Analysis Report: HEK293_OSMI2_2hA vs HEK293_DMSO_2hA
WANG Ziyi
Date: 18 5月 2022
Summary: HEK293_OSMI2_2hA vs HEK293_DMSO_2hA
1. Differentially Expressed Genes
Heatmap
No significcant DEGs were detected under the threshold of P<0.05
Volcano plot
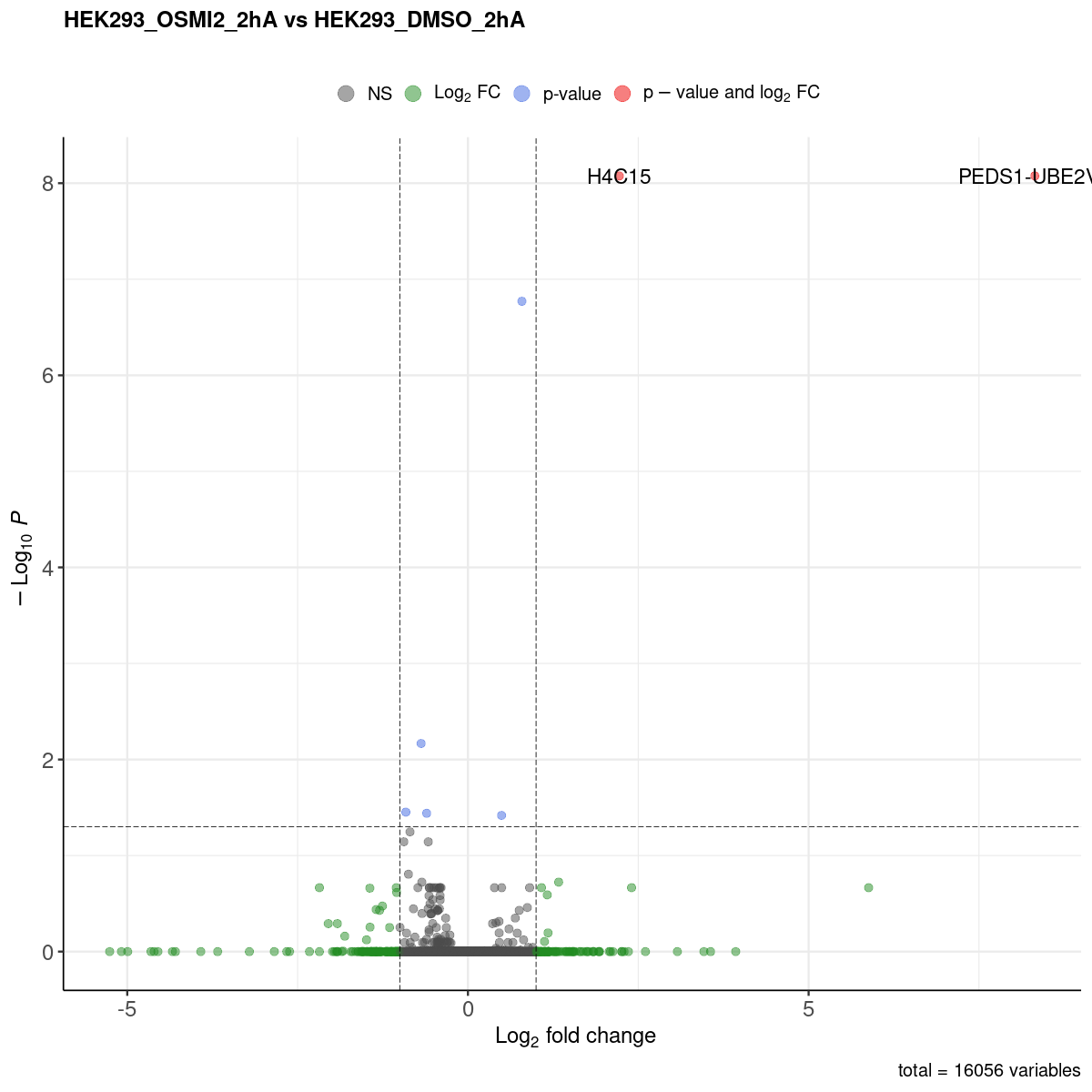
DEGs list
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_OSMI2_2hA | SE_Intercept_HEK293_OSMI2_2hA | Intercept_HEK293_DMSO_2hA | SE_Intercept_HEK293_DMSO_2hA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124208 | 387522 | PEDS1-UBE2V1 | protein_coding | I3L0A0 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; fatty acid metabolism. {ECO:0000256|ARBA:ARBA00004872}. | The TMEM189-UEV mRNA is an infrequent but naturally occurring read-through transcript of the neighboring TMEM189 and UBE2V1 genes. Ubiquitin-conjugating E2 enzyme variant proteins constitute a distinct subfamily within the E2 protein family. They have sequence similarity to other ubiquitin-conjugating enzymes but lack the conserved cysteine residue that is critical for the catalytic activity of E2s. The protein produced by this transcript has UEV1 B domains but the protein is localized to the cytoplasm rather than to the nucleus. The significance of this read-through mRNA and the function of its protein product has not yet been determined. [provided by RefSeq, Oct 2010]. | integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654]; fatty acid metabolic process [GO:0006631] | 20677014_Observational study of gene-disease association. (HuGE Navigator) | 30.85308 | 319.665405 | 8.320419 | 1.3573868 | 51.42673 | 7.431964e-13 | 8.395611e-09 | Yes | No | 55.50345 | 72.39902 | 0.2076871 | 0.2383429 | ||||
| ENSG00000270276 | 554313 | H4C15 | protein_coding | P62805 | FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. | 3D-structure;Acetylation;Chromosomal rearrangement;Chromosome;Citrullination;DNA-binding;Direct protein sequencing;Hydroxylation;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene encodes a replication-dependent histone that is a member of the histone H4 family. Some transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in a histone cluster on chromosome 1. This gene is one of four histone genes in the cluster that are duplicated; this record represents the telomeric copy. [provided by RefSeq, May 2020]. | hsa:121504;hsa:554313;hsa:8294;hsa:8359;hsa:8360;hsa:8361;hsa:8362;hsa:8363;hsa:8364;hsa:8365;hsa:8366;hsa:8367;hsa:8368;hsa:8370; | CENP-A containing nucleosome [GO:0043505]; chromosome, telomeric region [GO:0000781]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; membrane [GO:0016020]; nuclear chromosome [GO:0000228]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; DNA binding [GO:0003677]; protein domain specific binding [GO:0019904]; protein heterodimerization activity [GO:0046982]; RNA binding [GO:0003723]; DNA replication-dependent chromatin assembly [GO:0006335]; DNA replication-independent chromatin assembly [GO:0006336]; DNA-templated transcription, initiation [GO:0006352]; negative regulation of megakaryocyte differentiation [GO:0045653]; nucleosome assembly [GO:0006334]; telomere organization [GO:0032200] | ENSMUSG00000067455 | H4c11 | 194.64943 | 4.663242 | 2.221333 | 0.3093479 | 50.75625 | 1.045791e-12 | 8.395611e-09 | Yes | Yes | 202.12058 | 110.92566 | 46.2575503 | 19.3781117 |
- Note:
- The spreadsheet above contains the genes with significantly changes in expression (P-value < 0.05 and |log2FC| > 1). Please Click HERE to check all mapped genes in a Microsoft .excel file.
- The comparisons made can be identified from the website name. “HEK293_OSMI2_2hA vs HEK293_DMSO_2hA” means that “HEK293_DMSO_2hA” is considered the ground state and “HEK293_OSMI2_2hA” the changed state. This also means that a positive log2FC value indicates that the gene expression is increased in “HEK293_OSMI2_2hA” compared to “HEK293_DMSO_2hA”.
- Click HERE to check all parameters of DESeq2 model for all detected genes of the current samples set in a Microsoft .excel file. For more detials of the DESeq2 model, please Click HERE.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| protein_coding | 2 |
AS in non-DEGs
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_OSMI2_2hA | SE_Intercept_HEK293_OSMI2_2hA | Intercept_HEK293_DMSO_2hA | SE_Intercept_HEK293_DMSO_2hA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000003147 | 3382 | ICA1 | protein_coding | Q05084 | FUNCTION: May play a role in neurotransmitter secretion. {ECO:0000250}. | Alternative splicing;Cell junction;Cytoplasm;Cytoplasmic vesicle;Golgi apparatus;Membrane;Neurotransmitter transport;Reference proteome;Synapse;Transport | This gene encodes a protein with an arfaptin homology domain that is found both in the cytosol and as membrane-bound form on the Golgi complex and immature secretory granules. This protein is believed to be an autoantigen in insulin-dependent diabetes mellitus and primary Sjogren's syndrome. Several transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Feb 2013]. | hsa:3382; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; intracellular membrane-bounded organelle [GO:0043231]; secretory granule membrane [GO:0030667]; synaptic vesicle membrane [GO:0030672]; membrane curvature sensor activity [GO:0140090]; protein domain specific binding [GO:0019904]; neurotransmitter transport [GO:0006836]; regulation of insulin secretion [GO:0050796]; regulation of transport [GO:0051049] | 12409289_The exon A promoter exhibits greater activity in islet cells, whereas the exon B promoter more efficiently activates transcription in neuronal cells. 18187231_ICA69 as a novel Rab2 effector and its role in regulating the early transport of insulin secretory granule proteins 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20526340_Observational study of gene-disease association. (HuGE Navigator) 20962850_Observational study of gene-disease association. (HuGE Navigator) 22447927_polymorphisms within the NOD Ica1 core promoter may determine AIRE-mediated down-regulation of ICA69 expression in medullary thymic epithelial cells. 23836780_Novel association was found between intraocular pressure and a common variant at 7p21 near to GLCCI1 and ICA1. 24358315_C-terminal domain of ICA69 interacts with PICK1 and acts on trafficking of PICK1-PKCalpha complex and cerebellar plasticity. 31478943_Novel SRF-ICA1L Fusions in Cellular Myoid Neoplasms With Potential For Malignant Behavior | ENSMUSG00000062995 | Ica1 | 141.44729 | 0.9742445 | -0.0376442100 | 0.25402302 | 2.170273e-02 | 8.828807e-01 | 9.998360e-01 | No | Yes | 139.83811 | 24.804367 | 1.411834e+02 | 19.664364 | |
| ENSG00000005302 | 10943 | MSL3 | protein_coding | Q8N5Y2 | FUNCTION: Has a role in chromatin remodeling and transcriptional regulation (PubMed:20018852, PubMed:20657587, PubMed:20943666, PubMed:21217699, PubMed:30224647). Has a role in X inactivation (PubMed:21217699). Component of the MSL complex which is responsible for the majority of histone H4 acetylation at 'Lys-16' which is implicated in the formation of higher-order chromatin structure (PubMed:16227571, PubMed:20657587, PubMed:16543150, PubMed:30224647). Specifically recognizes histone H4 monomethylated at 'Lys-20' (H4K20Me1) in a DNA-dependent manner and is proposed to be involved in chromosomal targeting of the MSL complex (PubMed:20657587, PubMed:20943666). {ECO:0000269|PubMed:16227571, ECO:0000269|PubMed:16543150, ECO:0000269|PubMed:20018852, ECO:0000269|PubMed:20657587, ECO:0000269|PubMed:20943666, ECO:0000269|PubMed:21217699, ECO:0000269|PubMed:22547026, ECO:0000269|PubMed:30224647}. | 3D-structure;Alternative splicing;Chromatin regulator;DNA-binding;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene encodes a nuclear protein that is similar to the product of the Drosophila male-specific lethal-3 gene. The Drosophila protein plays a critical role in a dosage-compensation pathway, which equalizes X-linked gene expression in males and females. Thus, the human protein is thought to play a similar function in chromatin remodeling and transcriptional regulation, and it has been found as part of a complex that is responsible for histone H4 lysine-16 acetylation. This gene can undergo X inactivation. Alternative splicing results in multiple transcript variants. Related pseudogenes have been identified on chromosomes 2, 7 and 8. [provided by RefSeq, Jul 2010]. | hsa:10943; | histone acetyltransferase complex [GO:0000123]; MSL complex [GO:0072487]; NuA4 histone acetyltransferase complex [GO:0035267]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; histone acetyltransferase activity (H4-K16 specific) [GO:0046972]; methylated histone binding [GO:0035064]; heterochromatin assembly [GO:0031507]; histone acetylation [GO:0016573]; histone deacetylation [GO:0016575]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; histone H4-K16 acetylation [GO:0043984]; regulation of transcription, DNA-templated [GO:0006355] | 16227571_A multisubunit human histone acetylase complex that contains homologs of the Drosophila MSL proteins MOF, MSL1 (hampin A), MSL2, and MSL3 was described. This complex is responsible for histone H4 lysine-16 acetylation of all cellular chromosomes. 20943666_MSL3 plays an important role in targeting the male specific lethal complex to chromatin in both humans and flies by binding to H4K20Me(1). 30224647_Characterization of syndrome that allowed us to decipher the developmental importance of MSL3 in humans. 33173220_Defining the genotypic and phenotypic spectrum of X-linked MSL3-related disorder. | ENSMUSG00000031358 | Msl3 | 637.93647 | 1.0017242 | 0.0024854214 | 0.15155207 | 2.611552e-04 | 9.871065e-01 | 9.998360e-01 | No | Yes | 654.22328 | 86.981124 | 6.924525e+02 | 71.278770 | |
| ENSG00000007372 | 5080 | PAX6 | protein_coding | P26367 | FUNCTION: Transcription factor with important functions in the development of the eye, nose, central nervous system and pancreas. Required for the differentiation of pancreatic islet alpha cells (By similarity). Competes with PAX4 in binding to a common element in the glucagon, insulin and somatostatin promoters. Regulates specification of the ventral neuron subtypes by establishing the correct progenitor domains (By similarity). Acts as a transcriptional repressor of NFATC1-mediated gene expression (By similarity). {ECO:0000250, ECO:0000250|UniProtKB:P63015}. | 3D-structure;Alternative splicing;DNA-binding;Developmental protein;Differentiation;Disease variant;Homeobox;Nucleus;Paired box;Peters anomaly;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes paired box protein Pax-6, one of many human homologs of the Drosophila melanogaster gene prd. In addition to a conserved paired box domain, a hallmark feature of this gene family, the encoded protein also contains a homeobox domain. Both domains are known to bind DNA and function as regulators of gene transcription. Activity of this protein is key in the development of neural tissues, particularly the eye. This gene is regulated by multiple enhancers located up to hundreds of kilobases distant from this locus. Mutations in this gene or in the enhancer regions can cause ocular disorders such as aniridia and Peter's anomaly. Use of alternate promoters and alternative splicing results in multiple transcript variants encoding different isoforms. Interestingly, inclusion of a particular alternate coding exon has been shown to increase the length of the paired box domain and alter its DNA binding specificity. Consequently, isoforms that carry the shorter paired box domain regulate a different set of genes compared to the isoforms carrying the longer paired box domain. [provided by RefSeq, Mar 2019]. | hsa:5080; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; co-SMAD binding [GO:0070410]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; histone acetyltransferase binding [GO:0035035]; HMG box domain binding [GO:0071837]; protein kinase binding [GO:0019901]; R-SMAD binding [GO:0070412]; RNA binding [GO:0003723]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II core promoter sequence-specific DNA binding [GO:0000979]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; transcription coregulator binding [GO:0001221]; ubiquitin protein ligase binding [GO:0031625]; anatomical structure development [GO:0048856]; animal organ morphogenesis [GO:0009887]; astrocyte differentiation [GO:0048708]; axon guidance [GO:0007411]; blood vessel development [GO:0001568]; cell fate determination [GO:0001709]; cellular response to leukemia inhibitory factor [GO:1990830]; central nervous system development [GO:0007417]; cerebral cortex regionalization [GO:0021796]; commitment of neuronal cell to specific neuron type in forebrain [GO:0021902]; cornea development in camera-type eye [GO:0061303]; dorsal/ventral axis specification [GO:0009950]; embryonic camera-type eye morphogenesis [GO:0048596]; establishment of mitotic spindle orientation [GO:0000132]; eye development [GO:0001654]; eye photoreceptor cell development [GO:0042462]; forebrain dorsal/ventral pattern formation [GO:0021798]; forebrain-midbrain boundary formation [GO:0021905]; gene expression [GO:0010467]; glucose homeostasis [GO:0042593]; habenula development [GO:0021986]; iris morphogenesis [GO:0061072]; keratinocyte differentiation [GO:0030216]; lacrimal gland development [GO:0032808]; lens development in camera-type eye [GO:0002088]; negative regulation of epithelial cell proliferation [GO:0050680]; negative regulation of neural precursor cell proliferation [GO:2000178]; negative regulation of neurogenesis [GO:0050768]; negative regulation of neuron differentiation [GO:0045665]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neuron fate commitment [GO:0048663]; neuron migration [GO:0001764]; oligodendrocyte cell fate specification [GO:0021778]; pancreatic A cell development [GO:0003322]; pituitary gland development [GO:0021983]; positive regulation of core promoter binding [GO:1904798]; positive regulation of epithelial cell differentiation [GO:0030858]; positive regulation of gene expression [GO:0010628]; positive regulation of neuroblast proliferation [GO:0002052]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein localization to organelle [GO:0033365]; regulation of asymmetric cell division [GO:0009786]; regulation of cell migration [GO:0030334]; regulation of timing of cell differentiation [GO:0048505]; regulation of transcription by RNA polymerase II [GO:0006357]; response to wounding [GO:0009611]; retina development in camera-type eye [GO:0060041]; salivary gland morphogenesis [GO:0007435]; signal transduction involved in regulation of gene expression [GO:0023019]; smoothened signaling pathway [GO:0007224]; type B pancreatic cell differentiation [GO:0003309]; ventral spinal cord development [GO:0021517]; visual perception [GO:0007601] | 11069920_HoxB1 interacts with Pax6 and enhances its transcriptional activity. This interaction was modeled on a demonstrated interaction between zebrafish Pax6 and human HoxB1. 11069920_Pbx1 interacts with Pax6 and enhances its transcriptional activity. This interaction was modeled on a demonstrated interaction between zebrafish Pax6 and human Pbx1. 11850181_New 3' elements control Pax6 expression in the developing pretectum, neural retina and olfactory region 12043047_Observational study of gene-disease association. (HuGE Navigator) 12324464_mDia influences Pax6-induced transcriptional activity and axonal pathfinding in a way opposite from ROCK (Rho kinase) and that it may act via Pax6 to modulate early neuronal development 12325030_Independent modifying factors underlie the variability of the different phenotypic features of the PAX6 mutation in aniridia. 12721955_Mutations of the PAX6 gene were detected in patients with a variety of optic-nerve malformations. 12731001_in 24 humans heterozygous for defined PAX6 mutations, widespread structural abnormalities including absence of the pineal gland and unilateral polymicrogyria were demonstrated 12789139_We report for the first time the identification of PAX6 gene mutations in Indian aniridia patients. 12868034_PAX6 gene mutations are associated with aniridia 12953159_This study confirms that foveal hypoplasia in the so-called isolated form have a similar origin as in aniridia namely PAX6 mutation and that it is a symptom in all cases while the iris anomaly may be variable. 14872040_The possible role of PAX6 includes neurodevelopmental roles not only in visual and olfactory sensory domains but also in higher-order auditory processing. 15066147_The brain functional differences in humans with PAX6 mutation that are compatible both with anatomical abnormalities in the same subjects. 15079031_Patients with PAX6 gene mutations and agenesis of the anterior commissure performed more poorly on measures of working memory than those without this abnormality, suggesting the anterior commissure may play a role in cognitive processing 15086958_Study supports the hypothesis that a mutation in the PAX6 gene correlates with expression of aniridia. 15389894_These results are consistent with deficient auditory interhemispheric transfer in patients with a PAX6 mutation, which may be attributable to structural and/or functional abnormalities of the anterior commisure and corpus callosum. 15629294_The association of anterior segment anomalies and foveal hypoplasia with one of the slightest alterations of the PAX6 protein described to date confirms the association of variant phenotypes 15659382_Epidermal growth factor-induced proliferation requires down-regulation of Pax6 in corneal epithelial cells 15677484_Pax6(+5a) induces a developmental cascade in the prospective fovea, area centralis or visual streak region that leads to the formation of a retinal architecture bearing densely packed visual cells. 15735909_Transient overexpression of PAX6 via adenovirus suppressed cell growth by increasing the number of cells in G1 and by decreasing the number of cells in S-phase, and later on caused a dramatic level of cell death. 15757974_central roles in neural retina transdifferentiation 15889018_A novel PAX6 gene mutation was identified in a Chinese aniridia family. This mutation may also contribute to congenital cataracts in these aniridia patients. 15918896_The consistent association of truncating mutations with the aniridia phenotype, and the distribution of truncating mutations in the PAX6 open reading frame, suggests that nonsense-mediated decay acts on PAX6 mutant alleles 16098226_PAX6 interacts with HOMER3, DNCL1, and TRIM11. Three C-terminal PAX6 mutations, previously identified in patients with eye malformations, all reduced or abolished the interactions. 16115881_Pax6 regulation of Optimedin in the eye and brain may directly affect multiple developmental processes, including cell migration and axon growth 16407227_HIPK2 is an upstream protein kinase for Pax6 that may modulate Pax6-mediated transcriptional regulation 16511221_X-ray analysis of the Pax6 paired domain bound to the Pax6 gene enhancer 16543198_Truncating PAX6 mutations and ocular phenotypes is associated with aniridia 16604056_Observational study of genotype prevalence. (HuGE Navigator) 16604056_Two sequence variations in PAX6 gene. These missense mutations may uniquely alter structure and expression of PAX6 protein, resulting in distinct clinical phenotypes. 16712695_This review describes how cross regulation for PAX6, SOX2 and perhaps OTX2 has now been uncovered, pointing to the mechanisms that can fine-tune the expression of three such essential components in eye development. 16785853_Four novel mutations including c.141+1G>A, c.184-3C>G, c.542C>A (Ser181X), and c.562C>T (Gln188X) and one known mutation c.120C>A (Cys40X) were identified in PAX6 of five unrelated patients with aniridia. 16803629_New deletions and an insertion create frameshifts predicted to introduce premature termination codons into the PAX6 reading frame. The genetic alterations are predicted to lead to loss-of-function mutations segregating in autosomal dominant manner. 16873704_Observational study of gene-disease association. (HuGE Navigator) 17031679_potential effect of the PAX6 mutation on the mtDNA mutation rate 17148041_To our knowledge, this is the first mutation of PAX6 gene reported in association with a Gillespie-like syndrome. 17202185_the proliferation of cortical progenitors is sensitive to altered Pax6 levels 17318412_PAX6 over-expression in low PAX6-expressing glioma cells attenuated recovery of growth after detachment-induced stress, and intracellular reactive oxygen species levels increased following cell detachment. 17415970_Mutation of PAX6 gene can result in the occurrence of congenital aniridia. 17417613_We identified three mutations associated with aniridia phenotypes (Q179X, C40X, and V48fsX53). The three other mutations reported here cause non-aniridia ocular phenotypes associated in some cases with neurological anomalies. 17485622_Finds children with PAX6 mutations may have auditory interhemispheric transfer deficits and difficulty localizing sound and understanding speech in noisy backgrounds even when there is a normal audiogram. 17568989_PAX6 point mutations and deletions can cause aniridia. 17595013_Screening of PAX6 in patients with suspected Gillespie syndrome should be performed with up-to-date methodology. 17653045_Observational study of gene-disease association. (HuGE Navigator) 17679951_A significant fraction of familial aniridia patients appears to be caused by a 3' deletion to PAX6. 17893655_Two novel mutations in PAX6 are capable of causing the classic aniridia phenotype. PAX6 plays a role in hereditary aniridia. 17896318_PAX6 mutation, particularly predicted haploinsufficiency, may be associated with extreme refractive error. 17898260_Observational study of gene-disease association. (HuGE Navigator) 17898260_PAX6 and SOX2 are obvious candidates in RE genetic studies because of their biological roles and prior linkage studies. The present findings strongly suggest refractive error is not directly affected in this population by variants in either gene. 17901057_Pax-6 and c-Maf interact with G1 to activate basal expression of the glucagon gene 17908181_Pax6 is strongly expressed in the embryonic and postnatal olfactory systems, and regulates neuronal specification, migration and differentiation. 17947347_Strongly expressed in dorsal and ventral proliferative zones, mainly in proliferating radial glia (RG) cells, neuronal and intermediate progenitors, and sporadic deep cortical plate neurons; has a critical role in neurogenic regulation of RG cells. 17948041_Observational study of gene-disease association. (HuGE Navigator) 18070176_methylated PAX6- or TPEF-promoters could represent biomarkers for bladder cancer. 18241071_67 of 71 aniridia cases (94%) undergoing full mutation analysis had a mutation in the PAX6 genomic region. 18243151_These four cases provide evidence for genetic heterogeneity in aniridia. In aniridic patients without a PAX6 mutation, vision seems to be relatively well preserved. 18273794_The tumor suppressor role of PAX6 in human gliomas is not due to mutation in its coding and regulating regions. 18273794_the tumor suppressor role of PAX6, reported in previous studies on gliomas, is not due to mutation in its coding and regulating regions, suggesting the involvement of epigenetic mechanisms in the silencing of PAX6 in these tumors 18332330_The anterior and posterior segment anomalies suggested that the nystagmus was secondary to a panocular disorder with PAX6 as a candidate causative gene. 18334930_A novel de novo frameshift mutation in PAX6, which presumably occurred in the paternal gamete, was found in a family with autosomal dominant aniridia. 18393239_The genetic analysis suggests that the novel mutation in the PAX6 gene may be the cause of the classical aniridia phenotype. 18421978_Pax6 has multiple functions in the developing central nervous system and in postnatal neurogenesis. 18440259_PAX6 sequence changes in both families segregated with the anterior segment phenotype and were not observed in controls. Both mutations occur in the paired domain of the PAX6 gene. 18467663_Pax6 is a multifunctional player regulating proliferation and differentiation through the control of expression of different downstream molecules in a highly context-dependent manner [review] 18593849_concomitant expression of Pax6 and Pdx1 is important for glucose-dependent insulinotropic polypeptide expression 18595732_Pax6 increases neurogenesis from human fetal striatal neural stem cells; these cells retain a specific neuronal fate consistent with their region of origin. 18616618_The exclusion of these genes as likely candidates supports the hypothesis that the ocular phenotype associated with peters' anomaly segregating in this family is a distinct, new, autosomal dominant entity in the anterior segment dysgenesis spectrum. 18754095_Alterations of gastric PAX6 expression in Helicobacter pylori infection, incisural antralisation, and intestinal metaplasia. 18766996_PAX6 haploinsufficiency, the major cause of classic hereditary aniridia worldwide, is also associated with the phenotype in two different families from the Arabian Peninsula. 18776953_This study adds four novel mutations to the worldwide PAX6 mutational spectrum. 18973570_These data support recent reports that EMX2 but not PAX6 is more directly involved in arealization, highlighting that analysis of human development allows better spatio-temporal resolution than studies in rodents. 19034419_Function for PAX6 in the regulation of proinsulin processing and glucose metabolism via modulation of PC1/3 production. 19124844_Observational study of gene-disease association. (HuGE Navigator) 19124844_This study demonstrates the association of PAX6 variants with susceptibility to high myopia. The PAX6 locus may contain polymorphisms playing a role in high myopia in southern Han Chinese. 19142206_Observational study of gene-disease association. (HuGE Navigator) 19218613_Observational study of gene-disease association. (HuGE Navigator) 19218613_PAX6 mutations cause panocular malformations that vary considerably in pattern and severity. 19414065_In the human fetal eye, PAX6 was primarily localized to nuclei of the neural retina. Exprssion peaks at fetal days 51-60. 19607881_Observational study of gene-disease association. (HuGE Navigator) 19650334_Temporal and spatial distribution of PAX6 gene expression in the embryonic eye. 19651775_a model for a neoplastic pathway where expression of PAX6 from development activates the MET receptor 19686589_This study demonstrated that the quantitative changes in Pax6 protein expression during the preneurogenic to neurogenic transition and during the neurogenic cell cycle. 19790232_PAX6 may act as a prostate cancer repressor by interacting with androgen receptor (AR) and repressing the transcriptional activity and target gene expression of AR to regulate cell growth and regeneration. 19793656_This study supported the notion that assessing copy number variation of the PAX6 gene itself and also of flanking regions, may contribute to the molecular diagnosis of aniridia 19806578_The nonsense mutation of the PAX6 gene were associated with congenital aniridia and cataract. 19806579_Nonsense mutation Arg240Ter of the PAX6 is the genetic basis of the Chinese family with congenital aniridia. 19862335_Two novel PAX6 mutations in families with severe aniridia, were identified. 19898691_This study is aimed at exploring the role of PAX6 mutations in Taiwanese patients with congenital eye anomalies. 19907666_Observational study of gene-disease association. (HuGE Navigator) 19907666_These results revealed an association between high myopia and AC and AG dinucleotide repeat lengths in the PAX6 P1 promoter. 19917615_in the presence of Pax6(S), calcium channel beta(3) subunit is translocated from the cytoplasm to the nucleus 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20054790_Case report.Disturbances of brain development in two sib fetuses. Molecular analysis in both parents has shown different mutations in PAX6 gene and a compound heterozygosity for two PAX6 mutations in both fetuses. 20074565_Data report that either Pax6(+5a) or Pax6(-5a) was sufficient to promote, whereas their knockdown reduced the expression of delta-catenin (CTNND2), a neural specific member of the armadillo/beta-catenin superfamily. 20360993_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20396773_Data show that Pax6 expression in retina during active neurogenesis is stable and higher than in neocortex, and in primary cultures, Pax6 expression is retained and repeats that in native tissues. 20413449_Overexpression of transgenic Pax6 increases cell cycle length of apical progenitor cells and drives the system toward neurogenesis. 20523026_Data demonstrate by in situ hybridization and immunohistochemistry that the two homeobox genes Pax6 and MEIS2 are expressed during early fetal brain development in humans. 20577777_Data show that Pax6 and also other Pax family proteins display a strikingly low nuclear mobility compared to other transcriptional regulators. 20592023_Pax6 is critical for alpha cell function and differentiation through the transcriptional control of key genes involved in glucagon gene transcription, proglucagon processing, and alpha cell differentiation 20621053_Findings indicate that Pax6 is a transcriptional determinant of the human neuroectoderm and suggest that Pax6a and Pax6b coordinate with each other in determining the transition from pluripotency to the NE fate in human. 20664694_A novel deletion mutation in the PAX6 gene was identified in a Chinese family with aniridia and congenital cataract. 20713004_The molecular features of the DNA search mechanism were explored for two multidomain transcription factors: human Pax6 and Oct-1. 20801516_Observational study of genetic testing. (HuGE Navigator) 20806047_Mutations in several transcription factors associated with aniridia and congenital cataract, FOXE3, (PAX6), PITX2, and PITX3 genes, were examined. 21092858_Pax6 utilizes different DNA-binding domains to regulate survival of dopaminergic olfactory bulb neurons. 21169528_Silencing the Pax6 gene with short interfering RNA resulted in an inhibited growth and an increased apoptosis of cultured human retinoblastoma cells. 21271670_De novo duplication 11p13 involving the PAX6 gene in a patient with neonatal seizures, hypotonia, microcephaly, developmental disability and minor ocular manifestations. 21321669_analysis of a 556 kb deletion in the downstream region of the PAX6 gene that causes familial aniridia and other eye anomalies in a Chinese family 21348901_The present study did not find evidence of PAX6 polymorphisms being associated with supernumerary teeth in the population studied. 21353197_This high frequency of causal submicroscopic chromosomal aberrations in patients with congenital ocular malformation warrants implementation of array comparative genomic hybridization in the diagnostic work-up of these patients. 21397818_We report a child with aniridia and Peters anomaly associated with a PAX6 gene mutation. 21421876_In this study, a functional SNP was identified at the 3'UTR that influences the risk for extreme myopia. 21524647_PAX6-G36A and PAX6-G51A mutants were found to have higher ability to transactivate whereas PAX6(5a)-G36A and PAX6(5a)-G51A have lower transactivation potential compared to their respective wild type forms. 21589860_PAX6 haplotypes are associated with susceptibility to the development of high myopia in Chinese. The PAX6 locus plays a role in high myopia 21617155_Genome-wide identification of novel PAX6 targets, using Hidden Markov Modelling with published targets. Many predictions were empirically validated using zebrafish ChIP, and gene expression changes in knock-downs and in some mouse mutants. 21633710_study to identify the causative PAX6 mutation in a family with autosomal dominant aniridia; a novel PAX6 mutation in the donor splice site of intron 12 was identified in all three affected individuals from the family 21655361_3 non-pathogenic mutations have been identified in PAX6 gene analysis in irido-fundal coloboma in the North Indian patient cohort. 21691140_We examined the exon sequence of the PAX6 gene in a Chinese family and an unrelated individual with aniridia. The predicted outcome of both mutations is premature termination 21710692_Promoter hypermethylation of PAX6, BRCA2, PAX5, WT1, CDH13 and MSH6 seems to be a frequent early event in breast cancer 21816254_Ocular disease segregated with a novel PAX6 Q178X nonsense mutation with Western blot analysis suggesting that this led to haploinsufficiency of PAX6 protein. 21850189_The PAX6 mutation spectrum in Chinese aniridia patients is comparable to that reported in other ethnic groups. 21904390_Our findings further confirmed that different kind of mutations might cause different ocular phenotype, and clearly clinical phenotype classification might increase the mutation detection rate of the PAX6gene. 21922321_genetic association studies in Finnish population: An SNP (rs685428) upstream of PAX6 is associated with type 2 diabetes mellitus, insulin resistance, and reduced PAX6 and proprotein convertase 1 (PCSK1) expression in islets. [includes meta-analysis] 21935435_The results suggest that Pax6 represses AR activity by displacing and/or inhibiting recruitment of coactivators to AR target promoters. 21944253_after Pax6 was knocked down,tumor cells were arrested at G0/G1 phase. 21948554_overexpression of PAX6 in human retinoblastoma cells was associated with increased cell proliferation parallel to a reduced caspase-3-dependent apoptotic rate and a change in the p53 regulated cell cycle. 21985185_A small subset of aniridia cases is caused by rearrangements of PAX6 neighboring regions, and the so-called 'position effect' is considered to be the underlying pathogenic mechanism. 21997878_PARP-1 regulates PAX6 transcription during human embryonic stem cell neural differentiation. 22025896_A novel missense mutation in PAX6 (amino acid substitution of proline to glutamine at position 118 (p.P118Q) was identified in all affected individuals but not in asymptomatic members and 110 normal controls. 22103961_we report the absence of mutations in all studied genes in four families with phenotypes associating cataract, mental retardation and microcephaly. 22146551_Mutational screening and genotyping could not conclusively clarify a role for PAX6 in familial peripheral keratopathy phenotype, suggesting that it is a distinct clinical and genetic disease entity, not a variant of aniridia. 22171157_The identification of two new PAX6 gene mutations in Chinese aniridia patients, is reported. 22171686_This study expands the phenotypic spectrum of PAX6 mutation and enriched our knowledge of the genetic cause for microphthalmia and late-onset keratitis. 22361317_This study adds 2 novel PAX6 mutations to those previously reported, providing further evidence for genetic and phenotypic heterogeneity in aniridic ocular malformation. 22392277_data show structure-function relationship of wild-type PAX6 and its alternatively spliced isoform PAX6(5a); it indicates the mutation induced changes are not only confined to the place of mutation but are dispersed due to conformational changes induced by missense mutations 22393272_A recurrent PAX6 c.307C-->T mutation is associated with the variable phenotypes among Chinese family patients with aniridia and congenital progressive cataract. 22393275_Fourteen different kinds of mutations in the PAX6 gene were detected in 16 of 18 unrelated Korean families with congenital aniridia. 22447870_Down-regulation of PAX6 in RPE increased RPE proliferation, but reduced scleral cell proliferation. 22509105_A splice site mutation in the PAX6 gene resulting in exon skipping was found in a German family with autosomal dominant aniridia. 22550392_A novel heterozygous PAX6 deletion c.1251_1353del103 (p.Pro418Serfs*87) affecting exon 14 and the 3'-untranslated-region was identified in the congenital aniridia Chinese family. 22561546_Pax6 functions at multiple levels to integrate extracellular information and execute cell-intrinsic differentiation programs that culminate in the specification and differentiation of a distinct ocular lineage. 22583899_These data provide the first evidence of direct transactivation of the FABP7 proximal promoter by PAX6 and suggest a synergistic mechanism for PAX6 and other co-factor(s) in regulating FABP7 expression in malignant glioma. 22621390_We identified a novel PAX6 mutation in a family with severe ocular malformation. 22809227_Our results confirm that the PAX6, Lumican, and MYOC genes were not associated with high myopia in the Han Chinese in Northeastern China. 22815628_A nonsense mutation in PAX6 exon 9 (c.718C>T) was identified in the Northeastern Chinese patients with sporadic aniridia. 22860217_Controlling inhibition of bone morphogenetic protein modulates the number of SOX1 expressing cells, whereas PAX6, another neural precursor marker, remains the same. 22893676_We identified two novel and a known mutation of PAX6 in four probands with microcornea. 23044950_Herein, we report the first case series to describe the foveal characteristics on SD-OCT imaging of 5 affected individuals in a single family affected with autosomal recessive isolated foveal hypoplasia with an absent PAX6 mutation. 23146210_The first complete pituitary function assessment, together with glucose tolerance evaluations, in five related patients with a PAX6 mutation. 23211052_Results indicate positive expression of SOX2, SOX3, PAX6, OCT3/4, and NANOG in the CD105+ and CD105(-) cell subpopulations. 23213273_A significant association of PAX6 with high and extreme myopia has been found in Japanese patients. The A allele of rs644242 is a protective allele. 23566044_This is the first report of this particular constellation of ocular signs occurring in association with a PAX6 mutation. There was no association with anthropometric features, but affected subjects had worse glycaemia than controls 23734086_This study is to summarize the phenotypes and identify the underlying genetic cause of the paired box 6 (PAX6) gene responsible for aniridia in two three-generation Chinese families in northern China. 23931477_We described 3 cases with unusual ophthalmic manifestations of congenital aniridia. Three cases were negative for systemic involvement without any PAX6 mutation. 23942204_a novel PAX6 mutation P76R is associated with infantile nystagmus and presenile cataract in a large family. 23970099_In conclusion, the present study supports miR-223 and PAX6 as novel therapeutic targets for glioblastoma. 23990468_Pax6 binds endogenously to the proximal region of the tartrate acid phosphatase (TRAP) gene promoter and suppresses nuclear factor of activated T cells c1 (NFATc1)-induced TRAP gene expression. 24078574_A novel deletion mutation (c.957-958delCA) in exon 13 of the PAX6 gene was identified in a family with congenital aniridia. 24143217_functional role of Pax6 loss in SQM pathogenesis 24185687_Levels of PAX6 were increased, while the expression of miR-7 was reduced with malignancy of colorectal cancer. PAX6 was identified as a target of miR-7, and its expression was negatively regulated by miR-7 in human colorectal cancer cells. 24281366_Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. 24290376_Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. 24349436_The 681 kb large deletion of chromosome 11p13 downstream of PAX6 is the genetic cause of the familial ocular coloboma in this large Chinese family 24357251_the unique genotype identified in this study suggested that haploinsufficiencies of PAX6 or PRRG4 included in this region are candidate genes for severe developmental delay and autistic features characteristic of WAGR syndrome. 24390526_[review] A slightly impaired pituitary function is identified in an aniridia patient with a heterozygous PAX6 mutation. 24454925_PAX6 mRNA levels are significantly elevated in primary lung cancer tissues compared to their matched adjacent tissues. 24505629_frequent PIK3CA amplification and promoter methylation of RASSF1A and PAX6 genes in gastric cancers 24562376_PAX6 (5a) transcription factor expression, together with medium supplemented with fibronectin, is able to induce the differentiation of human adipose-tissue-derived stem cell into retinal progenitors. 24623969_transcriptional control of Trpm3/miR-204, is reported. 24637479_Meta-analysis of existing data revealed a suggestive association of PAX6 rs644242 with extreme and high myopia, which awaits validation in further studies. 24737507_this study is the first to report a PAX6 mutation in a Malaysian family. This mutation is the cause of the aniridia spectra observed in this family and of congenital cataract. 24787241_we identified a novel de novo duplication mutation of PAX6 in the aniridia and other ocular abnormalities family. This mutation has occurred de novo on a paternal chromosome by direct duplication. 24802670_PAX6 activates miR-135b to inhibit TGF-b and BMP signaling thereby differentiating hESC toward a neural phenotype. 24939714_In conclusion, our data demonstrate that the suppression of PAX6 increases proliferation and decreases apoptosis in human retinoblastoma cells by regulating several cell cycle and apoptosis biomarkers. 25029272_Dkk3/REIC3 expression is regulated by PAX6 in several human cell lines. 25030175_findings suggest a central role of the WNT7A-PAX6 axis in corneal epithelial cell fate determination, and point to a new strategy for treating corneal surface diseases 25051057_Trans-placental BPA exposure down-regulated gene expression of Sox2 and Pax6 potentially underlying the adverse effect on childhood neuronal development. [Meta-analysis] 25189681_Report of a novel duplication in the PAX6 gene capable of causing the classic aniridia phenotype. 25313118_This study contributes patient data on the prevalence and significance of ocular pathology in aniridia with PAX6 mutations. 25342853_Heterozygous PAX6 mutations are associated with congenital hypopituitarism. 25366758_novel mutation in PAX6 is likely responsible for the pathogenesis of the congenital aniridia and nystagmus in 3 affected family members. 25433656_PAX6 and PAX8 positivity was seen in of metastatic pancreatic neuroendocrine tumors to the liver 25542770_SOX2, OTX2 and PAX6 analysis in subjects with anophthalmia and microphthalmia 25555363_This is the first report demonstrating abnormal cone-driven electroretinography responses associated with this particular mutation of the PAX6 gene. 25578969_demonstrate that Sox-Pax partnerships have the potential to substantially alter DNA target specificities and likely enable the pleiotropic and context-specific action of these cell-lineage specifiers 25678763_11 novel PAX6 mutations causing aniridia complicated with other ocular abnormalities have been identified in Southern Indian patients. 25687215_Molecular analysis identified two compound heterozygous TYR mutations known to cause OCAIA and cosegregate with oculocutaneous albinism. In addition, we identified a novel heterozygous PAX6 mutation confirming the atypical aniridia phenotype. 25696017_the case of a Korean family with novel splice site mutation in the PAX6 gene in isolated aniridia inherited in an autosomal dominant manner, is reported. 25746674_A novel deletion mutation in the PAX6 gene resulting in an abnormal PAX6 COOH-terminal extension in the Chinese family with aniridia. 25804118_SOX4, SOX11, and PAX6 were significant for tumor type. 26045558_loss of PAX6 converts LSCs to epidermal stem cells, as demonstrated by a switch in the keratin gene expression profile and by the appearance of congenital dermoid tissue 26130484_congenital primary aphakia is associated with PAX6 mutation. 26295830_In post-mortem substantia nigra from Parkinson's disease patients, a reduced number of PAX6 expressing cells were found. 26345820_ChIP experiments confirmed that Pdx1 activates the expression of the downstream transcription factors, Ngn3 and Pax6, by combined with the promoter regions of insulin (Insulin-P), Ngn3 (Ngn3-P), and Pax6 (Pax6-P). 26394807_Variants in TRIM44 Cause Aniridia by Impairing PAX6 Expression 26439359_findings suggest that PAX6+/- is associated with smaller pineal size, lower melatonin secretion and greater parental report of sleep disturbances in children; further studies are needed to explore the potential use of melatonin replacement for improving sleep quality in patients with PAX6+/- 26440771_We analyzed the PAX6 gene in a Bosma arrhinia microphthalmia syndrome patient but found no variation or mutation that could constitute or establish a causal association in our patient 26535646_we reported the results of the clinical and molecular evaluation of a threegeneration Chinese family with aniridia and identified a rare heterozygous M1K mutation in PAX6. 26617874_We found that PAX6 gene was specifically methylated in non small cell lung cancer 26661695_Data indicate paired box gene 6 (aniridia, keratitis) protein (PAX6) haploinsufficiency as causal for aniridia. 26849621_It is important to establish the molecular diagnosis early to avoid repeated and long-term screening for Wilms tumor. Our work further emphasizes that a wide range of ocular phenotypes are associated with loss of function PAX6 mutations. 26879676_We report the first association study of the transcription factor PAX6 with HSCR and that its low expression levels may result in an aberrant neurogenesis, which is directly related with manifestation of HSCR phenotype. 26899008_we show that the two PAX6 isoforms differentially and cooperatively regulate the expression of genes specific to the structure and functions of the corneal epithelium, particularly keratin 3 (KRT3) and keratin 12 (KRT12). PAX6 isoform-a induced KRT3 expression by targeting its upstream region. KLF4 enhanced this induction. A combination of PAX6 isoform-b, KLF4, and OCT4 induced KRT12 expression 27110298_Data suggest that promoter hypermethylation of PAX6 is a common event in hepatocellular carcinoma and the association of PAX6 methylation in clinicopathological features is divergent with different viral status. 27124303_Fourteen of these mutations presented in the known aniridia genes; PAX6, FOXC1 and PITX2. 27229137_photoinduced excess electron transfer assay can be used for analysing cooperativity of proteins in transcription complex using cooperative binding of Pax6 to Sox2 on the regulatory DNA element (DC5 enhancer) as an example. 27431685_the present study identified a heterozygous deletion and a run-on mutation in PAX6 in two families with autosomal dominant aniridia. 27470361_restoration of Notch1 and PAX6 expression partially rescued the inhibition of cell proliferation and metastasis induced by miR-433 overexpression. 27583466_epigenetic factor CTCF-mediated chromatin remodeling regulates interactions between eye-specific PAX6 and those genes | ENSMUSG00000027168 | Pax6 | 318.21186 | 0.7970855 | -0.3271935860 | 0.20189806 | 2.741720e+00 | 9.775949e-02 | 9.998360e-01 | No | Yes | 342.87565 | 50.914885 | 4.078424e+02 | 46.769544 | |
| ENSG00000008130 | 65220 | NADK | protein_coding | O95544 | 3D-structure;ATP-binding;Alternative splicing;Kinase;Metal-binding;NAD;NADP;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase | NADK catalyzes the transfer of a phosphate group from ATP to NAD to generate NADP, which in its reduced form acts as an electron donor for biosynthetic reactions (Lerner et al., 2001 [PubMed 11594753]).[supplied by OMIM, Mar 2008]. | hsa:65220; | cytosol [GO:0005829]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; NAD+ kinase activity [GO:0003951]; ATP metabolic process [GO:0046034]; NAD metabolic process [GO:0019674]; NADP biosynthetic process [GO:0006741]; phosphorylation [GO:0016310] | 17855339_NAD kinase levels control the NADPH concentration in human cells 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21526340_Using purified human NAD kinase, we revealed a sigmoidal kinetic behavior toward ATP and the inhibitory effects of NADPH and NADH 26577408_Taken together, this indicates that Ser-188 (and perhaps the other residues) is an important phosphorylation site that can downregulate the NAD kinase activity of this critical enzyme. 26806015_Depletion of wild-type NADK in pancreatic ductal adenocarcinoma cells attenuates cancer cell growth. 28954733_Data indicate metabolic enzymes NAD kinase and ketohexokinase as candidate metabolic gene targets, and the chromatin remodeling protein INO80C as a tumor suppressor in KRAS(MUT) colorectal tumor xenograft. 30846598_These data indicate that Akt-mediated phosphorylation of NADK stimulates its activity to increase NADP(+) production through relief of an autoinhibitory function inherent to its amino terminus. 34133937_NADK is activated by oncogenic signaling to sustain pancreatic ductal adenocarcinoma. | ENSMUSG00000029063 | Nadk | 4844.65751 | 1.0333494 | 0.0473281888 | 0.09300632 | 2.596093e-01 | 6.103887e-01 | 9.998360e-01 | No | Yes | 4708.69483 | 343.231034 | 4.339185e+03 | 245.324436 | ||
| ENSG00000011105 | 10867 | TSPAN9 | protein_coding | O75954 | Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. Alternatively spliced transcripts encoding the same protein have been identified. [provided by RefSeq, Nov 2009]. | hsa:10867; | focal adhesion [GO:0005925]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; tetraspanin-enriched microdomain [GO:0097197] | 18795891_Findings suggest a role for Tspan9 in regulating platelet function in concert with other platelet tetraspanins and their associated proteins. 18799160_tetraspanin CD9 has a role in determining vascular smooth muscle cell injury phenotypes 19204726_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 26865714_TSPAN9 is an endosomal tetraspanin whose depletion has a pronounced effect on the entry of several viruses that fuse in the early endosome. 27177197_The proliferation, migration and invasion of human gastric cancer SGC7901 cells were significantly inhibited by overexpression of Tspan9. 31242895_expression of the membrane protein TSPAN9 regulates gastric cancer cell migration and invasion by inhibiting the FAK-RAS-ERK1/2 signaling pathway. The extracellular secretory protein EMILIN1 can increase this tumor suppressive effect by promoting the expression of TSPAN9. | ENSMUSG00000030352 | Tspan9 | 1503.59872 | 1.0806611 | 0.1119141018 | 0.12046011 | 8.830126e-01 | 3.473779e-01 | 9.998360e-01 | No | Yes | 1386.69577 | 152.819454 | 1.203151e+03 | 103.028080 | ||
| ENSG00000011426 | 54443 | ANLN | protein_coding | Q9NQW6 | FUNCTION: Required for cytokinesis (PubMed:16040610). Essential for the structural integrity of the cleavage furrow and for completion of cleavage furrow ingression. Plays a role in bleb assembly during metaphase and anaphase of mitosis (PubMed:23870127). May play a significant role in podocyte cell migration (PubMed:24676636). {ECO:0000269|PubMed:10931866, ECO:0000269|PubMed:12479805, ECO:0000269|PubMed:15496454, ECO:0000269|PubMed:16040610, ECO:0000269|PubMed:16357138, ECO:0000269|PubMed:23870127, ECO:0000269|PubMed:24676636}. | 3D-structure;Acetylation;Actin-binding;Alternative splicing;Cell cycle;Cell division;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Isopeptide bond;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes an actin-binding protein that plays a role in cell growth and migration, and in cytokinesis. The encoded protein is thought to regulate actin cytoskeletal dynamics in podocytes, components of the glomerulus. Mutations in this gene are associated with focal segmental glomerulosclerosis 8. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Oct 2014]. | hsa:54443; | actin cytoskeleton [GO:0015629]; actomyosin contractile ring [GO:0005826]; bleb [GO:0032059]; cell cortex [GO:0005938]; cytosol [GO:0005829]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; actin binding [GO:0003779]; cadherin binding [GO:0045296]; actomyosin contractile ring assembly [GO:0000915]; glomerular visceral epithelial cell migration [GO:0090521]; hematopoietic progenitor cell differentiation [GO:0002244]; mitotic cytokinesis [GO:0000281]; positive regulation of bleb assembly [GO:1904172]; regulation of exit from mitosis [GO:0007096]; septin ring assembly [GO:0000921]; septin ring organization [GO:0031106] | 15496454_anillin has a role in spatially regulating the contractile activity of myosin II during cytokinesis 16040610_Anillin is a substrate of APC/C that controls spatial contractility of myosin during late cytokinesis. 16203764_anillin is overexpressed in diverse common human tumors, but not simply because of its role in cell proliferation 16357138_Over-expression of ANLN is associated with pulmonary carcinogenesis 18158243_Human anillin contains a conserved C-terminal domain that is essential for its function and localization. 19995712_This study was to establish if ANLN was a molecular marker for pancreatic cancer. 21109967_Cytoplasmic anillin expression is a marker of favourable prognosis in renal cell carcinoma patients. 21849473_CIT-K is capable of physically and functionally interacting with the actin-binding protein anillin. 22514687_Data supports an analogous function for the anillin-Ect2 complex in human cells and one hypothesis is that this complex has functionally replaced the Drosophila anillin-RacGAP50C complex. 23547718_Antibody screening revealed 3 candidate prognostic markers in breast cancer: the Anillin (ANLN); PDZ-Binding Kinase (PBK); and, PDZ-Domain Containing 1 (PDZK1). 24088567_Supervillin concentrates activated and total myosin II at the furrow, and simultaneous knockdown of supervillin and anillin additively increases cell division failure. 24676636_Identified a missense mutation R431C in anillin (ANLN), an F-actin binding cell cycle gene, as a cause of FSGS. We screened 250 additional families with FSGS and found another variant, G618C, that segregates with disease in a second family with FSGS. 24809965_ANLN is positively associated with Wnt/beta-catenin signaling in gastric cancer.Elevated expression of ANLN is a predictor of intestinal type gastric cancer. ER-alpha is negatively associated with the expression of ANLN in gastric cancer. 24994938_The sequestration of anillin by astral microtubules might alter the organization of cortical proteins to polarize cells for cytokinesis 25223638_Data suggest that anillin (ANLN) could be involved in breast cancer progression and a potential target candidate in breast cancer. 25359885_these data demonstrate that a complex of p190RhoGAP-A and anillin modulates RhoA-GTP levels in the cytokinetic furrow to ensure progression of cytokinesis. 25809162_Anillin regulates intercellular adhesion in model human epithelia by mechanisms involving the suppression of JNK activity and controlling the assembly of the perijunctional cytoskeleton. 26135313_High nuclear expression of anillin (ANLN) is an independent predictor of poor disease-specific survival and it is a useful prognostic marker of urothelial carcinoma of the upper urinary tract (UCUT). 26319989_Identification 4 novel and 18 known exonic ANLN variants associated with carotid intima-media thickness at bifurcation. 26486082_Findings indicate that miR-497 is a potent tumor suppressor that inhibits cancer phenotypes by targeting ANLN and HSPA4L in NPC. 27062703_ANLN expression is elevated in colorectal cancer and has a strong potential to act as a biomarker for the prognosis of colorectal cancer. 27863473_Anillin (ANLN) expression in tumor cells is correlated to poor prognosis in breast cancer patients, independent of Ki-67 or tumor size. 28081137_S635 phosphorylation is essential for cytokinesis. 28600503_Bladder urothelial carcinoma patients with elevated ANLN expression had poorer survival prospects. 28931593_Authors found that decreasing Ran-GTP levels or tethering active Ran to the equatorial membrane affects anillin's localization and causes cytokinesis phenotypes. 29274368_Knockdown of ANLN in liver cells blocks cytokinesis and inhibits development of liver tumors. 30530503_High ANLN expression is associated with glioma. 30548429_Missense mutation in ANLN was identified as a cause of branchio-otic syndrome in a specific Chinese family. 31268619_Study suggests that the expression of ANLN in lung adenocarcinoma is associated with metastasis of cancer cells. ANLN may be involved in the metastasis of lung adenocarcinoma by promoting epithelial mesenchymal transformation of tumor cells. 31578580_concomitantly high expression of ANLN and KDR is associated with breast cancer. 31853940_Visualizing dynamic actin cross-linking processes driven by the actin-binding protein anillin. 31910867_Anillin regulates breast cancer cell migration, growth, and metastasis by non-canonical mechanisms involving control of cell stemness and differentiation. 32198770_LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. 32413362_High mobility group AT-hook 2 promotes tumorigenicity of pancreatic cancer cells via upregulating ANLN. 32880660_Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation. 33089886_Actin-binding protein Anillin promotes the progression of gastric cancer in vitro and in mice. 33862101_Molecular basis of functional exchangeability between ezrin and other actin-membrane associated proteins during cytokinesis. 34082790_ANLN promotes carcinogenesis in oral cancer by regulating the PI3K/mTOR signaling pathway. 34321459_Anillin propels myosin-independent constriction of actin rings. 34344861_Alternatively spliced ANLN isoforms synergistically contribute to the progression of head and neck squamous cell carcinoma. 34777569_ANLN Regulated by miR-30a-5p Mediates Malignant Progression of Lung Adenocarcinoma. 35340220_Comprehensive Analysis of ANLN in Human Tumors: A Prognostic Biomarker Associated with Cancer Immunity. | ENSMUSG00000036777 | Anln | 686.64038 | 0.8837642 | -0.1782666749 | 0.14464496 | 1.494558e+00 | 2.215105e-01 | 9.998360e-01 | No | Yes | 696.26146 | 119.339027 | 7.681640e+02 | 101.430450 | |
| ENSG00000013375 | 5238 | PGM3 | protein_coding | O95394 | FUNCTION: Catalyzes the conversion of GlcNAc-6-P into GlcNAc-1-P during the synthesis of uridine diphosphate/UDP-GlcNAc, a sugar nucleotide critical to multiple glycosylation pathways including protein N- and O-glycosylation. {ECO:0000303|PubMed:24589341, ECO:0000303|PubMed:24698316, ECO:0000303|PubMed:24931394}. | Acetylation;Alternative splicing;Carbohydrate metabolism;Disease variant;Isomerase;Magnesium;Metal-binding;Phosphoprotein;Reference proteome | PATHWAY: Nucleotide-sugar biosynthesis; UDP-N-acetyl-alpha-D-glucosamine biosynthesis; N-acetyl-alpha-D-glucosamine 1-phosphate from alpha-D-glucosamine 6-phosphate (route I): step 2/2. {ECO:0000303|PubMed:24589341, ECO:0000303|PubMed:24698316, ECO:0000303|PubMed:24931394}. | This gene encodes a member of the phosphohexose mutase family. The encoded protein mediates both glycogen formation and utilization by catalyzing the interconversion of glucose-1-phosphate and glucose-6-phosphate. A non-synonymous single nucleotide polymorphism in this gene may play a role in resistance to diabetic nephropathy and neuropathy. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Dec 2010]. | hsa:5238; | cytosol [GO:0005829]; magnesium ion binding [GO:0000287]; phosphoacetylglucosamine mutase activity [GO:0004610]; phosphoglucomutase activity [GO:0004614]; carbohydrate metabolic process [GO:0005975]; glucosamine metabolic process [GO:0006041]; glucose 1-phosphate metabolic process [GO:0019255]; hemopoiesis [GO:0030097]; protein N-linked glycosylation [GO:0006487]; protein O-linked glycosylation [GO:0006493]; spermatogenesis [GO:0007283]; UDP-N-acetylglucosamine biosynthetic process [GO:0006048] | 12174217_PGM(3) is identical to AGM(1). 20221814_Observational study of genotype prevalence. (HuGE Navigator) 20221814_Polymorphic analysis of the human phosphoglucomutase-3 gene. 20800603_Observational study of gene-disease association. (HuGE Navigator) 24589341_Autosomal recessive hypomorphic PGM3 mutations underlie a disorder of severe atopy, immune deficiency, autoimmunity, intellectual disability, and hypomyelination. 24698316_Impairment of PGM3 function leads to a novel primary (inborn) error of development and immunity because biallelic hypomorphic mutations are associated with impaired glycosylation and a hyper-IgE-like phenotype. 24931394_define PGM3-CDG as a treatable immunodeficiency, document the power of whole-exome sequencing in gene discoveries for rare disorders, and illustrate the utility of genomic analyses in studying combined and variable phenotypes 26482871_Data indicate the effect of the phosphoglucomutase 3 (PGM3) mutation for four immunodeficient siblings in a Swedish family. 26687240_PGM3 mutation identified in a patient with hyper IgE syndrome results in lack of glycosylation at Asn264 and altered glycosylation profile. 28543917_Novel PGM3 Mutation Is Associated With a Severe Phenotype of Bone Marrow Failure, Severe Combined Immunodeficiency, Skeletal Dysplasia, and Congenital Malformations. 28704707_study reports the first founder mutation in PGM3 gene (p.Glu340del) in twelve Tunisian PGM3 deficient patients belonging to three consanguineous families originating from a rural district in west central Tunisia 30578875_our findings demonstrate that defective glycosylation in PGM3-deficient patients results in reduced expression of unglycosylated gp130 protein and consequently, impaired gp130-dependent STAT3 phosphorylation. 31231132_Deficiency of phosphoglucomutase 3 (PGM3) is an autosomal recessive disorder of N- and O-glycosylation. 31707513_Compound Heterozygous PGM3 Mutations in a Thai Patient with a Specific Antibody Deficiency Requiring Monthly IVIG Infusions. 35011738_Targeting PGM3 as a Novel Therapeutic Strategy in KRAS/LKB1 Co-Mutant Lung Cancer. | ENSMUSG00000056131 | Pgm3 | 419.29178 | 1.0290845 | 0.0413615027 | 0.17977397 | 5.074172e-02 | 8.217776e-01 | 9.998360e-01 | No | Yes | 384.67843 | 71.480492 | 4.031313e+02 | 57.714489 |
| ENSG00000025423 | 8630 | HSD17B6 | protein_coding | O14756 | FUNCTION: NAD-dependent oxidoreductase with broad substrate specificity that shows both oxidative and reductive activity (in vitro). Has 17-beta-hydroxysteroid dehydrogenase activity towards various steroids (in vitro). Converts 5-alpha-androstan-3-alpha,17-beta-diol to androsterone and estradiol to estrone (in vitro). Has 3-alpha-hydroxysteroid dehydrogenase activity towards androsterone (in vitro). Has retinol dehydrogenase activity towards all-trans-retinol (in vitro). Can convert androsterone to epi-androsterone. Androsterone is first oxidized to 5-alpha-androstane-3,17-dione and then reduced to epi-andosterone. Can act on both C-19 and C-21 3-alpha-hydroxysteroids. {ECO:0000269|PubMed:10896656, ECO:0000269|PubMed:11360992, ECO:0000269|PubMed:11513953}. | Endoplasmic reticulum;Endosome;Glycoprotein;Lipid metabolism;Membrane;Microsome;NAD;Oxidoreductase;Reference proteome;Signal;Steroid metabolism | The protein encoded by this gene has both oxidoreductase and epimerase activities and is involved in androgen catabolism. The oxidoreductase activity can convert 3 alpha-adiol to dihydrotestosterone, while the epimerase activity can convert androsterone to epi-androsterone. Both reactions use NAD+ as the preferred cofactor. This gene is a member of the retinol dehydrogenase family. [provided by RefSeq, Aug 2013]. | hsa:8630; | early endosome membrane [GO:0031901]; endoplasmic reticulum [GO:0005783]; 17-beta-hydroxysteroid dehydrogenase (NAD+) activity [GO:0044594]; 17-beta-hydroxysteroid dehydrogenase (NADP+) activity [GO:0072582]; 5alpha-androstane-3beta,17beta-diol dehydrogenase activity [GO:0047024]; androstan-3-alpha,17-beta-diol dehydrogenase activity [GO:0047044]; androsterone dehydrogenase activity [GO:0047023]; catalytic activity [GO:0003824]; electron transfer activity [GO:0009055]; NAD-retinol dehydrogenase activity [GO:0004745]; oxidoreductase activity [GO:0016491]; testosterone 17-beta-dehydrogenase (NADP+) activity [GO:0047045]; testosterone dehydrogenase (NAD+) activity [GO:0047035]; androgen biosynthetic process [GO:0006702]; androgen catabolic process [GO:0006710]; brexanolone catabolic process [GO:0062175] | 17070195_Observational study of gene-disease association. (HuGE Navigator) 17070195_These data suggest that polymorphisms in the HSD17B6 gene are associated with PCOS and key clinical phenotypes of the disorder. 17289849_RoDH enzymes are expressed in tissues that have microsomal 3alpha-hydroxysteroid dehydrogenase/epimerase activities 19837928_Observational study of gene-disease association. (HuGE Navigator) 19837928_These replication data suggest a role for HSD17B6 in polycystic ovary syndrome 20200332_Observational study of gene-disease association. (HuGE Navigator) 21039282_Observational study of gene-disease association. (HuGE Navigator) 21039282_data suggest that there is no association of HSD17B6 and HSD17B5 variants with the occurrence of polycystic ovary syndrome in the Chinese population 22114194_17beta-hydroxysteroid dehydrogenase type 6 (17betaHSD6) converts the androgen DHT to the estrogen 3beta-Adiol, and this leads to activation of the ERbeta reporter. 24244276_the CYP11A1, CYP17A1, HSD3B2, SRD5A2, and HSD17B6 mRNA levels in metastases were significantly lower. 25422294_No significant difference was found in genotype or allele distributions of the polymorphisms rs12529 of HSD17B5 and rs898611 of HSD17B6 between patients with PCOS and controls. 32732174_Molecular genetic analysis of AKR1C2-4 and HSD17B6 genes in subjects 46,XY with hypospadias. | ENSMUSG00000025396 | Hsd17b6 | 119.58609 | 1.1859777 | 0.2460769091 | 0.27962615 | 7.699350e-01 | 3.802372e-01 | 9.998360e-01 | No | Yes | 122.86537 | 20.649902 | 1.158025e+02 | 15.320671 | |
| ENSG00000038210 | 55300 | PI4K2B | protein_coding | Q8TCG2 | FUNCTION: Together with PI4K2A and the type III PI4Ks (PIK4CA and PIK4CB) it contributes to the overall PI4-kinase activity of the cell. This contribution may be especially significant in plasma membrane, endosomal and Golgi compartments. The phosphorylation of phosphatidylinositol (PI) to PI4P is the first committed step in the generation of phosphatidylinositol 4,5-bisphosphate (PIP2), a precursor of the second messenger inositol 1,4,5-trisphosphate (InsP3). Contributes to the production of InsP3 in stimulated cells and is likely to be involved in the regulation of vesicular trafficking. {ECO:0000269|PubMed:11923287}. | 3D-structure;ATP-binding;Cytoplasm;Kinase;Lipid metabolism;Membrane;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase | This gene encodes a member of the type II PI4 kinase protein family. The encoded protein is primarily cytosolic and contributes to overall PI4-kinase activity along with other protein family members. This protein is involved in early T cell activation. [provided by RefSeq, Dec 2016]. | hsa:55300; | cytosol [GO:0005829]; endosome [GO:0005768]; membrane [GO:0016020]; plasma membrane [GO:0005886]; trans-Golgi network [GO:0005802]; 1-phosphatidylinositol 4-kinase activity [GO:0004430]; ATP binding [GO:0005524]; endosome organization [GO:0007032]; Golgi organization [GO:0007030]; phosphatidylinositol biosynthetic process [GO:0006661]; phosphatidylinositol phosphate biosynthetic process [GO:0046854] | 16337488_Type II PtdIns 4-kinase beta interacts with TCR-CD3 zeta chain. The C-terminal ITAM is critical for enzyme docking on the zeta chain. The association is tyrosyl phosphorylation dependent. Mutation of Y-151 & Y-142 disrupts interaction of the 2 proteins. 16949365_UV irradiation increases nuclear PtdIns5P levels via inhibition of the activity of the beta isoform of PtdIns5P 4-kinase (PIP4Kbeta), an enzyme that can phosphorylate and remove PtdIns5P. 18316730_LB-PI4K2B-1S-specific CD4(+) T cells contributed to the antitumor response by both directly eliminating malignant cells as effector cells and stimulating CD8(+) T cell immunity as helper cells. 19539307_Observational study of gene-disease association. (HuGE Navigator) 19539307_There is no evidence to suggest that PI4K2B is contributing to bipolar disorder in this family but a role for this gene in schizophrenia has not been excluded. 20583997_PIP4Kbeta interacts with and modulates nuclear localization of the PIP4Kalpha. 23619705_type II PtdIns 4-kinase beta is a key component in early T cell activation signaling cascades 24481753_Canonical tyrosine residues mutation in FcepsilonRIgamma ITAM (Y65 and Y76) reveals that these two tyrosine residues in gamma subunit are required for its interaction with type II PtdIns 4-kinases. 24972704_Results provide evidence that type II PtdIns 4-kinase b was indeed recruited to CD4- p56lck complex upon CD4 receptor cross linking, suggests a mechanism for its association and a role for it in CD4-mediated intracellular calcium release. 26143926_Here two crystal structures are presented: the structure of human PIK42A and the structure of PIK42B containing a nucleoside analogue. 27068535_RNA interference of the type II phosphatidylinositol 4-kinases PI4KIIa and PI4KIIa in primary human endothelial cells leads to formation of an increased proportion of short WPB with perturbed packing of VWF, as exemplified by increased exposure of antibody-binding sites. | ENSMUSG00000029186 | Pi4k2b | 186.04118 | 1.1475890 | 0.1986060516 | 0.24958271 | 6.344484e-01 | 4.257283e-01 | 9.998360e-01 | No | Yes | 208.40156 | 37.362010 | 1.864082e+02 | 25.897992 | |
| ENSG00000039319 | 9765 | ZFYVE16 | protein_coding | Q7Z3T8 | FUNCTION: May be involved in regulating membrane trafficking in the endosomal pathway. Overexpression induces endosome aggregation. Required to target TOM1 to endosomes. {ECO:0000269|PubMed:11546807, ECO:0000269|PubMed:14613930}. | 3D-structure;Alternative splicing;Cytoplasm;Endosome;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | This gene encodes an endosomal protein that belongs to the FYVE zinc finger family of proteins. The encoded protein is thought to regulate membrane trafficking in the endosome. This protein functions as a scaffold protein in the transforming growth factor-beta signaling pathway and is involved in positive and negative feedback regulation of the bone morphogenetic protein signaling pathway. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:9765; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; intracellular membrane-bounded organelle [GO:0043231]; 1-phosphatidylinositol binding [GO:0005545]; metal ion binding [GO:0046872]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; endosomal transport [GO:0016197]; protein targeting to lysosome [GO:0006622]; regulation of endocytosis [GO:0030100]; signal transduction [GO:0007165]; vesicle organization [GO:0016050] | 14613930_marked recruitment of TOM1 to endosomes was observed in cells overexpressing endofin or its carboxyl-terminal fragment 16775010_The key mode of action of Trx-SARA was to reduce the level of Smad2 and Smad3 in complex with Smad4 after TGF-beta1 stimulation. 17272273_Facilitates TGF-beta signaling as a scaffold protein to promote the R-Smad-Smad4 complex formation by bringing Smad4 to the proximity of the receptor complex. 17570516_our study is the first to identify and validate Endofin, DCBLD2, and KIAA0582 as part of a complex EGF phosphotyrosine signaling network 19887107_Disruption of proper localization to the endosomes and the Y515 phosphorylation of Endofin enhanced MAPK activation suggesting that Endofin negatively modulates EGFR signaling following receptor endocytosis. 26944198_Two SNPs located near the ZFYVE16 gene on chromosome 5 may have played a role in the early, multicompartment sacrocolpopexy failure. | 149.06121 | 1.0503947 | 0.0709315540 | 0.30648997 | 5.006646e-02 | 8.229477e-01 | 9.998360e-01 | No | Yes | 126.49897 | 24.061201 | 1.202133e+02 | 17.712440 | |||
| ENSG00000047578 | 23247 | KATNIP | protein_coding | O60303 | FUNCTION: May influence the stability of microtubules (MT), possibly through interaction with the MT-severing katanin complex. {ECO:0000269|PubMed:26714646}. | Cell projection;Ciliopathy;Coiled coil;Cytoplasm;Cytoskeleton;Joubert syndrome;Phosphoprotein;Reference proteome | This gene encodes a novel, evolutionarily conserved, ciliary protein. In human hTERT-RPE1 cells, the protein is found at the base of cilia, decorating the ciliary axoneme, and enriched at the ciliary tip. The protein binds to microtubules in vitro and regulates their stability when it is overexpressed. A null mutation in this gene has been associated with Joubert syndrome, a recessive disorder that is characterized by a distinctive mid-hindbrain and cerebellar malformation and is also often associated with wider ciliopathy symptoms. Consistently, in a serum-starvation ciliogenesis assay, human fibroblast cells derived from patients with the mutation display a reduced number of ciliated cells with abnormally long cilia. [provided by RefSeq, Feb 2016]. | hsa:23247; | cell projection [GO:0042995]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular space [GO:0005615]; cerebrospinal fluid circulation [GO:0090660] | 19727342_KIAA0056 (also known as mouse MGC31549) is a strong candidate gene for retinitis pigmentosa, RP22 (human 16p12.3-p12.1). Conclusion is based on a massive expression data set for mouse (103 strains in Genenetwork.org) and joint analysis of RetNet database. 26714646_KIAA0556 mutation identified in patients with Joubert syndrome. KIAA0556 binds to microtubules, p60/p80 katanins and genetically interacts with ARL13B. 27245168_Whole exome sequencing in a multiplex consanguineous family from India revealed a KIAA0556 homozygous single base pair deletion mutation (c.4420del; p.Met1474Cysfs*11). Knockdown of the gene in zebrafish resulted in a ciliopathy phenotype, rescued by co-injection of wildtype cDNA 30982090_homozygous truncating mutations in both ADGRG1/GPR56 and KIAA0556 are found in brothers displaying an overlap of polymicrogyria, hydrocephalus, and Joubert syndrome 31197031_Pathogenic germline variants of KIAA0556 associated with hypothalamic hamartoma were found. 32164589_Co-occurrence of mutations in KIF7 and KIAA0556 in Joubert syndrome with ocular coloboma, pituitary malformation and growth hormone deficiency: a case report and literature review. | ENSMUSG00000032743 | Katnip | 666.27829 | 0.9458791 | -0.0802723435 | 0.16350794 | 2.525811e-01 | 6.152635e-01 | 9.998360e-01 | No | Yes | 877.83263 | 100.642284 | 8.790799e+02 | 78.392980 | |
| ENSG00000048544 | 55173 | MRPS10 | protein_coding | P82664 | 3D-structure;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that belongs to the ribosomal protein S10P family. Pseudogenes corresponding to this gene are found on chromosomes 1q, 3p, and 9p. [provided by RefSeq, Jul 2008]. | hsa:55173; | mitochondrial inner membrane [GO:0005743]; mitochondrial small ribosomal subunit [GO:0005763]; mitochondrion [GO:0005739] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000034729 | Mrps10 | 1161.18396 | 1.0841164 | 0.1165197059 | 0.12576857 | 8.629772e-01 | 3.529069e-01 | 9.998360e-01 | No | Yes | 1221.78904 | 232.098726 | 1.240744e+03 | 181.708242 | ||
| ENSG00000049246 | 8863 | PER3 | protein_coding | P56645 | FUNCTION: Originally described as a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1, NR1D2, RORA, RORB and RORG, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. Has a redundant role with the other PER proteins PER1 and PER2 and is not essential for the circadian rhythms maintenance. In contrast, plays an important role in sleep-wake timing and sleep homeostasis probably through the transcriptional regulation of sleep homeostasis-related genes, without influencing circadian parameters. Can bind heme. {ECO:0000269|PubMed:17346965, ECO:0000269|PubMed:19716732, ECO:0000269|PubMed:24439663, ECO:0000269|PubMed:24577121, ECO:0000269|PubMed:26903630}. | Alternative splicing;Biological rhythms;Cytoplasm;Disease variant;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | This gene is a member of the Period family of genes and is expressed in a circadian pattern in the suprachiasmatic nucleus, the primary circadian pacemaker in the mammalian brain. Genes in this family encode components of the circadian rhythms of locomotor activity, metabolism, and behavior. This gene is upregulated by CLOCK/ARNTL heterodimers but then represses this upregulation in a feedback loop using PER/CRY heterodimers to interact with CLOCK/ARNTL. Polymorphisms in this gene have been linked to sleep disorders. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2014]. | hsa:8863; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; kinase binding [GO:0019900]; transcription cis-regulatory region binding [GO:0000976]; transcription corepressor binding [GO:0001222]; ubiquitin protein ligase binding [GO:0031625]; circadian regulation of gene expression [GO:0032922]; entrainment of circadian clock by photoperiod [GO:0043153]; negative regulation of transcription by RNA polymerase II [GO:0000122]; protein stabilization [GO:0050821]; regulation of circadian sleep/wake cycle, sleep [GO:0045187] | 12655319_Polymorphism of period 3 (647 Val/Gly) is implicated in tendency of diurnal preference in seasonal depression as revealed by self-reported morningness-eveningness scores, with higher scores found in individuals with at least one glycine allele. 14712925_Circadian oscillations of Per1, Per2, and Per3 mRNA expression were observed in serum-stimulated normal human fibroblasts. 14750904_PER3 was cloned & sequenced. Homology and conserved motifs were determined. Transcripts of hPer3 underwent circadian oscillation. hPER1 increased between 3 and 12 h from an apparent molecular mass of 188 to 204 kDa. 15475734_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15700718_Observational study of gene-disease association. (HuGE Navigator) 15790588_disturbances in PER gene expression may result in disruption of the control of the normal circadian clock, thus benefiting the survival of cancer cells and promoting carcinogenesis 16528748_Data do not provide statistically significant evidence for association of PER3 with bipolar disorders(BPAD), but are suggestive of their involvement in BPAD. 16528748_Observational study of gene-disease association. (HuGE Navigator) 16999817_Downregulated hPER3 expression in chronic myeloid leukemia is correlated with the inactivation of hPER3 by methylation 17309758_Observational study of gene-disease association. (HuGE Navigator) 17309758_an association between the PER3 coding-region variable number tandem repeat and diurnal preference;4-repeat allele was significantly more frequent in evening types, and the 5-repeat allele more frequent in morning types 17346965_Homozygosity for the longer allele PER3(5/5) had a considerable effect on sleep, including several markers of sleep homeostasis and activity during wakefulness and REM sleep were all increased in PER3(5/5) compared to PER3(4/4) individuals. 17346965_Observational study of gene-disease association. (HuGE Navigator) 17451453_Observational study of gene-disease association. (HuGE Navigator) 17451453_Our results suggest that the 54-nucleotide repeat polymorphism of hPer3 significantly associates with heroin dependence at the allele frequency level and may be a potential risk factor for the development of heroin dependence. 17512705_This study demonstrated that rare genetic variants of hper3 are significantly associated to a number of mood disorders features, such as age of onset, response to SSRIs treatment, circadian mood oscillations and characteristics of temperament. 17984998_Observational study of gene-disease association. (HuGE Navigator) 18444243_The expression level of PER3 was decreased in hepatocellular carcinoma. 18517031_Observational study of gene-disease association. (HuGE Navigator) 18517031_The correlation between sleep timing and PER3 expression was stronger in individuals homozygous for the variant of the PER3 polymorphism that is associated with morningness. 18714788_Observational study of gene-disease association. (HuGE Navigator) 18714788_This PER3 polymorphism differentially influences the effects of sleep deprivation on executive and non-executive function in the early morning. 18789374_A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. 18789374_Observational study of gene-disease association. (HuGE Navigator) 18835917_Observational study of gene-disease association. (HuGE Navigator) 18835917_PER3 polymorphism affects the sympathovagal balance in cardiac control in NREM sleep similar to the effect of sleep deprivation. 18990770_Observational study of gene-disease association. (HuGE Navigator) 19013183_There was no significant age-related phase difference in PER1 or PER2 rhythm with respect to sleep timing; however, PER3 expression pattern was altered in the older subjects. 19328558_Observational study of gene-disease association. (HuGE Navigator) 19360490_Observational study of gene-disease association. (HuGE Navigator) 19360490_PER3 VNTR genotype may have a role in age-related differences in self-reported diurnal preference 19444755_amplitude of the PER3 rhythm at baseline is significantly reduced with age but this did not affect the response of the PER3 rhythm to light. 19516903_Observational study of gene-disease association. (HuGE Navigator) 19516903_PER3 VNTR polymorphism is not associated with individual differences in neurobehavioral responses to partial sleep deprivation, although it was related to one marker of sleep homoeostatic response during PSD 19553435_Observational study of gene-disease association. (HuGE Navigator) 19553435_The homozygosity for a variable-number (4 or 5) tandem-repeat polymorphism in the coding region of the clock gene PERIOD3 confers vulnerability to sleep loss and circadian misalignment through its effects on sleep homeostasis. 19693801_Observational study of gene-disease association. (HuGE Navigator) 19716732_The association between sleep timing and the circadian rhythms of melatonin and PER3 RNA in leukocytes is stronger in PER3(5/5) than in PER3(4/4). 19839995_Observational study of gene-disease association. (HuGE Navigator) 19861640_PER3 levels were correlated with fasting plasma glucose (beta = -.29, p < .05) and shift work (beta = .31, p < .05). 19913121_Observational study of gene-disease association. (HuGE Navigator) 19926609_Observational study of gene-disease association. (HuGE Navigator) 19926609_Per3 clock gene polymorphisms have also been associated with circadian disruption and with increased cancer risk. 19934327_Observational study of gene-disease association. (HuGE Navigator) 19967263_The frequencies of the shorter allele (4 repeats) in the PER3 gene and the T allele in the CLOCK gene among Asians (0.86 and 0.84, respectively) were significantly higher than among Caucasians (0.69 and 0.71, respectively). 20072116_Observational study of gene-disease association. (HuGE Navigator) 20149345_Observational study of gene-disease association. (HuGE Navigator) 20174623_Observational study of gene-disease association. (HuGE Navigator) 20364331_Observational study of gene-disease association. (HuGE Navigator) 20364331_The T3111C (RS1801260) polymorphism of hClock gene is associated with schizophrenia, but it seems that the length polymorphism of 18 exon of hPer3 may not be associated with schizophrenia. 20469812_In mammals the circadian oscillation is driven by a negative feedback loop involving Per3 20469812_Observational study of gene-disease association. (HuGE Navigator) 20625127_Deletion of PER3 is directly related to tumor recurrence in patients with ER positive breast cancers treated with tamoxifen. Low expression of PER3 mRNA is associated with poor prognosis. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20978934_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21070773_Per3 is a checkpoint protein that plays important roles in checkpoint activation, cell proliferation and apoptosis. 21176035_Data show the association remained significant between poor sleep quality and the Per3 rs228727 polymorphism in patients with bipolar disorder. 21316201_We showed a relationship between Per3 polymorphism and postpartum depressive onset in bipolar disorder. 21411511_Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. 21459569_PER1 and PER3 may modulate apoptotic reactions to cisplatin in gingival cancer cells 21559414_polymorphisms in UTS2 and PER3 may have roles in glucose homeostasis and diabetes 21714069_Failure to replicate previous research in relation to PERIOD3 and CLOCK concurs with previous research suggesting that the effects of these genes are small and may be related to population composition. 21729600_Decitabine can induce the expression of hPer3 gene and cells apoptosis in acute myeloid leukemia. 21919721_found no association between the PER3 clock gene VNTR polymorphism and chronotype, indicating that the proposed role of PER3 needs further clarification 22169200_The polymorphism in the clock gene PER3 may contribute to interindividual differences in sleep and circadian physiology in older people. 22188742_We provide first evidence that humans homozygous for the PER3 5/5 allele are particularly sensitive to blue-enriched light, as indexed by the suppression of endogenous melatonin and waking theta activity. 22217103_treatment with isoprenaline or dexamethasone induces circadian expression of hPer1, hPer2, hPer3, and hBMAL1 22324552_Data suggest that the PER3 variable number tandem repeat specifically affects measures of sleep timing and may also modify the effects of sleep on health outcome measures. 22350604_Decreased PER3 expression was observed in colon cancer tissue and was associated with multiple clinicopathologic factors as well as patient overall survival. 22467995_PER3 genotype contributed unique variance in predicting insomnia severity in alcohol dependence patients 22543063_No significant association was found between the clock-gene PER3(5/5) genotype and postoperative cognitive dysfunction at 1 week after noncardiac surgery. 22689435_PER3 was down-regulated in human hepatocellular carcinoma, compared to peritumorous and healthy liver tissues. 22809120_A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. 22881285_The relationship between a polymorphism of PER3 and susceptibility and behavior of Crohn disease and ulcerative colitis are reported. 22942500_Studies report that polymorphisms in the PERIOD 3 (PER3) gene explains phenotypic variance in responses to total sleep deprivation. 22971169_These data suggest that white males of European descent participating in individual endurance sports in South Africa are more likely to be morning types, and PER3 tandem repeat may be one of the factors contributing to this observation. 22976125_the association study of the 54-nucleotide repeat in exon 18 of the hPer3 gene with a predisposition to opioid dependence among residents of the West Siberian region of Russia 23128810_Per3 variable number tandem repeat polymorphism is not a major risk factor for chronic heart failure (CHF) or a factor modulating the severity of the CHF in this population. 23171222_polymorphisms more behaviorally salient with increasing severity and/or duration of sleep restriction 23323702_These data document for the first time that the expression of BMAL1, PER3, PPARD and CRY2 genes is altered in gestational diabetes compared to normal pregnant women. deranged expression of clock genes may play a pathogenic role in GDM. 23524621_Cortisol secretion was modified among police officers with different PER3 VNTR clock gene variants. 23546644_Studies indicate that in the cytoplasm, PER3 protein heterodimerizes with PER1, PER2, CRY1, and CRY2 proteins and enters into the nucleus, resulting in repression of CLOCK-BMAL1-mediated transcription. 23606611_there is significant daily variation in PER2, PER3, and ARNTL1 expression with earlier timing of expression in women than in men 23969301_the association of rs934945 with 'morning alertness' and rs2640909 with 'morningness'. 24150227_PER3 genotype predicted circadian rhythm period lengthening by lithium, specifically among bipolar disorder cases. 24393525_PER3, at least partially, was targeted by miRNA-103, which might affect cells apoptosis in G2/M phase by modulating apoptosis-related gene in p53 pathway. 24439663_there is a greater detrimental impact of sleep deprivation in PER3(5/5) than PER3(4/4) carriers. the group with greater attentional performance impairment due to sleep deprivation (PER3(5/5) carriers) is superior at initiating sleep over the 24-h cycle. 24635757_there is significant association between diurnal preference and a polymorphism in PER3 and a novel nominally significant association between diurnal preference and a polymorphism in ARNTL2 24636202_No association with SNPs or haplotypes of the PER3 gene was observed in prophylactic lithium response. 24678593_The polymorphisms studied in PER3 did not demonstrate any relationship with Bone Mineral Density or the odds of osteoporosis in postmenopausal Korean women. 24821610_Five SNPs (rs228727, rs228644, rs228729, rs707467, rs104620202) in the period 3 (PER3) gene are significant correlation with genotype and allele frequency in lung cancer. 24866331_Per3 VNTR may contribute to modulation of cardiac functions and interindividual differences in development and progression of myocardial infarction 24893318_The data indicate that humans homozygous for the PER3(5/5) allele are more sensitive to NIF light effects, as indexed by specific changes in sleep EEG activity. 24903750_Women with the 4/5 or 5/5 PER3 genotype had a nonstatistically significant 33% increased odds of breast cancer. 25201053_The findings support the notion that PER3 polymorphisms could be a potential genetic marker for an individual's circadian and sleep phenotypes. 25344870_A wild genotype of rs228729 in PER3 was the primary risk factor contributing to Chinese hepatocellular carcinoma patients recurrence-free survival. 25390010_An association is outlined of Per3 five repeat allele with T2DM occurrence and suggest that individuals with five repeat allele may be at a greater risk for T2DM as compared to those carrying the four repeat allele. 25400784_Provide some evidence that circadian rhythm of flight cadets with the PER3 (5) allele are less likely to be affected compared to those with the PER3 (4) allele. 25501848_PERIOD3 clock gene length polymorphism may have a role in colorectal adenoma formation 25545397_length polymorphism in the Period 3 gene is associated with sleepiness and maladaptive circadian phase in night-shift workers 25550826_In patients with non-small cell lung cancer, those with lower expression of Per1, Per2 and Per3 had a shorter survival time than those with higher expression. 25775462_minor polymorphisms of PER3 may be associated with poor morning gastric motility, and may have a combinatorial effect with CLOCK. 25798540_evidence of an association between PER3 gene and planning performance in a sample of healthy subjects 25837749_no evidence supporting a global association of PER3 genetic variants with the incidence of cancer (Meta-Analysis) 25892098_PER3 variants may be associated with decreased risk for depressive symptoms, including subthreshold levels of depressive symptoms, in older adults. 25940842_PER3 long allele carriers were more vulnerable to sleep-loss associated attentional lapses than those with the short allele. 26102236_PER3 VNTR genotype does not explain the difference in chronotype between South African and Dutch marathon runners 26359345_This study shown a possible association between Per3 polymorphism and consciousness recovery level in Disorders of Consciousness patients. 26406960_This work suggests a role of PER3 rs10462020 in predicting a prognosis in Diffuse large B-cell lymphoma overall survival of patients. 26440425_PER3 polymorphism is associated with sleep quality after a mild traumatic brain injury. 26453284_There was no significant association between the PER3 VNTR polymorphism and the irregularity of the menstrual cycle in Korean adolescents. 26624862_The PER1 c.2884C > G polymorphism and PER3 54bp VNTR were associated with annual percent changes in bone mineral density of femoral neck after 1 year of hormone therapy. 26707349_PER3 VNTR polymorphism influences both response to antidepressant chronotherapeutics and total sleep time after the repeated sleep deprivation treatment in patients with bipolar depression. 26903630_A role for PER3 in modulating circadian clock and mood during seasonal changes 26922944_No association between cluster headache, PER3 VNTR polymorphism and chronotype was found in this study. 27335043_This study showed that Lack of Association between Genetic Polymorphism of PER3 gene with Late Onset Depression and Alzheimer's Disease in a Sample of a Brazilian Population 27373683_TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1 27593530_4 and 5 variable-number tandem-repeat polymorphisms appear to influence sensitivity to the effects of stressful urban environments on sleep 27996307_CLOCK rs1801260*C and PER3(4/4) influence myelination processes by regulating sleep quality and quantity. 28045078_these findings suggest that rhythmic expression of the circadian clock gene PER3 is associated with the amount of daily physical activity and physical fitness in older adults 28055273_there is a significant association between the PER3 polymorphism and the extraversion personality trait 28276850_The impact of PER3 polymorphisms on sleep cycle and insomnia in older adults is reported. 28614626_Study identified several CpG sites and specifically several CpGs in the PTPRS and PER3 genes differentially methylated between obese and non-obese children, suggesting a role for DNA methylation concerning development of childhood obesity. 28708003_PER3 polymorphisms links with chronotype and affective dimensions 28821614_Data suggest that three E box-like response elements are located upstream of PER3 transcription start sites; these response elements appear to additively contribute to cell-autonomous transcriptional oscillation of PER3 and other circadian proteins. 28860482_our results confirm that polymorphisms in PER3, a nonessential clock gene involved in regulating circadian period length, are linked to anxiety, supporting the previously suggested role of PER3 in regulating human mood 28900721_This study showed that PER3(5/5) carriers had poorer cognitive performance (attention, executive function, semantic memory, and verbal fluency) and lower cortical integrity (structural and functional) than PER3(4/4). 29055479_PER34/4 genotype may accelerate the course of disease in multiple sclerosis susceptible individuals. 29055480_data suggest that, PER3(4/4) genotype may accelerate the course of disease in multiple sclerosis susceptible individuals especially in women 29510794_genotype may play an important role in individual vulnerability to the different mechanisms of daytime sleep disturbance in night shift workers 30442327_Volunteers with the PER3 4/4 variant who live farther from the equator have a greater increase in their weekend sleep duration. 30575220_Loss of PER3 expression is associated with shortened circadian period length rather than a phase shift. 30670712_Enhanced expression of miR-181a in iBMSCs and PASCs produced a robust increase in adipogenesis through the direct targeting of the circadian factor period circadian regulator 3 (PER3). 31249322_PER3 variable number tandem repeat (VNTR) polymorphism modulates the circadian variation of the descending pain modulatory system in healthy subjects. 31328557_Clock gene PERIOD3 polymorphism is associated with susceptibility to Graves' disease but not to Hashimoto's thyroiditis. 31692380_The Per3 rs228697 CC genotype was associated with a higher sleep factor score when compared with the CG genotype. In addition, the rs228729 TC genotype was associated with a greater risk of suffering from excitement/agitation, akathisia and weight loss (p = 0.041, OR = 2.287) when compared with the CC genotype. Finally, the rs10746473 AA genotype patients were more likely to suffer from dizziness 32682347_A PERIOD3 variable number tandem repeat polymorphism modulates melatonin treatment response in delayed sleep-wake phase disorder. 32737280_Effects of Occupational Stress and Circadian CLOCK Gene Polymorphism on Sleep Quality of Oil Workers in Xinjiang, China. 32805476_Comparing expression levels of PERIOD genes PER1, PER2 and PER3 in chronic insomnia patients and medical staff working in the night shift. 32841827_Depression and anxiety symptoms correlate with diurnal preference, sleep habits, and Per3 VNTR polymorphism (rs57875989) in a non-clinical sample. 32849922_Period Family of Clock Genes as Novel Predictors of Survival in Human Cancer: A Systematic Review and Meta-Analysis. 33393850_Possible Association of PER2/PER3 Variable Number Tandem Repeat Polymorphism Variants with Susceptibility and Clinical Characteristics in Pancreatic Cancer. 33501709_Associations of PER3 polymorphisms with clopidogrel resistance among Chinese Han people treated with clopidogrel. 33741899_hPER3 promotes adipogenesis via hHSP90AA1-mediated inhibition of Notch1 pathway. 34060156_Association of occupational stress, period circadian regulator 3 (PER3) gene polymorphism and their interaction with poor sleep quality. 34112033_For whom the circadian clock ticks? Investigation of PERIOD and CLOCK gene variants in bipolar disorder. 34215556_Genetic association of the PERIOD3 (PER3) Clock gene with extreme obesity. 34275001_Dopamine adjusts the circadian gene expression of Per2 and Per3 in human dermal fibroblasts from ADHD patients. 34542826_Is There a Link between Circadian Clock Protein PERIOD 3 (PER3) (rs57875989) Variant and the Severity of COVID-19 Infection? 34604590_PER3 polymorphisms and their association with prostate cancer risk in Japanese men. 34732337_Alterations in CRY2 and PER3 gene expression associated with thalamic-limbic community structural abnormalities in patients with bipolar depression or unipolar depression. | ENSMUSG00000028957 | Per3 | 48.30911 | 1.1758094 | 0.2336542366 | 0.41990747 | 3.091700e-01 | 5.781900e-01 | 9.998360e-01 | No | Yes | 58.30787 | 12.148414 | 5.319571e+01 | 8.791920 | |
| ENSG00000049769 | 89801 | PPP1R3F | protein_coding | Q6ZSY5 | FUNCTION: Glycogen-targeting subunit for protein phosphatase 1 (PP1). {ECO:0000269|PubMed:21668450}. | Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a protein that has been identified as one of several type-1 protein phosphatase (PP1) regulatory subunits. One or two of these subunits, together with the well-conserved catalytic subunit, can form the PP1 holoenzyme, where the regulatory subunit functions to regulate substrate specificity and/or targeting to a particular cellular compartment. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:89801; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; protein phosphatase type 1 complex [GO:0000164]; glycogen binding [GO:2001069]; protein phosphatase 1 binding [GO:0008157]; protein phosphatase binding [GO:0019903]; regulation of glycogen (starch) synthase activity [GO:2000465]; regulation of glycogen biosynthetic process [GO:0005979] | 8938429_Maps this gene as HB2E at the DXS9823E marker sequence. 20398921_Observational study of gene-disease association. (HuGE Navigator) 20479760_Observational study of gene-disease association. (HuGE Navigator) 34145793_Methylation of three genes encoded by X chromosome in blood leukocytes and colorectal cancer risk. | ENSMUSG00000039556 | Ppp1r3f | 443.64065 | 0.9408095 | -0.0880255180 | 0.16980615 | 2.775452e-01 | 5.983147e-01 | 9.998360e-01 | No | Yes | 436.43395 | 56.432968 | 4.304388e+02 | 44.463695 | |
| ENSG00000052723 | 80143 | SIKE1 | protein_coding | Q9BRV8 | FUNCTION: Physiological suppressor of IKK-epsilon and TBK1 that plays an inhibitory role in virus- and TLR3-triggered IRF3. Inhibits TLR3-mediated activation of interferon-stimulated response elements (ISRE) and the IFN-beta promoter. May act by disrupting the interactions of IKBKE or TBK1 with TICAM1/TRIF, IRF3 and DDX58/RIG-I. Does not inhibit NF-kappa-B activation pathways. {ECO:0000269|PubMed:16281057}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;Reference proteome | SIKE interacts with IKK-epsilon (IKBKE; MIM 605048) and TBK1 (MIM 604834) and acts as a suppressor of TLR3 (MIM 603029) and virus-triggered interferon activation pathways (Huang et al., 2005 [PubMed 16281057]).[supplied by OMIM, Mar 2008]. | hsa:80143; | cytosol [GO:0005829]; protein kinase binding [GO:0019901]; small GTPase binding [GO:0031267] | 16281057_Overexpression of SIKE inhibits virus- and Toll-like receptor 3-triggered interferon-stimulated response elements (ISRE) but not NF-kappa B activation. 23649622_Data indicate that suppressor of IKKepsilon (SIKE) inhibits TANK-binding kinase 1 (TBK1)-mediated phosphorylation of interferon regulatory factor 3 (IRF3), which is essential to type I interferon production. 27249321_SIKE, a negative regulator of the interferon pathway, attenuates pathological cardiac hypertrophy. 30487159_mRNA expression of individual inhibitory kappaB kinases and SIKE are associated with unique prognostic significance and may act as valuable prognostic biomarkers and potential targets for future therapeutic interventions in gastric cancer. | ENSMUSG00000027854 | Sike1 | 560.63553 | 0.9803418 | -0.0286432962 | 0.16563702 | 3.133441e-02 | 8.594964e-01 | 9.998360e-01 | No | Yes | 874.87271 | 105.130609 | 9.289677e+02 | 86.428884 | |
| ENSG00000053747 | 3909 | LAMA3 | protein_coding | Q16787 | FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components.; FUNCTION: Laminin-5 is thought to be involved in (1) cell adhesion via integrin alpha-3/beta-1 in focal adhesion and integrin alpha-6/beta-4 in hemidesmosomes, (2) signal transduction via tyrosine phosphorylation of pp125-FAK and p80, (3) differentiation of keratinocytes. | Alternative splicing;Basement membrane;Cell adhesion;Coiled coil;Disulfide bond;Epidermolysis bullosa;Extracellular matrix;Glycoprotein;Laminin EGF-like domain;Reference proteome;Repeat;Secreted;Signal | The protein encoded by this gene belongs to the laminin family of secreted molecules. Laminins are heterotrimeric molecules that consist of alpha, beta, and gamma subunits that assemble through a coiled-coil domain. Laminins are essential for formation and function of the basement membrane and have additional functions in regulating cell migration and mechanical signal transduction. This gene encodes an alpha subunit and is responsive to several epithelial-mesenchymal regulators including keratinocyte growth factor, epidermal growth factor and insulin-like growth factor. Mutations in this gene have been identified as the cause of Herlitz type junctional epidermolysis bullosa and laryngoonychocutaneous syndrome. Alternative splicing and alternative promoter usage result in multiple transcript variants. [provided by RefSeq, Dec 2014]. | hsa:3909; | basement membrane [GO:0005604]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; laminin-5 complex [GO:0005610]; extracellular matrix structural constituent [GO:0005201]; integrin binding [GO:0005178]; structural molecule activity [GO:0005198]; animal organ morphogenesis [GO:0009887]; axon guidance [GO:0007411]; cell migration [GO:0016477]; cell-cell adhesion [GO:0098609]; endodermal cell differentiation [GO:0035987]; epidermis development [GO:0008544]; integrin-mediated signaling pathway [GO:0007229]; morphogenesis of a polarized epithelium [GO:0001738]; regulation of cell adhesion [GO:0030155]; regulation of cell migration [GO:0030334]; regulation of embryonic development [GO:0045995]; tissue development [GO:0009888] | 11775027_Characterization of morphological and cytoskeletal changes in MCF10A breast epithelial cells plated on laminin-5: comparison with breast cancer cell line MCF7. 12196012_Neurite outgrowth promoting sites have been identified on the laminin alpha 3 chain LG4 module. 12382139_In all developing organs investigated, the mRNA of the alpha3 chain of laminin is strictly of epithelial origin and the corresponding protein localised in the underlying basement membrane zones 12532327_These results suggest that the G3 domain of laminin 5 alpha 3 contains two distinct regions that differently regulate cell adhesion and migration. 12826666_laminin alpha 3 LG4 module may play an important role in tissue remodeling by inducing MMP-1 expression during wound healing 12943669_a missense mutation in the adhesion G domain of laminin-5 causes mild junctional epidermolysis bullosa 12947106_results demonstrate that the laminin alpha3 LG4/5 modules within unprocessed laminin-5 permit its cell binding activity through heparan and chondroitin sulfate chains of syndecan-1 14612440_proteolytic processing of laminin-5 influences its interaction with alpha3beta1 integrin 14695139_LAMA3 promoter methylation is associated with increased breast tumor stage and tumor size 15044476_Characterization of laminin 5B and NH2-terminal proteolytic fragment of its alpha3B chain 15149852_co-cultures of epithelial cells and fibroblasts were studied to analyse the processing of laminin 5 alpha3, beta3, and gamma2 chains 15316072_The G4/5 domain in the alpha3 chain facilitates deposition of precursor laminin 5 into the PBM in epidermal wounds 15373767_Premature termination codon mutations in both alleles of LAMB3 or LAMC2 genes were found in nine of the 11 H junctional epidermolysis bullosa patients 15695818_the LG4-5 domain synergistically enhances integrin signaling as it is released from the precursor LN5 15854126_the absolute mRNA levels generated from the laminin 5 genes do not determine the translated protein levels of the laminin 5 chains in keratinocytes; the expression of the laminin 5 genes may be controlled by common regulation mechanisms 16179086_The uncoordinated production of chains of ln-5 in allergic asthma could have a bearing on the poor epithelial cell anchorage in these patients. 16219677_Suppression of laminin-5 alpha 3-chain expression in human mammary epithelial cells results in loss of reconstituted extracellular matrix-mediated growth control and apoptosis. 16297184_The hinge region between subdomains G3 and G4 of laminin alpha 3 carries the proteolytic cleavage sites but proteolytic processing plays no role in kerocyte migration. 16537560_tumor cell migration on laminin-5 is inhibited by HYD1, a biologically active integrin-targeting peptide 16870608_Ln-5 is an important regulator of ADPKD cell proliferation and cystogenesis; Ln-5 gamma(2) chain and Ln-5-alpha(6)beta(4) integrin interaction both contribute to these phenotypic changes 17000025_The major contributions of laminin-5 to the resistance of the epidermis against frictional stress but also for basement membrane regeneration and repair of damaged skin are reflected by the phenotype of Herlitz junctional epidermolysis bullosa. 17071854_Laminin-5 may contribute to the development of bone tissues by promoting the proliferation and by suppressing the chondrogenic differentiation of MSCs. 17137774_Laminin-5 alone stimulates global changes in gene/protein expression in mesenchymal stem cells that lead to commitment of these cells to the osteogenic phenotype, and this correlates with extracellular matrix production. 17466943_These results suggest a new possible approach to repairing SCI and, in general, a model which will be useful for other multidisciplinary procedures for complex neurological situations. 17482449_LM-332 is a crucial motility-promoting factor for B-CLL lymphocytes and is a potential constituent favoring the dissemination of B-CLL lymphocytes through vascular basement membranes and possibly lymph node compartments. 17825249_These results suggest that Crk is required for early attachment to laminin, cell motility, and growth of glioblastoma cell line KMG4. 18603785_the alpha3 chain can assemble with only beta3-gamma2 heterodimer to form a heterotrimer via disulfide bonds 18664273_Smad4 mediates transcriptional regulation through three mechanisms: Smad4 binding to a functional SBE site in the LAMA3 promoter, Smad4 binding to AP1 (and Sp1) sites via interaction with AP1 family, and Smad4 impact on transcription of AP1 factors 19724895_Observational study of gene-disease association. (HuGE Navigator) 19752234_Alpha3- and alpha5-laminins, but not other laminin isoforms, mediate mast cell adhesion via alpha3beta1 integrin. 19773554_immunofluorescence analyses revealed a basement membrane staining in epithelial tissue for LaNt alpha3 and LaNt alpha3 localized along the substratum-associated surface of cultured keratinocytes. 19834535_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 19936049_a hypoxia related splice variant of LAMA3 may have a role in progression of head and neck cancers 19940114_Bisecting GlcNAc residues on laminin-332 down-regulate galectin-3-dependent keratinocyte motility. 19945619_the alpha3 subunit of laminins-332, -321, and -311 plays an important role in mediating epidermal-dermal integrity and is essential for the skin to withstand mechanical stresses 20036365_SEMA3F, CLEC16A, LAMA3, and PCSK2 variants have roles in myocardial infarction in Japanese individuals 20198315_Observational study of gene-disease association. (HuGE Navigator) 20372818_Observational study of gene-disease association. (HuGE Navigator) 21034821_Cell surface COL17 can interact with laminin 332 and, together, participate in the adherence of a cell to the extracellular matrix. 21276136_results suggest that Lm3B11/3B21 may be required for normal mature vessels and interfere with tumor angiogenesis 21940345_Expression of a3, beta3 and beta2 cannot predict the prognosis. However, high expression of beta2 mRNA in HCC/non-cancerous liver correlated significantly with the absence of complete encapsulation, which is an important tumor invasiveness factor. 22351752_the LAMA3 LG45 domain may trigger different signals toward keratinocytes depending on its interaction with syndecan-1 or -4. 22434185_Letter/Case Report: report novel LAMA3 mutations leading to enamel defects in epidermolysis bullosa. 22563463_Data show laminin-332 (alpha3ss3gamma2)(Lm332) matrix supported adhesion of keratinocytes much more strongly and stably than purified Lm332. 22673183_invasive breast cancer cells confer an anoikis-resistant phenotype on myofibroblasts during tissue remodeling by inducing laminin-332 upregulation and integrin beta4 neoexpression 22963541_A male patient with Herlitz junctional epidermolysis bullosa with a novel homozygosity for insertion LAMA3 Premature Termination Codon mutation is described. Both parents and his sibling were shown to be heterozygous carriers. 23869449_Results describe a new pedigree identifying a novel mutation of LAMA3 in LOC syndrome 23907728_High LAMA3 expression is associated with gastric cancer. 24675808_Two of the six genes (LAMA3 and DST) validated by quantitative RT-PCR for tumor-specific alternative splicing events 25188360_Loss of the normal basement membrane organization of alpha3 laminin observed in fibrotic regions from the lungs of patients with pulmonary fibrosis contributes to their disease progression. 25363238_We established the LAMA3 gene as novel potential susceptibility gene for atopic dermatitis. 25443231_LAMNA variants have been identified in atrial fibrillation cohort studies, demonstrating abnormalities in cardiac excitation - supra ventricular tachycardia and atrial fibrillation. 27375110_Seventeen percent of the patients were compound heterozygous or homozygous for mutations in the gene LAMA3, 59% carried mutations in both alleles of LAMB3, and 12% were homozygous for mutations in LAMC2. In nine patients with severe generalized JEB, detection of two mutations in one of the genes LAMA3, LAMB3, or LAMC2 was not possible, so the molecular basis of disease could not be clarified completely 27827380_Both parents had offspring affected with JEB and displayed subtle enamel pitting of secondary dentition without any sign of skin blistering. The reported enamel abnormality in LAMA3 mutation carriers could be attributed to a half dose effect of the laminin a3 chain 29077994_The authors describe the natural history of JEB in three children with the same LAMA3 splice-site mutation. In spite of residual laminin 332, two of them died prematurely. 29797489_The study has identified two mutations in two large consanguineous pedigrees. Identification of novel variants in the LAMA3 and PLEC genes will expand the mutation spectrum and also help in genetic counselling of patients in the Pakistani population. 29920840_We identified two pathogenic and 10 loss-of-function (LOF) candidate variants, accounting for 4.74% (12 out of 253) of all the. In burden tests, rare missense variants in three genes (CSF3R, DSP, and LAMA3) were identified that have a statistically significant relationship with Idiopathic pulmonary fibrosis 32767748_Loss of the laminin subunit alpha-3 induces cell invasion and macrophage infiltration in cutaneous squamous cell carcinoma. 33179081_Laminin332 mediates proliferation, apoptosis, invasion, migration and epithelialtomesenchymal transition in pancreatic ductal adenocarcinoma. 33992120_LAMA3 DNA methylation and transcriptome changes associated with chemotherapy resistance in ovarian cancer. 34134622_High expression of LAMA3/AC245041.2 gene pair associated with KRAS mutation and poor survival in pancreatic adenocarcinoma: a comprehensive TCGA analysis. 34796550_Gene expression profiling of laminin alpha3-blocked keratinocytes reveals an immune-independent mechanism of blistering. | ENSMUSG00000024421 | Lama3 | 393.51260 | 0.8187865 | -0.2884407106 | 0.18724007 | 2.259918e+00 | 1.327611e-01 | 9.998360e-01 | No | Yes | 477.25017 | 75.235969 | 5.400380e+02 | 66.072133 | |
| ENSG00000060138 | 8531 | YBX3 | protein_coding | P16989 | FUNCTION: Binds to the GM-CSF promoter. Seems to act as a repressor. Binds also to full-length mRNA and to short RNA sequences containing the consensus site 5'-UCCAUCA-3'. May have a role in translation repression (By similarity). {ECO:0000250}. | Acetylation;Alternative splicing;Cytoplasm;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation | hsa:8531; | bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; mRNA 3'-UTR binding [GO:0003730]; nucleic acid binding [GO:0003676]; polysome binding [GO:1905538]; RNA binding [GO:0003723]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; small GTPase binding [GO:0031267]; 3'-UTR-mediated mRNA stabilization [GO:0070935]; cellular hyperosmotic response [GO:0071474]; cellular response to tumor necrosis factor [GO:0071356]; fertilization [GO:0009566]; in utero embryonic development [GO:0001701]; male gonad development [GO:0008584]; negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress [GO:1902219]; negative regulation of necroptotic process [GO:0060546]; negative regulation of skeletal muscle tissue development [GO:0048642]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of cytoplasmic translation [GO:2000767]; positive regulation of organ growth [GO:0046622]; regulation of gene expression [GO:0010468]; spermatogenesis [GO:0007283] | 12239625_human dbpA has a role in accelerating hepatocarcinogenesis 14728692_identification of complexes, binding to the vascular endothelial growth factor mRNA 5'- and 3'-UTR, that contain cold shock domain and polypyrimidine tract binding RNA binding proteins 16508013_ZONAB regulates the transcription of genes that are important for G1/S-phase progression and links tight junctions to the transcriptional control of key cell cycle regulators and epithelial cell differentiation. 19240061_Observational study of gene-disease association. (HuGE Navigator) 19328795_The symplekin/ZONAB complex inhibits intestinal cell differentiation by the repression of AML1/Runx1. 19749785_The expression of human DBPA was upregulated in gastric cancer tissues and cell lines. siRNA treatment successfully silenced DBPA expression. 20133480_ZONAB is an important component of the mechanisms that sense epithelial density and participates in the complex transcriptional networks that regulate the switch between proliferation and differentiation 21473684_In human skeletal muscle cells, CSDA was upregulated during hypoxia when cells were damaged and apoptosis was induced. 22159460_DbpA promoter was methylated in 37.7% of HCC samples. 22711822_We demonstrate that ZONAB promotes cell survival in response to proinflammatory, hyperosmotic, and cytotoxic stress and that stress-induced ZONAB activation involves the Rho regulator GEF-H1. 24817634_In the prenatal brain, the cold shock domain protein A were found to be abundantly expressed in radial glial cells, neuroblasts and neurons. 24885929_Stratification according to Fuhrman grade disclosed higher YBX3 expression levels. 27430286_Data suggest that DNA binding protein A (dbpA) may be used as a therapeutic target in colorectal cancer (CRC). 27569444_dbpA knockdown in SW620 cells altered the expression of carcinogenesis-associated genes in Colorectal cancer. 29227750_Authors examined, for the first time in a Turkish population, the association between protamine gene alleles (PRM1 c.-190C>A, PRM1 c.197G>T, and PRM2 c.248C>T), and YBX2 (c.187T>C and c.1095 + 16A>G) and male infertility. 30784285_ZONAB is up-regulated in bladder cancer cell lines, which promotes invasion, demonstrating the important role it plays in tumorogenesis and cancer progression 31189097_YBX3 binds within the 3' UTR of SLC7A5 to stabilize the transcript. 31944153_YB-3 substitutes YB-1 in global mRNA binding. 32553055_Y-Box-Binding Protein 3 (YBX3) Restricts Influenza A Virus by Interacting with Viral Ribonucleoprotein Complex and Imparing its Function. 32652867_Knockdown of DNA-binding protein A enhances the chemotherapy sensitivity of colorectal cancer via suppressing the Wnt/beta-catenin/Chk1 pathway. | ENSMUSG00000030189 | Ybx3 | 10449.02226 | 1.0364161 | 0.0516032753 | 0.08760374 | 3.476806e-01 | 5.554290e-01 | 9.998360e-01 | No | Yes | 11073.83620 | 910.438833 | 1.049263e+04 | 666.583307 | ||
| ENSG00000061656 | 6676 | SPAG4 | protein_coding | Q9NPE6 | FUNCTION: Involved in spermatogenesis. Required for sperm head formation but not required to establish and maintain general polarity of the sperm head. Required for anchoring and organization of the manchette. Required for targeting of SUN3 and probably SYNE1 through a probable SUN1:SYNE3 LINC complex to the nuclear envelope and involved in accurate posterior sperm head localization of the complex. May anchor SUN3 the nuclear envelope. Involved in maintenance of the nuclear envelope integrity. May assist the organization and assembly of outer dense fibers (ODFs), a specific structure of the sperm tail. {ECO:0000250|UniProtKB:O55034, ECO:0000250|UniProtKB:Q9JJF2}. | Cell projection;Cilium;Coiled coil;Cytoplasm;Cytoskeleton;Differentiation;Flagellum;Membrane;Nucleus;Reference proteome;Spermatogenesis;Transmembrane;Transmembrane helix | The mammalian sperm flagellum contains two cytoskeletal structures associated with the axoneme: the outer dense fibers surrounding the axoneme in the midpiece and principal piece and the fibrous sheath surrounding the outer dense fibers in the principal piece of the tail. Defects in these structures are associated with abnormal tail morphology, reduced sperm motility, and infertility. In the rat, the protein encoded by this gene associates with an outer dense fiber protein via a leucine zipper motif and localizes to the microtubules of the manchette and axoneme during sperm tail development. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2015]. | hsa:6676; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; integral component of nuclear inner membrane [GO:0005639]; meiotic nuclear membrane microtubule tethering complex [GO:0034993]; motile cilium [GO:0031514]; nuclear envelope [GO:0005635]; protein-membrane adaptor activity [GO:0043495]; structural molecule activity [GO:0005198]; cell differentiation [GO:0030154]; nuclear envelope organization [GO:0006998]; spermatogenesis [GO:0007283] | 23602831_SPAG4 is an independent prognostic factor in renal cell carcinoma and plays a crucial role in cytokinesis to defend against hypoxia-induced tetraploid formation. 23818324_SPAG4 knockdown reduces the invasion capability of RCC cells. 29901114_SPAG4, in cooperation with Nesprin3, has a fundamental pathological function in the migration of lung carcinoma cells. 30817682_Our study revealed that SPAG4 was identified as a cancer biomarker for glioblastoma and might be a promising target for clinical diagnosis and intervention of glioblastoma. 31144711_Results indicate that sperm associated antigen 4 (SPAG4L/SPAG4Lbeta) transcript isoform interacts with spectrin repeat containing nuclear envelope protein 2 (Nesprin2) in the meiotic process. 34311595_Human sperm-associated antigen 4 as a potential prognostic biomarker of lung squamous cell carcinoma. | ENSMUSG00000038180 | Spag4 | 81.69468 | 1.0275452 | 0.0392019145 | 0.33195437 | 1.422136e-02 | 9.050747e-01 | 9.998360e-01 | No | Yes | 81.05350 | 12.679585 | 7.420673e+01 | 9.874296 | |
| ENSG00000063176 | 56848 | SPHK2 | protein_coding | Q9NRA0 | FUNCTION: Catalyzes the phosphorylation of sphingosine to form sphingosine-1-phosphate (SPP), a lipid mediator with both intra- and extracellular functions. Also acts on D-erythro-dihydrosphingosine, D-erythro-sphingosine and L-threo-dihydrosphingosine. Binds phosphoinositides (PubMed:19168031, PubMed:12954646). In contrast to prosurvival SPHK1, has a positive effect on intracellular ceramide levels, inhibits cells growth and enhances apoptosis (PubMed:16118219). In mitochondria, is important for cytochrome-c oxidase assembly and mitochondrial respiration. The SPP produced in mitochondria binds PHB2 and modulates the regulation via PHB2 of complex IV assembly and respiration (PubMed:20959514). In nucleus, plays a role in epigenetic regulation of gene expression. Interacts with HDAC1 and HDAC2 and, through SPP production, inhibits their enzymatic activity, preventing the removal of acetyl groups from lysine residues with histones. Up-regulates acetylation of histone H3-K9, histone H4-K5 and histone H2B-K12 (PubMed:19729656). In nucleus, may have an inhibitory effect on DNA synthesis and cell cycle (PubMed:12954646, PubMed:16103110). In mast cells, is the main regulator of SPP production which mediates calcium influx, NF-kappa-B activation, cytokine production, such as TNF and IL6, and degranulation of mast cells (By similarity). In dopaminergic neurons, is involved in promoting mitochondrial functions regulating ATP and ROS levels (By similarity). Also involved in the regulation of glucose and lipid metabolism (By similarity). {ECO:0000250|UniProtKB:Q9JIA7, ECO:0000269|PubMed:12954646, ECO:0000269|PubMed:16103110, ECO:0000269|PubMed:16118219, ECO:0000269|PubMed:19168031, ECO:0000269|PubMed:19729656, ECO:0000269|PubMed:20959514}. | ATP-binding;Alternative splicing;Cytoplasm;Endoplasmic reticulum;Kinase;Lipid metabolism;Lysosome;Membrane;Mitochondrion;Mitochondrion inner membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase | This gene encodes one of two sphingosine kinase isozymes that catalyze the phosphorylation of sphingosine into sphingosine 1-phosphate. Sphingosine 1-phosphate mediates many cellular processes including migration, proliferation and apoptosis, and also plays a role in several types of cancer by promoting angiogenesis and tumorigenesis. The encoded protein may play a role in breast cancer proliferation and chemoresistance. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2011]. | hsa:56848; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; ATP binding [GO:0005524]; D-erythro-sphingosine kinase activity [GO:0017050]; lipid kinase activity [GO:0001727]; NAD+ kinase activity [GO:0003951]; nucleosomal histone binding [GO:0031493]; small GTPase binding [GO:0031267]; sphinganine kinase activity [GO:0008481]; sphingosine-1-phosphate receptor activity [GO:0038036]; blood vessel development [GO:0001568]; brain development [GO:0007420]; cellular response to phorbol 13-acetate 12-myristate [GO:1904628]; epigenetic maintenance of chromatin in transcription-competent conformation [GO:0045815]; female pregnancy [GO:0007565]; histone H2A-K5 acetylation [GO:0043977]; histone H2B-K12 acetylation [GO:0043980]; negative regulation of cell growth [GO:0030308]; negative regulation of histone deacetylase activity [GO:1901726]; negative regulation of histone deacetylation [GO:0031064]; phosphorylation [GO:0016310]; positive regulation of apoptotic process [GO:0043065]; positive regulation of calcium ion import [GO:0090280]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of ceramide biosynthetic process [GO:2000304]; positive regulation of cytokine production involved in immune response [GO:0002720]; positive regulation of histone H3-K9 acetylation [GO:2000617]; positive regulation of interleukin-13 production [GO:0032736]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of mast cell activation involved in immune response [GO:0033008]; positive regulation of mast cell degranulation [GO:0043306]; positive regulation of protein kinase C signaling [GO:0090037]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of ATP biosynthetic process [GO:2001169]; regulation of cytochrome-c oxidase activity [GO:1904959]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; regulation of reactive oxygen species biosynthetic process [GO:1903426]; sphinganine-1-phosphate biosynthetic process [GO:0006669]; sphingolipid biosynthetic process [GO:0030148]; sphingolipid metabolic process [GO:0006665]; sphingosine biosynthetic process [GO:0046512]; sphingosine metabolic process [GO:0006670] | 14596938_although both sphingosine kinase type 1 (SphK1) and type 2 (SphK2) can phosphorylate FTY720 with low efficiency, SphK2 is much more effective than SphK1 15951439_Sphingosine kinase 2 plays an important role in migration of MDA-MB-453 cells toward EGF. 16103110_the N-terminal-extended form of sphingosine kinase 2 has a role in serum-dependent regulation of cell proliferation and apoptosis 17311928_hSphK2 is phosphorylated on Ser-351 and Thr-578 by ERK1 and phosphorylation of these residues is important for EGF-stimulated migration of MDA-MB-453 cells 17635916_PKD is a physiologically relevant enzyme for SPHK2 phosphorylation, which leads to its nuclear export for subsequent cellular signaling. 17895250_Sphingosine kinase 2, but not sphingosine kinase 1, acted in concert with SPP-1 to regulate recycling of sphingosine into ceramide. 18178871_These data suggest that differential formation of sphingosine-1-phosphate by SphK1 and SphK2 has distinct and important actions in human mast cells. 18263879_a novel mechanism of regulation of both SK1 and SK2 that is mediated by their interaction with eEF1A. 19168031_A lipid binding domain in Sphk2 that is important for the enzyme's sub-cellular localisation was identified. 19240026_Results indicate that SPHK1 and 2 isoforms and neutral sphingomyelinase contribute to the regulation of chemosensitivity by controlling ceramide formation and the downstream Akt pathway in human colon cancer cells. 19490468_SPHK2 may regulate the autonomous proliferation of synovial fibroblasts as one of the predisposing genes to rheumatoid arthritis 20197547_Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. 20237496_Observational study of gene-disease association. (HuGE Navigator) 21307639_pharmacological inhibition of Sphk2 with the orally bioavailable selective inhibitor, ABC294640, has therapeutic potential in the treatment of chemo- and endocrine therapy- resistant breast cancer. 21620961_SK2 functions as a pro-survival protein and is involved in promoting actin rearrangement into membrane ruffles/lamellipodia in response to sphingosine 1-phosphate in MCF-7 breast cancer cells. 23314175_Sphingosine kinase 2 might function simply to dynamically regulate sphingosine-S1P cycling. 23359503_The function of SphK1 and SphK2 can be interchangeable in mast cells and is species and cell type determined. 23881266_SphK2 plays a controversial role in arthritis. 23918304_the expression of SphK2 parallels the progression of NSCLC. The expression of SphK2 might represent a novel and potentially independent biomarker for the prognosis of patients with NSCLC. 24140934_these results implicate interrelated mechanisms and SPHK2 inhibition in the induction of PEL cell death by ABC294640 and rationalize evaluation of ABC294640 in clinical trials for the treatment of KSHV-associated lymphoma. 24385109_Silencing of sphingosine kinase 2 (SphK-2) also blocked HDL-induced COX-2/PGI-2 activation. 24422628_IGF1R mediates insulin-stimulated phosphorylation of both SphK1 and SphK2. 24486401_SK2 prevents the nuclear translocation of Y416 phosphorylated c-Src and this appears to be independent of S1P4 and might involve an intracellular S1P-dependent mechanism, which is resistant to exogenously added S1P. 24686171_Results demonstrated that SK2 regulates MYC, which has a pivotal role in hematologic malignancies. 24709100_SPHK2 regulates apoptosis in colon cancer cells. 24903384_Data show that sphingosine kinase SphK1 and sphingosine-1-phosphate (S1P) receptors S1P1, S1P2, S1P3, and S1P5 were expressed from primary, up to recurrent and secondary glioblastomas, with sphingosine kinase SphK2 levels were highest in primary tumors. 25010828_These data illuminate a novel survival mechanism and potential therapeutic target for KSHV-infected endothelial cells: SphK2-associated maintenance of viral latency 25455157_Down-regulation of SphK2 increased the effects of all-trans-retinoic acid on colon cancer cells. 25808826_Because of the unique relationship observed after serum depletion, we examined effects of siRNA for SPHK2, and found the role of SPHK2 as a growth or survival factor but not a cell proliferation inhibitor in FCS(-) culture 26209696_results define new mechanistic insights for EGF-mediated invasion and novel actions of SK2 26337959_Targeting of SphK2 by ABC294640 potently inhibits colorectal cancer cell growth both in vitro and in vivo. 26494858_ABC294640 may reduce the proliferative capacity of castration-resistant prostate cancer cells through inhibition of both sphingosine kinase 2 and dihydroceramide desaturase. 26628299_In this study, we describe the findings that overexpression of SphK2 promotes chemoresistance in non-small cell lung cancer (NSCLC) cells. Inhibition of SphK2 might be considered as a strategy in NSCLC treatment with gefitinib 26733171_the pathogenesis of CRC maybe mediated by SphK2, and SphK2 could represent a selective target for the molecularly targeted treatments of CRC 26822263_data suggest that in vitro inhibition of SK-2, can compromise the integrity of the EC monolayer 26886371_increased SK and SPL mRNA expression along with reduced sphingosine 1-phosphate levels were more commonly observed in hepatocellular carcinoma tissues. 27021309_The analysis suggests that the catalytic function and regulation of SPHK1 and SPHK2 might be dependent on their conformational mobility. (Review) 27223438_miR-613 functions as a tumor suppressor in papillary thyroid carcinoma and its suppressive effect is mediated by repressing SphK2 expression 27517489_Data show that inhibition of sphingosine kinase-2 by ABC294640 is synergistically cytotoxic with gemcitabine toward three pancreatic cancer cell lines, resulting in decreased expression of both ribonucleotide reductase regulatory subunit M2 (RRM2) and c-MYC protein (Myc) in all three cell lines. 27588496_this study shows that SK2 can have a direct role in promoting oncogenesis 28108260_SphKs/Sphingosine-1-phosphate signaling is critical for the growth and survival of estrogen receptor positive MCF-7 human breast cancer cells. 28465486_These results provide new insights into the biological functions of Sphk2 and the molecular mechanisms that underlie the Sphk2-mediated resistance to retinoid therapy. 28807595_sphingosine kinase-2 plays an important role in kidney fibrogenesis by modulating transforming growth factor-beta signaling 28950390_Results form study in non-small-cell lung cancer cells showed that SphK2 plays a critical role in doxorubicin-induced resistance by regulating key anti-apoptotic gene, survivin. 28975989_findings indicate that miR-577 is a potential tumor suppressor in Papillary Thyroid Cancer by targeting SphK2, and may be a potential therapeutic target in PTC. 29057430_This review focuses on the recent advances in research on SphK2 regulation and its potential roles in diseases, highlighting SphK2 may be a novel therapeutic strategy for diseases--{REVIEW} 29145490_results were consistent across all of the four serotypes of DENV infection, which supports the pro-apoptotic role of SPHK2 in DENV-infected liver cells 29321083_Colony formation and cell proliferation were suppressed by enhanced expression of miR-338-3p in LC cells. Moreover, miR-338-3p targeted sphingosine kinase 2 (SphK2). Silencing of SphK2 had an identical influence as overexpression of miR-338-3p in LC cells. 29615132_The results of this study suggested that a shift in the subcellular localization of the sphingosine 1-phosphate generating SphK2 may compromise the well established pro-survival cytosolic sphingosine 1-phosphate by favoring the production of nuclear sphingosine 1-phosphate associated with adverse effects in AD pathogenesis. 29678573_the study revealed that lncRNA DANCR might promote the proliferation of osteoarthritis chondrocytes and reduce apoptosis through the miR-577/SphK2 axis. 29697136_SPHK2 may promote glioma growth by stimulating tumor-associated macrophages to polarize M2-type macrophages 29748384_High SPHK2 expression is associated with lung cancer. 29955024_siRNA-mediated knockdown of Sphk2 but not SphK1 resulted in a reduction of cargo content in purified exosomes 29991026_these data demonstrate that SK-2 exerts an antiproliferative and apoptosis-sensitizing effect in renal mesangial cells which suggests that selective inhibitors of SK-2 may promote proliferation and reduce apoptosis 30021386_In colorectal cancer cell lines, inhibition of miR-363-3p elevated SphK2 levels. 30077738_Sphk2 gene silencing induced by nanoparticles in hepatocellular carcinoma (HCC) cells could reduce miRNA-21 sorting into exosomes, contributing to the inhibition of tumor cell migration and tumorigenic function of exosomes to normal liver cells. 30185808_Kinases SphK1 and SphK2 regulate IL-17 expression in human T cells. 30227139_Study demonstrated that increased SPHK1 expression and sphingosine-1-phosphate production, and conversely decreases in SPHK2 expression and dihydrosphingosine-1-phosphate production, are likely responsible for increased cathelicidin antimicrobial peptide levels in differentiated keratinocytes in parallel with endoplasmic reticulum stress. 30250299_Thus, these findings demonstrate a tumor-suppressive function of DYNC1I1, and uncover new mechanistic insights into SK2 regulation which may have implications in targeting this enzyme as a therapeutic strategy in glioblastoma 30251698_SPHK2 is highly expressed in the kidney interstitium of patients with renal fibrosis and highly correlates with disease progression. SPHK2 phosphorylates Fyn to activate downstream STAT3 and AKT, thereby promoting extracellular matrix synthesis, kidney fibroblast activation, and renal fibrosis. 30452878_Sphingosine kinase 2 deficiency in mice significantly prevented the loss of beta-cell mass. 30456838_We will also highlight the connections between SphK2 and the DNA damage response. Finally, we will provide our insight into the regulatory mechanisms of SphKs by two main categories, micro and long, noncoding RNAs as the novel players of cancer chemoresistance. 30463717_S1P produced by SphK1 and SphK2 may have different functions in pancreatic cancer. 30478856_These findings suggested that LINC00460 could function as a competing endogenous RNA to regulate SphK2 expression by sponging miR-613 in papillary thyroid carcinoma. 30869582_Normal CD8+ T-cell infiltration into the DENV-infected brain independent of SK2 or S1P. 31003959_SphK2 expression levels were down-regulated in the livers of human patients with alcoholic cirrhosis and hepatocellular carcinoma compared to healthy controls. 31128248_SPHK2 is degraded by autophagy at early time-points of macrophage activation. SPHK2 degradation is required for inflammatory cytokine production. 31157539_Serum S1P is decreased in PACI, which may be partly due to downregulation of pancreatic SPHK2 31247144_Increased SPHK2 levels in proliferating airway smooth muscle cells could be exploited to counteract airway smooth muscle thickening with synthetic substrates. 31477835_The specific antagonism of beta3-AR by SR59230A inhibits Neuroblastoma growth and tumor progression, by switching from stemness to cell differentiation both in vivo and in vitro through the specific blockade of SK2/S1P2 signaling. 31954175_Sphingosine kinase 1 and sphingosine kinase 2 have opposing roles in regulating p53-dependent function. siRNA knockdown of SK-2 reduces the expression of the pro-survival XBP-1s. 32075502_Long noncoding RNA LINC00520 accelerates progression of papillary thyroid carcinoma by serving as a competing endogenous RNA of microRNA-577 to increase Sphk2 expression. 32124438_Sphingosine kinase-2 is overexpressed in large granular lymphocyte leukaemia and promotes survival through Mcl-1. 32705254_Inhibition of sphingosine kinase 2 attenuates hypertrophic scar formation via upregulation of Smad7 in human hypertrophic scar fibroblasts. 33758320_METTL3-mediated m(6)A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. 35394045_Upregulated flotillins and sphingosine kinase 2 derail AXL vesicular traffic to promote epithelial-mesenchymal transition. | ENSMUSG00000057342 | Sphk2 | 1236.17773 | 1.0927061 | 0.1279053677 | 0.13027059 | 9.898900e-01 | 3.197693e-01 | 9.998360e-01 | No | Yes | 1327.60216 | 178.772976 | 1.105509e+03 | 115.621977 | |
| ENSG00000064225 | 10402 | ST3GAL6 | protein_coding | Q9Y274 | FUNCTION: Involved in the synthesis of sialyl-paragloboside, a precursor of sialyl-Lewis X determinant. Has a alpha-2,3-sialyltransferase activity toward Gal-beta1,4-GlcNAc structure on glycoproteins and glycolipids. Has a restricted substrate specificity, it utilizes Gal-beta1,4-GlcNAc on glycoproteins, and neolactotetraosylceramide and neolactohexaosylceramide, but not lactotetraosylceramide, lactosylceramide or asialo-GM1. | Alternative splicing;Glycoprotein;Glycosyltransferase;Golgi apparatus;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the sialyltransferase family. Members of this family are enzymes that transfer sialic acid from the activated cytidine 5'-monophospho-N-acetylneuraminic acid to terminal positions on sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins. This protein has high specificity for neolactotetraosylceramide and neolactohexaosylceramide as glycolipid substrates and may contribute to the formation of selectin ligands and sialyl Lewis X, a carbohydrate important for cell-to-cell recognition and a blood group antigen. [provided by RefSeq, Apr 2016]. | hsa:10402; | extracellular exosome [GO:0070062]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; beta-galactoside (CMP) alpha-2,3-sialyltransferase activity [GO:0003836]; beta-galactoside alpha-2,3-sialyltransferase activity [GO:0052798]; sialyltransferase activity [GO:0008373]; cellular protein modification process [GO:0006464]; cellular response to interleukin-6 [GO:0071354]; glycolipid metabolic process [GO:0006664]; keratan sulfate biosynthetic process [GO:0018146]; oligosaccharide biosynthetic process [GO:0009312]; oligosaccharide metabolic process [GO:0009311]; protein glycosylation [GO:0006486] | 18485915_Epigenetic changes in a group of glycosyltransferases including B4GALNT2 and ST3GAL6 represent a malignant phenotype of gastric cancer caused by silencing of the activity of these enzymes 19781661_Expression of ST3Gal IV in several gastrointestinal cell lines is correlated with the expression of sialyl Lewis x at the cell surface. 20532202_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21147760_the occurrence of CD75s- and iso-CD75s-gangliosides in tumor tissues is largely independent of the transcriptional expression of ST6GAL1 and ST3GAL6 25061176_The sialyltransferase ST3GAL6 influences homing and survival in multiple myeloma. 31914669_a cross-restoration of each of the three genes in ST3GAL6 KO cells showed that overexpression of ST3GAL6 sufficiently rescued the total alpha2,3-sialylation levels, cell morphology, and alpha2,3-sialylation of EGFR, whereas the alpha2,3-sialylation levels of beta1 were greatly enhanced by an overexpression of ST3GAL4 33649796_lncRNA ST3GAL6AS1 promotes invasion by inhibiting hnRNPA2B1mediated ST3GAL6 expression in multiple myeloma. | ENSMUSG00000022747 | St3gal6 | 95.89800 | 0.9161561 | -0.1263345972 | 0.32489680 | 1.488879e-01 | 6.996004e-01 | 9.998360e-01 | No | Yes | 76.59002 | 15.321914 | 8.212938e+01 | 12.700133 | |
| ENSG00000064607 | 10147 | SUGP2 | protein_coding | Q8IX01 | FUNCTION: May play a role in mRNA splicing. {ECO:0000305}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a member of the arginine/serine-rich family of splicing factors. The encoded protein functions in mRNA processing. Alternatively spliced transcript variants have been described. [provided by RefSeq, Feb 2009]. | hsa:10147; | nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; RNA splicing [GO:0008380] | ENSMUSG00000036054 | Sugp2 | 3158.67458 | 0.9831903 | -0.0244574393 | 0.12296934 | 3.978987e-02 | 8.418920e-01 | 9.998360e-01 | No | Yes | 3489.77214 | 317.000478 | 3.629113e+03 | 254.845529 | ||
| ENSG00000065357 | 1606 | DGKA | protein_coding | P23743 | FUNCTION: Diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:2175712, PubMed:15544348). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (PubMed:2175712, PubMed:15544348). Also plays an important role in the biosynthesis of complex lipids (Probable). Can also phosphorylate 1-alkyl-2-acylglycerol in vitro as efficiently as diacylglycerol provided it contains an arachidonoyl group (PubMed:15544348). Also involved in the production of alkyl-lysophosphatidic acid, another bioactive lipid, through the phosphorylation of 1-alkyl-2-acetyl glycerol (PubMed:22627129). {ECO:0000269|PubMed:15544348, ECO:0000269|PubMed:2175712, ECO:0000269|PubMed:22627129, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Calcium;Cytoplasm;Kinase;Lipid metabolism;Metal-binding;Nucleotide-binding;Reference proteome;Repeat;Transferase;Zinc;Zinc-finger | PATHWAY: Lipid metabolism; glycerolipid metabolism. {ECO:0000305|PubMed:15544348, ECO:0000305|PubMed:2175712, ECO:0000305|PubMed:22627129}. | The protein encoded by this gene belongs to the eukaryotic diacylglycerol kinase family. It acts as a modulator that competes with protein kinase C for the second messenger diacylglycerol in intracellular signaling pathways. It also plays an important role in the resynthesis of phosphatidylinositols and phosphorylating diacylglycerol to phosphatidic acid. Several transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Apr 2017]. | hsa:1606; | cytosol [GO:0005829]; membrane [GO:0016020]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; diacylglycerol kinase activity [GO:0004143]; kinase activity [GO:0016301]; lipid binding [GO:0008289]; NAD+ kinase activity [GO:0003951]; phospholipid binding [GO:0005543]; diacylglycerol metabolic process [GO:0046339]; glycerolipid metabolic process [GO:0046486]; intracellular signal transduction [GO:0035556]; lipid phosphorylation [GO:0046834]; phosphatidic acid biosynthetic process [GO:0006654]; platelet activation [GO:0030168]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205] | 14734770_Defects in both polymorphonuclear neutrophil (PMN) transendothelial migration and PMN diacylglycerol kinase alpha signaling are implicated as disordered functions in subjects with localized aggressive periodontitis. 15117825_PPARgamma agonists upregulate DGKalpha production.This suppresses the diacylglycerol/protein-kinase-C signaling pathway. 15870081_DGKalpha is crucial for the control of cell activation and also for the regulation of the secretion of lethal exosomes, which in turn controls cell death. 15928040_ALK-mediated alphaDGK activation is dependent on p60src tyrosine kinase, with which alphaDGK forms a complex; alphaDGK activation is involved in the control of ALK-mediated mitogenic properties. 17276726_These results strongly suggest that DGKalpha is a novel positive regulator of NF-kappaB, which suppresses TNF-alpha-induced melanoma cell apoptosis. 17911109_diacylglycerol kinase alpha-conserved domains have a role in membrane targeting in intact T cells 18004883_2,3-dioleoylglycerol binds to a site on the alpha and zeta isoforms of diacylglycerol kinase that is exposed as a consequence of the substrate binding to the active site. 18424699_Lck-dependent tyrosine phosphorylation of diacylglycerol kinase alpha regulates its membrane association in T cells.( 19751727_These results strongly suggest that DGKalpha positively regulates TNF-alpha-dependent NF-kappaB activation via the PKCzeta-mediated Ser311 phosphorylation of p65/RelA. 21252909_Diacylglycerol kinase alpha is a key regulator of the polarised secretion of exosomes. 21493725_findings further suggest that DGL-alpha and -beta may regulate neurite outgrowth by engaging temporally and spatially distinct molecular pathways 22048771_SAP-mediated inhibition of DGKalpha sustains diacylglycerol signaling, thereby regulating T cell activation 22271650_Antigen-specific CD8-positive T cells from DGKalpha-deficient transgenic mice show enhanced expansion and increased cytokine production after lymphocytic choriomeningitis virus infection, yet DGK-deficient memory CD8+ T cells exhibit impaired expansion. 22425622_DGKalpha is involved in hepatocellular carcinoma progression by activation of the MAPK pathway. 22573804_DGK-alpha was more highly expressed in CD8-tumor-infiltrating T cellscompared with that in CD8non-tumor kidney-infiltrating lymphocytes. 24158111_High diacylglycerol kinase alpha expression is associated with glioblastoma. 24887021_These data indicates the existence of a SDF-1alpha induced DGKalpha - atypical PKC - beta1 integrin signaling pathway, which is essential for matrix invasion of carcinoma cells. 25248744_DGKalpha generates phosphatidic acid to drive its own recruitment to tubular recycling endosomes via its interaction with MICAL-L1 25921290_Redundant and specialized roles for diacylglycerol kinases alpha and zeta in the control of T cell functions. 26420856_An abandoned compound that also inhibits serotonin receptors may have more translational potential as a DGKa inhibitor, but more potent and specific DGKa inhibitors are sorely needed 26964756_Decreased DNA methylation at this enhancer enables recruitment of the profibrotic transcription factor early growth response 1 (EGR1) and facilitates radiation-induced DGKA transcription in cells from patients later developing fibrosis. 27498782_LIPFDGKA might serve as a potential possible biomarkers for diagnosis of gastric cancer, and their downregulation may bring new perspective into the investigation of gastric cancer prognosis 27697466_Diacylglycerol kinases alpha and zeta are up-regulated in cancer in cancer, and contribute towards tumor immune evasion and T cells clonal anergy. (Review) 27731506_DGKalpha isoform is highly expressed in the nuclei of human erythroleukemia cell line K562, and its nuclear activity drives K562 cells through the G1/S transition during cell cycle progression. 29967261_This novel study demonstrates efficient ablation of diacylglycerol kinase in human CAR-T cells that leads to improved antitumor immunity and may have significant impact in human cancer immunotherapy 30653270_This study presents the first crystal structure of EF-hand domains of diacylglycerol kinase alpha in its Ca(2+) bound form and characterize Ca(2+) -induced conformational changes, which likely regulates intra-molecular interactions. 31766109_Upon neutrophil stimulation, DGK-alpha activation is necessary for migration and a productive response. This paper focuses on the role of DGK-alpha in obstructive respiratory diseases, including asthma and chronic obstructive pulmonary disease, but also rare genetic diseases such as alpha-1-antitrypsin deficiency. [review] 32341033_DGKA Provides Platinum Resistance in Ovarian Cancer Through Activation of c-JUN-WEE1 Signaling. 32345612_Diacylglycerol kinases regulate TRPV1 channel activity. 33608256_DGKA Mediates Resistance to PD-1 Blockade. 34293268_Diacylglycerol Kinase Inhibition Reduces Airway Contraction by Negative Feedback Regulation of Gq-Signaling. 35131384_DGKA interacts with SRC/FAK to promote the metastasis of non-small cell lung cancer. | ENSMUSG00000025357 | Dgka | 189.41865 | 0.8932703 | -0.1628313235 | 0.25050119 | 4.345101e-01 | 5.097842e-01 | 9.998360e-01 | No | Yes | 200.38646 | 31.249171 | 2.202748e+02 | 27.697027 |
| ENSG00000065485 | 10954 | PDIA5 | protein_coding | Q14554 | 3D-structure;Alternative splicing;Disulfide bond;Endoplasmic reticulum;Isomerase;Redox-active center;Reference proteome;Repeat;Signal | This gene encodes a member of the disulfide isomerase (PDI) family of endoplasmic reticulum (ER) proteins that catalyze protein folding and thiol-disulfide interchange reactions. The encoded protein has an N-terminal ER-signal sequence, three catalytically active thioredoxin (TRX) domains, a TRX-like domain, and a C-terminal ER-retention sequence. The N-terminal TRX-like domain is the primary binding site for the major ER chaperone calreticulin and possibly other proteins and substrates as well. Alternative splicing results in multiple protein- and non-protein-coding transcript variants. [provided by RefSeq, Dec 2016]. | hsa:10954; | endoplasmic reticulum lumen [GO:0005788]; endoplasmic reticulum membrane [GO:0005789]; oxidoreductase activity [GO:0016491]; protein disulfide isomerase activity [GO:0003756]; protein-disulfide reductase activity [GO:0015035]; electron transport chain [GO:0022900]; protein folding [GO:0006457] | 14627699_Three thioredoxin motifs (CXXC) of purified PDIR were found to contribute to its isomerase activity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20796029_Protein disulfide isomerases are enzymes that mediate oxidative protein folding in the endoplasmic reticulum. 22002444_protein disulfide isomerase 5 is deregulated in the mucopolysaccharidoses, although the unfolded protein response is not activated 24037032_This study demonistrated that Protein disulfide isomerase P5-immunopositive inclusions in patients with Alzheimer's disease. 24636989_PDIA5 is a key regulator ATF6alpha-mediated cellular functions in cancer. 29293453_Data suggest that upon activation of integrins, protein disulfide isomerase (PDI) is released from endothelial cells and forms a disulfide bond complex with alphaVbeta3 integrin. 33664747_PDIA5 is Correlated With Immune Infiltration and Predicts Poor Prognosis in Gliomas. | ENSMUSG00000022844 | Pdia5 | 920.25478 | 0.9736536 | -0.0385194664 | 0.12513947 | 9.371691e-02 | 7.595040e-01 | 9.998360e-01 | No | Yes | 931.74622 | 60.822780 | 9.712418e+02 | 49.835801 | ||
| ENSG00000065491 | 55633 | TBC1D22B | protein_coding | Q9NU19 | FUNCTION: May act as a GTPase-activating protein for Rab family protein(s). {ECO:0000250}. | 3D-structure;Acetylation;Direct protein sequencing;GTPase activation;Phosphoprotein;Reference proteome | hsa:55633; | 14-3-3 protein binding [GO:0071889]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; intracellular protein transport [GO:0006886] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23572552_Using affinity purification-mass spectrometry, we identified the putative Rab33 GTPase-activating proteins TBC1D22A and TBC1D22B as ACBD3-interacting factors. | ENSMUSG00000042203 | Tbc1d22b | 910.42112 | 0.9991074 | -0.0012882929 | 0.12675501 | 1.023322e-04 | 9.919288e-01 | 9.998360e-01 | No | Yes | 910.50878 | 73.751604 | 9.245407e+02 | 58.400021 | ||
| ENSG00000066697 | 91283 | MSANTD3 | protein_coding | Q96H12 | Alternative splicing;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:91283; | 28212443_MSANTD3 rearrangement as a recurrent event in salivary gland AcCC 30520817_we find the HTN3-MSANTD3 gene fusion to be a recurrent event in acinic cell carcinoma with prominent serous differentiation and an indolent clinical course | ENSMUSG00000039693 | Msantd3 | 566.92899 | 0.9055233 | -0.1431763920 | 0.15121387 | 8.774546e-01 | 3.488998e-01 | 9.998360e-01 | No | Yes | 565.51638 | 64.326230 | 5.968377e+02 | 52.804499 | ||||
| ENSG00000067113 | 8611 | PLPP1 | protein_coding | O14494 | FUNCTION: Magnesium-independent phospholipid phosphatase of the plasma membrane that catalyzes the dephosphorylation of a variety of glycerolipid and sphingolipid phosphate esters including phosphatidate/PA, lysophosphatidate/LPA, diacylglycerol pyrophosphate/DGPP, sphingosine 1-phosphate/S1P and ceramide 1-phosphate/C1P (PubMed:9305923, PubMed:9705349, PubMed:9607309, PubMed:10962286, PubMed:17379599). Also acts on N-oleoyl ethanolamine phosphate/N-(9Z-octadecenoyl)-ethanolamine phosphate, a potential physiological compound (PubMed:9607309). Through its extracellular phosphatase activity allows both the hydrolysis and the cellular uptake of these bioactive lipid mediators from the milieu, regulating signal transduction in different cellular processes (PubMed:10962286, PubMed:12909631, PubMed:15461590, PubMed:17379599). It is for instance essential for the extracellular hydrolysis of S1P and subsequent conversion into intracellular S1P (PubMed:17379599). Involved in the regulation of inflammation, platelets activation, cell proliferation and migration among other processes (PubMed:12909631, PubMed:15461590). May also have an intracellular activity to regulate phospholipid-mediated signaling pathways (By similarity). {ECO:0000250|UniProtKB:O08564, ECO:0000269|PubMed:10962286, ECO:0000269|PubMed:12909631, ECO:0000269|PubMed:15461590, ECO:0000269|PubMed:17379599, ECO:0000269|PubMed:9305923, ECO:0000269|PubMed:9607309, ECO:0000269|PubMed:9705349}. | Alternative splicing;Cell membrane;Glycoprotein;Hydrolase;Lipid metabolism;Membrane;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; phospholipid metabolism. {ECO:0000269|PubMed:10962286, ECO:0000269|PubMed:9305923, ECO:0000269|PubMed:9607309, ECO:0000269|PubMed:9705349}. | The protein encoded by this gene is a member of the phosphatidic acid phosphatase (PAP) family. PAPs convert phosphatidic acid to diacylglycerol, and function in synthesis of glycerolipids and in phospholipase D-mediated signal transduction. This enzyme is an integral membrane glycoprotein that plays a role in the hydrolysis and uptake of lipids from extracellular space. Alternate splicing results in multiple transcript variants of this gene. [provided by RefSeq, May 2013]. | hsa:8611; | apical plasma membrane [GO:0016324]; caveola [GO:0005901]; extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; membrane raft [GO:0045121]; plasma membrane [GO:0005886]; ceramide-1-phosphate phosphatase activity [GO:0106235]; diacylglycerol diphosphate phosphatase activity [GO:0000810]; lipid phosphatase activity [GO:0042577]; phosphatidate phosphatase activity [GO:0008195]; sphingosine-1-phosphate phosphatase activity [GO:0042392]; androgen receptor signaling pathway [GO:0030521]; ceramide metabolic process [GO:0006672]; intracellular steroid hormone receptor signaling pathway [GO:0030518]; negative regulation of cell population proliferation [GO:0008285]; phospholipid dephosphorylation [GO:0046839]; phospholipid metabolic process [GO:0006644]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205]; regulation of lipid metabolic process [GO:0019216]; signal transduction [GO:0007165]; sphingolipid biosynthetic process [GO:0030148]; sphingosine metabolic process [GO:0006670] | 12426308_this enzyme regulates interleukin-4-mediated STAT6 signaling. 12444089_This protein and phosphoprotein SET regulate androgen production by P450c17. 14506139_lipid phosphate phosphatase-1 has a role in regulating accumulation of lysophosphatidic acid in ovarian cancer 16950767_Lipopolysaccharide-induced up-regulation of cyclooxygenase-2 depends on the activity of the Mg(+2)-dependent phosphatidic acid phosphohydrolase 1 (PAP-1) 17005594_LPP1 and LPP3 are distributed in distinct lipid rafts that may provide unique microenvironments defining their non-redundant physiological functions. 17169329_These results suggest that the expression of PAP2a is directly regulated by p73. 18755152_These results suggest that LPP1a is important for the determination of plasma FTY720-P levels. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23426360_Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association 23936490_the first of the two transmembrane regions in human SAC1 (TM1) functions in Golgi localization | ENSMUSG00000021759 | Plpp1 | 292.08956 | 0.9553188 | -0.0659457953 | 0.19166427 | 1.174514e-01 | 7.318153e-01 | 9.998360e-01 | No | Yes | 282.54602 | 33.660301 | 3.336570e+02 | 31.101356 |
| ENSG00000068078 | 2261 | FGFR3 | protein_coding | P22607 | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. Plays an essential role in the regulation of chondrocyte differentiation, proliferation and apoptosis, and is required for normal skeleton development. Regulates both osteogenesis and postnatal bone mineralization by osteoblasts. Promotes apoptosis in chondrocytes, but can also promote cancer cell proliferation. Required for normal development of the inner ear. Phosphorylates PLCG1, CBL and FRS2. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. Plays a role in the regulation of vitamin D metabolism. Mutations that lead to constitutive kinase activation or impair normal FGFR3 maturation, internalization and degradation lead to aberrant signaling. Over-expressed or constitutively activated FGFR3 promotes activation of PTPN11/SHP2, STAT1, STAT5A and STAT5B. Secreted isoform 3 retains its capacity to bind FGF1 and FGF2 and hence may interfere with FGF signaling. {ECO:0000269|PubMed:10611230, ECO:0000269|PubMed:11294897, ECO:0000269|PubMed:11703096, ECO:0000269|PubMed:14534538, ECO:0000269|PubMed:16410555, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17145761, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17509076, ECO:0000269|PubMed:17561467, ECO:0000269|PubMed:19088846, ECO:0000269|PubMed:19286672, ECO:0000269|PubMed:8663044}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Cell membrane;Chromosomal rearrangement;Craniosynostosis;Cytoplasmic vesicle;Deafness;Disease variant;Disulfide bond;Dwarfism;Ectodermal dysplasia;Endoplasmic reticulum;Glycoprotein;Immunoglobulin domain;Kinase;Lacrimo-auriculo-dento-digital syndrome;Membrane;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | This gene encodes a member of the fibroblast growth factor receptor (FGFR) family, with its amino acid sequence being highly conserved between members and among divergent species. FGFR family members differ from one another in their ligand affinities and tissue distribution. A full-length representative protein would consist of an extracellular region, composed of three immunoglobulin-like domains, a single hydrophobic membrane-spanning segment and a cytoplasmic tyrosine kinase domain. The extracellular portion of the protein interacts with fibroblast growth factors, setting in motion a cascade of downstream signals, ultimately influencing mitogenesis and differentiation. This particular family member binds acidic and basic fibroblast growth hormone and plays a role in bone development and maintenance. Mutations in this gene lead to craniosynostosis and multiple types of skeletal dysplasia. [provided by RefSeq, Aug 2017]. | hsa:2261; | cell surface [GO:0009986]; endoplasmic reticulum [GO:0005783]; extracellular region [GO:0005576]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; transport vesicle [GO:0030133]; ATP binding [GO:0005524]; fibroblast growth factor binding [GO:0017134]; fibroblast growth factor-activated receptor activity [GO:0005007]; identical protein binding [GO:0042802]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; bone maturation [GO:0070977]; bone mineralization [GO:0030282]; bone morphogenesis [GO:0060349]; cell-cell signaling [GO:0007267]; chondrocyte differentiation [GO:0002062]; chondrocyte proliferation [GO:0035988]; endochondral bone growth [GO:0003416]; endochondral ossification [GO:0001958]; fibroblast growth factor receptor apoptotic signaling pathway [GO:1902178]; fibroblast growth factor receptor signaling pathway [GO:0008543]; MAPK cascade [GO:0000165]; negative regulation of developmental growth [GO:0048640]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of phosphatidylinositol 3-kinase activity [GO:0043552]; positive regulation of phospholipase activity [GO:0010518]; positive regulation of tyrosine phosphorylation of STAT protein [GO:0042531]; protein autophosphorylation [GO:0046777]; skeletal system development [GO:0001501]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11556601_G380R mutation of this gene is common mutation associated with achondroplasia 11731410_We identified a novel ETV6 partner gene, fibroblast growth factor receptor 3 (FGFR3), in a patient with peripheral T-cell lymphoma (PTCL) with a t(4;12)(p16;p13) translocation. 11754059_Two patients with clinical and radiological findings of achondroplasia, who had the most common FGFR3 missense mutations. 11759058_distribution in normal endocrine cells and related tumors of the gastroenteropancreatic system; immunoreactive in duodenal G cells 11827956_interacts with adapter protein SH2-B, and has a role in STAT5 activation 11906172_Identification and characterization of an alternatively spliced isoform 12009017_the G370C and S371C mutant receptors spontaneously dimerize in the correct spatial orientation required for effective signal transduction, whereas the 372-5 mutants, like the WT receptor, may achieve this orientation only on ligand binding 12048679_Nucleotide 1138 in transmembrane domain of FGFR3 gene is the hot point for mutation in ACH and hence its major pathologic cause. 12297284_phosphorylation is essential for FGFR3 ubiquitylation, but is not sufficient to induce downregulation of its internalization resistant mutants 12368157_there is an FGFR3 mutation with a demonstrated deregulatory mechanism and alternative splicing in the absence of t(4;14) in multiple myeloma patients 12368206_Parathyroid hormone receptor type 1/Indian hedgehog expression is preserved in the growth plate of human fetuses affected with activating mutations in this protein 12373339_Differences in spatial patterns of FGFR expression in normal skin may generate functional diversity in response to FGFs, and in wounded skin FGFs may function in wound healing via induced FGFRs. 12424440_strong correlation beween mutations of FGFR3 and disturbances of skeletal growth-REVIEW 12433679_data indicate that t(4;14)(p16;q32) and loss of fibroblast growth factor receptor 3 occurred at a very early stage of multiple myeloma and suggest that activation of multiple myeloma SET domain protein may be transforming event of this translocation 12461689_mutations in bladder cancer previously identified in non-lethal skeletal disorders 12624096_the importance of the immature FGFR3 proteins as mediators of an abnormal signaling in thanatophoric dysplasia type II 12664252_Cherubism was mapped to region 4p16.3. Because of the associated craniosynostosis, we excluded the FGFR3 gene as a candidate gene for cherubism. 12764678_The FGFR3-associated coronal synostosis syndrome (Muenke craniosynostosis) is caused by a point mutation (C749G) on the FGFR3 gene resulting in a Pro250Arg substitution. 12833394_A missense mutation in FGFR3 resulted in skeltal dysplasia distinct from thanatophoric dysplasia. 12921294_results give further support to the fact that the G380R mutation of FGFR-3 is the most common mutation causing achondroplasia in different populations 12929929_introduction of these mutated FGFR3s into ATDC5 cells downregulated PTHrP expression and induced apoptosis with reduction of Bcl-2 expression 14520460_FGFR3IIIS may regulate FGF and FGFR trafficking and function, possibly contributing to the development of a malignant phenotype 14562121_involvement of FGFR-3 in malignant hematopoiesis and FGFR-3 tyrosine kinase in CD34+ leukemic cells 14606518_IGF-1 prevents the apoptosis induced by FGFR3 mutation through the PI3K pathway and MAPK pathway 14678961_FGFR3 mutations were associated with low-stage, low-grade urothelial carcinomas of the blader. 14715624_Inhibition of FGFR3 in myeloma cell lines was associated with morphologic, phenotypic, and functional changes typical of plasma cell differentiation, including increase in light-chain secretion and expression of CD31, followed by apoptosis 14751560_defective differentiation of chondrocytes is the main cause of longitudinal bone growth retardation in FGFR3-related human chondrodysplasias 15026322_Mutations in growth factor receptor 3 is associated with the pathogenesis of urothelial cell carcinoma 15105428_fibroblast growth factor receptor 3 activation is regulated by cytoplasmic tyrosine kinase Pyk2 15221641_Observational study of genetic testing. (HuGE Navigator) 15292251_fibroblast growth factor receptor 3 has a role in trafficking and signaling 15517832_Acanthosis nigricans with achondroplasia due to Gly380Arg mutation in FGFR3. 15558020_Reciprocal relationship in gene expression between FGFR1 and FGFR3 in colorectal tissue plays an important role in the progression of the carcinomas to malignancy. 15701828_Over expression of FGFR3 is associated with urinary tract carcinoma progression 15772091_Activating mutations of FGFR3 are associated with benign skin tumors. 15788896_FGFR3 is an important cell survival and antiapoptotic factor for multiple myeloma cells 15869706_tumors with FGFR3 mutation appear to have distinctive clinical and biological characteristics that may help in defining a population of patients for FGFR3 mutation screening 15880580_Mutation in the FGFR3 is associated with progression of oral squamous cell car 15915095_Observational study of gene-disease association. (HuGE Navigator) 15915095_presence of a Pro250Arg mutation predisposed to an increased transcranial reoperation rate...on the basis of raised intracranial pressure...in apparently 'isolated' coronal synostosis 15940250_results indicate that FGFR 3 plays a crucial role in the accelerated proliferation of MM carrying t(4;14)(p16.3;q32) 16061860_FGFR3 and Tp53 mutations do not appear to be associated with progression of T1G3 transitional bladder carcinomas 16061860_Observational study of gene-disease association. (HuGE Navigator) 16091423_Data indicate that after endocytosis, fibroblast growth factor receptor (FGFR)4 and its bound ligand, FGF1, are sorted mainly to the recycling compartment, whereas FGFR1-3 with ligand are sorted mainly to degradation in the lysosomes. 16149130_Over-expression of FGFR3 may play an important role in liver carcinogenesis. 16210019_For the first time in humans, the expression of basic fibroblast growth factor (bFGF) and its receptors FGFR-2, FGFR-3, and FGFR-4 has been documented in ovaries of second- and third-trimester fetuses. 16274647_Review. The role of FGFR3 in endochondral ossification and mutations leading to chondrodysplasia are reviewed. 16278391_FGFR3 mutations do not seem to play a role in bladder cancer progression 16278395_fibroblast growth factor receptor 3 mutations have a role in development of bladder cancer 16288035_alternative splicing of FGFR3IIIb results in a secreted isoform that inhibits FGF1-induced proliferation 16384584_First quantitative measurement of mutant receptor tyrosine kinase (RTK) stabilization in the membrane domain environment of FGFR3 shows the profound effect of resultant increase in the dimer fraction on RTK-mediated signal transduction. 16410555_Results suggest a novel interaction between the SOCS1 and SOCS3 proteins and the FGFR3 signaling pathway. 16412606_FGFR3 expression is significantly associated with two important prognostic factors; stage and grade. Tumours with FGFR3+/p53- phenotype seem to have a distinctive pathway in bladder tumorigenesis. 16434832_Observational study of genetic testing. (HuGE Navigator) 16467200_PRO-001 antibody is a potent and specific inhibitor of FGFR3 and deserves further study for the treatment of FGFR3-expressing myeloma. 16532037_FGFR3 mutation status and loss of 9q is associated with early-stage bladder carcinomas 16570285_The detection of FGFR3 mutations in FUH (Flat Urothelial Hyperpalsias) supports the role of this lesion as precursor of papillary bladder cancer. 16634636_Gly380 --> Arg mutation does not alter dimerization energetics of FGFR3 transmembrane domain in detergent micelles or lipid bilayers. This indicates that pathogenesis in achondroplasia cannot be explained simply by higher dimerization of mutant FGFR3. 16685373_These results suggest that constitutive levels of both FGFR1 and FGFR3, but not FGFR4 are essential for FGF-stimulated anchorage-independent growth of SW-13 cells. 16778799_Mutations were detected in 23 of 27 (85%) adenoid seborrheic keratoses. R248C mutations were the most frequent mutation type. 16841094_The R248C mutation appears to be a hot spot for FGFR3 mutations in epidermal nevi. 16849642_MIP-1 alpha promoter function, gene expression, and protein secretion were each down-regulated following inhibition of FGFR3 signaling. 16877735_Observational study of gene-disease association. (HuGE Navigator) 16885334_PIK3CA mutations were strongly associated with FGFR3 mutations in superficial papillary bladder tumors. 17033969_identified a heterozygous missense mutation that is predicted to cause a p.R621H substitution in the tyrosine kinase domain and partial loss of FGFR3 function 17070479_Patients with TWIST gene mutations may have more ophthalmic abnormalities, including more strabismus, ptosis, NLDO, astigmatism, vertical deviations, and amblyopia compared with patients with FGFR3 gene mutations. 17114345_The present studies show that MUC1 associates with FGFR3. 17240035_The group of low-grade noninvasive papillary tumours is defined by the presence of an FGFR3 mutation 17255960_findings indicate that: (1) FGFR3 mutations occur in mosaicism and can cause epidermal nevi and (2) other genes are involved in epidermal nevi 17256796_double mutation in FGFR3 encoding GLY380LYS is responsible for Hypochondroplasia. 17320202_These results suggest that intracellular domain mutations define a distinct means by which mutated FGFR3 could disrupt bone development. 17343269_Observational study of gene-disease association. (HuGE Navigator) 17375526_FGFR3 gene mutation is found in thanatophoric dysplasia type 1 and bilateral cystic renal dysplasia. 17384684_the mutant S249C FGFR3 may have a role in bladder cancer 17392824_study showed that FGFR3 mutations appear to be common genetic alterations in multiple seborrheic keratoses with a varying interindividual mutation frequency but without specific intraindividual hot spots 17414280_no evidence that mosaicism for mutations, normally associated with syndromal forms of craniosynostosis, occur in single suture craniosynostosis 17507011_The cell model will be useful for the study of FGFR3 function in cartilage studies and future therapeutic approaches in chondrodysplasias. 17509076_Although mutation K650M induces defective targeting of receptor, different mechanism characterized by receptor retention at plasma membrane, excessive ubiquitylation and reduced degradation results from mutations of extracellular domain and stop codon. 17554105_the fibroblast growth factor family has a pleiotrophic function in human spermatogonia, which physiologically express FGFR3 17561467_Results suggest that a PLCgamma-STAT1 pathway mediates apoptotic signaling by FGFR3 in genetic dwarfism and chondrogenic cell lines. 17585316_Show preferential occurrence of FGFR3 mutations in seborrheic keratoses of the head and neck. Increased age appears to be a risk factor for these mutations. 17602067_CHEK2 mutation has a role in hereditary breast cancer 17621485_A child who has hypochondroplasia due to an N540K mutation and who has medial temporal lobe dysgenesis. 17624273_analysis of a Chinese family with autosomal dominant achondroplasia; disease locus was mapped to chromosome 4p16.3, where the FGFR3 gene is located; a novel Ser217Cys mutation in exon 5 of FGFR3 that causes autosomal dominant achondroplasia was identified 17668422_over-expression of FGFR3 protein in many tumours compared to normal bladder and ureteric controls. Increased expression was associated with mutation (85% of mutant tumours) 17683901_Observational study of genetic testing. (HuGE Navigator) 17785202_FGFR3 mediates hematopoietic transformation by activating RSK2 in a two-step fashion, promoting both the ERK-RSK2 interaction and subsequent phosphorylation of RSK2 by ERK. 17803960_Mutations were detected in 12 of 13 (92.3%) tumor tissues and 11 of 13 (84.6%) urine samples from patients with superficial bladder cancer. 17867592_FGFR3 may represent a prognostic marker of chromosomally stable bladder tumors with low malignant potential. 17867592_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17867603_Mutations are a possible prognostic tool in survival of urothelial carcinoma of the upper urinary tract. 17875876_previously undescribed, heterozygous lysine to threonine mutation at codon 650 of the FGFR3 gene in familial acanthosis nigricans 17950653_FGFR3 is a negative regulator of chondrocytes proliferation and differentiation in growth plate. 18000903_a K650Q mutation in the FGFR3 gene may have a role in Acanthosis nigricans [case report] 18000976_analysis of FGFR3 mutation in Muenke syndrome 18036184_MM Patients showing the t(4;14) chromosomal translocation at FGFR3 and MMSET genes had a significant elevation of serum crosslaps, reported to be the marker most reliably correlated with the extent of bone resorption 18072261_Strong immunohistochemical expression of FGFR3, a superficial staining pattern of CK20, and a low proliferative activity define those papillary urothelial neoplasms of low malignant potential that do not recur. 18166262_FGFR3 status allows a better risk stratification for patients with high-grade non-muscle-invasive UCC 18199430_Observational study of genetic testing. (HuGE Navigator) 18199430_The technique had a sensitivity and specificity of 100%. CONCLUSION: High-resolution melting analysis is a simple, rapid, and sensitive one tube assay for genotyping the FGFR3 gene. 18216705_No sequence variation was found, indicating that mutations in the 'hot spot' exons are not associated with nonsynostotic plagiocephaly. 18231572_In human uterine leiomyomas, FGFR3 were also overexpressed. 18231634_FGFR3 mutation frequency was significantly associated with tumor grade. 18336810_Pyridoxal-5'-phosphate-6-azophenyl-2', 4'-disulfonate reduced the tyrosine phosphorylation of FGF receptor type 3 triggered by FGF9 and the activation of the ERK1/2 pathway. 18485666_Results suggest that activated Spry2 may interfere with c-Cbl-mediated ubiquitination of FGFR3 by sequestering c-Cbl. 18503601_FGFR3 and PIK3CA germline mutations can be excluded as an underlying genetic basis, therefore alternative mechanisms have to contribute to familial Seborrheic Keratoses. 18528286_Aberrant FGFR3 staining may be seen infrequently in many forms of malignant lymphoma, although it is not the cause in most cases. 18567530_Results show that DNA methyltransferase 3B enhances polycomb protein 2-mediated transcriptional repression of FGFR3, and suggest that DNMT3B is a co-repressor of hPc2 in inducing transcriptional repression independent of DNA methylation. 18583390_A large pedigree with the clinical phenotype of Hypochondroplasia and Acanthosis nigricans due to a FGFR3 mutation, p.Lys650Thr. 18584939_FGFR3 mutation status might be used to select patients with invasive UCC who have a lower risk of death 18584939_Observational study of gene-disease association. (HuGE Navigator) 18642369_Mosaicism of FGFR3 caused an epidermal nevus syndrome with cerebral involvement. 18677770_Mutations in FGFR3 in melanoma by inhibition of nonsense-mediated mRNA decay. 18718050_Over-expression of fgfr3 gene exists in AL and CML patients, but expression was not related with chromosome abnormality. 18818193_The Muenke syndrome cohort showed significant low-frequency sensorineural hearing loss, and the Fgfr3(P244R) mice showed dominant penetrant hearing loss that was more severe than that in Muenke syndrome individuals. 18923003_FGFR3 and MAPK signaling in chondrocytes promote synchondrosis closure and fusion of ossification centers in human cases of homozygous achondroplasia and thanatophoric dysplasia. 18923003_Member of MAPK pathway, activation increases Bmp ligand mRNA expression. Premature synchondrosis associated with increased bone formation providing support for ACH and TD phenotype. 18945538_Observational study of gene-disease association. (HuGE Navigator) 19070887_The findings suggest that a thalidomide-based regimen may overcome the poor prognosis associated with a cyclin D1-negative or fibroblast growth factor receptor 3-positive phenotype. 19073250_Hearing loss in the Fgfr3(Y367C/+) mouse model demonstrates the crucial role of Fgfr3 in the development of the inner ear 19076977_results suggest that FGFR3 and PIK3CA mutations are involved in the pathogenesis of solar lentigo and occurrence of these mutations in both SL and SK suggests a common genetic basis. 19088846_STAT1 activation by six FGFR3 mutants associated with skeletal dysplasia undermines dominant role of STAT1 in FGFR3 signaling in cartilage. 19156776_Observational study of gene-disease association. (HuGE Navigator) 19215249_In Achondroplasia and Hypochondroplasia, genetic heterogeneity was present amongst the 70 clinically diagnosed patients with 5 different FGFR3 mutations identified. 19215249_Observational study of gene-disease association. (HuGE Navigator) 19255125_Amino acid substitution in FGFR3 inhibits growth plate cartilage and results in achondroplasias. 19286672_FGFR3 and p85 proteins interact in myeloma cells which express p85alpha and p85beta. 19287463_Point mutations of the FGFR3 gene were identified in 45% (9 of 20) of inverted papillomas 19327639_Observational study of gene-disease association. (HuGE Navigator) 19327639_frequency of mutations suggests a functional role for the FGFR3 receptor in the development of epithelial disorders 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19377444_FGFR3 does not seem to be central to the pathogenesis of prostate cancer, but it is significantly associated with a subgroup of low-grade prostate tumors, and with the finding of other tumors, mainly arising in bladder and skin. 19377444_Observational study of gene-disease association. (HuGE Navigator) 19381019_Results provide in vivo evidence demonstrating an oncogenic role of FGFR3 in bladder cancer and support antibody-based targeting of FGFR3 in hematologic and epithelial cancers driven by WT or mutant FGFR3. 19404844_elevated pERK expression occurs in urothelial carcinoma in the absence of B-Raf mutations and is not correlated with FGFR3 over-expression 19407216_Signaling through FGFR-3 sustains high levels of beta-catenin/Tcf-4-mediated transcriptional activity in Caco-2 cells. 19449410_Monozygotic twins had a de novo C749G mutation in the FGFr3 codon. 19449430_Thanatophoric dysplasia caused by double missense FGFR3 mutations is reported. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19551630_The G1138A mutation rate in the fibroblast growth factor receptor 3 (FGFR3) gene is increased in cells carrying the t (4; 14) translocation. 19594619_FGFR3 expression is not just an epiphenomenon in t(4;14) MM, but an important part of the malignant phenotype. 19621447_Observational study of gene-disease association. (HuGE Navigator) 19621447_mutations in the FGFR3 gene are not the earliest genetic alterations in bladder carcinogenesis and are associated with a hyperproliferative (hyperplastic) phenotype in the urothelium. 19694823_Point mutation of FGFR3 gene was identified in one case. 19722178_Observational study of gene-disease association. (HuGE Navigator) 19722178_Results show that occupational exposure to PAH influenced neither the frequency nor the spectrum of FGFR3 mutations and there was no direct relationship between these mutations and this occupational hazard. 19728793_The aim of this study was to investigate and compare FGFR expression in in vivo embryonic limb development and in vitro chondrogenesis of mesenchymal stem cells. 19729838_study uncovered a positive regulatory loop between FGFR3 and FOXN1 that underlies a benign versus malignant skin tumor phenotype 19752524_A mutation within the FGFR3 gene is associated with thanatophoric dysplasia type 1 in Chinese patients. 19761767_cell adaptation to activated FGFR3 include Sprouty4 activity, which silences the premature receptor signaling and suppress apoptosis. 19812598_Observational study of gene-disease association. (HuGE Navigator) 19821490_Study identified an overrepresentation of focal amplifications of known (FGFR3, CCND1, MYC, MDM2) and novel candidate genes (MYBL2, YWHAB and SDC4) in stage Ta bladder carcinoma. 19837276_AUthors report a novel case of amplification of the FGFR3 gene region, 4p16.3, in the absence of FGFR3-IGH@ fusion (i.e., no t(4;14) translocation) in a patient diagnosed with myeloma. 19838370_Observational study of gene-disease association and genetic testing. (HuGE Navigator) 19843069_Observational study of gene-disease association. (HuGE Navigator) 19843069_Urine FGFR3 mutation assay and cytological examination may be available in the future as complementary diagnostic modalities in postoperative management ofnon-muscle invasive bladder cancer. 19843843_Upregulation of FGFR3 is associated with bladder cancer. 19845664_Three of five stucco keratosis samples revealed a PIK3CA mutation(E542K, E545K), but no FGFR3 mutation was found. In contrast, both dermatosis papulosa nigra samples harboured an FGFR3 mutation (R248C, S249C) but no PIK3CA mutation. 19855393_A single cellular event for mutations arising in male germ cells that show a paternal age effect; screening of 30 spermatocytic seminomas for oncogenic mutations in 17 genes identified 2 mutations in FGFR3 and 5 mutations in HRAS. 19863427_We have analysed women with BRCA1/2-negative hereditary breast cancer to study whether these families might have mutations in Saethre-Chotzen-associated genes. 19888223_rhabdomyosarcoma cell lines include a minor subpopulation of FGFR3-positive sarcoma-initiating cells, which can be maintained indefinitely in culture and which is crucial for their malignancy. 19898608_many signaling pathways may be directly or indirectly altered by FGFR3, which has a crucial role of FGFR3 in the control of growth plate development 19901323_Results identify overlapping sets of co-modulated tyrosine phosphorylations to present an outline of an FGFR3 network in multiple myeloma. 19909015_Combined analysis of smoking, TP53, and FGFR3 mutations in Tunisian patients with invasive and superficial high-grade bladder tumors. 19909015_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20234367_mediates growth and migration in colorectal cancer 20393505_Ectopic expression of wild-type FGFR3 cooperates with MYC to accelerate development of B-cell lineage neoplasms. 20404005_Observational study of genetic testing. (HuGE Navigator) 20420824_FGFR3 mutation affects cell growth, apoptosis and attachment in keratinocytes 20538960_Observational study of gene-disease association. (HuGE Navigator) 20539070_FGFR3 contributes to malignancy in oral cancer cells and may be a useful biomarker fo early detection and da possible target for therapy. 20542753_Observational study of gene-disease association. (HuGE Navigator) 20542753_both mutations and overexpression of FGFR3 are correlated together, and are more prevalent in early stage (pTa and pT1) and low grade (G1 and G2) bladder tumors; survival analysis showed no contribution of changes in FGFR3 on patient survival 20634891_Observational study of gene-disease association. (HuGE Navigator) 20643727_Observational study of gene-disease association. (HuGE Navigator) 20643727_prevalence and complications of single gene and chromosomal disorders in craniosynostosis. 20646825_Observational study of gene-disease association. (HuGE Navigator) 20665023_New molecular data such as the distribution pattern of the fibroblast growth factor receptor 3 (FGFR3) has further supported the principle of low and high grade entities of urothelial carcinoma. 20673820_Observations support the notion that FGFR3 has a dual effect, as both a negative and a ositive regulator of the endochondral ossification process during post-natal bone development. 20711586_The R248C mutant keratinocytes revealed significantly enhanced cell growth compared with wildtype cells after reaching confluence. 20713021_Neu/V664E TM domain does not affect the phosphorylation levels of full-length FGFR3/A3 20717167_Observational study of gene-disease association. (HuGE Navigator) 20824703_Observational study of gene-disease association. (HuGE Navigator) 20824703_results suggest an inverse correlation between FGFR3 mutations and hypermethylation events, which may be used to improve noninvasive, DNA-based detection of bladder cancer 20890030_Loss-of-function mutation in FGFR3 causes achondroplasia/hypochondroplasia. (review) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21072204_Data show that of primary tumors 64% were mutant for FGFR3, 11% for RAS, 24% for PIK3CA, and 26% for p53. 21072204_Observational study of gene-disease association. (HuGE Navigator) 21116113_FGFR3 expression in invasive breast cancer was not found to be significantly associated with specific clinicopathological/molecular parameters, but might be used as a candidate marker for a poor prognosis. 21116115_Mutational activation of FGFR3 plays no important role in prostate carcinogenesis. 21119106_The extracellular domain of fibroblast growth factor receptor 3 inhibits ligand-independent dimerization. 21152424_Studies show that EGFR TKI-induced FGFR2 and FGFR3 signaling as a novel and rapid mechanism of acquired resistance to EGFR TKIs. 21225389_Hypochondroplasia due to FGFR3 gene mutation (N540K) 21264819_FGFR3 thanatophoric dysplasia mutations are detected homogenously in bladder carcinoma patients regardless of the tumor classification and tumor grade 21273290_fibroblast growth factor receptor 3 is a target of the homeobox transcription factor SHOX in limb development 21273588_Constitutively active FGFR3 with Lys650Glu mutation enhances bortezomib sensitivity in plasma cell malignancy. 21324899_FGFR3 heterodimerization in achondroplasia, the most common form of human dwarfism. 21356388_Fibroblast growth factor 1/fibroblast growth factor receptor 3 (FGF1/FGFR-3) signaling are mediators leading to the increased migration and invasion. 21383697_Findings suggest that the miR-99a-mTOR/FGFR3 pathway is crucial for controlling tumor growth in a wide range of human cancers that harbor upregulation of the Src-related oncogenic pathways. 21388956_Activation of the MAPK pathway by FGFR-3 is also required for the induction of Paneth cell markers in addition to and independent of the effect of FGFR-3 on TCF4/beta-catenin activity 21403567_The facial features of children with FGFR3Pro250Arg mutation (Muenke syndrome) differ from those with the other eponymous craniosynostotic disorders. We documented midfacial growth and forehead position after fronto-orbital advancement. 21451043_FGF signaling, through cooperation between Fgfr1 and Fgfr2 (but not Fgfr3), is required for initial generation of oligodendrocyte precursors in transgenic mouse ventral forebrain, with Fgfr1 being a stronger inducer than Fgfr2. 21459758_rs798766 was significantly associated with FGFR3 messenger RNA expression and results in bladder cancer. 21487019_establish FGFR3 as a strong Hsp90 client and suggest that modulating Hsp90 chaperone complexes may beneficially influence the stability and function of FGFR3 in disease. 21521775_FGFR3 was overexpressed in WM cells and its activation induced cell proliferation. 21536014_A391Emutation enhances the ligand-independent activation propensity of FGFR3. 21547910_results indicate that although FGFR3 mutations occur with lower frequency in high-grade tumours as compared to low-grade disease, FGFR3 mutation status defines a distinct morphological subtype of high-grade urothelial carcinoma 21577207_Our results provided novel information on which to base further mechanistic study of radiosensitization by inhibiting FGFR3 in human SCC cells 21683397_The sensitivity of tumor detection increased with tumor size. FGFR3 assay sensitivity depends on the number of shed tumor cells and improves by increasing urine volume 21779335_Data indicate that clathrin-mediated endocytosis is required for efficient internalization and downregulation of FGFR1 while FGFR3, however, is internalized by both clathrin-dependent and clathrin-independent mechanisms. 21792889_findings show FGFR3 expression is increased in tamoxifen resistant tumours compared with sensitive tumours and FGFR3 may play a role in promoting resistance to endocrine-therapy; hypothesise that FGFRs could play an integral part, not only in breast cancer development but also in resistance to endocrine-therapy 21815251_we have identified a novel submicroscopic duplication involving dosage sensitive genes TACC3, FGFR3, and LETM1. 21894939_FGFR3c, critical for bone development, elicit a similar response to Fgf1 and Fgf2 at low, but not at high, concentrations; results demonstrate the versatility of FGFR3c response to fgf1 and fgf2 and highlight the complexity in fgf signaling. 22038757_we have identified a previously undescribed FGFR3 mutation, p.Ala334Thr, in a family with mild craniosynostosis. 22045636_An array of vastly different diagnoses is caused by similar mutations in FGFR3, including syndromes affecting skeletal development, skin, and cancer. (Review) 22099989_FGFR3 mutations selectively identify patients with pT1 bladder cancer who have favorable disease characteristics. Further study may confirm that FGFR3 identifies those who would benefit from a conservative approach to the disease 22145492_Activating mutations of FGFR3 resulted in skeletal dysplasia and craniosynostosis in one twin brother. 22161110_The hereditary dwarfism featured by this family has been caused by hypochondroplasia due to a N540K mutation in the FGFR3 gene. 22188534_Mutations in fgfr3 proteins are present in benign lichenoid keratosis 22203473_The results argue against an involvement of mutational activation of FGFR3 in the development of RCC. 22285006_Studies indicate the prognostic impact of FGFR3 mutations in urothelial carcinoma (UC) as well as a potential target for therapeutics. 22329352_Data indicate that the level of FGFR3 expression provides additional prognostic information to t(4;14) in myeloma induction and consolidation therapy. 22375084_The researchers found an association between the FGFR3 gene and the prevalence of thanatophoric dysplasia and lethal osteogenesis imperfecta. 22401680_The author propose the acronym for Round and RAVEN Nevus and suggest the involvement of the FGFR3 gene in the pathogenesis. 22401682_A dozen similar case reports can be found in the medical literature, mostly under the term 'nevoid acanthosis nigricans'. Such a rash may be linked to postzygotic mosaicism with the same mutations of the FGFR3 gene. 22417847_we have analyzed the prevalence of somatic mutations in the FGFR3, PIK3CA, AKT1, KRAS, HRAS, and BRAF genes in bladder cancers 22422578_Highlight the crucial role of CDKN2A loss in the progression of non-muscle-invasive FGFR3-mutated bladder carcinomas and provide a potentially useful clinical marker for adapting the treatment of such tumours. 22448597_fibroblast growth factor receptor 3 gene mutation related to favorable bladder cancer prognosis 22529939_We find that this mutation does not increase FGFR3 phosphorylation and decreases FGFR3 cross-linking propensity, a finding which raises questions whether this mutation is indeed a genetic cause for human dwarfism. 22730329_Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. 22777346_in the adult human testis, FGFR3 expression is a feature of small clones of rarely dividing type A spermatogonia (Spg) which resemble 'undifferentiated' Spg, including the spermatogonial stem cells. 22837387_study reports that a small subset of glioblastoma multiforme tumors harbors oncogenic chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of fibroblast growth factor receptor genes(FGFR1 or FGFR3) to the transforming acidic coiled-coil coding domains of TACC1 or TACC3; the FGFR-TACC fusion protein displays oncogenic activity 22869148_The results show for the first time that acquired resistance to FGFR inhibitors can arise due to the emergence of a second site FGFR3V555M gatekeeper mutation. 22903874_Tthe 56.9% of hypochondroplasia patients with | ENSMUSG00000054252 | Fgfr3 | 3782.65826 | 0.9766887 | -0.0340292485 | 0.09262242 | 1.335405e-01 | 7.147890e-01 | 9.998360e-01 | No | Yes | 3190.44924 | 428.292358 | 2.946799e+03 | 305.446000 | |
| ENSG00000068079 | 3430 | IFI35 | protein_coding | P80217 | FUNCTION: Acts as a signaling pathway regulator involved in innate immune system response (PubMed:26342464, PubMed:29038465, PubMed:29350881). In response to interferon IFN-alpha, associates in a complex with signaling pathway regulator NMI to regulate immune response; the complex formation prevents proteasome-mediated degradation of IFI35 and correlates with IFI35 dephosphorylation (PubMed:10779520, PubMed:10950963). In complex with NMI, inhibits virus-triggered type I interferon/IFN-beta production (PubMed:26342464). In complex with NMI, negatively regulates nuclear factor NF-kappa-B signaling by inhibiting the nuclear translocation, activation and transcription of the NF-kappa-B subunit p65/RELA, resulting in the inhibition of endothelial cell proliferation, migration and re-endothelialization of injured arteries (PubMed:29350881). Beside its role as an intracellular signaling pathway regulator, also functions extracellularly as damage-associated molecular patterns (DAMPs) to promote inflammation when actively released by macrophage to the extracellular space during cell injury and pathogen invasion (PubMed:29038465). Macrophage-secreted IFI35 activates NF-kappa-B signaling in adjacent macrophages through Toll-like receptor 4/TLR4 activation, thereby inducing NF-kappa-B translocation from the cytoplasm into the nucleus which promotes the release of proinflammatory cytokines (PubMed:29038465). {ECO:0000269|PubMed:10779520, ECO:0000269|PubMed:10950963, ECO:0000269|PubMed:26342464, ECO:0000269|PubMed:29038465, ECO:0000269|PubMed:29350881}. | Alternative splicing;Cytoplasm;Direct protein sequencing;Immunity;Innate immunity;Nucleus;Phosphoprotein;Reference proteome;Secreted | hsa:3430; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular space [GO:0005615]; membrane [GO:0016020]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; innate immune response [GO:0045087]; macrophage activation involved in immune response [GO:0002281]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of NIK/NF-kappaB signaling [GO:1901223]; positive regulation of inflammatory response [GO:0050729]; positive regulation of innate immune response [GO:0045089]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of toll-like receptor 4 signaling pathway [GO:0034145] | 11911807_Dissociation of the IFN-induced protein IFP35 from NMI is a newly defined specific cytoplasmic event occurring during apoptosis. 17197158_The results provide a novel role of CKIP-1 in cytokine signaling response and the biochemical mechanism, by which two previously identified modulators IFP35 and Nmi are involved via interactions. 18305040_IFP35 can interact with the bovine Tas (BTas) regulatory protein of bovine foamy virus; overexpression of IFP35 disturbs the ability of BTas to activate viral-gene transcription and inhibits viral replication. 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 23226549_IRF-1 also activates IFP35 expression in an IFN-gamma-inducible manner. 24371060_IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. 25085622_Knockdown of IFP35 expression abolished pEGFP-N1-2C and pEGFP-N1-Nmi-induced activation of type I interferon promoters. 26342464_Trim21 regulates Nmi-IFI35 complex-mediated inhibition of innate antiviral response 27639618_Activation of TLR3 by poly IC induces the IFI35 expression in Mesangial Cells. Regional expression of IFI35 and its dysregulation may be involved in the pathogenesis of glomerular inflammation in CKD. 28064541_IFI35 enhances proliferation of mesangial cells and is regulated by MBD2 in lupus nephritis. 28109979_Study found that TLR3 signaling induces the expression of IFI35, and IFI35 negatively regulates IFN-beta-P-STAT1-RIG-I-CXCL10/CCL5 axis in U373MG cells. This suggests that IFI35 expressed in astrocytes may play an important role in regulating the innate immune system in astrocytoma cells. 29038465_Damage-associated molecular patterns (DAMP) are important mediators of innate immunity. Here the authors show that N-myc and STAT interactor (NMI) and interferon-induced protein 35 (IFP35) act as DAMPs to promote inflammation by activating macrophages via the Toll-like receptor 4 and NF-kappaB pathways. 29350881_inhibits both endothelial cell proliferation and migration by inhibiting the NF-kappaB/p65 pathway; functionally interacts with Nmi through its NID1 domain 32781067_IFI35 as a biomolecular marker of neuroinflammation and treatment response in multiple sclerosis. 34362845_IFP35 family proteins promote neuroinflammation and multiple sclerosis. | ENSMUSG00000010358 | Ifi35 | 105.75650 | 0.9530972 | -0.0693047302 | 0.29027278 | 5.713553e-02 | 8.110820e-01 | 9.998360e-01 | No | Yes | 94.81941 | 18.144197 | 9.333162e+01 | 14.944829 | ||
| ENSG00000068650 | 23250 | ATP11A | protein_coding | P98196 | FUNCTION: Catalytic component of a P4-ATPase flippase complex which catalyzes the hydrolysis of ATP coupled to the transport of aminophospholipids, phosphatidylserines (PS) and phosphatidylethanolamines (PE), from the outer to the inner leaflet of the plasma membrane (PubMed:25315773, PubMed:25947375, PubMed:26567335, PubMed:29799007, PubMed:30018401). Contributes to the maintenance of membrane lipid asymmetry with a specific role in morphogenesis of muscle cells. In myoblasts, mediates PS enrichment at the inner leaflet of plasma membrane, triggering PIEZO1-dependent Ca2+ influx and Rho GTPases signal transduction, subsequently leading to the assembly of cortical actomyosin fibers and myotube formation (PubMed:29799007). May be involved in the uptake of farnesyltransferase inhibitor drugs, such as lonafarnib. {ECO:0000269|PubMed:15860663, ECO:0000269|PubMed:25315773, ECO:0000269|PubMed:25947375, ECO:0000269|PubMed:26567335, ECO:0000269|PubMed:29799007, ECO:0000269|PubMed:30018401, ECO:0000305}. | ATP-binding;Cell membrane;Endoplasmic reticulum;Endosome;Lipid transport;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Translocase;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is an integral membrane ATPase. The encoded protein is probably phosphorylated in its intermediate state and likely drives the transport of ions such as calcium across membranes. [provided by RefSeq, Apr 2022]. | hsa:23250; | early endosome [GO:0005769]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; phospholipid-translocating ATPase complex [GO:1990531]; plasma membrane [GO:0005886]; recycling endosome [GO:0055037]; specific granule membrane [GO:0035579]; tertiary granule membrane [GO:0070821]; trans-Golgi network [GO:0005802]; ATP binding [GO:0005524]; ATPase-coupled intramembrane lipid transporter activity [GO:0140326]; magnesium ion binding [GO:0000287]; phosphatidylethanolamine flippase activity [GO:0090555]; phosphatidylserine flippase activity [GO:0140346]; phosphatidylserine floppase activity [GO:0090556]; in utero embryonic development [GO:0001701]; phospholipid translocation [GO:0045332]; positive regulation of myotube differentiation [GO:0010831] | 20043114_ATP11A is a useful predictive marker of metastasis in colorectal cancer patients. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20858683_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 27628304_The lipid flippases, ATP8B1 and ATP11A are novel elements of the innate immune response that are essential to attenuate the inflammatory response, possibly by mediating endotoxin-induced internalization of TLR4. 32596364_Disease Mutation Study Identifies Critical Residues for Phosphatidylserine Flippase ATP11A. 33808877_DNA Methylation at ATP11A cg11702988 Is a Biomarker of Lung Disease Severity in Cystic Fibrosis: A Longitudinal Study. 34403372_A sublethal ATP11A mutation associated with neurological deterioration causes aberrant phosphatidylcholine flipping in plasma membranes. 35278131_Autosomal dominant non-syndromic hearing loss maps to DFNA33 (13q34) and co-segregates with splice and frameshift variants in ATP11A, a phospholipid flippase gene. | ENSMUSG00000031441 | Atp11a | 453.80268 | 0.9619669 | -0.0559408441 | 0.18256632 | 8.726339e-02 | 7.676855e-01 | 9.998360e-01 | No | Yes | 469.41397 | 63.548046 | 4.464469e+02 | 46.799107 | |
| ENSG00000069020 | 375449 | MAST4 | protein_coding | O15021 | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:375449; | cytoplasm [GO:0005737]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cytoskeleton organization [GO:0007010]; intracellular signal transduction [GO:0035556]; peptidyl-serine phosphorylation [GO:0018105] | 17086981_high expression level of MAST4 in most normal human tissues, with an exception of in testis, small intestine, colon and peripheral blood leukocyte. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 29948376_Molecular studies incriminated different genes, mainly CACNA1H and MAST4. Since at least 2 susceptibility genes were likely shared by different populations, genetic factors involved in the majority of Tunisian Genetic generalized epilepsies families remain to be discovered. 33219327_Mast4 knockout shows the regulation of spermatogonial stem cell self-renewal via the FGF2/ERM pathway. 35064934_Estrogen-Responsive Gene MAST4 Regulates Myeloma Bone Disease. | ENSMUSG00000034751 | Mast4 | 270.56022 | 0.9725015 | -0.0402276531 | 0.21181763 | 3.620833e-02 | 8.490859e-01 | 9.998360e-01 | No | Yes | 581.51605 | 98.093284 | 5.776566e+02 | 75.903166 | |||
| ENSG00000069869 | 4734 | NEDD4 | protein_coding | P46934 | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Specifically ubiquitinates 'Lys-63' in target proteins (PubMed:23644597). Involved in the pathway leading to the degradation of VEGFR-2/KDFR, independently of its ubiquitin-ligase activity. Monoubiquitinates IGF1R at multiple sites, thus leading to receptor internalization and degradation in lysosomes. Ubiquitinates FGFR1, leading to receptor internalization and degradation in lysosomes. Promotes ubiquitination of RAPGEF2. According to PubMed:18562292 the direct link between NEDD4 and PTEN regulation through polyubiquitination described in PubMed:17218260 is questionable. Involved in ubiquitination of ERBB4 intracellular domain E4ICD. Involved in the budding of many viruses. Part of a signaling complex composed of NEDD4, RAP2A and TNIK which regulates neuronal dendrite extension and arborization during development. Ubiquitinates TNK2 and regulates EGF-induced degradation of EGFR and TNF2. Ubiquitinates BRAT1 and this ubiquitination is enhanced in the presence of NDFIP1 (PubMed:25631046). {ECO:0000269|PubMed:11598133, ECO:0000269|PubMed:17218260, ECO:0000269|PubMed:18305167, ECO:0000269|PubMed:18562292, ECO:0000269|PubMed:20086093, ECO:0000269|PubMed:21399620, ECO:0000269|PubMed:21765395, ECO:0000269|PubMed:23644597, ECO:0000269|PubMed:25631046}.; FUNCTION: (Microbial infection) Involved in the ubiquitination of Ebola virus protein VP40 which plays a role in viral budding. {ECO:0000269|PubMed:12559917}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Cytoplasm;Host-virus interaction;Membrane;Neurogenesis;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:23644597}. | This gene is the founding member of the NEDD4 family of HECT ubiquitin ligases that function in the ubiquitin proteasome system of protein degradation. The encoded protein contains an N-terminal calcium and phospholipid binding C2 domain followed by multiple tryptophan-rich WW domains and, a C-terminal HECT ubiquitin ligase catalytic domain. It plays critical role in the regulation of a number of membrane receptors, endocytic machinery components and the tumor suppressor PTEN. [provided by RefSeq, Jul 2016]. | hsa:4734; | apicolateral plasma membrane [GO:0016327]; cell cortex [GO:0005938]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendritic spine [GO:0043197]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; ubiquitin ligase complex [GO:0000151]; beta-2 adrenergic receptor binding [GO:0031698]; enzyme binding [GO:0019899]; phosphoserine residue binding [GO:0050815]; phosphothreonine residue binding [GO:0050816]; proline-rich region binding [GO:0070064]; protein domain specific binding [GO:0019904]; RNA polymerase binding [GO:0070063]; sodium channel inhibitor activity [GO:0019871]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; cellular response to UV [GO:0034644]; formation of structure involved in a symbiotic process [GO:0044111]; glucocorticoid receptor signaling pathway [GO:0042921]; lysosomal transport [GO:0007041]; negative regulation of sodium ion transmembrane transporter activity [GO:2000650]; negative regulation of sodium ion transport [GO:0010766]; negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage [GO:0010768]; negative regulation of vascular endothelial growth factor receptor signaling pathway [GO:0030948]; neuromuscular junction development [GO:0007528]; neuron projection development [GO:0031175]; positive regulation of nucleocytoplasmic transport [GO:0046824]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein catabolic process [GO:0045732]; progesterone receptor signaling pathway [GO:0050847]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein K63-linked ubiquitination [GO:0070534]; protein polyubiquitination [GO:0000209]; protein targeting to lysosome [GO:0006622]; protein ubiquitination [GO:0016567]; receptor catabolic process [GO:0032801]; receptor internalization [GO:0031623]; regulation of dendrite morphogenesis [GO:0048814]; regulation of ion transmembrane transport [GO:0034765]; regulation of macroautophagy [GO:0016241]; regulation of membrane potential [GO:0042391]; regulation of potassium ion transmembrane transporter activity [GO:1901016]; regulation of synapse organization [GO:0050807]; response to calcium ion [GO:0051592]; ubiquitin-dependent protein catabolic process [GO:0006511]; ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway [GO:0043162]; viral budding [GO:0046755] | 14645220_Endogenous Nedd4-2, but not Nedd4, negatively regulates the epithelial Na+ channel (ENaC) and is a component of a signaling pathway by which steroid hormones regulate ENaC. 15060076_Nedd4-mediated vascular endothelial growth factor receptor-2 degradation is prevented by Grb10 15126635_Nedd4.1 and Tsg101 act successively in the assembly process of HTLV-1 to ensure proper Gag trafficking through the endocytic pathway up to late endosomes where the late steps of retroviral release occur 15217910_Nedd4-2 acts on Na(v)1.5 by decreasing the channel density at the cell surface. the effect of Nedd4-2 requires the PY-motif of Nav1.5. 15252135_N4WBP5A acts as an adaptor to recruit Nedd4 family ubiquitin-protein ligases to the protein trafficking machinery 15548568_Data show that WWP2 and Nedd4-2 both bind to the cardiac sodium channel Na(v)1.5, but only Nedd4-2 robustly ubiquitinated and downregulated Na(v)1.5. 15703212_Both NEDD4 and ubiquitin protein ligase ubiquitin protein ligase Itch participate in the degradation of Melan-A 16867982_Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation 16885233_A new mechanistic model of DAT endocytosis is proposed whereby the PKC-induced ubiquitination of DAT mediated by Nedd4-2 leads to interaction of DAT with adaptor proteins. 17116753_These results demonstrate a novel mechanism by which the ubiquitin-ligase Nedd4, via interactions with GGA3 and cargo (LAPTM5), regulates cargo trafficking to the lysosome without requiring cargo ubiquitination. 17218260_NEDD4-1 is a potential proto-oncogene that negatively regulates PTEN via ubiquitination, a paradigm analogous to that of Mdm2 and p53. 17544362_G-protein-coupled receptor kinase 2 interacts not only with epithelial Na(+) channels, but also with both Nedd4 and Nedd4-2 17996703_Nedd4 is an E3 ligase that associates with and ubiquitylates RNAPII in response to UV-induced DNA damage in human cells. 18174164_Nedd4-2 reduces ENaC surface expression by altering its trafficking at two distinct sites in the endocytic pathway, inducing endocytosis of cleaved channels and targeting them for degradation. 18287095_a mechanistically novel function of ISG15 in the enhancement of the innate anti-viral response through specific inhibition of Nedd4 Ub-E3 activity 18305167_These data provide evidence of antiviral activity of ISG15 against Ebola virus and suggest a mechanism of action involving disruption of Nedd4 function and subsequent ubiquitination of VP40. 18307411_Results discuss the role of PTEN upon the E3 ubquitin ligase Nedd4 as a negative feedback regulator as well as a substrate. 18353951_UL56 regulates Nedd4 in HSV-2-infected cells, although deletion of UL56 had no apparent effect on viral growth in v 18544533_degradative fate of the beta(2)AR in the lysosomal compartments is dependent upon beta-arrestin2-mediated recruitment of Nedd4 to the activated receptor and Nedd4-catalyzed ubiquitination 18703514_decreased PMEPA1 expression frequently noted in prostate cancers may lead to increased AR functions and strengthen the biological role of the NEDD4-binding protein PMEPA1 in prostate cancers 18723765_The ubiquitin ligase Nedd4 mediates oxidized low-density lipoprotein-induced downregulation of insulin-like growth factor-1 receptor. 18772139_C2 domain of Nedd4 is ubiquitinated itself and as such is recruited by the ubiquitin-interacting motif of gamma2-adaptin for subsequent ubiquitin conjugation 18804462_Our results suggest human Nedd4 ligase inhibits yeast cell growth by disturbing the actin cytoskeleton, in part by increasing Las17p level, and that Nedd4 ubiquitination targets may include actin cytoskeleton-associated proteins conserved in evolution. 19024597_The increased expression of NEDD4-1 in malignant gastric and colorectal cells compared to their normal epithelial cells suggests that NEDD4-1 expression may play a role in colorectal and gastric cancer development. 19054764_NEDD4 is capable of binding to Spy1A and the dominant negative forms and knockdown of Nedd4 reduce ubiquitination and further degradation of Spy1A 19125695_Data found that hMTMR4 (human MTMR4) and Nedd4 co-immunoprecipitated and co-localized to late endosomes. 19366705_the Nedd4/proSP-C tandem is part of a larger protein complex containing a ubiquitinated component that further directs its transport 19602703_Nedd4-2 phosphorylation is emerging as a central convergence point for the regulation of epithelial Na(+) transport [REVIEW] 19670909_A low-energy barrier exists between different macrostates from the ubiquitin ligase Nedd4 fourth tryptophan (WW4) domain, placing it at the frontier of cooperative folding. 19812267_Here, the authors report that Vps4B and Nedd4.1 play critical roles in Marburg virus VP40-mediated budding. 19835589_These data suggest that UL56 regulates Nedd4 and functions to facilitate the cytoplasmic transport of virions from trans-Golgi network to the plasma membrane and/or release of virions from the cell surface. 19864419_Spry1 and Spry2, but not Spry3 or Spry4, associate with the HECT domain family E3 ubiquitin ligase, Nedd4. 19953087_Although several substrates were recognized by both Nedd4-1 and Nedd4-2, others were specific to only one, with several Tyr kinases preferred by Nedd4-1 and some ion channels by Nedd4- 20078934_NEDD4 gene polymorphisms are not associated with hypertension in Kazak Chinese population. 20078934_Observational study of gene-disease association. (HuGE Navigator) 20086093_These findings suggest an essential role of Nedd4-1 in regulation of EGFR degradation through interaction with and ubiquitination of ACK. 20172859_calcium-mediated membrane translocation through the C2 domain might be an activation mechanism of Nedd4 20332230_Results showed that overexpression of FoxM1B upregulated NEDD4-1, an E3 ligase that mediates the degradation and downregulation of phosphatase and tensin homologue (PTEN) in multiple cell lines. 20519395_Together these data support a model in which Alix recruits Nedd4-1 to facilitate HIV-1 release mediated through the LYPX(n)L/Alix budding pathway via a mechanism that involves Alix ubiquitination. 20530479_Nedd4-2 interacts with Na(v)1.6 via a Pro-Ser-Tyr(1945) motif in the C terminus of the channel and reduces Na(v)1.6 current density, and this regulation requires both the Pro-Gly-Ser-Pro motif in L1 and the Pro-Ser-Tyr motif in the C terminus 20559325_Data show that ARRDC3 interacts with NEDD4 through two conserved PPXY motifs and recruits NEDD4 to the activated beta2-adrenergic receptor. 20711176_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20819705_NEDD4, in combination with PTEN, provides signals for axonal branching in humans. 20855896_Nedd4-1 and beta-arrestin-1 as key regulators of NHE1 ubiquitylation, endocytosis, and function. 20889565_Data indicate that NEDD4-1 may play an important role in thedevelopment of NSCLC, and that NEDD4-1 overexpression may modulate the malignant phenotype of NSCLC cells, likely by regulating the intracellular levels of PTEN. 21308777_An E3 ligase, neural precursor cell expressed, developmentally down-regulated 4 (Nedd4), also was found as one of the important modulators of the p-Smad1 in both BMP-2 and TGF-beta1 action. 21338354_These data indicate that Nedd4 is the E3 ubiquitin ligase responsible for AMPA receptor ubiquitination 21765395_Nedd4-1 binds directly to and ubiquitylates activated FGFR1, by interacting primarily via its WW3 domain with a novel non-canonical sequence on FGFR1, regulating endocytosis & signalling during neuronal differentiation & embryonic development. 21936852_This present study described a regulatory link between Beclin 1 and the ubiquitin ligase Nedd4. 21953697_ubiquitination by Nedd4 targets alpha-synuclein to the endosomal-lysosomal pathway and, by reducing alpha-synuclein content, may help protect against the pathogenesis of Parkinson disease and other alpha-synucleinopathies 21986318_NEDD4 upregulated expressions of fibronectin and type 1 collagen and contributed to the excessive accumulation of extracellular matrix. 22096579_Data suggest a role for the ubiquitin ligase Nedd4 in membrane sorting of LAPTM4 proteins. 22467858_Data reveal a novel Rac1-dependent signalling pathway that, through Nedd4-mediated ubiquitylation of Dvl1, stimulates the maturation of epithelial cell-cell contacts. 22875931_This study demonistrated that the first time that zinc can induce IGF-1Rbeta protein degradation in neurons via NEDD4-mediated ubiquitination and the subsequent UPS mechanism. 22974840_The expression of NEDD4 did not influence the PTEN subcellular localization or protein level. 22992774_Insertion mutation in the pentanucleotide repeat of NEDD4 gene is associated with spinocerebellar ataxia type 31. 23106887_Nedd4 positively regulates the Hh pathway and provides a potential target for inhibiting Hh signaling in cancer therapy. 23180960_Letter: NEDD4-1 expression did not correlate with PTEN expression changes during gastric carcinogenesis. 23195959_Ubiquitination of phosphoAKT by NEDD4-1 is coupled to AKT activation at the plasma membrane by insulin-like growth factor-1 (IGF-1), which promotes phosphoAKT nuclear trafficking. 23262292_Understanding of the contribution of Nedd4 and Nedd4-2 in regulating key functions in the brain is shedding new light on the ubiquitination signal not only in orchestrating degradation events but also in protein trafficking 23460917_This suggests NEDD4-1 functions in conjunction with other post-translational mechanisms to regulate Thoc1 protein and THO activity. 23549616_NEDD4-1 overexpression leads to PPARgamma ubiquitination and reduced expression of PPARgamma. 23589096_DeltaNp63alpha-mediated suppression of nuclear PTEN in basal layer keratinocytes occurs through repression of NEDD4-1. 23644597_The donor ubiquitin, transferred from the E2, is bound to the Nedd4 C lobe with its C-terminal tail locked in an extended conformation, primed for catalysis. 23964096_Nedd4 as a regulator of noncoding RNAs that are generated by stalled RNA polII genome-wide. 24107629_NEDD4 directly interacts with LATS1, leading to ubiquitination and decreased levels of LATS1 and, thus, increased YAP localization in the nucleus, which subsequently increases the transcriptional activity of YAP. 24340059_NEDD4-1 regulates cell migration and invasion through ubiquitination of CNrasGEF in vitro. 24413081_Data show that NEDD4-1 E3 ligase as a novel component of the tumor suppressor protein p53/proto-oncogene proteins c-mdm2 regulatory feedback loop that controls p53 activity during stress responses. 24496234_Single nucleotide polymorphism of rs8032158 may influence keloid severity. 24657926_Findings revealed the CKI/SCF(beta-TRCP) signaling axis as the upstream negative regulator of NEDD4. 24662824_Results demonstrated that NEDD4 is a novel interaction partner of HER3 and the C-terminal tail of HER3 interacts with the WW domains of NEDD4. 24743017_Relevant interaction partners of GUCD1 were identified, direct interactions with NEDD4-1 (E3 ubiquitin protein ligase neural precursor cell expressed, developmentally downregulated gene 4) was revealed, the latest resulted in control of GUCD1 stability. 24746824_Nedd4-1 as a general E3 ubiquitin ligase controls the abundance of all the three major forms of Ras, revealing a fundamental role of Nedd4-1 in regulating Ras signaling. 24831002_Lys-63 ubiquitination by Nedd4-1 facilitates endosomal targeting of alpha-synuclein. 24907641_Findings suggest that the NEDD4-PTEN pathway is dysregulated in myotonic dystrophy type 2 skeletal muscle. 25216516_NEDD4-1 overexpression sensitizes cancer cells to etoposide-induced apoptosis by reducing SAG levels through targeted degradation. SAG is added to a growing list of NEDD4-1 substrates and mediates its biological function. 25292214_Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. 25395181_Nedd4-1 is an exceptional prognostic biomarker for GCA. 25438670_Studies on Smurf2 and Nedd4 show that the C2 domain has the potential to regulate E3 activity by maintaining the HECT domain in a low-activity state where its ability for transthiolation and noncovalent Ubiquitin binding are impaired. 25527121_NEDD4 has been shown to play a critical role in the regulation of a number of membrane receptors, endocytic machinery components and the tumour suppressor PTEN. [Review] 25692647_NEDD4 controls the Hippo pathway leading to coordinated cell proliferation and apoptosis 25809873_Nedd4 promoted autophagy and was associated with the mTOR signalling pathway in the prostate cancer cells. 25823820_sNEDD4 is a novel metastasis-associated gene, which prevents apoptosis under nutrient restriction conditions. This support the prognostic potential of sNEDD4 for Hepatocellular carcinoma. 25879670_Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. 25883222_Data show that silencing of PMEPA1 protein facilitates the growth of prostate cancer cells and modulates androgen receptor (AR) through NEDD4 ubiquitin protein ligase and PTEN protein. 26006083_Data show that human ABCB1 is a substrate for human NEDD4-1 and identified 8 lysine residues that were ubiquitinated by NEDD4-1. 26037357_rs3088077, rs7162435 and rs2303579 of the NEDD4 gene are associated with schizophrenia. 26193427_Study identifies NEDD4 as a putative oncogene in endometrial cancer that may augment activation of the IGF-1R/PI3K/Akt signaling pathway. 26263374_we demonstrate that the E3 ubiquitin ligase NEDD4 ubiquitinates IFITM3 in cells and in vitro. 26296893_HUWE1 and NEDD4-1 are two E3 ligases that are fundamental enzymes in the post-translational regulation of ABCG1 and ABCG4 protein levels and cellular cholesterol export activity 26503960_Elevated NEDD4 is implicated as the responsible ubiquitin E3 ligase for HSF1 degradation through ubiquitin-proteasome system. Aberrant HSF1 degradation is a key neurodegenerative mechanism underlying alpha-synucleinopathy. 26685112_These results show that the binding of the hNedd4-1 WW3* domain to alpha-hENaC is coupled to the folding equilibrium 26823285_both Nedd4-1 and Nedd4-2 are important regulators for hOAT1 ubiquitination, expression, and function 26854353_LRAD3 is a component of pathways that function effectively to modulate Itch and Nedd4 auto-ubiquitination and levels. 26904956_Upregulation of long non-coding RNA PAPAS in response to hypoosmotic stress does not increase H4K20me3 because of Nedd4-dependent ubiquitinylation and proteasomal degradation of Suv4-20h2. 27173227_we found that Ndfipl binds to Nedd4-1, and that increased expression of Ndfipl significantly reduced Itch expression. We also found that increased ubiquitination played a role in Ndfipl-mediated processes, and that Ndfipl and a-synuclein interact 27226107_this is the first demonstration that Nedd4-1 regulates hOAT1 ubiquitination, expression, and transport activity through its WW2 and WW3 domains 27467187_Data suggest a possible molecular mechanism might be that NEDD4 silence led to an increase in PTEN (phosphatase and tensin homologue) expression. 27494837_NEDD4 ligase regulates RTP801 and is sensitive to Parkinson disease-associated oxidative stress 28085563_SQSTM1 is ubiquitinated by NEDD4 while LC3 functions as an activator of NEDD4 ligase activity. 28126338_We concluded that USP15 attenuates IGF-I signaling by antagonizing Nedd4-induced IRS-2 ubiquitination. 28300060_Data indicate the mechanism for epigenetic regulation in cancer by inducing E3 ubiquitin ligase NEDD4-dependent histone H3 ubiquitination. 28320822_Two additional regulators of BST2 constitutive ubiquitylation and sorting to the lysosomes: the E3 ubiquitin ligases NEDD4 and MARCH8, are reported. 28379054_NEDD4 is largely involved in epithelial-mesenchymal transition features and chemoresistance of nasopharyngeal carcinoma cancer cells. 28405688_Nedd4-1 inhibited Rap2a activity, and promoted the migration and invasion of glioma cells. 28423617_Our results reveal a novel mechanism of upregulation of NEDD4 expression in pancreatic adenocarcinoma 28437168_In the present study, we identified that the adaptor protein Numb, which is demonstrated to be a novel binding partner of NEDD4-1, plays important roles in controlling PTEN ubiquitination through regulating NEDD4-1 activity and the association between PTEN and NEDD4-1. 28470758_Insights into links between autophagy and the ubiquitin system showed that LC3B-binding can steer intrinsic NEDD4 E3 ligase activity. 28475870_Describe an autoinhibitory mechanism for NEDD4-1 ubiquitin ligase involving a linker-HECT domain interaction. This intramolecular interaction traps the HECT enzyme in its inactive state and can be relieved by linker phosphorylation. 28627598_We further investigated whether curcumin exerts its antitumor activities via suppressing NEDD4 expression. We found that curcumin reduced the expression of NEDD4 and Notch1 and pAKT, leading to glioma cell growth inhibition, apoptosis, and suppression of migration and invasion. Moreover, deletion of NEDD4 expression enhanced the sensitivity of glioma cells to curcumin treatment 28666866_NEDD4-1 recognizes SERTA domain containing proline rich region of p34SEI-1. Residues of NEDD4-1 responsible for direct interaction with p34SEI-1 are identified by NMR titration experiments. 28714370_Our results demonstrate that NEDD4 may promote the acquired resistance of non-small-cell lung cancer cells to erlotinib by decreasing phosphatase and tensin homolog deleted on chromosome 10 protein expression. Targeted decrease in NEDD4 expression may be a potential therapeutic strategy for tyrosine kinase inhibitor-resistant non-small-cell lung cancer. 28733455_an oncogenic E3 ubiquitin ligase promotes loss of gap junctions and Cx43 degradation in human carcinoma cells. 28745938_NEDD4 exerts its oncogenic function partly due to regulation of PTEN and Notch-1 in bladder cancer cells. 29021346_NEDD4 is an autophagic E3 ubiquitin ligase that ubiquitylates SQSTM1, facilitating SQSTM1-mediated inclusion body autophagy. 29251248_NEDD4 participates in killing of intracellular bacterial pathogens via autophagy by sustaining the stability of BECN1. 29271375_NEDD4-1 may regulate the proliferation, invasion, migration, and chemoresistance of lung ADC cells through the PI3K/Akt pathway, suggesting that it may be regarded as a therapeutic target for the treatment of lung ADC. 29455656_NEDD4 mediates the EGFR lung cancer cell migration signaling through promoting lysosomal secretion of cathepsin B. 29463679_Authors conclude that recruitment of alpha arrestins to membrane receptors and aggregation of unstable proteins after heat shock may be physiologically relevant mechanisms for triggering ubiquitination by Nedd4 family HECT domain-containing E3 ligases. 29480061_NEDD4 may be involved in the HCC progression via regulating LATS1 associated signaling pathway. 29582580_Streptococcus pneumoniae is entrapped by selective autophagy in pneumolysin- and ubiquitin-p62-LC3 cargo-dependent manners. Streptococcus pneumoniae-containing autophagic vesicles (PcAV) were formed only in the presence of Rab41-positive intact Golgi apparatuses. Nedd4-1 was recruited to PcAV which played a pivotal role in K63-linked polyubiquitin chain (K63Ub) generation on PcAV and elimination of intracellular bacteria. 29851245_The results suggest that PIP5KA is a novel degradative substrate of NEDD4 and that the PIP5KA-dependent phosphatidylinositol 4,5-diphosphate pool contributing to breast cancer cell proliferation through PI3K/Akt activation is negatively controlled by NEDD4. 30273597_Study shows that the high frequency of risk allele C in rs8032158 in keloid patients is associated with a selectively higher expression of transcript variant 3 (TV3) of NEDD4 to activate the NF-kappaB pathway. Results suggest that NEDD4 TV3 is a potential diagnostic marker and therapeutic target for chronic skin diseases, including keloid. 30367623_Low HER3 expression is suggested to be a valuable prognostic biomarker to predict recurrence in HER2-amplified breast cancer. 30708959_We found that AMOTL1 could link together parainfluenza virus 5 M proteins and NEDD4 family proteins. 30854719_Distinguishing the optimal binding mechanism of an E3 ubiquitin ligase: Covalent versus noncovalent inhibition. 31159502_Alix serves as a co-factor for the interaction between the E3-ubiquitin ligase NEDD4-1 and the ABC transporter targets, ABCG1 and ABCG4. 31373553_These findings provide a model in which Nedd4 and beta-arrestin act together as a complex to regulate mGlu7 surface expression and function at presynaptic terminals. 31390487_Clinically, low NEDD4-1 expression has been linked to poor prognosis in patients with multiple myeloma (MM). Functionally, NEDD4-1 knockdown resulted in bortezomib resistance in MM cells in vitro and in vivo. 31423749_PPARgamma induces NEDD4 gene expression to promote autophagy and insulin action. 31451218_NEDD4 NEDD4 NEDD4 NEDD4 31578285_We found that in addition to influencing catalytic activities, the WW domain linker regions in NEDD4-1 and WWP2 can impact product distribution, including the degree of polyubiquitination and Lys-48 versus Lys-63 linkages. We show that allosteric activation by NDFIP1 or engineered ubiquitin variants is largely mediated by relief of WW domain linker autoinhibition. 31636332_Binding site plasticity in viral PPxY Late domain recognition by the third WW domain of human NEDD4. 31690112_NEDD4 regulates hydrogen peroxide-induced cell proliferation and death through inhibition of Hippo signaling. 31831018_Results suggest that NEDD4 could interact with p21, increases p21 ubiquitylation and target p21 protein for degradation. 31849325_In patients of Chinese Han population, polymorphisms in NEDD4 to both the pathogenesis of schizophrenia and cognitive dysfunction. 31856858_NEDD4 expression is elevated in BC and is associated with BC growth. NEDD4 correlated with clinicopathological parameters and predicts a poor prognosis. Thus, NEDD4 is a potential biomarker of poor prognosis and a potential therapeutic target for BC treatment. 31867777_NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. 31974380_Nedd4 role in the melanoma drug resistance to erastin.Erastin-induced resistance mediated by FOXM1-Nedd4-VDAC2/VDAC3 negative feedback loop in melanoma.Nedd4 ubiquitinates and degrades VDAC2 and VDAC3. 32101753_Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. 32171886_NEDD4 E3 ligase: Functions and mechanism in human cancer. 33037408_PRRG4 promotes breast cancer metastasis through the recruitment of NEDD4 and downregulation of Robo1. 33530829_NEDD4 triggers FOXA1 ubiquitination and promotes colon cancer progression under microRNA-340-5p suppression and ATF1 upregulation. 33836347_LAPTM4alpha is targeted from the Golgi to late endosomes/lysosomes in a manner dependent on the E3 ubiquitin ligase Nedd4-1 and ESCRT proteins. 33962630_NEDD4 regulates ubiquitination and stability of the cell adhesion molecule IGPR-1 via lysosomal pathway. 34512149_NEDD4L-induced beta-catenin ubiquitination suppresses the formation and progression of interstitial pulmonary fibrosis via inhibiting the CTHRC1/HIF-1alpha axis. 34638586_The E3 Ubiquitin Ligase NEDD4-1 Mediates Temozolomide-Resistant Glioblastoma through PTEN Attenuation and Redox Imbalance in Nrf2-HO-1 Axis. 35031788_Oncogenic E3 ubiquitin ligase NEDD4 binds to KLF8 and regulates the microRNA-132/NRF2 axis in bladder cancer. 35162941_The Ubiquitin E3 Ligase Nedd4 Regulates the Expression and Amyloid-beta Peptide Export Activity of P-Glycoprotein. 35167936_NEDD4 degrades TUSC2 to promote glioblastoma progression. 35409239_The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. | ENSMUSG00000032216 | Nedd4 | 97.44902 | 0.8299222 | -0.2689519638 | 0.35751540 | 5.123370e-01 | 4.741289e-01 | 9.998360e-01 | No | Yes | 127.79318 | 30.085904 | 1.328554e+02 | 24.150024 |
| ENSG00000071054 | 9448 | MAP4K4 | protein_coding | O95819 | FUNCTION: Serine/threonine kinase that may play a role in the response to environmental stress and cytokines such as TNF-alpha. Appears to act upstream of the JUN N-terminal pathway. Phosphorylates SMAD1 on Thr-322. {ECO:0000269|PubMed:21690388, ECO:0000269|PubMed:9890973}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Kinase;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene is a member of the serine/threonine protein kinase family. This kinase has been shown to specifically activate MAPK8/JNK. The activation of MAPK8 by this kinase is found to be inhibited by the dominant-negative mutants of MAP3K7/TAK1, MAP2K4/MKK4, and MAP2K7/MKK7, which suggests that this kinase may function through the MAP3K7-MAP2K4-MAP2K7 kinase cascade, and mediate the TNF-alpha signaling pathway. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:9448; | cytoplasm [GO:0005737]; focal adhesion [GO:0005925]; ATP binding [GO:0005524]; creatine kinase activity [GO:0004111]; microtubule binding [GO:0008017]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; intracellular signal transduction [GO:0035556]; MAPK cascade [GO:0000165]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell-matrix adhesion [GO:0001953]; neuron projection morphogenesis [GO:0048812]; positive regulation of ARF protein signal transduction [GO:0032014]; positive regulation of cell migration [GO:0030335]; positive regulation of focal adhesion assembly [GO:0051894]; positive regulation of focal adhesion disassembly [GO:0120183]; positive regulation of GTPase activity [GO:0043547]; positive regulation of keratinocyte migration [GO:0051549]; protein phosphorylation [GO:0006468]; regulation of JNK cascade [GO:0046328]; regulation of MAPK cascade [GO:0043408] | 12387898_interaction with GBP3 12612079_The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. 14966141_MAP4K4 is a putative effector of Rap2, a Ras family small GTP-binding protein, mediating the activation of JNK by Rap2 16537454_studies of the promigratory role of MAP4K4 showed that the knockdown of this transcript inhibited the migration of multiple carcinoma cell lines, indicating a broad role in cell motility 17227768_MAP4K4 silencing prevents TNFa-induced insulin resistance in human skeletal muscle and restores appropriate signaling inputs to enhance glucose uptake 18981001_MAP4K4 expression significantly correlated with overall and recurrence-free survival (P=0.025 and 0.004) independent of age, tumor size, differentiation, and stage. 19690174_Results identify MAP4K4 as a key upstream mediator of TNF-alpha action on the beta cell, preventing tumor necrosis factor-alpha-induced decrease of IRS-2 and inhibition of glucose-stimulated insulin secretion. 20857524_Data show that down-regulation of hepatocyte progenitor kinase-like kinase (HGK) can obviously inhibit the migration and invasion of HepG2 cells in vitro, and suggest that HGK may be a new therapeutic target for treatment of hepatocellular carcinoma. 21196414_MAP4K4 overexpression is an independent predictor of poor prognosis of Hepatocellular Carcinoma patients, and inhibition of its expression might be of therapeutic significance 22824148_Elevated MAP4K4 expression is closely associated with lung adenocarcinoma progression with independent prognostic value in predicting overall survival for patients with lung adenocarcinoma. 23094072_Common polymorphisms in MAP4K4 are associated with insulin resistance and beta-cell dysfunction, possibly via this gene's role in inflammatory signaling. 23543740_TRAF1.NIK is a central complex linking canonical and non-canonical pathways by disrupting the TRAF2-cIAP2 ubiquitin ligase complex 24233838_results reveal a key target of SOX2 expression and highlight the unexpected context-dependent role for MAP4K4, a pluripotent activator of several mitogen-activated protein kinase pathways, in regulating tumor cell survival. 24244164_that MAP4K4, a known mediator of inflammation, is involved in KS aetiology by regulating KSHV lytic reactivation 25799996_loss of MAP4K4 function suppressed pathological angiogenesis in disease models, identifying MAP4K4 as a potential therapeutic target 26549737_The results of the present study suggested that inhibition of MAP4K4 may be a therapeutic strategy for gastric cancer. 26722431_miR-194 regulated the progression of hepatocellular carcinoma through directly inhibiting the expression of MAP4K4 26918832_This report identifies HGK methylation/downregulation in T cells as a potential biomarker for non-obese type 2 diabetes 27010469_MAP4K4 promotes the epithelial-mesenchymal transition and invasiveness of hepatocellular carcinoma cells 27044870_Results reveal that endothelial Map4k4 is critical for lymphatic vascular development by regulating endothelial cells (EC)quiescence and lymphatic EC fate. 27174326_of the five variants, SNP rs2236935T/C was significantly associated with type 2 diabetes mellitus (T2DM) in this study population; conclude that MAP4K4 gene is associated with T2DM in a Chinese Han population, and MAP4K4 gene variants may contribute to the risk toward the development of T2DM 28061846_Interactions between MAP4K4 gene variants and environmental factors may contribute to MAP4K4 attenuation in T cells, leading to non-obese T2D. 28306189_these findings identify MAP4K4 as a novel MAPK/ERK pathway regulator in lung adenocarcinoma that is required for lung adenocarcinoma maintenance. 28771231_the close proximity between AcSDKP and FGFR1 was essential for the suppression of TGFbeta/smad signaling and EndMT associated with MAP4K4 phosphorylation (P-MAP4K4) in endothelial cells. 28855631_Results show that MAP4K4 is a target gene for the novel miR-TG. Its expression is up-regulated in papillary thyroid carcinoma (PTC) which is inversely correlated with that of miR-TG. 29138007_RBM4-SRSF3-MAP4K4 constitutes a novel mechanism for manipulating the metastasis of colorectal cancer cells through the JNK1 signaling pathway. 29461619_miR-200c could suppress cervical cancer cell proliferation and progression via regulating MAP4K4, which might provide a new target for cervical cancer diagnosis and therapy 29620289_an anti-cancer effect of microRNA-141 on breast cancer by cytotoxic CD4+ T cells through MAP4K4 expression. 29970191_These results suggest that downregulation of miR-98-5p promotes tumor development by downregulation of MAP4K4 and inhibition of the downstream MAPK/ERK signaling 30258193_TIIA triggered HGK/JNK1-dependent Jun activation and led to increased Jun recruitment to AP-1-binding site in the SESN2 promoter region. 30429233_Our findings showed that 5-FU inhibited malignant behavior of human colorectal cancer (CRC) cells in vitro and in vivo by enhancing the efficiency of miR-141 Our data suggested that targetting the miR-141/MAP4K4 signaling pathway could be a potential molecular target that may enhance chemotherapeutic efficacy in the treatment of CRC 30699345_These studies highlight a MAP4K4-initiated signaling cascade that induces motor neuron degeneration. 31570734_Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. 31913126_STRIPAK directs PP2A activity toward MAP4K4 to promote oncogenic transformation of human cells. 32220977_MAP4K4 negatively regulates CD8 T cell-mediated antitumor and antiviral immunity. 34112843_HGK promotes metastatic dissemination in prostate cancer. 34511598_MAP4K4 promotes pancreatic tumorigenesis via phosphorylation and activation of mixed lineage kinase 3. 34930918_MAP4K4 mediates the SOX6-induced autophagy and reduces the chemosensitivity of cervical cancer. 35026645_MiR-181c suppresses triple-negative breast cancer tumorigenesis by targeting MAP4K4. | ENSMUSG00000026074 | Map4k4 | 3692.21897 | 0.9628257 | -0.0546533892 | 0.11370698 | 2.336076e-01 | 6.288617e-01 | 9.998360e-01 | No | Yes | 3569.95206 | 388.350826 | 3.889872e+03 | 326.816969 | |
| ENSG00000072163 | 55679 | LIMS2 | protein_coding | Q7Z4I7 | FUNCTION: Adapter protein in a cytoplasmic complex linking beta-integrins to the actin cytoskeleton, bridges the complex to cell surface receptor tyrosine kinases and growth factor receptors. Plays a role in modulating cell spreading and migration. {ECO:0000269|PubMed:12167643}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Disease variant;LIM domain;Limb-girdle muscular dystrophy;Membrane;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Zinc | This gene encodes a member of a small family of focal adhesion proteins which interacts with ILK (integrin-linked kinase), a protein which effects protein-protein interactions with the extraceullar matrix. The encoded protein has five LIM domains, each domain forming two zinc fingers, which permit interactions which regulate cell shape and migration. A pseudogene of this gene is located on chromosome 4. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2011]. | hsa:55679; | cell-cell junction [GO:0005911]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; cell-cell adhesion [GO:0098609]; cell-cell junction organization [GO:0045216]; negative regulation of apoptotic process [GO:0043066]; negative regulation of hepatocyte proliferation [GO:2000346]; negative regulation of neural precursor cell proliferation [GO:2000178]; positive regulation of integrin-mediated signaling pathway [GO:2001046]; positive regulation of substrate adhesion-dependent cell spreading [GO:1900026] | 12167643_These results identify a novel nuclear and focal adhesion protein that associates with ILK and reveals an important role of PINCH-2 in the regulation of the PINCH-1-ILK interaction, cell shape change, and migration. 16959213_LIMS2 may be useful as a molecular biomarker and a therapeutic target by increasing its expression and activity in gastric cancer 20237496_Observational study of gene-disease association. (HuGE Navigator) 20500520_PINCH-2 mRNA is overexpressed in malignant mesothelioma 25346044_Results defined the functional role of copy number variations involving PINCH-2 in cancer progression based on the field cancerization effect; cell migration and invasion through autocrine and paracrine function as part of the field cancerization effect. 25589244_Data indicate compound heterozygous missense mutations that are predicted to be pathogenic in LIM and senescent cell antigen-like domains 2 protein (LIMS2). 27590440_Mammalian cells have two functional PINCH proteins, PINCH1 and PINCH2. PINCH not only binds to Nck2 and engages in the signaling of growth factor receptors, but also forms a ternary complex with ILK and parvin (IPP complex). 32752897_Fluid shear stress modulates endothelial inflammation by targeting LIMS2. | ENSMUSG00000024395 | Lims2 | 37.45649 | 1.2431129 | 0.3139573438 | 0.48365536 | 4.340513e-01 | 5.100077e-01 | 9.998360e-01 | No | Yes | 51.38591 | 13.764338 | 3.942174e+01 | 10.933330 | |
| ENSG00000073350 | 3993 | LLGL2 | protein_coding | Q6P1M3 | FUNCTION: Part of a complex with GPSM2/LGN, PRKCI/aPKC and PARD6B/Par-6, which may ensure the correct organization and orientation of bipolar spindles for normal cell division. This complex plays roles in the initial phase of the establishment of epithelial cell polarity. {ECO:0000269|PubMed:15632202}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Cytoplasm;Exocytosis;Phosphoprotein;Reference proteome;Repeat;WD repeat | The lethal (2) giant larvae protein of Drosophila plays a role in asymmetric cell division, epithelial cell polarity, and cell migration. This human gene encodes a protein similar to lethal (2) giant larvae of Drosophila. In fly, the protein's ability to localize cell fate determinants is regulated by the atypical protein kinase C (aPKC). In human, this protein interacts with aPKC-containing complexes and is cortically localized in mitotic cells. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:3993; | cortical actin cytoskeleton [GO:0030864]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; plasma membrane [GO:0005886]; GTPase activator activity [GO:0005096]; myosin II binding [GO:0045159]; PDZ domain binding [GO:0030165]; cell division [GO:0051301]; cortical actin cytoskeleton organization [GO:0030866]; establishment of spindle orientation [GO:0051294]; exocytosis [GO:0006887]; leucine transport [GO:0015820]; regulation of establishment or maintenance of cell polarity [GO:0032878]; regulation of Notch signaling pathway [GO:0008593]; regulation of protein secretion [GO:0050708] | 15632202_binding between Lgl2 and LGN play a role in mitotic spindle organization through regulating formation of the LGN.NuMA complex; Lgl2 forms a Lgl2.Par-6.aPKC.LGN complex, which responds to mitotic signaling to establish normal cell division 18155665_The identification and functional characterization of the promoter region ( approximately 1.2kb) of the Hugl-2 gene are reported. 18199550_Retention of Lgl2 expression is critical for the epithelial phenotype;its loss might be involved in metastasis. 19403135_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19407852_We propose that Lgl2 may be a potential marker to rule out gastric epithelial dysplasia and adenocarcinoma in diagnostic specimens. 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20233622_Lgl2 differentiates pancreatic intraepithelial neoplasia-3 and ductal adenocarcinoma of the pancreas from lower-grade pancreatic intraepithelial neoplasias. 20941506_There is a role of Lgl2 immunohistochemistry as an adjunct in the diagnosis of foveolar-type gastric dysplasia. 22580609_Hugl-2 induces MET and suppresses Snail tumorigenesis. 23110097_Hugl1 and Hugl2 play an essential role in the maintenance of breast epithelial polarity and differentiated cell morphology, as well as growth control. 23332925_Loss or aberrant Lgl2 staining was useful in identifying Barrett gastric foveolar dysplasia 24318287_It is correlated with lymphatic invasion and lymph node metastasis. 24513855_The crystal structures of the Dlg4 GK domain in complex with two phosphor-Lgl2 peptides reveal the molecular mechanism underlying the specific and phosphorylation-dependent Dlg/Lgl complex formation. 30996345_LLGL2 functions as a promoter of tumour growth and not as a tumour suppressor in ER(+) breast cancer; beyond breast cancer, adaptation to nutrient stress is critically important, and findings identify an unexpected role for LLGL2 in this process 31088962_The crystal structures of human lethal giant larvae homolog 2 (Lgl2) in both its unphosphorylated and atypical protein kinase C(aPKC) phosphorylated states are presented and investigated in this article. 31088964_Study and determination of the crystal structure of lethal giant larvae homolog 2 (Lgl2). 32385218_A human cell polarity protein Lgl2 regulates influenza A virus nucleoprotein exportation from nucleus in MDCK cells. | ENSMUSG00000020782 | Llgl2 | 1205.60573 | 0.9720422 | -0.0409091798 | 0.12614087 | 1.082151e-01 | 7.421854e-01 | 9.998360e-01 | No | Yes | 1273.59030 | 164.234280 | 1.211995e+03 | 121.789057 | |
| ENSG00000073614 | 5927 | KDM5A | protein_coding | P29375 | FUNCTION: Histone demethylase that specifically demethylates 'Lys-4' of histone H3, thereby playing a central role in histone code. Does not demethylate histone H3 'Lys-9', H3 'Lys-27', H3 'Lys-36', H3 'Lys-79' or H4 'Lys-20'. Demethylates trimethylated and dimethylated but not monomethylated H3 'Lys-4'. Regulates specific gene transcription through DNA-binding on 5'-CCGCCC-3' motif (PubMed:18270511). May stimulate transcription mediated by nuclear receptors. Involved in transcriptional regulation of Hox proteins during cell differentiation (PubMed:19430464). May participate in transcriptional repression of cytokines such as CXCL12. Plays a role in the regulation of the circadian rhythm and in maintaining the normal periodicity of the circadian clock. In a histone demethylase-independent manner, acts as a coactivator of the CLOCK-ARNTL/BMAL1-mediated transcriptional activation of PER1/2 and other clock-controlled genes and increases histone acetylation at PER1/2 promoters by inhibiting the activity of HDAC1 (By similarity). Seems to act as a transcriptional corepressor for some genes such as MT1F and to favor the proliferation of cancer cells (PubMed:27427228). {ECO:0000250|UniProtKB:Q3UXZ9, ECO:0000269|PubMed:11358960, ECO:0000269|PubMed:15949438, ECO:0000269|PubMed:17320160, ECO:0000269|PubMed:17320161, ECO:0000269|PubMed:17320163, ECO:0000269|PubMed:18270511, ECO:0000269|PubMed:19430464, ECO:0000269|PubMed:27427228}. | 3D-structure;Activator;Alternative splicing;Biological rhythms;Chromatin regulator;Chromosomal rearrangement;Developmental protein;Dioxygenase;Iron;Isopeptide bond;Metal-binding;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the Jumonji, AT-rich interactive domain 1 (JARID1) histone demethylase protein family. The encoded protein plays a role in gene regulation through the histone code by specifically demethylating lysine 4 of histone H3. The encoded protein interacts with many other proteins, including retinoblastoma protein, and is implicated in the transcriptional regulation of Hox genes and cytokines. This gene may play a role in tumor progression. [provided by RefSeq, Aug 2013]. | hsa:5927; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-DNA complex [GO:0032993]; chromatin DNA binding [GO:0031490]; DNA binding [GO:0003677]; histone binding [GO:0042393]; histone demethylase activity [GO:0032452]; histone H3-tri/di/monomethyl-lysine-4 demethylase activity [GO:0034647]; methylated histone binding [GO:0035064]; transcription cis-regulatory region binding [GO:0000976]; transcription coactivator activity [GO:0003713]; zinc ion binding [GO:0008270]; chromatin remodeling [GO:0006338]; circadian regulation of gene expression [GO:0032922]; histone H3-K4 demethylation [GO:0034720]; negative regulation of histone deacetylase activity [GO:1901726]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of DNA-binding transcription factor activity [GO:0051090] | 16645588_that RBP2-H1 exerts a broad tumor-suppressive function partially mediated by pRb modulation 17320161_Study shows that RBP2 is displaced from Hox genes during embryonic stem cell differentiation correlating with an increase of their di- and trimethylated histone 3 lysine 4 levels and expression. 17573780_RBP2 associates with MRG15 complex to maintain reduced H3K4 methylation at transcribed regions, which may ensure the transcriptional elongation state 18270511_ARID is required for RBP2 demethylase activity in cells and that DNA recognition is essential to regulate transcription 18722178_During differentiation, RBP2 exerts inhibitory effects on multiple genes through direct interaction with their promoters. 19430464_fusing an H3K4-trimethylation-binding PHD finger, such as the carboxy-terminal PHD finger of JARID1A, to a common fusion partner NUP98, generated potent oncoproteins that arrested haematopoietic differentiation and induced acute myeloid leukaemia 19762557_Mad1 recruits RBP2 to the hTERT promoter that, in turn, demethylates H3-K4, thereby contributing to a stable repression of the hTERT gene in normal or differentiated malignant cells. 19850045_RBP2 is overexpressed in gastric cancer, and its inhibition triggers senescence of malignant cells at least partially by derepressing its target genes cyclin-dependent kinase inhibitors. 21348942_that H3K4 tri- and dimethylation play an important role and JARID1A is the histone-demethylating enzyme responsible for removal of this mark 21562575_JARID1A or a locus in strong linkage disequilibrium with it is a positional candidate for susceptibility to AS. 21935404_Evidence was provided that chronic drug exposure generated drug-tolerant cells via epigenetic mechanisms involving molecules such as CD44 and KDM5A. 22615382_Jarid1a/b-mediated H3K4 demethylation contributes to silencing of retinoblastoma target genes in senescent cells, suggesting a mechanism by which retinoblastoma triggers gene silencing. 23093672_In terminally differentiated cells, common KDM5A and E2F4 gene targets were bound by the pRB-related protein p130, a DREAM complex component. 23266085_High expression of KDM3B and KDM5A is associated with a better prognosis (no recurrence after mastectomy p=0.005 and response to docetaxel p=0.005)in breast cancer patients. 23722541_our results establish an oncogenic role for RBP2 in lung tumorigenesis and progression and uncover novel RBP2 targets mediating this role. 23794145_MiR-212 directly regulates the expression of RBP2 and inhibits cell growth in gastric cancer, which may provide new clues to treatment. 23922798_RBP2 is overexpressed in HCC and negatively regulated by hsa-miR-212. The hsa-miR-212-RBP2-CDKI pathway may be important in the pathogenesis of HCC. 24068396_LSD1 is a more sensitive molecular marker than RBP2 on thyroid cancer diagnosis. 24069348_JARID1A, JMY, and PTGER4 polymorphisms are related to ankylosing spondylitis in Chinese Han patients. 24376841_RBP2 down-regulated the expression of E-cadherin, up-regulated the expression of N-cadherin and snail, and induced epithelial-mesenchymal transition in non-small cell lung cancer cells. 24442343_Epigenetic changes mediated by JARID1A, SMYD3 and DNA methylation may be responsible, at least in part, for the functional progesterone withdrawal that precipitates human labour. 24582965_RBP2 is critical for breast cancer metastasis to the lung in multiple in vivo models. Mechanistically, RBP2 promotes metastasis as a pleiotropic positive regulator of many metastasis genes, including TNC. 24716659_Overexpression of RBP2 and activation of VEGF might play important roles in human gastric cancer development and progression. 25015565_RBP2 may link chronic inflammation to tumor development. 25162518_RBP2 promotes HIF-1alpha-VEGF-induced angiogenesis of non-small cell lung cancer via the Akt pathway. 25190814_KDM5A and the NuRD complex cooperatively function to control developmentally regulated genes 25575817_Data indicate that lysine (K)-specific demethylase 5A RBP2 (JARID1A; KDM5A) epigenetically downregulated micrRNA-21 (miR-21) in blast transformation of chronic myeloid leukemia (CML). 25686748_KDM5A is regulated by its reader domain through a positive-feedback mechanism 26566863_Treatment-induced temozolimide resistance in glioblastoma cells involves KDM5A mediated epigenetic mechanisms. 26645689_Characterization of a Linked Jumonji Domain of the KDM5/JARID1 Family of Histone H3 Lysine 4 Demethylases. 27008505_promotes overexpression and activation of BCL2 in acute lymphoblastic leukemia development and progression 27224921_KDM5A 5A inhibitor blocks cancer cell growth and drug resistance 27253695_the radiation sensitivity observed following depletion of Jarid1A is not caused by a deficiency in repair of DNA double-strand breaks. 27282106_Ectopic overexpression of RBP2 can induce cancer stem cell-like (CSC) phenotypes through epithelial to mesenchymal transition in renal cell carcinoma cells by converting them to a more mesenchymal phenotype. 27453008_miR-34a promotes the osteogenic differentiation of human adipose-derived stem cells via the RBP2/NOTCH1/CYCLIN D1 coregulatory network. 27512956_KDM5A-mediated H3K4me3 modification participated in the etiology of osteoporosis and may provide new strategies to improve the clinical efficacy of BMP2 in osteoporotic conditions. 28138513_both KDM5A and KDM5B are involved in the lengthening of DICER1 28572115_KDM5A demethylates H3K4 to allow ZMYND8-NuRD to operate within damaged chromatin to repair DNA double strand breaks. 28582381_This study aimed to explore the expression level of RBP2 in hepatocellular carcinoma (HCC) and its prognostic significance. 28714030_KDM5A suppresses ovarian cancer cell apoptosis under paclitaxel treatment. 29059406_A point mutation in LxCxE motif of KDM5A/RBP2 renders it incapable of 130 kDa retinoblastoma-associated protein (p130)-interaction and hence, repression of E2F transcription factor 4 and (E2F)-regulated gene promoters. 29324315_ZEB1 play a crucial role in KDM5A induced function. 30472020_Findings indicate the importance of cellular phenotypic heterogeneity in therapeutic resistance and identify lysine demethylase 5A/B (KDM5A/B) as key regulators of this process. 30872526_inactivation of one of the JmjC-containing enzymes, lysine demethylase 5A (KDM5A), mimics hypoxia-induced cellular responses. 31160694_Reduction in H3K4me patterns due to aberrant expression of methyltransferases and demethylases in renal cell carcinoma: prognostic and therapeutic implications. 31374292_Histone demethylase RBP2 mediates the blast crisis of chronic myeloid leukemia through an RBP2/PTEN/BCR-ABL cascade. 31641207_A novel KDM5A/MPC-1 signaling pathway promotes pancreatic cancer progression via redirecting mitochondrial pyruvate metabolism. 31727771_establishes a role for KDM5A in SCLC tumorigenesis 31985200_The findings provide insights into H3K4-specific recognition by KDM5A, as well as how chromatin context can regulate KDM5A activity and H3K4 methylation status. 32208897_Effect of histone demethylase KDM5A on the odontogenic differentiation of human dental pulp cells. 32381579_The clinical and biological characteristics of NUP98-KDM5A in pediatric acute myeloid leukemia. 32918895_Human papillomavirus type 16 E7 oncoprotein-induced upregulation of lysine-specific demethylase 5A promotes cervical cancer progression by regulating the microRNA-424-5p/suppressor of zeste 12 pathway. 33314922_Exploring the Ligand Preferences of the PHD1 Domain of Histone Demethylase KDM5A Reveals Tolerance for Modifications of the Q5 Residue of Histone 3. 33350388_KDM5A mutations identified in autism spectrum disorder using forward genetics. 33596982_The emerging role of KDM5A in human cancer. 33621431_KDM5A silencing transcriptionally suppresses the FXYD3-PI3K/AKT axis to inhibit angiogenesis in hepatocellular cancer via miR-433 up-regulation. 34003252_Poly(ADP-ribose) binding and macroH2A mediate recruitment and functions of KDM5A at DNA lesions. 34109988_lncRNA NEAT1 facilitates the progression of colorectal cancer via the KDM5A/Cul4A and Wnt signaling pathway. 34184733_KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance. 34258103_Lysine Demethylase 5A is Required for MYC Driven Transcription in Multiple Myeloma. 34448811_KDM5A suppresses PML-RARalpha target gene expression and APL differentiation through repressing H3K4me2. 34678252_Identification of in vitro JMJD lysine demethylase candidate substrates via systematic determination of substrate preference. | ENSMUSG00000030180 | Kdm5a | 1101.18216 | 0.8554430 | -0.2252563355 | 0.17923517 | 1.617107e+00 | 2.034956e-01 | 9.998360e-01 | No | Yes | 1260.97882 | 265.968262 | 1.622149e+03 | 263.440652 | |
| ENSG00000074621 | 9187 | SLC24A1 | protein_coding | O60721 | FUNCTION: Critical component of the visual transduction cascade, controlling the calcium concentration of outer segments during light and darkness. Light causes a rapid lowering of cytosolic free calcium in the outer segment of both retinal rod and cone photoreceptors and the light-induced lowering of calcium is caused by extrusion via this protein which plays a key role in the process of light adaptation. Transports 1 Ca(2+) and 1 K(+) in exchange for 4 Na(+). {ECO:0000269|PubMed:10608890}. | Alternative splicing;Antiport;Calcium;Calcium transport;Congenital stationary night blindness;Glycoprotein;Ion transport;Membrane;Phosphoprotein;Reference proteome;Repeat;Sensory transduction;Signal;Symport;Transmembrane;Transmembrane helix;Transport;Vision | This gene encodes a member of the potassium-dependent sodium/calcium exchanger protein family. The encoded protein plays an important role in sodium/calcium exchange in retinal rod and cone photoreceptors by mediating the extrusion of one calcium ion and one potassium ion in exchange for four sodium ions. Mutations in this gene may play a role in congenital stationary night blindness. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Dec 2011]. | hsa:9187; | integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; neuronal cell body [GO:0043025]; outer membrane [GO:0019867]; plasma membrane [GO:0005886]; calcium channel activity [GO:0005262]; calcium, potassium:sodium antiporter activity [GO:0008273]; symporter activity [GO:0015293]; calcium ion import across plasma membrane [GO:0098703]; calcium ion transmembrane transport [GO:0070588]; calcium ion transport [GO:0006816]; cellular calcium ion homeostasis [GO:0006874]; ion transport [GO:0006811]; long-term synaptic depression [GO:0060292]; long-term synaptic potentiation [GO:0060291]; potassium ion transmembrane transport [GO:0071805]; response to light intensity [GO:0009642]; sodium ion transmembrane transport [GO:0035725]; visual perception [GO:0007601] | 20850105_A mutation in SLC24A1 is implicated in autosomal-recessive congenital stationary night blindness. 26822852_The index patient and his affected brother carry a homozygous single-nucleotide variants (SNVs) in sodium-calcium, potassium exchanger (SLC24A1) (c.2401G > T). 27624628_We report the association of many previously unreported variants with retinal disease, as well as new disease phenotypes associated with known genes, including the first association of the SLC24A1 gene with retinitis pigmentosa | ENSMUSG00000034452 | Slc24a1 | 374.27778 | 0.9088559 | -0.1378765650 | 0.18105950 | 5.681728e-01 | 4.509859e-01 | 9.998360e-01 | No | Yes | 372.10413 | 54.434762 | 3.710598e+02 | 42.262382 | |
| ENSG00000077782 | 2260 | FGFR1 | protein_coding | P11362 | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of embryonic development, cell proliferation, differentiation and migration. Required for normal mesoderm patterning and correct axial organization during embryonic development, normal skeletogenesis and normal development of the gonadotropin-releasing hormone (GnRH) neuronal system. Phosphorylates PLCG1, FRS2, GAB1 and SHB. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. Promotes phosphorylation of SHC1, STAT1 and PTPN11/SHP2. In the nucleus, enhances RPS6KA1 and CREB1 activity and contributes to the regulation of transcription. FGFR1 signaling is down-regulated by IL17RD/SEF, and by FGFR1 ubiquitination, internalization and degradation. {ECO:0000250|UniProtKB:P16092, ECO:0000269|PubMed:10830168, ECO:0000269|PubMed:11353842, ECO:0000269|PubMed:12181353, ECO:0000269|PubMed:1379697, ECO:0000269|PubMed:1379698, ECO:0000269|PubMed:15117958, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17623664, ECO:0000269|PubMed:18480409, ECO:0000269|PubMed:19224897, ECO:0000269|PubMed:19261810, ECO:0000269|PubMed:19665973, ECO:0000269|PubMed:20133753, ECO:0000269|PubMed:20139426, ECO:0000269|PubMed:21765395, ECO:0000269|PubMed:8622701, ECO:0000269|PubMed:8663044}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Chromosomal rearrangement;Craniosynostosis;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Disulfide bond;Dwarfism;Glycoprotein;Heparin-binding;Holoprosencephaly;Hypogonadotropic hypogonadism;Immunoglobulin domain;Kallmann syndrome;Kinase;Membrane;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transcription;Transcription regulation;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase;Ubl conjugation | The protein encoded by this gene is a member of the fibroblast growth factor receptor (FGFR) family, where amino acid sequence is highly conserved between members and throughout evolution. FGFR family members differ from one another in their ligand affinities and tissue distribution. A full-length representative protein consists of an extracellular region, composed of three immunoglobulin-like domains, a single hydrophobic membrane-spanning segment and a cytoplasmic tyrosine kinase domain. The extracellular portion of the protein interacts with fibroblast growth factors, setting in motion a cascade of downstream signals, ultimately influencing mitogenesis and differentiation. This particular family member binds both acidic and basic fibroblast growth factors and is involved in limb induction. Mutations in this gene have been associated with Pfeiffer syndrome, Jackson-Weiss syndrome, Antley-Bixler syndrome, osteoglophonic dysplasia, and autosomal dominant Kallmann syndrome 2. Chromosomal aberrations involving this gene are associated with stem cell myeloproliferative disorder and stem cell leukemia lymphoma syndrome. Alternatively spliced variants which encode different protein isoforms have been described; however, not all variants have been fully characterized. [provided by RefSeq, Jul 2008]. | hsa:2260; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; fibroblast growth factor binding [GO:0017134]; fibroblast growth factor-activated receptor activity [GO:0005007]; heparin binding [GO:0008201]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; receptor-receptor interaction [GO:0090722]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; cell migration [GO:0016477]; chordate embryonic development [GO:0043009]; epithelial to mesenchymal transition [GO:0001837]; fibroblast growth factor receptor signaling pathway [GO:0008543]; MAPK cascade [GO:0000165]; neuron migration [GO:0001764]; peptidyl-tyrosine phosphorylation [GO:0018108]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of blood vessel endothelial cell migration [GO:0043536]; positive regulation of cell differentiation [GO:0045597]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of endothelial cell chemotaxis to fibroblast growth factor [GO:2000546]; positive regulation of kinase activity [GO:0033674]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of phospholipase activity [GO:0010518]; positive regulation of phospholipase C activity [GO:0010863]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of vascular endothelial cell proliferation [GO:1905564]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of cell differentiation [GO:0045595]; regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001239]; skeletal system development [GO:0001501]; skeletal system morphogenesis [GO:0048705]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11693202_vitronectin increased the presence of all four growth factor receptors and most notably, VEGFR-1; in contrast, fibrin decreased all four receptors, especially FGFR-1 and FGFR-2 11746971_In the fusion of the FGFR1 and BCR genes in myeloproliferative disorder, it is likely that the dimerization properties of BCR lead to aberrant FGFR1 signaling and neoplastic transformation. 11759058_distribution in normal endocrine cells and related tumors of the gastroenteropancreatic system; immunoreactive in rare duodenal endocrine cells and in pancreatic A cells 11919391_REVIEW; The 8p11 myeloproliferative syndrome is a distinct clinical entity caused by constitutive activation of FGFR1. 12031912_overexpressed in acute myeloid luekemia while translocations associated with this gene are absent, and more frequently in patients with CD56 immunophenoytpe 12080186_Inhibiting expression of bFGF or FGFR-1 in only the melanoma cells is as effective in blocking tumor growth as simultaneously inhibiting bFGF or FGFR-1 synthesis in the melanoma cells and the melanoma cell-interspersing vasculature. 12121226_CD56 molecules on NK cells interact with fibroblast growth factor receptor 1 on Jurkat T cells to trigger IL-2 production. 12141425_Review of FGFR1 isoforms and structure-activity analysis [review] 12373339_Differences in spatial patterns of FGFR expression in normal skin may generate functional diversity in response to FGFs, and in wounded skin FGFs may function in wound healing via induced FGFRs. 12397010_Our results suggest an autocrine role of the FGF-FGFR-1 system in the pathogenesis of COPD-associated vascular remodeling. 12411316_Fibroblast growth factor receptor-1 is expressed by endothelial progenitor cells. 12440521_Recombinant FGFR1 was expressed on the surface of Sf9 insect cells. The peptide ValTyrMetSerProPhe can specifically bind to the hydrophobic surface of FGFR1. 12573278_alternatively spliced FGFR-1 isoforms induce differential signal transduction pathways 12594223_ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. 12604616_FGFR1 tyrosine phosphorylation is inhibited by sef protein 12614330_cAMP-induced nuclear accumulation of FGFR1 provides a signal that triggers molecular events leading to neuronal differentiation of neuronal progenitor cells 12627230_FGFR1 has a role in autosomal dominant Kallmann syndrome 12651930_expression system is involved in angiogenesis in inflamed synovial tissue in the temporomandibular joint 12746216_TSH stimulates FGFR1 but not FGF-2 expression and PKC activation stimulates FGF-2 synthesis and secretion, and TSH synergizes with PKC activators so increases in FGFR1 or FGF-2 or in both may contribute to goitrogenesis. 12791257_Results describe a direct interaction between neural cell adhesion molecule (fibronectin type III [F3] modules 1 and 2) and fibroblast growth factor receptor R1 (Ig modules 2 and 3) by surface plasmon resonance analysis. 12794748_HFGFR1 was expressed primarily in the ventricular zone embryologically 12799194_Tyrosine 463 is phosphorylated and able to transduce signals in response to FGF-2 treatment alone. Furthermore, FGFR-1 dimerization/kinase activation is stabilized by heparin. 14587039_involvement of a nuclear matrix bound FGFR1 in transcriptional and RNA processing events in the cell nucleus 14636241_Here we show that the TCR and fibroblast growth factor receptors co-localize during combined stimulation [which] synergistically enhances the activation of nuclear factors of activated T cells. 15001591_two novel intragenic FGFR1 mutations in two sporadic male cases in Kallmann syndrome 15096041_The weak binding affinity of the fibroblast growth factor receptor (FGFR) 1 interaction with heparin suggests that in this model, FGFR and heparan sulfate proteoglycan are unbound in the absence of FGF ligand on the cell surface. 15117958_Results suggest that active fibroblast growth factor receptor 1 kinase regulates the functions of nuclear 90-kDa ribosomal S6 kinase. 15273729_Although FGFR-1 dimerization achieved by fgfr-2 injection led to highly differentiated and smaller bladder tumors, no sign of reduction of tumor angiogenesis was observed, thus suggesting that endothelial cells are resistant to FGF. 15297314_fibrinogen binding of FGF-2 enhances EC proliferation through the coordinated effects of colocalized alpha(v)beta(3) and FGFR1 15316024_insulin receptor substrate-4 and ShcA have roles in signaling by the fibroblast growth factor receptor 15509650_conjoint endocytosis and trafficking is a novel mechanism for the coregulation of E-cadherin and FGFR1 during cell signaling and morphogenesis 15558020_The reciprocal relationship in gene expression between FGFR1 and FGFR3 in colorectal tissue plays an important role in the progression of the carcinomas to malignancy. 15564375_Recruitment of SRC to FRS2 leads to activation of signal attenuation pathways. 15613419_the reversal of hypogonadotropic hypogonadism in a proband carrying an FGFR1 mutation suggests a role of FGFR1 beyond embryonic GnRH neuron migration, and a loss of function mutation in the FGFR1 gene causing delayed puberty. 15618886_In 'undifferentiated' neurospheres of embryonic brain and spinal cord, transcripts from FGFR1 and FGFR2 were consistently detected. 15680705_Fibroblast growth factor trophic signaling to differentiated neurons could involve the release of astrocytic basic FGF acting on neuronal FGFR1 expression. 15774903_FGFR-1 is expressed in early hematopoietic/endothelial precursor cells, as well as in a subpool of endothelial cells in tumor vessels, and that it is critical for hematopoietic but not for vascular development 15817662_When used individually, FGFR1 partially prevented goiter and sVEGFR1 partially reduced vascular volume. 15929978_The interaction of FGFR1 with CREB binding protein allows activation of gene transcription and may play a role in cell differentiation. 15955231_involvement of a nuclear matrix bound FGFR1 in transcriptional and RNA processing events in the cell nucleus 16091423_Data indicate that after endocytosis, fibroblast growth factor receptor (FGFR)4 and its bound ligand, FGF1, are sorted mainly to the recycling compartment, whereas FGFR1-3 with ligand are sorted mainly to degradation in the lysosomes. 16186508_Two somatic mutations in kinase domain found in glioblastomas (N546K, R576W). 16188231_Stimulation of FGFR-1 results in a Ca2+ channel-independent change of gene expression in retinal pigment epithelial cells. 16305343_data describe percentage of bone marrow cells expressing receptors for interleukin-1, platelet-derived growth factor, fibroblast growth factor, transforming growth factor-beta, epidermal growth factor and c-Fos and c-Myc in untreated lung & breast cancer 16316338_the initial stimulus for renal inflammation, whether immune complex, hypersensitivity or rejection, did not alter expression patterns of FGF-1 or its receptor 16365308_FGF-receptor-mediated mitogen-activated protein kinase stimulation is potentiated in cells costimulated with ephrin-A1 16424058_up-regulation of the secreted FGF-BP1 protein during initiation of pancreas and colon neoplasia could make this protein a possible serum marker indicating the presence of high-risk premalignant lesions 16598308_The activation of FGF-2/FGFR1beta supports progression and chemoresistance in subsets of AML. Therefore, FGFR1 targeting may be of therapeutic benefit in subsets of AML. 16606836_Mutations leading to FGFR1 loss-of-function were found. 16685373_These results suggest that constitutive levels of both FGFR1 and FGFR3, but not FGFR4 are essential for FGF-stimulated anchorage-independent growth of SW-13 cells. 16757108_Paediatric phenotypic expression of FGFR1 loss of function mutations is highly variable, the severity of the oro-facial malformations at birth does not predict gonadotropic function at the puberty. 16764984_Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. 16807070_Analysis of FGFR1 protein revealed that a high FGFR1 gene expression is a distinct molecular feature of early OSCC indicating a participation in initial oral carcinogenesis. 16807244_analysis of heparan sulfate-related oligosaccharide binding to fibroblast growth factors 1 and 2 and their receptors 16876430_Data suggest that the relative concentrations of Anosmin-1 and FGF-2 modulate the migration of oligodendrocyte precursors during development through their interaction with FGFR1. 16949906_Immunohistochemical expression of FGFR1 in tumors was confirmed by real-time polymerase chain reaction. 17121884_FGFR1 signaling contributes to the survival of MDA-MB-134 cells. 17154279_Identification of 15 new FGFR1 sequence variants in 17 patients with Kallman Syndrome. 17255109_PP2A binding to Sprouty2 and phosphorylation changes are a prerequisite for ERK inhibition downstream of FGFR stimulation 17318851_Data suggest that preferential premolar agenesis is associated with FGFR and IRF6. 17318851_Observational study of gene-disease association. (HuGE Navigator) 17343269_Observational study of gene-disease association. (HuGE Navigator) 17360555_FGF3, FGF7, FGF10, FGF18, and FGFR1 may have roles in nonsyndromic cleft lip and palate 17366557_results of analysis of the genome-wide scan data was a 20 cM region at 8p11-23 in which markers had LODs > or =1.0. Linkage and association analyses of these SNPs yield suggestive results for markers in FGFR1 and BAG4. 17389761_ZNF198-FGFR1 activated both the AKT and mitogen activated protein kinase (MAPK) prosurvival signaling pathways, resulting in elevated phosphorylation of the AKT target FOXO3a at T32 and BAD at S112, respectively. 17397528_FGFR1 has a role in preventing progression of breast neoplasms 17414280_no evidence that mosaicism for mutations, normally associated with syndromal forms of craniosynostosis, occur in single suture craniosynostosis 17545628_FGF receptor 1 (FGFR1), which is expressed mainly in neoplastic thyroid cells, propagates MAPK activation and promotes tumor progression. In contrast, FGFR2 is down-regulated in neoplastic thyroid cells through DNA promoter methylation. 17671674_analysis of EGFR, HER2 and HER3 expression in esophageal primary tumours and corresponding metastases 17696196_data suggest an important role for FGFR1 and FGFR1-downstream genes in rhabdomyosarcoma (RMS) tumorigenesis and a possible association with the deregulation of proliferation and differentiation of skeletal myoblasts in RMS 17893707_Observational study of gene-disease association. (HuGE Navigator) 17963255_FGF and FGFR may have a role in cleft lip and cleft palate 18034870_Current evidence supports a heparan sulphate -dependent interaction between anosmin-1 and FGFR1, where anosmin-1 serves as a co-ligand activator enhancing the signal activity. (Review) 18042549_the involvement of FGFR-1 through FGF2 in eliciting PGE(2) angiogenic responses 18059337_Molecular analyses in salivary gland tumors revealed that ring formation consistently generated novel FGFR1-PLAG1 gene fusions in which the 5'-part of FGFR1 is linked to the coding sequence of PLAG1 18160472_KAL1 mutations result in a more severe reproductive phenotype than FGFR1/KAL2 mutations. 18171671_Basic fibroblast growth factor-induced neuronal differentiation of mouse bone marrow stromal cells requires FGFR-1, MAPK/ERK, and transcription factor AP-1 18216705_No sequence variation was found, indicating that mutations in the 'hot spot' exons are not associated with nonsynostotic plagiocephaly. 18216705_Observational study of genotype prevalence. (HuGE Navigator) 18231572_In human uterine leiomyomas, FGFR1 were also overexpressed. 18315732_Observational study of gene-disease association. (HuGE Navigator) 18411303_p38alpha MAPK has a critical role in the regulated translocation of exogenous FGF1 into the cytosol/nucleus 18415014_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18463157_12% of Kallman syndrome males have KAL1 deletions, but intragenic deletions of the FGFR1, GNRH1, GNRHR, GPR54 and NELF genes are uncommon in Idiopathic hypogonadotropic hypogonadism/Kallman syndrome. 18469019_Identification of two previously unreported SNPs in FGFR1 and FABP3 associated with BMD and a third SNP in TIMP2 related to risk for non-vertebral osteoporotic fractures. 18469019_Observational study of gene-disease association. (HuGE Navigator) 18480409_fibroblast growth factor receptor 1 ubiquitination is required for its intracellular sorting but not for its endocytosis 18563556_BRCA2-associated cancers were characterized by the higher relative expression of FGF1 and FGFR2 18665077_FGFR1-IIIb and FGFR1-IIIc are coexpressed, and the FGFR1-III isoforms are differentially regulated by growth factors and cyclin D1. 18669637_the FGF binding domain and the heparin binding domain are necessary for the hBP3 interaction with endogenous FGF and the activation of FGFR signaling in vivo 18682503_Observational study of gene-disease association. (HuGE Navigator) 18719360_Temporal-mediatd FGFR1 indepepndence: implications for targeting candidate molecules in prostate cancer are reported. 18723471_Observational study of gene-disease association. (HuGE Navigator) 18723471_no mutations found in Kallmann syndrome 18790757_These results indicate that EphA4 plays an important role in malignant phenotypes of glioblastoma by enhancing cell proliferation and migration through accelerating a canonical FGFR signaling pathway. 18829480_bFGF, FGFR1, and FGFR2 are frequently overexpressed in squamous cell carcinoma and adenocarcinoma of the lung and may have a role in neoplasm pathogenesis 18940940_Fibroblast growth factor receptor 1 activity is required for FGF-1 stimulated cell proliferation and priming in early adipogenic events. 18978678_Observational study of gene-disease association. (HuGE Navigator) 19082464_Overexpression of the FGFR-1 gene may thus be a useful predictor of liver metastasis in patients with colorectal cancer. 19096867_FGF-2 and FGFR1 expression is preserved in the motor system in end stage ALS. 19211868_Signaling of transgenic Fgfr1 and Fgfr2 is important for their potential ability to mediate axon-glial interaction in the peripheral sensory pain pathway, via influencing myelinating and nonmyelinating Schwann cell function. 19224897_sequential autophosphorylation of five tyrosines in the FGFR1 kinase domain is under kinetic control, mediated by both the amino acid sequence surrounding the tyrosines and their locations within the kinase structure 19258500_Results show the transforming activity of FGFR1 in mammary epithelial cells and identify RSK as a critical component of FGFR1 signaling in lobular carcinomas. 19330026_Data show that FGFR1 and DDHD2 at 8p12 cooperated functionally with MYC, whereas CCND1 and ZNF703 cooperated with a dominant negative form of TP53. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19458078_FGFR1 has significant effects on urothelial cell phenotype and may represent a useful therapeutic target in some cases of urothelial carcinoma. 19489874_Observational study of gene-disease association. (HuGE Navigator) 19574212_that the main role of heparin in FGF-induced signaling is to protect this naturally unstable protein against heat and/or proteolytic degradation and heparin is not essential for a direct FGF1-FGFR interaction and receptor activation 19658100_ZNF198-FGFR1 is associated with phosphorylation of several proteins including SSBP2, ABL, FLJ14235, CALM and TRIM4 proteins. 19663702_monoclonal antibody analysis of multiple FGFR1 isoforms, generated by alternative splicing and post-translational modifications through glycosylation 19665973_Study describes the crystal structure of the activated tyrosine kinase domain of FGFR1 in complex with a phospholipase Cgamma fragment. 19666467_a neurofascin intracellular domain activates FGFR1 for neurite outgrowth, whereas the extracellular domain functions as an additional, regulatory FGFR1 interaction domain in the course of development 19696444_binding of anosmin-1 to FGFR1 and heparin can play a dual role in assembly and activity of the ternary FGFR1.FGF2.heparin complex. 19727229_Observational study of gene-disease association. (HuGE Navigator) 19727229_There was no association among gene FGFR1 rs13317, p. E467K, p. M369I, p. S393S and gene FGF10 rs1448037 and nonsyndromic cleft lip with or without palate in Chinese population. 19728793_The aim of this study was to investigate and compare FGFR expression in in vivo embryonic limb development and in vitro chondrogenesis of mesenchymal stem cells. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19802384_syndecan-1 and FGF-2, but not FGFR-1 share a common transport route and co-localize with heparanase in the nucleus, and this transport is mediated by the RMKKK motif in syndecan-1 19820032_Loss-of-function mutations in FGFR1 underlie 7% of normosmic idiopathic hypogonadotropic hypogonadism with different degrees of impairment in vitro. 19822079_Data report that the bFGF, FGFR1/2 and syndecan 1-4 expressions are altered in bladder tumours. 19890272_depressed expression of the Klotho-FGFR1 complex in hyperplastic glands underlies the pathogenesis of secondary hyperparathyroidism and its resistance to extremely high FGF23 levels in uremic patients. 19920251_Data show that oncogenic forms of fibroblast growth factor receptor type 1 inhibit the pyruvate kinase M2 (PKM2) isoform by direct phosphorylation of PKM2 tyrosine residue 105 (Y(105)). 19920824_Endogenous SPARC expression can be modulated by FGFR1-III isoform expression. Endogenous SPARC expression in PANC-1 cells was increased in FGFR1-IIIb over-expressing cells, but decreased in FGFR1-IIIc over-expressing cells. 19923290_Loss of FGFR1 signaling provides evidence that extracellular signals regulate not simply the proliferation or survival of radial glial cells, but specifically their progression to intermediate progenitor cells during neurogenesis in vivo. 20024612_FGFR-1 amplification or protein overexpression in breast cancers may be an indicator for brivanib treatment, where it may have direct anti-proliferative effects in addition to its' anti-angiogenic effects. 20036812_Studies indicate communication between tumor cells and their microenvironment is through polypeptide growth factors EGF, FGF, PDGF and receptors for these growth factors. 20068287_Decreased expression of alpha-Klotho and FGFR1c in parallel with CaR expression and parathyroid cell growth may be involved in the pathogenesis of secondary hyperparathyroidism 20103604_brivanib is a dual inhibitor of vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1 kinases 20117945_Recent advances in interactions of Kallman Syndrome (KS)-associated molecules within the FGFR1 signalling complex are covered in this review, and linkage of autosomal dominant and sex-linked modes of inheritance are discussed [review] 20179196_Data suggest that amplification and overexpression of FGFR1 may be a major contributor to poor prognosis in luminal-type breast cancers, driving anchorage-independent proliferation and endocrine therapy resistance. 20362962_Mutations of FGFR1 underlie an autosomal dominant form of Kallmann syndrome. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20388777_amplified FGFR expression engages the STAT3 pathway 20389085_Role of FGFR1 mutations in Kallmann syndrome (Review) 20389169_Based on our results, it is possible that a subtle dysfunction (expression) of the FGFR1 gene is involved in the development of the most common male reproductive tract disorder - unilateral or bilateral cryptorchidism 20421966_the phosphorylation state of FLRT1, which is itself FGFR1 dependent, may play a critical role in the potentiation of FGFR1 signalling and may also depend on a SFK-dependent phosphorylation mechanism acting via the FGFR 20536592_Dental agenesis may be a clinical feature of Kallmann syndrome caused by a mutation in the FGFR1 gene. 20538960_Observational study of gene-disease association. (HuGE Navigator) 20544801_Observational study of gene-disease association. (HuGE Navigator) 20606682_The bFGF-FGFR1-PI3K-Rac1 pathway in the bone microenvironment may have a significant role in the invasion and metastasis of Ewing sarcoma. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20643727_Observational study of gene-disease association. (HuGE Navigator) 20672350_Observational study of gene-disease association. (HuGE Navigator) 20686451_FGFR1 expression levels in parathyroid glands were found to be positively correlated to renal function and significantly decreased over chronic kidney disase stages. 20717167_Observational study of gene-disease association. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 20855522_Ginsenoside-Rg1 induces angiogenesis via non-genomic crosstalk of glucocorticoid receptor and fibroblast growth factor receptor-1. 20889570_Data suggest that an FGFR1alpha-to-FGFR1beta isoform switch and increased FGF1-induced activation of FGFR1beta may result in a proliferative advantage that plays a key role during bladder tumor progression. 20932831_Grb14 was recruited to FGFR1 into a trimeric complex containing also phospholipase C gamma 21041608_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21160078_Focal FGFR1 amplification is common in squamous cell lung cancer and associated with tumor growth and survival, suggesting that FGFR inhibitors may be a viable therapeutic option in this cohort of patients. 21329481_FGFR1 is highly expressed in renal cell carcinoma patients 21330321_we identified a novel CUX1-FGFR1 fusion oncogene in a patient with the 8p11 myeloproliferative syndrome and demonstrated its transforming potential in the Ba/F3 cell line. 21430024_FGFR1 is a novel obesity gene that may promote obesity by influencing adipose tissue and the hypothalamic control of appetite. 21451043_FGF signaling, through cooperation between Fgfr1 and Fgfr2 (but not Fgfr3), is required for initial generation of oligodendrocyte precursors in transgenic mouse ventral forebrain, with Fgfr1 being a stronger inducer than Fgfr2. 21515689_amitriptyline-induced FGFR activation might occur by the MMP-dependent shedding of FGFR ligands, such as FGF-2, thus resulting in GDNF production 21527526_FGFR1/2 are expressed by bone-derived mesenchymal stem cells in vivo and in vitro and are developmentally regulated during their differentiation. 21621521_homodimerization of FGFR1 appear to be a fundamental mechanism for the agonist activity of all FGF ligands at least in the case of the MAPK signaling. 21666686_Case Report: identify a novel fusion partner FGFR1 for the known anchor gene FOXO1 in alveolar rhabdomyosarcoma. 21666749_These studies show that FGFR1 amplification is common in squamous cell lung cancer, and that FGFR1 may represent a promising therapeutic target in non-small cell lung cancer. 21682876_Comprehensive mutation analysis of all 7 known KS genes (KAL1, FGFR1, FGF8, PROK2, PROKR2, CHD7, and WDR11) in 30 well-phenotyped probands revealed mutations in KAL1 (3 men) and FGFR1 (all 5 women vs. 4/25 men), but not in other genes in Finland patients. 21712446_Data show that the majority of TN cell lines only modest sensitive to FGFR inhibition in growth but were highly sensitive in anchorage-independent conditions. 21739604_These results point to NCAM-mediated stimulation of FGFR as a novel mechanism underlying epithelial ovarian carcinoma malignancy and indicate that this interplay may represent a valuable therapeutic target. 21765395_Nedd4-1 binds directly to and ubiquitylates activated FGFR1, by interacting primarily via its WW3 domain with a novel non-canonical sequence on FGFR1, regulating endocytosis & signalling during neuronal differentiation & embryonic development. 21779335_Data indicate that clathrin-mediated endocytosis is required for efficient internalization and downregulation of FGFR1 while FGFR3, however, is internalized by both clathrin-dependent and clathrin-independent mechanisms. 21835001_FGFR1 is the major mediator with the degenerative potential in the presence of FGF-2 in human adult articular chondrocytes 21868365_inductiion of mammary tumorigenesis requires activation of the epidermal growth factor receptor 21885851_MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. 21942441_Levels of FGF2 and FGFR1 in saliva and serum from patients with salivary gland tumors were significantly higher than those from healthy control subjects suggesting their potential use as cancer biomarkers. 21952621_For the first time, our gene expression profiling experiment on archival tumour materials has identified upregulated FGFR1 expression to be associated with PC progression to the CR state. 21969607_LDH-A is tyrosine phosphorylated and activated by FGFR1 in cancer cells. 22030335_FGFR1 gene rearrangement is associated with systemic mastocytosis and myeloid/lymphoid neoplasm in blast crisis. 22179561_An increased FGFR1 gene copy number was found in 32 (32%) lung cancer patients 22247553_Fibronectin induces endothelial cell migration through beta1 integrin and Src-dependent phosphorylation of fibroblast growth factor receptor-1 at tyrosines 653/654 and 766. 22248288_Deletion of the D1 and the D1-D2 linker (the D1/linker region) from FGFR1c led to beta-Klotho-independent receptor activation by FGF21, suggesting that there may be a direct interaction between FGF21 and the D1/linker region-deficient FGFR1c. 22319038_genetic association studies in 103 patients from US and UK: Mutations in FGFR1, FGF8, or PROKR2 contributed to 7.8% of patients with combined pituitary hormone deficiency or septo-optic dysplasia. Data suggest genetic overlap with Kallmann syndrome. 22438050_The expression of FGFR1 in patients' biopsies may serve as a marker of response to chemoradiotherapy. 22442730_Differential specificity of endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in complex with KLB. 22499828_FGFR1 amplification is a common genetic event occurring at a frequency of 16% in L-SCCs. Moreover, lymph node metastases derived from FGFR1-amplified L-SCCs also exhibit FGFR1 amplification. 22514272_INFS/Nurr1 nuclear partnership provides a novel mechanism for TH gene regulation in mDA neurons and a potential therapeutic target in neurodevelopmental and neurodegenerative disorders. 22523080_KLB and FGFR1 form a 1:1 heterocomplex independent of the galectin lattice that transitions to a 1:2 complex upon the addition of FGF21. 22569333_FGFR1 initiates MAPK signaling, whereas S4-dependent FGFR1 macropinocytosis modulates the kinetics of MAPK activation 22573348_Targeting FGFR signaling is a promising new approach to treating aggressive prostate cancer 22648708_There were no significant associations between FGFR1 and clinicopathological parameters in lung cancer 22665522_FGFR1 localized to the nucleus specifically in invading cells in both clinical material and a three-dimensional model of breast cancer 22673519_proteins with LRR, IG, and FNIII are candidate regulators of the FGFRs. Here we identify leucine-rich repeat, immunoglobulin-like and transmembrane domain 3 (LRIT3) as a regulator of the FGFRs 22684217_Conclude that FGFR1 amplification is one of the most frequent therapeutically tractable genetic lesions in pulmonary carcinomas. 22701738_FGFR1 activation in urothelial carcinoma cell lines promotes epithelial-mesenchymal transition via coordinated activation of multiple signalling pathways and by promoting activation of prostaglandin synthesis. 22724017_The data shows that patients with congenital hypogonadotropic hypogonadism and a splice-site mutation in FGFR1 can undergo reversal. 22781593_Mutations in FGFR1 gene is associated with the development of myeloid and lymphoid malignancies. 22791819_results of the present study suggest that MUC4 promotes invasion and metastasis by FGFR1 stabilization through the N-cadherin upregulation. 22837387_study reports that a small subset of glioblastoma multiforme tumors harbors oncogenic chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of fibroblast growth factor receptor genes(FGFR1 or FGFR3) to the transforming acidic coiled-coil coding domains of TACC1 or TACC3; the FGFR-TACC fusion protein displays oncogenic activity 22842457_The novel findings reported in this study are expected to provide valuable clues toward a complete understanding of the other genetic diseases linked to mutations in the FGFR. 22863309_FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. 22903848_Report recycling/degradation pathways of FRG receptor 1 in human glioma cell line. 22955284_Nectin-1 Ig3 induced phosphorylation of FGFR1c in the same manner as the whole nectin-1 ectodomain, and promoted survival of cerebellar granule neurons induced to undergo apoptosis. 22989054_ErbB3 but not Fgfr1 mRNA levels are reduced in leukocytes of patients with major depressive disorders compared to healthy subjects. 23029290_Fibroblast growth factor receptor 1 activation leads to induction of CX3CL1 in a tumor setting. 23070782_FGFR1 polymorphisms, especially rs4647905, can have an important role in the normal human skull variation, primarily due to their influence in head length. 23082000_The incidence of FGFR1 amplification within Chinese patient non-small cell lung carcinoma tumors was 6 of 48 squamous origin and 5 of 76 adenocarcinoma. 23154428_demonstrate skeletal phenotypic characterization of patients presenting with Kallmann syndrome and FGFR1 mutations 23154548_FGFR1 amplification is associated with squamous cell carcinoma of the lung. 23171834_A novel mRNA in-frame fusion between exon 4 of the breakpoint cluster region (BCR) gene at chromosome 22q11 and exon 9 of FGFR1 gene on chromosome 8p11-12 was identified by reverse transcription polymerase chain reaction 23182986_Data indicate that FGFR1 amplification is an independent negative prognostic factor in surgically resected squamous cell carcinoma of the lung (SCCL) and is associated with cigarette smoking in a dose-dependent manner. 23243019_Data suggest for therapeutic targeting of the FGF-2/FGFR1/CEP57 axis in prostate cancer. 23276709_2 patients with congenital isolated hypogonadotropic hypogonadism were found to have mutations in FGFR1 (R254W and R254Q); both are loss-of-function mutations demonstrated by their reduced overall and cell surface expression suggesting a deleterious effect on receptor folding and stability 23296701_Upregulation of FGFR1 is associated with gastric cancer. 23334987_The role of the FGFR/klotho-axis remains still unclear in primary hyperparathyroidism. 23343422_the combination of dovitinib + NVP-BEZ235 or dovitinib + AEE788 results in strong inhibition of tumor growth and a block in metastatic spread. Only these combinations strongly down-regulate the FGFR/FRS2/Erk and PI3K/Akt/mTOR signaling pathways 23348274_In Thai Pfeiffer syndrome patients, FGFR1 mutations in exon 5 were identified. 23401445_FGFR1 amplification is much more common in squamous cell lung cancers (21%) than in lung adenocarcinoma (3.4%). Survival of FGFR1-amplified lung cancer cell lines was additionally shown to be dependent on overexpression of the FGFR1 kinase. 23434054_Copy number variations of the FGFR1 gene occur in a subset of OTSCC with approximately 10% of cases showing amplification of the gene. FGFR1 amplification may represent a therapeutic target in OTSCC 23468956_data demonstrate that FGFR1 and FGFR3 have largely non-overlapping roles in regulating invasion/metastasis and proliferation in distinct 'mesenchymal' and 'epithelial' subsets of human BC cells 23536707_FGF2 -FGFR1 activation through an autocrine loop is a novel mechanism of acquired resistance to EGFR-tyrosine kinase inhibitors. 23563700_Growth inhibition induced by ponatinib was associated with inactivation of FGFR1 and its downstream targets. 23564461_Phosphorylation of serine 779 in fibroblast growth factor receptor 1 and 2 by protein kinase C(epsilon) regulates Ras/mitogen-activated protein kinase signaling and neuronal differentiation. 23569208_KLF10 is an effective repressor of myoblast proliferation and | ENSMUSG00000031565 | Fgfr1 | 3433.91691 | 1.0074598 | 0.0107222593 | 0.10518491 | 1.040180e-02 | 9.187652e-01 | 9.998360e-01 | No | Yes | 3216.37936 | 274.792956 | 3.013943e+03 | 199.313593 | |
| ENSG00000078114 | 10529 | NEBL | protein_coding | O76041 | FUNCTION: Binds to actin and plays an important role in the assembly of the Z-disk. May functionally link sarcomeric actin to the desmin intermediate filaments in the heart muscle sarcomeres (PubMed:27733623). Isoform 2 might play a role in the assembly of focal adhesion (PubMed:15004028). {ECO:0000269|PubMed:15004028, ECO:0000269|PubMed:27733623}. | 3D-structure;Actin-binding;Alternative splicing;Cytoplasm;Methylation;Reference proteome;Repeat;SH3 domain | This gene encodes a nebulin like protein that is abundantly expressed in cardiac muscle. The encoded protein binds actin and interacts with thin filaments and Z-line associated proteins in striated muscle. This protein may be involved in cardiac myofibril assembly. A shorter isoform of this protein termed LIM nebulette is expressed in non-muscle cells and may function as a component of focal adhesion complexes. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Mar 2010]. | hsa:10529; | extracellular exosome [GO:0070062]; I band [GO:0031674]; stress fiber [GO:0001725]; Z disc [GO:0030018]; actin filament binding [GO:0051015]; cytoskeletal protein binding [GO:0008092]; filamin binding [GO:0031005]; structural constituent of muscle [GO:0008307]; tropomyosin binding [GO:0005523]; cardiac muscle thin filament assembly [GO:0071691] | 11822876_our data demonstrate the importance of this cardiac-specific nebulin isoform in myofibril organization and function. Our data demonstrate that nebulette plays a significant role in the structure and stability of the cardiac Z-line. 15004028_LIM-nebulette, Lasp-1, and zyxin may play an important role in the organization of focal adhesions 16385451_Observational study of gene-disease association. (HuGE Navigator) 17987659_filamin-C, a known component of striated muscle Z-lines, interacts with nebulette modules 18823973_the importance of the nebulette-TPM interactions in the maintenance and stability of the thin filaments. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20951326_Different mutations in nebulette transgene trigger a pathological cascade leading to endocardial fibroelastosis and dilated cardiomyopathy in mutant embryonic mouse hearts. 23340173_Report oncogenic potential of MLL-NEBL and NEBL-MLL fusion genes in acute myeloid leukemia. 23389630_Data indicate that lasp-2 interacts with the focal adhesion proteins vinculin and paxillin. 23985323_We identify the SH3 domains of nebulin and nebulette as novel ligands of proline-rich regions of Xin and XIRP2 24563210_predisposition to multibacillary leprosy in Vietnam is associated with CUBN and NEBL common variants in the chromosome 10p13 linkage region 28606091_LASP2 may play a significant role in suppressing CRC progression and provided a novel biomarker for CRC therapy 28667800_The levels of phosphorylated FAK (Tyr397 and Tyr925) were increased after overexpressing Lasp2 and were downregulated by transfecting Lasp2-siRNA. 29257257_Survival analysis suggested that overexpressed NEBL in patients with colorectal cancer was associated with a positive prognosis for overall survival. 30945341_Upregulation of LASP2 inhibits pancreatic cancer cell migration and invasion through suppressing TGF-beta-induced EMT. 31026088_Knockdown of LASP2 inhibits the proliferation, migration, and invasion of cervical cancer cells. 32325347_LASP2 is downregulated in human liver cancer and contributes to hepatoblastoma cell malignant phenotypes through MAPK/ERK pathway. 32635793_LASP2 inhibits trophoblast cell migration and invasion in preeclampsia through inactivation of the Wnt/beta-catenin signaling pathway. 33015783_LASP2 functions as a potential prognostic factor and therapeutic target in nasopharyngeal carcinoma. | ENSMUSG00000053702 | Nebl | 192.63206 | 1.5083184 | 0.5929410057 | 0.26908051 | 5.048231e+00 | 2.465110e-02 | 9.998360e-01 | No | Yes | 227.15148 | 55.152591 | 1.443052e+02 | 27.079310 | |
| ENSG00000078369 | 2782 | GNB1 | protein_coding | P62873 | FUNCTION: Guanine nucleotide-binding proteins (G proteins) are involved as a modulator or transducer in various transmembrane signaling systems. The beta and gamma chains are required for the GTPase activity, for replacement of GDP by GTP, and for G protein-effector interaction. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Mental retardation;Phosphoprotein;Reference proteome;Repeat;Transducer;WD repeat | Heterotrimeric guanine nucleotide-binding proteins (G proteins), which integrate signals between receptors and effector proteins, are composed of an alpha, a beta, and a gamma subunit. These subunits are encoded by families of related genes. This gene encodes a beta subunit. Beta subunits are important regulators of alpha subunits, as well as of certain signal transduction receptors and effectors. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:2782; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular vesicle [GO:1903561]; heterotrimeric G-protein complex [GO:0005834]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; photoreceptor disc membrane [GO:0097381]; plasma membrane [GO:0005886]; synapse [GO:0045202]; GTPase activity [GO:0003924]; GTPase binding [GO:0051020]; protein-containing complex binding [GO:0044877]; signaling receptor complex adaptor activity [GO:0030159]; adenylate cyclase-activating dopamine receptor signaling pathway [GO:0007191]; cell population proliferation [GO:0008283]; cellular response to catecholamine stimulus [GO:0071870]; cellular response to prostaglandin E stimulus [GO:0071380]; G protein-coupled acetylcholine receptor signaling pathway [GO:0007213]; G protein-coupled receptor signaling pathway [GO:0007186]; phospholipase C-activating G protein-coupled receptor signaling pathway [GO:0007200]; Ras protein signal transduction [GO:0007265]; retina development in camera-type eye [GO:0060041]; sensory perception of taste [GO:0050909]; signal transduction [GO:0007165] | 12887923_Directional sensing requires GNB1-mediated PAK1 and PIX alpha-dependent activation of Cdc42. 15105422_Data show that G protein inhibition of N-type calcium channels is critically dependent on two separate but adjacent approximately 20-amino acid regions of the Gbeta subunit, as examined with Gbetas 1 and 5 and Ggamma2. 15331550_in elderly subjects of similar ages, those with diabetes have 1.7-fold higher levels of Galpha(i2) and twofold higher levels of Gbeta(1). 16221676_G betagamma binds HDAC5 and inhibits its transcriptional co-repression activity 16368546_HSD-3.8 (SPAG1), interacts with G-protein beta 1 subunit and activates extracellular signal-regulated kinases 1 and 2 16371464_Gialpha and Gbeta subunits both define selectivity of G protein activation by alpha2-adrenergic receptors. 17110384_G protein betagamma subunits stimulate type V and VI adenylyl cyclases 17167406_No likely pathogenic GNB1 mutations have been found in any of 185 unrelated patients with autosomal dominant retinitis pigmentosa. 17167406_Observational study of genotype prevalence. (HuGE Navigator) 17356515_While digenic disease with the SP4 Asn306Ser and the GNB1 intronic variant alleles has not been established, neither has it been ruled out. This leaves open the possibility of a cooperative involvement of SP4 and GNB1 in the normal function of the retina. 17492941_Fission of transport carriers at the trans-Golgi network is dependent on specifically PLCbeta3, which is necessary to activate PKCeta and PKD in that Golgi compartment, via diacylglycerol production. 17548351_Gbetagamma mediates UVB-induced human keratinocyte apoptosis by augmenting the ectodomain shedding of HB-EGF, which sequentially activates EGFR and p38 18045877_signaling pathway by which G(i)-coupled receptor specifically induces Rac and Cdc42 activation through direct interaction of Gbetagamma with FLJ00018. 18596232_RACK1 regulates directional cell migration by acting on G betagamma at the interface with its effectors PLC beta and PI3K gamma 18729826_Results identify novel functions of beta-arrestin1 in binding to the beta1gamma2 subunits of heterotrimeric G-proteins and promoting G(betagamma)-mediated Akt signalling for NF-kappaB activation. 19110265_This protein has been found differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19727342_Strong candidate gene for severe retinitis pigmentosa, RP32 (human 1p34.3-p13.3). Conclusion is based on a massive expression data set for mouse (103 strains) and joint analysis of RetNet database. 19917775_Gbeta1-mediated Fyn activation integrates FAK with AJ, preventing persistent endothelial barrier leakiness. 20007712_Data show that activation of PLCbeta(2) by alpha(q) and beta1gamma2 differ from activation by Rac2 and from each other. 20048162_Gbetagamma subunits enter in a protein complex with activated Rap1a and its effector Radil; this complex is required downstream of receptor stimulation for the activation of integrins and the positive modulation of cell-matrix adhesiveness. 20181083_Data implicate the domain I-II linker region as an important contributor to voltage dependent Gbeta1/Ggamma2 modulation of Cav2.2 calcium channels. 20381070_This protein has been found differentially expressed in the anterior cingulate cortex from patients with schizophrenia 21679469_Gbetagamma inhibits Epac-induced Ca 2+ elevation in melanoma cells. Cross talk of Ca 2+ signaling between Gbetagamma & Epac plays a major role in melanoma cell migration. 22065575_WDR26 is a novel Gbetagamma-binding protein that is required for the efficacy of Gbetagamma signaling and leukocyte migration 23171003_This study provided evidence that GNB1 gene polymorphisms are related to rapid virological response in HCV-1 and HCV-2 infected patients. GNB1 may play an important role in activating the antiviral response prior to treatment. 23326349_Findings suggest a wide-ranging mechanism by which direct interaction of Gbetagamma with specific chromatin bound transcription factors regulates functional gene networks in response to GPCR activation in cells including the angiotensin II type 1 receptor. 23603342_GNB1 plays an important role in the mTOR-related anti-apoptosis pathway and can potentially be targeted in the treatment of human breast cancer. 23954591_During corticogenesis, a cilium-transduced, noncanonical IGF-1R-Gbetagamma-phospho(T94)Tctex-1 signaling pathway promotes the proliferation of neural progenitors through modulation of ciliary resorption and G1 length. 24462769_Data indicate that endogenous mTOR interacts with Gbetagamma. 25485910_GNB1 and GNB2 alterations confer transformed and resistance phenotypes across a range of human tumors and may be targetable with inhibitors of G protein signaling. 25675501_PhLP1 binding stabilizes the Gbeta fold, disrupting interactions with CCT and releasing a PhLP1-Gbeta dimer for assembly with Ggamma. 27108799_Germline De Novo Mutations in GNB1 Cause Severe Neurodevelopmental Disability, Hypotonia, and Seizures. 28087732_we demonstrate a pathogenic role of de novo and autosomal dominant mutations in GNB1 as a cause of Global developmental delay and provide insights how perturbation in heterotrimeric G protein function contributes to the disease 28650474_Through analysis of the genomic and proteomic profiles of resistant cells, we identified an acquired mutation in the GNB1 gene, K89M, as the most likely cause of the resistance 29174093_Mutation in the GNB1 gene is associated with neurodevelopmental disorder and cutaneous mastocytosis. 30194818_In the new cohort of 18 patients, 50% of males had genitourinary anomalies and 61% of patients had gastrointestinal anomalies, suggesting a possible association of these findings with variants in GNB1. 33427398_Genotype-phenotype correlation in GNB1-related neurodevelopmental disorder: Potential association of p.Leu95Pro with cleft palate. 34022220_Gbetagamma translocation to the Golgi apparatus activates ARF1 to spatiotemporally regulate G protein-coupled receptor signaling to MAPK. 35192430_Shikonin induces cell autophagy via modulating the microRNA -545-3p/guanine nucleotide binding protein beta polypeptide 1 axis, thereby disrupting cellular carcinogenesis in colon cancer. 35193469_AFAP1 antisense RNA 1 promotes retinoblastoma progression by sponging microRNA miR-545-3p that targets G protein subunit beta 1. | ENSMUSG00000029064 | Gnb1 | 14618.45811 | 1.0271457 | 0.0386408440 | 0.08912718 | 1.866521e-01 | 6.657177e-01 | 9.998360e-01 | No | Yes | 14350.56577 | 933.562183 | 1.427351e+04 | 717.747251 | |
| ENSG00000078487 | 55063 | ZCWPW1 | protein_coding | Q9H0M4 | FUNCTION: Dual histone methylation reader specific for PRDM9-catalyzed histone marks (H3K4me3 and H3K36me3) (PubMed:32744506, PubMed:20826339). Facilitates the repair of PRDM9-induced meiotic double-strand breaks (DSBs) (By similarity). Essential for male fertility and spermatogenesis (By similarity). Required for meiosis prophase I progression in male but not in female germ cells (By similarity). {ECO:0000250|UniProtKB:Q6IR42, ECO:0000269|PubMed:20826339, ECO:0000269|PubMed:32744506}. | 3D-structure;Alternative splicing;Chromosome;Coiled coil;Differentiation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Spermatogenesis;Zinc;Zinc-finger | hsa:55063; | chromosome [GO:0005694]; nucleus [GO:0005634]; methyl-CpG binding [GO:0008327]; methylated histone binding [GO:0035064]; zinc ion binding [GO:0008270]; cell differentiation [GO:0030154]; homologous chromosome pairing at meiosis [GO:0007129]; meiosis I [GO:0007127]; positive regulation of DNA recombination [GO:0045911]; positive regulation of double-strand break repair [GO:2000781]; spermatogenesis [GO:0007283] | 24743338_Of those 394 variants, 34 showed strong evidence of regulatory function (RegulomeDB score <3), and only 3 of them were genome-wide significant SNPs (ZCWPW1/rs1476679, CLU/rs1532278 and ABCA7/rs3764650). 26919393_PILRB and GATS expression levels, within the ZCWPW1 locus, were associated with Alzheimer's disease status. 26958812_rs1476679 polymorphism in ZCWPW1 is associated with late-onset Alzheimer's disease. 32744506_ZCWPW1 is recruited to recombination hotspots by PRDM9 and is essential for meiotic double strand break repair. 35217607_PRDM9 losses in vertebrates are coupled to those of paralogs ZCWPW1 and ZCWPW2. | ENSMUSG00000037108 | Zcwpw1 | 56.60331 | 1.6619521 | 0.7328787663 | 0.41590341 | 3.001543e+00 | 8.318523e-02 | 9.998360e-01 | No | Yes | 64.79012 | 11.532817 | 4.174725e+01 | 5.994340 | ||
| ENSG00000078674 | 5108 | PCM1 | protein_coding | Q15154 | FUNCTION: Required for centrosome assembly and function (PubMed:12403812, PubMed:15659651, PubMed:16943179). Essential for the correct localization of several centrosomal proteins including CEP250, CETN3, PCNT and NEK2 (PubMed:12403812, PubMed:15659651). Required to anchor microtubules to the centrosome (PubMed:12403812, PubMed:15659651). Also involved in cilium biogenesis by recruiting the BBSome, a ciliary protein complex involved in cilium biogenesis, to the centriolar satellites (PubMed:20551181, PubMed:24121310, PubMed:27979967). {ECO:0000269|PubMed:12403812, ECO:0000269|PubMed:15659651, ECO:0000269|PubMed:16943179, ECO:0000269|PubMed:20551181, ECO:0000269|PubMed:24121310, ECO:0000269|PubMed:27979967}. | 3D-structure;Acetylation;Alternative splicing;Cell projection;Chromosomal rearrangement;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Proto-oncogene;Reference proteome;Ubl conjugation | The protein encoded by this gene is a component of centriolar satellites, which are electron dense granules scattered around centrosomes. Inhibition studies show that this protein is essential for the correct localization of several centrosomal proteins, and for anchoring microtubules to the centrosome. Chromosomal aberrations involving this gene are associated with papillary thyroid carcinomas and a variety of hematological malignancies, including atypical chronic myeloid leukemia and T-cell lymphoma. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2015]. | hsa:5108; | apical part of cell [GO:0045177]; centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; ciliary transition zone [GO:0035869]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; nuclear membrane [GO:0031965]; pericentriolar material [GO:0000242]; protein-containing complex [GO:0032991]; identical protein binding [GO:0042802]; centrosome cycle [GO:0007098]; cilium assembly [GO:0060271]; cytoplasmic microtubule organization [GO:0031122]; interkinetic nuclear migration [GO:0022027]; intraciliary transport involved in cilium assembly [GO:0035735]; microtubule anchoring [GO:0034453]; microtubule anchoring at centrosome [GO:0034454]; negative regulation of neurogenesis [GO:0050768]; neuron migration [GO:0001764]; neuronal stem cell population maintenance [GO:0097150]; non-motile cilium assembly [GO:1905515]; positive regulation of intracellular protein transport [GO:0090316]; protein localization to centrosome [GO:0071539]; social behavior [GO:0035176] | 15659651_multiple processes involved in regulating the abundance of NIMA (never in mitosis gene a)-related kinase 2 kinase at the centrosome including microtubule binding, the centriolar satellite component PCM-1, and localized protein degradation 16034466_To study the rearrangement created by the t(8;9)(p22;p24)used dual-colour FISH on metaphases from patient cells using labelled-BAC clones centred on PCM1. 16091753_A genetic translocation in atypical chronic myeloid leukemia yields a new PCM1-JAK2 fusion gene. 16894060_Observational study of gene-disease association. (HuGE Navigator) 16894060_The PCM1 gene is implicated in susceptibility to schizophrenia and is associated with orbitofrontal gray matter volumetric deficits. 18594780_cytogenetic change of t(8;9)(p22;p24) may induce HLA-DR immunophenotypic switch and a coordination of the two evolutional changes may play a role in leukemic cell progression 18762586_PCM1 gene is implicated in schizophrenia. 18772192_CEP290 binds to PCM-1 and localizes to centriolar satellites in a PCM-1- and microtubule-dependent manner. 19048012_Observational study of gene-disease association. (HuGE Navigator) 19455170_decreased colocalization of DISC1 and its binding partner PCM1 after Phe607 DISC1 transfection 20152126_Hook3- and PCM1-mediated dynamic assembly of pericentriolar material is essential for interkinetic nuclear migration. 20360304_DISC1 coding variants modulate centrosomal PCM1 localization, highlight a role for DISC1 in glial function and provide a possible cellular mechanism contributing to the association of these DISC1 variants with psychiatric phenotypes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20468070_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20468070_These data provide further evidence that PCM1-though certainly not a major risk factor in the Northern Swedish population-cannot be ruled out as a contributor to schizophrenia risk and/or protection. 20734064_Observational study of gene-disease association. (HuGE Navigator) 21481569_The data failed to find a significant association between SNPs or haplotypes of the PCM1 gene and schizophrenia in the Japanese population (P>0.28). 21985783_WT HTT regulates ciliogenesis by interacting through huntingtin-associated protein 1 (HAP1) with pericentriolar material 1 protein (PCM1). 21998199_PCM1 interacts with Hook2 in a complex that regulates a limiting step required for further initiation of ciliogenesis after centriole maturation. 22100915_NEK7 is essential for PCM accumulation in a cell cycle stage-specific manner. 23110211_CEP90 physically interacts with PCM-1 at centriolar satellites, and this interaction is essential for centrosomal accumulation of the centriolar satellites and eventually for primary cilia formation. 23345402_these data suggest a mechanism whereby the recruitment of Plk1 to pericentriolar matrix by PCM1 plays a pivotal role in the regulation of primary cilia disassembly before mitotic entry. 23372669_Chromosomal translocation [t(8;9)(p22;p24)]/PCM1-JAK2 fusion protein activates SOCS2 and SOCS3 via STAT5 in a cutaneous T-cell lymphoma cell line. 24576429_no association between the PCM1 gene and schizophrenia in a Japanese population 24909162_a novel variant in the PCM1 3'UTR is significantly associated with ovarian cancer 24913195_Haematological neoplasms associated with t(8;9)(p22;p24); PCM1-JAK2 have features in common and we suggest that they should be recognized as a specific entity in the WHO classification 27146717_In the absence of PCM1, Mib1 destabilizes Talpid3 through poly-ubiquitylation and suppresses cilium assembly. 28620049_Data suggest that USP9X as an integral component of centrosome where it functions to stabilize PCM1 and CEP55 and to promote centrosome biogenesis; N-terminal domain of USP9X appears to be responsible for physical association of USP9X with PCM1 and CEP55. (USP9X = ubiquitin-specific protease 9X; PCM1 = pericentriolar material 1 protein; CEP55 = 55kDa centrosomal protein) 29567772_detecting PDGFRA, PDGFRB, FGFR1 and PCM1-JAK2 rearrangements is a prerequisite for up-to-date WHO classification, and an essential step in the differential diagnosis of neoplasms with eosinophilia. 30584065_study reveals that USP9X is a constituent of centriolar satellites and functions to maintain centriolar satellite integrity by stabilizing PCM1. 31671755_SNX17 Recruits USP9X to Antagonize MIB1-Mediated Ubiquitination and Degradation of PCM1 during Serum-Starvation-Induced Ciliogenesis. 33214552_PCM1 is necessary for focal ciliary integrity and is a candidate for severe schizophrenia. 34382410_LncRNA-ENST00000421645 promotes T cells to secrete IFN-gamma by sponging PCM1 in neurosyphilis. | ENSMUSG00000031592 | Pcm1 | 667.79024 | 0.7326073 | -0.4488879539 | 0.18061535 | 6.553913e+00 | 1.046538e-02 | 9.998360e-01 | No | Yes | 614.43399 | 91.219003 | 7.928645e+02 | 90.710617 | |
| ENSG00000080224 | 285220 | EPHA6 | protein_coding | Q9UF33 | FUNCTION: Receptor tyrosine kinase which binds promiscuously GPI-anchored ephrin-A family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling (By similarity). {ECO:0000250}. | ATP-binding;Alternative splicing;Glycoprotein;Kinase;Membrane;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | hsa:285220; | integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; ATP binding [GO:0005524]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; transmembrane-ephrin receptor activity [GO:0005005]; axon guidance [GO:0007411]; positive regulation of kinase activity [GO:0033674]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 19850283_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20011078_During development of the retinal vasculature, migration of ligand-bearing astrocytes is slowed along Eph-A6 expression gradient through repellent Eph-A6 - ephrin-A1 and -A4 signaling. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20950786_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 25583493_Two predominant genes, ephrin type A receptor 6 (EPHA6) and folliculin (FLCN), with mutations exclusive to African American CRCs, are by genetic and biological criteria highly likely African American CRC driver genes. 26041887_EphA6 mRNA expression is higher in 112 CaP tumor samples compared with benign tissues from 58 benign prostate hyperplasia patients. Positive correlation was identified between EphA6 expression and vascular invasion, neural invasion, PSA level, and TNM staging in CaP cases. 27582484_Gene-based analysis identified EPHA6 as the gene most significantly associated with paclitaxel-induced neuropathy...This first study sequencing EPHA genes revealed that low-frequency variants in EPHA6, EPHA5, and EPHA8 contribute to the susceptibility to paclitaxel-induced neuropathy 29208002_The EPHA6 rs4857055 C > T SNP is a novel candidate gene for hypertension in the Korean population. 31483918_our data imply that EPHA6 expression is beneficial for glioblastoma multiforme inhibition, particularly in combination with activation of BMP-2 signaling.These results suggest that EPHA6 expression or protein levels could be used as biomarkers for identification of subsets of glioblastoma multiforme patients who might benefit from BMP treatment. | ENSMUSG00000055540 | Epha6 | 67.63516 | 1.2134760 | 0.2791456178 | 0.36873951 | 5.401973e-01 | 4.623510e-01 | 9.998360e-01 | No | Yes | 111.34200 | 21.657898 | 9.771067e+01 | 14.807692 | ||
| ENSG00000080503 | 6595 | SMARCA2 | protein_coding | P51531 | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Binds DNA non-specifically (PubMed:22952240, PubMed:26601204). Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). {ECO:0000250|UniProtKB:Q6DIC0, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | 3D-structure;ATP-binding;Acetylation;Activator;Alternative splicing;Bromodomain;DNA-binding;Disease variant;Helicase;Hydrolase;Hypotrichosis;Isopeptide bond;Mental retardation;Neurogenesis;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Schizophrenia;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a member of the SWI/SNF family of proteins and is highly similar to the brahma protein of Drosophila. Members of this family have helicase and ATPase activities and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SNF/SWI, which is required for transcriptional activation of genes normally repressed by chromatin. Alternatively spliced transcript variants encoding different isoforms have been found for this gene, which contains a trinucleotide repeat (CAG) length polymorphism. [provided by RefSeq, Jan 2014]. | hsa:6595; | chromatin [GO:0000785]; intermediate filament cytoskeleton [GO:0045111]; intracellular membrane-bounded organelle [GO:0043231]; nBAF complex [GO:0071565]; npBAF complex [GO:0071564]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; SWI/SNF complex [GO:0016514]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATP-dependent activity, acting on DNA [GO:0008094]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; helicase activity [GO:0004386]; histone binding [GO:0042393]; transcription cis-regulatory region binding [GO:0000976]; transcription coactivator activity [GO:0003713]; chromatin remodeling [GO:0006338]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; nervous system development [GO:0007399]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; spermatid development [GO:0007286] | 11719516_Brm could also drive expression of CD44; Brm can compensate for BRG-1 loss as pertains to RB sensitivity 11850427_Brm-containing SWI/SNF complex subfamily (trithorax-G) and a complex including YY1 and HDACs (Polycomb-G) counteract each other to maintain transcription of exogenously introduced genes 12493776_human adrenal carcinoma cells can spontaneously transition between two subtypes by switching expression of BRG1 and Brm at the post-transcriptional level 12566296_This report provides supportive evidence that BRG1 and BRM act as tumor suppressor proteins and implicates a role for their loss in the development of non-small cell lung cancers. 12620226_BRG1 and BRM complexes may direct distinct cellular processes by recruitment to specific promoters through protein-protein interactions that are unique to each ATPase. 14657023_Cell culture in the presence of HDAC inhibitors facilitates the isolation of clones overexpressing Brm. 15240517_BRM and BRG1 participate in two distinct chromosome remodeling complexes that are functionally complementary in non-small cell lung cancer 16341228_on genes regulated by SWI/SNF, Brm contributes to the crosstalk between transcription and RNA processing by decreasing RNAPII elongation rate and facilitating recruitment of the splicing machinery to variant exons with suboptimal splice sites 16749937_Observational study of gene-disease association. (HuGE Navigator) 16749937_family-based and case-control association study suggest that there is no association between the trinucleotide repeat polymorphism within SMARCA2 and schizophrenia 17075831_Aberrant expression of BRM genes is associated with disease development and progression in prostate cancers. 17546055_Loss of BRM through epigenetic silencing is associated with neoplasms 17938176_p53 activity is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes 18006815_Brm is required for villin expression, a definitive marker of intestinal metaplasia and differentiation 18042045_at the TERT gene locus in human tumour cells containing a functional SWI/SNF complex, Brm, and possibly BRG1, in concert with p54(nrb), would initiate efficient transcription and could be involved in the subsequent splicing of TERT transcripts 18082132_C/EBPbeta and GATAs may developmentally regulate the expression of brm by mutually exclusive binding. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18923443_Hotspot mutation of Brahma in non-melanoma skin cancer. 19144648_BRM and BRG1 SWI/SNF complexes have roles in differentiation 19183483_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19363039_A missense polymorphism (rs2296212)induced a lower nuclear localization of BRM was associated with low SMARCA2 expression in the postmortem prefrontal cortex of schizophrenia patients. 19371634_Cdx2 regulates intestinal villin expression through recruiting Brm-type SWI/SNF complex to the villin promoter. 19488910_Loss of heterozygosity at the 9p21-24 region and identification of BRM as a candidate tumor suppressor gene in head and neck squamous cell carcinoma. 19784067_Data suggest that heterogeneous SWI/SNF complexes composed of either the BRG1 or BRM subunit promote expression of distinct and overlapping MITF target genes. 20011120_show that SWI/SNF activity favors (from subunits Brg1/Brm) loading of HP1 proteins to chromatin both in vivo and in vitro 20333683_Knockdown of either BRM or BRG1 resulted in an inhibition of cell proliferation in monolayer cultures. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20457675_The SWI/SNF chromatin-remodelin complex asa key component of the genetic architecture of schizophrenia. 20460684_REQ functions as an efficient adaptor protein between the SWI/SNF complex and RelB/p52 and plays important roles in noncanonical NF-kappaB transcriptional activation and its associated oncogenic activity. 20719309_The methylation levels of CpG islands within Brahma increased during spermatogenesis and decreased during oogenesis 21070662_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21079652_The hBrm/Brg1 switch is an indicator of the responsiveness of a gene to heat-shock or IFNgamma stimulation and may represent an 'on-off switch' of gene expression in vivo. 21092585_The expression of BRG1 and BRM correlates with the development of prostatic cancer. 21189327_results reveal that miR-199a and Brm form a double-negative feedback loop through Egr1, leading to the generation of two distinct cell types during carcinogenesis. 21646426_multiple distinct transcriptional patterns of GR and Brm interdependence 22366787_sequenced the exomes of ten individuals with Nicolaides-Baraitser syndrome and identified heterozygous variants in SMARCA2 in eight of them. 22721696_SWI/SNF chromatin remodeling complex catalytic subunits Brg1 and Brm modulate cisplatin cytotoxicity by facilitating efficient repair of the cisplatin DNA lesions. 23088494_Reduced expression of BRM may contribute to the carcinogenesis of hepatocellular carcinoma. 23163725_Loss of BRG1 and BRM was frequent in E-cadherin-low, TTF-1-low, and vimentin-high cases. 23276717_SMARCA2 rs2296212 and rs4741651 variants were associated with oligodendroglioma risk. 23322154_BRM expression was lost in 25% of cell lines and 16% of tumors. 23359823_findings suggest that BRM promoter polymorphism (BRM-1321) could regulate BRM expression and may serve as a potential marker for genetic susceptibility to HCC 23524580_the mitogen-activated protein kinase pathway regulates both BRM acetylation and BRM silencing as MAP kinase pathway inhibitors both induced BRM as well as caused BRM deacetylation. 23770848_loss of BRM epigenetically induces C/EBPbeta transcription, which then directly induces alpha5 integrin transcription to promote the malignant behavior of mammary epithelial cells 23872584_High SMARCA2 expression is associated with lung cancer 23897427_Data indicate that transient knockdown of BRG1 or BRM reduces hypoxia induction of several known HIF1 and HIF2 target genes in Hep3B cells. 23963727_Data suggest that Brg1 and Brm integrate various proinflammatory cues into cell adhesion molecule transactivation in endothelial injury. 24421395_A SMARCA2-containing residual SWI/SNF complex underlies the oncogenic activity of SMARCA4 mutant cancers. 24471421_loss of BRM expression is a common feature among poorly differentiated tumours in clear cell renal cell carcinomas. We hypothesize that loss of BRM expression is involved in tumor de-differentiation in clear cell RCCs. 24519853_Findings suggest that the BRM promoter double insertion homozygotes may be associated with an increased risk of early-stage UADT cancers independent of smoking status and histology. 24520176_Depletion of BRM in BRG1-deficient cancer cells leads to a cell cycle arrest, induction of senescence, and increased levels of global H3K9me3. 24913006_these data show that the mechanism of BRM silencing contributes to the pathogenesis of Rhabdoid tumors 25026375_Over-expression of BRM in melanoma cells that harbor oncogenic BRAF promoted changes in cell cycle progression and apoptosis consistent with a tumor suppressive role. 25081545_BAF complex gene SMARCA2 is mutated in Coffin-Siris syndrome patients. 25169058_Phenotype and genotype in Nicolaides-Baraitser syndrome patients with SMARCA2 mutations 25496315_We report, for the first time, co-inactivation and frequent mutations of SMARCB1, SMARCA2 and PBRM1 in MRTs. 25673149_The miR-199a/Brm/EGR1 axis is a determinant of anchorage-independent growth in epithelial tumor cell lines 25808524_Knockout of BRG1 or BRM using CRISPR/Cas9 technology resulted in the loss of viability, consistent with a requirement for both enzymes in triple negative breast cancer cells 26356327_SMARCA4 loss, either alone or with SMARCA2, is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type, restoration of either SWI/SNF ATPase can inhibit the growth of SCCOHT cell lines 26551623_this study shows for the first time novel SMARCA4-deficient and SMARCA2-deficient variants in undifferentiated gastrointestinal tract carcinomas 26564006_our data suggest that concomitant loss of SMARCA2 and SMARCA4 is another hallmark of small cell carcinoma of the ovary, hypercalcemic type-a finding that offers new opportunities for therapeutic interventions. 27264538_We conclude that their features better resemble Coffin-Siris syndrome, rather than Nicolaides-Baraitser syndrome and that these features likely arise from SMARCA2 over-dosage. Pure 9p duplications (not caused by unbalanced translocations) are rare. Copy number analysis in patients with features that overlap with Coffin-Siris syndrome is recommended to further determine their genetic aspects. 27487558_Two functional promoter BRM polymorphisms were not associated with pancreatic adenocarcinoma risk, but are strongly associated with survival. 27665729_This de novo SMARCA2 missense mutation c.3721C>G, p.Gln1241Glu is the only reported mutation on exon 26 outside the ATPase domain of SMARCA2 to be associated with Nicolaides-Baraitser syndrome and adds to chromatin remodeling as a pathway for epileptogenesis. 27827316_Epigenetic regulatory molecules bind to two BRM promoter sequence variants but not to their wild-type sequences. These variants are associated with adverse overall and progression-free survival. Decreased BRM gene expression, seen with these variants, is also associated with worse overall survival 28038711_SMARCA4 and SMARCA2 deficiency is observed in 5.1% and 4.8% of non-small cell lung cancer 28070921_BRM gene mutation, chromosome 9 monosomy or BRM deletion and CpG methylation contribute collectively to the loss of BRM expression in poorly differentiated clear cell renal cell carcinoma 28232072_BRG1/BRM and c-MYC have an antagonistic relationship regulating the expression of cardiac conduction genes that maintain contractility, which is reminiscent of their antagonistic roles as a tumor suppressor and oncogene in cancer. 28292935_We also demonstrate that tazemetostat, a potent and selective EZH2 inhibitor currently in phase II clinical trials, induces potent antiproliferative and antitumor effects in SCCOHT cell lines and xenografts deficient in both SMARCA2 and SMARCA4. These results exemplify an additional class of rhabdoid-like tumors that are dependent on EZH2 activity for survival. 28296015_Although Genome-Wide Association studies have not been carried out in the field of alcohol-related hepatocellular carcinoma (HCC), common single nucleotide polymorphisms conferring a small increase in the risk of liver cancer risk have been identified. Specific patterns of gene mutations including CTNNB1, TERT, ARID1A and SMARCA2 exist in alcohol-related HCC. [review] 28427211_two promoter BRM germline variants were associated with worse outcome in our esophageal adenocarcinoma (EAC) patients. This significantly poorer outcome was independent of TNM classification at diagnosis or other clinic-demographic variables. 28571677_BRM-741 and BRM-1321 insertion polymorphisms are associated with susceptibility to cancer. 28602977_BRM could activate JAK2/STAT3 pathway to promote pancreatic cancer growth and chemoresistance. 28678310_Expression of BRM and MMP2 in the thoracic aortic aneurysm and aortic dissection is very high, indicating that BRM and MMP2 may play important roles in the occurrence and development of thoracic aortic aneurysm and aortic dissection. 28706277_association of the BRG1/hBRM bromodomain with nucleosomes plays a regulatory rather than targeting role 28892201_Two BRM promoter polymorphisms were strongly associated with hepatocellular carcinoma (HCC) prognosis but were not associated with increased HCC susceptibility. 29087303_PRC2-mediated repression of SMARCA2 predicts EZH2 inhibitor activity in SWI/SNF mutant tumors. 29273066_at genes where BRG1 and BRM antagonize one another we observe a nearly complete rescue of gene expression changes in the combined BRG/BRM double knockdown 29391527_High expression of SMARCA2 is associated with benign differentiated tumors. 29848589_Here, the authors show that C-terminally truncated forms of both SMARCA2 and SMARCA4, produced by caspase-mediated cleavage, accumulate in cells infected with different RNA or DNA viruses. The levels of truncated SMARCA2 or SMARCA4 strongly correlate with the degree of cell damage and death observed after virus infection. 29894502_Indel polymorphisms in the promoter region of the Brahma gene is associated with colorectal cancer. 30287812_The sensitivity of SWI/SNF-deficient cells to DNA damage induced by UV irradiation and cisplatin treatment depends on GTF2H1 levels. 30447346_Inactivation of SMARCA2 by promoter hypermethylation is associated with lung cancer development. 30478150_targeting of BRM in combination with radiotherapy is supposed to improve the therapeutic outcome of lung cancer patients harboring BRG1 mutations.The present study shows that the moderate radioresponsiveness of NSCLC cells with BRG1 mutations can be increased upon BRM depletion that is associated with a prolonged Rad51-foci prevalence at DNA DSBs. 30522882_In both mouse keratinocytes and HaCaT cells, Brm deficiency led to an increased cell population growth following ultraviolet radiation (UVR) exposure compared to cells with normal levels of Brm. The loss of Brm in keratinocytes exposed to UVR enables them to resume proliferation while harboring DNA photolesions, leading to an increased fixation of mutations and, consequently, increased carcinogenesis. 30722027_Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1-associated atypical neurofibromas. 30790683_Transcriptional regulation of miR-302a-3p by BRM potentiates pancreatic cancer cell metastasis by epigenetically suppressing SOCS5 expression and activating STAT3 signaling. 30946989_BRM suppresses the migration and invasion of breast cancer cells via epigenetically activating the transcription of Claudins. 31288860_SMARCA2 variants in Nicolaides-Baraitser syndrome 31355511_Never-smokers who carry BRM homozygous variants have an increased chance of developing MPM, which results in worse prognosis. In contrast, in ever-smokers, there may be a protective effect, with no difference in overall survival. Mechanisms for the interaction between BRM and smoking require further study. 31375262_results demonstrate that SMARCA2 mutations cause impaired differentiation through enhancer reprogramming via inappropriate targeting of SMARCA4. 31406271_SMARCA2-deficiency confers sensitivity to targeted inhibition of SMARCA4 in esophageal squamous cell carcinoma cell lines. 31751681_SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. 31906887_This work describes, for the first time, loss of one of the SWI/SNF ATPase subunit proteins in a large number of adenocarcinomas of the oesophagus. 32019955_AATF and SMARCA2 are associated with thyroid volume in Hashimoto's thyroiditis patients. 32073734_SWI/SNF chromatin remodeling complex and glucose metabolism are deregulated in advanced bladder cancer. 32312722_Review of the role of SMARCA2 and SMARCA4 mutations in various neoplasms. 32376693_The transcription factor GLI1 cooperates with the chromatin remodeler SMARCA2 to regulate chromatin accessibility at distal DNA regulatory elements. 32502208_BRM-SWI/SNF chromatin remodeling complex enables functional telomeres by promoting co-expression of TRF2 and TRF1. 32657847_Two cases of Nicolaides-Baraitser syndrome, one with a novel SMARCA2 variant. 32694869_De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. 32855269_Loss of SWI/SNF Chromatin Remodeling Alters NRF2 Signaling in Non-Small Cell Lung Carcinoma. 33027072_SMARCA4/SMARCA2-deficient Carcinoma of the Esophagus and Gastroesophageal Junction. 33481850_The clinicopathological significance of SWI/SNF alterations in gastric cancer is associated with the molecular subtypes. 33602783_SMARCA2 Is a Novel Interactor of NSD2 and Regulates Prometastatic PTP4A3 through Chromatin Remodeling in t(4;14) Multiple Myeloma. 34289068_The Bromodomains of the mammalian SWI/SNF (mSWI/SNF) ATPases Brahma (BRM) and Brahma Related Gene 1 (BRG1) promote chromatin interaction and are critical for skeletal muscle differentiation. 34518526_SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by restricting IP3R3-mediated Ca(2+) flux to mitochondria. 35289322_SMARCA2 deficiency in NSCLC: a clinicopathologic and immunohistochemical analysis of a large series from a single institution. | ENSMUSG00000024921 | Smarca2 | 738.66638 | 0.7339510 | -0.4462442801 | 0.13649192 | 1.055592e+01 | 1.158168e-03 | 3.719110e-01 | No | Yes | 774.54128 | 96.111804 | 1.097328e+03 | 105.338002 | |
