Analysis Report: HEK293_TMG_6hB vs HEK293_DMSO_6hB
WANG Ziyi
Date: 19 5月 2022
Summary: HEK293_TMG_6hB vs HEK293_DMSO_6hB
1. Differentially Expressed Genes
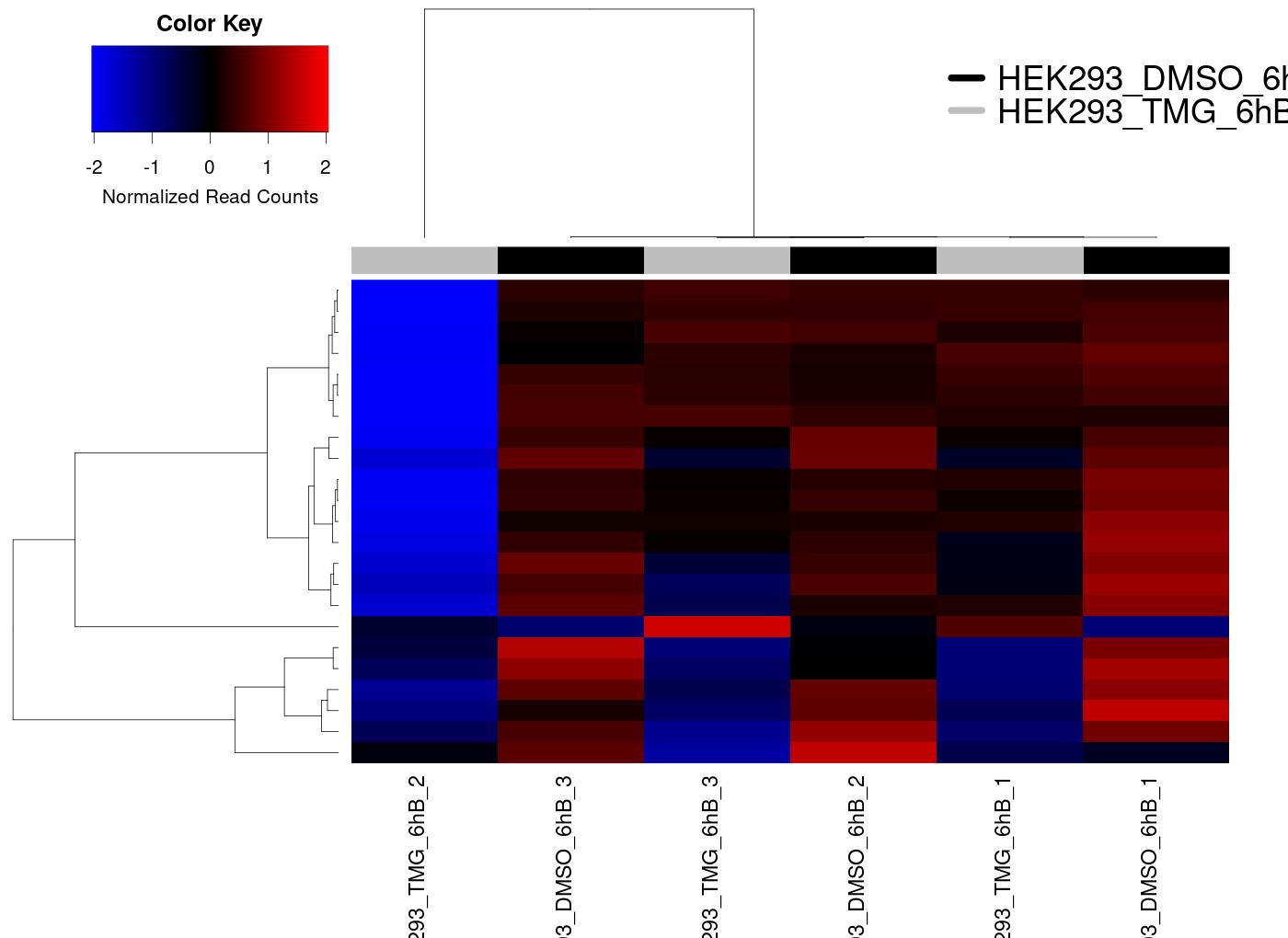
Heatmap
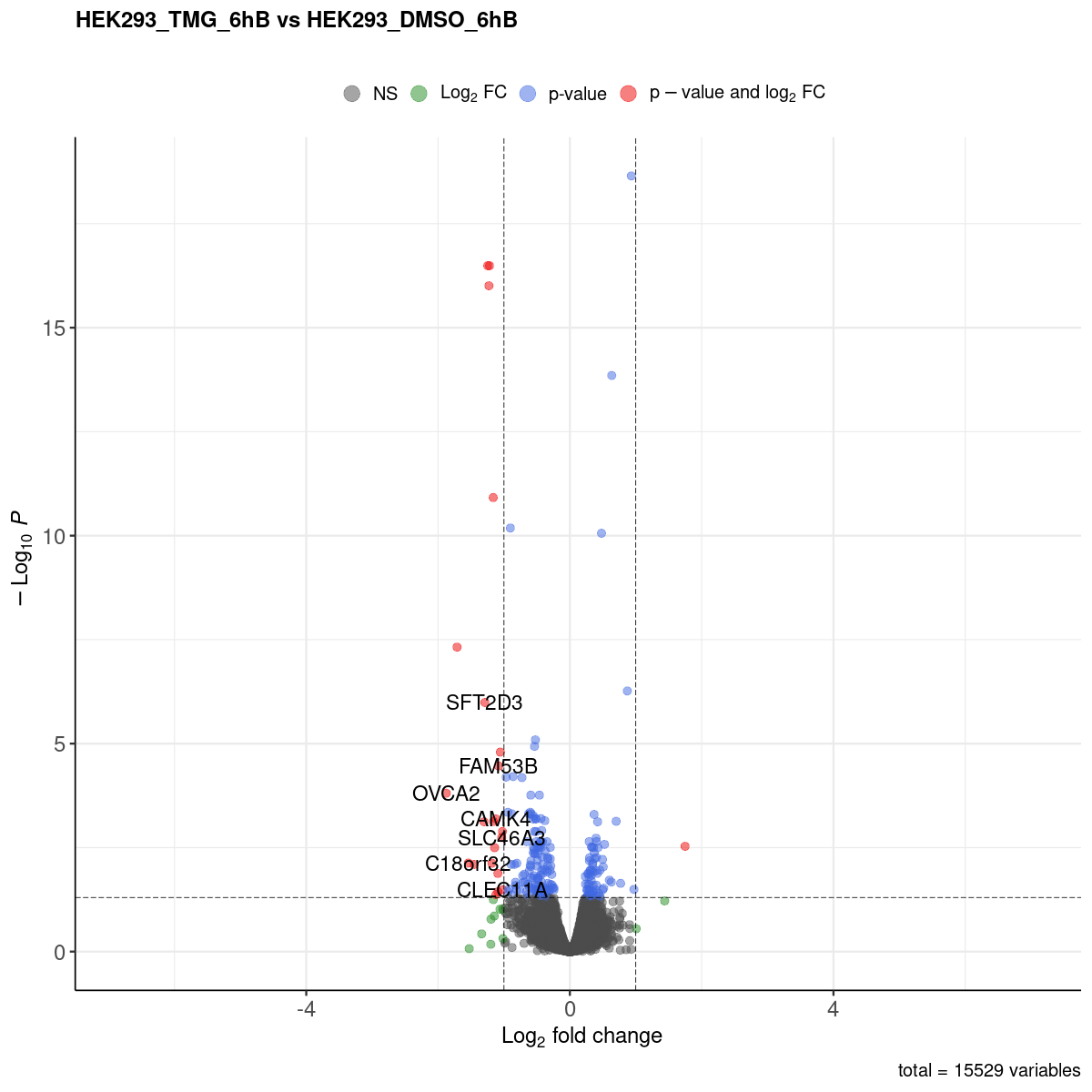
Volcano plot
DEGs list
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_TMG_6hB | SE_Intercept_HEK293_TMG_6hB | Intercept_HEK293_DMSO_6hB | SE_Intercept_HEK293_DMSO_6hB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000105472 | 6320 | CLEC11A | protein_coding | Q9Y240 | FUNCTION: Promotes osteogenesis by stimulating the differentiation of mesenchymal progenitors into mature osteoblasts (PubMed:27976999). Important for repair and maintenance of adult bone (By similarity). {ECO:0000250|UniProtKB:O88200, ECO:0000269|PubMed:27976999}. | Cytoplasm;Direct protein sequencing;Disulfide bond;Glycoprotein;Growth factor;Lectin;Osteogenesis;Reference proteome;Secreted;Signal | This gene encodes a member of the C-type lectin superfamily. The encoded protein is a secreted sulfated glycoprotein and functions as a growth factor for primitive hematopoietic progenitor cells. An alternative splice variant has been described but its biological nature has not been determined. [provided by RefSeq, Jul 2008]. | hsa:6320; | cytoplasm [GO:0005737]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; carbohydrate binding [GO:0030246]; growth factor activity [GO:0008083]; ossification [GO:0001503]; positive regulation of cell population proliferation [GO:0008284] | 11920266_SCGF is selectively produced by osseous and hematopoietic stromal cells, and can mediate their proliferative activity on primitive hematopoietic progenitor cells. 14746805_Results indicate that interleukin-4, together with recombinant human stem cell factor, can induce T cell maturation from cord blood progenitor cells, and that IL-4 increased the expression of FcepsilonRI on fetal liver mast cells. 15234225_Human vascular smooth muscle cells express stem cell factor(SCF) and its receptor, c-kit. SCF is released from its membrane-bound form via MMP-9. SCF/c-kit signaling may affect SMC function via an autocrine pathway. 19416273_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19528216_phagocytosis of hemozoin promotes a decrease in SCGF gene products, which may contribute to reduced erythropoiesis in children with severe malarial anemia. 19884328_The results presented here demonstrate that homozygous T at -539 in the SCGF promoter is associated with elevated SCGF production, enhanced erythropoiesis, and protection against severe malarial anemia. 21943129_These studies highlight a possible role of SCGFalpha in imatinib-induced changes of gastrointestinal stromal tumors structure, consistent with a therapeutic response. 23357302_This is the first report of SCGF beta in heart failure patients. 26440592_level of expression correlates with pain response in subjects with intervertebral disc disorders 26564003_Data show that asymptomatic patients with unstable plaques exhibited higher levels of endothelial microparticles (EMPs), CXCL9 chemokine and stem cell growth factor; lymphocyte secreted C-type lectin (SCGF-beta) compared to those with stable plaques. 27976999_Clec11a maintains the adult skeleton by promoting the differentiation of mesenchymal progenitors into mature osteoblasts. 34140410_The effect of parathyroid hormone on osteogenesis is mediated partly by osteolectin. | ENSMUSG00000004473 | Clec11a | 117.7521 | 0.4934360 | -1.019065 | 0.2901774 | 12.24201 | 4.672547e-04 | 3.275181e-02 | Yes | No | 141.73068 | 33.27312 | 275.26032 | 59.65727 | |
| ENSG00000112238 | 59336 | PRDM13 | protein_coding | Q9H4Q3 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Methyltransferase;Nucleus;Reference proteome;Repeat;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Zinc;Zinc-finger | hsa:59336; | nucleus [GO:0005634]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone methyltransferase activity [GO:0042054]; metal ion binding [GO:0046872]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; negative regulation of transcription by RNA polymerase II [GO:0000122]; neurogenesis [GO:0022008]; regulation of gene expression [GO:0010468] | 25546159_Findings indicate that Prdm13/Nkx2-1-mediated signaling in the DMC declines with advanced age, leading to decreased sleep quality and increased adiposity. 27777503_The duplication found in the RFS355 family is distinct from the previously reported duplication and provides additional support that dysregulation of PRDM13, not CCNC, is the cause of NCMD mapped to the MCDR1 locus. 29767251_The over-expression of PRDM13 upregulated deleted in liver cancer 1 (DLC1) to inhibit the proliferation and invasion of U87 cells. 30710461_These results prompt us to propose RECQL5 as a gene that would be worth to analyze in larger studies to explore its possible implication in BC susceptibility. 32476814_A unique PRDM13-associated variant in a Georgian Jewish family with probable North Carolina macular dystrophy and the possible contribution of a unique CFH variant. 34125159_North Carolina Macular Dystrophy: Phenotypic Variability and Computational Analysis of Disease-Associated Noncoding Variants. 34526759_A novel duplication involving PRDM13 in a Turkish family supports its role in North Carolina macular dystrophy (NCMD/MCDR1). 34730112_A recessive PRDM13 mutation results in congenital hypogonadotropic hypogonadism and cerebellar hypoplasia. | ENSMUSG00000040478 | Prdm13 | 122.9999 | 0.4692251 | -1.091648 | 0.3243540 | 11.86579 | 5.717440e-04 | 3.643263e-02 | Yes | No | 169.19442 | 39.23578 | 290.91997 | 61.81288 | ||
| ENSG00000139508 | 283537 | SLC46A3 | protein_coding | Q7Z3Q1 | Alternative splicing;Glycoprotein;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of a transmembrane protein family that transports small molecules across membranes. The encoded protein has been found in lysosomal membranes, where it can transport catabolites from the lysosomes to the cytoplasm. This protein has been shown to be an effective transporter of the cytotoxic drug maytansine, which is used in antibody-based targeting of cancer cells. [provided by RefSeq, Dec 2016]. | hsa:283537; | extracellular exosome [GO:0070062]; integral component of lysosomal membrane [GO:1905103]; transmembrane transporter activity [GO:0022857]; vacuolar transmembrane transport [GO:0034486] | 26631267_our results establish SLC46A3 as a direct transporter of maytansine-based catabolites from the lysosome to the cytoplasm | ENSMUSG00000029650 | Slc46a3 | 178.0252 | 0.4882665 | -1.034259 | 0.2263179 | 20.32070 | 6.548824e-06 | 1.814786e-03 | Yes | No | 131.62643 | 29.69794 | 262.67847 | 57.15198 | ||
| ENSG00000152495 | 814 | CAMK4 | protein_coding | Q16566 | FUNCTION: Calcium/calmodulin-dependent protein kinase that operates in the calcium-triggered CaMKK-CaMK4 signaling cascade and regulates, mainly by phosphorylation, the activity of several transcription activators, such as CREB1, MEF2D, JUN and RORA, which play pivotal roles in immune response, inflammation, and memory consolidation. In the thymus, regulates the CD4(+)/CD8(+) double positive thymocytes selection threshold during T-cell ontogeny. In CD4 memory T-cells, is required to link T-cell antigen receptor (TCR) signaling to the production of IL2, IFNG and IL4 (through the regulation of CREB and MEF2). Regulates the differentiation and survival phases of osteoclasts and dendritic cells (DCs). Mediates DCs survival by linking TLR4 and the regulation of temporal expression of BCL2. Phosphorylates the transcription activator CREB1 on 'Ser-133' in hippocampal neuron nuclei and contribute to memory consolidation and long term potentiation (LTP) in the hippocampus. Can activate the MAP kinases MAPK1/ERK2, MAPK8/JNK1 and MAPK14/p38 and stimulate transcription through the phosphorylation of ELK1 and ATF2. Can also phosphorylate in vitro CREBBP, PRM2, MEF2A and STMN1/OP18. {ECO:0000269|PubMed:10617605, ECO:0000269|PubMed:17909078, ECO:0000269|PubMed:18829949, ECO:0000269|PubMed:7961813, ECO:0000269|PubMed:8065343, ECO:0000269|PubMed:8855261, ECO:0000269|PubMed:8980227, ECO:0000269|PubMed:9154845}. | 3D-structure;ATP-binding;Adaptive immunity;Calcium;Calmodulin-binding;Cytoplasm;Glycoprotein;Immunity;Inflammatory response;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The product of this gene belongs to the serine/threonine protein kinase family, and to the Ca(2+)/calmodulin-dependent protein kinase subfamily. This enzyme is a multifunctional serine/threonine protein kinase with limited tissue distribution, that has been implicated in transcriptional regulation in lymphocytes, neurons and male germ cells. [provided by RefSeq, Jul 2008]. | hsa:814; | cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; glutamatergic synapse [GO:0098978]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; postsynapse [GO:0098794]; ATP binding [GO:0005524]; calcium-dependent protein serine/threonine kinase activity [GO:0009931]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; adaptive immune response [GO:0002250]; inflammatory response [GO:0006954]; intracellular signal transduction [GO:0035556]; long-term memory [GO:0007616]; myeloid dendritic cell differentiation [GO:0043011]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of transcription, DNA-templated [GO:0045893]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of osteoclast differentiation [GO:0045670]; regulation of T cell differentiation in thymus [GO:0033081]; signal transduction [GO:0007165] | 12065094_CaMKIV proteins were found in the nucleus of epithelial ovarian cancer tissue. CaMKIV expression was significantly associated with clinical stage (P<0.01), histological grade (P<0.01), and clinical outcome (P<0.01). 14701808_sequestration of CaMKK may be the molecular mechanism by which catalytically inactive mutants of CaMKIV exert their 'dominant-negative' functions within the cell 15143065_the Ca(2+)/CaM binding-autoinhibitory domain of CaMKIV is required for association of the kinase with PP2A 15591024_calcium/CaMKIV signaling pathway may play an important role in the excitation-mediated regulation of corticotropin releasing hormone synthesis 15665723_the function of CaMK II is essential for PAF-induced macrophage priming, while CaMK IV is not specific for priming by PAF and appears to have a direct link in TLR4-mediated events 15840651_CaMKIV is expressed in human sperm and may have a role in the regulation of human sperm motility 15841182_Results identify calcium/calmodulin-dependent kinase IV as being responsible for the increased expression of CREM and the decreased production of interleukin-2 in systemic lupus erythematosus T cells. 17909078_a novel link between TLR4 and a calcium-dependent signaling cascade comprising CaMKIV-CREB-Bcl-2 that is essential for DC survival. 18053176_Transgenic CaMKIV plays a modulatory role in the nucleus accumbens in anxiety-like behavior of adult CaMKIV variant mice. 18606955_Observational study of gene-disease association. (HuGE Navigator) 18660489_Observational study of gene-disease association. (HuGE Navigator) 18829949_CaMK-4 expression correlates positively with the ability to form long-term memory and implicates the decline of CaMKIV signaling mechanisms in age-related memory deficits. 19001277_CaMKIV plays a critical role in the development and persistence of cocaine-induced behaviors, through mechanisms dissociated from acute effects on gene expression and CREB-dependent transcription. 19017650_hnRNP L is an essential component of CaMKIV-regulated alternative splicing through CA repeats, with its phosphorylation likely playing a critical role. 19386606_a group of RNA elements are responsive to PKA and CaMKIV from in vivo selection 19436069_CaMKIV is a molecular link between Group I mGluRs and fragile X mental retardation protein in anterior cingulate cortex neurons 19506079_analysis of regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification 19538941_these data indicate that the B subunits alpha and delta are essential for the interaction of PP2A with CaMKIV. 19633294_Data show that RA-induced repression of the CaMKIV signaling pathway may represent an early event in retinoid-dependent neuronal differentiation. 20171262_These findings suggest that PLC/CAMK IV-NF-kappaB is involved in RAGE mediated signaling pathway in human endothelial cells. 20378615_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 21514275_The regulation of RORalpha activity by PKA as well as CaMK-IV provides a new link in the signalling network that regulates metabolic processes such as glycogen and lipid metabolism. 21612516_Prolongevity genes are activated by CAMKIV, the levels of which are influenced by rs10491334, a single-nucleotide polymorphism associated with human longevity. 22897820_study suggests that the mutations in CAMK4 may lead to abnormal semen parameters 23049845_CaMK4 regulates beta-cell proliferation and apoptosis in a CREB-dependent manner and CaMK4-induced IRS-2 expression is important in these processes 23103515_Phosphorylated Notch1-IC by CaMKIV increases Notch1-IC stability, which enhances osteoclast differentiation. 24442360_An imbalance of specific isoforms of CYFIP1, an FMRP interaction partner, and CAMK4, a transcriptional regulator of the FMRP gene, modulates risk for autism spectrum disorders. 24667640_CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. 25446257_Expression of CaMKIV inhibits autophosphorylation and activation of CaMKII, and elicits G0/G1cell cycle arrest,impairing cell proliferation. 26909912_The T-allele of rs10491334 in CAMK4 was associated with hypertension in the Uygur group. 27032767_Within the pH range 5.0-11.5, CAMK4 maintained both its secondary and tertiary structures, along with its function, whereas significant aggregation was observed at acidic pH (2.0-4.5). 27298345_hTau accumulation impairs synapse and memory by CaN-mediated suppression of nuclear CaMKIV/CREB signaling. 27659345_A positive association was not observed between rs10491334 in the CAMK4 gene and longevity in a Chinese population. 28734942_Genotype and allele frequencies of CAMKIV gene SNPs differed significantly between alcohol dependence patients and control subjects. The results of the present study suggest that CAMKIV might be a candidate alcohol dependence gene. 28744811_vanillin binds strongly to the active site cavity of CAMKIV and stabilized by a large number of non-covalent interactions. 29985166_CaMK4 is pivotal in immune and nonimmune podocyte injury and that its targeted cell-specific inhibition preserves podocyte structure and function and should have therapeutic value in lupus nephritis and podocytopathies, including focal segmental glomerulosclerosis. 30113881_Clinical disease severity directly correlates with calmodulin-dependent kinase IV (CaMKIV) activation, as does expression of proinflammatory cytokines and histologic features of colitis. In wild-type mice, CaMKIV activation is associated with increases in expression of 2 cell cycle proarrest signals: p53 and p21 30462889_CaMK4 could be responsible for glycolysis, which contributes to the production of IL-17, and CaMK4 may contribute to aberrant expression of GLUT1 in T cells from patients with active SLE. 31624237_MiR-129-5p inhibits liver cancer growth by targeting calcium calmodulin-dependent protein kinase IV (CAMK4). 31976761_rs2300782 of gene CAMK4 is associated with diabetic retinopathy incidence and severity among Chinese Hui population. 31978801_Comparative transcriptome analysis reveals a potential role for CaMK4 in gammadeltaT17 cells from systemic lupus erythematosus patients with lupus nephritis. 32460794_CAMKK2-CAMK4 signaling regulates transferrin trafficking, turnover, and iron homeostasis. 32572897_MiR-507 inhibits the growth and invasion of trophoblasts by targeting CAMK4. 32738170_CaMK4 promotes abortion-related Th17 cell imbalance by activating AKT/mTOR signaling pathway. 33784256_Aberrantly glycosylated IgG elicits pathogenic signaling in podocytes and signifies lupus nephritis. | ENSMUSG00000038128 | Camk4 | 232.1739 | 0.4607923 | -1.117811 | 0.2282276 | 22.89330 | 1.712475e-06 | 6.481476e-04 | Yes | No | 148.56184 | 47.28360 | 313.52115 | 97.69913 | |
| ENSG00000173349 | 84826 | SFT2D3 | protein_coding | Q587I9 | FUNCTION: May be involved in fusion of retrograde transport vesicles derived from an endocytic compartment with the Golgi complex. {ECO:0000250|UniProtKB:P38166}. | Membrane;Protein transport;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:84826; | integral component of membrane [GO:0016021]; protein transport [GO:0015031]; vesicle-mediated transport [GO:0016192] | ENSMUSG00000044982 | Sft2d3 | 390.3586 | 0.4090293 | -1.289724 | 0.2478782 | 37.43353 | 9.458219e-10 | 1.024583e-06 | Yes | No | 516.90657 | 143.52795 | 1081.08078 | 281.72562 | |||
| ENSG00000177576 | 497661 | C18orf32 | protein_coding | Q8TCD1 | FUNCTION: May activate the NF-kappa-B signaling pathway. {ECO:0000269|PubMed:12761501}. | Endoplasmic reticulum;Lipid droplet;Reference proteome | hsa:497661; | endoplasmic reticulum [GO:0005783]; lipid droplet [GO:0005811]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123] | ENSMUSG00000036299 | BC031181 | 135.1984 | 0.3437934 | -1.540386 | 0.3670042 | 16.32079 | 5.347392e-05 | 7.409247e-03 | Yes | No | 69.49664 | 31.83825 | 208.68133 | 93.04039 | |||
| ENSG00000180425 | 54494 | C11orf71 | protein_coding | Q6IPW1 | Alternative splicing;Reference proteome | hsa:54494; | nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634] | ENSMUSG00000042293 | Gm5617 | 122.2605 | 0.4408817 | -1.181537 | 0.2874409 | 16.27604 | 5.475204e-05 | 7.499142e-03 | Yes | No | 70.96031 | 11.57519 | 157.43721 | 23.90883 | ||||
| ENSG00000189319 | 9679 | FAM53B | protein_coding | Q14153 | FUNCTION: Acts as a regulator of Wnt signaling pathway by regulating beta-catenin (CTNNB1) nuclear localization. {ECO:0000269|PubMed:25183871}. | Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Wnt signaling pathway | hsa:9679; | nucleus [GO:0005634]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; protein import into nucleus [GO:0006606]; regulation of canonical Wnt signaling pathway [GO:0060828]; Wnt signaling pathway [GO:0016055] | 19727342_Strong candidate gene for age-related macular degeneration, ARMS2, (human 10q26.13). Conclusion is based on a massive expression data set for mouse (103 strains) and joint analysis of RetNet database. 23958962_First study to identify risk variants for cocaine dependence using GWAS; rs2629540 at the FAM53B identified as a risk factor in African- and European-Americans | ENSMUSG00000030956 | Fam53b | 408.8200 | 0.4726518 | -1.081150 | 0.1931551 | 29.98697 | 4.349592e-08 | 3.455316e-05 | Yes | No | 294.53144 | 50.04981 | 604.32575 | 99.00296 | ||
| ENSG00000189343 | 125208 | RPS2P46 | processed_pseudogene | 307.2564 | 0.4920024 | -1.023263 | 0.2485970 | 21.07069 | 4.426482e-06 | 1.299688e-03 | Yes | No | 490.96076 | 109.77427 | 803.01471 | 163.78497 | ||||||||||
| ENSG00000196656 | transcribed_processed_pseudogene | 3031.6616 | 0.4465900 | -1.162977 | 0.2117959 | 60.86680 | 6.107007e-15 | 1.212852e-11 | Yes | No | 5128.12625 | 1363.58927 | 10157.33845 | 2538.81215 | ||||||||||||
| ENSG00000198618 | PPIAP22 | processed_pseudogene | 3324.3010 | 0.4295179 | -1.219210 | 0.1989460 | 87.61705 | 7.943691e-21 | 3.236557e-17 | Yes | No | 5647.67188 | 1510.86634 | 11367.58922 | 2859.33006 | |||||||||||
| ENSG00000204934 | 401431 | ATP6V0E2-AS1 | lncRNA | 127.1156 | 0.4532655 | -1.141572 | 0.2617281 | 18.43453 | 1.758421e-05 | 3.174750e-03 | Yes | No | 102.91759 | 12.20610 | 219.22244 | 22.85261 | ||||||||||
| ENSG00000213442 | 390354 | RPL18AP3 | processed_pseudogene | 3928.2828 | 0.4213475 | -1.246917 | 0.2017396 | 87.56672 | 8.148432e-21 | 3.236557e-17 | Yes | No | 6792.52462 | 1840.79416 | 14072.77461 | 3575.42673 | ||||||||||
| ENSG00000223459 | 653199 | TCAF1P1 | unprocessed_pseudogene | 203.2553 | 0.4688205 | -1.092892 | 0.2973602 | 14.70998 | 1.253810e-04 | 1.322159e-02 | Yes | No | 312.55967 | 103.84873 | 571.06143 | 174.73527 | ||||||||||
| ENSG00000227097 | RPS28P7 | processed_pseudogene | 1585.4240 | 0.4816135 | -1.054052 | 0.2307187 | 31.61735 | 1.877449e-08 | 1.597977e-05 | Yes | No | 2676.33330 | 741.94796 | 4810.40842 | 1251.52466 | |||||||||||
| ENSG00000236552 | 728658 | RPL13AP5 | processed_pseudogene | 3914.3080 | 0.4053490 | -1.302763 | 0.2922218 | 22.23409 | 2.413505e-06 | 7.568244e-04 | Yes | No | 6683.27786 | 1909.62910 | 13855.08690 | 3734.63319 | ||||||||||
| ENSG00000244398 | 729362 | processed_pseudogene | 7985.4625 | 0.4271397 | -1.227220 | 0.1969315 | 84.80194 | 3.297999e-20 | 9.824739e-17 | Yes | No | 14210.13116 | 5609.87169 | 32129.33903 | 12031.79874 | |||||||||||
| ENSG00000256861 | 65082 | protein_coding | H3BMM5 | Mouse_homologues FUNCTION: Plays a role in vesicle-mediated protein trafficking to lysosomal compartments including the endocytic membrane transport and autophagic pathways. Believed to act as a core component of the putative HOPS and CORVET endosomal tethering complexes which are proposed to be involved in the Rab5-to-Rab7 endosome conversion probably implicating MON1A/B, and via binding SNAREs and SNARE complexes to mediate tethering and docking events during SNARE-mediated membrane fusion. The HOPS complex is proposed to be recruited to Rab7 on the late endosomal membrane and to regulate late endocytic, phagocytic and autophagic traffic towards lysosomes. The CORVET complex is proposed to function as a Rab5 effector to mediate early endosome fusion probably in specific endosome subpopulations. Required for fusion of endosomes and autophagosomes with lysosomes; the function is dependent on its association with VPS16 but not VIPAS39. The function in autophagosome-lysosome fusion implicates STX17 but not UVRAG. {ECO:0000250|UniProtKB:Q96AX1}. | Protein transport;Reference proteome;Transport | Mouse_homologues mmu:77573; | clathrin-coated vesicle [GO:0030136]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; protein transport [GO:0015031]; vesicle-mediated transport [GO:0016192] | Mouse_homologues 12538872_VPS33A is mutated in Hermansky-Pudlak syndrome and may have a role in melanogenesis 19254700_These studies suggest that loss of Purkinje neurons is the most obvious neurological atrophy in the buff mutant mouse, a structural change that generates motor coordination deficits and impaired postural phenotypes. 19419298_Knockdown of Vps33a expression reduced the lysosomal storage of RANKL and caused the accumulation of newly synthesized RANKL in the Golgi apparatus, indicating that Vps33a is involved in transporting RANKL from the Golgi apparatus to secretory lysosomes. 26259518_VPS33A(D251E) mutation plays dual roles by increasing the HOPS complex assembly and its association with the autophagic SNARE complex, which selectively affects the autophagosome-lysosome fusion. | ENSMUSG00000029434 | Vps33a | 116.2839 | 0.3656085 | -1.451629 | 0.3659051 | 16.08582 | 6.053556e-05 | 8.014908e-03 | Yes | No | 169.42406 | 289.94366 | 459.89469 | 754.18942 | |||
| ENSG00000262664 | 124641 | OVCA2 | protein_coding | Q8WZ82 | Hydrolase;Reference proteome | hsa:124641; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; hydrolase activity [GO:0016787]; response to retinoic acid [GO:0032526] | 11979432_is downregulated and degraded during retinoid-induced apoptosis 32182256_OVCA2 was confirmed as a serine hydrolase with a strong preference for long-chain alkyl ester substrates (>10-carbons) and high selectivity against a variety of short, branched, and substituted esters, homologous to FSH1 from S. cerevisiae 33715035_Human OVCA2 and its homolog FSH3-induced apoptosis in Saccharomyces cerevisiae. | ENSMUSG00000038268 | Ovca2 | 109.4564 | 0.2724585 | -1.875892 | 0.3680056 | 26.61520 | 2.482821e-07 | 1.557121e-04 | Yes | No | 105.70818 | 24.85682 | 325.37626 | 64.92346 | |||
| ENSG00000271122 | 101930085 | HERPUD2-AS1 | lncRNA | 188.0950 | 0.3052907 | -1.711745 | 0.2833422 | 43.81402 | 3.611091e-11 | 4.781084e-08 | Yes | No | 189.44611 | 56.73616 | 525.88885 | 146.27800 | ||||||||||
| ENSG00000274425 | lncRNA | 161.5378 | 0.4480482 | -1.158274 | 0.2432093 | 22.36497 | 2.254492e-06 | 7.462369e-04 | Yes | No | 179.34513 | 33.29190 | 382.40466 | 64.32540 | ||||||||||||
| ENSG00000287856 | protein_coding | A0A590UJK7 | Mouse_homologues FUNCTION: Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins. Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A. Also hydroxylates HIF2A. Has a preference for the CODD site for both HIF1A and HIF1B. Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex. Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes. EGLN1 is the most important isozyme under normoxia and, through regulating the stability of HIF1, involved in various hypoxia-influenced processes such as angiogenesis in retinal and cardiac functionality. Target proteins are preferentially recognized via a LXXLAP motif. {ECO:0000269|PubMed:18096761, ECO:0000269|PubMed:18500250, ECO:0000269|PubMed:21435465}. | Dioxygenase;Iron;Metal-binding;Oxidoreductase;Reference proteome;Signal | Mouse_homologues mmu:112405; | dioxygenase activity [GO:0051213]; iron ion binding [GO:0005506]; L-ascorbic acid binding [GO:0031418]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen [GO:0016705] | Mouse_homologues 16966370_Taken together, these data indicate that among all three PHD proteins, PHD2 is uniquely essential during mouse embryogenesis. 17044072_Egln1/2/3 play dual roles in chondrocyte growth, acting as oxygen sensors and mediators of late stage events in the cell cycle. 17646578_PHD2 is a major negative regulator for vascular growth in adult mice. 18056838_PHD1/3 double deficiency leads to erythrocytosis partly by activating the hepatic HIF-2alpha/erythropoietin pathway, whereas PHD2 deficiency leads to erythrocytosis by activating the renal Epo pathway 18096761_Mice lacking PHD2 exhibit premature mortality associated with marked venous congestion and dilated cardiomyopathy 18285459_through this novel pathway involving P1465 hydroxylation and Ser5 phosphorylation of Rbp1, pVHL may regulate tumor growth 18347341_Chronic hypoxia not only increases the pool of PHDs but also overactivates the three PHD isoforms. 18824759_Inhibition of PHD2 by shRNA led to significant improvement in angiogenesis and contractility by in vitro and in vivo experiments. 19217150_Haplodeficiency of PHD2 did not affect tumor vessel density or lumen size, but normalized the endothelial lining and vessel maturation. 19338032_Prolyl hydroxylase enzyme (PHD) is the key enzyme responsible for degrading HIF-1. HIF activation is an approach to increase vascularity and bone formation following skeletal trauma. 19546213_Data indicate that PHD2 protein stability is regulated by a ubiquitin-independent proteasomal pathway involving FKBP38 as adaptor protein that mediates proteasomal interaction. 19683511_the embryopathic effects are associated with knockdown of EGNL1 and the associated induction of Igfbp1 mRNA in the placenta, but not the embryo. 20400342_The findings indicate that the HIF-prolyl hydroxylase 2:erythropoietin pathway is robustly inducible in aging mice. 20574527_the underlying mechanism by which ET-1, through the regulation of PHD2, controls HIF-1alpha stability and thereby regulates angiogenesis and melanoma cell invasion 20733101_Sustained loss of PHD activity and subsequent hypoxia-inducible factor activation, as would occur in the setting of chronic ischemia, are sufficient to account for many of the changes in the hearts of individuals with chronic coronary artery disease 21270129_Cardiomyocyte-specific prolyl-4-hydroxylase domain 2 knock out protects from acute myocardial ischemic injury. 21435465_PHD2 mediates oxygen-induced retinopathy in neonatal mice 21983962_results unravel how PHD2 regulates arteriogenesis and artery homeostasis by controlling a specific differentiation state in macrophages and suggest new treatment options for ischaemic disorders 22354010_PHD2 inhibition is essential for the regulation of the anti-tumoral activity in mouse tumor cells and might bring some new insight in our understanding of tumor growth inhibition. 22420978_This study shows that inhibiting prolyl hydroxylase domain 2 increases the hepatocarcinogenesis and stimulates the development of cholangiocarcinoma. 22611156_Data show it is feasible to reactivate hepatic erythropoietin production using systemically delivered siRNAs targeting the EglN1, EglN2 and EglN3 prolyl hydroxylase mRNA specifically in the liver. 22615432_Alterations in the function of all 3 isoforms of prolyl-4-hydroxylase enzymes (PHD1-3) in the first 24 h following transient focal cerebral ischaemia are reported. 22933585_Neuron-specific prolyl-4-hydroxylase domain 2 knockout reduces brain injury after transient cerebral ischemia. 23264599_Conditional loss of PHD2 in mice leads to HIF-2alpha-dependent erythrocytosis, whereas HIF-1alpha protects these mice. 23616286_Phd2 downregulation by ANG1 promotes proarteriogenic macrophages. 23630130_Prolyl hydroxylase domain protein 2 plays a critical role in diet-induced obesity and glucose intolerance. 23690557_Heterozygous inactivation of the enzyme PHD2 is associated with markedly enhanced ventilatory sensitivity to hypoxia. 23778187_Phd2 deletion in myeloid lineage attenuates hypertensive cardiovascular hypertrophy and fibrosis, which may be mediated by decreased inflammation- and fibrosis-associated gene expression in macrophages 23798557_Egln1 deficiency only in keratinocytes and not in myeloid or endothelial cells was found to lead to faster wound closure, which involved enhanced migration of the hyperproliferating epithelium. 23913502_High PHD2 expression is associated with tumor development. 24121508_These studies formally prove that a missense mutation in PHD2 is the cause of the erythrocytosis, show that this occurs through haploinsufficiency, and point to multifactorial control of red cell mass by PHD2. 24376825_PHD2 silencing through siRNA treatment not only increased the expression of HIF1alpha and VEGFa, but also improved fibroblast proliferation leading to improvement in diabetic wound healing. 24753072_Phd2 plays an important role in regulating bone formation in part by modulating expression of Osx and bone formation marker genes. 24789921_Hif-p4h-2-deficient mice, whether fed normal chow or a high-fat diet, had less adipose tissue, smaller adipocytes, and less adipose tissue inflammation than their littermates. 25546437_PHD2 activity is a critical contributor to the high fat-diet-induced cardiac dysfunction. 25647640_Regulatory link was discovered between mitochondrial Txnrd and the JNK-PHD2-Hif-1alpha axis, which highlights how the loss of Txnrd2 and the resulting altered mitochondrial redox balance impairs tumor growth as well as tumor-related angiogenesis. 25832622_results suggest that HLV delivery of shPHD2 might become a promising treatment strategy to promote vascular regeneration in critical limb ischemia disease via enhancing innate endogenous pathways 26047609_a genetic link between EGLN1 and VWF in a constitution specific manner which could modulate thrombosis/bleeding susceptibility and outcomes of hypoxia, is reported. 26075818_combined deletion of Phd2 and Phd3 dramatically decreased expression of phospholamban (PLN), resulted in sustained activation of calcium/calmodulin-activated kinase II (CaMKII), and sensitized mice to chronic beta-adrenergic stress-induced myocardial injury 26323721_findings show hypoxia and loss of PHD2 revert cancer-associated fibroblast (CAF) activation; this reversion is associated with loss of matrix stiffening and consequently reduction in spontaneous metastases to lungs and liver 26324945_PHD2 in the liver has a role in survival in lactic acidosis by activating the Cori cycle 26452676_Key messages: HIF-P4H-2 deficiency protects skeletal muscle from ischemia-reperfusion injury. The mechanisms involved are mediated via normoxic HIF-1alpha and HIF-2alpha stabilization. HIF-P4H-2 deficiency increases capillary size but not number. HIF-P4H-2 deficiency maintains energy metabolism during ischemia-reperfusion. 26562260_Data (including data from studies in transgenic/knockout mice) suggest that expression of Phd2/Egln1 in chondrocytes plays key role in chondrogenesis, osteogenesis, and developmental gene expression regulation. 26848160_HIF-P4H-2 inhibition may be a novel strategy for protecting against the development of atherosclerosis. 26949511_Stabilized HIF-1alpha induced by PHD2 conditional knockout resulted in the transition of muscle fibers toward a slow fiber type via a calcineurin/NFATc1 signaling pathway. 26972007_Diminished degradation of FLNA upon PHD2 inactivation in hypoxia rearranges the actin cytoskeleton to reduce the number of dendritic spines, synapses. 26976644_Results found that loss of endothelial PHD2 induced pulmonary arterial hypertension and vascular remodeling in a HIF-2-dependent fashion. 27001147_brain tissue protection and increased angiogenesis upon sub-acute ischemic stroke was completely absent in Phd2 knockout mice that were additionally deficient for both Hif1a and Hif2a 27030384_miR-21 contributes to the protection of delayed ischemic preconditioning against renal ischemia reperfusion injury in mice, which is at least in part mediated by targeting of PHD2 and subsequently up-regulating HIF-1alpha/VEGF pathway. 27082941_data identify the PHD2:HIF-2alpha:EPO axis as a so far unknown regulator of osteohematology by controlling bone homeostasis. 27393382_We conclude that the activation of the HIF pathway induced by PHD2 deficiency enhances the effect of running training 27613846_expression of PHD2 in endothelial cells plays a critical role in preventing pulmonary arterial remodeling in mice 27614241_deleting Phd1-3 genes in osteoblasts increased osteoclast formation in vitro and in bone. 27683416_Epo transcription in brain pericytes was HIF-2 dependent and cocontrolled by PHD2 and PHD3, oxygen- and 2-oxoglutarate-dependent prolyl-4-hydroxylases that regulate HIF activity. 27720797_This study demonstrated that the Neuronal prolyl-4-hydroxylase 2 deficiency improves cognitive abilities in a murine model of cerebral hypoperfusion. 27775044_Phd2 expressed in chondrocytes inhibits endochondral ossification at the epiphysis by suppressing HIF signaling pathways. 27795296_Results identified a critical role of PHD2 for a reversible glycolytic reprogramming in macrophages with a direct impact on their function. 27821476_Notch ligand genes Jag1, Jag2, and Dll1 and target Hes1 became downregulated upon aging HIF-2alpha dependently. 27827416_aberrant hypoxic responses due to dysfunction of PHD2 caused both glomerular and tubular damages in HFD-induced obese mice. Phd2-inactivation provides a novel strategy against obesity-induced kidney injury 28266128_the expression of PHD2 in endothelial cells plays a critical role in renal fibrosis and vascular remodelling in adult mice. 28805660_Phd2 is the dominant HIF-hydroxylase in neutrophils under normoxic conditions; intrinsic regulation of glycolysis and glycogen stores is linked to the resolution of neutrophil-mediated inflammatory responses 28847650_PHD2 and PHD3 are essential for normal kidney development as the combined inactivation of stromal PHD2 and PHD3 resulted in renal failure that was associated with reduced kidney size, decreased numbers of glomeruli, and abnormal postnatal nephron formation. 29280872_PHD-2 knockdown mesenchymal stromal cells overexpressed HIF-1alpha and multiple angiogenic factors compared to control. Wounds treated with PHD-2 knockdown mesenchymal stromal cells healed at a significantly accelerated rate compared with wounds treated with shScramble mesenchymal stromal cells. 29688249_Data show that deletion of prolyl hydroxylase 2 (PHD2) in intestinal epithelial cells (IECs) did not lead to spontaneous enteritis or colitis in mice. 29967369_Oxygen sensing by PHD2 in osteocytes negatively regulates bone mass through epigenetic regulation of sclerostin and targeting PHD2 elicits an osteo-anabolic response in osteoporotic mouse models. 30339838_The inhibition of endothelial PHD2 activity may be a new therapeutic strategy for acute inflammatory diseases. 30452097_Inhibition of the Oxygen Sensor PHD2 Enhances Tissue-Engineered Endochondral Bone Formation. 30575721_Deletion of Phd2 in combination with expression of BRaf(V600E) in melanocytes (Tyr::CreER;Phd2(lox/lox);BRaf(CA)) leads to the development of melanoma with 100% penetrance and frequent lymph node metastasis. 31162141_findings have shown a new role for the PHD2/Hif2a couple in the reversible regulation of T cell and immune activity. 31356139_Depletion of PHD2 and PHD3 leads to dilated cardiomyopathy and exacerbates cardiac injury. 31390373_PHD2 may itself directly mediate increases in PHD2 may itself directly mediate increases in 5-hydroxymethylcytosine in chondrocyte and osteoblast genes. 31838134_exerts anti-cancer and anti-inflammatory effects in colon cancer xenografts via attenuating NF-kappaB activity 32296880_HIF-P4H-2 inhibition enhances intestinal fructose metabolism and induces thermogenesis protecting against NAFLD. 32413092_Prolyl hydroxylase domain 2 reduction enhances skeletal muscle tissue regeneration after soft tissue trauma in mice. 34610308_PHDs/CPT1B/VDAC1 axis regulates long-chain fatty acid oxidation in cardiomyocytes. 34658364_Developmental role of PHD2 in the pathogenesis of pseudohypoxic pheochromocytoma. 34863041_Prolyl-4-hydroxylases 2 and 3 control erythropoietin production in renin-expressing cells of mouse kidneys. | ENSMUSG00000031987 | Egln1 | 107.8160 | 3.3602455 | 1.748567 | 0.4016492 | 18.89392 | 1.381922e-05 | 2.940532e-03 | Yes | No | 216.89689 | 444.24974 | 64.79463 | 129.63227 | ||||
| ENSG00000288825 | 8337 | H2AC18 | protein_coding | Mouse_homologues FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. | Mouse_homologues Acetylation;Chromosome;Citrullination;DNA-binding;Hydroxylation;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H2A family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in a histone cluster on chromosome 1. This gene is one of four histone genes in the cluster that are duplicated; this record represents the centromeric copy. [provided by RefSeq, Aug 2015]. | Mouse_homologues mmu:15267;mmu:319192; | Mouse_homologues nucleosome [GO:0000786]; nucleus [GO:0005634]; DNA binding [GO:0003677]; protein heterodimerization activity [GO:0046982]; heterochromatin assembly [GO:0031507]; nucleosome assembly [GO:0006334] | 22980979_Ubiquitin-dependent signaling during the DNA damage response (DDR) to double-strand breaks is initiated by two E3 ligases, RNF8 and RNF168, targeting histone H2A and H2AX. Study shows that ubiquitin chains per se are insufficient for signaling, but RNF168 target ubiquitination is required for DDR damage. | ENSMUSG00000063954 | H2ac19 | 104.1500 | 0.4596085 | -1.121523 | 0.3363865 | 11.51068 | 6.919761e-04 | 4.250303e-02 | Yes | No | 200.41456 | 223.29149 | 408.03260 | 425.97975 |
- Note:
- The spreadsheet above contains the genes with significantly changes in expression (P-value < 0.05 and |log2FC| > 1). Please Click HERE to check all mapped genes in a Microsoft .excel file.
- The comparisons made can be identified from the website name. “HEK293_TMG_6hB vs HEK293_DMSO_6hB” means that “HEK293_DMSO_6hB” is considered the ground state and “HEK293_TMG_6hB” the changed state. This also means that a positive log2FC value indicates that the gene expression is increased in “HEK293_TMG_6hB” compared to “HEK293_DMSO_6hB”.
