial-mesenchymal transition of hepatoma cells, accelerating the malignant progression of HCC. 28677742_The expression of HOXB7 was upregulated in osteosarcoma tissues and cells compared with paired adjacent nontumor bone tissues and osteoblastic cells using reverse transcriptionquantitative polymer chain reaction and western blotting. HOXB7 knockdown dramatically suppressed cell viability, proliferation, migration and epithelialmesenchymal transition. 28749115_High expression of HOXB7 protein was associated with poor prognosis of patients with digestive system cancers, as well as clinicopathologic characteristics, including the tumor invasion, lymph node status, distant metastasis and TNM stage 28912272_These results suggested that HOXB7 stimulates ERK1/2 phosphorylation and provided evidence that HOXB7, besides its role in transcriptional regulation, also promotes cell motility and invasiveness. 29576613_HOXB7 upregulates several canonical embryonic stem cell/induced pluripotent stem cell markers and sustains the expansion of a subpopulation of cells with stem cell characteristics, through modulation of LIN28B, an emerging cancer gene and pluripotency factor, which is a direct target of HOXB7. 30067384_silencing of the HOXB7 has the mechanism of inactivating the Wnt/beta-catenin signaling pathway, thereby accelerating cell apoptosis and suppressing cell migration and invasion in cutaneous squamous cell carcinoma (CSCC), which could provide a candidate target for the CSCC treatment 30317675_High HOXB7 expression is associated with prostate cancer. 30379579_silencing of the HOXB7 has the mechanism of inactivating the Wnt/beta-catenin signaling pathway, thereby accelerating cell apoptosis and suppressing cell migration and invasion in cutaneous squamous cell carcinoma (CSCC), which could provide a candidate target for the CSCC treatment. 30388035_hsa_circ_0074362 plays a crucial role in glioma progression by regulating the miR-1236-3p/HOXB7 pathway. 30537478_High HOXB7 expression is associated with acute lymphoblastic leukemia. 30664713_upregulated MMP2, MMP9, VEGFa, and IL8 expression via the ERK pathway to accelerate the malignant progression of intrahepatic cholangiocarcinoma 30890185_HOXB7 increased AD-MSC proliferation potential, reduced senescence, and improved chondrogenesis together with a significant increase of basic fibroblast growth factor (bFGF) secretion. 30951946_HOXB7 upregulation reversed the effect of miR-384 by promoting CAP in vitro proliferation and migration. 31103063_inhibited trophoblast cell differentiation by down-regulating DKK1 expression 31568655_HOXB7 mediates cisplatin resistance in esophageal squamous cell carcinoma through involvement of DNA damage repair. 31609764_The let-7c/HoxB7 axis regulates the cell proliferation, migration and apoptosis in hepatocellular carcinoma. 32626939_CircRNA hsa_circ_0070934 functions as a competitive endogenous RNA to regulate HOXB7 expression by sponging miR12363p in cutaneous squamous cell carcinoma. 33155195_MiR-513a-3p inhibits EMT mediated by HOXB7 and promotes sensitivity to cisplatin in ovarian cancer cells. 33782039_Silencing of hsa_circ_0009035 Suppresses Cervical Cancer Progression and Enhances Radiosensitivity through MicroRNA 889-3p-Dependent Regulation of HOXB7. 34680970_Epigenetic Regulation of CDH1 Is Altered after HOXB7-Silencing in MDA-MB-468 Triple-Negative Breast Cancer Cells. | ENSMUSG00000038721 | Hoxb7 | 268.982390 | 1.3196762 | 0.400184022 | 0.16752921 | 5.673357e+00 | 1.722447e-02 | No | Yes | 340.038899 | 44.087394 | 254.449161 | 33.014050 | ||
| ENSG00000261342 | lncRNA | 125.223002 | 0.8363549 | -0.257812895 | 0.23669625 | 1.197974e+00 | 2.737269e-01 | No | Yes | 143.618600 | 20.632564 | 177.547748 | 25.103001 | |||||||||||||
| ENSG00000261556 | 100506060 | SMG1P7 | transcribed_unprocessed_pseudogene | 176.826030 | 2.0043402 | 1.003127431 | 0.21018196 | 2.264212e+01 | 1.951594e-06 | No | Yes | 229.558917 | 66.221800 | 116.220406 | 33.735902 | |||||||||||
| ENSG00000261570 | AGK-DT | lncRNA | 10.163741 | 1.4084282 | 0.494086024 | 0.80789795 | 3.782588e-01 | 5.385366e-01 | No | Yes | 12.096714 | 5.065657 | 8.370884 | 3.591582 | ||||||||||||
| ENSG00000262185 | 102724927 | LINC02861 | lncRNA | 16.082348 | 0.8508471 | -0.233028148 | 0.64372608 | 1.305897e-01 | 7.178214e-01 | No | Yes | 15.492836 | 4.808146 | 17.449176 | 5.303617 | |||||||||||
| ENSG00000263072 | ZNF213-AS1 | lncRNA | 333.455932 | 1.0564767 | 0.079260922 | 0.15416195 | 2.669196e-01 | 6.054056e-01 | No | Yes | 342.268171 | 33.439883 | 324.983308 | 31.767344 | ||||||||||||
| ENSG00000263146 | LINC01896 | lncRNA | 26.886132 | 0.7487313 | -0.417480027 | 0.50367067 | 6.836176e-01 | 4.083438e-01 | No | Yes | 20.214093 | 4.330490 | 28.952136 | 5.891908 | ||||||||||||
| ENSG00000264343 | 388677 | NOTCH2NLA | protein_coding | P0DPK4 | FUNCTION: Human-specific protein that promotes neural progenitor proliferation and evolutionary expansion of the brain neocortex by regulating the Notch signaling pathway (PubMed:29856954, PubMed:29856955, PubMed:29561261). Able to promote neural progenitor self-renewal, possibly by down-regulating neuronal differentiation genes, thereby delaying the differentiation of neuronal progenitors and leading to an overall final increase in neuronal production (PubMed:29856954). Acts by enhancing the Notch signaling pathway via two different mechanisms that probably work in parallel to reach the same effect (PubMed:29856954). Enhances Notch signaling pathway in a non-cell-autonomous manner via direct interaction with NOTCH2 (PubMed:29856954). Also promotes Notch signaling pathway in a cell-autonomous manner through inhibition of cis DLL1-NOTCH2 interactions, which promotes neuronal differentiation (By similarity). {ECO:0000250|UniProtKB:P0DPK3, ECO:0000269|PubMed:29561261, ECO:0000269|PubMed:29856954, ECO:0000269|PubMed:29856955}. | Calcium;Disulfide bond;EGF-like domain;Glycoprotein;Neurodegeneration;Notch signaling pathway;Reference proteome;Repeat;Secreted | hsa:388677; | extracellular region [GO:0005576]; calcium ion binding [GO:0005509]; cerebral cortex development [GO:0021987]; Notch signaling pathway [GO:0007219]; positive regulation of Notch signaling pathway [GO:0045747] | 29856954_the emergence of human-specific NOTCH2NL genes may have contributed to the rapid evolution of the larger human neocortex, accompanied by loss of genomic stability at the 1q21.1 locus and resulting recurrent neurodevelopmental disorders 32330268_Evolution of Human Brain Size-Associated NOTCH2NL Genes Proceeds toward Reduced Protein Levels. | 147.088904 | 1.5388244 | 0.621828567 | 0.31295264 | 3.724223e+00 | 5.362856e-02 | No | Yes | 169.662084 | 29.377289 | 113.638222 | 19.677142 | |||||
| ENSG00000265479 | 441263 | DTX2P1-UPK3BP1-PMS2P11 | lncRNA | This locus represents naturally-occurring readthrough transcription spanning multiple pseudogenes: DTX2P1 (DTX2 pseudogene 1), UPK3BP1 (uroplakin 3B pseudogene 1), PMS2P11 (PMS1 homolog 2, mismatch repair system component pseudogene 11). Some transcripts may also extend to PMS2P9 (PMS1 homolog 2, mismatch repair system component pseudogene 9). The readthrough transcripts likely do not encode functional proteins. [provided by RefSeq, Jan 2016]. | 185.003218 | 1.0132484 | 0.018987919 | 0.19717953 | 9.327621e-03 | 9.230603e-01 | No | Yes | 180.644880 | 20.207721 | 180.088135 | 19.945381 | ||||||||||
| ENSG00000266086 | 6426 | protein_coding | J3QKU1 | Mouse_homologues FUNCTION: Plays a role in preventing exon skipping, ensuring the accuracy of splicing and regulating alternative splicing (PubMed:28785060). Interacts with other spliceosomal components, via the RS domains, to form a bridge between the 5'- and 3'-splice site binding components, U1 snRNP and U2AF. Can stimulate binding of U1 snRNP to a 5'-splice site-containing pre-mRNA. Binds to purine-rich RNA sequences, either the octamer, 5'-RGAAGAAC-3' (r=A or G) or the decamers, AGGACAGAGC/AGGACGAAGC. Binds preferentially to the 5'-CGAGGCG-3' motif in vitro. Three copies of the octamer constitute a powerful splicing enhancer in vitro, the ASF/SF2 splicing enhancer (ASE) which can specifically activate ASE-dependent splicing (By similarity). Specifically regulates alternative splicing of cardiac isoforms of CAMK2D, LDB3/CYPHER and TNNT2/CTNT during heart remodeling at the juvenile to adult transition. The inappropriate accumulation of a neonatal and neuronal isoform of CAMKD2 in the adult heart results in aberrant calcium handling and defective excitation-contraction coupling in cardiomyocytes. May function as export adapter involved in mRNA nuclear export through the TAP/NXF1 pathway (PubMed:15652482). {ECO:0000250|UniProtKB:Q07955, ECO:0000269|PubMed:15652482, ECO:0000269|PubMed:28785060}. | Reference proteome | Mouse_homologues mmu:110809; | Mouse_homologues catalytic step 2 spliceosome [GO:0071013]; cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA topoisomerase binding [GO:0044547]; mRNA binding [GO:0003729]; protein kinase B binding [GO:0043422]; RNA binding [GO:0003723]; RS domain binding [GO:0050733]; alternative mRNA splicing, via spliceosome [GO:0000380]; cardiac muscle contraction [GO:0060048]; cellular response to interleukin-17 [GO:0097398]; in utero embryonic development [GO:0001701]; interleukin-17-mediated signaling pathway [GO:0097400]; mRNA 5'-splice site recognition [GO:0000395]; mRNA splicing, via spliceosome [GO:0000398]; mRNA stabilization [GO:0048255]; mRNA transport [GO:0051028]; positive regulation of RNA splicing [GO:0033120]; protein localization to nucleus [GO:0034504]; protein localization to P-body [GO:0110012]; regulation of RNA splicing [GO:0043484]; RNA splicing [GO:0008380]; signal transduction involved in regulation of gene expression [GO:0023019] | Mouse_homologues 11925564_Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1 15390079_Both hnRNP A1 and alternative splicing factor/splicing factor 2 contents rose in adenomas and during injury-induced hyperplasia compared to control lungs 18987029_These results highlight the requirement of Sfrs1-mediated alternative splicing for the survival of retinal neurons, with sensitivity defined by the window of time in which the neuron was generated. 20657585_Modulation of Xist RNA processing may be part of the stochastic process that determines which X chromosome will be inactivated. 20923760_analysis of the miRNA-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects cell senescence and apoptosis 21822258_Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). 23562324_Specific effects on regulated splicing by SR proteins SRSF1 and SRSF2 depends on a complex set of relationships with multiple other SR proteins in mammalian genomes. 23604122_RRP1B suppresses metastatic progression by altering the transcriptome through its interaction with splicing regulators such as SRSF1 23843040_Deletion of RRM1 eliminated the splicing activity of SRSF1 and thus cellular transformation. 24552714_The expression levels of three splicing factors, ESRP1, PTB and SF2/ASF, are significantly altered during cardiac hypertrophy in mice. 27616424_This study showed that the splicing factor kinase SRPK1 is a key regulator of spinal nociceptive processing in naive and nerve injured animals. We present evidence for a novel mechanism in which altered SRSF1 localization/function in neuropathic pain results in sensitization of spinal cord neurons. 28444861_LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin and SRSF1. 28799539_SRSF1 promotes vascular smooth muscle cell (VSMC) proliferation and injury-induced neointima formation. SRSF1 favors the induction of a truncated p53 isoform, Delta133p53, which has an equal proliferative effect and in turn transcriptionally activates Kruppel-like factor 5 (KLF5) via the Delta133p53-EGR1 complex, resulting in an accelerated cell-cycle progression and increased VSMC proliferation. 29120871_Authors showed that Mir505-3p was capable of inhibiting tumor proliferation driven by SRSF1 in two neural tumor cell lines, Neuro-2a (N2a) and U251, exclusively in serum-reduced condition. Authors observed that the protein level of SRSF1 was gradually promoted by increasing concentration of serum. 29233683_In addition, overexpression of SRSF1 in XRCC4-deficient cells restored the normal level of apoptosis, suggesting that SRSF1 functions downstream of XRCC4 in activating CAD. 29262322_SRSF1 is a key regulator of DBF4B pre-mRNA splicing dysregulation in colon cancer. SRSF1 is required for cancer cell proliferation. 31487268_Loss of SRSF1 limits the expression of the mTOR repressor PTEN, leading to aberrant T cell activation and the expression of systemic autoimmunity. 32810232_Splicing factor SRSF1 limits IFN-gamma production via RhoH and ameliorates experimental nephritis. 32888503_Splicing Factor SRSF1 Is Essential for Satellite Cell Proliferation and Postnatal Maturation of Neuromuscular Junctions in Mice. 32964995_LncRNA HOTAIR aggravates myocardial ischemia-reperfusion injury by sponging microRNA-126 to upregulate SRSF1. 33452620_Morphological study of TNPO3 and SRSF1 interaction during myogenesis by combining confocal, structured illumination and electron microscopy analysis. 33607522_The splicing factor SRSF1 stabilizes the mRNA of TSLP to enhance acute lung injury. 33981019_The long non-coding RNA PFI protects against pulmonary fibrosis by interacting with splicing regulator SRSF1. 34195277_lncRNA LINC01296 Promotes Oral Squamous Cell Carcinoma Development by Binding with SRSF1. 34233194_Splicing factor SRSF1 is indispensable for regulatory T cell homeostasis and function. 34338635_Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. 34522020_SRSF1 plays a critical role in invariant natural killer T cell development and function. 34839165_Splicing factor SRSF1 controls distinct molecular programs in regulatory and effector T cells implicated in systemic autoimmune disease. 35154164_SRSF1 Deficiency Impairs the Late Thymocyte Maturation and the CD8 Single-Positive Lineage Fate Decision. 35202586_SRSF1 governs progenitor-specific alternative splicing to maintain adult epithelial tissue homeostasis and renewal. | ENSMUSG00000018379 | Srsf1 | 1368.235728 | 1.0567357 | 0.079614586 | 0.09933418 | 6.307530e-01 | 4.270793e-01 | 7.619222e-01 | No | Yes | 1231.587118 | 226.567575 | 1160.459776 | 213.469483 | |||
| ENSG00000267673 | 112812 | FDX2 | protein_coding | Q6P4F2 | FUNCTION: Essential for heme A and Fe/S protein biosynthesis. {ECO:0000269|PubMed:20547883}. | 2Fe-2S;3D-structure;Alternative splicing;Disease variant;Electron transport;Iron;Iron-sulfur;Metal-binding;Mitochondrion;Primary mitochondrial disease;Reference proteome;Transit peptide;Transport | Mouse_homologues NA; + ;NA | This gene encodes a member of the ferredoxin family. The encoded protein contains a 2Fe-2S ferredoxin-type domain and is essential for heme A and Fe/S protein biosynthesis. Mutation in FDX1L gene is associated with mitochondrial muscle myopathy. [provided by RefSeq, Sep 2014]. | hsa:112812; | mitochondrial matrix [GO:0005759]; 2 iron, 2 sulfur cluster binding [GO:0051537]; electron transfer activity [GO:0009055]; metal ion binding [GO:0046872] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 23208207_unique conformational change is observed when holo hFd2 is warmed to physiological temperatures, or higher 24281368_Fdx2 is the second component of the Fe-S cluster biogenesis machinery, the first being IscU that is associated with isolated mitochondrial myopathy. 30010796_The phenotype was mapped to the same homozygous missense mutation in FDX2 (c.431C > T, p.P144L). The deleterious effect of the mutation was validated by real-time reverse transcription polymerase chain reaction and western blot analysis, which demonstrated normal expression of FDX2 mRNA but severely reduced expression of FDX2 protein in muscle tissue. 31395877_By binding zinc-free ISCU, iron drives persulfide uptake from NFS1 and allows persulfide reduction into sulfide by FDX2, thereby coordinating sulfide production with its availability to generate Fe-S clusters. FXN stimulates the whole process by accelerating persulfide transfer. This reconstitution recapitulates physiological conditions which provides a model for Fe-S cluster biosynthesis on the scaffold protein ISCU. | ENSMUSG00000111692+ENSMUSG00000079677 | Gm49373+Fdx2 | 510.398299 | 0.8542824 | -0.227215114 | 0.12078543 | 3.549749e+00 | 5.955447e-02 | 3.275496e-01 | No | Yes | 494.345095 | 72.308723 | 586.887711 | 85.598505 |
| ENSG00000268388 | 400550 | FENDRR | lncRNA | This gene produces a spliced long non-coding RNA transcribed bidirectionally with FOXF1 on the opposite strand. A similar gene in mouse is essential for normal development of the heart and body wall. The encoded transcript is thought to act by binding to polycomb repressive complex 2 (PRC2) and/or TrxG/MLL complexes to promote the methylation of the promoters of target genes, thus reducing their expression. It has been suggested that this transcript may play a role in the progression of gastric cancer. Alternatively spliced transcript variants have been identified. [provided by RefSeq, Mar 2015]. | 25167886_FENDRR plays an important role in the progression and metastasis of gastric cancer. 27577075_Results suggest that long noncoding RNA FOXF1-AS1 might regulate epithelial-mesenchymal transition (EMT), stemness and metastasis of non-small cell lung cancer (NSCLC) cells via EZH2, indicating it as a therapeutic target for future treatment of NSCLC. 29293945_These findings suggested the CNVR_3425.1 of FENDRR to be a possibly predictive biomarker for the risk of lung cancer and chronic obstructive pulmonary disease 29465000_FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. 29917257_lncRNAs MHRT, FENDRR and CARMEN show distinct expression profiles in hypertensive patients 30556873_Over-expression of FENDRR inhibited the malignant phenotypes of non-small cell lung carcinoma cells by binding competitively to miR-761. 30981768_FENDRR attenuates NSCLC cell stemness through inhibiting the HuR/MDR1 axis 30983519_FENDRR silences survivin via SETDB1-mediated H3K9 methylation, thereby represses cholangiocarcinoma cell proliferation, migration and invasion. 31502679_Screening of significant biomarkers related with prognosis of liver cancer by lncRNA-associated ceRNAs analysis. 31539119_FENDRR reduces tumor invasiveness in prostate cancer PC-3 cells by targeting CSNK1E. 31545237_long noncoding RNA FENDRR suppressed the progression of non-small cell lung cancer via binding to miR-761 and regulating TIMP2 expression. 32134466_Long non-coding RNA FENDRR inhibits migration and invasion of cutaneous malignant melanoma cells. 32251605_LncRNA FENDRR Upregulation Promotes Hepatic Carcinoma Cells Apoptosis by Targeting miR-362-5p Via NPR3 and p38-MAPK Pathway. 32537758_LncRNA Fendrr inhibits hypoxia/reoxygenation-induced cardiomyocyte apoptosis by downregulating p53 expression. 32590916_Integrated Analysis of lncRNAs and mRNAs Identifies a Potential Driver lncRNA FENDRR in Lung Cancer in Xuanwei, China. 32792604_Long non-coding RNA FENDRR regulates IFNgamma-induced M1 phenotype in macrophages. 32945076_Decreased long non-coding RNA lincFOXF1 indicates poor progression and promotes cell migration and metastasis in osteosarcoma. 33356998_FENDRR Sponges miR-424-5p to Inhibit Cell Proliferation, Migration and Invasion in Colorectal Cancer. 33692441_N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. 33739555_Lung-specific distant enhancer cis regulates expression of FOXF1 and lncRNA FENDRR. 33761608_FENDRR: A pivotal, cancer-related, long non-coding RNA. 34200642_LncRNA FENDRR Expression Correlates with Tumor Immunogenicity. 34445242_Long Noncoding RNA FENDRR Inhibits Lung Fibroblast Proliferation via a Reduction of beta-Catenin. 34697986_Long non-coding RNA FENDRR inhibits the stemenss of colorectal cancer cells through directly binding to Sox2 RNA. 35183135_An integrated analysis of the competing endogenous RNA network associated of prognosis of stage I lung adenocarcinoma. | 237.024809 | 0.8506472 | -0.233367164 | 0.17399749 | 1.780711e+00 | 1.820621e-01 | No | Yes | 239.075439 | 35.765166 | 280.186610 | 41.763707 | |||||||||
| ENSG00000268543 | TEC | 27.265403 | 0.6365648 | -0.651620645 | 0.51920098 | 1.525246e+00 | 2.168274e-01 | No | Yes | 22.591419 | 5.799944 | 34.893553 | 8.796026 | |||||||||||||
| ENSG00000269439 | 100507551 | PGLS-DT | lncRNA | 16.732850 | 1.5609367 | 0.642412061 | 0.67641083 | 8.553269e-01 | 3.550497e-01 | No | Yes | 16.968740 | 4.232408 | 10.795908 | 2.810083 | |||||||||||
| ENSG00000270580 | 105369154 | lncRNA | This locus represents naturally-occurring readthrough transcription between two unprocessed pseudogenes, PKD1P6 (polycystic kidney disease 1 (autosomal dominant) pseudogene 6) and NPIPP1 (nuclear pore complex interacting protein pseudogene 1). The individual pseudogene loci are not curated as transcribed regions. Readthrough transcripts likely do not encode functional proteins. [provided by RefSeq, Jan 2016]. | 531.567991 | 1.4361262 | 0.522182563 | 0.11955422 | 1.902495e+01 | 1.290204e-05 | 1.918951e-03 | No | Yes | 523.692061 | 113.733901 | 379.499001 | 82.454360 | ||||||||||
| ENSG00000271138 | IGLVIVOR22-1 | IG_V_pseudogene | 11.031343 | 1.2874093 | 0.364470787 | 0.83278798 | 1.966195e-01 | 6.574634e-01 | No | Yes | 9.895762 | 4.729513 | 7.961781 | 3.882061 | ||||||||||||
| ENSG00000271913 | 105378083 | TAGAP-AS1 | lncRNA | 75.220383 | 1.2200814 | 0.286977374 | 0.37152312 | 6.226424e-01 | 4.300670e-01 | No | Yes | 78.582259 | 15.832326 | 64.807846 | 13.058981 | |||||||||||
| ENSG00000272419 | LINC01145 | transcribed_unprocessed_pseudogene | 133.590374 | 0.8128572 | -0.298926089 | 0.24344356 | 1.534197e+00 | 2.154837e-01 | No | Yes | 147.121831 | 22.472339 | 182.419687 | 27.561218 | ||||||||||||
| ENSG00000272690 | 107986100 | LINC02018 | lncRNA | 28.513942 | 1.6281955 | 0.703273904 | 0.51190294 | 1.896601e+00 | 1.684593e-01 | No | Yes | 32.254400 | 8.265708 | 19.946839 | 5.171638 | |||||||||||
| ENSG00000272977 | lncRNA | 67.646878 | 1.4673978 | 0.553260060 | 0.32106375 | 2.950732e+00 | 8.583868e-02 | No | Yes | 70.072957 | 16.262753 | 50.173666 | 11.797826 | |||||||||||||
| ENSG00000273136 | 101060684 | NBPF26 | protein_coding | A0A494C1A6 | Reference proteome | 116.230877 | 1.8215235 | 0.865145596 | 0.33724966 | 6.582302e+00 | 1.029976e-02 | No | Yes | 127.519012 | 35.248761 | 70.837091 | 19.901537 | |||||||||
| ENSG00000274372 | LINC02804 | lncRNA | 21.544550 | 0.2461653 | -2.022300726 | 0.58348858 | 1.173608e+01 | 6.130004e-04 | No | Yes | 6.765937 | 2.339046 | 29.030825 | 8.933111 | ||||||||||||
| ENSG00000276043 | 29128 | UHRF1 | protein_coding | Q96T88 | FUNCTION: Multidomain protein that acts as a key epigenetic regulator by bridging DNA methylation and chromatin modification. Specifically recognizes and binds hemimethylated DNA at replication forks via its YDG domain and recruits DNMT1 methyltransferase to ensure faithful propagation of the DNA methylation patterns through DNA replication. In addition to its role in maintenance of DNA methylation, also plays a key role in chromatin modification: through its tudor-like regions and PHD-type zinc fingers, specifically recognizes and binds histone H3 trimethylated at 'Lys-9' (H3K9me3) and unmethylated at 'Arg-2' (H3R2me0), respectively, and recruits chromatin proteins. Enriched in pericentric heterochromatin where it recruits different chromatin modifiers required for this chromatin replication. Also localizes to euchromatic regions where it negatively regulates transcription possibly by impacting DNA methylation and histone modifications. Has E3 ubiquitin-protein ligase activity by mediating the ubiquitination of target proteins such as histone H3 and PML. It is still unclear how E3 ubiquitin-protein ligase activity is related to its role in chromatin in vivo. Plays a role in DNA repair by cooperating with UHRF1 to ensure recruitment of FANCD2 to interstrand cross-links (ICLs) leading to FANCD2 activation. {ECO:0000269|PubMed:10646863, ECO:0000269|PubMed:15009091, ECO:0000269|PubMed:15361834, ECO:0000269|PubMed:17673620, ECO:0000269|PubMed:17967883, ECO:0000269|PubMed:19056828, ECO:0000269|PubMed:21745816, ECO:0000269|PubMed:21777816, ECO:0000269|PubMed:22945642, ECO:0000269|PubMed:30335751}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Chromatin regulator;DNA damage;DNA repair;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a member of a subfamily of RING-finger type E3 ubiquitin ligases. The protein binds to specific DNA sequences, and recruits a histone deacetylase to regulate gene expression. Its expression peaks at late G1 phase and continues during G2 and M phases of the cell cycle. It plays a major role in the G1/S transition by regulating topoisomerase IIalpha and retinoblastoma gene expression, and functions in the p53-dependent DNA damage checkpoint. It is regarded as a hub protein for the integration of epigenetic information. This gene is up-regulated in various cancers, and it is therefore considered to be a therapeutic target. Multiple transcript variants encoding different isoforms have been found for this gene. A related pseudogene exists on chromosome 12. [provided by RefSeq, Feb 2014]. | hsa:29128; | chromatin [GO:0000785]; euchromatin [GO:0000791]; heterochromatin [GO:0000792]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; replication fork [GO:0005657]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; hemi-methylated DNA-binding [GO:0044729]; histone binding [GO:0042393]; identical protein binding [GO:0042802]; methyl-CpG binding [GO:0008327]; methylated histone binding [GO:0035064]; nucleosomal histone binding [GO:0031493]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; cell cycle [GO:0007049]; chromatin organization [GO:0006325]; DNA repair [GO:0006281]; histone monoubiquitination [GO:0010390]; histone ubiquitination [GO:0016574]; maintenance of DNA methylation [GO:0010216]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of cellular protein metabolic process [GO:0032270]; positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity [GO:2000373]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein autoubiquitination [GO:0051865]; protein ubiquitination [GO:0016567]; regulation of epithelial cell proliferation [GO:0050678]; ubiquitin-dependent protein catabolic process [GO:0006511] | 12838312_ICBP90 expression altered in cancer cell lines and upregulated by E2F-1 overexpression, with efficiency depending on cancer status of cell line 15033738_Apoptosis is dependent upon ICBP90 expression downregulation and ICBP90 exhibits anti-apoptotic properties. 15361834_ICBP90 is involved in cell proliferation by way of methylation-mediated regulation of certain genes. 15964557_ICBP90 down-regulation is a key event in G1 arrest preceding T cell death 16007129_ICBP90 overexpression is involved in the altered checkpoint controls occurring in cancerogenesis 16195352_General requirement for UHRF1 in tumor cell proliferation implicates the RING domain of UHRF1 as a functional determinant of growth regulation. 17065439_UHRF1 inhibits the late phase of oligodendrocyte differentiation. 17067204_These findings collectively indicate that the human NP95 gene is the functional orthologue of the murine Np95 gene. 17673620_data suggest that UHRF1 may help recruit DNMT1 to hemimethylated DNA to facilitate faithful maintenance of DNA methylation 17934516_A new role of ICBP90 in the relationship between histone ubiquitination and DNA methylation in the context of tumoral angiogenesis and tumour suppressor genes silencing. 17967883_ICBP90 is required for proper heterochromatin formation in mammalian cells 18005238_Parasite proliferation was suppressed in G1-arrested cells induced by UHRF1-siRNA, indicating the importance of the G2 phase via UHRF1-induced G1/S transition for T. gondii growth. 18772889_the 1.7 A crystal structure of the apo SRA domain of human UHRF1 and a 2.2 A structure of its complex with hemi-methylated DNA, revealing a previously unknown reading mechanism for methylated CpG sites (mCpG) 18945682_structural analysis and hemimethylated CpG binding of the SRA domain from human UHRF1 19056828_UHRF1 recruited and cooperated with G9a to inhibit the p21 promoter activity, which correlated with the elevated p21 protein level in human UHRF1 siRNA-transfected HeLa cells. 19077538_UHRF1 may bring two components (histones and DNA) carrying appropriate markers (on the tails of H3 and hemimethylated CpG sites) ready to be assembled into a nucleosome after replication 19270723_role of UHRF1 in human cervical cancer cells as a negative regulator of radiosensitivity. 19328461_ICBP90 might be a pivotal target for the ERK1/2 signaling pathway to control the proliferation of Jurkat T cells. 19491893_Significant overexpression of UHRF1 was observed in bladder cancer. The overexpression was correlated with the stage and grade of the cancer. 19798101_Np95, Dnmt3a and Dnmt3b have regulatory roles in mediating epigenetic silencing through histone modification followed by DNA methylation 19800870_Tip60 is a novel partner of the epigenetic integration platform interplayed by UHRF1, DNMT1 and HDAC1 involved in the epigenetic code replication. 19851296_Observational study of gene-disease association. (HuGE Navigator) 19943104_UHRF1 regulates BRCA1 transcription by inducing DNA methylation, histone modifications, and recruitment of transcriptional complex on the BRCA1 promoter in breast neoplasms. 20037778_Knockdown of UHRF1 activates MDR1 promoter activity and expression, attenuates the binding of UHRF1 and HDAC1 to the MDR1 promoter.Overexpression of UHRF1 in NCI/ADR-RES cells can induce deacetylation of histones H3 and H4 on the MDR1 promoter 20198315_Observational study of gene-disease association. (HuGE Navigator) 20613874_results reveal that the disruption of Dnmt1/PCNA/UHRF1 interactions acts as an oncogenic event and that one of its signatures (i.e. the low level of mMTase activity) is a molecular biomarker associated with a poor prognosis in GBM patients 21067293_UHRF1 confers radioresistance to human breast cancer cells 21214517_Data suggest that UHRF1 links epigenetic regulation with DNA replication. UHRF1 depletion/knockdown leads to caspase 8-mediated apoptosis, cell cycle arrest, and cell death. UHRF1 accumulates rapidly at sites of DNA injury. 21268065_Findings suggest that, by balancing Dnmt1 ubiquitination, Usp7 and Uhrf1 fine tune Dnmt1 stability. 21296067_down-regulation of ICBP90 induced the descended expression of Topo IIalpha protein which is the target enzyme of doxorubicin 21351083_UHRF1 should be considered, along with DNMTs, among the potential targets for cancer treatment and/or therapeutic stratification. 21489993_Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. 21539450_the downregulation of UHRF1 in both breast cancer cell lines significantly inhibited the colony formation capacity. 21611839_High UHRF1 is associated with tumor recurrence in non-muscle-invasive bladder cancer. 21777816_PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression 21808300_the PHD domain of UHRF1 is an epigenetic regulatory module dedicated to the recognition of an unmodified arginine residue (R2) on histone H3 22096602_The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. 22100450_UHRF1 PHD finger (including the preceding zinc-Cys4 knuckle) acts together with the adjacent double Tudor domain to specifically recognize the H3K9me3 mark. 22219067_High UHRF1 expression is associated with colorectal cancer. 22285022_The fusion of NRIP1 with UHRF1 involved in the unbalanced translocation between chromosomes 19 and 21 in a patient with an ALL-positive for a t(9;22) translocation. 22286757_UHRF1 coordinates peroxisome proliferator activated receptor gamma epigenetic silencing and mediates colorectal cancer progression. 22330138_Depletion of UHRF1 resulted in reactivation of several tumour suppressor genes. 22411829_A cell cycle-specific signaling event that relieves UHRF1 from its interaction with USP7, is reported. 22552622_UHRF1 promoted the proliferation of breast cancer cells by apoptosis inhibition, G1 phase shortage and promotion of tumor vessel formation 22837395_UHRF1 contains linked two-histone reader modules tethered by a 17-aa linker, which plays a role as a functional switch involved in multiple regulatory pathways such as maintenance of DNA methylation and transcriptional repression. 22945642_These findings suggest that UHRF1 promotes the turnover of PML protein 23022729_UHRF1 association with methylated histone H3 Lys9 (H3K9) is required for DNA methylation maintenance. 23023523_UHRF1 may be involved in cellular proliferation and molecular pathogenesis of colorectal cancer in the right hemicolon. 23107467_High UHRF1 DNA level in plasma was significantly associated with short progression-free survival. 23161542_H3K9me3 peptide induces conformational changes of TTD-PHD, which do not affect the autoubiquitination activity or hemimethylated DNA binding affinity of UHRF1 in vitro 23245651_From alchemical mutation free energy calculations the study finds that the binding affinity of fully-methylated DNA to UHRF1 is weaker by 17.9kJ/mol relative to the binding of hemi-methylated DNA. 23297342_UHRF1 degradation is accelerated in response to DNA damage. 23463006_UHRF1 can target DNMT1 for DNA maintenance methylation through binding either histone 3 lysine 9 methylation 2/3 or hemi-methylated CpG. 23537643_UHRF1 interacts with N-methylpurine DNA glycosylase (MPG) in cancer cells in vitro and displays a co-localization with MPG in the nucleoplasm. 23677994_UHRF1 is a critical negative regulator of TIP60 and suggest that UHRF1-mediated effects on p53 may contribute, at least in part, to its role in tumorigenesis. 23752590_linked tandem Tudor & PHD of UHRF1 operates as a functional unit in cells, providing a defined combinatorial readout of a heterochromatin signature within a single histone H3 tail, essential for UHRF1-directed epigenetic inheritance of DNA methylation 23943380_Inhibiting UHRF1 expression enhances radiosensitivity in human esophageal squamous cell carcinoma. 24005809_Increased expression of UHRF1 was an independent prognostic factor for LSCC. 24134914_reviews the principles how UHRF1 acts as an epigenetic reader and discusses the properties of UHRF1 to be a biomarker as well as a therapeutic target 24486181_UHRF1 overexpression drives DNA hypomethylation leading to hepatocellular carcinoma. 24637615_The global DNA hypomethylation induced by the DNMT1/PCNA/UHRF1 disruption is an oncogenic event of human tumorigenesis. 24706678_Results implicate UHRF1 as an oncogene and suggest that global DNA hypomethylation induced by UHRF1 overexpression induces a p53-mediated senescence program that is bypassed in hepatocellular carcinoma. 24756644_Our findings suggest that UHRF1 is involved in the proliferation and migration of GBC cells and may serve as a biomarker or even a therapeutic target for GBC. 24813945_Binding of the phosphatidylinositol phosphate to the polybasic region of UHRF1 results in a conformational rearrangement of the domains, allowing the tandem tudor domain to bind H3K9me3. 25189999_By DNA methyltransferase 1 under regulation by Uhrf1. 25207720_Luteolin induces UHRF1 and DNMT1 downregulation and stimulates p16INK4A reexpression in human colon cancer cell line BE. 25272010_These data demonstrated that upregulated UHRF1 increases bladder cancer cell invasion by epigenetic silencing of KiSS1. 25323766_Epigenetic repression of RGS2 by UHRF1 contributes to bladder cancer progression. UHRF1 inhibits RGS2 expression by increasing the methylation of CpG nucleotides of the RGS2 promoter. 25338120_Upregulation of HELLS and UHRF1 is essential for the tumor phenotype. Also, these epigenetic regulators are important for the regulation of SYK. 25416862_our study demonstrated that UHRF1 played an oncogenic role in the progression of pancreatic cancer, and UHRF1 might be a promising target for the treatment of pancreatic cancer. 25430639_Results show that UHRF1 is overexpressed in subgroups of patients with liver cancer and its repression through T3/TR signaling induces hepatoma cell growth inhibition. 25451918_UHRF1 interacts with TopoIIalpha and regulates its localization to hemimethylated DNA. TopoIIalpha decatenates the hemimethylated DNA following replication, which might facilitate the methylation of the nascent strand by DNMT1 25550546_CD47 regulates the epigenetic code by targeting UHRF1. 25572953_UHRF1 may play a pivotal role in suppressing the malignant alteration of cancer cells. 25641194_Findings demonstrated the inhibitory effect of MEG3 in vivo and vitro and illuminated that MEG3 could be a potential biomarker for the survival of hepatocellular carcinoma (HCC) patients. Its expression seemed to be regulated by UHRF1 in HCC. 25743796_High UHRF1 expression is associated with ovarian cancer. 25801034_describe a mechanism of interstrand crosslink (ICL) sensing and propose that UHRF1 is a critical factor that binds to ICLs. In turn, this binding is necessary for the subsequent recruitment of FANCD2, which allows the DNA repair process to initiate 25818288_UHRF1 promotes recruitment of lesion-processing activities via its affinity to recognize DNA damage 25940709_miR-9 could repress the expression of UHRF1, and function as a tumor-suppressive microRNA in colorectal cancer. 25982273_LASP-1 associated with UHRF1, G9a, Snail1 and di- and tri-methylated histoneH3 in a CXCL12-dependent manner based on immunoprecipitation and proximity ligation assays 26049093_Promoter hypermethylation and down-expression of let-7a-3 may play a role in DN by targeting UHRF1. 26070868_UHRF1 could act as a new oncogenic factor in ovarian cancer and be a potential molecular target for ovarian cancer gene therapy. 26102039_UHRF1 regulates p53 ubiquitination and p53-dependent cell apoptosis in clear cell renal cell carcinoma. 26147747_UHRF1 promotes proliferation of gastric cancer via mediating tumor suppressor gene hypermethylation. 26161699_UHRF1 is overexpressed in serum and gastric tissue of stomach cancer patients. 26239364_The downregulation of UHRF1 by Torin-1 was partially due to a decrease in the UHRF1 mRNA level. Torin-2 effectively inhibited hepatocellular carcinoma cell proliferation through induction of autophagy 26310391_miR-124 could control the fate of target gene UHRF1 mRNA by binding 3'-UTR 26368281_results are consistent with a 'reader' role, in which the SRA domain scans DNA sequences for hemimethylated CpG sites without perturbation of the structure of contacted nucleotides. 26464697_UHRF1 was over-expressed in hepatocellular carcinoma tissues compared to the adjacent normal tissues and UHRF1 expression shared significant relevance with distant metastasis, cancer area and HBV. 26497117_K-Ras drives UHRF1 expression, establishing a novel link between this oncogene and Nrf2-mediated cellular protection. 26548607_UHRF1 promotes osteosarcoma cell invasion by downregulating the expression of Ecadherin and increasing epithelialtomesenchymal transition in an Rb1dependent manner. 26597461_UHRF1 could form a complex with PRMT5 to contribute to the endometrial carcinogenesis. 26700964_Studies indicate that ubiquitin like with PHD and ring finger domains 1 protein (UHRF1) interacts with DNA interstrand crosslinks (ICLs) both in vitro and in vivo. 26727879_This study showed that UHRF1 interacts directly with BRCA1 and is involved in DNA double-strand break repair. 26752645_These findings identify ICBP90 as a key regulator of MIF transcription and provide functional insight into the regulation of the polymorphic MIF locus. 26768845_results suggest that UPAT and UHRF1 may be promising molecular targets for the therapy of colon cancer 26884069_Results showed that UHRF1 was overexpressed in almost all of the prostate cancer cell (PCa) lines. Its expression levels were correlated with some clinical features of PCa suggesting it as an independent prognostic factor for biochemical recurrence. 26896831_The overexpression of UHRF1 was correlated with the stage and grade of gastric cancer and is associated with the genotype distribution of MiR-146a rs2910164. 27045799_Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its H3 histone recognition. 27072587_our present data demonstrated that both strands of miR-145 (miR-145-5p: guide-strand and miR-145-3p: passenger-strand) play pivotal roles in bladder cancer cells by regulating UHRF1 27431502_Our study uncovered that UHRF1 overexpressed in cervical squamous cell carcinoma and silent UHRF1 gene can reduce tumorigenicity of the CaSki cells by decreasing its proliferation capacity and making it stagnate at G0 and G1 phases as well as accelerating its apoptosis 27449774_Down-regulation of UHRF1 by RNAi inhibited proliferation and clonogenic ability of medullobastoma cell lines with cell cycle arrest in G1/G2-phase. Cells transfected with lenti-shUHRF1 showed increased p16 expression and location shift of CDK4 in medulloblastoma cells. 27489107_results identify UHRF1 as a novel HSP90 client protein and shed light on the regulation of UHRF1 stability and function. 27507047_UHRF1 is a key epigenetic regulator of DNA methylation in esophageal squamous cell carcinoma and might be a potential target for cancer treatment 27595565_Hemi-methylated CpGs DNA recognition activates UHRF1 ubiquitylation towards multiple lysines on the H3 tail adjacent to the UHRF1 histone-binding site. 27839516_Results find the overexpression of the anti-apoptotic UHRF1 to coordinate the epigenetic silencing of several tumor suppressor genes in many human haematological and solid cancers causing apoptosis inhibition. [review] 27886214_Epigenetic enhancement of the post-replicative DNA mismatch repair of mammalian genomes by a Hemi-(m)CpG-Np95-Dnmt1 axis has been demonstrated for humans and mice. 28060737_Elevated UHRF1 expression contributes to poor prognosis by promoting cell proliferation and metastasis in hepatocellular carcinoma. 28100769_decreased UHRF1 expression is a key initial event in the suppression of DNMT1-mediated DNA methylation and in the consequent induction of senescence via increasing WNT5A expression 28128913_UHRF1 gene expression levels correlates with the major pathological characteristics of transitional cell carcinoma samples and with the clinical outcome of those patients. Determination of UHRF1 gene expression could have a potential to be used as a sensitive molecular marker in patients with urinary bladder cancer. 28285359_The results indicated that globular adiponectin inhibited the prooncogenic effects of leptin via AdipoR2-mediated suppression of UHRF1 28334952_Our results provide insights into the molecular basis of DNMT1 dysfunction in hereditary sensory and autonomic neuropathies with dementia and hearing loss patients and emphasize the importance of the TS domain in the regulation of DNA methylation in pluripotent and differentiating cells 28394343_Down-regulation of UHRF1 is an important mechanism of PIM1-mediated cellular senescence. 28467809_These results suggest that tumor-promoting functions of UHRF1 in retinoblastoma are largely independent of its role in DNA methylation. 28584306_UHRF1 deficiency leads to the induction of EMT by activating the CXCR4/AKT-JNK/IL-6 signaling pathway, thereby contributing to the expansion of cancer stem-like cells 28803780_A histone H3K9-like mimic within LIG1 is methylated by G9a and GLP and avidly binds UHRF1. Interaction with methylated LIG1 promotes the recruitment of UHRF1 to DNA replication sites and is required for DNA methylation maintenance. 28901428_The emodininduced downregulation of UHRF1 led to an increase in the level of DNA methyltransferase 3A. 28901497_Findings confirmed that UHRF1 is a gene driver in medulloblastoma (MB) and, revealed that UHRF1 is in a modulation axis downstream of miR-378. Its promoter region is targeted by miR-378 which negatively regulates its expression in MB cells. 28981102_The induced expression of RIP3 by UHRF1 RNAi depends on the presence of Sp1. Remarkably, the ectopic expression of RIP3 in RIP3-null cancer cells results in a decrease in tumor growth in mice. Therefore, our findings offer insights into RIP3 expression control in cancer cells and suggest an inhibitory effect of RIP3 on tumorigenesis. 29074623_Using an integrative approach that combines small angle X-ray scattering, NMR spectroscopy, and molecular dynamics simulations, we characterized the dynamics of the tandem tudor domain-plant homeodomain (TTD-PHD) histone reader module, including its 20-residue interdomain linker. 29157076_hese findings establish UBE2L6 as a novel target of UHRF1 that regulates the apoptosis function of UHRF1. 29268763_Our data demonstrate for the first time that TIP60 through its MYST domain directly interacts with UHRF1 29415984_UHRF1-mediated XRCC4 upregulation under pathophysiological conditions triggered by RB1 gene inactivation may confer protection against endogenous DNA damages that arise during retinoblastoma development. 29466696_UHRF1 controls both CDH1 and antisense-directed non coding RNA RCDH1-AS by directly binding this bidirectional promoter region. 29506131_Uncoupling of UHRF2 from the DNA methylation maintenance program is linked to differences in the molecular readout of chromatin signatures that connect UHRF1 to ubiquitination of histone H3. 29507645_Findings suggest that UHRF1 is critical for aberrant tumor suppressor genes silencing and sustained growth signaling in hepatoblastoma and that UHRF1 overexpression levels might serve as a prognostic biomarker. 29516630_These findings reveal that WDR79 is a novel UHRF1 regulator by maintaining UHRF1 stability. 29620240_Study provides evidence that elevated expression of UHRF1 plays an important role in melanoma cell proliferation and progression, and it can be used as a prognostic biomarker for melanoma. 29656054_Lys4 methylation on H3 peptide has an insignificant effect on combinatorial recognition of R2 and K9me2 on H3 by the UHRF1 TTD-PHD. We propose that subtle variations of key residues at the binding pocket determine status specific recognition of histone methyl-lysines by the reader domains. 29752436_a novel mechanism by which FOXM1 controls cancer stem cells and taxane resistance through a UHRF1-mediated signaling pathway 30104358_studies support a noncompetitive model for UHRF1 and DNMT1 chromatin recruitment to replicating chromatin and define a role for hemimethylated linker DNA as a regulator of UHRF1 ubiquitin ligase substrate selectivity. 30309512_Our novel findings strongly indicate that inactivation of the Notch signaling pathway impedes hepatocellular carcinoma progress via reduction of ICBP90. 30352833_These findings indicated that UHRF1 promoted the growth, migration and invasion of MGC803 and SGC7901 cells and inhibited apoptosis via a ROS-associated pathway 30357346_SET7-mediated UHRF1 methylation is also shown to be essential for cell viability against DNA damage. Our data revealed the regulatory mechanism underlying the UHRF1 methylation status by SET7 and LSD1 in double-strand break repair pathway 30392931_The UBL coordinates with other UHRF1 domains that recognize epigenetic marks on DNA and histone H3 to direct ubiquitin to H3. Results show UBLs from other E3s also have a conserved interaction with the E2, Ube2D, highlighting a potential prevalence of interactions between UBLs and E2s. 30639225_Data report the crystal structure of the UHRF1 tandem tudor domain (TTD) bound to a LIG1K126me3 peptide. The data explain the basis for the high TTD-binding affinity of LIG1K126me3 and reveal that the interaction may be regulated by phosphorylation. Binding of LIG1K126me3 switches the overall structure of UHRF1 from a closed to a flexible conformation, suggesting that auto-inhibition is relieved. 30710064_This study demonstrated that overexpressing UHRF1 was involved in vascular smooth cells proliferation through reducing inhibitory geminin protein levels to promote cell cycle as well as activating PI3K-Akt signaling. 30956060_Key domains of the DNMT1 targeting protein UHRF1 maintain the function of cancer-specific DNA methylation, contributing to key malignant properties of colorectal cancer. 30989656_UHRF1, regulated by miR-124-3p, acts as a tumor promoter by promoting cell proliferation in intrahepatic cholangiocarcinoma. 31043707_results confirm that UHRF1 has oncogenic function in RCC and UHRF1 may promote tumor progression through epigenetic regulation of TXNIP 31044388_Silencing UHRF1 Inhibits Cell Proliferation and Promotes Cell Apoptosis in Retinoblastoma Via the PI3K/Akt Signalling Pathway. 31058289_Results indicate that miR-202 inhibits the proliferation and invasion of colorectal cancer (CRC) via targeting ubiquitin-like with PHD and RING finger domain 1 (UHRF1). 31064417_Results suggest that maintenance of DNA methylation in colorectal cancer cells is highly dependent on UHRF1. 31238111_Role of UHRF1 in malignancy and its function as a therapeutic target for molecular docking towards the SRA domain. 31400111_SET8, a cell-cycle-regulated protein methyltransferase, controls protein stability of both UHRF1 and DNMT1 through methylation-mediated, ubiquitin-dependent degradation and consequently prevents excessive DNA methylation. 31428652_results showed that UHRF1 can promote proliferation of hADSCs after overexpression of UHRF1, while proliferation of human adipose-derived stem cells (hADSCs) was reduced through downregulation of UHRF1, and UHRF1 can control proliferation of hADSCs through transition from G1-phase to S-phase; besides, we found that UHRF1 negatively regulates adipogenesis of hADSCs via PPAR gamma . 31481468_UHRF1's SRA finger loop regulates its conformation and function. 31492965_This study reports a new parasite persistence strategy involving T. gondii rhoptry protein ROP16 secreted early during invasion, which targets the transcription factor UHRF1 (ubiquitin-like containing PHD and RING fingers domain 1), and leads to host cell cycle arrest. 31545467_Results find that UHRF1 can suppress KLF17 expression through the hypermethylation of its promoter in colorectal cancer. 31582837_UHRF1-KAT7-mediated regulation of TUSC3 expression via histone methylation/acetylation is critical for the proliferation of colon cancer cells. 31802345_We detected two SNPs in the UHRF1 gene showing a significant difference between the case and control groups. Two screened SNPs affected UHRF1 promoter activity, improving the understanding of the pathophysiology of oligozoospermia. 31873045_Survival analysis revealed that hsa-let-7b was the only miRNA that affected the overall survival in melanoma. Overexpressed hsa-let-7b could significantly inhibit the proliferation ability of A375 and A2058 cells, and this phenomenon was reversed after co-transfection with pLenti-UHRF1. In conclusion, hsa-let-7b regulates melanoma cells proliferation in vitro by targeting UHRF1. 31964471_PARP1 mediates the homologous recombination mechanism, which is regulated by UHRF1 methylation. 32034755_review of the regulation of UHRF1 in DNA methylation and histone methylation, and the potential epigenetic role of UHRF1 in angiogenesis [review] 32212938_UHRF1 is expressed at higher levels in both lung adenocarcinoma (AD) and squamous cell carcinoma (SQ); however, a meta-analysis showed that UHRF1 expression is correlated with worse survival in patients with AD but not in those with SQ. UHRF1 knockdown suppressed the growth of lung cancer cell lines through G1 cell cycle arrest in some cell lines. 32312517_UHRF1 silences gelsolin to inhibit cell death in early stage cervical cancer. 32379550_CircFAT1 Suppresses Colorectal Cancer Development Through Regulating miR-520b/UHRF1 Axis or miR-302c-3p/UHRF1 Axis. 32428527_Serine 298 Phosphorylation in Linker 2 of UHRF1 Regulates Ligand-Binding Property of Its Tandem Tudor Domain. 32495469_UHRF1 predicts poor prognosis by triggering cell cycle in lung adenocarcinoma. 32543137_[Effects of UHRF1 on Estrogen Receptor and Proliferation, Invasion and Migration of BCPAP Cells in Thyroid Papillary Carcinoma]. 32726623_Acetylation of UHRF1 Regulates Hemi-methylated DNA Binding and Maintenance of Genome-wide DNA Methylation. 32819559_Silencing UHRF1 enhances cell autophagy to prevent articular chondrocytes from apoptosis in osteoarthritis through PI3K/AKT/mTOR signaling pathway. 32927122_UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. 33097091_The histone and non-histone methyllysine reader activities of the UHRF1 tandem Tudor domain are dispensable for the propagation of aberrant DNA methylation patterning in cancer cells. 33170271_A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association. 33795533_UHRF1 Induces Methylation of the TXNIP Promoter and Down-Regulates Gene Expression in Cervical Cancer. 33844457_ICBP90 Regulates MIF Expression, Glucocorticoid Sensitivity, and Apoptosis at the MIF Immune Susceptibility Locus. 33922029_Thymoquinone Is a Multitarget Single Epidrug That Inhibits the UHRF1 Protein Complex. 34013373_UHRF1 regulates the transcriptional repressor HBP1 through MIF in T acute lymphoblastic leukemia. 34265399_UHRF1 promotes androgen receptor-regulated CDC6 transcription and anti-androgen receptor drug resistance in prostate cancer through KDM4C-Mediated chromatin modifications. 34465029_UHRF1 Suppresses HIV-1 Transcription and Promotes HIV-1 Latency by Competing with p-TEFb for Ubiquitination-Proteasomal Degradation of Tat. 34558642_TIP60 governs the autoubiquitination of UHRF1 through USP7 dissociation from the UHRF1/USP7 complex. 34897340_Tannin extract from maritime pine bark exhibits anticancer properties by targeting the epigenetic UHRF1/DNMT1 tandem leading to the re-expression of TP73. 35262949_UHRF1 shapes both the trophoblast invasion and decidual macrophage differentiation in early pregnancy. | ENSMUSG00000001228 | Uhrf1 | 5172.152527 | 0.9534909 | -0.068708995 | 0.05533086 | 1.545573e+00 | 2.137902e-01 | 5.861005e-01 | No | Yes | 4736.917289 | 441.552717 | 4956.249245 | 461.832194 |
| ENSG00000276141 | WHAMMP3 | transcribed_unprocessed_pseudogene | 89.835896 | 1.2591761 | 0.332480030 | 0.28218224 | 1.395230e+00 | 2.375239e-01 | No | Yes | 95.580556 | 27.105436 | 77.128925 | 21.937141 | ||||||||||||
| ENSG00000278259 | 80179 | MYO19 | protein_coding | Q96H55 | FUNCTION: Actin-based motor molecule with ATPase activity that localizes to the mitochondrion outer membrane (PubMed:19932026, PubMed:23568824, PubMed:25447992). Motor protein that moves towards the plus-end of actin filaments (By similarity). Required for mitochondrial inheritance during mitosis (PubMed:25447992). May be involved in mitochondrial transport or positioning (PubMed:23568824). {ECO:0000250|UniProtKB:Q5SV80, ECO:0000269|PubMed:19932026, ECO:0000269|PubMed:25447992, ECO:0000305|PubMed:23568824}. | ATP-binding;Actin-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Membrane;Mitochondrion;Mitochondrion outer membrane;Motor protein;Myosin;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat | hsa:80179; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; myosin complex [GO:0016459]; vesicle [GO:0031982]; actin binding [GO:0003779]; actin filament binding [GO:0051015]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; microfilament motor activity [GO:0000146]; myosin light chain binding [GO:0032027]; plus-end directed microfilament motor activity [GO:0060002]; actin filament organization [GO:0007015]; mitochondrion migration along actin filament [GO:0034642]; regulation of cytokinesis [GO:0032465]; regulation of mitochondrial fission [GO:0090140]; vesicle transport along actin filament [GO:0030050] | 19932026_These results suggest that this novel myosin functions as an actin-based motor for mitochondrial movement in vertebrate cells. 25447992_Myo19 is a novel regulator of cell division. 26659663_Myo19 is a stably attached OMM molecular motor. 27126804_these data indicate that the MyMOMA domain contains strong membrane-binding activity, and membrane targeting is mediated by a specific, basic region of the MYO19 tail with slow dissociation kinetics appropriate for its role(s) in mitochondrial network dynamics 28912602_we provide a model explaining how Myo19 translocation may be regulated by the local ATP/ADP ratio, coupled to the mitochondria presence in the filopodia. 30111583_Miro1 binds directly to a C-terminal fragment of the Myo19 tail region and that Miro1/2 recruit the Myo19 tail. 31479585_The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. 34013964_Coordination of mitochondrial and cellular dynamics by the actin-based motor Myo19. | ENSMUSG00000020527 | Myo19 | 1141.296994 | 1.0971565 | 0.133769297 | 0.09509918 | 1.952550e+00 | 1.623123e-01 | 5.236694e-01 | No | Yes | 1122.163057 | 110.757358 | 1032.828866 | 101.949191 | ||
| ENSG00000278903 | lncRNA | 142.918286 | 1.1464336 | 0.197152861 | 0.25219345 | 5.883309e-01 | 4.430653e-01 | No | Yes | 162.681619 | 34.507345 | 144.683888 | 30.774517 | |||||||||||||
| ENSG00000280734 | LINC01232 | lncRNA | 30.420882 | 1.4029644 | 0.488478401 | 0.50138354 | 9.066615e-01 | 3.410018e-01 | No | Yes | 33.341115 | 8.003039 | 23.373447 | 5.529288 | ||||||||||||
| ENSG00000281706 | 100507173 | LINC01012 | lncRNA | 144.832975 | 1.0467229 | 0.065879588 | 0.21758267 | 9.173090e-02 | 7.619881e-01 | No | Yes | 150.653530 | 20.710152 | 143.569734 | 19.830823 | |||||||||||
| ENSG00000283050 | 101926918 | GTF2IP12 | transcribed_unprocessed_pseudogene | 586.630058 | 1.1188345 | 0.161996652 | 0.12442339 | 1.694740e+00 | 1.929773e-01 | 5.634622e-01 | No | Yes | 692.591981 | 90.500029 | 632.180341 | 82.582039 | ||||||||||
| ENSG00000283341 | lncRNA | 70.568320 | 1.1308643 | 0.177425778 | 0.33319737 | 2.790565e-01 | 5.973203e-01 | No | Yes | 82.613841 | 25.244241 | 75.596835 | 22.969780 | |||||||||||||
| ENSG00000285219 | 100506207 | HULC | lncRNA | 87.051634 | 1.0881955 | 0.121937704 | 0.28733804 | 1.789007e-01 | 6.723197e-01 | No | Yes | 94.446143 | 15.807830 | 86.138347 | 14.411789 | |||||||||||
| ENSG00000286011 | lncRNA | 16.360020 | 0.8667559 | -0.206302296 | 0.63967732 | 1.026435e-01 | 7.486801e-01 | No | Yes | 11.230416 | 3.561479 | 12.845795 | 4.049698 | |||||||||||||
| ENSG00000288674 | 5664 | protein_coding | Q8NI60 | FUNCTION: Atypical kinase involved in the biosynthesis of coenzyme Q, also named ubiquinone, an essential lipid-soluble electron transporter for aerobic cellular respiration (PubMed:25498144, PubMed:21296186, PubMed:25540914, PubMed:27499294). Its substrate specificity is unclear: does not show any protein kinase activity (PubMed:25498144, PubMed:27499294). Probably acts as a small molecule kinase, possibly a lipid kinase that phosphorylates a prenyl lipid in the ubiquinone biosynthesis pathway, as suggested by its ability to bind coenzyme Q lipid intermediates (PubMed:25498144, PubMed:27499294). Shows an unusual selectivity for binding ADP over ATP (PubMed:25498144). {ECO:0000269|PubMed:25498144, ECO:0000269|PubMed:27499294, ECO:0000305|PubMed:21296186, ECO:0000305|PubMed:25540914}. | 3D-structure;ATP-binding;Alternative splicing;Direct protein sequencing;Disease variant;Kinase;Membrane;Mitochondrion;Neurodegeneration;Nucleotide-binding;Primary mitochondrial disease;Reference proteome;Transferase;Transit peptide;Transmembrane;Transmembrane helix;Ubiquinone biosynthesis | PATHWAY: Cofactor biosynthesis; ubiquinone biosynthesis. {ECO:0000269|PubMed:25498144}. | hsa:56997; | extrinsic component of mitochondrial inner membrane [GO:0031314]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; ADP binding [GO:0043531]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; phosphorylation [GO:0016310]; ubiquinone biosynthetic process [GO:0006744] | Mouse_homologues 11756438_These data reveal that a novel PS1/2-independent mechanism plays a partial role in Notch signal transduction 11817902_inhibits protein synthesis 11904448_Wild-type and mutated presenilins 2 trigger p53-dependent apoptosis and down-regulate presenilin 1 expression in HEK293 human cells and in murine neurons 12198112_PS2/gamma-secretase contains PEN-2 and requires it for presenilin expression 12646573_role in stability and maturation of nicastrin in mammalian brain 12684521_gamma-secretase activity in blastocysts from normal, heterozygote and knockout mice 12885769_presenilins are multifunctional proteins with catalytic activity as well as roles in the generation, stabilization, and transport of the gamma-secretase complex 14566063_gamma-secretase contains a presenilin dimer in its catalytic core, that binding of substrate is at a site separate from the active site, and that substrate is cleaved at the interface of two PS molecules 15122901_Results establish a critical role of presenilin 2 in myelopoiesis; partial loss of PS2 in mice leads to age-dependent myeloproliferative disease, mediated through its gamma-secretase activity. 15148382_presenilins are essential for the ongoing maintenance of cortical structures and function 15264228_Expression of mutant presenilin 2(M239V-PS2) caused neuronal cytotoxicity which could be inhibited by the combination of two clinically usable inhibitors of superoxide-generating enzymes, apocynin and oxypurinol 15287885_Our data demonstrate that presenilin-directed gamma-secretase inhibitors affect caspase 3 activity in a presenilin-independent manner 15322084_presenilin 1 (PS1)-derived fragments, mature nicastrin, APH-1, and PEN-2, associate with cholesterol-rich detergent insoluble membrane (DIM) domains of non-neuronal cells and neurons 15537629_results suggest that the proximal two-thirds of the PEN-2 TMD1 is functionally important for endoproteolysis of PS1 holoproteins and the generation of PS1 fragments, essential components of the gamma-secretase complex 15613480_PS1 and PS2 affect caveolin 1 trafficking in an indirect manner and are required for caveolae formation by controlling transport of intracellular caveolin 1 to the plasma membrane. 15649694_The metabolic profile, however, showed clear indicators of hypometabolism with age in the PS2APP mice: both N-acetyl-aspartate and glutamate were significantly reduced in the older animals. 15858415_presenilin 2 may be implicated in neuronal cell death by altering the antiapoptotic activity of the transcription factors 15866159_animals lacking all presenilin alleles have no somites 15922300_presenilin 1 and 2 in mouse embryonic fibroblasts have roles in responding to invasion and replication of Salmonella typhimurium 16103123_presenilin, nicastrin, APH-1, and PEN-2, are present and enriched on phagosome membranes from both murine macrophages and Drosophila S2 phagocytes 16204356_PS2 plays an important role in cardiac excitation-contraction coupling by interacting with RyR2 16384915_tyrosinase and related proteins as physiological substrates of presenilins and link gamma-secretase activity with intracellular protein transport 16641999_TMP21, a member of the p24 cargo protein family, is a component of presenilin complexes and differentially regulates gamma-secretase cleavage without affecting epsilon-secretase activity 16887746_the promoter of the mouse Presenilin-2 gene is bound and activated by CLOCK and BMAL1, transcription factors of the mammalian circadian clock 17121184_Findings indicate that PS2 is involved in the abnormalities of the lipid profile, which could cause or result in Alzheimer's disease. 17292944_C-terminal fragment of presenillin 2 is released by shedding into the extracellular compartment as a soluble form. This release is 4.07-fold increased during apoptosis. 17556541_PS2 is necessary for the proteolytic activity of gamma-secretase. 17614943_environmental enrichment effectively enhanced memory and partially rescued the forebrain atrophy of the Presenilin 1 and 2 conditional double knockout mice 17698590_These findings suggest a model in which PS-dependent Notch signaling influences positive selection and the development of alphabeta T cells by modifying T-cell receptor signal transduction. 17728018_Thus these results demonstrate up-regulation of PS2 protein expression by sex steroids, which in turn may influence PS2 associated brain functions. 17874292_Age and sex dependent alteration in presenilin expression. 18195359_B cells deficient in both PS1 and PS2 function have an unexpected and substantial deficit in both lipopolysaccharide and B cell antigen receptor-induced proliferation and signal transduction events 18293935_Mature integrin beta1 with increased expression level is delivered to the cell surface, which results in an increased cell surface expression level of mature integrin beta 1 in presenilin (PS)1 and PS2 double-deficient fibroblasts. 18440065_Data demonstrate facilitation of ryanodine receptor gating by presenilin-2 NTF1-87. 18803281_PS2 mutation causes reduction of anxiety 18822370_the oxidative stress status in PS1 and PS2 double-knockout (PS cDKO) mice using F(2)-isoprostanes (iPF(2alpha)-III) as the marker of lipid peroxidation 19297528_Met is processed in epithelial cells by presenilin-dependent regulated intramembrane proteolysis (PS-RIP) independently of ligand stimulation. 19376115_PS2beta may inhibit gamma-secretase activity by affecting the gamma-secretase complex assembly 19382908_Presenilin-2 dampens intracellular Ca2+ stores by increasing Ca2+ leakage and reducing Ca2+ uptake 19573580_Egr-1 over-expression and down-regulation, as well as in-vivo examination of Egr-1 and Psen2 expression during fear conditioning, showed that Egr-1 does not regulate the Psen2 promoter. 19834068_Data show that presenilins 1 and 2 are highly enriched in a subcompartment of the endoplasmic reticulum associated with mitochondria that forms a physical bridge between the two organelles. 20573903_These results indicate that disruption of ER Ca(2+) leak function of presenilin 1 and 2 may play an important role in Alzheimer disease pathogenesis. 20875805_partial, but highly significant, loss of mGlur2 receptors in amyloid-affected discrete brain regions of Alzheimer cases and PS2APP mice 20953898_These findings suggest that inflammation in the presenilin 1 (PS1) and presenilin 2 (PS2) double knockout mice could expand to the periphery, including the oral tissue, which could ultimately induce abnormalities in the periodontal and salivary tissues. 20974250_4-O-Methylhonokiol attenuates memory impairment in presenilin 2 mutant mice . 21084199_This study confirms the regulatory role of CXCR2 in APP processing, and poses it as a potential target for developing novel therapeutics for intervention in Alzheimer's disease. 21146496_this study shows that the contribution of PS2 on gamma-secretase activity is of less importance, explaining the mild phenotype of PS2-deficient mice. 21206757_PS2 is the predominant gamma-secretase in microglia and modulates release of proinflammatory cytokines 21296884_Study supports the hypothesis that Pen-2 is more than a structural component of the gamma-secretase complex and may contribute to the catalytic mechanism of the enzyme. 21414900_Endogenous wild-type presenilin 2 is necessary for proper protein degradation through the autophagosome-lysosome system by functioning at the lysosomal level. 21910732_Presenilin 2 has the ability to regulate nectin-1 processing. 21914807_Polar transmembrane-based amino acids in presenilin 1 are involved in endoplasmic reticulum localization, Pen2 protein binding, and gamma-secretase complex stabilization 22140537_Upregulation of PS1/gamma-secretase activity may be a risk factor for late onset sporadic Alzheimer's disease. 22170863_Inhibition of APP cleavage by gamma-secretase did not ameliorate synaptic/memory deficits of familial Danish dementia mice. 22311977_lack of evidence for presenilins as endoplasmic reticulum Ca2+ leak channels 22715045_Double knockout Psen1Psen2 leads to a decrease in immunoreactivity for drebrin A at both synaptic and nonsynaptic areas of CA1 hippocampus. 22723704_transcriptome studies of presenilin knockout mise mouse brains reveal, for the first time, a role for presenilins in regulating lysosomal biogenesis 22796216_Distribution and expression profile of Psen2 differs from Psen1 in the cerebral cortex during development. 23103503_These data indicate that alterations in lysosomal calcium in the absence of presenilins might be leading to disruptions in autophagy. 23359614_data delineate a promoter responsive element targeted by parkin that drives differential regulation of presenilin-1 and presenilin-2 transcription with functional consequences for gamma-secretase activity and cell death. 23370287_PS2 mutation significantly accelerates the onset of cognitive impairment in associative trace eyeblink memory. 23589300_Interactome analyses of mature gamma-secretase complexes reveal distinct molecular environments of presenilin (PS) paralogs and preferential binding of signal peptide peptidase to PS2. 23918386_mechanism by which PS regulates synaptic function and calcium homeostasis using acute hippocampal slices from PS1 and PS2 conditional knockouts and primary cultured postnatal hippocampal neurons 23952003_One mechanism by which PS2 works to reign in proinflammatory microglial behavior and PS2 dysfunction or deficiency could result in unchecked proinflammatory activation. contributing to neurodegeneration. 24145027_At the transcriptional level, Psen1/2 removal induced cyclic AMP response element-binding protein (CREB)/CREB-binding protein binding. 24858037_The loss of PS2 could have a critical role in lung tumor development through the upregulation of iPLA2 activity by reducing gamma-secretase. 25204494_This study presents novel evidence for the differential expression of PS proteins in a nongenetic model for aging, resulting in an overall increase of the PS2 to PS1 ratio. 25652771_Results showed that epigenetic mechanisms play a pivotal role in transcriptional regulation of PS1 and PS2 during cerebral cortical development 26081153_These observations illustrate a novel PS2-dependent means of modulating LPS-mediated immune responses and identify a functional distinction between PS1 and PS2 in innate immunity. 26593275_Results suggest that mutation of PS2 can lead to NF-kappaB mediate amyloidogensis, and this effect can be amplified by the absence of estrogen. 27177724_Study shows that the binding of HSF-1, Cdx1, Ets-1 and Sp1 to Presenilin 1 promoter and that of Nkx2.2, HFH-2, Cdx1 and NF-kappaB to Presenilin 2 promoter regulate their differential expression during brain development. 27239030_Presenilin 2 (PS2), mutations in which underlie familial Alzheimer's disease (FAD), promotes endoplasmic reticulum-mitochondria coupling only in the presence of mitofusin 2 (Mfn2). 27354375_ARF4 is required for Presenilin basal body localization, Notch signaling, and subsequent epidermal differentiation. 27889678_Although hyperactivity and hypersynchronicity were respectively detected in mice expressing the PS2-N141I or the APP Swedish mutant alone, the increase in cross-frequency coupling specifically characterized the 6-month-old PS2APP mice, just before the surge of the cognitive decline 28879407_The altered expressions of hippocampal microRNAs were associated to the imbalance between neurotoxic and neuroprotective functions and seemed to affect neurodegeneration in PSEN1/PSEN2 double knockout mice more severely than in wild-type mice. 28994238_PS1- and PS2-dependent substrate processing in murine cells lacking presenilins (PSs) or stably re-expressing human PS1 or PS2 in an endogenous PS-null (PSdKO) background, were studied. 30429473_Study uncovers a role of PS in presynaptic mechanisms, through APP cleavage and regulation of Syt7, that highlights aberrant synaptic vesicle processing as a possible new pathway in Alzheimer's disease. 31862541_Loss of presenilin 2 age-dependently alters susceptibility to acute seizures and kindling acquisition. 32961023_Neuron-specific deletion of presenilin enhancer2 causes progressive astrogliosis and age-related neurodegeneration in the cortex independent of the Notch signaling. 33494218_Calcium Signaling and Mitochondrial Function in Presenilin 2 Knock-Out Mice: Looking for Any Loss-of-Function Phenotype Related to Alzheimer's Disease. | ENSMUSG00000010609 | Psen2 | 137.128581 | 0.9389480 | -0.090882797 | 0.24054598 | 1.406231e-01 | 7.076624e-01 | No | Yes | 135.188462 | 31.809651 | 144.946997 | 34.066719 | |||
| ENSG00000288721 | 9477 | protein_coding | A0A804HK38 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. {ECO:0000256|ARBA:ARBA00025687, ECO:0000256|RuleBase:RU364152}. | Activator;Nucleus;Reference proteome;Transcription;Transcription regulation | Mouse_homologues mmu:56771; | Mouse_homologues mediator complex [GO:0016592]; nucleoplasm [GO:0005654]; ubiquitin ligase complex [GO:0000151]; transcription coactivator activity [GO:0003713]; protein ubiquitination [GO:0016567]; regulation of transcription by RNA polymerase II [GO:0006357]; skeletal muscle cell differentiation [GO:0035914] | Mouse_homologues 34233190_The Mediator subunit MED20 organizes the early adipogenic complex to promote development of adipose tissues and diet-induced obesity. | ENSMUSG00000092558 | Med20 | 59.483447 | 1.1042690 | 0.143091660 | 0.42716348 | 1.087860e-01 | 7.415305e-01 | No | Yes | 63.501462 | 26.097967 | 56.660905 | 23.378461 | ||||
| ENSG00000289486 | lncRNA | 29.979298 | 0.7592283 | -0.397394293 | 0.49271023 | 6.579114e-01 | 4.172983e-01 | No | Yes | 22.770170 | 5.677144 | 31.588499 | 7.716833 |
- Note:
- The spreadsheet above contains the non-differential expressed genes but with significantly changes in alternative splicing (q < 0.05 and dif > 0.05). Please Click HERE to check all mapped genes in a Microsoft .excel file.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| IG_V_pseudogene | 1 |
| lncRNA | 51 |
| protein_coding | 457 |
| TEC | 1 |
| transcribed_unitary_pseudogene | 4 |
| transcribed_unprocessed_pseudogene | 19 |
| unprocessed_pseudogene | 3 |
MA plot
PCA plot
No significcant DEGs were detected under the threshold of P<0.05
Co-expression Analysis
The co-expression pattern was determined using weighted gene co-expression network analysis (WGCNA) through WGCNA R software.