| ENSG00000080815 | 5663 | PSEN1 | protein_coding | P49768 | FUNCTION: Catalytic subunit of the gamma-secretase complex, an endoprotease complex that catalyzes the intramembrane cleavage of integral membrane proteins such as Notch receptors and APP (amyloid-beta precursor protein) (PubMed:15274632, PubMed:10545183, PubMed:10593990, PubMed:10206644, PubMed:10899933, PubMed:10811883, PubMed:12679784, PubMed:12740439, PubMed:25043039, PubMed:26280335, PubMed:30598546, PubMed:30630874, PubMed:28269784, PubMed:20460383). Requires the presence of the other members of the gamma-secretase complex for protease activity (PubMed:15274632, PubMed:25043039, PubMed:26280335, PubMed:30598546, PubMed:30630874). Plays a role in Notch and Wnt signaling cascades and regulation of downstream processes via its role in processing key regulatory proteins, and by regulating cytosolic CTNNB1 levels (PubMed:9738936, PubMed:10593990, PubMed:10899933, PubMed:10811883). Stimulates cell-cell adhesion via its interaction with CDH1; this stabilizes the complexes between CDH1 (E-cadherin) and its interaction partners CTNNB1 (beta-catenin), CTNND1 and JUP (gamma-catenin) (PubMed:11953314). Under conditions of apoptosis or calcium influx, cleaves CDH1 (PubMed:11953314). This promotes the disassembly of the complexes between CDH1 and CTNND1, JUP and CTNNB1, increases the pool of cytoplasmic CTNNB1, and thereby negatively regulates Wnt signaling (PubMed:9738936, PubMed:11953314). Required for normal embryonic brain and skeleton development, and for normal angiogenesis (By similarity). Mediates the proteolytic cleavage of EphB2/CTF1 into EphB2/CTF2 (PubMed:17428795, PubMed:28269784). The holoprotein functions as a calcium-leak channel that allows the passive movement of calcium from endoplasmic reticulum to cytosol and is therefore involved in calcium homeostasis (PubMed:25394380, PubMed:16959576). Involved in the regulation of neurite outgrowth (PubMed:15004326, PubMed:20460383). Is a regulator of presynaptic facilitation, spike transmission and synaptic vesicles replenishment in a process that depends on gamma-secretase activity. It acts through the control of SYT7 presynaptic expression (By similarity). {ECO:0000250|UniProtKB:P49769, ECO:0000269|PubMed:10206644, ECO:0000269|PubMed:10545183, ECO:0000269|PubMed:10593990, ECO:0000269|PubMed:10811883, ECO:0000269|PubMed:10899933, ECO:0000269|PubMed:11953314, ECO:0000269|PubMed:12679784, ECO:0000269|PubMed:12740439, ECO:0000269|PubMed:15004326, ECO:0000269|PubMed:15274632, ECO:0000269|PubMed:15341515, ECO:0000269|PubMed:16305624, ECO:0000269|PubMed:16959576, ECO:0000269|PubMed:17428795, ECO:0000269|PubMed:20460383, ECO:0000269|PubMed:25043039, ECO:0000269|PubMed:25394380, ECO:0000269|PubMed:26280335, ECO:0000269|PubMed:28269784, ECO:0000269|PubMed:30598546, ECO:0000269|PubMed:30630874, ECO:0000269|PubMed:9738936}. | 3D-structure;Alternative splicing;Alzheimer disease;Amyloidosis;Apoptosis;Cardiomyopathy;Cell adhesion;Cell junction;Cell membrane;Cell projection;Direct protein sequencing;Disease variant;Endoplasmic reticulum;Endosome;Golgi apparatus;Hydrolase;Membrane;Neurodegeneration;Notch signaling pathway;Phosphoprotein;Protease;Reference proteome;Synapse;Transmembrane;Transmembrane helix | Alzheimer's disease (AD) patients with an inherited form of the disease carry mutations in the presenilin proteins (PSEN1; PSEN2) or in the amyloid precursor protein (APP). These disease-linked mutations result in increased production of the longer form of amyloid-beta (main component of amyloid deposits found in AD brains). Presenilins are postulated to regulate APP processing through their effects on gamma-secretase, an enzyme that cleaves APP. Also, it is thought that the presenilins are involved in the cleavage of the Notch receptor, such that they either directly regulate gamma-secretase activity or themselves are protease enzymes. Several alternatively spliced transcript variants encoding different isoforms have been identified for this gene, the full-length nature of only some have been determined. [provided by RefSeq, Aug 2008]. | hsa:5663; | aggresome [GO:0016235]; apical plasma membrane [GO:0016324]; azurophil granule membrane [GO:0035577]; cell cortex [GO:0005938]; cell junction [GO:0030054]; cell surface [GO:0009986]; centrosome [GO:0005813]; ciliary rootlet [GO:0035253]; dendritic shaft [GO:0043198]; early endosome membrane [GO:0031901]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; gamma-secretase complex [GO:0070765]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; growth cone [GO:0030426]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; integral component of presynaptic membrane [GO:0099056]; kinetochore [GO:0000776]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; membrane raft [GO:0045121]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; neuromuscular junction [GO:0031594]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; rough endoplasmic reticulum [GO:0005791]; sarcolemma [GO:0042383]; smooth endoplasmic reticulum [GO:0005790]; synaptic vesicle [GO:0008021]; Z disc [GO:0030018]; aspartic endopeptidase activity, intramembrane cleaving [GO:0042500]; aspartic-type endopeptidase activity [GO:0004190]; ATPase binding [GO:0051117]; beta-catenin binding [GO:0008013]; cadherin binding [GO:0045296]; calcium channel activity [GO:0005262]; endopeptidase activity [GO:0004175]; growth factor receptor binding [GO:0070851]; PDZ domain binding [GO:0030165]; amyloid precursor protein catabolic process [GO:0042987]; amyloid precursor protein metabolic process [GO:0042982]; amyloid-beta formation [GO:0034205]; amyloid-beta metabolic process [GO:0050435]; astrocyte activation [GO:0048143]; astrocyte activation involved in immune response [GO:0002265]; autophagosome assembly [GO:0000045]; blood vessel development [GO:0001568]; brain morphogenesis [GO:0048854]; Cajal-Retzius cell differentiation [GO:0021870]; calcium ion transport [GO:0006816]; cell fate specification [GO:0001708]; cell-cell adhesion [GO:0098609]; cellular response to amyloid-beta [GO:1904646]; cellular response to DNA damage stimulus [GO:0006974]; cerebellum development [GO:0021549]; cerebral cortex cell migration [GO:0021795]; choline transport [GO:0015871]; dorsal/ventral neural tube patterning [GO:0021904]; embryonic limb morphogenesis [GO:0030326]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; epithelial cell proliferation [GO:0050673]; heart looping [GO:0001947]; hematopoietic progenitor cell differentiation [GO:0002244]; intracellular signal transduction [GO:0035556]; L-glutamate import across plasma membrane [GO:0098712]; learning or memory [GO:0007611]; membrane protein ectodomain proteolysis [GO:0006509]; membrane protein intracellular domain proteolysis [GO:0031293]; memory [GO:0007613]; mitochondrial transport [GO:0006839]; modulation of age-related behavioral decline [GO:0090647]; myeloid dendritic cell differentiation [GO:0043011]; negative regulation of apoptotic process [GO:0043066]; negative regulation of apoptotic signaling pathway [GO:2001234]; negative regulation of axonogenesis [GO:0050771]; negative regulation of core promoter binding [GO:1904797]; negative regulation of epidermal growth factor-activated receptor activity [GO:0007175]; negative regulation of gene expression [GO:0010629]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; negative regulation of ubiquitin-protein transferase activity [GO:0051444]; neural retina development [GO:0003407]; neuron apoptotic process [GO:0051402]; neuron development [GO:0048666]; neuron migration [GO:0001764]; neuron projection maintenance [GO:1990535]; Notch receptor processing [GO:0007220]; Notch signaling pathway [GO:0007219]; positive regulation of amyloid fibril formation [GO:1905908]; positive regulation of apoptotic process [GO:0043065]; positive regulation of catalytic activity [GO:0043085]; positive regulation of coagulation [GO:0050820]; positive regulation of dendritic spine development [GO:0060999]; positive regulation of gene expression [GO:0010628]; positive regulation of glycolytic process [GO:0045821]; positive regulation of L-glutamate import across plasma membrane [GO:0002038]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of phosphorylation [GO:0042327]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; positive regulation of protein binding [GO:0032092]; positive regulation of protein import into nucleus [GO:0042307]; positive regulation of receptor recycling [GO:0001921]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of tumor necrosis factor production [GO:0032760]; post-embryonic development [GO:0009791]; protein glycosylation [GO:0006486]; protein processing [GO:0016485]; protein transport [GO:0015031]; regulation of canonical Wnt signaling pathway [GO:0060828]; regulation of gene expression [GO:0010468]; regulation of neuron projection development [GO:0010975]; regulation of phosphorylation [GO:0042325]; regulation of resting membrane potential [GO:0060075]; regulation of synaptic plasticity [GO:0048167]; regulation of synaptic transmission, glutamatergic [GO:0051966]; response to oxidative stress [GO:0006979]; sequestering of calcium ion [GO:0051208]; skeletal system morphogenesis [GO:0048705]; skin morphogenesis [GO:0043589]; smooth endoplasmic reticulum calcium ion homeostasis [GO:0051563]; somitogenesis [GO:0001756]; synapse organization [GO:0050808]; synaptic vesicle targeting [GO:0016080]; T cell activation involved in immune response [GO:0002286]; T cell receptor signaling pathway [GO:0050852]; thymus development [GO:0048538] | 11110974_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11129109_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11140838_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11200686_Observational study of gene-disease association. (HuGE Navigator) 11389157_Observational study of gene-disease association. (HuGE Navigator) 11436125_Observational study of gene-disease association. (HuGE Navigator) 11444983_Binding of the APP protease inhibitor pepstatin to presenilin 1 (PS1) is decreased in cells of a heterozygous carrier of the PS1 exon 9 deletion, suggesting that the PS1 mutation alters the inhibitor binding site. 11524469_Observational study of genetic testing. (HuGE Navigator) 11568920_Observational study of gene-disease association. (HuGE Navigator) 11744687_Here we present a novel cell-based reporter gene assay for the quantification of PS-controlled gamma-secretase cleavage of the Alzheimer amyloid precursor protein (APP). 11755019_Observational study of gene-disease association. (HuGE Navigator) 11757955_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 11757955_variable expression not a major determinant of risk for late-onset Alzheimer's disease 11775232_Observational study of gene-disease association. (HuGE Navigator) 11799129_enhancement of amyloid beta protein by Herp, and endoplasmic reticulum stress-inducible protein 11836371_The PSEN1 mutation (P264L causes disease that begins with spastic paraparesis and is associated with dementia that is not of the Alzheimer type. 11876645_inhibition of endoproteolysis by gamma-secretase inhibitors 11891288_Notch receptor cleavage depends on but is not directly executed by presenilins 11904448_Wild-type and mutated presenilins 2 trigger p53-dependent apoptosis and down-regulate presenilin 1 expression in HEK293 human cells and in murine neurons 11905990_Neurotoxic mechanisms triggered by Alzheimer's disease-linked mutant M146L presenilin 1: involvement of NO synthase via a novel pertussis toxin target. 11912199_Accelerated plaque accumulation, associative learning deficits, and up-regulation of alpha 7 nicotinic receptor protein in transgenic mice co-expressing mutant human presenilin 1 and amyloid precursor proteins 11920851_It seems this novel missense substitution of serine for glycine, occurred at codon 266 in exon 8 of presenilin 1 is pathogenic, and findings provide a new clue to the etiology of the familial early onset dementia. 11927360_The presence of a human presenilin 1 gene, normal or with an Alzheimer's disease mutation, leads to enhanced plasticity in the mouse brain. 11943765_requirement for maturation and cell surface accumulation of nicastrin 11973477_Alternative transcripts of presenilin 1 are associated with sporadic frontotemporal dementia, deletions having been identified within the exon 4-8 region. 11987239_Expression of PS1 was found throughout myeloid development from CD34+ stem cells to platelets and neutrophils, and colocalized with amyloid precursor protein in cell-specific granules, suggesting a conserved function across different tissues. 12048239_mutations of leucine 166 in presenilin-1 equally affect the generation of the Notch and APP intracellular domains independent of their effect on Abeta 42 production 12048259_regulation by nicastrin and role in determining amyloid beta-peptide production via complex formation 12050157_expression induced by interleukin-1beta and amyloid beta 42 peptide is potentiated by hypoxia in primary human neural cells 12058025_interaction with GFAP epsilon 12111439_Observational study of gene-disease association. (HuGE Navigator) 12112163_Very early-onset familial Alzheimer's disease linked to G206V mutation in PS1 12119298_mutations in PSEN1 increase Abeta42 production, inhibit cleavage of APP and notch 12121968_presenilin 1 has a role in PI signal transduction, higher calcium, and apoptosis 12147673_Presenilins are targeted as a biologically active complex with Nicastrin through the secretory pathway to the cell surface, suggesting a dual function of Presenilins in gamma-secretase processing and in trafficking. 12192622_Observational study of genotype prevalence. (HuGE Navigator) 12192622_The genotype frequency of the Glu318Gly mutation in all Alzheimer's disease cases and controls in the Australian population was 2.8%. 12198112_PS1/gamma-secretase contains PEN-2 and requires it for presenilin expression 12218704_Presenilin-1 mutations alter K+ currents in the human neuroblastoma cell line, SH-SY5Y. 12230303_In PS1 human-mutant knockin mice, PS1 is expressed in microglia & the M146V mutation confers a heightened sensitivity to LPS, as indicated by superinduction of inducible nitric oxide synthase & activation of MAP kinases. 12297048_PS1 functions as a scaffold that rapidly couples beta-catenin phosphorylation through two sequential kinase activities independent of the Wnt-regulated Axin/CK1alpha complex. 12297508_These data indicate that mammalian APH-1 (mAPH-1) along with presenilin 1 and nicastrin is probably a functional component of the gamma-secretase complex required for the intramembrane proteolysis of APP and Notch 12374741_Presenilin1 mediates a dual intramembranous gamma-secretase cleavage of Notch-1. 12413003_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12413003_The PSEN1 gene -4,752 C/T polymorphism modifies the risk for AD. 12435726_regulates intracellular trafficking and cell surface delivery of beta-amyloid precursor protein 12444985_tetradecanoylphorbol acetate appears to activate the transcription of the PS1 gene by a mechanism which does not require the -10 Ets motif or the -6 CREB/AP1 motif 12460547_Presenilin 1 regulates the glycosylation and intracellular trafficking of APP and select membrane proteins. 12470641_Gamma-secretase activity is not involved in presenilin 1-mediated regulation of beta-catenin. 12495082_Observational study of gene-disease association. (HuGE Navigator) 12522139_regulation of proteolytic processing by PEN-2 and APH-1 12609057_The missense mutation in exon 5 of presenilin-1 (PS-1) gene, found in this Alzheimer's disease family, may be one of the responsible PS-1 gene mutations for familial Alzheimer disease in Chinese. 12629514_Our present findings suggest important implications for understanding CD44-dependent signal transduction and a potential role of PS/gamma-secretase activity in the functional regulation of adhesion molecules. 12660785_APH-1 stabilizes the presenilin holoprotein in the complex, whereas PEN-2 is required for endoproteolytic processing of presenilin and conferring gamma-secretase activity to the complex. 12668610_these studies provide evidence that the increased risk for AD associated with PSEN1 may result from genetic variations in the regulatory region, leading to altered expression levels of PSEN1 in neurons. 12686406_A novel mutation (L250V) was found in the presenilin 1 gene in a Japanese case of familial Alzheimer's disease with myoclonus and generalized convulsion 12742741_long-term potentiation in the CA1 was facilitated in the transgenic /presenilin 1/ rats with a different pattern of synaptic enhancement 12752408_This clinical and pathological heterogeneity in patients with the same PSEN-1 mutation suggests phenotype modulation by genetic and/or epigenetic factors. 12805290_GSK3beta activity increased in cells in the presence of mutant PS1 or in the absence of PS1 (PS1-/-). With increased GSK3beta activity, levels of kinesin light chain phosphorylation increased, and the amount of kinesin-I bound to organelles decreased 12817569_Observational study of genotype prevalence. (HuGE Navigator) 12821663_PS1 is essential for gamma-secretase activity 12846562_In vitro characterization of the presenilin-dependent gamma-secretase complex using a novel affinity ligand 12850546_In a comparison of presenilin-1 protein in SH-SY5Y neuroblastoma cell lines over-expressing either the wild type or mutant PS1 proteins, there is a decrease in the expression levels of both proteins at the level of the cell membrane. 12885573_Two novel presenilin-1 mutations (Y256S and Q222H) are associated with early-onset Alzheimer's disease. 12885769_presenilins are multifunctional proteins with catalytic activity as well as roles in the generation, stabilization, and transport of the gamma-secretase complex 12925374_Observational study of genotype prevalence. (HuGE Navigator) 12935881_These results suggest that PS1 associates intra-molecularly to form higher order complexes, which may be needed for endoproteolytic cleavage and/or gamma-secretase-associated activity. 12960155_Presenilin 1 is downregulated by notch, which directly engages gamma-secretase 13678586_PS1 has a role in N-Cad/CTF2 production and upregulating CREB-mediated transcription 14502086_PS1 is not the core of the enzyme responsible for Notch1 cleavage but is clearly part of a partly known gamma-secretase complex implicated in the cleavage of different type 1 transmembrane proteins, e.g., Notch1 and amyloid precursor protein. 14515347_Presenilin-1 is essential for efficient trafficking of N-cadherin from the endoplasmic reticulum to the plasma membrane. 14576165_phosphorylation/dephosphorylation at the caspase recognition site provides a mechanism to reversibly regulate properties of PS1 in apoptosis. 14581682_The authors describe a 28-year-old woman with histopathologically confirmed early onset Alzheimer disease characterized by severe frontal lobe involvement associated with a de novo mutation in the presenilin 1 gene (PSEN1) 14590205_Observational study of gene-disease association. (HuGE Navigator) 14625299_PS1 can regulate PLC activity and this function is gamma-secretase activity-dependent 14645205_PS1 mutants do not simply alter the preferred cleavage site for gamma-secretase, but rather they have complex effects on the regulation of gamma-secretase and its access to substrates. 14717705_Exon 9 deletion in presenilin 1 is associated with Alzheimer's disease 14741365_capable of cleaving transmembrane proteins such as presenilin 1. 14769392_Observational study of genotype prevalence. (HuGE Navigator) 14993906_PS1DeltaE9 molecules expressed in Spodoptera cells cells retain the ability to modulate amyloid beta protein levels. 15003276_Observational study of gene-disease association. (HuGE Navigator) 15004326_Clinical and cell culture phenotypes produced by these 2 codon 117 mutations are closely related suggesting that the pathogenic action of PS1 may involve effect on neurite outgrowth and endoproteolytic cleavage of the full-length protein. 15087467_Presenilin 1 stabilizes the C-terminal fragment(C99) of the amyloid precursor protein independently of gamma-secretase activity. 15115757_a sporadic early-onset patient with Alzheimer's disease, and this individual is a somatic mosaic for a mutation in the presenilin-1 gene, suggesting a novel molecular mechanism for AD 15119739_Three missense mutations in presenilin 1(Ala246Glu in exon 7, Pro267Leu in exon 8, and Leu424Arg in exon 12) are found in Polish families with early-onset Alzheimer's disease. 15123598_very low abundance, high mass (>/=670 kDa) heteromeric complexes of PS1 are associated with the highest gamma-secretase-specific activity 15123653_gamma-secretase/presenilin has a role in processing and function of notch oncoproteins 15147205_Data describe a synthetic peptide corresponding to the presenilin-1 exon 9 loop sequence, which was found to inhibit gamma-secretase in a cell-free enzymatic assay. 15159497_A new 6-nucleotide insertional mutation at 715 in highly conserved exon 3 of the presenilin 1 gene causes an aggressive dementia that maintains the usual regional hierarchy of AD pathology while extending abnormalities into more widespread brain areas. 15192701_PS1 activates P13K thus inhibiting GSK-3 activity and tau overphosphorylation: effects of Alzheimer disease mutations were studied. 15209417_PS1 is involved in neuronal differentiation by a mechanism likely independent of endoproteolysis of the protein 15210705_role of co-expression with nicastrin in rescuing loss of function mutant of APH-1 15240571_monoubiquitination and endocytosis of Notch are a prerequisite for its presenilin-dependent cleavage 15272895_Observational study of genotype prevalence. (HuGE Navigator) 15272895_This ssudy report a novel presenilin 1 (PSN1) mutation (Thr116Ile) in a woman with early onset Alzheimer's disease (AD). 15294909_C terminus of PS1 is an activation peptide ligand for the PDZ domain of Omi/HtrA2 and may regulate the protease activity of Omi/HtrA2 after its release from the mitochondria during apoptosis. 15308304_Observational study of gene-disease association. (HuGE Navigator) 15308304_the -48C/T polymorphism in the PSN1 gene promoter is involved in the modulation of amyloid beta load in human AD, and is associated with DS. 15322084_presenilin 1 (PS1)-derived fragments, mature nicastrin, APH-1, and PEN-2, associate with cholesterol-rich detergent insoluble membrane (DIM) domains of non-neuronal cells and neurons 15340889_Observational study of gene-disease association. (HuGE Navigator) 15469450_New V272A presenilin 1 mutation with very early onset subcortical dementia and parkinsonism. 15476169_Mutations in exon 6 of presenilin-1 existed in the patients with familial Alzheimer's disease and sporadic Alzheimer's disease, and the two missense mutations were probably pathological by nature. 15476169_Observational study of gene-disease association. (HuGE Navigator) 15480851_Observational study of gene-disease association. (HuGE Navigator) 15480879_Human presenilin 1 (PS1) is alternatively spliced, resulting in the presence or absence of a four-amino acid motif, VRSQ, in the PS1 N-terminus 15537629_results suggest that the proximal two-thirds of the PEN-2 TMD1 is functionally important for endoproteolysis of PS1 holoproteins and the generation of PS1 fragments, essential components of the gamma-secretase complex 15549135_Data show that the cytoplasmic tail of presenilin 1 (PS1) fulfills several functions required for complex formation, retention of unincorporated PS1 and gamma-secretase activity. 15569674_presenilin/gamma-secretase exists as a mature complex at the cell surface and has roles in processing many adhesion molecules and receptors required for cell-cell interaction or intercellular signaling 15591316_dimeric (NCSTN/APH-1) and trimeric (NCSTN/APH-1/PS1) intermediates of gamma-secretase complex assembly are retained within the ER and incorporation of the fourth binding partner (PEN-2) also occurs on immature NCSTN. 15622541_M146L mutant PS-1 may predispose to both Pick's disease and AD by affecting multiple intracellular pathways involving tau phosphorylation and amyloid metabolism. 15629423_knock down of anterior pharynx defective 1 homolog A (APH-1a), but not APH-1b, resulted in impaired maturation of nicastrin and reduced expression of presenilin 1, presenilin 2, and PEN-2 proteins 15703411_The FAD (familial Alzheimer's disease)-linked PS1 variant produces transcriptome changes primarily by gain of aberrant function in transfected mice. 15722417_aggressive Alzheimer-causing mutation in PS1 strongly reduced photolabeling by a transition-state analogue but not by helical peptides, providing biochemical evidence that the effect of this PS mutation is due to alteration of the active-site topography 15732120_the mutant PSEN1 might play an important role in the pathogenetic process of both aggregation of alpha-synuclein into Lewy bodies and deposition of beta-amyloid into cotton wool plaques. 15743767_GHR is subject to sequential proteolysis by metalloprotease and gamma-secretases, including PS1 15764367_analysis of interaction between presenilin 1 and APP (amyloid beta precursor) 15764596_PS1 mutations can lead to a wide spectrum of changes in the activity and specificity of gamma-secretase 15776278_Of the nine pathogenic mutations found in 12 cases, three were in APP, one in PSEN2, and five in PSEN1, including two novel Greek mutations (L113Q and N135S)in Alzheimer disease 15843437_Hydrophobic region 10 spans the membrane and the COOH terminal amino acids of PS1 lie in the extracytoplasmic space 15851849_This novel presenilin 1 (PS1) gene mutation is likely to be causative of Chinese familial Alzheimer's disease. 15866047_PS1 endoproteolysis occurs via intramolecular cleavage and does not require dimerization 15908021_PS1/I213T KI mice had retarded acquisition of place learning in the water maze and in the visible platform subtest, were display whole-body startle to an auditory stimulus and a tighter grip on a horizontal grid. 15917251_show that LRP colocalizes and interacts with endogenous PS1 and tested its role as a competitive substrate for gamma-secretase 15936948_Altogether, our results demonstrated that phosphorylated CTFs can be the substrates of the gamma-secretase and that an increase in the phosphorylation of APP-CTFs facilitates their processing by gamma-secretase. 15946688_The PS-1 mutations predominantly affect the CA regions with profound neurodegeneration, which contributes early and severe clinical features of familial Alzheimer's disease . 15951428_Wild-type PS1 transgenes expressed in the mouse CNS support Abeta40 or Abeta42 production. 15975068_Observational study of genotype prevalence. (HuGE Navigator) 15975090_results suggest that ubiquilin regulates presenilin fragment production 16006137_Ncam1 is altered in transgenic mouse brains expressing FAD-linked PS1 variants, suggesting that expression of dominantly inherited mutant PS1 genes interferes with the normal Ncam1 expression via the p75 signaling pathway. 16014629_Presenilin (PS)1 mutations interfere with PS2-mediated activity by reducing PS2 fragments 16046406_PS1 has nine TM domains and the C terminus locates to the lumen/extracellular space 16079160_Psen1 expression in neural progenitor cells is crucial for cortical development and reveal a novel role for neuroectodermal expression of Psen1 in development of the brain vasculature 16116115_study is consistent with findings in sporadic Alzheimer disease of early problems with memory, visuospatial function, and particularly with executive function in PS1 mutation carriers 16126725_cadherins mediate both the association of PS1 and beta-catenin and the effects of PS1 on the cellular levels of beta-catenin 16128583_strong evidence provided that hydrophobic regions 8 and 9 are indeed membrane spanning and the integration of HR 8 into the membrane is dependent on the presence of HR 9 16135086_endoproteolysis, N and C terminal fragment interactions, and the assembly and activity of gamma-secretase complexes are very conserved between PS1 and PS2 16139258_Observational study of genetic testing. (HuGE Navigator) 16234243_mutational analyses revealed that the 'NF' sequence within the TMD4 of PS1 is the minimal motif that is required for binding with PEN-2, promoting PS1 endoproteolysis and gamma-secretase activity 16234244_Pen-2 may contribute to the activation of the gamma-secretase complex by directly binding to the TMD4 of PS1 16267640_Clinical phenotypic heterogeneity of Alzheimer's disease associated with mutations of the presenilin-1 gene. 16344340_A novel PSEN1 mutation causes very-early-onset FAD with associated Lewy bodies. To our knowledge, this kindred has the earliest reported onset of pathologically confirmed FAD and dementia with Lewy bodies. 16376112_These results suggest that PS1 regulates the growth and differentiation of endothelial progenitor cells through its beta-catenin-binding region. 16388371_novel presenilin 1 L166H mutation in a pseudo-sporadic case of early-onset Alzheimer's disease. 16401857_The authors describe four members of a family with a novel P284S presenilin 1 mutation presenting a clinical phenotype characterized by early-onset dementia, paratetraparesis, dysarthria, dysphagia, and marked involvement of brain white matter. 16407539_Expression of familial-Azheimer-disease-linked mutant PS1DeltaE9 variants enhances the vulnerability of mouse neurons in the entorhinal cortex to perforant-pathway- lesion-induced cytotoxicity. 16423463_Observational study of gene-disease association. (HuGE Navigator) 16569643_the DAPT binding site as a novel functional domain within the PS C-terminal fragment that is distinct from the catalytic site or the substrate binding site 16574645_protective role of the wild-type PS1 against the early onset familial Alzheimer disease mutation-induced amyloid pathology through a partial loss-of-function mechanism 16605258_These results suggest that the precision of cleavage by the PS/gamma-secretase complex may be physiologically regulated by the subcellular location and conditions. 16620965_Familial Alzheimer's disease (FAD)-linked Presenilin mutants lower the Ca(2+) content of intracellular stores. 16628450_In conclusion, the Ala431Glu mutation in Presenilin 1 (PSEN1), is a prevalent cause of early-onset familial Alzheimer's disease in families from the State of Jalisco, Mexico. 16735229_Elevated APP and PS1 gene expression is associated with dementia but not especially with Alzheimer's disease. 16756946_We demonstrate the existence of two physically different forms of gamma-secretase-associated PS1, one that is relatively proteinase K-sensitive and one that is significantly more PK-resistant. 16814287_PS1 is an unprimed GSK-3beta substrate;important implications for regulation of PS1 function and the pathogenesis of Alzheimer's disease 16815845_Data show that ubiquilin 1 interacts both with presenilin 1 (PS1) holoprotein and heterodimer and that the interaction between PS1 and ubiquilin 1 takes place near the cell surface. 16846981_Data indicate that presenilin-1 has a nine-transmembrane domain topology with the COOH terminus exposed to the lumen/extracellular surface. 16897084_the Ala431Glu substitution in the PSEN1 gene is not an uncommon cause of early-onset autosomal dominant Alzheimer disease in persons of Mexican origin 16908988_PS1 mutations associated with FAD abolish production of the N-cadherin intracellular fragment 16916581_Our data reveal that the Presenilin-1 V97L variant can elevate Abeta42 levels both intracellularly and extracellularly, and was a potentially pathogenic mutation for this Chinese FAD pedigree. 16938285_Observational study of gene-disease association. (HuGE Navigator) 16941492_PSEN1 gene mutations resulting in large increases in secreted Abeta42 levels or loss of PSEN1 exons 8 or 9 may cause Alzheimer disease with spastic paraparesis and cotton wool plaques. 16952411_In a case-control association study in an Italian population there is a significant association in PSEN-1 carriers of a glutamic acid318glycine mutation and familial Alzheimer's disease. 16952411_Observational study of gene-disease association. (HuGE Navigator) 17088253_expression of mutant PS1 in cultured neurons depresses synaptic transmission by causing a physical reduction in the number of synapses 17099291_PS1 may be involved in neuritogenesis and morphological change in SH-SY5Y cells, and P117L mutation may linked to Alzheimer's disease by different mechanisms 17108181_Collectively, our data suggest that the active site of gamma-secretase resides in a catalytic pore filled with water within the lipid bilayer and is tapered around the catalytic aspartates. 17158800_Presenilin mutations linked to familial Alzheimer's disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. 17192785_Meta-analysis of gene-disease association. (HuGE Navigator) 17210196_The elevated Abeta42/Abeta40 ratio observed with mutant PS1-expressing cells may be due to reduced Abeta40 production not increased Abeta42 production. 17229472_provides comprehensive profile of non-cognitive behaviors of APP/PS1 transgenic mouse model; reveals behavior impairments that may be pertinent to behavior seen in AD patients 17259169_PS/gamma-secretase-mediated cleavage of LAR controls LAR-beta-catenin interaction, suggesting an essential role for PS/gamma-secretase in the regulation of LAR signaling 17268504_reduced presenilin proteolytic function leads to increased Abeta42/Abeta40 in Alzheimer disease (Review) 17268505_mutations in Alzheimer disease (Review) 17309564_RNA interference of PS1 or Notch1 can block Notch signaling and consequently induce growth inhibition of HeLa cells. 17320044_In this work, spectroscopic data is presented that does not correlate structure or stability of the proposed PS-I autoinhibitory module with pathogenicity. 17349981_PC12 cells were stably transfected with familial mutations and were examined for gamma-secretase-dependent proteolysis of p75NTR. The presence of the M146V mutation was shown to significantly increase cleavage of p75NTR. 17366635_results also suggest that PSEN1 mutations can cause Alzheimer's disease with a large range in age of onset, spanning both early- and late-onset Alzheimer's disease 17389597_abnormal activation of glycogen synthase kinase 3beta can reduce neuronal viability and synaptic plasticity via modulating Presenilin 1/N-cadherin/beta-catenin interaction and thus have important implications in the pathophysiology of Alzheimer disease. 17401156_Exogenous cholesterol and compartmentalization in neuroblastoma cells play a relevant role in regulating the transcription of presenilin 1. 17405936_APP and PS1 are closely associated in the centrosomes of the H4 cell 17412506_No association with late-onset form of Alzheimer's disease. 17412506_Observational study of gene-disease association. (HuGE Navigator) 17431506_Many familial Alzheimer disease mutations in presenilins are loss-of-function mutations affecting endoplasmic reticulum calcium (Ca2+) leak activity, consistent with the potential role of disturbed Ca2+ homeostasis in Alzheimer disease pathogenesis. 17489097_activation of PS1 transcription by ERM was eliminated with increasing levels of CHD3 17502474_Association of a presenilin 1 S170F mutation with a novel Alzheimer disease molecular phenotype. 17507029_The novel PSEN1 mutation identified in this patient adds to the diverse list of existing mutations causing early onset Alzheimer disease associated with spastic paraparesis. 17553989_Intense and focal positron emission tomography agent (Pittsburgh compound-B) retention is observed in the striatum of all presenilin-1 mutation carriers studied (aged 35-49 years and at high risk for Alzheimer disease). 17560791_all known gamma-secretase complexes are active in APP processing and that all combinations of APH-1 variants with either familial Alzheimer mutant PS1 or PS2 support pathogenic Abeta(42) production 17573346_aggressive presenilin-1 mutations cause insensitivity to Abeta42-lowering nonsteroidal anti-inflammatory drugs and gamma-secretase inhibitors 17588625_summarizes work focused on identifying and studying domains in PS1 that are critical for mediating gamma-secretase activity 17594345_A new mutation (496_498delCTT at codon 166) in the 3d transmembrane domain of exon 6 was associated with the 'typical' presentation of early-onset AD seen in most exon 6 mutations, but not in deletions at other sites. 17627113_Observational study of gene-disease association. (HuGE Navigator) 17630980_These data indicate that the presence of an FAD-associated mutant human PS1 transgene is associated with redistribution of the APP and APP CTFs in brain neurons toward TGN-enriched fractions. 17632280_We sequenced nine spastic paraparesis genes in three Alzheimer's disease families with PSEN1 exon 9 deletions. We did not observe any correlation of polymorphisms or mutations in the nine spastic paraparesis genes. 17645236_a single point mutation, p.L420R, is the mutation responsible for early onset Alzheimer disease, seizures and cotton wool plaques without spastic paraparesis in a family 17718701_We describe a case of a young patient suffering from a rapidly progressive cognitive decline, associated with delusions, myoclonus and seizures and with no family history for dementia anddemonstrated a de novo presenilin 1 mutation. 17719017_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17719017_Presenilin-1 2/2 genotype is a risk factor for late onset Alzheimer disease in the Spanish population, and probably, for Europeans. 17852592_Healthy carriers of the E280A presenilin-1 gene mutation scored significantly lower than noncarriers on naming of famous faces. Cognitive changes in lexical-semantic tasks can be detected before the clinical diagnosis of probable familial Alzheimer's. 17942194_the familial Alzheimer's disease-linked PS1 mutation accelerates the cleavage of caspase-4 under the endoplasmic reticulum stress 17962197_The amount of Abeta(42) produced by cells expressing 10 different familial Alzheimer disease is associated with mutations in presenilin 1. 17968601_Familial cases presenting very early onset autosomal dominant Alzheimer's disease with I143T in presenilin-1 gene: implication for genotype-phenotype correlation. 18024701_present study demonstrates that PSEN1 linked autosomal dominant early onset Alzheimer's disease has to be considered, even when ataxia precedes dementia, the Pro117Ala mutation being responsible for a predominant precocious ataxia 18028191_L286P is a novel mutation in PSEN1 that causes familial early-onset AD and brain haematomas related to amyloid angiopathy. 18090315_neurogenesis is decreased with degrees of Abeta pathology, and that there is no gender difference in their proliferation in APPswe/PS1dE9 transgenic mice 18174159_Overexpression of presenilin-1 increased HIF-1beta, suggesting that HIF is downstream of presenilin 18239458_In the PS1 familial Alzheimer's disease brain, cyclin D1 accumulation may occur and lead to neuronal apoptosis secondary to an abortive entry into the cell cycle. 18293935_Mature integrin beta1 with increased expression level is delivered to the cell surface, which results in an increased cell surface expression level of mature integrin beta 1 in presenilin (PS)1 and PS2 double-deficient fibroblasts. 18299393_PS1 can interact with AChE and influence its expression, supporting the notion of cholinergic-amyloid interrelationships. 18314228_This study identified two novel heterozygous mutations in two unrelated patients, Cys139Arg in the PGRN gene and Val412Ile in the PSEN1 gene. 18317569_Data show that a presenilin-1 280Glu-->Ala mutation alters C-terminal amyloid precursor protein processing, yielding longer amyloid beta peptides, and discuss the implications for Alzheimer's disease. 18320103_Data show that the E280A mutation in presenilin 1 is not associated with increased production of amyloid-beta in non-neuronal peripheral tissues, which is in contrast to the expectation in a gamma-secretase gain of function. 18350357_We report a late onset familial Alzheimer's disease: novel presenilin 2 mutation and PS1 E318G polymorphism. 18376127_Observational study of gene-disease association. (HuGE Navigator) 18376127_The tendency for a higher prevalence of headaches held for different PSEN1 and APP mutations but was not significant unless all families were combined. 18403054_Observational study of gene-disease association. (HuGE Navigator) 18403054_case-control study failed to detect any convincing risk factors of polymorphisms in PSEN1 affecting either the risk of developing Alzheimer's disease and/or cognitive decline 18430735_I213T mutant presenilin-1/gamma-secretase in cell models and knock-in mouse brains: familial Alzheimer disease-linked mutation impairs gamma-site cleavage of amyloid precursor protein C-terminal fragment beta 18437002_PSEN1 and APP gene mutations may not be uncommon in Korean patients with early-onset Alzheimer's disease 18445230_pathological levels of Abeta42 may be caused by the negative effects of a novel presenilin-1 mutant (V97L) on insulin-degrading enzyme expression and activity 18479822_Observational study of gene-disease association. (HuGE Navigator) 18479822_The P117A variant is a novel mutation in PSEN1, which causes early-onset AD in an autosomal dominant manner. 18525293_did not find any modification of the A beta(1-42)/A beta(1-40) ratio, suggesting that this double mutation might be involved in early-onset AD etiopathogenesis by affecting a PSEN-1 function other than gamma-secretase activity. 18579078_Enhanced Ca2+ release from the endoplasmic reticulum as a result of the specific interaction of mutan | ENSMUSG00000019969 | Psen1 | 1314.10468 | 0.9765084 | -0.0342956936 | 0.12271886 | 7.804454e-02 | 7.799651e-01 | 9.998360e-01 | No | Yes | 1068.03991 | 124.070371 | 1.199239e+03 | 107.606001 | |
| ENSG00000080823 | 5891 | MOK | protein_coding | Q9UQ07 | FUNCTION: Able to phosphorylate several exogenous substrates and to undergo autophosphorylation. Negatively regulates cilium length in a cAMP and mTORC1 signaling-dependent manner. {ECO:0000250|UniProtKB:Q9WVS4}. | ATP-binding;Alternative splicing;Cell projection;Cytoplasm;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene belongs to the MAP kinase superfamily. The gene was found to be regulated by caudal type transcription factor 2 (Cdx2) protein. The encoded protein, which is localized to epithelial cells in the intestinal crypt, may play a role in growth arrest and differentiation of cells of upper crypt and lower villus regions. Multiple alternatively spliced transcript variants encoding different isoforms have been observed for this gene. [provided by RefSeq, Dec 2012]. | hsa:5891; | ciliary base [GO:0097546]; cilium [GO:0005929]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; ATP binding [GO:0005524]; cyclin-dependent protein serine/threonine kinase activity [GO:0004693]; metal ion binding [GO:0046872]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; intracellular signal transduction [GO:0035556]; protein phosphorylation [GO:0006468]; signal transduction [GO:0007165] | 12777992_RAGE was demonstrated in all 8 yolk sac tumors and 21 of 26 embryonal carcinomas. In yolk sac tumors, RAGE reactivity was diffusely present throughout the tumors. In embryonal carcinomas, RAGE was identified only in yolk sac components 15327990_identification of MOK, a member of the mitogen-activated protein kinase superfamily, as one of the genes induced by a caudal-related homeobox transcription factor, Cdx2 15900605_May provide suitable targets for immunotherapy of renal cell carcinoma. 18685487_Compared RAGE and PAX-2 staining in metastatic clear renal cell carcinoma. 19060005_POLL is under genetic selection in Sub-Saharan African populations. 21264954_The RAGE pathway may play an important role in STAT3 induction in glioma-associated macrophages and microglia, a process that may be mediated through S100B. 21377387_PBMNC from type 2 diabetics were more sensitive to innate immune stimulation with LPS and monoclonal agonist anti-TLR4 than were cells from ND. The actions of LPS, anti-TLR4 and anti-RAGE potentiated the production of IL-6 and TNF-alpha in both groups. 21717246_Our results suggest that RAGE may be important in tumor invasion and could be a potential predictor for the prognosis of hepatocellular carcinoma patients. 24244486_the expressions of ICK/MAK/MOK proteins in the intestinal tract can be differentially and dynamically regulated, implicating a significant functional diversity within this group of protein kinases. 24529564_The findings indicate a statistically significant association of p.Gly82Ser polymorphism in RAGE with DR in T2DM patients. 25755699_our results suggest MOK promoter hypomethylation is a common event and contributes to MOK overexpression in acute myeloid leukemia 33639133_RAGE silencing deters CML-AGE induced inflammation and TLR4 expression in endothelial cells. | ENSMUSG00000056458 | Mok | 437.39402 | 0.9599826 | -0.0589198118 | 0.16244254 | 1.296582e-01 | 7.187867e-01 | 9.998360e-01 | No | Yes | 449.36854 | 51.939011 | 4.487035e+02 | 40.581474 | |
| ENSG00000080986 | 10403 | NDC80 | protein_coding | O14777 | FUNCTION: Acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity (PubMed:9315664, PubMed:12351790, PubMed:14654001, PubMed:14699129, PubMed:15062103, PubMed:15235793, PubMed:15239953, PubMed:15548592, PubMed:16732327, PubMed:30409912). Required for kinetochore integrity and the organization of stable microtubule binding sites in the outer plate of the kinetochore (PubMed:15548592, PubMed:30409912). The NDC80 complex synergistically enhances the affinity of the SKA1 complex for microtubules and may allow the NDC80 complex to track depolymerizing microtubules (PubMed:23085020). Plays a role in chromosome congression and is essential for the end-on attachment of the kinetochores to spindle microtubules (PubMed:25743205, PubMed:23891108). {ECO:0000269|PubMed:12351790, ECO:0000269|PubMed:14654001, ECO:0000269|PubMed:14699129, ECO:0000269|PubMed:15062103, ECO:0000269|PubMed:15235793, ECO:0000269|PubMed:15239953, ECO:0000269|PubMed:15548592, ECO:0000269|PubMed:16732327, ECO:0000269|PubMed:23085020, ECO:0000269|PubMed:23891108, ECO:0000269|PubMed:25743205, ECO:0000269|PubMed:30409912, ECO:0000269|PubMed:9315664}. | 3D-structure;Acetylation;Cell cycle;Cell division;Centromere;Chromosome;Coiled coil;Kinetochore;Mitosis;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a component of the NDC80 kinetochore complex. The encoded protein consists of an N-terminal microtubule binding domain and a C-terminal coiled-coiled domain that interacts with other components of the complex. This protein functions to organize and stabilize microtubule-kinetochore interactions and is required for proper chromosome segregation. [provided by RefSeq, Oct 2011]. | hsa:10403; | centrosome [GO:0005813]; chromosome, centromeric region [GO:0000775]; cytosol [GO:0005829]; kinetochore [GO:0000776]; membrane [GO:0016020]; Ndc80 complex [GO:0031262]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; outer kinetochore [GO:0000940]; cyclin binding [GO:0030332]; identical protein binding [GO:0042802]; attachment of mitotic spindle microtubules to kinetochore [GO:0051315]; attachment of spindle microtubules to kinetochore [GO:0008608]; cell division [GO:0051301]; centrosome duplication [GO:0051298]; chromosome segregation [GO:0007059]; establishment of mitotic spindle orientation [GO:0000132]; G2/MI transition of meiotic cell cycle [GO:0008315]; kinetochore organization [GO:0051383]; metaphase plate congression [GO:0051310]; mitotic cell cycle [GO:0000278]; mitotic sister chromatid segregation [GO:0000070]; mitotic spindle organization [GO:0007052]; positive regulation of mitotic cell cycle spindle assembly checkpoint [GO:0090267]; positive regulation of protein localization to kinetochore [GO:1905342]; regulation of protein stability [GO:0031647]; spindle assembly involved in female meiosis I [GO:0007057] | 12351790_role in spindle checkpoint signaling; required for the recruitment of Mps1 kinase and Mad1/Mad2 complexes to kinetochores 12386167_cell cycle-regulated serine phosphorylation of Hec1 by Nek2 is essential for faithful chromosome segregation 15548592_HEC1 is a core component of the kinetochore outer plate essential for organizing microtubule attachment sites. 15931389_Altered expression is associated with therapy failure and death in patients with multiple types of cancer. 15961401_the Spc24, Spc25, Nuf2, and Ndc80/Hec1 complex is a faithful copy of the endogenous Ndc80 complex 17129782_These findings reveal a key role for the Hec1 N terminus in controlling dynamic behavior of kinetochore microtubules. 17195848_Study describes the crystal structure of the most conserved region of HEC1, which lies at one end of the rod and near the N terminus of the polypeptide chain, it folds into a calponin-homology domain. 17822787_overexpression of either CREB or ATF4 enhanced the activation of the HEC1 promoter and overexpression of both of them had an additive effect on the activation of the HEC1 transcription. 18297113_that Hec1 binds to microtubule in low affinity and phosphorylation by NEK2A, which prevents aberrant kinetochore-microtubule connections in vivo, increases the affinity of the Ndc80 complex for microtubules in vitro 18455984_The 180 kDa Ndc80 complex is a direct point of contact between kinetochores and microtubules; its four subunits contain coiled coils and form an elongated rod structure with functional globular domains at either end. 18782526_Hec 1 was highhy expressed in cancer tissues with lymph node metastasis and poor differentiation. 18794333_Results suggest that the globular domain of the Ndc80 subunit binds strongly at the interface between tubulin dimers and weakly at the adjacent intradimer interface along the microtubule protofilament axis. 19553660_Data show that the kinetochore localization of PinX1 is dependent on Hec1 and CENP-E. 19776357_Results suggest that Hec1, through cooperation with Hice1, contributes to centrosome-directed microtubule growth to facilitate establishing a proper mitotic spindle. 19878654_cell growths of colorectal and gastric cancers after the siRNA-mediated knockdown of either CDCA1 or KNTC2 were significantly suppressed. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20944740_subnanometre-resolution cryo-electron microscopy reconstruction of the human Ndc80 complex bound to microtubules 20948316_Data provide evidence for a functional link between Hec1 expression and the pRb pathway. 21056971_CENP-U is a novel microtubule binding protein and plays an important role in kinetochore-microtubule attachment through its interaction with Hec1 21266467_Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. 21270439_These data suggest that the CH and tail domains of Hec1 generate essential contacts between kinetochores and microtubules in cells, whereas the Nuf2 CH domain does not. 21297979_N-terminally modified Hec1 promotes spindle pole fragmentation by CENP-E-mediated plus-end directed kinetochore pulling forces that disrupt the fine balance of kinetochore- and centrosome-associated forces regulating spindle bipolarity 21325630_In vertebrates there is a tripartite attachment point facilitating the interaction between Hec1/Ndc80 and microtubules. 21352579_Alteration in NDC80, were also detected in benign breast tumors. 21832156_Hec1 serine 165 (S165) is preferentially phosphorylated at kinetochores by Nek2 and it serves as an important mechanism in modulating spindle assemble checkpoint signaling and chromosome alignment. 22454517_The Ndc80 internal loop is essential for end-on microtubule attachment to kinetochores. 22561346_results reveal the molecular function of CENP-T proteins and demonstrate how the Ndc80 complex is anchored to centromeres in a manner that couples chromosome movement to spindle dynamics 22581055_results support the conclusion that Cdt1 binding to Hec1 is essential for an extended Ndc80 configuration and stable kinetochore-microtubule attachment 22908300_The Ndc80 kinetochore complex directly modulates microtubule dynamics. 23056589_NDC80, NUF2 and PTN were significantly aberrantly overexpressed in serous adenocarcinomas. 23085714_Ndc80's interaction with either growing or shrinking microtubule ends can be tuned by the phosphorylation state of its tail. 23474708_Hec1 expression was highest in paclitaxel-resistant A2780/Taxol cells. 23591767_Hec1 is critical in maintaining the in vitro and in vivo growth of gastric cancer cells; elevated Hec1 levels may occur at the early stage of gastric tumorigenesis 24129578_these findings demonstrated that three buried glutamic acid-lysine pairs, in concert with hydrophobic interactions of core residues, provide the major specificity and stability requirements for Hec1-Nuf2 dimerization 24187132_a novel role for Aurora B-Hec1-Mps1 signaling axis in governing accurate chromosome segregation in mitosis 24327015_Growth inhibition following knockdown of NDC80, CDK1 and PLK1. 24413531_The Ndc80 complex binds straight microtubules by recognizing the dimeric interface of tubulin. 24694948_Certain clinical subtypes of breast cancer more likely to respond to Hec1-targeted therapy were identified and these subtypes are the ones associated with poor prognosis. 25132262_N-terminus-modified Hec1 suppresses tumour growth by interfering with kinetochore-microtubule dynamics 25557589_Overproduction of Ndc80 in cancer cells may unfavourably absorb protein interactors through the internal loop domain and lead to a change in the equilibrium of microtubule-associated proteins. [Review] 25601404_Mechanisms of mitosis-specific assembly of the checkpoint platform Knl1/MIS12/NDC80 at human kinetochores. 25808492_Independent molecular binding events to microtubules (MTs) by individual NDC80 complexes, rather than their structured oligomers, regulate the dynamics and stability of kinetochore-MT attachments in dividing cells. 26382282_This study suggests that Ndc80 may play an important role in the process of hepatitis B virus-related hepatocellular carcinoma, and that it may be a potential biological treatment target in the future. 26612002_we conclude that Hec1 is consistently overexpressed in human PCa and Hec1 is closely linked with human PCa progression through the meditator LncRNA BX647187 26941333_Ndc80 complex bound to microtubules binds every tubulin monomer along the microtubule protofilament. 27173328_Expression of NDC80 in colon cancer cells and tissues was higher than that in controls. NDC80 promotes the proliferation and metastasis of colon cancer cells. 28012276_Whereas CENP-C recruits a single MIS12:NDC80 complex, the authors show here that CENP-T binds one MIS12:NDC80 and two NDC80 complexes upon phosphorylation by the mitotic CDK1:Cyclin B complex at three distinct CENP-T sites. 28479321_Study shows that Cdk1 phosphorylates Ska3 to promote its direct binding to the Ndc80 complex (Ndc80C), a core outer kinetochore component, also show that this phosphorylation occurs specifically during mitosis and is required for the kinetochore localization of the Ska complex. 28535377_Ska is recruited to kinetochores by clusters of Ndc80 proteins. 28537682_We provided novel evidence that NDC80 expression is upregulated in osteosarcoma tissues, where it is positively correlated with advanced tumor stage and distant metastasis 28552353_Hec1 tail phosphorylation tunes friction along polymerizing microtubules and yet does not compromise the kinetochore's ability to grip depolymerizing microtubules. 28611520_Elevated expression of NDC80 may play a role in promoting the development of hepatocellular carcinoma 28841134_The authors show that the Astrin-SKAP complex binds synergistically to microtubules with the Ndc80 complex to form an integrated interface. 29142109_Ndc80 recruits Bod1 to kinetochores which directly feeds forward to regulate Ndc80 29187526_Aurora A kinase regulates kinetochore-microtubule dynamics of metaphase chromosomes, and Hec1 S69, a previously uncharacterized phosphorylation target site in the Hec1 tail, is a critical Aurora A substrate for this regulation. 29341479_High NDC80 expression is associated with colorectal cancer. 29475948_These findings support the idea that dynein may control the function of the Ndc80 complex in stabilizing kinetochore-microtubule attachments directly by interfering with Ndc80-microtubule binding or indirectly by controlling the Rod-mediated inhibition of Ndc80. 29487209_Ska complex directly binds Ndc80 complex through interactions between the Ska3 unstructured C-terminal region and the coiled-coil regions of each Ndc80 complex subunit. 30044223_NDC80 binding is modulated in a chromosome autonomous fashion over prometaphase and metaphase, and is predominantly regulated by centromere tension. 30409912_Using pulldown assays with HeLa cell lysates and site-directed mutagenesis, this study shows that HEC1 is a bona fide substrate of the lysine acetyltransferase Tat-interacting protein, 60 kDa (TIP60) and that TIP60-mediated acetylation of HEC1 is essential for accurate chromosome segregation in mitosis. 30475206_The results indicate that Sf3A2 and Prp31 directly regulate interactions among kinetochores, spindle microtubules and the Ndc80 complex in both Drosophila and human cells. 30816525_nuclear division cycle 80, cyclin B2 and topoisomerase 2alpha may serve important roles in adrenocortical tumor development. 32219319_Mps1 dimerization and multisite interactions with Ndc80 complex enable responsive spindle assembly checkpoint signaling. 32401635_The Hec1/Ndc80 tail domain is required for force generation at kinetochores, but is dispensable for kinetochore-microtubule attachment formation and Ska complex recruitment. 32491969_Multisite phosphorylation determines the formation of Ska-Ndc80 macro-complexes that are essential for chromosome segregation during mitosis. 32795325_Bioinformatic analysis revealing mitotic spindle assembly regulated NDC80 and MAD2L1 as prognostic biomarkers in non-small cell lung cancer development. 33109182_Involvement of NEK2 and its interaction with NDC80 and CEP250 in hepatocellular carcinoma. 33988677_Chromosome oscillation promotes Aurora A-dependent Hec1 phosphorylation and mitotic fidelity. | ENSMUSG00000024056 | Ndc80 | 261.28595 | 1.0013517 | 0.0019487767 | 0.20063278 | 9.192515e-05 | 9.923502e-01 | 9.998360e-01 | No | Yes | 268.99744 | 46.395343 | 2.658238e+02 | 35.370658 | |
| ENSG00000082068 | 55100 | WDR70 | protein_coding | Q9NW82 | 3D-structure;Acetylation;Isopeptide bond;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;WD repeat | hsa:55100; | nucleus [GO:0005634]; site of double-strand break [GO:0035861]; enzyme binding [GO:0019899]; regulation of DNA double-strand break processing [GO:1903775]; regulation of histone H2B conserved C-terminal lysine ubiquitination [GO:2001173] | 27098497_CRL4(Wdr70) regulates H2B monoubiquitination and facilitates Exo1-dependent DNA repair resection. 29130659_Ovarian cancer had different expression of WDR70 and H2B monoubiquitination compared with normal tissue | ENSMUSG00000039828 | Wdr70 | 639.69790 | 0.8119542 | -0.3005298094 | 0.15744232 | 3.463637e+00 | 6.273220e-02 | 9.998360e-01 | No | Yes | 509.75116 | 84.038999 | 6.854898e+02 | 87.109052 | |||
| ENSG00000083544 | 81550 | TDRD3 | protein_coding | Q9H7E2 | FUNCTION: Scaffolding protein that specifically recognizes and binds dimethylarginine-containing proteins. In nucleus, acts as a coactivator: recognizes and binds asymmetric dimethylation on the core histone tails associated with transcriptional activation (H3R17me2a and H4R3me2a) and recruits proteins at these arginine-methylated loci. In cytoplasm, may play a role in the assembly and/or disassembly of mRNA stress granules and in the regulation of translation of target mRNAs by binding Arg/Gly-rich motifs (GAR) in dimethylarginine-containing proteins. {ECO:0000269|PubMed:15955813, ECO:0000269|PubMed:18632687, ECO:0000269|PubMed:21172665}. | 3D-structure;Alternative splicing;Chromatin regulator;Cytoplasm;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:81550; | cytosol [GO:0005829]; DNA topoisomerase III-beta-TDRD3 complex [GO:0140225]; Golgi apparatus [GO:0005794]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; methylated histone binding [GO:0035064]; RNA binding [GO:0003723]; transcription coactivator activity [GO:0003713]; chromatin organization [GO:0006325] | 18632687_Tudor domain of TDRD3 was required for its recruitment to cytoplasmic stress granules. 18664458_propose a contribution of Tdrd3 to FMRP-mediated translational repression and suggest that the loss of the FMRP-Tdrd3 interaction caused by the I304N mutation might contribute to the pathogenesis of Fragile X syndrome 21172665_The TDRD3 is an effector molecule that promotes transcription by binding methylarginine marks on histone tails. 22363433_The binding specificity and affinity of the Tudor domains of TDRD3, SMN and SPF30 proteins were characterized quantitatively. 27257063_Top3b proteins from several animals associate with polyribosomes, which are units of mRNA translation, whereas the Top3 homologs from E. coli and yeast lack the association. The Top3b-polyribosome association requires TDRD3, which directly interacts with Top3beta and is present in animals but not bacteria or yeast. 28176834_Structural basis of the interaction between TOP3B and the TDRD3 auxiliary factor has been reported. 28698590_Study reports TDRD3 as a novel regulator of cell proliferation and invasion in breast cancer cells. Its depletion inhibits tumor formation and metastasis to the lung in vivo. Furthermore, TDRD3 regulates the expression of a number of key genes associated with promotion of breast cancer tumorigenesis and disease progression at the level of translation. 29645362_Our work depicts the structural plasticity of the TDRD3 Tudor domain and paves the way for the subsequent structure-guided discovery of selective inhibitors targeting Tudor domains 34329467_TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. 35085371_TDRD3 is an antiviral restriction factor that promotes IFN signaling with G3BP1. | ENSMUSG00000022019 | Tdrd3 | 157.53635 | 0.7603356 | -0.3952917281 | 0.25722538 | 2.307663e+00 | 1.287375e-01 | 9.998360e-01 | No | Yes | 144.97444 | 26.236206 | 1.939663e+02 | 27.052565 | ||
| ENSG00000083812 | 25799 | ZNF324 | protein_coding | O75467 | FUNCTION: May be involved in transcriptional regulation. May be involved in regulation of cell proliferation. {ECO:0000305|PubMed:11779640}. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:25799; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cell population proliferation [GO:0008283]; G1/S transition of mitotic cell cycle [GO:0000082]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000004500 | Zfp324 | 730.07092 | 1.3229871 | 0.4037989841 | 0.16532058 | 6.220456e+00 | 1.262826e-02 | 9.998360e-01 | No | Yes | 1094.77220 | 166.473937 | 7.882450e+02 | 93.286489 | |||
| ENSG00000083838 | 55663 | ZNF446 | protein_coding | Q9NWS9 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:55663; | chromatin [GO:0000785]; extracellular space [GO:0005615]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 15936718_Overexpression of ZNF446 in COS-7 cells inhibits the transcriptional activities of SRE and AP-1. | ENSMUSG00000033961 | Zfp446 | 1805.52274 | 0.9900588 | -0.0144139171 | 0.11533076 | 1.605067e-02 | 8.991848e-01 | 9.998360e-01 | No | Yes | 1815.15462 | 198.943321 | 1.706411e+03 | 145.130818 | ||
| ENSG00000085840 | 4998 | ORC1 | protein_coding | Q13415 | FUNCTION: Component of the origin recognition complex (ORC) that binds origins of replication. DNA-binding is ATP-dependent. The DNA sequences that define origins of replication have not been identified yet. ORC is required to assemble the pre-replication complex necessary to initiate DNA replication. | 3D-structure;ATP-binding;Acetylation;DNA replication;DNA-binding;Disease variant;Dwarfism;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | The origin recognition complex (ORC) is a highly conserved six subunits protein complex essential for the initiation of the DNA replication in eukaryotic cells. Studies in yeast demonstrated that ORC binds specifically to origins of replication and serves as a platform for the assembly of additional initiation factors such as Cdc6 and Mcm proteins. The protein encoded by this gene is the largest subunit of the ORC complex. While other ORC subunits are stable throughout the cell cycle, the levels of this protein vary during the cell cycle, which has been shown to be controlled by ubiquitin-mediated proteolysis after initiation of DNA replication. This protein is found to be selectively phosphorylated during mitosis. It is also reported to interact with MYST histone acetyltransferase 2 (MyST2/HBO1), a protein involved in control of transcription silencing. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jun 2010]. | hsa:4998; | chromosome, telomeric region [GO:0000781]; cytosol [GO:0005829]; nuclear origin of replication recognition complex [GO:0005664]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; origin recognition complex [GO:0000808]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA replication origin binding [GO:0003688]; metal ion binding [GO:0046872]; DNA replication initiation [GO:0006270]; mitotic DNA replication checkpoint signaling [GO:0033314] | 11716535_a component of the origin recognition complex (ORC) that functions in DNA replication 11779870_ORC2 physically interacts with ORC1 on non-chromatin nuclear structures 11931757_Data suggest that hOrc1p is targeted for destruction by an SCF-Skp2 complex during S phase. 12909626_ORC1 regulates the status of the ORC complex in human nuclei by tethering ORCs 2-5 to nuclear structures 12909627_the ORC1 cycle in human cells is highly linked with cell cycle progression, allowing the initiation of replication to be coordinated with the cell cycle and preventing origins from refiring 12912926_The precise nucleotide of binding for ORC1 was identified near the start sites for leading-strand DNA synthesis. 15454574_HP1 has a role in the recruitment but not in the stable association of Orc1p with heterochromatin 17066079_BAH domain in human Orc1 facilitates its ability to activate replication origins in vivo by promoting association of ORC with chromatin. 18761675_Interacts with TRF2 to achieve stable binding in pre-replication complex assembly at telomeres. These data suggest that ORC might be involved in telomere homeostasis in human cells. 19197067_data suggest that Orc1 is a regulator of centriole and centrosome reduplication as well as the initiation of DNA replication 20800603_Observational study of gene-disease association. (HuGE Navigator) 21085491_Data show that Orc1 specifically binds hypo-phosphorylated Rb and that this interaction is competitive with the binding of Rb to E2F1. 21115485_in human cancer cells, RBX1 silencing causes the accumulation of DNA replication licensing proteins CDT1 and ORC1, leading to DNA double-strand breaks, DDR, G(2) arrest, and, eventually, aneuploidy 21358633_Mutations in ORC1, encoding a subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. These mutations disrupt ORC1 functions including pre-replicative complex formation and origin activation. 22045277_As cells moved through the cell cycle, the local-ization of ORC1 shifted, suggesting changes in thelocalization of ORC-bound origin sequences. 22333897_A lethal phenotype was seen in four individuals with compound heterozygous ORC1 mutations 22398447_results identify the BAH domain as a novel methyl-lysine-binding module, thereby establishing the first direct link between histone methylation and the metazoan DNA replication machinery, and defining a pivotal aetiological role for the canonical H4K20me2 mark, via ORC1, in primordial dwarfism 22589552_Data show that purified Epstein-Barr nuclear antigen 1 EBNA1 recruits purified Human Orc1 and Cdc6 onto replication origin oriP. 22855792_Orc1 harbors a PACT centrosome-targeting domain and a separate domain that differentially inhibits the protein kinase activities of Cyclin E-CDK2 and Cyclin A-CDK2 23187890_ORC1 associated with transcription start sites of coding or noncoding RNAs. Transcription levels at the ORC1 sites directly correlated with replication timing. 24003239_ORC1 harbors a G-rich RNA/ssDNA-binding domain, which may be involved in the preferential binding to G-quadruplex-formable RNA/DNA by ORC. Structure modeling predicts the structural similarity between the G-rich RNA/ssDNA-binding domain of ORC1 and part of mammalian DNA methyltransferase 1. 25784553_Orc1 acts as a nucleating center for origin recognition complex assembly and then pre-replication complex assembly by binding to mitotic chromosomes, followed by gradual removal from chromatin during the G1 phase. 27458800_The opposing effects of ORC1 (represor) and CDC6 (gene activator) in controlling the level of Cyclin E ensures genome stability and a mechanism for linking directly DNA replication and cell division commitment. 27906128_The authors have discovered that human cell lines in culture proliferate with intact chromosomal origins of replication after disruption of both alleles of ORC2 or of the ATPase subunit, ORC1. 31309634_Authors report the crystal structure of Cyclin A-CDK2 complex bound to a peptide derived from ORC1 at 2.54 a resolution; structure revealed that the ORC1 peptide interacts with a hydrophobic groove, termed cyclin binding groove, of Cyclin A via a KXL motif. 31866342_Origin recognition complex subunit 1 regulates cell growth and metastasis in glioma by altering activation of ERK and JNK signaling pathway. 33761311_Multiple, short protein binding motifs in ORC1 and CDC6 control the initiation of DNA replication. | ENSMUSG00000028587 | Orc1 | 2213.73686 | 0.9375925 | -0.0929670224 | 0.10495115 | 7.874456e-01 | 3.748737e-01 | 9.998360e-01 | No | Yes | 2219.69767 | 160.344629 | 2.420314e+03 | 135.512833 | |
| ENSG00000086289 | 54749 | EPDR1 | protein_coding | Q9UM22 | FUNCTION: Binds anionic lipids and gangliosides at acidic pH. {ECO:0000269|PubMed:30729188}. | 3D-structure;Alternative splicing;Disulfide bond;Glycoprotein;Lipid-binding;Lysosome;Reference proteome;Secreted;Signal | The protein encoded by this gene is a type II transmembrane protein that is similar to two families of cell adhesion molecules, the protocadherins and ependymins. This protein may play a role in calcium-dependent cell adhesion. This protein is glycosylated, and the orthologous mouse protein is localized to the lysosome. Alternative splicing results in multiple transcript variants. A related pseudogene has been identified on chromosome 8. [provided by RefSeq, Aug 2011]. | hsa:54749; | extracellular region [GO:0005576]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; calcium ion binding [GO:0005509]; ganglioside GM1 binding [GO:1905573]; identical protein binding [GO:0042802]; phospholipid binding [GO:0005543]; cell-matrix adhesion [GO:0007160]; myofibroblast contraction [GO:1990764] | 16954209_Shows that the mouse ortholog is a lysosomal protein. 18374504_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24089297_Results of this study suggest that EPDR1 gene can be added to a growing list of genes associated with Dupuytren's disease development. 27245865_Study results suggest functional involvement of EPDR1 in the etiology of Dupuytren's Disease. 27805251_EPDR1 exhibits a pattern of isoform abundance dependent on KRAS G13D and G12D mutations. 30729188_Crystal structures of human lysosomal EPDR1 reveal homology with the superfamily of bacterial lipoprotein transporters, such as LolA. 32111877_EPDR1 up-regulation in human colorectal cancer is related to staging and favours cell proliferation and invasiveness. 32682838_Evaluation of Primary Angle-Closure Glaucoma Susceptibility Loci for Estimating Angle Closure Disease Severity. 32935479_EPDR1 correlates with immune cell infiltration in hepatocellular carcinoma and can be used as a prognostic biomarker. 33902536_EPDR1 is related to stages and metastasize in bladder cancer and can be used as a prognostic biomarker. | ENSMUSG00000002808 | Epdr1 | 224.47657 | 1.1294337 | 0.1755995288 | 0.21797008 | 6.404936e-01 | 4.235321e-01 | 9.998360e-01 | No | Yes | 237.85603 | 34.052367 | 2.221479e+02 | 25.070757 | |
| ENSG00000087502 | 51290 | ERGIC2 | protein_coding | Q96RQ1 | FUNCTION: Possible role in transport between endoplasmic reticulum and Golgi. {ECO:0000250}. | Cytoplasm;ER-Golgi transport;Endoplasmic reticulum;Golgi apparatus;Membrane;Nucleus;Reference proteome;Transmembrane;Transmembrane helix;Transport | ERGIC2, or PTX1, is a ubiquitously expressed nuclear protein that is downregulated in prostate carcinoma (Kwok et al., 2001 [PubMed 11445006]).[supplied by OMIM, Aug 2008]. | hsa:51290; | COPII-coated ER to Golgi transport vesicle [GO:0030134]; cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleus [GO:0005634]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890] | 12932305_may play an important role in the growth and tumorigenicity of PC-3 prostate tumor cells 15308636_ERGIC-32 functions as a modulator of the hErv41-hErv46 complex by stabilizing hErv46 16989575_Ectopic expression of a partial sequence of PTX1 (Met84 - Leu225) as a VP22-fusion protein in prostate cancer cell line, PC-3, induced cellular senescence. 17980171_CDA14 participated in the elongation factor 1alpha regulated mechanisms 24303950_A variant of ERGIC2 with four coding bases deleted has an abrogated function as a transport shuttle, but does continue to upregulate the heme oxygenase 1 gene, suggesting that it may be involved in the oxidative stress pathway. | ENSMUSG00000030304 | Ergic2 | 300.61109 | 1.2463934 | 0.3177595111 | 0.29027190 | 9.751848e-01 | 3.233905e-01 | 9.998360e-01 | No | Yes | 302.10991 | 80.796560 | 2.753511e+02 | 56.595169 | |
| ENSG00000089022 | 8550 | MAPKAPK5 | protein_coding | Q8IW41 | FUNCTION: Tumor suppressor serine/threonine-protein kinase involved in mTORC1 signaling and post-transcriptional regulation. Phosphorylates FOXO3, ERK3/MAPK6, ERK4/MAPK4, HSP27/HSPB1, p53/TP53 and RHEB. Acts as a tumor suppressor by mediating Ras-induced senescence and phosphorylating p53/TP53. Involved in post-transcriptional regulation of MYC by mediating phosphorylation of FOXO3: phosphorylation of FOXO3 leads to promote nuclear localization of FOXO3, enabling expression of miR-34b and miR-34c, 2 post-transcriptional regulators of MYC that bind to the 3'UTR of MYC transcript and prevent MYC translation. Acts as a negative regulator of mTORC1 signaling by mediating phosphorylation and inhibition of RHEB. Part of the atypical MAPK signaling via its interaction with ERK3/MAPK6 or ERK4/MAPK4: the precise role of the complex formed with ERK3/MAPK6 or ERK4/MAPK4 is still unclear, but the complex follows a complex set of phosphorylation events: upon interaction with atypical MAPK (ERK3/MAPK6 or ERK4/MAPK4), ERK3/MAPK6 (or ERK4/MAPK4) is phosphorylated and then mediates phosphorylation and activation of MAPKAPK5, which in turn phosphorylates ERK3/MAPK6 (or ERK4/MAPK4). Mediates phosphorylation of HSP27/HSPB1 in response to PKA/PRKACA stimulation, inducing F-actin rearrangement. {ECO:0000269|PubMed:17254968, ECO:0000269|PubMed:17728103, ECO:0000269|PubMed:19166925, ECO:0000269|PubMed:21329882, ECO:0000269|PubMed:9628874}. | ATP-binding;Alternative splicing;Coiled coil;Cytoplasm;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Tumor suppressor | Mouse_homologues NA; + ;NA | The protein encoded by this gene is a tumor suppressor and member of the serine/threonine kinase family. In response to cellular stress and proinflammatory cytokines, this kinase is activated through its phosphorylation by MAP kinases including MAPK1/ERK, MAPK14/p38-alpha, and MAPK11/p38-beta. The encoded protein is found in the nucleus but translocates to the cytoplasm upon phosphorylation and activation. This kinase phosphorylates heat shock protein HSP27 at its physiologically relevant sites. Two alternately spliced transcript variants of this gene encoding distinct isoforms have been reported. [provided by RefSeq, Nov 2012]. | hsa:8550; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; calcium-dependent protein serine/threonine kinase activity [GO:0009931]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; MAP kinase kinase activity [GO:0004708]; mitogen-activated protein kinase binding [GO:0051019]; p53 binding [GO:0002039]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; cellular senescence [GO:0090398]; intracellular signal transduction [GO:0035556]; negative regulation of TOR signaling [GO:0032007]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of telomerase activity [GO:0051973]; positive regulation of telomere capping [GO:1904355]; positive regulation of telomere maintenance via telomerase [GO:0032212]; protein autophosphorylation [GO:0046777]; Ras protein signal transduction [GO:0007265]; regulation of signal transduction by p53 class mediator [GO:1901796]; regulation of translation [GO:0006417]; signal transduction [GO:0007165]; stress-induced premature senescence [GO:0090400] | 19166925_results imply that MK5 is involved in Hsp27-controlled F-actin dynamics in response to activation of the cAMP-dependent protein kinase pathway. 19473979_Data defined a novel MK5 interaction motif (FRIEDE) within both ERK4 and ERK3 that is essential for binding to the C-terminal region of MK5. 19484198_Several binding motifs for heat shock factor are dispersed in the mouse and rat promoter, and temperature shock transiently enhanced the MAP-kinase-activated kinase 5 transcript levels. 20849292_This review discusses the different characteristics of regulating the activity and subcellular localization of MK5 and RSK1 by PKA and the functional implications of these interactions. 21177870_Activation loop phosphorylation of ERK3/ERK4 by group I p21-activated kinases (PAKs) defines a novel PAK-ERK3/4-MAPK-activated protein kinase 5 signaling pathway. 21336308_Rheb inactivation by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1 22279049_IGF2BP1 promotes the velocity and directionality of tumor-derived cell migration by determining the cytoplasmic fate of two novel target mRNAs: MAPK4 and PTEN 23022185_these results firstly demonstrate that MK5 is degraded in response to doxorubicin and negatively regulates doxorubicin-induced apoptosis, providing novel insights into the molecular mechanism of doxorubicin resistance in hepatoma cells. 23685072_study shows Tip60 plays an essential role in oncogenic ras-induced senescence; revealed a cascade of posttranslational modifications involving p38, Tip60 and PRAK, 3 proteins essential for ras-induced senescence; these modifications are critical for prosenescent function of Tip60 and PRAK 24309468_Studies with specific phosphoantibodies indicate that MK5 phosphorylates Hsp40/DnaJB1 in vivo at Ser-149 or/and Ser-151 and Ser-171 in the C-terminal domain of Hsp40/DnaJB1. 24651460_Data indicate that the structurally most flexible regions during molecular dynamics (MD) simulations of the native mitogen-activated protein kinase-activated protein kinase MK5 model were the loop regions. 25383140_Data highlight that DJ-1 is the downstream interacting target for PRAK, and in response to oxidative stress PRAK may exert a cytoprotective effect by facilitating DJ-1 to sequester Daxx in the nucleus, thus preventing cell death. 26080319_Plasma MAPKAPK5 protein was found to positively associate with the 10-year change in paired associates learning assessment in asymptomatic older twins. 26758977_PRAK might be a potential therapeutic target of Alzheimer's disease involved in receptor for advanced glycation end products-mediated cell signaling induced by Abeta 28941148_MK5 is being discussed as a putative novel regulator of cardiac fibroblast function. (Review) 31578200_Study identified the serine/threonine kinase MK5 as a positive regulator of YAP. MK5 physically interacted with YAP and counteracted CK1delta/epsilon-mediated YAP ubiquitination and degradation independent of LATS1/2. MK5 upregulation was associated with high levels of YAP expression and poor prognosis in clinical tumor samples, confirming its important role for YAP activity in human cancer. 33442026_Biallelic truncating variants in MAPKAPK5 cause a new developmental disorder involving neurological, cardiac, and facial anomalies combined with synpolydactyly. 35468721_MAPKAPK5-AS1 drives the progression of hepatocellular carcinoma via regulating miR-429/ZEB1 axis. | ENSMUSG00000029454+ENSMUSG00000105340 | Mapkapk5+Gm42878 | 1095.66645 | 1.0126835 | 0.0181833519 | 0.14154705 | 1.625083e-02 | 8.985615e-01 | 9.998360e-01 | No | Yes | 1185.37646 | 164.349862 | 1.235053e+03 | 132.286679 |
| ENSG00000092203 | 9878 | TOX4 | protein_coding | O94842 | FUNCTION: Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell cycle progression during the transition from mitosis into interphase. {ECO:0000269|PubMed:20516061}. | Alternative splicing;DNA-binding;Methylation;Nucleus;Phosphoprotein;Reference proteome | hsa:9878; | chromatin [GO:0000785]; nucleus [GO:0005634]; PTW/PP1 phosphatase complex [GO:0072357]; chromatin DNA binding [GO:0031490]; regulation of transcription by RNA polymerase II [GO:0006357] | 19293638_the coordinated spatial and temporal regulation of LCP1 and PNUTS may be a novel mechanism to control the expression of genes that are critical for certain physiological and pathological processes. 20516061_mammalian Wdr82 functions in a variety of cellular processes; PTW/PP1 phosphatase complex (PNUTS, Tox4, Wdr82, PP1) has a role in the regulation of chromatin structure during the transition from mitosis into interphase 21184731_interaction between TOX4 and platinated DNA 22496870_Compared with TOX4, expression of TOX1, TOX2 and TOX3 in normal lung was 25, 44, and 88%lower, respectively, supporting the premise that reduced promoter activity confers increased susceptibility to methylation during lung carcinogenesis. 23127401_The formation of vasculogenic mimicry is inhibited by reducing the expression of Mig-7 in gastric cancer cells. 24312278_a regulation of LEDGF interaction with chromatin by cellular partners of its PWWP domain could be involved in several processes linked to LEDGF tethering properties, such as lentiviral integration, DNA repair or transcriptional regulation 34914893_TOX4, an insulin receptor-independent regulator of hepatic glucose production, is activated in diabetic liver. 35365735_TOX4 facilitates promoter-proximal pausing and C-terminal domain dephosphorylation of RNA polymerase II in human cells. | ENSMUSG00000016831 | Tox4 | 3496.16481 | 1.0914546 | 0.1262521872 | 0.13921165 | 8.155618e-01 | 3.664810e-01 | 9.998360e-01 | No | Yes | 3410.91765 | 341.079159 | 3.124326e+03 | 241.705563 | ||
| ENSG00000095574 | 64376 | IKZF5 | protein_coding | Q9H5V7 | FUNCTION: Transcriptional repressor that binds the core 5'GNNTGTNG-3' DNA consensus sequence (PubMed:10978333, PubMed:31217188). Involved in megakaryocyte differentiation. {ECO:0000269|PubMed:10978333, ECO:0000269|PubMed:31217188}. | DNA-binding;Disease variant;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Members of the Ikaros (ZNFN1A1; MIM 603023) family of transcription factors, which includes Pegasus, are expressed in lymphocytes and are implicated in the control of lymphoid development.[supplied by OMIM, Jul 2002]. | hsa:64376; | nucleus [GO:0005634]; protein-containing complex [GO:0032991]; chromatin binding [GO:0003682]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; protein domain specific binding [GO:0019904]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 31217188_IKZF5 is a novel transcriptional regulator of megakaryopoiesis and the eighth transcription factor associated with dominant thrombocytopenia in humans. 32419556_Highly impaired platelet ultrastructure in two families with novel IKZF5 variants. | ENSMUSG00000040167 | Ikzf5 | 231.03520 | 1.1929767 | 0.2545658263 | 0.22422467 | 1.250211e+00 | 2.635122e-01 | 9.998360e-01 | No | Yes | 218.16647 | 47.190999 | 1.956658e+02 | 32.638969 | |
| ENSG00000095794 | 1390 | CREM | protein_coding | Q03060 | FUNCTION: Transcriptional regulator that binds the cAMP response element (CRE), a sequence present in many viral and cellular promoters. Isoforms are either transcriptional activators or repressors. Plays a role in spermatogenesis and is involved in spermatid maturation (PubMed:10373550). {ECO:0000269|PubMed:10373550}.; FUNCTION: [Isoform 6]: May play a role in the regulation of the circadian clock: acts as a transcriptional repressor of the core circadian component PER1 by directly binding to cAMP response elements in its promoter. {ECO:0000250}. | Activator;Alternative promoter usage;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a bZIP transcription factor that binds to the cAMP responsive element found in many viral and cellular promoters. It is an important component of cAMP-mediated signal transduction during the spermatogenetic cycle, as well as other complex processes. Alternative promoter and translation initiation site usage allows this gene to exert spatial and temporal specificity to cAMP responsiveness. Multiple alternatively spliced transcript variants encoding several different isoforms have been found for this gene, with some of them functioning as activators and some as repressors of transcription. [provided by RefSeq, Jul 2008]. | hsa:1390; | ATF4-CREB1 transcription factor complex [GO:1990589]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; cAMP response element binding protein binding [GO:0008140]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; cell differentiation [GO:0030154]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; rhythmic process [GO:0048511]; signal transduction [GO:0007165]; spermatogenesis [GO:0007283] | 11988318_review of CREM transcription factor involvement with spermatogenesis 12370343_Increased expression of CREM in T cells from systemic lupus erythmatosus (SLE) patients results from increased transcriptional activity of the CREM gene, and its binding to IL-2 promoter is responsible for decreased production of IL-2 by SLE T cells. 12397208_5'-RACE on human testis cDNA indicated that exon theta2 is > or = 113 bp in size. In-vitro translation of CREM-theta1 and CREM-theta2 splice variants cloned from human testis yielded full length proteins and also shorter repressor products 12555239_Observational study of gene-disease association. (HuGE Navigator) 12626549_Direct binding of CREM to the CRE site of the IL-2 promoter endows CREM with a central role in repression of IL-2 gene expression: CREM binding promotes chromatin deacetylation, limits promoter accessibility and decreases its transcriptional activity. 14511788_expression of cAMP-responsive element modulator(CREM) activators is a prerequisite for normal spermatogenesis, and the lack of CREM activator expression results in male infertility 14754893_CREB-1/CREM-1 have roles as regulators of macrophage differentiation 15048659_These findings provide little additional evidence for a susceptibility locus for panic disorder either within the CREM gene or in a nearby region of chromosome 10p11 in our sample 15225122_Sperm nucleus PHGPx expression is mediated by the transcription factor CREM-tau, which acts as a cis-acting element localized in the first intron of the PHGPx gene. [CREM-tau] 15456763_down-regulation of CREMtau-mediated gene expression by GCNF 15474076_Lack of spermatid elongation was not due to defective CREM expression. Therefore, CREM did not play a pathogenetic role in the onset of SMA in humans. 15569686_that heart-directed expression of CREM-IbDeltaC-X leads to complex cardiac alterations, suggesting CREM as a central regulator of cardiac morphology, function, and gene expression 15691874_isoforms regulate discrete groups of genes in myometrium 15841182_Results identify calcium/calmodulin-dependent kinase IV as being responsible for the increased expression of CREM and the decreased production of interleukin-2 in systemic lupus erythematosus T cells. 16048633_CREM activator and repressor isoforms were found in all germ cell types, but not in Sertoli cells; data suggest a fine-tuning between CREM activator and repressor isoforms in normal germ cells that might be disturbed during impaired spermatogenesis 16103121_SRp40 regulates the switch in splicing from production of CREMtau(2)alpha to CREMalpha 16143638_Screening of a substantial number of patients would be required to clarify whether observed combinations of genetic changes in the CREM gene might explain some forms of male infertility. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16687568_The interaction between CREM and one haplotype of ACT (activator of CREM in the testis) was reduced by 45% in a yeast two-hybrid assay. 16893891_HNF4alpha, CREM, HNF1alpha, and C/EBPalpha have roles in transcriptional regulation of the glucose-6-phosphatase gene by cAMP/vasoactive intestinal peptide in the intestine 16894555_These results constitute the first demonstration of the transcriptional control of ATP1A4 gene expression by cAMP and by CREMtau, a transcription factor essential for male germ cell gene expression. 16899810_functional analysis of isoforms from the testes 17056544_CREMalpha exerts its repressor activity by a mechanism that involves recruitment of HDAC1, increased deacetylation of histones, and repression of promoter activity. 17211988_CREM-alpha mRNA levels were higher in T cells from patients with systemic lupus erythematosus than controls while CREB mRNA levels did not differ between the two groups 17332439_The positive-feedback loop described in this review is regulated by phosphodiesterase 3A and ICER and is pathologically important in adult hearts. 17340624_Data show that inducible cAMP early repressor splice variants ICER I and IIgamma both repress transcription of c-fos and chromogranin A. 17712720_Transcription factors of the CREB/CREM/ATF family have a moderate effect on human MC2-R promoter activity, but seem to play a minor role in transmitting stimulation of the cAMP pathway to increased MC2-R expression. 18202121_CREM is an essential regulator of NIS gene expression. 18700132_CREM has a role in CYP1A1 induction through ligand-independent activation pathway of aryl hydrocarbon receptor in HepG2 cells 18784739_CREB/ICER expression needs to be considered a pathogenetic feature in leukemogenesis 18922788_BDNF- and seizure-dependent phosphorylation of STAT3 cause the adenosine 3',5'-monophosphate (cAMP) response element-binding protein (CREB) family member ICER (inducible cAMP early repressor) to bind with phosphorylated CREB at the Gabra1:CRE site. 18984674_Observational study of gene-disease association. (HuGE Navigator) 19299714_CREMalpha is an important negative regulator of costimulation and APC-dependent T cell function 19434522_This review will bring together data on ICER and its functions in the brain, with a special emphasis on recent findings highlighting the involvement of ICER in the regulation of long-term plasticity underlying learning and memory. [REVIEW] 19531482_cAMP stringently regulates human cathelicidin antimicrobial peptide expression in the mucosal epithelial cells by activating cAMP-response element-binding protein, AP-1, and inducible cAMP early repressor 20237496_Observational study of gene-disease association. (HuGE Navigator) 20378615_Observational study of gene-disease association. (HuGE Navigator) 20413592_DNA cytosine methylation in the bovine leukemia virus promoter has a role in direct inhibition of cAMP-responsive element (CRE)-binding protein/CRE modulator/activation transcription factor binding 20488182_Results indicate that SPAG8 acts as a regulator of ACT and plays an important role in CREM-ACT-mediated gene transcription during spermatogenesis. 21097497_anscriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. 21325296_ICER mediates chemotherapy anticancer activity through DUSP1-p38 pathway activation and drives the cell program from survival to apoptosis 21507395_Patients with two types of male factor infertility display an increased abnormal methylation of CREM compared with control subjects. 21547497_Induction of ICER links oxidative stress to beta cell failure caused by oxidised LDL, and it can be effectively abrogated by antioxidant treatment. 21757709_AP-1-dependent up-regulation of the P2 promoter, SLE T cells fail to further increase their basal CREM levels upon T cell activation due to a decreased content of the AP-1 family member c-Fos 21767532_Phosphorylation of ICER on a discrete residue targeted ICER to be monoubiquitinated. 21953500_CREMalpha suppresses spleen tyrosine kinase expression in normal but not systemic lupus erythematosus T cells. 21976679_Data provide direct evidence that CREMalpha mediates silencing of the IL2 gene in SLE T cells though histone deacetylation and CpG-DNA methylation. 21998402_Impaired expression of ICER contributes to elevation in CREB target genes and, therefore, to the development of insulin resistance in obesity. 22019623_Common variants of the CREM gene are involved in the genetic component conferring general susceptibility to inflammatory bowel disease in the Tunisian population. 22025620_cAMP-responsive element modulator (CREM)alpha protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. 22093963_Transcription factor CREM is an important regulator of atrial growth implicated in the development of atrial fibrillation. 22184122_cAMP-responsive element modulator alpha (CREMalpha) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). 22198373_scriptive Statement The rsults of this study suggested the lack of influence of SNPs under investigation in the present study on the susceptibility to schizophrenia and on the response to antipsychotics. 22281835_Estrogen can modulate the expression of CREMalpha and lead to IL-2 suppression in human T lymphocytes, thus revealing a molecular link between hormones and the immune system in Systemic lupus erythematosus 22386572_The results of this study suggested that the single nucleotide polymorphisms of CREM do not influence diagnosis and treatment response in patients with major depressive disorder and bipolar disorder. 22510021_CREM expression is increased in thyroid cancer tissue and may play a role in the downregulation of sodium iodide symporter expression in thyroid cancer acting at the transcriptional level 23019580_Data indicate that CpG-DNA methylation and mRNA expression of CREM, IL2, and IL17A of systemic lupus erythematosus (SLE) T cells reflect the effector memory CD4+ T-cell phenotype. 23124208_Increased CREMalpha binding to the Notch-1 promoter resulted in significantly reduced Notch-1 promoter activity and gene transcription. 23929392_Data suggest that cyclic AMP response element modulator-1 (CREM-1) might play an important role in the regulation of tumor metastasis and invasion and serve as a tumor suppressor in esophageal squamous cell carcinoma (ESCC). 24047902_transcription factor cAMP-responsive element modulator alpha (CREMalpha), which is expressed at increased levels in T cells from systemic lupus erythematosus patients, contributes to transcriptional silencing of CD8A and CD8B. 24100545_In Alzheimer's brain, we found an increased cellular expression of CREM in dentate gyrus neurons as compared to normal aging brains. 24297179_CREMalpha orchestrates epigenetic remodeling of the CD8A,B through the recruitment of DNA methyltransferase (DNMT) 3a and histone methyltransferase G9a. 24667640_CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. 24672804_Data suggest ICER/CREM plays seminal role in down-regulation of expression/secretion of insulin by pancreatic beta-cell as an adaptive response to factors that promote diabetes; inappropriate induction of ICER leads to beta-cell dysfunction. [REVIEW] 24943041_These findings indicated that the polymorphisms of CREM gene were associated with nonobstructive azoospermia in the Chinese population and low CREM expression might be involved in the pathogenesis of spermatogenesis maturation arrest. 25401338_overexpressed in the nuclei of hepatocellular carcinoma cells 26601115_CREMalpha SNPs rs2295415 and rs1057108 may be novel genetic susceptibility factors for SLE, especially at haplotype level. 27822872_Data indicate a role for inducible cyclic AMP early repressor (ICER) in G1 checkpoint regulation in hematopoietic stem cells (HSCs). 27840176_this study shows an eventually involvement of CREM gene in the development of T1D pathology in Tunisian families. These facts are consistent with a major role for transcription factor genes involved in the immune pathways in the control of autoimmunity. 27904655_Study provides evidence that increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates CREMA in systemic lupus erythematosus. 28009602_Report a distinct group of myxoid mesenchymal neoplasms occurring in children or young adults with a predilection for intracranial locations with EWSR1-AFT1/CREB1/CREM fusions. 28439100_Genetic studies in seven independent human populations illustrate that a CREM promoter variant at rs12765063 is associated with impulsivity, hyperactivity and addiction-related phenotypes. 29788195_EWSR1 fusion with CREM has only been observed in 3 intracranial myxoid tumors 29925386_CREM drives an inflammatory phenotype of T cells in Juvenile idiopathic arthritis. 29975250_We describe a novel gene fusion, EWSR1-CREM, identified in 3 cases of clear cell carcinoma 30228239_Increased CREM expression is also observed in early E. histolytica infection 31305268_Report the phenotypic spectrum of mesenchymal tumors associated with the EWSR1-CREM fusion. 34049318_Cytokeratin-positive Malignant Tumor in the Abdomen With EWSR1/FUS-CREB Fusion: A Clinicopathologic Study of 8 Cases. 34261344_Expression of Transcription Factor CREM in Human Tissues. | ENSMUSG00000063889 | Crem | 125.17497 | 0.9486091 | -0.0761143885 | 0.28365191 | 7.390280e-02 | 7.857368e-01 | 9.998360e-01 | No | Yes | 117.24910 | 18.396501 | 1.285282e+02 | 15.735285 | |
| ENSG00000096070 | 27154 | BRPF3 | protein_coding | Q9ULD4 | FUNCTION: Scaffold subunit of various histone acetyltransferase (HAT) complexes, such as the MOZ/MORF and HBO1 complexes, which have a histone H3 acetyltransferase activity (PubMed:16387653, PubMed:26620551, PubMed:26677226). Plays a role in DNA replication initiation by directing KAT7/HBO1 specificity towards histone H3 'Lys-14' acetylation (H3K14ac), thereby facilitating the activation of replication origins (PubMed:26620551). Component of the MOZ/MORF complex which has a histone H3 acetyltransferase activity (PubMed:16387653). {ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:26620551, ECO:0000269|PubMed:26677226}. | 3D-structure;Acetylation;Alternative splicing;Bromodomain;Chromatin regulator;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:27154; | cytosol [GO:0005829]; extracellular region [GO:0005576]; histone acetyltransferase complex [GO:0000123]; MOZ/MORF histone acetyltransferase complex [GO:0070776]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; histone H3 acetylation [GO:0043966]; histone H3-K14 acetylation [GO:0044154]; positive regulation of DNA replication [GO:0045740] | Mouse_homologues 26677226_These results indicate that BRPF3 forms a functional tetrameric complex with HBO1 but is not required for mouse development and survival | ENSMUSG00000063952 | Brpf3 | 2026.48201 | 1.0480443 | 0.0676997214 | 0.11645264 | 3.290371e-01 | 5.662266e-01 | 9.998360e-01 | No | Yes | 2422.39735 | 209.155347 | 2.129659e+03 | 142.767043 | ||
| ENSG00000097046 | 8317 | CDC7 | protein_coding | O00311 | FUNCTION: Seems to phosphorylate critical substrates that regulate the G1/S phase transition and/or DNA replication. Can phosphorylate MCM2 and MCM3. {ECO:0000269|PubMed:12065429}. | 3D-structure;ATP-binding;Alternative splicing;Cell cycle;Cell division;Isopeptide bond;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation | This gene encodes a cell division cycle protein with kinase activity that is critical for the G1/S transition. The yeast homolog is also essential for initiation of DNA replication as cell division occurs. Overexpression of this gene product may be associated with neoplastic transformation for some tumors. Multiple alternatively spliced transcript variants that encode the same protein have been detected. [provided by RefSeq, Aug 2008]. | hsa:8317; | cytoplasm [GO:0005737]; intercellular bridge [GO:0045171]; mitotic spindle [GO:0072686]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; metal ion binding [GO:0046872]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell cycle phase transition [GO:0044770]; cell division [GO:0051301]; double-strand break repair via break-induced replication [GO:0000727]; G1/S transition of mitotic cell cycle [GO:0000082]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of G2/M transition of mitotic cell cycle [GO:0010971]; positive regulation of nuclear cell cycle DNA replication [GO:0010571] | 12065429_Drf1, a nuclear cell cycle-regulated protein binds to Cdc7 and activates the kinase 15466207_Down-regulation of Cdc7 by small interfering RNA in a variety of tumor cell lines causes an abortive S phase, leading to cell death by either p53-independent apoptosis or aberrant mitosis. 15474462_CR (periphilin) retards S-phase progression by modifying expression of Cdc7 and other genes involved in progression of DNA replication 15668232_ASKL1 in a complex with Cdc7 may play a role in normal progression of both S and M phases 16082200_CINP is part of the Cdc7-dependent mechanism of origin firing and a functional and physical link between Cdk2 and Cdc7 complexes at the origins 16446360_the majority of the Mcm2 isoforms phosphorylated by Cdc7 are not stably associated with chromatin 16826239_Cdc7-Dbf4 kinase efficiently phosphorylates p150. 16864800_Results describe the mapping of the sites in human Mcm2 protein that are phosphorylated by Cdc7. 16899510_Taken together, these results indicate that Cdc7/Dbf4 phosphorylation of MCM2 is essential for the initiation of DNA replication in mammalian cells. 17046832_MCM4 phosphorylation by Cdc7 kinase facilitates its interaction with Cdc45 on chromatin 17062569_Cdc7 kinase plays a role in maintaining cell viability during replication stress 17711849_CRM1 may down-regulate Cdc7 by masking its kinase domain 18286467_Cdc7/Dbf4 kinase activity inhibition affects specific phosphorylation sites on MCM2 in cancer cells 18625709_the ATR-dependent activation of the p38 MAP kinase is a major signaling pathway that induces apoptotic cell death after depletion of Cdc7 in cancer cells 18714392_increased Cdc7-Dbf4 abundance may be a common occurrence in human malignancies 19054765_chromatin-bound CDT1 is first stabilized and subsequently displaced by CDC7 activity, thereby ensuring the timely execution of DNA replication 19111665_These results indicate that Ddk functions as an upstream regulator to monitor S-phase checkpoint signaling. 19278428_differences in Cdc7 expression may account for some of the differences between malignant melanomas and benign melanocytic nevi. 19318489_Increased Cdc7 protein levels were significantly associated with arrested tumor differentiation, advanced clinical stage, genomic instability, and accelerated cell cycle progression. 19426592_Data show that CDC7L1 and CDC10 was up-regulated but the expression of CDK9, CDC20 and CLK3 was down- regulated in azoospermic testes. 19647517_Cdc7 phosphorylates Mcm2 promoting pre-replication complex assembly. 19864417_The interaction with LEDGF relieves autoinhibition of Cdc7-ASK kinase, imposed by the C terminus of ASK. 19896697_cell division cycle 7 is a replication associated protein with relationships to gene amplification and genomic instability in breast carcinomas. 20548946_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20647475_Inhibition of Cdc7 kinase activity in cancer cells restricts DNA replication and induces apoptotic cell death by an unprecedented molecular mechanism of action. 20707412_Data syggest important implications for the continued development of promising Cdc7-targeted cancer therapies. 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21536671_bipartite interaction between Cdc7 and Dbf4/ASK subunits facilitates ATP binding and substrate recognition by the Cdc7 kinase. 22528513_The results suggest that CDC7 expression level can be specific prognostic factors for Diffuse large B-cell lymphoma patients 22574151_cell death process induced by Cdc7 depletion 22806309_Significantly higher levels of apoptosis were detected in siCDC7-transfected cells. 23066029_Cdc7 phosphorylates and interacts with Tob to inhibit the Cul4-DDB1(Cdt2)-dependent Tob degradation. 23598722_The interaction between Claspin and Cdc7 is not dependent on Cdc7 kinase activity, but Claspin interaction with the DNA helicase subunit Mcm2 is lost upon Cdc7 inhibition. 23684929_Cdcy is universally up-regulated in oral squamous cell carcinoma is an independent prognostic marker, contributing to resistance to DNA damaging agents. 23716994_huCdc7 may play an important role in the development and progression of colorectal cancer. 25255219_MiR-630 promoted apoptosis by downregulation of CDC7. 25258324_The state of DUE-B phosphorylation is maintained by the equilibrium between Cdc7-dependent phosphorylation and PP2A-dependent dephosphorylation. 26208856_Our data show that Cdc7 is highly expressed in colorectal cancer 26383992_The presence of the index SNP rs1192415 (TGFBR3-CDC7) was associated with visual field progression in POAG (primary open-angle glaucoma) patients. 27105124_The data support a model where Cdc7 (de)phosphorylation is the molecular switch for the activation and inactivation of DNA replication in mitosis, directly connecting Cdc7 and PP1a/Cdk1 to the regulation of once-per-cell cycle DNA replication in mammalian cells. 27401717_Results suggest a new role of Claspin in initiation of DNA replication during normal S phase through the recruitment of Cdc7 that facilitates phosphorylation of Mcm proteins. 27407105_we propose that phosphorylation of TOP2A by CDC7/DBF4 in early S-phase prevents its localization and/or activity at centromeres, and inhibition of TOP2A function could be relevant to prevent premature separation of centromeric DNA. 27611229_p53-dependent control of CDC7 levels is essential for blocking G1/S cell-cycle transition upon genotoxic stress. 28448802_Both CDC7 and DBF4 promoters bind E2F, suggesting that increased E2F activity in RB1 mutant cancers promotes increased DDK expression. Surprisingly, increased DDK expression levels are also correlated with both increased chemoresistance and genome-wide mutation frequencies. 28887320_Increased Cdc7-dependent replication initiation is a hallmark of p53 gain-of function mutations in lung adenocarcinoma. 29209046_High CDC7 expression is associated with oral cancer. 29611806_Here, the authors show that, in human cells, cohesin loading onto chromosomes during early S phase requires the replicative helicase MCM2-7 and the kinase DDK. Cohesin and its loader SCC2/4 (NIPBL/MAU2 in humans) associate with DDK and phosphorylated MCM2-7. 29713760_Overexpression of Cdc7 promotes progression in oral squamous cell carcinoma (OSCC) and that inhibition of Cdc7 is a very promising therapy for OSCC patients. 30012245_Higher expression of CDC7 in odontogenic keratocyst (OKC) and radicular cyst was shown in comparison to dentigerous cyst, while radicular cyst and OKC groups showed no difference in CDC7 expression. 30157471_CDC7-DBF4 kinase (DDK) has a primary role in the replication checkpoint to promote single-stranded DNA accumulation at stalled forks, which is required to initiate a robust checkpoint response and cell cycle arrest to maintain genome integrity. 30392930_CDC7-bound PGK1 converts ADP to ATP, thereby abrogating the inhibitory effect of ADP on CDC7-ASK activity, promoting the recruitment of DNA helicase to replication origins, DNA replication, cell proliferation, and brain tumorigenesis. 30573684_Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. 31002360_the immunohistochemical results and The Cancer Genome Atlas (TCGA) database revealed that CDC7 was significantly upregulated in lung adenocarcinoma tissues, suggesting that miR888 may function as an oncogene in the progression of lung adenocarcinoma patients, and the miR888/CDC7 axis was not the dominant pathway for CDC7 regulation in patients with lung adenocarcinoma 31578454_Targeting CDC7 sensitizes resistance melanoma cells to BRAF(V600E)-specific inhibitor by blocking the CDC7/MCM2-7 pathway. 31819079_Cdc7 kinase stimulates Aurora B kinase in M-phase. 31889509_we show that Cdc7 is required for Claspin-Chk1 interaction in human cancer cells by phosphorylating CKBD (Chk1-binding-domain) of Claspin. 32496651_CDC7 kinase promotes MRE11 fork processing, modulating fork speed and chromosomal breakage. 32877678_ATR Restrains DNA Synthesis and Mitotic Catastrophe in Response to CDC7 Inhibition. 33599894_MiR-200a with CDC7 as a direct target declines cell viability and promotes cell apoptosis in Wilm's tumor via Wnt/beta-catenin signaling pathway. 33763482_Upregulation of CDC7 Associated with Cervical Cancer Incidence and Development. 34663432_Targeting CDC7 potentiates ATR-CHK1 signaling inhibition through induction of DNA replication stress in liver cancer. | ENSMUSG00000029283 | Cdc7 | 742.89849 | 1.0386684 | 0.0547351745 | 0.15393390 | 1.225733e-01 | 7.262601e-01 | 9.998360e-01 | No | Yes | 733.93576 | 134.579429 | 7.296299e+02 | 103.153188 | |
| ENSG00000099814 | 283638 | CEP170B | protein_coding | Q9Y4F5 | FUNCTION: Plays a role in microtubule organization. {ECO:0000250|UniProtKB:Q5SW79}. | Alternative splicing;Cytoplasm;Cytoskeleton;Microtubule;Phosphoprotein;Reference proteome | hsa:283638; | cytoplasm [GO:0005737]; microtubule [GO:0005874] | ENSMUSG00000072825 | Cep170b | 3565.23418 | 0.9842341 | -0.0229265566 | 0.11212319 | 4.267787e-02 | 8.363330e-01 | 9.998360e-01 | No | Yes | 4363.03300 | 510.791808 | 4.022317e+03 | 364.255538 | |||
| ENSG00000099840 | 113177 | IZUMO4 | protein_coding | Q1ZYL8 | Alternative splicing;Glycoprotein;Reference proteome;Secreted;Signal | hsa:113177; | extracellular region [GO:0005576]; nucleus [GO:0005634] | ENSMUSG00000055862 | Izumo4 | 95.29184 | 1.2493766 | 0.3212084223 | 0.30977382 | 1.038378e+00 | 3.081990e-01 | 9.998360e-01 | No | Yes | 125.05425 | 19.133602 | 9.609210e+01 | 11.923042 | ||||
| ENSG00000099889 | 421 | ARVCF | protein_coding | O00192 | FUNCTION: Involved in protein-protein interactions at adherens junctions. | Alternative splicing;Cell adhesion;Coiled coil;Methylation;Phosphoprotein;Reference proteome;Repeat | Armadillo Repeat gene deleted in Velo-Cardio-Facial syndrome (ARVCF) is a member of the catenin family. This family plays an important role in the formation of adherens junction complexes, which are thought to facilitate communication between the inside and outside environments of a cell. The ARVCF gene was isolated in the search for the genetic defect responsible for the autosomal dominant Velo-Cardio-Facial syndrome (VCFS), a relatively common human disorder with phenotypic features including cleft palate, conotruncal heart defects and facial dysmorphology. The ARVCF gene encodes a protein containing two motifs, a coiled coil domain in the N-terminus and a 10 armadillo repeat sequence in the midregion. Since these sequences can facilitate protein-protein interactions ARVCF is thought to function in a protein complex. In addition, ARVCF contains a predicted nuclear-targeting sequence suggesting that it may have a function as a nuclear protein. [provided by RefSeq, Jun 2010]. | hsa:421; | adherens junction [GO:0005912]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules [GO:0016339]; cell adhesion [GO:0007155]; cell-cell adhesion [GO:0098609]; cell-cell junction assembly [GO:0007043] | 15340358_Evidence was found for association of illness to rs165849 in ARVCF, and a stronger signal from three-marker haplotypes spanning the 3' portions of COMT and ARVCF. Val(108/158)Met was in linkage disequilibrium with the markers in ARVCF. 15456900_Interactions with zona occludens-1 and zona occludens-2, in particular, may mediate recruitment of ARVCF to the plasma membrane and the nucleus 15509897_Results implicate a very close association of ARVCF with migrating neurons from the ganglionic eminence. 16118784_Observational study of gene-disease association. (HuGE Navigator) 16118784_Two haplotypes covering the catechol-O-methyltransferase-ARVCF region show significant transmission disequilibrium in anorexia nervosa-restricting Israeli-Jewish families 18198266_Observational study of gene-disease association. (HuGE Navigator) 18600340_Differential expression pattern of protein ARVCF in nephron segments of human kidney. 19508883_Single nucleotide polymorphisms in the ARVGF gene are associated with the risk of schizophrenia. 19617637_Over-expression of TXNRD2, COMT and ARVCF affects incentive learning and working memory in transgenic mice. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20333729_Observational study of gene-disease association. (HuGE Navigator) 20333729_The functional variant rs165815, which affects a critical region of ARVCF, is a considerable source of the genetic variability associated with the risk of developing schizophrenia. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 21846818_Five SNPs were validated as being significantly associated with prostate cancer mortality, one each in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes. 22053977_Schizophrenic patients with more copies of the haplotype T-G-A-T-T-G-G-C-T-G-T (ARVCF-Hap1) have lower white matter integrity in caudate nucleus and greater perseverative errors. 24644279_Data indicate that armadillo repeat protein ARVCF interacts with the splicing factors the splicing factor SRSF1 (SF2/ASF), the RNA helicase p68 (DDX5), and the heterogeneous nuclear ribonucleoprotein hnRNP H2. 24819575_Carriage of the minor allele at rs2518824 in the armadillo repeat gene deleted in velocardiofacial syndrome (ARVCF) gene was associated with white matter abnormality. 25683624_observed ARVCF-dependent changes in small GTPase (mainly RhoA) activity in lung cancer cells. We confirmed that ARVCF plays an important role in the malignant phenotype 31827232_p53-induced ARVCF modulates the splicing landscape and supports the tumor suppressive function of p53. | ENSMUSG00000118669 | Arvcf | 857.30077 | 0.9808407 | -0.0279093175 | 0.14282915 | 3.963377e-02 | 8.421984e-01 | 9.998360e-01 | No | Yes | 824.13684 | 101.740795 | 7.708628e+02 | 74.197264 | |
| ENSG00000100150 | 9681 | DEPDC5 | protein_coding | O75140 | FUNCTION: As a component of the GATOR1 complex functions as an inhibitor of the amino acid-sensing branch of the TORC1 pathway. The GATOR1 complex strongly increases GTP hydrolysis by RRAGA and RRAGB within RRAGC-containing heterodimers, thereby deactivating RRAGs, releasing mTORC1 from lysosomal surface and inhibiting mTORC1 signaling. The GATOR1 complex is negatively regulated by GATOR2 the other GATOR subcomplex in this amino acid-sensing branch of the TORC1 pathway. {ECO:0000269|PubMed:23723238, ECO:0000269|PubMed:25457612, ECO:0000269|PubMed:29769719}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Epilepsy;GTPase activation;Lysosome;Membrane;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes a member of the IML1 family of proteins involved in G-protein signaling pathways. The mechanistic target of rapamycin complex 1 (mTORC1) pathway regulates cell growth by sensing the availability of nutrients. The protein encoded by this gene is a component of the GATOR1 (GAP activity toward Rags) complex which inhibits the amino acid-sensing branch of the mTORC1 pathway. Mutations in this gene are associated with autosomal dominant familial focal epilepsy with variable foci. A single nucleotide polymorphism in an intron of this gene has been associated with an increased risk of hepatocellular carcinoma in individuals with chronic hepatitis C virus infection. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:9681; | cytosol [GO:0005829]; GATOR1 complex [GO:1990130]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; perinuclear region of cytoplasm [GO:0048471]; GTPase activator activity [GO:0005096]; protein-containing complex binding [GO:0044877]; cellular response to amino acid starvation [GO:0034198]; intracellular signal transduction [GO:0035556]; negative regulation of TOR signaling [GO:0032007]; negative regulation of TORC1 signaling [GO:1904262]; regulation of autophagy [GO:0010506] | 21725309_Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. 23542697_Mutations in DEPDC5 cause familial focal epilepsy with variable foci. 23542701_Mutations in DEPDC5 cause familial focal epilepsy with variable foci. 24283814_A recurrent mutation in DEPDC5 predisposes to focal epilepsies in the French-Canadian population. 24585383_DEPDC5 mutations are associated with both lesional and nonlesional epilepsies. 24591017_Mutations in DEPDC5 are associated with childhood focal epilepsies. 24814846_DEPDC5 loss-of-function mutations may represent a part of the broader familial focal epilepsy with variable foci phenotype found in 30 European families with a presentation of autosomal dominant nocturnal frontal lobe epilepsy. 25032264_PAPL, IL10RB and DEPDC5 polymorphisms have an impact on progression of hepatitis B virus-related liver disease. 25194487_This chapter focuses on DEPDC5, a newly identified gene in autosomal dominant focal epilepsies [review] 25366275_The effects of 10 DEPDC5 variants identified in individuals with focal epilepsy and two DEPDC5 variants identified in serous ovarian tumors, on TORC1 signaling and GATOR-1 complex formation. 25551790_Genetic variations in DEPDC5 gene region may influence HCV-associated liver cirrhosis and/or hepatocellular carcinoma. development. 25599672_An association was made for DEPDC5 with sporadic focal cortical dysplasia and also hemimegalencephaly. 25623524_Truncating DEPDC5 mutations were found in all four French families with focal cortical dysplasia and focal epilepsy. 25764692_MICA and DEPDC5 SNPs were found to be strongly associated with HCV-induced HCC. 25964426_This revealed four patients to have two or more tumors that were clonally related, all of which lacked MED12 mutations. DEPDC5 was discovered as a novel tumor suppressor gene playing a role in the progression of uterine leiomyomas. 26216793_This study found a single DEPDC5 mutation in one of (2.2%) 45 families with genetic temporal lobe epilepsy, a proportion much lower than that reported in other inherited focal epilepsies. 26517016_DEPDC5 variants increase fibrosis progression in European subjects with chronic HCV infection. Our findings suggest that DEPDC5 down-regulation may contribute to HCV-related fibrosis by increasing MMP2 synthesis through the beta-catenin pathway 27173016_This cohort study showed DEPDC5 mutations were present in 8% of individuals with focal epilepsy, including one individual with focal cortical dysplasia. 28170089_Our finding suggests that DEPDC5 is not only the most common gene for familial focal epilepsy but also could be a significant gene for sporadic focal epilepsy. Since focal epilepsies account for more than 60% of all epilepsies, the effect of mTORC1 inhibitor on patients with focal epilepsy due to DEPDC5 mutations will be an important future direction of research. 28928439_This study showed that polymorphisms in MICA, but not in DEPDC5, HCP5 or PNPLA3, are associated with with chronic hepatitis C-related hepatocellular carcinoma development in Japanese patients with chronic hepatitis C virus infection. 28974734_Heterozygous mice appeared to be normal and we found no evidence of increased susceptibility to seizures or tumorigenesis. Together, these data support mTORC1 hyperactivation as the likely pathogenic mechanism that underpins DEPDC5 loss of function in humans and highlights the potential utility of mTORC1 inhibitors in the treatment of DEPDC5-associated epilepsy 29311600_the DEPDC5-KO HCC cells could acquire anti-oxidant ability through p62 accumulation and survive under leucine starvation, and that downregulated DEPDC5 expression was an independent predictive factor for patient outcome. 29708508_Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. 30087333_Knockout of either TSC1 or DEPDC5 led to enhanced HIV-1 reactivation in both a T-cell and a monocyte cell lines. 30093711_overall, 63 distinct variants were identified: 53 in DEPDC5, three in NPRL2 and seven in NPRL3 . Among these, 46 were novel (including 39 single nucleotide variants and seven CNVs) and 16 were newly defined as recurrent variants; 34 were loss-of-function (LoF) variants (nonsense, splice-site, frameshift indels and CNVs). 30683632_There was a significant correlation between DEPDC5 rs1012068A/C and HBV-related hepatocellular carcinoma in the Han Chinese population. A to C mutation increased the risk of the developing of HBV-related hepatocellular carcinoma. 30723271_A variant in the MICA gene is associated with liver fibrosis progression in chronic hepatitis C through TGF-beta1 dependent mechanisms. 31174205_our data provide the first evidence of behavioral alterations in mice with Depdc5 loss and support mTOR inhibition as a rational therapeutic strategy for DEPDC5-related epilepsy in humans. 31636198_findings of recurrent genomic alterations, together with functional data, validate the DEPDC5 as a bona fide tumor suppressor contributing to GIST progression and a biologically relevant target of the frequent chromosome 22q deletions 31835056_Sleep-related hypermotor epilepsy (SHE): Contribution of known genes in 103 patients. 32086284_Diagnostic exome sequencing in non-acquired focal epilepsies highlights a major role of GATOR1 complex genes. 32574724_DEPDC5 haploinsufficiency drives increased mTORC1 signaling and abnormal morphology in human iPSC-derived cortical neurons. 33461085_GATOR1-related focal cortical dysplasia in epilepsy surgery patients and their families: A possible gradient in severity? 34587683_[Genotype and phenotype of children with DEPDC5 gene variants related epilepsy]. 35429726_DEPDC5-related epilepsy: A comprehensive review. | ENSMUSG00000037426 | Depdc5 | 651.87990 | 1.0659195 | 0.0920985350 | 0.13759332 | 4.524981e-01 | 5.011510e-01 | 9.998360e-01 | No | Yes | 670.64714 | 62.611510 | 6.173043e+02 | 45.018250 | |
| ENSG00000100239 | 9701 | PPP6R2 | protein_coding | O75170 | FUNCTION: Regulatory subunit of protein phosphatase 6 (PP6). May function as a scaffolding PP6 subunit. Involved in the PP6-mediated dephosphorylation of NFKBIE opposing its degradation in response to TNF-alpha. {ECO:0000269|PubMed:16769727}. | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome | The protein encoded by this gene is a regulatory protein for the protein phosphatase-6 catalytic subunit. Together, these proteins act as a significant T-loop phosphatase for Aurora A, an essential mitotic kinase. Loss of function of either the regulatory or catalytic subunit of protein phosphatase-6 interferes with spindle formation and chromosome alignment. [provided by RefSeq, May 2017]. | hsa:9701; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; protein phosphatase binding [GO:0019903]; protein phosphatase regulator activity [GO:0019888]; regulation of phosphoprotein phosphatase activity [GO:0043666] | ENSMUSG00000036561 | Ppp6r2 | 4114.90959 | 0.9862644 | -0.0199536264 | 0.10049056 | 4.031499e-02 | 8.408660e-01 | 9.998360e-01 | No | Yes | 3989.39059 | 347.930547 | 3.812470e+03 | 257.537961 | ||
| ENSG00000100271 | 25809 | TTLL1 | protein_coding | O95922 | FUNCTION: Catalytic subunit of a polyglutamylase complex which modifies tubulin, generating side chains of glutamate on the gamma-carboxyl group of specific glutamate residues within the C-terminal tail of tubulin. Probably involved in the side-chain elongation step of the polyglutamylation reaction rather than the initiation step. Modifies both alpha- and beta-tubulins with a preference for the alpha-tail. Unlike most polyglutamylases of the tubulin--tyrosine ligase family, only displays a catalytic activity when in complex with other proteins as it is most likely lacking domains important for autonomous activity. Part of the neuronal tubulin polyglutamylase complex. Mediates cilia and flagella polyglutamylation which is essential for their biogenesis and motility. Involved in respiratory motile cilia function through the regulation of beating asymmetry. Essential for sperm flagella biogenesis, motility and male fertility. Involved in KLF4 glutamylation which impedes its ubiquitination, thereby leading to somatic cell reprogramming, pluripotency maintenance and embryogenesis. {ECO:0000250|UniProtKB:Q91V51}. | ATP-binding;Alternative splicing;Cell projection;Cilium;Cytoplasm;Cytoskeleton;Flagellum;Ligase;Magnesium;Metal-binding;Microtubule;Nucleotide-binding;Reference proteome | hsa:25809; | ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytoplasm [GO:0005737]; extracellular region [GO:0005576]; microtubule [GO:0005874]; motile cilium [GO:0031514]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; tubulin binding [GO:0015631]; tubulin-glutamic acid ligase activity [GO:0070740]; cerebellar Purkinje cell differentiation [GO:0021702]; immune response in nasopharyngeal-associated lymphoid tissue [GO:0002395]; microtubule cytoskeleton organization [GO:0000226]; mucociliary clearance [GO:0120197]; protein polyglutamylation [GO:0018095]; regulation of blastocyst development [GO:0120222]; sperm axoneme assembly [GO:0007288] | 19767753_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20564319_Meta-analysis of gene-disease association. (HuGE Navigator) | ENSMUSG00000022442 | Ttll1 | 229.22235 | 0.9883797 | -0.0168627871 | 0.20516911 | 6.780484e-03 | 9.343734e-01 | 9.998360e-01 | No | Yes | 226.00722 | 22.838394 | 2.438908e+02 | 19.711613 | ||
| ENSG00000100697 | 23405 | DICER1 | protein_coding | Q9UPY3 | FUNCTION: Double-stranded RNA (dsRNA) endoribonuclease playing a central role in short dsRNA-mediated post-transcriptional gene silencing. Cleaves naturally occurring long dsRNAs and short hairpin pre-microRNAs (miRNA) into fragments of twenty-one to twenty-three nucleotides with 3' overhang of two nucleotides, producing respectively short interfering RNAs (siRNA) and mature microRNAs. SiRNAs and miRNAs serve as guide to direct the RNA-induced silencing complex (RISC) to complementary RNAs to degrade them or prevent their translation. Gene silencing mediated by siRNAs, also called RNA interference, controls the elimination of transcripts from mobile and repetitive DNA elements of the genome but also the degradation of exogenous RNA of viral origin for instance. The miRNA pathway on the other side is a mean to specifically regulate the expression of target genes. {ECO:0000269|PubMed:15242644, ECO:0000269|PubMed:15973356, ECO:0000269|PubMed:16142218, ECO:0000269|PubMed:16271387, ECO:0000269|PubMed:16289642, ECO:0000269|PubMed:16357216, ECO:0000269|PubMed:16424907, ECO:0000269|PubMed:17452327, ECO:0000269|PubMed:18178619}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Disease variant;Endonuclease;Helicase;Host-virus interaction;Hydrolase;Magnesium;Manganese;Metal-binding;Nuclease;Nucleotide-binding;Phosphoprotein;RNA-binding;RNA-mediated gene silencing;Reference proteome;Repeat | This gene encodes a protein possessing an RNA helicase motif containing a DEXH box in its amino terminus and an RNA motif in the carboxy terminus. The encoded protein functions as a ribonuclease and is required by the RNA interference and small temporal RNA (stRNA) pathways to produce the active small RNA component that represses gene expression. This protein also acts as a strong antiviral agent with activity against RNA viruses, including the Zika and SARS-CoV-2 viruses. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2021]. | hsa:23405; | ARC complex [GO:0033167]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; RISC complex [GO:0016442]; RISC-loading complex [GO:0070578]; ATP binding [GO:0005524]; deoxyribonuclease I activity [GO:0004530]; DNA binding [GO:0003677]; double-stranded RNA binding [GO:0003725]; endoribonuclease activity [GO:0004521]; helicase activity [GO:0004386]; metal ion binding [GO:0046872]; pre-miRNA binding [GO:0070883]; protein domain specific binding [GO:0019904]; ribonuclease III activity [GO:0004525]; RNA binding [GO:0003723]; siRNA binding [GO:0035197]; apoptotic DNA fragmentation [GO:0006309]; conversion of ds siRNA to ss siRNA [GO:0036404]; conversion of ds siRNA to ss siRNA involved in RNA interference [GO:0033168]; miRNA loading onto RISC involved in gene silencing by miRNA [GO:0035280]; miRNA metabolic process [GO:0010586]; negative regulation of gene expression [GO:0010629]; negative regulation of Schwann cell proliferation [GO:0010626]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of tumor necrosis factor production [GO:0032720]; nerve development [GO:0021675]; neuron projection morphogenesis [GO:0048812]; NIK/NF-kappaB signaling [GO:0038061]; peripheral nervous system myelin formation [GO:0032290]; positive regulation of myelination [GO:0031643]; positive regulation of Schwann cell differentiation [GO:0014040]; pre-miRNA processing [GO:0031054]; production of miRNAs involved in gene silencing by miRNA [GO:0035196]; production of siRNA involved in RNA interference [GO:0030422]; RNA phosphodiester bond hydrolysis [GO:0090501]; RNA phosphodiester bond hydrolysis, endonucleolytic [GO:0090502]; siRNA loading onto RISC involved in RNA interference [GO:0035087]; targeting of mRNA for destruction involved in RNA interference [GO:0030423]; tRNA catabolic process [GO:0016078] | 12411504_cloning and expression of the 218 kDa human Dicer, and characterization of its ribonuclease activity and dsRNA-binding properties 12411505_purification and properties of a recombinant human Dicer 14576312_fragile X syndrome CGG repeats readily form RNA hairpins and is digested by the human Dicer enzyme, a step central to the RNA interference effect on gene expression 15242644_Dicer has a single RNA post-transcriptional processing center 15247924_Dicer is essential for formation of the heterochromatin structure in vertebrate cells. 15811921_various attributes of the 3' end structure, including overhang length and sequence composition, play a primary role in determining the position of Dicer cleavage in both dsRNA and unimolecular, short hairpin RNA 16380083_RNA containing the AU-rich element of GM-CSF is destabilized by dicer and positive charge of proteins 16582496_The C-terminal RNase III domain (RNase IIIb) of human Dicer was expressed, purified and crystallized by the sitting-drop vapour-diffusion method 17071602_Dicer up-regulation may explain an almost global increase of microRNA expression in prostate adenocarcinoma. 17303335_Up-regulation of TSSC3 occurred in Dicer knockdown cells. 17317629_Transcripts containing long hairpin structures composed of CNG repeats are another class of Dicer targets. 17332367_Overexpression of Dicer correlates with precancerous conditions for Lung adenocarcinoma. 17360756_These results suggest that, in the context of HIV replication, TRBP contributes mainly to the enhancement of virus production and that Dicer does not mediate HIV restriction by RNAi. 17379831_Maintenance and regulation of endogenous microRNA levels via Dicer-mediated processing is critical for endothelial cell gene expression and functions in vitro. 17452327_Results indicate that human TRBP and PACT directly interact with each other and associate with Dicer to stimulate the cleavage of double-stranded or short hairpin RNA to siRNA. 17482383_a unique amino acid sequence in human DICER protein is essential for binding to Argonaute family proteins 17663774_Results show that recombinant Dicer is capable of cleaving the TAR element in vitro and that TAR derived miRNA is present in HIV-1 infected cell lines and primary T-cell blasts. 18023283_a role for the PAZ domain of Dicer in binding ssRNAs. 18167183_DICER1 may play an important role in the development of cancer and the epigenetical regulation involved. 18178619_Ago2, Dicer, and TRBP comprise the RISC-loading complex (RLC) and assembles spontaneously in vitro from purified components 18239938_Abnormal immunoexpression of Dicer in aggressive mucoepidermoid carcinoma suggests a role for microRNA and microRNA machinery in tumor progression. 18289125_the role of Dicer, one of the central proteins of the miRNA processing machinery during apoptosis, and show that down-regulation of Dicer results in accelerated apoptosis of HeLa cells, triggered by TNFalpha (tumour necrosis factor alpha). 18325616_Our results suggest that HCV core protein may abrogate host cell RNA silencing defense by suppressing the ability of Dicer to process precursor dsRNAs into siRNAs. 18413723_Intact DICER is required to maintain full promoter DNA hypermethylation of select epigenetically silenced loci in human cancer cells. 18508075_These results suggest that the DExD/H-box domain likely disrupts the functionality of the Dicer active site until a structural rearrangement occurs, perhaps upon assembly with its molecular partners. 18637735_Chemical modifications patterns compatible with high potency dicer-substrate small interfering RNAs are reported. 18644891_Up-regulation of MICA and MICB is the result of DNA damage response activation caused by Dicer knockdown. RNAi is indirectly linked to the human innate immune system via the DNA damage pathway. 18700235_luciferase assay using a reporter carrying a putative target site in the 3' untranslated region of Dicer revealed that let-7 directly affects Dicer expression 18927112_Evidence that a Dicer helicase mutant is sensitive to the thermodynamic properties of the stems in microRNAs and short-hairpin RNAs, with thermodynamically unstable stems resulting in poor processing and a reduction in the levels of functional mi/siRNAs. 18978195_Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. 19017633_Dicer cleaved substrates containing short siRNA-like double-strand regions and extended 3' or 5' ssRNA overhangs in the adjacent ssRNA regions 19022417_Data delineated the smallest 5-lipoxygenase binding domain (5LObd) of Dicer to its C-terminal 140 amino acids comprising the double-stranded RNA (dsRNA) binding domain (dsRBD). 19047128_Observational study of gene-disease association. (HuGE Navigator) 19082437_knockdown of Dicer inhibits human breast carcinoma cell growth 19092150_Our findings indicate that levels of Dicer and Drosha mRNA in ovarian-cancer cells have associations with outcomes in patients with ovarian cancer. 19118902_Dicer is a stress response component and interferons are potentially important regulators of Dicer expression. 19127773_Data demonstrated a direct interaction between Dicer and Wig-1 and both may play a common role in dsRNA-related gene regulation. 19138993_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19155329_Phosphorylation of FMRP regulates its association with the miRNA pathway by modulating association with Dicer. 19393748_Dicer 1 gene is expressed in differentiating and terminally differentiated N-type cells. 19422693_Binding of Dicer to TRBPs is critical for RNAi function. 19520829_Results show that Dicer is responsible for the generation of the mature miR-222 and -339, which suppress ICAM-1 expression on tumor cells. 19556464_findings show that 11 multiplex pleuropulmonary blastoma families harbor heterozygous germline mutations in DICER1 19646895_Loss of Dicer within ovarian granulosa cells, luteal tissue, oocyte, oviduct and, potentially, the uterus renders females infertile [review] 19672267_Assessment of Dicer expression may facilitate prediction of distant metastases for patients suffering from breast cancer. 19678941_Hepatitis C virus (HCV) internal ribosome entry site (IRES) can be recognized and processed into small ribonucleic acid (RNA)s by human ribonuclease Dicer in vitro; domains II, III and VI of HCV IRES are potential in vitro substrates for Dicer. 19782670_Data show that Dicer mRNA expression is down-regulated in the majority of ovarian tumors when compared to normal tissues. 19806572_The proliferation activity and invasive ability were significantly enhanced in cells transfected with Dicer siRNA. 19836333_the three-dimensional structure of human Dicer bound to the protein TRBP at approximately 20 A resolution determined by negative-stain electron microscopy (EM) and single-particle analysis 19851984_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19914350_Upregulation of fibronectin-1 protects Dicer knockdown HEK293T cells against apoptosis. 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20210522_Report dysregulation of microRNA processing enzymes Drosha and Dicer in epithelial skin tumors when compared to healthy control samples. 20211803_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20232482_examination of Dicer protein levels in four head and neck squamous cell carcinoma (HNSCC) cell lines demonstrated that Dicer had between 4- and 24-fold higher expression levels when compared to normal human primary gingival epithelial cells. 20237275_Dicer is necessary for the developmental change in competence of the retinal progenitor cells. 20405249_reduced expression of Dicer may play an important role during the process of hepatocarcinogenesis. 20463238_loss of Dicer impairs Schwann cells (SCs)differentiation by disrupting the balance between stimulating and inhibiting pathways for the myelination process, suggest that SC-axon interactions are affected by SC-specific loss of miRNA-based gene regulation 20530258_Dicer-dependent generation of microRNAs affects homeostasis and function of epidermal Langerhans cells. 20550935_Targeted knockout of DICER is lethal to melanocytes, at least partly via DICER-dependent processing of the pre-miRNA-17 approximately 92 cluster thus targeting BIM, a known proapoptotic regulator of melanocyte survival. 20584909_Data show that HIV-1 suppresses the expression and function of DICER in macrophages, and that the presence of miRNAs in monocytes lacking DICER indicates that some miRNAs can be generated by proteins other than DICER. 20615407_The identification of a novel splice variant of human dicer gene, the first one bearing a modified coding sequence, is identified. 20721975_Observational study of gene-disease association. (HuGE Navigator) 20730047_Dicer functions in maintaining integrity of rDNA arrays 20732906_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20805302_Findings of growth promotion by silencing Dicer implies its potential use as therapeutic targets for neuroblastoma. 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20869963_Results suggest that the interaction of Dicer with HTLV-I Rex inhibits Dicer activity and thereby reduces the efficiency of the conversion of shRNA to siRNA. 20932845_Dicer is much faster at processing a pre-miRNA substrate compared to a pre-siRNA substrate under both single and multiple turnover conditions 20962848_TAp63, a p53 family member, suppresses tumorigenesis and metastasis, and coordinately regulates Dicer and miR-130b to suppress metastasis 20974746_Short RNA duplexes guide sequence-dependent cleavage by human dicer. 21029372_miR-107, an oncogene miRNA promoting gastric cancer metastasis through down-regulation of DICER1. 21036787_that DICER1 mutations are unlikely to have a major role in the aetiology of sporadic Wilms tumour. 21044367_Observational study of gene-disease association. (HuGE Navigator) 21048142_A transgenic mouse model with an inducible disruption of the Dicer1 gene in the adult forebrain shows microRNAs as key players in the learning and memory process of mammals. 21160068_Dicer expression may play an important role in the progression and prognosis of monoclonal gammopathies 21205968_DICER1 mutations are associated with both familial multinodular goiter (MNG) and MNG with Sertoli-Leydig cell tumor of the ovary, independent of pleuropulmonary blastoma 21266384_Haploinsufficiency of the Dicer1 gene is associated with a predisposition to a broad range of tumors. 21297615_findings reveal a miRNA-independent cell survival function for DICER1 involving retrotransposon transcript degradation 21297638_Exportin-5 controls Dicer1 expression post-transcriptionally. 21306637_The two-step cleavage of a hairpin RNA with 5' overhangs shows that DICER releases double-stranded RNAs after the first cleavage and binds them again in the inverse direction for a second cleavage. 21345667_Strong expression of the central microRNA biosynthesis enzyme Dicer predicts poor prognosis in patients with colorectal cancer 21346072_Dicer1 helps regulate tubal expression of steroid hormone receptors in a cycle-dependent manner and may contribute to tubal transport in humans 21425145_Lower DICER1 transcript levels were correlated with disease recurrence and worse DFS survival in patients with endometrioid endometrial cancer. 21501861_germline DICER1 mutations may be found in Pleuropulmonary blastoma-associated ovarian sex cord-stromal tumors (OSCST) cases. 21559780_This study revealed for the first time that expression alterations of Dicer and Drosha are present in endometrial cancer tissue and could be potentially responsible for altered microRNAs profiles observed in this malignancy. 21698147_deregulation of Dicer and its influence on miRNA maturation is needed to predict the susceptibility of melanoma patients to miRNA-based therapy in the future 21753850_5'-end recognition by Dicer is important for precise and effective biogenesis of miRNAs 21761362_microRNAs actively control some of the most distinguishing characteristics of the breast cancer subtypes, such as ERalpha itself, Dicer, and growth factor receptor levels 21769619_Our results suggest that Drosha, Dicer, and Ago2 are possibly implicated in colorectal carcinoma pathobiology 21827717_Data show that in liver metastases, Dicer was positively related to miR-141. 21858095_Data suggest NUP153 plays a crucial role in the nuclear localization of the DICER1 protein. 21878538_Dicer-overexpressing breast cancer cells enriched for cells with enhanced BCRP function. 21882293_We report DICER1 mutations in seven additional families that manifested uterine cervix embryonal rhabdomyosarcoma , primitive neuroectodermal tumor, Wilms tumor, pulmonary sequestration and juvenile intestinal polyp. 21889945_Data support the view that mutagens interfere with microRNA maturation by binding DICER. 21898071_Low dicer expression is associated with breast cancer. 21937200_Data suggest Dicer expression as a possible molecular marker in patients with primary cutaneous T cell lymphomas and apparently indicate that miRNA(s) might be of clinical relevance in CTCL. 21953080_Drosha and Dicer expression was dysregulation in nasopharyngeal carcinoma compared with healthy control samples. 22055188_MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. 22133720_The identification of neoplasia-associated germline mutations in DICER1 has focused translational research on components of the miRNA processing pathway. 22163034_AGO2, PACT and TRBP are required for the efficient functioning of Dicer in cells 22179432_Dicer is significantly downregulated in transitional cell carcinomas compared to paired normal adjacent tissues samples and normal samples, suggesting that reduced expression of Dicer may play an important role in bladder cancer 22180160_Germline DICER1 mutations were found in all four patients with familial pleuropulmonary blastoma and 2 of 52 sporadic embryonal rhabdomyosarcoma had somatic mutations. 22187960_Somatic missense mutations affecting the RNase IIIb domain of DICER1 are common in nonepithelial ovarian tumors. 22252463_A novel effect of reduced DICER1 function in cancer cells, is reported. 22310293_MiR-9 directly targets HuR and Dicer1 for posttranscriptional repression. 22320315_Our data suggest that Dicer expression may play an important role in the progression and prognosis of CLL. 22350415_miR-130a inhibited autophagy by reducing autophagosome formation, an effect mediated by downregulation of the genes ATG2B and DICER1. 22371183_mesenchymal stromal cells from MDS patients show low gene and protein expression of DICER1 and DROSHA which are involved in the microRNA biogenesis, as well as their target gene SBDS. 22381205_The results showed that genetic variants of DICER1 may modify the risk of abnormal semen parameters, which could result in male infertility. 22394132_results provide novel evidence that Dicer, is probably involved in the pathobiology of human smooth muscle neoplasms 22407237_study demonstrated that expression of miR-107 in gastric cancer tissues showed a significant negative association with DICER1 mRNA levels 22426548_Study describes a new domain localization strategy developed to determine the structure of human Dicer by electron microscopy.The helicase domains form a clamp-like structure adjacent to the RNase III active site, facilitating recognition of pre-miRNA loops or translocation on long dsRNAs. 22446887_The effect of introducing transcription factors into two distinct colorectal cancer (CRC) cell lines, HCT116 and DLD-1, in the presence and absence of Dicer 1, ribonuclease type III (Dicer1), a critical miRNA processing enzyme, was studied. 22479364_Data show that down-regulation of either Dicer or Drosha had no effect on the sensitivity of cells to irradiation. 22541070_Findings present a novel self-recognition immune response, whereby endogenous Alu RNA-induced NLRP3 inflammasome activation results from DICER1 deficiency in a nonimmune cell, retinal pigmented epithelium and is implicated in georaphic atrophy. 22546613_The results suggest that mutation of the clinically relevant residue D1709 in the RNase IIIB results in a uniquely miRNA-haploinsufficient state in which the let-7 family of tumor suppressor miRNAs is lost while a complement of 3p-derived miRNAs remains. 22615744_findings suggest loss of Dicer and failure of mature miRNA expression may be a feature of the pathophysiology of HS in patients with TLE 22647351_Drosha, Dicer, Argonaute 1, and Argonaute 2 are differentially expressed at different metastatic sites in ovarian carcinoma compared with primary carcinomas. 22689055_Knockdown of Sox4 induces a major change in the expression pattern of miRNAs in melanoma cells, mainly due to reduced expression of Dicer. 22716222_Our data suggest that reduced Dicer expression might contribute to tumour progression in colorectal cancer 22718634_The RNase activity of Dicer could be inhibited by substituting Ca 2 + for Mg 2 + . 22727743_Distinct interactions of each Dicer domain with different RNA structural features. 22737231_expression of human Dicer in Dicer-null mouse embryonic fibroblasts restores their reprogramming potential, demonstrating that microRNAs are indispensable for dedifferentiation reprogramming 22745131_functional deficiency of Dicer in chronic hypoxia is relevant to both HIF-alpha isoforms and hypoxia-responsive/HIF target genes, especially in the vascular endothelium. 22766726_Dicer, Drosha, and Exportin 5 genes were up-regulated in bladder urothelial carcinoma compared to normal urothelium. Silencing these genes induced cell proliferation inhibition and apoptosis in bladder urothelial carcinoma cells. 22808238_Dicer can process 7SL RNA. 22821364_Dicer protein plays a role in human breast cancer progression and behaviour. 22867905_study outlines a cancer syndrome caused by germline mutations in DICER1, responsible for microRNA processing; multiple endocrine manifestations of the DICER1 syndrome 22922797_MiR-3928 plays an important role in regulating the expression of Dicer. 22972926_Commitment to the T cell lineage requires transgenic Dicer expression in thymic epithelial cells. 22984192_Dicer binding protects the terminal loop from digestion by S1 nuclease, suggesting that Dicer interacts directly with the terminal loop region. 22999255_Depletion of Dicer and TRBP, proteins involved in miRNA biogenesis, reduced HCV RNA accumulation, mature duplex miR-122 abundance, and miR-122 directed mRNA translation suppression, suggesting roles in miR-122 processing. 23047949_The results define CLIMP-63 as a novel protein interactor and regulator of Dicer function, involved in maintaining Dicer protein levels in human cells. 23064648_DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. 23068969_The G allele for IL-10 (-1082 A/G), the C allele for IL-4 (+3 C/T) and the C and T alleles for IL-28B (rs12979860 and rs8099917, respectively) are associated with spontaneous viral clearance in hepatitis C infection 23071822_rs1057035 in 3'UTR of DICER was associated with a significantly decreased risk of oral cancer 23141545_Results suggest that Dicer recognizes the loop/bulge structure in addition to the ends of shRNAs/pre-miRNAs for accurate processing. 23143396_Data show that the miRNA-processing enzyme, DICER and the main miRNA effector, AGO2, are targeted for degradation as miRNA-free entities 23222681_the molecular mechanisms by which loss of Dicer leads to DNA damage, as well as the role of Dicer in tumorigenesis. 23251602_Dicer rs3742330 is associated with T cell lymphoma survival. 23266047_Data indicate that tumour sample showed a significantly higher Drosha expression versus normal mucosa, while Dicer levels did not differ. 23307239_Dicer1 was downregulated in nasopharyngeal carcinoma 23311378_Heat-induced regulation of Dicer expression occurs primarily post- transcriptionally, and the expression levels of Dicer protein are increased and often oscillate in response to fever-range hyperthermia in multiple mouse and human cells 23349018_Levels of DICER1, DROSHA and five different miRNAs were measured in NSCLC specimens. 23392577_DICER1 is a miR-130b target gene in human endometrial cancer cells.DICER1 induced abnormal expression of the epithelial mesenchymal transition related genes. 23418872_The results suggest that DICER1 exons 25 and 26 mutations may be very rare in human hematopoietic tumors. 23426981_DICER1 is upregulated by the hematopoietic transcription factor, GATA1, in acute myeloid leukemia. 23441612_homozygous CC genotype of DICER1, rs1057035, was significantly associated with decreased risk of colorectal cancer 23468895_Dicer can fine-tune the efficiency of SRP-mediated protein targeting via processing a proportion of 7SL RNA into fragments of different lengths. 23472152_Dicer is a direct target of miR-31, but also isomiRs display similar and disparate regulation of target genes in cell-based systems 23519840_Downregulation of DICER1 is associated with lymph node metastasis of gastric carcinoma. 23547758_germ-line or somatic DICER1 mutations do not haev a major role in the etiology of familial testicular germ cell tumours 23550157_RISC proteins PACT, TRBP, and Dicer, together with PKR, are steroid receptor RNA activator-binding nuclear receptor coregulators that are recruited to the promoters of hormone-regulated genes and regulate the expression of downstream target genes. 23620094_Biallelic DICER1 mutations occur in Wilms tumours. 23624860_Dicer traps pre-siRNAs in a nonproductive conformation, whereas interactions of Dicer with pre-miRNAs and dsRNA-binding proteins induce structural changes in the enzyme that enable productive substrate recognition in the central catalytic channel. 23625684_The DICER1 gene encodes an enzyme that is involved in the biogenesis of microRNAs. The entire tumor spectrum and the respective tumor risks are unknown. 23663110_It is a miRNA processing enzyme and altered in non-alcoholic fatty liver disease. 23728841_Genetic testing revealed a novel heterozygous premature termination mutation (c.1525C>T p.R509X) in the DICER1 gene. 23763383_confirmed DICER1 hot-spot mutations in Sertoli-Leydig cell tumours (15.4%) in Korean patients. 23849790_These results support a model in which Vpr complexes with human Dicer to boost its interaction with the CRL4(DCAF1) ubiquitin ligase complex and its subsequent degradation. 23851498_High Dicer expression is associated with prostate cancer growth and metastasis. 23868280_potentially fatal nature of pleuropulmonary blastoma argues that screening of DICER1 mutation carriers should be offered during the years of susceptibility 23868705_Polymorphism in DICER1 gene is associated with HBV-related hepatocellular carcinoma. 23882114_we demonstrate that human Dicer has the ability to shuttle between the nucleus and the cytoplasm. We conclude that Dicer is a shuttling protein whose steady-state localization is cytoplasmic. 24025624_Authors establish HIV-1 tat and rev raginine rich motifs as a novel viral motifs evolved to target the Dicer dependent RNAi pathway. 24065110_We report here the first characterized exonic deletion in DICER1, which are responsible for a rare cancer syndrome, including tumors that can co-occur with multinodular goiter. 24094887_Data show that the knockdown of Dicer significantly promoted cell proliferation, migration, and invasion. 24096488_DICER1-deficient colorectal cancer cells have an enhanced ability to initiate tumors and metastasis. 24136150_More than half (8/15) of Sertoli-Leydig cell tumours (SLCTs) harbour DICER1 mutations in the RNase IIIb domain, while mutations are rarely found in germ cell tumors. 24163352_Deletion or depletion of Dicer in mouse or human activated CD8+ T cells causes up-regulation of perforin, granzymes, and effector cytokines. 24204924_These specific inhibitors can simultaneously bind both Dicer and pre-miRNAs. 24220032_a clear relationship between mutant p53, TAp63, and Dicer that might contribute to the metastatic function of mutant p53 but, interestingly, also reveal TAp63-independent functions of mutant p53 in controlling Dicer activity. 24223844_Data suggest that miR-192 might be a key player in neuroblastoma (NB) by regulating Dicer1 expression. 24244599_Dicer expression exhibits a tissue-specific diurnal pattern that is lost during aging and in diabetes. 24287487_Induction of Dicer by cyclin D1 coordinates microRNA biogenesis in breast cancer. 24317682_Dicer functions as an indirect tumor suppressor, as silencing Dicer expression makes cancer cells more malignant. 24324261_The Hippo pathway effectors TAZ/YAP regulate dicer expression and microRNA biogenesis through Let-7. 24337369_Taken together, the present study contributes important new findings for the role of Dicer-mediated miRNA processing in collagen synthesis of hepatic stellate cells , which may serve as a foundation for RNAi study of liver fibrosis in vivo. 24386264_Deregulated Dicer expression is associated with aggressive tumour characteristics and is an independent prognostic factor for OS. 24462910_gDicer is required for follicle development, and structural differences in the helicase domain of two gDicer isoforms might contribute to their different roles in controlling granulosa cell apoptosis. 24481001_The genetic analyses confirm that DICER1 mutations are the major genetic event in the development of cystic nephroma. Further, cystic nephroma and pleuropulmonary blastoma have similar DICER1 loss of function and 'hotspot' missense mutation rates 24486018_analysis of the structure of the Dicer phosphate-binding pocket within the platform-PAZ-connector helix cassette 24574065_Low Dicer expression is associated with invasive breast carcinoma. 24617712_This report of somatic DICER1 mutations in differentiated thyroid carcinoma (DTC) strengthens the association between DTC and the DICER1 syndrome. 24668803_Data indicate that cigarette smoke decreases RNA endonuclease DICER protein levels in exposed cells. 24675358_Results highlight the role of RNase IIIb domain mutations in DICER1 along with TP53 inactivation in PPB pathogenesis. 24676357_Mutations affecting the metal binding sites of the DICER1 RNase IIIb domain alter the balance of 3p and 5p microRNAs leading to deregulation of these growth signalling pathways, causing a novel human overgrowth syndrome. 24676997_Down-regulation of Dicer expression is associated with cervical cancer. 24725360_The altered expression of Dicer and Drosha may serve as markers for disrupted miRNA biogenesis in Triple negative breast cancer 24769857_Association between recurrent pregnancy loss development and the polymorphism of the miRNA machinery gene RAN and combined genotype of DROSHA/DICER. 24787017_Dicer expression regulated by miR-130a is an important potential prognostic factor in cervical cancer. 24814348_Dicer restricts dsRNA accumulation in nuclei. 24817967_Dysregulation of microRNA biosynthesis enzyme Dicer plays an important role in gastric cancer progression. 24821309_DICER1 germline mutation is associated with pleuropulmonary blastoma. 24839956_findings show germ-line DICER1 mutations are a major contributor to pituitary blastoma (PitB); second somatic DICER1 'hits' occurring within the RNase IIIb domain also appear to be critical in PitB pathogenesis 24846461_Temporal separation of pro- and anti-apoptotic signaling induced by inhibition of Dicer1 is synergistic and synthetic lethal to low-dose 5-FU chemotherapy in p53-mutated HACAT cells. 24909177_Data indicate that in addition to frequent biallelic loss of TP53 protein, each case had compound disruption of DICER1 protein. 24913918_miR-581 promotes hepatitis B virus surface antigen expression by targeting Dicer and EDEM1 in HepG2 cells. 24987961_Discovery of a dicer-independent, cell-type dependent alternate targeting sequence generator: implications in gene silencing & pooled RNAi screens. 25022261_This study suggests that germ-line DICER1 mutations make a clinically significant contribution to pineoblastoma. 25031339_Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. 25035295_Importance of a weak G-U or U-G base pair at the top of the shRNA hairpin stem to hinder Dicer recognition. 25061173_High Dicer1 expression is associated with resistance to bortezomib therapy in myeloma. 25115815_Results show that the expression levels of Dicer1e influence the pathogenesis of oral cancer cells. 25118636_Pathogenic germline and somatic mutations of DICER1 in Nasal chondromesenchymal hamartoma (NCMH) establishes that the genetic etiology of NCMH is similar to pleuropulmonary blastoma (PPB), despite the disparate biological potential of these neoplasms. 25123850_expression in laryngeal squamous cell carcinoma associated with survival 25135428_High Dicer expression levels were associated with prostate cancer. 25176334_Both germline and somatic mutations in DICER1 are implicated in diverse types of cancer. [Review] 25190313_DICER1 RNase IIIB mutations preferentially impair processing of miRNAs deriving from the 5'-arm of pre-miRNA hairpins in Wilms tumors. 25195038_The aim of this study was to detect the expression levels of Dicer1, Drosha, DGCR8, and Ago2 messenger ribonucleic acids (mRNAs) in patients with primary gastrointestinal diffuse large B-cell lymphoma 25295740_Dicer is upregulated in gestational diabetes. 25349431_DICER1/Alu RNA dysmetabolism induces Caspase-8-mediated cell death in age-related macular degeneration. 25351418_DICER expression is suppressed by hypoxia through an epigenetic mechanism. 25361944_Down-regulation of Dicer1 promotes cellular senescence and decreases the differentiation and stem cell-supporting capacities of mesenchymal stromal cells in patients with myelodysplastic syndrome. 25416952_Report of transcriptome-wide map of Dicer targets; identify thousands of Dicer-binding sites in human cells and C. elegans; find known and hundreds of additional miRNAs; also report structural RNAs, promoter RNAs, and mitochondrial transcripts as Dicer targets; most Dicer-binding sites reside on mRNAs/lncRNAs and are not significantly processed into small RNAs. 25428210_We show that reduction of DICER1 in human female cells increases XIST transcripts without compromising the binding of the XIST and histone tail modifications on the Xi chromosome. 25439752_We aimed to evaluate the expression of the major components of microRNA biogenesis machinery including Drosha, Dicer and DiGeorge syndrome critical region gene 8 (DGCR8) in multiple sclerosis patients 25456905_Dysregulation of Dicer and possibly altered expression of miRNAs are associated with aggressive features in thyroid cancers. 25457613_These findings indicate that miRNA depletion in dicer deficiency is due to the combined loss of miRNA-generating activity and catabolic function of TN/TX. 25480859_Dicer expression is decreased and is mechanistically linked to increased expression of co-stimulatory molecule CD80 in B cells of patients with multiple sclerosis 25500911_define precise temporal and epithelial cell type-specific DICER1 functions in the developing lung and demonstrate that loss of these DICER1 functions is sufficient for the development of cystic pleuropulmonary blastoma 25526195_None of the observed variants appeared to be disease-related, suggesting that germline mutations in DICER1 are rare or absent in familial breast cancer patients. 25557550_Results show the crystal structure of the interface between microRNA biogenesis proteins Dicer and TRBP. Mutations in this interface prevent recruitment of TRBP to Dicer. 25640338_The present meta-analysis suggests that the C allele of the DICER rs1057035 polymorphism probably decreases cancer risk. However, this association may be Asian-specific. 25656609_EIF2C2, Dicer, and Drosha are more highly expressed in bladder carcinoma, promote the development of bladder cancer, and suggested a poor prognosis 25702703_Data show that increased p53, together with deceased cyclin D1 and endoribonuclease DICER1 in mammospheres were responsible for microRNA let-7 inhibition. 25752336_Data indicate a direct correlation betwe | ENSMUSG00000041415 | Dicer1 | 732.13748 | 0.7707748 | -0.3756186313 | 0.18319759 | 4.099835e+00 | 4.288739e-02 | 9.998360e-01 | No | Yes | 689.07619 | 141.453315 | 8.535663e+02 | 134.971833 | |
| ENSG00000101000 | 10544 | PROCR | protein_coding | Q9UNN8 | FUNCTION: Binds activated protein C. Enhances protein C activation by the thrombin-thrombomodulin complex; plays a role in the protein C pathway controlling blood coagulation. | 3D-structure;Blood coagulation;Disulfide bond;Glycoprotein;Hemostasis;Membrane;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a receptor for activated protein C, a serine protease activated by and involved in the blood coagulation pathway. The encoded protein is an N-glycosylated type I membrane protein that enhances the activation of protein C. Mutations in this gene have been associated with venous thromboembolism and myocardial infarction, as well as with late fetal loss during pregnancy. The encoded protein may also play a role in malarial infection and has been associated with cancer. [provided by RefSeq, Jul 2013]. | hsa:10544; | cell surface [GO:0009986]; centrosome [GO:0005813]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; focal adhesion [GO:0005925]; integral component of plasma membrane [GO:0005887]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; signaling receptor activity [GO:0038023]; blood coagulation [GO:0007596]; negative regulation of coagulation [GO:0050819] | 11552992_Observational study of gene-disease association. (HuGE Navigator) 11686350_A 23bp insertion in the endothelial protein C receptor (EPCR) gene impairs EPCR function. Since protein C activation depends on the concentration of EPCR, patients with the EPCR insertion could have a diminished protein C activation capacity. 11776299_Observational study of gene-disease association. (HuGE Navigator) 11776299_Our findings showed that the EPCR 23bp insertion is very rare in both patients with VTE and the general population and failed to support an association between the EPCR 23bp insertion and an increased risk of VTE. 12152660_Observational study of gene-disease association. (HuGE Navigator) 12152662_Observational study of gene-disease association. (HuGE Navigator) 12413583_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12439143_Observational study of gene-disease association. (HuGE Navigator) 12514663_Observational study of gene-disease association. (HuGE Navigator) 12529763_Observational study of gene-disease association. (HuGE Navigator) 12560236_Functional analysis of a hypersensitive site in the 5' flanking region of the hEPCR gene in endothelial cells identified multiple regions, containing high and low homology consensus Sp1 binding sequences, that were protected from methylation. 12586611_data suggest that activated protein C regulates calcium concentration in human brain endothelium and in human umbilical vein endothelial cells by binding to endothelial protein C receptor and signaling via protease activated receptor-1 12897745_Endothelial protein C receptor expressed by eosinophils activated with protein C or activated protein C arrests directed migration 14576048_Observational study of gene-disease association. (HuGE Navigator) 15080580_Endothelial protein C receptor may play an important inflammation-related role in plaque development. 15116250_Observational study of gene-disease association. (HuGE Navigator) 15116250_individuals carrying the 4600AG EPCR genotype have high sEPCR levels but do not have an increased risk of thrombosis, whereas individuals carrying the 4678CC genotype have higher APC levels and lower risk of venous thromboembolism 15150078_Anti-EPCR autoantibodies can be detected in antiphospholid syndrome patients and are independent risk factors for fetal death. 15178554_Review. TM, APC, & EPCR impact coagulation, inflammation, fibrinolysis, & cell proliferation. We review the functions of this complex multimolecular system & how its components maintain homeostasis under hypercoagulable &/or proinflammatory conditions. 15304035_Observational study of gene-disease association. (HuGE Navigator) 15304035_this study does not show a strong association between EPCR haplotypes and thrombosis risk, but low sEPCR levels may reduce the risk of deep venous thrombosis 15634335_protein C binding to sEPCR and phospholipids is broadly dependent on correct Gla domain folding, but can be selectively influenced by judicious mutation 15710622_endothelial protein C receptor ligation and sphingosine 1-phosphate receptor transactivation results in endothelial cell cytoskeletal rearrangement and barrier protection through activated protein C 15921688_EPCR-transfected cells suggested increased basal release of sEPCR from cells expressing the 219Gly EPCR phenotype. 15921688_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15954900_the vascular dysfunction characteristic of systemic lupus erythematosus may be related to a dramatically altered distribution of EPCR, both soluble and membrane-bound forms 15978102_protein C receptor autoantibodies may have a role in acute myocardial infarction in young women 16113830_EPCR 4600A/G and 4678G/C polymorphisms may be involved in venous thromboembolism in carriers of factor V Leiden 16113830_Observational study of gene-disease association. (HuGE Navigator) 16153429_CRP significantly decreases the expression of thrombomodulin and EPCR in human endothelial cells, thereby promoting thrombogenic conditions. 16354176_In vascular endotheliumt, binds activated protein C, generating an anti-inflammatory response. 16354200_In cultured keratinocytes, EPCR expression was upregulated by the addition of activated protein C and inhibited by tumor necrosis factor-alpha 16409473_the A3 haplotype does promote cellular shedding in either 293 or endothelial cells and therefore is likely directly contributory to the higher soluble EPCR levels seen in patients carrying this haplotype 16444434_Observational study of gene-disease association. (HuGE Navigator) 17027065_Observational study of gene-disease association. (HuGE Navigator) 17027065_study confirms that there is a strong association between A3 haplotype and elevated sEPCR levels; we suggest that elevated sEPCR levels might increase the risk of stroke at pediatric age 17049585_possible role for high levels of thrombomodulin as a marker of HCC development in patients with cirrhosis, whereas high levels of EPCR are a possible marker of worse HCC prognosis, being a sign of endothelial damage of large vessels 17054378_Gamma-carboxyglutamic acid--> Lysine mutation causes a significant reduction in the binding force between this protein and protein C. 17099142_Release of EPCR into the conditioned medium was inhibited by the metalloproteinase inhibitors TAPI and GM6001, indicating that the shedding of EPCR from the alveolar epithelium is mediated by a metalloproteinase. 17099210_Observational study of gene-disease association. (HuGE Navigator) 17099210_mutations in the EPCR are not a major cause of recurrent miscarriage although they may exert a modifier effect in combination with other variants 17155946_the EPCR sequence requirement for shedding is amino acids from residue 193 to residue 200. ADAM17 is responsible for EPCR shedding. 17170365_EPCR expression is detected in smooth muscle cells in the fibrous cap of human carotid artery plaques. 17327234_EPCR serves as a cellular binding site for FVII/FVIIa 17439632_the presence of anti-EPCR autoantibodies is a moderate risk factor for deep vein thrombosis in the general population 17454790_Homozygous 23-bp insertion of endothelial protein c receptor gene is associated with fatal sepsis 17570903_relationship between plasma sEPCR levels and development of arteriovenous fistula thromboses 17823308_when EPCR is bound by protein C, the PAR-1 cleavage-dependent protective signaling responses in endothelial cells can be mediated by either thrombin or APC. 17849044_Highlights the different mutations/polymorphisms reported in the EPCR gene and their association with the risk of thrombosis. 18073349_the sEPCRisoform could contribute to the regulatory effect of sEPCR in plasma. 18160602_Observational study of gene-disease association. (HuGE Navigator) 18278202_Failed to validate association of EPCR gene polymorphisms with venous thromboembolism amoung Californians of European ancestry. 18403391_Observational study of gene-disease association. (HuGE Navigator) 18403391_in 20210A carriers the venous thromboembolism risk is influenced both by the actual prothrombin levels and by the EPCR A3 haplotype, via its effect on sEPCR levels 18443268_tEPCR behaves as sEPCR generated by shedding of the cellular endothelial cell protein C receptor 18480081_Observational study of gene-disease association. (HuGE Navigator) 18539144_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18539144_The EPCR A1 haplotype reduced the risk for recurrent miscarriage in carriers of FV Leiden. 18555797_Expression of protein-C receptor in vascular endothelium is activated by aldesterone. 18604550_Possible role of soluble EPCR in the pathogenesis of Behcet's disease. Studies of mutations and polymorphisms in EPCR gene in patients with Behcet's disease might bring to light pathogenic mechanism of ocular and systemic vascular complications. 18612544_Protease activated receptor 1 (PAR-1) activation by thrombin is protective in human pulmonary artery endothelial cells if endothelial protein C receptor is occupied by its natural ligand, protein C. 18680534_Observational study of gene-disease association. (HuGE Navigator) 18680534_no association between PROCR Ser219Gly and risk of CHD, stroke, or mortality; the minor allele SNP, rs2069948, was associated with lymphoid PROCR mRNA expression and increased risk of incident stroke and all-cause mortality, and decreased healthy survival 18757851_Observational study of gene-disease association. (HuGE Navigator) 18757851_carriers of the A3 haplotype have a reduced risk of myocardial infarction, only in part due to increased soluble endothelial protein C levels 18977990_Observational study of gene-disease association. (HuGE Navigator) 19004141_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19129726_Protein C is activated on the surface of endothelial cells by the thrombin-thrombomodulin complex with the stimulation of the endothelial protein C receptor. 19222470_Observational study of gene-disease association. (HuGE Navigator) 19236779_A6936G polymorphism of EPCR can be detected in Chinese Han population which may be correlated with increasing risk of thrombosis in cerebral infarction patients. 19236779_Observational study of gene-disease association. (HuGE Navigator) 19238444_HuGE review of gene-disease association and gene-environment interaction. (HuGE Navigator) 19296056_negative relationship between sEPCR levels and the age of individuals 19436051_significantly increased expression of EPCR and TM in the valvular sinus endothelium as opposed to the vein lumenal endothelium, and the opposite pattern with VWF 19467228_Results suggest that the shedding of endothelial protein C receptors in HUVEC is effectively regulated by IL-1beta and TNF-alpha, and downstream by MAP kinase signaling pathways and metalloproteinases. 19587380_FVIIa or activated protein C binding to EPCR promotes EPCR endocytosis, and EPCR-mediated endocytosis may facilitate the transcytosis of FVIIa and its clearance from the circulation 19634686_Activated protein C protects endothelial cells from inflammatory mediators through an EPCR-dependent mechanism. 19689997_Soluble endothelial protein C receptor did not show any positive correlation between these proinflammatory cytokines (TNF-alpha, IL-1beta, IL-2, IL-6, and IL-8). 19696402_Higher soluble EPCR levels were confirmed for Gly allele carriers (P<0.0001). 19806250_polymorphism is associated with the risk of foetal loss in Mediterranean European couples 19860767_Observational study of gene-disease association. (HuGE Navigator) 19895674_Endothelial protein C receptor plays an unexpected role in supporting cell surface recruitment, PAR1 activation, and signaling by FXa. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20141580_Suggest that the PROCR 6936G allele when carried by the father may be a risk factor for iliac deep vein thrombosis during pregnancy. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20506163_Data show that occupancy of EPCR by protein C switches signaling specificity so that activation of PAR-1 by thrombin inhibits TNF-alpha-mediated synthesis of IL-6 and IL-8 and down-regulates TGF-beta-mediated expression of collagen type 4 and fibronectin. 20619102_EPCR gene was found in 91.7% of solid tumors. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21149441_EPCR interacts with the ternary TF coagulation initiation complex to enable PAR1,2 signaling. 21228323_Data suggest that activated protein C protects endothelial cells by attenuating cell adhesion molecule expression during inflammation, and that APC engages a regulatory crosstalk involving EPCR, ERK1/2, and NF-kappaB that impairs TNF signaling. 21252088_FVIIa, upon binding to EPCR on endothelial cells, activates endogenous protease activated receptor-1 (PAR1) and induces PAR1-mediated p44/42 mitogen-activated protein kinase (MAPK) activation. 21273788_Male gender is another parameter with A3 haplotype associated with elevated soluble EPCR levels in blood, and both parameters may contribute to selective regulatory mechanisms of EPCR release upon inflammation. 21362311_EPCR gene 6936A/G polymorphism is associated with increased plasma levels of sEPCR. Subjects carrying 6936AG likely have an increased risk of thrombosis. 21392254_High levels of protein C are determined by PROCR haplotype 3 21824644_The modest increase of sEPCR levels, together with the correlation with other endothelium activation/damage markers, suggest that it is more an innocent bystander of the endothelium activation/damage than an actor in PE. 21851971_These findings indicate that the H1 haplotype protects Behcet's patients from thrombosis, likely via increased levels of activated protein C. 21885613_Analysis of the binding of biotinylated, active-site blocked FXa, FVIIa & APC to human EPCR on ransfected CHO-EPCR & HUVECs revealed that little FXa was bound to EPCR on cell surfaces compared with FVIIa or APC. 21901231_genetic polymorphism is associated with early-onset myocardial infarction in the Italian population 22036807_EPCR 1651C/G polymorphism and elevated soluble EPCR levels but low soluble EPCR levels increase the risk of idiopathic RSM. 22251481_Studies indicate a significant association between the PROCR rs867186 variant and venous thromboembolism (VTE). 22370814_Human FVIIa binds efficiently to both murine and human EPCR. 22614534_in cancer patients, the levels of sEPCR are significantly higher than the normal range compared to healthy volunteers 22681753_data indicate that it is possible to overexpress EPCR at a sufficient level to provide protection against transplant-related thrombotic and inflammatory injury 22715361_Elevated levels of soluble endothelial protein C receptor, a sensitive marker of endothelial damage, indicated a low level of inflammation and coagulation activation in Maraviroc treated patients not picked up by other widely used markers. 22885985_Our results demonstrate how a gammadelta TCR mediates recognition of broadly stressed human cells by engaging endothelial protein C receptor 22885990_Among 78 Nepalese blood donors, no one had an EPCR 23 bp insertion. 22903729_FFAs inhibit TM-EPCR-Protein C system in endothelial cells through activating JNK signaling, which may be a mechanism for the prothrombotic state in metabolic syndrome. 23136988_findings show that in patients with coronary artery disease, circulating sEPCR levels are related to classical cardiovascular risk factors and renal impairment but are not related to long-term incidence of cardiovascular events 23342274_EPCR is expressed not only by a wide range of human malignant hematological cells but also the detection of plasma sEPCR levels provides a powerful insight into thrombotic risk assessment in cancer patients, especially when it surpasses 200 ng/mL. 23348224_EPCR and APC play a limited role in the host response during pulmonary tuberculosis. 23377316_The ratio of sEPCR levels to protein C activity is high, with a significant negative correlation in patients with STEMI. The sEPCR level has no relation to the development of no-reflow. 23408932_Under static conditions, human monocytes bind soluble EPCR in a concentration dependent manner, as demonstrated by flow cytometry. Binding can be inhibited by specific antibodies (anti-EPCR and anti-Mac-1). 23539451_Tissue factor promoted tumor growth may be attenuated by EPCR expression. 23555015_Rab GTPases regulate endothelial cell protein C receptor-mediated endocytosis and trafficking of factor VIIa. 23581230_EPCR was involved in regulating progression of human lung cancer cells. Manipulation of EPCR expression may be a potential therapeutic strategy for lung cancer. 23661674_Endothelial protease nexin-1 is a novel regulator of A disintegrin and metalloproteinase 17 maturation and endothelial protein C receptor shedding via furin inhibition. 23717683_conclude that urinary sEPCR could be a novel non-invasive biomarker of antibody mediated rejection in renal transplantation 23724628_These observations provide evidence that EPCR may be involved in the carcinogenesis of lung cancer. EPCR may be a useful biomarker for evaluating the clinical status of lung cancer 23739325_identification of endothelial protein C receptor (EPCR), which mediates the cytoprotective effects of activated protein C, as the endothelial receptor for Plasmodium falciparum 23741007_Loss of EPCR and Thrombomodulin at sites of malaria-infected erythrocytes cytoadherence is detectible in nonfatal cerebral malaria. 23875041_Increased EPCR-levels correlate with accelerated mortality in patients with melioidosis. 23877403_sEPCR acts on the innate immune response by decreasing effector cells such as natural killer and T helper cells (TH2, TH17 and TH21). 23881209_3 polymorphisms in the EPCR gene were genotyped in 389 critically ill Greek patients to assess genetic risk for developing severe sepsis or septic shock. H2 carriers had an excess of ss/ss vs. genotypes H1 and H3. 23993723_The study suggests a putative role of EPCR SNPs in the development of thrombosis in multiple myeloma patients. 24024878_EPCR expression in breast cancer cells, despite having initial growth advantage, may have a role in limiting cancer progression at an advanced stage 24051141_Our data suggest that mutations that impair PC-EPCR interactions may be associated with an increased risk of venous thromboembolism. 24127121_Piperlonguminine might have potential as an anti-sEPCR shedding reagent against PMA-mediated EPCR shedding. 24158116_The EPCR AA genotype was significantly more frequent in the healthy volunteers free of venous thrombosis. 24308805_results did not conclusively identify a direct role of sEPCR in HSP, but our findings warrant further investigations, especially in severe HSP cases characterized by gastrointestinal bleeding or renal involvement 24436369_PROCR H1 protects against venous thromboembolism. There is an increased risk of VTE associated with the H3 haplotype. 24495480_results demonstrate that EPCR is overexpressed and mediates the aggressive behavior of rheumatoid synovial fibroblasts, and is likely driven by group V secretory phospholipase A2 24627030_The degree of arteriovenous fistula stenosis was not correlated with serum EPCR 24635948_The association of PROCR rs867186 with severe malaria is examined in Thai population and showed significant association with protection from severe malaria. 24862151_Blocking antibodies to EPCR attenuate in vivo tumor growth and proliferation specifically of EPCR(+) cells on defined integrin matrices in vitro. 25220546_Upon ICU admission, sEPCR levels in initially non-septic critically-ill patients appear elevated in the subjects who will subsequently become septic 25246042_Low PROCR mRNA expression levels associated with a poor prognosis in patients with disseminated intravascular coagulation (DIC) represents an exhaustion of the natural anticoagulant system, and reflects the final decompensate stage of DIC. 25407022_Increased coagulation activity and genetic polymorphisms in the F5, F10 and EPCR genes are associated with breast cancer. 25425698_It was concluded that measuring EPCR levels at admission could provide an early biological marker of the outcome of cerebral malaria. 25541704_genetic variation in the PROCR gene in our study population does not influence susceptibility to major severe malaria phenotypes 25588409_Serum soluble EPCR is elevated in Alzheimer's disease. The degree of cognitive impairment is significantly correlated with serum sEPCR levels in Alzheimer's disease and mild cognitive impairment patients and healthy controls. 25667200_Report activation of protein C and down-regulation of EPCR in trophobolasts stimulated with TNF-alpha. 25760048_data suggest that either the lack of the protective EPCR 4678C allele or the presence of EPCR 4600G allele may be associated with earlier development of thrombosis. 25895599_This effect of EPCR may be dependent on PAR1. 26015475_Individuals exposed to high levels of Plasmodium falciparum PfEMP1 acquire antibodies to EPCR-binding CIDR domains early in life. 26118955_These data reveal a previously unknown functional heterogeneity in the interaction between Plasmodium falciparum PfEMP1 CIDRalpha1domain binding to EPCR and have major implications for understanding the distinct clinical pathologies of cerebral malaria. 26119044_Overall, these findings reveal a much greater complexity of how Plasmodium falciparum PfEMP1 CIDRalpha1 domain-expressing parasites may modulate malaria pathogenesis through EPCR adhesion. 26340463_These data suggest that 6936A/G polymorphism is a risk factor for deep venous thrombosis and is associated with elevated plasma levels of soluble EPCR, while 4678G/C polymorphism plays a role in protection against deep venous thrombosis. 26620701_In this Tanzanian population, neither PROCR haplotype nor level of soluble EPCR was associated with severe malaria. 26990852_analysis of renal endothelial PROCR expression and shedding during diabetic nephropathy 27207424_Observations that binding of P. falciparum erythrocyte membrane protein 1 to EPCR results in an acquired functional protein C system deficiency support the new paradigm that EPCR plays a central role in the pathogenesis of severe malaria. [review] 27255786_The EPCR rs867186-GG genotype is associated with increased soluble protein, and it could mediate protection against severe malaria. 27406562_This study shows that ICAM-1 is a coreceptor for a subset of EPCR-binding Plasmodium falciparum parasites and provides the first evidence of how EPCR and ICAM-1 interact to mediate parasite binding to both resting and TNF-alpha-activated primary brain and lung endothelial cells. 27561318_EPCR occupancy recruits G-protein coupled receptor kinase 5, thereby inducing beta-arrestin-2 biased PAR1 signaling by both APC and thrombin. In 27784899_Plasma levels of Ang2 were associated with markers of malaria severity and levels of var transcripts encoding P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1) containing Cysteine Rich Inter Domain Region alpha1 (CIDRalpha1) domains predicted to bind Endothelial Protein C receptor (EPCR). 28007459_Elevated endothelium-related mediators (vWF, E-selectin and EPCR) appear to participate in the development of pancreatic necrosis and may be a potential indicator of overall prognosis. 28103946_Analysis of a cohort of breast cancer patients revealed an association of high EPCR levels with adverse clinical outcome. Interestingly, EPCR knockdown did not affect cell growth kinetics in 2D but reduced cell growth in 3D cultures. EPCR silencing reduced primary tumor growth and secondary outgrowths at metastatic sites. EPCR via SPOCK1 confers a cell growth advantage in 3D promoting breast tumorigenesis and metastasis. 28219843_This study demonstrates that impaired EPCR function can be detected by thrombin generation and clot lysis assays on cells expressing thrombomodulin and EPCR. Deficiency in EPCR has procoagulant effects that lead to a delay in clot lysis. 28367652_The results showed that AS-IV could significantly inhibit PMA-induced EPCR shedding. 28415941_EPCR polymorphisms may be associated with increased risk of sepsis, but this has no effect on the release of soluble EPCR in patients with sepsis. 28480559_The data indicate that EPCR can regulate p63, is associated with highly proliferative keratinocytes, and is a potential human epidermal stem cell marker. 28538400_CORM-2 protects human umbilical vein endothelial cells from lipopolysaccharide-induced injury, by way of suppressing NF-kappaB activity, which downregulates TM and EPCR mRNAs. It also decreases MMP-2 expression and prevents the shedding of TM and EPCR from the surface of endothelial cells, so as to preserve their protective effect. 28756987_Our findings suggested that breast cancer patients with expression of PROCR is more prone to suffer from distant metastasis and bad clinical outcomes. 29217770_this is the first comprehensive study of PROCR signaling in breast cancer cells, and its findings also shed light on the molecular mechanisms of PROCR in stem cells in normal tissue. 29339517_analysis of DC8 and DC13 PfEMP1 variants reveals that Plasmodium falciparum-infected erythrocytes-endothelial protein C receptor interaction may be prevented by plasma components under physiological conditions 29461258_Recent studies provide a mechanistic basis to how EPCR contributes to PAR1-mediated biased signaling. EPCR may play a role in influencing a wide array of biological functions by binding to diverse ligands. 29484413_The activation of PAR-1 on the cell surface of SGC7901 and AGS cells was significantly reduced after the knockdown of EPCR. By contrast, blockade of PAR-1 reduced the proliferation and migration of gastric cells in vitro 29630665_Low EPCR expression is associated with meningococcal purpura fulminans. 29879294_As EPCR protein expression has not previously been shown in platelets, the presence of EPCR was confirmed in platelets using immunofluorescence, flow cytometry, immunoprecipitation, and mass spectrometry. This is the first demonstration that human platelets express EPCR. 30048851_PROCR H1 and H3 haplotypes were determined by genotyping 7014G/C and 1651C/G tag-polymorphisms, respectively. The PROCR H1 haplotype was less frequently found in antiphospholipid syndrome patients with arterial thrombosis, suggesting a protective effect of PROCR H1 against arterial thrombosis in these patients. 30281048_biophysical analysis of a possible mode of ligand recognition by the EPCR via the involvement of phosphatidylcholine within its hydrophobic groove 30365003_The acquisition of antibodies against EPCR-binding cysteine-rich interdomain region alpha1 domains of PfEMP1 after a severe malaria episode suggest that EPCR-binding PfEMP1 may have a role in the pathogenesis of severe malaria in Papua New Guinea. 30383853_PROCR, but not THBD, polymorphisms are associated with early-onset ischemic stroke in young Caucasians. There were no PROCR or THBD associations in African-Americans. 30384036_Plasma soluble EPCR was elevated in alpha-thalassemia intermedia. 31138740_EPCR and ICAM-1 are important in Plasmodium falciparum binding to a 3D human brain microvessel model 31645483_single nucleotide polymorphisms of endothelial protein C receptor are associated with Kawasaki disease susceptibility in a Chinese Han children 32003123_Down-regulation of endothelial protein C receptor promotes preeclampsia by affecting actin polymerization. 32007797_Elevated levels of soluble Endothelial protein C receptor in rheumatoid arthritis and block the therapeutic effect of protein C in collagen-induced arthritis. 32496885_Determining the association of thrombophilic gene polymorphisms with recurrent pregnancy loss in Iranian women. 34281556_Lentiviral CRISPR-guided RNA library screening identified Adam17 as an upstream negative regulator of Procr in mammary epithelium. 34989590_Decoding the Host-Parasite Protein Interactions Involved in Cerebral Malaria Through Glares of Molecular Dynamics Simulations. 35169126_Protein C receptor maintains cancer stem cell properties via activating lipid synthesis in nasopharyngeal carcinoma. 35264566_Elucidating mechanisms of genetic cross-disease associations at the PROCR vascular disease locus. | ENSMUSG00000027611 | Procr | 204.86930 | 1.0730928 | 0.1017748688 | 0.21621239 | 2.196787e-01 | 6.392848e-01 | 9.998360e-01 | No | Yes | 209.57568 | 21.228891 | 2.020179e+02 | 16.940988 | |
| ENSG00000101104 | 80336 | PABPC1L | protein_coding | Q4VXU2 | Mouse_homologues FUNCTION: Binds the poly(A) tail of mRNA. {ECO:0000256|RuleBase:RU362004}. | Alternative splicing;RNA-binding;Reference proteome;Repeat | This gene belongs to the polyadenylate-binding protein type-1 family of proteins. Members of this family bind to the polyA tails of mRNAs to regulate mRNA stability and translation. The mouse ortholog of this gene is required for female fertility. In human, expression of a functional protein is regulated by alternative splicing. The protein-coding splice variant for this gene is abundantly expressed in human oocytes, while a noncoding splice variant subject to nonsense-mediated decay is the predominant splice variant expressed in somatic tissues. [provided by RefSeq, Aug 2019]. | Mouse_homologues mmu:381404; | cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA 3'-UTR binding [GO:0003730]; poly(A) binding [GO:0008143]; poly(U) RNA binding [GO:0008266]; RNA binding [GO:0003723] | 18716053_Human EPAB mRNA is detected in ovaries, testes and several somatic tissues including pancreas, liver and thymus, and EPAB is the predominant poly(A) binding protein in immature (germinal vesicle) and mature (metaphase II) oocytes. 26843391_EPAB decreased expression in in infertile men with non-obstructive azoospermia | ENSMUSG00000054582 | Pabpc1l | 436.00368 | 0.9052150 | -0.1436676223 | 0.16597088 | 7.291783e-01 | 3.931497e-01 | 9.998360e-01 | No | Yes | 405.99779 | 76.753756 | 3.956954e+02 | 57.882909 | |
| ENSG00000101266 | 1457 | CSNK2A1 | protein_coding | P68400 | FUNCTION: Catalytic subunit of a constitutively active serine/threonine-protein kinase complex that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine (PubMed:11239457, PubMed:11704824, PubMed:16193064, PubMed:19188443, PubMed:20625391, PubMed:22406621, PubMed:24962073). Regulates numerous cellular processes, such as cell cycle progression, apoptosis and transcription, as well as viral infection (PubMed:12631575, PubMed:19387552, PubMed:19387551). May act as a regulatory node which integrates and coordinates numerous signals leading to an appropriate cellular response (PubMed:12631575, PubMed:19387552, PubMed:19387551). During mitosis, functions as a component of the p53/TP53-dependent spindle assembly checkpoint (SAC) that maintains cyclin-B-CDK1 activity and G2 arrest in response to spindle damage (PubMed:11704824, PubMed:19188443). Also required for p53/TP53-mediated apoptosis, phosphorylating 'Ser-392' of p53/TP53 following UV irradiation. Can also negatively regulate apoptosis (PubMed:11239457). Phosphorylates the caspases CASP9 and CASP2 and the apoptotic regulator NOL3 (PubMed:16193064). Phosphorylation protects CASP9 from cleavage and activation by CASP8, and inhibits the dimerization of CASP2 and activation of CASP8 (PubMed:16193064). Regulates transcription by direct phosphorylation of RNA polymerases I, II, III and IV. Also phosphorylates and regulates numerous transcription factors including NF-kappa-B, STAT1, CREB1, IRF1, IRF2, ATF1, ATF4, SRF, MAX, JUN, FOS, MYC and MYB (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551, PubMed:23123191). Phosphorylates Hsp90 and its co-chaperones FKBP4 and CDC37, which is essential for chaperone function (PubMed:19387550). Mediates sequential phosphorylation of FNIP1, promoting its gradual interaction with Hsp90, leading to activate both kinase and non-kinase client proteins of Hsp90 (PubMed:30699359). Regulates Wnt signaling by phosphorylating CTNNB1 and the transcription factor LEF1 (PubMed:19387549). Acts as an ectokinase that phosphorylates several extracellular proteins (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551). During viral infection, phosphorylates various proteins involved in the viral life cycles of EBV, HSV, HBV, HCV, HIV, CMV and HPV (PubMed:19387550, PubMed:12631575, PubMed:19387552, PubMed:19387551). Phosphorylates PML at 'Ser-565' and primes it for ubiquitin-mediated degradation (PubMed:20625391, PubMed:22406621). Plays an important role in the circadian clock function by phosphorylating ARNTL/BMAL1 at 'Ser-90' which is pivotal for its interaction with CLOCK and which controls CLOCK nuclear entry (By similarity). Phosphorylates CCAR2 at 'Thr-454' in gastric carcinoma tissue (PubMed:24962073). {ECO:0000250|UniProtKB:P19139, ECO:0000269|PubMed:11239457, ECO:0000269|PubMed:11704824, ECO:0000269|PubMed:16193064, ECO:0000269|PubMed:19188443, ECO:0000269|PubMed:20625391, ECO:0000269|PubMed:22406621, ECO:0000269|PubMed:23123191, ECO:0000269|PubMed:24962073, ECO:0000269|PubMed:30699359, ECO:0000303|PubMed:12631575, ECO:0000303|PubMed:19387549, ECO:0000303|PubMed:19387550, ECO:0000303|PubMed:19387551, ECO:0000303|PubMed:19387552}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Biological rhythms;Cell cycle;Disease variant;Kinase;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase;Wnt signaling pathway | Casein kinase II is a serine/threonine protein kinase that phosphorylates acidic proteins such as casein. It is involved in various cellular processes, including cell cycle control, apoptosis, and circadian rhythm. The kinase exists as a tetramer and is composed of an alpha, an alpha-prime, and two beta subunits. The alpha subunits contain the catalytic activity while the beta subunits undergo autophosphorylation. The protein encoded by this gene represents the alpha subunit. Multiple transcript variants encoding different protein isoforms have been found for this gene. [provided by RefSeq, Apr 2018]. | hsa:1457; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; NuRD complex [GO:0016581]; plasma membrane [GO:0005886]; protein kinase CK2 complex [GO:0005956]; Sin3 complex [GO:0016580]; ATP binding [GO:0005524]; Hsp90 protein binding [GO:0051879]; identical protein binding [GO:0042802]; kinase activity [GO:0016301]; protein N-terminus binding [GO:0047485]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; apoptotic process [GO:0006915]; cell cycle [GO:0007049]; chaperone-mediated protein folding [GO:0061077]; negative regulation of apoptotic signaling pathway [GO:2001234]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of cell growth [GO:0030307]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of protein catabolic process [GO:0045732]; positive regulation of Wnt signaling pathway [GO:0030177]; protein phosphorylation [GO:0006468]; regulation of cell cycle [GO:0051726]; regulation of chromosome separation [GO:1905818]; rhythmic process [GO:0048511]; signal transduction [GO:0007165]; Wnt signaling pathway [GO:0016055] | 11756447_uPA-dependent VSMC adhesion is a function of selective Vn phosphorylation by the ectoprotein kinase CK2 11827164_Generation of mutants of CK2alpha which are dependent on the beta-subunit for catalytic activity 11827166_HIV-1 Rev transactivator: a beta-subunit directed substrate and effector of protein kinase CK2 11827167_Protein kinase CK2: signaling and tumorigenesis in the mammary gland. 11827168_Response of cancer cells to molecular interruption of the CK2 signal. 11827170_Functional specialization of CK2 isoforms and characterization of isoform-specific binding partners 11827171_Characterization of CK2 holoenzyme variants with regard to crystallization 11827174_Transcriptional coordination of the genes encoding catalytic (CK2alpha) and regulatory (CK2beta) subunits of human protein kinase CK2. 11827176_Consequences of CK2 signaling to the nuclear matrix. 11827177_Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus 11940573_Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. 11956194_Unique activation mechanism of protein kinase CK2. The N-terminal segment is essential for constitutive activity of the catalytic subunit but not of the holoenzyme 12037680_Protein kinase CK2 dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TRCp and enhances beta-catenin degradation [UBC3B] 12145206_FGF-1 binds to both the catalytic alpha-subunit and to the regulatory beta-subunit of CK2. FGF-1 & CK2 alpha are shown to interact in vivo. A correlation between the mitogenic potential of FGF-1 mutants & their ability to bind to CK2 alpha was observed. 12568341_Data point to a particular role of the catalytic alpha and alpha' subunits of protein kinase CK2, which may be different from their roles in the holoenzyme. 12628923_This protein is associated with the COP9 signalosome. 12659875_HSF1 activation by heat is correlated with the thermal activation of nuclear CK2 and overexpression of CK2 activates HSF1 12748192_results indicate that protein kinase CKII may control IkappaBalpha and p27Kip1 degradation and thereby G1/S phase transition through the phosphorylation of threonine 10 within CKBBP1 protein 12860116_Crystal structure of a C-terminal deletion mutant of human protein kinase CK2 catalytic subunit 14962846_Evaluation of different pathways involved in death signaling suggest that the regulation of a critical proapoptotic step in HuH-7 cells by CK2alpha' is mediated by a JNK signaling cascade. 15108354_structure activity relationship: the PA 382-384 mutant exhibits an increased thermal and proteolytic stability 15218032_might play an important role in vivo in regulating the function and transport activity of ABCA1 and possibly of other members of the ABCA subfamily 15287743_Utilizing a kinase-driven assay biochemical purification, the authors identified casein kinase II (CKII) from HeLa cell nuclear extract as a cellular phosphoprotein pp32 kinase. 15342635_CK2 acts as an inhibitor of Cdk5 in the brain 15557471_Involvement of ubiquitous protein kinase CK2 in angiogenesis. Naturally derived CK2 inhibitors may be useful for treatment of proliferative retinopathies. 15659405_CK2 regulates the DNA-binding ability of SSRP1 and that this regulation may be responsive to specific cell stresses. 15740749_Data demonstrate that CK2alpha possesses sophisticated structural adaptations in favour of dual-cosubstrate specificity. 15818404_constitutive phosphorylation by CK2 may be required for maximal activation of Akt/PKB 15955816_CK2 may have the capacity to differentially regulate U1 and U6 transcription even though SNAP(C) is universally utilized for human snRNA gene transcription 16107342_multiple kinases, including CK2 and GSK3beta, participate in PTEN phosphorylation and GSK3beta may provide feedback regulation of PTEN 16133877_Protein kinase CK2 is characterized by an extremely high stability that might be due to its association with other intracellular proteins, enhanced half-life or lower vulnerability towards proteolytic degradation. 16157582_protein kinase CK2 is inactive in CCVs because of the fact that it is bound to the clathrin-coated vesicle (CCV) membrane via an interaction between phosphatidylinositol 4,5-bisphosphate in the CCV membrane and the active site in CK2 16225457_In vitro phosphorylation of eIF2beta also pointed to Ser2 as a preferred site for CK2 phosphorylation 16227438_CK2 may be involved in the regulation of cell cycle progression by associating with and phosphorylating a key molecule for translation initiation. 16308564_casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia 16342410_CK2 is potentially a highly plausible target for cancer therapy. 16426576_phosphorylation of the (T300) residue is dependent on CK2 and is a necessary and functional prerequisite for TSPY's transport into the nucleus 16540521_Dynamics of nucleolar reformation is ATP/GTP-dependent, sensitive to temperature, and creatin kinase-2 driven. 16581776_CK2 regulates NKX3-1 in prostate cells. 16651637_Based on its retinal localization, CK2 may be considered a new immunohistochemical astrocytic marker, and combination of CK2 inhibitors and octreotide may be a promising future treatment for proliferative retinopathies. 16751801_role of CK2beta for cell survival and might allow the design of novel proapoptotic agents targeting this protein 16806200_CK2 activity in Chinese Hamster Ovary (CHO) cells is entirely accounted for by the holoenzyme. 16837460_v-Src-dependent down-regulation of the Ste20-like kinase SLK through casein kinase II. 16874460_These studies suggested that CK2 might regulate SAG-SCF E3 ligase activity through modulating SAG's stability, rather than its enzymatic activity directly. 16880508_CK2 has the potential to regulate Pol I transcription at multiple levels, in preinitiation complex (PIC) formation, activation, and reinitiation of transcription. 17009010_CK2 plays an active role in NF-kappaB signaling in intestinal epithelial cell lines and may represent a possible target for intervention. 17106255_CK2-site phosphorylation of p53 is induced in DeltaNp63 expressing basal stem cells in UVB irradiated human skin 17113388_CK2 is required for the assembly and cycling of Wnt-enhancer complexes in vivo. 17524418_Results describe the 1.6 A resolution crystal structure of a fully active C-terminal deletion mutant of human CK2alpha liganded by two sulfate ions, and compare this structure systematically with representative structures of related CMGC kinases. 17545155_extracellular phosphorylation of collagen XVII by ecto-CK2 inhibits its shedding by TACE and represents novel mechanism to regulate adhesion and motility of epithelial cells 17629615_Suggest a novel role of CK2 in breast cancer resistance to antiestrogens. 17636126_cytotoxic adaptor function of NS1 was confirmed with fusion peptides, where the tropomyosin-binding domain of NS1 and CKIIalpha are physically linked 17670747_CK2 phosphorylation triggers an allosteric inhibition of the SNAP190 Myb DNA binding domain 17699575_EB2-mediated production of infectious EBV virions is regulated by CK2 phosphorylation at one or more of the serine residues Ser-55, -56, and -57 17894550_ABC50 N-terminal region interacts with eukaryotic initiation factor eIF2 and is a target for regulatory phosphorylation by CK2 17923166_The number of predicted CK2 sites correlated with the degree of in vitro and in vivo phosphorylation of NS5A by CK2. CK2-dependent phosphorylation of NS5A is heterogeneous among different HCV genotypes and clinical isolates. 17935135_CK2 may be a key mediator for HDAC1 and HDAC2 activation under hypoxia in tumor cells 18026141_splicing activity is significantly influenced by the CK2-hPrp3p interaction 18062282_There was a positive correlation between the expressions of CK2alpha and CK2beta in laryngeal squamous cell carcinoma. 18089732_Human herpesvirus 6Binduces p53 Ser392 phosphorylation by an atypical pathway independent of CK2 protein and p38 kinases. 18242640_The complex structure between a C-terminal deletion mutant of human CK2alpha and the ATP-competitive inhibitor emodin is studies in comparison with with a previously published complex structure of emodin and maize CK2alpha. 18347021_CK2-mediated phosphorylation of TRF1 plays an important role in modulating telomere length homeostasis by determining the levels of TRF1 at telomeres 18363813_Protease activated receptor 1 activation of platelet is associated with an increase in protein kinase CK2 activity 18548200_CK2 is tightly associated with TAFII250 and is the principal activity responsible for TAFII250-mediated phosphorylation of Mdm2. 18553055_Studies in yeast and in human cells demonstrate that the different forms of CK2 interact with a large number of cellular proteins. 18553059_Transcript amounts of the subunits CK2alpha and CK2beta and holoenzyme CK2 activity in 34 muscle biopsies of human patients with different muscle pathologies, were determined. 18560764_CK2 involvement in the phenomenon of the drug resistance was investigated. 18563533_Casein kinase II activity is increased by the binding of haptoglobin 1-1-hemoglobin to CD163. 18566753_CK2 activity was investigated both in vitro and in cultured cells by using anti-[pSer13]-Cdc37 antibody. 18566754_These data identify a key post-translational mechanism that controls PML protein levels in cancer cells and suggest that CK2 inhibitors may be beneficial anti-cancer drugs. 18574673_These results suggest that one of the modes of CK2-mediated modulation of apoptotic activity is via its impact on cellular inhibitors of apoptosis proteins. 18607692_A crystal of a C-terminally truncated variant of human CK2alpha to which two molecules of the inhibitor 5,6-dichloro-1-beta-D-ribo-furanosyl-benzimidazole (DRB) are bound, is analyzsed. 18614797_CK2 binds to the NHE3 C terminus and stimulates basal NHE3 activity by phosphorylating a separate single site on the NHE3 C terminus 18649047_CK2 may be involved in the regulation of cell cycle progression through the phosphorylation of a key molecule for translation initiation and of nuclear substrates upon activation of CK2 by itself. 18662771_CK2 catalyzes the phosphorylation of tyrosine residues in mammalian cells. 18682247_KIF5C, a member of the kinesin 1 heavy chain family, is a substrate for protein kinase CK2. 18710613_Mitoxantrone is a strong inhibitor of recombinant human protein kinase CK2 in vitro. Apoptosis induced by mitoxantrone in HL-60 cells has no correlation to intracellular CK2 activity. 18790693_InsP(6) specifically binds to CK2alpha and disrupts the interaction between CK2alpha and Nopp140 with an IC(50) value of 25 microM, thereby attenuating the Nopp140-mediated repression of CK2 activity. 18824508_study presents unbound three-dimensional structure of a CK2beta construct that is fully capable of CK2alpha recruitment and quantify its affinity to CK2alpha thermodynamically 19027835_DNA methylation-dependent down-regulation of transcription factors Sp1, Ets1 and NF-kappaB might be involved in silencing of the CKII alpha and CKII alpha' genes during cellular senescence. 19027835_DNA methylation-dependent down-regulation of transcription factors Sp1, Ets1 and NF-kappaB might be involved in silencing of the CKII alpha and CKII alpha' genes during cellular senescence.[CKII alpha'] 19035320_PrP protein can bind to protein kinase CK2 both in native and recombinant forms in vitro. 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 19282287_Ikaros is controlled by the CK2 and PP1 pathways and that a balance between these two signal transduction pathways is essential for normal cellular function and for the prevention of malignant transformation. 19361447_CK2beta dimer stabilizes the glycine-rich loop in the extended active conformation known from the majority of CK2alpha structures. 19398558_protein kinase CK2 phosphorylates Ser(361) on Sgt1, and this phosphorylation inhibits Sgt1 dimerization 19486891_Extracellular protein kinase CK2 binds to the extracellular domain of NRP1 which is also phosphorylated by CK2 both in vitro and in vivo. 19542537_CK2-dependent phosphorylation controls the nuclear localization, aggregation and stability of ataxin-3. 19556345_These studies reveal multisite phosphorylation of IGFBP-3 that both positively and negatively regulate its apoptotic potential. 19596613_The constitutive phosphorylation of XRCC1 in the chromatin and its DNA damage-induced recruitment to the nuclear matrix are critical for foci formation. 19616514_PDCD5 is phosphorylated in vitro by both CK2alpha subunit and by the CK2. 19723109_Data show that decreasing CK2 activity increases intracellular hydrogen peroxide, creating an intracellular environment conducive for death execution. 19726289_Protein kinase CK2 plays an important role in the radiosensitivity of the nasopharyngeal carcinoma cells. 19839995_Observational study of gene-disease association. (HuGE Navigator) 19855935_This study shows that the activation of the p53-p21(Cip1/WAF1) pathway acts as a major mediator of cellular senescence induced by CKII inhibition. 19904978_CK2 is the kinase responsible for phosphorylating Pax3 and oncogenic fusion protein Pax3-FOXO1 at serine205 in proliferating mouse primary myoblasts and in a variety of translocation-containing alveolar rhabdomyosarcoma cell lines. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19923321_These results highlight serines 11 and 92 as new players in Snail1 regulation and suggest the participation of CK2 and PKA in the modulation of Snail1 functionality. 19933278_MSK1 and MSK2 are differentially regulated by CK2 during the UV response and that MSK2 is the major protein kinase responsible for the UV-induced phosphorylation of p65 at Ser(276) that positively regulates NF-kappaB activity in MDA-MB-231 cells 19941816_The EGFR-ERK-CK2-mediated phosphorylation of alpha-catenin promotes beta-catenin transactivation and tumor cell invasion. 20021963_Protein kinase CK2alpha plays an important role in the proliferation and invasion of nasopharyngeal carcinoma. 20026644_CK-II-dependent PPARgamma phosphorylation at Ser16 and Ser21 is necessary for CRM1/Ran/RanBP3-mediated nucleocytoplasmic translocation of PPARgamma. 20157113_CK2-mediated pathways reversibly regulate purinosome assembly 20356928_The interface between apoptosis initiation and execution by determining caspase-8 activation, Bid cleavage and mitochondrial engagement (onset of mitochondrial depolarisation) in individual HeLa, was analysed. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20526659_These data show that Tax-1 is phosphorylated in vitro by the pleiotropic human serine/threonine kinase CK2 at three residues, Ser-336, Ser-344 and Thr-351, close to and within its C-terminal PDZ-binding motif. 20545769_CK2 plays a crucial role in the ATR-dependent checkpoint pathway through its ability to phosphorylate Ser-341 and Ser-387 of the Rad9 subunit of the 9-1-1 complex. 20610541_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20625391_Data show that the CSNK2A1P gene is a functional proto-oncogene in human cancers and its functional polymorphism appears to degrade PML differentially in cancer cells. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20664522_Herein, the authors have identified polo-like kinase 1 (Plk1) and casein kinase 2 (CK2) as two kinases of CLIP-170 and mapped S195 and S1318 as their respective phosphorylation sites. 20671611_The prognostic significance of the genes casein kinase 2 alpha subunit (CSNK2A1), anti-apoptosis clone-11 (AAC-11), and tumor metastasis suppressor NME1 in completely resected non-small cell lung cancer (NSCLC) patients, was analysed. 20711232_These results indicate that CK2 has an important role in the modulation of DNA-PKcs activity and its phosphorylation status providing important insights into the mechanisms by which DNA-PKcs is regulated in vivo. 20718998_Reduced Casein Kinase II enhances chemosensitivity to gemcitabine in pancreatic cancer. 20719947_an interaction between EBNA1 and the host CK2 kinase is crucial for EBNA1 to disrupt PML bodies and degrade PML proteins. 20807566_Protein kinase CK2 inhibition results in activation of the receptor mediated apoptosis pathway via the ER stress response highlight the importance of CK2 inhibitors. 20864032_The CK2 phospho-dependent interaction between TEL2 and the R2TP complex affects phosphatidylinositol 3-kinase-related kinase functions and is essential for mTOR and SMG1 stability in vivo. 21093442_Data presented the crystal structures of CK2alpha in complex with CX-4945 and adenylyl phosphoramidate at 2.7 and 1.3 A, respectively. 21162802_Protein kinase CK2alpha plays an important role in the apoptosis of human laryngeal carcinoma cells possibly by decreasing bcl-2/Bax. 21165564_Sox4 stimulates ss-catenin activity through induction of CK2. 21182307_CK2 is a central regulator of topo I hyperphosphorylation. 21270425_The effect of a mutation in the follicle-stimulating hormone receptor in a protein kinase-CK2 consensus site that alters degradation of follicle-stimulating hormone (FSH) is studied. 21359197_Overexpression of nuclear CK2alpha can be a useful marker for predicting the outcome of patients with colorectal cancer. 21486957_Findings identify CK2 as a crucial protein involved in the formation and clearance of aggresomes, and hence in cell viability in response to misfolded protein stress. 21518957_Data suggest that phospho-Ser81 progesterone receptors (PR) provides a platform for casein kinase 2 recruitment and regulation of selected PR-B target genes. 21527517_we report that the kinase CK2 is a novel interaction partner of JAKs and is essential for JAK-STAT activation 21559372_Twist expression can be regulated at the post-translational level through phosphorylation by CK2, which increases Twist stability in response to IL-6 stimulation 21559479_CK2 down regulation facilitates TNF-alpha-mediated chondrocyte death through apoptosis and autophagy 21576649_A novel mechanism in the development of cardiac hypertrophy is demonstrated when transgenic CK2alpha1 activates histone deacetylase 2 (HDAC2) via phosphorylating HDAC2 serine394. 21697493_Human papillomavirus type 38 E7 protein promotes keratinocyte proliferation by negatively regulating actin cytoskeleton fiber formation through the CK2-MEK-ERK-Rho pathway and by inhibiting eEF1A and its effects on actin cytoskeleton remodeling. 21702981_CK2alpha plays an essential role in the development of colorectal cancer. 21711111_CK2 inhibitors enhance the radiosensitivity of human non-small cell lung cancer cells through inhibition of stat3 activation 21735091_The Arabidopsis co-chaperone protein p23 is a CK2 target. 21735095_Persistence of high CK2 level in R (resistant)-CEM, as opposed to S (sensitive)-CEM, is accompanied by the presence of an immunospecific form of Cdc37. 21739153_In hsCK2alpha' an open conformation of the interdomain hinge/helix alphaD region that is critical for ATP-binding is found corresponding to an incomplete catalytic spine. In contrast hsCK2alpha often adopts the canonical, PKA-like version of the catalytic spine. 21739154_The nuclear localization signal sequence-dependent Grp94 kinase in the cell lysate is identical with CK2. 21750986_CK2 appears to play an important role in the biology of B-cell chronic lymphocytic leukemia. 21750987_Inhibition of CK2 activity by three different inhibitors led to a down-regulation of the level of cdc25C. 21750988_An interaction of the adiponectin receptor 1 with the tetrameric complex and protein kinase CK2 as a key player in adiponectin signaling, is reported. 21777522_These data suggest a novel cellular function of CK-2 as a transcriptional co-activator of AP-2alpha. 21871133_The primary mechanisms by which apigenin kill multiple myeloma cells is by targeting the trinity of CK2-Cdc37-Hsp90. 21936567_our results demonstrate that EEF1D is a bona fide physiological CK2 substrate for CK2 phosphorylation 21968188_CKII downregulation induces p53 stabilization by negatively regulating SIRT1 deacetylase activity during senescence 22027148_the pleotropic Serine/Threonine kinase CK2 is implicated in the regulation of IL-6 expression in a model of inflammatory breast cancer. 22184066_identify serine 118 in the transactivation domain of YY1 as the site of CK2alpha phosphorylation, proximal to a caspase 7 cleavage site 22186626_The unique flexibility of the CK2alpha catalytic subunit in the hinge region, the p-loop and the beta4 beta5 loop, show that there is no clear-cut correlation between the conformations of these flexible zones. 22190719_The human adenovirus type 5 E1B protein is phosphorylated by protein kinase CK2. 22209233_CK2 regulates vaccinia virus dissemination and actin tail formation. 22267120_The the CK2 catalytic subunit CK2alpha is modified by O-linked beta-N-acetyl-glucosamine (O-GlcNAc) on Ser347, proximal to a cyclin-dependent kinase phosphorylation site (Thr344). 22317921_The connection with proliferation and migration, as well as the activation of ZIP7 by CK2, a kinase that is antiapoptotic and promotes cell division 22351691_Results suggest CK2 as a novel regulator of the endoplasmic reticulum (ER) stress/unfolded protein response (UPR) cascades and HSP90 function in myeloma cells and offer the groundwork to design novel combination treatments for this disease. 22404984_Protein kinase CK2 co-localizes with gamma-H2AX to sites of genetic lesions and modulation of its expression levels affects the cellular DNA damage response. 22506723_A new crystal structure of the CK2 holoenzyme was determined that plays an essential role in the formation of inactive polymeric assemblies. 22562247_Unbalanced expression of CK2 subunits may drive epithelial-to-mesenchymal transition, thereby contributing to tumour progression. 22609407_ATF4 is involved in ER stress signalling through the AARE, which further supports our finding that CK2 inhibition provokes an amino acid induced response pathway. 22675025_CK2alpha-NCoR cascade selectively represses the transcription of IP-10 and promotes oncogenic signaling in human esophageal cancer cells. 22718630_Phosphorylation of S284 by protein kinase CK2 significantly decreases nuclear targeting of IPMK in HEK-293 cells. 22767590_that high intracellular PKCK2 activity confers anoikis resistance on esophageal cancer cells by inducing E- to N-cadherin switching. 22768056_The inhibition of CK2alpha down-regulates Hh/Gli signaling and subsequently reduces stem-like side population in human lung cancer cells. 22810236_Recovery of RNA polymerase III transcription from the glycerol-repressed state: revisiting the role of protein kinase CK2 in Maf1 phosphoregulation. 22814265_CK2 and human malignant tumor 22868753_analysis of crystal packing of human protein kinase CK2 catalytic subunit in complex with resorufin or other ligands 22904299_A CD5-CK2 signaling pathway represents a major signaling cascade initiated by CD5 that regulates the threshold of T cell activation and T helper (Th) cell differentiation. 23007634_our findings identify CK2 as a novel component of the autophagic machinery and underline the potential of its downregulation to kill glioblastoma cells by overcoming the resistance to multiple anticancer agents. 23017496_Functional proteomics strategy for validation of protein kinase inhibitors reveals new targets for a TBB-derived inhibitor of protein kinase CK2. 23137536_the present results show that miR-186, miR-216b, miR-337-3p, and miR-760 cooperatively promote cellular senescence through the p53-p21(Cip1/WAF1) pathway by CKII downregulation-mediated ROS production in HCT116 cells. 23153582_Casein kinase 2alpha inhibition decreases the total protein level of beta-catenin and cell growth. 23185622_CK2-mediated hyperphosphorylation of topoisomerase I targets serine 506, enhances topoisomerase I-DNA binding, and increases cellular camptothecin sensitivity. 23287549_Phosphorylation of Sec63 by CK2 enhanced its binding to Sec62 23317504_Brd4 association with p53 is modulated by casein kinase II (CK2)-mediated phosphorylation of a conserved acidic region in Brd4 23349870_These results suggest that Sec31 phosphorylation by CK2 controls the duration of COPII vesicle formation, which regulates ER-to-Golgi trafficking. 23360763_Data indicate that by altering the scaffold of CK2 inhibitors to obtain selective inhibitors of DYRK1A and PIM1. 23416530_Studies indicate systematic platforms for studying CK2 inhibitors. 23474121_crystal structure of CK2alpha with a 13-meric cyclic peptide derived from the C-terminal CK2beta segment 23492774_Activation of protein kinase CK2 attenuates FOXO3a functioning in a PML-dependent manner. 23523798_PI3K is involved in CKII inhibition-mediated cellular senescence in HCT116 cells. 23580615_Data indicate that protein S phosphorylation by casein kinase 1 (CK1) and casein kinase 2 (CK2) increased activated proten C cofactor activity. 23599180_CK2alpha' exhibits a striking preference for caspase-3 phosphorylation in cells as compared to CK2alpha and that CK2beta exhibits the capacity to abolish caspase-3 phosphorylation 23612983_Enzyme inhibition studies and mutational analyses demonstrated that protein kinase CK2-catalyzed phosphorylation of HDAC1 and -2 is crucial for the dissociation of these two enzymes. 23651443_inhibition of CK2alpha down-regulates Notch1 signalling and subsequently reduces a cancer stem-like cell population in human lung cancer cells 23732914_These results identify casein kinase 2 as a new target of PD-1 and reveal an unexpected mechanism by which PD-1 decreases PTEN protein expression while increasing PTEN activity, thereby inhibiting the PI3K/Akt signaling axis. 23816881_The study identifies casein kinase 2 as an NRF1-binding protein and finds that the knockdown of casein kinase 2 enhances the Nrf1-dependent expression of the proteasome subunit genes. 23885116_These findings identify CK2 as an upstream activating kinase of PAK1, providing a novel mechanism for PAK1 activation. 23979991_HspA1A also interacts with CK2 and enhances the kinase activities of CK2 during DNA repair. 24012109_knockdown of CK2alpha expression by siRNA restores the sensitivity of resistant LAMA84 cells to low imatinib concentrations. 24021586_upregulation of VEGF in primary RPE cells was blocked by a chemical inhibitor of protein kinase CK2 known to suppress induction of ATF4 and VEGF by OxPLs 24036851_The CSNK2A1 gene has gene dosage gains in glioblastoma, and is significantly associated with the classical glioblastoma subtype. 24098452_miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-alpha, CCNJ, and MEGF9. 24121351_crystal structure of CK2alpha with 21 loci of alternative conformations, including a niacin, 19 ethylene glycols and 346 waters, was determined at 1.06 A resolution to an Rwork of 14.0% 24175891_Asymmetric alpha2beta2 tetramers of casein kinase 2 are organized in trimeric rings that correspond to inactive forms of the enzyme. The new crystal structures presented here reveal the symmetric architecture of the isolated active tetramers. 24178769_The CFTR Ser511 can be phosphorylated by the combined action of tyrosine kinases and CK2 and disclose a new mechanism of hierarchical phosphorylation where the role of the priming kinase is that of removing negative determinants. 24218616_CK2 activity is inhibited by Nopp140 and reactivated by inositol hexakisphosphate by competitive binding at the substrate recognition site of CK2. 24268649_Phosphorylation of the CK2 site on SMRT significantly increased affinity for SHARP. 24297901_These data support a model in which activated Akt enhances rRNA synthesis both by preventing TIF-IA degradation and phosphorylating CK2alpha, which in turn phosphorylates TIF-IA. 24440309_Protein kinase CK2 alpha is involved in the phosphorylation of the ESCRT-III subunits CHMP3 and CHMP2B, as well as of VPS4B/SKD1, an ATPase that mediates ESCRT-III disassembly. 24457960_Phosphorylation by CK2 impairs Par-4 proapoptotic functions. 24486797_LSD1 is a novel substrate of protein kinase CK2alpha. 24616922_Data suggest that overexpression of CK2a (casein kinase 2, alpha) in liver promotes oncogenesis by mediating EGF/EGFR- (epidermal growth factor/EGF receptor)-induced HDAC2 (histone deacetylase 2) expression in hepatocarcinogenesis. 24686080_the inhibition of protein kinase CK2 transiently inhibits cell proliferation through the induction of G1 cell cycle arrest and attenuation of protein synthesis by phosphorylating eIF2 alpha. 24718935_HCV core up-regulates HAMP gene transcription via a complex signaling network that requires both SMAD/BMP and STAT3 pathways and CK2 involvement 24726840_Overexpression of CK2 (alpha, alpha' or beta subunits) by transient transfection resulted in decreased Stat3 Ser-727 phosphorylation. 24742922_we characterize CK2 as an essential component of the Jak/STAT pathway 24831064_Genetic suppression of CK2alpha enhances the apoptosis induced by cisplatin in laryngeal carcinoma cells. 24840952_Study revealved that RPL22 binds CK2alpha in lung cancer cells. 24897506_protein kinase CK2 has a role in bone marrow stromal cell-fueled multiple myeloma growth and osteoclastogenesis 24962073_CK2 alpha is an independent prognostic indicator for gastric carcinoma patients and is involved in tumorigenesis by regulating the phosphorylation of DBC1. 25052887_CagA(+) H. pylori upregulates cellular invasiveness and motility through casein kinase 2. 25120778_These findings highlight the potential role of dysregulated miRNA expression regulated by CK2 in breast cancer. 25241897_Results show that CK2alpha may have an important role in brain tumor-initiating cell maintenance through the regulation of beta-catenin in glioblastoma. 25379016_CK2 phosphorylates and inhibits TAp73 tumor suppressor function to promote expression of cancer stem cell genes and phenotype in head and neck cancer. 25422081_Over-expressed CK2alpha positively regulate Hh/Gli1 signaling in human mesothelioma. 25476899_JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. 25788269_CK2 is widely expressed in follicular, Burkitt and diffuse large B-cell lymphomas and may have role in malignant B-cell growth. 25805179_our findings provide new insights on the potential relevance of the CK2-mediated phosphorylation of B23/NPM in cancer cells, revealing at the same time the potentialities of its pharmacological manipulation for cancer therapy 25837326_CDK11 and CK2 expression are individually essential for breast cancer cell survival, including TNBC. 25872870_Data show that casein kinase 2 (CK2)-mediated phosphorylation of deubiquitylating enzyme OTUB1 at Ser16 causes nuclear accumulation of OTUB1. 25882195_These phosphopeptides include altogether 69 phosphoresidues, a large proportion of which (almost 50%) are generated by CK2, while the others do not conform to the CK2 consensus 25887626_CK2-phosphorylation of eIF3j at Ser127 promotes the assembly of the eIF3 complex, a crucial step in the activation of the translation initiation machinery. 25891901_Overall results, confirm that a balance of hydrophobic and electrostatic interactions contribute predominantly relative to possible intermolecular halogen/hydrogen bonding ,in binding of halogenated benzotriazoles to the ATP-binding site of hCK2alpha 25995454_Casein kinase 2-mediated phosphorylation of Hsp90beta and stabilization of PXR is a key mechanism in the regulation of MDR1 expression. 25998125_Phosphorylation of KCNQ2 and KCNQ3 anchor domains by protein kinase CK2 augments binding to AnkG. 26083323_ATG16L1 as a bona fide physiological CSNK2 and PPP1 substrate, which reveals a novel molecular link from CSNK2 to activation of the autophagy-specific ATG12-ATG5-ATG16L1 complex and autophagy induction 26160174_There is a major role of the CK2alpha-interacting protein CKIP-1 in activation of PAK1 for neoplastic prostate cells transformation. 26165401_The combination treatment of TRAIL and the CK2 inhibitor decreased p65 nuclear translocation... the treatment of a sub-dose of TRAIL, downregulation of CK2, | ENSMUSG00000074698 | Csnk2a1 | 4671.59279 | 1.0711342 | 0.0991392391 | 0.09013187 | 1.205065e+00 | 2.723118e-01 | 9.998360e-01 | No | Yes | 4380.45231 | 542.904557 | 4.490529e+03 | 429.447264 | |
| ENSG00000101955 | 8406 | SRPX | protein_coding | P78539 | FUNCTION: May be involved in phagocytosis during disk shedding, cell adhesion to cells other than the pigment epithelium or signal transduction. | Alternative splicing;Cell adhesion;Disulfide bond;Reference proteome;Repeat;Signal;Sushi | hsa:8406; | autophagosome [GO:0005776]; cell surface [GO:0009986]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum [GO:0005783]; membrane [GO:0016020]; autophagy [GO:0006914]; cell adhesion [GO:0007155]; negative regulation of cell proliferation involved in contact inhibition [GO:0060244]; phagolysosome assembly [GO:0001845]; positive regulation of extrinsic apoptotic signaling pathway in absence of ligand [GO:2001241]; response to endoplasmic reticulum stress [GO:0034976] | 209648_The SRPX protein contains the P-DUDES structural domain in its C-terminal region. This domain has significant albeit remote sequence similarity to thioredoxin-like domains, and is predicted to possess an oxidoreductase function. 12152160_expression of drs mRNA in well-differentiated, moderately differentiated, and poorly differentiated lung adenocarcinoma tissues (drs; down-regulated by v-scr) 12716466_down-regulation of drs mRNA is closely correlated with carcinomas which arise from adenomatous polyps in the course of the adenoma-carcinoma sequence, but most carcinomas arising de novo are independent of the tumor suppressor function of the drs gene 12874760_Down-regulation of drs mRNA is closely correlated with development of prostate carcinoma, suggesting a tumor-suppressor function of the drs gene in this cancer. 15021917_An apoptosis-inducing protein localized in the endoplasmic reticulum which increases efficiency of apoptosis 19424611_Downregulation of drs tumor suppressor gene in highly malignant human pulmonary neuroendocrine tumors. 19862339_This is the first report demonstrating overexpression of ETX1 in glaucomatous trabecular meshwork. 20406461_PELO is subcellularly localized at the actin cytoskeleton, interacts with HAX1, EIF3G and SRPX proteins and that this interaction occurs at the cytoskeleton; this interaction may facilitate PELO to detect and degrade aberrant mRNAs. 28478503_SRPX1 co-accumulated with Abeta deposits in cerebral blood vessels of all autopsied cases with severe cerebral amyloid angiopathy (CAA); no SRPX1 co-accumulated with Abeta deposits in senile plaques. Furthermore, we demonstrated that both Abeta40 and Abeta42 bound to SRPX1 in vitro and enhanced SRPX1 expression in primary cultures of cerebrovascular smooth muscle cells. SRPX1 enhanced caspase activity induced by Abeta40. 31638245_the findings of the present study suggest that targeting the Cancerassociated fibroblasts genes, SRPX and HMCN1, can inhibit ovarian cancer migration and invasion. | ENSMUSG00000090084 | Srpx | 93.46440 | 1.2329309 | 0.3020919598 | 0.32889239 | 8.588247e-01 | 3.540678e-01 | 9.998360e-01 | No | Yes | 97.17656 | 16.217526 | 8.103380e+01 | 10.910453 | ||
| ENSG00000103024 | 4832 | NME3 | protein_coding | Q13232 | FUNCTION: Major role in the synthesis of nucleoside triphosphates other than ATP. The ATP gamma phosphate is transferred to the NDP beta phosphate via a ping-pong mechanism, using a phosphorylated active-site intermediate. Probably has a role in normal hematopoiesis by inhibition of granulocyte differentiation and induction of apoptosis. | 3D-structure;ATP-binding;Apoptosis;Kinase;Magnesium;Metal-binding;Nucleotide metabolism;Nucleotide-binding;Reference proteome;Transferase | hsa:4832; | cytosol [GO:0005829]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; nucleoside diphosphate kinase activity [GO:0004550]; apoptotic process [GO:0006915]; CTP biosynthetic process [GO:0006241]; GTP biosynthetic process [GO:0006183]; UTP biosynthetic process [GO:0006228] | 17900511_Among genes correlated to nodal metastatic progression, we verified in vitro that NM23-H3 reduced cell motility. 20877624_Observational study of gene-disease association. (HuGE Navigator) 23765094_Down-regulated DR-nm23 expression is associated with invasion and metastasis in colorectal cancer. 26945015_Thus Tip60 interacts with RNR and NME3 to provide site-specific synthesis of dNTP for facilitating DNA repair in serum-deprived cells which contain low levels of dNTPs. 29523766_Proinflammatory signaling mediated by innate immunity engagement of flagellin-activated TLR5 in tumor cells results in antitumor effects through NME3 kinase, a positive downstream regulator of flagellin-mediated NFkappaB signaling, enhancing survival for several human cancers. 30111592_The ciliopathy-typical manifestations of NME3 depletion in two vertebrate in vivo models, the biochemical association of NME3 with validated NPHPs, and its localization to the basal body reveal a role for NME3 in ciliary function. We conclude that mutations in the NME3 gene may aggravate the ciliopathy phenotypes observed in humans. 30587587_Given the critical role of mitochondrial dynamics and energy requirements in neuronal development, the homozygous mutation in NME3 is linked to a fatal mitochondrial neurodegenerative disorder. 32708927_NME3 Regulates Mitochondria to Reduce ROS-Mediated Genome Instability. | ENSMUSG00000073435 | Nme3 | 4380.59720 | 0.9555163 | -0.0656476417 | 0.12214500 | 2.971495e-01 | 5.856750e-01 | 9.998360e-01 | No | Yes | 4645.58639 | 673.827132 | 4.431412e+03 | 497.126655 | ||
| ENSG00000103121 | 56942 | CMC2 | protein_coding | Q9NRP2 | FUNCTION: May be involved in cytochrome c oxidase biogenesis. {ECO:0000250}. | Disulfide bond;Mitochondrion;Reference proteome | hsa:56942; | mitochondrion [GO:0005739] | 19844255_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21424380_Single nucleotide polymorphisms in C16orf61 gene is associated with breast cancer. | 918.98464 | 0.9609900 | -0.0574066369 | 0.13100219 | 1.931744e-01 | 6.602876e-01 | 9.998360e-01 | No | Yes | 939.19037 | 98.036602 | 1.030043e+03 | 83.465310 | ||||
| ENSG00000104133 | 80208 | SPG11 | protein_coding | Q96JI7 | FUNCTION: May play a role in neurite plasticity by maintaining cytoskeleton stability and regulating synaptic vesicle transport. {ECO:0000269|PubMed:24794856}. | Alternative splicing;Amyotrophic lateral sclerosis;Cell projection;Charcot-Marie-Tooth disease;Cytoplasm;Hereditary spastic paraplegia;Neurodegeneration;Neuropathy;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a potential transmembrane protein that is phosphorylated upon DNA damage. Defects in this gene are a cause of spastic paraplegia type 11 (SPG11). Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2009]. | hsa:80208; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; dendrite [GO:0030425]; lysosomal membrane [GO:0005765]; nucleolus [GO:0005730]; plasma membrane [GO:0005886]; synapse [GO:0045202]; axo-dendritic transport [GO:0008088]; axon extension [GO:0048675]; chemical synaptic transmission [GO:0007268]; phagosome-lysosome fusion involved in apoptotic cell clearance [GO:0090389]; synaptic vesicle transport [GO:0048489]; walking behavior [GO:0090659] | 17322883_mutations in the SPG11 gene causes spastic paraplegia with thin corpus callosum 18067136_Frameshift, nonsense mutations, and splice mutations in SPG11. Mutations are major cause of autosomal recessive hereditary spastic paraplegia with thin corpus callosum associated with severe motor and cognitive impairment. 18079167_The study reveals the high frequency of SPG11 mutations in patients with HSP, a TCC and cognitive impairment, including in isolated patients, and extends the associated phenotype. 18332254_Autosomal recessive HSP-TCC is a frequent subtype of complicated HSP in Tunisia and is clinically and genetically heterogeneous. SPG11 and SPG15 are the major loci for this entity. 18337587_Mutations on KIAA1840 are frequent in complex autosomal recessive hereditary spastic paraplegia, but they are an infrequent cause of sporadic complex hereditary spastic paraplegia. 18361476_Mutations of the SPG11 gene encoding the spatacsin protein have been identified as a major cause of hereditary spastic paraplegia. 18408091_Genetic and phenotypic data on five patients from two Taiwanese/Chinese families with ARHSP-TCC. 18439221_SPG11 mutations should be suspected in patients with isolated or recessive HSP, thin corpus callosum and mental retardation. 18663179_Loss-of-function SPG11 mutations are the major cause of autosomal recessive hereditary spastic paraparesis with thin corpus callosum in Southern Europe, even in apparently sporadic cases. 18717728_5 new spatacsin mutations were found in complex autosomal recessive hereditary spastic paraplegia:p.C133LfsX154, p.Q1875X, p.K2386QfsX2393,c.2834 + 1G > T & c.6754 + 4insTG. 18835492_This study widens the mutation spectrum of the SPG11 gene and the mutations in the SPG11 gene are also the major causative gene for HSP-TCC in the Chinese Hans. 19040626_Abnormal MRI signal in the region of the forceps minor of the corpus callosum is a characteristic early imaging finding of HSP-TCC with SPG11 mutations. 19084844_ZFYVE26 is the second gene responsible for spastic paraplegia with thinning of the corpus callosum in the Italian population 19087158_This study confirms heterogeneity amongst Italian families with hereditary spastic paraplegia/thin corpus callosum and reports a new mutation predicted to affect splicing in the spatacsin gene. 19105190_While expanding the spectrum of mutations in SPG11, this larger series also corroborated the notion that even within apparently homogeneous population a molecular diagnosis cannot be achieved without full gene sequencing. 19194956_Degeneration of the central retina is a common and previously unrecognized feature in SPG11 related disease. 19196735_Findings expand the mutation spectrum of SPG11 and suggest that SPG11 mutations may occur more frequently in familial than sporadic forms of cHSP without TCC. 19224311_Evidence that parkinsonism may initiate SPG11-linked HSP TCC and that SPG11 may cause juvenile parkinsonism. 19513778_we report three novel and one known heterozygous compound SPG11 mutations in patients with hereditary spastic paraplegia with thin corpus callosum; these are the first cases of genetically confirmed SPG11 mutations in the Korean population. 19917823_phenotype and mutation frequency compared with SPG15 in complicated hereditary spastic paraplegia 20110243_Up to 12 sequence alterations in the spatacsin gene have been identified in unrelated pedigrees with autosomal recessive juvenile amyotrophic lateral sclerosis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20390432_analysis of SPG11 mutations in Asian kindreds and disruption of spatacsin function in the zebrafish 20669327_We identified genetic deficits in spatacsin that were associated with Levodopa responsive parkinsonism with pyramidal signs. 20971220_Data support the importance of SPG11 as a frequent cause for ARHSP-TCC, and expands the clinical SPG11 spectrum. 21035867_Retinal changes, an integral part of SPG11 mutations in this series of patients, are only observed once the paraplegia has become apparent. 21440262_SPG11, the most frequent gene associated with hereditary spastic paraplegia with thin corpus callosum (HSP-TCC) encodes spatacsin, demonstrating the extensive genetic heterogeneity of this condition. 21545838_Spatacsin was strongly expressed in cortical and spinal motor neurons and in embryos. It partially co-localized with multiple organelles, particularly with protein-trafficking vesicles, endoplasmic reticulum and microtubules. 22154821_This study confirmed that SPG11 as a genetic cause of juvenile amyotrophic lateral sclerosis and indicate that SPG11 mutations could be associated with 2 different clinical phenotypes within the same family. 22237444_The analysis shows that the high number of repeated elements in SPG11 together with the presence of recombination hotspots and the high intrinsic instability of the 15q locus all contribute toward making this genomic region more prone to large gene rearrangements. 22696581_mutations result in white and grey matter abnormalities 23121729_This study widens the spectrum of mutations in SPG11 23221952_There was a characteristic gradation in the reduction of microstructural integrity among fiber types and within the CC in patients with the SPG11 mutation 23438842_A novel homozygous nonsense mutation in exon 15 of the SPG11 gene (c.2678G>A; p.W893X) found in two Spanish siblings with a complicated forms of hereditary spastic paraplegia. 23825025_We propose AP-5, SPG15, SPG11 form a coat-like complex, with AP-5 involved in protein sorting, SPG15 facilitating docking of the coat onto membranes by interacting with PI3P via its FYVE domain, and SPG11 (possibly together with SPG15) forming a scaffold. 24085347_SPG11 mutations were identified in autosomal recessive juvenile Amyotrophic lateral sclerosis. 24090761_This study identified novel compound heterozygous mutations in the SPG11 gene of the patients as follows: a nonsense mutation c.6856C>T (p.R2286X) in exon 38 and a deletion mutation c.2863delG (p.Glu955Lysfs*8) in exon 16. 24112408_widespread accumulation of spatacsin observed in pathologic alpha-synuclein-containing inclusions suggests that spatacsin may be involved in the pathogenesis of alpha-synucleinopathies 24315199_We have identified an Hereditary spastic paraplegia patient who inherited the c.5121_5122insAG mutation from his mother and the c.6859C>T mutation from his father 24794856_Study provides evidence that SPG11 is implicated in axonal maintenance and cargo trafficking. 25365221_spastizin and spatacsin were essential components for the initiation of lysosomal tubulation. Together, these results link dysfunction of the autophagy/lysosomal biogenesis machinery to neurodegeneration. 25769290_SPG11 mutation has been identified in a Turkish familial hypobetalipoproteinemia family with hereditary spastic paraplegia. 26003865_novel compound heterozygous mutations in SPG11 are associated with HSP and lower motor neuron involvement, mild cerebellar signs and dysgenesis of the corpus callosum 26374131_SPG11 and CYP7B1 were the most common cause of autosomal recessive hereditary spastic paraplegia in Greece. A novel variant in SPG11, which led to disease with later onset was identified in the Greek population. 26556829_SPG11 is the causative gene of a wide spectrum of clinical features, including autosomal recessive axonal Charcot-Marie-Tooth disease. 1 26671123_a novel homozygous mutation in the splice site donor of intron 30 (c.5866+1G>A) in consanguineous Japanese SPG11 siblings showing late-onset spastic paraplegia (whole-exome sequencing) 27071356_Screening of large cohorts of hereditary spastic paraplegia (HSP) patients identified 83 alleles with 'small' mutations and 13 alleles with large genomic rearrangements. These findings widen the spectra of mutations and mutational mechanisms in SPG11, underscore the pivotal role played by Alu elements, and are of high diagnostic relevance for a wide spectrum of clinical phenotypes including the most frequent form of HSP. 27217339_SPG11 defects were found to be by far the commonest cause of complex hereditary spastic paraplegia in the UK, accounting for 30.9% of cases. 27256065_We identified two novel compound heterozygous mutations in SPG11 in 2 affected individuals with autosomal recessive hereditary spastic paraplegia (ARHSP) with thin corpus callosum 27544499_This study identified novel compound heterozygous mutations in SPG11 in a complex hereditary spastic paraplegia family with thin corpus callosum and severe axonal sensory-motor polyneuropathy as a late manifestation of the disease. 27900367_These hereditary spastic paraplegia patients are compound heterozygous for variants in the SPG11 gene, including the paternally inherited c.6856C>T (p.Arg2286 *) variant and the novel maternally inherited c.2316+5G>A splice-donor region variant. 28681766_Compound heterozygous mutations of the SPG11 gene were identified in the index patient and her younger brother, while the parents were carriers 28933964_SPG11 was suspected to be the most common subtype of autosomal recessive hereditary spastic paraplegia in China, whereas SPG15, SPG5 or SPG7 are rare. The core symptoms of Chinese SPG11 patients showed no difference when compared to SPG11 in western countries, and clinical heterogeneity also existed in our SPG11 patients. 29732542_increased levels of homocarnosine do not seem to be a biomarker for SPG11 in our patients 29946510_SPG11-related hereditary spastic paraplegia is characterized by selective neuronal vulnerability, in which a precocious and widespread white matter involvement is later followed by a restricted but clearly progressive grey matter degeneration. 30081747_ZFYVE26 and SPG11 are differently involved in autophagy and endocytosis. 31281085_Whereas, a previously reported variant c.5769delT (p.Ser1923Argfs*28) in the SPG11 gene was identified in family B manifesting clinical features of SPG11 in 3 affected individuals 31900114_Investigated spastic paraplegia type 11 (SPG11) mutations in Chinese patients with autosomal recessive hereditary spastic paraplegia. 32007496_Two types of recessive hereditary spastic paraplegia in Roma patients in compound heterozygous state; no ethnically prevalent variant found. 32007754_A novel variant in the spatacsin gene causing SPG11 in a Malian family. 32040826_Identification of a Mutation in SPG11 in an Iranian Patient with Spastic Paraplegia and Ears of the Lynx Sign. 32383541_Description of combined ARHSP/JALS phenotype in some patients with SPG11 mutations. 32593884_Hereditary spastic paraplegia type 11 (SPG11) is associated with obesity and hypothalamic damage. 32671691_Phenotypic and genotypic features of patients diagnosed with ALS in the city of Sakarya, Turkey. 33581793_Hereditary spastic paraplegia type 11: Clinicogenetic lessons from 339 patients. 34031922_Transactivation response DNA-binding protein of 43 kDa proteinopathy and lysosomal abnormalities in spastic paraplegia type 11. 34140036_Circ-SPG11 knockdown hampers IL-1beta-induced osteoarthritis progression via targeting miR-337-3p/ADAMTS5. | ENSMUSG00000033396 | Spg11 | 331.97802 | 1.0790536 | 0.1097664806 | 0.17874671 | 3.808173e-01 | 5.371661e-01 | 9.998360e-01 | No | Yes | 388.19153 | 46.406691 | 3.530367e+02 | 32.672726 | |
| ENSG00000104218 | 79848 | CSPP1 | protein_coding | Q1MSJ5 | FUNCTION: May play a role in cell-cycle-dependent microtubule organization. {ECO:0000269|PubMed:16826565}. | Alternative splicing;Ciliopathy;Coiled coil;Cytoplasm;Cytoskeleton;Joubert syndrome;Microtubule;Phosphoprotein;Reference proteome | This gene encodes a centrosome and spindle pole associated protein. The encoded protein plays a role in cell-cycle progression and spindle organization, regulates cytokinesis, interacts with Nephrocystin 8 and is required for cilia formation. Mutations in this gene result in primary cilia abnormalities and classical Joubert syndrome. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Apr 2014]. | hsa:79848; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; spindle [GO:0005819]; spindle pole [GO:0000922]; positive regulation of cell division [GO:0051781]; positive regulation of cytokinesis [GO:0032467] | 15580290_Novel centrosome/microtubule-associated coiled-coil protein (CSPP)is associated with centrosomes and microtubules and may play a role in the regulation of G(1)/S-phase progression and spindle assembly [CSPP]. 16826565_Taken together, CSPP and CSPP-L interact with centrosomes and microtubules and can differently affect microtubule organization. 16826565_Taken together, CSPP and CSPP-L interact with centrosomes and microtubules and can differently affect microtubule organization.[CSPP-L] 19129481_CSPP interacts with and recruits MyoGEF to the central spindle, where MyoGEF contributes to the spatiotemporal regulation of cytokinesis. 20519441_CSPP isoforms require their common C-terminal domain to interact with Nephrocystin 8 (NPHP8/RPGRIP1L) and to form a ternary complex with NPHP8 and NPHP4. 24360803_mutations in CSPP1 were associated with variable ciliopathy phenotypes ranging from Joubert syndrome to the more severe Meckel-Gruber syndrome with perinatal lethality and occipital encephalocele 24360807_Our data suggest that CSPP1 is required for proper primary cilium formation or stability and that CSPP1 mutations result in abnormal mid-hindbrain development. 24360808_CSPP1 mutations are a major cause of the Joubert-Jeune phenotype in humans. 24901235_Differential expression of a nuclear CSPP1 isoform identified biologically and clinically distinct subgroups of basal-like breast carcinoma. 26241740_Microtubule-independent but desmoplakin-dependent localization of CSPP-L to desmosomes occurs in apical-basal polarized epithelial cells. CSPP-L depletion promoted multi-lumen spheroid formation in Caco-2 cells. 26378239_propose that CSPP1 cooperates with CENP-H on kinetochores to serve as a novel regulator of kinetochore microtubule dynamics for accurate chromosome segregation 30965236_Tumor-promoting effect was inhibited after we transfected miR-1236-3p into circ-CSPP1 overexpressing OC cells. 32495924_Roles of circ-CSPP1 on the proliferation and metastasis of glioma cancer. | ENSMUSG00000056763 | Cspp1 | 226.00713 | 0.8727725 | -0.1963224588 | 0.21311282 | 8.453478e-01 | 3.578718e-01 | 9.998360e-01 | No | Yes | 192.57120 | 36.457384 | 2.028438e+02 | 29.674608 | |
| ENSG00000104219 | 51201 | ZDHHC2 | protein_coding | Q9UIJ5 | FUNCTION: Palmitoyltransferase that catalyzes the addition of palmitate onto various protein substrates and is involved in a variety of cellular processes (PubMed:18508921, PubMed:18296695, PubMed:19144824, PubMed:21343290, PubMed:22034844, PubMed:23793055). Has no stringent fatty acid selectivity and in addition to palmitate can also transfer onto target proteins myristate from tetradecanoyl-CoA and stearate from octadecanoyl-CoA (By similarity). In the nervous system, plays a role in long term synaptic potentiation by palmitoylating AKAP5 through which it regulates protein trafficking from the dendritic recycling endosomes to the plasma membrane and controls both structural and functional plasticity at excitatory synapses (By similarity). In dendrites, mediates the palmitoylation of DLG4 when synaptic activity decreases and induces synaptic clustering of DLG4 and associated AMPA-type glutamate receptors (By similarity). Also mediates the de novo and turnover palmitoylation of RGS7BP, a shuttle for Gi/o-specific GTPase-activating proteins/GAPs, promoting its localization to the plasma membrane in response to the activation of G protein-coupled receptors. Through the localization of these GTPase-activating proteins/GAPs, it also probably plays a role in G protein-coupled receptors signaling in neurons (By similarity). Also probably plays a role in cell adhesion by palmitoylating CD9 and CD151 to regulate their expression and function (PubMed:18508921). Palmitoylates the endoplasmic reticulum protein CKAP4 and regulates its localization to the plasma membrane (PubMed:18296695, PubMed:19144824). Could also palmitoylate LCK and regulate its localization to the plasma membrane (PubMed:22034844). {ECO:0000250|UniProtKB:P59267, ECO:0000250|UniProtKB:Q9JKR5, ECO:0000269|PubMed:18296695, ECO:0000269|PubMed:18508921, ECO:0000269|PubMed:19144824, ECO:0000269|PubMed:21343290, ECO:0000269|PubMed:22034844, ECO:0000269|PubMed:23793055}.; FUNCTION: (Microbial infection) Promotes Chikungunya virus (CHIKV) replication by mediating viral nsp1 palmitoylation. {ECO:0000269|PubMed:30404808}. | Acyltransferase;Cell junction;Cell membrane;Endoplasmic reticulum;Endosome;Golgi apparatus;Lipoprotein;Membrane;Palmitate;Phosphoprotein;Reference proteome;Synapse;Transferase;Transmembrane;Transmembrane helix | hsa:51201; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; postsynaptic recycling endosome [GO:0098837]; recycling endosome membrane [GO:0055038]; palmitoyltransferase activity [GO:0016409]; protein homodimerization activity [GO:0042803]; protein-cysteine S-myristoyltransferase activity [GO:0019705]; protein-cysteine S-palmitoyltransferase activity [GO:0019706]; protein-cysteine S-stearoyltransferase activity [GO:0140439]; cellular protein metabolic process [GO:0044267]; peptidyl-L-cysteine S-palmitoylation [GO:0018230]; positive regulation of AMPA glutamate receptor clustering [GO:1904719]; positive regulation of endosome to plasma membrane protein transport [GO:1905751]; positive regulation of long-term synaptic potentiation [GO:1900273]; protein localization to plasma membrane [GO:0072659]; protein localization to postsynaptic membrane [GO:1903539]; protein palmitoylation [GO:0018345]; protein targeting to membrane [GO:0006612]; regulation of cell-cell adhesion [GO:0022407]; regulation of neuronal synaptic plasticity [GO:0048168]; regulation of protein catabolic process [GO:0042176]; regulation of protein localization to plasma membrane [GO:1903076]; synaptic vesicle maturation [GO:0016188] | 18296695_Identification of CKAP4/p63 as a substrate of DHHC2, a putative tumor suppressor. 18508921_DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151 19144824_DHHC2-mediated palmitoylation of CKAP4 has a role in opposing cancer-related cellular behaviors. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21343290_Gi/o signaling and the palmitoyltransferase DHHC2 regulate palmitate cycling and shuttling of RGS7 family-binding protein. 21471008_The palmitoyl transferase DHHC2 targets a dynamic membrane cycling pathway: regulation by a C-terminal domain 22034844_DHHC2 localizes primarily to the endoplasmic reticulum and Golgi apparatus suggesting that it is involved in S-acylation of newly-synthesized or recycling Lck involved in T cell signalling. 23457560_reduced ZDHHC2 expression is associated with lymph node metastasis and independently predicts an unfavorable prognosis in gastric adenocarcinoma patients 24995331_These results suggest an important role for ZDHHC2 as a tumor suppressor in metastasis and recurrence of HCC. 32180374_Fine-mapping of ZDHHC2 identifies risk variants for schizophrenia in the Han Chinese population. 33488612_Zdhhc2 Is Essential for Plasmacytoid Dendritic Cells Mediated Inflammatory Response in Psoriasis. | ENSMUSG00000039470 | Zdhhc2 | 268.09084 | 1.2885765 | 0.3657782106 | 0.23926825 | 2.294528e+00 | 1.298307e-01 | 9.998360e-01 | No | Yes | 332.08565 | 54.845013 | 2.951475e+02 | 37.797497 | ||
| ENSG00000104231 | 79752 | ZFAND1 | protein_coding | Q8TCF1 | FUNCTION: Plays a role in the regulation of cytoplasmic stress granules (SGs) turnover. SGs are dynamic and transient cytoplasmic ribonucleoprotein assemblies important for cellular protein homeostasis when protein production is suspended after acute exogenous stress (PubMed:29804830). Associates with SGs and is involved in the efficient and specific arsenite-induced clearance process of SGs through the recruitment of the ubiquitin-selective ATPase VCP and the 26S proteasome (PubMed:29804830). This process requires both complexes for efficient degradation of damaged ubiquitinated SG proteins during recovery from arsenite stress, and hence avoiding aberrant cytoplasmic SGs degradation via autophagy (PubMed:29804830). {ECO:0000269|PubMed:29804830}. | Alternative splicing;Cytoplasm;Metal-binding;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:79752; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; proteasome binding [GO:0070628]; zinc ion binding [GO:0008270]; cellular response to arsenite ion [GO:1903843]; positive regulation of intracellular protein transport [GO:0090316]; stress granule disassembly [GO:0035617] | 29804830_By recruiting the 26S proteasome and the ubiquitin-selective ATPase p97 to arsenite-induced stress granules, ZFAND1 ensures their timely clearance and prevents their aberration. | ENSMUSG00000039795 | Zfand1 | 274.79253 | 1.0915462 | 0.1263731918 | 0.19068437 | 4.374268e-01 | 5.083670e-01 | 9.998360e-01 | No | Yes | 237.41875 | 49.276348 | 2.500069e+02 | 40.002148 | ||
| ENSG00000104691 | 7993 | UBXN8 | protein_coding | O00124 | FUNCTION: Involved in endoplasmic reticulum-associated degradation (ERAD) for misfolded lumenal proteins, possibly by tethering VCP to the endoplasmic reticulum membrane. May play a role in reproduction. {ECO:0000269|PubMed:21949850}. | Alternative splicing;Endoplasmic reticulum;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | p97 or VCP (valosin-containing protein) is a versatile ATPase complex, and many cofactors are required for the p97 functional diversity. This gene encodes one of the p97 cofactors. This cofactor is a transmembrane protein and localized in the endoplasmic reticulum (ER) membrane. It tethers p97 to the ER membrane via its UBX domain. The association of this cofactor with p97 facilitates efficient ER-associated degradation of misfolded proteins. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Aug 2013]. | hsa:7993; | endoplasmic reticulum [GO:0005783]; integral component of endoplasmic reticulum membrane [GO:0030176]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ubiquitin ligase complex [GO:0000151]; protein-macromolecule adaptor activity [GO:0030674]; single fertilization [GO:0007338]; ubiquitin-dependent ERAD pathway [GO:0030433] | 34921223_Epigenetic silencing of UBXN8 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. | ENSMUSG00000052906 | Ubxn8 | 216.36369 | 0.9654778 | -0.0506850030 | 0.21681044 | 5.512873e-02 | 8.143678e-01 | 9.998360e-01 | No | Yes | 206.14094 | 37.297682 | 2.453705e+02 | 34.278388 | |
| ENSG00000105647 | 5296 | PIK3R2 | protein_coding | O00459 | FUNCTION: Regulatory subunit of phosphoinositide-3-kinase (PI3K), a kinase that phosphorylates PtdIns(4,5)P2 (Phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Binds to activated (phosphorylated) protein-tyrosine kinases, through its SH2 domain, and acts as an adapter, mediating the association of the p110 catalytic unit to the plasma membrane. Indirectly regulates autophagy (PubMed:23604317). Promotes nuclear translocation of XBP1 isoform 2 in a ER stress- and/or insulin-dependent manner during metabolic overloading in the liver and hence plays a role in glucose tolerance improvement (By similarity). {ECO:0000250|UniProtKB:O08908, ECO:0000269|PubMed:23604317}. | 3D-structure;Disease variant;Phosphoprotein;Protein transport;Reference proteome;Repeat;SH2 domain;SH3 domain;Stress response;Transport;Ubl conjugation | hsa:5296; | cytosol [GO:0005829]; nucleus [GO:0005634]; phosphatidylinositol 3-kinase complex [GO:0005942]; phosphatidylinositol 3-kinase complex, class IA [GO:0005943]; 1-phosphatidylinositol-3-kinase regulator activity [GO:0046935]; phosphotyrosine residue binding [GO:0001784]; protein heterodimerization activity [GO:0046982]; protein phosphatase binding [GO:0019903]; receptor tyrosine kinase binding [GO:0030971]; cellular glucose homeostasis [GO:0001678]; cellular response to insulin stimulus [GO:0032869]; insulin receptor signaling pathway [GO:0008286]; negative regulation of MAPK cascade [GO:0043409]; phosphatidylinositol 3-kinase signaling [GO:0014065]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; positive regulation of protein import into nucleus [GO:0042307]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein transport [GO:0015031]; regulation of autophagy [GO:0010506]; regulation of phosphatidylinositol 3-kinase activity [GO:0043551]; response to endoplasmic reticulum stress [GO:0034976] | Mouse_homologues 16227599_Mice with cardiac deletion of both p85 subunits exhibit attenuated Akt signaling in the heart, reduced heart size, and altered cardiac gene expression. 17360811_PI3K activity is required for PMA-induced colocalization between AFAP-110 and cSrc and subsequent cSrc activation, and this signaling pathway promotes cell migration. 18621722_Complete loss of all PI3K regulatory subunits caused acute embryonic lethality at E11.5 due to hemorrhaging, whereas retention of a single p85alpha allele yielded viable mice that survived to adulthood 18704194_BCR-ABL leukemogenesis inis blocked by PI3K in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells 19190244_Involvement of p85beta in CD28-mediated activation and differentiation of antigen-stimulated T cells. 19747165_Data show that mesothelin is a potential target in reducing resistance to cytotoxic drugs, and mesothelin-treated cells revealed rapid tyrosine phosphorylation of the p85 subunit of PI3K. 19811262_These results indicate that the p85beta regulatory isoform has partially overlapping functions with p85alpha in B cells as well as a unique role in opposing BCR responses. 20348926_heterodimerization disrupted by insulin, increasing nuclear translocation of XBP-1 21383062_study shows that p110beta nuclear localization signal and p85beta nuclear export sequence regulate p85beta/p110beta nuclear localization, supporting the idea that nuclear, but not cytoplasmic, p110beta controls cell survival 22733740_p85beta phosphoinositide 3-kinase subunit regulates tumor progression 22867989_miR-126-mediated phosphoinositide-3-kinase regulation, not only fine-tunes VEGF-signaling, but it strongly enhances the activities of Ang-1 on vessel stabilization and maturation. 23330804_Activation of PI3K (p110beta and p85beta), and phospho-AKT (Ser473), participated in the regulation of activating macrophages by wear particles, ultimately resulting in the secretion of TNF-alpha. 27381007_p85beta deficiency does not alter NK cell differentiation and maturation in spleen or bone marrow. NK cells from p85beta(-/-) mice nonetheless produced more IFN-gamma and degranulated more effectively when stimulated with anti-NKG2D antibody. p85beta deficiency impaired NKG2D internalization, which could contribute to the activated phenotype. 32895527_Anthrax lethal factor cleaves regulatory subunits of phosphoinositide-3 kinase to contribute to toxin lethality. | ENSMUSG00000031834 | Pik3r2 | 5338.44772 | 1.0284907 | 0.0405287140 | 0.09826164 | 1.711741e-01 | 6.790704e-01 | 9.998360e-01 | No | Yes | 6027.53179 | 786.870041 | 5.470608e+03 | 551.741855 | ||
| ENSG00000105835 | 10135 | NAMPT | protein_coding | P43490 | FUNCTION: Catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, an intermediate in the biosynthesis of NAD. It is the rate limiting component in the mammalian NAD biosynthesis pathway. The secreted form behaves both as a cytokine with immunomodulating properties and an adipokine with anti-diabetic properties, it has no enzymatic activity, partly because of lack of activation by ATP, which has a low level in extracellular space and plasma. Plays a role in the modulation of circadian clock function. NAMPT-dependent oscillatory production of NAD regulates oscillation of clock target gene expression by releasing the core clock component: CLOCK-ARNTL/BMAL1 heterodimer from NAD-dependent SIRT1-mediated suppression (By similarity). {ECO:0000250|UniProtKB:Q99KQ4, ECO:0000269|PubMed:24130902}. | 3D-structure;Acetylation;Biological rhythms;Cytokine;Cytoplasm;Glycosyltransferase;Nucleus;Phosphoprotein;Pyridine nucleotide biosynthesis;Reference proteome;Secreted;Transferase | PATHWAY: Cofactor biosynthesis; NAD(+) biosynthesis; nicotinamide D-ribonucleotide from 5-phospho-alpha-D-ribose 1-diphosphate and nicotinamide: step 1/1. | This gene encodes a protein that catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, one step in the biosynthesis of nicotinamide adenine dinucleotide. The protein belongs to the nicotinic acid phosphoribosyltransferase (NAPRTase) family and is thought to be involved in many important biological processes, including metabolism, stress response and aging. This gene has a pseudogene on chromosome 10. [provided by RefSeq, Feb 2011]. | hsa:10135; | cell junction [GO:0030054]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nuclear speck [GO:0016607]; cytokine activity [GO:0005125]; identical protein binding [GO:0042802]; nicotinamide phosphoribosyltransferase activity [GO:0047280]; nicotinate-nucleotide diphosphorylase (carboxylating) activity [GO:0004514]; cell-cell signaling [GO:0007267]; circadian regulation of gene expression [GO:0032922]; NAD biosynthesis via nicotinamide riboside salvage pathway [GO:0034356]; NAD biosynthetic process [GO:0009435]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of nitric-oxide synthase biosynthetic process [GO:0051770]; positive regulation of transcription by RNA polymerase II [GO:0045944]; signal transduction [GO:0007165] | 15124023_PBEF1 is upregulated in neutrophils by IL-1beta and functions as a novel inhibitor of apoptosis in response to a variety of inflammatory stimuli. 15947248_These findings identify PBEF1 as a regulator of NAD+-dependent reactions in smooth muscle cells (SMCs), reactions that promote, among other potential processes, the acquisition of a mature SMC phenotype. 16021090_Although PBEF had no chemotaxic effects, it was antiapoptotic for both amniotic epithelial cells and fibroblasts and may protect these cells against apoptosis that is induced by chronic distension, labor, or infection. 16186392_visfatin plasma concentrations and visceral visfatin messenger RNA expression correlated with measures of obesity but not with visceral fat mass or waist-to-hip ratio 16188281_PBEF contributes to acute lung injury susceptibility 16234302_visfatin may play a role in the pathogenesis of type 2 diabetes mellitus 16394088_A true adipokine, but it is not regulated by thiazolidineidones and, thus unlikely to contribute to the insulin-sensitizing actions of these drugs. 16496121_increased macrophage population in obese human visceral white adipose tissue might be responsible for the enhanced production of chemokines as well as resistin and visfatin 16531748_Visfatin/pre-B-cell colony-enhancing factor appears to be preferentially produced by the visceral adipose tissue and has insulin mimetic actions 16636125_Observational study of gene-disease association. (HuGE Navigator) 16636125_data suggest that genetic variation in the visfatin gene may have a minor effect on visceral and subcutaneous visfatin messenger RNA expression profiles but does not play a major role in the development of obesity or type 2 diabetes mellitus 16701870_These data show that an inflammatory stimulus in the fetal membranes inducing NF-kappaB and AP-1 would up-regulate PBEF as well as IL-8. 16720654_findings show that, in human obesity, plasma visfatin is reduced, whereas visfatin mRNA is differentially regulated in adipose tissue 16736128_Circulating visfatin concentrations are increased by hyperglycaemia. This effect is suppressed by exogenous hyperinsulinaemia or somatostatin infusion. 16783377_FK866 is bound in a tunnel at the interface of the NMPRTase dimer, and mutations in this binding site can abolish the inhibition by FK866, providing a starting point for the development of new anticancer agents. 16802343_Pre-B cell colony-enhancing factor (PBEF) is regulated via IL-6 trans-signaling and the IL-6-related cytokine OSM. PBEF is also actively expressed during arthritis. 16814166_results show that there are decreased concentrations of plasma visfatin in gestational diabetes mellitus subjects and this may indicate that visfatin plays a role in the pathogenesis of gestational diabetes mellitus 16828081_visfatin is a new hypoxia-inducible gene of which expression is stimulated through the interaction of HIF-1 with HRE sites in its promoter region. 16868228_visfatin has a local metabolic role in the recovery period following exercise. 16922702_Observational study of gene-disease association. (HuGE Navigator) 16956691_The results indicate that hyperglycemia causes an increase in plasma visfatin levels and, as in people with diabetes mellitus type 2 but not with impaired glucose tolerance, this increase gets more prominent as the glucose intolerance worsens. 17003355_circulating visfatin is increased with progressive beta-cell deterioration. 17003359_A significant association was found between two SNPs of visfatin (rs9770242 and rs1319501), in perfect linkage disequilibrium, and fasting insulin levels (P = 0.002). 17003359_Observational study of gene-disease association. (HuGE Navigator) 17090638_visfatin is highly expressed in lean, more insulin-sensitive subjects and is attenuated in subjects with high intramyocellular lipids, low insulin sensitivity, and high levels of inflammatory markers 17177135_potential link with pathogenesis of glucose resistance and type 2 diabetes 17189536_Observational study of gene-disease association. (HuGE Navigator) 17237424_In patients with inflammatory bowel disease, plasma levels of visfatin are elevated and its mRNA expression is significantly increased in colonic tissue of Crohn's and ulcerative colitis patients 17261426_Circulating levels of the cytokine visfatin/PBEF-1 are influenced by renal function, but are not associated with fat mass or surrogate markers of insulin resistance in patients with chronic kidney disease. 17261426_Observational study of gene-disease association. (HuGE Navigator) 17284735_Visfatin is down-regulated by overfeeding 17307730_Nampt is a longevity protein that can add stress-resistant life to human smooth muscle cells by optimizing SIRT1-mediated p53 degradation 17327330_Serum visfatin concentration is significantly associated with parameters of iron metabolism, especially in subjects with altered glucose tolerance. 17334748_An increase in fasting visfatin, the levels of which correlate with both fasting and post-glucose-load insulin concentrations, accompanies worsening glucose tolerance in the third trimester of pregnancy. 17335820_Women with PCOS exhibit higher plasma visfatin levels than control subjects of similar body mass index. 17340225_the regulation of glucose uptake, proliferation, and type I collagen production by visfatin in human osteoblasts involves IR phosphorylation, the same signal-transduction pathway used by insulin 17392604_This review summarizes current knowledge of the various functions of PBEF/visfatin towards involvement in the pathophysiology of several diseases. 17408594_Taken together, these results demonstrate that visfatin promotes angiogenesis via activation of mitogen-activated protein kinase ERK-dependent pathway. 17429683_PBEF/NAMPT/visfatin level is an indicator of beneficial lipid profile in non-diabetic Caucasian subjects 17512309_Observational study of gene-disease association. (HuGE Navigator) 17556870_Plasma visfatin levels are increased in overweight and obese subjects with metabolic syndrome. 17582143_Visfatin has a role in insulin resistance and markers of hyperandrogenism in lean PCOS patients 17889652_During fasting, increased Nampt provides protection against cell death and requires an intact mitochondrial NAD(+) salvage pathway as well as the mitochondrial NAD(+)-dependent deacetylases SIRT3 and SIRT4. 17904242_Circulating visfatin may be related with some proinflammatory condition even in a nondiabetic state 17931620_Rosuvastatin induced a significant decrease in plasma visfatin levels in patients with primary hyperlipidemia. 17952840_Visfatin, TNF-alpha, and IL-6 mRNA expressions are increased in peripheral mononuclear-monocytic cells from women with type 2 diabetes. 17984105_Circulating levels of visfatin and adiponectin are associated with endothelial dysfunction in all stages of chronic kidney disease, independently of inflammation and insulin resistance. 18034779_Serum levels of the adipokine visfatin are increased in pre-eclampsia. 18083357_Severely obese women have higher than normal blood levels of visfatin. 18239648_As a good surrogate marker, plasma visfatin level can predict the visceral adipose tissue area in obese children. 18241953_Results show that visfatin levels may decrease with age and be related to the HDL metabolism in obese adolescents. 18248642_A probable new role of visfatin in inflammation reflected in PBMCs, in the context of obesity. 18252866_PBEF is shown to exert three distinct activities of central importance to cellular energetics and innate immunity.[REVIEW] 18270432_Observational study of gene-disease association. (HuGE Navigator) 18270432_The -1535 promoter variant of the visfatin gene is associated with serum triglyceride and HDL-cholesterol levels in Japanese subjects. 18272217_Chronic stretching of the amniotic epithelial cells increases PBEF expression, which protects them from apoptosis. 18356846_C-reactive protein, migration inhibitor factor (MIF), leptin, and visfatin levels decreased after weight loss. 18363885_Our results showed that serum visfatin was increased in patients with metabolic syndrome, especially in those with carotid plaques. Visfatin may be an inflammatory marker of MetS 18410550_Circulating visfatin is independently associated with diabetes & resistin concentration, but is not related to adiponectin multimers or other metabolic covariates. data are suggestive of a potential role of visfatin in subclinical inflammatory states. 18486613_These results may reveal a novel role of PBEF in the pathogenesis of acute lung injury; it interacts with ND1, IFITM3, and ferritin light chain involved in oxidative stress and inflammation. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18686225_change of plasma visfatin concentration by intensive glycemic control may be a compensatory mechanism to ameliorate insulin deficiency due to pancreatic beta-cell dysfunction in type 2 diabetes 18691043_analysis of the structure and function of visfatin and its relation to diabetes mellitus and other dysfunctions [review] 18714344_Circulating visfatin are significantly increased in HLA DR2 positive narcoleptic patients compared to controls. 18728403_PBEF1/NAmPRTase/Visfatin is a potential malignant astrocytoma/glioblastoma serum marker with prognostic value. 18728403_PBEF1/NAmPRTase/Visfatin may have a role in progression of malignant astrocytoma/glioblastoma 18787378_Results show that women with gestational diabetes mellitus have significantly decreased visfatin concentrations in the third trimester. 18823127_Although the weak energetic coupling of ATP hydrolysis appears to be a nonoptimized enzymatic function, closer analysis of this remarkable protein reveals an enzyme designed to capture NAM with high efficiency at the expense of ATP hydrolysis. 18940394_Observational study of gene-disease association. (HuGE Navigator) 18940394_The visfatin (PBEF1) G-948T gene polymorphism is associated with increased high-density lipoprotein cholesterol in obese subjects. 18957417_cartilage-specific gene expression in human chondrocytes is regulated by SirT1 and nicotinamide phosphoribosyltransferase 18996492_Nicotinamide phosphoribosyltransferase may play a critical role as an inflammatory cytokine in the development of pulmonary inflammation and dysregulation of pulmonary vascular endothelial and alveolar epithelial cell barriers. 19005759_Visfatin plasma concentrations could predict the presence of portal inflammation in non-alcoholic fatty liver disease patients. 19009499_Visfatin upregulate secretion of MCP-1and IL-6 in human umbilical vein endothelial cells. 19073361_After a lifestyle intervention in patients with metabolic syndrome plasma visfatin concentrations were directly associated with inflammatory markers. 19074645_Plasma levels of PBEF/Nampt/visfatin are decreased in patients with liver cirrhosis. 19087564_Elevated expression of VF mRNA has a strong correlation with gestational diabetes mellitus and obesity. 19093738_Circulating visfatin levels are not affected by the presence of chronic malnutrition in anorexia nervosa or binge/purge eating behavior in bulimia nervosa. 19095760_Visfatin levevns in human cerebrospinal fluid decrease with rising body fat, supporting the assumption that visfatin transport is impaired in obesity and that central nervous visfatin insufficiency is linked to pathogenetic mechanisms of obesity. 19099366_maternal gestational diabetes mellitus, as well as delivery of large for gestational age neonate were independently associated with a higher maternal plasma visfatin concentrations. 19100714_These data further suggest an integral role for visfatin-FGF-2 signaling axis in modulating endothelial angiogenesis. 19111298_Circulating visfatin levels were higher in patients with polycystic ovary syndrome than healthy controls 19166999_PBEF/visfatin induces secretion of MCP-1 in human endothelial cells: role in visfatin-induced angiogenesis 19185944_Circulating visfatin was significantly lower in the gestational diabetes mellitus than in the NGT subjects at term, although no differences in its mRNA expression in fat and placental tissues were observed. 19200966_The follicular fluid visfatin concentrations are correlated to the number of oocytes retrieved. 19222486_Data suggest that visfatin may play a role in the hormone stabilization process independent of anthropometric, inflammatory or insulin resistance variables. 19284295_median maternal plasma concentration of visfatin peaks between 19-26, has a nadir between 27-34 weeks of gestation. Normal and overweight/obese pregnant women differed in the pattern of changes in circulating visfatin concentrations with gestational age 19300429_Observational study of gene-disease association. (HuGE Navigator) 19300429_a visfatin gene (NAMPT/PBEF1) variant is associated with protection from obesity 19302375_Enhancing endothelial Nampt activity may thus be beneficial in scenarios requiring EC-based vascular repair and regeneration during aging and hyperglycemia, such as atherosclerosis and diabetes-related vascular disease. 19389865_Loss of renal function is accompanied by increased circulating active visfatin concentrations. Furthermore, decreased HDL cholesterol may hint at an increased probability of cardiovascular events in HD patients with elevated serum visfatin. 19402217_High serum visfatin is associated with endothelial dysfunction in kidney diseases. 19403191_Metabolic syndrome X is not significantly associated with serum visfatin. 19408173_plasma visfatin levels are associated with HDL-C and markers of hyperandrogenism, but it is not associated with proinflammatory markers and insulin resistance in lean women with polycystic ovary syndrome 19488934_Increased maternal levels of leptin and visfatin may be involved in the pathogenesis of pre-eclampsia. 19498324_The effects of hyper- and hypothyroidism on various metabolic parameters may be partly mediated by visfatin. 19513556_Nicotinamide phosphoribosyltransferase and prostaglandin H2 synthase 2 are up-regulated in human pancreatic adenocarcinoma cells after stimulation with interleukin-1. 19533482_Asian PCOS women had significantly higher serum visfatin. Their levels were significantly correlated with 2-hour post-load glucose and blood pressure in PCOS women and with fasting insulin and blood pressure in PCOS women with glucose intolerance. 19571557_plasma visfatin concentrations are lower in patients with type 1 DM and related to glycemic control reflected by HbA1c. 19572235_preterm labor with intra-amniotic infection is characterized by high maternal circulating visfatin concentrations 19583971_This is the first report demonstrating a potential role for this important cytokine/enzyme in inflammation-related bone disease. 19650786_The dynamics of Nampt/visfatin and high molecular weight adiponectin during oral glucose tolerance test appear to be linked with insulin and adiposity. 19654329_a novel role of PBEF in the pathogenesis of acute lung injury and other inflammatory disorders via AP-1 dependent mechanism 19666527_crystallographic analysis of nicotinamide phosphoribosyltransferase 19727662_Visfatin is a cause of vascular inflammation, a feature of atherothrombotic diseases linked to metabolic disorders.isfatin appears to be a direct contributor to vascular inflammation, a feature of atherothrombotic diseases linked to metabolic disorders. 19751774_Visfatin through STAT3 activation enhances IL-6 expression that promotes endothelial angiogenesis. 19765775_findings suggest that visfatin/PBEF/Nampt is a proinflammatory marker of adipose tissue associated with systemic insulin resistance and hyperlipidemia 19804767_results suggested that Visfatin -1535C>T polymorphism might be associated with reduced risk of coronary artery disease in a Chinese population 19811274_adiponectin, ghrelin and visfatin levels were higher in the patients with systemic lupus erythematosus than in controls 19819277_These data suggest a contributory and multifunctional role for visfatin in prostate cancer progression, with particular relevance and emphasis in an obese population. 19819904_crystal structure and reaction mechanism 19834878_high nampt serum levels are significantly associated with advanced carotid atherosclerosis in patients with type 2 diabetes mellitus. 19855187_The role of Nampt in regulating autophagy and potential mechanisms by which NAD(+) regulates autophagy in the heart, is discussed. 19887595_exercise increases skeletal muscle NAMPT expression and that NAMPT correlates with mitochondrial content 19900033_elevated in plasma of women with a small-for-gestational-age neonate; not significantly different from normal in women with pre-eclampsia 19912992_these results demonstrate that human hepatocytes are a potential source of circulating NAMPT. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19929281_The elevation of visfatin observed in children and adolescents with metabolic syndrome was proportionate to number of components of MetS but was not associated with insulin resistance. 19936064_activated, but not resting, T lymphocytes undergo massive NAD(+) depletion upon FK866-mediated Nampt inhibition 19948877_High visfatin in chronic kidney disease patients may constitute a counter-regulatory response to central visfatin resistance in uremia. 20019453_Plasma levels do not correlate with coronary calcification. 20046156_Observational study of gene-disease association. (HuGE Navigator) 20046156_Significant differences were found in LDL- & HDl-cholesterol, total cholesterol, & fasting serum insulin among visfatin genotypes (TT, GG, & GT). Circulating visfatin levels correlated with weight, BMI, hs-CRP & fasting insulin in the TT genotype. 20059442_Visfatin levels are higher in plasma of pregnant women with gestational diabetes, and in a pregestational diabetic state. 20085562_These findings suggest that visfatin/PBEF may play a role in the regulation of the complex and dynamic crosstalk between inflammation and metabolism during pregnancy. 20091460_These data suggest a differential role for visfatin and resistin in linking metabolic diseases to atherosclerosis with visfatin/PBEF/Nampt being more important in patients with type 2 diabetes and resistin in obese but nondiabetic human subjects. 20106640_Circulating levels of NAMPT and gene expression of NAMPT in human peripheral blood cells are increased in morbidly obese patients. 20121389_visfatin concentration was lower in umbilical cord blood than in maternal circulation, in normal pregnancy, Small for gestational age, and pre-eclampsia 20220292_Visfatin levels were significantly elevated in women with gestational diabetes mellitus and during the course of pregnancy and increased visfatin concentrations were reduced within 6 to 10 weeks after delivery. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20346233_Positive correlation between levels of visfatin and resistin may suggest that visfatin plays a role in inflammation in rheumatoid arthritis 20346239_In rheumatoid arthritis patients on TNF-alpha blocker treatment, circulating visfatin levels are unrelated to disease activity, adiposity or metabolic syndrome 20358348_visfatin/Nampt/PBEF1 does not have direct insulin-like action in human adipocytes 20383745_Observational study of gene-disease association. (HuGE Navigator) 20383745_the -1535C>T polymorphism of visfatin is associated with decreased plasma levels of inflammatory markers in coronary artery disease patients 20392873_sed on these findings, we suggest a role for decreased NAMPT/visfatin levels in hepatocyte apoptosis in NAFLD-related disease 20409603_Findings suggest that visfatin synthesis is activated from adipose tissue in a diabetic environment, induces NF-kappaB activation and leads to activation of pro-inflammatory cytokines and systemic inflammation. 20451405_Observational study of gene-disease association. (HuGE Navigator) 20451405_The involvement of the NAMPT gene in the development of type 2 diabetes (T2DM) in the Greek population, was evaluated. 20470278_Elevated visfatin related to markers of inflammation might represent a novel link between inflammation and adipocytokines in dialyzed patients. 20470283_Plasma visfatin levels are related to the endothelial functions, inflammation and the severity of proteinuria in diabetic nephropathy. Ramipril decreased visfatin levels and improved proteinuria, endothelial dysfunction and inflammation. 20482384_In patients with osteoarthritis, infrapatellar fat pad and synovium were important sources of visfatin, while osteophytes released largest amounts of visfatin. 20486883_higher in large-for-gestational age newborns 20506278_Data suggest that Nampt/SIRT1 pathway could be a novel therapeutic tool for the treatment of HIV-1 infection. 20586547_Visfatin levels in obese and overweight patients with polycystic ovary syndrome were higher than that found in females without concomitant disease with similar BMI, and visfatin had positive correlation with BMI, waist circumference and insulin resistance 20598369_Cyclic stretch/release of amniotic epithelial cells increased both intracellular and secreted PBEF. 20605615_both vaspin and visfatin/Nampt might play an important role in the pathogenesis of T2DM. 20606733_This is the first report demonstrating Nicotinamide phosphoribosyltransferase overexpression in ovarian serous adenocarcinomas 20608974_PPARgamma ligands increase visfatin gene expression in a PPARgamma-dependent manner in primary human resting macrophages and in adipose tissue macrophages, but not in adipocytes 20621376_investigation of the main source of circulating visfatin (apparently placenta) and visfatin's potential role in pathogenesis of gestational diabetes mellitus 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20712903_Observational study of gene-disease association. (HuGE Navigator) 20720266_A significant decline in visceral adipose tissue visfatin level was found to be associated with degree of steatosis in nonalcoholic fatty liver disease patients 20724478_analysis of inhibition of nicotinamide phosphoribosyltransferase and how cellular bioenergetics reveals a mitochondrial insensitive NAD pool 20817637_Provide evidence of Notch1-dependent endothelial FGF-2 induction by visfatin and of Notch1 activation in visfatin-stimulated endothelial angiogenesis. 20823688_Even though the expression of NPY, omentin and visfatin was comparable between obese individuals and controls, we have to consider differences in the total production rate of adipose tissue-derived factors 20848232_visfatin plays an important role in breast cancer progression 20850423_Decreased vaspin and increased visfatin serum levels were observed in asymptomatic patients with coronary artery disease. 20885390_Observational study of gene-disease association. (HuGE Navigator) 20951014_Suggest that increased circulating visfatin concentration is associated with insulin sensitivity improvement achieved after energy restricted diet intervention induced weight loss. 20956937_Our results suggest important roles of concomitant upregulation of NAMPT and SIRT1 along with increased FOXO3a protein level for prostate carcinogenesis 21061834_The administration of olmesartan improved blood pressure, insulin, HOMA, visfatin and lipid profile in hypertensive obese women. 21122936_Elevation of visfatin in T2DM is independent of obesity and insulin resistance and is mainly determined by fasting glucose and triglycerides. 21144000_CHS-828 and TP201565 are competitive inhibitors of NAMPT and that acquired resistance towards NAMPT inhibitors can be expected primarily to be caused by mutations in NAMPT. 21145308_these results identify visfatin as a gene oppositely regulated by the LXR and PPARgamma pathways in human macrophages. 21225497_Our study showed a positive association between visfatin and the fibrosis stage in nonalcoholic fatty liver disease. 21246369_synovial fluid visfatin might involved in cartilage matrix degradation. 21296365_LDL cholesterol and C-reactive protein levels are positively correlated with visfatin levels. Weight is negatively correlated with visfatin levels, in an independent way and adjusted by age, sex and dietary intake. 21298414_Leucocytes are a major source of enzymatically active NAMPT, which may serve as a biomarker or even mediator linking obesity, inflammation and insulin resistance. 21327328_visfatin enhances atheroma inflammation through the NAMPT-MAPK (p38, ERK1/2)-NF-kappaB-EMMPRIN/MMP-9 pathway 21332548_Serum visfatin was correlated with disease severity and metabolic syndrome in chronic hepatitis C patients. 21375135_Weight reduction after a 2 months on a hypocaloric diet is not associated with a significant change in circulating visfatin in morbidly obese patients. 21375196_The present study confirms the association of visfatin with chronic kidney disease (CKD), however further studies at molecular level to check its expression within renal tissue may clarify its definitive role in CKD 21406189_Nampt and RBP4 serum concentrations did not correlate with the maximum percentage of carotid stenosis 21484571_This chapter summarizes the various functional aspects of Nampt and discusses its potential roles in diseases, with special focus on type 2 diabetes mellitus--REVIEW 21492230_expression of NAMPT was generally high in the more aggressive malignant lymphomas, with >80% strong expression, whereas the expression in the more indolent follicular lymphoma was significantly lower 21494798_Visfatin levels were significantly lower in patients with Behcet's disease whose illnesses were active or inactive than the control group. 21500554_Visfatin may be involved in the pathogenesis of gestational diabetes mellitus. 21517777_role of NAMPT in human metabolism 21518975_Results demonstrate that PBEF can prime for PMN respiratory burst activity by promoting p40 and p47 translocation to the membrane. 21524918_In patients with active rheumatoid arthritis, serum visfatin levels are related to the number of B cells. 21542902_Hyperbaric oxygen (HBO) activates visfatin expression in cultured human coronary arterial endothelial cells. HBO-induced visfatin is mediated by TNF-alpha and at least in part through JNK pathway. 21625240_Elevations in serum visfatin and C-reactive protein (CRP) may contribute to accelerated atherogenic processes in a spinal cord-injured population. 21631570_data suggest that the NAMPT -3186C>T polymorphism is significantly associated with plasma levels of post-prandial serum insulin and total cholesterol in Chinese T2DM patients with repaglinide monotherapy 21634360_Similar to IGF-I levels, determination of visfatin levels can be a predictive marker of retinopathy of prematurity, but more studies are needed. 21664630_The expression of visfatin is closely associated with the expression of proinflammatory cytokines in FFA-induced inflammation and is significantly decreased by NF-kappaB inhibition in HepG2 cells. 21677044_Insulin and GLP-1 are responsible for the rapid suppression of visfatin levels upon an oral glucose uptake in healthy probands. 21687707_transcriptional regulation of FTO and NAMPT in preadipocytes and adipocytes by metabolic regulators 21694775_These findings suggest that visfatin may represent a pro-inflammatory cytokine that is influenced by insulin/insulin sensitivity via the NF-kappaB and JNK pathways. 21697093_IL-1beta induces dedifferentiation of articular chondrocytes by up-regulation of SIRT1 activity enhanced by both NAMPT and ERK signaling. 21712732_Results support an association between serum visfatin/nicotinamide phosphoribosyl-transferase and risk of postmenopausal breast cancer. 21726671_PBEF is partially responsible for the increased expression of inflammatory cytokines and apoptosis factors in infected human pulmonary microvascular endothelial cells in vitro. 21738955_Data indicating a possible role of leptin, adiponectin, visfatin, chemerin and vaspin in the pathogenesis of chronic hepatitis are summarized. 21777266_Compared with healthy girls, serum VISF concentrations are decreased in girls with anorexia nervosa. 21839801_visfatin activates pro-inflammatory cytokine release and phospholipid metabolism in human placenta via activation of the nuclear factor kappa B pathway 21840905_visfatin mRNA concentration in omental adipose tissue is closely correlated with BMI and insulin resistance 21906432_NAMPT rs9770242 and rs59744560 polymorphisms are not markers of disease susceptibility and cardiovascular disease disease in rheumatoid arthritis. 21939650_Visfatin level was increased in type 2 diabetes mellitus, possibly related to hyperglycemia 21963513_Results support a role for visfatin in the detection of subjects with many metabolic abnormalities, which result in increased CVD risk. 21975728_role in LPS-induced inflammatory responses of human monocytes 22024288_first-degree relatives of subjects with type 2 diabetes mellitus showed a significant association with lower visfatin levels which may suggest a pathophysiological role for visfatin in beta cell dysfunction in this group. 22090280_A regulatory impact of Nampt rhythmicity on glucose homeostasis. 22137121_This study indicates that plasma visfatin levels are significantly higher in ST-elevation myocardial infarction patients. 22147201_Report potential mechanistic link between the visfatin -1535C>T polymorphism and reduced coronary artery disease risk. 22151390_serum level increased in psoriasis 22167446_Plasma visfatin concentrations are lower in patients with gestational diabetes mellitus and related to glycemic control reflected by HbA1c. 22186408_These data demonstrate that curcumin down-regulates visfatin gene expression and suggest that visfatin may contribute to breast cancer cell invasion and link obesity to breast cancer development and progression. 22228719_association of the common variants of IL6, LEPR, and PBEF1 with obesity in Indian children. 22251423_NAMPT rs1319501 minor allele associates with increased MI risk in young women. In young men a protective effect of the AKT1 rs3730358 minor allele was suggested, possibly related to an attenuated inflammation 22261365_Further prospective and longitudinal studies are needed to determine whether serum visfatin could be used as a prognostic tool in the armamentarium of postmenopausal breast cancer monitoring and management in conjunction with other biomarkers. 22272940_Visfatin serum concentration seems to be increased in preeclampsia as compared with uncomplicated pregnancy. 22293189_these results showed that visfatin promoted IL-8 production by upregulation of TXAS, leading to angiogenic activation in endothelial cells. 22313145_Data suggest that serum visfatin levels in women with polycystic ovary syndrome (PCOS) correlate with risk factors for cardiovascular diseases but not with other metabolic factors in PCOS (i.e., obesity, insulin resistance). 22319029_Data suggest that increased serum NAMPT levels in patients with active acromegaly negatively correlate with adiposity in limbs/total body. NAMPT induces monocyte chemoattractant protein-1 in mature adipocytes suggesting role in adipose inflammation. 22377803_P2X7-dependent release in LPS-activated monocytes following treatment with ATP 22380724_expression of VEGF and visfatin was significantly decreased in the preeclampsia group compared with the normotensive control group 22397743_Plasma visfatin level is not a useful biomarker of insulin resistance and hyperandrogenism. 22399297_the proinflammatory actions of visfatin in chondrocytes involve regulation of IR signaling pathways, possibly through the control of Nampt enzymatic activity 22425808_Physical exercise was reflected by a significant increase in lactate (p=0.01), insulin (p=0.01), visfatin concentrations (p=0.01, p<0.05, respectively) and only during transitory phase in glucose (p=0.01) and resistin concentrations (p<0.05). 22430142_an overexpression of NAMPT led to a decreased expression of GADD45A, whereas, the inhibition of NAMPT by the known chemical inhibitor FK866 increased the expression of GADD45A in cells. 22467327_Serum visfatin concentration increases in patients with nonalcoholic fatty liver disease. 22490590_Elevated circulating visfatin levels in subjects with hyperthyroidism and hypothyroidism are possibly due to an increase of visfatin mRNA expression in visceral fat. 22541073_When expressio | ENSMUSG00000020572 | Nampt | 645.32106 | 1.0506032 | 0.0712178616 | 0.16430809 | 1.847045e-01 | 6.673611e-01 | 9.998360e-01 | No | Yes | 628.59001 | 119.202265 | 6.122982e+02 | 89.545445 |
| ENSG00000105854 | 5445 | PON2 | protein_coding | Q15165 | FUNCTION: Capable of hydrolyzing lactones and a number of aromatic carboxylic acid esters. Has antioxidant activity. Is not associated with high density lipoprotein. Prevents LDL lipid peroxidation, reverses the oxidation of mildly oxidized LDL, and inhibits the ability of MM-LDL to induce monocyte chemotaxis. {ECO:0000269|PubMed:11579088, ECO:0000269|PubMed:15772423}. | Alternative splicing;Calcium;Disulfide bond;Glycoprotein;Hydrolase;Membrane;Metal-binding;Reference proteome;Signal | This gene encodes a member of the paraoxonase gene family, which includes three known members located adjacent to each other on the long arm of chromosome 7. The encoded protein is ubiquitously expressed in human tissues, membrane-bound, and may act as a cellular antioxidant, protecting cells from oxidative stress. Hydrolytic activity against acylhomoserine lactones, important bacterial quorum-sensing mediators, suggests the encoded protein may also play a role in defense responses to pathogenic bacteria. Mutations in this gene may be associated with vascular disease and a number of quantitative phenotypes related to diabetes. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:5445; | extracellular region [GO:0005576]; lysosome [GO:0005764]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; acyl-L-homoserine-lactone lactonohydrolase activity [GO:0102007]; arylesterase activity [GO:0004064]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; aromatic compound catabolic process [GO:0019439]; response to oxidative stress [GO:0006979]; response to toxic substance [GO:0009636] | 11206400_Observational study of gene-disease association. (HuGE Navigator) 11257265_Observational study of gene-disease association. (HuGE Navigator) 11512679_Observational study of genotype prevalence. (HuGE Navigator) 11676977_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11676977_study suggested a gene-gene interaction between the PON1 and PON2 polymorphisms for CAD risk; may have linkage disequilibrium with a tightly linked PON3 locus or significant atherosclerotic alleles of nearby genes 11692002_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11768721_Observational study of gene-disease association. (HuGE Navigator) 11768721_association between left ventricular hypertrophy and the C825T allele of the G-protein beta3 subunit gene in Arabs. (G PROTEIN BETA3) 11803456_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11803456_PON2*S and apoE4 alleles have interactive effect on the development of the two most common forms of dementias AD and VD, and further support the hypothesis that cardiovascular factors contribute to the development of AD. 11918623_Observational study of gene-disease association. (HuGE Navigator) 11918623_diabetic microangiopathy is genetically heterogeneous; PON1 Leu/Leu increases the risk for retinopathy and PON2 Ser/Ser increases the risk for microalbuminuria 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12151850_REVIEW: The paraoxonase gene family and coronary heart disease 12433026_Observational study of gene-disease association. (HuGE Navigator) 12442067_Observational study of gene-disease association. (HuGE Navigator) 12454802_Association between the severity of angiographic coronary artery disease and paraoxonase gene polymorphisms in the National Heart, Lung, and Blood Institute-sponsored Women's Ischemia Syndrome Evaluation (WISE) study 12454802_Observational study of gene-disease association. (HuGE Navigator) 12561466_Observational study of gene-disease association. (HuGE Navigator) 12588779_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12778447_Observational study of gene-disease association. (HuGE Navigator) 12939804_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12955589_Observational study of gene-disease association. (HuGE Navigator) 12955589_Polymorphisms of paraoxonase 1 and 2 genes, alone or in combination is associated with with bone mineral density 14636952_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14741412_Observational study of gene-disease association. (HuGE Navigator) 14741412_These results suggested that the PON2 polymorphism might be a risk factor for LOAD independent of ApoE epsilon4 status in Chinese. 14984433_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14996478_Observational study of gene-disease association. (HuGE Navigator) 15001326_Meta-analysis of gene-disease association. (HuGE Navigator) 15039125_Observational study of gene-disease association. (HuGE Navigator) 15232408_Genotyping for polymorphisms of PON1 Q192R, PON2 A148G, and PON2 S311C finds no association between mother's PON1 and PON2 genotypes and preterm delivery, but finds infant's PON1 RR and PON2 CC genotypes are associated with preterm delivery. 15232408_Observational study of gene-disease association. (HuGE Navigator) 15256524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15345661_Observational study of gene-disease association. (HuGE Navigator) 15359538_Observational study of genotype prevalence. (HuGE Navigator) 15544923_PON2 stimulation may represent a compensatory mechanism against the increase in cellular superoxide anion production and atherogenesis 15772423_PON1, PON2, and PON3 are lactonases with overlapping and distinct substrate specificities 15776585_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 16030523_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16078734_Observational study of gene-disease association. (HuGE Navigator) 16080611_Observational study of gene-disease association. (HuGE Navigator) 16117861_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16135439_Observational study of gene-disease association. (HuGE Navigator) 16141008_Observational study of gene-disease association. (HuGE Navigator) 16164576_Observational study of gene-disease association. (HuGE Navigator) 16185677_Observational study of genetic testing. (HuGE Navigator) 16319130_Observational study of gene-disease association. (HuGE Navigator) 16411107_Observational study of gene-disease association. (HuGE Navigator) 16551349_Observational study of gene-disease association. (HuGE Navigator) 16614106_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16767666_Observational study of gene-disease association. (HuGE Navigator) 16767666_PON1 55 LM genotype and M allele, PON2 148 GG/AG genotype and G allele are the risk factors for coronary artery disease, and the activity of plasma PON is also markedly reduced in individuals with above genotypes. 16776623_Observational study of gene-disease association. (HuGE Navigator) 16822964_A haploblock of high linkage disequilibrium (LD) spanning PON2 and PON3 was associated with SALS. 16822964_Observational study of gene-disease association. (HuGE Navigator) 16822965_Observational study of gene-disease association. (HuGE Navigator) 16822965_the C311S polymorphism was associated with sALS in dominant and additive models. 16891303_Plays a protective role against atherosclerosis in vivo. 16926679_Observational study of gene-disease association. (HuGE Navigator) 17096118_Observational study of gene-disease association. (HuGE Navigator) 17096118_PON2 variants have, at best, a small effect on the risk of renal dysfunction in type 2 diabetes. 17137217_Observational study of gene-disease association. (HuGE Navigator) 17299970_Gene polymorphisms of PON1 55 Met/Leu, PON2 148 Ala/Gly and MnSOD 9 Ala/Val seemed to involve in the morbidity of CHD by influencing the plasma activities of PON and MnSOD. 17299970_Observational study of gene-disease association. (HuGE Navigator) 17309646_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17404154_Here, we analyzed the ROS-reducing capability of paraoxonase-2 (PON2) in different vascular cells and its involvement in the endoplasmic reticulum stress pathway known as the unfolded protein response. 17406108_Observational study of gene-disease association. (HuGE Navigator) 17406108_genotypes with the C allele of the PON2 gene C311S polymorphism is a risk factor for large vessel disease stroke in a Polish population 17412306_The rates of hydrolysis of estrogen esters by the paraoxonases is PON3 > PON1 > PON2, with the exception of 17beta estradiol, 3-acetate, 17-cyclopentylpropionate which is hydrolyzed at a slightly faster rate by PON2 compared to PON1. 17428620_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17436100_In Ashkenazi Jewish population, carriage of PON1 R192 allele may confer protection against the development of IBD. 17436100_Observational study of gene-disease association. (HuGE Navigator) 17557249_C311S polymorphism of PON2 has no significant correlation with stroke in Han people of Chinese Hunan area and allele C/S is not an independent risk factor for stroke,neither is G148A. 17557249_Observational study of gene-disease association. (HuGE Navigator) 17601350_Observational study of gene-disease association. (HuGE Navigator) 17664137_PON1/2/3 may have extracellular functions as part of the host response in inflammatory bowel diseases and celiac disease 17854416_Observational study of gene-disease association. (HuGE Navigator) 17916643_These observations demonstrate that the human intestine is preferentially endowed with a marked PON2 expression compared with the rat intestine and this expression shows a developmental and intracellular pattern of distribution. 17940058_Cys(311)Ser variant of PON2 may contribute to albumin excretion rate 18020951_PON2 expression is upregulated in unesterified cholesterol enriched macrophages through activation of the PI(3)K signal pathway 18063859_Observational study of gene-disease association. (HuGE Navigator) 18063859_There was no significant correlation between the C311S and G148A polymorphisms of PON2 and stroke in the Chinese population. 18258817_We demonstrate that PON2 mRNA and protein are decreased in plaques versus plaque-adjacent tissue, mammary arteries, and fetal carotids. Our data indicate that the protective effect of PON2 could fail during atherosclerosis exacerbation. 18347034_The hydrolytic effects of PON1, PON2, and PON3 on the key quorum sensing compound of P.aeruginosa are reported. 18361900_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18361900_results indicate that PON2-311 polymorphism is an independent risk factor of AMI 18413200_Observational study of gene-disease association. (HuGE Navigator) 18427977_Observational study of genetic testing. (HuGE Navigator) 18436804_Urokinase plasminogen activator upregulates paraoxonase 2 expression in macrophages via an NADPH oxidase-dependent mechanism. 18513389_Observational study of gene-disease association. (HuGE Navigator) 18569577_Observational study of gene-disease association. (HuGE Navigator) 18635682_Observational study of gene-disease association. (HuGE Navigator) 18691157_Data suggest that independent mechanisms mediate the degradation of paraoxonase-2 mRNA and protein after disturbance of calcium homoeostasis. 18695162_Observational study of gene-disease association. (HuGE Navigator) 18720901_Observational study of genotype prevalence. (HuGE Navigator) 18759523_Observational study of gene-disease association. (HuGE Navigator) 18759523_major effect of the paraoxonase-2 polymorphism on coronary artery disease risk in patients 18776646_This study revealed a significant association between Ser311Cys variation in the paraoxonase 2 gene and type 2 diabetes in northern Chinese. 18818748_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18977241_Observational study of gene-disease association. (HuGE Navigator) 18977341_Observational study of gene-disease association. (HuGE Navigator) 19019335_Observational study of gene-disease association. (HuGE Navigator) 19082953_The anti-atherogenic biological activities were studied in vitro using serum or cell cultures, and also in vivo, using PON 1/2/3 knockout or transgenic mice, as well as humans - healthy volunteers and atherosclerotic patients. 19091699_Data suggest that PON2 attenuates macrophage triglyceride accumulation and foam cell formation via inhibition of microsomal DGAT1 activity, which appears to be sensitive to oxidative state. 19131662_Meta-analysis of gene-disease association. (HuGE Navigator) 19151417_Observational study of gene-disease association. (HuGE Navigator) 19152805_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19166692_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19254215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19263529_Observational study of gene-disease association. (HuGE Navigator) 19371607_Observational study of gene-disease association. (HuGE Navigator) 19401157_Oxidative stress and proinflammatory agents selectively affect the expression of PONs. 19479237_Observational study of gene-disease association. (HuGE Navigator) 19497963_Urokinase activates macrophage PON2 gene transcription via the PI3K/ROS/MEK/SREBP-2 signalling cascade mediated by the PDGFR-beta. 19527514_Observational study of gene-disease association. (HuGE Navigator) 19540141_Observational study of gene-disease association. (HuGE Navigator) 19540141_a possible role for PON2 C311S polymorphism in the pathogenesis of cardiac ischemic damage 19546579_Observational study of gene-disease association. (HuGE Navigator) 19575027_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19587357_Meta-analysis of gene-disease association. (HuGE Navigator) 19654933_Observational study of gene-disease association. (HuGE Navigator) 19654933_the allele and genotype frequencies of PON polymorphisms were described in a south-western Korean population 19818126_Observational study of gene-disease association. (HuGE Navigator) 19840942_Observational study of gene-disease association. (HuGE Navigator) 19840942_impaired lactonase activity may play a role in innate immunity, atherosclerosis, and other diseases associated with the PON2 311 SNP. 19865538_Quercetin supplementation increases PON2 levels in cultured monocytes in vitro but not in human volunteers in vivo. 19878569_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19930448_Observational study of gene-disease association. (HuGE Navigator) 19930448_The PON2 311C allele is suggested as a possible predisposing factor for severe cases of ischemic stroke. 19939821_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20099504_Serum paraoxonase 1, arylesterase activities and total free sulphydryl group levels were significantly lower in endometrial cancer patients compared to controls 20381198_The data of this study suggested that PON2 polymorphisms are not involved in ALS pathogenesis in an Italian population. 20430392_Observational study of gene-disease association. (HuGE Navigator) 20458436_Observational study of gene-disease association. (HuGE Navigator) 20485444_Observational study of gene-disease association. (HuGE Navigator) 20503442_differences between PON2 mRNA and protein distributions could be due to missence mutations in the PON2 gene, causing nontranslation of mRNA to protein in some tissues 20529763_Observational study of gene-disease association. (HuGE Navigator) 20530481_PON2 prevents mitochondrial superoxide formation and apoptosis independent from its lactonase activity 20536507_Observational study of gene-disease association. (HuGE Navigator) 20536507_genetic variants, including Ala/Ala of SCYA11 (eotaxin) Ala23Thr, Cys/Cys or Cys/Ser of PON2 (paraoxonase 2) Ser311Cys and Arg/Arg of ADRB3 (beta3-adrenergic receptor) Trp64Arg, were independently associated with incident cardiac end-point. 20565774_Observational study of genotype prevalence. (HuGE Navigator) 20582942_Observational study of gene-disease association. (HuGE Navigator) 20582942_We now report that in genomic DNA from individuals with familial and sporadic amyotrophic lateral sclerosis, we have identified at least 7 gene mutations that are predicted to alter PON1, PON2, and PON3 function. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20839225_Observational study of gene-disease association. (HuGE Navigator) 20839225_These functional effects of PON1, PON2, PON3 variation should be considered in protecting vulnerable subpopulations from organophosphate pesticides and other inducers of oxidative stress. 20934178_paraoxonase components PON1, PON2, and PON3 are regulated by diverse nutritional molecules and pharmacological agents and pathophysiological events, such as oxidative stress and inflammation [review] 20980077_Low serum paraoxonase activity is a risk factor for Alzheimer disease; furthermore, multiple variants in PON influence serum paraoxonase activity and their effects may be synergistic. 20980077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21118365_in hemodialysis patients, frequency distribution of PON ratio showed 3 PON phenotypes: 74% oshowed PON1, 21% PON2, and 5% PON3. Compared to hemodialysis patients with PON1, patients with PON2 or 3 showed higher conversion rates for 4-nitrophenylacetate 21127310_the PON1 55M and PON1 192R alleles were associated with decreased sperm motility whereas the PON2 311C allele was associated with decreased concentration, supporting the significance of PON genes in semen quality. 21146823_This meta-analysis suggested no association between the PON2 polymorphisms and coronary heart disease risk. 21223581_PON2 genetic variation is significantly and independently associated with variation in serum PON activity and no obvious association between PON2 tagSNPs and SLE risk. 21368884_Deficiency of PON2 caused apoptosis of selective tumor cells. 21561808_Polymorphisms within Pon1, Pon2, and GSTM1 are associated with male infertility. 21620813_polymorphisms of pon1 gene affect PON1 paraoxonase activity while S311C polymorphism of pon2 gene might be associated with coronary heart disease 21672555_Both paraoxonases are expressed in the human frontal cortex; PON2 but not PON1 mRNA levels are up-regulated in Alzheimer disease. 21757906_the studied PON2 polymorphism is not associated with late-onset Alzheimer's disease 21765051_Single-nucleotide polymorphisms in PON2 is associated with chronic kidney disease in Type 2 diabetes. 22016051_The association of PON2, with Parkinson's disease among Indians may point toward an inherent population-specific genetic predisposition 22183305_The aim of this study was to perform a meta-analysis to investigate a more authentic association between PON2 Ser311Cys polymorphism and ischemic stroke. 22534874_PON2 acts as a QS-quenching factor in keratinocytes and may have an important role in cutaneous defense against bacterial infections. 22744335_Paraoxonase (PON) family members seem central to a wide variety of human illnesses, but appreciation of their antioxidative function in the gastrointestinal tract is in its infancy 22860094_lower PPARgamma and PON2 gene expression in the BALF of children with CF is associated specifically with P. aeruginosa infection and neutrophilia. 22964087_Data suggest that mRNA expression and activity of PON-2 enzyme in monocytes and macrophages can be modulated by dietary factors (here, traditional Brazilian beverages/tea made from green or roasted yerba mate). 23053877_In the presence of metabolic syndrome and diabetes, PON2-311CC was associated with an increased risk of significant coronary stenosis. 23225229_hypertension, HDL-cholesterol concentration, and the presence of C allele in PON2 gene were independently associated with atherothrombotic events; study sheds light on role of PON2 as a possible cofactor in determining risk of events together with the well-known risk markers HDL-cholesterol and hypertension 23327886_A case-control study of Chinese workers exposed to occupational noise, PON2 gene polymorphisms(rs7493, rs12026, rs7785846 and rs7786401)was associated with a higher risk to noise-induced hearing loss. No higher risk was found for genotype rs12704796. 23487294_The polymorphisms of the PON genes studied are not related to increased risk of MS in the Polish population. 23742759_Individuals homozygous for the R allele of the PON1 gene and the C allele of the PON2 gene are more likely to have an increased risk of coronary artery disease. 24088404_No significant differences were found for PON2-311 genotypes or allele frequencies in patients with dementia due to Alzheimer's disease. 24100645_The current metaanalysis highlighted results for the risk of association of PON155L with diabetic retinopathy (DR) and that PON2 gene polymorphisms, as well as PON1Q192R, may not confer major genetic risk to diabetic nephropathy or DR. 24301778_We conclude that the PON1 55M and PON2 311C alleles are independent risk factors for CAAD in essential hypertension patients from the Xinjiang Han population. 24421402_Data indicate that paraoxonase 2 (PON2) is a transmembrane protein that localizes to the plasma membrane in small intestinal tissue. 24636586_There were no statistically significant differences among all comparisons in pon2 mutation in patient with ischemic stroke. 24727057_S311C PON2 polymorphism is associated with the accelerated decline in kidney function in the chronic glomerulonephritis patients. 24807171_A modest dose-dependent allele effect between the PON2 c.311C > G polymorphism, increased triglyceride concentrations and decreased LDL particle size distribution. 24816800_changes in PON2 in the placenta in labour 24845160_Maternal genetic susceptibility GSTT1 and PON2 rs12026 could significantly modify the association of organic solvents with gestational age. 25038992_PON2 and PON3 (i) are associated with mitochondria and mitochondria-associated membranes, (ii) modulate mitochondria-dependent superoxide production, and (iii) prevent apoptosis. 25210784_Taken together, our data suggest a model by which DJ-1 exerts its antioxidant activities, at least partly through regulation of PON2. 25708945_PON2 levels reflect the cells' irradiation sensitivity. 25740199_D1-like receptors inhibit ROS production by altering PON2 distribution in membrane microdomains in the short-term, and by increasing PON2 expression in the long-term. 25913154_Newborns with PON2 148AG/GG genotype and exposed to high concentrations of MBP and MEHP had higher risks of LBW and SBL 25953737_Data suggest cigarette smoke leads to inhibition of hydrolytic activity of paraoxonase isoenzymes (PON1, PON2, PON3) by modification of thiol groups, by interactions with free radicals/heavy metals, or by decreasing HDL level in blood. [REVIEW] 25991559_Haplotype containing two risk alleles of PON1 and PON2 genes was significantly associated with disease diabetes mellitus, type 2. 26056385_PON2 is a central regulator of host cell responses to Pseudomonas aeruginosa N-(3-oxo-dodecanoyl)-L-homoserine lactone 26227792_PON2 SNPs rs12026 and rs7493 were associated with intracerebral hemorrhage. The C alleles of rs12026 and rs7493 contributed to a decreased risk of ICH. 26656916_Data indicate the relationship between ubiqutination of lysine (Lys) 168 and paraoxonase 2 (PON2) catalytic activity. 26978533_Thi study found that the CC genotype of the PON2 S311C polymorphism is a risk factor for ischemic stroke. 27322774_PON2 is a new potential biomarker for therapy resistance or a prognostic tumor marker. 27578362_Data show that the mRNA levels of both paraoxonases PON2 and PON3 were significantly upregulated whereas PON1's mRNA was absent in transgenic mouse hearts. 27609416_The PON2 Ser311Cys polymorphism is associated with coronary heart disease risk in Caucasians. 27623343_High soluble expression levels were achieved with a yield of 76 mg of fully human PON2 variants per liter of culture media 27771368_Studies indicate that three paraoxonases PON1, PON2, and PON3 genes are clustered on chromosome 7, and that PONs possess numerous atheroprotective properties. 28108734_Results suggest that valproic acid (VPA) reduces paraoxonase 2 (PON2) expression in glioblastoma multiforme (GBM) cells, which in turn increases reactive oxygen species (ROS) production and induces Bim protein production that inhibits cancer progression via the PON2-Bim cascade. 28430636_these results showed that PON2 could represent a molecular biomarker for bladder cancer and suggest a potential role of the enzyme as a prognostic factor for this neoplasm 28433610_Data suggest that paraoxonases (PON1, PON2, PON3) play roles in innate immunity, inflammatory response, and protection against oxidative stress; these factors are associated with the body's response to infectious diseases; low serum PON1 activities are associated with poor survival in patients with severe sepsis. [REVIEW] 28509526_No significant differences were observed between PON2 and PON3 gene expression in psoriatic lesional and non lesional skin compared with healthy controls 28566152_Study observed a genetic effect in ischemic stroke patients of the north area of the Gran Canaria island linked to the rs7493 variant in the PON2 gene with worse atherogenic ultrasonographic profile in ischemic stroke patients. Conversely, the Cys311Cys homozygosity, which is usually associated with an increased risk of stroke in the general population, was also related to a better ultrasonographic profile. 28637359_The decrease in monocyte/macrophage PON2 enzymatic activity observed in type 2 diabetes cannot be totally explained by abdominal obesity and insulin resistance. 28768768_PON-2 regulates ENaC activity by modulating its intracellular trafficking and surface expression 28803777_Paraoxonase 2 facilitates pancreatic ductal cancer growth and metastasis by stimulating GLUT1-mediated glucose transport. 28862184_In donor retina from patients with diabetes, all three PON1, PON2 and PON3 were expressed, and there was a significant increase in PON3 expression compared to control. This might be the reason for the increased thiolactonase activity observed in diabetic retina compared to control 29308836_Paraoxonase 2 (PON2) possesses antiatherogenic properties and is associated with lower Reactive Oxygen Species levels. PON2 is involved in the antioxidative and anti-inflammatory response in intestinal epithelial cells. 29439952_These experiments delineate a PON2 redox-dependent mechanism that regulates endothelial cell TF activity and prevents systemic coagulation activation and inflammation. 29531225_PON2 decreases OC cell proliferation by inhibiting insulin like growth factor-1 expression and signaling. 29729330_suggesting that PON2 is necessary for proper mitochondrial function 30138371_Microarray analysis revealed differential expression of three vitamin D associated genes in the aortic adventitia in rheumatoid arthritis (RA) and non-RA patients with coronary artery disease: while the expression of GADD45A and NCOR1 was higher, the expression of PON2 was lower in RA patients. 30607774_SOD2 A16V, but not PON2 S311C, polymorphism may be one of the genetic determinants for polycystic ovary syndrome in Chinese women 31338708_it is a oncogene which promotes gastric cancer. 31835890_Effect of High Glucose-Induced Oxidative Stress on Paraoxonase 2 Expression and Activity in Caco-2 Cells. 32306677_The rs7785846 (CT+TT) genotype carriers of PON2 gene are more susceptible to hearing impairment when exposed to high noise intensity. 32382056_WTAP and BIRC3 are involved in the posttranscriptional mechanisms that impact on the expression and activity of the human lactonase PON2. 33210737_Paraoxonase-2: A potential biomarker for skin cancer aggressiveness. 33531346_PON2 subverts metabolic gatekeeper functions in B cells to promote leukemogenesis. 34710487_Paraoxonase 2 protects against the CML mediated mitochondrial dysfunction through modulating JNK pathway in human retinal cells. 34987135_Insights into the role of paraoxonase 2 in human pathophysiology. 35092416_Insights into the role of paraoxonase 2 in human pathophysiology. 35286330_Airway epithelial Paraoxonase-2 in obese asthma. | ENSMUSG00000032667 | Pon2 | 423.13559 | 1.0765543 | 0.1064210411 | 0.17144342 | 3.784057e-01 | 5.384577e-01 | 9.998360e-01 | No | Yes | 416.60699 | 84.963789 | 4.052202e+02 | 63.758141 | |
| ENSG00000105983 | 64327 | LMBR1 | protein_coding | Q8WVP7 | FUNCTION: Putative membrane receptor. | Alternative splicing;Coiled coil;Membrane;Receptor;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a member of the LMBR1-like membrane protein family. Another member of this protein family has been shown to be a lipocalin transmembrane receptor. A highly conserved, cis-acting regulatory module for the sonic hedgehog gene is located within an intron of this gene. Consequently, disruption of this genic region can alter sonic hedgehog expression and affect limb patterning, but it is not known if this gene functions directly in limb development. Mutations and chromosomal deletions and rearrangements in this genic region are associated with acheiropody and preaxial polydactyly, which likely result from altered sonic hedgehog expression. [provided by RefSeq, Jul 2008]. | hsa:64327; | integral component of plasma membrane [GO:0005887]; transmembrane signaling receptor activity [GO:0004888]; embryonic digit morphogenesis [GO:0042733]; signal transduction [GO:0007165] | 12032320_mutation causes preaxial polydactyly 12491086_disruption is associated with preaxial polydactyly 17300748_C to T transition in LMBR1 results in the dysregulation of sonic hedgehog, which leads to the triphalangeal thumb-polysyndactyly syndrome found in this case 18698406_Intron 5 of LMBR1 was presumably subject to balancing selection during the evolution of modern human of various racial stocks. 20068592_Effect on transcription factor binding of a novel ZRS point mutation (463T>G) in a Pakistani family with preaxial polydactyly and triphalangeal thumb. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22495965_A novel 13 base pair insertion in the sonic hedgehog ZRS limb enhancer (ZRS/LMBR1) causes preaxial polydactyly with triphalangeal thumb. 22786669_report of a new point mutation within the ZRS in a family with digit malformations including triphalangeal thumb, pre-axial polydactyly and post-axial polydactyly; heterozygous C>A mutation at position 287 of the ZRS enhancer was detected in all affected subjects and is absent from four unaffected family members 23793141_Review of the literature and cases in a Chinese family confirm genetic homogeneity (duplication of ZRS wintin intron 5 of LMBR1) of syndactyly type IV and triphalangeal thumb-polysyndactyly syndrome. 24478176_Data describe extensive variation in limb phenotype in a large family and report on a novel sequence variation NG_009240.1: g.106737G>T (traditional nomenclature: ZRS404G>T) in the ZRS within the LMBR1 gene. 24777739_A novel ZRS mutation found in the Mexican population, 402C>T, suggests that a dosage effect exists for this mutation. 26749485_These include a patient with hypoplastic phalanges and absent hallux bilaterally with de novo deletion of 11.9 Mb on 7p21.1-22.1 spanning 63 genes including RAC1, another patient with severe Holt-Oram syndrome and a large de novo deletion 2.2 Mb on 12q24.13-24.21 spanning 20 genes including TBX3 and TBX5, and a third patient with acheiropodia who had a nullizygous deletion of 102 kb on 7q36.3 spanning LMBR1 31395945_Results identify a novel g.101779T>A variant segregating with all individuals affected with preaxial polydactyly type 1(PPDI). 32179704_Variable expression of subclinical phenotypes instead of reduced penetrance in families with mild triphalangeal thumb phenotypes. 32662247_Large duplication in LMBR1 gene in a large Chinese pedigree with triphalangeal thumb polysyndactyly syndrome. | ENSMUSG00000010721 | Lmbr1 | 688.51957 | 1.0600339 | 0.0841103714 | 0.15670369 | 2.872213e-01 | 5.920067e-01 | 9.998360e-01 | No | Yes | 633.99905 | 123.257193 | 6.685810e+02 | 100.141352 | |
| ENSG00000106049 | 11112 | HIBADH | protein_coding | P31937 | 3D-structure;Acetylation;Branched-chain amino acid catabolism;Direct protein sequencing;Mitochondrion;NAD;Oxidoreductase;Reference proteome;Transit peptide | PATHWAY: Amino-acid degradation; L-valine degradation. {ECO:0000269|PubMed:16466957}. | This gene encodes a mitochondrial 3-hydroxyisobutyrate dehydrogenase enzyme. The encoded protein plays a critical role in the catabolism of L-valine by catalyzing the oxidation of 3-hydroxyisobutyrate to methylmalonate semialdehyde. [provided by RefSeq, Nov 2011]. | hsa:11112; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; 3-hydroxyisobutyrate dehydrogenase activity [GO:0008442]; NAD binding [GO:0051287]; NADP binding [GO:0050661]; oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [GO:0016616]; valine catabolic process [GO:0006574] | 19834535_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 23423614_HIBADH is involved in the mitochondrial function of spermatozoa, and maintains sperm motility. It may serve as a sperm-motility marker. | ENSMUSG00000029776 | Hibadh | 808.24065 | 1.0669244 | 0.0934578979 | 0.13714519 | 4.698549e-01 | 4.930539e-01 | 9.998360e-01 | No | Yes | 806.78035 | 138.617632 | 8.795482e+02 | 116.630972 | |
| ENSG00000106100 | 10392 | NOD1 | protein_coding | Q9Y239 | FUNCTION: Enhances caspase-9-mediated apoptosis. Induces NF-kappa-B activity via RIPK2 and IKK-gamma. Confers responsiveness to intracellular bacterial lipopolysaccharides (LPS). Forms an intracellular sensing system along with ARHGEF2 for the detection of microbial effectors during cell invasion by pathogens. Required for RHOA and RIPK2 dependent NF-kappa-B signaling pathway activation upon S.flexneri cell invasion. Involved not only in sensing peptidoglycan (PGN)-derived muropeptides but also in the activation of NF-kappa-B by Shigella effector proteins IpgB2 and OspB. Recruits NLRP10 to the cell membrane following bacterial infection. {ECO:0000269|PubMed:11058605, ECO:0000269|PubMed:17054981, ECO:0000269|PubMed:19043560, ECO:0000269|PubMed:22672233}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Cell membrane;Cytoplasm;Immunity;Innate immunity;Leucine-rich repeat;Lipoprotein;Membrane;Nucleotide-binding;Palmitate;Reference proteome;Repeat;Ubl conjugation | This gene encodes a member of the nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) family of proteins. The encoded protein plays a role in innate immunity by acting as a pattern-recognition receptor (PRR) that binds bacterial peptidoglycans and initiates inflammation. This protein has also been implicated in the immune response to viral and parasitic infection. Major structural features of this protein include an N-terminal caspase recruitment domain (CARD), a centrally located nucleotide-binding domain (NBD), and 10 tandem leucine-rich repeats (LRRs) in its C terminus. The CARD is involved in apoptotic signaling, LRRs participate in protein-protein interactions, and mutations in the NBD may affect the process of oligomerization and subsequent function of the LRR domain. Mutations in this gene are associated with asthma, inflammatory bowel disease, Behcet disease and sarcoidosis in human patients. [provided by RefSeq, Aug 2017]. | hsa:10392; | anchored component of plasma membrane [GO:0046658]; apical plasma membrane [GO:0016324]; basolateral plasma membrane [GO:0016323]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; phagocytic vesicle [GO:0045335]; ATP binding [GO:0005524]; CARD domain binding [GO:0050700]; cysteine-type endopeptidase activator activity involved in apoptotic process [GO:0008656]; identical protein binding [GO:0042802]; pattern recognition receptor activity [GO:0038187]; peptidoglycan binding [GO:0042834]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; cellular response to muramyl dipeptide [GO:0071225]; defense response [GO:0006952]; defense response to bacterium [GO:0042742]; defense response to Gram-positive bacterium [GO:0050830]; detection of bacterium [GO:0016045]; detection of biotic stimulus [GO:0009595]; inflammatory response [GO:0006954]; innate immune response [GO:0045087]; intracellular signal transduction [GO:0035556]; pattern recognition receptor signaling pathway [GO:0002221]; positive regulation of cell death [GO:0010942]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; positive regulation of dendritic cell antigen processing and presentation [GO:0002606]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of interleukin-1 beta production [GO:0032731]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of interleukin-8 production [GO:0032757]; positive regulation of JNK cascade [GO:0046330]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of nitric-oxide synthase activity [GO:0051000]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of xenophagy [GO:1904417]; regulation of apoptotic process [GO:0042981]; signal transduction [GO:0007165] | 12775719_role in CARD6 modulation of NF-kappa B activation 12791997_results show that Nod1 specifically detects a unique diaminopimelate-containing N-acetylglucosamine-N-acetylmuramic acid (GlcNAc-MurNAc) tripeptide motif found in Gram-negative bacterial peptidoglycan, resulting in activation of the NF-kappaB pathway 12813035_NOD1/CARD4 is activated by interferon gamma in intestinal mucosal inflammation 12871942_Using a wide array of natural or modified muramyl peptides, it is shown that Nod1 and Nod2 have evolved divergent strategies to achieve peptidoglycan sensing 14977954_signaling through Nod1 is required for activating NF-kappaB in human intestinal epithelial cells infected with gram-negative enteric bacteria that can bypass TLR activation 15718249_NOD1 variation has a role in inflammatory bowel disease and childhood asthma 15718249_Observational study of gene-disease association. (HuGE Navigator) 15790594_inflammatory bowel disease susceptibility 15990792_Observational study of gene-disease association. (HuGE Navigator) 15990792_genetic variants within NOD1 are important determinants of atopy susceptibility. 16083881_Nod1 is demonstrated to be a client protein of the Hsp90 chaperone complex containing the Chp-1. 16115863_there is cross-talk between the Nod1 and Nod2 pathways; down-regulation of the Nod1/M-Tri(DAP) pathway may be associated with Crohn disease 16172124_analysis of the molecular mechanisms responsible for the detection of bacterial peptidoglycan by Nod1 16260731_IL-32 synergizes with nucleotide oligomerization domain (NOD) 1 and NOD2 ligands for IL-1beta and IL-6 production through a caspase 1-dependent mechanism. 16446438_absence of Nod1 correlates with tumor growth, an increased sensitivity to estrogen-induced cell proliferation, and a failure to undergo Nod1-dependent apoptosis 16464805_Finds somatic mutation in P-loop domains of proapoptotic NOD1 genes uncommon in colon cancers. 16493424_Review focuses on the molecular interactions by which NOD1 and NOD2 contribute to the maintenance of mucosal homeostasis and the induction of mucosal inflammation. 16741608_Observational study of gene-disease association. (HuGE Navigator) 16741608_Polymorphism observed in the NOD1/CARD4 gene is not genetic susceptibility factors for Crohn's disease or ulcerative colitis in Turkey. 16893397_Observational study of gene-disease association. (HuGE Navigator) 16893397_The results suggest that the analysed CARD4 mutations do not play a major role in the aetiology of coronary heart disease. 16918516_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16918516_Polymorphisms significantly modify the protective effect of exposure to a farming environment on allergies. 16935475_Observational study of gene-disease association. (HuGE Navigator) 16935475_The results indicate that impaired recognition of intracellular P. acnes through NOD1 affects the susceptibility to sarcoidosis in the Japanese population. 17005562_CENTB1 selectively down-regulates NF-kappaB activation via NODs pathways, creating a 'feedback' loop and suggesting a novel role of CENTB1 in innate immune responses to bacteria and inflammatory responses 17012967_Observational study of gene-disease association. (HuGE Navigator) 17030188_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17054981_high resolution NMR structure of NOD1 CARD; mutational analysis shows that interaction of NOD1 with RICK is critically dependent on 3 acidic residues on NOD1 CARD & 3 basic residues on RICK CARD & is likely to have a strong electrostatic component 17100974_Observational study of gene-disease association. (HuGE Navigator) 17156193_Both NOD1 and NOD2 were expressed by first trimester placental villi and localized to trophoblast cells 17285593_Observational study of gene-disease association. (HuGE Navigator) 17285593_This variant allele of NOD1/CARD4+32656 is not associated with a strong effect on susceptibility to IBD in children and adults in Northern Europe. 17309748_NOD1 gene polymorphism increases the risk of peptic ulceration in H. pylori-positive patients. 17309748_Observational study of gene-disease association. (HuGE Navigator) 17322292_lipophilic peptidoglycan-related molecules have roles in induction of Nod1-mediated immune responses 17403538_epithelial cells, did not secrete IL-6, IL-8 or monocyte chemoattractant protein-1 in response to NOD1 and NOD2 agonists; stimulation with NODs ligands induced beta-defensin 2 generation in all epithelial cells examined 17452051_These results suggest that anti-PR3 Abs prime human monocytic cells to produce cytokines upon stimulation with various bacterial components by up-regulating the TLR and NOD signaling pathway. 17521327_NOD1, but not NOD2 is a major PRR for C. jejuni in IEC. 17595233_Observational study of gene-disease association. (HuGE Navigator) 17613538_NOD2/CARD15 variant carriage had no influence on NOD1/CARD4 effect on inflammatory bowel disease susceptibility. 17613538_Observational study of gene-disease association. (HuGE Navigator) 17620097_Observational study of gene-disease association. (HuGE Navigator) 17907287_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17907287_Suggest population differences in the inheritance of NOD1 polymorphism and NOD2 mutations. Relationship between disease location and Nod-like receptor molecules was established. 17964870_Carriage of the NOD1 G796A mutation increases susceptibility for Crohn's disease in the Hungarian population. 17964870_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17970764_Nod1 localization at the plasma membrane in human cells is dependent on the integrity of the protein, on its signalling capacity and on an intact actin cytoskeleton. 18186648_homodimerization of Nod1_CARD is achieved by swapping the H6 helices at the carboxy termini and stabilized by forming an interchain disulfide bond between the Cys39 residues of the two monomers in solution and in the crystal. 18397186_Observational study of gene-disease association. (HuGE Navigator) 18426885_We conclude that the endogenous IL-8 response induced by C. trachomatis infection is dependent upon NOD1 signaling through RIP2 as part of a signal system requiring multiple inputs for optimal IL-8 induction. 18521924_Observational study of gene-disease association. (HuGE Navigator) 18536738_The NOD1 receptors may have a role in preventing disruption of the epithelial barrier in lung, during inflammatory states. 18573991_in innate immune responses to invading microbes, a combination of signaling through TLRs and NOD1/2 leads to the synergistic activation of antibacterial responses in the oral epithelium. 18574154_These data indicate that the NLR family members Nod1 and Nod2 have different functions in controlling inflammation, and that intracellular Nod1-Nod2 interactions may determine the severity of arthritis in this experimental model. 19043560_GEF-H1 is a critical component of cellular defenses forming an intracellular sensing system with NOD1 for the detection of microbial effectors during cell invasion by pathogens. 19050632_Observational study of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19120480_polymorphisms are associated with increased risk of developing atopic eczema and asthma 19122645_There is functional expression of the intracellular pattern recognition receptor NOD1 in human keratinocytes. 19167431_associated with atopic disease in farmers' children but not in children unexposed to a farming environment 19247692_Observational study of gene-disease association. (HuGE Navigator) 19260860_expression in the trophoblast is upregulated by bacterial lipopolysaccharide, through Toll-like receptor 4, in a NFkappaB-dependent manner 19264973_Observational study of gene-disease association. (HuGE Navigator) 19273470_Data show that NOD1 and 2 may have a role in innate immune protection in the uterus, and NOD2 may regulate inflammation associated with menstruation. 19327158_Observational study of gene-disease association. (HuGE Navigator) 19401779_both TLR2 and NOD1 may be involved in innate immune sensing of H. muridarum by epithelial cells 19406482_detected on all female reproductive tract tissues 19423540_Observational study of gene-disease association. (HuGE Navigator) 19538217_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19538217_Polymorphism in NOD1 G796A alone did not prove to be a risk factor for stroke in general, but in association with C. pneumoniae infection it appeared to be accompanied by an increased risk of the development of stroke. 19570976_Nod1-activating ligands entered cells through endocytosis with optimal pH ranging from 5.5 to 6.0. 19587002_Observational study of gene-disease association. (HuGE Navigator) 19587002_no association of gene polymorphism with periodontitis in North-European patients 19660916_no association of NOD1 Glu266Lys polymorphism has been revealed with the Japanese leprosy patients. 19667203_Data suggest a role for XIAP in regulating innate immune responses by interacting with NOD1 and NOD2 through interaction with RIP2. 19723304_Observational study of gene-disease association. (HuGE Navigator) 19723304_We propose that the location of mutations in the Exon 6 spanning the ATP and Mg2+ binding site of NBD in NOD1 gene may affect the process of oligomerization and subsequent function of the LRR domain 19818630_Studies provide molecular model for TLR and NLR signalling pathways. 19843337_Observational study of gene-disease association. (HuGE Navigator) 19843337_Variation in the innate immunity genes CARD4, CARD8 and CARD15 is unlikely to play a major role in the susceptibility to CRC in the German population. 19882212_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19882212_These results suggest that carriage of the NOD1 G796A mutation increases the susceptibility of gastric epithelial cells for intestinal metaplasia and atrophy when infected by CagA-positive Helicobacter pylori strains. 19898471_Results link bacterial sensing by Nod proteins to the induction of autophagy and provide a functional link between Nod2 and ATG16L1. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20002790_expression of NOD1, NOD2 and NLRP3 messenger RNA was determined in neutrophils 20007577_NOD1-dependent extracellular signal-related mitogen-activated kinase (MAPK) activation represents an additional signaling pathway through which Helicobacter pylori controls transcription of novel downstream target genes during infection. 20039881_This study demonstrates, the involvement of NOD1 and DEFB4 in direct killing of Helicobacter pylori bacteria by epithelial cells and confirms the importance of NOD1 in host defence mechanisms against cagPAI(+)Helicobacter pylori infection. 20124104_Data show that Nod1 and Nod2 were expressed highly in both human and mouse RTE cells. 20300534_Helicobacter pylori products, NOD1, ubiquitinated proteins and proteasome accumulate in a novel cytoplasmic structure 20384614_Nod1, Nod2, and Nalp3 receptors were found to be present in the human nose. The expression of Nod1 and Nalp3 were down-regulated during pollen season among patients with allergic rhinitis 20389019_NOD1 can activate the ISGF3 signaling pathway that is usually associated with protection against viral infection to provide robust type I IFN-mediated protection from H. pylori and possibly other mucosal infections 20395963_Observational study of gene-disease association. (HuGE Navigator) 20403997_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20470879_The results showed that Orientia tsutsugamushi infection activated the NOD1 pathway followed by IL-32 secretion, thus resulting in the production and expression of IL-1beta, IL-6, IL-8, and ICAM-1 endothelial cells. 20485444_Observational study of gene-disease association. (HuGE Navigator) 20519512_Characterization of natural human nucleotide-binding oligomerization domain protein 1 (Nod1) ligands from bacterial culture supernatant 20595247_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20646321_Observational study of gene-disease association. (HuGE Navigator) 20713621_We propose that alpha(5)beta(1) integrin, which is associated with cholesterol-rich microdomains at the host cell surface, is required for NOD1 recognition of peptidoglycan and subsequent induction of NF-kappaB-dependent responses to H. pylori. 20818820_Meta-analysis of gene-disease association. (HuGE Navigator) 20818820_an association between NOD1/CARD4 insertion/deletion polymorphism and inflammatory bowel disease in the younger age group at onset (< 40 years) in Caucasian populations. 20844241_Functional NLRs are expressed in human B cells. NOD1 and NOD2 have the ability to augment B cell receptor-induced activation independently of physical T cell help. 20846183_Klebsiella inhibits Rac1 activation; and inhibition of Rac1 activity triggers a NOD1-mediated CYLD and MKP-1 expression which in turn attenuates IL-1beta-induced IL-8 secretion. 21042538_findings suggest that NOD1 and NOD2 as well as TLRs are involved in regulating the differentiation of MSCs 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21241317_NOD1, NOD2, and NALP3 mRNA and protein were seen in all tissue specimens, but were higher in nasal polyps than in normal nasal mucosa 21263023_study shows that non-O1 non-O139 serogroups outer membrane vesicles elicit immune responses mediated by NOD1 and NOD2 in mammalian host cells 21310790_two nucleotide-binding domain and leucine-rich repeats employ different modes of activation and propose distinct models for activation of NOD1 and NOD2. 21330608_Nod1 ligands induce coronary inflammation. 21342182_NOD1, NOD2 and RIG-1/MDA-5 have a role in T-cell activation. 21677137_Functional NOD1 is expressed by trophoblast cells across gestation and may have a role in mediating bacterial infection-associated inflammation and prematurity. 21722177_Study reveals a polymorphism in intron 9 of the NOD1 gene associated with the severity of ulcerative colitis in North Indian patients. 21739538_NOD2 and NOD1 variants displayed antagonist effects on the risk of Crohn disease and anti-saccharomyces cerevisiae antibody level. 21745515_NOD1/CARD4 and NOD2/CARD15 gene polymorphisms may be associated with altered risk for a large variety of cancers in humans. (Review) 21757725_Tri-DAP interacts directly with the LRR domain of NOD1 and consequently increases RICK/NOD1 association and RICK phosphorylation activity 21864388_There was a tendency for NOD1 796G/G genotype to be associated with increased risk of high risk atrophic gastritis. 21924184_might participate in dental pulp inflammation through chemokine production via MAPK signaling pathways 21978001_Data show that eosinophils expressed NOD1 and NOD2 mRNA and protein, low levels of RIG-I and MDA-5, and NLRP3 was completely absent. 22062585_Here we describe the large-scale expression of recombinant NOD1 protein in non-adherent mammalian cells. 22143988_NOD-1 is strongly expressed in different cell types in the synovial tissue of patients with rheumatoid arthritis. 22218461_NOD1 and NOD2 are functionally expressed in human periodontal ligament cells and can trigger innate immune responses. 22507623_A haplotype, G-T-C-C, in the NOD1 gene, was associated with lower risk for Kawasaki disease development. 22563200_Genetic polymorphisms in NOD1 and NOD2 may interact with H. pylori infection and may play important roles in promoting the development of gastric cancer in the Chinese population. 22763410_Nod1, but not the ASC inflammasome, contributes to induction of IL-1beta secretion in human trophoblasts after sensing of Chlamydia trachomatis. 22795647_cooperative activation of DCs with NOD1 and NOD2 agonists and TLR7/8 ligands results in a synergistic release of pro-inflammatory mediators which promote the activation of IL-17-producing T cells. 22848741_NOD1 might function as an alternative costimulatory receptor in CD8 T cells 22870324_A key role for the endothelium in NOD1 mediated vascular inflammation: comparison to TLR4 responses. 22902391_ND1+32656 GG and IL-8-251 T/T allele may be associated with less reactivity to H. pylori infection, and may increase the risk of erosive esophagitis even in H. pylori infected Japanese population. 23062636_NOD1 gene silencing by siRNA reduced NOD1 ligand-induced MCP-1,IL-6, and IL-8 release and increased insulin-induced glucose uptake 23107019_These findings reveal a novel pathway whereby Helicobacter pylori CagL, via interaction with host integrins, can trigger pro-inflammatory responses independently of CagA translocation or NOD1 signalling. 23136938_NOD 1 polymorphism is closely correlated with H. pylori-associated gastric mucosal inflammation in the Korean population. 23182460_The present study aimed to determine the presence of NOD1 and NOD2 in human adipose cells as well as to assess their functionality. NOD1 activation reduced insulin-induced glucose uptake, leading to insulin resistance. 23275338_Vibrio cholerae O395 outer membrane vesicles modulate intestinal epithelial cells in a NOD1 protein-dependent manner and induce dendritic cell-mediated Th2/Th17 cell responses. 23300079_Data suggest that ubiquitin (Ub) binding provides a negative feedback loop upon NOD1 and NOD2 (nucleotide-binding oligomerization domain-containing proteins)-dependent activation of receptor-interacting protein kinase 2 (RIP2). 23353647_Genetic variation of human NOD1 protein is not associated with rheumatoid arthritis susceptibility or severity. 23382681_Cyclosporine A impairs nucleotide binding oligomerization domain (Nod1)-mediated innate antibacterial renal defenses in mice and human transplant recipients. 23460743_cross-talk between NOD1 and IFN-gamma signaling pathways contribute to H. pylori-induced inflammatory responses, potentially revealing a novel mechanism whereby virulent H. pylori strains promote more severe disease. 23711367_A novel function for Ankrd17 in Nod1 and Nod2 mediated anti-bacterial innate immune pathways. 23712977_Activation of NOD1 and NOD2 both significantly suppressed adipocyte differentiation of human adipose-derived adult stem cells, demonstrating the species specific effects of NOD activation. 23740944_NOD1 activation increases in laboring fetal membranes and myometrium and with bacterial infection 23839082_By forming a complex with NOD1, PSMA7 promotes NOD1 degradative polyubiquitination, decreases its protein expression and apoptosis induced by NOD1 activation. 23888881_No significant association could be identified with rs2907749 of NOD1 regarding hypersensitivity to beta-lactam and total immunoglobulin (Ig)E, despite association with inflammation and atopy. 23901396_NOD1 and NOD2 receptors appear to recognize both invasive and non-invasive forms of the bacteria. (Review) 23935490_Data suggest that Ssph2 (ubiquitin ligase effector from Salmonella typhimurium) significantly enhances human NOD1-mediated secretion of interleukin-8; SspH2 interacts with and ubiquitinates NOD1. 24018334_NOD1 and NOD2 mRNA expression were significantly up-regulated in monocytes from patients with type 2 diabetes. 24041848_NOD1 may have a role in mediating infection-associated inflammation 24053487_The expression profile of NLRs in head and neck cancer cells differed from that seen in healthy epithelial cells. 24143223_Helicobacter pylori cag pathogenicity island (cagPAI) involved in bacterial internalization and IL-8 induced responses via NOD1- and MyD88-dependent mechanisms in human biliary epithelial cells 24295830_bacterial infection-mediated activation of NOD1,2, together with IL-32gamma, can synergize the activation of eosinophils interacting with bronchial epithelial cells. 24366254_The Nod1/2-Rip2 axis was critical to induce optimal cytokine and chemokine responses to A. baumannii infection. 24444388_Monocyte expression and function of NOD1 and NOD2 in very preterm infants are intact and comparable/equivalent to term infants and adults. 24598002_Data indicate that the mutation of serine 7 does not affect nucleotide-binding oligomerization domain-containing protein 1 (NOD1) function. 24661094_NOD1 priming of human dendritic cells promoted a Th2 polarization profile that involved the production of CCL17 and CCL22 in nonallergic subjects but only CCL17 in allergic patients. 24690886_Human embryonic stem cell-derived endothelial cells are TLR4 deficient but respond to bacteria via NOD1. 24746552_NOD1 and RIP2 interact with bacterial peptidoglycan on endosomes to promote autophagy and inflammatory signaling. 24829218_NOD1 has a role in the development of insulin resistance and inflammation in pregnancies complicated by gestational diabetes mellitus. 24832447_Endosomal membranes are sites of NOD1/NOD2-dependent signal transduction. 24862550_Porphyromonas gingivalis induced NOD1 overexpression in endothelial cells and that NOD1 played an important role in the process of VCAM-1 and ICAM-1 expression in endothelial cells infected with Porphyromonas gingivalis through the NF-kappaB signaling pathway. 24934088_NOD1 has an active role in the heightened inflammatory environment associated with both experimental murine and human diabetic cardiac disease. 24958724_the NOD1-RIP2 signaling axis is more complex than previously assumed, that simple engagement of RIP2 is insufficient to mediate signaling 25012219_Data indicate that E3 ligase RNF34 is associated with nucleotide-binding oligomerization domain-containing protein 1 (NOD1). 25024364_This work reveals a role of Aggregatibacter actinomycetemcomitans outer membrane vesicles as a trigger of innate immunity via carriage of NOD1- and NOD2-active pathogen-associated molecular patterns (PAMPs). 25503380_We found that RNA interference-induced Caspase-12 silencing increased NOD1, hBD1 and hBD2 expression 25801093_NOD1 signaling results in the induction of the cellular degradation pathway of autophagy and the development of pro-inflammatory responses that activate the adaptive immune system. 25840495_The higher NOD1 and NOD2 were observed in villi from patients who experienced recurrent spontaneous abortion compared with those who experienced a normal pregnancy. 25854006_A significant association of the NOD1 +32656 GG insertion variant with protection against infection with Chlamydia trachomatis has been detected [p: 0.0057; OR: 0.52]. 25933107_NOD1 rs2709800, NOD2 rs718226, rs2111235, rs7205423 and interaction between rs718226 and H. pylori infection may be related to risk of gastric lesions. 26052894_Activation of the innate immune pathway via NOD1 may be partially responsible for the increased systemic inflammation and insulin resistance in metabolic syndrome. 26238283_NOD1/CARD4 gene may influence the diagnosis and treatment of lung cancer 26759244_findings illuminated a role for NOD1 signaling in attenuating H. pylori-induced Cdx2 expression in gastric epithelial cells 26880761_contributes to fetal growth retardation and death through vascular inflammation 26902715_The changes in the nucleotide-binding oligomerization domain-like receptors (NLRs) in human corneas with disease expression may reflect different susceptibility to infectious and non-infectious injuries in corneas with various diseases. 26915562_Brain pericytes can sense Gram-negative bacterial products by both NOD1 and TLR4 receptors, acting through distinct pathways. 26980698_over-expression of NOD1 and NOD2 may be involved in the pathogenesis of Vogt-Koyanagi-Harada (VKH) Syndrome syndrome. 27000222_NOD1 And NOD2 polymorphisms were associated with increased susceptibility to Guillain-Barre syndrome in a Northern Indian population. 27007849_NOD1 and NOD2, two members of the NOD-like receptor family of pattern recognition receptors, are important mediators of ER-stress-induced inflammation in mouse and human cells 27099311_findings show that NOD1, a PRR that normally senses bacterial peptidoglycans, is activated by HCV viral polymerase, probably through an interaction with dsRNA, suggesting that NOD1 acts as an RNA ligand recognition receptor. 27591899_Fusion of human SGT1 (hSGT1) to NOD1 LRR significantly enhanced the solubility, and the fusion protein was stabilized by coexpression of mouse Hsp90alpha. 27830463_this study reveals that LRRK2 is a new positive regulator of Rip2 and promotes inflammatory cytokine induction through the Nod1/2-Rip2 pathway. 27836539_Finally, NOD1 agonist increased the formation of cranial and subintestinal vessel plexus in zebrafish, and this effect was abrogated by concurrent PPARgamma activation. Overall, these findings identify a PPARgamma-miR-125a-NOD1 signaling axis in endothelial cells that is critical in the regulation of inflammation-mediated angiogenesis. 27856764_A role for NOD1 in HCMV control via RIPK2- IKKalpha-IRF3 signaling, NOD1 polymorphisms predict the risk of infection. 27885704_NOD1/2 gene variants are not linked with T2DM and IR. 27917621_the results suggest that the chronic activation of NOD1 and NOD2 receptors might play a role in the development of gastric cancer. 27936005_Nucleotide-binding oligomerization domain (NOD1) was the most significantly associated gene when analyzing exonic rare variants (RVs) in chromosome 7p to carotid bifurcation intima-media thickness (bIMT). 28114344_study provides structural and dynamic insights into the NOD1-RIP2 oligomer formation, which will be crucial in understanding the molecular basis of NOD1-mediated CARD-CARD interaction in higher and lower eukaryotes 28432285_Study proposes that NOD1 contributes to inflammation not only by promoting pro-inflammatory processes, but also by suppressing anti-inflammatory pathways. 28536249_Bronchial epithelial overexpression of TLR4 and NOD1 in severe/very severe stable COPD, associated with increased bronchial inflammation and P. aeruginosa bacterial load, may play a role in the pathogenesis of COPD 28592872_our work indicates that NOD1 plays a previously undetected protective role in larval survival through CD44a-mediated activation of the PI3K-Akt signaling. 28673961_Based on molecular docking studies using PG ligands, we propose few residues - G825, D826 and N850 in hNOD1-LRR and L904, G905, W931, L932 and S933 in hNOD2-LRR, evolutionarily conserved across different host species, which may play a major role in ligand recognition. 28768269_NOD1 (rs6958571) SNP was associated with gram-positive blood stream infection in Caucasian infants and extremely low birth weight infants. 29030987_NOD1 expression seems to be modulated by 5-HT and other immune receptors as TLR2 and TLR4. This study could clarify the relation between both the intestinal serotonergic system and innate immune system, and their implications in intestinal inflammation. 29211798_In transgenic mice expressing human NOD1 and deficient for the murine NOD1, we showed enhanced clearance of a lipl21- mutant of Leptospira interrogans compared to the complemented strain, or to what was observed in NOD1KO mice, suggesting that LipL21 facilitates escape from immune surveillance in humans. 29615116_Overexpression of either NOD1 or NOD2 reduces cell proliferation and increases clonogenic potential in vitro in breast cancer cell lines. 29760746_Upregulation of miR-495 ameliorates the high glucose-induced inflammatory, cell differentiation and extracellular matrix accumulation of human CFs by modulating both the NF-kappaB and TGF-beta1/Smad signaling pathways through downregulation of NOD1 expression. 30150286_this study reports the molecular and functional identification of an NLRP1 homolog, Danio rerio NLRP1 (DrNLRP1) from a zebrafish (D. rerio) model 30321637_NOD1 and NOD2 expression and induced release of pro-inflammatory mediators were impaired in infants, contributing to the high susceptibility of infants to infection 30332343_Two Single-Nucleotide Polymorphisms associated with susceptibility to develop dengue in NOD1 or RIPK2 genes were observed Children from Colombia. 30384473_results demonstrate that Evodiamine (Evo) could induce apoptosis remarkably and the inhibitory effect of Evo on Hepatocellular cancer cells may be through suppressing the NOD1 signal pathway in vitro and in vivo. 30685324_Nucleotide-binding oligomerization domain 1 (NOD1) modulates liver ischemia reperfusion through the expression adhesion molecules. 30717343_our results indicate that Columbianadin (CBN) suppressed the LPS-mediated inflammatory response by inhibiting NOD1/NF- kappa B activation. Further investigations are required to determine the mechanisms of action of CBN in the inhibition of NOD signaling: However, CBN may be employed as a therapeutic agent for multiple inflammatory diseases. 30874883_The NOD1 rs2075820 variant was associated with a higher childhood asthma risk and the NOD1 expression at mRNA and protein levels was significantly increased in asthma patients. 30903318_NOD1 and NOD2 mRNA was constitutively expressed in both tumor and adjacent healthy renal tissue, with NOD1 being significantly lower and in contrast NOD2 significantly higher expressed in tumor tissue compared to healthy tissues. 31123889_REVIEW: various mechanisms and pathways involved in H. pylori induction of NF-kappaB-dependent responses in gastric epithelial cells, including a 'state-of-the-art' review on the respective roles of NOD1 and ALPK1/TIFA pathways in these responses 31649195_ZDHHC5-mediated S-palmitoylation is indispensable for NOD1/2 recruitment to bacteria containing phagosomes. 31956962_Activation of the pattern recognition receptor NOD1 augments colon cancer metastasis. 32578848_Genetic variation in NOD1/CARD4 and NOD2/CARD15 immune sensors and risk of osteoporosis. 32677123_NOD1 and NOD2 in inflammatory and infectious diseases. 32927803_Deletion or Inhibition of NOD1 Favors Plaque Stability and Attenuates Atherothrombosis in Advanced Atherogenesis (dagger). 33293463_A small sustained increase in NOD1 abundance promotes ligand-independent inflammatory and oncogene transcriptional responses. 33503439_Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. 33529885_Decidual and placental NOD1 is associated with inflammation in normal and preeclamptic pregnancies. 33879265_NOD1 rs2075820 (p.E266K) polymorphism is associated with gastric cancer among individuals infected with cagPAI-positive H. pylori. 33942347_Cellular stress promotes NOD1/2-dependent inflammation via the endogenous metabolite sphingosine-1-phosphate. 34294014_Association of NOD1, NOD2, PYDC1 and PYDC2 genes with Behcet's disease susceptibility and clinical manifestations. 34825931_Activation of RIPK2-mediated NOD1 signaling promotes proliferation and invasion of ovarian cancer cells via NF-kappaB pathway. 35033591_Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-kappaB pathway. 35130902_Upregulation of NOD1 and NOD2 contribute to cancer progression through the positive regulation of tumorigenicity and metastasis in human squamous cervical cancer. 35225652_The AmiC/NlpD Pathway Dominates Peptidoglycan Breakdown in Neisseria meningitidis and Affects Cell Separation, NOD1 Agonist Production, and Infection. 35430507_NOD1 activation promotes cell apoptosis in papillary thyroid cancer. | ENSMUSG00000038058 | Nod1 | 227.68805 | 1.2093628 | 0.2742471143 | 0.20473739 | 1.797868e+00 | 1.799705e-01 | 9.998360e-01 | No | Yes | 268.22746 | 40.692182 | 2.115779e+02 | 25.153301 | |