- Click HERE to check all parameters of DESeq2 model for all detected genes of the current samples set in a Microsoft .excel file. For more detials of the DESeq2 model, please Click HERE.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| lncRNA | 3 |
| processed_pseudogene | 6 |
| protein_coding | 12 |
| transcribed_processed_pseudogene | 1 |
| unprocessed_pseudogene | 1 |
AS in non-DEGs
Spreadsheet
| Ensembl gene ID | Entrez gene ID | Gene symbol | Biotype | UniProtKBID | UniProtFunction | UniProtKeywords | UniProtPathway | RefSeqSummary | KEGG | GO | GeneRif | H.sapiens homolog ID | H.sapiens homolog symbol | baseMean | FoldChange | log2FoldChange | lfcSE | stat | pvalue | padj | Is.Sig. | Has.Sig.AS | Intercept_HEK293_TMG_6hB | SE_Intercept_HEK293_TMG_6hB | Intercept_HEK293_DMSO_6hB | SE_Intercept_HEK293_DMSO_6hB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000003096 | 90293 | KLHL13 | protein_coding | Q9P2N7 | FUNCTION: Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex required for mitotic progression and cytokinesis. The BCR(KLHL9-KLHL13) E3 ubiquitin ligase complex mediates the ubiquitination of AURKB and controls the dynamic behavior of AURKB on mitotic chromosomes and thereby coordinates faithful mitotic progression and completion of cytokinesis. {ECO:0000269|PubMed:14528312, ECO:0000269|PubMed:17543862, ECO:0000269|PubMed:19995937}. | Alternative splicing;Cell cycle;Cell division;Kelch repeat;Mitosis;Reference proteome;Repeat;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a BTB and kelch domain containing protein and belongs to the kelch repeat domain containing superfamily of proteins. The encoded protein functions as an adaptor protein that complexes with Cullin 3 and other proteins to form the Cullin 3-based E3 ubiquitin-protein ligase complex. This complex is necessary for proper chromosome segregation and completion of cytokinesis. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Mar 2010]. | hsa:90293; | Cul3-RING ubiquitin ligase complex [GO:0031463]; cytosol [GO:0005829]; midbody [GO:0030496]; cullin family protein binding [GO:0097602]; protein N-terminus binding [GO:0047485]; cell cycle [GO:0007049]; cell division [GO:0051301]; protein ubiquitination [GO:0016567]; regulation of cytokinesis [GO:0032465] | ENSMUSG00000036782 | Klhl13 | 454.28424 | 1.0822165 | 0.1139891150 | 0.14597942 | 6.112759e-01 | 4.343078e-01 | 7.817284e-01 | No | Yes | 607.131969 | 147.183335 | 512.915446 | 121.705771 | |
| ENSG00000004139 | 23098 | SARM1 | protein_coding | Q6SZW1 | FUNCTION: NAD(+) hydrolase, which plays a key role in axonal degeneration following injury by regulating NAD(+) metabolism (PubMed:25908823, PubMed:27671644, PubMed:28334607). Acts as a negative regulator of MYD88- and TRIF-dependent toll-like receptor signaling pathway by promoting Wallerian degeneration, an injury-induced form of programmed subcellular death which involves degeneration of an axon distal to the injury site (PubMed:15123841, PubMed:16964262, PubMed:20306472, PubMed:25908823). Wallerian degeneration is triggered by NAD(+) depletion: in response to injury, SARM1 is activated and catalyzes cleavage of NAD(+) into ADP-D-ribose (ADPR), cyclic ADPR (cADPR) and nicotinamide; NAD(+) cleavage promoting cytoskeletal degradation and axon destruction (PubMed:25908823, PubMed:28334607, PubMed:30333228, PubMed:31128467, PubMed:31439793, PubMed:32049506, PubMed:32828421, PubMed:31439792, PubMed:33053563). Also able to hydrolyze NADP(+), but not other NAD(+)-related molecules (PubMed:29395922). Can activate neuronal cell death in response to stress (PubMed:20306472). Regulates dendritic arborization through the MAPK4-JNK pathway (By similarity). Involved in innate immune response: inhibits both TICAM1/TRIF- and MYD88-dependent activation of JUN/AP-1, TRIF-dependent activation of NF-kappa-B and IRF3, and the phosphorylation of MAPK14/p38 (PubMed:16964262). {ECO:0000250|UniProtKB:Q6PDS3, ECO:0000269|PubMed:15123841, ECO:0000269|PubMed:16964262, ECO:0000269|PubMed:20306472, ECO:0000269|PubMed:25908823, ECO:0000269|PubMed:27671644, ECO:0000269|PubMed:28334607, ECO:0000269|PubMed:29395922, ECO:0000269|PubMed:30333228, ECO:0000269|PubMed:31128467, ECO:0000269|PubMed:31439792, ECO:0000269|PubMed:31439793, ECO:0000269|PubMed:32049506, ECO:0000269|PubMed:32828421, ECO:0000269|PubMed:33053563}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Cytoplasm;Differentiation;Hydrolase;Immunity;Innate immunity;Mitochondrion;NAD;Neurogenesis;Phosphoprotein;Reference proteome;Repeat;Synapse;Transit peptide | hsa:23098; | axon [GO:0030424]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dendrite [GO:0030425]; extrinsic component of mitochondrial outer membrane [GO:0031315]; microtubule [GO:0005874]; mitochondrion [GO:0005739]; synapse [GO:0045202]; identical protein binding [GO:0042802]; NAD(P)+ nucleosidase activity [GO:0050135]; NAD+ nucleosidase activity [GO:0003953]; NAD+ nucleotidase, cyclic ADP-ribose generating [GO:0061809]; signaling adaptor activity [GO:0035591]; cell differentiation [GO:0030154]; innate immune response [GO:0045087]; NAD catabolic process [GO:0019677]; negative regulation of MyD88-independent toll-like receptor signaling pathway [GO:0034128]; nervous system development [GO:0007399]; positive regulation of neuron death [GO:1901216]; regulation of apoptotic process [GO:0042981]; regulation of dendrite morphogenesis [GO:0048814]; regulation of neuron death [GO:1901214]; response to axon injury [GO:0048678]; response to glucose [GO:0009749]; signal transduction [GO:0007165] | 15893701_Candidate gene in the onset of hereditary infectious/inflammatory diseases. 16964262_TIR adaptor SARM is a negative regulator of Toll-like receptor signaling. 17804407_confirmed the co-localization of retinoschisin with Na/K ATPase and SARM1 in photoreceptors and bipolar cells of retina tissue 18089857_SARM1 deficiencies may uncover unexpected similarities between the ways in which neurons and immune cells sense and respond to danger. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20306472_SARM-mediated inhibition may not be exclusively directed at TRIF or MyD88, but that SARM may also directly inhibit MAPK phosphorylation 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 22145856_The N-terminal 27 amino acids (S27) of SARM, which is hydrophobic and polybasic, acts as a mitochondria-targeting signal sequence, associating SARM to the mitochondria. The S27 peptide has an inherent ability to bind to lipids and mitochondria. 23175186_SARM overexpression caused mitochondrial clustering which has also been observed in several cell death phenomenon. 23885119_These results indicate that association of PINK1 with SARM1 and TRAF6 is an important step for mitophagy. 23923041_The innate immunity adaptor SARM translocates to the nucleus to stabilize lamins and prevent DNA fragmentation in response to pro-apoptotic signaling. 24021647_Data found that the UXT isoforms elicit dual opposing regulatory effects on SARM-induced apoptosis. 24840802_Rapid Wallerian degeneration requires the pro-degenerative molecules SARM1. 26592460_Data show that sterile alpha- and armadillo-motif-containing protein (SARM) modulates MyD88 protein-mediated Toll-like receptors (TLRs) activation through BB-loop dependent interleukin-1 receptor (TIR) TIR-TIR interactions. 26844829_Active nerve degeneration requires SARM1 and MAP kinases, including DLK, while the NAD+ synthetic enzyme NMNAT2 prevents degeneration. 27671644_we identify a physical interaction between the autoinhibitory N terminus and the TIR domain of SARM1, revealing a previously unrecognized direct connection between these domains that we propose mediates autoinhibition and activation upon injury. 30333228_SARM1 is phosphorylated at Ser-548 by c-jun-N-terminal kinase under conditions of oxidative stress, and this increases SARM1's NADase activity, leading to inhibited mitochondrial respiration and decreased levels of NAD+ and ATP. 31278906_These results highlight the importance of oligomerization for SARM1 function and reveal critical epitopes for future targeted drug development. 31484833_Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. 32311648_The SARM1 axon degeneration pathway: control of the NAD(+) metabolome regulates axon survival in health and disease. 32428870_lncRNA OGFRP1 functions as a ceRNA to promote the progression of prostate cancer by regulating SARM1 level via miR-124-3p. 33053563_The NAD(+)-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. 33185189_Structural basis for SARM1 inhibition and activation under energetic stress. 33468661_Multiple domain interfaces mediate SARM1 autoinhibition. 33548217_SARM1 is required in human derived sensory neurons for injury-induced and neurotoxic axon degeneration. 33605409_Expression of sterile-alpha and armadillo motif containing protein (SARM) in rheumatoid arthritis monocytes correlates with TLR2-induced IL-1beta and disease activity. 33871822_Sarm1-mediated neurodegeneration within the enteric nervous system protects against local inflammation of the colon. 34213829_Acidic pH irreversibly activates the signaling enzyme SARM1. 34455132_SARM1-mediated wallerian degeneration: A possible mechanism underlying organophosphorus-induced delayed neuropathy. 34796871_Enrichment of SARM1 alleles encoding variants with constitutively hyperactive NADase in patients with ALS and other motor nerve disorders. 34935867_Sarm1 activation produces cADPR to increase intra-axonal Ca++ and promote axon degeneration in PIPN. 34991663_Constitutively active SARM1 variants that induce neuropathy are enriched in ALS patients. 35224705_SARM1 can be a potential therapeutic target for spinal cord injury. 35468924_TLR4 and SARM1 modulate survival and chemoresistance in an HPV-positive cervical cancer cell line. | ENSMUSG00000050132 | Sarm1 | 670.77786 | 0.8532808 | -0.2289074313 | 0.12849507 | 3.152294e+00 | 7.582029e-02 | 4.043888e-01 | No | Yes | 788.889395 | 87.754234 | 907.905198 | 98.415475 | ||
| ENSG00000005059 | 55013 | MCUB | protein_coding | Q9NWR8 | FUNCTION: Negatively regulates the activity of MCU, the mitochondrial inner membrane calcium uniporter, and thereby modulates calcium uptake into the mitochondrion. Does not form functional calcium channels by itself. Mitochondrial calcium homeostasis plays key roles in cellular physiology and regulates cell bioenergetics, cytoplasmic calcium signals and activation of cell death pathways. {ECO:0000250|UniProtKB:Q810S1}. | Calcium;Calcium transport;Coiled coil;Ion transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Transit peptide;Transmembrane;Transmembrane helix;Transport | hsa:55013; | calcium channel complex [GO:0034704]; integral component of mitochondrial inner membrane [GO:0031305]; intracellular membrane-bounded organelle [GO:0043231]; intrinsic component of membrane [GO:0031224]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; uniplex complex [GO:1990246]; calcium channel inhibitor activity [GO:0019855]; calcium import into the mitochondrion [GO:0036444]; mitochondrial calcium ion homeostasis [GO:0051560]; mitochondrial calcium ion transmembrane transport [GO:0006851] | 19773279_Observational study of gene-disease association. (HuGE Navigator) 20332099_Observational study of gene-disease association. (HuGE Navigator) 24231807_MCUb (also known as CCDC109b) is a paralogue of MCU (CCDC109a). MCUb physically resides within the mitochondrial uniporter complex (uniplex), which consists of the MCU, MCUb, EMRE, MICU1, and MICU2. 28754121_This study elucidated a role for CCDC109B as an oncogene and a prognostic marker in human gliomas. 31533452_MCUB-dependent changes in mitochondrial calcium uniporter stoichiometry are a prominent regulatory mechanism to modulate mitochondrial Ca(2+) uptake and cardiac myocyte cellular physiology. | ENSMUSG00000027994 | Mcub | 696.89399 | 0.9741465 | -0.0377893514 | 0.15155121 | 6.213098e-02 | 8.031590e-01 | 9.449080e-01 | No | Yes | 744.639553 | 152.571264 | 712.414297 | 142.807683 | ||
| ENSG00000005810 | 23077 | MYCBP2 | protein_coding | O75592 | FUNCTION: Atypical E3 ubiquitin-protein ligase which specifically mediates ubiquitination of threonine and serine residues on target proteins, instead of ubiquitinating lysine residues (PubMed:29643511). Shows esterification activity towards both threonine and serine, with a preference for threonine, and acts via two essential catalytic cysteine residues that relay ubiquitin to its substrate via thioester intermediates (PubMed:29643511). Interacts with the E2 enzymes UBE2D1, UBE2D3, UBE2E1 and UBE2L3 (PubMed:18308511, PubMed:29643511). Plays a key role in neural development, probably by mediating ubiquitination of threonine residues on target proteins (Probable). Involved in different processes such as regulation of neurite outgrowth, synaptic growth, synaptogenesis and axon degeneration (By similarity). Required for the formation of major central nervous system axon tracts (By similarity). Required for proper axon growth by regulating axon navigation and axon branching: acts by regulating the subcellular location and stability of MAP3K12/DLK (By similarity). Required for proper localization of retinogeniculate projections but not for eye-specific segregation (By similarity). Regulates axon guidance in the olfactory system (By similarity). Involved in Wallerian axon degeneration, an evolutionarily conserved process that drives the loss of damaged axons: acts by promoting destabilization of NMNAT2, probably via ubiquitination of NMNAT2 (By similarity). Catalyzes ubiquitination of threonine and/or serine residues on NMNAT2, consequences of threonine and/or serine ubiquitination are however unknown (PubMed:29643511). Regulates the internalization of TRPV1 in peripheral sensory neurons (By similarity). Mediates ubiquitination and subsequent proteasomal degradation of TSC2/tuberin (PubMed:18308511, PubMed:27278822). Independently of the E3 ubiquitin-protein ligase activity, also acts as a guanosine exchange factor (GEF) for RAN in neurons of dorsal root ganglia (PubMed:26304119). May function as a facilitator or regulator of transcriptional activation by MYC (PubMed:9689053). Acts in concert with HUWE1 to regulate the circadian clock gene expression by promoting the lithium-induced ubiquination and degradation of NR1D1 (PubMed:20534529). {ECO:0000250|UniProtKB:Q7TPH6, ECO:0000269|PubMed:18308511, ECO:0000269|PubMed:20534529, ECO:0000269|PubMed:26304119, ECO:0000269|PubMed:27278822, ECO:0000269|PubMed:29643511, ECO:0000269|PubMed:9689053}. | 3D-structure;Alternative splicing;Biological rhythms;Cell projection;Cytoplasm;Cytoskeleton;Disulfide bond;Guanine-nucleotide releasing factor;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:18308511, ECO:0000269|PubMed:29643511}. | This gene encodes an E3 ubiquitin-protein ligase and member of the PHR (Phr1/MYCBP2, highwire and RPM-1) family of proteins. The encoded protein plays a role in axon guidance and synapse formation in the developing nervous system. In mammalian cells, this protein regulates the cAMP and mTOR signaling pathways, and may additionally regulate autophagy. Reduced expression of this gene has been observed in acute lymphoblastic leukemia patients and a mutation in this gene has been identified in patients with a rare inherited vision defect. [provided by RefSeq, Mar 2017]. | hsa:23077; | axon [GO:0030424]; cytoplasm [GO:0005737]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; microtubule cytoskeleton [GO:0015630]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; small GTPase binding [GO:0031267]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; branchiomotor neuron axon guidance [GO:0021785]; central nervous system projection neuron axonogenesis [GO:0021952]; circadian regulation of gene expression [GO:0032922]; negative regulation of protein catabolic process [GO:0042177]; neuromuscular process [GO:0050905]; positive regulation of protein ubiquitination [GO:0031398]; protein ubiquitination [GO:0016567]; regulation of axon guidance [GO:1902667]; regulation of cytoskeleton organization [GO:0051493]; regulation of protein localization [GO:0032880] | 14559897_Pam, through its interaction with tuberin, could regulate the ubiquitination and proteasomal degradation of the tuberin-hamartin complex particularly in the CNS 15257286_PAM the longest lasting nontranscriptional regulator of adenylyl cyclase activity known to date and presents a novel mechanism for the temporal regulation of cAMP signaling 15470080_Identifies the region in PAM that inhibits the domain of type V adenylyl cyclase. 19000755_PAM protein activated by facilitating the GDP/GTP-exchange of RHEBL1 protein which is an activator of mTOR protein. 19240061_Observational study of gene-disease association. (HuGE Navigator) 20534529_Arf-bp1 and Pam are novel regulators of circadian gene expression that target Rev-erb alpha for degradation 25460509_Data show that epithelial-mesenchymal transition (EMT)-transcription factors can be dynamically degraded by an atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 (SPFFbxo45). 25634536_We describe a distinct excavated optic disc anomaly associated with high myopia and increased axial length. The condition appears to follow an autosomal dominant pattern and segregate with a deletion in MYCBP2. 25731699_In castration resistant prostate cancer, MYCBP2 is down-regulated at the mRNA and protein levels. 26517351_Data indicate an oncogenic role for an Ikaros protein/MYCBP2 protein/proto-oncogene protein c-MYC axis in adult acute lymphoblastic leukemia (ALL), providing a mechanism of target therapies that activate Ikaros in ALL. 29643511_identification of the neuron-associated E3 ligase MYCBP2 (also known as PHR1) as the apparent single member of a class of RING-linked E3 ligase with esterification activity and intrinsic selectivity for threonine over serine 30318507_MYCBP2, a member of the c-myc oncogene family, is a direct functional target of miR-1247. Furthermore, in colorectal cancer patients, MYCBP2 protein levels are associated with miR-1247 levels and survival 31285543_FBXO45-MYCBP2 regulates mitotic cell fate by targeting FBXW7 for degradation. | ENSMUSG00000033004 | Mycbp2 | 5971.90776 | 0.9359494 | -0.0954975117 | 0.07606222 | 1.572281e+00 | 2.098760e-01 | 6.029537e-01 | No | Yes | 5843.263426 | 1523.389293 | 5613.813327 | 1430.603323 |
| ENSG00000005884 | 3675 | ITGA3 | protein_coding | P26006 | FUNCTION: Integrin alpha-3/beta-1 is a receptor for fibronectin, laminin, collagen, epiligrin, thrombospondin and CSPG4. Integrin alpha-3/beta-1 provides a docking site for FAP (seprase) at invadopodia plasma membranes in a collagen-dependent manner and hence may participate in the adhesion, formation of invadopodia and matrix degradation processes, promoting cell invasion. Alpha-3/beta-1 may mediate with LGALS3 the stimulation by CSPG4 of endothelial cells migration. {ECO:0000269|PubMed:10455171, ECO:0000269|PubMed:15181153}.; FUNCTION: (Microbial infection) Integrin ITGA3:ITGB1 may act as a receptor for R.delemar CotH7 in alveolar epithelial cells, which may be an early step in pulmonary mucormycosis disease progression. {ECO:0000269|PubMed:32487760}. | Alternative splicing;Calcium;Cell adhesion;Cell junction;Cell membrane;Cell projection;Cleavage on pair of basic residues;Direct protein sequencing;Disease variant;Disulfide bond;Epidermolysis bullosa;Glycoprotein;Integrin;Lipoprotein;Membrane;Metal-binding;Palmitate;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The gene encodes a member of the integrin alpha chain family of proteins. Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain that function as cell surface adhesion molecules. The encoded preproprotein is proteolytically processed to generate light and heavy chains that comprise the alpha 3 subunit. This subunit joins with a beta 1 subunit to form an integrin that interacts with extracellular matrix proteins including members of the laminin family. Expression of this gene may be correlated with breast cancer metastasis. [provided by RefSeq, Oct 2015]. | hsa:3675; | basolateral plasma membrane [GO:0016323]; cell periphery [GO:0071944]; cell surface [GO:0009986]; excitatory synapse [GO:0060076]; external side of plasma membrane [GO:0009897]; extracellular exosome [GO:0070062]; filopodium membrane [GO:0031527]; focal adhesion [GO:0005925]; growth cone filopodium [GO:1990812]; integrin alpha3-beta1 complex [GO:0034667]; integrin complex [GO:0008305]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; synaptic membrane [GO:0097060]; collagen binding [GO:0005518]; fibronectin binding [GO:0001968]; integrin binding [GO:0005178]; laminin binding [GO:0043236]; metal ion binding [GO:0046872]; protease binding [GO:0002020]; protein domain specific binding [GO:0019904]; protein heterodimerization activity [GO:0046982]; cell-matrix adhesion [GO:0007160]; dendritic spine maintenance [GO:0097062]; exploration behavior [GO:0035640]; heart development [GO:0007507]; integrin-mediated signaling pathway [GO:0007229]; lung development [GO:0030324]; maternal process involved in female pregnancy [GO:0060135]; memory [GO:0007613]; mesodermal cell differentiation [GO:0048333]; negative regulation of cell projection organization [GO:0031345]; negative regulation of Rho protein signal transduction [GO:0035024]; nephron development [GO:0072006]; neuron migration [GO:0001764]; positive regulation of cell-substrate adhesion [GO:0010811]; positive regulation of epithelial cell migration [GO:0010634]; positive regulation of gene expression [GO:0010628]; positive regulation of neuron projection development [GO:0010976]; positive regulation of protein localization to plasma membrane [GO:1903078]; regulation of BMP signaling pathway [GO:0030510]; regulation of transforming growth factor beta receptor signaling pathway [GO:0017015]; regulation of Wnt signaling pathway [GO:0030111]; renal filtration [GO:0097205]; response to gonadotropin [GO:0034698]; response to xenobiotic stimulus [GO:0009410]; skin development [GO:0043588] | 11907260_Data demonstrate that multiple tetraspanin (transmembrane 4 superfamily) proteins such as CD151 are palmitoylated, and that palmitoylation is not required for cd151-alpha3beta1 integrin association. 11988844_Human prostate tumors display on their cell surface the alpha6beta1 and/or alpha3beta1 integrins 12372459_initial adhesion of endometrial cells to mesothelium is not mediated by alpha(3)beta(1)integrin 12452046_Integrin-alpha 3 sub-unit participates the process of regulating growth of meningiomas. 12615788_Observational study of gene-disease association. (HuGE Navigator) 12699087_expression of alpha3 integrin subunit is responsible for differentiation-associated changes in cells behavior in terminally differentiated oral keratinocytes 12717361_signaling through both constitutively expressed alpha2 integrin and Matrigel-induced alpha3 integrin expression is required to acquire a differentiated phenotype in Caco-2 cells. 12919677_Data show that macrophage stimulating protein (MSP) and its receptor (Ron) induce phosphorylation of both Ron and alpha6beta4 integrin, and result in activation of alpha3beta1 integrin. 14596610_Disulfide bonding patterns of the integrin alpha 3 chain are described; the alpha chain displays differences in the disulfide patterns of those bonds near their C-terminal regions linking the heavy and light chains. 14602071_Data demonstrate that alpha6beta4 integrin mediates pancreatic epithelial cell adhesion to Netrin-1, whereas recruitment of alpha6beta4 and alpha3beta1 regulate the migration of putative pancreatic progenitors on Netrin-1. 14607975_ligand-binding specificities of integrin alpha3beta1 and alpha6beta1 14612440_alpha3beta1 integrin binding to laminin-5 depends on its proteolytic processing 14645603_results suggest that a direct interaction occurs between the Adenovirus penton base protein and the integrin receptor alpha3beta1 in vitro and in vivo 14662754_results show how laterally associated EWI-2 might regulate alpha3beta1 function in disease and development, and demonstrate how tetraspanin proteins can assemble multiple nontetraspanin proteins into functional complexes 14666169_alpha3beta1, alpha4beta1 and alphaVbeta1 integrins may play an important role in the implantation process 14706682_Observational study of gene-disease association. (HuGE Navigator) 15024036_Data suggest that the early arrest of tumor cells in the pulmonary vasculature through interaction of alpha3beta1 integrin with laminin-5 in exposed basement membrane provides a basis for cell arrest during pulmonary metastasis. 15109703_Observational study of gene-disease association. (HuGE Navigator) 15194015_Observational study of gene-environment interaction. (HuGE Navigator) 15280429_Alpha 3 beta 1 integrin is necessary and sufficient for maximal keratinocyte survival on laminin-5. 15302884_cAMP-Epac-Rap1 pathway regulates cell spreading and cell adhesion to laminin-5 through the alpha3beta1 integrin but not the alpha6beta4 integrin 15346842_Observational study of gene-disease association. (HuGE Navigator) 15351855_Observational study of gene-disease association. (HuGE Navigator) 15677332_Results indicate that CD151 association modulates the ligand-binding activity of integrin alpha3beta1 through stabilizing its activated conformation not only with purified proteins but also in a physiological context. 15805105_Beta ig-h3 induces keratinocyte differentiation via modulation of involucrin and transglutaminase expression through the integrin alpha3beta1 and the phosphatidylinositol 3-kinase/Akt signaling pathway 15878864_KSHV gB can activate VEGFR-3 on the microvascular endothelium and modulate endothelial cell migration and proliferation via an interaction between the alpha3beta1 integrin and the VEGFR-3 receptor 15919367_alpha3beta1 integrin binding results in a suppression of the interleukin-1 signaling pathway leading to the activation of NF-(kappa)B 15983209_alpha3, alpha5, and alpha6 beta1 integrins are expressed in ductal cells at 8 weeks, before glucagon- and insulin-immunoreactive cells bud off in fetal pancrea. 16055706_the signaling network involving Smad-dependent TGFbeta, PKCdelta, and integrin alpha2beta1/alpha3beta1, regulates cell spreading, motility, and invasion of the SNU16mAd gastric carcinoma cell variant 16103120_phenotypic conversion and reversion of bladder cancer cells is controlled by a glycosynapse 3 microdomain through GM3-mediated interaction of alpha3beta1 integrin with CD9 16273094_localized expression of the integrin alpha3 protein is regulated at the level of RNA localization by MBNL1; integrin alpha3 transcripts are physically associated with MLP1 in cells and MLP1 binds to a specific ACACCC motif in the integrin alpha3 3' UTR 16307893_Thus, the integrin/Src pathway negatively regulates the induction of long-term plasticity at inhibitory synapses on a cerebellar Purkinje cells. 16339173_Data show that alpha6 and alpha3 integrin subunits interact with laminin 5 to increase expression of E-cadherin, and suggest that phosphoinositide 3-kinase (PI 3-kinase) activation plays a key role in this cross-talk. 16373174_Our data show that alpha3beta1 integrin function may be altered by glycosylation, that both subunits contribute to these changes, and glycosylation may be considered a newly found mechanism in the regulation of integrin function. 16390868_Results suggest that loss of E-cadherin function is linked to regulation of cell-cell and cell-matrix adhesion, based in part on cell surface expression of alpha2, alpha3 and beta1 integrins. 16459165_decreased podocyte expression of alpha3beta1 integrins is closely related with podocyte depletion, glomerular sclerosis, and daily protein loss in patients with primary focal glomerulosclerosis 16475024_TGF-beta1 stimulates hepatoma cells to express a higher level of alpha3 integrin by transcriptional upregulation via Ets transcription factors and to exhibit a more invasive phenotype. 16510444_This study supports matrix-induced clustering of alpha3beta1 integrin promotes uPAR/alpha3beta1 interaction; potentiating cellular signal transduction pathways culminating in activation of uPA expression and enhanced uPA-dependent invasive behavior. 16537545_EWI proteins EWI-2 and EWI-F, alpha3beta1 and alpha6beta4 integrins, and protein palmitoylation have contrasting effects on cell surface CD9 organization 16571677_Both integrin alpha3beta1- and alpha6beta4-dependent cell adhesion to laminin-5 were also impaired in CD151-silenced cells. 16707493_Slug regulates integrin alpha3, beta1, and beta4 expression and cell proliferation in human epidermal keratinocytes 16731529_integrin alpha3beta1 is a receptor for the alpha3NC1 domain and transdominantly inhibits integrin alphavbeta3 activation 16732726_Acquisition of multiple antitumor drugs was accompanied by a drastically reduced expression of alpha3beta1 in the adenocarcinoma cells. 16785564_Binding of Borrelia burgdorferi to integrin alpha 3 beta 1 results in release of inflammatory mediators from human chondrocytes and is proposed as a Toll-like receptor-independent pathway for activation of the innate immune response. 17265493_Chondrocytes with low chondrogenic capacity expressed higher levels of IGF-1, MMP-2, aggrecanase 2, while chondrocytes with high chondrogenic capacity expressed higher levels of CD44, CD151, and CD49c. 17475774_Integrin-mediated adhesion via alpha3beta1, but not alpha6beta4 integrin, supports cell survival through EGFR by signaling downstream to Erk. 17481915_These results indicate that binding of the alpha3beta1 integrin results in a suppression in the activation of the IL-1 induced intracellular signaling pathway from JNK to AP-1. 17498594_findings suggest an important role of integrin alpha2beta1, alpha3beta1, and alpha5beta1 in the architectural characteristics of ameloblastomas and adenomatoid odentogenic tumor 17630833_Beta1 and alpha3 integrins are functionally important receptors for type 1 pili-expressing bacteria within the urinary tract and possibly at other sites within the host. 17888902_identification of alpha3beta1 as the predominant integrin and laminin receptor in glioma cells, and as a brain invasion-mediating integrin 17997226_These results strongly suggest that enhanced expression of alpha3beta1 integrin on hepatocellular carcinoma cells is directly involved in their malignant phenotypes such as invasion and metastasis. 18224668_Increase in ITGA3 is associated with lymph node metastasis in squamous cell carcinoma of the tongue 18384059_Observational study of gene-disease association. (HuGE Navigator) 18464290_Both integrins, v3 and v5, are involved in the glioma cell radioresistance through the integrin-linked kinase (ILK) and the small GTPase RhoB, at least by regulating the radiation-induced mitotic cell death. 18492066_CD151 regulates integrin alpha3beta1 functions in two independent aspects: potentiation of integrin alpha3beta1-mediated cell adhesion and promotion of integrin alpha3beta1-stimulated signaling events involving tyrosine phosphorylation 18550570_Observational study of gene-disease association. (HuGE Navigator) 18695939_Results suggest that HAb18G/CD147 enhances the invasion and metastatic potentials of human hepatoma cells via integrin alpha3beta1-mediated FAK-paxillin and FAKPI3K-Ca(2+) signal pathways. 18839319_Src overexpression downregulates alpha3 integrin total protein expression and localization to the cell surface of HCT116 colon cancer cells. This indicates that Src activity may enhance metastasis by altering alpha3 integrin expression. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19130304_alpha 3, alpha 6A, and beta 1 integrin expression in cancer cells at the invasive front are related to the mode of invasion and prognosis in oral squamous cell carcinoma 19224172_Proliferation activity and malignant grade of meningiomas were increased with decreased expression of integrin-alpha(3), and down-regulation of integrin-alpha(3) mRNA was associated with the invasive biological behaviors in meningiomas. 19230171_Glomerular deposition of C5b-9 and a positive correlation with the intensity of tubular alpha3beta1-integrin suggests a possible implication in the development of tubulointerstitial fibrosis. 19297475_a cellular protein that appeared to be degraded by E1B55K in combination with the E4orf6 protein was a species of molecular mass approximately 130 kDa that was identified as the integrin alpha3 subunit 19339270_Observational study of gene-disease association. (HuGE Navigator) 19411307_alpha(2)-, alpha(3)-, and beta(1)-integrins and E-cadherin expression in normal human lung and bronchopulmonary sequestration and congenital cystic adenomatoid malformation were evaluated using Western blot and immunohistochemistry. 19451223_Nox1 knockdown led to a loss of directional migration which takes place through a RhoA-dependent alpha2/alpha3 integrin switch. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19752234_For the first time, alpha3- and alpha5-laminins are shown to promote both adhesion and migration of mast cells, and alpha1 integrin is shown to mediate cell migration. 19755493_Gal-3 promotes epithelial cell migration by cross-linking MGAT5-modified complex N-glycans on alpha3beta1 integrin. 19759146_Integrin alpha3 is a bona fide substrate of the adenovirus serotype 5 E4orf6 and E1B55K protein-formed cullin 5-based E3 ubiquitin ligase complex. 20352300_The results suggest that the production of MMP-9 by MKN1 cells was potentiated by the alpha3beta1 integrin-laminin-5 interaction, which facilitated their invasion via degradation of the matrix. 20412552_down-regulation of alpha3beta1 integrin recapitulates crucial events governing keratinocyte migration associated with wound healing and tissue repair. 20877569_U937 macrophage responses to the TLR2 ligand, Pam3CSK4, are dependent upon integrin alpha3beta1. 20926544_Observational study of gene-disease association. (HuGE Navigator) 20927591_CD151, c-Met, and integrin alpha3/alpha6 were all overexpressed in pancreatic ductal adenocarcinoma. CD151 and c-Met might be new molecular markers to predict the prognosis of pancreatic ductal adenocarcinoma patients. 21182210_Studies have shown that ITGA3 plays a key role during cortical development, involved in neuronal migration and placement, as well as cortical layering 21195710_Data suggest that the alpha5 laminins emerge as putative primary extracellular matrix mediators of melanoma invasion and metastasis via alpha3/alpha6beta1 and other integrin receptors. 21530503_With increased tension cytoskeletal stress fibers develop that contain alphaSMA and alphavbeta3 integrin that replaces alpha2beta1 integrin, consistent with cell switching from collagen to non-collagen proteins interactions. 22512483_We identified three patients with homozygous mutations in the integrin alpha(3) gene that were associated with disrupted basement-membrane structures and compromised barrier functions in kidney, lung, and skin. 22684562_Taken together, it was concluded that CD151 promotes the proliferation and migration of PC3 cells through the formation of CD151-integrin complex and the activation of phosphorylated ERK. 22843693_CD151 links alpha(3)beta(1)/alpha(6)beta(1) integrins to Ras, Rac1, and Cdc42 by promoting the formation of multimolecular complexes in the membrane, which leads to the up-regulation of adhesion-dependent small GTPase activation 22917688_Most actinic cheilitis cases showed reduced expression of integrin alpha 3 and superficially invasive squamous cell carcinoma lacked intergrin alpha 3 in the invasive front. 22986527_Data indicate that alpha3beta1 and the tetraspanin CD151 directly associate at the front and retracting rear of polarised migrating breast carcinoma cells. 23011394_Loss of alpha3 integrin-adenomatous poliposis coli interaction promotes endothelial apoptosis. 23074279_Data show that TIMP-2-mediated inhibition of vascular endothelial cell permeability involves an integrin alpha3beta1-Shp-1-cAMP/protein kinase A-dependent vascular endothelial cadherin cytoskeletal association. 23094960_These results suggest that Ets-1 is involved in transcriptional activation of the alpha3 integrin gene through its binding to the Ets-consensus sequence at -133 bp 23233127_Collectively, these findings suggest that integrin alpha3beta1 plays pivotal roles in regulating cell proliferation and migration that enhance the invasive type of p53-deficient NSCLC cells. 23248240_Data indicate that Slug siRNA suppressed the TGF-beta1-induced integrin alpha3beta1-mediated cell migration ability of squamous cell carcinoma HSC-4 cells. 23533596_CD151 is positively associated with the invasiveness of HGC, and CD151 or the combination of CD151 and integrin alpha3 is a novel marker for predicting the prognosis of HGC patient. 23590307_Suggest that fibronectin fine tunes LM332-mediated migration by boosting bronchiolar cell adhesion to substrate via integrin alpha3beta1 integrin. 23613949_Both CD9/CD81-silenced cells and CD151-silenced cells showed delayed alpha3beta1-dependent cell spreading on laminin-332. 23652300_Integrin alpha3 contributes to the invasive nature of glioma stem-like cell via ERK1/2, which renders integrin alpha3 a prime candidate for anti-invasion therapy for glioblastoma. 23686814_The interaction between ephrin-As, Eph receptors and integrin alpha3 is plausibly important for the crosstalk between Eph and integrin signalling pathways at the membrane protrusions and in the migration of brain cancer cells. 23687274_These results showed evidence of Borrelia burgdorferi BB0172 localization in the outer membrane, the orientation of the vWFA domain to the extracellular environment, and its function as a metal ion-dependent integrin-binding protein. 23786209_study concluded that ITGA3 is a potential molecular marker for cells undergoing enhanced epithelial-mesenchymal transition as well as for cancer cells with aggressive phenotypes; integrin alpha3 likely plays a crucial role in the progression of both cancer cells and fibroblastic cells in cancer microenvironments 24006899_Two types of metastatic trait were found in OSCC: locoregional dissemination, which was reflected by high-ITGA3/CD9, and distant metastasis through hematogenous dissemination, uniquely distinguished by high-ITGB4/JUP. 24084442_High integrin alpha 3 expression is associated with malignant pleural mesothelioma. 24220332_the calf-1 domain is required for the transport of alpha3 from the ER to the Golgi apparatus to maintain the integrity of epithelial tissues, and hence the impairment of the calf-1 domain by the R628P mutation leads to severe diseases of the kidneys, lungs, and skin. 24289209_Integrin alpha2beta1 and integrin alpha3beta1 suppress anoikis in undifferentiated cells. 24381140_ITGA3 gene polymorphism is associated with with susceptibility of osteosarcoma. 24434582_Integrin alpha3beta1 mediates regulation of COX2 mRNA stability in human breast cancer cells. 24616281_Exposure to magnesium ions facilitated hBMSC proliferation via integrin alpha2 and alpha3 expression and partly promoted differentiation into osteoblasts via the alteration of ALP expression and activity 24621570_integrin-alpha3 mutation confers major renal developmental defects 24675526_endothelial integrin alpha3beta1 stabilizes tumor/endothelial cell adhesion and induces the formation of macromolecular signaling complex activating several major signaling pathways in endothelial cells. 24950714_COX2 and alpha3 are correlated in invasive ductal carcinoma independently of hormone receptor status or other clinicopathologic features, supporting the hypothesis that integrin alpha3beta1 is a determinant of COX2 expression in human breast cancer. 24958723_Molecular modeling indicated that galactosylation occurred on the periphery of alpha2beta1 integrin interaction with alpha1(IV)382-393 but right in the middle of alpha3beta1 integrin interaction with alpha1(IV)531-543 24985492_CTGF blocks integrin alpha3beta1-dependent adhesion of cancer cells. 25078904_ITGA3 translocation to the plasma membrane suppressed by hypoxia through inhibition of glycosylation facilitated cell invasion in A431. 25096929_NOX4 is highly predictive of relapse in stage II left-side colon cancer, whereas integrin alpha 3 beta 1 (ITGA3) is predictive of relapse in stage II right-side colon cancer. 25356755_our study demonstrates that CD151-alpha3beta1 integrin complexes regulate ovarian tumor growth by repressing Slug-mediated epithelial to mesenchymal transition and Wnt signaling 25472585_ITGA3, ITGA6, ITGB3,ITGB4 and ITGB5 are associated with GC susceptibility (rs2675), ITGA3, ITGA6, ITGB3, ITGB4 and ITGB5 are associated with gastric cancer susceptibility tumor stage and lymphatic metastasis in Chinese Han population 25497870_Findings show that alpha3 integrin is essentially involved in head and neck squamous cell carcinoma (HNSCC) cell radioresistance. 26318455_studies suggest that the presence and spacing of the RGD and synergy sites modulate integrin alpha3beta1 binding to Fn 26350464_based on the interaction motifs in Sdc1 and Sdc4, called synstatins (SSTN210-240 and SSTN87-131) competitively displace the receptor tyrosine kinase and alpha3beta1 integrin from the syndecan with an IC50 of 100-300 nm 26377974_Data show that CD151 protein (CD151)-alpha3beta1 integrin complexes cooperated with epidermal growth factor receptor (EGFR) to drive tumor cell motility. 26418968_the role of the CD151-alpha3beta1 complex in carcinoma progression is context dependent, and may depend on the mode of tumor cell invasion. 26464707_In high grade DCIS, when stratified according to the HER2 status, in the HER2-negative subgroup, CD151 assessed in combination with alpha3beta1 was significantly correlated with prognosis. 26674523_Genetic and Immunohistochemical Expression of Integrins ITGAV, ITGA6, and ITGA3 As Prognostic Factor for Colorectal Cancer 26788840_Expression of the alpha3-Integrin splice variants in the brain 26854491_ITGA3 mutation is responsible in the development of epidermolysis bullosa. 26992919_these findings identify CD151 and its interactions with integrins a3 and a6 as potential therapeutic targets for inhibiting stemness-driving mechanisms and stem cell populations in Glioblastoma. 27340677_The results suggested that podocyte detachment during early stage of diabetic nephropathy is mediated through upregulation of alpha3beta1-integrin. 27509031_Results provide evidence that alpha3 and alpha6 integrins have significantly different internalization kinetics and that coordination exists between them for internalization in prostate cancer cells. 27562932_During the past decades, many studies have provided evidence for a role of laminin-binding integrins in tumorigenesis, and both tumor-promoting and suppressive activities have been identified. (Review) 27717396_This study reports a variant of ILNEB syndrome in two siblings differing from the previously reported patients in the lack of nephrotic impairment and survival beyond childhood; they are the first reported compound heterozygous for ITGA3 mutations. 27821163_our studies clearly provide evidence that aberrant expression of sFRP2 can contribute to the invasive and metastatic potential for osteosarcoma. 27974569_Competition binding and infection experiments and biochemical assays pointed out alphaVbeta1 and alpha3beta1 to be of importance for human adenovirus-37 infection of corneal tissue. 28102841_Our study elucidates the molecular mechanisms of miR-101/ITGA3 pathway in regulating nasopharyngeal carcinoma (NPC)metastasis and angiogenesis, and the systemic delivery of miR-101 provides a potent evidence for the development of a novel microRNA-targeting anticancer strategy for NPC patients. 28324890_High ITGA3 expression is associated with bladder cancer. 28416479_findings identify a novel physiological context for combinatorial integrin signaling, laying the foundation for therapeutic strategies that manipulate alpha9beta1 and/or alpha3beta1 during wound healing 28612520_Results showed that ITGA3 was directly regulated by miR-199 family in head and neck squamous cell carcinoma cells (HNSCC). Its knockdown significantly inhibited cancer cell migration and invasion by HNSCC cells. 28618934_Both in cell lines and in mouse model, the extracellular matrix receptors including the integrin ( ITGA3 and ITGA2B), collagen ( COL5A1), and laminin ( LAMA5) were significantly inhibited by curcumin at messenger RNA and protein levels. 29511671_these findings suggest that ITGA3 may play a role as a potential oncogene in intrahepatic cholangiocarcinoma (ICC)and suppression of ITGA3 expression may establish a novel target for guiding the therapy of ICC patients. 29758195_ITGA3 is a novel target of miR-124 in colorectal cancer cells. 30128883_Bioinformatics analysis identifies ITGA3 as an oncogene in human tongue cancer. 30466509_Results show that in the absence of integrin alpha3, keratinocytes of patients with interstitial lung disease, nephrotic syndrome and epidermolysis bullosa (ILNEB) produce a fibronectin-rich microenvironment and make use of fibronectin-binding integrin subunits alphav and alpha5. 30614803_Up-regulation of integrin subunit alpha 3 (ITGA3) is observed in bladder cancer (BC) clinical samples and tumor cell lines. Silencing ITGA3 enhances apoptosis, inhibits tumor cell migration and invasion, and therefore decreases viability and proliferation of BC cells. 30659093_both integrin alpha3beta1 and PI4KIIalpha co-localized to the trans-Golgi network, where they physically interacted with each other, and PI4KIIalpha specifically associated with integrin alpha3 but not alpha5 30941771_MiR-524-5p targeting on FOXE1 and ITGA3 prevents thyroid cancer progression through different pathways including cell cycling and autophagy. 30944668_Elevated serum sFRP2 levels are associated with primary tumor size, tumor stage, and lymph node metastases. 30971252_Levels of antibodies that inhibit the binding of children's IE to the receptors ICAM-1, integrin alpha3beta1 and laminin increased with age. The breadth of antibodies that inhibit ICAM-1 and laminin adhesion (defined as the proportion of IE isolates whose binding was reduced by >/= 50%) also significantly increased with age. 31101121_Downregulation of alpha3beta1 in a HER2-driven mouse model and in HER2-overexpressing human mammary carcinoma cells promotes progression and invasiveness of tumors. The invasion-suppressive role of alpha3beta1 was not observed in triple-negative mammary carcinoma cells, illustrating the tumor type-specific and complex function of alpha3beta1 in breast cancer. 31340144_Integrin-alpha3 Is a Functional Marker of Ex Vivo Expanded Human Long-Term Hematopoietic Stem Cells. 31421623_The results strongly suggest that ITGA3 is a key molecule for cellular attachment and entry of non-enveloped hepatitis E virus. 31987909_findings indicate that integrin alpha3 interacts with VASP and regulates its expression 32210347_Anatomy of a viral entry platform differentially functionalized by integrins alpha3 and alpha6. 32356431_Alterations in the immunoreactivity of laminin, type IV collagen and alpha3beta1 integrin in diabetic rat ovarian follicles. 32705201_MicroRNA199a5p suppresses cell proliferation, migration and invasion by targeting ITGA3 in colorectal cancer. 32851057_TMT-Based Quantitative Proteomic Analysis Identification of Integrin Alpha 3 and Integrin Alpha 5 as Novel Biomarkers in Pathogenesis of Acute Aortic Dissection. 33099569_Expression and Prognostic Analyses of ITGA3, ITGA5, and ITGA6 in Head and Neck Squamous Cell Carcinoma. 33173103_Paracoccidioides brasiliensis downmodulates alpha3 integrin levels in human lung epithelial cells in a TLR2-dependent manner. 33428143_Key Role of CD151-integrin Complex in Lung Cancer Metastasis and Mechanisms Involved. 33768705_Skin fragility, renal malformation and interstitial lung disease due to compound heterozygous ITGA3 mutations. 34023363_Homozygous ITGA3 Missense Mutation in Adults in a Family with Syndromic Epidermolysis Bullosa (ILNEB) without Pulmonary Involvement. 34102169_Detection of bladder cancer with aberrantly fucosylated ITGA3. 34242748_CD9 and ITGA3 are regulated during HIV-1 infection in macrophages to support viral replication. 34938358_miRNA-144-5p/ITGA3 Suppressed the Tumor-Promoting Behaviors of Thyroid Cancer Cells by Downregulating ITGA3. | ENSMUSG00000001507 | Itga3 | 124.97707 | 0.6357633 | -0.6534384390 | 0.26735579 | 5.874099e+00 | 1.536523e-02 | 2.009080e-01 | No | Yes | 103.553922 | 21.466158 | 161.719177 | 32.218057 | |
| ENSG00000006704 | 9569 | GTF2IRD1 | protein_coding | Q9UHL9 | FUNCTION: May be a transcription regulator involved in cell-cycle progression and skeletal muscle differentiation. May repress GTF2I transcriptional functions, by preventing its nuclear residency, or by inhibiting its transcriptional activation. May contribute to slow-twitch fiber type specificity during myogenesis and in regenerating muscles. Binds troponin I slow-muscle fiber enhancer (USE B1). Binds specifically and with high affinity to the EFG sequences derived from the early enhancer of HOXC8 (By similarity). {ECO:0000250, ECO:0000269|PubMed:11438732}. | 3D-structure;Alternative splicing;DNA-binding;Developmental protein;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Williams-Beuren syndrome | The protein encoded by this gene contains five GTF2I-like repeats and each repeat possesses a potential helix-loop-helix (HLH) motif. It may have the ability to interact with other HLH-proteins and function as a transcription factor or as a positive transcriptional regulator under the control of Retinoblastoma protein. This gene plays a role in craniofacial and cognitive development and mutations have been associated with Williams-Beuren syndrome, a multisystem developmental disorder caused by deletion of multiple genes at 7q11.23. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2010]. | hsa:9569; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription, DNA-templated [GO:0006355]; transcription by RNA polymerase II [GO:0006366]; transition between slow and fast fiber [GO:0014886] | 12475981_characterization and involvement in slow muscle-specific gene expression 12857748_role as a repressor of slow fiber-specific transcription through mechanisms involving direct interactions with MEF2C and the nuclear receptor co-repressor 12865760_GTF2IRD1 and GTF2I have roles in causing deficits on visual spatial functioning 15941713_human VEGFR-2 promoter is functionally counter-regulated by TFII-I and TFII-IRD1. 16293761_GTF2IRD1 is a genetic determinant of mammalian craniofacial and cognitive development 16494860_a regulator of slow fiber-specific genes 17346708_functional analysis of human GTF2IRD1 in regulation of three genes (HOXC8, GOOSECOID and TROPONIN I) 18326499_analysis of the consensus binding site for TFII-I family member BEN 19205026_GTF2IRD1 is associated with Williams syndrome facies and visual-spatial construction. 19240061_Observational study of gene-disease association. (HuGE Navigator) 19897463_functional hemizygosity for the GTF2I and GTF2IRD1 genes is the main cause of the neurocognitive profile and some aspects of the gestalt phenotype of Williams-Beuren syndrome 20007321_Data show the existence of a negative autoregulatory mechanism that controls the level of GTF2IRD1 transcription via direct binding of the GTF2IRD1 protein to a highly conserved region of the GTF2IRD1 promoter containing an array of three binding sites. 22198572_This study provided evidences that insufficiency of GTF2IRD1 protein contributes to abnormalities of facial development, motor function and specific behavioural disorders that accompany Williams-Beuren syndrome. 22608712_CLIP2 haploinsufficiency by itself does not lead to the physical or cognitive characteristics of the Williams-Beuren syndrome; GTF2IRD1 and GTF2I are the main genes causing the cognitive defects 23145142_GTF2IRD1 is SUMOylated by the SUMO E2 ligase UBC9 and the level of SUMOylation is enhanced by PIASxbeta. 26275350_GTF2IRD1 binding partners are mostly involved in chromatin modification and transcriptional regulation, whilst others indicate an unexpected role in connection with the primary cilium. 26320362_Study demonstrates a significant association between SLE in Chinese Han population and the GTF2I rs117026326 T allele/GTF2IRD1 rs4717901 C allele. 27239038_The mis-regulation of genes downstream of GTF2IRD1, including TbetaR2 and BMPR1b, also individually promoted mammary cancer development, and silencing of TbetaR2 suppressed GTF2IRD1-driven tumor promotion. 29884845_Among 110 SNPs within the 7q11.23 William's Syndrome (WS) chromosomal region, we found one associated locus located at GTF2IRD1, which has been implicated in animal models of WS. 31758608_Study showed that GTF2IRD1 was overexpressed due to copy number amplification at Ch.7q in colorectal cancer (CRC). The expression of GTF2IRD1 was positively associated with the malignant pathological phenotype. Furthermore, high expression of GTF2IRD1 was an independent poor prognostic factor in CRC. Mechanistically, GTF2IRD1 promoted cell cycle progression by downregulation of TGFbetaR2 in CRC. 32936232_GTF2IRD1 overexpression promotes tumor progression and correlates with less CD8+ T cells infiltration in pancreatic cancer. | ENSMUSG00000023079 | Gtf2ird1 | 559.65600 | 1.0601373 | 0.0842510692 | 0.13782831 | 3.738881e-01 | 5.408925e-01 | 8.407612e-01 | No | Yes | 655.698648 | 66.550293 | 595.058286 | 58.730317 | |
| ENSG00000006757 | 8228 | PNPLA4 | protein_coding | P41247 | FUNCTION: Has abundant triacylglycerol lipase activity (PubMed:15364929). Transfers fatty acid from triglyceride to retinol, hydrolyzes retinylesters, and generates 1,3-diacylglycerol from triglycerides (PubMed:17603008). {ECO:0000269|PubMed:15364929, ECO:0000269|PubMed:17603008}. | Alternative splicing;Hydrolase;Lipid degradation;Lipid metabolism;Mitochondrion;Primary mitochondrial disease;Reference proteome | This gene encodes a member of the patatin-like family of phospholipases. The encoded enzyme has both triacylglycerol lipase and transacylase activities and may be involved in adipocyte triglyceride homeostasis. Alternate splicing results in multiple transcript variants. A pseudogene of this gene is found on chromosome Y. [provided by RefSeq, Feb 2010]. | hsa:8228; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; lipid droplet [GO:0005811]; membrane [GO:0016020]; mitochondrion [GO:0005739]; all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity [GO:0047376]; retinyl-palmitate esterase activity [GO:0050253]; triglyceride lipase activity [GO:0004806]; lipid homeostasis [GO:0055088]; triglyceride catabolic process [GO:0019433] | 15364929_iPLA2epsilon (adiponutrin), iPLA2zeta (TTS-2.2), and iPLA2eta (GS2) are three novel TAG lipases/acylglycerol transacylases that likely participate in TAG hydrolysis and the acyl-CoA independent transacylation of acylglycerols 15955102_The GS2 gene (PNPLA4) encodes a keratinocyte retinyl ester hydrolase. The protein also catalyzes fatty acyl CoA-dependent and -independent retinol esterification, using triolein as substrate and generates diacylglyceride and free fatty acid. 16741517_we report the identity of an inhibitor, TIP47, which prevents retinylester hydrolysis catalyzed by GS2 lipase and hormone-sensitive lipase 19181555_GS2 promotes RE accumulation and may do so either as a catalyst or as a regulatory protein that enhances retinylesters formation catalyzed by other acyl transferases. 19390624_No association between genetic variants in PNPLA4 genes and childhood and adolescent obesity 19390624_Observational study of gene-disease association. (HuGE Navigator) 27317427_These results strongly suggest that PNPLA9, -6 and -4 play a key role in GPL turnover and homeostasis in human cells. A hypothetical model suggesting how these enzymes could recognize the relative concentration of the different GPLs is proposed | 209.28870 | 0.8522882 | -0.2305866618 | 0.20751789 | 1.226636e+00 | 2.680623e-01 | 6.587695e-01 | No | Yes | 237.898970 | 34.071799 | 269.707499 | 37.488288 | |||
| ENSG00000008838 | 9862 | MED24 | protein_coding | O75448 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. {ECO:0000269|PubMed:12218053, ECO:0000269|PubMed:16595664}. | 3D-structure;Activator;Alternative splicing;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | This gene encodes a component of the mediator complex (also known as TRAP, SMCC, DRIP, or ARC), a transcriptional coactivator complex thought to be required for the expression of almost all genes. The mediator complex is recruited by transcriptional activators or nuclear receptors to induce gene expression, possibly by interacting with RNA polymerase II and promoting the formation of a transcriptional pre-initiation complex. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:9862; | core mediator complex [GO:0070847]; mediator complex [GO:0016592]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; histone acetyltransferase activity [GO:0004402]; nuclear receptor coactivator activity [GO:0030374]; protein-containing complex binding [GO:0044877]; thyroid hormone receptor binding [GO:0046966]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; ubiquitin protein ligase activity [GO:0061630]; vitamin D receptor binding [GO:0042809]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; positive regulation of transcription, DNA-templated [GO:0045893] | 22037903_Study identified significantly white blood cell count (WBC) level associated SNPs of three separate genes GSDMA, MED24, and PSMD3 in European continent (EA) subjects. 27163155_this study shows that haplotypes consisting of single nucleotide polymorphisms harboring PSMD3, CSF3 and MED24 genes are associated with asthma in Slovenian patients | ENSMUSG00000017210 | Med24 | 3760.09242 | 1.0987244 | 0.1358294891 | 0.08055049 | 2.851060e+00 | 9.131373e-02 | 4.375885e-01 | No | Yes | 4183.297052 | 413.716479 | 3720.208115 | 359.772307 | |
| ENSG00000010803 | 22955 | SCMH1 | protein_coding | Q96GD3 | FUNCTION: Associates with Polycomb group (PcG) multiprotein complexes; the complex class is required to maintain the transcriptionally repressive state of some genes. {ECO:0000250}. | 3D-structure;Alternative splicing;Developmental protein;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation | hsa:22955; | chromocenter [GO:0010369]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; anterior/posterior pattern specification [GO:0009952]; chromatin remodeling [GO:0006338]; heterochromatin assembly [GO:0031507]; negative regulation of transcription, DNA-templated [GO:0045892]; spermatogenesis [GO:0007283] | 19266077_Observational study of gene-disease association. (HuGE Navigator) 20546612_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000000085 | Scmh1 | 754.89254 | 1.0443723 | 0.0626361566 | 0.12455485 | 2.532961e-01 | 6.147637e-01 | 8.754286e-01 | No | Yes | 915.101231 | 58.763507 | 852.950342 | 53.547042 | ||
| ENSG00000010810 | 2534 | FYN | protein_coding | P06241 | FUNCTION: Non-receptor tyrosine-protein kinase that plays a role in many biological processes including regulation of cell growth and survival, cell adhesion, integrin-mediated signaling, cytoskeletal remodeling, cell motility, immune response and axon guidance. Inactive FYN is phosphorylated on its C-terminal tail within the catalytic domain. Following activation by PKA, the protein subsequently associates with PTK2/FAK1, allowing PTK2/FAK1 phosphorylation, activation and targeting to focal adhesions. Involved in the regulation of cell adhesion and motility through phosphorylation of CTNNB1 (beta-catenin) and CTNND1 (delta-catenin). Regulates cytoskeletal remodeling by phosphorylating several proteins including the actin regulator WAS and the microtubule-associated proteins MAP2 and MAPT. Promotes cell survival by phosphorylating AGAP2/PIKE-A and preventing its apoptotic cleavage. Participates in signal transduction pathways that regulate the integrity of the glomerular slit diaphragm (an essential part of the glomerular filter of the kidney) by phosphorylating several slit diaphragm components including NPHS1, KIRREL1 and TRPC6. Plays a role in neural processes by phosphorylating DPYSL2, a multifunctional adapter protein within the central nervous system, ARHGAP32, a regulator for Rho family GTPases implicated in various neural functions, and SNCA, a small pre-synaptic protein. Participates in the downstream signaling pathways that lead to T-cell differentiation and proliferation following T-cell receptor (TCR) stimulation. Phosphorylates PTK2B/PYK2 in response to T-cell receptor activation. Also participates in negative feedback regulation of TCR signaling through phosphorylation of PAG1, thereby promoting interaction between PAG1 and CSK and recruitment of CSK to lipid rafts. CSK maintains LCK and FYN in an inactive form. Promotes CD28-induced phosphorylation of VAV1. In mast cells, phosphorylates CLNK after activation of immunoglobulin epsilon receptor signaling (By similarity). {ECO:0000250|UniProtKB:P39688, ECO:0000269|PubMed:11005864, ECO:0000269|PubMed:11162638, ECO:0000269|PubMed:11536198, ECO:0000269|PubMed:12788081, ECO:0000269|PubMed:14707117, ECO:0000269|PubMed:14761972, ECO:0000269|PubMed:15536091, ECO:0000269|PubMed:15557120, ECO:0000269|PubMed:16387660, ECO:0000269|PubMed:16841086, ECO:0000269|PubMed:17194753, ECO:0000269|PubMed:18056706, ECO:0000269|PubMed:18258597, ECO:0000269|PubMed:19179337, ECO:0000269|PubMed:19652227, ECO:0000269|PubMed:20028775, ECO:0000269|PubMed:20100835, ECO:0000269|PubMed:22080863, ECO:0000269|PubMed:7568038, ECO:0000269|PubMed:7822789}. | 3D-structure;ATP-binding;Adaptive immunity;Alternative splicing;Cell membrane;Cytoplasm;Developmental protein;Host-virus interaction;Immunity;Kinase;Lipoprotein;Manganese;Membrane;Metal-binding;Myristate;Nucleotide-binding;Nucleus;Palmitate;Phosphoprotein;Proto-oncogene;Reference proteome;SH2 domain;SH3 domain;Transferase;Tyrosine-protein kinase | This gene is a member of the protein-tyrosine kinase oncogene family. It encodes a membrane-associated tyrosine kinase that has been implicated in the control of cell growth. The protein associates with the p85 subunit of phosphatidylinositol 3-kinase and interacts with the fyn-binding protein. Alternatively spliced transcript variants encoding distinct isoforms exist. [provided by RefSeq, Jul 2008]. | hsa:2534; | actin filament [GO:0005884]; cell body [GO:0044297]; cytosol [GO:0005829]; dendrite [GO:0030425]; endosome [GO:0005768]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; glial cell projection [GO:0097386]; membrane raft [GO:0045121]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; perinuclear endoplasmic reticulum [GO:0097038]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; Schaffer collateral - CA1 synapse [GO:0098685]; alpha-tubulin binding [GO:0043014]; ATP binding [GO:0005524]; disordered domain specific binding [GO:0097718]; enzyme binding [GO:0019899]; ephrin receptor binding [GO:0046875]; growth factor receptor binding [GO:0070851]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; phospholipase activator activity [GO:0016004]; phospholipase binding [GO:0043274]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; scaffold protein binding [GO:0097110]; signaling receptor binding [GO:0005102]; tau protein binding [GO:0048156]; tau-protein kinase activity [GO:0050321]; transmembrane transporter binding [GO:0044325]; type 5 metabotropic glutamate receptor binding [GO:0031802]; activated T cell proliferation [GO:0050798]; adaptive immune response [GO:0002250]; axon guidance [GO:0007411]; calcium ion transport [GO:0006816]; cell differentiation [GO:0030154]; cellular response to amyloid-beta [GO:1904646]; cellular response to glycine [GO:1905430]; cellular response to L-glutamate [GO:1905232]; cellular response to platelet-derived growth factor stimulus [GO:0036120]; cellular response to transforming growth factor beta stimulus [GO:0071560]; dendrite morphogenesis [GO:0048813]; dendritic spine maintenance [GO:0097062]; detection of mechanical stimulus involved in sensory perception of pain [GO:0050966]; ephrin receptor signaling pathway [GO:0048013]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; feeding behavior [GO:0007631]; forebrain development [GO:0030900]; heart process [GO:0003015]; innate immune response [GO:0045087]; intracellular signal transduction [GO:0035556]; learning [GO:0007612]; leukocyte migration [GO:0050900]; modulation of chemical synaptic transmission [GO:0050804]; negative regulation of dendritic spine maintenance [GO:1902951]; negative regulation of gene expression [GO:0010629]; negative regulation of hydrogen peroxide biosynthetic process [GO:0010730]; negative regulation of inflammatory response to antigenic stimulus [GO:0002862]; negative regulation of oxidative stress-induced cell death [GO:1903202]; negative regulation of protein catabolic process [GO:0042177]; negative regulation of protein ubiquitination [GO:0031397]; neuron migration [GO:0001764]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of cysteine-type endopeptidase activity [GO:2001056]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of neuron death [GO:1901216]; positive regulation of neuron projection development [GO:0010976]; positive regulation of protein localization to membrane [GO:1905477]; positive regulation of protein localization to nucleus [GO:1900182]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of tyrosine phosphorylation of STAT protein [GO:0042531]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of calcium ion import across plasma membrane [GO:1905664]; regulation of cell shape [GO:0008360]; regulation of glutamate receptor signaling pathway [GO:1900449]; regulation of peptidyl-tyrosine phosphorylation [GO:0050730]; response to amyloid-beta [GO:1904645]; response to ethanol [GO:0045471]; response to hydrogen peroxide [GO:0042542]; response to singlet oxygen [GO:0000304]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; T cell costimulation [GO:0031295]; T cell receptor signaling pathway [GO:0050852]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11121167_Observational study of gene-disease association. (HuGE Navigator) 11943772_Association of Fyn and Lyn with the proline-rich domain of glycoprotein VI regulates intracellular signaling 12368035_Src-kinase p59(fyn) play a role in anergy induction in CD8+ T cells. 12370810_the distinct potential of Fyn and Lck to phosphorylate Sam68 is likely controlled by the interaction of the kinase SH3 domain with the linker and Sam68, possibly on a competitive binding basis. 12408980_identification of novel isoform fynDelta7, in which exon 7 is absent 12426371_The Fyn but not the Lck tyrosine kinase SH3 domain competes with CD2BP2 GYF-domain binding to the same CD2 proline-rich sequence in vitro. CD2BP2 is replaced by Fyn SH3 after CD2 is translocated into lipid rafts upon CD2 ectodomain clustering. 12450793_High expression of FYN is restricted to low-stage tumors and predicts long-term survival. In culture, expression of active Fyn kinase induces differentiation and growth arrest of neuroblastoma cells. 12545174_Data show that the SLAM-associated protein (SAP) SH2 domain binds to the SH3 domain of FynT and directly couples FynT to SLAM. 12670706_There was an increase in both total area Fyn mRNA signal (17.7%, P<0.05) and cellular mRNA content (15.7%, P<0.05) in schizophrenic brains relative to controls. 12788081_p250GAP is phosphorylated by Fyn in oligodendrocytes. 12917446_upon ligation of integrin beta6 with fibronectin, beta6 complexed with Fyn and activated it, activating a pathway leading to activation of the matrix metalloproteinase-3 gene, and promoting oral SCC cell proliferation and experimental metastasis in vivo 14647465_tr-kit promotes the formation of a multimolecular complex composed of Fyn, PLCgamma1 and Sam68, which allows phosphorylation of PLCgamma1 by Fyn, and may modulate RNA metabolism. 14662334_Two Src kinases are selectively activated by TPO signaling in primary megakaryocytes, Fyn and Lyn, but only Fyn expression is significantly upregulated during MK differentiation, suggesting variable gene regulation. 14675807_Observational study of gene-disease association. (HuGE Navigator) 14675807_Our results indicate a possible association of alcohol dependence with a genotype of the SNP T137346C of the PTK fyn, with C being the risk allele. 14999081_Tyrosine phosphorylated tau is distributed in Alzheimer disease brain differently from other phosphorylated tau. Evidence of differentially phosphorylated tau within degenerating neurons was found supporting a role for fyn in neurodegeneration 15082191_Observational study of gene-disease association. (HuGE Navigator) 15098360_Observational study of gene-disease association. (HuGE Navigator) 15190072_LYN and FYN are downregulated by CBL in osteoblast differentiation induced by constitutive FGFR2 activation 15514010_activation of Fyn is inhibited by NSAIDs and constitutively active Fyn reverses the NSAID-dependent stress kinase inhibition 15536091_determined that Fyn phosphorylated MAP-2c on tyrosine 67 15537652_Fyn kinase plays a key role in the UVB-induced phosphorylation of histone H3 at serine 10 15708437_Alterations in Fyn localization might be associated with neurofibrillary pathology and synapse loss in Alzheimer's disease. 15713745_PEDF downregulates Fyn through Fes, resulting in inhibition of FGF-2-induced capillary morphogenesis of endothelial cells 15872086_calcium activates PLC-gamma1 via increased PIP3 formation mediated by c-src- and fyn-activated PI3K 16115884_the Fyn-tau interaction has a role in neurodegeneration 16145685_independent of one another Fyn and MAP-2c are able to induce process outgrowth and in concert can initiate and enhance process outgrowth in an additive manner. 16237174_Increased Fyn expression is sufficient to trigger prominent neuronal deficits in FYN/human amyloid precursor protein transgenic mice in the context of even relatively moderate Abeta levels. 