Choosing the soft-thresholding power: analysis of network topology
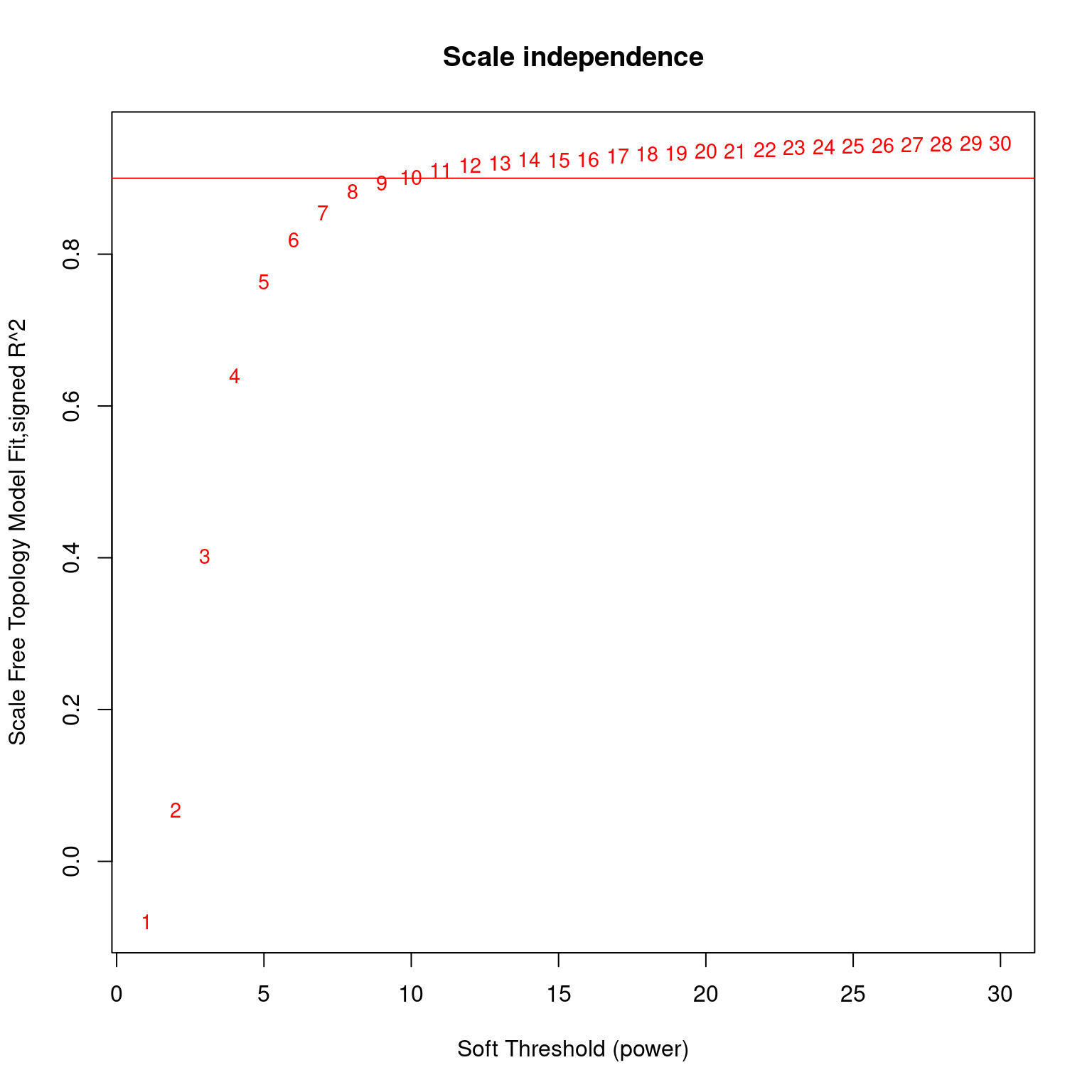
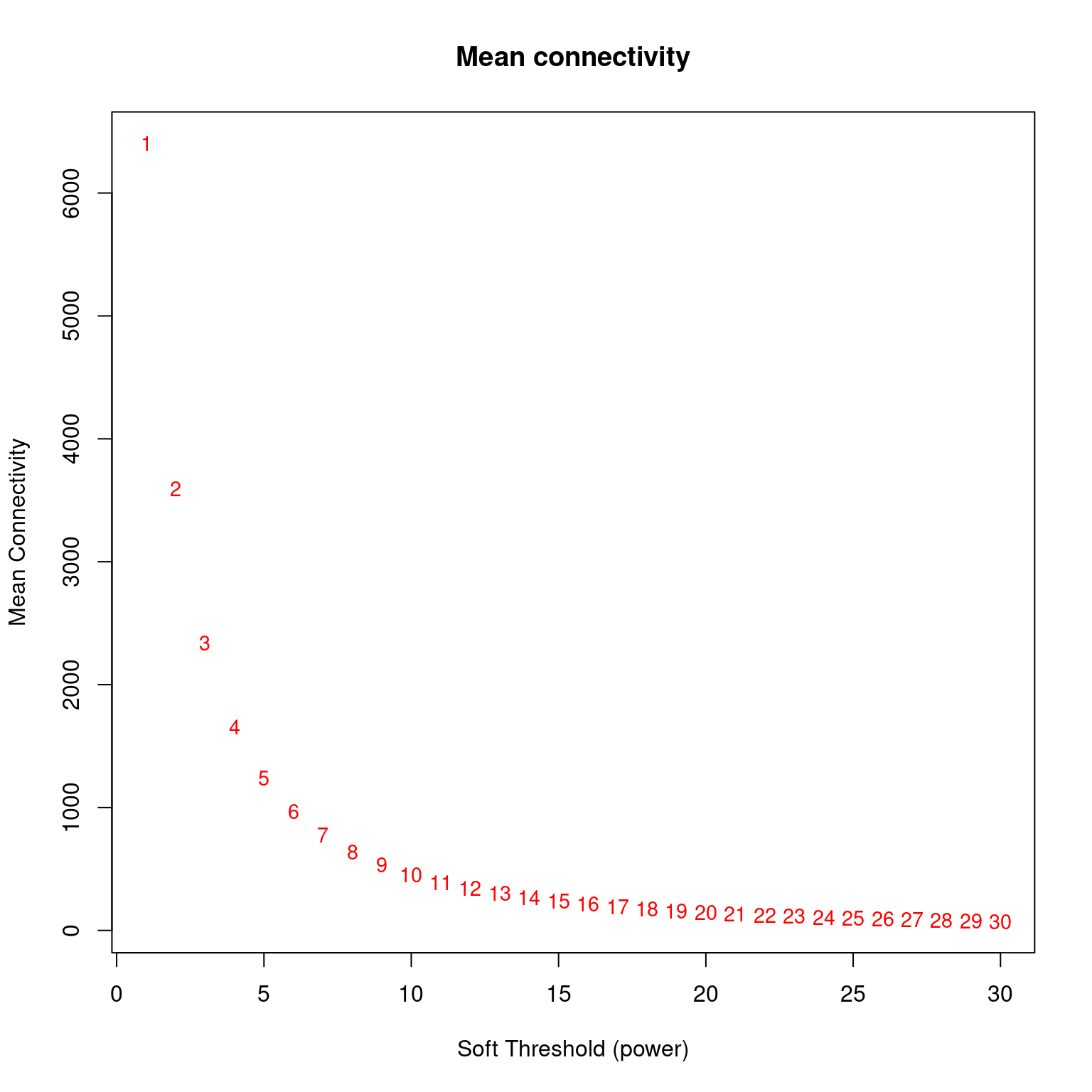
Quantifying module-trait associations
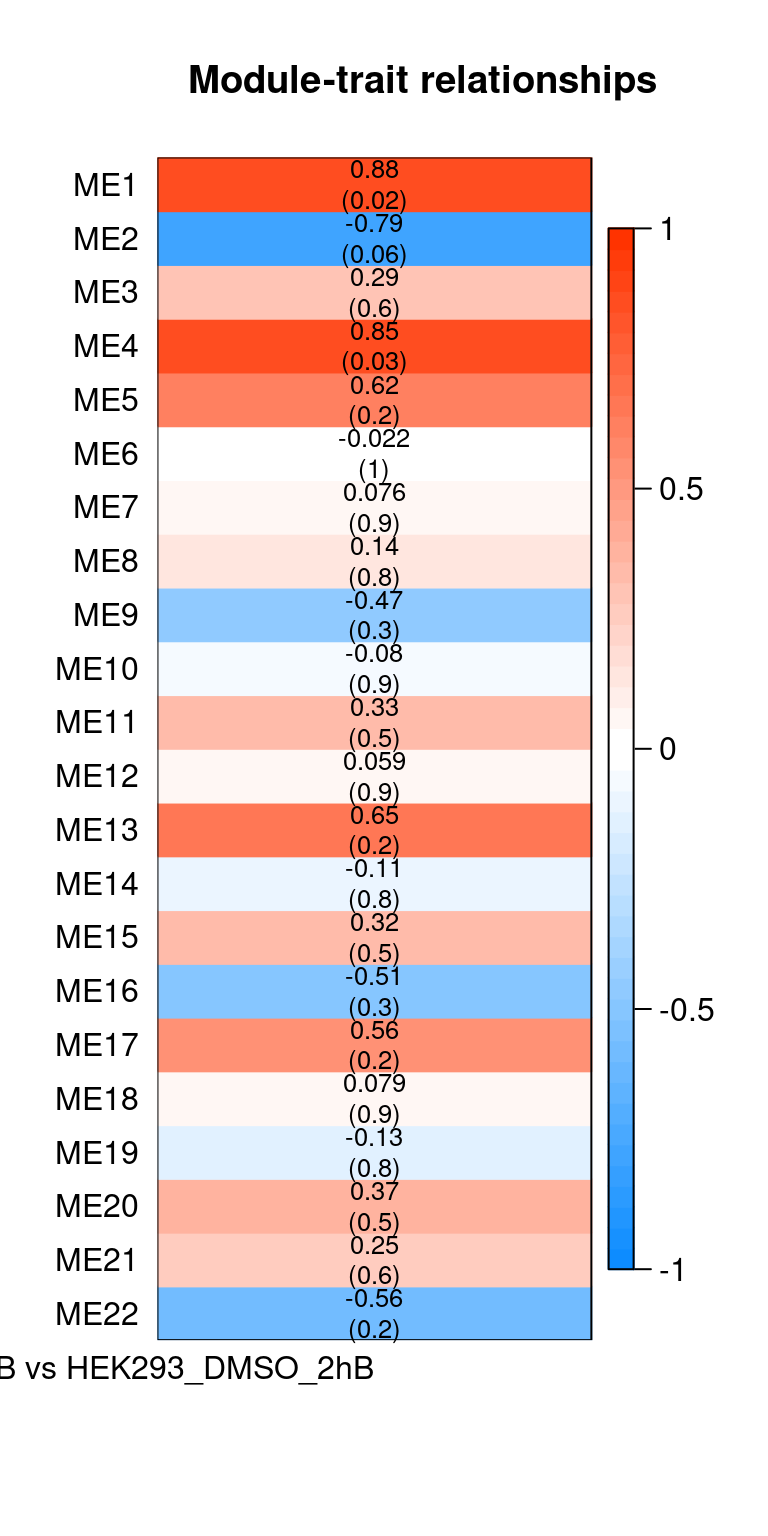
In the above figure, each cell contains the corresponding correlation (upper) and p-value (lower). ME, Module Eigengene.
Spreadsheet
| Cluster ID | Correlation | P-value | DEGs ID | DEGs Symbol |
|---|---|---|---|---|
| ME0 | 0.87666297 | 0.02187993 | ||
| ME1 | -0.79022393 | 0.06139330 | ||
| ME2 | 0.29475008 | 0.57067848 | ||
| ME3 | 0.85029816 | 0.03193850 | ||
| ME4 | 0.62454601 | 0.18498548 | ||
| ME5 | -0.02212905 | 0.96681185 | ||
| ME6 | 0.07556072 | 0.88687462 | ||
| ME7 | 0.14151686 | 0.78914179 | ||
| ME8 | -0.46828600 | 0.34891664 | ||
| ME9 | -0.07957762 | 0.88088553 | ||
| ME10 | 0.32613910 | 0.52813653 | ||
| ME11 | 0.05878418 | 0.91192530 | ||
| ME12 | 0.65213316 | 0.16046909 | ||
| ME13 | -0.11240164 | 0.83210759 | ||
| ME14 | 0.32276216 | 0.53266870 | ||
| ME15 | -0.51037004 | 0.30091491 | ||
| ME16 | 0.55898487 | 0.24885405 | ||
| ME17 | 0.07860541 | 0.88233472 | ||
| ME18 | -0.12607013 | 0.81189666 | ||
| ME19 | 0.37052740 | 0.46964386 | ||
| ME20 | 0.24507646 | 0.63974526 | ||
| ME21 | -0.56156807 | 0.24619558 |
Help
still working on
2. Functional Enrichment Analysis
2-1. Spreadsheet
GSEA
Please Click HERE to download a Microsoft .excel that contains all GSEA results.
GO: BP
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0007411 | axon guidance | 180 | -0.4477296 | -1.717832 | 0.0003625835 | 0.1687398 | 0.1651272 | 3711 | tags=43%, list=23%, signal=33% | CDH4||ROBO2||PRKCQ||NOTCH1||EPHA2||SEMA3D||RNF165||ERBB2||CRPPA||SEMA4G||LAMA1||IGSF9||CDK5||NCAM1||ALCAM||WNT5A||ROBO3||EPHA3||LAMA5||TUBB3||SEMA4C||GDNF||WNT3||EFNB2||EPHA4||EFNB3||EPHB4||LAMB2||GLI2||LHX1||UNC5C||EPHA8||FEZ1||APP||CXCL12||DAG1||EPHA5||TUBB2B||KALRN||NOG||NECTIN1||SEMA6B||GLI3||EPHB1||NTN1||PTCH1||B3GNT2||SEMA3F||UNC5A||PLXNC1||EFNB1||WNT3A||SEMA6D||NGFR||ETV1||BMPR2||EPHB6||RHOG||EPHB2||BSG||EFNA2||LHX9||SEMA4F||EFNA5||EFNA4||NFASC||LRP1||PLXNA2||KLF7||RET||EPHB3||NRP1||SLIT3||VSTM2L||SEMA7A||L1CAM||VAX1 | 4.6255068 | 5.1335028 | 4.5639284 | 5.097591 | 4.55996642 | 4.6050550 | 5.0656632 | 5.0782546 | 4.6698953 | 4.67043241 | 4.53181892 | 5.214201 | 5.0921197 | 5.0906707 | 4.6165168 | 4.5068608 | 4.5663214 | 5.122559 | 5.0587128 | 5.1107057 | 4.49810223 | 4.6260039 | 4.55293368 | 4.566732906 | 4.6279334 | 4.6197368 | 5.0788674 | 5.0327103 | 5.0848495 | 5.0552432 | 5.0885879 | 5.0906595 |
| GO:0032508 | DNA duplex unwinding | 80 | 0.5261477 | 1.841404 | 0.0013733857 | 0.2699887 | 0.2642085 | 5908 | tags=65%, list=37%, signal=41% | NAV2||HELB||POLQ||CHD9||ATRX||CHD7||CHD3||MNAT1||PIF1||CHD6||WRN||DHX36||MCM8||CHD2||CHD1||POT1||SETX||NBN||IGHMBP2||ASCC3||GTF2H2||FANCM||HFM1||CHD8||TOP2A||BLM||SMARCAD1||DDX11||CHD1L||MCM9||RAD54L2||CHTF18||RECQL5||DQX1||BRIP1||SUPV3L1||DDX3X||MCM4||HELQ||PURA||RAD54L||DHX9||TWNK||DDX1||MRE11||G3BP1||MCM3||GTF2F2||ERCC3||RAD54B||RFC2||CHTF8 | 5.7150154 | 6.2968269 | 5.9659787 | 6.390009 | 5.62850233 | 5.7452820 | 6.3524778 | 6.4706656 | 5.7146549 | 5.79739807 | 5.62802508 | 6.297578 | 6.2971489 | 6.2957536 | 6.1848829 | 5.8095480 | 5.8750354 | 6.357245 | 6.3544084 | 6.4560332 | 5.61464880 | 5.6673785 | 5.60265469 | 5.692618126 | 5.6927510 | 5.8450516 | 6.3496680 | 6.3453010 | 6.3624096 | 6.4575457 | 6.4849309 | 6.4693895 |
| GO:0030166 | proteoglycan biosynthetic process | 50 | -0.5623257 | -1.774132 | 0.0046314164 | 0.5617810 | 0.5497538 | 3291 | tags=54%, list=21%, signal=43% | PXYLP1||SLC2A10||CANT1||NDST3||CHSY1||HS6ST2||EXTL3||CHST10||CHST11||CHST8||CHPF2||GAL3ST4||HS6ST1||EXT2||B4GALT7||ENSG00000272916||B3GAT3||CHSY3||BMPR2||CHST12||B3GALT6||CHPF||CHST14||B3GAT1||CHST7||CSGALNACT1||FOXL1 | 3.7083717 | 4.5048803 | 4.2346688 | 4.475292 | 3.71555827 | 4.0547323 | 4.3885707 | 4.3642776 | 3.6736145 | 3.70239025 | 3.74812988 | 4.519711 | 4.5623982 | 4.4293498 | 4.2607019 | 4.2458605 | 4.1966682 | 4.450851 | 4.4964093 | 4.4782511 | 3.70433202 | 3.6426415 | 3.79558454 | 3.846169467 | 3.9676588 | 4.3095328 | 4.4231083 | 4.3982202 | 4.3432293 | 4.3612745 | 4.3808950 | 4.3504985 |
| GO:0048385 | regulation of retinoic acid receptor signaling pathway | 14 | -0.7358230 | -1.774415 | 0.0061703822 | 0.5943933 | 0.5816680 | 1466 | tags=36%, list=9%, signal=32% | CALR||CYP26A1||CYP26B1||KLF2||DHRS3 | 6.0938758 | 7.2765023 | 6.4830730 | 6.597277 | 6.03822471 | 6.1854867 | 7.0932918 | 6.5302623 | 6.0978523 | 6.23704387 | 5.93046639 | 7.310028 | 7.3227535 | 7.1932251 | 6.4705466 | 6.4907540 | 6.4878358 | 6.591353 | 6.6149745 | 6.5853313 | 6.09598508 | 6.0949458 | 5.91644332 | 6.270793555 | 5.9955107 | 6.2729037 | 7.1186376 | 7.0869852 | 7.0738849 | 6.5320805 | 6.5615409 | 6.4964290 |
| GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 75 | -0.4914171 | -1.662732 | 0.0064675671 | 0.5943933 | 0.5816680 | 4264 | tags=52%, list=27%, signal=38% | PCDH17||FAT1||PLXNB2||ESAM||ITGB1||PCDH10||DSG2||CDH4||ROBO2||SDK1||IGSF9||DCHS1||CDH2||PVR||ROBO3||DSC2||CDH3||CDHR1||CLSTN3||CLSTN1||NECTIN1||NECTIN2||CDH15||CADM1||PCDHGA7||BSG||CDH12||PCDH1||RET||CLSTN2||CEACAM1||VSTM2L||CDH26||PCDHB2||PCDHGA11||HMCN1||L1CAM||AMIGO2||PCDHGB7 | 4.1252782 | 4.6302347 | 4.6152759 | 4.618919 | 4.18675278 | 4.6609657 | 4.4596715 | 4.5682826 | 4.0386909 | 4.05258926 | 4.27243910 | 4.657361 | 4.6716711 | 4.5590883 | 4.5179452 | 4.7705632 | 4.5436965 | 4.639568 | 4.6285940 | 4.5880879 | 4.19577456 | 4.0159531 | 4.33129112 | 4.427473653 | 4.5991806 | 4.9135613 | 4.5090484 | 4.4514909 | 4.4169717 | 4.5632085 | 4.6394010 | 4.4988005 |
| GO:0032332 | positive regulation of chondrocyte differentiation | 17 | -0.7217597 | -1.825040 | 0.0064756672 | 0.5943933 | 0.5816680 | 2434 | tags=53%, list=15%, signal=45% | BMP6||GLI3||SOX9||POR||FGF18||LOXL2||SOX5||ZBTB16||RUNX2 | 2.7915782 | 3.0009451 | 3.2314174 | 3.084171 | 2.82017362 | 3.2200925 | 2.8579533 | 3.0575730 | 2.7175000 | 2.68647988 | 2.95547554 | 3.049077 | 3.0441989 | 2.9049705 | 3.1460114 | 3.3437368 | 3.1971330 | 3.075190 | 3.0721481 | 3.1049464 | 2.71068630 | 2.7313931 | 3.00002270 | 2.986640227 | 3.1502774 | 3.4790851 | 2.9319658 | 2.8008168 | 2.8378903 | 3.0568572 | 3.0200262 | 3.0948651 |
| GO:0002076 | osteoblast development | 13 | -0.7441513 | -1.768837 | 0.0065101598 | 0.5943933 | 0.5816680 | 2725 | tags=54%, list=17%, signal=45% | MEN1||GLI2||LRP5||ACHE||PTH1R||RUNX2||PTHLH | 3.7759899 | 3.8644834 | 4.0916649 | 3.750017 | 3.78166953 | 4.3337075 | 3.7590481 | 3.5940287 | 3.7071439 | 3.74051953 | 3.87479501 | 3.872284 | 3.9147962 | 3.8042197 | 3.9493647 | 4.2346207 | 4.0768179 | 3.839232 | 3.7115156 | 3.6949389 | 3.79388699 | 3.5579441 | 3.96454523 | 4.197087309 | 4.3207587 | 4.4702764 | 3.8208589 | 3.7351933 | 3.7190019 | 3.5786827 | 3.6540684 | 3.5472361 |
| GO:0007042 | lysosomal lumen acidification | 12 | -0.7595813 | -1.771511 | 0.0069033531 | 0.5943933 | 0.5816680 | 3070 | tags=67%, list=19%, signal=54% | PPT1||CLN6||TMEM106B||TMEM9||ATP6AP2||CLN5||TMEM199||GRN | 5.8510892 | 6.3500544 | 6.2965150 | 6.357931 | 5.90639046 | 6.1141704 | 6.1503191 | 6.2248732 | 5.8086063 | 5.84460643 | 5.89862905 | 6.432915 | 6.3718337 | 6.2386295 | 6.2246019 | 6.3505903 | 6.3114858 | 6.396846 | 6.3830814 | 6.2916129 | 5.90614772 | 5.8443477 | 5.96610770 | 5.936791549 | 5.9886945 | 6.3759903 | 6.2019258 | 6.1424375 | 6.1049318 | 6.2259104 | 6.2807293 | 6.1656865 |
| GO:0010829 | negative regulation of glucose transmembrane transport | 12 | 0.7971304 | 1.874018 | 0.0070897642 | 0.5943933 | 0.5816680 | 6 | tags=8%, list=0%, signal=8% | RSC1A1 | 4.7959876 | 5.4308989 | 5.0235669 | 5.330068 | 4.70940495 | 4.8511788 | 5.4468750 | 5.3348242 | 4.7820172 | 4.94303645 | 4.64772205 | 5.408765 | 5.4072361 | 5.4756326 | 5.2056756 | 4.9319073 | 4.9142337 | 5.283975 | 5.3218345 | 5.3826703 | 4.64689341 | 4.7719173 | 4.70669255 | 4.703166780 | 4.7862407 | 5.0421355 | 5.4668416 | 5.4217473 | 5.4516716 | 5.3360907 | 5.3656318 | 5.3020488 |
| GO:0051028 | mRNA transport | 121 | 0.4119344 | 1.542012 | 0.0088153085 | 0.6312885 | 0.6177732 | 5745 | tags=54%, list=36%, signal=35% | MVP||EIF5AL1||ZC3H11B||THOC1||NPIPA1||UPF3A||AGFG1||SMG1||THOC2||NUP214||NUP42||SETD2||EIF5A2||NCBP3||YTHDC1||NA||PARP11||SMG7||UPF2||IWS1||TNKS||NUP35||RANBP2||ENSG00000273784||TPR||UPF3B||DDX19B||NUTF2||CHTOP||AHCTF1||NUP58||RANBP17||ZC3H11A||ENY2||NXF1||NUP205||NUP98||IGF2BP1||SRSF1||MYO1C||NCBP2||PCID2||THOC7||XPO1||MCM3AP||IGF2BP2||NSUN2||NUP54||NXT2||NUP50||FYTTD1||CETN3||NUP160||KIF5C||SMG5||DHX9||THOC5||HNRNPA2B1||CASC3||POLDIP3||MAGOHB||FMR1||SENP2||NUP153||NUP155 | 6.4739734 | 7.0492737 | 6.5258501 | 7.031947 | 6.42911836 | 6.5366900 | 7.1027288 | 7.0953415 | 6.4399724 | 6.57017103 | 6.40654810 | 7.051340 | 7.0429560 | 7.0535042 | 6.7256697 | 6.3993536 | 6.4294928 | 7.009292 | 7.0028190 | 7.0823707 | 6.43302081 | 6.4301928 | 6.42412711 | 6.545940573 | 6.5221004 | 6.5419164 | 7.0966448 | 7.0970621 | 7.1144082 | 7.0744590 | 7.1122802 | 7.0990305 |
| GO:0071320 | cellular response to cAMP | 39 | -0.5630432 | -1.697042 | 0.0089199147 | 0.6312885 | 0.6177732 | 612 | tags=36%, list=4%, signal=35% | SLC5A5||HCN2||SLC8A1||APP||PTAFR||HSPA5||ASS1||KCNQ1||HCN4||SLC8A3||AANAT||RAPGEF3||PDE2A||AKAP6 | 4.4280649 | 5.4880139 | 4.6311021 | 5.379328 | 4.32639407 | 4.4686808 | 5.3891708 | 5.3584322 | 4.4523841 | 4.58075617 | 4.22940433 | 5.513190 | 5.5170621 | 5.4322083 | 4.8919945 | 4.4177851 | 4.5406502 | 5.362808 | 5.3748300 | 5.4000942 | 4.32868020 | 4.4224811 | 4.22098852 | 4.498996098 | 4.3790861 | 4.5238459 | 5.4032845 | 5.3934423 | 5.3705907 | 5.3595816 | 5.3839133 | 5.3313217 |
| GO:0051085 | chaperone cofactor-dependent protein refolding | 30 | -0.6042445 | -1.729254 | 0.0089633671 | 0.6312885 | 0.6177732 | 3608 | tags=47%, list=23%, signal=36% | HSPA1B||TOR1A||ERO1A||TOR2A||DNAJB5||HSPA13||HSPA5||HSPA2||SDF2L1||TOR1B||DNAJB13||DNAJB14||HSPA6||HSPA1L | 7.0002430 | 7.9261781 | 6.5594161 | 7.604809 | 6.98108535 | 6.5013923 | 7.9038823 | 7.6228608 | 7.0727323 | 7.19138903 | 6.69052641 | 7.982100 | 7.8997497 | 7.8950118 | 6.8911735 | 6.2194269 | 6.4875512 | 7.549531 | 7.5945551 | 7.6678616 | 7.02180010 | 7.0474042 | 6.86757317 | 6.563133282 | 6.5235677 | 6.4133306 | 7.9006448 | 7.9061407 | 7.9048556 | 7.6111766 | 7.6312536 | 7.6260769 |
| GO:0030903 | notochord development | 15 | -0.7148560 | -1.745776 | 0.0090162292 | 0.6312885 | 0.6177732 | 3911 | tags=73%, list=25%, signal=55% | TEAD2||EPHA2||WNT11||WNT5A||GLI2||COL13A1||NOG||SOX9||ID3||COL2A1||GLI1 | 4.1696590 | 4.0933550 | 3.8598745 | 3.450611 | 4.33712594 | 4.3492099 | 3.9105402 | 3.3780953 | 4.0086400 | 4.09919822 | 4.37539313 | 4.186619 | 4.0592619 | 4.0293029 | 3.7050134 | 3.9762630 | 3.8852766 | 3.492614 | 3.3759195 | 3.4804707 | 4.35413519 | 4.1079915 | 4.51981023 | 4.344766350 | 4.4986308 | 4.1874777 | 3.9259852 | 3.8754519 | 3.9295515 | 3.4027555 | 3.3609217 | 3.3702739 |
| GO:0042996 | regulation of Golgi to plasma membrane protein transport | 11 | 0.7461854 | 1.711433 | 0.0099371324 | 0.6388928 | 0.6252147 | 6 | tags=9%, list=0%, signal=9% | RSC1A1 | 9.0737713 | 8.5575215 | 8.8598412 | 8.503891 | 9.13698073 | 9.1385807 | 8.5220430 | 8.5018078 | 9.0146122 | 8.99535437 | 9.20217074 | 8.601061 | 8.5511855 | 8.5191346 | 8.6214585 | 8.9602931 | 8.9711338 | 8.568863 | 8.4473156 | 8.4928717 | 9.14614322 | 9.0782076 | 9.18458694 | 9.210066511 | 9.2461023 | 8.9406924 | 8.5152924 | 8.5354902 | 8.5152518 | 8.4981719 | 8.5029982 | 8.5042461 |
| GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 11 | -0.7664676 | -1.748244 | 0.0100571879 | 0.6388928 | 0.6252147 | 2692 | tags=55%, list=17%, signal=45% | MICA||HLA-E||HLA-B||CX3CR1||HLA-A||CEACAM1 | 5.2215497 | 5.4531964 | 5.5948163 | 5.241099 | 5.33335868 | 5.5992125 | 5.2466250 | 5.1254892 | 5.1284317 | 5.12822963 | 5.39165746 | 5.512477 | 5.4827372 | 5.3598823 | 5.4427105 | 5.7259821 | 5.6018477 | 5.307196 | 5.2014699 | 5.2122696 | 5.31892776 | 5.1790532 | 5.48572128 | 5.422798642 | 5.5512851 | 5.7980525 | 5.3332582 | 5.2245706 | 5.1775991 | 5.1334861 | 5.1898871 | 5.0496548 |
| GO:0015698 | inorganic anion transport | 108 | -0.4360969 | -1.557363 | 0.0103402895 | 0.6388928 | 0.6252147 | 3155 | tags=36%, list=20%, signal=29% | SLC4A3||TTYH3||CLCC1||SLC5A5||SLC37A4||GET3||SLC25A11||SLC39A14||SLC4A11||SLC4A4||TSPO||ANO10||LRRC8A||SLC26A11||ANO7||CLCN4||CLCN3||PTAFR||PACC1||PCYOX1||MFSD5||ADAMTS8||SLC1A4||LRRC8D||LRRC8E||SLC13A4||SLC12A5||SLC1A1||ATP8B1||CLDN4||GABRD||ABCB1||SLC37A1||SLC12A4||SLC4A5||KCNQ1||BEST1||GABRQ||SLC26A4 | 4.6043632 | 4.9608367 | 4.8173188 | 5.015825 | 4.57337716 | 4.6911038 | 4.9341376 | 5.0118224 | 4.5968839 | 4.65634454 | 4.55816221 | 4.964843 | 4.9774688 | 4.9399458 | 4.9028388 | 4.7536225 | 4.7912905 | 5.006335 | 4.9975316 | 5.0431997 | 4.57514874 | 4.5510911 | 4.59357723 | 4.628655061 | 4.6268381 | 4.8100360 | 4.9315687 | 4.9330307 | 4.9378060 | 4.9844215 | 5.0574722 | 4.9924558 |
| GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 19 | -0.6818680 | -1.762214 | 0.0106111220 | 0.6388928 | 0.6252147 | 3102 | tags=58%, list=19%, signal=47% | PVR||LAMP1||MICA||RASGRP1||HLA-E||NECTIN2||CADM1||HLA-B||HLA-A||CEACAM1||CD226 | 4.8507246 | 5.2659679 | 5.2624639 | 5.091314 | 4.93538224 | 5.2368964 | 5.0868298 | 5.0008215 | 4.7751091 | 4.77969895 | 4.98704957 | 5.299483 | 5.3076386 | 5.1876973 | 5.1453943 | 5.3776012 | 5.2550342 | 5.133443 | 5.0756551 | 5.0638805 | 4.93904339 | 4.8002285 | 5.05558320 | 5.072653475 | 5.1568523 | 5.4529782 | 5.1555833 | 5.0715696 | 5.0305063 | 5.0061513 | 5.0532113 | 4.9409019 |
| GO:0098656 | anion transmembrane transport | 163 | -0.3909246 | -1.480172 | 0.0107546820 | 0.6388928 | 0.6252147 | 4428 | tags=45%, list=28%, signal=33% | SLC12A9||SLC12A8||SLC9A6||SLC6A9||SLC16A1||SLC9A1||SLC25A4||ITGB1||SLC36A4||SLC25A42||ABCC4||SLC5A12||SLC39A8||SLC25A5||SLC38A7||ARL6IP5||SLC35B4||SLC27A1||SFXN2||SLC24A1||SLC38A3||PRNP||MFSD12||SLC4A3||SLC5A5||SLC37A4||PSEN1||SLC25A29||SLC24A4||SLC25A11||SLC7A1||PER2||SLC4A11||SLC4A4||SLC1A5||LRRC8A||SLC7A5||SLC26A11||ANO7||SLC35B2||SLC15A4||SLC43A1||CLCN3||PTAFR||SLC35A2||MFSD10||PCYOX1||PRAF2||ADAMTS8||SLC1A4||SLC66A1||LRRC8D||LRRC8E||SLC13A4||SLC19A1||SLC6A13||SLC12A5||SLC1A1||GABRD||SLC46A1||SLC43A2||ABCB1||SLC7A3||SLC37A1||SLC12A4||SLC4A5||SLC9A9||SLC16A7||CLTRN||BEST1||GABRQ||SLC9A2||SLC26A4 | 5.1844613 | 5.4314444 | 5.3096677 | 5.487074 | 5.16756069 | 5.2754896 | 5.3719893 | 5.4673766 | 5.1864402 | 5.25480012 | 5.10843066 | 5.449957 | 5.4578007 | 5.3854895 | 5.3145770 | 5.2861447 | 5.3279659 | 5.499147 | 5.4659320 | 5.4959107 | 5.17892262 | 5.1679010 | 5.15576547 | 5.259011942 | 5.2719220 | 5.2953001 | 5.3825430 | 5.3716083 | 5.3617415 | 5.4476537 | 5.5065306 | 5.4471339 |
| GO:0046599 | regulation of centriole replication | 20 | 0.6295221 | 1.678095 | 0.0116512656 | 0.6388928 | 0.6252147 | 3382 | tags=55%, list=21%, signal=43% | SPICE1||PLK4||KAT2B||CENPJ||MDM1||CEP295||ENSG00000285943||CDK5RAP2||CEP76||ALMS1||CEP120 | 5.5928830 | 7.6370344 | 5.9000144 | 7.707511 | 5.49054826 | 5.8518600 | 7.7348525 | 7.7778309 | 5.6590688 | 5.64958562 | 5.46152103 | 7.662662 | 7.5691336 | 7.6769488 | 6.6110784 | 5.4894480 | 5.1884583 | 7.642743 | 7.6744343 | 7.8004868 | 5.36097913 | 5.5637261 | 5.53859489 | 5.703326044 | 5.7136622 | 6.1018268 | 7.6814334 | 7.7202683 | 7.8002984 | 7.7719571 | 7.7536162 | 7.8074001 |
| GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 12 | -0.7317045 | -1.706496 | 0.0118343195 | 0.6388928 | 0.6252147 | 2438 | tags=67%, list=15%, signal=57% | TAP2||HLA-E||TAPBP||TAP1||ERAP1||HLA-B||HLA-C||HLA-A | 6.0443631 | 6.3528722 | 6.3906755 | 6.294118 | 6.14711410 | 6.3257296 | 6.1490282 | 6.1526023 | 5.9869824 | 5.95654816 | 6.17930159 | 6.395159 | 6.3802821 | 6.2805022 | 6.2509688 | 6.4993033 | 6.4108671 | 6.355609 | 6.3093390 | 6.2138009 | 6.12566594 | 6.0447613 | 6.26247258 | 6.165026976 | 6.2795433 | 6.5108574 | 6.2346248 | 6.1194565 | 6.0888753 | 6.1932877 | 6.1899624 | 6.0712378 |
| GO:0032239 | regulation of nucleobase-containing compound transport | 15 | 0.6849366 | 1.709457 | 0.0119736580 | 0.6388928 | 0.6252147 | 2976 | tags=40%, list=19%, signal=33% | RSC1A1||NRDE2||SETD2||IWS1||TPR||CPSF6 | 4.7918092 | 6.1034553 | 5.0337629 | 6.109421 | 4.63246124 | 4.9602083 | 6.1986190 | 6.1569531 | 4.7581350 | 4.95770592 | 4.64174060 | 6.062079 | 6.1123514 | 6.1349738 | 5.5251847 | 4.6590607 | 4.7525619 | 6.054491 | 6.0684300 | 6.2007772 | 4.58092288 | 4.6684488 | 4.64658040 | 4.973382564 | 5.0161177 | 4.8881993 | 6.1825332 | 6.2232720 | 6.1897233 | 6.1507703 | 6.1714146 | 6.1485643 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 22 | -0.6479322 | -1.728892 | 0.0120078740 | 0.6388928 | 0.6252147 | 4824 | tags=73%, list=30%, signal=51% | STT3B||DAD1||MGAT5||ST6GAL1||ALG5||ALG8||MGAT1||MGAT2||TUSC3||RPN1||RPN2||DDOST||NUDT14||STT3A||MAGT1||DERL3 | 6.6116829 | 7.3139997 | 6.9242151 | 7.178923 | 6.63326957 | 6.7067276 | 7.1774317 | 7.1336739 | 6.6068793 | 6.64451778 | 6.58298349 | 7.370854 | 7.3230223 | 7.2453521 | 6.9227322 | 6.9328671 | 6.9170014 | 7.170953 | 7.2191987 | 7.1456477 | 6.64712190 | 6.6167643 | 6.63575957 | 6.664915691 | 6.5335953 | 6.8978491 | 7.1956212 | 7.1900913 | 7.1460734 | 7.1217803 | 7.1711770 | 7.1072841 |
| GO:0002063 | chondrocyte development | 27 | -0.6061929 | -1.699627 | 0.0130451713 | 0.6388928 | 0.6252147 | 3354 | tags=44%, list=21%, signal=35% | SULF2||CHSY1||CHST11||SULF1||COMP||SOX9||BMPR2||FGF18||SERPINH1||RUNX2||SMPD3||PTHLH | 2.8657708 | 3.7252076 | 3.2208019 | 3.396283 | 2.94655956 | 3.2331712 | 3.6029695 | 3.2709053 | 2.7727194 | 2.89114743 | 2.92888308 | 3.698313 | 3.7942656 | 3.6804278 | 3.2200753 | 3.2504273 | 3.1912970 | 3.335441 | 3.4300929 | 3.4214307 | 2.93900804 | 2.8382137 | 3.05434063 | 3.094090990 | 3.0856978 | 3.4830070 | 3.6225681 | 3.6084463 | 3.5775269 | 3.2423866 | 3.2949760 | 3.2748660 |
| GO:1990034 | calcium ion export across plasma membrane | 11 | -0.7466991 | -1.703154 | 0.0134095839 | 0.6388928 | 0.6252147 | 1040 | tags=36%, list=7%, signal=34% | SLC24A4||RGS9||SLC8A2||SLC8A3 | 6.6651610 | 7.2415884 | 6.3821941 | 7.348912 | 6.57651181 | 6.4684691 | 7.2909318 | 7.4171957 | 6.7306058 | 6.80086482 | 6.43909639 | 7.264198 | 7.2133163 | 7.2467881 | 6.6479770 | 6.1698874 | 6.2845465 | 7.343321 | 7.3086360 | 7.3935157 | 6.52448995 | 6.7192785 | 6.47397858 | 6.516286876 | 6.5535023 | 6.3253954 | 7.2652764 | 7.2918952 | 7.3151916 | 7.4064689 | 7.4203411 | 7.4247144 |
| GO:0034103 | regulation of tissue remodeling | 42 | -0.5412658 | -1.653506 | 0.0136133800 | 0.6388928 | 0.6252147 | 4184 | tags=60%, list=26%, signal=44% | FLT4||SYK||SYT7||LTBP3||TMEM64||LEPR||CSK||PLEKHM1||TNFRSF11A||TGFB1||TNFAIP3||MDK||EGFR||CST3||ATP6AP1||DDR2||SFRP1||CEACAM1||THBS4||TMBIM1||TF||HAND2||PDK4||S1PR1||IL23A | 5.1663844 | 5.1147668 | 5.1987390 | 4.938486 | 5.19227395 | 5.3877730 | 5.0234475 | 4.8784059 | 5.1233470 | 5.06722206 | 5.29838969 | 5.137488 | 5.1449873 | 5.0603114 | 5.0246121 | 5.3470986 | 5.2064909 | 4.987837 | 4.9195455 | 4.9067524 | 5.17122435 | 5.0960158 | 5.30200685 | 5.284034084 | 5.4330627 | 5.4408726 | 5.0545008 | 5.0313481 | 4.9835897 | 4.8886233 | 4.9070873 | 4.8386406 |
| GO:1905146 | lysosomal protein catabolic process | 16 | -0.6890695 | -1.712405 | 0.0138394622 | 0.6388928 | 0.6252147 | 3042 | tags=56%, list=19%, signal=46% | LAPTM4B||LAMP2||TPP1||MGAT3||TMEM106B||GBA||ATP13A2||TMEM199||LRP1 | 4.4743619 | 5.4118493 | 5.1016235 | 5.458751 | 4.48942499 | 4.8285558 | 5.2739314 | 5.3569337 | 4.4254119 | 4.58248756 | 4.40874007 | 5.403508 | 5.4552328 | 5.3756750 | 5.1112586 | 5.1217534 | 5.0713701 | 5.401799 | 5.5139504 | 5.4583232 | 4.48500228 | 4.5019850 | 4.48120273 | 4.580117680 | 4.5155817 | 5.2649438 | 5.3054264 | 5.2868072 | 5.2284472 | 5.3366953 | 5.4054548 | 5.3273840 |
| GO:0030595 | leukocyte chemotaxis | 102 | -0.4342059 | -1.535589 | 0.0138941759 | 0.6388928 | 0.6252147 | 2718 | tags=33%, list=17%, signal=28% | CREB3||TMEM102||ADAM10||MDK||TRPV4||SLC8B1||CXCL12||TIRAP||PADI2||CXCL3||LGMN||MIF||PPIB||RIN3||TAFA4||CX3CR1||CALR||F2RL1||PLA2G7||BSG||GAS6||VEGFC||CHGA||IL6R||CXCR4||THBS4||EDNRB||C5||ITGB2||NBL1||TGFB2||C5AR1||S1PR1||CYP7B1 | 7.3666053 | 7.1001752 | 7.2054935 | 6.982405 | 7.37593515 | 7.3572092 | 6.9892693 | 6.9073958 | 7.3755464 | 7.30171110 | 7.42008921 | 7.187725 | 7.0620432 | 7.0465628 | 7.0495247 | 7.2924991 | 7.2625031 | 7.054691 | 6.9305467 | 6.9590279 | 7.38327577 | 7.2969582 | 7.44380806 | 7.325566540 | 7.4784279 | 7.2587671 | 7.0077905 | 6.9697797 | 6.9899871 | 6.9349063 | 6.8418032 | 6.9432949 |
| GO:0051180 | vitamin transport | 25 | -0.6154148 | -1.693485 | 0.0138943249 | 0.6388928 | 0.6252147 | 3537 | tags=52%, list=22%, signal=41% | SLC27A1||LMBRD1||SLC2A10||SLC2A1||CD320||SLC2A6||ABCG2||SLC19A1||SCARB1||SLC52A2||SLC46A1||SLC2A3||TCN2 | 4.6073981 | 4.7745881 | 5.0257002 | 4.802156 | 4.70922415 | 5.0679591 | 4.6235493 | 4.6886233 | 4.4942277 | 4.50688705 | 4.79989958 | 4.791757 | 4.8348830 | 4.6935081 | 4.9023462 | 5.1822365 | 4.9777969 | 4.832679 | 4.8178037 | 4.7548074 | 4.70129677 | 4.5413076 | 4.86672393 | 4.836714795 | 5.0057275 | 5.3193070 | 4.6998153 | 4.6166388 | 4.5502968 | 4.6764401 | 4.7727897 | 4.6120906 |
| GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 15 | -0.6928581 | -1.692054 | 0.0140252454 | 0.6388928 | 0.6252147 | 2669 | tags=53%, list=17%, signal=44% | LEAP2||H2BC12||CXCL3||FAM3A||ROMO1||H2BC4||ANG||H2BC11 | 10.3717309 | 9.8624093 | 10.1298141 | 9.810150 | 10.33583967 | 10.0393515 | 9.8411751 | 9.7143275 | 10.4321730 | 10.42570863 | 10.25001146 | 9.915139 | 9.8298207 | 9.8407654 | 10.0332333 | 10.0906622 | 10.2561607 | 9.841558 | 9.7842222 | 9.8040799 | 10.32016432 | 10.4839235 | 10.18817384 | 10.192761130 | 10.0719781 | 9.8300107 | 9.8266802 | 9.8520789 | 9.8446481 | 9.8165832 | 9.4192714 | 9.8668471 |
| GO:0016246 | RNA interference | 12 | -0.7253842 | -1.691756 | 0.0143984221 | 0.6412922 | 0.6275628 | 6 | tags=17%, list=0%, signal=17% | CLP1||RMRP | 4.9189727 | 5.8397469 | 5.4013988 | 5.827911 | 4.89226571 | 5.1928449 | 5.7728320 | 7.0870490 | 4.8598193 | 5.10037920 | 4.77684832 | 5.975098 | 5.7768034 | 5.7571117 | 5.6202896 | 5.2734944 | 5.2826746 | 5.873133 | 5.7193639 | 5.8853763 | 4.97926900 | 4.8827814 | 4.80972163 | 5.232568342 | 4.9846203 | 5.3388241 | 5.7681045 | 5.8116276 | 5.7378081 | 5.8369656 | 8.1901621 | 5.8741430 |
| GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 41 | 0.5184057 | 1.608973 | 0.0148606811 | 0.6516980 | 0.6377458 | 2543 | tags=34%, list=16%, signal=29% | SYNE4||DLG2||LIN7A||LIN7B||MYO9A||LRRC7||FOXF1||LIN7C||SH3BP1||ANK1||MTCL1||ERBIN||PARD3B||MARK2 | 5.1264385 | 5.3948308 | 5.2510019 | 5.388541 | 5.07689492 | 5.1689838 | 5.4316635 | 5.4369367 | 5.1157884 | 5.22250794 | 5.03486331 | 5.402847 | 5.3936084 | 5.3879979 | 5.3415102 | 5.1324993 | 5.2712181 | 5.355471 | 5.3428310 | 5.4642099 | 5.10735197 | 5.0735307 | 5.04921046 | 5.160373176 | 5.0923961 | 5.2498499 | 5.4249700 | 5.4243820 | 5.4455378 | 5.4217278 | 5.4567893 | 5.4320677 |
| GO:0019731 | antibacterial humoral response | 11 | -0.7380118 | -1.683339 | 0.0165647801 | 0.6552290 | 0.6412013 | 2505 | tags=64%, list=16%, signal=54% | H2BC12||HLA-E||H2BC4||HLA-A||ANG||H2BC11||TF | 8.6839128 | 8.1762492 | 8.4261856 | 8.195585 | 8.54923907 | 7.7907163 | 8.1707140 | 7.7547806 | 8.8516333 | 8.85500447 | 8.27042819 | 8.229507 | 8.1179645 | 8.1791165 | 8.4608289 | 8.1878760 | 8.5998995 | 8.165643 | 8.2512149 | 8.1682452 | 8.47768904 | 8.9509193 | 8.08993972 | 8.191537411 | 7.5575235 | 7.5201444 | 8.1165979 | 8.2165015 | 8.1772908 | 8.1777014 | 5.6516802 | 8.2673650 |
| GO:0015807 | L-amino acid transport | 42 | -0.5301731 | -1.619618 | 0.0173084403 | 0.6552290 | 0.6412013 | 4098 | tags=55%, list=26%, signal=41% | SERINC5||SLC47A1||ITGB1||SLC36A4||SLC38A7||ARL6IP5||SLC38A3||PSEN1||SLC25A29||SLC7A1||SERINC3||PER2||SLC1A5||SLC7A5||SLC43A1||CTNS||PRAF2||SLC1A4||SLC66A1||SLC1A1||SLC43A2||SLC7A3||CLTRN | 4.3925689 | 5.4533124 | 4.9114737 | 5.583252 | 4.43112736 | 4.8412208 | 5.3690321 | 5.5605974 | 4.3090897 | 4.41768931 | 4.44728746 | 5.456501 | 5.5121544 | 5.3886333 | 4.9378812 | 4.9702076 | 4.8221693 | 5.548894 | 5.6112227 | 5.5889485 | 4.42207670 | 4.3623927 | 4.50533123 | 4.703437345 | 4.7579295 | 5.0393172 | 5.3825955 | 5.3752275 | 5.3490584 | 5.5562547 | 5.6031541 | 5.5212102 |
| GO:0050829 | defense response to Gram-negative bacterium | 18 | -0.6549619 | -1.675304 | 0.0173194253 | 0.6552290 | 0.6412013 | 2505 | tags=56%, list=16%, signal=47% | H2BC12||ADGRB1||OPTN||F2RL1||ROMO1||MR1||CHGA||IL6R||H2BC11||IL23A | 8.8050576 | 8.6145978 | 8.5826182 | 8.447206 | 8.74412204 | 8.3563570 | 8.5695483 | 8.4761738 | 8.8948604 | 8.87110014 | 8.63520625 | 8.678561 | 8.5872720 | 8.5757454 | 8.5391167 | 8.5245470 | 8.6790899 | 8.494224 | 8.4064883 | 8.4395417 | 8.74103504 | 8.8759928 | 8.60237972 | 8.440029793 | 8.4005512 | 8.2189958 | 8.5887936 | 8.5486574 | 8.5709137 | 8.4622175 | 8.4670964 | 8.4989311 |
| GO:0050922 | negative regulation of chemotaxis | 41 | -0.5280411 | -1.606571 | 0.0176459182 | 0.6552290 | 0.6412013 | 4028 | tags=59%, list=25%, signal=44% | DPP4||PTPRO||DDT||GSTP1||ROBO2||NOTCH1||SEMA3D||SEMA4G||WNT5A||SEMA4C||WNT3||PADI2||MIF||SEMA6B||SEMA3F||RIN3||WNT3A||SEMA6D||SEMA4F||CORO1B||NRP1||SEMA7A||C5||NBL1 | 7.7241580 | 6.8448020 | 7.3389212 | 6.714561 | 7.81808093 | 7.8967550 | 6.6819899 | 6.5875174 | 7.6829648 | 7.48570581 | 7.96354979 | 6.988737 | 6.7611907 | 6.7729156 | 7.0184369 | 7.5329356 | 7.4165152 | 6.907425 | 6.6022685 | 6.6127359 | 7.82130938 | 7.5816219 | 8.01837637 | 7.811058908 | 8.1501565 | 7.6886209 | 6.6996344 | 6.6670115 | 6.6791350 | 6.6044772 | 6.5832086 | 6.5747033 |
| GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 18 | -0.6512892 | -1.665910 | 0.0177130486 | 0.6552290 | 0.6412013 | 4387 | tags=67%, list=27%, signal=48% | ALG6||MPDU1||ALG9||DOLPP1||ALG8||SRD5A3||ALG3||ALG12||DOLK||RFT1||ALG10||ALG1 | 5.1016698 | 5.6576576 | 5.5352964 | 5.646199 | 5.13799427 | 5.3376978 | 5.5288047 | 5.5767918 | 5.0339393 | 5.12034081 | 5.14828156 | 5.683394 | 5.7045204 | 5.5821144 | 5.5164604 | 5.5740041 | 5.5146311 | 5.611489 | 5.7004790 | 5.6250272 | 5.13955761 | 5.0732025 | 5.19850170 | 5.187733081 | 5.1949186 | 5.5922374 | 5.5491198 | 5.5468922 | 5.4896171 | 5.5630074 | 5.6378098 | 5.5273462 |
| GO:1902117 | positive regulation of organelle assembly | 61 | 0.4607780 | 1.540272 | 0.0179278716 | 0.6552290 | 0.6412013 | 4595 | tags=49%, list=29%, signal=35% | LCP1||PLK4||CENPJ||MAPK15||CCDC88A||CEP295||CEP135||FSCN1||PAN3||SPAG5||PIP4K2A||ZMYND10||DZIP1||GPSM2||HAP1||CEP120||DYNC1H1||CNOT1||HTT||ARHGAP35||ULK1||PIP4K2B||BBS4||PDCD6IP||CNOT6||CNOT6L||RAB3GAP2||BECN1||SDCBP||STX18 | 4.7587279 | 5.4586794 | 5.1057208 | 5.517788 | 4.73204124 | 4.9815117 | 5.5284726 | 5.5726858 | 4.7192207 | 4.86114723 | 4.68994258 | 5.441302 | 5.4770142 | 5.4575008 | 5.2791622 | 4.9930947 | 5.0276929 | 5.461152 | 5.4833331 | 5.6047012 | 4.75873369 | 4.7060376 | 4.73087058 | 4.922169858 | 4.8974944 | 5.1149057 | 5.5358336 | 5.5178747 | 5.5316485 | 5.5564626 | 5.5811735 | 5.5802854 |
| GO:0003094 | glomerular filtration | 17 | -0.6610382 | -1.671500 | 0.0180533752 | 0.6555560 | 0.6415212 | 4159 | tags=65%, list=26%, signal=48% | EMP2||PTPRO||CORO2B||SULF2||MCAM||XPNPEP3||SULF1||F2RL1||GAS6||CYBA||TMEM63C | 4.6890511 | 4.5352679 | 4.6997530 | 4.425750 | 4.69365032 | 4.8542557 | 4.3194660 | 4.2679720 | 4.7176863 | 4.49146937 | 4.83693369 | 4.667342 | 4.5553282 | 4.3673556 | 4.5129785 | 4.9227358 | 4.6322497 | 4.531659 | 4.3624945 | 4.3769276 | 4.67855166 | 4.5798064 | 4.81305758 | 4.709615839 | 4.8737934 | 4.9676365 | 4.3878939 | 4.3102146 | 4.2572905 | 4.2366240 | 4.2792127 | 4.2875636 |
| GO:0002686 | negative regulation of leukocyte migration | 22 | -0.6244528 | -1.666242 | 0.0183070866 | 0.6564069 | 0.6423539 | 4028 | tags=55%, list=25%, signal=41% | DPP4||DDT||PLCB1||HMOX1||CXCL12||PADI2||MIF||RIN3||HOXA7||PTGER4||C5||NBL1 | 8.2759333 | 7.4294819 | 7.8227892 | 7.247146 | 8.35700938 | 8.4511936 | 7.2511869 | 7.0953554 | 8.2408345 | 8.02800463 | 8.51707859 | 7.600085 | 7.3209178 | 7.3508578 | 7.4769026 | 8.0313307 | 7.9036600 | 7.467822 | 7.1296435 | 7.1157353 | 8.37595368 | 8.0861186 | 8.56861296 | 8.365832791 | 8.7203707 | 8.2206794 | 7.2611130 | 7.2405335 | 7.2518405 | 7.1172217 | 7.0908795 | 7.0776838 |
| GO:0007218 | neuropeptide signaling pathway | 33 | -0.5593472 | -1.631363 | 0.0187390201 | 0.6623060 | 0.6481267 | 1511 | tags=30%, list=9%, signal=27% | GPR37||NXPH3||NXPH4||UTS2R||PCSK1N||ECEL1||NPY1R||SORCS2||CYSLTR1||NPB | 3.4901478 | 3.3195081 | 3.3266624 | 3.105567 | 3.35969328 | 3.5203482 | 3.2031480 | 2.9472178 | 3.5155388 | 3.42323458 | 3.52937891 | 3.329609 | 3.3281286 | 3.3006024 | 3.2379236 | 3.4698942 | 3.2606568 | 3.189279 | 3.0986054 | 3.0240628 | 3.42666759 | 3.2087957 | 3.43254355 | 3.459677059 | 3.5779661 | 3.5209772 | 3.2660621 | 3.2025269 | 3.1380144 | 2.9494852 | 2.9854375 | 2.9056245 |
| GO:1904350 | regulation of protein catabolic process in the vacuole | 10 | -0.7432645 | -1.668434 | 0.0191743299 | 0.6623060 | 0.6481267 | 3042 | tags=60%, list=19%, signal=49% | LAPTM4B||MGAT3||GBA||CD81||ATP13A2||LRP1 | 6.4049199 | 6.5785669 | 6.6441710 | 6.545972 | 6.44570567 | 6.7321496 | 6.3848879 | 6.4233519 | 6.3295574 | 6.32188013 | 6.55135363 | 6.608533 | 6.6400328 | 6.4823553 | 6.4349814 | 6.7759381 | 6.6998416 | 6.568806 | 6.5866907 | 6.4801805 | 6.43955152 | 6.3483861 | 6.54262670 | 6.587443864 | 6.6967777 | 6.8952050 | 6.4285706 | 6.3803143 | 6.3445448 | 6.4119876 | 6.4918999 | 6.3632337 |
| GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA | 20 | -0.6388425 | -1.664085 | 0.0195063694 | 0.6623060 | 0.6481267 | 2347 | tags=25%, list=15%, signal=21% | ZMPSTE24||EGFR||BMP4||NCOR2||RMRP | 5.1724187 | 5.8621069 | 5.4060163 | 5.752690 | 5.17383303 | 5.3974061 | 5.7609716 | 6.6317799 | 5.1152281 | 5.20224453 | 5.19812404 | 5.961349 | 5.8064948 | 5.8131031 | 5.7160392 | 5.1913614 | 5.2516394 | 5.766978 | 5.6849951 | 5.8035539 | 5.21163145 | 5.0419581 | 5.25899837 | 5.369151986 | 5.3882926 | 5.4340020 | 5.7351271 | 5.7748347 | 5.7726091 | 5.7550086 | 7.5656110 | 5.7563013 |
| GO:0097529 | myeloid leukocyte migration | 94 | -0.4272067 | -1.491343 | 0.0197380907 | 0.6623060 | 0.6481267 | 2510 | tags=36%, list=16%, signal=31% | MDK||MCOLN2||TRPV4||CXCL12||TIRAP||CXCL3||LGMN||JAGN1||ROR2||MIF||PPIB||IRAK4||RIN3||CD81||CD9||TAFA4||CX3CR1||PLA2G7||RHOG||MYD88||BSG||VEGFC||SIRPA||CHGA||P2RX4||IL6R||PTGER4||THBS4||EDNRB||C5||ITGB2||NBL1||TGFB2||C5AR1 | 7.5214061 | 7.1401024 | 7.3495769 | 7.097168 | 7.53505004 | 7.5204422 | 7.0372413 | 7.0212364 | 7.5264742 | 7.44785913 | 7.58654008 | 7.233814 | 7.0943973 | 7.0873147 | 7.1830155 | 7.4420925 | 7.4100512 | 7.175090 | 7.0441647 | 7.0688680 | 7.54069471 | 7.4480460 | 7.61175129 | 7.480943135 | 7.6444214 | 7.4269802 | 7.0572121 | 7.0157975 | 7.0384164 | 7.0480023 | 6.9563824 | 7.0571869 |
| GO:0006405 | RNA export from nucleus | 75 | 0.4387012 | 1.518789 | 0.0198103267 | 0.6623060 | 0.6481267 | 5745 | tags=55%, list=36%, signal=35% | ZC3H11B||THOC1||NRDE2||AGFG1||SMG1||THOC2||NUP214||SETD2||YTHDC1||NA||SMG7||UPF2||TSC1||IWS1||TPR||CPSF6||PHAX||DDX19B||CHTOP||ZC3H11A||ENY2||NXF1||NCBP2||PCID2||THOC7||MCM3AP||NSUN2||SSB||FYTTD1||NUP160||NOL6||SMG5||DHX9||THOC5||HNRNPA2B1||NPM1||POLDIP3||XPO5||NUP153||XPOT||NUP155 | 6.7044174 | 7.7015643 | 6.8890279 | 7.704480 | 6.59244714 | 6.7355037 | 7.7725119 | 7.7604791 | 6.7109399 | 6.85930772 | 6.52348356 | 7.714982 | 7.6749782 | 7.7143696 | 7.2246905 | 6.6427396 | 6.7289562 | 7.666670 | 7.6747189 | 7.7697542 | 6.55019583 | 6.6767188 | 6.54658285 | 6.701993099 | 6.6573743 | 6.8407458 | 7.7517644 | 7.7670222 | 7.7983572 | 7.7509258 | 7.7602118 | 7.7702351 |
| GO:0045446 | endothelial cell differentiation | 82 | -0.4398865 | -1.507221 | 0.0206400303 | 0.6623060 | 0.6481267 | 2463 | tags=28%, list=15%, signal=24% | SOX18||BMP6||PDE4D||FSTL1||NOTCH4||BMP4||COL18A1||S1PR2||MYADM||BMPR2||F2RL1||MYD88||SCUBE1||GPX1||NRP1||CEACAM1||MESP1||CXCR4||CLDN1||ETV2||S1PR1||RAPGEF3||PDE2A | 5.0259013 | 5.1022383 | 4.9700307 | 5.042619 | 5.07972135 | 5.1448827 | 5.0366803 | 5.0171395 | 5.0058052 | 4.94728752 | 5.11928001 | 5.130847 | 5.1185296 | 5.0562347 | 4.9500034 | 4.9792682 | 4.9806135 | 5.061189 | 4.9948100 | 5.0706791 | 5.07258526 | 4.9916166 | 5.16945859 | 5.166219948 | 5.2589910 | 4.9973518 | 5.0660902 | 5.0147068 | 5.0287539 | 4.9944496 | 5.0465418 | 5.0099301 |
| GO:0032331 | negative regulation of chondrocyte differentiation | 17 | -0.6553793 | -1.657191 | 0.0208006279 | 0.6623060 | 0.6481267 | 3696 | tags=65%, list=23%, signal=50% | LTBP3||ADAMTS7||TGFBR1||EFEMP1||RARB||GLI2||SNAI2||BMP4||SOX9||WNT9A||PTHLH | 3.1919489 | 4.8484003 | 3.9005277 | 4.917607 | 3.00549610 | 3.0766967 | 4.8140186 | 4.9055413 | 3.2650622 | 3.42376105 | 2.82180714 | 4.864287 | 4.8881082 | 4.7910416 | 4.2369564 | 3.4693343 | 3.8943468 | 4.874803 | 4.9211727 | 4.9557044 | 2.91236720 | 3.2120632 | 2.86717128 | 2.990627263 | 2.8674736 | 3.3311834 | 4.8077326 | 4.8185723 | 4.8157290 | 4.8934908 | 4.9221754 | 4.9008034 |
| GO:0030201 | heparan sulfate proteoglycan metabolic process | 29 | -0.5694073 | -1.622134 | 0.0211650485 | 0.6623060 | 0.6481267 | 3354 | tags=48%, list=21%, signal=38% | SULF2||PXYLP1||NDST3||HS6ST2||EXTL3||SULF1||HS6ST1||EXT2||ENSG00000272916||B3GAT3||GPC1||B3GALT6||CSGALNACT1||HPSE | 3.9221333 | 4.8745778 | 4.5176538 | 4.931926 | 3.88986945 | 4.2315775 | 4.7753685 | 4.8410563 | 3.9014343 | 3.95212851 | 3.91234223 | 4.893428 | 4.9270682 | 4.8002659 | 4.6015658 | 4.4709036 | 4.4766780 | 4.907735 | 4.9595328 | 4.9280374 | 3.86409255 | 3.8591583 | 3.94474692 | 4.011271775 | 4.1243149 | 4.5104765 | 4.8096729 | 4.7729944 | 4.7426570 | 4.8433103 | 4.8413832 | 4.8384713 |
| GO:0042423 | catecholamine biosynthetic process | 10 | -0.7388762 | -1.658583 | 0.0213265506 | 0.6623060 | 0.6481267 | 1511 | tags=60%, list=9%, signal=54% | SNCA||INSM1||GPR37||HAND2||NR4A2||TGFB2 | 5.1354938 | 5.4521751 | 5.4983053 | 5.560233 | 4.84681568 | 4.4258751 | 5.4598886 | 5.4978024 | 5.3240336 | 5.46698755 | 4.40350838 | 5.461290 | 5.4397721 | 5.4553773 | 5.6186886 | 5.1724920 | 5.6555144 | 5.635768 | 5.4734454 | 5.5668998 | 4.63086605 | 5.3095135 | 4.45260265 | 4.483377856 | 4.1727390 | 4.5897173 | 5.4868958 | 5.4094730 | 5.4820016 | 5.4907155 | 5.4693935 | 5.5325804 |
| GO:0006357 | regulation of transcription by RNA polymerase II | 1964 | 0.2366733 | 1.126590 | 0.0216031836 | 0.6623060 | 0.6481267 | 3595 | tags=27%, list=23%, signal=24% | PRRX1||NLRC5||IFI27||ZNF114||ONECUT1||ZNF471||BHLHE41||ZNF492||HOXD3||BAZ2B||ZNF559-ZNF177||ZNF888||ZNF813||ZNF230||ZNF682||PRKN||ZNF404||ZNF586||APLN||HDAC9||NFIA||FOXD4L5||ZNF283||ENSG00000273046||ZNF17||ARID4A||ZNF587||FOSL2||ZNF345||ZNF780B||HOXD4||MAGEA2||CSRNP3||PLAC8||WWC3||SNIP1||MYSM1||ZNF571||ZNF608||CCDC62||SNAI3||MCIDAS||DACH2||ZNF548||HOXD9||ASH1L||FOXD2||GTF2IRD2||MTF1||ZNF442||ZNF611||LHX4||ZNF134||GATA5||NR2C2||CLOCK||ZNF525||HOXB7||KMT2D||KLF14||ZNF112||VDR||ZNF547||ATM||ZIC3||ZNF416||ZNF91||CREBRF||ZNF224||TET2||HNF4G||TMF1||HMBOX1||ZNF780A||ZNF223||ZNF516||ZNF502||DRD2||ZNF670||CCNL1||ONECUT2||NPAS3||ZNF782||HOXC8||ZNF329||ZBTB37||KMT2A||WT1||ZNF169||SIX4||EAF2||DMTF1||FOXF1||ZNF493||CCNT2||ZNF578||ZNF582||POU2F1||ZNF462||ZNF136||ARNTL||LPIN1||ZNF236||CCND1||MXD1||ZNF57||ZNF573||ZNF256||ZNF143||PPARGC1B||ZBTB17||ZNF417||ATRX||ZNF470||ZNF713||EMX2||OTX2||TCERG1||ZNF280D||ZNF550||ZNF852||CHD7||ZNF225||NFYB||TCF12||CAMTA2||ARID5B||ZNF304||USP3||ZNF791||BRD8||REL||ZNF675||ZNF506||BMI1||ZNF490||PLK3||ZNF551||LIME1||NKX2-2||MYT1||ZNF549||KAT2B||THAP1||STAT2||TGIF1||HDAC10||MLXIPL||MIDEAS||BRWD3||MC1R||ACVR2A||ZNF100||DPF3||TADA2A||PLAG1||GLI4||ZNF500||YEATS2||ZNF772||JDP2||RPS6KA5||RXRB||EPC1||ZNF382||MEF2C||RBM15||EYA1||HIVEP1||VGLL3||ZNF276||PPP1R12A||TGFB3||ZFHX4||URI1||MECP2||CHD3||MNAT1||ZNF641||RIF1||MAFF||N4BP2L2||BBX||BPTF||ZNF846||ZNF433||USP16||PKIA||ZNF510||HELZ2||ZNF354B||ATAD2B||ZNF543||ZNF680||KDM7A||BAZ1A||RFC1||ZNF333||EBF2||NFATC2IP||ZNF891||PPARGC1A||ZNF805||SP9||ZSCAN12||ZNF16||ZNF431||ZNF483||POGZ||ZNF639||ZNF613||MIER1||NR1D2||CRY2||SMARCA2||POU3F2||ZNF28||RREB1||FRK||MED21||ZNF449||ALX1||NCOA3||PIAS1||ZNF695||ZNF726||NR6A1||ZNF268||ZNF292||SUV39H2||GABPA||KLF6||ZNF248||ZKSCAN8||AUTS2||SP1||ZKSCAN1||ZNF117||ZBTB43||TET1||KDM3B||CHD6||ATF7IP||PAX6||ZNF430||MIER3||PAX3||KLF5||ZBTB11||AHR||NFAT5||SCAF8||ZNF776||PHF21A||NSD2||ASXL2||ZNF609||MED1||ZNF66||PRDM2||PGBD1||ZNF790||ZNF438||NFKBIA||ZIC5||OLIG1||DNMT1||DHX36||ZNF331||CCNL2||ZNF24||LPIN3||ZNF721||KAT7||ZNF391||HOXD10||EBF4||MGA||CRYM||TAF3||GTF2H4||ZNF684||IRF4||LARP7||LHX3||KLF11||PHIP||GABPB2||GNL3||ZNF770||PAXBP1||HOXA9||ZNF320||ZMIZ1||MAMLD1||ZGPAT||IKZF4||ZNF175||ZNF211||ZNF84||BRD9||FOXO1||NFATC4||MED13||RPS6KA3||ZMYM5||APBB2||ARID4B||TCF20||ATF7||LPIN2||EPAS1||HCFC1||SMAD5||HOXB8||HOXC9||STOX2||ZNF529||DICER1||TFAP2E||RSF1||GMEB1||ZNF507||ZNF567||ZNF217||ZNF263||PKD1||ATF2||NR1H2||FOXQ1||HOXB3||TFAP2C||ZNF180||ZNF407||LCOR||ECM1||ZBED5||TLE4||CDK12||ZNF655||POU4F1||ATXN7L3||CHD2||ESRRG||KAT6B||KMT2C||ZNF597||ACVR2B||FLCN||FOXN3||GATA6||ZNF577||MZF1||ZNF131||CHD1||VEZF1||CXXC5||EN1||ARNTL2||ZNF227||TTC21B||PPRC1||EFCAB7||HIF1A||SOX2||ELK4||ZNF367||ZNF79||PIK3R1||TNKS||GADD45A||ZNF514||TIAL1||TRAK1||TFCP2L1||MECOM||NFATC3||ZSCAN9||KLF3||ZNF704||ZNF653||JMJD1C||ZNF26||ATF1||CDK13||FOXC2||AGRN||HBP1||SETX||PHF20||ZNF692||TP53BP1||RLIM||ZBTB20||HIVEP2||GCN1||ZNF177||NSD1||LEO1||ZNF740||PYGO1||RTF1||ZSCAN26||BHLHE40||GMEB2||AR||TERT||KLF9||TPR||NIPBL||E2F8||AKNA||MLXIP||TBX15||TBX19||SMARCA5||SP3||HMGB1||ZNF518A||BMPR1B||ZFP91||CKS2||ZNF146||ZFX||E2F3||FOXD1||SIRT6||FOXK1||PSIP1||PPM1A||FGF2||ZKSCAN4||ZNF784||EHMT2||HMGN3||IRF2||ZNF829||RNASEL||ERCC6||KMT5A||NRIP1||HIPK2||ELOC||TOX||PITX2||ZBTB40||RYBP||KDM6B||CAPRIN2||ZNF121||ZBTB49||RORA||KLF12||BCL9||TCF7L2||SMAD3||HOXA1||MED11||ZNF420||MED12||ZNF841||ZNF669||OTX1||GRHL1||FOXO3||PPM1D||ZNF709||BRD7||ZNF587B||ZBTB24||MED24||ZBED3||ZNF155||JMY||ZHX3||FOXJ3||ZNF124||HMGB2||MLLT6||RFX7||TARBP1||BAZ1B||ZNF441||ZNF432||ZNF286A||PTPN2||ZNF845||MED19||ZNF397||ZSCAN5A||JARID2||ZNF614||HCFC2||MRTFB||ZEB2||ZNF202||CREBL2||FOXJ2||ZNF461||ZNF92||HMGB3||TAF6L||GTF2H2||ZNF266||IKBKG||PHF6||ZFP37||ZC3H8||ZNF23||CDX2||NFIB||SIM1||ARID1A||IKBKB||PHF20L1||NPAT||TCF25||MED6||CNOT1||TADA1||MAFG||HDX||ZNF485||LIN37||ENY2||SMARCC1||NFYA||NKX2-1||NKX6-1 | 5.1752107 | 5.4262003 | 5.2228527 | 5.427770 | 5.16218783 | 5.2347147 | 5.4537058 | 5.4688751 | 5.1590543 | 5.20328994 | 5.16287026 | 5.437099 | 5.4217994 | 5.4196397 | 5.2968630 | 5.1702710 | 5.1983389 | 5.420774 | 5.3899754 | 5.4713864 | 5.15410642 | 5.1547882 | 5.17754535 | 5.227143717 | 5.2327073 | 5.2442402 | 5.4549260 | 5.4463426 | 5.4598166 | 5.4602670 | 5.4801747 | 5.4661110 |
| GO:0051965 | positive regulation of synapse assembly | 39 | -0.5234418 | -1.577682 | 0.0224936979 | 0.6623060 | 0.6481267 | 2445 | tags=36%, list=15%, signal=30% | CLSTN3||NLGN1||CLSTN1||IL1RAP||ADGRB1||EPHB1||EPHB2||EPHB3||CLSTN2||LRRC4B||TPBG||PTPRD||AMIGO2||BHLHB9 | 2.2115433 | 3.1286019 | 3.0679391 | 3.104026 | 2.21107979 | 2.8971974 | 3.0855473 | 3.0938769 | 2.1536721 | 2.20656414 | 2.27195685 | 3.046562 | 3.2106758 | 3.1238914 | 3.1456190 | 3.1154522 | 2.9337766 | 3.074533 | 3.1370132 | 3.0998473 | 2.20967446 | 2.0612219 | 2.34810367 | 2.607898782 | 2.6912312 | 3.2919775 | 3.1205464 | 3.0714452 | 3.0639936 | 3.0510341 | 3.1670391 | 3.0606612 |
| GO:0021904 | dorsal/ventral neural tube patterning | 21 | -0.6214795 | -1.643682 | 0.0225474684 | 0.6623060 | 0.6481267 | 4340 | tags=57%, list=27%, signal=42% | PRKACA||TBC1D32||FKBP8||PSEN1||GPR161||GLI2||TCTN1||GLI3||PTCH1||BMP4||WNT3A||GSC | 4.8780302 | 4.7442342 | 4.9141958 | 4.617380 | 4.92121272 | 5.1959958 | 4.6737397 | 4.6093628 | 4.8008425 | 4.79123502 | 5.02921896 | 4.741374 | 4.7790124 | 4.7115228 | 4.8102193 | 5.0078377 | 4.9177617 | 4.688754 | 4.5349170 | 4.6243456 | 4.92961893 | 4.7926270 | 5.03148041 | 5.134414614 | 5.2952120 | 5.1529377 | 4.7063793 | 4.6687655 | 4.6454174 | 4.6117216 | 4.6597912 | 4.5546578 |
| GO:0060350 | endochondral bone morphogenesis | 49 | -0.4981310 | -1.566345 | 0.0231526143 | 0.6623060 | 0.6481267 | 2434 | tags=37%, list=15%, signal=31% | BMP6||TRPV4||ZMPSTE24||COMP||EXT2||COL13A1||BMP4||SOX9||POR||BMPR2||FGF18||ALPL||PHOSPHO1||COL2A1||SERPINH1||NPPC||RUNX2||SMPD3 | 3.4518360 | 4.0054435 | 3.8815182 | 3.911689 | 3.47339652 | 3.7992429 | 3.8987548 | 3.8640001 | 3.4022415 | 3.41409785 | 3.53537096 | 4.005355 | 4.0739548 | 3.9336088 | 3.8865809 | 3.9369647 | 3.8185672 | 3.871302 | 3.9274028 | 3.9355185 | 3.45436117 | 3.3894960 | 3.57047371 | 3.620300597 | 3.6446818 | 4.0840460 | 3.9490125 | 3.8923300 | 3.8533138 | 3.8318951 | 3.9334065 | 3.8241007 |
| GO:0085029 | extracellular matrix assembly | 35 | -0.5413123 | -1.598382 | 0.0231788079 | 0.6623060 | 0.6481267 | 3696 | tags=60%, list=23%, signal=46% | LTBP3||LAMB1||NOTCH1||ANTXR1||GPM6B||FBLN5||TGFB1||LAMB2||DAG1||THSD4||HAS3||FKBP10||SOX9||QSOX1||HAS2||LOX||GAS6||RAMP2||NTNG2||SMPD3||NTNG1 | 3.2851363 | 4.0064585 | 3.8707442 | 3.782111 | 3.32012614 | 3.7877705 | 3.8631732 | 3.6552717 | 3.2313876 | 3.18071694 | 3.43099766 | 4.003051 | 4.0604941 | 3.9538542 | 3.9006788 | 3.9638542 | 3.7384304 | 3.785943 | 3.7956944 | 3.7645199 | 3.27587132 | 3.1662562 | 3.49820108 | 3.518811861 | 3.6184891 | 4.1448112 | 3.9460966 | 3.8444825 | 3.7947955 | 3.6453538 | 3.7309285 | 3.5858356 |
| GO:0009154 | purine ribonucleotide catabolic process | 35 | -0.5392007 | -1.592147 | 0.0235683677 | 0.6623060 | 0.6481267 | 637 | tags=26%, list=4%, signal=25% | NUDT9||PDE4D||FITM2||ABCD1||MLYCD||NUDT18||PDE4C||PDE1A||PDE2A | 5.1962211 | 5.3895185 | 5.2705480 | 5.341771 | 5.14947202 | 4.9767675 | 5.3725156 | 5.3584292 | 5.2836003 | 5.26059715 | 5.03122179 | 5.430594 | 5.3630600 | 5.3739867 | 5.2851903 | 5.1973340 | 5.3261338 | 5.327594 | 5.3119761 | 5.3847233 | 5.17310515 | 5.2378757 | 5.02963368 | 5.000038101 | 4.7993368 | 5.1135664 | 5.3709761 | 5.3633382 | 5.3831631 | 5.3310450 | 5.3653131 | 5.3785145 |
| GO:0015868 | purine ribonucleotide transport | 22 | -0.6120945 | -1.633266 | 0.0240157480 | 0.6623060 | 0.6481267 | 4961 | tags=64%, list=31%, signal=44% | SLC35B3||SLC25A6||SLC33A1||SLC25A4||CD47||SLC25A42||ABCC4||SLC25A5||LRRC8A||SLC29A1||SLC35B2||LRRC8E||SLC19A1||ABCD1 | 6.8722711 | 6.5093923 | 6.7777562 | 6.575667 | 6.84141195 | 6.8084240 | 6.4128819 | 6.5804139 | 6.9169319 | 6.97029394 | 6.71746585 | 6.585036 | 6.5009388 | 6.4384431 | 6.6802667 | 6.7403219 | 6.9033488 | 6.648167 | 6.5082464 | 6.5671575 | 6.85382345 | 6.8981164 | 6.76934289 | 6.898539321 | 6.8838650 | 6.6270484 | 6.4271092 | 6.4113702 | 6.4000380 | 6.5725020 | 6.5960050 | 6.5726075 |
| GO:0051503 | adenine nucleotide transport | 22 | -0.6120945 | -1.633266 | 0.0240157480 | 0.6623060 | 0.6481267 | 4961 | tags=64%, list=31%, signal=44% | SLC35B3||SLC25A6||SLC33A1||SLC25A4||CD47||SLC25A42||ABCC4||SLC25A5||LRRC8A||SLC29A1||SLC35B2||LRRC8E||SLC19A1||ABCD1 | 6.8722711 | 6.5093923 | 6.7777562 | 6.575667 | 6.84141195 | 6.8084240 | 6.4128819 | 6.5804139 | 6.9169319 | 6.97029394 | 6.71746585 | 6.585036 | 6.5009388 | 6.4384431 | 6.6802667 | 6.7403219 | 6.9033488 | 6.648167 | 6.5082464 | 6.5671575 | 6.85382345 | 6.8981164 | 6.76934289 | 6.898539321 | 6.8838650 | 6.6270484 | 6.4271092 | 6.4113702 | 6.4000380 | 6.5725020 | 6.5960050 | 6.5726075 |