| ENSG00000106290 | 6878 | TAF6 | protein_coding | P49848 | FUNCTION: TAFs are components of the transcription factor IID (TFIID) complex, PCAF histone acetylase complex and TBP-free TAFII complex (TFTC). TIIFD is multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors.; FUNCTION: [Isoform 4]: Transcriptional regulator which acts primarily as a positive regulator of transcription (PubMed:20096117, PubMed:29358700). Recruited to the promoters of a number of genes including GADD45A and CDKN1A/p21, leading to transcriptional up-regulation and subsequent induction of apoptosis (PubMed:11583621). Also up-regulates expression of other genes including GCNA/ACRC, HES1 and IFFO1 (PubMed:18628956). In contrast, down-regulates transcription of MDM2 (PubMed:11583621). Acts as a transcriptional coactivator to enhance transcription of TP53/p53-responsive genes such as DUSP1 (PubMed:20096117). Can also activate transcription and apoptosis independently of TP53 (PubMed:18628956). Drives apoptosis via the intrinsic apoptotic pathway by up-regulating apoptosis effectors such as BCL2L11/BIM and PMAIP1/NOXA (PubMed:29358700). {ECO:0000269|PubMed:11583621, ECO:0000269|PubMed:18628956, ECO:0000269|PubMed:20096117, ECO:0000269|PubMed:29358700}. | 3D-structure;Alternative splicing;Apoptosis;Direct protein sequencing;Disease variant;Isopeptide bond;Mental retardation;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | Initiation of transcription by RNA polymerase II requires the activities of more than 70 polypeptides. The protein that coordinates these activities is transcription factor IID (TFIID), which binds to the core promoter to position the polymerase properly, serves as the scaffold for assembly of the remainder of the transcription complex, and acts as a channel for regulatory signals. TFIID is composed of the TATA-binding protein (TBP) and a group of evolutionarily conserved proteins known as TBP-associated factors or TAFs. TAFs may participate in basal transcription, serve as coactivators, function in promoter recognition or modify general transcription factors (GTFs) to facilitate complex assembly and transcription initiation. This gene encodes one of the smaller subunits of TFIID that binds weakly to TBP but strongly to TAF1, the largest subunit of TFIID. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2010]. | hsa:6878; | cytosol [GO:0005829]; MLL1 complex [GO:0071339]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; SAGA complex [GO:0000124]; SLIK (SAGA-like) complex [GO:0046695]; transcription factor TFIID complex [GO:0005669]; transcription factor TFTC complex [GO:0033276]; aryl hydrocarbon receptor binding [GO:0017162]; DNA binding [GO:0003677]; protein heterodimerization activity [GO:0046982]; RNA polymerase II general transcription initiation factor activity [GO:0016251]; transcription coactivator activity [GO:0003713]; apoptotic process [GO:0006915]; DNA-templated transcription, initiation [GO:0006352]; histone H3 acetylation [GO:0043966]; monoubiquitinated histone deubiquitination [GO:0035521]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; mRNA transcription by RNA polymerase II [GO:0042789]; negative regulation of cell cycle [GO:0045786]; negative regulation of cell population proliferation [GO:0008285]; positive regulation of apoptotic process [GO:0043065]; positive regulation of intrinsic apoptotic signaling pathway [GO:2001244]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; protein phosphorylation [GO:0006468]; regulation of transcription by RNA polymerase II [GO:0006357]; RNA polymerase II preinitiation complex assembly [GO:0051123]; transcription by RNA polymerase II [GO:0006366]; transcription initiation from RNA polymerase II promoter [GO:0006367] | 15328371_a novel protein-protein interaction was observed between TAFII70 (not TAFII80) and GADD45a 15601843_histone fold domain mediated interaction enhances the DNA binding activity of each of the TAF6-TAF9 and TAF4b-TAF12 pairs and of a histone-like octamer complex composed of the four TAFs 18628956_TAF6delta has a pivotal node in a signaling pathway that controls gene expression programs and apoptosis in the absence of p53 20096117_The transcriptome data uncovered novel links between TAF6delta expression and the Notch, oxidative stress response, integrin, p53, p53 feedback loop 2, and angiogenesis pathways. 22696218_TFIID TAF6-TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. 25025302_data point to several new RNA elements that can modulate TAF6delta and also reveal a role for RNA secondary structure in the selection of TAF6delta 29358700_BIM and NOXA contribute to TAF6delta-dependent cell death. 32030742_The third family with TAF6-related phenotype: Alazami-Yuan syndrome. | ENSMUSG00000036980 | Taf6 | 2268.33842 | 0.9711459 | -0.0422400703 | 0.10374592 | 1.682959e-01 | 6.816307e-01 | 9.998360e-01 | No | Yes | 2346.19716 | 259.759240 | 2.213594e+03 | 189.911525 | |
| ENSG00000106692 | 2218 | FKTN | protein_coding | O75072 | FUNCTION: Catalyzes the transfer of a ribitol-phosphate from CDP-ribitol to the distal N-acetylgalactosamine of the phosphorylated O-mannosyl trisaccharide (N-acetylgalactosamine-beta-3-N-acetylglucosamine-beta-4-(phosphate-6-)mannose), a carbohydrate structure present in alpha-dystroglycan (DAG1) (PubMed:26923585, PubMed:29477842, PubMed:27194101). This constitutes the first step in the formation of the ribitol 5-phosphate tandem repeat which links the phosphorylated O-mannosyl trisaccharide to the ligand binding moiety composed of repeats of 3-xylosyl-alpha-1,3-glucuronic acid-beta-1 (PubMed:17034757, PubMed:25279699, PubMed:26923585, PubMed:29477842, PubMed:27194101). Required for normal location of POMGNT1 in Golgi membranes, and for normal POMGNT1 activity (PubMed:17034757). May interact with and reinforce a large complex encompassing the outside and inside of muscle membranes (PubMed:25279699). Could be involved in brain development (Probable). {ECO:0000269|PubMed:17034757, ECO:0000269|PubMed:25279699, ECO:0000269|PubMed:26923585, ECO:0000269|PubMed:27194101, ECO:0000269|PubMed:29477842, ECO:0000305|PubMed:11115853}. | Alternative splicing;Cardiomyopathy;Congenital muscular dystrophy;Cytoplasm;Disease variant;Dystroglycanopathy;Glycoprotein;Golgi apparatus;Limb-girdle muscular dystrophy;Lissencephaly;Membrane;Nucleus;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:25279699, ECO:0000269|PubMed:26923585, ECO:0000269|PubMed:27194101, ECO:0000269|PubMed:29477842}. | The protein encoded by this gene is a putative transmembrane protein that is localized to the cis-Golgi compartment, where it may be involved in the glycosylation of alpha-dystroglycan in skeletal muscle. The encoded protein is thought to be a glycosyltransferase and could play a role in brain development. Defects in this gene are a cause of Fukuyama-type congenital muscular dystrophy (FCMD), Walker-Warburg syndrome (WWS), limb-girdle muscular dystrophy type 2M (LGMD2M), and dilated cardiomyopathy type 1X (CMD1X). Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Nov 2010]. | hsa:2218; | cis-Golgi network [GO:0005801]; endoplasmic reticulum [GO:0005783]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of Golgi membrane [GO:0030173]; nucleus [GO:0005634]; phosphotransferase activity, for other substituted phosphate groups [GO:0016780]; muscle organ development [GO:0007517]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of JNK cascade [GO:0046329]; nervous system development [GO:0007399]; protein glycosylation [GO:0006486]; protein O-linked glycosylation [GO:0006493]; protein O-linked mannosylation [GO:0035269]; regulation of protein glycosylation [GO:0060049] | 12172906_In Fukuyama congenital muscular dystrophy (FCMD) cases, expression of fukutin looked decreased. 14627679_Fukutin is associated with Walker-Warburg syndrome. 15103718_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 15213246_Data suggest that fukutin and fukutin-related protein (FKRP) may be involved at different steps in O-mannosylglycan synthesis of alpha-dystroglycan, and FKRP is most likely involved in the initial step in this synthesis. 17005282_Fukutin seems to bind to both the hypoglycosylated and fully glycosylated form of alpha-dystroglycan, and seems bind to the core area rather than the sugar chain of alpha-dystroglycan 17878207_Observational study of genotype prevalence. (HuGE Navigator) 18177472_Walker-Warburg syndrome carries a homozygous-single nucleotide insertion that produces a frameshift, or 2 mutations, a point mutation that produces an amino acid substitution, & deletion in 3'UTR that affects the polyadenylation signal of fukutin gene. 18752264_FCMD mutations are a more common cause of Walker-Warburg syndrome outside of the Middle East. 18834683_The homozygous nonsense mutations within the coding region identified in Turkish patients are predicted to cause a total loss of fukutin activity and are likely to produce a more severe phenotype which closely resembles WWS. 19015585_Observational study of gene-disease association. (HuGE Navigator) 19015585_The compound heterozygous FKTN mutation was a rare cause of dilated cardiomyopathy. Hyper-CKemia might be indicative of FKTN mutation in dilated cardiomyopathy. 19179078_Outside Japan, fukutinopathies are associated with a large spectrum of phenotypes from isolated hyperCKaemia to severe CMD, showing a clear overlap with that of FKRP. 19266496_an identical homozygous c.1167insA mutation in the FKTN gene on a common haplotype in four families and identified 2/299 (0.7%) carriers for the c.1167insA mutation among normal American Ashkenazi Jewish adults 19299310_Observational study of gene-disease association. (HuGE Navigator) 19342235_Our results provide further evidence for ethnic and allelic heterogeneity and the presence of milder phenotypes in FKTN-dystroglycanopathy despite a substantial degree of alpha-dystroglycan hypoglycosylation in skeletal muscle. 19396839_We found fukutin gene mutations in a 4.5-year-old Italian patient, with reduced alpha-dystroglycan expression, dystrophic features on muscle biopsy, hypotonia since birth, mild myopathy, but no brain involvement. 20620061_FKTN mutations are the most common genetic cause of congenital muscular dystrophies with defective alpha-dystroglycan glycosylation in Korea 20961758_four new non-Japanese patients with FKTN mutations and congenital muscular dystrophy 24530477_Mutation in the fukutin gene is associated with Fukuyama congenital muscular dystrophy and microcephaly. 26223471_Fukutin role in in tumor progression in gastric cancer 26923585_Fukutin and fukutin-related protein are sequentially acting Rbo5P transferases that use cytidine diphosphate ribitol. 27194101_ISPD and FKTN are essential for the incorporation of ribitol into alpha-dystroglycan. 28680109_the mutated fukutin protein was smaller than the normal protein, reflecting the truncation of fukutin due to a premature stop codon. Immunostaining analysis showed a decrease in the signal for the glycosylated form of alpha-dystroglycan. These findings indicated that this mutation is the second most prevalent loss-of-function mutation in Japanese Fukuyama congenital muscular dystrophy patients. 29416295_The results suggest that fukutin and FKRP not only participate in the synthesis of O-mannosyl glycans added to alpha-dystroglycan in the endoplasmic reticulum and Golgi complex, but that they could also play a role, that remains to be established, in the nucleus of retinal neurons. 29477842_Fukutin, FKRP, and TMEM5 form a complex while maintaining each of their enzyme activities. Data showed that endogenous fukutin and FKRP enzyme activities coexist with TMEM5 enzyme activity, and suggest the possibility that formation of this enzyme complex may contribute to specific and prompt biosynthesis of glycans that are required for dystroglycan function. 34830034_Fukutin Protein Participates in Cell Proliferation by Enhancing Cyclin D1 Expression through Binding to the Transcription Factor Activator Protein-1: An In Vitro Study. | ENSMUSG00000028414 | Fktn | 240.02914 | 0.6269539 | -0.6735687988 | 0.20865096 | 1.030990e+01 | 1.323183e-03 | 4.010737e-01 | No | Yes | 191.34009 | 35.149555 | 3.033079e+02 | 42.827709 |
| ENSG00000106829 | 7091 | TLE4 | protein_coding | Q04727 | FUNCTION: Transcriptional corepressor that binds to a number of transcription factors. Inhibits the transcriptional activation mediated by PAX5, and by CTNNB1 and TCF family members in Wnt signaling. The effects of full-length TLE family members may be modulated by association with dominant-negative AES. Essential for the transcriptional repressor activity of SIX3 during retina and lens development and for SIX3 transcriptional auto-repression (By similarity). {ECO:0000250}. | Acetylation;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;WD repeat;Wnt signaling pathway | hsa:7091; | beta-catenin-TCF complex [GO:1990907]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; chromatin binding [GO:0003682]; DNA-binding transcription factor binding [GO:0140297]; transcription corepressor activity [GO:0003714]; cellular response to leukemia inhibitory factor [GO:1990830]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of transcription by RNA polymerase II [GO:0000122]; Wnt signaling pathway [GO:0016055] | 17060451_Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors 18258796_Knockdown of TLE1 or TLE4 levels increased the rate of cell division of the AML1-ETO-expressing Kasumi-1 cell line, whereas forced expression of either TLE1 or TLE4 caused apoptosis and cell death. 19714205_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19893608_germ cell-specific RBMY and hnRNP G-T proteins were more efficient in stimulating TLE4-T incorporation than somatically expressed hnRNP G protein. 20816195_Observational study of gene-disease association. (HuGE Navigator) 20889853_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 22169276_Grg4 recruits the arginine methyltransferase PRMT5 to chromatin resulting in symmetric H4R3 dimethylation. 22927467_Displacing coactivators CREB-binding protein/p300 while promoting the recruitment of a corepressor, Grg4. 24099773_Groucho related gene Grg4 robustly activates the expression of a BMP reporter gene, as well as enhancing and sustaining the upregulation of the endogenous Id1 gene induced by BMP7. 25631048_PPM1B interacts with Groucho 4 and is localized to DNA in a Groucho-dependent manner, and phosphatase activity is required for transcriptional silencing. 26701208_the results suggested that TLE4, a potential prognostic biomarker for colorectal cancer, plays an important role in the development and progression of human colorectal cancer. 27486062_the loss of TLE4 confers proliferative advantage to leukemic cells, simultaneous with an upregulation of a pro- inflammatory signature mediated through aberrant increases in Wnt signaling activity 28262390_Genes TGFB1, TLE4 and MUC22 are associated with the risk of childhood asthma in Chinese population. 35099008_TLE4 regulates muscle stem cell quiescence and skeletal muscle differentiation. | ENSMUSG00000024642 | Tle4 | 1944.75137 | 1.0545381 | 0.0766112192 | 0.10548967 | 5.312626e-01 | 4.660765e-01 | 9.998360e-01 | No | Yes | 1936.24224 | 138.496983 | 1.859734e+03 | 103.158217 | ||
| ENSG00000107140 | 7016 | TESK1 | protein_coding | Q15569 | FUNCTION: Dual specificity protein kinase activity catalyzing autophosphorylation and phosphorylation of exogenous substrates on both serine/threonine and tyrosine residues (By similarity). Regulates the cellular cytoskeleton by enhancing actin stress fiber formation via phosphorylation of cofilin and by preventing microtubule breakdown via inhibition of TAOK1/MARKK kinase activity (By similarity). Inhibits podocyte motility via regulation of actin cytoskeletal dynamics and phosphorylation of CFL1 (By similarity). Positively regulates integrin-mediated cell spreading, via phosphorylation of cofilin (PubMed:15584898). Suppresses ciliogenesis via multiple pathways; phosphorylation of CFL1, suppression of ciliary vesicle directional trafficking to the ciliary base, and by facilitating YAP1 nuclear localization where it acts as a transcriptional corepressor of the TEAD4 target genes AURKA and PLK1 (PubMed:25849865). Probably plays a central role at and after the meiotic phase of spermatogenesis (By similarity). {ECO:0000250|UniProtKB:O70146, ECO:0000250|UniProtKB:Q63572, ECO:0000269|PubMed:15584898, ECO:0000269|PubMed:25849865}. | ATP-binding;Cell projection;Cytoplasm;Cytoskeleton;Kinase;Magnesium;Manganese;Metal-binding;Methylation;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Tyrosine-protein kinase | hsa:7016; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; lamellipodium [GO:0030027]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein C-terminus binding [GO:0008022]; protein kinase activity [GO:0004672]; protein kinase binding [GO:0019901]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; actin cytoskeleton organization [GO:0030036]; establishment of vesicle localization [GO:0051650]; glomerular visceral epithelial cell migration [GO:0090521]; negative regulation of cilium assembly [GO:1902018]; negative regulation of phosphorylation [GO:0042326]; negative regulation of protein autophosphorylation [GO:0031953]; negative regulation of protein serine/threonine kinase activity [GO:0071901]; positive regulation of protein localization to nucleus [GO:1900182]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of stress fiber assembly [GO:0051496]; positive regulation of substrate adhesion-dependent cell spreading [GO:1900026]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of protein localization [GO:0032880]; spermatogenesis [GO:0007283] | ENSMUSG00000028458 | Tesk1 | 1502.14249 | 1.0075266 | 0.0108179120 | 0.11065264 | 9.657916e-03 | 9.217142e-01 | 9.998360e-01 | No | Yes | 1364.88400 | 128.069181 | 1.253859e+03 | 92.102443 | |||
| ENSG00000107643 | 5599 | MAPK8 | protein_coding | P45983 | FUNCTION: Serine/threonine-protein kinase involved in various processes such as cell proliferation, differentiation, migration, transformation and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK8/JNK1. In turn, MAPK8/JNK1 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN, JDP2 and ATF2 and thus regulates AP-1 transcriptional activity (PubMed:18307971). Phosphorylates the replication licensing factor CDT1, inhibiting the interaction between CDT1 and the histone H4 acetylase HBO1 to replication origins (PubMed:21856198). Loss of this interaction abrogates the acetylation required for replication initiation. Promotes stressed cell apoptosis by phosphorylating key regulatory factors including p53/TP53 and Yes-associates protein YAP1 (PubMed:21364637). In T-cells, MAPK8 and MAPK9 are required for polarized differentiation of T-helper cells into Th1 cells. Contributes to the survival of erythroid cells by phosphorylating the antagonist of cell death BAD upon EPO stimulation (PubMed:21095239). Mediates starvation-induced BCL2 phosphorylation, BCL2 dissociation from BECN1, and thus activation of autophagy (PubMed:18570871). Phosphorylates STMN2 and hence regulates microtubule dynamics, controlling neurite elongation in cortical neurons. In the developing brain, through its cytoplasmic activity on STMN2, negatively regulates the rate of exit from multipolar stage and of radial migration from the ventricular zone. Phosphorylates several other substrates including heat shock factor protein 4 (HSF4), the deacetylase SIRT1, ELK1, or the E3 ligase ITCH (PubMed:20027304, PubMed:17296730, PubMed:16581800). Phosphorylates the CLOCK-ARNTL/BMAL1 heterodimer and plays a role in the regulation of the circadian clock (PubMed:22441692). Phosphorylates the heat shock transcription factor HSF1, suppressing HSF1-induced transcriptional activity (PubMed:10747973). Phosphorylates POU5F1, which results in the inhibition of POU5F1's transcriptional activity and enhances its proteosomal degradation (By similarity). Phosphorylates JUND and this phosphorylation is inhibited in the presence of MEN1 (PubMed:22327296). In neurons, phosphorylates SYT4 which captures neuronal dense core vesicles at synapses (By similarity). Phosphorylates EIF4ENIF1/4-ET in response to oxidative stress, promoting P-body assembly (PubMed:22966201). {ECO:0000250|UniProtKB:P49185, ECO:0000250|UniProtKB:Q91Y86, ECO:0000269|PubMed:10747973, ECO:0000269|PubMed:16581800, ECO:0000269|PubMed:17296730, ECO:0000269|PubMed:18307971, ECO:0000269|PubMed:18570871, ECO:0000269|PubMed:20027304, ECO:0000269|PubMed:21095239, ECO:0000269|PubMed:21364637, ECO:0000269|PubMed:21856198, ECO:0000269|PubMed:22327296, ECO:0000269|PubMed:22441692, ECO:0000269|PubMed:22966201}.; FUNCTION: JNK1 isoforms display different binding patterns: beta-1 preferentially binds to c-Jun, whereas alpha-1, alpha-2, and beta-2 have a similar low level of binding to both c-Jun or ATF2. However, there is no correlation between binding and phosphorylation, which is achieved at about the same efficiency by all isoforms. | 3D-structure;ATP-binding;Alternative splicing;Biological rhythms;Cell junction;Cytoplasm;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;S-nitrosylation;Serine/threonine-protein kinase;Synapse;Transferase | The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various cell stimuli, and targets specific transcription factors, and thus mediates immediate-early gene expression in response to cell stimuli. The activation of this kinase by tumor-necrosis factor alpha (TNF-alpha) is found to be required for TNF-alpha induced apoptosis. This kinase is also involved in UV radiation induced apoptosis, which is thought to be related to cytochrom c-mediated cell death pathway. Studies of the mouse counterpart of this gene suggested that this kinase play a key role in T cell proliferation, apoptosis and differentiation. Several alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Apr 2016]. | hsa:5599; | axon [GO:0030424]; basal dendrite [GO:0097441]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; ATP binding [GO:0005524]; enzyme binding [GO:0019899]; histone deacetylase binding [GO:0042826]; histone deacetylase regulator activity [GO:0035033]; JUN kinase activity [GO:0004705]; MAP kinase activity [GO:0004707]; protein phosphatase binding [GO:0019903]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine kinase binding [GO:0120283]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cellular response to amino acid starvation [GO:0034198]; cellular response to cadmium ion [GO:0071276]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to mechanical stimulus [GO:0071260]; cellular response to reactive oxygen species [GO:0034614]; cellular senescence [GO:0090398]; Fc-epsilon receptor signaling pathway [GO:0038095]; intracellular signal transduction [GO:0035556]; JNK cascade [GO:0007254]; JUN phosphorylation [GO:0007258]; negative regulation of apoptotic process [GO:0043066]; negative regulation of protein binding [GO:0032091]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cell killing [GO:0031343]; positive regulation of cyclase activity [GO:0031281]; positive regulation of deacetylase activity [GO:0090045]; positive regulation of gene expression [GO:0010628]; positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway [GO:1900740]; positive regulation of protein metabolic process [GO:0051247]; protein phosphorylation [GO:0006468]; regulation of circadian rhythm [GO:0042752]; regulation of DNA replication origin binding [GO:1902595]; regulation of DNA-binding transcription factor activity [GO:0051090]; regulation of macroautophagy [GO:0016241]; regulation of protein localization [GO:0032880]; response to mechanical stimulus [GO:0009612]; response to oxidative stress [GO:0006979]; response to UV [GO:0009411]; rhythmic process [GO:0048511]; stress-activated MAPK cascade [GO:0051403] | 11912216_Polycystin-1 activation of c-Jun N-terminal kinase and AP-1 is mediated by heterotrimeric G proteins 11931768_Results show that the N-CoR-HDAC3 complex inhibits JNK activation through the associated GPS2 subunit and thus could potentially provide an alternative mechanism for hormone-mediated antagonism of AP-1 function. 11971973_These data strongly suggest that in TNF-induced apoptosis, Hsp72 specifically interferes with the Bid-dependent apoptotic pathway via inhibition of JNK. 12058026_novel role for the I kappa B kinase complex-associated protein (IKAP) in the regulation of activation of the mammalian stress response via the c-Jun N-terminal kinase (JNK)-signaling pathway 12058028_role in stabilizing p21(Cip1) by phosphorylation 12079429_the NOx-induced cell proliferation via activation of JNK1 might contribute to lung tissue damage caused by NOx 12135322_Galpha13 can induce ppET-1 gene expression through a JNK-mediated pathway. 12140754_Elevated JNK activation contributes to the pathogenesis of human brain tumors 12143039_Jun kinase modulates tumor necrosis factor-dependent apoptosis in liver cells 12148599_Unimpaired activation of c-Jun NH2-terminal kinase (JNK) 1 upon CD40 stimulation in B cells of patients with X-linked agammaglobulinemia. 12206715_description of the signaling of JNK and p38 MAPK in apoptosis after stimulation by antioxidants 12296995_TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. 12354774_JNK has isoform-selective gene regulation and distinct JNK isoforms have a role in specific cellular responses 12359245_Relationship of Mcl-1 isoforms, ratio p21WAF1/cyclin A and this protein phosphorylation to apoptosis in human breast carcinomas. 12413764_Psoriatic epidermis shows selective activation of ERK and JNK, which might be related to hyperproliferation and abnormal differentiation of psoriatic epidermis. 12421945_JNK activation is predominantly involved in the induction of CD44 expression in monocytic cells via lipopolysaccharide-mediated signaling. 12478662_JNK-1 and p38 play a role in apoptosis induced by capsaicin in H-ras-transformed tumor cells 12514174_phosphorylation of JNK1 and WOX1 is necessary for their physical interaction and functional antagonism 12538493_JNK is required for growth of prostate carcinoma cells in vitro and in vivo 12592382_Western blot demonstrated that phosphorylation of JNK was induced only by TPA during 30 min to 1 h. 12646240_Data suggest that epidermal growth factor (EGF) stimulated c-Jun N-terminal kinase phosphorylation of c-Jun is uncoupled from protein kinase D suppression in cancer cells. 12707267_JNK1 has a role in the synergistic effect of TRAIL combined with DNA damage by mediating signals independent of p53 leading to apoptosis 12810082_Results suggest that tissue or plasma fibronectin may modulate the intestinal epithelial response to repetitive deformation through inhibted activation of p38 and jun kinases. 12847227_c-Jun N-terminal kinase plays a negative role in the production of IL-12 from human macrophages stimulated by lipopolysaccharide. 12859962_Data show that inhibition of arachidonate 5-lipoxygenase induces rapid activation of c-Jun N-terminal kinase (JNK) in human prostate cancer cells which is prevented by the 5-lipoxygenase metabolite, 5(S)-HETE. 12878610_Axin utilizes distinct regions for competitive MEKK1 and MEKK4 binding and JNK activation. 12902351_adipose cytokines and JNK are key mediators between obesity and hormone-resistant prostate cancer 12917434_Threonine 668 within the Amyloid beta protein precursor intracellular domain is indeed phosphorylated by JNK1; although JIP-1 can facilitate this phosphorylation, it is not required for this process. 14500675_Findings strongly suggest that the JNK/AP-1 transcription factor signaling pathway has little or no impact on the generation of inflammatory mediators in neutrophils. 14514687_late but not early JNK1 activation is associated with the induction of apoptosis 14532003_Results show that tumor necrosis factor alpha-mediated caspase 8 cleavage and apoptosis require a sequential pathway involving c-Jun N-terminal kinase, Bid, and Smac/DIABLO. 14557276_c-Jun N-terminal kinase activation in T cell receptor signaling is mediated by SH3 domain-containing adaptor HIP-55 14561739_JNK inhibition with SP600125 also blocked binding of Sp1 to the DR5/TRAIL-R2 promoter. 14637155_Fas-induced cell death and JNK activation are sensitive to Fas stimulation in cell lines carrying undetectable level of c-FLIP(L). 14688370_Stress-activated protein kinase 1 is involved in the control of monocyte chemoattractant protein-1-induced migration of MonoMac6 cells. 14699155_JNK activation is important for lipopolysacchairde-induced MCP-1 expression but not for TNF-alpha or IL-8 expression 14701702_data support an essential role for JNK signaling in the induction of growth inhibition and apoptosis by As(2)O(3) 14724588_activation of JNK is important for the induction of apoptosis following stresses that function at different cell cycle phases, and that basal JNK activity is necessary to promote proliferation and maintain diploidy in breast cancer cells 14729602_Calcium signaling in ovarian surface epithelial cells not only induces telomerase activity via JNK but also activates Pyk2. 14766760_JNK regulates the expression of HIPK3 in prostate cancer cells, which leads to increased resistance to Fas receptor-mediated apoptosis by reducing the interaction between FADD and caspase-8 14981905_Inducible expression of RbAp46 activated the c-Jun N-terminal kinase (JNK) signaling pathway and triggered apoptosis in Saos-2 cells xenografted into nude mice. 15013949_JNK signaling regulates the phosphorylation state of several kinases in skeletal muscle. JNK activation is unlikely to be the major mechanism by which contractile activity increases glycogen synthase activity in skeletal muscle. 15238629_The c-Jun-N-terminal kinase(JNK)cascade mediates the stimulatory effect Angiotensin II has on the proximal renin promoter in humans. 15456887_Data show that endogenous germinal center kinase is activated by agonists that require TRAF6 for c-Jun N-terminal kinase activation. 15474087_SAPKgamma/JNK1 and SAPKalpha/JNK2 may be important mediators of stress-induced responses in early implanting conceptuses that could mediate embryo loss. 15516492_Antiproliferative and prodifferentiation effects of BMP4 were Smad1 dependent with JNK also contributing to differentiation. 15527495_crucial role of JNK signalling pathway in N. meningitidis invasion in human brain microvascular endothelial cells 15528994_Cooperation of CD99 engagement with suboptimal TCR/CD3 signals resulted in enhanced CD4+ T cell proliferation, elevated expression of CD25 and GM1, increased apoptosis, augmented activation of JNK, and increased AP-1 activation 15542843_activation of p38 MAPK and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and TNFR1/TNFa expression 15569856_The ACE-inhibitor mediated activation of the c-Jun N-terminal kinase (JNK)/c-Jun pathway, results in an enhanced endothelial ACE expression 15629131_While JNK1 is a downstream effector of the Tumor Necrosis Factor (TNF) signaling, Zfra regulation of the TNF cytotoxic function is likely due to its interaction, in part, with JNK1 15637062_Caspase 3-cleaved SH3 domain of HIP-55 is likely involved in PRAM-1-mediated JNK activation upon arsenic trioxide-induced differentiation of NB4 cells. 15655348_ERK1/2 and JNK1/2 signaling is stimulated by radiation and can promote cell cycle progression in human colon cancer cells 15657352_Alpha-tocopheryl sulfsatse showed increased levels in prostate tumor cells. 15665513_TNF-alpha causes a net up-regulation of MUC2 gene expression in cultured colon cancer cells because NF-kappaB transcriptional activation of this gene is able to counter-balance the suppressive effects of the JNK pathway 15696159_JNK phosphorylates 14-3-3 proteins, which regulate nuclear targeting of c-Abl in the apoptotic response to DNA damage 15755722_PKD is a critical mediator in H2O2- but not TNF-induced ASK1-JNK signaling 15769735_raft-associated acid sphingomyelinase and JNK activation and translocation are induced by UV-C light on a nuclear signal 15778501_p53 participates in a feedback mechanism with JNK to regulate the apoptotic process and is oppositely regulated by JNK1 and JNK2. 15860507_Some green tea catechins induce pro-MMP-7 production via O2- production and the activation of JNK1/2. 15890690_JNK may act via c-Myc and Egr-1, which were shown to be important for B-lymphoma survival and growth. 15981086_rF1-induced JNK MAPK activity was correlated to the functional activation of macrophages by demonstrating the inhibition of NO, TNF-alpha production and microtubule polymerization 16086581_results suggest that activated JNK can, in turn, activate not only jun but also raf that, in turn, activates MEK that can then cross-activate JNK in a positive feedback loop 16105650_We concluded that JNK pathway might play an important role in mediating cisplatin-induced apoptosis in A2780 cells, and the duration of JNK activation might be critical in determining whether cells survive or undergo apoptosis. 16166642_JAMP is a membrane-anchored regulator of the duration of JNK1 activity in response to diverse stress stimuli 16176806_The activation of JNK1 is required for the triptolide-induced inhibition of tumor proliferation. 16243842_Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages 16260419_protein kinase Cdelta and JNK have roles in Ifn-alpha induced expression of phospholipid scramblase 1 through STAT1 16260609_c-Jun N-terminal kinase signaling is regulated by a stabilization-based feed-forward 16282329_Foxo1 is involved in the nucleocytoplasmic translocation of PDX-1 by oxidative stress and the JNK pathway 16283431_Results describe the opposite effect of ERK1/2 and JNK on p53-independent p21WAF1/CIP1 activation involved in the arsenic trioxide-induced human epidermoid carcinoma A431 cellular cytotoxicity. 16307741_gemcitabine-induced apoptosis in human non-small cell lung cancer H1299 cells requires activation of the JNK1 signaling pathway 16321971_in gastric epithelial cells, H. pylori up-regulates MMP-1 in a type IV secretion system-dependent manner via JNK and ERK1/2 16328781_Results suggest that VEGF induced by hyperbaric oxygen is through c-Jun/AP-1 activation, and through simultaneous activation of ERK and JNK pathways. 16339571_TNF-alpha induced PTX3 expression in human lung cell lines and primary epithelial cells; knockdown of either JNK1 or JNK2 with small interfering RNA also significantly reduced the regulated PTX3 expression 16381010_results indicate that the aberrant p-JNK1/2 expression and the co-expressed p-JNK1/2 and p-p38 in breast tissues may play a role in the carcinogenesis of breast infiltrating ductal carcinoma 16407310_Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1 16412424_Data report that cepharanthine induces apoptosis in HuH-7 cells through activation of JNK1/2 and the downregulation of Akt. 16434970_Both activation of JNK and inhibition of Akt play a role in translocation of Nur77 from the nucleus to the cytoplasm. 16465391_We investigated whether Jun-N-terminal kinase activation is increased in inflammatory bowel disease and analyzed the effects of SP600125, which decreases inflammatory cytokine synthesis by inhibiting the phosphorylation of this kinase. 16569638_matrix metalloproteinase-1 expression is regulated by JNK through Ets and AP-1 promoter motifs 16648634_Inhibition of JNK in epidermal keratinocytes is sufficient to initiate their differentiation program and suggest that augmenting JNK activity could be used to delay cornification and enhance wound healing. 16687404_JNK-mediated feedback phosphorylation of MLK3 regulates its activation and deactivation states by cycling between Triton-soluble and Triton-insoluble forms 16699726_ERK1/2, JNK1/2 and p38 mapk pathways are all required for B[a]P-induced G1/S transition 16760468_JNK is a critical component downstream of PI 3-kinase that may be involved in PDGF-stimulated chemotaxis presumably by modulating the integrity of focal adhesions by phosphorylating its components 16794185_Paclitaxel increases endothelial TF expression via its stabilizing effect on microtubules and selective activation of JNK 16802349_JNK (c-Jun N-terminal kinase) function might be modulated by targeting MKK-7 to suppress cytokine-mediated fibroblast-like synoviocytes (FLS) activation while leaving other stress responses intact. 16814421_Results show that activation of c-Jun N-terminal protein kinase, ERKs 1 and 2, and p38 MAP kinase is critical for Hs683 glioma cell migration induced by GDNF. 16815888_PP1-JNK pathway plays a role in H(2)O(2)-induced Sp1 phosphorylation in lung epithelial cells 16824735_JNK1 is associated with UV signal transduction in human epidermis and SCCA1 is a suppressor of this process. 16895791_TNF-alpha down-regulates human Cu/Zn superoxide dismutase 1 promoter via JNK/AP-1 signaling pathway 16912864_Leptin stimulates proliferation and inhibits apoptosis in colon cancer cells. This effect involves JAK2, PI3 kinase and JNK and activation of the oncogenic transcription factors signal transducer and activator of transcription 16927023_This study is the first to demonstrate that H2O2 induces a Rac1/JNK1/p38 signaling cascade, and that JNK and p38 activation is important for H2O2-induced apoptosis as well as apoptosis-inducing factor/Bax translocation of retinal pigment epithelial cells. 16972261_Data show that pharmacologic inhibitors of extracellular signal-regulated kinase (ERKs) and c-Jun NH(2)-terminal kinase (JNK) decrease glutathione content and sensitize human promonocytic leukemia cells to arsenic trioxide-induced apoptosis. 16983342_active JNK1 inhibits ubiquitination of C/EBPalpha possibly by phosphorylating in its DBD 17008315_STAT3 activation by G alpha(s) distinctively requires protein kinase A, JNK, and phosphatidylinositol 3-kinase 17054907_Collectively, our results suggest that the inhibition of the interaction between JNK and c-Jun may be an integral part of the mechanism underlying the negative regulation of the JNK signaling pathway by NO. 17074809_JNK1 and JNK2 differentially regulate TBP through Elk-1, controlling c-Jun expression and cell proliferation 17079291_Overall, these results demonstrate the importance of the JNK pathway for varicella-zoster virus replication. 17158878_PSK1-alpha is a bifunctional kinase that associates with microtubules, and JNK- and caspase-mediated removal of its C-terminal microtubule-binding domain permits nuclear translocation of the N-terminal region of PSK1-alpha and its induction of apoptosis 17178870_JNK1 is activated in response to collagen I, which increases tumorigenesis by up-regulating N-cadherin expression and by increasing motility. 17255354_Oxidative stress response regulates DKK1 expression through the JNK signaling cascade in multiple myeloma plasma cells. 17296730_Human Rev7 (hRev7)/MAD2B/MAD2L2 is an interaction partner for Elk-1 and hRev7 acts to promote Elk-1 phosphorylation by the c-Jun N-terminal protein kinase (JNK) MAP kinases. 17303384_These results suggest that in human tracheal smooth muscle cells, activation of p42/p44 MAPK, p38, and JNK pathways, at least in part, mediated through NF-kappaB, is essential for lipopolysaccharide-induced VCAM-1 gene expression. 17317777_Increased expression of stress-activated kinases and IKK and their phosphorylated forms in omental fat occurs in obesity. 17453826_The regulation of three major mitogen-activated protein kinases phosphorylation, ERKp44/p42, p38, and JNK, was determined. The influence of specific mitogen-activated protein kinase inhibitors on IL-1 beta protein levels during beta-endorphin stimulation. 17478078_Proinvasive activity of BMP7 through SMAD4/src-independent and ERK/Rac/JNK-dependent signaling pathways in colon cancer cells is reported. 17481915_These results indicate that binding of the alpha3beta1 integrin results in a suppression in the activation of the IL-1 induced intracellular signaling pathway from JNK to AP-1. 17496921_In this review, the interplay between NF-kappa B and JNK1 provides a paradigm that shows how crosstalk between different signaling pathways decides the function of the cell signaling circuitry. 17541429_Gemin5 functions as a scaffold protein for the ASK1-JNK1 signaling module and thereby potentiates ASK1-mediated signaling events. 17545598_Findings showed that TOPK positively modulated UVB-induced JNK1 activity and played a pivotal role in JNK1-mediated cell transformation induced by H-Ras. 17568996_IGF-1R and PDGFR co-inhibition caused an increased cell death in two human glioma cell lines and induced the radiosensitization of the JNK1 expressing cell line. 17584736_GCK is required for JNK and, unexpectedly, p38 activation by three bacterial PAMPs, lipopolysaccharide, peptidoglycan, and flagellin 17603935_TNF-alpha induced reactive oxygen species formation is mediated by JNK1, which regulates ferritin degradation and thus the level of highly reactive iron. 17620321_These results support a significant role for ALK1 as a negative regulator of endothelial cell migration and suggest the implication of JNK and ERK as mediators of this effect. 17626013_JNK and p38 mitogen-activated protein kinases were activated by TAK1. 17640761_This study reveals a novel pathway of gene regulation by alcohol which involves the activation of JNK and the consequent mRNA stabilization. 17652454_These results indicate that Ask1 oxidation is required at a step subsequent to activation for signaling downstream of Ask1 after H(2)O(2) treatment. 17690186_Data suggest that JNK activation and decreased expression of MKP-1 may play important roles in progression of urothelial carcinoma. 17693927_Studies suggest that the target of regulation by PP2A includes upstream kinases in the JNK MAPK pathway. 17699782_Findings suggest that the JNK/PTEN and NF-kappaB/PTEN pathways play a critical role in normal intestinal homeostasis and colon carcinogenesis. 17702750_long duration KOR antagonists disrupt KOR signaling by activating JNK 17703233_findings establish a major role for DAPk and its specific interaction with PKD in regulating the JNK signaling network under oxidative stress. 17704768_Results demonstrate that Fbl10 is a key regulator of c-Jun function. 17719653_Costimulation by anti-CD3 and anti-CD28 antibodies could activate JNK, p38 MAPK and NF-kappaB. The upregulation of IL-25 receptors were differentially regulated by intracellular JNK, p38 MAPK and NF-kappaB. 17785464_Study provides evidence during rolling and adhesion of platelets to vWF that platelet GPIb-vWF interaction triggers alphaIIbbeta3 activation in a JNK1-dependent manner; this was confirmed with a Glanzmann thrombastenic patient lacking alphaIIbbeta3. 17883418_Streptococcus intermedius histon-like DNA binding protein (Si-HLP) stimulation induced the activation of cell signal transduction pathways, extracellular signal-regulated kinase 1/2 (ERK1/2) and c-Jun N-terminal kinase (JNK). 17904874_Our data show that basic JNK activity plays an important role in the progression of the cell cycle at G2/M cell phase. 17908987_JNK1/2 activity is commonly increased in head and neck squamous cell carcinoma 17913539_These results collectively indicated that Chlamydophilal antigens induce foam cell formation mainly via Toll-like receptor 2 and c-Jun NH2 terminal kinase. 17933493_The induction of IFIT4 transcription by IFN-alpha depends upon sequential activation of PKCdelta, JNK and STAT1, and the influence of PKCdelta or JNK on IFN-alpha-mediated induction of IFIT4 is dependent upon the phosphorylation of STAT1 at Ser-727. 17942603_IFNalpha-induced apoptosis requires activation of ERK1/2, PKCdelta, and JNK downstream of PI3K and mTOR, and it can occur in a nucleus-independent manner, thus demonstrating that IFNalpha induces apoptosis in the absence of de novo transcription. 17967471_Critical role in cell transformation induced by EBV LMP1. 17982228_crude extract of D. farinae induces ICAM-1 expression in EoL-1 cells through signaling pathways involving both NF-kappaB and JNK 18025271_Cyclin G2 expression is modulated by HER2 signaling through multiple pathways including phosphoinositide 3-kinase, c-jun NH(2)-terminal kinase, and mTOR signaling. 18036196_Observational study of gene-disease association. (HuGE Navigator) 18036196_The G/G genotype of MAPK8 SNP-1066 did not affect T2DM susceptibility despite specific binding of AP2alpha. 18055217_We conclude that endogenous SOCS3 inhibits AP-1 activity through blocking of JNK phosphorylation. 18082745_Nonalcoholic steatohepatitis is specifically associated with (1) failure to generate sXBP-1 protein and (2) activation of JNK. 18086557_Activation of protease-activated receptors but not stimulation with lipopolysaccharides leads to ERK1/2 and JNK-mediated production of IL-8. 18087676_These results clearly indicate that CCDC134 is a novel member of the secretory family and down-regulates the Raf-1/MEK/ERK and JNK/ SAPK pathways. 18094581_The majority of acute myeloid leukemia cases did not show any levels of Mitogen-Activated Protein Kinases activation except for two cases, which were associated with an extremely high white blood cell count, chromosomal aberration. 18164704_Our data first suggest that JNK participates in bFGF-mediated surface cadherin downregulation. Loss of surface cadherins may affect the cell-cell interaction between endothelial cells and facilitate angiogenesis. 18181766_ERK and JNK are involved in PMA-mediated MD-2 gene expression during HL-60 cell differentiation. 18199680_JNK1 activation is necessary to phosphorylate Sp1 and to shield Sp1 from the ubiquitin-dependent degradation pathway during mitosis in tumor cell lines. 18212053_c-Jun amino-terminal kinase (JNK) was important for neurite outgrowth stimulated by both Wnt-3a and Dkk1. 18218857_corneal inflammation is significantly impaired in JNK1 knockout mice compared with control mice, and in mice treated with the JNK inhibitor compared with vehicle control. 18219313_These results identify NLRX1 as a NLR that contributes to the link between reactive oxygen species generation at the mitochondria and innate immune responses. 18249102_Our results suggest that clivorine has direct toxicity on HEK293 cells, and phosphorylated JNK may play some role in counteracting the toxicity of clivorine on HEK293 cells. 18253836_activator protein-1 (AP-1) was activated through phosphorylation of cJun and cFos, induced by JNK and p38, respectively. 18256527_Study shows that basal c-Jun N-terminal kinases (JNKs) are required for mitotic histone H3-S10 phosphorylation in human primary fibroblast IMR90 cells 18276794_Glutamate cysteine ligase iz induced by hydroxynonenal through the c-Jun N-terminal kinase (JNK) pathway in respiratory epithelium. 18286207_the activity of JIP1-JNK complexes is downregulated by VRK2 in response to interleukin-1beta 18288129_both constitutive and sIgM-induced phosphorylation of p38 and JNK is inhibited by LAIR-1 through an ITIM-dependent signal 18292600_PP2A and AIP1 cooperatively induce activation of ASK1-JNK signaling and vascular endothelial cell apoptosis. 18293403_Preventing the TAK1/JNK1 signaling cascade in astrocytes might provide a fruitful strategy for treating intractable neuropathic pain. 18297686_These results identify JNK, and not NFkappaB, as a critical mediator of TNF-alpha repressory effect on connexin 43 gene expression. 18316600_ERK and JNK MAPK/Elk-1/Egr-1 signal cascade is required for p53-independent transcriptional activation of p21(Waf1/Cip1) in response to curcumin in U-87MG human glioblastoma cells 18316603_c-Jun translocates B23 and ARF from the nucleolus after JNK activation by means of protein interactions 18325654_Oxidative stress-activated JNK signaling pathway is involved in METH-induced cell death. 18337589_Phosphorylation aids GR sumoylation and that cross talk of JNK and SUMO pathways fine tune GR transcriptional activity. 18344085_The protein expression rates of p-JNK and P-glycoprotein in gastric cancer were significantly higher than those in normal gastric tissue. 18348163_Data show that JNK modulates the effect of caspases and NF-kappaB in the TNF-alpha-induced down-regulation of Na+/K+ATPase in HepG2 cells. 18356158_H. pylori mediates CagA-independent signaling that promotes cell motility through the beta1 integrin-JNK pathway 18373696_Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in cervical cancer. 18401423_Although STAT1 phosphorylation required JNK and p38MAPK activation, only JNK activation was essential for IRF1 promoter activation by Tie2-R849W. 18405916_These data demonstrate the role of Cx43 in the proliferation and migration of human saphenous vein smooth muscle cells and angiotensin II-induced Cx43 expression via mitogen-activated protein kinases (MAPK)-AP-1 signaling pathway. 18429822_Study demonstrates that the serine/threonine kinase PKN1 plays a critical role in regulating constitutive IKK/JNK activity in unstimulated cells and report on the molecular mechanism. 18439101_These findings suggest that JNK1 and JNK2 are involved in TNF-alpha-induced neutrophil apoptosis and GM-CSF-mediated antiapoptotic effect on neutrophils, respectively. 18457359_is the key enzyme mediating melanogenesis in B16F10 cell. 18495129_inhibition of JNK and p38 activation interrupts CD40 induced endothelial cell activation and apoptosis 18506470_JNK is involved in regulation of proinflammatory mediators of endometrium 18524773_JNK1 is a critical transcriptional target of FoxM1 that contributes to FoxM1-regulated cell cycle progression, tumor cell migration, invasiveness, and anchorage-independent growth 18540881_Although JNK activation may be a primary inducing factor, further phosphorylation of tau is required for neuronal death and NFT formation in neurodegenerative diseases, including those characterized by tauopathy. 18541008_JNK, and in particular the JNK1 isoform, support the growth of melanoma cells, by controlling either cell cycle progression or apoptosis depending on the cellular context. 18547751_Nanosilver acts through ROS and JNK to induce apoptosis via the mitochondrial pathway. 18570871_JNK1 mediates starvation-induced Bcl-2 phosphorylation, Bcl-2 dissociation from Beclin 1, and autophagy activation 18573678_Data show that pokeweed antiviral protein (PAP) does not inhibit protein translation, but induces the activation of c-Jun NH2-terminal kinase (JNK), which was specific to rRNA depurination. 18594007_c-Jun NH2-terminal protein kinase activation caused by tubulin depolymerization and DNA damage has a crucial role in moscatilin-induced apoptosis in human colorectal cancer cells 18603327_Data show that M. bovis BCG-induced human beta-defensin mRNA expression in A549 cells is regulated at least in part through activation of signaling proteins of PKC, JNK and PI3K. 18620777_WWOX induces apoptosis and inhibits human hepatocellular carcinoma cell growth through a mechanism enhanced by JNK inhibition. 18636174_A functional analysis of JNK1 and M-RIP with RNA interference reveals a critical role for this cascade in the invasive behavior of cancer cells. 18651223_These findings suggest JNK to have an important pro-apoptotic function following ultraviolet rays B irradiation in human melanocytes, by acting upstream of lysosomal membrane permeabilization and Bim phosphorylation. 18663379_Shikonin-induced oxidative injury operates at a proximal point in apoptotic signaling cascades, and subsequently activates the stress-related JNK pathway, triggers mitochondrial dysfunction, cytochrome c release, and caspase activation. 18667537_necrosis factor-alpha-elicited stimulation of gamma-secretase is mediated by c-Jun N-terminal kinase-dependent phosphorylation of presenilin and nicastrin 18681908_SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes 18682391_Histamine-induced Egr-1 expression is dependent on the activation of the H1 receptor, and rapidly and transiently activates PKCdelta, ERK1/2, p38 kinase, and JNK prior to Egr-1 induction. 18703151_These results demonstrate that ERKs and JNKs are responsible for the decrease of cyclin D1 and CDK4 expression levels in human embryonic lung fibroblasts induced by silica. 18713649_These results suggest that phosphorylation of paxillin on Ser 178 by JNK is required for the association of paxillin with FAK, and subsequent tyrosine phosphorylation of paxillin. 18713996_JNK is differentially regulated by MKK4 and MKK7 depending on the stimulus. 18718914_15d-PGJ(2) induces vascular endothelial cell apoptosis through the signaling of JNK and p38 MAPK-mediated p53 activation both in vitro and in vivo 18723442_whether MAP kinase phosphatase (MKP)-1, a negative regulator of p38 and JNK, mediates the antiinflammatory effects of shear stress. 18757369_a link between RhoA, JNK, c-Jun, and MMP2 activity that is functionally involved in the reduction in osteosarcoma cell invasion by the statin. This suggests a novel strategy targeting RhoA-JNK-c-Jun signaling to reduce osteosarcoma cell tumorigenesis. 18769111_A speculative model for understanding the interrelationship between autophagy and apoptosis regulated by JNK1-mediated Bcl-2 phosphorylation was proposed. 18782768_pneumolysin selectively induced expression of MKP1 via a TLR4-dependent MyD88-TRAF6-ERK pathway, which inhibited the PAK4-JNK signaling pathway,leading to up-regulation of MUC5AC mucin production 18815275_These results establish a novel function of filamin B as a molecular scaffold in the JNK signaling pathway for type I IFN-induced apoptosis. 18818208_analysis of JNK-mediated phosphorylation of paxillin in adhesion assembly and tension-induced cell death by the adenovirus death factor E4orf4 18845538_a novel function for Parkin in modulating the expression of Eg5 through the Hsp70-JNK-c-Jun signaling pathway. 18922473_These results indicate that TGF-beta activates JNK and p38 through a mechanism similar to that operating in the interleukin-1beta/Toll-like receptor pathway. 18936517_JNK may play an important role in posttranscriptional control of LDL receptor expression, thus constituting a novel mechanism to enhance plasma LDL clearance by liver cells. 18950845_Observational study of gene-disease association. (HuGE Navigator) 18978303_dynasore may stimulate PAI-1 protein expression and enhance TGF-beta(1) activity through activation of JNK-mediated signaling in human pleural mesothelial cells 18979912_The changes of phospho-JNK expression after skin burned might correlate with wound healing. 18982452_Data show that an increase in JNK activation in the presence of NFkappaB inhibition significantly increased the expression of IGFBP6. 18989785_Results show that Docetaxel-induced apoptosis is mediated by induction of ER stress, through activation of JNK and downstream targets of JNK. 18996088_These results suggest that JNK, but not caspase 8, involves in Fas-mediated CH11-induced autophagy in HeLa cells, and this autophagy plays a protective role in CH11-induced cell death. 19033664_a mechanistic link between JNK activity and liver cell proliferation via p21 and c-Myc and suggest JNK1 targeting can be considered as a new therapeutic approach for HCC treatment. 19036714_Upon UVB-induced stress in keratinocytes, ROCK1 was activated, bound to JIP-3, and activated the JNK pathway 19037093_AMPK controls the molecular mechanism underlying the differential biological functions of JNK, providing a novel explanation for the antiapoptotic role of LKB1. 19041150_JNK1 plays important roles in the development of human HCC partially through the epigenetic mechanisms. 19052872_c-Abl and p53 are important for execution of the cell death program initiated in A2E-laden RPE cells exposed to blue light, while JNK might play an anti-apoptotic role 19056926_JNK1 stimulated and mediated the effects of IFN and TNF-alpha on XAF1 expression through transcriptional regulation by induction of IRF-1. 19060920_activation of JNK pathway can mediate Beclin 1 expression, which plays a key role in autophagic cell death in cancer cel | ENSMUSG00000021936 | Mapk8 | 285.25588 | 0.9655416 | -0.0505896254 | 0.19809821 | 6.415411e-02 | 8.000467e-01 | 9.998360e-01 | No | Yes | 230.64540 | 45.739391 | 2.615713e+02 | 39.985354 | |
| ENSG00000107798 | 3988 | LIPA | protein_coding | P38571 | FUNCTION: Catalyzes the deacylation of triacylglyceryl and cholesteryl ester core lipids of endocytosed low density lipoproteins to generate free fatty acids and cholesterol. {ECO:0000269|PubMed:15269241, ECO:0000269|PubMed:1718995, ECO:0000269|PubMed:7204383, ECO:0000269|PubMed:8112342, ECO:0000269|PubMed:9633819}. | 3D-structure;Alternative splicing;Direct protein sequencing;Disease variant;Glycoprotein;Hydrolase;Lipid degradation;Lipid metabolism;Lysosome;Reference proteome;Signal | This gene encodes lipase A, the lysosomal acid lipase (also known as cholesterol ester hydrolase). This enzyme functions in the lysosome to catalyze the hydrolysis of cholesteryl esters and triglycerides. Mutations in this gene can result in Wolman disease and cholesteryl ester storage disease. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jan 2014]. | hsa:3988; | cytosol [GO:0005829]; fibrillar center [GO:0001650]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; nucleoplasm [GO:0005654]; lipase activity [GO:0016298]; sterol esterase activity [GO:0004771]; cell morphogenesis [GO:0000902]; cell population proliferation [GO:0008283]; homeostasis of number of cells within a tissue [GO:0048873]; inflammatory response [GO:0006954]; lipid catabolic process [GO:0016042]; low-density lipoprotein particle clearance [GO:0034383]; lung development [GO:0030324]; sterol metabolic process [GO:0016125]; tissue remodeling [GO:0048771] | 15465627_Observational study of gene-disease association. (HuGE Navigator) 16013913_Observational study of gene-disease association. (HuGE Navigator) 16024911_macrophage cholesteryl ester hydrolase associates with its intracellular substrate (lipid droplets) and hydrolyzes cholesteryl esters more efficiently from mixed droplets 16131527_The human liver enzyme is expressed in hepatocytes, where it potentially regulates the synthesis of bile acids and thus the removal of cholesterol from the body. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16730122_Observational study of gene-disease association. (HuGE Navigator) 18796546_Although LAL contributed to the deesterification of DHEA-FAE, it was not solely responsible for the hydrolysis. 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19307143_the molecular characterization of three heterozygous patients with Cholesteryl Ester Storage Disease carrying the common LIPA gene mutation (c.894 G>A, del p.S275_Q298), in combination with two novel mutations resulting in null alleles, was reported. 19343046_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20602615_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21900179_Lysosomal acid lipase in myeloid cells plays a critical role in maintaining normal hematopoietic cell development and balancing immunosuppression and inflammation. 21963785_a novel paternally inherited c.482delA mutation in exon 5 of Lipase A that results in a frameshift mutation at amino acid 161 is reported in a fatal case of Wolman disease 22227072_study identified two novel mutations of LIPA gene in Wolman Disease patients which abolished the expression of LAL enzyme; also found that all cholesteryl ester storage disease patients carried the common mutant allele c.894G>A 22395809_LIPA polymorphisms contribute to the interindividual variability observed in obesity-related metabolic complications 22795295_The results show that lysosomal acid lipase E8SJM mutation carriers have an alteration in lipid profile with a Polygenic Hypercholesterolemia phenotype. 23164340_The expression of LIPA may be associated with increased phospholipid content in the brains of violent suicide completers. 23424026_CESD prevalence in African and Asian populations may require full-gene LIPA sequencing to determine heterozygote frequencies. CESD may be underdiagnosed in the general Caucasian and Hispanic populations. 23624251_used (1)H magnetic resonance (MR) spectroscopy to characterize the abnormalities in hepatic lipid content and composition in patients with LAL deficiency 23652569_Mutations in lysosomal acid lipase A result in two phenotypes depending on the extent of lysosomal acid lipase deficiency. [Review] 24069331_the rs1412444 and rs2246833 of the LIPA gene are shared susceptibility polymorphisms for CAD among different ethnicities. 24122380_To our knowledge, this is the first pediatric case of genetically and biopsy confirmed CESD without hepatomegaly, suggesting that this diagnosis can be easily missed. 24832708_Wolmans disease is a rare autosomal recessive lysosomal storage disease. 25620107_The observed loss-of-function phenotype in cholesteryl ester storage disease patients with the His295Tyr (H295Y) mutation in the LAL gene might arise from a combination of protein destabilization and the shift to a non-functional soluble aggregate. 25624737_Case Report: Mexican sisters with heterozygous mutations in exon 4: c.253C>A and c.294C>G resulting in lysosomal acid lipase deficiency. 26212911_lysosomal acid lipase in hepatocytes is a critical metabolic enzyme in controlling neutral lipid metabolism 26288848_These findings suggest a strong association between impaired LAL activity and Non-alcoholic fatty liver disease. 27219619_Study demonstrates that liver cirrhosis from any etiology is characterized by a significant reduction of LAL activity but no known c.894G>A SNP, which is likely on an acquired base and independent from the etiology of hepatic disease. 27354281_results indicate that LAL is the major acid RE hydrolase and required for functional retinoid homeostasis. 27423329_LIPA mutations may have a role in with a clinical diagnosis of familial hypercholesterolemia 27531897_LAL plays a critical role in regulating mesenchymal stem cells' ability to stimulate tumor growth and metastasis, which provides a mechanistic basis for targeting LAL in MSCs to reduce the risk of cancer metastasis 28279971_Coronary artery disease-associated coding variant rs1051338 causes reduced lysosomal LAL protein and activity because of increased LAL degradation. 28396038_Report a marked reduction of LAL activity in patients with cryptogenic cirrhosis. 28587063_LAL activity is significantly reduced in NAFLD, compared to that in HCV patients. This finding is particularly evident in the pre-cirrhotic stage of disease. LAL activity is also correlated with platelet and white blood cell count, suggesting an analytic interference of portal-hypertension-induced pancytopenia on DBS-determined LAL activity. 28881270_Report LIPA variants/phenotype in childhood-onset lysosomal acid lipase deficiency. 28882870_Use CRISPR/Cas9 techniques to knockout LIPA in human induced pluripotent stem cells and differentiate them to macrophages. 28958330_LIPA associated with Familial Hypercholesterolemia and Polygenic Hypercholesterolemia in patients with Acute Coronary Syndrome , age =65 years, and LDL-C levels >/=160 mg/dl. 29196158_Mutation in the lysosomal acid lipase gene is associated with cholesteryl ester storage disease in hypercholesterolemia. 30684275_Homozygous or compound heterozygous LIPA mutations were identified. Lysosomal acid lipase (LAL) activity in white blood cells is a validated tool for Lysosomal acid lipase deficiency (LAL-D) diagnosis. A cut-off below 12 pmol/min/mg protein might be useful to discriminate patients with LIPA mutations. 31131398_Most pathogenic mutations in LIPA gene (LAL) result in defective enzyme activity by affecting the normal folding of LAL. Nearly all mutations that affect the stability of the core domain result in endoplasmic reticulum (ER) stress. As a consequence, ER stress resulted in ER-associated degradation of the mutant protein. Rescue of mutant proteins by chemical chaperones did not restore enzymatic activity. 31182375_Mutations identified in a cohort of Mexican patients with lysosomal acid lipase deficiency. 31392821_Lysosomal acid lipase activity and liver fibrosis in the clinical continuum of non-alcoholic fatty liver disease. 31435171_Patients with non-alcoholic fatty liver disease show a significant, progressive reduction of LAL (LIPA) activity from simple steatosis to non-alcoholic steatohepatitis and cryptogenic cirrhosis. 31505261_LAL activity is reduced in non-alcoholic fatty liver disease patients, independently from disease progression. In vitro, impaired LAL activity induced by Fatty Acids loading was rescued by PPAR-alpha activation. 31582009_Lysosomal acid lipase deficiency is most common in Caucasians of European descent. It is due to a mutation in the LIPA gene. In North America, the prevalence of CESD is about 0.0008% in Caucasian and Hispanic populations. 31587363_Lysosomal acid lipase does not have a propeptide and should not be considered being a proprotein. 31645127_common LIPA exonic variants in the signal peptide are of minimal functional significance and suggest coronary artery disease risk is instead associated with increased LIPA function linked to intronic variants. Understanding the mechanisms and cell-specific contexts of LIPA function in the plaque is necessary to understand its association with cardiovascular risk. 32058863_LIPA gene mutations affect the composition of lipoproteins: Enrichment in ACAT-derived cholesteryl esters. 32101051_Association Between LOX-1, LAL, and ACAT1 Gene Single Nucleotide Polymorphisms and Carotid Plaque in a Northern Chinese Population. 32361002_Lysosomal acid lipase is the major acid retinyl ester hydrolase in cultured human hepatic stellate cells but not essential for retinyl ester degradation. 32463622_Reduced Lysosomal Acid Lipase Activity in Blood and Platelets Is Associated With Nonalcoholic Fatty Liver Disease. 32482718_Crystal structure of human lysosomal acid lipase and its implications in cholesteryl ester storage disease. 33792344_Low LAL (Lysosomal Acid Lipase) Expression by Smooth Muscle Cells Relative to Macrophages as a Mechanism for Arterial Foam Cell Formation. 33838322_Loss of function of lysosomal acid lipase (LAL) profoundly impacts osteoblastogenesis and increases fracture risk in humans. 33845949_Lysosomal acid lipase gene single nucleotide polymorphism and pulmonary tuberculosis susceptibility. 34476902_Effect of a common missense variant in LIPA gene on fatty liver disease and lipid phenotype: New perspectives from a single-center observational study. 34894393_Disturbance of lipid homeostasis in lysosomal lipase deficiency - pathomechanism, diagnosis and treatment', trans 'Zaburzenie homeostazy lipidowej w deficycie lizosomalnej lipazy - patomechanizm, diagnostyka i leczenie. 34957913_Lysosomal acid lipase promotes endothelial proliferation in cold-activated adipose tissue. 35192625_Interpreting coronary artery disease GWAS results: A functional genomics approach assessing biological significance. | ENSMUSG00000024781 | Lipa | 1256.84787 | 1.1342895 | 0.1817889057 | 0.14603786 | 1.533648e+00 | 2.155658e-01 | 9.998360e-01 | No | Yes | 1246.26435 | 299.272199 | 1.383836e+03 | 255.667706 | |
| ENSG00000107815 | 56652 | TWNK | protein_coding | Q96RR1 | FUNCTION: [Isoform 1]: Mitochondrial helicase involved in mtDNA replication and repair (PubMed:12975372, PubMed:15167897, PubMed:17324440, PubMed:18039713, PubMed:18971204, PubMed:25824949, PubMed:26887820, PubMed:27226550). Might have a role in mtDNA repair (PubMed:27226550). Has DNA strand separation activity needed to form a processive replication fork for leading strand synthesis which is catalyzed by the formation of a replisome complex with POLG and mtSDB (PubMed:12975372, PubMed:15167897, PubMed:18039713, PubMed:22383523, PubMed:26887820, PubMed:27226550). Preferentially unwinds DNA substrates with pre-existing 5'-and 3'- single-stranded tails but is also active on a 5'- flap substrate (PubMed:12975372, PubMed:15167897, PubMed:18039713, PubMed:22383523, PubMed:26887820, PubMed:27226550). Can dissociate the invading strand of immobile or mobile D-loop DNA structures irrespective of the single strand polarity of the third strand (PubMed:27226550). In addition to its DNA strand separation activity, also has DNA strand annealing, DNA strand-exchange and DNA branch migration activities (PubMed:22383523, PubMed:26887820, PubMed:27226550). {ECO:0000269|PubMed:12975372, ECO:0000269|PubMed:15167897, ECO:0000269|PubMed:17324440, ECO:0000269|PubMed:18039713, ECO:0000269|PubMed:18971204, ECO:0000269|PubMed:22383523, ECO:0000269|PubMed:25824949, ECO:0000269|PubMed:26887820, ECO:0000269|PubMed:27226550}.; FUNCTION: [Isoform 2]: Lack DNA unwinding and ATP hydrolysis activities (PubMed:18039713). Does not bind single-stranded or double-stranded DNA (PubMed:18039713). {ECO:0000269|PubMed:18039713}. | ATP-binding;Alternative splicing;DNA replication;Deafness;Disease variant;Helicase;Hydrolase;Mitochondrion;Mitochondrion nucleoid;Neurodegeneration;Neuropathy;Nucleotide-binding;Primary mitochondrial disease;Progressive external ophthalmoplegia;Reference proteome;Transit peptide | This gene encodes a hexameric DNA helicase which unwinds short stretches of double-stranded DNA in the 5' to 3' direction and, along with mitochondrial single-stranded DNA binding protein and mtDNA polymerase gamma, is thought to play a key role in mtDNA replication. The protein localizes to the mitochondrial matrix and mitochondrial nucleoids. Mutations in this gene cause infantile onset spinocerebellar ataxia (IOSCA) and progressive external ophthalmoplegia (PEO) and are also associated with several mitochondrial depletion syndromes. Alternative splicing results in multiple transcript variants encoding distinct isoforms.[provided by RefSeq, Aug 2009]. | hsa:56652; | mitochondrial matrix [GO:0005759]; mitochondrial nucleoid [GO:0042645]; mitochondrion [GO:0005739]; 5'-3' DNA helicase activity [GO:0043139]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA helicase activity [GO:0003678]; identical protein binding [GO:0042802]; protease binding [GO:0002020]; single-stranded DNA binding [GO:0003697]; cellular response to glucose stimulus [GO:0071333]; DNA unwinding involved in DNA replication [GO:0006268]; mitochondrial DNA replication [GO:0006264]; mitochondrial transcription [GO:0006390]; protein hexamerization [GO:0034214] | 12163192_Twinkle appears to be the most common gene associated with adPEO in Australian families. 12686611_both the mitochondrial transcription factor TFAM and mitochondrial single-stranded DNA-binding protein colocalize with Twinkle in intramitochondrial foci 12872260_A novel PEO1 mutation in a sporadic PEO patient with multiple mtDNA deletions. 15181170_mechanism whereby mutations in twinkle result in progressive accumulation of multiple mtDNA deletions in post- mitotic tissues 15668446_A novel heterozygous A to G transition at nucleotide 955 of C10Orf2 (Twinkle)ws found in siblings with sensory ataxic neuropathy. 16135556_the severe neurological phenotype observed in infantile onset spinocerebellar ataxia, suggests that the mutated Twinky and Twinkle play a crucial role in the maintenance and/or function of specific affected neuronal subpopulations [twinky] 16385451_Observational study of gene-disease association. (HuGE Navigator) 17324440_Mitochondrial DNA helicase belongs to the DnaB-like family of replicative DNA helicases. 17420318_Observational study of gene-disease association. (HuGE Navigator) 17614276_Direct sequencing showed a heteroplasmic mutation at nucleotide 7506 in the dihydrouridine stem of the tRNA(Ser(UCN)) gene. 17614277_We report a Spanish family showing a mild phenotype characterized by autosomal dominant ocular myopathy and morphological signs of mitochondrial dysfunction, that harboured a novel(p.R357P) mutation in the hot-spot linker region of the twinkle protein. 17620490_This finding broadens the clinical spectrum of Twinkle gene mutations and further implicates loss of mitochondrial DNA integrity in the pathogenesis of Parkinson disease. 17722119_Identifying other Twinkle mutations in mitochondrial DNA depletion and/or autosomal dominant progressive external ophthalmoplegia and studying their impact on proteins should help in understanding why some mutations are recessive and others are dominant. 17921179_study reports a new phenotype in two siblings with compound heterozygous Twinkle mutations (A318T and Y508C), characterized by severe early onset encephalopathy and signs of liver involvement 18039713_The N-terminal part of TWINKLE is required for efficient binding to single-stranded DNA. 18396044_We describe a new de novo mutation (1110C>A) in the PEO1 gene in patients with Ophthalmoplegia, Chronic Progressive External in a mother and her two sons. 18575922_Data show that PEO1 has a major role in determining familial progressive external ophthalmoplegia. 18575922_Observational study of gene-disease association. (HuGE Navigator) 18971204_MtDNA replication pausing is the consequence of Twinkle mutations that predisposes to progressive external ophthalmoplegia. 18973250_A heterozygous mutation predicting a R334Q substitution in Twinkle was associated with progressive external opthalmoplegia and/or Parkinsonism in several members of a family. 18989381_This study widens the mutation spectrum of PEO1 and is the first to report the PEO1 mutation in the Chinese population. 19425506_Association of the T(-365)C POLG1, G(-25)A ANT1 and G(-605)T PEO1 gene polymorphisms with diabetic polyneuropathy in patients with type 1 diabetes mellitus 19425506_Observational study of gene-disease association. (HuGE Navigator) 19428252_heterozygous c.907C>T (p.R303W) mutation found in the N-terminal domain of the human mitochondrial DNA helicase, Twinkle protein, associated with phenotypically mild autosomal dominant progressive external ophthalmoplegia 19513767_Twinkle variations in the linker domain alter cerebral function and further implicate disrupted mitochondrial DNA integrity in the pathogenesis of dementia 19853444_study of a Saudi Arabian family with autosomal dominant progressive external ophthalmoplegia & late onset multi-organ failure caused by a novel PEO1/Twinkle mutation (1078C > G + 1079T > G double nucleotide change predicting a Leu360Gly substitution) 20181062_Observational study of gene-disease association. (HuGE Navigator) 20479361_Our data suggest a shared clinical phenotype with variable mild multiorgan involvement, and that the contribution of PEO1 mutations as a cause of autosomal dominant progressive external ophthalmoplegia may well be underestimated. 20659899_Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis, and helicase activity. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20880070_We present a new French family of whom two members displayed autosomal dominant progressive external ophthalmoplegia associated with the R374Q mutation of the Twinkle gene 21540127_Multimeric unicircular mtDNA molecules are seen in cells expressing Twinkle. 21689831_PEO1 sequencing discloses novel mutations in exons 1 and 4 of the gene in chronic external ophthalmoplegia; this is the first report of a mutation occurring in exon 4. 22353293_A novel homozygous missense mutation c.1366C>G (L456V) in C10orf2 (the Twinkle gene) was identified in a family with infantile onset spinocerebellar ataxia. 22383523_the strand annealing activity of TWINKLE may play a role in recombination-mediated replication initiation found in the mitochondria of mammalian brain and heart or in replication fork regression during repair of damaged DNA replication forks. 22580846_analysis did not reveal disease causing POLG or PEO1 mutations in patients with atypical parkinsonism 22952820_Overexpression of d-mtDNA helicase containing either the K388A or A442P mutations causes a mitochondrial oxidative phosphorylation (OXPHOS) defect that significantly reduces cell proliferation. 23375728_A homozygous mutation in TWINKLE is the cause of multisystemic failure including renal tubulopathy in three consanguinity siblings. 24018892_16-year follow-up of autosomal dominant progressive external ophthalmoplegia (adPEO) due to the p.R357P gene mutation in PEO1; adPEO due to this mutation is a late-onset ocular myopathy beginning with ptosis and progressing slowly; ophthalmoparesis, if present, is mild 24218554_Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. 24524965_Mitochondrial DNA (mtDNA) content plays an important role in energy production and sustaining normal physiological function. 24816431_Identified compound heterozygous mutations of the C10orf2 gene as the cause of infantile-onset spinocerebellar ataxia with sensorimotor polyneuropathy and myopathy. 25193669_The mitochondrial replicative helicase Twinkle inefficiently unwinds well characterized intermolecular and intramolecular G-quadruplex DNA substrates, as well as a unimolecular G4 substrate. 25355836_Mutations in Twinkle were linked to Perrault syndrome with neurologic features. 25824949_An electron microscopy model of Twinkle reveals a hexameric two-layered ring comprising the zinc-binding domain and RNA polymerase domain in one layer and the RecA-like hexamerization C-terminal domain in another. 26689116_sequencing coding regions of C10orf2 revealed three variants in three different patients, of which two were novel (c.1964G>A/p.G655D; c.204G>A/p.G68G) variants and one was reported (c.1052A>G/p. N351S). 26838077_We identified a missense mutation in c10orf2 in an Iranian family with an association to progressive external ophthalmoplegia, myopathy, dysphagia, dysphonia, and behavior change. Early death was also a novel feature in affected family members. 26887820_Four DNA modifying activities of TWINKLE: strand-separation, strand-annealing, strand-exchange and branch migration suggest a dual role of TWINKLE in mitochondrial DNA maintenance. 26970254_ere we present eight families affected by Perrault syndrome. In five families we identified novel or previously reported variants in HSD17B4, LARS2, CLPP and C10orf2 27112570_It has been demonstrated that MTERF1 arrests mitochondrial DNA (mtDNA) replication with distinct polarity whereby MTERF1 acts as a directional contra-helicase, blocking mtDNA unwinding by the mitochondrial helicase TWINKLE. 27226550_studies shed new insight on the catalytic functions of Twinkle on the key DNA structures it would encounter during replication or possibly repair of the mitochondrial genome and how well it tolerates potential roadblocks to DNA unwinding 27600867_high mtDNA copy number is likely inheritable, which may act as a favorable factor to familial longevity through assuring adequate energy supply. 28178980_the newly identified for Perrault syndrome TWNK gene is involved in its pathogenesis. 29316893_A German cohort of 440 patients undergoing analysis for spinocerebellar ataxia (SCA) diagnosis was screened for DNA repeat expansions in SCA8, SCA10, SCA12, SCA36, FXTAS as well as for the pathogenic hexanucleotide repeat in the C9orf72 gene. Results found five patients showed 92 or more SCA8 CTA/CTG combined repeats. 30496414_Results provide strong evidence that defects in oligomerization resulting from mutations in the linker helix and primase domain of TWINKLE are a primary cause of the development of autosomal dominant progressive external ophthalmoplegia (adPEO) and potentially other life-threatening mitochondrial pathologies 30715486_These findings suggest that Twinkle is essential for RNA organization in granules, and that mtSSB is involved in the recently proposed GRSF1-mtRNA degradosome pathway, a route suggested to be particularly aimed at degradation of G-quadruplex prone long non-coding mtRNAs. 31455269_Two novel TWNK c.1186 C > T/ c.1844 G > C compound heterozygous mutations which were probably the disease-causing mutations of hepatocerebral form of Mitochondrial DNA depletion syndrome (MDS) and described the clinical manifestations of the proband, which expanded the phenotypic spectrum of MDS caused by variants in TWNK. 31455392_patients with Perrault syndrome caused by TWNK mutations will manifest neurological signs in adulthood. 32161153_Clinical, pathological and genetic spectrum in 89 cases of mitochondrial progressive external ophthalmoplegia. 32213598_Single-molecule level structural dynamics of DNA unwinding by human mitochondrial Twinkle helicase. 32387964_Characteristics of globus pallidus internus local field potentials in generalized dystonia patients with TWNK mutation. 33087282_Consequences of compromised mitochondrial genome integrity. 33396418_Progressive External Ophthalmoplegia in Polish Patients-From Clinical Evaluation to Genetic Confirmation. 33847968_Stimulation of Variant Forms of the Mitochondrial DNA Helicase Twinkle by the Mitochondrial Single-Stranded DNA-Binding Protein. | ENSMUSG00000025209 | Twnk | 4379.81619 | 0.9632486 | -0.0540198990 | 0.09519828 | 3.253428e-01 | 5.684144e-01 | 9.998360e-01 | No | Yes | 4941.77026 | 354.853162 | 5.168694e+03 | 287.841284 | |
| ENSG00000107897 | 91452 | ACBD5 | protein_coding | Q5T8D3 | FUNCTION: Acyl-CoA binding protein which acts as the peroxisome receptor for pexophagy but is dispensable for aggrephagy and nonselective autophagy. Binds medium- and long-chain acyl-CoA esters. {ECO:0000269|PubMed:24535825}. | 3D-structure;Acetylation;Alternative splicing;Autophagy;Coiled coil;Leukodystrophy;Lipid-binding;Membrane;Peroxisome;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the acyl-Coenzyme A binding protein family, known to function in the transport and distribution of long chain acyl-Coenzyme A in cells. This gene may play a role in the differentiation of megakaryocytes and formation of platelets. A related protein in yeast is involved in autophagy of peroxisomes. A mutation in this gene has been associated with autosomal dominant thrombocytopenia. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:91452; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; fatty-acyl-CoA binding [GO:0000062]; lipid binding [GO:0008289]; autophagy of peroxisome [GO:0030242] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 20626622_A mutation in the acyl-coenzyme A binding domain-containing protein 5 gene (ACBD5 ) may have a role in autosomal dominant thrombocytopenia 24905344_Yeast Atg37, which is homologous to human ACBD5, is involved in autophagy of peroxisomes. 25175022_Findings suggest that the ACBD5-RET rearrangement is causatively involved in the development of papillary thyroid cancer. 27899449_ACBD5 captures VLC-CoAs on the cytosolic side of the peroxisomal membrane so that the transport of VLC-CoAs into peroxisomes and subsequent beta-oxidation thereof can proceed efficiently. 28108524_ACBD5-VAPB interaction regulates peroxisome-endoplasmic reticulum associations. Loss of PO-ER association perturbs PO membrane expansion and increases PO movement. 28108526_VAP-ACBD5-mediated contact between the endoplasmic reticulum and peroxisomes mediate organelle maintenance and lipid homeostasis. 30589881_Peroxisomal (PO) long range movements were largely diminished in response to human ACBD5 overexpression in primary mouse hippocampal neurons. PO localization significantly changed in ACBD5-transfected neurons, PO numbers in neurites increased, while PO density in the soma was decreased. Alterations in PO motility and distribution in the hippocampal neurons were independent of the interaction between ACBD5 and mouse Vapb. 33427402_First reported adult patient with retinal dystrophy and leukodystrophy caused by a novel ACBD5 variant: A case report and review of literature. 35019937_Regulating peroxisome-ER contacts via the ACBD5-VAPB tether by FFAT motif phosphorylation and GSK3beta. | ENSMUSG00000026781 | Acbd5 | 111.18221 | 1.0952152 | 0.1312144267 | 0.28314930 | 2.110497e-01 | 6.459459e-01 | 9.998360e-01 | No | Yes | 102.85036 | 21.126946 | 9.626221e+01 | 15.299771 | |
| ENSG00000108021 | 54906 | TASOR2 | protein_coding | Q5VWN6 | Alternative splicing;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:54906; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | ENSMUSG00000033799 | Tasor2 | 1479.42901 | 0.9092912 | -0.1371857780 | 0.12338899 | 1.212721e+00 | 2.707938e-01 | 9.998360e-01 | No | Yes | 1923.91382 | 399.543790 | 1.848077e+03 | 295.595086 | ||||
| ENSG00000108094 | 8453 | CUL2 | protein_coding | Q13617 | FUNCTION: Core component of multiple cullin-RING-based ECS (ElonginB/C-CUL2/5-SOCS-box protein) E3 ubiquitin-protein ligase complexes, which mediate the ubiquitination of target proteins (PubMed:11384984, PubMed:26138980, PubMed:29779948, PubMed:29775578). CUL2 may serve as a rigid scaffold in the complex and may contribute to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme (PubMed:9122164, PubMed:10973499, PubMed:11384984, PubMed:12609982, PubMed:24076655). The E3 ubiquitin-protein ligase activity of the complex is dependent on the neddylation of the cullin subunit and is inhibited by the association of the deneddylated cullin subunit with TIP120A/CAND1 (PubMed:12609982, PubMed:24076655, PubMed:27565346). The functional specificity of the ECS complex depends on the substrate recognition component (PubMed:9122164, PubMed:10973499, PubMed:26138980, PubMed:29779948, PubMed:29775578). ECS(VHL) mediates the ubiquitination of hypoxia-inducible factor (HIF) (PubMed:9122164, PubMed:10973499). A number of ECS complexes (containing either KLHDC2, KLHDC3, KLHDC10, APPBP2, FEM1A, FEM1B or FEM1C as substrate-recognition component) are part of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation (PubMed:26138980, PubMed:29779948, PubMed:29775578). ECS complexes and ARIH1 collaborate in tandem to mediate ubiquitination of target proteins (PubMed:27565346). {ECO:0000269|PubMed:10973499, ECO:0000269|PubMed:11384984, ECO:0000269|PubMed:12609982, ECO:0000269|PubMed:24076655, ECO:0000269|PubMed:26138980, ECO:0000269|PubMed:27565346, ECO:0000269|PubMed:29775578, ECO:0000269|PubMed:29779948, ECO:0000269|PubMed:9122164}. | 3D-structure;Acetylation;Alternative splicing;Host-virus interaction;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:26138980, ECO:0000269|PubMed:29775578, ECO:0000269|PubMed:29779948}. | hsa:8453; | Cul2-RING ubiquitin ligase complex [GO:0031462]; cullin-RING ubiquitin ligase complex [GO:0031461]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; SCF ubiquitin ligase complex [GO:0019005]; VCB complex [GO:0030891]; protein-containing complex binding [GO:0044877]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; G1/S transition of mitotic cell cycle [GO:0000082]; intrinsic apoptotic signaling pathway [GO:0097193]; protein ubiquitination [GO:0016567]; SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [GO:0031146] | 15280393_Cullin selection is determined by specific Elongin C and Skp1 sequences 16385451_Observational study of gene-disease association. (HuGE Navigator) 17609271_the HPV16 E7-associated cullin 2 ubiquitin ligase complex contributes to aberrant degradation of the pRB tumor suppressor in HPV16 E7-expressing cells. 18372249_CUL2 is required for normal vasculogenesis, at least in part mediated by its regulation of HIF-mediated transcription. 18984674_Observational study of gene-disease association. (HuGE Navigator) 19760754_Observational study of gene-disease association. (HuGE Navigator) 20078552_Observational study of gene-disease association. (HuGE Navigator) 20078552_The data suggest that neither HIF1alpha nor CUL2 mutation may play a central role in HIF1alpha activation in gastric, colorectal, breast, lung and hepatocellular carcinomas, and acute leukemias. 21822215_The authors applied protein-complex purification strategies and identified PRAME as a substrate recognition subunit of a Cullin2-based E3 ubiquitin ligase. 22537386_Study finds that UBXN7 over-expression converts CUL2 to its neddylated form and causes the accumulation of non-ubiquitylated HIF1alpha. 22912744_a novel link between the oncoprotein PRAME and the conserved EKC complex 24453364_Nevertheless, the identification of the Cul2 box may allow prediction of Cullin specificity for all E4orf6-containing Adenoviridae. 25326537_Cullins are a family of NEDD8 targets important in the stabilization and degradation of proteins, such as hypoxia-inducible factor (HIF1A), via Cullin-2. 26751167_misfolded TDP-43 is cleared by VHL/CUL2 in a step-wise manner via fragmentation. 27153550_Results suggest that E7 recruited CUL2, driven by CUL2/E2F1/miR-424 regulatory loop, is overexpressed and accelerates HPV16-induced cervical carcinogenesis. 28591624_The crystal structure of a pentameric CRL2(VHL) complex, composed of Cul2, Rbx1, Elongin B, Elongin C, and pVHL reveals hydrophobic contact residues critical for the Cul2 interactions. 29367246_direct evidence that both A3A and HPV16 E7 interact with CUL2, suggesting that the E7-CUL2 complex formed during HPV infection may regulate A3A protein levels in the cell. 29844124_Twist1 bound the Cul2 promoter. 31898228_Roles of Cullin-2 E3 Ubiquitin Ligase Complex in Cancer 32918970_Integrating cullin2-RING E3 ligase as a potential biomarker for glioblastoma multiforme prognosis and radiosensitivity profiling. 33827988_ORF10-Cullin-2-ZYG11B complex is not required for SARS-CoV-2 infection. 34195792_Reconstitution of human CMG helicase ubiquitylation by CUL2LRR1 and multiple E2 enzymes. 34717191_Immunoexpression of HSPA9 and CUL2 in prostatic tissue and adenocarcinoma. | ENSMUSG00000024231 | Cul2 | 268.89258 | 0.8980338 | -0.1551582696 | 0.21118781 | 5.048678e-01 | 4.773690e-01 | 9.998360e-01 | No | Yes | 261.41910 | 40.617189 | 2.764418e+02 | 33.098225 | |
| ENSG00000108219 | 81619 | TSPAN14 | protein_coding | Q8NG11 | FUNCTION: Regulates maturation and trafficking of the transmembrane metalloprotease ADAM10 (PubMed:26668317, PubMed:23035126, PubMed:26686862). Negatively regulates ADAM10-mediated cleavage of GP6 (By similarity). Promotes ADAM10-mediated cleavage of CDH5 (By similarity). {ECO:0000250|UniProtKB:Q8QZY6, ECO:0000269|PubMed:23035126, ECO:0000269|PubMed:26668317, ECO:0000269|PubMed:26686862}. | Alternative splicing;Cell membrane;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:81619; | cell surface [GO:0009986]; endoplasmic reticulum lumen [GO:0005788]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; specific granule membrane [GO:0035579]; tertiary granule membrane [GO:0070821]; tetraspanin-enriched microdomain [GO:0097197]; enzyme binding [GO:0019899]; positive regulation of Notch signaling pathway [GO:0045747]; protein localization to plasma membrane [GO:0072659]; protein maturation [GO:0051604] | 19473719_genetic alterations of TSPAN14, SLC2A13 and PHF20 could play a role in non-small-cell lung cancer promotion 26668317_Data suggest that large extracellular loop of Tspan14 mediates interaction with ADAM10, promotes ADAM10 maturation/trafficking to cell surface, and affects ADAM10 substrate specificity; ADAM10/Tspan14 interact in platelets/vascular endothelial cells. | ENSMUSG00000037824 | Tspan14 | 1599.95421 | 1.0929759 | 0.1282615915 | 0.13002669 | 9.761807e-01 | 3.231436e-01 | 9.998360e-01 | No | Yes | 1604.79441 | 166.126753 | 1.366344e+03 | 109.946032 | ||
| ENSG00000108423 | 51174 | TUBD1 | protein_coding | Q9UJT1 | FUNCTION: Acts as a positive regulator of hedgehog signaling and regulates ciliary function. {ECO:0000250|UniProtKB:Q9R1K7}. | Alternative splicing;Cell projection;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Developmental protein;GTP-binding;Microtubule;Nucleotide-binding;Nucleus;Reference proteome | hsa:51174; | centriole [GO:0005814]; cilium [GO:0005929]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; microtubule [GO:0005874]; nucleoplasm [GO:0005654]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; structural constituent of cytoskeleton [GO:0005200]; cell projection organization [GO:0030030]; microtubule cytoskeleton organization [GO:0000226]; mitotic cell cycle [GO:0000278]; positive regulation of smoothened signaling pathway [GO:0045880] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20508983_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 28137601_our data suggest that TUBD1 mRNA isoform expression profile in peripheral blood could be an accessible biomarker for predicting the risk for diabetic retinopathy development. 28906251_Here, the authors report that centrioles in delta-tubulin and epsilon-tubulin null mutant human cells lack triplet microtubules and fail to undergo centriole maturation. | ENSMUSG00000020513 | Tubd1 | 198.27640 | 0.9775540 | -0.0327516301 | 0.24138246 | 1.766372e-02 | 8.942686e-01 | 9.998360e-01 | No | Yes | 186.66247 | 30.626424 | 1.936986e+02 | 24.569302 | ||
| ENSG00000108509 | 23125 | CAMTA2 | protein_coding | O94983 | FUNCTION: Transcription activator. May act as tumor suppressor. {ECO:0000269|PubMed:11925432}. | ANK repeat;Activator;Alternative splicing;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a member of the calmodulin-binding transcription activator protein family. Members of this family share a common domain structure that consists of a transcription activation domain, a DNA-binding domain, and a calmodulin-binding domain. The encoded protein may be a transcriptional coactivator of genes involved in cardiac growth. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Jan 2010]. | hsa:23125; | chromatin [GO:0000785]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; histone deacetylase binding [GO:0042826]; sequence-specific DNA binding [GO:0043565]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cardiac muscle hypertrophy in response to stress [GO:0014898]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 29110692_Mutation of CAMTA2 resulting in post-transcriptional inhibition of its own gene activity likely underlies a novel syndromic tremulous dystonia. | ENSMUSG00000040712 | Camta2 | 905.01913 | 1.0302553 | 0.0430019201 | 0.14060642 | 9.632420e-02 | 7.562861e-01 | 9.998360e-01 | No | Yes | 874.22956 | 148.698681 | 7.893549e+02 | 103.888669 | |
| ENSG00000108840 | 10014 | HDAC5 | protein_coding | Q9UQL6 | FUNCTION: Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Involved in muscle maturation by repressing transcription of myocyte enhancer MEF2C. During muscle differentiation, it shuttles into the cytoplasm, allowing the expression of myocyte enhancer factors. Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer. Serves as a corepressor of RARA and causes its deacetylation (PubMed:28167758). In association with RARA, plays a role in the repression of microRNA-10a and thereby in the inflammatory response (PubMed:28167758). {ECO:0000269|PubMed:24413532, ECO:0000269|PubMed:28167758}. | 3D-structure;Acetylation;Alternative splicing;Chromatin regulator;Cytoplasm;Hydrolase;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc | Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene belongs to the class II histone deacetylase/acuc/apha family. It possesses histone deacetylase activity and represses transcription when tethered to a promoter. It coimmunoprecipitates only with HDAC3 family member and might form multicomplex proteins. It also interacts with myocyte enhancer factor-2 (MEF2) proteins, resulting in repression of MEF2-dependent genes. This gene is thought to be associated with colon cancer. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:10014; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; histone deacetylase complex [GO:0000118]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor binding [GO:0140297]; histone deacetylase activity [GO:0004407]; histone deacetylase binding [GO:0042826]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; NAD-dependent histone deacetylase activity (H3-K14 specific) [GO:0032041]; protein deacetylase activity [GO:0033558]; protein kinase C binding [GO:0005080]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription corepressor binding [GO:0001222]; B cell activation [GO:0042113]; B cell differentiation [GO:0030183]; cellular response to insulin stimulus [GO:0032869]; chromatin organization [GO:0006325]; chromatin remodeling [GO:0006338]; heterochromatin assembly [GO:0031507]; histone deacetylation [GO:0016575]; inflammatory response [GO:0006954]; negative regulation of cell migration involved in sprouting angiogenesis [GO:0090051]; negative regulation of myotube differentiation [GO:0010832]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein deacetylation [GO:0006476]; regulation of gene expression, epigenetic [GO:0040029]; regulation of myotube differentiation [GO:0010830]; regulation of protein binding [GO:0043393] | 11929873_Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor 12019172_Histone deacetylase 5 is not a p53 target gene, but its overexpression inhibits tumor cell growth and induces apoptosis. 12242305_MITR, HDAC4, and HDAC5 associate with heterochromatin protein 1 (HP1), an adaptor protein that recognizes methylated lysines within histone tails and mediates transcriptional repression by recruiting histone methyltransferase 12626519_HDAC5 binds to Ca(2+)/calmodulin and inhibits MEF2a binding 15194749_ICP0 of herpes simplex virus Type 1 is able to overcome the HDAC5 amino-terminal- and MITR-induced MEF2A repression in gene reporter assays 15590418_The HDAC5, a class II HDAC involved in myogenesis, was not detected in the tissues. 16221676_G betagamma binds HDAC5 and inhibits its transcriptional co-repression activity 16236793_novel transcriptional pathway under the control of class II HDACs and suggest a role for these transcriptional repressors as signal-responsive regulators of antigen presentation 17975112_NO-dependent PP2A activation plays a key role in nuclear translocation of class II HDACs HDAC4 and HDAC5 18184930_AMP-activated protein kinase (AMPK) regulates GLUT4 transcription through the histone deacetylase (HDAC)5 transcriptional repressor. 18218981_Chronic upregulation/activation of CaMKIID, and PKD in heart failure shifts HDAC5 out of the nucleus, derepressing transcription of hypertrophic genes. 18288090_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18332134_Protein kinase D-HDAC5 pathway plays an important role in VEGF regulation of gene transcription and angiogenesis 19071119_These results indicate that HDAC4 and HDAC5 increase the transactivation function of HIF-1alpha by promoting dissociation of HIF-1alpha from FIH-1 and association with p300. 19351956_HDAC5 represses angiogenic genes, such as FGF2 and Slit2, which causally contribute to capillary-like sprouting of endothelial cells. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20042720_phosphorylation-dependent derepression of HDAC5 mediates flow-induced KLF2 and eNOS expression as well as flow anti-inflammation, and suggest that HDAC5 could be a potential therapeutic target for the prevention of atherosclerosis. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20716686_Findings identify HDAC5 as a substrate of PKA and reveal a cAMP/PKA-dependent pathway that controls HDAC5 nucleocytoplasmic shuttling and represses gene transcription. 21047791_differentiation-dependent GLUT4 gene expression in 3T3-L1 adipocytes is dependent on the nuclear concentration of a class II histone deacetylase (HDAC) protein, HDAC5 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21081666_Ser279 is a critical phosphorylation within the NLS involved in the nuclear import of HDAC5 21146494_in addition to activation of protein kinase D isozymes by phosphorylating Ser744 and Ser748 at their activation sites, PKCdelta may also play a role in the regulation of HDAC5 by phosphorylation of Ser259 21508384_Significantly increased methylation of the HDAC5 gene was associated with astrocytomas. 22243750_The results of this study suggested that suggest that HDAC5 provides a delayed braking mechanism on gene expression programs that support the development, but not expression, of cocaine reward behaviors. 22301920_HDAC5 in the maintenance/assembly of pericentric heterochromatin structure and demonstrate that class IIa HDAC5 can represent a potential target for anticancer therapies. 22914591_Loss of HDAC5 impairs memory function but has little impact in a transgenic mouse model of amyloid pathology. 22941650_The current study identified the class II deacetylase HDAC5 as a novel promoting factor of CTG*CAG expansions. 22991226_Data suggest that HDAC5 regulates muscle glucose metabolism and insulin action and that HDAC inhibitors can be used to modulate these parameters in muscle cells. 23297420_Dephosphorylation at a conserved SP motif governs cAMP sensitivity and nuclear localization of class IIa histone deacetylases HDAC4, 5 and 9 23364788_Nuclear calcium signaling is a regulator of nuclear export of HDAC4 and HDAC5. 23615070_Data indicate there was a link between baseline viral load, age (40 years), IL-28B (rs12979860), HDAC2 (rs3778216), HDAC3 (rs976552) and HDAC5 (rs368328) with sustained virological response (SVR). 23729589_HDAC5 is essential for the length maintenance of long telomeres and its depletion is required for sensitization of cancer cells with long telomeres to chemotherapy. 23812427_findings show N-Myc upregulated HDAC5 expression in neuroblastoma cells; HDAC5 repressed NEDD4 gene expression,increased Aurora A gene expression and consequently upregulated N-Myc protein expression;data identify HDAC5 as a novel co-factor in N-Myc oncogenesis 24092570_At the molecular level, we demonstrated that HDAC5 promoted mRNA expression of twist 1, which has been reported as an oncogene 24120667_These findings suggest that HDAC5 is a key determinant of p53-mediated cell fate decisions in response to genotoxic stress. 24191246_we show that Stat3 binds to the promoter region of PTPN13 and promotes its activity through recruiting HDAC5. Thus, our results suggest a previously unknown Stat3-PTPN13 molecular network controlling squamous cell lung carcinoma development 24594363_In erythroid cells, pull down experiments identified the presence of a novel complex formed by HDAC5, GATA1, EKLF and pERK which was instead undetectable in cells of the megakaryocytic lineage. 24706304_HDAC5 promoted the Six1 expression. 24732133_In C2C12 myoblasts, recombinant human HDAC5 phosphorylation by PKD regulated the expression of diverse metabolic genes and glucose metabolism. 24920159_Studied phosphorylation sites within functional HDAC5 domains, including the deacetylation domain (DAC, Ser755), nuclear export signal (NES, Ser1108), and an acidic domain (AD, Ser611). 25096223_Data reveal a novel role of HDAC5 in modulating the KLF2 transcriptional activation and eNOS expression. 25129440_mRNA and protein levels of HDAC5 were up-regulated in human hepatocellular carcinoma. 26059794_These results suggest a strong regulatory function of HDAC5 in the pro-inflammatory response of macrophages. 26261519_Formononetin-combined therapy may enhance the therapeutic efficacy of doxorubicin in glioma cells by preventing EMT through inhibition of HDAC5. 26747087_HDAC5 and HDAC6 were highly expressed in melanoma cells but exhibited low expression levels in normal skin cells. 26847592_HDAC5 promotes cellular proliferation through the upregulation of cMet, and may provide a novel therapeutic target for the treatment of patients with Wilms' tumor. 27177225_HDAC5 was extensively expressed in human BC tissues, and high HDAC5 expression was associated with an inferior prognosis. 27212032_these results suggest that HDAC5 is critical in regulating LSD1 protein stability through post-translational modification, and the HDAC5-LSD1 axis has an important role in promoting breast cancer development and progression. 27482699_The expression of HDAC5 was significantly increased in endothelial cells (ECs) from patients with systemic sclerosis (SSc) compared to healthy control endothelial cells. Silencing of HDAC5 in SSc ECs restored normal angiogenesis. HDAC5 knockdown followed by ATAC-seq assay in SSc ECs identified key HDAC5-regulated genes involved in angiogenesis and fibrosis, such as CYR61, PVRL2, and FSTL1. 27614433_HDAC5 inhibits hepatic lipogenic genes expression by attenuating the transcriptional activity of liver X receptor. 27900262_Hdac5 and Hdac6 expression are required for the adequate expression of Icer and adipocyte function. Altered adipose expression of the two Hdacs in obesity by hypoxia may contribute to the development of metabolic abnormalities. 28235630_HDAC5 is a negative predictor of disease-free and overall survival in pancreatic neuroendocrine tumor patients. 28414307_Interference with both glucose and glutamine supply in HDAC5-inhibited cancer cells significantly increases apoptotic cell death. 28440397_These findings demonstrate a novel mechanism for deregulation of HDAC5 in non-small cell lung cancer (NSCLC)and suggest that miR5895p/HDAC5 pathway may represent a new prognostic biomarker and therapeutic target against NSCLC. 28653891_the migration and invasion of hepatocellular carcinoma cells were impaired by knockdown of histone deacetylase 5 or hypoxia-inducible factor-1alpha but rescued when eliminating homeodomain-interacting protein kinase-2 in hepatocellular carcinoma cells, which suggested the critical role of histone deacetylase 5-homeodomain-interacting protein kinase-2-hypoxia-inducible factor-1alpha pathway in hypoxia-induced metastasis. 29223407_Latent autoimmune diabetes of adults (LADA) patients showed stronger binding of p-STAT3, HDAC5 and DNMT1 than controls. 29339432_Collectively, these data indicate that vIRF3 alters global gene expression and induces a hypersprouting formation in an HDAC5-binding-dependent and lymphatic endothelial cell-specific manner, ultimately contributing to Kaposi's sarcoma-associated herpesvirus-associated pathogenesis. 29508277_It identified BMAL1 as a cAMP-responsive coactivator of HDAC5 to regulate hepatic gluconeogenesis 29620443_HDAC5 expression was positively correlated with miR-2861 in lung cancer stem cells (LCSCs), and knockdown of miR-2861 decreased the expression of HDAC5, which implied that HDAC5 may be involved in the differentiation of LCSCs mediated by miR-2861. 29886060_HO-1 plays a key role in protecting tumor cells from apoptosis, in a process that involves Smad7 and HDAC4/5 in apoptosis of B-ALL cells 30013077_These results demonstrate a previously unknown negative epigenetic regulation of hematopoietic stem cells (HSC) homing and engraftment by HDAC5, and allow for a new and simple translational strategy to enhance HSC transplantation. 30066893_High HDAC5 expression is associated with invasion of lung cancer. 30110565_in hISMC from resected Crohn's strictures, we observed a significantly reduced contractile phenotype compared with patient-matched intrinsic controls that was associated with increased patient-specific expression of DNA methyltransferase 1, HDAC2, and HDAC5. 31052182_HDAC5 overexpression in urothelial carcinoma cell lines decreases cell proliferation but can promote epithelial-to-mesenchymal transition. 31690832_This study reveals that HDAC5 regulated by C-MYC is essential for SOX9 deacetylation and nuclear localisation, which is critical for tamoxifen resistance. These results indicate a potential therapy strategy for ER(+) breast cancer by targeting C-MYC/HDAC5/SOX9 axis. 31784580_HDAC4 and 5 repression of TBX5 is relieved by protein kinase D1. 33414476_METTL14-regulated PI3K/Akt signaling pathway via PTEN affects HDAC5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. 33419772_HDAC5 Loss Impairs RB Repression of Pro-Oncogenic Genes and Confers CDK4/6 Inhibitor Resistance in Cancer. 33673279_Whole Exome Sequencing Identifies APCDD1 and HDAC5 Genes as Potentially Cancer Predisposing in Familial Colorectal Cancer. 34036344_Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. 34125448_HDAC5 promotes intestinal sepsis via the Ghrelin/E2F1/NF-kappaB axis. 34502418_Neddylation Regulates Class IIa and III Histone Deacetylases to Mediate Myoblast Differentiation. 34740611_Histone deacetylase 5 deacetylates the phosphatase PP2A for positively regulating NF-kappaB signaling. 35265200_HDAC5 modulates PD-L1 expression and cancer immunity via p65 deacetylation in pancreatic cancer. | ENSMUSG00000008855 | Hdac5 | 2259.75519 | 1.1053936 | 0.1445601801 | 0.11483985 | 1.626379e+00 | 2.022046e-01 | 9.998360e-01 | No | Yes | 2534.51497 | 267.206000 | 2.161705e+03 | 176.900535 | |
| ENSG00000109381 | 1998 | ELF2 | protein_coding | Q15723 | FUNCTION: Isoform 1 transcriptionally activates the LYN and BLK promoters and acts synergistically with RUNX1 to transactivate the BLK promoter.; FUNCTION: Isoform 2 may function in repression of RUNX1-mediated transactivation. | Activator;Alternative splicing;DNA-binding;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | hsa:1998; | chromatin [GO:0000785]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; cell differentiation [GO:0030154]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357] | 11967990_angiopoietin-1 regulates expression of NERF2 and its own receptor in hypoxic cells. 12447867_Observational study of gene-environment interaction. (HuGE Navigator) 14970218_NERF/ELF-2 physically interacts with AML1 and mediates opposing effects on AML1-mediated transcription of the B cell-specific blk gene 17368566_These findings indicate that ELF2/NERF promotes VCP transcription and that ELF2/NERF-VCP pathway might be important for cell survival and proliferation under cytokine stress. 18544453_ELF2 transactivates VCP promoter through binding to two motifs, with a predominant contribution of the upstream one. 18754678_Affinity of the CAL PDZ domain for the cystic fibrosis transmembrane conductance regulator (CFTR) C-terminus is much weaker than that of the NHERF1/NHERF2 domains, enabling wild-type CFTR to avoid premature entrapment in the lysosomal pathway. 26968954_Our findings collectively support a potential role of triiodothyronine and its receptor in tumor growth inhibition through regulation of ELF2 28728844_PCAT7 contributed to the progression of nasopharyngeal carcinoma through regulating miR-134-5p/ELF2 signaling pathway. 29632131_Mutations in eIF2B genes cause vanishing white matter disease due to translation defects. | ENSMUSG00000037174 | Elf2 | 284.46513 | 0.9284918 | -0.1070388997 | 0.18829715 | 3.211631e-01 | 5.709095e-01 | 9.998360e-01 | No | Yes | 260.34427 | 37.996382 | 2.802572e+02 | 31.583445 | ||
| ENSG00000109458 | 2549 | GAB1 | protein_coding | Q13480 | FUNCTION: Adapter protein that plays a role in intracellular signaling cascades triggered by activated receptor-type kinases. Plays a role in FGFR1 signaling. Probably involved in signaling by the epidermal growth factor receptor (EGFR) and the insulin receptor (INSR). Involved in the MET/HGF-signaling pathway (PubMed:29408807). {ECO:0000269|PubMed:29408807}. | 3D-structure;Acetylation;Alternative splicing;Deafness;Disease variant;Non-syndromic deafness;Phosphoprotein;Reference proteome | The protein encoded by this gene is a member of the IRS1-like multisubstrate docking protein family. It is an important mediator of branching tubulogenesis and plays a central role in cellular growth response, transformation and apoptosis. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008]. | hsa:2549; | cell-cell junction [GO:0005911]; cytosol [GO:0005829]; actin cytoskeleton reorganization [GO:0031532]; angiogenesis [GO:0001525]; endothelial cell chemotaxis to vascular endothelial growth factor [GO:0090668]; epidermal growth factor receptor signaling pathway [GO:0007173]; insulin receptor signaling pathway [GO:0008286]; positive regulation of angiogenesis [GO:0045766]; positive regulation of cell migration by vascular endothelial growth factor signaling pathway [GO:0038089]; response to hepatocyte growth factor [GO:0035728]; vascular endothelial growth factor signaling pathway [GO:0038084] | 11606067_Unique phosphorylation mechanism of Gab1 using PI 3-kinase as an adaptor protein. 11701952_comparative FISH mapping of Gab1 and Gab2 genes in human, mouse and rat 11896055_ERK negatively regulates the epidermal growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase 12370245_Results indicate that Gab1 and SHP-2 promote the undifferentiated epidermal cell state by facilitating Ras/MAPK signaling. 12766170_Gab1 and the Met receptor interact in a novel manner, such that the activated kinase domain of Met and the negative charge of phosphotyrosine 1349 engage the Gab1 MBD as an extended peptide ligand 12808090_Gab1 is an integrator of cell death versus cell survival signals in oxidative stress 12855672_Gab1-SHP2 interaction plays a crucial role in gp130-dependent longitudinal elongation of cardiomyoctes and cardiac hypertrophy through activation of ERK5 14665621_results reveal that Gab1 protein recruits SHP2 protein tyrosine phosphatase to dephosphorylate paxillin 14701753_SHP-2/Gab1 association is critical for linking EGFR to NF-kappaB transcriptional activity via the PI3-kinase/Akt signaling axis in glioblastoma cells 15010462_Hck-mediated phosphorylation of Gab1 and Gab2 docking proteins in IL-6-induced proliferation and survival of multiple myeloma cells. 15351743_coupling of Gab1 to PI3K is important for biological responses in RET-expressing cells 15379552_extracellular signal-regulated kinases 1/2 modulate insulin action via Gab1 by targeting serine and threonine residues beside YXXM motifs 15665327_Gab1 tyrosine phosphorylation is stimulated by flow shear stress to mediate protein kinase B and endothelial nitric-oxide synthase activation in endothelial cells 15940252_RAI associates with the Grb 2-associated binder 1 (GAB 1) adapter. This association is constitutive, but, in the presence of RET oncoproteins, both RAI and GAB 1 are tyrosine-phosphorylated, and the stoichiometry of this interaction remarkably increases 15952937_Gab2 plays a pivotal role in the EGF-induced ERK activation pathway and that it can complement the function of Gab1 in the EGF signalling pathway; Gab1 and Gab2 are critical signalling threshold proteins for ERK activation by EGF. 16687399_By amplifying positive interactions between survival and mitogenic pathways, GAB1 plays the critical role in cell proliferation and tumorigenesis. 16787925_Gab1 appears as a primary actor in coupling VEGFR-2 to PI3K/Akt, recruited through an amplification loop involving PtdIns(3,4,5)P3 and its PH domain 16849525_Crk adaptor protein is required for the sustained phosphorylation of c-Met-docking protein Grb2-associated binder 1 (Gab1) in response to HGF, leading to the enhanced cell motility of human synovial sarcoma cell lines SYO-1, HS-SY-II, and Fuji. 17145761_Bisindolylmaleimide I abolishes the FGF2-mediated association of Shp2 tyrosine phosphatase with Frs2 and Gab1. 17178724_Gab1 is a novel critical regulatory component of endothelial cell migration and capillary formation with a key role in the activation of VEGF-evoked signaling pathways required for angiogenesis 17211494_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17625596_HGF signaling process from Gab1 to PI3K is negatively regulated by PKC-betaII, and its loss is critical for melanoma cells to gain invasive potential. 18003605_These results suggest that coupling of Grb2 to Gab1 mediates the hepatocyte growth factor-induced strong activation of the ERK pathway, which is required for the inhibition of HepG2 cell proliferation. 18192688_moderate level of Gab1 overexpression stimulated tumor growth 18577518_the Gab1-SHP2-ERK1/2 signaling pathway comprises an inhibitory axis for IGF-I-dependent myogenic differentiation. 19233262_These results underscore the non-redundant and essential roles of Gab1 and Gab2 in endothelial cells, and suggest major contributions of these proteins during in vivo angiogenesis. 19665053_Gab1 couples PI3K-mediated Erythropoietin signals with the Ras/Erk pathway and plays an important role in erythropoietin receptor-mediated signal transduction involved in the proliferation and survival of erythroid cells. 19881549_the phosphorylation of Gab1 by c-Src is important for hepatocyte growth factor -induced DNA synthesis 19913121_Observational study of gene-disease association. (HuGE Navigator) 20005866_The binding of Grb2 adaptor to its downstream partners Sos1 and Gab1 docker is under tight allosteric regulation. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20602450_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20602450_We could not confirm a major association between Gab1 SNP (rs3805246) and the predisposition to H. pylori infection and CAG in this study populat 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 20723025_PECAM-1-mediated inhibition of GPVI-dependent platelet responses result from recruitment of SHP-2-p85 complexes to tyrosine-phosphorylated PECAM-1, which diminishes the association of PI3K with activatory signaling molecules Gab1 and LAT 21282639_Gab1 is a critical upstream signaling component in VEGF-induced eNOS activation and tube formation, which is dependent on protein kinase A. 21782801_These data demonstrate that GAB1 is ubiquitinated by CBL and degraded by the proteasome, and plays a role in negative-feedback regulation of HGF/SF-MET signaling. 22366451_Met signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. 22536782_Data show that bivalent binding drives the formation of the Grb2-Gab1 signaling complex in a noncooperative manner. 22751113_that aberrant Gab1 signaling can directly contribute to breast cancer progression, and that negative feedback sites in docking proteins can be targeted by oncogenic mutations. 22851227_these data underscore the critical roles of Gab1 and Gab2 in IL-22-mediated HaCaT cell proliferation, migration, and differentiation. 22865653_GAB1 plays an important role as part of the mechanism of by which EGFR induces induced activation of the MAPK and AKT pathway. 22915589_an anti-apoptotic role of caspase-cleaved GAB1 in HGF/SF-MET signaling. 23334917_Although Sos1 and Gab1 recognize two non-overlapping sites within the Grb2 adaptor, allostery promotes the formation of two distinct pools of Grb2-Sos1 and Grb2-Gab1 binary signaling complexes in lieu of a composite Sos1-Grb2-Gab1 ternary complex. 23612964_the acquired substrate preference for GAB1 is critical for the ERBB2 mutant-induced oncogenesis. 23805312_The model showed agreement at several key nodes, involving scaffolding proteins Gab1, Gab2 and their complexes with Shp2. VEGFR2 recruitment of Gab1 is greater in magnitude, slower, and more sustained than that of Gab2. As Gab2 binds VEGFR2 complexes more transiently than Gab1, VEGFR2 complexes can recycle and continue to participate in other signaling pathways. 24312291_expression of Gab1, VEGFR-2, and MMP-9 are significantly related to the malignant biological behavior of hilar cholangiocarcinoma 24374147_Gab1 as major target in linoleic acid-induced enhancement of tumorigenesis. 24391994_The combined GRB2 and GAB1 protein expression was significantly associated with aggressive tumor progression and poor prognosis in patients with hepatocellular carcinoma. 24787006_miR-150 can influence the relative expression of GAB1 and FOXP1 and the signaling potential of the B-cell receptor 24872569_Gab1 is an essential component of NRG1-type III signaling during peripheral nerve development. 24998422_Upregulations of Gab1 and Gab2 proteins associate with tumor progression of human gliomas. 25041730_C-SH3 of Grb2 mediates the interaction with mutant Htt and this interaction being stronger could replace Gab1, with mutant Htt becoming the preferred partner. This would have immense effect on downstream signaling events. 25078664_Galphai1/3 proteins lie downstream of KGFR, but upstream of Gab1-mediated activation of PI3K-AKT-mTORC1 signaling. 25144631_Endometrial GAB1 protein and mRNA expression are reduced in women with PCOS, suggesting that the endometrium of PCOS women have a defect in insulin signaling due to GAB1 down-regulation. 25217982_This study suggests potential effects of SNPs of Gab1 on the onset and susceptibility of biliary tract cancer. 25569504_Studied the pleckstrin homology (PH) domain of GAB1 for cancer treatment. Using homology models we derived, high-throughput virtual screening of five million compounds resulted in five hits which exhibited strong binding affinities to GAB1 PH domain. 25596749_Results suggest that Gab1 is an essential regulator of the EGF-mediated mTORC pathways and may potentially be used as a biomarker for urothelial carcinoma 25969544_EGFR-activated Src family kinases maintain GAB1-SHP2 complexes distal from EGFR. 25991585_Data demonstrate that miR-409-3p is a metastatic suppressor, and post-transcriptional inhibition of the oncoprotein GAB1 is one of its mechanisms of action. 26014518_We found that expression of Gab1, VEGFR-2, and MMP-9 was highly and positively correlated with each other and with lymph node metastasis and TNM stage in intrahepatic cholangiocarcinoma tissues 26090437_Gab1 protein was upregulated in cyanotic compared to acyanotic hearts suggesting that Gab1 upregulation is a component of the survival program initiated by hypoxia in cyanotic children 26183772_CVB3 targets host GAB1 to generate a GAB1-N1-174 fragment that enhances viral infectivity, at least in part, via activation of the ERK pathway 26276357_Gab1 has a role in regulating SDF-1-induced progression via inhibition of apoptosis pathway induced by PI3K/AKT/Bcl-2/BAX pathway in human chondrosarcoma (CS). Gab1 can be recommended as a novel biomarker for diagnosis and prognosis in patients with CS. 26517531_These results demonstrate that cardiomyocyte Gab1 is a critical regulator of the compensatory cardiac response to aging and hemodynamic stress. 26871477_Knockdown of GAB1 mimicked the tumor-suppressive effects of miR-150 overexpression on HCC cells, whereas restoration of GAB1 expression partially abolished the inhibitory effects. 27241812_Gab1/SHP2/p38MAPK signaling pathway and Ang-2 have an essential role in regulating thrombin-induced monocyte adhesion and vascular leakage 27302321_Gab1 expression is correlated with poor prognosis of Epithelial ovarian cancer patients. 27475256_miR-5582-5p induced apoptosis and cell cycle arrest in cancer cells, but not in normal cells. GAB1, SHC1, and CDK2 were identified as direct targets of miR-5582-5p. 28081727_these data provide novel information for comprehending the tumor-suppressive role of miR-200a in HCC pathogenesis through inhibition of GAB1 translation. 28365441_onstitutive Gab1-dependent signalling in Jak2-V617F-expressing cells does not occur due to the constitutive association of Gab1 with PIP3 at the plasma membrane. 28619509_data suggested that miR-141-3p decreased the proliferation and migration of keloid fibroblasts by repressing GAB1 expression, providing a useful target for keloid management 28893350_These results suggested that Gab1 induced malignant progression of oral squamous carcinoma cells. 29408807_Modifier variant of METTL13 suppresses human GAB1-associated profound deafness. 29902453_Gab1 is required for H2O2-induced Akt activation in OB-6 osteoblasts. miR-29a inhibition upregulates Gab1 to protect OB-6cells from H2O2. Gab1 over-expression promotes Akt activation to inhibit H2O2-induced OB-6cell death. 30280777_Results identified GAB1 as a direct target of miR-590 in non-small cell lung cancer (NSCLC). Its knockdown had the same effect as overexpression of miR-590 in NSCLC. Moreover, its overexpression partially reversed the suppressive effect of miR-590 on NSCLC. 30665442_Our findings indicate that elevated expression of Gab1 promotes Breast cancer metastasis by dissociating the PAR complex that leads to EMT, implicating a role of Gab1 as a potential biomarker of metastatic Breast cancer. Moreover, inhibition of Gab1 expression might be a promising therapeutic strategy for Breast cancer metastasis 30697991_Gab1 is associated with angiogenesis function of EA.hy926 endothelium cells via PI3K-Akt signaling pathway 31443923_Gab1 expression in Ewing sarcoma, rhabdomyosarcoma and synovial sarcoma may have prognostic significance 31505074_Increased expression of GAB1 promotes inflammation and fibrosis in systemic sclerosis. 31651330_Gab1 enhances interleukin-6-induced MAPK-pathway activation in an SHP2-, Grb2-, and time-dependent manner 32650835_miR-183-3p suppresses proliferation and migration of keratinocyte in psoriasis by inhibiting GAB1. 32759334_Human Cytomegalovirus miR-US5-2 Downregulation of GAB1 Regulates Cellular Proliferation and UL138 Expression through Modulation of Epidermal Growth Factor Receptor Signaling Pathways. 32859628_Recurrent Fusion of the GRB2 Associated Binding Protein 1 (GAB1) Gene With ABL Proto-oncogene 1 (ABL1) in Benign Pediatric Soft Tissue Tumors. 32888745_Long non-coding RNA H19 promotes the proliferation, migration and invasion while inhibits apoptosis of hypertrophic scarring fibroblasts by targeting miR-3187-3p/GAB1 axis. 33323827_Loss of GRB2 associated binding protein 1 in arteriosclerosis obliterans promotes host autophagy. 33393621_Circ_0061012 contributes to IL-22-induced proliferation, migration and invasion in keratinocytes through miR-194-5p/GAB1 axis in psoriasis. 33786575_FoxO1-GAB1 axis regulates homing capacity and tonic AKT activity in chronic lymphocytic leukemia. 34732686_Association of Single-Nucleotide Polymorphisms of Gab1 Gene with Susceptibility to Meningioma in a Northern Chinese Han Population. | ENSMUSG00000031714 | Gab1 | 167.54620 | 1.1080917 | 0.1480772767 | 0.24812913 | 3.666218e-01 | 5.448514e-01 | 9.998360e-01 | No | Yes | 140.61288 | 29.875470 | 1.277361e+02 | 20.984025 | |
| ENSG00000109466 | 11275 | KLHL2 | protein_coding | O95198 | FUNCTION: Component of a cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex that mediates the ubiquitination of target proteins, such as NPTXR, leading most often to their proteasomal degradation (By similarity). Responsible for degradative ubiquitination of the WNK kinases WNK1, WNK3 and WNK4. Plays a role in the reorganization of the actin cytoskeleton. Promotes growth of cell projections in oligodendrocyte precursors. {ECO:0000250, ECO:0000269|PubMed:15715669, ECO:0000269|PubMed:23838290}. | 3D-structure;Actin-binding;Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Kelch repeat;Reference proteome;Repeat;Ubl conjugation pathway | hsa:11275; | actin cytoskeleton [GO:0015629]; Cul3-RING ubiquitin ligase complex [GO:0031463]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lamellipodium [GO:0030027]; ruffle [GO:0001726]; actin binding [GO:0003779]; identical protein binding [GO:0042802]; protein ubiquitination [GO:0016567] | 15735724_overexpression of Mayven may promote tumor growth through c-Jun and cyclin D1 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21549840_Results suggest a novel E3 ubiquitin ligase function of KLHL2, with NPCD as a substrate. 23838290_Co-expression of KLHL2 and Cullin3 decreases the abundance of WNK1, WNK3 and WNK4 within HEK293T cells. | ENSMUSG00000031605 | Klhl2 | 139.14174 | 1.0963207 | 0.1326698426 | 0.28337779 | 2.132230e-01 | 6.442529e-01 | 9.998360e-01 | No | Yes | 151.44012 | 30.916551 | 1.413096e+02 | 22.350560 | ||
| ENSG00000109501 | 7466 | WFS1 | protein_coding | O76024 | FUNCTION: Participates in the regulation of cellular Ca(2+) homeostasis, at least partly, by modulating the filling state of the endoplasmic reticulum Ca(2+) store (PubMed:16989814). Negatively regulates the ER stress response and positively regulates the stability of V-ATPase subunits ATP6V1A and ATP1B1 by preventing their degradation through an unknown proteasome-independent mechanism (PubMed:23035048). {ECO:0000269|PubMed:16989814, ECO:0000269|PubMed:23035048}. | Acetylation;Cataract;Cytoplasmic vesicle;Deafness;Diabetes insipidus;Diabetes mellitus;Disease variant;Endoplasmic reticulum;Glycoprotein;Membrane;Non-syndromic deafness;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein, which is located primarily in the endoplasmic reticulum and ubiquitously expressed with highest levels in brain, pancreas, heart, and insulinoma beta-cell lines. Mutations in this gene are associated with Wolfram syndrome, also called DIDMOAD (Diabetes Insipidus, Diabetes Mellitus, Optic Atrophy, and Deafness), an autosomal recessive disorder. The disease affects the brain and central nervous system. Mutations in this gene can also cause autosomal dominant deafness 6 (DFNA6), also known as DFNA14 or DFNA38. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:7466; | dendrite [GO:0030425]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum lumen [GO:0005788]; endoplasmic reticulum membrane [GO:0005789]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of synaptic vesicle membrane [GO:0030285]; secretory granule [GO:0030141]; ATPase binding [GO:0051117]; calcium-dependent protein binding [GO:0048306]; calmodulin binding [GO:0005516]; DNA-binding transcription factor binding [GO:0140297]; proteasome binding [GO:0070628]; ubiquitin protein ligase binding [GO:0031625]; calcium ion homeostasis [GO:0055074]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; endoplasmic reticulum unfolded protein response [GO:0030968]; ER overload response [GO:0006983]; glucose homeostasis [GO:0042593]; kidney development [GO:0001822]; negative regulation of apoptotic process [GO:0043066]; negative regulation of ATF6-mediated unfolded protein response [GO:1903892]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of programmed cell death [GO:0043069]; negative regulation of response to endoplasmic reticulum stress [GO:1903573]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of type B pancreatic cell apoptotic process [GO:2000675]; nervous system process [GO:0050877]; olfactory behavior [GO:0042048]; pancreas development [GO:0031016]; positive regulation of calcium ion transport [GO:0051928]; positive regulation of growth [GO:0045927]; positive regulation of protein metabolic process [GO:0051247]; positive regulation of protein ubiquitination [GO:0031398]; protein maturation by protein folding [GO:0022417]; protein stabilization [GO:0050821]; renal water homeostasis [GO:0003091]; response to endoplasmic reticulum stress [GO:0034976]; sensory perception of sound [GO:0007605]; ubiquitin-dependent ERAD pathway [GO:0030433]; visual perception [GO:0007601] | 11709537_Five different heterozygous missense mutations (T699M, A716T, V779M, L829P, G831D) in the WFS1 gene found in six low frequency sensorineural hearing loss (LFSNHL) families. 11709538_The causal relationship between WFS1 missense mutation and deafness was supported by two observations based on haplotype and mutation analysis of the kindred. 11916957_Observational study of gene-disease association. (HuGE Navigator) 11920861_Did not find evidence of an increased incidence of WFS carriers in the suicide panel and concluded that WFS1 carrier status is not a significant contributor to suicide in the general population. 12073007_Mutations in the WFS1 gene that cause low-frequency sensorineural hearing loss are small non-inactivating mutations. 12107816_Four new polymorphisms associated with Wolfram syndrome. Not all patients have full syndrome. 12605098_WFS1 is not a major susceptibility gene for the development of psychiatric disorders in subjects with Wolfram syndrome. 12650912_In all affected family members analysed, we detected a missense mutation in WFS1 (K705N) and therefore confirm the finding that the majority of mutations responsible for LFSNHI are missense mutations which localise to the C-terminal domain of the protein. 12707947_found a significantly higher frequency of the 611R/611R genotype in suicide completers. Scores of impulsivity and novelty seeking were higher in subjects with the associated genotype, suggesting a role for WFS1 in the pathophysiology of impulsive suicide 12754709_mutational analysis of the WFS1 coding region in 19 Italian Wolfram syndrome patients and 25 relatives, using a DHPLC-based protocol 12782971_This study does not support the involvement of tyrosine hydroxylase, catechol-O-methyl transferase and Wolfram syndrome 1 polymorphisms in mood disorders. 12913071_Here we investigate, for the first time, the molecular mechanisms that cause loss-of-function of wolframin in affected individuals. 12955714_Overview of the spectrum of WFS1 mutations in Wolfram syndrome, nonsyndromic low frequency sensorineural hearing impairment,diabetes mellitus, and psychiatric disease. 14527944_Wolframin has a role in the regulation of intracellular Ca2+ homeostasis 14968315_In this study we analyzed the phenotype of a large Hungarian family with LFSNHI and linkage to DFNA6. The family contains 14 affected persons. 15234338_Observational study of gene-disease association. (HuGE Navigator) 15277431_most causative changes identified in the WFS1 gene occurred in exon 8, and only one was identified outside this region in exon 4 in patients with Wolfram syndrome 15473915_Observational study of gene-disease association. (HuGE Navigator) 15473915_These results support the hypothesis that the WFS1 gene is involved in the genetic predisposition for mood disorders. 15852062_The relative risk of psychiatric hospitalization for depression was estimated to be 7.1 (95% CI 1.9-26.6) for carriers of a single wolframin mutation compared to noncarriers. 15912360_Mutations in one single gene, Wolfram syndrome 1 (WFS1), have been reported to account for most familial cases with low-frequency hearing loss. 16005363_A nine nucleotide insertional mutation in two members of a family with Wolfram syndrome. 16043233_This study presents a six-generation family from Hungary with nonsyndromic, post-lingual, bilateral, symmetric, progressive LFSNHI, that discloses positive linkage to the DFNA6 region. 16151413_Molecular analysis of WFS1 in seven families with Wolfram syndrome identified eight different mutations; one was a de novo mutation occurring independently in 2 families, whereas the remaining ones were inherited. 16195229_the pathogenesis of Wolfram syndrome involves chronic ER stress in pancreatic beta-cells caused by the loss of function of WFS1 16408729_Mutations in WFS1 are one cause of non-syndromic low frequency sensorineural hearing loss. 16550584_Missense mutations within a defined region are associated with dominant low-frequency hearing loss (DFNA6/14/38), while more severe mutations spanning WFS1 are found in Wolfram syndrome patients. 16806192_WFS1 mutations lead to drastically reduced steady-state levels of wolframin. 16876316_Observational study of gene-disease association. (HuGE Navigator) 16965966_WFS1 minimal promoter contains two DNA binding motifs (GC boxes) for the transcription factors Sp1/3/4 and binding of both Sp1 and Sp3 was demonstrated at both motifs in vitro and in vivo. 16989814_WFS1 protein participates in the regulation of cellular Ca(2+) homeostasis, at least partly, by modulating the filling state of the ER Ca(2+) store 17492394_one-third (3 out of 9) autosomal dominant low frequency sensorineural hearing loss(LFSNHL) families had mutations in WFS1, indicating that in non-syndromic hearing loss WFS1 is restrictively & commonly found within autosomal dominant LFSNHL families 17517145_a novel heterozygous missense mutation in exon 8 of WFS1 (i.e., Y669H) which is likely responsible for the low-frequency sensorineural hearing loss (LFSNHL) phenotype in a Taiwanese family was discovered 17568405_Results reported eight novel WFS1 mutations in Wolfram syndrome. 17603484_In a pooled analysis comprising 9,533 cases and 11,389 controls, SNPs in WFS1 were strongly associated with common type 2 diabetes risk. 17603484_Observational study of gene-disease association. (HuGE Navigator) 17719176_Observational study of gene-disease association. (HuGE Navigator) 17719176_The wolframin His611Arg polymorphism influences medication overuse headache. 17947299_Study identifies an interaction between Wolframin and Na+/K+ ATPase beta1 subunit in transfected Cos7 cells, and between endogenous proteins in placental, neuroblastoma and MIN6 pancreatic beta-cell lines. 18040659_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18040659_Replication of the previously reported associations between SNPs at this locus and the risk of type 2 diabetes. 18060660_Genome-wide association datase revealed that a strong linkage disequilibrium with the three WFS1 single nucleotide polymorphisms was associated with type 2 diabetes. 18060660_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18197395_May be a candidate gene for type 2 diabetes. 18426861_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18518985_a novel heterozygous missense mutation in exon 8 of WFS1 predicting a p.R685P amino acid substitution that is likely to underlie the low frequency sensorineural hearing loss phenotype in the American family 18544103_maternally inherited combination of diabetes mellitus and hearing impairment in three members of a family was found to be associated with autosomal dominant transmission of the E864K mutation of the WFS1 gene 18566338_The WFS1 gene is located on the short arm of chromosome 4 in Wolfram syndrome. 18568334_Observational study of gene-disease association. (HuGE Navigator) 18568334_Type 2 diabetes-associated risk alleles of WFS1 are associated with estimates of a decreased pancreatic beta cell function among middle-aged individuals with abnormal glucose regulation 18591388_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18597214_Observational study of gene-disease association. (HuGE Navigator) 18660851_This report of two novel WFS1 mutations expands the molecular spectrum of Wolfram syndrome. 18688868_A novel missense mutation in WFS1 was identified which caused Wolfram syndrome and may also be linked to autoimmune diseases. 18694974_Study show that polymorphisms in WFS1 were associated with type 2 diabetes risk in the studied population. 18806274_The WFS1 locus strongly contributes to juvenile-onset diabetes in Lebanon in Wolfram syndrome and non-syndromic non-autoimmune diabetes mellitus detected by linkage analysis; effect varies by allele. 18853134_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18984664_Observational study of gene-disease association. (HuGE Navigator) 18991055_Observational study of gene-disease association. (HuGE Navigator) 19020324_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19042979_WFS1 is the major gene involved in Wolfram syndrome in Brazilian patients and most mutations are concentrated in exon 8 19082521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19115052_Observational study of gene-disease association. (HuGE Navigator) 19139842_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19258404_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19258739_found no evidence for a substantial effect of WFS1 polymorphisms on risk of type 2 diabetes or clinical characteristics of diabetic subjects in Japanese population 19279076_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19324937_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19328217_Observational study of gene-disease association. (HuGE Navigator) 19328217_The H611R polymorphism of Wolframin gene was associated with mood disorders but not suicidal behavior, aggressive/impulsive traits or suicidality in first-degree relatives. 19330314_A common genetic variant in WFS1 specifically impairs GLP-1-induced insulin secretion independently of insulin sensitivity 19330314_Observational study of gene-disease association. (HuGE Navigator) 19380854_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19502414_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19598235_Observational study of gene-disease association. (HuGE Navigator) 19598235_WFS1 gene is associated with autistic traits, empathy and Asperger syndrome. 19602701_Meta-analysis and HuGE review of gene-disease association. (HuGE Navigator) 19720844_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19734900_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19741467_Observational study of gene-disease association. (HuGE Navigator) 19794065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19814620_Observational study of gene-disease association. (HuGE Navigator) 19833888_Observational study of gene-disease association. (HuGE Navigator) 19862325_Observational study of gene-disease association. (HuGE Navigator) 19877185_The family described with autosomal dominant inheritance of K836T of the WFS1 gene demonstrates a progressive hearing loss in the lower frequencies. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20028947_Observational study of gene-disease association. (HuGE Navigator) 20028947_six highly correlated single nucleotide polymorphisms that show strong and comparable associations with risk of type 2 diabetes 20069065_This study describes the phenotype of a family with autosomal dominant optic neuropathy and hearing impairment associated with a novel missense mutation in WFS1. 20160352_a role for WFS1 in the negative regulation of ER stress signaling and in the pathogenesis of diseases involving chronic, unresolvable ER stress, such as pancreatic beta cell death in diabetes. 20203524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20215779_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20361036_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20361036_the HNF4A and WFS1 risk alleles predispose to development of type 2 diabetes in an Ashkenazi Jewish population 20384434_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20490451_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20509872_Observational study of gene-disease association. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20580033_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20712903_Observational study of gene-disease association. (HuGE Navigator) 20802253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20816152_Observational study of gene-disease association. (HuGE Navigator) 20879858_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20889853_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21127832_The most frequent haplotype at the haplotype block containing the WFS1 gene modulated insulin secretion and was associated with an increased risk of type 2 diabetes. 21143470_Novel mutations in the wolframin gene are identified that are associated with non-immunogenic diabetes mellitus and a progressive atrophy of the optic nerve. 21356526_WFS1 is the gene responsible for autosomal dominant low-frequency sensorineural hearing loss in a Chinese family, as well as in a sporadic case. These WFS1 mutations are not present in unaffected control subjects. 21454619_ER stress induces Smurf1 degradation and WFS1 up-regulation 21538838_The p.A684V missense mutation in the WFS1 gene is a frequent cause of autosomal dominant optic atrophy and hearing impairment. 21564155_Nine different mutations in WFS1 (five of them novel) were identified in nine Wolfram syndrome patients. 21623591_Cataract could be a marker for the WFS1 heterozygosity in this family, namely the c.2431_2465dup35 mutation. 21632151_Their past medical history revealed diabetes mellitus and deafness since childhood. It was confirmed by molecular analysis, which evidenced composite WFS1 heterozygous mutations inherited from both their mother and father. 21713316_A WFS1 haplotype consisting of the minor alleles of rs752854, rs10010131, and rs734312 shows a protective role against type 2 diabetes in Russian patients 21823543_report a consanguineous family with three siblings affected by Wolfram syndrome. A homozygous single base pair deletion (c.877delC, L293fsX303) was found in the WFS1 gene in all three affected siblings 21968327_A new homozygous WFS1 mutation causing causing Wolfram syndrome is identified in a large inbred Turkish family. 22240535_genetic variation of Wolfram syndrome type 1 gene was a more crucial factor than other genes in causing hearing loss. 22498363_Two individuals who had heterozygosity of GJB2 mutations and heterozygosity of WFS1 mutations showed low-frequency hearing loss. One individual who had homozygosity of GJB2 mutation without WFS1 mutation had moderate, gradual high tone hearing loss. 22662265_In a family with MODY diabetes, three affected subjects had the mutation c.2107C-T/p.R703C. The affected amino acid is strongly conserved and the variant suggested to be probably damaging by prediction programs. The proband developed diabetes 14 years old with no type 1 auto-antibodies and required insulin. There was no familial hearing impairment. 22781099_report of male Wolfram patients with WFS1 mutations who have successfully fathered children 23035048_WFS1 has a specific interaction with the V1A subunit of H(+) ATPase; this interaction may be important both for pump assembly in the ER and for granular acidification. 23103830_Description of a novel missense mutation of the WFS1 gene in exon 4 of WFS1 gene in two Italian siblings with Wolfram syndrome. 23144361_In African Americans, seven of the 29 SNPs examined were found to be associated with T2D risk at P = 0.05, including 2 SNPs in the WFS1 gene (rs4689388 and rs1801214). 23257691_Data from case-control genome-wide association studies suggest that 2 SNPs in WFS1 (rs734312; rs10010131) are associated with type 2 diabetes; G allele of rs734312 and A allele of rs10010131 appear to have protective effects. [META-ANALYSIS] 23373429_The recognition of microspherophakia in two siblings carrying a novel WFS1 mutation expands the clinical and molecular spectrum of Wolfram syndrome. 23499253_Report on an efficient double-tube allele-specific amplification method in conjunction with ultrafast capillary gel electrophoresis for direct haplotyping analysis of the SNPs in two important miRNA-binding sites (rs1046322 and rs9457) in the WFS1 gene. 23531866_Identified a DNA substitution (c.1385A-to-G) in WFS1 exon. 23595122_The results support previous findings that genetic variation of WFS1 contributes to the risk of diabetes mellitus and sensorineural hearing impairment. 23650218_this is the first report describing a microRNA binding site polymorphism of the WFS1 gene and its association with human aggression based on a large, non-clinical sample 23845777_A homozygous insertion mutation in WFS1 may be associated with early onset of disease symptoms in Wolfram syndrome. 23903355_This represents the first compelling report of a mutation in WFS1 associated with dominantly inherited nonsyndromic adult-onset diabetes. 24117146_Two familial cases of Wolfram syndrome caused by a novel homozygous WFS1 missense mutation, are reported. 24588001_The decrease in wolframin expression in diabetic placenta suggests that this protein may participate in maintaining the physiologic glucose homeostasis in this organ. 24909696_WFS1 gene mutations are a rare cause of hearing impairment among Finnish children. 25074416_No association was found between wolframin gene H611R polymorphism and mood disorders. 25211237_This study emphasizes the clinical and genetic heterogeneity in patients with WFS. Genotype-phenotype correlations may exist in patients with WFS1 mutations, as demonstrated by the disease onset. 25250959_A novel missense mutation c.2389G > A (GAC -AAC) in WFS1 gene causes non-syndromic hearing loss in all, rather than in low or high, frequencies. 25255707_The analysis of our case, in the light of the most recent literature, suggests a possible role for WFS1 gene in the development of certain brain structures during the fetal period. 25274773_Results reveal a role for WFS1 in the negative regulation of SERCA and provide further insights into the function of WFS1 in calcium homeostasis. 25740874_Early-onset Central diabetes insipidus is associated with de novo mutations of the AVP gene and with hereditary WFS1 gene changes. 25764693_Here we review clinical features, molecular mechanisms and mutations of WFS1 gene that relate to this syndrome.[review] 25800097_Data show that Wolfram syndrome 1 (WFS1; wolframin) promoter activity was highest with the most frequent haplotype (H1; ATCGT) and lowest with second most frequent haplotype (H2; GATCG). 26169481_Data suggest that a novel mutation in WFS1 [c.13481350 del ins TAG (p.His450*)] causes Wolfram-like syndrome in homozygous daughter with maternal uniparental disomy of chromosome 4; heterozygous mother is unaffected. [CASE REPORT] 26426397_Micro-RNA Binding Site Polymorphisms in the WFS1 Gene Are Risk Factors of Diabetes Mellitus. 26773575_A mutation (c.376G>A, p.A126T) was found in all 5 family members affected with Wolfram syndrome in homozygous state and in both parents in heterozygous state. 26875006_WFS1 is a highly polymorphic gene and determining the mode of inheritance or the pathological significance of a specific WFS1 variant is not always straightforward, especially in singleton cases with no access to other family members. Our study has revealed an interesting association between dominant missense WFS1 mutations and distinct OPL lamination on spectral domain OCT, which was not observed in patients with recessi 26943604_Nonsense mutation in the WFS1 gene is associated with Wolfram syndrome. 27377286_provides genotyping protocols readily applicable in any multiplex SNP and VNTR analyses, moreover confirms and extends previous results about the role of WFS1 polymorphisms in the genetic risk of diabetes mellitus 27395765_In this study, we found that patients with isolated, autosomal recessive nonsyndromic optic atropy have biallelic mutations in WFS1. We found that a high percentage (15%) of autosomal recessive non-syndromic optic atropy in families is caused by WFS1 mutations 27412528_Four novel mutations of the WFS1 gene in Iranian Wolfram syndrome pedigrees identified. 27468121_Data show that mutations in Wolfram syndrome 1 (wolframin) protein (WFS1) gene were identified in three children with Wolfram syndrome. 28039263_We show for the first time the role of WFS1 in CAO and document a statistically significant interaction between increasing cumulative cisplatin dose and rs62283056 genotype. Our clinical translational results demonstrate that pretherapy patient genotyping to minimize ototoxicity could be useful when deciding between cisplatin-based chemotherapy regimens of comparable efficacy with different cumulative doses 28271504_WFS1 and GJB2 mutations were identified in eight of 74 cases of Low-Frequency Sensorineural Hearing Loss. Four cases had heterozygous WFS1 mutations; one had a heterozygous WFS1 mutation and a heterozygous GJB2 mutation; and three cases had biallelic GJB2 mutations. Three cases with WFS1 mutations were sporadic; two of them were confirmed to be caused by a de novo mutation based on the genetic analysis of their parents. 28419064_a novel mutation c.2614-2625delCATGGCGCCGTG in the WFS1-gene was identified in a family with autosomal-dominant hereditary hearing impairment 28468959_Specific dominant WFS1 mutations are a cause of a novel syndrome including neonatal/infancy-onset diabetes, congenital cataracts, and sensorineural deafness. 28802351_data extend the mutation spectrum of the WFS1 gene in Chinese individuals and may contribute to establishing a better genotype-phenotype correlation for LFSNHL. 28974383_A nonsynonymous mutation in the WFS1 gene causing late-onset sensorineural hearing impairment with audiogram configurations typical for age-related hearing impairment. 29258540_This is the second report to describe a pathological mutation in WFS1 among Korean patients and the second to describe the mutation in a different ethnic background. Given that the mutation was found in independent families, p.S807R possibly appears to be a 'hot spot' in WFS1, which is associated with LF-NSHL. 29357349_Protective role of wfs1 against stress and age-associated neurodegeneration. 29447883_findings strongly suggest that the c.2389G>A mutation in WFS1 is associated with all-frequency hearing loss, rather than low- or high-frequency loss 29529044_Study successfully identified eight previously reported mutations and five novel variants, and estimated the incidence of WFS1 variants to be 2.5% in Japanese families with presumably autosomal dominant or mitochondrial HL. Also, results found that some variants can occur as de novo change at the mutational hot spots in WFS1, resulting in an audiovestibular phenotype. 29626590_Altered expression of WFS1 and NOTCH2 genes may play a role in pathogenesis and development of DN in patients with T2DM. 30014265_analysis of a low-frequency coding variant in the WFS1 gene that is enriched in Ashkenazi Jewish individuals and causes a mild form of Wolfram syndrome characterized by young-onset diabetes and reduced penetrance for optic atrophy 30305294_Traditional genome-wide association studies have identified single-nucleotide polymorphisms in ACYP2 and WFS1 associated with cisplatin-induced hearing loss. 30957632_Ophthalmic, systemic, and genetic characteristics of patients with Wolfram syndrome. 31363008_Segregation of two variants suggests the presence of autosomal dominant and recessive forms of WFS1-related disease within the same family: expanding the phenotypic spectrum of Wolfram Syndrome. 31477210_Our study revealed that transcription and translation of WFS1, CRH, and UCN were altered during pregnancies complicated by early-onset intrahepatic cholestasis of pregnancy (ICP). This disrupted compensatory response mediated by WFS1 and CRH family peptides in early-onset ICP may play a significant role in the pathogenesis of sudden fetal death in acute fetal hypoxia. 31658956_Nonsyndromic diabetes with WSF1 mutations is not rare in Chinese. Its response to alternative treatments should be investigated. 31759989_This study revealed an association of SIRT1 and WFS1 with Type 2 Diabetes (T2D) risk. A positive association with TD2 risk was found for WFS1 rs6446482 (p = 0.046, Z = 1.994) under an additive model, and SIRT1 rs7896005 (p = 0.038, Z = 2.073) under the dominant model. 31937257_The mutational and phenotypic spectrum of Wolfram syndrome (WS) is broadened by our report of novel WFS1 mutation. Our results reveal the value of molecular analysis of WFS1 in the improvement of clinical diagnostics for WS. This study also confirms the role of WFS1 in type-2 diabetes mellitus (T2DM) 32219690_Effect of 4-phenylbutyrate and valproate on dominant mutations of WFS1 gene in Wolfram syndrome. 32382995_A comparative analysis of genetic hearing loss phenotypes in European/American and Japanese populations. 32567228_Recurrent de novo WFS1 pathogenic variants in Chinese sporadic patients with nonsyndromic sensorineural hearing loss. 32938580_Identification of Three Novel and One Known Mutation in the WFS1 Gene in Four Unrelated Turkish Families: The Role of Homozygosity Mapping in the Early Diagnosis 33179441_Autosomal-dominant WFS1-related disorder-Report of a novel WFS1 variant and review of the phenotypic spectrum of autosomal recessive and dominant forms. 33693650_Novel mutations in the WFS1 gene are associated with Wolfram syndrome and systemic inflammation. 33882198_Different clinical entities of the same mutation: a case report of three sisters with Wolfram syndrome and efficacy of dipeptidyl peptidase-4 inhibitor therapy. 34006618_WFS1 protein expression correlates with clinical progression of optic atrophy in patients with Wolfram syndrome. 34133072_Wolfram Syndrome: Cracking the Code to Better Therapies. 34258273_Missense Variant of Endoplasmic Reticulum Region of WFS1 Gene Causes Autosomal Dominant Hearing Loss without Syndromic Phenotype. 34360843_A Novel Genetic Variant in the WFS1 Gene in a Patient with Partial Uniparental Mero-Isodisomy of Chromosome 4. 34404380_Unique three-site compound heterozygous mutation in the WFS1 gene in Wolfram syndrome. 34792487_Prevalence and phenotypic features of diabetes due to recessive, non-syndromic WFS1 mutations. 34848728_WFS1 functions in ER export of vesicular cargo proteins in pancreatic beta-cells. 35018440_WFS1 Gene-associated Diabetes Phenotypes and Identification of a Founder Mutation in Southern India. | ENSMUSG00000039474 | Wfs1 | 2490.50804 | 1.0533713 | 0.0750141251 | 0.10752067 | 5.003311e-01 | 4.793547e-01 | 9.998360e-01 | No | Yes | 2703.96501 | 309.341681 | 2.379270e+03 | 210.942837 | |
| ENSG00000109680 | 55296 | TBC1D19 | protein_coding | Q8N5T2 | FUNCTION: May act as a GTPase-activating protein for Rab family protein(s). | Alternative splicing;GTPase activation;Reference proteome | hsa:55296; | GTPase activator activity [GO:0005096] | ENSMUSG00000039178 | Tbc1d19 | 47.63792 | 0.7830795 | -0.3527692467 | 0.42741729 | 6.471450e-01 | 4.211353e-01 | 9.998360e-01 | No | Yes | 45.43922 | 10.278340 | 5.957715e+01 | 10.413143 | |||
| ENSG00000109794 | 25854 | FAM149A | protein_coding | A5PLN7 | Alternative splicing;Reference proteome | hsa:25854; | 26600424_In total, 270,389 single nucleotide polymorphisms passed quality control, and 4 SNPs in the FAM149A gene were associated with Acute Mountain Sickness; however, in the validation cohorts, FAM149A was not associated with the presence or severity of AMS. 34303830_Whole exome sequencing identified FAM149A as a plausible causative gene for congenital hereditary endothelial dystrophy, affecting Nrf2-Antioxidant signaling upon oxidative stress. | ENSMUSG00000070044 | Fam149a | 150.12925 | 1.0490436 | 0.0690746769 | 0.24752599 | 7.861623e-02 | 7.791815e-01 | 9.998360e-01 | No | Yes | 158.63711 | 18.521198 | 1.434440e+02 | 13.846683 | ||||
| ENSG00000110075 | 55291 | PPP6R3 | protein_coding | Q5H9R7 | FUNCTION: Regulatory subunit of protein phosphatase 6 (PP6). May function as a scaffolding PP6 subunit. May have an important role in maintaining immune self-tolerance. {ECO:0000269|PubMed:11401438, ECO:0000269|PubMed:16769727}. | Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | Protein phosphatase regulatory subunits, such as SAPS3, modulate the activity of protein phosphatase catalytic subunits by restricting substrate specificity, recruiting substrates, and determining the intracellular localization of the holoenzyme. SAPS3 is a regulatory subunit for the protein phosphatase-6 catalytic subunit (PPP6C; MIM 612725) (Stefansson and Brautigan, 2006 [PubMed 16769727]).[supplied by OMIM, Nov 2010]. | hsa:55291; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein phosphatase binding [GO:0019903]; protein phosphatase regulator activity [GO:0019888]; regulation of phosphoprotein phosphatase activity [GO:0043666] | 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that PPP6R3 is a putative expression variability marker for the 3-day, 8-cell embryo stage. 27113271_Molecular analyses revealed the presence and amplification of the novel PPPR6-USP6 gene fusion, which resulted in USP6 mRNA transcriptional upregulation. These findings further support the oncogenic role of the USP6 protease in mesenchymal neoplasia and expand the biologic potential of Nodular fasciitis 28893231_we found a DPP9-PPP6R3 fusion transcript in one tumor showing a matching genomic 11;19-translocation. Another tumor had a rearrangement of DPP9 with PLIN3. Both rearrangements were associated with diminished expression of the 3' end of DPP9 corresponding to the breakpoints identified by RNA-seq. 31904830_The substitution of Ala in nine putative phosphorylation sites in SAPS3 was required to prevent CK2 activation of the phosphatase. Different CK2 chemical inhibitors equally increased phosphorylation of endogenous AURKA in living cells, consistent with reduction in PP6 activity. | ENSMUSG00000024908 | Ppp6r3 | 1049.89769 | 1.0621746 | 0.0870210001 | 0.13431781 | 4.124445e-01 | 5.207314e-01 | 9.998360e-01 | No | Yes | 1038.22454 | 150.564075 | 9.718122e+02 | 108.802626 | |
| ENSG00000110427 | 25758 | KIAA1549L | protein_coding | Q6ZVL6 | Alternative splicing;Coiled coil;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:25758; | integral component of membrane [GO:0016021] | 29192581_Four variants in KIAA1549 like (KIAA1549L), a gene previously associated with attempted suicide in bipolar patients, were suggestively associated with being an owl at p < 1.82E-05; post hoc analyses showed the top variant trending in both the Mexican Americans and American Indians cohorts at p = 2.50E-05 and p = .030, respectively | ENSMUSG00000068373 | D430041D05Rik | 360.46591 | 0.9099472 | -0.1361452108 | 0.18573674 | 5.379674e-01 | 4.632763e-01 | 9.998360e-01 | No | Yes | 323.62511 | 37.625602 | 3.276324e+02 | 29.761037 | |||
| ENSG00000110888 | 65981 | CAPRIN2 | protein_coding | Q6IMN6 | FUNCTION: Promotes phosphorylation of the Wnt coreceptor LRP6, leading to increased activity of the canonical Wnt signaling pathway (PubMed:18762581). Facilitates constitutive LRP6 phosphorylation by CDK14/CCNY during G2/M stage of the cell cycle, which may potentiate cells for Wnt signaling (PubMed:27821587). May regulate the transport and translation of mRNAs, modulating for instance the expression of proteins involved in synaptic plasticity in neurons (By similarity). Involved in regulation of growth as erythroblasts shift from a highly proliferative state towards their terminal phase of differentiation (PubMed:14593112). May be involved in apoptosis (PubMed:14593112). {ECO:0000250|UniProtKB:Q05A80, ECO:0000269|PubMed:14593112, ECO:0000269|PubMed:18762581, ECO:0000269|PubMed:27821587}. | 3D-structure;Alternative splicing;Calcium;Cell membrane;Coiled coil;Cytoplasm;Differentiation;Growth regulation;Membrane;Metal-binding;Mitochondrion;Phosphoprotein;Protein synthesis inhibitor;RNA-binding;Reference proteome | The protein encoded by this gene may regulate the transport of mRNA. It may play a role in the differentiation of erythroblasts. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2016]. | hsa:65981; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; signaling receptor binding [GO:0005102]; negative regulation of cell growth [GO:0030308]; negative regulation of translation [GO:0017148]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of dendritic spine morphogenesis [GO:0061003]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of protein binding [GO:0032092]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 14593112_regulated expression of EEG-1 is involved in the orchestrated regulation of growth that occurs as erythroblasts shift from a highly proliferative state toward their terminal phase of differentiation. 18762581_Caprin-2 promotes activation of the canonical Wnt signaling pathway by regulating LRP5/6 phosphorylation. 25331957_Caprin-2 C1q-related domain forms a flexible homotrimer mediated by calcium, and this trimeric assembly is required for the functioning of caprin-2. 27821587_findings revealed an unrecognized role of Caprin-2 in facilitating LRP5/6 constitutive phosphorylation at G2/M through forming a quaternary complex with CDK14, Cyclin Y, and LRP5/6. 32691935_LINC00941 promotes oral squamous cell carcinoma progression via activating CAPRIN2 and canonical WNT/beta-catenin signaling pathway. 34511033_Upregulated lncRNA Cyclin-dependent kinase inhibitor 2B antisense RNA 1 induces the proliferation and migration of colorectal cancer by miR-378b/CAPRIN2 axis. | ENSMUSG00000030309 | Caprin2 | 184.92865 | 0.7419193 | -0.4306657682 | 0.25375375 | 2.623197e+00 | 1.053120e-01 | 9.998360e-01 | No | Yes | 159.03778 | 33.343283 | 1.961794e+02 | 31.735199 | |
| ENSG00000111077 | 23371 | TNS2 | protein_coding | Q63HR2 | FUNCTION: Tyrosine-protein phosphatase which regulates cell motility proliferation and muscle-response to insulin (PubMed:15817639, PubMed:23401856). In muscles and under catabolic conditions, dephosphorylates IRS1 leading to its degradation and muscle atrophy (PubMed:23401856). Negatively regulates PI3K-AKT pathway activation (PubMed:15817639, PubMed:23401856). {ECO:0000269|PubMed:15817639, ECO:0000269|PubMed:23401856}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Hydrolase;Membrane;Metal-binding;Methylation;Phosphoprotein;Protein phosphatase;Reference proteome;SH2 domain;Zinc;Zinc-finger | The protein encoded by this gene belongs to the tensin family. Tensin is a focal adhesion molecule that binds to actin filaments and participates in signaling pathways. This protein plays a role in regulating cell migration. Alternative splicing occurs at this locus and three transcript variants encoding three distinct isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:23371; | dendrite [GO:0030425]; focal adhesion [GO:0005925]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; postsynapse [GO:0098794]; identical protein binding [GO:0042802]; kinase binding [GO:0019900]; metal ion binding [GO:0046872]; protein tyrosine phosphatase activity [GO:0004725]; cellular homeostasis [GO:0019725]; collagen metabolic process [GO:0032963]; intracellular signal transduction [GO:0035556]; kidney development [GO:0001822]; multicellular organism growth [GO:0035264]; multicellular organismal homeostasis [GO:0048871]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of insulin receptor signaling pathway [GO:0046627]; peptidyl-tyrosine dephosphorylation [GO:0035335]; response to muscle activity [GO:0014850] | 12470648_Interaction of Axl receptor tyrosine kinase with C1-TEN. 15817639_C1 domain-containing phosphatase and TENsin homologue (C1-TEN) appears to be a novel intracellular phosphatase that negatively regulates the Akt/Protein kinase B signaling cascade 17006924_variant 3 significantly promoted the cell growth and motility of HCC cells; clonal transfectants of variant 3 were more closely packed and resulted in a higher saturation density than in the control vector transfectants 19194507_Tensins may represent a novel group of metastasis suppressors in the kidney, the loss of which leads to greater tumor cell motility and consequent metastasis. 19440389_DLC1 interaction with tensin2 through this novel focal adhesion binding site contributes to the growth-suppressive activity of DLC1. 19747564_Differential proteolytic cleavage of Tensin2 can liberate domains with discrete localisations and functions, which has implications for the role of Tensins in cancer cell survival and motility. 19895840_Data suggest that the interaction between START-GAP1 and tensin2 occurs in a PTB domain-dependent manner. manner.ty=0 20069572_focal adhesion-localized tensin 2 negatively regulates DLC1 to permit Rho-mediated actomyosin contraction and remodeling of collagen fibers 20678486_Tensin2, in addition to regulating cytoskeletal dynamics, influences phosphoinositide-Akt signalling through its PTPase domain. 21461930_Here, we report complete chemical shift assignments of the SH2 domain of human tensin2 determined by triple resonance experiments. 21527831_Tensin2 is an important new mediator in TPO/c-Mpl pathway and has a positive affect on cellular growth, at least in part through its effect on the PI3K/Akt signaling. 21765928_Tensin2 SH2 domain was identified to interact with nonphosphorylated ligand (DLC-1) as well as phosphorylated ligand. 22019427_Results identified tensin2 as a Syk-binding protein. 22645138_The tumor suppressor DLC1 utilizes a novel binding site for tensin2 PTB domain interaction 23233134_Pulldown studies in human cells using Myc-tagged Tensin2 constructs revealed that DISC1 specifically interacts with the C-terminal PTB domain of Tensin2 in a phosphorylation-independent manner. 25101860_p62 expression increased but C1-Ten protein decreased during muscle differentiation, supporting a role for p62 as a physiological regulator of C1-Ten. 27203214_Patients with low TNS2 expression showed poor relapse-free survival rates for breast and lung cancers. These results strongly suggest a role of tensin2 in suppressing cell transformation and reduction of tumorigenicity. 28955049_Study observed significant increase in C1-Ten level in diabetic kidney and in high glucose-induced damaged podocytes. C1-Ten acts as a protein tyrosine phosphatase (PTPase) at the nephrin-PI3K binding site and renders PI3K for IRS-1, thereby activating mTORC1. These findings demonstrate the relationship between nephrin dephosphorylation and the mTORC1 pathway, mediated by C1-Ten PTPase activity. 30092354_Collectively,these findings suggest that the interaction between the C1-Ten/Tensin2 SH2 domain and PtdIns(3,4,5)P3 produces a negative feedback loop of insulin signaling through IRS-1. 30419905_Axl binds to and phosphorylates TNS2 and that Axl/TNS2/IRS-1 cross-talk may potentially play a critical role in glucose metabolism of cancer cells. 33753854_Novel candidate factors predicting the effect of S-1 adjuvant chemotherapy of pancreatic cancer. | ENSMUSG00000037003 | Tns2 | 78.53146 | 1.5775137 | 0.6576525077 | 0.36300428 | 3.185660e+00 | 7.428699e-02 | 9.998360e-01 | No | Yes | 66.72682 | 14.558959 | 4.019274e+01 | 7.042817 | |
| ENSG00000112033 | 5467 | PPARD | protein_coding | Q03181 | FUNCTION: Ligand-activated transcription factor. Receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Has a preference for poly-unsaturated fatty acids, such as gamma-linoleic acid and eicosapentanoic acid. Once activated by a ligand, the receptor binds to promoter elements of target genes. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the acyl-CoA oxidase gene. Decreases expression of NPC1L1 once activated by a ligand. {ECO:0000269|PubMed:1333051, ECO:0000269|PubMed:15604518}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Metal-binding;Nucleus;Receptor;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) family. The encoded protein is thought to function as an integrator of transcriptional repression and nuclear receptor signaling. It may inhibit the ligand-induced transcriptional activity of peroxisome proliferator activated receptors alpha and gamma, though evidence for this effect is inconsistent. Expression of this gene in colorectal cancer cells may be variable but is typically relatively low. Knockout studies in mice suggested a role for this protein in myelination of the corpus callosum, lipid metabolism, differentiation, and epidermal cell proliferation. Alternative splicing results in multiple transcript variants encoding distinct protein isoforms. [provided by RefSeq, Aug 2017]. | hsa:5467; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; linoleic acid binding [GO:0070539]; lipid binding [GO:0008289]; NF-kappaB binding [GO:0051059]; nuclear receptor activity [GO:0004879]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; steroid hormone receptor activity [GO:0003707]; transcription coactivator binding [GO:0001223]; zinc ion binding [GO:0008270]; adipose tissue development [GO:0060612]; apoptotic process [GO:0006915]; apoptotic signaling pathway [GO:0097190]; axon ensheathment [GO:0008366]; cell differentiation [GO:0030154]; cell population proliferation [GO:0008283]; cell-substrate adhesion [GO:0031589]; cellular response to hypoxia [GO:0071456]; cellular response to lipopolysaccharide [GO:0071222]; cholesterol metabolic process [GO:0008203]; decidualization [GO:0046697]; embryo implantation [GO:0007566]; fatty acid beta-oxidation [GO:0006635]; fatty acid catabolic process [GO:0009062]; fatty acid metabolic process [GO:0006631]; fatty acid transport [GO:0015908]; generation of precursor metabolites and energy [GO:0006091]; glucose metabolic process [GO:0006006]; glucose transmembrane transport [GO:1904659]; heart development [GO:0007507]; hormone-mediated signaling pathway [GO:0009755]; keratinocyte migration [GO:0051546]; keratinocyte proliferation [GO:0043616]; lipid metabolic process [GO:0006629]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell growth [GO:0030308]; negative regulation of cholesterol storage [GO:0010887]; negative regulation of collagen biosynthetic process [GO:0032966]; negative regulation of epithelial cell proliferation [GO:0050680]; negative regulation of inflammatory response [GO:0050728]; negative regulation of myoblast differentiation [GO:0045662]; negative regulation of pri-miRNA transcription by RNA polymerase II [GO:1902894]; negative regulation of smooth muscle cell migration [GO:0014912]; negative regulation of smooth muscle cell proliferation [GO:0048662]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; phospholipid biosynthetic process [GO:0008654]; positive regulation of epidermis development [GO:0045684]; positive regulation of fat cell differentiation [GO:0045600]; positive regulation of fatty acid metabolic process [GO:0045923]; positive regulation of fatty acid oxidation [GO:0046321]; positive regulation of gene expression [GO:0010628]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774]; positive regulation of myoblast proliferation [GO:2000288]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of skeletal muscle tissue regeneration [GO:0043415]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; proteoglycan metabolic process [GO:0006029]; regulation of lipid metabolic process [GO:0019216]; regulation of skeletal muscle satellite cell proliferation [GO:0014842]; regulation of transcription by RNA polymerase II [GO:0006357]; response to activity [GO:0014823]; response to glucose [GO:0009749]; response to lipid [GO:0033993]; response to vitamin A [GO:0033189]; vasodilation [GO:0042311]; vitamin A metabolic process [GO:0006776]; wound healing [GO:0042060] | 11980898_differential regulation of vascular endothelial growth factor expression in bladder cancer cells (peroxisome proliferative activated receptor, beta) 12009300_Results suggest that peroxisome proliferator-activated receptor beta overexpression is not an inherent property of breast cancer cell lines, but it may play a role through activation of downstream genes (PPARbeta). 12594814_Expression of peroxisome proliferator-activated receptors (PPARs) in human urinary bladder carcinoma and growth inhibition by its agonists. 12615676_4 polymorphisms were found: -409C/T (promoter, +73C/T (exon 1), +255A/G (exon 3), & +294T/C (exon 4). An interaction with the PPAR alpha L162V polymorphism was also detected for several lipid parameters. PPARD plays a role in cholesterol metabolism. 12909723_The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. 14641801_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14758356_results implicate PPAR-delta in the regulation of intestinal adenoma growth 14988273_Observational study of gene-disease association. (HuGE Navigator) 14988273_Positive associations of PPAR-delta polymorphisms with fasting plasma glucose and BMI detected in nondiabetic control subjects 15001550_gene regulation by PPARdelta in the uterine cells uniquely responds to SRC-2, N-CoR, SMRT, or RIP140, and these interactions may be operative during implantation when these cofactors are abundantly expressed. 15102088_PPAR-beta/delta activation stimulates keratinocyte differentiation, is anti-inflammatory, improves barrier homeostasis, and stimulates triglyceride accumulation in keratinocytes. 15128052_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15591138_11beta-HSD2 is an additional target for PPAR delta, which may regulate human placental function 15793256_This study was performed to determine whether specific activation of PPARdelta has direct effects on insulin action in skeletal muscle. 15811118_COX-2 immunopositivity was significantly associated with PPARbeta and PPARgamma immunoreactivity. Microvessel density was significantly higher among PPARbeta-immunoreactive squamous cell carcinomas. 15888456_the ligand-independent tight control of the position of the PPAR helix 12 provides an effective alternative for establishing an interaction with CoA proteins 15890193_PPARdelta signaling pathways are interconnected at the level of cross-regulation of their respective transcription factor mRNA levels 15979543_PPARdelta expression is up-regulated between the first and third trimester, indicating a role for this nuclear receptor in placental function 16053787_PPARdelta + 294T/C gene polymorphism in subjects with metabolic syndrome may be involved in the occurrence of obesity and dyslipidemia. 16091736_PPARdelta partially rescued prostate epithelial cells from growth inhibition and also dramatically inhibited sulindac sulfide-mediated p21WAF1/CIP1 upregulation. 16141797_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16285997_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16285997_PPARdelta +294T/C polymorphism has no influence on plasma lipoprotein concentrations, body mass index or atherosclerotic disease either in healthy subjects or in patients with DM-2, both in males and females. 16306381_Observational study of gene-disease association. (HuGE Navigator) 16306381_Single nucleotide polymorphisms of PPARD primarily affected insulin sensitivity by modifying glucose uptake in skeletal muscle but not in adipose tissue. 16344721_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16361076_The expression of PPARdelta gene in rectal cancers is not statistically different from normal mucosa. 16368717_Human platelets contain PPARbeta and that its selective activation inhibits platelet aggregation. 16387648_Data describe the activated form of the peroxisome proliferator-activated receptor-beta/delta using a ligand binding domain model. 16476973_overexpressed during the S phase of the cell cycle compared with the G0/1 phase 16511591_This review concludes that PPAR delta has emerged as a powerful metabolic regulator in diverse tissues including fat, skeletal muscle, and the heart. 16645156_PGI2 protects endothelial cells from H2O2-induced apoptosis by inducing PPARdelta binding to 14-3-3alpha promoter, thereby upregulating 14-3-3alpha protein expression. 16652134_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16652134_data provide further evidence for an involvement of PPARdelta in the regulation of BMI. 16752430_Skeletal muscle mRNA expression of PPAR delta increased in type 2 diabetic patients with an improved clinical profile following low-intensity exercise, but were unchanged in patients who did not show exercise-mediated improvements in clinical parameters. 16804087_Observational study of gene-disease association. (HuGE Navigator) 16804087_Single nucleotide polymorphisms in PPARD modify the conversion from glucose intolerance to type 2 diabetes. 16897074_Therefore, these results indicate that induction of fatty acid oxidation with PPARbeta activators during short-term exposition is not sufficient to correct for insulin resistance in muscle cells from type 2 diabetic patients. 16906219_Observational study of gene-disease association. (HuGE Navigator) 16953259_Observational study of gene-disease association. (HuGE Navigator) 16979821_Observational study of gene-disease association. (HuGE Navigator) 17068288_PPARbeta/delta is a novel regulator of endothelial cell proliferation and angiogenesis through VEGF. 17116180_Observational study of gene-disease association. (HuGE Navigator) 17116180_PPARD-87T/C polymorphism is associated with higher fasting plasma glucose concentrations in both normal glucose tolerant and diabetic subjects, largely due to impaired insulin sensitivity 17119917_PPAR-delta activation increases cholesterol export and represses inflammatory gene expression in macrophages and atherosclerotic lesions. 17148604_support the rationale for developing PPARdelta antagonists for prevention and/or treatment of cancer 17178883_PPARdelta induces COX-2 expression and the COX-2-derived PGE(2) further activates PPARdelta via cPLA(2)alpha. which forms a growth-promoting signaling that may play a role in hepatocarcinogenesis. 17254750_These studies demonstrate that ligand activation of PPARbeta/delta does not lead to an anti-apoptotic effect in either human or mouse keratinocytes, but rather, leads to inhibition of cell growth likely through the induction of terminal differentiation. 17259439_DNA sequence variation in the PPARdelta locus is a potential modifier of changes in cardiorespiratory fitness and plasma HDL-C in healthy individuals in response to regular exercise. 17259439_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17272748_Observational study of gene-disease association. (HuGE Navigator) 17294733_The expression of PPARdelta gene in rectal cancers is not statistically different from that in normal mucosa, and it is not correlated with cell differentiation, pathological categories and Dukes stages. 17303142_human aortic smooth muscle cells Prostacyclin 2 synthase(PGIS) gene transfer reduced peroxisome proliferator-activated receptor delta(PPAR-delta) expression and inhibited neointimal formation after balloon injury 17303761_PPAR delta activates 14-3-3 epsilon gene (YWHAE) promoter in human endothelial cells in a concentration and time-dependent manner. 17322100_The activated proinflammatory state of monocytes and MDM in low HDL-C subjects constitutes a novel parameter of risk associated with HDL deficiency, related to altered expression of metallothionein genes and the reciprocal regulation of PPARdelta. 17324937_analysis of the ligand-dependent control of peroxisome proliferator-activated receptor delta activity 17327385_Observational study of gene-disease association. (HuGE Navigator) 17327385_the minor serine-encoding allele of the Gly482Ser single-nucleotide polymorphism in PPARGC1A was associated with less increase in individual anaerobic threshold during lifestyle intervention 17355643_Observational study of genotype prevalence. (HuGE Navigator) 17356846_REVIEW: PPARbeta/delta is a transcriptional regulator of FA uptake and oxidation, mitochondrial respiration uncoupling, and glucose metabolism 17409576_New target for Metablic syndrome:PPARD. 17431579_Observational study of gene-disease association. (HuGE Navigator) 17431579_PPAED does not significantly affect the risk of metabolic disease in the population studied, such as type 2 diabetes. 17436029_In contrast to the PPARalpha, PPARdelta receptor isoforms were not activated by the epoxyalcohol. 17618585_High expression of PPARbeta may be related with the differentiation and metastasis of epithelial ovarian carcinoma. 17669420_PDK2, PDK3 and PDK4 are primary PPARbeta/delta target genes in humans underlining the importance of the receptor in the control of metabolism 17693664_PPARbeta/delta ligands do not potentiate tumorigenesis. 17705821_Alternative splicing of PPARD was studied in terms of translation efficiency and trans-activation ability. 17724132_Metabolism of endocannabinoids by the endothelial cell COX-2 coupled to the prostacyclin (PGI(2)) synthase activates the nuclear receptor peroxisomal proliferator-activated receptor delta, which negatively regulates the expression of tissue factor. 17855759_This study provides evidence that mRNA expression of PPARdelta in human skeletal muscle is under genetic control but also influenced by factors such as age, birth weight and central adiposity. 17926914_Observational study of gene-disease association. (HuGE Navigator) 18024853_PPARdelta agonist GW501516 reverses multiple abnormalities associated with the metabolic syndrome without increasing oxidative stress. 18037904_ternary complex consisting of PPARdelta, p65/RelA, and HDAC1 in keratinocytes PPARdelta repression 18048767_ligand activation of PPAR-delta (peroxisome proliferator-activated receptor delta ) in Endothelial cells has an antiinflammatory effect, perhaps via a binary mechanism involving the induction of antioxidative genes and the release of nuclear corepressors 18182682_review of role of activation of PPARalpha, -beta/delta, or -gamma or LXRs in skin physiology and cytology and disease 18240567_PPARD C allele is associated with predisposition to endurance performance. 18252792_Observational study of gene-disease association. (HuGE Navigator) 18252792_SNPs rs1053049, rs6902123, and rs2267668 in PPARD affect life style intervention-induced changes in overall adiposity, hepatic fat storage, and relative muscle mass. 18288282_Observational study of gene-disease association. (HuGE Navigator) 18305567_There is a direct link between type 1 IFN signaling and PPARdelta. The induction of PPARdelta by type 1 IFN contributes to the persistence of activated T cells in psoriasis skin lesions. 18337509_PPARdelta antagonizes multiple proinflammatory pathways 18379566_No significant effects of PPAR delta T+294C polymorphism or the interaction of the PPAR delta and PPAR gamma variants on adiposity-related phenotypes were observed in any age group or gender 18379566_Observational study of gene-disease association. (HuGE Navigator) 18388689_polymorphisms in peroxisome proliferator-activated receptors are critical susceptibility risk factors for dyslipidemias and diabetes [review] 18438697_Observational study of gene-disease association. (HuGE Navigator) 18511850_activation of COX-1/PGI(2)/PPARdelta pathway is an important mechanism underlying proangiogenic function of EPCs. 18566335_PPARbeta/delta is strongly expressed in the majority of lung cancers, and its activation induces proliferative and survival response in non-small cell lung cancer. 18606951_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18606951_PPARD gene is associated with bipolar disorder 18625220_These results demonstrate that epidermal growth factor induces PPARbeta/delta expression in a c-Jun-dependent manner and PPARbeta/delta plays a vital role in EGF-stimulated proliferation of HaCaT cells. 18627005_Novel mechanism by which PPARdelta regulates lipogenesis, suggesting potential therapeutic applications of PPARdelta modulators in obesity and type 2 diabetes, as well as related steatotic liver diseases. 18657264_We have found significant interaction between CAPN5 and PPARD genes that reduces risk for obesity in 55%. CAPN5 and PPARD gene products may also interact in vivo. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18676870_Observational study of gene-disease association. (HuGE Navigator) 18701481_up-regulation of peroxisome proliferator-activated receptor-delta is associated with thyroid tumors 18720000_The simultaneous expression of peroxisome proliferator-activated receptor delta and cyclooxygenase-2 may enhance angiogenesis and tumor venous invasion in tissues of colorectal cancers. 18797151_lower transactivity of PPAR delta for arachidonic acid in Caco-2 cells, compared with PPAR alpha, is associated with the binding activity of p300 to the receptor 18797401_Observational study of gene-disease association. (HuGE Navigator) 18937948_Hydrogen peroxide down-regulated the expression/activation of PPAR-beta, which played important roles in hydrogen peroxide-induced apoptosis in endothelial cells. 19056929_although acting through a ligand-dependent modality, PPARdelta is a negative regulator of VD-mediated monocyte differentiation 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19074989_ANGPTL4 is produced by human myotubes in response to long chain fatty acids and is of systemic relevance in humans. 19157507_Peroxisome proliferator-activated receptor expression in effusions from ovarian carcinomais associated with poor response to chemotherapy at disease recurrence and poor survival 19201410_PPARg, PPARd and LXRa are involved in the regulation of ABCA1 expression and HDL biogenesis in a cooperative signal transduction pathway 19208777_Observational study of gene-disease association. (HuGE Navigator) 19279199_Report PPAR/RXRalpha dysregulation in preterm and infection-driven infection labor. 19283612_Evaluation of PPAR gamma and delta expression is useful for predicting postoperative mortality in patients undergoing colorectal cancer surgery. 19303973_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19307598_Regulation of epithelial-mesenchymal IL-1 signaling by PPARbeta/delta is essential for skin homeostasis and wound healing. 19383774_Observational study of gene-disease association. (HuGE Navigator) 19383774_PPARdelta may affect height through a variety of mechanisms including altered metabolic efficiency or effects on osteoclast function. 19395102_PPARdelta attenuated CRP-induced pro-inflammatory effects may through CD32 and NF-kappaB pathway 19429679_ceramide, an important lipid component of epidermis, up-regulates ABCA12 expression via the PPARdelta-mediated signaling pathway, providing a substrate-driven, feed-forward mechanism for regulating this key lipid transporter 19435887_PGC-1alpha-induced fatty acid oxidation and mitochondrial biogenesis appear to be independent of PPARdelta 19453261_Observational study of gene-disease association. (HuGE Navigator) 19461048_PPARdelta-induced upregulation of extracellular matrix proteins exerts an antiapoptotic effect, thereby maintaining the stability of atherosclerotic plaques 19512923_review of evidence for role of PPAR[delta] in lipid & lipoprotein metabolism; genetic association studies of SNP's in PPARD gene have only provided negative or conflicting evidence for gross phenotypes: obesity, hyperlipidaemia & type 2 diabetes [review] 19527689_PPARalpha and PPARbeta/delta do not appear to modulate the alternative differentiation of human macrophages. 19538467_PPARdelta plays an important role in skin wound healing in vivo and that it functions by accelerating extracellular matrix-mediated cellular interactions in a process mediated by the TGF-beta1/Smad3 signaling-dependent or - independent pathway. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19615407_This review focuses particularly on the emerging roles of PPARbeta/delta as an alternative regulator of inflammation and inflammation-related disorders. [review] 19628794_Shear stress induces synthetic-to-contractile phenotypic modulation in smooth muscle cells via peroxisome proliferator-activated receptor alpha/delta activations by prostacyclin released by sheared endothelial cells. 19653005_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19666693_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19666693_a higher frequency of the peroxisome proliferator-activated receptor gamma coactivator 1 Gly/Gly + PPARD CC genotype is associated with elite-level endurance athletes 19681917_Meta-analysis of gene-disease association and gene-gene interaction. (HuGE Navigator) 19723884_results establish an important role for PPAR-delta in transcription of tumor-promoting genes, which can be specifically modulated by high-affinity RNA intramers in colon cancer cells 19756148_analysis of PPARgamma ligand modulated antagonism by genomic and non-genomic actions of PPARdelta 19805479_REVIEW: current status regarding the regulation, and the metabolic effects, of PPARdelta in skeletal muscle 19818126_Observational study of gene-disease association. (HuGE Navigator) 19838202_The data of this study show that genetic deficiency of PPAR- delays clearance of apoptotic cells and increases autoantibody production, leading to immune complex-mediated glomerulonephritis. 19863319_The majority of tumours showed strong p16, p21, p27, pRb and cyclin D1 staining and little or no p53 expression. Tumours harbouring dysplasia were significantly more likely to be p53-positive and exhibit up-regulated p21 and p27. 19878569_Observational study of gene-disease association. (HuGE Navigator) 19903700_important relationship between PPARdelta, PGC1alpha, and haem oxygenase-1, demonstrating that haem oxygenase-1 induction plays an important role in cytoprotective actions of PPARdelta ligands in vascular endothelium 19913121_Observational study of gene-disease association. (HuGE Navigator) 19935699_findings consistently indicate that PPAR-beta may facilitate differentiation and inhibit the cell-fibronectin adhesion of colon cancer. 19948841_PLA2-modified LDL transactivates macrophage PPARalpha and PPARdelta, but not PPARgamma. 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 19951549_EGF inhibits HaCaT apoptosis caused by TNF-alpha in a PPARbeta-dependent manner. 20044476_Observational study of gene-disease association. (HuGE Navigator) 20045185_PPARdelta may be involved in the pathophysiology of gestational diabes, preeclampsia, and IUGR. 20160399_Predominant role of PPARdelta in the transcriptional regulation of SIRT1 gene. 20200337_In subjects with and without type 2 diabetes, single polymorphism was associated with body mass index, high-density lipoprotein cholesterol, leptin, and TNFalpha and was dependent on gender 20200337_Observational study of gene-disease association. (HuGE Navigator) 20221637_PPARbeta/delta as an alternative COX-independent mechanism of VEGF induction in colorectal tumor cells. 20238044_The expression of genes involved in lipid metabolism and expression of the three isoforms of PPARs in an immortalized cell line of human retinal pigment epithelial cells, were investigated. 20308079_ACSL3 is a novel molecular target of PPARdelta in HepG2 cells; there is a regulatory mechanism for ACSL3 transcription in liver tissue 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20416077_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20424164_Fatty acid-binding protein 5 and PPARbeta/delta are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth 20453000_Observational study of gene-disease association. (HuGE Navigator) 20485444_Observational study of gene-disease association. (HuGE Navigator) 20512451_CYBA +242C/T and PPARD +294T/C variants modulate risk of coronary artery disease through their associations with atherogenic serum lipid profiles. 20512451_Observational study of gene-disease association. (HuGE Navigator) 20592029_PPARdelta induces IL-8 expression in nonstimulated endothelial cells via transcriptional as well as posttranscriptional mechanisms 20595396_Genome-wide transcriptional profiling that PPARbeta/delta and transforming growth factor-beta (TGFbeta) pathways functionally interact in human myofibroblasts and that a subset of these genes is cooperatively activated by TGFbeta and PPARbeta/delta. 20621019_Observational study of gene-disease association. (HuGE Navigator) 20621019_SNP and haplotypes of PPARD, PPARG and APM1 may underlie the genetic basis of the constitutions classified in traditional Chinese medicine. 20624891_PPAR-delta serves as an important molecular brake for the control of central nervous system autoimmune inflammation. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20645257_Observational study of gene-disease association. (HuGE Navigator) 20647061_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20811626_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20824502_The expression of both PPAR delta and COX-2 in tissues may lead to liver metastasis and consequent poor prognosis in colorectal cancer patients. 20840271_Observational study of gene-disease association. (HuGE Navigator) 20840271_The +294T/C polymorphism in the exon 4 of the PPAR-delta gene seems to cause an increase in fasting glucose levels. 20844743_Observational study of gene-disease association. (HuGE Navigator) 20855565_Observational study of gene-disease association. (HuGE Navigator) 21070867_Data describe the effects of the PPARbeta/delta agonist GW501516 on inflammation, and show that GW501516-induced PPARbeta/delta activation attenuates the inflammatory response induced in human cardiac AC16 cells exposed to palmitate and in mice fed a high-fat diet. 21176135_the association of PPARD +294T > C polymorphism and serum lipid levels is different between the Bai Ku Yao and Han populations 21283829_enabled the definition of 112 bona fide PPARbeta/delta target genes showing either of three distinct types of transcriptional response 21331343_myocyte expression of peroxisome proliferator-activated receptor delta and the adiponectin receptors 1 and 2 and T-cadherin are highly coordinated and this might be of relevance for human lipid metabolism in vivo 21352808_these results may lead to a better understanding of the molecular mechanisms underlying the anti-senescent effect of PPARdelta. 21400612_Expression of peroxisome proliferator-activated nuclear receptor beta/delta is lower in human and Apc(+/Min-FCCC) mouse colon tumors than in corresponding normal tissue. 21426320_Analysis of PPARdelta mRNA levels revealed a hypomorph expression of human PPARdelta in liver. 21487230_Gene-diet interactions were also found for for PPARdelta -87T-->C with the polyunsaturated/saturated fat ratio . 21496113_PPAR-delta agonists inhibit TGFalpha-induced MMP9 expression in human keratinocytes. This inhibition involves repression of c-fos binding at the MMP9 promoter. 21520069_The results support the important role of peroxisome proliferator-activated receptor beta/delta in neuronal differentiation strongly point towards peroxisome proliferator-activated receptor beta/delta as modulators of crucial pathways. 21531809_Increase of PPAR delta predicts favorable survival in the rectal cancer patients either with or without preoperative radiotherapy 21696362_[review] PPARbeta/delta activation exerts beneficial effects against organ-related ischemic events, which are among the most critical cardiovascular complications evoked by metabolic dysregulation. 21750705_Data show a novel pathway involving HL hydrolysis of VLDL that activates PPARdelta through generation of specific monounsaturated fatty acids. 21765467_PPARdelta and PPARgamma coordinately regulate cancer cell fate by controlling the balance between the cell death and survival 21834910_No significant associations between PPARD and T2D were found in either genotyped or imputed SNPs and no effect modification between exercise participation and PPARD genetic variation was found. 21896929_investigation of function of PPARD in regulating insulin secretion in hyperglycemia (i.e., high glucose): Data indicate that an endogenous ligand of PPARD (4-hydroxy-2-nonenal) mimics glucose in stimulating insulin secretion in a beta-cell line. 22021335_PPAR-gamma/delta signaling accounts in part for the vasodilating effect of prostacyclins in pulmonary arteries. 22072715_ligand-activated PPARdelta confers resistance to Ang II-induced senescence by up-regulation of PTEN and ensuing modulation of the PI3K/Akt signaling to reduce ROS generation in vascular cells. 22192471_there is no interaction between the PPARD +294T > C genotypes and alcohol consumption on serum lipid levels in alcohol drinkers. 22209682_Oleic acid activates GPR40-phospholipase C-calcium pathway to increase the expression of PPAR-delta and PPAR-delta further decreased the expression of PTEN to regulate insulin sensitivity in hepatic steatosis. 22247001_relationships between polymorphisms of the ACE, ACTN3, PPARD, and PPARGC1A genes and performance as measured by six fitness tests in sedentary adolescent girls 22277050_Polymorphic variants of PPARD and APOE genes are associated with coronary heart disease. 22310107_Gene variants in PPARD and PPARGC1A might be associated with timing of natural menopause, probably through direct actions on the ovaries, among the general Japanese population. 22509365_common variants in PPARD contribute to the risk of type 2 diabetes in Chinese Hans, and provided suggestive evidence of interaction between 25(OH)D levels and PPARD-rs6902123 on HbA1c 22606221_two residues (Val312 and Ile328) in the buried hormone binding pocket play special roles in PPARdelta selective binding 22683888_the reduction in PPARbeta/delta activity and SIRT1 expression caused by TNF-alpha stimulation through NF-kappaB helps perpetuate the inflammatory process in human adipocytes. 22944052_The C allele in the PPARD rs2016520 genotype is significantly associated with a lower rate of abdominal obesity. There is an interaction between PPARA, PPARD and PPARG SNPs on the incidence of abdominal obesity. 22945906_Peroxisome proliferator-activated receptor delta agonist attenuates nicotine suppression effect on human mesenchymal stem cell-derived osteogenesis and involves increased expression of heme oxygenase-1. 23023367_Activation of peroxisome proliferator-activated receptor delta inhibits human macrophage foam cell formation and the inflammatory response induced by very low-density lipoprotein. 23049921_PPARbeta/delta might be a central element in lung carcinogenesis controlling multiple pathways and representing a potential target for NSCLC treatment 23056264_Findings suggest that fatty acids (FAs)-PPARdelta/RXR-Angptl4 axis controls the lipoprotein lipase (LPL)-dependent uptake of FAs in myotubes. 23086933_beta-Catenin and peroxisome proliferator-activated receptor-delta coordinate dynamic chromatin loops for the transcription of vascular endothelial growth factor A gene in colon cancer cells 23109900_Several lipid-related gene polymorphisms interact with overweight/obesity to modulate blood pressure levels. 23208498_A PPARbeta/delta-ANGPTL4 pathway is involved in the regulation of tumor cell invasion. 23262340_PPAR-alpha, PPAR-delta and PPAR-gamma polymorphisms may contribute to the risk of hypertriglyceridemia independently and/or in an interactive manner. 23323702_These data document for the first time that the expression of BMAL1, PER3, PPARD and CRY2 genes is altered in gestational diabetes compared to normal pregnant women. deranged expression of clock genes may play a pathogenic role in GDM. 23512374_The PPARdelta +294T/C polymorphism was associated with the risk of ischemic stroke in Tunisian subjects. No interaction between diabetes and PPAR +294T/C polymorphism on the risk of ischemic stroke was found. 23525285_PPARdelta is regulated by miR-9 in monocytes. 23545576_rs2016520 and rs10865170 were associated with lower obesity risk. In addition, interaction was identified among rs2016520, rs9794, and rs10865170 in obesity. 23598720_Expression of PPARdelta is tightly correlated in both prostate tissues and cell lines and significantly higher in cancer vs. normal tissues. 23607100_These results suggest that PPAR delta may be a potential therapeutic target against the progression of vascular remodeling in Pulmonary arterial hypertension . 23639976_These results suggest that PPARdelta-mediated inhibition of MMP-1 secretion prevents some effects of photoaging. 23646170_PPARbeta/delta plays a role in the assembly of a cytoplasmic multi-protein complex containing TAK1, TAB1, HSP27 and PPARbeta/delta, and thereby participates in the NFkappaB response to IL-1beta. 23811234_PPARs play a role in placental nitric oxide (NO) and lipid homeostasis and can regulate NO production, lipid concentrations and lipoperoxidation in placentas from type 2 diabetic patients. 23832539_Data show that AP1 is the transcriptional factor that contributes to peroxisome proliferator-activated receptor delta gene (PPARdelta) expression in LoVo cells. 23842279_In the presence of PPARbeta/delta, Vpr induced a 3.3-fold increase in PPAR response element-driven transcriptional activity, a 1.9-fold increase in PDK4 protein expression, and a 1.6-fold increase in the phosphorylated pyruvate dehydrogenase subunit E1alpha. 23907334_Genetic variants of peroxisome proliferator-activated receptor delta are associated with gastric cancer. 23991827_role of peroxisome proliferator-activated receptor delta (PPARdelta) activation on global gene expression and mitochondrial fuel utilization in myotube 24060638_Down-regulation of PPAR delta gene is associated with Metabolic Syndrome. 24107399_Pleiotropic effects of peroxisome proliferator-activated receptor gamma and delta in vascular diseases. [review] 24118591_the PPARD A/C/C haplotype (rs2267668/rs2016520/rs1053049) was significantly underrepresented in athletes compared to controls, suggesting that the A/C/C haplotypes may influence elite athletic performance. 24252308_Results suggest that myelin-derived phosphatidylserine mediates PPAR-beat/delta activation in macrophages after myelin uptake 24391853_PPARD mutations are associated with colorectal cancer. 24508264_Data indicate that palmitic acid induced suppression of apolipoprotein M (APOM) expression is mediated via the peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) pathway. 24527778_PPARbeta activation in vitro and in vivo restores the endothelial function, preserving the insulin-Akt-eNOS pathway impaired by high glucose, at least in part, through PDK4 activation. 24610641_PPARdelta is required for hypoxic stress-mediated cytokine expression in colon cancer cells, resulting in promotion of angiogenesis, macrophage recruitment, and macrophage proliferation in the tumor microenvironment. 24692551_Data indicate that fatty acid-binding protein 5 (FABP5) is tuned to selectively stimulate peroxisome proliferation-activated receptor beta/delta transactivation in response to specific fatty acids based on their structural features. 24898257_new role for ER stress and UPR that attenuates H-RAS-induced senescence and suggest that PPARbeta/delta can repress this oncogene-induced ER stress to promote senescence in accordance with its | ENSMUSG00000002250 | Ppard | 1578.66390 | 1.0577349 | 0.0809780256 | 0.11138834 | 5.286129e-01 | 4.671906e-01 | 9.998360e-01 | No | Yes | 1775.51828 | 191.561170 | 1.553542e+03 | 130.320395 | |
| ENSG00000112096 | 6648 | SOD2 | protein_coding | P04179 | FUNCTION: Destroys superoxide anion radicals which are normally produced within the cells and which are toxic to biological systems. {ECO:0000269|PubMed:10334867}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Manganese;Metal-binding;Mitochondrion;Nitration;Oxidoreductase;Reference proteome;Transit peptide;Ubl conjugation | This gene is a member of the iron/manganese superoxide dismutase family. It encodes a mitochondrial protein that forms a homotetramer and binds one manganese ion per subunit. This protein binds to the superoxide byproducts of oxidative phosphorylation and converts them to hydrogen peroxide and diatomic oxygen. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), premature aging, sporadic motor neuron disease, and cancer. Alternative splicing of this gene results in multiple transcript variants. A related pseudogene has been identified on chromosome 1. [provided by RefSeq, Apr 2016]. | hsa:6648; | extracellular exosome [GO:0070062]; mitochondrial matrix [GO:0005759]; mitochondrial nucleoid [GO:0042645]; mitochondrion [GO:0005739]; DNA binding [GO:0003677]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; manganese ion binding [GO:0030145]; oxygen binding [GO:0019825]; superoxide dismutase activity [GO:0004784]; acetylcholine-mediated vasodilation involved in regulation of systemic arterial blood pressure [GO:0003069]; age-dependent response to reactive oxygen species [GO:0001315]; cellular response to ethanol [GO:0071361]; detection of oxygen [GO:0003032]; erythrophore differentiation [GO:0048773]; glutathione metabolic process [GO:0006749]; heart development [GO:0007507]; hemopoiesis [GO:0030097]; hydrogen peroxide biosynthetic process [GO:0050665]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; intrinsic apoptotic signaling pathway in response to oxidative stress [GO:0008631]; iron ion homeostasis [GO:0055072]; liver development [GO:0001889]; locomotory behavior [GO:0007626]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of fibroblast proliferation [GO:0048147]; negative regulation of membrane hyperpolarization [GO:1902631]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway [GO:1902176]; negative regulation of vascular associated smooth muscle cell proliferation [GO:1904706]; neuron development [GO:0048666]; oxygen homeostasis [GO:0032364]; positive regulation of cell migration [GO:0030335]; positive regulation of hydrogen peroxide biosynthetic process [GO:0010729]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; positive regulation of vascular associated smooth muscle cell apoptotic process [GO:1905461]; positive regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching [GO:1905932]; post-embryonic development [GO:0009791]; protein homotetramerization [GO:0051289]; regulation of blood pressure [GO:0008217]; regulation of catalytic activity [GO:0050790]; regulation of mitochondrial membrane potential [GO:0051881]; regulation of transcription by RNA polymerase II [GO:0006357]; release of cytochrome c from mitochondria [GO:0001836]; removal of superoxide radicals [GO:0019430]; respiratory electron transport chain [GO:0022904]; response to activity [GO:0014823]; response to axon injury [GO:0048678]; response to cadmium ion [GO:0046686]; response to cold [GO:0009409]; response to electrical stimulus [GO:0051602]; response to gamma radiation [GO:0010332]; response to hydrogen peroxide [GO:0042542]; response to hyperoxia [GO:0055093]; response to hypoxia [GO:0001666]; response to immobilization stress [GO:0035902]; response to isolation stress [GO:0035900]; response to L-ascorbic acid [GO:0033591]; response to lipopolysaccharide [GO:0032496]; response to magnetism [GO:0071000]; response to manganese ion [GO:0010042]; response to selenium ion [GO:0010269]; response to silicon dioxide [GO:0034021]; response to superoxide [GO:0000303]; response to xenobiotic stimulus [GO:0009410]; response to zinc ion [GO:0010043]; superoxide anion generation [GO:0042554]; superoxide metabolic process [GO:0006801] | 11124296_Observational study of gene-disease association. (HuGE Navigator) 11299047_Observational study of gene-disease association. (HuGE Navigator) 11323405_Observational study of gene-disease association. (HuGE Navigator) 11350569_Observational study of gene-disease association. (HuGE Navigator) 11481695_Observational study of gene-disease association. (HuGE Navigator) 11719088_Observational study of gene-disease association. (HuGE Navigator) 11721640_Review: Contribution of proteomics to the molecular analysis of renal cell carcinoma with an emphasis on manganese superoxide dismutase. 11756571_Cu,Zn-SOD and Mn-SOD are differently regulated by estrogen and progesterone in human endometrial stromal cells 11836586_Observational study of gene-disease association. (HuGE Navigator) 11836586_Polymorphism in MnSOD is associated with age among hispanics with colorectal carcinoma 11837748_Increased expression of manganese superoxide dismutase is associated with that of nitrotyrosine in myopathies with rimmed vacuoles. 11849743_Observational study of gene-disease association. (HuGE Navigator) 11853549_AP-2 down-regulates transcription of the human SOD2 gene via its interaction with Sp1 within the promoter region. 11912921_Transcription regulation of human manganese superoxide dismutase gene. 11912930_Catalytic pathway of manganese superoxide dismutase by direct observation of superoxide. 11977425_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12032830_NF-kappaB-dependent MnSOD expression protects adenocarcinoma cells from TNF-alpha-induced apoptosis 12063011_Observational study of gene-disease association. (HuGE Navigator) 12078513_Manganese superoxide dismutase transgenic mice: characteristics and implications 12127599_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12161520_Nuclear factor-kappa B is required for tumor necrosis factor-alpha-induced manganese superoxide dismutase expression in human endometrial stromal cells. 12239572_PKB-regulated Forkhead transcription factor FOXO3a (also known as FKHR-L1) protects quiescent cells from oxidative stress by directly increasing their quantities of manganese superoxide dismutase (MnSOD) messenger RNA and protein 12447859_Observational study of gene-disease association. (HuGE Navigator) 12447859_Val-Ala polymorphism in Mn-SOD influences neither susceptibility to alcohol-induced liver fibrosis nor alcohol-induced oxidative stress. 12469139_Manganese superoxide dismutase expression within tumor cells is closely related to mode of invasion in human gastric carcinoma 12517793_a new thiol-sensitive mutant form of the human mitochondrial enzyme 12540612_Gene transfer of this enzyme extends islet graft function in a mouse model of autoimmune diabetes. 12551919_This isoform, when expressed in PC-12 cells affects t-butylhydroperoxide-induced apoptosis differentially from its isoenzyme. 12590982_Observational study of gene-disease association. (HuGE Navigator) 12592389_In prostate cancer cells, one of the downstream mediators of the senescence-associated tumor suppression effect of mac25/IGFBP-rP1 is SOD-2. 12624725_Observational study of gene-disease association. (HuGE Navigator) 12627943_Replacement of His-30 with Val in human MnSOD results in a mutant with much decreased catalytic activity and highly susceptible to product inhibition compared to wild-type. 12644569_protein whose expression is deregulated in the epidermis of the elderly 12673575_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12683635_It is indicated that polymorphic mutations of Mn-SOD exist in human normal cells and that the deletions might be obtained in the course of malignant transformation of OSC although decrease in Mn-SOD activity is not involved in the transformation. 12684509_data suggest communication between the proximal promoter region and the TNF-responsive element which involves chromatin remodeling and histone acetylation during the induction process of Mn-SOD in response to TNF. 12700280_MnSOD may be a tumor suppressor gene in human pancreatic cancer 12711112_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12732398_Observational study of gene-disease association. (HuGE Navigator) 12815947_Higher frequencies of SOD2 allele Val and genotype Val/Val and of SOD3 allele Arg and genotype Arg/Arg were established for group DPN+. On this evidence, SOD2 and SOD3 were associated with DPN in DM type 1. 12815947_Observational study of gene-disease association. (HuGE Navigator) 12829021_Overexpression of the human MnSOD transgene in 32D cl 3 cells results in stabilization of the mitochondria and reduction in radiation-, TNF-alpha-, or IL-3 withdrawal-induced damage. 12880680_Observational study of gene-disease association. (HuGE Navigator) 12880680_no significant differences in the genotype, allele, and phenotype frequencies of MnSOD gene polymorphisms between patients with ankylosing spondylitis and controls 12946273_Elevated expression of MnSOD in neuronal ceroid lipofuscinosis. 12948282_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12960753_Observational study of gene-disease association. (HuGE Navigator) 12960753_the influence of a functional polymorphism of the dopamine D3 receptor, and its interaction with a Mn superoxide dismutase (MnSOD) polymorphism, in contributing to tardive dyskinesia in a chronic inpatient population with schizophrenia 12963120_No association of the Ala-9Val MnSOD polymorphism to the development of breast cancer 12963120_Observational study of gene-disease association. (HuGE Navigator) 14503839_Results suggest that the failure of manganese-superoxide dismutase mRNA induction by oxidative stress in peripheral lymphocytes may be involved in the development of gastric cancer. 14515147_Regulation of MnSOD by IL-1 in retinoic acid-differentiated neuroblastoma cells was mediated by the nuclear factor kappaB. 14578853_Cu/Zn-SOD in cytosol and Mn-SOD in mitochondria each are capable of protecting HepG2 cells expressing CYP2E1 against cytotoxicity induced by pro-oxidants. 14611903_The role of manganese superoxide dismutase (Mn SOD) genes polymorphisms in the pathogenesis of systemic lupus erythematosus (SLE). 14638684_His30 and Tyr166 in wild-type Mn-SOD have roles in prolonging the lifetime of the inhibited complex 14643949_Observational study of gene-disease association. (HuGE Navigator) 14687717_CYP1A1 4887A may be a risk factor for the development of reactive arthritis, especially in the presence of Mn SOD 1183T/T 14687717_Observational study of gene-disease association. (HuGE Navigator) 14688256_an active site mutant of human manganese-superoxide dismutase has anti-proliferative functions and demonstrates the signaling role of MnSOD 14704872_Observational study of gene-disease association. (HuGE Navigator) 14729580_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14744747_Observational study of gene-disease association. (HuGE Navigator) 15075214_Increase in hydroxyl radical concentration in the extracellular space of muscles from wild-type mice after the contraction protocol most likely results from degradation of hydrogen peroxide generated by MnSOD activity. 15087454_human SOD2 gene is induced by nucleophosmin and NF-kappaB 15088300_Observational study of gene-disease association. (HuGE Navigator) 15094225_Observational study of gene-disease association. (HuGE Navigator) 15094225_functional polymorphisms in MTP and MnSOD may be involved in determining susceptibility of non-alcoholic steatohepatitis 15131792_Observational study of gene-disease association. (HuGE Navigator) 15166009_Review. Blood vessels express 3 isoforms of superoxide dismutase, 1 of which is manganese SOD in mitochondria. This review will focus mainly on the role of individual SODs in relation to endothelium under normal conditions and in disease states. 15168344_Neoplastic cells in breast carcinomas retain their capability to produce MNSOD, thus are protected from possible cellular damage by reactive oxygen species. MNSOD content varies according to the degree of differentiation of breast carcinoma. 15184255_MnSOD Ala allele polymorphism increases risk of breast cancer in cigarette smokers. 15184255_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15196853_Observational study of gene-disease association. (HuGE Navigator) 15213518_Finds homozygous variant MnSOD genotype is associated with increased lung cancer risk among individuals with zero or 'low' asbestos exposure score and no association among the 'high' AES group. 15213518_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15217492_Observational study of gene-disease association. (HuGE Navigator) 15247771_Observational study of gene-disease association. (HuGE Navigator) 15266664_Observational study of gene-disease association. (HuGE Navigator) 15308628_Vascular endothelial growth factor is coupled to Mn-SOD expression through growth factor-specific reactive oxygen species. 15330761_results indicate that the PKC pathway leading to SOD2 induction proceeds at least in part through NF-kappaB 15331175_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15337840_Although (MnSOD) in DEV cells was significantly increased in acute infection with viral and nutritional stress, in persistent infection and nutritional stress, the expression of the MnSOD was drastically downregulated. 15338334_Cu, Zn- and MnSOD levels were significantly increased in frontal cortex and substantia innominata of the index group in schizophrenia 15345661_Observational study of gene-disease association. (HuGE Navigator) 15386537_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15455371_Observational study of gene-disease association. (HuGE Navigator) 15512788_Iindoleamine 2,3-dioxygenase, manganese superoxide dismutase, and interleukin-1 receptor antagonist protein transferred to isolated NOD mouse donor islets protect islet grafts from diabetogenic T cells. 15512801_Overexpression of manganese superoxide dismutase promotes the survival of prostate cancer cells exposed to hyperthermia 15534883_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15535847_Observational study of gene-disease association. (HuGE Navigator) 15581626_Data show that TNF-alpha increased the level of two antioxidant enzymes, thioredoxin and manganese superoxide dismutase, by an NF-kappaB-dependent mechanism in Ewing sarcoma cells. 15589819_Observational study of gene-disease association. (HuGE Navigator) 15591282_Observational study of gene-disease association. (HuGE Navigator) 15591282_the functional A16V MnSOD polymorphism affects the risk of cardiomyopathy related to iron overload and possibly to other known and unknown risk factors, and may represent an iron toxicity modifier 15598343_Observational study of genotype prevalence. (HuGE Navigator) 15610954_Observational study of gene-disease association. (HuGE Navigator) 15621215_Observational study of gene-disease association. (HuGE Navigator) 15673194_Reduced the oriental medicine-induced cytotoxicity and decreased the number of the oriental medicine-induced apoptotic cells. 15701646_Sod2 can serve as an alternative physiological source of the potent signaling molecule, H2O2 via PTEN oxidation 15705913_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15706661_HIV-1 Tat regulates the SOD2 basal promoter by altering Sp1/Sp3 binding activity 15734083_Observational study of gene-disease association. (HuGE Navigator) 15734485_Observational study of gene-disease association. (HuGE Navigator) 15734485_The association of the manganese superoxide dismutase polymorphisms at -102 C>T and the -9 T>C were not found to be associated with gastric cancer in a Polish case-control study. 15743756_MnSOD protects the redox-sensitive checkpoint which regulates the progression of fibroblasts from G0/G1 to S-phase. 15765450_Observational study of gene-disease association. (HuGE Navigator) 15767364_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15774926_Observational study of gene-disease association. (HuGE Navigator) 15781667_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15809720_hydrogen peroxide overload on p53-independent pathway due to MnSOD overexpression plus buthionine sulfoximine might increase apoptosis frequency without acceleration of apoptotic process of each cell 15817612_Mitochondria-localized MnSOD dramatically reduced the release of cytochrome c to cytosol by proline oxidase in colorectal cells. 15838728_Observational study of gene-disease association. (HuGE Navigator) 15869407_Study findings suggest a novel mechanism by which Tat modulates the repression of the MNSOD gene and establishes a link between HIV infection and liver cancer. 15878096_Observational study of gene-disease association. (HuGE Navigator) 15883815_Observational study of gene-disease association. (HuGE Navigator) 15883815_Polymorphism in the manganese superoxide dismutase gene is associated with breast cancer 15887859_Altered nuclear expression of Mn-SOD parallels, together with changes in other elements of the antioxidant protective system, the loss of differentiation occurring during the progression of thyroid tumors. 15894290_The data show that MnSOD affects sensitivity of cells to Pc 4-PDT-initiated apoptosis, and partly ceramide accumulation, suggesting that the processes are superoxide-mediated. 15908783_Epigenetic silencing of SOD2 in multiple myeloma increases cell proliferation. 15923250_Observational study of gene-disease association. (HuGE Navigator) 15933380_Observational study of gene-disease association. (HuGE Navigator) 15951095_Observational study of gene-disease association. (HuGE Navigator) 16006997_Observational study of gene-disease association. (HuGE Navigator) 16047490_Observational study of gene-disease association. (HuGE Navigator) 16076760_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16084535_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16107721_uncoupling of the electrochemical gradient by increased MnSOD activity gives rise to p53 up-regulation 16148556_Observational study of gene-disease association. (HuGE Navigator) 16157826_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16166634_Protein kinase D-mediated MnSOD expression promotes increased survival of cells upon release of mitochondrial reactive oxygen species 16170370_HIF-1alpha accumulation and VEGF expression could be modulated by the antioxidant enzyme MnSOD. 16179351_the increase in Mn-SOD expression in ovarian cancer is a cellular response to intrinsic ROS stress 16215873_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16281056_Endogenously generated matrix superoxide does not regulate UCP activity and in vivo energy expenditure; transgenic Sod2 demonstrates absence of effect on any UCP activities, and transgenic mice display normal resting metabolic rates 16324912_Observational study of gene-disease association. (HuGE Navigator) 16369462_Data show that manganese-dependent superoxide dismutase levels are much higher in the hippocampi of Alzheimer's diseased brains than in non-diseased control brains. 16423340_Ala-9Val polymorphism does not seem to be a significant predisposing factor for bronchial asthma in the Czech population 16423340_Observational study of gene-disease association. (HuGE Navigator) 16424062_Observational study of gene-disease association. (HuGE Navigator) 16458347_Observational study of gene-disease association. (HuGE Navigator) 16458347_We did not observe an association between the C47T polymorphism in the MnSOD gene and survival 16467073_Observational study of gene-disease association. (HuGE Navigator) 16475114_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16485861_Observational study of gene-disease association. (HuGE Navigator) 16485861_the Ala-9Val polymorphism in the MnSOD gene is not associated with genetic susceptibility in Alzheimer's disease patients 16510607_Observational study of gene-disease association. (HuGE Navigator) 16538174_Observational study of gene-disease association. (HuGE Navigator) 16540901_SODs might be important in the pathogenesis of NP; however, the roles these SOD isoforms play in both normal nasal mucosa and NP require further clarification. 16543247_Observational study of gene-disease association. (HuGE Navigator) 16626843_Observational study of gene-disease association. (HuGE Navigator) 16630148_Observational study of gene-disease association. (HuGE Navigator) 16740634_presence of p53 in a transcription complex of NPM and Sp1 or NF-kappaB at the promoter of the MnSOD gene was verified. p53 interacts with Sp1 to suppress expression of the MnSOD gene. 16769586_May be a genetic factor regulating the cellular redox state in determining the outcome of leukemia chemotherapy 16769586_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16780268_Observational study of gene-disease association. (HuGE Navigator) 16780879_that the localization of Cu/Zn- and Mn-SODs in adrenal tissues reflects the specificity of the adrenal cells that produce the tissue-specific hormones 16807759_Observational study of gene-disease association. (HuGE Navigator) 16807759_The results suggest that Ala/Val polymorphism of the SOD2 gene could be associated with the risk of developing methamphetamine psychosis. 16819819_These studies show that residues at the dimeric interface, such as Tyr169, have significantly less conformational freedom or mobility than do residues at the tetrameric interface, such as Tyr45. 16847469_Observational study of gene-disease association. (HuGE Navigator) 16847469_determination of MnSOD polymorphism and different genotypes of MnSOD associated with its antioxidant enzyme activity in chronic pelvic pain syndrome patients 16859522_Observational study of gene-disease association. (HuGE Navigator) 16868544_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16933053_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16933053_The MnSOD Ala-9Val polymorphism may contribute to an increase in breast cancer risk in the context of high alcohol consumption, however the polymorphism is not an overall risk factor for breast cancer in this primarily premenopausal population. 16945136_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16945136_Polymorphisms in the GPX-1 and MnSOD genes may have a role in development of breast cancer 16956821_Observational study of gene-disease association. (HuGE Navigator) 16956909_Observational study of gene-disease association. (HuGE Navigator) 16966185_Observational study of gene-disease association. (HuGE Navigator) 16966488_Phosphorylation of ERK1/2 led to an activation of NFkB, and increased mRNA levels of the antioxidant enzyme manganese-superoxide dismutase (MnSOD). 16969494_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16969494_the MnSOD Val16Ala variant allele may have a role in preventing development of breast cancer in women who never breast fed 17005595_Mean relative levels of SOD2 & its mRNA were significantly decreased in women > or =38 years, which may account for granulosa-cell changes associated with reproductive aging. 17018785_Observational study of gene-disease association. (HuGE Navigator) 17055157_Observational study of gene-disease association. (HuGE Navigator) 17085785_Binding of NF-kappaB members to the MnSOD gene leads to the induction of MnSOD mRNA and protein levels, leading to subsequent protection against oxidative stress-induced neuronal injury. 17142144_Observational study of gene-disease association. (HuGE Navigator) 17145829_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17149600_Observational study of gene-disease association. (HuGE Navigator) 17171548_Systemic activity of the enzymatic antioxidants (CuZn/SOD, MnSOD, GSH-Px, and CAT) as well as level of lipid peroxidation determined by MDA may not be increased in the course of immune-inflammatory processes associated with chronic idiopathic urticaria. 17186424_No significant association between MnSOD gene polymorphism and the risk of skin cancer. 17186424_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17188257_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17192491_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17192491_Smoking and homozygosity for the MnSOD Val allele is associated with an increased risk of diabetic nephropathy. 17211829_Activation of pattern recognition receptors of the innate immune system results in strong upregulation of SOD2 gene expression suggesting that SOD2 protects macrophages from oxidative stress during microbial infection. 17217237_Observational study of gene-disease association. (HuGE Navigator) 17277236_Observational study of gene-disease association. (HuGE Navigator) 17277236_SNPs involving the GSTP1, MnSOD and GPX2 genes were not associated with Barrett's esophagus or esophageal adenocarcinoma 17290392_An increased risk of pleural mesothelioma was found in those with the Ala/Ala genotypes at codon 16 within MnSOD. 17290392_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17291655_Observational study of gene-disease association. (HuGE Navigator) 17293864_Observational study of gene-disease association. (HuGE Navigator) 17296902_Extracellular SOD, endothelial NOS, and inducible NOS gene polymorphisms do not constitute a risk factor for developing BD (Behcet disease) in Japan. 17296902_Observational study of gene-disease association. (HuGE Navigator) 17299255_Observational study of gene-disease association. (HuGE Navigator) 17299970_Gene polymorphisms of PON1 55 Met/Leu, PON2 148 Ala/Gly and MnSOD 9 Ala/Val seemed to involve in the morbidity of CHD by influencing the plasma activities of PON and MnSOD. 17299970_Observational study of gene-disease association. (HuGE Navigator) 17331249_10T/9T polymorphism in the SOD2 intron 3 results in alternative splicing of exon 4; over-representation of 9T alleles indicates that this polymorphism may be involved in the altered susceptibilities to disorders that are more common in African-Americans 17331249_Observational study of genotype prevalence. (HuGE Navigator) 17336594_Ala-encoding MnSOD allele is represented equally in controls and patients with HCV-related cirrhosis, and it does not significantly influence the risks of liver iron overload, HCC, or death. 17336594_Observational study of gene-disease association. (HuGE Navigator) 17340208_No association between SOD polymorphism and bladder cancer incidence. 17340208_Observational study of gene-disease association. (HuGE Navigator) 17376152_Observational study of gene-disease association. (HuGE Navigator) 17376152_Under a dominant model, family-based association tests showed significant evidence for association of Alzheimer's disease with the first three loci of superoxide dismutase 2 in two candidate gene sets of families. 17400324_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17400324_study of the relationship between genetic polymorphisms of MnSOD, NQO1, GSTM1 and GSTT1 and the susceptibility to drug-induced liver injury (DILI); MnSOD mutant C allele may increase the susceptibility to DILI 17409931_Observational study of gene-disease association. (HuGE Navigator) 17409931_The common Val16Ala MnSOD polymorphism does not confer increased or reduced risk of lung cancer in Chinese in Hong Kong 17449559_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17453961_No major modifying role for the Mn-SOD gene polymorphism in patients with pseudoexfoliation syndrome. 17453961_Observational study of gene-disease association. (HuGE Navigator) 17459574_UDN glycoprotein regulated SOD2 activity and is a potential modulator of inflammatory signal pathways in a lipopolysaccharide-treated colonic cancer cell line. 17465268_Observational study of gene-disease association. (HuGE Navigator) 17465268_mutation of the MnSOD gene may be an important risk factor for prostate cancer 17473980_MnSOD plays a role in regulating tumor cell growth and invasive properties of estrogen-independent metastatic breast cancer cells via hydrogen peroxide. 17491681_Observational study of gene-disease association. (HuGE Navigator) 17548672_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17548864_Observational study of gene-disease association. (HuGE Navigator) 17548864_Polymorphisms of MnSOD and GSTP1 are not associated with chronic alcoholic pancreatitis. 17567676_Observational study of gene-disease association. (HuGE Navigator) 17567676_analysis of superoxide dismutase and catalase polymorphisms in smokers with COPD 17575500_Observational study of gene-disease association. (HuGE Navigator) 17577737_Observational study of gene-disease association. (HuGE Navigator) 17577737_Significant difference in the genotype frequency between healthy subjects and patients with sepsis. 17582511_Observational study of gene-disease association. (HuGE Navigator) 17582511_The distribution of the MnSOD genotypes and alleles was not significantly different between patients and controls. Logistic regression analysis also failed to reveal any association between MnSOD genotypes and TD. 17588204_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17601350_Observational study of gene-disease association. (HuGE Navigator) 17616699_that an O(2)(*-) signaling pathway regulates NAC-induced G(1) arrest by decreasing cyclin D1 protein levels and increasing MnSOD activity 17617122_During neoplastic transformation of cirrhotic liver, an increase in MnSOD activity may occur already during the precancerous phase, making this enzyme a probable malignancy-associated parameter. 17628794_Manganese superoxide dismutase gene polymorphisms is associated with urolithiasis 17628794_Observational study of gene-disease association. (HuGE Navigator) 17634480_Observational study of gene-disease association. (HuGE Navigator) 17646272_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17646272_These results suggest that the Ala variant of SOD2 is associated with moderately increased risk of prostate cancer. 17652337_p50 shows a negative effect on MnSOD induction upon repeated applications of TPA and provide an insight into a cause for the reduction of MnSOD expression during early stages of skin carcinogenesis. 17653087_Overexpression of MnSOD at moderate levels is able to protect cells from TRAIL-induced apoptosis. 17693525_Observational study of gene-disease association. (HuGE Navigator) 17719580_Data suggest that MnSOD may be useful in treating HER2/neu-mediated human breast tumor malignancy. 17822322_Increased activity of superoxide dismutase in advanced stages of head and neck squamous cell carcinoma with locoregional metastases 17879532_Clinical trial of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17922231_Observational study of gene-disease association, gene-gene interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17922231_the combined genotype group of SOD2-01 and GSTP1-01 was an independent predictor of progression free survival and breast cancer specific survival in patients with metastatic breast cancer 17936883_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17936883_study concludes that the Val9Ala MnSOD polymorphism does not influence ovarian cancer risk or survival 17965603_Epigenetic silencing of SOD2 constitutes one mechanism leading to the decreased expression of MnSOD observed in many breast cancers. 17967822_Observational study of gene-disease association. (HuGE Navigator) 17974967_These findings suggest that the association between increased Sod2 activity and poor prognosis in cancer can be attributed to alterations in their migratory and invasive capacity. 18023606_Findings provide further evidence of an association between the Ala-9Val MnSOD polymorphism and HCC occurrence in hepatitis C virus-infected Moroccan patients. 18023606_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18044968_emphasize the role of residue 66 in catalysis and inhibition and provide a structural explanation for differences in catalytic properties between human and certain bacterial forms of MnSOD 18057537_concludes that the VV genotype of the V16A polymorphism of the manganese superoxide dismutase(Mn-SOD) gene was associated with diabetic retinopathy in Caucasians with type 2 diabetes 18095014_Observational study of gene-disease association. (HuGE Navigator) 18095014_in systemic lupus erythematosus patients, MnSOD2 (superoxide dismutase 2)Val/Val genotype was associated with Raynaud's phenomenon and contributed to immunologic manifestations and autoantibody production 18167182_MnSOD was overexpressed in ESCC and its up-regulation in esophageal cancer cells was associated with apoptosis resistance. 18167310_Thus, long-term exposure of human cells to RES results in a highly specific upregulation of MnSOD, and this may be an important mechanism by which it elicits its effects in human cells. 18180754_Meta-analysis of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18180754_Meta-analysis shows a protective effect against antipsychotic-induced tardive dyskinesia for Ala9Val and Val carriers. 18205184_Observational study of gene-disease association. (HuGE Navigator) 18205184_Polymorphisms of the inflammatory pathway genes MPO -G463A and SOD2 Ala16Val are associated with elevated pancreatic cancer risk 18247479_Kinetic mechanism of manganese-containing superoxide dismutase. 18258609_Observational study of gene-disease association. (HuGE Navigator) 18291700_Manganese superoxide dismutase (MnSOD) activity has been used as an index of mitochondrial antioxidant defence in type 2 diabeates. 18296681_MnSOD is not associated with risk of prostate cancer, but MnSOD T to C (Val-9Ala, rs4880) polymorphism modifies associations between risk of clinically aggressive prostate cancer and dietary iron intake 18296681_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18305395_Observational study of gene-disease association. (HuGE Navigator) 18327668_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18373354_Glu162 at the tetrameric interface in human MnSOD supports stability and efficient catalysis and has a significant role in regulating product inhibition 18387669_Observational study of gene-disease association. (HuGE Navigator) 18387669_Polymorphisms of CuZn-SOD, MnSOD, GSTM1 and GSTT1 in the placental tissue were not associated with preeclampsia. 18396798_Observational study of gene-disease association. (HuGE Navigator) 18413745_Overexpression of MnSOD in breast epithelial cells alters stabilization of HIF-1 alpha under hypoxic conditions. 18423055_Observational study of gene-disease association. (HuGE Navigator) 18423055_genotype distribution of the SOD2 in patients with diabetes mellitus can differ from nondiabetic individuals 18434620_Shear stress influences spatial variations in vascul | ENSMUSG00000006818 | Sod2 | 2334.51943 | 1.1080434 | 0.1480143323 | 0.10937719 | 1.815080e+00 | 1.779000e-01 | 9.998360e-01 | No | Yes | 2804.39969 | 368.838317 | 3.008480e+03 | 305.341184 | |
| ENSG00000112425 | 7957 | EPM2A | protein_coding | O95278 | FUNCTION: Plays an important role in preventing glycogen hyperphosphorylation and the formation of insoluble aggregates, via its activity as glycogen phosphatase, and by promoting the ubiquitination of proteins involved in glycogen metabolism via its interaction with the E3 ubiquitin ligase NHLRC1/malin. Shows strong phosphatase activity towards complex carbohydrates in vitro, avoiding glycogen hyperphosphorylation which is associated with reduced branching and formation of insoluble aggregates (PubMed:16901901, PubMed:23922729, PubMed:26231210, PubMed:25538239, PubMed:25544560). Dephosphorylates phosphotyrosine and synthetic substrates, such as para-nitrophenylphosphate (pNPP), and has low activity with phosphoserine and phosphothreonine substrates (in vitro) (PubMed:11001928, PubMed:11220751, PubMed:11739371, PubMed:14532330, PubMed:16971387, PubMed:18617530, PubMed:22036712, PubMed:23922729, PubMed:14722920). Has been shown to dephosphorylate MAPT (By similarity). Forms a complex with NHLRC1/malin and HSP70, which suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system (UPS). Acts as a scaffold protein to facilitate PPP1R3C/PTG ubiquitination by NHLRC1/malin (PubMed:23922729). Also promotes proteasome-independent protein degradation through the macroautophagy pathway (PubMed:20453062). {ECO:0000250|UniProtKB:Q9WUA5, ECO:0000269|PubMed:11001928, ECO:0000269|PubMed:11220751, ECO:0000269|PubMed:11739371, ECO:0000269|PubMed:14532330, ECO:0000269|PubMed:14722920, ECO:0000269|PubMed:16901901, ECO:0000269|PubMed:16971387, ECO:0000269|PubMed:18070875, ECO:0000269|PubMed:18617530, ECO:0000269|PubMed:19036738, ECO:0000269|PubMed:20453062, ECO:0000269|PubMed:22036712, ECO:0000269|PubMed:23624058, ECO:0000269|PubMed:23922729, ECO:0000269|PubMed:25538239, ECO:0000269|PubMed:25544560, ECO:0000269|PubMed:26231210}.; FUNCTION: [Isoform 2]: Does not bind to glycogen (PubMed:18617530). Lacks phosphatase activity and might function as a dominant-negative regulator for the phosphatase activity of isoform 1 and isoform 7 (PubMed:18617530, PubMed:22036712). {ECO:0000269|PubMed:18617530, ECO:0000269|PubMed:22036712}.; FUNCTION: [Isoform 7]: Has phosphatase activity (in vitro). {ECO:0000269|PubMed:22036712}. | 3D-structure;Alternative initiation;Alternative splicing;Autophagy;Carbohydrate metabolism;Cell membrane;Cytoplasm;Disease variant;Endoplasmic reticulum;Epilepsy;Glycogen metabolism;Glycogen storage disease;Hydrolase;Membrane;Neurodegeneration;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome;Ubl conjugation | This gene encodes a dual-specificity phosphatase and may be involved in the regulation of glycogen metabolism. The protein acts on complex carbohydrates to prevent glycogen hyperphosphorylation, thus avoiding the formation of insoluble aggregates. Loss-of-function mutations in this gene have been associated with Lafora disease, a rare, adult-onset recessive neurodegenerative disease, which results in myoclonus epilepsy and usually results in death several years after the onset of symptoms. The disease is characterized by the accumulation of insoluble particles called Lafora bodies, which are derived from glycogen. [provided by RefSeq, Jan 2018]. | hsa:7957; | cytoplasm [GO:0005737]; cytoplasmic side of rough endoplasmic reticulum membrane [GO:0098556]; cytosol [GO:0005829]; dendrite [GO:0030425]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perikaryon [GO:0043204]; plasma membrane [GO:0005886]; carbohydrate binding [GO:0030246]; carbohydrate phosphatase activity [GO:0019203]; glycogen (starch) synthase activity [GO:0004373]; glycogen binding [GO:2001069]; phosphatase activity [GO:0016791]; protein dimerization activity [GO:0046983]; protein homodimerization activity [GO:0042803]; protein serine phosphatase activity [GO:0106306]; protein serine/threonine phosphatase activity [GO:0004722]; protein threonine phosphatase activity [GO:0106307]; protein tyrosine phosphatase activity [GO:0004725]; protein tyrosine/serine/threonine phosphatase activity [GO:0008138]; starch binding [GO:2001070]; autophagosome assembly [GO:0000045]; calcium ion transport [GO:0006816]; carbohydrate phosphorylation [GO:0046835]; dephosphorylation [GO:0016311]; glial cell proliferation [GO:0014009]; glycogen biosynthetic process [GO:0005978]; glycogen metabolic process [GO:0005977]; habituation [GO:0046959]; L-glutamate transmembrane transport [GO:0015813]; mitochondrion organization [GO:0007005]; negative regulation of cell cycle [GO:0045786]; negative regulation of dephosphorylation [GO:0035305]; negative regulation of gene expression [GO:0010629]; negative regulation of peptidyl-serine phosphorylation [GO:0033137]; negative regulation of phosphatase activity [GO:0010923]; peptidyl-tyrosine dephosphorylation [GO:0035335]; phosphorylated carbohydrate dephosphorylation [GO:0046838]; positive regulation of macroautophagy [GO:0016239]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein dephosphorylation [GO:0006470]; regulation of cell growth [GO:0001558]; regulation of glycogen (starch) synthase activity [GO:2000465]; regulation of proteasomal protein catabolic process [GO:0061136]; regulation of protein import into nucleus [GO:0042306]; regulation of protein kinase activity [GO:0045859]; regulation of protein localization to plasma membrane [GO:1903076]; regulation of protein ubiquitination [GO:0031396]; regulation of ubiquitin protein ligase activity [GO:1904666]; Wnt signaling pathway [GO:0016055] | 11883934_Laforin is an active phosphatase; therefore, isoforms targeted to different cellular compartments might dephosphorylate and regulate distinct cellular substrates. 12019207_identification of mutations in EPM2A with phenotypes of 22 patients (14 families) and identification of two subsyndromes 12782127_The EPM2AIP1 gene was identified and characterized in a screen for laforin-interacting proteins with a human brain cDNA library; the specificity of the interaction was confirmed; subcellular colocalization of laforin and EPM2AIP1 protein was demonstrated 12915448_Laforin interacts with HIRPI5. 14643920_Up to 20% cases of LD are not genetically linked to chromosome 6. We report two sisters affected from bioptically diagnosed LD but without evidence of EPM2A mutation 14706656_encodes a 331 amino acid protein that contains an N-terminal carbohydrate-binding domain (CBD) and a C-terminal dual-specificity phosphatase domain. The CBD of laforin targets the protein to Lafora inclusion bodies. 14722920_Six novel mutations were identified, one of which is the first mutation specific to the cytoplasmic laforin isoform, implicating this isoform in disease pathogenesis. 16115820_Laforin is a Glycogen-Synthase-Kinase-3 Ser 9 phosphatase, and therefore capable of inactivating GS through GSK3. 16901901_Laforin does not dephosphorylate GSK3B (beta) in vitro, but possesses the unique ability to utilize a phosphorylated complex carbohydrate as a substrate and this function may be necessary for the maintenance of normal cellular glycogen. 16959610_Inactivation of Epm2a resulted in increased Wnt signaling and tumorigenesis 16971387_results demonstrate a critical role of dimerization in Laforin function and suggest an important new dimension in protein phosphatase function and in molecular pathogenesis of Lafora's disease 17337485_Defects in laforin may lead to increased levels of misfolded and/or target proteins, which may eventually affect the physiological processes of the neuron, and likely to be the primary trigger in the physiopathology of lafora disease. 17509003_study concludse that considerable variability in the age at onset of Lafora disease can occur within families; identical mutations can be associated with the classic adolescent presentation, as well as late-onset cases 17646401_Laforin is conserved in all vertebrates and a small class of protists; it is not found in other organisms. Additionally, laforin is a functional equivalent of the plant phosphatase SEX4, and it may function to dephosphorylate complex carbohydrates. 18029386_Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. 18070875_Laforin acts as a scaffold that allows the E3 ubiquitin ligase malin to ubiquitinate protein targeting to glycogen (PTG). These results suggest an additional mechanism, involving laforin and malin, in regulating glycogen metabolism. 18311786_Results suggest that the altered subcellular localization of mutant proteins of the EPM2A and NHLRC1 genes could be one of the molecular bases of the Lafora disease phenotype. 18617530_EPM2A regulated by alternative splicing plays roles in Lafora progressive myoclonus epilepsy. 19036738_Laforin and malin interact with misfolded proteins and promote their degradation through the ubiquitin-proteasome system. 19171932_phosphorylation of R5/PTG at Ser-8 by AMPK accelerates its laforin/malin-dependent ubiquitination and subsequent proteasomal degradation, which results in a decrease of its glycogenic activity. 19267391_Meta-analysis of gene-disease association. (HuGE Navigator) 19403557_Mutations of EPM2A formed aggregates and elicited endoplasm reticulum stress in neuronal cell. 19529779_laforin and malin play a role protecting cells from ER-stress, likely contributing to the elimination of unfolded proteins 20453062_Laforin regulates autophagy. 20534808_These results suggest that the modification introduced by the laforin-malin complex could affect the subcellular distribution of AMPK beta subunits. 20738377_study described several novel mutations of EPM2A and NHLRC1 and brought additional data to genetic epidemiology of Lafora disease (LD); emphasized the high mutation rate in patients with classical LD as well as the high negativity rate of skin biopsy 21371719_Genetic analysis showed a novel c.659 T>A mutation on exon 3 of the EPM2A gene, in a patient with Lafora's disease. 21652633_Laforin and malin are defective in Lafora disease (LD), a neurodegenerative disorder associated with epileptic seizures 21728993_results of the present study suggest that phosphorylation of laforin-Ser(25) by AMPK provides a mechanism to modulate the interaction between laforin and malin 21887368_laforin monomer is the dominant form of the protein and that it contains phosphatase activity. 21930129_glycogen phosphorylation can be considered a catalytic error and laforin a repair enzyme. 22036712_alternative splicing could possibly be one of the mechanisms by which EPM2A may regulate the cellular functions of the proteins it codes for 22047982_This study identified that EPM2 gene mutations leading to Lafora disease in six Turkish families. 22364389_Studies indicate that laforin directly dephosphorylates glycogen. 22815132_Malin forms a functional complex with laforin. This complex promotes the ubiquitination of proteins involved in glycogen metabolism and misregulation of pathways involved in this process results in Lafora body formation. (Review) 23313408_A new novel mutation of the EPM2A gene is identified that causes Lafora disease. 23904258_This study identified the flexibility of K87A mutated laforin structure, with replacement of acidic amino acid to aliphatic amino acid in functional carbohydrate binding module domain, have more impact in abolishing glycogen binding that favors Lafora disease. 23922729_These results suggest that cysteine 329 is specifically involved in the dimerization process of laforin. 24068615_Polyglucosan body degradation requires a protein assembly that includes laforin and malin. 24770803_Studied the role of conformational changes in human laforin structure due to existing single mutation W32G and prepared double mutation W32G/K87A related to loss of glycogen binding. 25246353_This study suggest that variations in phenotypes of EPM2A-deficient Lafora disease. 25270369_This study identified some Mild Lafora disease have EPM2A mutation. 25538239_novel molecular determinants in the laforin active site that help decipher the mechanism of glucan phosphatase activity. 25544560_The crystal structure of laforin bound to phosphoglucan product, reveals its unique integrated tertiary and quaternary structure. 26102034_Lafora disease proteins laforin and malin negatively regulate the HIPK2-p53 cell death pathway. 26216881_laforin is responsible for glycogen dephosphorylation during exercise and acts during the cytosolic degradation of glycogen 26578817_Laforin-glycan interactions occur with a favourable enthalpic contribution counter-balanced by an unfavourable entropic contribution. 26648032_Laforin prevents the auto-degradation of malin by presenting itself as a substrate. Malin preferentially degrades the phosphatase-inactive laforin monomer. 27092952_rs702304 and rs2235481 within the EPM2A gene were associated with schizophrenia liability. 31758957_Laforin/malin complex interacts physically and co-localizes intracellularly with core components of the PI3KC3 complex (Beclin1, Vps34 and Vps15), and that this interaction is specific and results in the polyubiquitination of these proteins. 34147889_EPM2A in-frame deletion slows neurological decline in Lafora Disease. | ENSMUSG00000055493 | Epm2a | 102.04430 | 1.1109486 | 0.1517920890 | 0.29344535 | 2.705671e-01 | 6.029516e-01 | 9.998360e-01 | No | Yes | 112.77158 | 20.338665 | 1.050371e+02 | 14.772933 | |
| ENSG00000113272 | 54974 | THG1L | protein_coding | Q9NWX6 | FUNCTION: Adds a GMP to the 5'-end of tRNA(His) after transcription and RNase P cleavage. This step is essential for proper recognition of the tRNA and for the fidelity of protein synthesis (Probable). Also functions as a guanyl-nucleotide exchange factor/GEF for the MFN1 and MFN2 mitofusins thereby regulating mitochondrial fusion (PubMed:25008184, PubMed:27307223). By regulating both mitochondrial dynamics and bioenergetic function, it contributes to cell survival following oxidative stress (PubMed:25008184, PubMed:27307223). {ECO:0000269|PubMed:25008184, ECO:0000269|PubMed:27307223, ECO:0000305|PubMed:21059936}. | 3D-structure;Cytoplasm;Disease variant;GTP-binding;Magnesium;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Neurodegeneration;Nucleotide-binding;Nucleotidyltransferase;Reference proteome;Transferase;tRNA processing | The protein encoded by this gene is a mitochondrial protein that is induced by high levels of glucose and is associated with diabetic nephropathy. The encoded protein appears to increase mitochondrial biogenesis, which could lead to renal fibrosis. Another function of this protein is that of a guanyltransferase, adding GMP to the 5' end of tRNA(His). Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2015]. | hsa:54974; | cytosol [GO:0005829]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; transferase complex [GO:1990234]; ATP binding [GO:0005524]; GTP binding [GO:0005525]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; magnesium ion binding [GO:0000287]; nucleotidyltransferase activity [GO:0016779]; tRNA binding [GO:0000049]; tRNA guanylyltransferase activity [GO:0008193]; mitochondrial fusion [GO:0008053]; protein homotetramerization [GO:0051289]; response to oxidative stress [GO:0006979]; stress-induced mitochondrial fusion [GO:1990046]; tRNA modification [GO:0006400]; tRNA processing [GO:0008033] | 15459185_ICF45 is a highly conserved novel protein, which is expressed in a cell cycle-dependent manner and seemed to be involved in cell cycle progression and cell proliferation 18508967_induced in high glucose-1 (IHG-1)which increases in diabetic nephropathy, may enhance the actions of TGF-beta1 and contribute to the development of tubulointerstitial fibrosis 20877624_Observational study of gene-disease association. (HuGE Navigator) 21078997_The catalytic domain of Thg1 shares both a common architecture and a two-metal ion-dependent mechanism with canonical 5'-3' DNA polymerases. 21784897_IHG-1 increases mitochondrial biogenesis by promoting PGC-1alpha-dependent processes, potentially contributing to the pathogenesis of renal fibrosis 22136300_The high-resolution crystal structure of human Thg1 reveals remarkable structural similarity between canonical DNA/RNA polymerases and eukaryotic Thg1. 25008184_IHG-1 is a novel regulator of both mitochondrial dynamics and bioenergetic function and contributes to cell survival following oxidant stress. Increased IHG-1 expression may contribute to pathogenesis of diabetic kidney disease. 27307223_Study proposes that homozygosity for the p.Val55Ala mutation in tRNA-histidine guanylyltransferase 1 like (THG1L) is the cause of the abnormal mitochondrial network in the patient fibroblasts, likely by interfering with THG1L activity towards MFN2. 33682303_Severe epileptic encephalopathy associated with compound heterozygosity of THG1L variants in the Ashkenazi Jewish population. 33758037_Analysis of GTP addition in the reverse (3'-5') direction by human tRNA(His) guanylyltransferase. | ENSMUSG00000011254 | Thg1l | 447.81237 | 0.8900488 | -0.1680437020 | 0.15796920 | 1.124794e+00 | 2.888885e-01 | 9.998360e-01 | No | Yes | 461.19705 | 55.188251 | 5.216771e+02 | 48.498246 | |
| ENSG00000114491 | 7372 | UMPS | protein_coding | P11172 | 3D-structure;Acetylation;Alternative splicing;Decarboxylase;Disease variant;Glycosyltransferase;Lyase;Multifunctional enzyme;Phosphoprotein;Pyrimidine biosynthesis;Reference proteome;Transferase | PATHWAY: Pyrimidine metabolism; UMP biosynthesis via de novo pathway; UMP from orotate: step 1/2.; PATHWAY: Pyrimidine metabolism; UMP biosynthesis via de novo pathway; UMP from orotate: step 2/2. | This gene encodes a uridine 5'-monophosphate synthase. The encoded protein is a bifunctional enzyme that catalyzes the final two steps of the de novo pyrimidine biosynthetic pathway. The first reaction is carried out by the N-terminal enzyme orotate phosphoribosyltransferase which converts orotic acid to orotidine-5'-monophosphate. The terminal reaction is carried out by the C-terminal enzyme OMP decarboxylase which converts orotidine-5'-monophosphate to uridine monophosphate. Defects in this gene are the cause of hereditary orotic aciduria. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Mar 2010]. | hsa:7372; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; orotate phosphoribosyltransferase activity [GO:0004588]; orotidine-5'-phosphate decarboxylase activity [GO:0004590]; 'de novo' pyrimidine nucleobase biosynthetic process [GO:0006207]; 'de novo' UMP biosynthetic process [GO:0044205]; cellular response to xenobiotic stimulus [GO:0071466]; female pregnancy [GO:0007565]; lactation [GO:0007595]; nucleoside metabolic process [GO:0009116]; pyrimidine nucleobase biosynthetic process [GO:0019856]; UDP biosynthetic process [GO:0006225]; UMP biosynthetic process [GO:0006222] | 14562021_expression of OPRT gene and the OPRT/dihydropyrimidine dehydrogenase ratio might be useful as predictive parameters for the efficacy of fluoropyrimidine-based chemotherapy for metastatic colorectal cancer 15999119_thymidylate synthase and orotate phosphoribosyl transferase, but not dihydropyrimidine dehydrogenase, are more highly expressed in prostate cancer than in benign prostatic hyperplasia 16142362_orotate phosphoribosyl transferase has a role in lymph node metastasis of gastric cancer 16328050_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16328050_The investigated SNPs of OPRT may have no major influence on 5-FU sensitivity. 16818689_The OPRT Gly213Ala polymorphism seems to be a useful marker for predicting toxicity to bolus 5-FU chemotherapy. 17237621_Overexpression of the OPRT gene plays an important role in the antiproliferative effect of 5-FU. 17549346_High expression of OPRT is associated with the response to adjuvant chemotherapy in human pancreatic cancer 17607371_data suggest that OPRT is involved in early events of pancreatic and gallbladder carcinogenesis and invasion of hepatocellular carcinomas 17854773_decreased the sensitivities of the cultured tumor cells to 5-FU. These results suggest that the OPRT expression level in tumors is an additional determinant of the efficacy of 5-FU. 18597678_OPRT activity levels in tumor tissue may be prognostic factor for survival in colorectal carcinoma with radical resection and 5-FU chemotherapy;postoperative survival was significantly better in patients with high OPRT activity 18633253_determination of OPRT levels in gastric carcinoma tissue enables to predict the response to S-1-based neoadjuvant/adjuvant chemotherapy 18949394_orotate phosphoribosyltransferase is involved in the invasion and metastasis of colorectal carcinoma 19020740_decreased activity of OPRT plays an important role in the acquired resistance of gastric cancer cells towards 5-FU 19020767_head & neck, gastric, colorectal, breast, lung & pancreatic cancer were examined; findings show mRNA expression & protein level of thymidylate synthase, dihydropyrimidine dehydrogenase & orotate phosphoribosyltransferase differed according to cancer type 19074750_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19082440_Immunohistochemical staining for Orotate phosphoribosyl transferase (OPRT)revealed strong expression of OPRT in prostate cancer cells. There was a significant correlation between OPRT mRNA expression levels and the tumor pathological grade 19307741_High orotate phosphoribosyltransferase gene expression is associated with complete response to chemoradiotherapy in patients with squamous cell carcinoma of the esophagus 19332728_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19562503_[review] It is confirmed that the type I defect in hereditary orotic aciduria is caused by loss of uridine monophosphate (UMP)synthase activity 19724871_patients who developed distant recurrence of rectal cancer were found to have significantly higher intratumoral thymidylate synthase (TS), dihydropyrimidine dehydrogenase (DPD) and orotate phosphoribosyl transferase (OPRT) 20112501_Increased orotate phosphoribosyltransferase expression is associated with bladder cancers. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20527751_novel phosphoribosyltransferase transition states 20647221_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20647710_orotate phosphoribosyl transferase/DPD ratio has a relation to cancer staging and survival rate. 20665215_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21631301_Although no association was detected between UMPS variants and gastrointestinal cancer risk in Caucasians, polymerase chain reaction-RFLP with BsrI digestion and DHPLC set up at 59 degrees C are reliable and cost-effective methods to genotype UMPS. 22249354_The observed mutations, aberrant splicing and downregulation of UMPS represent novel mechanisms for acquired 5-FU resistance in colorectal cancer 22641663_High expression levels of orotate phosphoribosyl transferase and thymidylate synthase in colorectal cancer appear to be significantly involved in metastasis after curative surgery 22931617_OPRT expression in colorectal carcinoma tissues is not correlated with the toxicity of 5-FU, but OPRT expression in the normal tissues can help predict the toxicity associated with 5-FU. 24158442_OPRT transition state analogues identify crucial components of potent inhibitors targeting OPRT enzymes. 28205048_Partial UMPS-deficiency should be included in the differential diagnosis of mild orotic aciduria. 28347333_The UMPS 638 CC genotype might be a candidate biomarker predicting toxicity in patients receiving tegafur-uracil/leucovorin-based preoperative chemoradiation for locally advanced rectal cancer 32382150_LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. | ENSMUSG00000022814 | Umps | 2000.35104 | 0.7609819 | -0.3940659160 | 0.10841605 | 1.319493e+01 | 2.807076e-04 | 2.159080e-01 | No | Yes | 1378.38312 | 203.907986 | 2.039390e+03 | 232.760221 | |
| ENSG00000115041 | 30818 | KCNIP3 | protein_coding | Q9Y2W7 | FUNCTION: Calcium-dependent transcriptional repressor that binds to the DRE element of genes including PDYN and FOS. Affinity for DNA is reduced upon binding to calcium and enhanced by binding to magnesium. Seems to be involved in nociception (By similarity). {ECO:0000250|UniProtKB:Q9QXT8}.; FUNCTION: Regulatory subunit of Kv4/D (Shal)-type voltage-gated rapidly inactivating A-type potassium channels, such as KCND2/Kv4.2 and KCND3/Kv4.3. Modulates channel expression at the cell membrane, gating characteristics, inactivation kinetics and rate of recovery from inactivation in a calcium-dependent and isoform-specific manner. {ECO:0000269|PubMed:10676964, ECO:0000269|PubMed:12829703, ECO:0000269|PubMed:15485870, ECO:0000269|PubMed:16123112, ECO:0000269|PubMed:18957440}.; FUNCTION: May play a role in the regulation of PSEN2 proteolytic processing and apoptosis. Together with PSEN2 involved in modulation of amyloid-beta formation. {ECO:0000269|PubMed:11259376, ECO:0000269|PubMed:11988022, ECO:0000269|PubMed:9771752}. | 3D-structure;Alternative splicing;Apoptosis;Calcium;Cell membrane;Cytoplasm;Endoplasmic reticulum;Golgi apparatus;Ion channel;Ion transport;Isopeptide bond;Lipoprotein;Membrane;Metal-binding;Nucleus;Palmitate;Phosphoprotein;Potassium;Potassium channel;Potassium transport;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Transport;Ubl conjugation;Voltage-gated channel | hsa:30818; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; voltage-gated potassium channel complex [GO:0008076]; calcium ion binding [GO:0005509]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; potassium channel activity [GO:0005267]; potassium channel regulator activity [GO:0015459]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; voltage-gated ion channel activity [GO:0005244]; apoptotic process [GO:0006915]; negative regulation of transcription by RNA polymerase II [GO:0000122]; protein localization to plasma membrane [GO:0072659]; regulation of potassium ion transmembrane transport [GO:1901379]; signal transduction [GO:0007165] | Mouse_homologues 12207970_Calsenilin and presenilin 1 can interact at physiologic levels; calsenilin is a developmentally regulated protein that is expressed primarily in the cerebellum and hippocampus. 12531529_Results demonstrate that DREAM (DRE-antagonizing modulator) is a neural activity-stimulated late gene and suggest its involvement in adaptation to long-lasting neuronal activity. 12648752_Calsenilin is markedly decreased not only in the damaged cortex and CA3 region of hippocampus at 24 h after kainic acid-induced seizure but also in a cell-culture model of seizure-like activity. 12837631_calsenilin can be phosphorylated by casein kinase I and that its phosphorylation can be regulated by intracellular calcium 14534243_Abeta peptide levels are reduced in calsenilin knock-out mice, demonstrating that calsenilin affects presenilin-dependent gamma-cleavage in vivo. Long-term potentiation in the dentate gyrus of hippocampus is enhanced in calsenilin knock-out mice 15363885_KChIP3 transcripts were detected primarily in the layer V and deep layer VI of the cerebral cortex, the hippocampus, and the entire cerebellum. 16621893_Results demonstrate that prodynorphin transcription is regulated by DREAM in a calcium-dependent manner. 16787403_calsenilin and CtBP2 are present in synaptic vesicles and can interact in vivo 17241740_Expression of DREAM in various cell types coupled with the universal role of calcium raise the possibility that these factors may play similar role in other secretory cells. 18095322_A dense plexus of calsenilin-positive dendrites makes several basal contacts at cone pedicles, individual calsenilin-positive bipolar cell contacts five to seven cones, some calsenilin-positive dendrites also contact rod photoreceptors 18201103_Ca2+-induced dimerization of DREAM may partially block the putative DNA-binding site. 18579744_These results identify the PACAP-cAMP-Ca(2+)-DREAM cascade as a new pathway to activate GFAP gene expression during astrocyte differentiation. 19110430_Lack of DREAM protein enhances learning and memory and slows brain aging. 19223600_Male KChIP3 knockout (KO) mice showed significantly enhanced memory 24 hours after training and this study suggest a role for regulation of gene expression by KChIP3/DREAM/calsenilin in consolidation of contextual fear conditioning memories. 19299442_DREAM (downstream regulatory element antagonist modulator) modulates TSHR activity through a direct protein-protein interaction that promotes coupling between the receptor and Galphas. 20205763_Results demonstrate that DREAM plays a novel role in postsynaptic modulation of the NMDA receptor, and contributes to synaptic plasticity and behavioral memory. 21059893_Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. 21070824_Sumoylation regulates the nuclear localization of DREAM in differentiated neurons. 21167062_endogenous BDNF is involved in spinal sensitization following inflammation and blockade of BDNF induction in DREAM transgenic mice underlies the failure to develop spinal sensitization 21486818_In cardiomyocytes, [Ca2+]i-activated calmodulin kinase II (CaMKII) activates downstreamregulatory element (DRE) binding transcription factor DREAM, which consequently suppresses the expression of L-type calcium channel 1C-subunit gene (Cacna1c). 22451650_Restoring CANT1 levels in neuroblastoma clones recovered the phenotype, thus confirming a key role of CANT1, and of the regulation of its gene by DREAM, in the control of protein synthesis and degradation 23300953_GCM1-directed villous trophoblast differentiation is repressed by DREAM 23524266_Expression of DREAM in neuroblastoma cells enhances cisplatin mediated caspase-3 activity. 24248121_These results provide the first evidence that DR/C/K3 plays a timing-dependent role in estradiol regulation of learning, memory, and plasticity. 24366545_DREAM is a major master switch transcription factor. 24487321_studies define the critical opposing functions of DREAM and USF1 in inhibiting and inducing A20 expression 24857398_The protein DREAM decreases development of L-DOPA-induced dyskinesia in mice and reduces L-DOPA-induced expression of FosB, phosphoacetylated histone H3, and dynorphin-B in the striatum 25228688_NS5806 directly interacts with KChIP3 and modulates the interactions between this calcium-binding protein and the T1 domain of the Kv4.3 channels through reorientation of helix 10 on KChIP3 26020793_Results presented in this study show for the first time that down-regulation of DREAM promotes the degeneration of RGCs, amacrine cells, and bipolar cells. 26108881_The association of calcium-bound calmodulin (CaM) with DREAM is mediated by a short amphipathic amino acid sequence located between residues 29 and 44 on DREAM. 26896746_Genome-wide analysis of trigeminal neurons in daDREAM transgenic mice identified cathepsin L and the monoglyceride lipase as two new DREAM transcriptional targets related to pain. 26901070_Terbium(III) binding preserves the folding state of DREAM with minimal changes to overall structure of DREAM. 26928278_DREAM plays an important role in structural plasticity in the hippocampus. 27009418_association of PS1 carboxyl peptide (residues 445-467, HL9) with DREAM is calcium dependent and stabilized by a cluster of three aromatic residues: F462 and F465 from PS1 and F252 from DREAM. 27903531_Our findings demonstrate for the first time that DREAM plays an important role in thrombosis and hemostasis in mice. 29523177_DREAM inhibition markedly improves ATF6 processing in the hippocampus and that it might contribute to a delay in memory decline in Huntington mice. 29730765_calsenilin controls the activity of neuronal RyRs. 30399317_Pb(2+) binds to EF-hands in apo-DREAM (downstream regulatory element antagonist modulator) with a lower equilibrium dissociation constant. Based on the Trp169 emission and CD spectra, Pb(2+) association triggers changes in the protein secondary and tertiary structures that are analogous to those previously observed for Ca(2+)-bound protein. 31046050_These results suggest that DREAM and probably other members of the neuronal calcium sensor family bind Cd2+ with an affinity that is superior to that for Ca2+ and the interactions between toxic Cd2+ and DREAM and other neuronal calcium sensors provide novel insight into the molecular mechanism of Cd2+ neurotoxicity. 31329447_Proteomic Profile of a Chronic Binge Ethanol Exposure Model. | ENSMUSG00000079056 | Kcnip3 | 203.43703 | 1.0165994 | 0.0237512184 | 0.23091869 | 1.102809e-02 | 9.163641e-01 | 9.998360e-01 | No | Yes | 198.11573 | 29.022205 | 1.802194e+02 | 22.146159 | ||
| ENSG00000115568 | 7701 | ZNF142 | protein_coding | P52746 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000305}. | DNA-binding;Disease variant;Isopeptide bond;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene belongs to the Kruppel family of C2H2-type zinc finger proteins. It contains 31 C2H2-type zinc fingers and may be involved in transcriptional regulation. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jan 2013]. | hsa:7701; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000026135 | Zfp142 | 2863.69667 | 0.9907055 | -0.0134717715 | 0.12554497 | 1.155706e-02 | 9.143893e-01 | 9.998360e-01 | No | Yes | 2747.50031 | 269.688912 | 2.552567e+03 | 193.944334 | |
| ENSG00000116786 | 23207 | PLEKHM2 | protein_coding | Q8IWE5 | FUNCTION: Plays a role in lysosomes movement and localization at the cell periphery acting as an effector of ARL8B. Required for ARL8B to exert its effects on lysosome location, recruits kinesin-1 to lysosomes and hence direct their movement toward microtubule plus ends. Binding to ARL8B provides a link from lysosomal membranes to plus-end-directed motility (PubMed:28325809, PubMed:22172677, PubMed:25898167, PubMed:24088571). Critical factor involved in NK cell-mediated cytotoxicity. Drives the polarization of cytolytic granules and microtubule-organizing centers (MTOCs) toward the immune synapse between effector NK lymphocytes and target cells (PubMed:24088571). Required for maintenance of the Golgi apparatus organization (PubMed:22172677). May play a role in membrane tubulation (PubMed:15905402). {ECO:0000269|PubMed:15905402, ECO:0000269|PubMed:22172677, ECO:0000269|PubMed:24088571, ECO:0000269|PubMed:25898167, ECO:0000269|PubMed:28325809}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Lysosome;Membrane;Phosphoprotein;Reference proteome | This gene encodes a protein that binds the plus-end directed microtubule motor protein kinesin, together with the lysosomal GTPase Arl8, and is required for lysosomes to distribute away from the microtubule-organizing center. The encoded protein belongs to the multisubunit BLOC-one-related complex that regulates lysosome positioning. It binds a Salmonella effector protein called Salmonella induced filament A and is a critical host determinant in Salmonella pathogenesis. It has a domain architecture consisting of an N-terminal RPIP8, UNC-14, and NESCA (RUN) domain that binds kinesin-1 as well as the lysosomal GTPase Arl8, and a C-terminal pleckstrin homology domain that binds the Salmonella induced filament A effector protein. Naturally occurring mutations in this gene lead to abnormal localization of lysosomes, impaired autophagy flux and are associated with recessive dilated cardiomyopathy and left ventricular noncompaction. [provided by RefSeq, Feb 2017]. | hsa:23207; | endosome membrane [GO:0010008]; lysosomal membrane [GO:0005765]; kinesin binding [GO:0019894]; Golgi organization [GO:0007030]; lysosome localization [GO:0032418]; natural killer cell mediated cytotoxicity [GO:0042267]; positive regulation of membrane tubulation [GO:1903527]; regulation of protein localization [GO:0032880] | 15905402_A dynamic process of kinesin recruitment in Salmonella-infected cells is down-regulated by the SifA-mediated recruitment of SKIP on membranes [SKIP] 26464484_The association of PLEKHM2 mutation with dilated cardiomyopathy and left ventricular noncompaction supports the importance of autophagy for normal cardiac function. | ENSMUSG00000028917 | Plekhm2 | 4907.01224 | 0.9993513 | -0.0009361918 | 0.10996942 | 7.483845e-05 | 9.930976e-01 | 9.998360e-01 | No | Yes | 5807.52865 | 776.585970 | 5.361590e+03 | 554.044244 | |
| ENSG00000116793 | 10745 | PHTF1 | protein_coding | Q9UMS5 | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:10745; | cis-Golgi network [GO:0005801]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021] | 18305142_Observational study of gene-disease association. (HuGE Navigator) 20089178_Results supports the rs6679677 (PHTF1-PTPN22) SNP as a susceptibility factor for type 1 diabetes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000058388 | Phtf1 | 411.93021 | 1.0517459 | 0.0727862207 | 0.16913812 | 1.882136e-01 | 6.644076e-01 | 9.998360e-01 | No | Yes | 501.49268 | 66.597929 | 4.758951e+02 | 48.948955 | |||
| ENSG00000116922 | 54955 | C1orf109 | protein_coding | Q9NX04 | FUNCTION: May promote cancer cell proliferation by controlling the G1 to S phase transition. {ECO:0000269|PubMed:22548824}. | Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | hsa:54955; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634] | 22548824_our findings suggest that C1ORF109 may be the downstream target of protein kinase CK2 and involved in the regulation of cancer cell proliferation. 35354024_Labeling of heterochronic ribosomes reveals C1ORF109 and SPATA5 control a late step in human ribosome assembly. | ENSMUSG00000044730 | 9930104L06Rik | 941.24922 | 1.1626720 | 0.2174441580 | 0.14682889 | 2.159989e+00 | 1.416457e-01 | 9.998360e-01 | No | Yes | 930.54807 | 146.973288 | 9.213728e+02 | 112.390365 | ||
| ENSG00000117151 | 1486 | CTBS | protein_coding | Q01459 | FUNCTION: Involved in the degradation of asparagine-linked glycoproteins. Hydrolyze of N-acetyl-beta-D-glucosamine (1-4)N-acetylglucosamine chitobiose core from the reducing end of the bond, it requires prior cleavage by glycosylasparaginase. | Glycoprotein;Glycosidase;Hydrolase;Lysosome;Reference proteome;Signal | Chitobiase is a lysosomal glycosidase involved in degradation of asparagine-linked oligosaccharides on glycoproteins (Aronson and Kuranda, 1989 [PubMed 2531691]).[supplied by OMIM, Nov 2010]. | hsa:1486; | extracellular space [GO:0005615]; lysosome [GO:0005764]; chitin binding [GO:0008061]; chitinase activity [GO:0004568]; chitin catabolic process [GO:0006032]; oligosaccharide catabolic process [GO:0009313] | 16794344_biochemical behavior of di-N-acetylchitobiase indicates it has three subsites, -2, -1, +1, in which the reducing-end trimer of any sized chitooligosaccharide is bound. The +1 site is specific for an alpha-anomer. 26768631_Data indicate that infliximab changes the concentration of hexosaminidase (N-acetyl-beta-glucosaminidase; HEX) activity depending on the drug dose and time of administration. | ENSMUSG00000028189 | Ctbs | 62.54021 | 1.2956412 | 0.3736662768 | 0.38243739 | 1.001154e+00 | 3.170314e-01 | 9.998360e-01 | No | Yes | 72.31296 | 13.377212 | 5.838306e+01 | 8.597120 | |
| ENSG00000117245 | 57576 | KIF17 | protein_coding | Q9P2E2 | FUNCTION: Dendrite-specific motor protein which, in association with the Apba1-containing complex (LIN-10-LIN-2-LIN-7 complex), transports vesicles containing N-methyl-D-aspartate (NMDA) receptor subunit NR2B along microtubules. {ECO:0000250|UniProtKB:Q99PW8}. | ATP-binding;Alternative splicing;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Microtubule;Motor protein;Nucleotide-binding;Protein transport;Reference proteome;Transport | hsa:57576; | axoneme [GO:0005930]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; cytosol [GO:0005829]; dendrite cytoplasm [GO:0032839]; kinesin complex [GO:0005871]; microtubule [GO:0005874]; microtubule organizing center [GO:0005815]; neuron projection [GO:0043005]; periciliary membrane compartment [GO:1990075]; photoreceptor connecting cilium [GO:0032391]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; microtubule binding [GO:0008017]; microtubule motor activity [GO:0003777]; plus-end-directed microtubule motor activity [GO:0008574]; anterograde dendritic transport of neurotransmitter receptor complex [GO:0098971]; cell projection organization [GO:0030030]; microtubule-based movement [GO:0007018]; protein transport [GO:0015031]; vesicle-mediated transport [GO:0016192] | 14673085_KIF17b serves as a molecular motor component of a TB-RBP-mouse ribonucleoprotein complex transporting a group of specific CREM-regulated mRNAs. 19679349_The intense placental expression of KIFC1 in syncytiotrophoblast and KIF17 in vascular endothelium suggests that both proteins might be important in a cargo-transport system. KIFC1 and KIF17 expression are increased of both in preeclamptia and diabetes. 20378615_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20530208_Data show that the homodimeric kinesin-2 motor KIF17 is kept in an inactive state in the absence of cargo, and define two molecular mechanisms that contribute to autoinhibition of KIF17. 20646681_This study suggested that disruption of KIF17, although rare, could result in a schizophrenia phenotype and emphasize the possible involvement of rare de novo mutations in this disorder. 20696710_Depletion of KIF17 from cells growing in three-dimensional matrices results in aberrant epithelial cysts that fail to generate a single central lumen and to polarize apical markers. 24072717_although EB1 and KIF17-Tail may coordinate KIF17 catalytic activity, our data reveal a novel and direct role for KIF17 in regulating MT dynamics. 26421900_Expression of KIF17 in schizophrenic postmortem brains was significantly lower than controls. Both genotypic distribution and allelic frequency of rs2296225 polymorphism were significantly different between the chronic schizophrenia subjects and controls. 26759174_KIF17 can modify RhoA-GTPase signaling to influence junctional actin and the stability of the apical junctional complex of epithelial cells. 26823018_The first evidence of an interaction between septins and a nonmitotic kinesin is provided and it is suggested that SEPT9 modulates the interactions of KIF17 with membrane cargo. 28077622_In mammalian cells, KIF17 is dispensable for ciliogenesis and IFT-B trafficking but requires IFT-B, as well as its NLS, for its ciliary entry across the permeability barrier located at the ciliary base. 28761002_The rate of transport is set by an equilibrium between a faster state, where only kinesin family member 17 protein (KIF17) motors move the train, and a slower state, where at least one kinesin family member 3A/B protein (KIF3AB) motor on the train remains active in transport. 33922911_Biallelic Variants in KIF17 Associated with Microphthalmia and Coloboma Spectrum. | ENSMUSG00000028758 | Kif17 | 359.79420 | 0.9042094 | -0.1452711795 | 0.17253516 | 7.199721e-01 | 3.961530e-01 | 9.998360e-01 | No | Yes | 346.41141 | 46.692035 | 3.650660e+02 | 39.151339 | ||
| ENSG00000117859 | 114883 | OSBPL9 | protein_coding | Q96SU4 | Acetylation;Alternative splicing;Endosome;Golgi apparatus;Lipid transport;Lipid-binding;Membrane;Phosphoprotein;Reference proteome;Transport | This gene encodes a member of the oxysterol-binding protein (OSBP) family, a group of intracellular lipid receptors. Most members contain an N-terminal pleckstrin homology domain and a highly conserved C-terminal OSBP-like sterol-binding domain, although some members contain only the sterol-binding domain. This family member functions as a cholesterol transfer protein that regulates Golgi structure and function. Multiple transcript variants, most of which encode distinct isoforms, have been identified. Related pseudogenes have been identified on chromosomes 3, 11 and 12. [provided by RefSeq, Jul 2010]. | hsa:114883; | cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; late endosome membrane [GO:0031902]; membrane [GO:0016020]; sterol binding [GO:0032934]; sterol transporter activity [GO:0015248]; bile acid biosynthetic process [GO:0006699] | 16962287_Furthermore, mammalian target of rapamycin was implicated in ORP9L phosphorylation in HEK293 cells. These studies identify ORP9 as a PDK-2 substrate and negative regulator of Akt phosphorylation at the PDK-2 site. 19377067_MicroRNA-125a-5p may partly provide post-transcriptional regulation of the proinflammatory response, lipid uptake, and expression of ORP9 in oxLDL-stimulated monocyte/macrophages. 19413330_Includes data showing N-alpha terminal acetylation of this protein (OSBL9_HUMAN), which begins with nASIMEGPLSK following cleavage of the initiating Met-1 residue. 19554302_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20599956_The results identify ORP11 as an OSBP homologue distributing at the Golgi-LE interface and define the ORP9-ORP11 dimer as a functional unit that may act as an intracellular lipid sensor or transporter. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24190612_T allele of rs768529 may be a risk factor for the formation of the carotid vulnerable plaque in Chinese Hunan Han population 25255026_These studies identify ORP9 as a dual sterol/PI-4P binding protein that could regulate PI-4P in the Golgi apparatus. | ENSMUSG00000028559 | Osbpl9 | 1015.69769 | 0.8607857 | -0.2162740213 | 0.14198369 | 2.242485e+00 | 1.342651e-01 | 9.998360e-01 | No | Yes | 843.61600 | 167.975658 | 1.058276e+03 | 162.264227 | ||
| ENSG00000118217 | 22926 | ATF6 | protein_coding | P18850 | FUNCTION: [Cyclic AMP-dependent transcription factor ATF-6 alpha]: Precursor of the transcription factor form (Processed cyclic AMP-dependent transcription factor ATF-6 alpha), which is embedded in the endoplasmic reticulum membrane (PubMed:10564271, PubMed:11158310, PubMed:11779464). Endoplasmic reticulum stress promotes processing of this form, releasing the transcription factor form that translocates into the nucleus, where it activates transcription of genes involved in the unfolded protein response (UPR) (PubMed:10564271, PubMed:11158310, PubMed:11779464). {ECO:0000269|PubMed:10564271, ECO:0000269|PubMed:11158310, ECO:0000269|PubMed:11779464}.; FUNCTION: [Processed cyclic AMP-dependent transcription factor ATF-6 alpha]: Transcription factor that initiates the unfolded protein response (UPR) during endoplasmic reticulum stress by activating transcription of genes involved in the UPR (PubMed:10564271, PubMed:11163209, PubMed:11158310, PubMed:11779464). Binds DNA on the 5'-CCAC[GA]-3'half of the ER stress response element (ERSE) (5'-CCAAT-N(9)-CCAC[GA]-3') and of ERSE II (5'-ATTGG-N-CCACG-3') (PubMed:10564271, PubMed:11158310, PubMed:11779464). Binding to ERSE requires binding of NF-Y to ERSE. Could also be involved in activation of transcription by the serum response factor (PubMed:10564271, PubMed:11158310, PubMed:11779464). May play a role in foveal development and cone function in the retina (PubMed:26029869). {ECO:0000269|PubMed:10564271, ECO:0000269|PubMed:11158310, ECO:0000269|PubMed:11163209, ECO:0000269|PubMed:11779464, ECO:0000269|PubMed:26029869}. | Activator;DNA-binding;Disease variant;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Isopeptide bond;Membrane;Nucleus;Reference proteome;Signal-anchor;Transcription;Transcription regulation;Transmembrane;Transmembrane helix;Ubl conjugation;Unfolded protein response | This gene encodes a transcription factor that activates target genes for the unfolded protein response (UPR) during endoplasmic reticulum (ER) stress. Although it is a transcription factor, this protein is unusual in that it is synthesized as a transmembrane protein that is embedded in the ER. It functions as an ER stress sensor/transducer, and following ER stress-induced proteolysis, it functions as a nuclear transcription factor via a cis-acting ER stress response element (ERSE) that is present in the promoters of genes encoding ER chaperones. This protein has been identified as a survival factor for quiescent but not proliferative squamous carcinoma cells. There have been conflicting reports about the association of polymorphisms in this gene with diabetes in different populations, but another polymorphism has been associated with increased plasma cholesterol levels. This gene is also thought to be a potential therapeutic target for cystic fibrosis. [provided by RefSeq, Aug 2011]. | hsa:22926; | chromatin [GO:0000785]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of endoplasmic reticulum membrane [GO:0030176]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II transcription regulator complex [GO:0090575]; cAMP response element binding [GO:0035497]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; ubiquitin protein ligase binding [GO:0031625]; ATF6-mediated unfolded protein response [GO:0036500]; endoplasmic reticulum unfolded protein response [GO:0030968]; eye development [GO:0001654]; positive regulation of apoptotic process [GO:0043065]; positive regulation of ATF6-mediated unfolded protein response [GO:1903893]; positive regulation of autophagy [GO:0010508]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress [GO:1990440]; protein folding [GO:0006457]; regulation of transcription by RNA polymerase II [GO:0006357]; signal transduction [GO:0007165]; visual perception [GO:0007601] | 11805088_Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP 11821395_luminal domain senses endoplasmic reticulum stress and causes translocation of ATF6 from endoplasmic reticulum to Golgi apparatus 11909875_coordination of transcription and degradation by a domain shared with the viral transcription factor VP16 12014989_Distinct roles in transcription during the mammalian unfolded protein response 12076252_p38MAPK is activated by phosphorylated ATF6 and induces HSPA5 binding 12097557_stimulated by HCV replicons 12713871_The endoplasmic reticulum stress pathway mediated by ATF6 and by IRE1-XBP1 systems seems essential for the transformation-associated expression of the grp78 gene in hepatocarcinogenesis 12782636_the transport of ATF6 from the ER to the Golgi apparatus and that from the Golgi apparatus to the nucleus are distinct steps 14765107_ATF6 antagonizes SREBP2 to regulate the homeostasis of lipid and glucose 15063770_ATF6 and Ire1p signaling do not define the magnitude of UPR-dependent mRNA increases, even though they may be necessary for gene activation. 15299016_the bulky ATF6 luminal domain blocks its site-2 protease cleavage 15598891_ATF6 and XBP1 have roles in activation of endoplasmic reticulum stress-responsive cis-acting elements 16505252_Observational study of gene-disease association. (HuGE Navigator) 17092596_Our results clearly establish HBx as an inducer of UPR and the activator of the ATF6 and IRE1-XBP1 pathways of UPR. 17101776_Owing to the presence of intra- and intermolecular disulfide bridges formed between the two conserved cysteine residues in the luminal domain, ATF6 occurs in unstressed endoplasmic reticulum in monomer, dimer, and oligomer forms. 17327457_A study evaluting 64 single nucleotide polymorphisms (SNPs) spanning >213 kb in 95 people for their role in the development of type 2 diabetes and the prediabetic state is reported. 17327457_Observational study of gene-disease association. (HuGE Navigator) 17440018_Observational study of gene-disease association. (HuGE Navigator) 17440018_one or more variants in ATF6 are associated with disturbed glucose homeostasis and type 2 diabetes 17442311_Shorter ATF6alpha message lacks exon 7 and may have a regulatory role in the Unfolded protein response. 17522056_the relative levels of ATF6 alpha and -beta, may contribute to regulating the strength and duration of ATF6-dependent ERSR gene induction and cell viability 17686766_NUCB1 is the first-identified, Golgi-localized negative feedback regulator in the ATF6-mediated branch of the UPR 18022401_The aim was to study the activation of ATF6 and Grp78 in transfected human epithelial cells expressing the DeltaF508-CFTR protein. 18635891_Compared with COPD and donor lungs, protein levels of ER stress mediators, such as ATF-6 and ATF-4 and the apoptosis-inductor CHOP as well as XBP-1, were significantly elevated in lung homogenates and AECIIs of IPF lungs 18650380_ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo 18840095_Transcriptional induction of the human asparagine synthetase gene during the unfolded protein response does not require the ATF6 19122331_pXBP1(U) functions as a negative regulator of the unfolded protein response-specific transcription factors ATF6 and pXBP1(S). 19304306_These findings suggest that 8ab could modulate the unfolded protein response by activating ATF6 to facilitate protein folding and processing. 19420237_ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. 19667116_Observational study of gene-disease association. (HuGE Navigator) 19667116_The ATF6-Met[67]Val substitution is associated with increased plasma cholesterol levels. 19693772_unglycosylated ATF6beta may directly facilitate the expression of ERSR genes by losing its repressor function to ATF6alpha 19722195_Data show that GRP78 was upregulated following treatment with NAC or PEN, and both the ATF6 protein and XBP1 mRNA were processed, which facilitates the expression of C/EBP homologous protein CHOP. 19723703_Mechanistically, regulation of ER-stress-induced apoptosis by CerS6/C(16)-ceramide was linked to the activation of a specific arm, ATF6/CHOP, of the unfolded protein response pathway. 19822759_Using an in vitro budding reaction that recapitulates the ER-stress induced transport of ATF6, we show that no cytoplasmic proteins other than COPII are necessary for transport. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20219975_Studies show that ATF6, rather than being a soluble nuclear protein, was an active transcription factor and synthesized as a transmembrane protein embedded in the ER. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20732420_The ATF6alpha/Ras homolog enriched in brain (Rheb) pathway is altered in Huntington's disease, as the decrease in ATF6alpha processing is accompanied by a decrease in the accumulation of Rheb. 20800603_Observational study of gene-disease association. (HuGE Navigator) 21106748_The viral glycoprotein precursor (GPC)was responsible for the induction of ATF6 regulated branch of the host cell's unfolded protein response. 21211013_these data suggests common SNPs within DUSP12-ATF6 locus may not play a major role in glucose metabolism in the Chinese. 21385877_early DENV-2 infection triggers and then suppresses PERK-mediated eIF2alpha phosphorylation and that in mid and late DENV-2 infection, the IRE1-XBP1 and ATF6 pathways are activated, respectively 21521793_propose that a sensing mechanism operates within the lipid bilayer to trigger the selective activation of ATF6 21841196_Mcl-1 is a determinant of cell fate, and ATF6 mediates apoptosis via specific suppression of Mcl-1 through up-regulation of WBP1 21976666_activation of the ATF6 pathway of the UPR limits ATZ-dependent cell toxicity by selectively promoting ER-associated degradation of ATZ 22013072_Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network 22099811_activating transcription factor 6 single polymorphism has a role in increased body mass index after kidney transplantation 22102412_BMP2 induces osteoblast differentiation through Runx2-dependent ATF6 expression, which directly regulates Oc transcription. 22146569_Increased expression of Endoplasmic Reticulum (ER) stress markers, GRP78, ATF6 and CHOP, in endometrioid endometrial carcinomas suggests a role for ER stress in endometrial cancer. 22577136_ATF6alpha functions at least in part by recruiting to the endoplasmic reticulum stress response enhancer elements of ER stress response genes a collection of RNA polymerase II coregulatory complexes. 22872700_ATF6 activation is significantly higher for the CASPR2-D1129H compared with the wild-type protein. 22917505_one of three endoplasmic reticulum transmembrane protein sensors that signals the unfolded protein response [review] 22956602_This study indicates that selectively activating ATF6 or PERK prevents mutant rhodopsin from accumulating in retinal cells. 23011799_Data show that transfection of ORMDL3 in bronchial epithelial cells induced expression of MMP-9, ADAM-8, CCL-20, IL-8, CXCL-10, CXCL-11, oligoadenylate synthetases (OAS) genes, and selectively activated activating transcription factor 6 (ATF6). 23037953_results demonstrate that ER stress-related proteins, particularly ATF6 and its downstream molecule CHOP, are involved in ox-LDL-induced cholesterol accumulation and apoptosis in macrophages 23864652_Mediator subunit MED25 plays a critical role in this process and identify a MED25 domain that serves as a docking site on Mediator for the ATF6alpha transcription activation domain. 23924739_these results indicate that ATF6alpha can regulate de novo cholesterol synthesis through stimulation of cholesterogenic gene expression. 24043630_ATF6 represents a novel type of ERAD-Lm substrate requiring SEL1L for degradation despite its transmembrane nature. 24177270_Tick-borne encephalitis virus infection activates the IRE1 pathway and triggers expression of cleaved transcription factor 6 (ATF6) which suggest activation of ATF6 pathway. 24240056_Activation of the ATF6 pathways of the UPR is sustained along with PERK signaling in melanoma cells subjected to pharmacological ER stress, and that this plays an important role in protecting melanoma cells from ER stressinduced apoptosis. 24269637_ATF6 binds to the promoter of XBP1 and enhances the XBP1S expression in OA cartilage. 24302549_ATF6 mRNA expression was significantly decreased as the disease progressed 24636989_PDIA5 is a key regulator ATF6alpha-mediated cellular functions in cancer. 24664756_These studies indicate that activation of either IRE1, ATF6, or PERK prevents mutant rhodopsin from accumulating in the cells. 24726443_This study showed that the two compounds, polyP and isoquercitrin, have a co-enhancing effect on bone mineral formation and in turn might be of potential therapeutic value for prevention/treatment of osteoporosis. 25135476_Data shows that mutation of putative p38 MAPK phosphorylation sites in ATF6 suppresses its transcriptional induction of DAPK1. 25302688_The genetic variation in ATF6 is associated with pre-diabetes and has interactive effects with BMI on pre-diabetes in the Chinese Han population. 25444553_ATF6 preferentially reduces the secretion and extracellular aggregation of destabilized, aggregation-prone variants of the amyloidogenic protein TTR, as compared to stable WT TTR and nonamyloidogenic TTR variants. 25450523_our data demonstrate that CiC expression is activated during ER stress through the binding of ATF6alpha and XBP1 to an UPRE element located in the proximal promoter of Cic gene. 25593314_Data show that silver nanoparticles induce activating transcription factor-6 (ATF-6) degradation, leading to activation of the NLRP-3 inflammasome and pyroptosis. 25675914_protein expression were significantly higher in the placentas of women with early and late SPE than in the control women, whereas there were no differences in ATF6 and Ire1 mRNA and protein. 25754093_Defective podocyte insulin signaling through p85-XBP1 promotes ATF6-dependent maladaptive ER-stress response in diabetic nephropathy. 25976933_These results confirm that HIV infection activates stress-response components and that antiretroviral therapy contributes to changes in the unfolded protein response activation profile. 26029869_A crucial and unexpected role for ATF6A in human foveal development and cone function. 26063662_autosomal recessive achromatopsia is caused by a frameshift mutation in ATF6 in this Pakistani family 26261584_Endoplasmic reticulum stress related factor ATF6 and caspase-12 trigger apoptosis in neonatal hypoxic-ischemic encephalopathy. 26707144_We found that HCS identified compounds which inhibited ATF6 nuclear translocation with high specificity, as confirmed by the luciferase reporter assay and western blot analysis 26752648_results identify a role for DREAM silencing in the activation of ATF6 signaling, which promotes early neuroprotection in HD. 26925648_Genetic Variations in ATF6 (rs2070150) is not Associated with Hepatocellular Carcinoma in Thai Patients with Hepatitis B Virus Infection. 27085326_Authors observed the cleavage of ATF6, the phosphorylation of MRLC, and the expression of death-associated protein kinase (DAPK1) by western blotting; the transcription of DAPK1 by RT-PCR; and the subcellular localization of ATF6 and mAtg9 by immunofluorescence. 27117871_This review shown that the achromatopsia arising from genetic mutations in Activating Transcription Factor 6 (ATF6). 27461470_Activating transcription factor 6 (ATF6alpha) pathway and ER-associated protein degradation (ERAD) are elevated in salivary glands of Sjogren's syndrome patients (SS). 27563820_Findings indicate a central role for activating transcription factor 6 (ATF6alpha) in the establishment of morphological features of senescence in normal primary fibroblasts. 27590344_A transcription factor complex consisting of ATF6 (an endoplasmic reticulum-resident factor) and C/EBP-beta is required for the IFN-gamma-induced expression of DAPK1 IFN-gamma-induced proteolytic processing of ATF6 and phosphorylation of C/EBP-beta are obligatory for the formation of this transcriptional complex 28028229_Human ATF6 mutations interrupt distinct sequential steps of the ATF6 activation mechanism. 28105371_Three branches of the Unfolded Protein Response (UPR) have been described, including the activation of the inositol-requiring enzyme 1 (IRE1), the pancreatic ER kinase (PKR)-like ER kinase (PERK), and the activating transcription factor 6 (ATF6). 28157699_Low ATF6 expression is associated with cancer. 28629319_The expression of ASNS was significantly elevated when ATF6 was over expressed. The expressions of these 2 genes were both decreased in hepatocellular carcinoma (HCC)patients, and it was more significantly with ASNS. The mRNA levels of ASNS and ATF6 were positively correlated with each other. rs34050735 was associated with HCC in the case-control study and also an independent predictor of overall survival of HCC patients. 28803844_results support a critical role of ATF6alpha in the establishment and maintenance of cellular senescence in normal human fibroblasts via the up-regulation of a COX2/PGE2 intracrine pathway. 28812650_A novel homozygous c.1691A>G (p.(Asp564Gly)) ATF6 mutation was identified in two siblings with autosomal recessive cone-rod dystrophy. 28884228_ATF6 expression correlates with precancerous changes and low-grade dysplasia in ulcerative colitis-associated colorectal cancer 28958904_we have demonstrated that ATF6alpha expression in mASM and hASM cells in vitro plays an important role in ASM proliferation and contractility, which are key features of asthma 29061306_In summary, our study revealed a negative regulation of the UPR transducer ATF6 through post-translational SUMOylation. The information from this study will not only increase our understanding of the fine-tuning regulation of the UPR signaling but will also be informative to the modulation of the UPR for therapeutic benefits. 29386036_ATF6 plays a distinct role in viral protein stability and the host uses different cleavage strategies, rather than conventional cleavage by generating p50ATF6, to combat viral infection. 29440509_Findings indicate a role for activating transcription factor 6 (ATF6) during differentiation and identify a new strategy to generate mesodermal tissues through the modulation of the ATF6 arm of the unfolded protein response (UPR). 29483204_miR-103/107-Wnt3a/beta-catenin-ATF6 pathway is critical to the progression of apoptosis in preadipocytes, which suggested that approaches to activate miR-103/107 could potentially be useful as new therapies for treating obesity and metabolic syndrome-related disorders 29512699_The results demonstrated that high expression of activated ATF6 aggravates ER stressinduced VEC apoptosis through the mitochondrial apoptotic pathway. Furthermore, in response to ER stress, ATF6 upregulates the expression of caspase3, caspase9, CHOP, cytochrome c and Bax/Bcl2. 29851562_While PERK complexes shift to large complexes, ATF6alpha complexes are reduced to smaller complexes on endoplasmic reticulum (ER) stress. In contrast, IRE1alpha complexes were not significantly increased in size on ER stress, unless IRE1alpha is overexpressed. 30063110_Endoplasmic reticulum stress-related ATF6 upregulates CIP2A and contributes to the prognosis of colon cancer. 30063920_sustained intestinal activation of ATF6 in the colon to promote dysbiosis and microbiota-dependent tumorigenesis. 30084354_147 is a pro-drug that preferentially activates ATF6 signaling through a mechanism involving localized metabolic activation and selective covalent modification of endoplasmic reticulumresident proteins that regulate ATF6 activity. 30086303_Reporters constructed to monitor each mechanism show that phenobarbital-induced endoplasmic reticulum membrane expansion depends on transmembrane domain-induced ATF6 30287689_Overexpression of the transcriptionally active N-terminal domain of ATF6 reversed the increases in IRE1 levels. 30639234_Study in human proximal tubular cell line and mouse chronic kidney disease model demonstrated that ATF6alpha activation downregulates mitochondrial fatty acid beta-oxidation activity via suppression of PPARalpha, which subsequently results in intracellular lipid accumulation in PTCs and eventually leads to lipotoxicity-induced tubulointerstitial fibrosis. 30717233_Non-structural protein 2B of human rhinovirus16 induced an endoplasmic reticulum stress response through the PERK and ATF6 pathways rather than the IRE1 pathway. 31029032_Pharmacologic inhibition or knock-down of downstream targets of ATF6a, protein disulfide isomerases (PDI) and ERO1b, a thiol oxidase that is involved in the re-oxidation of PDIs also independently induced pronounced killing of osteosarcoma (OS) cells following chemotherapy. 31035281_ATF6 is associated with the destruction of ossicles. Our results suggest that certain ER stress-related genes are expressed in chronic otitis media-associated inflammation 31105062_Results define a molecular basis for the ATF6-dependent reduction in destabilized LC secretion and highlight the advantage for targeting this UPR-associated transcription factor to reduce secretion of destabilized, amyloidogenic proteins implicated in AL and related systemic amyloid diseases. 31227689_Study find that enforced activation of XBP1 and ATF6 results in reduction of stemness and proliferation. We expose a novel interaction between XBP1 and PERK-eIF2alpha signaling. 31237654_Our data demonstrate a near absence of cone structure in subjects harboring ATF6 mutations. This implicates ATF6 as having a major role in cone development and suggests that at least a subset of subjects with ATF6-ACHM have markedly fewer cellular targets for cone-directed gene therapies than do subjects with CNGA3- or CNGB3-ACHM. 31312025_In cancer with missense TP53 mutation, ATF6 activity is necessary for viability and invasion phenotypes. ATF6 inhibitors might be combined with mutant p53-targeting drugs to specifically sensitise cancer cells to endogenous or chemotherapy-induced ER stress. 31368601_Results uncover an unexpected layer of control over the activation of ATF6a at the level of Golgi processing with only a specific form of ATF6a being processed correctly to release soluble transcription factor. 31500833_The net outcome of Atf6alpha on cell survival and cell death depends on cell type and growth conditions, the presence and degree of ER stress, and the duration and intensity of Atf6alpha activation. 31506423_p53 expression could be regulated by the ATF6/XBP1/CHOP axis to promote the development of Chronic pancreatitis. 31852864_Reinvestigation of Disulfide-bonded Oligomeric Forms of the Unfolded Protein Response Transducer ATF6. 31884629_classification of mutations in ATF6 that interrupt distinct steps in the ATF6 signaling pathway 31900015_Homozygous ATF6 mutation is associated with Macular maldevelopment in ATF6-mediated retinal dysfunction. 31926341_Findings suggest that endoplasmic reticulum stress induces the ATF6 and C/EBPbeta binding, which may increase their DNA-binding affinity and inhibit the transcription activity of the PLK4 gene. 32046286_Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6alpha. 32138230_Designing Novel Therapies to Mend Broken Hearts: ATF6 and Cardiac Proteostasis. 32234634_Activating transcription factor 6 regulated cell growth, migration and inhibiteds cell apoptosis and autophagy via MAPK pathway in cervical cancer. 32271167_Multiexon deletion alleles of ATF6 linked to achromatopsia. 32514126_MiR-185-5p ameliorates endoplasmic reticulum stress and renal fibrosis by downregulation of ATF6. 32724472_ATF6 aggravates acinar cell apoptosis and injury by regulating p53/AIFM2 transcription in Severe Acute Pancreatitis. 32905769_Atf-6 Regulates Lifespan through ER-Mitochondrial Calcium Homeostasis. 33109440_The role of unfolded protein response in the pathogenesis of endometriosis: contribution of peritoneal fluid. 33397415_The role of activating transcription factor 6 in hydroxycamptothecin-induced fibroblast autophagy and apoptosis. 33545358_Enforced dimerization between XBP1s and ATF6f enhances the protective effects of the UPR in models of neurodegeneration. 33686769_OTUB1 facilitates bladder cancer progression by stabilizing ATF6 in response to endoplasmic reticulum stress. 33919712_The Epstein-Barr Virus Lytic Protein BMLF1 Induces Upregulation of GRP78 Expression through ATF6 Activation. 33960419_The Ca(2+) -binding protein sorcin stimulates transcriptional activity of the unfolded protein response mediator ATF6. 34186245_Comparative Host Interactomes of the SARS-CoV-2 Nonstructural Protein 3 and Human Coronavirus Homologs. 34561305_ATF6 is essential for human cone photoreceptor development. 34623328_Ubiquitination of ATF6 by disease-associated RNF186 promotes the innate receptor-induced unfolded protein response. 34664059_Targeting UPR branches, a potential strategy for enhancing efficacy of cancer chemotherapy. 34851146_ATF6-Mediated Unfolded Protein Response Facilitates Adeno-associated Virus 2 (AAV2) Transduction by Releasing the Suppression of the AAV Receptor on Endoplasmic Reticulum Stress. | ENSMUSG00000026663 | Atf6 | 967.05701 | 0.8519044 | -0.2312364801 | 0.12307523 | 3.505057e+00 | 6.118173e-02 | 9.998360e-01 | No | Yes | 906.05784 | 133.702259 | 1.136626e+03 | 129.317319 | |
| ENSG00000118369 | 57558 | USP35 | protein_coding | Q9P2H5 | 3D-structure;Alternative splicing;Hydrolase;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | This gene encodes a member of the peptidase C19 family of ubiquitin-specific proteases. These deubiquitinating enzymes (DUBs) catalyze the removal of ubiquitin proteins from other proteins. The encoded protein associates with polarized mitochondria and has been shown to inhibit NF-kappa B activation and delay PARK2-mediated degradation of mitochondria. Expression of this gene is upregulated by the let-7a microRNA and reduced expression has been observed in human tumor tissues. [provided by RefSeq, Jul 2017]. | hsa:57558; | cytosol [GO:0005829]; nucleus [GO:0005634]; cysteine-type endopeptidase activity [GO:0004197]; thiol-dependent deubiquitinase [GO:0004843]; protein deubiquitination [GO:0016579]; ubiquitin-dependent protein catabolic process [GO:0006511] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25915564_USP35 does not delay PARK2 recruitment 26348204_miR let-7a-regulated USP35 can inhibit NF-kappaB activation by deubiquitination and stabilization of ABIN-2 protein 29449677_findings suggest that USP35 regulates the stability and function of Aurora B by blocking APC(CDH1)-induced proteasomal degradation, thereby controlling mitotic progression 29685892_Two different USP35 isoforms that localise to different intracellular compartments and have distinct functions. 31723061_Genome-wide suppressor screen identifies USP35/USP38 as therapeutic candidates for ciliopathies. 32678307_Deubiquitinase USP35 restrains STING-mediated interferon signaling in ovarian cancer. 33931967_Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. 34131114_USP35, regulated by estrogen and AKT, promotes breast tumorigenesis by stabilizing and enhancing transcriptional activity of estrogen receptor alpha. 34438346_Regulation of survivin protein stability by USP35 is evolutionarily conserved. 34618999_USP35 mitigates endoplasmic reticulum stress-induced apoptosis by stabilizing RRBP1 in non-small cell lung cancer. | ENSMUSG00000035713 | Usp35 | 277.23050 | 1.0708083 | 0.0987001620 | 0.19665093 | 2.543645e-01 | 6.140185e-01 | 9.998360e-01 | No | Yes | 282.29623 | 37.894383 | 2.478508e+02 | 26.233222 | ||
| ENSG00000119328 | 54942 | ABITRAM | protein_coding | Q9NX38 | FUNCTION: Actin-binding protein that regulates actin polymerization, filopodia dynamics and increases the branching of proximal dendrites of developing neurons. {ECO:0000250|UniProtKB:Q80ZQ9}. | Actin-binding;Cell projection;Nucleus;Reference proteome | hsa:54942; | dendrite [GO:0030425]; filopodium tip [GO:0032433]; growth cone [GO:0030426]; lamellipodium [GO:0030027]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; actin filament binding [GO:0051015]; actin monomer binding [GO:0003785]; dendrite morphogenesis [GO:0048813]; regulation of actin filament polymerization [GO:0030833]; regulation of filopodium assembly [GO:0051489] | ENSMUSG00000038827 | Abitram | 278.73358 | 0.8857809 | -0.1749782398 | 0.19857559 | 7.545586e-01 | 3.850368e-01 | 9.998360e-01 | No | Yes | 272.09310 | 48.136719 | 3.173335e+02 | 43.303417 | |||
| ENSG00000119685 | 23093 | TTLL5 | protein_coding | Q6EMB2 | FUNCTION: Polyglutamylase which modifies tubulin, generating polyglutamate side chains on the gamma-carboxyl group of specific glutamate residues within the C-terminal tail of tubulin. Preferentially mediates ATP-dependent initiation step of the polyglutamylation reaction over the elongation step. Preferentially modifies the alpha-tubulin tail over a beta-tail (By similarity). Required for CCSAP localization to both polyglutamylated spindle and cilia microtubules (PubMed:22493317). Increases the effects of transcriptional coactivator NCOA2/TIF2 in glucocorticoid receptor-mediated repression and induction and in androgen receptor-mediated induction (PubMed:17116691). {ECO:0000250|UniProtKB:Q8CHB8, ECO:0000269|PubMed:17116691, ECO:0000269|PubMed:22493317}. | ATP-binding;Alternative splicing;Cell projection;Cilium;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Disease variant;Ligase;Magnesium;Metal-binding;Microtubule;Nucleotide-binding;Nucleus;Reference proteome;Transcription | This gene encodes a member of the tubulin tyrosine ligase like protein family. This protein interacts with two glucocorticoid receptor coactivators, transcriptional intermediary factor 2 and steroid receptor coactivator 1. This protein may function as a coregulator of glucocorticoid receptor mediated gene induction and repression. This protein may also function as an alpha tubulin polyglutamylase.[provided by RefSeq, Feb 2010]. | hsa:23093; | centrosome [GO:0005813]; cilium [GO:0005929]; cytosol [GO:0005829]; microtubule [GO:0005874]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; tubulin binding [GO:0015631]; tubulin-glutamic acid ligase activity [GO:0070740]; microtubule cytoskeleton organization [GO:0000226]; protein polyglutamylation [GO:0018095]; retina development in camera-type eye [GO:0060041] | 17116691_STAMP is an important new, downstream component of GR action in both gene activation and gene repression. 20374646_This study indicates that a physiological function of STAMP in several settings is to modify cell growth rates in a manner that can be independent of steroid hormones. 24791901_this study has performed exome sequencing in 28 individuals with a similar disease phenotype and subsequently used a casecontrol approach to identify mutations in TTLL5 as a cause of recessive retinal dystrophy. 28173158_5 homozygous variants [p.(Asp594fs), p.(Gln117*), p.(Met712fs), p.(Ile756Phe) and p.(Glu543Lys)] in TTLL5, in 8 patients from 6 families were identified. 2 male patients carrying truncating TTLL5 variants also displayed a reduction in sperm motility and infertility, whereas those carrying missense changes were fertile. TTLL has multiple viable isoforms, being highly expressed in retina, testis and spermatozoon flagellum. 28356705_in a study of 3 family members from 2 generations, identified in a previously misdiagnosed incomplete congenital stationary night blindness (icCSNB) case a splice-site mutation in intron 3 of TTLL5 (c.182-3_182-1delinsAA); reinvestigation of the clinical data corrected the diagnosis to cone dystrophy 30517872_Binding of CSAP to TTLL5 promotes relocalization of TTLL5 toward microtubules. 34203883_Novel TTLL5 Variants Associated with Cone-Rod Dystrophy and Early-Onset Severe Retinal Dystrophy. 35365235_Expanding the phenotype of TTLL5-associated retinal dystrophy: a case series. | 662.27310 | 1.1634103 | 0.2183599365 | 0.15275060 | 1.977260e+00 | 1.596794e-01 | 9.998360e-01 | No | Yes | 823.94002 | 128.161002 | 7.853900e+02 | 94.544531 | |||
| ENSG00000120088 | 104909134 | CRHR1 | protein_coding | P34998 | FUNCTION: G-protein coupled receptor for CRH (corticotropin-releasing factor) and UCN (urocortin). Has high affinity for CRH and UCN. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and down-stream effectors, such as adenylate cyclase. Promotes the activation of adenylate cyclase, leading to increased intracellular cAMP levels. Inhibits the activity of the calcium channel CACNA1H. Required for normal embryonic development of the adrenal gland and for normal hormonal responses to stress. Plays a role in the response to anxiogenic stimuli. {ECO:0000269|PubMed:18292205, ECO:0000269|PubMed:18801728, ECO:0000269|PubMed:23576434, ECO:0000269|PubMed:23863939}. | 3D-structure;Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;Endosome;G-protein coupled receptor;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transducer;Transmembrane;Transmembrane helix | This locus represents naturally occurring readthrough transcription between neighboring genes CRHR1-IT1, CRHR1 intronic transcript 1 (Gene ID: 147081) and CRHR1, corticotropin releasing hormone receptor 1 (Gene ID: 1394) on chromosome 17. The readthrough transcript encodes a protein that shares sequence identity with the product of the CRHR1 gene. [provided by RefSeq, Dec 2016]. | hsa:104909134;hsa:1394; | endosome [GO:0005768]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; intrinsic component of plasma membrane [GO:0031226]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; corticotrophin-releasing factor receptor activity [GO:0015056]; corticotropin-releasing hormone binding [GO:0051424]; corticotropin-releasing hormone receptor activity [GO:0043404]; G protein-coupled peptide receptor activity [GO:0008528]; peptide hormone binding [GO:0017046]; activation of adenylate cyclase activity [GO:0007190]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189]; adenylate cyclase-modulating G protein-coupled receptor signaling pathway [GO:0007188]; cell surface receptor signaling pathway [GO:0007166]; cellular response to corticotropin-releasing hormone stimulus [GO:0071376]; corticotropin secretion [GO:0051458]; female pregnancy [GO:0007565]; immune response [GO:0006955]; negative regulation of voltage-gated calcium channel activity [GO:1901386]; parturition [GO:0007567]; regulation of adenylate cyclase activity involved in G protein-coupled receptor signaling pathway [GO:0010578]; regulation of corticosterone secretion [GO:2000852] | Mouse_homologues 11988580_In mice lacking a functional CRH1 receptor, stress leads to enhanced and progressively increasing alcohol intake. It was associated with an up-regulation of the N-methyl-d-aspartate receptor subunit NR2B 12029097_binding sites in CRFR1 for sauvagine 12399950_CRHR1 is required for a normal chromaffin cell structure and function and deletion of this gene is associated with a significant impairment of epinephrine biosynthesis. 12832554_Treatment of CRFR2-deficient mice with the CRFR1 antagonist antalarmin decreased immobile time and increased swim time in both sexes. Significant behavioral differences between the sexes were noted 14526225_astrocytes and microglia are cellular targets of CRH, which could serve as a link between CRH and inflammatory responses in ischemic injury via CRH-R1. 14736736_significant increase in urocortin II mRNA levels in the skin, but not in skeletal muscle, of both corticotropin-releasing factor receptor 1 and 2-null mice 15001778_Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors.The central amygdala (CeA) plays a role in the relationship among stress, corticotropin-releasing factor (CRF), and alcohol abuse. 15001778_data indicate that CRF1 receptors mediate ethanol enhancement of GABAergic synaptic transmission in the central amygdala, and they suggest a cellular mechanism underlying involvement of CRF in ethanol's behavioral and motivational effects 15269266_The role of CRF1 in the defensive startle response in mice. 15496472_In CRF(1)(-/-) mice, the dendritic trees of hippocampal principal cells were exuberant. 15664114_Coexpression of the cannabinoid receptor type 1 with the corticotropin-releasing hormone receptor type 1 in distinct regions of the adult mouse forebrain. 15752579_Hypoxia-increased CRF, CRF mRNA, CRFR1 mRNA, and corticosterone were blocked by CRFR1 antagonist (CP-154,526). 16423327_Corticotropin releasing factor over-expression in the mouse brain is associated with down-regulation of CRF1 mRNA. 16769145_Results suggest that the selective anxiolytic properties of etifoxine appear unrelated to an antagonist activity at corticotropin-releasing factor (CRF)1 and CRF2 receptors. 16778399_CRF and urocortin promote the survival of cultured cerebellar GABAergic neurons through the type 1 CRF receptor 16953386_Results underline the importance of limbic CRF-R1 in modulating anxiety-related behavior. 17042789_brainstem and/or hypothalamic CRHR1 contribute to the suppression of basal corticosterone 17194724_CRFR1 is expressed in mouse colon and is overexpressed in altered behavioral responses to superimposed mild stressors. 17293846_Data suggest a novel, previously unknown role of the CRH/CRH-R1 system in modulating neurovascular gene expression and function. 17296558_The hypothalamus-pituitary-adrenal (HPA) system and brain extra-hypothalamic CRF/CRF1 receptor circuitry in somatic, molecular, and endocrine alterations induced by opiate withdrawal. 17316992_the activity of these receptor types might contribute to the development of the neuronal ability for plasticity like processes on the level of NMDAR subunit composition and GABAergic activity 17331244_Taken together, the results suggest that the presence of an intact CRFR1 receptor supports some aspects of nurturing behavior. 17444496_CRF1 and CRF2 are involved in peripheral stress adaptation processes, such as modulation of stress-induced analgesia and the mediation of visceral nociceptive information by CRF2. 17506983_urocortin modulates T-type Ca(2+) channel by interacting with CRF-R1 via the activation of PKC signal pathway in MN9D cells 17517441_Stress reduced CRHr in BALB/cByJ mice. In C57BL/6ByJ mice acute stress increased CRHr in portions of the orbital frontal cortex, whereas chronic stress reduced CRHr. Chronic stress increased CRH(1) mRNA expression in both mouse strains. 18166336_lung-associated corticotropin releasing hormone regulation through peripheral CRH and diverse CRH receptor expression by MHC II(+) antigen presenting cells in pneumonia 18195718_once the CCK system was sensitized by prior CRF(1) activation, it exhibited its anxiogenic effects, without influence by CRF(1), possibly because of its observed downregulation 18582531_In order to dissect the signal transduction cascades activated by CRH receptor type 1, a comparative proteome approach was performed in vitro utilizing murine corticotroph AtT-20 cells. 18591672_Mice lacking CRF1 receptors do not show psychomotor sensitization to ethanol, a phenomenon that was also absent in CRF1 + 2 receptor double-knockout mice. 18787023_Presence of CRH-R1 mRNA in a subset of lactotropes, gonadotropes, and thyrotropes establishes these cell types as novel sites of murine CRH-R1 expression in the pituitary. 18843268_results support an involvement of CRF receptors in the development of depression, such that elevated hippocampal CRF1 activity, in the absence of CRF2, produces a depression-dominated phenotype through the activation of the MEK/ERK pathway 18945225_Increased ethanol self-administration associated with the alcohol deprivation effect observed in a C57BL/6J mouse model is modulated by CRF-1 receptor signaling. 18955489_Residue 17 of sauvagine cross-links to the first transmembrane domain of corticotropin-releasing factor receptor 1 (CRFR1). 18973598_These results implicate CRFR1 in chronic stress-induced alterations in amygdala function and behavior. Furthermore, they show that CRFR1 antagonists can prevent changes induced by chronic stress, in particular in those animals that are highly anxious. 19003957_These results provide ultrastructural evidence for a primary involvement of CRF receptors in modulation of the postsynaptic excitability of CeA neurons, an effect that may be limited by the availability of CRF. 19020499_These results support an additive model of CRF1 and CRF2 receptor activation effects on potentiated startle. 19091975_Dopamine - CRF system interactions regulate excitatory transmission and plasticity in the bed nucleus of the stria terminalis. The CRF-R1-dependent enhancement of glutamatergic transmission here may be a common key feature of substances of abuse. 19151899_presynaptic CRF1 receptors play a critical role in permitting or mediating ethanol enhancement of GABAergic synaptic transmission in central amygdala 19302188_Data show that stress has opposing effects on CRHR1 and CRHR2 neuronal systems, and suggest that regulation of the relative contribution of the two CRH receptors to brain CRH pathways may be essential in coordinating physiological responses to stress. 19379774_suggesting that under basal conditions the circadian variation of hypothalamic orexin is not mediated by corticotropin-releasing hormone, at least not via CRH-R1 19407218_Cortagine, a CRF1 agonist, induces stresslike alterations of colonic function and visceral hypersensitivity in rodents primarily through peripheral pathways. 19539724_Compared with saline, however, mice receiving chronic morphine showed a significantly lower plasmalemmal, and greater cytoplasmic, density of CRFr immunogold in dendrites. 20032050_Permeation of urocortin across the blood-brain barrier is dependent on the level of CRHR1 expression in cerebral microvessels. 20052275_Corticotropin Releasing Factor and dynorphin/KOR systems may coordinate stress-induced anxiety behaviors and aversive behaviors via different mechanisms. 20080775_Presence of CRFR1 on beta cells adds another layer of complexity to the intricate network of paracrine and autocrine factors and their cognate receptors whose coordinated efforts can regulate beta cell proliferation. 20084060_reduced expression of CRFR1 mRNA levels in the basolateral amygdala mediates the anxiolytic effect of environmental enrichment on anxiety-like behavior 20206175_CRHR1 promotes intestinal inflammation, as well as endogenous and inflammatory angiogenesis whereas CRHR2 inhibits these activities. 20392192_CRFR1 is required for stress-induced weight loss, but hyper-reactivity of the hypothalamo-hypophyseal axis in RRS mice exposed to a subsequent novel stress is independent of it 20619419_This study presented a VSDI assay for the investigation of neuronal activity propagation through the HF and demonstrates that CRH, via CRHR1 21296667_forebrain CRHR1 deficiency prevented the down-regulation of hippocampal glucocorticoid receptor expression by chronic stress but induced increased body weight gain during persistent stress exposure. 21722209_Data suggest that corticotropin-releasing factor receptor types 1 and 2 mediation of stress-induced hippocampal tau phosphorylation may be limited to emotional stressors. 21723867_Present results are suggestive that CRF1 (but not CRF2) receptors play a crucial role in the anxiogenic and antinociceptive effects induced by CRF in the dPAG in mice. 21774994_CRH-R1 inhibition is not protective against pneumococcal disease induced by stress. 21885734_results define a bidirectional model for the role of CRHR1 in anxiety and suggest that an imbalance between CRHR1-controlled anxiogenic glutamatergic and anxiolytic dopaminergic systems might lead to emotional disorders 21895713_Our results confirm that corticotropin-releasing factor receptor type 1 plays a key role in binge drinking and identify CRF as the ligand critically involved in excessive alcohol consumption 21940453_results suggest that forebrain CRF1 is crucial for the programming of cognitive function by early-life stress. 22113086_CRHR1 does not have a role in basal alcohol intake or relapse-like drinking situations with a low stress load. 22131403_our data reveal a novel anxiolytic role for CRFR1 in the globus pallidus external. 22336193_Reductions in anxiety and corticosteroid levels conferred by heterozygosity of CRF receptor type-1 do not improve a deficit in working memory observed in APP/hAbeta/PS1 mice. 22451915_Data show that CRFR1 and CRFR2 double-KO mice did not exhibit repeated stress-induced alterations in tau-P or solubility. 22659651_CRH and CRHR1 are dynamic modulators of a variety of signal transduction mechanisms and cellular processes. 22672268_The findings highlight the importance of forebrain CRHR1 in modulating some of the anxiogenic effects of early-life stress. 22886732_The corticotropin-releasing hormone (CRH) and its type 1 receptor (CRHR1) play a central role in coordinating the endocrine, autonomic, and behavioral responses to stress. 23133512_CRHR1 agonism stimulates rat and mouse fetal testis steroidogenesis 23371389_ERK1/2 activation in response to CRH is biphasic, involving a first cAMP- and B-Raf-dependent early phase and a second phase that critically depends on CRHR1 internalization and beta-arrestin2. 23398267_Blockade of CRF1 receptors does not exert specific effects on ethanol intake in an animal model of binge drinking. 23458743_This study demonistrated that Expression of CRFR1 and Glu5R mRNA in different brain areas following repeated testing in mice that differ in habituation behaviour. 23576434_studies show that SAP97 interactions with CRFR1 attenuate CRFR1 endocytosis and that SAP97 is involved in coupling G protein-coupled receptors to the activation of the ERK1/2 signaling pathway. 23644483_the CRH-CRHR1 system interacts with the nectin-afadin complex to mediate the effects of stress on memory and structural plasticity. 23763790_Altered CRF1 receptor-mediated signaling in the ventral tegmental area promotes binge-like alcohol consumption in mice. 24054833_CRFR1 plays important roles in whole body energy homeostasis 24083959_Leptin regulates corticotropin-releasing factor and urocortin 2/3 mRNA in hypothalamic neurons. 24490859_Data (including data from studies in knockout mice) suggest that Crh/Crhr1 signal transduction in noradrenergic neurons in cardiac ventricles during morphine withdrawal contributes to cardiovascular dysfunction, mimicking stress-induced dysfunction. 24518397_Crhr1 in the basolateral amygdala is critically involved as plasticity gene in the bidirectional epigenetic rescue of extremes in trait anxiety. 24773341_data suggest that the gene x stress exposure interaction is based on naturally occurring genetic variations in the Crhr1 gene associated with enhanced CRHR1-mediated signaling. 24776847_cPLA2 participates in CRHR1-induced apoptosis and CRHR2-inhibited apoptosis. 24840018_The data supports a critical involvement of Crhr1 in environmentally-induced anxiolysis. 25015995_CRHR1 has a proinflammatory and therefore a protumorigenesis effect in terms of colitis associated cancer, which may be helpful to develop new therapeutic approaches for inflammation and cancer prevention and treatment 25125464_These data suggest critical roles for CRF and CRFR1 in tau-P and aggregation and may have implications for the development of Alzheimer disease cognitive decline. 25164654_CRFR1 signaling in the ventral tegmental area presents a target for convergent effects of both cue- and stress-induced cocaine-seeking pathways 25224546_Data suggest a physiologically relevant role for local corticotropin-releasing hormone signaling towards shaping the neuronal circuitry within the mouse olfactory bulb. 25238483_NPY and PYY are required for CRF1 modulation of feeding behavior during stress. 25358399_This study aims to investigate the cellular localization of CRF and UCNs in the ileum and to explore whether and how this cellular expression is altered in conditions of intestinal Schistosoma mansoni-induced inflammation. 25403725_Results reveal the influences of early postnatal stress on the development of medial prefrontal cortex (mPFC) pyramidal neurons, and highlights CRF1 as a key molecule in modulating the detrimental stress effects on the structural organizations of mPFC 25473847_it is shown that CRH and UCN upregulate galectin-1 expression in Ishikawa cell line and macrophages and this effect is mediated through CRHR1. 25582704_Sympathetic activity induced by naloxone-precipitated morphine withdrawal is blocked in genetically engineered mice lacking functional CRF1 receptor 25697705_Anatomical and biochemical findings demonstrate that accumulation of Abeta is significantly reduced in the brain of Alzheimer's disease model mice lacking CRFR1 providing support for a direct relationship between CRFR1 and Abeta production pathways 25830629_The genetic disruption of the CRF1R pathway decreased the period of extinction in males and females suggesting that CRF/CRF1R is implicated in the duration of aversive memory. 26052002_bladder smooth muscle contractility is unchanged by CsA; however, there are changes in the levels of the downstream transcription factors MEF-2 and NFAT. 26239581_these results indicate that NPS-NPSR system may regulate locomotion together with the CRF1 system in SN. 26306872_This study demonstrated that an anatomical mechanism for sex-differences in the convergent modulation of ventrolateral medulla catecholaminergic neurons by CRF and glutamate. 26576941_A role was found for extra-hypothalamic CRF-R1 in stress-escalated alcohol drinking. 26792004_Study characterized the influences of early-life stress on the developmental trajectory of hippocampal pyramidal neurons, and highlighted the critical role of CRHR1 in modulating these negative outcomes evoked by early-life stress. 26907806_Study demonstrated that CRF1 receptor-deficiency prolongs whereas CRF2 receptor-deficiency shortens the duration of recognition memory deficits induced by morphine discontinuation, unraveling opposite roles for the two known CRF receptor subtypes in cognitive dysfunction associated with opiate withdrawal. 27060334_CRF plays a marked anxiogenic role at CRF1 receptors in the amygdala of mice exposed to the elevated plus maze. 27211900_CRFR1, in a subset of AgRP neurons, plays a regulatory role that enables appropriate sympathetic nervous system activation and consequently protects the organism from hypothermia and hypoglycemia. 27303056_switch in G protein coupling for type 1 corticotropin-releasing factor receptors promotes excitability in epileptic brains 27402953_the cyclic AMP (cAMP) response of CRHR1 in physiologically relevant scenarios engages separate cAMP sources, involving the atypical soluble adenylyl cyclase (sAC) in addition to transmembrane adenylyl cyclases (tmACs). 28017470_Excitability of genetically isolated CRF-receptive (CRFR1) neurons in the central nucleus of the amygdala (CeA) is potently enhanced by CRF and that CRFR1 signaling in the CeA is critical for discriminative fear 28135239_found that corticotropin-releasing factor type 1 receptor within the paraventricular nucleus of the hypothalamus is an important central component of hypothalamic-pituitary-adrenal axis regulation that prepares the organism for successive exposure to stressful stimuli. 28492284_results show that in response to MS, CRHR1 mediates gut injury by promoting intestinal inflammation, increasing gut permeability, altering intestinal morphology, and modulating the intestinal microbiota. In contrast, CRHR2 activates intestinal stem cells and is important for gut repair. 28684600_These results revealed a prominent role for CRF1 signaling in mast cells as a positive modulator of stimuli-induced degranulation. 28750017_the beneficial effects of CRFR1 antagonism seen in transgenic mice may be mechanistically linked to the modulation of oxidative stress pathways 28823817_report a sexually dimorphic expression of CRFR1 within the rostral portion of the anteroventral periventricular nucleus of the hypothalamus 28993972_Expression of Crhr1 is decreased in the pituitary gland of corticotropin-releasing hormone knock-out mice exposed to repeat stress. 29115460_CRF-R1 expression levels were proportional to the severity of DSS-induced colitis. Activation of CRF-R1 aggravated inflammation, and inhibition of CRF-R1 ameliorates inflammation evaluated by the DAI and histological scores in the colon samples of the DSS-treated mice. 29417477_the miR-34-dependent modulation of CRFR1 expression may be involved in the dorsal raphe nuclei regulation of stress-coping strategies. 29934353_We found a profound effect on learning both at the cellular and behavioral levels without an effect on baseline motor skills after granular cell-specific ablation of CRFR1 30248435_In this study, mild blast-induced traumatic brain injury (mbTBI) did not alter CRFR1 gene expression in males or females. However, mbTBI disrupted CRFR2 gene expression in different limbic structures in males and females. 30282821_Study of transgenic HP5 mice overexpressing human SSTR5 in pituitary corticotrophs shows that SSTR5-induced miR-449c suppresses both CRHR1 expression and function and concludes that corticotroph SSTR5 attenuates hypothalamic-pituitary-adrenal axis responses via CRHR1 downregulation, suggesting a role for SSTR5 in the pathogenesis of secondary adrenal insufficiency. 30529655_Sevoflurane-induced nectin-1 and L-afadin expression decrease was mediated by CRHR1 signaling in the hippocampus. 30535142_Proton-induced CRHR1 signaling regulates ACTH production in response to an acidic microenvironment. 30704133_plasma extracellular vesicles from immune cells carry CRFRs as cargos and influence cell-cell communication in health and disease. 31055007_we report a sexually dimorphic distribution of CRFR1 expressing cells within the paraventricular hypothalamus (PVN; males > females). 31167849_Data provide important information about the neurobiology of Central Nucleus of the Amygdala corticotropin-releasing factor acting at cognate type 1 receptors (CRF1+) neurons that may have critical implications for their functional role under physiological and pathological conditions. 31489921_The data suggest that 5-HT2CR activation in the BLA contributes to neuropathic pain-related amygdala (CeA) activity by engaging CRF1 receptor signaling. 31715260_Activation of CRHR1 contributes to cerebral endothelial barrier impairment via cPLA2 phosphorylation in experimental ischemic stroke. 32006904_Anxiogenesis induced by social defeat in male mice: Role of nitric oxide, NMDA, and CRF1 receptors in the medial prefrontal cortex and BNST. 32041742_Corticotropin-Releasing Factor Receptor-1 Neurons in the Lateral Amygdala Display Selective Sensitivity to Acute and Chronic Ethanol Exposure. 32725663_The CRF1 receptor mediates social behavior deficits induced by opiate withdrawal. 33758173_Prenatally traumatized mice reveal hippocampal methylation and expression changes of the stress-related genes Crhr1 and Fkbp5. 34507241_Alterations in corticotropin-releasing factor receptor type 1 in the preoptic area and hypothalamus in mice during the postpartum period. 35122897_Corticotropin-releasing factor receptor 1 in infralimbic cortex modulates social stress-altered decision-making. | ENSMUSG00000018634 | Crhr1 | 18.02320 | 1.2488944 | 0.3206514714 | 0.69946538 | 2.293281e-01 | 6.320225e-01 | 9.998360e-01 | No | Yes | 15.69388 | 6.318139 | 1.239493e+01 | 4.103030 | |