16400523_Cells with single Src family kinase knockdown show that Src, Fyn and Yes kinases are all required for vascular endothelial growth factor (VEGF) mitogenic signaling in retinal microvascular endothelial cells. 16597701_Fibronectin rigidity response involves force-dependent Fyn phosphorylation of p130Cas with rigidity-dependent displacement. 16777849_Fyn suppresses the GTPase-activating protein (GAP) activity of wild-type brain-enriched Rho GTPase-activating protein TCGAP but not the Y406F mutant of TCGAP in a phosphorylation-dependent manner. 16782058_Staurosporine binds to the ATP-binding site of Fyn in a similar manner as in the Lck- and Csk-complexes 16860569_These results indicate that Fyn is activated by G-protein-coupled receptor stimulation and is responsible for transactivation of TrkA receptors on intracellular membranes. 16882656_extracellular matrix fragments produced by apoptotic EC initiate a state of resistance to apoptosis in fibroblasts via an alpha2beta1 integrin/SFK (Src and Fyn)/phosphatidylinositol 3-kinase (PI3K)-dependent pathway 16888650_Glucocorticoids causes dissociation of T-cell receptor associated protein complex containing LCK and FYN. 16891393_TNF-alpha regulates the pulmonary vascular endothelial paracellular pathway, in part, through fyn activation. 17130124_The extra-domain B binding D3 protein opens new biomedical opportunities for the in vivo imaging of solid tumorsand for the delivery of toxic agents to the tumoral vasculature. 17334644_Observational study of gene-disease association. (HuGE Navigator) 17417065_Observational study of gene-disease association. (HuGE Navigator) 17417065_Polymorphisms of the Fyn gene is related to the function of glutamatergic system, and a performance on neuropsychological test of prefrontal cortex activity in schizophrenic patients. 17599905_the mechanism of signal transduction by CD244 is to regulate FYN kinase recruitment and/or activity and the outcome of CD48/CD244 interactions is determined by which other receptors are engaged. 17701175_Fyn is weakly phosphorylated in normal B cells, but strongly phosphorylated in myeloma B cells. 17703099_Observational study of gene-disease association. (HuGE Navigator) 17703099_The results of this study may suggest a relationship between the FYN gene polymorphisms and allergic asthma 17943724_Chromosomal deletion, promoter hypermethylation and downregulation of FYN is associated with prostate cancer 18056706_engagement of the SH2 domain on PAG renders FynT insensitive to Csk negative regulation 18089558_Fyn, due in part to its effects on Dab1, regulates the phosphorylation, trafficking, and processing of APP and apoEr2. 18174230_Observational study of gene-disease association. (HuGE Navigator) 18180382_Up-regulation, in contrast to activation, of the ubiquitously expressed Src kinase, Fyn, by BCR-ABL1, is described. 18267011_NS5A binds to the Fyn SH3 domain with what can be considered a high affinity SH3 domain-ligand interaction (629 nM), and this binding did not require the presence of domain I of NS5A (amino acid residues 32-250). 18337055_DNMTs activity may have an indirect influence on the expression of Fyn without altering the methylation level of its promoter in Hut-78 T-lymphoma cells. 18564062_H2O2 significantly inhibited Cl(-)/OH(-) exchange activity in Caco-2 cells. H(2)O(2)-mediated inhibition of Cl(-)/OH(-) exchange activity involved the Src kinase Fyn and PI3K (phosphoinositide 3-kinase)-dependent pathways. 18628988_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18710921_PECAM-1 and Fyn are essential components of a PECAM-1-based mechanosensory complex in endothelial cells 18842137_Analysis reveals that the Fyn SH2 domain forms a noisy communication channel that couples residues located in the phosphopeptide and specificity binding sites and a number of residues at the other side of the domain. 18849153_Observational study of gene-disease association. (HuGE Navigator) 18922801_tyrosine phosphorylation of Neph1 mediated by Fyn results in significantly increased Neph1 and ZO-1 binding, suggesting a critical role for Neph1 tyrosine phosphorylation in reorganizing the Neph1-ZO-1 complex. 18978678_Observational study of gene-disease association. (HuGE Navigator) 19131339_Mutation of Sp1 and Egr1 binding sites within the essential region diminished Fyn promoter activity and identified Egr1 as conferring redox sensitivity. 19258392_Endocytosis of flotillins is regulated by the Src family kinase Fyn. 19258394_These results suggest that SFK trafficking is specified by the palmitoylation state in the SH4 domain. 19330793_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19330793_The results of the study demonstrated only marginal association between FYN polymorphisms and prophylactic lithium response in bipolar 19468241_The results suggest that the glutamatergic FYN gene may be associated with bipolar disorder, particularly with type I illness and early age of onset. 19501919_These results suggest novel deficits in Fyn function, manifested as the downregulation of Fyn protein or the altered transcription of the fyn gene, in patients with schizophrenia. 19542604_Mounting evidence summarized in this review suggests that tyrosine kinase Fyn and c-Abl are critical in the neurodegenerative process which occurs in tauopathies. 19567819_Increased expression of Fyn is associated with resistance to BCR-ABL inhibitors in chronic myelogenous leukemia. 19690143_a mechanism linking EGFR signaling with Fyn and Src activation to promote glioblastoma progression and invasion in vivo 19733625_Fyn is suggested to play an essential role in signaling events that implicate spindle assembly checkpoint pathway and hence in regulating the exit from metaphase in oocytes and zygote. 19805512_Results describe on a molecular level how Fyn kinase uses alternative splicing to adapt to different cellular environments. 19816407_brain-specific PTPRT regulates synapse formation through interaction with cell adhesion molecules, and this function and the phosphatase activity are attenuated through tyrosine phosphorylation by the synaptic tyrosine kinase Fyn. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19917775_Gbeta1-mediated Fyn activation integrates FAK with AJ, preventing persistent endothelial barrier leakiness. 19955046_Data confirm previous observations of higher expression of CD70 in CD4+ T cells from patients with SLE, and suggest that increased Fyn protein content in CD4+ T cells can be associated with high SLE disease activity. 19968749_Suggest a possible mechanism by which Fyn activity regulates cell proliferation and apoptosis that exerts an effect on pancreatic cancer metastasis. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20056645_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20506281_Data demonstrate that CRMP1 tyrosine 504 is a primary target of the Src family of tyrosine kinases (SFKs), specifically Fyn. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20658524_These findings indicate that phosphorylation of S21 within the pPKA recognition site (RxxS motif) of Fyn regulates its tyrosine kinase activity and controls focal adhesion targeting. 20682987_Activation of Fyn promoted SCC cell migration and its suppression thwarted SCC migration toward FN. These results indicate that the activation of Fyn kinase as well as local growth factor concentration modulate EMT in oral SCC. 20855525_Data describe how alpha6beta4 integrin selectively activates Fyn in response to receptor engagement, and show that both catalytic and noncatalytic functions of SHP2 are required for Fyn activation by alpha6beta4. 21098700_Results highlight an important role for the Fyn/ERK signaling pathway in imatinib-resistant cells that is driven by accumulation of intracellular SPARC. 21298565_Here we report the (1)H, (15)N and (13)C backbone and side-chain chemical shift assignments of the SH2 domain of the human protein tyrosine kinase Fyn. 21364031_Fyn plays an important role in prostate cancer biology by facilitating cellular growth and by regulating directed chemotaxis-a key component of metastasis. 21371426_Fyn, a member of Src family tyrosine kinases, plays a critical role in mediating TGF-beta1-induced down-regulation of E-cadherin in human A549 lung cancer cells. 21480388_Data show that the Ras/PI3K/Akt pathway can account for Fyn over-expression in cancers. 21513984_Fyn phosphorylates Tim-1 and binds Tim-1 in the absence of tyrosine 299 in 293T cells. Fyn is required for phosphorylation of Tim-1 in B cells. 21592972_Negative regulation of Gq-mediated pathways in platelets by G(12/13) pathways through Fyn kinase 21642356_High Fyn expression is associated with pancreatic cancer progression. 21692989_Pro216 in tau PXXP motif plays role in interaction of tau with Fyn-SH3; phosphorylation of tau at Tyr18 is important for mediating Fyn-SH2-tau interactions; tyrosine phosphorylation influences tau localization in cortical neurons which is Fyn-dependent 21749309_Data show that a higher percentage of classical Hodgkin lymphoma cases expressing Lyn, Fyn and Syk compared with previous reports. 21757751_CD5 glycoprotein-mediated T cell inhibition dependent on inhibitory phosphorylation of Fyn kinase 21799250_[review] Recent evidence cited in this review suggests that alterations in Fyn, a Src family kinase, might contribute to Alzheimer's disease pathogenesis. 21881001_TM4SF10, possibly through ADAP, may regulate Fyn activity 22161863_characterized the folding kinetics of a Fyn SH3 domain variant containing 5 amino acid substitutions that was computationally designed to optimize surface charge-charge interactions; results show that optimized Fyn SH3 domain is stabilized through an 8-fold acceleration in the folding rate. 22189847_Fyn is not required for the pathogenesis of Bcr-Abl-mediated leukemias. 22354875_Double RNA interference knockdown of Fyn and Lyn induced apoptosis accompanied by caspase-8 activation in NCI-H2052 and ACC-MESO-4 cells. 22403409_The protein phosphatase PP2A/Balpha binds to the microtubule-associated proteins Tau and MAP2 at a motif also recognized by the kinase Fyn. 22442244_X-ray data were collected to a resolution of 2.00 A for the unbound form and 1.40 A for the Fyn in complex with the phosphotyrosine peptide 22474169_Oxidative injury caused by arachidonic acid and iron enhanced Fyn phosphorylation at a tyrosine residue. 22689581_c-Src but not Fyn promotes proper spindle orientation in early prometaphase 22709448_Fyn mediates postsynaptic density protein- 95Y523 phosphorylation, which may be responsible for the excitotoxic signal cascades and neuronal apoptosis in brain ischemia and amyloid-beta peptide neurotoxicity. 22715382_these results demonstrate a Fyn kinase-dependent mechanism through which IFNgamma regulates E-cadherin stability and suggest a novel mechanism of disruption of epithelial cell contact, which could contribute to perturbed epithelial barrier function. 22833681_tau interacts in a phosphorylation-dependent manner with the PSD95-Fyn-NMDA receptor complex at the postsynaptic site 22843238_The results presented from this in silico study will open up new prospect for genetic analysis of FYN gene and their correlation with clinical data will be very useful in understanding the genetics of Alzheimer's disease. 22868769_analysis of binding of the Fyn SH3 domain to a peptide from the NS5A protein 22987042_Highlighted is recent evidence that FYN kinase mediates signal transduction downstream of the PrP(C)-ABETA oligomer complex in Alzheimer disease. [review] 23077515_results suggest that NS5A directly binds to the SH2 domain of Fyn in a tyrosine phosphorylation-dependent manner 23175838_we showed that gene dosage of Prnp regulates amyloid beta-induced Fyn/tau alterations 23188823_Nck may facilitate dynamic signaling events at the slit diaphragm by promoting Fyn-dependent phosphorylation of nephrin. 23250004_no association between FYN polymorphisms and schizophrenia risk or age at schizophrenia onset, was found. 23300847_the non-catalytic functions of the kinases Fyn and Pyk2 were required for late stage human T cell adhesion. 23405030_positively regulates IFN-lambda1 genes during viral infection 23438599_Fyn regulates the activity of the adipogenic transcription factor signal transducer and activator of transcription 5a (STAT5a) through enhancing its interaction with the GTPase phosphoinositide 3-kinase enhancer A (PIKE-A). 23497302_Expression of HPV type 16 E7 resulted in increase in Src and Yes proteins level, but did not alter the level of Fyn. 23606749_MicroRNA-125a-3p reduces cell proliferation and migration by targeting Fyn. 23805846_PrP(C) down-regulated tau via the Fyn pathway and the effect can be regulated by Abeta oligomers. 23836527_Fyn expression in mdMSCs contributes to basal cytoskeletal architecture and, when associated with FAs, functions as a proximal mechanical effector for environmental signals that influence MSC lineage allocation. 23851594_These results provided suggestive evidence that the FYN gene contributes toward the variance in human coping styles. 23915951_monomeric IgE, in the absence of antigen, induces VEGF production in MC and in vivo contributes to melanoma tumor growth through a Fyn kinase-dependent mechanism. 23918783_These data demonstrate a mechanism whereby Fyn and Lyn, redundantly mediate anticryptococcal killing by inducing the polarization of perforin-containing granules to the NK cell-cryptococcal synapse. 24413734_Mutations in FYN implicate SRC signaling, impaired DNA damage response, and escape from immune surveillance mechanisms in the pathogenesis of peripheral T cell lymphomas. 24852829_The SNPs in the selected regions of the Fyn gene are unlikely to confer the susceptibility of sAD in the Chinese Han population 24882577_Results indicate that FYN has an important role in tamoxifen resistance, and its subcellular localization in breast tumor cells may be an important novel biomarker of response to endocrine therapy in breast cancer. 24976598_Fyn inhibition may be an effective therapeutic approach in treating cSCC. 25130779_the main finding is that FYN polymorphisms were respon- sible for the variance in intermediate defense style(undoing)and mature defensestyle(suppression). 25503120_it is not just inactivation of Fyn that promotes multicellular spheroid formation but this must be coupled with the full length beta6 integrin. 25967238_Fyn, but not Lyn, was required for complete Pyk2 phosphorylation by thrombin. 26001218_Mutation of Fyn phosphorylation sites on PIKE-A, depletion of Fyn, or pharmacological inhibition of Fyn blunts the association between PIKE-A and AMPK, resulting in loss of its inhibitory effect on AMPK. 26125726_Our results establish that Fyn can arrest SW-induced apoptosis via the activity of Akt and its effective phosphorylation in 293T cells. 26365631_Fyn facilitates mitotic spindle formation through the increase in microtubule polymerization, resulting in the acceleration of M-phase progression. 26382759_Results show that Fyn differentially modulates Nav1.5 channel splice variants. It phosphorylates Nav1.5 variants Q-del and Q-pre resulting in there hyperpolarizing and depolarizing shift. Fyn's activity is abolished in the presence of both variants. 26384592_Data provide structural insight into the dimerization of Fyn SH2 both in solution and in crystalline conditions, providing novel crystal structures of both the dimer and peptide-bound structures of Fyn SH2. 26407913_our results provide no evidence that the Fyn -93A>G SNP contributes to the susceptibility to acute liver transplant rejection in a Caucasian population. 26561212_In this study, it is shown that the alternatively spliced FynT isoform is specifically up-regulated in the AD neocortex, with no change in FynB isoform. 26624980_Neuroendocrine differentiation in prostate cancer cells and visceral metastasis, are least in part, regulated by FYN kinase. 26646899_Results show that three CpG loci within FYN were hypermethylated in obese individuals, while obesity was associated with lower methylation of CpG loci within PIWIL4 and TAOK3. 26786295_Data suggest both KLF5 (Kruppel like factor 5) and FYN are important in regulation of migration in bladder cancer cells; KLF5 up-regulates cell migration, lamellipodia formation, FYN expression, and phosphorylation of FAK (focal adhesion kinase). 26848862_Data show that fyn proto-oncogene protein (FYN) expression is deregulated in acute myeloid leukemia and that higher expression of FYN, in combination with FLT3 protein-ITD mutation, resulted in enrichment of the STAT5 transcription factor signaling. 26888964_this study shows that p59(fyn), which is essential for activation of T cells through the T-cell receptor, is also critical for signal transduction through Toll-like receptors in T cells 26892111_Fyn expression fluctuated with the progress of normal pregnancy and was elevated in patients with recurrent spontaneous abortion 27040756_Results found that GluN2B subunit-containing NMDARs were dominant in induced pluripotent stem cell-derived neurons and that tyrosine-protein kinase Fyn potentiated the function of GluN2B subunit-containing NMDARs. 27193083_Study identified the binding site between tau and fyn-SH3 may facilitate the development of compounds that can inhibit tau-fyn interactions, which presents an alternative therapeutic strategy for Alzheimer's disease; and provide evidence that a physiological correlation between phosphorylated tau at S202, S262, and S396/404 and fyn is not present in Alzheimer's disease brain. 27349276_FYN was transcriptionally regulated by FOXO1. 27466485_These results indicate that the microenvironment and growth patterns in an multicellular spheroid are complex and require MAPK and FYN kinase 27520374_The data suggest that miR-106b inhibits Amyloid-beta (1-42)-induced tau phosphorylation at Tyrosine 18 by targeting Fyn. 27616741_upregulated in fibrotic kidneys 27692963_Study reveal that binding the phosphorylated tail of Fyn perturbs a residue cluster near the linker connecting the SH2 and SH3 domains of Fyn, which is known to be relevant in the regulation of the activity of Fyn. 27694211_a substantial fraction of unligated CD36 exists in nanoclusters, which not only promote TSP-1 binding but are also enriched with the downstream effector Fyn. 28033507_FYN expression is regulated according to AD status and regulatory region haplotype, and genetic variants may be instrumental in the development of neurofibrillary tangles in AD and other tauopathies. 28368000_Fyn and Lyn as important factors that promote Plasmacytoid dendritic cell responses. 28393199_Upon SMAD4 deletion, we detected high expression levels of FYN in vessel endothelial cells, suggesting the mechanism of the ovarian tumor cells cross the endothelial barrier and transform to an invasive phenotype 28560430_High FYN expression is associated with pancreatic cancer metastasis. 28811476_Fyn-dependent phosphorylation of SHP-1 serine 591 inactivates the phosphatase, enabling activatory immunoreceptor signaling. 28948209_Fyn binds to mGluR1a at a consensus binding motif located in the intracellular C-terminus (CT) of mGluR1a in vitro. Active Fyn phosphorylates mGluR1a at a conserved tyrosine residue in the CT region. In cerebellar neurons and transfected HEK293T cells, Fyn-mediated tyrosine phosphorylation of mGluR1a is constitutively active and facilitates surface expression of mGluR1a and potentiates mGluR1a postreceptor signaling. 29091353_Data suggest that Fyn tyrosine kinase (Fyn)-dependent phosphorylation at two critical tyrosines is a key feature of vertebrate plexin A1 (PlxnA1) and plexin A2 (PlxnA2) signal transduction. 29140740_Study was the first to demonstrate critical positive regulation of thyroid tumorigenesis by FYN, which could be a potential target gene for thyroid carcinoma treatment. 29348460_A pivotal role of FYN and its downstream effectors in maintaining the basal type features in breast cancer. 29486132_NRP1 can form complexes with FYN and have the correlation changes in odontoblast differentiation of dental pulp stem cells (DPSCs). Therefore, the study surmise that in the progress of dental caries, NRP1 interacts with FYN, by expanding inflammation and inhibition of odontoblast differentiation of DPSCs through nuclear factor kappa B signaling pathway. 29555370_Protein ISG15 mediates the tumorigenesis via c-MET/Fyn/beta-catenin pathway in Esophageal squamous cell carcinoma cell lines. 29734505_After binding, Fyn kinase phosphorylates tyrosine residues present in the N- and C-terminal regions of the subunit voltage-gated sodium channel subunit alpha Nav1.5 NaV 1.5 channel (Nav1.5). 29790812_Nav1.7 is a substrate for Fyn kinase. 30251698_SPHK2 is highly expressed in the kidney interstitium of patients with renal fibrosis and highly correlates with disease progression. SPHK2 phosphorylates Fyn to activate downstream STAT3 and AKT, thereby promoting extracellular matrix synthesis, kidney fibroblast activation, and renal fibrosis. 30266665_miR-381 induces sensitivity of breast cancer cells to doxorubicin by inactivation of MAPK signaling via FYN 30341149_We confirmed the interaction of endogenous SHROOM3 with FYN in podocytes via a critical Src homology 3-binding domain, distinct from its ROCK-binding domain. Shroom3-Fyn interaction was required in vitro and in vivo for activation of Fyn kinase and downstream nephrin phosphorylation in podocytes. 30482845_Results showed that the EF and BG loops in the Fyn SH2 domain are highly adaptable and evolvable. The extreme versatility of the EF and BG loops afford them the ability to encode the broad spectrum of specificity found in naturally occurring SH2 domains. 30978441_Disruption of NT5DC2 in GSCs markedly reduces the expression of Fyn, a Src family proto-oncogene that has been implicated in the regulation of GBM progression. The expression of NT5DC2 strongly correlated with increased aggression of human gliomas, but not that of other brain tumors. 31036561_In vivo experiments using A53T and viral-alphaSyn overexpression mouse models as well as human Parkinson's disease neuropathological results further confirm the role of Fyn in NLRP3 inflammasome activation. 31189776_Phosphorylated Fyn expression may affect the prognosis of patients with lung adenocarcinoma after lung resection. 31957823_Fyn stimulates the progression of pancreatic cancer via Fyn-GluN2b-AKT axis. 32291412_The phosphorylation of CD147 by Fyn plays a critical role for melanoma cells growth and metastasis. 32561075_Overexpression of FYN suppresses the epithelial-to-mesenchymal transition through down-regulating PI3K/AKT pathway in lung adenocarcinoma. 32606017_FYN and ABL Regulate the Interaction Networks of the DCBLD Receptor Family. 32611392_Fyn kinase inhibition reduces protein aggregation, increases synapse density and improves memory in transgenic and traumatic Tauopathy. 32811808_FYN is required for ARHGEF16 to promote proliferation and migration in colon cancer cells. 32814048_Fyn Kinase Controls Tau Aggregation In Vivo. 32929078_Chromatin accessibility mapping of the striatum identifies tyrosine kinase FYN as a therapeutic target for heroin use disorder. 32986360_Expression Analysis of Fyn and Bat3 Signal Transduction Molecules in Patients with Chronic Lymphocytic Leukemia. 33325804_Profibrotic epithelial TGF-beta1 signaling involves NOX4-mitochondria cross talk and redox-mediated activation of the tyrosine kinase FYN. 33380425_Disturbances in PP2A methylation and one-carbon metabolism compromise Fyn distribution, neuritogenesis, and APP regulation. 33409551_Tyrosine kinase Fyn promotes apoptosis after intracerebral hemorrhage in rats by activating Drp1 signaling. 33510346_Src-mediated tyrosine phosphorylation of PRC1 and kinastrin/SKAP on the mitotic spindle. 33787846_Synergy and allostery in ligand binding by HIV-1 Nef. 34140493_Fusion transcripts FYN-TRAF3IP2 and KHDRBS1-LCK hijack T cell receptor signaling in peripheral T-cell lymphoma, not otherwise specified. 34432266_Fyn Kinase Activity and Its Role in Neurodegenerative Disease Pathology: a Potential Universal Target? 35238864_Fyn and TOM1L1 are recruited to clathrin-coated pits and regulate Akt signaling. 35397600_A new finding in the key prognosis-related proto-oncogene FYN in hepatocellular carcinoma based on the WGCNA hub-gene screening trategy. | ENSMUSG00000019843 | Fyn | 1641.17788 | 1.0917546 | 0.1266486628 | 0.09403306 | 1.818607e+00 | 1.774792e-01 | 5.721976e-01 | No | Yes | 2394.208848 | 285.252359 | 2055.173448 | 239.469612 | |
| ENSG00000015133 | 440193 | CCDC88C | protein_coding | Q9P219 | FUNCTION: Required for activation of guanine nucleotide-binding proteins (G-proteins) during non-canonical Wnt signaling (PubMed:26126266). Binds to ligand-activated Wnt receptor FZD7, displacing DVL1 from the FZD7 receptor and leading to inhibition of canonical Wnt signaling (PubMed:26126266). Acts as a non-receptor guanine nucleotide exchange factor by also binding to guanine nucleotide-binding protein G(i) alpha (Gi-alpha) subunits, leading to their activation (PubMed:26126266). Binding to Gi-alpha subunits displaces the beta and gamma subunits from the heterotrimeric G-protein complex, triggering non-canonical Wnt responses such as activation of RAC1 and PI3K-AKT signaling (PubMed:26126266). Promotes apical constriction of cells via ARHGEF18 (PubMed:30948426). {ECO:0000269|PubMed:26126266, ECO:0000269|PubMed:30948426}. | Alternative splicing;Cell junction;Coiled coil;Cytoplasm;Disease variant;Guanine-nucleotide releasing factor;Neurodegeneration;Phosphoprotein;Reference proteome;Spinocerebellar ataxia;Wnt signaling pathway | This gene encodes a ubiquitously expressed coiled-coil domain-containing protein that interacts with the dishevelled protein and is a negative regulator of the Wnt signalling pathway. The protein encoded by this gene has a PDZ-domain binding motif in its C-terminus with which it interacts with the dishevelled protein. Dishevelled is a scaffold protein involved in the regulation of the Wnt signaling pathway. The Wnt signaling pathway plays an important role in embryonic development, tissue maintenance, and cancer progression. Mutations in this gene cause autosomal recessive, primary non-syndromic congenital hydrocephalus; a condition characterized by excessive accumulation of cerebrospinal fluid in the ventricles of the brain. [provided by RefSeq, Jan 2013]. | hsa:440193; | cell junction [GO:0030054]; centrosome [GO:0005813]; cytoplasm [GO:0005737]; dynein light intermediate chain binding [GO:0051959]; frizzled binding [GO:0005109]; G-protein alpha-subunit binding [GO:0001965]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; microtubule binding [GO:0008017]; PDZ domain binding [GO:0030165]; protein self-association [GO:0043621]; apical constriction [GO:0003383]; cytoplasmic microtubule organization [GO:0031122]; cytoskeleton-dependent intracellular transport [GO:0030705]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; non-canonical Wnt signaling pathway [GO:0035567]; protein destabilization [GO:0031648]; regulation of protein phosphorylation [GO:0001932]; small GTPase mediated signal transduction [GO:0007264]; stress-activated protein kinase signaling cascade [GO:0031098] | 23042809_Our data validate CCDC88C as causing autosomal recessive, primary non-syndromic congenital hydrocephalus, suggesting this gene may be an important cause of congenital hydrocephalus. 24772479_LDI-PCR revealed a fusion between CCDC88C exon 25 and PDGFRB exon 11. 25062847_A novel missense mutation in CCDC88C activates the JNK pathway and causes a dominant form of spinocerebellar ataxia. 25062847_Spinocerebellar ataxia 40 (SCA40) displays typical cerebellar ataxia signs and pontocerebellar atrophy. Whole-exome sequencing led to the identification of a novel missense mutation in the gene CCDC88C in all SCA40-affected individuals. Cell-based assays showed that the SCA40 mutation causes an up-regulation of the JNK stress kinase signaling cascade that subsequently triggers programmed cell death. 26126266_Thus, Daple activates Galphai proteins and enhances non-canonical Wnt signaling by Frizzled receptors, and its dysregulation can impact both tumor initiation and progression to metastasis. 26577606_we demonstrated the relevance of Daple expression to gastric cancer progression. 29021338_This work not only identifies Daple as a platform for cross-talk between Akt and the noncanonical Wnt pathway but also reveals the impact of such cross-talk on tumor cell phenotypes that are critical for cancer initiation and progression. 29341397_Our report further establishes CCDC88C as one of the few known recessive causes of severe prenatal-onset hydrocephalus. Recognition of this syndrome has important diagnostic and genetic implications for families identified in the future. 30398676_Whole-exome sequencing (WES) of the proband revealed a heterozygous substitution, c.127G>A, in the CCDC88C gene (NM_001080414) that resulted in a missense mutation, p.(Asp43Asn) 30575751_Detection of Daple transcripts in the peripheral blood (i.e., liquid biopsies) of patients with melanoma may serve as a prognostic marker and an effective strategy for non-invasive long-term follow-up of patients with melanoma. 31268831_DAPLE and MPDZ function as cooperative partners at apical junctions. 31431650_Two Isoforms of the Guanine Nucleotide Exchange Factor, Daple/CCDC88C Cooperate as Tumor Suppressors. 31511612_Fusion driven JMML: a novel CCDC88C-FLT3 fusion responsive to sorafenib identified by RNA sequencing. 31949046_DAPLE protein inhibits nucleotide exchange on Galphas and Galphaq via the same motif that activates Galphai. 32888647_The Daple-CK1epsilon complex regulates Dvl2 phosphorylation and canonical Wnt signaling. 34092257_Neuropathological hallmarks of fetal hydrocephalus linked to CCDC88C pathogenic variants. | ENSMUSG00000021182 | Ccdc88c | 442.23719 | 0.9173804 | -0.1244080888 | 0.14913785 | 6.942751e-01 | 4.047140e-01 | 7.637945e-01 | No | Yes | 498.266814 | 44.335979 | 524.639282 | 45.615428 | |
| ENSG00000021300 | 58473 | PLEKHB1 | protein_coding | Q9UF11 | Alternative splicing;Cytoplasm;Developmental protein;Membrane;Reference proteome | hsa:58473; | cytoplasm [GO:0005737]; integral component of membrane [GO:0016021]; phototransduction [GO:0007602]; regulation of cell differentiation [GO:0045595] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20301200_Phr1 is widely and abundantly expressed throughout mature olfactory neurons and other primary sensory neurons, but its absence does not appear to affect olfactory morphology, regeneration, sensory function, or adaptation. 20308990_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 24840802_Rapid Wallerian degeneration requires the pro-degenerative molecules PHR1. 26318175_The human PHR1, a PH domain-containing protein with low expression in the heart and high expression in the brain, interacts with coxsackievirus B3 VP1, a major structural protein of coxsackievirus B3. | ENSMUSG00000030701 | Plekhb1 | 94.59324 | 0.9463690 | -0.0795253325 | 0.29957783 | 7.034442e-02 | 7.908360e-01 | 9.403589e-01 | No | Yes | 118.434134 | 16.420882 | 118.821013 | 15.952913 | |||
| ENSG00000025039 | 58528 | RRAGD | protein_coding | Q9NQL2 | FUNCTION: Guanine nucleotide-binding protein that plays a crucial role in the cellular response to amino acid availability through regulation of the mTORC1 signaling cascade (PubMed:20381137, PubMed:24095279). Forms heterodimeric Rag complexes with RRAGA or RRAGB and cycles between an inactive GTP-bound and an active GDP-bound form (PubMed:24095279). In its active form participates in the relocalization of mTORC1 to the lysosomes and its subsequent activation by the GTPase RHEB (PubMed:20381137, PubMed:24095279). This is a crucial step in the activation of the TOR signaling cascade by amino acids (PubMed:20381137, PubMed:24095279). {ECO:0000269|PubMed:20381137, ECO:0000269|PubMed:24095279}. | 3D-structure;Alternative splicing;Cytoplasm;GTP-binding;Hydrolase;Lysosome;Nucleotide-binding;Nucleus;Reference proteome | RRAGD is a monomeric guanine nucleotide-binding protein, or G protein. By binding GTP or GDP, small G proteins act as molecular switches in numerous cell processes and signaling pathways.[supplied by OMIM, Apr 2004]. | hsa:58528; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; Gtr1-Gtr2 GTPase complex [GO:1990131]; intracellular membrane-bounded organelle [GO:0043231]; lysosome [GO:0005764]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; GTPase binding [GO:0051020]; protein heterodimerization activity [GO:0046982]; cellular protein localization [GO:0034613]; cellular response to amino acid stimulus [GO:0071230]; cellular response to leucine [GO:0071233]; cellular response to leucine starvation [GO:1990253]; cellular response to starvation [GO:0009267]; positive regulation of TOR signaling [GO:0032008]; positive regulation of TORC1 signaling [GO:1904263]; regulation of autophagy [GO:0010506] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24095279_FLCN and its binding partners, FNIP1/2, are Rag-interacting proteins with GAP activity for RagC/D, but not RagA/B. 28619945_demonstrating a key role of RagD in promoting tumor growth 28963468_BC-LI-0186 binds to the RagD interacting site of LRS, thereby inhibiting lysosomal localization of LRS and mTORC1 activity. 33434687_Ras related GTP binding D promotes aerobic glycolysis of hepatocellular carcinoma. 34607910_mTOR-Activating Mutations in RRAGD Are Causative for Kidney Tubulopathy and Cardiomyopathy. | ENSMUSG00000028278 | Rragd | 480.70461 | 0.9800080 | -0.0291345559 | 0.14053476 | 4.293306e-02 | 8.358514e-01 | 9.553711e-01 | No | Yes | 623.711528 | 81.256407 | 598.734196 | 76.241429 | |
| ENSG00000029725 | 9135 | RABEP1 | protein_coding | Q15276 | FUNCTION: Rab effector protein acting as linker between gamma-adaptin, RAB4A and RAB5A. Involved in endocytic membrane fusion and membrane trafficking of recycling endosomes. Involved in KCNH1 channels trafficking to and from the cell membrane (PubMed:22841712). Stimulates RABGEF1 mediated nucleotide exchange on RAB5A. Mediates the traffic of PKD1:PKD2 complex from the endoplasmic reticulum through the Golgi to the cilium (By similarity). {ECO:0000250|UniProtKB:O35551, ECO:0000269|PubMed:10698684, ECO:0000269|PubMed:11452015, ECO:0000269|PubMed:12773381, ECO:0000269|PubMed:22841712, ECO:0000269|PubMed:8521472}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Endocytosis;Endosome;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:9135; | early endosome [GO:0005769]; early endosome membrane [GO:0031901]; endocytic vesicle [GO:0030139]; endosome [GO:0005768]; intracellular membrane-bounded organelle [GO:0043231]; protein-containing complex [GO:0032991]; recycling endosome [GO:0055037]; growth factor activity [GO:0008083]; GTPase activator activity [GO:0005096]; protein domain specific binding [GO:0019904]; protein homodimerization activity [GO:0042803]; apoptotic process [GO:0006915]; endocytosis [GO:0006897]; Golgi to plasma membrane transport [GO:0006893]; membrane fusion [GO:0061025]; protein localization to ciliary membrane [GO:1903441]; protein transport [GO:0015031]; vesicle-mediated transport [GO:0016192] | 12505986_GGAs, a family of Arf-dependent clathrin adaptors involved in selection of TGN cargo, interact with the Rabaptin-5-Rabex-5 complex, a Rab4/Rab5 effector regulating endosome fusion 19500109_The authors determined that increased listeriolysin O-independent dissolution of vacuoles during RABEP1 knockdown required the Listeria monocytogenes broad-range phospholipase C (PC-PLC). 21271679_Breast cancer cell line studies showed that microRNA, miR-373, was capable of promoting breast cancer invasion and metastasis via translational inhibition of TXNIP and RABEP1 that were the direct target genes of miR-373. 22841712_Silencing of Rabaptin-5 induces down-regulation of recycling of K(V)10.1 channel in transfected cells and reduction of K(V)10.1 current density in cells natively expressing K(V)10.1, indicating a role of Rabaptin-5 in channel trafficking. 22975325_The disruption of Rabaptin-5 Ser407 phosphorylation reduces persistent cell migration in 2D and alphavbeta3-dependent invasion. 24957337_analysis of Rabex-5 GEF activation by Rabaptin-5 26430212_The results contradict the model of feedback activation of Rab5 and instead indicate that Rbpt5 is recruited by both Rabex5 recognizing ubiquitylated cargo and by Rab4 to activate Rab5 in a feed-forward manner. 26633357_ITSN2L interacts with RABEP1 and stimulates its degradation in regulation of endocytosis. 26680696_The pleckstrin homology domain (PH) of PLD1 itself promotes degradation of HIF-1alpha, then accelerates EGFR endocytosis via upregulation of rabaptin-5 and suppresses tumor progression. 28694205_SH2B1 and RABEP1 genetic variants are associated with worsening of LDL and glucose parameters in patients treated with psychotropic drugs | ENSMUSG00000020817 | Rabep1 | 3704.12162 | 1.0663006 | 0.0926141562 | 0.07805840 | 1.410850e+00 | 2.349153e-01 | 6.260906e-01 | No | Yes | 4284.485155 | 618.892878 | 3834.115809 | 541.572223 | ||
| ENSG00000041880 | 10039 | PARP3 | protein_coding | Q9Y6F1 | FUNCTION: Mono-ADP-ribosyltransferase that mediates mono-ADP-ribosylation of target proteins and plays a key role in the response to DNA damage (PubMed:16924674, PubMed:20064938, PubMed:21211721, PubMed:21270334, PubMed:25043379, PubMed:24598253). Mediates mono-ADP-ribosylation of glutamate, aspartate or lysine residues on target proteins (PubMed:20064938, PubMed:25043379). In contrast to PARP1 and PARP2, it is not able to mediate poly-ADP-ribosylation (PubMed:25043379). Associates with a number of DNA repair factors and is involved in the response to exogenous and endogenous DNA strand breaks (PubMed:16924674, PubMed:21211721, PubMed:21270334). Together with APLF, promotes the retention of the LIG4-XRCC4 complex on chromatin and accelerate DNA ligation during non-homologous end-joining (NHEJ) (PubMed:21211721). Cooperates with the XRCC5-XRCC6 (Ku80-Ku70) heterodimer to limit end-resection thereby promoting accurate NHEJ (PubMed:24598253). Involved in DNA repair by mediating mono-ADP-ribosylation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism, such as XRCC5 and XRCC6 (PubMed:16924674, PubMed:24598253). ADP-ribosylation follows DNA damage and appears as an obligatory step in a detection/signaling pathway leading to the reparation of DNA strand breaks (PubMed:16924674, PubMed:21211721, PubMed:21270334). May link the DNA damage surveillance network to the mitotic fidelity checkpoint (PubMed:16924674). In addition to proteins, also able to ADP-ribosylate DNA: mediates DNA mono-ADP-ribosylation of DNA strand break termini via covalent addition of a single ADP-ribose moiety to a 5'- or 3'-terminal phosphate residues in DNA containing multiple strand breaks (PubMed:29361132, PubMed:29520010). Acts as a negative regulator of immunoglobulin class switch recombination, probably by controlling the level of AICDA /AID on the chromatin (By similarity). {ECO:0000250|UniProtKB:Q3ULW8, ECO:0000269|PubMed:16924674, ECO:0000269|PubMed:20064938, ECO:0000269|PubMed:21211721, ECO:0000269|PubMed:21270334, ECO:0000269|PubMed:24598253, ECO:0000269|PubMed:25043379, ECO:0000269|PubMed:29361132, ECO:0000269|PubMed:29520010}. | 3D-structure;ADP-ribosylation;Alternative splicing;Chromosome;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;Glycosyltransferase;NAD;Nucleus;Reference proteome;Transferase | The protein encoded by this gene belongs to the PARP family. These enzymes modify nuclear proteins by poly-ADP-ribosylation, which is required for DNA repair, regulation of apoptosis, and maintenance of genomic stability. This gene encodes the poly(ADP-ribosyl)transferase 3, which is preferentially localized to the daughter centriole throughout the cell cycle. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:10039; | centriole [GO:0005814]; cytoplasm [GO:0005737]; intercellular bridge [GO:0045171]; nuclear body [GO:0016604]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; site of double-strand break [GO:0035861]; catalytic activity [GO:0003824]; NAD DNA ADP-ribosyltransferase activity [GO:0140294]; NAD+ ADP-ribosyltransferase activity [GO:0003950]; protein ADP-ribosylase activity [GO:1990404]; DNA ADP-ribosylation [GO:0030592]; DNA repair [GO:0006281]; double-strand break repair [GO:0006302]; negative regulation of isotype switching [GO:0045829]; negative regulation of telomerase RNA reverse transcriptase activity [GO:1905662]; positive regulation of DNA ligation [GO:0051106]; positive regulation of double-strand break repair via nonhomologous end joining [GO:2001034]; protein ADP-ribosylation [GO:0006471]; protein auto-ADP-ribosylation [GO:0070213]; protein localization to site of double-strand break [GO:1990166]; protein mono-ADP-ribosylation [GO:0140289]; protein poly-ADP-ribosylation [GO:0070212]; regulation of mitotic spindle organization [GO:0060236]; telomere maintenance [GO:0000723] | 16924674_PARP-3 is a nuclear protein involved in transcriptional silencing and in the cellular response to DNA damage 17203305_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20064938_the interaction between PARP-1 and PARP-3 is unrelated to DNA single-strand break repair 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21211721_PARP-3 has a role in chromosomal DNA double-strand break repair. 21264220_PARP3 gene occupancy in the human neuroblastoma cell line SK-N-SH occurs preferentially with developmental genes regulating cell fate specification, tissue patterning, craniofacial development and neurogenesis. 21270334_PARP3 is a critical player in the stabilization of the mitotic spindle and in telomere integrity by associating and regulating the mitotic components NuMA and tankyrase 1. 24528514_In some cancer cells, repression of PARP3 could be responsible for an increased telomerase activity. 24598253_PARP3 likely facilitates the recruitment of Ku80 to double strand breaks to antagonize DNA end resection but facilitate Ku-mediated accurate classical non-homologous end-joining. 25255219_MiR-630 reduced apoptosis by downregulating several apoptotic modulators, PARP3, DDIT4, and EP300. 25800440_Identification of ADP-ribosylation sites in PARP3 and the determination of the extent ofpoly(ADP-ribosyl)ated residues in this protein was performed. 26040766_we found that PARP3 interacted with FoxM1 to enhance its transcriptional activity and conferred glioblastoma cell radioresistance. Thus, our data suggest that PARP3 could be a therapeutic target to overcome radioresistance in glioblastoma 27530147_These data identify PARP3 as a molecular sensor of nicked nucleosomes. 27579892_PARP3 controls of TGFbeta-induced epithelial mesenchymal transformation and acquisition of stem-like cell features by stimulation transglutaminase 2/SNAI1 signaling. 27716488_In a process of single-strand DNA repair, PARP3 mono-ADP-ribosylates nucleosomal histone H2B. 29054115_Data indicate that PARP3, a DNA damage-activated ADP-ribosyltransferase, can mono-ADP-ribosylate double-stranded DNA ends. 29361132_PARP3 can PARylate and mono(ADP-ribosyl)ate (MARylate), respectively, 5'- and 3'-terminal phosphate residues at double- and single-strand break termini of a DNA molecule containing multiple strand breaks. 29520010_gapped DNA that was ADP-ribosylated by PARP3 could be ligated to double-stranded DNA by DNA ligases. Moreover, this ADP-ribosylated DNA could serve as a primed DNA substrate for PAR chain elongation by the purified proteins PARP1 and PARP2 as well as by cell-free extracts. 29676938_The integral roles of PARP3 in DNA damage repair. 30039287_Our results indicated this approach with PARP3 inhibitors and vinorelbine is unique and promising for breast cancer patients with metastases. This combination could significantly increase the survival of breast cancer patients with metastases 32785980_Progress and outlook in studying the substrate specificities of PARPs and related enzymes. 34066057_PARP Power: A Structural Perspective on PARP1, PARP2, and PARP3 in DNA Damage Repair and Nucleosome Remodelling. | ENSMUSG00000023249 | Parp3 | 219.32198 | 0.7768611 | -0.3642714543 | 0.21008946 | 2.977793e+00 | 8.441429e-02 | 4.233177e-01 | No | Yes | 215.722213 | 20.922172 | 275.371716 | 25.672991 | |
| ENSG00000049769 | 89801 | PPP1R3F | protein_coding | Q6ZSY5 | FUNCTION: Glycogen-targeting subunit for protein phosphatase 1 (PP1). {ECO:0000269|PubMed:21668450}. | Alternative splicing;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a protein that has been identified as one of several type-1 protein phosphatase (PP1) regulatory subunits. One or two of these subunits, together with the well-conserved catalytic subunit, can form the PP1 holoenzyme, where the regulatory subunit functions to regulate substrate specificity and/or targeting to a particular cellular compartment. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:89801; | integral component of membrane [GO:0016021]; membrane [GO:0016020]; protein phosphatase type 1 complex [GO:0000164]; glycogen binding [GO:2001069]; protein phosphatase 1 binding [GO:0008157]; protein phosphatase binding [GO:0019903]; regulation of glycogen (starch) synthase activity [GO:2000465]; regulation of glycogen biosynthetic process [GO:0005979] | 8938429_Maps this gene as HB2E at the DXS9823E marker sequence. 20398921_Observational study of gene-disease association. (HuGE Navigator) 20479760_Observational study of gene-disease association. (HuGE Navigator) 34145793_Methylation of three genes encoded by X chromosome in blood leukocytes and colorectal cancer risk. | ENSMUSG00000039556 | Ppp1r3f | 101.79124 | 0.7999766 | -0.3219703361 | 0.29157396 | 1.209684e+00 | 2.713947e-01 | 6.616079e-01 | No | Yes | 107.377107 | 15.543838 | 128.251911 | 17.813435 | |
| ENSG00000050767 | 91522 | COL23A1 | protein_coding | Q86Y22 | Alternative splicing;Cell membrane;Collagen;Membrane;Reference proteome;Repeat;Signal-anchor;Transmembrane;Transmembrane helix | COL23A1 is a member of the transmembrane collagens, a subfamily of the nonfibrillar collagens that contain a single pass hydrophobic transmembrane domain (Banyard et al., 2003 [PubMed 12644459]).[supplied by OMIM, Mar 2008]. | hsa:91522; | cell surface [GO:0009986]; collagen trimer [GO:0005581]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular space [GO:0005615]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; extracellular matrix structural constituent [GO:0005201]; heparin binding [GO:0008201]; identical protein binding [GO:0042802]; extracellular matrix organization [GO:0030198] | 12644459_identification and cloning; a new member of the transmembrane collagen family, showing structural homology with the transmembrane collagens XIII and XXV 16728390_analysis of collagen XXIII mRNA and protein 17627939_newly synthesized collagen XXIII either is cleaved inside the Golgi/trans-Golgi network or reaches the cell surface, where it becomes protected from processing by being localized in lipid rafts. 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20447926_High COL23A1 expression is associated with recurrent non-small cell lung cancer. 21652699_data suggest that extracellular membrane-bound CAIV, but not cytosolic CAII, augments transport activity of MCT2 in a non-catalytic manner, possibly by facilitating a proton pathway other than His-88 | ENSMUSG00000063564 | Col23a1 | 50.38033 | 1.5935329 | 0.6722287668 | 0.42052130 | 2.576731e+00 | 1.084454e-01 | No | Yes | 73.191434 | 19.920732 | 47.243286 | 12.596508 | |||
| ENSG00000051108 | 9709 | HERPUD1 | protein_coding | Q15011 | FUNCTION: Component of the endoplasmic reticulum quality control (ERQC) system also called ER-associated degradation (ERAD) involved in ubiquitin-dependent degradation of misfolded endoplasmic reticulum proteins (PubMed:16289116, PubMed:28827405). Could enhance presenilin-mediated amyloid-beta protein 40 generation. Binds to ubiquilins and this interaction is required for efficient degradation of CD3D via the ERAD pathway (PubMed:18307982). {ECO:0000269|PubMed:16289116, ECO:0000269|PubMed:18307982, ECO:0000269|PubMed:28827405}. | 3D-structure;Acetylation;Alternative splicing;Endoplasmic reticulum;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Unfolded protein response | The accumulation of unfolded proteins in the endoplasmic reticulum (ER) triggers the ER stress response. This response includes the inhibition of translation to prevent further accumulation of unfolded proteins, the increased expression of proteins involved in polypeptide folding, known as the unfolded protein response (UPR), and the destruction of misfolded proteins by the ER-associated protein degradation (ERAD) system. This gene may play a role in both UPR and ERAD. Its expression is induced by UPR and it has an ER stress response element in its promoter region while the encoded protein has an N-terminal ubiquitin-like domain which may interact with the ERAD system. This protein has been shown to interact with presenilin proteins and to increase the level of amyloid-beta protein following its overexpression. Alternative splicing of this gene produces multiple transcript variants encoding different isoforms. The full-length nature of all transcript variants has not been determined. [provided by RefSeq, Jan 2013]. | hsa:9709; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum quality control compartment [GO:0044322]; integral component of membrane [GO:0016021]; Lewy body core [GO:1990037]; membrane [GO:0016020]; transmembrane transporter binding [GO:0044325]; ubiquitin ligase-substrate adaptor activity [GO:1990756]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; endoplasmic reticulum unfolded protein response [GO:0030968]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236]; negative regulation of intrinsic apoptotic signaling pathway [GO:2001243]; positive regulation of ER-associated ubiquitin-dependent protein catabolic process [GO:1903071]; protein targeting to ER [GO:0045047]; regulation of ER-associated ubiquitin-dependent protein catabolic process [GO:1903069]; regulation of protein ubiquitination [GO:0031396]; response to endoplasmic reticulum stress [GO:0034976]; response to unfolded protein [GO:0006986]; retrograde protein transport, ER to cytosol [GO:0030970]; ubiquitin-dependent ERAD pathway [GO:0030433]; ubiquitin-dependent protein catabolic process [GO:0006511] | 11799129_enhances presnilin-mediated generation of amyloid beta protein 12153396_upregulation by Wnt-1 12370023_may associate through its ubiquitin-like domain with the 26S proteasome, in this way connecting the protein degradation machinery to the ER membrane and resulting in an efficient ERAD 14550564_The ubiquitin-like domain of Herp most likely plays a role in the regulation of the intracellular level of Herp under ER stress 16940180_Results report the identification of Herp, a gene involved in ER stress-associated protein degradation (ERAD), as a direct target of Luman. 17020760_Expression of Herp protein is up-regulated in response to ER stress, including homocysteine. 17020760_We identified the transcription factor binding site AARE (amino acid response element) by mutational analysis involved in Herp induction in SH-SY5Y cells, and the more significant role of the CREB binding site compared to AARE in HEK 293T cells. 18042451_Herp is in a complex with ubiquitinated proteins and with the 26S proteasome, suggesting that it plays a role in linking substrates with the proteasome. 18307982_Our data suggest that Herp binding to ubiquilin proteins plays an important role in the ERAD pathway and that ubiquilins are specifically involved in degradation of only a subset of ubiquitinated targets, including Herp-dependent ERAD substrates. 19251110_Type II alcoholic patients had a statistically significant higher expression of Herp mRNA due to upregulation of the expression of this neuroprotective cell non-chaperone by toxic effects of ethanol. 19447887_the underlying molecular mechanism(s) whereby Herp counteracts Ca(2+) disturbances will provide insights into the molecular cascade of cell death in dopaminergic neurons 19788048_Herp may play a role in the initiation of the well-known inflammation-like changes in Parkinson disease substantia nigra and serve as a molecular link between degeneration and neuroinflammation in Parkinson disease 20054003_Data show that that 4-trifluoromethyl-celecoxib can inhibit secretion but not transcription of IL-12 (p35/p40) and IL-23 (p40/p19 heterodimers), and that this is associated with HERP function in the endoplasmic reticulum. 20147634_Herp mimics structural determinants of DNA immunologically and can be immunogenic in vivo. Thus, Herp represents a candidate autoantigen for anti-DNA antibodies. 20604806_The results suggest that ERAD molecule Herp may delay the degradation of cytosolic proteins at the ubiquitination step. 21149444_binding of Herp to Hrd1-containing ERAD complexes positively regulates the ubiquitylation activity of these complexes, thus permitting survival of the cell during ER stress. 23372434_Together with histological grade, increased co-expression of MIF and MMP9 in tumor might be a valuable predictor for recurrence, especially for benign meningiomas 24120520_Herp operates as a relevant factor in the defense against glucose starvation by modulating autophagy levels. 25637874_The results indicate that Nrf1 is a transcriptional activator of Herpud1 expression during ER stress, and they suggest Nrf1 is a key player in the regulation of the ER stress response in cells. 26616647_concluded that the HERPUD1-mediated cytoprotective effect against oxidative stress depends on the ITPR and Ca(2+) transfer from the endoplasmic reticulum to mitochondria 26823705_HERPUD1 SNPs are highly associated with polypoidal choroidal vasculopathy. 27084451_Thus, our findings indicated that NQO1 could stabilize Herp protein expression via indirect regulation of synoviolin. 27405867_The authors found that the CREB3/Herp pathway limited the increase in cytosolic Ca2+ concentration and apoptosis early in poliovirus infection and this may reduce the extent of poliovirus-induced damage to the central nervous system during poliomyelitis. 28430789_results indicated that low expression of miR-9-3p results in a high level of Herpud1, which may protect against apoptosis in glioma 28954889_HERP plays an important role in the regulation of host innate immunity in response to ER stress during the infection of RNA viruses. 29295953_The cytosolic relocalization of endoplasmic reticulum chaperone BiP did not require the function of translocon and was counteracted by HERP, a component of endoplasmic reticulum-associated protein degradation. 29863080_This study showed that Herp stability was regulated by synoviolin through lysine ubiquitination-independent proteasomal degradation. 32488540_Herpud1 deficiency could reduce amyloid-beta40 expression and thereby suppress homocysteine-induced atherosclerosis by blocking the JNK/AP1 pathway. 34353909_The Brucella effector BspL targets the ER-associated degradation (ERAD) pathway and delays bacterial egress from infected cells. | ENSMUSG00000031770 | Herpud1 | 2257.99373 | 1.0245688 | 0.0350168275 | 0.09787991 | 1.282046e-01 | 7.203008e-01 | 9.171944e-01 | No | Yes | 2842.935244 | 260.024973 | 2674.871035 | 239.421538 | |
| ENSG00000055917 | 23369 | PUM2 | protein_coding | Q8TB72 | FUNCTION: Sequence-specific RNA-binding protein that acts as a post-transcriptional repressor by binding the 3'-UTR of mRNA targets. Binds to an RNA consensus sequence, the Pumilio Response Element (PRE), 5'-UGUANAUA-3', that is related to the Nanos Response Element (NRE) (, PubMed:21397187). Mediates post-transcriptional repression of transcripts via different mechanisms: acts via direct recruitment of the CCR4-POP2-NOT deadenylase leading to translational inhibition and mRNA degradation (PubMed:22955276). Also mediates deadenylation-independent repression by promoting accessibility of miRNAs (PubMed:18776931, PubMed:22345517). Acts as a post-transcriptional repressor of E2F3 mRNAs by binding to its 3'-UTR and facilitating miRNA regulation (PubMed:22345517). Plays a role in cytoplasmic sensing of viral infection (PubMed:25340845). Represses a program of genes necessary to maintain genomic stability such as key mitotic, DNA repair and DNA replication factors. Its ability to repress those target mRNAs is regulated by the lncRNA NORAD (non-coding RNA activated by DNA damage) which, due to its high abundance and multitude of PUMILIO binding sites, is able to sequester a significant fraction of PUM1 and PUM2 in the cytoplasm (PubMed:26724866). May regulate DCUN1D3 mRNA levels (PubMed:25349211). May support proliferation and self-renewal of stem cells. Binds specifically to miRNA MIR199A precursor, with PUM1, regulates miRNA MIR199A expression at a postranscriptional level (PubMed:28431233). {ECO:0000269|PubMed:18776931, ECO:0000269|PubMed:21397187, ECO:0000269|PubMed:22345517, ECO:0000269|PubMed:22955276, ECO:0000269|PubMed:25340845, ECO:0000269|PubMed:25349211, ECO:0000269|PubMed:26724866, ECO:0000269|PubMed:28431233}. | 3D-structure;Alternative splicing;Cytoplasm;Methylation;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Translation regulation | This gene encodes a protein that belongs to a family of RNA-binding proteins. The encoded protein functions as a translational repressor during embryonic development and cell differentiation. This protein is also thought to be a positive regulator of cell proliferation in adipose-derived stem cells. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:23369; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; nuclear membrane [GO:0031965]; perinuclear region of cytoplasm [GO:0048471]; miRNA binding [GO:0035198]; mRNA 3'-UTR binding [GO:0003730]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; positive regulation of gene silencing by miRNA [GO:2000637]; positive regulation of RIG-I signaling pathway [GO:1900246]; posttranscriptional regulation of gene expression [GO:0010608]; production of miRNAs involved in gene silencing by miRNA [GO:0035196]; regulation of chromosome segregation [GO:0051983]; regulation of gene silencing by miRNA [GO:0060964]; regulation of mRNA stability [GO:0043488]; regulation of translation [GO:0006417]; stress granule assembly [GO:0034063] | 12511597_PUM2 as a component of conserved cellular machinery that may be required for germ cell development. 15607425_PUM2 and DAZL, are capable of binding the same mRNA sequences in the 3'UTR of human SDAD1 mRNA. 15617101_P2P-R expression may be translationally regulated by PUM2 16967088_PUMILIO mediated translational regulation may be universally used in developmental processes of the human body. 17154300_Observational study of gene-disease association. (HuGE Navigator) 17154300_Variation in the PUMILIO2 gene is very low and it seems improbable that mutations of this gene significantly contribute to male infertility in humans. 19168546_Data found that PUMILIO2 interacts also with SNAPIN, a modulator of SNARE complex assembly, which is involved in vesicle trafficking. 21397187_Three distinct modes of target RNA binding by PUM2 around the fifth mRNA base were observed. 21589936_in mitosis, PUM2 physically associates with Aurora-A to ensure enough active Aurora-A at centrosomes for mitotic entry 21649561_PUM2 does not repress differentiation of adipose-derived stem cells (ASCs) but rather is involved in the positive control of ASCs division and proliferation. 21800163_NANOS1-PUMILIO2 complex, together with GEMIN3 and small noncoding RNAs, possibly regulate mRNA translation within the chromatoid body of the human germ cells. 22231398_Mammalian PUM2-Ago-eEF1A inhibited translation of nonadenylated and polyadenylated reporter mRNAs in vitro. 22345517_human Pumilio homologs Pum 1 and Pum 2 repress the translation of E2F3 by binding to the E2F3 3' untranslated region (UTR) and also enhance the activity of multiple E2F3 targeting microRNAs (miRNAs) 26517885_Pumilios (including PUM1 and PUM2) are RNA-binding proteins with Puf domains made up of 8 poorly conserved Puf repeats, 3 helix bundles arranged in rainbow architecture, where each repeat recognizes a single base of the RNA-binding sequence. [REVIEW] 26724866_Identification of NORAD-interacting proteins revealed that this lncRNA functions as a multivalent binding platform for PUM proteins, with the capacity to sequester a significant fraction of the total cellular pool of PUM1 and PUM2. Studies reveal unanticipated roles for a lncRNA and PUMILIO proteins in the maintenance of genomic stability. 27157388_A recent paper by the Mendell group identifies NORAD, a novel lncRNA that is regulated in response to DNA damage and plays a key role in maintaining genome integrity by modulating the activity the RNA binding proteins PUM2 and PUM1. 28138061_The study shows that pseudouridine and N(6)-methyladenosine weaken the binding of the human single-stranded RNA binding protein Pumilio 2 (hPUM2) to its consensus motif. 28232582_results reveal a novel regulatory pathway, underscoring a previously unknown and interconnected key role of PUM1/2 and FOXP1 in regulating normal hematopoietic stem/progenitor cell and leukemic cell growth. 29165587_Data indicate that RNAs-including mRNAs and non-coding RNAs-that are functionally regulated by Pumilio proteins, PUM1 and PUM2. 29386330_SAM68 is required for efficient recruitment of PUM2 to NORAD, regulation of Pum activity by NORAD, and proper chromosome segregation in mammalian cells. 30084199_PUM2 suppresses osteosarcoma progression via partly and competitively binding to STARD13 3'UTR with miRNAs 30269240_Using the novel PUM1 and PUM2 mRNA target SIAH1 as a model, this study shows mechanistic differences between PUM1 and PUM2 and between NANOS1, 2, and 3 paralogues in the regulation of SIAH1. 30333515_A transcriptome-wide approach was used to determine the binding profiles and inter-dependencies of Argonaute2 (AGO2), Pumilio (PUM1 and PUM2) sites on mRNA 3' untranslated regions (3'UTRs). 30642763_A PUM2-mediated layer of post-transcriptional regulation links altered Mff translation to mitochondrial dynamics and mitophagy, thereby mediating age-related mitochondrial dysfunctions. 30787206_PUM2 promote glioblastoma development via repressing BTG1 expression 30824182_circ_0075932 directly bound with the RNA-binding protein PUM2, which was reported to positively regulated AuroraA kinase, thus activating the NF-kappaB pathway. Moreover, either silencing PUM2, silencing AuroraA, or blockade of NF-kappaB activation, could abrogate the promoting effect of adipocyte-derived exosomal circ_0075932 on cell inflammation and apoptosis. 30909144_PUM2 could facilitate the stemness of breast cancer cells by competitively binding to NRP-1 3'UTR with miR-376a. 30914482_PUM2 protein cooperativity mediated by RNA structure has been reported. 31078383_We have used the RNA-MaP platform to directly measure equilibrium binding for thousands of designed RNAs and to construct a predictive model for RNA recognition by the human Pumilio proteins PUM1 and PUM2. 31595981_RNA-binding protein PUM2 regulates mesenchymal stem cell fate via repression of JAK2 and RUNX2 mRNAs. 31646551_LncRNA TUG1 aggravates the progression of cervical cancer by binding PUM2. 32316190_Characterization of RNP Networks of PUM1 and PUM2 Post-Transcriptional Regulators in TCam-2 Cells, a Human Male Germ Cell Model. 32437472_The results suggest that PUM1-2 affects the expression of pluripotency genes as well as the efficiency of the cardiac differentiation process. 32753408_Principles of mRNA control by human PUM proteins elucidated from multimodal experiments and integrative data analysis. 32945477_LncRNA TTNAS1 promotes endometrial cancer by sponging miR376a3p. 33397688_Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome. 33542625_CAFs-Derived Exosomal miRNA-130a Confers Cisplatin Resistance of NSCLC Cells Through PUM2-Dependent Packaging. 33576437_Downregulated expression levels of USP46 promote the resistance of ovarian cancer to cisplatin and are regulated by PUM2. | ENSMUSG00000020594 | Pum2 | 3741.77960 | 1.1922384 | 0.2536726948 | 0.09057204 | 7.885087e+00 | 4.984409e-03 | 1.172499e-01 | No | Yes | 5880.987969 | 1256.687645 | 4541.461414 | 949.090302 | |