| GO:0031640 | killing of cells of other organism | 15 | -0.6669749 | -1.628844 | 0.0240432779 | 0.6623060 | 0.6481267 | 4072 | tags=53%, list=26%, signal=40% | SYK||RPS19||H2BC12||F2RL1||MYD88||ROMO1||CHGA||H2BC11 | 10.1302746 | 9.6319766 | 9.9133097 | 9.547993 | 10.13196742 | 9.9752514 | 9.5980698 | 9.5716604 | 10.1461135 | 10.13646749 | 10.10797067 | 9.686065 | 9.6096327 | 9.5986515 | 9.7645248 | 9.9446079 | 10.0190666 | 9.604742 | 9.4863769 | 9.5504304 | 10.13721310 | 10.1760474 | 10.08106435 | 10.061031639 | 10.0544549 | 9.7945989 | 9.6014434 | 9.5939424 | 9.5988135 | 9.5624008 | 9.5598973 | 9.5924551 |
| GO:0050870 | positive regulation of T cell activation | 121 | -0.3975649 | -1.444949 | 0.0241076434 | 0.6623060 | 0.6481267 | 3802 | tags=37%, list=24%, signal=29% | NFKBID||IGF2||HES1||FADD||PRKCQ||BAD||CD276||CORO1A||CSK||NCK2||ICOSLG||HLX||SLC7A1||IL4R||CD55||ZBTB7B||RUNX3||EFNB3||RASGRP1||GLI2||MDK||HLA-DMB||HLA-E||IL6ST||GLI3||IGFBP2||PRKCZ||CD81||EFNB1||JAK3||SIRPA||HLA-DQB1||HLA-A||DOCK8||SOCS1||ITPKB||CD24||LGALS1||ZBTB16||HLA-DMA||HLA-DOA||IL23A||DUSP10||CCDC88B||IL4I1 | 6.2917749 | 6.2673944 | 6.1652291 | 6.278572 | 6.29007607 | 6.2026079 | 6.2265582 | 6.2797519 | 6.2973646 | 6.26915666 | 6.30851863 | 6.317105 | 6.2688641 | 6.2143853 | 6.0945384 | 6.1982042 | 6.2004281 | 6.302742 | 6.2500374 | 6.2824476 | 6.31066672 | 6.2470244 | 6.31159217 | 6.284935798 | 6.2016658 | 6.1162963 | 6.2332185 | 6.2269508 | 6.2194723 | 6.2806596 | 6.2738520 | 6.2847233 |
| GO:0014015 | positive regulation of gliogenesis | 42 | -0.5112529 | -1.561819 | 0.0243096072 | 0.6623060 | 0.6481267 | 3755 | tags=43%, list=24%, signal=33% | HES1||NOTCH1||UFL1||OLIG2||TGFB1||BIN1||SOX8||PTPRZ1||TSPO||MDK||EGFR||DAG1||IL6ST||SERPINE2||TP73||CXCR4||ZNF365||ETV5 | 5.3409583 | 5.7356715 | 5.4423752 | 5.661504 | 5.32970338 | 5.4425642 | 5.7282507 | 5.7023228 | 5.3287987 | 5.32173524 | 5.37182967 | 5.771736 | 5.7337068 | 5.7006953 | 5.4500719 | 5.4555194 | 5.4213005 | 5.662325 | 5.6220105 | 5.6991463 | 5.31640703 | 5.3023081 | 5.36952090 | 5.404328640 | 5.3719773 | 5.5454294 | 5.7288331 | 5.7270312 | 5.7288870 | 5.6777284 | 5.7382277 | 5.6903036 |
| GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 53 | 0.4676328 | 1.520640 | 0.0243648480 | 0.6623060 | 0.6481267 | 4085 | tags=34%, list=26%, signal=25% | MBNL2||RBM11||RBM5||CELF4||CELF6||CELF5||RBM15||NSRP1||YTHDC1||MBNL1||CELF1||RBFOX2||RBM25||DYRK1A||RAVER2||THRAP3||RBM10||MBNL3 | 6.6683705 | 7.4397611 | 6.8067333 | 7.376363 | 6.59444301 | 6.7431864 | 7.5015221 | 7.4344878 | 6.6500429 | 6.80263140 | 6.54036084 | 7.444342 | 7.4247320 | 7.4500874 | 7.1314174 | 6.5927685 | 6.6312083 | 7.340321 | 7.3508493 | 7.4359984 | 6.57711596 | 6.6231377 | 6.58263631 | 6.745365589 | 6.7098369 | 6.7736485 | 7.4882715 | 7.4953557 | 7.5207368 | 7.4140231 | 7.4441468 | 7.4450776 |
| GO:0051497 | negative regulation of stress fiber assembly | 24 | -0.5958291 | -1.621725 | 0.0252495596 | 0.6623060 | 0.6481267 | 262 | tags=8%, list=2%, signal=8% | S1PR1||TMEFF2 | 6.7624869 | 6.3369019 | 6.6235082 | 6.372291 | 6.77722817 | 6.9266785 | 6.3403662 | 6.3442232 | 6.7012387 | 6.74937637 | 6.83371647 | 6.363749 | 6.2891956 | 6.3565949 | 6.5136965 | 6.7120807 | 6.6378257 | 6.456531 | 6.2872368 | 6.3681309 | 6.81315918 | 6.6589542 | 6.85240831 | 6.930094815 | 7.0148007 | 6.8291665 | 6.3296106 | 6.3144934 | 6.3762734 | 6.3418590 | 6.3544950 | 6.3362549 |
| GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 15 | -0.6613742 | -1.615166 | 0.0258465237 | 0.6623060 | 0.6481267 | 4527 | tags=67%, list=28%, signal=48% | ELOVL1||ELOVL6||HSD17B12||HACD1||ELOVL5||ACSL3||ELOVL7||TECR||ELOVL4||ACSL6 | 4.9079816 | 5.5539919 | 5.1160129 | 5.629456 | 4.96886062 | 5.2501545 | 5.4141313 | 5.5318265 | 4.8436224 | 4.84258403 | 5.02959132 | 5.593097 | 5.5689813 | 5.4982204 | 5.0465778 | 5.2167591 | 5.0789854 | 5.616725 | 5.6428983 | 5.6286269 | 4.93743375 | 4.8862085 | 5.07618395 | 5.103239251 | 5.2429115 | 5.3900539 | 5.4267766 | 5.4243743 | 5.3909660 | 5.5414056 | 5.5784533 | 5.4736704 |
| GO:0006879 | cellular iron ion homeostasis | 47 | -0.4968613 | -1.548892 | 0.0262594826 | 0.6623060 | 0.6481267 | 4372 | tags=47%, list=27%, signal=34% | SLC6A9||FTH1||ABCB6||SLC39A8||SLC22A17||CYB561||MYC||HMOX1||SLC39A14||BMP6||ERFE||NDFIP1||ATP6V0A2||CYBRD1||ATP6AP1||STEAP2||ATP13A2||SLC40A1||TMEM199||CYB561A3||SLC46A1||TF | 6.8082926 | 6.6099323 | 6.6193278 | 6.478960 | 6.80797948 | 6.8346811 | 6.5597098 | 6.4302210 | 6.8090654 | 6.75805432 | 6.85609255 | 6.636973 | 6.5937675 | 6.5986675 | 6.5026635 | 6.6643369 | 6.6842005 | 6.530655 | 6.4234319 | 6.4807984 | 6.83923506 | 6.7260186 | 6.85527526 | 6.878576786 | 6.8880240 | 6.7322154 | 6.5785055 | 6.5337851 | 6.5664685 | 6.4198403 | 6.4417773 | 6.4289612 |
| GO:0009187 | cyclic nucleotide metabolic process | 25 | -0.5874236 | -1.616460 | 0.0264187867 | 0.6623060 | 0.6481267 | 736 | tags=44%, list=5%, signal=42% | ADCY9||EPHA2||ADCY5||CNP||GUCY1A2||PDE4D||NPPC||PDE4C||PDE1A||PTHLH||PDE2A | 2.7214308 | 3.0099520 | 3.1326845 | 2.930192 | 2.77745759 | 3.1855851 | 2.9789174 | 2.8669008 | 2.6136089 | 2.70092734 | 2.84061872 | 2.976217 | 3.0815052 | 2.9693732 | 3.1643559 | 3.1766357 | 3.0539242 | 2.939598 | 2.8829696 | 2.9667470 | 2.79755931 | 2.5770257 | 2.93537959 | 2.982863900 | 3.1014943 | 3.4337690 | 3.0101374 | 2.9660043 | 2.9600901 | 2.8557703 | 2.9674716 | 2.7706917 |
| GO:0030204 | chondroitin sulfate metabolic process | 25 | -0.5868644 | -1.614921 | 0.0264187867 | 0.6623060 | 0.6481267 | 3783 | tags=56%, list=24%, signal=43% | CHST3||IDUA||CHSY1||EGFLAM||CHST11||CHPF2||NDNF||CHSY3||CHST12||B3GALT6||CHPF||CHST7||CSGALNACT1||SPOCK3 | 3.0806426 | 3.7155721 | 3.5677923 | 3.509105 | 3.11571672 | 3.5363244 | 3.5762729 | 3.3285178 | 3.0418066 | 3.01446581 | 3.18013174 | 3.696186 | 3.7965322 | 3.6500903 | 3.5113194 | 3.6854235 | 3.4989999 | 3.486356 | 3.5239716 | 3.5167105 | 3.10107514 | 2.9647222 | 3.26558484 | 3.275038667 | 3.4391716 | 3.8358970 | 3.6147016 | 3.6008260 | 3.5111150 | 3.3225730 | 3.3862275 | 3.2745748 |
| GO:0071560 | cellular response to transforming growth factor beta stimulus | 183 | -0.3594842 | -1.380835 | 0.0266959225 | 0.6623060 | 0.6481267 | 3273 | tags=26%, list=21%, signal=21% | HTRA3||SLC2A10||C20orf27||ZNF703||RASL11B||WNT5A||TGFB1||HTRA1||COL4A2||FURIN||MEN1||STK16||CHST11||LTBP1||WWOX||CDKN2B||ZYX||LTBP4||CD109||HSPA5||SOX9||JUN||ZNF451||FOS||CX3CR1||LOX||CDKN1C||HYAL2||SFRP1||WFIKKN1||PMEPA1||IL17RD||ITGB5||HPGD||SOX5||DUSP15||SPRED3||VASN||CLDN1||DKK3||TGFB2||NRROS||GDF9||TGFB1I1||LRRC32||PDE2A||PEG10 | 4.7676920 | 5.2600644 | 4.8472183 | 5.286796 | 4.74942352 | 4.9383653 | 5.2632763 | 5.2899767 | 4.7388263 | 4.81329308 | 4.74983144 | 5.259488 | 5.2676366 | 5.2530315 | 5.0117474 | 4.7529690 | 4.7616913 | 5.283377 | 5.2482470 | 5.3276643 | 4.75313027 | 4.7091087 | 4.78502585 | 4.962705030 | 4.9761256 | 4.8741522 | 5.2655938 | 5.2583739 | 5.2658488 | 5.2927298 | 5.3022894 | 5.2747757 |
| GO:0045920 | negative regulation of exocytosis | 21 | 0.5875156 | 1.580122 | 0.0274969672 | 0.6623060 | 0.6481267 | 1070 | tags=14%, list=7%, signal=13% | RSC1A1||PRKN||FOXF1 | 3.6853205 | 4.5947678 | 3.8512326 | 4.593678 | 3.68641929 | 3.8267355 | 4.6205022 | 4.6382797 | 3.6494649 | 3.75056901 | 3.65364255 | 4.584366 | 4.6167817 | 4.5829004 | 4.1154050 | 3.7217063 | 3.6748539 | 4.557901 | 4.5692971 | 4.6519972 | 3.68386127 | 3.6711168 | 3.70408806 | 3.773362316 | 3.7651850 | 3.9352075 | 4.6129683 | 4.6236916 | 4.6248170 | 4.6317578 | 4.6401532 | 4.6429046 |
| GO:0006612 | protein targeting to membrane | 107 | -0.4029674 | -1.436532 | 0.0281848929 | 0.6623060 | 0.6481267 | 4522 | tags=42%, list=28%, signal=30% | RAB3IP||SEC61A2||CIB1||SSR1||STOM||SSR3||ZDHHC23||FIS1||ZDHHC20||ZDHHC9||PEX16||PEX3||HRAS||DMTN||ERBB2||PRNP||VPS37B||ICMT||CDK5||ARL6||ZDHHC4||ZFAND2B||GET3||ZDHHC22||VPS37D||MIEF2||SEC61A1||TRAM1||TRAM1L1||YIF1B||HSPA5||ZDHHC2||RN7SL3||SIL1||RN7SL1||SSR2||ZDHHC24||RN7SL2||ZDHHC12||SLC1A1||ZDHHC14||ZDHHC11B||GOLGA7B||ITGB2||NACAD | 5.9835062 | 6.4593257 | 6.0109336 | 6.374179 | 6.05926220 | 6.0583606 | 6.4137336 | 6.6841630 | 5.9308635 | 6.00349926 | 6.01472744 | 6.452614 | 6.4771188 | 6.4480743 | 6.1839112 | 5.9445109 | 5.8868928 | 6.357424 | 6.3757398 | 6.3891982 | 6.02737300 | 6.0031763 | 6.14331454 | 5.982026661 | 6.0891895 | 6.1009157 | 6.4142758 | 6.4028458 | 6.4240016 | 6.3855910 | 7.1281595 | 6.4093816 |
| GO:0045814 | negative regulation of gene expression, epigenetic | 45 | 0.4783180 | 1.513863 | 0.0283775447 | 0.6623060 | 0.6481267 | 4562 | tags=47%, list=29%, signal=33% | NRDE2||ZNF304||BMI1||TASOR||EPC1||BAZ2A||TET1||ATF7IP||DNMT1||ZNFX1||TPR||SMARCA5||PPM1D||EZH2||EZH1||SIRT1||CREBZF||PPHLN1||HMGA2||SETDB1||SETDB2 | 5.6066237 | 5.8118900 | 5.8026364 | 5.852248 | 5.59827328 | 5.8091180 | 5.8654267 | 5.8866973 | 5.5503642 | 5.60996126 | 5.65754956 | 5.814155 | 5.8180623 | 5.8034126 | 5.8693962 | 5.7568827 | 5.7791537 | 5.844934 | 5.8117557 | 5.8987157 | 5.58956601 | 5.5519890 | 5.65150960 | 5.743678232 | 5.7933094 | 5.8866944 | 5.8820952 | 5.8573374 | 5.8567019 | 5.8694235 | 5.9084392 | 5.8819536 |
| GO:0042752 | regulation of circadian rhythm | 97 | 0.3933712 | 1.426559 | 0.0284510011 | 0.6623060 | 0.6481267 | 4757 | tags=40%, list=30%, signal=28% | SIK1||CLOCK||PRKAA1||KCNA2||FBXL8||DRD2||OPN3||ARNTL||PPARGC1A||NR1D2||CRY2||SUV39H2||FBXL12||GSK3B||ARNTL2||BHLHE40||PRKAA2||RORA||RBM4B||CSNK1D||ROCK2||NKX2-1||TOP2A||FBXW7||THRAP3||PHLPP1||EZH2||ADRB1||SIN3A||MTOR||MAPK10||PRKDC||MAPK8||SRRD||USP7||CHML||PPP1CC||CDK1||FBXL3 | 5.3639015 | 5.8863744 | 5.5543018 | 5.870931 | 5.35769876 | 5.5597296 | 5.9454752 | 5.9174001 | 5.3160653 | 5.43984326 | 5.33264088 | 5.868930 | 5.8887977 | 5.9012123 | 5.6985124 | 5.4805909 | 5.4722046 | 5.830604 | 5.8399765 | 5.9396605 | 5.37547001 | 5.3125073 | 5.38406617 | 5.571861715 | 5.4970723 | 5.6080444 | 5.9362421 | 5.9436567 | 5.9564544 | 5.8928185 | 5.9400449 | 5.9189492 |
| GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 15 | -0.6571729 | -1.604906 | 0.0286515728 | 0.6623060 | 0.6481267 | 1429 | tags=47%, list=9%, signal=43% | SSTR2||HTR7||HTR6||CHRM3||PTH1R||CHRM4||NPY1R | -0.0427509 | 0.6950441 | 0.4962252 | 0.436894 | -0.06407109 | 0.3113214 | 0.6113996 | 0.2815151 | -0.1605464 | -0.07810152 | 0.09817248 | 0.674091 | 0.7790598 | 0.6277921 | 0.6332694 | 0.5465059 | 0.2868032 | 0.340557 | 0.5472601 | 0.4152256 | -0.05919687 | -0.2247892 | 0.07610032 | -0.008744817 | 0.1360125 | 0.7046157 | 0.6187293 | 0.6094401 | 0.6059994 | 0.3675001 | 0.2822581 | 0.1892867 |
| GO:0046718 | viral entry into host cell | 95 | -0.4152833 | -1.450932 | 0.0287838353 | 0.6623060 | 0.6481267 | 4098 | tags=44%, list=26%, signal=33% | ITGB1||DPP4||TRIM62||TPCN1||ITCH||HSPA1B||SCARB2||EPHA2||TRIM8||ITGAV||VAMP8||NCAM1||PVR||LAMP1||EFNB2||CD55||EFNB3||LY6E||SLC1A5||EGFR||TRIM5||DAG1||NECTIN1||GSN||NECTIN2||CTSB||CD81||P4HB||CTSL||PTX3||BSG||GAS6||HYAL2||SIVA1||ITGB5||SCARB1||SLC52A2||NRP1||LGALS1||SMPD1||CXCR4||CLDN1 | 7.0686136 | 6.8289667 | 6.9492108 | 6.645736 | 7.17196594 | 7.0553051 | 6.7051495 | 6.6116806 | 7.0297407 | 6.99180125 | 7.17755216 | 6.913888 | 6.8154603 | 6.7529728 | 6.7605783 | 7.0524035 | 7.0174209 | 6.691584 | 6.6135588 | 6.6308963 | 7.20369797 | 7.0718419 | 7.23520459 | 7.081992224 | 7.0306440 | 7.0528190 | 6.7266912 | 6.6969691 | 6.6915393 | 6.6057724 | 6.6216253 | 6.6075920 |
| GO:0003333 | amino acid transmembrane transport | 59 | -0.4615121 | -1.502508 | 0.0287893232 | 0.6623060 | 0.6481267 | 4536 | tags=58%, list=28%, signal=41% | SLC6A15||SLC47A1||SLC6A9||ITGB1||SLC36A4||SLC38A7||ARL6IP5||SFXN2||SLC38A3||MFSD12||PSEN1||SLC25A29||SLC7A1||PER2||SLC1A5||LRRC8A||SLC7A5||SLC16A2||SLC15A4||SLC43A1||SLC38A10||SLC38A6||PRAF2||SLC1A4||SLC66A1||SLC16A10||LRRC8D||LRRC8E||SLC6A13||SLC1A1||SLC43A2||SLC7A3||SLC22A4||CLTRN | 4.2733121 | 5.1909673 | 4.7699545 | 5.306058 | 4.31838756 | 4.7055355 | 5.1045240 | 5.2739666 | 4.1939836 | 4.27818717 | 4.34386226 | 5.198745 | 5.2448933 | 5.1268168 | 4.7718641 | 4.8373217 | 4.6972787 | 5.278446 | 5.3313595 | 5.3078815 | 4.31843176 | 4.2337476 | 4.39829399 | 4.559308354 | 4.6213922 | 4.9109843 | 5.1187137 | 5.1100349 | 5.0846062 | 5.2631374 | 5.3203539 | 5.2371481 |
| GO:0042133 | neurotransmitter metabolic process | 19 | -0.6239060 | -1.612417 | 0.0294753390 | 0.6623060 | 0.6481267 | 2942 | tags=47%, list=18%, signal=39% | BCHE||PRIMA1||GCHFR||COMT||GAD1||DAGLB||DAGLA||ACHE||MAOA | 3.9042647 | 4.1513782 | 4.0974492 | 4.117056 | 3.94840460 | 4.0785759 | 4.0727806 | 4.0892781 | 3.8627310 | 3.90795986 | 3.94103386 | 4.185203 | 4.1841771 | 4.0823538 | 4.0992295 | 4.0952394 | 4.0978759 | 4.134207 | 4.0934372 | 4.1232162 | 3.97015657 | 3.8860419 | 3.98700098 | 3.984715078 | 4.0717668 | 4.1730777 | 4.0829943 | 4.0888135 | 4.0461642 | 4.1018599 | 4.1115884 | 4.0537230 |
| GO:0034975 | protein folding in endoplasmic reticulum | 11 | -0.7069164 | -1.612413 | 0.0299743640 | 0.6623060 | 0.6481267 | 3474 | tags=64%, list=22%, signal=50% | ERO1A||CANX||PDIA3||HSP90B1||HSPA5||CALR||P4HB | 7.3188963 | 9.0322499 | 7.5300905 | 8.451728 | 7.24825621 | 7.4353310 | 8.8272285 | 8.3881460 | 7.3201263 | 7.43575739 | 7.19038376 | 9.045713 | 9.0852661 | 8.9630898 | 7.6353246 | 7.4918481 | 7.4568346 | 8.379976 | 8.5126335 | 8.4594948 | 7.28727133 | 7.2723928 | 7.18291136 | 7.486929424 | 7.3296448 | 7.4838806 | 8.8097833 | 8.8487536 | 8.8228756 | 8.3826232 | 8.4392722 | 8.3408486 |
| GO:0001774 | microglial cell activation | 21 | -0.6059978 | -1.602736 | 0.0300632911 | 0.6623060 | 0.6481267 | 3058 | tags=43%, list=19%, signal=35% | SYT11||IFNGR1||APP||CTSC||CX3CR1||GRN||CLU||ITGB2||C5AR1 | 4.0996795 | 5.2575513 | 4.6775765 | 5.276360 | 4.11280775 | 4.5245520 | 5.0902895 | 5.1770647 | 4.0401652 | 4.16934921 | 4.08654397 | 5.295117 | 5.3021754 | 5.1716646 | 4.7258147 | 4.6882798 | 4.6165051 | 5.295656 | 5.2813636 | 5.2517111 | 4.10088738 | 4.0668759 | 4.16878709 | 4.373929650 | 4.3640097 | 4.7929258 | 5.1301867 | 5.0832154 | 5.0564998 | 5.1899598 | 5.2265231 | 5.1123667 |
| GO:0071426 | ribonucleoprotein complex export from nucleus | 71 | 0.4280382 | 1.469877 | 0.0300751880 | 0.6623060 | 0.6481267 | 5745 | tags=56%, list=36%, signal=36% | ZC3H11B||THOC1||AGFG1||SMG1||THOC2||NOP9||NUP214||SDAD1||SETD2||YTHDC1||SMG7||UPF2||TSC1||IWS1||TPR||DDX19B||CHTOP||ZC3H11A||ENY2||NXF1||PCID2||THOC7||LSG1||ABCE1||XPO1||MCM3AP||LTV1||NSUN2||SSB||FYTTD1||NUP160||NOL6||SMG5||THOC5||HNRNPA2B1||NPM1||POLDIP3||RIOK2||XPOT||NUP155 | 7.3628861 | 7.6587553 | 7.2430406 | 7.689349 | 7.31198475 | 7.3397199 | 7.6984669 | 7.7383890 | 7.3839947 | 7.35812532 | 7.34627992 | 7.697557 | 7.6239016 | 7.6538552 | 7.3798313 | 7.1656566 | 7.1732384 | 7.690292 | 7.6419843 | 7.7342929 | 7.32495905 | 7.2641616 | 7.34559655 | 7.338636041 | 7.4358387 | 7.2379004 | 7.6806250 | 7.6902833 | 7.7241298 | 7.7369526 | 7.7336678 | 7.7445251 |
| GO:0019233 | sensory perception of pain | 60 | -0.4573841 | -1.492549 | 0.0301757066 | 0.6623060 | 0.6481267 | 2321 | tags=32%, list=15%, signal=27% | LXN||CXCL12||PTAFR||ADRA2C||EPHB1||COMT||MMP24||TAFA4||RETREG1||P2RX4||TRPA1||GRIN2D||CXCR4||TMEM120A||ZFHX2||EDNRB||PTGS2||KCND2||CHRNA4 | 4.2746799 | 4.2167968 | 4.4111106 | 4.131962 | 4.21230201 | 4.2813411 | 4.1944082 | 4.1220055 | 4.3030449 | 4.36342335 | 4.14920274 | 4.235060 | 4.2212377 | 4.1937868 | 4.4235122 | 4.3981293 | 4.4115786 | 4.168826 | 4.0841045 | 4.1416631 | 4.22828382 | 4.2480949 | 4.15901874 | 4.218262535 | 4.1706778 | 4.4404775 | 4.2339699 | 4.1817925 | 4.1665933 | 4.1209378 | 4.1485840 | 4.0960152 |
| GO:0002717 | positive regulation of natural killer cell mediated immunity | 13 | -0.6825937 | -1.622516 | 0.0301834681 | 0.6623060 | 0.6481267 | 3102 | tags=62%, list=19%, signal=50% | PVR||LAMP1||RASGRP1||HLA-E||NECTIN2||CADM1||CD226||RAET1G | 3.8445334 | 4.5938712 | 4.2693790 | 4.491291 | 3.84261295 | 4.1691330 | 4.4744279 | 4.4469017 | 3.8091836 | 3.85465490 | 3.86908634 | 4.576051 | 4.6536845 | 4.5498464 | 4.2850338 | 4.3125103 | 4.2085936 | 4.480356 | 4.5185822 | 4.4745373 | 3.88815017 | 3.7884851 | 3.84945824 | 4.035987990 | 4.0233827 | 4.4131057 | 4.5072573 | 4.4744207 | 4.4408413 | 4.4501312 | 4.4755747 | 4.4143439 |
| GO:0051384 | response to glucocorticoid | 96 | -0.4114520 | -1.440605 | 0.0303493033 | 0.6623060 | 0.6481267 | 2592 | tags=31%, list=16%, signal=26% | SSTR2||MDK||ABCA3||CDKN1A||GPR83||BMP6||EGFR||PTAFR||GBA||IGFBP2||ASS1||CDO1||PDX1||PCK2||IGFBP7||FOS||GLB1||FLT3||ALPL||SDC1||ANXA3||SPARC||SERPINF1||WNT7B||SLIT3||GJB2||CLDN1||PTGS2||HSD11B2||AANAT | 7.2928642 | 6.7069807 | 7.2459552 | 6.884987 | 7.38942406 | 7.4231499 | 6.7615876 | 6.8198336 | 7.2725068 | 7.21632797 | 7.38463822 | 6.527666 | 6.7726670 | 6.8049891 | 7.1902345 | 7.3277628 | 7.2161360 | 6.818782 | 6.9729748 | 6.8587316 | 7.44173825 | 7.2773275 | 7.44299504 | 7.364470356 | 7.3699151 | 7.5289413 | 6.7322540 | 6.7505079 | 6.8011157 | 6.8788010 | 6.7529112 | 6.8250291 |
| GO:2001251 | negative regulation of chromosome organization | 82 | 0.4103181 | 1.444322 | 0.0305020123 | 0.6623060 | 0.6481267 | 4488 | tags=45%, list=28%, signal=33% | ATM||CENPF||ATRX||SLX1B||MNAT1||XRN1||CTC1||APC||PIF1||ERCC4||GEN1||SPDL1||CDK5RAP2||NDC80||POT1||TNKS||TTK||EXOSC10||ESPL1||NBN||TPR||GNL3L||ZWILCH||WAPL||BUB1||TOP2A||XRCC3||AURKB||TNKS2||KNTC1||NAT10||PCID2||PTTG1||MAD2L1||FBXO5||MAD1L1||TENT4B | 6.2645072 | 6.9705230 | 6.4066889 | 6.947090 | 6.18781965 | 6.3167326 | 7.0164319 | 7.0004542 | 6.2569371 | 6.38609842 | 6.13997173 | 6.979374 | 6.9488860 | 6.9830654 | 6.6525312 | 6.1981867 | 6.3308340 | 6.917285 | 6.9068344 | 7.0146784 | 6.15807392 | 6.2489765 | 6.15440525 | 6.317940175 | 6.3118234 | 6.3204208 | 7.0143469 | 6.9972925 | 7.0373756 | 6.9885953 | 7.0175795 | 6.9950269 |
| GO:0030194 | positive regulation of blood coagulation | 11 | -0.7061604 | -1.610689 | 0.0305659633 | 0.6623060 | 0.6481267 | 4051 | tags=64%, list=25%, signal=48% | F12||PRDX2||DMTN||F3||ST3GAL4||ENPP4||HPSE | 6.5205040 | 5.8671651 | 6.3997208 | 5.863637 | 6.59613873 | 6.7333748 | 5.7705370 | 5.8268557 | 6.4769791 | 6.36451401 | 6.69963588 | 5.949765 | 5.8390704 | 5.8088129 | 6.1697865 | 6.5402500 | 6.4632469 | 5.937193 | 5.7827172 | 5.8668610 | 6.59062299 | 6.4765257 | 6.71167995 | 6.691681830 | 6.8511518 | 6.6493534 | 5.7963896 | 5.7737198 | 5.7409642 | 5.8251234 | 5.8390202 | 5.8163327 |
| GO:0060026 | convergent extension | 13 | -0.6815365 | -1.620003 | 0.0305780233 | 0.6623060 | 0.6481267 | 3430 | tags=69%, list=21%, signal=54% | WNT11||NPHP3||WNT5A||PTK7||NKD1||SFRP1||MESP1||LBX2||WNT5B | 6.3522413 | 6.0110009 | 6.3920837 | 6.065299 | 6.42478916 | 6.6662233 | 6.0102461 | 6.0827475 | 6.2483820 | 6.22018626 | 6.56254155 | 5.989280 | 6.0520417 | 5.9907878 | 6.2404452 | 6.4971663 | 6.4266044 | 6.151416 | 5.9873730 | 6.0523621 | 6.39644789 | 6.2840585 | 6.57845611 | 6.597279297 | 6.7602547 | 6.6360694 | 6.0711884 | 6.0003656 | 5.9568696 | 6.0756628 | 6.0986569 | 6.0737897 |
| GO:0060070 | canonical Wnt signaling pathway | 247 | -0.3341258 | -1.330414 | 0.0311270125 | 0.6623060 | 0.6481267 | 3751 | tags=31%, list=23%, signal=24% | FGFR2||LZTS2||TMEM64||ANKRD6||SIAH2||HHEX||NKD2||NOTCH1||WNT11||GPRC5B||SULF2||IGFBP4||GPC3||ZNF703||MAD2L2||FAM53B||NPHP3||CDH2||LGR5||WNT5A||TGFB1||SOX7||PSEN1||FRAT1||WNT3||PRDM15||CSNK1G2||TLE2||PTK7||NKX2-5||UBAC2||FZD7||CDH3||MDK||PORCN||EGFR||WLS||APC2||FZD6||EGR1||ROR2||APOE||FRMD8||NOG||SNAI2||GLI3||IGFBP2||TMEM9||DACT3||LRP5||ATP6AP2||FZD8||SOX9||AMFR||WNT3A||SDC1||NKD1||SHISA3||NOTUM||RSPO2||SFRP1||TMEM198||WNT7B||FZD2||FZD9||MESP1||GLI1||WNT9A||TPBG||TPBGL||WNT2B||DKK3||FZD10||TLE6||NR4A2||WNT5B||GPC5 | 5.5761686 | 5.5907674 | 5.6610524 | 5.555619 | 5.52903432 | 5.4112817 | 5.6090596 | 5.5681699 | 5.6724401 | 5.66733252 | 5.36821371 | 5.558743 | 5.6019040 | 5.6111146 | 5.6067688 | 5.6140523 | 5.7572878 | 5.575087 | 5.5426083 | 5.5489552 | 5.57404897 | 5.6986581 | 5.28356479 | 5.495070168 | 5.3400672 | 5.3943994 | 5.6067017 | 5.6196470 | 5.6007656 | 5.5663193 | 5.5652665 | 5.5729120 |
| GO:0071357 | cellular response to type I interferon | 39 | 0.4924713 | 1.513522 | 0.0313725490 | 0.6623060 | 0.6481267 | 1393 | tags=15%, list=9%, signal=14% | NLRC5||IFI27||IFIT1||ENSG00000249624||IFITM1||STAT2 | 5.7593803 | 6.1194180 | 5.8625504 | 6.212955 | 5.71082035 | 5.8681199 | 6.1630452 | 6.2437658 | 5.6916467 | 5.83404116 | 5.74888377 | 6.113918 | 6.1246735 | 6.1196424 | 6.0531868 | 5.7170446 | 5.7956252 | 6.208272 | 6.1556245 | 6.2725876 | 5.75595347 | 5.6614359 | 5.71352012 | 5.940807187 | 5.8987486 | 5.7585573 | 6.1710332 | 6.1486760 | 6.1693192 | 6.2529195 | 6.2472312 | 6.2310575 |
| GO:0048713 | regulation of oligodendrocyte differentiation | 28 | -0.5554801 | -1.569542 | 0.0317800741 | 0.6623060 | 0.6481267 | 3755 | tags=46%, list=24%, signal=36% | HES1||WDR1||NOTCH1||OLIG2||PTPRZ1||MDK||DAG1||TP73||DUSP15||CXCR4||ZNF365||DUSP10||HES5 | 5.4781731 | 6.0879921 | 5.7722656 | 6.058507 | 5.43547505 | 5.5471203 | 6.0559629 | 6.0804484 | 5.4842163 | 5.55187800 | 5.39409410 | 6.133940 | 6.0981902 | 6.0299169 | 5.8634127 | 5.6923303 | 5.7558458 | 6.064029 | 6.0444052 | 6.0669831 | 5.43826155 | 5.4706752 | 5.39653186 | 5.482071584 | 5.3544280 | 5.7725378 | 6.0673490 | 6.0602523 | 6.0401496 | 6.0523049 | 6.1160160 | 6.0722865 |
| GO:0030513 | positive regulation of BMP signaling pathway | 28 | -0.5553696 | -1.569230 | 0.0319750439 | 0.6623060 | 0.6481267 | 3755 | tags=50%, list=24%, signal=38% | PELO||HES1||NOTCH1||FKBP8||RNF165||GPC3||CCN1||FBXL15||SULF1||KCP||BMP4||ZNF423||BMPR2||HES5 | 4.5979587 | 4.4842683 | 4.6606882 | 4.467502 | 4.62205583 | 4.9006111 | 4.4339713 | 4.4679434 | 4.5582952 | 4.49940323 | 4.72645082 | 4.492787 | 4.5027957 | 4.4568182 | 4.5695876 | 4.7416171 | 4.6657214 | 4.501056 | 4.4276624 | 4.4728381 | 4.60967753 | 4.5309725 | 4.71929679 | 4.832568321 | 4.9622542 | 4.9040911 | 4.4773584 | 4.4249741 | 4.3984611 | 4.4569571 | 4.4985212 | 4.4478444 |
| GO:1905606 | regulation of presynapse assembly | 22 | -0.5963234 | -1.591183 | 0.0326771654 | 0.6623060 | 0.6481267 | 2884 | tags=50%, list=18%, signal=41% | GPC4||CLSTN3||NLGN1||APP||IL1RAP||LRFN4||LRFN3||WNT3A||LRRC4B||PTPRD||NTNG2 | 4.6300724 | 5.2803392 | 4.7736236 | 5.239317 | 4.59404384 | 4.9199165 | 5.2200116 | 5.1791079 | 4.5881025 | 4.63298222 | 4.66802144 | 5.271151 | 5.3190678 | 5.2499266 | 4.8410129 | 4.7575065 | 4.7196664 | 5.240159 | 5.2449189 | 5.2328467 | 4.59856640 | 4.5173528 | 4.66254999 | 4.868369214 | 4.9465967 | 4.9434325 | 5.2436435 | 5.2194085 | 5.1965991 | 5.1709110 | 5.2078622 | 5.1580864 |
| GO:0007608 | sensory perception of smell | 18 | 0.6020873 | 1.562031 | 0.0327169275 | 0.6623060 | 0.6481267 | 945 | tags=33%, list=6%, signal=31% | NCAM2||NAV2||OR2L2||ENSG00000262668||OR2A5||DRD2 | 2.4834856 | 3.4740001 | 2.9521316 | 3.466261 | 2.37665758 | 2.5812577 | 3.5323733 | 3.4670206 | 2.6231880 | 2.54010059 | 2.26303870 | 3.422846 | 3.4821057 | 3.5155264 | 3.2178447 | 2.8381618 | 2.7573876 | 3.329112 | 3.4758553 | 3.5826431 | 2.31411090 | 2.3700704 | 2.44289579 | 2.349353617 | 2.2368918 | 3.0257504 | 3.5977700 | 3.5885276 | 3.4025394 | 3.4222059 | 3.5878554 | 3.3826897 |
| GO:0042098 | T cell proliferation | 109 | -0.3969564 | -1.419663 | 0.0330082521 | 0.6623060 | 0.6481267 | 5267 | tags=52%, list=33%, signal=35% | CTNNB1||PNP||TFRC||SCRIB||IHH||TRAF6||IL27RA||GPAM||PSMB10||CD46||RPS3||BAX||CD1D||TNFSF9||RPS6||SOS2||CCND3||ZP3||CASP3||SYK||MAPK8IP1||PRDX2||ELF4||IGF2||HES1||NCSTN||FADD||PRKCQ||ITCH||ERBB2||CD276||CORO1A||PRNP||NCK2||ICOSLG||SLC7A1||CD55||ZBTB7B||RASGRP1||HLA-DMB||HLA-E||NDFIP1||IRF1||IL6ST||IGFBP2||BMP4||CD81||CD151||EFNB1||JAK3||HLA-A||DOCK8||CD24||IL23A||CCDC88B||IL4I1||LRRC32 | 6.7700756 | 7.0043161 | 6.6032426 | 6.935032 | 6.74715447 | 6.6069321 | 6.9342553 | 6.9594802 | 6.7900636 | 6.77239680 | 6.74744880 | 7.116508 | 6.9642306 | 6.9250421 | 6.5735618 | 6.5570726 | 6.6761839 | 6.961001 | 6.8979007 | 6.9454473 | 6.73446207 | 6.7605355 | 6.74634769 | 6.710391231 | 6.6426710 | 6.4558454 | 6.9249790 | 6.9369440 | 6.9407958 | 6.9285342 | 6.9681116 | 6.9812741 |
| GO:0045668 | negative regulation of osteoblast differentiation | 37 | -0.5128725 | -1.530259 | 0.0330932451 | 0.6623060 | 0.6481267 | 4876 | tags=54%, list=31%, signal=38% | BAMBI||NOCT||TWIST1||IGFBP5||TRPM4||TMEM64||CITED1||NOTCH1||MEN1||TWIST2||HDAC7||NOG||PTCH1||LRP5||SOX9||ID3||VEGFC||SFRP1||HAND2||CHRD | 3.8815774 | 4.0516006 | 3.8236242 | 3.747163 | 3.98937755 | 4.1607994 | 3.9467058 | 3.6719042 | 3.7687789 | 3.81168696 | 4.04828922 | 4.060969 | 4.0849398 | 4.0078168 | 3.7323548 | 3.9182401 | 3.8142486 | 3.770490 | 3.7232373 | 3.7473743 | 4.00767608 | 3.7923403 | 4.14628603 | 4.105152021 | 4.2223574 | 4.1524729 | 3.9824928 | 3.9271161 | 3.9298301 | 3.6808324 | 3.6855653 | 3.6490422 |
| GO:0060412 | ventricular septum morphogenesis | 37 | -0.5126114 | -1.529480 | 0.0332879112 | 0.6623060 | 0.6481267 | 3768 | tags=46%, list=24%, signal=35% | HEY1||HES1||FGFR2||ROBO2||TGFBR1||NOTCH1||WNT11||WNT5A||NKX2-5||ZFPM1||NOG||BMP4||BMPR2||FGFRL1||FZD2||SLIT3||TGFB2 | 3.1045044 | 4.1756563 | 3.4059912 | 4.245505 | 3.11549677 | 3.4650912 | 4.1720351 | 4.2212627 | 3.0670447 | 3.16679490 | 3.07757753 | 4.135929 | 4.2175449 | 4.1723352 | 3.6812390 | 3.2602370 | 3.2311928 | 4.177863 | 4.2385036 | 4.3167797 | 3.08424419 | 3.0526542 | 3.20506777 | 3.413661128 | 3.4149859 | 3.5615621 | 4.1615327 | 4.1892200 | 4.1651957 | 4.1821104 | 4.2408608 | 4.2400341 |
| GO:0098926 | postsynaptic signal transduction | 27 | -0.5608532 | -1.572504 | 0.0336838006 | 0.6623060 | 0.6481267 | 1566 | tags=30%, list=10%, signal=27% | LY6E||WNT3A||CHRM3||ACHE||PRR7||LYPD1||LY6G6D||CHRM4 | 5.1567918 | 5.3728433 | 5.2842300 | 5.293187 | 5.13095611 | 5.3550724 | 5.3770802 | 5.3083778 | 5.1343097 | 5.19647460 | 5.13875362 | 5.344320 | 5.4151836 | 5.3580431 | 5.3154398 | 5.2862408 | 5.2502715 | 5.328627 | 5.2357779 | 5.3134469 | 5.14775015 | 5.0796579 | 5.16407777 | 5.302883618 | 5.3474834 | 5.4127299 | 5.3720153 | 5.3715823 | 5.3875853 | 5.2893159 | 5.3417821 | 5.2934432 |
| GO:0060149 | negative regulation of posttranscriptional gene silencing | 19 | -0.6151146 | -1.589697 | 0.0337983887 | 0.6623060 | 0.6481267 | 6 | tags=16%, list=0%, signal=16% | ZMPSTE24||NCOR2||RMRP | 4.3755842 | 5.1557827 | 4.6035704 | 5.056346 | 4.44250000 | 4.7643694 | 4.9928984 | 6.3687733 | 4.2534398 | 4.54144705 | 4.31570168 | 5.293327 | 5.1069458 | 5.0561232 | 4.6604792 | 4.6060170 | 4.5417707 | 5.129940 | 4.9585857 | 5.0752411 | 4.59380505 | 4.2710691 | 4.44461172 | 4.871139073 | 4.6061725 | 4.8028633 | 4.9614295 | 5.0300311 | 4.9863972 | 5.0250776 | 7.5058104 | 5.0708142 |
| GO:0060967 | negative regulation of gene silencing by RNA | 19 | -0.6151146 | -1.589697 | 0.0337983887 | 0.6623060 | 0.6481267 | 6 | tags=16%, list=0%, signal=16% | ZMPSTE24||NCOR2||RMRP | 4.3755842 | 5.1557827 | 4.6035704 | 5.056346 | 4.44250000 | 4.7643694 | 4.9928984 | 6.3687733 | 4.2534398 | 4.54144705 | 4.31570168 | 5.293327 | 5.1069458 | 5.0561232 | 4.6604792 | 4.6060170 | 4.5417707 | 5.129940 | 4.9585857 | 5.0752411 | 4.59380505 | 4.2710691 | 4.44461172 | 4.871139073 | 4.6061725 | 4.8028633 | 4.9614295 | 5.0300311 | 4.9863972 | 5.0250776 | 7.5058104 | 5.0708142 |
| GO:0016579 | protein deubiquitination | 110 | 0.3753909 | 1.384822 | 0.0339525945 | 0.6623060 | 0.6481267 | 4971 | tags=49%, list=31%, signal=34% | PRKN||MYSM1||USP3||USP28||USP13||USP16||USP34||TANK||OTUD3||YOD1||MINDY2||USP49||USP54||USP32||USP24||UCHL5||ATXN7L3||TRRAP||OTUD6B||MINDY4||USP45||ATXN7||USP1||USP4||USP53||VCPIP1||USP31||USP42||SART3||ENY2||OTULIN||USP12||OTUD4||TRIM21||OTUD1||CYLD||BRCC3||WDR48||OTUD7A||ZRANB1||USP7||USP15||USP40||USP46||UIMC1||USP19||DESI2||CDK1||USP47||USP8||ABRAXAS1||USP36||BAP1||USP21 | 5.1189767 | 5.5050715 | 5.2361645 | 5.536566 | 5.05161887 | 5.1983843 | 5.5452514 | 5.5822083 | 5.1246338 | 5.16988747 | 5.06031219 | 5.503704 | 5.5113640 | 5.5001240 | 5.3246420 | 5.1708797 | 5.2084836 | 5.521523 | 5.4925265 | 5.5937561 | 5.05010925 | 5.0608196 | 5.04387685 | 5.169900661 | 5.2005095 | 5.2242290 | 5.5469129 | 5.5371882 | 5.5516158 | 5.5711473 | 5.5944099 | 5.5809730 |
| GO:0071402 | cellular response to lipoprotein particle stimulus | 22 | -0.5943120 | -1.585816 | 0.0340551181 | 0.6623060 | 0.6481267 | 2139 | tags=36%, list=13%, signal=32% | APOE||CD81||CD9||MYD88||ABCG1||ITGB2||TICAM1||SMPD3 | 5.1858776 | 5.3824031 | 5.3567219 | 5.450911 | 5.24771857 | 5.5596287 | 5.2573488 | 5.3775466 | 5.0951533 | 5.08008911 | 5.36430791 | 5.410046 | 5.4411591 | 5.2917342 | 5.1955373 | 5.4847664 | 5.3752110 | 5.461879 | 5.4897835 | 5.3995997 | 5.26138533 | 5.0764700 | 5.38837701 | 5.461401512 | 5.5986141 | 5.6140308 | 5.2808515 | 5.2599972 | 5.2307593 | 5.3801444 | 5.4320017 | 5.3182485 |
| GO:0007271 | synaptic transmission, cholinergic | 15 | -0.6488470 | -1.584573 | 0.0340613104 | 0.6623060 | 0.6481267 | 2598 | tags=47%, list=16%, signal=39% | NQO1||LYNX1||CHRNB1||APOE||ACHE||CHRNB4||CHRNA4 | 1.5309568 | 2.4978021 | 2.0731546 | 2.418289 | 1.41188351 | 1.6088476 | 2.4325826 | 2.3220121 | 1.5780093 | 1.62675103 | 1.37605651 | 2.502713 | 2.5050136 | 2.4856022 | 2.2882895 | 2.0802839 | 1.8116782 | 2.288100 | 2.5141349 | 2.4434535 | 1.37606572 | 1.4529352 | 1.40560532 | 1.303109271 | 1.3629089 | 2.0393524 | 2.4087421 | 2.4894540 | 2.3978040 | 2.3031594 | 2.3576713 | 2.3045331 |
| GO:0022011 | myelination in peripheral nervous system | 24 | 0.5542194 | 1.535655 | 0.0341303904 | 0.6623060 | 0.6481267 | 4224 | tags=54%, list=26%, signal=40% | PRX||ARHGEF10||CNTNAP1||FA2H||NF1||POU3F2||ITGB4||ADGRG6||DICER1||NDRG1||NTRK2||PALS1||PPP3R1 | 4.8760921 | 5.0822527 | 5.1871031 | 5.183261 | 4.86121189 | 5.0944156 | 5.1205159 | 5.1924611 | 4.8542685 | 4.95945587 | 4.81046176 | 5.073601 | 5.1007739 | 5.0722026 | 5.2425567 | 5.1615887 | 5.1555168 | 5.206113 | 5.1578994 | 5.1853663 | 4.85340982 | 4.8709733 | 4.85919696 | 5.026694139 | 4.9935295 | 5.2493826 | 5.1431270 | 5.0978068 | 5.1202580 | 5.1930739 | 5.1980675 | 5.1862173 |
| GO:0000070 | mitotic sister chromatid segregation | 157 | 0.3498083 | 1.358219 | 0.0346095609 | 0.6623060 | 0.6481267 | 5700 | tags=50%, list=36%, signal=32% | ATM||REC8||CENPF||ATRX||SLF1||CENPE||KIF14||APC||POGZ||KIF4B||GEN1||SPAG5||SPDL1||SGO1||CDK5RAP2||NDC80||SMC5||PRC1||TNKS||TTK||ESPL1||TPR||NIPBL||ZWILCH||NCAPG||KMT5A||SMC2||HECW2||KIF18A||NCAPD2||KIF23||SGO2||PIBF1||SMC4||BUB1||RMDN1||ANAPC1||PDS5A||XRCC3||AURKB||CDC23||CENPC||CHMP1B||DLGAP5||KNTC1||KIF2C||PCID2||PTTG1||MAD2L1||AKAP8||BECN1||FBXO5||SMC3||MAD1L1||CDC27||NCAPG2||BUB1B||CDC6||TEX14||PSRC1||KLHL22||CUL3||ANAPC5||DIS3L2||INO80||ANAPC7||INCENP||HASPIN||KPNB1||TACC3||RIOK2||MIS12||ANAPC4||SMC1A||CDC16||NCAPD3||PLK1||NSL1 | 5.7574080 | 6.2754398 | 5.8780192 | 6.293049 | 5.68537675 | 5.8197173 | 6.3229981 | 6.3828077 | 5.7371243 | 5.85100253 | 5.67874647 | 6.301883 | 6.2550296 | 6.2690053 | 6.0410739 | 5.7489299 | 5.8280190 | 6.269935 | 6.2519471 | 6.3551460 | 5.67718185 | 5.6956605 | 5.68322636 | 5.811174331 | 5.8273803 | 5.8205515 | 6.3269228 | 6.3171065 | 6.3249463 | 6.3698321 | 6.4030036 | 6.3753681 |
| GO:0034446 | substrate adhesion-dependent cell spreading | 89 | -0.4160993 | -1.439903 | 0.0346197503 | 0.6623060 | 0.6481267 | 4070 | tags=38%, list=25%, signal=29% | PARVB||MERTK||PARVA||ATRN||TRIOBP||LAMB1||DMTN||CSPG5||FN1||ITGAV||RAC3||ANTXR1||MELTF||LAMA5||LAMB2||FZD7||TESK1||MDK||MYADM||LIMS2||CALR||P4HB||HAS2||CASS4||EFNA5||FBLN1||EPHB3||NRP1||BVES||ITGA4||SRCIN1||NTNG2||NTNG1||TMEFF2 | 5.8240095 | 6.2041731 | 5.9465479 | 5.949301 | 5.77550525 | 5.8365947 | 6.1125575 | 5.9292750 | 5.8227555 | 5.91152825 | 5.73217330 | 6.237106 | 6.2185496 | 6.1556049 | 5.9253519 | 5.9374709 | 5.9763278 | 5.955521 | 5.9261356 | 5.9659525 | 5.77508134 | 5.8042510 | 5.74660759 | 5.805921821 | 5.7695151 | 5.9294353 | 6.1232635 | 6.1025125 | 6.1118218 | 5.9199640 | 5.9476263 | 5.9200581 |
| GO:0042417 | dopamine metabolic process | 23 | -0.5860134 | -1.578304 | 0.0349088056 | 0.6652905 | 0.6510473 | 2030 | tags=43%, list=13%, signal=38% | PNKD||COMT||GPR37||SLC1A1||MAOA||NR4A2||ITGB2||SNCAIP||SULT1A4||TGFB2 | 4.7672307 | 5.1009378 | 4.9906296 | 5.108447 | 4.64003374 | 4.4700452 | 5.0699790 | 5.0798953 | 4.8760149 | 4.94153509 | 4.43309788 | 5.123636 | 5.0943826 | 5.0845077 | 5.0620047 | 4.8130621 | 5.0816048 | 5.145279 | 5.0544383 | 5.1240648 | 4.50787664 | 4.8765526 | 4.50300070 | 4.412962642 | 4.4275949 | 4.5646728 | 5.0874948 | 5.0521281 | 5.0700974 | 5.0771459 | 5.0848825 | 5.0776444 |
| GO:0006878 | cellular copper ion homeostasis | 14 | -0.6596618 | -1.590755 | 0.0360270701 | 0.6652905 | 0.6510473 | 4139 | tags=71%, list=26%, signal=53% | ABCB6||ARF1||PRNP||SLC31A2||ATP7B||MT2A||COX19||APP||CCDC22||SCO2 | 6.5725534 | 6.5594308 | 6.5746066 | 6.441626 | 6.64854623 | 6.8011972 | 6.4312044 | 6.4275259 | 6.4993298 | 6.59325209 | 6.62223654 | 6.596160 | 6.5791077 | 6.5012613 | 6.4836426 | 6.6586790 | 6.5761911 | 6.503616 | 6.3819437 | 6.4367428 | 6.68316296 | 6.5242350 | 6.73025860 | 6.828951108 | 6.8119999 | 6.7617982 | 6.4624208 | 6.4214896 | 6.4091624 | 6.4251732 | 6.4561402 | 6.4007294 |
| GO:0003018 | vascular process in circulatory system | 172 | -0.3540585 | -1.350093 | 0.0365650970 | 0.6652905 | 0.6510473 | 3297 | tags=35%, list=21%, signal=28% | PLEC||SLC2A10||SLC4A3||SCPEP1||PTP4A3||TGFB1||SLC5A5||SLC8A1||SLC7A1||KCNMB4||PER2||SLC4A4||SLC1A5||SLC22A5||SLC29A1||SLC7A5||SLC16A2||BMP6||HMGCR||TRPV4||EGFR||COMP||ACTA2||SLC2A1||EXT2||APOE||SNTA1||ADRA2C||SLC6A17||LRP3||KCNJ8||FGFBP3||SLC1A4||KCNMA1||BMPR2||F2RL1||HTR7||ABCG2||ATP2A3||GPX1||SLC19A1||CHRM3||SLC6A13||SLC1A1||LRP1||MYLK3||SLC2A3||ABCB1||SLC7A3||UTS2R||CEACAM1||RAMP2||ADRB2||EDNRB||KLF2||ACE||PTGS2||SLC8A2||SLC16A7||PDE2A | 4.4056228 | 4.6577292 | 4.5309275 | 4.741127 | 4.44536480 | 4.6190672 | 4.6028899 | 4.7111847 | 4.3725265 | 4.36680099 | 4.47495363 | 4.658359 | 4.6860007 | 4.6282499 | 4.5204186 | 4.5640777 | 4.5076780 | 4.734592 | 4.7342089 | 4.7544882 | 4.43050758 | 4.3661985 | 4.53437594 | 4.551772340 | 4.6547107 | 4.6484284 | 4.6278366 | 4.5907607 | 4.5897448 | 4.7089375 | 4.7426411 | 4.6813213 |
| GO:0006953 | acute-phase response | 15 | -0.6459233 | -1.577433 | 0.0368663594 | 0.6652905 | 0.6510473 | 3331 | tags=53%, list=21%, signal=42% | FN1||TNFRSF11A||SIGIRR||ASS1||IL6R||F8||EDNRB||PTGS2 | 3.6074237 | 3.7707218 | 3.5097229 | 3.121979 | 3.64425090 | 3.6863988 | 3.6325320 | 3.0919219 | 3.6131825 | 3.47993128 | 3.71922845 | 3.858797 | 3.7482746 | 3.7004939 | 3.4214814 | 3.6047864 | 3.4969984 | 3.221391 | 3.0599750 | 3.0791209 | 3.63413344 | 3.5069265 | 3.77884589 | 3.642876700 | 3.7553912 | 3.6583309 | 3.6536370 | 3.6127995 | 3.6308689 | 3.0198052 | 3.1500598 | 3.1028986 |
| GO:0002446 | neutrophil mediated immunity | 17 | -0.6251709 | -1.580806 | 0.0368916797 | 0.6652905 | 0.6510473 | 2185 | tags=47%, list=14%, signal=41% | PTAFR||JAGN1||IRAK4||F2RL1||MYD88||ANXA3||ACE||ITGB2 | 4.3197641 | 4.5403450 | 4.4124146 | 4.441767 | 4.32405423 | 4.4028785 | 4.4797486 | 4.4395719 | 4.2826737 | 4.32454539 | 4.35124667 | 4.556003 | 4.5467038 | 4.5180575 | 4.3857466 | 4.4722919 | 4.3772759 | 4.438103 | 4.4108714 | 4.4755929 | 4.34056951 | 4.2311557 | 4.39561049 | 4.328094816 | 4.3362062 | 4.5347020 | 4.4890698 | 4.4932178 | 4.4566816 | 4.4041246 | 4.5127973 | 4.3989037 |
| GO:0046466 | membrane lipid catabolic process | 29 | -0.5428475 | -1.546471 | 0.0368932039 | 0.6652905 | 0.6510473 | 3254 | tags=41%, list=20%, signal=33% | HEXB||FUCA1||PPT1||MGST2||GLA||GBA||HEXA||SMPDL3B||NEU1||ENPP2||SMPD1||SMPD3 | 4.7939503 | 5.2241949 | 5.2892610 | 5.276842 | 4.75973708 | 4.9834834 | 5.1109561 | 5.1793045 | 4.7937682 | 4.88363186 | 4.69851669 | 5.285130 | 5.2402519 | 5.1435979 | 5.3397540 | 5.2663908 | 5.2602747 | 5.252936 | 5.3022394 | 5.2749289 | 4.78322063 | 4.7739174 | 4.72130367 | 4.817435623 | 4.7682185 | 5.3023278 | 5.1535648 | 5.0937776 | 5.0845482 | 5.1679728 | 5.2243986 | 5.1443659 |
| GO:0003341 | cilium movement | 95 | 0.3837186 | 1.388277 | 0.0370058873 | 0.6652905 | 0.6510473 | 2080 | tags=24%, list=13%, signal=21% | ZBBX||CFAP44||CFAP251||DNHD1||CFAP206||CFAP54||CFAP53||SPEF1||ASH1L||TCTE1||DNAAF4||FSIP2||SPEF2||TTLL3||DNAI3||TMF1||SLC9B1||DNAI4||CCDC65||ARMC2||CFAP221||ING2||DNAAF1 | 3.7573463 | 4.1716275 | 3.9797151 | 4.244442 | 3.59493879 | 3.5980343 | 4.2139220 | 4.2091507 | 3.8575809 | 3.85352844 | 3.53826334 | 4.171484 | 4.1673589 | 4.1760264 | 4.2217015 | 3.8179424 | 3.8648273 | 4.210229 | 4.2463848 | 4.2759612 | 3.61803851 | 3.6730322 | 3.48750471 | 3.489176196 | 3.4609172 | 3.8163119 | 4.2293306 | 4.2018903 | 4.2104083 | 4.2054702 | 4.2333318 | 4.1882915 |
| GO:0044827 | modulation by host of viral genome replication | 16 | 0.6168479 | 1.555337 | 0.0372168285 | 0.6652905 | 0.6510473 | 335 | tags=12%, list=2%, signal=12% | IFI27||PRKN | 7.2609098 | 8.0642357 | 7.1441591 | 7.841939 | 7.09475721 | 6.8350846 | 8.0318627 | 7.8603484 | 7.3499693 | 7.46105142 | 6.91628420 | 8.143565 | 8.0315054 | 8.0141886 | 7.2893456 | 6.9209906 | 7.1971932 | 7.795105 | 7.8290030 | 7.8997256 | 7.05989830 | 7.2920577 | 6.90593522 | 6.915693463 | 6.7110006 | 6.8706372 | 8.0319030 | 8.0348328 | 8.0288460 | 7.8408311 | 7.8755966 | 7.8643994 |
| GO:0045428 | regulation of nitric oxide biosynthetic process | 35 | -0.5116250 | -1.510722 | 0.0377873004 | 0.6652905 | 0.6510473 | 1798 | tags=29%, list=11%, signal=25% | ASS1||CX3CR1||PTX3||SIRPA||P2RX4||PTGIS||CLU||KLF2||PTGS2||TICAM1 | 6.2936987 | 6.9673854 | 6.3739123 | 7.028985 | 6.19822026 | 6.2888383 | 7.0232875 | 7.0530504 | 6.2715271 | 6.43946625 | 6.15594232 | 6.955594 | 6.9625556 | 6.9838565 | 6.6017100 | 6.1867407 | 6.3008652 | 7.006330 | 6.9935976 | 7.0853044 | 6.22900069 | 6.2181131 | 6.14615006 | 6.351144587 | 6.3204439 | 6.1898962 | 7.0203037 | 7.0169354 | 7.0325762 | 7.0611167 | 7.0419017 | 7.0560641 |
| GO:0006693 | prostaglandin metabolic process | 28 | -0.5455078 | -1.541365 | 0.0382140768 | 0.6652905 | 0.6510473 | 3258 | tags=39%, list=20%, signal=31% | ZADH2||TNFRSF1A||CBR1||PRXL2B||MIF||DAGLB||HPGD||PTGIS||PTGS2||ENSG00000258653||CYP2S1 | 8.3011381 | 7.5504127 | 7.9085027 | 7.424269 | 8.38308615 | 8.4314739 | 7.4183754 | 7.3408796 | 8.2689096 | 8.08987235 | 8.51316262 | 7.690649 | 7.4672563 | 7.4823596 | 7.6244414 | 8.0825186 | 7.9796196 | 7.585676 | 7.3142422 | 7.3579209 | 8.37948937 | 8.1828062 | 8.56209820 | 8.361287367 | 8.6736184 | 8.2218053 | 7.4156442 | 7.4136399 | 7.4258127 | 7.3543689 | 7.3395067 | 7.3286475 |
| GO:0000038 | very long-chain fatty acid metabolic process | 25 | -0.5614236 | -1.544914 | 0.0383561644 | 0.6652905 | 0.6510473 | 4586 | tags=60%, list=29%, signal=43% | ACOT2||ELOVL1||SLC27A4||ELOVL6||HACD3||HACD1||ELOVL5||SLC27A2||HSD17B4||ELOVL7||SLC27A5||TECR||ELOVL4||ACSL6||ABCD1 | 4.9040139 | 5.4446407 | 5.1704286 | 5.512032 | 4.94149510 | 5.1838092 | 5.3346884 | 5.4302930 | 4.8571799 | 4.86261493 | 4.98839032 | 5.445833 | 5.4897701 | 5.3968214 | 5.1756079 | 5.2154136 | 5.1186275 | 5.493063 | 5.5281854 | 5.5146318 | 4.91043223 | 4.8779921 | 5.03148566 | 5.011065030 | 5.1717666 | 5.3488001 | 5.3503215 | 5.3501220 | 5.3031108 | 5.4444312 | 5.4730502 | 5.3715065 |
| GO:0030282 | bone mineralization | 81 | -0.4205174 | -1.439480 | 0.0386355070 | 0.6652905 | 0.6510473 | 3202 | tags=35%, list=20%, signal=28% | GPC3||GPM6B||TGFB1||CCN1||SLC8A1||MINPP1||FBXL15||BMP6||ATRAID||ZMPSTE24||COMP||ROR2||BMP4||SOX9||BMPR2||PHOSPHO1||LOX||DDR2||NOTUM||RSPO2||FZD9||NELL1||ADRB2||PTH1R||PTGS2||S1PR1||SMPD3||PTHLH | 4.2157441 | 4.4259289 | 4.4147723 | 4.659623 | 4.17985686 | 4.3752056 | 4.4270684 | 4.6982270 | 4.2259630 | 4.28953757 | 4.12710785 | 4.461919 | 4.4245400 | 4.3904410 | 4.4767387 | 4.3670255 | 4.3983278 | 4.654798 | 4.6200188 | 4.7028522 | 4.15535986 | 4.2501609 | 4.13129362 | 4.371398781 | 4.2658243 | 4.4804139 | 4.4481256 | 4.4124292 | 4.4204068 | 4.6842023 | 4.7254231 | 4.6846663 |
| GO:0007610 | behavior | 396 | -0.2994415 | -1.248675 | 0.0386661937 | 0.6652905 | 0.6510473 | 3926 | tags=29%, list=25%, signal=22% | SNCA||ATP1B2||PRRT1||ATXN1||PCM1||THRB||FOXO6||PRKAR1B||NCSTN||HTRA2||NR4A3||LEPR||PLCB1||FADD||NLGN3||ARL6IP5||ALDH1A3||PPP1R9B||FOXB1||SDK1||PRNP||ADCY5||NPAS2||FGF12||ITPR3||HEXB||CDK5||PPT1||MAPK8IP2||SYT11||GRID1||BCHE||PSEN1||CNP||GDNF||SLC24A4||P2RX2||EPHA4||PTPRZ1||EFNB3||TMOD1||CLN6||TSPO||BRSK1||MDK||NRGN||ABHD12||HMGCR||NLGN1||APP||ZMPSTE24||EGFR||CHRNB1||BBS12||CXCL12||CLCN3||LGMN||EGR1||APOE||C1QL1||GBA||GLI3||OTOG||CTNS||COMT||PRKCZ||GAA||JUN||FIG4||SPTBN4||FOS||AMFR||C12orf57||GAD1||GPR37||ETV1||VLDLR||CX3CR1||SCN2A||EPHB2||MBD5||VWA1||SERPINF1||SLC12A5||SLC1A1||WFS1||P2RX4||ARC||NAGLU||NDRG4||GRIN2D||FZD9||CNTFR||PITX3||THBS4||RASD2||KCNQ1||ZFHX2||ASTN1||PAX5||TPBG||NRXN2||HAND2||PTGS2||SLC8A2||SHANK2||NR4A2||KCND2||APBA1||SLC8A3||TAFA2||GATM||CHRNA4||BHLHB9||NPB | 4.6709643 | 4.9730923 | 4.7850470 | 4.945287 | 4.67283106 | 4.8172956 | 4.9516438 | 4.9270911 | 4.6493234 | 4.68073570 | 4.68259212 | 4.982248 | 4.9864140 | 4.9503460 | 4.8284900 | 4.7570866 | 4.7685404 | 4.943457 | 4.9220904 | 4.9699168 | 4.65955132 | 4.6414978 | 4.71638133 | 4.756602864 | 4.8205575 | 4.8723980 | 4.9714848 | 4.9441733 | 4.9390623 | 4.9199707 | 4.9538890 | 4.9070065 |
| GO:0051604 | protein maturation | 216 | -0.3373472 | -1.323239 | 0.0386910143 | 0.6652905 | 0.6510473 | 4382 | tags=38%, list=27%, signal=28% | NAA10||SEC11C||PRKACA||ASPH||STOML2||TMEM98||CIAO2B||TYSND1||CASP3||F12||AIP||PISD||SEC11A||DOHH||SPPL3||NCSTN||HTRA2||BAK1||FADD||PCSK5||NKD2||BAD||APH1A||DHCR24||CPE||ERO1A||PRNP||PERP||LDLRAD3||FKRP||PCSK4||GAS1||F3||MELTF||BACE1||TMEM59||PSEN1||PSENEN||SPCS1||FURIN||PCSK6||DNAJB11||ADAM10||TSPAN33||XPNPEP3||ZMPSTE24||COMP||HM13||TSPAN15||LGMN||LMF2||SPCS3||GLI3||SPON1||GSN||CISD3||PTCH1||ATP6AP2||GALNT2||PRDX4||CTSH||SERPINE2||CALR||CTSL||CTSZ||ADAMTS2||NA||CLN5||RCE1||C1R||WFS1||TSPAN14||PLAT||SERPINH1||TESC||LMF1||PCSK1N||PRSS12||C1RL||ACE||ECEL1||AANAT||ASPRV1 | 5.9355703 | 6.1904368 | 6.0908159 | 6.127008 | 5.93000238 | 6.0251571 | 6.1213212 | 6.1013928 | 5.9259832 | 5.94062888 | 5.94005126 | 6.235889 | 6.1871068 | 6.1469382 | 6.0684884 | 6.1133666 | 6.0902436 | 6.127771 | 6.1083004 | 6.1447227 | 5.95995254 | 5.8936004 | 5.93567460 | 5.985345907 | 5.9637058 | 6.1213085 | 6.1372076 | 6.1112444 | 6.1153766 | 6.0948013 | 6.1198110 | 6.0893831 |
| GO:0043267 | negative regulation of potassium ion transport | 15 | 0.6249955 | 1.559857 | 0.0389143883 | 0.6652905 | 0.6510473 | 1194 | tags=33%, list=7%, signal=31% | RGS4||ACTN2||PTK2B||KCNH2||ANK3 | 2.8880162 | 4.5125171 | 3.1122953 | 4.658593 | 2.82048510 | 2.9260082 | 4.6439258 | 4.7643589 | 2.9669125 | 2.97050253 | 2.71168254 | 4.467412 | 4.5292795 | 4.5398047 | 3.7056955 | 2.7309035 | 2.6453699 | 4.501598 | 4.6991280 | 4.7624122 | 2.67748609 | 2.9501254 | 2.82098204 | 2.843772571 | 2.9115062 | 3.0174156 | 4.5904649 | 4.6658304 | 4.6740204 | 4.7316473 | 4.8018906 | 4.7586671 |
| GO:0070663 | regulation of leukocyte proliferation | 123 | -0.3797248 | -1.382772 | 0.0391864154 | 0.6652905 | 0.6510473 | 4168 | tags=41%, list=26%, signal=31% | ZP3||IKZF3||CASP3||SYK||MAPK8IP1||GSTP1||IGF2||HES1||FADD||PRKCQ||ITCH||HHEX||ERBB2||CD276||CORO1A||PRNP||CD40||PKN1||NCK2||ICOSLG||SLC7A1||CD55||ZBTB7B||TNFAIP3||CDKN1A||HLA-DMB||HLA-E||NDFIP1||IRF1||TIRAP||IL6ST||MIF||IGFBP2||BMP4||CD320||CD81||EFNB1||WNT3A||MYD88||EPHB2||JAK3||ENPP3||HLA-A||CD24||NFATC2||BST2||TICAM1||IL23A||CCDC88B||IL4I1||LRRC32 | 6.9552484 | 6.5075238 | 6.6844196 | 6.454671 | 7.00207026 | 6.9762749 | 6.4107557 | 6.4125067 | 6.9440276 | 6.82149884 | 7.08787535 | 6.606150 | 6.4770552 | 6.4337429 | 6.4810638 | 6.8038862 | 6.7481143 | 6.551713 | 6.3951544 | 6.4119719 | 7.01404084 | 6.8678142 | 7.11380745 | 6.980784073 | 7.1196776 | 6.8119767 | 6.4246854 | 6.4076193 | 6.3998504 | 6.4137254 | 6.4092351 | 6.4145540 |