| ENSG00000120334 | 91687 | CENPL | protein_coding | Q8N0S6 | FUNCTION: Component of the CENPA-CAD (nucleosome distal) complex, a complex recruited to centromeres which is involved in assembly of kinetochore proteins, mitotic progression and chromosome segregation. May be involved in incorporation of newly synthesized CENPA into centromeres via its interaction with the CENPA-NAC complex. {ECO:0000269|PubMed:16716197}. | Alternative splicing;Centromere;Chromosome;Nucleus;Phosphoprotein;Reference proteome | CENPL is a subunit of a CENPH (MIM 605607)-CENPI (MIM 300065)-associated centromeric complex that targets CENPA (MIM 117139) to centromeres and is required for proper kinetochore function and mitotic progression (Okada et al., 2006) [PubMed 16622420].[supplied by OMIM, Mar 2008]. | hsa:91687; | chromosome, centromeric region [GO:0000775]; cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 26698661_Study demonstrate that the CENP-L-N complex plays a crucial role at the core of the 16-subunit Constitutive Centromere-Associated Network through interactions with CENP-C and CENP-H-I-KM. 34457090_High mRNA Expression of CENPL and Its Significance in Prognosis of Hepatocellular Carcinoma Patients. 34607313_Highly expressed centromere protein L indicates adverse survival and associates with immune infiltration in hepatocellular carcinoma. | ENSMUSG00000026708 | Cenpl | 214.09778 | 1.0304855 | 0.0433241774 | 0.23861426 | 3.080221e-02 | 8.606824e-01 | 9.998360e-01 | No | Yes | 185.47117 | 38.026305 | 2.255943e+02 | 35.585680 | |
| ENSG00000120519 | 84068 | SLC10A7 | protein_coding | Q0GE19 | FUNCTION: Involved in teeth and skeletal development. Has an essential role in the biosynthesis and trafficking of glycosaminoglycans and glycoproteins, to produce a proper functioning extracellular matrix. Required for extracellular matrix mineralization (PubMed:30082715, PubMed:29878199). Also involved in the regulation of cellular calcium homeostasis (PubMed:30082715, PubMed:31191616). Does not show transport activity towards bile acids or steroid sulfates (including taurocholate, cholate, chenodeoxycholate, estrone-3-sulfate, dehydroepiandrosterone sulfate (DHEAS) and pregnenolone sulfate). {ECO:0000269|PubMed:17628207, ECO:0000269|PubMed:29878199, ECO:0000269|PubMed:30082715, ECO:0000269|PubMed:31191616}. | Alternative splicing;Amelogenesis imperfecta;Cell membrane;Disease variant;Dwarfism;Endoplasmic reticulum;Golgi apparatus;Ion transport;Membrane;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | hsa:84068; | cis-Golgi network [GO:0005801]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; Golgi medial cisterna [GO:0005797]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; intrinsic component of plasma membrane [GO:0031226]; plasma membrane [GO:0005886]; trans-Golgi network [GO:0005802]; bile acid transmembrane transporter activity [GO:0015125]; symporter activity [GO:0015293]; bone development [GO:0060348]; cellular calcium ion homeostasis [GO:0006874]; glycoprotein transport [GO:0034436]; Golgi vesicle transport [GO:0048193]; heparin biosynthetic process [GO:0030210]; sodium ion transport [GO:0006814] | 15932064_cloning and characterization; mapped to chromosome 4q31.2 and contains 12 exons; widely expressed in human tissues 17628207_Molecular characterization and expression analysis of a novel member of the SLC10 family, SLC10A7, previously known as C4orf13, is reported 19240061_Observational study of gene-disease association. (HuGE Navigator) 29878199_Mutation in SLC10A7 gene is associated with anxiety defective bone mineralization. 30082715_SLC10A7 is involved in glycosaminoglycan synthesis and specifically in skeletal development. SLC10A7 mutations cause a skeletal dysplasia with amelogenesis imperfecta. 32350310_The orphan solute carrier SLC10A7 is a novel negative regulator of intracellular calcium signaling. 33561315_Circular RNA circHECTD1 facilitates glioma progression by regulating the miR-296-3p/SLC10A7 axis. | ENSMUSG00000031684 | Slc10a7 | 90.23528 | 1.2105379 | 0.2756482688 | 0.32678541 | 7.469417e-01 | 3.874462e-01 | 9.998360e-01 | No | Yes | 96.72270 | 21.678105 | 8.674469e+01 | 15.035105 | ||
| ENSG00000120664 | 100507135 | SPART-AS1 | lncRNA | 12.94290 | 0.9705797 | -0.0430813360 | 0.80593865 | 2.730224e-03 | 9.583283e-01 | 9.998360e-01 | No | Yes | 18.04799 | 7.208911 | 1.819530e+01 | 5.778144 | ||||||||||
| ENSG00000120833 | 8835 | SOCS2 | protein_coding | O14508 | FUNCTION: SOCS family proteins form part of a classical negative feedback system that regulates cytokine signal transduction. SOCS2 appears to be a negative regulator in the growth hormone/IGF1 signaling pathway. Probable substrate recognition component of a SCF-like ECS (Elongin BC-CUL2/5-SOCS-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. | 3D-structure;Growth regulation;Isopeptide bond;Phosphoprotein;Reference proteome;SH2 domain;Signal transduction inhibitor;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a member of the suppressor of cytokine signaling (SOCS) family. SOCS family members are cytokine-inducible negative regulators of cytokine receptor signaling via the Janus kinase/signal transducer and activation of transcription pathway (the JAK/STAT pathway). SOCS family proteins interact with major molecules of signaling complexes to block further signal transduction, in part, by proteasomal depletion of receptors or signal-transducing proteins via ubiquitination. The expression of this gene can be induced by a subset of cytokines, including erythropoietin, GM-CSF, IL10, interferon (IFN)-gamma and by cytokine receptors such as growth horomone receptor. The protein encoded by this gene interacts with the cytoplasmic domain of insulin-like growth factor-1 receptor (IGF1R) and is thought to be involved in the regulation of IGF1R mediated cell signaling. This gene has pseudogenes on chromosomes 20 and 22. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2012]. | hsa:8835; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; phosphatidylinositol 3-kinase complex [GO:0005942]; 1-phosphatidylinositol-3-kinase regulator activity [GO:0046935]; growth hormone receptor binding [GO:0005131]; insulin-like growth factor receptor binding [GO:0005159]; JAK pathway signal transduction adaptor activity [GO:0008269]; cellular response to hormone stimulus [GO:0032870]; growth hormone receptor signaling pathway [GO:0060396]; intracellular signal transduction [GO:0035556]; lactation [GO:0007595]; mammary gland alveolus development [GO:0060749]; negative regulation of apoptotic process [GO:0043066]; negative regulation of multicellular organism growth [GO:0040015]; negative regulation of receptor signaling pathway via JAK-STAT [GO:0046426]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; positive regulation of neuron differentiation [GO:0045666]; protein ubiquitination [GO:0016567]; receptor signaling pathway via JAK-STAT [GO:0007259]; regulation of cell growth [GO:0001558]; regulation of signal transduction [GO:0009966]; response to estradiol [GO:0032355] | 11861294_SOCS-2 is overexpressed in advanced stages of chronic myeloid leukemia. It may be a component of a negative feedback mechanism that is functioning inadequately, induced by Bcr-Abl but unable to reverse its growth-promoting effects. 12552091_SOCS2 mediates the suppression of JAK2 phosphorylation by estrogen, which inhibits growth hormone signaling 15361843_SOCS1 and SOCS2 but not SOCS3 suppressed the growth of ovarian and breast cancer cells. 16406727_Association of single-nucleotide polymorphisms in the SOCS2 gene with type 2 diabetes in the Japanese was studied. 16406727_Observational study of gene-disease association. (HuGE Navigator) 16675548_Results describe the 1.9-A crystal structure of the ternary complex of SOCS2 with elongin C and elongin B. 16684815_SOCS2 only interacts with the Y1077 motif, but with higher binding affinity and can interfere with CIS and STAT5a prey recruitment at this site. 17008382_SOCS2 expression is regulated by STAT5. 17264307_Using RT-PCR, we demonstrated for the first time that neutrophils express mRNA for SOCS-2. 17325857_A defect in expression of SOCS-2 and SOCS-3 genes may be crucial for the IGF-I hypersensitivity and progressive increase in erythroid cell population size characteristic of Polycythemia vera. 17651480_Favorable prognostic value of high SOCS2 expression in primary mammary carcinomas. 17666591_analysis of STAT5, CIS, and SOCS2 interactions with the growth hormone receptor 17703412_Observational study of gene-disease association. (HuGE Navigator) 18769447_SOCS2 epigenetic downregulation might be an important second step in the genesis of cytokine-independent myeloproliferative disorder clones 18820827_difference in SOCS1, SOCS2 and SOCS3 transcript levels between normal individuals and SLE patients is not statistically significant 18844680_Acromegalic patients with active disease and hyperplastic polyps had higher levels of SOCS2 transcripts. 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19266077_Observational study of gene-disease association. (HuGE Navigator) 19279332_role of SOCS-2 in mediating HIV-1-induced immune evasion and dysregulation of IFNgamma signaling in primary human monocytes 19423540_Observational study of gene-disease association. (HuGE Navigator) 19779605_SOCS2 is required for appropriate TLR4 signaling in maturating human DCs via both the MyD88-dependent and -independent signaling pathway 19913121_Observational study of gene-disease association. (HuGE Navigator) 20136974_Probiotic administration increased expression of SOCS-2 and SOCS-3 in Helicobacter pylori infection to limit inflammatory signaling. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20503287_Observational study of gene-disease association. (HuGE Navigator) 20543098_Data show that knockdown of SOCS2 resulted in the accumulation of p-Pyk2(Tyr402) and blocked NK cell effector functions. 20546612_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20802378_Observational study of genetic testing. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21844389_We propose a model in which SOCS2 acts as a negative regulator of TLR-induced dendritic cell activation 22291912_LPS regulates SOCS2 transcription in a type I interferon dependent autocrine-paracrine loop. 22745742_Data demonstrated that suppressor of cytokine signaling 2 (SOCS2) protein level was distinctively increased by saponin, which in turn resulted in inhibition of HCV replication. 22768656_High SOCS2 is associated with idiopathic short stature. 22795647_Simultaneous stimulation of monocyte-derived DCs resulted in highly increased production of IL-1beta, IL-23 and SOCS2. 23475171_SOCS2 and SOCS6 expression are remarkably reduced in hepatocellular carcinoma and correlate with aggressive tumor progression and poor prognosis 23548639_SOCS2 associates with activated FLT3 through phosphotyrosine residues 589 and 919, and co-localizes with FLT3 in the cell membrane. 23567159_Estradiol can amplify GH intracellular signaling in human osteoblasts with an essential role played by the reduction of the SOCS2 mediated feedback loop. 23897481_crystal structure shows interaction between SOCS2-elongin BC and Cullin-5 24031028_SOCS2 mediates the cross talk between androgen and growth hormone signaling in prostate cancer. 24131863_methylation of SOCS1, SOCS2, SOCS3 and CISH is infrequent in Ph-ve MPN. 24280133_SOCS2 correlates with malignancy and exerts growth-promoting effects in prostate cancer. 24559289_our study indicates that high SOCS2 expression is associated with poor survival in pediatric AML. 24905066_The SOCS2 polymorphism (rs3782415) has an influence on the adult height of children with Turner syndrome and growth hormone deficiency after long-term therapy. 25025962_Stage-independent downregulation of SOCS2 and SOCS6 correlate with disease-free survival in colorectal cancer. 25283341_This study shows that over-expression of SOCS2 reduces the psychostimulant effects of amphetamine, enhances PPI, and alters mesolimbic dopaminergic activity. 25286386_This study showed that there was significantly increased levels of SOCS-2 mRNA in elderly and Alzheimer's disease brains. 25561270_miR-101 functions as a growth-suppressive miRNA in H. pylori related GC, and that its suppressive effects are mediated mainly by repressing SOCS2 expression. 26121586_crystals of SOCS2 in complex with its adaptor proteins, Elongin C and Elongin B, underwent a change in crystallographic parameters when treated with dimethyl sulfoxide during soaking experiments. 26709655_focus on SOCS2 and review its biological function as well as its implication in pathological processes 27071013_SOCS2 may improve outcome of TBI in mice by regulating aspects of the neuroinflammatory response 27330188_The induction of a tolerogenic phenotype in DCs by NPs was mediated by the AhR-dependent induction of Socs2, which resulted in inhibition of nuclear factor kappaB activation and proinflammatory cytokine production (properties of tolerogenic DCs). 27423467_this study shows that IL-7 induces the expression of SOCS2 through the JAK/STAT-5 pathway and that SOCS2 interacts with CD127 in early endosomes and direct the receptor complex to the proteasome for degradation 27465557_Overexpression of SOCS-2 is associated with hepatocellular carcinoma. 27566229_Long-term palmitate treatment up-regulates SOCS2 and reduces PI3K activity, thereby impairing glucose stimulated insulin secretion. 27900634_Despite of the key role of SOCS2 in the regulation of GH receptor signaling, study did not find any significant association between SOCS2 polymorphisms and acromegaly. 28011622_Targeting of SOCS2 by miR-194 resulted in derepression of the oncogenic kinases FLT3 and JAK2, leading to enhanced ERK and STAT3 signaling. 28216640_Study provides evidence for an inhibitory role of SOCS2 in TNFalpha induced NF-kappa B activation, identifies NDR1 as a novel substrate of SOCS2, and demonstrates that SOCS2 and TNFalpha induced NF-kappa B signaling are linked through NDR1. 28468588_Overexpression of SOCS2 inverted these phenotypes generated by hsv2-miR-H9-5p, indicating the potential roles of SOCS2 in Hsv2-miR-H9-5p-driven metastasis in lung cancers. The results highlighted that Hsv2-miR-H9-5p regulated and contributed to bone metastasis of lung cancers. We proposed that Hsv2-miR-H9-5p could be used as a potential target in lung cancer therapy 28496097_SOCS2 impairs IFN/JAK/STAT signaling through reducing the stability of tyrosine kinase 2 (TYK2), downregulating the expression of type I and III IFN receptors, attenuating the phosphorylation and nucleus translocation of STAT1. 28643757_SOCS-1, SOCS-2, and SOCS-3 proteins may directly or indirectly, have important roles in development and pathogenesis of papillary thyroid cancer 28666115_Single-cell RNA sequencing reveals enrichment of homeostatic modules in monocytes and dendritic cells from human metastatic melanoma. Suppressor-of-cytokine-2 (SOCS2) protein, a conserved program transcript, is expressed by mononuclear phagocytes infiltrating primary melanoma and is induced by IFNgamma. 28967904_Study in lung cancer BEAS-2B cells shows that SOCS2 binding to the growth hormone receptor (GHR) is impaired by a GHR threonine substitution at Pro 495. This results in decreased internalisation and degradation of the receptor. 29171881_Methyltransferase-like 3 represses SOCS2 expression in hepatocellular carcinoma through an m6A-YTHDF2-dependent mechanism. Our findings suggest an important mechanism of epigenetic alteration in liver carcinogenesis 29559623_Low expression of SOCS2 induces invasion and metastasis of human lung adenocarcinoma cells by regulating epithelial-mesenchymal transition both in vitro and in vivo, which is mainly dependent on the IGF1/IGF1R-stimulated STAT3 or STAT5 pathway. 29622769_SOCS2 expression was heterogeneously upregulated in some human colon cancers. Thus, SOCS2 was upregulated by p53 dysfunction and seemed to be associated with the tumorigenic potential of colon cancer. 29753737_Overexpression of miR-196b suppresses SOCS2 in human laryngeal squamous cell carcinoma resulting in tumor progression and poor prognosis. 30453988_High SOCS2 expression is associated with higher grade breast cancer. 30779065_miR-492 was over-expressed in prostate cancer (PCa) and exerted tumor-promoting function in PCa cells via repressing SOCS2 expression 31235852_SOCS2 is part of a highly prognostic 4-gene signature in AML and promotes disease aggressiveness. 31298374_MiR-875 can regulate the proliferation and apoptosis of non-small cell lung cancer cells via targeting SOCS2. 31398463_expression of SOCS2 was significantly higher in kidney allograft-rejected male patients compared to non-rejected group, however, such difference was not detected between female subjects 32006198_DNA methylation of SOCS1/2/3 predicts hepatocellular carcinoma recurrence after liver transplantation. 32266818_SOCS2 affects the proliferation, migration and invasion of nasopharyngeal carcinoma cells via regulating EphA1. 32344463_MiR-3613-3p from carcinoma-associated fibroblasts exosomes promoted breast cancer cell proliferation and metastasis by regulating SOCS2 expression. 32437330_LncRNA SOCS2-AS1 inhibits progression and metastasis of colorectal cancer through stabilizing SOCS2 and sponging miR-1264. 32495891_Gene polymorphisms of SOCS1 and SOCS2 and acute lymphoblastic leukemia. 32536038_LncRNA WDFY3-AS2 suppresses proliferation and invasion in oesophageal squamous cell carcinoma by regulating miR-2355-5p/SOCS2 axis. 32705223_m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. 34197875_Suppressor of cytokine signalling-2 controls hepatic gluconeogenesis and hyperglycemia by modulating JAK2/STAT5 signalling pathway. 34605350_MicroRNA -196b is related to the overall survival of patients with esophageal squamous cell carcinoma and facilitates tumor progression by regulating SOCS2 (Suppressor Of Cytokine Signaling 2). 34857742_Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands. 35037826_miR-3648 promotes lung adenocarcinoma-genesis by inhibiting SOCS2 (suppressor of cytokine signaling 2). | ENSMUSG00000020027 | Socs2 | 134.74266 | 0.9899540 | -0.0145665680 | 0.26321715 | 3.088117e-03 | 9.556837e-01 | 9.998360e-01 | No | Yes | 120.60012 | 20.149993 | 1.346551e+02 | 17.556685 |
| ENSG00000121957 | 29899 | GPSM2 | protein_coding | P81274 | FUNCTION: Plays an important role in mitotic spindle pole organization via its interaction with NUMA1 (PubMed:11781568, PubMed:15632202, PubMed:21816348). Required for cortical dynein-dynactin complex recruitment during metaphase (PubMed:22327364). Plays a role in metaphase spindle orientation (PubMed:22327364). Plays also an important role in asymmetric cell divisions (PubMed:21816348). Has guanine nucleotide dissociation inhibitor (GDI) activity towards G(i) alpha proteins, such as GNAI1 and GNAI3, and thereby regulates their activity (By similarity). {ECO:0000250|UniProtKB:Q8VDU0, ECO:0000269|PubMed:11781568, ECO:0000269|PubMed:15632202, ECO:0000269|PubMed:21816348, ECO:0000269|PubMed:22327364}. | 3D-structure;Cell cycle;Cell division;Cell membrane;Cytoplasm;Cytoskeleton;Deafness;Membrane;Mitosis;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Transport | The protein encoded by this gene belongs to a family of proteins that modulate activation of G proteins, which transduce extracellular signals received by cell surface receptors into integrated cellular responses. The N-terminal half of this protein contains 10 copies of leu-gly-asn (LGN) repeat, and the C-terminal half contains 4 GoLoco motifs, which are involved in guanine nucleotide exchange. This protein may play a role in neuroblast division and in the development of normal hearing. Mutations in this gene are associated with autosomal recessive nonsyndromic deafness (DFNB82). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2016]. | hsa:29899; | cell cortex [GO:0005938]; cell cortex region [GO:0099738]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; lateral cell cortex [GO:0097575]; lateral plasma membrane [GO:0016328]; mitotic spindle pole [GO:0097431]; protein-containing complex [GO:0032991]; dynein complex binding [GO:0070840]; G-protein alpha-subunit binding [GO:0001965]; GDP-dissociation inhibitor activity [GO:0005092]; identical protein binding [GO:0042802]; nucleotide binding [GO:0000166]; protein C-terminus binding [GO:0008022]; protein domain specific binding [GO:0019904]; protein self-association [GO:0043621]; cell division [GO:0051301]; establishment of mitotic spindle orientation [GO:0000132]; G protein-coupled receptor signaling pathway [GO:0007186]; maintenance of centrosome location [GO:0051661]; mitotic spindle organization [GO:0007052]; positive regulation of protein localization to cell cortex [GO:1904778]; positive regulation of spindle assembly [GO:1905832]; Ran protein signal transduction [GO:0031291]; regulation of mitotic spindle organization [GO:0060236] | 11832491_LGN is expressed in neuronal, astroglial, and microglial cultures 15632202_binding between Lgl2 and LGN play a role in mitotic spindle organization through regulating formation of the LGN.NuMA complex; Lgl2 forms a Lgl2.Par-6.aPKC.LGN complex, which responds to mitotic signaling to establish normal cell division 20056645_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20479129_Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20589935_Upregulation of GPSM2 is associated with breast cancer. 20602914_Identification of GPSM2 as essential to the development of normal hearing suggests dysregulation of cell polarity as a mechanism underlying hearing loss. 20933426_During mitosis, Pins is mislocalized to the apical surface in the absence of Par3 or by inhibition of atypical protein kinase C. 21348867_A novel truncating mutation of GPSM2 is associated with autosomal recessive non-syndromic hearing loss. 22074847_mInsc-LGN interaction is vital for stabilization of LGN and for intracellular localization of mInsc. 22578326_GPSM2 is required for orienting the mitotic spindle during cell division in multiple tissues, suggesting that the sensorineural hearing loss and characteristic brain malformations of CMS are due to defects in asymmetric cell divisions during development 22690686_Overexpression of LGN decreases the activity of cellular sGC, whereas knockdown of LGN mRNA and protein correlated with increased sGC activity 22977735_Studies indicate that the Inscuteable (Insc)and NuMA are mutually exclusive interactors of LGN. 22987632_Results show compound heterozygous mutations in the GPSM2 gene, in affected members of a family with Chudley-McCullough syndrome, co-segregate in the family as an autosomal recessive trait. 23389635_Data indicate that dynein- and astral microtubule-mediated transport of Galphai/LGN/nuclear mitotic apparatus (NuMA) complex from cell cortex to spindle poles. 23494849_one homozygous frameshift GPSM2 variants c.1473delG was identified in three Chudley-McCullough syndrome Dutch patients. 23907121_LGN is required for mitotic spindle rotation but not orientation maintenance. 24165937_Hepatocyte Par1b defines lumen position in concert with the position of the astral microtubule anchoring complex LGN-NuMA to yield the distinct epithelial division phenotypes. 25664792_A crystal structure of Frmpd4-bound LGN in an oxidized form is also reported, although oxidation does not appear to strongly affect the interaction with Frmpd4. 26398908_results fit a model whereby LGN influences interphase microtubule dynamics in endothelial cells to regulate migration, cell adhesion, and sprout extension, and reveal a novel non-mitotic role for LGN in sprouting angiogenesis 26662512_Kinocilium is essential for proper localization of Lgn, as well as Gai and aPKC, suggesting that cilium function plays a role in positioning of apical proteins critical for hearing. 26751642_This study determined the crystallographic structure of human Afadin in complex with LGN. 26987813_Data support the notion that the Galpha, but not Gbetagamma, arm of the Gi/o signalling is involved in TRPC4 activation and unveil new roles for RGS and LGN in fine-tuning TRPC4 activities. 27064331_A novel mutation (c.1093C > T; p.Arg365*) is described in a family with dizygotic twins with variable phenotype of Chudley-McCullough syndrome. 27180139_This mutation is predicted to abolish all four GoLoco domains in GPSM2 and this explains the bioinformatic prediction for this mutation to be functionally damaging. Full clinical and molecular accounts of the novel mutation are provided in this paper. 28045117_The results show how E-cadherin instructs the assembly of the LGN/NuMA complex at cell-cell contacts, and define a mechanism that couples cell division orientation to intercellular adhesion. 28347229_high expression of G-protein signaling modulator 2 was involved in the pathological processes of hepatocellular carcinoma through activation of the phosphatidylinositol 3-kinase/protein kinase B signaling pathway, which may provide an attractive potential diagnostic biomarker and therapeutic target for treatment of hepatocellular carcinoma. 28712573_Endothelial flow mechanotransduction through the junctional complex is mediated by a specific pool of VE-cadherin that is phosphorylated on cytoplasmic tyrosine Y658 and bound to LGN. 29523789_In mammary stem cells, the asymmetric domain of Insc bound to LGN:Galphai(GDP) suffices to drive asymmetric fate, and reverts aberrant symmetric divisions induced by p53 loss. 31101817_Study reports the crystal structure of NuMA:LGN hetero-hexamers, and unveil their role in promoting the assembly of active cortical dynein/dynactin motors that are required in orchestrating oriented divisions in polarized cells. 31732560_LGN has a role in cell cortex recruitment of NuMA 32020211_Downregulation of GPSM2 is associated with primary resistance to paclitaxel in breast cancer. 32058048_Loss of G-protein-signaling modulator 2 accelerates proliferation of lung adenocarcinoma via EGFR signaling pathway. 32812493_GPSM2 Serves as an Independent Prognostic Biomarker for Liver Cancer Survival. | ENSMUSG00000027883 | Gpsm2 | 226.38463 | 0.8400294 | -0.2514882469 | 0.21080986 | 1.426684e+00 | 2.323063e-01 | 9.998360e-01 | No | Yes | 215.63046 | 35.366658 | 2.535581e+02 | 32.057061 | |
| ENSG00000122417 | 57489 | ODF2L | protein_coding | Q9ULJ1 | FUNCTION: Acts as a suppressor of ciliogenesis, specifically, the initiation of ciliogenesis. {ECO:0000269|PubMed:28775150}. | Alternative splicing;Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Reference proteome | hsa:57489; | centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cell projection organization [GO:0030030]; negative regulation of cilium assembly [GO:1902018]; regulation of cilium assembly [GO:1902017] | ENSMUSG00000028256 | Odf2l | 172.51813 | 1.1021318 | 0.1402967918 | 0.29557128 | 2.196619e-01 | 6.392977e-01 | 9.998360e-01 | No | Yes | 183.85723 | 45.847818 | 1.352899e+02 | 26.032888 | |||
| ENSG00000122642 | 11328 | FKBP9 | protein_coding | O95302 | FUNCTION: PPIases accelerate the folding of proteins during protein synthesis. | Alternative splicing;Calcium;Endoplasmic reticulum;Glycoprotein;Isomerase;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Rotamase;Signal | hsa:11328; | endoplasmic reticulum [GO:0005783]; calcium ion binding [GO:0005509]; peptidyl-prolyl cis-trans isomerase activity [GO:0003755]; protein folding [GO:0006457] | 31780055_Increasing of FKBP9 can predict poor prognosis in patients with prostate cancer. 32111229_FKBP9 promotes the malignant behavior of glioblastoma cells and confers resistance to endoplasmic reticulum stress inducers. | ENSMUSG00000029781 | Fkbp9 | 1030.15076 | 0.9409014 | -0.0878845524 | 0.12993078 | 4.669646e-01 | 4.943869e-01 | 9.998360e-01 | No | Yes | 940.28632 | 90.588933 | 1.063569e+03 | 79.632494 | ||
| ENSG00000123124 | 11059 | WWP1 | protein_coding | Q9H0M0 | FUNCTION: E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Ubiquitinates ERBB4 isoforms JM-A CYT-1 and JM-B CYT-1, KLF2, KLF5 and TP63 and promotes their proteasomal degradation. Ubiquitinates RNF11 without targeting it for degradation. Ubiquitinates and promotes degradation of TGFBR1; the ubiquitination is enhanced by SMAD7. Ubiquitinates SMAD6 and SMAD7. Ubiquitinates and promotes degradation of SMAD2 in response to TGF-beta signaling, which requires interaction with TGIF. {ECO:0000269|PubMed:12535537, ECO:0000269|PubMed:15221015, ECO:0000269|PubMed:15359284}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Host-virus interaction;Membrane;Nucleus;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | WW domain-containing proteins are found in all eukaryotes and play an important role in the regulation of a wide variety of cellular functions such as protein degradation, transcription, and RNA splicing. This gene encodes a protein which contains 4 tandem WW domains and a HECT (homologous to the E6-associated protein carboxyl terminus) domain. The encoded protein belongs to a family of NEDD4-like proteins, which are E3 ubiquitin-ligase molecules and regulate key trafficking decisions, including targeting of proteins to proteosomes or lysosomes. Alternative splicing of this gene generates at least 6 transcript variants; however, the full length nature of these transcripts has not been defined. [provided by RefSeq, Jul 2008]. | hsa:11059; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ubiquitin ligase complex [GO:0000151]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; central nervous system development [GO:0007417]; ion transmembrane transport [GO:0034220]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of protein catabolic process [GO:0045732]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; signal transduction [GO:0007165]; viral entry into host cell [GO:0046718] | 12450395_First demonstration of ubiquitin-protein ligase WWP1 from human lung cDNA library recruited by penton base proteins of human adenovirus serotypes Ad2 and Ad3 in vitro and in vivo. 12450395_the interaction of nonenveloped viruses with ubiquitin-protein ligases of host cells. 12535537_Data show that the crystal structure of the HECT domain of the human ubiquitin ligase WWP1/AIP5 maintains a two-lobed structure like the HECT domain of the human ubiquitin ligase E6AP. 15221015_WWP1 negatively regulates TGF-beta signaling in cooperation with Smad7. 16223724_KLF5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells 16785210_Full-length expressed WWP1 could interact in vitro with the cytoplasmic domain of human Notch1, which also regulate the nuclear localization of WWP1. 16924229_these findings identify the first instance of a ubiquitin ligase that causes stabilization of p53 while inactivating its transcriptional activities. 17016436_WWP1 overexpression is a common mechanism involved in the inactivation of TGFbeta function in human cancer. 17330240_genomic aberrations of WWP1 may contribute to the pathogenesis of breast cancer 17609263_the interaction between Gag and WWP1 is required for functions other than Gag ubiquitination 18724389_WWP1 may promote cell proliferation and survival partially through suppressing RNF11-mediated ErbB2 and EGFR downregulation in human cancer cells. 18806757_WWP1 may have a context-dependent role in regulating cell survival through targeting different p63 proteins for degradation. 19035836_analysis of the interaction between ubiquitin ligase WWP1 and Nogo-A 19047365_WWP1 ubiquitinated and caused the degradation of HER4 but not of EGFR, HER2, or HER3. 19267401_Overexpression of WWP1 is associated with the estrogen receptor and insulin-like growth factor receptor 1 in breast carcinoma 19307600_Experiments suggest functions for WWP1 and SPG20 in the regulation of lipid droplet turnover and potential pathological mechanisms in Troyer syndrome. 19561640_WWP1 and its family members suppress the ErbB4 expression and function in breast cancer 19580544_SPG20 interacts with AIP4 and AIP5. 20951678_WWP1 regulates DeltaNp63 transcriptional activity, acting thus as a potential regulator of the proliferation and survival of epithelial-derived cells. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21191027_The authors show that WWP1 changes the ubiquitination status of ARRDC1, suggesting that the arrestins may provide a platform for ubiquitination in PPXY-dependent budding. 21480222_WWP1 depletion-induced apoptosis was rescued by the overexpression of the wild-type WWP1 but not the E3 ligase inactive WWP1-C890A mutant in MCF7 cells. 21795702_WWP1 markedly inhibited the replicative senescence induced by p27(Kip1) by promoting p27(Kip1) degradation. 21996799_WWP1 mutations are not a common cause of human dystroglycanopathy. 22045023_TAZ promotes breast cell growth partially through protecting KLF5 from WWP1-mediated degradation and enhancing KLF5's activities. 22051607_WWP1 may function as the E3 ligase for several PY motif-containing proteins, such as Smad2, KLF5, p63, ErbB4/HER4, RUNX2, JunB, RNF11, SPG20, and Gag, as well as several non-PY motif containing proteins, such as TbetaR1, Smad4, KLF2, and EPS15. Review. 22266093_WWP1 negatively regulates cell migration to CXCL12 by limiting CXCR4 degradation to promote breast cancer metastasis to bone. 23293026_WWP1 down-regulates AMPKalpha2 under high glucose culture conditions via the ubiquitin-proteasome pathway 23573293_LATS1 is critical in mediating WWP1-induced increased cell proliferation in breast cancer cells. 23849376_results reveal that WWP1 might play an oncogenic role in oral cancer cells. 24792179_Knocking down WWP1 promoted cleaved caspase3 protein and p53 expression in hepatocellular carcinoma cells, and caspase3 inhibition could prevent cell apoptosis induced by the knockdown of WWP1. 25051198_Results suggest that elevated transcription and expression levels of ubiquitin ligase E3s WWP1, Smurf1 and Smurf2 genes may be the mechanisms of occurrence, development and metastasis of prostate cancer. 25071155_Data show that WWP1, which specifically ubiquitinates and degrades DeltaNp73 heterodimerizes with another E3 ligase, WWP2. 25293520_Most notably, WWP1 downregulation both inactivated PTEN-Akt signaling pathway in MKN-45 and AGS cells. our findings suggest that WWP1 acts as an oncogenic factor and should be considered as a novel interfering molecular target for gastric carcinoma 25755729_miR-21 overexpression or WWP1 knockdown in endothelial progenitor cells significantly activates the TGFbeta signaling pathway and inhibits cell proliferation. 25814387_PTPN14, a Pez mammalian homolog, is degraded by overexpressed Su(dx) or Su(dx) homologue WWP1 in mammalian cells. 26152726_The cancer-driven alteration of WWP1 culminates in excessive TbetaRI degradation and attenuated TGFbeta1 cytostatic signaling, a consequence that could conceivably confer tumorigenic properties to WWP1. 26506518_Overexpression of WWP1 promotes tumorigenesis in patients with hepatocellular carcinoma. 27070713_Study shows that WWP1 was upregulated in prostate cancer (PCa) clinical specimens and contributed to cancer cell invasion, indicating that this target of Mir-452 functioned as an oncogene. 28426804_Knock-down of WWP1 abrogates DNA damage-induced down-regulation of DeltaNp63alphaand partially rescues cell apoptosis 28431583_Results show that WWP1 is frequently upregulated in gastric cancer (GC) tissues and cells, and that its 3'-UTR is a putative target of miR-584 which suppresses its protein expression by mRNA degradation. 28461335_results demonstrate that WWP1 catalyzes the formation of Ub chains through a sequential addition mechanism, in which Ub monomers are transferred in a successive fashion to the substrate, and that ubiquitination by WWP1 requires the presence of a low-affinity, noncovalent Ub-binding site within the HECT domain. 28475870_Describe an autoinhibitory mechanism for WWP1 ubiquitin ligase involving a linker-HECT domain interaction. This intramolecular interaction traps the HECT enzyme in its inactive state and can be relieved by linker phosphorylation. 28768865_study describes a physical and functional interaction between Ebola virus VP40 (eVP40) and WWP1, a host E3 ubiquitin ligase that ubiquitinates VP40 and regulates virus-like particles egress. 29209041_Provide molecular insights into the anti-cancer potential of WWP1 inhibition. 29635000_Study identified beta-dystroglycan as a substrate of WWP1 and found that the muscular dystrophy-causing mutation of WWP1 renders the enzyme hyperactive by relieving autoinhibition. 29888632_WWP1 expression was significantly associated with histological grade, invasion depth and lymph node metastasis in patients with cutaneous squamous cell carcinoma (CSCC). 30978403_WWP1 overexpression decreased miR-30a-5p expression and inhibited glioma cell malignant behaviors via inhibiting NF-kappaB p65. 31348766_RT-qPCR and Western blot showed that WWP1 was positively regulated by SNHG12 and negatively regulated by miR-129-5p at the mRNA level and protein level. Overexpression of WWP1 significantly increased proliferation and invasion of laryngeal cancer cells. 32459922_n this study involving patients with disorders resulting in a predisposition to the development of multiple malignant neoplasms without PTEN germline mutations, we confirmed the function of WWP1 as a cancer-susceptibility gene through direct aberrant regulation of the PTEN-PI3K signaling axis. 33113605_[Abnormal expression of WWP1 in chronic lymphocytic leukemia and its clinical significance]. 33198785_MicroRNA-15b shuttled by bone marrow mesenchymal stem cell-derived extracellular vesicles binds to WWP1 and promotes osteogenic differentiation. 33470109_Ubiquitin Ligase Activities of WWP1 Germline Variants K740N and N745S. 33516665_BAP1 antagonizes WWP1-mediated transcription factor KLF5 ubiquitination and inhibits autophagy to promote melanoma progression. 33648498_CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. 34035464_miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/beta-catenin signaling pathway. 34139860_Targeting E3 Ubiquitin Ligase WWP1 Prevents Cardiac Hypertrophy Through Destabilizing DVL2 via Inhibition of K27-Linked Ubiquitination. 34404733_AMOTL2 mono-ubiquitination by WWP1 promotes contact inhibition by facilitating LATS activation. 34404764_WWP1 targeting MUC1 for ubiquitin-mediated lysosomal degradation to suppress carcinogenesis. 34559578_Ubiquitin ligase Wwp1 gene deletion attenuates diastolic dysfunction in pressure-overload hypertrophy. 34907909_WWP1 inactivation enhances efficacy of PI3K inhibitors while suppressing their toxicities in breast cancer models. 35154481_alpha-Catulin promotes cancer stemness by antagonizing WWP1-mediated KLF5 degradation in lung cancer. | ENSMUSG00000041058 | Wwp1 | 228.77219 | 0.8191197 | -0.2878538421 | 0.24640272 | 1.248162e+00 | 2.639039e-01 | 9.998360e-01 | No | Yes | 170.64369 | 35.679292 | 2.216438e+02 | 35.632617 |
| ENSG00000124067 | 6560 | SLC12A4 | protein_coding | Q9UP95 | FUNCTION: Mediates electroneutral potassium-chloride cotransport when activated by cell swelling. May contribute to cell volume homeostasis in single cells. May be involved in the regulation of basolateral Cl(-) exit in NaCl absorbing epithelia (By similarity). Isoform 4 has no transport activity. {ECO:0000250}. | 3D-structure;Alternative splicing;Chloride;Glycoprotein;Ion transport;Membrane;Phosphoprotein;Potassium;Potassium transport;Reference proteome;Symport;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the SLC12A transporter family. The encoded protein mediates the coupled movement of potassium and chloride ions across the plasma membrane. This gene is expressed ubiquitously. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Jan 2013]. | hsa:6560; | integral component of plasma membrane [GO:0005887]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; plasma membrane [GO:0005886]; synapse [GO:0045202]; ammonium transmembrane transporter activity [GO:0008519]; potassium:chloride symporter activity [GO:0015379]; protein kinase binding [GO:0019901]; ammonium import across plasma membrane [GO:0140157]; cell volume homeostasis [GO:0006884]; chemical synaptic transmission [GO:0007268]; chloride ion homeostasis [GO:0055064]; chloride transmembrane transport [GO:1902476]; ion transport [GO:0006811]; potassium ion homeostasis [GO:0055075]; potassium ion import across plasma membrane [GO:1990573] | 12637262_human osteoblasts express functional K-Cl cotransporters in their cell membrane that seem to be able to induce the indirect activation of volume-sensitive Cl- channels by KCl through an increase in intracellular ions, water influx and cell swelling. 12902337_loss-of-function KCC mutant cervical cancer cells exhibit inhibited cell growth accompanied by decreased activity of cell cycle gene products 14976052_the KCC1 gene promoter lacks a TATA box and is composed of an initiator element (InR) and a downstream promoter element (DPE) 15039017_Hb polymerisation and sickling could be dissociated from the abnormal response of KCC to deoxygenation observed in HbS-containing red cells. 15262997_KCC activation by IGF-1 plays an important role in IGF-1 signaling to promote growth and spread of gynecological cancers. 19317253_IGF-II (insulin-like growth factors-2) can enhance KCC1 (KC1 co-transport-1) gene expression in cervical cancer cells through signal transduction pathways 19913121_Observational study of gene-disease association. (HuGE Navigator) 20602615_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21613606_The Wnk3 protein isoforms have a similar effect on SLC12 cotransporters. NKCC1/2 and NCC were inhibited, even in hypertonicity, while KCCs were activated, even in isotonic conditions. 21666489_Insulin-like growth factors I can induce the upregulation of KCC1 gene, and KCC1 gene participates in the invasion ability of HEC-1B cells through the ERK signaling pathway. 21733850_KCC3 is the dominant isoform in erythrocytes, with variable expression of KCC1 and KCC4 that could result in modulation of KCC activity 28093242_data indicate that Zn(2+) acting via ZnR/GPR39 has a direct role in controlling Cl(-) absorption via upregulation of basolateral KCC1 in the colon. Moreover, colonocytic ZnR/GPR39 and KCC1 reduce water loss during diarrhea and may therefore serve as effective drug targets. 31296230_The main goal of the review is to discuss the molecular features and tissue-specific functions of KCC1 from the hematological perspective for the greater part. As will be seen, the characterization of KCC1 has led the way to important findings and promising therapeutic avenues. [review] | ENSMUSG00000017765 | Slc12a4 | 365.25020 | 1.1380304 | 0.1865391199 | 0.17578565 | 1.150498e+00 | 2.834450e-01 | 9.998360e-01 | No | Yes | 378.23979 | 49.379078 | 3.124767e+02 | 32.334488 | |
| ENSG00000124275 | 4552 | MTRR | protein_coding | Q9UBK8 | FUNCTION: Key enzyme in methionine and folate homeostasis responsible for the reactivation of methionine synthase (MTR/MS) activity by catalyzing the reductive methylation of MTR-bound cob(II)alamin (PubMed:17892308). Cobalamin (vitamin B12) forms a complex with MTR to serve as an intermediary in methyl transfer reactions that cycles between MTR-bound methylcob(III)alamin and MTR bound-cob(I)alamin forms, and occasional oxidative escape of the cob(I)alamin intermediate during the catalytic cycle leads to the inactive cob(II)alamin species (Probable). The processing of cobalamin in the cytosol occurs in a multiprotein complex composed of at least MMACHC, MMADHC, MTRR and MTR which may contribute to shuttle safely and efficiently cobalamin towards MTR in order to produce methionine (PubMed:27771510). Also necessary for the utilization of methyl groups from the folate cycle, thereby affecting transgenerational epigenetic inheritance (By similarity). Also acts as a molecular chaperone for methionine synthase by stabilizing apoMTR and incorporating methylcob(III)alamin into apoMTR to form the holoenzyme (PubMed:16769880). Also serves as an aquacob(III)alamin reductase by reducing aquacob(III)alamin to cob(II)alamin; this reduction leads to stimulation of the conversion of apoMTR and aquacob(III)alamin to MTR holoenzyme (PubMed:16769880). {ECO:0000250|UniProtKB:Q8C1A3, ECO:0000269|PubMed:16769880, ECO:0000269|PubMed:17892308, ECO:0000269|PubMed:27771510, ECO:0000305|PubMed:19243433}. | 3D-structure;Alternative splicing;Amino-acid biosynthesis;Cytoplasm;Disease variant;FAD;FMN;Flavoprotein;Methionine biosynthesis;NADP;Oxidoreductase;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine | This gene encodes a member of the ferredoxin-NADP(+) reductase (FNR) family of electron transferases. This protein functions in the synthesis of methionine by regenerating methionine synthase to a functional state. Because methionine synthesis requires methyl-group transfer by a folate donor, activity of the encoded enzyme is important for folate metabolism and cellular methylation. Mutations in this gene can cause homocystinuria-megaloblastic anemia, cbl E type. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Dec 2015]. | hsa:4552; | cytosol [GO:0005829]; intermediate filament cytoskeleton [GO:0045111]; nucleoplasm [GO:0005654]; [methionine synthase] reductase activity [GO:0030586]; FAD binding [GO:0071949]; flavin adenine dinucleotide binding [GO:0050660]; FMN binding [GO:0010181]; NADPH binding [GO:0070402]; NADPH-hemoprotein reductase activity [GO:0003958]; oxidoreductase activity [GO:0016491]; oxidoreductase activity, acting on metal ions, NAD or NADP as acceptor [GO:0016723]; DNA methylation [GO:0006306]; folic acid metabolic process [GO:0046655]; homocysteine catabolic process [GO:0043418]; homocysteine metabolic process [GO:0050667]; methionine biosynthetic process [GO:0009086]; negative regulation of cystathionine beta-synthase activity [GO:1904042]; S-adenosylmethionine cycle [GO:0033353] | 10791559_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, gene-environment interaction, and healthcare-related. (HuGE Navigator) 11472746_Observational study of gene-disease association. (HuGE Navigator) 11592436_Observational study of gene-disease association. (HuGE Navigator) 11806787_Observational study of gene-disease association. (HuGE Navigator) 11807890_Observational study of gene-disease association. (HuGE Navigator) 12020105_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12375236_Variants influence the risk of spina bifida via the maternal rather than the embryonic genotype. 12416982_The kinetic and spectroscopic properties of the M22/S175 and I22/S175 and the I22/L175 and I22/S175 pairs of polymorphic variants of MSR have been compared. 12482550_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12590188_polymorphisms should be regarded as independent risk factors for spina bifida 12642343_Observational study of gene-disease association. (HuGE Navigator) 12649067_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12649067_results indicate that MTRR and MTR genes may interact to increase the infants' Neural tube defects risks 12716294_Observational study of gene-disease association. (HuGE Navigator) 12801615_No differences in mean homocysteine, prevalence of hyperhomocysteinemia and significant coronary artery disease between genotypes AA, AG, GG. 12801615_Observational study of gene-disease association. (HuGE Navigator) 12807760_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12810988_Results of screening mutations 2756A-->G and 66A-->G in MTR and MTRR genes respectively show that are might have an effect on NTDs incidence (neural tube defects). 12855226_Observational study of gene-disease association. (HuGE Navigator) 12876480_Methionine synthase polymorphism is a risk factor for Alzheimer disease. 12923861_Observational study of gene-disease association. (HuGE Navigator) 12923861_polymorphisms in methionine synthase reductase is associated with increased risk for Down syndrome 12939653_Observational study of gene-disease association. (HuGE Navigator) 14632302_Observational study of genotype prevalence. (HuGE Navigator) 14652285_Observational study of gene-disease association. (HuGE Navigator) 14716779_Observational study of genotype prevalence. (HuGE Navigator) 14967039_Common MSR polymorphisms, I22/L175 and M22/S175, which result in less effective methionine synthase activation, do not have effects on flavin potentials or electron transfer kinetics that would impinge on catalytic efficiency of the variants. 14977639_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 15059614_Observational study of gene-disease association. (HuGE Navigator) 15135249_Observational study of gene-disease association. (HuGE Navigator) 15159311_MTRR polymorhism in the genes for folate and methionine metabolism might play a role in the occurance in patient with ALL and NHL. 15159311_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15216546_Observational study of gene-disease association. (HuGE Navigator) 15347655_Results describe two common polymorphic variants of ATP:cob(I)alamin adenosyltransferase that are found in normal individuals, and their interactions with methionine synthase reductase. 15354395_Observational study of gene-disease association. (HuGE Navigator) 15514263_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15514263_findings indicate that when the homozygous variant for the MTHFR 677CT polymorphism coexists with the MTRR 66 AG genotype, plasma homocysteine is significantly elevated relative to the other genotype groups 15514969_Polymorphisms in the folate metabolic pathway were associated with a lower likelihood of Cervical intrepithelial neoplsia. 15612980_Observational study of gene-disease association. (HuGE Navigator) 15612980_SNPs in this enzyme affects homocysteine-regulating genes and may be important determinants of vitamin metabolism in heart transplantation. 15714522_3 new MTRR mutations (c.7A>T, c.1573C>T, and c.1953-6_1953-2del5) were detected. The identification of mutations in MTRR, & restoration of methionine synthesis following MTRR minigene expression confirms that MTRR gene defects cause cblE homocystinuria. 15797993_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15866085_Observational study of gene-disease association. (HuGE Navigator) 15889417_Observational study of gene-disease association. (HuGE Navigator) 15894670_Finds MTRR variant AA genotype associated with a significantly decreased squamous cell head and neck cancer risk. 15894670_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15979034_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16006998_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16013960_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16115349_Observational study of gene-disease association. (HuGE Navigator) 16268464_Observational study of gene-disease association. (HuGE Navigator) 16316363_Observational study of gene-disease association. (HuGE Navigator) 16351505_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16470748_Observational study of gene-disease association. (HuGE Navigator) 16485733_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16575899_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16580699_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16580699_the MTRR AA genotype acts to increase the micronuclei frequency resulting from cigarette smoking 16769880_we propose that MSR serves as a special chaperone for human methionine synthase and as an aquacobalamin reductase, rather than acting solely in the reductive activation of MS 16820193_Observational study of gene-disease association. (HuGE Navigator) 16861746_Observational study of gene-disease association. (HuGE Navigator) 16861746_we showed the first genetic evidence that MTHFR C677T, MS A2756G and MTRR A66G genotypes were independently associated with male infertility. Each SNP of the three enzymes may have a different impact on the folate cycle during spermatogenesis 16894458_Observational study of gene-disease association. (HuGE Navigator) 16947783_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16985020_Clinical trial of gene-disease association. (HuGE Navigator) 17009228_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17024475_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17024475_The MTRR polymorphism is a maternal risk factor for spina bifida. 17035141_Observational study of gene-disease association. (HuGE Navigator) 17074544_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17079868_66G/524C haplotype of the MTRR gene affect bone turn over rate. 17079868_Observational study of gene-disease association. (HuGE Navigator) 17087642_Data indicate that maternal MTRR 66A>G polymorphism is not a risk factor for congenital heart defects (CHD), but maternal MTRR 66GG genotype with compromised vitamin B(12) status may possibly result in increased CHD risk. 17087642_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17101561_Observational study of genetic testing. (HuGE Navigator) 17113603_Observational study of gene-disease association. (HuGE Navigator) 17119116_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17136115_Observational study of gene-disease association. (HuGE Navigator) 17152488_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17220339_Observational study of gene-disease association. (HuGE Navigator) 17311259_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17311260_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17376725_Distributions for the homozygous mutant form of MTRR were similar between cases and controls, polymorphisim did not increase the risk of placental abruption. 17376725_Observational study of gene-disease association. (HuGE Navigator) 17417062_Since MTR genes are located in 1q43 loci, our findings support the significance of chromosome 1q in etiopathogenesis of bipolar disorder and schizophrenia. 17436311_Observational study of gene-disease association. (HuGE Navigator) 17454638_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17461517_MTRR 2756A > G polymorphisms interact with elevated total homocysteine levels, leading to an increased risk of ischemic stroke. 17477549_Structure and function of activation and conformational heterogeneity of recombinant proteins. 17522601_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17522601_The interaction between low levels of serum cobalamin and MTHFR (C677T or A1298C) or MTRR A66G gene polymorphisms was associated with increased tHcy. 17533396_Observational study of gene-disease association. (HuGE Navigator) 17546637_MTRR polymorphism may play an important role in the risk of multiple myeloma. 17546637_Observational study of gene-disease association. (HuGE Navigator) 17553479_Observational study of gene-disease association. (HuGE Navigator) 17553479_These results not only highlight the involvement of the MSR and CBS genes in the etiology of cardiovascular disease, but also emphasize the strength of haplotype analyses in association studies. 17554763_methionine synthase reductase modulates the phenotype of a disease-causing mutation 17581676_Observational study of gene-disease association. (HuGE Navigator) 17581676_Results suggest that the alterations of folate metabolism related to MTHFR, MTR and MTRR polymorphisms are not involved in clefting in South Brazil. 17596206_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17611986_In our population of methotrexate (MTX)-treated rheumatoid arthritis patients the 2756GG genotype of the methionine synthase reductase gene was more common than expected and was associated with MTX-induced accelerated rheumatoid nodulosis. 17636160_Observational study of gene-disease association. (HuGE Navigator) 17655928_Observational study of gene-disease association. (HuGE Navigator) 17655928_similar frequencies of the MTHFR, the MTRR and the TYMS genotypes were seen in patients and controls 17822659_Observational study of gene-disease association. (HuGE Navigator) 17853476_Observational study of gene-disease association. (HuGE Navigator) 17853476_The risk of having a child with congenital malformation or FACS was three to four times higher for mothers who were MTHFR 677TT homozygotes compared with MTHFR 677CC homozygotes 17892308_Mechanism of coenzyme binding to human MSR revealed through the crystal structure of the FNR-like module and isothermal titration calorimetry. 17904392_Observational study of gene-disease association. (HuGE Navigator) 17925002_MTRR polymorphism is related with disease activity in Crohn's disease 17925002_Observational study of gene-disease association. (HuGE Navigator) 17934692_Meta-analysis of gene-disease association and gene-gene interaction. (HuGE Navigator) 17967524_Observational study of gene-disease association. (HuGE Navigator) 17967524_no association between prostate cancer and the MTRR A66C polymorphism 17993766_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17993766_These findings suggest that in obese subjects, homocysteine cycle efficiency is impaired by MTHFR, MTR, and MTRR inability to supply methyl-group donors, providing evidence that MTHFR, MTR, and MTRR gene polymorphisms are genetic risk factors for obesity. 18004208_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18023275_Observational study of gene-disease association. (HuGE Navigator) 18034637_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18034637_The MTRR 66GG genotype was found to be associated with 2.74-fold risk (95% CI: 1.73, 4.34) for deep vein thrombosis among South Indians. This risk is increased further to 3.46-fold (95% CI: 1.38, 8.63) in the presence of the MTHFR 677CT/1298AC genotype. 18061941_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18174236_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18174236_the MTHFR and MTRR polymorphisms are associated with individual susceptibility to breast cancer among postmenopausal women 18199722_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18204969_Observational study of gene-disease association. (HuGE Navigator) 18221906_MSR protein is restricted to the cytosol but, based on the Leal study, suggest that a similar protein may interact with MMAB to reduce the mitochondrial cobalamin substrate in the generation of adenosylcobalamin 18222012_Observational study of gene-disease association. (HuGE Navigator) 18226574_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18249021_Observational study of gene-disease association. (HuGE Navigator) 18257130_MTHFR and MTRR gene mutation alleles are related to Down syndrome, and CT, TT and GG gene mutation types increase the risk of Down syndrome 18257130_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18378576_Observational study of gene-disease association, gene-gene interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18386810_Observational study of gene-disease association. (HuGE Navigator) 18427977_Observational study of genetic testing. (HuGE Navigator) 18435414_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18483342_Observational study of gene-disease association. (HuGE Navigator) 18483342_The MTRR A66G and GG genotypes were significantly associated with risk of meningioma but not glioma. 18485163_Observational study of gene-disease association. (HuGE Navigator) 18510611_Observational study of gene-disease association. (HuGE Navigator) 18515090_Observational study of gene-disease association. (HuGE Navigator) 18515090_Study suggested a common missense SNP of the MTRR gene as a novel pancreatic cancer susceptibility factor with a functional significance in folate-related metabolism and the genome-wide methylation status. 18590621_Observational study and meta-analysis of genotype prevalence. (HuGE Navigator) 18607581_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18610829_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18610829_polymorphic variants of genes GSTT1, GSTM1, NAT2 and MTRR can modulate the risk of childhood acute leukemia, residents of European part of Russia. 18614746_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18614746_We found a significant interaction between MTHFR 677C-->T and MTRR 66A-->G on serum homocysteine concentrations among non-Hispanic whites. 18635682_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18682255_Observational study of gene-disease association. (HuGE Navigator) 18700049_Observational study of gene-disease association. (HuGE Navigator) 18771981_Observational study of gene-disease association. (HuGE Navigator) 18774170_Observational study of gene-disease association. (HuGE Navigator) 18774994_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18774994_The MTRR 66GG and MTHFR 1298 CC genotypes may confer protection against early nephropathy 18785313_Observational study of gene-disease association. (HuGE Navigator) 18792976_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18792976_study evaluated the importance of the polymorphisms in the MTHFR, MTRR, and CBS genes for hyperhomocysteinemia, considering B12 and folate levels; results confirm that genetic interactions can influence on the homocysteine status 18830263_Observational study of gene-disease association. (HuGE Navigator) 18836720_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18842997_Observational study of gene-disease association. (HuGE Navigator) 18843018_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18843018_Risk of pancreatic cancer was increased with alcohol consumption in subjects with the MTRR 66 G allele. 18936436_Observational study of genotype prevalence. (HuGE Navigator) 18978678_Observational study of gene-disease association. (HuGE Navigator) 18983896_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18988749_Observational study of genetic testing. (HuGE Navigator) 18992148_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19019492_Observational study of gene-disease association. (HuGE Navigator) 19020309_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19035314_The MTHFR 1298A>C and the MTRR 66A>G genotypes were associated with an increased risk of hepatocellular carcinoma in this Korean population. 19048631_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19064571_Observational study of gene-disease association. (HuGE Navigator) 19064578_Observational study of gene-disease association. (HuGE Navigator) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19112534_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19161160_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19254790_In a population with a high prevalence of the mutated T allele, maternal MTRR A66G, but not MTHFR, polymorphisms are associated with Down syndrome. 19254790_Observational study of gene-disease association. (HuGE Navigator) 19263808_relationship between occurrence of hyperlipidemia, plasma homocysteine and polymorphisms of methylenetetra hydrofolate reductase (MTHFR) gene and methionine synthase (MS) gene 19336370_Observational study of gene-disease association. (HuGE Navigator) 19336437_Observational study of gene-disease association. (HuGE Navigator) 19339913_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19339913_The MTRR 66A>G gene variant is not associated with peak elevated postoperative plasma total homocysteine after nitrous oxide anesthesia. 19348062_MTRR A66G polymorphism was not associated with the total homocysteine plasma concentration because the same genotype frequency distribution was detected in both patients and healthy individuals. 19348062_Observational study of gene-disease association. (HuGE Navigator) 19353223_Observational study of gene-disease association. (HuGE Navigator) 19394322_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19427504_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19461557_Observational study of gene-disease association. (HuGE Navigator) 19493349_Observational study of gene-disease association. (HuGE Navigator) 19533788_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19657388_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19706844_Meta-analysis of gene-disease association. (HuGE Navigator) 19729796_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19737740_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19760026_Observational study of gene-disease association. (HuGE Navigator) 19760026_there was no significant association between most SNPs in methionine synthase reductase , and the risk of cervical intraepithelial neoplasia and cervical cancer in Korean women. 19774638_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19776626_Polymorphisms in methionine synthase, methionine synthase reductase and serine hydroxymethyltransferase, folate and alcohol intake, and colon cancer risk. 19776634_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19777601_Observational study of gene-disease association. (HuGE Navigator) 19812220_Observational study of gene-disease association. (HuGE Navigator) 19837268_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19843671_Observational study of gene-disease association. (HuGE Navigator) 19852428_Molecular analysis of 12 genetic polymorphisms involved in the folate metabolism revealed that the mother is heterozygous for the MTHFR C677T and TC2 A67G polymorphisms, and homozygous for the mutant MTRR A66G polymorphism. 19858780_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19936946_Observational study of gene-disease association. (HuGE Navigator) 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20031554_Observational study of gene-disease association. (HuGE Navigator) 20047525_A66G polymorphism not significantly associated with increased risk for neural tube defects 20047525_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20056620_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20082058_Observational study of gene-disease association. (HuGE Navigator) 20085490_Meta-analysis of gene-disease association. (HuGE Navigator) 20099281_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20120036_A deep intronic mutation is the direct cause of MTRR pseudoexon inclusion and the pseudoexon is normally not recognized due to a suboptimal 5' splice site. 20140262_Observational study of gene-disease association. (HuGE Navigator) 20180013_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20209990_Observational study of gene-disease association. (HuGE Navigator) 20209990_Patients with hyperhomocysteinemia (HHcy) had statistically significant increase of allele MTHFR 677T and MTRR 66GG as compared both with the control group and with the group of patients without HHcy. 20373852_Observational study of gene-disease association. (HuGE Navigator) 20378615_Observational study of gene-disease association. (HuGE Navigator) 20386493_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20411324_Meta-analysis of gene-disease association. (HuGE Navigator) 20411324_Meta-analysis strongly suggests that MTRR A66G polymorphism is not associated with breast cancer risk, especially in Caucasians and Asians. 20437058_Observational study of genetic testing. (HuGE Navigator) 20447924_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20458436_Observational study of gene-disease association. (HuGE Navigator) 20466634_Data show that MTHFR 677C>T and MTRR 66A>G polymorphisms are two independent risk factors for DS pregnancies in young women, but RFC-1 80G>A and MTR 2756A>G polymorphism are not independent risk factor. 20466634_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20511665_Observational study of gene-disease association. (HuGE Navigator) 20544798_Observational study of gene-disease association. (HuGE Navigator) 20549016_Germ line polymorphisms in the folate- and methyl-associated genes MTHFR, MTR and MTRR, were analyzed in colorectal cancer patient cohort to find a possible link between these genetic variants and p16 hypermethylation. 20549016_Observational study of gene-disease association. (HuGE Navigator) 20615890_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20647221_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20718043_Observational study of gene-disease association. (HuGE Navigator) 20726305_study results do not demonstrate an association between A66G MTRR polymorphism and colorectal cancer or breast cancer in Romanian patients 20737570_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20852008_Observational study of gene-disease association. (HuGE Navigator) 20883119_Observational study of gene-disease association. (HuGE Navigator) 20888556_No association was found between MTHFR/MTRR genetic variants and sperm counts. 20888556_Observational study of gene-disease association. (HuGE Navigator) 20948192_Observational study of gene-disease association. (HuGE Navigator) 20955826_The MTHFR 677C>T SNP and the MTRR 66A >G SNP were identified as determinants of impaired bone mineral density (total body) in childhood acute lymphoblastic leukemia patients. 20960050_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20962453_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21041608_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21045269_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21055808_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 21070756_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21070756_the synergistic effects of polymorphisms in the folate metabolic pathway genes in Parkinson's disease susceptibility 21092627_These results suggest that MTRR G66A polymorphism may play a role in coronary artery disease susceptibility. 21204909_5-Fluorouracil-based chemotherapy for colorectal cancer and MTHFR/MTRR genotypes 21211571_No difference in the distribution between cases and controls was observed for the haplotypes based on the four polymorphisms in the MTRR gene. 21349258_The genotype and allele frequencies of the MTR Asp919Gly, MTHFR Ala222Val, MTHFD1 Arg653Gln and MTRR Ile22Met gene variants did not display statistical differences between patients with cervical cancer and controls. 21438757_Linkage disequilibrium (LD) analysis found possible protective role of MTRR 66 AA in sporadic colon cancer. The significant LD between two loci (MTHFR A1298C and MTRR A66G) located on different chromosomes indicates a selective force for their linkage. 21472912_ELDOR spectroscopy was used to identify multiple conformational intermediates in methionine synthase reductase and analyse their relative stabilities. 21547363_MTRR A66G polymorphism is a potential biomarker for cancer risk. 21603981_These data suggest that all polymorphisms coding for MTHFR, MTR, MTRR and TS have consistent roles in the increased risk of sporadic colorectal adenocarcinoma among the southeastern population of Brazil. 21774403_The heterozygous genotype MTRR 66AG was associated with the 5.56-fold increased CL/P risk (OR = 5.56) and for mothers with 2.6-fold increased risk of delivering a CL/P offspring (OR = 2.6). 21775772_MTRR polymorphisms did not appear to be an important genetic factor predisposing to idiopathic infertility in Brazilian men. 21780915_MTR A2756G polymorphism, but not MTRR A66G and MTHFR A1298C, is associated with risk of coronary heart disease for Europeans. 21947961_The variant allele and genotypic frequencies in MTRR A66G gene was significantly higher in patients with UC compared to healthy controls. 21987236_MTRR A66G and cSHMT C1420T polymorphisms influence CpG island methylator phenotype of BNIP3, thus epigenetically regulating BNIP3 in breast cancer 22005284_Constituents of the folate cycle could be involved in the etiology of idiopathic intellectual disability. 22057956_MTRR A66G and C524T polymorphisms are associated with increased risk of CHDs. 22097960_Data indicate the while W697 in MSR attenuates hydride transfer, it ensures coenzyme selectivity and accelerates FAD to FMN electron transfer (i.e., controls kinetics of interflavin electron transfer). 22179537_We have demonstrated that the MTRR c.56+781 A>C variant is an important genetic marker for increased congenital heart disease risk because this variant results in functionally reduced MTRR expression at the transcriptional level. 22236648_the single-nucleotide polymorphism A66G MTRR is not involved in the development of breast cancer. 22339686_Known common single-nucleotide polymorphisms in MTRR and BHMT genes may not be significant risk factors for cororonary artery disease. 22373582_Studies suggest that polymorphisms in methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) modulate the risk for breast cancer, particularly the A1298C polymorphism of the MTHFR gene. 22475273_The 66GG and AG genotypes were associated with decreased odds ratios for heart defects. This overall association was driven by decreased risk for ventricular septal defect for 66GG and AG and decreased odds ratio for aortic valve stenosis for 66AG. 22479380_Data indicate that genetic variants in folate pathway genes showed associations including infant MTRR 66G>A genotype and maternal MTHFR 677C>T genotype with IGF2 methylation. 22665368_studies suggest that SNPs in CBS and MTRR have sex-specific associations with aberrant methylation in the lung epithelium of smokers that could be mediated by the affected one-carbon metabolism and transsulfuration in the cells 22706675_MTHFR and MTRR genetic polymorphisms were not associated with childhood ALL. However, AL was positively associated with homozygosity for any of the MTHFR polymorphisms and carriership of both MTRR variant alleles. 22719222_Meta-analysis suggested that MTHFR 677T allele might provide protection against CRC in worldwide populations, while MTRR 66G allele might increase the risk of CRC in Caucasians. 22796266_Results indicate the importance of four gastric cancer susceptibility polymorphisms of IL-10, NOC3L, PSCA and MTRR in the Chinese Han population. 22813657_MTHFR and MTRR polymorphisms are associated with susceptibility to schizophrenia and support th | ENSMUSG00000034617 | Mtrr | 395.29991 | 1.0688984 | 0.0961247216 | 0.18848961 | 2.549538e-01 | 6.136084e-01 | 9.998360e-01 | No | Yes | 340.59145 | 69.530551 | 3.584694e+02 | 56.386045 | |
| ENSG00000124508 | 10385 | BTN2A2 | protein_coding | Q8WVV5 | FUNCTION: Inhibits the proliferation of CD4 and CD8 T-cells activated by anti-CD3 antibodies, T-cell metabolism and IL2 and IFNG secretion. {ECO:0000250}. | Alternative splicing;Coiled coil;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | Butyrophilin is the major protein associated with fat droplets in the milk. This gene is a member of the BTN2 subfamily of genes, which encode proteins belonging to the butyrophilin protein family. The gene is located in a cluster on chromosome 6, consisting of seven genes belonging to the expanding B7/butyrophilin-like group, a subset of the immunoglobulin gene superfamily. The encoded protein is a type I receptor glycoprotein involved in lipid, fatty-acid and sterol metabolism. Several alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2010]. | hsa:10385; | external side of plasma membrane [GO:0009897]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; signaling receptor binding [GO:0005102]; negative regulation of activated T cell proliferation [GO:0046007]; negative regulation of cytokine production [GO:0001818]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; negative regulation of phosphatidylinositol 3-kinase signaling [GO:0014067]; negative regulation of protein kinase B signaling [GO:0051898]; negative regulation of T cell receptor signaling pathway [GO:0050860]; positive regulation of regulatory T cell differentiation [GO:0045591]; regulation of cytokine production [GO:0001817]; T cell receptor signaling pathway [GO:0050852] | 26809444_Genes encoding butyrophilin-2A2 (BTN2A2) are regulated by the class II trans-activator and regulatory factor X, two transcription factors dedicated to major histocompatibility complex class II expression, suggesting a role in T cell immunity. | ENSMUSG00000053216 | Btn2a2 | 369.90365 | 1.1320270 | 0.1789083213 | 0.17342425 | 1.059826e+00 | 3.032548e-01 | 9.998360e-01 | No | Yes | 365.46244 | 61.926206 | 3.693310e+02 | 48.393796 | |
| ENSG00000124587 | 5190 | PEX6 | protein_coding | Q13608 | FUNCTION: Involved in peroxisome biosynthesis. Required for stability of the PTS1 receptor. Anchored by PEX26 to peroxisome membranes, possibly to form heteromeric AAA ATPase complexes required for the import of proteins into peroxisomes. | ATP-binding;Alternative splicing;Amelogenesis imperfecta;Cell projection;Cytoplasm;Deafness;Disease variant;Membrane;Methylation;Nucleotide-binding;Peroxisome;Peroxisome biogenesis;Peroxisome biogenesis disorder;Reference proteome;Repeat;Zellweger syndrome | This gene encodes a member of the AAA (ATPases associated with diverse cellular activities) family of ATPases. This member is a predominantly cytoplasmic protein, which plays a direct role in peroxisomal protein import and is required for PTS1 (peroxisomal targeting signal 1, a C-terminal tripeptide of the sequence ser-lys-leu) receptor activity. Mutations in this gene cause peroxisome biogenesis disorders of complementation group 4 and complementation group 6. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2015]. | hsa:5190; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; photoreceptor cell cilium [GO:0097733]; photoreceptor outer segment [GO:0001750]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; protein C-terminus binding [GO:0008022]; protein-containing complex binding [GO:0044877]; peroxisome organization [GO:0007031]; protein import into peroxisome matrix [GO:0016558]; protein import into peroxisome matrix, translocation [GO:0016561]; protein stabilization [GO:0050821]; protein targeting to peroxisome [GO:0006625] | 11873320_A PEX6-defective peroxisomal biogenesis disorder with severe phenotype in an infant, versus mild phenotype resembling Usher syndrome in the affected parents 16257970_Insufficient binding to Pex1p x Pex6p complexes, or mislocalization of patient-derived Pex26p mutants is most likely responsible for the complementation group impaired peroxisome biogenesis. 19105186_the relative fraction of disease-causing alleles that occur in the coding and splice junction sequences of PEX6 gene. 19433277_Leigh syndrome presenting the T8993G mutation in the ATPase 6 gene with variable heteroplasmic loads (44-98%) in a single Tunisian family is a novel finding. 19877282_We identified a total of 77 different mutations in Zellweger syndrome patients of which 47 mutations have not been reported previously, and 14 polymorphic variants. 21778671_hybrid exercise increases expression of eukaryotic translation initiation factor 5A (EIFSA), peroxisomal biogenesis factor 6 (PEX6) and histone cluster 1 H4 (HIST1H4), compared with electrical stimulation alone 22894767_increased incidence of Zellweger syndrome in French-Canadians of Lac-St-Jean region caused by a PEX6 founder mutation 25016021_results suggested that peroxisome biogenesis requires Pex1p- and Pex6p-regulated dissociation of Pex14p from Pex26p 26387595_Mutations in PEX6 gene is associated with Heimler Syndrome. 26476099_Our structural data suggest that the tilting of a central segment of a Pex1-Pex6 pair is responsible for polypeptide movement. 26593283_PEX6 is expressed in photoreceptor cilia and mutated in deafblindness with enamel dysplasia and microcephaly. 26669662_PEX6 identified as the 6p21 SCABD gene in a family with spinocerebellar ataxia with blindness and deafness. 27302843_Heimler syndrome is due to four novel and two known missense variants and an 8 bp deletion in PEX6 in five families. 27633571_As standard biochemical screening of blood for evidence of a peroxisomal disorder did not provide a diagnosis in the individuals with Heimler syndrome, patients with sensorineural hearing lossand retinal pigmentation should have mutation analysis of PEX1 and PEX6 genes. 28033303_the current study revealed novel expression quantitative trait loci (eQTLs) for SNHG5 and PEX6 genes in chromosome 6. Nucleotide substitutions of the eQTLs might be candidate factors for a variety of cancers by regulating expression of the 2 genes. 29047053_A novel homozygous PEX6 p.Ala94Pro mutation. 29220678_Overexpression models confirmed that the overrepresentation of the pathogenic PEX6 c.2578T variant compared to wild-type PEX6 c.2578C results in a peroxisome biogenesis defect and thus constitutes the cause of disease in the affected individuals. 29676688_Mutation in PAX6 gene is associated with Ophthalmic manifestations of Heimler syndrome. 29884772_This study provides evidence suggesting that monoubiquitinated PEX5 interacts directly with both PEX1 and PEX6 through its ubiquitin moiety and that the PEX5 polypeptide chain is globally unfolded during the ATP-dependent extraction event. 31374812_This article reviews the abundant records of missense mutations described in Peroxisome biogenesis disorders patients with the aim to classify and rationalize them by mapping them onto a homology model of the human Pex1/Pex6 complex. [review] 31831025_There are no significant differences between PEX1-, PEX6-, and PEX26-associated phenotypes inclinical and genetic spectrum of Heimler syndrome. 32214787_Exome sequencing identifies PEX6 mutations in three cases diagnosed with Retinitis Pigmentosa and hearing impairment. 32866347_The peroxisomal disorder spectrum and Heimler syndrome: Deep phenotyping and review of the literature. | ENSMUSG00000002763 | Pex6 | 400.13668 | 1.0405121 | 0.0572936918 | 0.16386138 | 1.218847e-01 | 7.269993e-01 | 9.998360e-01 | No | Yes | 410.63547 | 48.621693 | 3.623292e+02 | 33.780634 | |
| ENSG00000125787 | 2797 | GNRH2 | protein_coding | O43555 | FUNCTION: Stimulates the secretion of gonadotropins; it stimulates the secretion of both luteinizing and follicle-stimulating hormones. | Alternative splicing;Amidation;Cleavage on pair of basic residues;Hormone;Reference proteome;Secreted;Signal | This gene is a member of the gonadotropin-releasing hormone (GnRH) gene family. Proteins encoded by members of this gene family are proteolytically cleaved to form neuropeptides which, in part, regulate reproductive functions by stimulating the production and release of the gonadotropins follicle-stimulating hormone (FSH) and luteinizing hormone (LH). The human GNRH2 gene is predicted to encode a preproprotein from which a mature neuropeptide of 10 amino acids is cleaved. However, while the human genome retains the sequence for a functional GNRH2 decapeptide, translation of the human GNRH2 gene has not yet been demonstrated and the GNRH2 gene of chimpanzees, gorilla, and Sumatran orangutan have a premature stop at codon eight of the decapeptide sequence which suggests GNRH2 was a pseudogene in the hominid lineage. The GNRH2 gene is also believed to be a pseudogene in many other mammalian species such as mouse and cow. The receptor for this gene (GNRHR2) is predicted to be a pseudogene in human as well as many other mammalian species. The closely related GNRH1 and GNRHR1 genes are functional in human and other mammals and are generally functional in vertebrates. [provided by RefSeq, Mar 2019]. | hsa:2797; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; gonadotropin hormone-releasing hormone activity [GO:0005183]; gonadotropin-releasing hormone receptor binding [GO:0031530]; hormone activity [GO:0005179]; reproduction [GO:0000003]; signal transduction [GO:0007165] | 12447356_GnRH-II and GnRH-I interact directly with T cells and trigger gene transcription, adhesion, chemotaxis and homing to specific organs, which may be of clinical relevance. 12663744_a novel mechanism in stimulating basal human GnRH-II gene transcription mediated by cooperative actions of multiple regulatory elements within the untranslated first exon of the gene. 12770744_regulation of GnRH-I and GnRH-II gene expression in the ovary 12969578_gonadotropin releasing hormone-II is more effective than gonadotropin releasing hormone-I in stimulating leptin secretion 14726258_GnRH-II has been identified, together with a 'putative' type II GnRH receptor, both in the central nervous system and in peripheral structures, such as tissues of the reproductive tract (both normal and tumoral). 15001648_GnRH I and GnRH II have both common and discrete cellular distributions in the placenta and decidua and suggest that these two hormones are capable of eliciting their biological actions in an autocrine and/or paracrine manner 15562029_Progesterone receptor isoform p4 is a potent regulator of GnRHRI at the transcriptional level as well as GnRH I mRNA. 15578334_GnRH-II did not affect proliferation of normal B-cells alone, and did not alter the proliferative response to IL-2. 17202595_we observed that LHRH-(1-5), the specific processed peptide of LHRH-I, upregulates LHRH-II mRNA expression in Ishikawa cells, an endometrial cell line but does not exert any influence on LHRH-I mRNA levels 17296196_Expression of GnRH2 protein and receptor (GnRHR2) is confirmed in both leiomyoma and patient-matched myometrium. 17451432_these data demonstrate the differential roles of NF-kappaB p65 and RARalpha/RXRalpha, interacting with the same sequence in the promoter of the human GnRH II gene to influence gene expression in a cell-specific manner. 17914092_GnRH II is widely expressed in prostate cancer and is an AR-regulated gene. 18467526_analysis of how GNRH I and GNRH II inhibit cell growth 18477660_GnRH-I and -II induce apoptosis in human granulosa cells through GnRH-I receptors, which mediate the proteolytic caspase cascade involving caspase-8 (the initiator) and caspase-3 and -7 (the effectors). 18599546_p-CREB, C/EBPbeta, and CBP are recruited to the CRE of the GnRH-II promoter in a temporarily defined manner to enhance its transcription in JEG-3 and OVCAR-3 cells in response to cAMP 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19608641_These results provide evidence that EGF is an upstream regulator of the autocrine actions of GnRH-II on the invasive properties of ovarian cancer cells. 20018426_These preliminary data suggest that local GnRH-II may participate in the regulation of ovarian tumor growth in post-menopausal women. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21193558_results suggest GnRH-II-induced laminin receptor precursor expression increases 67-kDa nonintegrin laminin receptor, which appears to interact with laminin in the extracellular matrix to promote MMP-2 expression and enhance ovarian cancer cell invasion 21239435_concluded that GnRHII stimulates PI3K/Akt pathway, and the phosphorylation of glycogen synthase kinase3beta, thereby enhancing the beta-catenin-dependent up-regulation of MT1-MMP production which contributes to ovarian cancer metastasis 23287110_GnRHII expression correlated significantly with poor prognosis in breast cancer patients. 28941922_rs6051545 (GNRH2) genetic variation may result in inadequate suppression of serum testosterone during androgen deprivation therapy. This may lead to detrimental effects of androgen deprivation therapy on prognosis in men with metastatic prostate cancer. 32994424_The effects of common variants in MDM2 and GNRH2 genes on the risk and survival of osteosarcoma in Han populations from Northwest China. 33114333_Copper(II) Binding by the Earliest Vertebrate Gonadotropin-Releasing Hormone, the Type II Isoform, Suggests an Ancient Role for the Metal. | 11.63999 | 1.2707183 | 0.3456442544 | 0.91997749 | 1.333550e-01 | 7.149785e-01 | 9.998360e-01 | No | Yes | 11.75207 | 4.901899 | 7.343873e+00 | 2.453482 | |||
| ENSG00000125844 | 6238 | RRBP1 | protein_coding | Q9P2E9 | FUNCTION: Acts as a ribosome receptor and mediates interaction between the ribosome and the endoplasmic reticulum membrane. {ECO:0000250}. | Acetylation;Alternative splicing;Endoplasmic reticulum;Isopeptide bond;Membrane;Phosphoprotein;Protein transport;Reference proteome;Repeat;Translocation;Transmembrane;Transmembrane helix;Transport;Ubl conjugation | This gene encodes a ribosome-binding protein of the endoplasmic reticulum (ER) membrane. Studies suggest that this gene plays a role in ER proliferation, secretory pathways and secretory cell differentiation, and mediation of ER-microtubule interactions. Alternative splicing has been observed and protein isoforms are characterized by regions of N-terminal decapeptide and C-terminal heptad repeats. Splicing of the tandem repeats results in variations in ribosome-binding affinity and secretory function. The full-length nature of variants which differ in repeat length has not been determined. Pseudogenes of this gene have been identified on chromosomes 3 and 7, and RRBP1 has been excluded as a candidate gene in the cause of Alagille syndrome, the result of a mutation in a nearby gene on chromosome 20p12. [provided by RefSeq, Apr 2012]. | hsa:6238; | endoplasmic reticulum [GO:0005783]; integral component of endoplasmic reticulum membrane [GO:0030176]; membrane [GO:0016020]; ribosome [GO:0005840]; RNA binding [GO:0003723]; signaling receptor activity [GO:0038023]; osteoblast differentiation [GO:0001649]; protein transport [GO:0015031]; translation [GO:0006412] | 15184079_Results identify the ribosome receptor, p180, as a binding partner of the kinesin heavy chain isoform KIF5B. 17634287_These data suggest that p180 mediates interactions between the endoplasmic reticulum and microtubules mainly through the novel microtubule-binding and -bundling domain MTB-1. 18083570_over-expression of p180R failed to increase secretory pathway proteins calnexin, SEC61beta, and calreticulin, or ribosome biogenesis 19037105_results obtained demonstrate that p180 is both necessary & sufficient to induce a secretory phenotype in mammalian cells; findings support a central role for p180 in the terminal differentiation of secretory cells & tissues 19425502_the discovery overexpression of GPD1 and RRBP1 proteins and lack of expression for HNRNPH1 and SERPINB6 proteins which are new candidate biomarkers of colon cancer. 19932094_A novel function of p180-abundant endoplasmic reticulum on the trans-Golgi network expansion, both of which are highly developed in various professional secretory cells. 20647306_p180 plays crucial roles in enhancing collagen biosynthesis at the entry site of the secretory compartments by a novel mechanism that mainly involves facilitating ribosome association on the ER. 21111237_p180 is concentrated in ER sheets by interacting with polysomes and may play a role in ER morphology. 22679391_p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. 23318453_RRBP1 could alleviate ER stress and help cancer cell survive 24019514_Data indicate that p180 is required for the efficient targeting of placental alkaline phosphatase (ALPP) mRNA to the endoplasmic reticulum (ER). 26196185_High RRBP1 expression facilitates colorectal cancer progression and predicts an unfavourable post-operative prognosis. 30684972_RRBP1 expression was an independent prognostic factor for overall survival (OS) and disease-free survival (DFS) in patients with endometrial carcinoma (EC) (both P < 0.05). 31285390_Expression of RRBP1 in epithelial ovarian cancer and its clinical significance. 32289433_we found that E2F1 could bind to the RRBP1 promoter and promote the transcription of RRBP1, and the E2F1/RRBP1 axis played an important role in the process of high glucose promoting the proliferation, migration and invasion of HepG2 cells (hepatocellular carcinoma) 32536673_Correlation between reticulum ribosome-binding protein 1 (RRBP1) overexpression and prognosis in cervical squamous cell carcinoma. 33762722_RRBP1 rewires cisplatin resistance in oral squamous cell carcinoma by regulating Hippo pathway. 34445467_Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma. | ENSMUSG00000027422 | Rrbp1 | 6374.03424 | 0.9438383 | -0.0833883650 | 0.09801428 | 7.333532e-01 | 3.917984e-01 | 9.998360e-01 | No | Yes | 6246.05001 | 496.295333 | 6.317382e+03 | 388.191703 | |
| ENSG00000125875 | 128637 | TBC1D20 | protein_coding | Q96BZ9 | FUNCTION: GTPase-activating protein specific for Rab1 and Rab2 small GTPase families for which it can accelerate the intrinsic GTP hydrolysis rate by more than five orders of magnitude. | 3D-structure;Alternative splicing;GTPase activation;Host-virus interaction;Membrane;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a protein that belongs to a family of GTPase activator proteins of Rab-like small GTPases. The encoded protein and its cognate GTPase, Rab1, bind the nonstructural protein 5A (NS5A) of the hepatitis C virus (HCV) to mediate viral replication. Depletion of this protein inhibits replication of the virus and HCV infection. Mutations in this gene are associated with Warburg micro syndrome 4. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2014]. | hsa:128637; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; integral component of Golgi membrane [GO:0030173]; nuclear membrane [GO:0031965]; GTPase activator activity [GO:0005096]; small GTPase binding [GO:0031267]; acrosome assembly [GO:0001675]; COPII-coated vesicle cargo loading [GO:0090110]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; Golgi organization [GO:0007030]; lens fiber cell morphogenesis [GO:0070309]; lipid droplet organization [GO:0034389]; positive regulation by host of viral genome replication [GO:0044829]; positive regulation by virus of viral protein levels in host cell [GO:0046726]; positive regulation of ER to Golgi vesicle-mediated transport [GO:1902953]; positive regulation of GTPase activity [GO:0043547]; seminiferous tubule development [GO:0072520]; virion assembly [GO:0019068] | 17901050_TBC1D20 was found to be the first known GAP for Rab1, which is implicated in the regulation of anterograde traffic between the endoplasmic reticulum and the Golgi complex 22260459_These findings add TBC1D20 to the network of host factors regulating HIV replication cycle. 22491470_The NS5A interaction with TBC1D20 and Rab1 is essential for the viral life cycle. 23236136_The detailed molecular reaction mechanism of a complex between human Rab and RabGAP at the highest possible spatiotemporal resolution and in atomic detail, is described. 24239381_Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. 26063829_Warburg Micro syndrome is caused by TBC1D20 deficiency. 32162791_Martsolf syndrome with novel mutation in the TBC1D20 gene in a family from Iran. 32740904_Micro and Martsolf syndromes in 34 new patients: Refining the phenotypic spectrum and further molecular insights. | ENSMUSG00000027465 | Tbc1d20 | 2288.95761 | 1.0274242 | 0.0390319509 | 0.10016075 | 1.518098e-01 | 6.968118e-01 | 9.998360e-01 | No | Yes | 2384.84798 | 152.837269 | 2.211639e+03 | 110.487637 | |
| ENSG00000125962 | 64860 | ARMCX5 | protein_coding | Q6P1M9 | Reference proteome;Repeat | hsa:64860; | ENSMUSG00000072969 | Armcx5 | 131.64752 | 0.8584666 | -0.2201660277 | 0.27240661 | 6.429106e-01 | 4.226588e-01 | 9.998360e-01 | No | Yes | 120.69342 | 26.418661 | 1.537551e+02 | 25.887442 | |||||
| ENSG00000126653 | 84081 | NSRP1 | protein_coding | Q9H0G5 | FUNCTION: RNA-binding protein that mediates pre-mRNA alternative splicing regulation. {ECO:0000269|PubMed:21296756}. | Coiled coil;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ubl conjugation;mRNA processing;mRNA splicing | hsa:84081; | nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; developmental process [GO:0032502]; in utero embryonic development [GO:0001701]; mRNA processing [GO:0006397]; regulation of alternative mRNA splicing, via spliceosome [GO:0000381]; RNA splicing [GO:0008380] | 19047128_Observational study of gene-disease association. (HuGE Navigator) 25176346_We showed that NSRP70 is a novel lymphoblastic acute leukemia surrogate marker 26475744_Coiled coil domain-containing 55 gene may be involved in a functional bridging between the CNR1 activation and the disrupted in schizophrenia 1/Ran binding protein 9-associated pathways. 26797131_NSrp70 acts as a new molecular counterpart for alternative splicing of target RNA, counteracting SRSF1 and SRSF2 splicing activity. 34385670_Biallelic loss-of-function variants in the splicing regulator NSRP1 cause a severe neurodevelopmental disorder with spastic cerebral palsy and epilepsy. | ENSMUSG00000037958 | Nsrp1 | 304.82930 | 0.8803023 | -0.1839290456 | 0.18605404 | 9.904080e-01 | 3.196427e-01 | 9.998360e-01 | No | Yes | 311.81354 | 46.874427 | 3.294384e+02 | 38.284083 | ||
| ENSG00000127081 | 83744 | ZNF484 | protein_coding | Q5JVG2 | FUNCTION: May be involved in transcriptional regulation. | 3D-structure;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:83744; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 30.58060 | 1.8788371 | 0.9098399529 | 0.53201872 | 2.929235e+00 | 8.698870e-02 | 9.998360e-01 | No | Yes | 34.29527 | 7.447336 | 1.816769e+01 | 3.077093 | |||||
| ENSG00000127957 | 5387 | PMS2P3 | transcribed_unprocessed_pseudogene | 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) | 132.90490 | 1.2547085 | 0.3273522866 | 0.28233207 | 1.295517e+00 | 2.550338e-01 | 9.998360e-01 | No | Yes | 153.43444 | 30.790886 | 1.194334e+02 | 18.723578 | |||||||||
| ENSG00000129071 | 8930 | MBD4 | protein_coding | O95243 | FUNCTION: Mismatch-specific DNA N-glycosylase involved in DNA repair. Has thymine glycosylase activity and is specific for G:T mismatches within methylated and unmethylated CpG sites. Can also remove uracil or 5-fluorouracil in G:U mismatches. Has no lyase activity. Was first identified as methyl-CpG-binding protein. {ECO:0000269|PubMed:10097147, ECO:0000269|PubMed:10930409}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;DNA-binding;Hydrolase;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene is a member of a family of nuclear proteins related by the presence of a methyl-CpG binding domain (MBD). These proteins are capable of binding specifically to methylated DNA, and some members can also repress transcription from methylated gene promoters. This protein contains an MBD domain at the N-terminus that functions both in binding to methylated DNA and in protein interactions and a C-terminal mismatch-specific glycosylase domain that is involved in DNA repair. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2013]. | hsa:8930; | nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA N-glycosylase activity [GO:0019104]; endodeoxyribonuclease activity [GO:0004520]; pyrimidine-specific mismatch base pair DNA N-glycosylase activity [GO:0008263]; satellite DNA binding [GO:0003696]; depyrimidination [GO:0045008]; DNA repair [GO:0006281] | 11836615_MBD4 expression is associated with grade of malignancy in gliomas 12220634_results indicate the formation of a complex with the estradiol receptor 12430186_Frameshift mutations were found in 29% of gastric and 20% of colon MSI-H cancers, but not in any low-frequency microsatellite instability/microsatellite stable cancers. 12926109_MBD4 mutations were found in 15% MSI but not in MSS colrectal tumors. 15205355_Observational study of gene-disease association. (HuGE Navigator) 15899845_MBD4 acts as a repressor protein binding to hypermethylated promoters of the p16(INK4a) and hMLH1 genes. 16803845_MBD4 Glu346Lys polymorphism could be used as a marker for genetic susceptibility to adenocarcinoma of the lung. 16803845_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16831587_Mutations in MBD4 are unlikely to be implicated in HPS. 17049487_These results suggest that RFP is a mediator connecting several MBD proteins and allowing the formation of a more potent transcriptional repressor complex. 17285135_The overexpression of MBD4(tru) in Big Blue (lacI)-transfected, MSI human colorectal carcinoma cells doubled mutation frequency, indicating that the modest dominant negative effect on DNA repair can occur in living cells in short-term experiments. 17360956_MBD2 and MBD4 transcript overexpression and inverse correlations with DNA methylation indices indicate that both enzymes may really have a direct and active role on the genome-wide DNA hypomethylation observed in CD4+ T cells from SLE patients. 18162445_Truncation of MBD4 predisposes to reciprocal chromosomal translocations and alters the response to therapeutic agents in colon cancer cells. 18495292_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18495292_Single nucleotide polymorphisms in MBD4 are associated with lung cancer. 18519584_MBD4 efficiently processed T/G mismatches within the nucleosome. 18676680_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19395862_The methyl binding domain of MBD4/MED1 was found to specifically inhibit the activity of MBD4/MED1 as well as the glycosylase domain, when the G:IU mispairs were located in a methylated CpG context. 19403629_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19469655_Observational study of gene-disease association. (HuGE Navigator) 19469655_The Glu346Lys polymorphism and frameshift mutations of the Methyl-CpG Binding Domain 4 gene is associated with gastrointestinal cancer 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 20100009_Data suggest that methyl-CpG binding domain 4 polymorphism may not be a stratification marker to predict the susceptibility to immune thrombocytopenic purpura, at least in the Chinese population. 20100009_Observational study of gene-disease association. (HuGE Navigator) 20226869_Observational study of gene-disease association. (HuGE Navigator) 20453000_Observational study of gene-disease association. (HuGE Navigator) 20522537_Observational study of gene-disease association. (HuGE Navigator) 20574454_Observational study of gene-disease association. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20676650_MBD4-8666 and MBD4-9229, but not MBD4-1057, gene polymorphisms are related to rheumatoid arthritis in Chinese patients in Taiwan. 20731661_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21377502_decreased expression in patients with primary immune thrombocytopenia 21820404_the crystal structure of C-terminal glycosylase domain of human MBD4 was determined. 21971312_MBD 4--a potential substrate for protein kinase X 22560993_specificity of MBD4 for acting at CpG sites depends largely on its methyl-CpG-binding domain, which binds preferably to G.T mispairs in a methylated CpG site 22848106_Crystal structures of human MBD4(catalytic domain) reveal that MBD4 uses a base flipping mechanism to specifically recognize thymine and 5-hydroxymethyluracil. 23027038_MBD4 Glu346Lys polymorphism is associated with the risk of cervical cancer in a Chinese population. 23316048_the crystal structure of MBD4 bound to 5-hydroxymethylcytosine further demonstrates that MBDMBD4 is able to recognize a wide range of 5-methylcytosine modifications 24004570_ERCC4 rs1800124 and MBD4 rs10342 non-synonymous single nucleotide polymorphism variants were associated with DNA repair capacity. 24434851_Interaction between DNMT1 and MBD4 is involved in controlling gene expression and responding to oxidative stress. 25162968_MBD4 rs3138373 A>G and rs2005618 T>C single nucleotide polymorphisms were not associated with esophageal squamous cell carcinoma (ESCC) risk. rs3138355 GG genotype was associated with a decreased risk of ESCC among male patients and the elderly. 25358258_The ability of MBD4 to directly interact with and recruit USP7 to chromocenters implicates it as an additional factor that can potentially regulate Dnmt1 activity during cell proliferation. 26503472_these data suggest that MBD4 inactivation may contribute to tumorigenesis, acting as a modifier of mismatch repair-deficient cancer phenotype. 28542810_a novel molecular mechanism by which MBD4 inhibits GITR expression in a DNMT1-dependent manner 29018507_Results demonstrated that MBD4 was downregulated, which lead to the overexpression and promoter hypomethylation of CD70 in CD4+ T cells from patients from systemic lupus erythematosus (SLE). This study preliminarily revealed the role and mechanism of MBD4 in the pathogenesis of SLE. 29473320_These findings suggest that RNF144A is epigenetically silenced in breast cancer cells by promoter hypermethylation and MBD4. 30049810_Germ line MBD4 deficiency stimulates clonal hematopoiesis and guides the development of leukemia via recurrent mutations in DNMT3A. 31476572_MBD4 is sumoylated in vivo in a DNA damage-specific manner 32239153_Germline MBD4 Mutations and Predisposition to Uveal Melanoma. 32421892_Germline loss-of-function variants in MBD4 are rare in Finnish patients with uveal melanoma. 33871441_[Comparative Analysis of the Activity of the Polymorphic Variants of Human Uracil-DNA-Glycosylases SMUG1 and MBD4]. 34107280_Structural Insights into the Mechanism of Base Excision by MBD4. | ENSMUSG00000030322 | Mbd4 | 418.92334 | 0.9762639 | -0.0346568600 | 0.17052045 | 4.111766e-02 | 8.393110e-01 | 9.998360e-01 | No | Yes | 385.66164 | 66.406319 | 3.824881e+02 | 50.828221 | |
| ENSG00000129187 | 1635 | DCTD | protein_coding | P32321 | FUNCTION: Supplies the nucleotide substrate for thymidylate synthetase. | 3D-structure;Allosteric enzyme;Alternative splicing;Direct protein sequencing;Hydrolase;Metal-binding;Nucleotide biosynthesis;Phosphoprotein;Reference proteome;Zinc | The protein encoded by this gene catalyzes the deamination of dCMP to dUMP, the nucleotide substrate for thymidylate synthase. The encoded protein is allosterically activated by dCTP and inhibited by dTTP, and is found as a homohexamer. This protein uses zinc as a cofactor for its activity. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:1635; | cytosol [GO:0005829]; dCMP deaminase activity [GO:0004132]; identical protein binding [GO:0042802]; zinc ion binding [GO:0008270]; dTMP biosynthetic process [GO:0006231]; dUMP biosynthetic process [GO:0006226]; pyrimidine nucleotide metabolic process [GO:0006220] | 17602053_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20028759_Clinical trial of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20665488_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000031562 | Dctd | 1404.66952 | 1.0607575 | 0.0850948351 | 0.12387450 | 4.629838e-01 | 4.962328e-01 | 9.998360e-01 | No | Yes | 1323.12450 | 125.355203 | 1.372123e+03 | 100.945704 | |
| ENSG00000130558 | 10439 | OLFM1 | protein_coding | Q99784 | FUNCTION: Contributes to the regulation of axonal growth in the embryonic and adult central nervous system by inhibiting interactions between RTN4R and LINGO1. Inhibits RTN4R-mediated axon growth cone collapse (By similarity). May play an important role in regulating the production of neural crest cells by the neural tube (By similarity). May be required for normal responses to olfactory stimuli (By similarity). {ECO:0000250|UniProtKB:O88998, ECO:0000250|UniProtKB:Q9IAK4}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Coiled coil;Developmental protein;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Reference proteome;Secreted;Signal;Synapse | This gene product shares extensive sequence similarity with the rat neuronal olfactomedin-related ER localized protein. While the exact function of the encoded protein is not known, its abundant expression in brain suggests that it may have an essential role in nerve tissue. Several alternatively spliced transcripts encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:10439; | axon [GO:0030424]; axonal growth cone [GO:0044295]; endoplasmic reticulum [GO:0005783]; extracellular space [GO:0005615]; neuronal cell body [GO:0043025]; perikaryon [GO:0043204]; synapse [GO:0045202]; atrioventricular valve formation [GO:0003190]; cardiac epithelial to mesenchymal transition [GO:0060317]; negative regulation of gene expression [GO:0010629]; nervous system development [GO:0007399]; neuronal signal transduction [GO:0023041]; positive regulation of apoptotic process [GO:0043065]; positive regulation of epithelial to mesenchymal transition [GO:0010718]; positive regulation of gene expression [GO:0010628]; regulation of axon extension [GO:0030516] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21968811_Wnt activation suppresses Olfm-1 expression, and this may predispose a favorable microenvironment of the retained embryo in the Fallopian tube, leading to ectopic pregnancy in humans 25999259_Down-regulation of OLFM1 in fallopian tube epithelial cells enhances spheroid attachment to fallopian tube. 26121352_OLF domains from H. sapiens olfactomedin-1 and M. musculus gliomedin were biophysically, biochemically, and structurally characterized. 26377223_human chorionic gonadotropin stimulated miR-212, which down-regulated OLFM1 and CTBP1 expression in fallopian and endometrial epithelial cells to favor spheroid attachment 27555280_OLFM1 is a negative regulator of non-canonical NF-kappaB signalling by interacting with and inhibiting NIK. Thus, OLFM1 may serve as a valuable biomarker and therapeutic target for colorectal cancer (CRC) patients. | ENSMUSG00000026833 | Olfm1 | 119.81510 | 1.3611523 | 0.4448284693 | 0.27824865 | 2.586361e+00 | 1.077878e-01 | 9.998360e-01 | No | Yes | 145.60859 | 22.459086 | 1.060872e+02 | 13.042671 | |
| ENSG00000130997 | 353497 | POLN | protein_coding | Q7Z5Q5 | FUNCTION: DNA polymerase with very low fidelity that catalyzes considerable misincorporation by inserting dTTP opposite a G template, and dGTP opposite a T template (PubMed:16787914, PubMed:17118716). Is the least accurate of the DNA polymerase A family (i.e. POLG, POLN and POLQ) (PubMed:17118716). Can perform accurate translesion DNA synthesis (TLS) past a 5S-thymine glycol. Can perform efficient strand displacement past a nick or a gap and gives rise to an amount of product similar to that on non-damaged template. Has no exonuclease activity (PubMed:16787914). Error-prone DNA polymerase that preferentially misincorporates dT regardless of template sequence (PubMed:25775266). May play a role in TLS during interstrand cross-link (ICL) repair (PubMed:19908865). May be involved in TLS when genomic replication is blocked by extremely large major groove DNA lesions. May function in the bypass of some DNA-protein and DNA-DNA cross-links. May have a role in cellular tolerance to DNA cross-linking agents (PubMed:20102227). Involved in the repair of DNA cross-links and double-strand break (DSB) resistance. Participates in FANCD2-mediated repair. Forms a complex with HELQ helicase that participates in homologous recombination (HR) repair and is essential for cellular protection against DNA cross-links (PubMed:19995904). {ECO:0000269|PubMed:16787914, ECO:0000269|PubMed:17118716, ECO:0000269|PubMed:19908865, ECO:0000269|PubMed:19995904, ECO:0000269|PubMed:20102227, ECO:0000269|PubMed:25775266}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;DNA replication;DNA-binding;DNA-directed DNA polymerase;Nucleotidyltransferase;Nucleus;Reference proteome;Transferase | This gene encodes a DNA polymerase type-A family member. The encoded protein plays a role in DNA repair and homologous recombination. This gene shares its 5' exons with some transcripts from overlapping GeneID: 79441, which encodes an augmentin-like protein complex subunit. [provided by RefSeq, Dec 2014]. | hsa:353497; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cyclin binding [GO:0030332]; DNA binding [GO:0003677]; DNA-directed DNA polymerase activity [GO:0003887]; DNA-dependent DNA replication [GO:0006261]; double-strand break repair [GO:0006302]; double-strand break repair via homologous recombination [GO:0000724]; interstrand cross-link repair [GO:0036297]; translesion synthesis [GO:0019985] | 12794064_identification as homolog to Mus308 17118716_pol nu error rates for all 12 single base-base mismatches and for insertion and deletion errors 19908865_The DNA cross-linking agent mitomycin C hypersensitivity of PolN knockdown cells is rescued by overexpression of DNA polymerase-proficient PolN but not by DNA polymerase-deficient PolN. 19995904_The recently discovered polymerase POLN identified here is involved in repair of DNA cross-links. 20102227_Described is a novel translesion DNA synthesis substrate specificity of pol nu, demonstrating that it is able to bypass exceptionally large DNA lesions whose linkages are through the DNA major groove. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21671477_Our finding suggests that polymorphisms in DNA repair genes, POLN and PRKDC, were associated with increased melanoma risk in melanoma families with and without CDKN2A mutations. 22008035_The lower fidelity of POLN compared to that of Klenow fragment of Escherichia coli can be attributed to a much lower catalytic efficiency for correct dNTP incorporation, whereas both enzymes have similar kinetic parameters for G-dTTP misinsertion. 22262850_both yeast and human pol eta synthesize past the 3'-(m)C CPD in a >99% error-free manner, consistent with the highly water-exposed nature of the active site. 22623772_Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites 23045531_WRN improves the efficiency and fidelity of hpol eta to promote more effective replication of DNA. 25963146_Pol nu catalyzes both correct and mispair formation with high catalytic efficiency. 26740629_This is the first crystal structure of a DNA polymerase with an incoming rNTP opposite a DNA lesion. 26903512_The Proliferating Cell Nuclear Antigen (PCNA)-interacting Protein (PIP) Motif of DNA Polymerase eta Mediates Its Interaction with the C-terminal Domain of Rev1. 27226627_Five x-ray crystal structures of hpol eta ternary complexes were determined, three at the insertion and two at the extension stage. 27694439_The structural data are consistent with the observed tendency of hpol eta to insert both dC and dT opposite the O(6)-MeG lesion with similar efficiencies. 29330301_Data suggest that translesion DNA synthesis mediated by (1) POLI-dependent pathway (2) REV1- and POLN-dependent pathway, or (3) POLtheta-dependent pathway occur in predominantly error-free manner in human cells. (POLI = DNA polymerase iota; REV1 = DNA repair protein-REV1; POLN = DNA polymerase nu; POLtheta = DNA polymerase theta) 30368948_Crystallographic evidence for two-metal-ion catalysis in human pol eta. 30709915_the Poleta-oxaliplatin-GpG structure provides a structural basis for TLS-mediated bypass of the major oxaliplatin-DNA adducts and insights into resistance to platinum-based chemotherapy in humans. 30842261_These results suggest that hpol eta is one of the major reverse transcriptases involved in physiological processes in human cells. 32098870_The roles of polymerases nu and theta in replicative bypass of O (6)- and N (2)-alkyl-2'-deoxyguanosine lesions in human cells. 33175093_Translesion synthesis of the major nitrogen mustard-induced DNA lesion by human DNA polymerase eta. 34088846_Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction. 34461101_DNA polymerases eta and kappa bypass N(2)-guanine-O(6)-alkylguanine DNA alkyltransferase cross-linked DNA-peptides. | ENSMUSG00000045102 | Poln | 220.46047 | 0.7575345 | -0.4006165538 | 0.23690891 | 2.646601e+00 | 1.037716e-01 | 9.998360e-01 | No | Yes | 266.54651 | 44.540605 | 2.843732e+02 | 36.663052 | |
| ENSG00000131844 | 64087 | MCCC2 | protein_coding | Q9HCC0 | FUNCTION: Carboxyltransferase subunit of the 3-methylcrotonyl-CoA carboxylase, an enzyme that catalyzes the conversion of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA, a critical step for leucine and isovaleric acid catabolism. {ECO:0000269|PubMed:17360195}. | ATP-binding;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Ligase;Mitochondrion;Nucleotide-binding;Reference proteome;Transit peptide | PATHWAY: Amino-acid degradation; L-leucine degradation; (S)-3-hydroxy-3-methylglutaryl-CoA from 3-isovaleryl-CoA: step 2/3. {ECO:0000269|PubMed:17360195}. | This gene encodes the small subunit of 3-methylcrotonyl-CoA carboxylase. This enzyme functions as a heterodimer and catalyzes the carboxylation of 3-methylcrotonyl-CoA to form 3-methylglutaconyl-CoA. Mutations in this gene are associated with 3-Methylcrotonylglycinuria, an autosomal recessive disorder of leucine catabolism. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, May 2018]. | hsa:64087; | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial [GO:0002169]; cytosol [GO:0005829]; methylcrotonoyl-CoA carboxylase complex [GO:1905202]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; ATP binding [GO:0005524]; methylcrotonoyl-CoA carboxylase activity [GO:0004485]; coenzyme A metabolic process [GO:0015936]; leucine catabolic process [GO:0006552] | 16010683_factors other than the genotype at the MCCA and MCCB loci have a major influence on the phenotype of MCC deficiency 16023992_The amino-termini containing 20 amino acids (MCCbeta) were both necessary and sufficient for targeting. Structural requirements for mitochondrial import were defined by site-directed mutagenesis. 17876819_The Kd value of soraphen A for the BC domains of human ACC1 and ACC2 is 1 nM. This high binding affinity is mainly due to the extensive interactions between soraphen A and the human biotin carboxylase domain 17968484_Molecular analyses revealed novel mutations in one of the causative genes, MCCA or MCCB, in all five of the MCC deficiency patients 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 21071250_identified two novel MCCA and four novel MCCB mutant alleles from five MCC-deficient patients 22150417_study reports eight different mutant alleles of MCCC1 or MCCC2 including six novel mutations in Korean patients with 3-methylcrotonyl-CoA carboxylase (MCC) deficiency 22264772_Mutation in 3-methylcrotonyl CoA carboxylase 2 gene is associated with 3-methylcrotonyl-CoA carboxylase deficiency. 25382614_Novel mutation in MCCC2 gene was identified in Chinese population. 27601257_This study reports data mainly obtained from the Portuguese newborn screening program collected over a ten-year period. Analysis of the MCCC1 and MCCC2 genes yielded 26 previously unreported mutations and a variant of clinically unknown significance. 30895811_MCCC2 overexpression predicts an unfavorable prognosis and promotes cell proliferation in breast cancer 31901042_Mutations on MCCC1 and MCCC2 genes are the major genetic causes for the increased C5-OH in neonates 32205097_MCCC2 overexpression predicts poorer prognosis and promotes cell proliferation in colorectal cancer. 33423264_[Analysis of MCCC2 gene variant in a pedigree affected with 3-methylcrotonyl coenzyme A carboxylase deficiency]. | ENSMUSG00000021646 | Mccc2 | 4497.11599 | 1.0286563 | 0.0407609936 | 0.09798170 | 1.705743e-01 | 6.796019e-01 | 9.998360e-01 | No | Yes | 5129.84720 | 512.964345 | 5.226397e+03 | 403.835713 |
| ENSG00000133226 | 10250 | SRRM1 | protein_coding | Q8IYB3 | FUNCTION: Part of pre- and post-splicing multiprotein mRNP complexes. Involved in numerous pre-mRNA processing events. Promotes constitutive and exonic splicing enhancer (ESE)-dependent splicing activation by bridging together sequence-specific (SR family proteins, SFRS4, SFRS5 and TRA2B/SFRS10) and basal snRNP (SNRP70 and SNRPA1) factors of the spliceosome. Stimulates mRNA 3'-end cleavage independently of the formation of an exon junction complex. Binds both pre-mRNA and spliced mRNA 20-25 nt upstream of exon-exon junctions. Binds RNA and DNA with low sequence specificity and has similar preference for either double- or single-stranded nucleic acid substrates. {ECO:0000269|PubMed:10339552, ECO:0000269|PubMed:10668804, ECO:0000269|PubMed:11739730, ECO:0000269|PubMed:12600940, ECO:0000269|PubMed:12944400, ECO:0000269|PubMed:9531537}. | 3D-structure;Acetylation;Alternative splicing;Citrullination;DNA-binding;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Spliceosome;Ubl conjugation;mRNA processing;mRNA splicing | hsa:10250; | catalytic step 2 spliceosome [GO:0071013]; cytosol [GO:0005829]; nuclear matrix [GO:0016363]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; RNA binding [GO:0003723]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380]; RNA splicing, via transesterification reactions [GO:0000375] | 11991360_1H, 13C, and 15N resonance assignments and secondary structure of the PWI domain from SRm160 was determined using reduced dimensionality NMR. 12581738_Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins and plays a role in spliceosome assembly 12624182_two contiguous sequences that independently target SRm160 to nuclear matrix sites at splicing speckled domains: amino acids 300-350 and 351-688 15024032_Data show that SRm160, a splicing coactivator and component of the exon junction complex (EJC) involved in RNA export, has an adenosine triphosphate (ATP)-dependent mobility. 16159877_found that the majority of proteins identified in SRm160-containing complexes are associated with pre-mRNA processing. Interestingly, SRm160 is also associated with factors involved in chromatin regulation and sister chromatid cohesion 16354706_SRm160, a splicing coactivator, regulates CD44 alternative splicing in a Ras-dependent manner. 30590765_We discovered that two factors, SRRM1 and SF3B1, affect not only cis-SAGe chimeras, but also other types of chimeric RNAs in a genome-wide fashion. | ENSMUSG00000028809 | Srrm1 | 4453.81513 | 0.9567453 | -0.0637931470 | 0.09075771 | 4.920725e-01 | 4.830042e-01 | 9.998360e-01 | No | Yes | 4122.28489 | 346.033859 | 4.248299e+03 | 275.591409 | ||
| ENSG00000133872 | 51669 | SARAF | protein_coding | Q96BY9 | FUNCTION: Negative regulator of store-operated Ca(2+) entry (SOCE) involved in protecting cells from Ca(2+) overfilling. In response to cytosolic Ca(2+) elevation after endoplasmic reticulum Ca(2+) refilling, promotes a slow inactivation of STIM (STIM1 or STIM2)-dependent SOCE activity: possibly act by facilitating the deoligomerization of STIM to efficiently turn off ORAI when the endoplasmic reticulum lumen is filled with the appropriate Ca(2+) levels, and thus preventing the overload of the cell with excessive Ca(2+) ions. {ECO:0000269|PubMed:22464749}. | 3D-structure;Alternative splicing;Calcium;Calcium transport;Direct protein sequencing;Endoplasmic reticulum;Ion transport;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix;Transport | hsa:51669; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-plasma membrane contact site [GO:0140268]; integral component of endoplasmic reticulum membrane [GO:0030176]; calcium ion transport [GO:0006816]; regulation of store-operated calcium entry [GO:2001256] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22464749_Study shows that SARAF is an endoplasmic reticulum resident protein, which responds to cytosolic Ca2+ elevation after ER Ca2+ refilling by promoting a slow inactivation process of STIM2-dependent basal SOCE activity, as well as STIM1-mediated SOCE activity. 23049731_Store-operated Ca2+ entry is remodelled and controls in vitro angiogenesis in endothelial progenitor cells isolated from tumoral patients. 23816623_Deletion of STIM1(448-490) and, in particular, STIM1(447-460) and STIM1(475-490) markedly enhances interaction of SARAF with STIM1. 26817842_SARAF is constitutively expressed in the plasma membrane and interacts with Orai1. 27068144_SARAF overexpression attenuated store operated Ca2+ entry and the STIM1-Orai1 interaction in cells endogenously expressing STIM1 and Orai1 while RNAi-mediated SARAF silencing induced opposite effects. 27414851_These findings suggest that the surface location of SARAF is dependent on the expression of STIM1 in the plasma membrane. 27506849_SARAF modulates TRPC1, but not TRPC6, channel function in a STIM1-independent manner 28866365_This activation was sensitive to Src kinase inhibition, but not to CAMKII nor PKC inhibition, a result that sets STIM1 and SOCE as downstream targets of the axis Src-Raf-MEK-ERK, rather than upstream regulators. 29223474_SARAF also interacts with Orai1 and TRPC1 in cells endogenously expressing STIM1 and cells with a low STIM1 expression and modulates channel function. This review focuses on the modulation by SARAF of SOCE and other forms of Ca(2+) influx mediated by Orai1 and TRPC1 in order to provide spatio-temporally regulated Ca(2+) signals. 30481768_EFHB is a new store-operated Ca2+ entry regulator that modulates STIM1-SARAF interaction. 30975919_Y316F mutation altered the pattern of interaction between STIM1 and SARAF under resting conditions and upon Ca(2+) store depletion. Expression of the STIM1 Y316F mutant enhanced slow Ca(2+)-dependent inactivation (SCDI) as compared to STIM1 WT, an effect that was abolished by SARAF knockdown. 31082439_Described is the X-ray crystal structure of the SARAF luminal domain, SARAFL. This domain forms a novel 10-stranded beta-sandwich fold that includes a set of three conserved disulfide bonds, denoted the 'SARAF-fold.' The structure reveals a domain-swapped dimer in which the last two beta-strands (beta9 and beta10) are exchanged forming a region denoted the 'SARAF luminal switch' that is essential for dimerization. 31493399_Study shows that SARAF interacted with STIM1 to limit Ca2+ influx during physiological stimulation, but interaction of SARAF with STIM1 was transient during pathological stimulation of acinar cells, resulting in sustained, toxic Ca2+ influx. Transgenic expression of SARAF in mouse pancreatic acini reduced pathologic Ca2+ influx, prevented inflammation, and reduced tissue damage in mouse models of severe pancreatitis. 32079527_Chemoresistant ovarian cancer enhances its migration abilities by increasing store-operated Ca(2+) entry-mediated turnover of focal adhesions. 32394484_Temporal modulation of calcium sensing in hematopoietic stem cells is crucial for proper stem cell expansion and engraftment. 34705029_Bidirectional regulation of calcium release-activated calcium (CRAC) channel by SARAF. | ENSMUSG00000031532 | Saraf | 1558.57226 | 1.0924735 | 0.1275983136 | 0.13606838 | 8.936448e-01 | 3.444915e-01 | 9.998360e-01 | No | Yes | 1659.43868 | 261.709405 | 1.616140e+03 | 196.647471 | ||
| ENSG00000133943 | 80017 | DGLUCY | protein_coding | Q7Z3D6 | FUNCTION: D-glutamate cyclase that converts D-glutamate to 5-oxo-D-proline. {ECO:0000250|UniProtKB:Q8BH86}. | Alternative splicing;Lyase;Mitochondrion;Reference proteome;Transit peptide | hsa:80017; | mitochondrial matrix [GO:0005759]; D-glutamate cyclase activity [GO:0047820]; glutamate metabolic process [GO:0006536] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000021185 | Dglucy | 412.56284 | 0.9710847 | -0.0423309249 | 0.16514446 | 6.525075e-02 | 7.983815e-01 | 9.998360e-01 | No | Yes | 380.36370 | 51.057199 | 3.735858e+02 | 39.368098 | ||
| ENSG00000135116 | 8739 | HRK | protein_coding | O00198 | FUNCTION: Promotes apoptosis. {ECO:0000269|PubMed:15031724, ECO:0000269|PubMed:9130713}. | 3D-structure;Apoptosis;Membrane;Mitochondrion;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a member of the BCL-2 protein family. Members of this family are involved in activating or inhibiting apoptosis. The encoded protein localizes to intracellular membranes. This protein promotes apoptosis by interacting with the apoptotic inhibitors BCL-2 and BCL-X(L) via its BH3 domain. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Oct 2012]. | hsa:8739; | integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; apoptotic process [GO:0006915]; positive regulation of apoptotic process [GO:0043065]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of release of cytochrome c from mitochondria [GO:0090200] | 11796190_Hrk is involved in the induction of apoptosis in RGCs after optic nerve transection. 12217801_apoptosis inducers as diverse as oncoprotein inhibitors and cell death receptor activators trigger Hrk expression via blockade of DREAM in leukemia cells 12606589_Data report that human oocytes and fragmenting preimplantation embryos possess transcripts encoding Harakiri and caspase-3. 14695142_HRK is a target of epigenetic inactivation in colorectal and gastric cancer 15031724_The interaction between HRK and cellular protein p32 was studied. HRK-induced apoptosis was suppresssed by the expression of p32 mutants lacking the N-terminal sequences 74-282 and the C-terminal sequences 1-221. 18008329_HRK appears to be inactivated principally by promoter hypermethylation in prostate cancers and decreased expression may play an important role in tumor progression by modulating apoptotic cell death 18037991_in response to PAHs, Ahr-mediated activation of the harakiri, BCL2 interacting protein (contains only BH3 domain), was necessary for execution of cell death. 19641496_Aberrant 5'-CpG methylation status and loss of heterozygosity on 12q13.1 are associated with HRK expression in human malignancies, including prostate cancers, astrocytic tumors and primary central nervous system lymphomas. Review. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20855536_Observational study of gene-disease association. (HuGE Navigator) 21209886_analysis of a novel interaction between Bcl-2 members Diva and Harakiri 21731739_These results are used to propose a tentative structural model of how Harakiri works. 22773666_Data suggest that DP5 and PUMA/BBC3 (p53 up-regulated modulator of apoptosis/bcl-2-binding component 3) contribute to palmitate-induced apoptosis of pancreatic beta-cells via lipotoxic endoplasmic reticulum stress. 22964433_SUZ12 promotes the proliferation of human EOC cells by inhibiting apoptosis and HRK is a novel SUZ12 target gene whose upregulation contributes to apoptosis induced by SUZ12 knockdown. 23090478_The BH3-only protein harakiri (HRK) is transactivated by ATF4 in severe hypoxia through direct binding of ATF4 to the promoter region. 23192964_Diva binds peptides derived from the BH3 domain of several other proapoptotic Bcl-2 proteins, including mouse Harakiri, Bid, Bak and Bmf. 23580416_our findings suggest that induction of the BH3-only protein Hrk is a critical step in 2-ME activation of the JNK-induced apoptotic pathway, targeting mitochondria by liberating proapoptotic protein Bak. 27737950_miR-23a-3p, miR-23b-3p, and miR-149-5p, were downregulated by cytokines and selected for further studies. These miRNAs were found to regulate the expression of the proapoptotic Bcl-2 proteins DP5 and PUMA and consequent human beta-cell apoptosis. 28661478_The apoptosis phenotype was partly dependent on HRK upregulation, as HRK knockdown significantly abrogated the sensitization. KDM2B-silenced tumors exhibited slower growth in vivo. Taken together, our findings suggest a novel mechanism, where the key apoptosis components are under epigenetic control of KDM2B in glioblastoma multiforme cells. 31135059_In individuals with susceptible genetic backgrounds, Coxsackie B viral infection alters the epigenome to activate the pathological pathways leading to polymyositis-dermatomyositis via mitochondrial dysfunction and HRK up-regulation. 32620849_Characterization of an alternative BAK-binding site for BH3 peptides. 32900773_YAP-Mediated Repression of HRK Regulates Tumor Growth, Therapy Response, and Survival Under Tumor Environmental Stress in Neuroblastoma. 33174041_Upregulated lncHRK2:1 prompts nucleus pulposus cell senescence in intervertebral disc degeneration. | ENSMUSG00000046607 | Hrk | 308.85509 | 0.6736728 | -0.5698799543 | 0.22464583 | 6.621802e+00 | 1.007378e-02 | 9.998360e-01 | No | Yes | 400.07930 | 70.997690 | 5.869097e+02 | 81.026618 | |
| ENSG00000135241 | 50640 | PNPLA8 | protein_coding | Q9NP80 | FUNCTION: Calcium-independent and membrane-bound phospholipase, that catalyzes the esterolytic cleavage of fatty acids from glycerophospholipids to yield free fatty acids and lysophospholipids, hence regulating membrane physical properties and the release of lipid second messengers and growth factors (PubMed:10833412, PubMed:10744668, PubMed:15695510, PubMed:15908428, PubMed:17213206, PubMed:18171998, PubMed:28442572). Hydrolyzes phosphatidylethanolamine, phosphatidylcholine and probably phosphatidylinositol with a possible preference for the former (PubMed:15695510). Has also a broad substrate specificity in terms of fatty acid moieties, hydrolyzing saturated and mono-unsaturated fatty acids at nearly equal rates from either the sn-1 or sn-2 position in diacyl phosphatidylcholine (PubMed:10833412, PubMed:10744668, PubMed:15695510, PubMed:15908428). However, has a weak activity toward polyunsaturated fatty acids at the sn-2 position, and thereby favors the production of 2-arachidonoyl lysophosphatidylcholine, a key branch point metabolite in eicosanoid signaling (PubMed:15908428). On the other hand, can produce arachidonic acid from the sn-1 position of diacyl phospholipid and from the sn-2 position of arachidonate-containing plasmalogen substrates (PubMed:15908428). Therefore, plays an important role in the mobilization of arachidonic acid in response to cellular stimuli and the generation of lipid second messengers (PubMed:15695510, PubMed:15908428). Can also hydrolyze lysophosphatidylcholine (PubMed:15695510). In the mitochondrial compartment, catalyzes the hydrolysis and release of oxidized aliphatic chains from cardiolipin and integrates mitochondrial bioenergetics and signaling. It is essential for maintaining efficient bioenergetic mitochondrial function through tailoring mitochondrial membrane lipid metabolism and composition (PubMed:28442572). {ECO:0000250|UniProtKB:Q8K1N1, ECO:0000269|PubMed:10744668, ECO:0000269|PubMed:10833412, ECO:0000269|PubMed:15695510, ECO:0000269|PubMed:15908428, ECO:0000269|PubMed:17213206, ECO:0000269|PubMed:18171998, ECO:0000269|PubMed:28442572}. | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Hydrolase;Lipid degradation;Lipid metabolism;Membrane;Mitochondrion;Peroxisome;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Phospholipid metabolism. {ECO:0000305|PubMed:15908428}. | This gene encodes a member of the patatin-like phospholipase domain containing protein family. Members of this family are phospholipases which catalyze the cleavage of fatty acids from membrane phospholipids. The product of this gene is a calcium-independent phospholipase. Mutations in this gene have been associated with mitochondrial myopathy with lactic acidosis. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2015]. | hsa:50640; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; mitochondrial membrane [GO:0031966]; mitochondrion [GO:0005739]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; ATP binding [GO:0005524]; calcium-independent phospholipase A2 activity [GO:0047499]; lysophospholipase activity [GO:0004622]; phosphatidyl phospholipase B activity [GO:0102545]; phospholipase A1 activity [GO:0008970]; phospholipase activity [GO:0004620]; arachidonic acid metabolic process [GO:0019369]; arachidonic acid secretion [GO:0050482]; cardiolipin metabolic process [GO:0032048]; cell death [GO:0008219]; fatty acid metabolic process [GO:0006631]; linoleic acid metabolic process [GO:0043651]; lipid homeostasis [GO:0055088]; phosphatidylcholine catabolic process [GO:0034638]; phosphatidylethanolamine catabolic process [GO:0046338]; prostaglandin biosynthetic process [GO:0001516]; regulation of cellular response to oxidative stress [GO:1900407]; triglyceride homeostasis [GO:0070328] | 15695510_These results suggest distinct roles for iPLA2beta and iPLA2gamma in cellular homeostasis and signaling, a functional link between peroxisomal AA release and eicosanoid generation, and a potential contribution of iPLA2gamma to tumorigenesis. 18680539_iPLA2 activation is not sufficient for SOCE activation. iPLA2 may regulate basal phosphoinositide metabolism 19892409_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23258543_Thus, complement-mediated activation of iPLA(2)gamma is mediated via ERK and p38 pathways, and phosphorylation of Ser-511 and/or Ser-515 plays a key role in the catalytic activity and signaling of iPLA(2)gamma. 23429536_iPLA2gamma plays a cardioprotective role during the acute stage of Chagas' disease. 25313821_iPLA2gamma plays an important role in in vivo Thromboxane A2 production accompanied by thrombus formation. 29158256_Stable isotope kinetics revealed that in non-failing human hearts, cPLA2zeta metabolically channels arachidonic acid into EETs, whereas in failing hearts, increased iPLA2gamma activity channels AA into toxic HETEs. These results mechanistically identify the sequelae of pathological remodeling of human mitochondrial phospholipases in failing myocardium. 29681094_we report two unrelated individuals with variable but similar clinical features of microcephaly, severe global developmental delay, spasticity, lactic acidosis, and progressive cerebellar atrophy with biallelic loss-of-function variants in PNPLA8. 32161117_12-LOX catalyzes the oxidation of 2-arachidonoyl-lysolipids in platelets generating eicosanoid-lysolipids that are attenuated by iPLA2gamma knockout. | ENSMUSG00000036257 | Pnpla8 | 104.01071 | 0.8942083 | -0.1613170944 | 0.30706553 | 2.615711e-01 | 6.090428e-01 | 9.998360e-01 | No | Yes | 97.08151 | 17.976968 | 1.104010e+02 | 15.707724 |
| ENSG00000135299 | 22881 | ANKRD6 | protein_coding | Q9Y2G4 | FUNCTION: Recruits CKI-epsilon to the beta-catenin degradation complex that consists of AXN1 or AXN2 and GSK3-beta and allows efficient phosphorylation of beta-catenin, thereby inhibiting beta-catenin/Tcf signals. {ECO:0000250}. | ANK repeat;Alternative splicing;Coiled coil;Reference proteome;Repeat | hsa:22881; | cytoplasm [GO:0005737]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; positive regulation of JNK cascade [GO:0046330]; positive regulation of Wnt signaling pathway, planar cell polarity pathway [GO:2000096] | 19591803_Diversin, containing several nuclear localization signals, translocates to the nucleus, where it interacts with the transcription factor AF9. 22580979_ANKRD6 genetic variants were associated with the muscle size and strength response to resistance training and habitual physical activity levels. 24833088_diversin is overexpressed in human glioma and regulates glioma cell proliferation and invasion, possibly through MMP9 24858714_study demonstrated that diversin was overexpressed in human breast cancers. Diversin could contribute to breast cancer cell proliferation and invasion. 25200652_missense mutations in ANKRD6 are associated with neural tube defects. | ENSMUSG00000040183 | Ankrd6 | 184.01231 | 0.7457086 | -0.4233160316 | 0.27947653 | 2.161763e+00 | 1.414823e-01 | 9.998360e-01 | No | Yes | 177.92659 | 35.291586 | 2.397444e+02 | 36.909696 | ||
| ENSG00000135314 | 80759 | KHDC1 | protein_coding | Q4VXA5 | Mouse_homologues NA; + ;NA; + ;FUNCTION: Has pro-apoptotic activity. {ECO:0000269|PubMed:14657025}. | Alternative splicing;Membrane;RNA-binding;Reference proteome;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA; + ;NA | hsa:80759; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; RNA binding [GO:0003723]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919] | Mouse_homologues 20668163_Data suggest that KHDC1B, via its interaction with mCEPB1, may regulate translation of mRNA targets required for oocyte maturation.; + ;NA; + ;20668163_The paper describes the connection of this gene to translational machinery including CPEB | ENSMUSG00000085079+ENSMUSG00000041722+ENSMUSG00000067750 | Khdc1b+Khdc1c+Khdc1a | 116.53934 | 1.0720274 | 0.1003417584 | 0.27696718 | 1.310587e-01 | 7.173369e-01 | 9.998360e-01 | No | Yes | 140.11719 | 19.838942 | 1.317011e+02 | 16.090954 | |
| ENSG00000135316 | 10492 | SYNCRIP | protein_coding | O60506 | FUNCTION: Heterogenous nuclear ribonucleoprotein (hnRNP) implicated in mRNA processing mechanisms. Component of the CRD-mediated complex that promotes MYC mRNA stability. Isoform 1, isoform 2 and isoform 3 are associated in vitro with pre-mRNA, splicing intermediates and mature mRNA protein complexes. Isoform 1 binds to apoB mRNA AU-rich sequences. Isoform 1 is part of the APOB mRNA editosome complex and may modulate the postranscriptional C to U RNA-editing of the APOB mRNA through either by binding to A1CF (APOBEC1 complementation factor), to APOBEC1 or to RNA itself. May be involved in translationally coupled mRNA turnover. Implicated with other RNA-binding proteins in the cytoplasmic deadenylation/translational and decay interplay of the FOS mRNA mediated by the major coding-region determinant of instability (mCRD) domain. Interacts in vitro preferentially with poly(A) and poly(U) RNA sequences. Isoform 3 may be involved in cytoplasmic vesicle-based mRNA transport through interaction with synaptotagmins. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma activation assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation; seems not to be essential for GAIT complex function. {ECO:0000269|PubMed:11051545, ECO:0000269|PubMed:11134005, ECO:0000269|PubMed:11352648, ECO:0000269|PubMed:11574476, ECO:0000269|PubMed:19029303, ECO:0000269|PubMed:23071094}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Endoplasmic reticulum;Host-virus interaction;Isopeptide bond;Methylation;Microsome;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein;Spliceosome;Translation regulation;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a member of the cellular heterogeneous nuclear ribonucleoprotein (hnRNP) family. hnRNPs are RNA binding proteins that complex with heterogeneous nuclear RNA (hnRNA) and regulate alternative splicing, polyadenylation, and other aspects of mRNA metabolism and transport. The encoded protein plays a role in multiple aspects of mRNA maturation and is associated with several multiprotein complexes including the apoB RNA editing-complex and survival of motor neurons (SMN) complex. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene, and a pseudogene of this gene is located on the short arm of chromosome 20. [provided by RefSeq, Dec 2011]. | hsa:10492; | catalytic step 2 spliceosome [GO:0071013]; CRD-mediated mRNA stability complex [GO:0070937]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; GAIT complex [GO:0097452]; histone pre-mRNA 3'end processing complex [GO:0071204]; mCRD-mediated mRNA stability complex [GO:0106002]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA 5'-UTR binding [GO:0048027]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; cellular response to interferon-gamma [GO:0071346]; CRD-mediated mRNA stabilization [GO:0070934]; mRNA modification [GO:0016556]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900152]; negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:2000623]; negative regulation of translation [GO:0017148]; osteoblast differentiation [GO:0001649]; positive regulation of cytoplasmic translation [GO:2000767]; RNA processing [GO:0006396]; RNA splicing [GO:0008380] | 15340051_The overexpression of NSAP1 specifically enhanced HCV IRES-dependent translation, and knockdown of NSAP1 by use of a small interfering RNA specifically inhibited the translation of HCV mRNA. 15475564_SYNCRIP is transported within the dendrite as a component of mRNA granules 16765914_methylation of hnRNPQ is important for its nuclear localization 17010310_Results suggest hnRNPQ interacts via its acidic domain (AcD) with Apobec1 and that this interaction is regulated by AcD phosphorylation. 18794368_Data demonstrate that hnRNP Q is a splicing modulator of SMN2, further underscoring the potential of hnRNP Q as a therapeutic target for spinal muscular atrophy. 19137262_Results demonstrate that galectin-3 stabilizes hnRNP Q via complex formation, and reduction in the hnRNP Q level leads to slow proliferation and less susceptibility to 5-FU. 19232660_SYNCRIP participates in both RNA replication and translation in HCV life cycle. 19808671_Data demonstrate that expression levels of hnRNP A1, Q, K, R, and U influence HIV-1 production by persistently infected 23679954_These data indicate that hnRNP Q can stimulate the protein production of HIV-1 Rev-dependent mRNAs without changing mRNA levels and mRNA export, respectively. 23946508_The hnRNP Q is a novel substrate of SHP2 and the SHP2 activity may be also involved in RNA metabolisms via dephosphorylation of hnRNP Q. 23980891_The inhibitory effect of hnRNP Q on YB-1 mRNA translation can be explained by its ability to pro-mote YB-1 binding to the regulatory element within the YB-1 mRNA 3' UTR. 25100733_hnRNPQ6 is required for APOBEC1-enhanced IL8 production. 27081926_The acidic domain is a unique structural feature of the splicing factor SYNCRIP. 28079881_our data indicate that hnRNP Q1 is a novel trans-acting factor that binds to Aurora-A mRNA 5'-UTRs and regulates its translation, which increases cell proliferation and contributes to tumorigenesis in colorectal cancer 28436985_SYNCRIP is required for survival of leukemia cells. SYNCRIP controls the myeloid leukemia stem cell program. 29483512_A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. 29884818_The EGF/hnRNP Q1-induced translation of Aurora-A mRNA is mediated by the mTOR and ERK pathways. 30266814_Results indentified two candidate haploinsufficient genes contiguous at 6q14 SYNCRIP (encoding hnRNP-Q) and SNHG5 (that hosts snoRNAs), both involved in regulating RNA maturation and translation inT-cell acute lymphoblastic leukemia (T-ALL). 30755280_Identification of the frequent presence of hnRNP R and hnRNP Q in frontotemporal lobar degeneration (FTLD)-FUS inclusions suggests a potential role for these hnRNPs in FTLD-FUS pathogenesis and supports the role of dysfunctional RNA metabolism in FTLD. 31791586_ATP binds nucleic-acid-binding domains beyond RRM fold. 31907208_the terminal loop of pri-let-7a was shown to be the main contributor for its interaction with SYNCRIP. Functional studies demonstrated that the SYNCRIP RRM2-3 domain can promote the processing of pri-let-7a 32770626_A novel antisense lncRNA NT5E promotes progression by modulating the expression of SYNCRIP and predicts a poor prognosis in pancreatic cancer. 34157790_Further evidence for de novo variants in SYNCRIP as the cause of a neurodevelopmental disorder. | ENSMUSG00000032423 | Syncrip | 4820.47024 | 0.9270582 | -0.1092681736 | 0.09628984 | 1.270664e+00 | 2.596421e-01 | 9.998360e-01 | No | Yes | 4724.11443 | 702.071766 | 5.271656e+03 | 604.051329 | |
| ENSG00000135362 | 79899 | PRR5L | protein_coding | Q6MZQ0 | FUNCTION: Associates with the mTORC2 complex that regulates cellular processes including survival and organization of the cytoskeleton (PubMed:17461779). Regulates the activity of the mTORC2 complex in a substrate-specific manner preventing for instance the specific phosphorylation of PKCs and thereby controlling cell migration (PubMed:22609986). Plays a role in the stimulation of ZFP36-mediated mRNA decay of several ZFP36-associated mRNAs, such as TNF-alpha and GM-CSF, in response to stress (PubMed:21964062). Required for ZFP36 localization to cytoplasmic stress granule (SG) and P-body (PB) in response to stress (PubMed:21964062). {ECO:0000269|PubMed:17461779, ECO:0000269|PubMed:21964062, ECO:0000269|PubMed:22609986}. | Alternative splicing;Phosphoprotein;Reference proteome;Signal transduction inhibitor;Ubl conjugation | hsa:79899; | TORC2 complex [GO:0031932]; ubiquitin protein ligase binding [GO:0031625]; cellular response to oxidative stress [GO:0034599]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of signal transduction [GO:0009968]; positive regulation of intracellular protein transport [GO:0090316]; positive regulation of mRNA catabolic process [GO:0061014]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein phosphorylation [GO:0001934]; regulation of fibroblast migration [GO:0010762]; TORC2 signaling [GO:0038203] | 17461779_It was demonstrated that immunoprecipitation of Protor-1 or Protor-2 results in the co-immunoprecipitation of other mTORC2 subunits, but not Raptor, a specific component of mTORC1. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21964062_Protor-2 associates with tristetraprolin (TTP) to accelerate TTP-mediated mRNA turnover and functionally links the control of TTP-regulated mRNA stability to mTORC2 activity | ENSMUSG00000032841 | Prr5l | 62.60144 | 0.6442084 | -0.6344007093 | 0.38166386 | 2.693053e+00 | 1.007865e-01 | 9.998360e-01 | No | Yes | 46.23209 | 11.177688 | 6.996209e+01 | 13.342906 | ||
| ENSG00000135424 | 3679 | ITGA7 | protein_coding | Q13683 | FUNCTION: Integrin alpha-7/beta-1 is the primary laminin receptor on skeletal myoblasts and adult myofibers. During myogenic differentiation, it may induce changes in the shape and mobility of myoblasts, and facilitate their localization at laminin-rich sites of secondary fiber formation. It is involved in the maintenance of the myofibers cytoarchitecture as well as for their anchorage, viability and functional integrity. Isoform Alpha-7X2B and isoform Alpha-7X1B promote myoblast migration on laminin 1 and laminin 2/4, but isoform Alpha-7X1B is less active on laminin 1 (In vitro). Acts as Schwann cell receptor for laminin-2. Acts as a receptor of COMP and mediates its effect on vascular smooth muscle cells (VSMCs) maturation (By similarity). Required to promote contractile phenotype acquisition in differentiated airway smooth muscle (ASM) cells. {ECO:0000250, ECO:0000269|PubMed:10694445, ECO:0000269|PubMed:17641293, ECO:0000269|PubMed:9307969}. | ADP-ribosylation;Alternative splicing;Calcium;Cell adhesion;Cell shape;Cleavage on pair of basic residues;Congenital muscular dystrophy;Direct protein sequencing;Disulfide bond;Glycoprotein;Integrin;Membrane;Metal-binding;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene belongs to the integrin alpha chain family. Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain. They mediate a wide spectrum of cell-cell and cell-matrix interactions, and thus play a role in cell migration, morphologic development, differentiation, and metastasis. This protein functions as a receptor for the basement membrane protein laminin-1. It is mainly expressed in skeletal and cardiac muscles and may be involved in differentiation and migration processes during myogenesis. Defects in this gene are associated with congenital myopathy. Alternatively spliced transcript variants encoding different isoforms have been noted for this gene. [provided by RefSeq, Feb 2009]. | hsa:3679; | cell surface [GO:0009986]; integrin complex [GO:0008305]; plasma membrane [GO:0005886]; integrin binding [GO:0005178]; laminin binding [GO:0043236]; metal ion binding [GO:0046872]; cell-matrix adhesion [GO:0007160]; endodermal cell differentiation [GO:0035987]; heterotypic cell-cell adhesion [GO:0034113]; integrin-mediated signaling pathway [GO:0007229]; muscle organ development [GO:0007517]; regulation of cell shape [GO:0008360]; skeletal muscle tissue development [GO:0007519] | 12057917_conclude that secondary integrin alpha 7 deficiency is rather common in muscular dystrophy/myopathy of unknown etiology 15117962_FHL2 and FHL3, respectively, are colocalized with alpha(7)beta(1) integrin receptor at the periphery of Z-discs, suggesting a role in mechanical stabilization of muscle cells 17054947_alpha7-expressing fetal myoblasts are capable of differentiation to osteoblast lineage with a coordinated switch in integrin profiles and may represent a mechanism that promotes homing and recruitment of myogenic stem cells for tissue remodeling. 17551147_Integrin alpha7 mutations are associated with prostate cancer, liver cancer, glioblastoma multiforme, and leiomyosarcoma 17618648_analysis of how distinct acidic clusters and hydrophobic residues in the alternative splice domains X1 and X2 of alpha7 integrins define specificity for laminin isoforms 17641293_Alpha7B is a novel marker of the contractile phenotype, and alpha7 expression is essential for human airway smooth muscle cell maturation, which is a laminin-dependent process. 18940796_cleavage is a novel mechanism that regulates alpha7 integrin functions in skeletal muscle, and that the generation of such cleavage sites is another evolutionary mechanism for expanding and modifying protein functions. 19416897_laminin-111 (alpha(1), beta(1), gamma(1)), which is expressed during embryonic development but absent in normal or dystrophic skeletal muscle, increased alpha(7)-integrin expression in mouse and DMD patient myoblasts 20453000_Observational study of gene-disease association. (HuGE Navigator) 20460506_ILK interaction with MCM7 and MCM7 phosphorylation may be a critical event in ITGA7 signaling pathway, leading to tumor suppression. 20651226_This report provides a novel insight into the mechanism, involving interaction with high temperature requirement A2, by which ITGA7 acts as a tumor suppressor. 23800289_Digenic mutational inheritance of the integrin alpha 7 and the myosin heavy chain 7B genes causes congenital myopathy with left ventricular non-compact cardiomyopathy. [ITGA7] 23830872_ITGA7 binds to tissue inhibitor of metalloproteinase 3 (TIMP3) in prostate cancer cells. 24091324_Alpha7beta1D integrin modifies Ca2+ regulatory pathways and offers a means to protect the myocardium from ischemic injury. 24227711_The absence of either alpha7beta1 integrin or alpha6beta1 integrin impairs the ability of Schwann cells to spread and to bind laminin. 26011651_Data suggest that ITGA7 is an epigenetically regulated tumour suppressor gene and a prognostic factor in human malignant pleural mesothelioma. 26076707_Taken together, these results further support the use of a7 integrin as a potential therapy for Duchenne muscular dystrophy 26320193_Data indicate that S100 calcium binding protein P (S100P) increased lung cancer cell migration by binding integrin alpha7. 27924820_As knockdown of Integrin alpha7 (ITGA7) can effectively reduce the stemness of oesophageal squamous cell carcinoma (OSCC) cells, ITGA7 could be a potential therapeutic target in OSCC treatment. 28602620_targeting of ITGA7 by RNAi or blocking mAbs impaired laminin-induced signaling, and it led to a significant delay in tumor engraftment plus a strong reduction in tumor size and invasion. 29884889_we identified forkhead box C1 (FOXC1) as a novel regulator of colorectal cancer metastases. FOXC1 directly binds its target genes integrin alpha7 (ITGA7) and fibroblast growth factor receptor 4 (FGFR4) and activates their expression 29943828_circITGA7 inhibits the proliferation and metastasis of colorectal cancer cells by suppressing the Ras signalling pathway and promoting the transcription of ITGA7. 30089289_Speculate that the postnatal splicing of alpha7A to alpha7B and of beta1A to beta1D integrins is delayed, altering spontaneous descent of the testes in the first months of life. 31289310_Integrin alpha7 expression is increased in asthmatic patients and its inhibition reduces Kras protein abundance in airway smooth muscle cells. 31325216_Overexpression of ITGA7 is associated with breast cancer. 31418948_Overexpression of ITGA7 is associated with non-small-cell lung cancer. 31698037_Integrin alpha 7 correlates with poor clinical outcomes, and it regulates cell proliferation, apoptosis and stemness via PTK2-PI3K-Akt signaling pathway in hepatocellular carcinoma. 31713264_Integrin alpha7 is overexpressed and correlates with higher pathological grade, increased T stage, advanced TNM stage as well as worse survival in clear cell renal cell carcinoma patients: A retrospective study. 31789398_Integrin alpha7 correlates with worse clinical features and prognosis, and its knockdown inhibits cell proliferation and stemness in tongue squamous cell carcinoma. 31855276_Predictive value of integrin alpha7 for acute myeloid leukemia risk and its correlation with prognosis in acute myeloid leukemia patients. 31942970_Downregulating integrin subunit alpha 7 (ITGA7) promotes proliferation, invasion, and migration of papillary thyroid carcinoma cells through regulating epithelial-to-mesenchymal transition. 32963582_Circular RNA CircITGA7 Promotes Tumorigenesis of Osteosarcoma via miR-370/PIM1 Axis. 34253873_Transcriptome profiles of stem-like cells from primary breast cancers allow identification of ITGA7 as a predictive marker of chemotherapy response. 34743543_ITGA7 relates to disease risk, pathological feature, treatment response and survival in Ph(-) acute lymphoblastic leukemia. 34884422_Decrease in ITGA7 Levels Is Associated with an Increase in alpha-Synuclein Levels in an MPTP-Induced Parkinson's Disease Mouse Model and SH-SY5Y Cells. 35194895_ITGA7, CD133, ALDH1 are inter-correlated, and linked with poor differentiation, lymph node metastasis as well as worse survival in surgical cervical cancer. | ENSMUSG00000025348 | Itga7 | 384.06121 | 1.0350039 | 0.0496362166 | 0.17972616 | 7.775266e-02 | 7.803664e-01 | 9.998360e-01 | No | Yes | 345.29268 | 66.090092 | 3.085216e+02 | 45.794789 | |
| ENSG00000135945 | 51455 | REV1 | protein_coding | Q9UBZ9 | FUNCTION: Deoxycytidyl transferase involved in DNA repair. Transfers a dCMP residue from dCTP to the 3'-end of a DNA primer in a template-dependent reaction. May assist in the first step in the bypass of abasic lesions by the insertion of a nucleotide opposite the lesion. Required for normal induction of mutations by physical and chemical agents. {ECO:0000269|PubMed:10536157, ECO:0000269|PubMed:10760286, ECO:0000269|PubMed:11278384, ECO:0000269|PubMed:11485998, ECO:0000269|PubMed:22266823}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;DNA synthesis;DNA-binding;Magnesium;Metal-binding;Nucleotidyltransferase;Nucleus;Reference proteome;Transferase | This gene encodes a protein with similarity to the S. cerevisiae mutagenesis protein Rev1. The Rev1 proteins contain a BRCT domain, which is important in protein-protein interactions. A suggested role for the human Rev1-like protein is as a scaffold that recruits DNA polymerases involved in translesion synthesis (TLS) of damaged DNA. [provided by RefSeq, Mar 2016]. | hsa:51455; | nucleoplasm [GO:0005654]; damaged DNA binding [GO:0003684]; deoxycytidyl transferase activity [GO:0017125]; DNA-directed DNA polymerase activity [GO:0003887]; metal ion binding [GO:0046872]; DNA replication [GO:0006260]; error-free translesion synthesis [GO:0070987]; error-prone translesion synthesis [GO:0042276]; response to UV [GO:0009411] | 12529368_purified human REV1 and REV7 proteins form a heterodimer in solution, which is stable through intensive purification steps. 12930947_UV-induced mutant frequencies at the HPRT locus were reduced up to 75% in cells with reduced levels of REV1 mRNA and data support that targeting the mutagenic translesion DNA replication pathway can greatly reduce the frequency of induced mutations. 15189446_REV1 interacts with three Y-family DNA polymerases. 15380106_REV1 interacts with pol eta in translesion synthesis of damaged DNA 15609317_Observational study of gene-disease association. (HuGE Navigator) 16495473_REV1-dependent processes are important determinants of cisplatin-induced genomic instability and the development of resistance. 16803901_a novel biochemical activity of human REV1 protein, due to higher affinity for single-stranded DNA (ssDNA) than the primer terminus 16824193_Rev1 is a polypeptide associated with Poleta. The study results suggest that arrested replication forks strengthen interactions among Poleta, Rad18/Rad6 and Rev1, consistent with the requirement for effective TLS by Poleta at sites of DNA lesions. 18270339_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18470628_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18470628_Results support Phe257Ser and Ser257Ser genotypes are associated with a decreased risk for cervical carcinoma, while Asn373Ser and Ser373Ser genotypes increased the risk. 18498753_Data show that PCNA ubiquitination and REV1 play distinct roles in the coordination of DNA damage bypass that are temporally separated relative to replication fork arrest. 18591245_human REV1, apparently the slowest Y family polymerase, is kinetically highly tolerant to N(2)-adduct at G but not to O(6)-adducts. 18975621_plays a role in mutagenesis and translesin DNA synthesis. (review) 19074885_Observational study of gene-disease association. (HuGE Navigator) 19157994_Poleta-REV1 interactions prevent spontaneous mutations, probably by promoting accurate translesion DNA synthesis past endogenous DNA lesions 19390575_Observational study of gene-disease association. (HuGE Navigator) 19464298_Novel structural features are important for providing Rev1 greater latitude in promoting efficient and error-free translesion DNA synthesis through the diverse array of bulky and potentially carcinogenic N(2)-deoxyguanosine DNA adducts in human cells. 19628463_Translesional DNA synthesis through a C8-guanyl adduct of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in Vitro: REV1 inserts dC opposite the lesion, and DNA polymerase kappa potentially catalyzes extension reaction from the 3'-dC terminus. 19661089_Observational study of gene-disease association. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 20028736_REV1 and Polzeta facilitate repair of interstrand cross-links independently of PCNA monoubiquitination and Poleta, whereas RAD18 plus Poleta, REV1, and Polzeta are all necessary for replicative bypass of cisplatin intrastrand DNA cross-links. 20059978_The results suggest that the positive charge on R357 could prevent interaction of REV1 with dGTP. 20164194_the interaction between REV7 and REV3 creates a structural interface for REV1 binding 20453000_Observational study of gene-disease association. (HuGE Navigator) 20691646_WRN facilitates REV1-dependent translesion synthesis. 20888339_Results suggest that abasic sites might be bypassed by single B- and Y-family pols or combinations, possibly by REV1 and pols iota, eta, and delta/PCNA at the insertion step opposite the lesion and by pols eta and delta/PCNA at the subsequent extension step. 21690293_Hsp90 promotes folding of REV1 into a stable and/or functional form(s) to bind to monoubiquitinated proliferating cell nuclear antigen in the regulation of translesion DNA synthesis-mediated mutagenesis 21926160_The REV1/Polzeta complex maintains genomic stability by directly participating in DNA double-stranded break repair. 22266823_FAAP20 binding stabilizes Rev1 nuclear foci and promotes interaction of the Fanconi anemia core with PCNA-Rev1 DNA damage bypass complexes. 22303021_REV7 subunit of pol zeta mediated the interaction between REV3 and the REV1 C terminus. 22691049_a structural basis for understanding the recognition of the Rev1-CT by Y-family DNA polymerases 22761336_Findings indicate that miR-96 regulates DNA repair and chemosensitivity by repressing RAD51 and REV1. 22828282_the Rev1 C-terminal domain utilizes independent interaction interfaces to simultaneously bind a fragment of the 'inserter' poleta and Rev7 subunit of the 'extender' polvarsigma, thereby serving as a cassette that may accommodate several polymerases 22859296_analysis of the crystal structure of the ternary complex composed of the C-terminal domain of human REV1, REV7, and a REV3 fragment 22869133_ternary complex of the C-terminal domain of human REV1 in complex with REV7 bound to a REV3 fragment has been crystallized. The crystals belonged to space group P3(1)21, with unit-cell parameters a = b = 74.7, c = 124.5 A 23220741_the first structural insights into the regulation of human Rev1 for TLS polymerases. 23761444_Rev1 but not Poleta depletion is epistatic to the lack of PCNA ubiquitination. 24366879_The results show that human Rev1 disrupts G4 DNA structures and prevents refolding in vitro. 24409475_Structural studies suggest the possible involvement of XRCC1 and its associated repair factors, REV1 in post replication repair. 24956248_Our results suggest for the first time that REV1 and REV3L SNPs might serve as potential predictive markers of outcome of cisplatin-based chemotherapy 25080294_The molecular mechanism of 3-nitrobenzanthrone genotoxicity in HEK293 cells is reported. 25614517_show that REV1 is a novel binding partner of the tumor suppressor p53 and regulates its activity 26318859_Data suggest Rev1 protein recognition mechanism by Fanconi anemia-associated protein 20 (FAAP20). 26680302_Rev1 is indispensable for Translesion synthesis mediated by Poleta, Poliota, and Polkappa but is not required for TLS by Polzeta. 26795561_REV1 can promote PCNA monoubiquitylation after UV radiation through interacting with ubiquitylated RAD18. 26903512_Saccharomyces cerevisiae 26914252_the catalytic function of REV1 is moderately or slightly altered by at least nine genetic variations, and the G4 DNA processing function of REV1 is slightly enhanced by the N373S variation, which might provide the possibility that certain germline missense REV1 variations affect the individual susceptibility to carcinogenesis by modifying the capability of REV1 for replicative bypass past DNA lesions and G4 motifs derived 26982350_Data suggest that relatively high affinity binding of PolD3-RIR motif to Rev1-C-terminal domain displaces subunits from PolN, Pol-iota, or PolK from Rev1 complex and promotes formation of Rev1/PolZ4 assembly with PCNA for translesion DNA replication. 27095204_The data directly show that, in the human genome, DNA Pol-eta and Rev1 bypass cyclobutane pyrimidine dimers and 6-4PP at replication forks, while only 6-4PP are also tolerated by a Rev3L-dependent gap-filling mechanism, independent of S phase. 28498946_These data indicate that dysregulation of cellular Rev1 levels leads to the accumulation of mutations and suppression of cell death, which accelerates the tumorigenic activities of DNA-damaging agents. 29778604_Study reports the solution NMR structure of a 108-residue fragment of human REV1 encompassing the two putative ubiquitin-binding motifs UBM1 and UBM2 in complex with ubiquitin. While in mammals UBM1 and UBM2 are both required for optimal association of REV1 with replication factories after DNA damage, only REV1 UBM2 binds ubiquitin. 30111544_results provide insights into the structure of the Rev1/Polzeta TLS assembly and highlight the function of Rev7 homo- and heterodimerization. 30465533_RAD51 regulates REV1 recruitment to DNA double-strand break sites via pulsed laser microirradiation. 30963698_elevated expression of Rev1 alone is sufficient to confer enhanced UV-damage tolerance and that this tolerance depends on its physical interaction with monoubiquitinated PCNA and Polzeta but is independent of Poleta. 32066793_Exome Sequencing in Individuals with Isolated Biliary Atresia. 33555350_Human Rev1 relies on insert-2 to promote selective binding and accurate replication of stabilized G-quadruplex motifs. 34508659_REV1-Polzeta maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. 34725419_Genetic and physical interactions between Poleta and Rev1 in response to UV-induced DNA damage in mammalian cells. 35115490_REV1 promotes lung tumorigenesis by activating the Rad18/SERTAD2 axis. | ENSMUSG00000026082 | Rev1 | 268.21191 | 1.0415551 | 0.0587391915 | 0.22900931 | 6.542886e-02 | 7.981125e-01 | 9.998360e-01 | No | Yes | 216.12895 | 35.213878 | 2.107592e+02 | 26.522426 | |
| ENSG00000135976 | 375248 | ANKRD36 | protein_coding | A6QL64 | ANK repeat;Alternative splicing;Coiled coil;Reference proteome;Repeat | 34041646_CircANKRD36 Knockdown Suppressed Cell Viability and Migration of LPS-Stimulated RAW264.7 Cells by Sponging MiR-330. 35074490_Mechanism of CircANKRD36 regulating cell heterogeneity and endothelial mesenchymal transition in aortic valve stromal cells by regulating miR-599 and TGF-beta signaling pathway. | 49.38691 | 0.5937908 | -0.7519733501 | 0.42284916 | 3.121697e+00 | 7.725629e-02 | 9.998360e-01 | No | Yes | 39.91477 | 11.576529 | 6.781146e+01 | 14.980118 | |||||||
| ENSG00000136122 | 79866 | BORA | protein_coding | Q6PGQ7 | FUNCTION: Required for the activation of AURKA at the onset of mitosis. {ECO:0000269|PubMed:16890155}. | Alternative splicing;Cell cycle;Cell division;Mitosis;Phosphoprotein;Reference proteome | BORA is an activator of the protein kinase Aurora A (AURKA; MIM 603072), which is required for centrosome maturation, spindle assembly, and asymmetric protein localization during mitosis (Hutterer et al., 2006 [PubMed 16890155]).[supplied by OMIM, Mar 2008]. | hsa:79866; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; meiotic spindle [GO:0072687]; nucleus [GO:0005634]; protein kinase binding [GO:0019901]; activation of protein kinase activity [GO:0032147]; cell cycle [GO:0007049]; cell division [GO:0051301]; regulation of mitotic nuclear division [GO:0007088]; regulation of mitotic spindle organization [GO:0060236]; regulation of protein localization [GO:0032880] | 18378770_Study concludes that tight regulation of the Bora protein by its synthesis and degradation is critical for cell cycle progression. 18521620_Plk1 controls Aurora A localization and function by regulating cellular levels of hBora. 18566290_study reports that the synergistic action of Bora & the kinase Aurora A controls G2-M transition; Bora accumulates in G2 phase & promotes Aur-A-mediated activation of Polo-like kinase 1, leading to activation of cyclin-dependent kinase 1 & mitotic entry 19487276_Recent advances in the understanding of the functional crosstalk between Plk1 and Aurora-A before and during mitosis. 20923822_Observational study and genome-wide association study of gene-disease association and gene-environment interaction. (HuGE Navigator) 23592782_Following UV irradiation, ataxia telangiectasia-mutated- and Rad3-related protein phosphorylates Bora at Thr-501. The phosphorylated Thr-501 is subsequently recognized by the E3 ubiquitin ligase SCF-beta-TRCP, which targets Bora for degradation. 23970419_Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. 24338364_Activation of Plk1 by Aurora-A may function as a bistable switch 25742493_Bora was found to play a significant role in radiosensitivity through the regulation of MDC1 and DNA repair. 27068477_These results reveal a crucial and conserved role of phosphorylation of the N terminus of Bora for Plk1 activation and mitotic entry. 27831827_the mechanism of Plk1 activation and the potential role of Bora phosphorylation by Cdk1, is reported. 28402276_our findings demonstrated that Bora was overexpressed and served as an independent biomarker for poor prognosis in multiple adenocarcinomas. 33771996_Bora phosphorylation substitutes in trans for T-loop phosphorylation in Aurora A to promote mitotic entry. 34287649_Aurora A kinase activation: Different means to different ends. | ENSMUSG00000022070 | Bora | 320.21088 | 0.8416474 | -0.2487121980 | 0.19486491 | 1.619375e+00 | 2.031790e-01 | 9.998360e-01 | No | Yes | 294.21685 | 63.871846 | 3.703979e+02 | 61.837710 | |
| ENSG00000136754 | 10006 | ABI1 | protein_coding | Q8IZP0 | FUNCTION: May act in negative regulation of cell growth and transformation by interacting with nonreceptor tyrosine kinases ABL1 and/or ABL2. May play a role in regulation of EGF-induced Erk pathway activation. Involved in cytoskeletal reorganization and EGFR signaling. Together with EPS8 participates in transduction of signals from Ras to Rac. In vitro, a trimeric complex of ABI1, EPS8 and SOS1 exhibits Rac specific guanine nucleotide exchange factor (GEF) activity and ABI1 seems to act as an adapter in the complex. Regulates ABL1/c-Abl-mediated phosphorylation of ENAH. Recruits WASF1 to lamellipodia and there seems to regulate WASF1 protein level. In brain, seems to regulate the dendritic outgrowth and branching as well as to determine the shape and number of synaptic contacts of developing neurons. {ECO:0000269|PubMed:11003655, ECO:0000269|PubMed:18328268}. | Acetylation;Alternative splicing;Cell junction;Cell projection;Chromosomal rearrangement;Coiled coil;Cytoplasm;Cytoskeleton;Direct protein sequencing;Host-virus interaction;Nucleus;Phosphoprotein;Reference proteome;SH3 domain;Synapse | This gene encodes a member of the Abelson-interactor family of adaptor proteins. These proteins facilitate signal transduction as components of several multiprotein complexes, and regulate actin polymerization and cytoskeletal remodeling through interactions with Abelson tyrosine kinases. The encoded protein plays a role in macropinocytosis as a component of the WAVE2 complex, and also forms a complex with EPS8 and SOS1 that mediates signal transduction from Ras to Rac. This gene may play a role in the progression of several malignancies including melanoma, colon cancer and breast cancer, and a t(10;11) chromosomal translocation involving this gene and the MLL gene has been associated with acute myeloid leukemia. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene, and a pseudogene of this gene is located on the long arm of chromosome 14. [provided by RefSeq, Sep 2011]. | hsa:10006; | cytoskeleton [GO:0005856]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; extracellular exosome [GO:0070062]; filopodium tip [GO:0032433]; growth cone [GO:0030426]; lamellipodium [GO:0030027]; nucleus [GO:0005634]; postsynaptic density [GO:0014069]; SCAR complex [GO:0031209]; cadherin binding [GO:0045296]; cytoskeletal protein binding [GO:0008092]; protein tyrosine kinase activator activity [GO:0030296]; SH3 domain binding [GO:0017124]; signaling adaptor activity [GO:0035591]; actin polymerization or depolymerization [GO:0008154]; dendrite morphogenesis [GO:0048813]; lamellipodium morphogenesis [GO:0072673]; megakaryocyte development [GO:0035855]; negative regulation of cell population proliferation [GO:0008285]; peptidyl-tyrosine phosphorylation [GO:0018108]; somitogenesis [GO:0001756]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11034908_Human SSH3BP1 is a marker of macropinocytic vesicles and potential regulator of macropinocytosis in mammalian cells. SSH3BP1 may play a role in fusion of macropinocytic vesicles. 12547160_t(10;11)(p11.2;q23) involving MLL and this gene is associated with congenital acute monocytic leukemia 12672821_Abi-1 regulates c-Abl-mediated phosphorylation of Mena by interacting with both proteins 15048123_Abi1 interacts directly with the WHD domain of WAVE2, increases WAVE2 actin polymerization activity and mediates the assembly of a WAVE2-Abi1-Nap1-PIR121 complex. 15591787_Abi enhances Abl-mediated downregulation and phosphorylation of Cdc2 kinase 15657136_Abi-1-mediated coupling of Abl to WAVE2 promotes Abl-evoked WAVE2 tyrosine phosphorylation required to link WAVE2 with activated Rac and with actin polymerization and remodeling at the cell periphery. 15769844_We demonstrate a cell-type-dependent requirement for various WASP-related proteins in Listeria entry and InlB-induced membrane ruffling; Abi1, a key component of WAVE complexes, is recruited at the entry site 15893754_Abi-1 promotes Abl-mediated BCAP phosphorylation and suggest that Abi-1 in general coordinates kinase-substrate interactions 16182283_Overexpression of e3B1 in NIH3T3/EGFR cells sensitized EGF-induced activation of Rac1, whereas it had no impact on EGF-induced activation of p21Ras. 16225669_results suggest that the Eps8/Abi1 complex is capable of regulating the localization and/or activity of actin nucleators 16385451_Observational study of gene-disease association. (HuGE Navigator) 16899465_c-Abl activates WAVE2 via tyrosine phosphorylation to promote actin remodeling in vivo; Abi-1 forms the crucial link between these two factors 17101133_NESH (Abi-3), like Abi-1 and Abi-2, is a component of the Abi/WAVE complex, but likely plays a different role in the regulation of c-Abl. 17501982_Data show that Chlamydia trachomatis activates Rac and promotes its interaction with WAVE2 and Abi-1 to activate the Arp2/3 complex resulting in the induction of actin cytoskeletal rearrangements that are required for invasion. 17943163_by activating the Abi1 pathway, Bcr-Abl induces a translocation of MT1-MMP to a membrane-associated structural complex enriched with F-actin and adhesion molecules. 17951403_ABI-1 plays an important role in the spread of breast cancer and this role may be mediated via the phosphatidylinositol 3-kinase pathway. 18328268_c-Abl kinase inhibition mediated by a phosphotyrosine located in trans in the c-Abl substrate, Abi1. 19158278_These findings uncover a novel link between cadherin-mediated adhesion and the regulation of actin dynamics through the requirement for an Abi/Dia complex for the formation and stability of cell-cell junctions. 19554484_Downregulation of ABI1 expression in human gastric carcinoma may play a critical role in tumor progression and in determining patient prognosis. 19843640_Abl interactor 1 regulates Src-Id1-matrix metalloproteinase 9 axis and is required for invadopodia formation, extracellular matrix degradation and tumor growth of human breast cancer cells 20110575_Combination of experimental and computational interactome research was used for the analysis of protein-protein interactions between Abi-1 and VASP in human platelets. 20137729_Expression of Abi1 is up-regulated in hepatocellular carcinoma tissues compared with corresponding para carcinomatous liver tissue, and is significantly correlated with tumor number, capsular formation, venous invasion, grade and prognosis. 20193272_The ABI1 gene is down-regulated in gastric cancer cells. ABI1 overexpression effectively inhibits cell proliferation. 20479892_Abi1 isoforms differentially regulate macropinocytosis as a consequence of their different relative affinities for activated Rac1 in Wave 2 complex. 20598684_Macropinocytosis is regulated by interactions between Abi1 pY213 and the C-terminal SH2 domain of p85-thereby linking Abl kinase signaling to p85-dependent regulation of macropinocytosis. 21046228_Breast tumors expressing high levels of Abi1 are significantly associated with early recurrence and worse survival on multivariate analysis. 21118970_Study implicates that the integrity of SOS1/EPS8/ABI1 tri-complex is a determinant of ovarian cancer metastasis. 21320496_Data show that disruption of phosphorylation sits Y398 and Y213 significantly weakens the binding of Abi-1 to c-Abl. 21900237_CDK1-mediated phosphorylation of Abi1 attenuates Bcr-Abl-induced F-actin assembly and tyrosine phosphorylation of WAVE complex during mitosis 22014333_The phosphorylation and dephosphorylation cycle of VASP by the Abi-1-bridged mechanism regulates association of VASP with focal adhesions, which may regulate adhesion of Bcr-Abl-transformed leukaemic cells. 22430194_Down-regulation of E3B1/ABI-1 expression in human carcinomas may play a critical role in tumor progression and in determining disease prognosis. 22808230_a possible role for Abi1 as a marker for early KRAS mutation in hyperplastic polyps 23740246_Data indicate that Abi1 is activated by the c-Abl-Crk-associated substrate (CAS) pathway, and Abi1 reciprocally controls the activation of its upstream regulator c-Abl. 24196611_Abi1 is expressed at the invasive front of colorectal carcinomas and localizes to the leading edge of lamellipodia in cultured colorectal carcinoma cells. 24699303_These results indicate that the alpha4-Abi-1 signaling pathway may mediate acquisition of the drug-resistant phenotype of leukemic cells. 24913355_Our data indicate that phosphorylated Abi1 contributes to the invasive properties of colorectal cancer. 26193797_Suggest that Abi1 acts as a tumor-promoting gene in epithelial ovarian cancer progression, which may lead to unfavorable prognosis. 27551040_these data indicate that HCV exploits host Abi1 protein via NS5A to modulate MEK/ERK signaling pathway for its own propagation. 28339046_our findings indicated that ABI1 contributes to the development and progression of hepatocellular carcinoma (HCC) as an oncogene and may serve as a valuable prognostic marker for HCC patients. 29802737_The expression of ABI1 was suppressed by miR-181a/b, and ABI1 was validated as a direct target of miR-181a/b. miR-181a/b were significantly upregulated in aggressive neuroblastoma, which enhanced its tumorigenesis and progression by suppressing the expression of ABI1. 30089695_These studies demonstrate a requirement for human herpesvirus 5 pUL135 interactions with Abi-1 and CIN85 for regulation of EGFR and mechanistically link the regulation of EGFR to virus reactivation. 30213875_CD34(+) hematopoietic progenitors and granulocytes from patients with primary myelofibrosis showed decreased levels of ABI1 transcript as well as increased activity of Src family kinases , STAT3, and NF-kappaB. 31530281_ABI1 controls prostate tumor progression and epithelial plasticity through regulation of EMT-WNT pathway 32606387_Distinctive roles of Abi1 in regulating actin-associated proteins during human smooth muscle cell migration. 32673396_PTEN dephosphorylates Abi1 to promote epithelial morphogenesis. 32728066_PTEN suppresses epithelial-mesenchymal transition and cancer stem cell activity by downregulating Abi1. 33759281_RNA Splicing of the Abi1 Gene by MBNL1 contributes to macrophage-like phenotype modulation of vascular smooth muscle cell during atherogenesis. 34031495_The roles and prognostic significance of ABI1-TSV-11 expression in patients with left-sided colorectal cancer. 34369620_Acetylation of Abelson interactor 1 at K416 regulates actin cytoskeleton and smooth muscle contraction. | ENSMUSG00000058835 | Abi1 | 473.59188 | 1.1851936 | 0.2451227415 | 0.17215653 | 1.979142e+00 | 1.594809e-01 | 9.998360e-01 | No | Yes | 497.87417 | 86.622843 | 4.633328e+02 | 62.259225 | |
| ENSG00000136895 | 84253 | GARNL3 | protein_coding | Q5VVW2 | Alternative splicing;GTPase activation;Phosphoprotein;Reference proteome | hsa:84253; | cytoplasm [GO:0005737]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; regulation of small GTPase mediated signal transduction [GO:0051056] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000038860 | Garnl3 | 144.16873 | 1.4202990 | 0.5061947024 | 0.29035481 | 2.910992e+00 | 8.797775e-02 | 9.998360e-01 | No | Yes | 159.86214 | 37.215589 | 1.227781e+02 | 22.124390 | |||
| ENSG00000137216 | 55362 | TMEM63B | protein_coding | Q5T3F8 | FUNCTION: Acts as an osmosensitive calcium-permeable cation channel (By similarity). Mechanosensitive ion channel that converts mechanical stimuli into a flow of ion (By similarity). {ECO:0000250|UniProtKB:Q3TWI9}. | Alternative splicing;Calcium;Cell membrane;Ion channel;Ion transport;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:55362; | actin cytoskeleton [GO:0015629]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; calcium activated cation channel activity [GO:0005227]; mechanosensitive ion channel activity [GO:0008381]; osmolarity-sensing cation channel activity [GO:1990760] | 31243992_overexpression of TMEM63B in HEK293T cells significantly enhanced cell migration and wound healing. | ENSMUSG00000036026 | Tmem63b | 1213.07848 | 1.0526028 | 0.0739611512 | 0.11802718 | 3.948292e-01 | 5.297718e-01 | 9.998360e-01 | No | Yes | 1284.64718 | 107.827716 | 1.154407e+03 | 75.576006 | ||
| ENSG00000137343 | 79969 | ATAT1 | protein_coding | Q5SQI0 | FUNCTION: Specifically acetylates 'Lys-40' in alpha-tubulin on the lumenal side of microtubules. Promotes microtubule destabilization and accelerates microtubule dynamics; this activity may be independent of acetylation activity. Acetylates alpha-tubulin with a slow enzymatic rate, due to a catalytic site that is not optimized for acetyl transfer. Enters the microtubule through each end and diffuses quickly throughout the lumen of microtubules. Acetylates only long/old microtubules because of its slow acetylation rate since it does not have time to act on dynamically unstable microtubules before the enzyme is released. Required for normal sperm flagellar function. Promotes directional cell locomotion and chemotaxis, through AP2A2-dependent acetylation of alpha-tubulin at clathrin-coated pits that are concentrated at the leading edge of migrating cells. May facilitate primary cilium assembly. {ECO:0000255|HAMAP-Rule:MF_03130, ECO:0000269|PubMed:20829795, ECO:0000269|PubMed:21068373, ECO:0000269|PubMed:24097348, ECO:0000269|PubMed:24906155}. | 3D-structure;Acetylation;Acyltransferase;Alternative splicing;Cell junction;Cell projection;Coated pit;Cytoplasm;Cytoskeleton;Membrane;Methylation;Phosphoprotein;Reference proteome;Transferase | This gene encodes a protein that localizes to clathrin-coated pits, where it acetylates alpha tubulin on lysine 40. This process may be important in microtubule growth, for instance during chemotaxis and the formation of cilium. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:79969; | axon [GO:0030424]; clathrin-coated pit [GO:0005905]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; Golgi apparatus [GO:0005794]; microtubule [GO:0005874]; microtubule bundle [GO:0097427]; mitotic spindle [GO:0072686]; lysine N-acetyltransferase activity, acting on acetyl phosphate as donor [GO:0004468]; tubulin N-acetyltransferase activity [GO:0019799]; alpha-tubulin acetylation [GO:0071929]; cilium assembly [GO:0060271]; dentate gyrus development [GO:0021542]; neuron development [GO:0048666]; positive regulation of NLRP3 inflammasome complex assembly [GO:1900227]; regulation of fat cell differentiation [GO:0045598]; regulation of microtubule cytoskeleton organization [GO:0070507]; spermatogenesis [GO:0007283] | 19851445_Observational study of gene-disease association. (HuGE Navigator) 23071314_cysteine residues play important catalytic roles through a ternary complex mechanism. alphaTAT1 mutations have analogous effects on tubulin acetylation in vitro and in cells 23071318_analysis reveals a basic patch implicated in substrate binding and a conserved glutamine residue required for catalysis, demonstrating that the family of alpha-tubulin acetyltransferases uses a reaction mechanism different from other lysine acetyltransferases 24097348_microtubules contacting clathrin-coated pits become acetylated by alphaTAT1; in migrating cells, this mechanism ensures the acetylation of microtubules oriented towards the leading edge, thus promoting directional cell locomotion and chemotaxis 24760594_Results suggest that lithium chloride (LiCl) treatments activate alpha-tubulin N-acetyltransferase 1 (alphaTAT1) by the inhibition of glycogen synthase kinase 3 beta (GSK-3beta) and promote the alpha-tubulin acetylation, and then elongate the primary cilia. 24846647_Crystal structure of the catalytic core of human MEC-17 in complex with acetyl-CoA. MEC17 has large, conserved surface patch that is critical for enzymatic activity suggesting extensive interactions with alpha-tubulin. 24906155_Mechanistic underpinnings for TAT activity and its preference for microtubules with slow turnover; cocrystal structures constrain TAT action to the microtubule lumen with Lys40 engaged in a suboptimal active site; despite the confined location of Lys40, TAT efficiently scans the microtubule bidirectionally and acetylates stochastically without preference for ends. 25494100_cellular quiescence induces Mec17 to couple the production of acetylated microtubules and Myh10, whose accumulation overcomes the inhibitory role of Myh9 and initiates ciliogenesis 25602620_Data suggest that invariant residues Arg132 and Ser160 in catalytic domain of ATAT1 participate in stable interaction with CoA and acetyl-CoA; ATAT1 with mutation at either residue exhibits much faster intracellular degradation. 26227334_Studies indicate that alpha-tubulin acetylation and microtubule level is mainly governed by opposing actions of alpha-tubulin acetyltransferase 1 (ATAT1) and histone deacetylase 6 (HDAC6). 27752143_these results demonstrate alphaTAT1 enters the lumen of microtubules from open extremities and spreads K40 acetylation marks longitudinally along cellular microtubules; this mode of tip-directed microtubule acetylation may allow for selective acetylation of subsets of microtubules 27836544_These results suggest that alphaTAT1-mediated Wnt1 expression via microtubule acetylation is important for colon cancer progression. 28428427_Depletion of the tubulin acetyltransferase TAT1 led to a significant increase in the frequency of microtubule breakage. 29321169_the actin-MRTF-SRF circuit controls alpha-TAT1 transcription. INF2 regulates the circuit, and hence microtubule acetylation, in cell types where it has a prominent role in actin polymerization. 29869029_The specific distributions of ATAT1 through the cell cycle in fibroblasts suggest multiple functions of ATAT1, which could include acetylation of microtubules, RNA transcription activity, severing microtubules, and completion of cytokinesis. 30333138_The interactions of GCN5L1, RanBP2 and alphaTAT1 function in concert to control alpha-tubulin acetylation and may contribute towards the regulation of cellular lysosome positioning. 34199510_Microtubule Acetylation Controls MDA-MB-231 Breast Cancer Cell Invasion through the Modulation of Endoplasmic Reticulum Stress. 34508164_alphaTAT1-induced tubulin acetylation promotes ameloblastoma migration and invasion. | ENSMUSG00000024426 | Atat1 | 601.42909 | 0.9231074 | -0.1154295183 | 0.14949593 | 5.967491e-01 | 4.398211e-01 | 9.998360e-01 | No | Yes | 528.62408 | 52.200996 | 5.427314e+02 | 42.030851 | |
| ENSG00000137500 | 60492 | CCDC90B | protein_coding | Q9GZT6 | 3D-structure;Alternative splicing;Coiled coil;Membrane;Mitochondrion;Reference proteome;Transit peptide;Transmembrane;Transmembrane helix | hsa:60492; | integral component of membrane [GO:0016021]; mitochondrial membrane [GO:0031966]; mitochondrion [GO:0005739] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 30612859_Using MCUR1, the functionally characterized paralog of study the role of individual domains, results find that the conserved head domain in CCDC90B interacts directly with mitochondrial calcium uniporter and is destabilized upon Ca(2+) binding. | ENSMUSG00000030613 | Ccdc90b | 261.65910 | 1.2973963 | 0.3756192806 | 0.23438908 | 2.495928e+00 | 1.141411e-01 | 9.998360e-01 | No | Yes | 281.10574 | 57.060190 | 2.322909e+02 | 36.421083 | |||
| ENSG00000137507 | 2615 | LRRC32 | protein_coding | Q14392 | FUNCTION: Key regulator of transforming growth factor beta (TGFB1, TGFB2 and TGFB3) that controls TGF-beta activation by maintaining it in a latent state during storage in extracellular space (PubMed:19750484, PubMed:19651619, PubMed:22278742). Associates specifically via disulfide bonds with the Latency-associated peptide (LAP), which is the regulatory chain of TGF-beta, and regulates integrin-dependent activation of TGF-beta (PubMed:22278742). Able to outcompete LTBP1 for binding to LAP regulatory chain of TGF-beta (PubMed:22278742). Controls activation of TGF-beta-1 (TGFB1) on the surface of activated regulatory T-cells (Tregs) (PubMed:19750484, PubMed:19651619). Required for epithelial fusion during palate development by regulating activation of TGF-beta-3 (TGFB3) (By similarity). {ECO:0000250|UniProtKB:G3XA59, ECO:0000269|PubMed:19651619, ECO:0000269|PubMed:19750484, ECO:0000269|PubMed:22278742}. | 3D-structure;Cell membrane;Disease variant;Disulfide bond;Glycoprotein;Growth factor binding;Leucine-rich repeat;Membrane;Mental retardation;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a type I membrane protein which contains 20 leucine-rich repeats. Alterations in the chromosomal region 11q13-11q14 are involved in several pathologies. [provided by RefSeq, Jul 2008]. | hsa:2615; | cell surface [GO:0009986]; extracellular matrix [GO:0031012]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; nucleoplasm [GO:0005654]; transforming growth factor beta binding [GO:0050431]; negative regulation of activated T cell proliferation [GO:0046007]; negative regulation of cytokine production [GO:0001818]; positive regulation of gene expression [GO:0010628]; regulation of transforming growth factor beta activation [GO:1901388]; regulation of transforming growth factor beta3 activation [GO:1901398]; secondary palate development [GO:0062009]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 18628982_GARP is a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression 19453521_GARP is a key receptor controlling FOXP3 in T(reg) cells following T-cell activation in a positive feedback loop assisted by LGALS3 and LGMN 19651619_Platelets and activated Tregs co-express latent TGF-beta and GARP on their membranes. 19666573_Expression of GARP on activated regulatory T cells correlates with their suppressive capacity. 19750484_Data show that latent TGF-beta, i.e. both LAP and mature TGF-beta, binds to GARP, which is present on the surface of stimulated Treg clones but not on Th clones. 20237496_Observational study of gene-disease association. (HuGE Navigator) 21615933_the processing and expression of LRRC32 21907864_on chromosome 11q13.5 near the leucine-rich repeat containing 32 gene (LRRC32, also known as GARP) associated with asthma risk 22070912_There are 2 independent signals, one in C11orf30 and the other in LRRC32, that are strongly associated with serum IgE levels. C11orf30-LRRC32 region may represent a common locus for atopic diseases via pathways involved in regulation of serum IgE levels 22278742_Findings support the idea that GARP is a new latent TGFbeta-binding protein that regulates the bioavailability of TGFbeta and provides a cell surface platform for alphaV integrin-dependent TGFbeta activation. 23650616_we investigated in detail miR-142-3 pregulation of GARP expression in regulatory CD25(+) CD4 T cells 24098777_GARP is regulated by miRNAs and controls latent TGF-beta1 production by human regulatory T cells. 26357016_GARP deficiency leads to accumulation of sphingolipid synthesis intermediates, changes in sterol distribution, and lysosomal dysfunction. 26584734_since GARP functions as a transporter of transforming growth factor beta (TGFbeta), a cytokine with broad pleiotropic traits, GARP transcriptional attenuation by alternative promoters might provide a mechanism regulating peripheral TGFb 26885615_High GARP expression is associated with pancreatic cancer and liver metastases from colorectal cancer. 27095576_GARP is a surface molecule of regulatory T cells with roles in the regulatory function and TGF-beta releasing [review] 27248166_Data show that the Treg activation marker GARP (glycoprotein A repetitions predominant) is expressed on primary melanoma. 27884290_GARP plays an important role in the pathogenesis of atopic dermatitis. 27913437_these results define the oncogenic effects of the GARP-TGFbeta axis in the tumor microenvironment 28005267_LRRC32 expression is significantly upregulated in human masticatory mucosa during wound healing 28207945_CD4(+) CD25(+) GARP(+) Treg cells are defective in dilated cardiomyopathy patients and GARP seems to be a better molecular definition of the regulatory phenotype. 28607112_study showed that stimulated, human B lymphocytes produce active TGF-beta1 from surface GARP/latent TGF-beta1 complexes with isotype switching to IgA production. 29458436_This review summarizes the most important features of GARP biology described to date including gene regulation, protein expression and mechanism in activating latent TGF-beta, and the function of GARP in regulatory T cell biology and peripheral tolerance, as well as GARP's increasingly recognized roles in platelet-mediated cancer immune evasion. [review] 29618665_Study demonstrated that B cells are required for the induction of oral tolerance of T cell-dependent antigens via GARP and revealed for the first time that cell surface GARP-TGF-beta is an important checkpoint for regulating B cell peripheral tolerance, highlighting a mechanism of autoimmune disease pathogenesis. 30361387_This finding reveals how GARP exploits an unusual medley of interactions, including fold complementation by the amino terminus of TGF-beta1, to chaperone and orient the cytokine for binding and activation by alphaVbeta8. 30443770_Increased GARP expression in papillary thyroid cancer was positively correlated with increased expression of Foxp3, which is very important for development of Tregs. But, there is no significant association of elevated expression of GARP with lymph node metastasis in papillary thyroid cancer. 30976112_Data indicate homozygous stop-gain variant in leucine rich repeat containing 32 protein (LRRC32) (c.1630C>T; p.(Arg544Ter)) as a candidate disease-associated gene in two families with developmental delay, cleft palate, and proliferative retinopathy. 31033124_Novel biomarkers for primary biliary cholangitis to improve diagnosis and understand underlying regulatory mechanisms. 31357555_Our findings reveal that GARP, as an immunoregulatory molecule, is located on, as well as in, tumor cells of GB and low-grade glioma, inhibiting effector T cell function, and thus contributing to the immunosuppressive tumor microenvironment of primary brain tumors. 31812328_Genetic variants of the C11orf30-LRRC32 region are associated with childhood asthma in the Chinese population. 31915300_Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-beta. 32375042_The Parkinson's Disease Protein LRRK2 Interacts with the GARP Complex to Promote Retrograde Transport to the trans-Golgi Network. 33203838_GARP promotes the proliferation and therapeutic resistance of bone sarcoma cancer cells through the activation of TGF-beta. 33380925_Expression of Leucine-rich Repeat-containing Protein 32 Following Lymphocyte Stimulation in Patients with Non-IgE-mediated Gastrointestinal Food Allergies. 34059789_Dysregulated immunity in PID patients with low GARP expression on Tregs due to mutations in LRRC32. 34235533_Novel anti-GARP antibody DS-1055a augments anti-tumor immunity by depleting highly suppressive GARP+ regulatory T cells. 34421887_GARP Correlates With Tumor-Infiltrating T-Cells and Predicts the Outcome of Gastric Cancer. | ENSMUSG00000090958 | Lrrc32 | 22.96372 | 0.5564386 | -0.8457055899 | 0.61719070 | 1.863481e+00 | 1.722235e-01 | 9.998360e-01 | No | Yes | 15.73742 | 6.477997 | 2.678227e+01 | 8.932342 | |
| ENSG00000138434 | 6744 | ITPRID2 | protein_coding | P28290 | Actin-binding;Alternative splicing;Coiled coil;Cytoplasm;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:6744; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; actin filament binding [GO:0051015]; signaling receptor binding [GO:0005102] | 14673706_localized as a membrane-bound form with extracellular regions; results suggested KRAP might be involved in the regulation of filamentous actin and signals from the outside of the cells 17934691_results suggested that KRAP might be a cytoskeleton-associated protein involving the structural integrity and/or signal transductions in human cancers. 21457704_KRAP is involved in the proper regulation of IP3R-mediated Ca2+ release. 21501587_the critical region of KRAP protein for the regulation of IP(3)R was determined. 21873152_the predicted coiled-coil region & the region adjacent to the coiled-coil region of the carboxyl-terminus of KRAP may be crucial for its interaction with the cytoskeleton or directional targeting toward the apical pole in polarized epithelial cells 26947549_phosphorylation at Ser92 of the sperm-specific antigen 2 (SSFA2)[phospho-SSFA2 (pS92)], was related to poor prognosis. 30712887_These result reveals us that SSFA2 may act as oncogene to promote the progression of glioma 34301929_KRAP tethers IP3 receptors to actin and licenses them to evoke cytosolic Ca(2+) signals. 34787049_Circular RNA UBR1 promotes the proliferation, migration, and invasion but represses apoptosis of lung cancer cells via modulating microRNA-545-5p/SSFA2 axis. | ENSMUSG00000027007 | Itprid2 | 277.89736 | 1.0919817 | 0.1269487022 | 0.20489227 | 3.864507e-01 | 5.341710e-01 | 9.998360e-01 | No | Yes | 386.58818 | 77.661820 | 3.660272e+02 | 56.731220 | |||
| ENSG00000138442 | 55759 | WDR12 | protein_coding | Q9GZL7 | FUNCTION: Component of the PeBoW complex, which is required for maturation of 28S and 5.8S ribosomal RNAs and formation of the 60S ribosome. {ECO:0000255|HAMAP-Rule:MF_03029, ECO:0000269|PubMed:16043514, ECO:0000269|PubMed:17353269}. | 3D-structure;Acetylation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ribosome biogenesis;Ubl conjugation;WD repeat;rRNA processing | This gene encodes a member of the WD repeat protein family. WD repeats are minimally conserved regions of approximately 40 amino acids typically bracketed by gly-his and trp-asp (GH-WD), which may facilitate formation of heterotrimeric or multiprotein complexes. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation. This protein is highly similar to the mouse WD repeat domain 12 protein at the amino acid level. The protein encoded by this gene is a component of a nucleolar protein complex that affects maturation of the large ribosomal subunit.[provided by RefSeq, Dec 2008]. | hsa:55759; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; PeBoW complex [GO:0070545]; preribosome, large subunit precursor [GO:0030687]; ribonucleoprotein complex binding [GO:0043021]; maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [GO:0000466]; maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [GO:0000463]; Notch signaling pathway [GO:0007219]; regulation of cell cycle [GO:0051726]; ribosomal large subunit biogenesis [GO:0042273] | 16043514_WDR12 triggers accumulation of p53 in a p19ARF-independent manner in proliferating cells but not in quiescent cells. 17353269_Results describe the role of PeBoW-specific proteins Pes1, Bop1, and WDR12 in complex assembly and stability, nucleolar transport, and pre-ribosome association. 19198609_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20738937_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20971364_Observational study of gene-disease association. (HuGE Navigator) 25825154_The DDX27 can interact specifically with the Pes1 and Bop1 but fulfils critical function(s) for proper 3' end formation of 47S rRNA independently of the PeBoW-complex. 25915632_MI associated WDR12 allele was associated significantly with diastolic dysfunction and left atrial size. 32180229_Integrative genomic analyses identify WDR12 as a novel oncogene involved in glioblastoma. | ENSMUSG00000026019 | Wdr12 | 915.53276 | 1.0146176 | 0.0209360721 | 0.13422056 | 2.401755e-02 | 8.768403e-01 | 9.998360e-01 | No | Yes | 950.81729 | 170.663960 | 1.065346e+03 | 147.366460 | |
| ENSG00000138650 | 57575 | PCDH10 | protein_coding | Q9P2E7 | FUNCTION: Potential calcium-dependent cell-adhesion protein. | 3D-structure;Alternative splicing;Calcium;Cell adhesion;Cell membrane;Glycoprotein;Membrane;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene belongs to the protocadherin gene family, a subfamily of the cadherin superfamily. This family member contains 6 extracellular cadherin domains, a transmembrane domain and a cytoplasmic tail differing from those of the classical cadherins. The encoded protein is a cadherin-related neuronal receptor thought to function in the establishment of specific cell-cell connections in the brain. This gene plays a role in inhibiting cancer cell motility and cell migration. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jan 2015]. | hsa:57575; | integral component of plasma membrane [GO:0005887]; calcium ion binding [GO:0005509]; cell adhesion [GO:0007155]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156] | 16247458_Ectopic expression of PCDH10 strongly suppressed tumor cell growth, migration, invasion and colony formation. 18644894_These results suggest that OL-pc remodels the motility and adhesion machinery at cell junctions by recruiting the Nap1-WAVE1 complex to these sites and, in turn, promotes the migration of cells. 19084528_PCDH10 is a gastric tumor suppressor; its methylation at early stages of gastric carcinogenesis is an independent prognostic factor. 19262141_The role of OL-protocadherin-dependent striatal axon growth in neural circuit formation. 19681120_Showed aberrant promoter hypermethylation of PCDH10 is assoc'd with downreg'n of gene expression in cervical cancer, and the promoter methylation in the PCDH10 gene occurs at an earliest identifiable stage of low-grade squamous intraepithelial lesion. 19709077_field methylation of the PCDH10 gene specifically in the invasion stage of cervical carcinogenesis, which might be used to develop a highly specific diagnostic test for cervical scrapings. 20353276_epigenetic regulation of PCDH10 was associated with carcinogenesis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21237250_Sp1/Sp3 and CBF/NF-Y transcription factors play a crucial role in the basal expression of the human PCDH10 gene. 21314642_The decreased PCDH10 expression in prostate cancer cells was associated with the aberrant methylation of PCDH10 promoter CpG islands. 21597995_genetic and epigenetic deregulation of PCDH10 occurs in a significant portion of medulloblastoma patients 21960365_PCDH10 promoter hypermethylation was frequent in both B-cell (81.9%) and T-cell (80%) acute lymphoblastic leukemia (ALL). 22206871_Data show that expression of PCDH10 is markedly reduced in gastric cancer cells and tissues, and suggest that it functions as a tumor suppressor gene in gastric cancer. 22245948_PCDH10 gene is a target for epigenetic silencing in multiple myeloma and provides a link between the dysregulation of angiogenesis and DNA methylation. 22543497_The expression of PCDH10 was silenced in hepatocellular carcinoma via de novo DNA methylation. 23171734_PCDH10 methylation is closely associated with malignancy of bladder transitional cell carcinoma 23180019_PCDH10 is an important tumor suppression gene with key roles of suppressing cell proliferation, clonogenicity, and inhibiting cell invasion in the development of colorectal cancer. 23321168_PCDH10 promoter methylation was detected in 59/117 (50.4%) of patients with bladder cancer, and none of 37 (0%) controls. Methylation was significantly associated with advanced stage, high grade, tumour recurrence and larger tumour size. 23321465_PCDH10 is frequently downregulated by promoter methylation and may serve as a tumor suppressor gene in non-small cell lung cancer. 23569128_Downregulated PCDH10 levels correlated with malignant behaviour and poor overall survival in patients with bladder cancer. 23839493_There was a significant correlation between PCDH10 methylation in cfDNA and tumour tissue in patients with early CRC. 23929756_study suggests that PCDH10 methylation occurs early in lymphomagenesis. PCDH10 expression was down regulated via promoter hypermethylation in T- and B-cell lymphoma cell lines 24309322_These results indicate that promoter methylation status of PCDH10, SPARC, and UCHL1 may be used both as prognostic and predictive molecular marker for colorectal cancer patients 24406169_suppression of the expression of PCDH10 by RNA interference induces the growth arrest and apoptosis of glioblastoma cells in vitro 24740680_Loss of PCDH10 expression is associated with metastasis in colorectal cancer. 25085246_Findings establish a novel PCDH10-Wnt/beta-catenin-MALAT1 regulatory axis that contributes to EEC development. 25086586_Aberrant methylation of PCDH10 predicts worse biochemical recurrence-free survival in patients with prostate cancer after radical prostatectomy. 25260683_Gastric cancer patients with 5 or more methylated CpG sites of PCDH10 promoter had significantly poorer survival. 25590240_These results suggest an important role of p53 in regulating tumor cell migration through activating PCDH10 expression. 26081897_PCDH10 antagonized MM cell proliferation via the downregulation of Wnt/beta-catenin/BCL-9 signaling, whereas PCDH10 repressed the expression of AKT to promote the expression of GSK3beta and then to restrain the activation of beta-catenin 26276761_PCDH10 methylation is a potential biomarker that predicts a poor prognosis after curative resection of pathological stage I non-small-cell lung cancer. 26406945_Our present findings suggested that the hypermethylated CpG site counts of PCDH10 DNA promoter for evaluating the prognosis of GC was reasonable by using the D-BGS 26679037_Study found that hypermethylation of CpG probes in the promoter regions of HOXA9 and PCDH10 was associated with mature B-cell neoplasms. 26871474_we found that long noncoding RNA (lncRNA) MALAT1 binds EZH2, suppresses the tumor suppressor PCDH10, and promotes gastric cellular migration and invasion 26881880_PCDH10 methylation in serum is a potential prognostic biomarker for prostate cancer. 26927373_Data show that protocadherin 10 (PCDH10) over-expression could significantly induce cell apoptosis, and restrain proliferation, invasion and migration ability of BXPC-3 pancreatic cancer cells. 26970279_Results suggest that protocadherin 10 (PCDH10)-Dishevelled, EGL-10 and Pleckstrin domain containing 1 (DEPDC1)-caspase signaling may be a novel regulatory axis in endometrial endometrioid carcinoma (EEC) development. 27338109_These findings suggest that Pcdh10 may influence subcellular actin cytoskeletal organization and axon-axon interactions in the course of familial amyloidotic polyneuropathy 27659532_we found that the malignant character of GISTs was initiated and amplified by PCDH10 in a process regulated by HOTAIR. In summary, our findings imply that PCDH10 and HOTAIR may be useful markers of disease progression and therapeutic targets. 28498423_This study provides insights into the tumorigenesis and progression of hepatocellular carcinoma (HCC), and puts forward the novel hypothesis that PCDH10 could be a new biomarker for HCC, or that combined with other molecular markers could increase the specificity and sensitivity of diagnostic tests for HCC. Restoration of PCDH10 could be a valuable therapeutic target for HCC. 29202805_PCDH10 hypermethylation were found in 54.2% (58/107) of DLBCL cases, but only 12.5% (1/8) in reactive lymph node/follicular hyperplasia. 30213786_Study found elevated promoter methylation and decreased expression of PCDH10 and PCDH17 in advanced gastric lesions, suggesting that elevated PCDH10 and PCDH17 methylation may be an early event in gastric carcinogenesis. 31088413_Study shows for that the methylation status of PCDH10 can predict prognosis in pancreatic ductal adenocarcinoma (PDAC) patients with a significant impact on the outcome in terms of progression-free survival. High levels of PCDH10 promoter methylation could be useful to identify patients at high risk of PDAC disease progression. 31210316_PCDH10 inhibits invasion of lymphoma cells by regulating beta-catenin. 31651684_Quantitation Analysis of PCDH10 Methylation in Adolescent Idiopathic Scoliosis Using Pyrosequencing Study. 32535594_Disruption of PCDH10 and TNRC18 Genes due to a Balanced Translocation. 33271263_PCDH10 exerts tumor-suppressor functions through modulation of EGFR/AKT axis in colorectal cancer. 33527791_HOTAIR Induces Methylation of PCDH10, a Tumor Suppressor Gene, by Regulating DNMT1 and Sponging with miR-148b in Gastric Adenocarcinoma. | ENSMUSG00000049100 | Pcdh10 | 873.71415 | 1.0569330 | 0.0798839637 | 0.13900089 | 3.352334e-01 | 5.625936e-01 | 9.998360e-01 | No | Yes | 1059.41106 | 94.461186 | 1.008797e+03 | 70.558607 | |
| ENSG00000138785 | 57117 | INTS12 | protein_coding | Q96CB8 | FUNCTION: Component of the Integrator complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (PubMed:16239144). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267). {ECO:0000269|PubMed:16239144, ECO:0000269|PubMed:23904267}. | Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | hsa:57117; | integrator complex [GO:0032039]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; snRNA 3'-end processing [GO:0034472]; snRNA processing [GO:0016180] | ENSMUSG00000028016 | Ints12 | 351.49035 | 0.9677977 | -0.0472225400 | 0.18550365 | 6.223720e-02 | 8.029943e-01 | 9.998360e-01 | No | Yes | 335.56457 | 58.696564 | 3.737151e+02 | 50.397744 | |||
| ENSG00000139133 | 84920 | ALG10 | protein_coding | Q5BKT4 | FUNCTION: Adds the third glucose residue to the lipid-linked oligosaccharide precursor for N-linked glycosylation. Transfers glucose from dolichyl phosphate glucose (Dol-P-Glc) onto the lipid-linked oligosaccharide Glc(2)Man(9)GlcNAc(2)-PP-Dol. | Endoplasmic reticulum;Glycosyltransferase;Membrane;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene encodes a membrane-associated protein that adds the third glucose residue to the lipid-linked oligosaccharide precursor for N-linked glycosylation. That is, it transfers the terminal glucose from dolichyl phosphate glucose (Dol-P-Glc) onto the lipid-linked oligosaccharide Glc2Man9GlcNAc(2)-PP-Dol. The rat protein homolog was shown to specifically modulate the gating function of the rat neuronal ether-a-go-go (EAG) potassium ion channel. [provided by RefSeq, Jan 2010]. | hsa:84920; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; dolichyl pyrophosphate Glc2Man9GlcNAc2 alpha-1,2-glucosyltransferase activity [GO:0106073]; dolichol-linked oligosaccharide biosynthetic process [GO:0006488]; protein N-linked glycosylation [GO:0006487] | 19204726_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000075470 | Alg10b | 112.67777 | 1.7609332 | 0.8163401798 | 0.28366381 | 8.292053e+00 | 3.981894e-03 | 7.538800e-01 | No | Yes | 121.02690 | 24.181869 | 7.312466e+01 | 11.403495 |
| ENSG00000139324 | 160418 | TMTC3 | protein_coding | Q6ZXV5 | FUNCTION: Transfers mannosyl residues to the hydroxyl group of serine or threonine residues. The 4 members of the TMTC family are O-mannosyl-transferases dedicated primarily to the cadherin superfamily, each member seems to have a distinct role in decorating the cadherin domains with O-linked mannose glycans at specific regions. Also acts as O-mannosyl-transferase on other proteins such as PDIA3 (PubMed:28973932). Involved in the positive regulation of proteasomal protein degradation in the endoplasmic reticulum (ER), and the control of ER stress response. {ECO:0000269|PubMed:21603654, ECO:0000269|PubMed:28973932}. | Alternative splicing;Disease variant;Endoplasmic reticulum;Glycoprotein;Lissencephaly;Membrane;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:28973932}. | This gene encodes a protein that belongs to the transmembrane and tetratricopeptide repeat-containing protein family. [provided by RefSeq, May 2010]. | hsa:160418; | endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; dolichyl-phosphate-mannose-protein mannosyltransferase activity [GO:0004169]; mannosyltransferase activity [GO:0000030]; positive regulation of proteasomal protein catabolic process [GO:1901800]; protein O-linked mannosylation [GO:0035269]; response to endoplasmic reticulum stress [GO:0034976] | 21603654_that SMILE is involved in the endoplasmic reticulum stress response, by modulating proteasome activity and XBP-1 transcript expression. This function of SMILE may influence immune cell behavior in the context of transplantation. 21956870_SMILE is a novel transgene specifying both bronchial smooth muscle and lung alveolar myofibroblast lineages, contributing to the understanding of the biological control of the development of these cells. 28973932_O-mannosylation pathway dedicated to cadherins/ protocadherins orchestrated by the four TMTC1-4 genes 33436046_Conserved sequence motifs in human TMTC1, TMTC2, TMTC3, and TMTC4, new O-mannosyltransferases from the GT-C/PMT clan, are rationalized as ligand binding sites. 34794738_The deep infiltrating endometriosis tissue has lower T-cadherin, E-cadherin, progesterone receptor and oestrogen receptor than endometrioma tissue. | ENSMUSG00000036676 | Tmtc3 | 163.57541 | 1.2625105 | 0.3362953715 | 0.26698529 | 1.613209e+00 | 2.040413e-01 | 9.998360e-01 | No | Yes | 165.63944 | 37.238325 | 1.398812e+02 | 24.296107 |
| ENSG00000140044 | 122953 | JDP2 | protein_coding | Q8WYK2 | FUNCTION: Component of the AP-1 transcription factor that represses transactivation mediated by the Jun family of proteins. Involved in a variety of transcriptional responses associated with AP-1 such as UV-induced apoptosis, cell differentiation, tumorigenesis and antitumogeneris. Can also function as a repressor by recruiting histone deacetylase 3/HDAC3 to the promoter region of JUN. May control transcription via direct regulation of the modification of histones and the assembly of chromatin. {ECO:0000269|PubMed:12707301, ECO:0000269|PubMed:12903123, ECO:0000269|PubMed:16026868, ECO:0000269|PubMed:16518400}. | Alternative splicing;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:122953; | chromatin [GO:0000785]; nucleus [GO:0005634]; cAMP response element binding [GO:0035497]; chromatin binding [GO:0003682]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; leucine zipper domain binding [GO:0043522]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of histone deacetylation [GO:0031065]; regulation of transcription by RNA polymerase II [GO:0006357] | 14627710_c-Jun dimerization protein 2 inhibits cell transformation and has a role as a tumor suppressor gene 16026868_JDP2 is a cellular survival protein whose presence is necessary for normal cellular function 18396163_JDP2 acts as a repressor and could be functionally associated with HDAC3 to inhibit CHOP transcription 18671972_IRF2-BP1 is a JDP2-binding protein enhancing the polyubiquitination of JDP2 and represses ATF2-mediated transcriptional activation from a CRE-containing promoter. 19553667_A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. 20452405_3 SNPs (2 intronic: rs741846 & rs175646; & 1 in the untranslated region: rs8215) & their genotype distribution showed significant association in the Japanese & Korean but not Dutch intracranial aneurysm patients. 20452405_Observational study of gene-disease association. (HuGE Navigator) 20677166_JDP2 expression was downregulated in pancreatic carcinoma & this correlated with metastasis & decreased post-surgery survival. 20950777_The molecular mechanisms that underlie the action of JDP2 in cellular aging and replicative senescence by mediating the dissociation of polycomb repressive complexes from the p16(Ink4a)/Arf locus are discussed. 21525011_JDP2 is crucial to triggering reactivation from latency to lytic replication 22989952_the recruitment of multiple HDAC members to JDP2 and ATF3 is part of their transcription repression mechanism. 24120378_Preeclamptic plasma induces transcription modifications involving the AP-1 transcriptional regulator JDP2 in endothelial cells. 24232097_Results suggest that JDP2 is an integral component of the Nrf2-MafK complex and that it modulates antioxidant and detoxification programs by acting via the ARE. 28315425_In hepatocellular carcinoma, high expression of JDP2 is significantly correlated with smaller tumor size, early stage HCC and better survival. 29941549_These studies establish JDP2 as a novel oncogene in high-risk T cell acute lymphoblastic leukemia. 30721335_JDP2 was downregulated in myelodysplastic syndrome. Its expression was inversely related to disease aggressiveness and AML transformation. JDP2 suppression is a direct result of reduced PU.1. PU.1 and JDP2 expression correlate and are concurrently reduced with the extent of differentiation arrest and aggression/prognosis in MDS/AML. It was upregulated upon azacytidine treatment. 30877624_JDP2 is downregulated in HCC tissues and cells, and overexpressed JDP2 facilitated HCC cell invasion and EMT. 32150333_Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. 33108704_JDP2 is directly regulated by ATF4 and modulates TRAIL sensitivity by suppressing the ATF4-DR5 axis. | ENSMUSG00000034271 | Jdp2 | 127.67168 | 1.0812428 | 0.1126905765 | 0.27313662 | 1.675995e-01 | 6.822541e-01 | 9.998360e-01 | No | Yes | 107.35481 | 24.270603 | 1.064344e+02 | 18.920429 | ||
| ENSG00000140374 | 2108 | ETFA | protein_coding | P13804 | FUNCTION: Heterodimeric electron transfer flavoprotein that accepts electrons from several mitochondrial dehydrogenases, including acyl-CoA dehydrogenases, glutaryl-CoA and sarcosine dehydrogenase (PubMed:27499296, PubMed:15159392, PubMed:15975918, PubMed:9334218, PubMed:10356313). It transfers the electrons to the main mitochondrial respiratory chain via ETF-ubiquinone oxidoreductase (ETF dehydrogenase) (PubMed:9334218). Required for normal mitochondrial fatty acid oxidation and normal amino acid metabolism (PubMed:12815589, PubMed:1882842, PubMed:1430199). {ECO:0000269|PubMed:10356313, ECO:0000269|PubMed:12815589, ECO:0000269|PubMed:1430199, ECO:0000269|PubMed:15159392, ECO:0000269|PubMed:15975918, ECO:0000269|PubMed:27499296, ECO:0000269|PubMed:9334218, ECO:0000303|PubMed:17941859, ECO:0000305|PubMed:1882842}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Disease variant;Electron transport;FAD;Flavoprotein;Glutaricaciduria;Mitochondrion;Phosphoprotein;Reference proteome;Transit peptide;Transport | ETFA participates in catalyzing the initial step of the mitochondrial fatty acid beta-oxidation. It shuttles electrons between primary flavoprotein dehydrogenases and the membrane-bound electron transfer flavoprotein ubiquinone oxidoreductase. Defects in electron-transfer-flavoprotein have been implicated in type II glutaricaciduria in which multiple acyl-CoA dehydrogenase deficiencies result in large excretion of glutaric, lactic, ethylmalonic, butyric, isobutyric, 2-methyl-butyric, and isovaleric acids. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:2108; | electron transfer flavoprotein complex [GO:0045251]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; electron transfer activity [GO:0009055]; flavin adenine dinucleotide binding [GO:0050660]; oxidoreductase activity [GO:0016491]; cellular amino acid catabolic process [GO:0009063]; fatty acid beta-oxidation using acyl-CoA dehydrogenase [GO:0033539]; respiratory electron transport chain [GO:0022904] | 11756429_These studies indicate that a series of conformational changes occur during the assembly of the TMADH.ETF electron transfer complex and that the kinetics of assembly observed with mutant TMADH or ETF complexes are much slower 16510302_Tissue samples from 16 unrelated patients with ETF deficiency were analysed and the majority of the patients had mutations in the ETFA gene. 17689999_No mutations in electron-transfer-flavoprotein but maternal riboflavin deficiency led to multiple acyl-CoA dehydrogenation deficiency 19208393_Observational study of gene-disease association. (HuGE Navigator) 20674745_Data established structural hotspots within the ETF fold, and provided a rationale for the prediction of effects of mutations in ETF. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21219902_investigations are compatible with the notion that the ETFalpha-T171 variant displays an altered conformational landscape that results in reduced protein function under thermal stress 21308847_These results are consistent with the electron transfer flavoprotein alpha II domain adopting orientations in solution that deviate from the crystal structure of free ETF towards the active, substrate-bound orientation. 24394546_the mechanism of tert-butyl hydroperoxide-induced an apoptosis cascade and endoplasmic reticulum stress in hepatocyte cells by up-regulation of ETFA, providing a new mechanism for liver injury. 28320150_our results indicate that genetic variants in ETFA may modify individual susceptibility to non-GBM of glioma in the Han Chinese population and support the role of the ETFA genes in the occurrence of glioma. 29301933_Data suggest that ETF (heterodimer of ETFA and ETFB) catalyzes irreversible and pH-dependent oxidation of 8alpha-methyl group of FAD to form to 8-formyl-FAD (8f-FAD). 31418342_Molecular and Clinical Investigations on Portuguese Patients with Multiple acyl-CoA Dehydrogenase Deficiency. 31665600_Molecular Oxygen Binding in the Mitochondrial Electron Transfer Flavoprotein. 31996215_A case report of a mild form of multiple acyl-CoA dehydrogenase deficiency due to compound heterozygous mutations in the ETFA gene. 34405537_NBPF4 mitigates progression in colorectal cancer through the regulation of EZH2-associated ETFA. 34704421_Screening of multiple acyl-CoA dehydrogenase deficiency in newborns and follow-up of patients. | ENSMUSG00000032314 | Etfa | 2562.56470 | 0.9684585 | -0.0462378566 | 0.13316228 | 1.161324e-01 | 7.332677e-01 | 9.998360e-01 | No | Yes | 2344.58070 | 406.768637 | 2.893804e+03 | 386.894930 | |
| ENSG00000140386 | 49855 | SCAPER | protein_coding | Q9BY12 | FUNCTION: CCNA2/CDK2 regulatory protein that transiently maintains CCNA2 in the cytoplasm. {ECO:0000269|PubMed:17698606}. | Alternative splicing;Disease variant;Endoplasmic reticulum;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Retinitis pigmentosa;Zinc;Zinc-finger | hsa:49855; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleic acid binding [GO:0003676]; zinc ion binding [GO:0008270] | 17553665_An unstable [CCTG](n) repeat in the second intron of the ZNF291 gene, on chromosome 15q21-24, was identified. 17698606_Study describes the biochemically purification and identification of SCAPER, a novel protein that specifically interacts with cyclin A/Cdk2 in vivo. 18245951_By sequestering cyclin A/Cdk2, SCAPER is capable of directing the activity of this kinase complex away from the nucleus and regulating cyclin A/Cdk2 equilibrium in distinct subcellular compartments. 30561111_Unique to our patient's presentation is the absence of intellectual disability and attention-deficit/hyperactivity disorder, suggesting that SCAPER-associated retinitis pigmentosa can also present without systemic manifestations. 30723319_As SCAPER expression is known to peak at late G1 and S phase, overlapping the timing of ciliary resorption, our data suggest a possible role of SCAPER in ciliary dynamics and disassembly, also affecting microtubule-related mitotic progression. 31069901_Homozygous variants in the gene SCAPER cause syndromic intellectual disability. 31192531_Delineating the expanding phenotype associated with SCAPER gene mutation. 32510560_Male sterility and reduced female fertility in SCAPER-deficient mice. 32527956_Absence of SCAPER causes male infertility in humans and Drosophila by modulating microtubule dynamics during meiosis. | ENSMUSG00000034007 | Scaper | 131.05494 | 0.5557993 | -0.8473639602 | 0.29743651 | 7.856610e+00 | 5.063523e-03 | 8.037760e-01 | No | Yes | 109.04169 | 24.417381 | 1.924740e+02 | 33.286874 | ||
| ENSG00000140463 | 585 | BBS4 | protein_coding | Q96RK4 | FUNCTION: The BBSome complex is thought to function as a coat complex required for sorting of specific membrane proteins to the primary cilia. The BBSome complex is required for ciliogenesis but is dispensable for centriolar satellite function. This ciliogenic function is mediated in part by the Rab8 GDP/GTP exchange factor, which localizes to the basal body and contacts the BBSome. Rab8(GTP) enters the primary cilium and promotes extension of the ciliary membrane. Firstly the BBSome associates with the ciliary membrane and binds to RAB3IP/Rabin8, the guanosyl exchange factor (GEF) for Rab8 and then the Rab8-GTP localizes to the cilium and promotes docking and fusion of carrier vesicles to the base of the ciliary membrane. The BBSome complex, together with the LTZL1, controls SMO ciliary trafficking and contributes to the sonic hedgehog (SHH) pathway regulation. Required for proper BBSome complex assembly and its ciliary localization. Required for microtubule anchoring at the centrosome but not for microtubule nucleation. May be required for the dynein-mediated transport of pericentriolar proteins to the centrosome. {ECO:0000269|PubMed:15107855, ECO:0000269|PubMed:17574030, ECO:0000269|PubMed:22072986}. | 3D-structure;Alternative splicing;Bardet-Biedl syndrome;Cell membrane;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Disease variant;Flagellum;Membrane;Mental retardation;Obesity;Protein transport;Reference proteome;Repeat;Sensory transduction;TPR repeat;Transport;Vision | This gene is a member of the Bardet-Biedl syndrome (BBS) gene family. Bardet-Biedl syndrome is an autosomal recessive disorder characterized by severe pigmentary retinopathy, obesity, polydactyly, renal malformation and cognitive disability. The proteins encoded by BBS gene family members are structurally diverse. The similar phenotypes exhibited by mutations in BBS gene family members are likely due to the protein's shared roles in cilia formation and function. Many BBS proteins localize to the basal bodies, ciliary axonemes, and pericentriolar regions of cells. BBS proteins may also be involved in intracellular trafficking via microtubule-related transport. The protein encoded by this gene has sequence similarity to O-linked N-acetylglucosamine (O-GlcNAc) transferases in plants and archaebacteria and in human forms a multi-protein 'BBSome' complex with seven other BBS proteins. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Mar 2016]. | hsa:585; | BBSome [GO:0034464]; centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; ciliary membrane [GO:0060170]; ciliary transition zone [GO:0035869]; cilium [GO:0005929]; cytosol [GO:0005829]; motile cilium [GO:0031514]; non-motile cilium [GO:0097730]; nucleus [GO:0005634]; pericentriolar material [GO:0000242]; photoreceptor connecting cilium [GO:0032391]; photoreceptor inner segment [GO:0001917]; photoreceptor outer segment [GO:0001750]; alpha-tubulin binding [GO:0043014]; beta-tubulin binding [GO:0048487]; dynactin binding [GO:0034452]; protein-macromolecule adaptor activity [GO:0030674]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; adult behavior [GO:0030534]; brain morphogenesis [GO:0048854]; centrosome cycle [GO:0007098]; cerebral cortex development [GO:0021987]; cilium assembly [GO:0060271]; dendrite development [GO:0016358]; face development [GO:0060324]; fat cell differentiation [GO:0045444]; fat pad development [GO:0060613]; heart looping [GO:0001947]; hippocampus development [GO:0021766]; intracellular transport [GO:0046907]; maintenance of protein location in nucleus [GO:0051457]; melanosome transport [GO:0032402]; microtubule anchoring at centrosome [GO:0034454]; microtubule cytoskeleton organization [GO:0000226]; mitotic cytokinesis [GO:0000281]; negative regulation of actin filament polymerization [GO:0030837]; negative regulation of appetite by leptin-mediated signaling pathway [GO:0038108]; negative regulation of gene expression [GO:0010629]; negative regulation of GTPase activity [GO:0034260]; negative regulation of systemic arterial blood pressure [GO:0003085]; neural tube closure [GO:0001843]; neuron migration [GO:0001764]; non-motile cilium assembly [GO:1905515]; photoreceptor cell maintenance [GO:0045494]; photoreceptor cell outer segment organization [GO:0035845]; positive regulation of cilium assembly [GO:0045724]; positive regulation of multicellular organism growth [GO:0040018]; protein localization to centrosome [GO:0071539]; protein localization to cilium [GO:0061512]; protein localization to organelle [GO:0033365]; protein localization to photoreceptor outer segment [GO:1903546]; protein transport [GO:0015031]; regulation of cilium beat frequency involved in ciliary motility [GO:0060296]; regulation of cytokinesis [GO:0032465]; regulation of lipid metabolic process [GO:0019216]; regulation of non-motile cilium assembly [GO:1902855]; regulation of stress fiber assembly [GO:0051492]; retina homeostasis [GO:0001895]; retinal rod cell development [GO:0046548]; sensory perception of smell [GO:0007608]; sensory processing [GO:0050893]; social behavior [GO:0035176]; spermatid development [GO:0007286]; striatum development [GO:0021756]; ventricular system development [GO:0021591]; visual perception [GO:0007601] | 12016587_BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Evaluated the spectrum of mutations in the recently identified BBS4 gene with a combination of haplotype analysis and mutation screening. 12365916_The phenotype of patients with BBS4 mutations consists of severe retinitis pigmentosa, variable obesity, brachydactyly with variable polydactyly, small or missing teeth, genital hypoplasia, and cardiovascular disease. 12872256_A novel Frameshift Mutation between the splice donor site and exon 5 of BBS4 in a Bardet-Biedl syndrome patient and a novel heterozygous base substitution in both an affected mother and her affected daughter. 17003356_Observational study of gene-disease association. (HuGE Navigator) 18299575_BBS2 and BBS4 localized to cellular structures associated with motile cilia. 19077438_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20508983_Observational study of gene-disease association. (HuGE Navigator) 20801516_Observational study of genetic testing. (HuGE Navigator) 22219648_A novel missense mutation in BBS4 that co-segregates with Leber Congenital Amaurosis was identified in a consanguineous family from Saudi Arabia. 23554981_Ectopic expression of human BBS4 can rescue Bardet-Biedl syndrome phenotypes in Bbs4 null mice. 23716571_Findings indicate that Bbs proteins play a central role in the regulation of the actin cytoskeleton and control the cilia length through alteration of RhoA levels. 24681783_mediates endosomal recycling, sorting and signal transduction of Notch receptors 24691443_loss of BBS1, BBS4, or OFD1 led to decreased NF-kappaB activity and concomitant IkappaBbeta accumulation and that these defects were ameliorated with SFN treatment. 24867303_Results present evidence of a role for BBS4 in mediating the phosphorylation of TrkB by BDNF and its activation requires a proper localization to the ciliary axoneme. 25533820_a novel nonsense mutation in BBS4 gene in a Chinese family with Bardet-Biedl syndrome. This homozygous mutation was predicted to completely abolish the synthesis of the BBS4 protein; a rare heterozygous missense SNP in BBS10 gene was also detected 32759308_The BBSome assembly is spatially controlled by BBS1 and BBS4 in human cells. 32894499_BBS4 Is Essential for Nuclear Transport of Transcription Factors Mediating Neuronal ER Stress Response. 33860840_BBS4 protein has basal body/ciliary localization in sensory organs but extra-ciliary localization in oligodendrocytes during human development. 33964006_Nephroplex: a kidney-focused NGS panel highlights the challenges of PKD1 sequencing and identifies a founder BBS4 mutation. | ENSMUSG00000025235 | Bbs4 | 405.04566 | 0.9903095 | -0.0140486181 | 0.17031010 | 6.789984e-03 | 9.343276e-01 | 9.998360e-01 | No | Yes | 347.61587 | 64.902729 | 4.011346e+02 | 57.727348 | |
| ENSG00000140464 | 5371 | PML | protein_coding | P29590 | FUNCTION: Functions via its association with PML-nuclear bodies (PML-NBs) in a wide range of important cellular processes, including tumor suppression, transcriptional regulation, apoptosis, senescence, DNA damage response, and viral defense mechanisms. Acts as the scaffold of PML-NBs allowing other proteins to shuttle in and out, a process which is regulated by SUMO-mediated modifications and interactions. Isoform PML-4 has a multifaceted role in the regulation of apoptosis and growth suppression: activates RB1 and inhibits AKT1 via interactions with PP1 and PP2A phosphatases respectively, negatively affects the PI3K pathway by inhibiting MTOR and activating PTEN, and positively regulates p53/TP53 by acting at different levels (by promoting its acetylation and phosphorylation and by inhibiting its MDM2-dependent degradation). Isoform PML-4 also: acts as a transcriptional repressor of TBX2 during cellular senescence and the repression is dependent on a functional RBL2/E2F4 repressor complex, regulates double-strand break repair in gamma-irradiation-induced DNA damage responses via its interaction with WRN, acts as a negative regulator of telomerase by interacting with TERT, and regulates PER2 nuclear localization and circadian function. Isoform PML-6 inhibits specifically the activity of the tetrameric form of PKM. The nuclear isoforms (isoform PML-1, isoform PML-2, isoform PML-3, isoform PML-4 and isoform PML-5) in concert with SATB1 are involved in local chromatin-loop remodeling and gene expression regulation at the MHC-I locus. Isoform PML-2 is required for efficient IFN-gamma induced MHC II gene transcription via regulation of CIITA. Cytoplasmic PML is involved in the regulation of the TGF-beta signaling pathway. PML also regulates transcription activity of ELF4 and can act as an important mediator for TNF-alpha- and IFN-alpha-mediated inhibition of endothelial cell network formation and migration.; FUNCTION: Exhibits antiviral activity against both DNA and RNA viruses. The antiviral activity can involve one or several isoform(s) and can be enhanced by the permanent PML-NB-associated protein DAXX or by the recruitment of p53/TP53 within these structures. Isoform PML-4 restricts varicella zoster virus (VZV) via sequestration of virion capsids in PML-NBs thereby preventing their nuclear egress and inhibiting formation of infectious virus particles. The sumoylated isoform PML-4 restricts rabies virus by inhibiting viral mRNA and protein synthesis. The cytoplasmic isoform PML-14 can restrict herpes simplex virus-1 (HHV-1) replication by sequestering the viral E3 ubiquitin-protein ligase ICP0 in the cytoplasm. Isoform PML-6 shows restriction activity towards human cytomegalovirus (HHV-5) and influenza A virus strains PR8(H1N1) and ST364(H3N2). Sumoylated isoform PML-4 and isoform PML-12 show antiviral activity against encephalomyocarditis virus (EMCV) by promoting nuclear sequestration of viral polymerase (P3D-POL) within PML NBs. Isoform PML-3 exhibits antiviral activity against poliovirus by inducing apoptosis in infected cells through the recruitment and the activation of p53/TP53 in the PML-NBs. Isoform PML-3 represses human foamy virus (HFV) transcription by complexing the HFV transactivator, bel1/tas, preventing its binding to viral DNA. PML may positively regulate infectious hepatitis C viral (HCV) production and isoform PML-2 may enhance adenovirus transcription. Functions as an E3 SUMO-protein ligase that sumoylates (HHV-5) immediate early protein IE1, thereby participating in the antiviral response (PubMed:20972456, PubMed:28250117). Isoforms PML-3 and PML-6 display the highest levels of sumoylation activity (PubMed:20972456, PubMed:28250117). {ECO:0000269|PubMed:20972456, ECO:0000269|PubMed:28250117}. | 3D-structure;Acetylation;Activator;Alternative splicing;Antiviral defense;Apoptosis;Biological rhythms;Chromosomal rearrangement;Coiled coil;Cytoplasm;DNA-binding;Endoplasmic reticulum;Endosome;Host-virus interaction;Immunity;Innate immunity;Isopeptide bond;Membrane;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Tumor suppressor;Ubl conjugation;Zinc;Zinc-finger | PATHWAY: Protein modification; protein sumoylation. {ECO:0000269|PubMed:20972456, ECO:0000269|PubMed:28250117}. | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. This phosphoprotein localizes to nuclear bodies where it functions as a transcription factor and tumor suppressor. Its expression is cell-cycle related and it regulates the p53 response to oncogenic signals. The gene is often involved in the translocation with the retinoic acid receptor alpha gene associated with acute promyelocytic leukemia (APL). Extensive alternative splicing of this gene results in several variations of the protein's central and C-terminal regions; all variants encode the same N-terminus. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:5371; | chromatin [GO:0000785]; chromosome, telomeric region [GO:0000781]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extrinsic component of endoplasmic reticulum membrane [GO:0042406]; heterochromatin [GO:0000792]; nuclear matrix [GO:0016363]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; cobalt ion binding [GO:0050897]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; SMAD binding [GO:0046332]; SUMO binding [GO:0032183]; SUMO transferase activity [GO:0019789]; sumo-dependent protein binding [GO:0140037]; transcription coactivator activity [GO:0003713]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; branching involved in mammary gland duct morphogenesis [GO:0060444]; cell fate commitment [GO:0045165]; cellular response to interleukin-4 [GO:0071353]; cellular response to leukemia inhibitory factor [GO:1990830]; cellular senescence [GO:0090398]; circadian regulation of gene expression [GO:0032922]; common-partner SMAD protein phosphorylation [GO:0007182]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; entrainment of circadian clock by photoperiod [GO:0043153]; extrinsic apoptotic signaling pathway [GO:0097191]; fibroblast migration [GO:0010761]; innate immune response [GO:0045087]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress [GO:0070059]; intrinsic apoptotic signaling pathway in response to oxidative stress [GO:0008631]; maintenance of protein location in nucleus [GO:0051457]; myeloid cell differentiation [GO:0030099]; negative regulation of angiogenesis [GO:0016525]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of interleukin-1 beta production [GO:0032691]; negative regulation of mitotic cell cycle [GO:0045930]; negative regulation of telomerase activity [GO:0051974]; negative regulation of telomere maintenance via telomerase [GO:0032211]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of translation in response to oxidative stress [GO:0032938]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; oncogene-induced cell senescence [GO:0090402]; PML body organization [GO:0030578]; positive regulation of apoptotic process involved in mammary gland involution [GO:0060058]; positive regulation of defense response to virus by host [GO:0002230]; positive regulation of extrinsic apoptotic signaling pathway [GO:2001238]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of histone deacetylation [GO:0031065]; positive regulation of peptidyl-lysine acetylation [GO:2000758]; positive regulation of protein localization to chromosome, telomeric region [GO:1904816]; positive regulation of signal transduction by p53 class mediator [GO:1901798]; positive regulation of telomere maintenance [GO:0032206]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein import into nucleus [GO:0006606]; protein stabilization [GO:0050821]; protein targeting [GO:0006605]; protein-containing complex assembly [GO:0065003]; regulation of calcium ion transport into cytosol [GO:0010522]; regulation of cell adhesion [GO:0030155]; regulation of cell cycle [GO:0051726]; regulation of circadian rhythm [GO:0042752]; regulation of double-strand break repair [GO:2000779]; regulation of protein phosphorylation [GO:0001932]; regulation of transcription, DNA-templated [GO:0006355]; response to cytokine [GO:0034097]; response to gamma radiation [GO:0010332]; response to hypoxia [GO:0001666]; response to UV [GO:0009411]; retinoic acid receptor signaling pathway [GO:0048384]; suppression of viral release by host [GO:0044790]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 11891284_UBE1L is a retinoid target that triggers PML/RARalpha degradation and apoptosis in acute promyelocytic leukemia 11907221_contributes to innate immunity, defining host susceptibility to viral infections and immunopathology 12006491_Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. 12032336_variant-type PML-RAR(alpha) fusion transcript in acute promyelocytic leukemia 12032831_Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. 12060771_induced PML in a mouse model 12080044_represses A20-mediated transcription. 12093737_Of seven known PML isoforms, only PML IV is capable of causing premature senescence in PML +/+ murine and human primary fibroblasts but not in PML -/- murine cells, providing the first evidence for functional differences among these isoforms. 12167712_PML is a key stress-responsive regulator of eIF4E mRNA-dependent transport. PML inhibition of eIF4E depends on its association with the eIF4E nuclear body. 12186918_PML does not affect herpes simplex virus 1 replication or the changes in the localization of ICP0 through infection 12384561_PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. 12402044_PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2 12419228_SUMO-1 protease-1 regulates gene transcription through PML 12438698_Purified RINGs, including PML, self-assemble into supramolecular structures in vitro that resemble those they form in cells. Self-assembly controls and amplifies reduction of 5' mRNA cap affinity of eIF4E by PML. 12505266_Cryptic translocation of PML/RARA on 17q. A rare event in acute promyelocytic leukemia. 12506013_PML inhibited STAT3 activity in NIH3T3, 293T, HepG2, and 32D cells. PML formed a complex with STAT3 through B-box and COOH terminal regions in vitro and in vivo, thereby inhibiting its DNA binding activity. 12506025_In human cell lines, PML is not involved directly in the regulation of MHC class I expression. 12540841_PML induces apoptosis through repression of the NF-kappaB survival pathway. 12595526_Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. 12610143_herpes simplex virus type 1 ICP4 is recruited into foci juxtaposed to ND10 (PML) in live, infected cells 12647219_findings strongly suggest that increased promyelocytic leukemia protein expression is associated with growth inhibition and differentiation of human neuroblastoma cells 12727882_role of interaction in recruiting cyclin T1 to nuclear bodies 12773567_PML regulates TopBP1 functions by association and stabilization of the protein in response to IR-induced DNA damage 12810724_PML recruits Chk2 and p53 into the PML nuclear bodies and enhances p53/Chk2 interaction 12837286_PML protein colocalizes and interacts with RFN36. 12907596_PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. 12917339_We show here that ZIPK is present in PODs, where it colocalizes with and binds to proapoptotic protein Daxx. 14526201_alternative splicing and role implicated in interaction with HIV-1 14528266_Results suggest that adenovirus type 5 has evolved a unique strategy that leads to the sustained neutralization of promyelocytic leukaemia protein bodies throughout infection, thereby ensuring optimal viral proliferation. 14534537_Leukemia-associated translocation products able to activate RAS modify PML and render cells sensitive to arsenic-induced apoptosis. 14597622_PML-induced apoptosis by down-regulation of Survivin. 14597990_The ectopic expression of the acute promyelocytic leukemia-specific PML/RARalpha oncoprotein in U-937 cells results in induction of TF mRNA and promoter activity 14630830_DNA sequence variations resulting in a truncated PML protein was identified in acute promyelocytic leukemia cases that displayed retinoic acid resistance and a very poor prognosis 14663483_PML and the PML-NB act as molecular hubs for the induction and/or reinforcement of programmed cell death through a selective and dynamic regulation of proapoptotic transcriptional events. 14715247_Together our results suggest that PML may suppress prostate cancer cell growth by inhibiting AR transactivation and/or enhancing p53 activity. 14970191_Data suggest that promyelocytic leukemia (PML) bodies form in nuclear compartments of high transcriptional activity, but they do not directly regulate transcription of genes in these compartments. 14970276_PML protein expression is frequently lost in human cancers of various histologic origins, and its loss associates with tumor grade and progression in some tumor histotypes 14992722_PML is a direct p53 target that modulates p53 effector functions. 15060166_Results demonstrate that transcription activity associated with PML bodies is selectively repressed by the recruitment of Bach2 around PML bodies 15096541_acute promyelocytic leukemia cell differentiation parallels transcriptional activation through PML-RARA-RXR oligomers 15163746_interference by human Cytomegalovirus IE1 protein with both the sumoylation of PML and its repressor activity requires a physical interaction with PML that also leads to disruption of PML oncogenic domains (PODs). 15184504_PML plays a crucial role in modulating p73 function. 15273249_PML is a negative regulator of p38 kinase in tumor cells 15331439_PML I could act as a mediator for acute myeloid leukemia1 and its coactivator p300/CBP to assemble into functional complexes; a specific isoform PML I forms a complex with AML1 15359634_The 3 breakpoint cluster regions in the PML gene [Intron 6 (bcr1), exon 6 (bcr2) or intron 3 (bcr3)] were studied in 43 Mexican Mestizo APL patients. Bcr1=62.7%, bcr2=9.3% & bcr3=27.9%. Bcr1 was more prevalent than in Caucasians but similar to Asians. 15459016_TRAILprotein is a novel downstream transcriptional target of PML. 15467728_PML bodies control the distribution, dynamics and function of CHFR 15569683_SUMO-1 functions to tether proteins to PML-containing nuclear bodies through post-translational modification and noncovalent protein-protein interaction 15589835_INI1 is dispensable for retrovirus-induced cytoplasmic accumulation of PML and does not interfere with virus integration. 15601827_PML-retinoic acid receptor alpha activities are regulated by neutrophil elastase in early myeloid cells 15746941_CDDO-Im downregulates PML expression in acute promyleocytic leukemic cells. 15749021_PML3 has a direct role in the control of centrosome duplication through suppression of Aurora A activation to prevent centrosome reduplication 15809060_inhibition of monocyte differentiation all contribute to the oncogenic activity of PML-RARalpha 15855159_IEX-1 is organized in subnuclear structures and partially co-localizes with promyelocytic leukemia protein in HeLa cells 15922731_Isoforms of PML protein differ in their effects on ND10 organization. 15968309_Observational study of genetic testing. (HuGE Navigator) 16007146_findings strengthen the relevance of the cross talk between PML and the p53 family members, imply a new tumour suppressive function of PML and unveil a possible role for PML in epidermal morphogenesis and differentiation 16113082_loss of one copy of PU.1 through deletion, plus down-regulation of the residual allele caused by PML-RARalpha expression, synergizes to expand myeloid progenitors susceptible to transformation, increasing the penetrance of acute promyelocytic leukemia 16154611_These results indicate that PML isoforms which are expressed in a serum-dependent manner suppress the propagation of influenza virus at an early stage of infection. 16307818_Our work implies that the balance of promyelocytic leukemia protein and Epstein-Barr virus BZLF1 levels in cells may affect how each protein functions. 16432238_PML-RARalpha functions by recruiting an HDAC3-MBD1 complex that contributes to the establishment and maintenance of the silenced chromatin state 16449642_permanent transcriptional silencing is mediated by the association of PML-RAR with chromatin-modifying enzymes and by recruitment of the histone methyltransferase SUV39H1, responsible for trimethylation of lysine 9 of histone H3 16501113_Interaction between Adenovirus type 5 E4 Orf3 and PML isoform II is necessary for nuclear domain 10 (ND10) rearrangement to occur 16501610_PML stimulated hSUMO-1 modification in yeast, in a manner that was dependent upon PML's RING-finger domain. PML:RARalpha also stimulated hSUMO-1 conjugation in yeast. 16540467_analysis of cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha 16630218_frequencies of the PML-RARalpha transcripts and subtypes in a series of 32 APL patients from Northeast Brazil 16797070_The PML-RARA fusions can be identified by molecular analyses such as reverse transcriptase-polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH). 16818720_We show that expression of PML isoform IV leads to formation of distinct nuclear bodies enriched in components of the ubiquitin-proteasome system. These bodies recruit soluble mutant ataxin-7 and promote its degradation preventing aggregate formation 16873256_PML contributes to a cellular antiviral repression mechanism that is countered by the activity of ICP0. 16873257_PML functions as part of an intrinsic immune mechanism against cytomegalovirus infections. 16912307_Our results provide evidence of how poliovirus counteracts p53 antiviral activity by regulating PML and NBs, thus leading to p53 degradation. 16916642_The interaction of TGIF with cPML through c-Jun may negatively regulate TGF-beta signaling through controlling the localization of cPML and, consequently, the assembly of the cPML-SARA complex. 16924230_apoptin interacts directly with the promyelocytic leukemia protein (PML) in tumor cells and accumulates in PML nuclear bodies 16935935_In response to ATRA, PML/RARalpha is dissociated from CAK, leading to MAT1 degradation, G1 arrest, and decreased CAK phosphorylation of PML/RARalpha 17027752_SUMO-1, PML and ZNF198 colocalize to punctate structures, shown by immunocytochemistry to be PML bodies. 17030982_PML nuclear bodies increase in number following DNA damage and this response is regulated by NBS1 as well as the kinases ATM, Chk2 and ATR. 17062732_Histone deacetylase inhibitors blocked the increase of PML nuclear body number and suppressed up-regulation of PML mRNA and protein levels in several human cell lines and in normal diploid skin fibroblasts 17146439_establish PML-mediated destabilization of Myc and the derepression of cell cycle inhibitor genes as an important regulatory mechanism that allows cell differentiation 17172828_the role of PML cytoplasmic proteins in the regulation of p53. 17173041_Our studies identify PML and SATB1 as a regulatory complex that governs transcription by orchestrating dynamic chromatin-loop architecture. 17360386_The increased expression of SUMO-1 in rheumatoid arthritis (RA) synovial fibroblasts (SFs) contributes to the resistance of these cells against Fas-induced apoptosis through increased SUMOylation of nuclear PML protein. 17419608_Results demonstrate that the gamma 1 isoform of phospholipase C associates with nuclear promyelocytic leukemia protein. 17428679_PML may be involved in nucleolar functions of normal non-transformed or senescent cells. The absence of nucleolar PML compartment in rapidly growing tumor cells suggests that PML association with the nucleolus might be important for cell-cycle regulation. 17441430_PML may play a role in cancer repression but not in cancer metastasis in laryngocarcinoma and nasopharyngeal carcinoma. 17475621_the hormone-independent association between PML-RARalpha and coactivators contributes to its ability to regulate gene expression 17543368_Study recapitulated the effect of PML on early steps in infection by papillomavirus, it was found that replication and transcription of transfected paillomaviral genomes were not dependent on PML. 17562868_The type IV isoform of PML interacted with PU.1, promoted its association with p300, and then enhanced PU.1-induced transcription and granulocytic differentiation and PU.1 directly activates the transcription of the C/EBPepsilon gene. 17767548_expression of proto-oncogene PML and HLA class I molecules were concordantly upregulated in hepatocellular carcinoma (HCC) and the expression of PML gene might be one of the mechanisms that leads to the increased expression of class I antigen in HCC 17822314_Downregulation of the PML protein is associated with breast cancers 17878236_Only two specific PML splice variants (PML-I and PML-IV) are efficiently targeted to the nucleolus and the abundant PML-I isoform is required for the targeting of endogenous PML proteins to this organelle. 17942542_promyelocytic leukemia protein (PML) and hDaxx, mediate an intrinsic immune response against HCMV infection by contributing independently to the silencing of HCMV IE gene expression. 17943164_PML/RAR gene cytogenetic abnormalities in APL 17960172_The presence of a single PML-RARA isoform with exon 5 is associated with recurrence in acute promyelocytic leukemia 17991421_These results would shed new insights for the molecular mechanisms of PML-RARalpha-associated leukemogenesis. 17996922_Endogenous WRN and BLM proteins localize in nucleoli and in nuclear PML bodies defined by isoforms of the PML protein, which is a key regulator of cellular senescence. 18024792_The transcription factor PML-RARalpha regulates key cancer-related genes and pathways by inducing a repressed chromatin formation on its direct genomic target genes. 18039859_Binding to Pin1 results in degradation of PML in a phosphorylation-dependent manner. Degradation of PML due to Pin1 acts both to protect a breast cancer tumor cell line from hydrogen peroxide-induced death and to increase the rate of proliferation. 18056407_In alternative lengthening of telomere cells, TRF2 inactivation/silencing triggers cellular senescence and substantial loss of telomeric DNA upon stable TRF2 knockdown. 18158568_Overexpression of PML was associated with poor survival in gallbladder carcinomas. 18160400_the Hes6-CBP complex in PML-NB may influence the proliferation of cells via p53-dependent and -independent pathways. 18246125_Unexpected coupling of PML with NFAT reveals a novel mechanism underlying the diverse physiological functions of promyelocytic leukemia. 18246126_following DNA damage, PML facilitates Thr18 phosphorylation by recruiting p53 and CK1 into PML nuclear bodies, thereby protecting p53 from inhibition by Mdm2, leading to p53 activation. 18298799_Over-expression of PML-2KA mutant in the cytoplasm, which was generated by mutagenesis of the nuclear localization signals of PML, in MCF-7 breast cancer cells suppressed PKM2 activity and the accumulation of lactate. 18463162_PML sequesters HDAC7 to relieve repression and up-regulate gene expression 18480450_reorganization of the PML nuclear body by E4 ORF3 antagonizes an innate antiviral response mediated by both PML and Daxx. 18511908_PCTA defines a new component of the TGF-beta signalling pathway that functions to facilitate Smad2 phosphorylation through controlling the accumulation of cPML into the cytoplasm, and consequently, the assembly of Smad2-receptor complex. 18566754_These data identify a key post-translational mechanism that controls PML protein levels in cancer cells and suggest that CK2 inhibitors may be beneficial anti-cancer drugs. 18621739_PML is involved in trichostatin A-induced apoptosis 18625722_These results demonstrate a novel function of HDAC7 and provide a regulatory mechanism of PML sumoylation. 18636556_PML/RARalpha fusion protein mediates the unique sensitivity to arsenic cytotoxicity in acute promyelocytic leukemia cells. 18664490_A kinetics model for factor exchange at PML NBs and highlight potential mechanisms to regulate intranuclear trafficking of specific factors at these domains. 18689862_PML is a novel prognostic tool for ampullary cancer patients. 18716620_results delineate a previously unknown PML-DAXX-HAUSP molecular network controlling PTEN deubiquitinylation and trafficking, which is perturbed by oncogenic cues in human cancer 18755984_In a series of human AMKL samples from both Down syndrome and non-Down syndrome patients, mutations were identified within KIT, FLT3, JAK2, JAK3, and MPL genes, with a higher frequency in DS than in non-DS patients. 18809579_purified the PML complex and identified Fbxo3 (Fbx3), Skp1, and Cullin1 as novel components amd role in transcriptional regulation and leukemogenesis 18812519_Reduced PML Protein Expression Is associated with glioblastoma. 18833293_PML disruption by EBNA1 requires binding to the cellular ubiquitin specific protease, USP7 or HAUSP, but is independent of p53. 18856066_the increase of level of survivin in tumor tissues is not the result of decrease in content of its inhibitors SMAC and PML, as their content in tumor and normal cells is the same. 18945770_The results indicate that both sets of events mediated by ICP0, the degradation of PML and the blocking of silencing via the HDAC1/2-CoREST-REST complex, are interdependent and in large measure dependent on events in the ND10 nuclear bodies. 19015637_DNA damage-induced PML SUMOylation and are required for the ability of PML to cooperate with HIPK2 for the induction of cell death. 19023333_Promyelomonocytic leukemia activation is a critical early event that participates in the apoptotic demise of HIV-1-elicited syncytia. 19029980_whereas transcriptional activation of PML-RARA is likely to control differentiation, its catabolism triggers leukemia-initiating cell eradication and long-term remission of mouse acute promyelocytic leukemia 19074885_Observational study of gene-disease association. (HuGE Navigator) 19100514_Acute promyelocytic leukemia with insertion of PML exon 7a and partial deletion of exon 3 of RARA is reported. 19111660_The existence of a proapoptotic autoregulatory feedback loop between p73, YAP, and the promyelocytic leukemia (PML) tumor suppressor gene, is shown. 19150978_The physical interaction between PML3 and TIP60 protects TIP60 from Mdm2-mediated degradation, suggesting that PML3 competes with MDM2 for binding to TIP60. 19224461_We report three of 40 diagnosed APL cases that showed morphological, cytochemical, and immunophenotypic features of hypergranular APL, but did not show a PML/RARalpha fusion signal or any of its variants, on FISH. 19322209_Knock-in human PML-RARalpha alone can confer properties of self-renewal to committed murine hematopoietic progenitors before the onset of disease. 19339552_Results provide insight into a dynamic pool of cytoplasmic nucleoporins that form a complex with the tumor suppressor protein PML during the G1 phase of the cell cycle. 19380586_sumoylation- and Sumo binding domain-dependent PML oligomerization within nuclear bodies is sufficient for RNF4-mediated PML degradation 19487292_In the context of mutant p53, PML enhances its cancer-promoting activities. 19553342_The authors demonstrate that LANA2 relieves PML-mediated transcriptional repression of survivin, a protein that directly contributes to malignant progression of primary effusion lymphoma cells. 19597475_PML was found to be important for E1A-induced suppression of EGFR and subsequent killing of head and neck squamous cell carcinoma cells suggesting a novel pathway involving PML and p73 in the regulation of EGFR expression. 19703418_this is the first report of the variable susceptibility of influenza A virus subtypes/strains to human PML proteins. 19728758_Frequency of the c.2260G>C (p.Ala754Pro) variant in isoform IV of the PML gene was higher in patients with colon polyposis and cancer 19808906_PML in PML-NBs links the DNA damage response with HBV replication and may cooperate with HBV-core and HDAC1 on the HBV circular DNA basal core promoter to form a positive feedback loop for HBV exacerbation during chemotherapy and radiotherapy. 19851296_Observational study of gene-disease association. (HuGE Navigator) 19861416_Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. 19863682_alternative splicing of PML/RARalpha transcripts might be involved in nonsense-mediated decay 19865110_PML-RARalpha ligand-binding domain deletion mutations associated with reduced disease control and outcome after first relapse of acute promyelocytic leukemia. 20100838_Promyelocytic leukemia protein controls cell migration in response to hydrogen peroxide and insulin-like growth factor-1 20123968_Data show that the major histocompatibility complex class II gene DRA is relocated to promyelocytic leukemia (PML) nuclear bodies upon induction with IFN-gamma, and this topology is maintained long after transcription shut off. 20130140_During interphase PML-NBs adopt a spherical organization characterized by the assembly of PML and Sp100 proteins into patches within a 50- to 100-nm-thick shell. 20133705_PML/RARalpha transactivates the TF promoter through an indirect interaction with an element composed of a GAGC motif and the flanking nucleotides, independent of AP-1 binding. 20133893_Patients with decreased expression of PML protein have higher adverse clinical features in a subgroup of acute promyelocytic leukemia. 20155840_We studied 21 PML-RARA positive/RARA-PML negative cases by bubble PCR and multiplex long template PCR to identify the genomic breakpoints. 20159609_PML-RARalpha/RXR functions as a local chromatin modulator and that specific recruitment of histone deacetylase activities to genes important for hematopoietic differentiation, RAR signaling, and epigenetic control is crucial to its transforming potential. 20159610_Selective targeting of PU.1-regulated genes by PML/RARalpha is a critical mechanism for the pathogenesis of acute promyelocytic leukemia(APL). 20181954_SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. 20198315_Observational study of gene-disease association. (HuGE Navigator) 20228380_Promyelocytic leukemia protein (PML) was immunolocalized within nuclei of villus mesenchyme, but largely absent in trophoblast nuclei, with a trend for increased PML reactivity in preeclamptic placenta. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20501696_MORC3 colocalizes with PML by a two-step molecular mechanism. 20544846_PML breakpoints are associated with acute promyelocytic leukemia. 20574048_autophagic degradation contributes significantly both to the basal turnover as well as the therapy-induced proteolysis of PML/RARA 20578240_Data describe differences in promyelocytic leukemia protein expression between different types of soft tissue sarcoma and the corresponding normal surrounding tissue. 20625391_Data show that the CSNK2A1P gene is a functional proto-oncogene in human cancers and its functional polymorphism appears to degrade PML differentially in cancer cells. 20639899_results together with earlier work are consistent with the idea that SUMOylation regulates targeting of E1B-55K to PML-containing subnuclear structures, known to control transcriptional regulation, tumour suppression, DNA repair and apoptosis 20699642_Upregulation of the PML tumor suppressor in cellular senescence triggered by diverse drugs including clinically used anti-cancer chemotherapeutics relies on stimulation of PML transcription by JAK/STAT-mediated signaling. 20719947_an interaction between EBNA1 and the host CK2 kinase is crucial for EBNA1 to disrupt PML bodies and degrade PML proteins 20800603_Observational study of gene-disease association. (HuGE Navigator) 20826694_SUMOylation promotes PML degradation during encephalomyocarditis virus infection. 20972455_have demonstrated that the lymphoid leukemia-associated protein, PAX5-PML chimeric oncogenic protein, could bind to PAX5 response-element as homodimer to inhibit the transactivation of PAX5 target-genes 21037079_Pilocytic astrocytomas have telomere-associated promyelocytic leukemia bodies without alternatively lengthened telomeres. 21042751_Suppression of telomerase could activate the PML-dependent p53 signaling pathway and inhibit bladder cancer cell growth. 21057547_found that dominant-negative mutants of PML blocked AXIN-induced p53 activation, and that AXIN promotes PML sumoylation, a modification necessary for PML functions 21092142_The study emphasizes a role of the variable C-terminus in subcellular target selection and a general role of the N-terminal tripartite motif domain in promoting protein clustering. 21115099_PML functions as a positive regulator of IFNgamma signaling 21161613_Recent literature on the role of PML in the central nervous system. 21172801_PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. 21187718_Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein 21192925_These results suggest that SUMO interaction motif-mediated HIPK2 targeting to PML-NBs is crucial for HIPK2-mediated p53 activation and induction of apoptosis. 21198351_PML is induced by interferon, leading to a marked increase of expression of PML isoforms and the number of PML nuclear bodies [Review] 21205865_the mechanism by which PML induces a permanent cell cycle exit and activates p53 and senescence 21217775_data suggest the possible involvement of this fusion protein in the leukemogenesis of B-ALL in a dual dominant-negative manner and the possibility that ALL with PAX5-PML can be treated with ATO 21240990_the clinicopathological associations and prognostic implications of promyelocytic leukemia gene (PML) expressions in patients with esophageal squamous cell carcinomas 21245861_New insights that arose from these studies, in particular focussing on newly identified PML-RARalpha target genes, its interplay with RXR and deregulation of epigenetic modifications. 21304169_The antiviral functions of PML-nuclear bodies are inactivated through reorganization during normalBK virus infection. 21304940_The efficient sequestration of virion capsids in PML cages appears to be the outcome of a basic cytoprotective function of this distinctive category of PML-NBs in sensing and safely containing nuclear aggregates of aberrant proteins. 21305000_Silencing of USP7 was found to increase the number of PML-NBs, to increase the levels of PML protein and to inhibit PML polyubiquitylation in nasopharyngeal carcinoma cells 21310922_Depletion of S100A10 by RNA interference effectively blocked the enhanced fibrinolytic activity observed after induction of the PML-RAR-alpha oncoprotein 21364283_physiologic PML-RARA expression was sufficient to direct a hematopoietic progenitor self-renewal program in vitro and in vivo 21430051_These findings demonstrate that Us3 orthologues derived from distantly related alphaherpesviruses cause a disruption of PML protein nuclear bodies in a kinase- and proteasome-dependent manner but, do not target PML for degradation. 21489587_UL35 bodies form independently of PML and subsequently recruit PML, Sp100 and Daxx. 21529941_PML-ADAMTS17-RARA gene rearrangement is associated with pregnancy-related acute promyelocytic leukemia. 21613260_Data indicate that PML-RARA (PR)-WT in the soluble fraction was transferred to the insoluble fraction after treatment with AsO, but PR-B/L-mut was stably detected in fractions both with and without AsO. 21639834_PML interacts with WRN and regulates double-strand break repair in gamma-irradiation-induced DNA damage responses. 21678421_PML immunohistochemical results were correlated with prognosis and with radiological response to alkylating agents/antracycline-based first line therapy in soft tissue sarcoma. 21752469_The MPL immunohistochemical expression may represent a new marker for differential diagnosis between essential thrombocythemia and pre-fibrotic stage primary myelofibrosis. 21803845_PML functions in apoptosis regulation and tumor suppression are mediated by direct interaction with Fas. 21832009_Neither Daxx nor PML, the main players of ND10-based immunity, are required for the block to viral gene expression in the S/G2 phase. 21840486_study indicates that the KLHL20-mediated PML degradation and HIF-1alpha autoregulation play key roles in tumor progression 21934296_investigation of DNA breakpoints in PML-RARalpha V-form fusion transcripts in 7 Chinese acute promyelocytic leukemia patients (out of total of 134 APL patients); in 5 of these cases, the V-form transcripts were unique (i.e., not described previously) 21994459_The authors demonstrated that only isoform IV of PML interacted with encephalomyocarditis virus 3D polymerase (3Dpol) and sequestered it within nuclear bodies. 21998700_a novel functional connection between PML and the homologous recombination-mediated repair machinery 22022583_Loss of PML protein expression in gastric cancer cells contributes to increased IP-10 transcription. 22033920_a model in which Pin1 promotes PML degradation in an ERK2-dependent manner. 22045732_Assembly of a PML nuclear body occurs through multiple pathways and induces telomere elongation. 22155184_beta-catenin inhibits promyelocytic leukemia protein tumor suppressor function in colorectal cancer cells. 22167334_PML-RARa bcr1 fusion is not responsible for colorectal tumor development. 22237204_PML through its scaffold properties is able to control cell growth and survival at many different levels. (Review) 22296450_The 15q24 microdeletion may thus represent the first genetic hit to initiate leukaemogenesis and implicates PML and SUMO3 as novel components of the leukaemogenic network in TMD/AMKL. 22406621_PIAS1 was essential for PML degradation in non-small cell lung carcinoma (NSCLC) cells, and PML and PIAS1 were inversely correlated in NSCLC cell lines and primary specimens. 22498738_HK3 is: (1) directly activated by PU.1, (2) repressed by PML-RARA, and (3) functionally involved in neutrophil differentiation and cell viability of acute promyelocytic leukemia cells. 22573317_E4-ORF3 activity correlates with the inhibition of PML-mediated antiviral activity. 22589541_Data indicate that promyelocytic leukemia protein (PML) functions as a negative regulator in endothelial cell network formation and migration. 22711534_As IL6 is induced in response to various viral and genotoxic stresses, this cytokine may regulate autocrine/paracrine induction of PML under these pathophysiological states 22773875_Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation. 22875967_Herpes simplex virus 1 ICP0 has 2 distinct mechanisms of targeting PML: one dependent on SUMO modification and the other via SUMO-independent interaction with PML.I; conclude that the ICP0-PML.I interaction reflects a countermeasure to PML-related antiviral restriction 22886304_PML expression in breast cancer correlated strikingly with reduced time to recurrence, a gene signature of poor prognosis, and activated PPAR signaling. 22906876_results suggest that PML expression was positively associated with IL-6 expression in patients and was also related to tumor development and resistance to treatment in multiple myeloma. 22915647_PML-RARA rearrangement is associated with good response to therapy in acute myeloid leukemia. 22918509_Postulate that detection of PML cytoplasmic particles in patient fibroblasts could become a valuable marker for diagnosis of disease development in laminopathies. 22945642_These findings suggest that UHRF1 promotes the turnover of PML protein 22947142_PML is an essential regulator of TNFalpha signaling, and together they synergistically regulate cell adhesion by engaging multiple molecular mechanisms. 22982005_analysis of a cryptic PML-RARA translocation in a patient with acute promyelocytic leukemia [case report] 23007646_PML is required for efficient IFN-gamma-induced MHC II gene transcription through regulation of the class II transactivator (CIITA) 23064710_detection of double-stranded polymerase chain reaction (PCR) products of PML/RARalpha fusion gene in acute promyelocytic leukemia (APL) 23 | ENSMUSG00000036986 | Pml | 1472.11595 | 0.9950665 | -0.0071350938 | 0.11730464 | 3.736740e-03 | 9.512566e-01 | 9.998360e-01 | No | Yes | 1676.21217 | 186.223776 | 1.608761e+03 | 139.047276 |
| ENSG00000140543 | 55070 | DET1 | protein_coding | Q7L5Y6 | FUNCTION: Component of the E3 ubiquitin ligase DCX DET1-COP1 complex, which is required for ubiquitination and subsequent degradation of target proteins. The complex is involved in JUN ubiquitination and degradation. {ECO:0000269|PubMed:14739464}. | Alternative splicing;Nucleus;Reference proteome;Ubl conjugation pathway | hsa:55070; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; Cul4A-RING E3 ubiquitin ligase complex [GO:0031464]; cullin-RING ubiquitin ligase complex [GO:0031461]; nucleus [GO:0005634]; protein-containing complex binding [GO:0044877]; ubiquitin ligase-substrate adaptor activity [GO:1990756]; ubiquitin protein ligase binding [GO:0031625]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; protein ubiquitination [GO:0016567]; protein-containing complex assembly [GO:0065003] | 14739464_promotes ubiquitination and degradation of c-Jun by assembling a multisubunit ubiquitin ligase containing DNA Damage Binding Protein-1 (DDB1), cullin 4A (CUL4A), Regulator of Cullins-1 (ROC1), and constitutively photomorphogenic-1 17452440_These findings demonstrate that the conserved DET1 complex modulates Cul4A functions by a novel mechanism. 29360641_COP1/DET1/ETS axis regulates ERK transcriptome and sensitivity to MAPK inhibitors. | ENSMUSG00000030610 | Det1 | 390.51095 | 0.9196951 | -0.1207724319 | 0.16941663 | 5.103109e-01 | 4.750043e-01 | 9.998360e-01 | No | Yes | 375.46362 | 43.087246 | 4.364718e+02 | 38.952779 | ||
| ENSG00000140598 | 79631 | EFL1 | protein_coding | Q7Z2Z2 | FUNCTION: Involved in the biogenesis of the 60S ribosomal subunit and translational activation of ribosomes. Together with SBDS, triggers the GTP-dependent release of EIF6 from 60S pre-ribosomes in the cytoplasm, thereby activating ribosomes for translation competence by allowing 80S ribosome assembly and facilitating EIF6 recycling to the nucleus, where it is required for 60S rRNA processing and nuclear export. Has low intrinsic GTPase activity. GTPase activity is increased by contact with 60S ribosome subunits. {ECO:0000269|PubMed:21536732}. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Elongation factor;GTP-binding;Nucleotide-binding;Protein biosynthesis;Reference proteome;Ribosome biogenesis | hsa:79631; | ribonucleoprotein complex [GO:1990904]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; ribosome binding [GO:0043022]; translation elongation factor activity [GO:0003746]; GTP metabolic process [GO:0046039]; mature ribosome assembly [GO:0042256] | 20966410_Observational study of gene-disease association. (HuGE Navigator) 25015090_Downregulation of EFTUD1 induced cell-cycle arrest and apoptosis in gliomas by impairing ribosome biogenesis 25991726_Association of Elongation Factor-like 1 (EFL1) GTPase to SBDS did not modify the affinity for GTP but dramatically decreased that for GDP by increasing the dissociation rate of the nucleotide. 26479198_Upon EFL1 binding, SBDS is repositioned around helix 69, thus facilitating a conformational switch in EFL1 that displaces eIF6 by competing for an overlapping binding site on the 60S ribosomal subunit. 28331068_Mutations in EFL1 clinically manifest phenotypes of infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies. Mutant EFL1 proteins do not promote the release of yeast cytoplasmic Tif6 from the 60S subunit, likely preventing the formation of mature ribosomes. 30198570_Whole exome sequencing discloses heterozygous variants in the DNAJC21 and EFL1 genes but not in SRP54 in 6 out of 16 patients with Shwachman-Diamond Syndrome carrying biallelic SBDS mutations. 30545121_the effect of SBDS mutations on the interaction with EFL1were tested, and showed that all tested mutations disrupted the binding to EFL1. 31151987_we report biallelic mutations in EFL1 in 3 unrelated individuals with clinical features of SDS. Cellular defects in these individuals include impaired ribosomal subunit joining and attenuated global protein translation as a consequence of defective eIF6 eviction. 31838967_Exploring the role of elongation Factor-Like 1 (EFL1) in Shwachman-Diamond syndrome through molecular dynamics. 34115847_Somatic uniparental disomy mitigates the most damaging EFL1 allele combination in Shwachman-Diamond syndrome. | ENSMUSG00000038563 | Efl1 | 385.72741 | 0.8777159 | -0.1881740924 | 0.18511276 | 1.015730e+00 | 3.135340e-01 | 9.998360e-01 | No | Yes | 422.39317 | 78.016036 | 5.051310e+02 | 71.881304 | ||
| ENSG00000140859 | 3801 | KIFC3 | protein_coding | Q9BVG8 | FUNCTION: Minus-end microtubule-dependent motor protein. Involved in apically targeted transport (By similarity). Required for zonula adherens maintenance. {ECO:0000250, ECO:0000269|PubMed:19041755}. | 3D-structure;ATP-binding;Alternative splicing;Cell junction;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Membrane;Microtubule;Motor protein;Nucleotide-binding;Phosphoprotein;Reference proteome | This gene encodes a member of the kinesin-14 family of microtubule motors. Members of this family play a role in the formation, maintenance and remodeling of the bipolar mitotic spindle. The protein encoded by this gene has cytoplasmic functions in the interphase cells. It may also be involved in the final stages of cytokinesis. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jan 2016]. | hsa:3801; | centrosome [GO:0005813]; cytoplasmic vesicle membrane [GO:0030659]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; kinesin complex [GO:0005871]; microtubule [GO:0005874]; zonula adherens [GO:0005915]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; microtubule binding [GO:0008017]; microtubule motor activity [GO:0003777]; minus-end-directed microtubule motor activity [GO:0008569]; epithelial cell-cell adhesion [GO:0090136]; Golgi organization [GO:0007030]; microtubule-based movement [GO:0007018]; visual perception [GO:0007601]; zonula adherens maintenance [GO:0045218] | 19789344_High expression of KIFC3 is associated with docetaxel resistance in breast cancer. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23954441_KifC3 may play a regulatory role in minus-end directed peroxisomal transport for example by blocking the motor function of dynein at peroxisomes. 24275865_KIFC3 may guide free microtubules to their destination at the bridge and/or may slide and crosslink central bridge microtubules in order to stage the cells for abscission. 25253721_Data indicate that E-cadherin ubiquitination consistently increases after depletion of kinesin-like protein KIFC3 or ubiquitin-specific protease USP47. 26954557_Study found that in peripheral blood mononuclear cells the median expression of KIFC3, KIF1B, and KIF5C was much lower than the expression of dynactin subunits DCTN1 and DCTN3, in both sporadic amyotrophic lateral sclerosis and healthy cases 31481795_KIFC3 activity becomes the main driving force of centrosome cohesion to prevent premature spindle formation after linker dissolution as it counteracts the increasing EG5-driven pushing forces 32084403_Microtubule Minus-End Binding Protein CAMSAP2 and Kinesin-14 Motor KIFC3 Control Dendritic Microtubule Organization. | ENSMUSG00000031788 | Kifc3 | 434.62995 | 0.9391665 | -0.0905471624 | 0.17733674 | 2.691960e-01 | 6.038716e-01 | 9.998360e-01 | No | Yes | 424.16888 | 56.813530 | 4.152400e+02 | 44.254361 | |
| ENSG00000141068 | 8844 | KSR1 | protein_coding | Q8IVT5 | FUNCTION: Part of a multiprotein signaling complex which promotes phosphorylation of Raf family members and activation of downstream MAP kinases (By similarity). Independently of its kinase activity, acts as MAP2K1/MEK1 and MAP2K2/MEK2-dependent allosteric activator of BRAF; upon binding to MAP2K1/MEK1 or MAP2K2/MEK2, dimerizes with BRAF and promotes BRAF-mediated phosphorylation of MAP2K1/MEK1 and/or MAP2K2/MEK2 (PubMed:29433126). Promotes activation of MAPK1 and/or MAPK3, both in response to EGF and to cAMP (By similarity). Its kinase activity is unsure (By similarity). Some protein kinase activity has been detected in vitro, however the physiological relevance of this activity is unknown (By similarity). {ECO:0000250|UniProtKB:Q61097, ECO:0000269|PubMed:29433126}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Endoplasmic reticulum;Kinase;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Zinc;Zinc-finger | hsa:8844; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; membrane [GO:0016020]; protein-containing complex [GO:0032991]; ruffle membrane [GO:0032587]; 14-3-3 protein binding [GO:0071889]; ATP binding [GO:0005524]; MAP-kinase scaffold activity [GO:0005078]; metal ion binding [GO:0046872]; protein C-terminus binding [GO:0008022]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cAMP-mediated signaling [GO:0019933]; positive regulation of MAPK cascade [GO:0043410]; Ras protein signal transduction [GO:0007265]; regulation of cell population proliferation [GO:0042127]; regulation of MAP kinase activity [GO:0043405]; signal transduction [GO:0007165] | 15520853_KSR1 kinase activity is essential for anti-apoptotic protection of the intestinal epithelium [review] 15899786_KSR1-mediated regulation of ERK activity represents a novel determinant of CDDP sensitivity of cancer cells. 16732322_1,25D selectively increases the expression of the gene encoding kinase suppressor of Ras-1 (KSR-1) in HL60 cells 18332145_Human KSR1 proteins promote assembly of multivalent Raf.MEK complexes that are required for c-Raf kinase activation and functional coupling of active kinases to downstream substrates. 18426801_human 14-3-3gamma binds to the ERK1/2 molecular scaffold KSR1, which is mediated by the C-terminal stretch of 14-3-3gamma 18719367_Down-regulation of KSR1 in pancreatic cancer xenografts by antisense oligonucleotide correlates with tumor drug uptake. 19188442_KSR1 regulated the threshold required for MAPK activation in T cells without affecting the nature of the response. 19718030_DC-SIGN was constitutively associated with a signalosome complex consisting of the scaffold proteins LSP1, KSR1 and CNK and the kinase Raf-1. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20945381_Oncoprotein Cot1 represses kinase suppressors of Ras1/2 and 1,25-dihydroxyvitamin D3-induced differentiation of human acute myeloid leukemia cells. 21435442_KSR1 is overexpressed in endometrial carcinoma and regulates proliferation and TRAIL-induced apoptosis by modulating FLIP levels 21555152_Genetic disruption of the scaffolding protein, Kinase Suppressor of Ras 1 (KSR1), differentially regulates GM-CSF-stimulated hyperproliferation in hematopoietic progenitors expressing activating PTPN11 mutants D61Y and E76K. 21829671_simulations constitute a multi-dimensional exploration of how EGF-dependent EGFR endocytosis and ERK activation are dynamically affected by scaffolds KSR and MP1, co-regulated by Cbl-CIN85 and Endophilin A1 22752157_VRK2A can form a high molecular size complex with both MEK1 and KSR1; the KSR1 complex assembled and retained by VRK2A in the endoplasmic reticulum can have a modulatory effect on the signal mediated by MAPK,locally affecting the magnitude of its responses 23250398_the atypical C1 domain, the CC-SAM domain is required to target KSR-1 to the plasma membrane. 23416464_Elevated levels of phosphorylated KSR were detected in the nuclear fractions. 24095280_Data indicate that phosphorylation of BRAF by AMPK disrupts its association with KSR1. 24129246_Our findings integrate KSR1 into a network involving DBC1 and SIRT1, which results in the regulation of p53 acetylation and its transcriptional activity. 24909178_KSR1 stabilizes BRCA1 by reducing BRCA1 ubiquitination, inhibits tumor growth through BRCA1, and regulates BRCA1 stability via elevated BARD1 abundance and increased BRCA1-BARD1 interaction 25002533_Data show that caveolin-1 is necessary for optimal KSR1-dependent ERK activation by growth factors and oncogenic Ras. 25287073_KSR1 rs2241906 variants may predict survival in patients with advanced ERalpha+ BC treated with adjuvant TAM. 25608512_KSR1 is coordinately regulated with Notch signaling and oxidative phosphorylation in papillary thyroid cancer. 25962735_the relative mRNA copy values of KSR1, ERK1 and ERK2 in the cancer tissues were 2.43 +/- 0.49, 2.10 +/- 0.44 and 3.65 +/- 0.94..EGb 761 enhanced the chemotherapy sensitivity through suppression of the KSR1-mediated ERK1/2 pathway in gastric cancer 26549023_Data show that neurofibromin 2 (Merlin) suppresses proliferation and adhesion, at least partly, through inhibiting kinase suppressor of Ras 1 (KSR1) and DCAF1 protein. 26673620_Upregulation of KSR1 is associated with Colorectal cancer. 27195677_praja2 regulates KSR1 stability and mitogenic signaling. 28003362_These results suggest that Rap1 activation of ERKs requires PKA phosphorylation and KSR binding. 29433126_findings demonstrate that KSR-MEK complexes allosterically activate BRAF through the action of N-terminal regulatory region and kinase domain contacts and challenge the accepted role of KSR as a scaffold for MEK recruitment to RAF 29596465_Homozygous KSR1 deletion attenuates morbidity but does not prevent tumor development in a mouse model of RAS-driven pancreatic cancer 29980571_Erbin interacted with kinase suppressor of Ras 1 (KSR1) and displaced it from the RAF/MEK/ERK complex to prevent signal propagation..These findings establish the scaffold protein Erbin as a negative regulator of EMT and tumorigenesis in colorectal cancer through direct suppression of Akt and RAS/RAF signaling 30917119_Findings demonstrate a role for kinase suppressor of Ras 1 as a positive regulator of neurotensin secretion from human endocrine cells and indicate that this effect is mediated by the extracellular signal-regulated kinase 1 and 2 signaling pathway. 33461174_Praja2 suppresses the growth of gastric cancer by ubiquitylation of KSR1 and inhibiting MEK-ERK signal pathways. | ENSMUSG00000018334 | Ksr1 | 415.52451 | 0.8360172 | -0.2583954331 | 0.17204323 | 2.330654e+00 | 1.268486e-01 | 9.998360e-01 | No | Yes | 491.98014 | 80.713697 | 5.585268e+02 | 72.339525 | ||
| ENSG00000141524 | 11322 | TMC6 | protein_coding | Q7Z403 | FUNCTION: Probable ion channel. {ECO:0000250}. | Alternative splicing;Endoplasmic reticulum;Glycoprotein;Ion channel;Ion transport;Membrane;Methylation;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | Epidermodysplasia verruciformis (EV) is an autosomal recessive dermatosis characterized by abnormal susceptibility to human papillomaviruses (HPVs) and a high rate of progression to squamous cell carcinoma on sun-exposed skin. EV is caused by mutations in either of two adjacent genes located on chromosome 17q25.3. Both of these genes encode integral membrane proteins that localize to the endoplasmic reticulum and are predicted to form transmembrane channels. This gene encodes a transmembrane channel-like protein with 10 transmembrane domains and 2 leucine zipper motifs. [provided by RefSeq, Jul 2008]. | hsa:11322; | cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; nuclear membrane [GO:0031965]; plasma membrane [GO:0005886]; specific granule membrane [GO:0035579]; tertiary granule membrane [GO:0070821]; mechanosensitive ion channel activity [GO:0008381] | 12426567_Mutations in EVER1 are associated with epidermodysplasia verruciformis. 15042430_Nonsense mutations of EVER1gene is associated with epidermodysplasia verruciformis 16487602_Observational study of genotype prevalence. (HuGE Navigator) 16487602_four mutations in recurrent respiratory papillomatosis patients might indicate that EVER 1 alleles are not associated with susceptibility to RRP 17008061_The growing number of mutations in epidermodysplasia verruciformis (EV) pedigrees supports the hypothesis that EVER1 and EVER2 are the molecular basis of EV. 18158319_HPV16 E5 protein binds to EVER and ZnT-1 and blocks their negative regulation 19706093_epidermodysplasia verruciformis in a father and a son with typical histologic and clinical findings that occur in the absence of mutations in EVER1 21387292_Data support the involvement of the TMC6/8 region in cervix cancer susceptibility. 22761942_EVER proteins appear as key components of the activation-dependent regulation of Zn(2+) concentration in T cells. However, the impact of EVER-deficiency in T cells on EV pathogenesis remains to be elucidated 23534907_EV is also a rare autosomal recessive genodermatosis involving susceptibility to human papillomavirus (HPV) infections and squamous cell carcinoma, caused in most cases by homozygous mutations in EVER1 or EVER2. 23535066_We report a case of Merkel cell polyomavirus detection in the skin of a patient with epidermodysplasia verruciformis (EDV) and a family history remarkable for an unusual inheritance pattern for EDV. 25378492_Expression of both EVER1 and EVER2 in B cells is activated immediately after Epstein-Barr virus (EBV) infection, whereas at later stages, it is strongly repressed by latent membrane protein 1-activated NF-kappaB signaling. 26047157_TMC6 variants are associated with diminished age-of-onset of P. aeruginosa airway infection in children with cystic fibrosis. 26126409_Findings suggest that SNP in EVER 1 may be involved in the development of premalignant skin lesions that harbour beta-HPV, perhaps giving rise to SCC tumours that have lost beta-HPV gene expression during progression 26227733_TMC6/EVER1 and TMC8/EVER2 are known to be involved in the development of EV. 27097911_There were no differences in Ever1 SNPs between head and neck squamous cell carcinoma patients with human papilloma virus (HPV)-positive and HPV-negative tumors, and healthy controls. 27899077_The present study did not show any significant association of the EVER1/2 polymorphisms (rs2290907and rs16970849) with cervical cancer. 29893424_LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. 30068544_these findings suggest that the disruption of CIB1-EVER1-EVER2-dependent keratinocyte-intrinsic immunity underlies the selective susceptibility to beta-HPVs of epidermodysplasia verruciformis patients. 32917726_Expression of a TMC6-TMC8-CIB1 heterotrimeric complex in lymphocytes is regulated by each of the components. | ENSMUSG00000025572 | Tmc6 | 471.87745 | 0.8687335 | -0.2030143959 | 0.17357820 | 1.428972e+00 | 2.319322e-01 | 9.998360e-01 | No | Yes | 442.21081 | 68.931107 | 4.573909e+02 | 55.505486 | |
| ENSG00000142765 | 84958 | SYTL1 | protein_coding | Q8IYJ3 | FUNCTION: May play a role in vesicle trafficking (By similarity). Binds phosphatidylinositol 3,4,5-trisphosphate. Acts as a RAB27A effector protein and may play a role in cytotoxic granule exocytosis in lymphocytes (By similarity). {ECO:0000250, ECO:0000269|PubMed:11278853, ECO:0000269|PubMed:18266782}. | Alternative splicing;Cell membrane;Exocytosis;Membrane;Phosphoprotein;Reference proteome;Repeat | hsa:84958; | exocytic vesicle [GO:0070382]; extracellular exosome [GO:0070062]; extrinsic component of plasma membrane [GO:0019897]; melanosome [GO:0042470]; microvillus membrane [GO:0031528]; plasma membrane [GO:0005886]; neurexin family protein binding [GO:0042043]; small GTPase binding [GO:0031267]; exocytosis [GO:0006887]; intracellular protein transport [GO:0006886] | 11773082_Jfc1/synaptotagmin-like protein 1 (Slp1) contains an N-terminal Slp homology domain (SHD) (PMID: 11327731). The SHD of Slp1/Jfc1 specifically and directly binds the GTP-bound form of Rab27A (J. Biol. Chem. 277, (2002) 9212-9218; PMID: 11773082). 12137562_transcriptional activation of JFC1 by the TNFalpha/NF-kappaB pathway is significant in prostate carcinoma cell lines 16004602_evidence that JFC1 differentially regulates the secretion of PSAP and PSA, and that Rab27a and PI3K play a central role in the exocytosis of prostate-specific markers 17200145_Levels of mRNA and immunostaining of the antiproteases elafin and SLPI were enhanced strongly in inflamed versus noninflamed UC. Elafin and SLPI may be added to the list of defensin-like peptides with diminished induction in CD versus UC. 18266782_These results suggest that both Slp1 and Slp2-a may form part of a docking complex, capturing secretory lysosomes at the immunological synapse. 19528539_Slp1 inhibits dense granule secretion in platelets and that Rap1GAP2 modulates secretion by binding to Slp1. 22438581_during exocytosis, actin depolymerization commences near the secretory organelle, not the plasma membrane, and secretory granules use a JFC1- and GMIP-dependent molecular mechanism to traverse cortical actin. 23082202_Cdk2 also binds the N-terminal domain of Fbw7-gamma as well as SLP-1. 24285265_SLPI regulation and activity is altered in the nasal mucosa of smokers. 25441765_High Expression of secretory leukocyte protease inhibitor is associated with metastasis in colorectal cancer. 27018596_results reveal that MEIS1, through induction of SYTL1, promotes leukemogenesis and supports leukemic cell homing and engraftment, facilitating interactions between leukemic cells and bone marrow stroma. | ENSMUSG00000028860 | Sytl1 | 67.04677 | 0.8731240 | -0.1957414567 | 0.36431199 | 2.876192e-01 | 5.917503e-01 | 9.998360e-01 | No | Yes | 61.79164 | 11.624165 | 6.593505e+01 | 10.409546 | ||
| ENSG00000143036 | 126969 | SLC44A3 | protein_coding | Q8N4M1 | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:126969; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; choline transmembrane transporter activity [GO:0015220]; transmembrane transporter activity [GO:0022857]; phosphatidylcholine biosynthetic process [GO:0006656]; transmembrane transport [GO:0055085] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000039865 | Slc44a3 | 59.15657 | 1.1875594 | 0.2479996463 | 0.40329426 | 3.890125e-01 | 5.328189e-01 | 9.998360e-01 | No | Yes | 53.40218 | 10.861568 | 4.524618e+01 | 7.299746 | |||
| ENSG00000143158 | 25874 | MPC2 | protein_coding | O95563 | FUNCTION: Mediates the uptake of pyruvate into mitochondria. {ECO:0000269|PubMed:22628558}. | Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:25874; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; identical protein binding [GO:0042802]; pyruvate transmembrane transporter activity [GO:0050833]; mitochondrial acetyl-CoA biosynthetic process from pyruvate [GO:0061732]; mitochondrial pyruvate transmembrane transport [GO:0006850]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774] | 3022128_characterization of the rat ortholog 19536175_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 23933155_found no significant difference (P > 0.05) in either allele or genotype frequency in the SNPs between patients and controls. 25458841_Tumor cells expressing MPC1 and MPC2 display increased mitochondrial pyruvate oxidation, with no changes in cell growth in adherent culture. 26577410_These results reveal a novel post-translational regulation of MPC1 by Sirt3, which is important for its activity and colon cancer cell growth. 27460766_a significant association between MPC2 variant rs10489202 and Schizophrenia susceptibility in Han Chinese (Meta-Analysis) 27852261_Our study indicates that MPC1 and MPC2 expressions are of prognostic values in PCAs and that positive expression of MPC1 or MPC2 is a predictor of favorable outcome. 28263840_Results indicate mitochondrial pyruvate transporter (MPC) to be the key regulatory junction perturbed by virulent strains of Mycobacterium tuberculosis leading to alteration of mitochondrial metabolic flux and regulation of acetyl-CoA formation. 29472561_In contrast to MPC1, which co-purifies with a host chaperone, we demonstrated that MPC2 homo-oligomers promote efficient pyruvate transport into proteoliposomes. The derived functional requirements and kinetic features of MPC2 resemble those previously demonstrated for MPC in the literature 29845198_Hypoxia induces lactate secretion and glycolytic efflux by downregulating MPC1/MPC2 levels in HUVEC cells. 30087317_MPC2 rs10489202 was genome-wide significantly associated with schizophrenia. The expression quantitative trait loci analysis in lymphoblastoid cell lines from East Asian donors revealed that MPC2 rs10489202 was specifically and significantly associated with the expression of TIPRL gene. 30356033_The data demonstrated that CtBP1 directly bound to the promoters of MPC1 and MPC2 and transcriptionally repressed them, leading to increased levels of free NADH in the cytosol and nucleus, thus positively feeding back CtBP1's functions. 32403431_Characteristic Analysis of Homo- and Heterodimeric Complexes of Human Mitochondrial Pyruvate Carrier Related to Metabolic Diseases. 32664896_Mitochondrial pyruvate carrier: a potential target for diabetic nephropathy. 32708919_The Multifaceted Pyruvate Metabolism: Role of the Mitochondrial Pyruvate Carrier. 33791391_Decreased Expression of MPC2 Contributes to Aerobic Glycolysis and Colorectal Cancer Proliferation by Activating mTOR Pathway. 34664967_Structural Insights into the Human Mitochondrial Pyruvate Carrier Complexes. | ENSMUSG00000026568 | Mpc2 | 1346.17557 | 1.0909609 | 0.1255993481 | 0.13202086 | 8.947629e-01 | 3.441899e-01 | 9.998360e-01 | No | Yes | 1347.29055 | 200.289025 | 1.481912e+03 | 170.280477 | ||
| ENSG00000143493 | 25896 | INTS7 | protein_coding | Q9NVH2 | FUNCTION: Component of the Integrator (INT) complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Plays a role in DNA damage response (DDR) signaling during the S phase (PubMed:21659603). May be not involved in the recruitment of cytoplasmic dynein to the nuclear envelope by different components of the INT complex (PubMed:23904267). {ECO:0000269|PubMed:21659603, ECO:0000269|PubMed:23904267, ECO:0000305|PubMed:16239144}. | 3D-structure;Alternative splicing;Chromosome;Cytoplasm;DNA damage;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a subunit of the integrator complex. The integrator complex associates with the C-terminal domain of RNA polymerase II and mediates 3'-end processing of the small nuclear RNAs U1 and U2. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Dec 2010]. | hsa:25896; | chromosome [GO:0005694]; cytoplasm [GO:0005737]; integrator complex [GO:0032039]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cellular response to ionizing radiation [GO:0071479]; DNA damage checkpoint signaling [GO:0000077]; snRNA 3'-end processing [GO:0034472]; snRNA processing [GO:0016180] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000037461 | Ints7 | 1454.32649 | 0.9595604 | -0.0595544040 | 0.15299416 | 1.425056e-01 | 7.058027e-01 | 9.998360e-01 | No | Yes | 1418.42601 | 283.475778 | 1.740823e+03 | 267.890470 | |
| ENSG00000143537 | 8751 | ADAM15 | protein_coding | Q13444 | FUNCTION: Active metalloproteinase with gelatinolytic and collagenolytic activity. Plays a role in the wound healing process. Mediates both heterotypic intraepithelial cell/T-cell interactions and homotypic T-cell aggregation. Inhibits beta-1 integrin-mediated cell adhesion and migration of airway smooth muscle cells. Suppresses cell motility on or towards fibronectin possibly by driving alpha-v/beta-1 integrin (ITAGV-ITGB1) cell surface expression via ERK1/2 inactivation. Cleaves E-cadherin in response to growth factor deprivation. Plays a role in glomerular cell migration. Plays a role in pathological neovascularization. May play a role in cartilage remodeling. May be proteolytically processed, during sperm epididymal maturation and the acrosome reaction. May play a role in sperm-egg binding through its disintegrin domain. {ECO:0000269|PubMed:12091380, ECO:0000269|PubMed:15358598, ECO:0000269|PubMed:15818704, ECO:0000269|PubMed:17416588, ECO:0000269|PubMed:17575078, ECO:0000269|PubMed:18387333, ECO:0000269|PubMed:18434311}. | Alternative splicing;Angiogenesis;Cell adhesion;Cell junction;Cell projection;Cilium;Cleavage on pair of basic residues;Collagen degradation;Cytoplasmic vesicle;Disulfide bond;EGF-like domain;Flagellum;Glycoprotein;Hydrolase;Membrane;Metal-binding;Metalloprotease;Phosphoprotein;Protease;Reference proteome;SH3-binding;Signal;Transmembrane;Transmembrane helix;Zinc;Zymogen | The protein encoded by this gene is a member of the ADAM (a disintegrin and metalloproteinase) protein family. ADAM family members are type I transmembrane glycoproteins known to be involved in cell adhesion and proteolytic ectodomain processing of cytokines and adhesion molecules. This protein contains multiple functional domains including a zinc-binding metalloprotease domain, a disintegrin-like domain, as well as a EGF-like domain. Through its disintegrin-like domain, this protein specifically interacts with the integrin beta chain, beta 3. It also interacts with Src family protein-tyrosine kinases in a phosphorylation-dependent manner, suggesting that this protein may function in cell-cell adhesion as well as in cellular signaling. Multiple alternatively spliced transcript variants encoding distinct isoforms have been observed. [provided by RefSeq, Jul 2008]. | hsa:8751; | acrosomal vesicle [GO:0001669]; adherens junction [GO:0005912]; cell surface [GO:0009986]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; motile cilium [GO:0031514]; plasma membrane [GO:0005886]; integrin binding [GO:0005178]; metal ion binding [GO:0046872]; metalloendopeptidase activity [GO:0004222]; metallopeptidase activity [GO:0008237]; SH3 domain binding [GO:0017124]; angiogenesis [GO:0001525]; cardiac epithelial to mesenchymal transition [GO:0060317]; cell-matrix adhesion [GO:0007160]; cellular response to phorbol 13-acetate 12-myristate [GO:1904628]; collagen catabolic process [GO:0030574]; extracellular matrix disassembly [GO:0022617]; immune response to tumor cell [GO:0002418]; innate immune response [GO:0045087]; integrin-mediated signaling pathway [GO:0007229]; male gonad development [GO:0008584]; negative regulation of cell growth [GO:0030308]; negative regulation of cell migration [GO:0030336]; negative regulation of cell-matrix adhesion [GO:0001953]; negative regulation of receptor binding [GO:1900121]; response to hypobaric hypoxia [GO:1990910]; tissue regeneration [GO:0042246] | 11741929_These data demonstrate selective, phosphorylation-dependent interactions of ADAM15 with Src family PTKs and Grb2, which highlight the potential for integration of ADAM functions and cellular signaling 11839628_Atrial fibrillation is associated with an increase in the expression of ADAM15 in the heart atrium 11840679_structure determined by X ray chrystallography 11882657_functional classification based on a conserved motif for bining intergrinalpha9beta1 12091380_ADAM 15 IS involved in the restructuring of the mesangial matrix and in the migration of MC in disease. 12243749_Data suggest that ADAM15, whose expression may be driven by VE-cadherin, may be a component of adherens junctions and play a role in endothelial functions mediated by these cell contacts. 12777399_ADAM8, ADAM15, and MDC-L, but not ADAM17, catalyzed ectodomain shedding of CD23, the low affinity IgE receptor. 12897135_ADAM15 has a role in pathological neovascularization in mice 14707550_ADAM 15 was detected in perinatal cortical pyramidal cells; during aging there was also an increase in intracellular staining and the number of stained cells per volume (cell density). In AD brains ADAM 15 was seen in a few diffuse plaques 15263807_In humans, the cytoplasmic domain of ADAM15v2 strongly interacts with Lck and Hck and regulates leukocyte function. 15358598_The expression and the role of ADAMs in intestinal epithelial cells, including its role in wound healing in human cell lines and cultured colonic cells. 15384173_Altered regulation of alternative exon usage in ADAM15 gene may provide a useful target for cancer diagnostics development 15618016_ADAM15 decreases integrin alphavbeta3/vitronectin-mediated ovarian neoplasm cell adhesion and movement in an RGD-dependent fashion. 15756594_Lung carcinoma cell lines and tissues were frequently ADAM15 positive. 16756724_ADAM15 is generally overexpressed in adenocarcinoma and is highly associated with metastatic progression of prostate and breast cancers 16894352_ADAM15 is upregulated in epithelial cells during inflammatory bowel disease compared with the normal colon epithelial cells. 17080222_the ADAM-15 disintegrin-like domain and a number of mutants in which the RGD-containing loop was substituted by cognate regions from ADAM-2, -12 and -19 were tested in terms of integrin-binding activity 17416588_ADAM-15-mediated cell-cell interactions are involved in mechanisms of epithelial restitution and production of pro-inflammatory mediators 17575078_disintegrin domain of ADAM-15 inhibits ASMC adhesion and migration through the beta(1)-integrin, without modulating signaling pathways involved in ASMC migratory responses 17937806_the alternative exon use is a physiological post-transcriptional mechanism regulating ADAM15 expression in human tissues. 18281484_Loss of ADAM15 significantly attenuated the metastatic spread of PC-3 cells to bone. Data strongly support a functional role for ADAM15 in prostate tumor cell interaction with vascular endothelium and the metastatic progression of human prostate cancer. 18296648_Four ADAM-15 variants are differentially expressed in human mammary carcinoma tissues compared with normal breast. 18387333_This presents a novel mechanism by which ADAM15 regulates cell-matrix adhesion and migration. 18434311_ADAM15 catalyzes the cleavage of E-cadherin to generate a soluble fragment that in turn binds to and stimulates ErbB receptor signaling 18695922_Recombinant ADAM15 disintegrin domain inhibits melanoma cell proliferation partly through p38 kinase activation. 18774960_cytoplasmic tail of ADAM15 confers a modulatory effect on the autophosphorylation site Y397 of the focal adhesion kinase (FAK) during chondrocyte-collagen interaction 19054571_Observational study of gene-disease association. (HuGE Navigator) 19207106_results define key catalytic properties of ADAM15 in cells and its response to stimulators and inhibitors of ectodomain shedding. 19253070_involved in advanced atherosclerosis, in catalytically active form, most notably associated with cells of monocytic origin 19487280_Insights into the mechanism of how a splice variant linked to clinical agressiveness in breast cancer causes increased activity of ADAM15B. 19550037_analysis of the ADAM15 disintegrin domain and its binding properties 19718658_Alternative splicing of ADAM15 regulates its interactions with cellular SH3 proteins SNX33 and nephrocystin 20189953_The effects of ADAM15 on endothelial hyperpermeability and neutrophil transmigration are mediated by intracellular signalling involving Src and ERK1/2 activation. 20213810_ADAM15 conveys antiapoptotic properties to osteoarthritis chondrocytes that might sustain their potential to better resist the influence of death-inducing stimuli under pathophysiologic conditions 20851104_the downregulation of ADAM15 plays an important role in melanoma progression and ADAM15 act as a tumorsuppressor in melanoma. 21190186_Promoter methylation of ADAM15 was examined as well as the microsatellite instability status. Thirty-six percent of colorectal carcinomas displayed a reduced expression of ADAM15. 21196774_demonstrate the intrinsic promoter activity of ADAM15 in quiescent mesangial cells and show the involvement of Sp1 in its regulation 21728902_gene expressions for ADAM8 and ADAM15 were notably lower in ascending aorta as compared with aortic dissection 22505472_Exosomes rich in ADAM15 display enhanced binding affinity for integrin alphavbeta3 in an RGD-dependent manner and suppress vitronectin- and fibronectin-induced cell adhesion, growth, and migration, as well as in vivo tumor growth. 22544741_ADAM15 tail can transduce a percepted extracellular signal to enhance FAK and Src phosphorylation. 22621184_dispensable for cutaneous wound healing and B16F1 melanoma growth, but significantly contributes to metastasis formation 23365087_ADAM15 acts as a negative regulator of TRIF-mediated NF-kappaB and IFN-beta reporter gene activity via TLR3 and TLR4 signaling. 23688428_In conclusion, our data identified rhddADAM15 as a potent inhibitor of tumor growth and metastasis, making it a promising tool for use in anticancer treatment. 23910172_The severity of intrauterine adhesions positively correlates to the protein and transcript expression levels of ADAM-15 and ADAM-17 in uterine tissue. 23918525_ADAM15 contributes to apoptosis resistance in rheumatoid arthritis synovial fibroblasts by activating the Src/FAK pathway upon FasL exposure. 25208722_present a tumor suppressive mechanism for ADAM15 exosomes and provide insight into the functional significance of exosomes that generate tumor-inhibitory factors 25333931_these data suggest the potential role of miR147b in regulating endothelial barrier function by targeting ADAM15 expression. 25650586_findings indicated that silencing ADAM15 had antiinflammatory effects in FLSs and efficiently inhibited the development of CIA. 26323669_ADAM15 promotes lung cancer cell invasion through directly targeting MMP9 activation. 26930657_the results revealed an undescribed role of ADAM15 in the invasion of human bladder cancer and suggested that the ADAM15 catalytic domain may represent a viable therapeutic target in patients with advanced disease. 27554339_Data show that ADAM9 silencing affected MMP2 and ADAM15 expression. 28282546_The results of our study demonstrate that ADAM15 is strongly up regulated in a small but highly aggressive fraction of prostate cancers. 30003689_The antiapoptotic effect of ADAM15 in FasL-stimulated cells was demonstrated either by increased apoptosis of cells transfected with an ADAM15 construct lacking the cytoplasmic domain compared to cells transfected with full-length ADAM15 or by reduced apoptosis resistance of rheumatoid arthritis synovial fibroblasts upon RNA interference silencing of ADAM15. 30634456_The data show that ADAM15 plays a role in rheumatoid arthritis angiogenesis, suggesting that ADAM15 might be a potential target in inflammatory diseases such as rheumatoid arthritis. 30685994_ET-1 expression in the endothelial cells of diabetic foot ulcers is an important determinant of insulin resistance at the onset of disease and induces macrophages to produce NF-kappaB, which regulates inflammation. It is thought that ADAM 15 contributes to angiogenic effects as a means of stimulating endothelial cells 31271758_The absence of ADAM15 at low shear stress or static conditions may therefore lead to increased endothelial damage. 31467400_ADAM15 mediates upregulation of Claudin-1 expression in breast cancer cells. 32522006_ADAM (a Disintegrin and Metalloproteinase) 15 Deficiency Exacerbates Ang II (Angiotensin II)-Induced Aortic Remodeling Leading to Abdominal Aortic Aneurysm. 32677970_ADAM15 expression is increased in lung CD8(+) T cells, macrophages, and bronchial epithelial cells in patients with COPD and is inversely related to airflow obstruction. 33208450_ADAM15 Participates in Tick-Borne Encephalitis Virus Replication. 34426560_ADAM15 correlates with prognosis, immune infiltration and apoptosis in hepatocellular carcinoma. 34685689_A Novel Pro-Inflammatory Mechanosensing Pathway Orchestrated by the Disintegrin Metalloproteinase ADAM15 in Synovial Fibroblasts. | ENSMUSG00000028041 | Adam15 | 6655.98063 | 0.9946428 | -0.0077496439 | 0.11088892 | 5.032545e-03 | 9.434452e-01 | 9.998360e-01 | No | Yes | 6274.03267 | 647.548698 | 5.825094e+03 | 464.913326 | |
| ENSG00000143590 | 1944 | EFNA3 | protein_coding | P52797 | FUNCTION: Cell surface GPI-bound ligand for Eph receptors, a family of receptor tyrosine kinases which are crucial for migration, repulsion and adhesion during neuronal, vascular and epithelial development. Binds promiscuously Eph receptors residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Signal | This gene encodes a member of the ephrin (EPH) family. The ephrins and EPH-related receptors comprise the largest subfamily of receptor protein-tyrosine kinases and have been implicated in mediating developmental events, especially in the nervous system and in erythropoiesis. Based on their structures and sequence relationships, ephrins are divided into the ephrin-A (EFNA) class, which are anchored to the membrane by a glycosylphosphatidylinositol linkage, and the ephrin-B (EFNB) class, which are transmembrane proteins. This gene encodes an EFNA class ephrin. [provided by RefSeq, Jul 2008]. | hsa:1944; | anchored component of membrane [GO:0031225]; intrinsic component of plasma membrane [GO:0031226]; plasma membrane [GO:0005886]; ephrin receptor binding [GO:0046875]; transmembrane-ephrin receptor activity [GO:0005005]; axon guidance [GO:0007411]; cell-cell signaling [GO:0007267]; ephrin receptor signaling pathway [GO:0048013]; negative regulation of angiogenesis [GO:0016525]; positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process [GO:1902961] | 12907451_EphA2/ephrin-A3 interactions may play a role in the localization and network of Langerhans cells in the epithelium and in the regulation of their trafficking. 15901737_analysis of molecular surfaces in ephrin-A5 essential for a functional interaction with EphA3 17980912_Increasing ephrin-A expression enhances T-cell interactions not only with purified integrin ligands but also endothelial cells, while EphA activation down-regulates these interactions. 18417479_MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19823572_EphA3 mutants with constitutively-released kinase domains efficiently support shedding, even when their kinase is disabled. Our data suggest that this phosphorylation-activated conformational switch of EphA3 directly controls ADAM-mediated shedding. 23686814_The interaction between ephrin-As, Eph receptors and integrin alpha3 is plausibly important for the crosstalk between Eph and integrin signalling pathways at the membrane protrusions and in the migration of brain cancer cells. 25070915_The present study provides evidence that microglia upregulates endothelial ephrin-A3 and ephrin-A4 to facilitate in vitro angiogenesis of brain endothelial cells, which is mediated by microglia-released TNF-alpha. 25955218_Results show that EFNA3 serves as a tumor suppressor in malignant peripheral nerve sheath tumor cells and it may play a critical role in the FAK signaling and VEGF-associated tumor angiogenesis pathway. 28739548_E2F3 and ephrin A3 are putative targets of miR-210, and their protein expression was up-regulated in the angiosarcoma cells 32180353_MiR-210-3p-EphrinA3-PI3K/AKT axis regulates the progression of oral cancer. 32695810_OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. 34618621_Role of EphrinA3 in HIV-1 Neuropathogenesis. 35345980_LncRNA LINC01270 aggravates the progression of gastric cancer through modulation of miR-326/EFNA3 axis. | ENSMUSG00000028039 | Efna3 | 673.92403 | 0.8844579 | -0.1771345776 | 0.15977363 | 1.252443e+00 | 2.630865e-01 | 9.998360e-01 | No | Yes | 667.47142 | 81.900152 | 7.031624e+02 | 67.137331 | |
| ENSG00000143756 | 23219 | FBXO28 | protein_coding | Q9NVF7 | FUNCTION: Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation. {ECO:0000250}. | Alternative splicing;Centromere;Chromosome;Kinetochore;Phosphoprotein;Reference proteome;Ubl conjugation pathway | Members of the F-box protein family, such as FBXO28, are characterized by an approximately 40-amino acid F-box motif. SCF complexes, formed by SKP1 (MIM 601434), cullin (see CUL1; MIM 603134), and F-box proteins, act as protein-ubiquitin ligases. F-box proteins interact with SKP1 through the F box, and they interact with ubiquitination targets through other protein interaction domains (Jin et al., 2004 [PubMed 15520277]).[supplied by OMIM, Mar 2008]. | hsa:23219; | kinetochore [GO:0000776]; identical protein binding [GO:0042802]; protein polyubiquitination [GO:0000209] | 23776131_Study identified F-box protein, FBXO28 that controls MYC-dependent transcription by non-proteolytic ubiquitylation. Depletion of FBXO28 or overexpression of an F-box mutant unable to support MYC ubiquitylation results in an impairment of MYC-driven transcription, transformation and tumourigenesis. 24357076_Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. 27754753_Fbxo28 regulates topoisomerase IIalpha decatenation activity and plays an important role in maintaining genomic stability. 28179588_Expression levels of TP53BP2, FBXO28, and FAM53A genes were associated with patient survival specifically in ER-positive, TP53-mutated tumors. 29587369_It was shown on human primary islets that FBXO28 improves pancreatic beta-cell survival under diabetogenic conditions without affecting insulin secretion. 30160831_FBXO28 is a monogenic disease gene and contributes to the complex neurodevelopmental phenotype of the 1q41-q42 gene deletion syndrome. 31678254_SCF(FBXO28)-mediated self-ubiquitination of FBXO28 promotes its degradation. 33280099_FBXO28 causes developmental and epileptic encephalopathy with profound intellectual disability. | ENSMUSG00000047539 | Fbxo28 | 723.24689 | 1.1558566 | 0.2089624423 | 0.16108458 | 1.641038e+00 | 2.001831e-01 | 9.998360e-01 | No | Yes | 772.53832 | 144.559335 | 7.213327e+02 | 104.060437 | |
| ENSG00000143786 | 149111 | CNIH3 | protein_coding | Q8TBE1 | FUNCTION: Regulates the trafficking and gating properties of AMPA-selective glutamate receptors (AMPARs). Promotes their targeting to the cell membrane and synapses and modulates their gating properties by regulating their rates of activation, deactivation and desensitization. {ECO:0000269|PubMed:20805473}. | Cell junction;Cell membrane;Membrane;Postsynaptic cell membrane;Reference proteome;Synapse;Transmembrane;Transmembrane helix | hsa:149111; | AMPA glutamate receptor complex [GO:0032281]; dendrite [GO:0030425]; dendritic shaft [GO:0043198]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; ER to Golgi transport vesicle membrane [GO:0012507]; postsynaptic membrane [GO:0045211]; synapse [GO:0045202]; regulation of AMPA receptor activity [GO:2000311]; vesicle-mediated transport [GO:0016192] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23103966_Significant upregulation of CNIH-3 mRNA expression was found in schizophrenia. 26239289_Study's convergent findings in humans and mice support CNIH3 involvement in the pathophysiology of opioid dependence, complementing prior studies implicating the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate system | ENSMUSG00000026514 | Cnih3 | 235.04026 | 0.8751353 | -0.1924220636 | 0.20567726 | 8.937178e-01 | 3.444718e-01 | 9.998360e-01 | No | Yes | 225.98213 | 27.410716 | 2.440401e+02 | 24.047761 | ||
| ENSG00000144331 | 151126 | ZNF385B | protein_coding | Q569K4 | FUNCTION: May play a role in p53/TP53-mediated apoptosis. {ECO:0000269|PubMed:22945289}. | Alternative splicing;Apoptosis;Metal-binding;Nucleus;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:151126; | nucleus [GO:0005634]; nucleic acid binding [GO:0003676]; p53 binding [GO:0002039]; zinc ion binding [GO:0008270]; intrinsic apoptotic signaling pathway by p53 class mediator [GO:0072332] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 19401682_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20849254_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20849254_the relationship between ZNF533, environmental factors, and the etiology of nonsyndromic orofacial clefts 22945289_The direct binding of ZNF385B with p53 has suggested the involvement of ZNF385B in B-cell apoptosis via modulation of p53 transactivation 23029477_ZNF385B and VEGFA are strongly differentially expressed in serous ovarian carcinomas and correlate with survival. 24599690_The study found significant associations between autism and two SNPs of the ZNF533 gene. | ENSMUSG00000027016 | Zfp385b | 75.09352 | 0.8436094 | -0.2453530088 | 0.34801381 | 4.740361e-01 | 4.911362e-01 | 9.998360e-01 | No | Yes | 58.59372 | 17.967378 | 6.544524e+01 | 15.613219 | ||
| ENSG00000144354 | 83879 | CDCA7 | protein_coding | Q9BWT1 | FUNCTION: Participates in MYC-mediated cell transformation and apoptosis; induces anchorage-independent growth and clonogenicity in lymphoblastoid cells. Insufficient to induce tumorigenicity when overexpressed but contributes to MYC-mediated tumorigenesis. May play a role as transcriptional regulator. {ECO:0000269|PubMed:11598121, ECO:0000269|PubMed:15994934, ECO:0000269|PubMed:16580749, ECO:0000269|PubMed:23166294}. | Alternative splicing;Apoptosis;Cytoplasm;Disease variant;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene was identified as a c-Myc responsive gene, and behaves as a direct c-Myc target gene. Overexpression of this gene is found to enhance the transformation of lymphoblastoid cells, and it complements a transformation-defective Myc Box II mutant, suggesting its involvement in c-Myc-mediated cell transformation. Two alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Jul 2008]. | hsa:83879; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; apoptotic process [GO:0006915]; regulation of cell population proliferation [GO:0042127]; regulation of transcription, DNA-templated [GO:0006355] | 16580749_JPO1/CDCA7 is a unique transcription regulator whose expression is activated by E2F1 as well as c-Myc. 23166294_CDCA7 associates with MYC and this association is modulated in a phosphorylation-dependent manner. 24029427_The CDCA7 transcription factor regulates cell proliferation. 25385755_Data indicate that cell division cycle associated 7 protein (Cdca7) is strongly up-regulated in the hemogenic population during embryonic stem cell hematopoietic differentiation in a Notch-dependent manner. 26216346_Missense mutations in CDCA7 cause immunodeficiency-centromeric instability-facial anomalies syndrome type 3. 27466202_Transcriptome analysis identified Cdca7 as the top down-regulated gene in Zbtb24 homozygous mutant mESCs, which can be restored by ectopic ZBTB24 expression. Finally, we show that this regulation is conserved between species and that CDCA7 levels are reduced in patients carrying ZBTB24 nonsense mutations 29339483_HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex; immunodeficiency-centromeric instability-facial anomalies syndrome has a defective HELLS and CDCA7 bipartite nucleosome remodeling complex 29659838_Here, we performed a comparative analysis of perturbed DNA methylation landscapes in a cohort of ICF patients using an array-based assay to (i) evaluate the similarities and differences in methylation landscapes depending on patient genotypes as an index of a functional link between the four ICF factors, (ii) identify genomic regions that rely on DNMT3B, ZBTB24, CDCA7 or HELLS for their methylation status 29880607_CDCA7 was not required for anchorage-dependent growth of normal fibroblasts or non-malignant lymphocytes, it was essential but not sufficient for anchorage-independent growth of lymphoid tumor cells and for lymphomagenesis. 30010917_in contrast to Immunodeficiency, Centromeric instability and Facial anomalies syndrome type 1 (ICF1), the subtelomeric methylation patterns in cells of ICF2-4 patients do not differ significantly from those in normal cells, and ICF2-4 cells exhibit a normal telomeric phenotype. Also knocking down the expression of ZBTB24, CDCA7 and HELLS in normal human fibroblasts does not affect subtelomeric methylation. 30151890_Immunohistochemical analysis revealed that the CDCA7/EZH2 axis was clinical relevant. These findings suggest CDCA7 plays a crucial role in TNBC progression by transcriptionally upregulating EZH2. 30307408_the C-NHEJ defect alone did not cause CG hypomethylation, CDCA7 and HELLS are involved in maintaining CG methylation at centromeric and pericentromeric repeats. The defect in C-NHEJ may account for some common features of immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome cells, including centromeric instability, abnormal chromosome segregation, and apoptosis. 31030944_ZBTB24 activates the expression of CDCA7 in T cells. 31332696_The inhibiting effects of FGD5-AS1 knockdown on colorectal cancer (CRC) cell proliferation, migration, and invasion, and the promoting effects on CRC cell apoptosis could be revived by miR-302e suppression or CDCA7 upregulation. 31570276_the CDCA7 gene may play a tumor-promoting role in lung adenocarcinoma 32319649_High expression of CDCA7 predicts tumor progression and poor prognosis in human colorectal cancer. 33082427_CDCA7 and HELLS suppress DNA:RNA hybrid-associated DNA damage at pericentromeric repeats. 34339079_CDCA7-regulated inflammatory mechanism through TLR4/NF-kappaB signaling pathway in stomach adenocarcinoma. 34551671_Downregulation of cell division cycle-associated protein 7 (CDCA7) suppresses cell proliferation, arrests cell cycle of ovarian cancer, and restrains angiogenesis by modulating enhancer of zeste homolog 2 (EZH2) expression. | ENSMUSG00000055612 | Cdca7 | 1944.73095 | 1.0476706 | 0.0671851352 | 0.12303802 | 3.010532e-01 | 5.832229e-01 | 9.998360e-01 | No | Yes | 2079.11738 | 364.438265 | 2.064625e+03 | 278.995781 | |
| ENSG00000144677 | 10217 | CTDSPL | protein_coding | O15194 | FUNCTION: Recruited by REST to neuronal genes that contain RE-1 elements, leading to neuronal gene silencing in non-neuronal cells (By similarity). Preferentially catalyzes the dephosphorylation of 'Ser-5' within the tandem 7 residue repeats in the C-terminal domain (CTD) of the largest RNA polymerase II subunit POLR2A. Negatively regulates RNA polymerase II transcription, possibly by controlling the transition from initiation/capping to processive transcript elongation. {ECO:0000250, ECO:0000269|PubMed:12721286}. | 3D-structure;Alternative splicing;Hydrolase;Magnesium;Metal-binding;Nucleus;Protein phosphatase;Reference proteome | hsa:10217; | extracellular exosome [GO:0070062]; nucleus [GO:0005634]; metal ion binding [GO:0046872]; phosphoprotein phosphatase activity [GO:0004721]; protein serine phosphatase activity [GO:0106306]; protein threonine phosphatase activity [GO:0106307]; RNA polymerase II CTD heptapeptide repeat phosphatase activity [GO:0008420]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; negative regulation of protein phosphorylation [GO:0001933] | 15051889_analysis of RBSP3/HYA22located in the AP20 region, and evidence for tumor suppressor function 17085434_SCP3 acts as a phosphatase for regulatory phosphorylations in the linker regions of Smad1 and Smad2. 19016758_Frequent alterations of RBSP3 at chromosomal 3p22.3 region in early and late-onset breast carcinoma are reported with reference to clinical and prognostic significance. 19047128_Observational study of gene-disease association. (HuGE Navigator) 19140316_Down-regulation of RBSP3/CTDSPL, NPRL2/G21, RASSF1A, ITGA9, HYAL1 and HYAL2 genes in non-small cell lung cancer 19478941_This is the first report of high frequencies of somatic mutations in RASSF1 and RBSP3 in different cancers suggesting it may underlay the mutator phenotype of cancer. 19885927_In cervical intraepithelial neoplasms and uterine cervical carinoma, RBSP3 is oftne deletd, compared with other genes. 20193080_tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A are downregulated in primary non-small cell lung cancer 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20412120_dysregulation of hMLH1, ITGA9, and RBSP3 associated multiple cellular pathways are needed for the development of early dysplastic lesions of the head and neck. 21643017_identified RBSP3, a phosphatase-like tumor suppressor, as a bona fide target of miR-100 and validated that RBSP3 was involved in cell differentiation and survival in acute myeloid leukemia 27414789_CTDSPL and Rb directly interact and can be involved in the common mechanism of cell cycle regulation. 27458253_data suggest that overexpression of p-RB1 in basal-parabasal layers of normal cervical epithelium was due to methylation/low functional-linked non-synonimous SNPs of P16 and RBSP3. This pattern was maintained during cervical carcinogenesis by additional deletion/mutation 29382357_we have presented herein the novel finding that miR-181b contributes to cell cycle progression through depressing the expression of CTDSPL, which in turn activates the downstream effector E2F1 and promotes S-phase entry. 29672635_our data also suggests the importance ofLIMD1 and CDC25A in conjunction with HPV for use as diagnostic and prognostic markers of HNSCC, whereas RBSP3 as a prognostic marker only. 31774910_Tumor suppressor properties of the small C-terminal domain phosphatases in non-small cell lung cancer. | ENSMUSG00000047409 | Ctdspl | 1419.98463 | 1.0093667 | 0.0134503565 | 0.12218238 | 1.209451e-02 | 9.124292e-01 | 9.998360e-01 | No | Yes | 1625.51653 | 152.132164 | 1.708140e+03 | 123.919323 | ||
| ENSG00000144744 | 9039 | UBA3 | protein_coding | Q8TBC4 | FUNCTION: Catalytic subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M. Down-regulates steroid receptor activity. Necessary for cell cycle progression. {ECO:0000269|PubMed:10207026, ECO:0000269|PubMed:12740388, ECO:0000269|PubMed:9694792}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cell cycle;Ligase;Nucleotide-binding;Reference proteome;Ubl conjugation pathway | PATHWAY: Protein modification; protein neddylation. | The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, or E1s, ubiquitin-conjugating enzymes, or E2s, and ubiquitin-protein ligases, or E3s. This gene encodes a member of the E1 ubiquitin-activating enzyme family. The encoded enzyme associates with AppBp1, an amyloid beta precursor protein binding protein, to form a heterodimer, and then the enzyme complex activates NEDD8, a ubiquitin-like protein, which regulates cell division, signaling and embryogenesis. Multiple alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:9039; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; NEDD8 activating enzyme activity [GO:0019781]; NEDD8 transferase activity [GO:0019788]; protein heterodimerization activity [GO:0046982]; cellular protein modification process [GO:0006464]; endomitotic cell cycle [GO:0007113]; post-translational protein modification [GO:0043687]; protein modification by small protein conjugation [GO:0032446]; protein neddylation [GO:0045116]; proteolysis [GO:0006508]; regulation of cell cycle [GO:0051726] | 12646924_structure and mutational analysis of human APPBP1-UBA3, the heterodimeric E1 enzyme for NEDD8 12740388_Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. 14690597_Data report the structure of the quaternary complex between human APPBP1-UBA3, a heterodimeric E1, its ubl NEDD8, and ATP. 15694336_crystal structure of a complex between the C-terminal domain from NEDD8's heterodimeric E1 (APPBP1-UBA3) and the catalytic core domain of NEDD8's E2 (Ubc12) 18652489_X-ray crystallographic analysis of APPBP1-UBA3-NEDD8 shows that APPBP1-UBA3's preference for NEDD8's Ala72 appears to be indirect, due to proper positioning of UBA3's Arg190. 22821745_Demonstrated that Uba3-betaGD is an independently folded domain in solution and that residues involved in E2 binding are absent from the NMR spectrum, indicating that the E2-binding surface on Uba3-betaGD interconverts between multiple conformations. 24525735_Report role for neddylation via Nedd8-activating enzyme in the regulation of tumor angiogenesis. 24691136_The study demonstrated that two mutations in UBA3 which were not previously reported confer MLN4924 resistance to MLN4924, a selective NEDD8-activating enzyme inhibitor. 25229838_E1 (NAE1 and UBA3) and E2 (UBC12) enzymes, as well as global NEDD8 conjugation, were upregulated in over 2/3 of human intrahepatic cholangiocarcinoma | ENSMUSG00000030061 | Uba3 | 629.73758 | 0.9257576 | -0.1112935479 | 0.17397050 | 4.120241e-01 | 5.209440e-01 | 9.998360e-01 | No | Yes | 600.28226 | 101.805572 | 6.920902e+02 | 90.463409 |
| ENSG00000144824 | 90102 | PHLDB2 | protein_coding | Q86SQ0 | FUNCTION: Seems to be involved in the assembly of the postsynaptic apparatus. May play a role in acetyl-choline receptor (AChR) aggregation in the postsynaptic membrane (By similarity). {ECO:0000250, ECO:0000269|PubMed:12376540}. | Alternative splicing;Coiled coil;Cytoplasm;Membrane;Phosphoprotein;Reference proteome | hsa:90102; | basal cortex [GO:0045180]; cell leading edge [GO:0031252]; cytosol [GO:0005829]; intermediate filament cytoskeleton [GO:0045111]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; establishment of protein localization [GO:0045184]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of focal adhesion assembly [GO:0051895]; negative regulation of stress fiber assembly [GO:0051497]; negative regulation of wound healing, spreading of epidermal cells [GO:1903690]; positive regulation of basement membrane assembly involved in embryonic body morphogenesis [GO:1904261]; regulation of epithelial to mesenchymal transition [GO:0010717]; regulation of gastrulation [GO:0010470]; regulation of microtubule cytoskeleton organization [GO:0070507] | 16824950_LL5beta and ELKS can form a PIP3-regulated cortical platform to which CLASPs attach distal microtubule ends. 20236936_LL5beta directs the translocation of filamin A and SHIP2 to sites of phosphatidylinositol 3,4,5-triphosphate (PtdIns(3,4,5)P3) accumulation, and PtdIns(3,4,5)P3 localization is mutually modified by co-recruited SHIP2. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20513769_Signaling from laminin-integrin associations in epithelial cells attaches microtubule plus ends to the epithelial basal cell cortex via PHLDB1 and PHLDB2. 22111664_minor allele, A, intronic single nucleotide polymorphism 'rs951660'in PHLDB2 might induce a delayed splicing and increase the susceptibility to Vascular dementia 23525008_These and other LL5beta-interacting proteins are associated with conventional podosomes in macrophages and podosome-like invadopodia in fibroblasts, strengthening the close relationship between synaptic and non-synaptic podosomes. 24982445_Liprin-alpha1, ERC1a and LL5 also define new highly polarized and dynamic cytoplasmic structures uniquely localized near the protruding cell edge 27378169_Prickle1 localized to the membrane through its farnesyl moiety, and the membrane localization was necessary for Prickle1 to regulate migration, to bind to CLASPs and LL5beta, and to promote microtubule targeting of focal adhesions. 28392396_Collectively, our findings present the first to elucidate that miR-29c is a direct p53 target gene, and also identify PHLDB2 as an important miR-29c target gene involved in colon cancer metastasis. 33320958_Oncometabolite L-2-hydroxyglurate directly induces vasculogenic mimicry through PHLDB2 in renal cell carcinoma. 33452458_NOTCH3, a crucial target of miR-491-5p/miR-875-5p, promotes gastric carcinogenesis by upregulating PHLDB2 expression and activating Akt pathway. 34337001_RNA-seq-Based Screening in Coal Dust-Treated Cells Identified PHLDB2 as a Novel Lung Cancer-Related Molecular Marker. 34952201_PHLDB2 Mediates Cetuximab Resistance via Interacting With EGFR in Latent Metastasis of Colorectal Cancer. | ENSMUSG00000033149 | Phldb2 | 47.43783 | 0.8462708 | -0.2408086301 | 0.47870684 | 2.517794e-01 | 6.158249e-01 | 9.998360e-01 | No | Yes | 49.17521 | 12.370526 | 5.931783e+01 | 11.505234 | ||
| ENSG00000146094 | 79930 | DOK3 | protein_coding | Q7L591 | FUNCTION: DOK proteins are enzymatically inert adaptor or scaffolding proteins. They provide a docking platform for the assembly of multimolecular signaling complexes. DOK3 is a negative regulator of JNK signaling in B-cells through interaction with INPP5D/SHIP1. May modulate ABL1 function (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Cytoplasm;Membrane;Phosphoprotein;Reference proteome | hsa:79930; | cytoplasm [GO:0005737]; ficolin-1-rich granule membrane [GO:0101003]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; Ras protein signal transduction [GO:0007265]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 16436051_findings indicate that Dok-3 sequesters Grb2 from Shc and inhibits the Ras-Erk pathway downstream of PTKs 19682241_The novel platelet adapter Dok-3 is tyrosine phosphorylated in an Src kinase-independent manner downstream of alphaIIbbeta3 in human platelets, leading to an interaction with Grb2 and SHIP-1. 20139980_Identification of DOK genes as lung tumor suppressors. 22761938_absence of DOK3 increases LPS signaling, contributing to LPS-induced tolerance. Thus, DOK3 plays a role in TLR signaling during both naive and endotoxin-induced tolerant conditions 23223229_The Dok-3/Grb2 protein signal module attenuates Lyn kinase-dependent activation of Syk kinase in B cell antigen receptor microclusters 26585945_Mutations in DOK3 gene is associated with prostate cancer 27265473_DOK2 and DOK3 expression was significantly reduced in HTLV-1-infected T cells. 32193105_Elevated expression of DOK3 indicates high suppressive immune cell infiltration and unfavorable prognosis of gliomas. | ENSMUSG00000035711 | Dok3 | 353.78273 | 0.8239706 | -0.2793351941 | 0.17565325 | 2.474535e+00 | 1.157037e-01 | 9.998360e-01 | No | Yes | 277.63657 | 51.244886 | 2.875496e+02 | 40.989984 | ||