| ENSG00000061273 | 51564 | HDAC7 | protein_coding | Q8WUI4 | FUNCTION: Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Involved in muscle maturation by repressing transcription of myocyte enhancer factors such as MEF2A, MEF2B and MEF2C. During muscle differentiation, it shuttles into the cytoplasm, allowing the expression of myocyte enhancer factors (By similarity). May be involved in Epstein-Barr virus (EBV) latency, possibly by repressing the viral BZLF1 gene. Positively regulates the transcriptional repressor activity of FOXP3 (PubMed:17360565). Serves as a corepressor of RARA, causing its deacetylation and inhibition of RARE DNA element binding (PubMed:28167758). In association with RARA, plays a role in the repression of microRNA-10a and thereby in the inflammatory response (PubMed:28167758). {ECO:0000250|UniProtKB:Q8C2B3, ECO:0000269|PubMed:12239305, ECO:0000269|PubMed:17360565, ECO:0000269|PubMed:28167758}. | 3D-structure;Alternative splicing;Chromatin regulator;Cytoplasm;Hydrolase;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc | Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene has sequence homology to members of the histone deacetylase family. This gene is orthologous to mouse HDAC7 gene whose protein promotes repression mediated via the transcriptional corepressor SMRT. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:51564; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; histone deacetylase complex [GO:0000118]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 14-3-3 protein binding [GO:0071889]; chromatin binding [GO:0003682]; DNA-binding transcription factor binding [GO:0140297]; metal ion binding [GO:0046872]; NAD-dependent histone deacetylase activity (H3-K14 specific) [GO:0032041]; protein deacetylase activity [GO:0033558]; protein kinase binding [GO:0019901]; protein kinase C binding [GO:0005080]; SUMO transferase activity [GO:0019789]; transcription corepressor activity [GO:0003714]; cell-cell junction assembly [GO:0007043]; chromatin organization [GO:0006325]; negative regulation of interleukin-2 production [GO:0032703]; negative regulation of NIK/NF-kappaB signaling [GO:1901223]; negative regulation of osteoblast differentiation [GO:0045668]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of cell migration involved in sprouting angiogenesis [GO:0090050]; protein deacetylation [GO:0006476]; vasculogenesis [GO:0001570] | 10640276_Characterization of the mouse HDAC7 ortholog. 11929873_Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor 12753745_Interaction of HDAC7 with MEF2D is essential for repression of Nur77. 15166223_HDAC7 phosphorylation is mediated by calcium/calmodulin-dependent kinase I, which also promotes the association of HDAC7 with 14-3-3 and stabilizes HDAC7 15280364_HDAC7 increased transcriptional activity of HIF-1alpha through the formation of a complex with HIF-1alpha, HDAC7, and p300 15364908_HDAC7 is sequestered to the cytoplasm from mitochondrial and nuclear compartments upon initiation of apoptosis 15623513_Data indicate that protein kinase D1 regulates the expression of Nur77 during thymocyte activation at least in part by phosphorylating HDAC7. 15738054_a mutant of HDAC7 specifically deficient in phosphorylation by protein kinase D, inhibits T cell receptor-mediated apoptosis of T cell hybridomas 16860317_These results identify HDAC7 as a novel Androgen receptor corepressor whose subcellular and subnuclear compartmentalization can be regulated in an androgen-selective manner. 16980613_Class IIa histone deacetylases (HDACs) are subjected to signal-independent nuclear export that relies on their constitutive phosphorylation. EMK and C-TAK1, are identified as regulators of this process. 17947801_HDAC7 is a key modulator of endothelial cell migration and angiogenesis, at least in part, by regulating platelet-derived growth factor-B (PDGF-B) and its receptor PDGFR-beta gene expression. 17997710_Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner 18285338_HDAC7 has a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity 18339811_PP2A constitutively dephosphorylates the class IIa member HDAC7 to control its biological functions as a regulator of T cell apoptosis and endothelial cell functions. 18458084_Caspase-8 cleaves histone deacetylase 7 and abolishes its transcription repressor function. 18463162_PML sequesters HDAC7 to relieve repression and up-regulate gene expression 18506539_The data showed alteration of HDACs gene expression in pancreatic cancer. Increased expression of HDAC7 discriminates PA from other pancreatic tumors. 18509061_These results demonstrate that phosphorylation of HDAC7 serves as a molecular switch to mediate VEGF signaling and endothelial function. 18617643_VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating MT-MMP1/MMP10 expression and angiogenesis. 18625722_These results demonstrate a novel function of HDAC7 and provide a regulatory mechanism of PML sumoylation. 19240061_Observational study of gene-disease association. (HuGE Navigator) 19355988_recent developments in the crystal structure analysis of human HDAC4, HDAC7, and HDAC8 recent developments in the crystal structure analysis of human HDAC4, HDAC7, and HDAC8 [REVIEW] 19784544_Elevated HDAC7 expression in human osteoarthritis may contribute to cartilage degradation via promoting MMP-13 gene expression. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19917725_these data implicate a novel role for HDAC7 and FoxA1 in estrogen repression of RPRM. 19966789_histone deacetylase 7 has a role in function of misfolded CFTR in cystic fibrosis 20224040_HDAC7 interacts with beta-catenin keeping endothelial cells in a low proliferation stage. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20693714_The expression of HDAC7 protein plays an important role in the apoptosis and vascular tubulogenesis of hepatocellular carcinoma by the upregulation of p21 and HIF-1alpha and the downregulation of cyclin E and MMP10. 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21120446_Findings highlight for the first time an unrecognized link between HDAC7 and c-Myc and offer a novel mechanistic insight into the contribution of HDAC7 to tumor progression. 21324898_Data demonstrate that Mitf and HDAC7 interact in RAW 264 cells and osteoclasts. The transcriptional activity of Mitf is repressed by HDAC7. 22172637_HDAC7 reduction in COPD causes a defect of HIF-1alpha induction response to hypoxia with impaired VEGF gene expression. This poor cellular adaptation might play a role in the pathogenesis of COPD. 22584896_demonstrated for the first time that AKAP12 tumor/angiogenesis suppressor gene is an epigenetic target of HDAC7 23401860_VEGF and PKC promote degradation-independent protein ubiquitination of FLNB to control intracellular trafficking of HDAC7. 23696748_Our findings uncover a novel role for HDAC7 in maintaining the identity of a particular cell type by silencing lineage-inappropriate genes. 23853092_Histone deacetylase 7 promotes Toll-like receptor 4-dependent proinflammatory gene expression in macrophages. 23880157_Authors identified acetyltransferase p300 and deacetylase HDAC7 as enzymes modulating human T cell leukemia virus type 1 Tax protein acetylation. 23891737_Expression of JHDM2A was significantly increased but HDAC2, HDAC7, and SUV39H2 were significantly down-regulated in Systemic Sclerosis B cells relative to controls 24189120_endothelial progenitor cells involved in the angiogenesis might be controlled by VEGF-PKD1-HDAC7 axis, which regulates the EPCs angiogenesis by PKD1, but not the ERK and PI3K pathway 24239178_The transcriptional function of HCS was shown by in vitro pull down and in vivo co-immunoprecipitation assays to depend on its interaction with the histone deacetylases HDAC1, HDAC2 and HDAC7 25173798_Study identifies the miR-34a-HDAC1/HDAC7-HSP70 K246 axis as a novel molecular signature predictive of therapy resistance. 25511694_identify a new target of ROCK signaling via myosin phosphatase subunit (MYPT1) and histone deacetylase (HDAC7) at the nuclear level 26272600_This study demonstrated a simple and straightforward method of quantifying proneural/mesenchymal markers in glioblastoma. Of note, HDAC7 expression might be a novel therapeutic target in glioblastoma treatment. 26853466_silencing HDAC7 can reset the tumor suppressor activity of STAT3, independently of the EGFR/PTEN/TP53 background of the glioblastoma. 27796421_Study found increased HDAC7 expression in human pancreatic islets from type 2 diabetic compared with non-diabetic donors. HDAC7 expression correlated negatively with insulin secretion in human islets. 28178760_silencing induces apoptosis and autophagy in salivary mucoepidermoid carcinoma cells 28299580_High HDAC7 expression is associated with distant metastasis in gastric cancer. 28538176_Nur77 suppresses CD4(+) T cell proliferation and uncover a suppressive role for Irf4 in TH2 polarization; halving Irf4 gene-dosage leads to increases in GATA3(+) and IL-4(+) cells. 29071516_High HDAC7 expression is associated with recurrence and metastasis in colorectal cancer. 29126425_Study suggests that Hdac7 promotes lung tumorigenesis by inhibiting Stat3 activation via deacetylating Stat3. Also, high HDAC7 mRNA level was found to be correlated with poor prognosis of human lung cancer patients. 29490434_Our work demonstrates the molecular mechanism by which HDAC7 contributes to the angiogenic property of EPCs and provides a rational basis for specific targeting of antiangiogenic strategies in lung cancer. 29664401_The authors report that Histone Deacetylase 7 (HDAC7) controls the thymic effector programming of Natural Killer T (NKT) cells, and that interference with this function contributes to tissue-specific autoimmunity. 30628670_Histone deacetylase 7 (HDAC7) expression is increased in lung cancer samples and higher HDAC7 levels predict a poor outcome. HDAC7 promotes tumor growth and metastasis. Ectopic expression of HDAC7 suppresses mRNA and protein levels of plakoglobin in lung cancer cells, whereas silencing HDAC7 increases the the expression of plakoglobin. HDAC7 directly bind to the promoter region of plakoglobin in lung cancer. 30691485_High HDAC7 expression is associated with glioma. 30854734_miR-193b-5p regulates chondrocytes metabolism by directly targeting histone deacetylase 7 in interleukin-1beta-induced osteoarthritis. 31081251_HDAC7 levels are increased in RAS-transformed cells, in which this protein was required not only for proliferation and cancer stem-like cell growth, but also for invasive features. 31375747_results suggest that HDAC7 inactivation, directly or through inhibition of HDAC1 and HDAC3, can result in the inhibition of the cancer stem cell phenotype by downregulating multiple super enhancer-associated oncogenes 31527052_these results provide novel mechanistic insight into the essential role HDACs play in TGFbeta-mediated fibroblast activation via targeted gene repression. 31930344_Transcription Factor ZNF326 Upregulates the Expression of ERCC1 and HDAC7 and its Clinicopathologic Significance in Glioma. 32152265_miR-143 promotes angiogenesis and osteoblast differentiation by targeting HDAC7. 32284070_MiR-489 inhibited the development of gastric cancer via regulating HDAC7 and PI3K/AKT pathway. 32376822_HDAC7 promotes the oncogenicity of nasopharyngeal carcinoma cells by miR-4465-EphA2 signaling axis. 32709923_HDAC7 is an actionable driver of therapeutic antibody resistance by macrophages from CLL patients. 33262526_HDAC7 is a major contributor in the pathogenesis of infant t(4;11) proB acute lymphoblastic leukemia. 33734898_Histone Deacetylase 7 Gene Overexpression Is Associated with Poor Prognosis of Triple-Negative Breast Cancer Patients. 33878891_A regulative epigenetic circuit supervised by HDAC7 represses IGFBP6 and IGFBP7 expression to sustain mammary stemness. 34011384_Histone deacetylase 7 mediates endothelin-1-induced connective tissue growth factor expression in human lung fibroblasts through p300 and activator protein-1 activation. | ENSMUSG00000022475 | Hdac7 | 379.02899 | 1.1592877 | 0.2132386398 | 0.16502518 | 1.675760e+00 | 1.954889e-01 | 5.874899e-01 | No | Yes | 470.437286 | 63.124495 | 392.880835 | 51.498709 | |
| ENSG00000064225 | 10402 | ST3GAL6 | protein_coding | Q9Y274 | FUNCTION: Involved in the synthesis of sialyl-paragloboside, a precursor of sialyl-Lewis X determinant. Has a alpha-2,3-sialyltransferase activity toward Gal-beta1,4-GlcNAc structure on glycoproteins and glycolipids. Has a restricted substrate specificity, it utilizes Gal-beta1,4-GlcNAc on glycoproteins, and neolactotetraosylceramide and neolactohexaosylceramide, but not lactotetraosylceramide, lactosylceramide or asialo-GM1. | Alternative splicing;Glycoprotein;Glycosyltransferase;Golgi apparatus;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the sialyltransferase family. Members of this family are enzymes that transfer sialic acid from the activated cytidine 5'-monophospho-N-acetylneuraminic acid to terminal positions on sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins. This protein has high specificity for neolactotetraosylceramide and neolactohexaosylceramide as glycolipid substrates and may contribute to the formation of selectin ligands and sialyl Lewis X, a carbohydrate important for cell-to-cell recognition and a blood group antigen. [provided by RefSeq, Apr 2016]. | hsa:10402; | extracellular exosome [GO:0070062]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; beta-galactoside (CMP) alpha-2,3-sialyltransferase activity [GO:0003836]; beta-galactoside alpha-2,3-sialyltransferase activity [GO:0052798]; sialyltransferase activity [GO:0008373]; cellular protein modification process [GO:0006464]; cellular response to interleukin-6 [GO:0071354]; glycolipid metabolic process [GO:0006664]; keratan sulfate biosynthetic process [GO:0018146]; oligosaccharide biosynthetic process [GO:0009312]; oligosaccharide metabolic process [GO:0009311]; protein glycosylation [GO:0006486] | 18485915_Epigenetic changes in a group of glycosyltransferases including B4GALNT2 and ST3GAL6 represent a malignant phenotype of gastric cancer caused by silencing of the activity of these enzymes 19781661_Expression of ST3Gal IV in several gastrointestinal cell lines is correlated with the expression of sialyl Lewis x at the cell surface. 20532202_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21147760_the occurrence of CD75s- and iso-CD75s-gangliosides in tumor tissues is largely independent of the transcriptional expression of ST6GAL1 and ST3GAL6 25061176_The sialyltransferase ST3GAL6 influences homing and survival in multiple myeloma. 31914669_a cross-restoration of each of the three genes in ST3GAL6 KO cells showed that overexpression of ST3GAL6 sufficiently rescued the total alpha2,3-sialylation levels, cell morphology, and alpha2,3-sialylation of EGFR, whereas the alpha2,3-sialylation levels of beta1 were greatly enhanced by an overexpression of ST3GAL4 33649796_lncRNA ST3GAL6AS1 promotes invasion by inhibiting hnRNPA2B1mediated ST3GAL6 expression in multiple myeloma. | ENSMUSG00000022747 | St3gal6 | 337.02976 | 0.9987004 | -0.0018760972 | 0.17924945 | 1.096996e-04 | 9.916433e-01 | 9.979118e-01 | No | Yes | 441.860441 | 88.299074 | 440.299019 | 86.213122 | |
| ENSG00000064489 | BORCS8-MEF2B | protein_coding | Q96FH0 | FUNCTION: As part of the BORC complex may play a role in lysosomes movement and localization at the cell periphery. Associated with the cytosolic face of lysosomes, the BORC complex may recruit ARL8B and couple lysosomes to microtubule plus-end-directed kinesin motor. {ECO:0000305|PubMed:25898167}. | Alternative splicing;Lysosome;Membrane;Phosphoprotein;Reference proteome | hsa:729991; | BORC complex [GO:0099078]; lysosomal membrane [GO:0005765]; heart development [GO:0007507] | ENSMUSG00000079033 | Mef2b | 103.33925 | 0.9025730 | -0.1478844221 | 0.35504535 | 1.715634e-01 | 6.787261e-01 | 9.008142e-01 | No | Yes | 108.986939 | 46.209836 | 113.962298 | 47.189143 | ||||
| ENSG00000065150 | 3843 | IPO5 | protein_coding | O00410 | FUNCTION: Functions in nuclear protein import as nuclear transport receptor. Serves as receptor for nuclear localization signals (NLS) in cargo substrates. Is thought to mediate docking of the importin/substrate complex to the nuclear pore complex (NPC) through binding to nucleoporin and the complex is subsequently translocated through the pore by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to the importin, the importin/substrate complex dissociates and importin is re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus (By similarity). Mediates the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5. Binds to a beta-like import receptor binding (BIB) domain of RPL23A. In vitro, mediates nuclear import of H2A, H2B, H3 and H4 histones. Binds to CPEB3 and mediates its nuclear import following neuronal stimulation (By similarity). In case of HIV-1 infection, binds and mediates the nuclear import of HIV-1 Rev. {ECO:0000250|UniProtKB:Q8BKC5, ECO:0000269|PubMed:9687515}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Host-virus interaction;Nucleus;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport | Nucleocytoplasmic transport, a signal- and energy-dependent process, takes place through nuclear pore complexes embedded in the nuclear envelope. The import of proteins containing a nuclear localization signal (NLS) requires the NLS import receptor, a heterodimer of importin alpha and beta subunits also known as karyopherins. Importin alpha binds the NLS-containing cargo in the cytoplasm and importin beta docks the complex at the cytoplasmic side of the nuclear pore complex. In the presence of nucleoside triphosphates and the small GTP binding protein Ran, the complex moves into the nuclear pore complex and the importin subunits dissociate. Importin alpha enters the nucleoplasm with its passenger protein and importin beta remains at the pore. Interactions between importin beta and the FG repeats of nucleoporins are essential in translocation through the pore complex. The protein encoded by this gene is a member of the importin beta family. [provided by RefSeq, Jul 2008]. | hsa:3843; | cytoplasm [GO:0005737]; membrane [GO:0016020]; nuclear pore [GO:0005643]; nucleolus [GO:0005730]; nucleus [GO:0005634]; GTPase inhibitor activity [GO:0005095]; nuclear import signal receptor activity [GO:0061608]; nuclear localization sequence binding [GO:0008139]; RNA binding [GO:0003723]; small GTPase binding [GO:0031267]; cellular response to amino acid stimulus [GO:0071230]; negative regulation of cyclin-dependent protein serine/threonine kinase activity [GO:0045736]; NLS-bearing protein import into nucleus [GO:0006607]; positive regulation of protein import into nucleus [GO:0042307]; protein import into nucleus [GO:0006606]; ribosomal protein import into nucleus [GO:0006610] | 12620808_L1 major capsid protein of human papillomavirus type 11 interacts with Kap beta3 nuclear import receptors 15364420_The KPNB3 locus may contain a disease-causing variant for schizophrenia. 15507604_HPV16 L2 interacts via its NLSs with a network of karyopherins and can enter the nucleus via several import pathways mediated by Kapalpha(2)beta(1) heterodimers, Kapbeta(2), and Kapbeta(3). 16644122_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16644122_The present work suggests that the combination of the KPNA3 gene and the KPNB3 gene may increase a genetic risk for schizophrenia. 17005651_RanBP5 acts as an import factor for the newly synthesized influenza A virus RNA-dependent-RNA polymerase by targeting the PB1-PA dimer to the nucleus. 17458142_Observational study of gene-disease association. (HuGE Navigator) 18295457_Lack of association of the KPNB3 locus in schizophrenia. 18455505_HPV-16 E5 protein binds to karyopherin beta3. 18562802_These results suggest that karyopherin beta3 plays a crucial role in apo A-I secretion. 19581934_Impaired p53 binding to importin: a novel mechanism of cytoplasmic sequestration identified in oxaliplatin-resistant cells. 20542336_Observational study of gene-disease association. (HuGE Navigator) 20542336_The results of this study suggested that abnormal expression and alternative splicing of the IPO5 gene may be involved in the pathophysiology of schizophrenia. 20828572_importin beta3 is essential for the nuclear import of RPL7. The import is mediated via the multifaceted basic amino acid clusters present in the NH(2)-region of RPL7, and is RanGTP-dependent 21562121_The N-terminal of influenza A virus PB1 mediates its binding to host RanBP5. 23266416_In case of L7, importin beta2 or importin beta3 are preferentially used by clusters with a high import efficiency. 24196961_IQGAP1 interacts with human importin-beta5 in HEK 293T cells. 26488411_Single nucleotide polymorphism in IPO5 gene is associated with Peripheral Arterial Disease. 27094387_importin-alpha5, which is a key regulator of interferon signaling following Ebola virus infection, as one putative target of miRNA. 27528606_Importins, Impbeta, Kapbeta2, Imp4, Imp5, Imp7, Imp9, and Impalpha, show the H3 tail binding more tightly than the H4 tail. The H3 tail binds Kapbeta2 and Imp5 with KD values of 77 and 57 nm, respectively, and binds the other five Importins more weakly. 27703004_The nuclear importin IPO5 was identified as a novel interacting protein of SMAD1. Overexpression of IPO5 in various cell lines specifically increases nuclear localization of BMP receptor-activated SMADs (R-SMADs) confirming a functional relationship between IPO5 and BMP but not TGF-beta R-SMADs. 29127199_Results indicate that the interaction between FLIL33 and IPO5 is localized to a specific segment of the FLIL33 protein, is not required for nuclear localization of FLIL33, and protects FLIL33 from proteasome-dependent degradation. 31288861_IPO5 is an oncogene involved in CRC cell proliferation and migration. This highlights the significance of IPO5 in 5-fluorouracil-resistant CRC cells. The oncogenic function of IPO5 was mediated by promoting RAS signalling by increasing the nuclear translocation of RASAL2 31340999_DDX56 inhibits type I interferon by disrupting assembly of IRF3-IPO5 to inhibit IRF3 nucleus import. 32222384_X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking. 32373960_IPO5 promotes malignant progression of esophageal cancer through activating MMP7. | ENSMUSG00000030662 | Ipo5 | 16347.09843 | 1.0610010 | 0.0854259783 | 0.06875493 | 1.547565e+00 | 2.134953e-01 | 6.062746e-01 | No | Yes | 21576.394708 | 4288.605938 | 18913.498396 | 3675.271007 | |
| ENSG00000065328 | 55388 | MCM10 | protein_coding | Q7L590 | FUNCTION: Acts as a replication initiation factor that brings together the MCM2-7 helicase and the DNA polymerase alpha/primase complex in order to initiate DNA replication. Additionally, plays a role in preventing DNA damage during replication. Key effector of the RBBP6 and ZBTB38-mediated regulation of DNA-replication and common fragile sites stability; acts as a direct target of transcriptional repression by ZBTB38 (PubMed:24726359). {ECO:0000269|PubMed:11095689, ECO:0000269|PubMed:15136575, ECO:0000269|PubMed:17699597, ECO:0000269|PubMed:19608746, ECO:0000269|PubMed:24726359}. | Alternative splicing;Coiled coil;DNA damage;DNA replication;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is one of the highly conserved mini-chromosome maintenance proteins (MCM) that are involved in the initiation of eukaryotic genome replication. The hexameric protein complex formed by MCM proteins is a key component of the pre-replication complex (pre-RC) and it may be involved in the formation of replication forks and in the recruitment of other DNA replication related proteins. This protein can interact with MCM2 and MCM6, as well as with the origin recognition protein ORC2. It is regulated by proteolysis and phosphorylation in a cell cycle-dependent manner. Studies of a similar protein in Xenopus suggest that the chromatin binding of this protein at the onset of DNA replication is after pre-RC assembly and before origin unwinding. Alternatively spliced transcript variants encoding distinct isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:55388; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; replication fork protection complex [GO:0031298]; DNA replication origin binding [GO:0003688]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; single-stranded DNA binding [GO:0003697]; cell population proliferation [GO:0008283]; cellular response to DNA damage stimulus [GO:0006974]; DNA replication initiation [GO:0006270] | 11864598_Functional studies of the Xenopus homolog 12808023_Functional studies of the Drosophila homolog 15136575_human Mcm10 is temporarily recruited to the replication sites 30-60 min before they replicate and dissociates from chromatin after the activation of the prereplication complex 15195143_transcription of human MCM10 and TopBP1 is activated by transcription factors E2F1-3, but not by factors E2F4-7 16385451_Observational study of gene-disease association. (HuGE Navigator) 17699597_These results argue that cells can tolerate low levels of p180 as long as Mcm10 is present to 'recycle' it. 17761813_Mcm10 is required for chromatin loading of And-1. 17823614_Results show that MCM10 molecule is a ring-shaped hexamer with large central and smaller lateral channels and a system of inner chambers. 17975119_Observational study of gene-disease association. (HuGE Navigator) 17997977_MCM10 is essential for the efficient elongation step of chromosome replication. 17997981_These results indicate that MCM10 protein is essential for maintaining genome integrity as well as cell cycle progression. 19023099_Observational study of gene-disease association. (HuGE Navigator) 19608746_DNA and p180 binding to an Mcm10 construct that contains both the ID and CTD, provide the first mechanistic insight into how Mcm10 might use a handoff mechanism to load and stabilize pol alpha within the replication fork. 19696745_MCM10 interacts directly with RECQ4 and regulates its DNA unwinding activity. 19795399_Observational study of gene-disease association. (HuGE Navigator) 19805216_Assembly of the Cdc45-Mcm2-7-GINS complex requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. 20064936_High doses of ionizing gamma radiation and exposure to a combination of DNA-damaging chemicals do not decrease Mcm10 protein levels, demonstrating that Mcm10 down-regulation is triggered only by UV-specific damage 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22918587_Mcm10 utilizes a modular architecture to act as a replisome scaffold, which helps to define possible roles in origin DNA melting, Pol alpha recruitment and coordination of enzymatic activities during elongation. 23449222_This report shows that human Mcm10 is an acetylated protein regulated by SIRT1, which binds and deacetylates Mcm10 both in vivo and in vitro, and modulates Mcm10 stability and ability to bind DNA. 23750504_Data suggest that CDC45 and MCM10 (minichromosome maintenance complex component 10) directly interact and establish a mutual co-operation in DNA binding; key domains appear to interact and then interact with DNA inside cells or in cell-free systems. 24662891_Loss of Mcm10 engages checkpoint, DNA repair and SUMO-dependent rescue pathways that collectively counteract replication stress and chromosome breakage. [Review] 25602958_RecQL4-dependent association of Mcm10 and Ctf4 with replication origins appears to be the first important step controlled by S phase promoting kinases and checkpoint pathways for the initiation of DNA replication in human cells. 26032416_MCM10 is the natural substrate of the Cul4-DDB1[VprBP] E3 ubiquitin ligase whose degradation is regulated by VprBP, but Vpr enhances the proteasomal degradation of MCM10 by interacting with VprBP. 27780919_Results show that MCM10 is significantly upregulated in urothelial carcinoma (UC), and associated with tumor aggressiveness. Its knockdown significantly suppressed cell proliferation in UC cell lines. 28646110_Data suggest that interaction of Mcm10 with Mcm2-7 multimer requires Mcm10 domain that contains amino acids 530-655, which overlaps with domain required for stable retention of Mcm10 on chromatin; Mcm10 conserved domain (amino acids 200-482) is essential for DNA replication; both conserved domain and Mcm2-7-binding domain are required for full activity of Mcm10. 30095171_MCM10 was significantly upregulated in prostate cancer (PCa). We found increased MCM10 expression was significantly associated with advanced clinical stage and high Gleason score PCa. higher MCM10 expression was associated with a poorer patient prognosis in PCa. Furthermore, loss of function assays showed that MCM10 knockdown inhibited cell proliferation and colony formation, but promoted cell apoptosis. 30990947_decreased expression of beta-catenin and cyclin Dl was detected in MCM10 short hairpin RNA cells, implying that MCM10 might induce breast cancer metastasis via the Wnt/beta-catenin pathway.MCM10 can be defined as a potential diagnostic tool and a promising target for breast carcinoma. 31545501_High MCM10 expression is associated with lung adenocarcinoma. 31638210_MCM10 is highly expressed in lung cancer (LC) clinical specimens and significantly associated with recurrence, pathological stage and worse overall survival. MCM10 knockdown in A549 and H661 cell lines significantly suppressed cell viability, clone formation and induced G1 phase arrest by regulating the expression of CCND1. Results indicated a combined effect of MCM10CCND1 in predicting the prognosis of LC patients. 32030558_Knockdown of MCM10 Gene Impairs Glioblastoma Cell Proliferation, Migration and Invasion and the Implications for the Regulation of Tumorigenesis. 33340428_MCM10 compensates for Myc-induced DNA replication stress in breast cancer stem-like cells. 33712616_Bi-allelic MCM10 variants associated with immune dysfunction and cardiomyopathy cause telomere shortening. 34185429_Aberrant MCM10 SUMOylation induces genomic instability mediated by a genetic variant associated with survival of esophageal squamous cell carcinoma. 34350781_Prognostic significance and function of MCM10 in human hepatocellular carcinoma. 34645815_BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage. | ENSMUSG00000026669 | Mcm10 | 3569.57757 | 1.0307623 | 0.0437116368 | 0.09089583 | 2.315060e-01 | 6.304094e-01 | 8.816466e-01 | No | Yes | 4602.262104 | 823.252242 | 4145.747558 | 725.235098 | |
| ENSG00000065357 | 1606 | DGKA | protein_coding | P23743 | FUNCTION: Diacylglycerol kinase that converts diacylglycerol/DAG into phosphatidic acid/phosphatidate/PA and regulates the respective levels of these two bioactive lipids (PubMed:2175712, PubMed:15544348). Thereby, acts as a central switch between the signaling pathways activated by these second messengers with different cellular targets and opposite effects in numerous biological processes (PubMed:2175712, PubMed:15544348). Also plays an important role in the biosynthesis of complex lipids (Probable). Can also phosphorylate 1-alkyl-2-acylglycerol in vitro as efficiently as diacylglycerol provided it contains an arachidonoyl group (PubMed:15544348). Also involved in the production of alkyl-lysophosphatidic acid, another bioactive lipid, through the phosphorylation of 1-alkyl-2-acetyl glycerol (PubMed:22627129). {ECO:0000269|PubMed:15544348, ECO:0000269|PubMed:2175712, ECO:0000269|PubMed:22627129, ECO:0000305}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Calcium;Cytoplasm;Kinase;Lipid metabolism;Metal-binding;Nucleotide-binding;Reference proteome;Repeat;Transferase;Zinc;Zinc-finger | PATHWAY: Lipid metabolism; glycerolipid metabolism. {ECO:0000305|PubMed:15544348, ECO:0000305|PubMed:2175712, ECO:0000305|PubMed:22627129}. | The protein encoded by this gene belongs to the eukaryotic diacylglycerol kinase family. It acts as a modulator that competes with protein kinase C for the second messenger diacylglycerol in intracellular signaling pathways. It also plays an important role in the resynthesis of phosphatidylinositols and phosphorylating diacylglycerol to phosphatidic acid. Several transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Apr 2017]. | hsa:1606; | cytosol [GO:0005829]; membrane [GO:0016020]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; diacylglycerol kinase activity [GO:0004143]; kinase activity [GO:0016301]; lipid binding [GO:0008289]; NAD+ kinase activity [GO:0003951]; phospholipid binding [GO:0005543]; diacylglycerol metabolic process [GO:0046339]; glycerolipid metabolic process [GO:0046486]; intracellular signal transduction [GO:0035556]; lipid phosphorylation [GO:0046834]; phosphatidic acid biosynthetic process [GO:0006654]; platelet activation [GO:0030168]; protein kinase C-activating G protein-coupled receptor signaling pathway [GO:0007205] | 14734770_Defects in both polymorphonuclear neutrophil (PMN) transendothelial migration and PMN diacylglycerol kinase alpha signaling are implicated as disordered functions in subjects with localized aggressive periodontitis. 15117825_PPARgamma agonists upregulate DGKalpha production.This suppresses the diacylglycerol/protein-kinase-C signaling pathway. 15870081_DGKalpha is crucial for the control of cell activation and also for the regulation of the secretion of lethal exosomes, which in turn controls cell death. 15928040_ALK-mediated alphaDGK activation is dependent on p60src tyrosine kinase, with which alphaDGK forms a complex; alphaDGK activation is involved in the control of ALK-mediated mitogenic properties. 17276726_These results strongly suggest that DGKalpha is a novel positive regulator of NF-kappaB, which suppresses TNF-alpha-induced melanoma cell apoptosis. 17911109_diacylglycerol kinase alpha-conserved domains have a role in membrane targeting in intact T cells 18004883_2,3-dioleoylglycerol binds to a site on the alpha and zeta isoforms of diacylglycerol kinase that is exposed as a consequence of the substrate binding to the active site. 18424699_Lck-dependent tyrosine phosphorylation of diacylglycerol kinase alpha regulates its membrane association in T cells.( 19751727_These results strongly suggest that DGKalpha positively regulates TNF-alpha-dependent NF-kappaB activation via the PKCzeta-mediated Ser311 phosphorylation of p65/RelA. 21252909_Diacylglycerol kinase alpha is a key regulator of the polarised secretion of exosomes. 21493725_findings further suggest that DGL-alpha and -beta may regulate neurite outgrowth by engaging temporally and spatially distinct molecular pathways 22048771_SAP-mediated inhibition of DGKalpha sustains diacylglycerol signaling, thereby regulating T cell activation 22271650_Antigen-specific CD8-positive T cells from DGKalpha-deficient transgenic mice show enhanced expansion and increased cytokine production after lymphocytic choriomeningitis virus infection, yet DGK-deficient memory CD8+ T cells exhibit impaired expansion. 22425622_DGKalpha is involved in hepatocellular carcinoma progression by activation of the MAPK pathway. 22573804_DGK-alpha was more highly expressed in CD8-tumor-infiltrating T cellscompared with that in CD8non-tumor kidney-infiltrating lymphocytes. 24158111_High diacylglycerol kinase alpha expression is associated with glioblastoma. 24887021_These data indicates the existence of a SDF-1alpha induced DGKalpha - atypical PKC - beta1 integrin signaling pathway, which is essential for matrix invasion of carcinoma cells. 25248744_DGKalpha generates phosphatidic acid to drive its own recruitment to tubular recycling endosomes via its interaction with MICAL-L1 25921290_Redundant and specialized roles for diacylglycerol kinases alpha and zeta in the control of T cell functions. 26420856_An abandoned compound that also inhibits serotonin receptors may have more translational potential as a DGKa inhibitor, but more potent and specific DGKa inhibitors are sorely needed 26964756_Decreased DNA methylation at this enhancer enables recruitment of the profibrotic transcription factor early growth response 1 (EGR1) and facilitates radiation-induced DGKA transcription in cells from patients later developing fibrosis. 27498782_LIPFDGKA might serve as a potential possible biomarkers for diagnosis of gastric cancer, and their downregulation may bring new perspective into the investigation of gastric cancer prognosis 27697466_Diacylglycerol kinases alpha and zeta are up-regulated in cancer in cancer, and contribute towards tumor immune evasion and T cells clonal anergy. (Review) 27731506_DGKalpha isoform is highly expressed in the nuclei of human erythroleukemia cell line K562, and its nuclear activity drives K562 cells through the G1/S transition during cell cycle progression. 29967261_This novel study demonstrates efficient ablation of diacylglycerol kinase in human CAR-T cells that leads to improved antitumor immunity and may have significant impact in human cancer immunotherapy 30653270_This study presents the first crystal structure of EF-hand domains of diacylglycerol kinase alpha in its Ca(2+) bound form and characterize Ca(2+) -induced conformational changes, which likely regulates intra-molecular interactions. 31766109_Upon neutrophil stimulation, DGK-alpha activation is necessary for migration and a productive response. This paper focuses on the role of DGK-alpha in obstructive respiratory diseases, including asthma and chronic obstructive pulmonary disease, but also rare genetic diseases such as alpha-1-antitrypsin deficiency. [review] 32341033_DGKA Provides Platinum Resistance in Ovarian Cancer Through Activation of c-JUN-WEE1 Signaling. 32345612_Diacylglycerol kinases regulate TRPV1 channel activity. 33608256_DGKA Mediates Resistance to PD-1 Blockade. 34293268_Diacylglycerol Kinase Inhibition Reduces Airway Contraction by Negative Feedback Regulation of Gq-Signaling. 35131384_DGKA interacts with SRC/FAK to promote the metastasis of non-small cell lung cancer. | ENSMUSG00000025357 | Dgka | 65.97525 | 0.7815604 | -0.3555707408 | 0.36187407 | 9.561604e-01 | 3.281562e-01 | No | Yes | 83.004188 | 14.576984 | 106.839655 | 17.776966 | |
| ENSG00000065717 | 7089 | TLE2 | protein_coding | Q04725 | FUNCTION: Transcriptional corepressor that binds to a number of transcription factors. Inhibits the transcriptional activation mediated by CTNNB1 and TCF family members in Wnt signaling. The effects of full-length TLE family members may be modulated by association with dominant-negative AES (By similarity). {ECO:0000250}. | Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation;WD repeat;Wnt signaling pathway | hsa:7089; | extracellular space [GO:0005615]; focal adhesion [GO:0005925]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; transcription corepressor activity [GO:0003714]; animal organ morphogenesis [GO:0009887]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of transcription, DNA-templated [GO:0045892]; signal transduction [GO:0007165]; Wnt signaling pathway [GO:0016055] | 19939918_Immunofluorescence analysis showed that TLE2 and RTA were colocalized in the same nuclear compartment in KSHV-infected cells. | ENSMUSG00000034771 | Tle2 | 104.33082 | 1.2162594 | 0.2824509721 | 0.28785592 | 9.677444e-01 | 3.252434e-01 | 7.041518e-01 | No | Yes | 143.786127 | 22.066372 | 114.216705 | 17.099220 | ||
| ENSG00000069493 | 29121 | CLEC2D | protein_coding | Q9UHP7 | FUNCTION: Receptor for KLRB1 that protects target cells against natural killer cell-mediated lysis (PubMed:20843815, PubMed:16339513). Inhibits osteoclast formation (PubMed:14753741, PubMed:15123656). Inhibits bone resorption (PubMed:14753741). Modulates the release of interferon-gamma (PubMed:15104121). Binds high molecular weight sulfated glycosaminoglycans (PubMed:15123656). {ECO:0000269|PubMed:14753741, ECO:0000269|PubMed:15104121, ECO:0000269|PubMed:15123656, ECO:0000269|PubMed:16339513, ECO:0000269|PubMed:20843815}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Lectin;Membrane;Receptor;Reference proteome;Signal-anchor;Transmembrane;Transmembrane helix | Mouse_homologues NA; + ;NA; + ;NA; + ;NA; + ;NA | This gene encodes a member of the natural killer cell receptor C-type lectin family. The encoded protein inhibits osteoclast formation and contains a transmembrane domain near the N-terminus as well as the C-type lectin-like extracellular domain. Several alternatively spliced transcript variants have been identified for this gene. [provided by RefSeq, Oct 2010]. | hsa:29121; | cell surface [GO:0009986]; endoplasmic reticulum [GO:0005783]; external side of plasma membrane [GO:0009897]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; plasma membrane [GO:0005886]; carbohydrate binding [GO:0030246]; natural killer cell lectin-like receptor binding [GO:0046703]; transmembrane signaling receptor activity [GO:0004888]; cell surface receptor signaling pathway [GO:0007166] | 15104121_LLT1 induces Interferon Type II production by natural killer cells. 15123656_Data show that osteoclast inhibitory lectin (OCIL) binds a range of physiologically important glycosaminoglycans, and this property may modulate OCIL actions upon other cells [OCIL]. 16339512_Engagement of CD161 on NK cells with LLT1 expressed on target cells inhibited NK cell-mediated cytotoxicity and IFN-gamma secretion. LLT1/CD161 interaction in the presence of a TCR signal enhanced IFN-gamma production by T cells 16339513_LLT1 on target cells can inhibit NK cytotoxicity via interactions with CD161. LLT1 activates NFAT-GFP reporter cells expressing a CD3zeta-CD161 chimeric receptor; reciprocally, reporter cells with a CD3zeta-LLT1 chimeric receptor are stimulated by CD161 18453569_Expression of LLT1 on activated dendritic cells and B cells suggests that it might regulate the cross-talk between NK cells and APCs 18465072_Observational study of gene-disease association. (HuGE Navigator) 18465072_Women with a lysine (GG genotype) at position 19 of the OCIL protein displayed lower bone mineral density at femoral neck and at lumbar spine sites than women having an asparagine residue. 18593762_Observational study of gene-disease association. (HuGE Navigator) 20415786_LLT1 used Src-PTK, p38 and ERK signalling pathways, but not PKC, PI3K or calcineurin pathways, to increase production of IFN-gamma by human natural killer cells. 20843815_Data show that only CLEC2D isoform 1 (LLT1) is expressed on the cell surface. 21572041_Molecular basis for LLT1 protein recognition by human CD161 protein (NKRP1A/KLRB1). 21930700_LLT1 and CD161 have roles in modulating immune responses to pathogens; and interferon-gamma contributes to modulate immune responses 22664939_One polymorphism in LLT1 was found to be associated with our Crohn's Disease population (P<0.034).Our Ulcerative Colitis cohort was not associated with the variation in LLT1 (P=0.33) 25760607_The hexamer of glycosylated LLT1 consists of three classical dimers. The hexameric packing may indicate a possible mode of interaction of C-type lectin-like proteins in the glycosylated form. 26147876_In RA joints, LLT1 is expressed by cells of the monocyte/macrophage lineage. 26829983_these data suggest that LLT1-CD161 interactions play a novel and important role in B cell maturation within the Germinal center in humans. 27626681_Blocking LLT1-NKRP1A interaction will make prostate cancer cells susceptible to killing by NK cells, suggesting a therapeutic option for treatment of prostate cancer. 29212911_These results show that susceptibility of normal articular chondrocytes to lysis by NK cells is modulated by NKR-P1A/LLT1 interactions. Thus, NKR-P1A/LLT1 interaction might provide some novel target for therapeutic interventions in the course of pathological cartilage injury. 30955082_LLT1 strong expression was a significant risk factor for nodal metastasis in patients with head and neck cutaneous squamous cell carcinoma (cSCC) and for cSCC specific mortality. Strong LLT1 expression is an independent predictor of nodal metastasis and poor disease-specific survival and it might be helpful for risk stratification of patients with cSCC. 31002602_REVIEW: Biological and Clinical Significance of Human NKRP1A/LLT1 Receptor/Ligand Interactions 31859049_The authors identify Clec2d as a sensor for cell death through histone recognition and show that such interaction in macrophages shuttles histone-DNA complexes into endosomes to stimulate toll-like receptors. The consequent inflammation amplifies collateral tissue damage in a liver injury model. 32449811_Docetaxel suppresses immunotherapy efficacy of natural killer cells toward castration-resistant prostate cancer cells via altering androgen receptor-lectin-like transcript 1 signals. 33288556_Role of LLT1 and PCNA as Natural Killer Cell Immune Evasion Strategies of HCT 116 Cells. 35185935_LLT1-CD161 Interaction in Cancer: Promises and Challenges. | ENSMUSG00000030157+ENSMUSG00000030155+ENSMUSG00000000248+ENSMUSG00000030365+ENSMUSG00000030364 | Clec2d+Clec2e+Clec2g+Clec2i+Clec2h | 22.01242 | 1.5729633 | 0.6534850231 | 0.65299402 | 1.002813e+00 | 3.166307e-01 | No | Yes | 33.800775 | 17.866664 | 22.807991 | 12.002420 | |
| ENSG00000071242 | 6196 | RPS6KA2 | protein_coding | Q15349 | FUNCTION: Serine/threonine-protein kinase that acts downstream of ERK (MAPK1/ERK2 and MAPK3/ERK1) signaling and mediates mitogenic and stress-induced activation of transcription factors, regulates translation, and mediates cellular proliferation, survival, and differentiation. May function as tumor suppressor in epithelial ovarian cancer cells. {ECO:0000269|PubMed:16878154, ECO:0000269|PubMed:7623830}. | ATP-binding;Alternative splicing;Cytoplasm;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase;Tumor suppressor | This gene encodes a member of the RSK (ribosomal S6 kinase) family of serine/threonine kinases. This kinase contains two non-identical kinase catalytic domains and phosphorylates various substrates, including members of the mitogen-activated kinase (MAPK) signalling pathway. The activity of this protein has been implicated in controlling cell growth and differentiation. Alternative splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jan 2016]. | hsa:6196; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; meiotic spindle [GO:0072687]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; ribosomal protein S6 kinase activity [GO:0004711]; brain renin-angiotensin system [GO:0002035]; cardiac muscle cell apoptotic process [GO:0010659]; cellular response to carbohydrate stimulus [GO:0071322]; chemical synaptic transmission [GO:0007268]; heart contraction [GO:0060047]; heart development [GO:0007507]; intracellular signal transduction [GO:0035556]; negative regulation of cell cycle [GO:0045786]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of meiotic nuclear division [GO:0045835]; oocyte maturation [GO:0001556]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of apoptotic process [GO:0043065]; positive regulation of gene expression [GO:0010628]; regulation of protein processing [GO:0070613]; signal transduction [GO:0007165] | 12016217_Characterization of the terminal domain as a protein kinase 14646589_chronic activation CREB and p90RSK in the epileptic hippocampus may be closely associated with the histopathological changes of Ammon's horn sclerosis 15112576_overexpressed in breast tumors 15995633_The accumulation of S6K2 in the nuclei of cancer cells and the correlation with the expression of PCNA and Ki-67 suggest the involvement of S6K2 in the regulation of malignant growth 16621805_p90Rsk-mediated modulation of Hdm2 nuclear is linked to cytoplasmic shuttling with the diminished ability of p53 to regulate cell cycle checkpoints that ultimately leads to transformation 16626623_The large C-terminal domain of ERK5 is not required for binding or activation of RSK by ERK5. 16878154_The above results suggest that RPS6KA2 is a putative tumour suppressor gene to explain allele loss at 6q27. 16895915_there is a functional link between S6K1 II and CK2 signaling, which involves the regulation of S6K1 II nuclear export by CK2-mediated phosphorylation of Ser-17 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21035469_Data show that genetic variation in RPS6KA1, RPS6KA2, and PRS6KB2 were associated with risk of developing colon cancer while only genetic variation in RPS6KA2 was associated with altering risk of rectal cancer. 21035469_Observational study of gene-disease association. (HuGE Navigator) 21527514_p90RSK2 is dispensable for BCR-ABL-induced myeloid leukemia, but may be required for pathogenesis and lineage determination in FLT3-internal tandem duplication-induced hematopoietic transformation. 23564320_Data indicate that S6 kinase 2 (S6K2) can phosphorylate histone H3 at position Thr45, which may play a role during cell proliferation and/or differentiation. 23635776_Overexpression of RSK3 or RSK4 supports tumor cell proliferation upon PI3K inhibition both in vitro and in vivo therby contributing to drug resistance. 23727904_genetic association study in Han population in China: Data suggest that SNPs in RSK3 (rs2229712) and in MEK1 (rs28730804) demonstrate gene-gene interaction that affects antidepressant drug outcome in female patients with major depressive disorder. 24403857_Kinome screening revealed RPS6KA2 expression, in human pancreatic cancer cells, protects against erlotinib induced apoptosis. 26977024_RSK1 and 3 but not RSK2 are down-regulated in breast tumour and are associated with disease progression. RSK may be a key component in the progression and metastasis of breast cancer. 29144123_Data suggest that millisecond dynamic changes in PDZ1 domain conformation are responsible for higher affinity of scribble PDZ1 for phosphorylated ligands; oligopeptide fragments of RPS6KA2 and MCC were used as ligands in these nuclear magnetic resonance chemical shift experiments. (RPS6KA2 = ribosomal protein S6 kinase 2; MCC = mutated in colorectal cancer protein) 31937753_Elevated ribosomal protein S6 kinase A2 (RSK3) expression is responsible for BET bromodomain inhibitors (BETi) resistance. Proto-Oncogene Proteins c-jun (JunD)-dependent RSK3 transcription mediates BETi resistance. JunD/RSK3 signalling correlates to BET inhibition sensitivity. Loss of BRD4/FOXD3/miR-548d-3p axis enhances JunD/RSK3 signalling and determines BET inhibition resistance. 32869517_Genome-wide association study of cafe-au-lait macule number in neurofibromatosis type 1. 34550011_Multiomics analysis identifies key genes and pathways related to N6-methyladenosine RNA modification in ovarian cancer. | ENSMUSG00000023809 | Rps6ka2 | 693.27001 | 0.9732817 | -0.0390707097 | 0.12377136 | 9.948097e-02 | 7.524534e-01 | 9.279231e-01 | No | Yes | 932.528022 | 75.579872 | 926.978583 | 73.314028 | |
| ENSG00000072518 | 2011 | MARK2 | protein_coding | Q7KZI7 | FUNCTION: Serine/threonine-protein kinase (PubMed:23666762). Involved in cell polarity and microtubule dynamics regulation. Phosphorylates CRTC2/TORC2, DCX, HDAC7, KIF13B, MAP2, MAP4 and RAB11FIP2. Phosphorylates the microtubule-associated protein MAPT/TAU (PubMed:23666762). Plays a key role in cell polarity by phosphorylating the microtubule-associated proteins MAP2, MAP4 and MAPT/TAU at KXGS motifs, causing detachment from microtubules, and their disassembly. Regulates epithelial cell polarity by phosphorylating RAB11FIP2. Involved in the regulation of neuronal migration through its dual activities in regulating cellular polarity and microtubule dynamics, possibly by phosphorylating and regulating DCX. Regulates axogenesis by phosphorylating KIF13B, promoting interaction between KIF13B and 14-3-3 and inhibiting microtubule-dependent accumulation of KIF13B. Also required for neurite outgrowth and establishment of neuronal polarity. Regulates localization and activity of some histone deacetylases by mediating phosphorylation of HDAC7, promoting subsequent interaction between HDAC7 and 14-3-3 and export from the nucleus. Also acts as a positive regulator of the Wnt signaling pathway, probably by mediating phosphorylation of dishevelled proteins (DVL1, DVL2 and/or DVL3). Modulates the developmental decision to build a columnar versus a hepatic epithelial cell apparently by promoting a switch from a direct to a transcytotic mode of apical protein delivery. Essential for the asymmetric development of membrane domains of polarized epithelial cells. {ECO:0000269|PubMed:11433294, ECO:0000269|PubMed:12429843, ECO:0000269|PubMed:14976552, ECO:0000269|PubMed:15158914, ECO:0000269|PubMed:15324659, ECO:0000269|PubMed:15365179, ECO:0000269|PubMed:16775013, ECO:0000269|PubMed:16980613, ECO:0000269|PubMed:18626018, ECO:0000269|PubMed:20194617, ECO:0000269|PubMed:23666762}. | 3D-structure;ATP-binding;Alternative promoter usage;Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Developmental protein;Differentiation;Kinase;Lipid-binding;Magnesium;Membrane;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Wnt signaling pathway | This gene encodes a member of the Par-1 family of serine/threonine protein kinases. The protein is an important regulator of cell polarity in epithelial and neuronal cells, and also controls the stability of microtubules through phosphorylation and inactivation of several microtubule-associating proteins. The protein localizes to cell membranes. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2009]. | hsa:2011; | actin filament [GO:0005884]; cytoplasm [GO:0005737]; dendrite [GO:0030425]; lateral plasma membrane [GO:0016328]; membrane [GO:0016020]; microtubule bundle [GO:0097427]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; cadherin binding [GO:0045296]; lipid binding [GO:0008289]; magnesium ion binding [GO:0000287]; protein kinase activator activity [GO:0030295]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA binding [GO:0003723]; tau protein binding [GO:0048156]; tau-protein kinase activity [GO:0050321]; activation of protein kinase activity [GO:0032147]; autophagy of mitochondrion [GO:0000422]; axon development [GO:0061564]; establishment of cell polarity [GO:0030010]; establishment or maintenance of cell polarity regulating cell shape [GO:0071963]; establishment or maintenance of epithelial cell apical/basal polarity [GO:0045197]; intracellular signal transduction [GO:0035556]; microtubule cytoskeleton organization [GO:0000226]; mitochondrion localization [GO:0051646]; neuron migration [GO:0001764]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of neuron projection development [GO:0010976]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of axonogenesis [GO:0050770]; regulation of cytoskeleton organization [GO:0051493]; regulation of microtubule binding [GO:1904526]; regulation of microtubule cytoskeleton organization [GO:0070507]; Wnt signaling pathway [GO:0016055] | 15158914_Par1/Emk1 could have a role in the development of chronic allograft nephropathy in kidney allografts 15492257_Flot-2 binds to PAR-1, a known upstream mediator of major signal transduction pathways implicated in cell growth and metastasis, and may thereby influence tumor progression in melanoma. 16257959_GSK-3beta directly phosphorylates and activates MARK2/PAR-1 16472737_The X-ray structure of the catalytic and ubiquitin-associated domains of human MARK2. 16803889_analysis of variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2 16980613_Class IIa histone deacetylases (HDACs) are subjected to signal-independent nuclear export that relies on their constitutive phosphorylation. EMK and C-TAK1, are identified as regulators of this process. 17234589_Aberrant activation of PAR-1 may provide one of the molecular links in the pathogenic cascade of tauopathies. 18005242_We demonstrate that H. pylori causes the recruitment of MARK2 from the cytosol to the plasma membrane, where it colocalizes with the bacteria and interacts with CagA.[CagA in strain G27] 18315603_PAR-1 can be used with Breslow thickness and ulceration as a prognostic indicator for melanoma. 18760999_These results suggest that membrane accumulation of Par1b induced by Dvl is regulated by its phosphorylation status, which is important for Par1b to regulate the microtubule dynamics. 19011111_findings show protein kinase D phosphorylates Par-1b on S400 to positively regulate 14-3-3 binding and to negatively regulate membrane association 19530321_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19553522_Par1b functions in the establishment of T cell polarity following engagement to an APC 19945424_the 8th and 9th spectrin-like repeats (R8 and R9) of utrophin cooperatively form a PAR-1b-interacting domain, and that Ser1258 within R9 is specifically phosphorylated by PAR-1b. 20194617_These results reveal that GAKIN/KIF13B is a key intermediate linking Par1b to the regulation of axon formation. 21513698_These data suggest that Par1b-phosphorylation regulates turnover of GEF-H1 localization by regulating its interaction with microtubules, which may contribute to cell polarization. 22072711_Polarity-regulating kinase partitioning-defective 1b (PAR1b) phosphorylates guanine nucleotide exchange factor H1 (GEF-H1) to regulate RhoA-dependent actin cytoskeletal reorganization. 22238344_The results identify MARK2 as an upstream regulator of PINK1 and DeltaN-PINK1 and provide insights into the regulation of mitochondrial trafficking in neurons and neurodegeneration in PD. 22848487_automated image analysis of MT assembly dynamics identified MARK2 as a target regulated downstream of Rac1 that promotes oriented MT growth in the leading edge to mediate directed cell migration. 22883624_The scaffolding adaptor GAB1 interacts with two polarity proteins, PAR1 and PAR3. 24165937_Hepatocyte Par1b defines lumen position in concert with the position of the astral microtubule anchoring complex LGN-NuMA to yield the distinct epithelial division phenotypes. 24251416_The MARK2 binds to the N-terminal tail of Tau and selectively phosphorylates three major and five minor serine residues in the repeat domain and C-terminal tail. 24259665_Phosphorylation of RNF41 by Par-1b regulates basolateral membrane targeting of laminin-111 receptors. 24354359_Perturbation of PAR1b and SHP2 by CagA underlies the oncogenic potential of CagA. 24358023_induces asymmetric inheritance of plasma membrane domains via LGN-dependent mitotic spindle orientation in proliferating hepatocytes 25907283_MARK2 plays a role in promoting malignant phenotypes of lung cancer. 27445265_In this study, through quantitative analysis of the complex formation between CagA and PAR1b, the authors found that several CagA species have acquired elevated PAR1b-binding activity via duplication of the CagA multimerization motifs, while others have lost their PAR1b-binding activity. 27714636_In cell-based assays, Mark2 depletion indeed reduces Dvl gene expression and interrupts neural stem cell (NSCs) growth and differentiation, which are likely to be mediated through a decrease in class IIa HDAC phosphorylation and reduced H3K4ac and H3K27ac occupancies at the Dvl1/2 promoters. 27878245_In conclusion, baicalin and DDP were synergistic at inhibiting proliferation and invasion of human lung cancer cells at appropriate dosages and incubation time in the presence or absence of DDP resistance. The attenuation of DDP resistance was associated with downregulation of MARK2 and p-Akt. 28560405_Low expression of Mark2 is associated with uterine cervical neoplasms. 28711359_In the modeled structure of inactive MARK2, activation segment occludes the enzyme active site and assumes a relatively stable position. 28930676_HIV-1 did not stimulate widespread FEZ1 phosphorylation but, instead, bound microtubule (MT) affinity-regulating kinase 2 (MARK2) to stimulate FEZ1 phosphorylation on viral cores. 29941476_The authors uncovered a novel role for MARK2 in maintaining the mitotic spindle at the cell's geometric center. 30580666_Par1b-inhibition by CagA contributes to DNA Double Strand Breaks in H. pylori infected human primary gastric epithelial cells. 31080060_Clustering of the CD44 extracellular domain by high-molecular-weight hyaluronan leads to recruitment of the polarity-regulating kinase PAR1b by the CD44 intracellular domain, which results in disruption of the Hippo signaling-inhibitory PAR1b-MST complex. 31238822_Study shows that MARK2 is required to recentre spindles that are off-centred following actin disassembly, showing the close functional relationship between MARK2 and the actin network. These results suggest that, during both interphase and mitosis, MARK2 localizes at specialized membrane subdomains and coordinates actin and microtubule cytoskeletal changes, thus enabling normal cell division. 33596087_The cell polarity kinase Par1b/MARK2 activation selects specific NF-kB transcripts via phosphorylation of core mediator Med17/TRAP80. 33705388_MARK2 phosphorylates eIF2alpha in response to proteotoxic stress. 34793775_Long non-coding RNA ABHD11-AS1 facilitates the progression of cervical cancer by competitively binding to miR-330-5p and upregulating MARK2. 35192892_MARK2/4 promotes Warburg effect and cell growth in non-small cell lung carcinoma through the AMPKalpha1/mTOR/HIF-1alpha signaling pathway. | ENSMUSG00000024969 | Mark2 | 1858.33534 | 1.0948729 | 0.1307634259 | 0.10329226 | 1.606045e+00 | 2.050487e-01 | 5.969607e-01 | No | Yes | 2615.734394 | 321.969481 | 2287.416616 | 275.287584 | |
| ENSG00000075234 | 55020 | TTC38 | protein_coding | Q5R3I4 | Acetylation;Phosphoprotein;Reference proteome;Repeat;TPR repeat | hsa:55020; | extracellular exosome [GO:0070062] | 28371410_we proposed that four newly identified peripheral blood mononuclear cells-derived genes( DHRS3, TTC38, SAP30BP and LPIN2 )could be integrated with previously reported rheumatoid arthritis (RA)-associated genes to monitor and/or diagnose RA. | ENSMUSG00000035944 | Ttc38 | 1097.33979 | 1.0296827 | 0.0421999020 | 0.10503965 | 1.616030e-01 | 6.876850e-01 | 9.043290e-01 | No | Yes | 1368.214409 | 160.613563 | 1284.350060 | 147.419918 | |||
| ENSG00000075240 | 23151 | GRAMD4 | protein_coding | Q6IC98 | FUNCTION: Plays a role as a mediator of E2F1-induced apoptosis in the absence of p53/TP53 (PubMed:15565177). Plays a role as a mediator of E2F1-induced apoptosis in the absence of p53/TP53. Inhibits TLR9 response to nucelic acids and regulates TLR9-mediated innate immune response (By similarity). {ECO:0000250|UniProtKB:Q8CB44, ECO:0000269|PubMed:15565177}. | Alternative splicing;Apoptosis;Coiled coil;Endoplasmic reticulum;Membrane;Mitochondrion;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | GRAMD4 is a mitochondrial effector of E2F1 (MIM 189971)-induced apoptosis (Stanelle et al., 2005 [PubMed 15565177]).[supplied by OMIM, Jan 2011]. | hsa:23151; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; mitochondrial membrane [GO:0031966]; mitochondrion [GO:0005739]; apoptotic process [GO:0006915]; negative regulation of toll-like receptor 9 signaling pathway [GO:0034164]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280] | 15565177_DIP induces p53-independent caspase-dependent and -independent apoptosis. DIP is localized in the mitochondria. DIP accumulates upon E2F1 activation. 15565177_localizes to the mitochondria, upregulated following E2F1 induction 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21127500_GRAMD4 induces changes in Bcl-2 and Bax protein levels 23388340_The expression of GRAMD4 is up-regulated in hepatocellular carcinoma cell(HCC cell) lines and HCC tissues, and the increased expression is correlated with the clinicopathological characteristics of HCC. | ENSMUSG00000035900 | Gramd4 | 763.66213 | 1.0116663 | 0.0167335405 | 0.15234222 | 1.203223e-02 | 9.126540e-01 | 9.760532e-01 | No | Yes | 875.580656 | 104.927696 | 859.625181 | 100.567996 | |
| ENSG00000075826 | 25956 | SEC31B | protein_coding | Q9NQW1 | FUNCTION: As a component of the coat protein complex II (COPII), may function in vesicle budding and cargo export from the endoplasmic reticulum. {ECO:0000269|PubMed:16495487}. | Alternative splicing;Cytoplasm;Cytoplasmic vesicle;ER-Golgi transport;Endoplasmic reticulum;Membrane;Protein transport;Reference proteome;Repeat;Transport;Ubl conjugation;WD repeat | This gene encodes a protein of unknown function. The protein has moderate similarity to rat VAP1 protein which is an endosomal membrane-associated protein, containing a putative Ca2+/calmodulin-dependent kinase II phosphorylation site. [provided by RefSeq, Jul 2008]. | hsa:25956; | COPII vesicle coat [GO:0030127]; endoplasmic reticulum exit site [GO:0070971]; endoplasmic reticulum membrane [GO:0005789]; vesicle coat [GO:0030120]; structural molecule activity [GO:0005198]; COPII-coated vesicle cargo loading [GO:0090110]; endoplasmic reticulum organization [GO:0007029]; intracellular protein transport [GO:0006886] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 16495487_The SEC31B protein is an orthologue of Saccharomyces cerevisiae Sec31p, a component of the COPII vesicle coat that mediates vesicular traffic from the endoplasmic reticulum. | ENSMUSG00000051984 | Sec31b | 345.79319 | 1.2399843 | 0.3103218971 | 0.16230552 | 3.673019e+00 | 5.529993e-02 | 3.561913e-01 | No | Yes | 346.289688 | 83.658532 | 294.255768 | 69.504436 | |
| ENSG00000076513 | 88455 | ANKRD13A | protein_coding | Q8IZ07 | FUNCTION: Ubiquitin-binding protein that specifically recognizes and binds 'Lys-63'-linked ubiquitin. Does not bind 'Lys-48'-linked ubiquitin. Positively regulates the internalization of ligand-activated EGFR by binding to the Ub moiety of ubiquitinated EGFR at the cell membrane. {ECO:0000269|PubMed:22298428}. | ANK repeat;Cell membrane;Endosome;Membrane;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation | hsa:88455; | cytoplasm [GO:0005737]; late endosome [GO:0005770]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; ubiquitin-dependent protein binding [GO:0140036]; negative regulation of protein localization to endosome [GO:1905667]; negative regulation of receptor internalization [GO:0002091] | 22298428_overexpression of wild-type as well as truncated-mutant Ankrd 13A, 13B and 13D proteins strongly inhibited rapid endocytosis of ubiquitinated EGFR from the surface in EGF-treated cells 23620728_the microRNA miR-204 promotes both mesenchymal neural crest and lens cell migration and elongation by the direct targeting of the Ankrd13A gene. 26038114_We validated ANKRD13A as a novel constituent of BCR signalosome and showed that BCR-induced phosphorylation of RAB7A at S72 regulates its association with its effector proteins and with endo-lysosomal compartments. 26797118_The Ankrd13 family of ubiquitin-interacting motif (UIM)-containing proteins are novel VCP-interacting molecules in the endosome. 34694569_LncRNA USP30-AS1 promotes the survival of acute myeloid leukemia cells by cis-regulating USP30 and ANKRD13A. | ENSMUSG00000041870 | Ankrd13a | 1255.02293 | 0.9709772 | -0.0424906267 | 0.12395699 | 1.171886e-01 | 7.321039e-01 | 9.216229e-01 | No | Yes | 1418.285947 | 248.207887 | 1424.560285 | 243.714226 | ||
| ENSG00000076662 | 3385 | ICAM3 | protein_coding | P32942 | FUNCTION: ICAM proteins are ligands for the leukocyte adhesion protein LFA-1 (integrin alpha-L/beta-2) (PubMed:1448173). ICAM3 is also a ligand for integrin alpha-D/beta-2. In association with integrin alpha-L/beta-2, contributes to apoptotic neutrophil phagocytosis by macrophages (PubMed:23775590). {ECO:0000269|PubMed:1448173, ECO:0000269|PubMed:23775590}. | 3D-structure;Cell adhesion;Direct protein sequencing;Disulfide bond;Glycoprotein;Immunoglobulin domain;Membrane;Phagocytosis;Phosphoprotein;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the intercellular adhesion molecule (ICAM) family. All ICAM proteins are type I transmembrane glycoproteins, contain 2-9 immunoglobulin-like C2-type domains, and bind to the leukocyte adhesion LFA-1 protein. This protein is constitutively and abundantly expressed by all leucocytes and may be the most important ligand for LFA-1 in the initiation of the immune response. It functions not only as an adhesion molecule, but also as a potent signalling molecule. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Feb 2016]. | hsa:3385; | extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; integrin binding [GO:0005178]; signaling receptor binding [GO:0005102]; cell adhesion [GO:0007155]; cell-cell adhesion [GO:0098609]; phagocytosis [GO:0006909] | 11784723_A novel serine-rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting 11799126_identification of DC-SIGN binding sites 12021323_interactions with DC-SIGN does not promote DC-SIGN mediated HIV-1 transmission 12571844_Expression of DC-SIGN and its ligand, ICAM-3, is found in substantial amounts only in RA synovium, suggesting that their interaction is implicated in the additional activation of synovial macrophages that leads to the production of EMMPRIN and MMP-1. 12600815_the expression of ICAM-1 might involve both p38 MAPK and NF-kappaB activities, whereas the regulation of CD11b, CD18, and ICAM-3 expressions might be mediated through p38 MAPK but not NF-kappaB. 12743567_ICAM-3 is highly expressed on the surface of human eosinophils and has a role in the downregulation of GM-CSF production. 14704632_Relationship of intercellular adhesion molecule-3 and hepatocyte growth factor with amyloidosis A in chronic renal-failure patients. There was a higher density of intercellular adhesion molecule-3-positive cells in the patients with amyloidosis A. 14726630_ICAM-3 is expressed on human bone marrow endothelial cells and controls endothelial integrity via reactive oxygen species-dependent signaling. 14970226_soluble DC-SIGN bound to gp120-Fc more than 100- and 50-fold better than ICAM-2-Fc and ICAM-3-Fc, respectively. Binding sites are described. 15163761_acts as a costimulating molecule to increase HIV-1 transcription and viral replication, a process allowing productive infection of quiescent CD4+ T lymphocytes. 15880373_Expression of ICAM-3 can be used as a valuable biomarker to predict the radiation resistance in cervical cancer that occurs during radiotherapy. 15958383_the hybrid domain of integrin alphaL beta2 has different requirements of affinity states for ICAM-1 and ICAM-3 binding 17145745_The results suggest that ICAM-3 assists in the interaction of granulocytes with DC-SIGN of dendritic cells. 17570115_Observational study of gene-disease association. (HuGE Navigator) 17570115_Patients with SARS homozygous for ICAM3 Gly143 showed significant association with higher lactate dehydrogenase levels and lower total white blood cell counts. 17591777_talin induced an intermediate affinity alphaLbeta2 that adhered constitutively to its ligand intercellular adhesion molecule ICAM-1 but not ICAM-3. 17913807_Here we demonstrate that leukocyte function-associated antigen 1 (LFA-1), intercellular adhesion molecule 1 (ICAM-1), and ICAM-3 are enriched at the VS and that inhibition of these interactions influences conjugate formation and reduces VS assembly. 18261116_This is the first case of CD20 positive mycosis fungoides involving a lymph node to be reported in the literature. 18354203_extended alpha(L)beta(2) with an open headpiece is required for ICAM-3 adhesion 19225705_Lower expression of ICAM-3 and higher expression of ICAM-1 suggest that AMs may be involved in the pathogenesis of scleroderma. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19801714_Observational study of gene-disease association. (HuGE Navigator) 19801714_no significant risk association was found for SARS infection for the ICAM-3 Asp143Gly SNP. 19898481_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 19956847_ICAM-3 enhances the migratory and invasive potential of human non-small cell lung cancer cells by inducing MMP-2 and MMP-9 via Akt and CREB 20086017_CCR1 antagonist, BX471, did not significantly alter ICAM-3 expression in relapsing-remitting multiple sclerosis patients. 20331378_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21239057_Single nucleotide polymorphisms in ICAM3 gene is associated with lymphoma. 21381019_induction of morphological polarization in primary T lymphocytes and Jurkat cells enhances Kidins220/ARMS colocalization with ICAM-3 21712539_the cross-talk between neutrophils and NK cells is mediated by ICAM-3 and CD11d/CD18, respectively. 22117198_ICAM-3 may be an important adhesion molecule involved in chemotaxis to apoptotic human leukocytes. 22205703_the molecular basis of allergen-induced Th2 cell polarization 22396536_analysis of activated apoptotic cells induce dendritic cell maturation via engagement of Toll-like receptor 4 (TLR4), dendritic cell-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN), and beta2 integrins 22479382_Results indicate that the ICAM-3 gene promoter is negatively regulated by RUNX3. 23144795_Intercellular adhesion molecule (ICAM)-3 mRNA is upregulated in non-adherent endothelial forming cells. 23775590_ICAM3 acts as recognition receptors in the phagocytosis portals of macrophages for engulfment of apoptotic neutrophils. 24177012_this data clearly indicate that ICAM-3 promotes drug resistance via inhibition of apoptosis. 24474251_with larger patient groups and preferably detailed histopathological and clinical evaluations, are needed to explain the severity of ICAM-1, ICAM-2, and ICAM-3 molecules in Barrett's esophagus 27552332_Increased expression of PECAM-1, ICAM-3, and VCAM-1 in colonic biopsies from patients with inflammatory bowel disease (IBD) in clinical remission is associated with subsequent flares; this suggests that increases in the expression of these proteins may be early events that lead to flares in patients with IBD 29477378_we identify a potential CSC regulator and suggest a novel mechanism by which ICAM3 governs cancer cell stemness and inflammation. 29671117_Lewis-antigen-containing ICAM-2/3 on Jurkat leukemia cells interact with DC-SIGN to regulate DC functions. 29729315_exploration of the underlying mechanism demonstrated that ICAM3 not only binds to LFA-1 with its extracellular domain and structure protein ERM but also to lamellipodia with its intracellular domain which causes a tension that pulls cells apart (metastasis). 32013031_Changes in the Surface Expression of Intercellular Adhesion Molecule 3, the Induction of Apoptosis, and the Inhibition of Cell-Cycle Progression of Human Multidrug-Resistant Jurkat/A4 Cells Exposed to a Random Positioning Machine. 32371447_Vascular injury biomarkers and stroke risk: A population-based study. 33999358_Association of Circulating ICAM3 Concentrations with Severity and Short-term Outcomes of Acute Ischemic Stroke. | 51.62707 | 0.8012025 | -0.3197611903 | 0.42427101 | 5.673127e-01 | 4.513287e-01 | No | Yes | 62.946240 | 11.015250 | 77.612574 | 12.208505 | ||||