| GO:0010575 | positive regulation of vascular endothelial growth factor production | 19 | -0.6046712 | -1.562707 | 0.0393004520 | 0.6652905 | 0.6510473 | 410 | tags=42%, list=3%, signal=41% | SULF2||TGFB1||SULF1||IL6ST||PTGS2||C5||C5AR1||HPSE | 5.6965729 | 5.2691813 | 5.7634117 | 5.828367 | 5.64484381 | 5.7992986 | 5.3182064 | 5.9023888 | 5.7186595 | 5.83230165 | 5.52183004 | 5.340445 | 5.1964688 | 5.2670376 | 5.7810275 | 5.6823222 | 5.8232809 | 5.869205 | 5.7141905 | 5.8951515 | 5.60668493 | 5.8030713 | 5.50907980 | 5.875509387 | 5.7089943 | 5.8085454 | 5.3427426 | 5.2865793 | 5.3247286 | 5.9081123 | 5.8956679 | 5.9033588 |
| GO:0001977 | renal system process involved in regulation of blood volume | 10 | -0.7044082 | -1.581211 | 0.0393269419 | 0.6652905 | 0.6510473 | 4159 | tags=70%, list=26%, signal=52% | EMP2||PTPRO||CORO2B||F2RL1||GAS6||CYBA||HSD11B2 | 5.3952092 | 5.0394471 | 5.3650490 | 4.936573 | 5.40847069 | 5.5581541 | 4.7845689 | 4.7639515 | 5.4279943 | 5.16158145 | 5.56708381 | 5.203613 | 5.0611423 | 4.8291018 | 5.1078683 | 5.6259478 | 5.3137300 | 5.082354 | 4.8283493 | 4.8865812 | 5.39163724 | 5.2794872 | 5.54219821 | 5.401524112 | 5.6102582 | 5.6505048 | 4.8815726 | 4.7646114 | 4.7017156 | 4.7196380 | 4.7927916 | 4.7783885 |
| GO:0050678 | regulation of epithelial cell proliferation | 244 | -0.3283453 | -1.304648 | 0.0398341745 | 0.6687736 | 0.6544558 | 3775 | tags=31%, list=24%, signal=24% | IGF2||HES1||FGFR2||HRAS||LAMB1||NR4A3||WDR13||TGFBR1||NR4A1||BAD||NOTCH1||ERBB2||NKX3-1||SULF2||MYC||F3||HMOX1||GPC3||ZNF703||WNT5A||ZFP36||TGFB1||HTRA1||ESRP2||MEN1||RUNX3||TNFAIP3||FZD7||CDH3||MDK||BMP6||EGFR||SULF1||MYDGF||CXCL12||CDKN2B||APOE||NOG||SNAI2||CD109||EMC10||PTCH1||BMP4||SOX9||FMC1||LIMS2||WNT3A||EGFL7||NGFR||BMPR2||HAS2||CYBA||GPX1||CDKN1C||VEGFC||SPARC||SERPINF1||SFRP1||GRN||NRP1||ANG||ITGA4||CEACAM1||THBS4||GLI1||EDNRB||NUPR1||CLDN1||TGFB2||C5AR1||DUSP10||HES5||NKX2-8||TGFA||CYP7B1 | 5.5046540 | 5.6347777 | 5.5117431 | 5.612840 | 5.48851878 | 5.4569218 | 5.6145811 | 5.6057288 | 5.5231322 | 5.52150425 | 5.46866276 | 5.665143 | 5.6244158 | 5.6142703 | 5.5451926 | 5.4732200 | 5.5159101 | 5.631530 | 5.5749663 | 5.6312905 | 5.49283658 | 5.5207277 | 5.45114339 | 5.481891060 | 5.4015819 | 5.4857375 | 5.6346715 | 5.5913939 | 5.6173492 | 5.6059017 | 5.6098453 | 5.6014271 |
| GO:0006261 | DNA-dependent DNA replication | 143 | 0.3498914 | 1.340791 | 0.0400953619 | 0.6687736 | 0.6544558 | 5348 | tags=44%, list=34%, signal=30% | POLN||HELB||POLG||ZGRF1||POLQ||THOC1||TRAIP||ATRX||EME2||POLG2||BOD1L1||BAZ1A||RFC1||POLE||DBF4||GEN1||WRN||KAT7||TONSL||BRCA2||NOC3L||POLD3||ZNF830||NBN||POLE2||REV3L||E2F8||TOPBP1||DBF4B||RRM2B||MCM10||ATR||TIPIN||DDX21||FANCM||ETAA1||SRPK2||NUCKS1||BLM||GMNN||DDX11||MCM9||CHTF18||SMARCAL1||RECQL5||ORC6||SAMHD1||METTL4||FBXO5||MCM4||POLA1||CDC6||RTF2||PURA||POLB||TICRR||POLA2||TWNK||WDHD1||INO80||DDX23||MRE11||ORC3 | 5.6400983 | 6.0621693 | 5.8230227 | 6.096500 | 5.60392524 | 5.7310236 | 6.1068273 | 6.1801313 | 5.6195058 | 5.66797351 | 5.63237778 | 6.069563 | 6.0569276 | 6.0599868 | 5.9581043 | 5.7543691 | 5.7464651 | 6.078222 | 6.0517554 | 6.1574179 | 5.58750695 | 5.5779142 | 5.64542572 | 5.640932120 | 5.7018146 | 5.8428286 | 6.1152879 | 6.0910498 | 6.1140155 | 6.1607160 | 6.2109405 | 6.1682269 |
| GO:0010452 | histone H3-K36 methylation | 13 | 0.6464428 | 1.549895 | 0.0401378472 | 0.6687736 | 0.6544558 | 4359 | tags=69%, list=27%, signal=50% | ASH1L||SETD5||SETD2||NSD2||IWS1||NSD1||PAXIP1||BCOR||NSD3 | 3.9739756 | 4.9736042 | 4.6105105 | 5.133942 | 3.88190210 | 4.5553574 | 5.1161010 | 5.2474094 | 3.8274699 | 4.08803946 | 3.99444787 | 4.932355 | 5.0038026 | 4.9837171 | 4.9572079 | 4.4338713 | 4.3651279 | 5.041109 | 5.0924220 | 5.2591817 | 3.86636925 | 3.7646603 | 4.00458835 | 4.489774562 | 4.4953529 | 4.6732913 | 5.1348201 | 5.1351938 | 5.0775291 | 5.1842698 | 5.2813558 | 5.2745809 |
| GO:0035627 | ceramide transport | 12 | -0.6749251 | -1.574074 | 0.0402366864 | 0.6687736 | 0.6544558 | 1893 | tags=42%, list=12%, signal=37% | PSAP||PLTP||CPTP||ABCB1||MTTP | 5.0109358 | 5.4438198 | 5.5923051 | 5.394574 | 5.03664062 | 5.5076851 | 5.2726295 | 5.2441070 | 4.9266521 | 5.03249687 | 5.06986189 | 5.493426 | 5.4726262 | 5.3619762 | 5.4783356 | 5.7428662 | 5.5423356 | 5.419059 | 5.4011475 | 5.3629471 | 5.05325534 | 4.9519630 | 5.10072572 | 5.367029056 | 5.2363071 | 5.8460987 | 5.3057893 | 5.2693268 | 5.2420630 | 5.2748344 | 5.2544619 | 5.2020507 |
| GO:0034976 | response to endoplasmic reticulum stress | 230 | -0.3302548 | -1.302991 | 0.0404332130 | 0.6700865 | 0.6557406 | 5555 | tags=47%, list=35%, signal=31% | UFD1||UGGT2||SELENOK||SGTA||AIFM1||HERPUD1||MARCHF6||SCAMP5||BFAR||UBXN1||ATG10||DERL2||RNF5||CREB3L2||BRSK2||UBXN6||TNFRSF10B||TMTC4||FLOT1||LPCAT3||MBTPS2||DNAJB2||NR1H3||STT3B||RNF139||EEF2||CTH||ATF6||FAF2||ATF3||BAX||CDK5RAP3||TMCO1||SYVN1||SEL1L||BCL2||EDEM3||XBP1||TMEM117||RNF103||FOXRED2||TMUB2||ATF6B||TMED2||NIBAN1||CCDC47||BAK1||SELENOS||PML||FBXO2||CLGN||TOR1A||ERO1A||ERMP1||UFL1||FBXO44||DNAJB9||NCK2||RASGRF2||UBE4A||ERLEC1||OS9||ERLIN2||ATXN3||BCAP31||DERL1||SERINC3||TMBIM6||TMUB1||CREB3||UBAC2||ERP44||MANF||RHBDD2||ERP29||HM13||CANX||EDEM2||PDIA3||TRIB3||HSP90B1||TMEM33||TMEM129||DDRGK1||HSPA5||HYOU1||PDX1||JUN||BID||MAN1B1||AMFR||RCN3||CALR||P4HB||GRINA||ATP2A3||SRPX||SDF2L1||PDIA4||RNFT2||WFS1||BOK||DERL3||DNAJB14||THBS4||CREB3L4||CLU||NUPR1||CREB3L1 | 6.4122420 | 6.7712101 | 6.4865968 | 6.620391 | 6.42502997 | 6.5789341 | 6.6865281 | 6.6003375 | 6.3818735 | 6.41545941 | 6.43882555 | 6.784238 | 6.7971855 | 6.7313673 | 6.4475409 | 6.5055211 | 6.5059494 | 6.629462 | 6.6032634 | 6.6282966 | 6.43130711 | 6.3868880 | 6.45604553 | 6.585435483 | 6.5883444 | 6.5628880 | 6.6976156 | 6.6900178 | 6.6718293 | 6.5940453 | 6.6316591 | 6.5747259 |
| GO:0002021 | response to dietary excess | 14 | -0.6494440 | -1.566115 | 0.0414012739 | 0.6763364 | 0.6618567 | 2381 | tags=43%, list=15%, signal=36% | TRPV4||APOE||ACVR1C||PCSK1N||ADRB2||VGF | 3.0709215 | 3.1151029 | 2.9449196 | 2.804029 | 2.73250165 | 3.2166522 | 2.9350516 | 2.6351896 | 3.1742943 | 2.98918639 | 3.04294851 | 3.182721 | 3.0883400 | 3.0717468 | 2.8917148 | 3.1226526 | 2.8010717 | 2.858960 | 2.7866456 | 2.7647884 | 2.84493095 | 2.3756885 | 2.91908500 | 3.048380742 | 3.3488335 | 3.2369122 | 2.9740661 | 2.9152985 | 2.9149844 | 2.6306933 | 2.7036584 | 2.5680198 |
| GO:2001235 | positive regulation of apoptotic signaling pathway | 97 | -0.3997169 | -1.402176 | 0.0416587407 | 0.6766498 | 0.6621634 | 3998 | tags=35%, list=25%, signal=26% | FIS1||NACC2||SKIL||HTRA2||TGFBR1||FADD||PML||BAD||NKX3-1||NCK2||DEDD2||BCAP31||SERINC3||TIMP3||TNFRSF12A||STYXL1||WWOX||PDIA3||ITM2C||CTSC||CRADD||INHBB||BID||CTSH||NGFR||SRPX||HYAL2||SFRP1||BOK||RET||FAS||NUPR1||G0S2||SEPTIN4 | 8.1270089 | 8.1171362 | 7.9229557 | 8.034390 | 8.10351511 | 7.9209792 | 8.1063181 | 8.0565415 | 8.1347033 | 8.15706645 | 8.08840934 | 8.157989 | 8.0859760 | 8.1064866 | 7.8687743 | 7.9029408 | 7.9942196 | 8.055580 | 8.0122967 | 8.0349685 | 8.09659006 | 8.1593912 | 8.05256203 | 8.032606577 | 7.9288324 | 7.7913960 | 8.0987424 | 8.1059455 | 8.1142249 | 8.0481609 | 8.0399337 | 8.0811985 |
| GO:0030278 | regulation of ossification | 84 | -0.4120644 | -1.417742 | 0.0417217293 | 0.6766498 | 0.6621634 | 3100 | tags=33%, list=19%, signal=27% | WNT5A||GPM6B||CHSY1||TGFB1||CCN1||SLC8A1||MDK||BMP6||ATRAID||ZMPSTE24||COMP||BMP4||SOX9||BMPR2||PHOSPHO1||DDR2||NOTUM||KREMEN2||SFRP1||FZD9||NELL1||PTGER4||ADRB2||ZBTB16||DHRS3||TGFB2||S1PR1||RUNX2 | 3.6402179 | 4.1925560 | 3.8514947 | 4.049293 | 3.68010446 | 3.8420975 | 4.1561256 | 4.0466323 | 3.6009777 | 3.58366993 | 3.73143783 | 4.196909 | 4.2253373 | 4.1545437 | 3.8659536 | 3.8933387 | 3.7933502 | 4.019605 | 4.0599597 | 4.0678531 | 3.67172714 | 3.5940049 | 3.76922781 | 3.755959116 | 3.7842757 | 3.9759838 | 4.1826168 | 4.1513135 | 4.1340252 | 4.0382766 | 4.0788858 | 4.0221404 |
| GO:0048265 | response to pain | 16 | -0.6284606 | -1.561786 | 0.0419137999 | 0.6766498 | 0.6621634 | 2824 | tags=56%, list=18%, signal=46% | P2RX2||TSPO||COMT||VWA1||P2RX4||TRPA1||RET||THBS4||EDNRB | 4.6425627 | 4.3251616 | 4.5865023 | 4.092774 | 4.71093591 | 4.8535249 | 4.1955139 | 4.0524924 | 4.6038967 | 4.46660478 | 4.83319738 | 4.324016 | 4.3820601 | 4.2671192 | 4.3957500 | 4.7488507 | 4.5933303 | 4.210030 | 4.0011023 | 4.0590497 | 4.69620705 | 4.5564015 | 4.86374890 | 4.775838257 | 4.9496028 | 4.8296106 | 4.2411391 | 4.1691707 | 4.1751204 | 4.0790077 | 4.0976348 | 3.9779845 |
| GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 11 | -0.6835520 | -1.559121 | 0.0422007494 | 0.6766498 | 0.6621634 | 2920 | tags=45%, list=18%, signal=37% | EHD1||IL4R||ADGRB1||NFATC2||RAPGEF3 | 4.3939114 | 4.6080489 | 4.3773299 | 4.466575 | 4.40843305 | 4.5388517 | 4.5689797 | 4.4369968 | 4.3930639 | 4.38903511 | 4.39961528 | 4.618699 | 4.6033946 | 4.6019932 | 4.4449506 | 4.3435339 | 4.3410502 | 4.483226 | 4.3957404 | 4.5180224 | 4.41125709 | 4.3334260 | 4.47703935 | 4.520417974 | 4.5655504 | 4.5301950 | 4.6152754 | 4.5294191 | 4.5609340 | 4.4311976 | 4.4791426 | 4.3995348 |
| GO:2000273 | positive regulation of signaling receptor activity | 27 | -0.5474370 | -1.534889 | 0.0422507788 | 0.6766498 | 0.6621634 | 1386 | tags=30%, list=9%, signal=27% | ADRA2C||EPHB2||RGS9||ARC||NRP1||ADRB2||NEURL1||TGFA | 4.9657441 | 5.7288604 | 5.3597253 | 5.760545 | 4.78538420 | 4.7515787 | 5.7495421 | 5.7693374 | 5.0681339 | 5.19652001 | 4.55643475 | 5.740968 | 5.7252936 | 5.7202385 | 5.5595296 | 5.1186676 | 5.3673985 | 5.771803 | 5.7121962 | 5.7963435 | 4.70843321 | 5.0285317 | 4.58176401 | 4.727024254 | 4.5292413 | 4.9653306 | 5.7524048 | 5.7248297 | 5.7710179 | 5.7489751 | 5.7743724 | 5.7844336 |
| GO:0051480 | regulation of cytosolic calcium ion concentration | 178 | -0.3466001 | -1.326944 | 0.0431271793 | 0.6848692 | 0.6702069 | 3926 | tags=38%, list=25%, signal=29% | SNCA||MCOLN1||S1PR3||TPCN1||SPPL3||BAK1||TMEM64||PML||RYR1||SLC24A1||ERO1A||CORO1A||PRNP||DIAPH1||SELENON||TRPC1||ADCY5||ITGAV||ITPR3||GSTO1||CDK5||JPH1||WNT5A||NALF1||SLC8A1||BCAP31||SLC24A4||P2RX2||CD55||TMBIM6||CALCB||ADCYAP1R1||MCOLN2||LPAR1||CD19||TRPV4||SLC8B1||PDE4D||NMB||TMEM38A||PTGER2||CX3CR1||F2RL1||CYBA||P2RX4||TRPA1||LRP1||TRPC3||JSRP1||BOK||RYR2||GRIN2D||FZD9||CD24||PTGER4||CXCR4||PTH1R||EDNRB||SLC8A2||C1QTNF1||C5AR1||S1PR1||CCR10||CCKBR||SLC8A3||ACKR2||CYSLTR1||AKAP6 | 4.8547476 | 5.1346232 | 4.9826174 | 5.145448 | 4.82872549 | 4.9176410 | 5.1333715 | 5.1528713 | 4.8599787 | 4.88944902 | 4.81380959 | 5.155782 | 5.1339232 | 5.1138598 | 5.0270048 | 4.9754937 | 4.9441375 | 5.137291 | 5.1240452 | 5.1745322 | 4.83084356 | 4.8268107 | 4.82851936 | 4.854364737 | 4.8512937 | 5.0391323 | 5.1463490 | 5.1261729 | 5.1275042 | 5.1490023 | 5.1593510 | 5.1502383 |
| GO:0003230 | cardiac atrium development | 29 | -0.5309905 | -1.512692 | 0.0436893204 | 0.6863477 | 0.6716537 | 3546 | tags=38%, list=22%, signal=30% | NOTCH1||WNT11||NPHP3||WNT5A||CCN1||NKX2-5||ZFPM1||NOG||BMPR2||MESP1||TGFB2 | 2.8889031 | 3.4464158 | 3.3422136 | 3.557501 | 2.92276546 | 3.4425620 | 3.4657641 | 3.5647590 | 2.7940938 | 2.85617238 | 3.00797478 | 3.394022 | 3.5250925 | 3.4167044 | 3.3950216 | 3.3951366 | 3.2302902 | 3.478156 | 3.5211005 | 3.6664302 | 2.92331343 | 2.7242968 | 3.09671801 | 3.293034968 | 3.4380515 | 3.5821304 | 3.4692077 | 3.4676643 | 3.4604049 | 3.4657477 | 3.6674393 | 3.5539864 |
| GO:0030497 | fatty acid elongation | 11 | -0.6821188 | -1.555852 | 0.0437783475 | 0.6863477 | 0.6716537 | 4527 | tags=82%, list=28%, signal=59% | ELOVL1||ELOVL6||HACD3||HACD1||ELOVL5||ELOVL7||TECR||ELOVL4||ABCD1 | 5.3793352 | 5.9357108 | 5.6067287 | 6.019190 | 5.41967478 | 5.6670137 | 5.7777904 | 5.8917175 | 5.3249020 | 5.33524423 | 5.47306652 | 5.962855 | 5.9566381 | 5.8863968 | 5.5450842 | 5.6838287 | 5.5877530 | 6.014254 | 6.0354606 | 6.0077091 | 5.38824202 | 5.3555707 | 5.51054548 | 5.502587518 | 5.6399628 | 5.8385947 | 5.7964717 | 5.7838523 | 5.7526960 | 5.9129070 | 5.9287625 | 5.8316120 |
| GO:0043968 | histone H2A acetylation | 17 | 0.5930989 | 1.519707 | 0.0438238891 | 0.6863477 | 0.6716537 | 5224 | tags=71%, list=33%, signal=48% | MSL3P1||BRD8||EPC1||TRRAP||ING3||EP400||MEAF6||DMAP1||MSL3||ACTL6A||SPHK2||NAA40 | 6.0097914 | 6.6864484 | 6.1108744 | 6.561651 | 5.93209981 | 5.9644514 | 6.7317148 | 6.6173602 | 6.0158720 | 6.11389188 | 5.89098991 | 6.729084 | 6.6548587 | 6.6743728 | 6.3807231 | 5.9194576 | 5.9883413 | 6.533077 | 6.5222417 | 6.6273039 | 5.88415047 | 6.0025819 | 5.90680856 | 5.920299365 | 5.9484351 | 6.0226717 | 6.7318828 | 6.7103386 | 6.7526132 | 6.6136618 | 6.6086780 | 6.6296573 |
| GO:0051155 | positive regulation of striated muscle cell differentiation | 31 | -0.5224458 | -1.506025 | 0.0439432238 | 0.6863477 | 0.6716537 | 1845 | tags=26%, list=12%, signal=23% | BMP4||WNT3A||MAMSTR||MYLK3||CYP26B1||MESP1||NEK5||AKAP6 | 5.4727706 | 5.6387777 | 5.5826125 | 5.402405 | 5.45509856 | 5.4913810 | 5.6492588 | 5.4414642 | 5.4413138 | 5.49996119 | 5.47643396 | 5.672634 | 5.6354102 | 5.6075490 | 5.7166526 | 5.5275422 | 5.4934834 | 5.394974 | 5.3654114 | 5.4456832 | 5.46307134 | 5.4044364 | 5.49629284 | 5.344657558 | 5.4522783 | 5.6593044 | 5.6538829 | 5.6487412 | 5.6451389 | 5.4244596 | 5.4638897 | 5.4357571 |
| GO:0060740 | prostate gland epithelium morphogenesis | 21 | -0.5854416 | -1.548369 | 0.0443037975 | 0.6863477 | 0.6716537 | 3751 | tags=57%, list=23%, signal=44% | FGFR2||NOTCH1||NKX3-1||WNT5A||GLI2||SULF1||NOG||BMP4||SOX9||HOXB13||SFRP1||CYP7B1 | 2.3120918 | 3.0651056 | 2.6027561 | 3.141810 | 2.32715754 | 2.6693923 | 3.0691610 | 3.1158177 | 2.3010088 | 2.35175012 | 2.28262510 | 3.044928 | 3.1330939 | 3.0146550 | 2.7158446 | 2.5217826 | 2.5633264 | 3.089136 | 3.1729834 | 3.1618809 | 2.34960158 | 2.2532788 | 2.37573006 | 2.601037101 | 2.6211472 | 2.7793056 | 3.0393693 | 3.0509791 | 3.1159482 | 3.1055763 | 3.1300407 | 3.1117236 |
| GO:1900120 | regulation of receptor binding | 13 | -0.6596409 | -1.567957 | 0.0449792859 | 0.6887618 | 0.6740161 | 2052 | tags=69%, list=13%, signal=60% | RGMA||PTPRF||ADAM15||NOG||LOX||ATP2A3||PLCL1||NRP1||PHLDA2 | 4.6138337 | 5.6008228 | 5.1304191 | 5.704296 | 4.55024183 | 4.7272116 | 5.5780961 | 5.6607752 | 4.6242353 | 4.72105339 | 4.48664460 | 5.555448 | 5.6618575 | 5.5830383 | 5.3442550 | 4.9020647 | 5.1109918 | 5.667288 | 5.7710613 | 5.6721423 | 4.46073383 | 4.6674828 | 4.51445296 | 4.516892040 | 4.6330710 | 4.9888317 | 5.6121612 | 5.5800946 | 5.5411568 | 5.6850122 | 5.6771336 | 5.6192912 |
| GO:0071300 | cellular response to retinoic acid | 50 | -0.4621163 | -1.457973 | 0.0455422617 | 0.6887618 | 0.6740161 | 3911 | tags=44%, list=25%, signal=33% | TEAD2||FGFR2||HTRA2||WNT11||WNT5A||EPHA3||WNT3||PTK7||FZD7||NDUFA13||SOX9||WNT3A||MEIOSIN||SERPINF1||LTK||CYP26B1||RET||WNT7B||TESC||WNT9A||FZD10||WNT5B | 5.2594905 | 5.1436529 | 5.1427879 | 5.054717 | 5.30530100 | 5.2460064 | 5.0601765 | 5.0343657 | 5.2736391 | 5.14377927 | 5.35334472 | 5.236869 | 5.1242703 | 5.0644790 | 5.0506362 | 5.2071001 | 5.1661018 | 5.114252 | 4.9878195 | 5.0592978 | 5.27626859 | 5.2384016 | 5.39647277 | 5.281184482 | 5.3096418 | 5.1416515 | 5.0867782 | 5.0451261 | 5.0482509 | 5.0219522 | 5.0353767 | 5.0456701 |
| GO:0033619 | membrane protein proteolysis | 46 | -0.4712957 | -1.464280 | 0.0456842924 | 0.6887618 | 0.6740161 | 4997 | tags=59%, list=31%, signal=40% | MBTPS2||TIMP2||RGMA||CLPP||PSEN2||NAPSA||GPLD1||SPPL3||NCSTN||PRKCQ||APH1A||SPPL2A||BACE1||TGFB1||PSEN1||PSENEN||SNX33||TIMP3||FURIN||ADAM10||DAG1||HM13||APOE||ERAP1||CTSH||TIMP1||RET | 5.1770004 | 5.5633110 | 5.4543890 | 5.512303 | 5.19324889 | 5.4665657 | 5.4983232 | 5.4589503 | 5.1329129 | 5.16473975 | 5.23158409 | 5.595157 | 5.5780004 | 5.5155654 | 5.4537341 | 5.5084903 | 5.3988602 | 5.494007 | 5.5137781 | 5.5289134 | 5.19361319 | 5.1105155 | 5.27114890 | 5.335912308 | 5.4097909 | 5.6367105 | 5.5154232 | 5.4963211 | 5.4830415 | 5.4401524 | 5.5016739 | 5.4340500 |
| GO:0018027 | peptidyl-lysine dimethylation | 23 | 0.5451340 | 1.494665 | 0.0456863145 | 0.6887618 | 0.6740161 | 3739 | tags=43%, list=23%, signal=33% | ASH1L||KMT2D||KMT2A||CSKMT||SETD2||SUV39H2||SETD1A||EHMT2||SETD4||KMT2B | 4.7763412 | 4.9620240 | 5.1664815 | 4.998722 | 4.75629794 | 5.1178200 | 5.0426071 | 5.0616529 | 4.6711306 | 4.76707462 | 4.88300213 | 4.946728 | 4.9812221 | 4.9579070 | 5.2158472 | 5.1958999 | 5.0842409 | 4.987605 | 4.9810545 | 5.0270760 | 4.79473683 | 4.5963834 | 4.86454803 | 4.998434042 | 5.0488490 | 5.2891586 | 5.0681072 | 5.0471417 | 5.0120166 | 5.0414590 | 5.0998626 | 5.0428640 |
| GO:1990748 | cellular detoxification | 72 | -0.4229574 | -1.416541 | 0.0457328741 | 0.6887618 | 0.6740161 | 4560 | tags=44%, list=29%, signal=32% | HBQ1||CCS||ADH5||ABCB6||PTGES||GSTK1||PRXL2C||UBIAD1||PRDX2||GSTP1||NXN||SELENOS||AKR1A1||SELENOW||PRDX5||GSTO1||GPX3||NQO1||GPX8||FBLN5||MGST2||GLRX||MTARC2||PRXL2B||APOE||RDH11||GSTO2||PRDX4||ABCG2||GPX1||PTGS2||HBA1 | 7.1687647 | 6.7780180 | 7.0300250 | 6.900246 | 7.18564814 | 7.0891252 | 6.7270325 | 6.8884773 | 7.1930554 | 7.13358978 | 7.17898171 | 6.836057 | 6.7555014 | 6.7406586 | 6.9471174 | 7.0378368 | 7.1009881 | 6.929621 | 6.8583786 | 6.9117893 | 7.16746444 | 7.2006890 | 7.18859514 | 7.094476727 | 7.1857554 | 6.9797831 | 6.7429579 | 6.7145242 | 6.7234687 | 6.8901870 | 6.8805430 | 6.8946658 |
| GO:0006684 | sphingomyelin metabolic process | 13 | -0.6564599 | -1.560396 | 0.0461629513 | 0.6887618 | 0.6740161 | 1969 | tags=38%, list=12%, signal=34% | SGMS1||PEMT||SMPDL3B||SMPD1||SMPD3 | 4.4690138 | 4.4926554 | 4.8369415 | 4.471989 | 4.53450728 | 4.8969077 | 4.4400672 | 4.3499080 | 4.3837863 | 4.38730548 | 4.62272318 | 4.508114 | 4.5385036 | 4.4291518 | 4.7661455 | 4.9466231 | 4.7913572 | 4.500272 | 4.4552080 | 4.4600604 | 4.51796627 | 4.3835191 | 4.68606286 | 4.716635677 | 4.8480538 | 5.0994940 | 4.4929011 | 4.4123191 | 4.4134936 | 4.3577508 | 4.4013472 | 4.2883832 |
| GO:0061311 | cell surface receptor signaling pathway involved in heart development | 26 | -0.5461886 | -1.515034 | 0.0464209089 | 0.6887618 | 0.6740161 | 3768 | tags=46%, list=24%, signal=35% | HEY1||NOTCH1||WNT11||SNAI1||WNT5A||TGFB1||NOG||SNAI2||BMP4||WNT3A||MESP1||HAND2 | 3.3518707 | 4.6587868 | 3.8811719 | 4.695438 | 3.25228448 | 3.3689611 | 4.6019174 | 4.6351701 | 3.4173634 | 3.53333042 | 3.06473404 | 4.680879 | 4.6917421 | 4.6020907 | 4.1849869 | 3.5370822 | 3.8487696 | 4.657887 | 4.7058814 | 4.7217821 | 3.16481569 | 3.4209935 | 3.15497585 | 3.262078327 | 3.2085504 | 3.6036309 | 4.6187087 | 4.5942480 | 4.5926472 | 4.6181104 | 4.6605153 | 4.6265344 |
| GO:0008284 | positive regulation of cell population proliferation | 613 | -0.2768898 | -1.198707 | 0.0465358569 | 0.6887618 | 0.6740161 | 3702 | tags=28%, list=23%, signal=23% | HRAS||LTBP3||LAMB1||NR4A3||CITED1||TGFBR1||INSM1||FADD||PRKCQ||NR4A1||MALAT1||PML||BAD||NOTCH1||STX3||ERBB2||NKX3-1||CD276||FOXP1||IL13RA1||CORO1A||UFL1||SELENON||CD40||FN1||MST1R||ITGAV||MYC||NCK2||F3||HMOX1||ICOSLG||ZNF703||TNFRSF11A||WNT5A||MEIS3||DLL1||TGFB1||HLX||HTRA1||CCN1||ESRP2||ROGDI||GDNF||SLC7A1||EFNB2||CD55||TBX2||SOX8||NKX2-5||IRS1||GLI2||TSPO||TNFAIP3||LHX1||FZD7||ADAM10||CDH3||MDK||CDKN1A||HLA-DMB||HLA-E||BMP6||HMGCR||NES||EGFR||LBH||COMP||MYDGF||CXCL12||TIRAP||SHMT2||DISP3||IL6ST||LGMN||PTAFR||EGR1||MIF||NOG||NMB||GLI3||NTN1||DDRGK1||IGFBP2||EMC10||CGA||IRAK4||LRP5||BMP4||PRKCZ||CD320||CD81||SOX9||PDX1||S1PR2||JUN||CLEC11A||CTSH||RAD51B||EFNB1||WNT3A||ZNF423||EGFL7||CRLF1||NGFR||FLT3||CX3CR1||CALR||BMPR2||FGF18||HAS2||CTSZ||PDGFA||RHOG||MYD88||EPHB2||TIMP1||DDR2||ROMO1||TSPAN31||GAS6||CYBA||DMRTA2||VEGFC||JAK3||SFRP1||SERTAD1||GAREM1||ELL3||MAB21L1||GRN||HPGD||HLA-A||CRIP2||NRP1||MEGF10||FGF17||IL6R||ANG||FZD9||ITGA4||CD24||CNTFR||PITX3||PTH1R||THBS4||TBX6||GLI1||EPCAM||EDNRB||CTF1||NFATC2||CLDN1||BST2||PTGS2||SHANK2||TICAM1||CHRD||TGFB2||C5AR1||S1PR1||RUNX2||IL23A||GDF9||CCKBR||SMPD3||HPSE||HES5||CCDC88B||TGFA||PTHLH||CYP7B1||ETV5 | 6.4322721 | 6.5379759 | 6.3036561 | 6.479274 | 6.41368815 | 6.3230781 | 6.5131571 | 6.5026733 | 6.4566278 | 6.44807035 | 6.39124660 | 6.593269 | 6.5116626 | 6.5073616 | 6.2934863 | 6.2683287 | 6.3480001 | 6.502415 | 6.4416516 | 6.4930182 | 6.40295445 | 6.4481335 | 6.38931748 | 6.370863240 | 6.3719878 | 6.2212328 | 6.5095226 | 6.5120679 | 6.5178682 | 6.4949840 | 6.5011803 | 6.5118054 |
| GO:0051095 | regulation of helicase activity | 12 | 0.6565767 | 1.543583 | 0.0468369830 | 0.6887618 | 0.6740161 | 4298 | tags=67%, list=27%, signal=49% | IFIT1||MNAT1||POT1||CHTOP||GTF2H2||MSH3||MSH6||SIRT1 | 6.1590355 | 6.7985159 | 6.0429544 | 6.849038 | 6.04260338 | 5.9444157 | 6.8518393 | 6.9022576 | 6.1885224 | 6.28729584 | 5.98502614 | 6.804049 | 6.7802773 | 6.8110421 | 6.3688632 | 5.8657454 | 5.8289383 | 6.824524 | 6.8118990 | 6.9087559 | 6.01285241 | 6.1170799 | 5.99483510 | 5.985904881 | 5.9316590 | 5.9147212 | 6.8209149 | 6.8469871 | 6.8868503 | 6.9163562 | 6.8888710 | 6.9014145 |
| GO:0098698 | postsynaptic specialization assembly | 16 | -0.6192940 | -1.539006 | 0.0470541716 | 0.6887618 | 0.6740161 | 2387 | tags=56%, list=15%, signal=48% | NLGN1||IL1RAP||LRFN4||LRFN1||ZDHHC12||LRRC4B||PTPRD||NRXN2||NTNG2 | 3.2954640 | 3.5342457 | 3.5549193 | 3.323607 | 3.36393451 | 3.7729699 | 3.4031318 | 3.2385046 | 3.2478380 | 3.11575713 | 3.49658406 | 3.507444 | 3.5936404 | 3.4997570 | 3.4997392 | 3.7265603 | 3.4207381 | 3.367226 | 3.3421818 | 3.2592105 | 3.38087490 | 3.1213124 | 3.55673577 | 3.563398615 | 3.7942286 | 3.9369339 | 3.4501711 | 3.3950001 | 3.3628694 | 3.2095797 | 3.3439007 | 3.1554549 |
| GO:0051983 | regulation of chromosome segregation | 81 | 0.3943999 | 1.383263 | 0.0470613288 | 0.6887618 | 0.6740161 | 5695 | tags=57%, list=36%, signal=37% | ATM||CENPF||CENPE||MAPK15||APC||GEN1||MKI67||SPDL1||CDK5RAP2||NDC80||SMC5||TTK||ESPL1||TPR||ZWILCH||HECW2||WAPL||BUB1||ANAPC1||XRCC3||AURKB||CDC23||DLGAP5||KNTC1||KIF2C||PCID2||PTTG1||MAD2L1||PUM2||BECN1||FBXO5||MAD1L1||CDC27||BUB1B||CDC6||TEX14||RAD18||KLHL22||CUL3||ANAPC5||ANAPC7||TACC3||RIOK2||ANAPC4||CDC16||PLK1 | 5.6164549 | 6.1914280 | 5.7339786 | 6.239446 | 5.54008466 | 5.6765508 | 6.2541826 | 6.3372162 | 5.5941442 | 5.71131152 | 5.53847970 | 6.208240 | 6.1782274 | 6.1876530 | 5.9566314 | 5.5807031 | 5.6353436 | 6.206067 | 6.1988861 | 6.3106502 | 5.52790012 | 5.5470070 | 5.54526963 | 5.655439485 | 5.6834214 | 5.6905533 | 6.2538480 | 6.2513572 | 6.2573362 | 6.3238933 | 6.3638794 | 6.3235015 |
| GO:0097720 | calcineurin-mediated signaling | 36 | -0.4987212 | -1.481432 | 0.0471551052 | 0.6887618 | 0.6740161 | 642 | tags=17%, list=4%, signal=16% | LMCD1||NFATC2||SLC8A2||SULT1A4||NR5A2||AKAP6 | 4.8143731 | 4.9489014 | 4.9505846 | 4.952521 | 4.78984565 | 5.0387536 | 4.9850816 | 5.0080758 | 4.8042194 | 4.82848642 | 4.81030283 | 4.904525 | 4.9751477 | 4.9660140 | 4.9240470 | 4.9934379 | 4.9332801 | 4.951491 | 4.9029170 | 5.0014708 | 4.83279322 | 4.6848219 | 4.84642066 | 5.001197099 | 4.9716594 | 5.1378975 | 5.0156227 | 4.9812856 | 4.9577486 | 4.9826320 | 5.0341433 | 5.0069917 |
| GO:0071805 | potassium ion transmembrane transport | 124 | -0.3730481 | -1.360515 | 0.0474418605 | 0.6887618 | 0.6740161 | 3075 | tags=33%, list=19%, signal=27% | LRRC26||CD63||KCNN1||ATP1A1||HCN2||BIN1||SLC24A4||KCNMB4||KCNH4||KCNK13||KCNJ12||KCND3||CCDC51||KCNK5||TMEM38A||KCNJ8||KCNB2||HCN3||KCNMA1||KCNH3||KCNC1||KCNT2||KCNN3||SLC12A5||KCNS3||ATP1A3||KCNA3||KCNIP3||KCNN2||SLC12A4||KCNJ11||KCNQ1||KCNK1||SLC9A9||KCNQ3||HCN4||KCND2||KCNA6||KCNK7||SLC9A2||AKAP6 | 4.1274244 | 4.6034943 | 4.2776114 | 4.673232 | 4.08904073 | 4.2459108 | 4.5672929 | 4.6581411 | 4.1459296 | 4.17130850 | 4.06281650 | 4.622031 | 4.6236923 | 4.5639610 | 4.3808085 | 4.2535117 | 4.1920452 | 4.639538 | 4.6796447 | 4.6998607 | 4.07551327 | 4.0999411 | 4.09156108 | 4.178170206 | 4.1686669 | 4.3807918 | 4.5784497 | 4.5700155 | 4.5533000 | 4.6544342 | 4.6832312 | 4.6363704 |
| GO:0098719 | sodium ion import across plasma membrane | 12 | -0.6661626 | -1.553638 | 0.0475345168 | 0.6887618 | 0.6740161 | 454 | tags=67%, list=3%, signal=65% | SLC9A6||TRPM4||SLC9A1||HCN2||SLC8A1||SLC9A9||HCN4||SLC9A2 | 1.4909382 | 2.4688372 | 2.1513982 | 2.540168 | 1.46056335 | 1.9381839 | 2.4180166 | 2.5238138 | 1.4471285 | 1.47939345 | 1.54457837 | 2.407119 | 2.5409408 | 2.4552542 | 2.2753761 | 2.2139264 | 1.9437879 | 2.474467 | 2.6265042 | 2.5152089 | 1.42049264 | 1.3220869 | 1.62266341 | 1.736183964 | 1.8514863 | 2.1879077 | 2.4387519 | 2.4641110 | 2.3486519 | 2.5330037 | 2.6092689 | 2.4231247 |
| GO:0042744 | hydrogen peroxide catabolic process | 14 | -0.6393233 | -1.541709 | 0.0479697452 | 0.6887618 | 0.6740161 | 3926 | tags=57%, list=25%, signal=43% | HBQ1||SNCA||PRDX2||PRDX5||GPX3||PRDX4||GPX1||HBA1 | 8.3849171 | 7.7616153 | 8.2425870 | 7.963686 | 8.40265582 | 8.2958290 | 7.6681010 | 7.9637330 | 8.4172874 | 8.35054436 | 8.38614695 | 7.885099 | 7.7071444 | 7.6841023 | 8.1047055 | 8.2860568 | 8.3274102 | 7.995715 | 7.9062329 | 7.9874276 | 8.41156783 | 8.4215464 | 8.37442703 | 8.318588451 | 8.3560619 | 8.2088087 | 7.6814533 | 7.6621282 | 7.6606278 | 7.9705512 | 7.9527239 | 7.9678600 |
| GO:0036336 | dendritic cell migration | 13 | -0.6517961 | -1.549310 | 0.0481357270 | 0.6887618 | 0.6740161 | 1466 | tags=31%, list=9%, signal=28% | CALR||GAS6||DOCK8||CXCR4 | 6.9231947 | 7.9529161 | 7.1206548 | 7.686633 | 6.74746702 | 6.5708327 | 7.8962398 | 7.6854036 | 7.0063490 | 7.17368183 | 6.51019401 | 7.986685 | 7.9560007 | 7.9151716 | 7.2238315 | 6.9602014 | 7.1648673 | 7.652339 | 7.6803833 | 7.7262101 | 6.68279408 | 7.0054049 | 6.50972170 | 6.599624487 | 6.3828402 | 6.7109782 | 7.8915012 | 7.8886872 | 7.9084517 | 7.6920782 | 7.6542517 | 7.7093320 |
| GO:0072520 | seminiferous tubule development | 11 | 0.6673508 | 1.530620 | 0.0482660718 | 0.6887618 | 0.6740161 | 4266 | tags=64%, list=27%, signal=47% | REC8||ATRX||ING2||AR||KIF18A||WDR48||BRIP1 | 8.0849403 | 7.9036225 | 7.9516802 | 7.791762 | 7.98525567 | 7.5809519 | 7.8562285 | 7.8613876 | 8.0590903 | 8.19243410 | 7.99629158 | 8.037227 | 7.8230175 | 7.8406778 | 7.7989668 | 7.9370640 | 8.1029196 | 7.869854 | 7.7034196 | 7.7971986 | 8.07099799 | 8.0638770 | 7.80534546 | 7.793133766 | 7.2379634 | 7.6556691 | 7.8659885 | 7.8421918 | 7.8603978 | 7.8893649 | 7.8201128 | 7.8737739 |
| GO:0060997 | dendritic spine morphogenesis | 56 | -0.4450953 | -1.430016 | 0.0484615385 | 0.6887618 | 0.6740161 | 2845 | tags=27%, list=18%, signal=22% | DBNL||EPHA4||CAMK2B||LZTS3||ADAM10||NLGN1||EPHB1||DBN1||EPHB2||ARC||ITPKA||EPHB3||ZNF365||SRCIN1||BHLHB9 | 5.3844118 | 5.2756567 | 5.3751185 | 5.391083 | 5.44862381 | 5.5901036 | 5.2923920 | 5.4203237 | 5.3078501 | 5.36270576 | 5.47745528 | 5.262390 | 5.2873434 | 5.2771273 | 5.3203600 | 5.4353259 | 5.3673487 | 5.392096 | 5.3449892 | 5.4347664 | 5.45455697 | 5.3413478 | 5.54291363 | 5.554618150 | 5.6397474 | 5.5745648 | 5.3197284 | 5.2707516 | 5.2862610 | 5.4161775 | 5.4398708 | 5.4046994 |
| GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 52 | -0.4492527 | -1.430135 | 0.0489671002 | 0.6887618 | 0.6740161 | 4186 | tags=40%, list=26%, signal=30% | TMEM98||BCL3||CRACR2A||NFKBID||FOXP1||ENTPD7||HLX||IL4R||ZBTB7B||RUNX3||IL18R1||ZFPM1||PRKCZ||CTSL||JAK3||LOXL3||RELB||IL6R||SOCS1||PTGER4||IL23A | 3.3428463 | 4.6067941 | 3.5646670 | 4.580416 | 3.20861659 | 3.2663306 | 4.7015863 | 4.5929900 | 3.3720038 | 3.46186821 | 3.18052428 | 4.571450 | 4.6023382 | 4.6456307 | 3.9518316 | 3.3399819 | 3.3069280 | 4.514035 | 4.5859445 | 4.6385680 | 3.10478642 | 3.3197572 | 3.19318255 | 3.114232673 | 3.1269316 | 3.5199107 | 4.6964031 | 4.6897172 | 4.7184812 | 4.6382503 | 4.5085639 | 4.6285732 |
| GO:0000132 | establishment of mitotic spindle orientation | 25 | 0.5278670 | 1.477791 | 0.0490596893 | 0.6887618 | 0.6740161 | 4171 | tags=44%, list=26%, signal=33% | SPRY1||SPRY2||PAX6||SPDL1||CDK5RAP2||NDC80||GPSM2||NDEL1||HTT||PAFAH1B1||MAD2L1 | 4.9046131 | 5.6669090 | 5.1532744 | 5.765520 | 4.87663359 | 5.1476881 | 5.7287579 | 5.8037503 | 4.8675840 | 4.99483201 | 4.84693359 | 5.656794 | 5.6672460 | 5.6766191 | 5.3167615 | 5.0556700 | 5.0723099 | 5.719855 | 5.7301991 | 5.8432317 | 4.87053946 | 4.8766049 | 4.88273067 | 5.126910827 | 5.1175273 | 5.1973014 | 5.7281808 | 5.7421037 | 5.7158696 | 5.7951357 | 5.8088706 | 5.8072057 |
| GO:0051797 | regulation of hair follicle development | 13 | -0.6507147 | -1.546740 | 0.0491221148 | 0.6887618 | 0.6740161 | 3100 | tags=46%, list=19%, signal=37% | WNT5A||CDH3||NGFR||FST||TGFB2||HPSE | 4.3330326 | 4.0626804 | 4.2875705 | 4.108382 | 4.30919402 | 4.5334005 | 4.0457662 | 4.0717095 | 4.3140515 | 4.25389434 | 4.42584562 | 4.076339 | 4.0909849 | 4.0197403 | 4.1212209 | 4.4153976 | 4.3108381 | 4.145201 | 4.0874382 | 4.0917879 | 4.36457764 | 4.1535210 | 4.39754067 | 4.464571638 | 4.5640230 | 4.5692152 | 4.0854079 | 4.0351377 | 4.0158564 | 4.0646447 | 4.0999283 | 4.0500993 |
| GO:0060055 | angiogenesis involved in wound healing | 18 | -0.5970187 | -1.527093 | 0.0499901594 | 0.6887618 | 0.6740161 | 2926 | tags=56%, list=18%, signal=45% | XBP1||ADIPOR2||MCAM||TNFAIP3||DAG1||PRCP||NDNF||GPX1||CXCR4||HPSE | 6.2147158 | 5.6877831 | 5.9791922 | 5.570715 | 6.32337153 | 6.3650884 | 5.5044722 | 5.5135582 | 6.1669264 | 6.05621462 | 6.39947474 | 5.807832 | 5.6474349 | 5.5997560 | 5.8129807 | 6.0730112 | 6.0380334 | 5.637504 | 5.5175377 | 5.5544734 | 6.31033166 | 6.1718617 | 6.47222762 | 6.367935270 | 6.5419562 | 6.1601867 | 5.5476349 | 5.4653915 | 5.4992038 | 5.5068502 | 5.5459200 | 5.4872852 |
GO: MF
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0003678 | DNA helicase activity | 70 | 0.5304455 | 1.812412 | 0.003332747 | 0.1331817 | 0.1218409 | 5908 | tags=64%, list=37%, signal=41% | NAV2||HELB||POLQ||CHD9||ATRX||CHD7||CHD3||PIF1||CHD6||WRN||DHX36||MCM8||CHD2||CHD1||SETX||IGHMBP2||ASCC3||FANCM||HFM1||CHD8||BLM||SMARCAD1||DDX11||CHD1L||MCM9||RAD54L2||CHTF18||RECQL5||DQX1||BRIP1||SUPV3L1||DDX3X||MCM4||HELQ||RAD54L||DHX9||TWNK||MRE11||G3BP1||MCM3||GTF2F2||ERCC3||RAD54B||RFC2||CHTF8 | 5.606440 | 6.153593 | 5.952037 | 6.2479217 | 5.523164 | 5.681108 | 6.211893 | 6.338619 | 5.593290 | 5.690022 | 5.5315715 | 6.153681 | 6.160431 | 6.146633 | 6.141674 | 5.806698 | 5.886149 | 6.212123 | 6.2146037 | 6.3146495 | 5.516757 | 5.555384 | 5.496732 | 5.611749 | 5.5862861 | 5.832385 | 6.219637 | 6.201356 | 6.214625 | 6.3183422 | 6.3600928 | 6.3371202 |
| GO:0140658 | ATP-dependent chromatin remodeler activity | 32 | 0.6286136 | 1.852208 | 0.003955037 | 0.1521957 | 0.1392358 | 4550 | tags=66%, list=29%, signal=47% | CHD9||ERCC6L2||ATRX||CHD7||CHD3||SMARCA2||CHD6||CHD2||CHD1||SMARCA5||EP400||ERCC6||TTF2||CHD8||SMARCAD1||CHD1L||RAD54L2||SMARCAL1||BTAF1||SHPRH||HLTF | 4.138762 | 4.811132 | 4.640808 | 4.9801906 | 4.015192 | 4.480016 | 4.924076 | 5.004691 | 4.120413 | 4.166233 | 4.1292290 | 4.733929 | 4.854859 | 4.841585 | 4.885815 | 4.505534 | 4.495849 | 4.883793 | 4.9764840 | 5.0740212 | 4.023054 | 3.991026 | 4.031184 | 4.403738 | 4.3559395 | 4.661319 | 4.941748 | 4.934708 | 4.895340 | 4.9898204 | 5.0313525 | 4.9925237 |
| GO:0008201 | heparin binding | 83 | -0.4969793 | -1.709516 | 0.004738438 | 0.1747524 | 0.1598717 | 2623 | tags=42%, list=16%, signal=35% | PCSK6||COL25A1||MDK||APP||LXN||COMP||APOE||COL13A1||NDNF||FSTL1||APLP2||SMOC1||PTCH1||BMP4||FGFBP3||ADAMTS8||PCOLCE2||LRPAP1||ADAMTS5||SERPINE2||FGFRL1||APLP1||RSPO2||SFRP1||NRP1||ANG||RSPO4||NELL1||SLIT3||THBS4||RTN4R||ADAMTS15||CRISPLD2||FBLN7||ADAMTSL5 | 6.530156 | 6.385857 | 6.334810 | 6.3737440 | 6.492387 | 6.230217 | 6.341004 | 6.381575 | 6.550816 | 6.570741 | 6.4668156 | 6.423729 | 6.371869 | 6.361196 | 6.355883 | 6.292132 | 6.355486 | 6.381123 | 6.3505529 | 6.3892682 | 6.545843 | 6.568430 | 6.353356 | 6.434650 | 6.1375129 | 6.094161 | 6.340711 | 6.354594 | 6.327581 | 6.3881125 | 6.3807967 | 6.3757891 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 95 | -0.4665893 | -1.638526 | 0.006829824 | 0.2087114 | 0.1909390 | 2857 | tags=40%, list=18%, signal=33% | GLA||GANAB||MAN2B1||MAN1C1||OGG1||MACROD1||MANEAL||MAN2B2||GLB1L2||IL18R1||MOGS||DNPH1||MYORG||IL1RAP||TIRAP||EDEM2||GBA||OTOG||SIGIRR||GAA||MAN1B1||HEXA||GLB1||SMPDL3B||MYD88||NEU1||CLN5||MPG||NEIL1||CTBS||HYAL2||GLB1L||NAGLU||SMPD1||ADPRHL1||TICAM1||HPSE||AMY2B | 4.931881 | 5.301023 | 5.290077 | 5.2486936 | 4.966918 | 5.207074 | 5.219008 | 5.232126 | 4.901522 | 4.907931 | 4.9846974 | 5.333530 | 5.315151 | 5.253166 | 5.316072 | 5.329663 | 5.222136 | 5.253033 | 5.2349787 | 5.2579680 | 4.953247 | 4.900301 | 5.043555 | 5.046938 | 5.1445075 | 5.405569 | 5.262538 | 5.208359 | 5.185027 | 5.2107481 | 5.2901312 | 5.1936494 |
| GO:0008528 | G protein-coupled peptide receptor activity | 43 | -0.5651610 | -1.730160 | 0.008988334 | 0.2267337 | 0.2074267 | 2676 | tags=44%, list=17%, signal=37% | ADCYAP1R1||SSTR2||GPR83||AGTRAP||S1PR2||GPR37||CX3CR1||F2RL1||UTS2R||RAMP2||CXCR4||PTH1R||EDNRB||NPY1R||CCR10||CCKBR||SORCS2||ACKR2||CYSLTR1 | 3.751502 | 3.655741 | 3.864867 | 3.5733180 | 3.819804 | 4.050484 | 3.548929 | 3.394394 | 3.667907 | 3.684073 | 3.8915737 | 3.633534 | 3.709517 | 3.622610 | 3.675794 | 4.047454 | 3.847323 | 3.608168 | 3.5953351 | 3.5146825 | 3.865344 | 3.649204 | 3.930110 | 3.947755 | 4.0409214 | 4.155273 | 3.613047 | 3.524875 | 3.506604 | 3.4184766 | 3.4361110 | 3.3261955 |
| GO:0008227 | G protein-coupled amine receptor activity | 10 | -0.7862008 | -1.731078 | 0.009165367 | 0.2267337 | 0.2074267 | 2077 | tags=60%, list=13%, signal=52% | ADRA2C||HTR7||HTR6||CHRM3||ADRB2||CHRM4 | 1.465476 | 1.329704 | 1.566450 | 0.8373223 | 1.379955 | 1.709401 | 1.161910 | 0.505449 | 1.432513 | 1.334111 | 1.6155442 | 1.323664 | 1.339855 | 1.325538 | 1.443474 | 1.726215 | 1.514458 | 1.014316 | 0.7363528 | 0.7435486 | 1.402577 | 1.122109 | 1.578903 | 1.461444 | 1.7228877 | 1.909175 | 1.193472 | 1.174089 | 1.117082 | 0.5223486 | 0.5123174 | 0.4813654 |
| GO:0106310 | protein serine kinase activity | 324 | 0.3185317 | 1.342436 | 0.010737948 | 0.2373301 | 0.2171208 | 4547 | tags=41%, list=28%, signal=30% | RSKR||ALPK3||LRRK1||TRPM6||SIK1||PRKG2||ATM||MAPK13||PRKAA1||KSR1||PLK4||PIM1||DAPK2||TAOK1||PLK3||STK33||SMG1||CLK2||CDK3||CDK19||DAPK1||CDK10||MYO3A||RPS6KA5||MAP3K5||DYRK1B||STK11||MAPK15||MAP3K4||CDKL5||PASK||TP53RK||KSR2||MASTL||GSK3B||SGK1||RPS6KA3||ALPK1||MARK3||STK36||MAP2K4||CDK14||CDK12||CIT||MARK2||TLK1||TRIO||WNK1||CAMKK1||TTK||CSNK1G1||PHKG1||ROCK1||CDK13||MAP3K21||MAP3K1||STK4||PKN3||PRKCE||BRAF||RPS6KA6||CDC42BPA||UHMK1||PAK3||STK24||TNK2||LATS1||MAST2||PRKAA2||EIF2AK2||MAPK6||NEK11||MKNK1||OXSR1||HIPK1||HIPK2||CAMK1D||MARK1||SLK||CDC42BPB||MAP4K5||STK38L||ATR||MAP3K2||RIOK3||NUAK2||MAP3K3||CDK11A||CDK6||CSNK1D||ROCK2||DYRK2||CDK8||MAK||PIK3CA||IKBKB||MAP3K13||BUB1||ULK1||PRKX||AAK1||CHEK2||DYRK1A||SRPK2||CDK17||DCAF1||DMPK||PRKCA||MAP3K15||PDIK1L||AURKB||WNK2||PRPF4B||ERN1||TTBK2||SIK3||TRPM7||PIKFYVE||STK3||MTOR||CSNK1A1||MAPK10||WNK4||PRKDC||MAPK8||CAMK2D||PKN2||SRPK1||EIF2AK4||MAP3K8||STK17B||AKT3||RPS6KC1||CHEK1 | 4.406792 | 4.769352 | 4.684613 | 4.7569462 | 4.429787 | 4.748363 | 4.814029 | 4.807047 | 4.311767 | 4.386836 | 4.5144546 | 4.758396 | 4.780014 | 4.769564 | 4.759180 | 4.689168 | 4.601155 | 4.733351 | 4.7218591 | 4.8138754 | 4.437443 | 4.285486 | 4.553932 | 4.644199 | 4.7493981 | 4.844542 | 4.838560 | 4.803317 | 4.799892 | 4.7872509 | 4.8419901 | 4.7912516 |
| GO:0015464 | acetylcholine receptor activity | 11 | -0.7586600 | -1.703966 | 0.010948192 | 0.2373301 | 0.2171208 | 1199 | tags=45%, list=8%, signal=42% | CHRNB1||CHRM3||CHRM4||CHRNB4||CHRNA4 | 1.238559 | 2.726005 | 1.811757 | 2.7143386 | 1.092616 | 1.225124 | 2.663522 | 2.625017 | 1.365073 | 1.425777 | 0.8610773 | 2.736299 | 2.748360 | 2.692766 | 2.152542 | 1.694897 | 1.510173 | 2.565283 | 2.8183903 | 2.7476686 | 1.058633 | 1.212428 | 0.998253 | 0.883703 | 0.9758263 | 1.678700 | 2.614076 | 2.749402 | 2.623081 | 2.5928095 | 2.6805456 | 2.6000418 |
| GO:0001540 | amyloid-beta binding | 56 | -0.5139052 | -1.652636 | 0.012761905 | 0.2599925 | 0.2378535 | 3555 | tags=46%, list=22%, signal=36% | FBXO2||PRNP||LDLRAD3||APBA3||BACE1||MAPK8IP2||BCHE||EPHA4||COL25A1||NLGN1||CLSTN1||CST3||APOE||ITM2C||LRPAP1||NGFR||EPHB2||ATP1A3||ACHE||LRP1||SCARB1||ADRB2||CLU||ITGB2||TGFB2||APBA1 | 5.108737 | 5.401905 | 5.224367 | 5.4180662 | 5.083161 | 5.105246 | 5.332906 | 5.379891 | 5.142314 | 5.149164 | 5.0317494 | 5.400756 | 5.419483 | 5.385272 | 5.270982 | 5.220957 | 5.179713 | 5.414363 | 5.4418569 | 5.3976329 | 5.057579 | 5.174763 | 5.012230 | 5.020050 | 5.0705450 | 5.217759 | 5.359696 | 5.324936 | 5.313686 | 5.3862123 | 5.4057338 | 5.3471103 |
| GO:0017147 | Wnt-protein binding | 23 | -0.6412430 | -1.715708 | 0.014357780 | 0.2762543 | 0.2527305 | 2576 | tags=57%, list=16%, signal=47% | FZD7||PORCN||WLS||FZD6||ROR2||LRP5||FZD8||ROR1||SFRP1||FZD2||FZD9||FZD10||TRABD2A | 2.569411 | 3.809682 | 3.157106 | 3.6571098 | 2.528379 | 2.885036 | 3.687437 | 3.608848 | 2.531325 | 2.679148 | 2.4908806 | 3.847369 | 3.852558 | 3.725575 | 3.266165 | 3.119125 | 3.079240 | 3.622149 | 3.7030089 | 3.6449629 | 2.580161 | 2.478843 | 2.524347 | 2.715503 | 2.6012264 | 3.251792 | 3.696102 | 3.718878 | 3.646382 | 3.5993412 | 3.6656611 | 3.5595439 |
| GO:0005044 | scavenger receptor activity | 20 | -0.6615936 | -1.723461 | 0.015718999 | 0.2878897 | 0.2633751 | 3771 | tags=80%, list=24%, signal=61% | LOXL4||COLEC12||SCARB2||CXCL16||SCARF2||SCARA3||SSC4D||LGALS3BP||LRP1||LOXL2||SCARB1||MEGF10||LOXL3||ENPP2||PRSS12||ACKR2 | 2.817423 | 3.341300 | 3.131827 | 2.8839462 | 2.847592 | 3.162150 | 3.147306 | 2.723207 | 2.696970 | 2.838946 | 2.9083723 | 3.337013 | 3.378015 | 3.308015 | 3.007127 | 3.331486 | 3.033905 | 2.903004 | 2.9414559 | 2.8039109 | 2.883000 | 2.632284 | 3.003071 | 2.903427 | 3.0502625 | 3.471088 | 3.200875 | 3.137183 | 3.102117 | 2.7350497 | 2.7958465 | 2.6341210 |
| GO:0042165 | neurotransmitter binding | 11 | -0.7409282 | -1.664140 | 0.016422287 | 0.2878897 | 0.2633751 | 1199 | tags=45%, list=8%, signal=42% | CHRNB1||CHRM3||ACHE||CHRNB4||CHRNA4 | 3.353003 | 3.787252 | 3.522380 | 3.6046310 | 3.360163 | 3.471292 | 3.744162 | 3.618473 | 3.380465 | 3.526664 | 3.1233931 | 3.812713 | 3.767392 | 3.781278 | 3.522046 | 3.554221 | 3.490161 | 3.615110 | 3.5360169 | 3.6600439 | 3.408544 | 3.405107 | 3.262048 | 3.540334 | 3.1655866 | 3.662771 | 3.783702 | 3.683912 | 3.762963 | 3.6089148 | 3.6363089 | 3.6100274 |
| GO:0005540 | hyaluronic acid binding | 12 | -0.7233611 | -1.662654 | 0.016656869 | 0.2878897 | 0.2633751 | 1724 | tags=33%, list=11%, signal=30% | SUSD5||HAPLN3||HYAL2||HAPLN4 | 5.671847 | 6.057610 | 5.779523 | 6.2547326 | 5.399473 | 4.921439 | 6.066267 | 6.278316 | 5.791223 | 6.037311 | 4.9947890 | 6.158984 | 6.004153 | 6.004088 | 5.836061 | 5.509390 | 5.956853 | 6.232924 | 6.2183537 | 6.3111821 | 5.228090 | 5.814885 | 5.037926 | 4.901518 | 4.7391290 | 5.100851 | 6.026090 | 6.059043 | 6.112351 | 6.2433347 | 6.2660051 | 6.3243906 |
| GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | 24 | -0.6277443 | -1.694319 | 0.016870273 | 0.2878897 | 0.2633751 | 3512 | tags=46%, list=22%, signal=36% | DHCR24||TM7SF2||BLVRB||ZADH2||DPYD||SRD5A3||DHCR7||TECR||DHDH||ENSG00000258653||CYP2S1 | 6.325299 | 6.100203 | 6.389811 | 6.2147180 | 6.385407 | 6.569210 | 6.025755 | 6.091554 | 6.259906 | 6.260220 | 6.4475373 | 6.110399 | 6.121731 | 6.067925 | 6.227248 | 6.528569 | 6.397877 | 6.258891 | 6.1815536 | 6.2025969 | 6.393001 | 6.288950 | 6.468647 | 6.507379 | 6.5571530 | 6.639977 | 6.075579 | 6.001975 | 5.998389 | 6.0962115 | 6.1330517 | 6.0440145 |
| GO:0030374 | nuclear receptor coactivator activity | 56 | 0.4739609 | 1.560197 | 0.017255892 | 0.2878897 | 0.2633751 | 5342 | tags=59%, list=33%, signal=39% | ACTN2||CCDC62||TMF1||PPARGC1B||BRD8||HELZ2||CCAR1||PPARGC1A||NCOA3||MED1||ZMIZ1||SFR1||MED13||ATXN7L3||PPRC1||FGF2||MED12||MED24||ENY2||NCOA2||THRAP3||MED4||CNOT6||MED17||DCAF6||SLC30A9||RNF4||RBM14||NCOA6||CNOT9||PRKCB||PSMC3IP||ELK1 | 5.239857 | 5.529761 | 5.429663 | 5.5905858 | 5.241039 | 5.506527 | 5.589688 | 5.660480 | 5.183719 | 5.241204 | 5.2925933 | 5.510157 | 5.548367 | 5.530506 | 5.517687 | 5.424089 | 5.341841 | 5.589651 | 5.5443310 | 5.6363093 | 5.264999 | 5.144746 | 5.308429 | 5.445915 | 5.4986458 | 5.572225 | 5.616105 | 5.577302 | 5.575290 | 5.6600209 | 5.6846969 | 5.6363169 |
| GO:0052742 | phosphatidylinositol kinase activity | 17 | 0.6540829 | 1.668754 | 0.017486114 | 0.2878897 | 0.2633751 | 4068 | tags=65%, list=25%, signal=48% | ATM||PIK3CD||PI4KAP1||PIK3C2A||PIK3C2B||PI4KA||PIP5K1B||PIK3CA||PIP5K1A||PI4KAP2||PIKFYVE | 4.093095 | 4.383612 | 4.725927 | 4.5364928 | 4.064925 | 4.476856 | 4.501843 | 4.623976 | 4.019527 | 4.162263 | 4.0939647 | 4.394038 | 4.374530 | 4.382201 | 4.874806 | 4.671469 | 4.618646 | 4.496750 | 4.4858506 | 4.6228220 | 4.053819 | 3.982796 | 4.153075 | 4.235350 | 4.2839692 | 4.832193 | 4.532938 | 4.495046 | 4.476980 | 4.5861110 | 4.6603055 | 4.6245580 |
| GO:0070182 | DNA polymerase binding | 21 | -0.6485604 | -1.706455 | 0.017649341 | 0.2878897 | 0.2633751 | 6 | tags=14%, list=0%, signal=14% | ACD||LONP1||RMRP | 6.853620 | 7.900299 | 6.928565 | 8.0042844 | 6.757980 | 6.820738 | 7.968820 | 8.267172 | 6.856407 | 6.975147 | 6.7178465 | 7.917643 | 7.872656 | 7.910195 | 7.321945 | 6.607160 | 6.754338 | 7.962609 | 7.9639019 | 8.0829988 | 6.736065 | 6.824068 | 6.711358 | 6.891002 | 6.8444549 | 6.721484 | 7.942470 | 7.972746 | 7.990831 | 8.0605113 | 8.6066723 | 8.0631745 |
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 913 | 0.2620190 | 1.197849 | 0.018079959 | 0.2878897 | 0.2633751 | 3226 | tags=28%, list=20%, signal=24% | PRRX1||NLRC5||ZNF114||ONECUT1||ZNF471||BHLHE41||ZNF492||HOXD3||ZNF888||ZNF813||ZNF230||ZNF682||ZNF586||NFIA||FOXD4L5||ZNF283||ENSG00000273046||ZNF17||ZNF587||FOSL2||ZNF345||ZNF780B||HOXD4||ZNF571||SNAI3||DACH2||ZNF548||HOXD9||FOXD2||MTF1||ZNF611||ZNF134||GATA5||NR2C2||CLOCK||ZNF525||HOXB7||KLF14||ZNF112||VDR||ZNF547||ZIC3||ZNF416||ZNF91||ZNF224||HNF4G||ZNF780A||ZNF223||ZNF516||ZNF502||ZNF670||ONECUT2||ZNF782||ZBTB37||WT1||SIX4||DMTF1||FOXF1||ZNF493||ZNF578||POU2F1||ZNF136||ARNTL||ZNF236||MXD1||ZNF573||ZNF256||ZNF143||ZBTB17||ZNF417||ZNF470||ZNF713||EMX2||OTX2||ZNF280D||ZNF852||CHD7||ZNF225||NFYB||TCF12||ZNF304||USP3||REL||ZNF675||ZNF506||BMI1||ZNF490||ZNF551||NKX2-2||MYT1||ZNF549||THAP1||STAT2||TGIF1||MLXIPL||ZNF100||PLAG1||GLI4||ZNF500||ZNF772||JDP2||RXRB||ZNF382||MEF2C||HIVEP1||ZFHX4||ZNF641||MAFF||BPTF||ZNF354B||CCAR1||ZNF543||ZNF680||ZNF333||EBF2||ZNF805||SP9||ZSCAN12||ZNF16||ZNF431||ZNF483||ZNF639||ZNF613||NR1D2||POU3F2||ZNF28||RREB1||ZNF449||ZNF695||ZNF726||NR6A1||ZNF268||GABPA||KLF6||ZKSCAN8||SP1||ZKSCAN1||ZNF117||PAX6||ZNF430||HDAC6||PAX3||KLF5||ZBTB11||AHR||NFAT5||ZNF776||MED1||ZNF277||ZNF66||PGBD1||ZNF790||ZNF438||ZIC5||OLIG1||DHX36||ZNF331||ZNF24||ZNF721||HOXD10||EBF4||MGA||IRF4||LHX3||KLF11||ZNF770||HOXA9||ZNF320||ZGPAT||IKZF4||ZNF175||ZNF211||ZNF84||FOXO1||NFATC4||ATF7||EPAS1||SMAD5||HOXC9||TFAP2E||GMEB1||ZNF567||ZNF217||ZNF263||ATF2||NR1H2||FOXQ1||HOXB3||TFAP2C||ZNF655||POU4F1||CHD2||ESRRG||ZNF597||GATA6||ZNF577||MZF1||ZNF131||EN1||ARNTL2||ZNF227||HIF1A||SOX2||ELK4||ZNF367||ZNF514||TFCP2L1||MECOM||NFATC3||ZSCAN9||KLF3||ZNF704||ZNF26||ATF1||FOXC2||HBP1||ZNF692||ZBTB20||HIVEP2||NSD1||ZNF740||ZSCAN26||BHLHE40||GMEB2||AR||KLF9||E2F8||MLXIP||TBX15||TBX19||SP3||ZNF146||E2F3||FOXD1||FOXK1||ZKSCAN4||ZNF784||IRF2||ZNF829||NRIP1||PITX2||ZBTB40||KDM6B||ZNF121||ZBTB49||RORA||KLF12||TCF7L2||SMAD3||HOXA1||ZNF420||MED12||ZNF841||OTX1||GRHL1||FOXO3 | 3.881732 | 4.313721 | 4.131958 | 4.3405014 | 3.843948 | 4.063664 | 4.353367 | 4.366539 | 3.857593 | 3.966056 | 3.8174198 | 4.309472 | 4.316368 | 4.315313 | 4.268328 | 4.035825 | 4.081048 | 4.325910 | 4.3036956 | 4.3904831 | 3.819524 | 3.842931 | 3.868967 | 3.988910 | 4.0278765 | 4.167996 | 4.355701 | 4.347846 | 4.356538 | 4.3800435 | 4.3365197 | 4.3825901 |