| ENSG00000146918 | 54892 | NCAPG2 | protein_coding | Q86XI2 | FUNCTION: Regulatory subunit of the condensin-2 complex, a complex which establishes mitotic chromosome architecture and is involved in physical rigidity of the chromatid axis. {ECO:0000269|PubMed:14532007, ECO:0000269|PubMed:30609410}. | Alternative splicing;Cell cycle;Cell division;DNA condensation;Disease variant;Mitosis;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein that belongs to the Condensin2nSMC family of proteins. The encoded protein is a regulatory subunit of the condensin II complex which, along with the condensin I complex, plays a role in chromosome assembly and segregation during mitosis. A similar protein in mouse is required for early development of the embryo. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Aug 2013]. | hsa:54892; | condensed nuclear chromosome [GO:0000794]; condensin complex [GO:0000796]; membrane [GO:0016020]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; bHLH transcription factor binding [GO:0043425]; enzyme activator activity [GO:0008047]; histone deacetylase regulator activity [GO:0035033]; methylated histone binding [GO:0035064]; transmembrane receptor protein tyrosine kinase inhibitor activity [GO:0030293]; cell division [GO:0051301]; chromosome condensation [GO:0030261]; erythrocyte differentiation [GO:0030218]; inner cell mass cell proliferation [GO:0001833]; mitotic sister chromatid segregation [GO:0000070]; positive regulation of chromosome segregation [GO:0051984]; positive regulation of chromosome separation [GO:1905820]; transcription by RNA polymerase II [GO:0006366] | 14532007_The CAP-G2 subunit of the condensin II complex implicated in chromosome assembly and segregation 18718915_Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair 27862966_increased NCAPG2 expression could regulate cell proliferation and identified as a poor prognostic biomarker in lung adenocarcinoma. 30609410_data suggest that impaired function of NCAPG2 results in a severe condensinopathy, and they highlight the potential utility of examining candidate pathogenic lesions beyond the primary disease locus 31176678_that NCAPG2 is an important oncogene that contributes to HCC proliferation and metastasis. NCAPG2 can activate both the STAT3 and NF-kappaB pathways and activated STAT3 positively regulates NCAPG2 expression. 32897418_NCAPG2 facilitates glioblastoma cells' malignancy and xenograft tumor growth via HBO1 activation by phosphorylation. 34818025_microRNA-375 inhibits the malignant behaviors of hepatic carcinoma cells by targeting NCAPG2. | ENSMUSG00000042029 | Ncapg2 | 2481.53153 | 0.9707692 | -0.0427997980 | 0.10757355 | 1.562709e-01 | 6.926133e-01 | 9.998360e-01 | No | Yes | 2551.94950 | 417.609955 | 2.618153e+03 | 330.351295 | |
| ENSG00000147050 | 7403 | KDM6A | protein_coding | O15550 | FUNCTION: Histone demethylase that specifically demethylates 'Lys-27' of histone H3, thereby playing a central role in histone code (PubMed:17851529, PubMed:17713478, PubMed:17761849). Demethylates trimethylated and dimethylated but not monomethylated H3 'Lys-27' (PubMed:17851529, PubMed:17713478, PubMed:17761849). Plays a central role in regulation of posterior development, by regulating HOX gene expression (PubMed:17851529). Demethylation of 'Lys-27' of histone H3 is concomitant with methylation of 'Lys-4' of histone H3, and regulates the recruitment of the PRC1 complex and monoubiquitination of histone H2A (PubMed:17761849). Plays a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression (By similarity). {ECO:0000250|UniProtKB:O70546, ECO:0000269|PubMed:17713478, ECO:0000269|PubMed:17761849, ECO:0000269|PubMed:17851529, ECO:0000269|PubMed:18003914}. | 3D-structure;Chromatin regulator;Dioxygenase;Iron;Mental retardation;Metal-binding;Methylation;Nucleus;Oxidoreductase;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Zinc | This gene is located on the X chromosome and is the corresponding locus to a Y-linked gene which encodes a tetratricopeptide repeat (TPR) protein. The encoded protein of this gene contains a JmjC-domain and catalyzes the demethylation of tri/dimethylated histone H3. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Apr 2014]. | hsa:7403; | histone methyltransferase complex [GO:0035097]; MLL3/4 complex [GO:0044666]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin DNA binding [GO:0031490]; histone demethylase activity [GO:0032452]; histone H3-tri/di-methyl-lysine-27 demethylase activity [GO:0071558]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; chromatin remodeling [GO:0006338]; heart development [GO:0007507]; histone H3-K27 demethylation [GO:0071557]; histone H3-K4 methylation [GO:0051568]; regulation of gene expression [GO:0010468] | 17500065_UTX associates with MLL3- and MLL4-containing histone H3 K4 methyltransferase complex(es) that also include ASH2L, RBBP5, WDR5, hDPY-30, PTIP, PA1, NCOA6. 17713478_the human JmjC-domain-containing proteins UTX and JMJD3 demethylate tri-methylated Lys 27 on histone H3 17761849_study shows that UTX is a di- and trimethyl H3K27 demethylase; results suggest a concerted mechanism for transcriptional activation in which cycles of H3K4 methylation by MLL2/3 are linked with the demethylation of H3K27 through UTX 17851529_critical role for UTX in regulating H3K27 methylation at the HOX gene loci and in animal posterior development 18003914_The JmjC domain-containing protein UTX specifically demethylates mono-, di- and trimethylated K27 of histone H3 in vitro. 18003914_UTX and JMJD3 may function as H3K27 demethylases in vivo 19330029_Here, we describe inactivating somatic mutations in the histone lysine demethylase gene UTX, pointing to histone H3 lysine methylation deregulation in multiple tumor types 20054297_identification of inactivating mutations in two genes--SETD2, a histone H3 lysine 36 methyltransferase, and JARID1C, a histone H3 lysine 4 demethylase--as well as mutations in the histone H3 lysine 27 demethylase, UTX in clear cell renal cell carcinoma 20123895_UTX removes H3K27me3 and maintains expression of several RB-binding proteins, enabling cell cycle arrest 20442750_Observational study of gene-disease association. (HuGE Navigator) 21209387_The role of the H3K27 demethylases Jmjd3 and UTX in gene expression, is discussed. 21245294_KDM6A- and KDM6B-responsive Homeobox genes are expressed at significantly higher levels, suggesting that HPV16 E7 results in reprogramming of host epithelial cells 21515470_UXT is a potential interactor of HBV Pol. 21575637_inhibition measurements showed significant selectivity between KDM4C and KDM6A 21828135_Novel UTX, DNMT3A, and EZH2 mutations were found in 8%, 10%, and 5.5% of patients with chronic myelomonocytic leukemia. 21841772_H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency 21865393_Correlating with the loss of H3K27me3, human papillomavirus 16 E6/E7-expressing cells exhibited derepression of specific EZH2-, KMD6A-, and BMI1-targeted HOX genes. 22002947_clarified how UTX discriminates H3K27me3/2 from the other methyllysines with distinct roles 22197486_This study identifies KDM6A mutations as another cause of Kabuki syndrome and highlights the growing role of histone methylases and histone demethylases in multiple-congenital-anomaly and intellectual-disability syndromes. 22306297_demonstrate that UTX directly associates with the promoters of the Mll1, Runx1, and Scl genes and modulate their transcription by controlling H3K27me3 marks on respective promoter regions. 22589717_PAN RNA interacts with demethylases JMJD3 and UTX, and the histone methyltransferase MLL2 22801502_identification of Utx as a novel mediator with distinct functions during the re-establishment of pluripotency and germ cell development 22840376_Microdeletions and microduplications have not been identified in the MLL2 and KDM6A genes of a large cohort of patients with Kabuki syndrome. 22907667_KDM6A contributes to the activation of WNT3 and DKK1 at different differentiation stages when WNT3 and DKK1 are required for mesendoderm and definitive endoderm differentiation. 23184418_This study demonistrated that KDM6A mutations were most commonly identified in subgroups in medulloblastoma. 23266085_KDM6A is overexpressed in breast cancer patients with an unfavorable prognosis (mortality at 1 year, p=8.65E-7). 23365460_UTX regulates stem cell migration and hematopoiesis. 23527641_UTX histone demethylase plays important functional role in epigenetic alteration of HOX clusters during retinoic acid-induced neural differentiation. 23644518_PBRM1, KDM6A, SETD2 and BAP1 were unmethylated in all tumor and normal specimens. 23913813_The identification of novel KDM6A mutations in patients with Kabuki syndrome. 24123378_Both Ezh2 and Kdm6a were shown to affect expression of master regulatory genes involved in adipogenesis and osteogenesis. 24465480_results demonstrate that UTX is implicated in IL-4 mediated transcriptional activation of the ALOX15 gene 24491801_High levels of UTX or MLL4 are associated with poor prognosis in patients with breast cancer. 24527667_A report of novel KDM6A mutations in patients with Kabuki syndrome. 24739679_One girl had a novel splice-site mutation in KDM6A. 25071154_Results show that UTX interacts with the retinoic acid receptor alpha (RARalpha) and this interaction is essential for proper differentiation of leukemic U937 cells in response to retinoic acid. 25225064_This study is the first to identify frequent BAP1 and BRCA pathway alterations in bladder cancer, show TERT promoter alterations are independent of other bladder cancer gene alterations, and show KDM6A loss is a driver of the bladder cancer phenotype. 25281733_Mutations in KMT2D gene were identified in 10/16 (62%) of the patients, whereas none of the patients had KDM6A mutations. 25320243_H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia 25972376_Our results provide further support for the similar roles of KMT2D and KDM6A in the etiology of KS by using a vertebrate model organism to provide direct evidence of their roles in the development of organs and tissues affected in KS patients. 26049589_Kabuki syndrome may be caused by mutations in one of two histone methyltransferase genes: KMT2D and KDM6A. 26138514_The KDM6A gene is a histone demethylase specific for histone H3 Lysin 27 and regulates gene transcription [35]. In approximately 24% of urothelial carcinoma, KDM6A is altered. 26303947_UTX is a prominent tumour suppressor that functions as a negative regulator of EMT-induced Cancer Stem Cell-like properties by epigenetically repressing epithelial-mesenchymal transition -TFs. 26431949_Turner Syndrome subjects, who are predisposed to chronic ear infections, had reduced UTX expression in immune cells and decreased circulating CD4(+) CXCR5(+) T cell frequency. 26762983_The results define UTX as a bivalency-resolving histone modifier necessary for stem cell differentiation 26819089_UTX positively regulates E-cadherin expression in colon cancer cells. 26841933_Pathogenic variants in KMT2D resulting in protein truncation in 43% (6/14; of which 3 are novel) of all cases were detected, while analysis of KDM6A was negative. MLPA analysis was negative in all instances. 26898171_Mutations of the epigenetic genes KMT2D and KDM6A cause dysregulation of certain developmental genes and account for the multiple congenital anomalies of the syndrome 27028180_we identified a novel de novo deletion of KDM6A in a Chinese girl with KS. We consider her allergic skin manifestations to be part of the phenotypic spectrum of KS 27151432_Here, we discuss the roles of lysine 27 demethylases, JMJD3 and UTX, in cancer and potential therapeutic avenues targeting these enzymes. Despite a high degree of sequence similarity in the catalytic domain between JMJD3 and UTX, numerous studies revealed surprisingly contrasting roles in cellular reprogramming and cancer, particularly leukemia 27302555_Study presents a mutation screening of patients with Kabuki syndrome type 1 which identified 208 mutations in KMT2D. Two of the KDM6A mutations were maternally inherited and nine were shown to be de novo. 27533081_both UTX and UTY function as dose-dependent suppressors of urothelial bladder cancer development 27869828_Mutation in KDM6A gene is associated with cancer more frequently in males. 27983522_UTX gene expression in renal cell carcinoma and bladder cancer. 28197626_Kdm6a and Kdm6b were found to be significantly overexpressed in Malignant pleural mesothelioma (MPM) at the mRNA level. However, tests examining if targeting therapeutically Kdm6a/b using a specific small molecule inhibitor was potentially useful for treating MPM, revealed that members of the Kdm6 family may not be suitable candidates for therapy 28228601_inactivating mutations of KDM6A, which are common in urothelial bladder carcinoma, are potentially targetable by inhibiting EZH2. 28442529_Two novel missense mutations: p.G325A in the KDM6A gene responsible for Kabuki syndrome and p.G1877V in the SCN1A gene responsible for generalized epilepsy with febrile seizures plus were identified using the TruSight One sequencing panel. 28534508_Study identified a feed-forward loop between UTX and ER in the regulation of hormonally responsive breast carcinogenesis. 28968467_KDM6A and p21CIP1 expression are essential to curb E7 induced replication stress to levels that do not markedly interfere with cell viability 29045832_Rebalance of Histone h3 lysine 27 methylation 3 levels at specific genes through EZH2 inhibitors may be a therapeutic strategy in multiple myeloma cases harboring UTX mutations. 29136510_Data show that more mutations in the histone lysine demethylase KDM6A were present in non-invasive tumors from females than males. 29171124_Depletion of KDM6A inhibits the expression of SOX9, Col2a1, ACAN and results in increased H3K27me3 and decreased H3K4me3 levels. 29351209_High UTX expression is independently associated with a better prognosis in patients with esophageal squamous cell carcinoma (ESCC) and downregulation of UTX increases ESCC cell growth and decreases E-cadherin expression. Our results suggest that UTX may be a novel therapeutic target for patients with ESCC. 29846646_Study in Finnish Caucasian obesity patients demonstrated a different genome-wide DNA methylation pattern in autosomes and X-chromosome between males and females in human liver. Sex-specific differences in liver expression and methylation of KDM6A may contribute to higher HDL-cholesterol levels in females. 29902804_data suggest that haploinsufficiency for KDM6A due to mosaic X chromosome monosomy may be responsible for hyperinsulinism in Turner syndrome. 29907798_we retrospectively evaluated 100 infants with HI lacking a genetic diagnosis, for causative variants in KS genes. Molecular diagnoses of KS were established by identification of pathogenic variants in KMT2D (n = 5) and KDM6A (n = 4). 29973620_ut of twenty-seven genes culminating into leading hubs in the network, we identified two key regulators (KRs) i.e. KDM6A and BDNF 30006524_Lymphomas with low UTX expression express high levels of Efnb1, and cause significantly poor survival. 30107592_The mutation pattern of KMT2D and KDM6A in a cohort of 505 patients clinically diagnosed as Kabuki syndrome was reported. 30166694_The histone demethylase UTX/KDM6A is mutated in up to 10% of cases of multiple myeloma, activating genes by removing the H3K27me3 repressive histone mark, counteracting EZH2 activity. 30556125_KDM6A exhibited essential roles in human PDAC as a tumor suppressor and KDM6A deficiency could be a promising biomarker for unfavorable outcome in PDAC patients and a potential surrogate marker for response to HDAC inhibitors. 30556359_Three of the 27 children had a KDM6A mutation and had thesame hypermobility as the KMT2D subjects. Only one of the Kabuki syndrome chil-dren had a patellar luxation in here previous history and was not con-sidered hypermobile using both scores. 30718900_Study shows that UTX and 53BP1 directly interact and co-occupy promoters in human embryonic stem cells and differentiating neural progenitor cells. Data suggests that the 53BP1-UTX interaction supports the activation of key genes required for human neurodevelopment. 30753822_In clear cell renal cell carcinoma, KDM6A(UTX) is one of the most frequently mutated genes; In 101 cases of patient samples, 12 samples contained KDM6A mutations. Other studies provided additional evidence that KDM6A was highly mutated in different human solid tumors and leukemias 30872525_the H3K27 histone demethylase KDM6A/UTX, but not its paralog KDM6B, is oxygen sensitive. 30948420_targeting the KDM6A-KLF10 feedback loop may be beneficial to attenuate diabetes-induced kidney injury. 31097364_UTY is co-regulated with KDM6A. UTY compensates for KDM6A in eutherian males and is responsible for the association between the loss of the Y chromosome and poor prognosis in a range of cancers. [review] 31201358_our findings highlight KDM6A as a novel mediator of drug resistance in acute myeloid leukemia 31221981_This article reviewed the most recent findings regarding cancer-specific metabolic reprogramming and the tumor-suppressive roles of IDH1/2, JARID1C/KDM5C and UTX/KDM6A. [review] 31285428_EZH2 methyltransferase and JMJD3/UTX demethylases were deregulated during hepatic differentiation of human HepaRG cells. 31335488_Expression of UTX Indicates Poor Prognosis in Patients With Luminal Breast Cancer and is Associated With MMP-11 Expression. 31403472_the X escapee Kdm6a regulates multiple immune response genes, providing a mechanism for sex differences in autoimmune disease susceptibility 31654559_Prenatal and perinatal history in Kabuki Syndrome. 31685800_GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. 31883305_The phenotypic spectrum of Kabuki syndrome in patients of Chinese descent: A case series. 31935506_This study expands the number of naturally occurring KMT2D and KDM6A variants. The discovery of novel pathogenic variants will add to the knowledge on disease-causing variants and the relevance of missense variants in Kabuki syndrome. 31959746_Knock-out of KDM6s (JMJD3 and/or UTX) in human embryonic stem cells (hESCs) showed that KDM6s (JMJD3 and/or UTX)-deficient human ESCs exit pluripotency and commit to neural progenitor cells (NPC) differentiation normally, but the resulting NPCs fail to transit into neurons and glia due to a lack of accessibility at loci essential for neurogenesis. 32071397_Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization. 32125007_Coordinated demethylation of H3K9 and H3K27 is required for rapid inflammatory responses of endothelial cells. 32154941_HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. 32269126_KDM6A-Mediated Expression of the Long Noncoding RNA DINO Causes TP53 Tumor Suppressor Stabilization in Human Papillomavirus 16 E7-Expressing Cells. 32346926_Histone demethylase KDM6A promotes somatic cell reprogramming by epigenetically regulating the PTEN and IL-6 signal pathways. 32427586_Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. 32679064_UTX Regulates Human Neural Differentiation and Dendritic Morphology by Resolving Bivalent Promoters. 32732223_X- and Y-Linked Chromatin-Modifying Genes as Regulators of Sex-Specific Cancer Incidence and Prognosis. 32803813_Update of the genotype and phenotype of KMT2D and KDM6A by genetic screening of 100 patients with clinically suspected Kabuki syndrome. 32867456_[KDM6A mutation and expression in gastric cancer are associated with prognosis]. 32879445_Histone 3 lysine-27 demethylase KDM6A coordinates with KMT2B to play an oncogenic role in NSCLC by regulating H3K4me3. 32929331_Targeted inhibition of KDM6 histone demethylases eradicates tumor-initiating cells via enhancer reprogramming in colorectal cancer. 32977832_UTX/KDM6A suppresses AP-1 and a gliogenesis program during neural differentiation of human pluripotent stem cells. 32989154_Loss of UTX/KDM6A and the activation of FGFR3 converge to regulate differentiation gene-expression programs in bladder cancer. 33174323_Combination of lysine-specific demethylase 6A (KDM6A) and mismatch repair (MMR) status is a potential prognostic factor in colorectal cancer. 33253789_Histone demethylase UTX/KDM6A enhances tumor immune cell recruitment, promotes differentiation and suppresses medulloblastoma. 33314698_Clinical and molecular characterization study of Chinese Kabuki syndrome in Hong Kong. 33456567_KDM6A promotes imatinib resistance through YY1-mediated transcriptional upregulation of TRKA independently of its demethylase activity in chronic myelogenous leukemia. 33546721_Molecular mechanics and dynamic simulations of well-known Kabuki syndrome-associated KDM6A variants reveal putative mechanisms of dysfunction. 34051747_Significance of KDM6A mutation in bladder cancer immune escape. 34257820_hsa-miR-199b-3p Prevents the Epithelial-Mesenchymal Transition and Dysfunction of the Renal Tubule by Regulating E-cadherin through Targeting KDM6A in Diabetic Nephropathy. 34262032_SMARCA4 deficient tumours are vulnerable to KDM6A/UTX and KDM6B/JMJD3 blockade. 34465286_Overexpression of UTX promotes tumor progression in Oral tongue squamous cell carcinoma patients receiving surgical resection: a case control study. 34526716_UTX condensation underlies its tumour-suppressive activity. 34583087_KDM6A Regulates Cell Plasticity and Pancreatic Cancer Progression by Noncanonical Activin Pathway. 34661759_EGFR transcriptionally upregulates UTX via STAT3 in non-small cell lung cancer. 34667079_PROSER1 mediates TET2 O-GlcNAcylation to regulate DNA demethylation on UTX-dependent enhancers and CpG islands. 35022315_KDM6A Depletion in Breast Epithelial Cells Leads to Reduced Sensitivity to Anticancer Agents and Increased TGFbeta Activity. 35073341_Mutually exclusive mutation profiles define functionally related genes in muscle invasive bladder cancer. 35185915_Inhibiting KDM6A Demethylase Represses Long Non-Coding RNA Hotairm1 Transcription in MDSC During Sepsis. | ENSMUSG00000037369 | Kdm6a | 582.58353 | 0.9589913 | -0.0604104122 | 0.16527533 | 1.294304e-01 | 7.190233e-01 | 9.998360e-01 | No | Yes | 618.23390 | 100.735291 | 6.811901e+02 | 85.543980 | |
| ENSG00000147162 | 8473 | OGT | protein_coding | O15294 | FUNCTION: Catalyzes the transfer of a single N-acetylglucosamine from UDP-GlcNAc to a serine or threonine residue in cytoplasmic and nuclear proteins resulting in their modification with a beta-linked N-acetylglucosamine (O-GlcNAc) (PubMed:26678539, PubMed:23103939, PubMed:21240259, PubMed:21285374, PubMed:15361863). Glycosylates a large and diverse number of proteins including histone H2B, AKT1, ATG4B, EZH2, PFKL, KMT2E/MLL5, MAPT/TAU and HCFC1 (PubMed:19451179, PubMed:20200153, PubMed:21285374, PubMed:22923583, PubMed:23353889, PubMed:24474760, PubMed:26678539, PubMed:27527864). Can regulate their cellular processes via cross-talk between glycosylation and phosphorylation or by affecting proteolytic processing (PubMed:21285374). Probably by glycosylating KMT2E/MLL5, stabilizes KMT2E/MLL5 by preventing its ubiquitination (PubMed:26678539). Involved in insulin resistance in muscle and adipocyte cells via glycosylating insulin signaling components and inhibiting the 'Thr-308' phosphorylation of AKT1, enhancing IRS1 phosphorylation and attenuating insulin signaling (By similarity). Involved in glycolysis regulation by mediating glycosylation of 6-phosphofructokinase PFKL, inhibiting its activity (PubMed:22923583). Component of a THAP1/THAP3-HCFC1-OGT complex that is required for the regulation of the transcriptional activity of RRM1. Plays a key role in chromatin structure by mediating O-GlcNAcylation of 'Ser-112' of histone H2B: recruited to CpG-rich transcription start sites of active genes via its interaction with TET proteins (TET1, TET2 or TET3) (PubMed:22121020, PubMed:23353889). As part of the NSL complex indirectly involved in acetylation of nucleosomal histone H4 on several lysine residues (PubMed:20018852). O-GlcNAcylation of 'Ser-75' of EZH2 increases its stability, and facilitating the formation of H3K27me3 by the PRC2/EED-EZH2 complex (PubMed:24474760). Regulates circadian oscillation of the clock genes and glucose homeostasis in the liver. Stabilizes clock proteins ARNTL/BMAL1 and CLOCK through O-glycosylation, which prevents their ubiquitination and subsequent degradation. Promotes the CLOCK-ARNTL/BMAL1-mediated transcription of genes in the negative loop of the circadian clock such as PER1/2 and CRY1/2 (PubMed:12150998, PubMed:19451179, PubMed:20018868, PubMed:20200153, PubMed:21285374, PubMed:15361863). O-glycosylates HCFC1 and regulates its proteolytic processing and transcriptional activity (PubMed:21285374, PubMed:28584052, PubMed:28302723). Regulates mitochondrial motility in neurons by mediating glycosylation of TRAK1 (By similarity). Glycosylates HOXA1 (By similarity). O-glycosylates FNIP1 (PubMed:30699359). Promotes autophagy by mediating O-glycosylation of ATG4B (PubMed:27527864). {ECO:0000250|UniProtKB:P56558, ECO:0000250|UniProtKB:Q8CGY8, ECO:0000269|PubMed:12150998, ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:19451179, ECO:0000269|PubMed:20018852, ECO:0000269|PubMed:20018868, ECO:0000269|PubMed:20200153, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:22121020, ECO:0000269|PubMed:22923583, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:23353889, ECO:0000269|PubMed:24474760, ECO:0000269|PubMed:26678539, ECO:0000269|PubMed:27527864, ECO:0000269|PubMed:28302723, ECO:0000269|PubMed:28584052, ECO:0000269|PubMed:30699359}.; FUNCTION: [Isoform 2]: The mitochondrial isoform (mOGT) is cytotoxic and triggers apoptosis in several cell types including INS1, an insulinoma cell line. {ECO:0000269|PubMed:20824293}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Biological rhythms;Cell membrane;Cell projection;Chromatin regulator;Cytoplasm;Direct protein sequencing;Disease variant;Glycoprotein;Glycosyltransferase;Host-virus interaction;Lipid-binding;Membrane;Mental retardation;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:26678539}. | This gene encodes a glycosyltransferase that catalyzes the addition of a single N-acetylglucosamine in O-glycosidic linkage to serine or threonine residues. Since both phosphorylation and glycosylation compete for similar serine or threonine residues, the two processes may compete for sites, or they may alter the substrate specificity of nearby sites by steric or electrostatic effects. The protein contains multiple tetratricopeptide repeats that are required for optimal recognition of substrates. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Oct 2009]. | hsa:8473; | cell projection [GO:0042995]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; mitochondrial membrane [GO:0031966]; NSL complex [GO:0044545]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein N-acetylglucosaminyltransferase complex [GO:0017122]; protein-containing complex [GO:0032991]; acetylglucosaminyltransferase activity [GO:0008375]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; protein N-acetylglucosaminyltransferase activity [GO:0016262]; protein O-GlcNAc transferase activity [GO:0097363]; apoptotic process [GO:0006915]; chromatin organization [GO:0006325]; circadian regulation of gene expression [GO:0032922]; histone H3-K4 trimethylation [GO:0080182]; histone H4-K16 acetylation [GO:0043984]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; negative regulation of protein ubiquitination [GO:0031397]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of histone H3-K27 methylation [GO:0061087]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of proteolysis [GO:0045862]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein O-linked glycosylation [GO:0006493]; protein processing [GO:0016485]; regulation of dosage compensation by inactivation of X chromosome [GO:1900095]; regulation of gluconeogenesis [GO:0006111]; regulation of glycolytic process [GO:0006110]; regulation of insulin receptor signaling pathway [GO:0046626]; regulation of necroptotic process [GO:0060544]; regulation of Rac protein signal transduction [GO:0035020]; regulation of transcription by RNA polymerase II [GO:0006357]; response to insulin [GO:0032868]; response to nutrient [GO:0007584]; signal transduction [GO:0007165] | 11773972_We have delineated the complete genomic structure of human OGT spanning approx 43 kb of genomic DNA in Xq13.1 11846551_homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily 12136128_O-linked GlcNAc transferase participates in a hexosamine-dependent signaling pathway that is linked to insulin resistance and leptin production 14601650_a novel HLA-A0201-restricted cytotoxic T lymphocyte (CTL)-epitope (28-SLYKFSPFPL; FSP06) derived from a mutant OGT-protein 15336570_OGT can respond rapidly to heat stress through the enhancement of nucleocytoplasmic protein O-GlcNAcylation. 15561949_Staining of OGT in streptozotocin diabetic rat liver is clearly diminished, but it was substantially restored after 6 days of insulin treatment 15795231_By using a series of 4-methylumbelliferyl 2-deoxy-2-N-fluoroacetyl-beta-D-glucopyranoside substrates, Taft-like linear free energy analyses of these enzymes indicates that O-GlcNAcase uses a catalytic mechanism involving anchimeric assistance 15896326_Thus, stably transfected HeLa cells provide an abundant source of enzyme that can be used to study the structure, function, and regulation of OGT. 16105839_analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase 16966374_Overall, transcriptional inhibition is related to the integrated effect of O-GlcNAc by direct modification of critical elements of the transcriptome and indirectly through O-GlcNAc modification of the proteasome. 18174169_O-GlcNAc modification stimulated by glucose deprivation results from increased OGT and decreased O-GlcNAcase levels and that these changes affect cell metabolism, thus inactivating glycogen synthase. 18536723_The structure of an intact OGT homolog and kinetic analysis of human OGT variants reveal a contiguous superhelical groove that directs substrates to the active site. 18653473_the O-GlcNAc cycling enzymes associate with kinases and phosphatases at M phase to regulate the posttranslational status of vimentin 19073609_Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. 20068230_Data show that forced overexpression of OGT increased the inhibitory phosphorylation of CDK1 and reduced the phosphorylation of CDK1 target proteins. 20190804_OGT regulates breast cancer tumorignenesis and cancer growth through targeting FixM1. 20200153_THAP1 was found to bind HCF-1 in vitro and to associate with HCF-1 and OGT in vivo. 20206135_OGT could be a co-regulatory subunit shared by functionally distinct complexes supporting epigenetic regulation of MIP-1alpha gene promoter. 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20805223_regulating the amount of OGT during mitosis is important in ensuring correct chromosomal segregation during mitosis. 20824293_Enhanced OGT expression efficiently triggered programmed cell death. 20845477_Observational study of gene-disease association. (HuGE Navigator) 20876116_OGT deletion in infarcted mice significantly exacerbated cardiac dysfunction. 21240259_two crystal structures of human OGT, as a binary complex with UDP (2.8 A resolution) and as a ternary complex with UDP and a peptide substrate (1.95 A). 21327254_Data identify Tau as potential substrates for the O-beta-N-acetylglucosaminyltransferase (OGT). 21567137_Decrease in MGEA5 and increase in O-GlcNAc transferease expression in higher grade tumors suggests that increased O-GlcNAc modification may be implicated in breast tumor progression and metastasis. 22275356_as prostate cancer cells alter glucose and glutamine levels, O-GlcNAc modifications and OGT levels become elevated and are required for regulation of malignant properties 22294689_a 154-amino acid region of MIBP1 was necessary for its O-GlcNAc transferase binding and O-GlcNAcylation. 22311971_Data show that the interplay between O-GlcNAc and phosphorylation on proteins and indicate that these effects can be mediated by changes in hOGT and hOGA kinetic activity. 22371499_These studies identify a molecular mechanism of GR transrepression, and highlight the function of O-GlcNAc in hormone signaling. 22384635_O-GlcNAcylation may be an important regulatory modification involved in endometrial cancer pathogenesis but the actual significance of this modification for endometrial cancer progression needs to be investigated further. 22496241_Hsp90 is involved in the regulation of OGT and O-GlcNAc modification and that Hsp90 inhibitors might be used to modulate O-GlcNAc modification and reverse its adverse effects in human diseases. 22574218_AMPK functions as a physiological suppressor of 26S proteasomes through OGT 22783592_Analysis of urinary content of MGEA5 and OGT may be useful for bladder cancer diagnostics. 22883232_O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. 23103939_we describe structural snapshots of all species along the kinetic pathway for human O-linked beta-N-acetylglucosamine transferase (O-GlcNAc transferase), an intracellular enzyme that catalyzes installation of a dynamic post-translational modification 23103942_we define how human OGT recognizes the sugar donor and acceptor peptide and uses a new catalytic mechanism of glycosyl transfer, involving the sugar donor alpha-phosphate as the catalytic base as well as an essential lysine 23152511_The human respiratory syncytial virus-induced sequestration of p38-P in IBs resulted in a substantial reduction in the accumulation of a downstream signaling substrate, MAPK-activated protein kinase 2 (MK2). 23222540_The double epigenetic modifications on both DNA and histones by TET2 and OGT coordinate together for the regulation of gene transcription. 23487789_These studies identified OGT as a promising placental biomarker of maternal stress exposure that may relate to sex-biased outcomes in neurodevelopment. 23642195_Data suggest that changes in OGT (O-linked N-acetylglucosamine transferase) and OGA (peptide O-linked N-acetylglucosamine-beta-N-acetylglucosaminidase) expression are correlated with cancer prognosis. [REVIEW] 23700425_The backbone carbonyl oxygen of Leu653 and the hydroxyl group of Thr560 in OGT contribute to the recognition of sugar moieties via hydrogen bonds. 23720054_Expression of c-MYC and OGT was tightly correlated in human prostate cancer samples. 24256146_Data suggest that with multi-substrate enzymes, such as OGT, specific inhibition can rarely be achieved with ligands that compete solely with one of the substrates; OGT is inhibited by bisubstrate UDP-oligopeptide conjugates. 24311690_study reports the tetratricopeptide-repeat domain of O-GlcNAc transferase binds the carboxyl-terminal portion of an HCF-1 proteolytic repeat such that the cleavage region lies in the glycosyltransferase active site above uridine diphosphate-GlcNAc; protein glycosylation and HCF-1 cleavage occur in the same active site 24365779_Estrogen replacement therapy and plyometric training influence muscle OGT and OGA gene expression, which may be one of the mechanisms by which HRT and PT prevent aging-related loss of muscle mass. 24394411_OGT catalyzes the O-GlcNAcylation of TET3, promotes TET3 nuclear export, and, consequently, inhibits the formation of 5-hydroxymethylcytosine catalyzed by TET3. 24580054_Endogenous OTX2 from a medulloblastoma cell line is O-GlcNAcylated at several sites. 24928395_Instead, an adipogenesis-dependent increase in O-linked beta-N-acetylglucosamine (O-GlcNAc) glycosylation of EWS was observed. 25173736_Amino acid composition of splice variants, post-translational modifications, and stable associations with regulatory proteins influence subcellular distribution/substrate specificity of OGT and OGA (O-GlcNAcase beta-N-acetylglucosaminidase). [REVIEW] 25419848_O-linked beta-N-acetylglucosamine transferase mediates O-GlcNAcylation of the SNARE protein SNAP-29 and regulates autophagy in a nutrient-dependent manner. 25568311_Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). 25663381_Hexosamine biosynthetic pathway flux is increased in idiopathic pulmonary artery hypertension and drives OGT-facilitated pulmonary artery smooth muscle cell proliferation via specific proteolysis and direct activation of host cell factor-1. 25773598_Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. 25776937_miR4235p was associated with congestive heart failure and the expression levels of proBNP; in addition, OGT was found to be a direct target of miR4235p. 26041297_Inhibition of O-Linked N-Acetylglucosamine Transferase Reduces Replication of Herpes Simplex Virus and Human Cytomegalovirus. 26237509_This work reveals that although the N-terminal TPR repeats of OGT may have roles in substrate recognition, the sequence restriction imposed by the peptide-binding site makes a substantial contribution to O-GlcNAc site specificity. 26240142_Use of OGT(C917A) enhances O-GlcNDAz production, yielding improved cross-linking of O-GlcNDAz-modified molecules 26252736_These results suggested roles of O-GlcNAcylation in modulating serine phosphorylation, as well as in regulating PKM2 activity and expression. 26305326_These results demonstrate that distinct OGT-binding sites in HCF-1 promote proteolysis, and provide novel insights into the mechanism of this unusual protease activity. 26397041_We concluded that OGT plays a key role in gastric cancer proliferation and survival, and could be a potential target for therapy. 26399441_OGT expression is increased under hypoxic conditions. 26408091_a new function of histone O-GlcNAcylation in DNA damage response 26527687_E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner. 26707622_OGT inhibited the formation of the Ecadherin/catenin complex through reducing the interaction between p120 and Ecadherin. 26807597_Data suggest RNA polymerase II (POLR2A) is extensively modified on its unique C-terminal domain (CTD) by O-GlcNAc transferase (OGT); efficient O-GlcNAcylation requires a minimum of 20 heptad CTD repeats in POLR2A and more than half of NTD of OGT. 26854602_Together, these findings suggest that induction of SNO-OGT by Ab exposure is a pathogenic mechanism to cause cellular hypo-O-GlcNAcylation by which Ab neurotoxicity is executed 27060025_The findings suggest that OGT promotes the O-GlcNAc modification of HDAC1 in the development of progression hepatocellular carcinoma. 27129214_data indicate that O-GlcNAc-transferase activity is essential for RNA pol II promoter recruitment and that pol II goes through a cycling of O-GlcNAcylation at the promoter 27131860_O-GlcNAcylation expression and its nuclear expression were associated with the carcinogenesis and progression of gastric carcinoma. 27217568_These results support a model in which OGT modifies HIRA to regulate HIRA-H3.3 complex formation and H3.3 nucleosome assembly and reveal the mechanism by which OGT functions in cellular senescence. 27231347_the O-linked N-acetylglucosamine (O-GlcNAc) processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), interact with the (A)gamma-globin promoter at the -566 GATA repressor site 27294441_Beyond its well-known role in adding beta-O-GlcNAc to serine and threonine residues of nuclear and cytoplasmic proteins, OGT also acts as a protease in the maturation of the cell cycle regulator, HCF-1, and serves as an integral member of several protein complexes, many of them linked to gene expression. (Review) 27331873_Findings indicate O-linked N-acetylglucosamine transferase (OGT) as a cellular factor involved in human papillomaviruses type 16/18 E6 and E7 expressions and cervical cancer tumorigenesis, suggesting that targeting OGT in cervical cancer may have potential therapeutic benefit. 27505673_This work uncovers that URI-regulated OGT confers c-MYC-dependent survival functions in response to glucose fluctuations. 27705803_We identified two human PRC2 complexes and two PR-DUB deubiquitination complexes, which contain the O-linked N-acetylglucosamine transferase OGT1 and several transcription factors. 27845045_OGT functions in metastatic spread of HPV E6/E7-positive HeLa cells to xenografted lungs through E6/E7, HCF-1 and CXCR4 28100784_Reducing endogenous mitochondrial OGT expression leads to alterations in mitochondrial structure and function, including Drp1-dependent mitochondrial fragmentation, reduction in mitochondrial membrane potential, and a significant loss of mitochondrial content in the absence of mitochondrial reactive oxygen species. 28115479_The results of this study showed that the OGT is essential for sensory neuron survival and target innervation. 28232487_Fatty acid synthase fine-tunes the cell's response to stress and injury by remodeling cellular O-GlcNAcylation 28302723_Thus, a single amino acid substitution in the regulatory domain (the tetratricopeptide repeat domain) of OGT, which catalyzes the O-GlcNAc post-translational modification of nuclear and cytosolic proteins, appears causal for X-linked intellectual disability. 28347804_OGT, a unique glycosyltransferase enzyme, was identified to be upregulated in non-alcoholic fatty liver disease-associated hepatocellular carcinoma tissues by transcriptome sequencing. Here, we found that OGT plays a role in cancer by promoting tumor growth and metastasis in cell models. This effect is mediated by the induction of palmitic acid. 28450392_Data suggest that O-GlcNAc transferase 1 (OGT1) specifically binds to, O-GlcNAcylates, and stabilizes nonspecific lethal protein3 (NSL3); stabilization of NSL3 by OGT1 up-regulates global acetylation levels of histone 4 at Lys-5, Lys-8, and Lys-16. 28455227_conclusion, our results demonstrated that miR24 inhibits breast cancer cells invasion by targeting OGT and reducing FOXA1 stability. These results also indicated that OGT might be a potential target for the diagnosis and therapy of breast cancer metastasis. 28584052_Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability 28625484_Nrf1 is regulated by O-GlcNAc transferase. 28663241_The authors show that O-GlcNAcylation of KEAP1 by OGT at serine 104 is required for the efficient ubiquitination and degradation of NRF2. 28742148_Tax interacts with the host OGT/OGA complex and inhibits the activity of OGT-bound OGA. 28929346_These data predict that under conditions where O-GlcNAc levels are high (breast cancer) progesterone receptor (PR) through an interaction with the modifying enzyme OGT, will exhibit increased O-GlcNAcylation and potentiated transcriptional activity. Therapeutic strategies aimed at altering cellular O-GlcNAc levels may have profound effects on PR transcriptional activity in breast cancer 29059153_High OGT expression promotes cancer lipid metabolism via SREBP-1 regulation. 29208956_Findings demonstrate a novel role of Poleta O-GlcNAcylation by OGT in translesion DNA synthesis regulation and genome stability maintenance. 29465778_The O-GlcNAc transferase OGT interacts with and post-translationally modifies the transcription factor HOXA1. 29556021_a novel ASXL1-OGT axis and raise the possibility that this axis has a tumor-suppressor role in myeloid malignancies. 29577901_LXRalpha interacts with OGT in its N-terminal domain and ligand-binding domain (LBD) in a ligand-independent fashion. 29606577_The intellectual disability L254F mutation in OGT affects activity. The L254F mutation leads to shifts up to 12 A in the OGT structure. Thermal denaturing studies reveal reduction in tetratricopeptide repeat domain stability caused by L254F. L254F OGT mutation leads to conformational changes of the tetratricopeptide repeats and reduced activity, revealing the molecular mechanisms contributing to pathogenesis. 29769320_O-GlcNAc transferase missense mutations is associated with X-linked intellectual disability. 29788742_miR-483 inhibited the expression level of OGT mRNA by direct binding to its 3'-untranslated region. Expression of miR-483 was negatively correlated with OGT in gastric cancer tissues. 29967448_Study found that levels of placental Ogt determine sex differences in fetally derived placental trophoblast transcriptome profiles associated with key developmental processes and shape genome-wide patterning of the ubiquitous epigenetic transcriptional repressive mark, H3K27me3. 30069701_O-GlcNAc transferase (OGT) is a partner of the MCM2-7 complex and O-GlcNAcylation might regulate MCM2-7 complex by regulating the chromatin loading of MCM6 and MCM7 and stabilizing MCM/MCM interactions. 30106436_High OGT expression is associated with breast cancer. 30453909_O-GlcNAcylation and the expression of O-linked-beta-N-acetylglucosamine transferase (OGT) were upregulated in bladder cancer cell lines and tissue specimens. 30543776_these findings provide a mechanistic link between the OGT-mediated glucose metabolic pathway and antiviral innate immune signaling by targeting MAVS, and expand our current understanding of importance of glucose metabolic regulation in viral infection-associated diseases. 30550897_The ligand transport mechanism in the OGT enzymatic process is described here and is a great resource for designing inhibitors based on UDP or UDP-GlcNAc. 30555541_These findings suggest that down-regulation of OGT enhances cisplatin-induced autophagy via SNAP-29, resulting in cisplatin-resistant ovarian cancer. 30587575_novel regulatory mechanism for O-GlcNAcylation during FA complex formation, which thereby affects integrin activation and integrin-mediated functions such as cell adhesion and migration 30677218_our findings indicate that OGT promotes the stem-like cell potential of hepatoma cell through O-GlcNAcylation of eIF4E 30953348_HNF1A regulates ogt transcription in a time-dependent manner and that O-GlcNAcylation of HNF1A represses ogt transcription. 14 O-GlcNAc sites on HNF1A were revealed, six of which are predominantly modified, including Ser(303/304) , Ser(471) , Ser(560) and Thr(563/564). Loss of O-GlcNAcylation at Ser(303/304) or Thr(563/564) significantly elevates ogt transcription. 31149037_OGT is essential for proliferation of prostate cancer cells. OGT activity is required for the interaction between MYC and HCF-1 and expression of MYC-regulated mitotic proteins. 31296563_Pathogenic N567K mutation leads to loss of O-GlcNAcase activity and delayed differentiation down the neuronal lineage in mouse and Drosophila model. 31373491_Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. 31373757_silencing of OGT in HT29 cells upregulates E-cadherin (a major actor of epithelial-to-mesenchymal transition) and changes its glycosylation. On the other hand, OGT silencing perturbs biosynthesis of glycosphingolipids resulting in a decrease in gangliosides and an increase in globosides. 31527085_O-GlcNAcylation does not affect sOGT activity but does affect sOGT-interacting proteins. S56A bound to and hence glycosylated more proteins in contrast to T12A and WT sOGT. 31567281_Two cases of hand myoepithelioma showing unusual clinicopathologic features and novel OGT-FOXO3 gene fusions are described. 31627256_A missense mutation in the catalytic domain of O-GlcNAc transferase links perturbations in protein O-GlcNAcylation to X-linked intellectual disability. 31628985_Disease related single point mutations alter the global dynamics of a tetratricopeptide (TPR) alpha-solenoid domain. 31847126_findings in this report are the first to describe a role for the OGT/O-GlcNAc axis in modulating VEGF expression and vascularization in Idiopathic pulmonary arterial hypertension. 31974291_O-GlcNAc Transferase Regulates Cancer Stem-like Potential of Breast Cancer Cells. 32272438_Epigenetic activation of O-linked beta-N-acetylglucosamine transferase overrides the differentiation blockage in acute leukemia. 32310828_SIRT1 regulates O-GlcNAcylation of tau through OGT. 32471715_O-GlcNAc transferase affects the signal transduction of beta1 adrenoceptor in adult rat cardiomyocytes by increasing the O-GlcNAcylation of beta1 adrenoceptor. 32663610_Glucosamine regulates hepatic lipid accumulation by sensing glucose levels or feeding states of normal and excess. 32994395_Mutual regulation between OGT and XIAP to control colon cancer cell growth and invasion. 33006972_The O-GlcNAc transferase OGT is a conserved and essential regulator of the cellular and organismal response to hypertonic stress. 33215629_Inhibition of mechanistic target of rapamycin signaling decreases levels of O-GlcNAc transferase and increases serotonin release in the human placenta. 33307156_Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. 33333092_Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. 33419956_Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. 33801653_Feedback Regulation of O-GlcNAc Transferase through Translation Control to Maintain Intracellular O-GlcNAc Homeostasis. 33909326_The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. 34046694_Dual regulation of fatty acid synthase (FASN) expression by O-GlcNAc transferase (OGT) and mTOR pathway in proliferating liver cancer cells. 34288245_Upregulation of OGT by Caveolin-1 promotes hepatocellular carcinoma cell migration and invasion. 34502531_OGT Protein Interaction Network (OGT-PIN): A Curated Database of Experimentally Identified Interaction Proteins of OGT. 34510715_P53 suppresses the progression of hepatocellular carcinoma via miR-15a by decreasing OGT expression and EZH2 stabilization. 34608265_CEMIP, a novel adaptor protein of OGT, promotes colorectal cancer metastasis through glutamine metabolic reprogramming via reciprocal regulation of beta-catenin. 34638625_Regulation of O-Linked N-Acetyl Glucosamine Transferase (OGT) through E6 Stimulation of the Ubiquitin Ligase Activity of E6AP. 34943826_Inhibition of O-GlcNAc Transferase Alters the Differentiation and Maturation Process of Human Monocyte Derived Dendritic Cells. 34948036_TET3- and OGT-Dependent Expression of Genes Involved in Epithelial-Mesenchymal Transition in Endometrial Cancer. | ENSMUSG00000034160 | Ogt | 2205.02623 | 1.7315428 | 0.7920580841 | 0.12010409 | 4.407168e+01 | 3.165678e-11 | 1.694271e-07 | No | Yes | 2755.40500 | 574.876441 | 1.832242e+03 | 294.534312 |
| ENSG00000147650 | 29967 | LRP12 | protein_coding | Q9Y561 | FUNCTION: Probable receptor, which may be involved in the internalization of lipophilic molecules and/or signal transduction. May act as a tumor suppressor. {ECO:0000269|PubMed:12809483}. | Alternative splicing;Coated pit;Disulfide bond;Endocytosis;Glycoprotein;Membrane;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of the low-density lipoprotein receptor related protein family. The product of this gene is a transmembrane protein that is differentially expressed in many cancer cells. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Feb 2010]. | hsa:29967; | clathrin-coated pit [GO:0005905]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; low-density lipoprotein particle receptor activity [GO:0005041]; endocytosis [GO:0006897]; neuron migration [GO:0001764]; neuron projection development [GO:0031175]; regulation of growth [GO:0040008]; signal transduction [GO:0007165] | 12809483_ST7 is a novel low-density lipoprotein receptor-related protein (LRP) with a cytoplasmic tail that interacts with proteins related to signal transduction pathways. 20439316_As the preplate separates, Lrp12/Mig13a-positive neurons polarize in the radial plane and form a pseudocolumnar pattern, prior to moving to a deeper position within the emerging subplate layer. 26639854_Results indicate that low-density lipoprotein receptor-related protein 12 silencing during brain development results in cortical dyslamination and seizure sensitization. 27142828_Accumulation of functional promoter-associated allelic variants with impact on the transcriptional regulation of LRP12 provides a new pathomechanism for GGs, i.e. highly differentiated epileptogenic brain tumors 30029672_LRP12 DNA methylation as a powerful predictive marker for carboplatin resistance. 31332380_GGC repeat expansions in LRP12 gene is associated with oculopharyngodistal myopathy. 32493488_Oculopharyngodistal myopathy with coexisting histology of systemic neuronal intranuclear inclusion disease: Clinicopathologic features of an autopsied patient harboring CGG repeat expansions in LRP12. | ENSMUSG00000022305 | Lrp12 | 88.41990 | 1.1309665 | 0.1775562239 | 0.31987839 | 3.051162e-01 | 5.806927e-01 | 9.998360e-01 | No | Yes | 83.48962 | 15.290600 | 7.727457e+01 | 10.991212 | |
| ENSG00000147654 | 9166 | EBAG9 | protein_coding | O00559 | FUNCTION: May participate in suppression of cell proliferation and induces apoptotic cell death through activation of interleukin-1-beta converting enzyme (ICE)-like proteases. {ECO:0000269|PubMed:12054692, ECO:0000269|PubMed:12138241, ECO:0000269|PubMed:12672804}. | Alternative splicing;Apoptosis;Coiled coil;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | This gene was identified as an estrogen-responsive gene. Regulation of transcription by estrogen is mediated by estrogen receptor, which binds to the estrogen-responsive element found in the 5'-flanking region of this gene. The encoded protein is a tumor-associated antigen that is expressed at high frequency in a variety of cancers. Alternate splicing results in multiple transcript variants. A pseudogene of this gene has been defined on chromosome 10. [provided by RefSeq, Jul 2013]. | hsa:9166; | Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; secretory granule [GO:0030141]; peptidase activator activity involved in apoptotic process [GO:0016505]; regulation of cell growth [GO:0001558] | 11705872_Overrepresentation of EBAG9 may play a specific role in early stages of breast carcinogenesis. 11992411_High expression of tumor-associated antigen RCAS1 in pancreatic ductal adenocarcinoma is an unfavorable prognostic marker 12054692_RCAS1 is associated with ductal breast cancer progression 12672804_modulates surface expression of tumor-associated, normally cryptic O-linked glycan structures and contributes indirectly to the antigenicity of tumor cells 12774924_RCAS1 expressed on macrophages may play an important role in the induction of activated T-cell apoptosis in cases of HNL 12845666_Overexpression of EBAG9 is correlated with advanced pathologic stages of prostate cancer 12888828_RCAS1 may have role endometrial cancer invasiveness; overexpression associated with significantly poorer prognosis 14534714_RCAS1 gene or protein expression may not correlate with tumor progression in esophageal squamous cell carcinoma 14981953_The mean survival of patients who had primary hepatocellular carcinoma with high RCAS1 protein expression was significantly longer than that of patients with low expression. 15164121_wide distribution of EBAG9 and its relation to advanced disease suggest that this protein may play important roles in epithelial ovarian cancer 15254686_RCAS1 expression might be associated with progression of oral squamous cell carcinoma 15460847_Serum RCAS1 appears to be valuable as a diagnostic index for biliary carcinomas, as well as for evaluating the progression of cancers during therapy. 15635093_EBAG9 and Snapin have roles in controlling exocytosis processes 15813909_Macrophages may negatively regulate erythropoiesis at least in part through the production of RCAS1 molecules. 15867365_EBAG9 is a crucial regulator of tumor progression and a potential prognostic marker for RCC. 15904507_the estrogen-inducible EBAG9 gene-product and the 22-1-1 defined antigen are structurally and functionally separate antigens 16012715_RCAS1 expression is informative for the follow-up of malignant mesothelioma patients and sRCAS1 in pleural fluid may be useful for the diagnosis of malignant mesothelioma. 16112176_RCAS1 may contribute to acquisition of malignant uterine cervical phenotypic characteristics including invasion, metastasis, and tumor growth via connective tissue remodeling. 16113565_RCAS1 and CAP may play a role in the downregulation of the maternal immune response during pregnancy and may participate in the initiation of the labor 16175077_distribution of RCAS1 expression in normal female genital organs; significant positive correlation between age and RCAS1 expression; RCAS1 may affect metaplastic processes and tumor progression 16211275_expression of RCAS1 is correlated with recurrence not only in carcinomas, but also in mesenchymal tumors 16273616_metastatic lymph nodes from bile duct, gastric, colon and pancreatic cancer were investigated for RCAS1 expression 16595162_RCAS1 expression in gliomas may play roles in tumor progression and tumor immune escape. 16907986_The expression of RCAS1 by endometrial cells may favor the persistence of these cells in ectopic localization both in scar following cesarean section and in ovarian endometriosis. 17187007_Significantly higher RCAS1 expression was noticed in tumor in comparison to stroma in patients with the presence of lymph nodes metastases. No such difference was observed in patients without the metastases. 17187008_RCAS1 expression was simultaneous to the infiltration of activated immunological cells of tumor environment as well as decidua. The activity of immunological cells was selectively suppressed. 17466050_High-level expression of RCAS1 is involved in the malignant transformation of endometrium, and RCAS1 coexpression with ER-alpha may be associated with development and metastasis of endometrial carcinoma 17562271_The expression of RCAS1 in cervical cancer is significantly increased, and has correlation with malignant degree of cervical carcinoma. Some RCAS1-positive cervical cancer tissues are infected by HPV16. 17604121_possible role of the RCAS1 protein in the development of pre-eclampsia through an immunological pathway. 17717421_the drop of the RCAS1 level, which could be a result of an insufficiency of the compensatory immune response mechanisms in tubal mucosa (although these mechanisms are simultaneously preserved in endometrium), leads to tubal perforation. 17825484_Results show that the suppression of RCAS1 expression effectively recover T cell proliferation, reduce apoptosis and partially reverse the T cell function of IFN-gamma secretion. 17845206_The limited immune cells infiltration in decidua during severe pre-eclampsia is associated with increase in RCAS1 decidual level. 17849467_RCAS1 may be a pivotal regulator of tumor growth through angiogenesis. 17981616_RCAS1 is a bioactive marker that induces connective tissue remodeling and lymphocyte apoptosis [review] 18032910_The presence of an enhanced number of immune cells of higher activity in ectopic decidua during the final step of decidualization seems to be associated with an increase in the immunoreactivity level of RCAS1. 18292826_The lowest level of RCAS1 endometrial expression was found during the early secretory cycle phase, and significantly higher expression was found in the endometrium during the mid-secretory as compared to the early secretory cycle phase. 18688918_In rheumatoid arthritis, the lack of RCAS1 is thought to induce CTL infiltration through loss of the ability to evade immune attack, thus leading to apoptosis of the synovial lining cells. 19030177_EBAG9 may have a role in promoting progression of bladder cancer 19032612_The level of RCAS1 in the decidua seems to influence effectiveness of stillbirth induction. 19122463_The lack of alterations in the sRCAS1 blood serum concentration levels observed in patients with adenomyosis may favor the development of the condition 19337974_tissue RCAS1 expression...considered as an informative biomarker in several types of human malignancy 19574770_RCAS1 expression is correlated with a decreasing number of vimentin-positive stromal cells in cervical cancer. 19634109_RCAS1 has a role in advanced colonic neoplasms 19813143_We found expression of RCAS1 co-localizing with GFAP+ cells of gliomas and with CD68 and CD74 in large macrophages infiltrating metastatic and primary tumours and sometimes in cells which had morphological characteristics of microglia 19957811_RCAS1 can be used as a serum tumor marker for diagnosis of pancreatic cancer. 20079734_the RCAS1 putative receptor-expressing chronic myelogenous leukemia cell line K562 was co-cultured with SiSo, MCF-7, or soluble RCAS1 to follow RCAS1 secretion in apoptosis initiation 20164540_NASP and RCAS1 proteins were more frequently expressed in ovarian cancer tissues than with normal ovarian tissue and serous cystadenomas and MRE11 was less frequently expressed 20570965_EBAG9 acts as a negative regulator of a COPI-dependent ER-to-Golgi transport pathway in epithelial cells and shows pathogenetic principle in which interference with intracellular membrane trafficking results in the emergence of a tumor-associated glycome 20571277_analyzed RCAS-1 as a biomarker in the serum of patients with head and neck squamous cell carcinoma 20645939_We observed a statistically significant increase in the RCAS1-positive macrophage infiltration within the microenvironment of the molar lesions in patients with partial hydatidiform mole in comparison with those who exhibited complete hydatidiform mole. 21804460_Data suggest that RCAS1 may be involved in the early stages of tumor progression in mobile tongue squamous cell carcinoma. 21845402_The immunoreactivity of RCAS1 on the cells present in the ovarian cancer microenvironment, was analysed. 22460085_RCAS1 protein may participate in thyroid neoplastic transformation and could be considered as a useful biomarker to improve diagnostic scrutiny. 22530960_The intensity of the suppressive profile of the cervical cancer microenvironment indicated by the presence of both RCAS1 and B7H4 on the front of the tumor and in the macrophages and fibroblasts infiltrating the cancer stroma 23164108_RCAS1 may play an important role in the phenomenon of tumor escape from host immunological surveillance and in creating the immune tolerance for the tumor cells, as well as in the tumor microenvironment. 23563217_The membrane molecule RCAS1 induces immune cell apoptosis via the RCAS1-RCAS1R pathway. 23881387_sRCAS1 levels could have a clinical value for the diagnosis and management of lung cancer and could be used as a new tumor marker of lung cancer. 24119785_High EBAG9 expression is associated with tamoxifen resistance in breast cancer. 24720371_High EBAG9 expression is associated with malignant pleural effusion in patients with lung cancer. 24815841_RCAS1 could be a useful immunohistochemical biomarker, indicating not only tumor aggressiveness but also a poorer prognosis for patients with NSCLC. 24885040_suggests that RCAS1 has an apoptotic function via membranous/soluble expression pattern in OSCC cells. RCAS1 may thus affect tumor escape from immune surveillance in OSCC by inducing apoptosis 25177692_Given the significant correlation between tumor ADAM9 expression and serum RCAS1 concentration in both cervical and endometrial cancer as well as the role for ADAM9 in RCAS1 shedding. 25674852_Findings suggest that the histological effect of increased RCAS1 expression depends on its cellular source and that RCAS1 expression itself is a component of various signaling pathways in urothelial bladder cancer cells. 25773455_RCAS1 seems to be involved in creating tumor-induced inflammation in the tumor and its microenvironment 26438059_Data suggest that joint detection of receptor-binding cancer antigen expressed on SiSo cells (RCAS1) and carcinoembryonic antigen (CEA) can improve the diagnostic sensitivity and specificity. 30950002_Studied soluble cancer-associated surface antigen (srCaS1) levels as a biomarker for survival in endometrial cancer. Found elevated serum sRCAS1 to be associated with shortened overall survival in endometrial cancer patients. | ENSMUSG00000022339 | Ebag9 | 221.48211 | 0.7131958 | -0.4876299331 | 0.25022914 | 3.733723e+00 | 5.332440e-02 | 9.998360e-01 | No | Yes | 171.37970 | 36.859280 | 2.412923e+02 | 40.051819 | |
| ENSG00000148290 | 6834 | SURF1 | protein_coding | Q15526 | FUNCTION: Component of the MITRAC (mitochondrial translation regulation assembly intermediate of cytochrome c oxidase complex) complex, that regulates cytochrome c oxidase assembly. {ECO:0000269|PubMed:24027061, ECO:0000269|PubMed:9843204, ECO:0000305|PubMed:26321642}. | Alternative splicing;Charcot-Marie-Tooth disease;Disease variant;Leigh syndrome;Membrane;Mitochondrion;Mitochondrion inner membrane;Neurodegeneration;Neuropathy;Primary mitochondrial disease;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a protein localized to the inner mitochondrial membrane and thought to be involved in the biogenesis of the cytochrome c oxidase complex. The protein is a member of the SURF1 family, which includes the related yeast protein SHY1 and rickettsial protein RP733. The gene is located in the surfeit gene cluster, a group of very tightly linked genes that do not share sequence similarity, where it shares a bidirectional promoter with SURF2 on the opposite strand. Defects in this gene are a cause of Leigh syndrome, a severe neurological disorder that is commonly associated with systemic cytochrome c oxidase deficiency. [provided by RefSeq, Jul 2008]. | hsa:6834; | integral component of membrane [GO:0016021]; mitochondrial respirasome [GO:0005746]; cytochrome-c oxidase activity [GO:0004129]; aerobic respiration [GO:0009060]; mitochondrial cytochrome c oxidase assembly [GO:0033617]; respiratory chain complex IV assembly [GO:0008535] | 11955926_Three novel mutations of the SURF-1 gene were identified in Japanese patients with cytochrome c oxidase deficiency; loss of function of the SURF-1 protein; cytochrome c oxidase activity was decreased to less than 20% of the control mean. 12515039_new missense mutation of 574C>T in the SURF1 gene in Leigh's syndrome 12538779_Two novel pathogenic SURF1 mutations have been identified in a patient with Leigh syndrome. 12812953_Mutations in the nuclear SURF1 gene are specifically associated with cytochrome c oxidase (COX)-deficient Leigh syndrome. MR imaging abnormalities in three children with this condition involved the brain. 14557577_Four pathogenic mutations including a novel, in-frame, 15-bp tandem duplication (806-820) in exon 8 and a novel 751+1G>A splice site mutation in SURF1 in three cases of Leigh Syndrome with cytochrome c oxidase deficiency 14607829_study points to a role for surfeit 1(SURF1) in promoting the association of cytochrome c oxidase II with the cytochrome c oxidase I.cytochrome c oxidase subunit 4.cytochrome c oxidase subunit 5A subassembly 15214016_a SURF1 mutation may have a role in subacute encephalopathy 15764605_Surf1p plays a role in facilitating the insertion of heme a3 into the active site of cytochrome-c oxidase. 16083427_The consequences of SCO2 and SURF1 mutations suggest the existence of tissue-specific functional differences of these proteins that may serve different tissue-specific requirements for the regulation of COX biogenesis. 17908801_Histological and histochemical features of muscle of genetically homogenous SURF1-deficient LS were reproducible in detection of COX deficit. SURF1-deficient muscle assessed in the microscopy panel may be interpreted as normal if COX staining is not used. 18583168_Data show high prevalence of SURF1 c.845_846delCT mutation in Polish Leigh patients. 18583168_Observational study of genotype prevalence. (HuGE Navigator) 19295170_SURF1-deficient samples analyzed showed a tissue-specific copper deficiency similar to that of SCO-deficient samples, suggesting a role for Surf1 in copper homeostasis regulation 19625251_a direct role of Surf1 in heme a cofactor insertion into COX subunit I by providing a protein-bound heme a pool. 19780766_The presence of a missense mutation in the SURF1 gene may correlate with a milder course and longer survival of Leigh patients. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20436434_mutations (574-575insCTGT, 311-321del10insAT and IVS8-1G>) were also frequent in the Russian population. 20601676_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20624914_Analysis of mutations in the SURF1 homolog Shy1 revealed Coa4, a new member of the cytochrome oxidase assembly factor family. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 22410471_sequenced the SURF1 gene and identified two heterozygous mutations; c.49+1 G>T and c.752_753del in Case 1, and homozygous c.743 C>A in Case 2 22465034_Analysis of fibroblast cell lines from 9 patients with SURF1 mutations revealed a 70% decrease of the COX complex content to be associated with 32-54% upregulation of respiratory chain complexes I, III and V and accumulation of Cox5a subunit. 22488715_Study identified 21 patients with clinical features of Leigh syndrome who are either homozygous or compound heterozygous for SURF1 mutations. 22729384_This study suggested that hypertrophic olivary degeneration on magnetic resonance imaging in mitochondrial syndromes associated with POLG and SURF1 mutations. 24027061_Mutations in the SURF1 gene are a cause of Charcot-Marie-Tooth disease. 26804654_studies support the view that COX assembly is much more dependent on SURF1 in humans than in mice. 29481804_the MT-ATP6 and SURF1 gene screening in Tunisian patients affected with classical Leigh syndrome and the computational investigation of the effect of detected mutations on its structure and functions by clinical and bioinformatics analyses. 29715184_Role of SURF1 in etiology of Leigh syndrome in Slovakia. 29933018_SURF1 mutations may be associated with worse clinical outcome in Chinese patients with Leigh syndrome than other populations. 32380162_Novel p.P298L SURF1 mutation in thiamine deficient Leigh syndrome patients compromises cytochrome c oxidase activity. 33771987_Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. | ENSMUSG00000015790 | Surf1 | 1280.44141 | 0.9816239 | -0.0267577745 | 0.12443389 | 4.722654e-02 | 8.279616e-01 | 9.998360e-01 | No | Yes | 1291.98702 | 131.358330 | 1.234786e+03 | 97.949045 | |
| ENSG00000148334 | 80142 | PTGES2 | protein_coding | Q9H7Z7 | FUNCTION: Isomerase that catalyzes the conversion of PGH2 into the more stable prostaglandin E2 (PGE2) (in vitro) (PubMed:12804604, PubMed:18198127, PubMed:17585783). The biological function and the GSH-dependent property of PTGES2 is still under debate (PubMed:18198127, PubMed:17585783). In vivo, PTGES2 could form a complex with GSH and heme and would not participate in PGE2 synthesis but would catalyze the degradation of prostaglandin E2 H2 (PGH2) to 12(S)-hydroxy-5(Z),8(E),10(E)-heptadecatrienoic acid (HHT) and malondialdehyde (MDA) (PubMed:17585783) (By similarity). {ECO:0000250|UniProtKB:Q9N0A4, ECO:0000269|PubMed:12804604, ECO:0000269|PubMed:17585783, ECO:0000269|PubMed:18198127}. | Cytoplasm;Fatty acid biosynthesis;Fatty acid metabolism;Golgi apparatus;Isomerase;Lipid biosynthesis;Lipid metabolism;Membrane;Phosphoprotein;Prostaglandin biosynthesis;Prostaglandin metabolism;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; prostaglandin biosynthesis. | The protein encoded by this gene is a membrane-associated prostaglandin E synthase, which catalyzes the conversion of prostaglandin H2 to prostaglandin E2. This protein also has been shown to activate the transcription regulated by a gamma-interferon-activated transcription element (GATE). Multiple transcript variants have been found for this gene. [provided by RefSeq, Jun 2009]. | hsa:80142; | azurophil granule lumen [GO:0035578]; cytosol [GO:0005829]; extracellular region [GO:0005576]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; 12-hydroxyheptadecatrienoic acid synthase activity [GO:0036134]; DNA binding [GO:0003677]; glutathione binding [GO:0043295]; heme binding [GO:0020037]; lyase activity [GO:0016829]; prostaglandin-E synthase activity [GO:0050220]; cyclooxygenase pathway [GO:0019371]; glutathione metabolic process [GO:0006749]; lipid metabolic process [GO:0006629]; positive regulation of transcription, DNA-templated [GO:0045893]; secretion [GO:0046903] | 11847219_Up-regulation of prostaglandin E2 synthesis by interleukin-1beta in human orbital fibroblasts involves coordinate induction of prostaglandin-endoperoxide H synthase-2 and glutathione-dependent prostaglandin E2 synthase expression 12050152_role in regulating interferon-gamma-dependent gene expression 12804604_110Cys is essential in the active site of membrane-associated prostaglandin E synthase-2 15879117_Transactivators GBF1 and CCAAT/enhancer-binding protein-beta physically interact to induce interferon-gamma-regulated transcription; a 37-aa long peptide derived from the GBF1 protein can associate with C/EBP-beta in an IFN-inducible manner. 16714329_alphaTOS inhibits COX activity, thereby inhibiting PGE2 production in human lung epithelial cells 17266179_Observational study of gene-disease association. (HuGE Navigator) 17566096_Observational study of gene-disease association. (HuGE Navigator) 17566096_study obtained evidence from two Caucasian study populations that the His298-allele of PTGES2 Arg298His confers to reduced risk of type 2 diabetes 17697149_Carbonyl reductase-1 (CBR1), microsomal prostaglandin E synthase-1 and 2 (mPGES-1, mPGES-2), cytosolic prostaglandin E synthase (cPGES), aldoketoreductase (AKR1C1) and prostaglandin F synthase (AKR1C3) were all expressed in hair follicles. 17979097_Observational study of gene-disease association. (HuGE Navigator) 17979097_risk-reducing effects of the minor His allele of the prostaglandin E synthase 2 (PTGES2) Arg298His polymorphism could be mediated partly by lowered body mass index (BMI) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19268562_Observational study of gene-disease association. (HuGE Navigator) 19336370_Observational study of gene-disease association. (HuGE Navigator) 19347995_all three terminal prostaglandin synthases, mPGES-1, mPGES-2, and cPGES, are over-expressed in human gliomas 19371221_A marginal but significant influence of the PTGES2 298H single nucleotide polymorphism on BMI was found in a large population-based study. 19371221_Observational study of gene-disease association. (HuGE Navigator) 19412621_These results indicate that mPGES-1 and mPGES-2 may each play a role in colorectal cancer progression. 19664621_high immunoreactivity of mPGES-2 in pyramidal neurons of AD brains indicates that it might have a potential role in the functional replacement of cytosolic PGES or inactive mPGES-1 in later stages of Alzheimer disease 20877624_Observational study of gene-disease association. (HuGE Navigator) 24467603_Four single nucleotide polymorphisms in two genes in the PGE2 family, PTGES2 and PTGER4, were significantly associated with primary graft dysfunction after lung transplantation. 31113465_This suggests that the interaction with the microenvironment occurs at both early and late thyroid tumor stages, and favors tumor progression. The co-expression of PTGS2 gene and M2 markers in human thyroid carcinoma highlights the possibility to counteract tumor growth through COX-2 inhibition. 31726220_Parasitic load determination by differential expressions of 5-lipoxygenase and PGE2 synthases in visceral leishmaniasis. | ENSMUSG00000026820 | Ptges2 | 6966.35210 | 1.0062641 | 0.0090089981 | 0.11028372 | 6.886291e-03 | 9.338645e-01 | 9.998360e-01 | No | Yes | 7831.61145 | 1275.449673 | 7.231547e+03 | 908.996524 |
| ENSG00000148429 | 9712 | USP6NL | protein_coding | Q92738 | FUNCTION: Acts as a GTPase-activating protein for RAB5A and RAB43. Involved in receptor trafficking. In complex with EPS8 inhibits internalization of EGFR. Involved in retrograde transport from the endocytic pathway to the Golgi apparatus. Involved in the transport of Shiga toxin from early and recycling endosomes to the trans-Golgi network. Required for structural integrity of the Golgi complex. {ECO:0000269|PubMed:11099046, ECO:0000269|PubMed:17562788, ECO:0000269|PubMed:17684057}. | Acetylation;Alternative splicing;Cytoplasmic vesicle;GTPase activation;Golgi apparatus;Phosphoprotein;Reference proteome | hsa:9712; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; trans-Golgi network membrane [GO:0032588]; GTPase activator activity [GO:0005096]; small GTPase binding [GO:0031267]; activation of GTPase activity [GO:0090630]; Golgi organization [GO:0007030]; intracellular protein transport [GO:0006886]; plasma membrane to endosome transport [GO:0048227]; positive regulation of GTPase activity [GO:0043547]; regulation of Golgi organization [GO:1903358]; retrograde transport, plasma membrane to Golgi [GO:0035526]; virion assembly [GO:0019068] | 15152255_Rab5 signals to the actin cytoskeleton through RN-tre, a previously identified Rab5-specific GTPase-activating protein; RN-tre interacts with both F-actin and actinin-4, an F-actin bundling protein 17371873_RN-tre phosphorylation is critical for efficient hCdc14A association 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 28183528_Genome-wide significant (GWS) associations in single-nucleotide polymorphism (SNP)-based tests (P < 5 x 10(-8)) were identified for SNPs in PFDN1/HBEGF, USP6NL/ECHDC3, and BZRAP1-AS1. 29691252_Study reports that USP6NL is overexpressed in breast cancer, mainly of the basal-like/integrative cluster 10 subtype. Increased USP6NL levels were accompanied by gene amplification and were associated with worse prognosis in the METABRIC dataset. High levels of USP6NL in breast cancer cells delayed endocytosis and degradation of the EGFR, causing chronic AKT activation. 33054738_USP6NL mediated by LINC00689/miR-142-3p promotes the development of triple-negative breast cancer. | ENSMUSG00000039046 | Usp6nl | 263.64756 | 1.0218063 | 0.0311217119 | 0.20776947 | 2.233542e-02 | 8.811982e-01 | 9.998360e-01 | No | Yes | 302.45138 | 54.910111 | 3.178694e+02 | 44.568815 | ||
| ENSG00000149091 | 8525 | DGKZ | protein_coding | Q13574 | FUNCTION: Diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:9159104, PubMed:15544348, PubMed:18004883, PubMed:19744926, PubMed:22108654, PubMed:22627129, PubMed:23949095). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (PubMed:9159104, PubMed:15544348, PubMed:18004883, PubMed:19744926, PubMed:22108654, PubMed:22627129, PubMed:23949095). Also plays an important role in the biosynthesis of complex lipids (Probable). Does not exhibit an acyl chain-dependent substrate specificity among diacylglycerol species (PubMed:9159104, PubMed:19744926, PubMed:22108654). Can also phosphorylate 1-alkyl-2-acylglycerol in vitro but less efficiently and with a preference for alkylacylglycerols containing an arachidonoyl group (PubMed:15544348, PubMed:19744926, PubMed:22627129). The biological processes it is involved in include T cell activation since it negatively regulates T-cell receptor signaling which is in part mediated by diacylglycerol (By similarity). By generating phosphatidic acid, stimulates PIP5KIA activity which regulates actin polymerization (PubMed:15157668). Through the same mechanism could also positively regulate insulin-induced translocation of SLC2A4 to the cell membrane (By similarity). {ECO:0000250|UniProtKB:Q80UP3, ECO:0000269|PubMed:15157668, ECO:0000269|PubMed:15544348, ECO:0000269|PubMed:18004883, ECO:0000269|PubMed:19744926, ECO:0000269|PubMed:22108654, ECO:0000269|PubMed:22627129, ECO:0000269|PubMed:23949095, ECO:0000269|PubMed:9159104, ECO:0000305|PubMed:8626588}.; FUNCTION: [Isoform 1]: Regulates RASGRP1 activity. {ECO:0000269|PubMed:11257115}.; FUNCTION: [Isoform 2]: Does not regulate RASGRP1 activity. {ECO:0000269|PubMed:11257115}. | 3D-structure;ANK repeat;ATP-binding;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Kinase;Lipid metabolism;Membrane;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transferase;Zinc;Zinc-finger | PATHWAY: Lipid metabolism; glycerolipid metabolism. {ECO:0000305|PubMed:8626588}. | The protein encoded by this gene belongs to the eukaryotic diacylglycerol kinase family. It may attenuate protein kinase C activity by regulating diacylglycerol levels in intracellular signaling cascade and signal transduction. Alternative splicing occurs at this locus and multiple transcript variants encoding distinct isoforms have been identified. [provided by RefSeq, Nov 2010]. | hsa:8525; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; lamellipodium [GO:0030027]; nuclear speck [GO:0016607]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; diacylglycerol kinase activity [GO:0004143]; kinase activity [GO:0016301]; lipid kinase activity [GO:0001727]; metal ion binding [GO:0046872]; NAD+ kinase activity [GO:0003951]; protein C-terminus binding [GO:0008022]; cell migration [GO:0016477]; diacylglycerol metabolic process [GO:0046339]; glycerolipid metabolic process [GO:0046486]; intracellular signal transduction [GO:0035556]; lipid phosphorylation [GO:0046834]; mitotic G1 DNA damage checkpoint signaling [GO:0031571]; negative regulation of mitotic cell cycle [GO:0045930]; negative regulation of T cell receptor signaling pathway [GO:0050860]; phosphatidic acid biosynthetic process [GO:0006654]; platelet activation [GO:0030168]; positive regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity [GO:0090216]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205]; regulation of synaptic transmission, glutamatergic [GO:0051966] | 12015310_structural domain requirements for translocation and activity 12070163_Negative regulation of T cell receptor induced activation of the Ras-Erk1/2-AP1 pathway by DGKz 12890670_PKC alpha phosphorylates diacylglycerol kinase zeta in cells, and this phosphorylation inhibits its kinase activity to remove cellular diacylglycerol, thereby affecting cell growth. 14707140_role in controlling the induction of luteinizing hormone beta transcription by ERK1/2 15157668_DGKzeta generating PA, stimulates PIP5KIalpha activity to increase local PIP2, which regulates actin polymerization. 15632115_DGKzeta-derived phosphatidic acid acts as a mediator of mTOR signaling 16286473_DGKzeta may act in vivo as a downstream effector of pRB to regulate nuclear levels of diacylglycerol and phosphatidic acid 18004883_2,3-dioleoylglycerol binds to a site on the alpha and zeta isoforms of diacylglycerol kinase that is exposed as a consequence of the substrate binding to the active site. 18694729_PKD activation is induced by DGKzeta, suggesting DGK is an upstream regulator of oxidative stress-induced activation of the PKD signaling pathway in intestinal epithelial cells. 19211846_In DGKzeta-deficient fibroblasts PAK1 phosphorylation and Rac1-RhoGDI dissociation were attenuated, leading to reduced Rac1 activation after platelet-derived growth factor stimulation. 19308020_None of SNPs of diacylglycerol kinase zeta tested showed association with bipolar disorder in Sardinian sample. 21194521_Data show that Diacylglycerol that 2-arachidonoyl glycerol is a very poor substrate for either the epsilon or the zeta isoforms of diacylglycerol kinases. 21937721_DGK-zeta translocated rapidly to the plasma membrane at early stages of immunological synapse (IS) formation independent of enzyme activity; study highlights a DGKzeta-specific function for local diacylglycerol metabolism at the IS and offers new clues to its mode of regulation 21996351_Nucleosome assembly protein (NAP) 1-like 1 (NAP1L1) and NAP1-like 4 (NAP1L4) are identified as novel DGKzeta binding partners. 22271650_Antigen-specific CD8-positive T cells from DGKzeta-deficient transgenic mice show enhanced expansion and increased cytokine production after lymphocytic choriomeningitis virus infection, yet DGK-deficient memory CD8+ T cells exhibit impaired expansion. 22895365_DGK regulates melanogenesis via modulation of the posttranslational processing of tyrosinase, which may be related with the protein degradation machinery. 23723068_Data indicate that after P2Y6 receptor stimulation both phospholipase D (PLD) and DGKzeta enzymes are responsible for producing phosphatidic acid (PA). 24646293_Elevated DGKzeta expression contributes to increased Rho GTPase activation and the enhanced motility of metastatic cancer cells. 25450975_This study shows that DGKzeta knockdown facilitates degradation of IkappaB, followed by nuclear translocation of NF-kappaB p65 subunit. 25921290_Redundant and specialized roles for diacylglycerol kinases alpha and zeta in the control of T cell functions. 26452358_These results established the positive correlation between DGKzeta expression and gliomagrade. 26521214_DGKzeta knockdown engenders enhancement of NF-kappaB pathway in response to TNF-alpha. [review] 27697466_Diacylglycerol kinases alpha and zeta are up-regulated in cancer in cancer, and contribute towards tumor immune evasion and T cells clonal anergy. (Review) 27999176_these data suggest that the activation of DGKzeta downstream of antigen recognition provides a mechanism that ensures the activation of PA-dependent signaling as a direct result of the strength of TCR-dependent DAG mobilization. 28008152_Results show that DGKzeta is downregulated in bone marrow mononuclear cells and associated with the severity of aplastic anemia (AA). Also, DGKzeta is a downstream target gene of miR34a. Their dysregulation enhances T-cell activation in AA cells. 31087244_we showed that rs7951870-TT genotype was strongly associated with increased DGKZ expression level (P = 0.038). In conclusion, our findings revealed dysregulation of DGKZ in SCZ patients and a significant correction between the gene expression and DGKZ variant rs7951870. 31288898_knockdown of DGKZ can induce apoptosis and G2/M phase arrest in human acute myeloid leukemia HL-60 cells through the MAPK/survivin/caspase pathway. 31722116_Characterization of alpha-synuclein N-terminal domain as a novel cellular phosphatidic acid sensor. 32224048_DGKzeta depletion attenuates HIF-1alpha induction and SIRT1 expression, but enhances TAK1-mediated AMPKalpha phosphorylation under hypoxia. 33246984_Diacylglycerol kinase zeta limits IL-2-dependent control of PD-1 expression in tumor-infiltrating T lymphocytes. 33450306_Regulation of p53 and NF-kappaB transactivation activities by DGKzeta in catalytic activity-dependent and -independent manners. 33926342_Downregulation of Diacylglycerol kinase zeta (DGKZ) suppresses tumorigenesis and progression of cervical cancer by facilitating cell apoptosis and cell cycle arrest. 35115500_DGKZ promotes TGFbeta signaling pathway and metastasis in triple-negative breast cancer by suppressing lipid raft-dependent endocytosis of TGFbetaR2. | ENSMUSG00000040479 | Dgkz | 6521.10648 | 0.9970957 | -0.0041961627 | 0.09895620 | 1.850641e-03 | 9.656863e-01 | 9.998360e-01 | No | Yes | 6949.99038 | 956.328192 | 6.366519e+03 | 676.331452 |
| ENSG00000149313 | 60496 | AASDHPPT | protein_coding | Q9NRN7 | FUNCTION: Catalyzes the post-translational modification of target proteins by phosphopantetheine. Can transfer the 4'-phosphopantetheine moiety from coenzyme A, regardless of whether the CoA is presented in the free thiol form or as an acetyl thioester, to a serine residue of a broad range of acceptors including the acyl carrier domain of FASN. {ECO:0000269|PubMed:11286508, ECO:0000269|PubMed:12815048, ECO:0000269|PubMed:18022563, ECO:0000269|PubMed:19933275, ECO:0000269|PubMed:21238436}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Magnesium;Metal-binding;Phosphoprotein;Reference proteome;Transferase | The protein encoded by this gene is similar to Saccharomyces cerevisiae LYS5, which is required for the activation of the alpha-aminoadipate dehydrogenase in the biosynthetic pathway of lysine. Yeast alpha-aminoadipate dehydrogenase converts alpha-biosynthetic-aminoadipate semialdehyde to alpha-aminoadipate. It has been suggested that defects in the human gene result in pipecolic acidemia. [provided by RefSeq, Jul 2008]. | hsa:60496; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; holo-[acyl-carrier-protein] synthase activity [GO:0008897]; magnesium ion binding [GO:0000287]; lysine biosynthetic process via aminoadipic acid [GO:0019878]; pantothenate metabolic process [GO:0015939]; protein phosphopantetheinylation [GO:0018215] | 12815048_humans appear to utilize a single, broad specificity enzyme, 4'-phosphopantetheine transferase, for all posttranslational 4'-phosphopantetheinylation reactions 19933275_Our study identifies human PPT as the FDH-modifying enzyme and supports the hypothesis that mammals utilize a single enzyme for all phosphopantetheinylation reactions. | ENSMUSG00000025894 | Aasdhppt | 861.23894 | 1.1900667 | 0.2510424333 | 0.14915175 | 2.923224e+00 | 8.731327e-02 | 9.998360e-01 | No | Yes | 867.41348 | 143.609835 | 7.685125e+02 | 98.193411 | |
| ENSG00000150403 | 55002 | TMCO3 | protein_coding | Q6UWJ1 | FUNCTION: Probable Na(+)/H(+) antiporter. {ECO:0000250}. | Alternative splicing;Antiport;Coiled coil;Glycoprotein;Ion transport;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the monovalent cation:proton antiporter 2 (CPA2) family of transporter proteins. Members of this family typically couple the export of monovalent cations, such as potassium or sodium, to the import of protons across cellular membranes. Mutations in this gene have been identified in patients with a rare inherited vision defect, cornea guttata with anterior polar cataract. [provided by RefSeq, Mar 2017]. | hsa:55002; | integral component of membrane [GO:0016021]; solute:proton antiporter activity [GO:0015299] | 27484837_This study reveals, for the first time, that mutations in TMCO3 are associated with cornea guttata and anterior polar cataract, warranting further investigation into the pathogenesis of this disorder. | ENSMUSG00000038497 | Tmco3 | 264.44233 | 1.5030292 | 0.5878730806 | 0.21440691 | 7.751670e+00 | 5.366288e-03 | 8.052441e-01 | No | Yes | 278.77859 | 49.167065 | 2.096702e+02 | 28.674800 | |
| ENSG00000150556 | 130576 | LYPD6B | protein_coding | Q8NI32 | FUNCTION: Believed to act as a modulator of nicotinic acetylcholine receptors (nAChRs) activity. In vitro acts on nAChRs in a subtype- and stoichiometry-dependent manner. Modulates specifically alpha-3(3):beta-4(2) nAChRs by enhancing the sensitivity to ACh, decreasing ACh-induced maximal current response and increasing the rate of desensitization to ACh; has no effect on alpha-7 homomeric nAChRs; modulates alpha-3(2):alpha-5:beta-4(2) nAChRs in the context of CHRNA5/alpha-5 variant Asn-398 but not its wild-type sequence. {ECO:0000269|PubMed:26586467}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Lipoprotein;Membrane;Reference proteome;Signal | hsa:130576; | anchored component of membrane [GO:0031225]; extracellular region [GO:0005576]; plasma membrane [GO:0005886]; acetylcholine receptor regulator activity [GO:0030548] | 18360792_LYPD7 was especially highly expressed in testis, lung, stomach, and prostate. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 33019770_Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors. | ENSMUSG00000026765 | Lypd6b | 52.61512 | 0.9414785 | -0.0869999510 | 0.40123794 | 4.646679e-02 | 8.293295e-01 | 9.998360e-01 | No | Yes | 57.85160 | 10.077681 | 6.244469e+01 | 9.280101 | ||
| ENSG00000151500 | 29087 | THYN1 | protein_coding | Q9P016 | FUNCTION: Specifically binds 5-hydroxymethylcytosine (5hmC), suggesting that it acts as a specific reader of 5hmC. {ECO:0000250}. | 3D-structure;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein that is highly conserved among vertebrates and plant species and may be involved in the induction of apoptosis. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:29087; | nucleus [GO:0005634] | 12384300_Studies of the mouse counterpart. 15939300_HSPC144 was expressed and purified, and a stable fragment (residues 44-225) was identified by limited proteolysis method. 19237743_The 2.3A-resolution structure revealed only one 2-fold axis of rotational pseudosymmetry. A potential RNA-binding domain from human YTH-domain-containing protein 2 has the most similar 3-D fold to that of the DUF55 domain; it may be an RNA-related domain. | ENSMUSG00000035443 | Thyn1 | 901.76417 | 1.0114466 | 0.0164200974 | 0.13453172 | 1.522137e-02 | 9.018102e-01 | 9.998360e-01 | No | Yes | 916.25756 | 113.520668 | 9.668515e+02 | 92.876081 | |
| ENSG00000151576 | 79691 | QTRT2 | protein_coding | Q9H974 | FUNCTION: Non-catalytic subunit of the queuine tRNA-ribosyltransferase (TGT) that catalyzes the base-exchange of a guanine (G) residue with queuine (Q) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine). {ECO:0000255|HAMAP-Rule:MF_03043}. | 3D-structure;Alternative splicing;Cytoplasm;Membrane;Metal-binding;Mitochondrion;Mitochondrion outer membrane;Reference proteome;Zinc;tRNA processing | This gene encodes a subunit of tRNA-guanine transglycosylase. tRNA-guanine transglycosylase is a heterodimeric enzyme complex that plays a critical role in tRNA modification by synthesizing the 7-deazaguanosine queuosine, which is found in tRNAs that code for asparagine, aspartic acid, histidine, and tyrosine. The encoded protein may play a role in the queuosine 5'-monophosphate salvage pathway. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Feb 2012]. | hsa:79691; | cytoplasm [GO:0005737]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; protein-containing complex [GO:0032991]; transferase complex [GO:1990234]; metal ion binding [GO:0046872]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; queuine tRNA-ribosyltransferase activity [GO:0008479]; tRNA-guanine transglycosylation [GO:0101030] | 20354154_TGT is composed of a catalytic subunit, QTRT1, and QTRTD1, not USP14. QTRTD1 has been implicated as the salvage enzyme that generates free queuine from QMP. 21044367_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000022704 | Qtrt2 | 1350.48319 | 1.0802574 | 0.1113751488 | 0.12857422 | 7.591465e-01 | 3.835958e-01 | 9.998360e-01 | No | Yes | 1484.83892 | 247.894295 | 1.446259e+03 | 186.255144 | |
| ENSG00000151746 | 636 | BICD1 | protein_coding | Q96G01 | FUNCTION: Regulates coat complex coatomer protein I (COPI)-independent Golgi-endoplasmic reticulum transport by recruiting the dynein-dynactin motor complex. | Alternative splicing;Coiled coil;Golgi apparatus;Host-virus interaction;Reference proteome | This gene encodes an adaptor protein that belongs to the bicaudal D family of dynein cargo adaptors. The encoded protein acts as an intracellular cargo transport cofactor that regulates the microtubule-based loading of cargo onto the dynein motor complex. It also controls dynein motor activity and coordination. It has a domain architecture consisting of coiled-coil domains at the N- and C-termini that are highly conserved in other family members. Naturally occurring mutations in this gene are associated with short telomere length and emphysema. [provided by RefSeq, Aug 2017]. | hsa:636; | centrosome [GO:0005813]; cytoplasmic vesicle [GO:0031410]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; membrane [GO:0016020]; perinuclear region of cytoplasm [GO:0048471]; secretory vesicle [GO:0099503]; trans-Golgi network [GO:0005802]; cytoskeletal anchor activity [GO:0008093]; dynactin binding [GO:0034452]; dynein complex binding [GO:0070840]; dynein intermediate chain binding [GO:0045505]; protein kinase binding [GO:0019901]; proteinase activated receptor binding [GO:0031871]; small GTPase binding [GO:0031267]; structural constituent of cytoskeleton [GO:0005200]; anatomical structure morphogenesis [GO:0009653]; intracellular mRNA localization [GO:0008298]; microtubule anchoring at microtubule organizing center [GO:0072393]; minus-end-directed organelle transport along microtubule [GO:0072385]; negative regulation of phospholipase C activity [GO:1900275]; negative regulation of phospholipase C-activating G protein-coupled receptor signaling pathway [GO:1900737]; positive regulation of protein localization to centrosome [GO:1904781]; positive regulation of receptor-mediated endocytosis [GO:0048260]; protein localization to organelle [GO:0033365]; regulation of microtubule cytoskeleton organization [GO:0070507]; regulation of proteinase activated receptor activity [GO:1900276]; RNA processing [GO:0006396]; stress granule assembly [GO:0034063]; viral process [GO:0016032] | 16320833_Various processes in which BicD is involved during Drosophilian development (review) 17101644_findings show BICD1 localized to Chlamydia trachomatis inclusions in a biovar-specific manner and that EGFP-BICD1 is recruited to the inclusion in a microtubule- and Golgi apparatus-independent but chlamydial gene expression-dependent mechanism 17139249_These results imply that GSK-3beta may function in transporting centrosomal proteins to the centrosome by stabilizing the BICD1 and dynein complex, resulting in the regulation of a focused microtubule organization. 17707369_The brain-specific Rab6B via Bicaudal-D1 is linked to the dynein/dynactin complex, suggesting a regulatory role for Rab6B in the retrograde transport of cargo in neuronal cells. 18487243_BICD1 plays a similar role in telomere length homeostasis in humans. 20080650_Observational study of gene-disease association. (HuGE Navigator) 20164183_the protease-activated receptor-1 interactor, Bicaudal D1, regulates G protein signaling and internalization 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20686608_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20709820_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20709820_Variants in BICD1 are associated with length of telomeres, which suggests that a mechanism linked to accelerated aging may be involved in the pathogenesis of emphysema. 25792135_Graft BICD1 polymorphisms apart from the association with telomere length, affect early kidney function after transplantation 27496426_Kidney transplant recipients' polymorphisms of genes associated with telomere length, BICD1 and chromosome 18, but not hTERT, affect kidney allograft early and long-term function after transplantation. 28215293_Data suggest that BICD1 and BICD2 are highly expressed in the nervous system during development and are important in neuronal homeostasis. [REVIEW] 28410362_This study showed that rs2735940 hTERT CX-TT donor-recipient genotype pair was associated with almost five times higher odds (OR=4.82; 95% CI: 1.32-18; p=0.016) of delayed graft function (DGF), and that rs2735940 hTERT, rs2630578 BICD1, and rs7235755 chromosome 18 polymorphisms combined pairs were not associated with acute rejection (AR). 30464225_BICD1 mediates HIF1alpha nuclear translocation in mesenchymal stem cells during hypoxia adaptation. 31541015_The disrupting Bicd1/Fignl1 interaction induced motor axon pathfinding defects characteristic of Fignl1 gain or loss of function, respectively. 31846791_BICD1 genes may contribute to the decrease in forced vital capacity levels by interacting with PM10 exposure 32088084_BICD1 functions as a prognostic biomarker and promotes hepatocellular carcinoma progression. | ENSMUSG00000003452 | Bicd1 | 116.84400 | 0.8700093 | -0.2008973119 | 0.28059147 | 5.144866e-01 | 4.732031e-01 | 9.998360e-01 | No | Yes | 172.17904 | 31.415324 | 1.703355e+02 | 24.095566 | |
| ENSG00000152359 | 134359 | POC5 | protein_coding | Q8NA72 | FUNCTION: Essential for the assembly of the distal half of centrioles, required for centriole elongation. {ECO:0000269|PubMed:19349582}. | Acetylation;Alternative splicing;Cell cycle;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome;Repeat | hsa:134359; | centriole [GO:0005814]; centrosome [GO:0005813]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; cell cycle [GO:0007049] | 19349582_hPOC5, a conserved centrin-binding protein that contains Sfi1p-like repeats is characterized. 23844208_Data indicate that overexpression of the centrin interactor POC5 leads to the assembly of linear, centrin-dependent structures. 25642776_Mutations in the POC5 gene contribute to the occurrence of idiopathic scoliosis. 27185865_Depletion of CEP295 blocks the incorporation of POC5 and POC1B into the distal portion of centrioles and suppresses the post-translational modification of centriolar microtubules . Our study thus uncovers a new role for CEP295 during centriole elongation. 29189569_Common variant rs6892146 of POC5 is associated with the development of adolescent idiopathic scoliosis in the Chinese population. 29272404_The findings demonstrate that Poc5 is important for normal retinal development and function. Altogether, this study presents POC5 as a novel gene involved autosomal recessively inherited RP, and strengthens the hypothesis that mutations in centriolar proteins are important cause of retinal dystrophies. 30845169_POC5 mutation is associated with impairment of cell cycle, cilia length and centrosome protein interactions in Adolescent idiopathic scoliosis. 34356048_Prevalence of POC5 Coding Variants in French-Canadian and British AIS Cohort. | ENSMUSG00000021671 | Poc5 | 136.11814 | 1.1574916 | 0.2110016954 | 0.28685245 | 5.211847e-01 | 4.703368e-01 | 9.998360e-01 | No | Yes | 136.86478 | 28.861442 | 1.159301e+02 | 18.918805 | ||
| ENSG00000152380 | 167555 | FAM151B | protein_coding | Q6UXP7 | Reference proteome | hsa:167555; | extracellular space [GO:0005615] | Mouse_homologues 31949211_Fam151b, the mouse homologue of C.elegans menorin gene, is essential for retinal function. | ENSMUSG00000034334 | Fam151b | 22.26654 | 1.9104616 | 0.9339212886 | 0.64312060 | 1.802271e+00 | 1.794382e-01 | 9.998360e-01 | No | Yes | 26.24457 | 7.369554 | 1.528113e+01 | 3.378341 | |||
| ENSG00000153485 | 26175 | TMEM251 | protein_coding | Q8N6I4 | Alternative splicing;Disease variant;Dwarfism;Golgi apparatus;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:26175; | integral component of membrane [GO:0016021] | ENSMUSG00000046675 | Tmem251 | 254.07777 | 1.1433564 | 0.1932752050 | 0.23349298 | 6.986449e-01 | 4.032394e-01 | 9.998360e-01 | No | Yes | 239.31211 | 44.395647 | 2.420366e+02 | 34.748921 | ||||
| ENSG00000153922 | 1105 | CHD1 | protein_coding | O14646 | FUNCTION: ATP-dependent chromatin-remodeling factor which functions as substrate recognition component of the transcription regulatory histone acetylation (HAT) complex SAGA. Regulates polymerase II transcription. Also required for efficient transcription by RNA polymerase I, and more specifically the polymerase I transcription termination step. Regulates negatively DNA replication. Not only involved in transcription-related chromatin-remodeling, but also required to maintain a specific chromatin configuration across the genome. Is also associated with histone deacetylase (HDAC) activity (By similarity). Required for the bridging of SNF2, the FACT complex, the PAF complex as well as the U2 snRNP complex to H3K4me3. Functions to modulate the efficiency of pre-mRNA splicing in part through physical bridging of spliceosomal components to H3K4me3 (PubMed:18042460, PubMed:28866611). Required for maintaining open chromatin and pluripotency in embryonic stem cells (By similarity). {ECO:0000250|UniProtKB:P40201, ECO:0000269|PubMed:18042460, ECO:0000269|PubMed:28866611}. | 3D-structure;ATP-binding;Alternative splicing;Chromatin regulator;Cytoplasm;DNA-binding;Disease variant;Helicase;Hydrolase;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The CHD family of proteins is characterized by the presence of chromo (chromatin organization modifier) domains and SNF2-related helicase/ATPase domains. CHD genes alter gene expression possibly by modification of chromatin structure thus altering access of the transcriptional apparatus to its chromosomal DNA template. [provided by RefSeq, Jul 2008]. | hsa:1105; | cytoplasm [GO:0005737]; nuclear chromosome [GO:0000228]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; methylated histone binding [GO:0035064]; chromatin remodeling [GO:0006338]; positive regulation by host of viral transcription [GO:0043923] | 12890497_associates with NCoR and histone deacetylase as well as with RNA splicing proteins 16263726_yeast and human CHD1 have diverged in their ability to discriminate covalently modified histones and link histone modification-recognition and non-covalent chromatin remodeling activities within a single human protein 16372014_the structure of the tandem arrangement of the human CHD1 chromodomains, and its interactions with histone tails 17098252_analysis of chromodomains in human and fungal Chd1 19441106_overexpression of CHD1L could sustain tumor cell survival by preventing Nur77-mediated apoptosis 19625449_CENP-H-containing complex facilitates deposition of newly synthesized CENP-A into centromeric chromatin in cooperation with FACT and CHD1. 20363151_Observational study of genotype prevalence. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21979373_Mediator coactivator complex, which controls PIC assembly, is also necessary for CHD1 recruitment 22046413_that hPaf1/PD2 in association with MLL1 regulates methylation of H3K4 residues, as well as interacts and regulates nuclear shuttling of chromatin remodeling protein CHD1, facilitating its function in pancreatic cancer cells 22048254_CHD1, was the most 'dietary sensitive' genes, as methylation of their promoters was associated with intakes of at least two out of the eight dietary methyl factors examined. 22139082_findings collectively suggest that distinct CHD1-associated alterations of genomic structure evolve during and are required for the development of prostate cancer 22179824_findings suggest that CHD1 deletion may underlie cell invasiveness in a subset of prostate cancers, and indicate a possible novel role of altered chromatin remodeling in prostate tumorigenesis 23492366_CHD1 is the 5q21 tumor suppressor gene in prostate cancer 24735615_Data indicate that chromodomain-helicase-DNA-binding protein CHD1, neoplasm protein GREB1 and karyopherin alpha 2 protein KPNA2 as critical mediators of miR-26a and miR-26b elicited cell growth. 24853335_The double chromodomains of CHD1 adopt an 'open pocket' to interact with the free N-terminal amine of H3K4, and the open pocket permits the NS1 mimic to bind in a distinct conformation. 25175909_results demonstrate the ability of confocal microscopy and FISH to identify the cell-to-cell differences in common gene fusions such as TMPRSS2-ERG that may arise independently within the same tumor focus 25297984_CHD1 and CHD2 act as positive regulators of HIV-1 gene expression. 25770290_identify coordinate loss of MAP3K7 and CHD1 as a unique driver of aggressive prostate cancer development 25879624_We have identified CHD1 as the RUNX1 fusion partner in acute myeloid leukemia with t(5;21)(q21;q22). 26751641_These data link the assembly of methylated KDM1A and CHD1 with AR-dependent transcription and genomic translocations, thereby providing mechanistic insight into the formation of TMPRSS2-ERG gene fusions during prostate-tumor evolution. 26792750_These results indicate that CHD1 is a positive regulator of influenza virus multiplication and suggest a role for chromatin remodeling in the control of the influenza virus life cycle. 27591891_The authors have identified an additional conserved domain C-terminal to the SANT-SLIDE domain and determined its structure by multidimensional heteronuclear NMR spectroscopy. They have termed this domain the CHD1 helical C-terminal (CHCT) domain as it is comprised of five alpha-helices arranged in a variant helical bundle topology. 27596623_examined the role of CHD1 in DNA double-strand break (DSB) repair in prostate cancer cells; findings show that CHD1 is required for the recruitment of CtIP to chromatin and subsequent end resection during DNA DSB repair; data support a role for CHD1 in opening the chromatin around the DSB to facilitate the recruitment of homologous recombination (HR) proteins 28166537_In PTEN-deficient prostate and breast cancers, CHD1 depletion profoundly and specifically suppressed cell proliferation, cell survival and tumorigenic potential. 28383660_CHD1 loss is associated with an increased sensitivity to PARP inhibition and anti-cancer drugs that induce DNA intercross-strand links in prostate tumors. 28475736_CHD1 is required for the induction of osteoblast-specific gene expression, extracellular-matrix mineralization and ectopic bone formation in vivo. Genome-wide occupancy analyses revealed increased CHD1 occupancy around the transcriptional start site of differentiation-activated genes. 28646284_Increased CHD1L protein expression was significantly associated with poor overall survival in pancreatic cancer patients. 28866611_Our results suggest that variants in CHD1 can lead to diverse phenotypic outcomes; however, the neurodevelopmental phenotype appears to be limited to patients with missense variants, which is compatible with a dominant negative mechanism of disease. 29018037_CHD1 facilitates substrate handover from XPC to the downstream TFIIH (transcription factor IIH). 29529298_Identified a unique N-terminal region of CHD1 that inhibits the DNA binding, ATPase, and chromatin assembly and remodeling activities of CHD1. CHD1 lacking the N terminus was more active in rescuing the defects in gammaH2AX formation and CtIP recruitment in CHD1-KO cells than full-length CHD1, suggesting the N terminus is a negative regulator in cells. 30068710_SPOP-mutated mCRPCs are strongly enriched for CHD1 loss. These tumors appear highly sensitive to abiraterone treatment. 30930119_CHD1 functions as a tumor suppressor in the prostate by constraining AR binding/function to limit tumor progression. 31222142_The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans. 31667976_MATN1-AS1 promotes glioma progression through regulating miR-200b/c/429-CHD1 axis, suggesting MATN1-AS1 as a probable target for glioma treatment. 32220301_Loss of CHD1 Promotes Heterogeneous Mechanisms of Resistance to AR-Targeted Therapy via Chromatin Dysregulation. 33414516_CHD1 loss negatively influences metastasis-free survival in R0-resected prostate cancer patients and promotes spontaneous metastasis in vivo. 33593142_The Chromatin Remodeling Complex CHD1 Regulates the Primitive State of Mesenchymal Stromal Cells to Control Their Stem Cell Supporting Activity. 33846123_MAP3K7 Loss Drives Enhanced Androgen Signaling and Independently Confers Risk of Recurrence in Prostate Cancer with Joint Loss of CHD1. 34424918_Cell-cell adhesion regulates Merlin/NF2 interaction with the PAF complex. 34533858_SPOP and CHD1 alterations in prostate cancer: Relationship with PTEN loss, tumor grade, perineural infiltration, and PSA recurrence. | ENSMUSG00000023852 | Chd1 | 419.75385 | 0.7363727 | -0.4414919359 | 0.17614184 | 6.057581e+00 | 1.384673e-02 | 9.998360e-01 | No | Yes | 437.93455 | 99.340470 | 5.627996e+02 | 98.224978 | |
| ENSG00000154124 | 90268 | OTULIN | protein_coding | Q96BN8 | FUNCTION: Deubiquitinase that specifically removes linear ('Met-1'-linked) polyubiquitin chains to substrates and acts as a regulator of angiogenesis and innate immune response (PubMed:26997266, PubMed:23708998, PubMed:23746843, PubMed:23806334, PubMed:23827681, PubMed:27523608, PubMed:27559085, PubMed:24726323, PubMed:24726327, PubMed:28919039). Required during angiogenesis, craniofacial and neuronal development by regulating the canonical Wnt signaling together with the LUBAC complex (PubMed:23708998). Acts as a negative regulator of NF-kappa-B by regulating the activity of the LUBAC complex (PubMed:23746843, PubMed:23806334). OTULIN function is mainly restricted to homeostasis of the LUBAC complex: acts by removing 'Met-1'-linked autoubiquitination of the LUBAC complex, thereby preventing inactivation of the LUBAC complex (PubMed:26670046). Acts as a key negative regulator of inflammation by restricting spontaneous inflammation and maintaining immune homeostasis (PubMed:27523608). In myeloid cell, required to prevent unwarranted secretion of cytokines leading to inflammation and autoimmunity by restricting linear polyubiquitin formation (PubMed:27523608). Plays a role in innate immune response by restricting linear polyubiquitin formation on LUBAC complex in response to NOD2 stimulation, probably to limit NOD2-dependent proinflammatory signaling (PubMed:23806334). {ECO:0000269|PubMed:23708998, ECO:0000269|PubMed:23746843, ECO:0000269|PubMed:23806334, ECO:0000269|PubMed:23827681, ECO:0000269|PubMed:24726323, ECO:0000269|PubMed:24726327, ECO:0000269|PubMed:26670046, ECO:0000269|PubMed:26997266, ECO:0000269|PubMed:27523608, ECO:0000269|PubMed:27559085, ECO:0000269|PubMed:28919039}. | 3D-structure;Acetylation;Angiogenesis;Cytoplasm;Disease variant;Hydrolase;Immunity;Innate immunity;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation;Ubl conjugation pathway;Wnt signaling pathway | This gene encodes a member of the peptidase C65 family of ubiquitin isopeptidases. Members of this family remove ubiquitin from proteins. The encoded enzyme specifically recognizes and removes M1(Met1)-linked, or linear, ubiquitin chains from protein substrates. Linear ubiquitin chains are known to regulate the NF-kappa B signaling pathway in the context of immunity and inflammation. Mutations in this gene cause a potentially fatal autoinflammatory syndrome in human patients. [provided by RefSeq, Sep 2016]. | hsa:90268; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; LUBAC complex [GO:0071797]; cysteine-type peptidase activity [GO:0008234]; thiol-dependent deubiquitinase [GO:0004843]; innate immune response [GO:0045087]; negative regulation of inflammatory response [GO:0050728]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; nucleotide-binding oligomerization domain containing 2 signaling pathway [GO:0070431]; protein linear deubiquitination [GO:1990108]; protein ubiquitination [GO:0016567]; regulation of canonical Wnt signaling pathway [GO:0060828]; regulation of tumor necrosis factor-mediated signaling pathway [GO:0010803]; sprouting angiogenesis [GO:0002040]; Wnt signaling pathway [GO:0016055] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23708998_2.4 A and 2.8 A crystal structures of gumby (Fam105b) in the presence of free ubiquitin and linear diubiquitin substrate, respectively 23746843_reveal a previously unannotated human DUB, OTULIN (also known as FAM105B), which is exquisitely specific for Met1 linkages; data suggest that OTULIN regulates Met1-polyUb signaling. 23806334_OTULIN restricts Met1-ubiquitin formation after immune receptor stimulation to prevent unwarranted proinflammatory signaling. 24461064_cylindromatosis (CYLD) and OTULIN/Gumby/Fam105B, directly interact with the N-terminal PUB domain-containing region of HOIP. 24726323_Phosphorylation of OTULIN prevents HOIP binding, whereas unphosphorylated OTULIN is part of the endogenous LUBAC complex. 24726327_HOIP binding to OTULIN is required for the recruitment of OTULIN to the TNF receptor complex. 27523608_OTULIN is critical for restraining life-threatening spontaneous inflammation and maintaining immune homeostasis. 27559085_Biallelic hypomorphic mutations in OTULIN define otulipenia, an early-onset autoinflammatory disease. 28481361_results establish a role for the linear Ubiquitin coat around cytosolic S. Typhimurium as the local NF-kappaB signalling platform and provide insights into the function of OTULIN in NF-kappaB activation during bacterial pathogenesis 31541095_Authors define an additional, non-catalytic function of OTULIN in the regulation of SNX27-retromer assembly and recycling to the cell surface. 31825842_Post-translational Modification of OTULIN Regulates Ubiquitin Dynamics and Cell Death. 32231246_OTULIN protects the liver against cell death, inflammation, fibrosis, and cancer. 32543267_LUBAC and OTULIN regulate autophagy initiation and maturation by mediating the linear ubiquitination and the stabilization of ATG13. 32770022_ABL1-dependent OTULIN phosphorylation promotes genotoxic Wnt/beta-catenin activation to enhance drug resistance in breast cancers. | ENSMUSG00000046034 | Otulin | 499.39525 | 1.1591488 | 0.2130657288 | 0.17465646 | 1.491175e+00 | 2.220343e-01 | 9.998360e-01 | No | Yes | 552.79026 | 73.235726 | 5.285016e+02 | 54.185832 | |
| ENSG00000154814 | 92106 | OXNAD1 | protein_coding | Q96HP4 | NAD;Oxidoreductase;Reference proteome;Signal | hsa:92106; | oxidoreductase activity [GO:0016491] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000021906 | Oxnad1 | 366.87094 | 1.0157683 | 0.0225714270 | 0.18863630 | 1.418005e-02 | 9.052120e-01 | 9.998360e-01 | No | Yes | 356.58553 | 56.272680 | 3.996641e+02 | 48.734539 | |||
| ENSG00000154978 | 81552 | VOPP1 | protein_coding | Q96AW1 | FUNCTION: Increases the transcriptional activity of NFKB1 by facilitating its nuclear translocation, DNA-binding and associated apoptotic response, when overexpressed. {ECO:0000269|PubMed:15735698}. | Alternative splicing;Cytoplasmic vesicle;Membrane;Reference proteome;Signal;Transcription;Transcription regulation;Transmembrane;Transmembrane helix | hsa:81552; | cytoplasmic vesicle membrane [GO:0030659]; endosome [GO:0005768]; integral component of organelle membrane [GO:0031301] | 17515955_ECOP was used as a molecular marker in patients with chronic wounds to guide surgical debridement. 19525979_Results identify ECOP as a protein relevant to the biology of SCC. 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 21519330_overexpression in cancer participates in the control of the intracellular redox state; its loss leads to oxidative cellular injury leading to cell death by the intrinsic apoptotic pathway 23243056_Upregulation of ECOP is associated with glioma. 25398664_VOPP1 is overexpressed in gastric adenocarcinoma, which is involved in promoting cell proliferation and migration and thus might serve as a putative oncogene. 26860460_Candidate gene eQTL showed a trans-acting association between variants of G protein-coupled receptor kinase 5 gene, previously linked to altered BB response, and high expression of VOPP1. 28936884_miR-218 inhibits human lung adenocarcinoma cell migration and invasion via the suppression of Ecop and Robo1 expression 30229837_VOPP1 expression was negatively regulated by microRNA-218. 30285739_The findings emphasize the importance of the sequestration of WWOX by VOPP1 in addition to WWOX loss in breast tumors and define VOPP1 as a novel oncogene promoting breast carcinogenesis by inhibiting the anti-tumoral effect of WWOX. 32083568_The role of vesicular overexpressed in cancer pro-survival protein 1 in hepatocellular carcinoma proliferation. 33845647_RNA polymerase II subunit 3 regulates vesicular, overexpressed in cancer, prosurvival protein 1 expression to promote hepatocellular carcinoma. | ENSMUSG00000037788 | Vopp1 | 2896.72444 | 1.0696443 | 0.0971311445 | 0.10297930 | 8.951155e-01 | 3.440948e-01 | 9.998360e-01 | No | Yes | 3296.64739 | 270.482130 | 3.069196e+03 | 195.906540 | ||
| ENSG00000155256 | 118813 | ZFYVE27 | protein_coding | Q5T4F4 | FUNCTION: Key regulator of RAB11-dependent vesicular trafficking during neurite extension through polarized membrane transport (PubMed:17082457). Promotes axonal elongation and contributes to the establishment of neuronal cell polarity (By similarity). Involved in nerve growth factor-induced neurite formation in VAPA-dependent manner (PubMed:19289470). Contributes to both the formation and stabilization of the tubular ER network (PubMed:24668814). Involved in ER morphogenesis by regulating the sheet-to-tubule balance and possibly the density of tubule interconnections (PubMed:23969831). Acts as an adapter protein and facilitates the interaction of KIF5A with VAPA, VAPB, SURF4, RAB11A, RAB11B and RTN3 and the ZFYVE27-KIF5A complex contributes to the transport of these proteins in neurons. Can induce formation of neurite-like membrane protrusions in non-neuronal cells in a KIF5A/B-dependent manner (PubMed:21976701). {ECO:0000250|UniProtKB:Q3TXX3, ECO:0000269|PubMed:17082457, ECO:0000269|PubMed:19289470, ECO:0000269|PubMed:21976701, ECO:0000269|PubMed:23969831, ECO:0000269|PubMed:24668814}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Disease variant;Endoplasmic reticulum;Endosome;Hereditary spastic paraplegia;Membrane;Metal-binding;Neurodegeneration;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Zinc;Zinc-finger | This gene encodes a protein with several transmembrane domains, a Rab11-binding domain and a lipid-binding FYVE finger domain. The encoded protein appears to promote neurite formation. A mutation in this gene has been reported to be associated with hereditary spastic paraplegia, however the pathogenicity of the mutation, which may simply represent a polymorphism, is unclear. [provided by RefSeq, Mar 2010]. | hsa:118813; | axon [GO:0030424]; cytosol [GO:0005829]; dendrite [GO:0030425]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum tubular network [GO:0071782]; growth cone membrane [GO:0032584]; integral component of endoplasmic reticulum membrane [GO:0030176]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; recycling endosome membrane [GO:0055038]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; protein self-association [GO:0043621]; endoplasmic reticulum tubular network formation [GO:0071787]; neuron projection development [GO:0031175]; neurotrophin TRK receptor signaling pathway [GO:0048011]; positive regulation of axon extension [GO:0045773]; protein localization to plasma membrane [GO:0072659]; vesicle-mediated transport [GO:0016192] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 16826525_Mutation affects neuronal intracellular trafficking in the corticospinal tract, which is consistent with the pathology of hereditary spastic paraplegia. 17082457_protrudin regulates Rab11-dependent membrane recycling to promote the directional membrane trafficking required for neurite formation [protrudin] 18606302_The role of ZFYVE27/protrudin in hereditary spastic paraplegia is reported. 19289470_VAP-A is an important regulator both of the subcellular localization of protrudin and of its ability to stimulate neurite outgrowth. 21976701_Protrudin-KIF5 complex contributes to the vesicular transport in neurons. 22573551_findings indicate that protrudin interacts with spastin and induces axon formation through its N-terminal domain. Moreover, protrudin and spastin may work together to play an indispensable role in motor axon outg 23969831_SPG33 protein protrudin contains hydrophobic, intramembrane hairpin domains, interacts with tubular ER proteins, and functions in ER morphogenesis by regulating the sheet-to-tubule balance and possibly the density of tubule interconnections. 31772151_Protrudin modulates seizure activity through GABAA receptor regulation. 32479595_Protrudin-mediated ER-endosome contact sites promote MT1-MMP exocytosis and cell invasion. 32917905_Protrudin and PDZD8 contribute to neuronal integrity by promoting lipid extraction required for endosome maturation. 33154382_Protrudin functions from the endoplasmic reticulum to support axon regeneration in the adult CNS. | ENSMUSG00000018820 | Zfyve27 | 2374.73507 | 1.0613114 | 0.0858479894 | 0.10573004 | 6.745448e-01 | 4.114716e-01 | 9.998360e-01 | No | Yes | 2635.88143 | 226.437269 | 2.354007e+03 | 157.442403 | |
| ENSG00000156232 | 123720 | WHAMM | protein_coding | Q8TF30 | FUNCTION: Acts as a nucleation-promoting factor (NPF) that stimulates Arp2/3-mediated actin polymerization both at the Golgi apparatus and along tubular membranes. Its activity in membrane tubulation requires F-actin and interaction with microtubules. Proposed to use coordinated actin-nucleating and microtubule-binding activities of distinct WHAMM molecules to drive membrane tubule elongation; when MT-bound can recruit and remodel membrane vesicles but is prevented to activate the Arp2/3 complex. Involved as a regulator of Golgi positioning and morphology. Participates in vesicle transport between the reticulum endoplasmic and the Golgi complex. Required for RhoD-dependent actin reorganization such as in cell adhesion and cell migration. {ECO:0000269|PubMed:18614018, ECO:0000269|PubMed:23027905, ECO:0000269|PubMed:23087206}. | 3D-structure;Actin-binding;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Golgi apparatus;Membrane;Microtubule;Phosphoprotein;Reference proteome;Repeat | This gene encodes a protein that plays a role in actin nucleation, Golgi membrane association and microtubule binding. The encoded protein is a nucleation-promoting factor that regulates the Actin-related protein 2/3 complex. The activated complex initiates growth of new actin filaments by binding to existing actin filaments. The encoded protein also functions in regulation of transport from the endoplasmic reticulum to the Golgi complex and in maintenance of the Golgi complex near the centrosome. Four pseudogenes of this gene are present on the same arm of chromosome 15 as this gene. [provided by RefSeq, Aug 2013]. | hsa:123720; | cytoplasm [GO:0005737]; cytoplasmic vesicle membrane [GO:0030659]; cytosol [GO:0005829]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; Golgi membrane [GO:0000139]; microtubule [GO:0005874]; actin binding [GO:0003779]; Arp2/3 complex binding [GO:0071933]; microtubule binding [GO:0008017]; small GTPase binding [GO:0031267]; actin filament organization [GO:0007015]; actin filament reorganization [GO:0090527]; Arp2/3 complex-mediated actin nucleation [GO:0034314]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; focal adhesion assembly [GO:0048041]; lamellipodium assembly [GO:0030032]; plasma membrane tubulation [GO:0097320]; positive regulation of actin nucleation [GO:0051127] | 23027905_results give rise to a model whereby distinct MT-bound and actin-nucleating populations of WHAMM collaborate during membrane tubulation 23087206_Data show that RhoD binds the actin nucleation-promoting factor WHAMM (WASp homologue associated with actin Golgi membranes and microtubules), as well as the related filamin A-binding protein FILIP1. 26096974_WHAMM directs the Arp2/3 complex to the endoplasmic reticulum for autophagosome biogenesis through an actin comet tail mechanism. 31420534_WHAMM is required for autophagic lysosome reformation. WHAMM works as an actin nucleation promoting factor which promotes assembly of an actin scaffold on the surface of the autolysosome to promote autolysosome tabulation. 33036756_An actin-WHAMM interaction linking SETD2 and autophagy. 33872315_The actin nucleation factors JMY and WHAMM enable a rapid Arp2/3 complex-mediated intrinsic pathway of apoptosis. | ENSMUSG00000045795 | Whamm | 672.48812 | 0.9517252 | -0.0713829484 | 0.13955704 | 2.662084e-01 | 6.058866e-01 | 9.998360e-01 | No | Yes | 643.86939 | 99.165674 | 6.564504e+02 | 78.317085 | |
| ENSG00000156256 | 10600 | USP16 | protein_coding | Q9Y5T5 | FUNCTION: Specifically deubiquitinates 'Lys-120' of histone H2A (H2AK119Ub), a specific tag for epigenetic transcriptional repression, thereby acting as a coactivator. Deubiquitination of histone H2A is a prerequisite for subsequent phosphorylation at 'Ser-11' of histone H3 (H3S10ph), and is required for chromosome segregation when cells enter into mitosis. In resting B- and T-lymphocytes, phosphorylation by AURKB leads to enhance its activity, thereby maintaining transcription in resting lymphocytes. Regulates Hox gene expression via histone H2A deubiquitination. Prefers nucleosomal substrates. Does not deubiquitinate histone H2B. {ECO:0000255|HAMAP-Rule:MF_03062, ECO:0000269|PubMed:10077596, ECO:0000269|PubMed:17914355}. | 3D-structure;Activator;Alternative splicing;Cell cycle;Cell division;Chromatin regulator;Chromosomal rearrangement;Hydrolase;Isopeptide bond;Metal-binding;Mitosis;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Transcription;Transcription regulation;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | This gene encodes a deubiquitinating enzyme that is phosphorylated at the onset of mitosis and then dephosphorylated at the metaphase/anaphase transition. It can deubiquitinate H2A, one of two major ubiquitinated proteins of chromatin, in vitro and a mutant form of the protein was shown to block cell division. Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008]. | hsa:10600; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cysteine-type endopeptidase activity [GO:0004197]; histone binding [GO:0042393]; thiol-dependent deubiquitinase [GO:0004843]; transcription coactivator activity [GO:0003713]; ubiquitin binding [GO:0043130]; zinc ion binding [GO:0008270]; cell division [GO:0051301]; cellular response to DNA damage stimulus [GO:0006974]; chromatin organization [GO:0006325]; histone deubiquitination [GO:0016578]; histone H2A K63-linked deubiquitination [GO:0070537]; mitotic cell cycle [GO:0000278]; mitotic nuclear division [GO:0140014]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of translational elongation [GO:0045901]; protein deubiquitination [GO:0016579]; protein homotetramerization [GO:0051289]; regulation of cell cycle [GO:0051726]; regulation of transcription by RNA polymerase II [GO:0006357]; ubiquitin-dependent protein catabolic process [GO:0006511] | 17512543_Study reports the solution structure of the BUZ domain of Ubp-M, a ubiquitin-specific protease, and its interaction with ubiquitin; the Ubp-M BUZ domain features three zinc-binding sites consisting of 12 residues. 17914355_knockdown of Ubp-M in HeLa cells results in slow cell growth rates owing to defects in the mitotic phase of the cell cycle 18925961_a new cryptic USP16-RUNX1 fusion in chronic myelomonocytic leukemia 24013421_S552P disrupts the interaction between Ubp-M and nuclear export protein CRM1, increasing Ubp-M nuclear retention as cells progress into M phase, revealing a critical function for Ubp-M S552P. 24025767_in human tissues overexpression of USP16 reduces the expansion of normal fibroblasts and postnatal neural progenitors, whereas downregulation of USP16 partially rescues the proliferation defects of Down's syndrome fibroblasts 25305019_Data show that histone H2A deubiquitinase USP16 interacts with E3 ubiquitin-protein ligase HERC2, negatively regulates DNA damage-induced ubiquitin foci formation, and is required for termination of the ubiquitin signal. 26323689_The data unveil a unique mechanism by which Usp16 promotes the localization and maintenance of Plk1 on the kinetochores for proper chromosome alignment. 27586445_USP16 and TTC3 were dysregulated in all affected human cells and two mouse models of Down syndrome. 27633997_USP16 was frequently downregulated. 31135381_data reveal the physiological function of CNA ubiquitination and its deubiquitinase USP16 in peripheral T cells. 31888715_USP16 gene expression is tightly regulated at transcription level. 32129764_USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. 32999190_USP16 Regulates the Stability and Function of LDL receptor by Deubiquitination. 33546726_USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc. | ENSMUSG00000025616 | Usp16 | 458.76800 | 0.7837662 | -0.3515047425 | 0.16290541 | 4.487231e+00 | 3.414895e-02 | 9.998360e-01 | No | Yes | 398.14904 | 68.466980 | 4.880751e+02 | 64.599857 | |
| ENSG00000156531 | 84295 | PHF6 | protein_coding | Q8IWS0 | FUNCTION: Transcriptional regulator that associates with ribosomal RNA promoters and suppresses ribosomal RNA (rRNA) transcription. {ECO:0000269|PubMed:23229552}. | 3D-structure;Acetylation;Alternative splicing;Centromere;Chromosome;DNA-binding;Disease variant;Epilepsy;Hypotrichosis;Isopeptide bond;Kinetochore;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene is a member of the plant homeodomain (PHD)-like finger (PHF) family. It encodes a protein with two PHD-type zinc finger domains, indicating a potential role in transcriptional regulation, that localizes to the nucleolus. Mutations affecting the coding region of this gene or the splicing of the transcript have been associated with Borjeson-Forssman-Lehmann syndrome (BFLS), a disorder characterized by cognitive disability, epilepsy, hypogonadism, hypometabolism, obesity, swelling of subcutaneous tissue of the face, narrow palpebral fissures, and large ears. Alternate splicing results in multiple transcript variants, encoding different isoforms. [provided by RefSeq, Jun 2010]. | hsa:84295; | kinetochore [GO:0000776]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; enzyme binding [GO:0019899]; histone binding [GO:0042393]; histone deacetylase binding [GO:0042826]; metal ion binding [GO:0046872]; phosphoprotein binding [GO:0051219]; ribonucleoprotein complex binding [GO:0043021]; RNA binding [GO:0003723]; scaffold protein binding [GO:0097110]; tubulin binding [GO:0015631]; blastocyst hatching [GO:0001835]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 12415272_A novel, widely expressed zinc-finger (plant homeodomain[PHD]-like finger) gene had 8 different missense and truncation mutations in 7 familial and 2 sporadic cases of BFLS (p. 661). 14714741_...mutations within a novel widely expressed zinc-finger gene (PHF6) have been described in nine families with Borjesson-Forssman-Lehmann syndrome... p. 1208 14714754_The gene, PHF6, implicated in the Borjeson-Forssman-Lehmann syndrome has recently been identified. p. 1295 14756673_A study of 9 families with PHF6 muations revealed that the phenotype is milder and more variable than previously described and evolves with age; seven missense mutations and two truncation mutations were identifed 15241480_By finding a PHF-6 mutation in a family with Borjeson-Forssman-Lehmann syndrome it was speculated that there is a mutational hot spot in the gene. 15466013_novel mutation results in exon skipping and mild Borjeson-Forssman-Lehmann syndrome 15994862_Success of PHF6 screening in males suspected of having Borjeson-Forssman-Lehmann syndrome is markedly increased if there is a positive family history and/or skewed X-inactivation is found in the mother. 19264739_we describe a novel mutation that changes a residue within the first plant homeodomain zinc finger motif of PHF6 in a family with classical features of Borjeson-Forssman-Lehmann syndrome 20228800_these results identify PHF6 as a new X-linked tumor suppressor in T-ALL and point to a strong genetic interaction between PHF6 loss and aberrant expression of TLX transcription factors in the pathogenesis of this disease. 20806366_Borjeson-Forssman-Lehmann syndrome (BFLS) may represent a cancer predisposition syndrome and that mutations of PHF6 contribute to T-cell acute lymphoblastic leukemia. 21030981_PHF6 as a tumor suppressor gene mutated in acute myeloid leukemias (AML) and extend the role of this X-linked tumor suppressor gene in the pathogenesis of hematologic tumors. 21736506_Our data suggest that PHF6 mutation might play a role in tumorigenesis not only of T-cell acute lymphoblatic leukemia, but also acute myelogenous leukemia and hepatocellular carcinoma. 21880637_in T-cell acute lymphoblastic leukemia, PHF6 mutations are a recurrent genetic abnormality associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214. 22720776_PHF6 interacts with the nucleosome remodeling and deacetylation complex and is localized primarily in the nucleoplasm and nucleolus. 22928734_Data suggest that mutations of PHF6 are associated with chronic myeloid leukemia (CML) progression. 23229552_These results reveal that the key function of PHF6 is involved in regulating rRNA synthesis, which may contribute to its roles in cell cycle control, genomic maintenance, and tumor suppression. 24092917_The findings show that de novo mutations in PHF6 in females result in a recognisable phenotype which overlap with Borjeson-Forssman-Lehmann syndrome but also has additional distinct features, thus adding a new facet to this disorder. 24380767_Our report confirms that PHF6 loss in females results in a recognizable phenotype overlapping with Coffin-Siris syndrome and distinct from Borjeson-Forssman-Lehmann syndrome. 24554700_these data support the hypothesis that PHF6 may function as a transcriptional repressor using its ePHD domains binding to the promoter region of its repressed gene 24674452_Recurrent microdeletion was detected in Xq26.3, causing loss of PHF6 expression, a potential tumor suppressor gene, and the miR-424, which is involved in the development of acute myeloid leukemia. 24895337_The PHF6 tumor suppressor gene was targeted in acute lymphoblastic leukemia by microRNA-128-3p. 25099957_Female phenotypes of Borjeson-Forssman-Lehmann syndrome patients with PHF6 mutations 25601084_Our RBBP4-PHF6 complex structure provides insights into the molecular basis of PHF6-NuRD complex interaction and implicates a role for PHF6 in chromatin structure modulation and gene regulation. 25737277_Phf6 is a 'lineage-specific' cancer gene that plays opposing roles in developmentally distinct hematopoietic malignancies. 26561469_Our results suggest that PHF6 may function as an oncogenic factor in several types of cancer. We also hypothesize that PHF6 may also play its role in a tissue-specific manner. Our findings suggest further investigations regarding the exact role of PHF6 in tumor types. 27165002_PHF6 localizes to the sub-nucleolar fibrillar center where it binds to rDNA-coding sequences. PHF6 mediates the overall levels of ribosome biogenesis within a cell. 27479181_PHF6 defects most likely result in their loss of function and have a substantial effect on the evolution into the aggressive types of myeloid neoplasms, associated with other concomitant genetic defects including RUNX1 mutations 27633282_The mutations of the gene encoding plant homeodomain (PHD)-like finger protein 6 (PHF6) contribute to the pathogenesis of the X-linked intellectual disability disorder Borjeson-Forssman-Lehmann syndrome. 27885656_PHF6 mutations occur at a low frequency in pediatric acute myeloid leukemia in both female and male patients 28747286_EZH2 mutations coexisted with mutations of NOTCH1, IL7R, and PHF6 in the two Adult T-cell Acute Lymphoblastic Leukemia patients, and they responded poorly to chemotherapy and experienced difficult clinical histories and inferior outcomes 30530780_PHF6-mutated MPAL occurred in a younger patient cohort compared with DNMT3A-mutated cases. All 3 MPAL cases with both T- and B-lineage differentiation harbored PHF6 mutations. 30567843_PHF6 mutations are early events and drivers of leukemia stem cell activity in the pathogenesis of T-cell acute lymphoblastic leukemia. 30888215_PHF6 was significantly elevated in hepatocellular carcinoma (HCC) tissues and positively correlated with TNM stage, differentiation, and lymph node metastasis. Silencing PHF6 significantly inhibited cell proliferation, colony formation, and migration in HCC cells. Furthermore, silencing PHF6 obviously increased E-cadherin and decreased Vimentin expression. 31562564_Isothermal titration calorimetry and surface plasmon resonance experiments with PHF6 and full-length Tau (FL-Tau), respectively, indicated that Purpurin interacted with PHF6 predominantly via hydrophobic contacts and displayed a dose-dependent complexation with FL-Tau 31782600_PHF6 promotes non-homologous end joining and G2 checkpoint recovery. 32027463_Grandparental genotyping enhances exome variant interpretation. 32398456_Plant homeodomain finger protein 6 in the regulation of normal and malignant hematopoiesis. 32399860_A Novel Missense Variant in PHF6 Gene Causing Borjeson-Forssman-Lehman Syndrome. 32453232_Upregulated Plant Homeodomain Finger Protein 6 Promotes Extracellular Matrix Degradation in Intervertebral Disc Degeneration Based on Microarray Analysis. 32735658_The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. 33149206_Loss of PHF6 leads to aberrant development of human neuron-like cells. 33537816_Notoginsenoside R1 induces DNA damage via PHF6 protein to inhibit cervical carcinoma cell proliferation. 34473233_Nonleukemic T/B mixed phenotype acute leukemia with PHF6 and NOTCH1 mutations. 34520760_Gene body methylation safeguards ribosomal DNA transcription by preventing PHF6-mediated enrichment of repressive histone mark H4K20me3. 34859848_LINC00958/miR-3174/PHF6 axis is responsible for triggering proliferation, migration and invasion of endometrial cancer. | ENSMUSG00000025626 | Phf6 | 801.87247 | 0.9530172 | -0.0694257880 | 0.15053022 | 2.165705e-01 | 6.416655e-01 | 9.998360e-01 | No | Yes | 822.44219 | 173.141066 | 8.792744e+02 | 142.533112 | |
| ENSG00000156650 | 23522 | KAT6B | protein_coding | Q8WYB5 | FUNCTION: Histone acetyltransferase which may be involved in both positive and negative regulation of transcription. Required for RUNX2-dependent transcriptional activation. May be involved in cerebral cortex development. Component of the MOZ/MORF complex which has a histone H3 acetyltransferase activity. {ECO:0000269|PubMed:10497217, ECO:0000269|PubMed:11965546, ECO:0000269|PubMed:16387653}. | 3D-structure;Acetylation;Activator;Acyltransferase;Alternative splicing;Chromatin regulator;Chromosomal rearrangement;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a histone acetyltransferase and component of the MOZ/MORF protein complex. In addition to its acetyltransferase activity, the encoded protein has transcriptional activation activity in its N-terminal end and transcriptional repression activity in its C-terminal end. This protein is necessary for RUNX2-dependent transcriptional activation and could be involved in brain development. Mutations have been found in patients with genitopatellar syndrome. A translocation of this gene and the CREBBP gene results in acute myeloid leukemias. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2012]. | hsa:23522; | MOZ/MORF histone acetyltransferase complex [GO:0070776]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; acetyltransferase activity [GO:0016407]; DNA binding [GO:0003677]; histone acetyltransferase activity [GO:0004402]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coregulator activity [GO:0003712]; histone acetylation [GO:0016573]; histone H3 acetylation [GO:0043966]; negative regulation of transcription, DNA-templated [GO:0045892]; nucleosome assembly [GO:0006334]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription, DNA-templated [GO:0006355] | 15271374_role in regulating interleukin-5 expression 16385451_Observational study of gene-disease association. (HuGE Navigator) 17460191_MORF4 has a role in cellular aging, and MRG15 associates with both histone deacetylases and histone acetyl transferase complexes [review] 18794358_These findings indicate that BRPF proteins play a key role in assembling and activating MOZ/MORF acetyltransferase complexes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21804188_Data show that H3 acetylation by Myst4 is important for neural, craniofacial, and skeletal morphogenesis, mainly through its ability to specifically regulating the MAPK signaling pathway. 21880731_Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). 22077973_Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome 22265014_By exome sequencing, we found de novo heterozygous truncating mutations in KAT6B. 22265017_we identified de novo mutations of KAT6B in five individuals with Genitopatellar syndrome. 22715153_Propose that haploinsufficiency or loss of a function mediated by the C-terminal domain causes the common features, whereas gain-of-function activities would explain the features unique to genitopatellar syndrome. Review. 23063713_These data suggest that the tandem of plant homeodomain 1/2 fingers play a role in MOZ and MORF histone acetyltransferase association with histon H3 regions enriched in acetylated marks. 23436491_Our results confirm the implication of KAT6B mutations in typical SBBYS syndrome and emphasize the importance of genotype-phenotype correlations at the KAT6B locus. 24150941_MORF double PHD finger binds to Histone H3 in a manner that is enhanced by acetylation of the lysine residues K9 and K14 24294372_KAT5 and KAT6B regulate prostate cancer cell growth through PI3K-AKT signaling. 24444492_The study showed that markers rs11001178 (MYST4) showed weak associations. 24458743_Similar to those observed in other genetic disorders resulting from KAT6B mutations. 24698832_The relationship between MYO1C and KAT6B suggests that the two are interacting in chromatin remodelling for gene expression in human masseter muscle. This is the nuclear myosin1 (NM1) function of MYO1C. 25424711_KAT6B sequence variants are being reported in the Say-Barber-Biesecker type of blepharophimosis mental retardation syndromes (SBBS) and in the more severe genitopatellar syndrome. 25840828_This study shown MYST4 to be the risk factor for attention deficit/hyperactivity disorder. 26208904_Homozygous deletions of KAT6B and the loss of its mRNA occur in SCLC cell lines and primary tumors. KAT6B loss enhances cancer growth. Restoration induces tumor suppressor-like features involving a new type of histone H3 Lys23 acetyltransferase activity. 26334766_Kat6 c.3147G>A splice site mutation causes Say-Barber-Biesecker/Young-Simpson syndrome by inducing aberrant splicing. 27185879_Studies show that misregulation of MOZ/MORF results in tumorigenesis and developmental disorders. Results also provide evidence that these 2 proteins play important role in regulating cell proliferation and stem cell maintenance. [review] 27447113_chronic inflammation compromises unfolded protein response function through MORF-mediated-PERK transcription. 27452416_Our findings support that phenotypes associated with typical KAT6B disease-causing variants should be referred to as 'KAT6B spectrum disorders' or 'KAT6B related disorders', rather than their current SBBYSS and GTPTS classification 27880066_With ptosis, hypotonia, and developmental delay as the main diagnostic features of our patient, the effect of histone acetyltransferase-encoding KAT6B gene haploinsufficiency was suspected to have a significant role in determining the phenotype. 28286003_Study identified the double plant homeodomain finger (DPF) of the lysine acetyltransferase MORF as a reader of global histone H3K14 acylation. 28696035_KAT6B mutation is associated with Craniosynostosis. 29226580_The present report highlights the pivotal role of clinical genetics in avoiding clear-cut genotype-phenotype categories in syndromic forms of intellectual disability. In addition, it further supports the evidence that a continuum exists within the clinical spectrum of KAT6B-associated disorders 30569622_The clinical and genetic characterization of these patients could contribute to the understanding of the KAT6B-related disorders. 30921092_A novel pathogenic frameshift variant of KAT6B identified by clinical exome sequencing in a newborn with the Say-Barber-Biesecker-Young-Simpson syndrome. 31624313_Data indicate that the acetyltransferase function of the catalytic histone acetyltransferase KAT6B (MORF) subunit is positively regulated by the DPF domain of MORF (MORFDPF). 32424177_Further delineation of the clinical spectrum of KAT6B disorders and allelic series of pathogenic variants. 32448279_High methylation of lysine acetyltransferase 6B is associated with the Cobb angle in patients with congenital scoliosis. 32722658_CircKIAA0907 Retards Cell Growth, Cell Cycle, and Autophagy of Gastric Cancer In Vitro and Inhibits Tumorigenesis In Vivo via the miR-452-5p/KAT6B Axis. 33136874_Novel Approach of Mandibular Distraction to Avoid Tracheostomy in KAT6B-related Gene Disorders. 34464167_Low Expression of KAT6B May Affect Prognosis in Hepatocellular Carcinoma. | ENSMUSG00000021767 | Kat6b | 607.42693 | 0.9621550 | -0.0556587330 | 0.16686057 | 1.101918e-01 | 7.399258e-01 | 9.998360e-01 | No | Yes | 653.49849 | 118.134636 | 6.698287e+02 | 93.424127 | |
| ENSG00000156787 | 93594 | TBC1D31 | protein_coding | Q96DN5 | Alternative splicing;Coiled coil;Reference proteome;Repeat;WD repeat | hsa:93594; | centrosome [GO:0005813] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000022364 | Tbc1d31 | 242.56835 | 1.1545014 | 0.2072699753 | 0.20474181 | 1.001975e+00 | 3.168332e-01 | 9.998360e-01 | No | Yes | 255.16060 | 50.161465 | 2.357054e+02 | 35.759946 | |||
| ENSG00000156795 | 55093 | NTAQ1 | protein_coding | Q96HA8 | FUNCTION: Mediates the side-chain deamidation of N-terminal glutamine residues to glutamate, an important step in N-end rule pathway of protein degradation. Conversion of the resulting N-terminal glutamine to glutamate renders the protein susceptible to arginylation, polyubiquitination and degradation as specified by the N-end rule. Does not act on substrates with internal or C-terminal glutamine and does not act on non-glutamine residues in any position. Does not deaminate acetylated N-terminal glutamine. With the exception of proline, all tested second-position residues on substrate peptides do not greatly influence the activity. In contrast, a proline at position 2, virtually abolishes deamidation of N-terminal glutamine. {ECO:0000250|UniProtKB:Q80WB5}. | 3D-structure;Alternative splicing;Cytoplasm;Hydrolase;Nucleus;Reference proteome | hsa:55093; | cytosol [GO:0005829]; nucleus [GO:0005634]; protein-N-terminal asparagine amidohydrolase activity [GO:0008418]; protein-N-terminal glutamine amidohydrolase activity [GO:0070773]; cellular protein modification process [GO:0006464] | 25356641_the peptide backbone recognition patch of hNtaq1 forms nonspecific interactions with N-terminal peptides of substrate proteins | ENSMUSG00000022359 | Wdyhv1 | 188.75989 | 1.1272279 | 0.1727792123 | 0.23570640 | 5.314636e-01 | 4.659922e-01 | 9.998360e-01 | No | Yes | 179.41150 | 31.682762 | 1.755622e+02 | 24.097407 | ||
| ENSG00000157036 | 9941 | EXOG | protein_coding | Q9Y2C4 | FUNCTION: Endo/exonuclease with nicking activity towards supercoiled DNA, a preference for single-stranded DNA and 5'-3' exonuclease activity. {ECO:0000269|PubMed:18187503}. | 3D-structure;Alternative splicing;Endonuclease;Hydrolase;Membrane;Metal-binding;Mitochondrion;Mitochondrion inner membrane;Nuclease;Reference proteome;Transit peptide | This gene encodes an endo/exonuclease with 5'-3' exonuclease activity. The encoded enzyme catalyzes the hydrolysis of ester linkages at the 5' end of a nucleic acid chain. This enzyme is localized to the mitochondria and may play a role in programmed cell death. Alternatively spliced transcript variants have been described. A pseudogene exists on chromosome 18. [provided by RefSeq, Feb 2009]. | hsa:9941; | mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; 5'-3' exonuclease activity [GO:0008409]; endonuclease activity [GO:0004519]; endoribonuclease activity [GO:0004521]; metal ion binding [GO:0046872]; nucleic acid binding [GO:0003676]; single-stranded DNA endodeoxyribonuclease activity [GO:0000014]; apoptotic DNA fragmentation [GO:0006309] | 17415550_ENDOGL1 is a candidate disease-susceptibility gene for type 2 diabetes in a Japanese population 17415550_Observational study of gene-disease association. (HuGE Navigator) 18187503_Human endonuclease G-like-1 is a dimeric mitochondrial enzyme that displays 5'-3' exonuclease activity and further differs from EndoG in substrate specificity 20877624_Observational study of gene-disease association. (HuGE Navigator) 21768646_EXOG depletion induces persistent SSBs in the mtDNA, enhances ROS levels, and causes apoptosis in normal cells but not in mt genome-deficient rho0 cells. 23986585_These data indicate that HSV-1 UL12.5 deploys cellular proteins, including ENDOG and EXOG, to destroy mtDNA and contribute to a growing body of literature highlighting roles for ENDOG and EXOG in mtDNA maintenance. 28466855_Data suggest a role of exo/endonuclease G nuclease (hEXOG) in pathway for mitochondrial DNA repair. 30949702_ExoG has a role as a unique 5'-exonuclease to mediate the flap-independent RNA primer removal process during mtDNA replication to maintain mitochondrial genome integrity | ENSMUSG00000042787 | Exog | 627.37237 | 1.1447021 | 0.1949721587 | 0.15319408 | 1.599199e+00 | 2.060167e-01 | 9.998360e-01 | No | Yes | 526.45780 | 89.431287 | 5.441470e+02 | 71.348665 | |
| ENSG00000157193 | 7804 | LRP8 | protein_coding | Q14114 | FUNCTION: Cell surface receptor for Reelin (RELN) and apolipoprotein E (apoE)-containing ligands. LRP8 participates in transmitting the extracellular Reelin signal to intracellular signaling processes, by binding to DAB1 on its cytoplasmic tail. Reelin acts via both the VLDL receptor (VLDLR) and LRP8 to regulate DAB1 tyrosine phosphorylation and microtubule function in neurons. LRP8 has higher affinity for Reelin than VLDLR. LRP8 is thus a key component of the Reelin pathway which governs neuronal layering of the forebrain during embryonic brain development. Binds the endoplasmic reticulum resident receptor-associated protein (RAP). Binds dimers of beta 2-glycoprotein I and may be involved in the suppression of platelet aggregation in the vasculature. Highly expressed in the initial segment of the epididymis, where it affects the functional expression of clusterin and phospholipid hydroperoxide glutathione peroxidase (PHGPx), two proteins required for sperm maturation. May also function as an endocytic receptor. Not required for endocytic uptake of SEPP1 in the kidney which is mediated by LRP2 (By similarity). Together with its ligand, apolipoprotein E (apoE), may indirectly play a role in the suppression of the innate immune response by controlling the survival of myeloid-derived suppressor cells (By similarity). {ECO:0000250|UniProtKB:Q924X6, ECO:0000269|PubMed:12807892, ECO:0000269|PubMed:12899622, ECO:0000269|PubMed:12950167}. | 3D-structure;Alternative splicing;Calcium;Cell membrane;Disulfide bond;EGF-like domain;Endocytosis;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix;Ubl conjugation | This gene encodes a member of the low density lipoprotein receptor (LDLR) family. Low density lipoprotein receptors are cell surface proteins that play roles in both signal transduction and receptor-mediated endocytosis of specific ligands for lysosomal degradation. The encoded protein plays a critical role in the migration of neurons during development by mediating Reelin signaling, and also functions as a receptor for the cholesterol transport protein apolipoprotein E. Expression of this gene may be a marker for major depressive disorder. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jun 2011]. | hsa:7804; | axon [GO:0030424]; caveola [GO:0005901]; cell surface [GO:0009986]; dendrite [GO:0030425]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; microtubule associated complex [GO:0005875]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; receptor complex [GO:0043235]; amyloid-beta binding [GO:0001540]; apolipoprotein binding [GO:0034185]; calcium ion binding [GO:0005509]; calcium-dependent protein binding [GO:0048306]; cargo receptor activity [GO:0038024]; high-density lipoprotein particle binding [GO:0008035]; kinesin binding [GO:0019894]; low-density lipoprotein particle receptor activity [GO:0005041]; protein N-terminus binding [GO:0047485]; reelin receptor activity [GO:0038025]; transmembrane signaling receptor activity [GO:0004888]; very-low-density lipoprotein particle receptor activity [GO:0030229]; ammon gyrus development [GO:0021541]; cellular response to cholesterol [GO:0071397]; cellular response to growth factor stimulus [GO:0071363]; chemical synaptic transmission [GO:0007268]; cytokine-mediated signaling pathway [GO:0019221]; dendrite morphogenesis [GO:0048813]; endocytosis [GO:0006897]; layer formation in cerebral cortex [GO:0021819]; lipid metabolic process [GO:0006629]; modulation of chemical synaptic transmission [GO:0050804]; positive regulation of CREB transcription factor activity [GO:0032793]; positive regulation of dendrite development [GO:1900006]; positive regulation of dendritic spine morphogenesis [GO:0061003]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of protein tyrosine kinase activity [GO:0061098]; proteolysis [GO:0006508]; reelin-mediated signaling pathway [GO:0038026]; regulation of apoptotic process [GO:0042981]; regulation of innate immune response [GO:0045088]; response to xenobiotic stimulus [GO:0009410]; retinoid metabolic process [GO:0001523]; signal transduction [GO:0007165]; ventral spinal cord development [GO:0021517] | 12399018_Observational study of gene-disease association. (HuGE Navigator) 12621059_transmembrane domain and PXXP motif excludes it from carrying out clathrin-mediated endocytosis 12871934_Sequential proteolytic processing of murine ApoER2 results in the release of its intracellular domain by the protease gamma-secretase. The prior cleavage of its extracellular domain is determined by the glycosylation state of the receptor. 12950167_Data show that the apolipoprotein E receptor 2 binding domain of apolipoprotein E is in the 1-165 amino terminal region, whereas the carboxy terminal 230-299 region of apoE is required for efficient initial association with phospholipids. 15950758_The effects of apoE on receptor proteolysis were mediated by the ligand binding domain of the receptor. We suggest that signaling promoted by these receptors depends in part on these regulated proteolytic events. 16034672_Complete molecular structure examined by nuclear magnetic resonance. 16332682_ApoEr2 can form a multiprotein complex with NMDA receptor subunits and PSD95 16481437_Reelin signals by binding to two transmembrane receptors, apolipoprotein E receptor 2 (Apoer2) and very-low-density lipoprotein receptor. 16642433_In conclusion, results from the two independent samples of black women provide consistent evidence that SNP rs2297660 in LRP8 is associated with fetal growth. 16642433_Observational study of gene-disease association. (HuGE Navigator) 16951405_The effect of Dab1 on APP and apoEr2 processing in transfected cells and primary neurons is reported. 17470198_The presence of three splice variants of ApoER2 on platelets was confirmed by immuno-blotting, with ApoER2Delta4-5 being the most abundantly expressed splice variant. 17614163_However, this polymorphism increased the risk of AD conferred by the MAPK8IP1 G allele. 17614163_Observational study of gene-disease association. (HuGE Navigator) 17847002_A nonconservative substitution, R952Q, in LRP8 was significantly associated with susceptibility to premature CAD and/or MI by use of both population-based and family-based designs. 17847002_Observational study of gene-disease association. (HuGE Navigator) 18039658_the activity of PCSK9 and its binding affinity on VLDLR and ApoER2 does not depend on the presence of LDLR. 18089558_Fyn, due in part to its effects on Dab1, regulates the phosphorylation, trafficking, and processing of APP and apoEr2. 18489431_protein C and APC may directly promote cell signaling in other cells by binding to ApoER2 and/or GPIbalpha 18592168_Observational study of gene-disease association. (HuGE Navigator) 18592168_Study showed no evidence for association of genetic variants in the LRP8 gene with familial and sporadic myocardial infarction. 19116273_ligation of ApoER2 by APC signals via Dab1 phosphorylation and subsequent activation of PI3K and Akt and inactivation of GSK3beta, thereby contributing to APC's beneficial effects on cells. 19439088_Our data suggest that LRP8 R952Q variant may have an additive effect to APOE epsilon2/epsilon3/epsilon4 genotype in determining ApoE concentrations and risk of MI in an Italian population 19661487_apolipoprotein E receptor 2 (ApoER2, LRP8), a member of the LDL receptor family, is a platelet receptor for FXI. 19720620_ApoEr2 regulates cell movement, and both X11alpha and Reelin enhance this effect. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19948739_Differential functions of ApoER2 and very low density lipoprotein receptor in Reelin signaling depend on differential sorting of the receptors. 20208369_There were seven polymorphisms in apoE receptor 2 in Japanese sporadic Alzheimer disease patients, but no association of these polymorphisms with Alzheimers. 20331378_Observational study of gene-disease association. (HuGE Navigator) 20493228_the expression of ApoER2 may serve as a trait marker for major depressive disorder. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21157031_Deficient sLRP-amyloid-beta binding might precede and correlate later in disease with an increase in the tau/Abeta42 CSF ratio and global cognitive decline in mild cognitive impairment individuals converting into Alzheimer's disease. 21316997_LRP8 gene polymorphisms influence plasma cholesterol levels as well as size and composition of LDL particles 21347244_ApoEr2 plays important roles in structure and function of CNS synapses and dendritic spines 21601501_In later stages of cerebral cortical development, ApoER2 is expressed earlier than VLDLR in migrating neurons. 22170052_activation of VLDLR and apoER2 by reelin and apoE induces ABCA1 expression and cholesterol efflux via a Dab1-PI3K-PKCzeta-Sp1 signaling cascade. 22404453_genetic variant R952Q of LRP8 is associated with increased plasma TG levels in patients who are overweight and have premature CAD/MI and history of smoking. 22419519_Variation in genes encoding proteins at the gateway of Reelin signaling: ligands RELN and APOE, their common receptors APOER2 and VLDLR, and adaptor DAB1, was examined. 22589174_results identify LRP8 as a novel positive factor of canonical Wnt signaling pathway 22889673_In a Chinese unrelated Han population, variants within the LRP8 gene do not convey the risk of developing Parkinson's disease. 23524007_TCCGC haplotype at the 3'-terminal block of the LRP8 gene confers a protective role in the development of familial and early-onset coronary artery disease and/or myocardial infarction. 23739027_By incorporating the information from bioinformatics and RNA expression analyses, we identified at least two of the most promising risk genes for alcohol dependence: APOER2 and UBAP2 24076391_ectopic expression of HIC1 in U2OS and MDA-MB-231 cell lines decreases expression of the ApoER2 and VLDLR genes, encoding two canonical tyrosine kinase receptors for Reelin. 24344333_Data suggest that PS1/gamma-secretase-dependent processing of the reelin receptor ApoER2 inhibits reelin expression and may regulate its signaling. 24844606_results of this study demonstrated the presence of reelin, its receptors VLDLR and ApoER2 as well as Dab1 in the ENS and might indicate a novel role of the reelin system in regulating neuronal plasticity and pre-synaptic functions in the ENS. 24867879_A novel TACGC risk haplotype in the LRP8 gene that is present in patients with CAD and MI but not in normal controls. 26637325_LRP8 is a risk gene for psychosis. 26800564_ApoER2 contributes cooperatively with endothelial cell protein C receptor and protease activated receptor 1 to APC-initiated endothelial antiapoptotic and barrier protective signaling. 26817215_two SNPs (rs3737984 and rs2297660) in ApoER2 gene had significant association with dyslipidemia in Thai ethnic 26902204_These results reveal an association between ApoER2 isoform expression and Alzheimer's disease. 27853278_ABGL4, LRP8 and PCSK9 polymorphisms and gene interactions increase cardiometabolic risk. 28255385_The presence of reelin was elevated in junctional areas as in dysplastic nevi. VLDLR presented positive values in 16 cases (16/ 32) and ApoER2 was weak positive in 7 cases. 28446613_This study elucidates that the contracted-open conformation of ligand-bound ApoER2 at neutral pH resembles the contracted-closed conformation of ligand-unbound low-density lipoprotein receptor at acidic pH in a manner suggestive of being primed for ligand release even prior to internalization. 28495490_Further evidence for the association between LRP8 and schizophrenia. 29032149_Results show that none of the SNPs were significantly associated with myocardial infarction (MI). Interestingly, haplotype based association analysis showed TG and CG of rs10788952 and rs7546246 significantly associated with MI and in particular, haplotype TG was positively correlated with the risk of MI, as this increased the LDL and total cholesterol level in MI patients in south Indians. 29442527_ApoER2-CTF levels are decreased in AD brain at advanced Braak stages and after Abeta treatment. 30227220_Data indicate low-density lipoprotein receptor-related protein 8 (LRP8) as a therapeutic target for triple-negative breast cancer (TNBC) as inhibition of LRP8 can attenuate Wnt/beta-catenin signaling to suppress breast cancer stem cells (BCSCs). 30552869_CSF-ApoER2 fragments as a read-out of reelin signaling: Distinct patterns in sporadic and autosomal-dominant Alzheimer disease 30575334_Our findings suggest that new approaches targeting LRP8 may constitute promising treatments for hormone-negative breast cancers, those overexpressing HER2 and TNBCs. 32312520_Genome-scale CRISPR activation screening identifies a role of LRP8 in Sorafenib resistance in Hepatocellular carcinoma. 33054597_LRP8 activates STAT3 to induce PD-L1 expression in osteosarcoma. 33079740_LRP8 (rs5177) and CEP85L (rs11756438) are contributed to schizophrenia susceptibility in Iranian population. 33779882_miR-30b-5p inhibits cancer progression and enhances cisplatin sensitivity in lung cancer through targeting LRP8. | ENSMUSG00000028613 | Lrp8 | 610.58653 | 1.0765600 | 0.1064287497 | 0.17747463 | 3.646675e-01 | 5.459254e-01 | 9.998360e-01 | No | Yes | 627.52768 | 70.193219 | 5.697606e+02 | 49.381385 | |
| ENSG00000157224 | 9069 | CLDN12 | protein_coding | P56749 | FUNCTION: Plays a major role in tight junction-specific obliteration of the intercellular space, through calcium-independent cell-adhesion activity. {ECO:0000250}. | Cell junction;Cell membrane;Membrane;Phosphoprotein;Reference proteome;Tight junction;Transmembrane;Transmembrane helix | This gene encodes a member of the claudin family. Claudins are integral membrane proteins and components of tight junction strands. Tight junction strands serve as a physical barrier to prevent solutes and water from passing freely through the paracellular space between epithelial or endothelial cell sheets, and also play critical roles in maintaining cell polarity and signal transductions. This gene is expressed in the inner ear and bladder epithelium, and it is over-expressed in colorectal carcinomas. This protein and claudin 2 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Multiple alternatively spliced transcript variants encoding the same protein have been found.[provided by RefSeq, Sep 2011]. | hsa:9069; | bicellular tight junction [GO:0005923]; integral component of membrane [GO:0016021]; lateral plasma membrane [GO:0016328]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules [GO:0016338]; maintenance of blood-brain barrier [GO:0035633] | 17047970_Differential expression of genes encoding claudins in colorectal cancer suggests that these tight junction proteins may be associated to and involved in tumorigenesis. 18287530_These findings strongly suggest that claudin-2- and/or claudin-12-based tight junctions form paracellular Ca(2+) channels in intestinal epithelia, and they highlight a novel mechanism behind vitamin D-dependent calcium homeostasis. 25727011_Knockdown of claudin-3, claudin-4, and claudin-12, but not claudin-1, increased breast cancer MCF-7 cell migration with maximal effects observed in claudin-12 siRNA-transfected cells. 26926102_CLDN 12 expression could be clinically useful for predicting the survival of the estrogen receptor (ER)-negative subgroup of patients with breast cancer. 30339745_We found that COPII cargo CLDN12 is important for Hepatitis C Virus entry. 33917356_Reduced Claudin-12 Expression Predicts Poor Prognosis in Cervical Cancer. 33922921_Tight Junction Protein Claudin-12 Is Involved in Cell Migration during Metastasis. | ENSMUSG00000046798 | Cldn12 | 235.50983 | 1.2265611 | 0.2946190994 | 0.64930481 | 2.299497e-01 | 6.315611e-01 | 9.998360e-01 | No | Yes | 311.38697 | 73.171074 | 2.537148e+02 | 46.092246 | |
| ENSG00000157379 | 115817 | DHRS1 | protein_coding | Q96LJ7 | FUNCTION: NADPH-dependent oxidoreductase which catalyzes the reduction of steroids (estrone, androstene-3,17-dione and cortisone) as well as prostaglandin E1, isatin and xenobiotics in vitro (PubMed:30031147). May have a role in steroid and/or xenobiotic metabolism (PubMed:30031147). {ECO:0000269|PubMed:30031147}. | 3D-structure;Acetylation;Endoplasmic reticulum;Methylation;NAD;Oxidoreductase;Reference proteome | This gene encodes a member of the short-chain dehydrogenases/reductases (SDR) family. The encoded enzyme contains a conserved catalytic domain and likely functions as an oxidoreductase. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Nov 2008]. | hsa:115817; | endoplasmic reticulum [GO:0005783]; carbonyl reductase (NADPH) activity [GO:0004090]; oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [GO:0016616] | 12153138_novel human SDR-type dehydrogenase/reductase gene named Dehydrogenase/reductase (SDR family) member 1 (DHRS1) 20877624_Observational study of gene-disease association. (HuGE Navigator) 25052469_Data suggest that members of short chain dehydrogenase/reductase superfamily (human DHRS1; E coli galE) exhibit conserved residues and tertiary structure in active site domain that are essential for biocatalysis. 30031147_DHRS1 was proven to be an NADPH-dependent reductase that is able to catalyse the in vitro reductive conversion of some steroids (estrone, androstene-3,17-dione and cortisone), as well as other endogenous substances and xenobiotics. 34498498_Decreased DHRS1 expression is a novel predictor of poor survival in patients with hepatocellular carcinoma. | ENSMUSG00000002332 | Dhrs1 | 805.50143 | 0.9929722 | -0.0101747306 | 0.14214859 | 5.317844e-03 | 9.418670e-01 | 9.998360e-01 | No | Yes | 807.48042 | 86.786067 | 7.719602e+02 | 64.841794 | |
| ENSG00000157613 | 90993 | CREB3L1 | protein_coding | Q96BA8 | FUNCTION: Transcription factor involved in unfolded protein response (UPR). Binds the DNA consensus sequence 5'-GTGXGCXGC-3' (PubMed:21767813). In the absence of endoplasmic reticulum (ER) stress, inserted into ER membranes, with N-terminal DNA-binding and transcription activation domains oriented toward the cytosolic face of the membrane. In response to ER stress, transported to the Golgi, where it is cleaved in a site-specific manner by resident proteases S1P/MBTPS1 and S2P/MBTPS2. The released N-terminal cytosolic domain is translocated to the nucleus to effect transcription of specific target genes. Plays a critical role in bone formation through the transcription of COL1A1, and possibly COL1A2, and the secretion of bone matrix proteins. Directly binds to the UPR element (UPRE)-like sequence in an osteoblast-specific COL1A1 promoter region and induces its transcription. Does not regulate COL1A1 in other tissues, such as skin (By similarity). Required to protect astrocytes from ER stress-induced cell death. In astrocytes, binds to the cAMP response element (CRE) of the BiP/HSPA5 promoter and participate in its transcriptional activation (By similarity). Required for TGFB1 to activate genes involved in the assembly of collagen extracellular matrix (PubMed:25310401). {ECO:0000250|UniProtKB:Q9Z125, ECO:0000269|PubMed:12054625, ECO:0000269|PubMed:21767813, ECO:0000269|PubMed:25310401}.; FUNCTION: (Microbial infection) May play a role in limiting virus spread by inhibiting proliferation of virus-infected cells. Upon infection with diverse DNA and RNA viruses, inhibits cell-cycle progression by binding to promoters and activating transcription of genes encoding cell-cycle inhibitors, such as p21/CDKN1A (PubMed:21767813). {ECO:0000269|PubMed:21767813}. | Activator;Alternative splicing;DNA-binding;Developmental protein;Endoplasmic reticulum;Glycoprotein;Host-virus interaction;Isopeptide bond;Membrane;Nucleus;Osteogenesis imperfecta;Reference proteome;Signal-anchor;Transcription;Transcription regulation;Transmembrane;Transmembrane helix;Ubl conjugation;Unfolded protein response | The protein encoded by this gene is normally found in the membrane of the endoplasmic reticulum (ER). However, upon stress to the ER, the encoded protein is cleaved and the released cytoplasmic transcription factor domain translocates to the nucleus. There it activates the transcription of target genes by binding to box-B elements. [provided by RefSeq, Jun 2013]. | hsa:90993; | chromatin [GO:0000785]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; nucleus [GO:0005634]; cAMP response element binding [GO:0035497]; chromatin binding [GO:0003682]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; sequence-specific double-stranded DNA binding [GO:1990837]; SMAD binding [GO:0046332]; transcription cis-regulatory region binding [GO:0000976]; endoplasmic reticulum unfolded protein response [GO:0030968]; extracellular matrix constituent secretion [GO:0070278]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236]; negative regulation of fibroblast growth factor receptor signaling pathway [GO:0040037]; negative regulation of gene expression [GO:0010629]; negative regulation of sprouting angiogenesis [GO:1903671]; negative regulation of transcription, DNA-templated [GO:0045892]; osteoblast differentiation [GO:0001649]; positive regulation of collagen biosynthetic process [GO:0032967]; positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress [GO:1990440]; regulation of transcription by RNA polymerase II [GO:0006357] | 12054625_a transcriptional activator of CREB/ATF family with a transmembrane domain 17721195_Presence of FUS/CREB3L2 and FUS/CREB3L1 in low-grade fibromyxoid sarcoma and sclerosing epithelioid fibrosarcoma suggests these neoplasms may be related. 21041443_The human Creb3L1 can activate SPCG transcription in a heterologous system(Drosophila embryos), which suggests a general and direct role for this family of bZip transcription factors in mediating high-level secretory capacity. 21536545_FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. 21767813_CREB3L1 may play an important role in limiting virus spread by inhibiting proliferation of virus-infected cells. 21767813_Upon infection with diverse DNA and RNA viruses, CREB3L1 was proteolytically cleaved, allowing its NH(2) terminus to enter the nucleus and induce multiple genes encoding inhibitors of the cell cycle to block cell proliferation of infected cells. 21987447_Rapid amplification of cDNA ends revealed exon 6 of the cAMP-responsive element binding protein 3-like 1 gene (CREB3L1) fused in-frame to the EWSR1 exon 11. 22705851_OASIS is notably unstable proteins that are easily degraded via the ubiquitin-proteasome pathway under normal conditions. 23256041_CREB3L1 expression may be a useful biomarker in identifying cancer cells sensitive to doxorubicin. 23335989_OASIS is important for the ER stress response and maintenance of some extracellular matrix proteins in human glioma cells. 23383089_a novel regulatory mechanism for VEGFA transcription by OASIS in human retinal pigment epithelial cells 23588368_We report 2 cases of Low-grade fibromyxoid sarcoma serendipitously found to harbor a novel alternative EWSR1-CREB3L1 gene fusion. 23955342_Fbxw7 controls osteogenesis and chondrogenesis by targeting OASIS and BBF2H7, respectively, for degradation. 24126059_CREB3L1 plays an important role in suppressing tumorigenesis and that loss of expression is required for the development of a metastatic phenotype. 24242870_Temporally regulates the differentiation from neural precursor cells into astrocytes [review] 24441665_EWSR1-CREB3L1 gene fusions are predominant over FUS and CREB3L2 rearrangements in pure sclerosing epithelioid fibrosarcoma. 24896634_Case Report: low-grade fibromyxoid sarcoma of the kidney found to harbor the EWSR1-CREB3L1 gene fusion. 25310401_Cleavage of CREB3L1 releases its NH2-terminal domain from membranes, allowing it to enter the nucleus where it binds to Smad4 to activate transcription of genes encoding proteins required for assembly of collagen-containing extracellular matrix. 25353281_Case Reports: genetically confirmed primary renal sclerosing epithelioid fibrosarcoma with EWSR1-CREB3L1 gene fusion. 25625847_CREB3L1 mRNA expression is downregulated in human bladder cancer.CREB3L1 is epigenetically silenced in human bladder cancer facilitating tumor cell spreading and migration in vitro. 26110425_CREB3L1 was expressed in 19% of RCC, which is generally resistant to doxorubicin, but in 70% of diffuse large B-cell lymphoma that is sensitive to doxorubicin. 26810754_Our data further strengthens the role for CREB3L1 as a metastasis suppressor in breast cancer and demonstrates that epigenetic silencing is a major regulator of the loss of CREB3L1 expression 26917262_The results suggest that CREB3L1 is required for decidualization in mice and humans and may be linked to the pathogenesis of endometriosis in a progesterone-dependent manner. 27121396_These findings indicate that the miR-146a-CREB3L1-FGFBP1 signaling axis plays an important role in the regulation of angiogenesis in human umbilical vein endothelial cells. 27499293_Ceramide inverts the membrane orientation of TMS4SF20, creating a form of TM4SF20 that stimulates the cleavage of CREB3L1. 28750683_Identification of novel prostate cancer drivers, ERF, CREB3L1, and POU2F2, using RegNetDriver, a framework for integration of genetic and epigenetic alterations with tissue-specific regulatory network. 28817112_ConclusionThis report confirms that CREB3L1 is an OI-related gene and suggests the pathogenic mechanism of CREB3L1-associated OI involves the altered regulation of proteins involved in cellular secretion. 29093023_Our findings support a model in which CREB3L1 acts as a downstream effector of TSH to regulate the expression of cargo proteins, and simultaneously increases the synthesis of transport factors and the expansion of the Golgi to synchronize the rise in cargo load with the amplified capacity of the secretory pathway 29531016_There was a relationship between the expression levels of both proteins and survival time. CREB3L1 and PTN expression levels serve as biomarkers with utility in grading gliomas. Absence of CREB3L1 and presence of PTN in brain glioma cells correlate with survival time of the glioma patients. 30103710_These results suggest that CREB3L1 expression level may be used as a biomarker to identify TNBC patients who are more likely to benefit from doxorubicin-based chemotherapy. 30657919_Here, we presenta Turkish family, in which molecular analysis of the proband revealeda previously unreported homozygous missense variant(c.911C>T,p.(Ala304Val))of the CREB3L1 31207160_Study reports a novel homozygous CREB3L1 mutation in a large Indonesian family with osteogenesis imperfecta (OI); the homozygous affected members have survived to adulthood and they present a more severe phenotype than previously reported, expanding the clinical spectrum of OI for this gene. 32234057_Mutations in COL1A1/A2 and CREB3L1 are associated with oligodontia in osteogenesis imperfecta. 33150680_Transcription factor old astrocyte specifically induced substance is a novel regulator of kidney fibrosis. 34990868_A group of sclerosing epithelioid fibrosarcomas with low-level amplified EWSR1-CREB3L1 fusion gene in children. | ENSMUSG00000027230 | Creb3l1 | 72.34418 | 1.2554523 | 0.3282072338 | 0.35626902 | 8.830273e-01 | 3.473739e-01 | 9.998360e-01 | No | Yes | 89.25423 | 19.402524 | 6.768571e+01 | 12.061298 | |
| ENSG00000157653 | 257169 | C9orf43 | protein_coding | Q8TAL5 | Reference proteome | hsa:257169; | 20583170_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000058046 | 4933430I17Rik | 10.88904 | 0.7822965 | -0.3542126027 | 0.92204715 | 1.234888e-01 | 7.252809e-01 | 9.998360e-01 | No | Yes | 10.04860 | 4.216002 | 1.481501e+01 | 5.013567 | ||||
| ENSG00000157764 | 673 | BRAF | protein_coding | P15056 | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Allosteric enzyme;Cardiomyopathy;Cell membrane;Chromosomal rearrangement;Cytoplasm;Deafness;Direct protein sequencing;Disease variant;Ectodermal dysplasia;Isopeptide bond;Kinase;Membrane;Mental retardation;Metal-binding;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein belonging to the RAF family of serine/threonine protein kinases. This protein plays a role in regulating the MAP kinase/ERK signaling pathway, which affects cell division, differentiation, and secretion. Mutations in this gene, most commonly the V600E mutation, are the most frequently identified cancer-causing mutations in melanoma, and have been identified in various other cancers as well, including non-Hodgkin lymphoma, colorectal cancer, thyroid carcinoma, non-small cell lung carcinoma, hairy cell leukemia and adenocarcinoma of lung. Mutations in this gene are also associated with cardiofaciocutaneous, Noonan, and Costello syndromes, which exhibit overlapping phenotypes. A pseudogene of this gene has been identified on the X chromosome. [provided by RefSeq, Aug 2017]. | hsa:673; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; identical protein binding [GO:0042802]; MAP kinase kinase activity [GO:0004708]; MAP kinase kinase kinase activity [GO:0004709]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; scaffold protein binding [GO:0097110]; animal organ morphogenesis [GO:0009887]; cellular response to calcium ion [GO:0071277]; epidermal growth factor receptor signaling pathway [GO:0007173]; establishment of protein localization to membrane [GO:0090150]; MAPK cascade [GO:0000165]; negative regulation of apoptotic process [GO:0043066]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of gene expression [GO:0010628]; positive regulation of glucose transmembrane transport [GO:0010828]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; protein phosphorylation [GO:0006468]; trehalose metabolism in response to stress [GO:0070413] | 8621729_MEK1 interacts with B-Raf. 12068308_somatic missense mutations in 66% of malignant melanomas and at lower frequency in a wide range of human cancers 12198537_BRAF mutations in colorectal cancers occur only in tumours that do not carry mutations in a RAS gene known as KRAS, and BRAF mutation is linked to the proficiency of these tumours in repairing mismatched bases in DNA 12447372_High frequency of BRAF mutations in nevi 12619120_The V599E BRAF mutation appears to be a somatic mutation associated with melanoma development and/or progression in a proportion of affected individuals. 12644542_results demonstrate that the mutational status of BRAF and KRAS is distinctly different among histologic types of ovarian serous carcinoma, occurring most frequently in invasive micropapillary serous carcinomas and its precursors, serous borderline tumors 12670889_High prevalence of BRAF mutations in thyroid cancer is genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. 12697856_activating BRAF mutations may be an important event in the development of papillary thyroid cancer 12753285_cAMP activates ERK and increases proliferation of autosomal dominant polycystic kindey epithelial cells through the sequential phosphorylation of PKA, B-Raf and MAPK in a pathway separate from the classical receptor tyrosine kinase cascade 12778069_gene is mutated in skin melanoma, but not in uveal melanomas 12810628_13 germline BRAF variants, 4 of which were silent mutations in coding regions & 9 nucleotide substitutions in introns, were found in melanoma patients and melanoma family, but none appeared statistically likely to be a melanoma susceptibility gene. 12821662_B-raf is involved in adhesion-independent ERK1/2 signaling in melanocytes 12824225_Data suggest that BRAF T1796A activating mutation is not common in primary uveal melanoma. 12855697_B-Raf has a role in extracellular signal-regulated kinase (ERK) signaling in T cells and prevents antigen-presenting cell-induced anergy 12879021_BRAF has a role in in squamous cell carcinoma of the head and neck through uncommon mutations 12881714_The BRAF(V599E) mutation appears to be an alternative event to RET/PTC rearrangement rather than to RAS mutations, which are rare in PTC. BRAF(V599E) may represent an alternative pathway to oncogenic MAPK activation in PTCs without RET/PTC activation. 12893203_Mucinous ovarian cancers without a KRAS mutation have not sustained alternative activation of this signaling pathway through mutation of the BRAF oncogene. 12917419_3 cell lines derived from human choroidal melanoma express B-Raf containing the V599E mutation and showed a 10-fold increase in endogenous B-RafV599E kinase activity and a constitutive activation of the MEK/ERK pathway that is independent of Ras 12931219_Mutations are not detectable in plasma cell leukemia and multiple myeloma. 12970315_mutation of BRAF gene could be a potentially useful marker of prognosis of patients with advanced thyroid cancers 14501284_Our findings of a high frequency of BRAF mutations at codon 599 in benign melanocytic lesions of the skin indicate that this mutation is not sufficient by itself for malignant transformation. 14507635_Both BRAF and FBXW7 mutations functionally activate kinase effectors important in pancreatic cancer and extend potential options for therapeutic targeting of kinases in treatment of phenotypically distinct pancreatic adenocarcinoma subsets. 14513361_BRAF mutations, which are present in a variety of other human cancers, do not seem to be involved in gastric cancer development 14522897_Uceal melanomas arise independent of oncogenic BRAF and NRAS mutations. 14534542_BRAF mutations were seen in stomach neoplasms. 14602780_BRAF mutations are restricted to papillary carcinomas and poorly differentiated and anaplastic carcinomas arising from papillary carcinomas 14612909_BRAF is occasionally mutated in NHL, and BRAF mutation may contribute to tumor development in some NHLs 14618633_None of the cases of gastric cancer showed braf mutations 14639609_Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas 14668801_Missense mutation is marker of colonic but not gastric cancer. 14688025_Mutations were found in exon 15 in colorectal adenocarcinoma. 14691295_Our data indicate that BRAF gene mutations are rare to absent events in uveal melanoma of humans. 14695152_NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression 14695993_BRAF mutations are associated with proximal colon tumors with mismatch repair deficiency and MLH1 hypermethylation. 14719068_New enriched PCR-RFLP assay for detecting mutations of BRAF codon 599 mutation in pleural mesotheliomas. 14722037_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 14724583_RAS or BRAF mutations are detected in about 32% of all Barrett's adenocarcinomas; the disruption of the Raf/MEK/ERK (MAPK) kinase pathway is a frequent but also early event in the development of Barrett's adenocarcinoma 14734469_BRAF mutations are frequently present in sporadic colorectal cancer with methylated hMLH1 14961576_Mutations in BRAF gene is associated with malignant melanomas 14966563_These studies identify isoprenylcysteine carboxyl methyltransferase as a potential target for reducing the growth of K-Ras- and B-Raf-induced malignancies. 15001635_The lack or low prevalence of BRAF mutation in other thyroid neoplasms is consistent with the notion that other previously defined genetic alterations on the same signaling pathway are sufficient to cause tumorigenesis in most thyroid neoplasms. 15009714_possible cooperation between BRAF activation and PTEN loss in melanoma development. 15009715_mutations in the BRAF gene and to some extent in the N-ras gene represent early somatic events that occur in melanocytic nevi 15014028_BRAF mutation may be acquired during development of metastasis but is not a significant factor for primary melanoma development and disease outcome. 15077125_ovarian serous cystadenomas do not contain mutations in either BRAF or KRAS genes 15104286_These results suggest that the BRAF mutation is unlikely to be involved in gastric carcinogenesis. 15126572_BRAF(V599E) is more common genetic alteration found to date in adult sporadic papillary thyroid carcinomas (PTCs). It is unique for this thyroid cancer histotype, and it might drive the development of PTCs of classic papillary subtype. 15140228_The finding of tandem mutations in thin melanomas makes it more likely that they arise as a simultaneous rather than sequential event. 15145515_Radiation-induced tumors have a low prevalence of BRAF point mutations and high prevalence of RET/PTC rearrangements 15150271_B-Raf kinase activity regulation by tuberin and Rheb is mammalian target of rapamycin (mTOR)-independent 15161700_mucosal melanomas of the head and neck do not frequently harbor an activating mutation of BRAF 15179189_in contrast to cutaneous melanoma, BRAF does not appear to be involved in the pathogenesis of uveal melanoma 15186612_BRAF mutations are rather rare in solitary cold adenomas and adenomatous nodules and do not explain the molecular etiology of ras mutation-negative cold thyroid nodules. 15191558_activation of this gene may be one of the early events in the pathogenesis of some melanomas. 15263001_B-Raf and ERK are activated by cyclic AMP after calcium restriction 15273715_mutated in papillary thyroid cancer. 15277467_In this study, this BRAF mutation was demonstrated in some conjunctival melanoma tissue samples, suggesting that some conjunctival melanomas may share biological features in common with cutaneous melanoma. 15313890_Data suggest that SPRY2, an inhibitor of ERK signaling, may be bypassed in melanoma cells either by down-regulation of its expression in WT BRAF cells, or by the presence of the BRAF mutation. 15330192_Mutations within the BRAF gene are useful markers for the differential diagnosis between Spitz nevus and malignant melanoma. 15331929_we found 19 cases (38%) to harbor somatic B-raf exon 15 mutations. 15339934_Data provide evidence that B-Raf is a positive regulator of T cell receptor-mediated sustained ERK activation, which is required for NFAT activation and the full production of IL-2. 15373778_BRAF(V599E) mutation is seven times higher in lesions with structural changes and 13 times higher in growing lesions as compared with lesions without changes 15488754_REVIEW: our understanding of B-RAF as an oncogene and of its role in cancer 15489648_Mutations of BRAF or KRAS oncogenes are early events in the serrated polyp neoplasia pathway. CpG island methylation plays a role in serrated polyp progression to colorectal carcinoma. 15538400_mutated in childhood acute lymphoblastic leukemia. 15577314_BRAF mutations are associated with conjunctival neoplasms 15630448_AKAP9-BRAF fusion was preferentially found in radiation-induced papillary carcinomas developing after a short latency, whereas BRAF point mutations were absent in this group 15632082_Data suggest that Rit is involved in a novel pathway of neuronal development and regeneration by coupling specific trophic factor signals to sustained activation of the B-Raf/ERK and p38 MAP kinase cascades. 15653554_a novel Ras-independent ERK1/2 activation system in which p110gamma/Raf-1/MEK1/2 and PKA/B-Raf/MEK1/2 cooperate to activate ERK1/2. 15702478_We found mutations in p53, K-ras, and BRAF genes in 35%, 30%, and 4% of tumors, respectively, and observed a minimal or no co-presence of these gene alterations. 15705790_KSHV-infected cell lines expressed higher levels of B-Raf and VEGF-A; B-Raf-induced VEGF-A expression was demonstrated to be sufficient to enhance tubule formation in endothelial cells 15710605_autoinhibition was negatively regulated by acidic substitutions at phosphorylation sites within the activation loop 15765445_Mutations in the BRAF protooncogene (V599E)may be an alternative pathway of tumorigenesis of familial colorectal cancer. 15782118_BRAF mutations proved to be absent in tumors from hereditary nonpolyposis colorectal cancer syndrome (HNPCC) families with germline mutations in the MMR genes MLH1 and MSH2. 15791479_The data of this study suggest that activating mutations of B-RAF are not a frequent event in gliomas; nevertheless, when present they are associated with high-grade malignant lesions. 15791648_B-raf mutations surrounding Thr439 found in human cancers are unlikely to contribute to increased oncogenic properties of B-raf 15824163_Observational study of gene-disease association. (HuGE Navigator) 15842051_These results suggest that BRAF mutations do not have a role in tumorigenesis of neuroendocrine gastroenteropancreatic tumors. 15880523_Anaplastic thyroid carcinomas which are derived from papillary carcinomas are due to BRAF and p53 mutations 15904951_Observational study of gene-disease association. (HuGE Navigator) 15935100_B-raf V599E and V599K oncogenic mutations are likely to affect melanocyte-specific pathways controlling proliferation and differentiation 15968271_Observational study of gene-disease association. (HuGE Navigator) 15968271_The increasing frequency of BRAF mutations as a function of age could help account for the well documented but poorly understood observation that age is a relevant prognostic indicator for patients with papillary thyroid carcinoma. 15980887_BRAF mutation occurs later in thyroid tumor progression and is restricted mainly to papillary thyroid carcinoma and anaplastic thyroid carcinoma 15994075_Observational study of gene-disease association. (HuGE Navigator) 15998781_Role of BRAF mutation in facilitating metastasis and progression of papillary thyroid cancer in lymph nodes. 16007166_determination of mutation specific gene expression profiles in papillary thyroid carcinoma 16007203_Single-cell clones with efficient knockdown of (V 600 E)B-RAF could be propagated in the presence of basic fibroblast growth factor but underwent apoptosis or senescence-like growth arrest upon withdrawal of this growth factor 16015629_Observational study of gene-disease association. (HuGE Navigator) 16024606_Observational study of gene-disease association. (HuGE Navigator) 16079850_sustained BRAF(V600E) expression in human melanocytes induces cell cycle arrest, which is accompanied by the induction of both p16(INK4a) and senescence-associated acidic beta-galactosidase (SA-beta-Gal) activity, a commonly used senescence marker 16096377_BRAF mutation in melanoma is most likely to occur prior to the development of metastatic disease 16098042_Although BRAF and NRAS mutations are likely to be important for the initiation and maintenance of some melanomas, other factors might be more significant for proliferation and prognosis in subgroups of aggressive melanoma 16098042_Observational study of gene-disease association. (HuGE Navigator) 16123397_The results showed that conjunctival nevi, similar to skin nevi, have a high frequency of oncogenic BRAF mutations. 16129781_These data suggest that MITF is an anti-proliferation factor that is down-regulated by B-RAF signaling and that this is a crucial event for the progression of melanomas that harbor oncogenic B-RAF. 16143028_Observational study of gene-disease association. (HuGE Navigator) 16144912_Mutations of the BRAF gene are partly involved in the malignant transformation of the endometrium. 16144912_Observational study of gene-disease association. (HuGE Navigator) 16172610_selective reduction in catalytic activity and expression of B-Raf but not Raf-1 suggest that B-Raf may be playing an important role in altered ERK signaling in brain of suicide subjects, and thus in the pathophysiology of suicide 16174717_In patients with papillary thyroid cancer, BRAF mutation is associated with poorer clinicopathological outcomes and independently predicts recurrence. 16174717_Observational study of gene-disease association. (HuGE Navigator) 16179867_As the BRAF oncogene is frequently found to be mutated in human cutaneous melanomas, it may constitute a risk factor for melanoma formation within CMN and DMN. 16179870_The oncogenic B-raf mutations V599E and V599K, as early events in melanocyte transformation, persist throughout metastasis with important prognostic implications. 16181240_Observational study of gene-disease association. (HuGE Navigator) 16181547_Observational study of gene-disease association. (HuGE Navigator) 16199894_copy number gain may represent another mechanism of BRAF activation in thyroid tumors 16268813_Observational study of gene-disease association. (HuGE Navigator) 16354196_Observational study of gene-disease association. (HuGE Navigator) 16354196_The estimated proportion of attributable risk of melanoma due to variants in BRAF is 1.6%, but the burden of disease associated with this variant is greater than that associated with the major melanoma locus (CDKN2A) which has a risk of 0.2%. 16354586_Mutation and elevated expression of BRAF is associated with the development of testicular germ cell tumors 16361694_The authors have developed and run a high-throughput screen to find inhibitors of V600E BRAF using an enzyme cascade assay in which oncogenic BRAF activates MEK1, which in turn activates ERK2, which then phosphorylates the transcription factor ELK1. 16364920_Data suggest that B-RAF activates C-RAF through a mechanism involving 14-3-3 mediated heterooligomerization and C-RAF transphosphorylation. 16371460_V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. 16373964_activating mutations of PDGFR-alpha, c-kit and B-RAF are absent in gliosarcomas 16376942_Observational study of gene-disease association. (HuGE Navigator) 16376942_V599E BRAF mutation was uncommon in Japanese lung cancer. 16382052_aberrant B-Raf activity in angiomyolipomas leads to abnormal cellular differentiation and migration [review] 16397024_Observational study of genotype prevalence. (HuGE Navigator) 16413100_The most frequent B-RAF gene alterations are not involved in prostate carcinogenesis 16417232_BRAF mutation does not seem to be sufficient to produce MAPK activation in melanocytic nevi. 16424035_gain-of-function BRAF signaling is strongly associated with in vivo tumorigenicity 16439621_findings demonstrate that heterogeneous de novo missense mutations in three genes within the mitogen-activated protein kinase pathway, BRAF, MEK1 and MEK2 cause cardio-facio-cutaneous syndrome 16452469_wild-type B-Raf-mediated ERK1/2 activation plays a major role in proliferation and transformation of uveal melanocytes; Raf-1 is not involved in this activation 16452550_Observational study of gene-disease association. (HuGE Navigator) 16462768_NRAS and BRAF activating mutations can coexist in the same melanoma, but are mutually exclusive at the single-cell level 16474404_Cardio-facio-cutaneous (CFC) syndrome involves dysregulation of the RAS-RAF-ERK pathway. 16487015_Observational study of gene-disease association. (HuGE Navigator) 16537381_Merlin and MLK3 can interact in situ and merlin can disrupt the interactions between B-Raf and Raf-1 or those between MLK3 and either B-Raf or Raf-1. 16547495_Melanoma cells require either B-RAF or phosphoinositide-3 kinase activation for protection from anoikis. 16601293_BRAF V600E is associated with a high risk of recurrence and less differentiated papillary thyroid carcinoma due to the impairment of Na+/I- targeting to the membrane 16601293_Observational study of gene-disease association. (HuGE Navigator) 16618717_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16687919_Observational study of genotype prevalence. (HuGE Navigator) 16691193_Observational study of gene-disease association. (HuGE Navigator) 16691193_UV light is not necessarily required for the acquisition of the BRAF(V600E) mutation, and non-mutagenic effects of UV light to melanocytes may be more important in the nevogenesis 16721785_BRAF mutations are as uncommon as KRAS mutations in prostate adenocarcinoma 16728573_B-RAF (V600E) was confirmed to be associated with the papillary growth pattern, but not with poorer differentiated papillary thyroid carcinoma variants. 16728573_Observational study of gene-disease association. (HuGE Navigator) 16773193_among 23 melanomas located at body sites with chronic UV exposure, only a single tumour harboured the B-raf V599E mutation which was a significantly lower frequency in comparison to melanomas from sun-protected body sites 16786134_a BRAFT1799A mutation may have a role in poor differentiation of thyroid carcinoma 16799476_A subset of Spitz nevi, some with atypical histologic features, possess BRAF mutations. The BRAF mutational status does not separate all Spitz nevi from spitzoid melanomas and non-Spitz types of melanocytic proliferations, contrary to previous reports. 16803888_Rheb has a central role in the regulation of the Ras/B-Raf/C-Raf/MEK signaling network 16804544_CpG island methylator phenotype-positive colorectal tumors represent a distinct subset, encompassing almost all cases of tumors with BRAF mutation 16809487_findings show that MC1R variants are strongly associated with BRAF mutations in non-chronic sun-induced damage melanomas; in this subtype, risk for melanoma associated with MC1R is due to increase in risk of developing melanomas with BRAF mutations 16845322_BRAF mutation is associated with melanoma and melanocytic nevi. 16858395_Thus, we propose that the hitherto unidentified function of the B-Raf amino-terminal region is to mediate calcium-dependent activation of B-Raf and the following MEK activation, which may occur in the absence of Ras activation. 16858683_Aberrant methylation and hence silencing of TIMP3, SLC5A8, DAPK and RARbeta2, in association with BRAF mutation, may be an important step in PTC tumorigenesis and progression. 16879389_BRAF mutation was frequent in hyperplastic polyps (67%) and sessile serrated adenomas (81%). 16912199_B-RAF has been identified as the most mutated gene in invasive cells and therefore an attractive therapeutic target in melanoma. 16918136_BRAF mutations are associated with colorectal cancers 16918957_Observational study of gene-disease association. (HuGE Navigator) 16924241_Expression of p27Kip1 in melanoma is regulated by B-RAF at the mRNA level and via B-RAF control of Cks1/Skp2-mediated proteolysis. 16932278_Single nucleotide polymorphism found exclusively in papillary thyroid carcinoma. 16937524_BRAF, K-ras and BAT26 are expressed in colorectal polyps and stool 16937524_BRAF, K-ras and BAT26 are expressed in colorectal polyps and stool [BAT26] 16946010_Braf mutations in thyroid tumorigenesis. 16953233_Concomitant KRAS and BRAF mutations increased along progression of MSS colorectal cancer, suggesting that activation of both genes is likely to harbour a synergistic effect 16959844_BRAFV600E activates not only MAPK but also NF-kappaB signaling pathway in human thyroid cancer cells, leading to an acquisition of apoptotic resistance and promotion of invasion. 16960555_Expression of active mutants of B-Raf induces fibronectin. 16964379_Extracellular signal-regulated kinase-3 (ERK3/MAPK6) is highly expressed in response to BRAF signaling. 16973828_Observational study of gene-disease association. (HuGE Navigator) 16987295_BRAF T1976A mutation is present at high frequency in benign naevi such as Spitz and Reed. 17001349_data support a model in which mutational activation of BRAF in human melanomas contributes to constitutive induction of NF-kappaB activity and to increased survival of melanoma cells 17018604_Normally, BRAF alone is responsible for signaling to MEK. However, when RAS is mutated in melanoma, melanocytes switch their signaling from BRAF to CRAF. 17044028_Activating BRAF mutation is associated with papillary thyroid carcinoma 17060774_BRAF mutation remained a significant prognostic factor for lymph node metastasis (odds ratio = 10.8, 95% confidence interval, 3.5-34.0, P < 0.0001). 17074813_phosphorylation on both S365 and S429 participate in the differential regulation of B-Raf isoforms through distinct mechanisms 17097223_data provide evidence that oncogenic properties of BRAF contribute to the tumorigenesis of intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC), but at a lower frequency than KRAS 17119056_BRAF-V600E mutations are mainly involved in colorectal cancer families characterized by an increased risk of other common malignancies 17119447_Association with preexisting nevi and pronounced infiltration of lymphocytes was significantly higher in BRAF mutated melanoma tumours 17148775_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17148775_Previously identified associations between smoking and colon cancer, whether microsatellite unstable or stable, appear to be explained by the association of smoking with BRAF mutation. 17159915_BRAF(T1799A) mutation is associated with a lower rate of tumor proliferation. 17159915_Observational study of gene-disease association. (HuGE Navigator) 17170014_RASSF1A methylation was observed in a high frequency in endometrioid endometrial carcinoma whereas K-ras and B-raf mutations were observed in a low frequency 17179987_The role for BRAF activation in thyroid cancer development and establishing the potential therapeutic efficacy of BRAF-targeted agents in patients with thyroid cancerwill be reviewed. 17186541_BRAF mutation is associated with thyroid carcinogenesis 17186541_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17195912_Observational study of gene-disease association. (HuGE Navigator) 17195912_there is a subgroup of colorectal carcinomas which develop via the microsatellite instability pathway that carry an alteration of the BRAF gene 17199737_Absence of association between BRAF mutation and activation of MAPK pathway in papillary thyroid carcinoma suggests the presence of mechanisms that downregulate MAPK activation. 17227125_Copy gain of PDGFB occurs in a subset of tumors showing no evidence of mutated BRAF or rearranged ret, suggesting that copy gain of PDGFB may underlie the increased expression of platelet-derived growth factor described recently in the literature. 17270239_Observational study of gene-disease association. (HuGE Navigator) 17297294_characterization of the T1799-1801del and A1799-1816ins BRAF mutations in papillary thyroid cancer; the two new mutations resulted in constitutive activation of the BRAF kinase and caused NIH3T3 cell transformation 17302867_Overexpression of B-Raf mRNA and protein may be a feature of nonfunctioning pituitary adenomas, highlighting overactivity of the Ras-B-Raf-MAP kinase pathway in these tumors. 17309670_BRAF gene plays a 'gatekeeper' role but does not act as a predisposition gene in the development of low-grade ovarian serous carcinomas 17309670_Observational study of gene-disease association. (HuGE Navigator) 17312306_Observational study of genetic testing. (HuGE Navigator) 17315191_BRAFV600E represents a detectable marker in the plasma/serum from melanoma patients for monitoring but not diagnostic purposes 17318013_B-RAF mutations are a rare event in pituitary tumorigenesis. 17355635_The aim of this study was to identify the effect that BRAF oncogene has on post-transcriptional regulation in papillary thyroid carcinoma by using microRNA analysis. 17360030_findings show that RASSF1A hypermethylation and KRAS mutations and BRAF mutations are inversely correlated and play an important role in the development of cervical adenocarcinomas 17366577_mutational analysis of KRAS, BRAF, and MAP2K1/2 in 56 patients with CFC syndrome; comparison of the genotype-phenotype correlation of CFC with that of Costello syndrome suggest a significant clinical overlap but not genotype overlap. 17387744_BRAF(V600E) mutation is identified in a subset of cutaneous metastases from papillary thyroid carcinomas 17393356_Observational study of gene-disease association. (HuGE Navigator) 17393356_data suggest that BRAF mutations might be present less frequently than KRAS mutations in Greek patients with colorectal carcinomas 17440063_finding of a strong association between BRAF mutations and serrated histology in hyperplastic aberrant crypt foci supports the idea that these lesions are an early, sentinel, or a potentially initiating step on the serrated pathway to colorectal carcinoma 17453004_BRAF V600E mutation was occasionally observed in anaplastic carcinomas with papillary carcinoma. 17453358_Observational study of gene-disease association. (HuGE Navigator) 17454879_MSI is rare in UC-related neoplasia as well as non-neoplastic lesions, and does not contribute to the development of dysplasia. 17464312_prevalence of BRAF mutation and RET/PTC were determined in diffuse sclerosing variant of papillary thyroid carcinoma; none of the cases showed a BRAF mutation 17483702_Molecular diagnosis and careful observations should be considered in children with Cardio-facio-cutaneous syndrome because they have germline mutations in BRAF and might develop malignancy. 17487277_Observational study of gene-disease association. (HuGE Navigator) 17487504_c-kit expression is not alternative to BRAF and/or KRAS activation. 17488796_BRAF V600E mutation in PTCs is associated with reduced expression of key genes involved in iodine metabolism 17507627_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17507627_data indicate that both early-life UV exposure and nevus propensity contribute to occurrence of BRAF+ melanoma, whereas nevus propensity and later-life sun exposure influence the occurrence of NRAS+ melanoma 17516929_analysis of a BRAF mutation-associated gene expression signature in melanoma 17518771_low rate of RAS-RAF mutations (2/22, 9.1%) observed in Spitz melanocytic nevi suggests that these lesions harbor as yet undetected activating mutations in other components of the RAS-RAF-MEK-ERK-MAPK pathway 17520704_Meta-analysis of gene-disease association. (HuGE Navigator) 17520704_frequency of the BRAF mutation and the associations between BRAF mutation and clinicopathologic parameters in papillary thyroid carcinoma were evaluated by meta-analysis 17525723_T1790A BRAF mutation (L597Q) in childhood acute lymphoblastic leukemia is a functional oncogene 17535994_The heterogeneous distribution of BRAF mutations suggests that discrete tumor foci in multifocal PTC may occur as independent tumors. 17542667_Presence of BRAF V600E in very early stages of papillary thyroid carcinoma. 17548320_influence of B-RAF-specific RNA interference on the proliferation and apoptosis of gastric cancer BGC823 cell line 17566669_Observational study of gene-disease association. (HuGE Navigator) 17566669_We conclude that screening for BRAF 15 exon mutation is an efficient tool in the diagnostic strategy for HNPCC 17635919_In contrast to C-RAF that requires farnesylated H-Ras, cytosolic B-RAF associates effectively and with significantly higher affinity with both farnesylated and nonfarnesylated H-Ras. 17663506_KLF6 and p53 mutations are involved in the development of nonpolypoid colorectal carcinoma, whereas K-ras and B-raf mutations are not 17671688_PPARbeta/delta has a role in growth of RAF-induced lung adenomas 17685465_BRAF V600E mutation in papillary carcinoma of the thyroid may facilitate tumor cell growth and progression once seeded in the lymph nodes. 17693984_Observational study of genotype prevalence, gene-disease association, and genetic testing. (HuGE Navigator) 17693984_There was no coexistence of BRAF (V600E) mutation in papillary thyroid carcinoma. 17696195_data showed differences in gene expression between nevi with and without the V600E BRAF mutation. Moreover, nevi with mutations showed over-expression of genes involved in melanocytic senescence and cell cycle inhibition 17699719_RNA interference and pharmacologic approaches were used to assess the role of B-Raf activation in the growth of human melanomas and additionally determined if a similar role for mutant B-Raf is seen for colorectal carcinoma cell lines. 17704260_5 unreported mutations (T241P, Q262R, G464R, E501V, N581K) were found in cardio-facio-cutaneous syndrome. A hotspot in exon 6 at Q257 was found. 17714762_diffuse expression of wild-type and/or mutant B-Raf may be involved in the tumorigenic process 17717450_BRAF V600E mutation is primarily present in conventional papillary thyroid cancer; it is associated with an aggressive tumor phenotype and higher risk of recurrent and persistent disease in patients with conventional papillary thyroid cancer 17717450_Observational study of gene-disease association. (HuGE Navigator) 17721188_Develompment of malignant strumo ovarii with papillary thyroid carcinoma features is associated with BRAF mutations. 17727338_BRAF(V600E) mutation detected on fine-needle aspiration biopsy specimens, more than RET/PTC rearrangements, is highly specific for papillary thyroid carcinoma. 17785355_BRAF V600E mutation is associated with high-risk papillary thyroid carcinoma 17785355_Observational study of gene-disease association. (HuGE Navigator) 17786355_BRAFV600E mutations were found in 41.2% of the papillary thyroid carcinomas 17854396_Papillary thyroid cancers with no 131I uptake had a high frequency of BRAF mutations. 17878251_MEK inhibition is cytostatic in papillary thyroid cancer and anaplastic thyroid cancer cells bearing a BRAF mutation 17911174_effects of a MEK inhibitor, CI-1040, on thyroid cancer cells, some of which, particularly cell proliferation and tumor growth, seemed to be BRAF mutation or RAS mutation selective 17914558_BRAF mutation is associated as early as the hyperplastic polyp stage followed by microsatellite instability at the carcinoma stage 17924122_Examined associations between BRAF mutations, morphology, and apoptosis in early colorectal cancer. 17940185_BRAF mutation represents a novel indicator of the progression and aggressiveness of papillary thyroid cancer (Review) 17942568_BRAF interacts with PLCepsilon1 in nephrotic syndrome type 3. Both proteins are coexpressed and colocalize in developing and mature glomerular podocytes. 17962436_In this small study, the T1799A BRAF mutation was identified in almost half of the iris melanoma tissues samples examined. This finding suggests that there may be genetic as well as clinical differences between iris and posterior uvea | ENSMUSG00000002413 | Braf | 336.22556 | 0.9262902 | -0.1104638539 | 0.20523730 | 2.827464e-01 | 5.949067e-01 | 9.998360e-01 | No | Yes | 409.65078 | 74.659485 | 4.515193e+02 | 63.476444 | |
| ENSG00000157985 | 116987 | AGAP1 | protein_coding | Q9UPQ3 | FUNCTION: GTPase-activating protein for ARF1 and, to a lesser extent, ARF5. Directly and specifically regulates the adapter protein 3 (AP-3)-dependent trafficking of proteins in the endosomal-lysosomal system. {ECO:0000269|PubMed:12640130}. | 3D-structure;ANK repeat;Acetylation;Alternative splicing;Cytoplasm;GTP-binding;GTPase activation;Metal-binding;Nucleotide-binding;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport;Zinc;Zinc-finger | This gene encodes a member of an ADP-ribosylation factor GTPase-activating protein family involved in membrane trafficking and cytoskeleton dynamics. This gene functions as a direct regulator of the adaptor-related protein complex 3 on endosomes. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2011]. | hsa:116987; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; GTP binding [GO:0005525]; GTPase activator activity [GO:0005096]; GTPase activity [GO:0003924]; metal ion binding [GO:0046872]; phospholipid binding [GO:0005543]; protein transport [GO:0015031] | 12388557_overexpression inhibited formation of platelet-derived growth factor-induced ruffles and also induced loss of actin stress fibers 15381706_AGAP1 is a novel binding partner of nitric oxide-sensitive guanylyl cyclase 15892143_CENTG2 is an intriguing candidate gene that merits further scrutiny for its role in autism. 15892143_Observational study of gene-disease association. (HuGE Navigator) 16079295_The AGAP1 has been found to associate with the adaptor protein complex AP-3 and regulate the function of AP-3 endosomes. 18003747_AGAP1 is unique among the AZAPs in its specificity for Arf1, and this activity is dependent on its NH(2)-terminal GTPase-like domain. AGAP1 induce reciprocal activation of Arf6. 22453919_GTP-binding protein-like domain of AGAP1 is protein binding site that allosterically regulates ArfGAP protein catalytic activity. 27531749_The effect of AGAP1 Knockdown could be rescued by FLAG-AGAP1, but not by an AGAP1 mutant that did not bind Kif2A efficiently, ArfGAP1-HA or Kif2A-GFP. The results support the hypothesis that the Kif2A.AGAP1 complex contributes to control of cytoskeleton remodeling involved in cell movement. 28430047_2 Arf GAPs, ASAP1 and AGAP1, have been found to bind directly to and influence the activity of myosins and kinesins, motor proteins associated with filamentous actin and microtubules, respectively. 31785816_AGAP1 regulates subcellular localization of FilGAP and control cancer cell invasion. 33618745_CircAGAP1 promotes tumor progression by sponging miR-15-5p in clear cell renal cell carcinoma. 33830075_Crystal structure of the GTP-binding protein-like domain of AGAP1. | ENSMUSG00000055013 | Agap1 | 1765.23267 | 1.0346289 | 0.0491134309 | 0.11613848 | 1.781215e-01 | 6.729926e-01 | 9.998360e-01 | No | Yes | 1652.45465 | 149.422513 | 1.483894e+03 | 104.142750 | |
| ENSG00000158106 | 114822 | RHPN1 | protein_coding | Q8TCX5 | FUNCTION: Has no enzymatic activity. May serve as a target for Rho, and interact with some cytoskeletal component upon Rho binding or relay a Rho signal to other molecules. {ECO:0000250|UniProtKB:Q61085}. | Coiled coil;Phosphoprotein;Reference proteome | hsa:114822; | cytosol [GO:0005829]; signal transduction [GO:0007165] | 31982726_The positive feedback loop of RHPN1-AS1/miR-1299/ETS1 accelerates the deterioration of gastric cancer. 32435875_LncRNA RHPN1-AS1 accelerates proliferation, migration, and invasion via regulating miR-485-5p/BSG axis in hepatocellular carcinoma. | ENSMUSG00000022580 | Rhpn1 | 650.26455 | 0.9609924 | -0.0574031477 | 0.15341120 | 1.432511e-01 | 7.050701e-01 | 9.998360e-01 | No | Yes | 648.66878 | 101.760413 | 6.096011e+02 | 74.392416 | ||
| ENSG00000158552 | 130617 | ZFAND2B | protein_coding | Q8WV99 | FUNCTION: Plays a role in protein homeostasis by regulating both the translocation and the ubiquitin-mediated proteasomal degradation of nascent proteins at the endoplasmic reticulum. It is involved in the regulation of signal-mediated translocation of proteins into the endoplasmic reticulum. It also plays a role in the ubiquitin-mediated proteasomal degradation of proteins for which signal-mediated translocation to the endoplasmic reticulum has failed. May therefore function in the endoplasmic reticulum stress-induced pre-emptive quality control, a mechanism that selectively attenuates the translocation of newly synthesized proteins into the endoplasmic reticulum and reroutes them to the cytosol for proteasomal degradation (By similarity). By controlling the steady-state expression of the IGF1R receptor, indirectly regulates the insulin-like growth factor receptor signaling pathway (PubMed:26692333). {ECO:0000250|UniProtKB:Q91X58, ECO:0000269|PubMed:26692333}. | 3D-structure;Alternative splicing;Endoplasmic reticulum;Lipoprotein;Membrane;Metal-binding;Methylation;Phosphoprotein;Prenylation;Reference proteome;Repeat;Zinc;Zinc-finger | This gene encodes a protein containing AN1-type zinc-fingers and ubiquitin-interacting motifs. The encoded protein likely associates with the proteosome to stimulate the degradation of toxic or misfolded proteins. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2012]. | hsa:130617; | anchored component of membrane [GO:0031225]; cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; proteasome complex [GO:0000502]; K48-linked polyubiquitin modification-dependent protein binding [GO:0036435]; ubiquitin binding [GO:0043130]; zinc ion binding [GO:0008270]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein targeting to ER [GO:0045047]; regulation of insulin-like growth factor receptor signaling pathway [GO:0043567]; SRP-dependent cotranslational protein targeting to membrane, translocation [GO:0006616] | 19414486_An AIP-1 human homologue, ZFAND2B, has a similar protective effect against Abeta toxicity. 24160817_AIRAPL binds to p97 and forms a complex in the endoplasmic reticulum membrane. 24215362_Zinc finger proteins may be associated with pathophysiology of severe dry eye syndrome. 26337389_Results demonstrate that on specific translocation inhibition, a p97-AIRAPL complex directly binds and regulates the efficient processing of polyubiquitinated pQC substrates by the UPS. Results also demonstrate p97's role in pQC processing of preproinsulin in cases of naturally occurring mutations within the signal sequence of insulin. 26692333_Consistent with its proposed role as a tumor suppressor of myeloid transformation, AIRAPL expression is widely abrogated in human myeloproliferative disorders. | ENSMUSG00000026197 | Zfand2b | 1082.34528 | 0.9561532 | -0.0646863613 | 0.14247237 | 2.146887e-01 | 6.431170e-01 | 9.998360e-01 | No | Yes | 1151.24405 | 133.836698 | 1.123986e+03 | 101.797370 | |
| ENSG00000158615 | 84919 | PPP1R15B | protein_coding | Q5SWA1 | FUNCTION: Maintains low levels of EIF2S1 phosphorylation in unstressed cells by promoting its dephosphorylation by PP1. {ECO:0000269|PubMed:26159176, ECO:0000269|PubMed:26307080}. | 3D-structure;Diabetes mellitus;Disease variant;Dwarfism;Mental retardation;Phosphoprotein;Reference proteome;Translation regulation | This gene encodes a protein phosphatase I-interacting protein that promotes the dephosphorylation of eukaryotic translation initiation factor 2A to regulate translation under conditions of cellular stress. The transcribed messenger RNA contains two upstream open reading frames (ORFs) that repress translation of the main protein coding ORF under normal conditions, while the protein coding ORF is expressed at high levels in response to stress. Continual translation of the mRNA under conditions of eukaryotic translation initiation factor 2A inactivation is thought to create a feedback loop for reactivation of the gene during recovery from stress. In addition, it has been shown that this protein plays a role in membrane traffic that is independent of translation and that it is required for exocytosis from erythroleukemia cells. Allelic variants of this gene are associated with microcephaly, short stature, and impaired glucose metabolism. [provided by RefSeq, Feb 2016]. | Mouse_homologues mmu:108954; | endoplasmic reticulum [GO:0005783]; protein phosphatase type 1 complex [GO:0000164]; protein phosphatase regulator activity [GO:0019888]; ER overload response [GO:0006983]; negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation [GO:1903912]; negative regulation of PERK-mediated unfolded protein response [GO:1903898]; negative regulation of protein phosphorylation [GO:0001933]; peptidyl-serine dephosphorylation [GO:0070262]; positive regulation of phosphoprotein phosphatase activity [GO:0032516]; response to endoplasmic reticulum stress [GO:0034976]; response to hydrogen peroxide [GO:0042542] | 22915583_the mammalian traffic machinery co-opts p-eIF2alpha and CReP, regulators of translation initiation. 25325377_CIP2A regulates cancer metabolism and CREB phosphorylation in non-small cell lung cancer 26159176_the first homozygous mutation in the PPP1R15B gene encoding the regulatory subunit of an eIF2alpha-specific phosphatase in two siblings affected by a novel syndrome of diabetes of youth with short stature, intellectual disability, and microcephaly 26307080_Whole-exome sequencing identified a homozygous missense mutation, c.1972G>A; p.Arg658Cys, in protein phosphatase 1, regulatory subunit 15b (PPP1R15B) 29599191_p97-mediated degradation, together with a reduction in CReP synthesis, is essential for timely stress-induced reduction of CReP levels and, consequently, for robust eIF2alpha phosphorylation to enforce the stress response. 34847777_Substrate recognition determinants of human eIF2alpha phosphatases. | ENSMUSG00000046062 | Ppp1r15b | 1556.36757 | 1.0540340 | 0.0759214032 | 0.12946386 | 3.485522e-01 | 5.549338e-01 | 9.998360e-01 | No | Yes | 1610.17881 | 217.698612 | 1.593872e+03 | 166.317968 | |
| ENSG00000159140 | 6651 | SON | protein_coding | P18583 | FUNCTION: RNA-binding protein that acts as a mRNA splicing cofactor by promoting efficient splicing of transcripts that possess weak splice sites. Specifically promotes splicing of many cell-cycle and DNA-repair transcripts that possess weak splice sites, such as TUBG1, KATNB1, TUBGCP2, AURKB, PCNT, AKT1, RAD23A, and FANCG. Probably acts by facilitating the interaction between Serine/arginine-rich proteins such as SRSF2 and the RNA polymerase II. Also binds to DNA; binds to the consensus DNA sequence: 5'-GA[GT]AN[CG][AG]CC-3'. May indirectly repress hepatitis B virus (HBV) core promoter activity and transcription of HBV genes and production of HBV virions. Essential for correct RNA splicing of multiple genes critical for brain development, neuronal migration and metabolism, including TUBG1, FLNA, PNKP, WDR62, PSMD3, PCK2, PFKL, IDH2, and ACY1 (PubMed:27545680). {ECO:0000269|PubMed:20581448, ECO:0000269|PubMed:21504830, ECO:0000269|PubMed:27545680}. | Acetylation;Alternative splicing;Cell cycle;DNA-binding;Disease variant;Isopeptide bond;Mental retardation;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation;mRNA processing;mRNA splicing | This gene encodes a protein that contains multiple simple repeats. The encoded protein binds RNA and promotes pre-mRNA splicing, particularly of transcripts with poor splice sites. The protein also recognizes a specific DNA sequence found in the human hepatitis B virus (HBV) and represses HBV core promoter activity. There is a pseudogene for this gene on chromosome 1. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:6651; | nuclear speck [GO:0016607]; DNA binding [GO:0003677]; RNA binding [GO:0003723]; microtubule cytoskeleton organization [GO:0000226]; mitotic cytokinesis [GO:0000281]; mRNA processing [GO:0006397]; negative regulation of apoptotic process [GO:0043066]; regulation of cell cycle [GO:0051726]; regulation of mRNA splicing, via spliceosome [GO:0048024]; regulation of RNA splicing [GO:0043484]; RNA splicing [GO:0008380] | 18952841_SON is an indispensable factor for cell growth, and AML1-ETO binding to SON may trigger signals inhibiting leukemogenesis. 20053686_Data suggest that Son is essential for appropriate subnuclear organization of pre-mRNA splicing factors and for promoting normal cell cycle progression. 20581448_The identification of the RNA/DNA-binding protein SON as a component of spliceosome that plays pleiotropic roles during mitotic progression, is reported. 21504830_Results reveal a mechanism for controlling cell-cycle progression through SON-dependent constitutive splicing at suboptimal splice sites, with strong implications for its role in cancer and other human diseases. 22193954_The data show that Son-regulated splicing encompasses all known types of alternative splicing, the most common being alternative splicing of cassette exons. 23227827_Studied knockdown screening of 78 MAPK-associated molecules associated with proliferation of pancreatic cancer cells in vitro. Knockdown of SON substantially suppressed pancreatic cancer cell proliferation and survival in vitro and tumorigenicity in vivo. 23322776_SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster 24013217_identify the spliceosome-associated factor SON as a factor essential for the maintenance of hESCs 24030980_Here, we summarize available information from several early studies on SON, and highlight recent discoveries describing molecular mechanisms of SON-mediated gene regulation. 26990989_progenitors. Our findings define SON as a fine-tuner of the MLL-menin interaction and reveal short SON overexpression as a marker indicating aberrant transcriptional initiation in leukemia. 27256762_we have established that haploinsufficiency of SON causes a new recognizable syndrome of intellectual disability. SON is located within 21q22.11, a critical region for Braddock-Carey syndrome, therefore, we suggest that the intellectual disability observed in Braddock-Carey syndrome could be accounted for by haploinsufficiency of SON. 27545676_description of seven unrelated individuals with de novo variants in SON and propose that deleterious variants in SON are associated with a severe multisystem disorder characterized by developmental delay, persistent feeding difficulties, and congenital malformations, including brain anomalies 27545680_data highlight SON as a master regulator governing neurodevelopment and demonstrate the importance of SON-mediated RNA splicing in human development. 29133588_We found that SON and SC35 (also known as SRSF2) localize to the central region of the speckle, whereas MALAT1 and small nuclear (sn)RNAs are enriched at the speckle periphery. 31005274_SON haploinsufficiency causes impaired pre-mRNA splicing of CAKUT genes and heterogeneous renal phenotypes. 32484234_SON DNA-binding protein mediates macrophage autophagy and responses to intracellular infection. 32705777_Phenotypic expansion in Zhu-Tokita-Takenouchi-Kim syndrome caused by de novo variants in the SON gene. 33095160_SON and SRRM2 are essential for nuclear speckle formation. 33247227_SON inhibits megakaryocytic differentiation via repressing RUNX1 and the megakaryocytic gene expression program in acute megakaryoblastic leukemia. 34331327_ZTTK syndrome: Clinical and molecular findings of 15 cases and a review of the literature. 34406792_The SON RNA splicing factor is required for intracellular trafficking structures that promote centriole assembly and ciliogenesis. 34548489_SON drives oncogenic RNA splicing in glioblastoma by regulating PTBP1/PTBP2 switching and RBFOX2 activity. | ENSMUSG00000022961 | Son | 12085.13383 | 0.9095410 | -0.1367893696 | 0.10272855 | 1.777646e+00 | 1.824387e-01 | 9.998360e-01 | No | Yes | 14521.50308 | 1906.323690 | 1.557666e+04 | 1577.253424 | |
| ENSG00000159788 | 6002 | RGS12 | protein_coding | O14924 | FUNCTION: Regulates G protein-coupled receptor signaling cascades. Inhibits signal transduction by increasing the GTPase activity of G protein alpha subunits, thereby driving them into their inactive GDP-bound form. {ECO:0000250|UniProtKB:O08774}.; FUNCTION: [Isoform 5]: Behaves as a cell cycle-dependent transcriptional repressor, promoting inhibition of S-phase DNA synthesis. {ECO:0000269|PubMed:12024043}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Cytoplasm;GTPase activation;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Signal transduction inhibitor;Synapse;Transcription;Ubl conjugation | This gene encodes a member of the 'regulator of G protein signaling' (RGS) gene family. The encoded protein may function as a guanosine triphosphatase (GTPase)-activating protein as well as a transcriptional repressor. This protein may play a role in tumorigenesis. Multiple transcript variants encoding distinct isoforms have been identified for this gene. Other alternative splice variants have been described but their biological nature has not been determined. [provided by RefSeq, Jul 2008]. | hsa:6002; | condensed nuclear chromosome [GO:0000794]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite [GO:0030425]; nuclear matrix [GO:0016363]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; synapse [GO:0045202]; GTPase activator activity [GO:0005096]; GTPase activity [GO:0003924]; GTPase regulator activity [GO:0030695]; G protein-coupled receptor signaling pathway [GO:0007186]; negative regulation of signal transduction [GO:0009968]; regulation of G protein-coupled receptor signaling pathway [GO:0008277] | 9168931_shows chromosomal mapping information for human locus. 17042716_RGS12 is essential for the terminal differentiation of osteoclasts induced by RANKL. 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20627871_Observational study of gene-disease association. (HuGE Navigator) 26987813_Data support the notion that the Galpha, but not Gbetagamma, arm of the Gi/o signalling is involved in TRPC4 activation and unveil new roles for RGS and RGS12 in fine-tuning TRPC4 activities. 28611045_Findings identify RGS12 as a candidate tumor-suppressor gene in AA prostate cancer, which acts by decreasing expression of AKT and MNX1, establishing a novel oncogenic axis in this disparate disease setting. 33198557_RGS12 Represses Oral Cancer via the Phosphorylation and SUMOylation of PTEN. 33686240_RGS12 is a novel tumor suppressor in osteosarcoma that inhibits YAP-TEAD1-Ezrin signaling. 34796776_RGS12 Drives Macrophage Activation and Osteoclastogenesis in Periodontitis. 34931491_RGS12 inhibits the progression and metastasis of multiple myeloma by driving M1 macrophage polarization and activation in the bone marrow microenvironment. | ENSMUSG00000029101 | Rgs12 | 530.88289 | 0.8400586 | -0.2514381611 | 0.17794020 | 1.844890e+00 | 1.743789e-01 | 9.998360e-01 | No | Yes | 493.80511 | 58.687549 | 5.286772e+02 | 49.069912 | |
| ENSG00000159917 | 9310 | ZNF235 | protein_coding | Q14590 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene product belongs to the zinc finger protein superfamily, members of which are regulatory proteins characterized by nucleic acid-binding zinc finger domains. The encoded protein is a member of the Kruppel family of zinc finger proteins, and contains Kruppel-associated box (KRAB) A and B domains and 15 tandemly arrayed C2H2-type zinc fingers. It is an ortholog of the mouse Zfp93 protein. This gene is located in a cluster of zinc finger genes on 19q13.2. [provided by RefSeq, Jul 2008]. | hsa:9310; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 46.82826 | 0.9267301 | -0.1097788389 | 0.44521419 | 6.111015e-02 | 8.047498e-01 | 9.998360e-01 | No | Yes | 46.26356 | 12.841671 | 4.930751e+01 | 10.555096 | ||||
| ENSG00000160117 | 126549 | ANKLE1 | protein_coding | Q8NAG6 | FUNCTION: Endonuclease that probably plays a role in the DNA damage response and DNA repair. {ECO:0000269|PubMed:22399800, ECO:0000269|PubMed:27245214}. | ANK repeat;Alternative splicing;Cytoplasm;DNA damage;DNA repair;Endonuclease;Hydrolase;Nuclease;Nucleus;Reference proteome;Repeat | hsa:126549; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; endonuclease activity [GO:0004519]; DNA repair [GO:0006281]; negative regulation of mitotic recombination [GO:0045950]; nucleic acid phosphodiester bond hydrolysis [GO:0090305]; positive regulation of response to DNA damage stimulus [GO:2001022]; protein export from nucleus [GO:0006611]; regulation of lymphoid progenitor cell differentiation [GO:1905456]; regulation of myeloid progenitor cell differentiation [GO:1905453] | 20237496_Observational study of gene-disease association. (HuGE Navigator) 20852631_Observational study and genome-wide association study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20852633_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22399800_Ankle1 is conserved in metazoans and contains a unique C-terminal GIY-YIG motif that confers endonuclease activity in vitro and in vivo 27245214_the domains mediating nuclear import and export of Ankle1, were identified. 28362817_Expression of RCCD1 in whole blood was also suggestively associated with disease risk (p-value: 1.2x10-05), as were expression of ACAP1 (p-value: 1.9x10-05) and LRRC25 (p-value: 5.2x10-05). While genome-wide association studies (GWAS) have implicated RCCD1 and ANKLE1 in breast cancer risk, they have not identified the remaining three genes 28957321_We used The Cancer Genome Atlas breast cancer patient data to identify ANKLE1 and ZNF404 as the target genes of candidate TF binding site SNPs in the 19p13.11 and 19q13.31 GWAS-identified loci. These SNPs are associated with the expression of ZNF404 and ANKLE1 in breast tissue. 31509622_ANKLE1 N(6) -Methyladenosine-related variant is associated with colorectal cancer risk by maintaining the genomic stability. 32866453_Human ANKLE1 Is a Nuclease Specific for Branched DNA. 33927264_Investigation of triple-negative breast cancer risk alleles in an International African-enriched cohort. | ENSMUSG00000046295 | Ankle1 | 66.33424 | 0.5166811 | -0.9526539149 | 0.41381141 | 4.694535e+00 | 3.025869e-02 | 9.998360e-01 | No | Yes | 52.60321 | 13.711309 | 8.478419e+01 | 17.033460 | ||
| ENSG00000160298 | 54058 | C21orf58 | protein_coding | P58505 | Alternative splicing;Reference proteome | hsa:54058; | ENSMUSG00000009114 | 2610028H24Rik | 819.63532 | 1.0362536 | 0.0513771666 | 0.13421392 | 1.471451e-01 | 7.012787e-01 | 9.998360e-01 | No | Yes | 829.35394 | 105.682519 | 7.491800e+02 | 74.148724 | |||||
| ENSG00000160606 | 116238 | TLCD1 | protein_coding | Q96CP7 | FUNCTION: Regulates the composition and fluidity of the plasma membrane (PubMed:30509349). Inhibits the incorporation of membrane-fluidizing phospholipids containing omega-3 long-chain polyunsaturated fatty acids (LCPUFA) and thereby promotes membrane rigidity (PubMed:30509349). Does not appear to have any effect on LCPUFA synthesis (PubMed:30509349). {ECO:0000269|PubMed:30509349}. | Alternative splicing;Cell membrane;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:116238; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; lipid homeostasis [GO:0055088]; membrane assembly [GO:0071709]; phospholipid homeostasis [GO:0055091]; plasma membrane organization [GO:0007009]; regulation of membrane lipid distribution [GO:0097035] | 30509349_inhibition of FLD-1 or TLCD1/2 prevents lipotoxicity by allowing increased levels of membrane phospholipids that contain fluidizing long-chain polyunsaturated fatty acids. | ENSMUSG00000019437 | Tlcd1 | 537.54692 | 1.0762078 | 0.1059566030 | 0.14691064 | 5.238122e-01 | 4.692200e-01 | 9.998360e-01 | No | Yes | 586.35083 | 70.396608 | 5.134380e+02 | 48.731480 | ||
| ENSG00000160961 | 84449 | ZNF333 | protein_coding | Q96JL9 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:84449; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 12151103_dna sequence analysis of ZNF333 gene located on chromosome 19p13.1 15607024_ZNF333 recognized the specific DNA core binding sequence ATAAT | 172.10410 | 0.9496273 | -0.0745666215 | 0.24705591 | 8.780272e-02 | 7.669893e-01 | 9.998360e-01 | No | Yes | 230.47743 | 31.739655 | 2.393528e+02 | 25.500830 | ||||
| ENSG00000163249 | 151195 | CCNYL1 | protein_coding | Q8N7R7 | Alternative splicing;Cyclin;Phosphoprotein;Reference proteome | hsa:151195; | plasma membrane [GO:0005886]; cyclin-dependent protein serine/threonine kinase activator activity [GO:0061575]; protein kinase binding [GO:0019901]; flagellated sperm motility [GO:0030317]; regulation of canonical Wnt signaling pathway [GO:0060828]; regulation of protein kinase activity [GO:0045859]; spermatogenesis [GO:0007283] | Mouse_homologues 26305884_CCNYL1 regulates spermatogenesis through the interaction and modulation of CDK16. 27203244_Here we show that Cyclin Y-like 1 (Ccnyl1) and Cyclin Y (Ccny) have overlapping function and are crucial for mouse embryonic development and mammary stem/progenitor cell functions. Double knockout of Ccnys results in embryonic lethality at E16.5 | ENSMUSG00000070871 | Ccnyl1 | 341.61949 | 0.6256165 | -0.6766496398 | 0.17671444 | 1.433244e+01 | 1.532017e-04 | 1.892158e-01 | No | Yes | 292.48367 | 61.926287 | 4.453642e+02 | 72.731558 | |||
| ENSG00000163349 | 204851 | HIPK1 | protein_coding | Q86Z02 | FUNCTION: Serine/threonine-protein kinase involved in transcription regulation and TNF-mediated cellular apoptosis. Plays a role as a corepressor for homeodomain transcription factors. Phosphorylates DAXX and MYB. Phosphorylates DAXX in response to stress, and mediates its translocation from the nucleus to the cytoplasm. Inactivates MYB transcription factor activity by phosphorylation. Prevents MAP3K5-JNK activation in the absence of TNF. TNF triggers its translocation to the cytoplasm in response to stress stimuli, thus activating nuclear MAP3K5-JNK by derepression and promoting apoptosis. May be involved in anti-oxidative stress responses. Involved in the regulation of eye size, lens formation and retinal lamination during late embryogenesis. Promotes angiogenesis and to be involved in erythroid differentiation. May be involved in malignant squamous cell tumor formation. Phosphorylates PAGE4 at 'Thr-51' which is critical for the ability of PAGE4 to potentiate the transcriptional activator activity of JUN (PubMed:24559171). {ECO:0000269|PubMed:12702766, ECO:0000269|PubMed:12968034, ECO:0000269|PubMed:15701637, ECO:0000269|PubMed:16390825, ECO:0000269|PubMed:19646965, ECO:0000269|PubMed:24559171}. | ATP-binding;Alternative splicing;Cytoplasm;Isopeptide bond;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene belongs to the Ser/Thr family of protein kinases and HIPK subfamily. It phosphorylates homeodomain transcription factors and may also function as a co-repressor for homeodomain transcription factors. Alternative splicing results in four transcript variants encoding four distinct isoforms. [provided by RefSeq, Jul 2008]. | hsa:204851; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; ATP binding [GO:0005524]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; adherens junction assembly [GO:0034333]; anterior/posterior pattern specification [GO:0009952]; definitive hemopoiesis [GO:0060216]; embryonic camera-type eye morphogenesis [GO:0048596]; embryonic retina morphogenesis in camera-type eye [GO:0060059]; endothelial cell apoptotic process [GO:0072577]; extrinsic apoptotic signaling pathway [GO:0097191]; eye development [GO:0001654]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; iris morphogenesis [GO:0061072]; lens induction in camera-type eye [GO:0060235]; neuron differentiation [GO:0030182]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of angiogenesis [GO:0045766]; positive regulation of cell population proliferation [GO:0008284]; protein phosphorylation [GO:0006468]; regulation of tumor necrosis factor-mediated signaling pathway [GO:0010803]; retina layer formation [GO:0010842]; smoothened signaling pathway [GO:0007224] | 15701637_TNFalpha-induced desumoylation and cytoplasmic translocation of HIPK1 are critical in TNFalpha-induced ASK1-JNK/p38 activation 19646965_c-Myb appears to be phosphorylated by HIPK1 in its negative regulatory domain as supported by both in vivo and in vitro data. 22173032_These findings indicate that the control of HIPK1 stability by Mdm2-NORE1 has a major effect on cell behaviour, and epigenetic inactivation of NORE1 enables adenocarcinoma formation in vivo through HIPK1 stabilization. 23071292_HIPK1 expression was identified only in invasive breast cancer cells with three different patterns: cytoplasmic, nuclear, and both cytoplasmic and nuclear. 23565059_HIPK1 and HIPK2, are transcriptional corepressors that regulate TGF-beta-dependent angiogenesis during embryonic development. 23676219_HIPK1 drives p53 activation to limit colorectal cancer cell growth. 24559171_PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1. 25630557_Analysis of these mutants revealed that HIPK1, HIPK2 and HIPK3 but not HIPK4 are capable of autophosphorylating on other tyrosines 28987030_The data presented here con fi rmed that HIPK1 is involved in the increased expression of recombinant luciferase and the secreted recombinant GPC3-hFc from HEK 293 cells following transfection with miR-22-3p. 29793420_HIPK1, HIPK2 and HIPK3 interact with the components of the carbon catabolite repressor 4 (CCR4)-negative on TATA (NOT) complex, an important regulator of all the major steps in the mRNA metabolism. it has emerged that HIPKs and their related miRNAs are involved in diabetic nephropathy, gastric cancer chemoresistance, cervical cancer progression, and recombinant protein expression in cultured cells. [Review] 34723781_MicroRNA-889-3p restrains the proliferation and epithelial-mesenchymal transformation of lung cancer cells via down-regulation of Homeodomain-interacting protein kinase 1. 34785661_Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation. | ENSMUSG00000008730 | Hipk1 | 1024.78876 | 1.1939480 | 0.2557400635 | 0.15508229 | 2.844211e+00 | 9.170358e-02 | 9.998360e-01 | No | Yes | 1297.94377 | 204.836114 | 1.151025e+03 | 140.233462 | |
| ENSG00000163467 | 128229 | TSACC | protein_coding | Q96A04 | FUNCTION: Co-chaperone that facilitates HSP-mediated activation of TSSK6. {ECO:0000269|PubMed:20829357}. | Chaperone;Reference proteome | hsa:128229; | cytoplasm [GO:0005737]; chaperone binding [GO:0051087] | ENSMUSG00000010538 | Tsacc | 113.25671 | 1.0600127 | 0.0840815472 | 0.29327974 | 8.640472e-02 | 7.687986e-01 | 9.998360e-01 | No | Yes | 108.09811 | 20.778637 | 1.010157e+02 | 15.644341 | |||
| ENSG00000163516 | 55139 | ANKZF1 | protein_coding | Q9H8Y5 | FUNCTION: Plays a role in the cellular response to hydrogen peroxide and in the maintenance of mitochondrial integrity under conditions of cellular stress (PubMed:28302725). Involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway (By similarity). {ECO:0000250, ECO:0000269|PubMed:28302725}. | ANK repeat;Coiled coil;Cytoplasm;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:55139; | cytoplasm [GO:0005737]; membrane [GO:0016020]; metal ion binding [GO:0046872]; cellular response to hydrogen peroxide [GO:0070301]; ubiquitin-dependent ERAD pathway [GO:0030433] | 28302725_loss-of-function mutations in ANKZF1 result in deregulation of mitochondrial integrity, and this may play a pathogenic role in the development of infantile-onset inflammatory bowel disease 29632312_ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes 30244831_Rendering 60S RNCs resistant to Ptrh1 but susceptible to ANKZF1. 31257922_High ANKZF1 is an independent factor of poor survival (overall survival and recurrence-free survival) in colon cancer by taking part in angiogenesis and some cancer signaling pathways. | ENSMUSG00000026199 | Ankzf1 | 935.74202 | 1.0817933 | 0.1134249170 | 0.13619976 | 6.928671e-01 | 4.051909e-01 | 9.998360e-01 | No | Yes | 900.75240 | 124.658862 | 8.118321e+02 | 86.992002 | ||
| ENSG00000163635 | 6314 | ATXN7 | protein_coding | O15265 | FUNCTION: Acts as component of the STAGA transcription coactivator-HAT complex. Mediates the interaction of STAGA complex with the CRX and is involved in CRX-dependent gene activation. Necessary for microtubule cytoskeleton stabilization. {ECO:0000269|PubMed:22100762}. | 3D-structure;Alternative splicing;Cytoplasm;Cytoskeleton;Disease variant;Isopeptide bond;Neurodegeneration;Nucleus;Reference proteome;Spinocerebellar ataxia;Transcription;Transcription regulation;Triplet repeat expansion;Ubl conjugation | The autosomal dominant cerebellar ataxias (ADCA) are a heterogeneous group of neurodegenerative disorders characterized by progressive degeneration of the cerebellum, brain stem and spinal cord. Clinically, ADCA has been divided into three groups: ADCA types I-III. ADCAI is genetically heterogeneous, with five genetic loci, designated spinocerebellar ataxia (SCA) 1, 2, 3, 4 and 6, being assigned to five different chromosomes. ADCAII, which always presents with retinal degeneration (SCA7), and ADCAIII often referred to as the 'pure' cerebellar syndrome (SCA5), are most likely homogeneous disorders. Several SCA genes have been cloned and shown to contain CAG repeats in their coding regions. ADCA is caused by the expansion of the CAG repeats, producing an elongated polyglutamine tract in the corresponding protein. The expanded repeats are variable in size and unstable, usually increasing in size when transmitted to successive generations. This locus has been mapped to chromosome 3, and it has been determined that the diseased allele associated with spinocerebellar ataxia-7 contains 37-306 CAG repeats (near the N-terminus), compared to 4-35 in the normal allele. The encoded protein is a component of the SPT3/TAF9/GCN5 acetyltransferase (STAGA) and TBP-free TAF-containing (TFTC) chromatin remodeling complexes, and it thus plays a role in transcriptional regulation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2016]. | hsa:6314; | cytosol [GO:0005829]; microtubule cytoskeleton [GO:0015630]; nuclear matrix [GO:0016363]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; histone deubiquitination [GO:0016578]; histone H3 acetylation [GO:0043966]; microtubule cytoskeleton organization [GO:0000226]; monoubiquitinated histone deubiquitination [GO:0035521]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; negative regulation of microtubule depolymerization [GO:0007026]; nucleus organization [GO:0006997]; regulation of transcription by RNA polymerase II [GO:0006357]; visual perception [GO:0007601] | 11697524_CAG expansion in SCA7 locus is associated with Machado-Joseph disease 11709544_Activated caspase-3 was recruited into the inclusions in both the cell models and human SCA7 brain and its expression was upregulated in cortical neurones. 11804332_Observational study of genotype prevalence. (HuGE Navigator) 12070661_Expression of ataxin-7 in CNS and non-CNS tissue of normal and SCA7 individuals 12533095_identification of a novel ataxin-7 protein enriched in the central nervous system suggests that expression of multiple polyglutamine-containing proteins may play a role in the neurodegeneration patterns characteristic of SCA7 15115762_Ataxin-7 is the human orthologue of SGF73, which is a subunit of the yeast SAGA complex, a coactivator required for transcription of a subset of RNA Pol II-dependent genes. Ataxin-7 is a new subunit of the mammalian SAGA-like complexes, TFTC/STAGA. 15115762_Demonstrate here that ataxin-7 is the human orthologue of the yeast SAGA SGF73 subunit and is a bona fide subunit of the human TFTC-like transcriptional complexes. 15148151_Observational study of genotype prevalence. (HuGE Navigator) 15316811_This patient with Spinocerebellar Ataxia 7 due to unique instability of the CAG repeat. 15932940_Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration in Spinocerebellar ataxia type 7. 15936949_We show that transcription mediated by both CBP and RORalpha1 was repressed by expanded ataxin-7. Ataxin-7 may act as a repressor of transcription by inhibiting the acetylation activity of TFTC and STAGA. 16325416_Ataxin-90A aggregates differed morphologically from ataxin7 - 100Q aggregatesand were more toxic to mesencephalic neurons, suggesting that toxicity was determined by the type of aggregate rather than the cellular misfolding response. 16389595_Observational study of gene-disease association. (HuGE Navigator) 16962040_Trinucleotide repeat expansions of ataxin 7 may be involved in neurodegenerative diseases such as cerebellar ataxia. 17026624_Origin of the SCA7 gene mutation in South Africa and the possibility of a founder effect in the Black population 17254003_Massive SCA7 expansion detected in a 7-month-old male with hypotonia, cardiomegaly, and renal failure. 17720198_We present a pediatric patient with 13 and 70 trinucleotide CAG repeats within SCA7 gene and no family history, whose presentation mimicked Kearns-Sayre syndrome (KSS). 18182848_Observational study of gene-disease association. (HuGE Navigator) 18216249_One common pathogenic response in transgenic SCA1 and SCA7 mice reveals the importance of intercellular mechanisms in the pathogenesis of spinocerebellar ataxias. 18325672_expanded CAG-repeats in the SCA7 gene within members of a large Chinese family with spinocerebellar ataxia 19172503_The patients with genetically confirmed SCA 7 presented an early macular dysfunction, preceding any signs of abnormalities in fundus appearance 19235102_Observational study of gene-disease association. (HuGE Navigator) 19259763_Observational study of gene-disease association. (HuGE Navigator) 19789634_analysis of RNA hairpins selective for silencing the mutant ataxin-7 transcript 19843541_These results demonstrate an influence of SUMOylation on the multistep aggregation process of ATXN7 and implicate a role for ATXN7 SUMOylation in SCA7 pathogenesis. 20069235_Observational study of gene-disease association. (HuGE Navigator) 20600911_In response to polyglutamine toxicity, transgenic murine SCA7 rods go through a range of radically different cell fates correlating with the nature, level and ratio of mutant transgene ATXN7 species. 20634802_The solution structures of the SCA7 domain of both ATXN7 and ATXN7L3 reveal a new, common zinc-finger motif at the heart of two distinct folds, providing a molecular basis for the observed functional differences. 20732423_The interaction between APLP2 and ataxin-7 and proteolytic processing of APLP2 may contribute to the pathogenesis of spinocerebellar ataxia type 7. 20739808_This study suggested that the SCA7 gene alternation in SCA7 patient in Chinese Han family. 21078624_identified 118 protein interactions for CACNA1A and ATXN7 linking them to other ataxia-causing proteins and the ataxia network; ataxia network is significantly enriched for proteins that interact with known macular degeneration-causing proteins 21827908_The Trinucleotide Repeat Expansion mutation in ATXN7 related to Spinocerebellar ataxia type 7. 22072678_The results of this study indicated that SCA7 disease pathogenesis involves a convergence of alterations in a variety of different cell types to fully recapitulate the cerebellar degeneration. 22100762_ATXN7 distribution frequently shifts from the nucleus to the cytoplasm; cytoplasmic ATXN7 associates with microtubules (MTs); expression of ATXN7 stabilizes MTs; findings provide a novel physiological function of ATXN7 in regulation of cytoskeletal dynamics and suggest that abnormal cytoskeletal regulation may contribute to SCA7 disease pathology 22367614_Full-length and cleaved fragments of the SCA7 disease protein ataxin-7 (ATXN7) are differentially degraded in a spinocerebellar ataxia type 7 rat model. 22827889_The results of this study demonstrated that oxidative stress contributes to ATXN7 aggregation as well as toxicity. 22917585_Critical nuclear events lead to transcriptional alterations in polyglutamine diseases such as spinocerebellar ataxia type 7 (SCA7) and Huntington's disease (HD). 23100044_The results demonstrated that a common genetic variant in the ataxia-causing gene ATXN7 influences cerebellar grey matter volume in healthy young adults. 23226359_role of ataxin-7 in differentiation of photoreceptors and cerebellar neurons 23236151_Polyglutamine expansion decreased ATXN7 occupancy, which correlated with increased levels of histone H2B monoubiquitination, at the reelin promoter. 23368522_Epidemiological evidence of a SCA7 founder effect in a Mexican population with spinocerebellar ataxia. 23828024_Haplotype and phylogenetic analyses provide evidence showing that the relatively high frequency of SCA7 in Mexican population is the result of a founder mutation and that Mexican SCA7 carriers possess the Western European ancestry. 23892081_Sequestration of the ponsin splice variant R85FL by the polyglutamine-expanded Atx7 in cell is mediated by the specific SH3C-PRR interaction, which is implicated in the pathogenesis of spinocerebellar ataxia 7. 24129567_polyQ-expanded ataxin-7 directly bound the Gcn5 catalytic core of SAGA while in association with its regulatory proteins, Ada2 and Ada3. 24374739_analysis of the founder effect and ancestral origin of the spinocerebellar ataxia type 7 mutation in Mexican families 24859968_This study shown evidence in vivo, in the SCA7 KI mouse model, that progressive accumulation of mutant ataxin-7 impairs autophagy. 25643591_The proband exhibited a typical phenotype of SCA7, which includes cone dystrophy and spinocerebellar ataxia. 25755283_Results suggest that sequestration of both enzymatic centers in SAGA upon ATXN7 poly(Q) expansion likely contributes to spinocerebellar ataxia type 7 development and progression. 25869926_South American cohort did not confirm the effect of the four candidate loci as modifier of onset age: mithocondrial A10398G polymorphism and CAGn at RAI1, CACNA1A, ATXN3, and ATXN7 genes 25900954_Our study provided the clinico-genetic analysis of nine Indian SCA7 families and CAG repeat distribution analysis in diverse Indian populations showed occurrence of ATXN7-CAG intermediate alleles in a predisposed population 26195632_Data show that the aggregates formed by polyQ-expanded ataxin 7 sequester ubiquitin-specific protease (USP22) through specific interactions. 26210447_Two pathological polyglutamine proteins, truncated Ataxin-7 and full-length Ataxin-3, suggest that accumulation of insoluble aggregates beyond a critical threshold could be responsible for neurotoxicity. 27296891_Results identified a chromosomal translocation between Rad51C and Ataxin-7 in colorectal tumors. The in-frame fusion transcript results in a fusion protein with molecular weight of 110 KDa. In vitro 5-Azacytidine treatment of colorectal tumor cells showed expression of the fusion gene is regulated by promoter methylation. 27855399_ATXN7 may be a potential predictor of post-operative prognosis of Hepatitis B Virus-related hepatocellular carcinoma . 28585930_we observed that carriers of either ATXN7 or TBP alleles with relatively large CAG repeat sizes in both alleles had a substantially increased risk of lifetime depression. 28597910_The intronic SNP rs6798742 is associated with ATXN7 CAG-region expansion. 28645341_Genetic testing showed the presence of 48 CAG repeats within one ATXN7 gene for spinocerebellar ataxia type 7 (SCA7). 30559154_the SUMO pathway contributes to the clearance of aggregated ATXN7 and suggest that its deregulation might be associated with SCA7 disease progression. 31097749_Structural and dynamic studies reveal that the Ala-rich region of ataxin-7 initiates alpha-helix formation of the polyQ tract but suppresses its aggregation. 32558018_Deciphering the natural history of SCA7 in children. 33338633_Polyglutamine expanded Ataxin-7 induces DNA damage and alters FUS localization and function. 34012225_Clinical characterization and the improved molecular diagnosis of autosomal dominant cone-rod dystrophy in patients with SCA7. | ENSMUSG00000021738 | Atxn7 | 365.56707 | 1.1245669 | 0.1693695334 | 0.18023482 | 9.077924e-01 | 3.407008e-01 | 9.998360e-01 | No | Yes | 388.73896 | 59.698473 | 3.527605e+02 | 41.976196 | |
| ENSG00000163964 | 54965 | PIGX | protein_coding | Q8TBF5 | FUNCTION: Essential component of glycosylphosphatidylinositol-mannosyltransferase 1 which transfers the first of the 4 mannoses in the GPI-anchor precursors during GPI-anchor biosynthesis. Probably acts by stabilizing the mannosyltransferase PIGM (By similarity). {ECO:0000250}. | Alternative splicing;Endoplasmic reticulum;GPI-anchor biosynthesis;Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | PATHWAY: Glycolipid biosynthesis; glycosylphosphatidylinositol-anchor biosynthesis. | This gene encodes a type I transmembrane protein in the endoplasmic reticulum (ER). The protein is an essential component of glycosylphosphatidylinositol-mannosyltransferase I, which transfers the first of the four mannoses in the GPI-anchor precursors during GPI-anchor biosynthesis. Studies in rat indicate that the protein is translated from a non-AUG translation initiation site. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2009]. | hsa:54965; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; GPI anchor biosynthetic process [GO:0006506] | 15635094_mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I | ENSMUSG00000023791 | Pigx | 454.35730 | 1.1834989 | 0.2430584012 | 0.16753404 | 2.134778e+00 | 1.439913e-01 | 9.998360e-01 | No | Yes | 537.29812 | 61.195288 | 5.008133e+02 | 44.310606 |
| ENSG00000164076 | 79012 | CAMKV | protein_coding | Q8NCB2 | FUNCTION: Does not appear to have detectable kinase activity. | Alternative splicing;Calmodulin-binding;Cell membrane;Cytoplasmic vesicle;Membrane;Phosphoprotein;Reference proteome | hsa:79012; | cytoplasmic vesicle membrane [GO:0030659]; glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; peptidyl-serine phosphorylation [GO:0018105]; regulation of modification of postsynaptic structure [GO:0099159] | 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) | ENSMUSG00000032936 | Camkv | 763.15697 | 0.9061303 | -0.1422095493 | 0.14711577 | 9.410362e-01 | 3.320115e-01 | 9.998360e-01 | No | Yes | 643.27724 | 79.393451 | 6.541631e+02 | 63.063087 | ||
| ENSG00000164707 | 26266 | SLC13A4 | protein_coding | Q9UKG4 | FUNCTION: Sodium/sulfate cotransporter that mediates sulfate reabsorption in the high endothelial venules (HEV). {ECO:0000269|PubMed:10535998, ECO:0000269|PubMed:15607730}. | Ion transport;Membrane;Reference proteome;Sodium;Sodium transport;Symport;Transmembrane;Transmembrane helix;Transport | hsa:26266; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; sodium:sulfate symporter activity [GO:0015382]; anion transmembrane transport [GO:0098656]; sulfate transport [GO:0008272] | 15607730_Here, we characterized the functional properties of the human Na(+)-sulfate cotransporter (hNaS2), determined its tissue distribution, and identified its gene (SLC13A4) structure. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23453247_SLC13A4 and SLC26A2 were the most abundant sulfate transporter mRNAs, which localized to syncytiotrophoblast and cytotrophoblast cells, respectively. 23485456_To investigate the regulation of SLC13A4 gene expression, we analysed the transcriptional activity of the SLC13A4 5'-flanking region in the JEG-3 placental cell line using luciferase reporter assays. 28385533_Study found that despite differential expression of the two SLC13A4 transcripts, no detectable functional difference in the cellular sorting or sulfate transporting was found. However, some variants can influence both mechanism in specific cell membranes. This is like to have clinical implications based on the consequences of impaired sulfate transport during pregnancy in rodent models. 34840533_SLC13A4 Might Serve as a Prognostic Biomarker and be Correlated with Immune Infiltration into Head and Neck Squamous Cell Carcinoma. | ENSMUSG00000029843 | Slc13a4 | 196.78210 | 1.2268397 | 0.2949467603 | 0.24211078 | 1.539131e+00 | 2.147472e-01 | 9.998360e-01 | No | Yes | 229.50128 | 33.419659 | 1.907535e+02 | 22.148361 | ||
| ENSG00000164877 | 79778 | MICALL2 | protein_coding | Q8IY33 | FUNCTION: Effector of small Rab GTPases which is involved in junctional complexes assembly through the regulation of cell adhesion molecules transport to the plasma membrane and actin cytoskeleton reorganization. Regulates the endocytic recycling of occludins, claudins and E-cadherin to the plasma membrane and may thereby regulate the establishment of tight junctions and adherens junctions. In parallel, may regulate actin cytoskeleton reorganization directly through interaction with F-actin or indirectly through actinins and filamins. Most probably involved in the processes of epithelial cell differentiation, cell spreading and neurite outgrowth (By similarity). {ECO:0000250}. | Alternative splicing;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoskeleton;Endosome;LIM domain;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Tight junction;Zinc | hsa:79778; | bicellular tight junction [GO:0005923]; cell-cell junction [GO:0005911]; cytosol [GO:0005829]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; recycling endosome [GO:0055037]; stress fiber [GO:0001725]; actin filament binding [GO:0051015]; actinin binding [GO:0042805]; filamin binding [GO:0031005]; metal ion binding [GO:0046872]; small GTPase binding [GO:0031267]; actin cytoskeleton reorganization [GO:0031532]; actin filament polymerization [GO:0030041]; bicellular tight junction assembly [GO:0070830]; endocytic recycling [GO:0032456]; neuron projection development [GO:0031175]; positive regulation of protein targeting to mitochondrion [GO:1903955]; substrate adhesion-dependent cell spreading [GO:0034446] | 17891173_Involved in epithelial cell scattering. 25864591_Data indicate that MICAL-L2 may be an important regulator of epithelial-mesenchymal transition (EMT) in ovarian cancer cells. 28416812_We identified one new significant locus at 7p22.3 for the Stroop word interference time (rs11514810, P=3.42E-09 for discovery, P=0.01176 for replication and combined P=5.249E-09). Regulatory feature analysis and expression quantitative trait loci (eQTL) data showed that this locus contributes to MICALL2 expression in the human brain. 31034158_Up-regulation of MICAL-L2 is associated with gastric cancer cell migration. 33976349_JRAB/MICAL-L2 undergoes liquid-liquid phase separation to form tubular recycling endosomes. | ENSMUSG00000036718 | Micall2 | 217.60495 | 0.7885830 | -0.3426655108 | 0.21433425 | 2.612286e+00 | 1.060388e-01 | 9.998360e-01 | No | Yes | 208.30325 | 39.105368 | 2.335960e+02 | 34.182403 | ||
| ENSG00000164902 | 51808 | PHAX | protein_coding | Q9H814 | FUNCTION: A phosphoprotein adapter involved in the XPO1-mediated U snRNA export from the nucleus. Bridge components required for U snRNA export, the cap binding complex (CBC)-bound snRNA on the one hand and the GTPase Ran in its active GTP-bound form together with the export receptor XPO1 on the other. Its phosphorylation in the nucleus is required for U snRNA export complex assembly and export, while its dephosphorylation in the cytoplasm causes export complex disassembly. It is recycled back to the nucleus via the importin alpha/beta heterodimeric import receptor. The directionality of nuclear export is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus. Its compartmentalized phosphorylation cycle may also contribute to the directionality of export. Binds strongly to m7G-capped U1 and U5 small nuclear RNAs (snRNAs) in a sequence-unspecific manner and phosphorylation-independent manner (By similarity). Plays also a role in the biogenesis of U3 small nucleolar RNA (snoRNA). Involved in the U3 snoRNA transport from nucleoplasm to Cajal bodies. Binds strongly to m7G-capped U3, U8 and U13 precursor snoRNAs and weakly to trimethylated (TMG)-capped U3, U8 and U13 snoRNAs. Binds also to telomerase RNA. {ECO:0000250, ECO:0000269|PubMed:15574332, ECO:0000269|PubMed:15574333}. | 3D-structure;Acetylation;Cytoplasm;Nucleus;Phosphoprotein;Protein transport;RNA-binding;Reference proteome;Transport | hsa:51808; | Cajal body [GO:0015030]; centrosome [GO:0005813]; cytosol [GO:0005829]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA cap binding complex binding [GO:0140262]; RNA binding [GO:0003723]; toxic substance binding [GO:0015643]; protein transport [GO:0015031]; RNA stabilization [GO:0043489]; snRNA export from nucleus [GO:0006408] | 15574332_PHAX and CRM1 have roles in transporting U3 snoRNA to nucleoli 20430857_PHAX RNA-binding domain mediates auxiliary RNA contracts with small nuclear and mall nucleolar RNA substrates. 32759388_The RNA transport factor PHAX is required for proper histone H2AX expression and DNA damage response. | ENSMUSG00000008301 | Phax | 664.59969 | 0.8753218 | -0.1921145361 | 0.15194324 | 1.543891e+00 | 2.140395e-01 | 9.998360e-01 | No | Yes | 598.22818 | 106.947421 | 6.198423e+02 | 85.436088 | ||
| ENSG00000165322 | 94134 | ARHGAP12 | protein_coding | Q8IWW6 | FUNCTION: GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state. {ECO:0000250}. | Alternative splicing;GTPase activation;Phosphoprotein;Reference proteome;Repeat;SH3 domain | This gene encodes a member of a large family of proteins that activate Rho-type guanosine triphosphate (GTP) metabolizing enzymes. The encoded protein may be involved in suppressing tumor formation by regulating cell invasion and adhesion. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jul 2012]. | hsa:94134; | cytoplasm [GO:0005737]; phagocytic cup [GO:0001891]; GTPase activator activity [GO:0005096]; actin filament organization [GO:0007015]; morphogenesis of an epithelial sheet [GO:0002011]; negative regulation of small GTPase mediated signal transduction [GO:0051058]; phagocytosis, engulfment [GO:0006911]; regulation of GTPase activity [GO:0043087]; signal transduction [GO:0007165] | 11854031_molecular cloning and characterization of ARHGAP12 16385451_Observational study of gene-disease association. (HuGE Navigator) 18504429_ARHGAP12 inactivates RAC1, thereby impairing cell motility, invasion and adhesion to the extracellular matrix. The gene is transcriptionally suppressed by Hepatocyte Growth Factor in vitro. 18504429_Met-driven invasive growth involves transcriptional regulation of Arhgap12. 18984674_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000041225 | Arhgap12 | 84.97702 | 0.8329812 | -0.2636441954 | 0.34912944 | 5.439252e-01 | 4.608105e-01 | 9.998360e-01 | No | Yes | 96.05849 | 19.180530 | 1.141857e+02 | 17.642345 | |
| ENSG00000165626 | 222389 | BEND7 | protein_coding | Q8N7W2 | Alternative splicing;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:222389; | extracellular exosome [GO:0070062]; DNA binding [GO:0003677] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 28300201_These results validated the association of two previously known skin pigmentation genes, SLC24A5 (minimum p = 2.62 x 10(-14), rs1426654) and SLC45A2 (minimum p = 9.71 x 10(-10), rs16891982), and revealed the intergenic region of BEND7 and PRPF18 as a novel locus associated with this trait (minimum p = 4.58 x 10(-9), rs6602666). | ENSMUSG00000048186 | Bend7 | 403.03918 | 1.0090527 | 0.0130015839 | 0.16252979 | 6.432258e-03 | 9.360771e-01 | 9.998360e-01 | No | Yes | 465.08867 | 58.248338 | 4.377403e+02 | 42.593302 | |||
| ENSG00000166002 | 56935 | SMCO4 | protein_coding | Q9NRQ5 | Coiled coil;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:56935; | integral component of membrane [GO:0016021] | ENSMUSG00000058173 | Smco4 | 412.18618 | 0.9966331 | -0.0048656698 | 0.16306872 | 8.931025e-04 | 9.761589e-01 | 9.998360e-01 | No | Yes | 395.09872 | 51.937142 | 4.124147e+02 | 42.719047 | ||||
| ENSG00000166133 | 27079 | RPUSD2 | protein_coding | Q8IZ73 | Alternative splicing;Phosphoprotein;Reference proteome | hsa:27079; | pseudouridine synthase activity [GO:0009982]; RNA binding [GO:0003723]; enzyme-directed rRNA pseudouridine synthesis [GO:0000455]; mRNA pseudouridine synthesis [GO:1990481] | ENSMUSG00000027324 | Rpusd2 | 1473.34574 | 1.0862078 | 0.1193001455 | 0.10892021 | 1.195814e+00 | 2.741600e-01 | 9.998360e-01 | No | Yes | 1641.09501 | 168.158634 | 1.452018e+03 | 115.955818 | ||||
| ENSG00000166206 | 2562 | GABRB3 | protein_coding | P28472 | FUNCTION: Ligand-gated chloride channel which is a component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the brain (PubMed:18514161, PubMed:22303015, PubMed:26950270, PubMed:22243422, PubMed:24909990). Plays an important role in the formation of functional inhibitory GABAergic synapses in addition to mediating synaptic inhibition as a GABA-gated ion channel (PubMed:25489750). The gamma2 subunit is necessary but not sufficient for a rapid formation of active synaptic contacts and the synaptogenic effect of this subunit is influenced by the type of alpha and beta subunits present in the receptor pentamer (By similarity). The alpha1/beta3/gamma2 receptor exhibits synaptogenic activity (PubMed:25489750). The alpha2/beta3/gamma2 receptor shows very little or no synaptogenic activity (By similarity). Functions also as histamine receptor and mediates cellular responses to histamine (PubMed:18281286). Plays an important role in somatosensation and in the production of antinociception (By similarity). {ECO:0000250|UniProtKB:P63080, ECO:0000269|PubMed:18281286, ECO:0000269|PubMed:18514161, ECO:0000269|PubMed:22243422, ECO:0000269|PubMed:22303015, ECO:0000269|PubMed:24909990, ECO:0000269|PubMed:25489750, ECO:0000269|PubMed:26950270}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Chloride;Chloride channel;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Disulfide bond;Epilepsy;Glycoprotein;Ion channel;Ion transport;Ligand-gated ion channel;Membrane;Postsynaptic cell membrane;Receptor;Reference proteome;Signal;Synapse;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the ligand-gated ionic channel family. The encoded protein is one the subunits of a multi-subunit chloride channel that serves as the receptor for gamma-aminobutyric acid, a major inhibitory neurotransmitter of the mammalian nervous system. This gene is located on the long arm of chromosome 15 in a cluster with two other genes encoding related subunits of the family. This gene may be associated with the pathogenesis of several disorders including Angelman syndrome, Prader-Willi syndrome, nonsyndromic orofacial clefts, epilepsy and autism. Alternatively spliced transcript variants encoding distinct isoforms have been described. [provided by RefSeq, Jul 2013]. | hsa:2562; | chloride channel complex [GO:0034707]; cytoplasmic vesicle membrane [GO:0030659]; GABA-A receptor complex [GO:1902711]; integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; synapse [GO:0045202]; GABA-A receptor activity [GO:0004890]; GABA-gated chloride ion channel activity [GO:0022851]; identical protein binding [GO:0042802]; neurotransmitter receptor activity [GO:0030594]; cellular response to histamine [GO:0071420]; chemical synaptic transmission [GO:0007268]; chloride transmembrane transport [GO:1902476]; gamma-aminobutyric acid signaling pathway [GO:0007214]; inhibitory synapse assembly [GO:1904862]; ion transmembrane transport [GO:0034220]; nervous system process [GO:0050877]; regulation of membrane potential [GO:0042391]; roof of mouth development [GO:0060021]; signal transduction [GO:0007165] | 11140838_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11711165_Observational study of gene-disease association. (HuGE Navigator) 11810291_linkage disequilibrium in cleft lip +/- cleft palate 11920158_Association between a GABRB3 polymorphism and autism 12048673_Observational study of gene-disease association. (HuGE Navigator) 12189488_The presence of inherited insomnia in the family of the affected individual suggests a possible link between insomnia and the mutation beta3(R192H). 12225856_modeling of molecular configuration 12367595_using full-length or truncated chimeric subunits it was demonstrated that homologous sequences from beta 3 are important for assembly of GABA(A) receptors composed of alpha(1), beta(3), and gamma(2) subunits 12491987_Lack of anomalies in 15q11-q13 region may be related to small number of probands, heterogenity of studied group, and small number of studied loci and markers. 14503638_This report indicates pronounced adaptive changes in the expression of these GABA(A) receptor subunits related to seizure activity and indicates altered assembly of GABA(A) receptors in temporal lobe epilepsy. 15066703_Observational study of gene-disease association. (HuGE Navigator) 15296817_Observational study of gene-disease association. (HuGE Navigator) 15318112_Observational study of gene-disease association. (HuGE Navigator) 15337300_No significant difference in mRNA expression is found between the control and alcoholic case groups in either the superior frontal or motor cortex for the GABA A beta 3 isoform 15615769_Reduced expression of GABRB3 is associated with Rett, Angelman and autism 15767729_the expression of the GABAA receptor pi subunit may play an important role in the pathogenesis of pancreatic cancer 16023997_Our data indicated that the haplotype 'GACTCT' (p = 0.00215, frequency = 53.6%) was overtransmitted which suggests that GABRB2 is in linkage disequilibrium with schizophrenia in the Chinese Han population. 16080114_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16674551_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16835263_Observational study of gene-disease association. (HuGE Navigator) 16835263_Reduced expression of the GABRB3 gene could therefore be one potential cause for the development of Childhood Absence Epilepsy. 17124266_alpha4beta3gamma2L receptors have unique kinetic properties that limit the range of GABA applications to which they can respond maximally. 17215107_Observational study of gene-disease association. (HuGE Navigator) 17215107_study failed to replicate an association of the common GABRB3 exon 1a promoter SNP rs4906902 with childhood absence epilepsy. no evidence that the common functional C-variant confers a substantial epileptogenic effect to a broad spectrum of IGE syndromes 17225872_Altered crosstalk between RA, GABAergic, and TGF-beta signaling systems could be involved in human cleft palate fibroblast phenotype. 17230033_Observational study of gene-disease association. (HuGE Navigator) 17230033_finding suggested that single-nucleotide polymorphisms in GABRB3 may play a significant role in the genetic predisposition to autism spectrum disorders in the Korean population 17272867_Observational study of gene-disease association. (HuGE Navigator) 17326191_The results indicate that human hepatocellular carcinoma (HCC) tissues are depolarized compared with adjacent nontumor tissues, and hepatic GABAA-beta3 receptor expression is down-regulated in human HCC. 17339270_These results suggest that MeCP2 acts as a chromatin organizer for optimal expression of both alleles of GABRB3 in neurons. 17471287_Subjects with schizophrenia exhibited expression deficits in GABRB3. 17880575_Observational study of gene-disease association. (HuGE Navigator) 17880575_study found mutations of GABRA1, GABRB3, and GABRG2 appear not to play a major role in the development of familial primary dystonia 17957331_Observational study of gene-disease association. (HuGE Navigator) 17957331_observed no significant difference in allelic frequencies or genotypic distributions of the 11 SNPs of intron 3 of GABRB3 between patients and controls 18085588_In the SNr GABA(A) receptors contain alpha(1), alpha(3), beta(2,3), and gamma(2) subunits and are localized in a weblike network over the cell soma, dendrites, and spines of SNr parvalbumin-positive nonpigmented neurons. 18281286_histamine modulates heteromultimeric GABA(A) receptors and may thus represent an endogenous ligand for an allosteric site 18358985_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18358985_The G1- alleles of the GABRB3 in children of alcoholics were significantly higher than nonCOAs. 18452349_Observational study of gene-disease association. (HuGE Navigator) 18452349_gene is involved in the pathogenesis of cleft lip with or without cleft palate in the Japanese population 18514161_Mutated GABRB3 could cause absence seizures through a gain in glycosylation of mutated exon 1a and exon 2, affecting maturation and trafficking of GABAR from endoplasmic reticulum to cell surface and resulting in reduced GABA-evoked currents. 18650446_three distinct sets of amino acid residues in the N-terminal extracellular domain of the hbeta1 subunit, which when mutated to the homologous residue in hbeta3 allow expression as a functional homomeric receptor 18821008_Data show that GABRB3 is significantly altered in cerebellum and significant reductions in parietal cortex in subjects with autism. 18837046_GABRB3 may contribute differently to the cleft phenotype in Iowans and in Filipinos, with a stronger effect in cases with palate involvement in Iowa, versus an effect in cases with involvement only of the lip in the Philippines. 18978678_Observational study of gene-disease association. (HuGE Navigator) 19058789_Observational study of gene-disease association. (HuGE Navigator) 19078961_Observational study of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19268543_Observational study of gene-disease association. (HuGE Navigator) 19430570_Observational study of gene-disease association. (HuGE Navigator) 19430570_The data provide preliminary evidence that GABRB3 gene is associated with autism spectrum disorders in Korea. 19598235_Observational study of gene-disease association. (HuGE Navigator) 19717274_Observational study of gene-disease association. (HuGE Navigator) 19736351_Observational study of gene-disease association. (HuGE Navigator) 19874574_Observational study of gene-disease association. (HuGE Navigator) 19935738_This study provides evidence of an association between a specific GABA(A) receptor defect and autism, direct evidence that this defect causes synaptic dysfunction that is autism relevant and maternal risk effect in the 15q11-q13 autism duplication region. 19937600_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20468064_Observational study of gene-disease association. (HuGE Navigator) 20550555_Observational study of gene-disease association. (HuGE Navigator) 20634891_Observational study of gene-disease association. (HuGE Navigator) 20843900_Gaba (A) beta1 and beta3 receptor subunit mRNA levels in the dorsolateral prefrontal cortex are not altered in schizophrenia. 21908847_Our data suggest the K289M mutation in gamma2 confers GABA(A)Rs with enhanced sensitivity of their membrane diffusion to neuronal activity 22080424_In comparison to healthy donors, chronic hepatitis C patients were found to present an increase in the expression of gamma-aminobutyric acid A receptor alpha1 subunit and a decrease in the expression of beta3 subunit in their blood mononuclear leukocytes. 22243422_Photoaffinity labeling and pepetide mapping data suggest that a homologous etomidate/general anesthetic binding site exists at the beta3-beta3 subunit interface in alpha1beta3 GABA receptor type A, specifically involving beta3-Met227. 22303015_Our results suggest both a mechanism for mutation-induced hyperexcitability and a novel role for the GABRB3 subunit N-terminal alpha-helix in receptor assembly and gating. 22414661_results provide additional evidence that GABRB3 and MAOB/NDP gene regions might constitute risk factors for hallucinations and delusions in schizophrenia. 22812221_The haplotype of C-A in rs4906902 and rs8179184 loci in the promoter of GABRB3 gene may be maternally inherited and positively associated with schizophrenia. 23438326_With a weak association, data do not support the hypothesis that the GABRB3 variants are a cause of nonsyndromic cleft lip and/or palate 23677991_Specificity of intersubunit general anesthetic-binding sites in the transmembrane domain of the human alpha1beta3gamma2 gamma-aminobutyric acid type A (GABAA) receptor. 24199598_Data suggest that GABA(A) receptor subunits (alpha-1 and beta-3) are able to form homologous receptors with either delta (extrasynaptic) or gamma-2 subunits (synaptic); allosteric modulation appears similar in the presence of agonist etomidate. 24249596_Considering our Argentinean ASD sample, it can be inferred that GABRB3 would be involved in the etiology of autism through interaction with GABRD. These results support the hypothesis that GABAR subunit genes are involved in autism. 24755890_Findings provide genetic evidence for the involvement of the genes GABRB3 and GABRA5 in the susceptibility to panic disorder 24865167_Increased GABRB3 expression may confer an increased risk of schizophrenia. 25025424_GABRB3 might be associated with heroin dependence, and increased expression of GABRB3 might contribute to the pathogenesis of heroin dependence 25086038_Propofol, AziPm, and o-PD Inhibit [3H]Azietomidate and R-[3H]mTFD-MPAB Photolabeling of alpha1beta3 GABAAR. 25211390_1,2-dichlorohexafluorocyclobutane enhancement of GABRA5 activity is abolished by GABRB3 mutations. 28053010_Results indicate that GABRB3 mutations are associated with a broad phenotypic spectrum of epilepsies and that reduced receptor function causing GABAergic disinhibition represents the relevant disease mechanism 28544625_In this study, we performed exome sequencing in six patients with SCN1A-negative Dravet syndrome to identify other genes related to this disorder..the data in this study identify GABRB3 as a candidate gene for Dravet syndrome 29162865_three de novo missense mutations in the GABAA receptor beta3 subunit gene (GABRB3) identified in patients with early-onset epileptic encephalopathy, are reported. 29196882_rs4906902 and rs8179184 in the 5' promoter region of GABRB3 are associated with schizophrenia in Han Chinese. 29725984_Meta-analysis showed that GABRB3 polymorphisms are not significantly associated with autism. 29961870_Receptors containing the GABRB3 p.P301L variant were less sensitive to GABA and produced less GABA-evoked current. 30074174_Meta-analysis indicates that neither rs4906902 nor rs20317 are significantly associated with the risk of autism spectrum disorder. 30140029_cryo-EM structure of the human alpha5beta3 GABAA receptor 30545943_This work demonstrated that only two substitutions, Q89R and G152T, in beta3 GABAAR are sufficient to reconstitute GABA-mediated activation and suggests that Tyr(87) prevents inhibitory effects of GABA. 30908890_Haplotype C-A might increase the risk of schizophrenia. Multiple regulatory regions that affect GABRB3 expression in vitro were identified. 31435640_We found that receptors containing either GABRB3 (N328D) or GABRB3 (E357K) mutant subunits had reduced GABA-evoked peak current amplitude. we found that both the GABRB3 (N328D) and GABRB3 (E357K) mutations reduced total subunit expression in neurons but not in HEK293T cells. 31610743_The intracellular domain of alpha3 and beta3 each contain essential anterograde trafficking signals that are required to overcome ER retention of assembled GABAA homo- or heteropentamers. 32376074_Gene expression meta-analysis reveals the down-regulation of three GABA receptor subunits in the superior temporal gyrus of patients with schizophrenia. 32540960_Photoaffinity labeling identifies an intersubunit steroid-binding site in heteromeric GABA type A (GABAA) receptors. 34698933_GABRB3-related epilepsy: novel variants, clinical features and therapeutic implications. 34906499_Structural mapping of GABRB3 variants reveals genotype-phenotype correlations. 35383156_Gain-of-function and loss-of-function GABRB3 variants lead to distinct clinical phenotypes in patients with developmental and epileptic encephalopathies. | ENSMUSG00000033676 | Gabrb3 | 321.88875 | 1.1810486 | 0.2400683909 | 0.22417695 | 1.064783e+00 | 3.021268e-01 | 9.998360e-01 | No | Yes | 358.88793 | 59.139702 | 2.794217e+02 | 35.512106 | |
| ENSG00000166261 | 7753 | ZNF202 | protein_coding | O95125 | FUNCTION: Transcriptional repressor that binds to elements found predominantly in genes that participate in lipid metabolism. Among its targets are structural components of lipoprotein particles (apolipoproteins AIV, CIII, and E), enzymes involved in lipid processing (lipoprotein lipase, lecithin cholesteryl ester transferase), transporters involved in lipid homeostasis (ABCA1, ABCG1), and several genes involved in processes related to energy metabolism and vascular disease. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7753; | chromosome [GO:0005694]; nuclear body [GO:0016604]; nucleolus [GO:0005730]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; lipid metabolic process [GO:0006629]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 16289551_Observational study of gene-disease association. (HuGE Navigator) 16289551_This is the first study to suggest that ZNF202 could be a new candidate gene for ischemic heart disease and myocardial infarction in the general population. 16415175_Observational study of gene-disease association. (HuGE Navigator) 16467280_Observational study of gene-disease association. (HuGE Navigator) 16467280_findings show that genetic variation in ZNF202 is common in the general population. However, SNPs in the protein-coding region of ZNF202 do not make a major contribution to HDL cholesterol levels. 18523156_SOX17-Chromatin immunoprecipitation identified zinc finger protein 202 (Zfp202) as a direct target of SOX17 during endoderm differentiation of F9 embryonal carcinoma cells. 18652945_Homozygosity for a common functional promoter variant in ZNF202 predicts severe atherosclerosis and an increased risk of IHD. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19875103_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000025602 | Zfp202 | 542.02090 | 0.9860433 | -0.0202770250 | 0.15146325 | 1.790455e-02 | 8.935545e-01 | 9.998360e-01 | No | Yes | 550.55575 | 72.132667 | 6.067589e+02 | 61.445891 | ||
| ENSG00000166340 | 1200 | TPP1 | protein_coding | O14773 | FUNCTION: Lysosomal serine protease with tripeptidyl-peptidase I activity (PubMed:11054422, PubMed:19038966, PubMed:19038967). May act as a non-specific lysosomal peptidase which generates tripeptides from the breakdown products produced by lysosomal proteinases (PubMed:11054422, PubMed:19038966, PubMed:19038967). Requires substrates with an unsubstituted N-terminus (PubMed:19038966). {ECO:0000269|PubMed:11054422, ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967}. | 3D-structure;Alternative splicing;Autocatalytic cleavage;Calcium;Direct protein sequencing;Disease variant;Disulfide bond;Epilepsy;Glycoprotein;Hydrolase;Lysosome;Metal-binding;Neurodegeneration;Neuronal ceroid lipofuscinosis;Protease;Reference proteome;Serine protease;Signal;Spinocerebellar ataxia;Zymogen | This gene encodes a member of the sedolisin family of serine proteases. The protease functions in the lysosome to cleave N-terminal tripeptides from substrates, and has weaker endopeptidase activity. It is synthesized as a catalytically-inactive enzyme which is activated and auto-proteolyzed upon acidification. Mutations in this gene result in late-infantile neuronal ceroid lipofuscinosis, which is associated with the failure to degrade specific neuropeptides and a subunit of ATP synthase in the lysosome. [provided by RefSeq, Jul 2008]. | hsa:1200; | extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; lysosomal lumen [GO:0043202]; lysosome [GO:0005764]; melanosome [GO:0042470]; membrane raft [GO:0045121]; recycling endosome [GO:0055037]; endopeptidase activity [GO:0004175]; lysophosphatidic acid binding [GO:0035727]; metal ion binding [GO:0046872]; peptidase activity [GO:0008233]; peptide binding [GO:0042277]; serine-type endopeptidase activity [GO:0004252]; serine-type peptidase activity [GO:0008236]; sulfatide binding [GO:0120146]; tripeptidyl-peptidase activity [GO:0008240]; bone resorption [GO:0045453]; central nervous system development [GO:0007417]; epithelial cell differentiation [GO:0030855]; lipid metabolic process [GO:0006629]; lysosomal protein catabolic process [GO:1905146]; lysosome organization [GO:0007040]; nervous system development [GO:0007399]; neuromuscular process controlling balance [GO:0050885]; peptide catabolic process [GO:0043171]; protein catabolic process [GO:0030163]; protein localization to chromosome, telomeric region [GO:0070198]; proteolysis [GO:0006508] | 12125808_The clinical, biochemical, and molecular genetic aspects of lysosomal storage disorders are discussed in this review 12134079_Data show that three neuronal ceroid lipofuscinoses disease forms with similar tissue pathology are connected at the molecular level: CLN5 polypeptides directly interact with the CLN2 and CLN3 proteins 12376936_Missense mutations, R127Q, N286S, and T353P represent novel, previously not described alleles. 12488460_human tripeptidyl-peptidase I is processed by a serine protease to the mature, active form in vivo 12698559_a novel mutation in neuronal ceroid lipofuscinosis 12950156_CLN2 gene mutations may result in low cerebrospinal fluid pterin production in classical neuronal ceroid lipofuscinoses of late infantile onset. 14702339_human tripeptidyl-peptidase I must be N-glycosylated for folding, trafficking, and stability 14736728_mutant Asn286Ser CLN2 lacks one oligosaccharide chain resulting in enzymatic inactivation 15143070_intramolecular (unimolecular) mechanism of TPP I activation and autoprocessing 15158442_TPP-I is the predominant proteolytic enzyme responsible for the intracellular degradation of neuromedin B 15317752_Functional analyses of CLN2 mutations reveal transport disruption of tripeptidyl-peptidase I to lysosomes. 15582991_tripeptidyl-peptidase I activation, activity, and stability are regulated by glycosaminoglycans 15733845_Ser475 and Asp360, also Glu272, Asp276, and Asp327 are important for catalytic activity of tripeptidyl peptidase I 16091586_Substrate-binding cleft of TPP-I composed of only 6 subsites; TPP-I prefers bulky and hydrophobic amino acid residues at P(1) position and Ala, Arg, or Asp at P(2) position; hydrophilic interactions at the S(2) subsite are necessary for TPP-I. 16168594_Mutational screening of CLCN2 gene, revealed a homozygous mutation G2003C (exon 17), leading to a Ser/Thr substitution at the codon 668, in two of the three in malignant migrating partial seizures patients. 16518810_there is a close correlation between CLN2 and CLN1 expression and colorectal carcinoma progression and metastasis and suggest that they may be potential molecular targets 17690061_Clinical features, histological findings, and genetic study reveal that CLN2 type is the most common form of neuronal ceroid lipofuscinosis. There is male predominance of 90.1% in this part of the Arab world. 17959406_CLN2/TPP1 deficiency: the novel mutation IVS7-10A>G causes intron retention and is associated with a mild disease phenotype. 18411270_the tripeptidyl peptidase I prosegment is a potent, slow-binding inhibitor of its cognate enzyme 18552385_Lysosome-related genes, such as CLN2, CLN3, and HEXB, may be involved in the pathogenesis of adipose tissue hypertrophy in TED. 19038966_Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis. 19038967_Crystal structure and autoactivation pathway of the precursor form of human tripeptidyl-peptidase 1, the enzyme deficient in late infantile ceroid lipofuscinosis 19201763_Observational study of gene-disease association. (HuGE Navigator) 19246452_genetic deletion of the lysosomal serine protease CLN2 and the subsequent loss of its catalytic function confer resistance to TNF in non-neuronal somatic cells in a Bid-dependent manner, indicating that CLN2 plays a role in TNF-induced cell death 19748052_This novel deletion mutation in the CLN2 gene in a family of Arab origin from Israel sheds further light on the epidemiology of neuronal ceroid lipofuscinosis as a worldwide disease 20340139_Data show that most TPPI variants displayed obstructed transport to the lysosomes. 20672930_The authors conducted a phase I study of late infantile neuronal ceroid lipofuscinosis using an adenoassociated virus serotype 2 (AAV2) vector containing the deficient CLN2 gene (AAV2(CU)hCLN2). 20689811_the critical residues in the TPPI catalysis and its structure-function analysis 21784683_Intrathecal human tripeptidyl-peptidase 1 administration reduces lysosomal storage in a canine model of late infantile neuronal ceroid lipofuscinosis. 22016395_Studies indicate that TPP-I is the only member of the sedolisin family that has been shown to exhibit tripeptidyl peptidase activity and is related to the fatal hereditary disease, Batten disease. 22832778_This study demonistrated that the CLN2 gene 4 mutation in late infantile neuronal ceroid lipofuscinosis. 22989886_Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor alpha and may have implications in late infantile Batten disease therapy 23249249_To our knowledge, our results bring the first evidence of a mechanism that links TPP-1 deficiency and oxidative stress-induced changes in mitochondrial morphology. 23266810_The variant juvenile phenotype comprises approximately 50% of CLN2 in South America. The five most frequent South American mutations comprise 66% of pathological alleles. 23418007_hypothesize that loss of function variants abolishing TPP1 enzyme activity lead to CLN2 disease, whereas variants that diminish TPP1 enzyme activity lead to SCAR7 23587805_TPP1 mutants utilize the advantages of a zebrafish model for understanding the pathogenesis of late infantile (or classic late infantile neuronal ceroid lipofuscinosis) disease. 24271013_TPP1(CLN2) mutation is associated with neuronal ceroid lipofuscinosis. 27553878_To confirm clinical suspicion of CLN2 disease, the recommended gold standard for laboratory diagnosis is demonstration of deficient TPP1 enzyme activity (in leukocytes, fibroblasts, or dried blood spots) and the identification of causative mutations in each allele of the TPP1/CLN2 gene. 27840983_TPP1 is overexpressed in hepatocellular carcinoma tissues and significantly correlated with poor prognosis of hepatocellular carcinoma patients.RFX5 acts as a direct positive transcriptional regulator of TPP1 in hepatocellular carcinoma. 28079862_These studies indicate that optimal treatment outcomes for CLN2 disease may require delivery of TPP1 systemically as well as directly to the central nervous system. 29160297_The reports the crystal structure of the N-terminal domain of TIN2 in complex with TIN2-binding motifs from TPP1 and TRF2, revealing how TIN2 interacts cooperatively with TPP1 and TRF2. 29378960_TPP1 cleaves and destabilizes fibrillar amyloid-beta at multiple sites in a time- and pH-dependent manner. 29631617_The results demonstrate that these patient iPSC derived NCL NSCs are valid cell- based disease models with characteristic disease phenotypes that can be used for study of disease pathophysiology and drug development. 30541466_the study contributes four novel variants to the spectrum of PPT1 gene mutations and eight novel variants to the TPP1 gene mutation data in neuronal ceroid lipofuscinoses type I and type II 31059981_Study performed targeted next-generation sequencing in a cohort of patients with epilepsies. A homozygous missense mutation in TPP1 was identified in a family with two affected siblings. The authors further systematically reviewed all TPP1 mutations and analyzed the correlations between genotype and phenotype. 31256057_The TPP1 activity was increased more than 3-fold in mucinous cysts relative to nonmucinous cysts. Moreover, TPP1 activity is primarily associated with mucinous cysts that harbor high-grade dysplasia or invasive pancreatic carcinoma. 31283065_In cases of neuronal ceroid lipofuscinosis type 2 (CLN2 disease), TPP1 mutations c.509-1 G>C and c.622 C>T (p.(Arg208*)), collectively occur in 60% of affected individuals in the sample, and account for 50% of disease-associated alleles. At least 86 variants (66%) are private to single families. Homozygosity occurs in 45% of individuals where both alleles are known (87% of reported individuals). 31533043_A High-Content Screen Identifies TPP1 and Aurora B as Regulators of Axonal Mitochondrial Transport. 32146219_Symmetric Age Association of Retinal Degeneration in Patients with CLN2-Associated Batten Disease. 32631363_Targeted re-sequencing for early diagnosis of genetic causes of childhood epilepsy: the Italian experience from the 'beyond epilepsy' project. 32735728_Identification of three predictors of gastric cancer progression and prognosis. 33317560_Tripeptidyl peptidase I promotes human endometrial epithelial cell adhesive capacity implying a role in receptivity. 35418675_Structure of active human telomerase with telomere shelterin protein TPP1. | ENSMUSG00000030894 | Tpp1 | 3391.06912 | 1.0236581 | 0.0337339321 | 0.10374299 | 1.059577e-01 | 7.447940e-01 | 9.998360e-01 | No | Yes | 3564.78471 | 286.220483 | 3.284244e+03 | 204.236262 | |
| ENSG00000166352 | 119710 | IFTAP | protein_coding | Q86VG3 | FUNCTION: Seems to play a role in ciliary BBSome localization, maybe through interaction with IFT-A complex. {ECO:0000269|PubMed:30476139}. | Alternative splicing;Phosphoprotein;Reference proteome | This gene encodes a protein that was identified as a cellular interacting partner of non-structural protein 10 of the severe acute respiratory syndrome coronavirus (SARS-CoV). The encoded protein may function as a negative regulator of transcription. There is a pseudogene for this gene on chromosome 1. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2013]. | hsa:119710; | 9+0 non-motile cilium [GO:0097731]; cytosol [GO:0005829]; intraciliary transport particle A binding [GO:0120160]; acrosome reaction [GO:0007340]; spermatogenesis [GO:0007283] | 16157265_identification of HEPIS as a protein that binds nsp10 of SARS-CoV 19773279_Observational study of gene-disease association. (HuGE Navigator) 30476139_The C11ORF74, interacts with the IFT-A complex via the IFT122 subunit and is accumulated at the distal tip in the absence of an IFT-A subunit IFT139, suggesting that at least a fraction of C11ORF74 molecules can be transported towards the ciliary tip by associating with the IFT-A complex, although its majority might be out of cilia at steady state. | ENSMUSG00000027165 | Iftap | 266.75204 | 0.9994254 | -0.0008291825 | 0.20647718 | 1.611400e-05 | 9.967971e-01 | 9.998360e-01 | No | Yes | 273.24984 | 44.996724 | 2.821457e+02 | 35.993950 | |
| ENSG00000166402 | 7275 | TUB | protein_coding | P50607 | FUNCTION: Functions in signal transduction from heterotrimeric G protein-coupled receptors. Binds to membranes containing phosphatidylinositol 4,5-bisphosphate. Can bind DNA (in vitro). May contribute to the regulation of transcription in the nucleus. Could be involved in the hypothalamic regulation of body weight (By similarity). Contribute to stimulation of phagocytosis of apoptotic retinal pigment epithelium (RPE) cells and macrophages. {ECO:0000250, ECO:0000269|PubMed:19837063}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Membrane;Nucleus;Obesity;Phagocytosis;Reference proteome;Secreted;Sensory transduction | This gene encodes a member of the Tubby family of bipartite transcription factors. The encoded protein may play a role in obesity and sensorineural degradation. The crystal structure has been determined for a similar protein in mouse, and it functions as a membrane-bound transcription regulator that translocates to the nucleus in response to phosphoinositide hydrolysis. Two transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:7275; | cilium [GO:0005929]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular region [GO:0005576]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; G protein-coupled receptor binding [GO:0001664]; intraciliary transport particle A binding [GO:0120160]; protein-containing complex binding [GO:0044877]; intraciliary transport [GO:0042073]; phagocytosis, recognition [GO:0006910]; photoreceptor cell maintenance [GO:0045494]; positive regulation of phagocytosis [GO:0050766]; protein localization to cilium [GO:0061512]; protein localization to photoreceptor outer segment [GO:1903546]; receptor localization to non-motile cilium [GO:0097500]; regulation of G protein-coupled receptor signaling pathway [GO:0008277]; response to stimulus [GO:0050896]; retina development in camera-type eye [GO:0060041]; sensory perception of sound [GO:0007605] | 10591637_Crystal structure of the core domain of the mouse Tubby protein. 11375483_Mouse Tubby protein may function as a membrane-bound transcription regulator that translocates to the nucleus in response to phosphoinositide hydrolysis, providing a direct link between G-protein signaling and the regulation of gene expression. 16443771_Observational study of gene-disease association. (HuGE Navigator) 16443771_TUB could be an important factor in controlling the central regulation of body weight in humans. 17498679_found the presence of tubby protein both in normal weight and in obese subjects; however in the latter an isoelectric point shift toward the acidic end was observed 17955208_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17955208_TUB is a candidate gene for late-onset obesity in humans 18183286_Genetic variation of the TUB gene was associated with both body composition and macronutrient intake, suggesting that TUB might influence eating behavior 18183286_Observational study of gene-disease association. (HuGE Navigator) 19058789_Observational study of gene-disease association. (HuGE Navigator) 19077438_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19837063_Tubby and Tulp1 function as phagocytosis sigmals (eat-me) for retinal pigment epithelium cells and other phagocytes. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20080650_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20978472_Tubby and Tulp1 are bridging molecules with their N-terminal region as MERTK-binding domain and C-terminal region as phagocytosis prey-binding domain. 24664737_Tubby and Tulp1 mediated phagocytosis through MerTK-dependent signaling with non-muscle myosin II redistribution leading to colocalization of phagocytosed vesicles with rearranged NMMIIA. 28852204_Studied the expression and distribution patterns of tubby bipartite transcription factor (TUB) in the hypothalamus and in adipose tissue in obese and healthy controls. 32956375_Candidate variants in TUB are associated with familial tremor. 33577845_Lnc-OIP5-AS1 exacerbates aorta wall injury during the development of aortic dissection through upregulating TUB via sponging miR-143-3p. | ENSMUSG00000031028 | Tub | 2172.78651 | 1.0157091 | 0.0224872201 | 0.09992515 | 5.053002e-02 | 8.221436e-01 | 9.998360e-01 | No | Yes | 2090.40528 | 287.367781 | 2.232647e+03 | 236.941760 | |
| ENSG00000166439 | 254225 | RNF169 | protein_coding | Q8NCN4 | FUNCTION: Probable E3 ubiquitin-protein ligase that acts as a regulator of double-strand breaks (DSBs) repair following DNA damage. Functions in a non-canonical fashion to harness RNF168-mediated protein recruitment to DSB-containing chromatin, thereby contributing to regulation of DSB repair pathway utilization (PubMed:22492721, PubMed:30773093). Once recruited to DSB repair sites by recognizing and binding ubiquitin catalyzed by RNF168, competes with TP53BP1 and BRCA1 for association with RNF168-modified chromatin, thereby favouring homologous recombination repair (HRR) and single-strand annealing (SSA) instead of non-homologous end joining (NHEJ) mediated by TP53BP1 (PubMed:30104380, PubMed:30773093). E3 ubiquitin-protein ligase activity is not required for regulation of DSBs repair. {ECO:0000269|PubMed:22492721, ECO:0000269|PubMed:22733822, ECO:0000269|PubMed:22742833, ECO:0000269|PubMed:30104380, ECO:0000269|PubMed:30773093}. | 3D-structure;Chromosome;DNA damage;DNA repair;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | hsa:254225; | cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; site of double-strand break [GO:0035861]; K63-linked polyubiquitin modification-dependent protein binding [GO:0070530]; metal ion binding [GO:0046872]; nucleosome binding [GO:0031491]; transferase activity [GO:0016740]; cellular response to DNA damage stimulus [GO:0006974]; negative regulation of double-strand break repair [GO:2000780]; protein ubiquitination [GO:0016567] | 22492721_results show that RNF169 functions in a noncanonical fashion to harness RNF168-mediated protein recruitment to DSB-containing chromatin, thereby contributing to regulation of DSB repair pathway utilization. 22733822_RNF169 as a component in DNA damage signal transduction and adds to the complexity of regulatory ubiquitylation in genome stability maintenance. 22742833_RNF168, its paralog RNF169, RAD18, and the BRCA1-interacting RAP80 protein accumulate at DNA double strand break sites through the use of bipartite modules composed of ubiquitin binding domains. 28325877_USP7 is RNF169 interacting protein.Expression of USP7 and RNF169 positively correlated in breast cancer. 28406400_The authors establish that RNF169 binds to ubiquitylated histone H2A-Lys13/Lys15 in a manner that involves its canonical ubiquitin-binding helix and a pair of arginine-rich motifs that interact with the nucleosome acidic patch. 28506460_Ubiquitin ligases RNF168, RNF169, and RAD18 specifically bind histone H2A Lys13/15-ubiquitylated nucleosomes. 53BP1 chromatin recruitment may be activated by RNF168 and blocked by RNF169 and RAD18. 30104380_results highlight the interplay of RNF169 with 53BP1 in fine-tuning choice of DSB repair pathways. 30979931_A comprehensive proteomics-based interaction screen that links DYRK1A to RNF169 and to the DNA damage response. | ENSMUSG00000058761 | Rnf169 | 679.27001 | 0.6677376 | -0.5826468027 | 0.14205734 | 1.667283e+01 | 4.441264e-05 | 7.186896e-02 | No | Yes | 599.46367 | 100.889113 | 9.840743e+02 | 127.784077 | |
| ENSG00000166575 | 65084 | TMEM135 | protein_coding | Q86UB9 | FUNCTION: Involved in mitochondrial metabolism by regulating the balance between mitochondrial fusion and fission. May act as a regulator of mitochondrial fission that promotes DNM1L-dependent fission through activation of DNM1L. May be involved in peroxisome organization. {ECO:0000250|UniProtKB:Q5U4F4, ECO:0000250|UniProtKB:Q9CYV5}. | Alternative splicing;Membrane;Mitochondrion;Peroxisome;Reference proteome;Transmembrane;Transmembrane helix | hsa:65084; | integral component of membrane [GO:0016021]; lipid droplet [GO:0005811]; mitochondrial membrane [GO:0031966]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; peroxisome organization [GO:0007031]; regulation of mitochondrial fission [GO:0090140]; response to cold [GO:0009409]; response to food [GO:0032094] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32157776_TMEM135 regulates primary ciliogenesis through modulation of intracellular cholesterol distribution. | ENSMUSG00000039428 | Tmem135 | 153.83589 | 1.4946654 | 0.5798225262 | 0.24919294 | 5.493758e+00 | 1.908448e-02 | 9.998360e-01 | No | Yes | 175.23382 | 33.689093 | 1.350642e+02 | 20.130549 | ||
| ENSG00000166788 | 113174 | SAAL1 | protein_coding | Q96ER3 | FUNCTION: Plays a role in promoting the proliferation of synovial fibroblasts in response to proinflammatory stimuli. {ECO:0000269|PubMed:22127701}. | Nucleus;Phosphoprotein;Reference proteome | hsa:113174; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; positive regulation of synoviocyte proliferation [GO:1901647] | 22127701_Overexpression of SPACIA1/SAAL1, a newly identified gene that is involved in synoviocyte proliferation, accelerates the progression of synovitis in mice and humans. | ENSMUSG00000006763 | Saal1 | 1051.83170 | 0.8967590 | -0.1572077623 | 0.12520873 | 1.584537e+00 | 2.081084e-01 | 9.998360e-01 | No | Yes | 935.30273 | 137.220178 | 1.178215e+03 | 133.355096 | ||
| ENSG00000167220 | 84064 | HDHD2 | protein_coding | Q9H0R4 | 3D-structure;Alternative splicing;Coiled coil;Direct protein sequencing;Magnesium;Metal-binding;Reference proteome | hsa:84064; | extracellular exosome [GO:0070062]; enzyme binding [GO:0019899]; metal ion binding [GO:0046872]; phosphatase activity [GO:0016791] | ENSMUSG00000025421 | Hdhd2 | 215.42114 | 1.1203815 | 0.1639901223 | 0.21106156 | 6.088302e-01 | 4.352286e-01 | 9.998360e-01 | No | Yes | 198.79268 | 30.240656 | 1.917196e+02 | 22.706196 | ||||
| ENSG00000167363 | 64122 | FN3K | protein_coding | Q9H479 | FUNCTION: Fructosamine-3-kinase involved in protein deglycation by mediating phosphorylation of fructoselysine residues on glycated proteins, to generate fructoselysine-3 phosphate (PubMed:11016445, PubMed:11522682, PubMed:11975663). Fructoselysine-3 phosphate adducts are unstable and decompose under physiological conditions (PubMed:11522682, PubMed:11975663). Involved in intracellular deglycation in erythrocytes (PubMed:11975663). Involved in the response to oxidative stress by mediating deglycation of NFE2L2/NRF2, glycation impairing NFE2L2/NRF2 function (By similarity). Also able to phosphorylate psicosamines and ribulosamines (PubMed:14633848). {ECO:0000250|UniProtKB:Q9ER35, ECO:0000269|PubMed:11016445, ECO:0000269|PubMed:11522682, ECO:0000269|PubMed:11975663, ECO:0000269|PubMed:14633848}. | ATP-binding;Acetylation;Direct protein sequencing;Kinase;Nucleotide-binding;Reference proteome;Transferase | A high concentration of glucose can result in non-enzymatic oxidation of proteins by reaction of glucose and lysine residues (glycation). Proteins modified in this way, fructosamines, are less active or functional. This gene encodes an enzyme which catalyzes the phosphorylation of fructosamines which may result in deglycation. [provided by RefSeq, Feb 2012]. | hsa:64122; | cytosol [GO:0005829]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; protein-fructosamine 3-kinase activity [GO:0102194]; protein-ribulosamine 3-kinase activity [GO:0102193]; epithelial cell differentiation [GO:0030855]; fructosamine metabolic process [GO:0030389]; fructoselysine metabolic process [GO:0030393]; post-translational protein modification [GO:0043687]; protein deglycation [GO:0036525] | 11975663_involved in the removal of fructosamine residues from hemoglobin in erythrocytes. 15102834_The aim of this work was to identify the fructosamine residues on hemoglobin that are removed as a result of the action of FN3K in intact erythrocytes. 15381090_These data suggest that FN3K and FN3KRP act as protein repair enzymes and are expressed constitutively in human cells independently of some of the variables altered in the diabetic state. 16037310_Enzyme is a constitutive 'housekeeping' gene and that ig plays an important role in cell metabolism, possibly as a deglycating enzyme. 16523184_No significant correlation between FN3K activity and the levels of HbA1c, total glycated haemoglobin (GHb) and haemoglobin fructoselysine residues, either in the normoglycaemic or diabetic group. 16920277_In this paper we propose a resolution of both these quandaries by proposing that fructosamine-6-phosphates are deglycated by phosphorylation to fructosamine-3,6-bisphosphates catalyzed by FN3KRP and/or possibly FN3K. 19834870_G900C polymorphism associates with the level of HbA (1c) and the onset of type 2 diabetes mellitus, but not with either of the diabetic microvascular complications. 19834870_Observational study of gene-disease association. (HuGE Navigator) 20858683_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 21253391_These findings suggest that deglycating enzymes Glyoxalase I and fructosamine-3-kinase may be involved in the malignant transformation of colon mucosa. 21288167_two new mutations and additional variants within the FN3K gene in diabetic patients 23492569_The marginal association of rs1056534 of FN3K is located in exon 6 with diabetic nephropathy progression. 24908234_Report association of rs1056534 and rs3848403 of fructosamine 3-kinase gene with sRAGE in patients with diabetes. 26352355_FN3K could act in concert with other molecular mechanisms and may impact on gene expression and activity of other enzymes involved in deglycation process 27461879_In a multiple regression analysis, FN3K rs1056534, TF polymorphism and presence of diabetes mellitus were predictors for HHV-8 infection. 31398338_FN3K is a targetable modulator of NRF2 activity in cancer. 32636308_A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily. 33208304_FN3K expression in COPD: a potential comorbidity factor for cardiovascular disease. | ENSMUSG00000025175 | Fn3k | 40.40010 | 0.5707718 | -0.8090139832 | 0.47305110 | 2.857772e+00 | 9.093340e-02 | 9.998360e-01 | No | Yes | 29.26183 | 9.337722 | 4.783828e+01 | 11.647818 | |
| ENSG00000167476 | 126306 | JSRP1 | protein_coding | Q96MG2 | FUNCTION: Involved in skeletal muscle excitation/contraction coupling (EC), probably acting as a regulator of the voltage-sensitive calcium channel CACNA1S. EC is a physiological process whereby an electrical signal (depolarization of the plasma membrane) is converted into a chemical signal, a calcium gradient, by the opening of ryanodine receptor calcium release channels. May regulate CACNA1S membrane targeting and activity. {ECO:0000269|PubMed:22927026}. | Endoplasmic reticulum;Membrane;Reference proteome;Sarcoplasmic reticulum | The protein encoded by this gene is involved in excitation-contraction coupling at the sarcoplasmic reticulum. The encoded protein can interact with CACNA1S, CACNB1, and calsequestrin to help regulate calcium influx and efflux in skeletal muscle. [provided by RefSeq, Jul 2012]. | hsa:126306; | sarcoplasmic reticulum [GO:0016529]; sarcoplasmic reticulum membrane [GO:0033017]; regulation of ryanodine-sensitive calcium-release channel activity [GO:0060314]; skeletal muscle contraction [GO:0003009] | 16423849_The results may be explained by a modulatory effect of JP-45 related to its reported in vitro interaction with the dihydropyridine receptor and the SR Ca(2+) binding protein calsequestrin (CSQ). 20442750_Observational study of gene-disease association. (HuGE Navigator) 22927026_Data indicate that the presence of either one of these JP-45 variants decreased the sensitivity of the dihydropyridine receptor DHPR to activation. | ENSMUSG00000020216 | Jsrp1 | 17.61844 | 0.5947417 | -0.7496648654 | 0.70594416 | 9.656797e-01 | 3.257600e-01 | 9.998360e-01 | No | Yes | 10.93470 | 3.328667 | 2.098815e+01 | 5.116434 | |
| ENSG00000167523 | 124045 | SPATA33 | protein_coding | Q96N06 | Alternative splicing;Cytoplasm;Nucleus;Phosphoprotein;Reference proteome | hsa:124045; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634] | 30098056_the current study illustrated that alterations in SPATA33 gene, at least those found in this study, may not impair spermatogenesis in patients with nonobstructive azoospermia. | 829.82910 | 1.1338514 | 0.1812315514 | 0.12661818 | 2.046320e+00 | 1.525746e-01 | 9.998360e-01 | No | Yes | 1032.50354 | 82.077455 | 8.783199e+02 | 55.429376 | |||||
| ENSG00000167526 | 6137 | RPL13 | protein_coding | P26373 | FUNCTION: Component of the ribosome, a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell (PubMed:31630789, PubMed:23636399). The small ribosomal subunit (SSU) binds messenger RNAs (mRNAs) and translates the encoded message by selecting cognate aminoacyl-transfer RNA (tRNA) molecules (Probable). The large subunit (LSU) contains the ribosomal catalytic site termed the peptidyl transferase center (PTC), which catalyzes the formation of peptide bonds, thereby polymerizing the amino acids delivered by tRNAs into a polypeptide chain (Probable). The nascent polypeptides leave the ribosome through a tunnel in the LSU and interact with protein factors that function in enzymatic processing, targeting, and the membrane insertion of nascent chains at the exit of the ribosomal tunnel (Probable). As part of the LSU, it is probably required for its formation and the maturation of rRNAs (PubMed:31630789). Plays a role in bone development (PubMed:31630789). {ECO:0000269|PubMed:23636399, ECO:0000269|PubMed:31630789, ECO:0000305}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Disease variant;Dwarfism;Isopeptide bond;Phosphoprotein;Reference proteome;Ribonucleoprotein;Ribosomal protein;Ubl conjugation | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 60S subunit. The protein belongs to the L13E family of ribosomal proteins. It is located in the cytoplasm. This gene is expressed at significantly higher levels in benign breast lesions than in breast carcinomas. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. [provided by RefSeq, Jul 2011]. | hsa:6137; | cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; cytosolic ribosome [GO:0022626]; endoplasmic reticulum [GO:0005783]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleus [GO:0005634]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; blastocyst development [GO:0001824]; bone development [GO:0060348]; cytoplasmic translation [GO:0002181]; translation [GO:0006412] | 16786168_Ribosomal protein L13 plays an essential role in the progression of some gastrointestinal malignancies. 19034380_This protein has been found differentially expressed in the temporal lobe from patients with schizophrenia. 31625562_Using a combination of human genetics and cardiac model systems, RPL13 gene was identified as a new candidate for congenital heart pathogenesis. 31630789_The identified RPL13 variants cause a human ribosomopathy defined by a rare skeletal dysplasia. 32916022_Novel RPL13 Variants and Variable Clinical Expressivity in a Human Ribosomopathy With Spondyloepimetaphyseal Dysplasia. | ENSMUSG00000000740 | Rpl13 | 164397.10861 | 0.8384018 | -0.2542863482 | 0.11852240 | 4.560132e+00 | 3.272456e-02 | 9.998360e-01 | No | Yes | 167281.30827 | 31583.758939 | 1.847520e+05 | 26869.702467 | |
| ENSG00000167635 | 7705 | ZNF146 | protein_coding | Q15072 | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | hsa:7705; | cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; heparin binding [GO:0008201]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 15838871_OZF overexpression in tumours may alter the balance between hRap1 and other telomeric proteins; therefore OZF function may be linked to telomere regulation 15881673_OZF interacts with UBC9, the E2 enzyme involved in the covalent conjugation of the small ubiquitin-like modifier 1 (SUMO-1). 32267586_OZF is a Claspin-interacting protein essential to maintain the replication fork progression rate under replication stress. 33394291_LncRNA KCNQ1OT1 acts as miR-216b-5p sponge to promote colorectal cancer progression via up-regulating ZNF146. 35100360_ZNF146/OZF and ZNF507 target LINE-1 sequences. | ENSMUSG00000037029 | Zfp146 | 1748.46762 | 0.8087296 | -0.3062707289 | 0.12406882 | 5.955266e+00 | 1.467339e-02 | 9.998360e-01 | No | Yes | 1686.83742 | 283.369027 | 1.884279e+03 | 243.976376 | |||
| ENSG00000167733 | 374875 | HSD11B1L | protein_coding | Q7Z5J1 | Alternative splicing;NADP;Oxidoreductase;Reference proteome;Secreted;Signal | This gene is a member of the hydroxysteroid dehydrogenase family. The encoded protein is similar to an enzyme that catalyzes the interconversion of inactive to active glucocorticoids (e.g. cortisone). Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jun 2012]. | hsa:374875; | extracellular region [GO:0005576]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; oxidoreductase activity [GO:0016491] | 19436836_cloning, purification, and characterization of SCDR10B; highly expressed in brain (especially in hippocampal neurons); up-regulated in lung cancer cell lines and lung cancer tissue | 294.78596 | 0.9828629 | -0.0249379130 | 0.19645053 | 1.673240e-02 | 8.970777e-01 | 9.998360e-01 | No | Yes | 304.19666 | 41.650358 | 2.921799e+02 | 32.081972 | ||||
| ENSG00000168061 | 29901 | SAC3D1 | protein_coding | A6NKF1 | FUNCTION: Involved in centrosome duplication and mitotic progression. {ECO:0000250}. | Alternative splicing;Cell cycle;Cell division;Cytoplasm;Cytoskeleton;Mitosis;Phosphoprotein;Reference proteome | Mouse_homologues mmu:66406; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; spindle [GO:0005819]; cell division [GO:0051301]; centrosome duplication [GO:0051298]; spindle assembly [GO:0051225] | 18838617_SHD1 is a novel cytokine-inducible negative feedback regulator of STAT5a. 30353105_In this study, 3 independent cohorts (GSE10186, n = 80; TCGA, n = 330 and ICGC, n = 237) were used to assess SAC3D1 as a biomarker, which demonstrated SAC3D1 overexpression in hepatocellular carcinoma tissues when compared to the matched normal tissues. 32202298_Distinct effects on mRNA export factor GANP underlie neurological disease phenotypes and alter gene expression depending on intron content. 32319600_Upregulated expression of SAC3D1 is associated with progression in gastric cancer. | ENSMUSG00000024790 | Sac3d1 | 3072.50960 | 0.9873364 | -0.0183863748 | 0.13125513 | 2.047034e-02 | 8.862313e-01 | 9.998360e-01 | No | Yes | 3191.75701 | 476.060210 | 3.036105e+03 | 350.278416 | ||
| ENSG00000168214 | 3516 | RBPJ | protein_coding | Q06330 | FUNCTION: Transcriptional regulator that plays a central role in Notch signaling, a signaling pathway involved in cell-cell communication that regulates a broad spectrum of cell-fate determinations. Acts as a transcriptional repressor when it is not associated with Notch proteins. When associated with some NICD product of Notch proteins (Notch intracellular domain), it acts as a transcriptional activator that activates transcription of Notch target genes. Probably represses or activates transcription via the recruitment of chromatin remodeling complexes containing histone deacetylase or histone acetylase proteins, respectively. Specifically binds to the immunoglobulin kappa-type J segment recombination signal sequence. Binds specifically to methylated DNA (PubMed:21991380). Binds to the oxygen responsive element of COX4I2 and activates its transcription under hypoxia conditions (4% oxygen) (PubMed:23303788). Negatively regulates the phagocyte oxidative burst in response to bacterial infection by repressing transcription of NADPH oxidase subunits (By similarity). {ECO:0000250|UniProtKB:P31266, ECO:0000269|PubMed:21991380, ECO:0000269|PubMed:23303788}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Disease variant;Notch signaling pathway;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation | The protein encoded by this gene is a transcriptional regulator important in the Notch signaling pathway. The encoded protein acts as a repressor when not bound to Notch proteins and an activator when bound to Notch proteins. It is thought to function by recruiting chromatin remodeling complexes containing histone deacetylase or histone acetylase proteins to Notch signaling pathway genes. Several transcript variants encoding different isoforms have been found for this gene, and several pseudogenes of this gene exist on chromosome 9. [provided by RefSeq, Oct 2013]. | hsa:3516; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; MAML1-RBP-Jkappa- ICN1 complex [GO:0002193]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; protein N-terminus binding [GO:0047485]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific DNA binding [GO:0043565]; angiogenesis [GO:0001525]; aortic valve development [GO:0003176]; arterial endothelial cell fate commitment [GO:0060844]; atrioventricular canal development [GO:0036302]; auditory receptor cell fate commitment [GO:0009912]; B cell differentiation [GO:0030183]; blood vessel endothelial cell fate specification [GO:0097101]; blood vessel lumenization [GO:0072554]; blood vessel remodeling [GO:0001974]; cardiac left ventricle morphogenesis [GO:0003214]; club cell differentiation [GO:0060486]; defense response to bacterium [GO:0042742]; dorsal aorta morphogenesis [GO:0035912]; endocardium morphogenesis [GO:0003160]; epidermal cell fate specification [GO:0009957]; epithelial to mesenchymal transition [GO:0001837]; epithelial to mesenchymal transition involved in endocardial cushion formation [GO:0003198]; hair follicle maturation [GO:0048820]; humoral immune response [GO:0006959]; inflammatory response to antigenic stimulus [GO:0002437]; keratinocyte differentiation [GO:0030216]; labyrinthine layer blood vessel development [GO:0060716]; myeloid dendritic cell differentiation [GO:0043011]; negative regulation of cell differentiation [GO:0045596]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of ossification [GO:0030279]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; Notch signaling involved in heart development [GO:0061314]; Notch signaling pathway [GO:0007219]; outflow tract morphogenesis [GO:0003151]; pituitary gland development [GO:0021983]; positive regulation of BMP signaling pathway [GO:0030513]; positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment [GO:1901297]; positive regulation of cardiac muscle cell proliferation [GO:0060045]; positive regulation of cell proliferation involved in heart morphogenesis [GO:2000138]; positive regulation of ephrin receptor signaling pathway [GO:1901189]; positive regulation of ERBB signaling pathway [GO:1901186]; positive regulation of gene expression [GO:0010628]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription from RNA polymerase II promoter in response to hypoxia [GO:0061419]; positive regulation of transcription of Notch receptor target [GO:0007221]; pulmonary valve development [GO:0003177]; regulation of timing of cell differentiation [GO:0048505]; regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation [GO:0003256]; sebaceous gland development [GO:0048733]; secondary heart field specification [GO:0003139]; somatic stem cell population maintenance [GO:0035019]; somitogenesis [GO:0001756]; ventricular septum morphogenesis [GO:0060412]; ventricular trabecula myocardium morphogenesis [GO:0003222] | 12374742_SHARP is a novel component of the HDAC corepressor complex, recruited by RBP-Jkappa to repress transcription of target genes in the absence of activated Notch. 12832621_RBP-J kappa-mediated repression is therefore not essential for establishment of latent KHSV infection, but the RTA-mediated redirection of RBP-J kappa activity from repression to activation is critical for lytic viral replication. 14570916_Ku antigen interacts with RBP-Jkappa and NF-kappaB p50 may act as a positive regulator of p50 expression in gastric cancer AGS cells. 14645224_C promoter-binding factor 1 binding is required for Notch-1-mediated repression of activator protein-1 14701863_RBP-Jkappa activated a full transcriptional response but only demonstrated partial antiapoptotic activity 15187023_The ankyrin repeats were also the only domain required for up-regulation of RBP-Jkappa-dependent gene expression 15194757_regulates Kaposi's sarcoma-associated herpesvirus (KSHV) K14/vGPCR transcripts; this suggests the possibility that other modulators of Notch signaling might be able to induce expression of this RNA outside the context of lytic KSHV replication. 15987768_Notch promotes changes in hVSMC phenotype via activation of CBF-1/RBP-Jkappa-dependent pathways in vitro and contributes to the phenotypic response of VSMCs to cyclic strain-induced changes in VSMC differentiation. 16287852_a corepressor complex containing CtIP/CtBP facilitates RBP-Jkappa/SHARP-mediated repression of Notch target genes 16354684_P48 interacts with the RBP-L and RBP-J subunits primarily through two short conserved tryptophan-containing motifs, similar to the motif of the Notch intracellular domain (NotchIC) that interacts with RBP-J. 16378632_c-Myc can cooperate with E6/E7 in epithelial transformation and can substitute for CBF1-dependent signals generated by Notch1. 16399505_Notch/Rbp-j signaling prevents premature differentiation of pancreatic progenitor cells into endocrine and ductal cells during early development of the transgenic pancreas. 16439682_findings show that induction of FcRH5 by Epstein-Barr virus nuclear antigen 2 is strictly CBF1 dependent 16530044_Results report the crystal structure of a Notch transcriptional activation complex containing the ankyrin domain of human Notch1, the transcription factor CSL on cognate DNA, and a polypeptide from the coactivator Mastermind-like-1 (MAML-1). 16873269_EBNA2 trans-activates bfl-1, which requires CBF1 (or RBP-J kappa). 17055026_RTA activates the Kaposi's sarcoma-associated herpesvirus K8 promoter through an indirect binding mechanism, i.e. being recruited to the K8 promoter through interaction with RBP-Jkappa bound to an RBP-Jkappa motif in the promoter 17070841_These results support an emerging molecular mechanism for the displacement of co-repressors from DNA-bound CSL by intracellular domain of the Notch receptor . 17245125_The phosphatase and tensin homolog (PTEN) gene is a direct target of Notch-1 signal transduction, through binding of the Notch-activated transcription factor CBF-1 to the PTEN minimal promoter. 17284587_A number of Notch-regulated targets are characterized by paired CSL-binding sites in a head-to-head arrangement; cooperative formation of dimeric Notch transcription complexes on promoters with paired sites is required to activate transcription 17434929_Transcription factor Ying Yang 1 (YY1) indirectly regulates the C promoter-binding factor 1 (CBF1)-dependent Notch1 signaling via direct interaction with the Notch1 receptor. 17513037_an important relationship exists between zinc and the Notch1 signaling pathway, and that this relationship is intimately involved with the cytoplasmic retention of Notch and RBP-Jk 17513780_The incorporation of RBP-J kappa into CCAAT-enhancer-binding protein (CEBP) zeta complexes in human hepatoma Hep3B cells is solely to support the binding of CEBP zeta to the CEBP site. 18155729_the RBP-Jkappa-associated domain of Notch increases the effective concentration of the ankyrin domain for its binding site on CSL, enabling docking of the ankyrin domain and subsequent recruitment of the Mastermind-like coactivator. 18239137_Present a novel mechanism by which a balance between Notch-1/-2/-4 signaling, via CBF-1, and HRT-1/-2 activity determines the expression of smooth muscle differentiation markers including actin. 18332109_ETO is part of the endogenous RBP-Jkappa-containing corepressor complex 19147558_In gliomas, the TNC gene is transactivated by Notch2 in an RBPJk-dependent manner mediated by an RBPJk binding element in the TNC promoter. 19237563_EBNA3C regulation of transcription through RBP-Jkappa is critical to maintaining lymphoblastoid cell growth 19240061_Observational study of gene-disease association. (HuGE Navigator) 19247952_Studies indicate that the mechanisms that lead to Notch activity in the receiving cell include the cleavage of Notch at the cell membrane and the assembly of a nuclear complex with the transcription factors CSL. 19487031_Both CBF1 and C/EBP-beta bind the CR2 promoter in B cells raising the possibility that these factors facilitate or respond to alterations in chromatin structure to control the timing and/or level of CR2 transcription. 19776126_The authors confirmed that EBNA3C upregulates TCL1 and discovered that EBNA3C upregulates TCL1 through RBP-Jkappa, indicating a central role for EBNA3C interaction with RBP-Jkappa in mediating cell gene transcription. 19838210_Data support a model in which Notch-1 can activate the transcription of ERalpha-target genes via IKKalpha-dependent cooperative chromatin recruitment of Notch-CSL-MAML1 and ERalpha, which promotes the recruitment of p300. 19969318_Inhibition of methylation in EBV-infected cells results in reduced expression of the EBNA2-regulated viral gene LMP1, evidencing that methylation is a prerequisite for DNA-binding by EBNA2 via association with the transcription factor RBPJkappa. 20006367_The interactions of RBPJ with ORF47 and ORF50 human herpesvirus 8 proteins in transactivation are reported. 20028974_mechanism by which the PhiW PhiP motif of RAM and EBNA2 compete with one another for binding at the hydrophobic pocket of the beta-trefoil domain (BTD) of CSL using overlapping but specific interactions that are unique to each BTD ligand. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20453842_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20511547_Notch target gene activator RBP-J kappa transgene plays no role in Notch target gene repression in Ikaros double-positive thymocytes during leukemia development. 21440926_These results indicate that EBNA3 proteins interact with multiple RBP/CSL domains, but only N-terminal domain interactions are required for lymphoblastoid cell line growth. 21507979_The RBP-Jkappa binding site within the KSHV LANA promoter is crucial for HHV8 latency during early primary infection. 21518914_data strongly support a model in which EBNA2 association with NCoR-deficient RBPJ enhances transcription and EBNALP dismisses NCoR and RBPJ repressive complexes from enhancers 21558417_Collectively, our results demonstrate that APP intracellular domain functions as a negative regulator in Notch1 signaling through the promotion of Notch1 and RBP-Jk protein degradation 21737748_there is a conserved network of cis-regulatory factors that interacts with Notch1 to regulate gene expression in TLL cells, as well as unique classes of divergent RBPJ-only sites that also likely regulate transcription 21880753_DNA binding and tetramerization mutants of Rta fail to stimulate RBP-Jk binding to Kaposi's sarcoma-associated herpesvirus DNA. 21991380_RBP-J binds methylated DNA in the context of a mutated RBP-J consensus motif. 22253595_RBP-Jkappa binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation. 22279105_Bacterial translocation is evident in the colonic mucosa of transgenic RBP-J(DeltaIEC) mice before the onset of colitis, suggesting attenuated epithelial barrier functions in these mice. 22302987_results indicate that PS2 modulates the degradation of RBP-Jk through phosphorylation by p38 MAPK. 22325781_Association of CSL with NICD exerts remarkably little effect on the exchange kinetics of the ANK domain, whereas MAML1 binding greatly retards the exchange kinetics of ANK repeats 2-3. 22366521_Three consensus RBP-Jkappa-binding sites found in the ORF46 promoter are critical for the binding of RBP-Jkappa protein and conferring the ORF50 responsiveness. 22792167_RBPJ regulates rhabdomyosarcoma growth. 22883147_Unique mutations in RBPJ lead to impaired DNA binding in cases of Adams-Oliver syndrome. 22915591_Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. 23365452_Authors report that human papillomavirus type 8 E6 subverts NOTCH activation during keratinocyte differentiation by inhibiting RBPJ/MAML1 transcriptional activator complexes at NOTCH target DNA. 23696732_CBF1/CSL acts as a global hub which is used by the virus to coordinate the lytic cascade. 23775085_Data indicate that cAMP activates Notch signaling and increases the expression of recombinant recognition sequence binding protein at the Jkappa site (RBP-J) and transducin-like enhancer of Split (TLE). 24068939_Data suggest that, in B-lymphocytes infected with Epstein-Barr virus (EBV), EBNA3A (EBV nuclear antigen 3A) binds to CBF1-occupied intergenic enhancers located between CXCL10 and CXCL9 and displaces the transactivator EBNA2. 24549417_missense mutations and frameshift deletions identified in leukemia patients 24696491_KAP1, RBP-Jkappa, and HIF-1alpha play an essential role in KSHV pathogenesis. 25014791_Single nucleotide polymorphisms in RBPJ, IL1R1, REV3L, TRAF3IP2, IRF1 and ICOS showed association with rheumatoid arthritis in black South Africans. 25324571_DNA methylation-dependent binding of RBPJ to a GC repressor element can negatively regulate smooth muscl myosin heavy chain promoter activity and can inhibit marker gene expression in phenotypically modulated cells 25445601_RBP-J-interacting and tubulin-associated (RITA) mediates the nuclear export of RBP-J to tubulin fibers and downregulates Notch-mediated transcription. 25512468_Data indicate that RBPJ protein depletion induces Notch target genes in a Notch-independent fashion. 25589461_Our data thus suggest that shRNA intervention of RBPJ expression could be a promising therapeutic approach for treating human lung cancer. 26202358_data suggest that shRNA intervention of RBPJ expression could be a promising therapeutic approach for treating human prostate cancer 26302407_Cancer-associated fibroblast activation-stromal co-evolution is under convergent CSL-p53 control. 26496776_The role of CSL-dependent and independent Notch signaling pathways in cell apoptosis is described in normal tissue homeostasis and in tumorigenesis. (Review) 26604133_RBPJ polymorphism [rs874040(CC) allele] skews memory T cells toward a pro-inflammatory phenotype involving Notch signaling, thus increasing the susceptibility to develop rheumatoid arthritis. 26655998_Suggest that hypoxia promotes SMO transcription through upregulation of MAML3 and RBPJ to induce proliferation, invasiveness and tumorigenesis in pancreatic cancer. 26719268_These results support a model in which EBNA2 and EBNA3s compete for distinct subsets of RBPJ sites to regulate cell genes and where EBNA3 subset specificity is determined by interactions with other cell transcription factors. 26735629_our findings reconcile the 2 biological events and point to a multistep process of CAF activation under convergent CSL and p53 control. Activation of p53 provides a failsafe mechanism against consequences of compromised CSL activity in stromal cells. 27066863_we report that genetic removal of CSL in breast tumor cells caused accelerated growth of xenografted tumors. Loss of CSL unleashed a hypoxic response during normoxic conditions, manifested by stabilization of the HIF1alpha protein and acquisition of a polyploid giant-cell, cancer stem cell-like, phenotype. 27163456_The present findings indicate that p53, in turn, decreases CSL expression, which can serve to enhance p53 activity in acute DNA damage response of cells. 27322055_RBPJ links MYC and transcriptional control through CDK9 in brain tumors, providing potential nodes of fragility for therapeutic intervention, potentially distinct from NOTCH 27466498_These results suggest that PSK suppresses Hedgehog signaling through down-regulation of MAML3 and RBPJ transcription under hypoxia, inhibiting the induction of a malignant phenotype in pancreatic cancer. Our results may lead to development of new treatments for refractory pancreatic cancer using PSK as a Hedgehog inhibitor 27489358_Notch1 signaling plays an important role in the maintenance of the cancer stem-like phenotype in diffuse type gastric cancer through an RBP-Jkappa dependent pathway; inhibiting Notch1 signaling could be an effective therapy against CD133 positive diffuse type gastric cancers 27794430_RBP-J mediated by miR-133a probably contributed to the regulation of DCs maturation and activation in osteosarcoma 27926858_We show that GIT1, which also contains an ANK domain, inhibits the Notch1-Dll4 signaling pathway by competing with Notch1 ANK domain for binding to RBP-J in stalk cells 28487372_structural and biophysical studies demonstrate that RITA binds RBP-J similarly to the RAM (RBP-J-associated molecule) domain of Notch, our biochemical and cellular assays suggest that RITA interacts with additional regions in RBP-J. 28571041_Mean CBF1 expression is significantly increased in isocitrate dehydrogenase 1 (IDH1) R132H mutant glioblastoma. Hypoxic regions of glioblastoma have higher CBF1 activation and exposure to low oxygen can induce its expression in glioma cells in vitro. 28877478_The ULK3 Kinase Is Critical for Convergent Control of Cancer-Associated Fibroblast Activation by CSL and GLI 29030483_RBPJ interacts with L3MBTL3 to promote repression of Notch signaling via histone demethylase KDM1A. 29242273_data, taken together, indicate that RBPJ regulates inflammation during endometrial repair, which is essential for future pregnancy potential, and its dysregulation may serve as an unidentified contributor to unexplained recurrent pregnancy loss. 29757189_histone demethylase KDM6B is a direct CSL-negative target, with inverse roles of CSL in HKC and SCC proliferative capacity, tumorigenesis, and tumor-associated inflammatory reaction. CSL/KDM6B protein expression could be used as a biomarker of SCC development and indicator of cancer treatment. 29931229_Data suggest that RBP-Jkappa protein (RBPJ) and serum response factor (SRF) cooperate to regulate gene expression in aortic smooth muscle cells (SMC). 29971915_Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. 30030832_The DNA-binding protein CSL is the centrepiece of transcriptional regulation in the Notch pathway, acting as a molecular hub for interactions with either corepressors or coactivators to repress or activate, respectively, transcription. 30061220_RBPJ and MAML3 could be new therapeutic targets for SCLC. 30075508_Variants rs2270226 and rs2077777 in the RBPJ gene were associated with the risk of cerebral infarction diseases in the Chinese Han population. 30157580_this report tried to address the molecular basis for the direct interaction between CSL and SMRT. 30213224_Notch signaling through recombination signal binding protein for immunoglobulin kappa J region (RBPJ) controls both ovarian progesterone receptor and glucose transporter SLC2A1 expression during decidualization 30231994_CSL associates with p62/SQSTM1, which is required for CSL down-modulation by autophagy. Increasing cellular CSL levels stabilizes p62 and down-modulates the autophagic process. 30254149_the cyclin F-RBPJ axis has a key role in response to metabolic stress in cancer cells 30642633_SNW1 interacts with RBPJ to regulate the Notch signaling pathway in neuroblastoma 30856484_LANA upregulates let-7a and its primary transcripts in parallel with its reduction of RBPJ expression. 31467287_CSL controls telomere maintenance and genome stability in human dermal fibroblasts. 32130980_Circular RNA circ-0060428 may improve the proliferation and survival of osteosarcoma cells by sponging miR-375 to upregulate RBPJ expression 32271392_Regulation of Janus Kinase 2 by an Inflammatory Bowel Disease Causal Non-coding Single Nucleotide Polymorphism. 32567455_A Novel Flow Cytometric Assay to Identify Inhibitors of RBPJ-DNA Interactions. 32961937_CBF-1 Promotes the Establishment and Maintenance of HIV Latency by Recruiting Polycomb Repressive Complexes, PRC1 and PRC2, at HIV LTR. 33034023_Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response. 33311552_Merkel cell carcinoma-derived exosome-shuttle miR-375 induces fibroblast polarization by inhibition of RBPJ and p53. 33387451_Ageing promotes early T follicular helper cell differentiation by modulating expression of RBPJ. 33389283_LncRNA FTX Promotes Colorectal Cancer Cells Migration and Invasion by miRNA-590-5p/RBPJ Axis. 34655278_Colon cancer cells secreted CXCL11 via RBP-Jkappa to facilitated tumour-associated macrophage-induced cancer metastasis. | ENSMUSG00000039191 | Rbpj | 726.96771 | 1.1415344 | 0.1909743573 | 0.14517982 | 1.774840e+00 | 1.827842e-01 | 9.998360e-01 | No | Yes | 758.69036 | 171.090829 | 7.314629e+02 | 127.025088 | |
| ENSG00000168286 | 57215 | THAP11 | protein_coding | Q96EK4 | FUNCTION: Transcriptional repressor that plays a central role for embryogenesis and the pluripotency of embryonic stem (ES) cells. Sequence-specific DNA-binding factor that represses gene expression in pluripotent ES cells by directly binding to key genetic loci and recruiting epigenetic modifiers (By similarity). {ECO:0000250}. | 3D-structure;Coiled coil;Cytoplasm;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation;Triplet repeat expansion;Zinc;Zinc-finger | The protein encoded by this gene contains a THAP domain, which is a conserved DNA-binding domain that has striking similarity to the site-specific DNA-binding domain (DBD) of Drosophila P element transposases. [provided by RefSeq, Jul 2008]. | hsa:57215; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 19008924_Results suggest that THAP11 functions as a cell growth suppressor by negatively regulating the expression of c-Myc. 21400515_the induced THAP11 might be one of transcriptional regulators of c-Myc expression in CML cell. 22371484_identify THAP11 as a transcriptional regulator differentially expressed in human colon cancer 22673507_In HCC patients, the expression of THAP11 mRNA significantly correlated with PCBP1 mRNA expression. Our results suggest a novel role of THAP11 in CD44 alternative splicing and hepatoma invasion 23539139_HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. 24637716_that THAP11 reversibly regulated erythroid and megakaryocytic differentiation. 25437553_THAP11, ZNF143, and HCF-1 form a mutually dependent complex on chromatin, which is independent of E2F occupancy. 26975212_the crystal structure of the C-ter region of THAP11 forms a left-handed parallel homo-dimeric coiled-coil structure possessing several unusual features. 27576892_The M4 motif (ACTAYRNNNCCCR) is a functional regulatory bipartite cis-element, which engages a THAP11/HCF-1 complex via binding to the ACTAYR module, while the CCCRRNRNRC subsequence part constitutes a binding platform for Ikaros and NFKB1 28449119_We sequenced THAP11 by Sanger sequencing and discovered a potentially pathogenic, homozygous variant in THAP11, c.240C > G (p.Phe80Leu). Further, we provide functional data in model organisms that suggests that both HCFC1 and THAP11 are essential for normal brain development and neural precursor differentiation. 29269392_A transcriptional repressor network including THAP domain containing 11 protein (THAP11) was identified and negatively regulates endogenous PARKIN abundance. 31905202_THAP11F80L exhibited a strong effect on association with the MMACHC promoter and led to a decrease in MMACHC gene transcription, suggesting that the THAP11F80L mutation is directly responsible for the observed cobalamin disorder 31969497_THAP11 inhibited growth of esophageal cancer cells 32908912_THAP11 Functions as a Tumor Suppressor in Gastric Cancer through Regulating c-Myc Signaling Pathways. | ENSMUSG00000036442 | Thap11 | 3165.32685 | 1.0946907 | 0.1305233449 | 0.10055444 | 1.700433e+00 | 1.922313e-01 | 9.998360e-01 | No | Yes | 3488.19181 | 369.746180 | 3.036361e+03 | 249.669068 | |