| ENSG00000076685 | 22978 | NT5C2 | protein_coding | P49902 | FUNCTION: Broad specificity cytosolic 5'-nucleotidase that catalyzes the dephosphorylation of 6-hydroxypurine nucleoside 5'-monophosphates (PubMed:1659319, PubMed:9371705, PubMed:10092873, PubMed:12907246). In addition, possesses a phosphotransferase activity by which it can transfer a phosphate from a donor nucleoside monophosphate to an acceptor nucleoside, preferably inosine, deoxyinosine and guanosine (PubMed:1659319, PubMed:9371705). Has the highest activities for IMP and GMP followed by dIMP, dGMP and XMP (PubMed:1659319, PubMed:9371705, PubMed:10092873, PubMed:12907246). Could also catalyze the transfer of phosphates from pyrimidine monophosphates but with lower efficiency (PubMed:1659319, PubMed:9371705). Through these activities regulates the purine nucleoside/nucleotide pools within the cell (PubMed:1659319, PubMed:9371705, PubMed:10092873, PubMed:12907246). {ECO:0000269|PubMed:10092873, ECO:0000269|PubMed:12907246, ECO:0000269|PubMed:1659319, ECO:0000269|PubMed:9371705}. | 3D-structure;ATP-binding;Allosteric enzyme;Alternative splicing;Cytoplasm;Disease variant;Hereditary spastic paraplegia;Hydrolase;Magnesium;Metal-binding;Neurodegeneration;Nucleotide metabolism;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase | This gene encodes a hydrolase that serves as an important role in cellular purine metabolism by acting primarily on inosine 5'-monophosphate and other purine nucleotides. [provided by RefSeq, Oct 2011]. | hsa:22978; | cytosol [GO:0005829]; 5'-nucleotidase activity [GO:0008253]; IMP 5'-nucleotidase activity [GO:0050483]; metal ion binding [GO:0046872]; nucleotide binding [GO:0000166]; adenosine metabolic process [GO:0046085]; allantoin metabolic process [GO:0000255]; IMP catabolic process [GO:0006204]; IMP metabolic process [GO:0046040] | 15923058_cN-II has a role in protecting against progression of non-small cell lung cancer 16385451_Observational study of gene-disease association. (HuGE Navigator) 17350683_the expression level of cN-II mRNA might be a prognostic factor of high-risk MDS. 17405878_Data describe the crystal structure of human cytosolic 5'-nucleotidase II and discuss its allosteric regulation and substrate recognition. 18775979_In leukoblasts from 82 patients with acute myeloid leukemia, various extent and frequency of differential allelic expression in the CDA, DCK, NT5C2, NT5C3, and TP53 genes was observed. 19428333_The DCK/cN-II ratio was again proportional to ara-CTP production and to ara-C sensitivity. 20542020_Observational study of gene-disease association. (HuGE Navigator) 21045733_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 21396942_seven high-resolution structures of human cN-II, including a ligand-free form and complexes with various substrates and effectors, were presented. 22071413_Polymorphisms in the CYP17A1 and NT5C2 genes influence a reduction in both visceral and subcutaneous fat mass in Japanese women. 23223233_analysis of Drosophila and human 7-methyl GMP-specific nucleotidases 23377183_These results suggest that mutations in NT5C2 are associated with the outgrowth of drug-resistant clones in acute lymphoblastic leukemia 23377281_results highlight the prominent role of relapse-specific mutations in NT5C2 as a mechanism of resistance to 6-Mercaptopurine and a genetic driver of relapse in acute lymphoblastic leukemia 24861553_Four novel body mass index-associated loci near the KCNQ1(rs2237892), ALDH2/MYL2 (rs671, rs12229654), ITIH4 (rs2535633) and NT5C2 (rs11191580) genes are identified in East Asian-ancestry populations. 25811392_cN-II co-immunoprecipitated both with wild type Ipaf and its LRR domain after transfection with corresponding expression vectors, but not with Ipaf lacking the LRR domain. 25857773_Data indicate that type II cytosolic 5'-nucleotidase (cN-II) exerts a role in nucleotide and drug metabolism and regulating cell survival. 26259531_Leukemia Relapse-Associated Mutation of NT5C2 Gene is Rare in de Novo Acute Leukemias and Solid Tumors. 27004590_this study implicates altered neural expression of BORCS7, AS3MT, and NT5C2 in susceptibility to schizophrenia arising from genetic variation at the chromosome 10q24 locus 27424800_Using data from a large-scale genome-wide association study of schizophrenia, study identified several potentially functional variants relating to miRNA function with our top finding being a schizophrenia protective allele that disrupts miR-206s binding to NT5C2 thus leading to increased expression of this gene. 27756303_acute lymphoblastic leukemia-specific mutations affect regulation of cN-II 27901213_NT5C2 variant rs11191580 conferred susceptibility to schizophrenia and affected the clinical symptoms of schizophrenia in a South Chinese Han population. 28327087_The aberrantly spliced NT5C2 showed substantial reduction in expression level in the in-vitro study, indicating marked instability of the mutant NT5C2 protein. 28884889_NT5C2 mutations seem to have a broad clinical spectrum and should be sought in patients manifesting either as uncomplicated or complicated spastic paraplegia 29535428_intersubunit interaction forms structural basis of hyperactive NT5C2 in drug-resistant leukemia in which heterozygous NT5C2 mutation gave rise to hetero-tetramer mutant and WT proteins. 29990496_Results uncover dynamic mechanisms of enzyme regulation targeted by chemotherapy resistance-driving NT5C2 mutations in relapsed acute lymphoblastic leukemia. 30201983_Targeted sequencing of NT5C2 did not identify any missense variants associated with rs72846714 or wmEry-TGN/wmDNA-TG. 30803894_NT5C2 deletion in mice protects against high-fat diet-induced weight gain, adiposity, insulin resistance and associated hyperglycemia 31097295_We provide an extensive neurobiological characterization of the psychiatric risk gene NT5C2, describing its previously unknown role in the regulation of AMPK signaling and protein translation in neural stem cells and its association with Drosophila melanogaster motility behavior 31358663_NT5C2 mutations alter thiopurine metabolism and cellular disposition 31971569_Subclonal NT5C2 mutations are associated with poor outcomes after relapse of pediatric acute lymphoblastic leukemia. 32541135_NT5C2 Gene Polymorphisms and the Risk of Coronary Heart Disease. 32590138_Association between NT5C2 rs11191580 and autism spectrum disorder in the Chinese Han population. 32970317_Enhanced migration of breast and lung cancer cells deficient for cN-II and CD73 via COX-2/PGE2/AKT axis regulation. 32999320_NT5C2 methylation regulatory interplay between DNMT1 and insulin receptor in type 2 diabetes. 33124053_Effects of NT5C2 Germline Variants on 6-Mecaptopurine Metabolism in Children With Acute Lymphoblastic Leukemia. 33434633_CD73 and cN-II regulate the cellular response to chemotherapeutic and hypoxic stress in lung adenocarcinoma cells. 34209768_Cytosolic 5'-Nucleotidase II Silencing in Lung Tumor Cells Regulates Metabolism through Activation of the p53/AMPK Signaling Pathway. 34636169_NUDT15 polymorphism and NT5C2 and PRPS1 mutations influence thiopurine sensitivity in acute lymphoblastic leukaemia cells. 35295960_Associations between Gene-Gene Interaction and Overweight/Obesity of 12-Month-Old Chinese Infants. | ENSMUSG00000025041 | Nt5c2 | 2072.02520 | 1.0955496 | 0.1316548012 | 0.10249456 | 1.656141e+00 | 1.981251e-01 | 5.902057e-01 | No | Yes | 2685.546431 | 428.447316 | 2278.820800 | 355.708391 | |
| ENSG00000076826 | 57662 | CAMSAP3 | protein_coding | Q9P1Y5 | FUNCTION: Key microtubule-organizing protein that specifically binds the minus-end of non-centrosomal microtubules and regulates their dynamics and organization (PubMed:19041755, PubMed:23169647). Specifically recognizes growing microtubule minus-ends and autonomously decorates and stabilizes microtubule lattice formed by microtubule minus-end polymerization (PubMed:24486153). Acts on free microtubule minus-ends that are not capped by microtubule-nucleating proteins or other factors and protects microtubule minus-ends from depolymerization (PubMed:24486153). In addition, it also reduces the velocity of microtubule polymerization (PubMed:24486153). Required for the biogenesis and the maintenance of zonula adherens by anchoring the minus-end of microtubules to zonula adherens and by recruiting the kinesin KIFC3 to those junctional sites (PubMed:19041755). Required for orienting the apical-to-basal polarity of microtubules in epithelial cells: acts by tethering non-centrosomal microtubules to the apical cortex, leading to their longitudinal orientation (PubMed:27802168, PubMed:26715742). Plays a key role in early embryos, which lack centrosomes: accumulates at the microtubule bridges that connect pairs of cells and enables the formation of a non-centrosomal microtubule-organizing center that directs intracellular transport in the early embryo (By similarity). Couples non-centrosomal microtubules with actin: interaction with MACF1 at the minus ends of non-centrosomal microtubules, tethers the microtubules to actin filaments, regulating focal adhesion size and cell migration (PubMed:27693509). Plays a key role in the generation of non-centrosomal microtubules by accumulating in the pericentrosomal region and cooperating with KATNA1 to release non-centrosomal microtubules from the centrosome (PubMed:28386021). Through the microtubule cytoskeleton, also regulates the organization of cellular organelles including the Golgi and the early endosomes (PubMed:28089391). Through interaction with AKAP9, involved in translocation of Golgi vesicles in epithelial cells, where microtubules are mainly non-centrosomal (PubMed:28089391). Plays an important role in motile cilia function by facilitatating proper orientation of basal bodies and formation of central microtubule pairs in motile cilia (By similarity). {ECO:0000250|UniProtKB:Q80VC9, ECO:0000269|PubMed:19041755, ECO:0000269|PubMed:23169647, ECO:0000269|PubMed:24486153, ECO:0000269|PubMed:26715742, ECO:0000269|PubMed:27693509, ECO:0000269|PubMed:27802168, ECO:0000269|PubMed:28089391, ECO:0000269|PubMed:28386021}. | Alternative splicing;Cell junction;Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Microtubule;Phosphoprotein;Reference proteome | hsa:57662; | axoneme [GO:0005930]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; microtubule minus-end [GO:0036449]; motile cilium [GO:0031514]; nucleoplasm [GO:0005654]; zonula adherens [GO:0005915]; actin filament binding [GO:0051015]; calmodulin binding [GO:0005516]; microtubule minus-end binding [GO:0051011]; spectrin binding [GO:0030507]; cilium movement [GO:0003341]; cytoplasmic microtubule organization [GO:0031122]; embryo development ending in birth or egg hatching [GO:0009792]; epithelial cell-cell adhesion [GO:0090136]; establishment of epithelial cell apical/basal polarity [GO:0045198]; establishment or maintenance of microtubule cytoskeleton polarity [GO:0030951]; in utero embryonic development [GO:0001701]; microtubule anchoring [GO:0034453]; microtubule cytoskeleton organization [GO:0000226]; negative regulation of microtubule depolymerization [GO:0007026]; neuron projection development [GO:0031175]; protein transport along microtubule [GO:0098840]; regulation of cell migration [GO:0030334]; regulation of focal adhesion assembly [GO:0051893]; regulation of Golgi organization [GO:1903358]; regulation of microtubule cytoskeleton organization [GO:0070507]; regulation of microtubule polymerization [GO:0031113]; regulation of organelle organization [GO:0033043]; zonula adherens maintenance [GO:0045218] | 19041755_KIAA1543 is a non-centrosomal minus end binding protein. It is termed Nezha(a character in Chinese mythic novel (Journey to the West)). 19508979_The CKK domain binds microtubules and represents a domain that evolved with the metazoa. 26715742_These findings demonstrate that apically localized CAMSAP3 determines the proper orientation of microtubules, and in turn that of organelles, in mature mammalian epithelial cells. 27349180_data suggest that CDH23-C is a CAMSAP3/Marshalin-binding protein that can modify MT networks indirectly through its interaction with CAMSAP3/Marshalin. 27693509_ACF7, a member of the spectraplakin family of cytoskeletal crosslinking proteins, interacts with Nezha (also called CAMSAP3) at the minus ends of noncentrosomal microtubules and anchors them to actin filaments. 27802168_in mammalian intestinal epithelial cells, the spectraplakin ACF7 (also known as MACF1) specifically binds to CAMSAP3 and is required for the apical localization of CAMSAP3-decorated microtubule minus ends. 28089391_CAMSAP3-dependent Golgi vesicle clustering and graded microtubule dynamics 30282632_Findings suggest that CAMSAP3 functions to protect lung carcinoma cells against EMT by suppressing Akt activity via microtubule regulation and that CAMSAP3 loss promotes EMT in these cells. 33566684_Rab14/MACF2 complex regulates endosomal targeting during cytokinesis. | ENSMUSG00000044433 | Camsap3 | 647.15610 | 0.8520827 | -0.2309345601 | 0.12975174 | 3.150231e+00 | 7.591622e-02 | 4.043888e-01 | No | Yes | 718.316434 | 89.501278 | 819.709269 | 99.616778 | ||
| ENSG00000077684 | 79960 | JADE1 | protein_coding | Q6IE81 | FUNCTION: Scaffold subunit of some HBO1 complexes, which have a histone H4 acetyltransferase activity (PubMed:16387653, PubMed:19187766, PubMed:20129055, PubMed:24065767). Plays a key role in HBO1 complex by directing KAT7/HBO1 specificity towards histone H4 acetylation (H4K5ac, H4K8ac and H4K12ac), regulating DNA replication initiation, regulating DNA replication initiation (PubMed:20129055, PubMed:24065767). May also promote acetylation of nucleosomal histone H4 by KAT5 (PubMed:15502158). Promotes apoptosis (PubMed:16046545). May act as a renal tumor suppressor (PubMed:16046545). Negatively regulates canonical Wnt signaling; at least in part, cooperates with NPHP4 in this function (PubMed:22654112). {ECO:0000269|PubMed:15502158, ECO:0000269|PubMed:16046545, ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:19187766, ECO:0000269|PubMed:20129055, ECO:0000269|PubMed:22654112, ECO:0000269|PubMed:24065767}. | Acetylation;Activator;Alternative splicing;Apoptosis;Cell projection;Chromosome;Cytoplasm;Cytoskeleton;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:79960; | chromosome [GO:0005694]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; transcription coactivator activity [GO:0003713]; apoptotic process [GO:0006915]; histone H3 acetylation [GO:0043966]; histone H4-K12 acetylation [GO:0043983]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; histone modification [GO:0016570]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of cell growth [GO:0030308]; negative regulation of G1/S transition of mitotic cell cycle [GO:2000134]; regulation of cell cycle [GO:0051726]; regulation of cell growth [GO:0001558]; regulation of DNA biosynthetic process [GO:2000278]; regulation of DNA replication [GO:0006275]; regulation of transcription, DNA-templated [GO:0006355] | 12169691_Jade-1 protein is a novel candidate regulatory factor in Von Hippel Lindau-mediated renal tumor suppression 15502158_Jade-1 is a novel candidate transcriptional co-activator associated with HAT activity and may play a key role in the pathogenesis of renal cancer and von Hippel-Lindau disease 16046545_Jade-1 may suppress renal cancer cell growth in part by increasing apoptosis. 18684714_Jade-1/1L are crucial co-factors for HBO1-mediated histone H4 acetylation 18806787_The pVHL tumour suppressor and the Wnt tumorigenesis pathway are therefore directly linked through Jade-1. 22516360_RCC with a low expression of Jade-1 is associated with a poor outcome and decreased survival 22654112_The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate beta-catenin signaling. 23824745_Reduced Jade-1 expression is associated with clear-cell renal cell carcinomas. 24097061_lncRNA-JADE is a key functional link that connects the DDR to histone H4 acetylation, and that dysregulation of lncRNA-JADE may contribute to breast tumorigenesis. 24739512_Increase in the number of cells with cytoplasmic JADE1S correlated with activation of tubular cell proliferation and inversely correlated with the number of cells with nuclear JADE1S, supporting role of HBO1-JADE1 shuttling during organ regeneration. 25100726_Casein kinase 1 alpha specifically phosphorylates Jade-1 at an unprimed SLS phosphorylation site. 26151225_JADE1S protein localized to centrosomes in interphase and mitotic cells, and to the midbody during cytokinesis. 26919559_Interactome analyses revealed that the Jade-1S mutant unable to be phosphorylated by CK1alpha has an increased binding affinity to proteins involved in chromatin remodelling, histone deacetylation, transcriptional repression, and ribosome biogenesis. 27155521_JADE1 expression has a role in colon cancers, renal carcinomas, and in acute kidney injuries and regeneration. (Review) 27160456_identification of SNPs within the IQCJ, NXPH1, PHF17 and MYB genes partly explaining the large interindividual variability observed in plasma triglyceride levels in response to an n-3 fatty acid supplementation 28134766_A genome-wide association study (GWAS) identified loci associated with the plasma triglyceride (TG) response to omega-3 fatty acid (FA) supplementation in IQCJ, NXPH1, PHF17 and MYB. 29382722_results indicate that the N-terminal region of JADE1 functions as a platform that brings together the catalytic HBO1 subunit with its cognate H3-H4 substrate for histone acetylation 34719765_Genome-wide association study and functional validation implicates JADE1 in tauopathy. | ENSMUSG00000025764 | Jade1 | 1772.55968 | 1.0523644 | 0.0736343278 | 0.10897708 | 4.578503e-01 | 4.986304e-01 | 8.191825e-01 | No | Yes | 2147.063053 | 240.390692 | 1918.745752 | 210.388241 | ||
| ENSG00000078124 | 55331 | ACER3 | protein_coding | Q9NUN7 | FUNCTION: Endoplasmic reticulum and Golgi ceramidase that catalyzes the hydrolysis of unsaturated long-chain C18:1-, C20:1- and C20:4-ceramides, dihydroceramides and phytoceramides into sphingoid bases like sphingosine and free fatty acids at alkaline pH (PubMed:20068046, PubMed:26792856, PubMed:20207939, PubMed:11356846, PubMed:30575723). Ceramides, sphingosine, and its phosphorylated form sphingosine-1-phosphate are bioactive lipids that mediate cellular signaling pathways regulating several biological processes including cell proliferation, apoptosis and differentiation (PubMed:20068046). Controls the generation of sphingosine in erythrocytes, and thereby sphingosine-1-phosphate in plasma (PubMed:20207939). Through the regulation of ceramides and sphingosine-1-phosphate homeostasis in the brain may play a role in neurons survival and function (By similarity). By regulating the levels of proinflammatory ceramides in immune cells and tissues, may modulate the inflammatory response (By similarity). {ECO:0000250|UniProtKB:Q9D099, ECO:0000269|PubMed:11356846, ECO:0000269|PubMed:20068046, ECO:0000269|PubMed:20207939, ECO:0000269|PubMed:26792856, ECO:0000269|PubMed:30575723, ECO:0000303|PubMed:20068046}. | 3D-structure;Alternative splicing;Calcium;Disease variant;Endoplasmic reticulum;Golgi apparatus;Hydrolase;Leukodystrophy;Lipid metabolism;Membrane;Metal-binding;Reference proteome;Sphingolipid metabolism;Transmembrane;Transmembrane helix;Zinc | PATHWAY: Lipid metabolism; sphingolipid metabolism. {ECO:0000269|PubMed:20068046, ECO:0000269|PubMed:20207939, ECO:0000269|PubMed:30575723}. | hsa:55331; | endoplasmic reticulum membrane [GO:0005789]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of Golgi membrane [GO:0030173]; integral component of membrane [GO:0016021]; calcium ion binding [GO:0005509]; ceramidase activity [GO:0102121]; dihydroceramidase activity [GO:0071633]; N-acylsphingosine amidohydrolase activity [GO:0017040]; phytoceramidase activity [GO:0070774]; zinc ion binding [GO:0008270]; ceramide catabolic process [GO:0046514]; inflammatory response [GO:0006954]; myelination [GO:0042552]; phytosphingosine biosynthetic process [GO:0071602]; positive regulation of cell population proliferation [GO:0008284]; regulation of programmed cell death [GO:0043067]; sphingolipid biosynthetic process [GO:0030148]; sphingosine biosynthetic process [GO:0046512] | 19834535_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20068046_ACER3 catalyzes the hydrolysis of unsaturated long chain ceramides and dihydroceramides and coordinates with ACER2 to regulate cell proliferation and survival 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26792856_Homozygosity for the p.E33G mutation in the ACER3 gene results in inactivation of ACER3. 27470583_The ACER3 deficiency resulted in decreased cell growth and colony formation, elevated apoptosis, and lower AKT signaling of leukemia cells. This study indicates that ACER3 contributes to AML pathogenesis, and suggests that alkaline ceramidase inhibition is an option to treat acute myeloid leukemia. 30097213_Our study suggests that Acer3 contributes to hepatocellular carcinoma propagation 34281620_ACER3-related leukoencephalopathy: expanding the clinical and imaging findings spectrum due to novel variants. 34817752_LINC01087 indicates a poor prognosis of glioma patients with preoperative MRI. 35034572_Circular RNA circ_0001955 promotes hepatocellular carcinoma tumorigenesis by up-regulating alkaline ceramidase 3 expression through microRNA-655-3p. | ENSMUSG00000030760 | Acer3 | 1270.65847 | 1.0342573 | 0.0485951036 | 0.12585973 | 1.493607e-01 | 6.991470e-01 | 9.089064e-01 | No | Yes | 1620.752151 | 353.581746 | 1464.727511 | 312.718322 | |
| ENSG00000078403 | 8028 | MLLT10 | protein_coding | P55197 | FUNCTION: Probably involved in transcriptional regulation. In vitro or as fusion protein with KMT2A/MLL1 has transactivation activity. Binds to cruciform DNA. In cells, binding to unmodified histone H3 regulates DOT1L functions including histone H3 'Lys-79' dimethylation (H3K79me2) and gene activation (PubMed:26439302). {ECO:0000269|PubMed:17868029, ECO:0000269|PubMed:26439302}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a transcription factor and has been identified as a partner gene involved in several chromosomal rearrangements resulting in various leukemias. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2010]. | hsa:8028; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; nucleosome binding [GO:0031491]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 12127405_FISH analysis in infant AML-M5 showed a complex rearrangement between chromosomes 10 and 11, disrupting the MLL gene, a paracentric inversion of the 11q13-q23 fragment translocated to 10p12. AF10 was the fusion partner gene of MLL in this rearrangement. 12453419_ALL1 is involved in remodeling, acetylating, deacetylating, and methylating nucleosomes and/or free histones 12482966_data reveal new properties for the leucine zipper domain and thus might provide new clues to understanding the mechanisms by which AF10 fusion proteins in which the PHD domain is lost might trigger leukemias in humans 16215946_The patients with immunophenotype of Pre-B-acute lymphoblastic leukemia were found to carry: MLL/AF10. 17868029_Data show that the nuclear isoform of FLRG lacks an intrinsic transactivation domain, but enhances AF10-mediated transcription, probably through promoting the homo-oligomerization of AF10, thus facilitating the recruitment of co-activators. 18037964_CALM(PICALM)/AF10 fusion protein might interfere with normal Ikaros (IKZF1)function, and thereby block lymphoid differentiation in CALM/AF10 positive leukemias. 19443658_the CALM-AF10 fusion protein can also greatly reduce global H3K79 methylation in both human and murine leukemic cells by disrupting the AF10-mediated association of hDOT1L with chromatin 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21706055_In leukemia cells, full-length CALM-AF10 localized to the nucleus with no consistent effect on growth factor endocyctosis, and suppressed histone H3 lysine 79 methylation regardless of the presence of clathrin 21804547_a new susceptibility locus for meningioma at 10p12.31 (MLLT10, rs11012732, odds ratio = 1.46, P(combined) = 1.88 x 10(-14)) was identified. 22064352_results strongly indicate that the differential regulation of these three genes is not due to the break point effect but as a consequence of the CALM/AF10 fusion gene expression, though the mechanism of regulation is not well understood 23670296_detection of PICALM-MLLT10 fusion transcript occurs in 7% of children with T-lineage ALL and is not associated with a poorer outcome for patients treated with contemporary, intensive chemotherapy 23673860_In pediatric T-acute lymphoblastic leukemia, we have identified 2 RNA processing genes, that is, HNRNPH1/5q35 and DDX3X/Xp11.3 as new MLLT10 fusion partners. 24755950_we report that variants at the MLLT10 locus are unlikely to alter risk of glioma, and they have no prognostic value among patients with high-grade tumors (glioblastoma). 25464900_Results provide evidence that transformation driven by MLL fusions as well as the recurrent AML-associated NUP98-NSD1 fusion oncogene is critically dependent on the ability of AF10 to stimulate DOT1L activity. 26439302_The PZP domain of AF10 senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. 26480920_Our results provide evidence for new loci influencing abdominal visceral (BBS9, ADCY8, KCNK9) and subcutaneous (MLLT10/DNAJC1/EBLN1) fat, and confirmed a locus (THNSL2) previously reported to be associated with abdominal fat in women 27859216_In a retroviral transduction/transplantation mouse model, mice transplanted with MLL/AF10(OM-LZ) cells harboring PTPN11(wt) developed myelomonocytic leukemia. Those transplanted with cells harboring PTPN11(G503A) -induced monocytic leukemia in a shorter latency. Adding PTPN11(G503A) to MLL/AF10 affected cell proliferation, chemo-resistance, differentiation, in vivo BM recruitment/clonal expansion and faster progression. 30102091_In vitro, the overexpression of MLLT10 promoted colorectal cancer (CRC) cell migration and invasion, while after MLLT10 was knocked down, the opposite results were observed. In lung metastasis sites, the knockdown of MLLT10 in SW620 cells significantly inhibited Vimentin expression, whereas the E-Cadherin was increased. These results indicate that MLLT10 regulates the metastasis of CRC cells via EMT. 31527241_the oligomerization ability of the DOT1L-AF10 complex is essential for MLL1-AF10's leukemogenic function 32569758_MLLT10 in benign and malignant hematopoiesis. 33690798_A JAK/STAT-mediated inflammatory signaling cascade drives oncogenesis in AF10-rearranged AML. 34215314_AF10 (MLLT10) prevents somatic cell reprogramming through regulation of DOT1L-mediated H3K79 methylation. 34226546_The role of the PZP domain of AF10 in acute leukemia driven by AF10 translocations. | ENSMUSG00000026743 | Mllt10 | 1379.25562 | 1.1642784 | 0.2194360713 | 0.09762201 | 5.073618e+00 | 2.429263e-02 | 2.514952e-01 | No | Yes | 998.005556 | 95.247054 | 830.955639 | 77.843717 | |
| ENSG00000078549 | 117 | ADCYAP1R1 | protein_coding | P41586 | FUNCTION: This is a receptor for PACAP-27 and PACAP-38. The activity of this receptor is mediated by G proteins which activate adenylyl cyclase. May regulate the release of adrenocorticotropin, luteinizing hormone, growth hormone, prolactin, epinephrine, and catecholamine. May play a role in spermatogenesis and sperm motility. Causes smooth muscle relaxation and secretion in the gastrointestinal tract. | 3D-structure;Alternative splicing;Cell membrane;Developmental protein;Differentiation;Disulfide bond;G-protein coupled receptor;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Spermatogenesis;Transducer;Transmembrane;Transmembrane helix | This gene encodes type I adenylate cyclase activating polypeptide receptor, which is a membrane-associated protein and shares significant homology with members of the glucagon/secretin receptor family. This receptor mediates diverse biological actions of adenylate cyclase activating polypeptide 1 and is positively coupled to adenylate cyclase. Multiple alternatively spliced transcript variants encoding distinct isoforms have been identified. [provided by RefSeq, Dec 2010]. | hsa:117; | bicellular tight junction [GO:0005923]; caveola [GO:0005901]; cell surface [GO:0009986]; endosome [GO:0005768]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; rough endoplasmic reticulum [GO:0005791]; adenylate cyclase binding [GO:0008179]; G protein-coupled peptide receptor activity [GO:0008528]; neuropeptide binding [GO:0042923]; peptide hormone binding [GO:0017046]; signaling receptor activity [GO:0038023]; small GTPase binding [GO:0031267]; vasoactive intestinal polypeptide receptor activity [GO:0004999]; activation of phospholipase C activity [GO:0007202]; adenylate cyclase-modulating G protein-coupled receptor signaling pathway [GO:0007188]; cAMP-mediated signaling [GO:0019933]; cell differentiation [GO:0030154]; cell surface receptor signaling pathway [GO:0007166]; development of primary female sexual characteristics [GO:0046545]; multicellular organismal response to stress [GO:0033555]; negative regulation of cell death [GO:0060548]; positive regulation of calcium ion transport into cytosol [GO:0010524]; positive regulation of cAMP-mediated signaling [GO:0043950]; positive regulation of inositol phosphate biosynthetic process [GO:0060732]; positive regulation of small GTPase mediated signal transduction [GO:0051057]; response to estradiol [GO:0032355]; response to ethanol [GO:0045471]; response to xenobiotic stimulus [GO:0009410]; spermatogenesis [GO:0007283] | 11930171_PACAP receptor type 1 mRNA is expressed in the trigeminal, otic and superior cervical ganglia (prejunctional) and cerebral arteries (postjunctional). 12409223_HCT8 human colon tumor cells express PAC1, and PAC1 activation is coupled to adenylate cyclase, increase cytosolic [Ca(2+)](i), and cellular proliferation 12409233_Using GST pull-down assay, ARF6 binds to the hop1 but not the null domain of PAC1 receptor 12573802_possible role for PACAP in the regulation of expression of genes encoding catecholamine-synthesizing enzymes in intra-adrenal pheochromocytomas 12732341_receptors were positively coupled to adenylyl cyclase in human lung cancer, but the enzyme activity was impaired as compared to normal lung 12948842_Expression of functional PAC(1), VPAC(1) and VPAC(2) receptors in human prostate as well as its maintenance after malignant transformation 14742913_In colon cancer cell lines, PAC1 is expressed as the SV1 or HIP splice variant and is coupled to the activation only of cAMP but not of intracellular Ca2+. 15518890_PACAP, PACAP receptor and tyrosine hydroxylase (TH) mRNAs in neuroblastomas 16226889_Results show that the combination of different amino terminal and intracellular loop 3 splicing variants in the PAC1 receptor dictates the ability of agonists, including PACAP and VIP, to activate signaling pathways. 16372333_PAC(1)-R null variant is the most relevant isoform expressed in human prostaste cancer, and may be related with the events determining the outcome of prostate cancer. 16401644_Stimulation of endogenously expressed PAC1 receptors with PACAP in human neuroblastoma cells increased APPsalpha secretion. PAC1R activation stimulates ERK phosphorylation. 16572459_PAC1-R is the predominant PACAP receptor found in fetuses, and both PAC1-R and VPAC1-R are expressed in the mature cerebellum. 16823490_Data demonstrate that altered neuronal proliferation/apoptosis and disrupted ependymal cilia are the main factors contributing to hydrocephalus in PAC1-overexpressing mice. 16888199_PACAP may regulate the biological release of peptides and serotonin from BON cells 17470806_structure of the N-terminal EC domain of the hPAC1-RS receptor in complex with the PACAP (6'-38') peptide antagonist; identified hormone residues that are critical for binding to the N-terminal EC domain of PAC1-RS 18353507_Novel stable PACAP analogs with potent activity towards the PAC1 receptor are reported. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19269029_a novel mechanism of calmodulin in regulating PACAP signaling by possible interaction with the inactive state of PAC1 and VPAC2 receptors. 19926922_LH causes a transient stimulation of PACAP & PAC(1)-R expression & PACAP stimulates progesterone production in human luteinized granulosa cells, suggesting possible role of PACAP as a local ovarian regulator in luteinization. 20026142_Results indicate that VPAC1, but not VPAC2 or PAC1, up-regulation in macrophages is a common mechanism in response to acute and chronic pro-inflammatory stimuli. 20395540_VPAC2 and/or PAC1 receptor activation is involved in cutaneous active vasodilation in humans. 21185349_The present study constitutes the first characterization of the binding domains of PACAP to its specific receptor PAC1 and suggests heterogeneity within the binding mode of peptide ligands to class B GPCRs. 21350482_data suggest that perturbations in the PACAP-PAC1 pathway are involved in abnormal stress responses underlying post-traumatic stress disorder 21912390_A reported association between rs2267735 and PTSD could not be replicated in either African American or European American females in two large independent samples. 22539193_The increased expressions of PAC1 receptors on lactating breast may indicate a PACAP38/PAC1 interaction in the mammary gland during lactation. 22581436_PAC1 regulates PYK-2 tyrosine phosphorylation in a calcium-dependent manner in lung cancer cell lines. 22609358_Activation of Sp1 by the Ras/MAPK pathway might participate in neuron specific expression of the PAC1 gene. 22776899_PAC1 receptor (ADCYAP1R1) genotype is associated with dark-enhanced startle in both male and female children. 22961270_Conclude that VPAC2/PAC1 receptors require NO in series to effect cutaneous active vasodilation during heat stress in humans. 23147486_[review] The Adcyap1r1 receptor single nucleotide polymorphism (SNP) is located within a predicted estrogen response element (ERE). 23280952_Genetic variation at the ADCYAP1R1 locus interacts with child abuse to shape risk of later posttraumatic stress disorder (PTSD) in women. 23328528_Epigenetic and genetic variants in ADVYAP1R1 are associated with asthma in Puerto Rican children. 23394710_The findings suggest that the PACAP-PAC1 receptor pathway may play an important role in female human responses to traumatic stress 23505260_ADCYAP1R1 genotype may have a role in post-traumatic stress symptoms in highly traumatized African-American females 23818986_Data indicate that VIP and PACAP increased macrophage resistance to HIV-1 replication by inducing the synthesis of beta-chemokines CCL3 and CCL5 and IL-10 following preferential activation of the receptors VPAC2 and PAC1. 23972788_and its receptor (PAC1) are involved in stress response and anxiety. 23981011_association of variants with sudden infant death not supported 24220567_This review focuses on role of PAC1 receptor in brain development, behavior of transgenic animals and potential implication in human neurodevelopmental disorders. 24516127_Individual differences in ADCYAP1R1 genotype may contribute to dysregulated fear circuitry known to play a central role in posttraumatic stress disorder and other anxiety disorders. 24696141_PAC1/PACAP receptor endocytosis contributes to ERK acktivation. 25680674_Rsults suggest that impaired contextual conditioning in the hippocampal formation may mediate the association between type I receptor of the pituitary adenylate cyclase activating polypeptideand and posttraumatic stress disorder symptoms. 25918834_High child stress and an ADCYAP1R1 single-nucleotide polymorphism are associated with reduced bronchodilator response in children with asthma. 26334183_Further investigations of genetic factors for trauma-related psychopathology should include careful assessments of the social environment 26343289_Data show that glucocorticoid response genes NR3C1, ADCYAP1R1 and HSD11B2 were relatively hypomethylated whereas FKBP5 was hypermethylated. 26700245_Results confirmed for the first time that doxycycline specially targeted pituitary adenylate cyclase-activating polypeptide (PACAP) receptor type 1 (PAC1) imitating PACAP(30-37) and acted as an enhancer by facilitating the subsequent ligand binding and the activation of PAC1. 27959335_These findings lead to a model in which E2 induces the expression of ADCYAP1R1 through binding of ERalpha at the ERE as an adaptive response to stress. Inhibition of E2/ERalpha binding to the ERE containing the rs2267735 risk allele results in reduced expression of ADCYAP1R1, diminishing estrogen regulation as an adaptive stress response and increasing risk for PTSD. 28356352_Data suggest that GCGR (glucagon receptor) activation proceeds via a mechanism in which transmembrane helix 6 (TM6) is held in an inactive conformation by a conserved polar core and a hydrophobic lock (involving intracellular loop 3, IC3); mutations in the corresponding polar core of GCGR or PAC1R disrupt these inhibitory elements, allow TM6 to swing outward, and induce constitutive G protein signaling. 28610473_Studies suggest that the level of stress and circulating gonadal hormones may differentially regulate the PACAPergic system in males and females to influence anxiety-like behavior and may be one mechanism underlying the discrepancies in human psychiatric disorders. [Review Article] 28710390_Transitions between different conformational states of the G protein-coupled pituitary adenylate cyclase-activating polypeptide receptor (PAC1R) range between microseconds to milliseconds. 28746747_These results provide evidence for an association between ADCYAP1R1 and PTSD and indicate that there may indeed be sex differences. 28926110_that perturbations in the PACAP-PAC1R pathway may be involved in amyotrophic lateral sclerosis pathology 29734363_These results show that b-arrestin1 and b-arrestin2 exert differential actions on PAC1R internalization and PAC1R-dependent ERK1/2 activation, and suggest that the two b-arrestin isoforms may be involved in fine and precise tuning of the PAC1R signaling pathways. 30548314_Data indicate the localization of adenylate cyclase activating polypeptide 1 (PACAP), its receptor PACAP type I receptor (PAC1R) and epidermal growth factor receptor (EGFR) in human corneal endothelium. 30556326_PAC1R encodes a receptor for pituitary adenylate cyclase activating polypeptide (PACAP), a member of the vasoactive intestinal polypeptide family. 31910434_Circulating PACAP peptide and PAC1R genotype as possible transdiagnostic biomarkers for anxiety disorders in women: a preliminary study. 32047270_Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism. 32157248_Molecular structure of the human PAC1 receptor coupled to an engineered heterotrimeric G protein has been reported. 32590837_Impact of ADCYAP1R1 genotype on longitudinal fear conditioning in children: interaction with trauma and sex. 32964398_The G Protein-Coupled Receptor PAC1 Regulates Transactivation of the Receptor Tyrosine Kinase HER3. 33078390_GIP receptor suppresses PAC1receptor-mediated neuronal differentiation via formation of a receptor heterocomplex. 35409075_PACAP-38 and PAC1 Receptor Alterations in Plasma and Cardiac Tissue Samples of Heart Failure Patients. | ENSMUSG00000029778 | Adcyap1r1 | 278.62933 | 0.7353556 | -0.4434860559 | 0.18811555 | 5.496049e+00 | 1.905949e-02 | 2.246418e-01 | No | Yes | 277.461935 | 35.716084 | 376.448786 | 46.718587 | |
| ENSG00000079102 | 862 | RUNX1T1 | protein_coding | Q06455 | FUNCTION: Transcriptional corepressor which facilitates transcriptional repression via its association with DNA-binding transcription factors and recruitment of other corepressors and histone-modifying enzymes (PubMed:12559562, PubMed:15203199, PubMed:10688654). Can repress the expression of MMP7 in a ZBTB33-dependent manner (PubMed:23251453). Can repress transactivation mediated by TCF12 (PubMed:16803958). Acts as a negative regulator of adipogenesis (By similarity). The AML1-MTG8/ETO fusion protein frequently found in leukemic cells is involved in leukemogenesis and contributes to hematopoietic stem/progenitor cell self-renewal (PubMed:23812588). {ECO:0000250|UniProtKB:Q61909, ECO:0000269|PubMed:10688654, ECO:0000269|PubMed:10973986, ECO:0000269|PubMed:16803958, ECO:0000269|PubMed:23251453, ECO:0000269|PubMed:23812588, ECO:0000303|PubMed:12559562, ECO:0000303|PubMed:15203199}. | 3D-structure;Alternative splicing;Chromosomal rearrangement;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a member of the myeloid translocation gene family which interact with DNA-bound transcription factors and recruit a range of corepressors to facilitate transcriptional repression. The t(8;21)(q22;q22) translocation is one of the most frequent karyotypic abnormalities in acute myeloid leukemia. The translocation produces a chimeric gene made up of the 5'-region of the runt-related transcription factor 1 gene fused to the 3'-region of this gene. The chimeric protein is thought to associate with the nuclear corepressor/histone deacetylase complex to block hematopoietic differentiation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2010]. | hsa:862; | nuclear matrix [GO:0016363]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor binding [GO:0140297]; metal ion binding [GO:0046872]; transcription corepressor activity [GO:0003714]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of transcription, DNA-templated [GO:0045892]; transcription, DNA-templated [GO:0006351] | 11869944_no conclusive evidence as yet that the AML1/ETO chimeric gene is sufficient per se to induce leukemia. 11983111_Analysis of nuclear distribution of the AML1/ETO protein and its homology domains led to identification of domains in ETO responsible for intranuclear transport and subnuclear distribution of AML1/ETO. 11986950_Potential involvement of the AML1-MTG8 fusion protein in the granulocytic maturation characteristic of the t(8;21) acute myelogenous leukemia cell 12393523_data demonstrate the capacity of AML1-ETO to promote the self-renewal of human hematopoietic cells and therefore support a causal role for t(8;21) translocations in leukemogenesis 12427969_2 independent subnuclear targeting signals in the N- and C-terminal regions of ETO direct ETO to the same binding sites occupied by AML1ETO. This provides a molecular basis for aberrant subnuclear targeting of AML1ETO, the defect in t(8;21)-related AML. 12557226_ETO rearrangements leading to the AML1-ETO fusion gene are frequently the result of small hidden interstitial insertions. 12773394_In t(8;21) leukaemic cells expressing the aberrant fusion protein AML1-ETO, we demonstrate that this protein is part of a transcription factor complex binding to extended sequences of the c-FMS intronic regulatory region rather than the promoter. 12874834_Data identify ETO as a partner for Gfi-1 and Gfi-1B, and suggest that Gfi-1 proteins repress transcription through recruitment of histone deacetylase-containing complexes. 14551142_ETO is a bona fide corepressor that links the transcriptional pathogenesis of acute leukemias and B-cell lymphomas and offers a compelling target for transcriptional therapy of hematologic malignancies. 14751048_RT-PCR for the detection of AML1/ETO in children with acute non-lymphoid leukemia (ANLL) was quick, convenient and sensitive, and could be regarded as a useful method for the diagnosis and prognosis of ANLL. 15203865_Present in acute myeloid leukemia of M2 subtype. 15295650_cloning, expression, purification and crystallization of NHR3 domain 15298716_These findings suggest a central role of RUNX1-CBFA2T1 in the maintenance of the leukaemia. 15333839_study identifes E proteins as AML1-ETO targets whose dysregulation may be important for t(8;21) leukemogenesis 15377655_N-CoR utilizes repression domains I and III for interaction and co-repression with ETO 15676213_DiffeRential expression of the ETO homologues suggests that they may have a potential role in hematopoietic differentiation. 15723339_Alternative splicing in the AML1-MTG8 fusion gene occurs in leukemia cell lines as well as in cells of leukemia paatients with a(8;21) translocation. 15735013_Results suggest a novel mechanism for gene silencing mediated by RUNX1/MTG8 and support the combination of HDAC and DNMT inhibitors as a novel therapeutic approach for t(8;21) AML. 15829516_AML1 and ETO are fused together at the t8;21 translocation breakpoint, resulting in the expression of a chimeric protein called AML1-ETO, which acts as a constitutive transcriptional repressor. 16502583_Translocations in acute myeloid leukemia (AML) is the t(8;21) is characterized by AML1-MTG8 (ETO) mutation. 16616331_Oligomerization is also required for AML1/ETO's interactions with ETO, MTGR1, and MTG16, but not with other corepressor molecules. 16652140_Suppression of AML1/MTG8 results in the increased expression of genes associated with myeloid differentiation, such as AZU1, BPI, CTSG, LYZ and RNASE2 as well as of antiproliferative genes such as IGFBP7, MS4A3 and SLA both in blasts and in cell lines 16741927_Conditional expression of AML1-ETO by the ecdysone-inducible system dramatically increases the expression of connexin 43 together with growth arrest at G1 phase in leukemic U937 cells. 16892037_AML1T1, an alternatively spliced isoform of the t(8;21) transcript, promotes leukemogenesis. 16990610_The leukemia-associated fusion protein AML1-ETO could aberrantly transactivate the EEN gene through an AML1 binding site. 17058450_As a result, we identified 14 unique proteins deregulated in AML1-ETO-carrying leukemic cells, including 3 up-regulated such as hairy and enhancer of split 5 (HES5) and 11 down-regulated such as MAT1 (menage a trois-1. 17244680_AML1/ETO participates in a protein complex with the RA receptor alpha (RARalpha) at RA regulatory regions on RARbeta2. 17284535_Loss of p21(WAF1) facilitates AML1-ETO-induced leukemogenesis. 17560011_Presence of AML1-ETO fusion protein increases the susceptibility of cells to chemical carcinogens, which favors the development of additional genetic alterations. 17625612_AML1-ETO and JAK2 may have a role in leukemogenesis, as shown by a myeloproliferative syndrome progressing to acute myeloid leukemia [case report] 17649722_Isoform AML1/ETO9a was correlated to acute myeloid leukemia-M2 subtype. 17875504_Our study suggests that KIT activating mutations in AML with t(8; 21) are associated with diminished CD 19 and positive CD56 expression on leukemic blasts and, thus, can be phenotypically distinguished from AML1-ETO leukemias 17989718_constitutively and overexpressed AML1-ETO protein was cleaved to four fragments of 70, 49, 40 and 25 kDa by activated caspase-3 during apoptosis induction by extrinsic mitochondrial and death receptor signaling pathways. 17996649_miR-223 is a direct transcriptional target of AML1/ETO 18039847_MTG8/ETO and Mtg16 (ETO2) associated with TCF4 18156164_These results demonstrate that scl is an important mediator of the ability of AML1-ETO to reprogram hematopoietic cell fate decisions, suggesting that scl may be an important contributor to AML1-ETO-associated leukemia. 18183572_VP-16 causes the ETO gene repositioning which allows AML1 and ETO genes to be localized in the same nuclear layer; data corroborate the so called 'breakage first' model of the origins of recurrent reciprocal translocation 18205948_The interaction between SIN3B and ETO required an intact amino-terminus of ETO and the NHR2 domain. 18258796_Knockdown of TLE1 or TLE4 levels increased the rate of cell division of the AML1-ETO-expressing Kasumi-1 cell line, whereas forced expression of either TLE1 or TLE4 caused apoptosis and cell death. 18332109_In functional assays, corepressor ETO, but not AML1/ETO, augments SHARP-mediated repression in an histone deacetylase-dependent manner. 18519037_AML1-ETO promotes leukemogenesis by blocking cell differentiation through inhibition of Sp1 transactivity. 18548094_RUNX1/RUNX1T1 siRNAs compromise the engraftment and/or self-renewal capacities of t(8;21) leukaemia-initiating cells. 18586123_ETO family member-mediated oligomerization and repression can be distinct events and that interaction between ETO family members and hSIN3B or N-CoR may not necessarily strengthen transcriptional repression. 18687517_survivin gene acts as a critical mediator of AML1/ETO-induced late oncogeneic events. 18952841_the crucial role of the NHR4 domain in determination of cellular fate during AML1-ETO-associated leukemogenesis. 19001502_novel role for the leukemia-related AML1-ETO protein in epigenetic control of cell growth through upregulation of ribosomal gene transcription mediated by RNA Pol I. 19043539_a major role for the functional interaction of AML1/ETO with AML1 and HEB in transcriptional regulation determined by the fusion protein. 19100510_Four copies of RUNX1T1 were found. 19179469_CBFbeta is essential for TEL-AML1's ability to promote self-renewal of B cell precursors in vitro. 19289505_Data show that E proteins contain another conserved ETO-interacting region, termed DES, and that differential associations with AD1 and DES allow ETO to repress transcription through both chromatin-dependent and chromatin-independent mechanisms. 19448675_Findings indicate a role for replication-independent pathways in RUNX and RUNX1-ETO senescence, and show that the context-specific oncogenic activity of RUNX1 fusion proteins is mirrored in their distinctive interactions with fail-safe responses. 19458628_AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) AML-M2. 19581587_CalpainB is required for AML1-ETO-induced blood cell disorders in Drosophila.[AML1-ETO] 20090777_NHR4 domain mutations of ETO are probably very infrequent in AML1-ETO positive myeloid leukemia cells. 20430957_Existence of an essential structural motif (hot spot) at the NHR2 dimer-tetramer interface, suitable for a molecular intervention in t(8;21) leukemias. 20487545_The critical role for an evolutionary conserved GATA binding site in transcriptional regulation of the ETO gene in cells of erythroid/megakaryocytic potential. 20688956_Expression of AML1-ETO caused an expansion of hematopoietic precursors in Drosophila, which expressed high levels of reactive oxygen species 20708017_ETO nervy homology region (NHR) 3 domain-PKA(RIIalpha) protein interaction does not appear to significantly contribute to AML1-ETO's ability to induce leukemia. 21200020_N-Ras(G12D) induces features of stepwise transformation in preleukemic umbilical cord blood cultures expressing the AML1-ETO fusion gene. 21245488_Elevated levels of AES mRNA and protein were confirmed in AML1/ETO-expressing leukemia cells, as well as in other acute myeloid leukemia specimens. 21499216_Data show that low RUNX1T1 expression was highly associated with hepatic metastases. 21540640_SMAD4 knockdown accelerated this re-silencing process, suggesting that normal TGF-beta signaling is essential for the maintenance of RunX1T1 expression 21571369_RUNX1T1 point mutation may be rare and passenger mutations in acute leukemias, lung and breast cancers 21764752_study found AML1-ETO is acetylated by the transcriptional coactivator p300 in leukemia cells isolated from t(8;21) AML patients, and this acetylation is essential for its self-renewal-promoting effects in cord blood CD34(+) cells and its leukemogenicity 22025082_Submicroscopic deletion of RUNX1T1 gene confirmed by high-resolution microarray in acute myeloid leukemia with RUNX1/RUNX1T1 rearrangement 22101339_The cooperative effect of the expression of mutated KIT and AML1-ETO oncogenes is crucial for selective toxic action of binase on malignant cells. 22201794_In this review, we detail the structural features, functions, and models used to study both RUNX1 and RUNX1-ETO in hematopoiesis over the past two decades 22343733_selective removal of RUNX1/ETO leads to a widespread reversal of epigenetic reprogramming and a genome-wide redistribution of RUNX1 binding, resulting in the inhibition of leukemic proliferation and self-renewal, and the induction of differentiation. 22498736_PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. 22995345_Study showed that in a widely used AML cell line, Kasumi-1, the chimeric AML1/ETO gene, is transcribed from the promoter P2 of AML1 gene, and this process does not depend on the formation of distant interactions between this promoter and the fused segment of ETO gene. 23223432_Our results indicated a feedback circuitry involving miR-193a and AML1/ETO/DNMTs/HDACs, cooperating with the PTEN/PI3K signaling pathway and contributing to leukemogenesis. 23426948_Importance of the degree of dysregulation by AML1-ETO in cellular transformation. 24402164_t(8;21)/RUNX1-RUNX1T1-positive AML shows a high frequency of additional genetic alterations. 24456692_The AML1/ETO target gene LAT2 interferes with differentiation of normal hematopoietic precursor cells. 24586690_Runx1t1 epigenetically regulates the proliferation and nitric oxide production of microglia. 24616160_Discordant MRD results were observed in 10/71 (14%) of the samples: in three samples from two patients who relapsed, RUNX1-RUNX1T1 was detectable only on DNA, while RUNX1-RUNX1T1 was detectable only on RNA in seven samples. 24727677_A reduction in KIT mRNA levels was also observed in AE-silenced cells, but silencing KIT expression reduced cell growth but did not induce apoptosis. 24783204_Our data shows that upregulated of the RUNX1-RUNX1T1 gene set maybe an important factor contributing to the etiology of ccRCC. 24897507_RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors are rescued by activating c-Kit mutations 24973361_identify a high-frequency mutation in t(8;21)/RUNX1-RUNX1T1 acute myekoid leukemia (AML) and identify the need for future studies to investigate the clinical and biological relevance of ASXL2 mutations in this unique subset of AML 24976338_RUNX1T1 gene may participate in t(8;21)(q22;q22)-dependent leukemic transformation due to its multiple interactions in cell regulatory network particularly through synergistic or antagonistic effects in relation to activity of RUNX1-RUNX1T1 fusion gene. 25082877_minimal residual disease determined by RUNX1/RUNX1T1 transcript levels could identify allogeneic hematopoietic stem cell transplantation t(8;21) (q22;q22) acute myeloid leukemia patients who are at high risk for relapse, together with c-KIT mutations 25101977_In t(8;21) acute myelogenous leukemia (AML), the acquisition of AML1/ETO is not sufficient, and the subsequent upregulation of AML1/ETO and the additional c-KIT mutant signaling are critical steps for transformation into leukemic stem cells. 25928846_A substantial fraction of AML1-ETO/p300 co-localization occurs near TSSs in promoter regions associated with transcriptionally active loci. 26320575_Exon splicing patterns in the human RUNX1-RUNX1T1 fusion gene present in acute myeloid leukemia patients have been decoded. 26546158_AML1/ETO transactivates gene expression through recruiting AP-1 to the AML1/ETO-AML1 complex. 26706127_Data suggest that biosynthesis and folding of leukemogenic fusion oncoprotein AML1-ETO/RUNX1-RUNX1T1 is facilitated by interaction with the chaperonin TRiC/CCT1/TCP1 and HSP70 (heat shock protein 70). 27252013_The specific association of ZBTB7A mutations with t(8;21) rearranged acute myeloid leukaemia points towards leukemogenic cooperativity between mutant ZBTB7A and the RUNX1/RUNX1T1 fusion protein has been reported. 27276256_Study demonstrated that TRiC's contribution to the activity of the DNA-binding domain (AML1-175) of AML1-ETO is consistent with its contribution to the activity of full-length AML1-ETO and is mediated through its direct association with the DNA-binding domain 27522676_The data suggest that the tumor suppressor activity of both RUNX1t1 and TFF1 are mechanistically connected to CEBPB and that cross-regulation between CEBPB-RUNX1t1-TFF1 plays an important role in gastric carcinogenesis. 27725192_the AML1-ETO fusion protein increases the expression of SIRT1, possibly by binding to the promoter region of SIRT1 to activate its transcription in t(8;21) AML. 27770540_A feedback circuitry involving miR-9-1 and RUNX1-RUNX1T1. 27798886_Our data revealed a novel role for RUNX1T1 as a tumor-suppressor gene in colorectal cancer through modulation of multiple cellular pathways affecting the cell cycle, DNA damage, DNA replication, estrogen signaling, and drug resistance 28092997_PKM2 as a novel target of RUNX1-ETO and is specifically downregulated in RUNX1-ETO positive AML patients, indicating that PKM2 level might have a diagnostic potential in RUNX1-ETO associated AML. 28166825_RUNX1-RUNX1T1 transcript levels were measured in bone marrow samples collected from 208 patients at scheduled time points after transplantation. Over 90% of the 175 patients who were in continuous complete remission had a >/=3-log reduction in RUNX1-RUNX1T1 transcript levels from the time of diagnosis at each time point after transplantation and a >/=4-log reduction at >/=12 months. 28322996_The role of RUNX1T1 in t(8;21) acute myeloid leukemia and miRNA expression regulation 28539478_Altogether, these results revealed an unexpected and important epigenetic mini-circuit of AML1-ETO/THAP10/miR-383 in t(8;21) acute myeloid leukaemia, in which epigenetic suppression of THAP10 predicts a poor clinical outcome and represents a novel therapeutic target. 28611288_Data indicate miR-29b-1 as a regulator of the AML1-ETO protein (RUNX1-RUNX1T1), and that miR-29b-1 expression in t(8;21)-carrying leukemic cell lines partially rescues the leukemic phenotype. 28640846_RUNX1T1 serves as a common angiogenic driver for vaculogenesis and functionality of endothelial lineage cells 28650479_Leukaemogenesis by AML1-ETO requires enhanced C/D box snoRNA/RNP formation. 29413154_At levels of condFDR < 0.01 and conjFDR < 0.05; we identified 5 ADHD-associated loci, 3 of these being shared between ADHD and EA. 29472719_identified most of these factors as RUNX1-RUNX1T1 targets, with Ras Homolog Family Member (RHOB) overexpression being the core of this network 29568097_Minimal residual disease (MRD) monitoring and mutational landscape in AML with RUNX1-RUNX1T1. 30050054_In an analysis of an independent validation cohort, KIAA0125 again showed a significant influence with respect to the impact of the RUNX1/RUNX1T1 fusion in mediation of acute myeloid leukemia. 30093631_As most transcriptional repressor proteins do not comprise tetramerization domains, our results provide a possible explanation as to the reason that RUNX1 is recurrently found translocated to ETO family members 30499136_lncRNA RUNX1-IT1 was down regulated both in colorectal cancer tissues and cell lines; besides, lncRNA RUNX1-IT1 could serve as a potential diagnostic biomarker and play a tumour-suppressive role owing to its good diagnostic efficacy and inhibition of colorectal cancer cell proliferation and migration 30569130_Study reports differential regulation of gene expression by RUNX1-RUNX1T1 oncoprotein and MAPK1 as determined by genome-wide expression analysis responsible for the phenotypic features of acute myeloid leukaemia with karyotypic discernible translocation (t)(8;21)(q22;q22). 30637949_found that the binding capacity of RUNX1-ETO9a, a truncated RUNX1-ETO isoform, to the c-KIT promoter was stronger than that of RUNX1-ETO, suggesting RUNX1-ETO9a as another valuable therapeutic target in t(8;21) AML 31119862_Targeting miR-193a-AML1-ETO-beta-catenin axis by melatonin suppresses the self-renewal of leukaemia stem cells in leukaemia with t (8;21) translocation. 31247675_Negative CD19 expression is associated with inferior relapse-free survival in children with RUNX1-RUNX1T1-positive acute myeloid leukaemia: results from the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 study. 31645082_Long non-coding RNA EPB41L4A-AS2 suppresses progression of ovarian cancer by sequestering microRNA-103a to upregulate transcription factor RUNX1T1. 31699991_Findings highlight the role of EZH1 in non-histone lysine methylation, indicating that cooperation between AML1-ETO and EZH1 and AML1-ETO site-specific lysine methylation promote AML1-ETO transcriptional repression in leukemia. 31725954_Morphological characteristics, cytogenetic profile, and outcome of RUNX1-RUNX1T1-positive acute myeloid leukemia: Experience of an Indian tertiary care center. 31839036_fusion transcript level after the first induction therapy in RUNX1-RUNX1T1-positive AML children is an independent factor influencing the long-term prognosis 31869378_AML1-ETO as a single oncogenic hit in a non-mutated background blocks granulocytic differentiation, deregulates the gene program via altering the acetylome of the differentiating granulocytic cells, and induces t(8;21) AML associated leukemic characteristics. 32024815_LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. 32071087_Histone deacetylase 3 preferentially binds and collaborates with the transcription factor RUNX1 to repress AML1-ETO-dependent transcription in t(8;21) AML. 32311890_In acute myeloid leukemia patients with positive RUNX1-RUNX1T1 receiving intensive consolidation therapy, the white blood cell counts at onset of leukemia, C-KIT mutations in exon 17, and FLT3-ITD gene mutations suggest poor prognosis 33067946_[Clinical Prognostic Factors Analysis of Initially Treated AML Children with t(8;21)/RUNX1-RUNX1T1()]. 33084222_Identification of RUNX1T1 as a potential epigenetic modifier in small-cell lung cancer. 33152396_RUNX1/ETO and mutant KIT both contribute to programming the transcriptional and chromatin landscape in t(8;21) acute myeloid leukemia. 33217477_The RUNX1/RUNX1T1 network: translating insights into therapeutic options. 33382982_Definition of a small core transcriptional circuit regulated by AML1-ETO. 33431365_Relationships between AML1-ETO and MLL-AF9 fusion gene expressions and hematological parameters in acute myeloid leukemia. 33483506_RUNX1/RUNX1T1 mediates alternative splicing and reorganises the transcriptional landscape in leukemia. 33524443_Optimized clinical application of minimal residual disease in acute myeloid leukemia with RUNX1-RUNX1T1. 33971010_Myeloid lncRNA LOUP mediates opposing regulatory effects of RUNX1 and RUNX1-ETO in t(8;21) AML. 34120331_Droplet digital polymerase chain reaction assay for the detection of the minor clone of KIT D816V in paediatric acute myeloid leukaemia especially showing RUNX1-RUNX1T1 transcripts. 34264479_RUNX1T1, a potential prognostic marker in breast cancer, is co-ordinately expressed with ERalpha, and regulated by estrogen receptor signalling in breast cancer cells. 34331016_AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms. 34369622_Leukemia associated RUNX1T1 gene reduced proliferation and invasiveness of glioblastoma cells. | ENSMUSG00000006586 | Runx1t1 | 128.97997 | 0.8945245 | -0.1608071761 | 0.25571821 | 3.942414e-01 | 5.300783e-01 | 8.344676e-01 | No | Yes | 169.747600 | 48.528752 | 171.568752 | 48.195072 | |
| ENSG00000079435 | 3991 | LIPE | protein_coding | Q05469 | FUNCTION: Lipase with broad substrate specificity, catalyzing the hydrolysis of triacylglycerols (TAGs), diacylglycerols (DAGs), monoacylglycerols (MAGs), cholesteryl esters and retinyl esters (PubMed:8812477, PubMed:15955102, PubMed:15716583, PubMed:19800417). Shows a preferential hydrolysis of DAGs over TAGs and MAGs and preferentially hydrolyzes the fatty acid (FA) esters at the sn-3 position of the glycerol backbone in DAGs (PubMed:19800417). Preferentially hydrolyzes FA esters at the sn-1 and sn-2 positions of the glycerol backbone in TAGs (By similarity). Catalyzes the hydrolysis of 2-arachidonoylglycerol, an endocannabinoid and of 2-acetyl monoalkylglycerol ether, the penultimate precursor of the pathway for de novo synthesis of platelet-activating factor (By similarity). In adipose tissue and heart, it primarily hydrolyzes stored triglycerides to free fatty acids, while in steroidogenic tissues, it principally converts cholesteryl esters to free cholesterol for steroid hormone production (By similarity). {ECO:0000250|UniProtKB:P15304, ECO:0000250|UniProtKB:P54310, ECO:0000269|PubMed:15716583, ECO:0000269|PubMed:15955102, ECO:0000269|PubMed:19800417, ECO:0000269|PubMed:8812477}. | Alternative splicing;Cell membrane;Cholesterol metabolism;Cytoplasm;Diabetes mellitus;Hydrolase;Lipid degradation;Lipid droplet;Lipid metabolism;Membrane;Obesity;Phosphoprotein;Reference proteome;Steroid metabolism;Sterol metabolism | PATHWAY: Glycerolipid metabolism; triacylglycerol degradation. | The protein encoded by this gene has a long and a short form, generated by use of alternative translational start codons. The long form is expressed in steroidogenic tissues such as testis, where it converts cholesteryl esters to free cholesterol for steroid hormone production. The short form is expressed in adipose tissue, among others, where it hydrolyzes stored triglycerides to free fatty acids. [provided by RefSeq, Jul 2008]. | hsa:3991; | caveola [GO:0005901]; cytosol [GO:0005829]; lipid droplet [GO:0005811]; membrane [GO:0016020]; 1,2-diacylglycerol acylhydrolase activity [GO:0102259]; 1,3-diacylglycerol acylhydrolase activity [GO:0102258]; acylglycerol lipase activity [GO:0047372]; all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity [GO:0047376]; hormone-sensitive lipase activity [GO:0033878]; retinyl-palmitate esterase activity [GO:0050253]; sterol esterase activity [GO:0004771]; triglyceride lipase activity [GO:0004806]; cholesterol metabolic process [GO:0008203]; diacylglycerol catabolic process [GO:0046340]; ether lipid metabolic process [GO:0046485]; lipid catabolic process [GO:0016042]; protein phosphorylation [GO:0006468]; triglyceride catabolic process [GO:0019433] | 11574428_Observational study of gene-disease association. (HuGE Navigator) 11731226_Observational study of gene-disease association. (HuGE Navigator) 11850754_Observational study of gene-disease association. (HuGE Navigator) 11979403_genes of C3, hormone-sensitive lipase, and PPARgamma may exert a modifying effect on lipid and glucose metabolism in familial combined hypersensitivity 12514936_Observational study of gene-disease association. (HuGE Navigator) 12518034_overexpression of HSL, despite increased lipase activity, does not lead to enhanced lipolysis 12534454_HSL i6 A5 HOMOZYGOSITY IS A RISK FACTOR FOR BODY FAT ACCUMULATION 12534454_Observational study of gene-disease association. (HuGE Navigator) 12701046_high concentrations of estradiol significantly increased both hormone-sensitive lipase expression and glycerol release relative to control 12727985_lipolytic catecholamine resistance of sc adipocytes in polycystic ovary syndrome is probably due to a combination of decreased amounts of beta(2)-adrenergic receptors, the regulatory II beta-component of protein kinase A, and hormone-sensitive lipase 12730334_High adrenaline levels can stimulate hormone-sensitive lipase(HSL) activity regardless of metabolic milieu. Large increases in adrenaline during exercise are able to further stimulate contraction-induced increase in HSL activity. 12765952_The presence of a catalytically inactive variant of this enzyme is associated with decreased lipolysis in abdominal subcutaneous adipose tissue of obese subjects 12970365_role of cyclic GMP in natriuretic peptide-mediated phosphorylation in adipocytes 14984467_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15231718_AMPK is a major regulator of skeletal muscle HSL activity that can override beta-adrenergic stimulation 15260473_catalytic serine of hormone-sensitive lipase is highly reactive and similar behavior was also observed with lipases with no lid domain covering their active site, or with a deletion in the lid domain 15308678_5'AMP-activated protein kinase phosphorylates hormone-sensitive lipase on Ser565 in human skeletal muscle during exercise with reduced muscle glycogen. Apparently, HSL Ser565 phosphorylation by AMPK during exercise 15345679_mechanism of infertility in HSL-deficient males is cell autonomous and resides in postmeiotic germ cells, because HSL expression in these cells is in itself sufficient to restore normal fertility 15456755_a pre-lipolysis complex containing at least AFABP and HSL exists 15609025_Basal HSL is decreased in patients with type 2 diabetes mellitus, and this may be a consequence of elevated plasma insulin levels. 15871848_Observational study of gene-disease association. (HuGE Navigator) 16243839_Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase 16534522_Observational study of gene-disease association. (HuGE Navigator) 16534522_the HSL C-60G polymorphism is associated with increased waist circumference in non-obese subjects 16690773_adrenergic stimulation contributes to the increase in HSL activity that occurs in human skeletal muscle in the first minute of exercise at 65% and 90% VO2 peak 16752181_HSL gene expression shows a regulation according to obesity status and is associated with increased adipose tissue lipase activity. 16822962_although HSL expression and Ser(659) phosphorylation in skeletal muscle during exercise is sex specific, total muscle HSL activity measured in vitro was similar between sexes 16940551_maternal type 1 diabetes is associated with TG accumulation and increased EL and HSL gene expression in placenta 17026959_hormone-sensitive lipase 17074755_Adipose triglyceride lipase and hormone sensitive lipase are responsible for more than 95% of the triacylglycerol hydrolase activity present in murine white adipose tissue. 17318300_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17318300_The combined effect of LIPC, LIPE and ApoCIII gene polymorphisms may increase the likelihood of gestational hypertension, but seemingly not of preeclampsia. 17327373_Adipose triglyceride lipase (ATGL) is less important than hormone-sensitive lipase (HSL) in regulating catecholamine-induced lipolysis. Both lipases regulate basal lipolysis in human adipocytes. ATGL expression is not influenced by obesity or PCOS. 17356053_In obese subjects, insulin resistance and hyperinsulinemia are strongly associated with ATGL and HSL mRNA and protein expression, independent of fat mass 17587400_Dydrogesterone and norethisterone increase secretion of HSL from abdominaal adipocytes. 18249203_Observational study of gene-disease association. (HuGE Navigator) 18249203_variation of the HSL gene might be associated with a physiological effect on in vivo beta-adrenoceptor-mediated fat oxidation 18383440_Real-time PCR revealed that large adipocytes expressed higher mRNA levels of hormone sensitive lipase. 18398140_Obesity is accompanied by impaired fasting glycerol release, lower HSL protein expression, and serine phosphorylation. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18820256_Mapping of the hormone-sensitive lipase binding site on the adipocyte fatty acid-binding protein (AFABP). Identification of the charge quartet on the AFABP/aP2 helix-turn-helix domain. 18824087_analysis of human, mouse and ovine Hormone Sensitive Lipase 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19018281_PKA activates human HSL against lipid substrates in vitro primarily through phosphorylation of Ser649 and Ser650 19164092_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19164092_These results suggest that the associations between physical activity and body fat and plasma lipoprotein/lipid concentrations in men are dependent on the LIPE C-60G polymorphism,. 19336475_Observational study of gene-disease association. (HuGE Navigator) 19369647_Observational study of gene-disease association. (HuGE Navigator) 19433586_results suggest that ATGL/CGI-58 acts independently of HSL and precedes its action in the sequential hydrolysis of triglycerides in human hMADS adipocytes 19491387_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19695247_TNFalpha decreased ATGL and HSL protein content and triglycerides (TG)-hydrolase activity but increased basal lipolysis due to a marked reduction in perilipin (PLIN) protein content 19913121_Observational study of gene-disease association. (HuGE Navigator) 20017959_The present study aimed at comparing expression and subcellular distribution of perilipin and hormone-sensitive lipase in two abdominal adipose tissues of lean and obese women. 20495294_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20495294_Those findings indicate improvement and conservation of lifestyle depending on genetic predisposition in ADIPOQ, PLIN and LIPE should be encouraged. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20734064_Observational study of gene-disease association. (HuGE Navigator) 20855565_Observational study of gene-disease association. (HuGE Navigator) 20926921_Data show that hormone-sensitive lipase activity is reduced in adipose tissue of patients with and without diabetes, while lipoprotein lipase activity is reduces only in patients with diabetes. 21081692_It is concluded that ACTH via the PKA pathway stimulates expression of SF-1, which activates transcription of LIPE presumably by interaction with putative binding sequences within promoter A 21241784_Studies indicate tha HSL is regulated by reversible phosphorylation on five critical residues. 21498783_Data indicate that altered ATGL and HSL expression in skeletal muscle could promote DAG accumulation and disrupt insulin signaling and action. 21543206_Resveratrol increased adipose triglyceride lipase gene and protein expressions, an effect that was not observed for hormone-sensitive lipase in human SGBS adipocytes. 21680814_total lipase, ATGL and HSL activities were higher in visceral white adipose tissue of cancer patients compared with individuals without cancer and higher in cancer patients with cachexia compared with cancer patients without cachexia 21826994_LIPE C-60G variation can inhibit the decrease of LDL-C and the increases of HDL-C and apo A-I in young healthy males, and can inhibit the decrease of LDL-C and the increase of insulin in young healthy females induced by a high-carbohydrate diet. 21919688_suggests that genetic variation of HSL may be a risk factor for male infertility 21933124_Enzyme promiscuity in the hormone-sensitive lipase family of proteins. 22553833_M. leprae suppresses lipid degradation through inhibition of HSL expression. 23688034_Serum triglyceride was significantly up-regulated in men with the (CG + GG) genotype of HSL promoter polymorphism. 24848981_These findings indicate the physiological significance of HSL in adipocyte function and the regulation of systemic lipid and glucose homeostasis and underscore the severe metabolic consequences of impaired lipolysis. 25475467_Identification of a homozygous nonsense variant p.Ala507fsTer563 in hormone sensitive lipase as the likely cause of the lipodystrophy phenotype in siblings. 25819461_Despite reductions in intramyocellular lipolysis and HSL expression, overexpression of HSL did not rescue defects in insulin action in skeletal myotubes from obese type 2 diabetic subjects. 27862896_The homozygous null LIPE mutation could result in marked inhibition of lipolysis from some adipose tissue depots and thus may induce an extremely rare phenotype of MSL and partial lipodystrophy in adulthood associated with complications of insulin resistance, such as diabetes, hypertriglyceridemia and hepatic steatosis. 31150775_HSL regulates vitamin A metabolism in quiescent hepatic stellate cells. 33445064_Dietary fat content and adipose triglyceride lipase and hormone-sensitive lipase gene expressions in adults' subcutaneous and visceral fat tissues. | ENSMUSG00000003123 | Lipe | 92.07369 | 1.1353754 | 0.1831693629 | 0.29687942 | 3.817557e-01 | 5.366651e-01 | 8.381594e-01 | No | Yes | 136.327883 | 25.546082 | 116.379040 | 21.019956 |