| GO:0035198 | miRNA binding | 32 | 0.5423739 | 1.598103 | 0.018109908 | 0.2878897 | 0.2633751 | 6377 | tags=75%, list=40%, signal=45% | ZC3H7A||AGO3||SPOUT1||SOX2||MATR3||NEAT1||RC3H1||DDX21||ZNF346||AGO1||MATR3||RBM10||PUM2||TUT7||AGO4||HNRNPA2B1||TRIM71||FMR1||KCNQ1OT1||TARBP2||AGO2||HNRNPA1||TUT4||PUM1 | 6.785516 | 7.697708 | 6.904595 | 7.7097285 | 6.682667 | 6.889441 | 7.774202 | 7.749033 | 6.763833 | 6.953565 | 6.6196252 | 7.679301 | 7.693891 | 7.719643 | 7.239150 | 6.660910 | 6.743630 | 7.696962 | 7.6724321 | 7.7584259 | 6.655305 | 6.739459 | 6.651511 | 6.865618 | 6.8514288 | 6.949324 | 7.765896 | 7.759483 | 7.796948 | 7.7335155 | 7.7585693 | 7.7548877 |
| GO:0050840 | extracellular matrix binding | 34 | -0.5685022 | -1.654356 | 0.018237666 | 0.2878897 | 0.2633751 | 3296 | tags=56%, list=21%, signal=44% | THSD1||ITGB1||ITGA9||ITGAV||CCN1||NID1||FBLN2||DAG1||ITGA2B||SMOC1||ADAMTS5||OLFML2A||GPC1||OLFML2B||SPARC||ACHE||LGALS1||LYPD3||ADAMTS15 | 7.671173 | 6.944655 | 7.423217 | 6.8941467 | 7.749410 | 7.469746 | 6.864881 | 6.912620 | 7.645361 | 7.558038 | 7.7996721 | 7.085914 | 6.872626 | 6.864312 | 7.230224 | 7.491004 | 7.530375 | 6.935711 | 6.8425267 | 6.9026600 | 7.761837 | 7.680461 | 7.803241 | 7.598984 | 7.4278723 | 7.372616 | 6.858501 | 6.862912 | 6.873192 | 6.9121308 | 6.8769161 | 6.9479376 |
| GO:0005125 | cytokine activity | 61 | -0.4817433 | -1.570439 | 0.018564593 | 0.2878897 | 0.2633751 | 3100 | tags=49%, list=19%, signal=40% | CXCL16||WNT5A||TGFB1||BMP1||WNT3||GDF1||BMP6||CXCL12||CXCL3||MIF||FAM3C||BMP4||INHBB||WNT3A||CMTM3||CRLF1||TIMP1||GRN||WNT7B||SECTM1||INHBC||WNT9A||CTF1||WNT2B||C5||CMTM1||WNT5B||TGFB2||IL23A||GDF9 | 6.618196 | 6.112163 | 6.241935 | 5.9564464 | 6.692266 | 6.789868 | 6.004907 | 5.852701 | 6.588811 | 6.399644 | 6.8332339 | 6.233083 | 6.037224 | 6.058055 | 5.991760 | 6.417572 | 6.284218 | 6.105315 | 5.8659594 | 5.8856231 | 6.699286 | 6.459355 | 6.886613 | 6.684779 | 7.0275798 | 6.623577 | 6.010668 | 5.999064 | 6.004966 | 5.8824802 | 5.8367001 | 5.8384561 |
| GO:0015026 | coreceptor activity | 27 | -0.5991068 | -1.661732 | 0.019425019 | 0.2968029 | 0.2715293 | 3296 | tags=59%, list=21%, signal=47% | ITGAV||GPC4||PTK7||ROR2||NECTIN1||IGSF1||NECTIN2||LRP5||ROR1||NGFR||CSPG4||LRP1||NRP1||ITGA4||RAMP2||CXCR4 | 3.001997 | 3.901060 | 3.397275 | 3.8226487 | 3.044329 | 3.428881 | 3.799839 | 3.761210 | 2.948049 | 2.901385 | 3.1448238 | 3.917844 | 3.971966 | 3.808599 | 3.423011 | 3.451415 | 3.313767 | 3.807413 | 3.8544576 | 3.8055411 | 3.017320 | 2.955580 | 3.152967 | 3.245368 | 3.3923545 | 3.623529 | 3.819758 | 3.815492 | 3.763592 | 3.7455131 | 3.8028318 | 3.7343444 |
| GO:0003756 | protein disulfide isomerase activity | 18 | -0.6619991 | -1.687593 | 0.020946470 | 0.3064764 | 0.2803791 | 2631 | tags=67%, list=16%, signal=56% | TMX3||ERO1A||PIGK||ERP44||TXNDC5||PDIA3||CRELD1||CRELD2||PDIA6||QSOX1||P4HB||PDIA4 | 5.978994 | 7.431027 | 6.382115 | 7.0419346 | 5.896213 | 6.170668 | 7.228145 | 6.933755 | 5.985440 | 6.117513 | 5.8185476 | 7.454422 | 7.478513 | 7.357314 | 6.480106 | 6.305982 | 6.354618 | 6.993186 | 7.0960750 | 7.0346814 | 5.936989 | 5.915946 | 5.833649 | 6.124861 | 5.9619930 | 6.392138 | 7.220451 | 7.242992 | 7.220878 | 6.9233705 | 6.9857057 | 6.8905655 |
| GO:0001618 | virus receptor activity | 46 | -0.5129301 | -1.594403 | 0.021973859 | 0.3064764 | 0.2803791 | 3608 | tags=57%, list=23%, signal=44% | ITGB1||DPP4||HSPA1B||SCARB2||EPHA2||ITGAV||NCAM1||PVR||LAMP1||EFNB2||CD55||EFNB3||SLC1A5||EGFR||DAG1||NECTIN1||NECTIN2||CD81||BSG||HYAL2||SIVA1||ITGB5||SCARB1||SLC52A2||CXCR4||CLDN1 | 7.871601 | 7.538277 | 7.684366 | 7.3212897 | 7.986093 | 7.809257 | 7.407841 | 7.294667 | 7.835958 | 7.780931 | 7.9897177 | 7.639624 | 7.512024 | 7.457058 | 7.470391 | 7.797982 | 7.762847 | 7.369987 | 7.2852181 | 7.3073178 | 8.022370 | 7.882380 | 8.048079 | 7.857602 | 7.7936078 | 7.775260 | 7.432860 | 7.394380 | 7.395955 | 7.2894578 | 7.2981363 | 7.2963922 |
| GO:0005161 | platelet-derived growth factor receptor binding | 10 | 0.7277816 | 1.623029 | 0.022363562 | 0.3064764 | 0.2803791 | 242 | tags=20%, list=2%, signal=20% | PDGFRA||IL1R1 | 1.560152 | 2.900571 | 2.597319 | 3.2306661 | 1.519223 | 2.333873 | 2.976076 | 3.337336 | 1.513024 | 1.711309 | 1.4424910 | 2.896706 | 2.928470 | 2.876053 | 2.952608 | 2.434013 | 2.324641 | 3.121012 | 3.2291017 | 3.3340276 | 1.467479 | 1.465416 | 1.619311 | 1.931984 | 2.0461770 | 2.843745 | 3.025818 | 2.965601 | 2.935333 | 3.2969969 | 3.4084928 | 3.3037886 |
| GO:0015293 | symporter activity | 79 | -0.4432095 | -1.513851 | 0.023596575 | 0.3064764 | 0.2803791 | 4538 | tags=61%, list=28%, signal=44% | MFSD2A||SLC6A15||SLC33A1||SLC12A9||SLC12A8||SLC6A9||SLC16A1||SLC36A4||SLC10A7||SLC5A12||SLC39A8||SLC24A1||SLC38A3||MFSD12||SLC2A10||SLC5A5||SLC17A5||SLC16A6||SLC24A4||SLC39A14||SLC4A11||SLC4A4||SLC1A5||SLC22A5||SLC16A2||SLC15A4||SLC16A9||SLC6A17||CTNS||SLC16A14||MFSD4B||SLC1A4||MFSD3||SLC13A4||SLC6A13||SLC12A5||SLC1A1||SLC10A4||SLC10A3||SLC6A16||SLC12A4||SLC22A4||SLC4A5||SLC16A13||SLC16A4||SLC22A18||SLC16A7||SLC5A11 | 3.908813 | 4.618543 | 4.442965 | 4.7604727 | 3.924098 | 4.339704 | 4.552234 | 4.734136 | 3.865563 | 3.945620 | 3.9141304 | 4.594428 | 4.679669 | 4.579496 | 4.439323 | 4.505345 | 4.381564 | 4.723011 | 4.7986159 | 4.7587989 | 3.978225 | 3.815173 | 3.973020 | 4.192281 | 4.1894372 | 4.597987 | 4.554493 | 4.562306 | 4.539814 | 4.7161715 | 4.7823099 | 4.7026605 |
| GO:0042393 | histone binding | 218 | 0.3308948 | 1.340711 | 0.023656865 | 0.3064764 | 0.2803791 | 4844 | tags=41%, list=30%, signal=29% | CDYL2||MYSM1||KMT2D||RAG1||PHF1||KMT2A||FAM156B||ATRX||USP3||L3MBTL1||ZCWPW1||UHRF2||PHC3||DPF3||YEATS2||IPO9||BAZ2A||BPTF||USP16||ATAD2B||KDM7A||SMARCA2||KDM5A||ING2||USP49||KAT7||TONSL||PHIP||BRD9||RSF1||CHD2||KAT6B||KMT2C||CHD1||HJURP||TNKS||PHC1||PSME4||MLLT3||TP53BP1||ING3||PYGO1||SMARCA5||ING4||CKS2||SPIN2B||MORC3||NCAPD2||BRD7||MLLT6||BAZ1B||PHF6||SBNO1||SART3||SMARCC1||BRPF1||CHD8||RBBP5||SPIN3||NAP1L4||ZMYND11||DNAJC9||TRIM24||MTF2||ANP32E||IPO7||ATAD2||EZH1||TSPYL2||SAP30L||DNAJC2||MSH6||KAT6A||GLYR1||SIRT1||KDM1B||NASP||MCM3AP||TSPYL4||STAT1||SUZ12||TSPYL1||USP15||NCAPG2||UIMC1||TAF1||MSL3||DEK||SCMH1 | 5.325891 | 6.201242 | 5.545352 | 6.2220604 | 5.275555 | 5.511229 | 6.265126 | 6.288518 | 5.303873 | 5.380097 | 5.2921113 | 6.197124 | 6.189982 | 6.216490 | 5.786371 | 5.413490 | 5.402172 | 6.182170 | 6.1917290 | 6.2898035 | 5.252840 | 5.260320 | 5.312763 | 5.436111 | 5.4810969 | 5.610733 | 6.244301 | 6.257845 | 6.292797 | 6.2774258 | 6.2983268 | 6.2897251 |
| GO:0015298 | solute:cation antiporter activity | 27 | -0.5870747 | -1.628359 | 0.023892774 | 0.3064764 | 0.2803791 | 2888 | tags=44%, list=18%, signal=36% | SLC8A1||SLC24A4||SLC8B1||CLCN4||CLCN3||TMCO3||SLC22A4||SLC9A9||SLC8A2||SLC22A18||SLC8A3||SLC9A2 | 3.448766 | 4.140452 | 4.100309 | 4.2354816 | 3.499567 | 4.024890 | 4.070476 | 4.205406 | 3.324063 | 3.448768 | 3.5635396 | 4.138450 | 4.205938 | 4.073949 | 4.132705 | 4.173589 | 3.988123 | 4.217835 | 4.2751845 | 4.2125861 | 3.539129 | 3.327577 | 3.616705 | 3.904232 | 3.8969118 | 4.245368 | 4.073375 | 4.097447 | 4.040031 | 4.1727916 | 4.2943475 | 4.1446517 |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 336 | 0.3021483 | 1.275900 | 0.025753047 | 0.3190379 | 0.2918709 | 3226 | tags=30%, list=20%, signal=25% | PRRX1||ONECUT1||HOXD3||NFIA||FOSL2||ZNF345||ZNF780B||HOXD4||CSRNP3||FOXD2||MTF1||LHX4||NR2C2||CLOCK||HOXB7||ZNF112||ZIC3||ZNF91||CREBRF||ZNF224||HNF4G||ONECUT2||ZNF782||WT1||SIX4||DMTF1||FOXF1||ZNF493||ZNF143||ZBTB17||OTX2||ZNF225||NFYB||TCF12||REL||NKX2-2||PLAG1||RXRB||MEF2C||MAFF||EBF2||ZNF639||POU3F2||RREB1||ALX1||NR6A1||ZNF268||ZNF292||GABPA||KLF6||SP1||PAX6||KLF5||NFAT5||PRDM2||ZNF438||ZNF24||ZNF721||HOXD10||IRF4||LHX3||ZNF770||HOXA9||ZNF175||FOXO1||EPAS1||STOX2||TFAP2E||GMEB1||ATF2||NR1H2||HOXB3||TFAP2C||POU4F1||ESRRG||ZNF597||MZF1||VEZF1||ZNF227||HIF1A||SOX2||ELK4||TFCP2L1||MECOM||NFATC3||ATF1||FOXC2||GMEB2||AR||MLXIP||TBX19||E2F3||FOXD1||ZNF784||IRF2||SMAD3||HOXA1||ZNF841||OTX1||GRHL1||FOXO3 | 3.687290 | 3.960871 | 3.902201 | 4.0618363 | 3.685663 | 3.993177 | 4.015249 | 4.098594 | 3.642966 | 3.717742 | 3.7001061 | 3.954923 | 3.961958 | 3.965712 | 3.988531 | 3.877364 | 3.836383 | 4.067658 | 3.9987039 | 4.1167167 | 3.683584 | 3.625827 | 3.745110 | 3.926223 | 3.9648495 | 4.083749 | 4.029447 | 3.996738 | 4.019367 | 4.0923687 | 4.1092510 | 4.0941027 |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 15 | 0.6546565 | 1.623886 | 0.025793245 | 0.3190379 | 0.2918709 | 3739 | tags=53%, list=23%, signal=41% | ASH1L||KMT2D||KMT2A||SETD1A||KMT2C||SETD4||RBBP5||KMT2B | 4.487751 | 5.009249 | 5.133934 | 5.0933121 | 4.419544 | 4.948827 | 5.101024 | 5.165419 | 4.401256 | 4.586923 | 4.4689214 | 4.954482 | 5.044919 | 5.026767 | 5.274379 | 5.100799 | 5.014362 | 5.063141 | 5.0814921 | 5.1343523 | 4.463160 | 4.333272 | 4.458476 | 4.785719 | 4.7742178 | 5.236765 | 5.120711 | 5.119919 | 5.061652 | 5.1684602 | 5.1718465 | 5.1559003 |
| GO:0005509 | calcium ion binding | 394 | -0.3072726 | -1.282131 | 0.027027027 | 0.3244254 | 0.2967997 | 3574 | tags=31%, list=22%, signal=25% | NKD2||RYR1||CLGN||SYT2||NOTCH1||EFEMP1||PRNP||RCN1||TLL2||SELENON||LPCAT1||SULF2||LPCAT2||ITPR3||CANT1||DCHS1||EPDR1||CDH2||ADGRL3||DLL1||SYT11||NUCB2||PRKCSH||FBLN5||ANXA4||IDS||EHD1||BMP1||SLC8A1||SUSD1||EFHC1||EGFLAM||MAN1C1||RASGRP1||DSC2||CDH3||NID1||CDHR1||CLSTN3||DLL3||LTBP1||FBLN2||CALU||SRR||SULF1||CLSTN1||COMP||S100A11||DAG1||PADI2||CANX||EDEM2||FKBP10||HSP90B1||NUCB1||FSTL1||LTBP4||SMOC1||HSPA5||GSN||CDH15||CRELD1||NOTCH4||TUBB4A||CD320||RAB11FIP4||SYT3||MAN1B1||CRACR2B||PCDHGA7||CRELD2||RHBDL3||SDF4||PITPNM1||PLCD1||EGFL7||VLDLR||RCN3||CALR||NKD1||PROS1||GAS6||SCUBE1||CDH12||C1R||ANXA3||SPARC||CRTAC1||FBLN1||ARSA||PCDH1||LRP1||LOXL2||ENPP3||REPS2||RET||DLK2||RYR2||ADGRE5||CLSTN2||DNER||KCNIP3||TESC||ENPP2||NELL1||SLIT3||SYT15||CDH26||THBS4||GJB2||PCDHB2||MATN3||PCDHGA11||SPOCK3||FSTL4||RASGRP2||HMCN1||SYT17||PVALB||FBLN7||PRRG2||NCALD||PCDHGB7||PAMR1 | 4.826626 | 5.448337 | 4.924838 | 5.3245755 | 4.827636 | 4.928643 | 5.378546 | 5.320560 | 4.817222 | 4.846572 | 4.8158742 | 5.461110 | 5.470946 | 5.412273 | 4.968374 | 4.913683 | 4.891364 | 5.303859 | 5.3215833 | 5.3479426 | 4.807076 | 4.832985 | 4.842613 | 4.891935 | 4.9057769 | 4.986403 | 5.384180 | 5.380654 | 5.370770 | 5.2957002 | 5.3640337 | 5.3009373 |
| GO:0050431 | transforming growth factor beta binding | 17 | -0.6513890 | -1.638763 | 0.027621085 | 0.3254886 | 0.2977723 | 3696 | tags=65%, list=23%, signal=50% | LTBP3||TGFBR1||ITGAV||LTBP1||LTBP4||CD109||HYAL2||WFIKKN1||VASN||NRROS||LRRC32 | 2.499104 | 3.632152 | 3.362410 | 3.4357879 | 2.642598 | 3.221586 | 3.469251 | 3.335013 | 2.232250 | 2.501452 | 2.7222069 | 3.577556 | 3.727100 | 3.586881 | 3.285714 | 3.471048 | 3.323756 | 3.415981 | 3.4778097 | 3.4126348 | 2.566000 | 2.479579 | 2.854924 | 2.951961 | 3.0077784 | 3.609176 | 3.512605 | 3.488359 | 3.404576 | 3.3014879 | 3.4485388 | 3.2474328 |
| GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 10 | -0.7303920 | -1.608197 | 0.028666147 | 0.3254886 | 0.2977723 | 1866 | tags=60%, list=12%, signal=53% | LOXL1||LOX||LOXL2||LOXL3||MAOA||IL4I1 | 2.277528 | 2.602620 | 2.348974 | 2.0769958 | 2.324757 | 2.366692 | 2.458414 | 1.785211 | 2.302856 | 2.297851 | 2.2307589 | 2.659745 | 2.583232 | 2.563069 | 2.271378 | 2.470975 | 2.296260 | 2.121112 | 2.0685610 | 2.0401419 | 2.441070 | 2.213061 | 2.311044 | 2.247687 | 2.1204500 | 2.672474 | 2.440929 | 2.476484 | 2.457611 | 1.8420862 | 1.8100094 | 1.6997029 |
| GO:0004402 | histone acetyltransferase activity | 34 | 0.5149256 | 1.537859 | 0.029000626 | 0.3254886 | 0.2977723 | 5875 | tags=68%, list=37%, signal=43% | CLOCK||KAT2B||TADA2A||EPC1||NCOA3||KAT7||BRCA2||ATF2||KAT6B||MED24||EPC2||KAT6A||EP300||MCM3AP||TAF1||MSL3||SUPT7L||TAF9||GTF3C4||NAA40||KAT2A||CREBBP||KAT8 | 4.751224 | 5.409984 | 4.929008 | 5.3841807 | 4.744497 | 5.061012 | 5.476690 | 5.440775 | 4.721135 | 4.721876 | 4.8088841 | 5.370418 | 5.432304 | 5.426426 | 5.034657 | 4.918188 | 4.826625 | 5.378793 | 5.3357417 | 5.4362427 | 4.765018 | 4.635004 | 4.826873 | 4.986451 | 5.0590617 | 5.133762 | 5.483034 | 5.478100 | 5.468899 | 5.4312830 | 5.4602628 | 5.4305804 |
| GO:0008083 | growth factor activity | 75 | -0.4382527 | -1.486005 | 0.029556650 | 0.3254886 | 0.2977723 | 3451 | tags=43%, list=22%, signal=34% | EFEMP1||NRTN||CSPG5||FGF12||TGFB1||GFER||BMP1||GDNF||GDF1||MANF||MDK||BMP6||CXCL12||BMP4||CD320||INHBB||CLEC11A||FGF18||PDGFA||TIMP1||VEGFC||TYMP||GRN||FGF17||RABEP2||INHBC||THBS4||OSGIN1||TGFB2||GDF9||VGF||TGFA | 5.053083 | 4.838445 | 5.023100 | 4.6468873 | 5.114123 | 5.251340 | 4.752778 | 4.588090 | 4.980921 | 4.990709 | 5.1788675 | 4.852855 | 4.854022 | 4.807981 | 4.852572 | 5.142550 | 5.058951 | 4.695874 | 4.5895556 | 4.6532525 | 5.106034 | 5.020466 | 5.209640 | 5.165631 | 5.3069912 | 5.277574 | 4.786802 | 4.733128 | 4.737788 | 4.6052894 | 4.6414395 | 4.5145976 |
| GO:0072349 | modified amino acid transmembrane transporter activity | 17 | -0.6455166 | -1.623989 | 0.029760747 | 0.3254886 | 0.2977723 | 2466 | tags=41%, list=15%, signal=35% | SLC22A5||CTNS||SLC1A4||SLC19A1||SLC6A13||SLC46A1||SLC22A4 | 3.971691 | 4.672125 | 4.687597 | 4.7400496 | 4.078596 | 4.691021 | 4.592479 | 4.643486 | 3.832401 | 3.943467 | 4.1240801 | 4.602885 | 4.765331 | 4.643158 | 4.687087 | 4.773698 | 4.596573 | 4.722115 | 4.7829706 | 4.7140754 | 4.100823 | 3.836783 | 4.266101 | 4.448381 | 4.5597515 | 5.003424 | 4.654354 | 4.593521 | 4.526741 | 4.6246068 | 4.7105905 | 4.5926696 |
| GO:0015179 | L-amino acid transmembrane transporter activity | 38 | -0.5223297 | -1.557955 | 0.031028539 | 0.3310323 | 0.3028440 | 4610 | tags=55%, list=29%, signal=39% | SERINC5||SLC47A1||SLC6A9||SLC36A4||SLC38A7||SLC38A3||SLC25A29||SLC7A1||SERINC3||SLC1A5||SLC7A5||SLC15A4||SLC43A1||SLC38A6||CTNS||SLC1A4||SLC66A1||OCA2||SLC1A1||SLC43A2||SLC7A3 | 4.497518 | 5.243346 | 5.011307 | 5.3859285 | 4.538420 | 4.966047 | 5.160972 | 5.345279 | 4.406526 | 4.512442 | 4.5689026 | 5.231135 | 5.313785 | 5.182034 | 4.973998 | 5.113103 | 4.940979 | 5.361112 | 5.4068560 | 5.3894482 | 4.551688 | 4.431862 | 4.625148 | 4.830093 | 4.8680208 | 5.174718 | 5.178275 | 5.160491 | 5.143945 | 5.3384470 | 5.3889446 | 5.3072660 |
| GO:0015020 | glucuronosyltransferase activity | 15 | -0.6675336 | -1.621830 | 0.032245456 | 0.3317132 | 0.3034669 | 3053 | tags=80%, list=19%, signal=65% | LARGE1||LARGE2||CHSY1||CHPF2||EXT2||B3GAT3||CHSY3||B4GAT1||CHPF||B3GAT1||UGT3A2||CSGALNACT1 | 4.544688 | 4.667291 | 5.052618 | 4.6372298 | 4.607669 | 4.985737 | 4.454522 | 4.363487 | 4.496970 | 4.454884 | 4.6728290 | 4.677342 | 4.751029 | 4.567623 | 4.908374 | 5.178787 | 5.058026 | 4.669789 | 4.6413462 | 4.5996938 | 4.590006 | 4.487000 | 4.735179 | 4.763410 | 4.8521095 | 5.285897 | 4.512394 | 4.467025 | 4.381079 | 4.3706779 | 4.4116657 | 4.3061626 |
| GO:0008146 | sulfotransferase activity | 36 | -0.5270311 | -1.555330 | 0.033135415 | 0.3342495 | 0.3057872 | 2761 | tags=47%, list=17%, signal=39% | HS6ST2||CHST10||CHST11||CHST8||GAL3ST4||HS6ST1||TPST1||ENSG00000272916||CHST12||WSCD1||CHST14||CHST7||CHST2||CHST1||HS3ST3A1||SULT1A4||GAL3ST1 | 2.589080 | 3.243257 | 3.125035 | 3.1091630 | 2.623206 | 3.004508 | 3.135684 | 2.932654 | 2.563942 | 2.534991 | 2.6650508 | 3.213805 | 3.312740 | 3.200604 | 3.114083 | 3.207837 | 3.048739 | 3.061721 | 3.1576423 | 3.1065285 | 2.637863 | 2.463085 | 2.753923 | 2.735992 | 2.8822615 | 3.327672 | 3.177279 | 3.141001 | 3.087356 | 2.9422311 | 2.9113591 | 2.9441379 |
| GO:0050321 | tau-protein kinase activity | 22 | 0.5763639 | 1.561621 | 0.033853098 | 0.3382055 | 0.3094064 | 3967 | tags=64%, list=25%, signal=48% | PRKAA1||PHKG2||TAOK1||GSK3B||MARK3||MARK2||PHKG1||ROCK1||MARK1||CSNK1D||ROCK2||DYRK1A||TTBK2||SIK3 | 4.593853 | 4.882117 | 5.083075 | 4.8523573 | 4.586106 | 5.071982 | 4.976076 | 4.875092 | 4.510321 | 4.592689 | 4.6739120 | 4.861735 | 4.895491 | 4.888904 | 5.089986 | 5.145696 | 5.010345 | 4.834580 | 4.8057954 | 4.9144865 | 4.614619 | 4.389662 | 4.733203 | 4.886863 | 4.9996849 | 5.297629 | 5.032600 | 4.958108 | 4.935726 | 4.8576218 | 4.9115005 | 4.8554535 |
| GO:0015081 | sodium ion transmembrane transporter activity | 85 | -0.4211763 | -1.452575 | 0.034581037 | 0.3421876 | 0.3130494 | 1412 | tags=21%, list=9%, signal=19% | SCN2A||SLC13A4||SLC6A13||SLC1A1||ATP1A3||SLC10A4||SLC10A3||SCN4B||GRIK4||SLC4A5||KCNK1||SLC9A9||SLC8A2||HCN4||SLC8A3||SLC9A2||SLC5A11||SHROOM2 | 3.493869 | 4.292277 | 4.130613 | 4.4570454 | 3.484446 | 3.966553 | 4.229957 | 4.429310 | 3.431842 | 3.570185 | 3.4761021 | 4.279566 | 4.355723 | 4.239102 | 4.198384 | 4.146588 | 4.042527 | 4.395709 | 4.5173523 | 4.4555101 | 3.526284 | 3.401600 | 3.522014 | 3.809941 | 3.7427596 | 4.284505 | 4.234853 | 4.255760 | 4.198680 | 4.4106320 | 4.4830473 | 4.3926552 |
| GO:0005005 | transmembrane-ephrin receptor activity | 16 | -0.6525498 | -1.610823 | 0.035104923 | 0.3440945 | 0.3147939 | 3503 | tags=75%, list=22%, signal=59% | EPHA2||EPHA3||EPHA4||EFNB3||EPHB4||EPHA8||EPHA5||EPHB1||EPHB6||EPHB2||EFNA4||EPHB3 | 2.695031 | 3.185308 | 3.152433 | 2.9903073 | 2.778987 | 3.186452 | 3.024201 | 2.889017 | 2.572068 | 2.639015 | 2.8582511 | 3.162295 | 3.261306 | 3.129020 | 2.988501 | 3.328600 | 3.119651 | 3.050279 | 2.9508666 | 2.9678032 | 2.808521 | 2.541328 | 2.956837 | 3.033590 | 3.1171122 | 3.384897 | 3.100249 | 2.965420 | 3.003566 | 2.8466457 | 2.9625868 | 2.8548931 |
| GO:0005249 | voltage-gated potassium channel activity | 47 | -0.4840951 | -1.510115 | 0.036833112 | 0.3529077 | 0.3228567 | 2087 | tags=51%, list=13%, signal=45% | LRRC26||KCNN1||HCN2||KCNH4||KCNJ12||KCND3||KCNJ8||KCNB2||HCN3||KCNMA1||KCNH3||KCNC1||KCNT2||KCNN3||KCNS3||KCNA3||KCNN2||KCNJ11||KCNQ1||KCNK1||KCNQ3||HCN4||KCND2||KCNA6 | 1.337135 | 1.538578 | 1.806399 | 1.3868874 | 1.388523 | 1.822793 | 1.462596 | 1.297365 | 1.252322 | 1.246980 | 1.4976307 | 1.482796 | 1.613113 | 1.516634 | 1.843359 | 1.881168 | 1.687451 | 1.444333 | 1.3358856 | 1.3783681 | 1.412801 | 1.148225 | 1.573068 | 1.556054 | 1.7574862 | 2.101445 | 1.537024 | 1.452025 | 1.395197 | 1.3438862 | 1.3406231 | 1.2031623 |
| GO:0008028 | monocarboxylic acid transmembrane transporter activity | 31 | -0.5419363 | -1.548085 | 0.037422840 | 0.3529607 | 0.3229051 | 4299 | tags=68%, list=27%, signal=50% | MFSD2A||SLC16A1||SLC27A4||SLC10A7||ABCC4||SLC5A12||SLC27A1||SLC16A6||SLC27A2||SLC16A2||SLC16A9||SLC27A5||SLC16A14||ABCG2||SLC6A13||SLC10A4||SLC10A3||CEACAM1||SLC16A13||SLC16A4||SLC16A7 | 3.818675 | 4.507858 | 4.069227 | 4.6317701 | 3.847204 | 4.022633 | 4.465385 | 4.574144 | 3.799551 | 3.791488 | 3.8638899 | 4.499925 | 4.549256 | 4.473363 | 4.143481 | 4.074288 | 3.985582 | 4.605188 | 4.6538771 | 4.6358259 | 3.807888 | 3.833351 | 3.898840 | 3.864725 | 3.9897661 | 4.194096 | 4.484337 | 4.473683 | 4.437722 | 4.5821105 | 4.6186966 | 4.5199000 |
| GO:0003774 | cytoskeletal motor activity | 79 | 0.4072428 | 1.421549 | 0.037708026 | 0.3529607 | 0.3229051 | 5196 | tags=49%, list=33%, signal=33% | DNHD1||MYO9A||MYH15||MYO3A||KIF20B||KIF13A||CENPE||MYO5B||KIF14||KIF4B||KIF21B||KIF15||KIF6||MYH3||KIFC2||KIF11||KIF20A||DNAH5||KIF9||DNAH7||MYO19||KIF18A||KIF23||MYO9B||DYNC1H1||DNAH1||MYO1C||KIF2C||KIF5B||KIF3B||SMC3||MYO10||KIF21A||MYH10||KIF17||MYO5A||KIF27||KIF5C||APPBP2 | 5.415319 | 4.925922 | 5.419896 | 5.1675459 | 5.460880 | 5.464310 | 4.937580 | 5.194567 | 5.367478 | 5.396232 | 5.4798709 | 4.985938 | 4.898631 | 4.891259 | 5.382621 | 5.441565 | 5.434783 | 5.177286 | 5.1132674 | 5.2103999 | 5.449144 | 5.447868 | 5.485314 | 5.477970 | 5.4875077 | 5.426715 | 4.975800 | 4.931925 | 4.904107 | 5.1769155 | 5.2185328 | 5.1879291 |
| GO:0043177 | organic acid binding | 80 | -0.4189662 | -1.433559 | 0.039536210 | 0.3640254 | 0.3330277 | 2476 | tags=26%, list=16%, signal=22% | PC||SRR||SHMT2||GCHFR||PLOD1||P4HA2||P4HA1||ASS1||GAD1||ID3||SLC19A1||CYP26A1||SLC1A1||FTCD||SLC46A1||CYP26B1||NAGS||RYR2||FTCDNL1||GNMT||HBA1 | 4.891808 | 5.296771 | 5.026679 | 5.2304467 | 4.870718 | 5.035670 | 5.264285 | 5.194759 | 4.865802 | 4.937652 | 4.8708497 | 5.306869 | 5.306377 | 5.276864 | 5.100144 | 4.990735 | 4.986250 | 5.226418 | 5.1909958 | 5.2727593 | 4.864903 | 4.853719 | 4.893244 | 4.982466 | 4.9803163 | 5.138460 | 5.276520 | 5.243545 | 5.272567 | 5.1832011 | 5.2166870 | 5.1841361 |
| GO:0016209 | antioxidant activity | 52 | -0.4672236 | -1.487102 | 0.039590832 | 0.3640254 | 0.3330277 | 4112 | tags=44%, list=26%, signal=33% | PTGES||GSTK1||PRXL2C||UBIAD1||PRDX2||GSTP1||NXN||SELENOS||SELENOW||PRDX5||GSTO1||GPX3||NQO1||GPX8||MGST2||GLRX||PRXL2B||APOE||GSTO2||PRDX4||GPX1||PTGS2||HBA1 | 7.578758 | 7.072353 | 7.428588 | 7.2116844 | 7.601360 | 7.505127 | 7.016813 | 7.198875 | 7.601066 | 7.538138 | 7.5962244 | 7.138399 | 7.045755 | 7.030526 | 7.320951 | 7.447271 | 7.511098 | 7.250499 | 7.1658025 | 7.2174911 | 7.582369 | 7.613169 | 7.608353 | 7.511545 | 7.6118511 | 7.382891 | 7.036293 | 7.002977 | 7.010959 | 7.2053471 | 7.1875261 | 7.2036839 |
| GO:0003724 | RNA helicase activity | 66 | 0.4207329 | 1.421295 | 0.041261109 | 0.3668300 | 0.3355934 | 6871 | tags=71%, list=43%, signal=41% | DDX55||DDX42||HELZ2||DHX36||TDRD12||DHX33||DDX52||DHX29||DDX19B||DHX38||IGHMBP2||DDX21||FANCM||DDX41||DDX27||AQR||BRIP1||SUPV3L1||DDX51||DDX3X||DHX57||DHX15||SNRNP200||DDX20||YTHDC2||DHX58||DHX9||DHX40||DDX1||DDX23||DDX46||G3BP1||DHX35||DDX6||RAD54B||EIF4A2||DHX8||DDX17||DDX50||DDX24||DDX19A||MTREX||DDX59||EIF4A3||DDX56||DHX30||DHX37 | 6.176049 | 6.725926 | 6.517651 | 6.7879984 | 6.071587 | 6.220967 | 6.785473 | 6.848533 | 6.138190 | 6.345708 | 6.0258220 | 6.737832 | 6.726063 | 6.713784 | 6.718224 | 6.373478 | 6.437484 | 6.755134 | 6.7656741 | 6.8416347 | 6.106059 | 6.115541 | 5.989795 | 6.176697 | 6.0605289 | 6.404252 | 6.792034 | 6.787336 | 6.777008 | 6.8268330 | 6.8791610 | 6.8390838 |
| GO:0008514 | organic anion transmembrane transporter activity | 120 | -0.3801790 | -1.381732 | 0.041362530 | 0.3668300 | 0.3355934 | 4681 | tags=49%, list=29%, signal=35% | SLC35B1||SLC19A2||SERINC5||MFSD2A||SLC33A1||SLCO4C1||SLC47A1||SLC6A9||SLC16A1||SLC27A4||SLC25A4||SLC36A4||SLC25A42||ABCC4||SLC5A12||SLC39A8||SLC25A5||SLC38A7||SLC35B4||SLC27A1||SFXN2||SLC38A3||MFSD12||SLC2A10||SLC4A3||SLCO2A1||SLC25A29||SLC25A11||SLC7A1||SERINC3||SLC4A11||SLC4A4||SLC1A5||LRRC8A||SLC29A1||SLC7A5||SLC35B2||SLC15A4||SLC2A1||MFSD10||SLC38A6||CTNS||SLC2A6||SLC1A4||SLC66A1||ABCG2||SLC19A1||SLC6A13||OCA2||SLC1A1||SLCO5A1||SLC52A2||SLC46A1||SLC2A3||SLC7A3||SLC4A5||SLC16A7||BEST1||SLC26A4 | 5.341492 | 5.410764 | 5.449982 | 5.4856290 | 5.354114 | 5.491749 | 5.330075 | 5.445556 | 5.326538 | 5.378521 | 5.3186805 | 5.437259 | 5.443841 | 5.349273 | 5.385954 | 5.482972 | 5.478948 | 5.512690 | 5.4636183 | 5.4801463 | 5.363798 | 5.327667 | 5.370510 | 5.462242 | 5.5080294 | 5.504526 | 5.356112 | 5.326902 | 5.306784 | 5.4358146 | 5.4841923 | 5.4158005 |
| GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor | 11 | -0.6943527 | -1.559530 | 0.042033236 | 0.3668300 | 0.3355934 | 3904 | tags=73%, list=24%, signal=55% | SUMF1||SELENBP1||ERO1A||GFER||PCYOX1||PCYOX1L||QSOX1||P4HB | 5.687159 | 6.273457 | 5.986203 | 5.9800631 | 5.714396 | 6.013190 | 6.060864 | 5.837134 | 5.630154 | 5.706138 | 5.7234860 | 6.282413 | 6.330083 | 6.205126 | 5.865968 | 6.117483 | 5.963955 | 6.006459 | 5.9898787 | 5.9431055 | 5.764699 | 5.599186 | 5.772741 | 5.935984 | 5.8704464 | 6.210191 | 6.086928 | 6.051941 | 6.043351 | 5.8140466 | 5.8996527 | 5.7955507 |
| GO:0008484 | sulfuric ester hydrolase activity | 12 | -0.6818605 | -1.567265 | 0.042132079 | 0.3668300 | 0.3355934 | 3354 | tags=83%, list=21%, signal=66% | ARSB||SGSH||SULF2||IDS||GNS||SULF1||GALNS||ARSA||ARSJ||ARSG | 3.140892 | 3.873812 | 3.768078 | 3.8390355 | 3.158706 | 3.629984 | 3.731592 | 3.785261 | 3.083055 | 3.160362 | 3.1775160 | 3.811114 | 3.893675 | 3.914585 | 3.884268 | 3.814645 | 3.589096 | 3.790348 | 3.8471339 | 3.8782517 | 3.184562 | 3.024029 | 3.257721 | 3.406775 | 3.4901374 | 3.935344 | 3.715962 | 3.745044 | 3.733620 | 3.7934790 | 3.8207448 | 3.7404062 |
| GO:0140030 | modification-dependent protein binding | 147 | 0.3447084 | 1.324803 | 0.042387543 | 0.3668300 | 0.3355934 | 4893 | tags=42%, list=31%, signal=30% | CDYL2||PHF1||KMT2A||FAM156B||ATRX||L3MBTL1||ZCWPW1||YEATS2||BAZ2A||BPTF||ATAD2B||KDM7A||RNF169||KDM5A||ING2||HDAC6||MINDY2||SPRTN||PHIP||BRD9||PLCG2||RNF31||CHD1||PSME4||MLLT3||TP53BP1||ING3||PYGO1||ING4||TAB3||SPIN2B||MORC3||BRD7||IKBKG||CHD8||TAB2||RBBP5||SPIN3||AGL||DZIP3||ZMYND11||TRIM24||LYN||MTF2||BRCC3||ANKRD13D||OTUD7A||PRPF8||RAD23B||ZRANB1||DNAJC2||MSH6||GLYR1||SUZ12||USP15||NCAPG2||UIMC1||TAF1||MSL3||ZBTB1||IDE||ABRAXAS1 | 5.012685 | 5.421783 | 5.217507 | 5.5019321 | 4.980027 | 5.260773 | 5.465737 | 5.528311 | 4.987232 | 5.013517 | 5.0368797 | 5.415092 | 5.429644 | 5.420576 | 5.327257 | 5.166665 | 5.151965 | 5.483106 | 5.4562988 | 5.5641922 | 4.964791 | 4.930019 | 5.042943 | 5.200781 | 5.2422023 | 5.335990 | 5.475960 | 5.452239 | 5.468908 | 5.5254117 | 5.5398260 | 5.5196200 |
| GO:0051787 | misfolded protein binding | 28 | -0.5459467 | -1.525699 | 0.042573872 | 0.3668300 | 0.3355934 | 3608 | tags=50%, list=23%, signal=39% | HSPA1B||TOR1A||DNAJB9||DERL1||DNAJB11||HSPA13||RHBDD2||HSPA5||HSPA2||SDF2L1||DERL3||CLU||HSPA6||HSPA1L | 7.022466 | 7.772162 | 6.539916 | 7.2614073 | 7.036964 | 6.594434 | 7.702945 | 7.252822 | 7.096227 | 7.150103 | 6.7965306 | 7.834811 | 7.757720 | 7.721627 | 6.704321 | 6.336746 | 6.555210 | 7.221307 | 7.2501205 | 7.3113243 | 7.102258 | 7.040562 | 6.964793 | 6.641839 | 6.6319866 | 6.505513 | 7.721448 | 7.703808 | 7.683327 | 7.2367743 | 7.2728129 | 7.2486433 |
| GO:0005227 | calcium activated cation channel activity | 13 | -0.6657008 | -1.558570 | 0.043281527 | 0.3668300 | 0.3355934 | 3017 | tags=62%, list=19%, signal=50% | KCNN1||KCNMB4||ANO10||KCNMA1||KCNN3||TMEM63C||KCNN2||CATSPER2 | 1.921651 | 2.695007 | 2.756199 | 2.7683207 | 1.904949 | 2.513641 | 2.587247 | 2.717192 | 1.941313 | 1.944261 | 1.8784265 | 2.652929 | 2.739399 | 2.691391 | 2.955484 | 2.708785 | 2.578646 | 2.756989 | 2.8011441 | 2.7462397 | 1.938408 | 1.811973 | 1.960076 | 2.265700 | 2.2514485 | 2.918633 | 2.559543 | 2.637110 | 2.563761 | 2.7201624 | 2.7678679 | 2.6615829 |
| GO:0016307 | phosphatidylinositol phosphate kinase activity | 14 | 0.6333880 | 1.545213 | 0.044462574 | 0.3668300 | 0.3355934 | 4068 | tags=71%, list=25%, signal=53% | PIK3CD||PIK3C2A||PIK3C2B||PIPSL||PIP4K2A||PIP5K1B||PIK3CA||PIP4K2B||PIP5K1A||PIKFYVE | 3.281299 | 4.030146 | 3.922619 | 4.0674594 | 3.230264 | 3.633314 | 4.137021 | 4.099106 | 3.269460 | 3.461573 | 3.0887742 | 4.017415 | 4.027885 | 4.045003 | 4.075771 | 3.843219 | 3.835731 | 3.988912 | 4.0064904 | 4.1975591 | 3.282479 | 3.229366 | 3.177018 | 3.481361 | 3.2652901 | 4.039443 | 4.161284 | 4.110268 | 4.139057 | 4.0601813 | 4.1147512 | 4.1216025 |
| GO:0030547 | signaling receptor inhibitor activity | 15 | -0.6454872 | -1.568267 | 0.044948212 | 0.3668300 | 0.3355934 | 2598 | tags=60%, list=16%, signal=50% | LYNX1||LY6E||IGSF1||LRPAP1||WFIKKN1||LYPD1||LY6G6D||DKK3||FST | 4.887842 | 4.870715 | 5.234152 | 4.8700258 | 4.962373 | 5.223324 | 4.703890 | 4.774586 | 4.815488 | 4.800577 | 5.0352901 | 4.874992 | 4.929259 | 4.805221 | 5.076374 | 5.388379 | 5.220776 | 4.949095 | 4.8509869 | 4.8062757 | 4.998552 | 4.832599 | 5.047295 | 5.059639 | 5.2092366 | 5.382920 | 4.758418 | 4.676960 | 4.674706 | 4.8024625 | 4.8459152 | 4.6695870 |
| GO:0008200 | ion channel inhibitor activity | 29 | 0.5133855 | 1.483081 | 0.045342127 | 0.3668300 | 0.3355934 | 928 | tags=10%, list=6%, signal=10% | RSC1A1||ANKRD36C||SCN1B | 7.889937 | 7.489671 | 7.711180 | 7.4165151 | 7.931597 | 7.908703 | 7.474996 | 7.430853 | 7.852893 | 7.839807 | 7.9733221 | 7.524515 | 7.475951 | 7.467895 | 7.544254 | 7.770649 | 7.804982 | 7.475278 | 7.3534295 | 7.4182644 | 7.927997 | 7.901611 | 7.964491 | 7.967847 | 8.0041412 | 7.740196 | 7.468002 | 7.486595 | 7.470321 | 7.4209786 | 7.4369075 | 7.4346217 |
| GO:0000295 | adenine nucleotide transmembrane transporter activity | 15 | -0.6439086 | -1.564431 | 0.045534493 | 0.3668300 | 0.3355934 | 5437 | tags=73%, list=34%, signal=48% | SLC35B3||SLC25A6||SLC33A1||SLC25A4||SLC25A42||SLC25A5||LRRC8A||SLC29A1||SLC35B2||SLC19A1||ABCD1 | 7.376430 | 6.933433 | 7.259411 | 6.9888792 | 7.345584 | 7.305757 | 6.834909 | 6.995078 | 7.421280 | 7.473294 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.144792 | 7.227423 | 7.394688 | 7.074890 | 6.9098698 | 6.9770898 | 7.360061 | 7.399446 | 7.274427 | 7.402083 | 7.3879628 | 7.108681 | 6.851226 | 6.828845 | 6.824511 | 6.9867244 | 7.0075384 | 6.9908865 |
| GO:0005346 | purine ribonucleotide transmembrane transporter activity | 15 | -0.6439086 | -1.564431 | 0.045534493 | 0.3668300 | 0.3355934 | 5437 | tags=73%, list=34%, signal=48% | SLC35B3||SLC25A6||SLC33A1||SLC25A4||SLC25A42||SLC25A5||LRRC8A||SLC29A1||SLC35B2||SLC19A1||ABCD1 | 7.376430 | 6.933433 | 7.259411 | 6.9888792 | 7.345584 | 7.305757 | 6.834909 | 6.995078 | 7.421280 | 7.473294 | 7.2227886 | 7.011317 | 6.923123 | 6.861940 | 7.144792 | 7.227423 | 7.394688 | 7.074890 | 6.9098698 | 6.9770898 | 7.360061 | 7.399446 | 7.274427 | 7.402083 | 7.3879628 | 7.108681 | 6.851226 | 6.828845 | 6.824511 | 6.9867244 | 7.0075384 | 6.9908865 |
| GO:0019956 | chemokine binding | 11 | -0.6904217 | -1.550701 | 0.045552297 | 0.3668300 | 0.3355934 | 1475 | tags=64%, list=9%, signal=58% | ITGAV||ZFP36||CX3CR1||ITGA4||CXCR4||CCR10||ACKR2 | 4.504445 | 6.220215 | 4.527126 | 6.2291833 | 4.285490 | 4.043157 | 6.360628 | 6.254579 | 4.604040 | 4.668644 | 4.1968425 | 6.168468 | 6.212995 | 6.277109 | 5.202850 | 4.033529 | 3.996700 | 6.141750 | 6.2384115 | 6.3028493 | 4.088050 | 4.532702 | 4.197746 | 3.938999 | 3.9387248 | 4.231533 | 6.345146 | 6.349811 | 6.386570 | 6.3265134 | 6.1241338 | 6.3046620 |
| GO:0072341 | modified amino acid binding | 61 | -0.4410198 | -1.437684 | 0.045741627 | 0.3668300 | 0.3355934 | 1782 | tags=25%, list=11%, signal=22% | ADGRB1||SYT3||FCHO2||MFGE8||GAS6||SLC19A1||SCARB1||FTCD||SLC46A1||SYTL2||SYT15||FTCDNL1||GNMT||SYT17||SMPD3 | 5.571828 | 6.357221 | 5.548661 | 6.1983214 | 5.410774 | 5.339397 | 6.369927 | 6.203419 | 5.644826 | 5.730728 | 5.3058991 | 6.418080 | 6.323375 | 6.328221 | 5.758814 | 5.344326 | 5.512507 | 6.157317 | 6.1780684 | 6.2576248 | 5.359075 | 5.543264 | 5.319998 | 5.284066 | 5.2799651 | 5.447733 | 6.373056 | 6.366259 | 6.370460 | 6.2003280 | 6.1939591 | 6.2158824 |
| GO:0005253 | anion channel activity | 43 | -0.4816166 | -1.474401 | 0.045897877 | 0.3668300 | 0.3355934 | 3054 | tags=37%, list=19%, signal=30% | TTYH3||CLCC1||ANO10||LRRC8A||ANO7||CLCN4||CLCN3||PACC1||SLC1A4||LRRC8D||LRRC8E||SLC1A1||CLDN4||GABRD||BEST1||GABRQ | 4.445845 | 5.060410 | 4.587105 | 5.1778127 | 4.314712 | 4.253021 | 5.055412 | 5.184134 | 4.487096 | 4.625927 | 4.1909472 | 5.059803 | 5.066040 | 5.055368 | 4.817027 | 4.317728 | 4.583566 | 5.136467 | 5.1664793 | 5.2289442 | 4.279961 | 4.428093 | 4.228572 | 4.273129 | 4.1878216 | 4.295877 | 5.024418 | 5.067094 | 5.074223 | 5.1522381 | 5.2201177 | 5.1792355 |
| GO:0046906 | tetrapyrrole binding | 78 | -0.4136184 | -1.409798 | 0.046967104 | 0.3725101 | 0.3407898 | 2468 | tags=23%, list=15%, signal=20% | GUCY1A2||CYB5R4||CYB561D2||CD320||CYB5D2||CYP2U1||CYP2R1||CYBA||STC2||CYP26A1||CYP26B1||CYP4F22||PTGIS||TCN2||PTGS2||CYP7B1||CYP2S1||HBA1 | 7.329215 | 7.228353 | 8.058663 | 7.4501070 | 7.452413 | 7.867769 | 7.346706 | 7.317971 | 7.209502 | 7.317943 | 7.4501149 | 6.743131 | 7.414304 | 7.427535 | 8.002138 | 8.227169 | 7.929763 | 7.342600 | 7.6782813 | 7.2988371 | 7.501610 | 7.324707 | 7.522838 | 7.639344 | 7.6284241 | 8.245508 | 7.326940 | 7.386188 | 7.326164 | 7.4005871 | 7.2557413 | 7.2936475 |
| GO:0004190 | aspartic-type endopeptidase activity | 17 | -0.6107325 | -1.536479 | 0.048045127 | 0.3727987 | 0.3410539 | 4463 | tags=65%, list=28%, signal=47% | PSEN2||NAPSA||CASP3||SPPL3||NCSTN||SPPL2A||BACE1||PSEN1||HM13||CTSD||ASPRV1 | 5.280087 | 5.718090 | 5.725934 | 5.5394223 | 5.345391 | 5.706633 | 5.543370 | 5.432750 | 5.226975 | 5.206146 | 5.3992525 | 5.753933 | 5.759741 | 5.637320 | 5.641972 | 5.823555 | 5.706369 | 5.575286 | 5.5399594 | 5.5020934 | 5.346452 | 5.231708 | 5.449779 | 5.506517 | 5.5896023 | 5.978579 | 5.598628 | 5.535773 | 5.493775 | 5.4429645 | 5.4824067 | 5.3706606 |
| GO:0030983 | mismatched DNA binding | 13 | 0.6412819 | 1.535482 | 0.049196991 | 0.3727987 | 0.3410539 | 50 | tags=8%, list=0%, signal=8% | MLH3 | 4.304891 | 6.188329 | 4.632213 | 6.1504412 | 4.273737 | 4.343241 | 6.249916 | 6.257606 | 4.390691 | 4.361058 | 4.1513225 | 6.193557 | 6.178349 | 6.193030 | 5.149843 | 4.258238 | 4.304403 | 6.084422 | 6.1208897 | 6.2413014 | 4.074724 | 4.405836 | 4.320532 | 4.188638 | 4.4011686 | 4.428192 | 6.226144 | 6.262506 | 6.260808 | 6.2154227 | 6.3150905 | 6.2404330 |
| GO:0071837 | HMG box domain binding | 12 | 0.6502328 | 1.517280 | 0.049397836 | 0.3727987 | 0.3410539 | 1985 | tags=42%, list=12%, signal=37% | PRRX1||TCF12||SP1||PAX6||PAX3 | 2.935614 | 3.353859 | 3.544052 | 3.6360376 | 3.075318 | 3.751942 | 3.381133 | 3.512953 | 2.923094 | 2.997991 | 2.8834025 | 3.284125 | 3.387257 | 3.387747 | 3.787411 | 3.398355 | 3.411652 | 3.688467 | 3.5274028 | 3.6863996 | 3.133161 | 2.898729 | 3.178714 | 3.656800 | 3.6657037 | 3.917814 | 3.408348 | 3.354590 | 3.379959 | 3.5193454 | 3.4982097 | 3.5211906 |
GO: CC
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005788 | endoplasmic reticulum lumen | 213 | -0.5182108 | -2.029773 | 6.355733e-07 | 4.559924e-05 | 4.090254e-05 | 3730 | tags=61%, list=23%, signal=47% | SHISA5||COLGALT1||LAMB1||ADAMTS7||COL4A5||COL3A1||TOR1A||ERO1A||RCN1||DNAJB9||COL4A1||FN1||CRTAP||P3H1||IGFBP4||MELTF||GPC3||BACE1||CDH2||SDC2||WNT5A||GPX8||PRKCSH||TOR2A||COL14A1||ERLEC1||OS9||BCHE||CCN1||PTPRN2||WNT3||MINPP1||GANAB||TMEM43||COL4A2||MEN1||LAMB2||CLN6||SLC27A2||ERP44||DNAJB11||COL25A1||ADAM10||MANF||TSPAN33||CKAP4||LTBP1||APP||CALU||ERP29||TXNDC5||MYDGF||COL6A1||DAG1||ALG12||TSPAN15||CANX||EDEM2||CST3||PDIA3||APOE||COL13A1||FKBP10||HSP90B1||NUCB1||COL6A2||FSTL1||APLP2||CTSC||P4HA2||HSPA5||TOR3A||SPON1||P4HA1||PPIB||HYOU1||BMP4||POGLUT2||ERAP1||COL18A1||SUMF2||IGFBP7||SIL1||PDIA6||QSOX1||MFGE8||LRPAP1||WNT3A||ADAMTS5||FLT3||RCN3||CALR||P4HB||CTSZ||PDGFA||SERPING1||CHGB||TIMP1||GAS6||NOTUM||SELENOM||COL2A1||STC2||VWA1||SDF2L1||PDIA4||ARSA||WFS1||TOR1B||TSPAN14||COL26A1||WNT7B||SERPINH1||ARSJ||MXRA8||LGALS1||ARSG||F8||MTTP||CLU||TF||FAM20C||MATN3||ADAMTSL4||PTGS2||WNT5B||IL23A||VGF||TOR4A | 5.335248 | 6.307567 | 5.642522 | 6.019422 | 5.310843 | 5.519382 | 6.143691 | 5.935911 | 5.324554 | 5.370494 | 5.310003 | 6.315601 | 6.355059 | 6.250098 | 5.675527 | 5.654021 | 5.596875 | 5.988592 | 6.064666 | 6.003880 | 5.319452 | 5.298614 | 5.314379 | 5.446614 | 5.412001 | 5.684061 | 6.161040 | 6.153358 | 6.116278 | 5.934764 | 5.986607 | 5.884559 |
| GO:0070062 | extracellular exosome | 1491 | -0.3115837 | -1.427399 | 2.392766e-06 | 9.890101e-05 | 8.871427e-05 | 4340 | tags=33%, list=27%, signal=27% | PRKACA||TUBB4B||EIF6||MEGF8||PTTG1IP||ALDH2||SLC16A1||ACTB||STK10||H2AW||ATP1B1||ITM2B||RAB4A||STOM||ACP2||PHGDH||MGAT5||FAT1||NPC2||FUCA2||SLC9A1||ARL8A||AHCY||TMEM98||NAGA||FTH1||GNAI1||RPS28||SARS1||ABCB6||PSMB5||IDH2||LOXL4||PLXNB2||NAPSA||ITGB1||RTN4RL1||CD47||GSTK1||ANXA11||F12||RAB43||PLPP1||DPP4||GIPC1||ARF1||SYT7||ATP1B3||PODXL||TMEM132A||RPS14||MYL6B||PTPRO||SLC5A12||AKR7A2||FUZ||MDH2||PMVK||PRDX2||VCL||DDT||RPS9||SMPDL3A||HSPB1||ATRN||TUBB2A||ADAM15||CYB5R1||TSPAN3||GSTP1||PAM||SELENBP1||RPS19||TSPAN6||SFT2D2||DSG2||NIBAN1||TMEM256||GPLD1||BTD||C1GALT1C1||FMNL1||NCSTN||GUSB||DDR1||SNF8||GALNT7||VPS4B||CFD||LTBP3||LAMB1||SERPINB6||PLCB1||ROBO2||TM7SF3||WDR1||ITCH||GNB2||QPRT||RNH1||ACTR3||HSPA1B||RTN4RL2||GALM||QPCT||ALDH1A3||SND1||RYR1||SCARB2||CNTN1||TOLLIP||AKR1A1||TOR1A||CPE||STX3||MSRA||B4GALT3||IDUA||EFEMP1||KRT18||CORO1A||PPIC||PRNP||GPRC5B||SPPL2A||TUBB6||CSK||DNAJB9||CREG1||ACY1||DCTN2||CD40||REXO5||FN1||VPS37B||PLEC||ITGAV||PRDX5||P3H1||BLVRB||PLOD2||GSTO1||RAC3||RAB11B||MPST||HEXB||FUCA1||GPX3||ICOSLG||MELTF||ARMC9||SCAMP3||CANT1||RAB34||SLC44A2||VAMP8||TOM1L2||GPM6A||ARL6||SYPL1||LAMA4||GGH||ARHGAP23||ALCAM||WNT5A||MGAT1||SCPEP1||TMEM59||MAL2||LRRC26||PPT1||GNPTG||CD63||TTYH3||NUCB2||FBLN5||GRID1||SLC5A5||CUTA||LAMA5||ANXA4||RPS3A||TUBB3||ATP1A1||CNP||HTRA1||LAMP1||LAMP2||TAGLN2||EHD1||CRABP2||RPS26||GLRX||GPC4||ENPP4||GET3||PGAP6||ACTA1||GLA||DBNL||SYNGR2||WNT3||MINPP1||RRAS||IGSF8||GANAB||ARHGEF18||MAN2B1||EPS8L1||NAXE||VPS37D||MAN1C1||PLA2G15||COL4A2||CD55||PGLS||FURIN||TM9SF2||SLC4A4||EPHB4||FAM234A||H2AX||H4C3||LAMB2||DSC2||TSPO||TNFAIP3||RPS18||SLC27A2||ERP44||HDHD2||VIM||TUBA1A||ADAM10||NID1||MAN2B2||SLC1A5||METRNL||CBR1||SLC22A5||A1BG||SLC7A5||HLA-E||CKAP4||SLC26A11||CD19||SH3BGRL3||HSPA13||MOGS||PFKP||DNPH1||PSMB3||APP||GNS||ZMPSTE24||SLC9A3R2||TXNDC5||KIFC3||WLS||TPP1||GMPPA||COMP||ACTA2||SLC2A1||S100A11||GAL3ST4||CXCL12||COL6A1||DAG1||SHMT2||LSR||THSD4||PADI2||IL6ST||PRXL2B||LGMN||PRCP||CANX||EXT2||RPLP2||ITGA2B||CYBRD1||MIF||CST3||PDIA3||APOE||KALRN||NUDT9||ENDOD1||CPVL||HSP90B1||NUCB1||PLOD1||COL6A2||GBA||PHPT1||RHOC||FSTL1||APLP2||C1orf116||ITM2C||ATP6AP1||PLD3||FAM3C||GALNS||CTSC||EPHB1||HSPA5||IGFBP2||TOR3A||GALK1||CHID1||GSN||NECTIN2||CDH15||CTNS||PCYOX1||TKFC||H2AC20||TMEM109||COMT||PSAP||PPIB||HYOU1||TMEM38A||TUBB4A||TMED9||SCRN2||SIAE||PRKCZ||ATP6AP2||CTSB||ERAP1||COL18A1||ASS1||GPRC5C||NAAA||CD81||CD9||SSR4||RFTN1||B3GAT3||LGALS3BP||ATP6V1C2||GAA||MMP24||GSTO2||IGFBP7||GP6||RIMS2||SPTBN4||RPS5||PDIA6||PRDX4||SYTL1||QSOX1||DNASE2||CTSH||HEXA||GLB1||MFGE8||EFNB1||WNT3A||SLC1A4||SDF4||SMPDL3B||HLA-B||PLCD1||SERPINI1||CILP2||CALR||P4HB||CTSL||B4GAT1||CTSZ||HSPA2||ALPL||SDC1||H2BC5||RHOG||SERPING1||BSG||NA||HID1||TIMP1||PROS1||GAS6||RIPOR1||CFAP70||NEU1||CLN5||MBD5||KRT8||ASL||PYGM||GPC1||C2||EPHX2||CSPG4||AGAP2||C1R||VWA1||ANXA3||GNG7||LMAN2||SLC6A13||CRTAC1||SERPINF1||FBLN1||SIRPA||CORO1B||ARSA||SFRP1||SLC1A1||TOR1B||HLA-C||P2RX4||CHST14||GRN||ITGB5||HPGD||SCARB1||ENPP3||ATP6V0C||FTCD||H2BC4||NAGLU||CTSD||TRHDE||HLA-A||SLC2A3||H4C11||SERINC2||WNT7B||PLAT||ABCB1||H4C8||CTSF||RENBP||ADGRE5||FAS||CDKL1||MXRA8||ITGA4||ICAM3||ABHD8||CEACAM1||LGALS1||SECTM1||SMPD1||CXCR4||H2AJ||SEC14L2||VASN||THBS4||RTN4R||TMBIM1||C1RL||CLU||EPCAM||CD82||TF||FAM20C||PTPRD||H2AC12||PRPH||H2AC11||SLC46A3||ACE||BST2||H2BC15||C5||HSPA6||CRISPLD2||RAB5B||ITGB2||WNT5B||H2AC6||H2BC18||JADE2||TUBA4A||CLTRN||HMCN1||CNKSR2||MUC1||H4C14||H4-16||RAPGEF3||GATM||SHROOM2||H2AC17||SLC26A4||AMY2B||HBA1||ANGPTL6||BHLHB9 | 7.754341 | 7.723194 | 7.560115 | 7.660803 | 7.750217 | 7.627223 | 7.692944 | 7.672497 | 7.766511 | 7.764920 | 7.731318 | 7.772342 | 7.699892 | 7.696061 | 7.519462 | 7.538006 | 7.620843 | 7.688702 | 7.620005 | 7.672809 | 7.748547 | 7.784686 | 7.716615 | 7.704401 | 7.678159 | 7.489751 | 7.689622 | 7.692806 | 7.696396 | 7.672211 | 7.657887 | 7.687244 |
| GO:0030176 | integral component of endoplasmic reticulum membrane | 142 | -0.5311919 | -1.967354 | 2.717250e-05 | 7.775517e-04 | 6.974643e-04 | 5573 | tags=70%, list=35%, signal=46% | INSIG1||DPAGT1||ACER3||ARL6IP1||MARCHF6||SLC35B3||ERGIC1||SGMS2||BFAR||TRAM2||DERL2||SPPL2B||EMC6||STIM1||SARAF||EMC9||DPM2||ATF6||RRBP1||SLC35B1||SLC37A2||TMCO1||SYVN1||WDR83OS||RTN1||ELOVL1||EMC4||CAMLG||ATF6B||ELOVL6||SLC37A3||ESYT1||PREB||EMC3||CCDC47||SPPL3||TAP2||SELENOS||SLC35B4||FKBP8||TM7SF2||SPPL2A||EMC7||XXYLT1||HACD3||ZFYVE27||DOLPP1||SGMS1||HACD1||ELOVL5||SLC37A4||BCAP31||DERL1||SPCS1||EMC1||PIGK||SLC27A2||HLA-E||RTN2||SLC35B2||ZMPSTE24||RHBDD2||PORCN||SEC61A1||PIGT||HM13||DHCR7||CANX||TAPBP||PDIA3||ERGIC3||TRAM1||TRAM1L1||TMEM33||DOLK||FITM2||ELOVL7||TAP1||HSPA5||PIGU||SLC27A5||EMC10||TECR||DPM3||AMFR||HLA-B||CALR||RETREG1||ELOVL4||LRRC8E||GPAA1||RCE1||HLA-DQB1||STING1||WFS1||HLA-C||HLA-A||DERL3||SLC37A1 | 5.856196 | 6.541186 | 6.164860 | 6.405940 | 5.879356 | 6.086349 | 6.397409 | 6.335143 | 5.817990 | 5.839736 | 5.909278 | 6.564221 | 6.584952 | 6.471891 | 6.137879 | 6.214191 | 6.141215 | 6.383190 | 6.444939 | 6.388880 | 5.872637 | 5.831706 | 5.931960 | 5.969827 | 6.003326 | 6.267151 | 6.423000 | 6.400985 | 6.367704 | 6.335911 | 6.379686 | 6.288387 |
| GO:0043202 | lysosomal lumen | 74 | -0.6057729 | -2.025745 | 1.985468e-04 | 4.616213e-03 | 4.140746e-03 | 4964 | tags=82%, list=31%, signal=57% | PPT2||ASAH1||GYG1||ARSB||CTSA||NCAN||RNASET2||GPC6||SGSH||PLBD2||CTSV||ACP2||NPC2||GUSB||SCARB2||IDUA||CSPG5||HEXB||FUCA1||GPC3||EPDR1||SDC2||PPT1||LAMP2||IDS||SDC3||GPC4||GLA||MAN2B1||MAN2B2||GNS||TXNDC5||LIPA||TPP1||LGMN||GBA||PLD3||GALNS||CHID1||PSAP||CTSB||NAAA||GAA||HEXA||IFI30||GLB1||ATP13A2||GPC2||CTSL||SDC1||NEU1||GPC1||CSPG4||ARSA||NAGLU||CTSD||CTSF||SMPD1||TCN2||HPSE||GPC5 | 5.581463 | 6.511787 | 5.892748 | 6.441427 | 5.442316 | 5.579253 | 6.440900 | 6.390105 | 5.617507 | 5.743676 | 5.356640 | 6.576752 | 6.503008 | 6.452903 | 6.025414 | 5.801896 | 5.840921 | 6.400596 | 6.448417 | 6.474302 | 5.432004 | 5.532233 | 5.357359 | 5.445782 | 5.389404 | 5.856246 | 6.451178 | 6.442015 | 6.429425 | 6.388553 | 6.409763 | 6.371747 |
| GO:0000172 | ribonuclease MRP complex | 10 | -0.8681134 | -1.918261 | 2.370301e-04 | 5.203604e-03 | 4.667635e-03 | 6 | tags=20%, list=0%, signal=20% | RPP25L||RMRP | 5.943331 | 5.958048 | 5.746393 | 5.766090 | 5.992286 | 5.884799 | 5.745444 | 7.181343 | 5.903187 | 5.952808 | 5.973104 | 6.181873 | 5.851146 | 5.811796 | 5.714552 | 5.725124 | 5.798062 | 5.934940 | 5.591920 | 5.750951 | 6.003603 | 5.923923 | 6.046656 | 5.943665 | 5.961683 | 5.738581 | 5.779826 | 5.749484 | 5.706071 | 5.705603 | 8.369904 | 5.712209 |
| GO:0140534 | endoplasmic reticulum protein-containing complex | 115 | -0.5197819 | -1.868355 | 3.710598e-04 | 7.887672e-03 | 7.075246e-03 | 5573 | tags=67%, list=35%, signal=44% | INSIG1||UFD1||ARL6IP1||PIGM||MARCHF6||SPCS2||UBXN1||DERL2||EMC6||PIGH||NBAS||EMC9||DPM2||STT3B||KRTCAP2||DAD1||RNF139||FAF2||SEC63||SYVN1||SEL1L||SEC61A2||EMC4||SEC11C||CAMLG||ORMDL2||SPTLC1||ELOVL6||HSD17B12||SEC11A||EMC3||TAP2||SELENOS||RYR1||PIGC||PIGA||EMC7||PRKCSH||OS9||GET3||DERL1||GANAB||SPCS1||EMC1||PIGK||RHBDD2||SEC61A1||PIGT||TUSC3||HM13||TAPBP||EXT2||SPCS3||PDIA3||RPN1||RPN2||HSP90B1||DDOST||SRPRB||STT3A||TAP1||HSPA5||PIGU||MAGT1||P4HA1||EMC10||PPIB||HYOU1||SSR4||PDIA6||AMFR||CALR||P4HB||GPAA1||SDF2L1||HLA-A||DERL3 | 6.827336 | 7.484077 | 7.052437 | 7.263255 | 6.839109 | 6.958832 | 7.349913 | 7.211815 | 6.823243 | 6.849803 | 6.808659 | 7.508672 | 7.515868 | 7.425974 | 7.038785 | 7.068445 | 7.049924 | 7.245957 | 7.297556 | 7.245629 | 6.840203 | 6.826268 | 6.850754 | 6.911243 | 6.885818 | 7.072265 | 7.367800 | 7.352787 | 7.328886 | 7.214265 | 7.245824 | 7.174470 |