| ENSG00000168487 | 649 | BMP1 | protein_coding | P13497 | FUNCTION: Metalloprotease that plays key roles in regulating the formation of the extracellular matrix (ECM) via processing of various precursor proteins into mature functional enzymes or structural proteins (PubMed:33206546). Thereby participates in several developmental and physiological processes such as cartilage and bone formation, muscle growth and homeostasis, wound healing and tissue repair (PubMed:33169406, PubMed:32636307). Roles in ECM formation include cleavage of the C-terminal propeptides from procollagens such as procollagen I, II and III or the proteolytic activation of the enzyme lysyl oxidase LOX, necessary to formation of covalent cross-links in collagen and elastic fibers (PubMed:31152061, PubMed:33206546). Additional substrates include matricellular thrombospondin-1/THBS1 whose cleavage leads to cell adhesion disruption and TGF-beta activation (PubMed:32636307). {ECO:0000269|PubMed:31152061, ECO:0000269|PubMed:32636307, ECO:0000269|PubMed:33169406, ECO:0000269|PubMed:33206546}.; FUNCTION: [Isoform BMP1-3]: Plays an important role in bone repair by acting as a coactivator of BMP7. {ECO:0000269|PubMed:21453682}. | 3D-structure;Alternative splicing;Calcium;Chondrogenesis;Cleavage on pair of basic residues;Cytokine;Developmental protein;Differentiation;Disease variant;Disulfide bond;EGF-like domain;Extracellular matrix;Glycoprotein;Golgi apparatus;Growth factor;Hydrolase;Metal-binding;Metalloprotease;Methylation;Osteogenesis;Osteogenesis imperfecta;Protease;Reference proteome;Repeat;Secreted;Signal;Zinc;Zymogen | This gene encodes a protein that is capable of inducing formation of cartilage in vivo. Although other bone morphogenetic proteins are members of the TGF-beta superfamily, this gene encodes a protein that is not closely related to other known growth factors. This gene is expressed as alternatively spliced variants that share an N-terminal protease domain but differ in their C-terminal region. [provided by RefSeq, Aug 2008]. | hsa:649; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; vesicle [GO:0031982]; calcium ion binding [GO:0005509]; cytokine activity [GO:0005125]; growth factor activity [GO:0008083]; identical protein binding [GO:0042802]; metalloendopeptidase activity [GO:0004222]; metallopeptidase activity [GO:0008237]; peptidase activity [GO:0008233]; serine-type endopeptidase activity [GO:0004252]; zinc ion binding [GO:0008270]; cartilage condensation [GO:0001502]; cell differentiation [GO:0030154]; collagen fibril organization [GO:0030199]; multicellular organism development [GO:0007275]; ossification [GO:0001503]; positive regulation of cartilage development [GO:0061036]; proteolysis [GO:0006508]; skeletal system development [GO:0001501] | 11986329_bone morphogenetic protein-1 (BMP-1), which exhibits procollagen C-proteinase activity, cleaves the C-terminal propeptide from human procollagen VII 12218058_Post-translational modification is required for secretion and stability of the protein. 12637537_the minimal domain structure for PCP activity is considerably shorter than expected and comprises the metalloproteinase domain and the CUB1 and CUB2 domains of BMP-1 12637569_Pro-BMP-1 is cleaved in the trans-Golgi network 15225209_Dermal wound healing in red Duroc pigs show unique mRNA expression of HSP47,BMP-1,TIMP1-3 and hypercontracted,hyperpigmented scars. 15591058_cleaves LG3 from recombinant endorepellin at the physiologically relevant site and cleaves LG3 from endogenous perlecan in cultured mouse and human cells 15817489_chordinase activity of BMP1 is not enhanced by PCPE-1 16507574_tolloid-like 1 binds procollagen C-proteinase enhancer protein 1 and differs from bone morphogenetic protein 1 in the functional roles of homologous protein domains 17071617_bone morphogenetic protein 1 is inhibited by native and altered forms of alpha2-macroglobulin 17255107_the BMP1 prodomain specifically binds and regulates signaling by BMP2 and BMP4 17407447_By mutating residues flanking the cleavage site of collagen type V alpha 1, we showed that the aspartate residue at position P2' is essential for BMP-1 activity. 17516847_data support the concept that the C-terminal domains of BMP1 are important for substrate recognition and for controlling and restricting its proteolytic activity via exosite binding 17548836_BMP1 processes PRL to a 17-kDa anti-angiogenic factor. 18056036_vascular Bmp Msx2 Wnt signaling and oxidative stress have roles in arterial calcification [review] 18349123_Expression of BMP1, BMP6, BMP7, and BMP-receptor 2 was significantly increased in advanced stages of myelofibrosis compared and enhanced levels of BMP6 expression were already evident in prefibrotic stages of primary myelofibrosis. 18824173_The crystal structures of the protease domains of human BMP-1 and the closely related Tolloid-like protease 1 (TLL-1), are reported. 19305382_BMP-1 expression was significantly higher in thyroid tumors with psammoma bodies or with stromal calcification. 19323056_Regulation of alternative splicing of mRNA for procollagen C-endopeptidase in leiomyomas and myometrium depends mainly on the hormonal status of women 19617627_FN binds BMP1-like proteinases in vivo and that FN is an important determinant of the in vivo activity levels of BMP1-like proteinases. 19801683_Data show that only those containing both PCPE1 CUB1 and CUB2 were capable of enhancing BMP-1 activity and binding to a mini-procollagen substrate with nanomolar affinity. 20026052_Three isoforms of BMP1 ranging from the shortest BMP1-5 to the longest (mTLD, inefficient at processing procollagen in vitro) were all shown to be capable of removing the highly conserved propeptides from both proDCN. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20955454_Observational study of gene-disease association. (HuGE Navigator) 21258932_Disruption of BMPR1A-mediated BMP1 signalling during the narrow window of early embryogenesis may interfere with normal VBW formation, causing omphalocele phenotype in the Cd chick model. 21415150_Circulating bone morphogenetic protein 1-3 isoform increases renal fibrosis. 21453682_BMP1-3 is a novel systemic regulator of bone repair. 21697095_Bone morphogenetic protein-1 processes insulin-like growth factor-binding protein 3. 21897187_Excluding anterior cervical fusions, there are no significant differences between spinal fusion procedures with and without BMP-associated overall complications. 21946044_High expression of BMP pathway genes are associated with atypical teratoid/rhabdoid tumors. 22052668_We conclude that BMP1 is an additional gene mutated in autosomal recessive osteogenesis imperfecta (AR-OI). 22482805_The molecular and cellular bases of BMP1-dependent osteogenesis were defined. The importance of BMP1 for bone formation and stability were shown in humans and zebrafish. 22860217_Controlling inhibition of bone morphogenetic protein (BMP1) modulates the number of SOX1 expressing cells, whereas PAX6, another neural precursor marker, remains the same. 23584484_miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27kip1 23640157_Sequence analysis of BMP1 genes did not reveal any putative mutations for hyperostosis cranialis interna to chromosome 8p21 24042462_Loss of bone morphogenetic protein is associated with prostate cancer. 24984282_High BMP1 expression is associated with type-1 diabetes. 25158199_mutations of the DSP-PP P4 to P4' cleavage site can block, impair or accelerate dentin sialoprotein phosphophoryn cleavage, and suggest that its Bone morphogenic protein 1 cleavage site is conserved in order to regulate its cleavage efficiency 25214535_Frequent bone fracture in children is cause by BMP1-1 deficiency. 25402547_Two novel variants in the BMP1 gene: c.808A>G and c.1297G>T care associated with osteogenesis imperfecta. 25656619_study thus highlights the severe and progressive nature of BMP1-associated OI in adults and broadens insights into the functional consequences of BMP1/mTLD-deficiency on ECM organization. 25701650_Data indicate that BMP-1 can simultaneously trigger matrix assembly and boost the synthesis of matrix proteins via a direct effect on growth factors in the contexts of development, growth and tissue repair. [review] 25944709_BMP-1 accelerates the connective tissue growth factor production dependently on cellular internalization in human dental pulp cells, indicating a novel property of BMP-1 which potentially enhances bone-like reparative dentin formation. 26592459_Studies indicate crosstalk between Notch receptor and Wnt protein, Hedgehog protein, hypoxia and transforming growth factor beta (TGFbeta)/bone morphogenetic protein (BMP) pathways. 26944735_a previously unknown O-glycosylation site and Asn-hydroxylation site, indicating a novel feature of BMP-1 in the EGF domain. The study clearly outlines the benefit of in-depth characterization of overexpressed proteins to deduce important protein modifications. 27576954_Given the association of BMP1-related Osteogenesis Imperfecta (OI) with very high bone material density, concerns remain whether anti-resorptive therapy is indicated in this ultra-rare form of OI 27782377_Data show that the KKN1 fragment generated by BMP1-cleavage of WFIKKN1 protein contributes most significantly to the observed enhancer activity. 28365001_For meprin beta a reduction and for BMP-1 an increase in activity was reported under increasing calcium concentrations. 28513615_BMP1 c.941G>A (p.(R314H)) variant was identified in the family with Chiari malformation type 1. 28883005_Results indicate the significance of follistatin-like protein 1 (FSTL1) in driving oncogenesis and metastasis in esophageal squamous cell carcinoma (ESCC) by coordinating NF-kappa B (NFkappaB) and bone morphogenetic proteins (BMP) pathway control. 28944874_Mechanical stress affects the osteogenic differentiation of human ligamentum flavum cells via the BMP-Smad1 signaling pathway. 29720137_BMP1 is upregulated in gastric cancer and is correlated with poor patient survival. 30062502_investigate the effects of the functional 5'UTR + 104 (T/C) variant of BMP1 on serum ApoA1 and HDL levels and risk of coronary heart disease. Our results indicated that the BMP1 5'UTR + 104 (T/C) variation may affect the serum ApoA1 and lipoprotein levels depending on statin therapy so that contributes to the development of coronary heart disease. 30638953_NEAT1 promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells by regulating miR-29b-3p/BMP1 axis. 31155610_Overexpression of BMP1 reflects poor prognosis in clear cell renal cell carcinoma. 31388055_Proteolysis of the low density lipoprotein receptor by bone morphogenetic protein-1 regulates cellular cholesterol uptake. 31600776_Bone morphogenetic protein 1 cleaves the linker region between ligand-binding repeats 4 and 5 of the LDL receptor and makes the LDL receptor non-functional. 31819067_Bone secreted factors induce cellular quiescence in prostate cancer cells. 32636307_BMP-1 disrupts cell adhesion and enhances TGF-beta activation through cleavage of the matricellular protein thrombospondin-1. 33085247_[Role of bone morphogenetic protein 1/tolloid proteinase family in the development of teeth and bone]. 33169406_Proteinase bone morphogenetic protein 1, but not tolloid-like 1, plays a dominant role in maintaining periodontal homeostasis. 35013172_Bone morphogenetic protein 1.3 inhibition decreases scar formation and supports cardiomyocyte survival after myocardial infarction. | ENSMUSG00000022098 | Bmp1 | 730.72569 | 0.9998589 | -0.0002036019 | 0.14549519 | 2.008491e-06 | 9.988692e-01 | 9.998360e-01 | No | Yes | 736.91349 | 81.185181 | 6.839023e+02 | 58.911426 | |
| ENSG00000168542 | 1281 | COL3A1 | protein_coding | P02461 | FUNCTION: Collagen type III occurs in most soft connective tissues along with type I collagen. Involved in regulation of cortical development. Is the major ligand of ADGRG1 in the developing brain and binding to ADGRG1 inhibits neuronal migration and activates the RhoA pathway by coupling ADGRG1 to GNA13 and possibly GNA12. | 3D-structure;Alternative splicing;Aortic aneurysm;Calcium;Collagen;Direct protein sequencing;Disease variant;Disulfide bond;Ehlers-Danlos syndrome;Extracellular matrix;Glycoprotein;Hydroxylation;Metal-binding;Reference proteome;Repeat;Secreted;Signal | This gene encodes the pro-alpha1 chains of type III collagen, a fibrillar collagen that is found in extensible connective tissues such as skin, lung, uterus, intestine and the vascular system, frequently in association with type I collagen. Mutations in this gene are associated with Ehlers-Danlos syndrome types IV, and with aortic and arterial aneurysms. Two transcripts, resulting from the use of alternate polyadenylation signals, have been identified for this gene. [provided by R. Dalgleish, Feb 2008]. | hsa:1281; | collagen type III trimer [GO:0005586]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular matrix structural constituent [GO:0005201]; extracellular matrix structural constituent conferring tensile strength [GO:0030020]; integrin binding [GO:0005178]; metal ion binding [GO:0046872]; platelet-derived growth factor binding [GO:0048407]; protease binding [GO:0002020]; SMAD binding [GO:0046332]; aorta smooth muscle tissue morphogenesis [GO:0060414]; cell-matrix adhesion [GO:0007160]; cellular response to amino acid stimulus [GO:0071230]; cerebral cortex development [GO:0021987]; chondrocyte differentiation [GO:0002062]; collagen fibril organization [GO:0030199]; digestive tract development [GO:0048565]; elastic fiber assembly [GO:0048251]; endochondral bone morphogenesis [GO:0060350]; extracellular matrix organization [GO:0030198]; fibroblast proliferation [GO:0048144]; heart development [GO:0007507]; in utero embryonic development [GO:0001701]; integrin-mediated signaling pathway [GO:0007229]; limb joint morphogenesis [GO:0036022]; lung development [GO:0030324]; multicellular organism growth [GO:0035264]; negative regulation of immune response [GO:0050777]; negative regulation of neuron migration [GO:2001223]; peptide cross-linking [GO:0018149]; platelet activation [GO:0030168]; positive regulation of Rho protein signal transduction [GO:0035025]; response to angiotensin [GO:1990776]; response to cytokine [GO:0034097]; response to radiation [GO:0009314]; skin development [GO:0043588]; supramolecular fiber organization [GO:0097435]; tissue homeostasis [GO:0001894]; transforming growth factor beta receptor signaling pathway [GO:0007179]; transforming growth factor beta1 production [GO:0032905]; wound healing [GO:0042060] | 11577371_mutational analysis in Ehlers-Danlos syndrome type IV 11973338_keratinocyte growth factor (KGF), a key stimulator of epithelial cell proliferation during wound healing, preferentially binds to collagens I, III, and VI. 12140670_COL3A1 mutations appear not to be a major cause of isolated spontaneous cervical artery dissections 12140670_Observational study of gene-disease association. (HuGE Navigator) 12149201_Observational study of gene-disease association. (HuGE Navigator) 12149201_variants in the COL3A1 gene modulate the risk of coronary artery disease and could also modulate the response to antithrombotic therapy 12631068_collagen type III was up-regulated by high glucose, but not by TGF-beta1 in renal fibroblasts 12880417_Proportion of collagen III relative to collagen I increased significantly up to 6 weeks after initial injury and remained elevated up to 6 months, at which time the proportion of collagen III was 70% above baseline values 14633859_Although connective tissue growth factor alone had no effect on collagen secretion, combined stimulation with IGF-I enhanced collagen accumulation. 14970208_fibroblasts from Ehlers-Danlos syndrome patients, with mutations in COL5A1 and COL3A1, synthesize aberrant types V and III collagen and show defective organization of these proteins into the extracellular matrix and reduction of alpha(2)beta(1) integrin 15193836_Association between COL3A1 collagen gene exon 31 and risk of floppy mitral valve/mitral valve prolapse among the Chinese population of Taiwan 15193836_Observational study of gene-disease association. (HuGE Navigator) 15227656_The lower collagen content in the endopelvic fascia and skin of women with SUI is not due to reduced collagen synthesis or selective reduction in synthesis of either collagen I or collagen III. 15365990_Predicted rates of AA substitution for Gly are compared with missense mutations known to cause disease. Any Gly replacement causes disease. The level of triple-helix destabilization determines outcome. More destabilizing mutations were seen than expected. 15514164_hnRNP A1 & K ARE positive effectors of collagen synthesis acting at the post-transcriptional level by interaction with the 3'-untranslated region (3'-UTR) of 3A1 mRNAs. 15838180_Observational study of genotype prevalence. (HuGE Navigator) 15894390_characterization of the proximal promoter of the COL3A1 gene; segment from -96 to -34 necessary for activation of transcription; multiple proteins depending on cell types, found to form the DNA-protein complex at -79 to -63 15944607_Observational study of gene-disease association. (HuGE Navigator) 16043429_analysis of binding between collagen type III and integrins alpha1beta1 and alpha2beta1 16088212_collagen III, and probably fibronectin, are degraded extracellularly in smooth muscle cells from varicose veins by a mechanism involving MMPs, and maybe MMP3 by a direct or an indirect pathway 16259598_Antisense oligodeoxynuclotides down-regulate collagen type III gene expression. 16356540_demonstrates, for the first time, that BIRC3 (anti-apoptotic protein), COL3A1 (matrix protein synthesis), and CXCL3 (chemokine) were up-regulated in the thrombin-stimulated human umbilical vein endothelial cells 16521042_The expression of precursor proteins and mRNA of type I and type III collagens is increased in usual interstitial pneumonia and sarcoidosis, reflecting mainly active synthesis of these collagens in different areas of the lung. 16681691_Type III collagen was expressed significantly higher in valvular cardiomyopathy. 16838047_data showed the complexity of the regulation of the COL3A1 gene (human alpha1(III) collagen) involving several transcription factors. 17146610_Altered expression of decorin mRNA in the different dermal strata and a decrease in the collagen-to-decorin ratio inflicted by both age and ultraviolet irradiation affect collagen bundle diameter and subsequently the mechanical properties of human skin. 17396208_The results suggest that a high level of decorin mRNA might be associated with the reduced content of collagen type III, resulting in a less flexible form of extracellular matrix in the connective tissue in stress urinary incontinence and prolapse. 18089612_COL3A1 was overexpressed in uterine fibroids. 18389341_Case report of a novel COL3A1 gene mutation in patient with aortic dissected aneurysm and cervical artery dissections. 18401458_High glucose levels downregulate mRNA levels in dermal fibroblast cell cultures. 18521510_Data reveal a more critical role for membrane cholesterol in collagen type III-induced than in VWF-induced Ca(2+) signalling. 18642782_Observational study of gene-disease association. (HuGE Navigator) 18722615_Observational study of gene-disease association. (HuGE Navigator) 18722615_There may be an association between COL3A1 genotype and risk of pelvic organ prolapse 18805790_the human type III collagen Gly991-Gly1032 cystine knot-containing peptide has both 7/2 and 10/3 triple helical symmetries 18818748_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19000145_Observational study of gene-disease association. (HuGE Navigator) 19000145_the constructs containing the 'G' allele of rs3106796 appear to exert lower transcriptional activity of COL3A1 than the 'A' allele, depending on the promoter types. 19019335_Observational study of gene-disease association. (HuGE Navigator) 19056482_Observational study of gene-disease association. (HuGE Navigator) 19152942_COL3A1 exon 31 polymorphism may have a role in determining the risk of pelvic organ prolapse in women 19152942_Observational study of gene-disease association. (HuGE Navigator) 19180518_Observational study of gene-disease association. (HuGE Navigator) 19389097_A dose-dependent increase in viable cells was demonstrated after the IPL irradiation. There was no significant change in mRNA levels of collagen I and fibronectin. 19398000_Results describe the mechanism of interaction and cleavage of human type III collagen by fibroblast MMP-1 by using a panel of recombinant human type III collagens (rhCIIIs) containing engineered sequences in the vicinity of the cleavage site. 19398442_COL3A1 is a disease-associated gene in both paediatric and adult gastroesophageal reflux and COL3A1 is genetically associated with hiatal hernia in adult males. 19398442_Observational study of gene-disease association. (HuGE Navigator) 19424605_the Aright curved arrow G (Ile1205Val) polymorphism of COL3A1 were associated (P<0.05) with the prevalence of CKD in high- subjects. 19444361_COL3A1 (collagen type III alpha 1) 2209G>A is a predictor of pelvic organ prolapse 19527514_Observational study of gene-disease association. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19639654_Observational study of gene-disease association. (HuGE Navigator) 19764567_The expression of collagen III and CTGF in pPROM group were decreased significantly when compared to their control groups. The expression of collagen III in pPROM group appeared significantly decreased when compared to that in tPROM group. 19790048_Hypoxia-inducible factor 1alpha inhibits the fibroblast-like markers type I and type III collagen during hypoxia-induced chondrocyte redifferentiation. 19858036_HGF inhibited TGF-beta1 mRNA expression and reduced collagen III secretion. 19893454_Data show that the expression of collagen types I, III and fibronectin was significantly higher in pancreatic cancer, and the expression of collagen type IV, laminin and vitronectin was significantly lower in pancreatic cancer. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19932771_The catalytic domain of MMP-12 binds to the triple helix and cleaves the typical sites -Gly(775)-Leu(776)- in alpha-2 type I collagen and -Gly(775)-Ile(776)- in alpha-1 type I and type III collagens and at multiple other sites in both collagen types. 20039537_Through alveolar macrophage mediation SiO2 can accelerate the expression of TIMP-1 and collagen III, and inhibit the expression of MMP-1 in human lung fibroblasts. 20063990_Data suggest that integrin alpha(2)beta(1), glycoprotein Ib and vWf interactions with collagen I and III contribute to platelet adhesion under high shear flow. 20140262_Observational study of gene-disease association. (HuGE Navigator) 20227120_Patients presenting a significant increase in maximum ascending aortic aneurysm size have a significantly higher type III procollagen level than those demonstrating no or limited growth in maximum aneurysm diameter. 20452482_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20631730_meprins could be important players in several remodeling processes involving collagen fiber deposition 20648054_Physical and laboratory examinations revealed that true haploinsufficiency of COL3A1, COL5A2, and MSTN, but not that of SLC40A1, leads to a clinical phenotype. 20673868_Observational study of gene-disease association. (HuGE Navigator) 20699621_In addition, among nondiabetic kidney disease, elevation of u-IVc was observed in patients with membranous nephropathy and ANCA. 20704113_Tendon sheath fibroblasts can transfect with antisense TGF-beta1 gene successfully and can decrease production of collagen I, collagen III, and TGF-beta1, which were factors of tendon adhere formation. 20720362_A novel point mutation is demonstrated at c.2528 G>A (p.Gly843Glu) in the COL3A1 gene of a Ehlers-Danlos syndrome type IV patient. 21044884_identification of the GVMGFO discoidin domain receptor binding motif on collagen III 21086191_cases of segmental mediolytic arteriopathy and traumatic subarachnoid hemorrhage undergo genetic testing for COL3A1 mutations 21343850_Data show that TGF-beta1 stimulated cell proliferation, enhanced the gene expression of type I and III collagen, and alpha-SMA. 21558934_RT-PCR showed that COL3A1 from osteoblasts from Pfeiffer syndrome grown on PLPG acid plates were downregulated after 30 days. 21637106_COL3A1 haploinsufficiency results in a variety of Ehlers-Danlos syndrome type IV with delayed onset of complications and longer life expectancy 21858035_Data suggest that combination of HA, PIIINP, TGF-ss1 may provide a potential useful tool to assess liver fibrosis in adult HCV patients. 21941774_Alveolar macrophages exposed to SiO2 can induce elevated type III procollagen and collagen III expression levels in human lung fibroblasts. 21946467_Type IV, or vascular, Ehlers-Danlos Syndrome is a life-threatening, autosomal dominant condition. It is caused by mutation of the COL3A1 gene. 22001912_Data show that homozygous and compound heterozygous changes found in PLOD1 and SLC2A10 may confer autosomal recessive effects, and three MYH11, ACTA2 and COL3A1 heterozygous variants were considered as putative pathogenic gene alterations. 22019127_Letter: Describe COL3A1 mutations and clinical features of vascular type Ehlers-Danlos syndrome in Italian patients. 22019950_procollagen III N-terminal peptide has a role in HBeAg loss in patients with chronic hepatitis B during entecavir therapy 22238662_Disease-associated mutations prevent GPR56-collagen III interaction. 22241462_study found that allele A of SNP rs1800255 conferred a 1.71-fold increased risk for intracranial aneurysms (IAs) and results in an amino acid change of Ala698Thr, which led to a lower thermal stability of the peptide; results support the view that the functional variant of COL3A1 is genetic risk factors for IAs in the Chinese population 22573319_The minimum type III sequence necessary for cleavage by the MMP1 and MMP13 was 5 GXY triplets, including 4 residues before and 11 residues after the cleavage site (P4-P11'). 22788708_Data suggest that cardiac fibrosis, as assessed by serum extracellular matrix protein biomarkers (including PIIINP, procollagen Type III-N-terminal peptide), develops early in hypertensive patients and is predictive of cardiovascular events or death. 22836729_This study indicates that SP, mediated via NK-1 R, increases collagen remodeling and leads to increased MMP3 mRNA and protein expression that is further enhanced by cyclic mechanical loading. 22851868_IL-17, IL-23 and PIIINP have an inverse correlation with vitamin D in their involvement in the immune response in patients with HCV-4-related liver diseases in Egypt 22884488_The present study reveals that serum levels of PEDF are independently associated with P-III-P levels, suggesting that PEDF level is a novel biomarker of liver fibrosis in patients with nonalcoholic fatty liver disease. 23013106_COL3A1 rs1800255, COL6A1 rs35796750 and COL12A1 rs970547 were not significantly associated with sit-and-reach, straight leg raise or total shoulder rotation range of motion 23040566_serum PICP and PIIINP levels correlate with the presence of left atrial fibrosis and may act as predictors for post-operative atrial fibrillation (AF) even in the absence of previous history of AF 23344861_Arteriovenous malformation patients exhibit changes in the Col I/Col III ratio and elastic fibers in the vasculature, which may compromise the structural integrity of cerebral vessels. 23645670_analysis of collagen folding in Vascular Ehlers-Danlos syndrome mutations 23688910_Case Report: Ehlers-Danlos syndrome IV due to a mutation in intron 14 of the COL3A1 gene leading to venous manifestations without affecting arterial vessels at clinical presentation. 23749290_A2AR activation downstream signaling for Col1 and Col3 expression proceeds via two distinct pathways with varying sensitivity to cAMP activation. 24038457_either selective knock-down of S1PR1 or silencing S1PR3 induced collagen alpha1(I) and collagen alpha1(III) expression in mesenchymal stem cells 24088220_The objective of this study was to determine the concentrations of types I, II and III collagen in six distinct regions of the supraspinatus tendon. 24286194_Data indicate that in dedifferentiating chondrocytes, alpha5beta1 integrin was found to be involved in the induction of type I (COL1A1) and type III procollagen (COL3A1) expression. 24361166_results suggest that mitofusin-2 protein may affect the synthesis of procollagen of fibroblasts in postmenopausal patients with pelvic organ prolapse (POP); changes in Mfn2 and procollagen expression may play a role in the development of POP 24421219_The results point to the fact that the injury and/or mandible fracture increase the collagen type III metabolism and its dynamics depends on the type of the used bone fixation. 24503541_Gal-1 decreased the expression of collagen genes COL3A1 and COL5A1 but increased the expression of fibronectin and laminin 5. 24641356_Expression of COL3A1 mRNA encoding key fibrotic extracellular matrix molecules was down-regulated by pre-miRNA-29b. 24650746_Arterial pathology in vascular Ehlers-Danlos syndrome individuals is related to the underlying COL3A1 mutation type. 24664438_Variants of the COL3A1 gene are associated with a risk of stroke recurrence and prognosis. 24700772_As a result of the translocations, COL3A1-PLAG1, the constitutively active promoter of the partner gene drives the ectopic expression of PLAG1. 24760181_rs1800255, COL3A1 2209 G>A polymorphism not associated with pelvic organ prolapse in Dutch population 25073002_The first main finding of this study was that the rare COL3A1 rs1800255 AA genotype was associated with increased risk of anterior cruciate ligament injuries in the Polish cohort but not the South African cohort. 25193015_These results suggest that let-7d may suppress renal cell carcinoma growth, metastasis, and tumor macrophage infiltration at least partially through targeting COL3A1 and CCL7. 25231012_The production and purification of rhCOL3A1 described in this study offer a new method for obtaining high level of rhCOL3A1 in relatively pure form, which is suitable for biomedical materials application. 25344368_During heart valve development, Krox20-mediated activation of fibrillar Col1a1 and Col3a1 genes is crucial to avoid postnatal degeneration of the aortic valve leaflets. 25420629_novel missense mutation c.2176G>C in Chinese family with vascular Ehlers-Danlos syndrome 25432063_intracellular S1P plays a crucial role in the TGF-beta1-induced expression of Col alpha1(I) and Col alpha1(III), which is required for human fibrosis development. 25559610_Data indicate that N-terminal propeptide of type III procollagen (PIIINP) is a highly effective means to evaluate left ventricular (LV) end-diastolic pressure (EDP) in patients with acute coronary syndrome (ACS). 25758994_the clinical phenotype of Ehlers-Danlos syndrome patients is influenced by the type of COL3A1 variant. 25786138_miR-29a and miR-29b enhance cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. 25846194_compare reported phenotypes for patients with missense variants in the C-propeptide domain for other human collagen disorders including COL1A1 and COL1A2 (osteogenesis imperfecta). 25893343_Report dysregulated expression of COL3A1 in disc degeneration. 26017485_In familial AAA we found one pathogenic and segregating variant (COL3A1 p.Arg491X), one likely pathogenic and segregating (MYH11 p.Arg254Cys), and fifteen VUS. 26258650_TGFbeta target genes including TGFBI, BAMBI, COL3A1 and SERPINE1 are significantly increased in Diamond Blackfan Anemia induced pluripotent stem cells 26406420_High serum Collagen Type III is associated with ovarian and breast cancer. 26497932_A novel missense mutation in COL3A1 was found in a young patient with cervical artery dissection as the single manifestation of Ehlers-Danlos syndrome. 26741506_High COL3A1 expression is associated with colorectal carcinoma. 27363273_Data show that emodin can block pulmonary fibroblast proliferation and differentiation into myofibroblasts, and reduce the synthesis of collagen type 1 (Col1) and collagen type 3 (Col3). 27442361_High serum procollagen type III N-terminal peptide expression is associated with non-alcoholic fatty liver disease. 27498063_Col3A may be a potential adjunct marker for both differentiating fibroadenoma from phyllodes tumor and assessing malignant potential in PTs. 27636223_Abnormal regulation of COL1 and COL3 may contribute to the early predisposition to POP in premenopausal women. 27648120_The findings indicate the critical role of CatB in regulating the expression of collagens III and IV by fibroblasts via prolonging TLR2/NF-kappaB activation and oxidative stress. 27655637_Kaplan-Meir analysis of GSE7696 indicate that COL3A1 and SNAP91 correlated with survival. 27889474_von Willebrand factor A1 domain-collagen binding is independent of gain- or loss-of-function phenotype and under shear stress, platelet translocation pause times on collagen-bound A1A2A3 are either normal or shorter depending on whether A1 is concertedly bound with the A3 domain to collagen. 28183226_Case Report: pathogenetic heterozygous COL3A1 mutation c.3140 G>A, p. Gly1047Asp in Ehlers-Danlos syndrome vascular type with different phenotypes in the same family. 28258187_Brain MRI in the affected siblings as well as in the two previously reported individuals with bi-allelic COL3A1 mutations showed a brain phenotype similar to that associated with mutations in GPR56. 28367737_Hepatitis C virus/Hepatitis B Virus co-infected patients with significant fibrosis did not show any significant difference (P > 0.05) from HCV mono-infected patients with respect to HCV-NS4, collagen III and MMP-1. 28481042_In conclusion, based on serological collagen formation and degradation makers, penetrating Crohn's disease is associated with increased matrix metalloproteinase-9 mediated breakdown of type III collagen 28742248_We identified biallelic COL3A1 variants in two unrelated families. In a 3-year-old female with developmental delay the nonsense variant c.1282C>T, p.(Arg428*) was detected in combination the c.2057delC, p.(Pro686Leufs*105) frame shift variant. 29191827_the role of cortisol regenerated by 11beta-hydroxysteroid dehydrogenase 1 (11beta-HSD1) in collagen III degradation in human amnion fibroblasts, was investigated. 29216800_Case Report: novel missense COL3A1 mutation in vascular Ehlers-Danlos syndrome patient presenting with pulmonary complications and iliac arterial dissection. 29263043_Reduced expression of types I and III collagen and TIMP-1 as well as the increased expression of MMP-1 and MMP-8 in the anterior vaginal wall tissues play important roles in the onset of pelvic organ prolapse. 29346445_provide a picture of the gene expression changes in vascular Ehlers-Danlos syndrome skin fibroblasts and highlight that dominant negative mutations in COL3A1 also affect post-translational modifications and deposition into the ECM of several structural proteins crucial to the integrity of soft connective tissues 29376591_Given the high specificity of the polymorphism at the rs1800255 locus of the COL3A1 gene, determined by the Sanger sequencing, it can be concluded that there is an association between this polymorphism and urinary incontinence and pelvic organ prolapse in women. 29471595_Elevated circulating PIIINP levels are associated with type 2 diabetes mellitus individuals with adipose tissue expansion and systemic proinflammatory profile suggestive for adipose tissue dysfunction. 29498185_Analysis of polymorphisms of COL3A1 gene (rs1800255) did not show differences in allele distribution between women with and without SD. 29533249_a variant of COL3A1 (rs3134646) is associated with the risk of developing colonic diverticulosis in white men, whereas rs1800255 (COL3A1) and rs1800012 (COL1A1) were not associated with this condition after adjusting for confounding factors. 30111669_High Aminoterminal Propeptide of Type III Procollagen level is associated with increase in Cardiovascular Events in Patients Undergoing Hemodialysis. 30213581_Association Between Plasma Level of Collagen Type III Alpha 1 Chain and Development of Strictures in Pediatric Patients With Crohn's Disease. 30229812_Serum levels of collagen Type III and miRNA-98 in patients with osteoarthritis were significantly higher compared to that of healthy volunteers. 30267195_Increased collagen III in the proximal portion of the long head of the biceps tendon is associated with glenohumeral arthritis. 30273997_CITP, PIIINP, and P4NP 7S do not reflect myocardial collagen mRNA expression but presumably reflect extra-cardiac organ injury in heart failure. 30474650_the frequency of COL3A1 mosaicism 30518744_Findings suggested a tumor-suppressor role of the miR-29 family in control of MTX resistance and cell apoptosis through regulating COL3A1 or MCL1. 30545625_a triple helix fragment of hCOL3A1, Gly489-Gly510, contained multiple charged residues, as well as representative Glu-Lys-Gly and Glu-Arg-Gly charged triplets. 30550979_Significant decrease in the expression of the genes HOXA13 and COL3A in the uterosacral ligaments in advanced stages of prolapse, indicates that these gene expressions may play a role in the development of uterine prolapse. 30837697_The three different Glu>Lys variants point toward a new variant type in COL3A1 causative of vEDS, which has consistent clinical features. This is important knowledge for COL3A1 variant interpretation 31041498_Polymorphism rs1800255 from COL3A1 gene and the risk for pelvic organ prolapse. 31075413_Mutations in the COL3A1 gene cause the vascular type of Ehlers-Danlos syndrome. It is the most serious form of EDS, since patients often die suddenly due to a rupture of large arteries. Most of the glycine mutations lead to the synthesis of type III collagen with reduced thermal stability, which is more susceptible for proteinases. Intracellular accumulation of this normally secreted protein is also found. [review] 31533654_COL3A1 expression was associated with shorter overall survival in the four major histotypes of epithelial ovarian carcinoma patients 31575845_2 novel COL3A1 gene mutations were identified in two vascular Ehlers-Danlos syndrome patients. 31600821_Application of the 2017 criteria for vascular Ehlers-Danlos syndrome in 50 patients ascertained according to the Villefranche nosology. 31816141_Human skin fibrosis: up-regulation of collagen type III gene transcription in the fibrotic skin nodules of lower limb lymphoedema. 31833208_Amniotic band sequence in paternal half-siblings with vascular Ehlers-Danlos syndrome. 31903873_Diagnostic role of collagen-III and matrix metalloproteinase-1 for early detection of hepatocellular carcinoma. 32168427_Extracellular vesicle-mediated crosstalk between NPCE cells and TM cells result in modulation of Wnt signalling pathway and ECM remodelling. 32692893_Collagen I and collagen III polymorphisms in women with pelvic organ prolapse. 32757451_Collagen Type III Alpha 1 chain regulated by GATA-Binding Protein 6 affects Type II IFN response and propanoate metabolism in the recurrence of lower grade glioma. 32945508_Prognostic significance of abnormal matrix collagen remodeling in colorectal cancer based on histologic and bioinformatics analysis. 32999266_COL3A1, COL6A3, and SERPINH1 Are Related to Glucocorticoid-Induced Osteoporosis Occurrence According to Integrated Bioinformatics Analysis. 33280513_Role of COL3A1 and POSTN on Pathologic Stages of Esophageal Cancer. 33472700_Lnc-GULP1-2:1 affects granulosa cell proliferation by regulating COL3A1 expression and localization. 33819468_miR-29a-3p-dependent COL3A1 and COL5A1 expression reduction assists sulforaphane to inhibit gastric cancer progression. 33930075_COL3A1 rs1800255 polymorphism is associated with pelvic organ prolapse susceptibility in Caucasian individuals: Evidence from a meta-analysis. 34047934_Identification of COL3A1 variants associated with sporadic thoracic aortic dissection: a case-control study. 34252356_Data mining-based study of collagen type III alpha 1 (COL3A1) prognostic value and immune exploration in pan-cancer. 34261485_Turnover of type I and III collagen predicts progression of idiopathic pulmonary fibrosis. 34587764_COL3A1 Missense Variant in a Patient Presenting With Hemoptysis. 34691289_Collagen Family Genes Associated with Risk of Recurrence after Radiation Therapy for Vestibular Schwannoma and Pan-Cancer Analysis. 35047627_COL3A1 and Its Related Molecules as Potential Biomarkers in the Development of Human Ewing's Sarcoma. | ENSMUSG00000026043 | Col3a1 | 21.58362 | 0.5110723 | -0.9684006038 | 0.64022540 | 1.977949e+00 | 1.596067e-01 | 9.998360e-01 | No | Yes | 17.91966 | 6.373105 | 2.951837e+01 | 8.328829 | |
| ENSG00000168876 | 54851 | ANKRD49 | protein_coding | Q8WVL7 | FUNCTION: Induces HBG1 expression (PubMed:16131492, PubMed:11162141). May have a role in spermatogenesis where it promotes autophagy in response to serum starvation, via the NF-kappaB pathway (By similarity). {ECO:0000250|UniProtKB:Q8VE42, ECO:0000269|PubMed:11162141, ECO:0000269|PubMed:16131492}. | ANK repeat;Differentiation;Nucleus;Phosphoprotein;Reference proteome;Repeat;Spermatogenesis | hsa:54851; | cytosol [GO:0005829]; nucleus [GO:0005634]; cell differentiation [GO:0030154]; positive regulation of transcription, DNA-templated [GO:0045893]; spermatogenesis [GO:0007283] | 19773279_Observational study of gene-disease association. (HuGE Navigator) 28694302_The research findings implicate that ANKRD49 promotes the proliferation of human malignant glioma cells. 29865034_The results of the present study highlighted an important role of the ANKRD49 protein in the progression of gastric cancer. The ANKRD49 protein could act as a potential biomarker for prognosis evaluation of gastric cancer and may be used as a molecular target for gastric cancer treatment. 30798416_these results suggest that ANKRD49 plays an important role in reducing intrinsic apoptosis of GC-1 cells by modulating the NF-kappaB signaling pathway. | ENSMUSG00000031931 | Ankrd49 | 131.60824 | 0.8166055 | -0.2922888338 | 0.27918716 | 1.042701e+00 | 3.071941e-01 | 9.998360e-01 | No | Yes | 115.08786 | 27.199771 | 1.575287e+02 | 28.522152 | ||
| ENSG00000168916 | 57507 | ZNF608 | protein_coding | Q9ULD9 | FUNCTION: Transcription factor, which represses ZNF609 transcription. {ECO:0000250|UniProtKB:Q56A10}. | Alternative splicing;Coiled coil;Isopeptide bond;Metal-binding;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:57507; | nucleus [GO:0005634]; metal ion binding [GO:0046872]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 27166999_Data indicate RNA Binding Protein with Multiple Splicing (RBPMS), Regulator of Chromosome Condensation and POZ Domain Containing Protein 1 (RCBTB1), and Zinc Finger protein 608 (ZNF608) as miR-21-3p target genes. | ENSMUSG00000052713 | Zfp608 | 74.06338 | 1.4009228 | 0.4863774514 | 0.34306188 | 1.984262e+00 | 1.589421e-01 | 9.998360e-01 | No | Yes | 90.74859 | 25.407198 | 8.019798e+01 | 17.394055 | ||
| ENSG00000169228 | 53917 | RAB24 | protein_coding | Q969Q5 | FUNCTION: May be involved in autophagy-related processes. {ECO:0000250}. | Autophagy;Cytoplasm;GTP-binding;Lipoprotein;Membrane;Nucleotide-binding;Prenylation;Protein transport;Reference proteome;Transport | RAB24 is a small GTPase of the Rab subfamily of Ras-related proteins that regulate intracellular protein trafficking (Olkkonen et al., 1993 [PubMed 8126105]).[supplied by OMIM, Aug 2009]. | hsa:53917; | autophagosome [GO:0005776]; cytosol [GO:0005829]; endocytic vesicle [GO:0030139]; endomembrane system [GO:0012505]; endosome [GO:0005768]; plasma membrane [GO:0005886]; secretory granule membrane [GO:0030667]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; autophagy [GO:0006914]; intracellular protein transport [GO:0006886] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 23387408_Rab24 modulates several mitotic events, including chromosome segregation and cytokinesis, perhaps through the interaction with microtubules 27550070_Our work provides new insights into the molecular function of Rab24 in the last steps of the endosomal degradative pathway. 32694843_Hepatic Rab24 controls blood glucose homeostasis via improving mitochondrial plasticity. | ENSMUSG00000034789 | Rab24 | 2184.58981 | 0.9327324 | -0.1004649233 | 0.11345942 | 7.995367e-01 | 3.712319e-01 | 9.998360e-01 | No | Yes | 2190.58315 | 215.884630 | 2.166375e+03 | 165.933240 | |
| ENSG00000169718 | 64118 | DUS1L | protein_coding | Q6P1R4 | FUNCTION: Catalyzes the synthesis of dihydrouridine, a modified base found in the D-loop of most tRNAs. {ECO:0000250}. | FMN;Flavoprotein;Oxidoreductase;Reference proteome;tRNA processing | hsa:64118; | flavin adenine dinucleotide binding [GO:0050660]; tRNA dihydrouridine synthase activity [GO:0017150] | ENSMUSG00000025155 | Dus1l | 14140.43683 | 0.9584746 | -0.0611878675 | 0.09005179 | 4.687564e-01 | 4.935598e-01 | 9.998360e-01 | No | Yes | 15203.89232 | 1639.352897 | 1.476815e+04 | 1229.807713 | |||
| ENSG00000169955 | 65988 | ZNF747 | protein_coding | Q9BV97 | Mouse_homologues NA; + ;NA | Alternative splicing;Metal-binding;Reference proteome;Repeat;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | hsa:65988; | regulation of transcription, DNA-templated [GO:0006355] | Mouse_homologues NA; + ;NA | ENSMUSG00000045757+ENSMUSG00000078580 | Zfp764+E430018J23Rik | 2005.82748 | 1.2036226 | 0.2673830735 | 0.12036587 | 4.953994e+00 | 2.603047e-02 | 9.998360e-01 | No | Yes | 2344.65011 | 280.336081 | 1.817530e+03 | 169.219308 | |
| ENSG00000170873 | 9788 | MTSS1 | protein_coding | O43312 | FUNCTION: May be related to cancer progression or tumor metastasis in a variety of organ sites, most likely through an interaction with the actin cytoskeleton. | 3D-structure;Actin-binding;Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome;Tumor suppressor | hsa:9788; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; endocytic vesicle [GO:0030139]; intrinsic component of the cytoplasmic side of the plasma membrane [GO:0031235]; ruffle [GO:0001726]; actin binding [GO:0003779]; actin monomer binding [GO:0003785]; identical protein binding [GO:0042802]; phospholipid binding [GO:0005543]; signaling receptor binding [GO:0005102]; actin cytoskeleton organization [GO:0030036]; adherens junction maintenance [GO:0034334]; cell adhesion [GO:0007155]; cellular response to fluid shear stress [GO:0071498]; epithelial cell proliferation involved in renal tubule morphogenesis [GO:2001013]; glomerulus morphogenesis [GO:0072102]; microspike assembly [GO:0030035]; negative regulation of epithelial cell proliferation [GO:0050680]; nephron tubule epithelial cell differentiation [GO:0072160]; plasma membrane organization [GO:0007009]; positive regulation of actin filament bundle assembly [GO:0032233]; renal tubule morphogenesis [GO:0061333]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 12082544_a potential metastasis suppressor gene in bladder cancer; data suggest that it may be involved in cytoskeletal organization 14752106_missing in metastasis protein and insulin receptor tyrosine kinase substrate p53 have a conserved novel actin bundling/filopodium-forming domain 15488640_Our data indicate that down-regulation of MIM and/or MIM-B expression can occur in bladder cancer cell lines but is not associated with increased invasive behaviour. 15545630_MTSS1, also known as MIM, is a hedgehog inducible protein expressed in carcinomas that potentiates Gli transcription factor activity 15684034_MTSS1, also known as MIM, is a modular hedgehog inducible protein that contains a dimerization domain and a distinct activation/scaffolding domain that binds RPTPdelta and induces membrane projections. 16280553_MIM-B is unlikely to be a metastasis suppressor but acts as a scaffold protein that interacts with Rac, actin and actin-associated proteins to modulate lamellipodia formation. 16921485_The expression of metastasis suppressor MIM is regulated by DNA methylation of a CpG island within its promoter region. 17442377_Elevated MIM-B expression may influence the development of hepatocellular carcinoma and may possibly be a powerful indicator for the disease at an early stage. 19328678_MTSS1 is a prognostic indicator of disease-free survival in breast cancer patients and demonstrates the ability to play a role in governing the metastatic nature of breast cancer cells. 20080650_Observational study of gene-disease association. (HuGE Navigator) 20201926_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20712855_Downregulation of metastasis suppressor 1 is associated with nodal metastasis and gastric cancer. 21196400_the current knowledge of MTSS1's role in cancer metastasis, carcinogenesis, and development is discussed. 21696600_MTSS1 serves as a potential prognostic indicator in human oesophageal squamous cell carcinoma and may be an important target for cancer therapy. 21909138_MTSS1 was characterized as a novel tumor suppressor gene in hepatocellular carcinoma 21927027_at high cell densities, Mtss1 impacts negatively on EGF signaling and this suggests why it inhibits metastasis. 21965727_MTSS1 is expressed at low levels or is absent from human bladder cancer cells. 22100162_Anthrax edema toxin induced transendothelial cell tunnels are resealed by MIM via Arp2/3-driven actin polymerization. 22455847_MiR-23a expression promotes colon carcinoma cell growth, invasion and metastasis through inhibition of the MTSS1 gene. 22479308_Data show that Mtss1 drives enhanced junction formation specifically by elevating Rac-GTP. 22681717_Compared with normal tissue, miR-182 was up-reg'd and MTSS1 was down-reg'd in HCC tissues. Over-expression of miR-182 was correlated with intrahepatic metastasis. There was a negative correlation between miR-182 and MTSS1 expression in both HCC tissues. 22721729_the dimeric configuration is essential for MIM-mediated membrane remodelling and serves as a proper target to develop antagonists specifically against an I-BAR-domain-containing protein. 23017832_Data show that the expression of the metastasis suppressor 1 (MTSS1) gene was regulated by miR-135a overexpression and knockdown. 23350348_Loww MTSS1 expression is associated with kidney cancer. 23383207_MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. 23474751_In breast tumor samples, miR-182 induction is linked to downregulation of MIM, RhoA activation and poor prognosis. 23852458_MTSS1 expression was analyzed in normal and cancerous tissue specimens from Chinese patients with Hilar cholangiocarcinoma. 24318128_Study provides a novel molecular mechanism for the negative regulation of MTSS1 by beta-TRCP in cancer cells. 24632752_MTSS1 is a metastasis driver in a subset of human melanomas. 25287652_The absence of MIM led to PTPdelta-mediated activation of SRC. 25385572_These results implicate that the role of MTSS1 suppresses cell migration and invasion by inhibiting expression of CTTN and as a prognosis biomarker in Glioblastomas 25625275_Data shows that MTSS1 is highly expressed in non-small-cell lung carcinomas compared with non-neoplastic lung tissues and may serve as an independent prognostic factor. 25783158_EGF induces microRNAs that target suppressors of cell migration: miR-15b targets MTSS1 in breast cancer. 25996952_our findings suggest that MTSS1 might have a context-dependent function and could act as a tumor suppressor, which is pharmacologically targetable in AML patients. 26119942_human breast tumors data sets the MTSS1/p63 co-expression is a negative prognostic factor on patient survival, suggesting that the MTSS1/p63 axis might be functionally important to regulate breast tumor progression 26198729_Down-regulation of MTSS1 expression is associated with lymph node metastasis in Pancreatic Cancer. 26316204_MTSS1 plays an essential inhibitory role in the development and progression of ovarian cancers 26621336_Low expression of Mtss1 is associated with Chronic myeloid leukemia. 27164937_MTSS1 is a new authentic target of miR-96 in prostate carcinoma. 27181205_Data suggest that overexpression of MAEL, caused by gene amplification and/or decreased miR-186, has a critical oncogenic role in UCB pathogenesis by downregulation of MTSS1. 27634022_MTSS1 suppressed H1299 cell migration and invasion, and its expression level can be used as a new independent factor for determining the prognosis of non-small cell lung cancer. 27693783_in contrast to MIM, which is a prototype of I-BAR proteins and does not contain an SH3 domain, IRTKS promoted serum-induced cell migration along with enhanced phosphorylation of mitogen activated kinases Erk1/2 and p38 28068324_Data identify MTSS1 as a new Akt2-regulated gene, and point to suppression of MTSS1 as a key step in the metastasis-promoting effects of Akt2 in CRC cells. 28146435_Overexpression of MTSS1 in PDAC cell lines leads to a loss of migratory potential in vitro and an increase in overall survival in vivo. Data provide insight into an important role for MTSS1 in suppressing tumor cell invasion and migration driven by the tumor microenvironment. 28159994_Missing in metastasis B, regulated by DNMT1, functions as a putative cancer suppressor in human lung giant-cell carcinoma. 28264927_this study shows a previously unknown property of MIM that establishes the linkage of protein ubiquitylation with Rab-guided trafficking of CXCR4 in endocytic vesicles 28739745_Overexpression of MTSS1 reduced BUCC cell proliferation, cell-cycle progression and colony formation, but had no influence on BUCC cell apoptosis. 29175021_MTSS1 is stabilized by the protein phosphatase activity of the tumor suppressor PTEN. Our data show that PTEN loss in PDAC cells results in both increased metastatic potential and decreased MTSS1 expression. Furthermore, we show that ectopic MTSS1 expression rescues this effect. 29218652_In vitro data suggests MTSS1 regulates lung adenocarcinoma through augmentation of cell invasion and migration. 29453719_These data suggested that miR-411 played as oncogene in the osteosarcoma partly by inhibiting the MTSS1 expression. . 29497041_SCAMP1 enhances Mtss1 anti-invasive function in HER2+/ER-/PR- breast cancer, by promoting its protein trafficking leading to elevated levels of RAC1-GTP and increased cell-cell adhesions. 30246429_Low MTSS1 expression is associated with Epithelial-mesenchymal transition in gastric carcinoma. 30808710_deletion of the SH3 domain in IRTKS abolished the IRTKS-RAB11 interaction and promoted CXCR4 degradation. Furthermore, the SH3 domain was required for selective targeting of MIM-IRTKS fusion proteins by both RAB7 and RAB11. 30949866_MicroRNA-182-5p Modulates Oral Squamous Cell Carcinoma Migration and Invasion Via Targeting MTSS1 Gene. 31541465_MTSS1 hypermethylation is associated with prostate cancer progression. 31545491_MEG3 was a promising tumor suppressor that may upregulate MTSS1 through the miR-96-5p pathway, subsequently suppressing the growth and metastasis of glioma. 32052679_Metastasis suppressor 1 acts as a tumor suppressor by inhibiting epithelial-to-mesenchymal transition in triple-negative breast cancer. 32645138_miR-28-5p targets MTSS1 to regulate cell proliferation and apoptosis in esophageal cancer. 32729377_Circular RNA SFMBT2 Inhibits the Proliferation and Metastasis of Glioma Cells Through Mir-182-5p/Mtss1 Pathway. 32929338_A reciprocal feedback of Myc and lncRNA MTSS1-AS contributes to extracellular acidity-promoted metastasis of pancreatic cancer. 33649808_MTSS1 inhibits colorectal cancer metastasis by regulating the CXCR4/CXCL12 signaling axis. 33686422_Circ_002059 suppresses cell proliferation and migration of gastric cancer via miR-182/MTSS1 axis. 33782537_Downregulation of MTSS1 in acute myeloid leukemia is associated with a poor prognosis, chemotherapy resistance, and disease aggressiveness. 35122005_MTSS1 suppresses mammary tumor-initiating cells by enhancing RBCK1-mediated p65 ubiquitination. | ENSMUSG00000022353 | Mtss1 | 112.85219 | 0.9463861 | -0.0794992696 | 0.28887738 | 7.433215e-02 | 7.851306e-01 | 9.998360e-01 | No | Yes | 103.86089 | 17.079187 | 1.171627e+02 | 15.142215 | ||
| ENSG00000171045 | 203062 | TSNARE1 | protein_coding | Q96NA8 | Alternative splicing;Coiled coil;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:203062; | endomembrane system [GO:0012505]; integral component of membrane [GO:0016021]; SNARE complex [GO:0031201]; SNAP receptor activity [GO:0005484]; SNARE binding [GO:0000149]; intracellular protein transport [GO:0006886]; vesicle docking [GO:0048278]; vesicle fusion [GO:0006906] | 24166486_GWAS meta analysis identifies TSNARE1 as a novel Schizophrenia / Bipolar susceptibility locus 25471352_Study provides the first evidence that the minor allele of TSNARE1 rs10098073 is significantly associated with a reduced risk of schizophrenia in a Han Chinese population, suggesting TSNARE1 may represent a susceptibility gene for this disease | 344.71580 | 1.0240582 | 0.0342977398 | 0.20163033 | 3.031066e-02 | 8.617872e-01 | 9.998360e-01 | No | Yes | 390.85569 | 48.148686 | 3.583431e+02 | 34.961734 | |||||
| ENSG00000171109 | 55669 | MFN1 | protein_coding | Q8IWA4 | FUNCTION: Mitochondrial outer membrane GTPase that mediates mitochondrial clustering and fusion (PubMed:12475957, PubMed:12759376, PubMed:27920125, PubMed:28114303). Membrane clustering requires GTPase activity (PubMed:27920125). It may involve a major rearrangement of the coiled coil domains (PubMed:27920125, PubMed:28114303). Mitochondria are highly dynamic organelles, and their morphology is determined by the equilibrium between mitochondrial fusion and fission events (PubMed:12475957, PubMed:12759376). Overexpression induces the formation of mitochondrial networks (in vitro) (PubMed:12759376). Has low GTPase activity (PubMed:27920125, PubMed:28114303). {ECO:0000269|PubMed:12475957, ECO:0000269|PubMed:12759376, ECO:0000269|PubMed:27920125, ECO:0000269|PubMed:28114303}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;GTP-binding;Hydrolase;Membrane;Mitochondrion;Mitochondrion outer membrane;Nucleotide-binding;Reference proteome;Transmembrane;Transmembrane helix;Ubl conjugation | The protein encoded by this gene is a mediator of mitochondrial fusion. This protein and mitofusin 2 are homologs of the Drosophila protein fuzzy onion (Fzo). They are mitochondrial membrane proteins that interact with each other to facilitate mitochondrial targeting. [provided by RefSeq, Jul 2008]. | hsa:55669; | integral component of membrane [GO:0016021]; integral component of mitochondrial outer membrane [GO:0031307]; intrinsic component of mitochondrial outer membrane [GO:0031306]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; outer mitochondrial membrane protein complex [GO:0098799]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; identical protein binding [GO:0042802]; GTP metabolic process [GO:0046039]; mitochondrial fusion [GO:0008053]; mitochondrial membrane fusion [GO:1990613]; mitochondrion localization [GO:0051646]; positive regulation of mitochondrial membrane potential [GO:0010918] | 11950885_Results show that Fzo homologs mitofusin 1 and 2 are ubiquitous mitochondrial membrane proteins that interact with each other to facilitate mitochondrial targeting. 12475957_Mfn1 mediates mitochondrial fusion in human cells 15509649_OPA1 functionally requires mitofusin 1 to regulate mitochondrial fusion 15961417_Mfn1, Mfn2, NRF-2 and COX IV mRNA were increased 24 h post-exercise in skeletal muscle 17718388_Precise interactions between a few proteins are required for mitochondrial fusion and division. Among them Drp1, Mfn1, Mfn2 and Opal are considered the most important. 18832378_Fis1 and Mfn1 activities influence mitochondrial signal generation thereby insulin exocytosis. 19168699_IL-6 induces Bcl-2 expression to perform cytoprotective functions in response to oxygen toxicity, and that this effect is mediated by alterations in the interactions between Bak and Mfn1/Mfn2. Bcl-2 inhibited the interaction between Bak and Mfn1. 19754948_Observational study of gene-disease association. (HuGE Navigator) 20103533_lack of MARCH5 results in mitochondrial elongation, which promotes cellular senescence by blocking Drp1 activity and/or promoting accumulation of Mfn1 at the mitochondria 20339081_the NIC-Akt-Mfn signaling cascade identifies a pathway regulating cell-survival, independent of canonical functions associated with NIC activity 20661427_MFN1 is required for both the virus-induced redistribution of IPS-1 and IFN production. 20871098_Ubiquitination of several mitochondrial proteins, including mitofusin 1 and mitofusin 2 were reduced following the silencing of parkin or PINK1. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20974255_Patterned Purkinje cell degeneration is dependent on caspase activation, leading to the marked decrease of mitofusion 1 in the transgenic Harlequin cerebellum. 20981029_Gbeta2 also regulated the mobility of Mfn1 on the surface of the mitochondrial membrane and affected the mitochondrial fusion. 21173115_Mitofusin degradation by mitochondria-associated Parkin inhibits the fusion of damaged mitochondria with healthy mitochondria to facilitate the selective elimination of the former by autophagy. 21408142_The impact of mutations in endogenous PINK1 and Parkin on the ubiquitination of mitochondrial fusion and fission factors and the mitochondrial network structure, was investigated. 21615408_Our data supports a model whereby the translocation of parkin to damaged mitochondria induces the degradation of mitofusin 1 leading to impaired mitochondrial fusion 21839087_mitochondrial dynamics, particularly those mediated by the mitofusins, play a role in endothelial cell function and viability. 22484496_These results collectively suggest a role for Mfn1 in regulating the activation of Bax on the outer mitochondrial membrane in a GTPase-dependent manner. 22649485_Knock-out of mitofusin protein Mfn1 increased the frequency of mitochondrial fission with increased lifetime of unpaired events whereas deletion of both Mfn1 and Mfn2 resulted in an instable dynamics. 23427266_A novel role for the endoplasmic reticulum-associated Gp78 ubiquitin ligase and the Mfn1 mitochondrial fusion factor in mitophagy. 23713734_In a amyotrophic lateral sclerosis transgenic mouse model, Mfn1 is significantly increased in spinal cord. 24722297_A fine balance of Mfn1 levels is maintained by MARCH5-mediated quality control on acetylated Mfn1. 24824927_miR-19b targets 3'UTR sequences of Mfn1 genes inhibit the expression of Mfn1 25033890_Improper transcriptional (in)activation of mitofusin-1 and dynamin-related protein 1 during early in vitro embryo development is associated with a decrease in mitochondrial membrane potential and with embryo fragmentation. 27609161_MFN1-positive expression could be seen mainly in ganglion cells after 1 week of minus lens intervention, and with time extension, more and more positive cells appeared in the rod-cone cell and bipolar cell layer, and this phenomenon could not be found in the normal control eyes. 27713096_Regulation of Mfn1 by MGRN1 and the proteasome modulates mitochondrial fusion. 27920125_These results suggest that MFN tethers apposing membranes, likely through nucleotide-dependent dimerization. 28057766_SLC25A46 is a new component in mitochondrial dynamics that serves as a regulator for MFN1/2 oligomerization. 28114303_crystal structures of engineered human MFN1 containing the GTPase domain and a helical domain during different stages of GTP hydrolysis; mechanistic model for MFN1-mediated mitochondrial tethering is proposed; results shed light on the molecular basis of mitochondrial fusion and mitofusin-related human neuromuscular disorders 28669827_mitochondria elongation under hypoxic condition is regulated through SIRT1-mediated MFN1 deacetylation and accumulation. 28758339_The results show that a metabolic shift from glycolysis in young to mitochondrial respiration in old normal human fibroblasts occurs during chronological lifespan, and MFN1 and OPA1 regulate this process. 29212658_The results lead to a revised understanding of Mfn 1 as single-spanning outer membrane proteins with an Nout-Cin orientation, providing functional insight into the IMS contribution to redox-regulated fusion events. 29483649_Structural basis for how MFN1 mediates homotypic membrane fusion. 29661855_heptad repeat domain of Mitofusin proteins induces membrane fusion and possesses a conserved amphipathic helix that folds upon interaction with the lipid bilayer surface. 30082159_The results may suggest that TP53BP1 and MFN1 frameshift mutations and their intratumoral heterogeneity (ITH) could contribute to cancer development by inhibiting the TSG activities. 30527740_This study elucidates an essential role of MFN1 in stem cell fate determination to mediate EMT-associated stemness. 30635120_Human Mfn1 contains internal N-Terminal mitochondrial targeting sequence (MTS) localized in the loop region between the two helices of the conserved transmembrane domain. Internal MTS is a shared feature of all opisthokont mitofusins. It facilitates anchoring and targeting of these proteins to the mitochondrial membrane. 30873819_The mRNA upregulation of Mitofusin 1 and 2 provides first insight into the complex changes of mitochondrial dynamics in cardiomyocytes of patients with reversible heart failure due to tachycardiomyopathy. 31819189_Our results reveal a critical involvement of mitochondrial dynamics in hepatocellular carcinoma (HCC) metastasis via modulating glucose metabolic reprogramming. MFN1 may serve as a novel potential therapeutic target for HCC 31947947_Mitochondrial Fusion Via OPA1 and MFN1 Supports Liver Tumor Cell Metabolism and Growth. 32708307_The GDP-Bound State of Mitochondrial Mfn1 Induces Membrane Adhesion of Apposing Lipid Vesicles through a Cooperative Binding Mechanism. 32802180_Lentinan-functionalized Selenium Nanoparticles target Tumor Cell Mitochondria via TLR4/TRAF3/MFN1 pathway. 33762690_Cardiac mitofusin-1 is reduced in non-responding patients with idiopathic dilated cardiomyopathy. 34364121_Analysis of mitochondrial regulatory transcripts in publicly available datasets with validation in placentae from pre-term, post-term and fetal growth restriction pregnancies. 34370506_Mitochondrial Fusion Mediated by Mitofusin 1 Regulates Macrophage Mycobactericidal Activity by Enhancing Autophagy. 34416390_Role of the lipid transport protein StarD7 in mitochondrial dynamics. 34425738_Associations between OPA1, MFN1, and MFN2 polymorphisms and primary open angle glaucoma in Polish participants of European ancestry. 34431132_Miro1 functions as an inhibitory regulator of MFN at elevated mitochondrial Ca(2+) levels. 34600274_The effect of gestational diabetes on the expression of mitochondrial fusion proteins in placental tissue. | ENSMUSG00000027668 | Mfn1 | 262.73178 | 0.9815161 | -0.0269161123 | 0.21725330 | 1.454554e-02 | 9.040040e-01 | 9.998360e-01 | No | Yes | 209.50154 | 42.597391 | 2.236334e+02 | 35.050543 | |
| ENSG00000171469 | 93134 | ZNF561 | protein_coding | Q8N587 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | hsa:93134; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | Mouse_homologues 21530259_Zfy2 (but not the closely related Zfy1) is sufficient to reinstate the apoptotic response to the X univalent meiotic cell. 22407129_Studies identified a Zfy2 transcript that predominates in spermatids, and a Zfy1 transcript, lacking an exon encoding approximately half of the acidic domain. 24496622_The expression of Y-linked Zfy2 in XY mouse oocytes leads to frequent meiosis 2 defects, a high incidence of subsequent early cleavage stage arrest and infertility. 24967676_Zfy2 proved to be essential for the efficient operation of a 'checkpoint' during the first meiotic division that identifies and kills cells that would otherwise produce sperm with an unbalanced chromosome set. 26719889_Zfy2 is essential for sperm formation and is essential for sperm function in assisted fertilization procedure. 26765744_Mouse Y-Encoded Transcription Factor Zfy2 Is Essential for Sperm Head Remodelling and Sperm Tail Development 27742779_we show that in mice, Zfy genes(Zfy1 and Zfy2 ) are also necessary for efficient meiotic sex chrand the sex chromosomes are not correctly silenced in Zfy-deficient spermatocytes. 28114340_Zfy2-KO mice did not show any significant phenotypic alterations. Zfy1/2-DKO mice were infertile and displayed abnormal sperm morphology and fertilization failure.; + ;22407129_Studies identified a Zfy2 transcript that predominates in spermatids, and a Zfy1 transcript, lacking an exon encoding approximately half of the acidic domain. 24967676_genetic information on the short arm of the Y chromosome promotes meiosis II, and by transgene addition Zfy1 and Zfy2 were identified as the genes responsible. 27742779_we show that in mice, Zfy genes(Zfy1 and Zfy2 ) are also necessary for efficient meiotic sex chrand the sex chromosomes are not correctly silenced in Zfy-deficient spermatocytes. 28114340_Zfy1-KO mice did not show any significant phenotypic alterations. Zfy1/2-DKO mice were infertile and displayed abnormal sperm morphology and fertilization failure. 28251288_our findings show that RNAi-mediated disruption of Zfx/Zfy in mouse testis affected X/Y spermatogenesis. Additionally, results suggest that the paralogous genes Zfx/Zfy play an important role in the process of X and Y sperm development. The individual interference of Zfx/Zfy may predict the outcome of X and Y haploid sperms. Presented herein is an advanced method developed to control mouse X/Y spermatogenesis and sex rati | ENSMUSG00000000103+ENSMUSG00000053211 | Zfy2+Zfy1 | 314.11739 | 0.9759105 | -0.0351792934 | 0.18524209 | 3.515776e-02 | 8.512655e-01 | 9.998360e-01 | No | Yes | 225.02443 | 40.468597 | 2.552234e+02 | 35.355496 | |
| ENSG00000171566 | 5356 | PLRG1 | protein_coding | O43660 | FUNCTION: Involved in pre-mRNA splicing as component of the spliceosome (PubMed:28502770, PubMed:28076346). Component of the PRP19-CDC5L complex that forms an integral part of the spliceosome and is required for activating pre-mRNA splicing (PubMed:11101529, PubMed:11544257). {ECO:0000269|PubMed:11101529, ECO:0000269|PubMed:11544257, ECO:0000269|PubMed:28076346, ECO:0000269|PubMed:28502770}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Spliceosome;WD repeat;mRNA processing;mRNA splicing | This gene encodes a core component of the cell division cycle 5-like (CDC5L) complex. The CDC5L complex is part of the spliceosome and is required for pre-mRNA splicing. The encoded protein plays a critical role in alternative splice site selection. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:5356; | catalytic step 2 spliceosome [GO:0071013]; Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; fibrillar center [GO:0001650]; nuclear membrane [GO:0031965]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Prp19 complex [GO:0000974]; spliceosomal complex [GO:0005681]; U2-type catalytic step 2 spliceosome [GO:0071007]; mRNA splicing, via spliceosome [GO:0000398]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; protein localization to nucleus [GO:0034504] | 9765207_Study includes the isolation of a cDNA encoding the human PRL1 homolog. 14576297_the interaction between CDC5L and PLRG1 is essential for pre-mRNA splicing 20467437_A central region in hnRNP-M is required for interaction with CDC5L/PLRG1. 33239170_Crystal structure of the WD40 domain of human PLRG1. | ENSMUSG00000027998 | Plrg1 | 914.61125 | 0.7352180 | -0.4437559806 | 0.13508184 | 1.051455e+01 | 1.184385e-03 | 3.728722e-01 | No | Yes | 804.87091 | 185.264469 | 1.223980e+03 | 216.733280 | |
| ENSG00000171695 | 198437 | LKAAEAR1 | protein_coding | Q8TD35 | Alternative splicing;Reference proteome | hsa:198437; | ENSMUSG00000045794 | Lkaaear1 | 23.22129 | 0.6385732 | -0.6470760389 | 0.62156952 | 1.014854e+00 | 3.137428e-01 | 9.998360e-01 | No | Yes | 22.03623 | 7.632824 | 3.495069e+01 | 12.783374 | |||||
| ENSG00000171954 | 126410 | CYP4F22 | protein_coding | Q6NT55 | FUNCTION: A cytochrome P450 monooxygenase involved in epidermal ceramide biosynthesis. Hydroxylates the terminal carbon (omega-hydroxylation) of ultra-long-chain fatty acyls (C28-C36) prior to ceramide synthesis (PubMed:26056268). Contributes to the synthesis of three classes of omega-hydroxy-ultra-long chain fatty acylceramides having sphingosine, 6-hydroxysphingosine and phytosphingosine bases, all major lipid components that underlie the permeability barrier of the stratum corneum (PubMed:26056268). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase) (PubMed:26056268). {ECO:0000269|PubMed:26056268}. | Endoplasmic reticulum;Heme;Ichthyosis;Iron;Lipid metabolism;Membrane;Metal-binding;Microsome;Monooxygenase;Oxidoreductase;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This gene is part of a cluster of cytochrome P450 genes on chromosome 19 and encodes an enzyme thought to play a role in the 12(R)-lipoxygenase pathway. Mutations in this gene are the cause of ichthyosis lamellar type 3. [provided by RefSeq, Jul 2008]. | hsa:126410; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; heme binding [GO:0020037]; iron ion binding [GO:0005506]; monooxygenase activity [GO:0004497]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen [GO:0016705]; ceramide biosynthetic process [GO:0046513]; icosanoid metabolic process [GO:0006690] | 21540472_REVIEW: genetic analyses have identified a wide spectrum of mutations in the CYP4V2gene from patients suffering from Bietti's crystalline corneoretinal dystrophy, and mutations in theCYP4F22 gene have been linked to lamellar ichthyosis 22209317_Letter: CYP4F22 is highly expressed at the site and timing of onset of keratinization during skin development. 23871423_description of CYP4F22 mutations from a Japanese collodion baby with lamellar ichthyosis [case report] 26646773_We report two cases of Congenital Ichthyosiform Erythroderma showing homozygous mutations in the gene CYP4F22. 27735052_patients carrying one or two truncating CYP4F22 mutations affecting the SBRs tend to develop collodion membrane at birth 30011118_we provide an up-to-date overview of all published and novel CYP4F22 mutations and point out possible mutation hot spots. We discuss the molecular and clinical findings, the genotype-phenotype correlations and consequences on genetic testing. 31020658_Study reports a consanguineous family from Southern Tunisia including three members affected with congenital ichthyosis likely due to a novel missense mutation c.728G>T (p.Arg243Leu) in exon 8 of CYP4F2. 32069299_The present study reports five CYP4F22 mutations, two of them novel, increasing the number of CYP4F22 mutations currently listed. Additionally, our results suggest that the recurrent c.1303C>T change has a founder effect in Spanish population and c.1303C>T carrier families originated from a single ancestor with probable African ancestry. 33067036_Impaired production of the skin barrier lipid acylceramide by CYP4F22 ichthyosis mutations. | ENSMUSG00000061126 | Cyp4f39 | 23.31109 | 1.2817304 | 0.3580928188 | 0.63203402 | 3.486610e-01 | 5.548721e-01 | 9.998360e-01 | No | Yes | 26.79906 | 8.271720 | 1.951192e+01 | 5.201176 | |
| ENSG00000172086 | 51315 | KRCC1 | protein_coding | Q9NPI7 | Coiled coil;Reference proteome | hsa:51315; | 29351065_By observing the data obtained from the isothermal titration calorimetry assay, both of the human proteins (KRCC1 and ZFAND6) were demonstrated to bind to their respective Toxoplasma gondii SAG1 and SAG2 proteins. 31908025_KRCC1: A potential therapeutic target in ovarian cancer. | ENSMUSG00000053012 | Krcc1 | 120.32117 | 1.3477934 | 0.4305993823 | 0.27480019 | 2.437773e+00 | 1.184443e-01 | 9.998360e-01 | No | Yes | 133.81307 | 20.784533 | 1.044958e+02 | 12.747633 | ||||
| ENSG00000172366 | 84331 | MCRIP2 | protein_coding | Q9BUT9 | Acetylation;Alternative splicing;Cytoplasm;Methylation;Nucleus;Phosphoprotein;Reference proteome | hsa:84331; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; nucleus [GO:0005634] | Mouse_homologues 34088848_Regulation of cold-induced thermogenesis by the RNA binding protein FAM195A. | ENSMUSG00000025732 | Mcrip2 | 2149.22747 | 0.8563234 | -0.2237723703 | 0.14395850 | 2.408667e+00 | 1.206651e-01 | 9.998360e-01 | No | Yes | 2206.30438 | 336.832977 | 2.379793e+03 | 281.148529 | |||
| ENSG00000172828 | 23491 | CES3 | protein_coding | Q6UWW8 | FUNCTION: Involved in the detoxification of xenobiotics and in the activation of ester and amide prodrugs. Shows low catalytic efficiency for hydrolysis of CPT-11 (7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin), a prodrug for camptothecin used in cancer therapeutics. | Alternative splicing;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Hydrolase;Reference proteome;Serine esterase;Signal | Mouse_homologues NA; + ;NA | This gene encodes a member of the carboxylesterase large family. The family members are responsible for the hydrolysis or transesterification of various xenobiotics, such as cocaine and heroin, and endogenous substrates with ester, thioester, or amide bonds. They may participate in fatty acyl and cholesterol ester metabolism, and may play a role in the blood-brain barrier system. This gene is expressed in several tissues, particularly in colon, trachea and in brain, and the protein participates in colon and neural drug metabolism. Multiple alternatively spliced transcript variants encoding distinct isoforms have been reported, but the biological validity and/or full-length nature of some variants have not been determined.[provided by RefSeq, Jun 2010]. | hsa:23491; | cytosol [GO:0005829]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; carboxylic ester hydrolase activity [GO:0052689]; methyl indole-3-acetate esterase activity [GO:0080030]; low-density lipoprotein particle clearance [GO:0034383]; xenobiotic metabolic process [GO:0006805] | 18794728_pharmacogenomic characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes 19508181_CES3 seems to have a lower catalytic efficiency than the other two CESs for selected substrates. 20422440_Studies indicate that CES3 mRNA isoform expression in several tissues, particularly in colon, trachea and in brain. 20529763_Observational study of gene-disease association. (HuGE Navigator) 22700792_This study provides the first evidence of functional compensation whereby increased expression of CES3 restores intracellular cholesteryl ester hydrolytic activity and free cholesterol efflux in CES1-deficient cells. | ENSMUSG00000062181+ENSMUSG00000069922 | Ces3b+Ces3a | 566.50481 | 0.9012779 | -0.1499560389 | 0.14549911 | 1.067893e+00 | 3.014219e-01 | 9.998360e-01 | No | Yes | 486.56222 | 80.489466 | 5.029406e+02 | 64.425075 |
| ENSG00000173275 | 203523 | ZNF449 | protein_coding | Q6P9G9 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a nuclear protein that likely functions as a transcription factor. The protein includes an N-terminal SCAN domain, and seven C2H2-type zinc finger motifs. [provided by RefSeq, May 2010]. | hsa:203523; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357]; spermatogonial cell division [GO:0007284] | 16636917_ZNF449 is a member of the zinc-finger family and it may function as a transcription factor 25546433_During chondrogenic differentiation of human mesenchymal stem cells, ZNF449 was increased at an early stage, and its overexpression enhanced SOX9 and SOX6 only at the initial stage of the differentiation. | ENSMUSG00000073176 | Zfp449 | 86.21123 | 1.0297713 | 0.0423239153 | 0.32751044 | 1.653793e-02 | 8.976743e-01 | 9.998360e-01 | No | Yes | 85.75109 | 18.224641 | 8.497071e+01 | 13.978645 | |
| ENSG00000173517 | 79834 | PEAK1 | protein_coding | Q9H792 | FUNCTION: Probable catalytically inactive kinase. Scaffolding protein that regulates the cytoskeleton to control cell spreading and migration by modulating focal adhesion dynamics (PubMed:23105102, PubMed:20534451). Acts as a scaffold for mediating EGFR signaling (PubMed:23846654). {ECO:0000269|PubMed:20534451, ECO:0000269|PubMed:23105102, ECO:0000269|PubMed:23846654}. | 3D-structure;Cell junction;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | This gene encodes a non-receptor tyrosine kinase that is a member of the new kinase family three (NFK3) family. In migrating cells, the encoded protein is associated with the actin cytoskeleton and focal adhesions and promotes developing focal adhesion elongation. This protein may play a role in the regulation of cell migration, proliferation and cancer metastasis. [provided by RefSeq, Mar 2014]. | hsa:79834; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; focal adhesion [GO:0005925]; identical protein binding [GO:0042802]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase activity [GO:0004672]; cell migration [GO:0016477]; focal adhesion assembly [GO:0048041]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of focal adhesion assembly [GO:0051893]; substrate adhesion-dependent cell spreading [GO:0034446] | 12471243_Also known as SgK269. Contains a divergent and possibly inactive eukaryotic protein kinase domain. Has one human paralog, known as SgK223 or pragmin. These two constitute the NKF3 subfamily of protein kinases, found in vertebrates and echinoderms 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22589274_Here, we report that the novel tyrosine kinase PEAK1 is upregulated in human malignancies, including human PDACs and pancreatic intraepithelial neoplasia (PanIN). 23105102_phosphorylation of tyrosine 665 in pseudopodium-enriched atypical kinase 1 (PEAK1) is essential for the regulation of cell migration and focal adhesion turnover 23378338_results define a novel signaling pathway in basal breast cancer involving Lyn and SgK269 that offers clinical opportunities for therapeutic intervention 25261239_eIF5A proteins utilize PEAK1 as a downstream effector to drive pancreatic ductal adenocarcinoma (PDAC) pathogenesis. 25445115_Suggest that loss of PEAK1/E-cadherin may play an important role in carcinogenesis and development of gastric cancer through activating epithelial to mesenchymal transition. 26267863_report is the first to provide evidence that PEAK1 mediates signaling cross talk between TGFbeta receptors and integrin/Src/MAPK pathways and that PEAK1 is an important molecular regulator of TGFbeta-induced tumor progression and metastasis in breast cancer 26297948_this regard, we have demonstrated that PEAK1 is necessary for TGFbeta to induce ZEB1-mediated EMT in the context of fibronectin/ITGB3 activation. 27531744_the critical role of the CH region in SgK269/SgK223 association. Importantly, although SgK269 bridged SgK223 to Grb2, it was unable to activate Stat3 or efficiently enhance migration in SgK223 knock-out cells generated by CRISPR/Cas9. 28381547_Findings indicate that eIF5A-PEAK1-YAP signaling contributes to PDAC development by regulating an STF program associated with increased tumorigenicity. 29212708_PEAK1's kinase cleft is occluded, and its newly identified SHED region may promote an unexpected dimerization mode. Similarities of PEAK1 with the active kinase PINK1 may reclassify the latter as a member of the new kinase family 3 group. 29321164_PEAK1 protein level is increased during pancreatic ductal adenocarcinoma progression with the highest levels of expression observed in metastatic cell populations.Kras protein expression is controlled by a self-regulating feedforward mechanism mediated by eIF5A-PEAK1. 29449544_PEAK1 functions as a tumor promoter in colorectal cancer cells, and its expression is regulated by the EGFR/KRas signaling axis and miR-181d. 30038287_PEAK1 overexpression could induce epithelial-to-mesenchymal transition (EMT) and the expression of matrix metalloproteinase-2 (MMP2) and MMP9 both in vitro and in vivo, whereas PEAK1 knockout had the opposite effects. 30472186_PEAK1-PPP1R12B axis inhibits colorectal tumorigenesis and metastasis through deactivation of the Grb2/PI3K/Akt pathway. 32681075_AXL confers cell migration and invasion by hijacking a PEAK1-regulated focal adhesion protein network. 33742335_LncRNA NORAD, sponging miR-363-3p, promotes invasion and EMT by upregulating PEAK1 and activating the ERK signaling pathway in NSCLC cells. 34273398_BioID-Screening Identifies PEAK1 and SHP2 as Components of the ALK Proximitome in Neuroblastoma Cells. 34365903_Pseudopodium enriched atypical kinase 1(PEAK1) promotes invasion and of melanoma cells by activating JAK/STAT3 signals. | ENSMUSG00000074305 | Peak1 | 708.32133 | 0.9857630 | -0.0206872727 | 0.48902257 | 1.755831e-03 | 9.665763e-01 | 9.998360e-01 | No | Yes | 665.34110 | 129.897495 | 6.780962e+02 | 102.042749 | |
| ENSG00000173715 | 79703 | C11orf80 | protein_coding | Q8N6T0 | FUNCTION: [Isoform 3]: Component of a topoisomerase 6 complex specifically required for meiotic recombination. Together with SPO11, mediates DNA cleavage that forms the double-strand breaks (DSB) that initiate meiotic recombination. The complex promotes relaxation of negative and positive supercoiled DNA and DNA decatenation through cleavage and ligation cycles. {ECO:0000250|UniProtKB:J3QMY9}. | Alternative splicing;Chromosome;Disease variant;Meiosis;Reference proteome | hsa:79703; | chromosome [GO:0005694]; meiotic DNA double-strand break formation [GO:0042138]; reciprocal meiotic recombination [GO:0007131] | 27529678_Top association findings suggested that the bipolar disorder risk allele at SNP rs10896235 in C11orf80 may be associated with increased risk of headache, including migraine. | ENSMUSG00000071691 | Gm960 | 285.46230 | 0.9762275 | -0.0347107388 | 0.24782214 | 2.053896e-02 | 8.860420e-01 | 9.998360e-01 | No | Yes | 220.17695 | 50.531040 | 2.419585e+02 | 43.100738 | ||
| ENSG00000174151 | 284613 | CYB561D1 | protein_coding | Q8N8Q1 | FUNCTION: Probable transmembrane reductase that may use ascorbate as an electron donor and transfer electrons across membranes to reduce monodehydro-L-ascorbate radical and iron cations Fe(3+) in another cellular compartment. {ECO:0000305}. | Alternative splicing;Electron transport;Heme;Iron;Membrane;Metal-binding;Reference proteome;Translocase;Transmembrane;Transmembrane helix;Transport | hsa:284613; | integral component of membrane [GO:0016021]; heme binding [GO:0020037]; metal ion binding [GO:0046872]; oxidoreductase activity [GO:0016491]; transmembrane ascorbate ferrireductase activity [GO:0140571]; transmembrane monodehydroascorbate reductase activity [GO:0140575] | ENSMUSG00000048796 | Cyb561d1 | 706.30639 | 1.0545651 | 0.0766480998 | 0.13751119 | 3.143134e-01 | 5.750453e-01 | 9.998360e-01 | No | Yes | 760.21491 | 75.883594 | 6.749745e+02 | 52.920542 | |||
| ENSG00000174516 | 246330 | PELI3 | protein_coding | Q8N2H9 | FUNCTION: E3 ubiquitin ligase catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins. Involved in the TLR and IL-1 signaling pathways via interaction with the complex containing IRAK kinases and TRAF6. Mediates 'Lys-63'-linked polyubiquitination of IRAK1. Can activate AP1/JUN and ELK1. Not required for NF-kappa-B activation. {ECO:0000269|PubMed:12874243, ECO:0000269|PubMed:17675297}. | Alternative splicing;Phosphoprotein;Reference proteome;Transferase;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene is a scaffold protein and an intermediate signaling protein in the innate immune response pathway. The encoded protein helps transmit the immune response signal from Toll-like receptors to IRAK1/TRAF6 complexes. Several transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Jul 2011]. | hsa:246330; | cytosol [GO:0005829]; ubiquitin protein ligase activity [GO:0061630]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of tumor necrosis factor-mediated signaling pathway [GO:0010804]; protein K63-linked ubiquitination [GO:0070534]; protein polyubiquitination [GO:0000209]; regulation of Toll signaling pathway [GO:0008592] | 15917247_Pellino3 is a novel upstream regulator of p38 MAPK that activates CREB in a p38-dependent manner 18326498_suggest that Pellino 3b acts as a negative regulator for IL-1 signaling by regulating IRAK degradation through its ubiquitin protein ligase activity [Pellino 3b] 23892723_Our findings identify RIP2 as a substrate for Pellino3 and Pellino3 as an important mediator in the Nod2 pathway and regulator of intestinal inflammation. 25027698_Peptide PEL3 derived from the interleukin-1 receptor-associated kinase (IRAK)1-binding motif reveals a distinct phosphothreonine peptide binding preference. 26310831_Pellino-3 is involved in endotoxin tolerance and functions as a negative regulator of Toll-like receptor 2/4 signaling. 27302665_The combination of low Pellino3 levels together with high and inducible Pellino1 expression may be an important determinant of the degree of inflammation triggered upon Toll-like receptor 2 engagement by Helicobacter pylori and/or its components, contributing to Helicobacter pylori-associated pathogenesis by directing the incoming signal toward an NF-kB-mediated proinflammatory response. 28011711_Two protective, low-frequency, non-synonymous variants were significantly associated with a decrease in age-related macular degeneration (AMD)risk: A307V in PELI3 and N1050Y in CFH .We also identified a strong protective signal for a common variant (rs8056814) near CTRB1 associated with a decrease in AMD risk (logistic regression: OR = 0.71, P = 1.8 x 10-07). 31520466_PELI3 gene expression in blood and ovarian cancer. 32556677_Long Non-coding RNA MIAT Mediates Non-small Cell Lung Cancer Development Through Regulating the miR-128-3p/PELI3 Axis. 32635799_miR-365a-5p suppresses gefitinib resistance in non-small-cell lung cancer through targeting PELI3. | ENSMUSG00000024901 | Peli3 | 710.74815 | 0.9784916 | -0.0313686837 | 0.15146545 | 4.460621e-02 | 8.327298e-01 | 9.998360e-01 | No | Yes | 745.95523 | 97.893751 | 7.247683e+02 | 74.643744 |
| ENSG00000176623 | 51115 | RMDN1 | protein_coding | Q96DB5 | Alternative splicing;Cytoplasm;Cytoskeleton;Microtubule;Reference proteome;Repeat;TPR repeat | hsa:51115; | cytoplasm [GO:0005737]; mitotic spindle pole [GO:0097431]; spindle microtubule [GO:0005876]; microtubule binding [GO:0008017]; attachment of mitotic spindle microtubules to kinetochore [GO:0051315]; mitotic spindle organization [GO:0007052] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000028229 | Rmdn1 | 371.64755 | 1.0738277 | 0.1027625367 | 0.19706246 | 2.787593e-01 | 5.975156e-01 | 9.998360e-01 | No | Yes | 365.47094 | 71.456439 | 3.688518e+02 | 55.667781 | |||
| ENSG00000176731 | 401466 | RBIS | protein_coding | Q8N0T1 | FUNCTION: Trans-acting factor in ribosome biogenesis required for efficient 40S and 60S subunit production. {ECO:0000269|PubMed:26711351}. | Acetylation;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome | hsa:401466; | cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ribosome biogenesis [GO:0042254] | ENSMUSG00000078784 | Rbis | 905.05241 | 0.9176094 | -0.1240479868 | 0.13816434 | 8.099860e-01 | 3.681244e-01 | 9.998360e-01 | No | Yes | 890.09150 | 174.773039 | 1.011747e+03 | 153.136063 | |||
| ENSG00000176809 | 374819 | LRRC37A3 | protein_coding | O60309 | Mouse_homologues NA; + ;NA | Glycoprotein;Leucine-rich repeat;Membrane;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA | hsa:374819; | integral component of membrane [GO:0016021] | Mouse_homologues NA; + ;NA | ENSMUSG00000034239+ENSMUSG00000078632 | Gm884+Lrrc37a | 142.37957 | 0.8460281 | -0.2412225221 | 0.25431002 | 8.900430e-01 | 3.454657e-01 | 9.998360e-01 | No | Yes | 137.38425 | 23.617384 | 1.568024e+02 | 20.953108 | |
| ENSG00000176853 | 157769 | FAM91A1 | protein_coding | Q658Y4 | FUNCTION: As component of the WDR11 complex acts together with TBC1D23 to facilitate the golgin-mediated capture of vesicles generated using AP-1. {ECO:0000269|PubMed:29426865}. | Cytoplasmic vesicle;Golgi apparatus;Phosphoprotein;Reference proteome | hsa:157769; | cytoplasmic vesicle [GO:0031410]; trans-Golgi network [GO:0005802]; intracellular protein transport [GO:0006886]; vesicle tethering to Golgi [GO:0099041] | ENSMUSG00000037119 | Fam91a1 | 343.55677 | 1.4791694 | 0.5647873329 | 0.23183797 | 6.201165e+00 | 1.276662e-02 | 9.998360e-01 | No | Yes | 401.29671 | 76.279226 | 2.846884e+02 | 41.846409 | |||
| ENSG00000177058 | 153129 | SLC38A9 | protein_coding | Q8NBW4 | FUNCTION: Lysosomal amino acid transporter involved in the activation of mTORC1 in response to amino acid levels. Probably acts as an amino acid sensor of the Rag GTPases and Ragulator complexes, 2 complexes involved in amino acid sensing and activation of mTORC1, a signaling complex promoting cell growth in response to growth factors, energy levels, and amino acids (PubMed:25561175, PubMed:25567906, PubMed:29053970). Following activation by amino acids, the Ragulator and Rag GTPases function as a scaffold recruiting mTORC1 to lysosomes where it is in turn activated. SLC38A9 mediates transport of amino acids with low capacity and specificity with a slight preference for polar amino acids (PubMed:25561175, PubMed:25567906). Acts as an arginine sensor (PubMed:25567906, PubMed:29053970). Following activation by arginine binding, mediates transport of leucine, tyrosine and phenylalanine with high efficiency, and is required for the efficient utilization of these amino acids after lysosomal protein degradation (PubMed:29053970). {ECO:0000269|PubMed:25561175, ECO:0000269|PubMed:25567906, ECO:0000269|PubMed:29053970}. | 3D-structure;Alternative splicing;Amino-acid transport;Disulfide bond;Endosome;Glycoprotein;Lysosome;Membrane;Metal-binding;Reference proteome;Sodium;Transmembrane;Transmembrane helix;Transport | hsa:153129; | integral component of lysosomal membrane [GO:1905103]; late endosome [GO:0005770]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; amino acid transmembrane transporter activity [GO:0015171]; L-arginine transmembrane transporter activity [GO:0061459]; L-leucine transmembrane transporter activity [GO:0015190]; metal ion binding [GO:0046872]; amino acid transmembrane transport [GO:0003333]; cellular response to amino acid stimulus [GO:0071230]; positive regulation of TOR signaling [GO:0032008] | 24762746_SNPs within SLC38A9 are correlated with inflammatory bowel disease patients' 6-thioguanine nucleotide blood concentrations. 25561175_SLC38A9 is a physical and functional component of the amino acid sensing machinery that controls the activation of mTOR 25567906_SLC38A9 functions upstream of the Rag GTPases and is an excellent candidate for being an arginine sensor for the mTORC1 pathway. 25963655_Data suggest that amino acid transporter SLC38A9 controls mTORC1 activity through binding to the Rag-Ragulator complex at the lysosome upon amino acid availability. 29053970_Study validates that SLC38A9 is an arginine sensor for the mTORC1 pathway, and we uncover an unexpectedly central role for SLC38A9 in amino acid homeostasis. SLC38A9 mediates the transport, in an arginine-regulated fashion, of many essential amino acids out of lysosomes, including leucine, which mTORC1 senses through the cytosolic Sestrin proteins. 30181260_Ragulator and SLC38A9 act on the Rag GTPases to activate the mTORC1 pathway in response to nutrient sufficiency. 32868926_Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9. | ENSMUSG00000047789 | Slc38a9 | 257.80090 | 1.2250533 | 0.2928445443 | 0.20816232 | 2.009725e+00 | 1.562936e-01 | 9.998360e-01 | No | Yes | 275.28777 | 50.156554 | 2.606697e+02 | 36.768283 | ||
| ENSG00000177169 | 8408 | ULK1 | protein_coding | O75385 | FUNCTION: Serine/threonine-protein kinase involved in autophagy in response to starvation (PubMed:18936157, PubMed:21460634, PubMed:21795849, PubMed:23524951, PubMed:25040165, PubMed:31123703). Acts upstream of phosphatidylinositol 3-kinase PIK3C3 to regulate the formation of autophagophores, the precursors of autophagosomes (PubMed:18936157, PubMed:21460634, PubMed:21795849, PubMed:25040165). Part of regulatory feedback loops in autophagy: acts both as a downstream effector and negative regulator of mammalian target of rapamycin complex 1 (mTORC1) via interaction with RPTOR (PubMed:21795849). Activated via phosphorylation by AMPK and also acts as a regulator of AMPK by mediating phosphorylation of AMPK subunits PRKAA1, PRKAB2 and PRKAG1, leading to negatively regulate AMPK activity (PubMed:21460634). May phosphorylate ATG13/KIAA0652 and RPTOR; however such data need additional evidences (PubMed:18936157). Plays a role early in neuronal differentiation and is required for granule cell axon formation (PubMed:11146101). May also phosphorylate SESN2 and SQSTM1 to regulate autophagy (PubMed:25040165). Phosphorylates FLCN, promoting autophagy (PubMed:25126726). Phosphorylates AMBRA1 in response to autophagy induction, releasing AMBRA1 from the cytoskeletal docking site to induce autophagosome nucleation (PubMed:20921139). Phosphorylates ATG4B, leading to inhibit autophagy by decreasing both proteolytic activation and delipidation activities of ATG4B (PubMed:28821708). {ECO:0000269|PubMed:11146101, ECO:0000269|PubMed:18936157, ECO:0000269|PubMed:20921139, ECO:0000269|PubMed:21460634, ECO:0000269|PubMed:21795849, ECO:0000269|PubMed:23524951, ECO:0000269|PubMed:25040165, ECO:0000269|PubMed:25126726, ECO:0000269|PubMed:28821708, ECO:0000269|PubMed:31123703}. | 3D-structure;ATP-binding;Acetylation;Autophagy;Cytoplasm;Kinase;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Ubl conjugation | hsa:8408; | Atg1/ULK1 kinase complex [GO:1990316]; autophagosome [GO:0005776]; axon [GO:0030424]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; extrinsic component of autophagosome membrane [GO:0097635]; extrinsic component of omegasome membrane [GO:0097629]; extrinsic component of phagophore assembly site membrane [GO:0097632]; mitochondrial outer membrane [GO:0005741]; phagophore assembly site [GO:0000407]; phagophore assembly site membrane [GO:0034045]; recycling endosome [GO:0055037]; ATP binding [GO:0005524]; GTPase binding [GO:0051020]; identical protein binding [GO:0042802]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein-containing complex binding [GO:0044877]; small GTPase binding [GO:0031267]; autophagosome assembly [GO:0000045]; autophagy [GO:0006914]; autophagy of mitochondrion [GO:0000422]; axon extension [GO:0048675]; cellular response to nutrient levels [GO:0031669]; late nucleophagy [GO:0044805]; macroautophagy [GO:0016236]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of collateral sprouting [GO:0048671]; negative regulation of protein-containing complex assembly [GO:0031333]; neuron projection development [GO:0031175]; neuron projection regeneration [GO:0031102]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; piecemeal microautophagy of the nucleus [GO:0034727]; positive regulation of autophagosome assembly [GO:2000786]; positive regulation of autophagy [GO:0010508]; protein autophosphorylation [GO:0046777]; protein localization [GO:0008104]; protein phosphorylation [GO:0006468]; regulation of macroautophagy [GO:0016241]; regulation of protein lipidation [GO:1903059]; response to starvation [GO:0042594]; reticulophagy [GO:0061709]; signal transduction [GO:0007165] | 16940348_Starvation-induced autophagy, which requires mAtg9, may rely on an alteration of the steady-state trafficking of mAtg9, in a Atg1-dependent manner. 17595159_ULK1 knockdown inhibited rapamycin-induced autophagy consistent with a role downstream of mTOR 18936157_The functions of ULK1 and ULK2 are controlled by autophosphorylation and conformational changes involving exposure of the C-terminal domain and interaction with the putative human homologue of Atg13. 19211835_mTORC1 suppresses autophagy through direct regulation of the approximately 3-MDa ULK1-Atg13-FIP200 complex. 19225151_The ULK-Atg13-FIP200 complexes are direct targets of mTOR and important regulators of autophagy in response to mTOR signaling. 19258318_ATG13 and ULK1 are phosphorylated by the mTOR pathway in a nutrient starvation-regulated manner, indicating that the ULK1.ATG13.FIP200 complex acts as a node for integrating incoming autophagy signals into autophagosome biogenesis. 19287211_The identification of the novel protein, Atg101, and the validation of Atg13 and Atg101 as ULK1-interacting proteins, suggests an Atg1 complex is involved in the induction of macroautophagy in mammalian cells. 19690328_Studies elucidated the inhibitory mechanism in which mTORC1 phosphorylates the autophagy regulatory complex containing unc-51-like kinase 1 (ULK1), the mammalian Atg13 protein, and focal adhesion kinase interacting protein of 200 kD (FIP200). 21072212_Results suggest that AMPK association with ULK1 plays an important role in autophagy induction. 21205641_findings uncover a conserved mechanism coupling nutrient status with autophagy and cell survival; loss of AMPK or ULK1 result in defective mitophagy;phosphorylation of ULK1 by AMPK required for mitochondrial homeostasis and cell survival during starvation 21258367_Data show that under nutrient sufficiency, high mTOR activity prevents Ulk1 activation by phosphorylating Ulk1 Ser 757 and disrupting the interaction between Ulk1 and AMPK. 21460630_ULK1 contributes to mTORC1 inhibition through hindrance of substrate docking to Raptor. 21460634_Phosphorylation of AMPK by Ulk1 represents a negative feedback circuit. 21475306_In response to DNA damage, ULK1 and ULK2 are upregulated by p53. The upregulation of ULK1 (ULK2)/ATG13 complex by p53 is necessary for the sustained autophagy activity induced by DNA damage. 21518141_ULK1 represents a novel and clinically useful biomarker for esophageal squamous cell carcinoma (ESCC) patients and plays an important role during the progression of ESCC. 21560199_study found frequency of the T-allele of tSNP rs12303764 in ULK1 was significantly higher in Crohn's disease versus contrtols 21795849_ULK1 negatively regulates the kinase activity of mTORC1 and cell proliferation in a manner independent of Atg5 and TSC2 21819378_our results suggest that ULK1 may act as a major node for regulation by multiple kinases including AMPK and Akt that play both stimulatory and inhibitory roles in regulating autophagy. 22539723_glycogen synthase kinase-3 (GSK3), when deinhibited by default in cells deprived of growth factors, activates acetyltransferase TIP60 through phosphorylating TIP60-Ser86, which acetylates and stimulates the protein kinase ULK1, which is required for autophagy 22585231_Decreased expression of ULK1 is associated with breast cancer progression, together with closely related to decreased autophagic capacity. 22613832_The ULK1 and Atg9 are found on Rab11- and transferrin (Tfn) receptor (TfnR)-positive recycling endosomes. 22885598_Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy 23027865_amino acid starvation regulates autophagy in part through an increase in cellular Ca(2+) that activates a CaMKK-beta-AMPK pathway and inhibits mTORC1, which results in ULK1 stimulation 23078367_Data show that the autophagy-initiating kinase ULK1 (UNC51-like kinase 1) is a direct transcriptional target of ATF4 (activating transcription factor 4), which drives the expression of ULK1 mRNA and protein in severe hypoxia and ER stress. 23291726_ULK1-dependent autophagy degrades BNIP3 via MTORC1 and AMPK 23524951_proposed that mTOR, besides its negative regulation of ULK1 through its direct phosphorylation, may also indirectly inhibit ULK1 stability and activity by modifying AMBRA1 and impairing its E3-ligase-related functions 23716696_Findings show that Ypt1/Rab1 infindings show that Ypt1/Rab1 interacts with Atg1/Ulk1 in yeast and mammalian cells. 23849170_ULK1 is a hypoxia regulated gene and is associated with hypoxia tolerance and a worse clinical outcome. 23863160_In order for cells to control cellular homoeostasis during growth, there is close signalling interplay between mTORC1 and two other protein kinases, AMPK and ULK1 24013547_ULK1 complex forms puncta associated with the ER and sporadically with mitochondria 24066173_ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. 24119841_After activation and trafficking, STING is phosphorylated by UNC-51-like kinase (ULK1). This occurs following ULK1 dissociation from its repressor AMPK and was found to be triggered by cGAS-generated cyclic dinucleotides. 24306881_Single nucleotide polymorphism in ATG1 gene is associated with Hodgkin disease therapy induced second malignancy. 24671035_FUNDC1 regulates ULK1 recruitment to damaged mitochondria, where FUNDC1 phosphorylation by ULK1 is crucial for mitophagy. 24955726_silencing of EEF2K promotes autophagic survival via activation of the AMPK-ULK1 pathway in colon cancer cells 25126726_The FLCN-GABARAP association is modulated by the presence of either folliculin-interacting protein (FNIP)-1 or FNIP2 and further regulated by ULK1. 25266655_Data show that under basal and stressed conditions, AMPK and ULK1 are differentially required for Atg2A S761 phosphorylation. 25429619_Reactive oxygen species inhibited autophagy by downregulating the p70S6K/p53/ULK1 axis in selenite-treated NB4 cells. 25541949_The results show that nitric oxide stabilizes SIRT1 by regulating 26S proteasome functionality through ULK1 and O-linked-GlcNAc transferase, but not autophagy, in endothelial cells. 25678702_Physical exercise increased phosphorylation at Ser(555); phosphorylation at Ser(757) was not affected by exercise. 25686248_Maximal activation of selective autophagy during stress is attained by the ability of Huntingtin to bind ULK1, a kinase that initiates autophagy, which releases ULK1 from negative regulation by mTOR. 25701603_In silico analysis of human ULK1 for biomarker autophagy identification showed its evolutionary relationships with other species, revealed the sites for post-translational modifications. 25714809_the examination of ULK1 expression by IHC method, could serve as an effective additional tool for predicting therapeutic response and patients' survival outcome in NPC patients. 25723488_our data provide mechanistic insights into the regulation of selective autophagy by ULK1 and p62 upon proteotoxic stress. 25749095_ULK1-mediated autophagy has a role in retinoic acid-induced IgG production in TLR9-activated human primary B cells 25761126_The computational model provides testable predictions about the behavior of the AMPK-MTORC1-ULK1 network, which plays a central role in maintaining cellular energy and nutrient homeostasis. 25792739_stimulation of STING-dependent IRF3 activation by UV is due to apoptotic signaling-dependent disruption of ULK1 (Unc51-like kinase 1), a pro-autophagic protein that negatively regulates STING 25892232_ULK1 activity mediates transcriptional activation of IFN-stimulated genes and activation of p38 MAPK 25909887_Findings highlight a cytoprotective role of p32 under starvation conditions by regulating ULK1 stability, and uncover a crucial role of the p32-ULK1-autophagy axis in coordinating stress response, cell survival and mitochondrial homeostasis. 25925668_a novel signaling pathway identified whereby starvation-induced activation of ULK leads to phosphorylation of endogenous DENND3, with subsequent activation of Rab12 and initiation of membrane trafficking events required for autophagy 25980607_conclude that AMPK-dependent phosphorylation of ULK1 is critical for translocation of ULK1 to mitochondria and for mitophagy in response to hypoxic stress 26018823_MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy 26103054_ROS-AMPK-ULK1 mechanism that couples T3-induced mitochondrial turnover with activity, wherein mitophagy is necessary not only for removing damaged mitochondria but also for sustaining efficient OXPHOS 26183158_ULK1 could inhibit p70S6K in starvation-induced autophagy, and further identified that miR-4487 and miR-595 were novel ULK1 target miRNAs 26207339_the inhibition of deubiquitinases by the compound WP1130 leads to increased ULK1 ubiquitination, the transfer of ULK1 to aggresomes, and the inhibition of ULK1 activity. 26282792_KIM-1-/TIM-1-mediated phagocytosis links ATG5-/ULK1-dependent clearance of apoptotic cells to antigen presentation. 26299883_Of several factors examined, bone metastasis, liver metastasis, and ULK1 expression were shown to have significant effects on the response to mTOR inhibitors. 26299944_Structure of the human Atg13-Atg101 HORMA heterodimer in the ULK1 complex that controls autophagy has been described. 26310906_Concurrent mTORC1 inactivation and PP2A-B55alpha stimulation fuel ULK1-dependent autophagy. 26687681_Study identifies a key role of Cul3-KLHL20 in autophagy termination by controlling autophagy-dependent turnover of ULK1 and VPS34 complex subunits and reveals the pathophysiological functions of this autophagy termination mechanism. 27023913_Knockdown of either ULK1 or DLP1 expression with shRNAs suppresses LRRK2 G2019S expression-induced mitochondrial clearance, suggesting that LRRK2 G2019S expression induces mitochondrial fission through DLP1 followed by mitophagy via an ULK1 dependent pathway. 27046250_These results define a key molecular event for the starvation-induced activation of the ATG14-containing PtdIns3K complex by ULK1. 27068414_review the diverse roles of ULK1, with special focus on its importance to type I IFN signaling, and highlight important future study questions. 27153534_ULK1/2 function as a bifurcate-signaling node that sustains glucose metabolic fluxes besides initiation of autophagy in response to nutritional deprivation. 27334615_As a Rab1a effector, C9orf72 controls initiation of autophagy by regulating the Rab1a-dependent trafficking of the ULK1 autophagy initiation complex to the phagophore. 27387056_These results show that the SiMoA technology can detect quantitatively low levels of endogenous biomarkers with the ability to detect the loss of pSer(318)-Atg13 upon ULK1 inhibition. 27485354_A strong association of rs12297124, a noncoding ULK1 SNP, with LTBI and a role for ULK1 regulation of TNF secretion. 27510922_the nucleation of autophagosomes occurs in endoplasmic reticulum tubulovesicular regions, where the ULK1 complex coalesces with ER and the ATG9 compartment 27629431_despite the significant upregulation of mRNA of the essential autophagy initiation gene ULK1, its protein level is rapidly reduced under starvation. 27679555_High expression of ULK1 concomitant with high expression of LRPPRC may serve as useful markers for shorter biochemical progression (BCP)-free survival and overall survival in patients with metastatic prostate cancer (PCa) after androgen deprivation therapy (ADT). 27687210_Lack of mitochondrial DNA impairs chemical hypoxia-induced autophagy in liver tumor cells through reactive oxygen species-AMPK-ULK1 signaling dysregulation independently of HIF-1A. 27783190_The newly developed ULK1 PCR assay was applied to genotype samples from 100 healthy individuals of North Indian origin. Genotype frequencies were 9, 34 and 57 % for GG, GT and TT, respectively. Allele frequencies were 0.26 and 0.74 for G and T, respectively. The allele frequencies were in Hardy-Weinberg's equilibrium (p = 0.2443). 27846372_ULK1 has a role in RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells 27864418_These findings revealed that prosurvival autophagy was one of the mechanisms involved in the resistance acute myeloid leukemia (AML) leukemia stem cells to JQ1. Targeting the AMPK/ULK1 pathway or inhibition of autophagy could be an effective therapeutic strategy for combating resistance to BET inhibitors in AML and other types of cancer 27871932_Here, the authors demonstrate that S100A10 is required for ULK1 localization to autophagosome formation sites. Silencing of S100A10 reduces IFN-gamma-induced autophagosome formation. 27932573_These results thus place NEDD4L and ULK1 in a key position to control oscillatory activation of autophagy during prolonged stress to keep the levels of this process under a safe and physiological threshold. 27934868_phosphorylation of mATG9 at Tyr8 by Src and at Ser14 by ULK1 functionally cooperate to promote interactions between mATG9 and the AP1/2 complex. 28011640_These results demonstrate a novel mechanism by which STAT1 negatively regulates ULK1 expression and autophagy. 28032867_These findings reveal that Endoplasmic reticulum stress engages the GSK3beta-TIP60-ULK1 pathway to increase autophagy. 28069524_we found that ATG14 interacted with Ulk1 and LC3, and knock down of Ulk1 prevented the lipidation of LC3 and autophagy in HeLa-ATG14 cells. We also identified a phosphatidylethanolamine (PE) binding region in ATG14, and the addition of Ulk1 to Hela-ATG14 cells decreased the ATG14-PE interaction. 28073914_Ulk1 promoted the degradation of Hsp90-Cdc37 client kinases, resulting in increased cellular sensitivity to Hsp90 inhibitors. Thus, our study provides evidence for an anti-proliferative role of Ulk1 in response to Hsp90 inhibition in cancer cells 28079894_Our findings demonstrate for the first time that miR-26a/b can promote apoptosis and sensitize Hepatocellular carcinoma (HCC) to chemotherapy via suppressing the expression of autophagy initiator ULK1, and provide the reduction of miR-26a/b in HCC as a novel mechanism of tumor chemoresistance. 28195531_While focusing on the role of SMCR8 during autophagy initiation, we found that kinase activity and gene expression of ULK1 are increased upon SMCR8 depletion. The latter phenotype involved association of SMCR8 with the ULK1 gene locus. 28410240_we demonstrate that Ulk1 over-expression in human gastric cancer is pro-survival. Its over-expression is associated with patients' T classification and cancer relapse. 28430962_Activation of autophagy can attenuate accumulation of LPL, thereby limiting fatty acid excess, and prevent cardiac dysfunction in obese hearts via ULK1. 28486929_Overexpression of unc-51 like autophagy activating kinase causes elevated autophagy and aggregation of the ER exit sites(ERES), a region of the ER dedicated for the budding of COPII vesicles. Transport of cargo proteins is therefore inhibited and is retained at the ERES. 28498429_our results show that inhibition of Ulk1 suppresses Non-small cell lung cancer (NSCLC)cell growth and sensitizes NSCLC cells to cisplatin by modulating both autophagy and apoptosis pathways, and that Ulk1 might be a promising target for NSCLC treatment. 28614291_CLDN1 activates autophagy through up-regulation of ULK1 phosphorylation and promotes drug resistance of non-small cell lung cancer cells to cisplatin. 28639909_these results demonstrate the effective anti-autophagic of NRAGE in non-small-cell lung cancer cells through AMPK/Ulk1/Atg13 autophagy signaling pathways. Therefore, NRAGE could be used as a potential therapeutic target for lung cancer. 28653878_results from western blotting assays and immunoprecipitation assays displayed that sirtuin 6 specifically interacted with ULK1 and positively regulated its activity by inhibiting its upstream factor mammalian target of rapamycin activity. 28667165_rs9652059 variation (C-->T) could increase AS susceptibility and haplotypes of rs9652059(C)-rs4964879(G), rs9652059(C)-rs11616018(T) and rs9652059(T)-rs11616018(T) may be associatd with AS in a Chinese Han population. 28677209_Downregulation of ULK1 inhibited the overexpression effects of miR-372, and upregulation of ULK1 reversed the effects of overexpressed miR-372 in human pancreatic adenocarcinoma cells. 28791376_this study found that upregulation of MACC1 in ESCC was associated with lymph node metastasis of patients, and MACC1 regulated ESCC cell proliferation, apoptosis, migration and invasion mainly through AMPK-ULK1 induced autophagy 28820317_ULK1 phosphorylation at 3 different sites on the same ULK1 target region for NEDD4L is preparatory for its ubiquitylation and subsequent degradation. 28821708_Data show that ULK1, a protein kinase activated at the autophagosome formation site, phosphorylates human ATG4B on serine 316. 29091866_Simultaneous high expression of ULK1 (and LC3B) had a poorer overall survival rate in hepatocellular cancer patients. 29109831_Authors conclude that miR-93 is involved in hypoxia-induced autophagy by regulating ULK1. Results provide a new angle to understand the complicated regulation of the key autophagy kinase ULK1 during different stress conditions. 29128638_ULK1 played a crucial role in ALDH2-offered protective effect against high glucose exposure-induced cardiomyocyte injury through regulation of autophagy 29148569_all four ULK1 variants were more common in the schizophrenia cases than controls 29313410_that BECN1 Ser30 is a ULK1 target site whose phosphorylation activates the ATG14-containing PIK3C3 complex and stimulates autophagosome formation in response to amino acid starvation, hypoxia, and MTORC1 inhibition. 29470986_silencing of HOTAIR decreased drug resistance of Non-Small Cell Lung Cancer cells to Crizotinib through inhibition of autophagy via suppressing phosphorylation of ULK1. 29487085_our results demonstrate a role of the USP20 deubiquitinating enzyme in acting as a critical regulator of ULK1 and show a novel mechanism of the autophagy process in terms of ULK1 stability. 29929428_Overexpression of wild-type or dominant-negative ULK1 constructs does not affect virus replication, indicating that ULK1 degradation may be a side effect of the ULK1-independent signaling mechanism used by Poliovirus. 30001707_Study finds that ULK1 protein expression is up-regulating at the posttranscriptional level by PVT1 inducing autophagy of pancreatic ductal adenocarcinoma. 30021155_These findings implicate mTOR and ULK1 as regulators of MR activity. 30046135_The Unc-51-like kinase 1 (ULK1) serves as a protein kinase that is activated upon autophagy stimulation and is critical for the recruitment of other autophagy-related proteins to the autophagosome formation site. 30078736_Our results suggest that ULK1 is upregulated in Clear Cell Renal Carcinoma tumors and may be a potential therapeutic target. 30103670_Knockdown of PRPF8 significantly impairs mitophagosome formation and subsequent mitochondrial clearance through the aberrant mRNA splicing of ULK1, which mediates macroautophagy/autophagy initiation. 30459284_IFN-gamma receptor stimulation requires ULK1-mediated activation of MLK3 and ERK5 30497781_The ULK1 plays a pivotal role in the regulation of Wnt/beta-catenin signaling pathway by phosphorylating Dsh. 30517873_ULK1 O-GlcNAcylation is crucial for binding and phosphorylation of ATG14L, allowing the activation of lipid kinase VPS34. 30545560_ULK-1 with EGFR can predict early impairment in DN while PDCN can highlight progressive diabetic nephropathy (DN) risk EGFR and PDCN may interact synergistically with ULK-1 in autophagy dysregulation as a pathogenic mechanism of DN induction and progression. 30596474_Here, the authors show that the F-box protein FBXW5 targets SEC23B, a component of COPII, for proteasomal degradation and that this event limits the autophagic flux in the presence of nutrients. In response to starvation, ULK1 phosphorylates SEC23B on Serine 186, preventing the interaction of SEC23B with FBXW5 and, therefore, inhibiting SEC23B degradation. 30782972_The data presented here provide structural models and chemical starting points for the development of ULK1/2 dual inhibitors with improved selectivity for future exploitation of autophagy inhibition. 30808462_These results demonstrated that ULK1 polymorphisms have significant associations with susceptibility to pulmonary tuberculosis in a Chinese Han population 30853401_The capability of NDP52 to induce mitophagy is dependent on its interaction with the FIP200/ULK1 complex, which is facilitated by TBK1. Ectopically tethering ULK1 to cargo bypasses the requirement for autophagy receptors and TBK1. 30907226_Dual roles of ULK1 (unc-51 like autophagy activating kinase 1) in cytoprotection against lipotoxicity. 30957634_The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. 31160490_Study reevaluated the hyperosmotic-stress induced autophagy using human T24 cells and unexpectedly revealed that this unique type of autophagy proceeds independently of the Ulk1 complex, which is considered essential for the initiation of starvation-induced macroautophagy. 31208283_GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction. 31267703_ULK1 phosphorylates ATG16L1 on S278 31291454_Autophagy-related protein 1 homolog (ULK1) phosphorylates the spindle assembly checkpoint (SAC) protein mitotic arrest deficient 1-like 1 protein (Mad1) at Ser546 to recruit Mad1 to kinetochores. 31311432_we strongly believe that these ULK1 antagonists could be novel and potent drug candidates for future cancer therapeutics. 31524222_we reported upregulated miR-302d-3p and decreased ULK1 mRNA expression levels in the cartilaginous tissue of OA patients. 31732843_Klotho-mediated changes in the expression of Atg13 alter formation of ULK1 complex and thus initiation of ER- and Golgi-stress response mediated autophagy. 31821132_mTORC1 restricts hepatitis C virus RNA replication through ULK1-mediated suppression of miR-122 and facilitates post-replication events. 31913283_ULK1 phosphorylation inhibits Exo70 homo-oligomerization as well as its assembly to the exocyst complex, which are needed for cell protrusion formation and matrix metalloproteinases secretion during cell invasion. 31923418_Circ_0009910 accelerated imatinib-resistance in chronic myeloid leukemia (CML) cells by modulating ULK1-induced autophagy via targeting miR-34a-5p, providing a potential target in imatinib resistance of CML 31959741_NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. 31986961_The role of the key autophagy kinase ULK1 in hepatocellular carcinoma and its validation as a treatment target. 32035621_The unc-51 like autophagy activating kinase 1-autophagy related 13 complex has distinct functions in tunicamycin-treated cells. 32125086_miR-373 inhibits autophagy and further promotes apoptosis of cholangiocarcinoma cells by targeting ULK1. 32203415_Copper is an essential regulator of the autophagic kinases ULK1/ULK2 to drive lung adenocarcinoma. 32317083_Serine 389 phosphorylation of 3-phosphoinositide-dependent kinase 1 by UNC-51-like kinase 1 affects its ability to regulate Akt and p70 S6kinase. 32320653_The Autophagy-Initiating Kinase ULK1 Controls RIPK1-Mediated Cell Death. 32393312_ULK1 inhibition as a targeted therapeutic strategy for FLT3-ITD-mutated acute myeloid leukemia. 32516310_the positive function of ULK1-ATG13 and their phosphorylation by CDK1 in mitotic autophagy regulation, is reported. 32516362_ULK complex organization in autophagy by a C-shaped FIP200 N-terminal domain dimer. 32705220_Long noncoding RNA growth arrestspecific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. 32773036_The autophagy adaptor NDP52 and the FIP200 coiled-coil allosterically activate ULK1 complex membrane recruitment. 32854424_Sestrin2 Phosphorylation by ULK1 Induces Autophagic Degradation of Mitochondria Damaged by Copper-Induced Oxidative Stress. 32913252_A pan-cancer assessment of alterations of the kinase domain of ULK1, an upstream regulator of autophagy. 33040463_ULK1 inhibitor induces spindle microtubule disorganization and inhibits phosphorylation of Ser10 of histone H3. 33078654_Mir214-3p and Hnf4a/Hnf4alpha reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. 33149253_Coxsackievirus infection induces a non-canonical autophagy independent of the ULK and PI3K complexes. 33172148_Autophagy Genes for Wet Age-Related Macular Degeneration in a Finnish Case-Control Study. 33201521_NPM1 mutant maintains ULK1 protein stability via TRAF6-dependent ubiquitination to promote autophagic cell survival in leukemia. 33213267_MAPK1/3 kinase-dependent ULK1 degradation attenuates mitophagy and promotes breast cancer bone metastasis. 33328309_The Autophagy-Initiating Protein Kinase ULK1 Phosphorylates Human Cytomegalovirus Tegument Protein pp28 and Regulates Efficient Virus Release. 33463048_Inhibition of ULK1 promotes the death of leukemia cell in an autophagy irrelevant manner and exerts the antileukemia effect. 33531625_Inhibition of CAMKK2 impairs autophagy and castration-resistant prostate cancer via suppression of AMPK-ULK1 signaling. 33572255_Negative Regulation of ULK1 by microRNA-106a in Autophagy Induced by a Triple Drug Combination in Colorectal Cancer Cells In Vitro. 33669246_Lifelong Ulk1-Mediated Autophagy Deficiency in Muscle Induces Mitochondrial Dysfunction and Contractile Weakness. 33735626_Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192-5p/ULK1 ceRNA pathway. 33932238_TFG binds LC3C to regulate ULK1 localization and autophagosome formation. 33988678_Atlastin 2/3 regulate ER targeting of the ULK1 complex to initiate autophagy. 34096600_An essential role of the autophagy activating kinase ULK1 in snRNP biogenesis. 34107300_Glucose starvation induces autophagy via ULK1-mediated activation of PIKfyve in an AMPK-dependent manner. 34162658_Identification of a Locus Near ULK1 Associated With Progression-Free Survival in Ovarian Cancer. 34204950_(V600E)BRAF Inhibition Induces Cytoprotective Autophagy through AMPK in Thyroid Cancer Cells. 34224814_MAPK15-ULK1 signaling regulates mitophagy of airway epithelial cell in chronic obstructive pulmonary disease. 34259310_Investigation of the specificity and mechanism of action of the ULK1/AMPK inhibitor SBI-0206965. 34345207_Crosstalk between hypoxia-sensing ULK1/2 and YAP-driven glycolysis fuels pancreatic ductal adenocarcinoma development. 34369843_An AMPK-ULK1-PIKFYVE signaling axis for PtdIns5P-dependent autophagy regulation upon glucose starvation. 34382905_AMPK-activated ULK1 phosphorylates PIKFYVE to drive formation of PtdIns5P-containing autophagosomes during glucose starvation. 34421918_ULK1 Inhibition as a Targeted Therapeutic Strategy for Psoriasis by Regulating Keratinocytes and Their Crosstalk With Neutrophils. 34454184_LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by Mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. 34555439_MiR-BART2-3p targets Unc-51-like kinase 1 and inhibits cell autophagy and migration in Epstein-Barr virus-associated gastric cancer. 34578425_Nonstructural Protein NS1 of Influenza Virus Disrupts Mitochondrial Dynamics and Enhances Mitophagy via ULK1 and BNIP3. 34592149_ULK1 phosphorylation of striatin activates protein phosphatase 2A and autophagy. 34654847_ULK1 promotes mitophagy via phosphorylation and stabilization of BNIP3. 35246531_Oxygen-sensitive methylation of ULK1 is required for hypoxia-induced autophagy. | ENSMUSG00000029512 | Ulk1 | 2398.30341 | 0.9314931 | -0.1023830392 | 0.09984552 | 1.055472e+00 | 3.042503e-01 | 9.998360e-01 | No | Yes | 2507.81575 | 259.276957 | 2.442222e+03 | 195.493919 | ||
| ENSG00000177873 | 285267 | ZNF619 | protein_coding | Q8N2I2 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:285267; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 239.19504 | 0.9953071 | -0.0067863769 | 0.20822382 | 1.082795e-03 | 9.737497e-01 | 9.998360e-01 | No | Yes | 226.51206 | 26.874874 | 2.366074e+02 | 21.889309 | |||||
| ENSG00000178752 | 151176 | ERFE | protein_coding | Q4G0M1 | FUNCTION: Iron-regulatory hormone that acts as an erythroid regulator after hemorrhage: produced by erythroblasts following blood loss and mediates suppression of hepcidin (HAMP) expression in the liver, thereby promoting increased iron absorption and mobilization from stores. Promotes lipid uptake into adipocytes and hepatocytes via transcriptional up-regulation of genes involved in fatty acid uptake. {ECO:0000250|UniProtKB:Q6PGN1}. | Disulfide bond;Glycoprotein;Hormone;Reference proteome;Secreted;Signal | hsa:151176; | extracellular region [GO:0005576]; extracellular space [GO:0005615]; hormone activity [GO:0005179]; identical protein binding [GO:0042802]; cellular iron ion homeostasis [GO:0006879]; negative regulation of gluconeogenesis [GO:0045721]; positive regulation of fatty acid transport [GO:2000193]; positive regulation of glucose import [GO:0046326]; positive regulation of insulin receptor signaling pathway [GO:0046628]; regulation of fatty acid metabolic process [GO:0019217] | 22351773_Myonectin links skeletal muscle to lipid homeostasis in liver and adipose tissue in response to alterations in energy state, revealing a novel myonectin-mediated metabolic circuit. 26978524_In hemodialysis patients DA and CERA increased levels of ERFE that regulated hepcidin 25 and led to iron mobilization from body stores during erythropoiesis. 27146013_Data suggest hepcidin is the master regulator of systemic iron homeostasis; hepcidin levels are suppressed when erythropoiesis is stimulated; the erythroid-derived hormone erythroferrone appears to be a convincing candidate for link between increased erythropoiesis and hepcidin suppression. [REVIEW] 27540014_FAM132b expression is upregulated in Congenital Dyserythropoietic Anemia type II. 28666715_From a clinical point of view, erythroferrone could become a useful biological marker of iron metabolism and a therapeutic target. [review] 29419424_serum ERFE levels acutely increase in response to EPO in the setting of normal or impaired kidney function 30303269_Serum CTRP15 concentrations were associated with the key components of MetS and insulin resistance. 31292266_The expression of the variant ERFE transcript that was restricted to SF3B1-mutated erythroblasts decreased in lenalidomide-responsive anemic patients, identifying variant ERFE as a specific biomarker of clonal erythropoiesis. 31400017_A recurrent low-frequency variant, A260S, in the ERFE gene was found in 12.5% of congenital dyserythropoietic anemia type II patients with a severe phenotype. This variant leads to increased levels of ERFE, with subsequently marked impairment of iron regulation pathways at the hepatic level. It modified iron overload by impairing the BMP/SMAD pathway. 31608708_Higher serum level of CTRP15 in patients with coronary artery disease is associated with disease severity, body mass index and insulin resistance. 31834456_using the ERFE gene expression, combined with serum hepcidin estimation, can substantiate the role of estimated TS% as a promising tool in screening for iron overload in beta-TM patients. 31965030_Implications of C1q/TNF-related protein superfamily in patients with coronary artery disease. 32314610_Association of decreased myonectin levels with metabolic and hormonal disturbance in polycystic ovary syndrome. 32324886_Interplay of erythropoietin, fibroblast growth factor 23, and erythroferrone in patients with hereditary hemolytic anemia. 32866306_Disordered serum erythroferrone and hepcidin levels as indicators of the spontaneous abortion occurrence during early pregnancy in humans. 33372284_Erythroferrone structure, function, and physiology: Iron homeostasis and beyond. 33486765_High erythroferrone expression in CD71(+) erythroid progenitors predicts superior survival in myelodysplastic syndromes. 33676229_The impacts of C1q/TNF-related protein-15 and adiponectin on Interleukin-6 and tumor necrosis factor-alpha in primary macrophages of patients with coronary artery diseases. 34236433_Umbilical Cord Erythroferrone Is Inversely Associated with Hepcidin, but Does Not Capture the Most Variability in Iron Status of Neonates Born to Teens Carrying Singletons and Women Carrying Multiples. 34283879_Differentiating iron-loading anemias using a newly developed and analytically validated ELISA for human serum erythroferrone. 34310730_Erythroferrone and hepcidin as mediators between erythropoiesis and iron metabolism during allogeneic hematopoietic stem cell transplant. 34376791_Serum erythroferrone levels during the first month of life in premature infants. | ENSMUSG00000047443 | Erfe | 1074.86844 | 0.9770935 | -0.0334314687 | 0.12603952 | 7.232078e-02 | 7.879872e-01 | 9.998360e-01 | No | Yes | 1150.14906 | 91.146035 | 1.123762e+03 | 70.138169 | ||
| ENSG00000179295 | 5781 | PTPN11 | protein_coding | Q06124 | FUNCTION: Acts downstream of various receptor and cytoplasmic protein tyrosine kinases to participate in the signal transduction from the cell surface to the nucleus (PubMed:10655584, PubMed:18559669, PubMed:18829466, PubMed:26742426, PubMed:28074573). Positively regulates MAPK signal transduction pathway (PubMed:28074573). Dephosphorylates GAB1, ARHGAP35 and EGFR (PubMed:28074573). Dephosphorylates ROCK2 at 'Tyr-722' resulting in stimulation of its RhoA binding activity (PubMed:18559669). Dephosphorylates CDC73 (PubMed:26742426). Dephosphorylates SOX9 on tyrosine residues, leading to inactivate SOX9 and promote ossification (By similarity). {ECO:0000250|UniProtKB:P35235, ECO:0000269|PubMed:10655584, ECO:0000269|PubMed:18559669, ECO:0000269|PubMed:18829466, ECO:0000269|PubMed:26742426, ECO:0000269|PubMed:28074573}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Deafness;Disease variant;Hydrolase;Nucleus;Phosphoprotein;Protein phosphatase;Reference proteome;Repeat;SH2 domain | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP contains two tandem Src homology-2 domains, which function as phospho-tyrosine binding domains and mediate the interaction of this PTP with its substrates. This PTP is widely expressed in most tissues and plays a regulatory role in various cell signaling events that are important for a diversity of cell functions, such as mitogenic activation, metabolic control, transcription regulation, and cell migration. Mutations in this gene are a cause of Noonan syndrome as well as acute myeloid leukemia. [provided by RefSeq, Aug 2016]. | hsa:5781; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; cadherin binding [GO:0045296]; cell adhesion molecule binding [GO:0050839]; insulin receptor binding [GO:0005158]; non-membrane spanning protein tyrosine phosphatase activity [GO:0004726]; peptide hormone receptor binding [GO:0051428]; phosphoprotein phosphatase activity [GO:0004721]; phosphotyrosine residue binding [GO:0001784]; protein kinase binding [GO:0019901]; protein tyrosine kinase binding [GO:1990782]; protein tyrosine phosphatase activity [GO:0004725]; receptor tyrosine kinase binding [GO:0030971]; signaling receptor complex adaptor activity [GO:0030159]; abortive mitotic cell cycle [GO:0033277]; atrioventricular canal development [GO:0036302]; axonogenesis [GO:0007409]; Bergmann glial cell differentiation [GO:0060020]; brain development [GO:0007420]; cellular response to epidermal growth factor stimulus [GO:0071364]; cerebellar cortex formation [GO:0021697]; cytokine-mediated signaling pathway [GO:0019221]; DNA damage checkpoint signaling [GO:0000077]; ephrin receptor signaling pathway [GO:0048013]; epidermal growth factor receptor signaling pathway [GO:0007173]; ERBB signaling pathway [GO:0038127]; face morphogenesis [GO:0060325]; fibroblast growth factor receptor signaling pathway [GO:0008543]; genitalia development [GO:0048806]; glucose homeostasis [GO:0042593]; heart development [GO:0007507]; homeostasis of number of cells within a tissue [GO:0048873]; hormone metabolic process [GO:0042445]; hormone-mediated signaling pathway [GO:0009755]; inner ear development [GO:0048839]; integrin-mediated signaling pathway [GO:0007229]; intestinal epithelial cell migration [GO:0061582]; megakaryocyte development [GO:0035855]; microvillus organization [GO:0032528]; multicellular organism growth [GO:0035264]; multicellular organismal reproductive process [GO:0048609]; negative regulation of cell adhesion mediated by integrin [GO:0033629]; negative regulation of chondrocyte differentiation [GO:0032331]; negative regulation of cortisol secretion [GO:0051463]; negative regulation of growth hormone secretion [GO:0060125]; negative regulation of insulin secretion [GO:0046676]; neurotrophin TRK receptor signaling pathway [GO:0048011]; organ growth [GO:0035265]; peptidyl-tyrosine dephosphorylation [GO:0035335]; platelet formation [GO:0030220]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of glucose import [GO:0046326]; positive regulation of hormone secretion [GO:0046887]; positive regulation of insulin receptor signaling pathway [GO:0046628]; positive regulation of interferon-beta production [GO:0032728]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of mitotic cell cycle [GO:0045931]; positive regulation of ossification [GO:0045778]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of cell adhesion mediated by integrin [GO:0033628]; regulation of protein export from nucleus [GO:0046825]; regulation of protein-containing complex assembly [GO:0043254]; regulation of type I interferon-mediated signaling pathway [GO:0060338]; T cell costimulation [GO:0031295]; triglyceride metabolic process [GO:0006641] | 11743164_upon translocation, CagA perturbs cellular functions by deregulating SHP-2 11896619_SHP-2 modulates phosphorylation of PDGF receptors, thereby controls RasGTP recruitment and Ras/MAP kinase signaling in the heterodimeric configuration of the PDGF receptors 11956229_Specific SHP-2 partitioning in raft domains triggers integrin-mediated signaling via Rho activation 11986327_interacts with siglec-11 11992261_Observational study of gene-disease association. (HuGE Navigator) 11992261_PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. 12070037_Band 3 is an anchor protein for and a target for SHP-2 tyrosine phosphatase in human erythrocytes. 12161469_PTPN11 mutations are responsible for Noonan syndrome in a substantial fraction of patients and that relatively infrequent features of Noonan syndrome, such as sensory deafness and bleeding diathesis, can also result from mutations of PTPN11. 12161596_Some PTPN11 mutations (e.g., Y279C) are associated with both the Noonan syndrome phenotype and with skin pigmentation anomalies, such as multiple lentigines or cafe au lait spots (LEOPARD syndrome). 12270932_SHP-2 is a dual-specificity protein phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues and plays an important role in modulating STAT function in gene regulation 12325025_Mutations in PTPN11/SHP2 underlie a common form of Noonan syndrome and confirm that the disease exhibits both allelic and locus heterogeneity. 12370245_Results indicate that Gab1 and SHP-2 promote the undifferentiated epidermal cell state by facilitating Ras/MAPK signaling. 12384786_absence of mutation in cases of cardiofaciocutaneous syndrome 12399420_activation state of alphaVbeta3 integrin is an important regulator of the duration of insulin-like growth factor I receptor phosphorylation and this regulation is mediated through changes in the subcellular localization of SHP-2 12403768_These data suggest, that there are two, largely distinct modes of negative regulation of gp130 activity, despite the fact that both SOCS3 and SHP2 are recruited to the same site within gp130. 12529707_We sequenced the entire coding region of the PTPN11 gene in ten well-characterised CFC patients and found no base changes. We also studied PTPN11 cDNA in our patients and demonstrated that there are no interstitial deletions either. 12531430_SHP-2 may function as an adaptor molecule downstream of the the prolactin receptor and highlight a new recruitment mechanism of SHP-2 substrates. 12543077_SHP2 positively regulates IL-2 induced MAPK activation in malignant T cells. SHP2 may not be involved in the activation of Stat3 or Stat5 in cutaneous T-cell lymphoma cells. 12552462_The CagA protein of Helicobacter pylori is translocated into epithelial cells and binds to SHP-2 in human gastric mucosa 12676785_During platelet activation, a functionally active complex between SHIP-2, filamin, actin, and GPIb-IX-V may orchestrate the localized hydrolysis of PtdIns(3,4,5)P3 and thereby regulate cortical and submembraneous actin. 12707331_SHP-2 catalytic activity plays a direct role in the inhibitory function of killer cell Ig-like receptors, and SHP-2 inhibits NK cell activation in concert with SHP-1. 12717436_Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. 12752577_PTPN11 mutations do not cause Costello syndrome 12959980_Required for RetM918T-induced Akt activation. Downstream mediator of mutated receptors RetC634Y and RetM918T. Acts as limiting factor in Ret-associated endocrine tumors, in neoplastic syndromes multiple endocrine neoplasia types 2A and 2B. 14522994_SHP-2 has a role as a positive regulator of cytokine receptor signaling by regulating ubiquitination/degradation pathways 14644997_SHP-2 is an important cellular PTPase that is mutated in myeloid malignancies 14665621_results reveal that Gab1 protein recruits SHP2 protein tyrosine phosphatase to dephosphorylate paxillin 14701753_SHP-2/Gab1 association is critical for linking EGFR to NF-kappaB transcriptional activity via the PI3-kinase/Akt signaling axis in glioblastoma cells 14982869_wider role of PTPN11 lesions in leukemogenesis, but also a lineage-related and differentiation stage-related contribution of these lesions to clonal expansion. 14991917_A missense mutation (836A-->G; Tyr279Cys) in exon 7 of PTPN11 gene was identified in the patient with LEOPARD syndrome, whereas no mutation in PTPN11 gene was detected in the father or in additional family members 15001945_Observational study of gene-disease association. (HuGE Navigator) 15187115_inhibition of NK cell cytotoxicity by KIR2DL5 was blocked by dominant-negative SHP-2, but not dominant-negative SHP-1, whereas both dominant-negative phosphatases can block inhibition by KIR3DL1. 15240615_Observational study of gene-disease association. (HuGE Navigator) 15240615_PTPN11 mutations account for approximately 40% of Noonan syndrome patients. Type of cardiovascular lesions and occurrence of hematological abnormalities are different in mutation-positive and mutation-negative patients. 15282682_In contrast to childood MDS and AML, mutations in PTPN11 make little or no contribution to the pathogenesis of adult MDS and AML. 15385933_PTPN11 missense mutations are associated with acute myeloid leukemia 15521065_Aspartate 61 plays a major role for proper down-regulation of the protein tyrosine phosphatase activity of SHP-2; the D61Del variant is predicted to have lower stability of the D'EF loop of the N-terminal SH2 domain compared to the wild-type 15539800_Most common mutation was A922G in exon 8. In exon 4 a mutation encoded C-SH2 domain of PTPN11 gene in two patients. A 218C-->T mutation was found in exon 3 in a patient with Noonan syndrome and mild juvenile myelomonocytic leukaemia. 15539800_Observational study of genotype prevalence. (HuGE Navigator) 15556604_SHP2 binds CAT and acquires a hydrogen peroxide-resistant phosphatase activity via integrin-signaling. 15563458_SHP-2 has a role in regulating IL-1-induced Ca2+ flux and ERK activation via phosphorylation of PLCgamma1 15644411_data support the hypothesis that PTPN11 mutations induce hematopoietic progenitor hypersensitivity to GM-CSF due to hyperactivation of the Ras signaling axis 15708852_Tyr-992 and Tyr-1173 are required for phosphorylation of the epidermal growth factor receptor by ionizing radiation and modulation by SHP2 15723289_Observational study of genotype prevalence. (HuGE Navigator) 15725481_Mutations are rare in adult myelodysplastic syndromes and chronic myelomonocytic leukemia. 15747776_10 genes were down-regulated following treatment of the T-ALL cells with 0.15 and 1.5 microg/mL of metal ores at 72 h 15834506_Pathogenesis of Noonan syndrome and leukemia is associated with enhanced phosphatase activity of mutant SHP-2 15888547_SHPS-1 functions as an anchor protein that recruits both Shc and SHP-2, whose recruitment is necessary for IGF-I-dependent Shc phosphorylation 15928039_Observational study of gene-disease association. (HuGE Navigator) 15928039_data suggest a genotype/phenotype correlation in the spectrum of PTPN11 mutations found in patients with juvenile myelomonocytic leukemia, Noonan syndrome/myeloproliferative disease, and Noonan syndrome 15940693_Results show that PTPN11 mutations are rarely found in two isolated forms of congenital heart disease. 15951301_PTPN11 mutations are rare in adult myeloid malignancies [review] 15956085_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15956085_Presence of PTPN11 mutations in patients with Noonan syndrome indicates a reduced growth response to long-term human growth hormone treatment. 15985432_an important role for a PECAM1-SHP2-Tie2 pathway in flow-mediated signal transduction. 15985475_SHP-2 mutations in Noonan syndrome cause mild GH resistance by postreceptor signaling defect, partially compensated for by elevated GH secretion. May contribute to short stature phenotype in children with SHP-2 mutations and poor response to rhGH. 15987685_Modeling analyses show that different SHP2 mutants can affect basal activation, SH2 domain-phosphopeptide affinity, and/or substrate specificity to varying degrees. 16030196_aberrantly increased expression of Shp2 may contribute, collaboratively with other factors, to leukemogenesis 16032704_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16053901_Missense mutations and the role of SHP-2 in signal transduction, development and hematopoiesis, as well as on the consequences of SHP-2 gain-of-function. Review. 16208280_cardiovascular anomalies and hematologic abnormalities are predominant in mutation positive patients 16250012_SHP-2 tyrosine phosphorylation on residue Y542 promoted IL-1beta mediated focal adhesion maturation. 16306077_SHP-2 recruitment to p85 is required for IGF-I-stimulated association of the p85/p110 complex with insulin receptor substrate-1 and for the subsequent activation of the PI-3 kinase pathway leading to increased cell migration 16339483_Type I collagen limits VEGFR-2 signaling by a SHP2 protein-tyrosine phosphatase-dependent mechanism 1. 16354697_Inhibition of focal adhesion kinase by SHP-2 plays a crucial role in the morphogenetic activity of Helicobacter pylori CagA. 16358218_Diversity of germline and somatic PTPN11 mutations were studied in Noonan syndrome, LEOPARD syndrome and leukemia. 16360206_The phosphorylation of SHP-2 by GM-CSF promotes the binding of SHP-2 to the GM-CSF receptor to the disadvantage of CLECSF6. 16380919_Observational study of genotype prevalence. (HuGE Navigator) 16488201_data suggests that mutations in the PTPN11 gene are not a cause of hypertrophic cardiomyopathy in the absence of Noonan/LEOPARD syndromes 16498234_A PTPN11 gene mutation (Y63C) causing Noonan syndrome is not associated with short stature in the general population. 16518851_Our data suggest that mutations of PTPN11 as well as RAS play a role in the pathogenesis of not only myeloid hematological malignancies but also a subset of RMS malignancies 16523510_The clinical suspicion of LEOPARD syndrome may be confirmed by molecular screening for PTPN11 mutations. 16533526_Observational study of genotype prevalence. (HuGE Navigator) 16598312_The PTPN11 protein mutation can be acquired during progression of myelodysplastic syndrome JMML. 16684964_Recruitment of SHP2 to speicific sites of autophosphorylation contributes to FLT3-mediated Erk activation and proliferation. 16762922_both SHP-1 and SHP-2 have a positive role in epidermal growth factor-induced ERK1/2 activation and they act cooperatively rather than antagonistically. 16825188_Upon IGF-I stimulation, a complex assembles on SHPS-1 that contains SHP-2, c-Src, and Shc wherein Src phosphorylates Shc, a signaling step that is necessary for an optimal mitogenic response 16905534_Tyr-542 of SHP-2 modulates IL-1-induced Ca2+ signals and association of the ER with focal adhesions 16914719_ICSBP tyrosine phosphorylation is necessary for the activation of NF1 transcription. ICSBP is a substrate for SHP2 protein tyrosine phosphatase (SHP2-PTP). 16920701_Our results suggest that DCA differentially regulates focal adhesion complexes and that tyrosine phosphatase ShP2 has a role in DCA signaling. 17020470_Mutation of the PTPN11 gene is the main causal factor in LEOPARD syndrome, and it also plays a role in neurofibromatosis-Noonan syndrome. 17020470_Observational study of genotype prevalence. (HuGE Navigator) 17028265_uPAR expression of lung airway epithelial cells is regulated at the level of mRNA stability by inhibition of protein tyrosine phosphatase-mediated dephosphorylation of uPAR mRNA binding proteins and demonstrate that the process involves SHP2. 17052965_1226 G-->C causes amino acid substitution G409A and results in Noonan Syndrome. Mutations in this particular region of SHP-2 may have effects on the protein that differ from those of the classical mutations. 17053061_PTPN11 as the first proto-oncogene that encodes a cytoplasmic tyrosine phosphatase with Shp2 domain; diseases caused by mutations; role in hematopoiesis 17177198_support the view that an increase in the affinity of SHP-2 for its binding partners, caused by destabilization of the closed, inactive conformation 17185494_The significance of exercise-induced alterations in cytosolic SHP2 and insulin-stimulated Akt pSer(473) on the improvement in insulin sensitivity requires further elucidation. 17211446_This short review discusses the physiological role of Shp2 in the molecular switch governing embryonic stem cell self-renewal versus differentiation. 17211494_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17214991_Association of single tSNPs with both apoB and LDL cholesterol as well as interactions between the two genes suggest that variants influencing SHP-2 activity may modulate the acute pathway by which insulin regulates these lipids 17235629_Observational study of gene-disease association. (HuGE Navigator) 17272397_novel mechanism regulating GH signaling in which estrogen receptor modulators enhance GH activation of the JAK2/STAT5 pathway in a cell-type-dependent manner by attenuating protein tyrosine phosphatase activities 17330262_An unselected series of 140 patients with therapy-related MDS or AML were investigated for mutations of PTPN11 in Exons 3, 4, 8, and 13. Four cases had mutations of the gene; three of these had deletions or loss of chromosome arm 7q. 17339163_Observational study of genotype prevalence. (HuGE Navigator) 17339163_PTPN11 mutations are responsible for Noonan syndrome in Taiwanese patients 17403678_association of the PDGFRbeta with lipid raft microdomains renders it susceptible to LXA(4)-mediated dephosphorylation by possible reactivation of oxidatively inactivated SHP-2. 17453145_understanding of Noonan syndrome and the LEOPARD syndrome pathogenesis will require solving the paradox that mutations oppositely influencing the biochemical activity of SHP-2 result in similar syndromes 17497712_Compund heterozygosity for Noonan syndrome-causing mutations in the PTPN11 gene, documenting association with early fetal death, is reported. 17515436_Observational study of gene-disease association. (HuGE Navigator) 17515436_The most prevalent links to PTPN11 gene dysfunction in cardiac development in patients with a PTPN11 mutation are pulmonary valve stenosis, atrial septal defect, ostium secundum type, and stenosis of the peripheral pulmonary arteries. 17546245_Missense mutations were identified in 48.5% of the NS patients. There was a positive correlation between the presence of PTPN11 mutations and pulmonary stenosis frequency in NS patients. 17546245_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17561098_SHP-1 and SHP-2, two structurally related cytoplasmic protein-tyrosine phosphatases with different cellular functions and cell-specific expression patterns, were compared for their intrinsic susceptibility to oxidation by H(2)O(2). 17664338_Results describe the neurite outgrowth multiadaptor RhoGAP, NOMA-GAP, and show that it regulates neurite extension through SHP2 and Cdc42. 17672918_occurrence of an unusual TG 3' splice site in intron 10 17717529_Virus attachment induced tyrosine phosphorylation of PVR; this permitted the association of PVR with SHP-2, a protein tyrosine phosphatase whose activation was required for entry and infection 17875892_FIRST CASE TO OUR KNOWLEDGE OF LS SYNDROME FEATURING A NEW PTPN11 GENE MUTATION 17910045_Frequently mutated in high hyperdiploid childhood acute lymphoblastic leukemia. 17910045_Observational study of gene-disease association. (HuGE Navigator) 17942397_Shp2E76K induces cytokine-independent survival of TF-1 cells by a novel mechanism involving up-regulation of Bcl-XL through the Erk1/2 pathway. 17962719_SHP-2 regulates endothelial cell survival through PI3-K-Akt and mitogen-activated protein kinase pathways thereby strongly affecting new vessel formation. 17972951_correlation of PTPN11 mutations with NPM1 mutations and FLT3/ITD among adult AML patients 18159945_The finding that defective Shp2 signaling induced cell movement defects as early as gastrulation may have implications for the monitoring and diagnosis of Noonan and LEOPARD syndrome. 18203203_There is little phenotype-genotype correlation with electrocardiographic findings in Noonan syndrome patients. 18223690_analysis of the gain-of-function SHP-2 mutants with oncogenic RAS-like transforming activity from solid tumors 18241070_Observational study of gene-disease association. (HuGE Navigator) 18246201_activation of SHP2 protein tyrosine phosphatase synergized with ICSBP haploinsufficiency to facilitate cytokine-induced myeloproliferation, apoptosis resistance, and rapid progression to AML in a murine bone marrow transplantation model. 18331608_Mutations in the PTPN11 gene are commonly involved in the pathogenesis of Noonan syndrome but are not a common cause of idiopathic short stature. 18331608_Observational study of gene-disease association. (HuGE Navigator) 18372317_L282V substitution perturbs the stability of SHP2's closed conformation and T42A substitution promotes phosphopeptide-binding affinity. 18381291_Shp2 binds most strongly when both of the NPXY motifs in LRP1 are phosphorylated 18543080_Our results demonstrate putative roles of SHP-1 and SHP-2 in the progression of both Condyloma acuminatum and cervical cancer after HPV infection. 18559669_Shp2 mediates dephosphorylation of ROCKII and, therefore, regulates RhoA-induced cell rounding, indicating that Shp2 couples with RhoA signaling to control ROCKII activation during deadhesion. 18562489_In 22 children with a mutation in PTPN11 mean gain in H-SDS for National standards was +1.3, not different from the mean gain in the five children without a mutation in PTPN11+1.3. 18564921_Observational study of gene-disease association. (HuGE Navigator) 18577518_the Gab1-SHP2-ERK1/2 signaling pathway comprises an inhibitory axis for IGF-I-dependent myogenic differentiation. 18593762_Observational study of gene-disease association. (HuGE Navigator) 18617527_Cdk2-associated complexes, by targeting SHP-1 for proteolysis, counteract the ability of SHP-1 to block cell cycle progression of intestinal epithelial cells 18640765_Mutations in PTPN11, which encodes the protein tyrosine phosphatase Shp2, are commonly found in juvenile myelomonocytic leukemia. 18643929_SHP2 is a widely overexpressed signalling protein in infiltrating ductal carcinoma breast tumours. 18647419_Data show that Shp2 activation promotes the dispersal of pre-patterned acetylcholine receptors (AChRs)clusters to facilitate the selective accumulation of AChRs at developing neuromuscular junction. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18712962_Observational study of gene-disease association. (HuGE Navigator) 18712962_Single nucleotide polymorphism at intron 3 of PTPN11 gene is associated with a lower risk of gastric atrophy. 18728972_higher nuclear SHP2 expression in B-cell lymphoma cases 18758896_Hodgkin's lymphoma in a patient with Noonan syndrome with germ-line PTPN11 mutations. 18827006_SHP-2 is a novel target of Abl kinases during cell proliferation 18829466_Shp-2 retains TERT in the nucleus by regulating tyrosine 707 phosphorylation. 18832710_SHP2 activation induced by HCMV infection is responsible for the down-regulation of IFN-gamma-induced STAT1 tyrosine phosphorylation. 19020799_Mutations in the PTPN11 are associated with Noonan syndrome. 19020799_Observational study of gene-disease association. (HuGE Navigator) 19047918_Juvenile myelomonocytic leukemia with PTPN11 mutation might be a distinct subgroup with specific clinical characteristics and poor outcome. 19047918_Observational study of gene-disease association. (HuGE Navigator) 19066472_Protein Tyrosine Phosphatase, Non-Receptor Type 11 modulates Cyclin-Dependent Kinase Inhibitor p27 stability and contributes to Cyclin-Dependent Kinase Inhibitor p27 -mediated cell cycle progression. 19077116_Observational study of gene-disease association. (HuGE Navigator) 19096001_Downregulation of platelet responsiveness upon contact with LDL by the protein-tyrosine phosphatases SHP-1 and SHP-2. 19120036_Mutation analysis of PTPN11 was conducted on patient with severe form of Noonan Syndrome and Cafe-Au-Lait spots, using DNA sequencing. Results suggest additive effect of F285L mutation in PTPN11 may be associated Cafe-au-Lait spots. 19133257_Results demonstrate that SHP-2 plays an important role in uPA-directed signaling and functional control of human VSMC and suggest that this phosphatase might contribute to the pathogenesis of the uPA-related vascular remodeling. 19133693_Observational study of gene-disease association. (HuGE Navigator) 19160029_Observational study of gene-disease association. (HuGE Navigator) 19160029_genetic polymorphisms are biomarkers for ulcerative colitis susceptibility in the Japanese population 19166311_The kinetic and mechanistic details of reversible oxidation of SHP-1 and SHP-2, were investigated. 19189703_In more than half the patients with Noonan Syndrome a mutation in the PTPN11 gene was identified. 19206169_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19261604_SHP2 promotes HER2-induced signaling and transformation at least in part by dephosphorylating a negative regulatory autophosphorylation site. 19275884_These results suggest that SHP-2 regulates tyrosine phosphorylation of Cas-L, hence opposing the effect of kinases, and SHP-2 is a negative regulator of cell migration mediated by Cas-L. 19287004_JAK2-SOCS1 and SHP2 reciprocally regulate ASK1 phosphorylation and stability in response to cytokines. 19290061_results reveal a common signaling mechanism shared by human and mouse embryonic stem cells via Shp2 modulation of overlapping and divergent pathways 19336370_Observational study of gene-disease association. (HuGE Navigator) 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19352411_analysis of SOS1 and PTPN11 mutations in five cases of Noonan syndrome with multiple giant cell lesions 19427850_The activation of SHP-2 phosphorylation at Tyr542 in glioblastoma cell lines results in increased PTPase activity and distinct mechanisms of cell cycle progression and SHP-2. Its PTPase activity plays a critical role in EGFRvIII-mediated transformation. 19449407_Independent NF1 and PTPN11 mutations in a family with neurofibromatosis-Noonan syndrome are reported. 19509418_Stat3 is an essential signaling component potentially contributing to the pathogenesis of Noonan syndrome and juvenile myelomonocytic leukemia caused by PTPN11 gain-of-function mutations 19528235_Results suggest that increased c-Jun expression due to mutant Shp2-induced Ras hyperactivation, and reduced GATA2 expression promote aberrant monocytic differentiation induced by activating PTPN11 mutants. 19582499_This study describes the association between the Gln510Glu mutation of the PTPN11 gene and lethal progressive hypertrophic cardiomyopathy (HCM) in a newborn with the NS phenotype. 19589142_PTPN11 polymorphism is associated with a lower risk of severe gastric atrophy in Helicobacter pylori infections, but it is not associated with a decreased risk of gastric cancer in Japanese. 19589868_SHP-2 is a component of the IGF signaling pathway that is required for normal placental growth. 19681119_Investigated the prevalence of mutations in a relatively large cohort of primary embryonal Rhabdomyosarcoma (RMS) tumors. PTPN11 was found mutated in one tumor specimen, confirming that somatic defects in this gene are relatively uncommon in RMS. 19690960_Data indicate that gastric cancers display a higher expression of PTPN11 protein than the normal cells, suggesting that neo-expression of this positive signaling protein in the cells might play a role in the cancer development. 19706677_These data establish a novel role for SHP-2 phosphatase in the dopamine-mediated regulation of VEGFR-2 phosphorylation. 19725129_This is the first report of mutations in both FBN1 and PTPN11 with combined phenotypes of Marfan and LEOPARD syndromes. 19735729_Grb2 and FGFR2 interaction controls the activity of Shp2 toward FGFR2 which might suggest that signalling is influenced by factors other than simply growth factor engagement. 19737548_Observational study of genetic testing. (HuGE Navigator) 19768645_two unrelated LEOPARD syndrome cases with a common PTPN11 mutation Y279C and with completely different clinical features 19773259_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19776760_Observational study of gene-disease association. (HuGE Navigator) 19776760_PTPN11 mutations in childhood acute lymphoblastic leukemia occur as a secondary event associated with high hyperdiploidy. 19822020_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19822020_Polymorphisms in NQO1, IL-10 and PTPN11, in combination with Helicobacter pylori status, could be used to identify individuals who are more likely to develop intestinal metaplasia and therefore gastric cancer. 19864201_reports 4 cases of LEOPARD syndrome with point mutation of PTPN11 gene 19874312_assessment of PTPN11 mutations in juvenile myelomonocytic leukaemia 19915046_our studies have identified a new, KIR-independent role for SHP-2 in dampening NK cell activation in response to tumor target cells in a concentration-dependent manner 20004456_The authors find that activity of G protein coupled receptor protein of human herpesvirus 8 results in phosphorylation of regulatory tyrosines in Shp2 and that in turn, Shp2 is required for vGPCR-mediated activation of MEK, NFkappaB, and AP-1. 20006740_Observational study of gene-disease association. (HuGE Navigator) 20064076_Prenatal diagnosis of Noonan syndrome in a woman carrying a PTPN11 gene mutation. 20080972_PRL diminished IGF-I-induced IGF-IR internalization, which may result from reduced SHP-2 association with IGF-IR, because we demonstrated an essential role for SHP-2 in IGF-IR internalization 20170098_Data show that the structure of SHP2 in complex with inhibitor II-B08 reveals molecular determinants that can be exploited for the acquisition of more potent and selective SHP2 inhibitors. 20186801_Observational study of gene-disease association. (HuGE Navigator) 20237281_Shp2 is specifically required for the initiation of retinal neurogenesis but not for the maintenance of the retinal differentiation program. 20237506_PTPN11 mutations are associated with relapsed childhood high hyperdiploid acute lymphoblastic leukemia. 20302979_Observational study of gene-disease association. (HuGE Navigator) 20308328_Data show that PTPN11 mutations causing Leopard syndrome (LS) facilitate EGF-induced PI3K/AKT/GSK-3beta stimulation through impaired GAB1 dephosphorylation, resulting in deregulation of a novel signaling pathway that could be involved in LS pathology. 20398180_These results suggest that calpain-dependent cleavage of SHP-1 and SHP-2 may contribute to protein tyrosine dephosphorylation in Jurkat T cell death induced by Entamoeba histolytica. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20472558_IL-1-induced signaling through focal adhesions leading to MMP3 release and interactions between SHP-2 and PTPalpha are dependent on the integrity of the catalytic domains of PTPalpha. 20512931_These findings highlight a novel negative regulatory role for integrin beta4 in endothelial cell inflammatory responses involving SHP-2-mediated MAPK signaling 20543023_Observational study of gene-disease association. (HuGE Navigator) 20577567_identified an 11 bp deletion in exon four of PTPN11, which alters frame, results in premature translation termination, and co-segregates with the phenotype. 20578946_Patients with Noonan syndrome and a PTPN11 mutation presented significantly higher prevalence of short stature (p = 0.03) and pulmonary valve stenosis (p = 0.01), and lower prevalence of hypertrophic cardiomyopathy (p = 0.01. 20631249_Data suggest a model for the involvement of SHP-2 in PECAM-1-dependent motility in which SHP-2, recruited by its interaction with PECAM-1, targets paxillin to ultimately activate the MAPK pathway and downstream events required for cell motility. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20671117_Tyr251 and Tyr301 of IL-22R1 are required for Shp2 binding and IL-22-induced Erk1/2 activation. 20673499_The expression rate of SHP2 is high and closely correlated to lymphnode metastasis in NSCLC, which implies the occurrence and development of lung cancer maybe related to SHP2, and SHP2 maybe a new marker and therapeutic targets for lung cancer. 20700123_Identification of the tumour-suppressive miRNA miR-489 and its target, PTPN11, might provide new insights into the underlying molecular mechanisms of hypopharyngeal squamous cell carcinoma 20718194_first report on molecular analysis and clinical features of a Turkish mother and son diagnosed with Noonan syndrome.The analysis revealed an A --> G transition at position 923 in exon 8 of the PTPN11 gene, indicating an Asn308Ser substitution. 20723025_PECAM-1-mediated inhibition of GPVI-dependent platelet responses result from recruitment of SHP-2-p85 complexes to tyrosine-phosphorylated PECAM-1, which diminishes the association of PI3K with activatory signaling molecules Gab1 and LAT 20734064_Observational study of gene-disease association. (HuGE Navigator) 20829714_Diagnosis of the syndrome was confirmed by the identification of earlier reported germline missense mutations in the PTPN11 gene. 20840817_The enhancement of SHP2 expression in non-small cell lung cancer is correlated with tobacco smoking. 20855525_Data describe how alpha6beta4 integrin selectively activates Fyn in response to receptor engagement, and show that both catalytic and noncatalytic functions of SHP2 are required for Fyn activation by alpha6beta4. 20880116_Observational study of gene-disease association. (HuGE Navigator) 20954246_The aim of this study was to evaluate phenotypic characteristics, PTPN11 gene mutations, and hematological and coagulation parameters in 30 clinically diagnosed cases of Noonan syndrome. 20958258_it was possible to identify candidate genes that could be involved in the molecular mechanisms discriminating PTPN11 and RAS mutations in ALL 21047965_The authors show that vGPCR contains a bona fide immunoreceptor tyrosine-based inhibitory motif (ITIM) that binds and constitutively activates Shp2. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21060863_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21068439_These findings illustrate an essential role for SHP-2 in skeletal growth and remodeling. 21106241_Germline PTPN11 mutation affecting exon 8 is associated with syndromic juvenile myelomonocytic leukemia. 21145937_The role of protein tyrosine phosphatases in the regulation of steroid biosynthesis, relating them to steroidogenic acute regulatory protein, arachidonic acid metabolism and mitochondrial rearrangement, is described. 21169408_SHP-2 as an essential component of tumor suppression and anoikis mediated by SIRPalpha1 in human breast carcinoma cells as well as in v-Src-transformed cells. 21365683_Combination of computational and experimental methods are used to investigate the structural mechanism of opening of SHP2 and the impact of three gain-of-function mutants, D61G, E76K, and N308D, on the opening mechanism. 21407260_study provides first evidence of an increased risk of cancer in patients with Noonan syndrome and a PTPN11 mutation, compared with that in the general population 21419049_The NS1 protein was observed to be expressed in 293T cells. 21528083_Data suggest that SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are potential prognostic markers in colorectal cancer. 21531714_SPRED1 is a likely substrate of SHP2, whose tyrosine dephosphorylation is required to attenuate the inhibitory action of SPRED1 in the Ras/ERK pathway. 21533187_Heterozygous loss-of-function mutations in PTPN11 are a frequent cause of metachondromatosis. | ENSMUSG00000043733 | Ptpn11 | 2159.94455 | 1.0009769 | 0.0014086274 | 0.12567171 | 1.227321e-04 | 9.911609e-01 | 9.998360e-01 | No | Yes | 2329.34422 | 438.837478 | 2.321357e+03 | 336.962306 | |
| ENSG00000179348 | 2624 | GATA2 | protein_coding | P23769 | FUNCTION: Transcriptional activator which regulates endothelin-1 gene expression in endothelial cells. Binds to the consensus sequence 5'-AGATAG-3'. | 3D-structure;Activator;Alternative splicing;DNA-binding;Disease variant;Isopeptide bond;Metal-binding;Methylation;Nucleus;Phagocytosis;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the GATA family of zinc-finger transcription factors that are named for the consensus nucleotide sequence they bind in the promoter regions of target genes. The encoded protein plays an essential role in regulating transcription of genes involved in the development and proliferation of hematopoietic and endocrine cell lineages. Alternative splicing results in multiple transcript variants.[provided by RefSeq, Mar 2009]. | hsa:2624; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; C2H2 zinc finger domain binding [GO:0070742]; chromatin binding [GO:0003682]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription coactivator binding [GO:0001223]; transcription coregulator binding [GO:0001221]; zinc ion binding [GO:0008270]; cell differentiation in hindbrain [GO:0021533]; cell fate commitment [GO:0045165]; cell fate determination [GO:0001709]; cell maturation [GO:0048469]; central nervous system neuron development [GO:0021954]; cochlea development [GO:0090102]; commitment of neuronal cell to specific neuron type in forebrain [GO:0021902]; definitive hemopoiesis [GO:0060216]; embryonic placenta development [GO:0001892]; eosinophil fate commitment [GO:0035854]; GABAergic neuron differentiation [GO:0097154]; homeostasis of number of cells within a tissue [GO:0048873]; inner ear morphogenesis [GO:0042472]; negative regulation of endothelial cell apoptotic process [GO:2000352]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of fat cell proliferation [GO:0070345]; negative regulation of gene expression [GO:0010629]; negative regulation of macrophage differentiation [GO:0045650]; negative regulation of neural precursor cell proliferation [GO:2000178]; negative regulation of Notch signaling pathway [GO:0045746]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neuron migration [GO:0001764]; phagocytosis [GO:0006909]; pituitary gland development [GO:0021983]; positive regulation of angiogenesis [GO:0045766]; positive regulation of blood vessel endothelial cell migration [GO:0043536]; positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis [GO:1903589]; positive regulation of cell migration involved in sprouting angiogenesis [GO:0090050]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of erythrocyte differentiation [GO:0045648]; positive regulation of gene expression [GO:0010628]; positive regulation of mast cell degranulation [GO:0043306]; positive regulation of megakaryocyte differentiation [GO:0045654]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of phagocytosis [GO:0050766]; positive regulation of phagocytosis, engulfment [GO:0060100]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of forebrain neuron differentiation [GO:2000977]; regulation of histone acetylation [GO:0035065]; regulation of primitive erythrocyte differentiation [GO:0010725]; response to lipid [GO:0033993]; semicircular canal development [GO:0060872]; somatic stem cell population maintenance [GO:0035019]; urogenital system development [GO:0001655]; vascular wound healing [GO:0061042]; ventral spinal cord interneuron differentiation [GO:0021514] | 7541039_The GATA-2 transcription factor interacts with the human eNOS promoter at a region which includes a putative GATA site [-230 to -226]. 11877047_GATA-2 transcripts was highly expressed in leukemia patients and had no change in remission. 11964310_Interactions of GATA-2 with the promyelocytic leukemia zinc finger (PLZF) protein, its homologue FAZF, and the t(11;17)-generated PLZF-retinoic acid receptor alpha oncoprotein. 12045236_Essential and instructive roles of GATA factors in eosinophil development. 12073612_regulates hematopoiesis 12145700_GATA-1 and GATA-2 gene expression is related to the severity of dysplasia in myelodysplastic syndrome. 12432220_REVIEW: Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage 12750312_Suppression of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. 15001660_GATA-2 exists as an acetylated protein in immature precursor cells; GATA-2 was acetylated in vitro by p300 and GCN5 at multiple acetylation sites, increasing its DNA-binding activity. 15254248_there is functional cross talk between RA and GATA-2-dependent pathways 15328158_Erythropoietin (Epo) gene expression is under the control of hypoxia-inducible factor 1 (HIF-1), and is negatively regulated by GATA2. 15632071_Results show that in addition to its previously recognized function in suppressing PPARgamma transcriptional activity, interaction of GATA-2 and -3 with C/EBP is necessary for their ability to negatively regulate adipogenesis. 15837948_insulin induces GATA2 phosphorylation on serine 401 in a PI-3K/Akt-dependent manner, impairing GATA2 translocation to the nucleus and its DNA binding activity 16153155_GATA-2 and HNF-3beta regulate the human alcohol dehydrogenase 1A (ADH1A) gene. 16672344_Gata2 can operate independently of neuronal differentiation 16934006_Observational study of gene-disease association. (HuGE Navigator) 16934006_identification of 5 single nucleotide polymorphisms significantly associated with early-onset coronary artery disease; observations identify GATA2 as a novel susceptibility gene for coronary artery disease 17095623_a lack of integrin engagement leads to the induction of cellular markers associated with myeloid differentiation 17654061_GATA-2 expression in bone marrow stromal cells from chronic aplastic anemia was significantly lower than controls. Expression levels of GATA genes may influence hematopoiesis in BM microenvironment & relate to the pathogenesis/development of AA. 18078130_GATA-1 and GATA-2 were expressed at higher levels in patients with Monge's disease than in controls. 18250304_data strongly suggest that GATA-2 mutations may play a role in acute myeloid transformation in a subset of CML patients 18308945_Introducing GATA2 into microvascular endothelial cells resulted in dedifferentiation-like transcriptome reprogramming, with hematopoietic stem cell genes (such as ANGPT1) being up and endothelial genes (such as EPHB2) being down. 18452556_Evi-1 promotes hematopoietic stem/progenitor expansion at the embryonic stage through up-regulation of GATA-2 and repression of TGF-beta signaling. 18720385_Hypoxic regulation of Ang-2 is HIF-dependent and demonstrate that HIF-1alpha binds in human microvascular endothelial cells (HMVEC) to an evolutionary conserved Hypoxia-Responsive Element (HRE) located in the first intron of the Ang-2 gene. 19097174_Gata-2 overexpression in AML was associated with a low percentage of blasts in bone marrow and males. 19168794_GATA-2 activity inhibits cell cycle in vitro & in vivo. It is a molecular entry point into the transcriptional program regulating quiescence in human hematopoietic stem and progenitor cells. 19212333_results suggest that GATA-1 and/or GATA-2 binding to a GATA site of the 3' enhancer of WT1 played an important role in WT1 gene expression 19304323_GATA-2 L359 V is exclusively associated with CML progression but not other hematological malignancies and P250A is a new single nucleotide polymorphism. 19304323_Observational study of gene-disease association. (HuGE Navigator) 19453261_Observational study of gene-disease association. (HuGE Navigator) 19522008_Data found that Gfi-1/GATA-2 in immature progenitors and Gfi-1B/GATA-1 in erythroblasts are bound to the Gfi-1B and c-myc promoter. The switch is associated to an increase in Gfi-1B transcription whereas it triggers repression of c-myc transcription. 19528235_Results suggest that increased c-Jun expression due to mutant Shp2-induced Ras hyperactivation, and reduced GATA2 expression promote aberrant monocytic differentiation induced by activating PTPN11 mutants. 19684615_upregulation of GATA-2 may contribute to the progression to aggressive prostate cancer through modulation of expression of androgen receptor and key androgen-regulated genes. 19706030_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19714312_Results established that GATA2 and c-Jun/c-Fos act additively in modulating the SIRT3-VNTR enhancer function. 19772889_Data show that GATA-2 expression by MSCs from AA patients was significantly lower than in normal subjects, and conversely, expression of PPARgamma was significantly higher in AA patients. 19860791_Observational study of gene-disease association. (HuGE Navigator) 19864173_GATA2 polymorphism is not an important risk factor for sporadic Parkinson disease in Caucasians 19864173_Observational study of gene-disease association. (HuGE Navigator) 19885677_Data suggest that GATA2 does not contribute to the development of angiographic CAD among sporadic cases. 19885677_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19941826_Chromatin immunoprecipitation-sequencing was used to define GATA-1 and GATA-2 occupancy genome-wide in erythroid cells. 20363750_acetylation of EVI1 at Lys(564) by P/CAF enhances the DNA binding capacity of EVI1 and thereby contributes to the activation of GATA2 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20634887_The chromatin modifications were determined at five PRC2 targets commonly underexpressed in multiple myeloma (CIITA, CXCL12, GATA2, CDH6 and ICSBP/IRF8). The selected genes were confirmed to be underexpressed in MM compared to normal plasma cells. 20838640_T3 increas the affinity of TR interaction with GATA2. 21297973_Forced re-expression of Gata2 was not compatible with sustained growth of leukaemic cells thus suggesting a previously unrecognised role for Gata2 in downregulation during the development of AML 21571218_The genome-wide binding sites for the GATA2 in primary human megakaryocytes to identify the essential regulator of complex mammalian differentiation processes. 21666600_Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. 21670465_Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection syndrome. 21765025_Mutations in GATA2 are associated with dendritic cell, monocyte, B and NK lymphoid deficiency. 21808000_This work establishes a link between a GATA factor and inflammatory genes, mechanistic insights underlying GATA-2-AP-1 cooperativity and a rigorous genetic framework for understanding GATA-2 function in normal and pathophysiological vascular states 21816832_Nonmyeloablative hematopoietic stem cell transplantation in GATA2 deficiency results in reconstitution of the severely deficient monocyte, B-cell, and NK-cell populations and reversal of the clinical phenotype. 21892158_Haploinsufficiency of GATA2 underlies primary lymphedema and predisposes to acute myeloid leukemia in Emberger syndrome. 21892162_GATA2 is a new predisposition gene for familial myelodysplastic syndrome-acute myeloid leukemia. 21904383_we report a large series of patients in which overexpression of GATA2 is a recurrent event associated with a poor outcome in AML. 22021428_Discoverery of a novel reverse regulation within the traditional PTEN/AKT signaling pathway, whereby AKT induces GATA2 with consequent decreased PTEN transcription in breast cancer. 22147895_A crucial role for GATA2 in lymphatic vascular development. 22271902_Germline GATA2 mutations predispose to familial myelodysplastic syndrome/acute myeloid leukemia; monosomy 7 and ASXL1 mutations may be recurrent secondary genetic abnormalities triggering overt malignancy. 22284968_GATA2 CPG island methylation predicts progression in bladder cancer. 22541434_Of 26 non-small cell lung cancer cell lines, 14 were KRAS mutant and 12 were wild type; short-term GATA2 knockdown led to specific decreased cell viability and increased apoptosis in KRAS mutant NSCLC cells, comparable to the effect seen with loss of KRAS itself. 22649106_mutational screening detected novel GATA2 ZF1 mutations in 13 of 33 biallelic CEBPA-positive cytogenetically normal-AML patients 22786876_High GATA2 expression is a novel prognostic marker for poor outcome in pediatric AML patients. 22814295_GATA2 mutations with with biallelic CEBPA mutations are associated are frequent in intermediate-risk karyotype acute myeloid leukemia. 22865859_GATA2 and GATA1 positively and negatively control human ST2 gene transcription, respectively 22942019_We used this new approach to analyse exome data from 24 patients with primary immunodeficiencies. Our analysis identified two novel causative deletions in the genes GATA2 and DOCK8 22996659_A heterozygous deletion in a GATA2 cis-element consisting of an E-box & a GATA motif which was found in a patient with mycobacterial infection & myelodysplasia caused embryo lethality when induced in mice. 23028422_GATA-2 directly regulates HOXB4 expression in hematopoietic stem cells, which may play an important role in the development and/or progression of aplastic anemia. 23223431_High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. 23322776_SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster 23327922_Transcriptional regulators cooperate to establish or maintain primitive stem cell-like signatures in leukemic cells. 23365437_Authors show that the cytomegalovirus UL144 gene is expressed during latent infection in two cell types of the myeloid lineage, CD34(+) and CD14(+) monocytes, and that the UL144 protein is functional in latently infected monocytes. 23365458_GATA2 is required for the maturation of human natural killer cells and the maintenance of the CD56(bright) pool in the periphery. 23423786_Colorectal cancer patients with high levels of GATA2 expression demonstrate worse disease-free survival outcomes than those with low GATA2 levels. 23502222_GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. 23521373_A large percentage of CEBPA double-mutated acute myeloid leukaemia cases harboured additional alterations in other molecular markers and that at least GATA2 and TET2 alterations demonstrated a substantial impact on prognosis in these patients. 23560626_CEBPA and GATA2 mutant levels indicated that both mutations were likely to be early events in leukaemogenesis 23563236_GATA2 mutation is associated with myelodysplastic syndromes. 23892628_GATA2 and Lmo2 cooperatively regulate VEGF-induced angiogenesis and lymphangiogenesis via NRP2. 24033149_GATA2 mutations are associated with a favourable outcome in paediatric AML. 24077845_Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. 24227816_GATA2 deficiency is associated with disorders of hematopoiesis, lymphatics, and immunity. 24345756_GATA2 mutation is associated with myelodysplastic syndromes. 24448395_GATA2 gene silencing in human prostate cancer LNCaP cells led to a marked reduction in cell migration, tissue invasion, focal adhesion disassembly and to a dramatic change in cell transcriptomes. 24498120_RNAi-mediated knockdown of GATA2 expression significantly enhanced proliferation. 24509415_Overexpression of GATA-2 in K562 cells led to reduced TGFbeta-induced erythroid differentiation while knockdown of GATA-2 enhanced TGFbeta-induced erythroid differentiation. 24514424_low-GATA2-expressing specimens (GATA2(low)) exhibit allele-specific expression (ASE) (skewing) in more than half of acute myeloid leukemia patients. 24603652_GATA6, which is hypomethylated and abundant in endometriotic cells, potently blocked hormone sensitivity, repressed GATA2, and induced markers of endometriosis when expressed in healthy endometrial cells. 24614497_The results of this study showed that GATA2 regulates the development of cochlear hair cells and the inferior colliculus (IC), which are important in tonotopic mapping. 24639354_we have demonstrated that PU.1, GATA1, and GATA2 are involved in the expression of FcepsilonRI in a human mast cell line and primary human mast cells using siRNA with high transfection efficiency, and by ChIP assay. 24703711_3q inversion and translocation reposition a distal GATA2 enhancer to ectopically activate EVI1 and simultaneously confer GATA2 functional haploinsufficiency, previously identified as the cause of sporadic familial acute myeloid leukemia/myelodysplastic syndrome. 24703906_Authors identify a mechanism whereby a GATA2 distal hematopoietic enhancer (G2DHE or -77-kb enhancer) is brought into close proximity to the EVI1 gene in inv(3)(q21;q26) inversions, leading to leukemogenesis. 24754962_study reported a case of familial AML-MDS with two novel GATA2 mutations; literature reviewed 24786211_This study points to an association of GATA2 at both SNP and haplotype levels with important metabolic risk traits for atherosclerosis. 24807155_GATA2 is epigenetically repressed in human and mouse lung tumors and its further inhibition is not a valid therapeutic strategy for KRAS mutant lung cancer. 25056917_p38 also induced multisite phosphorylation of wild-type GATA-2, which required a single phosphorylated residue (S192). Phosphorylation of GATA-2, but not T354M, stimulated target gene expression. 25092790_one of the targets of p38 in this context is another GATA transcription factor, GATA3, recently demonstrated to promote HSC cycling and restrict their long-term reconstitutive potential downstream of p38 in stress-induced hematopoiesis [9]. 25140787_MonoMAC syndrome is a newly discovered immune deficiency syndrome caused by GATA-2 mutation, which is an autosomal dominant genetic disease [review] 25150255_GATA2 plays an important role in regulating the differentiation potential of BM-MSC and contributes to hematopoietic supporting capacity. 25230694_GATA1 and GATA2 are involved in clear cell renal cell carcinoma biology possibly affecting tumor development and aggressiveness. 25241285_Data suggest that the original GATA binding protein 2 (GATA2) mutations might be lost during disease progression in GATA2-mutated patients, while novel GATA2 mutations might be acquired at relapse in GATA2-wild patients. 25340772_The results demonstrate that hGATA-1 and hGATA-2 expression in hippocampus is sufficient to cause depressive like behaviors. 25359990_GATA2 deficiency analysis is important for the differential diagnosis of idiopathic aplastic anemia 25489091_propose pharmacological inhibition of GATA2 as a first-in-field approach to target AR expression and function and improve outcomes in CRPC 25509816_we explain the new function of GATA2, and describe the clinical phenotypes, laboratory findings, pathology, genetic anomalies and etiology. 25611491_Homozygous CEPBA mutations have a similar gene expression signature as CEPBA double mutations and thus may be considered as equivalent in the presence of GATA2 mutations. 25624456_The study showed that human GATA2 binds to its own promoter, activating its transcription, and that the p.Arg396Gln mutation impairs the transcription of GATA2. 25670080_These studies reveal a GATA2-IGF2 aggressiveness axis in lethal prostate cancer and identify a therapeutic opportunity in this challenging disease. 25670854_Ubiquitin-dependent degradation of GATA 2 is promoted by Fbw7, is cyclin B-CDK1-mediated Thr176 phosphorylation-dependent, and influences hematopoietic cell differentiation. 25676417_GATA2 germline mutation in a pedigree presenting with myelodysplastic syndrome/acute myeloid leukemia is associated with concurrent thrombocytopenia. 25707267_the molecular biology, clinical, hematological and immunological features of patients with GATA2 mutations (Review) 25707769_High GATA2 promotes glioma progression through EGFR/ERK/Elk-1 pathway. 25810277_Altered binding of GATA2 (and Sp1/TBP) mediates aberrant MAOA expression under pathophysiological conditions. 25907033_Transgenic human GATA2 appeared to induce growth via downstream activation of Nmyc and Hoxa9 in mice. GATA2 overexpression at low level confers self-renewal capacity to myeloid progenitors and is relevant to myeloid leukemia development. 26161748_Data show that the Lim domain only 2 (LMO2) regulatory element (element-25) region consists of transcription factor GATA2-binding myeloid ennhancer and RUNX protein-binding T-cell repressor. 26214525_data unveil essential roles for GATA2 in the lymphatic vasculature and explain why a select catalogue of human GATA2 mutations results in lymphedema. 26264606_In children and adults with severe GM-CSF negative PAP a close cooperation between pneumologists and hemato-oncologists is needed to diagnose the underlying diseases, some of which are caused by mutations of transcription factor GATA2. 26287967_results demonstrated that GATA2 single nucleotide polymorphism rs2335052 is an independent predictor for prognosis of colorectal cancer patients. 26325290_we screened 995 transcription factor genes and revealed that CITED2 acts as a GATA-2 activator in human hematopoietic cells. These results provide novel insights into and further identify the regulatory mechanism of GATA-2 26710799_GATA2 deficiency is associated with impaired membrane expression and chemotactic dysfunctions of CXCR4. 26751772_the distinct mechanisms by which GATA2 and FOXA1 regulate AR cistrome and suggest that FOXA1 acts upstream of GATA2 and AR in determining hormone-dependent gene expression in prostate cancer. 26766440_GATA2-to-GATA1 switch is prevalent at dynamic enhancers and drives erythroid enhancer commissioning 26767875_Letter/Case Report: frameshift mutation in GATA2 resulting in Emberger syndrome in Korean patient. 27013649_we suggest screening for GATA2 mutations in pediatric myelodysplastic syndrome, preferentially in patients with impaired B-cell homeostasis in bone marrow and peripheral blood (low number of progenitors, intronRSS-Kde recombination excision circles and naive cells. 27157394_GATA2 was differentially expressed between Fibromyalgia patients and healthy controls. 27169477_Studied GATA2 Deficiency With Severe Primary Epstein-Barr Virus (EBV) Infection and EBV-associated Cancers. Seven patients with GATA2 deficiency developed severe EBV disease. Also found GATA2 to be the first gene associated with EBV hydroa vacciniforme-like lymphoma. 27375010_in our study on a large cohort of CEBPAmut AML patients, we found a high coincidence of GATA2mut, in particular within the subgroup of patients with CEBPAbi mutations 27389056_decline of GATA2 resulting from mutations contributes to the erythroid commitment, differentiation and the development of AEL 27416790_analysis of two separate cases of pediatric AML/MDS with underlying GATA2 mutations who underwent a successful umbilical cord hematopoietic stem cell transplantation using two different conditioning regimens [case report/review] 27460045_the results of this study demonstrated that high expression of GATA2 is correlated with worse survival outcomes in KRAS mutant colorectal cancer patients 27481672_Timely follow-up of a GATA2 deficiency patient allows successful treatment. 27528231_Study identified the novel role of transcription factors GATA-2 and GATA-3 in suppressing MICA/B expression in HBV-infected human hepatoma cells. 27545880_p38-dependent mechanism that phosphorylates GATA-2 and increases GATA-2 target gene activation has been demonstrated. This mechanism establishes a growth-promoting chemokine/cytokine circuit in acute myeloid leukemia cells. 27617961_we demonstrated that miR-22 promoted monocyte/macrophage differentiation, and MECOM (EVI1) mRNA is a direct target of miR-22 and MECOM (EVI1) functions as a negative regulator in the differentiation.The miR-22-mediated MECOM degradation increased c-Jun but decreased GATA2 expression, which results in increased interaction between c-Jun and PU.1 27651453_Knockdown of GATA-2 caused NGB expression to drop dramatically, indicating GATA-2 as an essential transcription factor for the activation of NGB expression via binding to a novel distal regulatory element of NGB. 27783953_Gata2 regulates a key regulatory network of gene expression for progesterone signaling at the early pregnancy stage. 28038451_Androgen receptor transcriptionally regulates semaphorin 3C in a GATA2-dependent manner in prostate tumor cells. 28093780_GATA2 mutation-related immunodeficiency may predispose to Epstein-Barr virus-associated subacute demyelinating polyradiculoneuropathy by both viral susceptibility and immune dysregulation 28114350_reducing GATA2 expression or inhibition of its transcription activity can relieve the drug resistance of acute myeloid leukemia cells and it would be helpful for eliminating the leukemia cells in patients 28179280_germ line mutations in GATA2 are associated with GATA2 deficiency syndrome, whereas acquired mutations are seen in myelodysplastic syndrome, acute myeloid leukemia, and in blast crisis transformation of chronic myeloid leukemia. 28209719_adaptive NK cells can persist in patients with GATA2 mutation, even after NK-cell progenitors expire. 28271814_we report a family with a null mutation in GATA2 and Emberger syndrome with variable expressivity and incomplete penetrance. This case emphasizes the need of genetic interactions to produce specific phenotypes in the clinical spectrum of GATA2 deficiency, where no precise genotype-phenotype correlation has been found. 28373026_this report describes a patient with WILD syndrome and de novo deletion mutation in the GATA2 gene 28381408_although relatively infrequent in patients with hypertrophic cardiomyopathy, GATA2, 4 and 6 transcription factors may represent a novel insight into the molecular mechanisms related to the pathogenesis of hypertrophic cardiomyopathy 28569748_endothelial SENP1-mediated SUMOylation drives graft arteriosclerosis by regulating the synergistic effect of GATA2 and NF-kappaB and consequent endothelial dysfunction. 28642594_Two GATA2 mutants (gT354M and gC373R) bound the key hematopoietic differentiation factor PU.1. 28752392_The absence of deleterious mutations in this large cohort of familial aggregations of hematological malignancies may strengthen the hypothesis that GATA2 mutations are an important predisposing factor, although as a secondary genetic event, required for the development of overt malignant disease 28937943_Gata2 and miR-124-3p are potential novel reporter biomolecules for ovarian cancer. 29106391_Low GATA2 expression due to methylation is associated with gastric cancer. 29156497_These findings suggest that GATA2 deficient patients may have an increased risk of melanoma and should be observed closely for new or changing skin lesions. 29217535_Our novel finding of dynamic pulsatile expression of Gata2 suggests a highly unstable genetic state in single cells concomitant with their transition to hematopoietic fate. This reinforces the notion that threshold levels of Gata2 influence fate establishment and has implications for transcription factor-related hematologic dysfunctions. 29275211_Data suggest that the stress of intermittent hypoxia up-regulates expression of mRNA and protein for POMC and CART in neuronal cell lines; GATA2 and GATA3 appear to be involved in these stress mechanisms. (POMC = proopiomelanocortin; CART = cocaine- and amphetamine-regulated transcript protein; GATA2 = endothelial transcription factor GATA-2; GATA3 = T-cell-specific transcription factor GATA-3) 29532200_To analyze the outcomes of COS in women with GATA2 deficiency. 29666442_Study identifies the GATA2 transcription factor as a critical regulator of matrix stiffness induced transcriptional program in the in lymphatic endothelial cells (LECs). As opposed to the previously reported activation of GATA2 by increased mechanical stimulus upon exposure of LECs to stiff matrix or oscillatory flow, study found that GATA2 expression is increased in LECs grown on a soft matrix. 29724903_patients with GATA2 missense mutations have a high risk of developing leukemia and this may be prevented by early HSCT with the help of new markers 29861167_The lack of GATA2 requirement for generation of HE and non-HE indicates the critical role of GATA2-independent pathways in specification of these two distinct endothelial lineages. 30030275_A novel disease-causing synonymous exonic mutation in GATA2 affecting RNA splicing. 30047422_GATA2 deficiency is an immunodeficiency and bone marrow failure disorder caused by pathogenic variants in GATA2. It is inherited in an autosomal-dominant pattern or can be due to de novo sporadic germline mutation. 30190467_GATA2 ZF1 mutations are associated with distinct clinico-biological features and predict better prognosis, different from ZF2 mutations, in AML patients. 30232126_This work describes 3 families with germline GATA2 mutations in which 1 member was transplanted with hematopoietic stem cell using a mutation-positive healthy related donor resulting in donor-derived posttransplant myelodysplastic syndrome (MDS)/acute myeloid leukemia (AML), recurrent MDS/AML, or fatal infection due to impaired immune reconstitution. 30245028_The rs2523393 A allele creates a GATA2 binding site in a progesterone-responsive distal enhancer that loops to the HLA-F promoter. 30478525_A Novel GATA2 Mutation. 30564229_In a primary immunodeficiency and the third report of HLH in GATA2-haploinsufficiency. 30578959_The findings suggest that abnormal clonal hematopoiesis is a common event in symptomatic germline mutated GATA2 patients with MDS and also in those with hypocellular marrows without overt morphologic evidence of dysplasia, possibly indicating a pre-MDS stage warranting close monitoring for disease progression. 30659233_Circulating GATA2 mRNA is decreased in women with established preeclampsia and decreased up to 12 weeks preceding onset of disease. Circulating mRNAs of endothelial origin may be a novel source of biomarker discovery for preeclampsia. 30710465_GATA2 hypomorphism leads to a hyperreactive defense response to infections, and this reaction is attributed to a unique intrinsic cell defect in the regulation of myeloid expansion that increases the risk of hematological neoplasm transformation. 30714451_The family study revealed that these twin sisters shared a de novo GATA2 mutation but showed different phenotypes (MDS in the younger sister and dysmegakaryopoiesis only in the older sister), possibly due to the critical role of epigenetic changes 30833300_The novel mechanisms of AR-V and GATA2 regulation in advanced prostate cancer. 31035956_clinicians should considerGATA2 deficiency in patients with myelodysplasia and long-standing Mycobacterium kansasii infection 31246134_Phenotypic heterogeneity associated with germline GATA2 haploinsufficiency: a comprehensive kindred study. 31296150_increasing GATA2 levels are linked to prostate cancer progression and aggressiveness 31340620_identify germline GATA2 mutations have a high prevalence in older pediatric patients with monosomy 7 31402335_GATA2 bound to regulatory regions, and repressed the expression of cardiac development-related genes. 31434974_Inhibition of GATA2 restrains cell proliferation and enhances apoptosis and chemotherapy mediated apoptosis in human GATA2 overexpressing AML cells. 31468074_Case-control study on the association between the GATA2 gene and premature myocardial infarction in the Iranian population.', trans 'Fall-Kontroll-Studie zur Assoziation zwischen GATA2-Gen und fruhzeitigem Myokardinfarkt in der iranischen Bevolkerung. 31501863_Our findings provide mechanistic insight into the future combination of GATA2 inhibitors and enzalutamide for improved AR-targeted therapy. 31582413_GATA2 controls lymphatic endothelial cell junctional integrity and lymphovenous valve morphogenesis through miR-126 31591264_Our study reveals a novel LSD1-GATA2 axis, which regulates human trophoblast syncytialization. 31785092_GATA2 mutations in myeloid malignancies: Two zinc fingers in many pies. 31933136_Cardiac Tamponade in Gorham-Stout Syndrome Associated with GATA2 Mutation. 32205587_GATA2 +9.5 enhancer: from principles of hematopoiesis to genetic diagnosis in precision medicine. 32250729_Urine Extracellular Vesicle GATA2 mRNA Discriminates Biopsy Result in Men with Suspicion of Prostate Cancer. 32330454_C/EBPalpha and GATA-2 Mutations Induce Bilineage Acute Erythroid Leukemia through Transformation of a Neomorphic Neutrophil-Erythroid Progenitor. 32335672_GATA2 and Progesterone Receptor Interaction in Endometrial Stromal Cells Undergoing Decidualization. 32430494_Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA). 32555368_Synonymous GATA2 mutations result in selective loss of mutated RNA and are common in patients with GATA2 deficiency. 32556109_Somatic genetic rescue in hematopoietic cells in GATA2 deficiency. 32558139_Humoral deficiency in a novel GATA2 mutation: A new clinical presentation successfully treated with hematopoietic stem cell transplantation. 32593672_The transcription factor NFE2 enhances expression of the hematopoietic master regulators SCL/TAL1 and GATA2. 32770553_Prevalence of germline GATA2 and SAMD9/9L variants in paediatric haematological disorders with monosomy 7. 32865708_Donor-derived myelodysplastic syndrome after allogeneic stem cell transplantation in a family with germline GATA2 mutation. 32960960_Human GATA2 mutations and hematologic disease: how many paths to pathogenesis? 33038986_Germline predisposition in myeloid neoplasms: Unique genetic and clinical features of GATA2 deficiency and SAMD9/SAMD9L syndromes. 33193444_Efferocytic Defects in Early Atherosclerosis Are Driven by GATA2 Overexpression in Macrophages. 33417088_Inherited GATA2 Deficiency Is Dominant by Haploinsufficiency and Displays Incomplete Clinical Penetrance. 33513878_LncRNA H19-Derived miR-675-5p Accelerates the Invasion of Extravillous Trophoblast Cells by Inhibiting GATA2 and Subsequently Activating Matrix Metalloproteinases. 33570623_Somatic GATA2 mutations define a subgroup of myeloid malignancy patients at high risk for invasive fungal disease. 33759087_A novel germline GATA2 frameshift mutation with a premature stop codon in a family with congenital sensory hearing loss and myelodysplastic syndrome. 33831168_Allele-specific expression of GATA2 due to epigenetic dysregulation in CEBPA double-mutant AML. 33856550_GATA2 regulates the CAD susceptibility gene ADTRP rs6903956 through preferential interaction with the G allele. 33957466_High penetrance of myeloid neoplasia with diverse clinical and cytogenetic features in three siblings with a familial GATA2 deficiency. 34040617_Case Report: Hemophagocytic Lymphohistiocytosis and Non-Tuberculous Mycobacteriosis Caused by a Novel GATA2 Variant. 34075404_Disruption of a GATA2-TAL1-ERG regulatory circuit promotes erythroid transition in healthy and leukemic stem cells. 34125173_Tumor suppressor function of Gata2 in acute promyelocytic leukemia. 34193836_Clinical and biological characteristics and prognostic impact of somatic GATA2 mutations in myeloid malignancies: a single institution experience. 34348000_Role of GATA2 in Human NK Cell Development. 34374210_GATA2 rs2335052 and GATA2 rs78245253 single-nucleotide polymorphisms in Chinese patients with acute myelocytic leukemia. 34387894_GATA2 deficiency syndrome: A decade of discovery. 34469508_Association of unbalanced translocation der(1;7) with germline GATA2 mutations. 34576178_Impairment of the Hypothalamus-Pituitary-Thyroid Axis Caused by Naturally Occurring GATA2 Mutations In Vitro. 34592889_Tumor suppr | ENSMUSG00000015053 | Gata2 | 2038.61287 | 0.9989369 | -0.0015345121 | 0.11260361 | 1.837690e-04 | 9.891841e-01 | 9.998360e-01 | No | Yes | 2162.22230 | 202.152725 | 2.012691e+03 | 146.180187 | |
| ENSG00000180011 | 284273 | ZADH2 | protein_coding | Q8N4Q0 | FUNCTION: Functions as 15-oxo-prostaglandin 13-reductase and acts on 15-keto-PGE1, 15-keto-PGE2, 15-keto-PGE1-alpha and 15-keto-PGE2-alpha with highest efficiency towards 15-keto-PGE2-alpha. Overexpression represses transcriptional activity of PPARG and inhibits adipocyte differentiation. {ECO:0000250|UniProtKB:Q8BGC4}. | 3D-structure;Acetylation;Alternative splicing;Lipid metabolism;NADP;Oxidoreductase;Peroxisome;Phosphoprotein;Reference proteome | hsa:284273; | peroxisome [GO:0005777]; 13-prostaglandin reductase activity [GO:0036132]; 15-oxoprostaglandin 13-oxidase activity [GO:0047522]; zinc ion binding [GO:0008270]; negative regulation of fat cell differentiation [GO:0045599] | 19690890_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 30372681_In patients with late-onset Alzheimer disease, a number of differentially methylated postitions were hypomethylated in ZADH2 compared with controls. 34143546_Thermal proteome profiling identifies PIP4K2A and ZADH2 as off-targets of Polo-like kinase 1 inhibitor volasertib. | ENSMUSG00000049090 | Zadh2 | 596.48402 | 0.8438309 | -0.2449741847 | 0.16783962 | 2.084835e+00 | 1.487683e-01 | 9.998360e-01 | No | Yes | 681.31888 | 71.270686 | 8.316112e+02 | 67.454790 | ||
| ENSG00000180035 | 197407 | ZNF48 | protein_coding | Q96MX3 | FUNCTION: May be involved in transcriptional regulation. | Acetylation;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:197407; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000045598 | Zfp553 | 2704.50787 | 1.0539957 | 0.0758690244 | 0.10662840 | 5.181132e-01 | 4.716476e-01 | 9.998360e-01 | No | Yes | 3018.09729 | 391.949401 | 2.672320e+03 | 268.963104 | |||
| ENSG00000180448 | 23526 | ARHGAP45 | protein_coding | Q92619 | FUNCTION: Contains a GTPase activator for the Rho-type GTPases (RhoGAP) domain that would be able to negatively regulate the actin cytoskeleton as well as cell spreading. However, also contains N-terminally a BAR-domin which is able to play an autoinhibitory effect on this RhoGAP activity. {ECO:0000269|PubMed:24086303}.; FUNCTION: Precursor of the histocompatibility antigen HA-1. More generally, minor histocompatibility antigens (mHags) refer to immunogenic peptide which, when complexed with MHC, can generate an immune response after recognition by specific T-cells. The peptides are derived from polymorphic intracellular proteins, which are cleaved by normal pathways of antigen processing. The binding of these peptides to MHC class I or class II molecules and its expression on the cell surface can stimulate T-cell responses and thereby trigger graft rejection or graft-versus-host disease (GVHD) after hematopoietic stem cell transplantation from HLA-identical sibling donor. GVHD is a frequent complication after bone marrow transplantation (BMT), due to mismatch of minor histocompatibility antigen in HLA-matched sibling marrow transplants. Specifically, mismatching for mHag HA-1 which is recognized as immunodominant, is shown to be associated with the development of severe GVHD after HLA-identical BMT. HA-1 is presented to the cell surface by MHC class I HLA-A*0201, but also by other HLA-A alleles. This complex specifically elicits donor-cytotoxic T-lymphocyte (CTL) reactivity against hematologic malignancies after treatment by HLA-identical allogenic BMT. It induces cell recognition and lysis by CTL. {ECO:0000269|PubMed:12601144, ECO:0000269|PubMed:8260714, ECO:0000269|PubMed:8532022, ECO:0000269|PubMed:9798702}. | 3D-structure;Alternative splicing;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Direct protein sequencing;GTPase activation;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | hsa:23526; | azurophil granule lumen [GO:0035578]; cytosol [GO:0005829]; extracellular region [GO:0005576]; membrane [GO:0016020]; ruffle membrane [GO:0032587]; secretory granule lumen [GO:0034774]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; activation of GTPase activity [GO:0090630]; intracellular signal transduction [GO:0035556]; regulation of small GTPase mediated signal transduction [GO:0051056] | 11920221_The intensity of the tetramer-staining of the HA-1/HA-2-specific cytotoxic T cells strongly correlates with their capability to recognize mHag positive target cells. 12091347_HA-1-specific CTLs restricted by nonself HLA-A2 molecules can be generated in an HLA-A2-mismatched setting. 15350465_In bone marrow transplant recipients and their genetically HLA-identical siblings, the presence of different alleles of two minor histocompatibility antigen genes is studied. 15498856_Pre-existing HA-1-specific T cells are observed in cord blood samples. Both circulating and ex vivo-generated HA-1-specific T cells show specific and hematopoietic restricted lysis of HLA-A2+/HA-1+ target cells, including leukemic cells. 15593299_Observational study of gene-disease association. (HuGE Navigator) 15593299_the incidence of the HA-1 168His allele is significantly lower in Sjogren's syndrome patients than in controls 16984283_Observational study of gene-disease association. (HuGE Navigator) 17580157_There was no difference in acute rejection rates between the HA-1-matched and -mismatched groups in kidney transplantation. 18093280_Observational study of gene-disease association. (HuGE Navigator) 18414982_targeting mHags encoded not only by HMHA1, whose aberrant expression in solid tumors has been reported, but also BCL2A1 may bring about beneficial selective graft-versus-tumor effects 19234124_study examined antigenic presentation & T-cell recognition of HA-1, a prototypic autosomal mHag derived from single nucleotide dimorphism (HA-1(H) versus HA-1(R)) in the HMHA1 gene; results define the molecular mechanisms governing immunogenicity of HA-1 19913121_Observational study of gene-disease association. (HuGE Navigator) 20509834_the information on allele and genotype frequencies of HA-1 and HA-2 in a Taiwanese population 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24086303_study shows that HMHA1 acts as a RhoGAP to regulate GTPase activity, cytoskeletal remodeling and cell spreading, which are crucial functions in normal hematopoietic and cancer cells 26095815_Placental HA-1 expression is regulated by oxygen and is increased in the syncytial nuclear aggregates and syncytiotrophoblast of preeclamptic as compared to control placentas. 28939173_HMHA1 significantly promotes melanoma cells proliferation, invasion and migration, and prevents cell apoptosis. 29174013_ArhGAP45 acts as a Rac-GAP contributing to the balance between formation and disruption of endothelial junctions, which is required for the dynamic regulation of vascular permeability. | ENSMUSG00000035697 | Arhgap45 | 113.50900 | 0.9285505 | -0.1069476671 | 0.28949160 | 1.385028e-01 | 7.097739e-01 | 9.998360e-01 | No | Yes | 46.29436 | 13.624033 | 4.598311e+01 | 10.520041 | ||
| ENSG00000182054 | 3418 | IDH2 | protein_coding | P48735 | FUNCTION: Plays a role in intermediary metabolism and energy production. It may tightly associate or interact with the pyruvate dehydrogenase complex. | 3D-structure;Acetylation;Alternative splicing;Disease variant;Glyoxylate bypass;Magnesium;Manganese;Metal-binding;Mitochondrion;NADP;Oxidoreductase;Reference proteome;Transit peptide;Tricarboxylic acid cycle | Isocitrate dehydrogenases catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. These enzymes belong to two distinct subclasses, one of which utilizes NAD(+) as the electron acceptor and the other NADP(+). Five isocitrate dehydrogenases have been reported: three NAD(+)-dependent isocitrate dehydrogenases, which localize to the mitochondrial matrix, and two NADP(+)-dependent isocitrate dehydrogenases, one of which is mitochondrial and the other predominantly cytosolic. Each NADP(+)-dependent isozyme is a homodimer. The protein encoded by this gene is the NADP(+)-dependent isocitrate dehydrogenase found in the mitochondria. It plays a role in intermediary metabolism and energy production. This protein may tightly associate or interact with the pyruvate dehydrogenase complex. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2014]. | hsa:3418; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; peroxisome [GO:0005777]; isocitrate dehydrogenase (NADP+) activity [GO:0004450]; magnesium ion binding [GO:0000287]; NAD binding [GO:0051287]; 2-oxoglutarate metabolic process [GO:0006103]; carbohydrate metabolic process [GO:0005975]; glyoxylate cycle [GO:0006097]; isocitrate metabolic process [GO:0006102]; NADP metabolic process [GO:0006739]; tricarboxylic acid cycle [GO:0006099] | 17854715_IDPm may play an important role in regulating apoptosis induced by TNF-alpha and anticancer drugs and the sensitizing effect of IDPm siRNA on the apoptotic cell death of HeLa cells offers the possibility of developing a modifier of cancer chemotherapy. 18096511_These results indicate that IDPm may play an important role in regulating the apoptosis induced by heat shock. 18484410_HOCl-mediated damage to IDPm may result in the perturbation of the cellular antioxidant defense mechanisms and subsequently lead to a pro-oxidant condition 19228619_Mutations of NADP(+)-dependent isocitrate dehydrogenases encoded by IDH1 and IDH2 occur in a majority of several types of malignant gliomas. 19350208_Mutations of IDH2 is not detected in brain metastases of colorectal cancer. 19469031_Observational study of gene-disease association. (HuGE Navigator) 19530255_IDH2 codon 172 mutation is not associated with cancers. 19554337_In patients with glioma, IDH2 mutations predominantly occur in oligodendroglial tumors. 19554337_Observational study of gene-disease association. (HuGE Navigator) 19667985_Studies indicate that mutations in IDH1/IDH2 are specific for diffuse gliomas. 19765000_IDH1 or IDH2 mutation plays a role in early tumor progression of several types of glioma 19765000_Observational study of gene-disease association. (HuGE Navigator) 19915015_IDH1 and 2 mutations are very rare in paragangliomas and pheochromocytomas and do not appear to play an important role in oncogenic HIF activation known to be present in these tumors. 19915484_testing for IDH1/2 mutations can be effectively performed in a clinical setting and can enhance the accuracy of diagnosis of gliomas 19933982_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20131059_Observational study of gene-disease association. (HuGE Navigator) 20142433_IDH2 mutation confer an enzymatic gain of function that dramatically increases 2-hydroxyglutarate in acute myelogenous leukemia 20160062_In 46% of cases with anaplastic oligodendroglioma an IDH1 mutation was found and only one IDH2 mutation was identified. 20171147_Some patients with cytogenetically normal acute myeloid leukemia and elevated 2-hydroxyglutarate possessed IDH2 mutations. 20367200_Mutations in IDH seem to play an important role in the formation of specific subtypes of gliomas. 20368543_IDH1 and IDH2 mutations are recurrent in de novo cytogenetically normal acute myeloid leukemia and have an unfavorable impact on outcome. 20376084_Mutations of IDH2 genes is associated with early and accelerated phases of myelodysplastic syndromes and myeloproliferative neoplasms. 20376084_Observational study of gene-disease association. (HuGE Navigator) 20410924_IDH2 gene mutation is associated with blast-phase myeloproliferative neoplasms. 20410924_Observational study of gene-disease association. (HuGE Navigator) 20421455_IDH2 mutations are frequently found in cytogenetically normal acute myeloid leukemia, but in our analysis these mutations did not influence treatment outcome. 20421455_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20427748_Observational study of gene-disease association. (HuGE Navigator) 20427748_This study identified the IDH2 mutation status of a large series of gliomas analyzed by array-based comparative genomic hybridization (aCGH). We investigated whether the occurrence of IDH2 mutation correlates with 1p19q status. 20465388_O(6)-methylguanine DNA methyltransferase (MGMT) status, and mutations of isocitrate dehydrogenases 1 and 2 (IDH1/IDH2) are currently the three most pertinent markers in diffuse gliomas [Review] 20485375_IDH2 mutations are associated with myelodysplastic syndrome and acute myeloid leukemia. 20494930_Observational study of gene-disease association. (HuGE Navigator) 20508616_IDH2 mutations are associated with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. 20508616_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20510884_IDH1 and IDH2 mutations are relevant to the progression of gliomas, prognosis and treatment of patients with gliomas harboring the mutation [review] 20538800_In AML, IDH1 and IDH2 mutations are more common among AML with normal karyotype and NPM1(mutant) genotypes. 20538800_Observational study of gene-disease association. (HuGE Navigator) 20567020_IDH1 and IDH2 mutations are recurring genetic changes in acute myeloid leukemia (AML); they constitute a poor prognostic factor in cytogenetically normal-AML with mutated NPM1 without FLT3-internal tandem duplication 20567020_Observational study of gene-disease association. (HuGE Navigator) 20603105_Observational study of gene-disease association. (HuGE Navigator) 20603105_Studies on the cell-lineages of tumors with IDH1/2 mutations may help clarify the role of these mutations in the development of brain tumors. 20625116_IDH2m were associated with a low WBC count at diagnosis and poor prognosis-lower response to chemotherapy and higher relapse and lower survival. 20625116_Observational study of gene-disease association. (HuGE Navigator) 20659156_Observational study of gene-disease association. (HuGE Navigator) 20659156_serum 2-hydroxyglutarate concentrations were substantially increased in AML patients with both IDH1 and IDH2 mutations 20678218_Mutations in IDH2 are associated with acute myeloid leukemias. 20678218_Observational study of gene-disease association. (HuGE Navigator) 20692206_IDH1 and IDH2 mutations are associated glioma and acute myeloid leukemia cases.[Review] 20713124_Results suggested that the suppression of HIF-1alpha accumulation by IDPm knockdown in PC3 cells was due to an inhibition of HIF-1alpha transcription. 20847235_study detected heterozygous germline mutations in IDH2 that alter enzyme residue Arg(140) in 15 unrelated patients with d-2-hydroxyglutaric aciduria 20861910_a possible association between IDH mutations and trisomy 8 in myelodysplastic syndromes and acute myeloid leukemia. 20877624_Observational study of gene-disease association. (HuGE Navigator) 20880116_Observational study of gene-disease association. (HuGE Navigator) 20929316_Correlation between IDH2 mutations and disease status in acute myeloid leukemia. 20929327_IDH2 mutations in patients with acute myeloid leukemia: missense p.R140 mutations are linked to disease status. 20929327_Observational study of gene-disease association. (HuGE Navigator) 20946881_Observational study of gene-disease association. (HuGE Navigator) 20946881_The frequency of IDH1 and IDH2 missense mutations in Chinese AML patients reached 5.9% and 8.3%, respectively. 20962861_Observational study of gene-disease association. (HuGE Navigator) 20962862_IDH2 somatic mutations in chronic myeloid leukemia patients in blast crisis. 20972461_review summarizes current understanding of the recently identified mutations in IDH1 and IDH2 and provide several potential molecular mechanisms linking them to malignant transformation [review] 20975057_IDH mutation appears to be a significant marker of positive prognosis and chemosensitivity in low-grade gliomas, independently of 1p-19q codeletion, whereas its impact on the course of untreated tumors seems to be limited. 20975057_Observational study of gene-disease association. (HuGE Navigator) 21075857_Observational study of gene-disease association. (HuGE Navigator) 21079611_Observational study of gene-disease association. (HuGE Navigator) 21079611_Serial analyses of IDH2 mutations at both diagnosis and relapse in 121 patients confirmed high stability of IDH2 mutations. IDH2 mutation is a stable marker during disease evolution and confers favorable prognosis. 21080178_Absence of IDH2 mutations in low grade gliomas (LGGs) identifies a novel entity of LGGs with distinctive location, infiltrative behavior, specific molecular alterations, and dismal outcome. 21080178_Observational study of gene-disease association. (HuGE Navigator) 21130701_Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. 21173122_We conclude that IDH1(R)(1)(3)(2) and IDH2(R)(1)(2) mutations occur most often in cytogenetically normal acute myeloid leukemia cases with an overall frequency of approximately 11.8%. 21225914_IDH1 codon 132 or IDH2 codon 172 mutations or elevated 2-hydroxyglutarate levels do not play a role in the biology of sporadic Wilms tumors. 21233841_Although uncommon in pediatric myeloid malignancies, IDH1 and IDH2 mutations, particularly IDH2 mutations, could contribute to the advanced phenotype of AML 21284999_the IDH mutations are not the origin of gliomas but a subsequent protective mechanism that interferes with the metabolism of the tumour cells, making these cells fragile and susceptible to cell death 21289278_N-acetyl-aspartyl-glutamate (NAAG), a common dipeptide in brain, was 50-fold reduced in cells expressing IDH1 mutants and 8.3-fold reduced in cells expressing IDH2 mutants 21294161_IDH mutations are observed in approximately 70-80% of grade II/III gliomas and the majority of secondary glioblastomas, but only 10% of primary glioblastomas. 21307773_Studies indicate that additional mutations in genes which appear to affect the epigenome of MPN patients have been discovered including mutations in TET2, IDH1/ 2, EZH2, and ASXL1. 21326614_The fact that the gain of the enzymatic activity to produce 2 hydroxyglutarate is a shared feature of the IDH1 and IDH2 mutations suggests that this is an important function for these mutants in driving cancer pathogenesis. 21346257_IDH2 mutation is asssociated with chronic myelogenous leukemia. 21383741_IDH1 AND IDH2 mutations have different prognostic effects when compared between AML and glioma 21454467_Differential prognosis impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. 21480859_Acute myeloid leukaemia with cuplike nuclei is associated with a high incidence of IDH mutations (either IDH1 or IDH2). 21504050_IDH2 mutations were detected in 4/180 pediatric AML patients. 21596855_The presence of an IDH2(R172) mutation was associated with a significantly worse outcome than IDH2(R140), and relapse in FLT3/ITD(WT)NPM1(WT)IDH2(R172) patients was comparable with adverse-risk cytogenetics patients (76% and 72%, respectively). 21598255_report of IDH1 (R132) and IDH2 (R172) heterozygous mutations in conventional central and periosteal cartilaginous tumours; the mutations are restricted to these specific subtypes of cartilaginous tumours and not found in other connective tissue neoplasms 21625441_IDH2 mutation is not associated with glioblastoma. 21643842_IDH2 mutation is associated with brain tumors. 21643985_we assessed the actual impact of IDH1 and IDH2 mutations in patients harboring WHO grade II and III gliomas. 21647152_analysis identified somatic IDH1/2 mutations in 4% of cases and the minor allele of single-nucleotide polymorphism (SNP) rs11554137 in 47 children (10.2%). IDH mutations were associated with an intermediate age, FAB M1/M2 and nucleophosmin1 mutations 21647154_Analysis of primary leukemic blasts confirmed high levels of 2-HG in AMLs with IDH1/IDH2 mutations. Interestingly, 3/5 AMLs with IDH2 mutations had FLT3-activating mutations, raising the possibility that these mutations cooperate in leukemogenesis. 21690245_Results suggest that TET2 promoter methylation, but not TET2 mutation, may be an alternative mechanism of pathogenesis in a small fraction of low-grade diffuse gliomas lacking IDH1/2 mutations. 21741430_Results suggest that the suppression of IDPm activity resulted in the disruption of cellular redox balance and subsequently exacerbates EGCG-induced apoptotic cell death in HeLa cells. 21867611_IDH point mutations occur in acute myeloid leukemia patients with normal karyotypes but not in patients with abnormal karyotypes. 21874255_IDH2 mutations were associated with prolonged overall survival in glioma patients exclusive of pilocytic astrocytoma 21885076_IDH1, more rarely IDH2, is mutated in 40% of gliomas (roughly 70% of low-grade gliomas, 50% of grade III, and 5 to 10% of primary glioblastomas--{REVIEW} 21889589_High IDH2 is associated with d-2-hydroxyglutaric aciduria type II. 21904853_Data show that TET2 and ASXL1 pathogenic mutations are found in 8% of myeloproliferative neoplasms lacking JAK2 and MPL mutations, whereas IDH1, IDH2, and c-CBL mutations are not detected in this subset of patients. 21996744_These data broaden our understanding of how IDH mutations may contribute to cancer through either neomorphic R(-)-2HG production or reduced wild-type enzymatic activity, and highlight the potential value of metabolite screening in identifying IDH-mutated tumors associated with elevated oncometabolite levels. 21997850_Data suggest that IDH1/2 mutations are recurrent but rare molecular aberrations in Chinese acute myeloid leukemia and myelodysplastic syndromes. 22002076_mechanisms of IDH mutations in gliomagenesis, and their value as diagnostic, prognostic marker and therapeutic target [review] 22020636_IDH aberrations and IDH1 codon 105 SNP occur in about 30% of younger patients with acute myeloid leukemia, mostly with diploid karyotype. 22033490_study examined the phenotypic and prognostic effects of IDH1 and IDH2 mutations among 277 patients with myelodysplastic syndromes (MDS); IDH mutations were detected in 34 cases: 26 IDH2 (all R140Q) and 8 IDH1 (6 R132S and 2 R132C); presence of IDH2R140Q did not affect the overall or leukemia-free survival 22034964_in patients with secondary glioblastoma,IDH mutation, MGMT promoter methylation, and 1p19q codeletion were associated with prolonged progression-free survival in univariate and multivariate analysis;IDH mutation and MGMT promoter methylation were correlated with a higher rate of objective response to temozolomide 22057234_Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome 22057236_The findings are compatible with a model in which IDH1 or IDH2 mutations represent early post-zygotic occurrences in individuals with these Ollier disease and Maffucci syndrome 22072542_Our detailed study of genetic aberrations in oligodendroglioma suggests a functional interaction between CIC mutation, IDH1/2 mutation, and 1p/19q co-deletion 22106302_Data show that the increased isocitrate dehydrogenase IDH2-dependent carboxylation of glutamine-derived alpha-ketoglutarate in hypoxia is associated with a concomitant increased synthesis of 2-hydroxyglutarate (2HG) in cells with wild-type IDH1 and IDH2. 22136423_Mutations of IDH1 and IDH2 were found in 73 (29%) and 2 (1%) cases, respectively 22147457_catalyzes the oxidative decarboxylation of isocitrate into alpha-KG and its mutation have been frequently found in some types of gliomas. (review) 22166653_Extraordinary high rates of somatic mutations in isocitrate dehydrogenase-1/2 occur in the majority of World Health Organization grade II and grade III gliomas as well as grade IV secondary glioblastomas[review] 22180306_Mutations in IDH1 and IDH2 were found only in cholangiocarcinomas of intrahepatic origin (nine of 40, 23%) and in none of the 22 extrahepatic cholangiocarcinomas and none of the 25 gallbladder carcinomas. 22215888_Data indicte that IDH2 mutations were identified in approximately 20% of angioimmunoblastic T-cell lymphomas (AITLs), but not in other peripheral T-cell lymphoma entities. 22217666_In WHO grade II astrocytomas, IDH1/2 mutations mostly occur in tumors infiltrating the frontal lobe. 22270848_No IDH2 mutations were found in 26 patients with gliosarcoma 22323113_IDH1/2 mutation status could be valuable for distinguishing intracranial chondrosarcomas from chordomas 22343901_Introduction of either mutant IDH or cell-permeable 2HG was associated with repression of the inducible expression of lineage-specific differentiation genes and a block to differentiation. 22360629_This study used formalin-fixed paraffin-embedded and multiplex PCR and single-base extension to dection IDH 2 mutation in gliomas. 22360810_Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. 22385606_90 adults with oral squamous cell carcinoma (OSCC)and 31 children with acute lymphoblastic leukemia (ALL) were scanned for IDH1 and IDH2 mutation hot spots; concluded that mutations of IDH are uncommon in ALL and OSCC 22397365_IDH2 mutation is associated with acute myeloid leukemia. 22399191_IDH1/2 mutational status defines biologically different subgroups among gliomas[review] 22410704_IDH2 mutation is associated with low-grade astrocytomas and their consecutive secondary high-grade gliomas. 22415316_IDH2 mutation followed by neuroglial developmental is associated with lower grade diffuse astrocytic glioma. 22416140_SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status 22427879_Gliomas of frontal origin had significantly higher incidence of 1p/19q co-deletion and IDH1/2 mutation than those of non-frontal origin, while gliomas of temporal origin had significantly lower incidence of 1p/19q co-deletion and IDH1/2 mutation than those of non-temporal origin. 22494415_Data indicate there was a strong association of IDH2 mutation with NPM1 mutations and a trend with FLT3-internal-tandem duplication. 22503487_The results of this study show for the first time that isocitrate dehydrogenase 1 and 2 genes are mutated in cholangiocarcinoma 22520341_IDH1 and IDH2 mutations are common genetic alterations in normal karyotype acute myeloid leukemia. 22616558_Studies suggest that screening of IDH1/2 mutations could help to identify patients at high risk within some subsets. 22683334_These results suggest that IDH1 and IDH2 have roles in production of D-2-hydroxyglutarate in cells. 22687971_IDH1/2 gene mutations affected tumorigenesis 22781800_The incidence of IDH1 and IDH2 mutation is higher in patients with de novo acute myeloid leukemia. IDH2 mutations are more frequent. 22790483_monitoring of the IDH1/2 status could be of value to predict the development of glioblastomas in patients with oligodendroglial tumors 22824796_elucidated the consequences of IDH1 and IDH2 mutations on DNA methylation and gene expression in intrahepatic cholangiocarcinomas and glioblastomas; identified several genes with both increased DNA methylation and decreased gene expression that may represent candidate tumor suppressors 22825915_IDH2 mutation was a predictive factor of response to chemoradiotherapy in grade II gliomas 22890969_IDH2 mutations are associated with grade II gliomas. 22917530_These data suggest that IDH1/2 mutations constitute a distinct mutational class in acute myeloid leukemia[review] 22922872_associations were observed for astrocytomas with mutated IDH1 or IDH2 but not for astrocytomas with wild-type IDH1 and IDH2. 22929312_Patients with ASXL1 mutations did not harbor IDH1, FLT3, or CEBPA mutations, and a combination of ASXL1 and IDH2 mutations was found only in one patient with acute myeloid leukemia. 23015095_IDH2 mutations are frequent in secondary high-grade gliomas. 23038259_Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT). 23039322_Type and location of isocitrate dehydrogenase mutations influence clinical characteristics and disease outcome of acute myeloid leukemia. 23071358_Abnormal histone and DNA methylation are emerging as a common feature of tumors with IDH1 and IDH2 mutations and may cause altered stem cell differentiation and eventual tumorigenesis. 23074281_Data indicate that 2-hydroxyglutarate (2-HG) levels are significantly higher in isocitrate dehydrogenase (IDH1 and IDH2) mutant patients. 23111198_Mutations in IDH1 codon 132 or IDH2 codon 172 were identified in 31.2% of all screened cases and 46.2% of screened World Health Organization grade I to IV gliomas with mutations. 23115158_Overexpression of IDH2 mutant protein renders glioma cells more sensitive to radiation. 23135354_epigenetics-modifying gene (DNMT3a, TET2 and IDH1/2) mutations had no change between diagnosis and relapse samples, and may become minimal residual disease marker 23187294_IDH1/2 and DNMT3A mutation status was independent of GADD45A hyper-methylation in predicting acute myeloid leukemia survival. 23192014_mutations of IDH1/2 are of the same type and occur at the same frequency in therapy-related and de novo MDS and AML 23232569_The emerging concept is that IDH mutations result in tumor formation by epigenetic alterations that affect gene expression and result in inhibition of cellular differentiation. 23264629_data demonstrate allelic and subcellular compartment differences can regulate the potential for IDH mutations to produce 2-hydroxyglutarate (2HG); consequences of 2HG elevation are dose-dependent and the non-equivalent 2HG accumulation resulting from IDH1 and IDH2 mutations may underlie their differential prognosis and prevalence in various cancers 23330999_There are distinct IDH1/IDH2 and consequently distinct 'triplenegative' patterns in purely insular versus paralimbic Grade II gliomas. 23361564_IDH2 mutation is associated with glioma. 23365461_Data indicate there were two patients carried ASXL1 mutations, both with t(8;21), 2 had DNMT3A mutations, 2 had IDH1 mutations, 1 had IDH2 mutation, and 3 had TET2 mutations. 23373447_knowledge of the IDH mutation status has had significant translational implications, and diagnostic tools are being used to monitor its expression and function[Review] 23391413_Mutations of several oncogenes were identified in less than 50% of cholangiocarcinomas but when combined with IDH1/2 testing, more than 90% had a detectable mutation. 23410661_IDH2 mutation is associated with gliomas. 23451940_An IDH1 mutation is detected by molecular genetics in 37% (21/57) of glioma cases and no IDH2 mutations are detected. 23494632_suggest that IDH1/2 mutations have an impact on the glioma history of secondary glioblastoma with different genetic pathway 23512379_A review of IDH2 mutations includes a discussion of how the mutations alter the catalytic properties. 23532369_Mutations in isocitrate dehydrogenase (IDH) 2 occur in the vast majority of low-grade gliomas and secondary high-grade gliomas. 23598960_IDH1/2 mutation analysis appears to be a promising biomarker for the distinction of chondrosarcoma from chondroblastic osteosarcoma 23641016_a firm association between IDH1, 2 mutations and serum 2HG concentration in AML 23681562_The combination of IDH1/2 and 1p/19q codeletion is able to stratify anaplastic oligodendroglioma. 23689617_Transcriptomic analyses revealed CIMP-specific gene expression signatures, indicating the impact of genetic status (IDH mutation, 1p/19q codeletion, TP53 mutation) on gene expression, and pointing to candidate biomarkers in oligodendrogliomas. 23877318_A review of the role of IDH2 mutations in the deregulation of cellular metabolism in glioma. 23894344_are potential prognostic biomarkers for gliomas 23918605_IDH2 mutation is associated with response to therapy in glioblastoma. 23996483_TET2 and IDH mutations did not have any significant impact on OS, while TET2 mutations were significantly associated with progression to sAML. 23999441_This review discusses how mutations in IDH2 affect the leukemia epigenome, hematopoietic differentiation, and clinical outcome. 24065766_neomorphic IDH2 mutations can be oncogenic in mesenchymal cells 24149775_study found 53.2 and 1.5 perc of anaplastic glioma patients in the study carried IDH1 and IDH2 mutation, respectively; there was higher proportion of MGMT promoter methylation, frontal lobe location and better outcome and lower proportion of temporal location in IDH-mutated samples; confirmed IDH mutation was a good prognosis marker for better outcome 24295421_Mutations in IDH1 and IDH2 have been discovered in glioma, acute myeloid leukemia and other solid tumors 24311631_IDH2 mutation is associated with gliomas and not malignant peripheral nerve sheath tumors. 24333121_Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. 24374336_We found significant signals of parallel evolution in IDH2 among these three groups 24403254_Isocitrate dehydrogenase 2 mutation is a frequent event in osteosarcoma. 24418992_Isocitrate dehydrogenase2 R140Q mutation induces myeloid and lymphoid neoplasms. 24443894_IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. 24460285_IDH2 mutations are not associated with glioma. 24478380_This study indicates that circulating 2HG may be a surrogate biomarker of IDH1 or IDH2 mutation status in intrahepatic cholangiocarcinoma and that circulating 2HG levels may correlate directly with tumor burden 24532263_we observed two mutations out of the usual hotspots at IDH1 and IDH2 genes in samples of pilocytic astrocytomas and grade-II astrocytomas pediatric patients. 24549719_These results indicated lithium chloride could decrease the proliferation and migration potential of C6 glioma cells harboring IDH2 mutation. 24569570_Hotspot mutations in IDH isoforms 1 or 2 occur in approximately 15% of intrahepatic cholangiocarcinomas. 24606448_the frequency of DNMT3A and IDH1 mutation is lower to the worldwide incidence, while that of IDH2 is comparable. 24699305_the presence of DNMT3A, IDH1 or IDH2 mutations may confer sensitivity to novel therapeutic approaches, including the use of demethylating agents [review] 24716838_IDH2 levels correlate with less aggressive tumor behavior in hepatocellular carcinoma. 24722048_Mutations in the IDH2 occurred mostly in astrocytomas, but were uncommon in glioblastomas. 24860178_Isocitrate dehydrogenase 2 mutation is associated with infiltrative glioma. 24867810_The recent discoveries reported here help understanding of the role of the IDH1/2 mutations in chondrosarcomas. IDH mutational status serves as a molecular signature to differentiate chondrosarcomas from other sarcomas with cartilaginous element. 24868540_IDH1/IDH2 but not TP53 mutations together with other prognostic factors such as age might be applied in clinical practice for prediction of outcome in patients with glioblastomas 24877111_These data refine current knowledge on IDH mutation prognostic impact and genotype-phenotype associations 24880135_Understanding of the biochemical consequences of IDH1/2 mutations in oncogenesis and survival prolongation will yield valuable information for rational therapy design [review] 24887488_Taken together, the hotspot mutations of IDH1, IDH2, DNMT3A, and MYD88 gene were absent in CRC. Aberrant mRNA expression of IDH1, DNMT3A, and MYD88 gene might be actively involved in the development of CRC. 24898068_This is the first report to describe IDH mutations in giant cell tumors of bone (GCTBg), and MsMab-1 can be anticipated for use in immunohistochemical determination of IDH1/2 mutation-bearing GCTB. 24936872_IDH2 mutations are associated with myelodysplastic syndromes. 24993250_IDH2 mutation is associated with response to therapy in anaplastic astrocytoma. 24995286_we propose the TCM compounds, precatorine and abrine, as potential candidates as lead compounds for further study in drug development process with the IDH2 R140Q mutant protein against cancer 25008158_Its mutation is strongly associated with pathological subtypes, genetic profiles, and clinical features in gliomas.(review) 25040869_IDH1 (R132) and IDH2 (R140, R172) mutations were not identified in any of the SCSTs: the mean coverage of the hotspot regions was 808 reads for IDH1 (R132), 944 for IDH2 (R140) and 1339 for IDH2 (R172) 25043045_transgenic mouse models expressing mutant human IDH in the adult liver show an aberrant response to hepatic injury, characterized by HNF-4alpha silencing, impaired hepatocyte differentiation, and markedly elevated levels of cell proliferation 25078896_A review summarizes the function of mutated vs. wild-type IDH enzymes and the role of IDH2 mutations in gliomas. 25251602_When normalized with the respective WT-IDH cells, the general metabolic shifts of MT-IDH1 and IDH2 were almost opposite. Lactate level was lower in MT-IDH2 cells which produced more 2-HG than MT-IDH1 cells. 25355558_IDH2 gene SNPs exhibited significant association with death risk in hepatocellular carcinoma patients. 25486927_The data suggest that IDH mutations are rare in the preleukemic disorders and may not be the major initial step in acute myeloid leukemia leukemogenesis. 25495392_report of clinicopathological characteristics of the gliomas with IDH2 mutations including two cases of primary GBM carrying a novel missense IDH2 mutation (c. 484C>T, p. P162S) 25524848_IDH1/2 mutation occurs more frequently in low-grade glioma patients with seizure as an initial symptom, suggesting a potential relationship between this genetic phenotype and clinical seizure presentation 25586680_The study of OxPhos-related genes revealed that an imbalance between the expression of IDH1 and IDH2, defined as overexpression of one isoform in relation to the other, was associated with worse prognosis in colorectal cancer patients. 25651001_IDH1 and IDH2 mutations may predict a favorable response to DNA methyltransferase inhibitors in patients with acute myeloid leukemia. 25652153_Isocitrate dehydrogenase (IDH) mutation is a valuable prognostic marker and a tool for decision-making for glioma treatment. 25783747_DNA-based molecular profiling of WHO grade II and III gliomas distinguishes biologically distinct tumor groups and provides prognostically relevant information beyond histological classification as well as IDH1/2 mutation and 1p/19q co-deletion status. 25811801_our data show that both mutant IDH1 and mutant IDH2 that, respectively, are expressed in cytosol and mitochondria, lead to decreased mitochondrial respiratory reserve 25818003_G6PD, GGCT, IDH1, isocitrate dehydrogenase 2 (NADP+,mitochondrial) (IDH2) and glutathione S-transferase pi 1(GSTP1), five of the critical components of GSH pathway, contribute to chemoresistance. 25836588_data demonstrate important clinical and biological differences between IDH1(MT) and IDH2(MT) myeloid neoplasms 25987093_IDH2 mutations are associated with normal cytogenetics and type A NPM1 mutations in Acute Myeloid Leukaemia. 26007236_Authors suggest 2HG as an analytic marker (in serum, urine, or biopsies) predicting malignancy of breast cancer in all patients. 26016821_The treatment outcome in acute myeloid leukemia patients with IDH1 and IDH2 mutations has been characterized. 26046462_suggest that mutations in isocitrate dehydrogenases IDH1/2 lead to a local block in osteogenic differentiation during skeletogenesis causing the development of benign cartilaginous tumors. 26061753_Gliomas were classified into five principal groups on the basis of three tumor markers including IDH2. The groups had different ages at onset, overall survival, and associations with germline variants. 26125858_Our data could not confirm that mutations in IDH1/IDH2 are indicative of malignancy and prognosis. 26147657_IDH2 mutations and their functions in human tumors (review) 26158269_Data suggest that long-term survival in glioblastoma (GBM) patients is if at all only weakly correlated to isocitrate dehydrogenase 1 or 2 (IDH1/2) mutations. 26188014_mechanistic studies of IDH2 mutations in gliomas (review) 26189213_IDH1 and IDH2 mutations are negative prognostic markers in AML patients. 26194445_Glioblastomas with IDH1-m should be considered a different entity from the IDH1-wt, as their natural history and prognosis differ. In the near future we should be classified glioblastomas based on the presence of the IDH1 mutation 26228814_IDH2 mutations are associated with myelodysplastic syndromes. 26268241_IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. 26314843_transcriptase (TERT) promoter mutations may predict enhanced sensitivity to genotoxic therapies in isocitrate dehydrogenase 1/2 (IDH1/2) wild-type WHO grade II and III diffuse gliomas. 26331834_The variant allelic frequencies in both IDH1 and IDH2 were equally low. 26338964_Classification based on IDH1/2 mutation status and Ki-67 expression level could be more convenient for clinical application and guide personalized treatment in malignant gliomas. 26486081_Dat | ENSMUSG00000030541 | Idh2 | 5769.38278 | 1.0039274 | 0.0056549154 | 0.09666885 | 3.465363e-03 | 9.530578e-01 | 9.998360e-01 | No | Yes | 6018.37751 | 672.615828 | 5.767923e+03 | 498.854308 | |
| ENSG00000182372 | 2055 | CLN8 | protein_coding | Q9UBY8 | FUNCTION: Could play a role in cell proliferation during neuronal differentiation and in protection against cell death. {ECO:0000269|PubMed:19431184}. | Disease variant;Endoplasmic reticulum;Epilepsy;Membrane;Mental retardation;Neurodegeneration;Neuronal ceroid lipofuscinosis;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein belonging to a family of proteins containing TLC domains, which are postulated to function in lipid synthesis, transport, or sensing. The protein localizes to the endoplasmic reticulum (ER), and may recycle between the ER and ER-Golgi intermediate compartment. Mutations in this gene are associated with a disorder characterized by progressive epilepsy with cognitive disabilities (EPMR), which is a subtype of neuronal ceroid lipofuscinoses (NCL). Patients with mutations in this gene have altered levels of sphingolipid and phospholipids in the brain. [provided by RefSeq, Jul 2017]. | hsa:2055; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; presynapse [GO:0098793]; ceramide binding [GO:0097001]; adult walking behavior [GO:0007628]; age-dependent response to oxidative stress [GO:0001306]; associative learning [GO:0008306]; cellular protein catabolic process [GO:0044257]; ceramide biosynthetic process [GO:0046513]; ceramide metabolic process [GO:0006672]; cholesterol metabolic process [GO:0008203]; glutamate reuptake [GO:0051935]; lipid biosynthetic process [GO:0008610]; lipid homeostasis [GO:0055088]; lipid transport [GO:0006869]; lysosome organization [GO:0007040]; mitochondrial membrane organization [GO:0007006]; musculoskeletal movement [GO:0050881]; negative regulation of apoptotic process [GO:0043066]; negative regulation of proteolysis [GO:0045861]; nervous system development [GO:0007399]; neurofilament cytoskeleton organization [GO:0060052]; neuromuscular process controlling balance [GO:0050885]; neuromuscular process controlling posture [GO:0050884]; phospholipid metabolic process [GO:0006644]; photoreceptor cell maintenance [GO:0045494]; protein catabolic process [GO:0030163]; regulation of cell size [GO:0008361]; retina development in camera-type eye [GO:0060041]; social behavior [GO:0035176]; somatic motor neuron differentiation [GO:0021523]; visual perception [GO:0007601] | 17129765_patients with CLN8 mutations from Italy. In these patients, the onset of epilepsy occurred between 3 and 6 years of age, with myoclonic, tonic-clonic, and atypical absence seizures. Electroencephalograms revealed focal and/or generalized abnormalities. 19201763_Observational study of gene-disease association. (HuGE Navigator) 19431184_CLN8 plays a role in cell proliferation during neuronal differentiation and in protection against cell death. 19807737_a novel, large CLN8 gene deletion c.544-2566_590del2613 in a Turkish family with a slightly more severe phenotype of neuronal ceroid lipofuscinose was described. 22388998_CLN8 is a candidate modifier gene for GD1. Increased expression may protect against severe GD1.It may function as a protective sphingolipid sensor and/or in glycosphingolipid trafficking. 22964447_A missense mutation at the CLN8 gene (763C>G)has been identified in 3 consanguineous Israeli-Arab patients. The phenotype in 2 of them is milder than that of their cousin who has typical neuronal ceroid lipofuscinosis. 23160995_This study highlights a close interaction between CLN5/CLN8 proteins, and their role in sphingolipid metabolism. Our findings suggest that CLN5p/CLN8p most likely are positive modulators of CerS1 and/or CerS2. 26443629_Novel missense mutation in CLN8 in late infantile neuronal ceroid lipofuscinosis 26657971_This study does not support a contribution of rare missense CLN8 variations to ASD susceptibility in the Japanese population. 27844444_Whole-exome sequencing and homozygosity mapping revealed a novel homozygous CLN8 mutation, c.677T>C (p.Leu226Pro in 5 relatives from a large Turkish consanguineous family 30397314_CLN8 recruits lysosomal soluble proteins in the endoplasmic reticulum (ER), delivers them to the Golgi apparatus via COPII-coated vesicles, and recycles back to the ER via COPI-coated vesicles. CLN8 interacts with the lysosomal soluble proteins through its large luminal loop. The export signal of CLN8 (261VDWNF265) is localized in its cytosolic C-terminus. CLN8 deficiency results in depletion of enzymes at the lysosome. 30453012_the phosphorylation levels of several substrates of PP2A, namely Akt, S6 kinase, and GSK3beta, were decreased in CLN8 disease patient fibroblasts. 30919163_The Neuronal Ceroid Lipofuscinoses-Linked Loss of Function CLN5 and CLN8 Variants Disrupt Normal Lysosomal Function. 32597833_CLN8 associates with CLN6 to form the EGRESS complex (ER-to-Golgi Relaying of Enzymes of the lySosomal System), the functional unit responsible for the recruitment of newly synthesized lysosomal enzymes in the endoplasmic reticulum and their transfer to the Golgi complex. 34044364_miR-3074-5p/CLN8 pathway regulates decidualization in recurrent miscarriage. 34201538_CLN8 Mutations Presenting with a Phenotypic Continuum of Neuronal Ceroid Lipofuscinosis-Literature Review and Case Report. | ENSMUSG00000026317 | Cln8 | 490.59031 | 0.9771362 | -0.0333683921 | 0.16600864 | 3.962046e-02 | 8.422245e-01 | 9.998360e-01 | No | Yes | 481.35991 | 63.069284 | 4.965467e+02 | 50.430993 | |
| ENSG00000182511 | 2242 | FES | protein_coding | P07332 | FUNCTION: Tyrosine-protein kinase that acts downstream of cell surface receptors and plays a role in the regulation of the actin cytoskeleton, microtubule assembly, cell attachment and cell spreading. Plays a role in FCER1 (high affinity immunoglobulin epsilon receptor)-mediated signaling in mast cells. Acts down-stream of the activated FCER1 receptor and the mast/stem cell growth factor receptor KIT. Plays a role in the regulation of mast cell degranulation. Plays a role in the regulation of cell differentiation and promotes neurite outgrowth in response to NGF signaling. Plays a role in cell scattering and cell migration in response to HGF-induced activation of EZR. Phosphorylates BCR and down-regulates BCR kinase activity. Phosphorylates HCLS1/HS1, PECAM1, STAT3 and TRIM28. {ECO:0000269|PubMed:11509660, ECO:0000269|PubMed:15302586, ECO:0000269|PubMed:15485904, ECO:0000269|PubMed:16455651, ECO:0000269|PubMed:17595334, ECO:0000269|PubMed:18046454, ECO:0000269|PubMed:19001085, ECO:0000269|PubMed:19051325, ECO:0000269|PubMed:20111072, ECO:0000269|PubMed:2656706, ECO:0000269|PubMed:8955135}. | 3D-structure;ATP-binding;Alternative splicing;Cell junction;Cell membrane;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Golgi apparatus;Kinase;Lipid-binding;Membrane;Nucleotide-binding;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;Transferase;Tumor suppressor;Tyrosine-protein kinase | This gene encodes the human cellular counterpart of a feline sarcoma retrovirus protein with transforming capabilities. The gene product has tyrosine-specific protein kinase activity and that activity is required for maintenance of cellular transformation. Its chromosomal location has linked it to a specific translocation event identified in patients with acute promyelocytic leukemia but it is also involved in normal hematopoiesis as well as growth factor and cytokine receptor signaling. Alternative splicing results in multiple variants encoding different isoforms.[provided by RefSeq, Jan 2009]. | hsa:2242; | cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; focal adhesion [GO:0005925]; Golgi apparatus [GO:0005794]; microtubule cytoskeleton [GO:0015630]; ATP binding [GO:0005524]; immunoglobulin receptor binding [GO:0034987]; microtubule binding [GO:0008017]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; phosphatidylinositol binding [GO:0035091]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; signaling receptor binding [GO:0005102]; cell adhesion [GO:0007155]; cell differentiation [GO:0030154]; cellular response to vitamin D [GO:0071305]; centrosome cycle [GO:0007098]; chemotaxis [GO:0006935]; innate immune response [GO:0045087]; microtubule bundle formation [GO:0001578]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of actin cytoskeleton reorganization [GO:2000251]; positive regulation of microtubule polymerization [GO:0031116]; positive regulation of monocyte differentiation [GO:0045657]; positive regulation of myeloid cell differentiation [GO:0045639]; positive regulation of neuron projection development [GO:0010976]; protein autophosphorylation [GO:0046777]; regulation of cell adhesion [GO:0030155]; regulation of cell differentiation [GO:0045595]; regulation of cell motility [GO:2000145]; regulation of cell population proliferation [GO:0042127]; regulation of cell shape [GO:0008360]; regulation of mast cell degranulation [GO:0043304]; regulation of vesicle-mediated transport [GO:0060627]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 11994747_Closing in on the biological functions of Fps/Fes and Fer. A review. 12653561_Fes naturally adopts an inactive conformation in vivo, and maintenance of the inactive structure requires the coiled-coil and SH2 domains. 12871378_Fps/Fes and Fer are expressed in human and mouse platelets, and are activated following stimulation with collagen and collagen-related peptide (CRP), suggesting a role in GPVI receptor signaling 15003822_Fes transduces inductive signals for terminal macrophage and granulocyte differentiation, and this biological activity is mediated through the activation of lineage-specific transcription factors. 15099290_FPS mediates enhanced sensitization to VEGF and PDGF signaling in ECs; this hypersensitization contributes to excessive angiogenic signaling and underlies the observed hypervascular phenotype of human myristoylated FPS expressed in transgenic mice. 15485904_c-Fes is a regulator of the tubulin cytoskeleton and may contribute to Fes-induced morphological changes in myeloid hematopoietic and neuronal cells 15630569_NMR assignment of the SH2 domain 15713745_PEDF downregulates Fyn through Fes, resulting in inhibition of FGF-2-induced capillary morphogenesis of endothelial cells 15929003_NMR analysis of Src homology 2 domain from the human feline sarcoma oncogene Fes 16455651_FES has a growth suppressive function in colorectal neoplasms. 16792528_A novel Fes-KAP-1 interaction is reported, suggesting a dual role for KAP-1 as both a Fes activator and downstream effector. 17521372_These novel results indicate the involvement of Fes in VEGF-A-induced cellular responses by cultured endothelial cells. 17595334_shows a major function of FES downstream of activated KIT receptor and thereby points to FES as a novel target in KIT-related pathologi 18046454_Ezrin/Fes interaction at cell-cell contacts plays an essential role in hepatocyte growth factor-induced cell scattering and implicates Fes in the cross-talk between cell-cell and cell-matrix adhesion. 18775312_The SH2 and catalytic domains of active Fes and Abl pro-oncogenic kinases form integrated structures essential for effective tyrosine kinase signaling. 19001085_study shows Fes phosphorylates C-terminal tyrosine residues in HS1 implicated in actin stabilization; coordinated action of F-BAR & SH2 domains of Fes allow for coupling to FcepsilonRI signaling & potential regulation of actin reorganization in mast cells 19051325_Study of promoter methylation as important mechanism responsible for downregulation of FES gene expression in colorectal cancer cells. Treatment with DNA methyltransferase inhibitor resulted in expression of functional FES transcripts in CRC cell lines. 19082481_Downregulation of the c-Fes protein-tyrosine kinase inhibits the proliferation of renal carcinoma. 19382747_c-Fes oligomerization is independent of activation; data suggest that conformational changes, rather than oligomerization, govern c-Fes kinase activation and downstream signaling in vivo. 19464057_Observational study of gene-disease association. (HuGE Navigator) 19865112_Observational study of gene-disease association. (HuGE Navigator) 19885553_FGF-2 activates Fes via the second coiled-coil domain, leading to lamellipodium formation and chemotaxis by endothelial cells 19913121_Observational study of gene-disease association. (HuGE Navigator) 20030920_c-fes gene expression was found in myeloid leukemias, whereas low or no expression in lymphocytic leukemias. 20117079_FES kinase is a mediator of wild-type KIT signalling implicated in cell migration. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23404507_analysis of the JAK-Fes-phospholipase D signaling pathway that is enhanced in highly proliferative breast cancer cells 31573955_Identification of FES as a Novel Radiosensitizing Target in Human Cancers. 31746983_This study supports the critical role of FER and FES tyrosine kinase fusions in the pathogenesis of follicular T-cell lymphoma and provides additional evidence that these can drive follicular T-cell lymphoma in the absence of RHOA mutations. | ENSMUSG00000053158 | Fes | 49.83958 | 0.9226912 | -0.1160802309 | 0.41247411 | 7.960972e-02 | 7.778270e-01 | 9.998360e-01 | No | Yes | 50.61170 | 11.163917 | 5.003651e+01 | 9.160288 | |
| ENSG00000182648 | 100506380 | LINC01006 | lncRNA | 29060927_LINC01006 levels in cancer tissues were significantly lower than those in adjacent normal tissues and healthy control tissues. 33336752_LncRNA LncOGD-1006 alleviates OGD-induced ischemic brain injury regulating apoptosis through miR-184-5p/CAAP1 axis. | 88.90563 | 1.0524340 | 0.0737296949 | 0.33905744 | 4.350337e-02 | 8.347803e-01 | 9.998360e-01 | No | Yes | 96.01391 | 17.999206 | 8.465192e+01 | 13.052903 | |||||||||
| ENSG00000182871 | 80781 | COL18A1 | protein_coding | P39060 | FUNCTION: Probably plays a major role in determining the retinal structure as well as in the closure of the neural tube. {ECO:0000269|PubMed:10942434}.; FUNCTION: [Non-collagenous domain 1]: May regulate extracellular matrix-dependent motility and morphogenesis of endothelial and non-endothelial cells; the function requires homotrimerization and implicates MAPK signaling. {ECO:0000269|PubMed:11257123}.; FUNCTION: [Endostatin]: Potently inhibits endothelial cell proliferation and angiogenesis (PubMed:9459295). May inhibit angiogenesis by binding to the heparan sulfate proteoglycans involved in growth factor signaling (By similarity). Inhibits VEGFA-induced endothelial cell proliferation and migration. Seems to inhibit VEGFA-mediated signaling by blocking the interaction of VEGFA to its receptor KDR/VEGFR2. Modulates endothelial cell migration in an integrin-dependent manner implicating integrin ITGA5:ITGB1 and to a lesser extent ITGAV:ITGB3 and ITGAV:ITGB5 (By similarity). May negatively regulate the activity of homotrimeric non-collagenous domain 1 (PubMed:11257123). {ECO:0000250|UniProtKB:P39061, ECO:0000269|PubMed:11257123, ECO:0000269|PubMed:9459295}. | 3D-structure;Alternative promoter usage;Alternative splicing;Basement membrane;Cell adhesion;Collagen;Disulfide bond;Extracellular matrix;Glaucoma;Glycoprotein;Hydroxylation;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal;Zinc | This gene encodes the alpha chain of type XVIII collagen. This collagen is one of the multiplexins, extracellular matrix proteins that contain multiple triple-helix domains (collagenous domains) interrupted by non-collagenous domains. A long isoform of the protein has an N-terminal domain that is homologous to the extracellular part of frizzled receptors. Proteolytic processing at several endogenous cleavage sites in the C-terminal domain results in production of endostatin, a potent antiangiogenic protein that is able to inhibit angiogenesis and tumor growth. Mutations in this gene are associated with Knobloch syndrome. The main features of this syndrome involve retinal abnormalities, so type XVIII collagen may play an important role in retinal structure and in neural tube closure. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2014]. | hsa:80781; | basement membrane [GO:0005604]; collagen trimer [GO:0005581]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular matrix structural constituent [GO:0005201]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; angiogenesis [GO:0001525]; animal organ morphogenesis [GO:0009887]; cell adhesion [GO:0007155]; endothelial cell morphogenesis [GO:0001886]; extracellular matrix organization [GO:0030198]; negative regulation of cell population proliferation [GO:0008285]; visual perception [GO:0007601] | 11700031_Hypoxia down-regulates endostatin production by microvascular endothelial cells and pericytes 11781696_An increase of about one-third of normal endostatin serum levels may represent an effective therapeutic dose to significantly inhibit many solid tumours. 11815623_Endostatin causes G1 arrest of endothelial cells through inhibition of cyclin D1. 11911017_Endostatin plays a crucial role in endothelial tubule stabilization, maintenance and integrity during angiogenesis. 11937714_type XV and XVIII collagens are expressed in specialized basement membranes. 12174873_production in head and neck carcinoma modulated by hsp47 12209593_Serum levels of angiogenin, basic fibroblast growth factor and endostatin in patients receiving intensive chemotherapy for acute myelogenous leukemia. 12231538_There is a positive correlation between the levels of tissue endostatin and malignancy grades in gliomas. The endostatin may be released near the tumor blood vessels with hyperplasia to counteract angiogenic stimuli in malignant gliomas. 12237301_Data are consistent with a model of endostatin with two binding sites: one mainly to laminin in the basement membrane and the other to heparan sulfates on the cell surface. 12408231_Collagen XVIII is a heparan sulfate proteoglycan associated with vascular amyloid beta and senile plaques in Alzheimer Disease. 12415512_Molecular analysis of collagen XVIII reveals novel mutations, presence of a third isoform, and possible genetic heterogeneity in Knobloch syndrome 12458056_Accumulation of endostatin/collagenXVIII in brains of patients who died with cerebral malaria. 12479859_Observational study of gene-disease association. (HuGE Navigator) 12486154_Aberrant neuronal and paracellular deposition of endostatin in brains of patients with Alzheimer's disease. 12682293_endostatin competes with fibronectin/RGD cyclic peptide to bind alpha 5 beta 1 integrin 12693719_Observational study of gene-disease association. (HuGE Navigator) 12839947_A recombinant fusion version of this protein may be a therapeutic agent for breast cancer. 12857600_level of endostatin was lower in the group of chronic lymphocytic leukemia patients with progressive disease as compared to patients with stable disease 14514474_Data indicate that serum endostatin levels are higher in multiple myeloma patients than in healthy controls, with the highest values found during the active phase of the disease. 14519482_analyses of age-matched study groups revealed elevated medians of endostatin concentration for severely and mildly preeclamptic patients 14695535_Analysis of knobloch syndrome-associated mutations in COL18A1 and genetic variation in endostatin. 14732364_Plasma endostatin increased after surgical removal of primary tumor. Decreased intratumoral microvessels in patients with higher endostatin levels may be due to apoptosis-inducing effect of endostatin on microvascular endothelial cells. 14760864_Pleural mesothelial cells play a key role in the antiangiogenesis process by producing endostatin in the pleural space 14973128_there is a putative integrin-binding sequence with anti-migratory activity within endostatin 15014038_Lentivirus-mediated gene transfer might represent an effective strategy for expression of angioinhibitory peptides to achieve inhibition of human bladder cancer proliferation and tumor progression. 15148373_E-selectin is required for the antiangiogenic activity of endostatin in vivo and ex vivo and confers endostatin sensitivity to nonresponsive human endothelial cells in vitro 15296943_Collagen XVIII and enamelysin were co-localized in the developing enamel matrix and stratum intermedium and in the enamel-like tumor matrix of odontogenic tumors. 15328173_Expression of collagen XVIII in tumor tissue is strongly associated with a poorer outcome in non-small-cell lung cancer and correlates with elevated levels of circulating serum endostatin. 15334690_Naked endostatin plasmid intratumoral injection can get a similar gene transfection efficiency to liposome-DNA complex when used in situ in inhibiting cell growth. 15374730_Hypoxia downregulates the concentration of endostatin in the culture media of human endometrial stromal cells. 15464359_Spatial and temporal expression of endostatin/collagen XVIII in cartilaginous tissue and its inactivation of VEGF signalling suggests that it is important for development and maintenance of avascular zones in cartilage and fibrocartilage. 15540202_Endostatin diminishes the levels of nitric oxide (NO) whereas NO donors enhanced VEGF expression and collagen XVIII expression. 15605080_Increased endostatin/collagen XVIII expression correlated with hepatoma progression and predicted poor prognosis for patients with hepatocellular carcinoma. 15662127_Observational study of gene-disease association. (HuGE Navigator) 15694132_angiostatic function is heparan sulfate-dependent, and in situ-binding of endostatin to endothelial cells is increased by heparan sulfates 15735323_Observational study of gene-disease association. (HuGE Navigator) 15950618_Biologically active endostatin, generated by matrix metalloproteases from collagen XVIII, may participate in the inhibition of endothelial cell proliferation, migration and angiogenesis. 15985216_results demonstrate that there is a direct connection between dependence of endostatin activity on heparin-like glycosaminoglycans and its ability to antagonize bFGF 16008891_Overexpression of endostatin effectively inhibits the growth of ovarian cancer. 16025479_Elevated serum levels of endostatin were observed in patients with idiopathic pulmonary fibrosis 16320826_Mechanically induced VEGF is involved in the destruction and endostatin in the maintenance of avascular tissues of the bone and joint system. (review) 16358965_Significantly higher serum concentrations of endostatin are associated with lung carcinoma 16466644_data show that inhibition of PI3K/protein kinase B (PKB) increased endostatin-induced apoptosis, and that endostatin-induced cell death is physiologically linked to PKB-mediated cell survival through caspase-8 16793908_The results indicate that the antiangiogenic functions of endostatin(ES) critically depend on the mode of delivery and the site of expression. 16807676_Homozygous COL18A1 polymorphism is a determinant of inherited breast cancer risk. 16807676_Observational study of gene-disease association. (HuGE Navigator) 16866623_An increase of endostatin/collagen XVIII-positive macrophages/microglial cells is observed, but not astrocytes, up to day 14 and a consequent decrease to day 16 in patients with post-traumatic brain injury. 16998835_Results suggest for the first time that an elevated serum level of endostatin at the diagnosis of metastatic gastric cancer could be predictive of a poor outcome. 17011029_Significantly higher endostatin levels were associated with childhood acute lymphoblastic leukemia 17067672_Polymorphism in endostatin is associated with acute and chronic myeloid leukaemia 17482599_Observational study of gene-disease association. (HuGE Navigator) 17482599_Serum endostatin levels but not genetic polymorphisms were negatively correlated with endometriosis. 17526870_Recombinant human endostatin inhibits choroidal neovascularization lesion size and vascular leakage in mutant mice lacking collagen XVIII as a structural membrane component. 17544408_Zn(II)-binding contributes to the structural integrity of endostatin. 17546652_This report confirms that endostatin plays a role in neuronal migration. 17587451_Endostatin 4349A allele is associated with invasive breast cancer. The Endostatin 4349G > A polymorphism however does not appear to be associated with breast cancer susceptibility or severity in invasive disease. 17587451_Observational study of gene-disease association. (HuGE Navigator) 17653045_Observational study of gene-disease association. (HuGE Navigator) 18006826_Endostatin binding to ovarian cancer cells inhibits peritoneal attachment and dissemination 18054814_D104N polymorphism of the COL18A gene may be an important inherited determinant of the LC susceptibility. 18345385_COL18A1 expression increases during human adipogenic differentiation. This genotype, after controlling for cholesterol, LDL cholesterol, and triglycerides, was independently associated with obesity, and increases the chance of obesity in 2.8 times. 18345385_Observational study of gene-disease association. (HuGE Navigator) 18360823_patients with pancreas cancer have increased circulating levels of endostatin and that levels are normalized after surgery or intraperitoneal chemotherapy. Findings indicate that endostatin could be used as a biomarker for pancreas cancer progression 18381814_VDAC1 plays a vital role in modulating endostatin-induced endothelial cell apoptosis 18382662_a frizzled module in cell surface collagen 18 inhibits Wnt/beta-catenin signaling 18478248_High endostatin expression in epididymis may protect this organ against tumor development 18676680_Observational study of gene-disease association. (HuGE Navigator) 18818971_Endostatin over expression is associated with thyroid cancer. 19074676_Endostatin and angiostatin are increased in diabetic patients with coronary artery disease and associated with impaired coronary collateral formation. 19084038_In Lewis lung cancer model, a dose of 10 mg/kg endostatin microencapsulated into poly(lactic-co-glycolic acid microspheres was just as effective in suppressing tumor growth as a dose of 2 mg/kg/day free endostatin for 35 days (total dose 70 mg/kg). 19127216_Airway concentrations of angiopoietin-1 and endostatin in ventilated extremely premature infants are decreased after funisitis and unbalanced with bronchopulmonary dysplasia/death. 19160445_A homozygous COL18A1 two base pair deletion segregating with neurological disorders was identified. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19223770_Results indicate that a pVAX1-mediated combinatorial antiangiogenic and proapoptotic gene therapy approach involving endostatin and sTRAIL can be an effective novel form of treatment for human liver cancer. 19298526_Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and beta-catenin levels 19390655_Identified four novel mutations in COL18A1 of Knobloch Syndrome patients, including a large deletion involving exon 41. Skin biopsies from Knobloch Syndrome patients revealed lack of type XVIII collagen in epithelial basement membranes and blood vessels. 19431146_Observational study of gene-disease association. (HuGE Navigator) 19431146_Polymorphism in endostatin is not associated with prostate cancer. 19542224_analysis of the endostatin interaction network 19585274_The transfected endostatin could inhibit endothelial proliferation and migration. 19615205_Elevated preoperative serum levels of endostatin in patients with clear cell renal cell carcinoma are associated with higher stage and grade. 19622587_Observational study of gene-disease association. (HuGE Navigator) 19622587_Polymorphisms in PAR-1, ES, and IL-8 may serve as independent molecular prognostic markers in patients with localized gastric adenocarcinoma 19625176_Observational study of gene-disease association. (HuGE Navigator) 19631658_Results report the isolation, characterization, and crystal structure of the trimerization domain of human type XVIII collagen, a member of the multiplexin family. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19877579_Arginine clusters in the heparin binding motif of endostatin significantly contribute to its interaction with receptor nucleolin and mediate the antiangiogenic and antitumor activities of endostatin. 19961619_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20027339_The mean serum level of endostatin in healthy individuals was significantly lower than in the total group of patients with bone tumors. 20063989_Data show that ADP significantly increased VEGF, but not endostatin, release from platelets, and that both P2Y(1) and P2Y(12) receptor antagonism inhibited this release. 20156196_Endostatin and TG-2 are co-localized in the extracellular matrix secreted by endothelial cells under hypoxia, which stimulates angiogenesis. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20361288_high collagen XVIII expression may serve a useful biomarker for predicting survival time and time to progression in patients with metastatic gastric cancer 20396759_Direct correlations between the initial levels of VEGF and endostatin and an inverse correlation between VEGF and TNF-beta concentrations were detected in patients with benign neuroendocrine tumors. 20616167_suggest that collagen XVIII/endostatin preserves the integrity of the extracellular matrix and capillaries in the kidney, protecting against progressive glomerulonephritis 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21085708_Data show that hypertriglyceridemia occurs in Knobloch patients lacking the vascular form of Col18. 21193414_Lack of collagen XVIII long isoforms affects kidney podocytes, whereas the short form is needed in the proximal tubular basement membrane 21457400_Endostatin can effectively inhibit rat peritoneum neoangiogenesis and the effect was dose-dependent. 21486700_The aim of the study was to evaluate and compare the circulating concentrations of OPN, E-selectin and END of severely obese patients with metabolic syndrome before and after vertical banded gastroplasty. 21527992_Common polymorphisms in four candidate genes (COL11A1, COL18A1, FBN1 and PLOD1) were unlikely to play important roles in the genetic susceptibility to high myopia. 21815140_study demonstrated significantly elevated endostatin levels in serum and urine samples of bladder cancer patients and show that high endostatin serum concentrations significantly correlate with the presence of lymph node metastasis 21896479_Cysteine cathepsins S and L modulate anti-angiogenic activities of human endostatin. 21963851_a potential role for collagen XXIII in mediating metastasis by facilitating cell-cell and cell-matrix adhesion as well as anchorage-independent cell growth. 21968021_Peripheral blood osteopontin (OPN) and endostatin (END) levels were studied in 35 patients with adrenal cortex tumors and 10 patients with pheochromocytoma before unilateral adrenalectomy, as well as in 10 healthy subjects (controls). 22009965_In osteosarcoma, expression of endostatin was positively correlated with that of CD34, and was greater in poorly differentiated osteosarcoma than in highly differentiated osteosarcoma. Expression of endostatin correlated with osteosarcoma metastasis. 22303445_Soluble FZC18 and Wnt3a physically interact in a cell-free system and soluble FZC18 binds the frizzled 1 and 8 receptors. 22431315_The detection of VEGF mRNA and endostatin mRNA appears to be suitable for distinguishing carcinoma cells from reactive mesothelial cells in pleural effusions, they could be useful to diagnose the pleural micrometastasis. 22461898_Findings indicate that COL18A1 rs7499 may contribute to the risk of Hepatocellular carcinoma (HCC) in Han Chinese. 22512651_Binding of VEGF-C and endostatin to recombinant VEGFR-3 is competitive. 22733742_Endostatin lowers blood pressure via nitric oxide and prevents hypertension associated with VEGF inhibition 22819906_Report pre/postoperative endostatin levels in surgically treated gastrointestinal cancers. 23605255_indicative of good tumor blood supply and permeability of vasculature, are associated with high levels of collagen XVIII and VEGF expression 23621901_These results suggest genetic variation in COL15A1 and COL18A1 can modify the age of onset of both early and late onset POAG. 23641912_common germline variants in Endostatin and eNOS genes have predictive significance for clinical outcome and survivality in advanced gastric cancer patients treated with first-line chemotherapy. 23667181_data suggest that KS is only caused by biallelic mutations in COL18A1 23912396_Hepatocellular carcinoma shows significantly elevated serum levels of angiogenic factors VEGF and Ang-2 and of anti-angiogenic factors endostatin and angiostatin. 23958573_Higher endostatin levels are associated with increased risk of early onset pre-eclampsia 24021340_Transcription levels of VEGF and endostatin by RT-PCR may be an adjunct to cytology screening for early detection of cervical carcinomas and may determine the progressive potentiality of individual lesions, especially in high-risk patients. 24030549_High serum endostatin was associated with increased mortality risk in 2 independent community-based cohorts of the elderly. 24089677_Our preliminary findings provide new evidence that endostatin/collagen XVIII concentration in the CSF increases substantially in patients with sTBI 24393402_Female type 2 diabetes mellitus patients have higher serum endostatin levels than males and exercise increases levels in both sexes. 24600079_Serum endostatin is raised in older men who have symptoms of intermittent claudication. 24998915_The ratio of VEGF/endostatin levels was significantly higher in operable non-small cell lung cancer patients than in normal controls. 25339042_High endostatin expression is associated with malignant pleural effusions in lung cancer. 25424718_Genebased analyses revealed associations of the COL18A1 gene with longitudinal Blood Pressure phenotypes, associations with essential hypertension, Blood Pressure salt sensitivity, preeclampsia, or preclinical stages of atherosclerosis. 25481287_ADAM8 and endostatin play a role in osteosarcoma progression. 25489667_In 2 large cohort studies using serum endostatin as a marker for disease severity and mortality in pulmonary arterial hypertension, a variant in the gene encoding ES, Col18a1, was linked independently with reduced mortality. 25788476_Study demonstrates that endostatin has novel ATPase activity, which mediates its antiangiogenic and antitumor activities. 25840998_the intima+media of IPAH vessels, collagens (COL4A5, COL14A1, and COL18A1), matrix metalloproteinase (MMP) 19, and a disintegrin and metalloprotease (ADAM) 33 were higher expressed, whereas MMP10, ADAM17, TIMP1, and TIMP3 were less abundant. 26275402_promoter methylation significantly increased in nasal polyp tissues 26315510_Data show that endostatin levels were elevated in patients with systemic sclerosis (SSc) and mixed connective tissue disease (MCTD). 26617886_The homozygous DN and NN genotypes of COL18A1 D104N were associated with the risk of osteosarcoma. 26771970_Collaterals formation seems to be associated with the activation of pro-inflammatory factors, growth factors and endostatin in chronic heart failure. 27214260_Endostatin is increased in patients with primary sjogren's syndrome patients with interstitial lung disease. 27261002_We further show that the SAGA deubiquitinase-activated gene Multiplexin (Mp) is required in glia for proper photoreceptor axon targeting. Mutations in the human ortholog of Mp, COL18A1, have been identified in a family with a SCA7-like progressive visual disorder, suggesting that defects in the expression of this gene in SCA7 patients could play a role in the retinal degeneration that is unique to this ataxia. 27746220_Structural and biological functions of COL18A1, such as its requirement in the maintenance of basement membrane integrity and its emerging roles in regulating cell survival, stem or progenitor cell maintenance and differentiation and inflammation have been reported. (Review) 27921229_a protease capable of producing endostatin from human Col XVIII, is reported 27930387_Blood endostatin wasdddddddddddddddddddddddddf significantly higher in chronic hepatitis C patients with liver fibrosis compared to those without fibrosis. 28189572_Lower endostatin levels or a predominance of VEGF over endostatin are predictors of poor short-term prognosis of intracranial atherosclerotic stenosis. 28428451_endostatin may have a role in subclinical atherosclerosis as shown in a study of a healthy Japanese population 28602933_Two COL18A1 variants co-segregated with familial epilepsy and anterior polymicrogyria in a family. 29528011_Increased serum levels of endostatin were associated with loss of graft function in kidney transplant recipients. 29532123_Our study suggests lack of association between DNA polymorphisms rs2236479 of COL18A1 and rs2862296 of LOXL-4 with advanced POP [pelvic organ prolapse] in this population [Brazil]. 29781737_results suggested that common polymorphisms in these two candidate genes were unlikely to play major roles in the genetic susceptibility to HM. Nevertheless, to avoid filtering real myopia genes, the role of COL11A1 and COL18A1 in the pathogenesis of myopia requires more refinement in both animal models and human genetic epidemiological studies 29860903_This study showed that endostatin might be a potential marker of vasculogenesis because of its significant increase after autologous cell therapy in diabetic patients with critical limb ischemia in contrast to those undergoing percutaneous transluminal angioplasty. This increase may be a sign of a protective feedback mechanism of this anti-angiogenic factor. 29893146_Higher serum endostatin was associated with left ventricular dysfunction and an increased heart failure risk in two community-based cohorts of elderly patients. 30007336_The role of COL18A1 in pathogenesis of angle closure was found by observation of COL18A1 mutations in primary angle-closure suspect-affected individuals of two additional unrelated families. 30055135_Combination of DESI2 and endostatin gene therapy in the mouse models significantly improves antitumor efficacy by accumulating DNA lesions, inducing apoptosis and inhibiting angiogenesis. 30474426_Higher endostatin predicted incident myocardial infarction predominantly in women but not independently of C-reactive protein. 31554264_our study demonstrated that down-regulation of COL18A1 in vitro resulted in inhibition of keratinization of epithelial cells. Moreover, deletion of Col18a1 in vivo led to an inadequate keratinization phenotype of oral mucosa in the Col18-KO mice, as demonstrated by a suppressed KRT10 expression and remarkably smaller size of KHG in the granular layer of epithelial tissue in those mice. 31922883_COL18A1/ES expression is markedly increased in remodeled pulmonary vessels in PAH. 32158205_Serum Endostatin Is a Novel Marker for COPD Associated with Lower Lung Function, Exacerbation and Systemic Inflammation. 32178553_Three cases of molecularly confirmed Knobloch syndrome. 32700429_Knobloch syndrome in a patient from Chile. 33238767_Variable phenotype of Knobloch syndrome due to biallelic COL18A1 mutations in children. 33296124_Collagen type XVIII alpha 1 chain (COL18A1) variants affect the risk of anti-tuberculosis drug-induced hepatotoxicity: A prospective study. 33341801_Elevated Endostatin Expression Is Regulated by the pIgA Immune Complex and Associated with Disease Severity of IgA Nephropathy. 34680907_Knobloch Syndrome Associated with Novel COL18A1 Variants in Chinese Population. | ENSMUSG00000001435 | Col18a1 | 5540.63657 | 0.9280362 | -0.1077469472 | 0.10350048 | 1.096616e+00 | 2.950100e-01 | 9.998360e-01 | No | Yes | 5552.99518 | 604.248889 | 5.492974e+03 | 461.857554 | |
| ENSG00000182899 | 6165 | RPL35A | protein_coding | P18077 | FUNCTION: Required for the proliferation and viability of hematopoietic cells. Plays a role in 60S ribosomal subunit formation. The protein was found to bind to both initiator and elongator tRNAs and consequently was assigned to the P site or P and A site. {ECO:0000269|PubMed:18535205}. | 3D-structure;Acetylation;Diamond-Blackfan anemia;Disease variant;RNA-binding;Reference proteome;Ribonucleoprotein;Ribosomal protein;tRNA-binding | Ribosomes, the organelles that catalyze protein synthesis, consist of a small 40S subunit and a large 60S subunit. Together these subunits are composed of 4 RNA species and approximately 80 structurally distinct proteins. This gene encodes a ribosomal protein that is a component of the 60S subunit. The protein belongs to the L35AE family of ribosomal proteins. It is located in the cytoplasm. The rat protein has been shown to bind to both initiator and elongator tRNAs, and thus, it is located at the P site, or P and A sites, of the ribosome. Although this gene was originally mapped to chromosome 18, it has been established that it is located at 3q29-qter. Alternative splicing results in multiple transcript variants. As is typical for genes encoding ribosomal proteins, there are multiple processed pseudogenes of this gene dispersed through the genome. [provided by RefSeq, Oct 2015]. | hsa:6165; | cytosol [GO:0005829]; cytosolic large ribosomal subunit [GO:0022625]; cytosolic ribosome [GO:0022626]; extracellular exosome [GO:0070062]; membrane [GO:0016020]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; tRNA binding [GO:0000049]; cytoplasmic translation [GO:0002181]; ribosomal large subunit biogenesis [GO:0042273]; rRNA processing [GO:0006364]; translation [GO:0006412] | 12175552_Inhibition of cell death by ribosomal protein L35a. 18535205_analysis of 2 Diamond-Blackfan anemia (DBA) patients with chromosome 3q deletions identified RPL35A as a potential DBA gene 19773262_Observational study of gene-disease association. (HuGE Navigator) 20378560_Observational study of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) 22045982_Studies identified deletions at known Diamond-Blackfan anemia (DBA)-related ribosomal protein gene loci in 17% (9 of 51) of patients without an identifiable mutation, including RPS19, RPS17, RPS26, and RPL35A. 22262766_Data show 1 proband with an RPL5 deletion, 1 patient with an RPL35A deletion, 3 with RPS17 deletions, and 1 with an RPS19 deletion. 28432740_We identified 85 overlapping deletions, of which six included the RPL35A gene and all should be had Diamond-Blackfan anemia (DBA).we sequenced the remaining RNF168 gene and examined her fibroblast culture for a DNA double strand break repair deficiency. These results were normal, indicating that the immunodeficiency is unlikely to result from a RNF168 deficiency. 32241839_Genotype-phenotype association and variant characterization in Diamond-Blackfan anemia caused by pathogenic variants in RPL35A. | ENSMUSG00000060636 | Rpl35a | 15786.24988 | 1.0878281 | 0.1214505482 | 0.08919867 | 1.838215e+00 | 1.751603e-01 | 9.998360e-01 | No | Yes | 15648.42798 | 3181.320423 | 1.876078e+04 | 2937.028130 | |
| ENSG00000183475 | 140460 | ASB7 | protein_coding | Q9H672 | FUNCTION: Probable substrate-recognition component of a SCF-like ECS (Elongin-Cullin-SOCS-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. {ECO:0000250, ECO:0000269|PubMed:16325183}. | ANK repeat;Alternative splicing;Reference proteome;Repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | The protein encoded by this gene belongs to a family of ankyrin repeat proteins that, along with four other protein families, contains a C-terminal SOCS box motif. Growing evidence suggests that the SOCS box acts as a bridge between specific substrate-binding domains and the more generic proteins that comprise a large family of E3 ubiquitin protein ligases. In this way, SOCS box containing proteins may regulate protein turnover by targeting proteins for polyubiquination and, therefore, for proteasome-mediated degradation. Two alternative transcripts encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:140460; | cytosol [GO:0005829]; intracellular signal transduction [GO:0035556]; protein ubiquitination [GO:0016567] | 27697924_these data indicate that ASB7 plays a crucial role in regulating spindle dynamics and genome integrity by controlling the expression of DDA3. 29630609_High ASB7 expression is associated with endoplasmic reticulum stress. | ENSMUSG00000030509 | Asb7 | 329.48961 | 0.9832254 | -0.0244059253 | 0.18216562 | 1.818170e-02 | 8.927387e-01 | 9.998360e-01 | No | Yes | 326.78200 | 54.347184 | 3.645896e+02 | 46.857987 |
| ENSG00000183479 | 11219 | TREX2 | protein_coding | Q9BQ50 | FUNCTION: Exonuclease with a preference for double-stranded DNA with mismatched 3' termini. May play a role in DNA repair. {ECO:0000269|PubMed:11279105}. | 3D-structure;Alternative splicing;DNA damage;DNA repair;Exonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease;Nucleus;Reference proteome | This gene encodes a nuclear protein with 3' to 5' exonuclease activity. The encoded protein participates in double-stranded DNA break repair, and may interact with DNA polymerase delta. [provided by RefSeq, Nov 2012]. | hsa:11219; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; 3'-5'-exodeoxyribonuclease activity [GO:0008296]; exodeoxyribonuclease III activity [GO:0008853]; magnesium ion binding [GO:0000287]; nucleic acid binding [GO:0003676]; protein homodimerization activity [GO:0042803]; DNA catabolic process, exonucleolytic [GO:0000738]; DNA metabolic process [GO:0006259]; DNA repair [GO:0006281] | 15581481_Polymorphisms exist in prostatic cancer patients. 15661738_analysis of human TREX2 3' -> 5'-exonuclease structure and description of the mechanism for efficient nonprocessive DNA catalysis 17426129_Results suggest that TREX2 plays an important function during DNA metabolism and cellular proliferation. 18092167_Observational study of gene-disease association. (HuGE Navigator) 18534978_analysis of cooperative DNA binding and communication across the dimer interface in the TREX2 3' --> 5'-exonuclease 19094998_Trex2 deletion caused high levels of Robertsonian translocations (RbTs) showing Trex2 is important for chromosomal maintenance. 19321497_a model for DNA binding and 3' hydrolysis for the TREX2 dimer. 21546543_Trex2 does not enable DSB repair and prompt a new model that posits Trex2 suppresses the formation of broken chromosomes. 23591820_TREX-2 is an NPC-associated complex in mammalian cells. 24896180_human TREX-2 complex prevents genome instability, as determined by the accumulation of gamma-H2AX and 53BP1 foci and single-cell electrophoresis in cells depleted of the TREX-2 subunits PCID2, GANP and DSS1 26090614_Altogether, data provide new insights in the molecular mechanisms of TREX2 activity and establish cell autonomous and non-cell autonomous functions of TREX2 in the UVB-induced skin response. 30054449_the scaffold subunit of TREX-2, GANP, positively regulates DNA repair through homologous recombination (HR). In contrast, DUBm adaptor subunits ENY2 and ATXNL3 are required to limit unscheduled HR. 31053176_Significant methylation loss at an intragenic site of TREX2 was a frequent trait in a cohort of patients with laryngeal cancer. Methylation loss correlated with increased expression of TREX2 in laryngeal tumors and improved overall survival. These data highlight a regulatory role of TREX2 DNA methylation for gene expression which might affect incidence and survival of laryngeal cancer. 32917881_Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. 33357432_TREX2 Exonuclease Causes Spontaneous Mutations and Stress-Induced Replication Fork Defects in Cells Expressing RAD51(K133A). | ENSMUSG00000031372 | Trex2 | 292.32039 | 0.6567727 | -0.6065338492 | 0.20972117 | 8.354895e+00 | 3.846497e-03 | 7.440887e-01 | No | Yes | 230.62582 | 39.220361 | 3.176149e+02 | 42.858566 | |
| ENSG00000183570 | 54039 | PCBP3 | protein_coding | P57721 | FUNCTION: Single-stranded nucleic acid binding protein that binds preferentially to oligo dC. {ECO:0000250}. | Alternative splicing;Cytoplasm;DNA-binding;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein | This gene encodes a member of the KH-domain protein subfamily. Proteins of this subfamily, also referred to as alpha-CPs, bind to RNA with a specificity for C-rich pyrimidine regions. Alpha-CPs play important roles in post-transcriptional activities and have different cellular distributions. The protein encoded by this gene lacks the nuclear localization signals found in other subfamily members. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jan 2017]. | hsa:54039; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; DNA binding [GO:0003677]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; mRNA metabolic process [GO:0016071]; regulation of gene expression [GO:0010468]; regulation of RNA metabolic process [GO:0051252] | 19914360_Study examined splicing factors located on chromosome 21 for their effect on tau exon 10 and discovered that one of them, hnRNPE3 (PCBP3), modestly activates splicing of exon 10 by interacting with its proximal downstream intron around position +19. 30275197_Low PCBP3 expression is associated with Pancreatic ductal adenocarcinoma. | ENSMUSG00000001120 | Pcbp3 | 225.95215 | 1.0435760 | 0.0615356292 | 0.20649466 | 8.774460e-02 | 7.670643e-01 | 9.998360e-01 | No | Yes | 227.28565 | 29.209271 | 2.073683e+02 | 21.163228 | |
| ENSG00000184381 | 8398 | PLA2G6 | protein_coding | O60733 | FUNCTION: Calcium-independent phospholipase involved in phospholipid remodeling with implications in cellular membrane homeostasis, mitochondrial integrity and signal transduction. Hydrolyzes the ester bond of the fatty acyl group attached at sn-1 or sn-2 position of phospholipids (phospholipase A1 and A2 activity respectively), producing lysophospholipids that are used in deacylation-reacylation cycles (PubMed:9417066, PubMed:10092647, PubMed:10336645, PubMed:20886109). Hydrolyzes both saturated and unsaturated long fatty acyl chains in various glycerophospholipid classes such as phosphatidylcholines, phosphatidylethanolamines and phosphatidates, with a preference for hydrolysis at sn-2 position (PubMed:10092647, PubMed:10336645, PubMed:20886109). Can further hydrolyze lysophospholipids carrying saturated fatty acyl chains (lysophospholipase activity) (PubMed:20886109). Upon oxidative stress, contributes to remodeling of mitochondrial phospholipids in pancreatic beta cells, in a repair mechanism to reduce oxidized lipid content (PubMed:23533611). Preferentially hydrolyzes oxidized polyunsaturated fatty acyl chains from cardiolipins, yielding monolysocardiolipins that can be reacylated with unoxidized fatty acyls to regenerate native cardiolipin species (By similarity). Hydrolyzes oxidized glycerophosphoethanolamines present in pancreatic islets, releasing oxidized polyunsaturated fatty acids such as hydroxyeicosatetraenoates (HETEs) (By similarity). Has thioesterase activity toward fatty-acyl CoA releasing CoA-SH known to facilitate fatty acid transport and beta-oxidation in mitochondria particularly in skeletal muscle (PubMed:20886109). Plays a role in regulation of membrane dynamics and homeostasis. Selectively hydrolyzes sn-2 arachidonoyl group in plasmalogen phospholipids, structural components of lipid rafts and myelin (By similarity). Regulates F-actin polymerization at the pseudopods, which is required for both speed and directionality of MCP1/CCL2-induced monocyte chemotaxis (PubMed:18208975). Targets membrane phospholipids to produce potent lipid signaling messengers. Generates lysophosphatidate (LPA, 1-acyl-glycerol-3-phosphate), which acts via G-protein receptors in various cell types (By similarity). Has phospholipase A2 activity toward platelet-activating factor (PAF, 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine), likely playing a role in inactivation of this potent proinflammatory signaling lipid (By similarity). In response to glucose, amplifies calcium influx in pancreatic beta cells to promote INS secretion (By similarity). {ECO:0000250|UniProtKB:A0A3L7I2I8, ECO:0000250|UniProtKB:P97570, ECO:0000250|UniProtKB:P97819, ECO:0000269|PubMed:10092647, ECO:0000269|PubMed:10336645, ECO:0000269|PubMed:18208975, ECO:0000269|PubMed:20886109, ECO:0000269|PubMed:23533611, ECO:0000269|PubMed:9417066}.; FUNCTION: [Isoform Ankyrin-iPLA2-1]: Lacks the catalytic domain and may act as a negative regulator of the catalytically active isoforms. {ECO:0000269|PubMed:9417066}.; FUNCTION: [Isoform Ankyrin-iPLA2-2]: Lacks the catalytic domain and may act as a negative regulator of the catalytically active isoforms. {ECO:0000269|PubMed:9417066}. | ANK repeat;Alternative splicing;Calmodulin-binding;Cell membrane;Cell projection;Chemotaxis;Cytoplasm;Disease variant;Dystonia;Hydrolase;Lipid metabolism;Membrane;Mitochondrion;Neurodegeneration;Parkinson disease;Parkinsonism;Pharmaceutical;Phospholipid metabolism;Reference proteome;Repeat;Transmembrane;Transmembrane helix | The protein encoded by this gene is an A2 phospholipase, a class of enzyme that catalyzes the release of fatty acids from phospholipids. The encoded protein may play a role in phospholipid remodelling, arachidonic acid release, leukotriene and prostaglandin synthesis, fas-mediated apoptosis, and transmembrane ion flux in glucose-stimulated B-cells. Several transcript variants encoding multiple isoforms have been described, but the full-length nature of only three of them have been determined to date. [provided by RefSeq, Dec 2010]. | hsa:8398; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; mitochondrion [GO:0005739]; plasma membrane [GO:0005886]; pseudopodium [GO:0031143]; 1-alkyl-2-acetylglycerophosphocholine esterase activity [GO:0003847]; ATP-dependent protein binding [GO:0043008]; calcium-independent phospholipase A2 activity [GO:0047499]; calmodulin binding [GO:0005516]; hydrolase activity [GO:0016787]; identical protein binding [GO:0042802]; lysophospholipase activity [GO:0004622]; myristoyl-CoA hydrolase activity [GO:0102991]; palmitoyl-CoA hydrolase activity [GO:0016290]; phosphatidyl phospholipase B activity [GO:0102545]; phospholipase A2 activity [GO:0004623]; protein kinase binding [GO:0019901]; serine hydrolase activity [GO:0017171]; antibacterial humoral response [GO:0019731]; cardiolipin acyl-chain remodeling [GO:0035965]; cardiolipin biosynthetic process [GO:0032049]; chemotaxis [GO:0006935]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; maternal process involved in female pregnancy [GO:0060135]; memory [GO:0007613]; negative regulation of synaptic transmission, glutamatergic [GO:0051967]; phosphatidic acid metabolic process [GO:0046473]; phosphatidylcholine catabolic process [GO:0034638]; phosphatidylethanolamine catabolic process [GO:0046338]; platelet activating factor metabolic process [GO:0046469]; positive regulation of arachidonic acid secretion [GO:0090238]; positive regulation of ceramide biosynthetic process [GO:2000304]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of exocytosis [GO:0045921]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774]; positive regulation of protein kinase C signaling [GO:0090037]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of release of cytochrome c from mitochondria [GO:0090200]; regulation of store-operated calcium channel activity [GO:1901339]; response to endoplasmic reticulum stress [GO:0034976]; urinary bladder smooth muscle contraction [GO:0014832]; vasodilation [GO:0042311] | 12208880_activation during apoptosis promotes the exposure of membrane lysophosphatidylcholine leading to binding by natural immunoglobulin M antibodies and complement activation 12423354_stimulation of three isoforms of PLA2 by thapsigargin liberates free AA that, in turn, induces capacitative calcium influx in human T-cells 14749286_Arachidonic acid produced by iPLA(2)beta-catalyzed hydrolysis of their substrates induces release of Ca(2+) from ER stores, an event thought to participate in glucose-stimulated insulin secretion. 15052324_The GVIB iPLA2 is widely expressed in human tissues but is enriched in heart, placenta, and skeletal muscle. 15249229_Here we show that the C-terminal region of human iPLA(2)gamma is responsible for the enzymatic activity. 15252038_iPLA2 may be dispensable for the apoptotic process to occur 15318030_Observational study of gene-disease association. (HuGE Navigator) 15364929_iPLA2epsilon (adiponutrin), iPLA2zeta (TTS-2.2), and iPLA2eta (GS2) are three novel TAG lipases/acylglycerol transacylases that likely participate in TAG hydrolysis and the acyl-CoA independent transacylation of acylglycerols 15385540_truncated iPLA(2) proteins associate with active iPLA(2) and down-regulate its activity during G(1) 15573142_Detailed characterization of group VIA phospholipase A2 beta suggests that the pancreatic islet beta-cells express multiple isoforms of iPLA2beta; the hypothesis in this review is that these isozymes participate in different cellular functions. 16585943_This study reviews the evidence and discusses the potential roles of phospholipase A2 Group 6A for schizophrenia with particular emphasis on published association studies. 16783378_mapped a locus for infantile neuroaxonal dystrophy (INAD) and neurodegeneration with brain iron accumulation (NBIA) to chromosome 22q12-q13 and identified mutations in PLA2G6, encoding a group VI phospholipase A2, in NBIA, INAD and Karak syndrome 16943248_Increase in iPLA(2) and accumulation of membrane phospholipid-derived metabolites in HCAEC exposed to hypoxia or thrombin have important implications in inflammation and arrhythmogenesis in atherosclerosis/thrombosis and myocardial ischemia. 16966332_iPLA2-VIA is a novel regulator of endothelial cell S phase progression, cell cycle residence, and angiogenesis 17003039_The role of calcium influx factor and PLA2G6 in the activation of CRAC channels and calcium entry in rat tumor cell lines is reported. 17033970_The disease gene was mapped to a 1.17-Mb locus on chromosome 22q13.1. 17082190_Transient receptor potential subfamily M member 8 (TRPM8) channel is stimulated by the Ca2+-independent phospholipase A2 (iPLA2) signaling pathway with its end products, lysophospholipids, acting as its endogenous ligands. 17188740_human coronary artery endothelial cells exposed to thrombin or tryptase stimulation demonstrated an increase in iPLA2 activity and arachidonic acid release 17254819_Cerebellar atrophy without cerebellar cortex hyperintensity in infantile neuroaxonal dystrophy (INAD) due to PLA2G6 mutation. 17275398_Secretion and activity of sPLA(2) were found to be similar in granulocyte-like PLB cells expressing or lacking cPLA(2)alpha, indicating that they are not under cPLA(2)alpha regulation. 17459165_oxytocin stimulation of uterine PGF2alpha production is mediated, at least in part, by up-regulation of PLA2G6 expression and activity 18208975_iPLA(2)beta and cPLA(2)alpha regulate monocyte migration from different intracellular locations, with iPLA(2)beta acting as a critical regulator of the cellular compass. 18562188_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18570303_PLA2G6 mutations are associated with infantile neuroaxonal dystrophy and have been reported previously to cause early cerebellar signs, and the syndrome was classified as neurodegeneration with brain iron accumulation (type 2). 18676680_Observational study of gene-disease association. (HuGE Navigator) 18775417_This review discusses the role of iPLA2 in cell growth with special emphasis placed on its role in cell signaling; the putative lipid signals involved are also discussed. 18790994_Tryptase stimulation of human small airway epithelial cells increased membrane-associated, calcium-independent phospholipase A(2)gamma (iPLA(2)gamma) activity, resulting in increased arachidonic acid and PGE(2) release. 18799783_Observational study of gene-disease association. (HuGE Navigator) 18799783_PLA2G6 mutations are associated with nearly all cases of classic infantile neuroaxonal dystrophy. 18826942_H2O2-mediated hyperoxidation of Prdx6 induces cell cycle arrest at the G2/M transition through up-regulation of iPLA2 activity. 19059366_iPLA2 activity is responsible for membrane phospholipid hydrolysis in response to tryptase or thrombin stimulation in pulmonary vascular endothelial cells 19087156_Different and even identical PLA2G6 mutations may cause neurodegenerative diseases with heterogeneous clinical manifestations, including dystonia-parkinsonism. 19138334_The nine novel mutations identified in this study suggest the uniqueness of the PLA2G6 mutation spectrum in Chinese patients 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19225567_Chromofungin or Catestatin , penetrate into PMNs, inducing extracellular calcium entry by a CaM-regulated iPLA2 pathway. 19556238_The region with the greatest change upon lipid binding in phospholipase A2 group VI was region 708-730. 19578365_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20053941_iPLA2 expression is increased in neutrophils from people with diabetes and mediates superoxide generation, presenting an alternate pathway independent of protein kinase C and phosphatidic acid phosphohydrolase-1 hydrolase signaling. 20171194_These data demonstrate the novel findings that iPLA2 inhibition activates p38 by inducing reactive species, and further suggest that this signaling kinase is involved in p53 activation, cell cycle arrest and cytostasis. 20186954_Observational study of gene-disease association. (HuGE Navigator) 20186954_Our data suggest that PLA2G6 mutations are unlikely to be an important cause of the common garden variety of Parkinson's disease patients with dystonia or a positive family history 20219570_Results characterize a pathway leading to NOX2 activation in which iPLA(2)-regulated p38 MAPK activity is a key regulator of S100A8/A9 translocation via S100A9 phosphorylation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20464283_Observational study of gene-disease association. (HuGE Navigator) 20584031_Compound heterozygosity for a large intragenic deletion and a nonsense mutation was found in one patient with infantile neuroaxonal dystrophy while the other is carrying two novel splice-site mutations 20619503_This report further defines the clinical features and neuropathology of PLA2G6 related childhood and adult onset dystonia-parkinsonism . 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20629144_Genetically determined NBIA cases from the Indian subcontinent suggest presence of unusual phenotypes of PANK2 and novel mutations 20647408_Observational study of gene-disease association. (HuGE Navigator) 20669327_We identified genetic deficits in PLA2G6 that were associated with Levodopa responsive parkinsonism with pyramidal signs. 20813170_This study demonistrated that pLA2G6 has a dynamic expression pattern both in terms of the location of expression and the differentiation state of expressing cells. 20881058_The A isoform of PLA2G6 was shown to play a role in controlling the architecture of the endoplasmic reticulum-golgi intermediate compartment proteins in a way that impacts transport. 20886109_Catalytic function of PLA2G6 impaired by mutations is associated with infantile neuroaxonal dystrophy. 20938027_Observational study of gene-disease association. (HuGE Navigator) 20938027_The results of this study suggested that PLA2G6 mutations should be considered in patients with early-onset l-dopa-responsive parkinsonism and dementia with frontotemporal lobar atrophy. 21191104_These results indicate that cardiac endothelial cell PAF production is dependent on iPLA(2)beta activation and that both iPLA(2)beta and iPLA(2)gamma may be involved in PGI(2) release. 21368765_PLA2G6 mutations are unlikely to be the major causes or risk factors of Parkinson's disease at least in Asian populations 21482170_data indicate that PLA2G6 mutation may not play a major role in general frontotemporal type of dementia 21700586_tHIS STUDY DEMONISTRATED THAT the PLA2G6 gene allocated PARK14 locus and is associated with autosomal recessive early-onset parkinsonism. 21812034_A possible involvement of calcium-independent group VI phospholipase A2 (iPLA2-VI) in the pathogenesis of Parkinson's disease has been proposed. 22213678_PLA2G6 mutations are associated with PARK14-linked young-onset parkinsonism and sporadic Parkinson's disease 22218592_acts as an inhibitory modulator of NKCC2 activity in thick ascending limb 22406380_Our result indicated that PLA2G6 mutations might not be a main cause of Chinese sporadic early-onset parkinsonism. 22459563_The results of this study suggested that PLA2G6 is not a susceptibility gene for parkinson disease in our population. 22549787_These data confirm the role of iPLA(2)beta as an essential mediator of endogenous store operated calcium entry. 22680611_The present study confirms the involvement of iPLA(2)-VIA in efficient retinal pigment epithelium phagocytosis of photoreceptor outer segments. 22903185_Neuronal phospholipid deacylation is essential for axonal and synaptic integrity through the action of iPLA2 and NTE. (Review) 23007400_membrane composition and the presence of nucleotides play key roles in recruiting and modulating GVIA-iPLA(2) activity in cells 23043102_Orai1 and PLA2g6 are involved in adhesion formation, whereas STIM1 participates in both adhesion formation and disassembly. 23074238_Findings reveal for the first time expression of iPLA(2)beta protein in human islet beta-cells and that induction of iPLA(2)beta during endoplasmic reticulum stress contributes to human islet beta-cell apoptosis. 23182313_identified four rare PLA2G6 mutations in 250 PD patients in Chinese population with Parkinson's disease 23196729_Mutations in PLA2G6 is often associated with rapidly progressive parkinsonism and with additional features including pyramidal signs, cognitive decline and loss of sustained Levodopa responsiveness. 23277130_The association with bipolar disorder of the iPLA2beta (PLA2G6) its genetic interaction with type 2 transient receptor potential channel gene TRPM2, was examined. 23587695_This study demonistrated that PLA2g8 expression was significantly decreased in patients treated with antipsychotic drug. 24130795_the phenotype of neurodegeneration associated with PLA2G6 mutations 24512906_our findings indicate that PRDX6 promotes lung tumor growth via increased glutathione peroxidase and iPLA2 activities 24522175_clinical findings may be helpful in distinguishing PLA2G6-related neurodegeneration from the other major cause of NBIA, recessive PANK2 mutations. 24745848_Novel PLA2G6 mutations were identified in all patients with Phospholipase A2 associated neurodegeneration. 24791136_The significance of calcium-independent phospholipase A, group VIA (iPLA2-VIA), in retinal pigment epithelial cell survival, was investigated. 24858037_The loss of PS2 could have a critical role in lung tumor development through the upregulation of iPLA2 activity by reducing gamma-secretase. 25004092_IL-1beta and IFNgamma induces mSREBP-1 and iPLA2beta expression and induce beta-cell apoptosis. 25207958_genetic association studies in a population of Han Chinese: Data suggest that SNPs in PLA2G6 (rs132984; rs2284060) are associated with type 2 diabetes and hypertriglyceridemia in the population studied. [Meta-Analysis included] 25348461_A homozygous novel mutation at position c.2277-1G>C in PLA2G6 gene presumed to give rise to altered splicing, was detected, thus confirming the diagnosis of infantile Neuroaxonal Dystrophy (INAD). 25482049_Stimulation of adrenoreceptors causes increased iPLA2 expression via MAP kinase/ERK 1/2. 25668476_Mutations in PANK2 and CoASY lead, respectively, to PKAN and CoPAN forms of Neurodegeneration with brain iron accumulation . Mutations in PLA2G6 lead to PLAN. Mutations in C19orf12 lead to MPAN 26001724_our findings demonstrate that loss of normal PLA2G6 gene activity leads to lipid peroxidation, mitochondrial dysfunction and subsequent mitochondrial membrane abnormalities. 26160611_Results demonstrated no significant impact of PLA2G6 and PLA2G4C gene polymorphisms on attenuated niacin skin flushing in schizophrenia patients. 26446356_Three catalytically active cPLA2, iPLA2, and sPLA2 are expressed in different areas within the human spermatozoon cell body. Spermatozoa with a significant low motility showed strong differences both in terms of total specific activity and of different intracellular distribution, compared with normal spermatozoa. Phospholipases could be potential biomarkers of asthenozoospermia. 26525102_genetic association study in Quebec City population: Data suggest total plasma n-6 fatty acid phospholipid levels and C-reactive protein are modulated by SNPs in PLA2G4A and PLA2G6 alone or in combination with fish oil dietary supplementation. 26668131_Mutations in PLA2G6 altered Golgi morphology, O-linked glycosylation and sialylation of protein in patients with neurodegeneration 26755131_Analysis of the cells from idiopathic Parkinson's disease patients reveals a significant deficiency in store-operated PLA2g6-dependent Ca(2+) signalling 27030050_This study demonstrated that elevated expression of alphaSyn/PalphaSyn in mitochondria appears to be the early response to PLA2G6-deficiency in neurons. 27196560_PLA2G6 mutations in Indian patients with infantile neuroaxonal dystrophy and atypical late-onset neuroaxonal dystrophy 27196560_PLA2G6 were identified in patients with a spectrum of neurodegenerative conditions, such as infantile neuroaxonal dystrophy, atypical late-onset neuroaxonal dystrophy and dystonia parkinsonism complex in Indian families 27268037_Finding suggest the broadness of the clinical spectrum of group VI phospholipases A2 (PLA2G6)-related neurodegeneration. 27317427_These results strongly suggest that PNPLA9, -6 and -4 play a key role in GPL turnover and homeostasis in human cells. A hypothetical model suggesting how these enzymes could recognize the relative concentration of the different GPLs is proposed 27513994_This study identifies a novel PLA2G6 mutation that is the possible genetic cause of FCMTE in this Chinese family. 27709683_This exome sequencing in a family and identified compound-heterozygous PLA2G6 mutations in 2 affected sisters. 27942883_Study performed direct sequencing and investigated copy number variations (CNVs) of this gene in 109 Japanese patients with parkinsonism. Results suggest that CNV in PLA2G6 is rare in parkinsonism, at least in the Japanese population, in contrast to the reports of its frequency in neurodegeneration associated with brain iron accumulation. 28091863_performed a longitudinal brain volumetry study in a couple of bicorial twins with PLA2G6-positive infantile neuronal axonal dystrophy 28150298_PLA2G6-associated neurodegeneration was caused by paternal isodisomy of the chromosome 22 (carrying c.680C>T mutation) following in vitro fertilization 28213071_the association of PLA2G6 with the pathogenesis of idiopathic PD, in addition to PARK14. 28295203_PLA2G6 gene mutations in 3 families, are reported. 28651698_We found no significant influence of the PLA2G6 and PLA2G4C polymorphisms on mean age at first hospital admission (P > 0.05) and that the investigated polymorphisms significantly influenced the clinical psychopathology only in male patients. The PLA2G4C polymorphism accounted for approximately 12% of negative symptom severity. 28821231_A novel missense mutation in PLA2G6 gene (c.3G > T:p.M1I) in one and half-year-old boy with muscle weakness and neurodevelopmental regression (speech, motor and cognition). 29325618_This study showed that PANK2 genes account for disease of patients diagnosed with an Neurodegeneration with brain iron accumulation disorder. 29342349_A lipidomics-based LC/MS assay was used to define the specificity of cPLA2, iPLA2, and sPLA2 toward a variety of phospholipids. A unique hydrophobic binding site for the cleaved fatty acid dominates each enzyme's specificity rather than its catalytic residues and polar headgroup binding site. 29454663_PLA2G6 mutations are associated with a continuous clinical spectrum from phospholipase A2-associated neurodegeneration to hereditary spastic paraplegia. 29472584_The catalytic domains of iPLA2beta form a tight dimer and surrounded by ankyrin repeat domains that adopt an outwardly flared orientation, poised to interact with membrane proteins. 29739362_A novel variant (NM_003560.2 c.1427 + 2 T > C) acting on a splice donor site and predicted to lead to skipping of exon 10 was found in PLA2G6 in two siblings diagnosed with infantile neuroaxonal dystrophy. This mutation seems to be pathogenic and was found in a homozygous state in the two patients and homozygous reference or heterozygous in five healthy family members. 29753029_PLA2G6 inherited genotype may influence melanoma BRAF/NRAS subtype development. 29909971_This study shows a hereto unknown role of PLA2G6/PARK14 in sphingolipid homeostasis and endolysosomal function. The suppression of PLA2G6 expression causes a reduction in Vps26 and Vps35 and leads to an elevation of ceramides and other sphingolipid intermediates in neuroblastoma cell line cells. 30232368_Two missense variants, p.R53C and p.T319M in PLA2G6 gene may contribute to Parkinson's disease susceptibility in Chinese population. 30302010_PLA2G6 mutation is associated with neurodegeneration presenting as a complicated form of hereditary spastic paraplegia. 30707893_Impaired iPLA2beta activity affects iron uptake and storage without iron accumulation: An in vitro study excluding decreased iPLA2beta activity as the cause of iron deposition in PLAN. 30772976_report a PLA2G6 compound complicated mutation in an atypical neuroaxonal dystrophy Chinese family, that is the p. A80T and p.D331Y mutation 31196701_A novel compound heterozygous mutation of the PLA2G6 gene, c.1648delC and c.991G>T, is associated with adult onset ataxia. 31277247_Proportion of calcium-independent (i)PLA2beta-containing mucosal mast cells (MCs) and the expression intensity of sPLA2-IIA was increased in Crohn's disease (CD). In vitro study showed that (i)PLA2beta is involved in the secretion of secretory phospholipases (sPLA2) from human MC line HMC-1. These results suggest that iPLA2beta-mediated release of sPLA2 from intestinal MCs may contribute to CD pathophysiology. 31492433_Study demonstrated significant, though weak, effects of the PLA2G6 polymorphism on the risk of nicotine dependence, as well as a significant, though weak, influence of the PLA2G6-smoking interaction on age of schizophrenia onset, with both effects manifested in a gender-specific fashion. 31493991_Genetic testing of the PLA2G6 confirmed presence of compound heterozygous novel mutations in Malaysian siblings with infantile neuroaxonal dystrophy 1 31506141_immunohistochemistry showed a reduction in the protein expression of PLAG6 in the muscular tissue of this child 31689548_Infantile onset progressive cerebellar atrophy and anterior horn cell Degeneration-A novel phenotype associated with mutations in the PLA2G6 gene. 31922589_The compound variants of c.668C>A (p.Pro223Gln) and c.2266C>T (p.Gln756Ter) of the PLA2G6 gene probably underlies infantile neuroaxonal dystrophy in the child 32771225_PLA2G6 variants associated with the number of affected alleles in Parkinson's disease in Japan. 33080873_Metabolic Effects of Selective Deletion of Group VIA Phospholipase A2 from Macrophages or Pancreatic Islet Beta-Cells. 33087576_PLA2G6 guards placental trophoblasts against ferroptotic injury. 33279242_Association of rare heterozygous PLA2G6 variants with the risk of Parkinson's disease. 33576074_Whole genome sequencing reveals biallelic PLA2G6 mutations in siblings with cerebellar atrophy and cap myopathy. 33766980_Typical MRI features of PLA2G6 mutation-related phospholipase-associated neurodegeneration (PLAN)/infantile neuroaxonal dystrophy (INAD). 34131139_iPLA2beta-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4. 34207793_iPLA2beta Contributes to ER Stress-Induced Apoptosis during Myocardial Ischemia/Reperfusion Injury. 34622992_Dissecting the Phenotype and Genotype of PLA2G6-Related Parkinsonism. 35092705_Novel insertion mutation in the PLA2G6 gene in an Iranian family with infantile neuroaxonal dystrophy. 35247231_Genome sequencing reveals novel noncoding variants in PLA2G6 and LMNB1 causing progressive neurologic disease. | ENSMUSG00000042632 | Pla2g6 | 360.01560 | 1.0521505 | 0.0733410873 | 0.17258966 | 1.833266e-01 | 6.685299e-01 | 9.998360e-01 | No | Yes | 299.89366 | 49.119016 | 2.551451e+02 | 32.551545 | |
| ENSG00000185024 | 2972 | BRF1 | protein_coding | Q92994 | FUNCTION: General activator of RNA polymerase which utilizes different TFIIIB complexes at structurally distinct promoters. The isoform 1 is involved in the transcription of tRNA, adenovirus VA1, 7SL and 5S RNA. Isoform 2 is required for transcription of the U6 promoter. | Activator;Alternative splicing;Direct protein sequencing;Disease variant;Dwarfism;Mental retardation;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes one of the three subunits of the RNA polymerase III transcription factor complex. This complex plays a central role in transcription initiation by RNA polymerase III on genes encoding tRNA, 5S rRNA, and other small structural RNAs. The gene product belongs to the TF2B family. Several alternatively spliced variants encoding different isoforms, that function at different promoters transcribed by RNA polymerase III, have been identified. [provided by RefSeq, Jun 2011]. | hsa:2972; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription factor TFIIIB complex [GO:0000126]; transcription preinitiation complex [GO:0097550]; metal ion binding [GO:0046872]; RNA polymerase III general transcription initiation factor activity [GO:0000995]; RNA polymerase III type 3 promoter sequence-specific DNA binding [GO:0001006]; TBP-class protein binding [GO:0017025]; DNA-templated transcription, initiation [GO:0006352]; positive regulation of transcription by RNA polymerase III [GO:0045945]; rRNA transcription [GO:0009303]; transcription by RNA polymerase III [GO:0006383]; transcription initiation from RNA polymerase III promoter [GO:0006384]; transcription preinitiation complex assembly [GO:0070897]; tRNA transcription [GO:0009304] | 11997511_CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. 12016223_human small nuclear RNA gene-specific transcription factor IIIB complex de novo on and off promoter 12198173_BRF1 accelerated mRNA decay and antagonized the stabilizing effect of PI3-kinase, while mutation of the zinc fingers abolished both function and ARE-binding activity. This approach identifies BRF1 as an essential regulator of ARE-dependent mRNA decay. 15538381_Data report that protein kinase B (PKB/Akt) stabilizes ARE transcripts by phosphorylating butyrate response factor (BRF1) at serine 92. 16982688_These results suggest a direct role of an RNA polymerase III transcription factor in the targeting process. 17369404_depletion of endogenous TTP and BRF-1 proteins, or overexpression of dominant-negative mutant TTP proteins, impairs the localization of reporter AU-rich element mRNAs 17499043_Maf1 occupancy of Pol III genes is inversely correlated with that of the initiation factor TFIIIB (subunit Brf1) and Pol III. 17877750_the hypo-phosphorylated Rb appeared to be largely sequestered into a complex with Brf1, which resulted in the blockage of Rb function to repress E2F1 transactivation 17968325_Brf1 gene was identified in the genome-wide loss-of-function genetic screen as putative tumor suppressor located at 14q32.33. 18326031_MK2-mediated inhibition of BRF1 requires phosphorylation at S54, S92, and S203. 18700021_deregulation of Brf1 and Brf2 expression could be a key mechanism responsible for the observed deregulation of RNA pol III transcription in cancer cells 20154270_results identify a human Pol III isoform and isoform-specific functions in the regulation of cell growth and transformation 21044367_Observational study of gene-disease association. (HuGE Navigator) 21106530_Alcohol induces RNA polymerase III-dependent transcription through c-Jun by co-regulating TATA-binding protein (TBP) and Brf1 expression. 21832157_these observations are in favor of a cell- and context-dependent regulation of Tis11b by hypoxia, which then contributes to modulation of angiogenesis. 24978456_hnRNP F is a co-factor in a subset of tristetraprolin/BRF1/BRF2-mediated mRNA decay. 25561519_BRF1 mutations that reduce protein activity cause neurodevelopmental anomalies, suggesting that BRF1-mediated Pol III transcription is required for normal cerebellar and cognitive development. 26701855_Brf1 expression is increased in human HCC cases, which is correlated with shorter survival times. 27708140_Site-directed mutagenesis combined with kinase assays and specific phosphosite immunodetection identified Ser-54 (S54) and Ser-334 (S334) as PKA target amino acids in vitro and in vivo. Phosphomimetic mutation of the C-terminal S334 markedly increased TIS11b half-life and, unexpectedly, enhanced TIS11b activity on mRNA decay. 27748960_Mutations in BRF1 cause severe short stature, remarkably delayed bone age, dysmorphic features, cerebellar hypoplasia and cognitive dysfunction inherited in an autosomal recessive pattern. 28912018_In an analysis of families with a history of colorectal cancer, we associated germline mutations in BRF1 with predisposition to colorectal cancer. Seven of the identified variants (1 detected in 2 families) affected BRF1 mRNA splicing, protein stability, or expression and/or function. 28972307_These results indicate an interaction between Brf1 and ER alpha, which synergistically regulates the transcription of Pol III genes. 30413534_These findings uncover a novel mechanism for the regulation of BRF1 and reveal RNF12 as an important regulator of Pol III-dependent transcription. 30914800_A derivative of TIS11b called R9-ZnC(S334D), by combining N-terminal domain deletion, serine-to-aspartate substitution at position 334 to enhance the function of the protein and fusion to the cell-penetrating peptide polyarginine R9. R9-ZnC(S334D) not only blunted secretion of vascular endothelial growth factor (VEGF) but also inhibited proliferation, migration, invasion, and anchorage-independent growth of breast cancer. 30962286_results provide evidence that AMD surveils poly(A)(+) Replication-dependent histone mRNAs via BRF1-mediated degradation under physiological conditions. 31740786_BRF1 accelerates prostate tumourigenesis and perturbs immune infiltration. 31781337_Recent studies have demonstrated that Brf1 is overexpressed in most ER+ (estrogen receptor positive) cases of breast cancer and the change in cellular levels of Brf1 reflects the therapeutic efficacy and prognosis of this disease. It suggests that Brf1 may be a potential diagnosis biomarker and a therapeutic target of alcohol-associated breast cancer. 32115405_The transcription factor Sp1 modulates RNA polymerase III gene transcription by controlling BRF1 and GTF3C2 expression in human cells. 32198086_Runx2 mediates the deregulation of Brf1 and Pol III genes and its abnormal expression predicts the worse prognosis of breast cancer. 32896090_Expanding the phenotype of cerebellar-facial-dental syndrome: Two siblings with a novel variant in BRF1. 33645901_Cerebellofaciodental syndrome in an adult patient: Expanding the phenotypic and natural history characteristics. 33925358_Comprehensive Analysis of Prognostic and Genetic Signatures for General Transcription Factor III (GTF3) in Clinical Colorectal Cancer Patients Using Bioinformatics Approaches. 34928935_tRNA biogenesis and specific aminoacyl-tRNA synthetases regulate senescence stability under the control of mTOR. | ENSMUSG00000011158 | Brf1 | 2941.87897 | 1.0012264 | 0.0017682784 | 0.09784942 | 3.284599e-04 | 9.855404e-01 | 9.998360e-01 | No | Yes | 3085.40909 | 330.018276 | 2.826026e+03 | 234.309838 | |
| ENSG00000185189 | 340371 | NRBP2 | protein_coding | Q9NSY0 | FUNCTION: May regulate apoptosis of neural progenitor cells during their differentiation. {ECO:0000250}. | Alternative splicing;Cytoplasm;Neurogenesis;Phosphoprotein;Reference proteome | hsa:340371; | cytoplasm [GO:0005737]; endomembrane system [GO:0012505]; ATP binding [GO:0005524]; protein serine/threonine kinase activity [GO:0004674]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; intracellular signal transduction [GO:0035556]; negative regulation of macroautophagy [GO:0016242]; negative regulation of neuron apoptotic process [GO:0043524]; neuron differentiation [GO:0030182]; protein phosphorylation [GO:0006468] | 18619852_Data show that NRBP2 protein levels increase as neural progenitor cells differentiate, and its down regulation renders neural progenitor cells more vulnerable to apoptosis, which suggest a role in stem cell survival. 31009519_The altered expression of NRBP2 and CALCOCO2 is associated with left ventricular dysfunction parameters in human dilated cardiomyopathy. | ENSMUSG00000075590 | Nrbp2 | 294.73237 | 0.9422556 | -0.0858096612 | 0.20199821 | 1.717745e-01 | 6.785395e-01 | 9.998360e-01 | No | Yes | 244.84954 | 55.133378 | 2.292979e+02 | 39.911976 | ||
| ENSG00000185453 | 374920 | ZSWIM9 | protein_coding | Q86XI8 | Alternative splicing;Isopeptide bond;Reference proteome;Ubl conjugation | hsa:374920; | hematopoietic progenitor cell differentiation [GO:0002244] | 19773279_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000070814 | Zswim9 | 484.23427 | 1.0364859 | 0.0517004253 | 0.18894667 | 7.296665e-02 | 7.870653e-01 | 9.998360e-01 | No | Yes | 462.13328 | 59.175631 | 4.165972e+02 | 41.972132 | |||
| ENSG00000185684 | 347918 | EP400P1 | transcribed_unprocessed_pseudogene | Q6ZTU2 | Alternative splicing;Reference proteome | 693.57942 | 0.9024771 | -0.1480377979 | 0.14648227 | 1.018869e+00 | 3.127875e-01 | 9.998360e-01 | No | Yes | 917.63639 | 108.713649 | 9.433320e+02 | 86.805151 | ||||||||
| ENSG00000185760 | 56479 | KCNQ5 | protein_coding | Q9NR82 | FUNCTION: Associates with KCNQ3 to form a potassium channel which contributes to M-type current, a slowly activating and deactivating potassium conductance which plays a critical role in determining the subthreshold electrical excitability of neurons. Therefore, it is important in the regulation of neuronal excitability. May contribute, with other potassium channels, to the molecular diversity of a heterogeneous population of M-channels, varying in kinetic and pharmacological properties, which underlie this physiologically important current. Insensitive to tetraethylammonium, but inhibited by barium, linopirdine and XE991. Activated by niflumic acid and the anticonvulsant retigabine. As the native M-channel, the potassium channel composed of KCNQ3 and KCNQ5 is also suppressed by activation of the muscarinic acetylcholine receptor CHRM1. {ECO:0000269|PubMed:10787416, ECO:0000269|PubMed:11159685, ECO:0000269|PubMed:28669405}. | 3D-structure;Alternative splicing;Cell membrane;Disease variant;Ion channel;Ion transport;Membrane;Mental retardation;Phosphoprotein;Potassium;Potassium channel;Potassium transport;Reference proteome;Transmembrane;Transmembrane helix;Transport;Voltage-gated channel | This gene is a member of the KCNQ potassium channel gene family that is differentially expressed in subregions of the brain and in skeletal muscle. The protein encoded by this gene yields currents that activate slowly with depolarization and can form heteromeric channels with the protein encoded by the KCNQ3 gene. Currents expressed from this protein have voltage dependences and inhibitor sensitivities in common with M-currents. They are also inhibited by M1 muscarinic receptor activation. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2009]. | hsa:56479; | clathrin coat [GO:0030118]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; voltage-gated potassium channel complex [GO:0008076]; calmodulin binding [GO:0005516]; delayed rectifier potassium channel activity [GO:0005251]; voltage-gated potassium channel activity [GO:0005249]; potassium ion transmembrane transport [GO:0071805]; regulation of ion transmembrane transport [GO:0034765] | 15304482_Src associates with KCNQ2-5 subunits but phosphorylates only KCNQ3-5. 17237198_In conclusion, this work demonstrates that inactivation is a key regulatory mechanism of Kv7.4 and Kv7.5 channels. 18786918_among the allowed assembly conformations are KCNQ3/4 and KCNQ4/5 heteromers. 19910673_While KCNE1 slows activation and suppresses inward rectification, KCNE3 drastically inhibits KCNQ5 currents. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21730298_Data show that KCNQ1 mRNA expression was increased and KCNQ5 decreased in the preterm preeclamptic women. 22190306_The results of this study indicated that Kv7.5 contributes to the spatial regulation of KCNE3. 22347474_characterized the cell-type specific spatial organization of the kcnq5 gene locus mediated by CTCF in detail using chromosome conformation capture (3C) and 3C-derived techniques 24297175_Differential protein kinase C-dependent modulation of Kv7.4 and Kv7.5 subunits of vascular Kv7 channels. 24855057_Kv7.1/Kv7.5 form heterotetrameric channels increasing the diversity of structures which fine-tune blood vessel reactivity. The lipid raft localization of Kv7.1/Kv7.5 heteromers provides efficient spatial and temporal regulation of smooth muscle function. 25127363_rs9351963 in KCNQ5 is a possible predictive factor of incidence of diarrhea in cancer patients treated with irinotecan chemotherapy. 25672891_suggestive loci for periodontitis: KCNQ5 on chromosome 6q13 in a Japanese population. study should contribute to further understanding of genetic factors for enhanced susceptibility to periodontitis. 26969140_Tannic acid activates Kv7.4 and Kv7.3/7.5 K(+) channels resulting in vasodilation. 28283479_These findings provide the first evidence linking PKC activation to suppression of Kv7 currents, membrane depolarization, and Ca(2+) influx via L-type voltage-sensitive Ca(2+) channels as a mechanism for histamine-induced bronchoconstriction. 28669405_both loss-of-function and gain-of-function KCNQ5 mutations, associated with increased excitability and decreased repolarization reserve, lead to pathophysiology. 28884119_Our data support the involvement of KCNQ5 gene polymorphisms in the genetic susceptibility to high myopia and further exploration of KCNQ5 as a risk factor for high myopia. 29325454_identified P2RX2, KCNQ5, ERBB3 and SOCS3 to be associated with the progression of age-related hearing impairment 29748663_Phylogenetic analysis, electrostatic potential mapping, in silico docking, electrophysiology, and radioligand binding assays reveal that the anticonvulsant binding pocket evolved to accommodate endogenous neurotransmitters including gamma-aminobutyric acid, which directly activates KCNQ5 and KCNQ3 via W265. 30061510_Phosphorylation of S53 on the amino terminus of Kv7.5 is essential for protein kinase A-dependent enhancement of channel activity in response to beta adrenergic receptor activation in vascular and airway smooth muscle cells. 31871302_Distinct functional domains were identified that confer differential sensitivities of Kv7.5 and Kv7.4 to stimulatory and inhibitory signaling. 32825637_Remodeling of Kv7.1 and Kv7.5 Expression in Vascular Tumors. | ENSMUSG00000028033 | Kcnq5 | 233.40563 | 1.1352528 | 0.1830135982 | 0.21565791 | 6.997908e-01 | 4.028540e-01 | 9.998360e-01 | No | Yes | 291.60649 | 50.724244 | 2.539917e+02 | 34.276723 | |
| ENSG00000185986 | 728609 | SDHAP3 | transcribed_unprocessed_pseudogene | 377.78613 | 0.9982074 | -0.0025885616 | 0.18887103 | 1.878611e-04 | 9.890643e-01 | 9.998360e-01 | No | Yes | 331.70388 | 70.024753 | 2.918985e+02 | 47.637456 | ||||||||||
| ENSG00000186020 | 57711 | ZNF529 | protein_coding | Q6P280 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:57711; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 199.71605 | 1.0519978 | 0.0731317000 | 0.24420130 | 9.357096e-02 | 7.596856e-01 | 9.998360e-01 | No | Yes | 181.65586 | 35.773462 | 1.704634e+02 | 25.958304 | |||||
| ENSG00000186073 | 84529 | CDIN1 | protein_coding | Q9Y2V0 | FUNCTION: Plays a role in erythroid cell differentiation. {ECO:0000269|PubMed:31191338}. | Alternative splicing;Congenital dyserythropoietic anemia;Cytoplasm;Disease variant;Hereditary hemolytic anemia;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a protein with two predicted helix-turn-helix domains. Mutations in this gene were found in families with congenital dyserythropoietic anemia type Ib. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Mar 2014]. | hsa:84529; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; erythrocyte differentiation [GO:0030218] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 29031773_Mutation in C!%ORF41 gene is associated with congenital dyserythropoietic anemia. 32239177_A complex comprising C15ORF41 and Codanin-1: the products of two genes mutated in congenital dyserythropoietic anaemia type I (CDA-I). 32293259_Characterization of the interactions between Codanin-1 and C15Orf41, two proteins implicated in congenital dyserythropoietic anemia type I disease. 32518175_Genetic and functional insights into CDA-I prevalence and pathogenesis. 33159567_Congenital dyserythropoietic anemia types Ib, II, and III: novel variants in the CDIN1 gene and functional study of a novel variant in the KIF23 gene. | ENSMUSG00000040282 | Cdin1 | 132.25757 | 1.0021433 | 0.0030888120 | 0.28072699 | 1.205763e-04 | 9.912388e-01 | 9.998360e-01 | No | Yes | 140.77664 | 26.485788 | 1.383500e+02 | 20.241395 | |
| ENSG00000186792 | 8372 | HYAL3 | protein_coding | O43820 | FUNCTION: Facilitates sperm penetration into the layer of cumulus cells surrounding the egg by digesting hyaluronic acid. Involved in induction of the acrosome reaction in the sperm. Involved in follicular atresia, the breakdown of immature ovarian follicles that are not selected to ovulate. Induces ovarian granulosa cell apoptosis, possibly via apoptotic signaling pathway involving CASP8 and CASP3 activation, and poly(ADP-ribose) polymerase (PARP) cleavage. Has no hyaluronidase activity in embryonic fibroblasts in vitro. Has no hyaluronidase activity in granulosa cells in vitro. {ECO:0000250|UniProtKB:Q8VEI3}. | Alternative splicing;Cell membrane;Cytoplasmic vesicle;Disulfide bond;EGF-like domain;Endoplasmic reticulum;Endosome;Fertilization;Glycoprotein;Glycosidase;Hydrolase;Membrane;Reference proteome;Secreted;Signal | This gene encodes a member of the hyaluronidase family. Hyaluronidases are endoglycosidase enzymes that degrade hyaluronan, one of the major glycosaminoglycans of the extracellular matrix. The regulated turnover of hyaluronan plays a critical role in many biological processes including cell proliferation, migration and differentiation. The encoded protein may also play an important role in sperm function. This gene is one of several related genes in a region of chromosome 3p21.3 associated with tumor suppression, and the expression of specific transcript variants may be indicative of tumor status. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene, and some isoforms may lack hyaluronidase activity. This gene overlaps and is on the same strand as N-acetyltransferase 6 (GCN5-related), and some transcripts of each gene share a portion of the first exon. [provided by RefSeq, Jan 2011]. | hsa:8372; | acrosomal membrane [GO:0002080]; acrosomal vesicle [GO:0001669]; cytoplasmic vesicle [GO:0031410]; early endosome [GO:0005769]; endoplasmic reticulum [GO:0005783]; extracellular region [GO:0005576]; lysosome [GO:0005764]; plasma membrane [GO:0005886]; sperm midpiece [GO:0097225]; hyaluronoglucuronidase activity [GO:0033906]; hyalurononglucosaminidase activity [GO:0004415]; carbohydrate metabolic process [GO:0005975]; cartilage development [GO:0051216]; cellular response to interleukin-1 [GO:0071347]; cellular response to tumor necrosis factor [GO:0071356]; cellular response to UV-B [GO:0071493]; hyaluronan catabolic process [GO:0030214]; inflammatory response [GO:0006954]; negative regulation of ovarian follicle development [GO:2000355]; ovarian follicle atresia [GO:0001552]; penetration of zona pellucida [GO:0007341]; positive regulation of acrosomal vesicle exocytosis [GO:2000368]; response to antibiotic [GO:0046677]; response to virus [GO:0009615] | 11929860_Characterization of the murine hyaluronidase gene region reveals complex organization and cotranscription of Hyal1 with downstream genes, Fus2 and Hyal3. 12084718_Alternative mRNA splicing controls cellular expression of enzymatically active hyaluronidase. 12684632_Down-regulation of HYAL3 is associated with small cell lung cancer and glioma 20586096_Hyal3 null sperm showed delayed cumulus penetration and reduced acrosomal exocytosis. 20849445_Nodular basal cell carcinoma is associated with increased levels of hyaluronic acid concomitant with upregulation of gene expression of HAS3, HYAL3 and RHAMM, when compared with normal adjacent skin. 20849597_Overexpression of HYAL3 is associated with colorectal cancer. 22960332_polymorphisms in HYAL3 are potentially involved in glaucomatous neurodegeneration. 23549009_mutations of the HYAL3 gene are rare in Chinese lung squamous cell carcinoma patients and might contribute to lymph node metastasis 25138066_the relative expression of hyalurosome (CD44, HAS3, HB-EGF) genes was found to be reduced in patients prior to topical treatment and to be notably increased following treatment. 31758257_Expression and activity of hyaluronidases HYAL-1, HYAL-2 and HYAL-3 in the human intervertebral disc. | ENSMUSG00000036091 | Hyal3 | 721.07864 | 0.9616308 | -0.0564450516 | 0.13492532 | 1.770479e-01 | 6.739226e-01 | 9.998360e-01 | No | Yes | 786.15430 | 101.141633 | 7.389061e+02 | 74.229559 | |