| ENSG00000079819 | 2037 | EPB41L2 | protein_coding | O43491 | FUNCTION: Required for dynein-dynactin complex and NUMA1 recruitment at the mitotic cell cortex during anaphase (PubMed:23870127). {ECO:0000269|PubMed:23870127}. | Acetylation;Actin-binding;Alternative splicing;Cell cycle;Cell division;Cell membrane;Cytoplasm;Cytoskeleton;Direct protein sequencing;Isopeptide bond;Membrane;Mitosis;Phosphoprotein;Reference proteome;Transport;Ubl conjugation | hsa:2037; | cell cortex [GO:0005938]; cell junction [GO:0030054]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; spectrin [GO:0008091]; actin binding [GO:0003779]; PH domain binding [GO:0042731]; spectrin binding [GO:0030507]; structural molecule activity [GO:0005198]; actomyosin structure organization [GO:0031032]; cell cycle [GO:0007049]; cell division [GO:0051301]; cortical actin cytoskeleton organization [GO:0030866]; positive regulation of protein localization to cell cortex [GO:1904778] | 15138281_PTA-1(CD226 antigen) is localized to membrane rafts and binds the carboxyl-terminal domain of isoforms of the actin-binding protein 4.1G and also can bind human discs large providing the structural basis for a regulated molecular adhesive complex 18023480_An alternatively spliced 4.1G product is shown to be associated with increased Fc gamma RI binding in yeast two-hybrid assays, and to be selectively enriched in most immune cells at the transcript level. 20189245_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22003208_A phosphoserine-dependent tethering role for protein 4.1G in lipid rafts provides insight into the unique phosphoserine-based regulation of FcgammaRI receptor signaling. 23201780_4.1G protein depresses PTH-related protein receptor-mediated Gs signaling pathway by suppression of adenylyl cyclase-mediated cAMP production. 23354586_Ca(2+)/CaM binding to GHP, and more specifically to pepG, has profound effects on other functional domains of 4.1G 31383768_Data found that direct binding of 4.1G to the N-terminus of AC6 (AC6-N) contributes to the plasma membrane association of AC6-N and the resulting suppression of AC6 cyclase activity. 34022068_Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. | ENSMUSG00000019978 | Epb41l2 | 3860.08865 | 1.0138023 | 0.0197762788 | 0.07918925 | 6.240423e-02 | 8.027355e-01 | 9.449080e-01 | No | Yes | 4877.959152 | 702.041672 | 4528.222901 | 637.240574 | ||
| ENSG00000080709 | 3781 | KCNN2 | protein_coding | Q9H2S1 | FUNCTION: Forms a voltage-independent potassium channel activated by intracellular calcium (PubMed:10991935, PubMed:9287325). Activation is followed by membrane hyperpolarization. Thought to regulate neuronal excitability by contributing to the slow component of synaptic afterhyperpolarization. {ECO:0000269|PubMed:10991935, ECO:0000269|PubMed:9287325}. | 3D-structure;Alternative splicing;Calmodulin-binding;Ion channel;Ion transport;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | Action potentials in vertebrate neurons are followed by an afterhyperpolarization (AHP) that may persist for several seconds and may have profound consequences for the firing pattern of the neuron. Each component of the AHP is kinetically distinct and is mediated by different calcium-activated potassium channels. The protein encoded by this gene is activated before membrane hyperpolarization and is thought to regulate neuronal excitability by contributing to the slow component of synaptic AHP. This gene is a member of the KCNN family of potassium channel genes. The encoded protein is an integral membrane protein that forms a voltage-independent calcium-activated channel with three other calmodulin-binding subunits. Alternate splicing of this gene results in multiple transcript variants. [provided by RefSeq, May 2013]. | hsa:3781; | cell surface [GO:0009986]; dendritic spine [GO:0043197]; integral component of membrane [GO:0016021]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; plasma membrane [GO:0005886]; Z disc [GO:0030018]; alpha-actinin binding [GO:0051393]; calcium-activated potassium channel activity [GO:0015269]; calmodulin binding [GO:0005516]; inward rectifier potassium channel activity [GO:0005242]; protein domain specific binding [GO:0019904]; protein homodimerization activity [GO:0042803]; small conductance calcium-activated potassium channel activity [GO:0016286]; ion transport [GO:0006811]; membrane repolarization during atrial cardiac muscle cell action potential [GO:0098914]; potassium ion transmembrane transport [GO:0071805]; potassium ion transport [GO:0006813]; regulation of potassium ion transmembrane transport [GO:1901379] | 12778407_RT-PCR analysis showed strong expression of SK2 mRNA in the normal human colon. 13679367_Because of the marked differential expression of SK2 channels in the heart, specific ligands for Ca2+-activated K+ currents may offer a unique therapeutic opportunity to modify atrial cells without interfering with ventricular myocytes 15362045_SK2 plays an important role in mediating the increase in transepithelial secretion due to increases in intracellular Ca 2+. SK2 channels, therefore, may represent a target for pharmacologic modulation of bile flow. 16396931_The subtype SK2 channels were up-regulated under hypoxia, shown with pharmacological tools and with mRNA analysis. 17110593_Functions of SK2 channels in atrial myocytes are critically dependent on the normal expression of Ca(v)1.3 Ca(2+) channels. 17452806_Results suggest that SK2-channel activation may largely contribute to the sustained Ca2+ influx in the G0/G1 phase in comparison of that in the G2/M phase in Jurkat T-lymphocytes. 19139040_Present study directly defines the functional roles of SK2 channels in transgenic mice using a genetically engineered model, and provides a possible link between abnormalities in cardiac SK2 channels and cardiac arrhythmias. 19815520_demonstrate that proper membrane localization of a small-conductance Ca(2+)-activated K(+) channel (SK2 or K(Ca)2.2) is dependent on its interacting protein, alpha-actinin2, a major F-actin crosslinking protein. 22154908_Decreased expression of small-conductance Ca2+-activated K+ channels SK1 and SK2 in human chronic atrial fibrillation 23525437_Increase in both Ca(2+) sensitivity and SK2 protein expression contributes to the IKAS upregulation in failing human ventricles. 23677057_KCNN2 gene can have an important role in the development of coronary artery aneurysms in Kawasaki disease. 24434522_Differentiated dopaminergic neurons expressed low levels of SK2 channels and high levels of SK1 and SK3 channels. 26586570_The ER SK2 channel activation preserves ER Ca(2+) uptake and retention which determines cell survival in conditions where sustained ER stress contributes to progressive neuronal death. 27357310_Study establishes the distribution profile of SK2 channel protein in human brain.The expression of SK2 human isoform b in brain could explain the variability of electrophysiological findings observed with SK2 channels. 27442679_There was a significant association between the KCNN2 variants and clinically significant VTa. These findings suggest an association between KCNN2 and VTa; it also appears that KCNN2 variants may be adjunctive markers for risk stratification in patients susceptible to SCD. 27779751_These results provide new insights into the regulation of SK2 channel trafficking by the cytoskeletal proteins FLNA and alpha-actinin2, involving distinct recycling pathways 29055091_Junctophilin 2, as junctional membrane complex (JMC) protein, is an important regulator of the cardiac SK channels 29737974_SK current is increased via the enhanced activation of CaMKII in patients with atrial fibrillation. 30922569_Immuno-visualization of the subcellular localization of SK2 and SK3 subunits showed a high degree of colocalization, consistent with the formation of heteromeric SK2/SK3 channels. 32212350_KCNN2 mutation in autosomal-dominant tremulous myoclonus-dystonia. 32860835_SK2 channel regulation of neuronal excitability, synaptic transmission, and brain rhythmic activity in health and diseases. 33242881_Variants in the SK2 channel gene (KCNN2) lead to dominant neurodevelopmental movement disorders. 33310041_HDAC2-dependent remodeling of KCa2.2 (KCNN2) and KCa2.3 (KCNN3) K(+) channels in atrial fibrillation with concomitant heart failure. | ENSMUSG00000054477 | Kcnn2 | 50.88315 | 0.6260400 | -0.6756731916 | 0.40197497 | 2.782990e+00 | 9.527017e-02 | No | Yes | 46.213621 | 9.072200 | 69.960453 | 12.763690 | ||
| ENSG00000084676 | 8648 | NCOA1 | protein_coding | Q15788 | FUNCTION: Nuclear receptor coactivator that directly binds nuclear receptors and stimulates the transcriptional activities in a hormone-dependent fashion. Involved in the coactivation of different nuclear receptors, such as for steroids (PGR, GR and ER), retinoids (RXRs), thyroid hormone (TRs) and prostanoids (PPARs). Also involved in coactivation mediated by STAT3, STAT5A, STAT5B and STAT6 transcription factors. Displays histone acetyltransferase activity toward H3 and H4; the relevance of such activity remains however unclear. Plays a central role in creating multisubunit coactivator complexes that act via remodeling of chromatin, and possibly acts by participating in both chromatin remodeling and recruitment of general transcription factors. Required with NCOA2 to control energy balance between white and brown adipose tissues. Required for mediating steroid hormone response. Isoform 2 has a higher thyroid hormone-dependent transactivation activity than isoform 1 and isoform 3. {ECO:0000269|PubMed:10449719, ECO:0000269|PubMed:12954634, ECO:0000269|PubMed:7481822, ECO:0000269|PubMed:9223281, ECO:0000269|PubMed:9223431, ECO:0000269|PubMed:9296499, ECO:0000269|PubMed:9427757}. | 3D-structure;Acetylation;Activator;Acyltransferase;Alternative splicing;Chromosomal rearrangement;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene acts as a transcriptional coactivator for steroid and nuclear hormone receptors. It is a member of the p160/steroid receptor coactivator (SRC) family and like other family members has histone acetyltransferase activity and contains a nuclear localization signal, as well as bHLH and PAS domains. The product of this gene binds nuclear receptors directly and stimulates the transcriptional activities in a hormone-dependent fashion. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:8648; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; RNA polymerase II transcription regulator complex [GO:0090575]; transcription regulator complex [GO:0005667]; aryl hydrocarbon receptor binding [GO:0017162]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; enzyme binding [GO:0019899]; estrogen receptor binding [GO:0030331]; histone acetyltransferase activity [GO:0004402]; nuclear receptor binding [GO:0016922]; nuclear receptor coactivator activity [GO:0030374]; protein dimerization activity [GO:0046983]; protein N-terminus binding [GO:0047485]; transcription coactivator activity [GO:0003713]; cellular response to hormone stimulus [GO:0032870]; cellular response to Thyroglobulin triiodothyronine [GO:1904017]; histone H4 acetylation [GO:0043967]; labyrinthine layer morphogenesis [GO:0060713]; mRNA transcription by RNA polymerase II [GO:0042789]; positive regulation of apoptotic process [GO:0043065]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription from RNA polymerase II promoter by galactose [GO:0000435]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of thyroid hormone mediated signaling pathway [GO:0002155] | 11773079_CBP/p300 and NcoA/SRC1a may function in a common pathway to regulate STAT3 transcriptional activity. 11818499_Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation 12114525_CAR antagonizes ER-mediated transcriptional activity by squelching limiting amounts of p160 coactivators, such as SRC-1 and GRIP-1. 12118039_mutation of the AF2 transactivation domain on Glucocorticoid Receptors disrupts the direct interaction of GR with steroid receptor coactivator 1 (SRC-1) 12138096_STAT6 has a protein binding motif that controls the interaction with NcoA-1 in transcriptional activation 12163482_Data show that steroid receptor coactivator-1 (SRC-1) enhanced ligand-independent activation of the AR by IL-6 to the same magnitude as that obtained via ligand-dependent activation, and that activation required MAPK. 12403846_Overexpression of SRC-1 (NCOA1) and TIF-2 (NCOA2) increases estrogen-induced gene expression. 12482985_our results indicate an important role for Mediator, as well as its functional interplay with p300/CBP-SRC, in the enhancement of ERalpha-dependent transcription with chromatin templates 12529333_Sumoylation was shown to increase progesterone receptor-SRC-1 interaction and to prolong SRC-1 retention in the nucleus. 12933903_is a coactivator of MHC class II genes that stimulates their interferon gamma (IFNgamma) and class II transactivator (CIITA)-mediated expression. 12954634_NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain 14715875_differential recruitment of steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors by estrogen receptor-alpha and beta in breast cancer may be central to the response of the tumor to endocrine treatment 14757047_surface complementarity between the hydrophobic faces of the STAT6 fragment and of the NCoA-1 PAS-B domain almost exclusively defines the binding specificity between the two proteins 15231721_TNF-alpha impairs progesterone-stimulated PR-B-mediated transactivation, and these effects appear to be due, in part, to reduced expression of SRC-1 and -2 15313887_A novel translocation t(2;2)(q35;p23), which generates a fusion protein composed of PAX3 and the NCOA1was identified in rhabdomyosarcoma. 15456935_Data show that steroid receptor coactivator-1 (SRC-1) overexpressing cells are more responsive to Po mRNA induction by dihydroprogesterone (DHP) than SRC-1-deficient cells, and that DHP treatment increases not only Po but also SRC-1 mRNA levels. 15530426_The conserved STAT3 region from 752 to 761, called STAT3 CR2, plays critical roles in STAT3-dependent transcription by recruiting SRC-1 and allowing Ser727 phosphorylation. 15615775_The SRC-1 has been shown to be involved in HIF-1alpha-mediated activation of transcription. 15862975_SRC-1 and SMRT content change in a tissue-specific manner in the rat brain during the estrous cycle. 16269961_Observational study of gene-disease association. (HuGE Navigator) 16606617_ASXL1 is a novel coactivator of RAR that cooperates with SRC-1 16723356_SRC1-deficient P19 cells showed severely compromised retinoid-induced responses, in agreement with the supposed role of SRC1 as a retinoic acid recepetor coactivator. 16775354_HPV E7 proteins dysregulate hormone-dependent gene expression by association with and relocalization of SRC-1. 16882880_These results establish the importance of coactivators PGC1alpha and SRC1 for the hepatic expression of human P450s and uncover a new HNF4alpha-dependent regulatory mechanism to constitutively control the CYP1A1/2 cluster. 17135255_Binding and dissociation of full-length steroid receptor coactivator-1a (SRC1a) from full-length estrogen receptor alpha or beta. 17429319_not only the Q158K variant found in Chinese, but also in native pregnane X receptor variants in other ethnic groups (D163G, A370T, R381W, and I403V) affect CYP3A4 induction by altering steroid receptor coactivator-1 recruitment 17919607_Expression of androgen receptor co-regulators in the testes of men with azoospermia. 18284209_The main transactivation function of the androgen receptor interacts with both the C-terminal domain of coactivator SRC-1a and the general transcription factor RAP74/TFIIF large subunit. 18660489_Observational study of gene-disease association. (HuGE Navigator) 18845648_important role for androgen receptor-associated coactivators in urothelial carcinoma of the bladder 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19095746_The small interfering RNA depletion of SRC-1 did not inhibited growth of MCF-7 cells as well as a reduction in ERalpha transcriptional activity. 19183483_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19203349_STAT3 transactivates its target genes by the recruitment of CBP/p300 co-activators and this process generally does not require the contribution of SRC-1. 19255064_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19276281_SRC-1 is a strong independent predictor of reduced disease free survival, whereas the interactions of the p160 proteins with estrogen receptor alpha can predict the response of patients to endocrine treatment. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19485965_Age-induced loss of PPARgamma/SRC-1 interactions increased the binding of PPARgamma to the promoter of the adipogenic gene aP2. 19763509_describes in detail the BRET(2) assay as it is used to examine the physical interaction between the nuclear receptor ERalpha and the transcriptional coactivator SRC-1 19913121_Observational study of gene-disease association. (HuGE Navigator) 19934375_Clomiphene citrate may inhibit E2-induced endometrial epithelial cell proliferation and ERE transactivation by inhibiting the recruitment of SRC-1 to ER alpha. 19953635_Studies expand knowledge of PAX3 variant translocations in rhabdomyossarcoma with identification of a novel PAX3-NCOA2 fusion. 20003447_Observational study of gene-disease association. (HuGE Navigator) 20145129_HOXC11 and SRC-1 cooperate to regulate expression of the calcium-binding protein S100beta in resistant breast cancer cells. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20682316_NR box-2 and -3 are the essential binding targets for the SRC-1-induced stimulation of LXR transactivity; the competitive in vitro binding of NR box-2 and -3 to LXR was observed. 21059860_Distinctive functions of p160 steroid receptor coactivators in proliferation of an estrogen-independent, tamoxifen-resistant breast cancer cell line. 21760925_The induced AF1 conformation facilitates its interaction with SRC-1, and subsequent AF1-mediated transcriptional activity. 22072566_findings suggest that SRC-1 switches steroid-responsive tumors to a steroid-resistant state in which the SRC-1 target gene ADAM22 has a critical role 22142990_Results demonstrate that estradiol induces growth of human astrocytoma cell lines through ERalpha and its interaction with SRC-1 and SRC-3 and also suggest differential roles of ERalpha on cell growth depending on astrocytoma grade. 22174377_We have identified a functional genetic variant of SRC-1 with decreased activity, resulting, at least in part, from the loss of a GSK3beta phosphorylation site, which was also associated with decreased bone mineral density in tamoxifen-treated women. 22328528_Binding of the ERalpha and ARNT1 AF2 domains to exon 21 of the SRC1 isoform SRC1e is essential for estrogen- and dioxin-transcription. 22371500_Our data demonstrate that SIP is a novel regulator in caspase-independent and AIF-mediated apoptosis. 24189439_IL-6 secreted by ovarian cancer cells may contribute to the refractoriness of these cells to tamoxifen via the estrogen receptor alpha isoform and SRC-1 (NCOA1). 24438340_SRC-1 responds to cellular energy stress in tumor cells to promote the expression of genes responsible for oxidative phosphorylation. 24586072_NCOA1 plays a necessary role in E2-induced CXCL12 expression and NCOA2 is required for P4 to inhibit the E2-induced CXCL12 production in normal and ectopic endometrium. 24648347_Elucidation of how SRC-1 controls transcription in breast cancers. 24652666_SRC-1 polymorphisms may be the underlying cause of coronary artery aneurysms in children with Kawasaki disease. 24719557_SRC1 and Twist1 expression are associated with poor survival in breast cancer. 24769444_In a cohort of 453 human breast tumors, NCOA1 and CSF1 levels correlated positively with disease recurrence, higher tumor grade, and poor prognosis. 24875297_Our study clearly demonstrated differentiation-dependant expression of SRC-1 and SRC-3 in chondrosarcoma 25053412_Data show that disrupting the steroid receptor coactivator-1 (SRC1) binding site on retinoid X receptor alpha (RXRalpha) alters the transactivation by CAR:RXR. 26066330_miR-137 has a role in targeting p160 steroid receptor coactivators SRC1, SRC2, and SRC3 and inhibits cell proliferation 26203193_Data indicate that finerenone inhibits mineralocorticoid receptor (MR), steroid receptor coactivator-1 binding at the regulatory sequence. 26267537_Data suggest that over-stimulating the steroid seceptor coactivators SRC-1, SRC-2, and SRC-3 oncogenic program can be an effective strategy to kill cancer cells. 26287601_NCOA1 potentiates breast cancer angiogenesis through upregulating HIF1alpha and AP-1-mediated VEGFa expression 26371783_Report novel PAX3-NCOA1 gene fusions in biphenotypic sinonasal sarcomas with focal rhabdomyoblastic differentiation. 26920453_The pregnane X receptor down-regulates organic cation transporter 1 (SLC22A1) in human hepatocytes by competing for ('squelching') SRC-1 coactivator. 27239967_IL-6-mediated AR antagonism induced by cypermethrin is related to repress the recruitment of co-regulators SRC-1 and SMRT to the AR in a ligand-independent manner 27255895_Our findings directly associate the AR/NCOA1 complex with PRKD1 regulation and cellular migration and support the concept of therapeutic inhibition of NCOA1 in prostate cancer. 27842582_miR-4443 acts in a tumor-suppressive manner by down-regulating TRAF4 and NCOA1 downstream of MEK-C/EBP-mediated leptin and insulin signaling, and that insulin and/or leptin resistance (e.g. in obesity) may suppress this pathway and increase the risk of metastatic CRC. 28060733_Results provide the first evidence that NCOA1 is a direct target of miR-105-1 suggesting that NCOA1 and miR-105-1 may have potential prognostic value and may be useful as tumor biomarkers for the diagnosis of HCC patients. 28264017_Our study indicated that genotype and allele frequencies of rs79480871 showed strong associations with MM patients (pa = 3.5x10-4 and pa = 1.5x10-4), and the rs6457327 genotype was more readily associated with MM patients than with controls 28286232_SRC-1 can impact osteoblast function in an estrogen receptor-independent manner. 28390937_Data suggest that steroid receptor coactivators (NCOA1, NCOA2, NCOA3) are over-expressed in a number of hormone-dependent cancers where they promote tumor growth, invasion, metastasis, and chemo-resistance; with their multiple roles in cancer, steroid receptor coactivators are promising targets for development of antineoplastic agents that can interfere with their function. [REVIEW] 28396297_The mechanism of IL-6-induced AR activation is mediated through enhancing AR-SRC-1 interaction and inhibiting AR-SMRT interaction. 29367763_SRC-1 can partner with STAT1 independently of the estrogen receptor to initiate a transcriptional cascade and control regulation of key endocrine resistant genes in breast cancer. 29717026_NCOA1 interacts with NF-kappaB to increase VEGFC levels in human thyroid cancer. 30275017_FXR and RXR bind to the transcriptional coregulator steroid receptor coactivator 1 with higher affinity when they are part of the heterodimer complex than when they are in their respective monomeric states. 30973923_Src-1 and Twist1 were aberrantly upregulated in nasopharyngeal carcinoma (NPC) tissues, and associated with advanced tumor stage, distant metastasis and unfavorable prognosis. Knockdown of Src-1 or Twist1 in NPC cell line CNE-1 suppressed colony formation, anchorage-independent growth, cell migration, invasion and tumor xenografts growth. 30979869_fifteen rare heterozygous variants in SRC-1 found in severely obese individuals impair leptin-mediated Pomc reporter activity in cells, whilst four variants found in non-obese controls do not 32187044_SRF-FOXO1 and SRF-NCOA1 gene fusion occurs in a subset of well differentiated rhabdomyosarcoma of the neck muscles. 32963012_A mutant form of ERalpha associated with estrogen insensitivity affects the coupling between ligand binding and coactivator recruitment. 33090636_Steroid receptor coactivator-1 enhances the stemness of glioblastoma by activating long noncoding RNA XIST/miR-152/KLF4 pathway. 33574497_Recurrent MEIS1-NCOA2/1 fusions in a subset of low-grade spindle cell sarcomas frequently involving the genitourinary and gynecologic tracts. 33576087_Ependymoma-like tumor with mesenchymal differentiation harboring C11orf95-NCOA1/2 or -RELA fusion: A hitherto unclassified tumor related to ependymoma. 34272753_PRRX1-NCOA1-rearranged fibroblastic tumour: a clinicopathological, immunohistochemical and molecular genetic study of six cases of a potentially under-recognised, distinctive mesenchymal tumour. | ENSMUSG00000020647 | Ncoa1 | 1141.79194 | 1.1474352 | 0.1984126771 | 0.13098135 | 2.304830e+00 | 1.289724e-01 | 5.042108e-01 | No | Yes | 1536.627565 | 253.797956 | 1262.579958 | 204.045549 | |
| ENSG00000085382 | 57531 | HACE1 | protein_coding | Q8IYU2 | FUNCTION: E3 ubiquitin-protein ligase involved in Golgi membrane fusion and regulation of small GTPases. Acts as a regulator of Golgi membrane dynamics during the cell cycle: recruited to Golgi membrane by Rab proteins and regulates postmitotic Golgi membrane fusion. Acts by mediating ubiquitination during mitotic Golgi disassembly, ubiquitination serving as a signal for Golgi reassembly later, after cell division. Specifically interacts with GTP-bound RAC1, mediating ubiquitination and subsequent degradation of active RAC1, thereby playing a role in host defense against pathogens. May also act as a transcription regulator via its interaction with RARB. {ECO:0000269|PubMed:15254018, ECO:0000269|PubMed:21988917, ECO:0000269|PubMed:22036506}. | ANK repeat;Alternative splicing;Cell cycle;Chromosomal rearrangement;Cytoplasm;Disease variant;Endoplasmic reticulum;Golgi apparatus;Hereditary spastic paraplegia;Membrane;Neurodegeneration;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a HECT domain and ankyrin repeat-containing ubiquitin ligase. The encoded protein is involved in specific tagging of target proteins, leading to their subcellular localization or proteasomal degradation. The protein is a potential tumor suppressor and is involved in the pathophysiology of several tumors, including Wilm's tumor. [provided by RefSeq, Mar 2016]. | hsa:57531; | cytoplasm [GO:0005737]; endoplasmic reticulum [GO:0005783]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; nuclear body [GO:0016604]; nucleus [GO:0005634]; small GTPase binding [GO:0031267]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin-protein transferase activity [GO:0004842]; cell cycle [GO:0007049]; Golgi organization [GO:0007030]; membrane fusion [GO:0061025]; positive regulation of protein catabolic process [GO:0045732]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein K48-linked ubiquitination [GO:0070936]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; Rac protein signal transduction [GO:0016601]; regulation of cell migration [GO:0030334]; ubiquitin-dependent protein catabolic process [GO:0006511] | 15254018_Hace1 as a novel ubiquitin-protein ligase with very low expression Wilms' tumors; hypermethylation of two upstream CpG islands correlates with low HACE1 expression in tumor samples. 17694067_HACE1 is a candidate chromosome 6q21 tumor-suppressor gene involved in multiple cancers. 18630515_Aberrant methylation of the HACE1 gene was detected in 9 out of the 32 (28%) primary colon carcinomas. 19528486_Aberrant methylation of the HACE1 gene is associated with gastric carcinoma. 19965620_Deregulation of the tumor suppressor HACE1 is associated with natural killer/T-cell lymphoma, nasal type. 20546612_Observational study of gene-disease association. (HuGE Navigator) 21326284_The 6q21-22 region was confirmed as a celiac disease susceptibility locus and harbors multiple independent associations, some of which may implicate ubiquitin-pathways in celiac disease susceptibility, such as mutations in gene HACE1. 22036506_The HACE1 expression increases the ubiquitylation of Rac1, when the GTPase is activated by point mutations or by the GEF-domain of Dbl. 22614015_HACE1 is an antagonist of cell migration through its ability to degrade active Rac1. 22941191_common variants in HACE1 and LIN28B influence neuroblastoma susceptibility and indicate that both genes likely have a role in disease progression. 23142381_HACE1 is tumor suppressor gene located within the 6q21 region, and loss of function of multiple tumor suppressor genes within 6q21 may be a critical determinant of NK cell lymphomagenesis. 23732777_All stages of HCCs presented HACE1 methylation, indicating that the HACE1 gene has been methylated from the early stages of HCCs. 24190883_Ubiquitylation and activation of RAB11a is promoted by a beta2 adrenergic receptor-HACE1 complex. 24516159_HACE1 reduces oxidative stress and mutant Huntingtin toxicity by promoting the NRF2 response. 25026213_HACE1-OPTN axis synergistically suppresses growth and tumorigenicity of lung cancer cells. 25203537_contrary to previous report, overexpression of HACE1 by transduction of recombinant protein did not affect proliferation or survival of NKTCL cell lines. We therefore conclude that HACE1 is not directly involved in NKTCL pathophysiology 25659579_supports a critical role for HACE1 in breast cancer progression and identifies patients that may benefit from Rac-targeted therapies 26343856_HACE1 as a specific E3 ligase that polyubiquitinates YB-1 through non-canonical K27 linked ubiquitin chains. 26424145_Biallelic loss-of-function mutations in HACE1 are associated with an autosomal recessive neurodevelopmental syndrome. 27107267_HACE1 can act as a haploinsufficient tumor suppressor gene in most B-cell lymphomas and can be downregulated by deacetylation of its promoter region chromatin 27213432_Mechanistic studies showed that HACE1 exerts its inhibitory role on virus-induced signaling by disrupting the MAVS-TRAF3 complex. 27805249_Hepatocellular carcinoma patients with low HACE1 expression levels exhibited poorer overall survival and HACE1 was found to be an independent prognostic factor for survival. 28317937_A functional interplay between HACE1 and Rac1 in cancer, is reported. 28651105_3-deazaneplanocin A treatment increased HACE1 gene expression which was further increased by the addition of trichostatine A (TSA), a promising therapeutic compound for the treatment of human B-Lymphoma 29243987_Results indicated that HACE1 rs9404576 polymorphism may be associated with Wilms' tumor susceptibility in the Chinese population. 29362425_Results identified HACE1 Ser-385 as a pivotal amino acid residue that is phosphorylated in response to CNF1 or VEGF treatments and that expression of activated Rac1 is sufficient to induce HACE1 phosphorylation and controlling its oligomerization state. 29515254_HACE1 promoted the K27 ubiquitination of fibronectin and regulated its secretion. Secreted fibronectin regulated ITGAV and ITGB1 expression, as well as melanoma cell adhesion and migration. 29673126_our studies unveiled a suppressive role of HACE1 in tumor growth and migration of gastric cancer, and it might help to provide novel insights into the blockage of tumorigenesis and malignant process of early stage of gastric cancer. 30362561_HACE1-YAP1 axis had an important part in the Colorectal cancer development and progression. 30622235_Ectopic expression of HACE1 markedly inhibited anchorage-independent growth and cell motility of HACE1 osteosarcoma cell lines, HACE1 overexpression blocked osteosarcoma xenograft growth and dramatically reduced pulmonary metastases. These findings point to a potential tumor suppressor function for HACE1 in osteosarcoma. 30659265_FIH interacts with the ankyrin repeat domain of HACE1.HACE1 is hydroxylated at asparagine 191 by FIH.FIH-dependent hydroxylation of HACE1 inhibits HACE1 ability to ubiquitinate Rac1.HACE1 expression inversely correlates with breast cancer progression. 30787155_The present study demonstrates that KSHV infection induces the E3 ligase HACE1 protein to regulate KSHV-induced oxidative stress by promoting the activation of Nrf2 and nuclear translocation. 30827236_Loss of the Tumor Suppressor HACE1 Contributes to Cancer Progression. 32616021_meQTL and ncRNA functional analyses of 102 GWAS-SNPs associated with depression implicate HACE1 and SHANK2 genes. 33421097_Ubiquitin-independent proteasomal degradation of Spindlin-1 by the E3 ligase HACE1 contributes to cell-cell adhesion. 33455098_Correlations of HACE1 expression with pathological stages, CT features and prognosis of hepatocellular carcinoma patients. 33603169_HACE1 blocks HIF1alpha accumulation under hypoxia in a RAC1 dependent manner. 34593974_HACE1 negatively regulates neuroinflammation through ubiquitylating and degrading Rac1 in Parkinson's disease models. 34815381_HACE1-mediated NRF2 activation causes enhanced malignant phenotypes and decreased radiosensitivity of glioma cells. | ENSMUSG00000038822 | Hace1 | 511.50638 | 0.9707725 | -0.0427948604 | 0.14302742 | 8.937623e-02 | 7.649716e-01 | 9.326662e-01 | No | Yes | 615.582976 | 104.967368 | 613.816020 | 102.565571 |
| ENSG00000085719 | 8895 | CPNE3 | protein_coding | O75131 | FUNCTION: Calcium-dependent phospholipid-binding protein that plays a role in ERBB2-mediated tumor cell migration in response to growth factor heregulin stimulation (PubMed:20010870). {ECO:0000269|PubMed:20010870}. | Calcium;Cell junction;Cell membrane;Cytoplasm;Direct protein sequencing;Membrane;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat | Calcium-dependent membrane-binding proteins may regulate molecular events at the interface of the cell membrane and cytoplasm. This gene encodes a protein which contains two type II C2 domains in the amino-terminus and an A domain-like sequence in the carboxy-terminus. The A domain mediates interactions between integrins and extracellular ligands. [provided by RefSeq, Aug 2008]. | hsa:8895; | azurophil granule membrane [GO:0035577]; cell junction [GO:0030054]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; focal adhesion [GO:0005925]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; calcium-dependent phospholipid binding [GO:0005544]; calcium-dependent protein binding [GO:0048306]; metal ion binding [GO:0046872]; protein serine/threonine kinase activity [GO:0004674]; receptor tyrosine kinase binding [GO:0030971]; RNA binding [GO:0003723]; cellular response to calcium ion [GO:0071277]; cellular response to growth factor stimulus [GO:0071363]; ERBB2 signaling pathway [GO:0038128]; positive regulation of cell migration [GO:0030335] | 18264096_Genome-wide association study of gene-disease association. (HuGE Navigator) 18264096_Susceptibility genes CPNE3, IL16 and CDH13 with moderate effects associated with susceptibility to prostate cancer. 20010870_Data suggest that Copine-III is a novel player in the regulation of ErbB2-dependent cancer cell motility. 23062752_This study demonistrated that dysregulation of cpne3 3'UTR in brain of patient with schizophrenia. 23713811_our results indicate that CPNE3 could play a critical role in NSCLC metastasis. 26719032_Copine3 binding to ErbB2 increases when Jab1 is overexpressed in SKBr3 breast cancer cells. Copine3 and Jab1 binding regulates the ErbB2 signaling pathway. 28670859_CPNE3(high) is an adverse prognostic biomarker for acute myeloid leukemia. 29297177_Patients who have low CPNE3 expression in peripheral blood are more likely to suffer from AMI than those with stable CAD. Low expression of CPNE3 gene serves as an potential independent risk factor of AMI. 29867202_upregulation of EMP1 significantly increases cancer cell migration that leads to tumor metastasis 30078189_Exosomal CPNE3 levels are associated with the tumor extent and may serve as a diagnostic biomarker in colorectal cancer patients. 33725213_Upregulation of CPNE3 suppresses invasion, migration and proliferation of glioblastoma cells through FAK pathway inactivation. 33767297_CPNE3 moderates the association between anxiety and working memory. | ENSMUSG00000028228 | Cpne3 | 7241.27720 | 1.1202202 | 0.1637822842 | 0.08300117 | 3.910623e+00 | 4.798179e-02 | 3.312579e-01 | No | Yes | 10744.605739 | 2713.586373 | 8650.894566 | 2135.819013 | |
| ENSG00000085982 | 55230 | USP40 | protein_coding | Q9NVE5 | FUNCTION: May be catalytically inactive. | Alternative splicing;Hydrolase;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway | Modification of cellular proteins by ubiquitin is an essential regulatory mechanism controlled by the coordinated action of multiple ubiquitin-conjugating and deubiquitinating enzymes. USP40 belongs to a large family of cysteine proteases that function as deubiquitinating enzymes (Quesada et al., 2004 [PubMed 14715245]).[supplied by OMIM, Mar 2008]. | hsa:55230; | cytosol [GO:0005829]; nucleus [GO:0005634]; cysteine-type endopeptidase activity [GO:0004197]; thiol-dependent deubiquitinase [GO:0004843]; protein deubiquitination [GO:0016579]; regulation of protein stability [GO:0031647]; ubiquitin-dependent protein catabolic process [GO:0006511] | 16917932_Da suggest that genetic variants in USP40 affect the risk for late-onset Parkinson disease (PD), which is consistent with the predicted role of the ubiquitination pathway in PD etiology. 16917932_Observational study of gene-disease association. (HuGE Navigator) 20302855_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20302855_USP24 alone plays a role in PD susceptibility among Taiwanese people >or=60 years of age, or acting synergistically with USP40 and UCHL1 in the total subjects. 22923019_present study is the first to report a lack of association between SNPs of USP40 and parkinson disease in Han Chinese patients. Other 31046799_Our findings show the de-ubiquitinase USP40 regulates the ubiquitination and degradation of CFLARL; and GMEB1 acts as a bridge protein for USP40 and CFLARL. Mechanistically, we found GMEB1 inhibits the activation of CASP8 by modulating ubiquitination and degradation of CFLARL | ENSMUSG00000005501 | Usp40 | 653.75215 | 1.2171261 | 0.2834786230 | 0.13164880 | 4.655707e+00 | 3.095049e-02 | 2.785889e-01 | No | Yes | 803.044554 | 104.829531 | 651.340035 | 83.040735 | |
| ENSG00000087206 | 51720 | UIMC1 | protein_coding | Q96RL1 | FUNCTION: Ubiquitin-binding protein (PubMed:24627472). Specifically recognizes and binds 'Lys-63'-linked ubiquitin (PubMed:19328070, Ref.38). Plays a central role in the BRCA1-A complex by specifically binding 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. Also weakly binds monoubiquitin but with much less affinity than 'Lys-63'-linked ubiquitin. May interact with monoubiquitinated histones H2A and H2B; the relevance of such results is however unclear in vivo. Does not bind Lys-48'-linked ubiquitin. May indirectly act as a transcriptional repressor by inhibiting the interaction of NR6A1 with the corepressor NCOR1. {ECO:0000269|PubMed:12080054, ECO:0000269|PubMed:17525340, ECO:0000269|PubMed:17525341, ECO:0000269|PubMed:17525342, ECO:0000269|PubMed:17621610, ECO:0000269|PubMed:17643121, ECO:0000269|PubMed:19015238, ECO:0000269|PubMed:19202061, ECO:0000269|PubMed:19261748, ECO:0000269|PubMed:19328070, ECO:0000269|PubMed:24627472, ECO:0000269|Ref.38}. | 3D-structure;Alternative splicing;Chromatin regulator;DNA damage;DNA repair;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a nuclear protein that interacts with Brca1 (breast cancer 1) in a complex to recognize and repair DNA lesions. This protein binds ubiquitinated lysine 63 of histone H2A and H2AX. This protein may also function as a repressor of transcription. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2015]. | hsa:51720; | BRCA1-A complex [GO:0070531]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; histone binding [GO:0042393]; K63-linked polyubiquitin modification-dependent protein binding [GO:0070530]; metal ion binding [GO:0046872]; chromatin organization [GO:0006325]; double-strand break repair [GO:0006302]; histone H2A K63-linked deubiquitination [GO:0070537]; mitotic G2 DNA damage checkpoint signaling [GO:0007095]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of DNA repair [GO:0045739]; response to ionizing radiation [GO:0010212] | 12080054_a novel nuclear protein that interacts with the retinoid-related testis-associated receptor 17525340_Abraxas and RAP80 were required for DNA damage resistance, G(2)-M checkpoint control, and DNA repair. 17525341_data support a model wherein ubiquitin chains at DNA damage sites are used as a targeting mechanism by specific BRCA1 complexes; RAP80 may represent a new class of DNA repair proteins that uses tandem UIM domains as part of its recruitment to DSBs 17525342_identification of receptor-associated protein 80 (RAP80) as a BRCA1-interacting protein in humans 17562356_RAP80/UIMC1, the protein highly expressed in testis, was identified as a new cancer-associated antigen. 17621610_the ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response 17698038_RAP80 interacts with the SUMO-conjugating enzyme UBC9 and is a novel target for sumoylation 18077395_the human Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage 18270812_Mutational analysis in 168 multiple-case breast/ovarian cancer families, negative for mutations in BRCA1 or BRCA2, suggests that RAP80 does not play an important role as a high penetrance breast cancer susceptibility gene. 18270812_Observational study of gene-disease association. (HuGE Navigator) 18306035_Observational study of gene-disease association. (HuGE Navigator) 18306035_Truncating mutations of the RAP80 gene do not seem to be a cause of familial breast cancer. A novel RAP80 haplotype or rare missense mutations (p.Ala342Thr, p.Met353Thr and p.Tyr575Asp) may be associated with a modest increased risk of breast cancer. 18519686_UV irradiation induces translocation of RAP80 to DNA damage foci that colocalize with gamma-H2AX 18550271_Depletion of RAP80 or RNF8 impairs the translocation of BRCA1 to DNA damage sites and results in defective cell cycle checkpoint control and DSB repair 18695986_Observational study of gene-disease association. (HuGE Navigator) 18695986_it seems unlikely that moderate to highly penetrant alleles of either RAP80 or Abraxas, confer a significantly high relative risk of breast cancer. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19261746_MERIT40 represents a novel factor that links BRCA1-Rap80 complex integrity, DSB recognition, and ubiquitin chain hydrolytic activities to the DNA damage response. 19305427_critical constitutional mutations in RAP80 abrogate DNA damage responses (DDR) function and may be involved in genetic predisposition to cancer. 19328070_findings show how the sequence between the Rap80 ubiquitin interacting motifs positions the domains for efficient avid polyubiquitin binding across a single K63 linkage, thus defining selectivity 19415121_RAP80 was a significant factor for survival in patients treated according to BRCA1 levels 19433585_Data show that it can function in an autoregulatory loop consisting of RAP80, HDM2, and the p53 master regulatory network, implpying an important role for this loop in genome stability and oncogenesis. 19448621_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21335604_a model in which the BRCA1-RAP80 complex limits nuclease accessibility to DSBs, thus preventing excessive end resection and potentially deleterious homology-directed DSB repair mechanisms that can impair genome integrity. 21406551_RAP80/BRCA1 complexes suppress excessive double-strand break end processing, HR-type double-strand break repair, and overt chromosomal instability 21622030_The interaction between MDC1 and RAP80 requires the tandem BRCT domain of MDC1 and the ubiquitin-interacting motifs of RAP80 21857162_MDC1 is required for the recruitment of RAP80 to DNA double-strand breaks. 22426463_APC/C(Cdc20) or APC/C(Cdh1) complexes regulate RAP80 stability during mitosis to the G(1) phase, and these events are critical for a novel function of RAP80 in mitotic progression. 22689573_a model in which SUMO and Ub modification is coordinated to recruit Rap80 and BRCA1 to DNA damage sites. 22742833_We show that RNF168, its paralog RNF169, RAD18, and the BRCA1-interacting RAP80 protein accumulate at DNA double strand break sites through the use of bipartite modules composed of ubiquitin binding domains. 22792303_Loss of RAP80 abolishes the recruitment of the BRCA1-A complex to DNA lesions in response to DNA damage. 23211528_connect ubiquitin- and SUMO-dependent DSB recognition, revealing that RNF4-synthesized hybrid SUMO-ubiquitin chains are recognized by RAP80 to promote BRCA1 recruitment and DNA repair. 23264621_post-translational phosphorylation of RAP80 by the Cdk1-cyclin B(1) complex is important for RAP80 functional sensitivity to IR and G(2)/M checkpoint control. 24627472_Data indicate that a single point deletion (DeltaE81) in RAP80 abrogates multivalent interactions with polyubiquitin. 25132264_A new role of FANCG in Homologous recombination repair of interstrand crosslinks through K63Ub-mediated interaction with the Rap80-BRCA1 complex. 25164908_patients with low RAP80 expression received gemcitabine/cisplatin, those with intermediate/high RAP80 expression and low/intermediate BRCA1 expression received docetaxel/cisplatin 26446986_Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. 26719330_Data suggest that RAP80 SIM (SUMO interacting motif) binds SUMO-2; both specificity and affinity are enhanced through phosphorylation of canonical CK2 (casein kinase 2) site within the SIM. 26748910_RAP80 is a critical gatekeeper in impeding epithelial-mesenchymal transition-induced metastasis and malignant phenotypes of cancer as well as preserving DNA integrity. 26781088_TRAIP/RNF206 is required for recruitment of RAP80 to sites of DNA damage.( 27443420_Low RAP80 mRNA expression correlates with sporadic high-grade serous ovarian carcinoma. 28569838_RAP80-BRCA1 complex foci formation is regulated by USP13.USP13 interacts with and deubiquitinates RAP80.RAP80 role in the DNA damage response. 28842250_The RAP80 deficiency reduces the protein level of p32 and p32 dependent mitochondrial translating proteins such as Rieske and COX1. 29396516_RAP80 positively regulated the stability of USP13 to promote cell proliferation of esophageal cancer cells. 31253574_Here we show that in the BRCA1-A complex structure, ABRAXAS integrates the DNA repair protein RAP80 and provides a high-affinity binding site that sequesters the tumor suppressor BRCA1 away from the break site. In the BRISC structure, ABRO1 binds SHMT2alpha, a metabolic enzyme enabling cancer growth in hypoxic environments, which we find prevents BRCC36 from binding and cleaving ubiquitin chains 31932421_BRCA1 poly-ADP ribosylation (PARsylation) and the presence of BRCA1-bound RAP80 are critical for the normal interaction of BRCA1 with some of its partners (e.g., CtIP and BACH1). 35108530_RAP80 suppresses the vulnerability of R-loops during DNA double-strand break repair. | ENSMUSG00000025878 | Uimc1 | 1427.67791 | 0.9275584 | -0.1084900113 | 0.09881099 | 1.202032e+00 | 2.729158e-01 | 6.617959e-01 | No | Yes | 1600.840663 | 209.381942 | 1648.506438 | 210.804672 | |
| ENSG00000088387 | 23348 | DOCK9 | protein_coding | Q9BZ29 | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Overexpression induces filopodia formation. {ECO:0000269|PubMed:12172552, ECO:0000269|PubMed:19745154}. | 3D-structure;Alternative splicing;Coiled coil;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;Repeat | hsa:23348; | cytosol [GO:0005829]; endomembrane system [GO:0012505]; membrane [GO:0016020]; cadherin binding [GO:0045296]; guanyl-nucleotide exchange factor activity [GO:0005085]; positive regulation of GTPase activity [GO:0043547]; small GTPase mediated signal transduction [GO:0007264] | 12172552_Sequence comparison combined with mutational analysis identified a new domain, which we named CZH2, that mediates direct interaction with Cdc42 17728666_DOCK9 contributes to both risk and increased illness severity in bipolar disorder. 17728666_Observational study of gene-disease association. (HuGE Navigator) 17935486_novel functions for the N-terminal region of zizimin1. 18056264_DOCK2 and DOCK9 specifically recognize Rac2 and Cdc42 through their switch 1 as well as beta2-beta3 regions and the mode of recognition via switch 1 appears to be conserved among diverse Rac-specific DHR-2 GEFs 18729074_interaction between Smad2/3 and the Cdc42 guanine nucleotide exchange factor, Zizimin1, in response to TGF-beta1 19745154_through structural analysis of DOCK9-Cdc42 complexes, we identify a nucleotide sensor within the alpha10 helix of the DHR2 domain that contributes to release of guanine diphosphate (GDP) and then to discharge of the activated GTP-bound Cdc42 19809089_Studies indicate that that many of the mechanistic principles of the exchange process are conserved in the DOCK9-catalyzed reaction. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 26641546_c.2262A>C substitution in DOCK9 leads to a splicing aberration. However, because the mutation effect was observed in vitro, a definitive relationship between DOCK9 and KTCN phenotype could not be established. 34741163_Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. | ENSMUSG00000025558 | Dock9 | 606.72631 | 0.9894753 | -0.0152644730 | 0.15266525 | 9.982257e-03 | 9.204148e-01 | 9.774351e-01 | No | Yes | 668.010209 | 91.865314 | 640.973333 | 86.038993 | ||
| ENSG00000088812 | 8455 | ATRN | protein_coding | O75882 | FUNCTION: Involved in the initial immune cell clustering during inflammatory response and may regulate chemotactic activity of chemokines. May play a role in melanocortin signaling pathways that regulate energy homeostasis and hair color. Low-affinity receptor for agouti (By similarity). Has a critical role in normal myelination in the central nervous system (By similarity). {ECO:0000250, ECO:0000269|PubMed:9736737}. | Alternative splicing;Cell membrane;Direct protein sequencing;Disulfide bond;EGF-like domain;Glycoprotein;Inflammatory response;Kelch repeat;Laminin EGF-like domain;Lectin;Membrane;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | This gene encodes both membrane-bound and secreted protein isoforms. A membrane-bound isoform exhibits sequence similarity with the mouse mahogany protein, a receptor involved in controlling obesity. A secreted isoform is involved in the initial immune cell clustering during inflammatory responses that may regulate the chemotactic activity of chemokines. [provided by RefSeq, Apr 2016]. | hsa:8455; | basement membrane [GO:0005604]; cytoplasm [GO:0005737]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; carbohydrate binding [GO:0030246]; signaling receptor activity [GO:0038023]; animal organ morphogenesis [GO:0009887]; cell migration [GO:0016477]; cerebellum development [GO:0021549]; inflammatory response [GO:0006954]; myelination [GO:0042552]; pigmentation [GO:0043473]; regulation of multicellular organism growth [GO:0040014]; response to oxidative stress [GO:0006979]; substrate adhesion-dependent cell spreading [GO:0034446]; tissue development [GO:0009888] | 9628581_Through use of PCR from hamster-human somatic cell hybrids, localizes the EST AB011120 (KIAA0548) to human chromosome 20p13. AB011120 encodes the central to 3' UTR of the membrane form of human attractin. 10086355_The product of the murine 'Mahogany' gene controls energy metabolism and is the murine orthologof human membrane attractin. 10086356_The product of the murine 'Mahogany' locus, controlling agouti-mediated melanocyte pigmentation and control of energy metabolism, is the murine ortholog of human attractin. 10811918_Provides the genomic basis for production of a soluble and membrane form of attractin by alternative splicing. In the process, by sequencing from positionally-determined BAC clones, confirms the localization of attractin to chromosome 20p13. 12675230_This protein is isolated from human blood and characterized. 16835316_It is the first time expression of attractin on monocytes and provide first data suggesting that drugs directed to DP IV-like enzyme activity could affect monocyte function via attractin inhibition. 19320733_Observational study of gene-disease association. (HuGE Navigator) 19384953_Observational study of gene-disease association. (HuGE Navigator) 19535787_associated with spermatozoa 20198559_The levels of both dipeptidyl-peptidase 4 and attractin in circulating monocytes were significantly higher in obese subjects as compared with levels in lean controls or in subjects with type 2 diabetes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25318887_Data indicate that Attractin was involved in male reproduction in both human and BALB/c mice, but it exerted a different expression profile in different species. 28493104_ATRN encodes Attractin, which was previously shown to play a critical role in central myelination | ENSMUSG00000027312 | Atrn | 2694.55318 | 0.9242207 | -0.1136906337 | 0.08825380 | 1.655584e+00 | 1.982006e-01 | 5.902057e-01 | No | Yes | 3131.286357 | 275.219791 | 3237.092922 | 278.137146 | |
| ENSG00000089050 | 10741 | RBBP9 | protein_coding | O75884 | FUNCTION: Serine hydrolase whose substrates have not been identified yet (PubMed:19329999, PubMed:20080647). May negatively regulate basal or autocrine TGF-beta signaling by suppressing SMAD2-SMAD3 phosphorylation (PubMed:20080647). May play a role in the transformation process due to its capacity to confer resistance to the growth-inhibitory effects of TGF-beta through interaction with RB1 and the subsequent displacement of E2F1 (PubMed:9697699). {ECO:0000269|PubMed:19329999, ECO:0000269|PubMed:20080647, ECO:0000269|PubMed:9697699}. | 3D-structure;Alternative splicing;Hydrolase;Reference proteome | The protein encoded by this gene is a retinoblastoma binding protein that may play a role in the regulation of cell proliferation and differentiation. Two alternatively spliced transcript variants of this gene with identical predicted protein products have been reported, one of which is a nonsense-mediated decay candidate. [provided by RefSeq, Jul 2008]. | hsa:10741; | nucleoplasm [GO:0005654]; hydrolase activity [GO:0016787]; regulation of cell population proliferation [GO:0042127] | 12296629_RBBP10 was expressed widely in various human tissues, and the expression level is somewhat higher in tumor tissues than in normal tissues. 19004028_The crystal structure of human RBBP9 has been determined at 1.72 A resolution by the seleno-methionyl single-wavelength anomalous diffraction method. 20080647_identify RBBP9 as a tumor-associated serine hydrolase that displays elevated activity in pancreatic carcinomas. RBBP9 promotes pancreatic carcinogenesis. 21689726_Data identified RBBP4 and RBBP9 as required for maintenance of multiple PS cell types, and both RBBPs were bound to RB in PS cells. 21933118_Structure- function studies of RBBP9 suggest possible routes for novel cancer drug discovery programs. 22430187_blood samples of lateral sclerosis patients were found to have significantly different levels of expression of CyFIP2 and RbBP9 compared to the levels of expression in control subjects. | ENSMUSG00000027428 | Rbbp9 | 658.58124 | 1.0832922 | 0.1154224503 | 0.13456861 | 7.372114e-01 | 3.905556e-01 | 7.530680e-01 | No | Yes | 881.170922 | 152.181704 | 770.012858 | 130.041662 | |
| ENSG00000091073 | 113878 | DTX2 | protein_coding | Q86UW9 | FUNCTION: Regulator of Notch signaling, a signaling pathway involved in cell-cell communications that regulates a broad spectrum of cell-fate determinations. Probably acts both as a positive and negative regulator of Notch, depending on the developmental and cell context. Mediates the antineural activity of Notch, possibly by inhibiting the transcriptional activation mediated by MATCH1. Functions as a ubiquitin ligase protein in vitro, suggesting that it may regulate the Notch pathway via some ubiquitin ligase activity. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Metal-binding;Methylation;Notch signaling pathway;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. | DTX2 functions as an E3 ubiquitin ligase (Takeyama et al., 2003 [PubMed 12670957]).[supplied by OMIM, Nov 2009]. | hsa:113878; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; Notch signaling pathway [GO:0007219]; protein ubiquitination [GO:0016567] | 12670957_It is reported that BBAP and the human family of DTX proteins (DTX1, DTX2, and DTX3) function as E3 ligases based on their capacity for self-ubiquitination. 17286044_DTX2 is a gene encoding a WWE-RING-finger protein and involved in regulating heart development and heart functions. | ENSMUSG00000004947 | Dtx2 | 296.91742 | 0.9067604 | -0.1412067552 | 0.17316524 | 6.626426e-01 | 4.156285e-01 | 7.705183e-01 | No | Yes | 307.854712 | 42.299620 | 326.017056 | 43.556134 |
| ENSG00000095794 | 1390 | CREM | protein_coding | Q03060 | FUNCTION: Transcriptional regulator that binds the cAMP response element (CRE), a sequence present in many viral and cellular promoters. Isoforms are either transcriptional activators or repressors. Plays a role in spermatogenesis and is involved in spermatid maturation (PubMed:10373550). {ECO:0000269|PubMed:10373550}.; FUNCTION: [Isoform 6]: May play a role in the regulation of the circadian clock: acts as a transcriptional repressor of the core circadian component PER1 by directly binding to cAMP response elements in its promoter. {ECO:0000250}. | Activator;Alternative promoter usage;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Developmental protein;Differentiation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a bZIP transcription factor that binds to the cAMP responsive element found in many viral and cellular promoters. It is an important component of cAMP-mediated signal transduction during the spermatogenetic cycle, as well as other complex processes. Alternative promoter and translation initiation site usage allows this gene to exert spatial and temporal specificity to cAMP responsiveness. Multiple alternatively spliced transcript variants encoding several different isoforms have been found for this gene, with some of them functioning as activators and some as repressors of transcription. [provided by RefSeq, Jul 2008]. | hsa:1390; | ATF4-CREB1 transcription factor complex [GO:1990589]; chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; cAMP response element binding protein binding [GO:0008140]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; cell differentiation [GO:0030154]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; rhythmic process [GO:0048511]; signal transduction [GO:0007165]; spermatogenesis [GO:0007283] | 11988318_review of CREM transcription factor involvement with spermatogenesis 12370343_Increased expression of CREM in T cells from systemic lupus erythmatosus (SLE) patients results from increased transcriptional activity of the CREM gene, and its binding to IL-2 promoter is responsible for decreased production of IL-2 by SLE T cells. 12397208_5'-RACE on human testis cDNA indicated that exon theta2 is > or = 113 bp in size. In-vitro translation of CREM-theta1 and CREM-theta2 splice variants cloned from human testis yielded full length proteins and also shorter repressor products 12555239_Observational study of gene-disease association. (HuGE Navigator) 12626549_Direct binding of CREM to the CRE site of the IL-2 promoter endows CREM with a central role in repression of IL-2 gene expression: CREM binding promotes chromatin deacetylation, limits promoter accessibility and decreases its transcriptional activity. 14511788_expression of cAMP-responsive element modulator(CREM) activators is a prerequisite for normal spermatogenesis, and the lack of CREM activator expression results in male infertility 14754893_CREB-1/CREM-1 have roles as regulators of macrophage differentiation 15048659_These findings provide little additional evidence for a susceptibility locus for panic disorder either within the CREM gene or in a nearby region of chromosome 10p11 in our sample 15225122_Sperm nucleus PHGPx expression is mediated by the transcription factor CREM-tau, which acts as a cis-acting element localized in the first intron of the PHGPx gene. [CREM-tau] 15456763_down-regulation of CREMtau-mediated gene expression by GCNF 15474076_Lack of spermatid elongation was not due to defective CREM expression. Therefore, CREM did not play a pathogenetic role in the onset of SMA in humans. 15569686_that heart-directed expression of CREM-IbDeltaC-X leads to complex cardiac alterations, suggesting CREM as a central regulator of cardiac morphology, function, and gene expression 15691874_isoforms regulate discrete groups of genes in myometrium 15841182_Results identify calcium/calmodulin-dependent kinase IV as being responsible for the increased expression of CREM and the decreased production of interleukin-2 in systemic lupus erythematosus T cells. 16048633_CREM activator and repressor isoforms were found in all germ cell types, but not in Sertoli cells; data suggest a fine-tuning between CREM activator and repressor isoforms in normal germ cells that might be disturbed during impaired spermatogenesis 16103121_SRp40 regulates the switch in splicing from production of CREMtau(2)alpha to CREMalpha 16143638_Screening of a substantial number of patients would be required to clarify whether observed combinations of genetic changes in the CREM gene might explain some forms of male infertility. 16385451_Observational study of gene-disease association. (HuGE Navigator) 16687568_The interaction between CREM and one haplotype of ACT (activator of CREM in the testis) was reduced by 45% in a yeast two-hybrid assay. 16893891_HNF4alpha, CREM, HNF1alpha, and C/EBPalpha have roles in transcriptional regulation of the glucose-6-phosphatase gene by cAMP/vasoactive intestinal peptide in the intestine 16894555_These results constitute the first demonstration of the transcriptional control of ATP1A4 gene expression by cAMP and by CREMtau, a transcription factor essential for male germ cell gene expression. 16899810_functional analysis of isoforms from the testes 17056544_CREMalpha exerts its repressor activity by a mechanism that involves recruitment of HDAC1, increased deacetylation of histones, and repression of promoter activity. 17211988_CREM-alpha mRNA levels were higher in T cells from patients with systemic lupus erythematosus than controls while CREB mRNA levels did not differ between the two groups 17332439_The positive-feedback loop described in this review is regulated by phosphodiesterase 3A and ICER and is pathologically important in adult hearts. 17340624_Data show that inducible cAMP early repressor splice variants ICER I and IIgamma both repress transcription of c-fos and chromogranin A. 17712720_Transcription factors of the CREB/CREM/ATF family have a moderate effect on human MC2-R promoter activity, but seem to play a minor role in transmitting stimulation of the cAMP pathway to increased MC2-R expression. 18202121_CREM is an essential regulator of NIS gene expression. 18700132_CREM has a role in CYP1A1 induction through ligand-independent activation pathway of aryl hydrocarbon receptor in HepG2 cells 18784739_CREB/ICER expression needs to be considered a pathogenetic feature in leukemogenesis 18922788_BDNF- and seizure-dependent phosphorylation of STAT3 cause the adenosine 3',5'-monophosphate (cAMP) response element-binding protein (CREB) family member ICER (inducible cAMP early repressor) to bind with phosphorylated CREB at the Gabra1:CRE site. 18984674_Observational study of gene-disease association. (HuGE Navigator) 19299714_CREMalpha is an important negative regulator of costimulation and APC-dependent T cell function 19434522_This review will bring together data on ICER and its functions in the brain, with a special emphasis on recent findings highlighting the involvement of ICER in the regulation of long-term plasticity underlying learning and memory. [REVIEW] 19531482_cAMP stringently regulates human cathelicidin antimicrobial peptide expression in the mucosal epithelial cells by activating cAMP-response element-binding protein, AP-1, and inducible cAMP early repressor 20237496_Observational study of gene-disease association. (HuGE Navigator) 20378615_Observational study of gene-disease association. (HuGE Navigator) 20413592_DNA cytosine methylation in the bovine leukemia virus promoter has a role in direct inhibition of cAMP-responsive element (CRE)-binding protein/CRE modulator/activation transcription factor binding 20488182_Results indicate that SPAG8 acts as a regulator of ACT and plays an important role in CREM-ACT-mediated gene transcription during spermatogenesis. 21097497_anscriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. 21325296_ICER mediates chemotherapy anticancer activity through DUSP1-p38 pathway activation and drives the cell program from survival to apoptosis 21507395_Patients with two types of male factor infertility display an increased abnormal methylation of CREM compared with control subjects. 21547497_Induction of ICER links oxidative stress to beta cell failure caused by oxidised LDL, and it can be effectively abrogated by antioxidant treatment. 21757709_AP-1-dependent up-regulation of the P2 promoter, SLE T cells fail to further increase their basal CREM levels upon T cell activation due to a decreased content of the AP-1 family member c-Fos 21767532_Phosphorylation of ICER on a discrete residue targeted ICER to be monoubiquitinated. 21953500_CREMalpha suppresses spleen tyrosine kinase expression in normal but not systemic lupus erythematosus T cells. 21976679_Data provide direct evidence that CREMalpha mediates silencing of the IL2 gene in SLE T cells though histone deacetylation and CpG-DNA methylation. 21998402_Impaired expression of ICER contributes to elevation in CREB target genes and, therefore, to the development of insulin resistance in obesity. 22019623_Common variants of the CREM gene are involved in the genetic component conferring general susceptibility to inflammatory bowel disease in the Tunisian population. 22025620_cAMP-responsive element modulator (CREM)alpha protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. 22093963_Transcription factor CREM is an important regulator of atrial growth implicated in the development of atrial fibrillation. 22184122_cAMP-responsive element modulator alpha (CREMalpha) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). 22198373_scriptive Statement The rsults of this study suggested the lack of influence of SNPs under investigation in the present study on the susceptibility to schizophrenia and on the response to antipsychotics. 22281835_Estrogen can modulate the expression of CREMalpha and lead to IL-2 suppression in human T lymphocytes, thus revealing a molecular link between hormones and the immune system in Systemic lupus erythematosus 22386572_The results of this study suggested that the single nucleotide polymorphisms of CREM do not influence diagnosis and treatment response in patients with major depressive disorder and bipolar disorder. 22510021_CREM expression is increased in thyroid cancer tissue and may play a role in the downregulation of sodium iodide symporter expression in thyroid cancer acting at the transcriptional level 23019580_Data indicate that CpG-DNA methylation and mRNA expression of CREM, IL2, and IL17A of systemic lupus erythematosus (SLE) T cells reflect the effector memory CD4+ T-cell phenotype. 23124208_Increased CREMalpha binding to the Notch-1 promoter resulted in significantly reduced Notch-1 promoter activity and gene transcription. 23929392_Data suggest that cyclic AMP response element modulator-1 (CREM-1) might play an important role in the regulation of tumor metastasis and invasion and serve as a tumor suppressor in esophageal squamous cell carcinoma (ESCC). 24047902_transcription factor cAMP-responsive element modulator alpha (CREMalpha), which is expressed at increased levels in T cells from systemic lupus erythematosus patients, contributes to transcriptional silencing of CD8A and CD8B. 24100545_In Alzheimer's brain, we found an increased cellular expression of CREM in dentate gyrus neurons as compared to normal aging brains. 24297179_CREMalpha orchestrates epigenetic remodeling of the CD8A,B through the recruitment of DNA methyltransferase (DNMT) 3a and histone methyltransferase G9a. 24667640_CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. 24672804_Data suggest ICER/CREM plays seminal role in down-regulation of expression/secretion of insulin by pancreatic beta-cell as an adaptive response to factors that promote diabetes; inappropriate induction of ICER leads to beta-cell dysfunction. [REVIEW] 24943041_These findings indicated that the polymorphisms of CREM gene were associated with nonobstructive azoospermia in the Chinese population and low CREM expression might be involved in the pathogenesis of spermatogenesis maturation arrest. 25401338_overexpressed in the nuclei of hepatocellular carcinoma cells 26601115_CREMalpha SNPs rs2295415 and rs1057108 may be novel genetic susceptibility factors for SLE, especially at haplotype level. 27822872_Data indicate a role for inducible cyclic AMP early repressor (ICER) in G1 checkpoint regulation in hematopoietic stem cells (HSCs). 27840176_this study shows an eventually involvement of CREM gene in the development of T1D pathology in Tunisian families. These facts are consistent with a major role for transcription factor genes involved in the immune pathways in the control of autoimmunity. 27904655_Study provides evidence that increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates CREMA in systemic lupus erythematosus. 28009602_Report a distinct group of myxoid mesenchymal neoplasms occurring in children or young adults with a predilection for intracranial locations with EWSR1-AFT1/CREB1/CREM fusions. 28439100_Genetic studies in seven independent human populations illustrate that a CREM promoter variant at rs12765063 is associated with impulsivity, hyperactivity and addiction-related phenotypes. 29788195_EWSR1 fusion with CREM has only been observed in 3 intracranial myxoid tumors 29925386_CREM drives an inflammatory phenotype of T cells in Juvenile idiopathic arthritis. 29975250_We describe a novel gene fusion, EWSR1-CREM, identified in 3 cases of clear cell carcinoma 30228239_Increased CREM expression is also observed in early E. histolytica infection 31305268_Report the phenotypic spectrum of mesenchymal tumors associated with the EWSR1-CREM fusion. 34049318_Cytokeratin-positive Malignant Tumor in the Abdomen With EWSR1/FUS-CREB Fusion: A Clinicopathologic Study of 8 Cases. 34261344_Expression of Transcription Factor CREM in Human Tissues. | ENSMUSG00000063889 | Crem | 253.90905 | 0.9848581 | -0.0220122750 | 0.19085868 | 1.328355e-02 | 9.082436e-01 | 9.740734e-01 | No | Yes | 299.039839 | 47.265537 | 286.949308 | 44.439878 | |