| GO:0005765 | lysosomal membrane | 334 | -0.3645772 | -1.495812 | 2.511356e-03 | 4.557192e-02 | 4.087804e-02 | 4273 | tags=39%, list=27%, signal=29% | STARD3||SORT1||STOM||ACP2||ARL8A||GNAI1||ABCB6||DPP4||SYT7||VPS16||LAMTOR2||SLC39A8||MCOLN1||NDUFC2||TMEM179B||BORCS6||VPS33A||TPCN1||GPLD1||RRAGA||NCSTN||MEAK7||TMEM30A||ABCA5||GNB2||AP2S1||SCARB2||CDIP1||LMBRD1||CYB561||MARCHF9||RDH14||SNX14||SPPL2A||LPCAT1||MFSD12||TMEM9B||ABHD6||SLC49A4||BLOC1S1||SLC44A2||VAMP8||TMEM79||GLMP||PLEKHM1||WDR24||TMEM59||CD63||SYT11||LAPTM4B||SLC17A5||PSEN1||LAMP1||LAMP2||PGAP6||SYNGR1||SLC39A14||BORCS8||EEF1A2||LRRC8A||ABCA3||HLA-DMB||SLC7A5||CKAP4||SLC26A11||SLC29A3||ATRAID||ATP6V0A2||SLC15A4||CLCN4||COL6A1||GPR137C||CLCN3||PRCP||CYBRD1||TMEM106B||SURF4||GBA||DDOST||ITM2C||PLD3||MAGT1||TMEM9||CTNS||HPS6||LAPTM4A||LAMP3||PSAP||ATP6AP2||ATP6V1C2||GAA||MFSD1||SPNS1||LAMTOR4||TM9SF1||SLC2A6||ATP13A2||SLC66A1||VLDLR||TMEM74||TECPR1||ATP6V0B||LRRC8E||NEU1||CLN5||DAGLB||SLC30A4||OCA2||HLA-DQB1||CYB561A3||P2RX4||ZNRF2||GRN||LRP1||SCARB1||MARCHF2||ATP6V0C||CTSD||SLC12A4||BORCS5||VASN||TMBIM1||ABCD1||OSTM1||SLC46A3||BST2||HLA-DMA||HLA-DOA||DRAM1||HPSE | 5.815015 | 6.457881 | 5.940828 | 6.421337 | 5.762284 | 5.809330 | 6.404065 | 6.397234 | 5.841345 | 5.868001 | 5.732135 | 6.538009 | 6.441753 | 6.389949 | 6.033229 | 5.827209 | 5.954600 | 6.436820 | 6.393082 | 6.433697 | 5.743729 | 5.807826 | 5.734179 | 5.774899 | 5.765202 | 5.884805 | 6.390977 | 6.416336 | 6.404771 | 6.414507 | 6.356294 | 6.420040 |
| GO:0046658 | anchored component of plasma membrane | 35 | -0.6601658 | -1.946172 | 4.023490e-03 | 6.507557e-02 | 5.837283e-02 | 1464 | tags=57%, list=9%, signal=52% | RTN4RL2||CNTN1||PRNP||GAS1||MELTF||GPC3||GPC4||GPC2||GPC1||ULBP3||HYAL2||ULBP1||EFNA5||CD24||RTN4R||ULBP2||LYPD3||NTNG2||GPC5||NTNG1 | 3.512314 | 3.962782 | 3.928122 | 3.955259 | 3.529727 | 3.770013 | 3.845623 | 3.769913 | 3.447125 | 3.552674 | 3.534942 | 3.969978 | 4.032043 | 3.882421 | 3.964784 | 3.922780 | 3.895967 | 3.994745 | 3.978161 | 3.890720 | 3.497217 | 3.507277 | 3.583147 | 3.631505 | 3.654512 | 3.994646 | 3.890326 | 3.828561 | 3.816898 | 3.779834 | 3.791017 | 3.738354 |
| GO:0005796 | Golgi lumen | 48 | -0.5772902 | -1.801491 | 4.936396e-03 | 7.651414e-02 | 6.863323e-02 | 3598 | tags=67%, list=23%, signal=52% | PCSK5||MMP11||CSPG5||GPC3||SDC2||WNT5A||TGFB1||SDC3||GPC4||WNT3||FURIN||PCSK6||APP||DAG1||PODXL2||CGA||LRPAP1||WNT3A||SDF4||GPC2||PDGFA||SDC1||PROS1||GAS6||GPC1||CSPG4||WNT7B||MUC3A||F8||WNT5B||MUC1||GPC5 | 4.148066 | 4.772249 | 4.634809 | 4.692430 | 4.161365 | 4.516937 | 4.603952 | 4.579978 | 4.067836 | 4.175329 | 4.197718 | 4.789384 | 4.815706 | 4.709550 | 4.600824 | 4.722371 | 4.576978 | 4.713741 | 4.711085 | 4.651612 | 4.200666 | 4.045767 | 4.230907 | 4.369092 | 4.359239 | 4.781111 | 4.674548 | 4.575378 | 4.559205 | 4.575100 | 4.654075 | 4.507002 |
| GO:0030173 | integral component of Golgi membrane | 55 | -0.5418618 | -1.732812 | 5.810684e-03 | 8.822753e-02 | 7.914015e-02 | 5487 | tags=69%, list=34%, signal=45% | ACER3||SLC35B3||ERGIC1||SGMS2||SLC35A4||ENTPD4||SYS1||SLC35B1||BET1L||LARGE1||UBIAD1||ZDHHC20||PCSK5||SLC35A5||SLC35B4||PCSK4||ATP2C1||RER1||SGMS1||MAN1C1||SLC39A13||FURIN||SLC35B2||TVP23C||B4GALNT1||POMGNT1||SLC35A2||TPST1||ERGIC3||YIF1B||STEAP2||GALNT2||QSOX1||CHST12||B4GAT1||YIF1A||UNC50||CSGALNACT1 | 4.984117 | 5.298469 | 5.349722 | 5.265967 | 5.048823 | 5.341960 | 5.165843 | 5.147096 | 4.907610 | 4.957704 | 5.081446 | 5.308335 | 5.349895 | 5.234832 | 5.282488 | 5.444782 | 5.316777 | 5.270504 | 5.280786 | 5.246395 | 5.073052 | 4.918045 | 5.146064 | 5.208148 | 5.237990 | 5.553733 | 5.206481 | 5.161375 | 5.128611 | 5.141318 | 5.200397 | 5.097731 |
| GO:0005791 | rough endoplasmic reticulum | 61 | -0.5251236 | -1.709810 | 6.387376e-03 | 9.504415e-02 | 8.525465e-02 | 4736 | tags=59%, list=30%, signal=42% | ARSB||SEC63||PLOD3||SEC61A2||RPS21||RPS28||F12||RANGRF||SNCA||CCDC47||SPPL3||RPL18||FKRP||PLOD2||PSEN1||RPS26||TMEM97||ADCYAP1R1||CKAP4||NAT8L||APP||SEC61A1||EPHA5||HM13||CANX||RPN1||RPN2||NUCB1||PLOD1||MACO1||CDKAL1||SRPRB||SYNE3||SSR4||LRPAP1||MTTP | 8.995496 | 8.978231 | 8.760103 | 8.900597 | 8.991781 | 8.741241 | 8.944818 | 8.891782 | 9.057273 | 8.992390 | 8.934197 | 8.994198 | 8.972836 | 8.967521 | 8.743622 | 8.723570 | 8.811634 | 8.942042 | 8.878233 | 8.880604 | 8.985499 | 9.050911 | 8.936650 | 8.904214 | 8.779411 | 8.512867 | 8.947689 | 8.944633 | 8.942125 | 8.891348 | 8.852363 | 8.930575 |
| GO:0005604 | basement membrane | 66 | -0.5086174 | -1.675403 | 6.615007e-03 | 9.650127e-02 | 8.656170e-02 | 3869 | tags=52%, list=24%, signal=39% | ATRN||TMEFF1||LAMB1||COL4A5||COL4A1||FN1||LAMA1||LAMA4||LAMA5||EGFLAM||COL4A2||LAMB2||NID1||DAG1||SMOC1||NTN1||LOXL1||COL18A1||CD151||APLP1||TIMP1||COL2A1||VWA1||SPARC||SERPINF1||FBLN1||ACHE||LOXL2||ANG||THBS4||NTNG2||HMCN1||NTNG1||TMEFF2 | 3.580942 | 3.819085 | 4.027938 | 3.677263 | 3.609887 | 3.959348 | 3.684569 | 3.565062 | 3.534790 | 3.500052 | 3.699980 | 3.796850 | 3.895734 | 3.761286 | 4.017766 | 4.110866 | 3.950685 | 3.685131 | 3.696120 | 3.650140 | 3.583840 | 3.501676 | 3.734418 | 3.739768 | 3.839696 | 4.247146 | 3.724322 | 3.699664 | 3.627994 | 3.583585 | 3.612285 | 3.496827 |
| GO:0030667 | secretory granule membrane | 199 | -0.3822570 | -1.481444 | 7.466764e-03 | 1.068322e-01 | 9.582851e-02 | 4121 | tags=42%, list=26%, signal=32% | TMX3||CD47||RAB24||ABCC4||SLC30A5||LAMTOR2||SNCA||NDUFC2||TMED2||CYB5R1||TMEM179B||PAM||RAB3A||NCSTN||SERPINB6||TMEM30A||SYT2||STX3||CYB561||DIAPH1||DEGS1||RND2||LPCAT1||PCSK4||ITGAV||ITPR3||SLC44A2||VAMP8||CD63||PSEN1||LAMP1||LAMP2||ENPP4||PTPRN2||AGPAT2||TMED10||SYNGR1||CD55||SLC27A2||ADAM10||ABCA3||CKAP4||SLC15A4||PTAFR||PRCP||ITGA2B||SURF4||APLP2||DDOST||CD109||MAGT1||LAMP3||PSAP||RAB9B||ATP6AP2||CD9||GAA||MFGE8||HLA-B||DGAT1||RHOG||BSG||CYBA||SPARC||SIRPA||STING1||HLA-C||TSPAN14||NFASC||ATP6V0C||SLC2A3||ADGRE5||CEACAM1||MMP25||TMBIM1||CFAP65||STBD1||PTPRB||BST2||RAB5B||RAB4B||ITGB2||CHRNB4||C5AR1 | 5.382151 | 5.824744 | 5.618660 | 5.778662 | 5.382034 | 5.548305 | 5.730047 | 5.726054 | 5.377612 | 5.402262 | 5.366344 | 5.845672 | 5.853808 | 5.773403 | 5.610378 | 5.645461 | 5.599742 | 5.780659 | 5.784553 | 5.770739 | 5.372590 | 5.359004 | 5.413941 | 5.451902 | 5.468806 | 5.709550 | 5.758292 | 5.721990 | 5.709412 | 5.720191 | 5.762400 | 5.694762 |
| GO:0071682 | endocytic vesicle lumen | 11 | -0.7739126 | -1.755544 | 8.172796e-03 | 1.126030e-01 | 1.010049e-01 | 2139 | tags=64%, list=13%, signal=55% | APOE||HSP90B1||HYOU1||CALR||CTSL||SPARC||HBA1 | 6.548360 | 8.407925 | 7.024090 | 7.986095 | 6.452453 | 6.712941 | 8.294168 | 7.961895 | 6.548718 | 6.656518 | 6.431043 | 8.423064 | 8.438683 | 8.360858 | 7.210967 | 6.956181 | 6.884351 | 7.911439 | 8.009172 | 8.034759 | 6.477355 | 6.471433 | 6.407536 | 6.672486 | 6.544272 | 6.899426 | 8.279914 | 8.306930 | 8.295533 | 7.960677 | 7.988291 | 7.936246 |
| GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane | 64 | -0.5028347 | -1.646069 | 9.048068e-03 | 1.223957e-01 | 1.097890e-01 | 3902 | tags=53%, list=24%, signal=40% | ERGIC2||STX5||CNIH3||TMED5||TMED2||SPPL3||TAP2||MPPE1||MGAT1||BCAP31||CNIH1||TMED10||TMED3||CD55||VMP1||TAPBP||SURF4||ERGIC3||ATP6AP1||CTSC||YIF1B||TAP1||TMED9||TMED1||CALR||KDELR1||CTSZ||YIF1A||TMEM199||LMAN2||STING1||AZIN2||F8||TGFA | 6.305578 | 6.833246 | 6.566200 | 6.587072 | 6.348399 | 6.530435 | 6.668145 | 6.516939 | 6.248984 | 6.282876 | 6.381563 | 6.856984 | 6.883729 | 6.755900 | 6.460858 | 6.654594 | 6.576594 | 6.584190 | 6.620760 | 6.555525 | 6.352807 | 6.276923 | 6.412282 | 6.450492 | 6.460960 | 6.669144 | 6.704134 | 6.666183 | 6.633246 | 6.518461 | 6.563870 | 6.466854 |
| GO:0000786 | nucleosome | 48 | -0.5436774 | -1.696599 | 1.006265e-02 | 1.336895e-01 | 1.199196e-01 | 1611 | tags=35%, list=10%, signal=32% | H2AC20||H1-2||H2BC5||H2BC17||H2BC4||H4C11||H4C8||H2BC11||H2AJ||H2AC12||H2AC11||H2BC15||H2AC6||H2BC18||H4C14||H4-16||H2AC17 | 6.040878 | 6.271642 | 6.244724 | 6.193609 | 5.926593 | 5.771782 | 6.283362 | 6.217637 | 6.100219 | 6.237862 | 5.739915 | 6.280463 | 6.256044 | 6.278292 | 6.355001 | 6.018191 | 6.336768 | 6.184069 | 6.161119 | 6.234655 | 5.857554 | 6.091711 | 5.814849 | 5.712095 | 5.666674 | 5.923370 | 6.272912 | 6.267285 | 6.309524 | 6.260776 | 6.157674 | 6.232503 |
| GO:0034358 | plasma lipoprotein particle | 16 | -0.7047206 | -1.742449 | 1.080247e-02 | 1.385696e-01 | 1.242970e-01 | 2220 | tags=56%, list=14%, signal=48% | LSR||APOC1||APOE||PCYOX1||VLDLR||PLA2G7||PLTP||APOBR||CLU | 4.799166 | 5.320209 | 5.004096 | 5.102638 | 4.749710 | 4.932811 | 5.228609 | 5.022764 | 4.810143 | 4.810862 | 4.776222 | 5.278829 | 5.375867 | 5.304166 | 5.067052 | 5.014747 | 4.927035 | 5.092914 | 5.084598 | 5.129995 | 4.770870 | 4.754727 | 4.723125 | 4.924486 | 4.852368 | 5.016858 | 5.254085 | 5.213623 | 5.217776 | 5.036746 | 5.052557 | 4.977921 |
| GO:0043025 | neuronal cell body | 368 | -0.3226120 | -1.335624 | 1.375419e-02 | 1.539245e-01 | 1.380704e-01 | 3269 | tags=27%, list=20%, signal=22% | ITPR3||RAC3||NQO1||GPM6A||CDK5||BACE1||ALCAM||PPT1||MAPK8IP2||SYT11||ELOVL5||KCNN1||TUBB3||PSEN1||LAMP1||NDUFS7||ROGDI||SLC8A1||DBNL||P2RX2||EFHC1||EPHA4||EEF1A2||ADAM10||GRK4||NRGN||UNC5C||FEZ1||APP||SRR||EPHA5||IL6ST||CANX||ROR2||APOE||ADRA2C||KCND3||TUBB4A||PRKCZ||RIN3||KCNB2||ASS1||UNC5A||HCN3||SPTBN4||AMFR||SLC1A4||ATP13A2||SERPINI1||NGFR||CX3CR1||MUL1||BMPR2||EPHB2||EFNA2||CYBA||PJVK||KCNC1||ANXA3||SEMA4F||SERPINF1||KCNN3||SLC12A5||SLC1A1||ATP1A3||AZIN2||P2RX4||LRP1||ARC||GABRD||SLC2A3||RET||NRP1||DNER||KCNN2||ANG||ITGA4||CPLX1||KCNJ11||SYNPO||RTN4R||KCNQ1||ASTN1||KCNK1||SKOR1||PRPH||SEPTIN4||SLC8A2||SRCIN1||SHANK2||SNCAIP||NEURL1||KCND2||TGFB2||SLC8A3||PVALB||CNKSR2||PDE1A||L1CAM||SORCS2||CHRNA4 | 6.088386 | 6.064478 | 6.143699 | 6.064380 | 6.088676 | 6.170687 | 6.061084 | 6.052170 | 6.055216 | 6.072169 | 6.136491 | 6.048338 | 6.082234 | 6.062661 | 6.108822 | 6.167064 | 6.154562 | 6.073336 | 6.054406 | 6.065335 | 6.095114 | 6.050134 | 6.119914 | 6.136132 | 6.150867 | 6.223536 | 6.065745 | 6.063231 | 6.054252 | 6.062128 | 6.050009 | 6.044315 |
| GO:0045121 | membrane raft | 235 | -0.3538722 | -1.400449 | 1.413744e-02 | 1.539245e-01 | 1.380704e-01 | 4281 | tags=40%, list=27%, signal=30% | ATP1B1||STOM||STOML2||SLC9A1||GNAI1||EMP2||ITGB1||RTN4RL1||CASP3||PLPP1||DPP4||ATP1B3||PODXL||LAMTOR2||RANGRF||SLC25A5||TGFBR1||FADD||RTN4RL2||SORBS1||CNTN1||PRNP||CSK||HMOX1||RGS19||BACE1||CDH2||TNFRSF1A||GPM6B||MAL2||PPT1||DLL1||ERLIN2||PSEN1||ATP1A1||LAMP2||CD55||FURIN||ADCYAP1R1||IRS1||CLN6||TUBA1A||CD19||APP||EGFR||SULF1||TPP1||SLC2A1||DAG1||IL6ST||GASK1A||EPHB1||CDH15||PTCH1||PRKCZ||UNC5A||RFTN1||GP6||MYADM||LAMTOR4||EFNB1||KCNMA1||BMPR2||HAS2||ABCG2||BSG||GPC1||HYAL2||EFNA5||SLC1A1||ORAI1||STOML1||ARC||SCARB1||KCNA3||CTSD||RET||BVES||SERPINH1||FAS||CD24||PTGIS||RTN4R||CD226||KCNQ1||EDNRB||BST2||PTGS2||ITGB2||KCND2||CAVIN4||S1PR1||HPSE||AKAP6 | 6.149780 | 6.211214 | 6.255736 | 6.249522 | 6.172197 | 6.312804 | 6.164023 | 6.246442 | 6.104961 | 6.190470 | 6.152637 | 6.248869 | 6.214154 | 6.169523 | 6.179639 | 6.297549 | 6.287092 | 6.281048 | 6.196845 | 6.269240 | 6.204933 | 6.126550 | 6.183972 | 6.343815 | 6.269183 | 6.324378 | 6.170378 | 6.161617 | 6.160054 | 6.244058 | 6.262938 | 6.232163 |
| GO:0034702 | ion channel complex | 159 | -0.3879298 | -1.462711 | 1.427526e-02 | 1.539245e-01 | 1.380704e-01 | 2477 | tags=28%, list=16%, signal=24% | LRRC8A||ABHD12||KCNH4||PORCN||CHRNB1||PDE4D||KCND3||CCDC51||PACC1||KCNJ8||KCNB2||SMDT1||KCNMA1||HSPA2||SCN2A||LRRC8D||LRRC8E||KCNC1||KCNS3||ORAI1||CLDN4||TRPC3||GABRD||KCNA3||SCN4B||RYR2||GRIK4||KCNIP3||GRIN2D||KCNJ11||OLFM2||KCNQ1||KCNK1||KCNQ3||HCN4||KCND2||CHRNB4||KCNA6||CACNG6||CATSPERE||BEST1||GABRQ||CATSPER2||AKAP6||CHRNA4 | 3.732001 | 3.979938 | 3.930328 | 3.968268 | 3.755962 | 3.903276 | 3.986664 | 3.991506 | 3.704058 | 3.784980 | 3.705471 | 3.969486 | 3.984144 | 3.986126 | 3.953376 | 3.953358 | 3.883117 | 3.975626 | 3.937215 | 3.991426 | 3.763657 | 3.700374 | 3.802034 | 3.823291 | 3.831802 | 4.043749 | 3.988304 | 3.991499 | 3.980165 | 3.978242 | 4.025389 | 3.970270 |
| GO:0031093 | platelet alpha granule lumen | 37 | -0.5537815 | -1.649472 | 1.530902e-02 | 1.604212e-01 | 1.438979e-01 | 3775 | tags=57%, list=24%, signal=43% | IGF2||CFD||MAGED2||FN1||TGFB1||A1BG||APP||PCYOX1L||QSOX1||PDGFA||SERPING1||TIMP1||PROS1||GAS6||TEX264||VEGFC||SPARC||F8||CLU||TGFB2||TOR4A | 6.455978 | 6.067589 | 6.225215 | 6.052273 | 6.495753 | 6.595885 | 5.997925 | 6.011824 | 6.445736 | 6.312899 | 6.595434 | 6.092561 | 6.084268 | 6.025001 | 6.092165 | 6.327989 | 6.245646 | 6.105500 | 5.989270 | 6.059680 | 6.479190 | 6.406549 | 6.595214 | 6.559323 | 6.728697 | 6.489009 | 6.012709 | 5.992107 | 5.988843 | 5.996404 | 6.034516 | 6.004271 |
| GO:0005681 | spliceosomal complex | 183 | 0.3586746 | 1.419955 | 1.591512e-02 | 1.643665e-01 | 1.474368e-01 | 6220 | tags=51%, list=39%, signal=31% | SNIP1||ZRSR2||RBM5||SRRM2||SREK1||SLU7||ZCCHC8||PRPF38B||LUC7L2||BUD13||WBP4||YJU2||ZNF830||CDC40||ENSG00000273784||ZCRB1||RBM22||SRRM1||PRKRIP1||PNN||DHX38||RBM28||SNRNP48||ZMAT2||CWF19L2||SF1||TTF2||FRG1||DDX41||PRPF40A||CDC5L||LSM5||CCDC12||LSM8||SRSF1||TRA2A||PRPF4B||CWC22||SNRPD3||PRPF8||AQR||PPWD1||PPP1R8||PDCD7||SNRPG||DQX1||NCL||MYEF2||HTATSF1||HNRNPR||TRA2B||API5||GPATCH1||PRPF39||PRPF38A||AKAP17A||DHX15||SF3A1||SMNDC1||PRPF18||CWC25||HNRNPH1||WBP11||SNRNP200||ADAR||SUGP1||SYNCRIP||XAB2||DDX23||SF3B3||SNRPE||HNRNPA2B1||SYF2||SF3B6||CASC3||LSM3||MAGOHB||DHX35||SNRPD1||C9orf78||RBM17||MFAP1||SNRPF||PRPF3||BCAS2||HNRNPA3||SNRNP35||SF3B1||RBM3||DHX8||HNRNPU||PPIE||HNRNPA1 | 7.104224 | 7.698157 | 7.198791 | 7.699457 | 7.019311 | 7.046866 | 7.744581 | 7.767768 | 7.121561 | 7.213042 | 6.967475 | 7.712279 | 7.686777 | 7.695299 | 7.423040 | 7.025046 | 7.117623 | 7.683254 | 7.671305 | 7.742792 | 6.985366 | 7.093440 | 6.976155 | 7.030759 | 7.007742 | 7.100474 | 7.733677 | 7.743919 | 7.756059 | 7.751403 | 7.779380 | 7.772374 |
| GO:0043083 | synaptic cleft | 13 | -0.7102575 | -1.670466 | 1.656920e-02 | 1.643665e-01 | 1.474368e-01 | 2986 | tags=69%, list=19%, signal=56% | LAMA5||EGFLAM||LAMB2||NLGN1||APOE||C1QL1||SLC1A1||ACHE||PRSS12 | 2.184061 | 2.669485 | 3.238747 | 2.270868 | 2.160663 | 2.767716 | 2.507872 | 2.142336 | 2.313763 | 1.816342 | 2.361564 | 2.572762 | 2.726862 | 2.704087 | 3.317982 | 3.327032 | 3.055038 | 2.236170 | 2.406578 | 2.158595 | 2.026584 | 2.028755 | 2.394704 | 2.027167 | 2.400705 | 3.469132 | 2.598599 | 2.478519 | 2.441797 | 2.109426 | 2.276402 | 2.030128 |
| GO:0098945 | intrinsic component of presynaptic active zone membrane | 13 | -0.7107340 | -1.671586 | 1.656920e-02 | 1.643665e-01 | 1.474368e-01 | 3173 | tags=62%, list=20%, signal=49% | GPM6A||CDH2||SYT11||CANX||NECTIN1||LRFN3||NTNG2||NTNG1 | 3.957734 | 6.404142 | 4.172257 | 6.120434 | 3.927657 | 4.143892 | 6.248890 | 6.108178 | 3.976419 | 4.071018 | 3.814164 | 6.374227 | 6.478347 | 6.356835 | 4.647914 | 3.822205 | 3.894536 | 5.998043 | 6.202594 | 6.152866 | 3.824054 | 4.108346 | 3.832035 | 4.222705 | 4.185397 | 4.015205 | 6.236471 | 6.260062 | 6.250041 | 6.115929 | 6.141818 | 6.065753 |
| GO:0031526 | brush border membrane | 33 | -0.5678438 | -1.650040 | 1.726428e-02 | 1.664761e-01 | 1.493291e-01 | 2466 | tags=39%, list=15%, signal=33% | SLC22A5||MFSD10||CYBRD1||PEMT||ABCG2||SLC19A1||ATP8B1||SLC46A1||PTH1R||MTTP||KCNK1||SHANK2||SLC26A4 | 5.322974 | 5.840624 | 5.542628 | 5.998322 | 5.238539 | 5.430938 | 5.871495 | 5.992134 | 5.270284 | 5.444081 | 5.246379 | 5.847459 | 5.839788 | 5.834597 | 5.767806 | 5.342592 | 5.484561 | 5.992744 | 5.953350 | 6.047326 | 5.282493 | 5.257300 | 5.173584 | 5.509898 | 5.415705 | 5.363372 | 5.886428 | 5.860365 | 5.867567 | 6.005840 | 5.994239 | 5.976167 |
| GO:0000776 | kinetochore | 131 | 0.3860043 | 1.460108 | 1.745314e-02 | 1.664761e-01 | 1.493291e-01 | 4951 | tags=45%, list=31%, signal=31% | REC8||CENPF||KAT2B||SKA3||ZNF276||PPP1R12A||CENPE||BOD1L1||APC||CENPI||CENPU||SPOUT1||MTBP||SPAG5||SUGT1||CENPQ||SPDL1||SGO1||DYNC1LI2||HJURP||CENPN||NDC80||TTK||TP53BP1||TPR||ITGB3BP||ZWILCH||CKAP5||SKA2||KIF18A||KNL1||NDEL1||SGO2||AHCTF1||PHF6||CLASP2||BUB1||NUP98||AURKB||KANSL1||CENPC||KNTC1||KIF2C||PAFAH1B1||TRAPPC12||SIN3A||MAD2L1||DCTN6||CSNK1A1||XPO1||MEAF6||MAD1L1||DCTN4||PPP1CC||CENPO||BUB1B||SEPTIN2||TEX14||NUP160 | 5.560569 | 6.219375 | 5.721311 | 6.261534 | 5.506113 | 5.620849 | 6.277896 | 6.340345 | 5.544315 | 5.643152 | 5.490043 | 6.229958 | 6.208856 | 6.219234 | 5.921241 | 5.580421 | 5.638776 | 6.228672 | 6.225319 | 6.328224 | 5.475269 | 5.539889 | 5.502450 | 5.576153 | 5.623125 | 5.661989 | 6.276911 | 6.276888 | 6.279886 | 6.320064 | 6.363214 | 6.337431 |
| GO:0016607 | nuclear speck | 382 | 0.2978530 | 1.273136 | 2.209302e-02 | 2.054651e-01 | 1.843023e-01 | 4273 | tags=35%, list=27%, signal=26% | CCNB3||PRKN||PRKAA1||TENM1||CCNL1||WT1||BASP1||EAF2||THOC1||AFDN||RBM11||NRDE2||TCERG1||NFKBIZ||TCF12||SON||THOC2||CLK2||SRRM2||PLAG1||DDX42||MEF2C||RBM15||ENSG00000275740||DENND1B||BAZ2A||AP5Z1||PNISR||SREK1||CRY2||RREB1||SLU7||PIAS1||SRSF10||DNAAF1||WRN||SPRTN||DHX36||CCNL2||WAC||NCBP3||GTF2H4||SGK1||ZC3H13||NSRP1||LUC7L2||SETD1A||AFF2||YTHDC1||PPP4R3A||PPIG||NFATC4||ERBIN||TRIP12||RBBP6||NOC3L||EPAS1||ZNF217||CDK12||WBP4||PPP4R3B||SCAPER||HIF1A||HAUS6||SMC5||GADD45A||MECOM||PSME4||CDK13||HBP1||ZNF830||CDC40||RBM25||AR||TERT||ACIN1||RBM39||FNBP4||CPSF6||PRKAA2||SRRM1||YLPM1||CARMIL1||PNN||EHMT2||EP400||AK6||HIPK1||NRIP1||FAM193B||RADX||CHTOP||RBM27||RBM4B||ASCC3||FAM76B||RUFY1||GTF2H2||SMC4||ZNF638||SART3||FTO||NXF1||PATL1||VIRMA||DAZAP2||ZC3H14||PRPF40A||CDC5L||DYRK1A||SRPK2||PIP5K1A||PIAS2||THRAP3||JADE1||SRSF1||SUMO1||PRPF4B||CWC22||RBM10||PRPF8||ZNF106||BCLAF1||GPATCH2||CIR1||PPP1R8||TEPSIN||MSL1||THOC7||KAT6A||EAF1||CSNK1A1 | 5.865260 | 6.445080 | 6.005981 | 6.417063 | 5.839672 | 5.936139 | 6.497895 | 6.491932 | 5.864389 | 5.927728 | 5.800877 | 6.461216 | 6.433690 | 6.440193 | 6.202953 | 5.888633 | 5.904118 | 6.384376 | 6.385743 | 6.479014 | 5.836205 | 5.838331 | 5.844467 | 5.884948 | 5.871835 | 6.045104 | 6.496430 | 6.489176 | 6.508016 | 6.473170 | 6.510306 | 6.492081 |
| GO:0035097 | histone methyltransferase complex | 57 | 0.4430355 | 1.462866 | 3.394200e-02 | 3.006291e-01 | 2.696645e-01 | 5158 | tags=49%, list=32%, signal=33% | HDAC9||KMT2D||KMT2A||MGA||SETD1A||HCFC1||KMT2C||PHF20||KDM6B||SENP3||JARID2||HCFC2||CHD8||RBBP5||PAXIP1||KMT2B||AEBP2||KANSL1||EZH2||EZH1||KDM6A||TAF4||PRDM4||SUZ12||TAF1||RNF2||TAF9||NCOA6 | 5.718651 | 6.149748 | 5.897540 | 6.125622 | 5.696651 | 5.859102 | 6.175842 | 6.183203 | 5.680314 | 5.755246 | 5.719419 | 6.166967 | 6.148681 | 6.133400 | 6.008375 | 5.834471 | 5.843030 | 6.122074 | 6.083434 | 6.170053 | 5.676211 | 5.675379 | 5.737480 | 5.812533 | 5.825026 | 5.936491 | 6.174276 | 6.174269 | 6.178976 | 6.162506 | 6.209213 | 6.177495 |
| GO:0016324 | apical plasma membrane | 214 | -0.3367659 | -1.319973 | 3.726931e-02 | 3.092964e-01 | 2.774390e-01 | 2941 | tags=31%, list=18%, signal=26% | PSEN1||ATP1A1||SLC24A4||SLC7A1||P2RX2||ARHGEF18||SLC39A14||C2CD2L||SLC4A11||SHROOM3||SLC22A5||SLC29A1||SLC7A5||SLC16A2||TRPV4||SLC9A3R2||EGFR||SLC2A1||FZD6||CYBRD1||PDE4D||OTOG||IGFBP2||AJM1||PRKCZ||ATP6AP2||CTSB||CD9||MFSD4B||SLC39A4||KCNMA1||BMPR2||MIP||CTSL||ABCG2||DDR2||CYBA||SLC19A1||CSPG4||HYAL2||SLC1A1||ATP8B1||CLDN4||ENPP3||SIPA1L3||SLC46A1||ABCB1||IL6R||SLC22A4||CEACAM1||CNTFR||SLC4A5||ADRB2||PTH1R||KCNQ1||EPCAM||KCNK1||TF||CLDN1||BST2||NHS||PARD6A||SLC22A18||SHANK2||MUC1||SHROOM2||SLC26A4 | 4.635046 | 5.200993 | 4.857120 | 5.227888 | 4.620413 | 4.856124 | 5.158821 | 5.191650 | 4.586591 | 4.659312 | 4.658040 | 5.216153 | 5.224250 | 5.161779 | 4.948551 | 4.826457 | 4.791602 | 5.216039 | 5.218717 | 5.248679 | 4.634086 | 4.566062 | 4.659481 | 4.821585 | 4.832456 | 4.912608 | 5.174494 | 5.157451 | 5.144361 | 5.182246 | 5.221622 | 5.170584 |
| GO:0010008 | endosome membrane | 440 | -0.2949305 | -1.243402 | 3.785653e-02 | 3.092964e-01 | 2.774390e-01 | 4278 | tags=33%, list=27%, signal=25% | ITM2B||RAB4A||STARD3||SORT1||THSD1||ARL8A||IRAK1||PIP5K1C||GOLIM4||ABCB6||ITGB1||SNX8||PARM1||YIPF1||COMMD1||VPS16||LAMTOR2||SBF2||MCOLN1||VPS33A||TPCN1||NCSTN||SNF8||VPS4B||ABCA5||ITCH||AP2S1||PML||SCARB2||ANTXR2||APH1A||CDIP1||ERBB2||STEAP3||SNX14||SPPL2A||RFFL||WIPI1||VPS37B||RAB11B||ANTXR1||TMEM9B||ZFYVE27||SCAMP3||ABHD6||VAMP8||BACE1||PLEKHM1||DTNBP1||TMEM59||CD63||SYT11||LAPTM4B||KIAA0319||PSEN1||PSENEN||LAMP1||LAMP2||EHD1||CHMP4C||SNX17||VPS37D||SLC39A14||EPHA4||STEAP1||FURIN||TM9SF2||LEPROT||FZD7||MCOLN2||ABCA3||HLA-DMB||HLA-E||EPHA8||SLC29A3||NDFIP1||HSD17B6||ATP6V0A2||EGFR||SLC15A4||WLS||CLCN4||TSPAN15||CLCN3||SUN2||PDIA3||TMEM106B||ATP6AP1||PLD3||EPHB1||STEAP2||TMEM9||MARCHF3||HPS6||ZDHHC2||LAPTM4A||LAMP3||IRAK4||CD320||ATP6AP2||RAB11FIP4||OPTN||PRAF2||FCGRT||FIG4||AP1G2||SLC39A4||LAMTOR4||WNT3A||ATP13A2||HLA-B||FLT3||CD207||MYD88||ATP6V0B||CPTP||SLC30A4||DAGLA||KREMEN2||OCA2||HLA-DQB1||CYB561A3||SLC1A1||MR1||HLA-C||PMEPA1||ZNRF2||STOML1||ARC||MARCHF2||ATP6V0C||SCAMP4||BOK||CTSD||HLA-A||RET||ADRB2||TMBIM1||TF||YIPF2||SLC9A9||HLA-DMA||RAB5B||TICAM1||HLA-DOA||PDLIM4||SORCS2 | 5.594003 | 5.796665 | 5.676522 | 5.757041 | 5.569940 | 5.596806 | 5.761073 | 5.744374 | 5.592019 | 5.622091 | 5.567377 | 5.812857 | 5.805094 | 5.771714 | 5.662104 | 5.659786 | 5.707178 | 5.761910 | 5.745658 | 5.763489 | 5.562752 | 5.597397 | 5.549244 | 5.575722 | 5.546928 | 5.665119 | 5.767223 | 5.761363 | 5.754607 | 5.744123 | 5.752603 | 5.736351 |
| GO:0031010 | ISWI-type complex | 11 | 0.6831360 | 1.566677 | 3.804236e-02 | 3.092964e-01 | 2.774390e-01 | 2937 | tags=55%, list=18%, signal=45% | BPTF||BAZ1A||HMGXB4||RSF1||CECR2||SMARCA5 | 5.357093 | 6.442555 | 5.467658 | 6.286114 | 5.225091 | 5.199817 | 6.478306 | 6.388474 | 5.424981 | 5.521098 | 5.090752 | 6.482613 | 6.406955 | 6.437089 | 5.821414 | 5.071874 | 5.411830 | 6.214612 | 6.255905 | 6.382477 | 5.166739 | 5.385804 | 5.107615 | 5.224101 | 5.171179 | 5.203676 | 6.447949 | 6.476436 | 6.509868 | 6.345135 | 6.425616 | 6.393536 |
| GO:0042581 | specific granule | 97 | -0.4041767 | -1.415464 | 3.824522e-02 | 3.092964e-01 | 2.774390e-01 | 4286 | tags=45%, list=27%, signal=33% | STK10||STOM||CD47||ANXA11||CRACR2A||LAMTOR2||DNASE1L1||VCL||TMEM30A||QPCT||TOLLIP||STX3||DEGS1||ITGAV||CANT1||SLC44A2||VAMP8||CNN2||GGH||AGPAT2||SLC27A2||ERP44||ADAM10||CKAP4||SLC15A4||CLCN3||GHDC||QSOX1||DGAT1||CTSZ||PTX3||NEU1||CYBA||ANXA3||TSPAN14||CTSD||SLC2A3||CEACAM1||MMP25||TMBIM1||PTPRB||ITGB2||CHRNB4||HPSE | 5.140466 | 5.611174 | 5.373704 | 5.588076 | 5.116500 | 5.253952 | 5.558000 | 5.548498 | 5.137039 | 5.170737 | 5.113038 | 5.617411 | 5.634401 | 5.581200 | 5.414158 | 5.355190 | 5.350895 | 5.574216 | 5.591366 | 5.598537 | 5.091923 | 5.144487 | 5.112604 | 5.167834 | 5.196744 | 5.387284 | 5.588468 | 5.555608 | 5.529316 | 5.538477 | 5.584838 | 5.521429 |
| GO:0030669 | clathrin-coated endocytic vesicle membrane | 45 | -0.4853929 | -1.497952 | 3.844697e-02 | 3.092964e-01 | 2.774390e-01 | 3601 | tags=36%, list=23%, signal=28% | AP2S1||SYT2||SCARB2||VAMP8||WNT5A||KIAA0319||EGFR||ROR2||APOE||CD9||CD207||HLA-DQB1||FZD2||ADRB2||TF||TGFA | 4.877200 | 5.006330 | 5.070948 | 5.031236 | 4.876745 | 5.046074 | 4.985476 | 5.037038 | 4.883386 | 4.816738 | 4.929261 | 5.005131 | 5.046995 | 4.965720 | 5.049918 | 5.078783 | 5.083912 | 5.038498 | 5.002303 | 5.052444 | 4.883927 | 4.844038 | 4.901667 | 5.010320 | 5.010415 | 5.114940 | 4.985538 | 4.993200 | 4.977647 | 5.028000 | 5.033976 | 5.049057 |
| GO:0031166 | integral component of vacuolar membrane | 10 | -0.7088336 | -1.566302 | 3.907776e-02 | 3.092964e-01 | 2.774390e-01 | 2932 | tags=60%, list=18%, signal=49% | LAMP2||LRRC8A||SLC15A4||ATP13A2||LRRC8E||SLC46A3 | 3.882848 | 4.865354 | 4.509787 | 4.998031 | 3.912317 | 4.437452 | 4.731325 | 4.836717 | 3.823713 | 3.929843 | 3.892983 | 4.840272 | 4.957924 | 4.792822 | 4.593347 | 4.539209 | 4.389128 | 4.965483 | 5.038188 | 4.989468 | 3.876465 | 3.810058 | 4.040571 | 4.226843 | 4.290666 | 4.739117 | 4.782818 | 4.724192 | 4.685291 | 4.828803 | 4.913590 | 4.763842 |
| GO:0042470 | melanosome | 95 | -0.4015911 | -1.403074 | 4.070531e-02 | 3.154661e-01 | 2.829733e-01 | 4255 | tags=49%, list=27%, signal=37% | STOM||AHCY||ABCB6||ITGB1||ANXA11||ATP1B3||NCSTN||SND1||STX3||MFSD12||SYPL1||GGH||DTNBP1||CD63||ATP1A1||CNP||LAMP1||TMED10||GANAB||SYNGR1||RAB32||SLC1A5||CALU||ERP29||TPP1||SLC2A1||CANX||PDIA3||GCHFR||RPN1||HSP90B1||TMEM33||HSPA5||CTNS||PPIB||CTSB||PDIA6||SYTL1||SLC1A4||P4HB||BSG||PDIA4||SERPINF1||OCA2||CTSD||SYTL2||RAB5B | 7.010296 | 7.833017 | 7.160667 | 7.769717 | 6.909859 | 6.978208 | 7.762656 | 7.769167 | 7.046093 | 7.145319 | 6.820559 | 7.876651 | 7.837294 | 7.783595 | 7.269641 | 7.061548 | 7.143156 | 7.736626 | 7.768613 | 7.803144 | 6.914484 | 6.998737 | 6.810190 | 6.980262 | 6.882413 | 7.066104 | 7.757252 | 7.768207 | 7.762489 | 7.767406 | 7.782845 | 7.757135 |
| GO:0098576 | lumenal side of membrane | 24 | -0.5579233 | -1.509343 | 4.245554e-02 | 3.256383e-01 | 2.920978e-01 | 3742 | tags=58%, list=23%, signal=45% | SPPL3||SPPL2A||BCAP31||HLA-E||HM13||CANX||TAPBP||NUCB1||HLA-B||CALR||HLA-DQB1||HLA-C||HLA-A||BDH1 | 7.098981 | 7.962694 | 7.260715 | 7.544408 | 7.024173 | 7.048568 | 7.818049 | 7.498862 | 7.121898 | 7.197615 | 6.968030 | 8.034320 | 7.969210 | 7.880428 | 7.239394 | 7.251208 | 7.291034 | 7.522752 | 7.557667 | 7.552560 | 7.007601 | 7.080379 | 6.982745 | 6.990595 | 6.955265 | 7.188709 | 7.837147 | 7.819565 | 7.797159 | 7.492556 | 7.531000 | 7.472414 |
| GO:0031233 | intrinsic component of external side of plasma membrane | 10 | -0.7048170 | -1.557426 | 4.298554e-02 | 3.263392e-01 | 2.927265e-01 | 3415 | tags=60%, list=21%, signal=47% | PRNP||F3||HYAL2||EFNA5||CD24||RTN4R | 3.332812 | 4.440187 | 4.058442 | 4.524013 | 3.402302 | 3.523824 | 4.322973 | 4.320987 | 3.179984 | 3.619809 | 3.149070 | 4.396505 | 4.560309 | 4.355514 | 4.134941 | 3.999764 | 4.037223 | 4.523665 | 4.604180 | 4.439498 | 3.297790 | 3.592363 | 3.296158 | 3.374845 | 3.149234 | 3.931761 | 4.365849 | 4.316475 | 4.285452 | 4.350808 | 4.331794 | 4.279415 |
| GO:1902562 | H4 histone acetyltransferase complex | 31 | 0.5128531 | 1.497573 | 4.510961e-02 | 3.306150e-01 | 2.965619e-01 | 4944 | tags=58%, list=31%, signal=40% | MSL3P1||BRD8||EPC1||ATF2||TRRAP||PHF20||ING3||KANSL2||EP400||PHF20L1||EPC2||KANSL1||MSL1||MEAF6||DMAP1||MSL3||ACTL6A||MSL2 | 7.075678 | 6.987043 | 6.817963 | 6.871040 | 7.067263 | 7.038000 | 6.979550 | 6.908346 | 7.052456 | 7.095717 | 7.078532 | 7.005960 | 6.981012 | 6.973962 | 6.775540 | 6.837551 | 6.839880 | 6.910678 | 6.797458 | 6.902250 | 7.113135 | 7.013926 | 7.073007 | 7.101807 | 7.009235 | 7.000765 | 6.982319 | 6.980272 | 6.976051 | 6.912660 | 6.912163 | 6.900181 |
| GO:0099056 | integral component of presynaptic membrane | 40 | -0.4882356 | -1.474865 | 4.674140e-02 | 3.306150e-01 | 2.965619e-01 | 4474 | tags=60%, list=28%, signal=43% | PTPRS||NPTN||GRIK5||SLC6A9||PCDH17||SYT7||NCSTN||APH1A||GPM6A||CDH2||SYT11||PSEN1||EFNB2||EPHA4||EFNB3||CANX||NECTIN1||LRFN3||SCN2A||EPHB2||KCNC1||KCNA3||GRIK4||PTPRD | 3.610413 | 5.143713 | 4.090730 | 4.937398 | 3.634082 | 4.021684 | 4.998610 | 4.894020 | 3.546521 | 3.627177 | 3.655342 | 5.126003 | 5.216851 | 5.085117 | 4.221529 | 4.096187 | 3.940825 | 4.856836 | 4.999799 | 4.951909 | 3.601155 | 3.582551 | 3.714947 | 3.915728 | 3.946062 | 4.187599 | 5.007725 | 5.006775 | 4.981172 | 4.898340 | 4.931903 | 4.850663 |
| GO:0098685 | Schaffer collateral - CA1 synapse | 59 | -0.4455830 | -1.444464 | 4.804636e-02 | 3.340794e-01 | 2.996694e-01 | 4474 | tags=41%, list=28%, signal=29% | PTPRS||SLC30A1||NPTN||ACTB||ITGB1||GIPC1||PRKAR1B||STX3||CDK5||DTNBP1||SYT11||EPHA4||TAMALIN||TUBB2B||PRKCZ||SYN1||PLAT||SYP||CPLX1||SYN2||PTPRD||APBA1||NTNG2||NTNG1 | 7.017200 | 6.545018 | 6.536901 | 6.480494 | 7.040540 | 6.921391 | 6.494004 | 6.468929 | 6.970165 | 7.014665 | 7.065201 | 6.541294 | 6.557670 | 6.536001 | 6.347160 | 6.618510 | 6.627808 | 6.547737 | 6.393707 | 6.495806 | 7.136971 | 6.922458 | 7.054127 | 7.070193 | 6.887637 | 6.792409 | 6.494905 | 6.511291 | 6.475596 | 6.469571 | 6.482053 | 6.455037 |
| GO:0030670 | phagocytic vesicle membrane | 60 | -0.4421085 | -1.435994 | 4.913728e-02 | 3.357663e-01 | 3.011825e-01 | 4048 | tags=42%, list=25%, signal=31% | RAB43||SYT7||PIK3R4||MCOLN1||TAP2||CORO1A||RAB11B||RAB34||VAMP8||LAMP1||RAB32||HLA-E||ATP6V0A2||TAPBP||TAP1||RAB9B||HLA-B||CALR||RAB20||ATP6V0B||CYBA||ANXA3||HLA-C||ATP6V0C||HLA-A | 5.873794 | 6.262405 | 6.062628 | 5.982838 | 5.905715 | 6.091466 | 6.092183 | 5.898822 | 5.850058 | 5.824989 | 5.943607 | 6.316206 | 6.290979 | 6.176207 | 5.943169 | 6.172875 | 6.062706 | 6.022430 | 5.974994 | 5.950153 | 5.916694 | 5.826329 | 5.970460 | 6.021160 | 6.067648 | 6.180880 | 6.124620 | 6.085173 | 6.066136 | 5.895489 | 5.943174 | 5.856495 |
| GO:0097431 | mitotic spindle pole | 34 | 0.4920388 | 1.467045 | 4.932546e-02 | 3.357663e-01 | 3.011825e-01 | 4462 | tags=47%, list=28%, signal=34% | CNTRL||MAPKBP1||ASPM||KIF20B||NIN||SPAG5||CDK5RAP2||TNKS||GPSM2||EML1||RMDN1||AURKB||FAM83D||SMC3||FAM161A||MAD1L1 | 4.901417 | 5.631067 | 5.184197 | 5.708531 | 4.845653 | 5.165152 | 5.709392 | 5.804958 | 4.826175 | 4.943945 | 4.931261 | 5.623476 | 5.637453 | 5.632238 | 5.395992 | 5.069230 | 5.061500 | 5.680394 | 5.678756 | 5.764757 | 4.842815 | 4.784394 | 4.907136 | 5.091749 | 5.157892 | 5.241885 | 5.718161 | 5.703923 | 5.706049 | 5.801101 | 5.825302 | 5.788225 |
| GO:0045211 | postsynaptic membrane | 182 | -0.3414459 | -1.309904 | 4.993621e-02 | 3.357663e-01 | 3.011825e-01 | 3213 | tags=33%, list=20%, signal=27% | ABHD6||SIGMAR1||CDH2||GABBR2||DTNBP1||GRID1||CACNG4||SLC8A1||SEMA4C||DBNL||EPHA4||TMUB1||EFNB3||ADAM10||TAMALIN||NRGN||CLSTN3||NLGN1||CLSTN1||CHRNB1||DAG1||CANX||SNTA1||COL13A1||ADRA2C||LRFN4||KCND3||DBN1||COMT||ATP6AP2||LRFN3||KCNMA1||EPHB2||DAGLB||KCNC1||LRFN1||DAGLA||SEMA4F||CHRM3||RGS9||PRR7||SYNE1||ARC||GABRD||KCNA3||GRIK4||NRP1||CLSTN2||GRIN2D||LRRC4B||CHRM4||SHANK2||KCND2||CHRNB4||SLC8A3||CNKSR2||PDLIM4||GABRQ||SORCS2||CHRNA4 | 4.144982 | 5.128552 | 4.240252 | 4.951631 | 4.017960 | 4.097804 | 5.071921 | 4.933420 | 4.175887 | 4.298755 | 3.937209 | 5.177698 | 5.129319 | 5.076878 | 4.421762 | 4.088666 | 4.189853 | 4.910060 | 4.959346 | 4.984495 | 3.998788 | 4.085136 | 3.967363 | 4.035187 | 4.062784 | 4.190628 | 5.081424 | 5.073675 | 5.060587 | 4.926799 | 4.956911 | 4.916240 |
KEGG
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa04142 | Lysosome | 115 | -0.4749959 | -1.739090 | 0.001003240 | 0.1037088 | 0.09505074 | 3720 | tags=52%, list=26%, signal=39% | SORT1||CTSV||ACP2||NPC2||FUCA2||NAGA||NAPSA||MCOLN1||SUMF1||GUSB||SCARB2||IDUA||HEXB||FUCA1||DMXL1||PPT1||GNPTG||CD63||LAPTM4B||CTSO||SLC17A5||LAMP1||LAMP2||IDS||GLA||MAN2B1||PLA2G15||GNS||ATP6V0A2||LIPA||TPP1||LGMN||GBA||ATP6AP1||GALNS||CTSC||CTNS||LAPTM4A||LAMP3||PSAP||CTSB||GAA||AP1G2||DNASE2||CTSH||HEXA||GLB1||CTSL||CTSZ||ATP6V0B||NEU1||CLN5||HYAL2||ARSA||ATP6V0C||NAGLU||CTSD||CTSF||ARSG||SMPD1 | 5.467940 | 5.794602 | 5.847021 | 5.811989 | 5.481333 | 5.762974 | 5.662281 | 5.718441 | 5.434193 | 5.464340 | 5.504426 | 5.830638 | 5.820942 | 5.730111 | 5.811361 | 5.903655 | 5.824304 | 5.811159 | 5.830577 | 5.793998 | 5.480723 | 5.430026 | 5.531466 | 5.627734 | 5.654944 | 5.979122 | 5.694653 | 5.660171 | 5.631321 | 5.723594 | 5.757407 | 5.673076 |
| hsa04080 | Neuroactive ligand-receptor interaction | 109 | -0.4852600 | -1.766460 | 0.002109456 | 0.1037088 | 0.09505074 | 2265 | tags=39%, list=16%, signal=33% | CALCB||ADCYAP1R1||TSPO||LYNX1||SSTR2||ADORA2B||GPR83||LPAR1||CHRNB1||PTAFR||ADRA2C||NMB||GPR50||CGA||S1PR2||PTGER2||GRM4||F2RL1||HTR7||HTR6||CHRM3||P2RX4||GABRD||GRIK4||GRIN2D||UTS2R||PTGER4||ADRB2||PTH1R||EDNRB||CHRM4||LYPD6B||C5||NPY1R||CHRNB4||C5AR1||S1PR1||CCKBR||GABRQ||VGF||CYSLTR1||CHRNA4||NPB | 2.485832 | 2.357086 | 2.479080 | 2.091528 | 2.497752 | 2.611350 | 2.277638 | 2.001408 | 2.452800 | 2.357261 | 2.633659 | 2.356186 | 2.380979 | 2.333705 | 2.403981 | 2.605237 | 2.419196 | 2.132570 | 2.079084 | 2.061985 | 2.497650 | 2.351923 | 2.630278 | 2.507427 | 2.655172 | 2.666073 | 2.332762 | 2.261257 | 2.237173 | 1.990721 | 2.052693 | 1.959237 |
| hsa04145 | Phagosome | 99 | -0.4766086 | -1.713389 | 0.003384095 | 0.1037088 | 0.09505074 | 2500 | tags=35%, list=18%, signal=29% | TUBB3||LAMP1||LAMP2||TUBA1A||HLA-DMB||HLA-E||ATP6V0A2||COMP||SEC61A1||CANX||TUBB2B||DYNC1I1||ATP6AP1||TAP1||TUBB4A||ATP6V1C2||HLA-B||CALR||MRC2||CTSL||ATP6V0B||CYBA||C1R||HLA-DQB1||HLA-C||ITGB5||SCARB1||ATP6V0C||HLA-A||THBS4||HLA-DMA||RAB5B||ITGB2||HLA-DOA||TUBA4A | 7.628790 | 7.374468 | 7.524253 | 7.222731 | 7.693626 | 7.731110 | 7.282890 | 7.208163 | 7.558754 | 7.616005 | 7.707686 | 7.405377 | 7.390687 | 7.326112 | 7.323788 | 7.637197 | 7.592302 | 7.275562 | 7.164417 | 7.226068 | 7.749972 | 7.588620 | 7.736790 | 7.800856 | 7.688738 | 7.701095 | 7.298162 | 7.282181 | 7.268171 | 7.217111 | 7.222326 | 7.184767 |
| hsa04613 | Neutrophil extracellular trap formation | 100 | -0.4683168 | -1.683150 | 0.004078165 | 0.1037088 | 0.09505074 | 2217 | tags=30%, list=16%, signal=25% | H2AX||H4C3||HDAC11||H2BC12||CLCN4||MAPK11||CLCN3||HDAC7||ITGA2B||H2AC20||H2BC5||CYBA||H2BC17||H2BC4||H4C11||H4C8||H2BC11||H2AJ||H2AC12||H2AC11||H2BC15||C5||ITGB2||H2AC6||H2AC18||H2BC18||C5AR1||H4C14||H4-16||H2AC17 | 7.038784 | 6.841004 | 6.825600 | 6.801365 | 7.004127 | 6.868386 | 6.829824 | 6.817625 | 7.043993 | 7.115883 | 6.951798 | 6.852408 | 6.833969 | 6.836565 | 6.791123 | 6.768794 | 6.912689 | 6.835026 | 6.737838 | 6.829186 | 7.040045 | 7.006107 | 6.965258 | 6.957310 | 6.837626 | 6.805758 | 6.824364 | 6.827590 | 6.837487 | 6.835317 | 6.791607 | 6.825588 |
| hsa04141 | Protein processing in endoplasmic reticulum | 159 | -0.4034042 | -1.543600 | 0.005107084 | 0.1037088 | 0.09505074 | 4323 | tags=44%, list=30%, signal=31% | DNAJB2||SEC23B||STT3B||DAD1||SEC13||ATF6||RRBP1||BAX||SEC63||SYVN1||SEL1L||BCL2||SEC61A2||SSR1||EDEM3||XBP1||SSR3||ATF6B||UBE2D4||PREB||SAR1B||BAK1||SELENOS||HSPA1B||FBXO2||ERO1A||HSPBP1||DNAJC1||PRKCSH||ERLEC1||OS9||ATXN3||BCAP31||DERL1||GANAB||MAN1C1||DNAJB11||CKAP4||MOGS||ERP29||TXNDC5||SEC61A1||TUSC3||CANX||EDEM2||PDIA3||RPN1||RPN2||HSP90B1||DDOST||TRAM1||TRAM1L1||STT3A||HSPA5||HYOU1||SSR4||MAN1B1||SIL1||PDIA6||AMFR||SSR2||CALR||P4HB||HSPA2||LMAN2||PDIA4||WFS1||DERL3||HSPA6||HSPA1L | 6.759139 | 7.466288 | 6.850926 | 7.235934 | 6.753035 | 6.788649 | 7.375111 | 7.219846 | 6.764908 | 6.833252 | 6.674897 | 7.499361 | 7.479646 | 7.418636 | 6.911478 | 6.800703 | 6.838388 | 7.206977 | 7.238038 | 7.262255 | 6.774405 | 6.757001 | 6.727305 | 6.793481 | 6.738353 | 6.832564 | 7.386167 | 7.377159 | 7.361903 | 7.215312 | 7.243071 | 7.200835 |
| hsa04514 | Cell adhesion molecules | 80 | -0.4804087 | -1.665318 | 0.005364250 | 0.1037088 | 0.09505074 | 3201 | tags=54%, list=23%, signal=42% | CDH4||NLGN3||CNTN1||CD276||CD40||ITGAV||ICOSLG||NCAM1||CDH2||ALCAM||SDC2||PVR||SDC3||CD99L2||CDH3||HLA-DMB||HLA-E||NLGN1||JAM2||NECTIN1||NECTIN2||CDH15||CADM1||HLA-B||SDC1||HLA-DQB1||HLA-C||CLDN4||NFASC||HLA-A||ITGA4||ICAM3||LRRC4B||102723996||CD226||NRXN2||CLDN1||HLA-DMA||ITGB2||HLA-DOA||NTNG2||L1CAM||NTNG1 | 4.146090 | 4.518703 | 4.513493 | 4.417971 | 4.217774 | 4.493803 | 4.351786 | 4.302158 | 4.098828 | 4.073195 | 4.259125 | 4.545877 | 4.571826 | 4.434762 | 4.407290 | 4.640980 | 4.482256 | 4.442572 | 4.432559 | 4.377949 | 4.229398 | 4.072248 | 4.339267 | 4.348197 | 4.412242 | 4.696648 | 4.405405 | 4.346965 | 4.301091 | 4.316395 | 4.353906 | 4.233557 |
| hsa04360 | Axon guidance | 161 | -0.3924903 | -1.503078 | 0.008242664 | 0.1298507 | 0.11901021 | 3009 | tags=34%, list=21%, signal=27% | EPHA2||SEMA3D||PAK4||TRPC1||NCK2||SEMA4G||RAC3||CDK5||WNT5A||ROBO3||EPHA3||SEMA4C||RRAS||EFNB2||EPHA4||EFNB3||EPHB4||CAMK2B||UNC5C||EPHA8||CXCL12||EPHA5||SEMA6B||EPHB1||NTN1||PTCH1||SEMA3F||PRKCZ||UNC5A||PLXNC1||EFNB1||SEMA6D||BMPR2||EPHB6||EPHB2||EFNA2||SEMA4F||EFNA5||EFNA4||PLXNA2||TRPC3||EPHB3||NRP1||SLIT3||CXCR4||FES||SEMA7A||NFATC2||PARD6A||WNT5B||ABLIM3||NTNG2||L1CAM||NTNG1 | 4.782213 | 4.803769 | 4.816522 | 4.840771 | 4.808555 | 4.954890 | 4.811167 | 4.863859 | 4.720088 | 4.784887 | 4.839201 | 4.794507 | 4.822989 | 4.793617 | 4.803297 | 4.836322 | 4.809735 | 4.846260 | 4.788513 | 4.885881 | 4.816708 | 4.728058 | 4.877020 | 4.916890 | 4.987283 | 4.959626 | 4.824391 | 4.805834 | 4.803184 | 4.851843 | 4.882331 | 4.857220 |
| hsa05168 | Herpes simplex virus 1 infection | 376 | 0.2953473 | 1.322964 | 0.008955224 | 0.1298507 | 0.11901021 | 2038 | tags=25%, list=14%, signal=22% | ZNF114||ZNF471||ZNF492||HLA-DOB||ZNF888||ZNF813||ZNF230||ZNF682||ZNF404||ZNF283||ZNF17||ZNF587||ZNF780B||ZNF766||ZNF571||ZNF548||ZNF442||ZNF611||ZNF112||ZNF547||ZNF416||ZNF91||ZNF224||ZNF780A||ZNF223||ZNF670||HLA-DPB1||ZNF782||ZNF169||PILRB||ZNF578||ZNF582||POU2F1||ZNF136||ZNF57||ZNF573||ZNF256||ZNF417||ZNF470||ZNF713||ZNF550||ZNF225||ZNF304||PIK3CD||ZNF791||ZNF675||ZNF506||ZNF490||ZNF551||ZNF549||STAT2||SOCS3||ZNF100||ZNF772||ZNF382||ZNF641||ZNF846||ZNF433||TSC2||ZNF510||ZNF354B||ZNF543||ZNF680||ZNF333||ZNF891||ZNF805||ZNF431||ZNF613||ZNF726||ZNF268||ZNF248||ZNF200||ZNF430||ZNF790||NFKBIA||CFP||ZNF331||ZNF721||ZNF684||ZNF320||ZNF175||ZNF211||ZNF84||HCFC1||ZNF529||ZNF567||ZNF180||ZNF597||ZNF577||ZNF227||TSC1||TRAF5||ZNF79||PIK3R1 | 4.388954 | 5.025723 | 4.546492 | 4.833528 | 4.378123 | 4.492159 | 4.987082 | 4.848860 | 4.372601 | 4.424142 | 4.369463 | 5.044975 | 5.034870 | 4.996879 | 4.667011 | 4.483299 | 4.481168 | 4.815203 | 4.820150 | 4.864713 | 4.366461 | 4.355440 | 4.411848 | 4.427392 | 4.461171 | 4.583216 | 4.994740 | 4.978205 | 4.988253 | 4.835197 | 4.877310 | 4.833647 |
| hsa05034 | Alcoholism | 113 | -0.4183903 | -1.529800 | 0.013376228 | 0.1552311 | 0.14227174 | 2279 | tags=28%, list=16%, signal=24% | CREB3||H2AX||H4C3||HDAC11||H2BC12||ADORA2B||SLC29A1||SLC29A3||HDAC7||H2AC20||SHC2||H2BC5||GNG7||H2BC17||H2BC4||H4C11||H4C8||GRIN2D||H2BC11||H2AJ||CREB3L4||H2AC12||H2AC11||CREB3L1||H2BC15||MAOA||H2AC6||H2AC18||H2BC18||H4C14||H4-16||H2AC17 | 6.081292 | 6.312998 | 6.203495 | 6.296332 | 6.035621 | 6.066784 | 6.329420 | 6.327174 | 6.084533 | 6.207338 | 5.939596 | 6.318456 | 6.308019 | 6.312499 | 6.267141 | 6.088087 | 6.248624 | 6.304355 | 6.246149 | 6.337027 | 5.995385 | 6.110528 | 5.997929 | 6.035454 | 6.030588 | 6.132024 | 6.323480 | 6.323340 | 6.341367 | 6.342897 | 6.301839 | 6.336450 |
| hsa03013 | Nucleocytoplasmic transport | 98 | 0.3938433 | 1.481801 | 0.013381995 | 0.1552311 | 0.14227174 | 5167 | tags=53%, list=36%, signal=34% | THOC1||UPF3A||THOC2||IPO9||NUP214||NUP42||UPF2||TNPO2||NUP35||RANBP2||ACIN1||TPR||UPF3B||TNPO1||PHAX||SRRM1||PNN||DDX19B||XPO6||AHCTF1||NUP58||RANBP17||NXF1||NUP205||NUP98||KPNA5||SUMO1||IPO7||NCBP2||THOC7||XPO1||XPO4||XPO7||NUP54||NXT2||NUP50||NUP160||UBE2I||THOC5||CASC3||MAGOHB||KPNB1||XPO5||SENP2||NUP153||XPOT||NUP155||RNPS1||NMD3||TNPO3||CSE1L||NUP43 | 6.634100 | 7.685674 | 6.767342 | 7.668316 | 6.465975 | 6.373296 | 7.699191 | 7.710681 | 6.693915 | 6.814767 | 6.355271 | 7.767399 | 7.648530 | 7.637462 | 7.056428 | 6.410580 | 6.763325 | 7.663388 | 7.627110 | 7.713156 | 6.425037 | 6.647286 | 6.304425 | 6.372836 | 6.284857 | 6.457060 | 7.661140 | 7.717789 | 7.717903 | 7.730295 | 7.639917 | 7.759165 |
| hsa05231 | Choline metabolism in cancer | 81 | 0.3916914 | 1.430069 | 0.027494667 | 0.2899438 | 0.26573793 | 2229 | tags=22%, list=16%, signal=19% | PDGFRA||PLA2G4B||PLD1||PLA2G4C||SLC22A1||JMJD7-PLA2G4B||PIK3CD||TSC2||SP1||DGKD||PLCG1||DGKE||CHKB||HIF1A||TSC1||PIK3R1||PIP5K1B||PCYT1A | 5.032919 | 5.249679 | 5.113171 | 5.108189 | 5.057964 | 5.281131 | 5.248483 | 5.137128 | 4.986448 | 4.985203 | 5.122730 | 5.256880 | 5.253134 | 5.238962 | 5.124556 | 5.130136 | 5.084390 | 5.127520 | 5.054597 | 5.140962 | 5.054853 | 4.968467 | 5.145161 | 5.202630 | 5.332058 | 5.305492 | 5.275118 | 5.234711 | 5.235246 | 5.124970 | 5.161767 | 5.124327 |
| hsa04020 | Calcium signaling pathway | 145 | -0.3556399 | -1.343168 | 0.045151616 | 0.3808983 | 0.34909913 | 1216 | tags=19%, list=9%, signal=18% | PLCD1||FGF18||HTR7||PDGFA||HTR6||ATP2A3||ORAI2||VEGFC||CHRM3||ORAI1||P2RX4||MYLK3||ITPKA||RET||RYR2||FGF17||GRIN2D||ITPKB||ADRB2||CXCR4||MYLK2||EDNRB||SLC8A2||ORAI3||CCKBR||SLC8A3||PDE1A||CYSLTR1 | 5.203155 | 5.281541 | 5.184341 | 5.246700 | 5.164563 | 5.227205 | 5.266210 | 5.277192 | 5.207760 | 5.294706 | 5.100455 | 5.299405 | 5.282556 | 5.262424 | 5.211856 | 5.118982 | 5.220017 | 5.271143 | 5.198329 | 5.269438 | 5.171691 | 5.176830 | 5.144966 | 5.257002 | 5.273877 | 5.147502 | 5.266994 | 5.269165 | 5.262462 | 5.256128 | 5.296764 | 5.278398 |
| hsa04014 | Ras signaling pathway | 170 | 0.3132744 | 1.282515 | 0.047213622 | 0.3808983 | 0.34909913 | 2124 | tags=20%, list=15%, signal=17% | PDGFRA||PLA2G4B||GNG2||PLD1||PLA2G4C||FGF5||CALML4||SYNGAP1||RIN1||JMJD7-PLA2G4B||RASA4||KSR1||RASAL1||AFDN||GNB3||PIK3CD||REL||RALGAPA1||CSF1||FGF22||NF1||RASA2||INSR||KSR2||PLCE1||RASAL2||PLCG2||PLCG1||FGF7||GNG11||NTRK1||PIK3R1||RAB5A||STK4 | 4.761771 | 4.980848 | 4.803409 | 4.931546 | 4.774280 | 4.934217 | 5.012591 | 4.974793 | 4.715112 | 4.773554 | 4.795457 | 4.962952 | 4.989976 | 4.989452 | 4.844316 | 4.781260 | 4.783764 | 4.933330 | 4.877798 | 4.981641 | 4.780179 | 4.704951 | 4.834773 | 4.885120 | 4.968421 | 4.947811 | 5.022804 | 5.005840 | 5.009071 | 4.950135 | 4.993474 | 4.980429 |
| hsa04072 | Phospholipase D signaling pathway | 105 | 0.3547018 | 1.349670 | 0.047468354 | 0.3808983 | 0.34909913 | 2182 | tags=23%, list=15%, signal=19% | PDGFRA||PLA2G4B||PLD1||PLA2G4C||PTK2B||RAPGEF4||JMJD7-PLA2G4B||DNM1||PIK3CD||GRM8||TSC2||INSR||ADCY7||GRM2||AGTR1||DGKD||PLCG2||PLCG1||DGKE||TSC1||PIK3R1||GNA13||PLCB4||PIP5K1B | 5.316572 | 5.419658 | 5.430767 | 5.261938 | 5.339656 | 5.589753 | 5.409541 | 5.298150 | 5.247865 | 5.327250 | 5.371877 | 5.413518 | 5.443124 | 5.402021 | 5.402771 | 5.476576 | 5.411826 | 5.291808 | 5.192194 | 5.299354 | 5.359483 | 5.226257 | 5.426107 | 5.551513 | 5.590116 | 5.626651 | 5.422538 | 5.406657 | 5.399331 | 5.287558 | 5.324249 | 5.282281 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 80 | 0.3749959 | 1.364912 | 0.049254085 | 0.3808983 | 0.34909913 | 3752 | tags=29%, list=26%, signal=21% | PLA2G4B||PLD1||PLA2G4C||JMJD7-PLA2G4B||PIK3CD||VAV3||ACTR3C||PLCG2||PLCG1||PIK3R1||PRKCE||PIP5K1B||PIK3CA||PIP5K1A||PRKCA||LYN||PLA2G6||GAB2||LAT||SPHK1||AKT3||WAS||MYO10 | 5.851749 | 5.748980 | 5.785355 | 5.764440 | 5.875050 | 5.962222 | 5.771819 | 5.798459 | 5.803969 | 5.861062 | 5.888920 | 5.765619 | 5.745214 | 5.735948 | 5.763563 | 5.809804 | 5.782322 | 5.791688 | 5.704043 | 5.795741 | 5.882932 | 5.801026 | 5.937914 | 5.952796 | 6.005196 | 5.927583 | 5.782275 | 5.758512 | 5.774569 | 5.791509 | 5.810801 | 5.792985 |
ORA: All DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: All DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
ORA: Down-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Down-regulated DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
ORA: Up-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Up-regulated DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
Help
still working on
2-2. Summary Plot
GSEA
GO: BP
GO: MF
GO: CC
KEGG
ORA: All DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Down-regulated DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Up-regulated DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Overlapped Terms
GO: BP
Summary Plot
No results under this category
Venn Plot
No results under this category
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: MF
Summary Plot
No results under this category
Venn Plot
No results under this category
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: CC
Summary Plot
No results under this category
Venn Plot
No results under this category
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
KEGG
Summary Plot
No results under this category
Venn Plot
No results under this category
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
Help
still working on
3. Isoform Switch Analysis
Alternative Splicing Events
Amount of Event
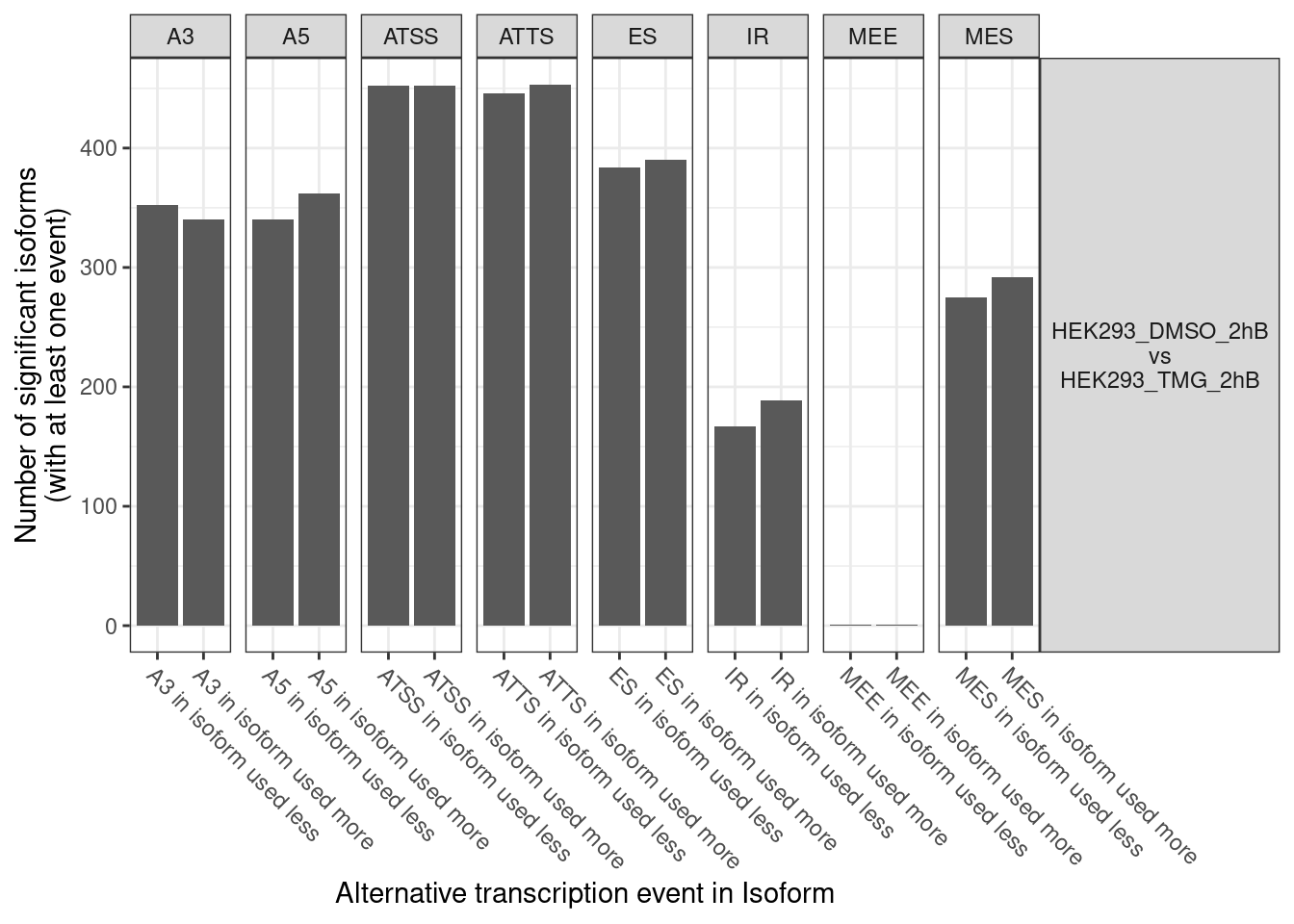
Enrichment Analysis
This below result summarizes the uneven usage within each comparison by for each alternative splicing type calculate the fraction of events being gains (as opposed to loss) and perform a statistical analysis of this fraction.