| ENSG00000186951 | 5465 | PPARA | protein_coding | Q07869 | FUNCTION: Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety. Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. May be required for the propagation of clock information to metabolic pathways regulated by PER2. {ECO:0000269|PubMed:10195690, ECO:0000269|PubMed:24043310, ECO:0000269|PubMed:7629123, ECO:0000269|PubMed:7684926, ECO:0000269|PubMed:9556573}. | 3D-structure;Activator;Alternative splicing;Biological rhythms;DNA-binding;Lipid-binding;Metal-binding;Nucleus;Receptor;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | Peroxisome proliferators include hypolipidemic drugs, herbicides, leukotriene antagonists, and plasticizers; this term arises because they induce an increase in the size and number of peroxisomes. Peroxisomes are subcellular organelles found in plants and animals that contain enzymes for respiration and for cholesterol and lipid metabolism. The action of peroxisome proliferators is thought to be mediated via specific receptors, called PPARs, which belong to the steroid hormone receptor superfamily. PPARs affect the expression of target genes involved in cell proliferation, cell differentiation and in immune and inflammation responses. Three closely related subtypes (alpha, beta/delta, and gamma) have been identified. This gene encodes the subtype PPAR-alpha, which is a nuclear transcription factor. Multiple alternatively spliced transcript variants have been described for this gene, although the full-length nature of only two has been determined. [provided by RefSeq, Jul 2008]. | hsa:5465; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; lipid binding [GO:0008289]; MDM2/MDM4 family protein binding [GO:0097371]; NFAT protein binding [GO:0051525]; nuclear receptor activity [GO:0004879]; phosphatase binding [GO:0019902]; protein domain specific binding [GO:0019904]; protein-containing complex binding [GO:0044877]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific DNA binding [GO:0043565]; steroid hormone receptor activity [GO:0003707]; transcription coactivator binding [GO:0001223]; ubiquitin conjugating enzyme binding [GO:0031624]; zinc ion binding [GO:0008270]; behavioral response to nicotine [GO:0035095]; cell differentiation [GO:0030154]; cellular response to starvation [GO:0009267]; circadian regulation of gene expression [GO:0032922]; enamel mineralization [GO:0070166]; epidermis development [GO:0008544]; fatty acid metabolic process [GO:0006631]; heart development [GO:0007507]; hormone-mediated signaling pathway [GO:0009755]; lipid localization [GO:0010876]; lipoprotein metabolic process [GO:0042157]; negative regulation of appetite [GO:0032099]; negative regulation of blood pressure [GO:0045776]; negative regulation of cell growth involved in cardiac muscle cell development [GO:0061052]; negative regulation of cholesterol storage [GO:0010887]; negative regulation of cytokine production involved in inflammatory response [GO:1900016]; negative regulation of glycolytic process [GO:0045820]; negative regulation of hepatocyte apoptotic process [GO:1903944]; negative regulation of inflammatory response [GO:0050728]; negative regulation of leukocyte cell-cell adhesion [GO:1903038]; negative regulation of macrophage derived foam cell differentiation [GO:0010745]; negative regulation of neuron death [GO:1901215]; negative regulation of pri-miRNA transcription by RNA polymerase II [GO:1902894]; negative regulation of protein binding [GO:0032091]; negative regulation of protein kinase B signaling [GO:0051898]; negative regulation of reactive oxygen species biosynthetic process [GO:1903427]; negative regulation of sequestering of triglyceride [GO:0010891]; negative regulation of signaling receptor activity [GO:2000272]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transforming growth factor beta receptor signaling pathway [GO:0030512]; positive regulation of ATP biosynthetic process [GO:2001171]; positive regulation of fatty acid beta-oxidation [GO:0032000]; positive regulation of fatty acid metabolic process [GO:0045923]; positive regulation of fatty acid oxidation [GO:0046321]; positive regulation of gluconeogenesis [GO:0045722]; positive regulation of lipid biosynthetic process [GO:0046889]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of viral genome replication [GO:0045070]; regulation of cellular ketone metabolic process [GO:0010565]; regulation of circadian rhythm [GO:0042752]; regulation of fatty acid metabolic process [GO:0019217]; regulation of fatty acid transport [GO:2000191]; regulation of lipid metabolic process [GO:0019216]; response to ethanol [GO:0045471]; response to hypoxia [GO:0001666]; response to insulin [GO:0032868]; response to lipid [GO:0033993]; wound healing [GO:0042060] | 11119019_Observational study of gene-disease association. (HuGE Navigator) 11409711_Observational study of gene-disease association. (HuGE Navigator) 11466580_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11668221_Observational study of gene-disease association. (HuGE Navigator) 11840500_polymorphism is unrelated to schizophrenia or alcoholism. 11845213_crystal structure of a ternary complex containing the peroxisome proliferator-activated receptor-alpha ligand-binding domain bound to the antagonist GW6471 and a SMRT co-repressor motif 11852057_Peroxisome proliferator-activated receptor (PPARalpha and PPARgamma) agonists decrease lipoprotein lipase secretion and glycated LDL uptake by human macrophages. 11864924_Observational study of gene-disease association. (HuGE Navigator) 11864924_Variation in the PPAR gene influences human left ventricular growth in response to exercise and hypertension 11897617_cPLA(2) plays a critical role in PPAR-mediated gene transcription in HepG2 cells 11897821_A Val227Ala polymorphism in the peroxisome proliferator activated receptor alpha (PPARalpha) gene is associated with variations in serum lipid levels. 11914252_Observational study of gene-disease association. (HuGE Navigator) 11934839_activation of PPARalpha in human CD4-positive T cells limits the expression of proinflammatory cytokines, such as IFNgamma 11940516_The results suggest that homocysteine may enhance vascular constrictive remodeling by inactivating PPAR-alpha and -gamma in ECs and PPAR-gamma in SMCs. 11980898_differential regulation of vascular endothelial growth factor expression in bladder cancer cells (peroxisome proliferative activated receptor, beta) 11981036_the organization of the 5'-flanking and untranslated region of the hPPARalpha gene was characterized and the hPPARalpha promoter region has been identified 12006394_Observational study of gene-disease association. (HuGE Navigator) 12006394_PPARA L162V polymorphism is associated with increase in plasma LDL cholesterol levels. 12023905_PPARalpha binds to apolipoprotein a enhancers and influences estrogen-mediated transcription 12042669_Observational study of gene-environment interaction. (HuGE Navigator) 12048138_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12055195_regulation of human ASBT gene by PPARalpha 12118000_turnover is predicted by the ubiquitin-proteasome system which controls the ligand-induced expression level of its target genes 12118038_redundancy in the functions of PPARs alpha and delta as transcriptional regulators of fatty acid homeostasis and suggest that in skeletal muscle high levels of the delta-subtype can compensate for deficiency of PPAR alpha 12161442_PPARalpha is regulated by fatty acids in human cells 12163133_PPARalpha binding characteristics by FRET. 12203367_Our results established the presence of PPARalpha in human breast cancer cell lines and showed for the first time that activation of PPARalpha in human breast cancer cells promoted proliferation. 12395215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12408750_5'-flanking region of this gene is transcriptionally active and binds PPARalpha, we characterized the peroxisome proliferator-responsive element in the proximal promoter of the CPT II gene, which appears to be a novel PPRE. 12468272_Observational study of gene-disease association. (HuGE Navigator) 12468272_PPARA gene is a modifier of the familial combined hyperlipidemia phenotype 12544508_Observational study of gene-environment interaction. (HuGE Navigator) 12554753_These results provide molecular evidence for a cross-talk between the FXR and PPARalpha pathways in humans. 12594295_The PPAR alpha subtype localizes to the nuclear and perinuclear regions of human airway smooth muscle cells. 12594814_Expression of peroxisome proliferator-activated receptors (PPARs) in human urinary bladder carcinoma and growth inhibition by its agonists. 12615366_TR2 and TR4 can have distinct functions. Existence of differential and bi-directional regulation between PPAR alpha and TR2/TR4. Possible roles in PPAR alpha signaling pathway in human keratinocytes. 12655356_decrease in cardiac PPARalpha transcription factor gene expression observed in the failing human heart could play an important role in a reduction in fatty acid utilisation by the adult heart during cardiac hypertrophy 12709436_PPARalpha regulates the human apolipoprotein AV gene 12745064_Molecular characterisation of six alternatively spliced variants and a novel promoter in peroxisome proliferator-activated receptor alpha. 12810707_PPARA has a role as an important modulator of the metabolism of endobiotics and xenobiotics in human hepatocytes 12835617_Observational study of gene-disease association. (HuGE Navigator) 12835617_There seems to be an association between the polymorphism of the PPARA gene and decreased fasting serum triglyceride levels in glucose tolerant subjects. 12847522_These results identify hepatic activation of peroxisome proliferator-activated receptor-alpha as a mechanism underlying glucocorticoid-induced insulin resistance. 12855749_Observational study of gene-disease association. (HuGE Navigator) 12855749_peroxisome proliferator-activated receptor alpha V162 allele is associated with reduced adiposity 12897377_ARA70, a coactivator of the androgen receptor and PPARgamma, was identified as a ligand-enhanced coactivator of peroxisome proliferator-activated receptor alpha in a prostate cancer cell line. 12914524_PPARalpha/RXRalpha complex was counteracted by the expression of ERRalpha in HeLa cells. 12938026_Observational study of gene-disease association. (HuGE Navigator) 14515181_agonists of PPAR alpha increased basal and insulin-stimulated PAI-1 antigen release with a mechanism involving gene transcription and requiring signaling through the ERK1/2 signaling pathway 14519597_In humans skeletal muscle PPARalpha expression and genes regulating lipid metabolism are tightly linked, but there was no association between both insulin sensitivity and BMI with PPARalpha expression in skeletal muscle. 14523052_PPARalpha transcription factor as a major factor governing hepatic CoA levels by specific modulation of PANK1alpha gene expression 14633846_Increases in PGC-1 and PPAR-alpha levels may play an important role in changes in muscle mitochondria content, oxidative phenotype, and sensitivity to insulin induced by endurance training. 14641801_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14671555_Observational study of gene-disease association. (HuGE Navigator) 14677049_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14730379_In type 2 diabetes there is a significant gene to gene interaction between the Ala55Val polymorphism in the uncoupling protein 2 gene ( UCP2) and the 161C>T polymorphism in the exon 6 of ppargamma. 14764586_chronic treatment with the PPARalpha agonist fully prevents the acute phase response gene expression in wild-type but not in PPARalpha-deficient mice 15001458_PPARalpha protects EC from glucose-mediated monocyte adhesion, in part through regulation of IL-6 production. 15001550_Only SRC-2 serves as a true coactivator for PPARdelta, whereas all SRC members could enhance PPARalpha-induced transcriptional activation. 15051727_PPARalpha and CITED2 proteins may participate in signaling cascades of hypoxic response and angiogenesis 15067378_PPARalpha and HNF-4 competitively affect the human angiotensinogen promoter through the C region 15083308_Observational study of gene-disease association. (HuGE Navigator) 15083308_Our findings argue against a significant contribution of PPARalpha variations to the genetic basis of psoriasis. 15111510_Observational study of gene-disease association. (HuGE Navigator) 15128052_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15131257_PPARalpha S179A-S230A protein displays an impaired ligand-induced transactivation activity and an enhanced trans-repression activity 15196699_Genistein activated transcriptional activity of PPARalpha in fatty acid catabolism. 15199365_Observational study of gene-disease association. (HuGE Navigator) 15199365_data suggest that the PPARalpha polymorphism L162V might protect against the development of atherosclerosis or coronary heart disease in patients with diabetes mellitus type 2 15231516_The antiinflammatory effects of fish oil may result from the inhibitory effects of oxidized omega-3 fatty acids on NF-kappaB activation via a PPARalpha-dependent pathway. 15258199_BF may enhance the capacity of human hepatocytes to direct PC into bile canaliculi via PPARalpha-mediated redistribution of ABCB4 to the canalicular membrane 15309680_Observational study of gene-disease association. (HuGE Navigator) 15318936_PPAR-alpha and PPAR-gamma ligands induced apoptotic and antiproliferative responses in human breast cancer cell lines, respectively, which were associated with specific changes in gene expression. 15456881_Data suggest that ERRalpha serves as a nodal point in the regulatory circuitry downstream of PGC-1alpha to direct the transcription of genes involved in mitochondrial energy-producing pathways in cardiac and skeletal muscle. 15497675_This paper focuses on the functions of PPAR-alpha in fatty acid beta-oxidation, lipid metabolism, and vascular inflammation. PPAR-alpha genetics, the clinical use of PPAR-alpha activators and their future perspective are also discussed. 15500444_CLOCK plays an important role in lipid homoeostasis by regulating the transcription of a key protein, PPARalpha 15521013_new cytochrome P450 1A1 induction pathway involving peroxisome proliferator-activated receptor-alpha and 2 peroxisome proliferator response element sites. 15539630_PPAR-alpha activation increases intracellular concentrations of ROS, through the activation of NADPH oxidase. 15549499_Observational study of genotype prevalence. (HuGE Navigator) 15608561_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15608561_Response to procetofen is associated with PPARA in subjects with type 2 diabetes. 15642120_Activation of both PPARgamma and PPARalpha may contribute to the improvement in insulin sensitivity 15677519_Observational study of gene-disease association. (HuGE Navigator) 15677519_PPARalpha gene intron variants influence age at diagnosis of typw 2 diabetes. 15685545_PPARalpha is one mechanism underlying the pathogenesis of hepatitis C infection. 15699244_Observational study of gene-environment interaction. (HuGE Navigator) 15699916_Observational study of gene-disease association. (HuGE Navigator) 15735069_Effect of the L162V polymorphism on plasma TG and apoC-III concentrations depends on the dietary PUFA, with a high intake triggering lower TG in carriers of the 162V allele. 15735069_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15759454_Clinical trial of gene-disease association. (HuGE Navigator) 15797250_PPARalpha has a role in CYP4X1 regulation, and the glucocorticoid and progesterone receptors have roles in CYP4Z1 gene activation 15811118_Less likely to occur in squamous cell carcinoma and actinic keratosis than in normal skin. 15888456_the ligand-independent tight control of the position of the PPAR helix 12 provides an effective alternative for establishing an interaction with CoA proteins 15935279_Phe273Ala mutation dramatically reduced the binding affinity of ligands to PPARalpha; solvent effect may be concluded as the major source of the decrease of binding affinity and thereby of the decrease of its transcriptional activation activity 15940190_Observational study of gene-disease association. (HuGE Navigator) 16043164_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16043164_variant frequencies of ICAM1, APOE, PPARA and PAI-1 genes in coronary artery disease patients and healthy blood donors; specific arrangement of polymorphic variants which differentiate both groups 16162941_NPC1 and NPC2 mRNA depletion using small interfering RNA, abolished ABCA1-dependent cholesterol efflux induced by PPARalpha activators 16221474_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16221474_a L162V polymorphism at the peroxisome proliferator activated receptor alpha locus modulates the risk of cardiovascular events associated with insulin resistance and diabetes mellitus 16226051_Data show that estrogen receptor alpha (ERa) reduces levels of peroxisome proliferator-activated receptor alpha (PPARa) in breast cancer cell lines, and that reduction of ERa by sodium butyrate results in an increase in PPARa expression. 16239970_PPAR alpha controls vascular smooth muscle cell-cycle progression at the G1/S transition by targeting cyclin-dependent kinase inhibitor and tumor suppressor p16INK4a 16271724_a conserved functional PPAR responsive element downstream of the transcriptional start site of the human CPT1A gene is localized; this sequence is fundamental for fatty acids or PGC1-induced transcriptional activation of the CPT1A gene 16275545_Observational study of gene-disease association. (HuGE Navigator) 16285997_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16288935_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16288935_Our findings implicate PPARalpha in the lipid lowering associated with diets high in PUFA and suggests that genetic variation at the PPARA locus may determine the lipid response to changes in PUFA intake 16288986_data show that PPARalpha activation causes an increased nephrin expression by a dual action, on the one hand by stimulating nephrin promoter activity and on the other hand by reducing nephrin mRNA degradation 16297361_Observational study of gene-disease association. (HuGE Navigator) 16309557_Observational study of gene-disease association. (HuGE Navigator) 16377806_structural differences between human and mouse PPARalpha are responsible for the differential susceptibility to the peroxisome proliferator-induced hepatocarcinogenesis 16416313_Observational study of gene-disease association. (HuGE Navigator) 16492688_Thioredoxin inhibited the binding of PPARalpha to the PPAR-response element. 16506057_In conclusion, PPARalpha intron 7 G/C polymorphism was associated with physical performance in Russian athletes/ 16506057_Observational study of gene-disease association. (HuGE Navigator) 16511589_This review focuses on the mechanisms of action of PPAR alpha in metabolic diseases and their associated vascular pathologies. 16630553_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 16637234_Observational study of genotype prevalence. (HuGE Navigator) 16652134_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16763159_Observational study of gene-disease association. (HuGE Navigator) 16822823_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16847426_Observational study of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 16847426_PPARalpha-V227A is a major polymorphism in the Japanese population, and its activity may be greater compared to wild-type, but decreased by alcohol drinking. 16875506_Diminished lymphocytic expression of PPAR alpha and its activity may contribute to the inflammatory processes that are observed in cystic fibrosis. 16956579_myeloperoxidase is regulated by LXR and PPARalpha ligands 17001213_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17008383_metabolism of a parent compound, beta-carotene, may alternatively activate (9-cisRA) or inhibit (beta-apo-14'-carotenal) specific RXR and PPAR responses 17084382_In conclusion, CEOOH present in oxidized LDL increase CD36 gene expression in a pathway involving PPARalpha. 17129741_Observational study of gene-disease association. (HuGE Navigator) 17150915_Selected genes (long-chain fatty-acid-CoA ligase (FACL1), carnitine palmitoyltransferase 1A (CPT1A), adipose differentiation-related protein (ADRP) and aquaporin 3 (AQP3) were down-regulated by PPARalpha siRNA in a kidney tumor cell line. 17181634_Differential transcription occurring early in atopic dermatitis skin was indicated for CCL18, CCL13, IFNalpha2, PPARalpha and PPARgamma. 17184146_PPAR-alpha agonist fenofibrate did not significantly affect insulin sensitivity or resistin/adiponectin concentrations in obese subjects with type 2 diabetes mellitus. 17200111_Residues in PPAR alpha AF-2 transcription factor domain determine the positioning of helix 12 in the active conformation in the absence of a ligand. 17244467_The PPAR-alpha Val227Ala polymorphism is associated with non-alcoholic fatty liver disease. 17272748_Observational study of gene-disease association. (HuGE Navigator) 17300045_Observational study of gene-disease association. (HuGE Navigator) 17317762_Single nuscleotide polymorphisms of PPARA increase the risk of type 2 diabetes alone and in combination with the SNPs of other genes acting closely with PPAR-alpha. 17331954_PPARalpha transgenic mice that target constitutively activated PPARalpha specifically to hepatocytes do not develop hepatocellular carcinomas, even though they exhibit peroxisome proliferation and hepatocyte proliferation. 17342071_Combined with abdominal obesity, epistasis in the VLDL pathway (lipoprotein lipase, apolipoprotein CIII, hepatic lipase, PPARalpha, PPARgamma, and apo E genes) has a deleterious effect on fasting triglycerides and coronary artery disease risk profile 17355223_Data suggest that activator protein 2alpha and peroxisome-proliferator-activated receptor alpha may be especially involved in the ozone-inducible up-regulation mechanism of bombesin receptor subtype 3 expression. 17356846_REVIEW: PPARalpha has been shown to control transcriptional expression of key enzymes that are involved in fatty acid (FA) uptake and oxidation, triglyceride synthesis, mitochondrial respiration uncoupling, and glucose metabolism. 17363697_Transgenic mice with cardiac-restricted overexpression of PPAR alpha exhibit myocyte lipid accumulation and cardiac dysfunction. 17363837_Data suggest that PPARalpha variants may modulate the risk of cardiovascular disease by influencing both fasting and postprandial lipid concentrations. 17431031_Cytochrome P450 eicosanoids are agonists of PPAR alpha. 17436029_Formation of a ligand for the nuclear receptor PPARalpha may be one possibility by which 12R-LOX and eLOX3 contribute to epidermal differentiation. 17492134_Peroxisome proliferator-activated receptor alpha/gamma ligands can down-regulate iNOS DNA methylation 17608096_Observational study of gene-disease association. (HuGE Navigator) 17616429_Interleukin-6 inhibits human peroxisome proliferator activated receptor alpha gene expression via CCAAT/enhancer-binding proteins in hepatocytes. 17646210_PPARalpha controls gene expression in human white adipocytes 17655842_PPAR-alpha is not involved in the early regulation of delta5 desaturase gene by simvastatin, in THP-1 cells. 17700210_Observational study of gene-disease association. (HuGE Navigator) 17705849_Observational study of gene-disease association. (HuGE Navigator) 17705849_Our results on association of PPARalpha and triglycerides in males showed a much larger effect of the V allele than previously reported in older and less healthy populations 17850927_Observational study of gene-disease association. (HuGE Navigator) 17850927_We found no significant differences in genotype or allele distributions between AD patients and controls. None of the PPAR-alpha gene variants influenced markers for AD. 17926914_Observational study of gene-disease association. (HuGE Navigator) 17951966_Finds influence of drinking, aging or exercise on TC, LDL-C and HDL-C levels in A227 carriers may be different from those in PPARA-WT subjects in Japan. 17951966_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17962186_PPARalpha ligands not only serve as PPARalpha agonists but possibly act as CAR antagonists. 17963696_Hence the anti-inflammatory effect of PPARalpha overrides the pro-inflammatory effect of AhR. 17991667_PBMC gene expression profiles reflect nutrition-related metabolic changes such as fasting and that part of the fasting-induced changes are likely regulated by PPARalpha. 17998026_PPAR-alpha mRNA expression is down-regulated in the liver of rats with chronic kidney failure and this down-regulation may play a role in the development of dyslipidemia. 18003597_Ethanol and acetaldehyde inhibited PPAR-alpha transactivation. 18004978_PPARalpha,gamma DNA methylation induced by Hcy may represent an important mechanism to explain atherosclerosis. 18061194_Observational study of gene-disease association. (HuGE Navigator) 18061194_Two PPARA SNPs, L162V and rs4253728 (intronic), are less prevalent in African-Americans than in Caucasians and in African-Americans only are associated with higher apoCIII and TG levels. 18172578_Both mouse and human melanoma cells produced more PPARalpha and PPARgamma protein compared to melanocytes. 18182682_review of role of activation of PPARalpha, -beta/delta, or -gamma or LXRs in skin physiology and cytology and disease 18184928_Hepatic PPARalpha activation may provide an explanation for telmisartan's antidyslipidemic actions observed in recent clinical trials. 18219093_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18245819_fibrates simultaneously decreased PCSK9 expression while increasing PC5/6A and furin expression, indicating a broad action of PPARalpha activation in proprotein convertase-mediated lipid homeostasis. 18255343_Dehydroepiandrosterone sulfate inhibits endothelial inflammatory processes by a PPAR-alpha mechanism. 18268046_PPAR alpha and PPAR gamma activation stimulates neoangiogenesis in umbilical veins through a VEGF-dependent mechanism. 18292238_investigation of the functional significance of the V227A substitution affecting PPARalpha hinge region; results provide first indication that defective function of a natural PPARalpha variant was due at least partially to increased corepressor binding 18313368_Current findings of PPARalpha and gamma open up the possibility of developing new therapeutic agents that modulate these nuclear receptors to control various inflammatory diseases. 18336366_Observational study of gene-disease association. (HuGE Navigator) 18388689_polymorphisms in peroxisome proliferator-activated receptors are critical susceptibility risk factors for dyslipidemias and diabetes [review] 18394939_Stable structures and electronic properties for the complexes of PPARalpha and phthalate as well as adipate esters, were investigated. 18398047_Observational study of gene-disease association. (HuGE Navigator) 18440986_Overexpression of human truncated peroxisome proliferator-activated receptor alpha induces apoptosis in HL-1 cardiomyocytes. 18518955_PPARalpha in human PBMCs regulates fatty acid and amino acid metabolism and PBMCs are a suitable model to study changes in PPARalpha activation in healthy humans. 18541586_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18541586_PPARA 3'UTR SNPs modulate the association between lipid concentrations and dietary n-6 fatty acid intake in whites and long-chain n-3 fatty acid intake in blacks. 18549840_Gene-wide variations in PPARA and TLR4 genes were associated with MI. The minor allele of the PPARA SNP, rs4253623, was associated with a higher risk of MI. 18549840_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18586686_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18586686_variants of PPARA modify the association between breast cancer and aspirin use. 18652775_Results show that a persistent upregulation of PPAR-alpha binding activity and protein expression occurred in injured cortex after traumatic brain injury, which peaked 24 - 72 hours post-injury. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18680716_FGF21 circulates in human plasma and increases by extreme fasting and PPARalpha activation. 18713766_Report activation of human PPARalpha by perfluoroalkyl acids of different functional groups and chain lengths. 18725353_Observational study of gene-disease association. (HuGE Navigator) 18725353_The results of genetic variants analysis revealed that L162V PPARalpha polymorphism would not be present among Senegalese black population, and consequently, should not be involved in diabetes onset. 18726867_L162V SNP does not have a strong impact on the pathogenesis of type 2 diabetes or obesity 18726867_Observational study of gene-disease association. (HuGE Navigator) 18727927_PAR-5359, a well-balanced PPARalpha/gamma dual agonist, exhibits equivalent antidiabetic and hypolipidemic activities in vitro and in vivo. 18786524_Observational study of gene-disease association. (HuGE Navigator) 18786524_PPAR-alpha and PGC-1A polymorphisms are associated with alcohol consumption in the Mediterranean population 18787507_Observational study of gene-disease association. (HuGE Navigator) 18797151_lower transactivity of PPAR delta for arachidonic acid in Caco-2 cells, compared with PPAR alpha, is associated with the binding activity of p300 to the receptor 18848838_stimulation of PPARalpha in human macrophages might reduce arterial inflammation through differential regulation of the Trx-1 and VDUP-1 gene expression 18853997_Observational study of gene-disease association. (HuGE Navigator) 18853997_PPAR-alpha val227ala polymorphism may be involved in the pathogenesis of non-alcoholic fatty liver disease and play a protective role in obesity. 18855529_Interaction between PPARA genotype and beta-blocker treatment influences clinical outcomes following acute coronary syndromes. 18855529_Observational study of gene-disease association. (HuGE Navigator) 18955051_Endogenous PPARalpha regulates PDZK1 expression. 18977277_Observational study of gene-disease association. (HuGE Navigator) 18981151_the response of the pleural mesothelium to LTB4 is the result of a balance between the activation of receptors for LTB4 with a proinflammatory outcome (BLT2) and the activation of a different receptor for LTB4 with an anti-inflammatory outcome (PPAR). 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19070893_Term human labour is associated with changes in expression and activity of PPAR isoforms and its transcription partner, RXRalpha. 19115207_A-FABP is a candidate progression marker of human transitional cell carcinoma of the bladder that is differentially regulated by PPAR in urothelial cancer cells 19118026_PPARgamma will preferentially bind with retinoid X receptor alpha and signal antiproliferative, antiangiogenic, and prodifferentiation pathways in several tissue types, thus making it a highly useful target for down-regulation of carcinogenesis 19119483_Peroxisome proliferator-activated receptor alpha L162V polymorphism tends to occur in HBV-induced hepatocellular carcinoma and is absent in HCV-related hepatocellular carcinoma. 19141539_The present data indicate that down-regulation of MMP and proteasome activities constitutes a novel mechanism of PPAR-induced protections against HIV-induced disruption of brain endothelial cells. 19157507_Peroxisome proliferator-activated receptor expression in effusions from ovarian carcinomais associated with poor response to chemotherapy at disease recurrence and poor survival 19208777_Observational study of gene-disease association. (HuGE Navigator) 19217440_Observational study of gene-disease association. (HuGE Navigator) 19217440_Polymorphsms may increase risk of myocardial infarct. 19223982_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19254215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19255064_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19279199_Report PPAR/RXRalpha dysregulation in preterm and infection-driven infection labor. 19357976_Dynamic Bayesian transcriptome networks to reveal PPARalpha-dependent and -independent pathways. 19362162_These findings are consistent with the presence of a functional PPAR-binding element in the promoter of the human APOA5 gene; this element is however degenerate and non-functional in the corresponding mouse Apoa5 sequence. 19367093_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19386311_Peroxisome proliferator-activated receptor-alpha has a role in obesity, diabetes and cardiovascular disease [review] 19420105_Observational study of gene-disease association. (HuGE Navigator) 19422369_P633H, a novel dual agonist at peroxisome proliferator-activated receptors alpha and gamma, with different anti-diabetic effects in db/db and KK-Ay mice. 19422653_Data indicate that a higher frequency of the PPAR alpha GG genotype may be associated with top-level endurance athletes. 19422653_Observational study of gene-disease association. (HuGE Navigator) 19433068_results indicate that human apoA-IV is regulated directly by PPARalphavia the -2979/-2967 PPRE 19451226_Transcriptional profiling demonstrated that Bcl3 activates genes involved in diverse pathways including a subset involved in cellular energy metabolism known to be regulated by PGC-1alpha, ERRalpha, and a second nuclear receptor, PPARalpha. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19458633_Peroxisome proliferator-activated receptor-alpha is a functional target of p63 in adult human keratinocytes. 19527689_PPARalpha and PPARbeta/delta do not appear to modulate the alternative differentiation of human macrophages. 19560472_Observational study of gene-disease association. (HuGE Navigator) 19628794_Shear stress induces synthetic-to-contractile phenotypic modulation in smooth muscle cells via peroxisome proliferator-activated receptor alpha/delta activations by prostacyclin released by sheared endothelial cells. 19653005_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19660836_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19681917_Meta-analysis of gene-disease association and gene-gene interaction. (HuGE Navigator) 19706994_The regulation and adjustment of PPARalpha activation underlying the balance of molecular cascades might resolve the progression of alcoholic liver diseases (Review) 19710929_results suggest that PPARalpha activation has a major impact on gene regulation in human hepatocytes. Importantly, the role of PPARalpha as master regulator of hepatic lipid metabolism is generally well-conserved between mouse and human 19733654_liver up-regulation of SREBP-1c and down-regulation of PPAR-alpha occur in obese patients, with enhancement in the SREBP-1c/PPAR-alpha ratio associated with n-3 LCPUFA depletion and IR 19748481_PPARalpha regulates the expression of human SLC25A20 via the peroxisome proliferator responsive element. 19776627_The PPARalpha L162V polymorphism may contribute to the interindividual variability in the cardiovascular disease risk factor response to n-3 PUFAs. 19780876_Identified the effect of post-transcriptional regulation of miRNA-10b on PPAR-alpha expression to influence steatosis level in hepatocytes, which may contribute to pathogenesis of non-alcoholic fatty liver disease. 19789836_Levels of mRNA and protein of peroxisome proliferator-activated receptor a, which regulates b-oxidation of fatty acid, were lower in Japanese patients infected with hepatitis C virus with steatosis than in those without steatosis 19822141_Observational study of gene-disease association. (HuGE Navigator) 19823578_CYP2J2 activates the nuclear receptor PPARalpha in vitro and in vi | ENSMUSG00000022383 | Ppara | 278.89628 | 1.0014699 | 0.0021190814 | 0.22656121 | 8.560079e-05 | 9.926180e-01 | 9.998360e-01 | No | Yes | 411.88411 | 68.816695 | 3.726131e+02 | 48.246619 | |
| ENSG00000187446 | 11261 | CHP1 | protein_coding | Q99653 | FUNCTION: Calcium-binding protein involved in different processes such as regulation of vesicular trafficking, plasma membrane Na(+)/H(+) exchanger and gene transcription. Involved in the constitutive exocytic membrane traffic. Mediates the association between microtubules and membrane-bound organelles of the endoplasmic reticulum and Golgi apparatus and is also required for the targeting and fusion of transcytotic vesicles (TCV) with the plasma membrane. Functions as an integral cofactor in cell pH regulation by controlling plasma membrane-type Na(+)/H(+) exchange activity. Affects the pH sensitivity of SLC9A1/NHE1 by increasing its sensitivity at acidic pH. Required for the stabilization and localization of SLC9A1/NHE1 at the plasma membrane. Inhibits serum- and GTPase-stimulated Na(+)/H(+) exchange. Plays a role as an inhibitor of ribosomal RNA transcription by repressing the nucleolar UBF1 transcriptional activity. May sequester UBF1 in the nucleoplasm and limit its translocation to the nucleolus. Associates to the ribosomal gene promoter. Acts as a negative regulator of the calcineurin/NFAT signaling pathway. Inhibits NFAT nuclear translocation and transcriptional activity by suppressing the calcium-dependent calcineurin phosphatase activity. Also negatively regulates the kinase activity of the apoptosis-induced kinase STK17B. Inhibits both STK17B auto- and substrate-phosphorylations in a calcium-dependent manner. {ECO:0000269|PubMed:10593895, ECO:0000269|PubMed:11350981, ECO:0000269|PubMed:15035633, ECO:0000269|PubMed:8901634}. | 3D-structure;Calcium;Cell membrane;Cytoplasm;Cytoskeleton;Disease variant;Endoplasmic reticulum;Lipoprotein;Membrane;Metal-binding;Myristate;Neurodegeneration;Nucleus;Phosphoprotein;Protein kinase inhibitor;Protein transport;Reference proteome;Repeat;Transport | This gene encodes a phosphoprotein that binds to the Na+/H+ exchanger NHE1. This protein serves as an essential cofactor which supports the physiological activity of NHE family members and may play a role in the mitogenic regulation of NHE1. The protein shares similarity with calcineurin B and calmodulin and it is also known to be an endogenous inhibitor of calcineurin activity. [provided by RefSeq, Jul 2008]. | hsa:11261; | cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; Golgi membrane [GO:0000139]; microtubule cytoskeleton [GO:0015630]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; transport vesicle [GO:0030133]; calcium ion binding [GO:0005509]; calcium-dependent protein binding [GO:0048306]; kinase binding [GO:0019900]; microtubule binding [GO:0008017]; potassium channel regulator activity [GO:0015459]; protein kinase inhibitor activity [GO:0004860]; cellular response to acidic pH [GO:0071468]; cytoplasmic microtubule organization [GO:0031122]; membrane docking [GO:0022406]; membrane fusion [GO:0061025]; membrane organization [GO:0061024]; microtubule bundle formation [GO:0001578]; negative regulation of calcineurin-NFAT signaling cascade [GO:0070885]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of phosphatase activity [GO:0010923]; negative regulation of protein autophosphorylation [GO:0031953]; negative regulation of protein import into nucleus [GO:0042308]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of protein ubiquitination [GO:0031397]; positive regulation of phospholipid biosynthetic process [GO:0071073]; positive regulation of protein glycosylation [GO:0060050]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of protein transport [GO:0051222]; positive regulation of sodium:proton antiporter activity [GO:0032417]; potassium ion transport [GO:0006813]; protein export from nucleus [GO:0006611]; protein stabilization [GO:0050821]; regulation of intracellular pH [GO:0051453]; regulation of neuron death [GO:1901214]; small GTPase mediated signal transduction [GO:0007264] | 12226101_serum-independent activation of NHE1 by bound CHP2 is one of the key mechanisms for the maintenance of high pH(i) and the resistance to serum deprivation-induced cell death in malignantly transformed cells 15035633_CHP is important for pH(i)-dependent regulation of Na+/H+ exchanger 1 (NHE1) via tightly bound Ca2+ ions which serve as a crucial structural elements required for this role. 16511206_crystallographic analysis of the human calcineurin homologous protein CHP2 bound to the cytoplasmic region of the Na+/H+ exchanger NHE1 17050540_helix formation of the cytoplasmic region of NHE1 by calcineurin B homologous protein 1 is a prerequisite for generating the active form of NHE1 18628988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18815128_CHP2 has a role in tumorigenesis and as an activator of the calcineurin/NFAT signaling pathway 19165527_Using shotgun mass spectrometry, we found this protein differentially expressed in the dorsolateral prefrontal cortex from patients with schizophrenia. 21085126_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 30846317_GPAT4 activity requires CHP1 to be N-myristoylated, forming a key molecular interface between the two proteins, regulating glycerolipid metabolism. 32787936_Novel CHP1 mutation in autosomal-recessive cerebellar ataxia: autopsy features of two siblings. 34108458_Structure and mechanism of the human NHE1-CHP1 complex. | ENSMUSG00000014077 | Chp1 | 2385.48366 | 1.0562404 | 0.0789382654 | 0.10790589 | 5.333257e-01 | 4.652120e-01 | 9.998360e-01 | No | Yes | 2408.18064 | 258.321273 | 2.356240e+03 | 195.817035 | |
| ENSG00000187486 | 3767 | KCNJ11 | protein_coding | Q14654 | FUNCTION: This receptor is controlled by G proteins. Inward rectifier potassium channels are characterized by a greater tendency to allow potassium to flow into the cell rather than out of it. Their voltage dependence is regulated by the concentration of extracellular potassium; as external potassium is raised, the voltage range of the channel opening shifts to more positive voltages. The inward rectification is mainly due to the blockage of outward current by internal magnesium. Can be blocked by extracellular barium (By similarity). Subunit of ATP-sensitive potassium channels (KATP). Can form cardiac and smooth muscle-type KATP channels with ABCC9. KCNJ11 forms the channel pore while ABCC9 is required for activation and regulation. {ECO:0000250, ECO:0000269|PubMed:17855752, ECO:0000269|PubMed:28842488, ECO:0000269|PubMed:9831708}. | 3D-structure;Alternative splicing;Diabetes mellitus;Disease variant;Ion channel;Ion transport;Membrane;Phosphoprotein;Potassium;Potassium transport;Reference proteome;Transmembrane;Transmembrane helix;Transport;Voltage-gated channel | Potassium channels are present in most mammalian cells, where they participate in a wide range of physiologic responses. The protein encoded by this gene is an integral membrane protein and inward-rectifier type potassium channel. The encoded protein, which has a greater tendency to allow potassium to flow into a cell rather than out of a cell, is controlled by G-proteins and is found associated with the sulfonylurea receptor SUR. Mutations in this gene are a cause of familial persistent hyperinsulinemic hypoglycemia of infancy (PHHI), an autosomal recessive disorder characterized by unregulated insulin secretion. Defects in this gene may also contribute to autosomal dominant non-insulin-dependent diabetes mellitus type II (NIDDM), transient neonatal diabetes mellitus type 3 (TNDM3), and permanent neonatal diabetes mellitus (PNDM). Multiple alternatively spliced transcript variants that encode different protein isoforms have been described for this gene. [provided by RefSeq, Oct 2009]. | hsa:3767; | integral component of plasma membrane [GO:0005887]; inward rectifying potassium channel [GO:0008282]; plasma membrane [GO:0005886]; T-tubule [GO:0030315]; ankyrin binding [GO:0030506]; ATP binding [GO:0005524]; ATP-activated inward rectifier potassium channel activity [GO:0015272]; ATPase-coupled cation transmembrane transporter activity [GO:0019829]; inward rectifier potassium channel activity [GO:0005242]; potassium ion binding [GO:0030955]; transmembrane transporter binding [GO:0044325]; voltage-gated potassium channel activity [GO:0005249]; glucose metabolic process [GO:0006006]; inorganic cation transmembrane transport [GO:0098662]; negative regulation of insulin secretion [GO:0046676]; nervous system process [GO:0050877]; potassium ion import across plasma membrane [GO:1990573]; potassium ion transmembrane transport [GO:0071805]; regulation of insulin secretion [GO:0050796]; regulation of ion transmembrane transport [GO:0034765]; regulation of membrane potential [GO:0042391]; response to ATP [GO:0033198]; response to xenobiotic stimulus [GO:0009410] | 11318841_Observational study of gene-disease association. (HuGE Navigator) 11424233_Observational study of gene-disease association. (HuGE Navigator) 11825905_Assembly limits the pharmacological complexity of ATP-sensitive potassium channels 12196481_Observational study of gene-disease association. (HuGE Navigator) 12196481_The prevalent Glu23Lys polymorphism in the potassium inward rectifier 6.2 (KIR6.2) gene is associated with impaired glucagon suppression in response to hyperglycemia. 12199344_ABCC8 (SUR1) and KCNJ11 (KIR6.2) mutations in persistent hyperinsulinemic hypoglycemia of infancy and evaluation of different therapeutic measures. May account for the different therapeutic responses. 12213829_the MDR-like core of SUR is linked with the K(IR) pore in KATP channels 12351459_the E23K mutation in the KIR6.2 gene is not associated with detectable alterations in glucose-stimulated insulin secretion 12356945_down-regulation of this channel may facilitate myometrial function during late pregnancy 12388475_Localization of ATP-sensitive K+ (KATP) channels in human skeletal muscle and the functional importance of these channels for human muscle K+ distribution at rest and during muscle activity 12540637_E23K variant is associated with type 2 diabetes. 12540637_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 12540638_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 12540638_The E23K variant associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. 12819904_No association for NIDDM susceptibility polymorphism in Kir6.2. 12860923_Binding of the alpha phosphate group of ATP to R201 then stabilizes the closed state. R50 on the N-terminus controls ATP binding by facilitating the interaction of the beta phosphate group of ATP with K185 to destabilize the open state. 12934053_In corporal smooth muscle is composed of Kir6.1-Kir6.2 construct expressed with SUR2B.K(ATP) channel in corporal smooth muscle cells is composed of heteromultimers of Kir6.1 and Kir6.2 with the ratio of 3 : 1 or 4 : 0 and SUR2B. 14514649_E23K/I337V polymorphism may have a diabetogenic effect via increased KATP channel activity in response to endogenous levels of LC-CoAs in tissues involved in the maintenance of glucose homeostasis. 14551916_Observational study of gene-disease association. (HuGE Navigator) 14656703_a compensatory increase in I(Ca) counteracts a mild activation of ATP-insensitive K(ATP) channels to maintain the action potential duration and elevate the inotropic state of transgenic hearts 14681552_binding of ATP to Kir6.2 alters the interaction between the N- and C-terminal domains 14871556_In this study of acute myocardial infarct patients, sudden cardiac death was not related to polymorphisms in the KCNJ11 gene 14871556_Observational study of gene-disease association. (HuGE Navigator) 14988278_Association of this gene's single nucleotide polymorphism with type 2 diabetes. 15111507_SUR1/Kir6.2 gene region contributes to risk of type 2 diabetes and encodes targets for hypoglycemic medications. Link between mechanism of disease and targets for pharmacological treatment. 15115830_Heterozygous activating mutations in the gene encoding Kir6.2 cause permanent neonatal diabetes and may also be associated with developmental delay, muscle weakness, and epilepsy. 15448106_Insulin-dependent patients with mutations in Kir6.2 may be managed on an oral sulfonylurea with sustained metabolic control rather than insulin injections. 15448107_Kir6.2 mutations are a common cause (53%) of permanent neonatal diabetes in Caucasians. 15504982_KCNJ11 mutations are rare in patients diagnosed with type 1 diabetes, the identification of a KCNJ11 mutation may have important treatment implications. 15562009_Mutations can yield partially functioning channels, including cases of hyperinsulinism that are fully responsive to diazoxide. 15579791_Observational study of gene-disease association. (HuGE Navigator) 15579791_Polymorphisms of SUR1 gene predicted conversion from impaired glucose tolerance to type 2 diabetes, and the effect of these polymorphisms on diabetes risk was additive with E23K polymorphism of Kir6.2 gene. 15580558_KCNJ11 mutations are a common cause of permanent neonatal diabetes mellitus either in isolation or associated with developmental delay 15583126_mutations in the slide helix of Kir6.2 (V59G) influence the channel kinetics, providing evidence that this domain is involved in Kir channel gating 15718250_mutations in KCNJ11 are the first genetic cause for remitting as well as permanent diabetes 15735229_Heterozygote mutation in a patient with severe diabetic ketoacidosis, was nevertheless able to tolerate glibenclamide despite transitory diarrhea. 15760904_the epoxyeicosatrienoic acid-Kir6.2 interaction may allosterically change the ATP binding site on Kir6.2, reducing the channel sensitivity to ATP 15797964_Amino acid substituion polymorphism increases the risk of type 2 diabetes. 15797964_Observational study of gene-disease association. (HuGE Navigator) 15838686_an R201H missense mutation, and sulfonylurea-treatable diabetes in a newborn 15842514_Meta-analysis of gene-disease association. (HuGE Navigator) 15855351_E23K variant in muscular K(ATP) channels affects systemic glucose homeostasis and poses an important risk factor for type 2 diabetes and obesity. 15930170_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15956217_Kir6.2 and INS VNTR variants may have roles in glucose homeostasis in young obese 15956217_Observational study of gene-disease association. (HuGE Navigator) 15962003_Mutations in Kir6.2 altered Kir6.2/SUR1 interactions. 15983208_Effects of side-chain length and the degree of saturation of various acyl CoAs on channel activity. 16087682_KCNJ11 mutations cause neonatal diabetes, and increase the current magnitude of heterozygous K(ATP) channels in two ways: by increasing MgATP activation and by decreasing ATP inhibition. 16142506_Observational study of gene-disease association. (HuGE Navigator) 16320083_Observational study of gene-disease association. (HuGE Navigator) 16320083_The E23K polymorphism of KCNJ11 seems to predispose to gestational diabetes mellitus (GDM) in Scandinavian women. 16332676_identification of a novel KCNJ11 mutation associated with congenital hyperinsulinism that renders a missense mutation, F55L, in the Kir6.2 protein. 16339180_The greater ATP inhibition of mutant Kir6.2/SUR2A than of Kir6.2/SUR1 can explain why gain-of-function Kir6.2 mutations manifest effects in brain and beta-cells but not in the heart. 16367885_Observational study of gene-disease association. (HuGE Navigator) 16416420_analysis of mutations in Kir6.2 (KCNJ11) and SUR1 (ABCC8), the spectrum of phenotypes, and the implications for treatment when patients are diagnosed with mutations in these genes [review] 16455067_E23K gene polymorphism in Kir6.2 gene appeared to be related to high susceptibility to coronary heart disease 16455067_Observational study of gene-disease association. (HuGE Navigator) 16595597_Observational study of gene-disease association. (HuGE Navigator) 16609879_KCNJ11 mutations are a common cause of permanent diabetes diagnosed in the first 6 months and all patients diagnosed in this age group should be tested. 16609879_Observational study of genotype prevalence. (HuGE Navigator) 16636122_inwardly rectifying potassium channel Kir6.2 mutations greatly reduce fetal insulin secretion and hence fetal growth, but this is independent of mutation severity 16670688_The severe developmental Delay, Epilepsy and Neonatal Diabetes syndrome was seen with the novel C166F KCNJ11 mutation and mild developmental delay with the V59M mutation. 16731860_Observational study of gene-disease association. (HuGE Navigator) 16732049_Neonatal hyperglycemia caused bsy an amino acid substitustion ins this protein. 16733889_Observational study of gene-disease association. (HuGE Navigator) 16873704_Observational study of gene-disease association. (HuGE Navigator) 17020404_Combining information from several known common risk polymorphisms allows the identification of population subgroups with markedly differing risks of developing type 2 diabetes compared to those obtained using single polymorphisms. 17021801_Heterozygous activating mutations in Kir6.2 (KCNJ11) are a common cause of neonatal diabetes 17047922_Sulfonylurea treatment can result in prolonged, excellent glycaemic control and may improve motor features, but not mental features, associated with KCNJ11 mutations. 17065345_R201H mutation pf the kir6.2 channel is able to cause neonatal diabetes in patients. 17137217_Observational study of gene-disease association. (HuGE Navigator) 17192350_Successful transfer off insulin to sulfonylurea is feasible in adults in neonatal diabetes due to KCNJ1-activating mutations. 17192490_Observational study of gene-disease association. (HuGE Navigator) 17213273_Mutation carriers with neonatal diabetes mellitus may be successflly transferred from insulin to sulfonylurea agents. 17257281_Observational study of gene-disease association. (HuGE Navigator) 17259376_results demonstrate that ATP-binding site mutations of KCNJ11 can cause Developmental delay, epilepsy, and neonatal diabetes (DEND) 17259403_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17259403_We conclude that the lysine variant in KCNJ11 E23K leads to diminished insulin secretion in individuals with IGT. 17296510_Kir6.2 is found in pancreatic beta-cells, brain, heart and skeletal muscle and in patients with permanent neonatal diabetes mellitus, mutations in 30 to 50% of the cases. [REVIEW] 17316607_showed, in two Italian patients, two new heterozygous mutations which result in the appearance of premature translation termination codons resulting in the premature end of Kir6.2 17327377_KCNJ11 mutations can arise either during gametogenesis or embryogenesis 17342155_Observational study of gene-disease association. (HuGE Navigator) 17342155_these data (involving >4600 subjects) provide no evidence that common variants of the KCNJ11 E23K polymorphism have a major influence on polycystic ovary syndrome susceptibility, though modest effect sizes (OR<1.25) cannot be excluded. 17378627_Observational study of gene-disease association. (HuGE Navigator) 17380317_Human fetal pancreas produces all key elements of the glucose-sensing apparatus, including KIR6.2, which may contribute to poor secretory responses in early life. 17395632_The C terminus of KIR6.2 contains F333 and is involved in more than one type of functional interaction with SUR, and that F333 interacts differentially with SUR1 and SUR2. 17431820_The result suggests that the Kir6.2/KCNJ11 gene is not related to sudden cardiac death in this family. 17446535_study has shown that mutations in the KCNJ11 and ABCC8 are a major cause of transient neonatal diabetes mellitus, accounting for 29% of all cases and 89% of non-6q24 transient neonatal diabetes mellitus 17463248_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17475937_Amino acid substitution predicts permanent neonatal diabetes. 17535866_The hyperactive Glu23Lys variant of the K(ATP) channel subunit Kir6.2 may cause defective glucose sensing in several tissues and impaired glycaemic control in children with type 1 diabetes 17570749_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17601994_Observational study of gene-disease association. (HuGE Navigator) 17635943_Both the 6q24 abnormality and KCNJ11 mutation are major causes of neonatal diabetes mellitus in Japanese patients 17652641_We screened the DNA of a 3-year-old patient with neonatal diabetes, severe developmental delay, and epilepsy for mutations. We identified a novel Kir6.2 mutation causing DEND syndrome. 17659066_most patients with neonatal diabetes caused by mutations in the KCNJ11 gene can be successfully managed with a sulfonylurea agent without the need for insulin 17720745_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17720745_the common E23K genetic variant at the KCNJ11 gene locus was significantly associated with cardiovascular function 17823772_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17823772_Significant associations between eight SNPs, including the KCNJ11 E23K and ABCC8 S1369A variants, and T2D were found. 17825556_Kir6.2/SUR2A deficiency is associated with impaired muscle function in K(+)-depleted rats and in hypoPP. 17855752_This study showed that F333 in Kir6.2 interacts functionally with SUR2A to modulate channel rundown and MgADP activation. 17890419_This article reports a girl who developed infantile spasms and early onset diabetes mellitus at the age of 3 months and revealed DEND syndrome with a heterozygous activating mutation in Kir6.2. 17894829_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17898091_KCNJ11 E23K polymorphism is associated with an increased risk ratio for type 2 diabetes. 17901525_Genetic testing enabled successful glibenclamide treatment as early as 3 months to a newborn with KCNJ11 mutation. 17919178_Both mutations reduced the sensitivity of the K(ATP) channel to inhibition by MgATP and enhanced whole-cell K(ATP) currents. In pancreatic beta cells, such an increase in the K(ATP) current would reduce insulin secretion and thereby cause diabetes. 17922473_Observational study of gene-disease association. (HuGE Navigator) 17922473_Our results report for the first time a positive association of the E23K variant with type 2 diabetes in an Arab population. 17923772_Studies reported showing activating mutations in the KCNJ11 gene, encoding the Kir6.2 subunit of the pancreatic KATP channel, in patients with permanent neonatal diabetes mellitus. 17965292_Effectiveness of sulfonylrea therapy for permanent neonatal diabetes in an adult patient carrying the G53D mutation in the KCNJ11 gene. 17965318_Kir6.2 E23K polymorphism is an independent genetic risk factor for diabetes in the general Japanese population. 17965318_Observational study of gene-disease association. (HuGE Navigator) 17976307_Carriers of the predisposing Kir6.2 E23K K allele showed no increased risk of either type of diabetes mellitus development. 17976307_Observational study of gene-disease association. (HuGE Navigator) 17977958_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17978456_a rare case of permanent neonatal diabetes due to R20IC mutation in KCNJ11 gene. 17994213_Observational study of gene-disease association. (HuGE Navigator) 17994213_PPARG and KCNJ11 variants that are known to influence individual predisposition to type 2 diabetes do not appear to have pleiotropic effect on early growth 18073297_The G53D mutation in Kir6.2 (KCNJ11) is associated with neonatal diabetes and motor dysfunction in adulthood. 18159846_This report describes two cases with PNDM due to the R201H mutation, treated with the same sulphonylurea (glipizide GITS). We conclude that to achieve normoglycemia, patients with PNDM should refrain from eating high glycemic-index products. 18162506_Observational study of genotype prevalence. (HuGE Navigator) 18162508_Observational study of gene-disease association. (HuGE Navigator) 18162508_The association of 6 loci with type 2 diabetes risk in Japanese patients is reported. 18221420_Case of an 18-month-old infant with permanent neonatal diabetes due to an activating KCNJ11 mutation who successfully transitioned from subcutaneous insulin therapy to oral sulfonylurea therapy in the outpatient setting. 18243136_The down-regulation of AK1 expression by hyperglycemia may contribute to the defective coupling of glucose metabolism to K-ATP channel activity in type 2 diabetes. 18281290_K(IR)6.2-based channels with diabetogenic receptors reveal that MgATP-dependent hyper-stimulation of mutant SUR can compromise the ability of K(ATP) channels to function as metabolic sensors 18290324_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18290324_The polymorphism in KCNJ11 is implicated in the persistent hyperglycemia hypolycemia of infancy disorder. 18335204_Mutations in this protein explain the inability of sulfonylureas to ameliorate the diabetes of affected patients. 18426861_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18498634_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18504616_Kir6.2 K23 as a risk factor for adverse subclinical myocardial remodeling. 18504616_Observational study of gene-disease association. (HuGE Navigator) 18516622_Observational study of gene-disease association. (HuGE Navigator) 18544102_a novel heterozygous mutation (c. 679C-->G and c. 680A-->T) was identified, resulting in a GAG-->CTG (E227L) substitution in KCNJ11 in a family with variable phenotypes of dominantly inherited diabetes mellitus 18556340_Case of diabetes without ketoacidosis diagnosed on the fourth day of life of a girla, with a nove R265H mutation of the KCNJ11 in her and her unaffected father. 18559200_KCNJ11 gene was implicated in gestational diabetes. 18566517_Mutations in the pore-forming K(ATP) channel subunit cause neonatal diabetes & discusses recent advances in understanding of clinical features of neonatal diabetes, its underlying molecular mechanisms & their impact on treatment[review] 18591388_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18596924_The mutation of KIR6.2 channel is recessively inherited. 18597214_Observational study of gene-disease association. (HuGE Navigator) 18598350_Observational study of gene-disease association. (HuGE Navigator) 18662362_We identified a mutation in KCNJ11 in 14 patients from 12 families with permanent neonatal diabetes mellitus. 18664331_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18664331_The KCNJ11 E23K variant was associated with the therapeutic effect of repaglinide 18710329_Observational study of gene-disease association. (HuGE Navigator) 18719881_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18752747_Underexpression of Kir6.2 decreased the proliferation and invasion of the HepG2 cells. 18758683_KCNJ11 E23K and ABCC8 exon 31 variants contribute to susceptibility to susceptibility to type 2 diabeetes, glucose intolerance and altered insulin secretion in a Russian population. 18758683_Observational study of gene-disease association. (HuGE Navigator) 18767144_Mutations of the gene in diabetes mellitus and hyperinsulinism (Review) 18796522_Observational study of gene-disease association. (HuGE Navigator) 18796522_variation marked by the Kir6.2 E23K and sulfonylurea receptor A1369S mutations is associated with alterations in glucose-stimulated insulin secretion but not with other measures of glucose homeostasis in an African-American population 18958766_Observational study of gene-disease association. (HuGE Navigator) 18958766_The rs7903146 variant of the TCF7L2 gene might influence PCOS predisposition, while no association is observed between the E23K variant of KCNJ11 and susceptibility to PCOS and related traits. 18972257_Four candidate polymorphisms in the three genes TCF7L2 (rs12255372 and rs7903146), PPARG (rs1801282), KCNJ11 (rs5219) and traditional risk factors were studied 18984664_Observational study of gene-disease association. (HuGE Navigator) 18998097_DNA is extracted from peripheral blood leukocytes and amplification of the KCNJ11 gene by the PCR, sequencing, and mutation detection are provided. 19002430_Observational study of gene-disease association. (HuGE Navigator) 19020323_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020324_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19033397_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19033397_Type 2 diabetes susceptibility of KCNJ11 was confirmed in Japanese. 19082521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19139106_Analysis of two KCNJ11 neonatal diabetes mutations, V59G and V59A, and the analogous KCNJ8 I60G substitution: differences between the channel subtypes formed with SUR1. 19139842_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19214942_Data suggest that patients with type 2 diabetes carrying the K variant of the E23K polymorphism in KCNJ11 have reduced response to sulfonylurea therapy, resulting in increased HbA(1c)and in lower risk for severe sulfonylurea-induced hypoglycemia. 19214942_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19225753_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19247372_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19254908_Recessive inactivating mutations in ABCC8 and KCNJ11 are the most common cause of CHI. 19258437_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19264985_Observational study of gene-disease association. (HuGE Navigator) 19279076_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19324937_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19336475_Observational study of gene-disease association. (HuGE Navigator) 19345438_A girl with celiac disease and KCNJ11 mutation was transferred to glibenclamide when 19.8 years old. When her compliance to the gluten free diet worsened, her metabolic control deteriorated. 19357197_Kir6.2 contains a di-acidic endoplasmic reticulum exit signal, which promotes endoplasmic reticulum exit via a process that requires Sar1. 19368707_Observational study of gene-disease association. (HuGE Navigator) 19380854_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19401414_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19435956_Tooth discoloration is a novel side effect of sulfonylurea therapy in patients with permanent neonatal diabetes due to mutations in KCNJ11. 19475716_Observational study of gene-disease association. (HuGE Navigator) 19481058_caveolin-3 negatively regulates Kir6.2/SUR2A channel function. 19491206_An amino acid substitution variant leads to overactivity of the K(ATP) channel, resulting in reduced insulin secretion and results in type 2 diabetes. 19491206_Observational study of gene-disease association. (HuGE Navigator) 19498446_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19498446_The common E23K variant of KCNJ11 is considered as a strong candidate for type 2 diabetes susceptibility across different ethnicities. 19502414_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19587354_Rare activating mutations cause neonatal diabetes, whereas the common variants, E23K in KCNJ11 and S1369A in ABCC8, are in strong linkage disequilibrium, constituting a haplotype that predisposes to type 2 diabetes 19592620_Observational study of gene-disease association. (HuGE Navigator) 19602701_Meta-analysis and HuGE review of gene-disease association. (HuGE Navigator) 19685080_Blunted heart rate response during exercise is a risk factor for mortality in patients with heart failure, establishing the clinical relevance of Kir6.2 E23K as a biomarker for impaired stress performance 19685080_Observational study of gene-disease association. (HuGE Navigator) 19692135_A patient diagnosed at the age of 12 weeks that showed islet cell antibodies at diagnosis, but not with positive result for KCNJ11 mutation. 19720844_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19766903_Observational study of gene-disease association. (HuGE Navigator) 19794065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19808892_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19862325_Observational study of gene-disease association. (HuGE Navigator) 19862325_there is an association between PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 and type 2 diabetes in the Chinese population 19876004_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19921246_tight metabolic regulation of K(ATP) (Kir6.2) activity in the pancreatic beta-cell is critical in normal excitation-secretion coupling such that abnormally elevated KATP currents suppress normal insulin release[review] 20036918_Osteoblast-like cells might contain mitoKATP channels in which Kir6.2 is the pore-forming subunit, though in extremely low abundance. 20043145_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20054294_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20054294_The polymorphism KCNJ11 Lys23Glu is associated with a heightened risk of developing type 2 diabetes mellitus in a Chinese Han population. 20075150_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 20079163_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20079163_Study replicated the association of rs5219 in KCNJ11 with type 2 diabetes in Chinese Han population in Beijing. 20161779_Observational study of gene-disease association. (HuGE Navigator) 20164212_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20203524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20215779_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20361036_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20384434_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20401705_KCNJ11 activating mutation in an Indian family with remitting and relapsing diabetes. 20424228_Observational study of gene-disease association. (HuGE Navigator) 20431170_TND has been shown to be genetically heretogenous and mutations in KCNJ11 have been shown to be associated with it. This is the first report from India describing TND due to E227K missense mutation in KCNJ11 gene 20437825_Observational study of gene-disease association. (HuGE Navigator) 20490451_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20503258_Observational study of gene-disease association. (HuGE Navigator) 20540435_every person diagnosed with diabetes before six months of life should be tested for KCNJ11 K(ATP) mutations 20540670_Observational study of gene-disease association. (HuGE Navigator) 20550665_Observational study of gene-disease association. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20595581_in mice with human Kir6.2 mutation targeted to either muscle or nerve, data show motor impairment originates in central nervous system rather than muscle or peripheral nerves;identifed motor hyperactivity as a feature of KATP channel overactivity 20597906_Observational study of gene-disease association. (HuGE Navigator) 20616309_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20682687_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20685672_Data show that ABCC8 or KCNJ11 defects were found in 82% of the CHI cases. 20712903_Observational study of gene-disease association. (HuGE Navigator) 20802253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20816152_Observational study of gene-disease association. (HuGE Navigator) 20863361_Observational study of gene-disease association. (HuGE Navigator) 20879858_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20922570_Mutations in the K(ATP) channel which result in permanent neonatal diabetes. 20923526_Meta-analysis and uncategorized study of gene-disease association. (HuGE Navigator) 20929593_Observational study of gene-disease association. (HuGE Navigator) 20980454_mutation V290M in the pore-forming Kir6.2 subunit was identified in patients with congenital hyperinsulinism 21352428_genetic basis of two Cypriot patients who developed diabetes before 6 months of age; both carried mutations of the KCNJ11 gene -one the R201H mutation and the other the R50Q mutation 21378087_Novel mutations in KCNJ11 are found in 32% of children with congenital hyperinsulism. 21418633_Down-regulation of Kir6.1 and Kir6.2 expression in myometrium may contribute to the enhanced uterine contractility associated with the onset of labour. 21422196_Mutations in ABCC8 and KCNJ11 are the most common causes of congenital hyperinsulinism in Korean patients 21540348_A novel heterozygous mutation, W68R, is found in the Kir6.2 subunit of the ATP-sensitive potassium (KATP) channel, in a patient with transient neonatal diabetes. 21544516_Data ound mutations in KCNJ11, INS and ABCC8 and GCK genes in permanent diabetes mellitus with onset in the first 12 months of age. 21573802_Meta-analysis verified that single nucleotide polymorphisms of KCNJ11 gene are significantly associated with the risk of type 2 diabetes mellitus in East Asian populations. 21674179_The phenotype associated with dominant ABCC8/KCNJ11 mutations ranges from asymptomatic macrosomia to persistent hyperinsulinaemic hypoglycaemia in childhood. 21682153_identified presence of the de nova V59M and E322K activating mutations in the KCNJ11 gene in two children with neonatal diabetes mellitus 21710463_findings provided evidence that the KCNJ11 gene plays a role in the pathogenesis of decreased insulin sensitivity in essential hypertension patients 21765448_Four common A190A, E23K, I337V and 3'UTR +62 G/A polymorphisms were found in KCNJ11. 21820692_Osteogenic differentiation strongly up-regulated Kir6.2 mRNA whereas Kir6.1 showed no significant change in expression. 21871684_Brief Report: 18-month follow-up of switching from insulin to sulfonylurea in a mother and daughter both carrying KCNJ11 gene activating mutation for permanent neonatal diabetes mellitus. 21981029_mutations in KCNJ11 or ABCC8 genes were identified in 197 patients diagnosed with diabetes before 6 months (48.6%), 3 infants diagnosed between 6 and 9 months (4.2%) and none diagnosed after 9 months; K(ATP) channel mutations are an uncommon cause of diabetes in infants presenting after 6 months 22082043_KCNJ11 polymorphism is associated with type 2 diabetes. 22133355_post-transcriptional events determine Kir6.2 protein expression in the left ventricle of patients with severe mitral dysfunction and low venous PO(2) 22163043_the KCNJ11 E23K variant has a role in glycaemic progression in Chinese, with its effect being more evident in the early stage of T2DM, as the subjects progressed from normal glucose tolerance to prediabetes 22264780_study found a significant association between a common KCNJ11 SNP and the risk of developing new-onset diabetes after transplantation among heart and kidney transplanted patients treated with tacrolimus 22308858_As a r | ENSMUSG00000096146 | Kcnj11 | 289.23967 | 1.1029108 | 0.1413161220 | 0.18889874 | 5.716199e-01 | 4.496159e-01 | 9.998360e-01 | No | Yes | 318.26504 | 39.457994 | 2.708815e+02 | 26.527796 | |
| ENSG00000187634 | 148398 | SAMD11 | protein_coding | Q96NU1 | FUNCTION: May play a role in photoreceptor development. {ECO:0000250}. | Alternative promoter usage;Alternative splicing;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:148398; | nucleus [GO:0005634]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; negative regulation of transcription, DNA-templated [GO:0045892] | 23978614_SAMD11 was found to be widely expressed in many cell lines and ocular tissues and its transcription was not regulated by CRX, OTX2 or NR2E3 proteins. 26204995_SEZ6L, HISPPD1, FEZF1, SAMD11 gene variants may be associated with autism spectrum disorder. 27734943_analysis of a homozygous nonsense mutation in SAMD11 in five individuals diagnosed with adult-onset retinitis pigmentosa; SAMD11 interacts with CRX and is expressed in retina 34546569_CircSAMD11 facilitates progression of cervical cancer via regulating miR-503/SOX4 axis through Wnt/beta-catenin pathway. | ENSMUSG00000096351 | Samd11 | 3524.29848 | 1.0403224 | 0.0570306314 | 0.11265314 | 2.589980e-01 | 6.108094e-01 | 9.998360e-01 | No | Yes | 3741.64609 | 429.841110 | 3.275738e+03 | 291.368727 | ||
| ENSG00000187730 | 2563 | GABRD | protein_coding | O14764 | FUNCTION: GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel. | Cell junction;Cell membrane;Chloride;Chloride channel;Disease variant;Disulfide bond;Epilepsy;Glycoprotein;Ion channel;Ion transport;Membrane;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Signal;Synapse;Transmembrane;Transmembrane helix;Transport | Gamma-aminobutyric acid (GABA) is the major inhibitory neurotransmitter in the mammalian brain where it acts at GABA-A receptors, which are ligand-gated chloride channels. Chloride conductance of these channels can be modulated by agents such as benzodiazepines that bind to the GABA-A receptor. The GABA-A receptor is generally pentameric and there are five types of subunits: alpha, beta, gamma, delta, and rho. This gene encodes the delta subunit. Mutations in this gene have been associated with susceptibility to generalized epilepsy with febrile seizures, type 5. Alternatively spliced transcript variants have been described for this gene, but their biological validity has not been determined. [provided by RefSeq, Jul 2008]. | hsa:2563; | axon [GO:0030424]; chloride channel complex [GO:0034707]; dendrite [GO:0030425]; GABA-A receptor complex [GO:1902711]; GABA-ergic synapse [GO:0098982]; integral component of plasma membrane [GO:0005887]; integral component of postsynaptic membrane [GO:0099055]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; synapse [GO:0045202]; chloride channel activity [GO:0005254]; GABA-A receptor activity [GO:0004890]; neurotransmitter receptor activity [GO:0030594]; transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential [GO:1904315]; chemical synaptic transmission [GO:0007268]; chloride transmembrane transport [GO:1902476]; ion transmembrane transport [GO:0034220]; nervous system process [GO:0050877]; regulation of membrane potential [GO:0042391]; signal transduction [GO:0007165] | 16023832_Observational study of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19289452_Lower delta mRNA levels in schizophrenia might reflect a reduced number of alpha(1)beta(x)delta GABA(A) receptors that could contribute to deficient tonic inhibition and prefrontal cortical dysfunction in schizophrenia. 19874574_Observational study of gene-disease association. (HuGE Navigator) 20561060_Observational study of gene-disease association. (HuGE Navigator) 20561060_These findings point to the GABRD gene as a susceptibility gene for COMD. 21795619_In recombinant human cDNA experiments in HEK293 cells, delta subunit coexpression leads to receptors activated by nanomolar THIP concentrations. 23756480_Genome-wide association studies identify GABRD mutation releated to juvenile myoclonic epilepsy 24249596_Considering our Argentinean ASD sample, it can be inferred that GABRB3 would be involved in the etiology of autism through interaction with GABRD. These results support the hypothesis that GABAR subunit genes are involved in autism. 29020412_This study found that increased methylation of the promoter of the delta subunit GABAA receptor was associated with reduced mRNA and protein levels in the cerebellum of alcohol use disorder subjects. 31545245_In IDH wild-type diffuse low-grade glioma patients preserved GABRD expression was independently associated with longer overall survival and reduced tumor infiltration macrophages(TIM). CpG methylation of cg13916816 showed a moderately negative correlation with GABRD expression. 33887383_Association between GABA receptor delta subunit gene polymorphisms and heroin addiction. 34194536_Research for Expression and Prognostic Value of GABRD in Colon Cancer and Coexpressed Gene Network Construction Based on Data Mining. | ENSMUSG00000029054 | Gabrd | 163.03722 | 0.8555312 | -0.2251075855 | 0.24438489 | 8.650598e-01 | 3.523266e-01 | 9.998360e-01 | No | Yes | 152.89647 | 26.314498 | 1.657745e+02 | 22.404844 | |
| ENSG00000187735 | 6917 | TCEA1 | protein_coding | P23193 | FUNCTION: Necessary for efficient RNA polymerase II transcription elongation past template-encoded arresting sites. The arresting sites in DNA have the property of trapping a certain fraction of elongating RNA polymerases that pass through, resulting in locked ternary complexes. Cleavage of the nascent transcript by S-II allows the resumption of elongation from the new 3'-terminus. | 3D-structure;Acetylation;Alternative splicing;Chromosomal rearrangement;DNA-binding;Direct protein sequencing;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:6917; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription factor TFIID complex [GO:0005669]; DNA binding [GO:0003677]; zinc ion binding [GO:0008270]; positive regulation of transcription by RNA polymerase II [GO:0045944]; transcription by RNA polymerase II [GO:0006366]; transcription, DNA-templated [GO:0006351] | 12914699_Results provide a model of the RNA polymerase II-transcription elongation factor TFIIS complex from X-ray diffraction data to 3.8 A resolution. 14506279_transcription factor IIF, hepatitis delta antigen, and stimulatory factor II control human RNA polymerase II (RNAP II) pausing and transcript cleavage 15351637_TFIIS has a role in suppression of transient pausing, which is the most important contribution of TFIIS to elongation from a stall position 16630816_Data shpw that SII is a major component of chromatin transcription and strongly synergizes with p300 (histone acetylation) at a step subsequent to preinitiation complex formation. 16736500_PLAG1 protein is overexpressed in epithelial, myoepithelial, and mesenchymal-like tumor cells in tumors with fusions to CHCHD7 and TCEA1. 20729154_a role for TFIIS in transcription recovery and re-establishment of the balance between hypo- and hyper-phosphorylated RNAPII after DNA damage repair 21070792_Results indicate that TFIIS is not limiting for the repair of transcription-blocking DNA lesions and thus the present work does not support a role for TFIIS in TC-NER. 21596312_RNF20, presumably via H2Bub, selectively represses oncogenic genes by interfering with chromatin recruitment of TFIIS, a factor capable of relieving stalled RNA polymerase II. RNF20 inhibits the interaction between TFIIS and the PAF1 complex. 31519522_A mutant form of TFIIS increased RNAPII pausing and backtracking results in R-loop formation and genome instability. 34822292_A ubiquitous disordered protein interaction module orchestrates transcription elongation. | ENSMUSG00000033813 | Tcea1 | 940.31101 | 1.1397684 | 0.1887406418 | 0.15178350 | 1.557583e+00 | 2.120195e-01 | 9.998360e-01 | No | Yes | 974.40424 | 172.899304 | 9.196658e+02 | 125.851505 | ||
| ENSG00000187764 | 10507 | SEMA4D | protein_coding | Q92854 | FUNCTION: Cell surface receptor for PLXNB1 and PLXNB2 that plays an important role in cell-cell signaling (PubMed:20877282). Regulates GABAergic synapse development (By similarity). Promotes the development of inhibitory synapses in a PLXNB1-dependent manner (By similarity). Modulates the complexity and arborization of developing neurites in hippocampal neurons by activating PLXNB1 and interaction with PLXNB1 mediates activation of RHOA (PubMed:19788569). Promotes the migration of cerebellar granule cells (PubMed:16055703). Plays a role in the immune system; induces B-cells to aggregate and improves their viability (in vitro) (PubMed:8876214). Induces endothelial cell migration through the activation of PTK2B/PYK2, SRC, and the phosphatidylinositol 3-kinase-AKT pathway (PubMed:16055703). {ECO:0000250|UniProtKB:O09126, ECO:0000269|PubMed:16055703, ECO:0000269|PubMed:19788569, ECO:0000269|PubMed:20877282, ECO:0000269|PubMed:8876214}. | 3D-structure;Alternative splicing;Cell membrane;Developmental protein;Differentiation;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Neurogenesis;Phosphoprotein;Receptor;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:10507; | extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; chemorepellent activity [GO:0045499]; identical protein binding [GO:0042802]; semaphorin receptor binding [GO:0030215]; signaling receptor activity [GO:0038023]; signaling receptor binding [GO:0005102]; transmembrane signaling receptor activity [GO:0004888]; axon guidance [GO:0007411]; cell adhesion [GO:0007155]; immune response [GO:0006955]; leukocyte aggregation [GO:0070486]; negative chemotaxis [GO:0050919]; negative regulation of alkaline phosphatase activity [GO:0010693]; negative regulation of apoptotic process [GO:0043066]; negative regulation of axon extension involved in axon guidance [GO:0048843]; negative regulation of cell adhesion [GO:0007162]; negative regulation of osteoblast differentiation [GO:0045668]; negative regulation of peptidyl-tyrosine phosphorylation [GO:0050732]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neural crest cell migration [GO:0001755]; ossification involved in bone maturation [GO:0043931]; positive regulation of cell migration [GO:0030335]; positive regulation of collateral sprouting [GO:0048672]; positive regulation of GTPase activity [GO:0043547]; positive regulation of inhibitory synapse assembly [GO:1905704]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein phosphorylation [GO:0001934]; regulation of cell projection organization [GO:0031344]; regulation of cell shape [GO:0008360]; regulation of dendrite morphogenesis [GO:0048814]; semaphorin-plexin signaling pathway [GO:0071526]; semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis [GO:1900220] | 12406905_Leukemic & normal CD5+ B cells express CD100; upon interaction between CD100 & Plexin-B1, both increase their proliferative activity & life span. CD100/Plexin-B1 crosstalk is not malignancy related but reproduces a mechanism used by normal CD5+ B cells. 14707103_Soluble CD100 induces a progressive decrease in process extension of oligodendrocytes, followed by their death and the death of multipotent neural precursors. 15613544_up-modulation of the survival receptor CD100 is restricted to proliferating B-cell leukemia cells 15632204_demonstrate that Sema4D is angiogenic in vitro and in vivo and that this effect is mediated by its high-affinity receptor, Plexin B1, and that biologic effects elicited by Plexin B1 require coupling and activation of the Met tyrosine kinase 16055703_Semaphorin 4D/plexin-B1 induces endothelial cell migration through the activation of PYK2, Src, and the phosphatidylinositol 3-kinase-Akt pathway 16754882_Data show that semaphorin 4D (Sema4D), a protein originally shown to regulate axonal growth cone guidance in the developing central nervous system, is highly expressed in cell lines derived from head and neck squamous cell carcinomas. 17244710_semaphorin 4D has a role in platelet responses to vascular injury 17520683_Multivariate analysis revealed that CD100 expression and tumor size were independent prognosticators for overall survival. 17786190_CD100-CD72 interaction can be the mechanism by which NK cell communicate with B cells. 18025083_a novel mechanism by which plexin-mediated signaling can be regulated and explains how Sema4D can exert different biological activities through the differential association of its receptor with ErbB-2 and Met. 19805522_show here that activation of plexin-B1 by Sema4D and its subsequent tyrosine phosphorylation creates docking sites for the SH2 domains of phospholipase Cgamma. 19957197_Observational study of gene-disease association. (HuGE Navigator) 19957197_neither Sema3A nor Sema4D likely influence the susceptibility to Alzheimer's disease 20237496_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877282_crystal structures of cognate complexes of the semaphorin-binding regions of plexins B1 and A2 with semaphorin ectodomains (human PLXNB1(1-2)-SEMA4D(ecto) and murine PlxnA2(1-4)-Sema6A(ecto)), plus unliganded structures of PlxnA2(1-4) and Sema6A(ecto) 21244334_dysregulations in CD100 expression and release could play a role in SSc development and/or maintenance. 21538148_Rho-mediated activation of PI(4)P5K and lipid second messengers is necessary for promotion of angiogenesis by Semaphorin 4D. 21812859_Sema4D potentiates the invasiveness of pancreatic cancer cells. The binding of Sema4D to plexinB1 induced small GTPase Ras homolog gene family, member A activation and resulted in the phosphorylation of MAPK and Akt. 21925246_From the data obtained in this study, SEMA4D may have a role in more aggressive and potentially metastatic breast tumours. 22111667_CD72 mRNA expression level correlates with Sema4D expression in peripheral blood mononuclear cells in immune thrombocytopenia. 22189792_Sema4D, the ligand for Plexin B1, suppresses c-Met activation and migration and promotes melanocyte survival and growth. 22652457_The expression of semaphorin 4D (SEMA4D), which is under the control of the HIF-family of transcription factors, cooperates with VEGF to promote tumor growth and vascularity in oral squamous cell carcinoma (OSCC). 23335257_copy number loss of the Sema4D gene region may play a role in the etiology of acetabular dysplasia 23376478_Lycorine hydrochloride suppressed the expression of several key angiogenic genes, including VE-cadherin and Sema4D, and reduced Akt phosphorylation in Hey1B cells. 23564909_The membrane-proximal cytoplasmic domain of Sema4D contains a binding site for calmodulin within the polybasic region Arg762-Lys779, that regulates Sema4D exodomain shedding in platelets. 23741311_There was an increased level of plasma soluble Sema4 in the Sema4D(high) population of T-cells suggesting a potential role of these T-cells in heart failure. 24040126_Elevated plasma soluble Sema4D/CD100 levels are associated with disease severity in patients of hemorrhagic fever with renal syndrome. 24098722_CD100 may have a role in atherosclerotic plaque development, and may possibly be employed in targeted treatments of these atheromas. 24131822_results suggest that the contribution of Sema4D in platelets applies to ITAM-containing receptors as a class 24289594_SEMA4D might possibly serve as a reliable tool for early and accurate prediction of EOC poor prognosis. 24603190_Sema4D contributes to enhanced invasion and tumor progression through increased motility of cervical cancer and VEGF-C/-D-mediated lymphangiogenesis. 25108441_Data suggest that CD100 seems to be involved in hepatitis C virus (HCV) clearance by natural killer (NK) cells. 25135716_Sema4D could play an important role in promoting tumor proliferation, migration and metastasis in the NSCLC, by influencing the Akt protein phosphorylation. Inhibition of Sema4D may be a useful approach for the treatment of NSCLC. 25707877_A positive feedback loop involving sSema4D/IL-6 and TNFalpha/ADAMTS-4 may contribute to the pathogenesis of rheumatoid arthritis. 25717256_Suggest that HIF-1alpha and Sema4D expression correlates with histological tumor type, TNM stage, and lymphatic metastasis in colorectal carcinoma. 26035216_Results show that decreased expression of Sema4D, plexin-B1 and -B2 was associated with local recurrence and poor prognosis of breast neoplasm. 26051877_Plexin-B1 induces cutaneous squamous cell carcinoma cell proliferation, migration, and invasion by interacting with Sema4D. Plexin-B1 might serve as a useful biomarker and/or as a novel therapeutic target for cSCC. 26275342_Blocking of CD100, plexin B1 and/or B2 in adhesion experiments have shown that both CD100 and plexins act as adhesion molecules involved in monocyte-endothelial cell binding. 26389656_Tax and semaphorin 4D released from lymphocytes infected with human lymphotropic virus type 1 and inhibit neurite growth in a neuron cell line. 26417899_Serum sSema4D levels are increased in patients with atrial fibrillation and are independently associated with atrial remodeling 26566052_This assay specifically and reproducibly measured cSEMA4D saturation and expression levels. Evaluation of the SEMA4D-specific PD markers were critical in determining the clinical saturation threshold of cSEMA4D by VX15/2503 26718213_sema 4D was the direct target of miR-214 and was negatively regulated by miR-214 in ovarian cancer cells 26740106_this study describes a novel immunosuppressive role for Sema4D in head and neck squamous cell carcinoma through induction of myeloid derived suppressor cells 26910109_Semaphorin 4D Promotes Skeletal Metastasis in Breast Cancer. 27456345_The positive expression of both Sema4D and PlexinB1 was found to be an independent risk factor for a worse survival in colorectal cancer. 27554682_In this review, we summarized the current findings on neuroimmune Sema4A and Sema4D molecules in chronic inflammation underlying many diseases and discussed their positive or negative impacts on the implicated molecular and cellular processes 27720716_A critical pathogenic engagement of Semaphorin 4D produced by gamma delta T cells in the development of medication-related osteonecrosis of the jaw 27787840_describe the transfection, purification, and visualization of Fc-tagged SEMA4D (semaphorin 4D) recombinant protein 27787869_describe the application of in vitro migration and tubulogenesis assays and the directed in vivo angiogenesis assay (DIVAA) in the measurement of the angiogenic potential of cell-derived and soluble SEMA4D 28222623_Interferon-alpha-induced CD100 expression on naive CD8(+) T cells enhances antiviral responses to hepatitis C infection through CD72 signal transduction. 28296344_We identified a novel genome-wide significant African-specific locus for BMI (SEMA4D, rs80068415). A novel variant in SEMA4D was significantly associated with body mass index. Carriers of the C allele were 4.6 BMI units heavier than carriers of the T allele. 28336906_Our results identified FGL2, GAL, SEMA4D, SEMA7A, and IDO1 as new candidate genes that could be involved in MSCs-mediated immunomodulation. FGL2, GAL, SEMA4D, SEMA7A, and IDO1 genes appeared to be differentially transcribed in the different MSC populations. Moreover, these genes were not similarly modulated following MSCs-exposure to inflammatory signals 28416516_Serum levels of soluble SEMA4D were elevated in patients with ANCA-associated vasculitis. Cell-surface expression of SEMA4D was downregulated, a consequence of proteolytic cleavage of membrane SEMA4D. Soluble SEMA4D exerted pro-inflammatory effects on endothelial cells. Membranous SEMA4D on neutrophils bound to plexin B2 on endothelial cells, and this interaction decreased NET formation. 28508207_The results obtained make significant adjustments in understanding of Seam4D effects in lymphoid cells. 28656278_knockdown of Sema4D in Head and neck squamous cell carcinomas (HNSCCs)cells inhibited tumor growth and decreased the number of osteoclasts in a mouse xenograft model. Taken together, IGF-I-driven production of Sema4D in HNSCCs promotes osteoclastogenesis and bone invasion. 28927892_Study data showed that cooperation of CD100 and PlxnB2 promoted the inflammatory responses in keratinocytes by activating NF-kappaB and the NLRP3 inflammasome and participated in the pathogenesis of psoriasis. 29054606_in bullous pemphigoid pathogenesis, membrane-bound ADAM10 expressed on activated CD15+ granulocytes in circulation, along with soluble ADAM10 in local lesions, causes Sema4D cleavage from the membrane of CD15+ granulocytes and results in a high level of circulating sSema4D. 29308068_SEMA4D coordinates with VEGF during angiogenesis via plexin-B1 in epithelial ovarian cancer. 29434448_Combined detection of tumor-associated macrophages markers, CD68 and Sema4D, in gastric carcinoma tissue shows potential to predict the trend of gastric carcinoma progression. 29748532_Our data suggest that Sema4D is elevated in MM patients and correlate with adverse myeloma features and increased bone resorption, providing a possible target for novel therapeutic approaches in MM. 29763494_Advanced peri-implantitis lesions showed higher levels of gene expression for Sem3A and Sem4D and lower levels of Sem4A in comparison to tissues obtained from a healthy dental implant. 29939944_The crosstalk between sema4D and Met could transactivate Met to promote trophoblast cell invasion and differentiation, and decreased expression of sema4D and Plexin-B1 may be responsible for the deficiency in Met signalling and the development of preeclampsia. 30538782_Despite no difference in Sema4D levels between rheumatoid arthritis (RA) patients and controls, its baseline levels were significantly higher in those with radiographic progression than those without progression. These data suggest that the Sema4D level might be a useful marker to identify RA patients with subsequent radiographic progression and that Sema4D may be an active mediator involved in RA-induced joint damage. 30674231_All these data suggest that Sema4D promotes cell proliferation and metastasis in bladder cancer in vivo and in vitro. The oncogenic behavior of Sema4D is achieved by activating the PI3K/AKT pathway. 30762724_SEMA4D could be a prospective biomarker for prognostic prediction of various malignancies except breast cancer. For Caucasian breast cancer patients, SEMA4D's high affinity receptor Plexin-B1 showed a significant positive correlation with survival 30937968_Sema4D/PlexinB1 promotes endothelial differentiation of dental pulp stem cells via activation of AKT and ERK1/2 signaling. 30967271_LINC01061 sponges miR-612 to increase SEMA4D expression. 31651222_SEMA4D under the posttranscriptional regulation of HuR and miR-4319 boosts cancer progression in esophageal squamous cell carcinoma. 31693461_REVIEW: data indicating the presence of an alternative mechanism for the regulatory activity of Sema4D that involves the functioning of membrane semaphorin as a receptor ensuring the outside-in signaling; cell signaling pathways mediated by the membrane Sema4D and their contribution to the Sema4D-dependent regulation of cell functions are discussed 32149403_Insufficient CD100 shedding contributes to suppression of CD8(+) T-cell activity in non-small cell lung cancer. 32244055_Sema4D and 4A seem to play a critical role in the pathogenesis of some autoimmune diseases, such as multiple sclerosis. 32382577_Sema4D Aggravated LPS-Induced Injury via Activation of the MAPK Signaling Pathway in ATDC5 Chondrocytes. 32416689_Potential Prognostic Predictors and Molecular Targets for Skin Melanoma Screened by Weighted Gene Co-expression Network Analysis. 32450646_Serum Sema4D can be used as an indicator of disease monitoring and prognosis evaluation in rheumatoid arthritis patients 32888127_Inhibition of semaphorin 4D enhances chemosensitivity by increasing 5-fluorouracile-induced apoptosis in colorectal cancer cells. 32945351_Effects of NRP1 on angiogenesis and vascular maturity in endothelial cells are dependent on the expression of SEMA4D. 32948333_Signaling from membrane semaphorin 4D in T lymphocytes. 33013906_Semaphorin 4D Induces an Imbalance of Th17/Treg Cells by Activating the Aryl Hydrocarbon Receptor in Ankylosing Spondylitis. 33381571_Neutrophil-Derived Semaphorin 4D Induces Inflammatory Cytokine Production of Endothelial Cells via Different Plexin Receptors in Kawasaki Disease. 33593275_CD100 modulates cytotoxicity of CD8(+) T cells in patients with acute myocardial infarction. 33627483_A heterozygous germline CD100 mutation in a family with primary sclerosing cholangitis. 33649851_Semaphorin 4D is a potential biomarker in pediatric leukemia and promotes leukemogenesis by activating PI3K/AKT and ERK signaling pathways. 33776991_Soluble Sema4D in Plasma of Head and Neck Squamous Cell Carcinoma Patients Is Associated With Underlying Non-Inflamed Tumor Profile. 33910443_Assessment of the relationship between semaphorin4D level and recurrence after catheter ablation in paroxysmal atrial fibrillation. 33974462_Sema4D correlates with tumour immune infiltration and is a prognostic biomarker in bladder cancer, renal clear cell carcinoma, melanoma and thymoma. 34337054_SEMA4D Knockdown Attenuates beta-Catenin-Dependent Tumor Progression in Colorectal Cancer. 34854398_Sema4D/Plexin-B1 promotes the progression of osteosarcoma cells by activating Pyk2-PI3K-AKT pathway. 35401878_Soluble Sema4D Level Is Positively Correlated with Sema4D Expression in PBMCs and Peripheral Blast Number in Acute Leukemia. | ENSMUSG00000021451 | Sema4d | 583.57728 | 0.8354190 | -0.2594280568 | 0.14512572 | 3.190512e+00 | 7.406679e-02 | 9.998360e-01 | No | Yes | 485.84672 | 63.173563 | 5.716807e+02 | 57.576695 | ||
| ENSG00000187954 | 50626 | CYHR1 | protein_coding | Q6ZMK1 | Alternative splicing;Cytoplasm;Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:50626; | nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; zinc ion binding [GO:0008270] | 26676980_High CYHR1 expression is associated with Esophageal Squamous Cell Carcinoma. | ENSMUSG00000053929 | Cyhr1 | 3267.29329 | 0.9572184 | -0.0630799363 | 0.11343417 | 3.202667e-01 | 5.714474e-01 | 9.998360e-01 | No | Yes | 3681.15012 | 382.718692 | 3.570445e+03 | 287.789423 | |||
| ENSG00000188070 | 65998 | ZFTA | protein_coding | C9JLR9 | Chromosomal rearrangement;Isopeptide bond;Reference proteome;Ubl conjugation | hsa:65998; | negative regulation of transcription, DNA-templated [GO:0045892] | 23672313_Based on a recurrent translocation t(11;16)(q13;p13), the C11orf95-MKL2 fusion gene has been found in eight further cases of chondroid lipomas. 24562983_C11orf95-RELA fusions represent a unique novel mechanism leading to pathological activation of NF-kappaB signaling in supratentorial ependymomas. 25388523_C11orf95-RELA fusion transcript is thought to be the driver genetic alteration leading to activation of the NFkB pathway in supratentorial ependymomas. 33221956_Ependymoma with C11orf95-MAML2 fusion: presenting with granular cell and ganglion cell features. 33576087_Ependymoma-like tumor with mesenchymal differentiation harboring C11orf95-NCOA1/2 or -RELA fusion: A hitherto unclassified tumor related to ependymoma. 33685520_C11orf95-RELA fusion drives aberrant gene expression through the unique epigenetic regulation for ependymoma formation. | ENSMUSG00000053080 | Zfta | 2303.85781 | 0.9224298 | -0.1164889549 | 0.10556330 | 1.205808e+00 | 2.721640e-01 | 9.998360e-01 | No | Yes | 2207.77738 | 151.417907 | 2.337362e+03 | 124.868481 | |||
| ENSG00000188283 | 163087 | ZNF383 | protein_coding | Q8NA42 | FUNCTION: May function as a transcriptional repressor, suppressing transcriptional activities mediated by MAPK signaling pathways. {ECO:0000269|PubMed:15964543}. | Cytoplasm;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene is a KRAB-related zinc finger protein that inhibits the transcription of some MAPK signaling pathway genes. The repressor activity resides in the KRAB domain of the encoded protein. [provided by RefSeq, Sep 2016]. | hsa:163087; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity [GO:0003700]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 15964543_Overexpression of ZNF383 in cells inhibits the transcriptional activities of AP-1 and SRE, suggesting that ZNF383 may act as a negative regulator in MAPK-mediated signaling pathways. | ENSMUSG00000099689 | Zfp383 | 62.53485 | 1.1698766 | 0.2263563231 | 0.44212867 | 2.420992e-01 | 6.226940e-01 | 9.998360e-01 | No | Yes | 74.56974 | 17.507591 | 6.278246e+01 | 11.286549 | |
| ENSG00000189007 | 134637 | ADAT2 | protein_coding | Q7Z6V5 | FUNCTION: Probably participates in deamination of adenosine-34 to inosine in many tRNAs. {ECO:0000250}. | 3D-structure;Alternative splicing;Hydrolase;Metal-binding;Reference proteome;Zinc;tRNA processing | hsa:134637; | nucleoplasm [GO:0005654]; tRNA-specific adenosine-34 deaminase activity [GO:0052717]; zinc ion binding [GO:0008270]; tRNA wobble adenosine to inosine editing [GO:0002100] | 20581866_data indicate that ATAD2 overexpression in somatic cells, by acting on basic properties of chromatin, may contribute to malignant transformation 28703578_Transfer RNAs (tRNAs) are among the most heavily modified RNA species. Data confirm that ADAT2 exhibits substrate specificity that includes tRNA(Ala-AGC) and tRNA(Thr-AGU), deaminating adenosine-34 to inosine-34 as post-transcriptional modifications. 32763916_Identification and rescue of a tRNA wobble inosine deficiency causing intellectual disability disorder. | ENSMUSG00000019808 | Adat2 | 360.31028 | 1.0287795 | 0.0409338580 | 0.18127762 | 5.043734e-02 | 8.223040e-01 | 9.998360e-01 | No | Yes | 387.21310 | 73.318425 | 3.593340e+02 | 52.497326 | ||
| ENSG00000189114 | 388552 | BLOC1S3 | protein_coding | Q6QNY0 | FUNCTION: Component of the BLOC-1 complex, a complex that is required for normal biogenesis of lysosome-related organelles (LRO), such as platelet dense granules and melanosomes. In concert with the AP-3 complex, the BLOC-1 complex is required to target membrane protein cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals. The BLOC-1 complex, in association with SNARE proteins, is also proposed to be involved in neurite extension. Plays a role in intracellular vesicle trafficking. {ECO:0000269|PubMed:16385460, ECO:0000269|PubMed:17182842}. | Albinism;Cytoplasm;Hermansky-Pudlak syndrome;Phosphoprotein;Reference proteome | This gene encodes a protein that is a component of the BLOC1 multi-subunit protein complex. This complex is necessary for the biogenesis of specialized organelles of the endosomal-lysosomal system, including platelet dense granules and melanosomes. Mutations in this gene cause Hermansky-Pudlak syndrome 8, a disease characterized by lysosomal storage defects, bleeding due to platelet storage pool deficiency, and oculocutaneous albinism. [provided by RefSeq, Jul 2008]. | hsa:388552; | axon cytoplasm [GO:1904115]; BLOC-1 complex [GO:0031083]; cytosol [GO:0005829]; transport vesicle [GO:0030133]; protein transmembrane transporter activity [GO:0008320]; anterograde axonal transport [GO:0008089]; anterograde synaptic vesicle transport [GO:0048490]; endosome to melanosome transport [GO:0035646]; eye development [GO:0001654]; melanosome organization [GO:0032438]; melanosome transport [GO:0032402]; neuron projection development [GO:0031175]; pigmentation [GO:0043473]; platelet activation [GO:0030168]; platelet dense granule organization [GO:0060155]; positive regulation of natural killer cell activation [GO:0032816]; response to xenobiotic stimulus [GO:0009410]; secretion of lysosomal enzymes [GO:0033299] | 16385460_A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). 20460622_Observational study, meta-analysis, and genome-wide association study of gene-disease association and gene-gene interaction. (HuGE Navigator) 30387913_BLOC-2 and BLOC-3 were destabilized due to the mutation of these Hermansky-Pudlak syndrome (HPS) genes which are so far the only reported causative genes in Chinese HPS patients 32687635_Novel variants in the BLOC1S3 gene in patients presenting a mild form of Hermansky-Pudlak syndrome. | ENSMUSG00000057667 | Bloc1s3 | 1273.88066 | 1.0429640 | 0.0606893329 | 0.15181132 | 1.693706e-01 | 6.806717e-01 | 9.998360e-01 | No | Yes | 1499.59554 | 177.270004 | 1.354153e+03 | 124.904348 | |
| ENSG00000196268 | 284443 | ZNF493 | protein_coding | Q6ZR52 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:284443; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 60.10589 | 1.5542222 | 0.6361927737 | 0.40653833 | 2.362162e+00 | 1.243098e-01 | 9.998360e-01 | No | Yes | 75.19586 | 20.851649 | 6.035208e+01 | 12.906668 | |||||
| ENSG00000196369 | 647135 | SRGAP2B | protein_coding | P0DMP2 | FUNCTION: May regulate cell migration and differentiation through interaction with and inhibition of SRGAP2 (PubMed:31822692, PubMed:31822692). In contrast to SRGAP2C, it is not able to induce long-lasting changes in synaptic density throughout adulthood (PubMed:31822692). {ECO:0000269|PubMed:31822692, ECO:0000305|PubMed:22559944, ECO:0000305|PubMed:31822692}. | Coiled coil;Neurogenesis;Reference proteome | This locus encodes a member of the SLIT-ROBO Rho GTPase activating protein family. This human-specific locus resulted from incomplete segmental duplication of the SLIT-ROBO Rho GTPase activating protein 2 locus. The encoded protein lacks the GTPase activating protein domain compared to proteins encoded by SLIT-ROBO Rho GTPase activating protein 2. The functionality of the protein encoded by this locus has been questioned, as several normal individuals with homozygous deletions for this locus have been identified, and the expression of this locus appears to be much lower than the similar SLIT-ROBO Rho GTPase activating protein 2C (SRGAP2C) locus. The SRGAP2C locus has been shown to encode a protein that functions antagonistically to SLIT-ROBO Rho GTPase activating protein 2 in cortical neuron development. [provided by RefSeq, Dec 2012]. | Mouse_homologues mmu:14270; | cytoplasm [GO:0005737]; negative regulation of cell migration [GO:0030336]; nervous system development [GO:0007399] | Mouse_homologues 19137586_Data show that srGAP2 is expressed in zones of neuronal differentiation in many different tissues of the central nervous system. 19737524_Study reports that srGAP2 negatively regulates neuronal migration and induces neurite outgrowth and branching through the ability of its F-BAR domain to induce filopodia-like membrane protrusions resembling those induced by I-BAR domains. 23505444_The inverse F-BAR domain protein srGAP2 acts through srGAP3 to modulate neuronal differentiation and neurite outgrowth of mouse neuroblastoma cells. 27966608_Results show that the effects of Srgap2 expression modulation in the murine OS cell lines support the hypothesis that SRGAP2 may have a role as a suppressor of metastases in osteosarcoma. 29242313_SRGAP2a protects podocytes in diabetic nephropathy by suppressing podocyte migration. 31880824_Rac1 Inhibition Via Srgap2 Restrains Inflammatory Osteoclastogenesis and Limits the Clastokine, SLIT3. | ENSMUSG00000026425 | Srgap2 | 292.99554 | 1.0311243 | 0.0442182713 | 0.23069186 | 3.516685e-02 | 8.512465e-01 | 9.998360e-01 | No | Yes | 280.06927 | 56.101562 | 2.624361e+02 | 40.580658 | |
| ENSG00000196422 | 9858 | PPP1R26 | protein_coding | Q5T8A7 | FUNCTION: Inhibits phosphatase activity of protein phosphatase 1 (PP1) complexes. May positively regulate cell proliferation. {ECO:0000269|PubMed:16053918, ECO:0000269|PubMed:19389623}. | Nucleus;Phosphoprotein;Protein phosphatase inhibitor;Reference proteome | hsa:9858; | nucleolus [GO:0005730]; protein phosphatase inhibitor activity [GO:0004864]; negative regulation of phosphatase activity [GO:0010923] | 16053918_we showed that KIAA0649 mRNA is widely expressed in human multiple tissues and cell lines. We have also demonstrated that KIAA0649 has oncogenic characteristics: it enhances colony formation, allows anchorage-independent growth. 26442585_identified NRBE3 as a novel ubiquitin E3 ligase for RB that might play a role as a potential oncoprotein in human cancers 30262434_Results show that up-regulation of NRBE3 is correlated with lymphatic metastasis in human breast cancer (BC) tissues. Ectopic expression of NRBE3 promotes migration and invasion in BC cells. Accordingly, its knockdown inhibits migration and invasion in BC cells and lung metastasis in vivo. | ENSMUSG00000035829 | Ppp1r26 | 1203.68440 | 1.1395381 | 0.1884492223 | 0.12959598 | 2.165796e+00 | 1.411115e-01 | 9.998360e-01 | No | Yes | 1367.76334 | 135.383535 | 1.156982e+03 | 89.368393 | ||
| ENSG00000196502 | 6817 | SULT1A1 | protein_coding | P50225 | FUNCTION: Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) as sulfonate donor to catalyze the sulfate conjugation of a wide variety of acceptor molecules bearing a hydroxyl or an amine groupe. Sulfonation increases the water solubility of most compounds, and therefore their renal excretion, but it can also result in bioactivation to form active metabolites. Displays broad substrate specificity for small phenolic compounds. Plays an important role in the sulfonation of endogenous molecules such as steroid hormones and 3,3'-diiodothyronin (PubMed:16221673, PubMed:12471039, PubMed:22069470, PubMed:21723874, PubMed:10199779, PubMed:7834621). Mediates the sulfate conjugation of a variety of xenobiotics, including the drugs acetaminophen and minoxidil (By similarity). Mediates also the metabolic activation of carcinogenic N-hydroxyarylamines leading to highly reactive intermediates capable of forming DNA adducts, potentially resulting in mutagenesis (PubMed:7834621). {ECO:0000250|UniProtKB:P17988, ECO:0000269|PubMed:10199779, ECO:0000269|PubMed:12471039, ECO:0000269|PubMed:16221673, ECO:0000269|PubMed:21723874, ECO:0000269|PubMed:22069470, ECO:0000269|PubMed:7834621}. | 3D-structure;Alternative splicing;Catecholamine metabolism;Cytoplasm;Direct protein sequencing;Lipid metabolism;Phosphoprotein;Reference proteome;Steroid metabolism;Transferase | Sulfotransferase enzymes catalyze the sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds. These cytosolic enzymes are different in their tissue distributions and substrate specificities. The gene structure (number and length of exons) is similar among family members. This gene encodes one of two phenol sulfotransferases with thermostable enzyme activity. Multiple alternatively spliced variants that encode two isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:6817; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; 3'-phosphoadenosine 5'-phosphosulfate binding [GO:0050656]; aryl sulfotransferase activity [GO:0004062]; flavonol 3-sulfotransferase activity [GO:0047894]; steroid sulfotransferase activity [GO:0050294]; sulfotransferase activity [GO:0008146]; 3'-phosphoadenosine 5'-phosphosulfate metabolic process [GO:0050427]; amine metabolic process [GO:0009308]; catecholamine metabolic process [GO:0006584]; estrogen metabolic process [GO:0008210]; ethanol catabolic process [GO:0006068]; flavonoid metabolic process [GO:0009812]; sulfation [GO:0051923]; xenobiotic metabolic process [GO:0006805] | 11156380_Observational study of gene-disease association. (HuGE Navigator) 11207031_Observational study of genotype prevalence. (HuGE Navigator) 11219777_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11692076_Observational study of gene-disease association. (HuGE Navigator) 11692076_The high activity SULT1A1*1 allozyme protects against dietary and/or environmental chemicals involved in the pathogenesis of colorectal cancer. 11804685_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11804685_genetic polymorphism in SULT1A1 gene may be associated with increased lung cancer risk 12162852_7-OH-flavone sulfotransferase followed Michaelis-Menten kinetics 12165038_Observational study of gene-disease association. (HuGE Navigator) 12165038_The SULT1A1 R213H polymorphism is not linked with colorectal cancer in an elderly Australian population. 12402313_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12419790_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12419790_There is an an association between genotype and survival of breast cancer patients receiving tamoxifen therapy 12419832_Observational study of gene-disease association. (HuGE Navigator) 12455060_Observational study of gene-disease association. (HuGE Navigator) 12455060_SULT1A1 his(213) allele is important in the development of esophageal cancer in men. 12468438_Observational study of gene-disease association. (HuGE Navigator) 12469224_Observational study of gene-disease association. (HuGE Navigator) 12469224_the relationship among the polymorphisms of this enzyme and SULT1A2 in different types of cancers in Taiwanese 12471039_the crystal structure of SULT1A1 provides the molecular basis for substrate inhibition and how the enzyme sulfonates a wide variety of lipophilic compounds 12725421_Observational study of gene-disease association. (HuGE Navigator) 12761191_the differential substrate specificity of the two enzymes M-PST and P-PST for the thirteen drug compounds tested 12814450_Observational study of genotype prevalence. (HuGE Navigator) 12867416_study of arginine residues in the active site of human phenol sulfotransferase 12867492_P. 1038 '...clearly expressed...SULT1A1...' P. 1040 '...SULT1A1...which belongs to phase II metabolism,...significantly expressed in HepG2.' 14520706_Observational study of gene-environment interaction. (HuGE Navigator) 14618622_Observational study of gene-environment interaction. (HuGE Navigator) 14618622_We conclude that smoking increases risk of colorectal adenomas and that SULT1A1 and NAT2 only modestly modify this association. 14642079_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14643027_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14643027_Sulfotransferase 1A1 polymorphism is associated with bladder cancer 14648207_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14648207_SULT1A1 slow acetylator genotype might modulate the effect of carcinogenic arylamines contained in tobacco smoke, and has a tendency to present a higher risk for highly differentiated tumors among heavy-smokers. 14688021_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14688021_SULT1A1 polymorphism is associated with susceptibility to lung cancer in relation to tobacco smoking 14871892_In SULT1A1, substrate inhibition occurs with pNP as the substrate but not with dopamine; residue Phe-247 of SULT1A1, which interacts with both p-nitrophenol molecules in the active site, is important for substrate inhibition 14973106_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15090717_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15093672_Observational study of gene-disease association. (HuGE Navigator) 15093672_SULT1A1 His allele was positively associated with the risk of breast cancer in Chinese women 15122594_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15318931_Observational study of gene-disease association. (HuGE Navigator) 15377847_Observational study of gene-disease association. (HuGE Navigator) 15383623_SULT1A1 promoter was shown to be dependent on the presence of Sp1 and Ets transcription factor binding sites. 15455371_Observational study of gene-disease association. (HuGE Navigator) 15516026_Observational study of gene-disease association. (HuGE Navigator) 15604994_Observational study of gene-disease association. (HuGE Navigator) 15632378_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15743503_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15894657_No significant association found for breast cancer risk. 15894657_Observational study of gene-disease association. (HuGE Navigator) 15949571_Observational study of gene-disease association. (HuGE Navigator) 15952058_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15970794_Observational study of gene-disease association. (HuGE Navigator) 15987423_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16006997_Observational study of gene-disease association. (HuGE Navigator) 16080486_Observational study of gene-disease association. (HuGE Navigator) 16103451_Observational study of gene-disease association. (HuGE Navigator) 16133548_Observational study of genotype prevalence. (HuGE Navigator) 16133548_SULT1A1*2 and SULT1A2*2 are the major allelic variants in the Korean population. 16137826_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16141802_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16175316_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16175316_Postmenopausal women carrying the variant SULT1A1 His allele may be more susceptible to estrogen-induced carcinogenesis in mammary tissue. 16221673_x-ray crystallographic structure of human SULT1A1 with estradiol 16232327_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16272171_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16280036_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16284375_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16317586_Observational study of genotype prevalence. (HuGE Navigator) 16317586_SULT1A1 genotype did not correlate with any prognostic or predictive markers associated with breast cancer. 16328031_Progressive SULT1A1 methylation within the promoter area of the gene occurs during breast carcinogenesis. 16380991_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16395669_Observational study of gene-disease association. (HuGE Navigator) 16402077_Observational study of gene-disease association. (HuGE Navigator) 16418064_Sulfation of resveratrol in human liver: evidence of a major role for the sulfotransferases SULT1A1. 16425401_Observational study of gene-disease association. (HuGE Navigator) 16504378_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16575574_Observational study of gene-disease association. (HuGE Navigator) 16637266_Observational study of gene-disease association. (HuGE Navigator) 16875543_Observational study of gene-disease association. (HuGE Navigator) 16926176_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16985032_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16985250_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16985250_The SULT1A1*2 allele and long-term use of estrogen replacement therapy were associated with statistically significantly higher risk of endometrial cancer 17013894_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17074589_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17074589_Results indicate that the SULT1A1 genotype may play an important role in the risk of developing lung cancer, especially in cigarette smokers. 17189289_The presence of SULT1A1 gene deletions and duplications, representing an additional source of variability in the metabolic activity of this enzyme, is demonstrated. 17244352_Clinical trial of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17274372_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17293380_Apigenin, epicatechin, and resveratrol exhibited SULT1a1 allele-specific variation in sulfation, with SULT1A1*1 and *3 acting as normal-activity allozymes and SULT1A*2 as low-activity allozyme. 17372239_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17372243_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17479406_Observational study of gene-disease association. (HuGE Navigator) 17603900_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17605044_Observational study of gene-disease association. (HuGE Navigator) 17605044_SULT1A1 polymorphism is associated with primary brain tumors 17619904_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17619904_SULT1A1 polymorphism is associated with urothelial cancer 17912498_Increased generation of urinary 8-hydroxy-2'-deoxyguanosine was found in betel-quid chewers with SULT1A1 and GSTP1 genotypes that affect susceptibility to DNA damage. 17912498_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17947222_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17996038_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18006944_Observational study of gene-disease association. (HuGE Navigator) 18259693_Observational study of gene-disease association. (HuGE Navigator) 18259693_SULT1A1/SULT1A2 gene complex showed suggestive haplotypic association in the family-based cardiovascular disease study, with the greatest increase in risk conferred by the SULT1A2 235T allele 18264785_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18318428_Observational study of gene-disease association. (HuGE Navigator) 18318428_a decreased single nucleotide polymorphism of CYP1A1 and an increased single nucleotide polymorphism for SULT1A1 and SULT1E1 genes may be risk factors for endometrial cancer in Caucasians. 18365755_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18365755_The aim of the study was to investigate NAT1, NAT2, GSTM1, GSTT1, GSTP1, SULT1A1, XRCC1, XRCC3 and XPD genetic polymorphisms, coffee consumption and risk of bladder cancer (BC) through a hospital-based case-control study. 18368507_Observational study of gene-disease association. (HuGE Navigator) 18368507_The study showed a null association of SULT1A1 polymorphism with familial prostate cancer risk in the Japanese population. 18447907_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18497059_Observational study of gene-disease association. (HuGE Navigator) 18497059_SULT1A1 polymorphism is associated with ovarian cancer 18499698_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18499698_The CYP1A2 and NAT1 but not SULT1A1 and NAT2 genotypes showed significant interactions with heavy smoking in women not men 18632753_Observational study of gene-disease association. (HuGE Navigator) 18794456_SULT1A1 mRNA is expressed in human skin, at similar levels in men and women. SULT1A1 levels are not altered by topical 17-beta-estradiol treatment. 18854828_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18854828_The SULT1A1*2 revealed contrasting risk association for upper aerodigestive tract cancers and conferred significant increased risk of breast cancer to Asian but not Causcasian women in patients with multiple tobacco-related cancers 18990750_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18990750_Red meat intake was associated with increased colorectal cancer regardless of genotype, but the SULT1A1-638G>A variant modified the association between meat doneness and cancer risk. 19120511_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19120511_SULT1A1 Arg213His polymorphism is associated with the development of urothelial cancer, especially among cigarette smokers exposed to hazardous chemicals. 19126640_Observational study of gene-disease association. (HuGE Navigator) 19126640_SULT1A1 gene is a risk modifier on environmental carcinogen in OSCC and the association of SULT1A1 haplotypes with the risk of OSCC might be modified by betel quid chewing. 19237513_he importance of human SULT1A1 as a homodimer was to maintain its structural stability, and the change of secondary structure was responsible for alternating its quaternary structure 19307236_Observational study of gene-disease association. (HuGE Navigator) 19322015_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19322015_SULT1A1 genetic polymorphisms might modify the arsenic methylation profile and urothelial carcinoma progression. 19339270_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19350537_Observational study of gene-disease association. (HuGE Navigator) 19350537_The variant SULT1A1 genotype (SULT1A1*1/SULT1A1*2 or SULT1A1*2/SULT1A1*2) was associated with a significantly increased lung cancer risk in cases. 19424794_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19484729_human CYP2E1 and SULT1A1 activate an endogenous cellular molecule or a medium component to become mutagenic 19575027_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19706609_para-Nitrophenyl sulfate activation of human sulfotransferase 1A1 is consistent with intercepting the E[middle dot]PAP complex and reformation of E[middle dot]PAPS. 19776291_Observational study of gene-disease association. (HuGE Navigator) 19789190_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19863350_Observational study of gene-disease association. (HuGE Navigator) 19904771_study demonstrates that the loss of SULT1A1 appears to be a characteristic molecular signature of hepatocellular carcinoma. 19906068_The interaction between SULT1A1 and CYP1A2 can play an important role in hepatocarcinogenesis in the Chinese population. 19949855_Meta-analysis did not find a significant general relationship between SULT1A1 R213H polymorphism & the risk of breast cancer, but ethnic population analysis revealed a significantly increased breast cancer risk for HH allele carriers among Asians. 19949855_Meta-analysis of gene-disease association. (HuGE Navigator) 19956635_Uncategorized study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20032816_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20142249_Observational study of gene-disease association. (HuGE Navigator) 20192879_Observational study of genetic testing. (HuGE Navigator) 20204402_Observational study of gene-disease association. (HuGE Navigator) 20214802_Observational study of gene-disease association. (HuGE Navigator) 20309015_Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20417180_the structures of SULT1A2 and an allozyme of SULT1A1, SULT1A1 *3, bound with 3'-phosphoadenosine 5'-phosphate at 2.4 and 2.3A resolution, respectively, were determined. 20437850_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20505544_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20505990_Meta-analysis of gene-disease association. (HuGE Navigator) 20528568_Observational study of gene-disease association. (HuGE Navigator) 20529763_Observational study of gene-disease association. (HuGE Navigator) 20565970_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20620409_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628863_Only NAT1 showed a significant lower DNA methylation rate in the control group than in the tamoxifen-resistant breast cancer group, and no significant difference in methylation was found in COMT, CYP1A1, CYP2D6, and SULT1A1 genes. 20634891_Observational study of gene-disease association. (HuGE Navigator) 20663177_Meta-analysis of gene-disease association. (HuGE Navigator) 20663177_Polymorphism of SULT1A1 Arg213His is associated with breast cancer. 20734064_Observational study of gene-disease association. (HuGE Navigator) 20881232_Observational study of gene-disease association. (HuGE Navigator) 20881232_SULT1A1 1/2 does not contribute to the variation in SULT1A1 enzymatic activity when the 3'-UTR SNPs are included in the statistical model 20936502_Observational study of genotype prevalence. (HuGE Navigator) 20936502_the statistical hypothesis that SULT1A1 and SULT1A2 alleles are independently distributed was rejected; a strongly positive linkage was detected between SULT1A1*2 and SULT1A2*2 alleles in Turkey population 21072184_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21111704_mechanism of SULT1A1-catalyzed sulfation of adenosine 3',5'-diphosphate by para-nitrophenyl sulfate 21670965_SULT1A1 Arg213His polymorphism, ethnicity, and smoking may modulate environment-related cancer risk. 21695180_This meta-analysis demonstrates that there is no association between the SULT1A1 R213H polymorphism and colorectal cancer. 21977969_Women in Siberia with SNPS in CYP1A1 gene, in CYP1A2 gene,and in the SULT1A1 gene have an increased risk of development of breast cancer . 22011087_SULT1A1 Arg213His polymorphism is associated with breast cancer. 22081606_Stp1 is important for appropriate regulation of Stk1 function, hemolysin activity, autolysis, and GBS virulence 22524828_Arg213His polymorphism is not associated with lung cancer. 22678655_discussion of possible association between lower-activity of SULT1A1 with sudden cardiac death (both holiday sudden cardiac death in older adults and sudden infant death syndrome) [REVIEW] 22708928_We observed a previously unreported association between the SULT1A1 rs9282861 genotype and overall survival of breast cancer patients treated with adjuvant chemotherapy or tamoxifen. 23080433_The functional significance of the single nucleotide polymorphisms/haplotypes located upstream of the SULT1A1 start codon. 23157889_SULT1A1 variant allele increases breast cancer risk among subjects who were exposed to high smoked meat intake. 23358261_resequenced the SULT1A2 and SULT1A2 genes and identified 51 variations in ethnic Koreans, including 9 that were previously unknown. Allele frequencies, haplotype structures, LD blocks, and haplotype-tagging SNPs were determined. 23711090_Association between BRCA2 mutation and SULT1A1 gene deletion in male breast cancer emerged. 24010997_The SULT1A1 genetic variability is associated with cancer risk and response to therapy (review). 24039991_Data indicate that protein-ligand interaction energy by using docking Quantitative Structure-Activity Relationships(QSAR) models showed accuracy of 67.28%, 78.00% and 75.46%, for the isoforms SULT1A1, SULT1A3 and SULT1E1, respectively. 24307569_The SNP rs9282861 with GG genotype of SULT1A1 was associated with an elevated risk of total neural tube defects. 24763827_The results of this meta-analysis indicate that the SULT1A1 Arg213His polymorphism is associated with the risk bladder cancer under a recessive model. 25103078_The present study provided epidemiological evidence for a significantly increased risk of UCB in ever smokers with the Ala/Ala genotype of the GSTO1 gene and the Arg/Arg genotype of the SULT1A1 gene. 25194687_that SULT1A1 Arg213His polymorphism is associated with bladder cancer risk. 25225888_the SULT1A1 Arg213His polymorphism may contribute UADT cancer risk, but didn't show any association with breast cancer. 25314023_3'-Phosphoadenosine 5'-phosphosulfate binds antisynergistically to the subunits of the SULT1A1 dimer. 25370010_SULT1A1 is responsible for bioactivation of food genotoxicants 5-hydroxymethylfurfural and furfuryl alcohol. 25385181_Results indicate that SULT1A1 Arg(213)His may act as a low-penetrance risk allele for developing MBC and could be associated with a specific tumor subtype associated with HER2 overexpression. 25654087_The aim of the study was to assess whether selected single nucleotide polymorphisms of CYP1A1 and 2E1, GSTM1, GSTT1, and SULT1A1 influence susceptibility towards hepatocellular carcinoma. 25771868_Sulfo-conjugation of the multi-hydroxylated metabolites of benzene by human SULT1A1 may represent an important detoxifying pathway. 25819444_Data suggest that the substrate specificities of SULT1E1 and SULT1A1*1 include metabolites of tamoxifen (endoxifen, 4-hydroxytamoxifen, and N-desmethyltamoxifen); these metabolites are weak inhibitors of sulfation of estradiol by SULT1E1/SULT1A1*1. 26022216_Higher SULT1A1 levels were observed in rats as well as in humans exposed to high altitude, when compared to sea-level controls 26067475_A systematic analysis showed that three of the twelve human SULTs, SULT1A1, SULT1A3 and SULT1C4, displayed the strongest sulphating activity towards acetaminophen. 26169578_It metabolizes breast cancer drugs like afimoxifene and endoxifen by sulfation. 26340710_Sulfotransferase 1A1 Substrate Selectivity: A Molecular Clamp Mechanism. 26455829_The polymorphism of SULT1A1*2 is not associated with esophageal squamous cell carcinoma risk. 26906565_Cytosolic sulfotransferase 1A1 regulates HIV-1 minus-strand DNA elongation in primary human monocyte-derived macrophages. 27207664_We conclude from these studies that SULT1A1 is involved in the bioactivation of AA-I through the sulfonation of AL-I-NOH, contributing significantly to the toxicities of AA observed in vivo. 27221864_SULT1A1 high Copy Number Variation on high Estrogen Concentration and Tamoxifen-Associated Adverse Drug Reactions in Premenopausal Thai Breast Cancer. 27300114_SULT1A1 gene copy number affected the minor allele frequency for each single nucleotide polymorphisms tested. Before administration of exogenous hormones, increasing number of G alleles at rs9282861 was associated with earlier age at menopause, lower frequency of night sweats, and less severe insomnia. Variability in onset of menopause and symptoms before initiation of hormone therapy is partly due to SULT1A1 variation. 27356022_Epigallocatechin gallate (EGCG) displays high affinity and specificity for SULT1A1. The allosteric network is shown to involve 14 distinct complexes. ECGG binds both the allosteric site and, relatively weakly, the active site of SULT1A1. 28160022_SULT1A1 role in the metabolic detoxification of heterocyclic aromatic amines 28326452_The importance of SULT1A1 genotype for hepatic methyleugenol DNA adducts in humans, and a strong impact of SULT1A1 copy number variations on SULT1A1 hepatic phenotype. 28523759_Silencing the SULT1A1 gene led to changes in resveratrol metabolism, with higher intracellular accumulation of the nonmetabolized resveratrol. 28589969_the NQO1 Pro187Ser or SULT1A1 Arg213His polymorphism combination with smoking significantly confer susceptibility to BC. [META-ANALYSIS] 28867356_SULT1A1 copy number variation was negatively correlated with estrone-sulfate to estrone ratio predominantly in males (E1S/E1; p=0.03, r=-0.21) and may be associated with increased risk for common allergies. 28887105_No significant difference was observed in the RNA levels of CYP1A1 and SULT1A1 between the two groups. The frequency of expression of the CYP17 T/C variant tended to be higher and the A allele of COMT polymorphism together with down-regulation of its mRNA expression may be more frequent in Chinese women with idiopathic POI 29110586_This study demonstrates that the presence of His allele and Gln allele in case of SULT1A1 rs9282861 and XRCC1 rs25487, respectively, involve in lung cancer prognosis in Bangladeshi population. 29233949_SULT1A1 Arg213His (rs9282861) polymorphism might be associated with breast cancer risk, especially among Asian population. 29790428_Different ethnic and racial populations have varying degrees of SULT1A1-mediated sulfation activity. 29908303_Suggest SULT1A1/1A2 play a central role in furfuryl alcohol bioactivation and the formation of hemoglobin adducts. 30120701_Patients were categorized in three groups depending on the decreased SULT1A1 activity due to rs6839 and rs1042157: low activity group (rs6839 (GG) and rs1042157 (TT)); high activity group (rs6839 (AA) and rs1042157 (CC)); and medium activity group (all the other combinations of rs6839 and rs1042157). Associations between SULT1A1 phenotypes and clinical outcome (RFS) were explored 31671219_Prediagnostic circulating inflammation biomarkers and esophageal squamous cell carcinoma: A case-cohort study in Japan. 31835852_These findings highlight the key insights of structural consequences caused by R213H mutation, which would enrich the understanding regarding the role of SULT1A1 mutation in cancer development. 32152050_Interaction of the Brain-Selective Sulfotransferase SULT4A1 with Other Cytosolic Sulfotransferases: Effects on Protein Expression and Function. 33049293_Evaluation of a conserved tryptophanyl residue in donor substrate binding and catalysis by a phenol sulfotransferase (SULT1A1). 33297275_Investigating the molecular mechanism of hydroxylated bromdiphenyl ethers to inhibit the thyroid hormone sulfotransferase SULT1A1. 34162941_Insights into the substrate binding mechanism of SULT1A1 through molecular dynamics with excited normal modes simulations. | ENSMUSG00000000739 | Sult5a1 | 260.48971 | 1.0219786 | 0.0313649776 | 0.20698471 | 2.370673e-02 | 8.776335e-01 | 9.998360e-01 | No | Yes | 283.28606 | 35.183444 | 2.710124e+02 | 27.219673 | |
| ENSG00000196550 | 729533 | FAM72A | protein_coding | Q5TYM5 | FUNCTION: May play a role in the regulation of cellular reactive oxygen species metabolism. May participate in cell growth regulation. {ECO:0000269|PubMed:21317926}. | Alternative splicing;Cytoplasm;Mitochondrion;Reference proteome | hsa:729533; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrion [GO:0005739] | 18676834_Using deletion constructs, the authors find that Ugene binds to the first 25 amino acids of the UNG2 NH(2) terminus. They suggest that Ugene induction in cancer may contribute to the cancer phenotype by interacting with the BER pathway. 19755123_this study provides an additional possible mechanism of neurotoxicity in Alzheimer's disease, the induction of p17(FAM72B), through which Abeta acts to induce apoptosis and exhibit other Alzheimer's disease characteristics. 21317926_It was found that Ugene, designated herein as LMP1-induced protein (LMPIP), was induced, in a time-dependent manner, in EBV-infected peripheral blood mononuclear cells and LMP1-transfected 293 cells. 23900679_Data indicate that p17 (p17 amyloid-beta peptide-induced protein; known as Ugene, LMPIP, or FAM72A/B) drives the cell cycle into the G0/G1 phase and enhances survival of proliferating cells. 26206078_An epistemological characterization of the human tumorigenic neuronal paralogous FAM72 gene loci (FAM72A, FAM72B, FAM72C, FAM72D). 34819670_FAM72A antagonizes UNG2 to promote mutagenic repair during antibody maturation. | ENSMUSG00000055184 | Fam72a | 389.31115 | 1.1020886 | 0.1402402207 | 0.20034003 | 5.027537e-01 | 4.782926e-01 | 9.998360e-01 | No | Yes | 402.30667 | 64.878412 | 3.646614e+02 | 45.617336 | ||
| ENSG00000196693 | 7582 | ZNF33B | protein_coding | Q06732 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the zinc finger family of proteins. This gene shows decreased expression in cumulus cells derived from patients undergoing controlled ovarian stimulation. This gene is present in a gene cluster with several related zinc finger genes in the pericentromeric region of chromosome 10. Pseudogenes have been identified on chromosomes 7 and 10. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2015]. | hsa:7582; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385451_Observational study of gene-disease association. (HuGE Navigator) | 248.49165 | 0.8805949 | -0.1834496273 | 0.24613509 | 4.926069e-01 | 4.827667e-01 | 9.998360e-01 | No | Yes | 292.61765 | 39.886118 | 3.316900e+02 | 35.025117 | |||
| ENSG00000196711 | 389658 | ALKAL1 | protein_coding | Q6UXT8 | FUNCTION: Ligand for receptor tyrosine kinase LTK and perhaps receptor tyrosine kinase ALK; activation of ALK is reported conflictingly. {ECO:0000269|PubMed:25331893, ECO:0000269|PubMed:26418745, ECO:0000269|PubMed:26630010}. | 3D-structure;Alternative splicing;Reference proteome;Secreted;Signal | hsa:389658; | extracellular region [GO:0005576]; receptor signaling protein tyrosine kinase activator activity [GO:0030298]; receptor tyrosine kinase binding [GO:0030971]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of ERK5 cascade [GO:0070378]; positive regulation of neuron projection development [GO:0010976] | 25331893_Only two related secreted factors, FAM150A and FAM150B (family with sequence similarity 150 member A and member B), stimulated LTK phosphorylation. 26418745_In conclusion, these data show that ALK is robustly activated by the FAM150A/B ligands. 26630010_Data show that leukocyte tyrosine kinase (LTK) ligand FAM150A (augmentor-beta; AUG-beta) is specific for LTK and only weakly binds to anaplastic lymphoma kinase (ALK). 29317532_activation of ALK/LTK family receptors by small ALKAL proteins (FAM150, AUG) conserved in vertebrates | ENSMUSG00000087247 | Alkal1 | 43.80234 | 1.2561502 | 0.3290089538 | 0.46346785 | 5.379878e-01 | 4.632678e-01 | 9.998360e-01 | No | Yes | 45.55355 | 10.224816 | 3.668992e+01 | 6.716793 | ||
| ENSG00000196715 | 154807 | VKORC1L1 | protein_coding | Q8N0U8 | FUNCTION: Involved in vitamin K metabolism. Can reduce inactive vitamin K 2,3-epoxide to active vitamin K (in vitro), and may contribute to vitamin K-mediated protection against oxidative stress. Plays a role in vitamin K-dependent gamma-carboxylation of Glu residues in target proteins. {ECO:0000269|PubMed:21367861, ECO:0000269|PubMed:23928358, ECO:0000269|PubMed:24532791}. | Alternative splicing;Disulfide bond;Endoplasmic reticulum;Membrane;Oxidoreductase;Quinone;Redox-active center;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes an enzyme important in the vitamin K cycle, which is involved in the carboxylation of glutamate residues present in vitamin K-dependent proteins. The encoded enzyme catalyzes the de-epoxidation of vitamin K 2,3-epoxide. Oxidative stress may upregulate expression of this gene and the encoded protein may protect cells and membrane proteins form oxidative damage. This gene and a related gene (Gene ID: 79001) may have arisen by gene duplication of an ancestral gene. [provided by RefSeq, Oct 2016]. | hsa:154807; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; quinone binding [GO:0048038]; vitamin-K-epoxide reductase (warfarin-sensitive) activity [GO:0047057]; cellular response to oxidative stress [GO:0034599]; peptidyl-glutamic acid carboxylation [GO:0017187]; vitamin K metabolic process [GO:0042373] | 14765194_molecular cloning 17996924_VKORC1L1 does not affect the variability of warfarin dose requirement in a Japanese patient population 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21367861_VKORC1L1 is responsible for driving vitamin K-mediated intracellular antioxidation pathways critical to cell survival. 23928358_The involvement of VKORC1L1 in VKOR activity partly explains the low susceptibility of some extrahepatic tissues to vitamin K antagonists. 24532791_a concerted action of the four conserved cysteines of VKORC1L1 for active site regeneration 29054760_This study showed that polymorphisms of EPHX1 and VKORC1L1 could be determinants of stable warfarin doses. 29581108_we have established KO HEK 293T cell lines that express either endogenously VKORC1 or VKORC1L1, and found that VKORC1 is more sensitive to OACs than VKORC1L1, whereas rodenticides apparently inhibit both VKOR enzymes equally 29743176_The disputed loop sequence between the 1st and 2d transmembrane domain of VKOR in the cytoplasm. Using molecular dynamics simulations, a T-shaped stacking interaction between warfarin and Tyr 139, within the proposed TY139A warfarin-binding motif, was seen. Y25, A26, and Y139) were essential for warfarin binding to VKOR, with a reversible dynamic warfarin-binding pocket opening and conformational changes . | ENSMUSG00000066735 | Vkorc1l1 | 1390.35465 | 1.2677146 | 0.3422300055 | 0.14702743 | 5.050776e+00 | 2.461491e-02 | 9.998360e-01 | No | Yes | 1693.65341 | 203.930356 | 1.345107e+03 | 125.009418 | |
| ENSG00000196810 | CTBP1-DT | lncRNA | 548.57910 | 1.0632047 | 0.0884193966 | 0.15113192 | 3.403684e-01 | 5.596166e-01 | 9.998360e-01 | No | Yes | 576.49504 | 65.217880 | 4.820649e+02 | 42.672047 | |||||||||||
| ENSG00000196843 | 10865 | ARID5A | protein_coding | Q03989 | FUNCTION: Binds to AT-rich stretches in the modulator region upstream of the human cytomegalovirus major intermediate early gene enhancer. May act as repressor and down-regulate enhancer-dependent gene expressison (PubMed:8649988). May positively regulate chondrocyte-specific transcription such as of COL2A1 in collaboration with SOX9 and positively regulate histone H3 acetylation at chondrocyte-specific genes. May stimulate early-stage chondrocyte differentiation and inhibit later stage differention (By similarity). Can repress ESR1-mediated transcriptional activation; proposed to act as corepressor for selective nuclear hormone receptors (PubMed:15941852). As RNA-binding protein involved in the regulation of inflammatory response by stabilizing selective inflammation-related mRNAs, such as IL6, STAT3 and TBX21. Binds to stem loop structures located in the 3'UTRs of IL6, STAT3 and TBX21 mRNAs; at least for STAT3 prevents binding of ZC3H12A to the mRNA stem loop structure thus inhibiting its degradation activity. Contributes to elevated IL6 levels possibly implicated in autoimmunity processes. IL6-dependent stabilization of STAT3 mRNA may promote differentiation of naive CD4+ T-cells into T-helper Th17 cells. In CD4+ T-cells may also inhibit RORC-induced Th17 cell differentiation independently of IL6 signaling. Stabilization of TBX21 mRNA contributes to elevated interferon-gamma secretion in Th1 cells possibly implicated in the establishment of septic shock (By similarity). Stabilizes TNFRSF4/OX40 mRNA by binding to the conserved stem loop structure in its 3'UTR; thereby competing with the mRNA-destabilizing functions of RC3H1 and endoribonuclease ZC3H12A (By similarity). {ECO:0000250|UniProtKB:Q3U108, ECO:0000269|PubMed:15941852, ECO:0000269|PubMed:8649988}. | Activator;Alternative splicing;DNA-binding;Immunity;Innate immunity;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | Members of the ARID protein family, including ARID5A, have diverse functions but all appear to play important roles in development, tissue-specific gene expression, and regulation of cell growth (Patsialou et al., 2005 [PubMed 15640446]).[supplied by OMIM, Mar 2008]. | hsa:10865; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; androgen receptor binding [GO:0050681]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; estrogen receptor binding [GO:0030331]; identical protein binding [GO:0042802]; retinoid X receptor binding [GO:0046965]; RNA stem-loop binding [GO:0035613]; sequence-specific DNA binding [GO:0043565]; thyroid hormone receptor binding [GO:0046966]; transcription cis-regulatory region binding [GO:0000976]; transcription corepressor activity [GO:0003714]; transcription factor binding [GO:0008134]; cellular response to estrogen stimulus [GO:0071391]; cellular response to lipopolysaccharide [GO:0071222]; chondrocyte differentiation [GO:0002062]; innate immune response [GO:0045087]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of histone acetylation [GO:0035066]; positive regulation of interferon-gamma production [GO:0032729]; positive regulation of interleukin-17 production [GO:0032740]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of T-helper 1 cell cytokine production [GO:2000556]; positive regulation of T-helper 17 type immune response [GO:2000318]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of transcription by RNA polymerase II [GO:0006357] | 24782182_AT-rich-interactive domain-containing protein 5A functions as a negative regulator of retinoic acid receptor-related orphan nuclear receptor gammat-induced Th17 cell differentiation. 29044508_Overexpression of ARID5A resulted in more number of cells in G0/G1 phase of cell cycle | ENSMUSG00000037447 | Arid5a | 856.24925 | 0.9784564 | -0.0314205605 | 0.12607447 | 6.267286e-02 | 8.023202e-01 | 9.998360e-01 | No | Yes | 869.39155 | 113.877144 | 8.329809e+02 | 85.271139 | |
| ENSG00000196873 | 445571 | CBWD3 | protein_coding | Q5JTY5 | ATP-binding;Alternative splicing;Nucleotide-binding;Reference proteome | hsa:445571; | cytoplasm [GO:0005737]; ATP binding [GO:0005524] | ENSMUSG00000024878 | Cbwd1 | 201.32680 | 1.0450486 | 0.0635700315 | 0.24609589 | 6.721599e-02 | 7.954343e-01 | 9.998360e-01 | No | Yes | 153.45643 | 36.180675 | 1.426356e+02 | 25.929088 | ||||
| ENSG00000197008 | 7697 | ZNF138 | protein_coding | P52744 | FUNCTION: May be involved in transcriptional regulation as a repressor. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7697; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 124.49743 | 1.1156357 | 0.1578660016 | 0.26915013 | 3.452249e-01 | 5.568287e-01 | 9.998360e-01 | No | Yes | 258.41870 | 34.023336 | 2.349053e+02 | 24.060768 | |||||
| ENSG00000197062 | 7741 | ZSCAN26 | protein_coding | Q16670 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000305}. | Alternative splicing;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7741; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | ENSMUSG00000022228 | Zscan26 | 159.61764 | 0.7616695 | -0.3927630504 | 0.24483117 | 2.492114e+00 | 1.144179e-01 | 9.998360e-01 | No | Yes | 136.13310 | 26.552152 | 1.872568e+02 | 28.045018 | |||
| ENSG00000197302 | 124411 | KRBOX5 | protein_coding | Q7Z2F6 | Alternative splicing;Reference proteome | hsa:124411; | regulation of transcription, DNA-templated [GO:0006355] | 165.71196 | 0.9641078 | -0.0527336216 | 0.25381900 | 4.331520e-02 | 8.351329e-01 | 9.998360e-01 | No | Yes | 190.65116 | 27.811992 | 2.010804e+02 | 22.908291 | ||||||
| ENSG00000197380 | 147906 | DACT3 | protein_coding | Q96B18 | FUNCTION: May be involved in regulation of intracellular signaling pathways during development. Specifically thought to play a role in canonical and/or non-canonical Wnt signaling pathways through interaction with DSH (Dishevelled) family proteins. {ECO:0000269|PubMed:18538736}. | Coiled coil;Methylation;Phosphoprotein;Reference proteome;Wnt signaling pathway | hsa:147906; | cytoplasm [GO:0005737]; delta-catenin binding [GO:0070097]; identical protein binding [GO:0042802]; protein kinase A binding [GO:0051018]; protein kinase C binding [GO:0005080]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of cell growth [GO:0030308]; negative regulation of epithelial to mesenchymal transition [GO:0010719]; negative regulation of Wnt signaling pathway [GO:0030178]; Wnt signaling pathway [GO:0016055] | 18538736_Epigenetic repression of DACT3 leads to aberrant Wnt-beta-catenin signaling in colorectal cancer cells. 33054518_Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. | ENSMUSG00000078794 | Dact3 | 237.89896 | 0.9619540 | -0.0559601298 | 0.20688475 | 7.311160e-02 | 7.868590e-01 | 9.998360e-01 | No | Yes | 303.43487 | 49.853953 | 2.790765e+02 | 37.496298 | ||
| ENSG00000197451 | 3182 | HNRNPAB | protein_coding | Q99729 | FUNCTION: Binds single-stranded RNA. Has a high affinity for G-rich and U-rich regions of hnRNA. Also binds to APOB mRNA transcripts around the RNA editing site. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ubl conjugation | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are produced by RNA polymerase II and are components of the heterogeneous nuclear RNA (hnRNA) complexes. They are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene, which binds to one of the components of the multiprotein editosome complex, has two repeats of quasi-RRM (RNA recognition motif) domains that bind to RNAs. Two alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jul 2008]. | hsa:3182; | cytoplasm [GO:0005737]; dendrite [GO:0030425]; messenger ribonucleoprotein complex [GO:1990124]; neuronal ribonucleoprotein granule [GO:0071598]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; RNA polymerase II transcription regulator complex [GO:0090575]; mRNA binding [GO:0003729]; protein-containing complex binding [GO:0044877]; RNA binding [GO:0003723]; sequence-specific double-stranded DNA binding [GO:1990837]; cellular response to amino acid stimulus [GO:0071230]; epithelial to mesenchymal transition [GO:0001837]; mRNA modification [GO:0016556]; negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [GO:2000623]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of gene expression [GO:0010468]; regulation of intracellular mRNA localization [GO:1904580] | 12692135_physical association of p63alpha and ABBP1 led to a specific shift of FGFR-2 alternative splicing toward the K-SAM isoform essential for epithelial differentiation 24638979_Findings define HNRNPAB as an activator of EMT and metastasis in HCC that predicts poor clinical outcomes. 28763864_In HCC [hepatocellular carcinoma ] cells, hnRNPAB and Kap1 form protein complexes. The expression levels of hnRNPAB alone or in combination with Kap1 in HCC patients are important because they provide not only a predictor for HCC prognosis but also a therapeutic target for future studies. 31090062_findings offer new insights into the regulatory mechanisms that underlie HNRNPAB cancer-promoting activities and demonstrate that lnc-ELF209 is a HNRNPAB-regulated lncRNA that may play an important role in the inhibition of hepatocellular carcinoma progression 34396427_MicroRNA8063 targets heterogeneous nuclear ribonucleoprotein AB to inhibit the selfrenewal of colorectal cancer stem cells via the Wnt/betacatenin pathway. | ENSMUSG00000020358 | Hnrnpab | 25332.63610 | 0.9307992 | -0.1034581574 | 0.08659907 | 1.419074e+00 | 2.335559e-01 | 9.998360e-01 | No | Yes | 25034.93256 | 1754.587527 | 2.750212e+04 | 1488.860767 | |
| ENSG00000197943 | 5336 | PLCG2 | protein_coding | P16885 | FUNCTION: The production of the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) is mediated by activated phosphatidylinositol-specific phospholipase C enzymes. It is a crucial enzyme in transmembrane signaling. {ECO:0000269|PubMed:23000145}. | 3D-structure;Calcium;Disease variant;Hydrolase;Lipid degradation;Lipid metabolism;Phosphoprotein;Reference proteome;Repeat;SH2 domain;SH3 domain;Transducer | The protein encoded by this gene is a transmembrane signaling enzyme that catalyzes the conversion of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate to 1D-myo-inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) using calcium as a cofactor. IP3 and DAG are second messenger molecules important for transmitting signals from growth factor receptors and immune system receptors across the cell membrane. Mutations in this gene have been found in autoinflammation, antibody deficiency, and immune dysregulation syndrome and familial cold autoinflammatory syndrome 3. [provided by RefSeq, Mar 2014]. | hsa:5336; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; intracellular vesicle [GO:0097708]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; phosphatidylinositol phospholipase C activity [GO:0004435]; phospholipase C activity [GO:0004629]; phosphorylation-dependent protein binding [GO:0140031]; phosphotyrosine residue binding [GO:0001784]; protein kinase binding [GO:0019901]; protein tyrosine kinase binding [GO:1990782]; scaffold protein binding [GO:0097110]; activation of store-operated calcium channel activity [GO:0032237]; antifungal innate immune response [GO:0061760]; B cell differentiation [GO:0030183]; B cell receptor signaling pathway [GO:0050853]; calcium-mediated signaling [GO:0019722]; cell activation [GO:0001775]; cellular response to calcium ion [GO:0071277]; cellular response to lectin [GO:1990858]; cellular response to lipid [GO:0071396]; Fc-epsilon receptor signaling pathway [GO:0038095]; follicular B cell differentiation [GO:0002316]; inositol trisphosphate biosynthetic process [GO:0032959]; intracellular signal transduction [GO:0035556]; lipopolysaccharide-mediated signaling pathway [GO:0031663]; macrophage activation involved in immune response [GO:0002281]; negative regulation of programmed cell death [GO:0043069]; phosphatidylinositol biosynthetic process [GO:0006661]; phospholipid catabolic process [GO:0009395]; platelet activation [GO:0030168]; positive regulation of calcium-mediated signaling [GO:0050850]; positive regulation of cell cycle G1/S phase transition [GO:1902808]; positive regulation of dendritic cell cytokine production [GO:0002732]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of gene expression [GO:0010628]; positive regulation of I-kappaB phosphorylation [GO:1903721]; positive regulation of interleukin-10 production [GO:0032733]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-2 production [GO:0032743]; positive regulation of interleukin-23 production [GO:0032747]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of macrophage cytokine production [GO:0060907]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of neuroinflammatory response [GO:0150078]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NLRP3 inflammasome complex assembly [GO:1900227]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of phagocytosis, engulfment [GO:0060100]; positive regulation of reactive oxygen species biosynthetic process [GO:1903428]; positive regulation of receptor internalization [GO:0002092]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of type I interferon production [GO:0032481]; regulation of calcineurin-NFAT signaling cascade [GO:0070884]; regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043122]; regulation of lipid metabolic process [GO:0019216]; release of sequestered calcium ion into cytosol [GO:0051209]; response to axon injury [GO:0048678]; response to yeast [GO:0001878]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; T cell receptor signaling pathway [GO:0050852]; toll-like receptor signaling pathway [GO:0002224]; Wnt signaling pathway [GO:0016055] | 12049640_collagen receptor glycoprotein VI and alphaIIbbeta3 trigger distinct patterns of receptor signalling in platelets, leading to tyrosine phosphorylation of PLCgamma2 (integrin alphaiibbeta3) 12181444_Two tyrosine residues in regulating the activity of PLCgamma2 12359094_full-length cDNA for human PLCgamma2 and expressed it in E. coli using the expression vector pT5T 12813055_PLCG2 has a signaling role in platelet glycoprotein Ib alpha calcium flux and cytoskeletal reorganization 14606067_in gastric cancer, protein translocation of PLCgamma2 and PKCalpha is critical event in the process of apoptosis induction. 15509800_PLC-gamma2 is phosphorylated on Y753, Y759, and Y1217 in response to engagement of the B-cell receptor 15744341_The PLCgamma2 is present in the majority of mediastinal B cell lymphomas. 15972651_PLC-gamma1 and PLC-gamma2 both regulate the functions of ITAM-containing receptors, whereas only PLC-gamma2 regulates the function of DAP10-coupled receptors. 16172125_novel mechanism of PLCgamma(2) activation by Rac GTPases involving neither protein tyrosine phosphorylation nor PI3K-mediated generation of PtdInsP(3) 17023658_observations suggest a model in which TFII-I suppresses agonist-induced calcium entry by competing with TRPC3 for binding to phospholipase C-gamma 18000612_intracellular mediators and pathways activated by leptin downstream of JAK2 were found to include phosphatidylinositol-3 kinase, phospholipase Cgamma2 and protein kinase C, as well as the p38 MAP kinase-phospholipase A(2) axis. 18022864_Plasmacytoid dendritic cells express a signalosome consisting of Lyn, Syk, Btk, Slp65 (Blnk) and PLCgamma2. Triggering CD303 leads to tyrosine phosphorylation of Syk, Slp65, PLCgamma2 & cytoskeletal proteins. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18728011_rac regulates its effector phospholipase Cgamma2 through interaction with a split pleckstrin homology domain 19086053_Observational study of gene-disease association. (HuGE Navigator) 19308021_Observational study of gene-disease association. (HuGE Navigator) 19965664_Data show that RTX treatment results in a time-dependent inhibition of the BCR-signaling cascade involving Lyn, Syk, PLC gamma 2, Akt, and ERK, and calcium mobilization. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20068106_SYK, together with phospholipase Cgamma2, may serve as potential biomarkers to predict dasatinib therapeutic response in patients. 20086178_Data show that bile acid reflux present in patients with BE may increase reactive oxygen species production and cell proliferation via activation of PI-PLCgamma2, ERK2 MAP kinase, and NADPH oxidase NOX5-S, thereby contributing to the development of EA. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21245382_Rac2 binding in the absence of lipid surfaces was not able to activate phospholipase C gamma 2. 21506110_Data indicate a role for PLCgamma2 and Ca(2+) signaling through the modulation of MEK/ERK in IL3/GM-csf stimulated human hematopoietic stem/progenitor cells. 22236196_Genomic deletions in PLCG2 cause gain of PLCgamma(2) function, leading to signaling abnormalities in multiple leukocyte subsets and a phenotype encompassing both excessive and deficient immune function. 22837484_PLCgamma2 participates in T cell receptor (TCR) signal transduction and plays a role in T cell selection in a transgenic mouse model. 23000145_Overexpression of the altered p.Ser707Tyr protein and ex vivo experiments using affected individuals' leukocytes showed clearly enhanced PLCgamma2 activity. 23039362_Associations between treatment response and Lyn, Syk, PLCgamma2 and ERK were not found. 23555801_BANK1 and BLK have roles in B-cell signaling through phospholipase C gamma 2 24080446_Single-nucleotide polymorphisms in PLCG2 gene is associated with breast cancer risk after menopausal hormone replacement therapy. 24127488_down-regulation of PLCgamma2-beta-catenin pathway occurs in mice and humans and leads to myeloid-derived suppressor cells-mediated tumor expansion. 24166973_early Ca(2+) fluxing provides feed-forward signal amplification by promoting anchoring of the PLCgamma2 C2 domain to phospho-SLP65. 24489640_The relationship between upstream tyrosine kinase SYK and its target, PLCgamma2, is maximally predictive and sufficient to distinguish chronic lymphocytic leukemia from healthy controls. 24868545_amarogentin prevents platelet activation through the inhibition of PLC gamma2-PKC cascade and MAPK pathway 24869598_identified three distinct mutations in PLCgamma2 in two patients resistant to ibrutinib 25012946_Data show that phospholipase Cgamma2 (PLCgamma2) is strongly expressed in B cell non-Hodgkin lymphoma and especially in a large subset of Diffuse large B-cell lymphoma (DLBCL). 25227611_The autoinhibitory C-terminal SH2 domain of phospholipase C-gamma2 stabilizes B cell receptor signalosome assembly. 25349203_PLCG2 missense mutation is a risk factor in the development of steroid sensitive nephrotic syndrome in childhood. 25972157_Characterization of the effect of missense point-mutation at R665W in PLCG2 on signaling mechanisms of ibrutinib resistance in chronic lymphocytic leukemia cells. 27196803_The results suggest a new mechanism of PLCgamma activation with unique thermodynamic features and assign a novel regulatory role to its spPH domain. 27442322_Ocular manifestations of phospholipase-Cgamma2-associated antibody deficiency and immune dysregulation show mutations in the PLC[gamma]2 gene leading to aberrant function of immune cells and overproduction of interleukin-1 [beta] (IL-1[beta]). 27542411_R665W and L845F be referred to as allomorphic rather than hypermorphic mutations of PLCG2 Rerouting of the transmembrane signals emanating from BCR and converging on PLCgamma2 through Rac in ibrutinib-resistant CLL cells may provide novel drug treatment strategies to overcome ibrutinib resistance mediated by PLCG2 mutations or to prevent its development in ibrutinib-treated CLL patients. 28366935_finding that mutations or polymorphisms in two putative calcium-regulated domains of PLCG2 are associated with ibrutinib-resistant CLL adds to the evidence supporting complex regulatory shifts in the PLCG2 protein likely occurring during the development of resistance 28714976_Data show that protein-altering changes are in PLCG2, ABI3, and TREM2 genes highly expressed in microglia and highlight an immune-related protein-protein interaction network in Alzheimer's disease. 28786489_Syk-induced signals in bone marrow stromal cell lines are mediated by phospholipase C gamma1 (PLCgamma1) in osteogenesis and PLCgamma2 in adipogenesis. 29381098_While BTK/PLCG2 mutations have characteristics suggesting that they can drive ibrutinib resistance, this conclusion remains formally unproven until specific inhibition of such mutations is shown to cause regression of ibrutinib-resistant chronic lymphocytic leukemia . Data suggest that alternative mechanisms of resistance do exist in some patients. 30326945_suggest distinct effects of the microglial genes, ABI3 and PLCG2 in neurodegenerative diseases that harbor significant vs. low/no amyloid ss pathology 30705288_Study shows that rare coding variants in TREM2, PLCG2, and ABI3 modulate susceptibility to Alzheimer disease in populations from Argentina, and they may have a European heritage. 31131421_The effect of the PLCG2 rs72824905-G on 7 neurodegenerative diseases and longevity, was studied in 53,627 patients, 3,516 long-lived individuals and 149,290 study-matched controls. It was associated with reduced Alzheimer disease, Lewy-body dementia, and frontotemporal dementia. It had no effect on Parkinson disease, amyotrophic lateral sclerosis or multiple sclerosis. It was also associated with longevity. 31560769_We performed pathway analyses on the largest available collection of advanced age-related macular cases and controls in the world. Eight genes strongly contributed to significant pathways from the three larger databases, and one gene (PLCG2) was central to significant pathways from all four databases. This is, to our knowledge, the first study to identify PLCG2 as a candidate gene for AMD based solely on genetic burden. 32014489_A new report of autoinflammation and PLCG2-associated antibody deficiency and immune dysregulation (APLAID) with a homozygous pattern from Iran. 32166339_PLCG2 protective variant p.P522R modulates tau pathology and disease progression in patients with mild cognitive impairment. 32184360_Noncatalytic Bruton's tyrosine kinase activates PLCgamma2 variants mediating ibrutinib resistance in human chronic lymphocytic leukemia cells. 32232486_Novel BCL2 mutations in venetoclax-resistant, ibrutinib-resistant CLL patients with BTK/PLCG2 mutations. 32514138_Alzheimer's-associated PLCgamma2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. 32671674_Severe Autoinflammatory Manifestations and Antibody Deficiency Due to Novel Hypermorphic PLCG2 Mutations. 32894242_Examination of the Effect of Rare Variants in TREM2, ABI3, and PLCG2 in LOAD Through Multiple Phenotypes. 32917267_The Alzheimer's disease-associated protective Plcgamma2-P522R variant promotes immune functions. 33089525_A novel somatic PLCG2 variant associated with resistance to BTK and SYK inhibition in chronic lymphocytic leukemia. 33092647_Association of ABI3 and PLCG2 missense variants with disease risk and neuropathology in Lewy body disease and progressive supranuclear palsy. 33523007_PLCG2 rs72824905 Variant Reduces the Risk of Alzheimer's Disease and Multiple Sclerosis. 33645887_Neutrophil Phospholipase Cgamma2 Drives Autoantibody-Induced Arthritis Through the Generation of the Inflammatory Microenvironment. 33823896_TREM2/PLCgamma2 signalling in immune cells: function, structural insight, and potential therapeutic modulation. 34093563_Case Report: A Rare Case of Autoinflammatory Phospholipase Cgamma2 (PLCgamma2)-Associated Antibody Deficiency and Immune Dysregulation Complicated With Gangrenous Pyoderma and Literature Review. 34157287_The role of PLCgamma2 in immunological disorders, cancer, and neurodegeneration. 34607960_Phospholipase Cgamma2 regulates endocannabinoid and eicosanoid networks in innate immune cells. 34615897_PLCgamma2 regulates TREM2 signalling and integrin-mediated adhesion and migration of human iPSC-derived macrophages. 34653364_Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. 35180881_PLCG2 is associated with the inflammatory response and is induced by amyloid plaques in Alzheimer's disease. 35196427_Mechanisms of Resistance to Noncovalent Bruton's Tyrosine Kinase Inhibitors. | ENSMUSG00000034330 | Plcg2 | 514.20556 | 0.6987916 | -0.5170659207 | 0.16465139 | 9.545147e+00 | 2.004788e-03 | 5.109345e-01 | No | Yes | 447.41269 | 48.217223 | 6.057141e+02 | 51.068364 | |
| ENSG00000198130 | 26275 | HIBCH | protein_coding | Q6NVY1 | FUNCTION: Hydrolyzes 3-hydroxyisobutyryl-CoA (HIBYL-CoA), a saline catabolite. Has high activity toward isobutyryl-CoA. Could be an isobutyryl-CoA dehydrogenase that functions in valine catabolism. Also hydrolyzes 3-hydroxypropanoyl-CoA. {ECO:0000269|PubMed:8824301}. | 3D-structure;Acetylation;Alternative splicing;Branched-chain amino acid catabolism;Disease variant;Hydrolase;Mitochondrion;Phosphoprotein;Reference proteome;Transit peptide | PATHWAY: Amino-acid degradation; L-valine degradation. | This gene encodes the enzyme responsible for hydrolysis of both HIBYL-CoA and beta-hydroxypropionyl-CoA. Mutations in this gene have been associated with 3-hyroxyisobutyryl-CoA hydrolase deficiency. Alternative splicing results in multiple transcript variants.[provided by RefSeq, May 2010]. | hsa:26275; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; 3-hydroxyisobutyryl-CoA hydrolase activity [GO:0003860]; branched-chain amino acid catabolic process [GO:0009083]; valine catabolic process [GO:0006574] | 17160907_Molecular analysis in both patients uncovered mutations in the HIBCH gene, including one missense mutation in a conserved part of the protein and two mutations affecting splicing. 20877624_Observational study of gene-disease association. (HuGE Navigator) 24299452_findings demonstrated a novel homozygous pathogenic missense mutation c.950G | ENSMUSG00000041426 | Hibch | 508.40762 | 0.8981198 | -0.1550201413 | 0.15423027 | 9.999664e-01 | 3.173186e-01 | 9.998360e-01 | No | Yes | 482.79149 | 80.404323 | 5.334896e+02 | 68.608271 |
| ENSG00000198160 | 57708 | MIER1 | protein_coding | Q8N108 | FUNCTION: Transcriptional repressor regulating the expression of a number of genes including SP1 target genes. Probably functions through recruitment of HDAC1 a histone deacetylase involved in chromatin silencing. {ECO:0000269|PubMed:12482978}. | Alternative splicing;Cytoplasm;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a protein that was first identified in Xenopus laevis by its role in a mesoderm induction early response (MIER). The encoded protein functions as a transcriptional regulator. Alternatively spliced transcript variants encode multiple isoforms, some of which lack a C-terminal nuclear localization signal. [provided by RefSeq, May 2013]. | hsa:57708; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; transcription repressor complex [GO:0017053]; histone deacetylase binding [GO:0042826]; transcription corepressor activity [GO:0003714]; chromatin remodeling [GO:0006338]; histone deacetylation [GO:0016575]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; regulation of transcription, DNA-templated [GO:0006355] | 12242014_Results demonstrate that alternate use of a facultative intron regulates the subcellular localization of hMI-ER1 proteins and this may have important implications for hMI-ER1 function. 12482978_we investigated the role of hMI-ER1alpha and hMI-ER1beta in the regulation of transcription.We demonstrate that this repressor activity is due to interaction and recruitment of a trichostatin A-sensitive histone deacetylase 1 (HDAC1). 15117948_the association of hMI-ER1 with Sp1 represents a novel mechanism for the negative regulation of Sp1 target promoters 18665173_Loss of nuclear MI-ER1 alpha might contribute to the development of invasive breast carcinoma 22384264_Differential splicing alters subcellular localization of the alpha but not beta isoform of the MIER1 transcriptional regulator in breast cancer cells 23277184_the first immunohistochemical study of the MIER1alpha protein expression pattern in human tissues, is reported. 24376786_nuclear targeting of MIER1alpha requires an intact ELM2 domain and is dependent on interaction with HDAC1/2 26281834_Insulin and IGF-1 alter the subcellular localization of MIER1alpha in breast carcinoma cells. 26938916_Biochemical analysis of the BAHD1-associated multiprotein complex identifies MIER proteins as novel partners of BAHD1 and suggests that BAHD1-MIER interaction forms a hub for histone deacetylases and methyltransferases 28046085_Histone deacetylase assays confirmed that MIER2, but not MIER3 complexes, have associated deacetylase activity. | ENSMUSG00000028522 | Mier1 | 290.41779 | 0.9863234 | -0.0198673794 | 0.21343019 | 8.343899e-03 | 9.272185e-01 | 9.998360e-01 | No | Yes | 289.65105 | 54.261803 | 3.001255e+02 | 43.384339 | |
| ENSG00000198176 | 7027 | TFDP1 | protein_coding | Q14186 | FUNCTION: Can stimulate E2F-dependent transcription. Binds DNA cooperatively with E2F family members through the E2 recognition site, 5'-TTTC[CG]CGC-3', found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication (PubMed:8405995, PubMed:7739537). The E2F1:DP complex appears to mediate both cell proliferation and apoptosis. Blocks adipocyte differentiation by repressing CEBPA binding to its target gene promoters (PubMed:20176812). {ECO:0000269|PubMed:20176812, ECO:0000269|PubMed:7739537, ECO:0000269|PubMed:8405995}. | 3D-structure;Acetylation;Activator;Alternative splicing;Cell cycle;Cytoplasm;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of a family of transcription factors that heterodimerize with E2F proteins to enhance their DNA-binding activity and promote transcription from E2F target genes. The encoded protein functions as part of this complex to control the transcriptional activity of numerous genes involved in cell cycle progression from G1 to S phase. Alternative splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 1, 15, and X.[provided by RefSeq, Jan 2009]. | hsa:7027; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; Rb-E2F complex [GO:0035189]; RNA polymerase II transcription regulator complex [GO:0090575]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; protein domain specific binding [GO:0019904]; anoikis [GO:0043276]; epidermis development [GO:0008544]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of fat cell proliferation [GO:0070345]; positive regulation of DNA-binding transcription factor activity [GO:0051091]; positive regulation of G1/S transition of mitotic cell cycle [GO:1900087]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of DNA biosynthetic process [GO:2000278]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription involved in G1/S transition of mitotic cell cycle [GO:0000083]; transcription by RNA polymerase II [GO:0006366] | 12029633_TFDP1, CUL4A, and CDC16 are probable targets of an amplification mechanism and therefore may be involved, together or separately, in development and/or progression of some hepatocellular carcinomas 12607600_expression of the transcription factor DP-1 and its heterodimeric partner E2F-1 in non-Hodgkin lymphoma 14618416_TFDP1 may have a role in progression of some hepatocellular carcinomas by promoting growth of the tumor cells 15863509_DP-1alpha is a novel isoform of DP-1 that acts as a dominant-negative regulator of cell cycle progression 16135794_DP1 is a critical direct target of ARF. 18687693_SOCS-3 acts as a negative regulator of the cell cycle progression under E2F/DP-1 control by interfering with heterodimer formation between DP-1 and E2F 19738611_Observational study of gene-disease association. (HuGE Navigator) 19995430_13q34 amplification may be of relevance in tumor progression of breast cancers by inducing overexpression of CUL4A and TFDP1, important in cell cycle regulation. These genes were also overexpressed in non-basal-like tumor samples. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20513349_the DP-1 'Stabilon' domain was a C-terminal acidic motif and was quite important for DP-1 stability. 21715488_The authors demonstrate that adenovirus E1A binds to E2F/DP-1 complexes through a direct interaction with DP-1 and may selectively activate a subset of E2F-regulated cellular genes during infection. 23934193_somatic mutations in DP-1 uncouple normal control of the E2F pathway, and thus define a new mechanism that could contribute to aberrant proliferation in tumor cells 25133581_The TFDP1 indel84 mutation generates a gain-of-function phenotype by increasing cell proliferation, migration, and invasion of colorectal cancer cells. 26684807_Amplification of CUL4A, IRS2, and TFDP1 genes showed a significant difference in disease-free survival by both univariate and multivariate survival analyses in intrahepatic cholangiocarcinoma. 27323154_According to our study results, the gene TFDP1 and the cell cycle pathway are strongly associated with high-grade glioblastoma multiforme (GBM); this result may provide new insights into the pathogenesis of GBM. 27802335_role for E2F1 and TFDP1 in the transcriptional regulation of PITX1 in articular chondrocytes 27825926_Here, the authors show that an acidic region of DP1, whose function has remained elusive, binds to the plekstrin homology (PH) domain of the p62 subunit of TFIIH that contributes to transcriptional activation. 27871936_COMMD9 participates in TFDP1/E2F1 activation and plays a critical role in non-small cell lung cancer. 30365067_DILC may mediate the crosstalk between the cascades of IL6/STAT3 and TNFalpha signaling, indicating that DILC may act as a prognostic biomarker of sepsis, and may serve as a potential therapeutic target for the treatment of sepsis. 30638096_We observed a downregulation of TFDP1 in the endometrium cells of women with deep infiltrating endometriosis when compared to the controls 31783876_It uncovered E2F1 and TFDP1 as transport substrates of KPNA2 being retained in the cytoplasm upon KPNA2 ablation, thereby resulting in reduced STMN1 expression. 32066912_miR-4711-5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. | ENSMUSG00000038482 | Tfdp1 | 3232.90756 | 0.9466553 | -0.0790889475 | 0.10092065 | 6.047010e-01 | 4.367900e-01 | 9.998360e-01 | No | Yes | 4315.23063 | 344.524162 | 4.669547e+03 | 288.255653 | |
| ENSG00000198324 | 144717 | PHETA1 | protein_coding | Q8N4B1 | FUNCTION: Plays a role in endocytic trafficking. Required for receptor recycling from endosomes, both to the trans-Golgi network and the plasma membrane. {ECO:0000269|PubMed:21233288}. | 3D-structure;Alternative splicing;Cytoplasmic vesicle;Endosome;Golgi apparatus;Phosphoprotein;Reference proteome | This gene encodes a protein that localizes to the endosome and interacts with the enzyme, inositol polyphosphate 5-phosphatase OCRL-1. Alternate splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:144717; | clathrin-coated vesicle [GO:0030136]; cytosol [GO:0005829]; early endosome [GO:0005769]; recycling endosome [GO:0055037]; trans-Golgi network [GO:0005802]; protein homodimerization activity [GO:0042803]; endosome organization [GO:0007032]; receptor recycling [GO:0001881]; retrograde transport, endosome to Golgi [GO:0042147] | 20133602_Two closely related endocytic proteins, Ses1 and Ses2, which interact with OCRL, were identified. The interaction is mediated by a short amino acid motif similar to that used by the rab-5 effector APPL1. 21233288_Two novel OCRL1-binding proteins, termed inositol polyphosphate phosphatase interacting protein of 27 kDa (IPIP27)A and B (also known as Ses1 and 2), that also bind the related 5-phosphatase Inpp5b, were identified. 32152089_Deficiency in the endocytic adaptor proteins PHETA1/2 impairs renal and craniofacial development. | ENSMUSG00000044134 | Pheta1 | 1098.06656 | 1.1112169 | 0.1521404312 | 0.13356680 | 1.319770e+00 | 2.506333e-01 | 9.998360e-01 | No | Yes | 1189.53450 | 161.178669 | 9.794508e+02 | 103.680273 | |
| ENSG00000198382 | 7405 | UVRAG | protein_coding | Q9P2Y5 | FUNCTION: Versatile protein that is involved in regulation of different cellular pathways implicated in membrane trafficking. Involved in regulation of the COPI-dependent retrograde transport from Golgi and the endoplasmic reticulum by associating with the NRZ complex; the function is dependent on its binding to phosphatidylinositol 3-phosphate (PtdIns(3)P) (PubMed:16799551, PubMed:18552835, PubMed:20643123, PubMed:24056303, PubMed:28306502). During autophagy acts as regulatory subunit of the alternative PI3K complex II (PI3KC3-C2) that mediates formation of phosphatidylinositol 3-phosphate and is believed to be involved in maturation of autophagosomes and endocytosis. Activates lipid kinase activity of PIK3C3 (PubMed:16799551, PubMed:20643123, PubMed:24056303, PubMed:28306502). Involved in the regulation of degradative endocytic trafficking and cytokinesis, and in regulation of ATG9A transport from the Golgi to the autophagosome; the functions seems to implicate its association with PI3KC3-C2 (PubMed:16799551, PubMed:20643123, PubMed:24056303). Involved in maturation of autophagosomes and degradative endocytic trafficking independently of BECN1 but depending on its association with a class C Vps complex (possibly the HOPS complex); the association is also proposed to promote autophagosome recruitment and activation of Rab7 and endosome-endosome fusion events (PubMed:18552835, PubMed:28306502). Enhances class C Vps complex (possibly HOPS complex) association with a SNARE complex and promotes fusogenic SNARE complex formation during late endocytic membrane fusion (PubMed:24550300). In case of negative-strand RNA virus infection is required for efficient virus entry, promotes endocytic transport of virions and is implicated in a VAMP8-specific fusogenic SNARE complex assembly (PubMed:24550300). {ECO:0000269|PubMed:18552835, ECO:0000269|PubMed:20643123, ECO:0000269|PubMed:24056303, ECO:0000269|PubMed:28306502, ECO:0000305}.; FUNCTION: Involved in maintaining chromosomal stability. Promotes DNA double-strand break (DSB) repair by association with DNA-dependent protein kinase complex DNA-PK and activating it in non-homologous end joining (NHEJ) (PubMed:22542840). Required for centrosome stability and proper chromosome segregation (PubMed:22542840). {ECO:0000269|PubMed:22542840}. | 3D-structure;Alternative splicing;Centromere;Chromosome;Coiled coil;Cytoplasmic vesicle;DNA damage;DNA repair;Endoplasmic reticulum;Endosome;Lysosome;Phosphoprotein;Reference proteome | This gene complements the ultraviolet sensitivity of xeroderma pigmentosum group C cells and encodes a protein with a C2 domain. The protein activates the Beclin1-PI(3)KC3 complex, promoting autophagy and suppressing the proliferation and tumorigenicity of human colon cancer cells. Chromosomal aberrations involving this gene are associated with left-right axis malformation and mutations in this gene have been associated with colon cancer. [provided by RefSeq, Jul 2008]. | hsa:7405; | autophagosome membrane [GO:0000421]; centrosome [GO:0005813]; chromosome, centromeric region [GO:0000775]; cytoplasm [GO:0005737]; early endosome [GO:0005769]; endoplasmic reticulum [GO:0005783]; endosome [GO:0005768]; late endosome [GO:0005770]; lysosome [GO:0005764]; lytic vacuole [GO:0000323]; midbody [GO:0030496]; phagocytic vesicle [GO:0045335]; phosphatidylinositol 3-kinase complex, class III [GO:0035032]; SH3 domain binding [GO:0017124]; SNARE binding [GO:0000149]; autophagosome maturation [GO:0097352]; autophagy [GO:0006914]; centrosome cycle [GO:0007098]; chromosome segregation [GO:0007059]; DNA repair [GO:0006281]; double-strand break repair via classical nonhomologous end joining [GO:0097680]; maintenance of Golgi location [GO:0051684]; multivesicular body sorting pathway [GO:0071985]; phosphatidylinositol-3-phosphate biosynthetic process [GO:0036092]; positive regulation of autophagosome maturation [GO:1901098]; protein phosphorylation [GO:0006468]; receptor catabolic process [GO:0032801]; regulation of autophagy [GO:0010506]; regulation of cytokinesis [GO:0032465]; regulation of protein serine/threonine kinase activity [GO:0071900]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890]; SNARE complex assembly [GO:0035493]; spindle organization [GO:0007051]; viral entry into host cell [GO:0046718] | 17106237_UVRAG: a new player in autophagy and tumor cell growth. 18495205_Frameshift mutations in the polyadenine tract in UVRAG gene are present in gastric carcinomas. 18552835_Data suggest that a Beclin1-binding autophagic tumour suppressor, UVRAG, interacts with the class C Vps complex, a key component of the endosomal fusion machinery. 18641390_Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1 18843052_These results suggest that mammalian cells have at least two distinct class III PI3-kinase complexes, and that beclin 1 interacts distinctly with mammalian Atg14 and UVRAG in two of these complexes. 20163458_Observational study of gene-disease association. (HuGE Navigator) 20163458_There is an association of UVRAG polymorphisms with susceptibility to non-segmental vitiligo in a Korean sample. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20643123_A specific sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates both receptor degradation and cytokinesis, whereas ATG14L, a PI3K-III subunit involved in autophagy, is not required. 20724836_Data indicate that mechanisms other than autophagy contribute to the tumorigenicity of microsatellite unstable colon carcinomas with monoallelic UVRAG mutation. 21597469_UVRAG has an essential role in the intrinsic mitochondrial pathway of apoptosis by regulating the localization of Bax. 21606679_UVRAG has cytoprotective functions in the cytosol that control the localization of Bax in tumor cells exposed to apoptotic stimuli 22314358_analysis of how the Beclin1 coiled-coil domain interface regulates homodimer and heterodimer formation with Atg14L and UVRAG 22493499_in Escherichia coli-containing phagosomes of mouse macrophages, Slamf1 interacts with the class III PI3K Vps34 in a complex with Beclin-1 and UVRAG 22542840_UVRAG promotes DNA double-strand-break repair by directly binding and activating DNA-PK in nonhomologous end joining. Disruption of UVRAG increases genetic instability and sensitivity of cells to irradiation. 23200933_Akt1 may inhibit autophagy by decreasing UVRAG expression, which also sensitizes cancer cells to UV irradiation. 24550300_UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. 24882360_The reports that Grb2, when in excess, interacts with ultraviolet radiation resistance-associated gene protein (UVRAG) under excess conditions of AICD-Grb2 or Grb2. 24956373_Beclin 1 and UVRAG confer protection against radiation-induced DNA DNA double strand breaks and may maintain centrosome stability in established tumor cells. 25533187_Dephosphorylation of UVRAG facilitates the lysosomal degradation of epidermal growth factor receptor (EGFR), reduces EGFR signaling, and suppresses cancer cell proliferation and tumor growth. 25807108_HCV, by differentially inducing the expression of Rubicon and UVRAG, temporally regulated the autophagic flux to enhance its replication. 26139536_tubulation requires mTOR activity, and we identified two direct mTOR phosphorylation sites on UVRAG (S550 and S571) that activate VPS34. 26234763_UVRAG frameshift is expressed as a truncated protein in colorectal cancer with microsatellite instability and promotes tumorigenesis. UVRAG(FS) expression renders cells more sensitive to standard chemotherapy regimen due to a DNA repair defect. 26717041_Under-expression of UVRAG is associated with colorectal cancer. 27203177_UVRAG is a regulator of CRL4(DDB2)-mediated nucleotide excision repair and its expression levels may influence melanoma predisposition. 27439570_UVRAG plays an essential role in protecting cells from UV-induced DNA damage by activating the nucleotide excision repair pathway 28982601_miR-216b enhances the efficacy of vemurafenib and reverses drug resistance by targeting Beclin-1, UVRAG and ATG5 in melanoma. 29277783_Our findings suggest that up-regulation of UVRAG by HDAC1 inhibition potentiates DNA-damage-mediated cell death in colorectal cancer cells 29710477_our study showed that the expression of miR-125b-5p is downregulated in peripheral blood mononuclear cells of systemic lupus erythematosus patients, accompanied by increased UVRAG expression and autophagy activation 29866835_To induce the redistribution of Beclin 1 among its self-associated form or Atg14L/UVRAG-containing complexes enhances both autophagy and endolysosomal trafficking. 30061422_results establish UVRAG as an important effector for melanocytes' response to alpha-MSH signaling as a direct target of MITF and reveal the molecular basis underlying the association between oncogenic BRAF and compromised UV protection in melanoma. 30221685_miR183 inhibits starvationinduced autophagy and apoptosis by targeting UVRAG in human gastric cancer cells. 30686098_UVRAG is ubiquitinated by SMURF1 at lysine residues 517 and 559, which decreases the association of UVRAG with RUBCN and promotes autophagosome maturation. 30894053_GORASP2/GRASP55 collaborates with the PtdIns3K UVRAG complex to facilitate autophagosome-lysosome fusion. 34646380_TNF-alpha-dependent neuronal necroptosis regulated in Alzheimer's disease by coordination of RIPK1-p62 complex with autophagic UVRAG. | ENSMUSG00000035354 | Uvrag | 370.88936 | 1.0040514 | 0.0058331035 | 0.17988675 | 1.041537e-03 | 9.742545e-01 | 9.998360e-01 | No | Yes | 324.71496 | 75.091851 | 3.463037e+02 | 61.680131 | |
| ENSG00000198496 | 10230 | NBR2 | lncRNA | This gene was identified by its close proximity on chromosome 17 to tumor suppressor gene BRCA1. Experimental evidence indicates that the two genes share a bi-directional promoter. Transcription for either gene is controlled individually by distinct transcriptional repressor factors. A short (112 amino acid) open reading frame is observed which includes a region derived from a LINE1 element. A strong Kozak signal is not observed for the putative ORF and the stop codon is more than 55 nucleotides upstream of the last splice site for the transcript, suggesting that the transcript is subject to nonsense-mediated decay. Therefore, this gene does not appear to encode a protein. Glucose starvation induces the expression of this gene and the long non-coding RNA transcribed by it functions with AMP-activated protein kinase in mediating the energy stress response. [provided by RefSeq, Aug 2016]. | 26999735_Data indicate that long non-coding RNAs (lncRNAs) NBR2 (neighbour of BRCA1 gene 2) interacts with AMP-activated kinase (AMPK) and promotes AMPK kinase activity during energy stress. 27792451_Data indicate that long non-coding RNAs (lncRNAs) NBR2 (neighbour of BRCA1 gene 2) modulates cancer cell sensitivity to phenformin through NBR2 regulation of GLUT1 expression at transcriptional level. 27792451_NBR2-GLUT1 axis may serve as an adaptive response in cancer cells to survive in response to phenformin treatment. 28249151_High NBR2 expression is associated with breast cancer. 28586153_data suggested that miR-19a negatively controlled the autophagy of hepatocytes attenuated in D-GalN/LPS-stimulated hepatocytes via regulating NBR2 and AMPK/PPARalpha signaling. 31599420_LncRNA NBR2 inhibits EMT progression by regulating Notch1 pathway in NSCLC. 31888414_This study revealed a novel mechanism by which long noncoding RNA NBR2 mediates curcumin suppression of colorectal cancer proliferation by activating adenosine monophosphate-activated protein kinase and inactivating the mTOR signaling pathway. 33378941_LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. 33651649_LncRNA NBR2 aggravates hepatoblastoma cell malignancy and promotes cell proliferation under glucose starvation through the miR-22/TCF7 axis. 34076984_NBR2 promotes the proliferation of glioma cells via inhibiting p15 expression. 34506209_Long non-coding RNA NBR2 suppresses the progress of colorectal cancer in vitro and in vivo by regulating the polarization of TAM. | 92.37740 | 0.9081186 | -0.1390474312 | 0.33541834 | 1.752879e-01 | 6.754544e-01 | 9.998360e-01 | No | Yes | 65.99342 | 13.917961 | 7.019664e+01 | 11.513038 | ||||||||
| ENSG00000198538 | 7576 | ZNF28 | protein_coding | P17035 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7576; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 47.81401 | 1.3557120 | 0.4390507043 | 0.42858501 | 1.094543e+00 | 2.954667e-01 | 9.998360e-01 | No | Yes | 62.65265 | 14.319874 | 4.454545e+01 | 7.804834 | |||||
| ENSG00000198554 | 11169 | WDHD1 | protein_coding | O75717 | FUNCTION: Acts as a replication initiation factor that brings together the MCM2-7 helicase and the DNA polymerase alpha/primase complex in order to initiate DNA replication. {ECO:0000269|PubMed:19805216}. | 3D-structure;Acetylation;Alternative splicing;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;WD repeat | The protein encoded by this gene contains multiple N-terminal WD40 domains and a C-terminal high mobility group (HMG) box. WD40 domains are found in a variety of eukaryotic proteins and may function as adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly. HMG boxes are found in many eukaryotic proteins involved in chromatin assembly, transcription and replication. Alternative splicing results in two transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:11169; | cytoplasm [GO:0005737]; nuclear replication fork [GO:0043596]; nucleoplasm [GO:0005654]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA repair [GO:0006281]; DNA-dependent DNA replication [GO:0006261]; mitotic cell cycle [GO:0000278] | 17761813_Mcm10 is required for chromatin loading of And-1. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19439411_AND-1 coordinates multiple cellular events in S phase and G2 phase. 19805216_Assembly of the Cdc45-Mcm2-7-GINS complex requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. 20089864_in human cells, hCtf4 plays an essential role in DNA replication and its ability to stimulate the replicative DNA polymerases may contribute to this effect 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21725360_Data show that And-1 forms a complex with both histone H3 and Gcn5. 23093411_Human And-1 facilitates loading of the MCM2-7 helicase onto chromatin during the assembly of prereplicative complex. 23184928_And-1 together with HJURP regulates the assembly of new CENP-A onto centromeres. 24255107_The Ctf4-CMG complex contains a homodimeric Ctf4 that acts as a platform linking polymerase alpha to the monomeric hCMG complex. 25602958_RecQL4-dependent association of Mcm10 and Ctf4 with replication origins appears to be the first important step controlled by S phase promoting kinases and checkpoint pathways for the initiation of DNA replication in human cells. 26082189_And-1 coordinates with Claspin for efficient Chk1 activation in response to replication stress. 27037360_Results provide additional evidence suggesting that WDHD1 is involved in the mismatch repair pathway. 27099318_The WDHD1 gene, one of the genes that is upregulated in human papillomavirus E7-expressing cells, was found to play an important role in E7-induced G1 checkpoint abrogation and rereplication 27940552_And-1 interacts with CtIP and that these interactions are required for DNA damage checkpoint maintenance, thereby linking DNA processing with prolonged cell cycle arrest to allow sufficient time for DNA repair. 27940557_important role of And-1 in homologous recombination repair by regulating CtIP-mediated DNA double-strand break end resection 28381552_findings provide important insights into the mechanistic details of human AND-1 function, advancing our understanding of replisome formation during eukaryotic replication. 30082684_AND-1 depletion is incompatible with proliferation, owing to cells accumulating in G2 with activated DNA damage checkpoint 30314946_The study indicated that miR-494 overexpression suppresses epithelial-mesenchymal transition, tumor formation and lymph node metastasis while promoting cholangiocarcinoma (CCA) cell apoptosis through inhibiting WDHD1 in CCA. 32883234_WDHD1 facilitates G1 checkpoint abrogation in HPV E7 expressing cells by modulating GCN5. 33221821_WDHD1 is essential for the survival of PTEN-inactive triple-negative breast cancer. 34999279_The clinical significance of transcription factor WD repeat and HMG-box DNA binding protein 1 in laryngeal squamous cell carcinoma and its potential molecular mechanism. 35065237_Targeted inhibition of acidic nucleoplasmic DNA-binding protein 1 enhances radiosensitivity of non-small cell lung cancer. | ENSMUSG00000037572 | Wdhd1 | 317.73688 | 0.6870121 | -0.5415926508 | 0.20084140 | 6.912694e+00 | 8.558596e-03 | 9.998360e-01 | No | Yes | 303.01583 | 70.737428 | 3.925289e+02 | 70.410814 | |
| ENSG00000203875 | 387066 | SNHG5 | lncRNA | This gene represent a snoRNA host gene and produces a long non-coding RNA. This RNA may regulate gene expression by acting as a sponge for microRNAs. This transcript may also stabilize mRNAs by blocking degradation by staufen double-stranded RNA binding protein 1. [provided by RefSeq, Dec 2017]. | 26440365_the upregulation of serum levels of SNHG5 in patients with malignant melanoma suggest that SNHG55 levels may be a new tumor marker of malignant melanoma 27065326_This study is the first to demonstrate that SNHG5 is a critical and powerful regulator that is involved in GC progression through trapping MTA2 in the cytosol. 27871067_A positive correlation between KLF4 and SNHG5 expression levels were detected in gastric cancer cells. SNHG5 could directly bind to miR-32 and effectively act as a sponge for miR-32 to modulate the suppression of KLF4. 28004750_Mechanistically, we suggest that SNHG5 stabilizes the target transcripts by blocking their degradation by STAU1. Accordingly, depletion of STAU1 rescues the apoptosis induced after SNHG5 knockdown. Hence, we characterize SNHG5 as a lncRNA promoting tumour cell survival in colorectal cancer. 28033303_the current study revealed novel expression quantitative trait loci (eQTLs) for SNHG5 and PEX6 genes in chromosome 6. Nucleotide substitutions of the eQTLs might be candidate factors for a variety of cancers by regulating expression of the 2 genes. 28731158_Mechanistic investigations demonstrated that SNHG3 functioned as a competing endogenous RNA (ceRNA) to 'sponge' miR-182-5p, thus leading to the release of c-Myc from miR-182-5p and modulating the expression of c-Myc. In conclusion, SNHG3 promoted CRC progression via sponging miR-182-5p and upregulating c-Myc and its target genes 29409014_Results show that the levels of small nucleolar RNA host gene 5 (SNHG5) and SRY (sex determining region Y)-box 2 protein (SOX2) were significantly downregulated in osteoarthritis (OA) tissues, while the level of miR-26a was upregulated. 29592872_SNHG5 expression is down-regulated in lung adenocarcinoma patients with acquired gefitinib resistance. 30114643_SNHG5 acts as an oncogene in osteosarcoma via the SNHG5-miR-26a-ROCK1 axis. 30227213_Overexpressed METase promoted cell apoptosis and autophagy via up-regulating the expression of SNHG5 and down-regulating miR-20a in GC 30242892_study suggested that SNHG5 promotes trophoblast cell proliferation, invasion, and migration at least partly via regulating the miR-26a-5p/N-cadherin axis. 30266814_Results indentified two candidate haploinsufficient genes contiguous at 6q14 SYNCRIP (encoding hnRNP-Q) and SNHG5 (that hosts snoRNAs), both involved in regulating RNA maturation and translation inT-cell acute lymphoblastic leukemia (T-ALL). 30395767_High SNHG5 expression is associated with proliferation, metastasis and migration of colorectal cancer. 31173289_LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. 31292168_Long non-coding RNA SNHG5 promotes glioma progression via miR-205/E2F3 axis. 31506418_downregulated SNHG5 could play a key role in chronic myelogenous leukemia progression. 31657621_lncRNA small nucleolar RNA host gene 5 (SNHG5) is highly expressed in glioblastoma (GBM). SNHG5 promotes GBM cell proliferation and inhibits cell apoptosis in GBM. Yin Yang 1 (YY1) acts as transcriptional activator of SNHG5 in GBM. SNHG5 plays the oncogenic role in GBM by activating p38/MAPK signaling pathway. 31884339_Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. 32131767_LncRNA SNHG5 promotes nasopharyngeal carcinoma progression by regulating miR-1179/HMGB3 axis. 32145584_LncRNA SNHG5 regulates cell apoptosis and inflammation by miR-132/PTEN axis in COPD. 32266420_LncRNA SNHG5 regulates SOX4 expression through competitive binding to miR-489-3p in acute myeloid leukemia. 32281285_The lncRNA SNHG5-mediated miR-205-5p downregulation contributes to the progression of clear cell renal cell carcinoma by targeting ZEB1. 32329834_Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-363-3p/RNF38 axis. 32360843_The functional role, molecular mechanism, and clinical significance of SNHG5 in human cancers are described (review). 32960357_miR-23a-3p regulated by LncRNA SNHG5 suppresses the chondrogenic differentiation of human adipose-derived stem cells via targeting SOX6/SOX5. 33095847_SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer. 33170429_Long non-coding RNA SNHG5 suppresses the development of acute respiratory distress syndrome by targeting miR-205/COMMD1 axis. 33176855_si-SNHG5-FOXF2 inhibits TGF-beta1-induced fibrosis in human primary endometrial stromal cells by the Wnt/beta-catenin signalling pathway. 33318617_LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. 33622094_LncRNA SNHG5 promotes cervical cancer progression by regulating the miR-132/SOX4 pathway. 33865958_Correlations of SNHG5 genetic polymorphisms with susceptibility and prognosis to gastric cancer in a Chinese population. 34185955_Based on bioinformatics analysis lncrna SNHG5 modulates the function of vascular smooth muscle cells through mir-205-5p/SMAD4 in abdominal aortic aneurysm. 34351577_LncRNA SNHG5 promotes the glycolysis and proliferation of breast cancer cell through regulating BACH1 via targeting miR-299. 34369033_SNHG5 protects chondrocytes in interleukin-1beta-stimulated osteoarthritis via regulating miR-181a-5p/TGFBR3 axis. 34891212_Long Non-Coding RNA Small Nucleolar RNA Host Gene 5 (SNHG5) Regulates Renal Tubular Damage in Diabetic Nephropathy via Targeting MiR-26a-5p. 35077478_SPATS2 is positively activated by long noncoding RNA SNHG5 via regulating DNMT3a expression to promote hepatocellular carcinoma progression. 35259061_LncRNA small nucleolar RNA host gene 5 inhibits trophoblast autophagy in preeclampsia by targeting microRNA-31-5p and promoting the transcription of secreted protein acidic and rich in cysteine. 35346361_Long non-coding RNA SNHG5 promotes the osteogenic differentiation of bone marrow mesenchymal stem cells via the miR-212-3p/GDF5/SMAD pathway. | 5257.86372 | 0.9241264 | -0.1138379185 | 0.10882489 | 1.098451e+00 | 2.946062e-01 | 9.998360e-01 | No | Yes | 5460.13860 | 1074.940135 | 6.783483e+03 | 1028.731339 | ||||||||
| ENSG00000204054 | LINC00963 | lncRNA | 577.94675 | 1.0387596 | 0.0548617517 | 0.14321503 | 1.477519e-01 | 7.006930e-01 | 9.998360e-01 | No | Yes | 636.94722 | 72.157497 | 5.754174e+02 | 51.703848 | |||||||||||
| ENSG00000204147 | 653308 | ASAH2B | protein_coding | P0C7U1 | Alternative splicing;Reference proteome | hsa:653308; | extracellular region [GO:0005576]; N-acylsphingosine amidohydrolase activity [GO:0017040]; ceramide catabolic process [GO:0046514]; long-chain fatty acid biosynthetic process [GO:0042759]; sphingosine biosynthetic process [GO:0046512] | 102.38353 | 1.2190613 | 0.2857707227 | 0.30436526 | 8.855497e-01 | 3.466861e-01 | 9.998360e-01 | No | Yes | 96.23004 | 18.147144 | 8.395207e+01 | 12.331991 | ||||||
| ENSG00000204920 | 7711 | ZNF155 | protein_coding | Q12901 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7711; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 54.29779 | 0.7581224 | -0.3994973584 | 0.40246612 | 9.454968e-01 | 3.308682e-01 | 9.998360e-01 | No | Yes | 42.89138 | 9.239299 | 5.700213e+01 | 9.408988 | |||||
| ENSG00000205084 | 79583 | TMEM231 | protein_coding | Q9H6L2 | FUNCTION: Transmembrane component of the tectonic-like complex, a complex localized at the transition zone of primary cilia and acting as a barrier that prevents diffusion of transmembrane proteins between the cilia and plasma membranes. Required for ciliogenesis and sonic hedgehog/SHH signaling (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Disease variant;Glycoprotein;Joubert syndrome;Meckel syndrome;Membrane;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein, which is a component of the B9 complex involved in the formation of the diffusion barrier between the cilia and plasma membrane. Mutations in this gene cause Joubert syndrome (JBTS). Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jan 2013]. | hsa:79583; | ciliary membrane [GO:0060170]; ciliary transition zone [GO:0035869]; integral component of membrane [GO:0016021]; MKS complex [GO:0036038]; camera-type eye development [GO:0043010]; cilium assembly [GO:0060271]; embryonic digit morphogenesis [GO:0042733]; in utero embryonic development [GO:0001701]; neuroepithelial cell differentiation [GO:0060563]; regulation of protein localization [GO:0032880]; smoothened signaling pathway [GO:0007224]; vasculature development [GO:0001944] | 23012439_mutations in TMEM231 cause JBTS, reinforcing the relationship between this condition and the disruption of the barrier at the ciliary transition zone. 23349226_TMEM231 represents a novel MKS locus. The very recent identification of TMEM231 mutations in Joubert syndrome supports the growing appreciation of the overlap in the molecular pathogenesis between these two ciliopathies. 25869670_Tmem231 is critical for organizing the Meckel syndrome complex and controlling ciliary composition, defects in which cause OFD3 and MKS. 27449316_Results identified a rare gene conversion event in TMEM231, leading to loss of exon 4, which in combination with c.712G>A missense mutation caused Joubert syndrome and in combination with c.334T>G missense mutation caused Meckel-Gruber syndrome. 31663672_Long-read nanopore sequencing resolves a TMEM231 gene conversion event causing Meckel-Gruber syndrome. | ENSMUSG00000031951 | Tmem231 | 551.65950 | 1.2238096 | 0.2913791256 | 0.15073963 | 3.646607e+00 | 5.618357e-02 | 9.998360e-01 | No | Yes | 555.81643 | 69.785956 | 4.870099e+02 | 47.594651 | |
| ENSG00000205269 | 100113407 | TMEM170B | protein_coding | Q5T4T1 | FUNCTION: Negatively regulates the canonical Wnt signaling in breast cancer cells. Exerts an inhibitory effect on breast cancer growth by inhibiting CTNNB1 stabilization and nucleus translocation, which reduces the activity of Wnt targets (PubMed:29367600). {ECO:0000269|PubMed:29367600}. | Cell membrane;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Wnt signaling pathway | hsa:100113407; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; Wnt signaling pathway [GO:0016055] | 29367600_these results highlight TMEM170B as a novel tumor suppressor target in association with the beta-catenin pathway. | ENSMUSG00000087370 | Tmem170b | 138.28092 | 1.2220780 | 0.2893364057 | 0.26063406 | 1.223942e+00 | 2.685884e-01 | 9.998360e-01 | No | Yes | 155.42818 | 32.759937 | 1.332453e+02 | 21.733980 | ||
| ENSG00000205309 | 56953 | NT5M | protein_coding | Q9NPB1 | FUNCTION: Dephosphorylates specifically the 5' and 2'(3')-phosphates of uracil and thymine deoxyribonucleotides, and so protects mitochondrial DNA replication from excess dTTP. Has only marginal activity towards dIMP and dGMP. {ECO:0000269|PubMed:10899995}. | 3D-structure;Hydrolase;Magnesium;Metal-binding;Mitochondrion;Nucleotide metabolism;Nucleotide-binding;Reference proteome;Transit peptide | This gene encodes a 5' nucleotidase that localizes to the mitochondrial matrix. This enzyme dephosphorylates the 5'- and 2'(3')-phosphates of uracil and thymine deoxyribonucleotides. The gene is located within the Smith-Magenis syndrome region on chromosome 17. [provided by RefSeq, Jul 2008]. | hsa:56953; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; 5'-nucleotidase activity [GO:0008253]; metal ion binding [GO:0046872]; nucleotidase activity [GO:0008252]; nucleotide binding [GO:0000166]; DNA replication [GO:0006260]; dUMP catabolic process [GO:0046079]; pyrimidine deoxyribonucleotide catabolic process [GO:0009223] | 12124385_Overproduction in cultured cells and functional aspects 12352955_structures of dNT-2 in complex with bound phosphate and beryllium trifluoride plus thymidine as model for a phosphoenzyme-product complex 16004879_substrate specificity analysis of the human mitochondrial deoxyribonucleotidase 20877624_Observational study of gene-disease association. (HuGE Navigator) 24506201_An alternative splice variant of the mdN gene containing an 18-nucleotide insertion encoding 6 amino acids (GKWPAT) at the 3'-end of the penultimate exon 4 has been characterized. | ENSMUSG00000032615 | Nt5m | 349.63980 | 1.0370686 | 0.0525113234 | 0.18758737 | 8.119615e-02 | 7.756829e-01 | 9.998360e-01 | No | Yes | 364.01480 | 45.990690 | 3.317927e+02 | 34.010478 | |
| ENSG00000205352 | 54458 | PRR13 | protein_coding | Q9NZ81 | FUNCTION: Negatively regulates TSP1 expression at the level of transcription. This down-regulation was shown to reduce taxane-induced apoptosis. {ECO:0000269|PubMed:16847352}. | Alternative splicing;Nucleus;Reference proteome;Transcription;Transcription regulation | hsa:54458; | cytosol [GO:0005829]; nucleoplasm [GO:0005654] | 5642.58542 | 1.0223216 | 0.0318490647 | 0.09393854 | 1.171321e-01 | 7.321660e-01 | 9.998360e-01 | No | Yes | 5743.75051 | 546.977453 | 5.502522e+03 | 406.250220 | |||||
| ENSG00000205683 | 8110 | DPF3 | protein_coding | Q92784 | FUNCTION: Belongs to the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a post-mitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to post-mitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). Muscle-specific component of the BAF complex, a multiprotein complex involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Specifically binds acetylated lysines on histone 3 and 4 (H3K14ac, H3K9ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac). In the complex, it acts as a tissue-specific anchor between histone acetylations and methylations and chromatin remodeling. It thereby probably plays an essential role in heart and skeletal muscle development. {ECO:0000250, ECO:0000269|PubMed:18765789}.; FUNCTION: [Isoform 2]: Acts as a regulator of myogenesis in cooperation with HDGFL2 (PubMed:32459350). Mediates the interaction of HDGFL2 with the BAF complex (PubMed:32459350). HDGFL2-DPF3a activate myogenic genes by increasing chromatin accessibility through recruitment of SMARCA4/BRG1/BAF190A (ATPase subunit of the BAF complex) to myogenic gene promoters (PubMed:32459350). {ECO:0000269|PubMed:32459350}. | 3D-structure;Activator;Alternative splicing;Chromatin regulator;Isopeptide bond;Metal-binding;Myogenesis;Neurogenesis;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the D4 protein family. The encoded protein is a transcription regulator that binds acetylated histones and is a component of the BAF chromatin remodeling complex. Alternate splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2013]. | hsa:8110; | nBAF complex [GO:0071565]; nucleoplasm [GO:0005654]; histone binding [GO:0042393]; transcription coregulator activity [GO:0003712]; zinc ion binding [GO:0008270]; chromatin organization [GO:0006325]; negative regulation of transcription, DNA-templated [GO:0045892]; nervous system development [GO:0007399]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 16109180_Observational study of gene-disease association. (HuGE Navigator) 18765789_DPF3 adds a further layer of complexity to the BAF complex by representing a tissue-specific anchor between histone acetylations as well as methylations and chromatin remodeling. 18978678_Observational study of gene-disease association. (HuGE Navigator) 20613843_three-dimensional solution structures and biochemical analysis of DPF3b highlight the molecular basis of the integrated tandem PHD finger, which acts as one functional unit in the sequence-specific recognition of lysine-14-acetylated histone H3 22334708_by linking RelA and p50 to the SWI/SNF complex, DPF3a/b induces the transactivation of NF-kappaB target gene promoters in relatively inactive chromatin contexts. 24155890_These findings provide insights in the STAT5 dependent transcriptional regulation of Dpf3. 24271036_DFP3 protein rs10129954 SNP is associated with poor sperm morphology. 25120773_Immunohistochemical staining showed that intensive DPF3 staining occurred in proximal anastomosis and the positive staining was hardly observed in stenotic segment in colon specimens from patients with Hirschsprung's disease 26582913_Activation of DPF3a upon hypertrophic stimuli in cardiac hypertrophy switches cardiac fetal gene expression from being silenced by HEY to being activated by BRG1. 27232852_SLC1A1 and DPF3 were strongly associated with idiopathic male infertility and were significantly correlated with semen quality alterations 27402533_the high resolution crystal structure of the tandem PHD fingers of DPF3b in complex with an H3K14ac peptide. 28975488_DPF3 single nucleotide polymorphisms were significantly correlated with male infertility in a Japanese population. 31076105_downregulation of DPF3 plays an indispensable function in the progression of breast cancer 34390653_Altered regulation of DPF3, a member of the SWI/SNF complexes, underlies the 14q24 renal cancer susceptibility locus. 35148991_The renal cancer risk allele at 14q24.2 activates a novel hypoxia-inducible transcription factor-binding enhancer of DPF3 expression. | ENSMUSG00000021221 | Dpf3 | 151.82888 | 0.8534273 | -0.2286598450 | 0.25383201 | 8.434327e-01 | 3.584169e-01 | 9.998360e-01 | No | Yes | 131.50978 | 21.840182 | 1.496128e+02 | 19.279179 | |
| ENSG00000205771 | 440278 | CATSPER2P1 | transcribed_unprocessed_pseudogene | Catsper genes belong to a family of putative cation channels that are specific to spermatozoa and localize to the flagellum. This gene is part of a tandem repeat on chromosome 15q15; this copy of the gene is thought to be a pseudogene. [provided by RefSeq, Oct 2008]. | 27.38819 | 0.9185197 | -0.1226174338 | 0.55243515 | 5.121882e-02 | 8.209559e-01 | 9.998360e-01 | No | Yes | 45.07761 | 10.777447 | 4.685039e+01 | 9.932944 | |||||||||
| ENSG00000205903 | 100131017 | ZNF316 | protein_coding | A6NFI3 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000250}. | Acetylation;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:100131017; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; sequence-specific DNA binding [GO:0043565]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000046658 | Zfp316 | 2538.44885 | 0.9780481 | -0.0320226862 | 0.15557830 | 4.098546e-02 | 8.395660e-01 | 9.998360e-01 | No | Yes | 2166.27065 | 269.503653 | 1.984576e+03 | 191.088890 | |||
| ENSG00000211455 | 23012 | STK38L | protein_coding | Q9Y2H1 | FUNCTION: Involved in the regulation of structural processes in differentiating and mature neuronal cells. {ECO:0000250, ECO:0000269|PubMed:15037617, ECO:0000269|PubMed:15067004}. | 3D-structure;ATP-binding;Acetylation;Actin-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Direct protein sequencing;Kinase;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:23012; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; actin binding [GO:0003779]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; intracellular signal transduction [GO:0035556]; peptidyl-serine phosphorylation [GO:0018105]; protein phosphorylation [GO:0006468]; regulation of cellular component organization [GO:0051128] | 15067004_NDR1 and NDR2 serine-threonine kinases are regulated by mob proteins 15582665_The NDR1 and NDR2 kinases were incorporated into HIV-1 particles and were cleaved by the HIV-1 protease. 16314523_Activation of NDR2 is a multistep process involving phosphorylation of the hydrophobic motif site Thr444/2 by MST3, autophosphorylation of Ser281/2, and binding of MOB1A. 16488889_NDR2 is an upstream kinase of ARK5 that plays an essential role in tumor progression through ARK5 19062280_findings identify NDR1/2 as novel proapoptotic kinases and key members of the RASSF1A/MST1 signaling cascade 19240061_Observational study of gene-disease association. (HuGE Navigator) 21262772_These findings establish a novel MST3-NDR-p21 axis as an important regulator of G(1)/S progression of mammalian cells. 23435566_NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by triggering the switch in binding specificity of Rabin8 from PS to Sec15. 23897809_cyclin D1 has a role in promoting cell cycle progression by enhancing NDR1/2 kinase activity independent of Cdk4 28556638_It was found that STK38L*I/D genotype had positive association with longevity in the ethnically homogeneous group of Tatars from the Republic of Bashkortostan, Russia. 29108249_This study shows that STK38L is co-amplified with oncogenic KRAS in pancreatic cancer cell lines, where it regulates LATS2 gene expression. 30504095_These results suggest that NDR2 may play important roles in IL-17-associated inflammation by promoting Smurf1-mediated MEKK2 ubiquitination and degradation. 30714141_Study identified rs10842893 as a novel independent SNP in the gene STK38L that was significantly associated with glioma risk in Chinese population, and furthermore confirmed previously reported associations of rs498872 and rs6010620 with glioma risk in European descent population and Chinese Han population. 30775439_NDR2 positively regulated an antiviral innate immune response in a kinase activity-independent manner. NDR2 interacts with RIG-I and promotes TRIM25-mediated RIG-I ubiquitination, which is required for downstream signaling activation and type I IFN production. 31427083_The results demonstrate that NDR2 is a reversible acetylated kinase regulated by SIRT1 and p300/CBP. | ENSMUSG00000001630 | Stk38l | 212.33434 | 1.4417132 | 0.5277841524 | 0.22409620 | 5.362444e+00 | 2.057492e-02 | 9.998360e-01 | No | Yes | 227.30062 | 58.428873 | 1.559001e+02 | 30.890712 | ||
| ENSG00000213347 | 83463 | MXD3 | protein_coding | Q9BW11 | FUNCTION: Transcriptional repressor. Binds with MAX to form a sequence-specific DNA-binding protein complex which recognizes the core sequence 5'-CAC[GA]TG-3'. Antagonizes MYC transcriptional activity by competing for MAX and suppresses MYC dependent cell transformation (By similarity). {ECO:0000250}. | Alternative splicing;DNA-binding;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a member of the Myc superfamily of basic helix-loop-helix leucine zipper transcriptional regulators. The encoded protein forms a heterodimer with the cofactor MAX which binds specific E-box DNA motifs in the promoters of target genes and regulates their transcription. Disruption of the MAX-MXD3 complex is associated with uncontrolled cell proliferation and tumorigenesis. Transcript variants of this gene encoding different isoforms have been described.[provided by RefSeq, Dec 2008]. | hsa:83463; | chromatin [GO:0000785]; RNA polymerase II transcription regulator complex [GO:0090575]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 22808009_Increased level and/or duration of MXD3 expression ultimately reduces cell numbers via increased cell death and cell cycle arrest in medulloblastoma cells. 25053245_Acute MXD3 activation results in a transient increase in DAOY cell proliferation while persistent activation of MXD3 results in an overall decrease in cell number, suggesting that the time course of MXD3 expression dictates the cellular outcome. 25554682_The results suggest that MXD3 is important for survival of Reh preB ALL cells, possibly as an anti-apoptotic factor. 28419087_results indicate that MXD3 is a potential new target and that the use of MXD3 siRNA nanocomplexes is a novel therapeutic approach for neuroblastoma 34943942_MXD3 Promotes Obesity and the Androgen Receptor Signaling Pathway in Gender-Disparity Hepatocarcinogenesis. | ENSMUSG00000021485 | Mxd3 | 1136.26768 | 1.0003404 | 0.0004910191 | 0.13188046 | 1.417487e-05 | 9.969960e-01 | 9.998360e-01 | No | Yes | 1288.37463 | 152.447830 | 1.179715e+03 | 108.623892 | |
| ENSG00000213963 | 100130691 | lncRNA | 21.75562 | 1.4410117 | 0.5270820980 | 0.70772515 | 4.606364e-01 | 4.973267e-01 | 9.998360e-01 | No | Yes | 21.04283 | 6.332448 | 1.637247e+01 | 3.865040 | |||||||||||
| ENSG00000214022 | 29803 | REPIN1 | protein_coding | Q9BWE0 | FUNCTION: Sequence-specific double-stranded DNA-binding protein required for initiation of chromosomal DNA replication. Binds on 5'-ATT-3' reiterated sequences downstream of the origin of bidirectional replication (OBR) and a second, homologous ATT sequence of opposite orientation situated within the OBR zone. Facilitates DNA bending. | Acetylation;Alternative splicing;DNA replication;DNA-binding;Direct protein sequencing;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | hsa:29803; | chromosome [GO:0005694]; nuclear origin of replication recognition complex [GO:0005664]; nucleoplasm [GO:0005654]; chromatin insulator sequence binding [GO:0043035]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; DNA replication [GO:0006260]; regulation of transcription by RNA polymerase II [GO:0006357] | 16924111_AP4 and geminin act as a repressor complex distinct from REST/NRSF to negatively regulate the expression of target genes in nonneuronal cells. 20727851_a potential role for Repin1 as regulator of adipocyte size was shown.Repin1 seems to be involved in the regulation of genes involved in adipogenesis. 24285725_there is a p53-induced double-negative feedback loop involving miR-15a/16-1 and AP4 that stabilizes epithelial and mesenchymal states, respectively, which may determine metastatic prowess in colorectal cancer 25266805_AP4 is transcriptionally activated by TGFB1 to inhibit phosphorylation of SMAD2 as a negative regulator of an activated TGFB receptor signaling on cell growth inhibition in non-small cell lung cancer 26037074_Results show that breast cancer cells underwent epithelial-mesenchymal transition when AP4 was upregulated, and that AP4 regulated p53 expression level suggesting AP4 can regulate cell migration via the activity of p53. 29337428_AP4 and LAPTM4B are highly coexpressed in hepatocellular carcinoma tissues, and their coexpression may be a marker of poor prognosis. 29915365_data suggest that genetic variation in human REPIN1 plays a role in glucose and lipid metabolism by differentially affecting the expression of REPIN1 target genes including glucose and fatty acid transporters 29979259_Increased expression of hsa-mir-127 and decreased expression of REPIN1 were both associated with poor overall survival 31922994_A Human REPIN1 Gene Variant: Genetic Risk Factor for the Development of Nonalcoholic Fatty Liver Disease. 33339069_Overexpression of miR-127 Predicts Poor Prognosis and Contributes to the Progression of Papillary Thyroid Cancer by Targeting REPIN1. | ENSMUSG00000052751 | Repin1 | 6006.40159 | 0.9856564 | -0.0208432178 | 0.09488160 | 4.866099e-02 | 8.254098e-01 | 9.998360e-01 | No | Yes | 6983.25304 | 841.072502 | 6.343553e+03 | 590.572767 | ||
| ENSG00000214026 | 6150 | MRPL23 | protein_coding | Q16540 | 3D-structure;Mitochondrion;Reference proteome;Ribonucleoprotein;Ribosomal protein | Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein. The gene is biallelically expressed, despite its location within a region of imprinted genes on chromosome 11. [provided by RefSeq, Jul 2008]. | hsa:6150; | fibrillar center [GO:0001650]; mitochondrial inner membrane [GO:0005743]; mitochondrial large ribosomal subunit [GO:0005762]; mitochondrion [GO:0005739]; RNA binding [GO:0003723]; structural constituent of ribosome [GO:0003735]; mitochondrial translation [GO:0032543]; translation [GO:0006412] | 17242401_5-FU treatment triggers a ribosomal stress response so that ribosomal proteins L5, L11, and L23 are released from ribosome to activate p53 by ablating the MDM2-p53 feedback circuit 19304784_Meta-analysis of gene-disease association. (HuGE Navigator) 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000037772 | Mrpl23 | 8978.32070 | 0.9554926 | -0.0656834267 | 0.11688013 | 3.243111e-01 | 5.690283e-01 | 9.998360e-01 | No | Yes | 9571.27807 | 1480.556317 | 9.352740e+03 | 1116.901224 | ||
| ENSG00000214182 | 150928 | PTMAP5 | transcribed_processed_pseudogene | 21.21085 | 0.9601799 | -0.0586232849 | 0.63348650 | 8.993845e-03 | 9.244452e-01 | 9.998360e-01 | No | Yes | 21.74281 | 6.798726 | 2.437072e+01 | 6.010704 | ||||||||||
| ENSG00000214413 | 92482 | BBIP1 | protein_coding | A8MTZ0 | FUNCTION: The BBSome complex is thought to function as a coat complex required for sorting of specific membrane proteins to the primary cilia. The BBSome complex is required for ciliogenesis but is dispensable for centriolar satellite function. This ciliogenic function is mediated in part by the Rab8 GDP/GTP exchange factor, which localizes to the basal body and contacts the BBSome. Rab8(GTP) enters the primary cilium and promotes extension of the ciliary membrane. Firstly the BBSome associates with the ciliary membrane and binds to RAB3IP/Rabin8, the guanosyl exchange factor (GEF) for Rab8 and then the Rab8-GTP localizes to the cilium and promotes docking and fusion of carrier vesicles to the base of the ciliary membrane. Required for primary cilia assembly and BBSome stability. Regulates cytoplasmic microtubule stability and acetylation. {ECO:0000269|Ref.4}. | 3D-structure;Alternative splicing;Bardet-Biedl syndrome;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Obesity;Protein transport;Reference proteome;Transport | This gene encodes one of eight proteins that form the BBSome complex and is essential for its assembly. The BBSome complex is involved in trafficking signal receptors to and from the cilia. Mutations in this gene result in Bardet-Biedl syndrome 18. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2014]. | hsa:92482; | BBSome [GO:0034464]; ciliary membrane [GO:0060170]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; cilium assembly [GO:0060271]; eating behavior [GO:0042755]; protein transport [GO:0015031]; receptor localization to non-motile cilium [GO:0097500] | 24026985_Identification of BBIP1 as the 18th BBS gene (BBS18), BBSome assembly may represent a unifying pathomechanism for Bardet-Biedl syndrome. | ENSMUSG00000084957 | Bbip1 | 145.57694 | 0.9385236 | -0.0915349951 | 0.27018733 | 1.214990e-01 | 7.274143e-01 | 9.998360e-01 | No | Yes | 140.58571 | 29.969783 | 1.551163e+02 | 25.616398 | |
| ENSG00000214655 | 23053 | ZSWIM8 | protein_coding | A7E2V4 | FUNCTION: Substrate recognition component of a SCF-like E3 ubiquitin-protein ligase complex that promotes target-directed microRNA degradation (TDMD), a process that mediates degradation of microRNAs (miRNAs) (PubMed:33184234, PubMed:33184237). The SCF-like E3 ubiquitin-protein ligase complex acts by catalyzing ubiquitination and subsequent degradation of AGO proteins (AGO1, AGO2, AGO3 and/or AGO4), thereby exposing miRNAs for degradation (PubMed:33184234, PubMed:33184237). Specifically recognizes and binds AGO proteins when they are engaged with a TDMD target (PubMed:33184234). May also acts as a regulator of axon guidance: specifically recognizes misfolded ROBO3 and promotes its ubiquitination and subsequent degradation (PubMed:24012004). {ECO:0000269|PubMed:24012004, ECO:0000269|PubMed:33184234, ECO:0000269|PubMed:33184237}. | Alternative splicing;Cytoplasm;Metal-binding;Phosphoprotein;Reference proteome;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:33184234, ECO:0000269|PubMed:33184237}. | hsa:23053; | Cul2-RING ubiquitin ligase complex [GO:0031462]; Cul3-RING ubiquitin ligase complex [GO:0031463]; cytosol [GO:0005829]; ubiquitin ligase-substrate adaptor activity [GO:1990756]; zinc ion binding [GO:0008270]; positive regulation of miRNA catabolic process [GO:2000627]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein ubiquitination [GO:0016567] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 33184234_A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. 33184237_The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. 34686700_ZSWIM8 is a myogenic protein that partly prevents C2C12 differentiation. | ENSMUSG00000021819 | Zswim8 | 2185.36217 | 0.9254675 | -0.1117457707 | 0.12486333 | 8.163110e-01 | 3.662609e-01 | 9.998360e-01 | No | Yes | 1921.98420 | 225.144096 | 1.906534e+03 | 172.782193 | |
| ENSG00000214717 | 9189 | ZBED1 | protein_coding | O96006 | FUNCTION: Functions as an E3-type small ubiquitin-like modifier (SUMO) ligase which sumoylates CHD3/Mi2-alpha, causing its release from DNA (PubMed:27068747). This results in suppression of CHD3/Mi2-alpha transcription repression, increased recruitment of RNA polymerase II to gene promoters and positive regulation of transcription including H1-5 and ribosomal proteins such as: RPS6, RPL10A, and RPL12 (PubMed:12663651, PubMed:17209048, PubMed:17220279, PubMed:27068747). The resulting increased transcriptional activity drives cell proliferation (PubMed:12663651, PubMed:17220279). Binds to 5'-TGTCG[CT]GA[CT]A-3' consensus sequences in gene promoters of ribosomal proteins (PubMed:12663651, PubMed:17209048, PubMed:17220279, PubMed:27068747). {ECO:0000269|PubMed:12663651, ECO:0000269|PubMed:17209048, ECO:0000269|PubMed:17220279, ECO:0000269|PubMed:27068747}.; FUNCTION: (Microbial infection) Binds to human adenovirus gene promoters and contributes to transcriptional repression and virus growth inhibition during early stages of infection. {ECO:0000269|PubMed:25210186}. | 3D-structure;DNA-binding;Host-virus interaction;Metal-binding;Nucleus;Reference proteome;Transcription;Transcription regulation;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein sumoylation. {ECO:0000269|PubMed:27068747}. | This gene is located in the pseudoautosomal region 1 (PAR1) of X and Y chromosomes. It was earlier identified as a gene with similarity to Ac transposable elements, however, was found not to have transposase activity. Later studies show that this gene product is localized in the nucleus and functions as a transcription factor. It binds to DNA elements found in the promoter regions of several genes related to cell proliferation, such as histone H1, hence may have a role in regulating genes related to cell proliferation. Alternatively spliced transcript variants with different 5' untranslated region have been found for this gene. [provided by RefSeq, Jan 2010]. | hsa:9189; | centrosome [GO:0005813]; chromatin [GO:0000785]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; SUMO ligase activity [GO:0061665]; SUMO transferase activity [GO:0019789]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; negative regulation by host of viral genome replication [GO:0044828]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein autosumoylation [GO:1990466]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12663651_expression of hDREF/KIAA0785 may have a role in regulation of human genes related to cell proliferation 17209048_self-association of hDREF via the hATC domain is necessary for its nuclear accumulation and DNA binding. 17220279_hDREF is an important transcription factor for cell proliferation which plays roles in cell cycle-dependent regulation of a number of ribosomal protein genes 25210186_These findings identify DREF as a novel adenovirus E1A C terminus binding partner and provide evidence supporting a role for DREF in viral replication. 26279084_The pseudoautosomal region of human X and Y chromosome encoded ZBED1 protein was predicted to interact with at least 10 proteins encoded on the somatic chromosomes. 30304065_ZBED1 is not a cell proliferation-associated factor such as Drosophila DREF, and our study adds to the cumulative understanding of the functions of ZBED1 in human cells and tissues. 35052473_ZBED1 Regulates Genes Important for Multiple Biological Processes of the Placenta. | 2384.37020 | 1.0793246 | 0.1101288564 | 0.10491475 | 1.095588e+00 | 2.952364e-01 | 9.998360e-01 | No | Yes | 2626.28037 | 281.405598 | 2.263098e+03 | 188.183457 | ||
| ENSG00000215067 | 100506713 | ALOX12-AS1 | lncRNA | 34174394_Antisense overlapping long non-coding RNA regulates coding arachidonate 12-lipoxygenase gene by translational interference. | 172.90998 | 0.9663061 | -0.0494478019 | 0.23529393 | 4.362104e-02 | 8.345603e-01 | 9.998360e-01 | No | Yes | 196.66993 | 20.434453 | 1.934100e+02 | 16.267367 | |||||||||
| ENSG00000215784 | 728833 | FAM72D | protein_coding | Q6L9T8 | Mouse_homologues FUNCTION: May play a role in the regulation of cellular reactive oxygen species metabolism. May participate in cell growth regulation (By similarity). {ECO:0000250}. | Reference proteome | hsa:728833; | cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231] | 26206078_An epistemological characterization of the human tumorigenic neuronal paralogous FAM72 gene loci (FAM72A, FAM72B, FAM72C, FAM72D). 31221779_The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation. | ENSMUSG00000055184 | Fam72a | 335.77377 | 1.1342481 | 0.1817362277 | 0.22562782 | 6.935042e-01 | 4.049750e-01 | 9.998360e-01 | No | Yes | 364.72132 | 67.782197 | 3.246649e+02 | 46.647243 | ||
| ENSG00000217128 | 96459 | FNIP1 | protein_coding | Q8TF40 | FUNCTION: Binding partner of the GTPase-activating protein FLCN: involved in the cellular response to amino acid availability by regulating the mTORC1 signaling cascade controlling the MiT/TFE factors TFEB and TFE3 (PubMed:17028174, PubMed:18663353, PubMed:24081491). In low-amino acid conditions, component of the lysosomal folliculin complex (LFC) on the membrane of lysosomes, which inhibits the GTPase-activating activity of FLCN, thereby inactivating mTORC1 and promoting nuclear translocation of TFEB and TFE3 (By similarity). Upon amino acid restimulation, disassembly of the LFC complex liberates the GTPase-activating activity of FLCN, leading to activation of mTORC1 and subsequent cytoplasmic retention of TFEB and TFE3 (By similarity). Required to promote FLCN recruitment to lysosomes and interaction with Rag GTPases (PubMed:24081491). Together with FLCN, regulates autophagy: following phosphorylation by ULK1, interacts with GABARAP and promotes autophagy (PubMed:25126726). In addition to its role in mTORC1 signaling, also acts as a co-chaperone of HSP90AA1/Hsp90: following gradual phosphorylation by CK2, inhibits the ATPase activity of HSP90AA1/Hsp90, leading to activate both kinase and non-kinase client proteins of HSP90AA1/Hsp90 (PubMed:27353360, PubMed:30699359). Acts as a scaffold to load client protein FLCN onto HSP90AA1/Hsp90 (PubMed:27353360). Competes with the activating co-chaperone AHSA1 for binding to HSP90AA1, thereby providing a reciprocal regulatory mechanism for chaperoning of client proteins (PubMed:27353360). Also acts as a core component of the reductive stress response by inhibiting activation of mitochondria in normal conditions: in response to reductive stress, the conserved Cys degron is reduced, leading to recognition and polyubiquitylation by the CRL2(FEM1B) complex, followed by proteasomal (By similarity). Required for B-cell development (By similarity). {ECO:0000250|UniProtKB:Q68FD7, ECO:0000250|UniProtKB:Q9P278, ECO:0000269|PubMed:17028174, ECO:0000269|PubMed:18663353, ECO:0000269|PubMed:24081491, ECO:0000269|PubMed:25126726, ECO:0000269|PubMed:27353360, ECO:0000269|PubMed:30699359}. | Alternative splicing;Cytoplasm;Glycoprotein;Isopeptide bond;Lysosome;Membrane;Oxidation;Phosphoprotein;Reference proteome;Ubl conjugation | This gene encodes a protein that binds to the tumor suppressor protein folliculin and to AMP-activated protein kinase (AMPK). The encoded protein participates in the regulation of cellular metabolism and nutrient sensing by modulating the AMPK and target of rapamycin signaling pathways. This gene has a closely related paralog that encodes a protein with similar binding activities. Both related proteins also associate with the molecular chaperone heat shock protein-90 (Hsp90) and negatively regulate its ATPase activity and facilitate its association with folliculin. [provided by RefSeq, Jul 2017]. | hsa:96459; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; lysosomal membrane [GO:0005765]; ATPase inhibitor activity [GO:0042030]; chaperone binding [GO:0051087]; enzyme binding [GO:0019899]; guanyl-nucleotide exchange factor activity [GO:0005085]; cellular response to starvation [GO:0009267]; immature B cell differentiation [GO:0002327]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; negative regulation of TOR signaling [GO:0032007]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of B cell apoptotic process [GO:0002904]; positive regulation of peptidyl-serine phosphorylation [GO:0033138]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of TOR signaling [GO:0032008]; regulation of pro-B cell differentiation [GO:2000973]; regulation of protein phosphorylation [GO:0001932]; TOR signaling [GO:0031929] | 17028174_Results suggest that FLCN, mutated in Birt-Hogg-Dube syndrome, and its interacting partner FNIP1 may be involved in energy and/or nutrient sensing through the AMPK and mTOR signaling pathways. 18804346_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 22709692_The FLCN-FNIP complex deregulated in Birt-Hogg-Dube syndrome is absolutely required for B-cell differentiation. 30099721_Based on previous studies and gene ontology database, we found that POLQ encoding DNA polymerase theta enzyme and FNIP1 encoding tumor suppressor folliculin-interacting protein might have contributed to the Interdigitating dendritic cell sarcoma (IDCS). Our study provides potential causative genetic factors of IDCS and plays a role in advancing the understanding of IDCS pathogenesis 30699359_casein kinase-2 phosphorylation of the co-chaperone folliculin-interacting protein 1 (FNIP1) on priming serine-938 and subsequent relay phosphorylation on serine-939, 941, 946, and 948 promotes its gradual interaction with Hsp90. 32181500_Mutations of the gene FNIP1 associated with a syndromic autosomal recessive immunodeficiency with cardiomyopathy and pre-excitation syndrome. 32905580_Absent B cells, agammaglobulinemia, and hypertrophic cardiomyopathy in folliculin-interacting protein 1 deficiency. 32941802_A Cellular Mechanism to Detect and Alleviate Reductive Stress. 33459596_Loss of FLCN-FNIP1/2 induces a non-canonical interferon response in human renal tubular epithelial cells. | ENSMUSG00000035992 | Fnip1 | 112.03191 | 1.1799030 | 0.2386683034 | 0.31276561 | 6.191667e-01 | 4.313571e-01 | 9.998360e-01 | No | Yes | 120.74482 | 26.322214 | 1.108610e+02 | 18.722702 | |
| ENSG00000219545 | 729852 | UMAD1 | protein_coding | C9J7I0 | Mouse_homologues NA; + ;NA | Reference proteome | Mouse_homologues NA; + ;NA | hsa:729852; | Mouse_homologues NA;NA | 32737115_Association of the RPA3-UMAD1 locus with interstitial lung diseases complicated with rheumatoid arthritis in Japanese. | ENSMUSG00000089862+ENSMUSG00000107705 | Umad1+Gm45062 | 119.05217 | 1.5732570 | 0.6537543834 | 0.30304506 | 4.844270e+00 | 2.773813e-02 | 9.998360e-01 | No | Yes | 155.29179 | 31.109080 | 1.017809e+02 | 15.884023 | |
| ENSG00000221817 | 101929145 | PPP3CB-AS1 | lncRNA | 53.54224 | 1.3726682 | 0.4569829546 | 0.41822233 | 1.219299e+00 | 2.694982e-01 | 9.998360e-01 | No | Yes | 59.03672 | 12.569003 | 4.223697e+01 | 7.114966 | ||||||||||
| ENSG00000223478 | 100506100 | ZDHHC12-DT | lncRNA | 45.93384 | 1.2928329 | 0.3705357691 | 0.43562435 | 6.981402e-01 | 4.034093e-01 | 9.998360e-01 | No | Yes | 46.23507 | 10.449466 | 4.334931e+01 | 8.108001 | ||||||||||
| ENSG00000223745 | CCDC18-AS1 | lncRNA | 51.66221 | 1.0350441 | 0.0496922206 | 0.41043721 | 1.523584e-02 | 9.017638e-01 | 9.998360e-01 | No | Yes | 60.75399 | 14.021516 | 5.992153e+01 | 10.837302 | |||||||||||
| ENSG00000223960 | 101927027 | CHROMR | lncRNA | 104.43782 | 1.0359233 | 0.0509172388 | 0.32979194 | 2.513870e-02 | 8.740219e-01 | 9.998360e-01 | No | Yes | 110.32750 | 20.885838 | 1.115650e+02 | 16.488903 | ||||||||||
| ENSG00000224046 | 101927420 | DMTF1-AS1 | lncRNA | 49.92193 | 0.7360026 | -0.4422172243 | 0.41613018 | 1.156729e+00 | 2.821450e-01 | 9.998360e-01 | No | Yes | 50.26309 | 12.053749 | 6.976685e+01 | 13.349577 | ||||||||||
| ENSG00000224846 | NQO2-AS1 | lncRNA | 20.57239 | 1.7044051 | 0.7692682877 | 0.65778959 | 1.251486e+00 | 2.632688e-01 | 9.998360e-01 | No | Yes | 25.80827 | 8.545396 | 1.726678e+01 | 4.898798 | |||||||||||
| ENSG00000224940 | 401399 | PRRT4 | protein_coding | C9JH25 | Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:401399; | integral component of membrane [GO:0016021] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000079654 | Prrt4 | 212.05564 | 1.0696002 | 0.0970716078 | 0.23238696 | 1.743381e-01 | 6.762849e-01 | 9.998360e-01 | No | Yes | 214.08924 | 36.142895 | 1.742658e+02 | 23.233841 | |||
| ENSG00000225096 | 101927293 | lncRNA | 151.74647 | 1.4417563 | 0.5278272791 | 0.25590833 | 4.079399e+00 | 4.340908e-02 | 9.998360e-01 | No | Yes | 208.11769 | 26.309238 | 1.474025e+02 | 15.043277 | |||||||||||
| ENSG00000226686 | 101927667 | LINC01535 | lncRNA | 31273925_Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. 32329845_LINC01535 promotes proliferation and inhibits apoptosis in esophageal squamous cell cancer by activating the JAK/STAT3 pathway. 33174047_lncRNA LINC01535 upregulates BMP2 expression levels to promote osteogenic differentiation via sponging miR36195p. | 103.68589 | 0.8865850 | -0.1736692129 | 0.29912046 | 3.434441e-01 | 5.578479e-01 | 9.998360e-01 | No | Yes | 120.43217 | 17.929739 | 1.323052e+02 | 17.666361 | |||||||||
| ENSG00000227372 | 57212 | TP73-AS1 | transcribed_unitary_pseudogene | 20477830_PDAM knockdown induces cisplatin resistance in glioma cells harboring wild-type p53. The finding suggests that PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes (BCL2 and BCL2L1). 20477830_The study suggested that PDAM might function as a non-protein-coding RNA.And PDAM deregulation play a role in oligodendroglial tumors development, PDAM may possess the capacity to modulate apoptosis via regulation of p53-dependent anti-apoptotic genes. 22388545_Report expression of TP73 isoforms in human cell lines. 26410378_KIAA0495 methylation was detected in none of both primary myeloma samples at diagnosis (N = 61) and at relapse/progression (N = 16). KIAA0495 methylation appeared unimportant in the pathogenesis or progression of myeloma. 26799587_High expression level of long non-coding RNA TP73-AS1 is associated with esophageal squamous cell carcinoma. 28379612_Study showed that TP73-AS1 was specifically upregulated in brain glioma tissues and cell lines, and was associated with poorer prognosis in patients with glioma. TP73-AS1 knock down suppressed human brain glioma cell proliferation and invasion in vitro. TP73-AS1 seems to act as an oncogenic lncRNA promoting brain glioma proliferation and invasion. 28639399_Study showed that TP73-AS1 was specifically upregulated in BC tissues and breast cancer (BC) cell lines and was correlated to a poor prognosis. Its knocking down suppressed BC cell proliferation in vitro through regulation of TFAM. Also, data revealed that TP73-AS1 could regulate miR-200a through direct targeting. 28857253_Data show a direct binding between TP73-AS1, miR-200a and ZEB1. TP73-AS1 promoted ZEB1 expression via competing with its 3'UTR for miR-200a binding. ZEB1 binds the promoter region of TP73-AS1 activating its expression. TP73-AS1 and ZEB1 expression was increased, whereas miR-200a expression was low in breast cancer (BC). These regulating loop TP73-AS1/miR-200a/ZEB1 in BC promotes cancer cell invasion and migration. 29412778_TP73-AS1 inhibited the brain glioma growth and metastasis as a competing endogenous RNA (ceRNA) through miR-124-dependent iASPP regulation. 29474580_miR-941 sponge region of nine high-affinity miR-941 binding sites clustering within 1 kb 29625110_lncRNA TP73-AS1 may serve as a tumor suppressor participated in bladder cancer progression, which provided a promising therapy strategy for patients with bladder cancer. 29750302_TP73-AS1 may be an oncogenic lncRNA that promotes the proliferation of ovarian cancer cells 29803931_An explicit oncogenic role of TP73-AS1 in the NSCLC tumorigenesis, suggesting a TP73-AS1-miR-449a-EZH2 axis and providing new insight for NSCLC tumorigenesis. 29864904_TP73-AS1 could function as a sponge of miR-142 to positively regulate Rac1 in osteosarcoma cells. 29904939_Study found that TP73-AS1 was upregulated in the ovarian cancer tissues and ovarian cancer cells, and correlated with poor prognosis. it promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. 29966969_TP73-AS1 was upregulated in cholangiocarcinoma, facilitating migration and invasive potential of tumor cells. 30010111_this study demonstrated that TP73-AS1 regulated Colorectal cancer (CRC)progression by acting as a competitive endogenous RNA to sponge miR-194 to modulate the expression of TGFa 30016766_TP73-AS1 knockdown activated PI3K/Akt/mTOR signaling pathway, while overexpression of TP73-AS1 induced inhibition of PI3K/Akt/mTOR pathway and these effects could be partly abolished by overexpression of KISS1. 30118890_Data showed that TP73-AS1 was enhanced in gastric cancer (GC) tissues and cells, and tightly associated with tumor size, TNM stage, and overall survival. Decreased TP73-AS1 could restrain cell growth and promoted apoptosis partly by regulating Bcl-2/caspase-3 pathway. Importantly, TP73-AS1 could promote xenograft growth in vivo. 30193732_Twist-related protein 1 (TWIST1) was a target of miR-490-3p and participated in long non-coding RNA TP73 antisense RNA 1 (TP73-AS1)/miR-490-3p-modulated MDA-MB-231cell vasculogenic mimicry (VM) formation. 30279010_TP73-AS1 was upregulated in gastric cancer tissues and cell lines. Furthermore, TP73-AS1 exerted oncogenic role in gastric cancer through promoting cell growth and metastasis. In addition, TP73-AS1 was certified as a ceRNA by regulating miR-194-5p/SDAD1 axis. 30472379_we showed that TP73AS1 inhibits CRC cell growth by functioning as a ceRNA (competing endogenous RNAs) to regulate PTEN levels. Our findings provide new insights into the underlying molecular mechanisms of TP73AS1-mediated CRC. 30541897_LncRNA TP73-AS1 promoted the progression of lung adenocarcinoma via PI3K/AKT pathway. 30643007_LncRNA TP73 antisense RNA 1T (TP73-AS1) positively regulates 3-hydroxybutyrate dehydrogenase type 2 (BDH2) by regulating miR-141. 30867410_results reveal that high TP73-AS1 predicts poor prognosis in primary glioblastoma multiform cohorts and that this lncRNA promotes tumor aggressiveness and temozolomide resistance in glioblastoma cancer stem cells. 31015368_LncRNA TP73-AS1 down-regulates miR-139-3p to promote retinoblastoma cell proliferation. 31081944_The lncRNA TP73-AS1 is a prognostic marker. 31129247_Results found that P73-AS1 was upregulated in non-small cell lung cancer (NSCLC) tissues and predicted poor survival. TP73-AS1 is a potential upstream positive regulator of miRNA to promote NSCLC cell migration and invasion. 31210297_LncRNA TP73-AS1 promotes malignant progression of hepatoma by regulating microRNA-103. 31232491_LncRNA TP73-AS1 is a novel regulator in cervical cancer via miR-329-3p/ARF1 axis. 31432138_the findings suggested that upregulation of TP73AS1 promoted cervical cancer progression by promoting CCND2 via the suppression of miR607 expression. 31549851_Association of TP73-AS1 gene polymorphisms with the risk and survival of gastric cancer in a Chinese Han Population 31652459_Role of long non-coding RNA TP73-AS1 in cancer. 31663517_LncRNA TP73-AS1 promoted proliferation of cervical cancer cell lines by targeting miR-329-3p to regulate the expression of the SMAD2 gene. A regulatory network was formed between lncRNA TP73-AS1, miR-329-3p, and SMAD2. 31678571_role of TP73-AS1 lncRNA in human cancers 31901156_Knockdown of long noncoding RNA TP73-AS1 suppresses the malignant progression of breast cancer cells in vitro through targeting miRNA-125a-3p/metadherin axis. 31978409_Long non-coding RNA TP73-AS1 in cancers. 32412786_lncRNA TP73-AS1 Regulates miR-21/PTEN Axis to Affect Cell Proliferation in Acute Myeloid Leukemia. 32818670_LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-beta1 signaling. 32901838_Long noncoding RNA TP73AS1 accelerates the progression and cisplatin resistance of nonsmall cell lung cancer by upregulating the expression of TRIM29 via competitively targeting microRNA34a5p. 33400379_LncRNA TP73-AS1 regulates miR-495 expression to promote migration and invasion of nasopharyngeal carcinoma cells through junctional adhesion molecule A. 33589576_Long non-coding RNA TP73-AS1 is a potential immune related prognostic biomarker for glioma. 33683827_LncRNA TP73-AS1 enhances the malignant properties of pancreatic ductal adenocarcinoma by increasing MMP14 expression through miRNA -200a sponging. 34007135_Long non-coding RNA TP73-AS1 promotes pancreatic cancer growth and metastasis through miRNA-128-3p/GOLM1 axis. 34103010_LncRNA TP73-AS1 promotes oxidized low-density lipoprotein-induced apoptosis of endothelial cells in atherosclerosis by targeting the miR-654-3p/AKT3 axis. 34115613_TP73-AS1 is induced by YY1 during TMZ treatment and highly expressed in the aging brain. | 2336.19759 | 1.0549596 | 0.0771878159 | 0.11993748 | 4.304517e-01 | 5.117674e-01 | 9.998360e-01 | No | Yes | 2615.23315 | 214.301975 | 2.373135e+03 | 151.008053 | |||||||||
| ENSG00000228175 | GEMIN8P4 | processed_pseudogene | 42.30186 | 0.8971272 | -0.1566155650 | 0.46585100 | 1.116542e-01 | 7.382687e-01 | 9.998360e-01 | No | Yes | 42.88627 | 8.639501 | 5.132298e+01 | 8.263966 | |||||||||||
| ENSG00000228288 | 100506696 | PCAT6 | lncRNA | 27458097_PCAT6 is a novel potential oncogenic lncRNA in lung cancer. Combined with the results that high PCAT6 expression positively correlated with metastasis and showed poor overall survival in lung cancer patients, our studies suggest that PCAT6 may be a potential molecular therapeutic target or a prognostic marker for patients with lung cancer. 30314898_mechanistic investigation demonstrated that the oncogenic activity of PCAT6 is partially attributable to its repression of LATS2 via association with the epigenetic repressor EZH2 (Enhancer of zeste homolog 2). 30569799_Our data highlighted that PCAT6 acts as a key activator of ARC expression by forming a complex with EZH2, inhibiting cell apoptosis and contributing to colon cancer progression. These findings elucidated that PCAT6 may be a novel prognostic predictor and therapeutic target of colon cancer 30915737_Long noncoding RNA PCAT6 regulates cell growth and metastasis via Wnt/beta-catenin pathway and is a prognosis marker in cervical cancer. 30938104_we demonstrate that the abnormal PCAT6 overexpression inhibits miR-204 expression in CRC, thereby promoting HMGA2/PI3K signaling activity, ultimately enhancing the chemoresistance of CRC cells to 5-FU; PCAT6 represents a promising target for dealing with CRC chemoresistance. 31646553_LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. 31676070_PCAT6, acting as a competitive endogenous RNA, upregulated expression of TGFBR1 and TGFBR2 to activate TGF-beta pathway via sponging miR-185-5p. 32039820_Silencing of long non-coding RNA PCAT6 restrains gastric cancer cell proliferation and epithelial-mesenchymal transition by targeting microRNA-15a. 32064636_LncRNA PCAT6 predicts poor prognosis in hepatocellular carcinoma and promotes proliferation through the regulation of cell cycle arrest and apoptosis. 32339330_PCAT6 mediates cellular biological functions in gastrointestinal stromal tumor via upregulation of PRDX5 and activation of Wnt pathway. 32908134_M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. 32990800_LncRNA PCAT6 Regulated by YY1 Accelerates the Progression of Glioblastoma via miR-513/IGF2BP1. 33090394_LncRNA PCAT6 aggravates the progression of bladder cancer cells by targeting miR-513a-5p. 33125146_Long noncoding RNA PCAT6 promotes the development of osteosarcoma by increasing MDM2 expression. 33142195_LncRNA PCAT6: A potential biomarker for diagnosis and prognosis of bladder cancer. 33761616_Promising role of long non-coding RNA PCAT6 in malignancies. 34185427_m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. 35081872_Long non-coding RNA prostate cancer-associated transcript 6 inhibited gefitinib sensitivity of non-small cell lung cancer by serving as a competing endogenous RNA of miR-326 to up-regulate interferon-alpha receptor 2. | 40.04523 | 1.3210923 | 0.4017312216 | 0.47568070 | 7.090506e-01 | 3.997592e-01 | 9.998360e-01 | No | Yes | 48.01176 | 12.099745 | 3.447979e+01 | 7.287331 | |||||||||
| ENSG00000228315 | 91316 | GUSBP11 | lncRNA | This transcribed pseudogene is similar to two functional genes. The 5' portion of the pseudogene is related to glucuronidase, beta, and the 3' portion is related to immunoglobulin lambda-like polypeptide 1. [provided by RefSeq, Jul 2011]. | 1244.56726 | 0.9883641 | -0.0168854504 | 0.12974347 | 1.726097e-02 | 8.954739e-01 | 9.998360e-01 | No | Yes | 1247.66426 | 161.376156 | 1.202506e+03 | 120.446489 | |||||||||
| ENSG00000231074 | 414777 | HCG18 | lncRNA | 30426533_Low HCG18 expression is associated with proliferation and migration of bladder cancer. 31841193_LncRNA HCG18 contributes to nasopharyngeal carcinoma development by modulating miR-140/CCND1 and Hedgehog signaling pathway. 32663252_LncRNA HCG18 contributes to the progression of hepatocellular carcinoma via miR-214-3p/CENPM axis. 32725768_Long noncoding RNA HCG18 up-regulates the expression of WIPF1 and YAP/TAZ by inhibiting miR-141-3p in gastric cancer. 33176682_Long noncoding RNA HCG18 inhibits the differentiation of human bone marrow-derived mesenchymal stem cells in osteoporosis by targeting miR-30a-5p/NOTCH1 axis. 33596783_Identification of HCG18 and MCM3AP-AS1 That Associate With Bone Metastasis, Poor Prognosis and Increased Abundance of M2 Macrophage Infiltration in Prostate Cancer. 33798913_LncRNA-HCG18 regulates the viability, apoptosis, migration, invasion and epithelial-mesenchymal transition of papillary thyroid cancer cells via regulating the miR-106a-5p/PPP2R2A axis. 34041718_Knockdown of HCG18 Inhibits Cell Viability, Migration and Invasion in Pediatric Osteosarcoma by Targeting miR-188-5p/FOXC1 Axis. 34523356_LncRNA HCG18 suppresses CD8(+) T cells to confer resistance to cetuximab in colorectal cancer via miR-20b-5p/PD-L1 axis. 34618022_Long non-coding RNA HLA complex group 18 promotes gastric cancer progression by targeting microRNA-370-3p expression. 34708395_lncRNA HLA Complex Group 18 (HCG18) Facilitated Cell Proliferation, Invasion, and Migration of Prostate Cancer Through Modulating miR-370-3p/DDX3X Axis. 34878161_Long noncoding RNA HCG18 facilitates the progression of laryngeal and hypopharyngeal squamous cell carcinoma by upregulating FGFR1 via miR133b. 34983361_LncRNA HCG18 upregulates TRAF4/TRAF5 to facilitate proliferation, migration and EMT of epithelial ovarian cancer by targeting miR-29a/b. 34991445_LncRNA HCG18 promotes osteosarcoma growth by enhanced aerobic glycolysis via the miR-365a-3p/PGK1 axis. 35240920_Long non-coding RNA HCG18 promotes gastric cancer progression by regulating miRNA-146a-5p/tumor necrosis factor receptor-associated factor 6 axis. 35389764_Long non-coding RNA human leucocyte antigen complex group-18 HCG18 (HCG18) promoted cell proliferation and migration in head and neck squamous cell carcinoma through cyclin D1-WNT pathway. | 466.53243 | 1.0324111 | 0.0460175216 | 0.16544178 | 7.719046e-02 | 7.811416e-01 | 9.998360e-01 | No | Yes | 584.35322 | 67.139726 | 6.298511e+02 | 56.105745 | |||||||||
| ENSG00000232677 | 100506930 | LINC00665 | lncRNA | 29728556_LINC00665 may be involved in the regulation of cell cycle pathways in hepatocellular carcinoma through ten identified hub genes. 30692511_Transcription factor SP1 induced the transcription of linc00665 in LUAD cells, which exerted its oncogenic role by functioning as competing endogenous RNA (ceRNA) for miR-98 and subsequently activating downstream AKR1B10-ERK signaling pathway. 31736127_LINC00665 induces gastric cancer progression through activating Wnt signaling pathway. 31907362_LINC00665 promotes breast cancer progression through regulation of the miR-379-5p/LIN28B axis. 32083756_Inflammation-Induced Long Intergenic Noncoding RNA (LINC00665) Increases Malignancy Through Activating the Double-Stranded RNA-Activated Protein Kinase/Nuclear Factor Kappa B Pathway in Hepatocellular Carcinoma. 32271427_Long non-coding RNA LINC00665 promotes metastasis of breast cancer cells by triggering EMT. 32403047_Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. 32423800_Long non-coding RNA linc00665 interacts with YB-1 and promotes angiogenesis in lung adenocarcinoma. 32776307_LINC00665/miR-9-5p/ATF1 is a novel axis involved in the progression of colorectal cancer. 33090388_LINC00665 facilitates the progression of osteosarcoma via sponging miR-3619-5p. 33650673_LINC00665 functions as a competitive endogenous RNA to regulate AGTR1 expression by sponging miR34a5p in glioma. 33658535_LINC00665 promotes the progression of acute myeloid leukemia by regulating the miR-4458/DOCK1 pathway. 33865827_LINC00665 activates Wnt/beta-catenin signaling pathway to facilitate tumor progression of colorectal cancer via upregulating CTNNB1. 33903885_LINC00665 promotes HeLa cell proliferation, migration, invasion and epithelial-mesenchymal transition by activating the WNT-CTNNB1/betacatenin signaling pathway. 34171521_LINC00665 promotes the viability, migration and invasion of T cell acute lymphoblastic leukemia cells by targeting miR-101 via modulating PI3K/Akt pathway. 34232917_Downregulation of LINC00665 suppresses the progression of lung adenocarcinoma via regulating miR-181c-5p/ZIC2 axis. 34306997_LINC00665 Facilitates the Malignant Processes of Osteosarcoma by Increasing the RAP1B Expression via Sponging miR-708 and miR-142-5p. | 372.51142 | 0.9422150 | -0.0858717721 | 0.17857920 | 2.364256e-01 | 6.267998e-01 | 9.998360e-01 | No | Yes | 403.14377 | 31.903595 | 4.239396e+02 | 27.198553 | |||||||||
| ENSG00000234684 | 100507495 | SDCBP2-AS1 | lncRNA | 34218800_Long non-coding RNA SDCBP2-AS1 delays the progression of ovarian cancer via microRNA-100-5p-targeted EPDR1. | 82.76249 | 0.9542620 | -0.0675427085 | 0.34021811 | 4.175060e-02 | 8.380959e-01 | 9.998360e-01 | No | Yes | 128.40871 | 19.925644 | 1.303493e+02 | 16.075398 | |||||||||
| ENSG00000236088 | 100874058 | COX10-DT | lncRNA | 34323174_Regulating COX10-AS1 / miR-142-5p / PAICS axis inhibits the proliferation of non-small cell lung cancer. | 143.81476 | 0.9548261 | -0.0666900903 | 0.28451860 | 5.826159e-02 | 8.092650e-01 | 9.998360e-01 | No | Yes | 156.12244 | 26.370290 | 1.573567e+02 | 20.682083 | |||||||||
| ENSG00000236104 | 9278 | ZBTB22 | protein_coding | O15209 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:9278; | chromatin [GO:0000785]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357] | 19851445_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000051390 | Zbtb22 | 835.65298 | 1.0410805 | 0.0580816060 | 0.13996555 | 1.780270e-01 | 6.730743e-01 | 9.998360e-01 | No | Yes | 897.60061 | 103.417963 | 8.078642e+02 | 72.829178 | ||
| ENSG00000236901 | 81571 | MIR600HG | lncRNA | 32270866_MIR600HG suppresses metastasis and enhances oxaliplatin chemosensitivity by targeting ALDH1A3 in colorectal cancer. | 346.38388 | 1.0504417 | 0.0709960929 | 0.18810237 | 1.458159e-01 | 7.025663e-01 | 9.998360e-01 | No | Yes | 410.99085 | 43.095497 | 3.811378e+02 | 32.442834 | |||||||||
| ENSG00000237523 | 439990 | LINC00857 | lncRNA | 26862852_Overexpression of LINC00857 increased cancer cell proliferation, colony formation and invasion. Mechanistic analyses indicated that LINC00857 mediates tumor progression via cell cycle regulation. 32138762_Long non-coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple-negative breast cancer. 32918541_LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. 33657224_LINC00857 contributes to proliferation and lymphomagenesis by regulating miR-370-3p/CBX3 axis in diffuse large B-cell lymphoma. 33661995_LncRNA LINC00857 strengthens the malignancy behaviors of pancreatic adenocarcinoma cells by serving as a competing endogenous RNA for miR-340-5p to upregulate TGFA expression. 33833823_Long Noncoding RNA LINC00857 Promotes Proliferation, Migration, and Invasion of Colorectal Cancer Cell through miR-1306/Vimentin Axis. 34753396_LINC00857 promotes colorectal cancer progression by sponging miR-150-5p and upregulating HMGB3 (high mobility group box 3) expression. 35293286_Knockdown of long noncoding RNA HUMT inhibits the proliferation and metastasis by regulating miR-455-5p/LRP4 axis in hepatocellular carcinoma. | 106.97722 | 0.8597995 | -0.2179278279 | 0.29040899 | 5.626136e-01 | 4.532091e-01 | 9.998360e-01 | No | Yes | 94.30710 | 15.284595 | 1.204909e+02 | 15.879964 | |||||||||
| ENSG00000240230 | 90639 | COX19 | protein_coding | Q49B96 | FUNCTION: Required for the transduction of an SCO1-dependent redox signal from the mitochondrion to ATP7A to regulate cellular copper homeostasis (PubMed:23345593). May be required for the assembly of mitochondrial cytochrome c oxidase (By similarity). {ECO:0000250|UniProtKB:Q3E731, ECO:0000269|PubMed:23345593}. | Acetylation;Cytoplasm;Disulfide bond;Mitochondrion;Reference proteome | COX19 encodes a cytochrome c oxidase (COX)-assembly protein. The S. cerevisiae Cox19 protein may play a role in metal transport to the mitochondrial intermembrane space and assembly of complex IV of the mitochondrial respiratory chain (Sacconi et al., 2005 [PubMed 16212937]).[supplied by OMIM, Mar 2008]. | hsa:90639; | cytosol [GO:0005829]; mitochondrial intermembrane space [GO:0005758]; mitochondrion [GO:0005739]; cellular copper ion homeostasis [GO:0006878]; mitochondrial cytochrome c oxidase assembly [GO:0033617] | 16212937_Both hCox18p and hCox19p present significant amino acid identity with the corresponding yeast polypeptides and reveal highly conserved functional domains. 23345593_COX19 is necessary for the transduction of a SCO1-dependent mitochondrial redox signal that regulates ATP7A-mediated cellular copper efflux. | ENSMUSG00000045438 | Cox19 | 680.10855 | 0.8014790 | -0.3192633824 | 0.14121932 | 5.121245e+00 | 2.363466e-02 | 9.998360e-01 | No | Yes | 553.38570 | 73.525835 | 6.773660e+02 | 69.614124 | |
| ENSG00000240583 | 358 | AQP1 | protein_coding | P29972 | FUNCTION: Forms a water-specific channel that provides the plasma membranes of red cells and kidney proximal tubules with high permeability to water, thereby permitting water to move in the direction of an osmotic gradient. {ECO:0000269|PubMed:1373524}. | 3D-structure;Alternative splicing;Blood group antigen;Cell membrane;Direct protein sequencing;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene encodes a small integral membrane protein with six bilayer spanning domains that functions as a water channel protein. This protein permits passive transport of water along an osmotic gradient. This gene is a possible candidate for disorders involving imbalance in ocular fluid movement. [provided by RefSeq, Aug 2016]. | hsa:358; | apical part of cell [GO:0045177]; apical plasma membrane [GO:0016324]; basal plasma membrane [GO:0009925]; basolateral plasma membrane [GO:0016323]; brush border [GO:0005903]; brush border membrane [GO:0031526]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; nuclear membrane [GO:0031965]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; sarcolemma [GO:0042383]; ammonium transmembrane transporter activity [GO:0008519]; carbon dioxide transmembrane transporter activity [GO:0035379]; glycerol transmembrane transporter activity [GO:0015168]; identical protein binding [GO:0042802]; intracellular cGMP-activated cation channel activity [GO:0005223]; nitric oxide transmembrane transporter activity [GO:0030184]; potassium channel activity [GO:0005267]; potassium ion transmembrane transporter activity [GO:0015079]; transmembrane transporter activity [GO:0022857]; water channel activity [GO:0015250]; water transmembrane transporter activity [GO:0005372]; ammonium transport [GO:0015696]; carbon dioxide transmembrane transport [GO:0035378]; carbon dioxide transport [GO:0015670]; cell volume homeostasis [GO:0006884]; cellular homeostasis [GO:0019725]; cellular hyperosmotic response [GO:0071474]; cellular response to cAMP [GO:0071320]; cellular response to copper ion [GO:0071280]; cellular response to dexamethasone stimulus [GO:0071549]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to hypoxia [GO:0071456]; cellular response to inorganic substance [GO:0071241]; cellular response to mechanical stimulus [GO:0071260]; cellular response to mercury ion [GO:0071288]; cellular response to nitric oxide [GO:0071732]; cellular response to retinoic acid [GO:0071300]; cellular response to salt stress [GO:0071472]; cellular response to UV [GO:0034644]; cellular water homeostasis [GO:0009992]; cerebrospinal fluid secretion [GO:0033326]; cGMP-mediated signaling [GO:0019934]; defense response to Gram-negative bacterium [GO:0050829]; establishment or maintenance of actin cytoskeleton polarity [GO:0030950]; glycerol transport [GO:0015793]; hyperosmotic response [GO:0006972]; lateral ventricle development [GO:0021670]; multicellular organismal water homeostasis [GO:0050891]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043154]; nitric oxide transport [GO:0030185]; odontogenesis [GO:0042476]; pancreatic juice secretion [GO:0030157]; positive regulation of angiogenesis [GO:0045766]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of saliva secretion [GO:0046878]; potassium ion transport [GO:0006813]; renal water homeostasis [GO:0003091]; renal water transport [GO:0003097]; transepithelial water transport [GO:0035377]; water transport [GO:0006833] | 11884383_transmembrane biogenesis is cotranslational in intact mammalian cells 11909995_A novel role for aquaporin-1 as a gated ion channel reshapes our current views of this ancient family of transmembrane channel proteins. 11922632_These data suggest that the transcription of the AQP1 by hypertonicity in renal cells is upregulated by the interaction with putative DNA binding proteins to a novel HRE located at -54 to -46 in the AQP1 gene. 12002613_This study showed the distinct localization of AQP1 in the mesangial cells of human glomeruli, suggesting its role in water movement through these cells. 12027013_visualization of the water-selective pathway at 3.7A resolution in a three-dimenional density map (REVIEW) 12237771_In astrocytomas, aquaporin 1 expressed in microvessel endothelia and neoplastic astrocytes. In metastatic carcinomas, aquaporin 1 present in microvessel endothelia and reactive astrocytes. Aquaporin 1 may participate in formation of brain tumour oedema. 12399631_Data suggest that variations found in plasma osmolarity during hemodialysis may induce aquaporin 1 expression on the membrane of intact red blood cells. 12498798_Data present a detailed comparison between the cryo-electron microscopy and X-ray crystallography model structures of the human and bovine water channel aquaporin-1 (AQP1). 12781664_Transmembrane helices at the periphery of the hAQP1 tetramer exhibited smaller extraction forces than helices at the interface between hAQP1 monomers. 14514735_substantial and striking upregulation of AQP-1 in the glomeruli of most diseased kidneys. 14592814_presence of AQP-1 in endothelia and water-transporting epithelia and new locations: mammary epithelium, articular chondrocytes, synoviocytes, and synovial microvessels where it may be involved in milk, chondrocyte volume, synovial fluid, and homeostasis. 14701836_lack of significant cyclic guanine nucleotide ion channel activity rules out a secondary role of aquaporin 1 water channels in cellular signal transduction 14753493_Increased expression. Over-expression occurred on astrocytic processes. Induction of AQP1 and AQP4 on reactive astrocytes in subarachnoid hemorrhage. May be involved in brain edema formation or resolution. 14753494_Intense upregulation of AQP1 expression was found in all glioblastomas. Expression of aquaporins in glioblastomas suggests pathologic role. Selective AQP inhibition might be new therapeutic option for tumor-associated cerebral edema. 15024704_AQP1 has a role in the movement of extracellular matrix and metabolic water across the membranes of chondrocytes and synoviocytes 15135660_AQP1 co-localizes with t-tubular and caveolar proteins 15502805_Aquaporin-1 is a reliable marker for clear cell renal cell carcinomas of lower grades but not for higher grades. 15563082_The expression of AQP-1 mRNA was positively correlated with Bcl-2 mRNA expression in nasal polyps. AQP-1 contributes to the survival of eosinophils in nasal polyps. 15667881_AQP-1 may be a possible critical reabsorption factor, acting to reduce abnormal fluid retention in endotubular cells and the extracellular matrix and, to a lesser extent, in Leydig cells. 15783300_Observational study of genotype prevalence. (HuGE Navigator) 15809704_increased AQP1 expression in some human adenocarcinomas may be a consequence of angiogenesis and important for the formation or clearance of tumor edema 15847654_Observational study of genetic testing. (HuGE Navigator) 16300893_AQP1 expression heavely in cystic hemangioblastomas. 16481371_Aquaporin (AQP) 1 is predominantly situated in the apical plasma membrane domain of the human choroid plexus, although distinct basolateral and endothelial immunoreactivity is also observed. 16515633_high AQP1 expression may play an important role in ovarian carcinogenesis, disease progression, and ascites formation. 16534779_AQPs are differentially expressed in the peripheral versus central nervous system and that channel-mediated water transport mechanisms may be involved in peripheral neuronal activity by regulating water homeostasis in nerve plexuses and bundles. 16574458_Review. In Colton (null) RBC ghosts, lack of AQP1 resulted in about 30% reduction of the alkalinization rates. 16698771_aquaporin-1-mediated CO(2) permeation is to be expected only in membranes with a low intrinsic CO(2) permeability 16814974_These results in human fetal brain lend morphological support to the previous findings that aquaporin-1 and aquaporin-4 play different roles in the regulation of the water homeostasis of the brain. 16871401_Significant increased expression levels of AQP1 and AQP4 were seen in Creutzfeldt-Jakob disease, but not in advanced Alzheimer's disease and diffuse Lewy body disease. 17012249_AQP1 is responsible for 60% of the high P(CO2) of red cells and that another, so far unidentified, CO2 pathway is present in this membrane that may account for at least 30% of total P(CO2). 17077939_In hemangioblastomas, expression of AQP1 was predominantly localized on membranes of stromal cells. The expression level of AQP1 in cystic group of hemangioblastomas is much higher than that of solid group. 17219999_Inhibiting AQP-1 with acetazolamide may significantly induce apoptosis of Hep-2 cells. 17273788_The deregulation of aquaporin-1 in menorrhagia may be involved in abnormal endometrial vascular growth and permeability. 17408468_Observational study of gene-disease association. (HuGE Navigator) 17409744_AQP1 is expressed at the endomysial capillary endothelial cell and further AQP1 may be expressed at the human skeletal myofiber plasma membrane. 17511167_AQP1 mRNA and protein expression level in laryngeal tumor tissues is remarkably stronger than that in normal tissues. 17545093_Data show that AQP-1, 3, 8, 9 mRNA expression was detected in both amnion and chorion and can be associated with intramembranous transport and volume regulation of amniotic fluid. 17549682_Only the presence of AQP1, but not AQP4, enhanced cell growth and migration, typical properties of gliomas, while AQP4 enhanced cell adhesion suggesting differential biological roles for AQP1 and AQP4 in glioma cell biology. 17632520_In AQP1, Asn49 and Lys51 interact with Asp185 at the C terminus of TM5 to form a polar, quaternary structural motif that influences multiple stages of folding. 17854859_AQP-1 is up-regulated in biliary dysplasia...and down-regulation of AQP-1 is associated with mucin production and aggressive progression of intrahepatic cholangiocarcinoma. 17890385_These results establish the nature and determinants of AQP1 diffusion in cell plasma membranes and demonstrate long-range nonanomalous diffusion of AQP1. 17894331_a detailed mechanism for ion exclusion in aquaporin-1 (AQP1) at an atomistic level is investigated by calculating the free energy for transport of ions in AQP1 17898873_Increased AQP1 expression may relieve intracellular acidosis and edema in highly glycolytic glioma cells. 18052958_Aquaporin 1 may be involved in the pathophysiology of migraine. 18067501_Observational study of genetic testing. (HuGE Navigator) 18080132_These data illustrate the potential of the peritoneal membrane as an experimental model in the investigation of the role of AQP1--REVIEW 18202181_For small solutes permeating through AQP1, a remarkable anticorrelation between permeability and solute hydrophobicity is observed, rendering AQP1 a selective filter for small polar solutes. 18247144_The aim of this study was to use immunohistochemsitry to investigate the expression of aquaporins 1, 2 and 3 within the human intervertebral disc. 18275976_AQP1 is involved in hypoxia-inducible angiogenesis in retinal vascular endothelial cells through a mechanism that is independent of the VEGF signaling pathway. 18280225_AQP1 is not expressed in human airway epithelial cells from the A549 adeoncarcinoma cell line. 18282122_The genomic, structural and functional aspects of AQP1 are briefly described. Its role in human tumors and, in particular, those of the kidney is discussed. Review. 18313673_AQP1 may be involved in the tumorigenesis and progression of endometrioid adenocarcinoma by promoting angiogenesis, and AQP1 level may be both a tumor indicator and a new therapeutic target. 18392839_Our study shows that AQP4 downregulation can occur in muscular dystrophies with either normal or disrupted expression of dystrophin-associated proteins, and that this might be associated with upregulation of AQP1. 18509662_These observations suggest the possible association of astrocytic AQP1 with Abeta deposition in Alzheimer disease brains. 18510579_Observational study of genetic testing. (HuGE Navigator) 18538351_Modulation of AQP1 expression by maternal hormones may regulate invasion and fetal-placental-amnion water homeostasis during gestation. 18544259_We show high expression of AQP1 water channels in intractable epilepsy and suggest two mechanisms to explain this finding. Increased AQP1 expression of astrocytes may be a cause or a consequence of IE. 18563339_AQP1 and HIF1 interact each other and regulate the oncogenesis of breast cancer 18575775_The objective of this study was to evaluate the AQP1 expression in endometrial blood vessels during normal cycle and after mifepristone treatment. 18841368_AQP1 expression in invasive breast carcinomas is associated with a basal-like phenotype and poor prognosis. 19080511_The expression level of AQP1 of patients with preeclampsia increases in placenta and peritoneum and decreases in embryolemma. 19253825_Hypoxia regulates the expression of AQP1 in vascular endothelial cells. 19306058_AQP1 expression is a new characteristic feature of a particularly aggressive subgroup of basal-like breast carcinomas. 19424603_AQP1-specific siRNA knockdown impaired water permeability of ARPE-19 cells. 19472194_These results provide evidence that NPA motifs are important for water permeation but not essential for the expression, intracellular processing and the basic structure of human aquaporin 1. 19522191_The expression of AQP1 in tumor cells and micro-angiogenesis of primary laryngeal carcinoma are higher than normal. 19545896_Alteration of aquaporin 1 and aquaporin 3 expression in fetal membranes and placenta may be important in the pathophysiology of isolated oligohydramnios. 19619954_AQP-1 was overexpressed in hepatitis B virus associated cirrhotic liver tissues. AQP-1, similar to CK19, might be a more specific and more sensitive marker than CK7 for the identification of HPCs. 19670620_The expression of AQP1 and VEGF in laryngeal carcinoma was significantly higher than that in vocal cord polyps and normal controls. 19726340_The expression of AQP1 mRNA was significantly lower in oligohydramnios placenta than in normal pregnancy placenta at term. 19772916_TXA(2) receptor mediates water influx through aquaporins in astrocytoma cells via TXA(2) receptor-mediated activation of G alpha(12/13), Rho A, Rho kinase and Na(+)/H(+)-exchanger. 19787701_AQP1 activity of cell membrane affects HT20 colon cancer cell migration. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20063900_An increase in the level of water transport in response to changing osmotic conditions in the cellular environment may be due to a protein kinase C-dependent increase in AQP1 membrane localization. 20101282_Observational study of gene-disease association. (HuGE Navigator) 20101282_There was no association between common sequence variants in the AQP1 or SLC4A10 genes and primary open-angle glaucoma in the Caucasian population. 20137115_Inhibiting AQP1 expression with siRNA can inhibit the proliferation and induce apoptosis of K562 cells. 20149606_Potential role for synovial AQP1 and other aquaporins in joint swelling and vasogenic edema. 20360993_Observational study of gene-disease association. (HuGE Navigator) 20409716_Aquaporin 1 is localized in the dura mater following chronic subdural hematoma; the outer membrane might be the source of increased fluid accumulation responsible for chronic hematoma enlargement. 20424473_Observational study of gene-disease association. (HuGE Navigator) 20431033_Observational study of genetic testing. (HuGE Navigator) 20461409_Expression of AQP1, AQP4, or AQP6 mRNA did not differ in vestibular endorgans from patients with Meniere's disease. 20578142_AQP-1 enhances osmotic water permeability and FGF-induced dynamic membrane blebbing in liver endothelial cell and thereby drives invasion and pathological angiogenesis during cirrhosis. 20628061_Data identify miR-320a as a potential modulator of aquaporin 1 and 4 and explore the possibility of using miR-320a to alter the expression of aquaporin 1 and 4 in normal and ischemic conditions. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20739606_Compared with human AQP1, zebrafish Aqp1a has about twice the selectivity for CO(2) over NH(3). 20795314_The physiology and molecular basis of the Colton blood group antigen AQP1 is discussed. Review. 20806077_The purpose of this study was to determine the effects of down-regulation of Aquaporin 1 (AQP1) and Aquaporin 5 (AQP5) on cell proliferation and migration in human corneal endothelial (HCEC) and human corneal epithelial (CEPI17) cell lines, respectively. 20828513_The expression level of aquaporin-1 in nasopharyngeal cancer tissue could be involved in tumour migration. 20965731_Expression of AQP1 in pediatric brain tumors. Increased in ependymomas in posterior fossa and in pilocytic astrocytomas. Not expressed in medulloblastoma, primitive neuroectodermal tumor, germinoma. 20969805_This review outlines newly emerging evidence indicating that AQP-1 plays an important role in pain signal transduction and migraine. 21063116_After sequencing analysis of the coding regions and exon-intron boundaries one single variation, no significant mutation in AQP 1 was found. 21063116_Observational study of gene-disease association. (HuGE Navigator) 21107133_No difference is found between aquaporin 1 expression in Alzheimer's disease brain and its expression in cerebral amyloid angiopathy. 21237499_Expression patterns of aquaporin 1, 3, and 5 in lung cancer cells are mostly associated with cellular differentiation. 21244858_AQP1 and AQP5 might play an important role in the development of lung edema and lung injury 21252246_The late rise of AQP1 suggests a role in corpus luteum formation. 21271497_The researchers found an association between carriers of the AQP1 single nucleotide polymorphism and greater fluid loss in long-distance running. 21360438_An important role of AQP1 in tumor angiogenesis is sustained by the abundant expression of this protein in the endothelia of tumor capillaries. 21373963_Water flux through human aquaporin 1 was studied under osmotic challenges and the inhibition by intracellular furosemide. 21395179_AQP1 and VEGF had a higher expression in nasopharyngeal carcinoma tissues than in non-tumor tissues. 21538271_AQP-1 expression is increased in colorectal carcinoma while the expression of AQP-3 is not. There is no correlation between the expression of AQP-1 and AQP-3 in CRC. 21551254_AQP1 is a promising oncogene candidate for ACC and is transcriptionally regulated by promoter hypomethylation. 21612401_hypoxia-induced expression of AQP1 requires transcriptional activation, and the HIF-1 binding site of the 5'-promoter is necessary for transcriptional activation 21760919_Negative stain transmission electron microscopy and single particle analysis of KCC4 and the aquaporin-1 AQP1 water channel, revealed the expected quaternary structures within homogeneous preparations, and thus correct protein folding and assembly. 21784068_REVIEW: summarizes literature data concerning the involvement of AQP-1 and -4 in human brain tumor growth and edema formation 21793635_our results suggest, for the first time, that the rs1049305 (C/G, UTR3) AQP1 polymorphism could be involved in the genetic susceptibility to develop water retention in patients with liver cirrhosis. 21839760_This study demonistrated that AQP1 in inner ear plays an important role in the development of motion sickmness, and might be a potential target for the prevention or management of motion sickness. 21896312_The mean concentration of AQP1 in CSF was significantly elevated in patients with BM (BM: 3.8+/-3.4ng/ml, controls 22006723_Phosphorylation of tyrosine Tyr253 in the carboxyl terminal domain, acts as a master switch regulating responsiveness of AQP1 ion channels to cGMP, and the tetrameric central pore is the ion permeation pathway. 22093331_Aberrant expressions of AQP1 in periportal sinusoidal regions in human cirrhotic liver indicate the proliferation of arterial capillaries directly connected to the sinusoids, contributing to microvascular resistance in cirrhosis. 22269467_AQP1 is normally expressed in the temporomandibular joint (TMJ) disc with a role in the maintenance of TMJ homeostasis 22310126_regulates microvessel permeability and barrier function and contributes to blood vessel formation 22334691_novel pathway in mammalian cells whereby a hypotonic stimulus directly induces intracellular calcium elevations through transient receptor potential channels, which trigger AQP1 translocation 22348807_A new AQP1 null allele identified in a Gypsy woman who developed an anti-CO3 during her first pregnancy. 22372348_We found AQP1 and AQP4 in lung cancer cell extravasation and spread, which may provide a functional explanation for the expression of AQP1 and AQP4 in lung cancer tissues and lung cancer cell lines. 22472942_expression of aquaporin 1 and 5 was higher in uterine leiomyomata than in unaffected uteri 22901156_AQP1 expression significantly increased in advanced stage of cervical cancer, deeper infiltration, metastatic lymph nodes and larger tumor volume. 22901921_Exosomal AQP1 is downregulated after the release of ureteropelvic junction obstruction. This may be due to locally increased TGF-beta-1 in the postobstructed kidney. 22964306_Data indicate reconstitution of AQP-1 into cholesterol-containing vesicles leads to drastic increases in P(CO2). 23029502_Selective expression of AQP1 in the trigeminal neurons innervating the oral mucosa indicates an involvement of AQP1 in oral sensory transduction. 23219802_Stomatin interacts with GLUT1/SLC2A1, band 3/SLC4A1, and aquaporin-1 in human erythrocyte membrane domains 23220481_This study demonstrated that laminar shear stress stimulates the endothelial expression of AQP1 that plays a role in wound healing. 23268390_K562 cells show a significant increase of AQP1 expression after retinoic acid-induced erythroid differentiation. 23276695_AQP1(+) cells immediately beneath the epithelial basement membrane may be stromal niche-like cells that directly interact with N-cad(+) limbal basal epithelial progenitor cells. 23313295_High AQP1 expression is associated with malignant pleural mesothelioma. 23317544_These findings suggest that NKCC1 and AQP1 participate in meningioma biology and invasion 23332061_hAQP1 is a constitutively open channel that closes mediated by membrane-tension increments. 23361277_AQP1-immunoexpression had a good correlation with high-grade tumors. 23393355_AQP1 expression correlates with the grade of malignancy in astrocytoma and is associated with angiogenesis, as well as with invasion of grade IV tumour in areas of tumour infiltration 23450058_Estrogen induces AQP1 expression by activating ERE in the promoter of the Aqp1 gene, resulting in tubulogenesis of vascular endothelial cells. 23549977_AQP-1 appears to be involved in the arterial capillary proliferation in the cirrhotic liver. [Review] 23928039_AQP1 and 4 gene expression levels did not differ between mesial temporal sclerosis patients and control groups 23949237_Aquaporin-1 is associated with arterial capillary proliferation and hepatic sinusoidal transformation contributing to portal hypertension in primary biliary cirrhosis. 23974882_The data suggest that the increased expression of AQP1 and AQP3 in pterygial tissues may be involved in the pathogenesis of pterygia 24014128_Cox-regression analyses revealed the AQP1 -783G/C genotype status as an independent prognostic-factor when jointly considering other predictors of survivalin glioblastoma multiforme. 24028651_Aquaporin-1 is induced in leukocytes of patients with sepsis and exhibits higher expression in septic shock. 24086369_Anti-AQP1 autoantibodies are present in a subgroup of patients with chronic demyelination in the central nervous sytem and similarities with anti-AQP4-seronegative neuromyelitis optica spectrum disorders. 24169407_Over expression of aquaporin 1 causes increased intracranial pressure, and downregulation reduces pressure and alleviates the symptomatology and complications of idiopathic intracranial hypertension. 24252214_Report is the first to establish astrocytic water channel loss in a subset of human central pontine myelinolysis (CPM) cases and suggests AQP1 and AQP4 may be involved in the pathogenesis of CPM 24333057_the prevalence of this AQP1 c.601delG allele in the ethnic minority of Romani 24493792_AQP1 promoter hypomethylation is common in ACC, and AQP1 tends to be overexpressed in these tumors. Increased AQP1 methylation is associated with improved prognosis. 24688444_the expression of AQP1, at both the mRNA and protein levels, in membranes from PVR and ERM. 24777974_Results show that in AQP1, helix 3 inverts its orientation in the membrane after the initial insertion, whereas this does not occur in the homologous AQP4. 24803718_Freshly obtained urine samples from renal cell carcinoma patients are positive for AQP1 whereas archived samples and fresh healthy control samples are negative. 24918928_Aquaporin 1 and 3 were upregulated in cervical cancer compared to mild cervicitis and cervical intraepithelial neoplasia 2-3 (P<0.05). 25252847_AQP1 over-expression effectively inhibited cell proliferation and induced cell growth arrest in G1 phase of K562 cells. 25299207_the reduction of aquaporins 1 and 3 may be a factor resulting in lumbar intervertebral disc degeneration. 25495276_The AQP1 rs1049305 single nucleotide polymorphism is associated with running performance, but not relative body weight change, during South African Ironman Triathlons. 25582271_Through molecular genotyping we also identified polymorphisms in RhCE, Kell, Duffy, Colton, Lutheran and Scianna loci in donors and patients. 25609088_CAII increases water conductance through AQP1 by a physical interaction between the two proteins. 25721378_Study found a significant correlation between AQP1, AQP3, and AQP5 overexpression and lymph node metastasis in patients with surgically resected colon cancer. 25987071_High AQP1 expression is associated with recurrence in bladder uroepithelial cell carcinoma . 26074259_AQP1 was found to be a marker of a subgroup of aggressive basaloid-like squamous cell carcinomas 26115524_the benign subependymomas aquaporins 1 and 4 are dramatically redistributed and upregulated. 26151179_The results suggest that AQP1 may facilitate lung cancer cell proliferation and migration in an MMP-2 and-9-dependent manner 26176849_AQP1 knockdown can effectively inhibit cell proliferation, adhesion, invasion and tumorigenesis by targeting TGF-beta signaling pathway and focal adhesion genes. 26178576_PBEF regulates the expression of inflammatory factors and AQP1 through the MAPK pathways. 26181025_Results demonstrate the clinical utility, specificity, and sensitivity of urine AQP1 and PLIN2 to diagnose renal cell carcinoma. 26261083_The study demonstrated the abnormal expression pattern of AQP1, AQP2, AQP3, and AQP4 in the kidney tissues of patients with nephrotic syndrome, providing a basis for an improved understanding of the role of aquaporins in the pathogenesis of this disease. 26549133_the downregulation of AQP1 impedes extracellular to intracellular fluid transportation. 26707370_No significant relationship was found between gallbladder mucosa aquaporin expression and the presence or absence of gallbladder stones. 26717516_These data provide support for a continuous role of KLF2 in stabilizing the vessel wall via co-temporal expression of eNOS and AQP1 both preceding and during the pathogenesis of atherosclerosis. 26773069_The role of AQP1 in malignant pleural mesothelioma. 26786101_Based on a comparative mutagenic analysis of AQP1, AQP 3, and AQP 4, the results suggest that loop D interactions may provide a general structural framework for tetrameric assembly within the AQP family. 26812884_High cytoplasmic expression of AQP1 is associated with breast cancer progression. 26823734_Suggest that sputum AQP1 and AQP5 could be used as diagnostic markers in mild-to-moderate adult-onset asthma. 26838488_AQP1 was overexpressed in cystic cholangiocytes of patients with polycystic liver disease. A tendency of increased AQP1 protein expression in correlation with the cyst size was also found. 26848795_The BRAF V600 mutations were significantly associated with AQP1 expression (P=0.014). Long-term follow-up indicated a reduced progression-free survival (P=0.036) and overall survival (P=0.017) for the AQP1-positive cutaneous melanoma patients. 26919570_Findings suggest that a defined population of AQP4- and AQP1-expressing reactive astrocytes may modify alpha-syn deposition in the neocortex of patients with Parkinson's Disease. 26923194_Study provides evidence that Mef2c cooperated with Sp1 to activate human AQP1 transcription by binding to its proximal promoter in human umbilical cord vein endothelial cells indicating that AQP1 is a direct target of Mef2c in regulating angiogenesis and vasculogenesis of endothelial cells. 26972451_We demonstrated that AQP1-like immunoreactivity and expression are significantly increased in plaques of Peyronie's disease patients Vs controls, implying that AQP1 overexpression might be the consequence of a localized maladaptive response of the connective tissue to repeated mechanical trauma 27143047_Strong aquaporin 1 protein expression was an independent adverse prognosticator in hilar cholangiocarcinoma. 27157540_Aquaporin1 overexpression restored anti-cell proliferation and metastatic effect of si-Gli1. 27295912_Urinary AQP1 is significantly lower in patients with clear cell renal carcinoma than in controls. Urinary AQP1 increases after nephrectomy in clear cell renal carcinoma patients. 27465156_AQP1 expression was significantly lower in hereditary spherocytosis patients and silent carriers than in the simultaneously processed normal control, while no AQP1 decrease was detected in the autoimmune hemolytic anemia sample. The decreased AQP1 expression could contribute to explain variable degrees of anemia in hereditary spherocytosis. 27497833_results suggest regulation of AQP1 and AQP5 at the post-translational level and support previous observations on the implication of AQP1 and 5 in maintenance of lens transparency 27817002_AQP-1 overexpression is associated with prostate cancer progression. 27835672_Data show that the expression of aquaporin (AQP) 1, AQP3, AQP5, epithelial Na+ channel (ENaC) and sodium potassium ATPase (Na-K-ATPase) are altered in patients with acute respiratory failure (ARF) due to diffuse alveolar damage (DAD), and the cause of DAD does not seem to influence the level of impairment of these channels. 28072927_The native AQP1 gene, normally latent in human salivary gland cells, can be activated by both promoter-independent artificial transcription and epigenetic editing of the promoter. 28129470_AQP1 is expressed in atherosclerotic lesion neovasculature in human and mouse arteries and AQP1 deficiency augments lesion development in angiotensin II-promoted atherosclerosis in mice. 28409279_data suggest an important functional role of AQP1 in the pathobiology of hypoxia-induced pulmonary hypertension 28596233_function of AQP1 in tonicity response could be coupled or correlated to its function in band 3-mediated CO2/HCO3(-) exchange 28708257_The expression of AQP1 and 3 is a manifestation of physiological adaptation of functionally mature chondrocytes able to respond to the change of their internal environment influenced by extracellular matrix. 28766180_AQP1, 3 and 5 expression through the stages of human salivary gland morphogenesis, was examined. 28827734_Integrative epigenomics, transcriptomics and proteomics of patients chondrocytes from hip or knee osteoarthritis (OA) identified AQP1, COL1A1 and CLEC3B as significantly and differentially regulated suggesting they play an important role in OA pathogenesis. 29087027_Adenovirus-mediated human AQP1 expression in hepatocytes improves lipopolysaccharide-induced cholestasis in rats. 29104239_AQP1 may interact with VEGFA and play a role in vasculogenic mimicry, especially under hypoxic conditions, but the heterogeneity of malignant mesothelioma cells may result in different dominant pathways between patients. 29257313_AQP1 may serve a role in glioblastoma multiforme migration, invasion and vasculogenic mimicry formation 29258764_indicate pivotal roles for aquaporin-1 and -5 in the aggressive growth and metastatic potential of soft tissue sarcomas 29495596_The present study represents further confirmation of the hypothesized prognostic role of AQP1, which seems a reliable prognostic indicator. 29538612_Data show that miR-320 negatively regulates aquaporin 1 (AQP1) expression by targeting its 3' untranslated regions (3'-UTR). 29554889_Study demonstrated that a region upstream of the AQP1 gene (- 2300 to - 2200 bp) contains an enhancer required for pH-mediated regulation of AQP1 gene expression. 29650961_Case-control analyses reveal significant overrepresentation of rare variants in ATP13A3, AQP1 and SOX17, and provide independent validation of a critical role for GDF2 in heritable pulmonary arterial hypertension. 29769447_conclude that AQP1 is a major regulator of the platelet procoagulant response, able to modulate coagulation after injury or pathologic stimuli without affecting other platelet functional responses or normal hemostasis 29916035_used solid-state MAS NMR to investigate the binding of a lead compound [1-(4-methylphenyl)1H-pyrrole-2,5-dione] to AqpZ, through mapping of chemical shift perturbations in the presence of the compound. 30021355_H19 acted as AQP1 ceRNA in regulating miR-874. 30270117_Dermal fibroblasts and endothelial cells highly expressed AQP1 in systemic sclerosis lesional skin, and AQP1 expression in dermal fibroblasts and endothelial cells positively correlated with the degrees of tissue fibrosis and edema, respectively. 30378011_Aquaporin-1 Protein Expression of the Primary Tumor May Predict Cerebral Progression of Cutaneous Melanoma. 30467901_Aquaporin 1 and perilipin 2 possess high sensitivity and specificity for detecting clear cell and papillary renal cell carcinoma. Use of these markers might compliment renal mass biopsy in the characterization of small renal masses. 30531392_positive AQP1 expression is associated with the tumorigenesis and progression of pancreatic ductal adenocarcinoma; AQP1 is a diagnostic marker of pancreatic ductal adenocarcinoma and a predictive marker of poor prognosis in pancreatic ductal adenocarcinoma patients 30739527_AQP1 transcript expression was decreased in colorectal carcinoma (CRC) compared to normal mucosa, and this was associated with AQP1 promoter hypermethylation. AQP1 transcript expression increased with advanced disease but was not an independent prognostic indicator. 30840592_AQP1 may play a role in the risk of malignant mesothelioma. 31039249_We defined histologically the expression of AQP1 in the ChP from 6 nonsurvival preterm-pregnancy infants ranging ages between 20 and 25 gestational weeks in which AQP1 was mainly expressed at the apical pole of the We also defined histologically the expression of AQP1 in the ChP from 6 nonsurvival preterm-pregnancy infants ranging ages between 20 and 25 gestational epithelium in control and lissencephalic patients 31254364_homeostasis and physiological function of AQP1 in endothelial health are maintained by the MEF2C and miR-133a-3p.1 regulatory circuit 31261369_Aquaporin1-3 expression in normal and hydronephrotic kidneys in the human fetus. 31305898_To our knowledge, this work represents the first demonstration of AQP1/AQP3 expression in human melanocytic skin tumors 31311397_The expression AQP-1 protein was significantly reduced after transfected with AQP-1 silencing vector. Inhibition of miR-223 expression could suppress proliferation and promote apoptosis of SW579, and its mechanism i | ENSMUSG00000004655 | Aqp1 | 39.96530 | 1.3729692 | 0.4572992796 | 0.46378434 | 9.601068e-01 | 3.271600e-01 | 9.998360e-01 | No | Yes | 44.59054 | 10.442605 | 3.136061e+01 | 6.453433 | |
| ENSG00000241015 | 147804 | TPM3P9 | transcribed_processed_pseudogene | 121.38484 | 0.9857394 | -0.0207218512 | 0.28862674 | 4.817583e-03 | 9.446642e-01 | 9.998360e-01 | No | Yes | 114.66760 | 19.611012 | 1.316736e+02 | 17.481867 | ||||||||||
| ENSG00000243766 | HOTTIP | lncRNA | 15.31905 | 0.8978927 | -0.1553850439 | 0.72965560 | 4.394804e-02 | 8.339503e-01 | 9.998360e-01 | No | Yes | 18.17416 | 5.472685 | 1.979400e+01 | 4.931744 | |||||||||||
| ENSG00000244486 | 91179 | SCARF2 | protein_coding | Q96GP6 | FUNCTION: Probable adhesion protein, which mediates homophilic and heterophilic interactions. In contrast to SCARF1, it poorly mediates the binding and degradation of acetylated low density lipoprotein (Ac-LDL) (By similarity). {ECO:0000250}. | Alternative splicing;Cell adhesion;Disease variant;Disulfide bond;EGF-like domain;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is similar to SCARF1/SREC-I, a scavenger receptor protein that mediates the binding and degradation of acetylated low density lipoprotein (Ac-LDL). This protein has only little activity of internalizing modified low density lipoproteins (LDL), but it can interact with SCARF1 through its extracellular domain. The association of this protein with SCARF1 is suppressed by the presence of scavenger ligands. Alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Jul 2008]. | hsa:91179; | focal adhesion [GO:0005925]; integral component of membrane [GO:0016021]; scavenger receptor activity [GO:0005044]; heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules [GO:0007157] | 12154095_SRECII binds to SRECI and has a similar tissue distribution pattern 20887961_Mutations in SCARF2 are responsible for Van Den Ende-Gupta syndrome. 24478002_the full VDEGS phenotype may include sclerocornea resulting from homozygosity or compound heterozygosity for loss of function variants in SCARF2. 33783941_Further delineation of van den Ende-Gupta syndrome: Genetic heterogeneity and overlap with congenital heart defects and skeletal malformations syndrome. | ENSMUSG00000012017 | Scarf2 | 2768.37545 | 0.8597695 | -0.2179781723 | 0.10293835 | 4.465610e+00 | 3.458375e-02 | 9.998360e-01 | No | Yes | 2750.03497 | 407.035353 | 2.881693e+03 | 330.022816 | |
| ENSG00000244627 | TPTEP2 | transcribed_unprocessed_pseudogene | 123.48449 | 0.9914927 | -0.0123259715 | 0.27176333 | 2.047729e-03 | 9.639066e-01 | 9.998360e-01 | No | Yes | 128.29946 | 19.986399 | 1.292542e+02 | 15.778356 | |||||||||||
| ENSG00000244649 | LINC02086 | lncRNA | 14.07926 | 0.6694286 | -0.5789979967 | 0.76446263 | 5.306464e-01 | 4.663352e-01 | 9.998360e-01 | No | Yes | 13.35367 | 4.911389 | 1.772197e+01 | 5.343898 | |||||||||||
| ENSG00000245849 | 100505648 | RAD51-AS1 | lncRNA | 26230935_results identify TPIP as a novel E2F1 co-activator, suggest a similar role for other TPTEs, and indicate that the TODRA lncRNA affects RAD51 dysregulation and RAD51-dependent DSB repair in malignancy 28667302_Overexpression of RAD51-AS1 promoted EOC cell proliferation, while silencing of RAD51-AS1 inhibited EOC cell proliferation, delayed cell cycle progression and promoted apoptosis in vitro and in vivo. 29749376_Corylin increases the sensitivity of hepatocellular carcinoma cells to chemotherapy through long noncoding RNA RAD51-AS1-mediated inhibition of DNA repair. 33314669_LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed proliferation, invasion and glycolysis of colorectal cancer. | 93.98888 | 1.1797814 | 0.2385195355 | 0.31168858 | 5.793944e-01 | 4.465498e-01 | 9.998360e-01 | No | Yes | 79.04387 | 16.776696 | 6.876952e+01 | 11.287563 | |||||||||
| ENSG00000247081 | 105369147 | BAALC-AS1 | lncRNA | 67.98483 | 0.9783408 | -0.0315910262 | 0.36363129 | 7.180524e-03 | 9.324697e-01 | 9.998360e-01 | No | Yes | 79.89108 | 13.956984 | 8.077498e+01 | 10.909013 | ||||||||||
| ENSG00000247828 | 100505894 | TMEM161B-DT | lncRNA | 34046994_TMEM161B-AS1 suppresses proliferation, invasion and glycolysis by targeting miR-23a-3p/HIF1AN signal axis in oesophageal squamous cell carcinoma. | 195.20724 | 0.9234926 | -0.1148276764 | 0.22097046 | 2.682092e-01 | 6.045356e-01 | 9.998360e-01 | No | Yes | 259.02310 | 34.780980 | 2.974031e+02 | 30.970555 | |||||||||
| ENSG00000248049 | 550112 | UBA6-DT | lncRNA | 34951345_Long noncoding RNA UBA6-AS1 inhibits the malignancy of ovarian cancer cells via suppressing the decay of UBA6 mRNA. 35137449_Amino acid restriction induces a long non-coding RNA UBA6-AS1 to regulate GCN2-mediated integrated stress response in breast cancer. | 389.70965 | 1.1038250 | 0.1425114052 | 0.17429022 | 6.910026e-01 | 4.058236e-01 | 9.998360e-01 | No | Yes | 469.09692 | 57.874479 | 4.444965e+02 | 42.778922 | |||||||||
| ENSG00000249115 | 23354 | HAUS5 | protein_coding | O94927 | FUNCTION: Contributes to mitotic spindle assembly, maintenance of centrosome integrity and completion of cytokinesis as part of the HAUS augmin-like complex. {ECO:0000269|PubMed:19369198, ECO:0000269|PubMed:19427217}. | Acetylation;Alternative splicing;Cell cycle;Cell division;Coiled coil;Cytoplasm;Cytoskeleton;Microtubule;Mitosis;Phosphoprotein;Reference proteome | HAUS5 is 1 of 8 subunits of the 390-kD human augmin complex, or HAUS complex. The augmin complex was first identified in Drosophila, and its name comes from the Latin verb 'augmentare,' meaning 'to increase.' The augmin complex is a microtubule-binding complex involved in microtubule generation within the mitotic spindle and is vital to mitotic spindle assembly (Goshima et al., 2008 [PubMed 18443220]; Uehara et al., 2009 [PubMed 19369198]).[supplied by OMIM, Jun 2010]. | hsa:23354; | centrosome [GO:0005813]; cytosol [GO:0005829]; HAUS complex [GO:0070652]; mitotic spindle microtubule [GO:1990498]; cell division [GO:0051301]; centrosome cycle [GO:0007098]; spindle assembly [GO:0051225] | 28596487_Our results suggest that GSCs differentially rely on ZNF131-dependent expression of HAUS5 as well as the Augmin/HAUS complex activity to maintain the integrity of centrosome function and viability. | ENSMUSG00000078762 | Haus5 | 2004.63990 | 0.9203226 | -0.1197883843 | 0.10354185 | 1.332551e+00 | 2.483520e-01 | 9.998360e-01 | No | Yes | 1810.62997 | 224.742944 | 1.807123e+03 | 173.584067 | |
| ENSG00000251357 | protein_coding | H7C1H1 | Glycoprotein;Membrane;Proteomics identification;Reference proteome;Transmembrane;Transmembrane helix | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; transmembrane transporter activity [GO:0022857] | 45.99312 | 1.0491574 | 0.0692311773 | 0.48440663 | 1.853560e-02 | 8.917062e-01 | 9.998360e-01 | No | Yes | 55.41565 | 13.258667 | 4.673326e+01 | 9.276331 | |||||||||
| ENSG00000253352 | 55000 | TUG1 | protein_coding | A0A6I8PU40 | Cytoplasm;Membrane;Mitochondrion;Nucleus;Reference proteome;Signal;Transmembrane;Transmembrane helix | Mouse_homologues 15797018_taurine upregulated gene 1 is necessary for the proper formation of photoreceptors in the developing rodent retina 25871529_Knockdown of lncRNA TUG1 expression resulted in an increased apoptosis ratio and decreased insulin secretion in beta cells. 26785829_Results suggest that the long noncoding RNA TUG1 (lncRNA TUG1) exerts a protective effect against cold-induced liver damage by inhibiting apoptosis and for the prevention of cold-induced liver damage in liver transplantation. 27760046_Tug1 regulates mitochondrial function in podocytes by epigenetic targeting of expression of the transcription factor PPARgamma coactivator 1alpha (PGC-1alpha, encoded by Ppargc1a). 27760051_These findings indicate that a direct interaction between PGC-1alpha and Tug1 modulates mitochondrial bioenergetics in podocytes in the diabetic milieu. 29268138_TUG1 knockdown ameliorates atherosclerosis by modulating FGF1 via miR-133a in ApoE knockout mice. 30066872_The present study demonstrated that TUG1, along with miR29c, may contribute to cardiac FMT activation and promote fibrosis in chronic hypoxia. 30468486_The overexpression of TUG1 can promote neuronal death after cerebral infarction in mice by competitive binding to microRNA-9 and promotion of FOXO3 expression 30657572_TUG1 was downregulated in TNF-alpha-treated primary mouse interstitial cells of Cajal (ICC). TUG1 overexpression protected ICC from TNF-alpha-induced apoptosis and pro-inflammatory cytokines expression. MiR-127 was negatively regulated by TUG1 and implicated in the action of TUG1 in ICC. Mechanistically, TUG1 inhibited TNF-alpha-induced activation of NF-kappaB and Notch pathways in ICC by down-regulating miR-127. 31115515_these results demonstrated that LPSinduced podocyte injury could be alleviated by the TUG1/miR197/MAPK1 axis. 31145943_TUG1, by targeting miR-142-3p and up-regulating HMGB1 and Rac1, plays a central role in stimulating autophagic cell apoptosis in ischemia/hypoxia-challenged cardiomyocytes. Down-regulating TUG1 or up-regulating miR-142-3p may ameliorate myocardial injury and protect against acute myocardial infarction. 31183682_TUG1 functioned as a positive modulator of cardiac hypertrophy via sponging miR-29b-3p, indicating that TUG1 might serve as a potential target for the treatment of cardiac hypertrophy and even heart failure. 31539141_LncRNA TUG1 inhibits the proliferation and fibrosis of mesangial cells in diabetic nephropathy via inhibiting the PI3K/AKT pathway. 31679623_UG1 knockdown significantly prevented the development of PAH in vivo. Moreover, TUG1 promoted the proliferative responses of HPASMCs, including cell viability, 5-bromodeoxyuridine incorporation, the expression of proliferating cell nuclear antigen, and cell-cycle progression. All these functions of TUG1 were likely to be associated with miR-328. 31697952_the overexpression of TUG1 significantly up-regulated the levels of MMP-14, VEGF, p-p38 mitogen-activated protein kinase (p-p38 MAPK) and p-HSP27 (heat shock protein 27), and promoted the proliferation, invasion and EMT of Cc cells 31709761_Long noncoding RNA TUG1 contributes to cerebral ischaemia/reperfusion injury by sponging mir-145 to up-regulate AQP4 expression. 31787746_The long non-coding RNA TUG1-miR-9a-5p axis contributes to ischemic injuries by promoting cardiomyocyte apoptosis via targeting KLF5. 31858814_H2O2 or ischemia-reperfusion-induced TUG1, by sponging microRNA 132-3p, activated histone deacetylase 3, which in turn targeted multiple protective genes, stimulated intracellular reactive oxygen species accumulation 31907757_LncRNA TUG1 promoted osteogenic differentiation through promoting bFGF ubiquitination. 31930775_The protection of NF-kappaB inhibition on kidney injury of systemic lupus erythematosus mice may be correlated with lncRNA TUG1. 32057178_LncRNA TUG1 alleviates cardiac hypertrophy by targeting miR-34a/DKK1/Wnt-beta-catenin signalling. 32467232_Role for carbohydrate response element-binding protein (ChREBP) in high glucose-mediated repression of long noncoding RNA Tug1. 32577774_Knockdown of long non-coding RNA TUG1 depresses apoptosis of hippocampal neurons in Alzheimer's disease by elevating microRNA-15a and repressing ROCK1 expression. 32597038_LncRNA TUG1 regulates ApoM to promote atherosclerosis progression through miR-92a/FXR1 axis. 32841583_LncRNA TUG1 reverses LPS-induced cell apoptosis and inflammation of macrophage via targeting MiR-221-3p/SPRED2 axis. 32869325_The mechanisms of lncRNA Tug1 in islet dysfunction in a mouse model of intrauterine growth retardation. 32888158_LncRNA TUG1 reduces inflammation and enhances insulin sensitivity in white adipose tissue by regulating miR-204/SIRT1 axis in obesity mice. 32894169_The Tug1 lncRNA locus is essential for male fertility. 33047284_LncRNA TUG1 regulates the balance of HuR and miR-29b-3p and inhibits intestinal epithelial cell apoptosis in a mouse model of ulcerative colitis. 33059750_TUG1 enhances high glucose-impaired endothelial progenitor cell function via miR-29c-3p/PDGF-BB/Wnt signaling. 33125086_lncRNA TUG1 promotes the brown remodeling of white adipose tissue by regulating miR204targeted SIRT1 in diabetic mice. 33135476_Silencing of the lncRNA TUG1 attenuates the epithelial-mesenchymal transition of renal tubular epithelial cells by sponging miR-141-3p via regulating beta-catenin. 33164260_LncRNA TUG1 competitively binds to miR-340 to accelerate myocardial ischemia-reperfusion injury. 33640857_Long non-coding RNA TUG1 promotes airway remodeling and mucus production in asthmatic mice through the microRNA-181b/HMGB1 axis. 33660806_SP1-induced lncRNA TUG1 regulates proliferation and apoptosis in islet cells of type 2 diabetes mellitus via the miR-188-3p/FGF5 axis. 33971184_LncRNA TUG1 silencing enhances proliferation and migration of ox-LDL-treated human umbilical vein endothelial cells and promotes atherosclerotic vascular injury repairing via the Runx2/ANPEP axis. 34153134_Knockdown of lncRNA TUG1 attenuates cerebral ischemia/reperfusion injury through regulating miR-142-3p. 34380028_PGC1alpha is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites. 34547172_LncRNA TUG1 attenuates ischaemia-reperfusion-induced apoptosis of renal tubular epithelial cells by sponging miR-144-3p via targeting Nrf2. 34743197_Functional delivery of lncRNA TUG1 by endothelial progenitor cells derived extracellular vesicles confers anti-inflammatory macrophage polarization in sepsis via impairing miR-9-5p-targeted SIRT1 inhibition. 34851489_Interfering TUG1 Attenuates Cerebrovascular Endothelial Apoptosis and Inflammatory injury After Cerebral Ischemia/Reperfusion via TUG1/miR-410/FOXO3 ceRNA Axis. 35084431_The Long-Noncoding RNA TUG1 Regulates Oxygen-Induced Retinal Neovascularization in Mice via MiR-299. 35298636_Role of lncRNA TUG1 in Adenomyosis and its Regulatory Mechanism in Endometrial Epithelial Cell Functions. 35498127_Long Noncoding RNA TUG1 Aggravates Cerebral Ischemia/Reperfusion Injury by Acting as a ceRNA for miR-3072-3p to Target St8sia2. | ENSMUSG00000056579 | Tug1 | 2072.65401 | 0.8762594 | -0.1905700428 | 0.12303360 | 2.380099e+00 | 1.228898e-01 | 9.998360e-01 | No | Yes | 2167.04819 | 384.560102 | 2.454649e+03 | 335.661922 | |||||
| ENSG00000253771 | TPTE2P1 | transcribed_unprocessed_pseudogene | 22.75445 | 0.8688533 | -0.2028154166 | 0.62930384 | 1.136681e-01 | 7.360061e-01 | 9.998360e-01 | No | Yes | 29.76326 | 8.159906 | 3.443445e+01 | 7.656186 | |||||||||||
| ENSG00000254806 | 767557 | SYS1-DBNDD2 | protein_coding | F2Z2A3 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | This locus represents naturally occurring read-through transcription from the neighboring SYS1 Golgi-localized integral membrane protein homolog and dysbindin domain containing 2 (DBNDD2) genes. The read-through transcript includes the majority of exons from each individual gene, but it would be subject to nonsense-mediated mRNA decay (NMD) and is therefore predicted to be non-coding. [provided by RefSeq, Oct 2010]. | integral component of membrane [GO:0016021] | ENSMUSG00000090996 | Gm20458 | 204.10480 | 1.1338896 | 0.1812801771 | 0.23389144 | 6.134954e-01 | 4.334748e-01 | 9.998360e-01 | No | Yes | 250.82826 | 53.440400 | 2.131453e+02 | 35.917786 | ||||
| ENSG00000254860 | 493900 | TMEM9B-AS1 | lncRNA | 91.30902 | 1.0623466 | 0.0872544879 | 0.35667270 | 5.176386e-02 | 8.200220e-01 | 9.998360e-01 | No | Yes | 116.51800 | 23.267304 | 9.642432e+01 | 15.231745 | ||||||||||
| ENSG00000254995 | 100534593 | STX16-NPEPL1 | protein_coding | H3BU86 | Mouse_homologues FUNCTION: SNARE involved in vesicular transport from the late endosomes to the trans-Golgi network. {ECO:0000250}. | Coiled coil;Reference proteome | This locus represents naturally occurring read-through transcription between the neighboring syntaxin 16 (STX16) and aminopeptidase-like 1 (NPEPL1) genes on chromosome 20. The read-through transcript is a candidate for nonsense-mediated mRNA decay (NMD), and is thus unlikely to produce a protein product. [provided by RefSeq, Mar 2011]. | Mouse_homologues mmu:228960; | SNARE complex [GO:0031201]; SNAP receptor activity [GO:0005484]; intracellular protein transport [GO:0006886]; vesicle-mediated transport [GO:0016192] | Mouse_homologues 17317779_Mice genetically altered to carry the equivalent of STX16del4-6 contributing to pseudohypoparathyroidism type 1b. 17852734_Syntaxin 16 may thus play a role in neurite outgrowth and perhaps other specific dendritic anterograde/retrograde traffic. | ENSMUSG00000027522 | Stx16 | 81.42480 | 0.7437331 | -0.4271430815 | 0.37898112 | 1.195536e+00 | 2.742158e-01 | 9.998360e-01 | No | Yes | 96.23052 | 42.299917 | 1.035745e+02 | 34.964141 | |
| ENSG00000255284 | 171391 | lncRNA | 247.37392 | 0.9965548 | -0.0049788997 | 0.20011435 | 6.197761e-04 | 9.801385e-01 | 9.998360e-01 | No | Yes | 257.24796 | 25.195040 | 2.512692e+02 | 19.847541 | |||||||||||
| ENSG00000255495 | lncRNA | 24.84463 | 1.3778854 | 0.4624559332 | 0.66342819 | 5.449531e-01 | 4.603872e-01 | 9.998360e-01 | No | Yes | 28.90918 | 9.416125 | 2.123117e+01 | 5.652578 | ||||||||||||
| ENSG00000256683 | 59348 | ZNF350 | protein_coding | Q9GZX5 | FUNCTION: Transcriptional repressor. Binds to a specific sequence, 5'-GGGxxxCAGxxxTTT-3', within GADD45 intron 3. {ECO:0000269|PubMed:11090615}. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:59348; | nuclear body [GO:0016604]; nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription repressor complex [GO:0017053]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding [GO:0001162]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12872252_ZBRK1 mutation found in BRCA1 and BRCA2 mutation negative probands with breast or ovarian cancer. 14517299_ZBRK1 is a novel target for DNA damage-induced degradation 14660588_ZBRK1 zinc fingers have dual roles in sequence-specific DNA-binding and BRCA1-dependent transcriptional repression 15113441_Observational study of gene-disease association. (HuGE Navigator) 15496401_the CTRD is a novel protein interaction surface responsible for directing homotypic and heterotypic interactions necessary for ZBRK1-directed transcriptional repression 15596820_interacts with BBLF2/3 to provide a tethering point on oriLyt for the EBV replication complex 16485136_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16997916_KRAB-induced transcriptional repression is robust and active over a variety of genomic contexts that include at least the wide range of sites targeted by lentiviral integration 17557904_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17764113_Genetic variants and haplotypes of the ZBRK1 gene is associated with breast and ovarian cancer 17764113_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18950845_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19279087_KRAB/KAP1 regulation is fully functional within the context of episomal DNA 19484476_BRCA1 and ZNF350 may jointly contribute to individuals' susceptibility of breast cancer in Chinese women. 19484476_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19996286_Our findings suggest that ZBRK1 acts to inhibit metastasis of cervical carcinoma, perhaps by modulating MMP9 expression. 20007691_Data found that HMGA2, along with a dozen of other genes, was co-repressed by ZBRK1, BRCA1, and CtIP. 20150366_Observational study of gene-disease association. (HuGE Navigator) 20221260_KRAB/KAP1 recruitment induces long-range repression through the spread of heterochromatin. 20306497_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20644561_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20713352_RB.E2F1 complex plays a critical role in ZBRK1 transcriptional repression, and loss of this repression may contribute to cellular sensitivity of DNA damage, ultimately leading to carcinogenesis. 20926453_KRAB-containing zinc-finger transcriptional regulator, ZBRK1, an interaction partner of the SCA2 gene product ataxin-2. 21791101_high levels of gene activity in the genomic environment and the pre-deposition of repressive histone marks within a gene increase its susceptibility to KRAB/KAP1-mediated repression 22975076_ZBRK1 negatively regulates the HIV-1 LTR 23151675_promoter assays in two breast cancer cell lines identified two haplotypes (H11 and H12) stimulating significantly the expression of ZNF350 transcript compared with the common haplotype H8 23991171_results indicate that a loss of ZBRK1 contributes to the increased expression of KAP1, potentiating its role to enhance metastasis and invasion 24924633_The secondary structure of the ZBRK1-DNA complex is found to be significantly altered from the standard B-DNA conformation. 25107531_MAGE proteins bind to KAP1, a gene repressor and ubiquitin E3 ligase which also binds KRAB domain containing zinc finger transcription factors (KZNFs), and MAGE expression may affect KZNF mediated gene regulation. 25749518_ZBRK1 suppresses renal cancer progression perhaps by regulating VHL expression through formation of a complex with VHL and p300 in renal cancer 26658611_villin directly interacts with a transcriptional corepressor and ligand of the Slug promoter, ZBRK1. 27586871_Results indicated that SNP rs2278414 at ZNF350 may in fl uence an individual's susceptibility to age-related cataract (ARC) by affecting the binding af fi nity of miR-21-3p and miR-150-5p and regulating expression levels of the mRNAs, resulting in different levels of cellular DNA breaks, thus contributing to ARC. 29653063_Results indicate ZNF350 as an important gene mammary oncogenesis. 30061100_These data indicate that the KAP1-KZNF pathway contributes to genome stability and innate immune control in adult human cells. 30714292_BRCA1 forms a co-repressor complex with ZBRK1 that coordinately represses GOT2 expression via a ZBRK1 recognition element in the promoter of GOT2. 31289231_The work identifies the interaction interfaces in the KAP1 tripartite motif responsible for self-association and KRAB binding and establishes their role in retrotransposon silencing. 32470015_PFKP is transcriptionally repressed by BRCA1/ZBRK1 and predicts prognosis in breast cancer. 33407485_Correlation between ZBRK1/ZNF350 gene polymorphism and breast cancer. 34685484_Role of the Transcriptional Repressor Zinc Finger with KRAB and SCAN Domains 3 (ZKSCAN3) in Retinal Pigment Epithelial Cells. | 37.80450 | 2.1062898 | 1.0747039211 | 0.47991979 | 5.107748e+00 | 2.381922e-02 | 9.998360e-01 | No | Yes | 67.45705 | 16.978761 | 3.256732e+01 | 6.239712 | ||||
| ENSG00000257556 | LINC02298 | lncRNA | 31.11448 | 0.8940549 | -0.1615646054 | 0.53471034 | 9.192218e-02 | 7.617476e-01 | 9.998360e-01 | No | Yes | 49.36222 | 13.085618 | 5.082067e+01 | 11.269281 | |||||||||||
| ENSG00000257702 | 151534 | LBX2-AS1 | lncRNA | 30824187_LncRNA LBX2-AS1 promoted cell migration and epithelial-mesenchymal transition process in esophageal squamous cell carcinoma. LBX2-AS1 interacting with HNRNPC to promote ZEB1/2 mRNA stability. 31512745_LBX2-AS1 might be associated with periodontitis 31539129_Novel long non-coding RNA LBX2-AS1 indicates poor prognosis and promotes cell proliferation and metastasis through Notch signaling in non-small cell lung cancer. 31673844_LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. 32143907_Long noncoding RNA LBX2-AS1 drives the progression of hepatocellular carcinoma by sponging microRNA-384 and thereby positively regulating IRS1 expression. 32559617_LncRNA LBX2-AS1 facilitates abdominal aortic aneurysm through miR-4685-5p/LBX2 feedback loop. 33099720_ELK1 activated-long noncoding RNA LBX2-AS1 aggravates the progression of ovarian cancer through targeting miR-4784/KDM5C axis. 33342041_LBX2-AS1 promotes ovarian cancer progression by facilitating E2F2 gene expression via miR-455-5p and miR-491-5p sponging. 33637977_Silencing of lncRNA LBX2-AS1 suppresses glioma cell proliferation and metastasis through the Akt/GSK3beta pathway in vitro. 33733923_Overexpression of Long Noncoding RNA LBX2-AS1 Promotes the Proliferation of Colorectal Cancer. | 111.70670 | 1.3787474 | 0.4633582103 | 0.29921893 | 2.399783e+00 | 1.213521e-01 | 9.998360e-01 | No | Yes | 198.64372 | 43.892577 | 1.277357e+02 | 24.319902 | |||||||||
| ENSG00000258311 | protein_coding | F8W036 | FUNCTION: May negatively regulate aerobic respiration through mitochondrial protein lysine-acetylation. May counteract the action of the deacetylase SIRT3 by acetylating and regulating proteins of the mitochondrial respiratory chain including ATP5F1A and NDUFA9. {ECO:0000256|ARBA:ARBA00003284}. | Coiled coil;Reference proteome | BLOC-1 complex [GO:0031083]; lysosomal membrane [GO:0005765]; mitochondrial intermembrane space [GO:0005758]; cellular localization [GO:0051641] | 258.43501 | 1.5186774 | 0.6028153983 | 0.20227830 | 9.143142e+00 | 2.496520e-03 | 5.809295e-01 | No | Yes | 277.63009 | 56.508727 | 1.815653e+02 | 28.774347 | ||||||||
| ENSG00000258366 | 51750 | RTEL1 | protein_coding | Q9NZ71 | FUNCTION: ATP-dependent DNA helicase implicated in telomere-length regulation, DNA repair and the maintenance of genomic stability. Acts as an anti-recombinase to counteract toxic recombination and limit crossover during meiosis. Regulates meiotic recombination and crossover homeostasis by physically dissociating strand invasion events and thereby promotes noncrossover repair by meiotic synthesis dependent strand annealing (SDSA) as well as disassembly of D loop recombination intermediates. Also disassembles T loops and prevents telomere fragility by counteracting telomeric G4-DNA structures, which together ensure the dynamics and stability of the telomere. {ECO:0000255|HAMAP-Rule:MF_03065, ECO:0000269|PubMed:18957201, ECO:0000269|PubMed:23453664, ECO:0000269|PubMed:24009516}. | 4Fe-4S;ATP-binding;Alternative splicing;DNA damage;DNA repair;DNA-binding;Disease variant;Dyskeratosis congenita;Helicase;Hydrolase;Iron;Iron-sulfur;Metal-binding;Nucleotide-binding;Nucleus;Reference proteome | This gene encodes a DNA helicase which functions in the stability, protection and elongation of telomeres and interacts with proteins in the shelterin complex known to protect telomeres during DNA replication. Mutations in this gene have been associated with dyskeratosis congenita and Hoyerall-Hreidarsson syndrome. Read-through transcription of this gene into the neighboring downstream gene, which encodes tumor necrosis factor receptor superfamily, member 6b, generates a non-coding transcript. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Sep 2013]. | hsa:51750; | chromosome, telomeric region [GO:0000781]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 4 iron, 4 sulfur cluster binding [GO:0051539]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; DNA polymerase binding [GO:0070182]; metal ion binding [GO:0046872]; DNA duplex unwinding [GO:0032508]; DNA repair [GO:0006281]; mitotic telomere maintenance via semi-conservative replication [GO:1902990]; negative regulation of DNA recombination [GO:0045910]; negative regulation of t-circle formation [GO:1904430]; negative regulation of telomere maintenance in response to DNA damage [GO:1904506]; positive regulation of telomere capping [GO:1904355]; positive regulation of telomere maintenance [GO:0032206]; positive regulation of telomere maintenance via telomere lengthening [GO:1904358]; positive regulation of telomeric loop disassembly [GO:1904535]; regulation of double-strand break repair via homologous recombination [GO:0010569]; replication fork processing [GO:0031297]; strand displacement [GO:0000732]; telomere maintenance [GO:0000723]; telomere maintenance in response to DNA damage [GO:0043247]; telomeric loop disassembly [GO:0090657] | 18957201_Study finds that rtel-1 mutant worms and RTEL1-depleted human cells share characteristic phenotypes with yeast srs2 mutants: lethality upon deletion of the sgs1/BLM homolog, hyperrecombination, and DNA damage sensitivity.[RTEL1] 19578366_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19578366_On 9p21, rs1412829 near CDKN2B had discovery P = 3.4 x 10(-8), replication P = 0.0038 and combined P = 1.85 x 10(-10). On 20q13.3, rs6010620 intronic to RTEL1 had discovery P = 1.5 x 10(-7), replication P = 0.00035 and combined P = 3.40 x 10(-9). 19578367_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20368557_Polymorphisms in the LIG4, BTBD2, HMGA2, and RTEL1 genes, which are involved in the double-strand break repair pathway, are associated with glioblastoma multiforme survival. 20462933_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20610542_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20847058_Observational study of gene-disease association. (HuGE Navigator) 21350045_SUSCEPTIBILITY LOCI FOR GLIOMA AND GLIOBLASTOMA RISK IN A CHINESE POPULATION INCLUDED RTEL1 21920947_rs6010620 a genetic variant of RTEL1, was one of 3 genetic variants implicated in a pool of US epidemiologic studies of glioma risk 23115063_rs6010620 (RTEL1) was associated with an increased risk of glioma when restricting to cases with family history of brain tumours. 23329068_The nonsense mutations both cause truncation of the RTEL1 protein, resulting in loss of the PIP box; this may abrogate an important protein-protein interaction. These findings implicate a new telomere biology gene, RTEL1, in the etiology of DC. 23453664_These biallelic RTEL1 mutations are responsible for a major subgroup ( approximately 29%) of HHS. 23591994_results identify RTEL1 as a novel Hoyeraal-Hreidarsson syndrome-causing gene and highlight its role as a genomic caretaker in humans. 23683922_Two significant Ten candidate tagging SNPs in RTEL1 gene were observed to be associated with glioma risk 23812731_Results indicate the potential roles of regulator of telomere elongation helicase 1 (RTEL1) and telomerase reverse transcriptase (TERT) in astrocytoma development. 23959892_Data show that RTEL1 interacts with the shelterin protein TRF1, indicating a potential recruitment mechanism of RTEL1 to telomeres. 24009516_The molecular data and the patterns of inheritance are consistent with a hypomorphic mutation in RTEL1 as the underlying basis of the clinical and cellular phenotypes. 24130156_The C-terminal extension of RTEL1, downstream of its catalytic domain and including several HHS-associated mutations, contains a yet unidentified tandem of harmonin-N-like domains. 24523019_suggested that RTEL1 rs6010620 polymorphism is likely to be associated with increased glioma risk, which lends further biological plausibility to these findings 24561255_Authors propose that RTEL1 serves as a human analog of Srs2 to inhibit (CTGCAG) repeat expansions and fragility, likely by unwinding problematic hairpins. 25227808_rs6010620 polymorphism in the RTEL1 gene is associated with increased risk of glioma in both Caucasians and Asians. [Meta-Analysis] 25556444_Association between the RTEL1 rs6010620 polymorphism and glioma risk was significant. [Meta-Analysis] 25607374_Rare loss-of-function variants in RTEL1 represent a newly defined genetic predisposition for FIP, supporting the importance of telomere-related pathways in pulmonary fibrosis. 25620558_The shelterin protein TRF2 recruits RTEL1 to telomeres in S phase, which is required to prevent catastrophic t-loop processing by structure-specific nucleases. 25628358_This work unravels completely unanticipated roles for RTEL1 in RNP trafficking and strongly suggests that defects in RNP biogenesis pathways contribute to the pathology of Hoyeraal-Hreidarsson syndrome 25848748_PARN and RTEL1 mutation carriers had shortened leukocyte telomere lengths. 26014354_RTEL1 single nucleotide polymorphisms are associated with decreased susceptibility to pediatric brain astrocytoma. 26022962_Heterozygous RTEL1 mutations are responsible for familial pulmonary fibrosis (FPF) and, thereby, extend the clinical spectrum of RTEL1 deficiency. Thus, RTEL1 enlarges the number of telomere-associated genes implicated in FPF. 26025130_A homozygous mutation of RTEL1 in a child presenting with an apparently isolated natural killer cell deficiency. 26156397_results suggest a significant association between the RETL1, TREH, and PHLDB1 genes and GBM development in the Han Chinese population 26581417_Telomere length is associated with Esophageal squamous cell carcinoma risk in a U-shaped pattern and demonstrates that TL-SNPs may not be important in carcinogenesis in Chinese population. 26803811_Deletion in the RTEL1 gene is associated with metastatic glioblastoma. 26839018_Genetic risk variants in the RTEL1 gene is associated with somatic biomarkers in glioma. 26939676_This meta-analysis demonstrates that the RTEL1 rs2297440 polymorphism plays a moderate, but significant role in the risk of glioma. 27485611_promoter methylated in 51.4% of lung cancer patients and in 8.8% of healthy individuals 27540018_Pulmonary fibrosis patients with mutations in telomerase reverse transcriptase, telomerase RNA component, regulator of telomere elongation helicase 1 and poly(A)-specific ribonuclease were identified and clinical data were analysed. Genetic mutations in telomere related genes lead to a variety of interstitial lung disease diagnoses that are universally progressive. 27765928_Findings suggest a potential association between regulator of telomere elongation helicase 1 (RTEL1) polymorphisms and lung cancer (LC) risk in a Chinese Han population. 28360516_SNPs in the RTEL1 are associated with COPD in a Chinese Han population. It is possible that these variants are COPD risk factors. 28495916_Observation are firstly, heterozygous LOF RTEL1 variants are associated with myelodysplasia and liver disease in adulthood. Secondly, biallelic RTEL1 variants can present with just bone marrow failure in adulthood. Thirdly, many heterozygous variants, and even some biallelic RTEL1 variants, are bystanders. 28953687_The allele 'G' of rs6089953 and rs6010621 and the allele 'A' of rs2297441 were associated with decreased risk of High altitude pulmonary edema, haplotype 'GG, GT, AT' of rs6089953-rs6010621 were detected significantly associated with High altitude pulmonary edema risk in the Chinese population. 29151059_a novel association signal in the RTEL1 gene (intronic single nucleotide poly morphism (SNP) rs2297439; P=2.82x10(-7)) that is independent of previously reported Telomere-associated SNPs in this region. 29344583_heterozygous RTEL1 variants were associated with marrow failure, and telomere length measurement alone may not identify patients with telomere dysfunction carrying RTEL1 variants. 29522136_hRTEL1 contributes to the maintenance of long telomeres by preserving long G-overhangs, thereby facilitating POT1 binding and elongation by telomerase. 30345460_RTEL1 SNPs were associated with relative telomere length. Shorter relative telomere length was associated with an increased risk of stroke. 30462709_Study demonstrates that previously identified loci in RTEL1 are confirmed to have an association with increased risk of adult gliomas. Moreover, two coding variants (rs6062302 and rs115303435) were found to confer independent risk for glioma in RTEL1. A novel missense SNP (rs77086616, T434M) was also observed only in Korean glioma samples. 30523160_Regulator of telomere length 1 (RTEL1) mutations are associated with heterogeneous pulmonary and extra-pulmonary phenotypes. 30623606_RTEL1 SNPs may have a protective role against coronary heart disease risk. 31677132_Homozygous RTEL1 variant is associated with telomeropathies. 31721021_The rs6010620 (RTEL1), rs4977756 (CDKN2A/B), and rs498872 (PHLDB1) are associated with glioma risk in the Portuguese population. The GA genotype of the rs6010620 (RTEL1) was associated with a decreased risk of glioblastomas (OR 0.45). 32398827_RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. 32398829_SLX4 interacts with RTEL1 to prevent transcription-mediated DNA replication perturbations. 32460026_Synthetic Lethality between DNA Polymerase Epsilon and RTEL1 in Metazoan DNA Replication. 32542379_Full length RTEL1 is required for the elongation of the single-stranded telomeric overhang by telomerase. 32561545_Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. 33200790_miR-4530 inhibits the malignant biological behaviors of human glioma cells by directly targeting RTEL1. 34021146_RTEL1 influences the abundance and localization of TERRA RNA. 34479523_Pulmonary fibrosis in dyskeratosis congenita: a case report with a PRISMA-compliant systematic review. | ENSMUSG00000038685 | Rtel1 | 1824.15236 | 0.9802323 | -0.0288043630 | 0.11689245 | 6.274945e-02 | 8.022019e-01 | 9.998360e-01 | No | Yes | 1563.82521 | 178.595512 | 1.454082e+03 | 128.778648 | |
| ENSG00000258526 | lncRNA | 12.14763 | 1.2623438 | 0.3361049259 | 0.83067549 | 1.736652e-01 | 6.768747e-01 | 9.998360e-01 | No | Yes | 13.67930 | 5.007915 | 1.102684e+01 | 3.245156 | ||||||||||||
| ENSG00000260537 | 55308 | protein_coding | F6QDS0 | ATP-binding;Helicase;Hydrolase;Nucleotide-binding;Proteomics identification;Reference proteome | Mouse_homologues mmu:234733; | ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; nucleic acid binding [GO:0003676]; RNA helicase activity [GO:0003724] | ENSMUSG00000033658 | Ddx19b | 795.13194 | 1.1398243 | 0.1888114400 | 0.16372494 | 1.246053e+00 | 2.643076e-01 | 9.998360e-01 | No | Yes | 454.69131 | 81.080342 | 4.051162e+02 | 56.273686 | |||||
| ENSG00000260923 | LINC02193 | lncRNA | 24.18814 | 0.8496230 | -0.2351052890 | 0.60672011 | 1.306925e-01 | 7.177151e-01 | 9.998360e-01 | No | Yes | 23.13605 | 6.006869 | 2.827602e+01 | 5.734347 | |||||||||||
| ENSG00000262580 | lncRNA | 165.63270 | 0.8787386 | -0.1864940204 | 0.24050014 | 5.996720e-01 | 4.387032e-01 | 9.998360e-01 | No | Yes | 122.38246 | 21.172975 | 1.294293e+02 | 17.559899 | ||||||||||||
| ENSG00000263020 | 58496 | protein_coding | Q5SRQ3 | FUNCTION: Regulatory subunit of casein kinase II/CK2. As part of the kinase complex regulates the basal catalytic activity of the alpha subunit a constitutively active serine/threonine-protein kinase that phosphorylates a large number of substrates containing acidic residues C-terminal to the phosphorylated serine or threonine. Participates in Wnt signaling. {ECO:0000256|RuleBase:RU361268}. | Proteomics identification;Reference proteome;Wnt signaling pathway | protein kinase CK2 complex [GO:0005956]; protein kinase regulator activity [GO:0019887]; Wnt signaling pathway [GO:0016055] | 455.73456 | 0.8518632 | -0.2313062855 | 0.16888234 | 1.884516e+00 | 1.698218e-01 | 9.998360e-01 | No | Yes | 442.28736 | 52.156491 | 4.911823e+02 | 46.082396 | |||||||
| ENSG00000263843 | 100287042 | MIF4GD-DT | lncRNA | 103.78268 | 0.9206288 | -0.1193085005 | 0.33580378 | 1.304661e-01 | 7.179493e-01 | 9.998360e-01 | No | Yes | 99.55620 | 16.751069 | 1.008109e+02 | 13.549200 | ||||||||||
| ENSG00000267191 | 118827818 | ZNF45-AS1 | lncRNA | 9.49673 | 1.3664019 | 0.4503818620 | 0.94106077 | 2.036186e-01 | 6.518158e-01 | 9.998360e-01 | No | Yes | 11.73970 | 6.981574 | 8.979521e+00 | 4.330340 | ||||||||||
| ENSG00000267248 | 100996660 | lncRNA | 45.05616 | 0.9331563 | -0.0998092952 | 0.45876912 | 4.722465e-02 | 8.279650e-01 | 9.998360e-01 | No | Yes | 44.62236 | 8.802526 | 4.403323e+01 | 6.929296 | |||||||||||
| ENSG00000267260 | 728485 | lncRNA | 51.82390 | 0.9487430 | -0.0759107922 | 0.40949216 | 3.557065e-02 | 8.504049e-01 | 9.998360e-01 | No | Yes | 51.30141 | 9.681263 | 5.525804e+01 | 8.519454 | |||||||||||
| ENSG00000267757 | 100287177 | EML2-AS1 | lncRNA | 58.05589 | 1.1002024 | 0.1377689389 | 0.38491972 | 1.301831e-01 | 7.182422e-01 | 9.998360e-01 | No | Yes | 76.94797 | 14.601833 | 6.474823e+01 | 10.527168 | ||||||||||
| ENSG00000268350 | 29057 | FAM156A | protein_coding | Q8NDB6 | Acetylation;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:29057;hsa:727866; | integral component of membrane [GO:0016021]; nuclear envelope [GO:0005635]; methylated histone binding [GO:0035064] | ENSMUSG00000041353 | Tmem29 | 529.21716 | 1.2879006 | 0.3650212616 | 0.65600324 | 3.485828e-01 | 5.549164e-01 | 9.998360e-01 | No | Yes | 682.56816 | 167.758080 | 5.262895e+02 | 99.727576 | ||||
| ENSG00000270049 | 101927837 | ATP6V0D1-DT | lncRNA | 83.97120 | 1.1573793 | 0.2108616988 | 0.35410039 | 3.577507e-01 | 5.497581e-01 | 9.998360e-01 | No | Yes | 86.74429 | 22.550817 | 7.689530e+01 | 15.801437 | ||||||||||
| ENSG00000271147 | ARMCX5-GPRASP2 | lncRNA | 64.50687 | 0.8995663 | -0.1526985142 | 0.36752249 | 1.683247e-01 | 6.816051e-01 | 9.998360e-01 | No | Yes | 64.88636 | 13.392490 | 7.884183e+01 | 12.619185 | |||||||||||
| ENSG00000272690 | 107986100 | LINC02018 | lncRNA | 38.12364 | 0.7839989 | -0.3510763782 | 0.51876422 | 4.710750e-01 | 4.924930e-01 | 9.998360e-01 | No | Yes | 36.73750 | 9.090668 | 4.668432e+01 | 9.222015 | ||||||||||
| ENSG00000273162 | lncRNA | 52.69504 | 0.9268197 | -0.1096394501 | 0.40917488 | 7.069074e-02 | 7.903337e-01 | 9.998360e-01 | No | Yes | 45.83796 | 9.077741 | 5.603614e+01 | 8.667630 | ||||||||||||
| ENSG00000274372 | LINC02804 | lncRNA | 21.54431 | 0.7521961 | -0.4108191750 | 0.67988007 | 4.027277e-01 | 5.256839e-01 | 9.998360e-01 | No | Yes | 20.72938 | 6.578144 | 2.741058e+01 | 6.777559 | |||||||||||
| ENSG00000274602 | 728233 | PI4KAP1 | transcribed_unprocessed_pseudogene | 1443.27845 | 0.9095214 | -0.1368205498 | 0.13547859 | 9.957625e-01 | 3.183380e-01 | 9.998360e-01 | No | Yes | 1335.26022 | 177.155936 | 1.344872e+03 | 137.949065 | ||||||||||
| ENSG00000276234 | 6871 | TADA2A | protein_coding | A0A087WZS1 | Mouse_homologues FUNCTION: Component of the ATAC complex, a complex with histone acetyltransferase activity on histones H3 and H4 (By similarity). Required for the function of some acidic activation domains, which activate transcription from a distant site (By similarity). Binds double-stranded DNA (PubMed:16299514). Binds dinucleosomes, probably at the linker region between neighboring nucleosomes (PubMed:16299514). Plays a role in chromatin remodeling (By similarity). May promote TP53/p53 'Lys-321' acetylation, leading to reduced TP53 stability and transcriptional activity (By similarity). May also promote XRCC6 acetylation thus facilitating cell apoptosis in response to DNA damage (By similarity). {ECO:0000250|UniProtKB:O75478, ECO:0000269|PubMed:16299514}. | Metal-binding;Proteomics identification;Reference proteome;Zinc;Zinc-finger | Many DNA-binding transcriptional activator proteins enhance the initiation rate of RNA polymerase II-mediated gene transcription by interacting functionally with the general transcription machinery bound at the basal promoter. Adaptor proteins are usually required for this activation, possibly to acetylate and destabilize nucleosomes, thereby relieving chromatin constraints at the promoter. The protein encoded by this gene is a transcriptional activator adaptor and has been found to be part of the PCAF histone acetylase complex. Several alternatively spliced transcript variants encoding different isoforms of this gene have been described, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Oct 2009]. | Mouse_homologues mmu:217031; | Mouse_homologues ATAC complex [GO:0140672]; chromosome [GO:0005694]; mitotic spindle [GO:0072686]; nucleus [GO:0005634]; SAGA complex [GO:0000124]; SAGA-type complex [GO:0070461]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; metal ion binding [GO:0046872]; transcription coactivator activity [GO:0003713]; chromatin remodeling [GO:0006338]; histone H3 acetylation [GO:0043966]; mitotic cell cycle [GO:0000278]; positive regulation of histone acetylation [GO:0035066]; regulation of cell cycle [GO:0051726]; regulation of cell division [GO:0051302]; regulation of embryonic development [GO:0045995]; regulation of histone deacetylation [GO:0031063]; regulation of protein phosphorylation [GO:0001932]; regulation of protein stability [GO:0031647]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of tubulin deacetylation [GO:0090043] | 16299514_The three-dimensional solution structure of the SWIRM domain from the human transcriptional adaptor ADA2alpha was reported. 18059173_hADA2a and hADA3 as crucial cofactors of beta-catenin that are likely involved in the assembly of transactivation-competent beta-catenin complexes at Wnt target genes. 20602615_Observational study of gene-disease association. (HuGE Navigator) 22644376_CCDC134 increased the PCAF-dependent K320 acetylation of p53 and p53 protein stability in the presence of hADA2a overexpression. 25492086_Single nucleotide polymorphism in ADA2 gene is associated with type 1 diabetes. 26468280_results thus demonstrate that the catalytic activity of GCN5 is stimulated by subunits of the ADA2a- or ADA2b-containing HAT modules and is further increased by incorporation of the distinct HAT modules in the ATAC or SAGA holo-complexes 34296306_Circular RNA TADA2A promotes proliferation and migration via modulating of miR638/KIAA0101 signal in nonsmall cell lung cancer. | ENSMUSG00000018651 | Tada2a | 389.99422 | 1.0134598 | 0.0192888393 | 0.17062843 | 1.296334e-02 | 9.093514e-01 | 9.998360e-01 | No | Yes | 376.02167 | 68.069825 | 3.951651e+02 | 55.183268 | |
| ENSG00000276550 | 400322 | HERC2P2 | transcribed_unprocessed_pseudogene | 30809632_Cell colony formation, Transwell and cell scratch tests were performed to evaluate the role of HERC2P2 in glioblastoma growth. Furthermore, we overexpressed HERC2P2 in U87 cells and established a mouse intracranial glioma model to examine the function of HERC2P2 in vivo | 792.66161 | 0.9101124 | -0.1358833905 | 0.16496803 | 6.540081e-01 | 4.186833e-01 | 9.998360e-01 | No | Yes | 781.06333 | 182.765045 | 7.552013e+02 | 136.073393 | |||||||||
| ENSG00000277072 | STAG3L2 | transcribed_unprocessed_pseudogene | 467.34756 | 1.0983254 | 0.1353054792 | 0.16887062 | 6.426815e-01 | 4.227415e-01 | 9.998360e-01 | No | Yes | 478.42839 | 71.496432 | 4.102165e+02 | 47.683780 | |||||||||||
| ENSG00000278932 | lncRNA | 80.08914 | 0.7215508 | -0.4708270321 | 0.33568610 | 1.872456e+00 | 1.711941e-01 | 9.998360e-01 | No | Yes | 99.13588 | 27.187311 | 1.375809e+02 | 29.399105 | ||||||||||||
| ENSG00000281706 | 100507173 | LINC01012 | lncRNA | 108.51369 | 1.0169244 | 0.0242124232 | 0.29029155 | 7.175478e-03 | 9.324934e-01 | 9.998360e-01 | No | Yes | 122.28355 | 16.942277 | 1.179092e+02 | 12.846798 | ||||||||||
| ENSG00000285533 | 105369347 | RELA-DT | lncRNA | 104.46356 | 1.1135404 | 0.1551538398 | 0.29469932 | 2.707312e-01 | 6.028417e-01 | 9.998360e-01 | No | Yes | 131.82230 | 25.174867 | 1.066406e+02 | 16.575972 | ||||||||||
| ENSG00000285565 | lncRNA | 173.58952 | 1.6735428 | 0.7429054733 | 0.69165144 | 8.988621e-01 | 3.430870e-01 | 9.998360e-01 | No | Yes | 241.94394 | 67.156291 | 1.584823e+02 | 33.998589 | ||||||||||||
| ENSG00000286140 | 113455421 | DERPC | protein_coding | P0CG12 | FUNCTION: Potential tumor suppressor. Inhibits prostate tumor cell growth, when overexpressed. {ECO:0000269|PubMed:12477976}. | Methylation;Nucleus;Phosphoprotein;Reference proteome | Mouse_homologues mmu:214987; | extracellular exosome [GO:0070062]; nucleoplasm [GO:0005654] | 12477976_decreased expression of DERPC may be implicated in tumorigenesis of prostate and renal tumors | ENSMUSG00000117748 | Derpc | 5787.82670 | 1.0435311 | 0.0614735527 | 0.08779946 | 4.888260e-01 | 4.844515e-01 | 9.998360e-01 | No | Yes | 6307.83205 | 773.122047 | 5.671044e+03 | 537.139715 | ||
| ENSG00000286905 | 246243 | protein_coding | E5KN15 | FUNCTION: Endonuclease that specifically degrades the RNA of RNA-DNA hybrids. {ECO:0000256|PIRNR:PIRNR036852}. | Endonuclease;Hydrolase;Magnesium;Metal-binding;Nuclease | hsa:246243; | magnesium ion binding [GO:0000287]; nucleic acid binding [GO:0003676]; RNA-DNA hybrid ribonuclease activity [GO:0004523] | Mouse_homologues 12667461_RNase H1 is involved in generation of mitochondrial DNA 20823270_translational organization of RNase H1 allows tight control of expression of RNase H1 in mitochondria, where its excess or absence can lead to cell death, without affecting the expression of the nuclear RNase H1 26162680_the essential role of RNase H1 in mitochondrial DNA replication is the removal of primers at the origin of replication 27131367_RNase H1 is necessary for the activity of DNA-like ASOs. During liver regeneration, a clone of hepatocytes that expressed RNase H1 developed and partially restored mitochondrial and liver function. | ENSMUSG00000020630 | Rnaseh1 | 162.75078 | 0.9292028 | -0.1059346037 | 0.26680196 | 1.608777e-01 | 6.883497e-01 | 9.998360e-01 | No | Yes | 150.65975 | 29.247082 | 1.710298e+02 | 25.728005 | |||
| ENSG00000287750 | 102724156 | lncRNA | 29.70707 | 0.9383951 | -0.0917325738 | 0.52898161 | 2.893089e-02 | 8.649387e-01 | 9.998360e-01 | No | Yes | 49.56719 | 10.868138 | 5.444701e+01 | 10.891202 | |||||||||||
| ENSG00000288271 | lncRNA | 119.53854 | 1.0921895 | 0.1272231400 | 0.33549491 | 1.507450e-01 | 6.978244e-01 | 9.998360e-01 | No | Yes | 115.32768 | 35.971678 | 9.125362e+01 | 22.031120 | ||||||||||||
| ENSG00000289088 | 101929898 | lncRNA | 48.64275 | 0.7862095 | -0.3470143208 | 0.42406159 | 6.879300e-01 | 4.068694e-01 | 9.998360e-01 | No | Yes | 49.59491 | 13.548909 | 5.790455e+01 | 13.089752 |
- Note:
- The spreadsheet above contains the non-differential expressed genes but with significantly changes in alternative splicing (q < 0.05 and dif > 0.05). Please Click HERE to check all mapped genes in a Microsoft .excel file.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| lncRNA | 55 |
| processed_pseudogene | 1 |
| protein_coding | 398 |
| transcribed_processed_pseudogene | 2 |
| transcribed_unitary_pseudogene | 1 |
| transcribed_unprocessed_pseudogene | 9 |
MA plot
PCA plot
No significcant DEGs were detected under the threshold of P<0.05
Co-expression Analysis
The co-expression pattern was determined using weighted gene co-expression network analysis (WGCNA) through WGCNA R software.