| ENSG00000096063 | 6732 | SRPK1 | protein_coding | Q96SB4 | FUNCTION: Serine/arginine-rich protein-specific kinase which specifically phosphorylates its substrates at serine residues located in regions rich in arginine/serine dipeptides, known as RS domains and is involved in the phosphorylation of SR splicing factors and the regulation of splicing. Plays a central role in the regulatory network for splicing, controlling the intranuclear distribution of splicing factors in interphase cells and the reorganization of nuclear speckles during mitosis. Can influence additional steps of mRNA maturation, as well as other cellular activities, such as chromatin reorganization in somatic and sperm cells and cell cycle progression. Isoform 2 phosphorylates SFRS2, ZRSR2, LBR and PRM1. Isoform 2 phosphorylates SRSF1 using a directional (C-terminal to N-terminal) and a dual-track mechanism incorporating both processive phosphorylation (in which the kinase stays attached to the substrate after each round of phosphorylation) and distributive phosphorylation steps (in which the kinase and substrate dissociate after each phosphorylation event). The RS domain of SRSF1 binds first to a docking groove in the large lobe of the kinase domain of SRPK1. This induces certain structural changes in SRPK1 and/or RRM2 domain of SRSF1, allowing RRM2 to bind the kinase and initiate phosphorylation. The cycles continue for several phosphorylation steps in a processive manner (steps 1-8) until the last few phosphorylation steps (approximately steps 9-12). During that time, a mechanical stress induces the unfolding of the beta-4 motif in RRM2, which then docks at the docking groove of SRPK1. This also signals RRM2 to begin to dissociate, which facilitates SRSF1 dissociation after phosphorylation is completed. Isoform 2 can mediate hepatitis B virus (HBV) core protein phosphorylation. It plays a negative role in the regulation of HBV replication through a mechanism not involving the phosphorylation of the core protein but by reducing the packaging efficiency of the pregenomic RNA (pgRNA) without affecting the formation of the viral core particles. Isoform 1 and isoform 2 can induce splicing of exon 10 in MAPT/TAU. The ratio of isoform 1/isoform 2 plays a decisive role in determining cell fate in K-562 leukaemic cell line: isoform 2 favors proliferation where as isoform 1 favors differentiation. {ECO:0000269|PubMed:10049757, ECO:0000269|PubMed:10390541, ECO:0000269|PubMed:11509566, ECO:0000269|PubMed:12134018, ECO:0000269|PubMed:14555757, ECO:0000269|PubMed:15034300, ECO:0000269|PubMed:16122776, ECO:0000269|PubMed:16209947, ECO:0000269|PubMed:18155240, ECO:0000269|PubMed:18687337, ECO:0000269|PubMed:19240134, ECO:0000269|PubMed:19477182, ECO:0000269|PubMed:19886675, ECO:0000269|PubMed:20708644, ECO:0000269|PubMed:8208298, ECO:0000269|PubMed:9237760}. | 3D-structure;ATP-binding;Alternative splicing;Chromosome;Chromosome partition;Cytoplasm;Differentiation;Direct protein sequencing;Endoplasmic reticulum;Host-virus interaction;Kinase;Microsome;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;mRNA processing;mRNA splicing | hsa:6732; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; nuclear matrix [GO:0016363]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; RNA binding [GO:0003723]; chromosome segregation [GO:0007059]; innate immune response [GO:0045087]; intracellular signal transduction [GO:0035556]; negative regulation of viral genome replication [GO:0045071]; positive regulation of viral genome replication [GO:0045070]; protein phosphorylation [GO:0006468]; regulation of mRNA processing [GO:0050684]; regulation of mRNA splicing, via spliceosome [GO:0048024]; RNA splicing [GO:0008380]; sperm chromatin condensation [GO:0035092]; spliceosomal complex assembly [GO:0000245] | Mouse_homologues 15256051_SRPK1 expression might be an important prognostic indicator for the chemoresponsiveness of nonseminomatous male germ cell tumors. 24703948_SRPK1 overexpression is also tumorigenic because excess SRPK1 squelches PHLPP1. 25774502_In 2 independent murine models of breast tumor metastasis, stable shRNA-based SRPK1 knockdown suppressed metastasis to distant organs, including lung, liver, and spleen, and inhibited focal adhesion reorganization. 29474929_Depletion of endogenous SRPK1 enhanced the development of C3H10T1/2 cells toward brown adipocytes. 33080171_Functional Diversification of SRSF Protein Kinase to Control Ubiquitin-Dependent Neurodevelopmental Signaling. 34524595_LncRNA Neat1/miR-298-5p/Srpk1 Contributes to Sevoflurane-Induced Neurotoxicity. | ENSMUSG00000004865 | Srpk1 | 6252.77920 | 1.0094599 | 0.0135836204 | 0.08384340 | 2.624617e-02 | 8.713006e-01 | 9.664356e-01 | No | Yes | 7968.251514 | 1379.840482 | 7437.658467 | 1259.354715 | ||
| ENSG00000099840 | 113177 | IZUMO4 | protein_coding | Q1ZYL8 | Alternative splicing;Glycoprotein;Reference proteome;Secreted;Signal | hsa:113177; | extracellular region [GO:0005576]; nucleus [GO:0005634] | ENSMUSG00000055862 | Izumo4 | 60.47965 | 0.9177013 | -0.1239034766 | 0.36780329 | 1.130223e-01 | 7.367292e-01 | No | Yes | 61.751846 | 10.334477 | 67.882720 | 11.041764 | |||||
| ENSG00000100362 | 5816 | PVALB | protein_coding | P20472 | FUNCTION: In muscle, parvalbumin is thought to be involved in relaxation after contraction. It binds two calcium ions. | 3D-structure;Acetylation;Calcium;Direct protein sequencing;Metal-binding;Muscle protein;Phosphoprotein;Reference proteome;Repeat | The protein encoded by this gene is a high affinity calcium ion-binding protein that is structurally and functionally similar to calmodulin and troponin C. The encoded protein is thought to be involved in muscle relaxation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Oct 2015]. | hsa:5816; | axon [GO:0030424]; cuticular plate [GO:0032437]; cytoplasm [GO:0005737]; neuronal cell body [GO:0043025]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; stereocilium [GO:0032420]; synapse [GO:0045202]; calcium ion binding [GO:0005509]; identical protein binding [GO:0042802]; protein-containing complex binding [GO:0044877]; cochlea development [GO:0090102]; excitatory chemical synaptic transmission [GO:0098976]; gene expression [GO:0010467]; inhibitory chemical synaptic transmission [GO:0098977] | 12867516_In subjects with schizophrenia a reduction in parvalbumin [PV] and GAD67 mRNA expression in prefrontal cortex neurons was noted 15059934_Slow relaxation caused by alpha-Tm mutants can be corrected by modifying calcium handling with parvalbumin. 15257133_In dopaminergic cells of the substantia nigra in Parkinson disease an increased parvalbumin content is detected reflecting a natural protective mechanism against putative increase of intracellular calcium caused by excitotoxic injury and oxidative stress. 17405923_demonstration of a reduced number of parvalbumin-immunoreactive projection neurons in the mammillary bodies in schizophrenia 17850980_There is no correlation between total neuronal loss and PV-ir neuronal loss in any of the hippocampal fields in epileptic patient. 18297277_The results of this study indicate a significant reduction in the number of PV interneurons within layer 2 of the multiple sclerosis primary motor cortex 18469313_sequencing of PVALB was performed in 132 cases of Gitelman's syndrome in whom only one or no (N = 79) mutant SLC12A3 allele was found 20093724_parvalbumin has a role in calcium handling in cardiac diastolic dysfunction [review] 20926528_PVALB is a novel diagnostic marker for Hurthle ademonas of the thyroid. 22272358_Data show calbindin (CB)- and tyrosine hydroxylase (TH)-cells were distributed in the three striatal territories, and the density of calretinin (CR) and parvalbumin (PV) interneurons were more abundant in the associative and sensorimotor striatum. 23254904_A subpopulation of projection neurons containing calcium-binding protein parvalbumin (PV) is identified in a precise mapping of the GABAergic cortical distribution. 23764361_This study demonistrated that loss of parvalbium immunoreactive axons in anterolateral columns of spinal cord in patient of lateral sclerosis spinal cords 23825418_Parvalbumin neurons decrease the drive of the input to the visual cortex in contrast sensitivity. 23891794_mRNA labeling at the single cell level shows a significant decrease in parvalbumin expression in Parkinson's (PD) cases; however, neuronal density of parvalbumin-positive neurons was not significantly different between PD patients and controls. 24217255_This study showed that the parvalbumin basket cell inputs in the dorsolateral prefrontal cortex were lower in patient with schizophrenia. 24628518_This study demonistrated that differentially expressed in PV neurons in subjects with schizophrenia, including genes associated with WNT (wingless-type), NOTCH, and PGE2 (prostaglandin E2) signaling. 24764033_TRPV1-, TRPV2-, P2X3-, and parvalbumin-immunoreactivity neurons in the human nodose ganglion innervate the pharynx and epiglottis through the pharyngeal branch and superior laryngeal nerve 26081613_Increased numbers of PVALB neurons and fiber labeling in focal cortical dysplasia compared to nondysplastic epileptic temporal neocortex and postmortem controls may be related to cortical malformation. 27173597_This study demonstrated that A reduction of PV-positive cells and PV-immunoreactivity was observed exclusively in FCD type I/III specimens compared with cryptogenic tissue from control patients with a poor postsurgical outcome. 27444795_excitatory synapse density is lower selectively on parvalbumin interneurons in schizophrenia and predicts the activity-dependent down-regulation of parvalbumin and GAD67 27458244_Our data support the hypothesis that the loss of PVALB plays a role in the pathogenesis of thyroid tumors. 28074478_The results of this study suggested that innervation from PV-containing thalamic nuclei extends across superficial and middle layers of the human dorsolateral prefrontal cortex. 28835159_These results demonstrate a specific association between elevated PVALB methylation and METH-induced psychosis. 28859333_This study showed that the density of parvalbumin was significantly diminished in the basolateral complex in patients with Parkinson Disease. 29688033_These results provide the first evidence that PVALB promoter methylation is abnormal in schizophrenia and suggest that this epigenetic finding may relate to the reduction of PV expression seen in the disease. 30545133_the Calreticulin deficiency-mediated increase in cell death was not prevented by calbindin-D28k or Parvalbumin. 31588185_PVALB promoter methylation in major depressive disorder and its correlation with major depressive disorder severity indicating a role for epigenetics in this psychiatric disorder. 31981822_Age-related changes in the number of cresyl-violet-stained, parvalbumin and NMDAR 2B expressing neurons in the human spiral ganglion. 32497062_GluN2D-mediated excitatory drive onto medial prefrontal cortical PV+ fast-spiking inhibitory interneurons. 32514083_Transcriptional and imaging-genetic association of cortical interneurons, brain function, and schizophrenia risk. 32524185_Parvalbumin immunohistochemical expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. 33386125_Propofol sedation-induced alterations in brain connectivity reflect parvalbumin interneurone distribution in human cerebral cortex. 34439824_Strontium Binding to alpha-Parvalbumin, a Canonical Calcium-Binding Protein of the ''EF-Hand'' Family. | ENSMUSG00000005716 | Pvalb | 20.11193 | 1.2931563 | 0.3708967097 | 0.63010005 | 3.475018e-01 | 5.555307e-01 | No | Yes | 29.705491 | 7.320524 | 21.241606 | 5.272410 | ||
| ENSG00000100505 | 114088 | TRIM9 | protein_coding | Q9C026 | FUNCTION: E3 ubiquitin-protein ligase which ubiquitinates itself in cooperation with an E2 enzyme UBE2D2/UBC4 and serves as a targeting signal for proteasomal degradation. May play a role in regulation of neuronal functions and may also participate in the formation or breakdown of abnormal inclusions in neurodegenerative disorders. May act as a regulator of synaptic vesicle exocytosis by controlling the availability of SNAP25 for the SNARE complex formation. {ECO:0000269|PubMed:20085810}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Synapse;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:20085810}. | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein localizes to cytoplasmic bodies. Its function has not been identified. Alternate splicing of this gene generates two transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:114088; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; dendrite [GO:0030425]; synaptic vesicle [GO:0008021]; protein domain specific binding [GO:0019904]; protein homodimerization activity [GO:0042803]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161] | 20085810_These results suggest that TRIM9 plays an important role in the regulation of neuronal functions and participates in pathological process of Lewy body disease through its ligase activity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25190485_TRIM9 is a brain-specific negative regulator of the NF-kappaB pro-inflammatory signalling pathway. 26915459_TRIM9s undergoes Lys-63-linked auto-polyubiquitination and serves as a platform to bridge GSK3beta to TBK1, leading to the activation of IRF3 signaling. 28701345_Authors demonstrate that tripartite motif protein 9 (TRIM9)-dependent ubiquitination of DCC blocks the interaction with and phosphorylation of FAK. 29669288_TRIM9s promotes the K63-linked ubiquitination of MKK6 at Lys82, thus inhibiting the degradative K48-linked ubiquitination of MKK6 at the same lysine. MKK6 could also stabilize TRIM9s by promoting the phosphorylation of TRIM9s at Ser76/80 via p38, thereby blocking the ubiquitin-proteasome pathway. 30970257_Findings reveal that TRIM9 is essential for resolving NF-kappaB-dependent neuroinflammation to promote recovery and repair after brain injury. 32679146_TRIM9 overexpression promotes uterine leiomyoma cell proliferation and inhibits cell apoptosis via NF-kappaB signaling pathway. 34676875_Knockdown of TRIM9 attenuates irinotecaninduced intestinal mucositis in IEC6 cells by regulating DUSP6 expression via the P38 pathway. | ENSMUSG00000021071 | Trim9 | 421.30168 | 0.9505012 | -0.0732396119 | 0.15343265 | 2.273197e-01 | 6.335184e-01 | 8.828768e-01 | No | Yes | 472.710469 | 68.175694 | 475.078420 | 66.797851 |
| ENSG00000100605 | 3705 | ITPK1 | protein_coding | Q13572 | FUNCTION: Kinase that can phosphorylate various inositol polyphosphate such as Ins(3,4,5,6)P4 or Ins(1,3,4)P3 (PubMed:11042108, PubMed:8662638). Phosphorylates Ins(3,4,5,6)P4 at position 1 to form Ins(1,3,4,5,6)P5 (PubMed:11042108). This reaction is thought to have regulatory importance, since Ins(3,4,5,6)P4 is an inhibitor of plasma membrane Ca(2+)-activated Cl(-) channels, while Ins(1,3,4,5,6)P5 is not. Also phosphorylates Ins(1,3,4)P3 on O-5 and O-6 to form Ins(1,3,4,6)P4, an essential molecule in the hexakisphosphate (InsP6) pathway (PubMed:11042108, PubMed:8662638). Also acts as an inositol polyphosphate phosphatase that dephosphorylates Ins(1,3,4,5)P4 and Ins(1,3,4,6)P4 to Ins(1,3,4)P3, and Ins(1,3,4,5,6)P5 to Ins(3,4,5,6)P4. May also act as an isomerase that interconverts the inositol tetrakisphosphate isomers Ins(1,3,4,5)P4 and Ins(1,3,4,6)P4 in the presence of ADP and magnesium. Probably acts as the rate-limiting enzyme of the InsP6 pathway. Modifies TNF-alpha-induced apoptosis by interfering with the activation of TNFRSF1A-associated death domain (PubMed:11909533, PubMed:12925536, PubMed:17616525). Plays an important role in MLKL-mediated necroptosis. Produces highly phosphorylated inositol phosphates such as inositolhexakisphosphate (InsP6) which bind to MLKL mediating the release of an N-terminal auto-inhibitory region leading to its activation. Essential for activated phospho-MLKL to oligomerize and localize to the cell membrane during necroptosis (PubMed:17616525). {ECO:0000269|PubMed:11042108, ECO:0000269|PubMed:11909533, ECO:0000269|PubMed:12925536, ECO:0000269|PubMed:17616525, ECO:0000269|PubMed:8662638}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Hydrolase;Isomerase;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Transferase | This gene encodes an enzyme that belongs to the inositol 1,3,4-trisphosphate 5/6-kinase family. This enzyme regulates the synthesis of inositol tetraphosphate, and downstream products, inositol pentakisphosphate and inositol hexakisphosphate. Inositol metabolism plays a role in the development of the neural tube. Disruptions in this gene are thought to be associated with neural tube defects. A pseudogene of this gene has been identified on chromosome X. [provided by RefSeq, Jul 2016]. | hsa:3705; | apical plasma membrane [GO:0016324]; cytosol [GO:0005829]; ATP binding [GO:0005524]; catalytic activity [GO:0003824]; hydrolase activity [GO:0016787]; inositol tetrakisphosphate 1-kinase activity [GO:0047325]; inositol tetrakisphosphate 6-kinase activity [GO:0000825]; inositol-1,3,4-trisphosphate 5-kinase activity [GO:0052726]; inositol-1,3,4-trisphosphate 6-kinase activity [GO:0052725]; isomerase activity [GO:0016853]; magnesium ion binding [GO:0000287]; blood coagulation [GO:0007596]; inositol phosphorylation [GO:0052746]; inositol trisphosphate metabolic process [GO:0032957]; necroptotic process [GO:0070266]; neural tube development [GO:0021915]; signal transduction [GO:0007165] | 12925536_5/6-kinase modifies TNFalpha-induced apoptosis by interfering with the activation of TNF-R1-associated death domain 17616525_analysis of inositol phosphate signaling pathway integration via human ITPK1 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22308441_HEK293 cells stably expressing acetylated ITPK1 had reduced levels of the higher phosphorylated forms of inositol, compared with the levels seen in cells expressing unacetylated ITPK1 22928859_Activation of PLC by an endogenous cytokine (GBP) in Drosophila S3 cells and its application as a model for studying inositol phosphate signalling through ITPK1. 24465924_The results suggested that the maternal rs3783903 of ITPK1 might be associated with spina bifida, and the allele G of rs3783903 might affect the binding of AP-1 and the decrease of maternal plasma inositol hexakisphosphate concentration. 31754032_ITPK1 role in the lipid-independent synthesis of inositol phosphates. | ENSMUSG00000057963 | Itpk1 | 1467.23480 | 1.0760062 | 0.1056863888 | 0.10396755 | 1.034869e+00 | 3.090178e-01 | 6.902549e-01 | No | Yes | 2207.648592 | 287.045563 | 1974.652890 | 250.854960 | |
| ENSG00000100897 | 80344 | DCAF11 | protein_coding | Q8TEB1 | FUNCTION: May function as a substrate receptor for CUL4-DDB1 E3 ubiquitin-protein ligase complex. {ECO:0000269|PubMed:16949367, ECO:0000269|PubMed:16964240}. | Alternative splicing;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation pathway;WD repeat | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a WD repeat-containing protein that interacts with the COP9 signalosome, a macromolecular complex that interacts with cullin-RING E3 ligases and regulates their activity by hydrolyzing cullin-Nedd8 conjugates. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Jul 2009]. | hsa:80344; | Cul4-RING E3 ubiquitin ligase complex [GO:0080008]; nucleoplasm [GO:0005654]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein ubiquitination [GO:0016567] | 19273594_Includes the functional characterization of a similar protein in C. elegans and comparison to the human protein. 27203182_CRL4(WDR23) is required for efficient histone mRNA 3' end processing to produce mature histone mRNAs for translation. CRL4(WDR23) binds and ubiquitylates SLBP in vitro and in vivo, and this modification activates SLBP function in histone mRNA 3' end processing without affecting its protein levels. 27254819_CRL4-DCAF11 mediates the degradation of SLBP at the end of S phase and this degradation is essential for the viability of cells. 28446751_Here, the authors demonstrated that CUL4B forms an E3 ligase with RBX1 (RING-box 1), DDB1 (DNA damage binding protein 1), and DCAF11 (DDB1 and CUL4 associated factor 11) that promotes the ubiquitination of p21(Cip1) and regulates cell cycle progression in human osteosarcoma cells. 28453520_ur results identify WDR23 as an alternative regulator of NRF2 proteostasis and uncover a cellular pathway that regulates NRF2 activity and capacity for cytoprotection independently of KEAP1. 29377600_results support a model in which the activation of the TNF-alpha/NF-kappaB axis contributes to an increase in CRL4B(DCAF)(11) activity and a decrease in p21(Cip1) protein levels, thereby controlling cell cycle progression in human osteosarcoma cells. 31586112_TFEB activates Nrf2 by repressing its E3 ubiquitin ligase DCAF11 and promoting phosphorylation of p62. 32839090_WDR23 regulates the expression of Nrf2-driven drug-metabolizing enzymes. 33783207_DCAF11 Supports Targeted Protein Degradation by Electrophilic Proteolysis-Targeting Chimeras. | ENSMUSG00000022214 | Dcaf11 | 1559.92750 | 1.0282029 | 0.0401249197 | 0.11520845 | 1.212231e-01 | 7.277117e-01 | 9.204344e-01 | No | Yes | 2083.796498 | 212.306959 | 1960.853209 | 195.157377 |
| ENSG00000102316 | 10916 | MAGED2 | protein_coding | Q9UNF1 | FUNCTION: Regulates the expression, localization to the plasma membrane and function of the sodium chloride cotransporters SLC12A1 and SLC12A3, two key components of salt reabsorption in the distal renal tubule. {ECO:0000269|PubMed:27120771}. | Acetylation;Alternative splicing;Bartter syndrome;Direct protein sequencing;Disease variant;Phosphoprotein;Reference proteome;Tumor antigen | This gene is a member of the MAGED gene family. The MAGED genes are clustered on chromosome Xp11. This gene is located in Xp11.2, a hot spot for X-linked intellectual disability (XLID). Mutations in this gene cause a form of transient antenatal Bartter's syndrome. This gene may also be involved in several types of cancer, including breast cancer and melanoma. The protein encoded by this gene is progressively recruited from the cytoplasm to the nucleoplasm during the interphase and after nucleolar stress and is thus thought to play a role in cell cycle regulation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2017]. | hsa:10916; | cytosol [GO:0005829]; extracellular region [GO:0005576]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; platelet alpha granule lumen [GO:0031093]; female pregnancy [GO:0007565]; negative regulation of transcription by RNA polymerase II [GO:0000122]; renal sodium ion absorption [GO:0070294] | 11856887_Expression pattern and further characterization of human MAGED2 and identification of rodent orthologues 15162511_Maged2 is mainly expressed in tissues of mesodermal origin 15465002_Results identified a cDNA clone, corresponding to MAGE D2 mRNA, from primary human bronchial epithelial cells which exhibits increased expression in vitro after treatment with all-trans retinoic acid. 17912449_MAGED2, a novel protein, is a p53-dissociator. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20237496_Observational study of gene-disease association. (HuGE Navigator) 22791814_these results identify the expression of MAGE-D2 suppresses TRAIL receptor 2 and protects againstas TRAIL-induced apoptosis 25743330_Increased expression of MAGE-D2 mRNA was associated with distant metastasis in Gastric Cancer. 26705694_MAGE-D2 is a dynamic protein whose shuttling properties could suggest a role in cell cycle regulation. 27120771_We found that MAGED2 mutations caused X-linked polyhydramnios with prematurity and a severe but transient form of antenatal Bartter's syndrome. 29146702_MAGED2 mutations explained 9% of cases of antenatal Bartter syndrome in a French cohort, and accounted for 38% of patients without other characterized mutations and for 44% of male probands of negative cases. 29677005_MAGED2 loss of function is the cause of an X-linked transient form of antenatal Bartter's syndrome. Moreover, our findings showed that MAGE-D2 promotes the biogenesis of kidney membrane transporters. 34775100_MAGED2 controls vasopressin-induced aquaporin-2 expression in collecting duct cells. | ENSMUSG00000025268 | Maged2 | 3609.93349 | 0.9308344 | -0.1034036077 | 0.07375506 | 1.959890e+00 | 1.615251e-01 | 5.523860e-01 | No | Yes | 4242.334314 | 331.060808 | 4404.970552 | 335.861169 | |
| ENSG00000102781 | 84056 | KATNAL1 | protein_coding | Q9BW62 | FUNCTION: Regulates microtubule dynamics in Sertoli cells, a process that is essential for spermiogenesis and male fertility. Severs microtubules in an ATP-dependent manner, promoting rapid reorganization of cellular microtubule arrays (By similarity). Has microtubule-severing activity in vitro (PubMed:26929214). {ECO:0000250|UniProtKB:Q8K0T4, ECO:0000269|PubMed:26929214}. | 3D-structure;ATP-binding;Acetylation;Cytoplasm;Cytoskeleton;Isomerase;Microtubule;Nucleotide-binding;Phosphoprotein;Reference proteome | hsa:84056; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; spindle [GO:0005819]; spindle pole [GO:0000922]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; identical protein binding [GO:0042802]; isomerase activity [GO:0016853]; microtubule binding [GO:0008017]; microtubule-severing ATPase activity [GO:0008568]; microtubule severing [GO:0051013]; spermatogenesis [GO:0007283] | 20519956_KL1-mediated microtubule severing is utilized to generate microtubule seeds within the poles and loss of this activity alters the normal balance of motor-generated forces that determine spindle length. 24664804_microdeletion 13q12.3 represents a novel clinically recognizable condition and that the microtubule severing gene KATNAL1 and the chromatin-associated gene HMGB1 are candidate genes for intellectual disability inherited in an autosomal dominant pattern. 24913027_KATNAL1 gene sequence variants are not associated with azoospermia. | ENSMUSG00000041298 | Katnal1 | 496.91365 | 0.8416454 | -0.2487155266 | 0.15840936 | 2.449045e+00 | 1.175964e-01 | 4.875707e-01 | No | Yes | 529.752298 | 93.235521 | 616.602185 | 106.028845 | ||
| ENSG00000102910 | 83752 | LONP2 | protein_coding | Q86WA8 | FUNCTION: ATP-dependent serine protease that mediates the selective degradation of misfolded and unassembled polypeptides in the peroxisomal matrix. Necessary for type 2 peroxisome targeting signal (PTS2)-containing protein processing and facilitates peroxisome matrix protein import (By similarity). May indirectly regulate peroxisomal fatty acid beta-oxidation through degradation of the self-processed forms of TYSND1. {ECO:0000255|HAMAP-Rule:MF_03121, ECO:0000269|PubMed:22002062}. | ATP-binding;Acetylation;Alternative splicing;Hydrolase;Nucleotide-binding;Peroxisome;Protease;Reference proteome;Serine protease | In human, peroxisomes function primarily to catalyze fatty acid beta-oxidation and, as a by-product, produce hydrogen peroxide and superoxide. The protein encoded by this gene is an ATP-dependent protease that likely plays a role in maintaining overall peroxisome homeostasis as well as proteolytically degrading peroxisomal proteins damaged by oxidation. The protein has an N-terminal Lon N substrate recognition domain, an ATPase domain, a proteolytic domain, and, in some isoforms, a C-terminal peroxisome targeting sequence. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jan 2017]. | hsa:83752; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; peroxisomal matrix [GO:0005782]; peroxisome [GO:0005777]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; ATP-dependent peptidase activity [GO:0004176]; enzyme binding [GO:0019899]; peptidase activity [GO:0008233]; protease binding [GO:0002020]; serine-type endopeptidase activity [GO:0004252]; peroxisome organization [GO:0007031]; protein import into peroxisome matrix [GO:0016558]; protein processing [GO:0016485]; protein quality control for misfolded or incompletely synthesized proteins [GO:0006515]; protein targeting to peroxisome [GO:0006625]; regulation of fatty acid beta-oxidation [GO:0031998]; response to organic cyclic compound [GO:0014070] | 22002062_The proteolytic activity of oligomeric Tysnd1 is in turn controlled by self-cleavage of Tysnd1 and degradation of Tysnd1 cleavage products by PsLon. 29502128_Findings suggest that LONP2 promotes cervical tumorigenesis via oxidative stress. | ENSMUSG00000047866 | Lonp2 | 2633.95581 | 1.1062565 | 0.1456858831 | 0.11310863 | 1.661831e+00 | 1.973563e-01 | 5.894511e-01 | No | Yes | 4132.008480 | 547.425747 | 3523.549440 | 456.602529 | |
| ENSG00000102984 | 55565 | ZNF821 | protein_coding | O75541 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;Coiled coil;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a protein with two C2H2 zinc finger motifs and a score-and-three (23)-amino acid peptide repeat (STPR) domain. The STPR domain of the encoded protein binds to double stranded DNA and may also contain a nuclear localization signal, suggesting that this protein interacts with chromosomal DNA. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:55565; | nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000031728 | Zfp821 | 180.97481 | 0.8602836 | -0.2171157201 | 0.21640454 | 1.001174e+00 | 3.170265e-01 | 6.973764e-01 | No | Yes | 196.706405 | 19.879146 | 218.711465 | 21.353317 | |
| ENSG00000103034 | 65009 | NDRG4 | protein_coding | Q9ULP0 | FUNCTION: Contributes to the maintenance of intracerebral BDNF levels within the normal range, which is necessary for the preservation of spatial learning and the resistance to neuronal cell death caused by ischemic stress (By similarity). May enhance growth factor-induced ERK1 and ERK2 phosphorylation, including that induced by PDGF and FGF. May attenuate NGF-promoted ELK1 phosphorylation in a microtubule-dependent manner. {ECO:0000250, ECO:0000269|PubMed:12755708}. | Alternative splicing;Cytoplasm;Phosphoprotein;Reference proteome | This gene is a member of the N-myc downregulated gene family which belongs to the alpha/beta hydrolase superfamily. The protein encoded by this gene is a cytoplasmic protein that is required for cell cycle progression and survival in primary astrocytes and may be involved in the regulation of mitogenic signalling in vascular smooth muscles cells. Alternative splicing results in multiple transcripts encoding different isoforms.[provided by RefSeq, Jun 2011]. | hsa:65009; | basolateral plasma membrane [GO:0016323]; cell projection membrane [GO:0031253]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; mitochondrion [GO:0005739]; brain development [GO:0007420]; cardiac muscle cell proliferation [GO:0060038]; cell differentiation [GO:0030154]; cell migration involved in heart development [GO:0060973]; embryonic heart tube development [GO:0035050]; heart looping [GO:0001947]; negative regulation of platelet-derived growth factor receptor signaling pathway [GO:0010642]; negative regulation of smooth muscle cell migration [GO:0014912]; negative regulation of smooth muscle cell proliferation [GO:0048662]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of neuron projection development [GO:0010976]; regulation of endocytic recycling [GO:2001135]; signal transduction [GO:0007165]; vesicle docking [GO:0048278]; visual learning [GO:0008542] | 11936845_Cloning and expression of the gene; specifically expressed in brain and heart 12755708_smap8 is involved in the regulation of mitogenic signalling in vascular smooth muscle cells, possibly in response to a homocysteine-induced injury [SMAP8] 16408304_NDRG4 overexpression enhances ERK activation 19305408_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 19535783_NDRG4 is a candidate tumor suppressor gene in colorectal cancer whose expression is frequently inactivated by promoter methylation. 19592488_NDRG4 is required for cell cycle progression and survival, thereby diverging in function from its tumor suppressive family member NDRG2 in astrocytes and GBM cells 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22399192_expression of NDRG4 is downregulated in human gliomas. The glioma patients with lower NDRG4 expression have a poor prognosis. 22489821_Over expression of NDRG4 inhibited proliferation of GBM cells. 22869042_NDRG4 is involved in modulating cell proliferation, invasion, migration and angiogenesis in meningioma, and may play a valuable role as a molecular target in its treatment 23725756_summarizes the current research on NDRG3 and NDRG4,including the molecular structure, cellular and tissue distribution, biological function, and function in cancer. 24993691_Methylation level in stool decreases dramatically following colorectal cancer resection 25241110_A homozygous variant in NDRG4 may be the causative variant of the autosomal recessive form of infantile myofibromatosis. 25520251_Two downregulated genes, NDRG4 and GINS3, have been located in a genomic interval associated with cardiac repolarization in published GWASs and zebra fish knockout models 26053091_These findings bring novel insight to the roles of NDRG4 in meningioma progression 26515606_Data indicate that patients with tumor of reduced NDRG family member 4 protein (NDRG4) mRNA level had unfavorable disease-free and overall survival. 26976975_Study shows that NDRG4 was significantly up-regulated in glioblastomas (GBM) and seems to play a role in GBM prognosis. These results indicate that NDRG4 gene in MGMT-methylated cells is a putative tumor-suppressor gene and an oncogene in cells with unmethylated MGMT. 27175791_NDRG4 might play an important role during early pregnancy. 28042954_NDRG4 promoter hypermethylation contributed to the risk of gastric cancer and predicted a poor prognosis in Chinese gastric cancer patients. 28044229_NDRG4 - the biomarker of a currently in-use multi-target stool DNA test was commonly expressed in tumor tissue specimens, independent of Fecal Immunochemical Test result. 28524415_NDRG4 is exclusively expressed by central, peripheral and enteric neurons/nerves, suggesting a neuronal-specific role of this protein 29500881_Results indicate that NDRG family member 4 protein (NDRG4) could be a potential tumor suppressor and prognostic marker of gastric cancer. 32166572_NDRG4 Alleviates Abeta1-40 Induction of SH-SY5Y Cell Injury via Activation of BDNF-Inducing Signalling Pathways. 32187107_MiR-433 Regulates Myocardial Ischemia Reperfusion Injury by Targeting NDRG4 Via the PI3K/Akt Pathway. 32196834_TFPI2 and NDRG4 gene promoter methylation analysis in peripheral blood mononuclear cells are novel epigenetic noninvasive biomarkers for colorectal cancer diagnosis. 33211401_A loss-of-function mutation p.T256M in NDRG4 is implicated in the pathogenesis of pulmonary atresia with ventricular septal defect (PA/VSD) and tetralogy of Fallot (TOF). 33298240_Hypermethylation-mediated silencing of NDRG4 promotes pancreatic ductal adenocarcinoma by regulating mitochondrial function. 33667214_Identification of NDRG Family Member 4 (NDRG4) and CDC28 Protein Kinase Regulatory Subunit 2 (CKS2) as Key Prognostic Genes in Adrenocortical Carcinoma by Transcriptomic Analysis. 33890711_Loss of enteric neuronal Ndrg4 promotes colorectal cancer via increased release of Nid1 and Fbln2. 34564976_Highly sensitive fecal DNA testing of NDRG4 12b methylation is a promising marker for detection of colorectal precancerosis. | ENSMUSG00000036564 | Ndrg4 | 78.68323 | 0.4795183 | -1.0603421538 | 0.35210868 | 8.740806e+00 | 3.111669e-03 | 9.556353e-02 | No | Yes | 52.034855 | 7.976746 | 100.147559 | 14.451400 | |
| ENSG00000103253 | 84264 | HAGHL | protein_coding | Q6PII5 | FUNCTION: Hydrolase acting on ester bonds. {ECO:0000305}. | Alternative splicing;Hydrolase;Metal-binding;Reference proteome;Zinc | hsa:84264; | hydroxyacylglutathione hydrolase activity [GO:0004416]; metal ion binding [GO:0046872] | 20237496_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000061046 | Haghl | 164.72185 | 1.0225906 | 0.0322287168 | 0.27966761 | 1.327624e-02 | 9.082687e-01 | 9.740734e-01 | No | Yes | 180.441239 | 28.794565 | 172.903117 | 26.919472 | ||
| ENSG00000103353 | 56061 | UBFD1 | protein_coding | O14562 | FUNCTION: May play a role as NF-kappa-B regulator. {ECO:0000269|PubMed:19285159}. | Reference proteome | hsa:56061; | cadherin binding [GO:0045296]; RNA binding [GO:0003723] | ENSMUSG00000030870 | Ubfd1 | 5032.68245 | 1.1060622 | 0.1454325794 | 0.07197811 | 4.095757e+00 | 4.299097e-02 | 3.164178e-01 | No | Yes | 6147.777067 | 527.410137 | 5383.252182 | 451.722423 | |||
| ENSG00000103528 | 51760 | SYT17 | protein_coding | Q9BSW7 | FUNCTION: Plays a role in dendrite formation by melanocytes (PubMed:23999003). {ECO:0000269|PubMed:23999003}. | 3D-structure;Differentiation;Membrane;Phosphoprotein;Reference proteome;Repeat | hsa:51760; | exocytic vesicle [GO:0070382]; plasma membrane [GO:0005886]; trans-Golgi network [GO:0005802]; calcium ion binding [GO:0005509]; calcium-dependent phospholipid binding [GO:0005544]; clathrin binding [GO:0030276]; phosphatidylserine binding [GO:0001786]; SNARE binding [GO:0000149]; syntaxin binding [GO:0019905]; calcium-ion regulated exocytosis [GO:0017156]; cell differentiation [GO:0030154]; cellular response to calcium ion [GO:0071277]; positive regulation of dendrite extension [GO:1903861]; regulation of calcium ion-dependent exocytosis [GO:0017158]; regulation of dopamine secretion [GO:0014059]; vesicle-mediated transport [GO:0016192] | 16672768_The results suggest that B/K proteins play a role as potential substrates for PKA in the area where they are expressed. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | ENSMUSG00000058420 | Syt17 | 17.54151 | 0.5009235 | -0.9973377112 | 0.69030227 | 2.025931e+00 | 1.546341e-01 | No | Yes | 11.279718 | 3.115001 | 21.333019 | 5.416597 | |||
| ENSG00000104044 | 4948 | OCA2 | protein_coding | Q04671 | FUNCTION: Could be involved in the transport of tyrosine, the precursor to melanin synthesis, within the melanocyte. Regulates the pH of melanosome and the melanosome maturation. One of the components of the mammalian pigmentary system. Seems to regulate the post-translational processing of tyrosinase, which catalyzes the limiting reaction in melanin synthesis. May serve as a key control point at which ethnic skin color variation is determined. Major determinant of brown and/or blue eye color. {ECO:0000269|PubMed:11310796, ECO:0000269|PubMed:15262401, ECO:0000269|PubMed:18252222, ECO:0000269|PubMed:22234890, ECO:0000269|PubMed:7601462}. | Albinism;Alternative splicing;Disease variant;Glycoprotein;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes the human homolog of the mouse p (pink-eyed dilution) gene. The encoded protein is believed to be an integral membrane protein involved in small molecule transport, specifically tyrosine, which is a precursor to melanin synthesis. It is involved in mammalian pigmentation, where it may control skin color variation and act as a determinant of brown or blue eye color. Mutations in this gene result in type 2 oculocutaneous albinism. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:4948; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; melanosome membrane [GO:0033162]; L-tyrosine transmembrane transporter activity [GO:0005302]; transporter activity [GO:0005215]; cell population proliferation [GO:0008283]; eye pigment biosynthetic process [GO:0006726]; melanin biosynthetic process [GO:0042438]; melanocyte differentiation [GO:0030318]; spermatid development [GO:0007286] | 8875191_Human DNA sequences;hair colour 12028586_role of P protein and tyrosinase in oculocutaneous albinism 12163334_P gene, in part, determiontes normal phenotypic variation in human eye color and may represent an inherited biomarkers of cutaneous cancer risk 12469324_A 122.5-kilobase deletion of the P gene underlies the high prevalence of oculocutaneous albinism type 2 in the Navajo population 12579416_We show that OCA2 has measurable effects on skin pigmentation differences between the west African and west European parental populations. 12727022_two missense substitutions, A481T and Q799H in the P gene in oculocutaneous albinism 12817591_A candidate gene for pigmentation. 15712365_9 novel mutations and 12 novel polymorphisms associated with oculocutaneous albinism type II are reported. 15889046_Observational study of gene-disease association. (HuGE Navigator) 15889046_we show that MM and OCA2 are associated (p value=0.030 after correction for multiple testing). 16417222_Observational study of genotype prevalence. (HuGE Navigator) 16453125_The macular hypoplasia has to be considered a concerted interaction with compound heterozygous mutations in the P gene manifesting a mild form of oculocutaneous albinism. 17236130_Differences within the 5' proximal regulatory control region of the OCA2 gene alter expression or messenger RNA-transcript levels and may be responsible for eye-color and other pigmentary trait associations. 17236130_Observational study of gene-disease association. (HuGE Navigator) 17568986_Observational study of genotype prevalence. (HuGE Navigator) 17568986_These findings suggest that OCA2 481Thr arose in a region of low ultraviolet radiation and thereafter spread to neighboring populations. 17570052_Observational study of genotype prevalence. (HuGE Navigator) 17619204_results confirm that OCA2 is the major human iris color gene and suggest that using an empirical database-driven system, genotypes from a modest number of SNPs within this gene can be used to accurately predict iris melanin content from DNA 17767372_3 different haplotypes (TAGCT, TAGTT and TAGCC with frequencies of 0.66, 0.28 and 0.06, respectively) associated with the mutation in the 53 OCA2 patients, while 11 different haplotypes were observed in the control group 17952075_Genome-wide association study of gene-disease association. (HuGE Navigator) 17960121_Pink-eye-dilution gene mutations underlie oculocutaneous albinism in this family. Two known mutations in MC1R caused red hair color in one family member. No modifier effect of MC1R on P mutations could be deduced. 18093281_Variation present in the OCA2 gene and perhaps some other pigment related genes must be taken into account in order to explain the high phenotypic variation in iris colour. 18252221_Genome-wide association study of gene-disease association. (HuGE Navigator) 18252222_Observational study of gene-disease association. (HuGE Navigator) 18326704_Most patients with AROA (autosomal recessive ocular albinism) represent phenotypically mild variants of oculocutaneous albinism , well over half of which is OCA1. 18463683_Observational study of gene-disease association. (HuGE Navigator) 18528436_OCA2 and HERC2 have roles in hair color in Australian adolescents 18528436_Observational study of gene-disease association. (HuGE Navigator) 18563784_Observational study of gene-disease association. (HuGE Navigator) 18636124_Observational study of gene-disease association. (HuGE Navigator) 18650849_strong correlations in MATP-L374F, OCA2, and melanocortin-1 receptor with skin, eye, and hair color variation, respectively 18676680_Observational study of gene-disease association. (HuGE Navigator) 18680187_Oculocutaneous albinism phenotype (OCA2) can be modified by mutation in TYRP1. 18683857_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18839200_Observational study of genotype prevalence. (HuGE Navigator) 19060277_Observational study of gene-disease association. (HuGE Navigator) 19060277_TYR is the major OCA (oculocutaneous albinism) gene in Denmark, but several patients do not have mutations in the investigated genes. 19116314_OCA2 is targeted to and functions within melanosomes but that residence within melanosomes may be regulated by secondary or alternative targeting to lysosomes. 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19208107_It is concluded that OCA2 rs1800407 is associated with eye colour. 19208107_Observational study of gene-disease association. (HuGE Navigator) 19320733_Observational study of gene-disease association. (HuGE Navigator) 19320733_The variant allele of OCA2 R419Q (rs1800407) is associated with increased risk of malignant melanoma. 19340012_Observational study and genome-wide association study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19382693_Observational study of genotype prevalence. (HuGE Navigator) 19382693_Polymorphism of pigmentation genes (OCA2 and ASIP) in some populations of Russia 19384953_Observational study of gene-disease association. (HuGE Navigator) 19384953_The OCA2 Arg419Gln is associated with basal cell carcinoma 19472299_Observational study of gene-disease association. (HuGE Navigator) 19472299_Three single nucleotide polymorphisms found within intron 1 of the OCA2 gene (rs7495174, rs4778241, rs4778138). 30 UTR region (rs1129038) of the HERC2 gene 19625176_Observational study of gene-disease association. (HuGE Navigator) 19626598_In 5 israeli families a P gene mutation was detected. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19710684_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19865097_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20019752_Inheritance of a novel mutated allele of the OCA2 gene associated with high incidence of oculocutaneous albinism in a Polynesian community. 20221248_Observational study of gene-disease association. (HuGE Navigator) 20221248_The non-synonymous polymorphism rs1800414 (His615Arg) located within the OCA2 gene is significantly associated with skin pigmentation in this sample. 20308648_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20457063_). Sequence variations in rs11636232 and rs7170852 in HERC2, rs1800407 in OCA2 and rs16891982 in MATP showed additional association with eye colours 20457063_Observational study of gene-disease association. (HuGE Navigator) 20585627_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20629734_Observational study of gene-disease association. (HuGE Navigator) 20629734_role in pigmentation characteristics in Spanish population 20801516_Observational study of genetic testing. (HuGE Navigator) 20861488_TYR gene mutations represent a relevant cause of oculocutaneous albinism in Italy, whereas mutations in P present a lower frequency. Clinical analysis revealed that the severity of the ocular manifestations depends on the degree of retinal pigmentation. 21085994_Using quantitative multiplex fluorescent PCR and very high-resolution array-CGH focussed on the OCA2 gene and surrounding regions in 15q12, study identified 2 new gene deletions and 1 duplication in Oculocutaneous albinism type 2 patients. 21541274_TYR gene mutations have a more severe effect on pigmentation than mutations in OCA2 and the GPR143 gene. Nevertheless, mutations in these genes affect the development of visual function either directly or by interaction with other genes like MC1R. 21979861_In this paper I shall discuss the anatomy and genetics of normal eye colour, together with a wide and diverse range of conditions that may produce an alteration in normal iris pigmentation or form. 22042571_Three mutational alleles, R278X and R52I of the TYR gene and C229Y of the SLC45A2 gene, are added to the mutational spectra of Korean patients with oculocutaneous albinism (OCA) 23063908_Although variants within OCA2 were tested for association, the 2.7kb deletion allele of OCA2 was not tested. This led us to hypothesize that the deletion allele may confer resistance to susceptibility 23103111_The discovery of this novel OCA2 variant adds to the body of evidence on the detrimental effects of OCA2 gene mutations on pigmentation and supports existing GWAS data on the relevance of the OCA2 gene in melanoma predisposition. 23165166_We examined the association between 12 variants of four pigmentation-related genes (TYR, OCA2, SLC45A2, MC1R) and variations in the melanin index of 456 Japanese females using a multiple regression analysis. 23601698_given a particular HERC2/OCA2 genotype, males are more prone to have lighter eye colors than predicted by their genotypes, while females tend to have darker eye colors than predicted 23824587_The most disease-associated mutation of R305W which corresponds to OCA2 results showed prominent loss of stability and rise in mutant flexibility values in 3D space. 24361966_Functional characterization of two novel splicing mutations in the OCA2 gene associated with oculocutaneous albinism type II. c.2139G>A represents the first exonic splicing mutation identified in an OCA2 gene. 24387780_REVIEW: current hypotheses and the available data on the mechanism of OCA2 transcriptional regulation and how this is influenced by genetic variation 25119903_4 heterozygous mutations of the P gene were found two Chinese families affected with oculocutaneous albinism type . 25455140_individuals from ''El Santuario and Marinilla, Antioquia'', genetically isolated northwestern towns in Colombia well known for its high albinism prevalence [7], were all homozygous for the previously reported p.A787T mutation in OCA2 gene 25469862_variations in OCA2 might have developed by diversifying selection. 25809079_Two nonsynonymous OCA2 polymorphisms (rs1800414 and rs74653330) are independently associated with normal skin pigmentation variation in East Asian populations and have very different frequency distributions in East Asia. 25919014_The two mutations (c.1114delG in the TYR gene and c.1426A>G in the OCA2 gene) may be responsible for partial clinical manifestations of Oculocutaneous albinism. 26744415_We analyzed five SNPs located in the 44.2 kb region and three SNPs outside this region with strong selective signals. Under the additive genetic model, we detected a significant association of rs1800414 with skin pigmentation for both exposed and unexposed areas consistent with a previous association analysis of rs1800414 No significant association was observed for the other seven SNPs (P > 0.05) 28081795_Evaluated the association of seven OCA2-HERC2 SNPs and haplotypes with pigmentation characteristics (eye, skin, hair and freckles) in the highly admixed and phenotypically heterogeneous Brazilian population. Such SNPs and haplotypes could be deemed as good predictors for the presence of freckles and for skin, eye and hair pigmentation in the Brazilian population. 28456133_The results of this study is the first to show an association between OCA2 variants and time to first cSCC post-transplant. 31345173_Novel compound heterozygous mutations in OCA2 gene associated with non-syndromic oculocutaneous albinism in a Chinese Han patient. 31813138_The main type of ocular albinism is oculocutaneous albinism type II in China's Liuzhou region, where the most common variations of the P gene were c.803-3C>G and c.1327G>A (p.Val443Ile). 32963319_The distinctive geographic patterns of common pigmentation variants at the OCA2 gene. 32966289_Germline and somatic albinism variants in amelanotic/hypomelanotic melanoma: Increased carriage of TYR and OCA2 variants. 33800529_Genetic Causes of Oculocutaneous Albinism in Pakistani Population. 33974259_[Analysis of genetic variation for a child affected with congenital insensitivity to pain with anhidrosis and albinism by whole genome sequencing]. 34246199_A custom capture sequence approach for oculocutaneous albinism identifies structural variant alleles at the OCA2 locus. 34795370_Further insight into the global variability of the OCA2-HERC2 locus for human pigmentation from multiallelic markers. 35328057_Delineating Novel and Known Pathogenic Variants in TYR, OCA2 and HPS-1 Genes in Eight Oculocutaneous Albinism (OCA) Pakistani Families. | ENSMUSG00000030450 | Oca2 | 165.64456 | 1.1776398 | 0.2358983859 | 0.22677072 | 1.086759e+00 | 2.971903e-01 | 6.814908e-01 | No | Yes | 215.409601 | 28.606838 | 180.964947 | 23.672959 | |
| ENSG00000104218 | 79848 | CSPP1 | protein_coding | Q1MSJ5 | FUNCTION: May play a role in cell-cycle-dependent microtubule organization. {ECO:0000269|PubMed:16826565}. | Alternative splicing;Ciliopathy;Coiled coil;Cytoplasm;Cytoskeleton;Joubert syndrome;Microtubule;Phosphoprotein;Reference proteome | This gene encodes a centrosome and spindle pole associated protein. The encoded protein plays a role in cell-cycle progression and spindle organization, regulates cytokinesis, interacts with Nephrocystin 8 and is required for cilia formation. Mutations in this gene result in primary cilia abnormalities and classical Joubert syndrome. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Apr 2014]. | hsa:79848; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; spindle [GO:0005819]; spindle pole [GO:0000922]; positive regulation of cell division [GO:0051781]; positive regulation of cytokinesis [GO:0032467] | 15580290_Novel centrosome/microtubule-associated coiled-coil protein (CSPP)is associated with centrosomes and microtubules and may play a role in the regulation of G(1)/S-phase progression and spindle assembly [CSPP]. 16826565_Taken together, CSPP and CSPP-L interact with centrosomes and microtubules and can differently affect microtubule organization. 16826565_Taken together, CSPP and CSPP-L interact with centrosomes and microtubules and can differently affect microtubule organization.[CSPP-L] 19129481_CSPP interacts with and recruits MyoGEF to the central spindle, where MyoGEF contributes to the spatiotemporal regulation of cytokinesis. 20519441_CSPP isoforms require their common C-terminal domain to interact with Nephrocystin 8 (NPHP8/RPGRIP1L) and to form a ternary complex with NPHP8 and NPHP4. 24360803_mutations in CSPP1 were associated with variable ciliopathy phenotypes ranging from Joubert syndrome to the more severe Meckel-Gruber syndrome with perinatal lethality and occipital encephalocele 24360807_Our data suggest that CSPP1 is required for proper primary cilium formation or stability and that CSPP1 mutations result in abnormal mid-hindbrain development. 24360808_CSPP1 mutations are a major cause of the Joubert-Jeune phenotype in humans. 24901235_Differential expression of a nuclear CSPP1 isoform identified biologically and clinically distinct subgroups of basal-like breast carcinoma. 26241740_Microtubule-independent but desmoplakin-dependent localization of CSPP-L to desmosomes occurs in apical-basal polarized epithelial cells. CSPP-L depletion promoted multi-lumen spheroid formation in Caco-2 cells. 26378239_propose that CSPP1 cooperates with CENP-H on kinetochores to serve as a novel regulator of kinetochore microtubule dynamics for accurate chromosome segregation 30965236_Tumor-promoting effect was inhibited after we transfected miR-1236-3p into circ-CSPP1 overexpressing OC cells. 32495924_Roles of circ-CSPP1 on the proliferation and metastasis of glioma cancer. | ENSMUSG00000056763 | Cspp1 | 1236.40900 | 1.0022048 | 0.0031772925 | 0.12176645 | 6.808541e-04 | 9.791830e-01 | 9.950724e-01 | No | Yes | 1465.571856 | 281.181606 | 1452.879811 | 272.794000 | |