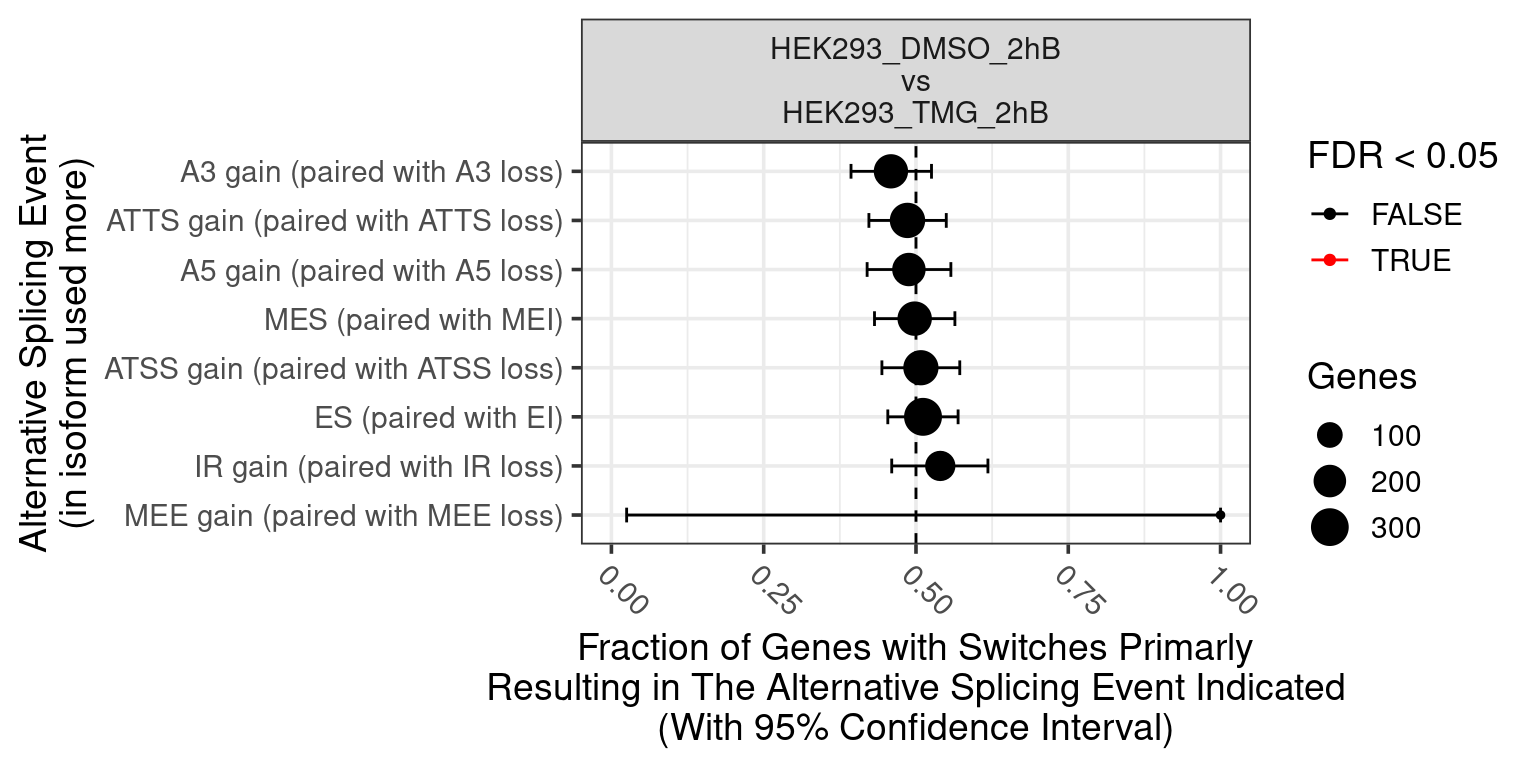
Comparison Analysis
This below result answered the question: How does the isoform usage of all isoforms utilizing a particular splicing type change - in other words is all isoforms or only a subset of isoforms that are affected.
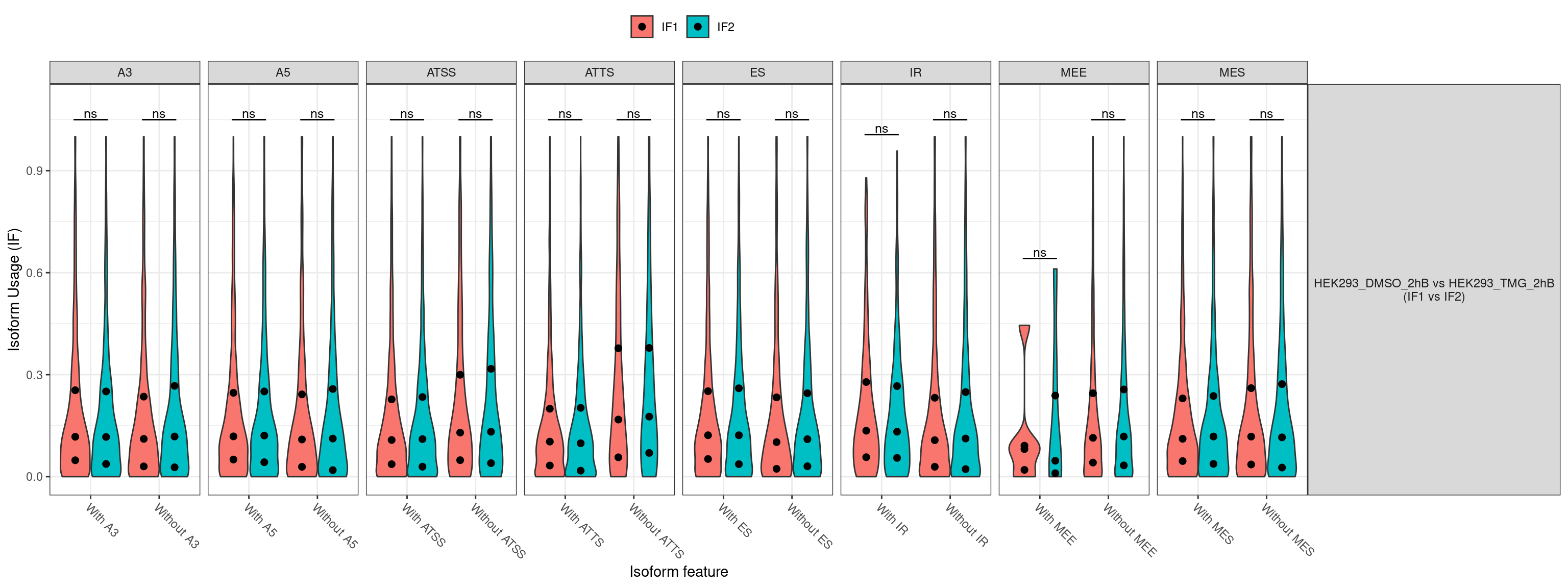
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- IR : Intron Retention.
- A5 : Alternative 5’ donor site (changes in the 5’end of the upstream exon).
- A3 : Alternative 3’ acceptor site (changes in the 3’end of the downstream exon).
- ATSS : Alternative Transcription Start Site.
- ATTS : Alternative Transcription Termination Site.
- ES : Exon Skipping (EI means Exon Inclusion).
- MES : Multiple Exon Skipping. Skipping of >1 consecutive exons. (MEI means Multiple Exon Inclusion).
- MEE : Mutually Exclusive Exons.
Alternative Splicing Consequence
Amount of Event
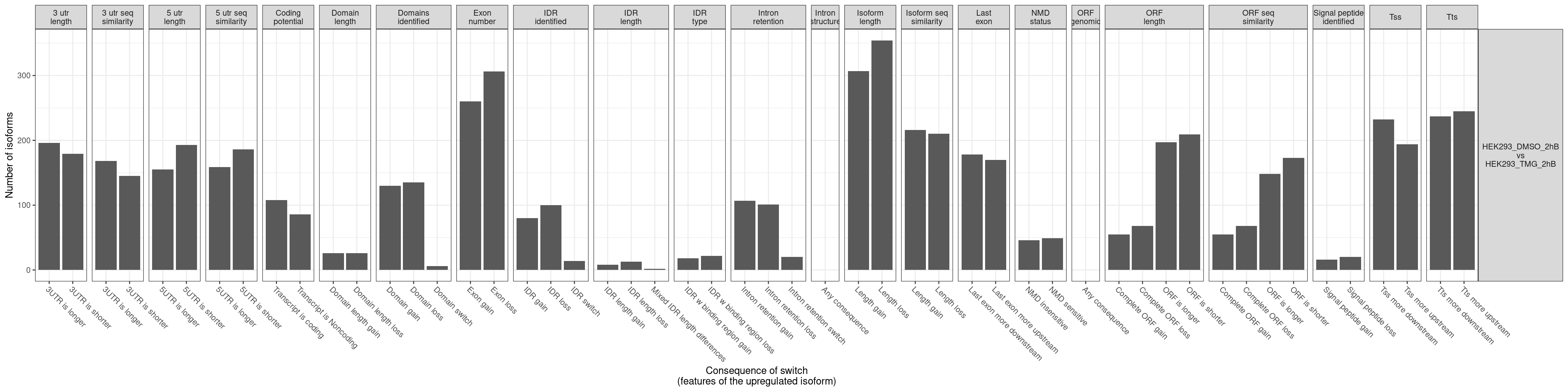
Enrichment Analysis
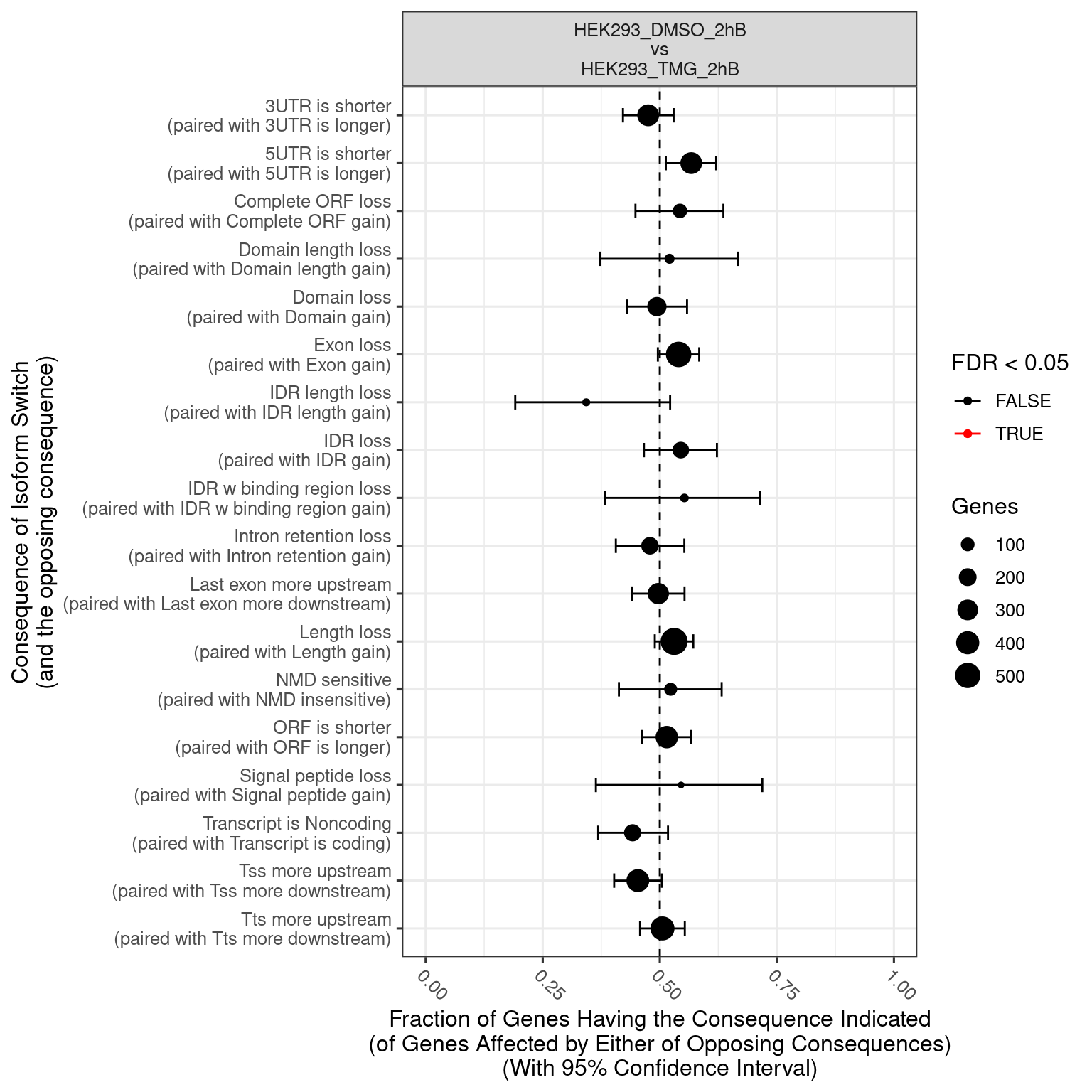
Comparison Analysis
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- tss : Test transcripts for whether they use different Transcription Start Site (TSS) (more than ntCutoff from each other).
- tts : Test transcripts for whether they use different Transcription Termination Site (TTS) (more than ntCutoff from each other).
- last_exon : Test whether transcripts utilizes different last exons (defined as the last exon of each transcript is non-overlapping).
- isoform_seq_similarity : Test whether the isoform nucleotide sequences are different (as described above). Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff.
- isoform_length : Test transcripts for differences in isoform length. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Please * note that this is a less powerful analysis than implemented in ‘isoform_seq_similarity’ as two equally long sequences might be very different.
- exon_number : Test transcripts for differences in exon number.
- intron_structure : Test transcripts for differences in intron structure, e.g. usage of exon-exon junctions. This analysis corresponds to analyzing whether all introns in one isoform is also found in the other isoforms.
- intron_retention : Test for differences in intron retentions (and their genomic positions). Require that analyzeIntronRetention have been run.
- isoform_class_code : Test transcripts for differences in the transcript classification provide by cufflinks. For a updated list of class codes see http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/#transfrag-class-codes.
- coding_potential : Test transcripts for differences in coding potential, as indicated by the CPAT or CPC2 analysis. Requires that analyzeCPAT or analyzeCPC2 have been used to add external conding potential analysis to the switchAnalyzeRlist.
- ORF_seq_similarity : Test whether the amino acid sequences of the ORFs are different (as described above). Reported as different if the measured JCsim is smaller than AaJCsimCutoff and the length difference of the aligned and combined region is larger than AaCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_genomic : Test transcripts for differences in genomic position of the Open Reading Frames (ORF). Requires that least one of the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and settingaddAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_length : Test transcripts for differences in length of Open Reading Frames (ORF). Note that this is a less powerful analysis than implemented in ORF_seq_similarity as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 5’ UnTranslated Region (UTR) are different (as described above). The 5’UTR is defined as the region from the transcript start to the ORF start. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_length : Test transcripts for differences in the length of the 5’ UnTranslated Region (UTR). The 5’UTR is defined as the region from the transcript start to the ORF start. Note that this is a less powerful analysis than implemented in ‘5_utr_seq_similarity’ as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 3’ UnTranslated Region (UTR) are different (as described above). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_length : Test transcripts for differences in the length of the 3’ UnTranslated Regions (UTR). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Note that this is a less powerful analysis than implemented in 3_utr_seq_similarity as two equally long sequences might be very different. Requires that identifyORF have been used to predict NMD sensitivity or that the ORF was imported though one of the dedicated import functions implemented in isoformSwitchAnalyzeR. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- NMD_status : Test transcripts for differences in sensitivity to Nonsense Mediated Decay (NMD). Requires that both the isoforms have been annotated with PTC either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domains_identified : Test transcripts for differences in the name and order of which domains are identified by the Pfam in the transcripts. Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domain_length : Test transcripts for differences in the length of overlapping domains of the same type (same hmm_name) thereby enabling analysis of protein domain truncation. Do however note that a small difference in length is will likely not truncate the protein domain. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff .Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- genomic_domain_position : Test transcripts for differences in the genomic position of the domains identified by the Pfam analysis (Will be different unless the two isoforms have the same domains at the same genomic location). Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
- IDR_identified : Test for differences in isoform IDRs. Specifically the two isoforms are analyzed for whether they contain IDRs which do not overlap in genomic coordinates. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_length : Test for differences in the length of overlapping (in genomic coordinates) IDRs. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_type : Test for differences in IDR type. Specifically the two isoforms are tested for overlapping IDRs (genomic coordinates) and overlapping IDRs are compared with regards to their IDR type (IDR vs IDR w binding site). Only available if analyzeIUPred2A was used to add external IDR analysis to the switchAnalyzeRlist.
- signal_peptide_identified : Test transcripts for differences in whether a signal peptide was identified or not by the SignalP analysis. Requires that analyzeSignalP have been used to add external SignalP analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via analyzeORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
Volcano Plot
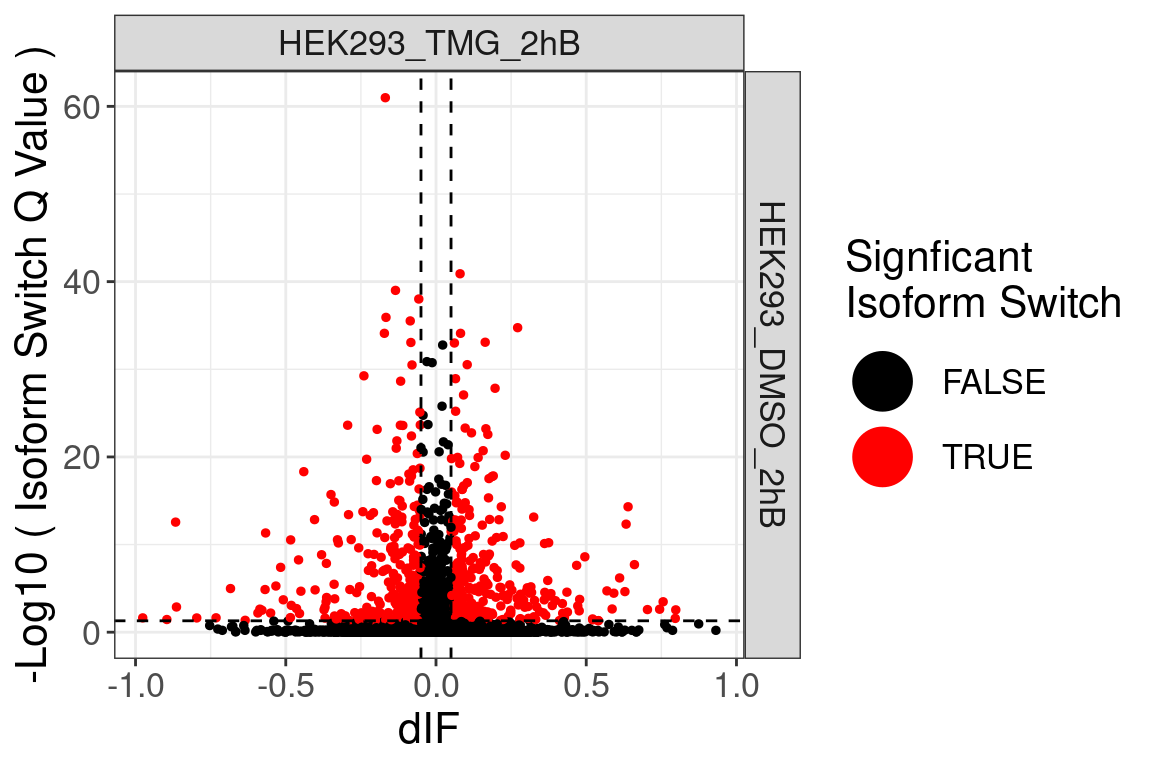
Spreadsheet
| iso_ref | gene_ref | isoform_id | gene_id | gene_name | condition_1 | condition_2 | IF1 | IF2 | dIF | isoform_switch_q_value | switchConsequencesGene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| isoComp_00245441 | geneComp_00048448 | MSTRG.4473.27 | ENSG00000107521 | HPS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.169 | 0.000 | -0.169 | 1.028645e-61 | TRUE |
| isoComp_00331066 | geneComp_00057970 | MSTRG.4456.7 | ENSG00000171307 | ZDHHC16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.080 | 0.080 | 1.260712e-41 | TRUE |
| isoComp_00225957 | geneComp_00046552 | ENST00000498648 | ENSG00000082146 | STRADB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.135 | 0.000 | -0.135 | 9.936147e-40 | TRUE |
| isoComp_00225784 | geneComp_00046530 | MSTRG.23731.9 | ENSG00000081307 | UBA5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.057 | 0.000 | -0.057 | 9.605411e-39 | TRUE |
| isoComp_00290832 | geneComp_00053213 | ENST00000590061 | ENSG00000141646 | SMAD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.166 | 0.000 | -0.166 | 1.195713e-36 | TRUE |
| isoComp_00308269 | geneComp_00055200 | ENST00000379632 | ENSG00000158019 | BABAM2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.086 | 0.000 | -0.086 | 3.025356e-36 | TRUE |
| isoComp_00290723 | geneComp_00053204 | ENST00000269392 | ENSG00000141577 | CEP131 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.272 | 0.272 | 1.785983e-35 | TRUE |
| isoComp_00242559 | geneComp_00048128 | ENST00000474049 | ENSG00000105443 | CYTH2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.172 | 0.000 | -0.172 | 7.896801e-35 | TRUE |
| isoComp_00260821 | geneComp_00050006 | MSTRG.18205.22 | ENSG00000119812 | FAM98A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.082 | 0.082 | 7.896801e-35 | TRUE |
| isoComp_00245440 | geneComp_00048448 | MSTRG.4473.26 | ENSG00000107521 | HPS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.164 | 0.164 | 8.206437e-34 | TRUE |
| isoComp_00276648 | geneComp_00051786 | MSTRG.568.20 | ENSG00000133226 | SRRM1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.084 | 0.000 | -0.084 | 8.727576e-34 | TRUE |
| isoComp_00292564 | geneComp_00053419 | ENST00000469074 | ENSG00000143319 | ISG20L2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.061 | 0.061 | 1.005909e-33 | TRUE |
| isoComp_00332222 | geneComp_00058141 | ENST00000398888 | ENSG00000172046 | USP19 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.104 | 0.104 | 2.965061e-31 | TRUE |
| isoComp_00272982 | geneComp_00051404 | ENST00000679103 | ENSG00000130816 | DNMT1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.080 | 0.000 | -0.080 | 3.200003e-31 | |
| isoComp_00290730 | geneComp_00053204 | ENST00000575907 | ENSG00000141577 | CEP131 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.240 | 0.000 | -0.240 | 5.583605e-30 | TRUE |
| isoComp_00219656 | geneComp_00046009 | ENST00000651988 | ENSG00000067955 | CBFB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.065 | 0.065 | 1.181696e-29 | |
| isoComp_00282137 | geneComp_00052357 | MSTRG.19392.3 | ENSG00000136536 | MARCHF7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.118 | 0.000 | -0.118 | 2.275862e-29 | TRUE |
| isoComp_00362992 | geneComp_00062718 | MSTRG.27919.15 | ENSG00000204599 | TRIM39 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.197 | 0.197 | 1.467931e-28 | TRUE |
| isoComp_00294747 | geneComp_00053643 | ENST00000412591 | ENSG00000144524 | COPS7B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.092 | 0.092 | 8.489981e-28 | TRUE |
| isoComp_00257493 | geneComp_00049633 | ENST00000444547 | ENSG00000116698 | SMG7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.065 | 0.065 | 6.132527e-26 | TRUE |
| isoComp_00242019 | geneComp_00048072 | MSTRG.17169.10 | ENSG00000105221 | AKT2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.054 | 0.000 | -0.054 | 7.752443e-26 | |
| isoComp_00292568 | geneComp_00053419 | MSTRG.2374.22 | ENSG00000143319 | ISG20L2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.052 | 0.000 | -0.052 | 2.252044e-24 | TRUE |
| isoComp_00253452 | geneComp_00049261 | MSTRG.22411.8 | ENSG00000113851 | CRBN | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 2.410223e-24 | |
| isoComp_00268237 | geneComp_00050877 | MSTRG.648.7 | ENSG00000126705 | AHDC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.294 | 0.000 | -0.294 | 2.410223e-24 | TRUE |
| isoComp_00259659 | geneComp_00049886 | ENST00000686381 | ENSG00000118873 | RAB3GAP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.110 | 0.000 | -0.110 | 2.564406e-24 | TRUE |
| isoComp_00237914 | geneComp_00047693 | MSTRG.8615.15 | ENSG00000102710 | SUPT20H | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.097 | 0.097 | 5.103529e-24 | TRUE |
| isoComp_00371644 | geneComp_00065445 | MSTRG.6604.8 | ENSG00000226210 | WASH8P | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.166 | 0.166 | 6.056091e-24 | TRUE |
| isoComp_00311149 | geneComp_00055525 | MSTRG.33625.5 | ENSG00000160360 | GPSM1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.196 | 0.000 | -0.196 | 7.187179e-24 | TRUE |
| isoComp_00265824 | geneComp_00050593 | ENST00000480585 | ENSG00000124596 | OARD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.118 | 0.118 | 1.794776e-23 | TRUE |
| isoComp_00388765 | geneComp_00072263 | ENST00000507287 | ENSG00000251474 | RPL32P3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.172 | 0.172 | 2.709468e-23 | TRUE |
| isoComp_00229700 | geneComp_00046884 | ENST00000372858 | ENSG00000090621 | PABPC4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.082 | 0.000 | -0.082 | 4.090551e-23 | TRUE |
| isoComp_00264034 | geneComp_00050390 | ENST00000520374 | ENSG00000123124 | WWP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.130 | 0.000 | -0.130 | 1.465762e-22 | TRUE |
| isoComp_00368845 | geneComp_00064375 | MSTRG.13715.21 | ENSG00000221926 | TRIM16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.132 | 0.000 | -0.132 | 1.009637e-21 | TRUE |
| isoComp_00316545 | geneComp_00056183 | MSTRG.24145.2 | ENSG00000163872 | YEATS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.157 | 0.157 | 1.937109e-21 | TRUE |
| isoComp_00257516 | geneComp_00049633 | MSTRG.2724.6 | ENSG00000116698 | SMG7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.062 | 0.000 | -0.062 | 4.086528e-21 | TRUE |
| isoComp_00361375 | geneComp_00062456 | ENST00000391839 | ENSG00000203667 | COX20 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.230 | 0.230 | 6.510940e-21 | TRUE |
| isoComp_00221277 | geneComp_00046150 | ENST00000322269 | ENSG00000071889 | FAM3A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.072 | 0.072 | 1.126396e-20 | TRUE |
| isoComp_00359359 | geneComp_00062107 | ENST00000607800 | ENSG00000198276 | UCKL1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.140 | 0.140 | 1.186505e-20 | TRUE |
| isoComp_00258673 | geneComp_00049763 | ENST00000649106 | ENSG00000117593 | DARS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.052 | 0.052 | 1.559889e-20 | |
| isoComp_00385525 | geneComp_00071079 | ENST00000671605 | ENSG00000245849 | RAD51-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.231 | 0.000 | -0.231 | 1.842076e-20 | TRUE |
| isoComp_00280975 | geneComp_00052237 | ENST00000472019 | ENSG00000135924 | DNAJB2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.079 | 0.079 | 5.530249e-20 | TRUE |
| isoComp_00214794 | geneComp_00045582 | ENST00000692002 | ENSG00000044090 | CUL7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.129 | 0.129 | 1.264474e-19 | TRUE |
| isoComp_00234849 | geneComp_00047370 | ENST00000553548 | ENSG00000100650 | SRSF5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.053 | 0.000 | -0.053 | 1.933326e-19 | |
| isoComp_00274770 | geneComp_00051588 | ENST00000343304 | ENSG00000132128 | LRRC41 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.000 | -0.076 | 2.944411e-19 | FALSE |
| isoComp_00393819 | geneComp_00074805 | ENST00000239165 | ENSG00000260027 | HOXB7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.560 | -0.440 | 4.818046e-19 | TRUE |
| isoComp_00331035 | geneComp_00057970 | ENST00000414567 | ENSG00000171307 | ZDHHC16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 8.934247e-19 | TRUE |
| isoComp_00368982 | geneComp_00064396 | ENST00000420217 | ENSG00000222011 | FAM185A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.191 | 0.191 | 1.465063e-18 | TRUE |
| isoComp_00330866 | geneComp_00057943 | ENST00000477070 | ENSG00000171163 | ZNF692 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.082 | 0.000 | -0.082 | 1.502228e-18 | TRUE |
| isoComp_00272218 | geneComp_00051336 | ENST00000595519 | ENSG00000130529 | TRPM4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.187 | 0.187 | 1.737423e-18 | TRUE |
| isoComp_00330140 | geneComp_00057835 | ENST00000606082 | ENSG00000170634 | ACYP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.176 | 0.176 | 2.961860e-18 | TRUE |
| isoComp_00212194 | geneComp_00045328 | ENST00000548917 | ENSG00000015153 | YAF2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.198 | 0.000 | -0.198 | 5.030800e-18 | TRUE |
| isoComp_00297794 | geneComp_00053995 | MSTRG.34539.1 | ENSG00000147180 | ZNF711 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.124 | 0.000 | -0.124 | 5.282059e-18 | TRUE |
| isoComp_00331033 | geneComp_00057970 | ENST00000370846 | ENSG00000171307 | ZDHHC16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.088 | 0.000 | -0.088 | 5.760927e-18 | TRUE |
| isoComp_00326977 | geneComp_00057416 | ENST00000372886 | ENSG00000168734 | PKIG | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.104 | 0.104 | 8.548400e-18 | TRUE |
| isoComp_00253849 | geneComp_00049297 | MSTRG.24302.4 | ENSG00000114331 | ACAP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.152 | 0.000 | -0.152 | 1.131174e-17 | TRUE |
| isoComp_00285118 | geneComp_00052670 | MSTRG.4550.19 | ENSG00000138111 | MFSD13A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.092 | 0.092 | 2.044110e-17 | TRUE |
| isoComp_00286479 | geneComp_00052801 | ENST00000681992 | ENSG00000138796 | HADH | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.056 | 0.000 | -0.056 | 4.541763e-17 | TRUE |
| isoComp_00267266 | geneComp_00050773 | ENST00000398782 | ENSG00000125844 | RRBP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.087 | 0.087 | 5.022601e-17 | TRUE |
| isoComp_00313098 | geneComp_00055723 | ENST00000361837 | ENSG00000162062 | TEDC2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.087 | 0.087 | 5.158581e-17 | TRUE |
| isoComp_00259624 | geneComp_00049884 | ENST00000505609 | ENSG00000118816 | CCNI | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.052 | 0.052 | 1.003610e-16 | |
| isoComp_00304033 | geneComp_00054708 | ENST00000413169 | ENSG00000153531 | ADPRHL1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.349 | 0.000 | -0.349 | 1.960337e-16 | TRUE |
| isoComp_00308266 | geneComp_00055200 | ENST00000361704 | ENSG00000158019 | BABAM2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.063 | 0.063 | 2.697064e-16 | TRUE |
| isoComp_00381226 | geneComp_00069474 | MSTRG.4817.5 | ENSG00000237489 | C10orf143 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.175 | 0.175 | 4.708723e-16 | TRUE |
| isoComp_00228657 | geneComp_00046789 | MSTRG.20302.25 | ENSG00000088888 | MAVS | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.124 | 0.000 | -0.124 | 8.793736e-16 | TRUE |
| isoComp_00277701 | geneComp_00051890 | ENST00000559085 | ENSG00000134138 | MEIS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.120 | 0.000 | -0.120 | 9.720046e-16 | TRUE |
| isoComp_00209843 | geneComp_00045127 | MSTRG.13681.4 | ENSG00000006740 | ARHGAP44 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.338 | 0.000 | -0.338 | 1.407750e-15 | TRUE |
| isoComp_00308093 | geneComp_00055181 | ENST00000508274 | ENSG00000157869 | RAB28 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.071 | 0.071 | 1.407750e-15 | TRUE |
| isoComp_00317336 | geneComp_00056255 | ENST00000642042 | ENSG00000164073 | MFSD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.097 | 0.097 | 1.681432e-15 | TRUE |
| isoComp_00267935 | geneComp_00050839 | ENST00000585821 | ENSG00000126249 | PDCD2L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.068 | 0.068 | 1.716857e-15 | TRUE |
| isoComp_00229933 | geneComp_00046903 | ENST00000261183 | ENSG00000091039 | OSBPL8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.061 | 0.000 | -0.061 | 3.171949e-15 | FALSE |
| isoComp_00214918 | geneComp_00045591 | MSTRG.33919.7 | ENSG00000046653 | GPM6B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.112 | 0.000 | -0.112 | 3.988499e-15 | TRUE |
| isoComp_00248934 | geneComp_00048777 | ENST00000534083 | ENSG00000110080 | ST3GAL4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.072 | 0.000 | -0.072 | 4.530031e-15 | TRUE |
| isoComp_00270154 | geneComp_00051098 | ENST00000249440 | ENSG00000128652 | HOXD3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.639 | 0.639 | 4.866991e-15 | TRUE |
| isoComp_00316006 | geneComp_00056124 | MSTRG.23149.3 | ENSG00000163686 | ABHD6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.217 | 0.217 | 4.866991e-15 | TRUE |
| isoComp_00260325 | geneComp_00049957 | ENST00000509153 | ENSG00000119599 | DCAF4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.086 | 0.086 | 9.012678e-15 | TRUE |
| isoComp_00337747 | geneComp_00058848 | ENST00000578311 | ENSG00000175970 | UNC119B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.110 | 0.110 | 1.015181e-14 | TRUE |
| isoComp_00230392 | geneComp_00046950 | ENST00000560875 | ENSG00000092098 | RNF31 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.056 | 0.000 | -0.056 | 1.208179e-14 | |
| isoComp_00396750 | geneComp_00076315 | MSTRG.14748.1 | ENSG00000266086 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.243 | 0.000 | -0.243 | 1.776172e-14 | TRUE | |
| isoComp_00287806 | geneComp_00052964 | ENST00000267522 | ENSG00000140006 | WDR89 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.144 | 0.000 | -0.144 | 1.811770e-14 | TRUE |
| isoComp_00229946 | geneComp_00046904 | ENST00000413936 | ENSG00000091073 | DTX2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.208 | 0.000 | -0.208 | 2.305259e-14 | TRUE |
| isoComp_00238998 | geneComp_00047785 | ENST00000566609 | ENSG00000103227 | LMF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.129 | 0.000 | -0.129 | 3.623557e-14 | TRUE |
| isoComp_00303800 | geneComp_00054680 | ENST00000421804 | ENSG00000153208 | MERTK | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.291 | 0.000 | -0.291 | 3.847332e-14 | TRUE |
| isoComp_00241283 | geneComp_00047995 | ENST00000593431 | ENSG00000104870 | FCGRT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.052 | 0.000 | -0.052 | 4.101902e-14 | |
| isoComp_00362561 | geneComp_00062655 | ENST00000470063 | ENSG00000204406 | MBD5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.219 | 0.000 | -0.219 | 4.705394e-14 | TRUE |
| isoComp_00260493 | geneComp_00049977 | MSTRG.9942.21 | ENSG00000119685 | TTLL5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.112 | 0.112 | 4.780783e-14 | TRUE |
| isoComp_00315134 | geneComp_00056020 | ENST00000355499 | ENSG00000163374 | YY1AP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.128 | 0.000 | -0.128 | 5.426221e-14 | |
| isoComp_00342791 | geneComp_00059626 | ENST00000590390 | ENSG00000180479 | ZNF571 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.325 | 0.325 | 7.297012e-14 | TRUE |
| isoComp_00273466 | geneComp_00051449 | ENST00000671907 | ENSG00000131089 | ARHGEF9 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.114 | 0.000 | -0.114 | 7.370526e-14 | TRUE |
| isoComp_00234151 | geneComp_00047301 | MSTRG.22207.9 | ENSG00000100413 | POLR3H | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.054 | 0.054 | 1.322438e-13 | |
| isoComp_00263206 | geneComp_00050299 | ENST00000570372 | ENSG00000122390 | NAA60 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.069 | 0.000 | -0.069 | 1.338254e-13 | TRUE |
| isoComp_00385521 | geneComp_00071079 | ENST00000499988 | ENSG00000245849 | RAD51-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.179 | 0.179 | 1.358831e-13 | TRUE |
| isoComp_00405103 | geneComp_00080947 | MSTRG.5410.5 | ENSG00000283341 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.404 | 0.000 | -0.404 | 1.426875e-13 | TRUE | |
| isoComp_00298659 | geneComp_00054094 | ENST00000277082 | ENSG00000148019 | CEP78 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.209 | 0.209 | 1.463680e-13 | TRUE |
| isoComp_00338112 | geneComp_00058915 | ENST00000540936 | ENSG00000176371 | ZSCAN2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.140 | 0.000 | -0.140 | 1.493132e-13 | TRUE |
| isoComp_00298533 | geneComp_00054081 | MSTRG.32374.12 | ENSG00000147854 | UHRF2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.064 | 0.064 | 1.560396e-13 | TRUE |
| isoComp_00318886 | geneComp_00056469 | ENST00000433212 | ENSG00000164828 | SUN1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.084 | 0.084 | 1.574756e-13 | TRUE |
| isoComp_00357788 | geneComp_00061902 | MSTRG.15879.29 | ENSG00000197563 | PIGN | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 1.815586e-13 | TRUE |
| isoComp_00260278 | geneComp_00049951 | ENST00000644624 | ENSG00000119537 | KDSR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.163 | 0.000 | -0.163 | 1.987462e-13 | TRUE |
| isoComp_00314304 | geneComp_00055897 | ENST00000462374 | ENSG00000162885 | B3GALNT2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.056 | 0.056 | 2.143596e-13 | |
| isoComp_00326532 | geneComp_00057363 | MSTRG.15487.2 | ENSG00000168461 | RAB31 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.114 | 0.000 | -0.114 | 2.524855e-13 | TRUE |
| isoComp_00275577 | geneComp_00051665 | ENST00000254898 | ENSG00000132561 | MATN2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.867 | 0.000 | -0.867 | 2.748278e-13 | TRUE |
| isoComp_00219114 | geneComp_00045964 | MSTRG.28923.7 | ENSG00000066651 | TRMT11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.069 | 0.069 | 3.409669e-13 | |
| isoComp_00270321 | geneComp_00051119 | ENST00000417247 | ENSG00000128805 | ARHGAP22 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.135 | 0.000 | -0.135 | 4.423056e-13 | TRUE |
| isoComp_00384401 | geneComp_00070748 | ENST00000566076 | ENSG00000243708 | PLA2G4B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.633 | 0.633 | 4.686156e-13 | TRUE |
| isoComp_00317319 | geneComp_00056255 | ENST00000641590 | ENSG00000164073 | MFSD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.074 | 0.000 | -0.074 | 6.062122e-13 | TRUE |
| isoComp_00373565 | geneComp_00066227 | ENST00000686363 | ENSG00000228409 | CCT6P1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.154 | 0.154 | 6.062122e-13 | TRUE |
| isoComp_00219854 | geneComp_00046027 | ENST00000622231 | ENSG00000068400 | GRIPAP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.084 | 0.084 | 1.438762e-12 | TRUE |
| isoComp_00293108 | geneComp_00053473 | ENST00000366890 | ENSG00000143498 | TAF1A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.060 | 0.060 | 2.076428e-12 | TRUE |
| isoComp_00372700 | geneComp_00065872 | ENST00000649413 | ENSG00000227372 | TP73-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.061 | 0.000 | -0.061 | 2.248885e-12 | TRUE |
| isoComp_00226650 | geneComp_00046611 | ENST00000497370 | ENSG00000084072 | PPIE | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.060 | 0.060 | 3.142508e-12 | |
| isoComp_00242612 | geneComp_00048134 | ENST00000522889 | ENSG00000105483 | CARD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.050 | 0.050 | 3.191151e-12 | TRUE |
| isoComp_00321344 | geneComp_00056793 | ENST00000370162 | ENSG00000166169 | POLL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.058 | 0.000 | -0.058 | 3.303585e-12 | TRUE |
| isoComp_00359700 | geneComp_00062155 | MSTRG.14406.2 | ENSG00000198496 | NBR2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.196 | 0.000 | -0.196 | 4.628107e-12 | TRUE |
| isoComp_00208308 | geneComp_00044992 | ENST00000542785 | ENSG00000002016 | RAD52 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.060 | 0.000 | -0.060 | 4.650488e-12 | |
| isoComp_00301215 | geneComp_00054360 | ENST00000280115 | ENSG00000150556 | LYPD6B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.567 | 0.000 | -0.567 | 4.759665e-12 | TRUE |
| isoComp_00225653 | geneComp_00046514 | ENST00000261441 | ENSG00000081019 | RSBN1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.126 | 0.000 | -0.126 | 5.478977e-12 | FALSE |
| isoComp_00367139 | geneComp_00063789 | ENST00000690171 | ENSG00000214765 | SEPTIN7P2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.079 | 0.000 | -0.079 | 6.933861e-12 | TRUE |
| isoComp_00266367 | geneComp_00050665 | MSTRG.14430.1 | ENSG00000125319 | HROB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.131 | 0.131 | 1.007490e-11 | TRUE |
| isoComp_00268226 | geneComp_00050877 | ENST00000642245 | ENSG00000126705 | AHDC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.223 | 0.223 | 1.215082e-11 | TRUE |
| isoComp_00397981 | geneComp_00076823 | ENST00000343376 | ENSG00000267673 | FDX2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.073 | 0.000 | -0.073 | 1.409729e-11 | TRUE |
| isoComp_00216261 | geneComp_00045715 | ENST00000557084 | ENSG00000054654 | SYNE2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.201 | 0.201 | 1.571064e-11 | TRUE |
| isoComp_00353627 | geneComp_00061363 | ENST00000305783 | ENSG00000189050 | RNFT1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.170 | 0.000 | -0.170 | 1.571064e-11 | TRUE |
| isoComp_00285907 | geneComp_00052761 | MSTRG.25331.12 | ENSG00000138658 | ZGRF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.138 | 0.000 | -0.138 | 1.624355e-11 | TRUE |
| isoComp_00365920 | geneComp_00063475 | ENST00000436437 | ENSG00000213722 | DDAH2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.110 | 0.110 | 2.025055e-11 | TRUE |
| isoComp_00335667 | geneComp_00058573 | ENST00000690127 | ENSG00000174405 | LIG4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.282 | 0.000 | -0.282 | 2.709589e-11 | TRUE |
| isoComp_00300167 | geneComp_00054242 | MSTRG.6291.1 | ENSG00000149289 | ZC3H12C | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.328 | 0.000 | -0.328 | 2.829662e-11 | TRUE |
| isoComp_00309024 | geneComp_00055302 | ENST00000374552 | ENSG00000158813 | EDA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.484 | 0.000 | -0.484 | 2.933046e-11 | TRUE |
| isoComp_00280061 | geneComp_00052142 | ENST00000553089 | ENSG00000135452 | TSPAN31 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.067 | 0.067 | 3.751160e-11 | TRUE |
| isoComp_00311629 | geneComp_00055571 | ENST00000691742 | ENSG00000160766 | GBAP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.187 | 0.187 | 4.001634e-11 | TRUE |
| isoComp_00308496 | geneComp_00055234 | ENST00000404115 | ENSG00000158290 | CUL4B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.052 | 0.000 | -0.052 | 4.461250e-11 | TRUE |
| isoComp_00337136 | geneComp_00058754 | MSTRG.27354.4 | ENSG00000175414 | ARL10 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.052 | 0.000 | -0.052 | 5.992026e-11 | |
| isoComp_00392339 | geneComp_00073988 | ENST00000547834 | ENSG00000258325 | ITFG2-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.375 | 0.375 | 6.151557e-11 | TRUE |
| isoComp_00393821 | geneComp_00074805 | ENST00000567101 | ENSG00000260027 | HOXB7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.279 | 0.279 | 6.467567e-11 | TRUE |
| isoComp_00290097 | geneComp_00053157 | ENST00000590956 | ENSG00000141391 | PRELID3A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.325 | 0.000 | -0.325 | 6.563160e-11 | TRUE |
| isoComp_00327449 | geneComp_00057480 | ENST00000382448 | ENSG00000168970 | JMJD7-PLA2G4B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.361 | 0.361 | 7.622518e-11 | TRUE |
| isoComp_00319199 | geneComp_00056498 | MSTRG.31960.6 | ENSG00000164933 | SLC25A32 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.060 | 0.000 | -0.060 | 9.254906e-11 | TRUE |
| isoComp_00315158 | geneComp_00056021 | MSTRG.23206.1 | ENSG00000163376 | KBTBD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.069 | 0.069 | 1.020324e-10 | TRUE |
| isoComp_00248207 | geneComp_00048709 | ENST00000515420 | ENSG00000109572 | CLCN3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.054 | 0.000 | -0.054 | 1.152308e-10 | TRUE |
| isoComp_00246727 | geneComp_00048563 | MSTRG.13807.3 | ENSG00000108448 | TRIM16L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.106 | 0.106 | 1.236832e-10 | TRUE |
| isoComp_00229949 | geneComp_00046904 | ENST00000430490 | ENSG00000091073 | DTX2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.262 | 0.262 | 1.240242e-10 | TRUE |
| isoComp_00265856 | geneComp_00050600 | ENST00000373188 | ENSG00000124615 | MOCS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.140 | 0.000 | -0.140 | 1.534499e-10 | TRUE |
| isoComp_00274623 | geneComp_00051569 | ENST00000555965 | ENSG00000131966 | ACTR10 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.150 | 0.000 | -0.150 | 1.947101e-10 | TRUE |
| isoComp_00367447 | geneComp_00063879 | ENST00000315614 | ENSG00000215041 | NEURL4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.094 | 0.094 | 2.229903e-10 | TRUE |
| isoComp_00287831 | geneComp_00052967 | MSTRG.10012.5 | ENSG00000140022 | STON2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.257 | 0.000 | -0.257 | 2.382024e-10 | TRUE |
| isoComp_00287558 | geneComp_00052923 | MSTRG.8240.8 | ENSG00000139697 | SBNO1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.093 | 0.000 | -0.093 | 2.399894e-10 | TRUE |
| isoComp_00256793 | geneComp_00049563 | ENST00000616884 | ENSG00000116138 | DNAJC16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.080 | 0.080 | 3.113992e-10 | TRUE |
| isoComp_00337416 | geneComp_00058793 | MSTRG.33060.18 | ENSG00000175611 | LINC00476 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.151 | 0.000 | -0.151 | 3.250224e-10 | TRUE |
| isoComp_00375963 | geneComp_00067284 | MSTRG.1915.3 | ENSG00000231365 | WARS2-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.085 | 0.000 | -0.085 | 3.486812e-10 | TRUE |
| isoComp_00298674 | geneComp_00054094 | ENST00000645398 | ENSG00000148019 | CEP78 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.137 | 0.000 | -0.137 | 5.500702e-10 | TRUE |
| isoComp_00275256 | geneComp_00051629 | ENST00000571360 | ENSG00000132386 | SERPINF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.112 | 0.000 | -0.112 | 7.108709e-10 | TRUE |
| isoComp_00243232 | geneComp_00048205 | ENST00000594553 | ENSG00000105738 | SIPA1L3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.145 | 0.000 | -0.145 | 8.162686e-10 | TRUE |
| isoComp_00294645 | geneComp_00053633 | MSTRG.19899.5 | ENSG00000144451 | SPAG16 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.137 | 0.000 | -0.137 | 8.511421e-10 | TRUE |
| isoComp_00311621 | geneComp_00055571 | ENST00000687347 | ENSG00000160766 | GBAP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.226 | 0.000 | -0.226 | 1.112624e-09 | TRUE |
| isoComp_00283241 | geneComp_00052481 | ENST00000502297 | ENSG00000137177 | KIF13A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.085 | 0.085 | 1.164070e-09 | TRUE |
| isoComp_00355608 | geneComp_00061633 | MSTRG.15840.16 | ENSG00000196628 | TCF4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.178 | 0.178 | 1.230822e-09 | TRUE |
| isoComp_00225757 | geneComp_00046526 | MSTRG.26568.10 | ENSG00000081189 | MEF2C | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.206 | 0.000 | -0.206 | 1.328308e-09 | TRUE |
| isoComp_00306298 | geneComp_00054986 | ENST00000519804 | ENSG00000156170 | NDUFAF6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.103 | 0.261 | 0.158 | 1.348569e-09 | |
| isoComp_00248188 | geneComp_00048707 | MSTRG.25307.5 | ENSG00000109534 | GAR1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.304 | 0.182 | -0.122 | 1.361501e-09 | TRUE |
| isoComp_00332309 | geneComp_00058145 | ENST00000540845 | ENSG00000172059 | KLF11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.381 | 0.000 | -0.381 | 1.446808e-09 | TRUE |
| isoComp_00307643 | geneComp_00055136 | ENST00000561186 | ENSG00000157450 | RNF111 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.071 | 0.000 | -0.071 | 1.527481e-09 | TRUE |
| isoComp_00286213 | geneComp_00052782 | MSTRG.25053.5 | ENSG00000138756 | BMP2K | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.084 | 0.084 | 1.757815e-09 | TRUE |
| isoComp_00213880 | geneComp_00045490 | ENST00000520903 | ENSG00000034677 | RNF19A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.174 | 0.174 | 2.046252e-09 | TRUE |
| isoComp_00296398 | geneComp_00053828 | ENST00000676634 | ENSG00000145907 | G3BP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.065 | 0.000 | -0.065 | 2.182780e-09 | TRUE |
| isoComp_00267027 | geneComp_00050740 | ENST00000615753 | ENSG00000125740 | FOSB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.495 | 0.495 | 2.502809e-09 | TRUE |
| isoComp_00226224 | geneComp_00046573 | ENST00000496703 | ENSG00000082781 | ITGB5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 2.525330e-09 | TRUE |
| isoComp_00276785 | geneComp_00051797 | ENST00000410396 | ENSG00000133316 | WDR74 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.183 | 0.000 | -0.183 | 2.822064e-09 | TRUE |
| isoComp_00308897 | geneComp_00055284 | MSTRG.27894.7 | ENSG00000158691 | ZSCAN12 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.124 | 0.124 | 3.032240e-09 | TRUE |
| isoComp_00347480 | geneComp_00060369 | MSTRG.20949.5 | ENSG00000184402 | SS18L1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.136 | 0.000 | -0.136 | 4.067209e-09 | TRUE |
| isoComp_00317997 | geneComp_00056343 | ENST00000308394 | ENSG00000164305 | CASP3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.085 | 0.085 | 5.339477e-09 | TRUE |
| isoComp_00240040 | geneComp_00047886 | ENST00000261866 | ENSG00000104133 | SPG11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.812 | 0.354 | -0.457 | 5.615400e-09 | TRUE |
| isoComp_00360072 | geneComp_00062202 | MSTRG.20603.25 | ENSG00000198646 | NCOA6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 5.985510e-09 | TRUE |
| isoComp_00213630 | geneComp_00045470 | ENST00000417477 | ENSG00000032219 | ARID4A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.113 | 0.113 | 7.307094e-09 | TRUE |
| isoComp_00341392 | geneComp_00059413 | ENST00000599712 | ENSG00000179134 | SAMD4B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.069 | 0.000 | -0.069 | 8.453083e-09 | |
| isoComp_00380597 | geneComp_00069257 | ENST00000457331 | ENSG00000236869 | ZKSCAN7-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.165 | 0.165 | 9.660866e-09 | TRUE |
| isoComp_00238412 | geneComp_00047740 | ENST00000566987 | ENSG00000102984 | ZNF821 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.147 | 0.147 | 1.152786e-08 | TRUE |
| isoComp_00266948 | geneComp_00050733 | ENST00000443289 | ENSG00000125703 | ATG4C | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.072 | 0.072 | 1.157963e-08 | TRUE |
| isoComp_00343440 | geneComp_00059735 | MSTRG.3553.2 | ENSG00000181192 | DHTKD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.081 | 0.081 | 1.330933e-08 | TRUE |
| isoComp_00381614 | geneComp_00069626 | ENST00000622171 | ENSG00000237945 | LINC00649 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.365 | 0.000 | -0.365 | 1.410799e-08 | TRUE |
| isoComp_00401510 | geneComp_00078785 | ENST00000668298 | ENSG00000274372 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.117 | 0.777 | 0.660 | 1.941295e-08 | TRUE | |
| isoComp_00316093 | geneComp_00056130 | ENST00000696815 | ENSG00000163702 | IL17RC | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 2.023217e-08 | TRUE |
| isoComp_00350749 | geneComp_00060864 | MSTRG.1130.1 | ENSG00000186564 | FOXD2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.267 | 0.267 | 2.084769e-08 | TRUE |
| isoComp_00385112 | geneComp_00071013 | ENST00000505213 | ENSG00000244754 | N4BP2L2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.066 | 0.066 | 2.206732e-08 | |
| isoComp_00261215 | geneComp_00050057 | ENST00000517946 | ENSG00000120159 | CAAP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.083 | 0.083 | 2.284351e-08 | TRUE |
| isoComp_00274191 | geneComp_00051522 | ENST00000253925 | ENSG00000131626 | PPFIA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.000 | -0.076 | 2.290431e-08 | TRUE |
| isoComp_00405102 | geneComp_00080947 | ENST00000693635 | ENSG00000283341 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.468 | 0.468 | 2.364802e-08 | TRUE | |
| isoComp_00290725 | geneComp_00053204 | ENST00000450824 | ENSG00000141577 | CEP131 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.214 | 0.000 | -0.214 | 2.603269e-08 | TRUE |
| isoComp_00301739 | geneComp_00054419 | ENST00000612776 | ENSG00000151150 | ANK3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.087 | 0.087 | 3.271257e-08 | TRUE |
| isoComp_00327611 | geneComp_00057497 | ENST00000475218 | ENSG00000169062 | UPF3A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.148 | 0.148 | 3.271257e-08 | TRUE |
| isoComp_00278950 | geneComp_00052030 | ENST00000481069 | ENSG00000134897 | BIVM | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.076 | 0.076 | 3.374245e-08 | TRUE |
| isoComp_00322621 | geneComp_00056931 | ENST00000409481 | ENSG00000166762 | CATSPER2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.517 | 0.000 | -0.517 | 3.982968e-08 | TRUE |
| isoComp_00245964 | geneComp_00048501 | MSTRG.3688.4 | ENSG00000107890 | ANKRD26 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.193 | 0.193 | 4.248246e-08 | TRUE |
| isoComp_00395869 | geneComp_00075822 | ENST00000571963 | ENSG00000263072 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.050 | 0.000 | -0.050 | 4.291999e-08 | TRUE | |
| isoComp_00403096 | geneComp_00079677 | ENST00000622055 | ENSG00000278259 | MYO19 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.053 | 0.000 | -0.053 | 4.510869e-08 | |
| isoComp_00302594 | geneComp_00054517 | ENST00000684105 | ENSG00000151892 | GFRA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.279 | 0.279 | 4.808501e-08 | TRUE |
| isoComp_00317325 | geneComp_00056255 | ENST00000641743 | ENSG00000164073 | MFSD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.095 | 0.000 | -0.095 | 4.892350e-08 | TRUE |
| isoComp_00266328 | geneComp_00050657 | MSTRG.9036.4 | ENSG00000125247 | TMTC4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.127 | 0.127 | 4.945128e-08 | TRUE |
| isoComp_00385001 | geneComp_00070982 | ENST00000662474 | ENSG00000244625 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.162 | 0.000 | -0.162 | 6.248061e-08 | TRUE | |
| isoComp_00233230 | geneComp_00047214 | MSTRG.22119.22 | ENSG00000100206 | DMC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.063 | 0.063 | 6.512247e-08 | |
| isoComp_00390780 | geneComp_00073261 | MSTRG.5599.1 | ENSG00000255508 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.122 | 0.122 | 6.637002e-08 | TRUE | |
| isoComp_00326654 | geneComp_00057375 | MSTRG.15472.5 | ENSG00000168502 | MTCL1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.144 | 0.144 | 6.846362e-08 | TRUE |
| isoComp_00277693 | geneComp_00051890 | ENST00000314177 | ENSG00000134138 | MEIS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.100 | 0.000 | -0.100 | 8.202895e-08 | TRUE |
| isoComp_00262609 | geneComp_00050235 | MSTRG.742.22 | ENSG00000121753 | ADGRB2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.082 | 0.082 | 8.352537e-08 | TRUE |
| isoComp_00320116 | geneComp_00056638 | ENST00000553526 | ENSG00000165521 | EML5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.172 | 0.000 | -0.172 | 8.374458e-08 | TRUE |
| isoComp_00222200 | geneComp_00046218 | ENST00000644523 | ENSG00000073584 | SMARCE1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.089 | 0.089 | 8.504457e-08 | TRUE |
| isoComp_00346251 | geneComp_00060171 | MSTRG.21712.8 | ENSG00000183506 | PI4KAP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.075 | 0.075 | 8.504457e-08 | TRUE |
| isoComp_00358988 | geneComp_00062060 | ENST00000395867 | ENSG00000198105 | ZNF248 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.225 | 0.000 | -0.225 | 8.876590e-08 | TRUE |
| isoComp_00223642 | geneComp_00046345 | ENST00000592945 | ENSG00000076662 | ICAM3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.796 | 1.000 | 0.204 | 9.321926e-08 | TRUE |
| isoComp_00289550 | geneComp_00053114 | ENST00000564853 | ENSG00000141013 | GAS8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.054 | 0.054 | 1.161547e-07 | TRUE |
| isoComp_00274612 | geneComp_00051567 | MSTRG.16922.4 | ENSG00000131944 | FAAP24 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.177 | 0.000 | -0.177 | 1.161636e-07 | TRUE |
| isoComp_00243018 | geneComp_00048183 | MSTRG.16757.21 | ENSG00000105676 | ARMC6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.051 | 0.051 | 1.469452e-07 | |
| isoComp_00333139 | geneComp_00058271 | MSTRG.4116.9 | ENSG00000172731 | LRRC20 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.206 | 0.000 | -0.206 | 1.674333e-07 | TRUE |
| isoComp_00223639 | geneComp_00046345 | ENST00000587992 | ENSG00000076662 | ICAM3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.141 | 0.000 | -0.141 | 1.988032e-07 | TRUE |
| isoComp_00335204 | geneComp_00058509 | MSTRG.18039.6 | ENSG00000173960 | UBXN2A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.093 | 0.093 | 1.994241e-07 | TRUE |
| isoComp_00216802 | geneComp_00045762 | MSTRG.24116.33 | ENSG00000058056 | USP13 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 2.323455e-07 | TRUE |
| isoComp_00317357 | geneComp_00056257 | ENST00000476105 | ENSG00000164076 | CAMKV | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.059 | 0.000 | -0.059 | 2.349661e-07 | TRUE |
| isoComp_00318995 | geneComp_00056478 | MSTRG.29364.9 | ENSG00000164877 | MICALL2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.141 | 0.000 | -0.141 | 2.686803e-07 | TRUE |
| isoComp_00289557 | geneComp_00053114 | ENST00000568705 | ENSG00000141013 | GAS8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.080 | 0.000 | -0.080 | 3.046746e-07 | TRUE |
| isoComp_00377911 | geneComp_00068096 | MSTRG.20113.12 | ENSG00000233611 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.103 | 0.103 | 3.046746e-07 | TRUE | |
| isoComp_00244014 | geneComp_00048287 | MSTRG.30019.15 | ENSG00000106133 | NSUN5P2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.110 | 0.110 | 3.807418e-07 | TRUE |
| isoComp_00325235 | geneComp_00057246 | MSTRG.11857.5 | ENSG00000167972 | ABCA3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.062 | 0.000 | -0.062 | 3.851262e-07 | TRUE |
| isoComp_00334131 | geneComp_00058374 | ENST00000482512 | ENSG00000173230 | GOLGB1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.073 | 0.000 | -0.073 | 4.591799e-07 | TRUE |
| isoComp_00267519 | geneComp_00050804 | ENST00000376112 | ENSG00000125968 | ID1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.961 | 0.889 | -0.072 | 5.206062e-07 | TRUE |
| isoComp_00344306 | geneComp_00059883 | ENST00000497460 | ENSG00000182185 | RAD51B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.074 | 0.000 | -0.074 | 5.507200e-07 | TRUE |
| isoComp_00282912 | geneComp_00052451 | ENST00000520751 | ENSG00000136997 | MYC | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.163 | 0.087 | -0.075 | 5.540203e-07 | |
| isoComp_00259748 | geneComp_00049895 | MSTRG.18024.13 | ENSG00000118961 | LDAH | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.204 | 0.204 | 5.554956e-07 | TRUE |
| isoComp_00267518 | geneComp_00050804 | ENST00000376105 | ENSG00000125968 | ID1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.039 | 0.111 | 0.072 | 5.554956e-07 | TRUE |
| isoComp_00232814 | geneComp_00047172 | MSTRG.21844.13 | ENSG00000100068 | LRP5L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.117 | 0.117 | 6.107563e-07 | TRUE |
| isoComp_00355569 | geneComp_00061633 | ENST00000356073 | ENSG00000196628 | TCF4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.128 | 0.000 | -0.128 | 6.413539e-07 | TRUE |
| isoComp_00406465 | geneComp_00081648 | ENST00000650829 | ENSG00000286011 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.611 | 0.611 | 6.445275e-07 | TRUE | |
| isoComp_00341678 | geneComp_00059452 | ENST00000552138 | ENSG00000179364 | PACS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.117 | 0.037 | -0.080 | 6.578091e-07 | |
| isoComp_00286689 | geneComp_00052822 | ENST00000550863 | ENSG00000139117 | CPNE8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.068 | 0.068 | 7.857683e-07 | TRUE |
| isoComp_00246835 | geneComp_00048572 | ENST00000381311 | ENSG00000108509 | CAMTA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.160 | 0.160 | 8.269994e-07 | TRUE |
| isoComp_00345885 | geneComp_00060118 | ENST00000330551 | ENSG00000183250 | LINC01547 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.070 | 0.000 | -0.070 | 9.084609e-07 | TRUE |
| isoComp_00334164 | geneComp_00058374 | ENST00000695039 | ENSG00000173230 | GOLGB1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.056 | 0.056 | 9.353397e-07 | TRUE |
| isoComp_00240225 | geneComp_00047898 | ENST00000519219 | ENSG00000104228 | TRIM35 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.087 | 0.000 | -0.087 | 1.016163e-06 | TRUE |
| isoComp_00242608 | geneComp_00048134 | ENST00000521437 | ENSG00000105483 | CARD8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.053 | 0.000 | -0.053 | 1.025936e-06 | TRUE |
| isoComp_00269263 | geneComp_00050991 | ENST00000248437 | ENSG00000127824 | TUBA4A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.628 | 1.000 | 0.372 | 1.252232e-06 | TRUE |
| isoComp_00398364 | geneComp_00077005 | ENST00000671115 | ENSG00000268388 | FENDRR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.142 | 0.000 | -0.142 | 1.389717e-06 | TRUE |
| isoComp_00350787 | geneComp_00060868 | ENST00000672461 | ENSG00000186575 | NF2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.094 | 0.094 | 1.585067e-06 | TRUE |
| isoComp_00228891 | geneComp_00046810 | ENST00000434275 | ENSG00000089123 | TASP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.058 | 0.000 | -0.058 | 1.619986e-06 | |
| isoComp_00243474 | geneComp_00048230 | ENST00000440636 | ENSG00000105866 | SP4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 1.868321e-06 | TRUE |
| isoComp_00243903 | geneComp_00048277 | ENST00000445054 | ENSG00000106078 | COBL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.158 | 0.000 | -0.158 | 1.906699e-06 | TRUE |
| isoComp_00400848 | geneComp_00078399 | MSTRG.1963.42 | ENSG00000273136 | NBPF26 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.167 | 0.167 | 2.167797e-06 | TRUE |
| isoComp_00400849 | geneComp_00078399 | MSTRG.1963.43 | ENSG00000273136 | NBPF26 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.167 | 0.167 | 2.167797e-06 | TRUE |
| isoComp_00269960 | geneComp_00051073 | ENST00000468477 | ENSG00000128536 | CDHR3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.164 | 0.164 | 3.253974e-06 | TRUE |
| isoComp_00366895 | geneComp_00063724 | ENST00000689804 | ENSG00000214439 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.339 | 0.000 | -0.339 | 3.396708e-06 | TRUE | |
| isoComp_00250049 | geneComp_00048882 | MSTRG.6670.15 | ENSG00000111203 | ITFG2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.066 | 0.003 | -0.062 | 3.455939e-06 | TRUE |
| isoComp_00225742 | geneComp_00046526 | ENST00000510942 | ENSG00000081189 | MEF2C | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.166 | 0.166 | 3.897835e-06 | TRUE |
| isoComp_00229435 | geneComp_00046858 | ENST00000570099 | ENSG00000090238 | YPEL3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.173 | 0.173 | 3.939874e-06 | TRUE |
| isoComp_00321698 | geneComp_00056831 | ENST00000617744 | ENSG00000166321 | NUDT13 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.245 | 0.245 | 3.963268e-06 | TRUE |
| isoComp_00250714 | geneComp_00048949 | ENST00000341550 | ENSG00000111653 | ING4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.054 | 0.054 | 4.494051e-06 | TRUE |
| isoComp_00384339 | geneComp_00070735 | MSTRG.18558.1 | ENSG00000243667 | DNAAF10 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.073 | 0.073 | 4.527051e-06 | |
| isoComp_00405864 | geneComp_00081299 | ENST00000646377 | ENSG00000285219 | HULC | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.148 | 0.148 | 5.221132e-06 | TRUE |
| isoComp_00402155 | geneComp_00079164 | ENST00000624301 | ENSG00000276043 | UHRF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 5.340255e-06 | FALSE |
| isoComp_00299770 | geneComp_00054216 | ENST00000610573 | ENSG00000149054 | ZNF215 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.061 | 0.061 | 5.360113e-06 | TRUE |
| isoComp_00267018 | geneComp_00050740 | ENST00000353609 | ENSG00000125740 | FOSB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.533 | 0.000 | -0.533 | 5.380311e-06 | TRUE |
| isoComp_00306475 | geneComp_00055006 | ENST00000642395 | ENSG00000156313 | RPGR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.254 | 0.000 | -0.254 | 6.386379e-06 | TRUE |
| isoComp_00399531 | geneComp_00077649 | MSTRG.21844.24 | ENSG00000271138 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.316 | 0.316 | 6.630828e-06 | TRUE | |
| isoComp_00223947 | geneComp_00046369 | MSTRG.12331.4 | ENSG00000077238 | IL4R | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.083 | 0.000 | -0.083 | 6.635183e-06 | TRUE |
| isoComp_00241927 | geneComp_00048068 | ENST00000221801 | ENSG00000105202 | FBL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.372 | 0.280 | -0.092 | 7.275138e-06 | |
| isoComp_00359982 | geneComp_00062191 | ENST00000545671 | ENSG00000198598 | MMP17 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.149 | 0.000 | -0.149 | 7.360285e-06 | TRUE |
| isoComp_00402203 | geneComp_00079191 | MSTRG.10312.5 | ENSG00000276141 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.213 | 0.213 | 7.496312e-06 | TRUE | |
| isoComp_00283687 | geneComp_00052524 | ENST00000379586 | ENSG00000137434 | C6orf52 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.111 | 0.111 | 7.828519e-06 | TRUE |
| isoComp_00388685 | geneComp_00072224 | MSTRG.17798.6 | ENSG00000251369 | ZNF550 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.155 | 0.155 | 8.005044e-06 | TRUE |
| isoComp_00395092 | geneComp_00075495 | ENST00000573141 | ENSG00000261556 | SMG1P7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.217 | 0.217 | 8.199274e-06 | TRUE |
| isoComp_00223384 | geneComp_00046321 | MSTRG.4496.3 | ENSG00000075891 | PAX2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.304 | 0.304 | 8.522794e-06 | TRUE |
| isoComp_00333073 | geneComp_00058261 | MSTRG.3885.10 | ENSG00000172671 | ZFAND4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.133 | 0.133 | 9.176584e-06 | TRUE |
| isoComp_00378220 | geneComp_00068242 | ENST00000453008 | ENSG00000234028 | EIF2AK3-DT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.684 | 0.000 | -0.684 | 1.045755e-05 | TRUE |
| isoComp_00287053 | geneComp_00052871 | ENST00000550295 | ENSG00000139354 | GAS2L3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.118 | 0.000 | -0.118 | 1.068785e-05 | TRUE |
| isoComp_00209272 | geneComp_00045079 | MSTRG.4990.19 | ENSG00000005801 | ZNF195 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.069 | 0.000 | -0.069 | 1.259255e-05 | |
| isoComp_00252884 | geneComp_00049203 | ENST00000514284 | ENSG00000113369 | ARRDC3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.110 | 0.110 | 1.266256e-05 | TRUE |
| isoComp_00287144 | geneComp_00052880 | ENST00000549550 | ENSG00000139437 | TCHP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.083 | 0.083 | 1.329092e-05 | TRUE |
| isoComp_00391449 | geneComp_00073541 | ENST00000600703 | ENSG00000256683 | ZNF350 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.287 | 0.000 | -0.287 | 1.352057e-05 | TRUE |
| isoComp_00291877 | geneComp_00053338 | ENST00000404795 | ENSG00000142700 | DMRTA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.431 | -0.569 | 1.355020e-05 | TRUE |
| isoComp_00347442 | geneComp_00060365 | ENST00000430886 | ENSG00000184381 | PLA2G6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.062 | 0.000 | -0.062 | 1.369009e-05 | TRUE |
| isoComp_00375395 | geneComp_00067005 | ENST00000656041 | ENSG00000230590 | FTX | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.103 | 0.000 | -0.103 | 1.417552e-05 | TRUE |
| isoComp_00335706 | geneComp_00058579 | ENST00000567816 | ENSG00000174442 | ZWILCH | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.066 | 0.066 | 1.433023e-05 | TRUE |
| isoComp_00354788 | geneComp_00061533 | ENST00000692468 | ENSG00000196338 | NLGN3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.402 | 0.000 | -0.402 | 1.484170e-05 | TRUE |
| isoComp_00313348 | geneComp_00055749 | ENST00000294187 | ENSG00000162241 | SLC25A45 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.253 | 0.253 | 1.609955e-05 | TRUE |
| isoComp_00299214 | geneComp_00054146 | MSTRG.33617.3 | ENSG00000148411 | NACC2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.093 | 0.000 | -0.093 | 1.638849e-05 | TRUE |
| isoComp_00375398 | geneComp_00067005 | ENST00000657327 | ENSG00000230590 | FTX | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.010 | 0.139 | 0.129 | 1.642264e-05 | TRUE |
| isoComp_00218463 | geneComp_00045905 | MSTRG.13637.1 | ENSG00000065320 | NTN1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.050 | 0.000 | -0.050 | 1.660295e-05 | TRUE |
| isoComp_00232851 | geneComp_00047178 | ENST00000423024 | ENSG00000100083 | GGA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.025 | 0.086 | 0.062 | 1.889371e-05 | TRUE |
| isoComp_00291878 | geneComp_00053338 | ENST00000418121 | ENSG00000142700 | DMRTA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.569 | 0.569 | 1.923820e-05 | TRUE |
| isoComp_00273906 | geneComp_00051494 | MSTRG.17550.2 | ENSG00000131400 | NAPSA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.450 | 0.000 | -0.450 | 2.100285e-05 | TRUE |
| isoComp_00253423 | geneComp_00049259 | MSTRG.24222.8 | ENSG00000113838 | TBCCD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 2.146546e-05 | TRUE |
| isoComp_00396240 | geneComp_00076037 | ENST00000479995 | ENSG00000264343 | NOTCH2NLA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.046 | 0.347 | 0.301 | 2.146546e-05 | TRUE |
| isoComp_00346761 | geneComp_00060253 | MSTRG.10278.10 | ENSG00000183828 | NUDT14 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.116 | 0.023 | -0.094 | 2.220682e-05 | |
| isoComp_00404441 | geneComp_00080679 | ENST00000625254 | ENSG00000280734 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.629 | 0.629 | 2.332045e-05 | TRUE | |
| isoComp_00276293 | geneComp_00051741 | ENST00000477277 | ENSG00000132964 | CDK8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.142 | 0.142 | 2.400633e-05 | TRUE |
| isoComp_00211467 | geneComp_00045262 | ENST00000471976 | ENSG00000011332 | DPF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.066 | 0.066 | 2.570810e-05 | TRUE |
| isoComp_00247923 | geneComp_00048686 | ENST00000507338 | ENSG00000109255 | NMU | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.065 | 0.065 | 2.711853e-05 | TRUE |
| isoComp_00335354 | geneComp_00058533 | MSTRG.24443.3 | ENSG00000174137 | FAM53A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.436 | 0.436 | 2.711853e-05 | TRUE |
| isoComp_00212230 | geneComp_00045331 | ENST00000568281 | ENSG00000015413 | DPEP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.362 | 0.362 | 2.813748e-05 | TRUE |
| isoComp_00245025 | geneComp_00048398 | ENST00000374079 | ENSG00000106948 | AKNA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.318 | 0.318 | 2.906556e-05 | TRUE |
| isoComp_00293130 | geneComp_00053476 | ENST00000608996 | ENSG00000143502 | SUSD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.264 | 0.000 | -0.264 | 3.045557e-05 | TRUE |
| isoComp_00229963 | geneComp_00046904 | ENST00000492339 | ENSG00000091073 | DTX2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.053 | 0.000 | -0.053 | 3.203431e-05 | TRUE |
| isoComp_00366107 | geneComp_00063549 | ENST00000571460 | ENSG00000213918 | DNASE1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.066 | 0.066 | 3.203431e-05 | TRUE |