Choosing the soft-thresholding power: analysis of network topology
Quantifying module-trait associations
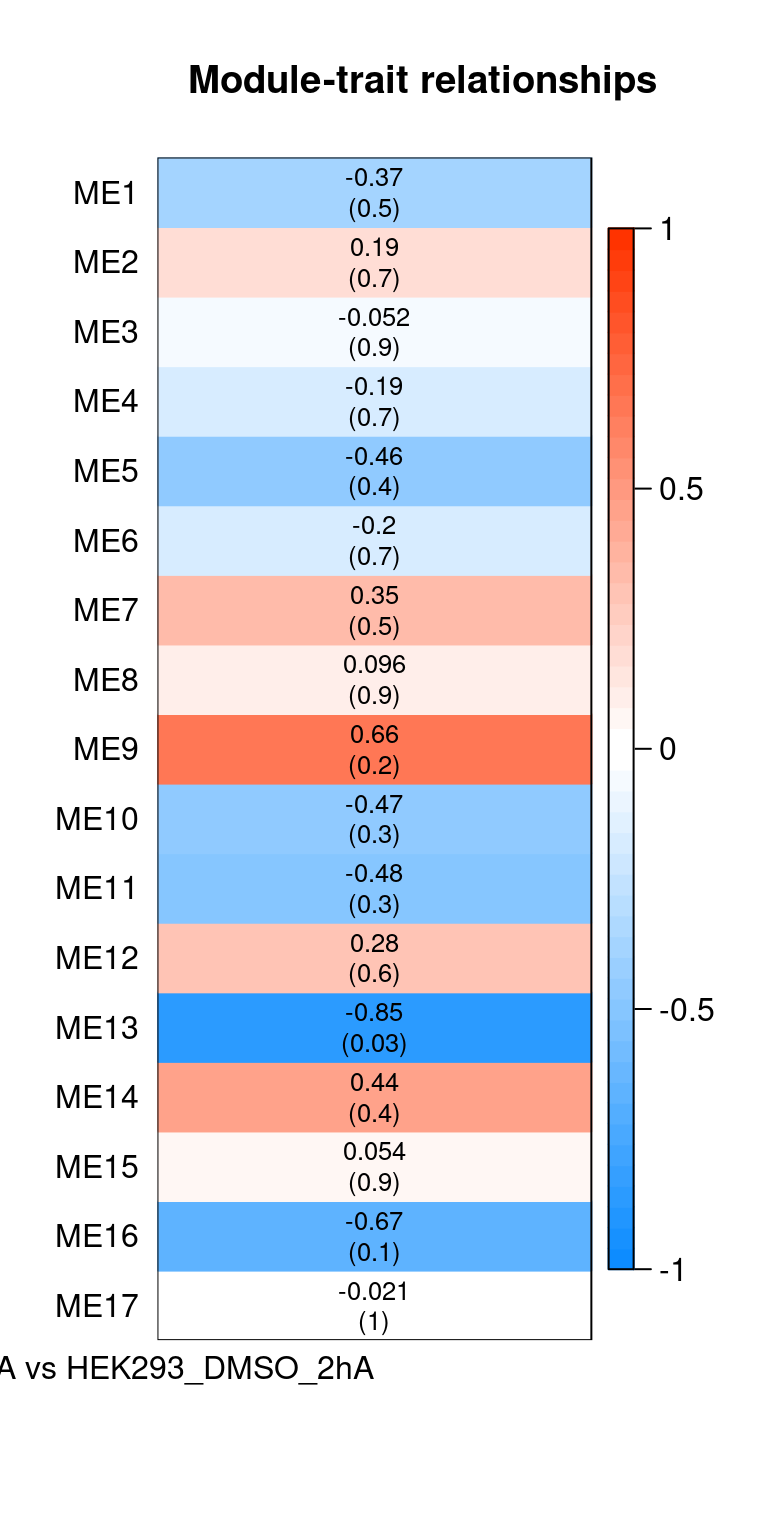
In the above figure, each cell contains the corresponding correlation (upper) and p-value (lower). ME, Module Eigengene.
Spreadsheet
| Cluster ID | Correlation | P-value | DEGs ID | DEGs Symbol |
|---|---|---|---|---|
| ME0 | -0.37151190 | 0.46837038 | ||
| ME1 | 0.18743710 | 0.72213693 | ||
| ME2 | -0.05239051 | 0.92148614 | ||
| ME3 | -0.19428718 | 0.71223615 | ||
| ME4 | -0.46411959 | 0.35380792 | ||
| ME5 | -0.19705252 | 0.70824696 | ||
| ME6 | 0.35219146 | 0.49355552 | ||
| ME7 | 0.09613465 | 0.85624225 | ||
| ME8 | 0.65649536 | 0.15672717 | ||
| ME9 | -0.46862641 | 0.34851808 | ENSG00000124208 | PEDS1-UBE2V1 |
| ME10 | -0.48015092 | 0.33512179 | ||
| ME11 | 0.28193591 | 0.58830138 | ||
| ME12 | -0.84574535 | 0.03385654 | ||
| ME13 | 0.44385362 | 0.37794049 | ||
| ME14 | 0.05419663 | 0.91878465 | ||
| ME15 | -0.67222563 | 0.14354667 | ||
| ME16 | -0.02128145 | 0.96808265 | ENSG00000270276 | H4C15 |
Help
still working on
2. Functional Enrichment Analysis
2-1. Spreadsheet
GSEA
Please Click HERE to download a Microsoft .excel that contains all GSEA results.
GO: BP
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0032508 | DNA duplex unwinding | 79 | -0.5219304 | -1.665705 | 0.003156234 | 1 | 1 | 2315 | tags=28%, list=14%, signal=24% | MCM6||XRCC5||RAD54L||DDX1||POLQ||NAV2||FANCM||CHD6||DHX36||CHD2||MRE11||ERCC6L||BLM||CHD9||RAD50||RAD54B||CHD7||CHD1||MCM8||HELQ||WRN||MNAT1 | 5.715015 | 6.296827 | 5.965979 | 6.390009 | 5.628502 | 5.745282 | 6.352478 | 6.470666 | 5.714655 | 5.797398 | 5.628025 | 6.297578 | 6.297149 | 6.295754 | 6.184883 | 5.809548 | 5.875035 | 6.357245 | 6.354408 | 6.456033 | 5.614649 | 5.667379 | 5.602655 | 5.692618 | 5.692751 | 5.845052 | 6.349668 | 6.345301 | 6.362410 | 6.457546 | 6.484931 | 6.469389 |
| GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 10 | 0.8094783 | 1.755139 | 0.005755396 | 1 | 1 | 1723 | tags=40%, list=11%, signal=36% | SLC35D3||TMEM30A||EDEM1||SORL1 | 7.117005 | 6.919455 | 7.362000 | 6.925550 | 7.172971 | 7.378534 | 6.749864 | 6.867203 | 7.028314 | 7.002908 | 7.300542 | 6.973389 | 6.964864 | 6.814654 | 7.144745 | 7.503631 | 7.413927 | 6.945211 | 6.978096 | 6.850313 | 7.163357 | 7.059422 | 7.287111 | 7.279743 | 7.357815 | 7.490157 | 6.779679 | 6.762612 | 6.706281 | 6.857446 | 6.927409 | 6.814497 |
| GO:0030317 | flagellated sperm motility | 50 | -0.5680518 | -1.678623 | 0.008349973 | 1 | 1 | 1450 | tags=28%, list=9%, signal=26% | SPEF2||TTLL3||ASH1L||TMF1||TPGS1||IQCG||TEKT2||CATSPER3||MNS1||CELF3||CATSPER2||QRICH2||CFAP69||DNAH11 | 4.070786 | 4.473719 | 4.298454 | 4.546458 | 3.858772 | 3.814021 | 4.505455 | 4.496005 | 4.181592 | 4.203162 | 3.791627 | 4.476091 | 4.469532 | 4.475525 | 4.550882 | 4.121844 | 4.184435 | 4.518525 | 4.554705 | 4.565724 | 3.908495 | 3.973766 | 3.677563 | 3.695033 | 3.662147 | 4.051689 | 4.508830 | 4.491161 | 4.516259 | 4.495004 | 4.515870 | 4.476876 |
| GO:0032757 | positive regulation of interleukin-8 production | 32 | 0.6229562 | 1.766581 | 0.009556907 | 1 | 1 | 2230 | tags=41%, list=14%, signal=35% | DDX58||TLR5||HSPA1A||F3||LGALS9||HSPA1B||BCL10||F2RL1||HYAL2||NOD1||AFAP1L2||CD58||MYD88 | 6.193407 | 6.675882 | 5.828182 | 6.192691 | 6.328540 | 5.812416 | 6.660389 | 6.155266 | 6.258940 | 6.321583 | 5.976850 | 6.681989 | 6.671116 | 6.674521 | 5.916004 | 5.706693 | 5.853915 | 6.191892 | 6.151338 | 6.233667 | 6.419771 | 6.322275 | 6.237820 | 5.864867 | 5.772644 | 5.798158 | 6.698575 | 6.635527 | 6.646274 | 6.178223 | 6.106362 | 6.179998 |
| GO:0002551 | mast cell chemotaxis | 11 | 0.7746115 | 1.714378 | 0.010144928 | 1 | 1 | 1596 | tags=36%, list=10%, signal=33% | RAC2||VEGFC||KIT||VEGFA | 4.676936 | 4.958654 | 4.536017 | 4.769253 | 4.630246 | 4.658660 | 4.957081 | 4.776210 | 4.669463 | 4.779616 | 4.574409 | 5.002662 | 4.920981 | 4.951134 | 4.621149 | 4.401422 | 4.576265 | 4.753430 | 4.747181 | 4.806411 | 4.632225 | 4.657178 | 4.600781 | 4.646639 | 4.678546 | 4.650587 | 4.968829 | 4.941646 | 4.960633 | 4.783542 | 4.756107 | 4.788766 |
| GO:0051573 | negative regulation of histone H3-K9 methylation | 11 | -0.7998671 | -1.730183 | 0.010634184 | 1 | 1 | 484 | tags=45%, list=3%, signal=44% | SMARCB1||DNMT1||PAX5||BRCA1||KDM3A | 5.715437 | 5.854169 | 6.002217 | 5.863733 | 5.691422 | 5.916908 | 5.901695 | 5.882977 | 5.637451 | 5.757369 | 5.748426 | 5.858879 | 5.846908 | 5.856693 | 6.058505 | 5.972051 | 5.974402 | 5.898860 | 5.801647 | 5.888725 | 5.732230 | 5.602162 | 5.735893 | 5.903625 | 5.815286 | 6.024169 | 5.911480 | 5.893537 | 5.900012 | 5.869137 | 5.895331 | 5.884344 |
| GO:0010935 | regulation of macrophage cytokine production | 11 | -0.7977049 | -1.725506 | 0.011020882 | 1 | 1 | 787 | tags=45%, list=5%, signal=43% | SPON2||TGFB3||HLA-G||PLCG2||TGFB2 | 3.715189 | 3.440599 | 3.429881 | 3.236336 | 3.745123 | 3.815708 | 3.402379 | 3.156727 | 3.708361 | 3.571470 | 3.852088 | 3.449413 | 3.437958 | 3.434382 | 3.360561 | 3.501054 | 3.424593 | 3.259842 | 3.150120 | 3.295114 | 3.707108 | 3.647706 | 3.871171 | 3.777186 | 3.974282 | 3.679908 | 3.444458 | 3.358817 | 3.402590 | 3.107956 | 3.195636 | 3.165220 |
| GO:0045661 | regulation of myoblast differentiation | 32 | -0.6276340 | -1.711668 | 0.012041497 | 1 | 1 | 111 | tags=19%, list=1%, signal=19% | MEF2C||PRICKLE1||TRIP4||PLCB1||CAPN3||KLHL41 | 4.199800 | 4.287589 | 4.210677 | 4.061901 | 4.287788 | 4.395809 | 4.211100 | 4.058662 | 4.112206 | 4.192155 | 4.289561 | 4.319064 | 4.284878 | 4.258179 | 4.190097 | 4.245019 | 4.196285 | 4.094373 | 3.993491 | 4.095480 | 4.294312 | 4.157026 | 4.401646 | 4.351914 | 4.431458 | 4.402932 | 4.239030 | 4.180677 | 4.213000 | 4.046844 | 4.091105 | 4.037467 |
| GO:2001257 | regulation of cation channel activity | 116 | -0.4474449 | -1.506032 | 0.012041975 | 1 | 1 | 2755 | tags=26%, list=17%, signal=22% | KCNG1||ATPSCKMT||GALR2||CACNB2||CACNB4||HOMER1||CNIH3||NLGN2||MEF2C||CRBN||PTK2B||CACNG5||NLGN3||AKAP6||ADRB2||SHISA7||GPR35||ABCC8||NLGN1||CBARP||DRD4||SHISA8||PLCG2||HPCA||FGF12||JSRP1||S100A1||LRRC26||NOS1AP||PPARGC1A | 4.525423 | 4.895474 | 4.646889 | 4.886305 | 4.486052 | 4.586802 | 4.896749 | 4.900193 | 4.549629 | 4.597656 | 4.423419 | 4.907010 | 4.894645 | 4.884679 | 4.694826 | 4.630568 | 4.614004 | 4.879308 | 4.861034 | 4.917987 | 4.519181 | 4.495741 | 4.442157 | 4.547208 | 4.498319 | 4.706571 | 4.900210 | 4.890472 | 4.899545 | 4.878741 | 4.931169 | 4.890141 |
| GO:0046599 | regulation of centriole replication | 20 | -0.7003538 | -1.733689 | 0.012608205 | 1 | 1 | 3246 | tags=60%, list=20%, signal=48% | KAT2A||MDM1||CDK5RAP2||TRIM37||STIL||KAT2B||CEP295||ENSG00000285943||PLK4||ALMS1||BRCA1||SPICE1 | 5.592883 | 7.637034 | 5.900014 | 7.707511 | 5.490548 | 5.851860 | 7.734853 | 7.777831 | 5.659069 | 5.649586 | 5.461521 | 7.662662 | 7.569134 | 7.676949 | 6.611078 | 5.489448 | 5.188458 | 7.642743 | 7.674434 | 7.800487 | 5.360979 | 5.563726 | 5.538595 | 5.703326 | 5.713662 | 6.101827 | 7.681433 | 7.720268 | 7.800298 | 7.771957 | 7.753616 | 7.807400 |
| GO:0032836 | glomerular basement membrane development | 10 | -0.8081453 | -1.710056 | 0.012847966 | 1 | 1 | 208 | tags=30%, list=1%, signal=30% | MYO1E||COL4A4||COL4A3 | 4.373399 | 4.609314 | 4.882903 | 4.456527 | 4.417680 | 4.857746 | 4.525393 | 4.344772 | 4.330139 | 4.319946 | 4.465489 | 4.620230 | 4.638996 | 4.567774 | 4.937169 | 4.946610 | 4.757144 | 4.451756 | 4.516704 | 4.398697 | 4.412440 | 4.286046 | 4.543106 | 4.636767 | 4.752337 | 5.135703 | 4.568416 | 4.527110 | 4.479275 | 4.350826 | 4.339661 | 4.343806 |
| GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 14 | 0.7153001 | 1.679210 | 0.012967998 | 1 | 1 | 2270 | tags=43%, list=14%, signal=37% | WNT2B||LHX1||GDNF||SOX9||VEGFA||SIX1 | 4.490414 | 4.498680 | 4.606550 | 4.509528 | 4.509450 | 4.703315 | 4.540171 | 4.516592 | 4.436740 | 4.508619 | 4.524379 | 4.486599 | 4.534101 | 4.474653 | 4.626359 | 4.613221 | 4.579669 | 4.507511 | 4.465158 | 4.554528 | 4.539003 | 4.426784 | 4.559072 | 4.643446 | 4.603970 | 4.850220 | 4.561772 | 4.529750 | 4.528746 | 4.500391 | 4.568223 | 4.479668 |
| GO:0090657 | telomeric loop disassembly | 11 | -0.7841458 | -1.696177 | 0.014501160 | 1 | 1 | 1037 | tags=55%, list=6%, signal=51% | POT1||TERF1||RECQL4||BLM||SLX1B||WRN | 4.459681 | 4.931730 | 4.979441 | 4.857542 | 4.479054 | 4.860181 | 4.966312 | 4.898262 | 4.382509 | 4.330784 | 4.645617 | 4.939405 | 4.952912 | 4.902401 | 5.105121 | 4.994721 | 4.824759 | 4.822146 | 4.828516 | 4.919877 | 4.448261 | 4.302958 | 4.663045 | 4.499258 | 4.800619 | 5.195639 | 5.004350 | 4.974350 | 4.918940 | 4.901928 | 4.902503 | 4.890322 |
| GO:0007608 | sensory perception of smell | 16 | -0.7260524 | -1.709558 | 0.014773599 | 1 | 1 | 259 | tags=38%, list=2%, signal=37% | UBR3||NAV2||NCAM2||CFAP69||B3GNT2||SLC24A4 | 2.483486 | 3.474000 | 2.952132 | 3.466261 | 2.376658 | 2.581258 | 3.532373 | 3.467021 | 2.623188 | 2.540101 | 2.263039 | 3.422846 | 3.482106 | 3.515526 | 3.217845 | 2.838162 | 2.757388 | 3.329112 | 3.475855 | 3.582643 | 2.314111 | 2.370070 | 2.442896 | 2.349354 | 2.236892 | 3.025750 | 3.597770 | 3.588528 | 3.402539 | 3.422206 | 3.587855 | 3.382690 |
| GO:0070588 | calcium ion transmembrane transport | 193 | -0.3937133 | -1.412810 | 0.016041905 | 1 | 1 | 2399 | tags=20%, list=15%, signal=17% | CACNB4||CACNA1B||PLCG1||CACNA2D2||ATP2C1||SESTD1||ATP2A3||ITPR1||THADA||PTK2B||CACNG5||AKAP6||PMPCB||GPR35||GJC2||MCUB||CAV3||PRKD1||CACNG6||CBARP||CHD7||CACNA1C||DRD4||ATP2B1||ITPR2||SLC8A3||PLCG2||CATSPER3||GRIN2C||HPCA||CATSPER2||JSRP1||S100A1||GRIN3B||NOS1AP||TRPM6||SLC24A4||RYR1||CAPN3 | 4.757783 | 5.120097 | 4.870185 | 5.143505 | 4.725059 | 4.812746 | 5.096249 | 5.153394 | 4.775890 | 4.806338 | 4.688545 | 5.149419 | 5.122145 | 5.088073 | 4.935606 | 4.844303 | 4.828307 | 5.134756 | 5.116552 | 5.178503 | 4.737192 | 4.729471 | 4.708359 | 4.772468 | 4.750667 | 4.909885 | 5.107491 | 5.094507 | 5.086671 | 5.137187 | 5.188163 | 5.134192 |
| GO:0043201 | response to leucine | 13 | -0.7478347 | -1.679022 | 0.016516228 | 1 | 1 | 521 | tags=54%, list=3%, signal=52% | MTOR||LARS1||SESN1||UBR2||UBR1||PIK3C3||PPARGC1A | 3.684900 | 4.634703 | 4.166731 | 4.722119 | 3.675384 | 4.205711 | 4.674708 | 4.730457 | 3.576952 | 3.739723 | 3.732244 | 4.621544 | 4.687632 | 4.593302 | 4.336832 | 4.138237 | 4.005756 | 4.705384 | 4.663781 | 4.794105 | 3.668366 | 3.529827 | 3.813967 | 4.140280 | 4.233300 | 4.241381 | 4.652734 | 4.707530 | 4.663272 | 4.693585 | 4.757418 | 4.739619 |
| GO:0007144 | female meiosis I | 10 | -0.7903263 | -1.672350 | 0.017325287 | 1 | 1 | 604 | tags=30%, list=4%, signal=29% | FBXO5||MEIOC||MLH3 | 8.282961 | 8.120456 | 8.146903 | 7.998808 | 8.182585 | 7.797381 | 8.076632 | 8.063378 | 8.259457 | 8.390972 | 8.191262 | 8.251551 | 8.037830 | 8.062372 | 8.019234 | 8.122182 | 8.286608 | 8.082477 | 7.900842 | 8.007355 | 8.262058 | 8.262020 | 8.009208 | 8.001262 | 7.487235 | 7.856249 | 8.086095 | 8.058640 | 8.084993 | 8.089335 | 8.027331 | 8.072756 |
| GO:0010469 | regulation of signaling receptor activity | 105 | -0.4432865 | -1.475281 | 0.018738181 | 1 | 1 | 2452 | tags=23%, list=15%, signal=19% | PHLDA2||HOMER1||CNIH3||NLGN2||MEF2C||BICD1||ADRA2C||PTEN||PTK2B||CACNG5||NLGN3||ADRB2||SHISA7||CAV3||DKK3||NLGN1||AKAP9||ESR2||GPRC5A||JAK2||SHISA8||TGFA||SLC24A4||PPARGC1A | 4.293084 | 4.825128 | 4.617211 | 4.840012 | 4.253722 | 4.409355 | 4.791049 | 4.820051 | 4.285478 | 4.368480 | 4.221523 | 4.833431 | 4.844343 | 4.797190 | 4.701009 | 4.555271 | 4.591330 | 4.857797 | 4.811221 | 4.850585 | 4.236981 | 4.275778 | 4.248130 | 4.319105 | 4.335562 | 4.560569 | 4.814921 | 4.774047 | 4.783862 | 4.812768 | 4.853174 | 4.793567 |
| GO:0030199 | collagen fibril organization | 45 | -0.5503249 | -1.601619 | 0.019472247 | 1 | 1 | 1043 | tags=24%, list=6%, signal=23% | EMILIN1||ADAMTS2||DDR2||FOXC2||LOX||TNXB||EFEMP2||COL11A2||TGFB2||COL3A1||COL12A1 | 3.902525 | 4.525365 | 4.291559 | 4.275594 | 3.951325 | 4.295064 | 4.389670 | 4.131100 | 3.819943 | 3.909995 | 3.973523 | 4.541371 | 4.571005 | 4.461507 | 4.267655 | 4.354420 | 4.250435 | 4.271913 | 4.281910 | 4.272938 | 3.959272 | 3.832183 | 4.053972 | 4.160791 | 4.163000 | 4.529376 | 4.433120 | 4.389403 | 4.345146 | 4.122772 | 4.181274 | 4.087700 |
| GO:0071426 | ribonucleoprotein complex export from nucleus | 70 | -0.4899856 | -1.534682 | 0.020614892 | 1 | 1 | 4472 | tags=44%, list=28%, signal=32% | TSC1||NMD3||NUP160||HNRNPA2B1||XPOT||ZC3H11A||XPO1||AKAP8L||SUPT6H||SARNP||ABCE1||NUP155||FYTTD1||SMG6||YTHDC1||NSUN2||NUP133||RIOK2||SETD2||AGFG1||LTV1||SSB||DDX19B||NUP107||TPR||SDAD1||IWS1||UPF2||THOC2||THOC1||C12orf50 | 7.362886 | 7.658755 | 7.243041 | 7.689349 | 7.311985 | 7.339720 | 7.698467 | 7.738389 | 7.383995 | 7.358125 | 7.346280 | 7.697557 | 7.623902 | 7.653855 | 7.379831 | 7.165657 | 7.173238 | 7.690292 | 7.641984 | 7.734293 | 7.324959 | 7.264162 | 7.345597 | 7.338636 | 7.435839 | 7.237900 | 7.680625 | 7.690283 | 7.724130 | 7.736953 | 7.733668 | 7.744525 |
| GO:0008643 | carbohydrate transport | 106 | 0.4256963 | 1.474778 | 0.021739130 | 1 | 1 | 4123 | tags=42%, list=26%, signal=31% | SLC35D3||MFSD4A||C3||MFSD4B||OPN3||AQP1||SLC2A4||ARPP19||PLS1||SLC5A3||SLC2A3||OSBPL8||CLIP3||SLC17A5||MFSD2B||SLC45A3||IRS1||PRKCI||SLC35A5||SLC2A10||OSTN||C1QTNF12||NR4A3||AQP11||SMPD3||SLC37A2||TMEM144||SLC35A2||SLC35D1||STXBP4||RFT1||CREBL2||SLC2A6||EDNRA||APPL1||SLC35B4||SELENOS||PEA15||RAP1A||EZR||INPP5K||OCLN||MAPK14||PRKAG2 | 4.067728 | 4.603316 | 4.481574 | 4.654156 | 4.098509 | 4.416416 | 4.573956 | 4.592612 | 4.003992 | 4.084798 | 4.112212 | 4.592728 | 4.632689 | 4.584062 | 4.548696 | 4.501119 | 4.390364 | 4.623737 | 4.643619 | 4.694196 | 4.087781 | 4.017844 | 4.185004 | 4.268240 | 4.334638 | 4.621454 | 4.592995 | 4.566268 | 4.562412 | 4.587029 | 4.626629 | 4.563470 |
| GO:0021545 | cranial nerve development | 39 | -0.5723291 | -1.624586 | 0.022809558 | 1 | 1 | 3010 | tags=31%, list=19%, signal=25% | PLXNA3||NRP2||SLC1A3||HOXB3||TBX1||EGR2||NAV2||CHD7||NTRK1||ATOH7||KCNA2||SLC24A4 | 5.804785 | 6.427598 | 5.147250 | 6.408699 | 5.596955 | 5.023904 | 6.403166 | 6.434967 | 5.969458 | 5.943638 | 5.442736 | 6.565864 | 6.314758 | 6.390556 | 5.440645 | 4.605001 | 5.268637 | 6.430874 | 6.351882 | 6.441686 | 5.431631 | 5.905070 | 5.396343 | 5.236440 | 5.199000 | 4.534333 | 6.393097 | 6.356821 | 6.457762 | 6.398767 | 6.407570 | 6.496522 |
| GO:0002070 | epithelial cell maturation | 14 | 0.6935198 | 1.628079 | 0.023216900 | 1 | 1 | 4001 | tags=71%, list=25%, signal=54% | PGR||CDKN1A||HIF1A||RFX3||FOXA1||HOXA5||FEM1B||TMEM79||HOXB13||NKX6-1 | 5.691642 | 5.647916 | 5.798885 | 5.611478 | 5.693983 | 5.760625 | 5.654982 | 5.605400 | 5.671085 | 5.709409 | 5.694173 | 5.654042 | 5.647803 | 5.641876 | 5.752822 | 5.807278 | 5.835337 | 5.618388 | 5.575049 | 5.640234 | 5.720994 | 5.668534 | 5.691943 | 5.805026 | 5.652106 | 5.818879 | 5.654088 | 5.659888 | 5.650957 | 5.589451 | 5.614234 | 5.612384 |
| GO:0051450 | myoblast proliferation | 12 | -0.7419804 | -1.638250 | 0.024012393 | 1 | 1 | 290 | tags=33%, list=2%, signal=33% | MYOD1||PAXBP1||ATOH8||KLHL41 | 6.591417 | 5.590463 | 6.112698 | 5.420077 | 6.753272 | 6.761611 | 5.388810 | 5.392556 | 6.528048 | 6.327538 | 6.866577 | 5.751742 | 5.501129 | 5.503902 | 5.825322 | 6.275897 | 6.197908 | 5.544132 | 5.302073 | 5.403748 | 6.720201 | 6.557714 | 6.954188 | 6.797160 | 7.052144 | 6.350342 | 5.439705 | 5.332247 | 5.392471 | 5.367045 | 5.402507 | 5.407778 |
| GO:0150172 | regulation of phosphatidylcholine metabolic process | 10 | 0.7443718 | 1.613973 | 0.024049332 | 1 | 1 | 789 | tags=20%, list=5%, signal=19% | FABP3||RAB38 | 4.947319 | 5.359463 | 5.126044 | 5.237293 | 4.956951 | 5.251235 | 5.224216 | 5.118486 | 4.948221 | 4.913380 | 4.979595 | 5.341247 | 5.390787 | 5.345832 | 5.103175 | 5.234939 | 5.032666 | 5.271250 | 5.248988 | 5.190439 | 4.959161 | 4.833335 | 5.068762 | 5.152392 | 5.193561 | 5.395812 | 5.270768 | 5.175534 | 5.224774 | 5.115246 | 5.194688 | 5.041451 |
| GO:0070166 | enamel mineralization | 10 | -0.7704539 | -1.630300 | 0.025111933 | 1 | 1 | 1761 | tags=20%, list=11%, signal=18% | TBX1||SLC24A4 | 4.134193 | 4.217583 | 4.623655 | 4.209853 | 4.145896 | 4.614894 | 4.177041 | 4.084412 | 4.088516 | 3.959155 | 4.330072 | 4.142634 | 4.304307 | 4.201143 | 4.490601 | 4.785657 | 4.578615 | 4.279447 | 4.204710 | 4.142121 | 4.068735 | 4.032478 | 4.319260 | 4.329365 | 4.541980 | 4.912194 | 4.227171 | 4.220508 | 4.078617 | 4.085616 | 4.177685 | 3.983397 |
| GO:0007215 | glutamate receptor signaling pathway | 35 | -0.5781134 | -1.605896 | 0.025881057 | 1 | 1 | 479 | tags=29%, list=3%, signal=28% | SLC1A1||GRM4||HOMER1||PTK2B||GRIN2C||SULT1A4||GRIN3B||GRM8||PLCB1||FRRS1L | 3.151331 | 4.259795 | 3.724803 | 4.281027 | 3.118149 | 3.537725 | 4.163780 | 4.217397 | 3.078851 | 3.267012 | 3.100697 | 4.261687 | 4.314293 | 4.201186 | 3.945340 | 3.588136 | 3.612663 | 4.271068 | 4.313665 | 4.257755 | 3.121491 | 3.059816 | 3.170993 | 3.416304 | 3.388283 | 3.775434 | 4.207583 | 4.171091 | 4.111024 | 4.212023 | 4.263384 | 4.175429 |
| GO:0051965 | positive regulation of synapse assembly | 37 | -0.5700074 | -1.603322 | 0.025976385 | 1 | 1 | 2241 | tags=43%, list=14%, signal=37% | CLSTN2||ADNP||CUX2||NLGN2||ADGRL2||ADGRB1||SEMA4D||NLGN3||SLITRK3||OXTR||NA||FLRT3||NLGN1||NTRK1||PTPRD||BHLHB9 | 2.211543 | 3.128602 | 3.067939 | 3.104026 | 2.211080 | 2.897197 | 3.085547 | 3.093877 | 2.153672 | 2.206564 | 2.271957 | 3.046562 | 3.210676 | 3.123891 | 3.145619 | 3.115452 | 2.933777 | 3.074533 | 3.137013 | 3.099847 | 2.209674 | 2.061222 | 2.348104 | 2.607899 | 2.691231 | 3.291977 | 3.120546 | 3.071445 | 3.063994 | 3.051034 | 3.167039 | 3.060661 |
| GO:1900543 | negative regulation of purine nucleotide metabolic process | 14 | -0.7150475 | -1.631286 | 0.026048650 | 1 | 1 | 881 | tags=21%, list=5%, signal=20% | H19||HPCA||PPARGC1A | 5.239451 | 5.547673 | 5.555209 | 5.693632 | 5.176475 | 5.626166 | 5.566487 | 5.675930 | 5.228694 | 5.413458 | 5.053780 | 5.500593 | 5.572622 | 5.568674 | 5.740607 | 5.406484 | 5.497549 | 5.722605 | 5.614892 | 5.740236 | 5.197950 | 5.228573 | 5.099786 | 5.794275 | 5.635531 | 5.425091 | 5.585337 | 5.519780 | 5.593222 | 5.667358 | 5.691820 | 5.668481 |
| GO:0009409 | response to cold | 40 | -0.5601491 | -1.595382 | 0.026263358 | 1 | 1 | 3105 | tags=42%, list=19%, signal=34% | ACADVL||EIF2AK4||SLC9A1||MTCO2P12||VGF||DNAJC3||HSPD1||HSP90AA1||PLAC8||ADRB2||PLIN1||ATP2B1||SLC25A27||LPL||PCSK1N||UCP3||PPARGC1A | 9.042013 | 8.495550 | 9.401369 | 8.859587 | 9.122633 | 9.382336 | 8.626819 | 8.741444 | 8.898807 | 8.985278 | 9.222236 | 7.978722 | 8.688088 | 8.707586 | 9.225255 | 9.586446 | 9.369334 | 8.763326 | 9.094401 | 8.687877 | 9.167891 | 9.004428 | 9.188618 | 9.233160 | 9.255985 | 9.623674 | 8.598751 | 8.659501 | 8.621552 | 8.815098 | 8.708896 | 8.697395 |
| GO:0007611 | learning or memory | 177 | -0.3854346 | -1.368795 | 0.026715040 | 1 | 1 | 3088 | tags=22%, list=19%, signal=18% | NF1||EIF2AK4||AGER||ADNP||KCNK2||CUX2||GALR2||PIAS1||SLC1A1||UBE3A||SNAP25||ITGA3||MEF2C||AFF2||PRKN||PTEN||PAFAH1B1||NLGN3||EGR2||OXTR||CEBPB||SHISA7||APOE||FOXO6||UBA6||C1QL1||ABCC8||SLC8A3||ATP8A1||MME||SHANK2||PTCHD1||NTRK1||CAMK4||PTPRZ1||PLCB1||DNAH11||SCN2A||BHLHB9 | 4.301716 | 4.881181 | 4.555364 | 4.857336 | 4.292216 | 4.504657 | 4.854895 | 4.855548 | 4.271724 | 4.364009 | 4.267334 | 4.877976 | 4.909428 | 4.855631 | 4.649344 | 4.479876 | 4.531609 | 4.845432 | 4.845905 | 4.880393 | 4.288243 | 4.272289 | 4.315782 | 4.427494 | 4.449328 | 4.628594 | 4.874780 | 4.855700 | 4.833915 | 4.853626 | 4.887472 | 4.824865 |
| GO:0045653 | negative regulation of megakaryocyte differentiation | 11 | 0.7256998 | 1.606126 | 0.026915114 | 1 | 1 | 1334 | tags=18%, list=8%, signal=17% | H4C15||H4C8 | 6.049987 | 5.482801 | 5.971996 | 5.461753 | 6.117977 | 6.216745 | 5.401195 | 5.613185 | 5.940400 | 5.948233 | 6.240599 | 5.626098 | 5.422343 | 5.388328 | 5.799650 | 6.101654 | 5.998539 | 5.559602 | 5.373461 | 5.446070 | 6.108669 | 5.968952 | 6.261462 | 6.160958 | 6.316615 | 6.167220 | 5.412996 | 5.357048 | 5.432482 | 5.463612 | 5.822811 | 5.527130 |
| GO:0016246 | RNA interference | 12 | 0.7072440 | 1.603387 | 0.027284002 | 1 | 1 | 3 | tags=8%, list=0%, signal=8% | RMRP | 4.918973 | 5.839747 | 5.401399 | 5.827911 | 4.892266 | 5.192845 | 5.772832 | 7.087049 | 4.859819 | 5.100379 | 4.776848 | 5.975098 | 5.776803 | 5.757112 | 5.620290 | 5.273494 | 5.282675 | 5.873133 | 5.719364 | 5.885376 | 4.979269 | 4.882781 | 4.809722 | 5.232568 | 4.984620 | 5.338824 | 5.768105 | 5.811628 | 5.737808 | 5.836966 | 8.190162 | 5.874143 |
| GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 12 | -0.7337836 | -1.620152 | 0.027691712 | 1 | 1 | 1073 | tags=17%, list=7%, signal=16% | CHRM3||PLCB1 | 4.893457 | 5.115609 | 5.088606 | 5.059752 | 4.886372 | 5.161139 | 5.165361 | 5.057316 | 4.829588 | 4.948236 | 4.900084 | 5.040534 | 5.189051 | 5.113419 | 5.073886 | 5.129321 | 5.061708 | 5.102710 | 4.997508 | 5.076965 | 4.928269 | 4.768477 | 4.955390 | 5.070630 | 5.061744 | 5.334118 | 5.189280 | 5.159110 | 5.147369 | 5.025409 | 5.081349 | 5.064619 |
| GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 17 | -0.6794072 | -1.621527 | 0.027793642 | 1 | 1 | 4008 | tags=65%, list=25%, signal=49% | EGFR||CACNG8||CACNG4||ADORA2A||NLGN2||PTK2B||CACNG5||NLGN3||OXTR||NLGN1||NTRK1 | 3.467660 | 3.917247 | 4.023339 | 3.834123 | 3.404444 | 3.737706 | 3.929246 | 3.789645 | 3.438506 | 3.684468 | 3.246450 | 3.866786 | 3.975335 | 3.907529 | 4.110530 | 3.968191 | 3.987113 | 3.832726 | 3.788689 | 3.879523 | 3.508755 | 3.424877 | 3.269579 | 3.584371 | 3.300261 | 4.183845 | 3.934954 | 3.913398 | 3.939254 | 3.784469 | 3.780712 | 3.803648 |
| GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 18 | 0.6421357 | 1.600968 | 0.028142192 | 1 | 1 | 2854 | tags=50%, list=18%, signal=41% | ITGA4||ZDHHC21||KLF4||CHST2||ALOX5||FUT4||PTAFR||ETS1||NFAT5 | 6.094106 | 5.668700 | 5.946838 | 5.451057 | 6.112902 | 6.174617 | 5.602646 | 5.420637 | 6.042523 | 6.064820 | 6.171648 | 5.679248 | 5.682707 | 5.643824 | 5.806154 | 6.014304 | 6.010390 | 5.496388 | 5.375825 | 5.478056 | 6.152415 | 6.019986 | 6.161984 | 6.171526 | 6.250063 | 6.098265 | 5.614204 | 5.589607 | 5.604022 | 5.433257 | 5.436141 | 5.392094 |
| GO:0003356 | regulation of cilium beat frequency | 11 | -0.7446474 | -1.610738 | 0.029389018 | 1 | 1 | 363 | tags=27%, list=2%, signal=27% | ODAD2||DNAAF1||DNAH11 | 3.097986 | 4.102964 | 3.342625 | 4.117059 | 2.868357 | 2.547258 | 4.170561 | 4.132036 | 3.354167 | 3.295930 | 2.495515 | 4.111158 | 4.084684 | 4.112877 | 3.702107 | 2.976106 | 3.255480 | 4.011768 | 4.123740 | 4.208916 | 2.792029 | 3.101790 | 2.677198 | 2.594116 | 2.165375 | 2.809752 | 4.225910 | 4.208987 | 4.071897 | 4.096235 | 4.198558 | 4.098941 |
| GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA | 20 | 0.6246936 | 1.594768 | 0.030076792 | 1 | 1 | 3 | tags=5%, list=0%, signal=5% | RMRP | 5.172419 | 5.862107 | 5.406016 | 5.752690 | 5.173833 | 5.397406 | 5.760972 | 6.631780 | 5.115228 | 5.202245 | 5.198124 | 5.961349 | 5.806495 | 5.813103 | 5.716039 | 5.191361 | 5.251639 | 5.766978 | 5.684995 | 5.803554 | 5.211631 | 5.041958 | 5.258998 | 5.369152 | 5.388293 | 5.434002 | 5.735127 | 5.774835 | 5.772609 | 5.755009 | 7.565611 | 5.756301 |
| GO:0032682 | negative regulation of chemokine production | 15 | 0.6668412 | 1.595288 | 0.030119222 | 1 | 1 | 2122 | tags=33%, list=13%, signal=29% | LGALS9||C1QTNF3||F2RL1||KLF4||MAP2K5 | 7.042144 | 5.969350 | 6.801771 | 5.983931 | 7.178230 | 7.166479 | 5.863181 | 5.958760 | 6.977385 | 6.844745 | 7.270838 | 6.021229 | 5.965938 | 5.919068 | 6.516508 | 6.968072 | 6.882061 | 6.095274 | 5.874694 | 5.973343 | 7.129682 | 7.052187 | 7.337535 | 7.099190 | 7.373492 | 7.000422 | 5.902849 | 5.825324 | 5.860325 | 5.970826 | 5.953891 | 5.951485 |
| GO:0070286 | axonemal dynein complex assembly | 25 | -0.6159321 | -1.602355 | 0.030314302 | 1 | 1 | 779 | tags=32%, list=5%, signal=30% | LRRC49||DNAAF2||DNAAF4||TEKT2||ODAD2||SPAG1||DNAAF1||DNAH17 | 1.965163 | 2.486936 | 2.392256 | 2.617401 | 2.043233 | 2.250927 | 2.536143 | 2.654755 | 1.952084 | 1.965455 | 1.977836 | 2.422353 | 2.555768 | 2.479576 | 2.574841 | 2.347506 | 2.233224 | 2.566197 | 2.618557 | 2.665731 | 2.037722 | 1.941685 | 2.143242 | 2.155106 | 2.033666 | 2.518839 | 2.541198 | 2.546431 | 2.520671 | 2.619430 | 2.679438 | 2.664721 |
| GO:1901264 | carbohydrate derivative transport | 62 | 0.4722873 | 1.511699 | 0.030939476 | 1 | 1 | 5260 | tags=55%, list=33%, signal=37% | SLC35D3||SLC25A24||SLC10A7||GJA1||VLDLR||SLC33A1||ADORA1||SLC15A4||SLC35A5||CD47||ENSG00000261832||ABCG1||SLC37A2||SLC35A2||SLC35D1||RFT1||SLC25A17||G6PC3||SLC35B4||LRRC8C||SLC35B3||ABCC6||SLC25A5||SLC35E3||SLC35B1||SLC25A6||SLC25A4||NPC2||SLC29A3||SCARB1||SLC35A4||SLC25A19||SLC37A4||SLC50A1 | 6.199161 | 5.964336 | 6.252716 | 5.966508 | 6.217881 | 6.298529 | 5.838792 | 5.913297 | 6.190638 | 6.232173 | 6.174049 | 6.026962 | 5.971292 | 5.891551 | 6.135743 | 6.299603 | 6.316007 | 6.023343 | 5.924545 | 5.949799 | 6.226994 | 6.193210 | 6.233120 | 6.304625 | 6.319828 | 6.270697 | 5.867036 | 5.830690 | 5.818204 | 5.910628 | 5.943493 | 5.885177 |
| GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 58 | 0.4778900 | 1.511042 | 0.031081081 | 1 | 1 | 3340 | tags=29%, list=21%, signal=23% | KLF7||EPHA5||CLTRN||PTPRN2||HIF1A||NR1D1||GPLD1||C1QTNF12||GPR27||SELENOT||ABCG1||HMGN3||SYBU||HMGCR||STXBP4||FKBP1B||MPC2 | 5.384732 | 5.688461 | 5.448932 | 5.578498 | 5.344677 | 5.353543 | 5.688918 | 5.585226 | 5.405443 | 5.444271 | 5.300687 | 5.679474 | 5.693278 | 5.692589 | 5.513417 | 5.367243 | 5.462342 | 5.584865 | 5.550566 | 5.599625 | 5.309216 | 5.365919 | 5.358244 | 5.274089 | 5.365026 | 5.417865 | 5.708428 | 5.693202 | 5.664785 | 5.586831 | 5.597807 | 5.570914 |
| GO:1905269 | positive regulation of chromatin organization | 13 | -0.7148665 | -1.605003 | 0.032072210 | 1 | 1 | 2022 | tags=54%, list=13%, signal=47% | ATF7IP||MPHOSPH8||TPR||RESF1||TAL1||SETDB2||ZNF304 | 6.261364 | 6.201783 | 6.368359 | 6.336681 | 6.317380 | 6.588352 | 6.261699 | 6.372262 | 6.128751 | 6.160281 | 6.469867 | 6.165700 | 6.243688 | 6.194883 | 6.360790 | 6.383906 | 6.360256 | 6.372571 | 6.286527 | 6.349576 | 6.291206 | 6.185258 | 6.462066 | 6.519869 | 6.683177 | 6.556884 | 6.296020 | 6.260207 | 6.228070 | 6.363279 | 6.379797 | 6.373660 |
| GO:0035020 | regulation of Rac protein signal transduction | 20 | 0.6184723 | 1.578886 | 0.032209898 | 1 | 1 | 1044 | tags=20%, list=7%, signal=19% | OGT||KRAS||KBTBD6||KBTBD7 | 5.013166 | 5.334232 | 5.153032 | 5.211367 | 5.029493 | 5.155393 | 5.325128 | 5.165876 | 4.990107 | 5.033242 | 5.015822 | 5.342166 | 5.318265 | 5.342134 | 5.289150 | 5.069996 | 5.089602 | 5.184994 | 5.162987 | 5.283256 | 5.066433 | 4.954274 | 5.064929 | 5.191598 | 5.118987 | 5.154680 | 5.348269 | 5.308304 | 5.318511 | 5.152910 | 5.182401 | 5.162158 |
| GO:0006312 | mitotic recombination | 23 | -0.6262143 | -1.595937 | 0.032264116 | 1 | 1 | 2168 | tags=43%, list=14%, signal=38% | GEN1||NSMCE2||RAD51||SMC5||SMC6||MRE11||RAD50||RAD54B||MSH3||ANKLE1 | 4.004976 | 4.778981 | 4.282758 | 4.820938 | 3.983334 | 4.201971 | 4.825171 | 4.854420 | 3.985911 | 4.020368 | 4.008438 | 4.740049 | 4.794946 | 4.801168 | 4.533342 | 4.191275 | 4.084751 | 4.718159 | 4.832034 | 4.906419 | 3.969613 | 3.911190 | 4.065003 | 4.075788 | 4.198162 | 4.321504 | 4.825336 | 4.818766 | 4.831382 | 4.814900 | 4.921601 | 4.824323 |
| GO:0061001 | regulation of dendritic spine morphogenesis | 44 | -0.5346475 | -1.550215 | 0.032825535 | 1 | 1 | 2800 | tags=27%, list=17%, signal=23% | PPFIA2||CUX2||UBE3A||DNM1L||PTEN||TANC2||PAFAH1B1||SRCIN1||DHX36||NLGN1||CAPRIN2||BHLHB9 | 5.566168 | 5.321422 | 5.488638 | 5.445423 | 5.637284 | 5.763623 | 5.332782 | 5.468298 | 5.483560 | 5.529407 | 5.678281 | 5.300435 | 5.339018 | 5.324549 | 5.380528 | 5.588478 | 5.489421 | 5.452653 | 5.399007 | 5.483348 | 5.647482 | 5.516804 | 5.738966 | 5.739923 | 5.831996 | 5.716344 | 5.366074 | 5.302721 | 5.328849 | 5.470880 | 5.483432 | 5.450389 |
| GO:0050830 | defense response to Gram-positive bacterium | 27 | -0.5960409 | -1.570777 | 0.036113179 | 1 | 1 | 51 | tags=11%, list=0%, signal=11% | IL12A||LYG1||H2BC6 | 7.962656 | 7.535785 | 7.728311 | 7.545887 | 7.870942 | 7.308301 | 7.523527 | 7.219699 | 8.100271 | 8.075282 | 7.674098 | 7.589917 | 7.484700 | 7.530807 | 7.763851 | 7.545485 | 7.858033 | 7.539830 | 7.575805 | 7.521494 | 7.799347 | 8.182693 | 7.561943 | 7.595598 | 7.208204 | 7.068187 | 7.483292 | 7.552670 | 7.533731 | 7.539566 | 6.050367 | 7.606132 |
| GO:0006700 | C21-steroid hormone biosynthetic process | 14 | -0.6925831 | -1.580036 | 0.038115304 | 1 | 1 | 1075 | tags=21%, list=7%, signal=20% | DKK3||EGR1||PPARGC1A | 5.960250 | 5.324623 | 5.893316 | 5.221694 | 6.005673 | 6.048056 | 5.270919 | 5.172167 | 5.898326 | 5.853555 | 6.115093 | 5.340361 | 5.348367 | 5.284302 | 5.678640 | 6.022140 | 5.956624 | 5.311774 | 5.141784 | 5.206400 | 6.025680 | 5.891386 | 6.092723 | 6.087327 | 6.056231 | 5.999230 | 5.319502 | 5.238651 | 5.253311 | 5.178738 | 5.192060 | 5.145303 |
| GO:0035176 | social behavior | 34 | -0.5583399 | -1.540964 | 0.038645565 | 1 | 1 | 1909 | tags=47%, list=12%, signal=42% | MECP2||GRID1||DLG4||EN1||ATXN1L||MTOR||KCNQ1||PTEN||TBX1||SEPTIN5||NLGN3||OXTR||DRD4||PCM1||SHANK2||PTCHD1 | 3.663829 | 4.011690 | 4.067352 | 4.028096 | 3.608490 | 4.034047 | 4.058403 | 4.032097 | 3.641531 | 3.673756 | 3.675944 | 3.944500 | 4.055230 | 4.032977 | 4.122924 | 4.079840 | 3.996439 | 3.978518 | 4.029384 | 4.074781 | 3.627727 | 3.495798 | 3.694888 | 3.914031 | 3.918376 | 4.244272 | 4.102899 | 4.076028 | 3.994066 | 4.014378 | 4.066552 | 4.014735 |
| GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 12 | 0.6862416 | 1.555772 | 0.039685821 | 1 | 1 | 4135 | tags=67%, list=26%, signal=50% | HSPA1A||JUN||VHL||DNAJB1||CITED2||NCK1||HIF1AN||TMBIM6 | 5.527106 | 6.349108 | 6.063614 | 6.135238 | 5.547824 | 5.782047 | 6.275673 | 6.052226 | 5.569942 | 5.656375 | 5.336312 | 6.391899 | 6.349690 | 6.304408 | 6.159351 | 5.982407 | 6.043451 | 6.119552 | 6.131697 | 6.154250 | 5.595395 | 5.556148 | 5.489967 | 5.627281 | 5.430384 | 6.180443 | 6.278320 | 6.275345 | 6.273350 | 6.043409 | 6.088751 | 6.023745 |
| GO:0062009 | secondary palate development | 20 | -0.6332966 | -1.567693 | 0.039706436 | 1 | 1 | 801 | tags=30%, list=5%, signal=29% | TBX1||CHD7||TGFB3||COL11A2||LRRC32||TGFB2 | 2.187038 | 3.267017 | 2.727517 | 3.417801 | 2.089292 | 2.695690 | 3.285790 | 3.431951 | 2.151435 | 2.139060 | 2.267130 | 3.244855 | 3.302433 | 3.253088 | 2.882448 | 2.688847 | 2.596330 | 3.378844 | 3.381859 | 3.489901 | 2.037860 | 1.947958 | 2.263495 | 2.512447 | 2.582823 | 2.952133 | 3.276833 | 3.289162 | 3.291332 | 3.416809 | 3.442962 | 3.435957 |
| GO:0042551 | neuron maturation | 34 | 0.5317121 | 1.523654 | 0.039842382 | 1 | 1 | 2849 | tags=26%, list=18%, signal=22% | GLDN||NTN4||B4GALT6||C3||CLN5||BCL2||SPTBN4||ACTL6B||RB1 | 4.796422 | 5.018079 | 4.888009 | 5.029738 | 4.840709 | 5.063494 | 4.950421 | 4.984217 | 4.735018 | 4.739627 | 4.907815 | 5.042103 | 5.033901 | 4.977375 | 4.930240 | 4.884355 | 4.848259 | 5.059501 | 5.002292 | 5.026849 | 4.835892 | 4.748369 | 4.932013 | 5.029392 | 5.134361 | 5.024025 | 5.003500 | 4.923764 | 4.922497 | 4.961039 | 5.039037 | 4.950954 |
| GO:0090382 | phagosome maturation | 22 | 0.5926439 | 1.545950 | 0.041095890 | 1 | 1 | 3749 | tags=50%, list=23%, signal=38% | RAB38||SRPX||RAB43||RAB39A||MREG||RAB14||ENSG00000261832||PIKFYVE||ARL8B||RAB20||SPG11 | 3.848994 | 4.401109 | 4.156967 | 4.348051 | 3.865656 | 4.082045 | 4.412053 | 4.403773 | 3.810223 | 3.896581 | 3.838832 | 4.414829 | 4.435255 | 4.351942 | 4.271936 | 4.115112 | 4.076336 | 4.326275 | 4.323531 | 4.393260 | 3.865711 | 3.820239 | 3.909633 | 3.981130 | 3.961008 | 4.281208 | 4.417398 | 4.418229 | 4.400462 | 4.386833 | 4.459322 | 4.363423 |
| GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 14 | -0.6873768 | -1.568159 | 0.041371385 | 1 | 1 | 562 | tags=14%, list=4%, signal=14% | LEAP2||H2BC6 | 10.371731 | 9.862409 | 10.129814 | 9.810150 | 10.335840 | 10.039351 | 9.841175 | 9.714328 | 10.432173 | 10.425709 | 10.250011 | 9.915139 | 9.829821 | 9.840765 | 10.033233 | 10.090662 | 10.256161 | 9.841558 | 9.784222 | 9.804080 | 10.320164 | 10.483923 | 10.188174 | 10.192761 | 10.071978 | 9.830011 | 9.826680 | 9.852079 | 9.844648 | 9.816583 | 9.419271 | 9.866847 |
| GO:0042541 | hemoglobin biosynthetic process | 11 | 0.7009946 | 1.551448 | 0.041407867 | 1 | 1 | 2060 | tags=45%, list=13%, signal=40% | INHA||KLF4||HIF1A||SLC25A37||ABCB10 | 6.157395 | 6.077549 | 6.239225 | 6.100326 | 6.209660 | 6.407124 | 6.062616 | 6.144101 | 6.036463 | 6.132333 | 6.291785 | 6.146088 | 6.049195 | 6.034806 | 6.159510 | 6.336922 | 6.215504 | 6.138913 | 6.047216 | 6.113304 | 6.199009 | 6.082593 | 6.336190 | 6.350867 | 6.455191 | 6.413408 | 6.076854 | 6.043782 | 6.067013 | 6.128990 | 6.117536 | 6.184873 |
| GO:0006012 | galactose metabolic process | 10 | -0.7413816 | -1.568782 | 0.042047888 | 1 | 1 | 33 | tags=20%, list=0%, signal=20% | GALK1||PPARGC1A | 4.601123 | 4.638810 | 4.962626 | 4.639183 | 4.668680 | 4.940274 | 4.472523 | 4.482255 | 4.534675 | 4.550119 | 4.711820 | 4.654160 | 4.655913 | 4.605798 | 4.932858 | 5.029734 | 4.922853 | 4.645514 | 4.583228 | 4.686923 | 4.686734 | 4.541336 | 4.768840 | 4.717460 | 4.758415 | 5.275462 | 4.530479 | 4.437731 | 4.447548 | 4.537856 | 4.491727 | 4.414500 |
| GO:0048143 | astrocyte activation | 16 | 0.6364439 | 1.546641 | 0.042171190 | 1 | 1 | 1776 | tags=25%, list=11%, signal=22% | CNTF||C5AR1||TTBK1||NR1D1 | 4.433361 | 5.473583 | 5.033802 | 5.480401 | 4.465865 | 4.946278 | 5.280764 | 5.369367 | 4.332158 | 4.480803 | 4.482034 | 5.514488 | 5.539730 | 5.360054 | 5.033452 | 5.091999 | 4.973523 | 5.518119 | 5.488901 | 5.432885 | 4.475795 | 4.368303 | 4.547862 | 4.774102 | 4.787277 | 5.229431 | 5.343694 | 5.273303 | 5.222731 | 5.392789 | 5.428184 | 5.283197 |
| GO:0051602 | response to electrical stimulus | 28 | -0.5795667 | -1.538567 | 0.042875866 | 1 | 1 | 2244 | tags=39%, list=14%, signal=34% | DISC1||OPA1||PALM||SLC9A1||AKAP12||PTEN||CD14||AKAP9||HPCA||NTRK1||PPARGC1A | 8.582416 | 8.463599 | 9.393716 | 8.697277 | 8.721939 | 9.186392 | 8.613624 | 8.541907 | 8.440156 | 8.582227 | 8.712070 | 7.836918 | 8.679699 | 8.715466 | 9.335559 | 9.571512 | 9.255044 | 8.561124 | 8.969816 | 8.516334 | 8.786035 | 8.573729 | 8.795350 | 8.949207 | 8.921495 | 9.586212 | 8.580904 | 8.659760 | 8.599020 | 8.644500 | 8.461001 | 8.513989 |
| GO:0051983 | regulation of chromosome segregation | 81 | -0.4437201 | -1.420982 | 0.043280977 | 1 | 1 | 4098 | tags=41%, list=26%, signal=30% | KIF2C||IK||HNRNPU||FBXO5||GEN1||SPDL1||NSMCE2||ZWILCH||BUB1||CDC26||CUL3||CDK5RAP2||ANAPC1||RIOK2||KNTC1||CDC6||ZW10||SMC5||CDCA2||CDC16||APC||SMC6||TPR||MKI67||RAD18||WAPL||DLGAP5||HECW2||ANAPC4||CENPE||TTK||CDC27||CENPF | 5.616455 | 6.191428 | 5.733979 | 6.239446 | 5.540085 | 5.676551 | 6.254183 | 6.337216 | 5.594144 | 5.711312 | 5.538480 | 6.208240 | 6.178227 | 6.187653 | 5.956631 | 5.580703 | 5.635344 | 6.206067 | 6.198886 | 6.310650 | 5.527900 | 5.547007 | 5.545270 | 5.655439 | 5.683421 | 5.690553 | 6.253848 | 6.251357 | 6.257336 | 6.323893 | 6.363879 | 6.323502 |
| GO:0032892 | positive regulation of organic acid transport | 19 | 0.6028065 | 1.522987 | 0.045793397 | 1 | 1 | 1288 | tags=21%, list=8%, signal=19% | FABP3||CLTRN||ARL6IP1||PTGES | 2.695721 | 4.799709 | 3.323080 | 4.835255 | 2.700453 | 2.816093 | 4.749491 | 4.885729 | 2.662775 | 2.895598 | 2.501389 | 4.834321 | 4.822743 | 4.740253 | 3.744188 | 2.977028 | 3.129296 | 4.745909 | 4.867204 | 4.888587 | 2.560059 | 2.918138 | 2.595543 | 2.699069 | 2.672239 | 3.046067 | 4.734778 | 4.764060 | 4.749485 | 4.870122 | 4.936611 | 4.848997 |
| GO:0060999 | positive regulation of dendritic spine development | 41 | -0.5220161 | -1.493357 | 0.046553919 | 1 | 1 | 2232 | tags=22%, list=14%, signal=19% | DNM1L||ITSN1||PAFAH1B1||APOE||FOXO6||DHX36||NLGN1||CAPRIN2||BHLHB9 | 5.288196 | 5.791525 | 5.258798 | 5.740947 | 5.326114 | 5.475010 | 5.809049 | 5.845508 | 5.222789 | 5.348336 | 5.290729 | 5.788131 | 5.799110 | 5.787303 | 5.316739 | 5.274750 | 5.181609 | 5.707981 | 5.692158 | 5.819343 | 5.355654 | 5.213501 | 5.402538 | 5.517056 | 5.489546 | 5.416567 | 5.803247 | 5.804847 | 5.819002 | 5.805955 | 5.910329 | 5.817965 |
| GO:0031445 | regulation of heterochromatin assembly | 15 | -0.6721043 | -1.556055 | 0.046734342 | 1 | 1 | 2022 | tags=40%, list=13%, signal=35% | ATF7IP||MPHOSPH8||TPR||RESF1||SETDB2||ZNF304 | 6.104767 | 6.062530 | 6.233787 | 6.191771 | 6.159415 | 6.441855 | 6.123011 | 6.229200 | 5.974985 | 6.009775 | 6.306092 | 6.023659 | 6.105166 | 6.057601 | 6.227319 | 6.251363 | 6.222512 | 6.225084 | 6.141530 | 6.207362 | 6.136298 | 6.027321 | 6.301357 | 6.371393 | 6.523178 | 6.426888 | 6.157189 | 6.120080 | 6.091002 | 6.219178 | 6.239142 | 6.229211 |
| GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 17 | -0.6483944 | -1.547509 | 0.047020750 | 1 | 1 | 1487 | tags=29%, list=9%, signal=27% | AKAP6||ABCC8||AKAP9||LRRC26||NOS1AP | 4.454294 | 4.638083 | 4.421043 | 4.670839 | 4.368681 | 4.526067 | 4.587778 | 4.594187 | 4.473899 | 4.460921 | 4.427668 | 4.643778 | 4.653230 | 4.616997 | 4.461779 | 4.427781 | 4.372153 | 4.636212 | 4.719229 | 4.655764 | 4.407701 | 4.294003 | 4.401530 | 4.488576 | 4.516900 | 4.571490 | 4.603080 | 4.593882 | 4.566116 | 4.598420 | 4.628438 | 4.554752 |
| GO:0050901 | leukocyte tethering or rolling | 17 | 0.6191803 | 1.524742 | 0.047167825 | 1 | 1 | 2275 | tags=47%, list=14%, signal=40% | ITGA4||CHST2||FUT4||SELPLG||PTAFR||GOLPH3||MADCAM1||GCNT1 | 3.168380 | 3.892211 | 3.402067 | 3.866991 | 3.190157 | 3.475377 | 3.817888 | 3.780068 | 3.104987 | 3.151602 | 3.245011 | 3.898658 | 3.923560 | 3.853543 | 3.391467 | 3.467867 | 3.344158 | 3.811753 | 3.888965 | 3.898691 | 3.198832 | 3.045395 | 3.313743 | 3.348792 | 3.471478 | 3.595334 | 3.833285 | 3.818968 | 3.801232 | 3.801041 | 3.785681 | 3.753067 |
| GO:0002313 | mature B cell differentiation involved in immune response | 15 | -0.6713061 | -1.554207 | 0.047692013 | 1 | 1 | 1773 | tags=33%, list=11%, signal=30% | PTK2B||DOCK11||PLCG2||BCL3||MFNG | 3.843939 | 4.021886 | 3.977241 | 4.052598 | 3.784100 | 3.834860 | 3.987061 | 4.026186 | 3.817085 | 3.960604 | 3.745767 | 4.072530 | 4.053055 | 3.936347 | 4.075665 | 3.929598 | 3.921170 | 4.025442 | 4.078818 | 4.053040 | 3.838366 | 3.771788 | 3.740407 | 3.692530 | 3.670792 | 4.099626 | 4.024041 | 3.984257 | 3.951981 | 3.997417 | 4.108513 | 3.968824 |
| GO:0045910 | negative regulation of DNA recombination | 36 | -0.5377578 | -1.504424 | 0.048366490 | 1 | 1 | 3000 | tags=39%, list=19%, signal=32% | FOXP3||ZRANB3||TP53BP1||H1-10||SHLD2||MSH2||POLQ||MSH6||BLM||THOC1||MSH3||FANCB||H1-4||ANKLE1 | 4.683774 | 5.165161 | 4.867947 | 5.128843 | 4.622100 | 4.860593 | 5.169313 | 5.167346 | 4.684674 | 4.669948 | 4.696578 | 5.202055 | 5.140756 | 5.151931 | 4.983255 | 4.816019 | 4.797203 | 5.099420 | 5.096384 | 5.188800 | 4.631308 | 4.519880 | 4.708882 | 4.788627 | 4.823699 | 4.963465 | 5.184760 | 5.153930 | 5.169085 | 5.121769 | 5.260849 | 5.114661 |
| GO:0060969 | negative regulation of gene silencing | 22 | 0.5809143 | 1.515353 | 0.048587329 | 1 | 1 | 3 | tags=5%, list=0%, signal=5% | RMRP | 4.287492 | 5.038475 | 4.554407 | 4.945828 | 4.350161 | 4.694356 | 4.891639 | 6.201103 | 4.171335 | 4.445511 | 4.231005 | 5.162213 | 4.996612 | 4.947742 | 4.606749 | 4.562706 | 4.491425 | 5.012771 | 4.851184 | 4.968732 | 4.494712 | 4.181912 | 4.356912 | 4.781582 | 4.525265 | 4.762370 | 4.864872 | 4.923812 | 4.885612 | 4.919838 | 7.315288 | 4.962686 |
| GO:0034502 | protein localization to chromosome | 87 | -0.4319347 | -1.395385 | 0.048677990 | 1 | 1 | 4365 | tags=36%, list=27%, signal=26% | XRCC4||CCT8||CTCF||IK||TNKS2||TNKS||TINF2||CCT2||ATRX||SPDL1||CCT5||ZWILCH||ACD||CCT4||SETD2||KNTC1||EZH2||XRCC5||ZW10||SMC5||CCT6A||GNL3||CENPQ||ATR||MSH2||WAPL||ESCO2||TTK||TERT||MCM8||SLF1 | 5.969102 | 6.724512 | 6.224802 | 6.780559 | 5.842056 | 5.951419 | 6.770401 | 6.859688 | 5.990759 | 6.129738 | 5.763363 | 6.759612 | 6.702870 | 6.710394 | 6.422374 | 6.056574 | 6.170854 | 6.740873 | 6.743659 | 6.854222 | 5.842690 | 5.927066 | 5.751045 | 5.934711 | 5.843988 | 6.066818 | 6.766598 | 6.768213 | 6.776372 | 6.843356 | 6.879394 | 6.856081 |
| GO:0048814 | regulation of dendrite morphogenesis | 59 | -0.4751759 | -1.449806 | 0.049221407 | 1 | 1 | 3504 | tags=32%, list=22%, signal=25% | OPA1||CDKL5||ID1||PARP6||LZTS1||SS18L2||RAPGEF2||CUX2||TNIK||DNM1L||PAFAH1B1||SEMA4D||NEDD4||HECW2||DHX36||CHRNA3||CAPRIN2||PTPRD||BHLHB9 | 4.269563 | 4.797434 | 4.461055 | 4.764316 | 4.267072 | 4.442967 | 4.832017 | 4.805539 | 4.211893 | 4.285863 | 4.309155 | 4.774430 | 4.817949 | 4.799593 | 4.556560 | 4.420824 | 4.400750 | 4.730902 | 4.742288 | 4.818189 | 4.276311 | 4.182677 | 4.338004 | 4.341664 | 4.405510 | 4.571838 | 4.859318 | 4.800291 | 4.835830 | 4.785880 | 4.846690 | 4.783148 |
| GO:0051383 | kinetochore organization | 23 | -0.5962378 | -1.519540 | 0.049521666 | 1 | 1 | 2910 | tags=39%, list=18%, signal=32% | SMC2||SENP6||CENPN||KNTC1||NUF2||DLGAP5||CENPE||SMC4||CENPF | 5.265995 | 5.593190 | 5.419495 | 5.759069 | 5.291712 | 5.499246 | 5.665301 | 5.864575 | 5.198369 | 5.204856 | 5.386727 | 5.609066 | 5.577635 | 5.592697 | 5.610676 | 5.331438 | 5.295295 | 5.675063 | 5.739840 | 5.856417 | 5.226373 | 5.218208 | 5.421260 | 5.357683 | 5.622909 | 5.504956 | 5.672576 | 5.662791 | 5.660507 | 5.824036 | 5.899352 | 5.869342 |
| GO:1902808 | positive regulation of cell cycle G1/S phase transition | 40 | -0.5205981 | -1.482735 | 0.049990944 | 1 | 1 | 4027 | tags=40%, list=25%, signal=30% | CCND1||EGFR||CUL4A||ADAM17||ANXA1||CDK10||EZH2||CDC6||CDC73||ANKRD17||UBE2E2||PLRG1||TERT||PLCG2||GLI1||PLCB1 | 5.734904 | 5.976365 | 5.759741 | 5.933613 | 5.726478 | 5.825695 | 5.982069 | 5.945318 | 5.706578 | 5.750072 | 5.747650 | 6.002643 | 5.966245 | 5.959836 | 5.780248 | 5.749130 | 5.749624 | 5.940598 | 5.881466 | 5.977162 | 5.735224 | 5.678676 | 5.764226 | 5.788372 | 5.820785 | 5.866847 | 6.009006 | 5.961734 | 5.975055 | 5.933020 | 5.965105 | 5.937619 |
GO: MF
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005516 | calmodulin binding | 152 | -0.4476465 | -1.561215 | 0.003358522 | 0.4013186 | 0.3947929 | 1448 | tags=25%, list=9%, signal=23% | MYLK||SLC9A1||KCNQ4||CAMSAP3||CAMSAP2||CDK5RAP2||SCN5A||STRN||MYO5B||DDX5||MYO15A||CFAP221||AKAP12||MAP6||PNCK||IQGAP2||RGS2||INVS||PHKB||OBSCN||FAS||MYLK2||KCNN1||CACNA1C||ATP2B1||ASPM||IQCG||SLC8A3||MBP||MYH15||MAP2||MYO3A||CAMK4||KCNN3||PLCB1||SLC24A4||RYR1||SPATA17 | 4.747095 | 4.955480 | 4.840337 | 4.883896 | 4.734901 | 4.868611 | 4.959657 | 4.885179 | 4.719767 | 4.770825 | 4.750236 | 4.970425 | 4.945669 | 4.950225 | 4.976658 | 4.775600 | 4.758389 | 4.890315 | 4.849762 | 4.910940 | 4.729215 | 4.685183 | 4.788441 | 4.805676 | 4.886409 | 4.911638 | 4.983324 | 4.939953 | 4.955358 | 4.876566 | 4.919783 | 4.858499 |
| GO:0005524 | ATP binding | 1223 | -0.3006505 | -1.247260 | 0.003862547 | 0.4013186 | 0.3947929 | 4115 | tags=28%, list=26%, signal=23% | PTK2||MSH5||KIF2C||ADCY5||STK11||DDX47||DDX11||EPRS1||MYO6||MAP2K4||CHUK||ABCA7||GRK3||CLK2||HNRNPU||GAK||EGFR||TOP2A||PALS1||HELLS||MTPAP||PAPOLG||PIM3||MYO5C||NEK11||DHX32||ULK1||CAD||KARS1||KIF5C||PSMC2||MYH3||MYO9B||IARS1||MYLK4||IDE||FARSB||YTHDC2||NVL||DNAJA2||GMPS||CCT2||RECQL||PRKCB||PSMC6||BRAF||KIF9||ATRX||UBA3||MAP3K15||CLPX||KIF2A||ACACA||ACVR2A||HLTF||BMPR1A||DCAF1||KIF3B||AFG3L2||FES||MYO1E||DCLK2||HSPA5||CDKL5||CDK11B||CDC42BPB||DDX20||DGKQ||MTOR||CCT5||SWAP70||DHX15||LIG3||PKMYT1||RAD51||RARS2||PIM1||DNAJA1||PRKCG||KIF20B||YME1L1||ABCE1||SUCLA2||CHD3||DMPK||BUB1||AK8||FGFR4||MAP3K2||KATNAL1||RFC4||RIOK1||HSPA9||ATP10D||RPS6KA6||HSPA12B||KIF3A||ATP11B||ERCC6L2||SPEG||CSNK1A1||KIF4A||TNK2||CAMKV||OLA1||EIF2AK4||MAST1||RPS6KC1||EPHA10||TLK2||BRIP1||KIF17||LARS1||CDK10||MAP3K7||MYLK||SLC27A2||SMC2||HSPA14||SGK3||RPS6KA3||PANK3||NMRK1||NARS1||MARK1||CNNM2||ATP2A1||BAZ1B||ZRANB3||PNPLA8||DGKA||CDC42BPA||TTLL11||CCT4||THG1L||WNK3||RIOK2||TARS3||TRIO||WNK1||SMARCA1||DDX50||TOP2B||NEK10||TRNT1||QRSL1||MAPK13||KATNA1||AKT3||MYO5B||DDX5||TNIK||ABCB7||MYO15A||TYK2||MDN1||PNKP||ABCG4||DDX18||NSF||CACNA1B||DNAH6||KIFC2||MCM6||XRCC5||HSPD1||BMS1||ADK||ERCC6||ATP5F1D||NEK8||DDX21||PAPOLA||CDC6||CDKL1||ATP2C1||SMC5||CCT6A||AATK||GK5||FLT1||TARS1||ATP2A3||RAD54L||DDX19B||KIF5A||GPHN||RARS1||PNCK||VRK2||KIF16B||HACL1||ATAD5||HSPA4||GSK3A||MAGI1||STK17A||ACSS3||RAD51B||KIF26B||SMC6||HSPA4L||KALRN||KIF11||VWA8||ATR||AK3||MASTL||TSSK3||HELZ2||TAOK3||MAP3K6||PTK2B||HSPH1||YARS2||HSP90B1||CSNK1G1||ATAD2||DYRK1B||DDX1||WNK4||MAP3K8||RRM1||HSP90AA1||MKI67||BTAF1||KSR1||ACTA1||ROCK1||MSH2||POLQ||UBE2S||PLK3||UBA5||EPHB6||KIF13B||PRPF4B||NAV2||DNAH14||FANCM||ROCK2||IKBKE||TTLL3||DDX10||SMC3||DYRK4||ABCC11||MAP4K3||MYO9A||CHD6||DDX55||STARD9||SMARCA5||UBA6||SLFN5||OBSCN||CENPE||GK||TPK1||MSH6||MAB21L1||ATAD3B||MAP3K5||DHX36||CHD2||NUAK2||PRKD1||TTK||PLK4||ABCC8||DDX59||ERCC6L||DDR2||KIF15||BLM||MYLK2||AQR||SMC4||DICER1||AFG1L||CHD9||UBE2E2||PIK3CA||RAD50||DHX29||DDX52||KIF18A||MST1R||ALPK3||ABCB1||LIG4||RAD54B||CHD7||SLK||PCCA||TEC||KSR2||RFC1||DGKH||TK2||BMP2K||DDX46||CHD1||SMARCA2||SLC22A4||RSKR||JAK2||ATP2B1||MTREX||ATP8A1||ACVRL1||ULK4||MCM8||MSH3||HELQ||ABCA5||WRN||H1-4||FICD||PPIP5K2||FIGN||ABCC2||ALPK1||NTRK1||TTLL7||MYH15||KATNAL2||FN3K||CLCN4||ACVR1C||AK5||ACVR2B||MYO3A||MLH3||CAMK4||UBA7||ACSBG1||TRPM6||DNAH11||PIK3C3||RYR1||DNAH17 | 5.621334 | 5.918010 | 5.693265 | 5.913823 | 5.586657 | 5.665527 | 5.930838 | 5.948967 | 5.603199 | 5.673469 | 5.585834 | 5.936757 | 5.913115 | 5.903961 | 5.772357 | 5.643519 | 5.660492 | 5.900106 | 5.874303 | 5.965522 | 5.620920 | 5.557620 | 5.580717 | 5.662487 | 5.612029 | 5.720039 | 5.937437 | 5.926240 | 5.928814 | 5.934348 | 5.971707 | 5.940567 |
| GO:0034062 | 5'-3' RNA polymerase activity | 44 | 0.5947832 | 1.775745 | 0.005574136 | 0.4455021 | 0.4382580 | 556 | tags=9%, list=3%, signal=9% | RMRP||POLR2M||POLR2J2||PRIM1 | 6.441752 | 6.202221 | 6.445229 | 6.298622 | 6.481083 | 6.538085 | 6.172812 | 6.659760 | 6.415942 | 6.407526 | 6.499966 | 6.255975 | 6.191867 | 6.157070 | 6.367403 | 6.492235 | 6.472944 | 6.382469 | 6.205185 | 6.302762 | 6.470189 | 6.396598 | 6.571126 | 6.502665 | 6.593742 | 6.516165 | 6.195820 | 6.174956 | 6.147249 | 6.302244 | 7.172080 | 6.325426 |
| GO:0140658 | ATP-dependent chromatin remodeler activity | 32 | -0.6620929 | -1.795428 | 0.008305998 | 0.5076431 | 0.4993885 | 4305 | tags=66%, list=27%, signal=48% | SMARCA4||EP400||HELLS||ATRX||HLTF||CHD3||ERCC6L2||ZRANB3||SMARCA1||ERCC6||RAD54L||BTAF1||CHD6||SMARCA5||CHD2||ERCC6L||CHD9||RAD54B||CHD7||CHD1||SMARCA2 | 4.138762 | 4.811132 | 4.640808 | 4.980191 | 4.015192 | 4.480016 | 4.924076 | 5.004691 | 4.120413 | 4.166233 | 4.129229 | 4.733929 | 4.854859 | 4.841585 | 4.885815 | 4.505534 | 4.495849 | 4.883793 | 4.976484 | 5.074021 | 4.023054 | 3.991026 | 4.031184 | 4.403738 | 4.355940 | 4.661319 | 4.941748 | 4.934708 | 4.895340 | 4.989820 | 5.031353 | 4.992524 |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 1019 | -0.2981397 | -1.225297 | 0.011939070 | 0.5133450 | 0.5049978 | 3306 | tags=26%, list=21%, signal=22% | ZNF182||POU3F3||CEBPG||ZBTB7C||ZBTB26||KLF12||ZNF92||MKX||ZNF557||ZNF140||GATA5||ZNF696||SNAI1||E2F6||PRDM16||ZNF550||ZNF772||ZNF267||GLI2||NFKB2||ZNF302||ZNF444||NFX1||ZNF44||PKNOX1||SREBF1||ZNF83||EGR3||ZNF891||ZNF367||ZNF148||JUND||FOXP3||ZSCAN30||ZNF23||NFIL3||ZNF101||HIVEP1||NKX2-4||ZNF225||ADNP||BATF3||ENSG00000273046||CC2D1B||DMRTA2||MYOD1||ZNF440||FOXD1||MNT||ZNF345||NKX3-2||PURG||NEUROG2||ZKSCAN5||NHLH1||CUX2||HSF4||ZNF121||MESP1||ZNF417||ZNF789||NKX2-8||MNX1||SOX6||NFATC3||ZBTB16||NFIB||PBX4||KLF16||ZNF467||TFAP2C||NR6A1||EPAS1||PURB||ZNF391||ZNF580||ZFHX4||ZBTB46||ZNF33B||HIC1||SATB1||ZNF107||LBX2||TCF7L2||GSC||IRF7||ZNF473||ZNF787||SP9||ZBED5||NFXL1||ZNF837||MEF2C||ZNF616||ETV6||GBX1||NR5A1||LBX1||IRX3||ZNF394||ZBTB7A||HIVEP2||ZNF775||ZNF555||HLX||GLI4||ZFP90||GLIS2||HOXB3||LHX2||ZNF124||ZEB1||ERFL||NFATC1||GBX2||ZNF773||ATF6||ZNF254||ZNF426||FOXD4L5||ZNF223||NR2F6||POU3F1||ZNF846||ZNF414||ZNF184||HMX2||TBX1||VAX1||OTX2||ZNF287||STOX1||ZFP1||SKOR1||ZNF784||ZNF443||ZNF75A||ZNF674||LHX3||RLF||TAL1||EGR2||ZNF197||ZNF35||GLIS3||ZNF502||SOX18||ZNF565||CEBPB||RARA||ETV2||ZNF26||ZNF143||KLF11||ZNF320||ZNF701||ZBTB11||ZNF175||CDC5L||ZNF57||ZNF585A||FOXO6||ZNF549||ZNF146||ZNF708||POU6F1||ZNF222||ETV1||IRF9||ZNF778||ZNF416||ZNF654||PAX6||HOXD9||PITX3||ZNF266||ZNF136||ZNF470||ELK4||ZNF354A||IRX4||ZEB2||ZNF562||BBX||ZNF280D||ZIK1||PLAG1||ZNF675||FOXC2||ZNF45||GTF2IRD2B||ZNF521||HES7||TCF12||ZSCAN26||ZNF423||CLOCK||ZNF134||SP110||ZNF155||ZNF226||ZNF514||ZNF260||ZNF230||ZNF771||ZNF546||ESR2||ZNF615||ZNF543||HMX1||ZIC1||ZFP14||ZNF628||TLX3||ZNF438||ZNF780B||ZNF14||EGR1||ZNF160||YY2||ZNF586||RORB||HOXD3||ZNF551||ZNF117||UNCX||IRF1||ZNF594||ZNF304||ZNF836||ZNF799||NKX6-2||ZNF583||ZNF585B||KLF2||NR2C1||ZNF606||ZNF25||OLIG1||ZNF709||TBX19||ZNF816||GLI1||ZNF256||HOXC12||HES4||ZNF547||ZNF224||ATOH8||ZNF559-ZNF177||ZNF525||ZNF577||ATOH7||ZNF570||ZNF91||ZNF814||ZNF112||ELF3||ZNF461||ZNF17||ZNF782||ZNF829||ZNF724||SOX5||ZNF214||TSHZ2||ZNF284 | 3.684632 | 3.853136 | 3.861134 | 3.886372 | 3.695094 | 3.947585 | 3.881516 | 3.914947 | 3.621323 | 3.675383 | 3.754093 | 3.859268 | 3.855025 | 3.845079 | 3.898607 | 3.873231 | 3.810132 | 3.894299 | 3.831600 | 3.931457 | 3.709581 | 3.598650 | 3.771752 | 3.874228 | 3.937405 | 4.027026 | 3.897360 | 3.869110 | 3.877932 | 3.907879 | 3.926506 | 3.910385 |
| GO:0005227 | calcium activated cation channel activity | 12 | -0.7825204 | -1.721455 | 0.013487476 | 0.5133450 | 0.5049978 | 1670 | tags=42%, list=10%, signal=37% | KCNMA1||TMEM63C||KCNN1||CATSPER2||KCNN3 | 1.921651 | 2.695007 | 2.756199 | 2.768321 | 1.904949 | 2.513641 | 2.587247 | 2.717192 | 1.941313 | 1.944261 | 1.878427 | 2.652929 | 2.739399 | 2.691391 | 2.955484 | 2.708785 | 2.578646 | 2.756989 | 2.801144 | 2.746240 | 1.938408 | 1.811973 | 1.960076 | 2.265700 | 2.251449 | 2.918633 | 2.559543 | 2.637110 | 2.563761 | 2.720162 | 2.767868 | 2.661583 |
| GO:0016887 | ATP hydrolysis activity | 225 | -0.3849208 | -1.402446 | 0.014450398 | 0.5133450 | 0.5049978 | 3920 | tags=35%, list=24%, signal=27% | DHX32||KIF5C||MYH3||ACIN1||YTHDC2||NVL||RECQL||KIF9||ATRX||CLPX||KIF2A||KIF3B||MYO1E||HSPA5||DDX20||DHX15||KIF20B||CHD3||HSPA9||ATP10D||KIF3A||ATP11B||OLA1||BRIP1||KIF17||HSPA14||ATP6V1H||ATP2A1||DDX50||DDX5||RNF213||ABCG4||DDX18||NSF||KIFC2||MCM6||HSPD1||ERCC6||DDX21||ATP2C1||ATP2A3||RAD54L||DDX19B||KIF5A||KIF16B||VWA8||ATAD2||DDX1||HSP90AA1||KIF13B||NAV2||FANCM||DDX10||CHD6||DDX55||SMARCA5||DHX36||CHD2||DDX59||ERCC6L||KIF15||BLM||AQR||AFG1L||CHD9||DHX29||DDX52||KIF18A||CHD7||DDX46||CHD1||ATP2B1||MTREX||ATP8A1||MCM8||HELQ||WRN||MLH3 | 5.766938 | 6.481253 | 5.963415 | 6.456550 | 5.706497 | 5.770197 | 6.503083 | 6.496978 | 5.754469 | 5.905743 | 5.627087 | 6.503776 | 6.478329 | 6.461338 | 6.167238 | 5.822951 | 5.875722 | 6.410514 | 6.441641 | 6.515472 | 5.745987 | 5.716522 | 5.655511 | 5.724020 | 5.659101 | 5.915047 | 6.510847 | 6.501278 | 6.497090 | 6.481535 | 6.521457 | 6.487620 |
| GO:0003774 | cytoskeletal motor activity | 80 | -0.4870704 | -1.553758 | 0.015164874 | 0.5133450 | 0.5049978 | 4306 | tags=50%, list=27%, signal=37% | MYO10||KIF27||KIFC3||KIF2C||MYO6||MYO5C||KLC1||KIF5C||MYH3||MYO9B||APPBP2||KIF9||KIF2A||KIF3B||MYO1E||KIF20B||KIF3A||KIF4A||KIF17||MYO5B||MYO15A||DNAH6||KIFC2||MYL3||KIF5A||KIF16B||KIF26B||KIF11||KIF13B||DNAH14||SMC3||MYO9A||STARD9||CENPE||KIF15||KIF18A||MYH15||MYO3A||DNAH11||DNAH17 | 5.415319 | 4.925922 | 5.419896 | 5.167546 | 5.460880 | 5.464310 | 4.937580 | 5.194567 | 5.367478 | 5.396232 | 5.479871 | 4.985938 | 4.898631 | 4.891259 | 5.382621 | 5.441565 | 5.434783 | 5.177286 | 5.113267 | 5.210400 | 5.449144 | 5.447868 | 5.485314 | 5.477970 | 5.487508 | 5.426715 | 4.975800 | 4.931925 | 4.904107 | 5.176915 | 5.218533 | 5.187929 |
| GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity | 13 | 0.7219106 | 1.671910 | 0.017478152 | 0.5133450 | 0.5049978 | 711 | tags=23%, list=4%, signal=22% | PIK3R5||CISH||SOCS6 | 2.436292 | 2.967339 | 2.549375 | 3.091392 | 2.510318 | 2.852851 | 3.019378 | 3.081521 | 2.416151 | 2.402340 | 2.488880 | 2.949437 | 3.030517 | 2.919772 | 2.622891 | 2.554840 | 2.466121 | 3.046989 | 3.013387 | 3.206347 | 2.514966 | 2.368440 | 2.635222 | 2.851296 | 2.954656 | 2.744988 | 3.056872 | 2.988760 | 3.011668 | 3.108609 | 3.085014 | 3.050347 |
| GO:0070182 | DNA polymerase binding | 22 | 0.6417365 | 1.669690 | 0.017761609 | 0.5133450 | 0.5049978 | 3 | tags=5%, list=0%, signal=5% | RMRP | 6.853620 | 7.900299 | 6.928565 | 8.004284 | 6.757980 | 6.820738 | 7.968820 | 8.267172 | 6.856407 | 6.975147 | 6.717847 | 7.917643 | 7.872656 | 7.910195 | 7.321945 | 6.607160 | 6.754338 | 7.962609 | 7.963902 | 8.082999 | 6.736065 | 6.824068 | 6.711358 | 6.891002 | 6.844455 | 6.721484 | 7.942470 | 7.972746 | 7.990831 | 8.060511 | 8.606672 | 8.063174 |
| GO:0000400 | four-way junction DNA binding | 17 | -0.7061136 | -1.676647 | 0.018774891 | 0.5133450 | 0.5049978 | 1932 | tags=35%, list=12%, signal=31% | RAD51B||MSH2||FANCM||MSH6||BLM||WRN | 5.715366 | 6.951842 | 5.839354 | 6.921715 | 5.584281 | 5.608621 | 7.090857 | 6.985262 | 5.704955 | 5.849273 | 5.579191 | 6.906915 | 6.956316 | 6.991059 | 6.381839 | 5.509903 | 5.419850 | 6.852645 | 6.911539 | 6.997284 | 5.472982 | 5.697942 | 5.573093 | 5.444947 | 5.564074 | 5.794688 | 7.068251 | 7.083298 | 7.120520 | 7.020992 | 6.936030 | 6.997437 |
| GO:0008308 | voltage-gated anion channel activity | 13 | -0.7485326 | -1.672532 | 0.019822941 | 0.5281035 | 0.5195162 | 188 | tags=46%, list=1%, signal=46% | CLCN3||VDAC2||CLCN2||GPR89A||CLCN4||CLCNKB | 5.601355 | 6.282037 | 5.663506 | 6.427706 | 5.389773 | 5.243107 | 6.311515 | 6.456295 | 5.677125 | 5.840969 | 5.215642 | 6.282452 | 6.274024 | 6.289592 | 6.017237 | 5.181498 | 5.672364 | 6.371830 | 6.413300 | 6.495240 | 5.348517 | 5.574109 | 5.224723 | 5.320985 | 5.264655 | 5.137614 | 6.268426 | 6.318869 | 6.346175 | 6.421578 | 6.473759 | 6.472930 |
| GO:0005355 | glucose transmembrane transporter activity | 11 | 0.7501854 | 1.671326 | 0.021302999 | 0.5398492 | 0.5310709 | 2096 | tags=55%, list=13%, signal=47% | MFSD4A||MFSD4B||SLC2A4||SLC5A3||SLC2A3||SLC2A10 | 3.197576 | 3.561758 | 3.835014 | 3.770803 | 3.300661 | 3.817801 | 3.405161 | 3.600768 | 3.083627 | 3.137364 | 3.356973 | 3.591809 | 3.629227 | 3.458728 | 3.806711 | 3.968098 | 3.719089 | 3.786996 | 3.772295 | 3.752916 | 3.311514 | 3.072844 | 3.487823 | 3.590672 | 3.640958 | 4.152404 | 3.486791 | 3.412409 | 3.310898 | 3.563697 | 3.703408 | 3.529243 |
| GO:0051393 | alpha-actinin binding | 16 | -0.7078352 | -1.658283 | 0.022627876 | 0.5597706 | 0.5506684 | 2983 | tags=44%, list=19%, signal=36% | CACNA1D||ADORA2A||MAGI1||PDLIM3||RARA||CACNA1C||PDLIM4 | 4.237835 | 4.169846 | 4.220394 | 3.977593 | 4.238727 | 4.367988 | 4.091302 | 3.943981 | 4.151133 | 4.226208 | 4.330522 | 4.178250 | 4.183672 | 4.147352 | 4.174751 | 4.309140 | 4.173021 | 4.004023 | 3.977388 | 3.950879 | 4.281188 | 4.095492 | 4.329098 | 4.339769 | 4.330085 | 4.431906 | 4.125007 | 4.085232 | 4.062980 | 3.955578 | 3.976636 | 3.898602 |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 15 | -0.7182948 | -1.661139 | 0.023637057 | 0.5711373 | 0.5618502 | 2596 | tags=53%, list=16%, signal=45% | SETD1B||KMT2D||SETD1A||KMT2A||SETMAR||KMT2B||KMT2C||ASH1L | 4.487751 | 5.009249 | 5.133934 | 5.093312 | 4.419544 | 4.948827 | 5.101024 | 5.165419 | 4.401256 | 4.586923 | 4.468921 | 4.954482 | 5.044919 | 5.026767 | 5.274379 | 5.100799 | 5.014362 | 5.063141 | 5.081492 | 5.134352 | 4.463160 | 4.333272 | 4.458476 | 4.785719 | 4.774218 | 5.236765 | 5.120711 | 5.119919 | 5.061652 | 5.168460 | 5.171847 | 5.155900 |
| GO:0019003 | GDP binding | 62 | 0.4805459 | 1.526371 | 0.024544022 | 0.5795736 | 0.5701494 | 4246 | tags=45%, list=26%, signal=33% | KRAS||NRAS||RAB10||RAP2A||RAP2C||RASEF||GEM||RAB9A||RAB21||ARL3||RAP1B||RAB14||RAB18||RAP2B||RAB5C||RHOB||RHEB||RAB2A||ARL8B||RAB28||RAB5B||DIRAS1||RAP1A||RIT1||GNAI3||NME2||ARF1||RAB8B | 6.580800 | 6.578569 | 6.514481 | 6.569068 | 6.528452 | 6.335800 | 6.571506 | 6.617578 | 6.636917 | 6.686923 | 6.402926 | 6.620706 | 6.553442 | 6.560608 | 6.478216 | 6.454546 | 6.606033 | 6.594774 | 6.499640 | 6.610319 | 6.562516 | 6.640385 | 6.369108 | 6.495456 | 6.184204 | 6.310659 | 6.559321 | 6.570297 | 6.584788 | 6.606261 | 6.612794 | 6.633538 |
| GO:0043138 | 3'-5' DNA helicase activity | 15 | -0.7148983 | -1.653284 | 0.025543271 | 0.5860972 | 0.5765669 | 2315 | tags=40%, list=14%, signal=34% | MCM6||NAV2||FANCM||BLM||HELQ||WRN | 5.564711 | 5.972845 | 5.881028 | 6.163702 | 5.510709 | 5.688522 | 6.005312 | 6.229604 | 5.489824 | 5.610677 | 5.590746 | 5.977101 | 5.982614 | 5.958712 | 6.041268 | 5.788035 | 5.799342 | 6.153280 | 6.128277 | 6.208381 | 5.514852 | 5.444064 | 5.570437 | 5.554034 | 5.587674 | 5.898314 | 6.038214 | 5.986924 | 5.990226 | 6.216724 | 6.246084 | 6.225849 |
| GO:0042162 | telomeric DNA binding | 36 | -0.5681693 | -1.580314 | 0.026171303 | 0.5860972 | 0.5765669 | 3245 | tags=36%, list=20%, signal=29% | ACD||SMG6||TP53BP1||NCL||UPF3A||XRCC5||CTC1||HMBOX1||BLM||UPF2||RAD50||TERT||WRN | 6.956435 | 7.826831 | 7.038201 | 7.817559 | 6.849239 | 7.034672 | 7.898482 | 7.845281 | 6.929669 | 7.118141 | 6.804066 | 7.806137 | 7.810109 | 7.863534 | 7.414556 | 6.739476 | 6.868409 | 7.774711 | 7.794627 | 7.881109 | 6.828122 | 6.896706 | 6.821683 | 7.061675 | 7.064985 | 6.975587 | 7.873817 | 7.892651 | 7.928443 | 7.830090 | 7.861268 | 7.844316 |
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 910 | -0.2928262 | -1.196594 | 0.027406789 | 0.5932428 | 0.5835963 | 3306 | tags=26%, list=21%, signal=22% | ZNF182||POU3F3||CEBPG||ZBTB7C||ZBTB26||KLF12||ZNF92||MKX||ZNF140||GATA5||SNAI1||E2F6||PRDM16||ZNF772||KDM6B||ZNF267||GLI2||NFKB2||ZNF302||ZNF444||PKNOX1||SREBF1||ZNF83||EGR3||ZNF367||ZNF148||JUND||FOXP3||ZSCAN30||ZNF23||NFIL3||ZNF707||ZNF296||HIVEP1||NKX2-4||ZNF225||BATF3||ENSG00000273046||USP3||CC2D1B||DMRTA2||MYOD1||FOXD1||MNT||ZNF345||NKX3-2||NEUROG2||ZKSCAN5||NHLH1||CUX2||HSF4||ZNF121||MESP1||ZNF417||ZNF789||NKX2-8||SOX6||NFATC3||ZBTB16||NFIB||PBX4||KLF16||ZNF467||TFAP2C||NR6A1||EPAS1||ZNF580||ZFHX4||ZNF33B||HIC1||SATB1||ZNF107||TCF7L2||EZH2||SLC2A4RG||GSC||IRF7||ZNF473||SP9||ZNF837||MEF2C||ZNF616||ETV6||NR5A1||IRX3||ZNF394||ZBTB7A||HIVEP2||ZNF775||GLI4||GLIS2||HOXB3||LHX2||ZEB1||NFATC1||ZNF773||ATF6||ZNF254||ZNF426||FOXD4L5||ZNF223||NR2F6||POU3F1||ZNF184||TBX1||VAX1||OTX2||ZNF287||ZFP1||SKOR1||ZNF784||TRIM24||LHX3||TAL1||EGR2||ZNF197||GLIS3||ZNF502||SOX18||ZNF565||CEBPB||RARA||ETV2||ZNF26||ZNF143||KLF11||ZNF564||ZNF320||BPTF||ZNF701||ZBTB11||ZNF175||ZNF585A||FOXO6||ZNF549||ZNF146||NRIP1||ZNF708||POU6F1||ZNF222||ETV1||IRF9||ZNF613||ZNF416||CCAR1||PAX6||HOXD9||PITX3||ZNF136||ZNF470||MUC1||DHX36||CHD2||ELK4||ZNF354A||IRX4||ZEB2||ZNF562||ZNF280D||ZIK1||PLAG1||ZNF675||FOXC2||ZNF45||ZNF521||HES7||TCF12||ZSCAN26||ZNF423||CLOCK||ZNF134||ZNF155||ZNF226||ZNF514||ZNF260||ZNF230||ZNF771||CHD7||ZNF704||ZNF546||ESR2||ZNF615||ZNF543||ZIC1||ZFP14||ZNF628||TLX3||ZNF438||ZNF780B||EGR1||ZNF160||ZNF567||YY2||ZNF586||RORB||HOXD3||ZNF551||ZNF117||IRF1||ZNF594||ZNF304||ZNF836||NKX6-2||ZNF583||ZNF585B||KLF2||NR2C1||ZNF606||OLIG1||TBX19||ZNF816||GLI1||ZNF256||HES4||ZNF547||ZNF224||ATOH8||ZNF525||ZNF577||ATOH7||ZNF570||ZNF91||ZNF814||ZNF112||ELF3||ZNF461||ZNF17||ZNF782||ZNF829||ZNF724||SOX5||IKZF1||ZNF214||ZNF284 | 3.881732 | 4.313721 | 4.131958 | 4.340501 | 3.843948 | 4.063664 | 4.353367 | 4.366539 | 3.857593 | 3.966056 | 3.817420 | 4.309472 | 4.316368 | 4.315313 | 4.268328 | 4.035825 | 4.081048 | 4.325910 | 4.303696 | 4.390483 | 3.819524 | 3.842931 | 3.868967 | 3.988910 | 4.027876 | 4.167996 | 4.355701 | 4.347846 | 4.356538 | 4.380044 | 4.336520 | 4.382590 |
| GO:0005518 | collagen binding | 44 | -0.5342623 | -1.542332 | 0.030813848 | 0.6210981 | 0.6109987 | 2545 | tags=25%, list=16%, signal=21% | COCH||ITGA3||CD44||ITGA1||DDR2||LOX||TNXB||ITGA10||CTSK||CCBE1||ADGRG6 | 4.877180 | 5.561416 | 5.120605 | 5.284837 | 4.878563 | 4.980652 | 5.381587 | 5.165234 | 4.891964 | 4.938340 | 4.797696 | 5.590402 | 5.604682 | 5.486292 | 5.052053 | 5.179323 | 5.127606 | 5.264661 | 5.326692 | 5.262227 | 4.912116 | 4.913850 | 4.807156 | 4.987431 | 4.752275 | 5.171831 | 5.423473 | 5.377044 | 5.343114 | 5.163526 | 5.213461 | 5.117106 |
| GO:0005245 | voltage-gated calcium channel activity | 26 | -0.6098424 | -1.577977 | 0.032866480 | 0.6443070 | 0.6338302 | 3793 | tags=62%, list=24%, signal=47% | TPCN2||CACNG8||TPCN1||CACHD1||CACNA2D1||CACNA1D||CACNG4||CACNB2||CACNB4||CACNA1B||CACNA2D2||CACNG5||CACNG6||CACNA1C||CATSPER3||RYR1 | 2.567080 | 2.714660 | 2.964097 | 2.741886 | 2.603872 | 2.971730 | 2.655835 | 2.638114 | 2.479940 | 2.522816 | 2.689857 | 2.740106 | 2.760117 | 2.640957 | 2.958846 | 3.027371 | 2.903396 | 2.734838 | 2.747090 | 2.743703 | 2.668243 | 2.403848 | 2.720017 | 2.817404 | 2.891990 | 3.179897 | 2.715090 | 2.635284 | 2.615186 | 2.603404 | 2.687957 | 2.621602 |
| GO:0008191 | metalloendopeptidase inhibitor activity | 11 | -0.7319092 | -1.577215 | 0.036191213 | 0.6848248 | 0.6736891 | 2062 | tags=45%, list=13%, signal=40% | RECK||SPOCK2||LXN||SPOCK3||COL4A3 | 3.982341 | 3.358634 | 4.096355 | 2.894167 | 4.035197 | 4.200806 | 3.182792 | 2.810184 | 4.016783 | 3.799898 | 4.112784 | 3.315214 | 3.441265 | 3.315734 | 3.911269 | 4.222283 | 4.137869 | 2.866176 | 2.943198 | 2.871842 | 4.010944 | 3.997256 | 4.095419 | 4.018628 | 4.162508 | 4.395933 | 3.211975 | 3.158531 | 3.177361 | 2.766990 | 2.947174 | 2.705312 |
| GO:0004722 | protein serine/threonine phosphatase activity | 82 | 0.4291008 | 1.428417 | 0.036262204 | 0.6848248 | 0.6736891 | 1456 | tags=13%, list=9%, signal=12% | LRRC39||DUSP19||PPP1CB||LRRK1||DUSP26||PDP2||DUSP10||PPM1K||CTDSPL2||PPM1B||MTMR6 | 5.054175 | 5.316418 | 5.058123 | 5.287918 | 5.061920 | 5.170255 | 5.326364 | 5.321694 | 5.022564 | 5.047689 | 5.091429 | 5.344401 | 5.303682 | 5.300757 | 5.051337 | 5.087128 | 5.035416 | 5.293358 | 5.244350 | 5.324906 | 5.061139 | 4.985561 | 5.135184 | 5.144318 | 5.180429 | 5.185667 | 5.327434 | 5.321321 | 5.330323 | 5.302671 | 5.340079 | 5.322091 |
| GO:0005509 | calcium ion binding | 393 | -0.3272354 | -1.258684 | 0.036910670 | 0.6848248 | 0.6736891 | 2839 | tags=20%, list=18%, signal=17% | ZZEF1||LRP2||RAPGEF2||SYT17||ATP2A1||ENPP2||DGKA||ACER3||COLEC11||CAPN2||ADGRV1||EML1||PLCB4||CALML4||MCTP1||CDH23||TNNC1||CACNA1B||FBN3||LRP4||PLCG1||HEG1||PLA2G4A||PLCH1||NUCB2||C2CD5||ITSN1||USP32||MYL3||NOTCH4||ITPR1||SCUBE1||SPOCK2||LRP1||HSP90B1||CELSR1||FAT3||SYT6||AOC3||CDH22||LTBP3||CDH26||CELSR3||LPCAT2||TBC1D8B||PRRG2||EFCAB12||CDH12||EFEMP1||EFHC1||S100A16||MEGF6||RAB11FIP4||ADAMTS13||RASGRP1||HSPG2||PLA2G4C||ITPR2||HMCN1||EFCAB10||NPNT||RASGRP2||C1S||SPOCK3||ITSN2||PCDH18||LPL||HPCA||H1-4||EFEMP2||CCBE1||PCDHGB7||S100A1||BGLAP||PLA2G4B||PLCB1||RYR1||ASAH2||CAPN3 | 4.826626 | 5.448337 | 4.924838 | 5.324575 | 4.827636 | 4.928643 | 5.378546 | 5.320560 | 4.817222 | 4.846572 | 4.815874 | 5.461110 | 5.470946 | 5.412273 | 4.968374 | 4.913683 | 4.891364 | 5.303859 | 5.321583 | 5.347943 | 4.807076 | 4.832985 | 4.842613 | 4.891935 | 4.905777 | 4.986403 | 5.384180 | 5.380654 | 5.370770 | 5.295700 | 5.364034 | 5.300937 |
| GO:0008201 | heparin binding | 80 | -0.4475756 | -1.427769 | 0.040380885 | 0.7233748 | 0.7116123 | 3889 | tags=35%, list=24%, signal=27% | NRP1||THBS3||THBS4||LTBP2||NELL2||PTCH1||FGFR4||CRISPLD2||RSPO2||PTPRS||NRP2||ADAMTS3||CEL||LXN||PAFAH1B1||COLQ||NAV2||APOE||ZNF146||ADAMTSL5||EVA1C||TNXB||CCN2||ADAMTS5||NDNF||LPL||EFEMP2||CCDC80 | 6.530156 | 6.385857 | 6.334810 | 6.373744 | 6.492387 | 6.230217 | 6.341004 | 6.381575 | 6.550816 | 6.570741 | 6.466816 | 6.423729 | 6.371869 | 6.361196 | 6.355883 | 6.292132 | 6.355486 | 6.381123 | 6.350553 | 6.389268 | 6.545843 | 6.568430 | 6.353356 | 6.434650 | 6.137513 | 6.094161 | 6.340711 | 6.354594 | 6.327581 | 6.388112 | 6.380797 | 6.375789 |
| GO:0005125 | cytokine activity | 67 | 0.4429259 | 1.427843 | 0.043448590 | 0.7367779 | 0.7247975 | 2577 | tags=27%, list=16%, signal=23% | CNTF||WNT2B||INHBC||CCL5||TNFSF4||C5||INHBB||WNT9A||INHA||VEGFA||GDF9||WNT4||BMP8A||IL23A||INHBE||CKLF||CMTM8||GDF15 | 6.618196 | 6.112163 | 6.241935 | 5.956446 | 6.692266 | 6.789868 | 6.004907 | 5.852701 | 6.588811 | 6.399644 | 6.833234 | 6.233083 | 6.037224 | 6.058055 | 5.991760 | 6.417572 | 6.284218 | 6.105315 | 5.865959 | 5.885623 | 6.699286 | 6.459355 | 6.886613 | 6.684779 | 7.027580 | 6.623577 | 6.010668 | 5.999064 | 6.004966 | 5.882480 | 5.836700 | 5.838456 |
| GO:0030020 | extracellular matrix structural constituent conferring tensile strength | 28 | -0.5780890 | -1.519767 | 0.044205052 | 0.7367779 | 0.7247975 | 1031 | tags=32%, list=6%, signal=30% | COL23A1||COL6A5||COL11A2||COL16A1||COL4A4||COL3A1||COL7A1||COL4A3||COL12A1 | 2.468383 | 2.829255 | 3.153573 | 2.609938 | 2.461283 | 3.028681 | 2.705913 | 2.512613 | 2.388703 | 2.394245 | 2.610894 | 2.782412 | 2.931858 | 2.767711 | 3.101732 | 3.317978 | 3.024773 | 2.691603 | 2.585216 | 2.549167 | 2.428696 | 2.264275 | 2.662889 | 2.711981 | 2.876868 | 3.403833 | 2.756762 | 2.697272 | 2.662115 | 2.508544 | 2.627163 | 2.392599 |
| GO:0042577 | lipid phosphatase activity | 12 | 0.6856551 | 1.559025 | 0.044887781 | 0.7367779 | 0.7247975 | 425 | tags=25%, list=3%, signal=24% | PLPP6||SGPP1||PLPP7 | 2.528486 | 3.182254 | 2.818890 | 3.169308 | 2.581931 | 2.883676 | 3.101278 | 3.123119 | 2.468105 | 2.551848 | 2.563637 | 3.139211 | 3.214241 | 3.192281 | 2.802683 | 2.894237 | 2.756320 | 3.150965 | 3.180372 | 3.176413 | 2.652539 | 2.458839 | 2.626846 | 2.765751 | 2.819571 | 3.049684 | 3.113961 | 3.118918 | 3.070464 | 3.121677 | 3.124353 | 3.123325 |
| GO:0004197 | cysteine-type endopeptidase activity | 75 | -0.4460598 | -1.408284 | 0.046232575 | 0.7367779 | 0.7247975 | 3855 | tags=36%, list=24%, signal=27% | CASP8||USP36||USP28||CAPN15||USP34||USP10||CTSZ||SENP6||SENP1||CAPN2||USP12||USP33||USP7||CASP6||USP1||USP9X||USP47||USP2||USP24||USP49||USP25||USP16||CAPN7||USP48||CTSK||USP8||CAPN3 | 5.129985 | 5.721290 | 5.308326 | 5.642894 | 5.101098 | 5.322380 | 5.671699 | 5.643129 | 5.111396 | 5.169422 | 5.108310 | 5.723192 | 5.748225 | 5.691903 | 5.373895 | 5.285680 | 5.263008 | 5.618687 | 5.632034 | 5.677305 | 5.115968 | 5.066866 | 5.119857 | 5.259590 | 5.298515 | 5.405075 | 5.665435 | 5.673468 | 5.676173 | 5.631568 | 5.672450 | 5.624908 |
| GO:0051880 | G-quadruplex DNA binding | 10 | -0.7305039 | -1.534845 | 0.047122441 | 0.7367779 | 0.7247975 | 1125 | tags=40%, list=7%, signal=37% | DHX36||BLM||RAD50||WRN | 8.190800 | 7.935968 | 8.100158 | 7.916746 | 8.144063 | 7.755853 | 7.916091 | 7.953067 | 8.279860 | 8.260441 | 8.017590 | 7.966630 | 7.892919 | 7.947345 | 8.023835 | 8.066440 | 8.203995 | 7.962902 | 7.850277 | 7.934694 | 8.212162 | 8.320117 | 7.861055 | 8.018576 | 7.385630 | 7.793721 | 7.926074 | 7.883601 | 7.938032 | 7.933711 | 7.956088 | 7.969180 |
| GO:0030291 | protein serine/threonine kinase inhibitor activity | 32 | 0.5233844 | 1.472750 | 0.049727372 | 0.7367779 | 0.7247975 | 4432 | tags=62%, list=28%, signal=45% | SPRED1||INKA1||PKIB||PKIA||PRKAR1A||CDKN1A||CDKN1C||PRKAR2A||MACROH2A1||HEXIM2||PRKAR2B||CDKN1B||HEXIM1||SPRY2||INKA2||PRKAR1B||PRKAG2||CDKN2B||CDKN2C||INCA1 | 5.326243 | 5.263788 | 5.297892 | 5.130422 | 5.304605 | 5.431623 | 5.236943 | 5.075909 | 5.322816 | 5.303128 | 5.352359 | 5.314421 | 5.246810 | 5.228711 | 5.278306 | 5.312032 | 5.303127 | 5.181894 | 5.055364 | 5.151011 | 5.324230 | 5.200128 | 5.383437 | 5.376011 | 5.469374 | 5.447835 | 5.247841 | 5.208392 | 5.254170 | 5.068967 | 5.110438 | 5.047614 |
| GO:0017002 | activin-activated receptor activity | 10 | -0.7263979 | -1.526218 | 0.049826188 | 0.7367779 | 0.7247975 | 564 | tags=30%, list=4%, signal=29% | ACVRL1||ACVR1C||ACVR2B | 1.125785 | 2.956182 | 2.041774 | 2.974482 | 1.108387 | 1.889486 | 2.927555 | 2.981152 | 1.014281 | 1.277302 | 1.072351 | 2.920522 | 2.996925 | 2.950068 | 2.337884 | 1.969532 | 1.757248 | 2.925340 | 3.007399 | 2.989423 | 1.053910 | 1.086241 | 1.181921 | 1.769484 | 1.599796 | 2.224896 | 2.909851 | 2.988469 | 2.882227 | 2.920461 | 3.098482 | 2.916942 |
GO: CC
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0000172 | ribonuclease MRP complex | 10 | 0.8759736 | 1.883243 | 0.0007979671 | 0.4104909 | 0.4006917 | 3 | tags=10%, list=0%, signal=10% | RMRP | 5.943331 | 5.958048 | 5.746393 | 5.766090 | 5.992286 | 5.884799 | 5.745444 | 7.181343 | 5.9031874 | 5.952808 | 5.973104 | 6.181873 | 5.851146 | 5.811796 | 5.714552 | 5.725124 | 5.798062 | 5.934940 | 5.591920 | 5.750951 | 6.003603 | 5.9239234 | 6.046656 | 5.943665 | 5.961683 | 5.738581 | 5.779826 | 5.749484 | 5.706071 | 5.705603 | 8.369904 | 5.712209 |
| GO:0098644 | complex of collagen trimers | 15 | -0.7871802 | -1.840067 | 0.0011049553 | 0.4104909 | 0.4006917 | 389 | tags=40%, list=2%, signal=39% | COL11A2||COL16A1||COL4A4||COL3A1||COL7A1||COL4A3 | 2.238087 | 2.406861 | 2.856092 | 2.254233 | 2.205097 | 2.703671 | 2.365542 | 2.159532 | 2.1108297 | 2.231763 | 2.360830 | 2.303724 | 2.538444 | 2.368112 | 2.919428 | 2.937652 | 2.699207 | 2.276373 | 2.272500 | 2.212953 | 2.143484 | 2.0446362 | 2.403049 | 2.439842 | 2.571191 | 3.031200 | 2.396957 | 2.383064 | 2.315288 | 2.105698 | 2.294069 | 2.068556 |
| GO:0042405 | nuclear inclusion body | 12 | -0.7943470 | -1.759375 | 0.0096043027 | 0.5674104 | 0.5538652 | 1735 | tags=42%, list=11%, signal=37% | TPR||RAD18||ATXN3||RANBP2||SLF1 | 5.904212 | 6.038142 | 6.053321 | 6.046952 | 5.933783 | 6.107478 | 6.096334 | 6.093583 | 5.8910711 | 5.799111 | 6.014344 | 6.024914 | 6.036025 | 6.053345 | 6.185325 | 6.011401 | 5.952998 | 6.033794 | 5.999271 | 6.105737 | 5.887344 | 5.8538081 | 6.052277 | 6.007576 | 6.141102 | 6.168665 | 6.125637 | 6.094471 | 6.068323 | 6.074598 | 6.140563 | 6.064397 |
| GO:0032809 | neuronal cell body membrane | 15 | -0.7218830 | -1.687432 | 0.0165273556 | 0.5674104 | 0.5538652 | 664 | tags=33%, list=4%, signal=32% | FLRT1||ATP2B1||HPCA||KCNA2||SLC4A8 | 1.066423 | 2.602139 | 1.938149 | 2.623805 | 1.079581 | 1.866128 | 2.548336 | 2.471245 | 0.8893178 | 1.094109 | 1.198866 | 2.508999 | 2.731169 | 2.556652 | 1.988654 | 2.057337 | 1.750850 | 2.549520 | 2.704501 | 2.613174 | 1.164831 | 0.8078774 | 1.231164 | 1.498034 | 1.680411 | 2.294788 | 2.587297 | 2.556686 | 2.499657 | 2.505730 | 2.469498 | 2.437705 |
| GO:0033017 | sarcoplasmic reticulum membrane | 26 | -0.6273552 | -1.639844 | 0.0204848713 | 0.5674104 | 0.5538652 | 658 | tags=19%, list=4%, signal=18% | ITPR2||JSRP1||NOS1AP||RYR1||KLHL41 | 5.230838 | 5.440056 | 5.480456 | 5.510080 | 5.221127 | 5.374881 | 5.420564 | 5.523051 | 5.2263309 | 5.321031 | 5.139430 | 5.453104 | 5.457422 | 5.409149 | 5.438894 | 5.515491 | 5.485951 | 5.506431 | 5.494988 | 5.528620 | 5.262381 | 5.1887263 | 5.211283 | 5.312966 | 5.182253 | 5.597682 | 5.437570 | 5.424044 | 5.399824 | 5.499820 | 5.552246 | 5.516589 |
| GO:0000775 | chromosome, centromeric region | 191 | -0.3891934 | -1.392885 | 0.0207774799 | 0.5674104 | 0.5538652 | 4223 | tags=39%, list=26%, signal=29% | NGDN||CTCF||NUP160||CLASP2||KIF2C||HNRNPU||TOP2A||HELLS||XPO1||PPP2CA||NCAPD2||REC8||GPATCH11||ATRX||SKA2||STAG1||SPDL1||CENPU||SUV39H2||SEPTIN7||ZWILCH||KAT7||SUGT1||ORC2||BUB1||PPP2CB||SNAI1||CSNK1A1||ZNF276||CBX3||BAZ1B||PDS5A||NUP133||CLASP1||TP53BP1||CENPN||SIN3A||PPP2R5A||KNTC1||ZW10||CKAP5||SMC5||CENPQ||HJURP||NUP107||APC||SMC6||NUF2||SGO2||PDS5B||PAFAH1B1||TPR||SPAG5||BOD1L1||WAPL||CEBPB||SMC3||UHRF2||KMT5B||KAT2B||NCAPG||CENPE||ESCO2||CLIP1||TTK||ERCC6L||SMC4||MIS18BP1||KIF18A||CENPF||DYNLT3||SYCP2L||AHCTF1||STAG3||IKZF1 | 5.366433 | 6.096000 | 5.559790 | 6.143111 | 5.312483 | 5.471341 | 6.158274 | 6.222578 | 5.3407100 | 5.450384 | 5.304160 | 6.100004 | 6.089282 | 6.098690 | 5.763534 | 5.423733 | 5.467932 | 6.104046 | 6.109466 | 6.213179 | 5.288502 | 5.3326340 | 5.315970 | 5.431356 | 5.459191 | 5.521977 | 6.153189 | 6.156611 | 6.164997 | 6.205565 | 6.243418 | 6.218493 |
| GO:0005813 | centrosome | 567 | -0.3150158 | -1.249429 | 0.0212157689 | 0.5674104 | 0.5538652 | 3652 | tags=27%, list=23%, signal=22% | KIF2A||PIBF1||CKAP2||TUBGCP6||RBBP6||KIF3B||PHF1||DENND1C||CENPU||IQCB1||HAUS5||CEP57||PCGF5||CDKL5||HAUS6||ID1||CCT5||IL4R||B9D2||CEP350||KIF20B||ORC2||CTDP1||CEP55||ZFYVE26||PCNT||CHD3||KAT2A||KATNAL1||KIF3A||MDM1||ERCC6L2||CSNK1A1||CEP41||CNTROB||OLA1||CUL3||ERC1||TXNDC9||CAMSAP3||CEP162||IFT81||HAUS8||CAMSAP2||CLASP1||IFT88||CDK5RAP2||CCT4||CNTLN||CEP170||CEP135||KATNA1||LRRC45||PPP4R3B||CEP152||PPP2R5A||TUBGCP4||USP33||KIAA0753||PHAX||CEP72||STIL||CSPP1||NEK8||PDE4DIP||ATP6V1D||BICD1||CEP250||C2CD5||CKAP5||CCDC57||LEO1||CDC16||HMBOX1||CLUAP1||APC||MASTL||SDCCAG8||USP2||LRRCC1||PAFAH1B1||CCDC77||DIS3L||GPSM2||STOX1||TAF1D||CEP95||CEP112||CEP78||CYLD||CEP290||TRIP4||SPAG5||PLK3||BOD1L1||RPGR||DYNC2I1||RAD18||CIR1||CCDC14||WAPL||DLGAP5||AKNA||ROCK2||HMMR||IFT74||TAPT1||CCDC85B||UPF3B||KAT2B||CNTRL||RNF19A||CEP295||CCDC66||CLIP1||ENSG00000285943||CKAP2L||PDZD2||PLK4||KIAA0586||KIF15||CTAG2||PLAG1||TUBGCP5||CAPN7||DTL||KIZ||FAM161A||EFHC1||RAB11FIP4||AKAP9||LRRC7||CDC27||CAPRIN2||CENPF||TMEM67||CCDC88A||PCM1||ALMS1||TPGS1||OFD1||ASPM||BBS9||DNAAF4||SLF1||ITSN2||AHI1||WRN||PATJ||ALPK1||CCDC15||CHODL||AK5||SPICE1 | 5.462069 | 6.050077 | 5.506813 | 6.022614 | 5.437734 | 5.505328 | 6.074471 | 6.065576 | 5.4625390 | 5.517464 | 5.403972 | 6.079776 | 6.021179 | 6.048681 | 5.658514 | 5.405881 | 5.442937 | 6.002414 | 5.984499 | 6.079166 | 5.424252 | 5.4422094 | 5.446644 | 5.470264 | 5.511919 | 5.533096 | 6.076661 | 6.060971 | 6.085672 | 6.046729 | 6.081201 | 6.068586 |
| GO:0030018 | Z disc | 85 | -0.4716433 | -1.512196 | 0.0214902237 | 0.5674104 | 0.5538652 | 1187 | tags=15%, list=7%, signal=14% | SYNE2||PDLIM3||TCAP||OBSCN||CAV3||CACNA1C||NEXN||PPP1R12B||S100A1||PDLIM4||NOS1AP||RYR1||CAPN3 | 4.909574 | 5.134408 | 4.959062 | 5.145273 | 4.894943 | 4.965203 | 5.125455 | 5.161548 | 4.9171634 | 4.937605 | 4.873205 | 5.171279 | 5.129509 | 5.101583 | 4.996384 | 4.939161 | 4.940904 | 5.147443 | 5.110777 | 5.176840 | 4.885260 | 4.8749009 | 4.924200 | 4.906400 | 4.923891 | 5.060344 | 5.145752 | 5.117467 | 5.112927 | 5.148519 | 5.181878 | 5.154025 |
| GO:0005778 | peroxisomal membrane | 61 | 0.4872813 | 1.544464 | 0.0233655358 | 0.5674104 | 0.5538652 | 4246 | tags=46%, list=26%, signal=34% | TMEM135||PJVK||PEX12||PEX13||PXMP4||FNDC5||ACSL4||ATAD1||TMEM35A||PECR||GDAP1||FAR1||CAT||HMGCR||SLC25A17||DHRS7B||PLAAT3||ACBD5||PEX7||MGST1||USP30||PEX11B||HSD17B4||MAVS||ABCD4||PEX16||ARF1||RAB8B | 5.522930 | 5.609937 | 5.551536 | 5.526961 | 5.545375 | 5.644969 | 5.561869 | 5.524164 | 5.5050408 | 5.548200 | 5.515195 | 5.637902 | 5.610554 | 5.580789 | 5.514284 | 5.591108 | 5.548186 | 5.534130 | 5.484416 | 5.561288 | 5.564351 | 5.4930893 | 5.577268 | 5.669120 | 5.608979 | 5.656115 | 5.576028 | 5.552011 | 5.557458 | 5.512838 | 5.566325 | 5.492313 |
| GO:0005637 | nuclear inner membrane | 47 | 0.5123292 | 1.547041 | 0.0252969644 | 0.5674104 | 0.5538652 | 2425 | tags=28%, list=15%, signal=24% | ARL6IP6||DPY19L4||LBR||DPY19L1||SMAD1||ZMPSTE24||P2RX5||MATR3||NEMP1||DPY19L3||LEMD3||UNC50||TMEM120A | 5.096232 | 5.942963 | 5.384339 | 6.041041 | 5.151501 | 5.410629 | 5.944907 | 6.031480 | 5.0366411 | 5.108571 | 5.141499 | 5.937505 | 5.957916 | 5.933349 | 5.500663 | 5.364178 | 5.279498 | 5.994886 | 6.046042 | 6.080898 | 5.138214 | 5.0646131 | 5.245894 | 5.303019 | 5.410622 | 5.510773 | 5.952401 | 5.931278 | 5.950945 | 6.015448 | 6.060212 | 6.018344 |
| GO:0097440 | apical dendrite | 15 | -0.6992522 | -1.634531 | 0.0258358663 | 0.5674104 | 0.5538652 | 2491 | tags=40%, list=16%, signal=34% | SLC1A1||ITSN1||GSK3A||PTK2B||MAP2||PPARGC1A | 5.064917 | 4.699547 | 5.333213 | 4.610146 | 5.042554 | 5.432146 | 4.749519 | 4.617690 | 4.9902741 | 5.046760 | 5.152963 | 4.655994 | 4.706701 | 4.734843 | 5.202820 | 5.457651 | 5.327913 | 4.658497 | 4.552845 | 4.617138 | 5.048620 | 4.8767670 | 5.185745 | 5.313080 | 5.410950 | 5.561498 | 4.822017 | 4.725950 | 4.697623 | 4.616353 | 4.648161 | 4.587927 |
| GO:1904724 | tertiary granule lumen | 31 | 0.5644830 | 1.581867 | 0.0261241970 | 0.5674104 | 0.5538652 | 3036 | tags=32%, list=19%, signal=26% | PTPN6||PTX3||CFP||QPCT||METTL7A||GOLGA7||B2M||GGH||ALDOC||IDH1 | 7.650277 | 7.256939 | 7.423058 | 7.185989 | 7.632285 | 7.678359 | 7.197750 | 7.137430 | 7.6465562 | 7.615223 | 7.688124 | 7.281852 | 7.245373 | 7.243264 | 7.273137 | 7.481352 | 7.503669 | 7.250036 | 7.129394 | 7.175964 | 7.650845 | 7.5834071 | 7.661368 | 7.687083 | 7.766312 | 7.575327 | 7.209193 | 7.179416 | 7.204464 | 7.132337 | 7.162885 | 7.116683 |
| GO:0034703 | cation channel complex | 118 | -0.4239769 | -1.427333 | 0.0274162762 | 0.5674104 | 0.5538652 | 2983 | tags=34%, list=19%, signal=28% | KCNQ1||CACHD1||SCN9A||CACNA2D1||CACNA1D||CACNG4||KCNQ4||KCNK2||ATP2A1||KCNG1||SCN5A||CACNB2||CACNB4||CACNA1B||CNIH3||CACNA2D2||PTK2B||KCNMA1||CACNG5||AKAP6||SHISA7||MCUB||ABCC8||KCND3||KCNJ14||CACNG6||AKAP9||UNC80||KCNN1||CACNA1C||SHISA8||CATSPER3||GRIN2C||CATSPER2||KCNA2||CATSPERE||LRRC26||GRIN3B||RYR1||SCN2A | 3.822535 | 3.991580 | 4.001125 | 3.955467 | 3.853467 | 4.017729 | 4.016274 | 3.998274 | 3.7928274 | 3.870043 | 3.803515 | 3.977629 | 3.988849 | 4.008096 | 4.018693 | 4.029612 | 3.953914 | 3.973591 | 3.912068 | 3.979776 | 3.869031 | 3.7855976 | 3.903249 | 3.940397 | 3.966207 | 4.138443 | 4.019075 | 4.015173 | 4.014570 | 3.987775 | 4.022842 | 3.983884 |
| GO:0030315 | T-tubule | 40 | -0.5556212 | -1.572777 | 0.0291064145 | 0.5674104 | 0.5538652 | 1487 | tags=22%, list=9%, signal=20% | AKAP6||CAV3||CACNG6||TGFB3||FXYD1||CACNA1C||NOS1AP||SCN2A||CAPN3 | 4.218942 | 4.648111 | 4.502344 | 4.801953 | 4.097256 | 4.246239 | 4.589258 | 4.764065 | 4.2391588 | 4.391732 | 3.999088 | 4.671896 | 4.672551 | 4.598644 | 4.522063 | 4.465978 | 4.518314 | 4.774588 | 4.798645 | 4.832048 | 4.188612 | 4.1739838 | 3.912840 | 4.213646 | 3.936649 | 4.527751 | 4.596731 | 4.586060 | 4.584953 | 4.747976 | 4.782279 | 4.761733 |
| GO:0031941 | filamentous actin | 25 | -0.6115474 | -1.585466 | 0.0302916275 | 0.5674104 | 0.5538652 | 2328 | tags=20%, list=14%, signal=17% | EHBP1||PDLIM3||NCKAP1||PDLIM4||MYO3A | 7.513819 | 6.826721 | 6.827795 | 6.757482 | 7.548350 | 7.326784 | 6.786033 | 6.761331 | 7.4539965 | 7.491596 | 7.592297 | 6.820661 | 6.820922 | 6.838507 | 6.599629 | 6.934020 | 6.925233 | 6.821704 | 6.663471 | 6.782603 | 7.657938 | 7.3999983 | 7.575221 | 7.533426 | 7.327534 | 7.084628 | 6.788897 | 6.792856 | 6.776295 | 6.763017 | 6.765990 | 6.754965 |
| GO:0000922 | spindle pole | 157 | -0.3913483 | -1.364095 | 0.0313739011 | 0.5674104 | 0.5538652 | 4088 | tags=38%, list=25%, signal=28% | DDX11||IK||HNRNPU||TNKS||BCCIP||NEDD9||WDR62||PPP2CA||BRCC3||KIF2A||CKAP2||TUBGCP6||STAG1||SPDL1||TTC28||TPX2||KIF20B||CTDP1||PPP2CB||KATNAL1||CUL3||HAUS8||CDK5RAP2||PRC1||TOPBP1||KATNA1||EML1||BIRC6||TUBGCP4||KNTC1||CSPP1||CEP104||CDC6||ZW10||CKAP5||SMC6||KIF11||GPSM2||CEP95||SPAG5||PLK3||BOD1L1||DYNC2I1||DLGAP5||SMC3||TOPORS||MTCL1||CNTRL||CKAP2L||PLK4||TUBGCP5||FAM161A||EFHC1||CENPF||ALMS1||ASPM||ALPK1||KATNAL2||DNAAF1 | 5.487054 | 6.510484 | 5.608836 | 6.544246 | 5.435549 | 5.623009 | 6.568425 | 6.610358 | 5.4775020 | 5.550764 | 5.430337 | 6.526238 | 6.481708 | 6.523078 | 5.914015 | 5.438529 | 5.417468 | 6.507513 | 6.513436 | 6.609502 | 5.392620 | 5.4576879 | 5.455398 | 5.572652 | 5.629085 | 5.665769 | 6.541955 | 6.563892 | 6.598856 | 6.594402 | 6.620561 | 6.615976 |
| GO:0005901 | caveola | 60 | -0.4946845 | -1.503332 | 0.0318918919 | 0.5674104 | 0.5538652 | 1670 | tags=20%, list=10%, signal=18% | KCNMA1||AKAP6||MYOF||CAV3||LIPE||KIF18A||FXYD1||ADRA1B||JAK2||TFPI||NOS1AP||ASAH2 | 4.344208 | 4.918158 | 4.703642 | 4.977787 | 4.333913 | 4.574809 | 4.848473 | 4.941678 | 4.3121149 | 4.424336 | 4.292642 | 4.959774 | 4.943098 | 4.849155 | 4.779204 | 4.652941 | 4.675617 | 4.964942 | 4.986246 | 4.982084 | 4.350337 | 4.3056239 | 4.345363 | 4.498396 | 4.446195 | 4.759886 | 4.867887 | 4.837515 | 4.839819 | 4.931210 | 4.974092 | 4.919152 |
| GO:0098993 | anchored component of synaptic vesicle membrane | 10 | -0.7388259 | -1.570747 | 0.0363214838 | 0.5997081 | 0.5853919 | 1675 | tags=20%, list=10%, signal=18% | RAB26||RAB3C | 4.043962 | 4.362095 | 4.130280 | 4.281540 | 4.074339 | 4.273257 | 4.386651 | 4.288021 | 4.0116609 | 3.971732 | 4.142886 | 4.332567 | 4.441864 | 4.308317 | 4.111518 | 4.187067 | 4.090456 | 4.313940 | 4.269006 | 4.261110 | 4.083356 | 3.9073455 | 4.215816 | 4.240857 | 4.289778 | 4.288597 | 4.385229 | 4.396701 | 4.377962 | 4.291774 | 4.289091 | 4.283185 |
| GO:0036126 | sperm flagellum | 65 | -0.4750652 | -1.461578 | 0.0412352731 | 0.6382877 | 0.6230505 | 1592 | tags=26%, list=10%, signal=24% | IL4I1||IFT88||SLC9B2||HSPD1||TCTE1||SPATA6L||RPGR||SPEF2||TTLL3||CCDC181||PMFBP1||IQCG||MNS1||CATSPERE||QRICH2||CFAP69||DNAH17 | 5.251010 | 5.802204 | 5.437501 | 5.921977 | 5.126909 | 5.219442 | 5.814367 | 5.914327 | 5.2831148 | 5.382165 | 5.070395 | 5.830217 | 5.780556 | 5.795388 | 5.601719 | 5.331175 | 5.364245 | 5.875643 | 5.908660 | 5.979662 | 5.193513 | 5.1637858 | 5.017335 | 5.218092 | 5.064528 | 5.360546 | 5.803868 | 5.795772 | 5.843017 | 5.915950 | 5.910028 | 5.916992 |
| GO:0089717 | spanning component of membrane | 12 | -0.7036334 | -1.558457 | 0.0430272762 | 0.6414388 | 0.6261265 | 2241 | tags=50%, list=14%, signal=43% | SLC24A1||NLGN2||NLGN3||NPC1L1||NLGN1||SLC24A4 | 2.228984 | 3.701217 | 3.332728 | 3.918645 | 2.115245 | 2.670392 | 3.720709 | 3.930440 | 2.2629864 | 2.317019 | 2.098033 | 3.729180 | 3.750850 | 3.620256 | 3.640435 | 3.084732 | 3.212741 | 3.827728 | 4.044384 | 3.874713 | 2.006413 | 2.2146904 | 2.117120 | 2.227762 | 2.337127 | 3.226670 | 3.732886 | 3.747067 | 3.681348 | 3.889888 | 4.027966 | 3.868205 |
| GO:0016605 | PML body | 92 | -0.4358959 | -1.414718 | 0.0431654676 | 0.6414388 | 0.6261265 | 3887 | tags=37%, list=24%, signal=28% | LRCH4||AKAP8L||KAT6A||ATRX||MTOR||CALCOCO2||NSMCE2||RAD51||CHD3||RNF111||ZBTB16||PIAS1||TOPBP1||SATB1||TCF7L2||USP7||SMC5||HMBOX1||SMC6||PTEN||ATR||RNF6||WDFY3||ISG20||MORC3||IKBKE||TOPORS||ZNF451||MRE11||BLM||TERT||NR2C1||ZMYM2||PPARGC1A | 4.842707 | 5.590820 | 5.104883 | 5.605622 | 4.779030 | 4.869922 | 5.645653 | 5.646414 | 4.8621646 | 4.988680 | 4.658191 | 5.566885 | 5.593568 | 5.611656 | 5.287148 | 4.954102 | 5.052847 | 5.561452 | 5.573646 | 5.678857 | 4.722832 | 4.8754348 | 4.733746 | 4.830773 | 4.776456 | 4.993592 | 5.638561 | 5.638248 | 5.660043 | 5.634593 | 5.657489 | 5.647070 |
| GO:0005746 | mitochondrial respirasome | 82 | 0.4224719 | 1.404167 | 0.0465816918 | 0.6521582 | 0.6365899 | 4643 | tags=38%, list=29%, signal=27% | NDUFC2-KCTD14||NDUFA4L2||NDUFA9||NDUFB1||COX7A2L||UQCRH||COX15||NDUFB6||NDUFA4||SDHD||NNT||SDHC||UQCR11||NDUFB5||NDUFA1||COX7C||STMP1||NDUFC2||NDUFA7||MT-CO1||SDHB||COX7B||MT-ND3||NDUFA8||NDUFB7||COX5B||COX8A||NDUFC1||MT-ND4L||NDUFAF1||COX6A1 | 9.720776 | 9.288178 | 10.183148 | 9.557713 | 9.791767 | 10.071043 | 9.346980 | 9.425188 | 9.6745915 | 9.703727 | 9.781865 | 8.934063 | 9.433533 | 9.440876 | 10.100281 | 10.308661 | 10.131636 | 9.481517 | 9.767208 | 9.397932 | 9.828530 | 9.7103452 | 9.833101 | 9.922813 | 9.914407 | 10.334712 | 9.337767 | 9.373930 | 9.328846 | 9.512605 | 9.365791 | 9.392902 |
| GO:0000235 | astral microtubule | 10 | -0.7235743 | -1.538322 | 0.0467542504 | 0.6521582 | 0.6365899 | 1745 | tags=50%, list=11%, signal=45% | PAFAH1B1||MAP9||FAM161A||KIF18A||DYNLT3 | 3.979139 | 5.074795 | 4.506628 | 5.178350 | 3.894630 | 4.372741 | 5.134981 | 5.268415 | 3.9311424 | 4.085820 | 3.914180 | 5.030852 | 5.105592 | 5.086897 | 4.774924 | 4.360410 | 4.341743 | 5.105301 | 5.171363 | 5.254506 | 3.883331 | 3.8876772 | 3.912708 | 4.260310 | 4.321394 | 4.523219 | 5.107316 | 5.138152 | 5.159006 | 5.260369 | 5.282561 | 5.262209 |
| GO:0099056 | integral component of presynaptic membrane | 39 | -0.5308595 | -1.496417 | 0.0473977695 | 0.6521582 | 0.6365899 | 696 | tags=13%, list=4%, signal=12% | ADAM23||ATP2B1||PTPRD||KCNA2||SCN2A | 3.610413 | 5.143713 | 4.090730 | 4.937398 | 3.634082 | 4.021684 | 4.998610 | 4.894020 | 3.5465213 | 3.627177 | 3.655342 | 5.126003 | 5.216851 | 5.085117 | 4.221529 | 4.096187 | 3.940825 | 4.856836 | 4.999799 | 4.951909 | 3.601155 | 3.5825510 | 3.714947 | 3.915728 | 3.946062 | 4.187599 | 5.007725 | 5.006775 | 4.981172 | 4.898340 | 4.931903 | 4.850663 |
KEGG
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa05168 | Herpes simplex virus 1 infection | 370 | -0.3749466 | -1.456548 | 0.001314388 | 0.1537833 | 0.1537833 | 1640 | tags=21%, list=12%, signal=19% | ZNF555||ZFP90||ZNF124||ZNF773||ZNF254||ZNF426||ZNF223||ZNF846||ZNF184||HCFC2||ZFP1||ZNF443||ZNF674||ZNF565||ZNF26||ZNF564||ZNF320||IKBKE||PILRB||ZNF701||ZNF175||ZNF57||ZNF585A||ZNF549||ZNF708||ZNF222||IRF9||ZNF778||ZNF613||ZNF416||ZNF136||ZNF470||ZNF354A||FAS||ZNF562||ZIK1||ZNF675||ZNF45||PIK3CA||ZNF155||ZNF226||ZNF514||ZNF230||ZNF546||ZNF615||ZNF543||ZFP14||JAK2||ZNF780B||HLA-G||ZNF14||ZNF160||ZNF567||ZNF551||ZNF304||ZNF836||ZNF799||ZNF583||ZNF585B||ZNF606||ZNF25||163051||ZNF816||ZNF256||ZNF547||ZNF224||ZNF577||ZNF570||ZNF91||ZNF814||ZNF112||ZNF461||ZNF17||ZNF782||ZNF829||ZNF214||ZNF284 | 4.388954 | 5.025723 | 4.546492 | 4.833528 | 4.378123 | 4.492159 | 4.987082 | 4.848860 | 4.372601 | 4.424142 | 4.369463 | 5.044975 | 5.034870 | 4.996879 | 4.667011 | 4.483299 | 4.481168 | 4.815203 | 4.820150 | 4.864713 | 4.366461 | 4.355440 | 4.411848 | 4.427392 | 4.461171 | 4.583216 | 4.994740 | 4.978205 | 4.988253 | 4.835197 | 4.877310 | 4.833647 |
| hsa05017 | Spinocerebellar ataxia | 121 | -0.4507640 | -1.550926 | 0.006238004 | 0.3649232 | 0.3649232 | 3297 | tags=30%, list=23%, signal=23% | ULK1||PSMC2||NFYA||PRKCB||NOP56||PSMC6||ATXN1L||AFG3L2||OPA1||MTOR||PSMD14||PRKCG||PIK3R1||MYOD1||ATP2A1||AKT3||PLCB4||PSMA6||ATP2A3||ITPR1||PSMD1||OMA1||ATG2B||ATXN3||SPTBN2||RB1CC1||MAP3K5||KCND3||PIK3CA||ITPR2||GRIN2C||DAB1||GRIN3B||PLCB1||PIK3C3||RYR1 | 6.541779 | 6.635936 | 6.590437 | 6.748216 | 6.468867 | 6.398964 | 6.645912 | 6.798934 | 6.578091 | 6.643483 | 6.392131 | 6.678554 | 6.613546 | 6.614745 | 6.668767 | 6.479657 | 6.616343 | 6.759987 | 6.707434 | 6.776331 | 6.456755 | 6.550561 | 6.395019 | 6.423882 | 6.357705 | 6.414419 | 6.650907 | 6.640822 | 6.645989 | 6.792327 | 6.803983 | 6.800468 |
| hsa00190 | Oxidative phosphorylation | 118 | 0.4298434 | 1.556929 | 0.011758418 | 0.4585783 | 0.4585783 | 3777 | tags=37%, list=27%, signal=28% | NDUFC2-KCTD14||NDUFA4L2||COX6B2||NDUFA9||ATP6V1A||ATP6V1G2||NDUFB1||ATP5F1E||COX7A2L||UQCRH||ATP6V0A2||ATP6V1G1||COX15||NDUFB6||NDUFA4||SDHD||SDHC||NDUFB5||NDUFA1||ATP5ME||COX7C||COX11||NDUFC2||MT-CO1||ATP5MC3||SDHB||COX7B||MT-ND3||LHPP||ATP5MC2||ATP6V1C1||ATP6V1C2||NDUFA8||ATP6V1F||ATP5MC1||NDUFB7||COX6B1||COX5B||ATP5F1B||ATP5MG||COX8A||NDUFC1||ATP6V0E2||MT-ND4L | 10.022035 | 9.478658 | 10.462432 | 9.820110 | 10.096000 | 10.376440 | 9.560072 | 9.693546 | 9.946739 | 9.997150 | 10.116885 | 9.094780 | 9.630618 | 9.645434 | 10.349612 | 10.603921 | 10.421726 | 9.740565 | 10.036663 | 9.654724 | 10.136587 | 10.004387 | 10.142833 | 10.242821 | 10.235103 | 10.617399 | 9.543148 | 9.586456 | 9.550236 | 9.777633 | 9.643996 | 9.655161 |
ORA: All DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: All DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0045653 | negative regulation of megakaryocyte differentiation | 1/1 | 18/20870 | 0.000862482 | 0.006796039 | H4C15 | 1 | 6.049987 | 5.482801 | 5.971996 | 5.461753 | 6.117977 | 6.216745 | 5.401195 | 5.613185 | 5.940400 | 5.948233 | 6.240599 | 5.626098 | 5.422343 | 5.388328 | 5.799650 | 6.101654 | 5.998539 | 5.559602 | 5.373461 | 5.446070 | 6.108669 | 5.968952 | 6.261462 | 6.160958 | 6.316615 | 6.167220 | 5.412996 | 5.357048 | 5.432482 | 5.463612 | 5.822811 | 5.527130 | |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 1/1 | 34/20870 | 0.001629133 | 0.006796039 | H4C15 | 1 | 5.171712 | 6.002626 | 5.413545 | 5.948303 | 5.126661 | 5.283335 | 6.026839 | 6.106002 | 5.111963 | 5.257440 | 5.141602 | 6.015067 | 5.997426 | 5.995303 | 5.667672 | 5.251594 | 5.283133 | 5.914926 | 5.916353 | 6.011492 | 5.126511 | 5.087835 | 5.164617 | 5.200474 | 5.247989 | 5.394391 | 6.005323 | 6.006487 | 6.067816 | 6.022282 | 6.263774 | 6.017986 | |
| GO:0034723 | DNA replication-dependent nucleosome organization | 1/1 | 34/20870 | 0.001629133 | 0.006796039 | H4C15 | 1 | 5.171712 | 6.002626 | 5.413545 | 5.948303 | 5.126661 | 5.283335 | 6.026839 | 6.106002 | 5.111963 | 5.257440 | 5.141602 | 6.015067 | 5.997426 | 5.995303 | 5.667672 | 5.251594 | 5.283133 | 5.914926 | 5.916353 | 6.011492 | 5.126511 | 5.087835 | 5.164617 | 5.200474 | 5.247989 | 5.394391 | 6.005323 | 6.006487 | 6.067816 | 6.022282 | 6.263774 | 6.017986 | |
| GO:0006336 | DNA replication-independent nucleosome assembly | 1/1 | 36/20870 | 0.001724964 | 0.006796039 | H4C15 | 1 | 6.276754 | 6.930258 | 6.486784 | 6.850898 | 6.242141 | 6.203981 | 6.984665 | 6.836890 | 6.287344 | 6.431049 | 6.091928 | 6.893572 | 6.940001 | 6.956466 | 6.760136 | 6.209948 | 6.436862 | 6.818530 | 6.829232 | 6.903442 | 6.165577 | 6.354841 | 6.198846 | 6.120023 | 6.135964 | 6.344873 | 6.971507 | 6.965927 | 7.016036 | 6.947002 | 6.616241 | 6.924365 | |
| GO:0034724 | DNA replication-independent nucleosome organization | 1/1 | 37/20870 | 0.001772880 | 0.006796039 | H4C15 | 1 | 6.264050 | 6.946672 | 6.481378 | 6.876479 | 6.222985 | 6.192213 | 7.004600 | 6.864720 | 6.274237 | 6.421507 | 6.075676 | 6.907178 | 6.958171 | 6.973829 | 6.766679 | 6.195511 | 6.424118 | 6.841308 | 6.856999 | 6.929584 | 6.146494 | 6.336449 | 6.178769 | 6.111340 | 6.122460 | 6.331963 | 6.990598 | 6.986409 | 7.036261 | 6.969550 | 6.656437 | 6.947513 | |
| GO:0045652 | regulation of megakaryocyte differentiation | 1/1 | 37/20870 | 0.001772880 | 0.006796039 | H4C15 | 1 | 5.844312 | 5.762053 | 5.861604 | 5.702518 | 5.879855 | 5.967918 | 5.783963 | 5.823364 | 5.759164 | 5.778602 | 5.984223 | 5.789733 | 5.755130 | 5.740858 | 5.887553 | 5.893704 | 5.801735 | 5.735876 | 5.643858 | 5.726061 | 5.839952 | 5.798742 | 5.993492 | 5.889451 | 6.031769 | 5.978960 | 5.787007 | 5.763552 | 5.801079 | 5.762286 | 5.928200 | 5.773578 | |
| GO:0030219 | megakaryocyte differentiation | 1/1 | 64/20870 | 0.003066603 | 0.010075981 | H4C15 | 1 | 5.265483 | 5.369258 | 5.308828 | 5.335000 | 5.293259 | 5.419309 | 5.405155 | 5.447812 | 5.177253 | 5.213468 | 5.396066 | 5.378749 | 5.370688 | 5.358264 | 5.363847 | 5.325656 | 5.233906 | 5.346640 | 5.282091 | 5.374710 | 5.258515 | 5.207416 | 5.406376 | 5.345064 | 5.483727 | 5.425783 | 5.405722 | 5.386923 | 5.422598 | 5.397150 | 5.533150 | 5.409170 | |
| GO:0045638 | negative regulation of myeloid cell differentiation | 1/1 | 106/20870 | 0.005079061 | 0.014602300 | H4C15 | 1 | 6.174098 | 5.981335 | 6.139208 | 6.001427 | 6.143290 | 5.914926 | 5.950575 | 6.036772 | 6.226754 | 6.235375 | 6.052924 | 6.050914 | 5.936928 | 5.953516 | 6.073716 | 6.117883 | 6.221985 | 6.047812 | 5.938902 | 6.015411 | 6.193669 | 6.254763 | 5.965624 | 6.076815 | 5.692347 | 5.949448 | 5.961736 | 5.920478 | 5.969036 | 6.003727 | 6.070303 | 6.035518 | |
| GO:0006352 | DNA-templated transcription, initiation | 1/1 | 142/20870 | 0.006804025 | 0.015566603 | H4C15 | 1 | 5.526734 | 5.866982 | 5.548106 | 5.886052 | 5.566092 | 5.692639 | 5.918271 | 5.941398 | 5.465711 | 5.503005 | 5.607704 | 5.868000 | 5.865446 | 5.867500 | 5.597158 | 5.553598 | 5.491617 | 5.872631 | 5.847678 | 5.936390 | 5.545987 | 5.486531 | 5.660319 | 5.639811 | 5.750997 | 5.684934 | 5.921678 | 5.909769 | 5.923328 | 5.941242 | 5.957333 | 5.925442 | |
| GO:0006334 | nucleosome assembly | 1/1 | 147/20870 | 0.007043603 | 0.015566603 | H4C15 | 1 | 6.150069 | 7.095473 | 6.331647 | 7.057708 | 6.101249 | 6.231562 | 7.160069 | 7.108922 | 6.156997 | 6.230727 | 6.057248 | 7.085394 | 7.076396 | 7.124180 | 6.660161 | 6.111004 | 6.157187 | 7.014918 | 7.025243 | 7.130125 | 6.044606 | 6.145867 | 6.111442 | 6.153445 | 6.187934 | 6.345921 | 7.129534 | 7.145612 | 7.203991 | 7.122914 | 7.076187 | 7.127112 | |
| GO:0031497 | chromatin assembly | 1/1 | 171/20870 | 0.008193579 | 0.015566603 | H4C15 | 1 | 6.131194 | 6.999264 | 6.290442 | 6.971021 | 6.086622 | 6.210389 | 7.069811 | 7.021027 | 6.131780 | 6.202123 | 6.055979 | 6.986115 | 6.982307 | 7.028903 | 6.589398 | 6.098185 | 6.129615 | 6.930110 | 6.938725 | 7.041542 | 6.033980 | 6.121602 | 6.102816 | 6.137944 | 6.170213 | 6.316654 | 7.044080 | 7.055511 | 7.109007 | 7.037262 | 6.984536 | 7.040601 | |
| GO:0032200 | telomere organization | 1/1 | 172/20870 | 0.008241495 | 0.015566603 | H4C15 | 1 | 6.527224 | 7.318417 | 6.722691 | 7.351486 | 6.424548 | 6.482130 | 7.369899 | 7.397102 | 6.550322 | 6.687990 | 6.319632 | 7.328375 | 7.296327 | 7.330297 | 7.009867 | 6.456790 | 6.645758 | 7.310837 | 7.318517 | 7.422391 | 6.396083 | 6.530065 | 6.340883 | 6.506180 | 6.430487 | 6.508367 | 7.353876 | 7.359842 | 7.395624 | 7.399303 | 7.377155 | 7.414603 | |
| GO:0034728 | nucleosome organization | 1/1 | 188/20870 | 0.009008146 | 0.015566603 | H4C15 | 1 | 6.103363 | 6.888427 | 6.253774 | 6.859059 | 6.059458 | 6.209572 | 6.951407 | 6.909873 | 6.097879 | 6.168160 | 6.041245 | 6.879068 | 6.871821 | 6.914038 | 6.517029 | 6.083734 | 6.119293 | 6.826648 | 6.822624 | 6.925532 | 6.026714 | 6.074127 | 6.076981 | 6.156421 | 6.181135 | 6.287757 | 6.926296 | 6.937081 | 6.990035 | 6.920015 | 6.885343 | 6.923950 | |
| GO:0006333 | chromatin assembly or disassembly | 1/1 | 199/20870 | 0.009535218 | 0.015566603 | H4C15 | 1 | 6.092644 | 6.854650 | 6.229507 | 6.829187 | 6.054348 | 6.195426 | 6.924455 | 6.879249 | 6.082930 | 6.150189 | 6.042765 | 6.840244 | 6.841391 | 6.881923 | 6.485451 | 6.074117 | 6.090260 | 6.795078 | 6.793775 | 6.896291 | 6.017875 | 6.066012 | 6.078451 | 6.139843 | 6.162675 | 6.279817 | 6.901318 | 6.911134 | 6.960221 | 6.893736 | 6.848901 | 6.894637 | |
| GO:0006323 | DNA packaging | 1/1 | 221/20870 | 0.010589363 | 0.015566603 | H4C15 | 1 | 6.186009 | 6.894821 | 6.314623 | 6.871020 | 6.158097 | 6.299544 | 6.960808 | 6.935335 | 6.169614 | 6.231414 | 6.155873 | 6.892235 | 6.876381 | 6.915577 | 6.553947 | 6.172278 | 6.184124 | 6.831380 | 6.837288 | 6.941702 | 6.118969 | 6.157500 | 6.196773 | 6.243036 | 6.281630 | 6.370965 | 6.941159 | 6.946773 | 6.993907 | 6.943090 | 6.910678 | 6.951911 | |
| GO:0065004 | protein-DNA complex assembly | 1/1 | 226/20870 | 0.010828941 | 0.015566603 | H4C15 | 1 | 5.860695 | 6.712449 | 6.029192 | 6.692678 | 5.819164 | 5.945211 | 6.780882 | 6.746567 | 5.856938 | 5.925007 | 5.797310 | 6.706431 | 6.694152 | 6.736436 | 6.315810 | 5.851670 | 5.870765 | 6.649948 | 6.662552 | 6.762860 | 5.767450 | 5.843870 | 5.844815 | 5.861747 | 5.914731 | 6.052386 | 6.759813 | 6.766512 | 6.815673 | 6.757041 | 6.720160 | 6.762139 | |
| GO:0045637 | regulation of myeloid cell differentiation | 1/1 | 251/20870 | 0.012026833 | 0.016240890 | H4C15 | 1 | 5.871456 | 6.042320 | 5.806211 | 5.888195 | 5.909507 | 5.735793 | 6.035442 | 5.901316 | 5.892709 | 5.926512 | 5.791763 | 6.067840 | 6.025306 | 6.033461 | 5.832649 | 5.771640 | 5.813669 | 5.899156 | 5.849795 | 5.914840 | 5.960730 | 5.934143 | 5.830386 | 5.810031 | 5.626862 | 5.764250 | 6.055617 | 6.012823 | 6.037565 | 5.889414 | 5.910245 | 5.904209 | |
| GO:0071824 | protein-DNA complex subunit organization | 1/1 | 268/20870 | 0.012841399 | 0.016240890 | H4C15 | 1 | 6.024187 | 6.727371 | 6.147060 | 6.722548 | 5.959877 | 6.038323 | 6.797698 | 6.771240 | 6.040361 | 6.116508 | 5.908030 | 6.716735 | 6.710566 | 6.754422 | 6.392397 | 5.970900 | 6.041701 | 6.687337 | 6.694729 | 6.783577 | 5.893976 | 6.050443 | 5.930530 | 5.967054 | 6.011283 | 6.131569 | 6.779027 | 6.782602 | 6.830881 | 6.780672 | 6.748603 | 6.784180 | |
| GO:0006338 | chromatin remodeling | 1/1 | 280/20870 | 0.013416387 | 0.016240890 | H4C15 | 1 | 6.205154 | 6.730183 | 6.303999 | 6.704723 | 6.175350 | 6.278770 | 6.772589 | 6.746820 | 6.197807 | 6.256505 | 6.159491 | 6.730423 | 6.716486 | 6.743513 | 6.460356 | 6.195641 | 6.241927 | 6.688258 | 6.659323 | 6.764532 | 6.167272 | 6.176529 | 6.182211 | 6.253404 | 6.227629 | 6.352255 | 6.758404 | 6.759092 | 6.799879 | 6.752287 | 6.737163 | 6.750963 | |
| GO:0071103 | DNA conformation change | 1/1 | 316/20870 | 0.015141351 | 0.017412554 | H4C15 | 1 | 6.042827 | 6.738844 | 6.216321 | 6.756773 | 5.993302 | 6.139361 | 6.804486 | 6.826204 | 6.029098 | 6.106137 | 5.990845 | 6.736256 | 6.725306 | 6.754815 | 6.467447 | 6.056722 | 6.087519 | 6.716362 | 6.724283 | 6.827013 | 5.960031 | 6.004748 | 6.014542 | 6.080122 | 6.110568 | 6.223413 | 6.786973 | 6.794207 | 6.831872 | 6.826616 | 6.815987 | 6.835939 | |
| GO:1903706 | regulation of hemopoiesis | 1/1 | 444/20870 | 0.021274557 | 0.021849545 | H4C15 | 1 | 5.765715 | 5.848857 | 5.740161 | 5.735975 | 5.787985 | 5.707612 | 5.835409 | 5.751759 | 5.774577 | 5.797709 | 5.723874 | 5.880644 | 5.831966 | 5.833429 | 5.747792 | 5.716056 | 5.756323 | 5.749478 | 5.695336 | 5.762239 | 5.822780 | 5.791193 | 5.749034 | 5.745613 | 5.656974 | 5.718823 | 5.851530 | 5.818864 | 5.835648 | 5.740737 | 5.763707 | 5.750741 | |
| GO:0006325 | chromatin organization | 1/1 | 445/20870 | 0.021322472 | 0.021849545 | H4C15 | 1 | 6.077880 | 6.507844 | 6.146370 | 6.509754 | 6.052385 | 6.166631 | 6.559351 | 6.556265 | 6.058536 | 6.111954 | 6.062535 | 6.500928 | 6.499657 | 6.522828 | 6.289175 | 6.054709 | 6.083727 | 6.492453 | 6.464773 | 6.569966 | 6.041605 | 6.035122 | 6.080018 | 6.136915 | 6.153185 | 6.208804 | 6.547538 | 6.547840 | 6.582397 | 6.559398 | 6.551215 | 6.558169 | |
| GO:0030099 | myeloid cell differentiation | 1/1 | 456/20870 | 0.021849545 | 0.021849545 | H4C15 | 1 | 6.515962 | 6.640869 | 6.329068 | 6.533120 | 6.504832 | 6.310351 | 6.593443 | 6.552470 | 6.542514 | 6.558800 | 6.443913 | 6.718924 | 6.604950 | 6.595431 | 6.342247 | 6.266924 | 6.375885 | 6.551835 | 6.489820 | 6.556745 | 6.498630 | 6.545161 | 6.469699 | 6.417873 | 6.323429 | 6.179855 | 6.598441 | 6.581253 | 6.600557 | 6.530582 | 6.562048 | 6.564534 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0046982 | protein heterodimerization activity | 1/1 | 337/20678 | 0.01629751 | 0.01629751 | H4C15 | 1 | 5.931334 | 6.15762 | 6.030139 | 6.199901 | 5.878559 | 5.901048 | 6.152002 | 6.224369 | 5.94361 | 6.03475 | 5.806547 | 6.19077 | 6.146048 | 6.135443 | 6.088654 | 5.93635 | 6.060897 | 6.203936 | 6.15508 | 6.239446 | 5.862078 | 5.933971 | 5.837889 | 5.883679 | 5.846293 | 5.970351 | 6.159284 | 6.139225 | 6.157413 | 6.232434 | 6.210858 | 6.229719 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0000786 | nucleosome | 1/1 | 112/21916 | 0.005110422 | 0.01368863 | H4C15 | 1 | 6.040878 | 6.271642 | 6.244724 | 6.193609 | 5.926593 | 5.771782 | 6.283362 | 6.217637 | 6.100219 | 6.237862 | 5.739915 | 6.280463 | 6.256044 | 6.278292 | 6.355001 | 6.018191 | 6.336768 | 6.184069 | 6.161119 | 6.234655 | 5.857554 | 6.091711 | 5.814849 | 5.712095 | 5.666674 | 5.923370 | 6.272912 | 6.267285 | 6.309524 | 6.260776 | 6.157674 | 6.232503 | |
| GO:0044815 | DNA packaging complex | 1/1 | 120/21916 | 0.005475452 | 0.01368863 | H4C15 | 1 | 5.954709 | 6.263016 | 6.176568 | 6.211908 | 5.844387 | 5.740177 | 6.285084 | 6.262950 | 6.004341 | 6.142993 | 5.678208 | 6.269988 | 6.249891 | 6.269078 | 6.302815 | 5.955413 | 6.247880 | 6.185480 | 6.185617 | 6.263224 | 5.781094 | 5.992624 | 5.746978 | 5.680456 | 5.644594 | 5.883774 | 6.275823 | 6.270935 | 6.308210 | 6.297365 | 6.212771 | 6.277364 | |
| GO:0000781 | chromosome, telomeric region | 1/1 | 186/21916 | 0.008486950 | 0.01368863 | H4C15 | 1 | 6.134933 | 6.616700 | 6.290595 | 6.653338 | 6.093597 | 6.173372 | 6.655028 | 6.697925 | 6.130570 | 6.209720 | 6.060650 | 6.625055 | 6.603170 | 6.621777 | 6.455084 | 6.149060 | 6.250626 | 6.628773 | 6.613838 | 6.715309 | 6.071594 | 6.116238 | 6.092614 | 6.162274 | 6.144360 | 6.212612 | 6.646155 | 6.641376 | 6.677289 | 6.707983 | 6.671723 | 6.713711 | |
| GO:0032993 | protein-DNA complex | 1/1 | 207/21916 | 0.009445154 | 0.01368863 | H4C15 | 1 | 6.015182 | 6.696574 | 6.235620 | 6.698442 | 5.918405 | 5.934967 | 6.735271 | 6.759565 | 6.053975 | 6.150438 | 5.821641 | 6.715227 | 6.669990 | 6.704121 | 6.447156 | 6.022897 | 6.205256 | 6.673546 | 6.659692 | 6.760027 | 5.879047 | 6.016227 | 5.854627 | 5.897595 | 5.860118 | 6.040820 | 6.721054 | 6.719454 | 6.764844 | 6.760027 | 6.743777 | 6.774724 | |
| GO:0000228 | nuclear chromosome | 1/1 | 250/21916 | 0.011407191 | 0.01368863 | H4C15 | 1 | 5.730873 | 6.265150 | 5.876322 | 6.239559 | 5.687217 | 5.788818 | 6.305189 | 6.285961 | 5.713430 | 5.811160 | 5.664129 | 6.267556 | 6.262902 | 6.264988 | 6.031300 | 5.756340 | 5.827009 | 6.225346 | 6.197282 | 6.294313 | 5.681769 | 5.687620 | 5.692242 | 5.759689 | 5.755215 | 5.849573 | 6.306885 | 6.290882 | 6.317673 | 6.302075 | 6.255758 | 6.299583 | |
| GO:0098687 | chromosomal region | 1/1 | 386/21916 | 0.017612703 | 0.01761270 | H4C15 | 1 | 5.768083 | 6.341013 | 5.933470 | 6.380348 | 5.721943 | 5.833797 | 6.389579 | 6.444249 | 5.753803 | 5.844181 | 5.702692 | 6.347329 | 6.331019 | 6.344637 | 6.110665 | 5.797590 | 5.873376 | 6.351011 | 6.342677 | 6.445095 | 5.700382 | 5.740730 | 5.724431 | 5.810084 | 5.814764 | 5.875612 | 6.383931 | 6.381525 | 6.403184 | 6.441663 | 6.440121 | 6.450941 |
KEGG
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa00565 | Ether lipid metabolism | 1/2 | 49/8146 | 0.01199500 | 0.04946178 | PEDS1-UBE2V1 | 1 | 3.901025 | 4.579184 | 4.343802 | 4.642578 | 3.956054 | 4.301113 | 4.523417 | 4.577604 | 3.831360 | 3.920579 | 3.948555 | 4.542667 | 4.644727 | 4.547850 | 4.384590 | 4.359527 | 4.285459 | 4.605653 | 4.659436 | 4.661946 | 3.946237 | 3.882257 | 4.035549 | 4.097509 | 4.235391 | 4.535190 | 4.570156 | 4.521019 | 4.477589 | 4.529895 | 4.632283 | 4.568777 | |
| hsa05322 | Systemic lupus erythematosus | 1/2 | 136/8146 | 0.03311390 | 0.04946178 | H4C15 | 1 | 6.321517 | 6.470763 | 6.451806 | 6.480259 | 6.267784 | 6.143901 | 6.478292 | 6.522252 | 6.354829 | 6.462215 | 6.127557 | 6.504658 | 6.449156 | 6.457854 | 6.551703 | 6.278809 | 6.510198 | 6.480561 | 6.446902 | 6.512567 | 6.198438 | 6.388119 | 6.208804 | 6.083071 | 6.119473 | 6.225346 | 6.476118 | 6.469098 | 6.489583 | 6.546568 | 6.483946 | 6.535471 | |
| hsa05034 | Alcoholism | 1/2 | 187/8146 | 0.04538788 | 0.04946178 | H4C15 | 1 | 6.081292 | 6.312998 | 6.203495 | 6.296332 | 6.035621 | 6.066784 | 6.329420 | 6.327174 | 6.084533 | 6.207338 | 5.939596 | 6.318456 | 6.308019 | 6.312499 | 6.267141 | 6.088087 | 6.248624 | 6.304355 | 6.246149 | 6.337027 | 5.995385 | 6.110528 | 5.997929 | 6.035454 | 6.030588 | 6.132024 | 6.323480 | 6.323340 | 6.341367 | 6.342897 | 6.301839 | 6.336450 | |
| hsa04613 | Neutrophil extracellular trap formation | 1/2 | 190/8146 | 0.04610743 | 0.04946178 | H4C15 | 1 | 7.038784 | 6.841004 | 6.825600 | 6.801365 | 7.004127 | 6.868386 | 6.829824 | 6.817625 | 7.043993 | 7.115883 | 6.951798 | 6.852408 | 6.833969 | 6.836565 | 6.791123 | 6.768794 | 6.912689 | 6.835026 | 6.737838 | 6.829186 | 7.040045 | 7.006107 | 6.965258 | 6.957310 | 6.837626 | 6.805758 | 6.824364 | 6.827590 | 6.837487 | 6.835317 | 6.791607 | 6.825588 | |
| hsa05203 | Viral carcinogenesis | 1/2 | 204/8146 | 0.04946178 | 0.04946178 | H4C15 | 1 | 6.119194 | 6.324710 | 6.152361 | 6.370672 | 6.099044 | 6.207099 | 6.325810 | 6.411415 | 6.097899 | 6.171332 | 6.086881 | 6.363155 | 6.307925 | 6.302261 | 6.185922 | 6.121732 | 6.148707 | 6.379203 | 6.310376 | 6.420305 | 6.113889 | 6.084145 | 6.098946 | 6.205132 | 6.188296 | 6.227598 | 6.330195 | 6.310430 | 6.336675 | 6.393171 | 6.441876 | 6.398704 |
ORA: Down-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Down-regulated DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
ORA: Up-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Up-regulated DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0045653 | negative regulation of megakaryocyte differentiation | 1/1 | 18/20870 | 0.000862482 | 0.006796039 | H4C15 | 1 | 6.049987 | 5.482801 | 5.971996 | 5.461753 | 6.117977 | 6.216745 | 5.401195 | 5.613185 | 5.940400 | 5.948233 | 6.240599 | 5.626098 | 5.422343 | 5.388328 | 5.799650 | 6.101654 | 5.998539 | 5.559602 | 5.373461 | 5.446070 | 6.108669 | 5.968952 | 6.261462 | 6.160958 | 6.316615 | 6.167220 | 5.412996 | 5.357048 | 5.432482 | 5.463612 | 5.822811 | 5.527130 | |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 1/1 | 34/20870 | 0.001629133 | 0.006796039 | H4C15 | 1 | 5.171712 | 6.002626 | 5.413545 | 5.948303 | 5.126661 | 5.283335 | 6.026839 | 6.106002 | 5.111963 | 5.257440 | 5.141602 | 6.015067 | 5.997426 | 5.995303 | 5.667672 | 5.251594 | 5.283133 | 5.914926 | 5.916353 | 6.011492 | 5.126511 | 5.087835 | 5.164617 | 5.200474 | 5.247989 | 5.394391 | 6.005323 | 6.006487 | 6.067816 | 6.022282 | 6.263774 | 6.017986 | |
| GO:0034723 | DNA replication-dependent nucleosome organization | 1/1 | 34/20870 | 0.001629133 | 0.006796039 | H4C15 | 1 | 5.171712 | 6.002626 | 5.413545 | 5.948303 | 5.126661 | 5.283335 | 6.026839 | 6.106002 | 5.111963 | 5.257440 | 5.141602 | 6.015067 | 5.997426 | 5.995303 | 5.667672 | 5.251594 | 5.283133 | 5.914926 | 5.916353 | 6.011492 | 5.126511 | 5.087835 | 5.164617 | 5.200474 | 5.247989 | 5.394391 | 6.005323 | 6.006487 | 6.067816 | 6.022282 | 6.263774 | 6.017986 | |
| GO:0006336 | DNA replication-independent nucleosome assembly | 1/1 | 36/20870 | 0.001724964 | 0.006796039 | H4C15 | 1 | 6.276754 | 6.930258 | 6.486784 | 6.850898 | 6.242141 | 6.203981 | 6.984665 | 6.836890 | 6.287344 | 6.431049 | 6.091928 | 6.893572 | 6.940001 | 6.956466 | 6.760136 | 6.209948 | 6.436862 | 6.818530 | 6.829232 | 6.903442 | 6.165577 | 6.354841 | 6.198846 | 6.120023 | 6.135964 | 6.344873 | 6.971507 | 6.965927 | 7.016036 | 6.947002 | 6.616241 | 6.924365 | |
| GO:0034724 | DNA replication-independent nucleosome organization | 1/1 | 37/20870 | 0.001772880 | 0.006796039 | H4C15 | 1 | 6.264050 | 6.946672 | 6.481378 | 6.876479 | 6.222985 | 6.192213 | 7.004600 | 6.864720 | 6.274237 | 6.421507 | 6.075676 | 6.907178 | 6.958171 | 6.973829 | 6.766679 | 6.195511 | 6.424118 | 6.841308 | 6.856999 | 6.929584 | 6.146494 | 6.336449 | 6.178769 | 6.111340 | 6.122460 | 6.331963 | 6.990598 | 6.986409 | 7.036261 | 6.969550 | 6.656437 | 6.947513 | |
| GO:0045652 | regulation of megakaryocyte differentiation | 1/1 | 37/20870 | 0.001772880 | 0.006796039 | H4C15 | 1 | 5.844312 | 5.762053 | 5.861604 | 5.702518 | 5.879855 | 5.967918 | 5.783963 | 5.823364 | 5.759164 | 5.778602 | 5.984223 | 5.789733 | 5.755130 | 5.740858 | 5.887553 | 5.893704 | 5.801735 | 5.735876 | 5.643858 | 5.726061 | 5.839952 | 5.798742 | 5.993492 | 5.889451 | 6.031769 | 5.978960 | 5.787007 | 5.763552 | 5.801079 | 5.762286 | 5.928200 | 5.773578 | |
| GO:0030219 | megakaryocyte differentiation | 1/1 | 64/20870 | 0.003066603 | 0.010075981 | H4C15 | 1 | 5.265483 | 5.369258 | 5.308828 | 5.335000 | 5.293259 | 5.419309 | 5.405155 | 5.447812 | 5.177253 | 5.213468 | 5.396066 | 5.378749 | 5.370688 | 5.358264 | 5.363847 | 5.325656 | 5.233906 | 5.346640 | 5.282091 | 5.374710 | 5.258515 | 5.207416 | 5.406376 | 5.345064 | 5.483727 | 5.425783 | 5.405722 | 5.386923 | 5.422598 | 5.397150 | 5.533150 | 5.409170 | |
| GO:0045638 | negative regulation of myeloid cell differentiation | 1/1 | 106/20870 | 0.005079061 | 0.014602300 | H4C15 | 1 | 6.174098 | 5.981335 | 6.139208 | 6.001427 | 6.143290 | 5.914926 | 5.950575 | 6.036772 | 6.226754 | 6.235375 | 6.052924 | 6.050914 | 5.936928 | 5.953516 | 6.073716 | 6.117883 | 6.221985 | 6.047812 | 5.938902 | 6.015411 | 6.193669 | 6.254763 | 5.965624 | 6.076815 | 5.692347 | 5.949448 | 5.961736 | 5.920478 | 5.969036 | 6.003727 | 6.070303 | 6.035518 | |
| GO:0006352 | DNA-templated transcription, initiation | 1/1 | 142/20870 | 0.006804025 | 0.015566603 | H4C15 | 1 | 5.526734 | 5.866982 | 5.548106 | 5.886052 | 5.566092 | 5.692639 | 5.918271 | 5.941398 | 5.465711 | 5.503005 | 5.607704 | 5.868000 | 5.865446 | 5.867500 | 5.597158 | 5.553598 | 5.491617 | 5.872631 | 5.847678 | 5.936390 | 5.545987 | 5.486531 | 5.660319 | 5.639811 | 5.750997 | 5.684934 | 5.921678 | 5.909769 | 5.923328 | 5.941242 | 5.957333 | 5.925442 | |
| GO:0006334 | nucleosome assembly | 1/1 | 147/20870 | 0.007043603 | 0.015566603 | H4C15 | 1 | 6.150069 | 7.095473 | 6.331647 | 7.057708 | 6.101249 | 6.231562 | 7.160069 | 7.108922 | 6.156997 | 6.230727 | 6.057248 | 7.085394 | 7.076396 | 7.124180 | 6.660161 | 6.111004 | 6.157187 | 7.014918 | 7.025243 | 7.130125 | 6.044606 | 6.145867 | 6.111442 | 6.153445 | 6.187934 | 6.345921 | 7.129534 | 7.145612 | 7.203991 | 7.122914 | 7.076187 | 7.127112 | |
| GO:0031497 | chromatin assembly | 1/1 | 171/20870 | 0.008193579 | 0.015566603 | H4C15 | 1 | 6.131194 | 6.999264 | 6.290442 | 6.971021 | 6.086622 | 6.210389 | 7.069811 | 7.021027 | 6.131780 | 6.202123 | 6.055979 | 6.986115 | 6.982307 | 7.028903 | 6.589398 | 6.098185 | 6.129615 | 6.930110 | 6.938725 | 7.041542 | 6.033980 | 6.121602 | 6.102816 | 6.137944 | 6.170213 | 6.316654 | 7.044080 | 7.055511 | 7.109007 | 7.037262 | 6.984536 | 7.040601 | |
| GO:0032200 | telomere organization | 1/1 | 172/20870 | 0.008241495 | 0.015566603 | H4C15 | 1 | 6.527224 | 7.318417 | 6.722691 | 7.351486 | 6.424548 | 6.482130 | 7.369899 | 7.397102 | 6.550322 | 6.687990 | 6.319632 | 7.328375 | 7.296327 | 7.330297 | 7.009867 | 6.456790 | 6.645758 | 7.310837 | 7.318517 | 7.422391 | 6.396083 | 6.530065 | 6.340883 | 6.506180 | 6.430487 | 6.508367 | 7.353876 | 7.359842 | 7.395624 | 7.399303 | 7.377155 | 7.414603 | |
| GO:0034728 | nucleosome organization | 1/1 | 188/20870 | 0.009008146 | 0.015566603 | H4C15 | 1 | 6.103363 | 6.888427 | 6.253774 | 6.859059 | 6.059458 | 6.209572 | 6.951407 | 6.909873 | 6.097879 | 6.168160 | 6.041245 | 6.879068 | 6.871821 | 6.914038 | 6.517029 | 6.083734 | 6.119293 | 6.826648 | 6.822624 | 6.925532 | 6.026714 | 6.074127 | 6.076981 | 6.156421 | 6.181135 | 6.287757 | 6.926296 | 6.937081 | 6.990035 | 6.920015 | 6.885343 | 6.923950 | |
| GO:0006333 | chromatin assembly or disassembly | 1/1 | 199/20870 | 0.009535218 | 0.015566603 | H4C15 | 1 | 6.092644 | 6.854650 | 6.229507 | 6.829187 | 6.054348 | 6.195426 | 6.924455 | 6.879249 | 6.082930 | 6.150189 | 6.042765 | 6.840244 | 6.841391 | 6.881923 | 6.485451 | 6.074117 | 6.090260 | 6.795078 | 6.793775 | 6.896291 | 6.017875 | 6.066012 | 6.078451 | 6.139843 | 6.162675 | 6.279817 | 6.901318 | 6.911134 | 6.960221 | 6.893736 | 6.848901 | 6.894637 | |
| GO:0006323 | DNA packaging | 1/1 | 221/20870 | 0.010589363 | 0.015566603 | H4C15 | 1 | 6.186009 | 6.894821 | 6.314623 | 6.871020 | 6.158097 | 6.299544 | 6.960808 | 6.935335 | 6.169614 | 6.231414 | 6.155873 | 6.892235 | 6.876381 | 6.915577 | 6.553947 | 6.172278 | 6.184124 | 6.831380 | 6.837288 | 6.941702 | 6.118969 | 6.157500 | 6.196773 | 6.243036 | 6.281630 | 6.370965 | 6.941159 | 6.946773 | 6.993907 | 6.943090 | 6.910678 | 6.951911 | |
| GO:0065004 | protein-DNA complex assembly | 1/1 | 226/20870 | 0.010828941 | 0.015566603 | H4C15 | 1 | 5.860695 | 6.712449 | 6.029192 | 6.692678 | 5.819164 | 5.945211 | 6.780882 | 6.746567 | 5.856938 | 5.925007 | 5.797310 | 6.706431 | 6.694152 | 6.736436 | 6.315810 | 5.851670 | 5.870765 | 6.649948 | 6.662552 | 6.762860 | 5.767450 | 5.843870 | 5.844815 | 5.861747 | 5.914731 | 6.052386 | 6.759813 | 6.766512 | 6.815673 | 6.757041 | 6.720160 | 6.762139 | |
| GO:0045637 | regulation of myeloid cell differentiation | 1/1 | 251/20870 | 0.012026833 | 0.016240890 | H4C15 | 1 | 5.871456 | 6.042320 | 5.806211 | 5.888195 | 5.909507 | 5.735793 | 6.035442 | 5.901316 | 5.892709 | 5.926512 | 5.791763 | 6.067840 | 6.025306 | 6.033461 | 5.832649 | 5.771640 | 5.813669 | 5.899156 | 5.849795 | 5.914840 | 5.960730 | 5.934143 | 5.830386 | 5.810031 | 5.626862 | 5.764250 | 6.055617 | 6.012823 | 6.037565 | 5.889414 | 5.910245 | 5.904209 | |
| GO:0071824 | protein-DNA complex subunit organization | 1/1 | 268/20870 | 0.012841399 | 0.016240890 | H4C15 | 1 | 6.024187 | 6.727371 | 6.147060 | 6.722548 | 5.959877 | 6.038323 | 6.797698 | 6.771240 | 6.040361 | 6.116508 | 5.908030 | 6.716735 | 6.710566 | 6.754422 | 6.392397 | 5.970900 | 6.041701 | 6.687337 | 6.694729 | 6.783577 | 5.893976 | 6.050443 | 5.930530 | 5.967054 | 6.011283 | 6.131569 | 6.779027 | 6.782602 | 6.830881 | 6.780672 | 6.748603 | 6.784180 | |
| GO:0006338 | chromatin remodeling | 1/1 | 280/20870 | 0.013416387 | 0.016240890 | H4C15 | 1 | 6.205154 | 6.730183 | 6.303999 | 6.704723 | 6.175350 | 6.278770 | 6.772589 | 6.746820 | 6.197807 | 6.256505 | 6.159491 | 6.730423 | 6.716486 | 6.743513 | 6.460356 | 6.195641 | 6.241927 | 6.688258 | 6.659323 | 6.764532 | 6.167272 | 6.176529 | 6.182211 | 6.253404 | 6.227629 | 6.352255 | 6.758404 | 6.759092 | 6.799879 | 6.752287 | 6.737163 | 6.750963 | |
| GO:0071103 | DNA conformation change | 1/1 | 316/20870 | 0.015141351 | 0.017412554 | H4C15 | 1 | 6.042827 | 6.738844 | 6.216321 | 6.756773 | 5.993302 | 6.139361 | 6.804486 | 6.826204 | 6.029098 | 6.106137 | 5.990845 | 6.736256 | 6.725306 | 6.754815 | 6.467447 | 6.056722 | 6.087519 | 6.716362 | 6.724283 | 6.827013 | 5.960031 | 6.004748 | 6.014542 | 6.080122 | 6.110568 | 6.223413 | 6.786973 | 6.794207 | 6.831872 | 6.826616 | 6.815987 | 6.835939 | |
| GO:1903706 | regulation of hemopoiesis | 1/1 | 444/20870 | 0.021274557 | 0.021849545 | H4C15 | 1 | 5.765715 | 5.848857 | 5.740161 | 5.735975 | 5.787985 | 5.707612 | 5.835409 | 5.751759 | 5.774577 | 5.797709 | 5.723874 | 5.880644 | 5.831966 | 5.833429 | 5.747792 | 5.716056 | 5.756323 | 5.749478 | 5.695336 | 5.762239 | 5.822780 | 5.791193 | 5.749034 | 5.745613 | 5.656974 | 5.718823 | 5.851530 | 5.818864 | 5.835648 | 5.740737 | 5.763707 | 5.750741 | |
| GO:0006325 | chromatin organization | 1/1 | 445/20870 | 0.021322472 | 0.021849545 | H4C15 | 1 | 6.077880 | 6.507844 | 6.146370 | 6.509754 | 6.052385 | 6.166631 | 6.559351 | 6.556265 | 6.058536 | 6.111954 | 6.062535 | 6.500928 | 6.499657 | 6.522828 | 6.289175 | 6.054709 | 6.083727 | 6.492453 | 6.464773 | 6.569966 | 6.041605 | 6.035122 | 6.080018 | 6.136915 | 6.153185 | 6.208804 | 6.547538 | 6.547840 | 6.582397 | 6.559398 | 6.551215 | 6.558169 | |
| GO:0030099 | myeloid cell differentiation | 1/1 | 456/20870 | 0.021849545 | 0.021849545 | H4C15 | 1 | 6.515962 | 6.640869 | 6.329068 | 6.533120 | 6.504832 | 6.310351 | 6.593443 | 6.552470 | 6.542514 | 6.558800 | 6.443913 | 6.718924 | 6.604950 | 6.595431 | 6.342247 | 6.266924 | 6.375885 | 6.551835 | 6.489820 | 6.556745 | 6.498630 | 6.545161 | 6.469699 | 6.417873 | 6.323429 | 6.179855 | 6.598441 | 6.581253 | 6.600557 | 6.530582 | 6.562048 | 6.564534 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0046982 | protein heterodimerization activity | 1/1 | 337/20678 | 0.01629751 | 0.01629751 | H4C15 | 1 | 5.931334 | 6.15762 | 6.030139 | 6.199901 | 5.878559 | 5.901048 | 6.152002 | 6.224369 | 5.94361 | 6.03475 | 5.806547 | 6.19077 | 6.146048 | 6.135443 | 6.088654 | 5.93635 | 6.060897 | 6.203936 | 6.15508 | 6.239446 | 5.862078 | 5.933971 | 5.837889 | 5.883679 | 5.846293 | 5.970351 | 6.159284 | 6.139225 | 6.157413 | 6.232434 | 6.210858 | 6.229719 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0000786 | nucleosome | 1/1 | 112/21916 | 0.005110422 | 0.01368863 | H4C15 | 1 | 6.040878 | 6.271642 | 6.244724 | 6.193609 | 5.926593 | 5.771782 | 6.283362 | 6.217637 | 6.100219 | 6.237862 | 5.739915 | 6.280463 | 6.256044 | 6.278292 | 6.355001 | 6.018191 | 6.336768 | 6.184069 | 6.161119 | 6.234655 | 5.857554 | 6.091711 | 5.814849 | 5.712095 | 5.666674 | 5.923370 | 6.272912 | 6.267285 | 6.309524 | 6.260776 | 6.157674 | 6.232503 | |
| GO:0044815 | DNA packaging complex | 1/1 | 120/21916 | 0.005475452 | 0.01368863 | H4C15 | 1 | 5.954709 | 6.263016 | 6.176568 | 6.211908 | 5.844387 | 5.740177 | 6.285084 | 6.262950 | 6.004341 | 6.142993 | 5.678208 | 6.269988 | 6.249891 | 6.269078 | 6.302815 | 5.955413 | 6.247880 | 6.185480 | 6.185617 | 6.263224 | 5.781094 | 5.992624 | 5.746978 | 5.680456 | 5.644594 | 5.883774 | 6.275823 | 6.270935 | 6.308210 | 6.297365 | 6.212771 | 6.277364 | |
| GO:0000781 | chromosome, telomeric region | 1/1 | 186/21916 | 0.008486950 | 0.01368863 | H4C15 | 1 | 6.134933 | 6.616700 | 6.290595 | 6.653338 | 6.093597 | 6.173372 | 6.655028 | 6.697925 | 6.130570 | 6.209720 | 6.060650 | 6.625055 | 6.603170 | 6.621777 | 6.455084 | 6.149060 | 6.250626 | 6.628773 | 6.613838 | 6.715309 | 6.071594 | 6.116238 | 6.092614 | 6.162274 | 6.144360 | 6.212612 | 6.646155 | 6.641376 | 6.677289 | 6.707983 | 6.671723 | 6.713711 | |
| GO:0032993 | protein-DNA complex | 1/1 | 207/21916 | 0.009445154 | 0.01368863 | H4C15 | 1 | 6.015182 | 6.696574 | 6.235620 | 6.698442 | 5.918405 | 5.934967 | 6.735271 | 6.759565 | 6.053975 | 6.150438 | 5.821641 | 6.715227 | 6.669990 | 6.704121 | 6.447156 | 6.022897 | 6.205256 | 6.673546 | 6.659692 | 6.760027 | 5.879047 | 6.016227 | 5.854627 | 5.897595 | 5.860118 | 6.040820 | 6.721054 | 6.719454 | 6.764844 | 6.760027 | 6.743777 | 6.774724 | |
| GO:0000228 | nuclear chromosome | 1/1 | 250/21916 | 0.011407191 | 0.01368863 | H4C15 | 1 | 5.730873 | 6.265150 | 5.876322 | 6.239559 | 5.687217 | 5.788818 | 6.305189 | 6.285961 | 5.713430 | 5.811160 | 5.664129 | 6.267556 | 6.262902 | 6.264988 | 6.031300 | 5.756340 | 5.827009 | 6.225346 | 6.197282 | 6.294313 | 5.681769 | 5.687620 | 5.692242 | 5.759689 | 5.755215 | 5.849573 | 6.306885 | 6.290882 | 6.317673 | 6.302075 | 6.255758 | 6.299583 | |
| GO:0098687 | chromosomal region | 1/1 | 386/21916 | 0.017612703 | 0.01761270 | H4C15 | 1 | 5.768083 | 6.341013 | 5.933470 | 6.380348 | 5.721943 | 5.833797 | 6.389579 | 6.444249 | 5.753803 | 5.844181 | 5.702692 | 6.347329 | 6.331019 | 6.344637 | 6.110665 | 5.797590 | 5.873376 | 6.351011 | 6.342677 | 6.445095 | 5.700382 | 5.740730 | 5.724431 | 5.810084 | 5.814764 | 5.875612 | 6.383931 | 6.381525 | 6.403184 | 6.441663 | 6.440121 | 6.450941 |
KEGG
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa00565 | Ether lipid metabolism | 1/2 | 49/8146 | 0.01199500 | 0.04946178 | PEDS1-UBE2V1 | 1 | 3.901025 | 4.579184 | 4.343802 | 4.642578 | 3.956054 | 4.301113 | 4.523417 | 4.577604 | 3.831360 | 3.920579 | 3.948555 | 4.542667 | 4.644727 | 4.547850 | 4.384590 | 4.359527 | 4.285459 | 4.605653 | 4.659436 | 4.661946 | 3.946237 | 3.882257 | 4.035549 | 4.097509 | 4.235391 | 4.535190 | 4.570156 | 4.521019 | 4.477589 | 4.529895 | 4.632283 | 4.568777 | |
| hsa05322 | Systemic lupus erythematosus | 1/2 | 136/8146 | 0.03311390 | 0.04946178 | H4C15 | 1 | 6.321517 | 6.470763 | 6.451806 | 6.480259 | 6.267784 | 6.143901 | 6.478292 | 6.522252 | 6.354829 | 6.462215 | 6.127557 | 6.504658 | 6.449156 | 6.457854 | 6.551703 | 6.278809 | 6.510198 | 6.480561 | 6.446902 | 6.512567 | 6.198438 | 6.388119 | 6.208804 | 6.083071 | 6.119473 | 6.225346 | 6.476118 | 6.469098 | 6.489583 | 6.546568 | 6.483946 | 6.535471 | |
| hsa05034 | Alcoholism | 1/2 | 187/8146 | 0.04538788 | 0.04946178 | H4C15 | 1 | 6.081292 | 6.312998 | 6.203495 | 6.296332 | 6.035621 | 6.066784 | 6.329420 | 6.327174 | 6.084533 | 6.207338 | 5.939596 | 6.318456 | 6.308019 | 6.312499 | 6.267141 | 6.088087 | 6.248624 | 6.304355 | 6.246149 | 6.337027 | 5.995385 | 6.110528 | 5.997929 | 6.035454 | 6.030588 | 6.132024 | 6.323480 | 6.323340 | 6.341367 | 6.342897 | 6.301839 | 6.336450 | |
| hsa04613 | Neutrophil extracellular trap formation | 1/2 | 190/8146 | 0.04610743 | 0.04946178 | H4C15 | 1 | 7.038784 | 6.841004 | 6.825600 | 6.801365 | 7.004127 | 6.868386 | 6.829824 | 6.817625 | 7.043993 | 7.115883 | 6.951798 | 6.852408 | 6.833969 | 6.836565 | 6.791123 | 6.768794 | 6.912689 | 6.835026 | 6.737838 | 6.829186 | 7.040045 | 7.006107 | 6.965258 | 6.957310 | 6.837626 | 6.805758 | 6.824364 | 6.827590 | 6.837487 | 6.835317 | 6.791607 | 6.825588 | |
| hsa05203 | Viral carcinogenesis | 1/2 | 204/8146 | 0.04946178 | 0.04946178 | H4C15 | 1 | 6.119194 | 6.324710 | 6.152361 | 6.370672 | 6.099044 | 6.207099 | 6.325810 | 6.411415 | 6.097899 | 6.171332 | 6.086881 | 6.363155 | 6.307925 | 6.302261 | 6.185922 | 6.121732 | 6.148707 | 6.379203 | 6.310376 | 6.420305 | 6.113889 | 6.084145 | 6.098946 | 6.205132 | 6.188296 | 6.227598 | 6.330195 | 6.310430 | 6.336675 | 6.393171 | 6.441876 | 6.398704 |
Help
still working on
2-2. Summary Plot
GSEA
GO: BP
GO: MF
GO: CC
KEGG
ORA: All DEGs
GO: BP
GO: MF
GO: CC
KEGG
ORA: Down-regulated DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Up-regulated DEGs
GO: BP
GO: MF
GO: CC
KEGG
ORA: Overlapped Terms
GO: BP
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | GO:0045653||GO:0006335||GO:0034723||GO:0006336||GO:0034724||GO:0045652||GO:0030219||GO:0045638||GO:0006352||GO:0006334||GO:0031497||GO:0032200||GO:0034728||GO:0006333||GO:0006323||GO:0065004||GO:0045637||GO:0071824||GO:0006338||GO:0071103||GO:1903706||GO:0006325||GO:0030099 |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: MF
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | GO:0046982 |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: CC
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | GO:0000786||GO:0044815||GO:0000781||GO:0032993||GO:0000228||GO:0098687 |
| All ∪ Down-Regulated ∪ Up-Regulated |
KEGG
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | |
| All ∪ Up-Regulated | hsa00565||hsa05322||hsa05034||hsa04613||hsa05203 |
| All ∪ Down-Regulated ∪ Up-Regulated |
Help
still working on
3. Isoform Switch Analysis
Alternative Splicing Events
Amount of Event
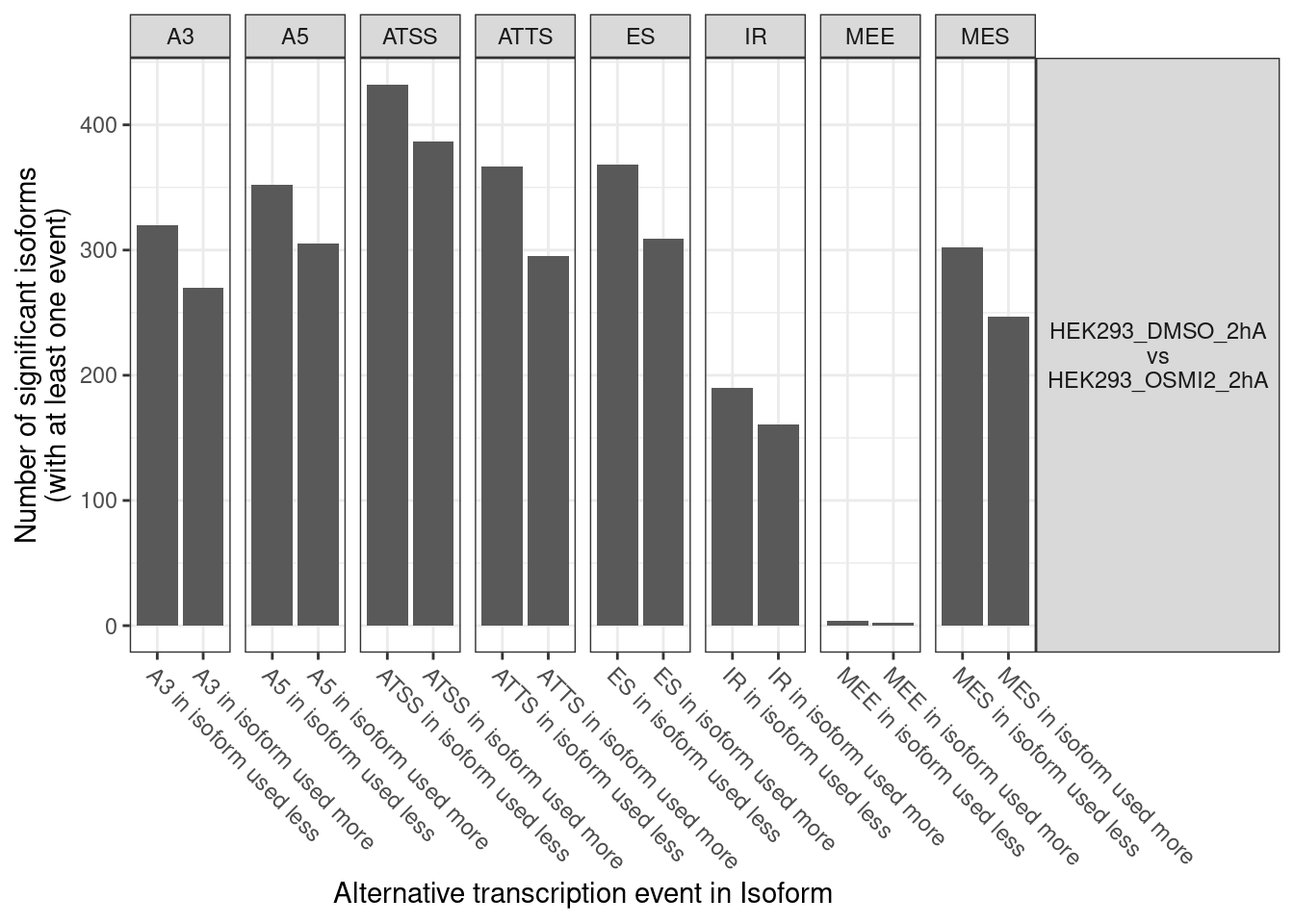
Enrichment Analysis
This below result summarizes the uneven usage within each comparison by for each alternative splicing type calculate the fraction of events being gains (as opposed to loss) and perform a statistical analysis of this fraction.
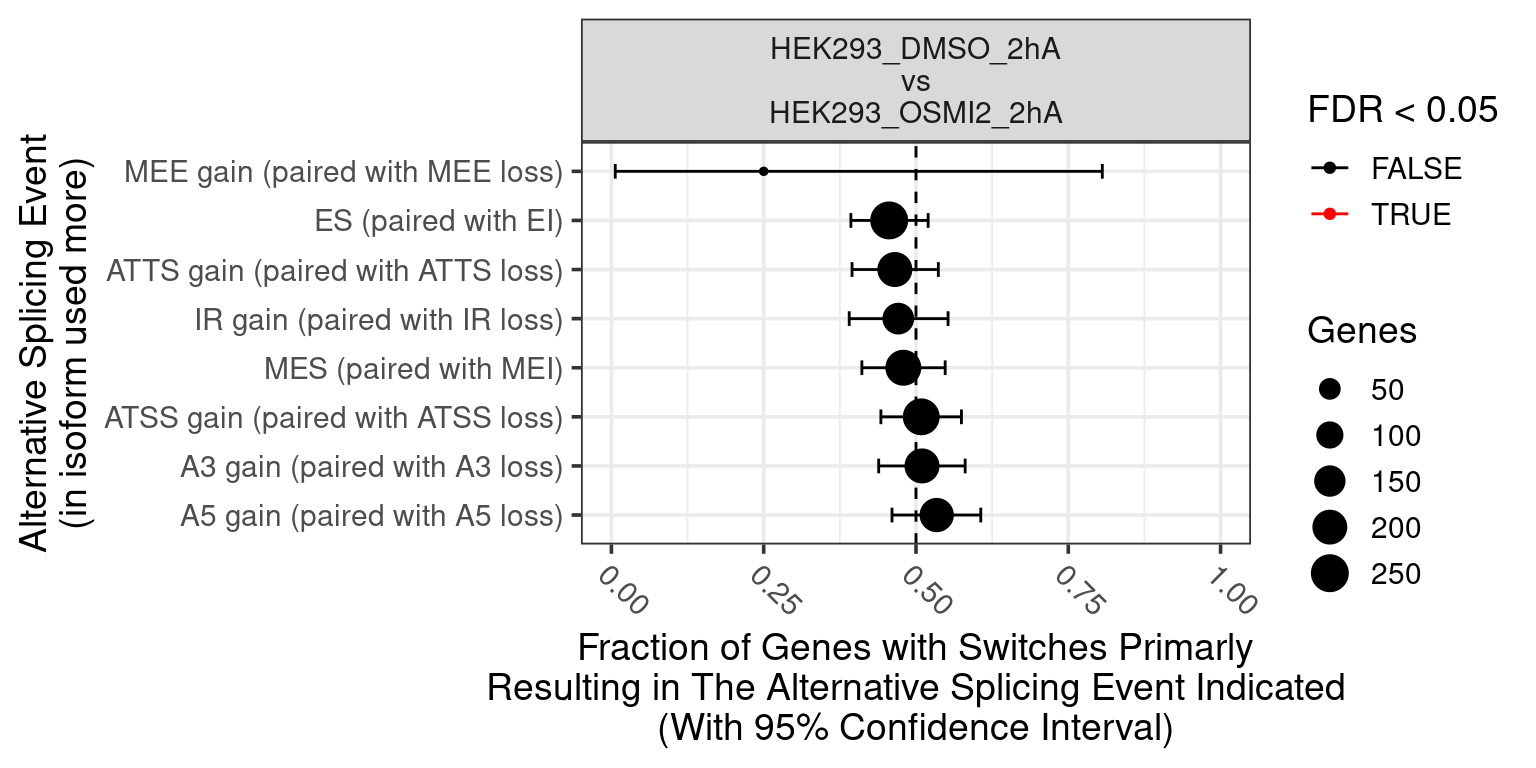
Comparison Analysis
This below result answered the question: How does the isoform usage of all isoforms utilizing a particular splicing type change - in other words is all isoforms or only a subset of isoforms that are affected.
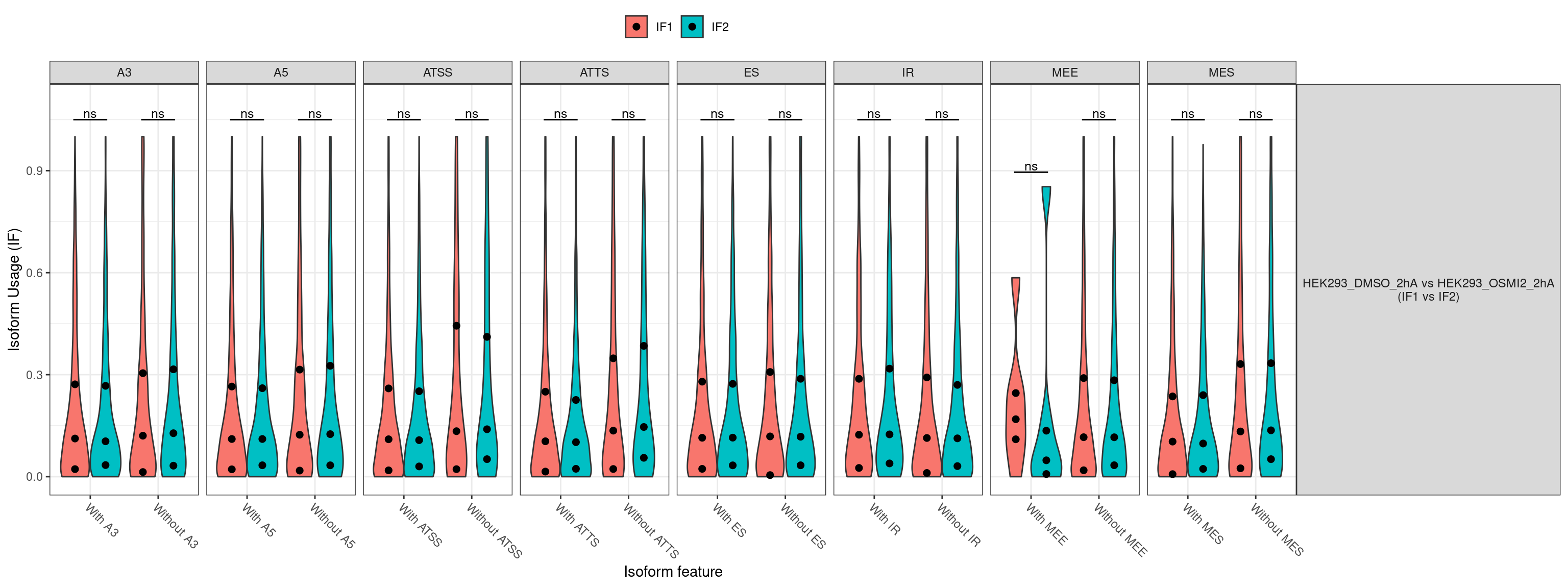
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- IR : Intron Retention.
- A5 : Alternative 5’ donor site (changes in the 5’end of the upstream exon).
- A3 : Alternative 3’ acceptor site (changes in the 3’end of the downstream exon).
- ATSS : Alternative Transcription Start Site.
- ATTS : Alternative Transcription Termination Site.
- ES : Exon Skipping (EI means Exon Inclusion).
- MES : Multiple Exon Skipping. Skipping of >1 consecutive exons. (MEI means Multiple Exon Inclusion).
- MEE : Mutually Exclusive Exons.
Alternative Splicing Consequence
Amount of Event
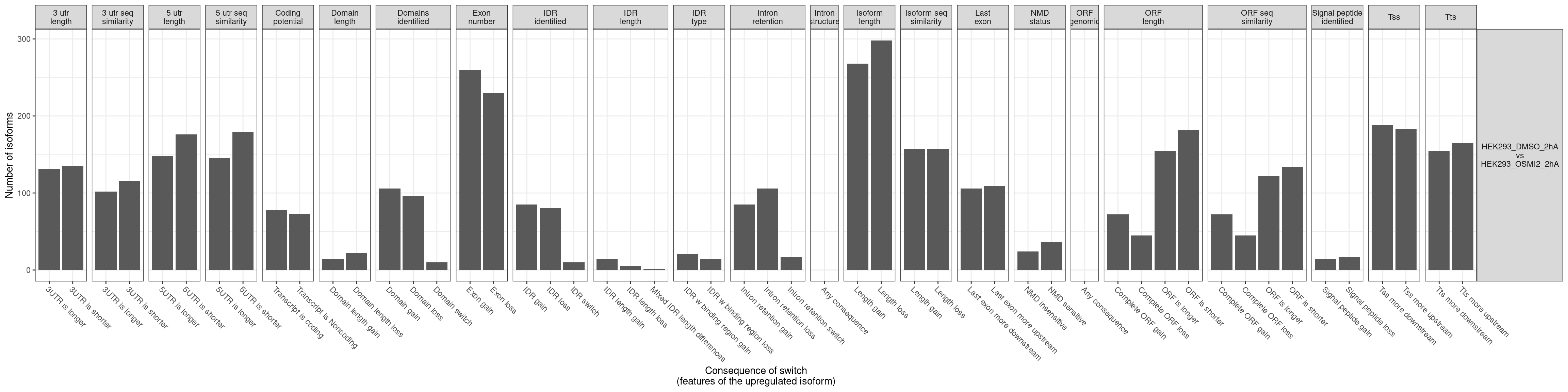
Enrichment Analysis
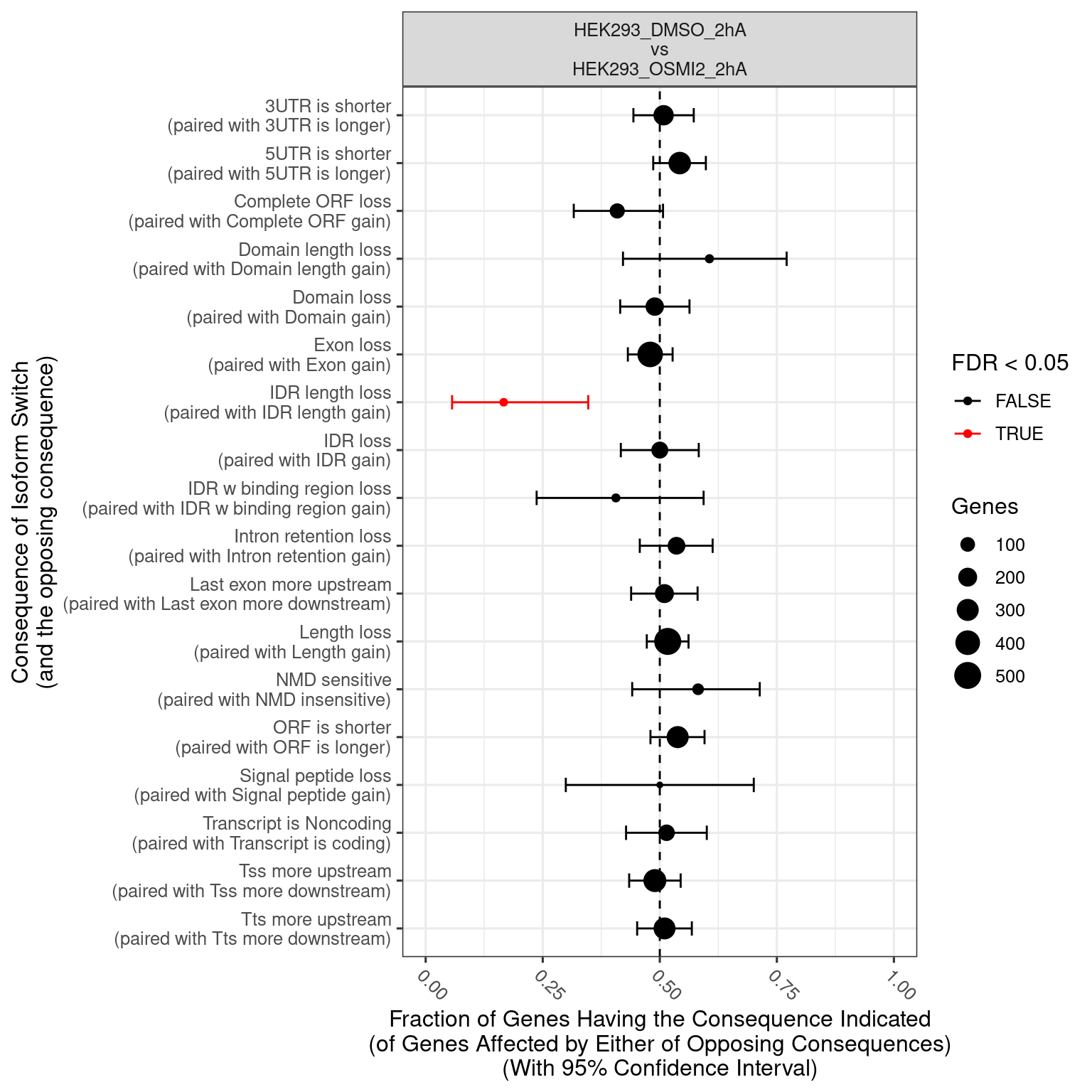
Comparison Analysis
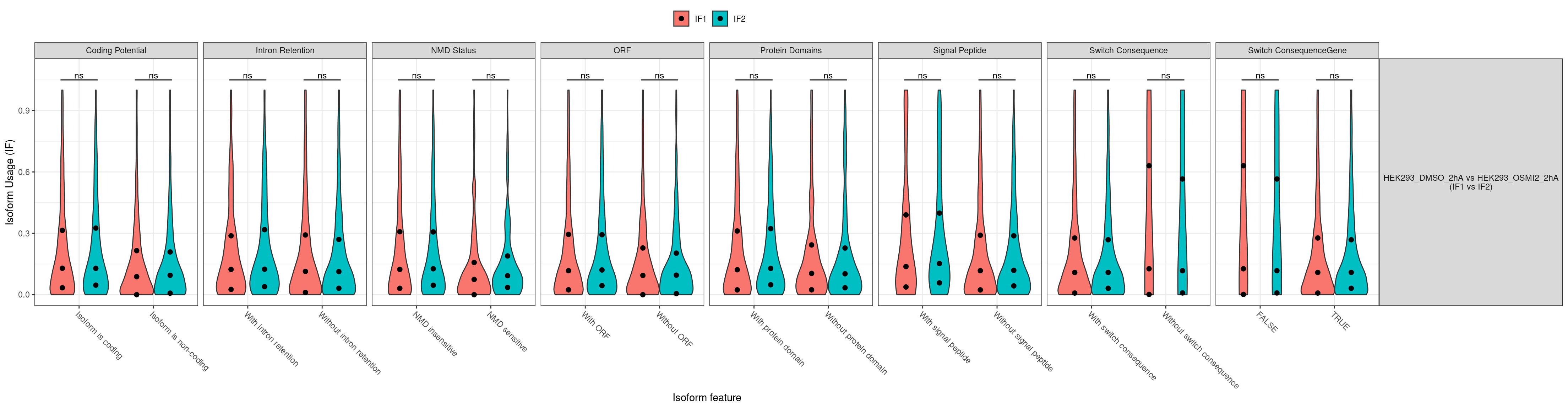
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- tss : Test transcripts for whether they use different Transcription Start Site (TSS) (more than ntCutoff from each other).
- tts : Test transcripts for whether they use different Transcription Termination Site (TTS) (more than ntCutoff from each other).
- last_exon : Test whether transcripts utilizes different last exons (defined as the last exon of each transcript is non-overlapping).
- isoform_seq_similarity : Test whether the isoform nucleotide sequences are different (as described above). Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff.
- isoform_length : Test transcripts for differences in isoform length. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Please * note that this is a less powerful analysis than implemented in ‘isoform_seq_similarity’ as two equally long sequences might be very different.
- exon_number : Test transcripts for differences in exon number.
- intron_structure : Test transcripts for differences in intron structure, e.g. usage of exon-exon junctions. This analysis corresponds to analyzing whether all introns in one isoform is also found in the other isoforms.
- intron_retention : Test for differences in intron retentions (and their genomic positions). Require that analyzeIntronRetention have been run.
- isoform_class_code : Test transcripts for differences in the transcript classification provide by cufflinks. For a updated list of class codes see http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/#transfrag-class-codes.
- coding_potential : Test transcripts for differences in coding potential, as indicated by the CPAT or CPC2 analysis. Requires that analyzeCPAT or analyzeCPC2 have been used to add external conding potential analysis to the switchAnalyzeRlist.
- ORF_seq_similarity : Test whether the amino acid sequences of the ORFs are different (as described above). Reported as different if the measured JCsim is smaller than AaJCsimCutoff and the length difference of the aligned and combined region is larger than AaCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_genomic : Test transcripts for differences in genomic position of the Open Reading Frames (ORF). Requires that least one of the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and settingaddAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_length : Test transcripts for differences in length of Open Reading Frames (ORF). Note that this is a less powerful analysis than implemented in ORF_seq_similarity as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 5’ UnTranslated Region (UTR) are different (as described above). The 5’UTR is defined as the region from the transcript start to the ORF start. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_length : Test transcripts for differences in the length of the 5’ UnTranslated Region (UTR). The 5’UTR is defined as the region from the transcript start to the ORF start. Note that this is a less powerful analysis than implemented in ‘5_utr_seq_similarity’ as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 3’ UnTranslated Region (UTR) are different (as described above). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_length : Test transcripts for differences in the length of the 3’ UnTranslated Regions (UTR). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Note that this is a less powerful analysis than implemented in 3_utr_seq_similarity as two equally long sequences might be very different. Requires that identifyORF have been used to predict NMD sensitivity or that the ORF was imported though one of the dedicated import functions implemented in isoformSwitchAnalyzeR. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- NMD_status : Test transcripts for differences in sensitivity to Nonsense Mediated Decay (NMD). Requires that both the isoforms have been annotated with PTC either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domains_identified : Test transcripts for differences in the name and order of which domains are identified by the Pfam in the transcripts. Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domain_length : Test transcripts for differences in the length of overlapping domains of the same type (same hmm_name) thereby enabling analysis of protein domain truncation. Do however note that a small difference in length is will likely not truncate the protein domain. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff .Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- genomic_domain_position : Test transcripts for differences in the genomic position of the domains identified by the Pfam analysis (Will be different unless the two isoforms have the same domains at the same genomic location). Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
- IDR_identified : Test for differences in isoform IDRs. Specifically the two isoforms are analyzed for whether they contain IDRs which do not overlap in genomic coordinates. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_length : Test for differences in the length of overlapping (in genomic coordinates) IDRs. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_type : Test for differences in IDR type. Specifically the two isoforms are tested for overlapping IDRs (genomic coordinates) and overlapping IDRs are compared with regards to their IDR type (IDR vs IDR w binding site). Only available if analyzeIUPred2A was used to add external IDR analysis to the switchAnalyzeRlist.
- signal_peptide_identified : Test transcripts for differences in whether a signal peptide was identified or not by the SignalP analysis. Requires that analyzeSignalP have been used to add external SignalP analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via analyzeORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
Volcano Plot
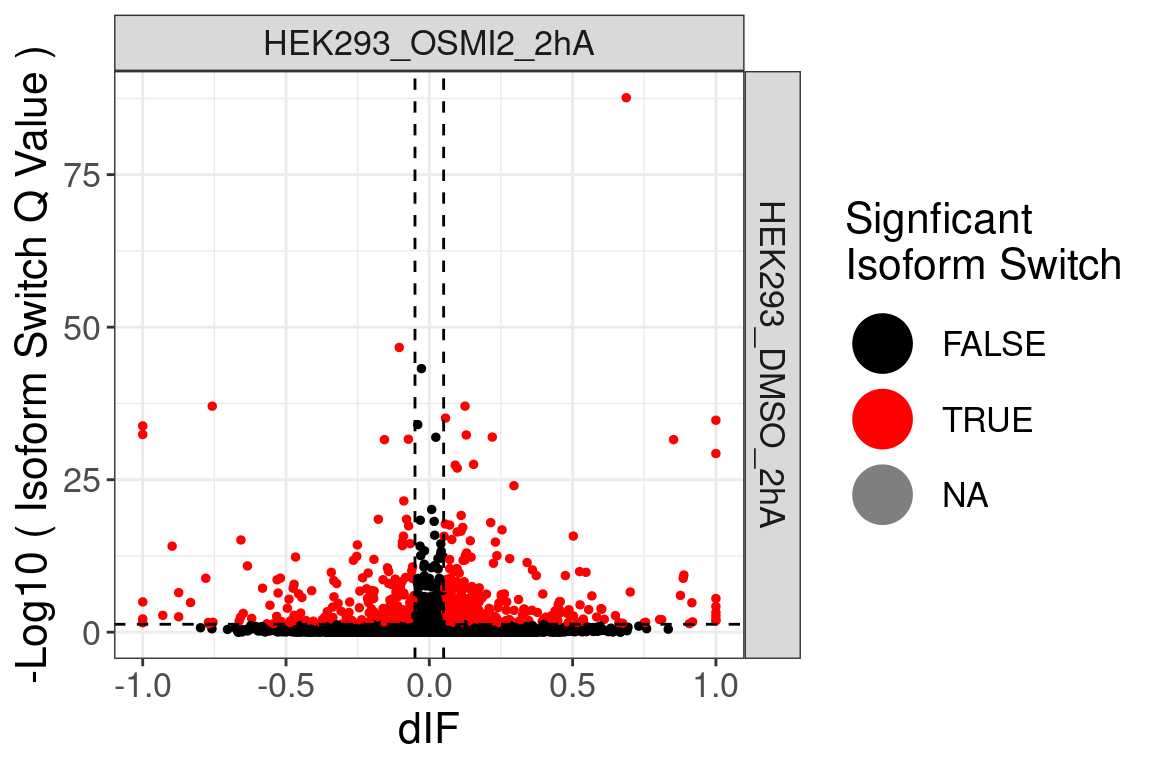
Spreadsheet
| iso_ref | gene_ref | isoform_id | gene_id | gene_name | condition_1 | condition_2 | IF1 | IF2 | dIF | isoform_switch_q_value | switchConsequencesGene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| isoComp_00035364 | geneComp_00003256 | ENST00000222572 | ENSG00000105854 | PON2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.687 | 0.687 | 2.452278e-88 | FALSE |
| isoComp_00094081 | geneComp_00009492 | ENST00000392595 | ENSG00000151500 | THYN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.105 | 0.000 | -0.105 | 2.128896e-47 | TRUE |
| isoComp_00035379 | geneComp_00003256 | ENST00000633531 | ENSG00000105854 | PON2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.757 | 0.000 | -0.757 | 8.847407e-38 | FALSE |
| isoComp_00119858 | geneComp_00012569 | MSTRG.27382.9 | ENSG00000169228 | RAB24 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.125 | 0.125 | 8.847407e-38 | TRUE |
| isoComp_00101392 | geneComp_00010366 | ENST00000470533 | ENSG00000159140 | SON | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.056 | 0.056 | 8.206103e-36 | TRUE |
| isoComp_00172555 | geneComp_00024298 | ENST00000449175 | ENSG00000236901 | MIR600HG | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 1.797131e-35 | FALSE |
| isoComp_00172556 | geneComp_00024298 | ENST00000545631 | ENSG00000236901 | MIR600HG | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.000 | -1.000 | 1.553099e-34 | FALSE |
| isoComp_00165249 | geneComp_00021164 | ENST00000380526 | ENSG00000228175 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.000 | -1.000 | 3.879418e-33 | TRUE | |
| isoComp_00097464 | geneComp_00009921 | MSTRG.4459.14 | ENSG00000155256 | ZFYVE27 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.129 | 0.129 | 4.661579e-33 | |
| isoComp_00007894 | geneComp_00000712 | ENST00000466657 | ENSG00000052723 | SIKE1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.220 | 0.220 | 1.035219e-32 | TRUE |
| isoComp_00066437 | geneComp_00006583 | ENST00000683429 | ENSG00000131844 | MCCC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.073 | 0.000 | -0.073 | 2.330736e-32 | TRUE |
| isoComp_00191159 | geneComp_00032481 | ENST00000612061 | ENSG00000270276 | H4C15 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.852 | 0.852 | 2.722269e-32 | TRUE |
| isoComp_00084220 | geneComp_00008416 | MSTRG.2532.7 | ENSG00000143158 | MPC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.156 | 0.000 | -0.156 | 2.754245e-32 | TRUE |
| isoComp_00165250 | geneComp_00021164 | MSTRG.1568.1 | ENSG00000228175 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 5.148823e-30 | TRUE | |
| isoComp_00086998 | geneComp_00008701 | MSTRG.22724.6 | ENSG00000144677 | CTDSPL | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.154 | 0.154 | 3.094396e-28 | TRUE |
| isoComp_00068593 | geneComp_00006816 | MSTRG.568.6 | ENSG00000133226 | SRRM1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.090 | 0.090 | 4.096586e-28 | TRUE |
| isoComp_00086386 | geneComp_00008649 | ENST00000410019 | ENSG00000144354 | CDCA7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.097 | 0.097 | 1.252613e-27 | TRUE |
| isoComp_00159026 | geneComp_00018810 | ENST00000381223 | ENSG00000214717 | ZBED1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.295 | 0.295 | 9.647391e-25 | TRUE |
| isoComp_00143749 | geneComp_00016075 | ENST00000616016 | ENSG00000187634 | SAMD11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.089 | 0.000 | -0.089 | 3.032591e-22 | TRUE |
| isoComp_00016110 | geneComp_00001420 | ENST00000397113 | ENSG00000077782 | FGFR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.111 | 0.111 | 7.146172e-20 | TRUE |
| isoComp_00075520 | geneComp_00007545 | ENST00000319027 | ENSG00000137343 | ATAT1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.178 | 0.000 | -0.178 | 3.075598e-19 | TRUE |
| isoComp_00080461 | geneComp_00008055 | ENST00000268058 | ENSG00000140464 | PML | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.079 | 0.000 | -0.079 | 3.075598e-19 | TRUE |
| isoComp_00018485 | geneComp_00001629 | ENST00000196482 | ENSG00000083812 | ZNF324 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.214 | 0.214 | 1.079816e-18 | TRUE |
| isoComp_00100723 | geneComp_00010299 | ENST00000475533 | ENSG00000158552 | ZFAND2B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.056 | 0.056 | 1.821352e-18 | TRUE |
| isoComp_00193552 | geneComp_00033875 | MSTRG.21595.16 | ENSG00000274602 | PI4KAP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.071 | 0.071 | 2.772699e-18 | TRUE |
| isoComp_00193553 | geneComp_00033875 | MSTRG.21595.17 | ENSG00000274602 | PI4KAP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.072 | 0.000 | -0.072 | 3.641426e-18 | TRUE |
| isoComp_00077541 | geneComp_00007757 | MSTRG.19811.6 | ENSG00000138442 | WDR12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.117 | 0.117 | 6.562402e-18 | TRUE |
| isoComp_00020077 | geneComp_00001772 | ENST00000548909 | ENSG00000087502 | ERGIC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.254 | 0.254 | 1.684245e-17 | TRUE |
| isoComp_00164616 | geneComp_00020902 | ENST00000423764 | ENSG00000227372 | TP73-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.111 | 0.111 | 2.801610e-17 | TRUE |
| isoComp_00184313 | geneComp_00029032 | ENST00000508582 | ENSG00000258366 | RTEL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.097 | 0.097 | 3.680151e-17 | TRUE |
| isoComp_00046711 | geneComp_00004412 | ENST00000360990 | ENSG00000115041 | KCNIP3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.502 | 0.502 | 1.740983e-16 | TRUE |
| isoComp_00123367 | geneComp_00013063 | ENST00000503251 | ENSG00000171566 | PLRG1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.090 | 0.000 | -0.090 | 1.799852e-16 | TRUE |
| isoComp_00059225 | geneComp_00005803 | MSTRG.20406.35 | ENSG00000125844 | RRBP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.052 | 0.052 | 1.874191e-16 | TRUE |
| isoComp_00080474 | geneComp_00008055 | ENST00000565898 | ENSG00000140464 | PML | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.078 | 0.078 | 6.428825e-16 | TRUE |
| isoComp_00073018 | geneComp_00007275 | ENST00000258428 | ENSG00000135945 | REV1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.657 | 0.000 | -0.657 | 7.662421e-16 | TRUE |
| isoComp_00059309 | geneComp_00005816 | ENST00000680050 | ENSG00000125875 | TBC1D20 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.143 | 0.143 | 1.043137e-15 | TRUE |
| isoComp_00119853 | geneComp_00012569 | ENST00000478234 | ENSG00000169228 | RAB24 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.094 | 0.000 | -0.094 | 1.556560e-15 | TRUE |
| isoComp_00097271 | geneComp_00009895 | ENST00000428648 | ENSG00000154978 | VOPP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.231 | 0.231 | 1.699570e-15 | TRUE |
| isoComp_00059221 | geneComp_00005803 | MSTRG.20406.24 | ENSG00000125844 | RRBP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.067 | 0.000 | -0.067 | 3.105591e-15 | TRUE |
| isoComp_00081587 | geneComp_00008152 | ENST00000580430 | ENSG00000141068 | KSR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.251 | 0.000 | -0.251 | 4.890892e-15 | TRUE |
| isoComp_00143844 | geneComp_00016097 | ENST00000521604 | ENSG00000187735 | TCEA1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.992 | 0.898 | -0.094 | 6.474699e-15 | TRUE |
| isoComp_00189451 | geneComp_00031654 | ENST00000590750 | ENSG00000267260 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.897 | 0.000 | -0.897 | 7.946070e-15 | TRUE | |
| isoComp_00003215 | geneComp_00000280 | MSTRG.6631.5 | ENSG00000011105 | TSPAN9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.130 | 0.130 | 1.030861e-13 | TRUE |
| isoComp_00012957 | geneComp_00001160 | MSTRG.18958.24 | ENSG00000071054 | MAP4K4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.126 | 0.126 | 2.818816e-13 | TRUE |
| isoComp_00055526 | geneComp_00005361 | ENST00000490776 | ENSG00000122642 | FKBP9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.236 | 0.236 | 2.858906e-13 | TRUE |
| isoComp_00187793 | geneComp_00030845 | ENST00000409691 | ENSG00000263020 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.253 | 0.000 | -0.253 | 3.727615e-13 | TRUE | |
| isoComp_00046713 | geneComp_00004412 | ENST00000468529 | ENSG00000115041 | KCNIP3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.930 | 0.463 | -0.467 | 4.713407e-13 | TRUE |
| isoComp_00075521 | geneComp_00007545 | ENST00000329992 | ENSG00000137343 | ATAT1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.146 | 0.146 | 5.021877e-13 | TRUE |
| isoComp_00061432 | geneComp_00006041 | ENST00000422064 | ENSG00000127957 | PMS2P3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.280 | 0.280 | 8.718435e-13 | TRUE |
| isoComp_00025474 | geneComp_00002271 | ENST00000266254 | ENSG00000100271 | TTLL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.807 | -0.193 | 1.148402e-12 | TRUE |
| isoComp_00148107 | geneComp_00016722 | ENST00000497920 | ENSG00000196843 | ARID5A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.071 | 0.071 | 1.207987e-12 | TRUE |
| isoComp_00132826 | geneComp_00014365 | ENST00000473274 | ENSG00000178752 | ERFE | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.124 | 0.124 | 1.495634e-12 | TRUE |
| isoComp_00014507 | geneComp_00001289 | ENST00000505666 | ENSG00000074621 | SLC24A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.265 | 0.000 | -0.265 | 1.634927e-12 | TRUE |
| isoComp_00142916 | geneComp_00015933 | ENST00000450982 | ENSG00000186792 | HYAL3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.118 | 0.118 | 1.672765e-12 | TRUE |
| isoComp_00113934 | geneComp_00011892 | ENST00000299563 | ENSG00000166439 | RNF169 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.656 | 0.998 | 0.341 | 3.791962e-12 | TRUE |
| isoComp_00077823 | geneComp_00007789 | ENST00000264360 | ENSG00000138650 | PCDH10 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.224 | 0.224 | 4.995181e-12 | TRUE |
| isoComp_00059430 | geneComp_00005830 | ENST00000479502 | ENSG00000125962 | ARMCX5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.635 | 0.000 | -0.635 | 1.364537e-11 | TRUE |
| isoComp_00049784 | geneComp_00004707 | ENST00000472584 | ENSG00000116922 | C1orf109 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.058 | 0.000 | -0.058 | 1.934506e-11 | |
| isoComp_00155790 | geneComp_00017922 | ENST00000389022 | ENSG00000205309 | NT5M | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.146 | 0.000 | -0.146 | 2.771112e-11 | TRUE |
| isoComp_00035746 | geneComp_00003301 | ENST00000428288 | ENSG00000106049 | HIBADH | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.071 | 0.071 | 3.683159e-11 | TRUE |
| isoComp_00050898 | geneComp_00004824 | ENST00000462759 | ENSG00000117859 | OSBPL9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.095 | 0.095 | 3.748953e-11 | TRUE |
| isoComp_00037677 | geneComp_00003512 | ENST00000370228 | ENSG00000107815 | TWNK | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.097 | 0.097 | 4.362530e-11 | TRUE |
| isoComp_00091161 | geneComp_00009178 | ENST00000277575 | ENSG00000148429 | USP6NL | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.360 | 0.360 | 6.091263e-11 | TRUE |
| isoComp_00144323 | geneComp_00016183 | ENST00000338498 | ENSG00000188070 | ZFTA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.061 | 0.000 | -0.061 | 9.018916e-11 | |
| isoComp_00143839 | geneComp_00016097 | ENST00000396401 | ENSG00000187735 | TCEA1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.088 | 0.088 | 1.103757e-10 | TRUE |
| isoComp_00161058 | geneComp_00019495 | ENST00000443631 | ENSG00000223478 | ZDHHC12-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.858 | -0.142 | 1.103757e-10 | TRUE |
| isoComp_00073019 | geneComp_00007275 | ENST00000393445 | ENSG00000135945 | REV1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.524 | 0.524 | 1.160408e-10 | TRUE |
| isoComp_00130462 | geneComp_00014017 | MSTRG.14912.7 | ENSG00000176809 | LRRC37A3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.547 | 0.547 | 1.525458e-10 | TRUE |
| isoComp_00113935 | geneComp_00011892 | ENST00000527301 | ENSG00000166439 | RNF169 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.342 | 0.000 | -0.342 | 1.550435e-10 | TRUE |
| isoComp_00022505 | geneComp_00001987 | ENST00000455138 | ENSG00000092203 | TOX4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.104 | 0.104 | 2.007093e-10 | TRUE |
| isoComp_00106890 | geneComp_00011018 | ENST00000581168 | ENSG00000163249 | CCNYL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.214 | 0.000 | -0.214 | 2.007093e-10 | TRUE |
| isoComp_00126184 | geneComp_00013416 | ENST00000339249 | ENSG00000173275 | ZNF449 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.888 | 0.888 | 4.208620e-10 | TRUE |
| isoComp_00199964 | geneComp_00037765 | ENST00000693426 | ENSG00000287750 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.475 | 0.475 | 5.231425e-10 | TRUE | |
| isoComp_00151469 | geneComp_00017159 | ENST00000528420 | ENSG00000198382 | UVRAG | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.373 | 0.373 | 5.254365e-10 | TRUE |
| isoComp_00148105 | geneComp_00016722 | ENST00000467498 | ENSG00000196843 | ARID5A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.083 | 0.000 | -0.083 | 1.073118e-09 | TRUE |
| isoComp_00097268 | geneComp_00009895 | ENST00000285279 | ENSG00000154978 | VOPP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.990 | 0.758 | -0.233 | 1.216134e-09 | TRUE |
| isoComp_00028139 | geneComp_00002517 | ENST00000400217 | ENSG00000101266 | CSNK2A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.074 | 0.074 | 1.221576e-09 | TRUE |
| isoComp_00043775 | geneComp_00004127 | ENST00000611340 | ENSG00000112425 | EPM2A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.520 | 0.000 | -0.520 | 1.345886e-09 | TRUE |
| isoComp_00071405 | geneComp_00007120 | ENST00000453144 | ENSG00000135241 | PNPLA8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.780 | 0.000 | -0.780 | 1.440227e-09 | TRUE |
| isoComp_00189454 | geneComp_00031654 | ENST00000665759 | ENSG00000267260 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.886 | 0.886 | 1.470320e-09 | TRUE | |
| isoComp_00011066 | geneComp_00000996 | ENST00000489377 | ENSG00000066697 | MSANTD3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.059 | 0.000 | -0.059 | 1.572702e-09 | TRUE |
| isoComp_00014175 | geneComp_00001252 | ENST00000540156 | ENSG00000073614 | KDM5A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.119 | 0.000 | -0.119 | 1.965773e-09 | TRUE |
| isoComp_00158801 | geneComp_00018748 | ENST00000423273 | ENSG00000214413 | BBIP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.161 | 0.000 | -0.161 | 2.468849e-09 | TRUE |
| isoComp_00011647 | geneComp_00001045 | ENST00000481110 | ENSG00000068078 | FGFR3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.062 | 0.062 | 2.533355e-09 | TRUE |
| isoComp_00151465 | geneComp_00017159 | ENST00000356136 | ENSG00000198382 | UVRAG | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.908 | 0.377 | -0.531 | 2.572484e-09 | TRUE |
| isoComp_00113685 | geneComp_00011866 | ENST00000643516 | ENSG00000166340 | TPP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.055 | 0.055 | 2.672134e-09 | TRUE |
| isoComp_00151073 | geneComp_00017113 | MSTRG.9109.17 | ENSG00000198176 | TFDP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.001 | 0.062 | 0.061 | 3.099190e-09 | TRUE |
| isoComp_00187598 | geneComp_00030778 | ENST00000572730 | ENSG00000262580 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.333 | 0.000 | -0.333 | 3.547938e-09 | TRUE | |
| isoComp_00023493 | geneComp_00002072 | ENST00000534400 | ENSG00000096070 | BRPF3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.068 | 0.068 | 3.681374e-09 | |
| isoComp_00116712 | geneComp_00012211 | ENST00000577701 | ENSG00000167733 | HSD11B1L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.095 | 0.095 | 4.925694e-09 | TRUE |
| isoComp_00012950 | geneComp_00001160 | MSTRG.18958.16 | ENSG00000071054 | MAP4K4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.095 | 0.000 | -0.095 | 4.941302e-09 | TRUE |
| isoComp_00038108 | geneComp_00003548 | MSTRG.3486.20 | ENSG00000108021 | TASOR2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.122 | 0.122 | 5.374076e-09 | TRUE |
| isoComp_00136532 | geneComp_00014954 | ENST00000637083 | ENSG00000182372 | CLN8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.142 | 0.000 | -0.142 | 1.011750e-08 | TRUE |
| isoComp_00195349 | geneComp_00034881 | MSTRG.21120.11 | ENSG00000278932 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.323 | 0.000 | -0.323 | 1.044655e-08 | TRUE | |
| isoComp_00134715 | geneComp_00014654 | ENST00000586378 | ENSG00000180448 | ARHGAP45 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.472 | 0.000 | -0.472 | 1.395358e-08 | TRUE |
| isoComp_00169032 | geneComp_00022784 | ENST00000667789 | ENSG00000232677 | LINC00665 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.130 | 0.000 | -0.130 | 1.648677e-08 | TRUE |
| isoComp_00094190 | geneComp_00009500 | ENST00000493014 | ENSG00000151576 | QTRT2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.059 | 0.059 | 1.812104e-08 | |
| isoComp_00027591 | geneComp_00002454 | MSTRG.20617.1 | ENSG00000101000 | PROCR | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.148 | 0.148 | 3.312618e-08 | TRUE |
| isoComp_00017492 | geneComp_00001535 | ENST00000520266 | ENSG00000080823 | MOK | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.074 | 0.074 | 3.472962e-08 | TRUE |
| isoComp_00078356 | geneComp_00007826 | ENST00000394735 | ENSG00000138785 | INTS12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.101 | 0.000 | -0.101 | 3.584926e-08 | TRUE |
| isoComp_00125318 | geneComp_00013326 | ENST00000303334 | ENSG00000172828 | CES3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.118 | 0.118 | 3.881710e-08 | TRUE |
| isoComp_00158962 | geneComp_00018796 | ENST00000466568 | ENSG00000214655 | ZSWIM8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.083 | 0.083 | 5.017663e-08 | TRUE |
| isoComp_00182555 | geneComp_00028203 | MSTRG.4920.3 | ENSG00000255284 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.054 | 0.054 | 5.312669e-08 | TRUE | |
| isoComp_00162561 | geneComp_00020077 | MSTRG.28453.8 | ENSG00000225096 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.174 | 0.174 | 5.331690e-08 | TRUE | |
| isoComp_00155835 | geneComp_00017929 | ENST00000429243 | ENSG00000205352 | PRR13 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.581 | 0.663 | 0.082 | 5.921241e-08 | TRUE |
| isoComp_00050066 | geneComp_00004735 | ENST00000370625 | ENSG00000117151 | CTBS | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.582 | 0.000 | -0.582 | 6.155796e-08 | TRUE |
| isoComp_00084085 | geneComp_00008398 | ENST00000271227 | ENSG00000143036 | SLC44A3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.475 | 0.000 | -0.475 | 6.259798e-08 | TRUE |
| isoComp_00014044 | geneComp_00001243 | ENST00000167462 | ENSG00000073350 | LLGL2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.056 | 0.056 | 6.438388e-08 | |
| isoComp_00053271 | geneComp_00005116 | ENST00000367710 | ENSG00000120334 | CENPL | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.219 | 0.000 | -0.219 | 7.563173e-08 | TRUE |
| isoComp_00110931 | geneComp_00011508 | MSTRG.29364.6 | ENSG00000164877 | MICALL2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.163 | 0.163 | 8.658269e-08 | TRUE |
| isoComp_00127296 | geneComp_00013565 | ENST00000393709 | ENSG00000174151 | CYB561D1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.144 | 0.144 | 9.658182e-08 | TRUE |
| isoComp_00080718 | geneComp_00008077 | MSTRG.11476.22 | ENSG00000140543 | DET1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.112 | 0.112 | 9.755226e-08 | TRUE |
| isoComp_00085031 | geneComp_00008501 | ENST00000612340 | ENSG00000143493 | INTS7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.081 | 0.081 | 1.212400e-07 | TRUE |
| isoComp_00153779 | geneComp_00017569 | ENST00000654131 | ENSG00000204054 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.115 | 0.115 | 1.467699e-07 | TRUE | |
| isoComp_00018487 | geneComp_00001629 | ENST00000593925 | ENSG00000083812 | ZNF324 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.923 | 0.723 | -0.200 | 1.510200e-07 | TRUE |
| isoComp_00098721 | geneComp_00010074 | ENST00000650232 | ENSG00000156650 | KAT6B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.410 | 0.000 | -0.410 | 1.577978e-07 | TRUE |
| isoComp_00140470 | geneComp_00015575 | MSTRG.32231.10 | ENSG00000185189 | NRBP2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.192 | 0.000 | -0.192 | 1.799470e-07 | TRUE |
| isoComp_00115491 | geneComp_00012084 | ENST00000588183 | ENSG00000167220 | HDHD2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.259 | 0.016 | -0.242 | 1.826569e-07 | TRUE |
| isoComp_00038297 | geneComp_00003562 | MSTRG.4276.2 | ENSG00000108219 | TSPAN14 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.075 | 0.075 | 1.861957e-07 | TRUE |
| isoComp_00049631 | geneComp_00004687 | ENST00000369604 | ENSG00000116793 | PHTF1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.116 | 0.000 | -0.116 | 2.524878e-07 | TRUE |
| isoComp_00090273 | geneComp_00009090 | ENST00000424843 | ENSG00000147650 | LRP12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.701 | 0.701 | 2.565762e-07 | TRUE |
| isoComp_00134328 | geneComp_00014581 | ENST00000528032 | ENSG00000180035 | ZNF48 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.167 | 0.167 | 2.738250e-07 | TRUE |
| isoComp_00126187 | geneComp_00013416 | MSTRG.34935.4 | ENSG00000173275 | ZNF449 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.874 | 0.000 | -0.874 | 3.273219e-07 | TRUE |
| isoComp_00100773 | geneComp_00010307 | ENST00000689921 | ENSG00000158615 | PPP1R15B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.111 | 0.111 | 3.357082e-07 | TRUE |
| isoComp_00059316 | geneComp_00005816 | ENST00000681129 | ENSG00000125875 | TBC1D20 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.091 | 0.000 | -0.091 | 3.421031e-07 | TRUE |
| isoComp_00173220 | geneComp_00024517 | ENST00000422847 | ENSG00000237523 | LINC00857 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.528 | 0.000 | -0.528 | 3.781029e-07 | TRUE |
| isoComp_00057583 | geneComp_00005606 | ENST00000482536 | ENSG00000124508 | BTN2A2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.168 | 0.168 | 4.405860e-07 | TRUE |
| isoComp_00010069 | geneComp_00000904 | MSTRG.16756.30 | ENSG00000064607 | SUGP2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.058 | 0.058 | 4.575228e-07 | TRUE |
| isoComp_00181071 | geneComp_00027474 | ENST00000569384 | ENSG00000253352 | TUG1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.089 | 0.000 | -0.089 | 4.579320e-07 | TRUE |
| isoComp_00026837 | geneComp_00002404 | ENST00000527416 | ENSG00000100697 | DICER1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.196 | 0.000 | -0.196 | 4.962579e-07 | TRUE |
| isoComp_00035339 | geneComp_00003252 | ENST00000680823 | ENSG00000105835 | NAMPT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.262 | 0.262 | 5.180054e-07 | TRUE |
| isoComp_00019185 | geneComp_00001699 | ENST00000371566 | ENSG00000085840 | ORC1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.064 | 0.064 | 5.210124e-07 | |
| isoComp_00156201 | geneComp_00017994 | ENST00000557704 | ENSG00000205683 | DPF3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.456 | 0.000 | -0.456 | 5.219731e-07 | TRUE |
| isoComp_00165339 | geneComp_00021203 | ENST00000690240 | ENSG00000228288 | PCAT6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.396 | 0.396 | 5.452185e-07 | TRUE |
| isoComp_00098643 | geneComp_00010065 | ENST00000625464 | ENSG00000156531 | PHF6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.199 | 0.199 | 5.720728e-07 | TRUE |
| isoComp_00064181 | geneComp_00006371 | ENST00000539529 | ENSG00000130558 | OLFM1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.251 | 0.251 | 6.924052e-07 | TRUE |
| isoComp_00113752 | geneComp_00011875 | ENST00000446510 | ENSG00000166352 | IFTAP | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.089 | 0.000 | -0.089 | 7.750692e-07 | TRUE |
| isoComp_00192819 | geneComp_00033447 | ENST00000609242 | ENSG00000273162 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.876 | 0.876 | 9.368368e-07 | TRUE | |
| isoComp_00082378 | geneComp_00008217 | ENST00000392467 | ENSG00000141524 | TMC6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.053 | 0.000 | -0.053 | 9.370465e-07 | TRUE |
| isoComp_00050068 | geneComp_00004735 | ENST00000465118 | ENSG00000117151 | CTBS | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.567 | 0.567 | 1.108081e-06 | TRUE |
| isoComp_00071523 | geneComp_00007130 | ENST00000447838 | ENSG00000135299 | ANKRD6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.252 | 0.252 | 1.197650e-06 | TRUE |
| isoComp_00009863 | geneComp_00000892 | ENST00000394162 | ENSG00000064225 | ST3GAL6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.333 | 0.000 | -0.333 | 1.577435e-06 | TRUE |
| isoComp_00200830 | geneComp_00038384 | ENST00000684887 | ENSG00000289088 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.791 | -0.209 | 1.581259e-06 | TRUE | |
| isoComp_00148436 | geneComp_00016769 | ENST00000440598 | ENSG00000197008 | ZNF138 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.114 | 0.003 | -0.111 | 1.668911e-06 | TRUE |
| isoComp_00071709 | geneComp_00007145 | ENST00000532578 | ENSG00000135362 | PRR5L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.444 | 0.000 | -0.444 | 2.072112e-06 | TRUE |
| isoComp_00161059 | geneComp_00019495 | MSTRG.33468.2 | ENSG00000223478 | ZDHHC12-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.142 | 0.142 | 2.178780e-06 | TRUE |
| isoComp_00004973 | geneComp_00000450 | ENST00000555159 | ENSG00000025423 | HSD17B6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.184 | 0.184 | 2.290658e-06 | TRUE |
| isoComp_00025291 | geneComp_00002260 | ENST00000427222 | ENSG00000100239 | PPP6R2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.060 | 0.060 | 2.375188e-06 | TRUE |
| isoComp_00074668 | geneComp_00007456 | MSTRG.33408.10 | ENSG00000136895 | GARNL3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.216 | 0.000 | -0.216 | 2.808188e-06 | TRUE |
| isoComp_00023633 | geneComp_00002091 | MSTRG.1591.3 | ENSG00000097046 | CDC7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.098 | 0.098 | 2.906004e-06 | TRUE |
| isoComp_00194317 | geneComp_00034307 | MSTRG.10313.13 | ENSG00000276550 | HERC2P2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.091 | 0.091 | 2.974636e-06 | TRUE |
| isoComp_00124030 | geneComp_00013153 | ENST00000269703 | ENSG00000171954 | CYP4F22 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 3.009406e-06 | TRUE |
| isoComp_00085533 | geneComp_00008555 | MSTRG.3112.4 | ENSG00000143756 | FBXO28 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.108 | 0.108 | 3.011287e-06 | TRUE |
| isoComp_00194511 | geneComp_00034423 | MSTRG.30062.26 | ENSG00000277072 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.089 | 0.089 | 3.029053e-06 | TRUE | |
| isoComp_00016590 | geneComp_00001457 | MSTRG.31218.5 | ENSG00000078674 | PCM1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.194 | 0.000 | -0.194 | 3.064287e-06 | TRUE |
| isoComp_00107022 | geneComp_00011043 | ENST00000369553 | ENSG00000163349 | HIPK1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.133 | 0.133 | 3.095165e-06 | TRUE |
| isoComp_00047895 | geneComp_00004510 | MSTRG.19965.5 | ENSG00000115568 | ZNF142 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.097 | 0.097 | 3.176387e-06 | TRUE |
| isoComp_00036147 | geneComp_00003334 | MSTRG.30328.69 | ENSG00000106290 | TAF6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.063 | 0.000 | -0.063 | 3.320480e-06 | |
| isoComp_00089701 | geneComp_00009020 | MSTRG.34438.5 | ENSG00000147162 | OGT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.150 | 0.022 | -0.128 | 3.320480e-06 | TRUE |
| isoComp_00153965 | geneComp_00017594 | ENST00000643851 | ENSG00000204147 | ASAH2B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.490 | 0.000 | -0.490 | 3.894071e-06 | TRUE |
| isoComp_00006113 | geneComp_00000555 | MSTRG.24704.1 | ENSG00000038210 | PI4K2B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.311 | 0.311 | 4.355267e-06 | TRUE |
| isoComp_00010439 | geneComp_00000937 | ENST00000552903 | ENSG00000065357 | DGKA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.213 | 0.000 | -0.213 | 4.355267e-06 | |
| isoComp_00122590 | geneComp_00012942 | MSTRG.32164.12 | ENSG00000171045 | TSNARE1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.138 | 0.000 | -0.138 | 5.254349e-06 | TRUE |
| isoComp_00182160 | geneComp_00028017 | MSTRG.20763.5 | ENSG00000254806 | SYS1-DBNDD2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.176 | 0.176 | 6.060968e-06 | TRUE |
| isoComp_00198572 | geneComp_00036764 | ENST00000519520 | ENSG00000286140 | DERPC | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.941 | 0.884 | -0.057 | 6.197308e-06 | TRUE |
| isoComp_00107278 | geneComp_00011076 | ENST00000466306 | ENSG00000163467 | TSACC | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.063 | 0.063 | 6.249486e-06 | TRUE |
| isoComp_00127781 | geneComp_00013629 | ENST00000526296 | ENSG00000174516 | PELI3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.089 | 0.089 | 7.452854e-06 | TRUE |
| isoComp_00148020 | geneComp_00016713 | ENST00000357591 | ENSG00000196810 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.123 | 0.123 | 7.564459e-06 | TRUE | |
| isoComp_00038210 | geneComp_00003554 | ENST00000690092 | ENSG00000108094 | CUL2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.151 | 0.151 | 9.016941e-06 | TRUE |
| isoComp_00140203 | geneComp_00015535 | ENST00000327359 | ENSG00000185024 | BRF1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.064 | 0.064 | 9.069842e-06 | |
| isoComp_00151012 | geneComp_00017107 | ENST00000355356 | ENSG00000198160 | MIER1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.457 | 0.457 | 9.622248e-06 | TRUE |
| isoComp_00156202 | geneComp_00017994 | ENST00000614862 | ENSG00000205683 | DPF3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.454 | 0.454 | 9.784387e-06 | TRUE |
| isoComp_00075749 | geneComp_00007570 | ENST00000527495 | ENSG00000137500 | CCDC90B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.125 | 0.125 | 1.023460e-05 | TRUE |
| isoComp_00036933 | geneComp_00003422 | MSTRG.32903.22 | ENSG00000106829 | TLE4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.067 | 0.000 | -0.067 | 1.054640e-05 | TRUE |
| isoComp_00124031 | geneComp_00013153 | ENST00000601005 | ENSG00000171954 | CYP4F22 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.000 | -1.000 | 1.079540e-05 | TRUE |
| isoComp_00177464 | geneComp_00026109 | MSTRG.10570.4 | ENSG00000245849 | RAD51-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.270 | 0.270 | 1.080946e-05 | TRUE |
| isoComp_00151761 | geneComp_00017204 | ENST00000567693 | ENSG00000198554 | WDHD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.278 | 0.000 | -0.278 | 1.124538e-05 | TRUE |
| isoComp_00007517 | geneComp_00000682 | ENST00000466508 | ENSG00000049769 | PPP1R3F | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.173 | 0.173 | 1.198176e-05 | TRUE |
| isoComp_00171935 | geneComp_00024010 | ENST00000662986 | ENSG00000236088 | COX10-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.212 | 0.000 | -0.212 | 1.364712e-05 | TRUE |
| isoComp_00192822 | geneComp_00033447 | MSTRG.4520.3 | ENSG00000273162 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.833 | 0.000 | -0.833 | 1.373223e-05 | TRUE | |
| isoComp_00163631 | geneComp_00020497 | ENST00000693353 | ENSG00000226266 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.916 | 0.916 | 1.481140e-05 | TRUE | |
| isoComp_00147816 | geneComp_00016692 | MSTRG.29938.4 | ENSG00000196715 | VKORC1L1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.243 | 0.243 | 1.590442e-05 | TRUE |
| isoComp_00144165 | geneComp_00016150 | ENST00000530374 | ENSG00000187954 | CYHR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.089 | 0.021 | -0.068 | 1.686620e-05 | TRUE |
| isoComp_00197940 | geneComp_00036394 | ENST00000691526 | ENSG00000285533 | RELA-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.106 | 0.106 | 1.686620e-05 | TRUE |
| isoComp_00073375 | geneComp_00007309 | ENST00000651376 | ENSG00000136122 | BORA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.318 | 0.000 | -0.318 | 1.817132e-05 | TRUE |
| isoComp_00002272 | geneComp_00000202 | ENST00000341991 | ENSG00000008130 | NADK | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.048 | 0.208 | 0.160 | 1.836226e-05 | TRUE |
| isoComp_00016511 | geneComp_00001449 | ENST00000479315 | ENSG00000078487 | ZCWPW1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.450 | 0.450 | 2.987880e-05 | TRUE |
| isoComp_00183386 | geneComp_00028571 | ENST00000599258 | ENSG00000256683 | ZNF350 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.526 | 0.526 | 3.281600e-05 | TRUE |
| isoComp_00003853 | geneComp_00000334 | ENST00000605602 | ENSG00000013375 | PGM3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.127 | 0.127 | 3.415616e-05 | TRUE |
| isoComp_00016587 | geneComp_00001457 | MSTRG.31218.12 | ENSG00000078674 | PCM1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.127 | 0.127 | 3.770653e-05 | TRUE |
| isoComp_00155765 | geneComp_00017915 | ENST00000379426 | ENSG00000205269 | TMEM170B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.444 | -0.556 | 3.804357e-05 | TRUE |
| isoComp_00145507 | geneComp_00016385 | MSTRG.29077.12 | ENSG00000189007 | ADAT2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.115 | 0.115 | 3.841930e-05 | TRUE |
| isoComp_00099363 | geneComp_00010141 | MSTRG.30203.22 | ENSG00000157224 | CLDN12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.111 | 0.000 | -0.111 | 4.830618e-05 | TRUE |
| isoComp_00137178 | geneComp_00015056 | ENST00000651438 | ENSG00000182871 | COL18A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.789 | 0.871 | 0.082 | 5.352859e-05 | TRUE |
| isoComp_00077521 | geneComp_00007755 | ENST00000470129 | ENSG00000138434 | ITPRID2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.138 | 0.000 | -0.138 | 5.759726e-05 | TRUE |
| isoComp_00185322 | geneComp_00029564 | ENST00000664246 | ENSG00000259456 | ADNP-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 5.956971e-05 | TRUE |
| isoComp_00080225 | geneComp_00008039 | ENST00000563246 | ENSG00000140386 | SCAPER | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.337 | 0.000 | -0.337 | 7.556840e-05 | TRUE |
| isoComp_00100771 | geneComp_00010307 | ENST00000367188 | ENSG00000158615 | PPP1R15B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.895 | 0.760 | -0.135 | 7.588234e-05 | TRUE |
| isoComp_00138243 | geneComp_00015212 | ENST00000400309 | ENSG00000183570 | PCBP3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.136 | 0.136 | 8.073226e-05 | TRUE |
| isoComp_00141927 | geneComp_00015767 | ENST00000684972 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.062 | 0.062 | 8.444258e-05 | |
| isoComp_00111027 | geneComp_00011518 | ENST00000514725 | ENSG00000164902 | PHAX | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.063 | 0.063 | 8.474423e-05 | |
| isoComp_00103304 | geneComp_00010579 | ENST00000394933 | ENSG00000160606 | TLCD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.016 | 0.074 | 0.059 | 8.852685e-05 | TRUE |
| isoComp_00099462 | geneComp_00010158 | MSTRG.9254.3 | ENSG00000157379 | DHRS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.073 | 0.005 | -0.068 | 9.250332e-05 | TRUE |
| isoComp_00200276 | geneComp_00038008 | MSTRG.33001.5 | ENSG00000288271 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.240 | 0.240 | 9.250332e-05 | TRUE | |
| isoComp_00023408 | geneComp_00002063 | ENST00000489388 | ENSG00000095794 | CREM | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.124 | 0.124 | 9.485462e-05 | TRUE |
| isoComp_00011662 | geneComp_00001046 | ENST00000546325 | ENSG00000068079 | IFI35 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.129 | 0.000 | -0.129 | 9.562536e-05 | TRUE |
| isoComp_00024211 | geneComp_00002150 | ENST00000406522 | ENSG00000099889 | ARVCF | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.134 | 0.134 | 9.863683e-05 | TRUE |
| isoComp_00023626 | geneComp_00002091 | ENST00000234626 | ENSG00000097046 | CDC7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.809 | 0.565 | -0.244 | 9.903259e-05 | TRUE |
| isoComp_00102419 | geneComp_00010498 | ENST00000394458 | ENSG00000160117 | ANKLE1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.495 | 0.000 | -0.495 | 1.167896e-04 | TRUE |
| isoComp_00146983 | geneComp_00016599 | ENST00000604351 | ENSG00000196422 | PPP1R26 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.069 | 0.069 | 1.177687e-04 | TRUE |
| isoComp_00035887 | geneComp_00003311 | MSTRG.29665.12 | ENSG00000106100 | NOD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.209 | 0.209 | 1.252580e-04 | TRUE |
| isoComp_00038785 | geneComp_00003602 | MSTRG.13490.22 | ENSG00000108509 | CAMTA2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.015 | 0.123 | 0.108 | 1.274053e-04 | TRUE |
| isoComp_00099912 | geneComp_00010198 | MSTRG.30832.5 | ENSG00000157764 | BRAF | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.161 | 0.000 | -0.161 | 1.314415e-04 | TRUE |
| isoComp_00041871 | geneComp_00003902 | ENST00000379902 | ENSG00000111077 | TNS2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.599 | 0.599 | 1.386756e-04 | TRUE |
| isoComp_00089478 | geneComp_00008998 | ENST00000683021 | ENSG00000147050 | KDM6A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.110 | 0.000 | -0.110 | 1.432513e-04 | TRUE |
| isoComp_00055980 | geneComp_00005420 | MSTRG.31767.2 | ENSG00000123124 | WWP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.202 | 0.000 | -0.202 | 1.569618e-04 | TRUE |
| isoComp_00173223 | geneComp_00024517 | ENST00000671081 | ENSG00000237523 | LINC00857 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.331 | 0.331 | 1.663344e-04 | TRUE |
| isoComp_00189415 | geneComp_00031644 | ENST00000658811 | ENSG00000267248 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.330 | 0.330 | 1.670225e-04 | TRUE | |
| isoComp_00157369 | geneComp_00018342 | MSTRG.27382.25 | ENSG00000213347 | MXD3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 1.685264e-04 | TRUE |
| isoComp_00032131 | geneComp_00002925 | ENST00000678362 | ENSG00000104218 | CSPP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.154 | 0.000 | -0.154 | 1.697386e-04 | TRUE |
| isoComp_00136742 | geneComp_00014980 | ENST00000444422 | ENSG00000182511 | FES | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.602 | 0.602 | 1.711174e-04 | TRUE |
| isoComp_00006971 | geneComp_00000633 | MSTRG.12333.1 | ENSG00000047578 | KATNIP | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.093 | 0.093 | 1.719041e-04 | TRUE |
| isoComp_00126784 | geneComp_00013496 | MSTRG.5802.17 | ENSG00000173715 | C11orf80 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.133 | 0.133 | 1.828485e-04 | TRUE |
| isoComp_00155766 | geneComp_00017915 | MSTRG.27629.2 | ENSG00000205269 | TMEM170B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.556 | 0.556 | 1.975943e-04 | TRUE |
| isoComp_00007173 | geneComp_00000656 | ENST00000053468 | ENSG00000048544 | MRPS10 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.877 | 0.767 | -0.110 | 2.017483e-04 | TRUE |
| isoComp_00158619 | geneComp_00018680 | ENST00000393073 | ENSG00000214182 | PTMAP5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.487 | 0.487 | 2.064912e-04 | TRUE |
| isoComp_00085141 | geneComp_00008512 | MSTRG.2296.30 | ENSG00000143537 | ADAM15 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.061 | 0.117 | 0.056 | 2.138714e-04 | |
| isoComp_00135978 | geneComp_00014882 | MSTRG.11521.2 | ENSG00000182054 | IDH2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.148 | 0.055 | -0.093 | 2.166283e-04 | |
| isoComp_00158351 | geneComp_00018633 | ENST00000397298 | ENSG00000214026 | MRPL23 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.922 | 0.811 | -0.111 | 2.229462e-04 | TRUE |
| isoComp_00199254 | geneComp_00037229 | ENST00000658393 | ENSG00000286905 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.331 | 0.331 | 2.239765e-04 | TRUE | |
| isoComp_00020705 | geneComp_00001829 | MSTRG.8038.6 | ENSG00000089022 | MAPKAPK5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.051 | 0.000 | -0.051 | 2.273095e-04 | TRUE |
| isoComp_00071537 | geneComp_00007130 | MSTRG.28650.5 | ENSG00000135299 | ANKRD6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.172 | 0.172 | 2.315123e-04 | TRUE |
| isoComp_00158348 | geneComp_00018633 | ENST00000381519 | ENSG00000214026 | MRPL23 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.062 | 0.173 | 0.111 | 2.333601e-04 | TRUE |
| isoComp_00162332 | geneComp_00019981 | ENST00000429319 | ENSG00000224846 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.129 | 0.000 | -0.129 | 2.378585e-04 | TRUE | |
| isoComp_00017271 | geneComp_00001513 | MSTRG.23324.1 | ENSG00000080224 | EPHA6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.269 | 0.269 | 2.439259e-04 | TRUE |
| isoComp_00098334 | geneComp_00010029 | MSTRG.21206.4 | ENSG00000156256 | USP16 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.085 | 0.000 | -0.085 | 2.500283e-04 | TRUE |
| isoComp_00031993 | geneComp_00002916 | ENST00000560299 | ENSG00000104133 | SPG11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.158 | 0.158 | 2.510323e-04 | TRUE |
| isoComp_00197968 | geneComp_00036412 | MSTRG.28053.12 | ENSG00000285565 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.358 | 0.358 | 2.550136e-04 | TRUE | |
| isoComp_00025046 | geneComp_00002232 | ENST00000642696 | ENSG00000100150 | DEPDC5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.099 | 0.099 | 2.550913e-04 | TRUE |
| isoComp_00187602 | geneComp_00030778 | MSTRG.15222.3 | ENSG00000262580 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.021 | 0.317 | 0.296 | 2.628371e-04 | TRUE | |
| isoComp_00040065 | geneComp_00003731 | ENST00000505913 | ENSG00000109458 | GAB1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.456 | 0.456 | 2.888315e-04 | TRUE |
| isoComp_00086983 | geneComp_00008701 | ENST00000273179 | ENSG00000144677 | CTDSPL | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.197 | 0.097 | -0.100 | 2.919276e-04 | TRUE |
| isoComp_00200829 | geneComp_00038384 | ENST00000684814 | ENSG00000289088 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.209 | 0.209 | 3.017233e-04 | TRUE | |
| isoComp_00013482 | geneComp_00001196 | ENST00000494613 | ENSG00000072163 | LIMS2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.439 | 0.439 | 3.199541e-04 | TRUE |
| isoComp_00030642 | geneComp_00002794 | ENST00000569187 | ENSG00000103121 | CMC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.221 | 0.149 | -0.071 | 3.256185e-04 | |
| isoComp_00118511 | geneComp_00012397 | MSTRG.31243.3 | ENSG00000168487 | BMP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.067 | 0.067 | 3.434978e-04 | TRUE |
| isoComp_00167679 | geneComp_00022213 | ENST00000438412 | ENSG00000231074 | HCG18 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.070 | 0.000 | -0.070 | 3.562412e-04 | |
| isoComp_00159954 | geneComp_00019057 | ENST00000400889 | ENSG00000215784 | FAM72D | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.989 | 0.847 | -0.142 | 3.619815e-04 | TRUE |
| isoComp_00019414 | geneComp_00001716 | ENST00000199448 | ENSG00000086289 | EPDR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.803 | 0.496 | -0.308 | 3.620358e-04 | TRUE |
| isoComp_00163952 | geneComp_00020647 | ENST00000666743 | ENSG00000226686 | LINC01535 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.060 | 0.000 | -0.060 | 3.651353e-04 | TRUE |
| isoComp_00053520 | geneComp_00005144 | ENST00000493739 | ENSG00000120664 | SPART-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.340 | 0.340 | 3.937626e-04 | TRUE |
| isoComp_00155623 | geneComp_00017876 | ENST00000693457 | ENSG00000205084 | TMEM231 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 3.943149e-04 | TRUE |
| isoComp_00040280 | geneComp_00003749 | ENST00000505206 | ENSG00000109680 | TBC1D19 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.356 | 0.000 | -0.356 | 4.257114e-04 | TRUE |
| isoComp_00186195 | geneComp_00030077 | MSTRG.12927.11 | ENSG00000260537 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.984 | 0.864 | -0.120 | 4.312727e-04 | TRUE | |
| isoComp_00069383 | geneComp_00006889 | ENST00000428926 | ENSG00000133943 | DGLUCY | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.067 | 0.067 | 4.422121e-04 | TRUE |
| isoComp_00159418 | geneComp_00018916 | ENST00000573939 | ENSG00000215067 | ALOX12-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.093 | 0.093 | 4.450008e-04 | TRUE |
| isoComp_00156388 | geneComp_00018042 | ENST00000382252 | ENSG00000205903 | ZNF316 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.610 | 0.418 | -0.192 | 4.953253e-04 | TRUE |
| isoComp_00017313 | geneComp_00001519 | ENST00000357248 | ENSG00000080503 | SMARCA2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.095 | 0.095 | 4.961271e-04 | TRUE |
| isoComp_00198571 | geneComp_00036764 | ENST00000306585 | ENSG00000286140 | DERPC | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.059 | 0.116 | 0.057 | 5.170028e-04 | TRUE |
| isoComp_00080832 | geneComp_00008086 | ENST00000561340 | ENSG00000140598 | EFL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.121 | 0.121 | 5.546466e-04 | TRUE |
| isoComp_00034838 | geneComp_00003200 | MSTRG.16724.10 | ENSG00000105647 | PIK3R2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.035 | 0.143 | 0.108 | 5.750794e-04 | TRUE |
| isoComp_00037908 | geneComp_00003532 | ENST00000396271 | ENSG00000107897 | ACBD5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.377 | 0.377 | 5.771701e-04 | TRUE |
| isoComp_00099788 | geneComp_00010186 | ENST00000621158 | ENSG00000157613 | CREB3L1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.720 | -0.280 | 5.800038e-04 | TRUE |
| isoComp_00114514 | geneComp_00011956 | ENST00000504030 | ENSG00000166736 | HTR3A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 5.800038e-04 | TRUE |
| isoComp_00003478 | geneComp_00000298 | MSTRG.29734.3 | ENSG00000011426 | ANLN | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.104 | 0.104 | 6.034509e-04 | TRUE |
| isoComp_00085285 | geneComp_00008529 | ENST00000498667 | ENSG00000143590 | EFNA3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.331 | 0.121 | -0.210 | 6.641144e-04 | TRUE |
| isoComp_00002017 | geneComp_00000180 | ENST00000639034 | ENSG00000007372 | PAX6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.103 | 0.000 | -0.103 | 6.761491e-04 | TRUE |
| isoComp_00025477 | geneComp_00002271 | ENST00000440761 | ENSG00000100271 | TTLL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.138 | 0.138 | 6.913675e-04 | TRUE |
| isoComp_00130788 | geneComp_00014064 | ENST00000396865 | ENSG00000177058 | SLC38A9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.087 | 0.087 | 7.126762e-04 | TRUE |
| isoComp_00040076 | geneComp_00003732 | ENST00000421009 | ENSG00000109466 | KLHL2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.449 | 0.000 | -0.449 | 7.225216e-04 | TRUE |
| isoComp_00079801 | geneComp_00008002 | ENST00000559060 | ENSG00000140044 | JDP2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.456 | 0.456 | 7.298527e-04 | TRUE |
| isoComp_00097099 | geneComp_00009877 | ENST00000435829 | ENSG00000154814 | OXNAD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.101 | 0.000 | -0.101 | 7.298527e-04 | TRUE |
| isoComp_00078943 | geneComp_00007894 | ENST00000266712 | ENSG00000139324 | TMTC3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.351 | -0.649 | 8.174822e-04 | TRUE |
| isoComp_00018422 | geneComp_00001622 | MSTRG.8847.6 | ENSG00000083544 | TDRD3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.073 | 0.556 | 0.482 | 8.547366e-04 | TRUE |
| isoComp_00032784 | geneComp_00002987 | ENST00000615729 | ENSG00000104691 | UBXN8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.058 | 0.000 | -0.058 | 1.011968e-03 | |
| isoComp_00151629 | geneComp_00017185 | ENST00000356906 | ENSG00000198496 | NBR2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.195 | 0.195 | 1.062329e-03 | TRUE |
| isoComp_00059085 | geneComp_00005782 | ENST00000245983 | ENSG00000125787 | GNRH2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.447 | 0.447 | 1.212138e-03 | TRUE |
| isoComp_00008016 | geneComp_00000728 | ENST00000399516 | ENSG00000053747 | LAMA3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.151 | 0.151 | 1.232962e-03 | TRUE |
| isoComp_00184260 | geneComp_00029012 | MSTRG.7421.2 | ENSG00000258311 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.162 | 0.162 | 1.243898e-03 | TRUE | |
| isoComp_00143935 | geneComp_00016105 | ENST00000420670 | ENSG00000187764 | SEMA4D | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.110 | 0.000 | -0.110 | 1.259269e-03 | TRUE |
| isoComp_00098331 | geneComp_00010029 | MSTRG.21206.1 | ENSG00000156256 | USP16 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.096 | 0.096 | 1.303662e-03 | TRUE |
| isoComp_00050132 | geneComp_00004747 | ENST00000477167 | ENSG00000117245 | KIF17 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.122 | 0.000 | -0.122 | 1.354698e-03 | TRUE |
| isoComp_00029052 | geneComp_00002631 | ENST00000378533 | ENSG00000101955 | SRPX | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.859 | -0.141 | 1.425907e-03 | TRUE |
| isoComp_00153967 | geneComp_00017594 | ENST00000647317 | ENSG00000204147 | ASAH2B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.325 | 0.325 | 1.435151e-03 | TRUE |
| isoComp_00090795 | geneComp_00009147 | ENST00000371974 | ENSG00000148290 | SURF1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.444 | 0.319 | -0.125 | 1.546301e-03 | TRUE |
| isoComp_00052429 | geneComp_00005007 | MSTRG.9942.18 | ENSG00000119685 | TTLL5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.110 | 0.110 | 1.557131e-03 | |
| isoComp_00062517 | geneComp_00006177 | ENST00000515266 | ENSG00000129071 | MBD4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.095 | 0.095 | 1.592412e-03 | TRUE |
| isoComp_00071886 | geneComp_00007161 | ENST00000553893 | ENSG00000135424 | ITGA7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.101 | 0.101 | 1.659446e-03 | TRUE |
| isoComp_00130297 | geneComp_00013984 | MSTRG.31768.7 | ENSG00000176623 | RMDN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.061 | 0.061 | 1.659446e-03 | TRUE |
| isoComp_00182989 | geneComp_00028394 | ENST00000663068 | ENSG00000255920 | CCND2-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.930 | 0.000 | -0.930 | 1.720289e-03 | TRUE |
| isoComp_00081579 | geneComp_00008152 | ENST00000398982 | ENSG00000141068 | KSR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.068 | 0.000 | -0.068 | 1.722830e-03 | TRUE |
| isoComp_00115926 | geneComp_00012132 | ENST00000593238 | ENSG00000167476 | JSRP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.472 | 0.000 | -0.472 | 1.722830e-03 | TRUE |
| isoComp_00012103 | geneComp_00001079 | ENST00000434115 | ENSG00000069020 | MAST4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.053 | 0.053 | 1.725115e-03 | TRUE |
| isoComp_00011217 | geneComp_00001011 | ENST00000307259 | ENSG00000067113 | PLPP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.797 | 0.623 | -0.175 | 1.781403e-03 | TRUE |
| isoComp_00113841 | geneComp_00011883 | ENST00000299506 | ENSG00000166402 | TUB | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.064 | 0.000 | -0.064 | 1.796882e-03 | TRUE |
| isoComp_00100167 | geneComp_00010224 | ENST00000304032 | ENSG00000157985 | AGAP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.119 | 0.003 | -0.116 | 1.800122e-03 | TRUE |
| isoComp_00136922 | geneComp_00015013 | ENST00000435354 | ENSG00000182648 | LINC01006 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.108 | 0.000 | -0.108 | 1.807499e-03 | TRUE |
| isoComp_00006253 | geneComp_00000568 | ENST00000338008 | ENSG00000039319 | ZFYVE16 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.427 | 0.427 | 1.879761e-03 | TRUE |
| isoComp_00078948 | geneComp_00007894 | MSTRG.7735.6 | ENSG00000139324 | TMTC3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.649 | 0.649 | 1.879761e-03 | TRUE |
| isoComp_00191033 | geneComp_00032405 | ENST00000656196 | ENSG00000270049 | ATP6V0D1-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.184 | 0.184 | 1.952350e-03 | TRUE |
| isoComp_00060145 | geneComp_00005905 | ENST00000612959 | ENSG00000126653 | NSRP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.110 | 0.110 | 1.970365e-03 | TRUE |
| isoComp_00174638 | geneComp_00025100 | ENST00000441328 | ENSG00000240583 | AQP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.502 | 0.502 | 2.093886e-03 | TRUE |
| isoComp_00109303 | geneComp_00011287 | MSTRG.22971.27 | ENSG00000164076 | CAMKV | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.005 | 0.097 | 0.092 | 2.116566e-03 | TRUE |
| isoComp_00141501 | geneComp_00015713 | ENST00000628967 | ENSG00000185760 | KCNQ5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.201 | 0.000 | -0.201 | 2.193749e-03 | TRUE |
| isoComp_00168892 | geneComp_00022722 | ENST00000424736 | ENSG00000232499 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 2.241635e-03 | TRUE | |
| isoComp_00117856 | geneComp_00012333 | ENST00000348160 | ENSG00000168214 | RBPJ | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.110 | 0.000 | -0.110 | 2.324026e-03 | TRUE |
| isoComp_00193449 | geneComp_00033815 | MSTRG.2061.12 | ENSG00000274372 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.533 | 0.000 | -0.533 | 2.484501e-03 | TRUE | |
| isoComp_00017874 | geneComp_00001580 | ENST00000504564 | ENSG00000082068 | WDR70 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.080 | 0.000 | -0.080 | 2.490890e-03 | TRUE |
| isoComp_00016240 | geneComp_00001432 | ENST00000473616 | ENSG00000078114 | NEBL | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.229 | 0.229 | 2.593487e-03 | TRUE |
| isoComp_00146521 | geneComp_00016544 | MSTRG.16846.7 | ENSG00000196268 | ZNF493 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.290 | 0.000 | -0.290 | 2.673409e-03 | TRUE |
| isoComp_00018504 | geneComp_00001634 | MSTRG.17859.3 | ENSG00000083838 | ZNF446 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.431 | 0.291 | -0.140 | 2.895488e-03 | TRUE |
| isoComp_00041649 | geneComp_00003883 | ENST00000454014 | ENSG00000110888 | CAPRIN2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.119 | 0.000 | -0.119 | 2.954924e-03 | TRUE |
| isoComp_00182703 | geneComp_00028285 | ENST00000528514 | ENSG00000255495 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.875 | 0.000 | -0.875 | 2.959415e-03 | TRUE | |
| isoComp_00087181 | geneComp_00008722 | ENST00000640181 | ENSG00000144824 | PHLDB2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.584 | 0.584 | 3.203029e-03 | TRUE |
| isoComp_00073122 | geneComp_00007285 | ENST00000494336 | ENSG00000135976 | ANKRD36 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.364 | 0.000 | -0.364 | 3.206042e-03 | TRUE |
| isoComp_00088602 | geneComp_00008892 | ENST00000509310 | ENSG00000146094 | DOK3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.102 | 0.102 | 3.206042e-03 | TRUE |
| isoComp_00094391 | geneComp_00009528 | ENST00000395758 | ENSG00000151746 | BICD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.114 | 0.000 | -0.114 | 3.272605e-03 | TRUE |
| isoComp_00099282 | geneComp_00010135 | ENST00000460214 | ENSG00000157193 | LRP8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.056 | 0.056 | 3.272605e-03 | TRUE |
| isoComp_00158167 | geneComp_00018602 | MSTRG.19570.23 | ENSG00000213963 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.406 | 0.406 | 3.372491e-03 | TRUE | |
| isoComp_00051140 | geneComp_00004851 | MSTRG.2469.6 | ENSG00000118217 | ATF6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 3.474734e-03 | TRUE |
| isoComp_00143106 | geneComp_00015964 | MSTRG.22333.7 | ENSG00000186951 | PPARA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.124 | 0.124 | 3.509103e-03 | TRUE |
| isoComp_00183940 | geneComp_00028845 | ENST00000603175 | ENSG00000257702 | LBX2-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.659 | 0.000 | -0.659 | 3.538216e-03 | TRUE |
| isoComp_00085629 | geneComp_00008562 | ENST00000496372 | ENSG00000143786 | CNIH3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.050 | 0.000 | -0.050 | 3.621811e-03 | TRUE |
| isoComp_00134319 | geneComp_00014578 | MSTRG.15916.1 | ENSG00000180011 | ZADH2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.059 | 0.007 | -0.052 | 3.803222e-03 | TRUE |
| isoComp_00126442 | geneComp_00013453 | MSTRG.11281.16 | ENSG00000173517 | PEAK1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.131 | 0.131 | 3.909805e-03 | TRUE |
| isoComp_00162462 | geneComp_00020021 | ENST00000446477 | ENSG00000224940 | PRRT4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.452 | 0.116 | -0.336 | 3.916347e-03 | TRUE |
| isoComp_00107411 | geneComp_00011093 | ENST00000409849 | ENSG00000163516 | ANKZF1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.073 | 0.073 | 4.085419e-03 | TRUE |
| isoComp_00024151 | geneComp_00002143 | ENST00000497445 | ENSG00000099840 | IZUMO4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.107 | 0.007 | -0.100 | 4.195847e-03 | TRUE |
| isoComp_00038540 | geneComp_00003588 | ENST00000588009 | ENSG00000108423 | TUBD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.100 | 0.100 | 4.234480e-03 | TRUE |
| isoComp_00040018 | geneComp_00003725 | MSTRG.25481.3 | ENSG00000109381 | ELF2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.071 | 0.000 | -0.071 | 4.326880e-03 | TRUE |
| isoComp_00010524 | geneComp_00000943 | MSTRG.28225.2 | ENSG00000065491 | TBC1D22B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 4.346630e-03 | TRUE |
| isoComp_00093154 | geneComp_00009390 | ENST00000409029 | ENSG00000150556 | LYPD6B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.158 | 0.158 | 4.354220e-03 | TRUE |
| isoComp_00030402 | geneComp_00002776 | ENST00000563498 | ENSG00000103024 | NME3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.273 | 0.108 | -0.166 | 4.720917e-03 | TRUE |
| isoComp_00145666 | geneComp_00016408 | ENST00000592910 | ENSG00000189114 | BLOC1S3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.100 | 0.030 | -0.070 | 4.720917e-03 | TRUE |
| isoComp_00130402 | geneComp_00014004 | ENST00000619594 | ENSG00000176731 | RBIS | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.317 | 0.407 | 0.090 | 4.743069e-03 | TRUE |
| isoComp_00190239 | geneComp_00032027 | ENST00000622323 | ENSG00000268350 | FAM156A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.071 | 0.071 | 4.937377e-03 | TRUE |
| isoComp_00141369 | geneComp_00015696 | ENST00000613241 | ENSG00000185684 | EP400P1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.018 | 0.131 | 0.112 | 5.200041e-03 | TRUE |
| isoComp_00150535 | geneComp_00017039 | MSTRG.13112.11 | ENSG00000197943 | PLCG2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.352 | 0.587 | 0.235 | 5.200041e-03 | TRUE |
| isoComp_00040824 | geneComp_00003803 | ENST00000527403 | ENSG00000110075 | PPP6R3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.009 | 0.196 | 0.187 | 5.399237e-03 | TRUE |
| isoComp_00141060 | geneComp_00015659 | ENST00000571004 | ENSG00000185527 | PDE6G | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.620 | 0.000 | -0.620 | 5.431823e-03 | TRUE |
| isoComp_00017580 | geneComp_00001542 | ENST00000574096 | ENSG00000080986 | NDC80 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.065 | 0.065 | 5.640083e-03 | TRUE |
| isoComp_00112174 | geneComp_00011689 | ENST00000689161 | ENSG00000165626 | BEND7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.089 | 0.089 | 5.658173e-03 | TRUE |
| isoComp_00117865 | geneComp_00012333 | ENST00000509158 | ENSG00000168214 | RBPJ | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.086 | 0.086 | 5.785072e-03 | TRUE |
| isoComp_00024105 | geneComp_00002137 | ENST00000556508 | ENSG00000099814 | CEP170B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.192 | 0.083 | -0.109 | 5.822522e-03 | TRUE |
| isoComp_00057275 | geneComp_00005575 | ENST00000507414 | ENSG00000124275 | MTRR | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.069 | 0.069 | 5.853894e-03 | TRUE |
| isoComp_00062652 | geneComp_00006193 | ENST00000357067 | ENSG00000129187 | DCTD | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.613 | 0.480 | -0.133 | 5.853894e-03 | TRUE |
| isoComp_00096216 | geneComp_00009768 | ENST00000508756 | ENSG00000153922 | CHD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.065 | 0.000 | -0.065 | 5.853894e-03 | |
| isoComp_00147369 | geneComp_00016640 | ENST00000470041 | ENSG00000196550 | FAM72A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.062 | 0.000 | -0.062 | 5.970746e-03 | TRUE |
| isoComp_00090868 | geneComp_00009156 | ENST00000338961 | ENSG00000148334 | PTGES2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.858 | 0.790 | -0.068 | 6.181616e-03 | TRUE |
| isoComp_00001029 | geneComp_00000097 | ENST00000361672 | ENSG00000005302 | MSL3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.066 | 0.066 | 6.214228e-03 | |
| isoComp_00116105 | geneComp_00012145 | ENST00000467736 | ENSG00000167526 | RPL13 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.422 | 0.316 | -0.106 | 6.256058e-03 | TRUE |
| isoComp_00088267 | geneComp_00008851 | ENST00000652664 | ENSG00000145861 | C1QTNF2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.000 | -1.000 | 6.593864e-03 | TRUE |
| isoComp_00040103 | geneComp_00003735 | ENST00000673642 | ENSG00000109501 | WFS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.127 | 0.038 | -0.089 | 6.595210e-03 | TRUE |
| isoComp_00098306 | geneComp_00010025 | MSTRG.11397.3 | ENSG00000156232 | WHAMM | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.087 | 0.087 | 6.985341e-03 | TRUE |
| isoComp_00053384 | geneComp_00005130 | ENST00000432059 | ENSG00000120519 | SLC10A7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.537 | 0.537 | 7.110857e-03 | TRUE |
| isoComp_00133574 | geneComp_00014476 | MSTRG.23640.10 | ENSG00000179348 | GATA2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.003 | 0.068 | 0.064 | 7.144028e-03 | |
| isoComp_00059301 | geneComp_00005816 | ENST00000354200 | ENSG00000125875 | TBC1D20 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.478 | 0.632 | 0.155 | 7.289541e-03 | TRUE |
| isoComp_00011849 | geneComp_00001062 | ENST00000487903 | ENSG00000068650 | ATP11A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.091 | 0.091 | 7.344239e-03 | TRUE |
| isoComp_00143100 | geneComp_00015964 | ENST00000496865 | ENSG00000186951 | PPARA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.095 | 0.000 | -0.095 | 7.522965e-03 | TRUE |
| isoComp_00057719 | geneComp_00005619 | ENST00000244546 | ENSG00000124587 | PEX6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.091 | 0.006 | -0.085 | 7.671523e-03 | TRUE |
| isoComp_00137205 | geneComp_00015064 | ENST00000485439 | ENSG00000182899 | RPL35A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.589 | 0.672 | 0.084 | 7.671523e-03 | TRUE |
| isoComp_00123210 | geneComp_00013037 | ENST00000483768 | ENSG00000171469 | ZNF561 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.014 | 0.146 | 0.132 | 7.676231e-03 | |
| isoComp_00075322 | geneComp_00007520 | MSTRG.28354.5 | ENSG00000137216 | TMEM63B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.030 | 0.096 | 0.066 | 7.678491e-03 | TRUE |
| isoComp_00149286 | geneComp_00016881 | ENST00000391916 | ENSG00000197380 | DACT3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.921 | 0.548 | -0.372 | 7.691122e-03 | TRUE |
| isoComp_00138123 | geneComp_00015196 | ENST00000338525 | ENSG00000183479 | TREX2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.161 | 0.771 | 0.611 | 7.804872e-03 | TRUE |
| isoComp_00112173 | geneComp_00011689 | ENST00000688845 | ENSG00000165626 | BEND7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.070 | 0.070 | 7.829905e-03 | TRUE |
| isoComp_00170695 | geneComp_00023497 | ENST00000653665 | ENSG00000234684 | SDCBP2-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.052 | 0.000 | -0.052 | 7.858816e-03 | |
| isoComp_00049614 | geneComp_00004685 | ENST00000375799 | ENSG00000116786 | PLEKHM2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.129 | 0.003 | -0.126 | 8.020958e-03 | TRUE |
| isoComp_00143504 | geneComp_00016034 | ENST00000334660 | ENSG00000187446 | CHP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.878 | 0.795 | -0.083 | 8.073671e-03 | TRUE |
| isoComp_00142004 | geneComp_00015777 | ENST00000586115 | ENSG00000186020 | ZNF529 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.108 | 0.000 | -0.108 | 8.106343e-03 | TRUE |
| isoComp_00090279 | geneComp_00009091 | ENST00000337573 | ENSG00000147654 | EBAG9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.470 | 0.749 | 0.279 | 8.127260e-03 | TRUE |
| isoComp_00151728 | geneComp_00017198 | ENST00000464469 | ENSG00000198538 | ZNF28 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.399 | 0.399 | 8.127260e-03 | TRUE |
| isoComp_00156829 | geneComp_00018132 | ENST00000545470 | ENSG00000211455 | STK38L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.180 | 0.180 | 8.359829e-03 | TRUE |
| isoComp_00096448 | geneComp_00009796 | MSTRG.25956.2 | ENSG00000154124 | OTULIN | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.099 | 0.099 | 8.370927e-03 | TRUE |
| isoComp_00053099 | geneComp_00005079 | ENST00000314537 | ENSG00000120088 | CRHR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.804 | 0.804 | 8.407677e-03 | TRUE |
| isoComp_00147365 | geneComp_00016640 | ENST00000367128 | ENSG00000196550 | FAM72A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.484 | 0.683 | 0.199 | 8.412054e-03 | TRUE |
| isoComp_00196596 | geneComp_00035798 | ENST00000687535 | ENSG00000281706 | LINC01012 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.117 | 0.000 | -0.117 | 8.506058e-03 | TRUE |
| isoComp_00053443 | geneComp_00005135 | ENST00000475691 | ENSG00000120555 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.811 | 0.811 | 8.635731e-03 | TRUE | |
| isoComp_00098816 | geneComp_00010084 | MSTRG.32053.14 | ENSG00000156795 | NTAQ1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.091 | 0.091 | 8.647671e-03 | TRUE |
| isoComp_00116404 | geneComp_00012177 | ENST00000443387 | ENSG00000167635 | ZNF146 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.307 | 0.441 | 0.134 | 8.917141e-03 | TRUE |
| isoComp_00142040 | geneComp_00015783 | ENST00000565792 | ENSG00000186073 | CDIN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.177 | 0.177 | 8.917141e-03 | TRUE |
| isoComp_00088266 | geneComp_00008851 | ENST00000393975 | ENSG00000145861 | C1QTNF2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 9.173283e-03 | TRUE |
| isoComp_00099112 | geneComp_00010114 | ENST00000453767 | ENSG00000157036 | EXOG | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.056 | 0.056 | 9.173283e-03 | TRUE |
| isoComp_00139375 | geneComp_00015395 | ENST00000332509 | ENSG00000184381 | PLA2G6 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.066 | 0.066 | 9.173283e-03 | |
| isoComp_00140902 | geneComp_00015636 | MSTRG.17423.1 | ENSG00000185453 | ZSWIM9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.258 | 0.022 | -0.236 | 9.173283e-03 | TRUE |
| isoComp_00121030 | geneComp_00012711 | ENST00000395094 | ENSG00000169955 | ZNF747 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.631 | 0.502 | -0.128 | 9.182227e-03 | TRUE |
| isoComp_00081147 | geneComp_00008115 | ENST00000379655 | ENSG00000140859 | KIFC3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.204 | 0.405 | 0.201 | 9.240397e-03 | TRUE |
| isoComp_00102101 | geneComp_00010461 | ENST00000338806 | ENSG00000159788 | RGS12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.095 | 0.095 | 9.369422e-03 | TRUE |
| isoComp_00146849 | geneComp_00016579 | MSTRG.1989.4 | ENSG00000196369 | SRGAP2B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.155 | 0.155 | 9.419977e-03 | TRUE |
| isoComp_00035882 | geneComp_00003311 | ENST00000467706 | ENSG00000106100 | NOD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.093 | 0.000 | -0.093 | 9.801941e-03 | TRUE |
| isoComp_00094911 | geneComp_00009600 | ENST00000506164 | ENSG00000152359 | POC5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.118 | 0.118 | 9.929231e-03 | TRUE |
| isoComp_00181479 | geneComp_00027628 | ENST00000692136 | ENSG00000253771 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.218 | 0.218 | 9.999602e-03 | TRUE | |
| isoComp_00113501 | geneComp_00011846 | ENST00000530393 | ENSG00000166261 | ZNF202 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.053 | 0.053 | 1.022133e-02 | TRUE |
| isoComp_00156333 | geneComp_00018014 | ENST00000692045 | ENSG00000205771 | CATSPER2P1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.234 | 0.234 | 1.038908e-02 | TRUE |
| isoComp_00056876 | geneComp_00005517 | ENST00000575857 | ENSG00000124067 | SLC12A4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.092 | 0.092 | 1.054382e-02 | TRUE |
| isoComp_00159955 | geneComp_00019057 | MSTRG.1977.2 | ENSG00000215784 | FAM72D | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.011 | 0.153 | 0.142 | 1.084144e-02 | TRUE |
| isoComp_00143512 | geneComp_00016034 | MSTRG.10595.7 | ENSG00000187446 | CHP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.035 | 0.120 | 0.084 | 1.111830e-02 | TRUE |
| isoComp_00114607 | geneComp_00011967 | ENST00000524803 | ENSG00000166788 | SAAL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.517 | 0.611 | 0.093 | 1.182838e-02 | |
| isoComp_00176959 | geneComp_00026013 | MSTRG.22113.38 | ENSG00000244627 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.022 | 0.138 | 0.116 | 1.188290e-02 | TRUE | |
| isoComp_00149120 | geneComp_00016857 | MSTRG.12553.29 | ENSG00000197302 | KRBOX5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.002 | 0.055 | 0.052 | 1.205691e-02 | TRUE |
| isoComp_00071539 | geneComp_00007132 | ENST00000257765 | ENSG00000135314 | KHDC1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.468 | 0.015 | -0.453 | 1.208996e-02 | TRUE |
| isoComp_00191164 | geneComp_00032481 | MSTRG.2133.4 | ENSG00000270276 | H4C15 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.525 | 0.094 | -0.430 | 1.208996e-02 | TRUE |
| isoComp_00023280 | geneComp_00002052 | MSTRG.4765.4 | ENSG00000095574 | IKZF5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.143 | 0.143 | 1.237102e-02 | TRUE |
| isoComp_00037122 | geneComp_00003444 | ENST00000620767 | ENSG00000107140 | TESK1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.240 | 0.470 | 0.230 | 1.238702e-02 | TRUE |
| isoComp_00184258 | geneComp_00029012 | ENST00000550412 | ENSG00000258311 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.886 | 0.617 | -0.269 | 1.242465e-02 | TRUE | |
| isoComp_00102228 | geneComp_00010475 | ENST00000291182 | ENSG00000159917 | ZNF235 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.347 | 0.347 | 1.249047e-02 | TRUE |
| isoComp_00071279 | geneComp_00007107 | ENST00000552092 | ENSG00000135116 | HRK | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.069 | 0.069 | 1.267685e-02 | TRUE |
| isoComp_00057582 | geneComp_00005606 | ENST00000472507 | ENSG00000124508 | BTN2A2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.054 | 0.000 | -0.054 | 1.275658e-02 | TRUE |
| isoComp_00041134 | geneComp_00003837 | ENST00000321505 | ENSG00000110427 | KIAA1549L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.268 | 0.268 | 1.280974e-02 | TRUE |
| isoComp_00161389 | geneComp_00019601 | ENST00000669506 | ENSG00000223745 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.106 | 0.000 | -0.106 | 1.292989e-02 | TRUE | |
| isoComp_00186546 | geneComp_00030251 | ENST00000621799 | ENSG00000260923 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.192 | 0.192 | 1.308477e-02 | TRUE | |
| isoComp_00161630 | geneComp_00019678 | ENST00000453026 | ENSG00000223960 | CHROMR | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.268 | 0.268 | 1.323582e-02 | TRUE |
| isoComp_00089698 | geneComp_00009020 | MSTRG.34438.1 | ENSG00000147162 | OGT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.092 | 0.022 | -0.071 | 1.325537e-02 | TRUE |
| isoComp_00198783 | geneComp_00036890 | ENST00000665268 | ENSG00000286340 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 1.000 | 1.000 | 1.335358e-02 | TRUE | |
| isoComp_00170001 | geneComp_00023191 | ENST00000415434 | ENSG00000233806 | LINC01237 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.448 | 0.448 | 1.353886e-02 | TRUE |
| isoComp_00130975 | geneComp_00014082 | MSTRG.8325.2 | ENSG00000177169 | ULK1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.003 | 0.069 | 0.066 | 1.376640e-02 | |
| isoComp_00186470 | geneComp_00030222 | ENST00000566085 | ENSG00000260874 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.515 | -0.485 | 1.379051e-02 | TRUE | |
| isoComp_00146981 | geneComp_00016599 | ENST00000356818 | ENSG00000196422 | PPP1R26 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.103 | 0.018 | -0.084 | 1.435064e-02 | TRUE |
| isoComp_00143522 | geneComp_00016039 | ENST00000682350 | ENSG00000187486 | KCNJ11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.072 | 0.072 | 1.456108e-02 | TRUE |
| isoComp_00177835 | geneComp_00026192 | ENST00000521383 | ENSG00000247081 | BAALC-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.023 | 0.337 | 0.314 | 1.480015e-02 | TRUE |
| isoComp_00107784 | geneComp_00011137 | MSTRG.23192.3 | ENSG00000163635 | ATXN7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.151 | 0.000 | -0.151 | 1.496116e-02 | TRUE |
| isoComp_00155486 | geneComp_00017838 | ENST00000270014 | ENSG00000204920 | ZNF155 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.747 | 0.083 | -0.664 | 1.496642e-02 | TRUE |
| isoComp_00160188 | geneComp_00019145 | ENST00000510461 | ENSG00000217128 | FNIP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.538 | 0.538 | 1.508013e-02 | TRUE |
| isoComp_00086371 | geneComp_00008647 | ENST00000410066 | ENSG00000144331 | ZNF385B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.706 | 0.977 | 0.271 | 1.541216e-02 | TRUE |
| isoComp_00118124 | geneComp_00012353 | ENST00000303596 | ENSG00000168286 | THAP11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.755 | 0.350 | -0.405 | 1.568128e-02 | TRUE |
| isoComp_00080171 | geneComp_00008036 | ENST00000267950 | ENSG00000140374 | ETFA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.154 | 0.103 | -0.051 | 1.629330e-02 | |
| isoComp_00030400 | geneComp_00002776 | ENST00000219302 | ENSG00000103024 | NME3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.497 | 0.653 | 0.156 | 1.644959e-02 | TRUE |
| isoComp_00040452 | geneComp_00003768 | ENST00000514956 | ENSG00000109794 | FAM149A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.079 | 0.079 | 1.657887e-02 | TRUE |
| isoComp_00098802 | geneComp_00010082 | MSTRG.32041.3 | ENSG00000156787 | TBC1D31 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.099 | 0.099 | 1.657887e-02 | TRUE |
| isoComp_00118125 | geneComp_00012353 | MSTRG.12849.1 | ENSG00000168286 | THAP11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.245 | 0.650 | 0.405 | 1.668004e-02 | TRUE |
| isoComp_00095959 | geneComp_00009734 | ENST00000283534 | ENSG00000153485 | TMEM251 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.298 | 0.534 | 0.235 | 1.757132e-02 | TRUE |
| isoComp_00180589 | geneComp_00027246 | ENST00000433835 | ENSG00000251357 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.879 | 0.362 | -0.517 | 1.765535e-02 | TRUE | |
| isoComp_00174809 | geneComp_00025197 | MSTRG.17658.6 | ENSG00000241015 | TPM3P9 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.038 | 0.432 | 0.394 | 1.800988e-02 | TRUE |
| isoComp_00032176 | geneComp_00002929 | ENST00000520604 | ENSG00000104231 | ZFAND1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.061 | 0.004 | -0.057 | 1.809574e-02 | TRUE |
| isoComp_00178163 | geneComp_00026244 | ENST00000664462 | ENSG00000247828 | TMEM161B-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.063 | 0.005 | -0.058 | 1.861296e-02 | TRUE |
| isoComp_00124295 | geneComp_00013182 | ENST00000347055 | ENSG00000172086 | KRCC1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.964 | 0.815 | -0.148 | 1.871615e-02 | TRUE |
| isoComp_00191487 | geneComp_00032682 | MSTRG.34617.4 | ENSG00000271147 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.086 | 0.086 | 1.953213e-02 | TRUE | |
| isoComp_00190003 | geneComp_00031896 | ENST00000591087 | ENSG00000267757 | EML2-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.809 | 0.600 | -0.208 | 2.002587e-02 | TRUE |
| isoComp_00111820 | geneComp_00011622 | ENST00000375250 | ENSG00000165322 | ARHGAP12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.191 | 0.000 | -0.191 | 2.016050e-02 | TRUE |
| isoComp_00141351 | geneComp_00015694 | ENST00000423306 | ENSG00000185674 | LYG2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.918 | 0.918 | 2.016217e-02 | TRUE |
| isoComp_00069394 | geneComp_00006889 | ENST00000520328 | ENSG00000133943 | DGLUCY | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.068 | 0.004 | -0.065 | 2.037407e-02 | TRUE |
| isoComp_00141925 | geneComp_00015767 | ENST00000515467 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.065 | 0.065 | 2.075094e-02 | |
| isoComp_00099817 | geneComp_00010190 | ENST00000374165 | ENSG00000157653 | C9orf43 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.469 | 1.000 | 0.531 | 2.097672e-02 | TRUE |
| isoComp_00110594 | geneComp_00011472 | ENST00000354042 | ENSG00000164707 | SLC13A4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.040 | 0.329 | 0.289 | 2.127647e-02 | TRUE |
| isoComp_00012277 | geneComp_00001102 | ENST00000648451 | ENSG00000069869 | NEDD4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.061 | 0.000 | -0.061 | 2.139343e-02 | TRUE |
| isoComp_00036778 | geneComp_00003407 | ENST00000644273 | ENSG00000106692 | FKTN | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.227 | 0.000 | -0.227 | 2.202175e-02 | TRUE |
| isoComp_00108878 | geneComp_00011251 | ENST00000392391 | ENSG00000163964 | PIGX | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.316 | 0.140 | -0.177 | 2.206249e-02 | TRUE |
| isoComp_00092227 | geneComp_00009280 | ENST00000278618 | ENSG00000149313 | AASDHPPT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.953 | 0.852 | -0.100 | 2.241440e-02 | TRUE |
| isoComp_00115737 | geneComp_00012111 | ENST00000574496 | ENSG00000167363 | FN3K | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.248 | 0.248 | 2.255189e-02 | TRUE |
| isoComp_00007333 | geneComp_00000670 | ENST00000361923 | ENSG00000049246 | PER3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.518 | 0.518 | 2.309329e-02 | TRUE |
| isoComp_00176860 | geneComp_00025981 | MSTRG.21679.3 | ENSG00000244486 | SCARF2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.054 | 0.002 | -0.053 | 2.331822e-02 | |
| isoComp_00118638 | geneComp_00012415 | ENST00000304636 | ENSG00000168542 | COL3A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.245 | 1.000 | 0.755 | 2.363571e-02 | TRUE |
| isoComp_00118639 | geneComp_00012415 | ENST00000317840 | ENSG00000168542 | COL3A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.755 | 0.000 | -0.755 | 2.387415e-02 | TRUE |
| isoComp_00144554 | geneComp_00016225 | ENST00000586658 | ENSG00000188283 | ZNF383 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.425 | 0.425 | 2.409988e-02 | TRUE |
| isoComp_00062653 | geneComp_00006193 | ENST00000438320 | ENSG00000129187 | DCTD | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.196 | 0.299 | 0.103 | 2.429819e-02 | TRUE |
| isoComp_00083858 | geneComp_00008373 | ENST00000616558 | ENSG00000142765 | SYTL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.270 | 0.270 | 2.502331e-02 | TRUE |
| isoComp_00090272 | geneComp_00009090 | ENST00000276654 | ENSG00000147650 | LRP12 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.936 | 0.165 | -0.771 | 2.510989e-02 | TRUE |
| isoComp_00130514 | geneComp_00014024 | ENST00000334705 | ENSG00000176853 | FAM91A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.904 | 0.691 | -0.213 | 2.523195e-02 | TRUE |
| isoComp_00178234 | geneComp_00026259 | ENST00000514109 | ENSG00000248049 | UBA6-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 2.545429e-02 | |
| isoComp_00054771 | geneComp_00005290 | ENST00000264126 | ENSG00000121957 | GPSM2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.110 | 0.110 | 2.606965e-02 | TRUE |
| isoComp_00153766 | geneComp_00017569 | ENST00000444184 | ENSG00000204054 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.127 | 0.008 | -0.119 | 2.632857e-02 | TRUE | |
| isoComp_00186469 | geneComp_00030222 | ENST00000565827 | ENSG00000260874 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.485 | 0.485 | 2.632857e-02 | TRUE | |
| isoComp_00117613 | geneComp_00012303 | ENST00000652489 | ENSG00000168061 | SAC3D1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.404 | 0.244 | -0.161 | 2.648326e-02 | TRUE |
| isoComp_00183944 | geneComp_00028845 | ENST00000689056 | ENSG00000257702 | LBX2-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.662 | 0.662 | 2.658645e-02 | TRUE |
| isoComp_00009083 | geneComp_00000822 | ENST00000228251 | ENSG00000060138 | YBX3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.631 | 0.486 | -0.145 | 2.668397e-02 | TRUE |
| isoComp_00199963 | geneComp_00037765 | ENST00000692550 | ENSG00000287750 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.639 | 0.152 | -0.487 | 2.669785e-02 | TRUE | |
| isoComp_00147775 | geneComp_00016690 | ENST00000358543 | ENSG00000196711 | ALKAL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.778 | 1.000 | 0.222 | 2.681123e-02 | TRUE |
| isoComp_00147711 | geneComp_00016685 | MSTRG.3829.4 | ENSG00000196693 | ZNF33B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.077 | 0.077 | 2.708576e-02 | TRUE |
| isoComp_00149400 | geneComp_00016902 | ENST00000506259 | ENSG00000197451 | HNRNPAB | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.101 | 0.044 | -0.057 | 2.710743e-02 | |
| isoComp_00180590 | geneComp_00027246 | MSTRG.21801.15 | ENSG00000251357 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.121 | 0.638 | 0.517 | 2.710743e-02 | TRUE | |
| isoComp_00103947 | geneComp_00010635 | ENST00000601134 | ENSG00000160961 | ZNF333 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.055 | 0.055 | 2.762810e-02 | TRUE |
| isoComp_00071403 | geneComp_00007120 | ENST00000436062 | ENSG00000135241 | PNPLA8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.093 | 0.843 | 0.750 | 2.799420e-02 | TRUE |
| isoComp_00159433 | geneComp_00018916 | MSTRG.13539.17 | ENSG00000215067 | ALOX12-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.071 | 0.071 | 2.810328e-02 | TRUE |
| isoComp_00046011 | geneComp_00004348 | ENST00000232607 | ENSG00000114491 | UMPS | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.276 | 0.376 | 0.100 | 2.874968e-02 | TRUE |
| isoComp_00045855 | geneComp_00004333 | ENST00000395144 | ENSG00000114378 | HYAL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 1.000 | 0.000 | -1.000 | 2.892856e-02 | TRUE |
| isoComp_00113157 | geneComp_00011809 | ENST00000315616 | ENSG00000166133 | RPUSD2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.884 | 0.812 | -0.072 | 2.910365e-02 | TRUE |
| isoComp_00009320 | geneComp_00000843 | MSTRG.20632.3 | ENSG00000061656 | SPAG4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.015 | 0.174 | 0.159 | 2.911215e-02 | TRUE |
| isoComp_00171946 | geneComp_00024014 | ENST00000418724 | ENSG00000236104 | ZBTB22 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.922 | 0.803 | -0.120 | 2.911215e-02 | TRUE |
| isoComp_00010518 | geneComp_00000942 | ENST00000472319 | ENSG00000065485 | PDIA5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.095 | 0.024 | -0.071 | 2.947527e-02 | TRUE |
| isoComp_00053791 | geneComp_00005173 | ENST00000547229 | ENSG00000120833 | SOCS2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.080 | 0.000 | -0.080 | 2.948399e-02 | TRUE |
| isoComp_00055216 | geneComp_00005332 | ENST00000696300 | ENSG00000122417 | ODF2L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.102 | 0.000 | -0.102 | 3.009686e-02 | TRUE |
| isoComp_00044676 | geneComp_00004218 | MSTRG.27229.1 | ENSG00000113272 | THG1L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.416 | 0.669 | 0.252 | 3.014362e-02 | TRUE |
| isoComp_00124613 | geneComp_00013237 | ENST00000307650 | ENSG00000172366 | MCRIP2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.461 | 0.529 | 0.068 | 3.014362e-02 | TRUE |
| isoComp_00017437 | geneComp_00001531 | ENST00000557356 | ENSG00000080815 | PSEN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.060 | 0.060 | 3.014829e-02 | TRUE |
| isoComp_00002271 | geneComp_00000202 | ENST00000341426 | ENSG00000008130 | NADK | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.696 | 0.530 | -0.166 | 3.079251e-02 | TRUE |
| isoComp_00151339 | geneComp_00017144 | ENST00000683047 | ENSG00000198324 | PHETA1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.882 | 0.650 | -0.231 | 3.114043e-02 | TRUE |
| isoComp_00192399 | geneComp_00033207 | ENST00000610109 | ENSG00000272690 | LINC02018 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.107 | 0.107 | 3.223711e-02 | TRUE |
| isoComp_00153621 | geneComp_00017537 | ENST00000671650 | ENSG00000203875 | SNHG5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.322 | 0.446 | 0.124 | 3.248732e-02 | TRUE |
| isoComp_00184480 | geneComp_00029102 | ENST00000666503 | ENSG00000258526 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.410 | 0.000 | -0.410 | 3.290256e-02 | TRUE | |
| isoComp_00035587 | geneComp_00003281 | ENST00000415428 | ENSG00000105983 | LMBR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.076 | 0.076 | 3.327197e-02 | TRUE |
| isoComp_00119265 | geneComp_00012492 | ENST00000513986 | ENSG00000168916 | ZNF608 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.300 | 0.300 | 3.337807e-02 | TRUE |
| isoComp_00122318 | geneComp_00012908 | ENST00000524243 | ENSG00000170873 | MTSS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.241 | 0.014 | -0.227 | 3.379925e-02 | TRUE |
| isoComp_00182704 | geneComp_00028285 | MSTRG.31172.1 | ENSG00000255495 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.675 | 0.675 | 3.433767e-02 | TRUE | |
| isoComp_00094925 | geneComp_00009602 | ENST00000502608 | ENSG00000152380 | FAM151B | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.350 | 0.350 | 3.445781e-02 | TRUE |
| isoComp_00071557 | geneComp_00007134 | ENST00000676630 | ENSG00000135316 | SYNCRIP | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.312 | 0.190 | -0.122 | 3.459445e-02 | TRUE |
| isoComp_00113001 | geneComp_00011783 | ENST00000298966 | ENSG00000166002 | SMCO4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.591 | 0.802 | 0.211 | 3.459445e-02 | TRUE |
| isoComp_00123540 | geneComp_00013087 | ENST00000302096 | ENSG00000171695 | LKAAEAR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.710 | 0.269 | -0.441 | 3.481438e-02 | TRUE |
| isoComp_00155848 | geneComp_00017929 | MSTRG.7365.17 | ENSG00000205352 | PRR13 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.237 | 0.117 | -0.120 | 3.481966e-02 | TRUE |
| isoComp_00131935 | geneComp_00014220 | ENST00000429348 | ENSG00000177873 | ZNF619 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.113 | 0.113 | 3.518844e-02 | TRUE |
| isoComp_00174425 | geneComp_00025023 | ENST00000457254 | ENSG00000240230 | COX19 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.095 | 0.008 | -0.087 | 3.536180e-02 | TRUE |
| isoComp_00027779 | geneComp_00002469 | MSTRG.20760.5 | ENSG00000101104 | PABPC1L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.006 | 0.093 | 0.086 | 3.573635e-02 | TRUE |
| isoComp_00043239 | geneComp_00004053 | MSTRG.28182.4 | ENSG00000112033 | PPARD | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.004 | 0.072 | 0.068 | 3.601925e-02 | TRUE |
| isoComp_00074310 | geneComp_00007416 | ENST00000359188 | ENSG00000136754 | ABI1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 3.601925e-02 | |
| isoComp_00191031 | geneComp_00032405 | ENST00000635000 | ENSG00000270049 | ATP6V0D1-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.154 | 0.000 | -0.154 | 3.625054e-02 | TRUE |
| isoComp_00007174 | geneComp_00000656 | MSTRG.28277.6 | ENSG00000048544 | MRPS10 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.120 | 0.209 | 0.089 | 3.690654e-02 | TRUE |
| isoComp_00037468 | geneComp_00003492 | ENST00000374182 | ENSG00000107643 | MAPK8 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.112 | 0.000 | -0.112 | 3.698076e-02 | TRUE |
| isoComp_00065169 | geneComp_00006460 | ENST00000503371 | ENSG00000130997 | POLN | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.092 | 0.092 | 3.698793e-02 | TRUE |
| isoComp_00087085 | geneComp_00008710 | ENST00000349511 | ENSG00000144744 | UBA3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.110 | 0.031 | -0.079 | 3.700026e-02 | TRUE |
| isoComp_00000386 | geneComp_00000038 | ENST00000406470 | ENSG00000003147 | ICA1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.075 | 0.000 | -0.075 | 3.780215e-02 | TRUE |
| isoComp_00075809 | geneComp_00007574 | ENST00000464145 | ENSG00000137507 | LRRC32 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.534 | 0.000 | -0.534 | 3.844387e-02 | TRUE |
| isoComp_00093044 | geneComp_00009378 | ENST00000434316 | ENSG00000150403 | TMCO3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.738 | 0.530 | -0.209 | 3.887512e-02 | TRUE |
| isoComp_00032152 | geneComp_00002926 | ENST00000262096 | ENSG00000104219 | ZDHHC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.290 | 0.081 | -0.209 | 3.904089e-02 | TRUE |
| isoComp_00120635 | geneComp_00012664 | MSTRG.15330.22 | ENSG00000169718 | DUS1L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.124 | 0.198 | 0.074 | 3.922821e-02 | |
| isoComp_00039364 | geneComp_00003664 | MSTRG.14432.6 | ENSG00000108840 | HDAC5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.550 | 0.389 | -0.161 | 3.928145e-02 | TRUE |
| isoComp_00199881 | geneComp_00037708 | ENST00000662935 | ENSG00000287670 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.908 | 0.908 | 3.965258e-02 | TRUE | |
| isoComp_00158323 | geneComp_00018632 | ENST00000444957 | ENSG00000214022 | REPIN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.005 | 0.201 | 0.196 | 3.970713e-02 | FALSE |
| isoComp_00113159 | geneComp_00011809 | ENST00000616318 | ENSG00000166133 | RPUSD2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.073 | 0.155 | 0.081 | 4.009378e-02 | TRUE |
| isoComp_00194185 | geneComp_00034241 | ENST00000622085 | ENSG00000276234 | TADA2A | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.008 | 0.082 | 0.074 | 4.009378e-02 | TRUE |
| isoComp_00043324 | geneComp_00004064 | MSTRG.29276.7 | ENSG00000112096 | SOD2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.308 | 0.216 | -0.092 | 4.014498e-02 | TRUE |
| isoComp_00057595 | geneComp_00005606 | MSTRG.27798.6 | ENSG00000124508 | BTN2A2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.369 | 0.024 | -0.345 | 4.014498e-02 | TRUE |
| isoComp_00116063 | geneComp_00012141 | MSTRG.13280.27 | ENSG00000167523 | SPATA33 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.008 | 0.103 | 0.095 | 4.014498e-02 | TRUE |
| isoComp_00165376 | geneComp_00021216 | MSTRG.21793.20 | ENSG00000228315 | GUSBP11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.002 | 0.058 | 0.056 | 4.014498e-02 | TRUE |
| isoComp_00188049 | geneComp_00030980 | ENST00000585075 | ENSG00000263843 | MIF4GD-DT | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.765 | 1.000 | 0.235 | 4.014498e-02 | TRUE |
| isoComp_00114284 | geneComp_00011931 | ENST00000340353 | ENSG00000166575 | TMEM135 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.607 | 0.145 | -0.463 | 4.022518e-02 | TRUE |
| isoComp_00182309 | geneComp_00028091 | MSTRG.20905.25 | ENSG00000254995 | STX16-NPEPL1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.150 | 0.150 | 4.041012e-02 | TRUE |
| isoComp_00176965 | geneComp_00026017 | ENST00000478824 | ENSG00000244649 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.544 | 0.000 | -0.544 | 4.055305e-02 | TRUE | |
| isoComp_00089357 | geneComp_00008985 | ENST00000356309 | ENSG00000146918 | NCAPG2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.505 | 0.382 | -0.123 | 4.069872e-02 | TRUE |
| isoComp_00037659 | geneComp_00003510 | ENST00000371837 | ENSG00000107798 | LIPA | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.083 | 0.024 | -0.059 | 4.109203e-02 | |
| isoComp_00060600 | geneComp_00005954 | ENST00000375495 | ENSG00000127081 | ZNF484 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.973 | 0.406 | -0.566 | 4.156878e-02 | TRUE |
| isoComp_00113387 | geneComp_00011835 | ENST00000400188 | ENSG00000166206 | GABRB3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.057 | 0.057 | 4.206849e-02 | TRUE |
| isoComp_00100307 | geneComp_00010244 | MSTRG.32197.8 | ENSG00000158106 | RHPN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.067 | 0.067 | 4.220919e-02 | TRUE |
| isoComp_00168287 | geneComp_00022443 | ENST00000618253 | ENSG00000231752 | EMBP1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.769 | 0.000 | -0.769 | 4.262952e-02 | TRUE |
| isoComp_00069331 | geneComp_00006882 | ENST00000256255 | ENSG00000133872 | SARAF | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.493 | 0.603 | 0.110 | 4.300699e-02 | TRUE |
| isoComp_00160467 | geneComp_00019295 | ENST00000482067 | ENSG00000219545 | UMAD1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.044 | 0.224 | 0.180 | 4.321729e-02 | TRUE |
| isoComp_00148145 | geneComp_00016731 | ENST00000619322 | ENSG00000196873 | CBWD3 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.055 | 0.004 | -0.051 | 4.331455e-02 | TRUE |
| isoComp_00122701 | geneComp_00012953 | ENST00000471841 | ENSG00000171109 | MFN1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.542 | 0.805 | 0.263 | 4.406864e-02 | TRUE |
| isoComp_00147170 | geneComp_00016623 | ENST00000314752 | ENSG00000196502 | SULT1A1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.782 | 0.937 | 0.155 | 4.423686e-02 | TRUE |
| isoComp_00009615 | geneComp_00000868 | MSTRG.17444.14 | ENSG00000063176 | SPHK2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.130 | 0.008 | -0.122 | 4.432161e-02 | TRUE |
| isoComp_00078644 | geneComp_00007855 | ENST00000266483 | ENSG00000139133 | ALG10 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.700 | 0.438 | -0.263 | 4.432161e-02 | TRUE |
| isoComp_00102887 | geneComp_00010540 | ENST00000472607 | ENSG00000160298 | C21orf58 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.101 | 0.029 | -0.072 | 4.432161e-02 | TRUE |
| isoComp_00161701 | geneComp_00019703 | ENST00000446309 | ENSG00000224046 | DMTF1-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.567 | 0.263 | -0.304 | 4.432161e-02 | TRUE |
| isoComp_00178916 | geneComp_00026560 | ENST00000592291 | ENSG00000249115 | HAUS5 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.100 | 0.026 | -0.074 | 4.432161e-02 | TRUE |
| isoComp_00055198 | geneComp_00005332 | ENST00000370566 | ENSG00000122417 | ODF2L | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.146 | 0.146 | 4.449923e-02 | TRUE |
| isoComp_00148565 | geneComp_00016786 | ENST00000623276 | ENSG00000197062 | ZSCAN26 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.123 | 0.000 | -0.123 | 4.458299e-02 | TRUE |
| isoComp_00183853 | geneComp_00028803 | ENST00000659265 | ENSG00000257556 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.152 | 0.152 | 4.500849e-02 | TRUE | |
| isoComp_00084217 | geneComp_00008416 | MSTRG.2532.4 | ENSG00000143158 | MPC2 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.414 | 0.582 | 0.168 | 4.515036e-02 | TRUE |
| isoComp_00123541 | geneComp_00013087 | ENST00000308906 | ENSG00000171695 | LKAAEAR1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.290 | 0.731 | 0.441 | 4.540865e-02 | TRUE |
| isoComp_00150977 | geneComp_00017097 | MSTRG.19668.7 | ENSG00000198130 | HIBCH | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.192 | 0.358 | 0.166 | 4.601808e-02 | TRUE |
| isoComp_00091749 | geneComp_00009250 | ENST00000456247 | ENSG00000149091 | DGKZ | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.138 | 0.072 | -0.066 | 4.630573e-02 | |
| isoComp_00080452 | geneComp_00008054 | ENST00000565160 | ENSG00000140463 | BBS4 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.067 | 0.067 | 4.634407e-02 | TRUE |
| isoComp_00016447 | geneComp_00001444 | ENST00000378609 | ENSG00000078369 | GNB1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.654 | 0.521 | -0.133 | 4.643257e-02 | TRUE |
| isoComp_00051241 | geneComp_00004867 | ENST00000528910 | ENSG00000118369 | USP35 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.111 | 0.111 | 4.675499e-02 | TRUE |
| isoComp_00119159 | geneComp_00012478 | MSTRG.6179.8 | ENSG00000168876 | ANKRD49 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.070 | 0.000 | -0.070 | 4.759484e-02 | TRUE |
| isoComp_00051944 | geneComp_00004956 | ENST00000322940 | ENSG00000119328 | ABITRAM | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.646 | 0.880 | 0.234 | 4.763274e-02 | TRUE |
| isoComp_00138116 | geneComp_00015194 | ENST00000332783 | ENSG00000183475 | ASB7 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.802 | 0.949 | 0.147 | 4.790003e-02 | TRUE |
| isoComp_00182207 | geneComp_00028041 | ENST00000532599 | ENSG00000254860 | TMEM9B-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.265 | 0.020 | -0.245 | 4.810716e-02 | TRUE |
| isoComp_00133492 | geneComp_00014468 | ENST00000639857 | ENSG00000179295 | PTPN11 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.007 | 0.091 | 0.084 | 4.813183e-02 | TRUE |
| isoComp_00160630 | geneComp_00019378 | ENST00000687563 | ENSG00000221817 | PPP3CB-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.207 | 0.207 | 4.828288e-02 | TRUE |
| isoComp_00176421 | geneComp_00025796 | ENST00000618195 | ENSG00000243766 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.375 | 0.000 | -0.375 | 4.885137e-02 | TRUE | |
| isoComp_00189361 | geneComp_00031614 | ENST00000690695 | ENSG00000267191 | ZNF45-AS1 | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.000 | 0.237 | 0.237 | 4.925345e-02 | TRUE |
| isoComp_00143824 | geneComp_00016096 | ENST00000378585 | ENSG00000187730 | GABRD | HEK293_DMSO_2hA | HEK293_OSMI2_2hA | 0.589 | 0.317 | -0.272 | 4.967793e-02 | TRUE |
- Note:
- The comparisons made can be identified as “from ‘condition_1’ to ‘condition_2’”, meaning ‘condition_1’ is considered the ground state and ‘condition_2’ the changed state. This also means that a positive dIF value indicates that the isoform usage is increased in ‘condition_2’ compared to ‘condition_1’. Since the ‘isoformFeatures’ entry is the most relevant part of the switchAnalyzeRlist object, the most-used standard methods have also been implemented to work directly on isoformFeatures.
4. Circular RNA Analysis
Volcano Plot
No significantly changed circRNA was detected.
Details of sig. circRNA
No significantly changed circRNA was detected.
Annotation of sig. circRNA
No significantly changed circRNA was detected.