| isoComp_00242284 | geneComp_00048099 | ENST00000519811 | ENSG00000105339 | DENND3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.312 | 0.312 | 3.489183e-05 | TRUE |
| isoComp_00378222 | geneComp_00068242 | ENST00000688818 | ENSG00000234028 | EIF2AK3-DT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.592 | 0.592 | 3.540619e-05 | TRUE |
| isoComp_00352856 | geneComp_00061238 | ENST00000479340 | ENSG00000188508 | KRTDAP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.382 | 0.382 | 3.925497e-05 | TRUE |
| isoComp_00364164 | geneComp_00062947 | MSTRG.12343.17 | ENSG00000205609 | EIF3CL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.168 | 0.168 | 3.966274e-05 | TRUE |
| isoComp_00348463 | geneComp_00060536 | ENST00000578118 | ENSG00000185158 | LRRC37B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.000 | -0.076 | 4.203315e-05 | TRUE |
| isoComp_00277931 | geneComp_00051921 | ENST00000410092 | ENSG00000134278 | SPIRE1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.470 | 0.751 | 0.281 | 4.645876e-05 | TRUE |
| isoComp_00391365 | geneComp_00073512 | MSTRG.5570.8 | ENSG00000256591 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.059 | 0.000 | -0.059 | 4.667027e-05 | ||
| isoComp_00300798 | geneComp_00054309 | ENST00000441250 | ENSG00000149761 | NUDT22 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.221 | 0.083 | -0.138 | 4.718779e-05 | TRUE |
| isoComp_00258700 | geneComp_00049770 | ENST00000412742 | ENSG00000117602 | RCAN3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.182 | 0.182 | 4.775295e-05 | TRUE |
| isoComp_00336333 | geneComp_00058670 | ENST00000566413 | ENSG00000174943 | KCTD13 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.058 | 0.058 | 4.793742e-05 | TRUE |
| isoComp_00272276 | geneComp_00051345 | ENST00000302995 | ENSG00000130584 | ZBTB46 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.144 | 0.000 | -0.144 | 5.088460e-05 | TRUE |
| isoComp_00383015 | geneComp_00070224 | ENST00000464420 | ENSG00000241316 | SUCLG2-DT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.240 | 0.240 | 5.270193e-05 | TRUE |
| isoComp_00211934 | geneComp_00045306 | ENST00000432425 | ENSG00000013441 | CLK1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.091 | 0.179 | 0.088 | 5.615638e-05 | TRUE |
| isoComp_00404947 | geneComp_00080894 | ENST00000686466 | ENSG00000283050 | GTF2IP12 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.053 | 0.053 | 6.329023e-05 | TRUE |
| isoComp_00282474 | geneComp_00052399 | ENST00000497048 | ENSG00000136826 | KLF4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.228 | 0.228 | 6.486637e-05 | TRUE |
| isoComp_00311145 | geneComp_00055525 | ENST00000440944 | ENSG00000160360 | GPSM1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.509 | 0.732 | 0.222 | 7.035978e-05 | TRUE |
| isoComp_00239219 | geneComp_00047805 | ENST00000568402 | ENSG00000103326 | CAPN15 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.046 | 0.106 | 0.061 | 7.312497e-05 | TRUE |
| isoComp_00357612 | geneComp_00061892 | ENST00000553916 | ENSG00000197535 | MYO5A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.058 | 0.058 | 7.622597e-05 | TRUE |
| isoComp_00302368 | geneComp_00054486 | ENST00000432036 | ENSG00000151690 | MFSD6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.380 | 0.380 | 7.723798e-05 | TRUE |
| isoComp_00313813 | geneComp_00055819 | ENST00000476933 | ENSG00000162600 | OMA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.052 | 0.000 | -0.052 | 8.148108e-05 | |
| isoComp_00366740 | geneComp_00063669 | MSTRG.14715.6 | ENSG00000214226 | C17orf67 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.292 | 0.077 | -0.215 | 8.646051e-05 | TRUE |
| isoComp_00267849 | geneComp_00050831 | ENST00000469302 | ENSG00000126216 | TUBGCP3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.067 | 0.000 | -0.067 | 9.632151e-05 | |
| isoComp_00226341 | geneComp_00046579 | ENST00000237163 | ENSG00000083097 | DOP1A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 9.892598e-05 | TRUE |
| isoComp_00257239 | geneComp_00049607 | ENST00000677042 | ENSG00000116539 | ASH1L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.122 | 0.000 | -0.122 | 1.010020e-04 | TRUE |
| isoComp_00330698 | geneComp_00057917 | ENST00000690857 | ENSG00000171084 | FAM86JP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.199 | 0.199 | 1.017585e-04 | TRUE |
| isoComp_00312610 | geneComp_00055659 | ENST00000638348 | ENSG00000161551 | ZNF577 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.114 | 0.000 | -0.114 | 1.060467e-04 | TRUE |
| isoComp_00253163 | geneComp_00049236 | ENST00000524093 | ENSG00000113645 | WWC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.057 | 0.000 | -0.057 | 1.071732e-04 | |
| isoComp_00392338 | geneComp_00073988 | ENST00000547794 | ENSG00000258325 | ITFG2-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.955 | 0.591 | -0.364 | 1.089169e-04 | TRUE |
| isoComp_00273186 | geneComp_00051420 | MSTRG.291.5 | ENSG00000130940 | CASZ1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.156 | 0.000 | -0.156 | 1.199518e-04 | TRUE |
| isoComp_00249151 | geneComp_00048800 | ENST00000533742 | ENSG00000110330 | BIRC2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.067 | 0.067 | 1.250325e-04 | |
| isoComp_00328021 | geneComp_00057552 | ENST00000468028 | ENSG00000169249 | ZRSR2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.057 | 0.057 | 1.250325e-04 | TRUE |
| isoComp_00388216 | geneComp_00072064 | ENST00000502433 | ENSG00000250802 | ZBED3-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.109 | 0.109 | 1.254693e-04 | TRUE |
| isoComp_00301699 | geneComp_00054417 | ENST00000357167 | ENSG00000151136 | BTBD11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.270 | 0.270 | 1.356423e-04 | TRUE |
| isoComp_00328391 | geneComp_00057606 | ENST00000304058 | ENSG00000169570 | DTWD2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.126 | 0.000 | -0.126 | 1.372950e-04 | TRUE |
| isoComp_00241773 | geneComp_00048051 | MSTRG.16392.1 | ENSG00000105088 | OLFM2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.116 | 0.000 | -0.116 | 1.379534e-04 | TRUE |
| isoComp_00289690 | geneComp_00053125 | ENST00000576915 | ENSG00000141086 | CTRL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.337 | 0.000 | -0.337 | 1.555008e-04 | TRUE |
| isoComp_00336422 | geneComp_00058685 | MSTRG.4789.17 | ENSG00000175029 | CTBP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.094 | 0.000 | -0.094 | 1.677111e-04 | |
| isoComp_00405335 | geneComp_00081081 | MSTRG.21831.8 | ENSG00000284128 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.478 | 0.478 | 1.801373e-04 | TRUE | |
| isoComp_00408570 | geneComp_00083090 | MSTRG.28272.7 | ENSG00000288721 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.237 | 0.237 | 1.859857e-04 | TRUE | |
| isoComp_00215270 | geneComp_00045630 | ENST00000646411 | ENSG00000048707 | VPS13D | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.092 | 0.000 | -0.092 | 1.873773e-04 | TRUE |
| isoComp_00409243 | geneComp_00083635 | ENST00000686810 | ENSG00000289486 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.508 | 0.000 | -0.508 | 2.011726e-04 | TRUE | |
| isoComp_00297557 | geneComp_00053971 | ENST00000491907 | ENSG00000147082 | CCNB3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.360 | 0.360 | 2.035026e-04 | TRUE |
| isoComp_00335021 | geneComp_00058487 | MSTRG.1058.4 | ENSG00000173846 | PLK3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.292 | 0.292 | 2.179372e-04 | TRUE |
| isoComp_00384556 | geneComp_00070811 | ENST00000431553 | ENSG00000243970 | PPIEL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.085 | 0.000 | -0.085 | 2.253373e-04 | TRUE |
| isoComp_00215985 | geneComp_00045684 | ENST00000512986 | ENSG00000052795 | FNIP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.051 | 0.000 | -0.051 | 2.269201e-04 | TRUE |
| isoComp_00389910 | geneComp_00072818 | ENST00000492556 | ENSG00000254413 | CHKB-CPT1B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.004 | 0.086 | 0.082 | 2.301329e-04 | TRUE |
| isoComp_00352703 | geneComp_00061208 | ENST00000620404 | ENSG00000188338 | SLC38A3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.052 | 0.052 | 2.323956e-04 | TRUE |
| isoComp_00369002 | geneComp_00064402 | ENST00000409526 | ENSG00000222020 | HDAC4-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.328 | 0.328 | 2.364655e-04 | TRUE |
| isoComp_00332304 | geneComp_00058145 | ENST00000305883 | ENSG00000172059 | KLF11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.569 | 0.967 | 0.397 | 2.516405e-04 | TRUE |
| isoComp_00293106 | geneComp_00053473 | ENST00000350027 | ENSG00000143498 | TAF1A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.104 | 0.000 | -0.104 | 2.593209e-04 | TRUE |
| isoComp_00238397 | geneComp_00047739 | ENST00000602727 | ENSG00000102981 | PARD6A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.191 | 0.000 | -0.191 | 2.887934e-04 | TRUE |
| isoComp_00249913 | geneComp_00048869 | ENST00000552093 | ENSG00000111052 | LIN7A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.157 | 0.157 | 3.056743e-04 | TRUE |
| isoComp_00351081 | geneComp_00060920 | ENST00000476653 | ENSG00000186866 | POFUT2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.037 | 0.122 | 0.084 | 3.349452e-04 | TRUE |
| isoComp_00245664 | geneComp_00048470 | ENST00000470317 | ENSG00000107738 | VSIR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.756 | 0.756 | 3.354667e-04 | TRUE |
| isoComp_00345590 | geneComp_00060065 | ENST00000329078 | ENSG00000183018 | SPNS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.634 | -0.366 | 3.394120e-04 | TRUE |
| isoComp_00260111 | geneComp_00049933 | ENST00000688065 | ENSG00000119397 | CNTRL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.078 | 0.078 | 3.791815e-04 | TRUE |
| isoComp_00261653 | geneComp_00050124 | ENST00000681208 | ENSG00000120697 | ALG5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.062 | 0.062 | 3.955724e-04 | TRUE |
| isoComp_00354184 | geneComp_00061466 | ENST00000543610 | ENSG00000196118 | CFAP119 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.083 | 0.000 | -0.083 | 4.161929e-04 | TRUE |
| isoComp_00350147 | geneComp_00060762 | MSTRG.5201.6 | ENSG00000186104 | CYP2R1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.055 | 0.055 | 4.190390e-04 | TRUE |
| isoComp_00395917 | geneComp_00075831 | ENST00000573860 | ENSG00000263146 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.136 | 0.000 | -0.136 | 4.204524e-04 | TRUE | |
| isoComp_00283405 | geneComp_00052492 | MSTRG.28329.10 | ENSG00000137221 | TJAP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.020 | 0.072 | 0.052 | 4.286728e-04 | |
| isoComp_00299768 | geneComp_00054216 | ENST00000529903 | ENSG00000149054 | ZNF215 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.089 | 0.000 | -0.089 | 4.443288e-04 | TRUE |
| isoComp_00221088 | geneComp_00046135 | MSTRG.25584.3 | ENSG00000071205 | ARHGAP10 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.119 | 0.119 | 4.782235e-04 | TRUE |
| isoComp_00352749 | geneComp_00061214 | ENST00000336517 | ENSG00000188372 | ZP3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.996 | 0.879 | -0.117 | 4.974890e-04 | TRUE |
| isoComp_00257394 | geneComp_00049622 | ENST00000367501 | ENSG00000116668 | SWT1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.421 | 0.421 | 5.465884e-04 | TRUE |
| isoComp_00342321 | geneComp_00059538 | ENST00000527197 | ENSG00000179950 | PUF60 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.148 | 0.087 | -0.061 | 5.769185e-04 | |
| isoComp_00320558 | geneComp_00056692 | ENST00000683007 | ENSG00000165731 | RET | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 6.699371e-04 | TRUE |
| isoComp_00265955 | geneComp_00050620 | MSTRG.27595.2 | ENSG00000124782 | RREB1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.186 | 0.000 | -0.186 | 6.744188e-04 | TRUE |
| isoComp_00307179 | geneComp_00055084 | ENST00000489813 | ENSG00000157036 | EXOG | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.051 | 0.122 | 0.071 | 6.758440e-04 | TRUE |
| isoComp_00360602 | geneComp_00062281 | ENST00000648262 | ENSG00000198825 | INPP5F | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.000 | -0.076 | 7.276599e-04 | TRUE |
| isoComp_00255158 | geneComp_00049424 | ENST00000233331 | ENSG00000115274 | INO80B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.759 | 0.627 | -0.131 | 7.507311e-04 | TRUE |
| isoComp_00348166 | geneComp_00060483 | MSTRG.22120.6 | ENSG00000184949 | FAM227A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.274 | 0.274 | 7.791985e-04 | TRUE |
| isoComp_00313946 | geneComp_00055833 | ENST00000462149 | ENSG00000162631 | NTNG1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.482 | 0.000 | -0.482 | 8.643331e-04 | TRUE |
| isoComp_00259334 | geneComp_00049843 | ENST00000503942 | ENSG00000118420 | UBE3D | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.051 | 0.000 | -0.051 | 8.666021e-04 | TRUE |
| isoComp_00210853 | geneComp_00045213 | ENST00000313375 | ENSG00000009950 | MLXIPL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.124 | 0.000 | -0.124 | 8.777634e-04 | |
| isoComp_00298242 | geneComp_00054048 | ENST00000422166 | ENSG00000147576 | ADHFE1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.387 | 0.387 | 9.052876e-04 | TRUE |
| isoComp_00256537 | geneComp_00049536 | ENST00000612082 | ENSG00000115966 | ATF2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.036 | 0.090 | 0.054 | 9.627990e-04 | TRUE |
| isoComp_00403372 | geneComp_00079835 | ENST00000688828 | ENSG00000278903 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.061 | 0.061 | 9.628381e-04 | TRUE | |
| isoComp_00380457 | geneComp_00069210 | ENST00000447904 | ENSG00000236753 | MKLN1-AS | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.104 | 0.104 | 1.007097e-03 | TRUE |
| isoComp_00287864 | geneComp_00052972 | ENST00000559773 | ENSG00000140044 | JDP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.472 | 0.472 | 1.015891e-03 | TRUE |
| isoComp_00357472 | geneComp_00061874 | ENST00000402802 | ENSG00000197461 | PDGFA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.774 | 0.989 | 0.216 | 1.026228e-03 | TRUE |
| isoComp_00363278 | geneComp_00062766 | MSTRG.5706.5 | ENSG00000204710 | SPDYC | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.060 | 0.000 | -0.060 | 1.108360e-03 | TRUE |
| isoComp_00305659 | geneComp_00054907 | ENST00000369626 | ENSG00000155380 | SLC16A1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.742 | 0.662 | -0.080 | 1.115222e-03 | TRUE |
| isoComp_00267127 | geneComp_00050747 | MSTRG.20319.1 | ENSG00000125772 | GPCPD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.071 | 0.071 | 1.119610e-03 | TRUE |
| isoComp_00285709 | geneComp_00052739 | ENST00000261847 | ENSG00000138593 | SECISBP2L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.139 | 0.139 | 1.124899e-03 | TRUE |
| isoComp_00350257 | geneComp_00060780 | MSTRG.6394.4 | ENSG00000186174 | BCL9L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 1.134697e-03 | TRUE |
| isoComp_00395527 | geneComp_00075689 | ENST00000657721 | ENSG00000262185 | LINC02861 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.306 | 0.306 | 1.174958e-03 | TRUE |
| isoComp_00313682 | geneComp_00055802 | ENST00000294521 | ENSG00000162522 | KIAA1522 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.079 | 0.195 | 0.116 | 1.235506e-03 | TRUE |
| isoComp_00399296 | geneComp_00077509 | MSTRG.12130.3 | ENSG00000270580 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.004 | 0.059 | 0.055 | 1.287718e-03 | TRUE | |
| isoComp_00284963 | geneComp_00052659 | ENST00000433140 | ENSG00000138080 | EMILIN1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.335 | 0.335 | 1.299712e-03 | TRUE |
| isoComp_00355204 | geneComp_00061592 | ENST00000405201 | ENSG00000196498 | NCOR2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.140 | 0.260 | 0.120 | 1.316578e-03 | TRUE |
| isoComp_00385546 | geneComp_00071083 | ENST00000685108 | ENSG00000245904 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.864 | 0.000 | -0.864 | 1.328586e-03 | TRUE | |
| isoComp_00235416 | geneComp_00047406 | ENST00000216797 | ENSG00000100906 | NFKBIA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.964 | 0.875 | -0.090 | 1.343283e-03 | |
| isoComp_00367618 | geneComp_00063932 | ENST00000342314 | ENSG00000215252 | GOLGA8B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.096 | 0.000 | -0.096 | 1.358173e-03 | TRUE |
| isoComp_00360064 | geneComp_00062202 | MSTRG.20603.17 | ENSG00000198646 | NCOA6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.092 | 0.092 | 1.389926e-03 | TRUE |
| isoComp_00373044 | geneComp_00066028 | ENST00000453790 | ENSG00000227885 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.369 | 0.000 | -0.369 | 1.411918e-03 | TRUE | |
| isoComp_00354439 | geneComp_00061502 | ENST00000490889 | ENSG00000196220 | SRGAP3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.296 | 0.296 | 1.418155e-03 | TRUE |
| isoComp_00309225 | geneComp_00055322 | ENST00000360515 | ENSG00000158985 | CDC42SE2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.426 | 0.195 | -0.230 | 1.446978e-03 | TRUE |
| isoComp_00337623 | geneComp_00058826 | ENST00000572043 | ENSG00000175826 | CTDNEP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.094 | 0.193 | 0.099 | 1.461765e-03 | TRUE |
| isoComp_00240058 | geneComp_00047886 | ENST00000682065 | ENSG00000104133 | SPG11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.143 | 0.143 | 1.522857e-03 | TRUE |
| isoComp_00260892 | geneComp_00050014 | MSTRG.28520.3 | ENSG00000119899 | SLC17A5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 1.532468e-03 | TRUE |
| isoComp_00280649 | geneComp_00052198 | ENST00000566626 | ENSG00000135740 | SLC9A5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.087 | 0.000 | -0.087 | 1.725819e-03 | TRUE |
| isoComp_00336541 | geneComp_00058691 | MSTRG.23841.5 | ENSG00000175066 | GK5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.100 | 0.000 | -0.100 | 1.758065e-03 | TRUE |
| isoComp_00295438 | geneComp_00053713 | MSTRG.24376.5 | ENSG00000145016 | RUBCN | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.104 | 0.000 | -0.104 | 1.908420e-03 | TRUE |
| isoComp_00337373 | geneComp_00058788 | ENST00000574781 | ENSG00000175595 | ERCC4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.026 | 0.098 | 0.072 | 1.929761e-03 | TRUE |
| isoComp_00298036 | geneComp_00054023 | MSTRG.31337.12 | ENSG00000147421 | HMBOX1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.095 | 0.000 | -0.095 | 2.005381e-03 | TRUE |
| isoComp_00306354 | geneComp_00054992 | ENST00000561483 | ENSG00000156218 | ADAMTSL3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.464 | 0.000 | -0.464 | 2.005381e-03 | TRUE |
| isoComp_00321537 | geneComp_00056813 | ENST00000448775 | ENSG00000166250 | CLMP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.414 | 1.000 | 0.586 | 2.236955e-03 | TRUE |
| isoComp_00360450 | geneComp_00062263 | ENST00000628418 | ENSG00000198791 | CNOT7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.205 | 0.144 | -0.062 | 2.283166e-03 | |
| isoComp_00229235 | geneComp_00046845 | ENST00000463905 | ENSG00000089820 | ARHGAP4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.068 | 0.068 | 2.337218e-03 | TRUE |
| isoComp_00321538 | geneComp_00056813 | ENST00000529128 | ENSG00000166250 | CLMP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.586 | 0.000 | -0.586 | 2.337218e-03 | TRUE |
| isoComp_00300244 | geneComp_00054248 | MSTRG.6268.6 | ENSG00000149308 | NPAT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.086 | 0.086 | 2.355719e-03 | TRUE |
| isoComp_00404840 | geneComp_00080833 | ENST00000631443 | ENSG00000282772 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.744 | 0.744 | 2.394339e-03 | TRUE | |
| isoComp_00261884 | geneComp_00050146 | ENST00000547743 | ENSG00000120868 | APAF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.052 | 0.052 | 2.517643e-03 | TRUE |
| isoComp_00297845 | geneComp_00054003 | ENST00000276185 | ENSG00000147234 | FRMPD3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.704 | 0.704 | 2.615054e-03 | TRUE |
| isoComp_00240603 | geneComp_00047933 | ENST00000519409 | ENSG00000104450 | SPAG1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.058 | 0.000 | -0.058 | 2.687415e-03 | TRUE |
| isoComp_00385430 | geneComp_00071067 | ENST00000531805 | ENSG00000245680 | ZNF585B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.096 | 0.096 | 2.687415e-03 | TRUE |
| isoComp_00245767 | geneComp_00048485 | ENST00000465383 | ENSG00000107819 | SFXN3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.145 | 0.145 | 2.708280e-03 | TRUE |
| isoComp_00305672 | geneComp_00054907 | MSTRG.1834.8 | ENSG00000155380 | SLC16A1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.240 | 0.318 | 0.078 | 2.745728e-03 | TRUE |
| isoComp_00353377 | geneComp_00061324 | ENST00000354422 | ENSG00000188859 | FAM78B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.373 | 0.000 | -0.373 | 2.766378e-03 | TRUE |
| isoComp_00251545 | geneComp_00049059 | MSTRG.28673.9 | ENSG00000112234 | FBXL4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.061 | 0.000 | -0.061 | 2.794494e-03 | |
| isoComp_00374390 | geneComp_00066537 | ENST00000664818 | ENSG00000229267 | SNHG31 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.798 | 0.798 | 2.813833e-03 | TRUE |
| isoComp_00276261 | geneComp_00051735 | ENST00000382909 | ENSG00000132950 | ZMYM5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.069 | 0.069 | 2.816032e-03 | TRUE |
| isoComp_00225948 | geneComp_00046552 | ENST00000194530 | ENSG00000082146 | STRADB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.747 | 0.834 | 0.088 | 2.958017e-03 | TRUE |
| isoComp_00228422 | geneComp_00046768 | ENST00000449796 | ENSG00000088387 | DOCK9 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.064 | 0.000 | -0.064 | 2.972330e-03 | TRUE |
| isoComp_00224784 | geneComp_00046440 | ENST00000518832 | ENSG00000079102 | RUNX1T1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.249 | 0.249 | 3.062324e-03 | TRUE |
| isoComp_00287826 | geneComp_00052967 | ENST00000555447 | ENSG00000140022 | STON2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.142 | 0.142 | 3.084884e-03 | TRUE |
| isoComp_00306201 | geneComp_00054976 | ENST00000502655 | ENSG00000156096 | UGT2B4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.579 | 0.000 | -0.579 | 3.104261e-03 | TRUE |
| isoComp_00400484 | geneComp_00078177 | ENST00000667045 | ENSG00000272690 | LINC02018 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.099 | 0.000 | -0.099 | 3.555727e-03 | TRUE |
| isoComp_00364217 | geneComp_00062956 | ENST00000475605 | ENSG00000205643 | CDPF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.237 | 0.120 | -0.117 | 3.560105e-03 | TRUE |
| isoComp_00378457 | geneComp_00068348 | ENST00000671292 | ENSG00000234350 | ERICH2-DT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.477 | 0.477 | 3.660476e-03 | TRUE |
| isoComp_00272231 | geneComp_00051338 | ENST00000432404 | ENSG00000130540 | SULT4A1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.105 | 0.105 | 3.894624e-03 | TRUE |
| isoComp_00240224 | geneComp_00047898 | ENST00000305364 | ENSG00000104228 | TRIM35 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.799 | 0.958 | 0.159 | 3.962120e-03 | TRUE |
| isoComp_00383318 | geneComp_00070338 | ENST00000409586 | ENSG00000241852 | C8orf58 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.223 | 0.012 | -0.212 | 4.144314e-03 | TRUE |
| isoComp_00279764 | geneComp_00052115 | ENST00000527487 | ENSG00000135362 | PRR5L | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.194 | 0.000 | -0.194 | 4.242916e-03 | TRUE |
| isoComp_00315749 | geneComp_00056094 | MSTRG.24581.2 | ENSG00000163612 | FAM86KP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.742 | 0.991 | 0.248 | 4.410396e-03 | TRUE |
| isoComp_00357875 | geneComp_00061914 | MSTRG.1108.4 | ENSG00000197587 | DMBX1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.345 | 0.345 | 4.424368e-03 | TRUE |
| isoComp_00302462 | geneComp_00054498 | ENST00000652176 | ENSG00000151746 | BICD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.009 | 0.123 | 0.114 | 4.544120e-03 | TRUE |
| isoComp_00384785 | geneComp_00070893 | MSTRG.12164.86 | ENSG00000244257 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.064 | 0.000 | -0.064 | 4.570501e-03 | TRUE | |
| isoComp_00318865 | geneComp_00056466 | ENST00000461576 | ENSG00000164818 | DNAAF5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.010 | 0.076 | 0.066 | 4.734796e-03 | TRUE |
| isoComp_00225335 | geneComp_00046483 | MSTRG.23324.6 | ENSG00000080224 | EPHA6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.106 | 0.106 | 4.766731e-03 | TRUE |
| isoComp_00357530 | geneComp_00061879 | MSTRG.3429.7 | ENSG00000197472 | ZNF695 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.208 | 0.380 | 0.171 | 4.766731e-03 | TRUE |
| isoComp_00249094 | geneComp_00048796 | ENST00000532529 | ENSG00000110318 | CEP126 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.041 | 0.478 | 0.437 | 5.021583e-03 | TRUE |
| isoComp_00282191 | geneComp_00052364 | ENST00000470571 | ENSG00000136603 | SKIL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.240 | 0.115 | -0.125 | 5.093916e-03 | TRUE |
| isoComp_00357796 | geneComp_00061903 | ENST00000538570 | ENSG00000197565 | COL4A6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.079 | 0.000 | -0.079 | 5.348125e-03 | TRUE |
| isoComp_00215415 | geneComp_00045644 | ENST00000407925 | ENSG00000049323 | LTBP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.177 | 0.329 | 0.152 | 5.625519e-03 | TRUE |
| isoComp_00276260 | geneComp_00051735 | ENST00000382907 | ENSG00000132950 | ZMYM5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.175 | 0.422 | 0.248 | 5.756587e-03 | TRUE |
| isoComp_00226112 | geneComp_00046563 | ENST00000542398 | ENSG00000082458 | DLG3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.285 | 0.457 | 0.172 | 5.853684e-03 | TRUE |
| isoComp_00280968 | geneComp_00052237 | ENST00000392086 | ENSG00000135924 | DNAJB2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.621 | 0.463 | -0.159 | 5.853684e-03 | TRUE |
| isoComp_00398419 | geneComp_00077035 | MSTRG.17839.6 | ENSG00000268543 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.270 | 0.000 | -0.270 | 5.872378e-03 | TRUE | |
| isoComp_00380598 | geneComp_00069257 | ENST00000685649 | ENSG00000236869 | ZKSCAN7-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.230 | 0.013 | -0.218 | 5.987416e-03 | TRUE |
| isoComp_00317713 | geneComp_00056304 | ENST00000296585 | ENSG00000164171 | ITGA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.906 | 0.720 | -0.186 | 6.030993e-03 | TRUE |
| isoComp_00315748 | geneComp_00056094 | ENST00000509817 | ENSG00000163612 | FAM86KP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.258 | 0.009 | -0.248 | 6.118600e-03 | TRUE |
| isoComp_00381610 | geneComp_00069626 | ENST00000599421 | ENSG00000237945 | LINC00649 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.492 | 0.849 | 0.357 | 6.128957e-03 | TRUE |
| isoComp_00364728 | geneComp_00063078 | ENST00000479422 | ENSG00000206530 | CFAP44 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.115 | 0.115 | 6.172306e-03 | TRUE |
| isoComp_00279351 | geneComp_00052078 | ENST00000552920 | ENSG00000135119 | RNFT2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.131 | 0.000 | -0.131 | 6.267340e-03 | TRUE |
| isoComp_00373132 | geneComp_00066065 | ENST00000430844 | ENSG00000228010 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.593 | 0.000 | -0.593 | 6.437284e-03 | TRUE | |
| isoComp_00324989 | geneComp_00057220 | ENST00000525128 | ENSG00000167861 | HID1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 6.456440e-03 | |
| isoComp_00317714 | geneComp_00056304 | ENST00000503810 | ENSG00000164171 | ITGA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.094 | 0.280 | 0.186 | 6.462821e-03 | TRUE |
| isoComp_00317384 | geneComp_00056260 | ENST00000467110 | ENSG00000164078 | MST1R | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.550 | 0.000 | -0.550 | 6.556166e-03 | TRUE |
| isoComp_00309025 | geneComp_00055302 | ENST00000374553 | ENSG00000158813 | EDA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.156 | 0.578 | 0.422 | 6.930349e-03 | TRUE |
| isoComp_00377290 | geneComp_00067851 | ENST00000657232 | ENSG00000232931 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.070 | 0.070 | 7.068778e-03 | TRUE | |
| isoComp_00307440 | geneComp_00055115 | ENST00000644160 | ENSG00000157259 | GATAD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.040 | 0.102 | 0.062 | 7.094844e-03 | |
| isoComp_00329496 | geneComp_00057740 | ENST00000465122 | ENSG00000170248 | PDCD6IP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.123 | 0.178 | 0.054 | 7.184564e-03 | |
| isoComp_00355301 | geneComp_00061600 | ENST00000378099 | ENSG00000196511 | TPK1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.155 | 0.155 | 7.346025e-03 | TRUE |
| isoComp_00249095 | geneComp_00048796 | ENST00000670091 | ENSG00000110318 | CEP126 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.454 | 0.000 | -0.454 | 7.377397e-03 | TRUE |
| isoComp_00339325 | geneComp_00059097 | ENST00000543884 | ENSG00000177406 | NINJ2-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.249 | 0.249 | 7.594440e-03 | TRUE |
| isoComp_00307486 | geneComp_00055124 | MSTRG.12929.6 | ENSG00000157350 | ST3GAL2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.036 | 0.104 | 0.068 | 7.690302e-03 | TRUE |
| isoComp_00312161 | geneComp_00055621 | ENST00000341533 | ENSG00000161048 | NAPEPLD | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.315 | 0.000 | -0.315 | 7.761968e-03 | TRUE |
| isoComp_00294600 | geneComp_00053631 | ENST00000434469 | ENSG00000144426 | NBEAL1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.123 | 0.000 | -0.123 | 7.924276e-03 | TRUE |
| isoComp_00369301 | geneComp_00064507 | ENST00000590789 | ENSG00000223573 | TINCR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.122 | 0.000 | -0.122 | 7.926745e-03 | TRUE |
| isoComp_00208565 | geneComp_00045012 | ENST00000490965 | ENSG00000003402 | CFLAR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.057 | 0.057 | 7.963396e-03 | TRUE |
| isoComp_00369698 | geneComp_00064648 | ENST00000663803 | ENSG00000223960 | CHROMR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.085 | 0.008 | -0.078 | 7.998831e-03 | TRUE |
| isoComp_00255487 | geneComp_00049450 | ENST00000427708 | ENSG00000115392 | FANCL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.173 | 0.099 | -0.074 | 8.510650e-03 | TRUE |
| isoComp_00311362 | geneComp_00055548 | MSTRG.13976.1 | ENSG00000160602 | NEK8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.119 | 0.119 | 8.678268e-03 | TRUE |
| isoComp_00241753 | geneComp_00048047 | ENST00000594035 | ENSG00000105072 | C19orf44 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.121 | 0.309 | 0.188 | 9.475909e-03 | TRUE |
| isoComp_00352616 | geneComp_00061195 | ENST00000586658 | ENSG00000188283 | ZNF383 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.089 | 0.000 | -0.089 | 9.679705e-03 | TRUE |
| isoComp_00379861 | geneComp_00068935 | ENST00000422825 | ENSG00000235944 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.122 | 0.122 | 9.729838e-03 | TRUE | |
| isoComp_00266826 | geneComp_00050721 | ENST00000409656 | ENSG00000125637 | PSD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.169 | 0.169 | 9.762511e-03 | TRUE |
| isoComp_00266946 | geneComp_00050733 | ENST00000371120 | ENSG00000125703 | ATG4C | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.255 | 0.345 | 0.090 | 1.013434e-02 | TRUE |
| isoComp_00320553 | geneComp_00056692 | ENST00000498820 | ENSG00000165731 | RET | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.218 | 0.000 | -0.218 | 1.051272e-02 | TRUE |
| isoComp_00349174 | geneComp_00060639 | MSTRG.11686.11 | ENSG00000185596 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.109 | 0.221 | 0.111 | 1.059219e-02 | TRUE | |
| isoComp_00349677 | geneComp_00060691 | ENST00000572995 | ENSG00000185813 | PCYT2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.153 | 0.091 | -0.062 | 1.089502e-02 | |
| isoComp_00297127 | geneComp_00053922 | ENST00000459688 | ENSG00000146648 | EGFR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.000 | -0.076 | 1.099608e-02 | TRUE |
| isoComp_00245542 | geneComp_00048462 | MSTRG.3948.5 | ENSG00000107643 | MAPK8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.057 | 0.119 | 0.062 | 1.100536e-02 | |
| isoComp_00277239 | geneComp_00051844 | ENST00000401424 | ENSG00000133794 | ARNTL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.082 | 0.000 | -0.082 | 1.113542e-02 | TRUE |
| isoComp_00208812 | geneComp_00045031 | MSTRG.16988.4 | ENSG00000004777 | ARHGAP33 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.087 | 0.017 | -0.070 | 1.114948e-02 | |
| isoComp_00389975 | geneComp_00072847 | MSTRG.5899.18 | ENSG00000254469 | XNDC1N | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.098 | 0.000 | -0.098 | 1.139880e-02 | TRUE |
| isoComp_00233945 | geneComp_00047283 | ENST00000443735 | ENSG00000100362 | PVALB | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.208 | 0.000 | -0.208 | 1.166242e-02 | TRUE |
| isoComp_00319200 | geneComp_00056498 | MSTRG.31960.7 | ENSG00000164933 | SLC25A32 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.048 | 0.112 | 0.065 | 1.166242e-02 | TRUE |
| isoComp_00398763 | geneComp_00077206 | ENST00000596643 | ENSG00000269439 | PGLS-DT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.365 | 0.365 | 1.169686e-02 | TRUE |
| isoComp_00234210 | geneComp_00047308 | ENST00000459821 | ENSG00000100425 | BRD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.057 | 0.000 | -0.057 | 1.170381e-02 | |
| isoComp_00384390 | geneComp_00070747 | ENST00000667620 | ENSG00000243701 | DUBR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.126 | 0.000 | -0.126 | 1.170381e-02 | TRUE |
| isoComp_00302150 | geneComp_00054463 | ENST00000281187 | ENSG00000151502 | VPS26B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.651 | 0.482 | -0.170 | 1.182095e-02 | TRUE |
| isoComp_00290861 | geneComp_00053216 | ENST00000585949 | ENSG00000141664 | ZCCHC2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.069 | 0.069 | 1.206247e-02 | TRUE |
| isoComp_00395093 | geneComp_00075495 | ENST00000581050 | ENSG00000261556 | SMG1P7 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.935 | 0.724 | -0.211 | 1.240683e-02 | TRUE |
| isoComp_00333889 | geneComp_00058336 | ENST00000682272 | ENSG00000173064 | HECTD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.129 | 0.291 | 0.161 | 1.242260e-02 | TRUE |
| isoComp_00229129 | geneComp_00046835 | ENST00000586269 | ENSG00000089639 | GMIP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.119 | 0.000 | -0.119 | 1.254598e-02 | TRUE |
| isoComp_00389532 | geneComp_00072598 | ENST00000429698 | ENSG00000253771 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.262 | 0.000 | -0.262 | 1.254637e-02 | TRUE | |
| isoComp_00209410 | geneComp_00045087 | ENST00000592253 | ENSG00000005961 | ITGA2B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.240 | 0.000 | -0.240 | 1.265489e-02 | TRUE |
| isoComp_00272782 | geneComp_00051386 | MSTRG.17389.4 | ENSG00000130751 | NPAS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.057 | 0.057 | 1.300192e-02 | TRUE |
| isoComp_00218135 | geneComp_00045875 | ENST00000502849 | ENSG00000064651 | SLC12A2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.041 | 0.094 | 0.053 | 1.315288e-02 | TRUE |
| isoComp_00215922 | geneComp_00045679 | ENST00000607619 | ENSG00000051825 | MPHOSPH9 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.020 | 0.074 | 0.054 | 1.332051e-02 | |
| isoComp_00306556 | geneComp_00055017 | ENST00000515351 | ENSG00000156453 | PCDH1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.059 | 0.059 | 1.347495e-02 | TRUE |
| isoComp_00367156 | geneComp_00063794 | MSTRG.29794.11 | ENSG00000214783 | POLR2J4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.134 | 0.134 | 1.347495e-02 | TRUE |
| isoComp_00335013 | geneComp_00058487 | ENST00000372201 | ENSG00000173846 | PLK3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.519 | 0.217 | -0.302 | 1.352198e-02 | TRUE |
| isoComp_00339015 | geneComp_00059050 | MSTRG.15532.8 | ENSG00000177150 | FAM210A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.094 | 0.041 | -0.053 | 1.364492e-02 | |
| isoComp_00339260 | geneComp_00059081 | ENST00000523766 | ENSG00000177335 | LINC02904 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.299 | 0.039 | -0.260 | 1.368738e-02 | TRUE |
| isoComp_00260371 | geneComp_00049964 | ENST00000553945 | ENSG00000119638 | NEK9 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.239 | 0.132 | -0.107 | 1.390618e-02 | |
| isoComp_00305430 | geneComp_00054882 | ENST00000404789 | ENSG00000155100 | OTUD6B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.853 | 0.515 | -0.338 | 1.405263e-02 | TRUE |
| isoComp_00228794 | geneComp_00046803 | ENST00000541652 | ENSG00000089053 | ANAPC5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.024 | 0.085 | 0.061 | 1.408827e-02 | TRUE |
| isoComp_00220025 | geneComp_00046040 | ENST00000377497 | ENSG00000068831 | RASGRP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.010 | 0.251 | 0.241 | 1.478345e-02 | TRUE |
| isoComp_00213628 | geneComp_00045470 | ENST00000355431 | ENSG00000032219 | ARID4A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.854 | 0.592 | -0.261 | 1.531927e-02 | TRUE |
| isoComp_00318909 | geneComp_00056469 | MSTRG.29357.12 | ENSG00000164828 | SUN1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.003 | 0.121 | 0.118 | 1.651956e-02 | TRUE |
| isoComp_00364157 | geneComp_00062945 | ENST00000433760 | ENSG00000205593 | DENND6B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.098 | 0.098 | 1.670926e-02 | TRUE |
| isoComp_00331402 | geneComp_00058028 | ENST00000411860 | ENSG00000171551 | ECEL1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.221 | 0.019 | -0.202 | 1.717894e-02 | TRUE |
| isoComp_00369003 | geneComp_00064402 | ENST00000413029 | ENSG00000222020 | HDAC4-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.326 | 0.326 | 1.724045e-02 | TRUE |
| isoComp_00315404 | geneComp_00056051 | ENST00000492486 | ENSG00000163482 | STK36 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.036 | 0.094 | 0.058 | 1.764645e-02 | TRUE |
| isoComp_00215127 | geneComp_00045616 | ENST00000545540 | ENSG00000048028 | USP28 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.112 | 0.053 | -0.059 | 1.765889e-02 | |
| isoComp_00346784 | geneComp_00060260 | ENST00000476565 | ENSG00000183856 | IQGAP3 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.066 | 0.138 | 0.071 | 1.881981e-02 | TRUE |
| isoComp_00351058 | geneComp_00060917 | ENST00000436322 | ENSG00000186854 | TRABD2A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.327 | 0.000 | -0.327 | 1.915109e-02 | TRUE |
| isoComp_00355788 | geneComp_00061656 | MSTRG.12914.8 | ENSG00000196696 | PDXDC2P-NPIPB14P | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.120 | 0.120 | 1.945771e-02 | TRUE |
| isoComp_00296333 | geneComp_00053823 | ENST00000394370 | ENSG00000145868 | FBXO38 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.152 | 0.066 | -0.086 | 2.117443e-02 | TRUE |
| isoComp_00317168 | geneComp_00056244 | ENST00000478171 | ENSG00000164050 | PLXNB1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.058 | 0.058 | 2.118281e-02 | TRUE |
| isoComp_00401507 | geneComp_00078785 | ENST00000614663 | ENSG00000274372 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.319 | 0.000 | -0.319 | 2.136856e-02 | TRUE | |
| isoComp_00253003 | geneComp_00049219 | ENST00000451528 | ENSG00000113532 | ST8SIA4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.030 | 0.457 | 0.427 | 2.141458e-02 | TRUE |
| isoComp_00329708 | geneComp_00057771 | ENST00000425046 | ENSG00000170364 | SETMAR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.462 | 0.258 | -0.204 | 2.142804e-02 | TRUE |
| isoComp_00396525 | geneComp_00076222 | ENST00000584900 | ENSG00000265479 | DTX2P1-UPK3BP1-PMS2P11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.841 | 0.614 | -0.226 | 2.142804e-02 | TRUE |
| isoComp_00318283 | geneComp_00056384 | MSTRG.27318.4 | ENSG00000164463 | CREBRF | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.430 | 0.215 | -0.215 | 2.147190e-02 | TRUE |
| isoComp_00394068 | geneComp_00074935 | ENST00000667063 | ENSG00000260288 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.485 | 0.000 | -0.485 | 2.147190e-02 | TRUE | |
| isoComp_00238459 | geneComp_00047744 | MSTRG.12733.7 | ENSG00000103021 | CCDC113 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.133 | 0.023 | -0.110 | 2.210782e-02 | TRUE |
| isoComp_00269575 | geneComp_00051016 | ENST00000297273 | ENSG00000127995 | CASD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.934 | 0.728 | -0.206 | 2.231136e-02 | TRUE |
| isoComp_00403349 | geneComp_00079821 | MSTRG.21052.10 | ENSG00000278878 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.732 | 0.000 | -0.732 | 2.272433e-02 | TRUE | |
| isoComp_00400728 | geneComp_00078322 | ENST00000609475 | ENSG00000272977 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.966 | 0.658 | -0.307 | 2.289979e-02 | TRUE | |
| isoComp_00379328 | geneComp_00068694 | ENST00000450990 | ENSG00000235257 | ITGA9-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.027 | 0.128 | 0.102 | 2.313409e-02 | TRUE |
| isoComp_00295860 | geneComp_00053767 | ENST00000347063 | ENSG00000145428 | RNF175 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.976 | 0.000 | -0.976 | 2.318815e-02 | TRUE |
| isoComp_00226343 | geneComp_00046579 | ENST00000369739 | ENSG00000083097 | DOP1A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.055 | 0.055 | 2.320328e-02 | TRUE |
| isoComp_00253371 | geneComp_00049254 | MSTRG.24204.3 | ENSG00000113790 | EHHADH | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.132 | 0.132 | 2.322819e-02 | TRUE |
| isoComp_00303247 | geneComp_00054614 | ENST00000426626 | ENSG00000152672 | CLEC4F | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.204 | -0.796 | 2.360974e-02 | TRUE |
| isoComp_00285965 | geneComp_00052766 | ENST00000507538 | ENSG00000138670 | RASGEF1B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.056 | 0.056 | 2.405453e-02 | TRUE |
| isoComp_00325498 | geneComp_00057262 | ENST00000541367 | ENSG00000168010 | ATG16L2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.446 | 0.256 | -0.190 | 2.446066e-02 | TRUE |
| isoComp_00318032 | geneComp_00056348 | ENST00000354891 | ENSG00000164318 | EGFLAM | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.369 | 0.000 | -0.369 | 2.456178e-02 | TRUE |
| isoComp_00274707 | geneComp_00051581 | ENST00000346736 | ENSG00000132016 | BRME1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.234 | 0.103 | -0.131 | 2.569256e-02 | TRUE |
| isoComp_00277706 | geneComp_00051890 | ENST00000560570 | ENSG00000134138 | MEIS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.088 | 0.249 | 0.161 | 2.604878e-02 | TRUE |
| isoComp_00385795 | geneComp_00071134 | MSTRG.27398.2 | ENSG00000246596 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.007 | 0.071 | 0.064 | 2.610349e-02 | TRUE | |
| isoComp_00400729 | geneComp_00078322 | MSTRG.21848.27 | ENSG00000272977 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.034 | 0.342 | 0.307 | 2.614298e-02 | TRUE | |
| isoComp_00284930 | geneComp_00052655 | MSTRG.18114.10 | ENSG00000138074 | SLC5A6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.145 | 0.085 | -0.060 | 2.615511e-02 | |
| isoComp_00297122 | geneComp_00053922 | ENST00000275493 | ENSG00000146648 | EGFR | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.913 | 0.996 | 0.083 | 2.647682e-02 | TRUE |
| isoComp_00227901 | geneComp_00046727 | ENST00000565682 | ENSG00000087263 | OGFOD1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.178 | 0.097 | -0.081 | 2.648516e-02 | TRUE |
| isoComp_00399875 | geneComp_00077857 | ENST00000665423 | ENSG00000271913 | TAGAP-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.077 | 0.077 | 2.658376e-02 | TRUE |
| isoComp_00329417 | geneComp_00057725 | ENST00000306324 | ENSG00000170166 | HOXD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.885 | -0.115 | 2.664548e-02 | TRUE |
| isoComp_00303246 | geneComp_00054614 | ENST00000272367 | ENSG00000152672 | CLEC4F | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.796 | 0.796 | 2.667740e-02 | TRUE |
| isoComp_00262373 | geneComp_00050208 | ENST00000240727 | ENSG00000121413 | ZSCAN18 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.108 | 0.018 | -0.090 | 2.668453e-02 | TRUE |
| isoComp_00305876 | geneComp_00054928 | ENST00000569592 | ENSG00000155666 | KDM8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.060 | 0.060 | 2.737662e-02 | TRUE |
| isoComp_00221208 | geneComp_00046145 | MSTRG.17982.1 | ENSG00000071575 | TRIB2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 2.746368e-02 | TRUE |
| isoComp_00231399 | geneComp_00047031 | ENST00000478093 | ENSG00000095777 | MYO3A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.090 | 0.000 | -0.090 | 2.756015e-02 | TRUE |
| isoComp_00275100 | geneComp_00051615 | ENST00000254654 | ENSG00000132323 | ILKAP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.781 | 0.692 | -0.090 | 2.775639e-02 | |
| isoComp_00390274 | geneComp_00073011 | MSTRG.5134.3 | ENSG00000254860 | TMEM9B-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.568 | 0.244 | -0.324 | 2.796661e-02 | TRUE |
| isoComp_00331889 | geneComp_00058093 | ENST00000591380 | ENSG00000171827 | ZNF570 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.104 | 0.000 | -0.104 | 2.818834e-02 | TRUE |
| isoComp_00373049 | geneComp_00066028 | ENST00000666996 | ENSG00000227885 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.281 | 0.281 | 2.823395e-02 | TRUE | |
| isoComp_00219546 | geneComp_00045999 | ENST00000468310 | ENSG00000067606 | PRKCZ | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.055 | 0.004 | -0.052 | 2.958590e-02 | TRUE |
| isoComp_00218670 | geneComp_00045920 | ENST00000439399 | ENSG00000065609 | SNAP91 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.111 | 0.000 | -0.111 | 2.969535e-02 | TRUE |
| isoComp_00394914 | geneComp_00075401 | ENST00000567905 | ENSG00000261342 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.905 | -0.095 | 2.990819e-02 | TRUE | |
| isoComp_00289685 | geneComp_00053125 | ENST00000572144 | ENSG00000141086 | CTRL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.515 | 0.823 | 0.308 | 3.027111e-02 | TRUE |
| isoComp_00372243 | geneComp_00065690 | ENST00000437593 | ENSG00000226864 | ATE1-AS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.798 | -0.202 | 3.027111e-02 | TRUE |
| isoComp_00408501 | geneComp_00083064 | ENST00000676884 | ENSG00000288674 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.668 | -0.332 | 3.027111e-02 | TRUE | |
| isoComp_00253309 | geneComp_00049248 | ENST00000265087 | ENSG00000113739 | STC2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.631 | 0.724 | 0.092 | 3.046048e-02 | TRUE |
| isoComp_00373131 | geneComp_00066065 | ENST00000366167 | ENSG00000228010 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.322 | 0.843 | 0.521 | 3.058797e-02 | TRUE | |
| isoComp_00245472 | geneComp_00048452 | MSTRG.4703.3 | ENSG00000107560 | RAB11FIP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.064 | 0.182 | 0.118 | 3.077934e-02 | TRUE |
| isoComp_00309237 | geneComp_00055322 | MSTRG.26883.9 | ENSG00000158985 | CDC42SE2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.440 | 0.606 | 0.166 | 3.135264e-02 | TRUE |
| isoComp_00282483 | geneComp_00052401 | ENST00000319107 | ENSG00000136828 | RALGPS1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.125 | 0.007 | -0.118 | 3.207285e-02 | TRUE |
| isoComp_00329418 | geneComp_00057725 | MSTRG.19554.4 | ENSG00000170166 | HOXD4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.115 | 0.115 | 3.230279e-02 | TRUE |
| isoComp_00330135 | geneComp_00057835 | ENST00000394666 | ENSG00000170634 | ACYP2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.598 | 0.408 | -0.190 | 3.252052e-02 | TRUE |
| isoComp_00337617 | geneComp_00058826 | ENST00000318988 | ENSG00000175826 | CTDNEP1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.368 | 0.261 | -0.107 | 3.261005e-02 | TRUE |
| isoComp_00282923 | geneComp_00052454 | ENST00000469050 | ENSG00000137038 | DMAC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.187 | 0.251 | 0.063 | 3.292206e-02 | |
| isoComp_00255791 | geneComp_00049467 | ENST00000405984 | ENSG00000115507 | OTX1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.206 | 0.046 | -0.161 | 3.300280e-02 | TRUE |
| isoComp_00266194 | geneComp_00050647 | ENST00000688026 | ENSG00000125122 | LRRC29 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.132 | 0.011 | -0.121 | 3.325744e-02 | TRUE |
| isoComp_00234784 | geneComp_00047362 | ENST00000525399 | ENSG00000100614 | PPM1A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.479 | 0.361 | -0.118 | 3.347341e-02 | TRUE |
| isoComp_00394915 | geneComp_00075401 | MSTRG.16127.6 | ENSG00000261342 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.095 | 0.095 | 3.382217e-02 | TRUE | |
| isoComp_00237971 | geneComp_00047703 | ENST00000476666 | ENSG00000102786 | INTS6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.004 | 0.086 | 0.082 | 3.414935e-02 | TRUE |
| isoComp_00302015 | geneComp_00054449 | MSTRG.26704.5 | ENSG00000151422 | FER | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.267 | 0.117 | -0.149 | 3.441438e-02 | TRUE |
| isoComp_00387602 | geneComp_00071773 | ENST00000671468 | ENSG00000249898 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.235 | 0.000 | -0.235 | 3.448529e-02 | TRUE | |
| isoComp_00361376 | geneComp_00062456 | ENST00000411948 | ENSG00000203667 | COX20 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.553 | 0.297 | -0.256 | 3.462483e-02 | TRUE |
| isoComp_00304932 | geneComp_00054819 | ENST00000517387 | ENSG00000154548 | SRSF12 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.121 | 0.019 | -0.101 | 3.511115e-02 | TRUE |
| isoComp_00313155 | geneComp_00055729 | ENST00000318782 | ENSG00000162073 | PAQR4 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.603 | 0.492 | -0.111 | 3.538967e-02 | TRUE |
| isoComp_00381545 | geneComp_00069601 | ENST00000668985 | ENSG00000237877 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.279 | 0.279 | 3.538967e-02 | TRUE | |
| isoComp_00324717 | geneComp_00057174 | MSTRG.13365.4 | ENSG00000167703 | SLC43A2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 3.541635e-02 | TRUE |
| isoComp_00395108 | geneComp_00075500 | ENST00000567503 | ENSG00000261570 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.223 | 0.223 | 3.548061e-02 | TRUE | |
| isoComp_00408500 | geneComp_00083064 | ENST00000366779 | ENSG00000288674 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.332 | 0.332 | 3.554729e-02 | TRUE | |
| isoComp_00275895 | geneComp_00051696 | ENST00000255039 | ENSG00000132702 | HAPLN2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 1.000 | 0.104 | -0.896 | 3.568675e-02 | TRUE |
| isoComp_00251870 | geneComp_00049100 | MSTRG.28140.13 | ENSG00000112511 | PHF1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.007 | 0.062 | 0.055 | 3.586414e-02 | TRUE |
| isoComp_00356116 | geneComp_00061691 | ENST00000372874 | ENSG00000196839 | ADA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.648 | 0.505 | -0.144 | 3.595154e-02 | |
| isoComp_00346901 | geneComp_00060283 | ENST00000567649 | ENSG00000183971 | NPW | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.075 | 0.000 | -0.075 | 3.652146e-02 | TRUE |
| isoComp_00359285 | geneComp_00062099 | MSTRG.26323.1 | ENSG00000198237 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.172 | 0.172 | 3.702145e-02 | TRUE | |
| isoComp_00400245 | geneComp_00078064 | ENST00000614510 | ENSG00000272419 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.149 | 0.000 | -0.149 | 3.715283e-02 | TRUE | |
| isoComp_00225375 | geneComp_00046489 | ENST00000357248 | ENSG00000080503 | SMARCA2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.251 | 0.122 | -0.130 | 3.717560e-02 | TRUE |
| isoComp_00289244 | geneComp_00053088 | ENST00000564386 | ENSG00000140905 | GCSH | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.106 | 0.163 | 0.057 | 3.737390e-02 | TRUE |
| isoComp_00324158 | geneComp_00057114 | ENST00000579650 | ENSG00000167525 | PROCA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.044 | 0.275 | 0.231 | 3.777443e-02 | TRUE |
| isoComp_00242083 | geneComp_00048077 | ENST00000595741 | ENSG00000105245 | NUMBL | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.050 | 0.000 | -0.050 | 3.780291e-02 | TRUE |
| isoComp_00404655 | geneComp_00080768 | ENST00000685096 | ENSG00000281706 | LINC01012 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.070 | 0.000 | -0.070 | 3.796451e-02 | TRUE |
| isoComp_00240986 | geneComp_00047967 | ENST00000381733 | ENSG00000104763 | ASAH1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.202 | 0.286 | 0.083 | 3.797944e-02 | TRUE |
| isoComp_00372008 | geneComp_00065617 | ENST00000591116 | ENSG00000226686 | LINC01535 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.167 | 0.000 | -0.167 | 3.797944e-02 | TRUE |
| isoComp_00359401 | geneComp_00062114 | ENST00000683047 | ENSG00000198324 | PHETA1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.898 | 0.784 | -0.114 | 3.894182e-02 | TRUE |
| isoComp_00384877 | geneComp_00070935 | ENST00000306376 | ENSG00000244405 | ETV5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.478 | 1.000 | 0.522 | 3.897475e-02 | TRUE |
| isoComp_00221560 | geneComp_00046168 | ENST00000452101 | ENSG00000072195 | SPEG | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.227 | 0.060 | -0.166 | 3.973094e-02 | TRUE |
| isoComp_00342874 | geneComp_00059648 | MSTRG.4364.1 | ENSG00000180628 | PCGF5 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.538 | 0.304 | -0.235 | 3.980080e-02 | TRUE |
| isoComp_00270710 | geneComp_00051162 | ENST00000527884 | ENSG00000129173 | E2F8 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.281 | 0.431 | 0.150 | 4.054997e-02 | TRUE |
| isoComp_00283651 | geneComp_00052521 | ENST00000676266 | ENSG00000137411 | VARS2 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.154 | 0.100 | -0.054 | 4.131814e-02 | |
| isoComp_00221280 | geneComp_00046150 | ENST00000369643 | ENSG00000071889 | FAM3A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.030 | 0.103 | 0.073 | 4.136692e-02 | TRUE |
| isoComp_00405097 | geneComp_00080947 | ENST00000532864 | ENSG00000283341 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.061 | 0.000 | -0.061 | 4.295211e-02 | TRUE | |
| isoComp_00244468 | geneComp_00048338 | ENST00000674539 | ENSG00000106477 | CEP41 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.076 | 0.009 | -0.067 | 4.350496e-02 | TRUE |
| isoComp_00302366 | geneComp_00054486 | ENST00000281416 | ENSG00000151690 | MFSD6 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.743 | 0.108 | -0.635 | 4.384620e-02 | TRUE |
| isoComp_00314258 | geneComp_00055888 | ENST00000407071 | ENSG00000162849 | KIF26B | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.785 | 1.000 | 0.215 | 4.393882e-02 | TRUE |
| isoComp_00324543 | geneComp_00057153 | ENST00000513264 | ENSG00000167653 | PSCA | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.288 | 0.000 | -0.288 | 4.466690e-02 | TRUE |
| isoComp_00334443 | geneComp_00058418 | MSTRG.22886.21 | ENSG00000173473 | SMARCC1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.281 | 0.186 | -0.095 | 4.466690e-02 | TRUE |
| isoComp_00232574 | geneComp_00047147 | ENST00000382363 | ENSG00000099999 | RNF215 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.675 | 0.571 | -0.104 | 4.478524e-02 | TRUE |
| isoComp_00237125 | geneComp_00047603 | ENST00000481776 | ENSG00000101966 | XIAP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.168 | 0.075 | -0.093 | 4.478524e-02 | TRUE |
| isoComp_00218195 | geneComp_00045880 | ENST00000509154 | ENSG00000064692 | SNCAIP | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.344 | 0.000 | -0.344 | 4.511379e-02 | TRUE |
| isoComp_00314698 | geneComp_00055955 | ENST00000413057 | ENSG00000163075 | CFAP221 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.131 | 0.131 | 4.702252e-02 | TRUE |
| isoComp_00310894 | geneComp_00055505 | ENST00000397743 | ENSG00000160282 | FTCD | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.411 | 0.411 | 4.709039e-02 | TRUE |
| isoComp_00271526 | geneComp_00051266 | ENST00000592312 | ENSG00000130159 | ECSIT | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.026 | 0.078 | 0.052 | 4.734497e-02 | TRUE |
| isoComp_00310148 | geneComp_00055429 | ENST00000445068 | ENSG00000159761 | C16orf86 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.540 | 0.540 | 4.746717e-02 | TRUE |
| isoComp_00364384 | geneComp_00062984 | ENST00000381680 | ENSG00000205771 | CATSPER2P1 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.086 | 0.000 | -0.086 | 4.786691e-02 | TRUE |
| isoComp_00340995 | geneComp_00059350 | ENST00000325509 | ENSG00000178860 | MSC | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.460 | 1.000 | 0.540 | 4.827064e-02 | TRUE |
| isoComp_00291792 | geneComp_00053324 | ENST00000478210 | ENSG00000142632 | ARHGEF19 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.226 | 0.000 | -0.226 | 4.832259e-02 | TRUE |
| isoComp_00338242 | geneComp_00058937 | ENST00000554998 | ENSG00000176473 | WDR25 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.238 | 0.119 | -0.119 | 4.845289e-02 | TRUE |
| isoComp_00370620 | geneComp_00065047 | MSTRG.28453.4 | ENSG00000225096 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.375 | 0.173 | -0.202 | 4.892914e-02 | TRUE | |
| isoComp_00355704 | geneComp_00061650 | MSTRG.27476.5 | ENSG00000196670 | ZFP62 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.032 | 0.099 | 0.067 | 4.933130e-02 | TRUE |
| isoComp_00240061 | geneComp_00047886 | ENST00000682495 | ENSG00000104133 | SPG11 | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.009 | 0.068 | 0.059 | 4.959913e-02 | TRUE |
| isoComp_00272129 | geneComp_00051325 | MSTRG.16698.2 | ENSG00000130477 | UNC13A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.000 | 0.076 | 0.076 | 4.967498e-02 | TRUE |
| isoComp_00325552 | geneComp_00057265 | ENST00000466875 | ENSG00000168026 | TTC21A | HEK293_DMSO_2hB | HEK293_TMG_2hB | 0.151 | 0.000 | -0.151 | 4.970482e-02 | TRUE |
- Note:
- The comparisons made can be identified as “from ‘condition_1’ to ‘condition_2’”, meaning ‘condition_1’ is considered the ground state and ‘condition_2’ the changed state. This also means that a positive dIF value indicates that the isoform usage is increased in ‘condition_2’ compared to ‘condition_1’. Since the ‘isoformFeatures’ entry is the most relevant part of the switchAnalyzeRlist object, the most-used standard methods have also been implemented to work directly on isoformFeatures.
4. Circular RNA Analysis
Volcano Plot
No significantly changed circRNA was detected.
Details of sig. circRNA
No significantly changed circRNA was detected.
Annotation of sig. circRNA
No significantly changed circRNA was detected.