| ENSG00000104365 | 3551 | IKBKB | protein_coding | O14920 | FUNCTION: Serine kinase that plays an essential role in the NF-kappa-B signaling pathway which is activated by multiple stimuli such as inflammatory cytokines, bacterial or viral products, DNA damages or other cellular stresses (PubMed:30337470). Acts as part of the canonical IKK complex in the conventional pathway of NF-kappa-B activation. Phosphorylates inhibitors of NF-kappa-B on 2 critical serine residues. These modifications allow polyubiquitination of the inhibitors and subsequent degradation by the proteasome. In turn, free NF-kappa-B is translocated into the nucleus and activates the transcription of hundreds of genes involved in immune response, growth control, or protection against apoptosis. In addition to the NF-kappa-B inhibitors, phosphorylates several other components of the signaling pathway including NEMO/IKBKG, NF-kappa-B subunits RELA and NFKB1, as well as IKK-related kinases TBK1 and IKBKE (PubMed:11297557, PubMed:20410276). IKK-related kinase phosphorylations may prevent the overproduction of inflammatory mediators since they exert a negative regulation on canonical IKKs. Phosphorylates FOXO3, mediating the TNF-dependent inactivation of this pro-apoptotic transcription factor (PubMed:15084260). Also phosphorylates other substrates including NCOA3, BCL10 and IRS1 (PubMed:17213322). Within the nucleus, acts as an adapter protein for NFKBIA degradation in UV-induced NF-kappa-B activation (PubMed:11297557). Phosphorylates RIPK1 at 'Ser-25' which represses its kinase activity and consequently prevents TNF-mediated RIPK1-dependent cell death (By similarity). Phosphorylates the C-terminus of IRF5, stimulating IRF5 homodimerization and translocation into the nucleus (PubMed:25326418). {ECO:0000250|UniProtKB:O88351, ECO:0000269|PubMed:11297557, ECO:0000269|PubMed:15084260, ECO:0000269|PubMed:17213322, ECO:0000269|PubMed:19716809, ECO:0000269|PubMed:20410276, ECO:0000269|PubMed:20434986, ECO:0000269|PubMed:20797629, ECO:0000269|PubMed:21138416, ECO:0000269|PubMed:25326418, ECO:0000269|PubMed:30337470}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Disease variant;Host-virus interaction;Hydroxylation;Isopeptide bond;Kinase;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;S-nitrosylation;SCID;Serine/threonine-protein kinase;Transferase;Ubl conjugation | The protein encoded by this gene phosphorylates the inhibitor in the inhibitor/NF-kappa-B complex, causing dissociation of the inhibitor and activation of NF-kappa-B. The encoded protein itself is found in a complex of proteins. Several transcript variants, some protein-coding and some not, have been found for this gene. [provided by RefSeq, Sep 2011]. | hsa:3551; | CD40 receptor complex [GO:0035631]; cytoplasm [GO:0005737]; cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; IkappaB kinase complex [GO:0008385]; membrane raft [GO:0045121]; nucleus [GO:0005634]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; IkappaB kinase activity [GO:0008384]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; protein kinase activity [GO:0004672]; protein kinase binding [GO:0019901]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; scaffold protein binding [GO:0097110]; transferrin receptor binding [GO:1990459]; antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent [GO:0002479]; cellular response to tumor necrosis factor [GO:0071356]; cortical actin cytoskeleton organization [GO:0030866]; Fc-epsilon receptor signaling pathway [GO:0038095]; I-kappaB kinase/NF-kappaB signaling [GO:0007249]; inflammatory response [GO:0006954]; innate immune response [GO:0045087]; interleukin-1-mediated signaling pathway [GO:0070498]; negative regulation of bicellular tight junction assembly [GO:1903347]; negative regulation of myosin-light-chain-phosphatase activity [GO:0035509]; peptidyl-serine phosphorylation [GO:0018105]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; protein localization to plasma membrane [GO:0072659]; protein phosphorylation [GO:0006468]; regulation of establishment of endothelial barrier [GO:1903140]; regulation of phosphorylation [GO:0042325]; regulation of tumor necrosis factor-mediated signaling pathway [GO:0010803]; response to virus [GO:0009615]; stimulatory C-type lectin receptor signaling pathway [GO:0002223]; stress-activated MAPK cascade [GO:0051403]; T cell receptor signaling pathway [GO:0050852]; tumor necrosis factor-mediated signaling pathway [GO:0033209] | 11815618_difference of kinetic mechanisms of heterodimer with IKK-1 compared with IKK and TBK-1 11839743_IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. 11864612_Tnf-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. 11945026_Endothelial IKK beta signaling is required for monocyte adhesion under laminar flow conditions. 11954826_Hyperoxia prolongs tumor necrosis factor-alpha-mediated activation of NF-kappaB: role of IkappaB kinase. 12210728_FANCA may function to recruit IKK2, thus providing the cell a means of rapidly responding to stress. 12393548_results demonstrate that IKK2 is essential for dendritic cell activation induced by CD40L or contact with allogeneic T cells, but not by LPS, whereas NIK is not required for any of these signals 12411322_activity is comparable in normal B cells and B-CLL lymphocytes and is not involved in up-regulation of TRAF1 in B-CLL 12547194_TNFalpha activation of the NF-kappaB pathway is associated with the inducible binding of TAK1 to TRAF2 and both IKKalpha and IKKbeta 12589056_IKKalpha and IKKbeta may alter their substrate and signaling specificities to regulate mitogen-induced DNA synthesis through distinct mechanisms 12637324_identified the alpha-chain of the GMCSF receptor as interaction partner of IkappaB kinase beta; direct interaction of IKKbeta and GMRalpha in cells was verified. 12645577_IKKbeta has a role in TNFalpha-induced ICAM-1 expression 12657630_identification of inducible phosphoacceptors in IKKgamma/NEMO subunit 12842894_IkappaB and NF-kappaB are substrates for the IKK complex in the activation of NF-kappaB 12890679_ligand-induced homotypic interactions between IKKbeta molecules result in IKKbeta phosphorylation and consequently IKK activation 14514672_determination of signal induced ubiquitination 14585846_noncoordinate expression of I kappa B kinases plays a role in determining the cell type-specific role of Akt in NF-kappa B activation. 14585847_PP2Cbeta down-regulates cytokine-induced NF-kappaB activation by altering IKK activity. 14625285_IKBK has a role in CXCL16 signaling that induces cell-cell adhesion and aortic smooth muscle cell proliferation 14715628_Virtually all TNF-alpha-inducible genes were dependent on I kappa B kinase 2 (IKK2)/NF-kappa B activation, whereas a minor number was additionally modulated by p38; genes suppressed by IKK2/NF-kappa B were newly identified 14990741_required for Human Cytomegalovirus-induced NF-kappaB activation, as well as the replication of different HCMV strains 15128824_Phosphorylation of serine 536, the main T-cell-inducible phosphorylation site of the NF-kappa B p65 subunit, is mediated by IKK beta and occurs within the cytosolic p50/p65/I kappa B alpha complex. 15140882_NF-kappaB-inducing kinase induces p100 processing by docking IkappaB kinase alpha (IKKalpha) to p100 15217951_NF-kappaB dysregulation and uPA overexpression have roles in a more aggressive tumor behavior in hepatocellular carcinoma, and IKKbeta plays a critical role in the HBx-activated NF-kappaB signaling pathway 15226300_IKKbeta-IKKgamma complexes are involved in mainstream NF-kappaB activation cascades because they can be activated by tumor necrosis factor 15254232_IkappaBalpha kinase beta is inhibited by dehydroascorbic acid 15319427_Ubiquitin-like domain as a critical functional domain specific for IKKbeta that might play a role in dissociating IKKbeta from p65. 15489227_IL-1-inducible phosphorylation of p65 NFkB is mediated by multiple protein kinases including IKKalpha, IKKbeta, IKKepsilon, TBK1, and an unknown kinase and couples p65 to TAFII31-mediated IL-8 transcription 15492226_activation through a proteasome-independent mechanism via effect of TIFA on TRAF6 15564333_IKK-beta/NF-kappa B transcription pathway is a key regulator of IL-6, IL-8, and TNF-alpha release from adipose tissue and skeletal muscle 15837793_results identify IKK as an important factor in triggering influenza virus-induced inflammatory reactions in pulmonary epithelium 15880043_Allogenic dendritic cells, rendered immature by IKK2 transfection, induce in vitro differentiation of naive T cells into CD4+ T-regulatory cells which might prevent graft rejection. 15951441_NIBP is a NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation 16046471_IKKbeta phosphorylates multiple p65 sites, as well as in an IkappaB-p65 complex, and S468 phosphorylation slightly reduces TNF-alpha- and IL-1beta-induced NF-kappaB activation 16082226_Ser15 phosphorylation is important in regulating the oncogenic function of mutant p53 apoptosis induction in the context of the NF-kappaB/IkappaB signaling pathway 16123045_IKKbeta has an important role in TNF-alpha-mediated mucus production in airway epithelium in vitro and in vivo 16126728_PP2A plays a positive rather than a negative role in the regulation of IKKbeta 16207722_results indicate that 3-phosphoinositide-dependent protein kinase-1 is a critical regulator of cell survival by modulating the IkappaB kinase (IKK)/nuclear factor-kappaB pathway 16267042_chronic phosphorylation of IKKbeta at Ser-177/Ser-181 leads to monoubiquitin attachment at nearby Lys-163, which in turn modulates the phosphorylation status of IKKbeta at select C-terminal serines 16280329_LMP1 utilizes two distinct pathways to activate NF-kappaB: a major one through CTAR2/TRAF6/TAK1/IKKbeta (canonical pathway) and a minor one through CTAR1/TRAF3/NIK/IKKalpha (noncanonical pathway) 16583354_IKK-2 NBD (NEMO binding domain), encompasses LDWSWL from amino acids 735-745, showed modest binding to NEMO. 16728640_proposed that Yersinia YopJ blocks signaling of the and NFkappaB pathway by binding and acetylating critical residues in the activation loop of IKKbeta thereby preventing these residues from being phosphorylated 16938294_These findings indicate that parasite-induced IkappaB kinase (IKK) activity does not require functional Hsp90. 16966325_IKKbeta provides inhibitory signals for cell mobility and growth 16982623_COX-2 induction by nickel compounds occurs via an IKKbeta/p65 NF-kappaB-dependent but IKKalpha- and p50-independent pathway and plays a crucial role in antagonizing nickel-induced cell apoptosis in Beas-2B cells 16989899_Protein kinase R is involved in NF-kappaB mediated gene transcription of pro-inflammatory cytokines via IkappaB kinase-beta. 16998237_CDDO and CDDO-Me directly block IKKbeta activity and thereby the NF-kappaB pathway by interacting with Cys-179 in the IKKbeta activation loop 17016640_This novel finding indicated that IKKbeta could mediate cytoplasmic p21 accumulation via activation of its downstream target Akt, which was known to phosphorylate p21 and lead to cytoplasmic localization of p21. 17079871_Cys-179 of IKKbeta plays a critical role in enzyme activation by promoting phosphorylation of activation-loop serines and interaction with ATP. 17114296_hypoxia releases repression of NFkappaB activity through decreased prolyl hydroxylase-dependent hydroxylation of IKKbeta 17145747_IKK beta is predominantly relocalized with Tax in Jurkat T cell perinuclear hot spots. 17237423_IKK alpha and IKKbeta are distinctly involved in MUC5AC induction by S. pneumoniae via an ERK1-dependent mechanism 17244613_IL1 induced NF-kappaB activation is NEMO-dependent but does not require IKKbeta 17363905_This study provides the biochemical and genetic evidence that phosphorylation of IKKalpha/beta and ubiquitination of NEMO are regulated by two distinct pathways upon T- cell receptor stimulation. 17419715_There is an essential role of virus-induced IKK/NF-kappaB activity to trigger both viral IE gene expression and productive replication in quiescent cells. 17466952_Activation of NF-kappa B was correlated with higher inhibition of IKBKB activity. 17616684_IKK alpha controls mTOR kinase activity in Akt-active, PTEN-null prostate cancer cells, with less involvement by IKK beta 17684021_FAF1 robustly suppresses NF-kappaB activation through the inhibition of IKK activation in combination with previously reported cytoplasmic retention of NF-kappaB p65 17693255_Expression of activated IKK beta is associated with tuberous sclerosis complex 1 protein phosphorylation and vascular endothelial growth factor production in multiple tumor types. 17703412_Observational study of gene-disease association. (HuGE Navigator) 17939994_These data support a function for IKK2 as an antagonist of Aurora A signaling during mitosis. 17947699_IKK2 is also involved in the IFN-gamma-stimulated release of the CXCR3 ligands through a novel mechanism that is independent NF-kappaB 17997719_Pellino isoforms may be the E3 ubiquitin ligases that mediate the IL-1-stimulated formation of K63-pUb-IRAK1 in cells, which may contribute to activation of IKKbeta and transcription factor NF-kappaB, as well as signalling pathways dependent on IRAK1/4 18037881_MUC1 is important for physiological activation of IKKbeta and that overexpression of MUC1, as found in human cancers, confers sustained induction of the IKKbeta-NF-kappaB p65 pathway. 18163512_IKKalpha and IKKbeta exert differential roles in ECM remodeling and endochondral ossification, which are events characteristic of hypertrophic chondrocytes in osteoarthritis 18260825_The canonical IKK2/IkappaBalpha pathway of NF-kappaB activation mediates the up-regulation of RGS4 expression in response to IL-1beta. 18266324_Data show that a longer region of the IKKbeta C-terminal region provides high affinity for NEMO and that the longer IKKbeta C-terminal region forms a 2:2 stoichiometirc complex with NEMO. 18308615_IkappaB kinase beta is a key molecule in signaling to the transcription factor NF-kappaB [review] 18316610_a TAB1:TAK1:IKK beta:NF-kappaB signaling axis forms aberrantly in breast cancer cells and, consequently, enables oncogenic signaling by TGF-beta 18408758_Identification of the microRNA hsa-miR-199a as a regulator of IKKbeta expression. 18411264_TAp63gamma by IKKbeta stabilizes the TAp63gamma protein by blocking ubiquitylation-dependent degradation of this protein. 18434448_Observational study of gene-disease association. (HuGE Navigator) 18490760_results demonstrate that an insulin or TNF response that uses Akt-dependent signaling activates IKKalpha to induce mTOR activity, whereas a TNF response that does not involve Akt activates IKKbeta to activate mTOR 18571841_Atypical protein kinase C (PKC-iota/-zeta) phosphorylates IKKalphabeta in transformed non-malignant and malignant prostate cell survival. 18600306_Data demonstrated that the NF-kappaB pathway components, p65 and IKK-2, are expressed in hMSCs. The data provide evidence that this signal transduction pathway is implicated in TNF-alpha-mediated invasion and proliferation of hMSCs. 18636537_Blocking NF-kappaB with IKKbeta- or RelA siRNA substantially sensitized Adriamycin-induced cytotoxicity 18657515_In the absence of its carboxy-terminal protein structural motifs the human IKKbeta subunit kinase domain exhibits a CK2-like phosphorylation specificity. 18952604_S6K1 directly phosphorylates IRS-1 on Ser-270 to promote insulin resistance in response to TNF-(alpha) signaling through IKK2. 18990758_Observational study of gene-disease association. (HuGE Navigator) 19050262_IKK beta phosphorylation is involved in decoy receptor 3 expression in AsPC-1 pancreatic adenocarcinoma cells. 19104039_NF-kappaB pathway activation by CARD11 or tumor necrosis factor-alpha, compensatory IKKalpha activity was also observed with IKKbeta 19109741_These results suggest the anti-inflammatory activity of platonin is a result of reduced NF-kappaB activity due to inhibition of Akt and IKKbeta. 19141566_A novel PDGF response in human preadipocytes that involves the pro-inflammatory kinase IKKbeta and demonstrate that it is required for the inhibition of adipogenesis. 19202066_propose a novel mechanism for the enhancement of NF-kappaB activity by loss of p53, which evokes positive feedback regulation from enhanced glucose metabolism to IKK in oncogenesis 19243472_Results suggest that loss of IKKbeta activation is important for the enhancement of p53 stability, leading to p21 expression and cell cycle arrest and apoptosis of tumour cells. 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19270264_IKKbeta inhibitor blocks canonical pathway and growth of MM cell lines but does not inhibit the noncanonical NF-kappaB pathway. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19488402_IKKalpha and IKKbeta regulation of DNA damage-induced cleavage of huntingtin 19526344_Results suggest that sulindac induces apoptosis in susceptible human breast cancer cells through, at least in part, the inhibition of IKKbeta and the subsequent p38 MAPK-dependent activation of JNK1. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19591457_These results suggest that TPCK inhibits NF-kappaB activation by directly modifying thiol groups on two different targets: Cys-179 of IKKbeta and Cys-38 of p65/RelA. 19594441_PP2Ceta-2 inhibits the IL-1-NF-kappaB signalling pathway by selectively dephosphorylating IKKbeta. 19656241_The CSN complex dissociates from IKK's allowing full and rapid activation of the NF-kappaB pathway by the concerted action of interacting protein complexes. 19675099_Ro52 down-regulates Tax-induced NF-kappaB signalling by monoubiquitinating IKKbeta and by reducing the level of Tax 19716809_ARD1 is indeed a bona fide substrate of IKKbeta. 19728335_IKKalpha, IKKbeta, RANK, Maspin, c-FLIP, Cip2 and cyclinD1 were found to show significant differences between hepatocellular tumor tissue and its corresponding adjacent tissue. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 19786027_PHD3 appears to be a tumor suppressor in colorectal cancer cells that inhibits IKKbeta/NF-kappaB signaling, independent of its hydroxylase activity. 19818716_Dysregulation of KEAP1-mediated IKKbeta ubiquitination may contribute to tumorigenesis. 20007573_IkappaB kinase beta is required for neuroprotection, suppression of inflammation, limitation of T helper (Th)1 type lymphocyte accumulation, and enhancement of natural killer (NK) cell recruitment in experimental autoimmune encephalomyelitis. 20051109_EBNA1 inhibits the canonical NF-kappaB pathway in carcinoma lines by inhibiting the phosphorylation of IKKalpha/beta. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20068038_we report the identification of two regulatory phosphorylation sites at Thr-516 and Ser-520 within the kinase activation loop that is essential for MEKK3-mediated IkappaB kinase beta (IKKbeta)/NF-kappaB activation 20145131_IKK activation represents one mechanism by which levels of DeltaNp63alpha can be reduced, thereby rendering cells susceptible to cell death in the face of cellular stress or DNA damage. 20152798_these results reveal the novel role of IKKbeta in P16 phosphorylation and broaden our understanding of the regulation of P16. 20176108_Showed IKK-2 mutants based on induced-fit docking of a selective IKK-2 inhibitor, PHA-408, into the homology model of IKK-2; functional characterization of these mutants shows the role of a Gly-rich loop residue of a kinase in binding kinetics. 20331378_Observational study of gene-disease association. (HuGE Navigator) 20347815_IKKbeta regulates gastric carcinogenesis via IL-1alpha expression, which is associated with anti-apoptotic signaling and cell proliferation. 20371626_[review] molecular mechanisms by which JNK1 and IKKbeta mediate obesity-induced metabolic stress 20410276_Data identify a new RIG-I/MAVS/TRAF6/IKKbeta/p65Ser536 pathway placed under the control of NOX2, thus characterizing a novel regulatory pathway involved in NF-kappaB-driven proinflammatory response in the context of RSV infection. 20421348_Report of an interaction between a homeobox protein and IkappaBbeta in endothelial cells and suggest that MEOX2 modulates the activity of the RelA complex through direct interaction with its components. 20448038_PP2A plays an important role in the termination of LPA-mediated NF-kappaB activation through dephosphorylating and inactivating MEKK3 20448286_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20465575_In addition to having a pivotal role in the up-regulation of IL-2 and IL-2RA gene expression, IKK controls the expression of cyclin D3, cyclin E and CDK2, and the stability SKP2 and its co-factor CKS1B, through mechanisms independent of IL-2. 20503287_Observational study of gene-disease association. (HuGE Navigator) 20529849_CC2D1A activates NF-kappaB through the canonical IKK pathway 20568250_Observational study of gene-disease association. (HuGE Navigator) 20627395_Ro52-mediated monoubiquitination is involved in the subcellular translocation of active IKK beta to autophagosomes. 20657549_Here, the authors demonstrate that both the NF-kappaB family of transcription factors and their upstream kinase IKK can regulate Claspin levels by controlling its mRNA expression. 20693425_Although Homer3 is dispensable for T cell receptor-induced Ikappa B kinase (IKK) activation, association with the IkappaB kinase (IKK) complex is essential for Homer3 localization at the immune synapse. 20803413_The anti-adipogenic effect of macrophage-conditioned medium requires the activation of preadipocyte IKKbeta/NF-kappaB pathway. 20856938_Interaction of TRAF1 with I-kappa B kinase-2 and TRAF2 is important for regulation of NF-kappa B activity. 20925653_regulatory subunit of PP5 G4-1 functions as an adaptor to recruit PP5 to the phosphorylated C-terminus of activated IKKbeta and to down-regulate the activation of IKKbeta. 20933503_miR-218 plays an important role in preventing the invasiveness of glioma cells, and this results present a novel mechanism of miRNA-mediated direct suppression of IKK-beta/NF-kappaB pathway in gliomas. 20977779_NF-kappaB targeting by way of IKK inhibition sensitizes lung cancer cells to adenovirus delivery of TRAIL 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 21248029_gamma(1)34.5 recruits both IKKalpha/beta and protein phosphatase 1, forming a complex that dephosphorylates two serine residues within the catalytic domains of IkappaB kinase. 21317297_IKKalpha may function as a tumor suppressor gene 21390216_Modeling of the IKK-2 kinase domain, virtual screening and activity assays. 21399639_Data show that the IKKbeta-dependent modification of a specific amino acid in RPS3 promoted specific NF-kappaB functions that underlie the molecular pathogenetic mechanisms of E. coli O157:H7. 21482671_findings show IKKbeta increases DeltaNp73alpha protein stability independently of its ability to activate NF-kappaB; IKKbeta associates with and phosphorylates DeltaNp73alpha leading to its accumulation in the nucleus, where it binds and represses several p53-regulated genes 21606193_Data show that IkappaB kinase beta (IKKbeta) can be activated in the nucleus by Nkx3.2 in the absence of exogenous IKK-activating signals, allowing constitutive nuclear degradation of IkappaB-alpha. 21798539_Hepatocyte-specific IKKbeta expression activates NFkappaB and aggravates atherosclerosis development in APOE*3-Leiden mice. 21810613_our data indicate that EV71 2C protein inhibits IKKbeta activation and thus blocks NF-kappaB activation 21911292_This study compares four models of human IKKB and evaluate their performance in both broad and focused library docking studies. 21985298_Conditional IKKbeta transgene deletion from lung epithelium transiently decreases alveolar type I and type II cells and myofibroblasts and delays alveolar formation. 21987722_IKKbeta induced AKT activity, whereas IKKbeta-driven NF-kappaB transcription was required for GLUT1 surface localization downstream of AKT. 21993219_IL-1beta-induced CCL27 gene expression in normal human keratinocytes is regulated through the p38 MAPK/MSK1/Mnk1+2 as well as the IKKbeta/NF-kappaB signalling pathways. 22262057_both IKKalpha and IKKbeta coimmunoprecipitated with Bcl-2 and in vitro kinase assay proved the ability of IKK to phosphorylate Bcl-2. 22331067_in human primary CD4+ T cells, IKKbeta is required for the activation of NF-kappaB and AP-1 transcription factors in response to engagement of CD3 and CD28 coreceptor 22511786_that IKKbeta activity, as well as the subsequent IFNbeta-stimulated activation of the JAK-STAT1/2 signaling pathway, are essential for the production of IFNalpha by TLR9 ligands. 22637744_findings demonstrate that IkBbeta is a master regulator of mitochondrial retrograde signaling pathway and that the retrograde signaling plays a role in tumor growth in vivo 22766331_Diosgenin ameliorated endothelial dysfunction involved in insulin resistance through an IKKbeta/IRS-1-dependent manner. 22824620_Expressions of Caveolin-1, IKKbeta, and COX-2 were significantly higher in patients from endemic areas of blackfoot disease compared to patients from non-endemic areas whereas eNOS was significantly lower. 22848663_HBx deregulates TSC1/mTOR signaling through IKKbeta, which is crucially linked to HBV-associated HCC development 22891964_H5N1 virus NS1 not only blocks IKKbeta-mediated phosphorylation and degradation of IkappaBalpha in the classical pathway but also suppresses IKKalpha-mediated processing of p100 to p52 in the alternative pathway. 22955948_The pro-death function of IKK-beta under oxidative stress was mediated by p85 S6K1, but not p70 S6K1 through a rapamycin-insensitive and mammalian target of rapamycin complex 1 kinase-independent mechanism. 23016877_The inhibitor of IKK complex responds to changes in iron levels within the cell. 23032264_Inhibition of IKK beta partially rescued p53 levels, while concomitant IKK beta inhibition fully rescued p53 and regulates MDM2 Ser166 phosphorylation. 23069812_Epithelial-mesenchymal transformation, along with acquisition of a tumor stem cell-like phenotype mediated by IKKbeta/IkappaBalpha/RelA signal pathway via Snail, contributes to a low concentration of arsenite-induced tumorigenesis. 23090968_activation of IKKbeta within the AKAP-Lbc complex promotes NF-kappaB-dependent production of interleukin-6 23108365_Minocycline modulates cytokine and chemokine production in lipopolysaccharide-stimulated THP-1 monocytic cells by inhibiting IkappaB kinase alpha/beta phosphorylation. 23178494_roles for p53 and IKKalpha/IKKbeta in non-canonical Notch signaling and IL-6 as a novel non-canonical Notch target gene 23349803_Data indicate that the IKK-2/NF-kappaB(p65) signalling pathway does not play a prominent role in the inflammatory response to cigarette smoke (CS) exposure and that this pathway may not be important in COPD pathogenesis. 23386606_the CD91/IKK/NF-kappaB signaling cascade is involved in secreted HSP90alpha-induced TCF12 expression, leading to E-cadherin down-regulation and enhanced CRC cell migration/invasion 23589370_Data indicate that IKKbeta inhibited the binding between TAp63gamma and p300, a co-activator of TAp63gamma, and consequently counteracted the positive effect of p300 on TAp63gamma transcriptional activity. 23743204_Data indicate the impact of IkappaB kinase 2 (IKK-2) in TNFalpha dependent mesenchymal stem cell (hMSC) recruitment. 23776406_IKK2 dimers transiently associate with one another through interaction surfaces to promote trans auto-phosphorylation. 23792959_observed domain movements in the structures of IKKbeta may represent trans-phosphorylation steps that accompany IKKbeta activation 23817200_IKK2 overexpression significantly increased the phosphorylation level of MLC Ser19 but not the regulatory subunit of MLCP myosin phosphatase target subunit 1 Thr696 in human aortic myocytes. 23872070_IKK interacts with rictor and regulates the function of mTORC2 including phosphorylation of AKT (at Serine473) and organization of actin cytoskeleton. 23963361_we investigated the in vitro and in vivo antilymphoma activity and associated molecular mechanism of action of a novel compound, 13-197, a quinoxaline analog that specifically perturbs IkappaB kinase (IKK) b, a key regulator of the NF-kappaB pathway 23974204_These results identify a novel interaction between Aurora and IKK kinases and show that these pathways can cooperate to promote TRAIL resistance in multiple myeloma. 24086395_IkBalpha kinase (IKK)-signaling is necessary to allow ZNF395 to activate transcription and simultaneously enhances its proteolytic degradation. 24155403_study provides convinced morphologic evidence that VMAT2 is not present in beta cells 24240172_IKKgamma facilitates RhoA activation via a guanine nucletotide exchange factor, which in turn activates ROCK to phosphorylate IKKbeta, leading to NF-kappaB activation that induced the chemokine expression and cell migration upon TGF-beta1. 24266532_Data suggest that all seven cysteines (4 in zinc finger domain) of NEMO (NF-kappaB essential modulator protein) can be simultaneously mutated to alanine without affecting binding affinity of NEMO for I-kappa B kinase beta catalytic subunit. 24337575_Both IKKbeta activity and EGR-1 expression are required for the increased IL-8 expression induced by proteasome inhibition in ovarian cancer cells. 24369075_Although Ikk2 deficiency is lethal in mouse embryos, our observations suggest a more restricted, unique role of IKK2-NF-kappaB signaling in humans. 24386391_RTK-mediated Tyr phosphorylation of IKKbeta has the potential to directly regulate NFkappaB transcriptional activation. 24450414_2-MS specifically inhibits JAK and IKKbeta kinase activities but has little effect on activities of other kinases tested 24550137_High IKKbeta expression is associated with non-small cell lung cancer. 24553260_PHLPP2 loss enhances Bcl10-MALT1 complex formation, NEMO ubiquitination and subsequent IKKbeta phosphorylation, resulting in increased NF-kappaB-dependent transcription of multiple target genes. 24586253_the requirement of IKKbeta for VEEV replication 24611898_Activation via trans autophosphorylation of activation loop serines in IKK2 requires transient assembly of higher-order oligomers when a scaffold dimerization domain is removed. 24613833_Taken together, these results reveal that aspirin up-regulates the expression of APC via the suppression of IKKbeta. 24721172_NLK functions as a pivotal negative regulator of NF-kappaB via disrupting the interaction of TAK1 with IKKbeta. 24755559_Gain-of function or knockdown of miR-200c in leiomyoma smooth muscle cells (LSMC) regulated IL8 mRNA and protein expression through direct targeting of IKBKB and alteration of NF-kB activity. 24811176_NME1L down-regulates IKKbeta signaling by blocking IKKbeta-mediated IkappaB degradation. 24854552_Expression of IKBKB gene reduced the cisplain sensitivity of A549 cells. 24865276_The activation of NF-kappaB induced translocation of AMAP1 to cytoplasm from cell membrane and nucleus, which resulted in augmented interaction of AMAP1 and IKKbeta 24911653_Data suggest that TAK1 (MAP kinase kinase kinase 7) and IKKbeta phosphorylate different serines of IKKbeta; TAK1-catalyzed phosphorylation of IKKbeta at Ser177 is priming event that enables IKKbeta to activate itself by phosphorylating Ser181. 24981452_A macrophage-specific constitutively active IkappaB kinase transgenic mouse model demonstrates that activated macrophages can increase proinflammatory gene expression and can reduce the response of fetal lung macrophages in bronchopulmonary dysplasia. 25039491_IKKbeta regulates endothelial thrombomodulin in a Klf2-dependent manner 25049379_An unexpected role for the bromodomain and extraterminal domain (BET) proteins BRD2 and BRD4 in maintaining oncogenic IKK activity in activated B-cell-like diffuse large B-cell lymphoma. 25066210_Demonstrate the essential role of EGFR/Akt/IkappaBbeta/NF-kappaB pathway in the inhibitory effect of PA-MSHA on invasion and metastasis of HCC through suppressing EMT. 25096806_Suppression of PKK expression by RNA interference inhibits phosphorylation of IKKalpha and IKKbeta as well as activation of NF-kappaB in human cancer cell lines; thus, PKK regulates NF-kappaB activation by modulating activation of IKKalpha and IKKbeta. 25107905_Transcription activator-like effector nuclease-based knock-in mutagenesis provides evidence from a B lymphoid context that K171E IKKbeta contributes to lymphomagenesis. 25139357_A nonsense mutation in IKBKB causes combined immunodeficiency. 25286246_the rescuing of the binding affinity implies that a preordered IKK-binding region of NEMO is compatible with IKK binding, and the conformational heterogeneity observed in NEMO(44-111) may be an artifact of the truncation. 25326418_IKKbeta activates two 'master' transcription factors of the innate immune system, IRF5 and NF-kappaB 25326420_IKKbeta is an IRF5 kinase that instigates inflammation 25326706_IKBKB-rs3747811AT single nucleotide polymorphism was associated with a significantly increased risk of developing wheezing. 25432706_Studies indicate that transcription factor NF-kappaB plays a key role in numerous physiological processes, and its activation is tightly controlled by a kinase complex, IkappaB kinase (IKK). 25630970_High IKBKB expression is associated with inflammation in heart Valve Diseases. 25652452_These studies not only reinforce the significance of maintaining a homeostatic balance of eNOS and IKKbeta within the cell system that regulates NO production, but they also confirm that the IKKbeta-Hsp90 interaction is favored in a high-glucose environment, leading to impairment of the eNOS-Hsp90 interaction, which contributes to endothelial dysfunction and vascular complications in diabetes. 26020802_we demonstrate that one miR-497, is a likely negative regulator of IKKbeta 26106822_identified IKK-beta as a kinase capable of phosphorylating threonine 3 in N-terminal hungtingtin fragments 26167925_findings indicate that the IKBKB and POLB SNPs confer no genetic predisposition to SLE risk in this Chinese Han population 26267322_Combining bortezomib with IKK inhibitor is effective in treating ovarian cancer. 26301506_In cells with functional KEAP1, RTA 405 increased NRF2 levels, but not IKKb or BCL2 levels, and did not increase cell proliferation or survival. 26334375_IFIT5 promotes SeV-induced IKK phosphorylation and NF-kappaB activation by regulating the recruitment of IKK to TAK1. 26427514_Down-regulation of IKBKB expression and NFkappaB signaling in microglia/macrophages infiltrating glioblastoma correlates with defective expression of immune/inflammatory genes and M2 polarization that may result in the global impairment of anti-tumor immune responses in glioblastoma. 26433127_our results demonstrate that miR-200b, a transcriptional target of NF-kappaB, suppresses breast cancer cell growth and migration, and NF-kappaB activation, through downregulation of IKBKB, indicating that miR-200b has potential as a therapeutic target in breast cancer patients. 26603838_IKK-beta suppresses GLI1 ubiquitination. 26647777_DAT stabilized IkBa by inhibiting the phosphorylation of Ika by the IkB kinase (IKK) complex. DAT induced proteasomal degradation of TRAF6, and DAT suppressed IKKb-phosphorylation through downregulation of TRAF6 26669856_MyD88s is positively regulated by IKKbeta and CREB and negatively regulated by ERK1/2 signaling pathways. 26718331_Survivin overexpression activates NFkappaB p65, which is important in the acquisition and maintenance of the oncogenic characteristics of esophageal squamous cell carcinoma. 26757982_Rare variants in IKBKB are associated with decreased waist-to-hip ratio in European-Americans. 26859364_Smad7 expression in necrotizing enterocolitis macrophages interrupts TGF-beta signaling and promotes NF-kappaB-mediated inflammatory signaling in these cells through increased expression of IKK-beta 26895469_EGFR/PI3K/Akt/mTOR/IKK-beta/NF-kappaB signaling promotes head and neck cancer progression. 26914121_Over-expressed IKK-Beta inhibits cell apoptosis in laryngeal squamous cell carcinoma. 26945075_N-acetyl-seryl-aspartyl-lysyl-proline inhibition of TNF-alpha activation of canonical, i.e., IKK-beta-dependent, NF-kappaB pathway and subsequent decrease in ICAM-1 expression is achieved via inhibition of IKK-beta. 27196294_Cis- and trans-gnetin H suppress cytokine | ENSMUSG00000031537 | Ikbkb | 1080.96581 | 1.0992277 | 0.1364902471 | 0.11026140 | 1.536184e+00 | 2.151866e-01 | 6.080541e-01 | No | Yes | 1326.855150 | 138.111895 | 1203.202761 | 122.444664 | |
| ENSG00000104983 | 729440 | CCDC61 | protein_coding | Q9Y6R9 | FUNCTION: Microtubule-binding centrosomal protein required for centriole cohesion, independently of the centrosome-associated protein/CEP250 and rootletin/CROCC linker (PubMed:31789463). In interphase, required for anchoring microtubule at the mother centriole subdistal appendages and for centrosome positioning (PubMed:31789463). During mitosis, may be involved in spindle assembly and chromatin alignment by regulating the organization of spindle microtubules into a symmetrical structure (PubMed:30354798). Has been proposed to play a role in CEP170 recruitment to centrosomes (PubMed:30354798). However, this function could not be confirmed (PubMed:31789463). Plays a non-essential role in ciliogenesis (PubMed:31789463, PubMed:32375023). {ECO:0000269|PubMed:30354798, ECO:0000269|PubMed:31789463, ECO:0000269|PubMed:32375023}. | 3D-structure;Acetylation;Alternative splicing;Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:729440; | centriolar satellite [GO:0034451]; centriolar subdistal appendage [GO:0120103]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; microtubule organizing center [GO:0005815]; identical protein binding [GO:0042802]; microtubule binding [GO:0008017]; cell projection organization [GO:0030030]; centriole assembly [GO:0098534]; mitotic spindle assembly [GO:0090307] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 30354798_Ccdc61 controls centrosomal localization of Cep170 and is required for spindle assembly and symmetry. 31789463_hVFL3/CCDC61 is the human orthologue of proteins required for anchoring distinct sets of cytoskeletal fibers to centrioles in unicellular eukaryotes. 32375023_CCDC61/VFL3 Is a Paralog of SAS6 and Promotes Ciliary Functions. | ENSMUSG00000074358 | Ccdc61 | 45.91359 | 0.9887113 | -0.0163787583 | 0.42459573 | 1.489249e-03 | 9.692166e-01 | No | Yes | 57.598482 | 11.159517 | 58.885696 | 10.908484 | |||
| ENSG00000105048 | 7138 | TNNT1 | protein_coding | P13805 | FUNCTION: Troponin T is the tropomyosin-binding subunit of troponin, the thin filament regulatory complex which confers calcium-sensitivity to striated muscle actomyosin ATPase activity. | Alternative splicing;Muscle protein;Nemaline myopathy;Phosphoprotein;Reference proteome | This gene encodes a protein that is a subunit of troponin, which is a regulatory complex located on the thin filament of the sarcomere. This complex regulates striated muscle contraction in response to fluctuations in intracellular calcium concentration. This complex is composed of three subunits: troponin C, which binds calcium, troponin T, which binds tropomyosin, and troponin I, which is an inhibitory subunit. This protein is the slow skeletal troponin T subunit. Mutations in this gene cause nemaline myopathy type 5, also known as Amish nemaline myopathy, a neuromuscular disorder characterized by muscle weakness and rod-shaped, or nemaline, inclusions in skeletal muscle fibers which affects infants, resulting in death due to respiratory insufficiency, usually in the second year. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:7138; | cytosol [GO:0005829]; troponin complex [GO:0005861]; tropomyosin binding [GO:0005523]; troponin T binding [GO:0031014]; muscle contraction [GO:0006936]; negative regulation of muscle contraction [GO:0045932]; sarcomere organization [GO:0045214]; skeletal muscle contraction [GO:0003009]; slow-twitch skeletal muscle fiber contraction [GO:0031444]; transition between fast and slow fiber [GO:0014883] | 15665378_Data suggest that inefficient incorporation into myofilament is responsible for the instability of mutant slow troponin T in Amish nemaline myopathy. 18579801_Report adaptation by alternative RNA splicing of slow troponin T isoforms in type 1 but not type 2 Charcot-Marie-Tooth disease. 19326042_slow TnT was encoded by two different transcripts in significantly different ratios in myotonic dystrophy type 1 and myotonic dystrophy type 2 muscles. 19541721_Troponin T may have a role in pulmonary embolism progeresion to death 19690080_TNT is a biochemical marker of susceptibility to hypoxia in infants of type 1 diabetic mothers. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19916752_The occurrence of myocardial infarction is associated with elevated troponin T levels. 20038417_troponin-T mutations were responsible for 3% of the hypertrophic cardiomyopathy cases in our study population 20380359_Among athletees, faster runners demonstrate significantly stronger cardoac TnT releases and inflammation signs. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20889975_analysis of order and disorder in troponin C, T and I 21111984_Elevated serum troponin T levels are associated with different conditions related to the severity of hypertrophic cardiomyopathy. 21448949_Cardiac troponin T and creatine kinase have roles in infarct size and left ventricular function after acute myocardial infarction 21683708_the hypertrophic phenotype associated with the TnT mutations can be characterized by a significant increase in disorder of rigor cross-bridges. 21729325_In heart failure patients with normal ejection fraction, highly sensitive troponin T and heart fatty acid binding protein are elevated independent of coronary artery disease. 21784424_carotid-femoral pulse wave and office pulse pressure are associated with minimally elevated hsTnT levels in the elderly 22239123_baseline cTnT levels are higher in patients with MPI evidence of reversible myocardial ischaemia than those without reversible ischaemia 22448368_analysis of parameters of oxygen-dependent metabolism of neutrophils by NBT test and levels of vWF antigen in the serum can be used for predicting the risk of unfavorable outcome in patients with ACS and normal troponin T 22977240_Human slow skeletal troponin T (HSSTnT) isoforms, despite being homologues of cardiac TnT may display distinct functional properties in muscle regulation. 23244308_TNNT1 DNA methylation levels were positively correlated with mean HDL particle size, HDL-phospholipid, HDL-apolipoprotein AI, HDL-C and TNNT1 expression levels. 24020864_Troponin T1 blood levels had a positive association with increased risk for hypertrophic cardiomyopathy. 24625749_troponin T and creatinine kinase isoenzyme (CK-MB) have roles in combined renal and myocardial injuries in asphyxiated infants 24781421_Biopsy-proven acute and viral myocarditis is associated with elevated concentrations of hs-TnT. 26296490_Nemaline body myopathy Palestinian patients were found to have a novel mutation in troponin T1. 26774798_Three homologous genes have evolved in vertebrates to encode three muscle type-specific TnT isoforms: TNNT1 for slow skeletal muscle TnT, TNNT2 for cardiac muscle TnT, and TNNT3 for fast skeletal muscle TnT. 26950807_TNNT1 genetic and epigenetic variations are associated with HDL-C levels and coronary artery disease. 27429059_pathogenesis of TNNT1 myopathies 27903076_Copeptin and troponin T measurement could potentially improve the prehospital diagnostic and prognostic classification of patients with a suspected AMI. 28530094_Data suggest that mutations in troponin C (TnC, A8V) and troponin T (TnT, delta14-TnT) found in patients with hypertrophic cardiomyopathy together fully stabilize the active M state of regulated actin (the actin-tropomyosin-troponin complex). 28923663_investigated the effects of one of these mutations, K247R of TnT, on the picosecond dynamics of the Tn core domain (Tn-CD), consisting of TnC, TnI and TnT2 (183-288 residues of TnT), by carrying out the quasielastic neutron scattering measurements on the reconstituted Tn-CD containing either the wild-type TnT2 (wtTn-CD) or the mutant TnT2 (K247R-Tn-CD) in the absence and presence of Ca(2+) 29178646_This study describes the first TNNT1 mutation that transmits in an autosomal dominant fashion to cause nemaline myopathy. 29880121_In a nationwide cohort in Sweden, patients with a first myocardial infarction had increased levels of Troponin T. 29931346_Similar functional and histological phenotypes were observed in other human cohorts and two transgenic murine models (Tnnt1-/- and Tnnt1 c.505G>T). These findings have implications for emerging molecular therapies, including the suitably of TNNT1 gene replacement for newborns with 'Amish' nemaline myopathy or other TNNT1-associated myopathies. 30031058_High TNNT1 expression is associated with breast cancer. 31512553_These findings indicated that TNNT1 may promote the progression of colon adenocarcinoma, mediating epithelial-mesenchymal transition process, and thus shed a novel light on colon adenocarcinoma therapeutic treatments. 31830337_TNNT1, negatively regulated by miR-873, promotes the progression of colorectal cancer. 31970803_Three adults and 1 child shared a novel missense homozygous variant in the TNNT1 gene (NM_003283.6: c.287T > C; p.Leu96Pro). This study expands the phenotypic spectrum of TNNT1 myopathy. 32994279_Clinical phenotype and loss of the slow skeletal muscle troponin T in three new patients with recessive TNNT1 nemaline myopathy. 33051334_Troponin T but not C reactive protein is associated with future surgery for aortic stenosis: a population-based nested case-referent study. 35053326_Biomarkers-in-Cardiology 8 RE-VISITED-Consistent Safety of Early Discharge with a Dual Marker Strategy Combining a Normal hs-cTnT with a Normal Copeptin in Low-to-Intermediate Risk Patients with Suspected Acute Coronary Syndrome-A Secondary Analysis of the Randomized Biomarkers-in-Cardiology 8 Trial. 35165004_TNNT1 myopathy with novel compound heterozygous mutations. | ENSMUSG00000064179 | Tnnt1 | 131.66789 | 1.0841419 | 0.1165536616 | 0.26218610 | 1.980188e-01 | 6.563248e-01 | 8.920163e-01 | No | Yes | 175.956107 | 24.020528 | 156.891208 | 20.812901 | |
| ENSG00000105202 | 2091 | FBL | protein_coding | P22087 | FUNCTION: S-adenosyl-L-methionine-dependent methyltransferase that has the ability to methylate both RNAs and proteins (PubMed:24352239, PubMed:30540930, PubMed:32017898). Involved in pre-rRNA processing by catalyzing the site-specific 2'-hydroxyl methylation of ribose moieties in pre-ribosomal RNA (PubMed:30540930). Site specificity is provided by a guide RNA that base pairs with the substrate (By similarity). Methylation occurs at a characteristic distance from the sequence involved in base pairing with the guide RNA (By similarity). Probably catalyzes 2'-O-methylation of U6 snRNAs in box C/D RNP complexes (PubMed:32017898). U6 snRNA 2'-O-methylation is required for mRNA splicing fidelity (PubMed:32017898). Also acts as a protein methyltransferase by mediating methylation of 'Gln-105' of histone H2A (H2AQ104me), a modification that impairs binding of the FACT complex and is specifically present at 35S ribosomal DNA locus (PubMed:24352239, PubMed:30540930). {ECO:0000250|UniProtKB:P15646, ECO:0000269|PubMed:24352239, ECO:0000269|PubMed:30540930, ECO:0000269|PubMed:32017898}. | 3D-structure;Acetylation;Isopeptide bond;Methylation;Methyltransferase;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ribonucleoprotein;S-adenosyl-L-methionine;Transferase;Ubl conjugation;rRNA processing | This gene product is a component of a nucleolar small nuclear ribonucleoprotein (snRNP) particle thought to participate in the first step in processing preribosomal RNA. It is associated with the U3, U8, and U13 small nuclear RNAs and is located in the dense fibrillar component (DFC) of the nucleolus. The encoded protein contains an N-terminal repetitive domain that is rich in glycine and arginine residues, like fibrillarins in other species. Its central region resembles an RNA-binding domain and contains an RNP consensus sequence. Antisera from approximately 8% of humans with the autoimmune disease scleroderma recognize fibrillarin. [provided by RefSeq, Jul 2008]. | hsa:2091; | box C/D RNP complex [GO:0031428]; Cajal body [GO:0015030]; extracellular exosome [GO:0070062]; fibrillar center [GO:0001650]; granular component [GO:0001652]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; small-subunit processome [GO:0032040]; ATPase binding [GO:0051117]; histone-glutamine methyltransferase activity [GO:1990259]; RNA binding [GO:0003723]; rRNA methyltransferase activity [GO:0008649]; TFIID-class transcription factor complex binding [GO:0001094]; box C/D RNA 3'-end processing [GO:0000494]; histone glutamine methylation [GO:1990258]; osteoblast differentiation [GO:0001649]; rRNA methylation [GO:0031167]; rRNA processing [GO:0006364]; snoRNA localization [GO:0048254] | 17461797_Increased levels of exogenous BRAG2 in nucleoli result in redistribution of FBL to the nucleolar periphery. 17603021_Our data suggest that fibrillarin would play a critical role in the maintenance of nuclear shape and cellular growth. 18292223_Evidence that BIG1 and nucleolin, but not fibrillarin, can be present with p62 at the nuclear envelope confirms the presence of BIG1 and nucleolin in dynamic molecular complexes that change in composition while moving through nuclei 19331828_Data demonstrate that fibrillarin and Nop56 directly interact in vivo, and that this interaction is indispensable for the association of both proteins with the box C/D snoRNPs. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21536856_p32 is a new rRNA maturation factor involved in the remodeling from pre-90S particles to pre-40S and pre-60S particles that requires the exchange of FBL for Nop52. 22909121_NS1 protein of the human H3N2 virus interacts primarily via the C-terminal NLS2/NoLS and to a minor extent via the N-terminal NLS1 with the main nucleolar proteins, nucleolin, B23 and fibrillarin. 24029231_FBL overexpression contributes to tumorigenesis and is associated with poor survival in patients with breast cancer. 27682166_Nucleolar Methyltransferase Fibrillarin: Evolution of Structure and Functions. 30190478_Fibrillarin acts as a central node in a regulatory network engaged in imparting immunity against bacterial pathogens. 30540930_SIRT7-dependent deacetylation impacts nucleolar activity by an FBL-driven circuitry that mediates cell-cycle-dependent fluctuation of ribosomal DNA transcription. 32011831_This is the first report on the clinical aspect of ribosome biogenesis in pediatric BCP-ALL [B-cell precursor acute lymphoblastic leukemia ], and it shows that overexpression of CMYC and C/D box nucleoproteins FBL and NOP56 is an antecedent event in patients who subsequently relapse. 32384686_Fibrillarin Ribonuclease Activity is Dependent on the GAR Domain and Modulated by Phospholipids. 33522570_Phase transition of fibrillarin LC domain regulates localization and protein interaction of fibrillarin. 33795875_A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation. | ENSMUSG00000046865 | Fbl | 11334.45002 | 1.0108604 | 0.0155837369 | 0.07911975 | 3.880512e-02 | 8.438352e-01 | 9.583418e-01 | No | Yes | 14769.628925 | 1857.528088 | 13726.665828 | 1687.947645 | |
| ENSG00000105298 | 58509 | CACTIN | protein_coding | Q8WUQ7 | FUNCTION: Involved in the regulation of innate immune response (PubMed:20829348). Acts as negative regulator of Toll-like receptor, interferon-regulatory factor (IRF) and canonical NF-kappa-B signaling pathways (PubMed:20829348, PubMed:26363554). Contributes to the regulation of transcriptional activation of NF-kappa-B target genes in response to endogenous proinflammatory stimuli (PubMed:20829348, PubMed:26363554). {ECO:0000269|PubMed:20829348, ECO:0000269|PubMed:26363554}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;Developmental protein;Immunity;Innate immunity;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Spliceosome;Ubl conjugation;mRNA processing;mRNA splicing | hsa:58509; | catalytic step 2 spliceosome [GO:0071013]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; RNA binding [GO:0003723]; cellular response to interleukin-1 [GO:0071347]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to tumor necrosis factor [GO:0071356]; innate immune response [GO:0045087]; mRNA cis splicing, via spliceosome [GO:0045292]; mRNA splicing, via spliceosome [GO:0000398]; negative regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043124]; negative regulation of innate immune response [GO:0045824]; negative regulation of interferon-beta production [GO:0032688]; negative regulation of interleukin-8 production [GO:0032717]; negative regulation of lipopolysaccharide-mediated signaling pathway [GO:0031665]; negative regulation of NF-kappaB transcription factor activity [GO:0032088]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of toll-like receptor signaling pathway [GO:0034122]; negative regulation of tumor necrosis factor production [GO:0032720]; negative regulation of type I interferon-mediated signaling pathway [GO:0060339] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 26033110_results indicate that PKC-mediated cortactin phosphorylation might be implicated in the maintenance of growth cone 26363554_TRIM39 negatively regulates the NFkappaB signaling pathway possibly via stabilization of cactin. 28062851_cellular complexes comprising cactin, DHX8 and SRRM2 sustain precise chromosome segregation, genome stability and cell proliferation by allowing faithful splicing of specific pre-mRNAs. | ENSMUSG00000034889 | Cactin | 1166.58211 | 1.0755187 | 0.1050326166 | 0.10810293 | 9.456460e-01 | 3.308300e-01 | 7.080048e-01 | No | Yes | 1501.991731 | 175.983476 | 1352.101610 | 154.902375 | ||
| ENSG00000105419 | 56917 | MEIS3 | protein_coding | Q99687 | FUNCTION: Transcriptional regulator which directly modulates PDPK1 expression, thus promoting survival of pancreatic beta-cells. Also regulates expression of NDFIP1, BNIP3, and CCNG1. {ECO:0000250|UniProtKB:P97368}. | Alternative splicing;DNA-binding;Homeobox;Nucleus;Reference proteome | This gene encodes a homeobox protein and probable transcriptional regulator. The orthologous protein in mouse controls expression of 3-phosphoinositide dependent protein kinase 1, which promotes survival of pancreatic beta-cells. [provided by RefSeq, Sep 2016]. | hsa:56917; | chromatin [GO:0000785]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; animal organ morphogenesis [GO:0009887]; brain development [GO:0007420]; embryonic pattern specification [GO:0009880]; eye development [GO:0001654]; negative regulation of apoptotic signaling pathway [GO:2001234]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 20553494_This work suggests that the transcriptional activity of all members of the Meis/Prep Hth protein family is subject to autoinhibition by their Hth domains, and that the Meis3.2 splice variant encodes a protein that bypasses this autoinhibitory effect. 26354419_Findings support a model in which Meis3 is required for neural crest proliferation, migration into, and colonization of the gut such that its loss leads to severe defects in enteric nervous system development. 33376716_Inhibition of MEIS3 Generates Cetuximab Resistance through c-Met and Akt. | ENSMUSG00000041420 | Meis3 | 106.43939 | 1.0166550 | 0.0238301154 | 0.29244815 | 6.635829e-03 | 9.350757e-01 | 9.829183e-01 | No | Yes | 130.341026 | 17.775089 | 124.390149 | 16.367886 | |
| ENSG00000105649 | 5864 | RAB3A | protein_coding | P20336 | FUNCTION: Small GTP-binding protein that plays a central role in regulated exocytosis and secretion. Controls the recruitment, tethering and docking of secretory vesicles to the plasma membrane (By similarity). Upon stimulation, switches to its active GTP-bound form, cycles to vesicles and recruits effectors such as RIMS1, RIMS2, Rabphilin-3A/RPH3A, RPH3AL or SYTL4 to help the docking of vesicules onto the plasma membrane (By similarity). Upon GTP hydrolysis by GTPase-activating protein, dissociates from the vesicle membrane allowing the exocytosis to proceed (By similarity). Stimulates insulin secretion through interaction with RIMS2 or RPH3AL effectors in pancreatic beta cells (By similarity). Regulates calcium-dependent lysosome exocytosis and plasma membrane repair (PMR) via the interaction with 2 effectors, SYTL4 and myosin-9/MYH9 (PubMed:27325790). Acts as a positive regulator of acrosome content secretion in sperm cells by interacting with RIMS1 (PubMed:22248876, PubMed:30599141). Plays also a role in the regulation of dopamine release by interacting with synaptotagmin I/SYT (By similarity). Interacts with MADD (via uDENN domain); the GTP-bound form is preferred for interaction (By similarity). {ECO:0000250|UniProtKB:P63011, ECO:0000250|UniProtKB:P63012, ECO:0000269|PubMed:22248876, ECO:0000269|PubMed:27325790, ECO:0000269|PubMed:30599141}. | Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Exocytosis;GTP-binding;Lipoprotein;Lysosome;Membrane;Methylation;Nucleotide-binding;Phosphoprotein;Prenylation;Protein transport;Reference proteome;Synapse;Transport | hsa:5864; | acrosomal vesicle [GO:0001669]; anchored component of synaptic vesicle membrane [GO:0098993]; axon [GO:0030424]; clathrin-sculpted acetylcholine transport vesicle membrane [GO:0060201]; clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane [GO:0061202]; clathrin-sculpted glutamate transport vesicle membrane [GO:0060203]; clathrin-sculpted monoamine transport vesicle membrane [GO:0070083]; cytosol [GO:0005829]; endosome [GO:0005768]; extracellular vesicle [GO:1903561]; lysosome [GO:0005764]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; postsynapse [GO:0098794]; presynapse [GO:0098793]; presynaptic active zone [GO:0048786]; protein-containing complex [GO:0032991]; secretory granule membrane [GO:0030667]; synaptic vesicle [GO:0008021]; terminal bouton [GO:0043195]; ATPase activator activity [GO:0001671]; ATPase binding [GO:0051117]; GDP-dissociation inhibitor binding [GO:0051021]; GTP binding [GO:0005525]; GTP-dependent protein binding [GO:0030742]; GTPase activity [GO:0003924]; myosin V binding [GO:0031489]; protein C-terminus binding [GO:0008022]; acrosomal vesicle exocytosis [GO:0060478]; axonogenesis [GO:0007409]; constitutive secretory pathway [GO:0045054]; evoked neurotransmitter secretion [GO:0061670]; exocytosis [GO:0006887]; lung development [GO:0030324]; lysosome localization [GO:0032418]; maintenance of presynaptic active zone structure [GO:0048790]; mitochondrion organization [GO:0007005]; neuromuscular synaptic transmission [GO:0007274]; plasma membrane repair [GO:0001778]; positive regulation of exocytosis [GO:0045921]; positive regulation of regulated secretory pathway [GO:1903307]; post-embryonic development [GO:0009791]; protein localization to plasma membrane [GO:0072659]; protein secretion [GO:0009306]; regulated exocytosis [GO:0045055]; regulation of dopamine secretion [GO:0014059]; regulation of exocytosis [GO:0017157]; regulation of plasma membrane repair [GO:1905684]; regulation of short-term neuronal synaptic plasticity [GO:0048172]; regulation of synaptic vesicle exocytosis [GO:2000300]; regulation of synaptic vesicle fusion to presynaptic active zone membrane [GO:0031630]; regulation of synaptic vesicle priming [GO:0010807]; respiratory system process [GO:0003016]; response to electrical stimulus [GO:0051602]; sensory perception of touch [GO:0050975]; synaptic vesicle clustering [GO:0097091]; synaptic vesicle exocytosis [GO:0016079]; synaptic vesicle maturation [GO:0016188]; synaptic vesicle recycling [GO:0036465]; synaptic vesicle transport [GO:0048489]; vesicle docking involved in exocytosis [GO:0006904] | 12937130_a possible role in glomerulopathies 15005721_Mutation screening of the RAB3A gene in 47 individuals with autism provided no evidence that DNA variants in this gene are associated with autism. 16099449_Cholesterol content regulates acrosomal exocytosis by enhancing Rab3A plasma membrane association. 16584842_In conclusion, we have found no evidence for RAB3A conferring susceptibility on mental retardation in the Han Chinese population. 16584842_Observational study of gene-disease association. (HuGE Navigator) 17625073_Initiates exocytosis in acrosome when prenylated and activated with guanosine triphosphate (GTP). 18559336_Rab3GEP has a role as the non-redundant guanine nucleotide exchange factor for Rab27a in melanocytes 19546222_Epac activates the small G proteins Rap1 and Rab3A to achieve exocytosis 19923287_The maturation of amyloid precursor protein (APP) transport vesicles, including recruitment of conventional kinesin, requires Rab3A GTPase activity. 21349835_Myo5a and Rab3A are direct binding partners and interact on synaptic vesicles and the Myo5a/Rab3A complex is involved in transport of neuronal vesicles 22753498_Rab27 and Rab3 sequentially regulate human sperm dense-core granule exocytosis. 23344955_alpha-Synuclein membrane association is regulated by the Rab3a recycling machinery and presynaptic activity 24652202_Five compounds which possess good inhibitory activity and may act as potential high affinity inhibitors against Rab3A active site were identified 24965146_Rab3a accelerates cell proliferation by increasing cyclin D1 expression, enhances anti-cancer drug resistance, and increases tumorigenicity and self-renewal of glioma cells. 25713146_Data indicate that exocytic stimuli promote ADP ribosylation factor 6 (ARF6) activation, which accomplishes exocytosis by stimulating Rab3A GTP-Binding Protein. 27613869_Rab3A-22A blocks exocytosis at a stage downstream intra-acrosomal calcium release. 30599141_Rab27/Rabphilin3a/GRAB/Rab3 constitutes a signaling module in sperm exocytosis. 31698103_Unveiling the interaction between the molecular motor Myosin Vc and the small GTPase Rab3A. | ENSMUSG00000031840 | Rab3a | 141.99526 | 0.8211990 | -0.2841962601 | 0.25288337 | 1.253810e+00 | 2.628261e-01 | 6.536914e-01 | No | Yes | 156.737614 | 25.006136 | 182.123053 | 28.103993 | ||
| ENSG00000105875 | 29062 | WDR91 | protein_coding | A4D1P6 | FUNCTION: Functions as a negative regulator of the PI3 kinase/PI3K activity associated with endosomal membranes via BECN1, a core subunit of the PI3K complex. By modifying the phosphatidylinositol 3-phosphate/PtdInsP3 content of endosomal membranes may regulate endosome fusion, recycling, sorting and early to late endosome transport (PubMed:26783301). It is for instance, required for the delivery of cargos like BST2/tetherin from early to late endosome and thereby participates indirectly to their degradation by the lysosome (PubMed:27126989). May play a role in meiosis (By similarity). {ECO:0000250|UniProtKB:Q7TMQ7, ECO:0000269|PubMed:26783301, ECO:0000269|PubMed:27126989}. | 3D-structure;Alternative splicing;Coiled coil;Endosome;Membrane;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:29062; | cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extrinsic component of endosome membrane [GO:0031313]; late endosome membrane [GO:0031902]; phosphatidylinositol 3-kinase regulator activity [GO:0035014]; early endosome to late endosome transport [GO:0045022]; regulation of cellular protein catabolic process [GO:1903362]; regulation of phosphatidylinositol 3-kinase activity [GO:0043551] | 27126989_suggest a role for the WDR81-WDR91 complex in the fusion of endolysosomal compartments and the absence of WDR81 leads to impaired receptor trafficking and degradation 28860274_WDR91 serves as a Rab7 effector that is essential for neuronal development by facilitating endosome conversion in the endosome-lysosome pathway. | ENSMUSG00000058486 | Wdr91 | 1219.69616 | 1.2128717 | 0.2784269278 | 0.10319784 | 7.315164e+00 | 6.837518e-03 | 1.360198e-01 | No | Yes | 1589.020185 | 140.821254 | 1302.142246 | 112.866266 | ||
| ENSG00000105928 | 1687 | GSDME | protein_coding | O60443 | FUNCTION: [Gasdermin-E]: Precursor of a pore-forming protein that converts non-inflammatory apoptosis to pyroptosis (PubMed:27281216, PubMed:28459430). This form constitutes the precursor of the pore-forming protein: upon cleavage, the released N-terminal moiety (Gasdermin-E, N-terminal) binds to membranes and forms pores, triggering pyroptosis (PubMed:28459430). {ECO:0000269|PubMed:27281216, ECO:0000269|PubMed:28459430}.; FUNCTION: [Gasdermin-E, N-terminal]: Pore-forming protein produced by cleavage by CASP3 or granzyme B (GZMB), which converts non-inflammatory apoptosis to pyroptosis or promotes granzyme-mediated pyroptosis, respectively (PubMed:27281216, PubMed:28459430, PubMed:32188940). After cleavage, moves to the plasma membrane, homooligomerizes within the membrane and forms pores of 10-15 nanometers (nm) of inner diameter, triggering pyroptosis (PubMed:28459430, PubMed:32188940). Binds to inner leaflet lipids, bisphosphorylated phosphatidylinositols, such as phosphatidylinositol (4,5)-bisphosphate (PubMed:28459430). Cleavage by CASP3 switches CASP3-mediated apoptosis induced by TNF or danger signals, such as chemotherapy drugs, to pyroptosis (PubMed:27281216, PubMed:28459430, PubMed:32188940). Mediates secondary necrosis downstream of the mitochondrial apoptotic pathway and CASP3 activation as well as in response to viral agents (PubMed:28045099). Exhibits bactericidal activity (PubMed:27281216). Cleavage by GZMB promotes tumor suppressor activity by triggering robust anti-tumor immunity (PubMed:21522185, PubMed:32188940). Suppresses tumors by mediating granzyme-mediated pyroptosis in target cells of natural killer (NK) cells: cleavage by granzyme B (GZMB), delivered to target cells from NK-cells, triggers pyroptosis of tumor cells and tumor suppression (PubMed:32188940, PubMed:31953257). May play a role in the p53/TP53-regulated cellular response to DNA damage (PubMed:16897187). {ECO:0000269|PubMed:16897187, ECO:0000269|PubMed:21522185, ECO:0000269|PubMed:27281216, ECO:0000269|PubMed:28045099, ECO:0000269|PubMed:28459430, ECO:0000269|PubMed:31953257, ECO:0000269|PubMed:32188940}. | Alternative splicing;Cell membrane;Cytoplasm;Deafness;Membrane;Necrosis;Non-syndromic deafness;Reference proteome;Transmembrane;Transmembrane beta strand;Tumor suppressor | Hearing impairment is a heterogeneous condition with over 40 loci described. The protein encoded by this gene is expressed in fetal cochlea, however, its function is not known. Nonsyndromic hearing impairment is associated with a mutation in this gene. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:1687; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; plasma membrane [GO:0005886]; cardiolipin binding [GO:1901612]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; wide pore channel activity [GO:0022829]; cell death [GO:0008219]; cellular response to tumor necrosis factor [GO:0071356]; cellular response to virus [GO:0098586]; granzyme-mediated programmed cell death signaling pathway [GO:0140507]; inner ear receptor cell differentiation [GO:0060113]; necrotic cell death [GO:0070265]; negative regulation of cell population proliferation [GO:0008285]; positive regulation of immune response to tumor cell [GO:0002839]; positive regulation of intrinsic apoptotic signaling pathway [GO:2001244]; positive regulation of MAPK cascade [GO:0043410]; pyroptosis [GO:0070269]; sensory perception of sound [GO:0007605] | 12461698_no significant linkage between age-related hearing impairment (ARHI) and microsatellite markers from the DFNA5 region; there exists no strong association between DFNA5 and ARHI 14559215_Here, we report another mutation in DFNA5, a CTT deletion in the polypyrimidine tract of intron 7. 14676472_A novel DFNA5 mutation was found in a Dutch family, of which 37 members were examined. 16897187_These results suggest that DFNA5 plays a role in the p53-regulated cellular response to genotoxic stress probably by cooperating with p53. 17427029_description of a DFNA5 mutation: the insertion of a cytosine at nucleotide position 640 (AF073308.1:_c.640insC, AAC69324.1:_p. Thr215HisfsX8) which does not lead to hearing impairment 17616391_GCs induce dfna5 mRNA and its expression appears to be repressed in the basal state. Induction of dfna5 mRNA correlates with GC-dependent apoptosis of CEM cells, though dfna5 expression alone is not sufficient for apoptosis. 17868390_DFNA5-associated hearing loss is caused by a very specific gain-of-function mutation. 18223688_DFNA5 is a novel tumor suppressor gene in CRC and a valuable molecular marker for human cancer 18346456_These data implicate DFNA5 promoter methylation as a novel molecular biomarker in human breast cancer. 19911014_A founder effect was demonstrated for the mutation of the DFNA5 gene casusing hearing loss in East Asians. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20403915_Observational study of gene-disease association. (HuGE Navigator) 21522185_DFNA5 is composed of two domains, separated by a hinge region. The first region induces apoptosis when transfected in HEK293T cells, the second region masks and probably regulates this apoptosis inducing capability 21805831_A mutation in DFNA5 leads to a type of hearing loss that closely resembles the frequently observed age-related hearing impairment. 24154762_DFNA5 protein expression in hepatocellular carcinoma cells was significantly lower than that in normal cells. 24506266_DFNA5 deletion mutation is associated with autosomal dominant hereditary hearing loss in Japanese families. 24933359_We identified a novel c.991-2A>G mutation in DFNA5 which again may lead to exon 8 skipping at the mRNA level. 26365971_Study identified a novel DFNA5 mutation IVS8+1 delG in a Chinese family which led to skipping of exon 8. This is the sixth DFNA5 mutation relates to hearing loss and the second one in DFNA5 intron 8. 26400775_Genetic variations in the EYA4, GRHL2 and DFNA5 genes and their interactions with occupational noise exposure may play an important role in the incidence of noise-induced hearing loss (NIHL). 28404884_DFNA5 methylation shows strong potential as a biomarker for detection of breast cancer. Slightly increased methylation in histologically normal breast tissue surrounding the tumor suggests that it may be a good early detection marker. 28459430_findings suggest that caspase-3 activation can trigger necrosis by cleaving GSDME and offer new insights into cancer chemotherapy 29183726_In conclusion, our findings firstly revealed that GSDME switches chemotherapy drug-induced caspase-3 dependent apoptosis into pyroptosis in gastric cancer cells. 29266521_the first exonic mutations in DFNA5 to cause deafness, are reported. 29682089_DFNA5 methylation and expression were significantly different between breast cancer and normal breast samples. DFNA5 methylation, in 10 out of 22 CpGs, and expression were significantly higher in lobular compared to ductal breast cancers. 29849037_specific gain-of-function mechanism of DFNA5 related hearing loss 30061362_These results pinpoint GSDME-dependent pyroptosis as a previously unrecognized mechanism of action for molecular targeted agents to eradicate oncogene-addicted neoplastic cells, which may have important implications for the clinical development and optimal application of anticancer therapeutics. 30091681_DFNA5 variant is associated with tobacco- and HPV-mediated oral oncogenesis. 30564238_The function of GSDME in regulating membrane permeabilization and cell disassembly during apoptosis may be more limited. 30710195_The levels of secondary necrosis/pyroptosis correlated with the levels of active caspase-3 and GSDME-NT. 30976076_Study demonstrated that in addition to its pyroptotic activity, GSDME augments caspase-3/7 activation and apoptotic cell death by targeting the mitochondria and releasing cytochrome c. Like Bid, cleavage of GSDME by death receptor signaling bridges the extrinsic to the intrinsic apoptotic pathway. 30993897_Results found differential methylation in all 22 GSDME CpGs between colorectal cancer (GC) tumor and normal tissues, and in 18 CpGs between the left- and right-sided groups. Although the methylation of 5 distinct probes was a good predictor of gene expression, no association was found between GSDME methylation and its expression. However, a combination of 2 CpGs was found to discriminate between cancer and normal tumor. 31953257_Gasdermin E-mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. 32188940_tumor GSDME acts as a tumor suppressor by activating pyroptosis, enhancing anti-tumor immunity 32296846_Gasdermine E-Dependent Mitochondrial Pyroptotic Pathway in Dermatomyositis: A Possible Mechanism of Perifascicular Atrophy. 32486382_DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot? 32839451_GSDME enhances Cisplatin sensitivity to regress non-small cell lung carcinoma by mediating pyroptosis to trigger antitumor immunocyte infiltration. 33004249_Ultraviolet B induces proteolytic cleavage of the pyroptosis inducer gasdermin E in keratinocytes. 33186472_GSDME and its role in cancer: From behind the scenes to the front of the stage. 33422423_GSDME: A Potential Ally in Cancer Detection and Treatment. 33542198_Gasdermin E deficiency attenuates acute kidney injury by inhibiting pyroptosis and inflammation. 33689708_Dihydroartemisinin induces pyroptosis by promoting the AIM2/caspase-3/DFNA5 axis in breast cancer cells. 33979579_Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes Gasdermin-E-dependent pyroptosis in barrier epithelial cells. 34133932_Gasdermin-E-mediated pyroptosis participates in the pathogenesis of Crohn's disease by promoting intestinal inflammation. 34256094_Photodynamic therapy induces human esophageal carcinoma cell pyroptosis by targeting the PKM2/caspase-8/caspase-3/GSDME axis. 34273529_Caspase-3 and gasdermin E detection in peri-implantitis. 34369076_Caspase-3-mediated GSDME induced Pyroptosis in breast cancer cells through the ROS/JNK signalling pathway. 34433433_Prognostic role of DFNA5 in head and neck squamous cell carcinoma revealed by systematic expression analysis. 34476497_Long noncoding RNA nuclear paraspeckle assembly transcript 1 regulates ionizing radiationinduced pyroptosis via microRNA448/gasdermin E in colorectal cancer cells. 34480835_Attenuation of Rheumatoid Arthritis Through the Inhibition of Tumor Necrosis Factor-Induced Caspase 3/Gasdermin E-Mediated Pyroptosis. 34571081_STAT3beta disrupted mitochondrial electron transport chain enhances chemosensitivity by inducing pyroptosis in esophageal squamous cell carcinoma. 35002520_Apoptin induces pyroptosis of colorectal cancer cells via the GSDME-dependent pathway. 35292781_Gasdermin E mediates resistance of pancreatic adenocarcinoma to enzymatic digestion through a YBX1-mucin pathway. | ENSMUSG00000029821 | Gsdme | 48.56903 | 0.7693900 | -0.3782129580 | 0.40413003 | 8.683511e-01 | 3.514122e-01 | No | Yes | 48.709732 | 9.840766 | 59.720048 | 11.423801 | ||
| ENSG00000106069 | 1124 | CHN2 | protein_coding | P52757 | FUNCTION: GTPase-activating protein for p21-rac. Insufficient expression of beta-2 chimaerin is expected to lead to higher Rac activity and could therefore play a role in the progression from low-grade to high-grade tumors. | 3D-structure;Alternative splicing;GTPase activation;Membrane;Metal-binding;Reference proteome;SH2 domain;Zinc;Zinc-finger | This gene encodes a guanosine triphosphate (GTP)-metabolizing protein that contains a phorbol-ester/diacylglycerol (DAG)-type zinc finger, a Rho-GAP domain, and an SH2 domain. The encoded protein translocates from the cytosol to the Golgi apparatus membrane upon binding by diacylglycerol (DAG). Activity of this protein is important in cell proliferation and migration, and expression changes in this gene have been detected in cancers. A mutation in this gene has also been associated with schizophrenia in men. Alternative transcript splicing and the use of alternative promoters results in multiple transcript variants. [provided by RefSeq, May 2014]. | hsa:1124; | cytosol [GO:0005829]; membrane [GO:0016020]; synapse [GO:0045202]; GTPase activator activity [GO:0005096]; metal ion binding [GO:0046872]; intracellular signal transduction [GO:0035556]; positive regulation of GTPase activity [GO:0043547]; regulation of GTPase activity [GO:0043087]; regulation of small GTPase mediated signal transduction [GO:0051056] | 15863513_beta 2 chimerin has a role in rac-GAP-dependent inhibition of breast cancer cell proliferation 16352660_Data demonstrate that beta2-chimaerin provides a novel, diacylglycerol-dependent mechanism for Rac regulation in T cells and suggest a functional role for this protein in Rac-mediated cytoskeletal remodeling. 16525710_Beta2-chimaerin regulates the proliferation and migration of vascular smooth muscle cells downstream of growth factor signaling pathway implicating human atherogenesis. 17560670_results suggest Tyr-21 phosphorylation of beta2-chimaerin as a novel, Src-family kinase-dependent mechanism that negatively regulates beta2-chimaerin Rac-GAP activity 17803461_the interaction of diacylglycerol kinase gamma with the Src homology 2 and C1 domains of beta2-chimaerin is induced synergistically by Phorbol ester and hydrogen peroxide 18249095_Identify chimaerins as candidates for the downmodulation of Rac1 in T-lymphocytes and, in addition, uncover a novel regulatory mechanism that mediates their activation in T-cells. 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19201754_Tyr-153 is the Lck-dependent phosphorylation residue and its phosphorylation negatively regulates membrane stabilization of beta2-chimaerin, decreasing its GAP activity to Rac 19306875_results suggest that beta2-chimaerin is activated by EphA receptors and mediates the EphA receptor-dependent regulation of cell migration 19720790_Likely digenic cause of insulin resistance and growth deficiency resulting from the combined heterozygous disruption of INSR and CHN2, implicating CHN2 for the first time as a key element of proximal insulin signaling in vivo. 19911011_Observational study of gene-disease association. (HuGE Navigator) 20335173_the Rac-GAP beta2-chimaerin is negatively regulated by protein kinase Cdelta-mediated phosphorylation 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20460425_Observational study of gene-disease association. (HuGE Navigator) 20602751_Observational study of gene-disease association. (HuGE Navigator) 20818722_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21911749_genetic association studies in a Chinese population with type 2 diabetes: An SNP in CHN2 (rs39059) is associated with diabetic retinopathy in a Chinese population of type 2 diabetic patients. 23941981_A significant association of the novel rs186911567 polymorphism was found for the CHN2 gene with smoking. 24854763_genetic association studies in a population in Taiwan: Data suggest that an SNP in CHN2 (rs1002630) is associated with non-proliferative diabetic retinopathy in patients with diabetes type 2 in the Han Chinese population studied. 25057852_CHN2, ABCB1 and PPP1R9A expression on chromosome 7 is implicated in the pathogenesis of hepatosplenic T-cell lymphoma distinguishing it from other malignancies. 26315110_CHN2 is consistently hypermethylated and downregulated in small bowel adenocarcinoma with diagnostic potential 27058424_our data redefine the role of beta2-chimaerin as tumor suppressor and provide the first in vivo evidence of a dual function in breast cancer, suppressing tumor initiation but favoring tumor progression. 28160556_this study shows that serum chemerin is an independent risk factor of prognosis of non-small cell lung cancer patients 31594917_The findings indicate that differential methylation at CpG sites upstream of the CHN2 and JAK2 TSS regions are possible peripheral predictors of antidepressant treatment response. 34089273_Chemerin regulates formation and function of brown adipose tissue: Ablation results in increased insulin resistance with high fat challenge and aging. | ENSMUSG00000004633 | Chn2 | 39.19825 | 0.8308160 | -0.2673990852 | 0.45577182 | 3.424041e-01 | 5.584448e-01 | No | Yes | 43.632817 | 18.202286 | 56.215915 | 22.414018 | ||
| ENSG00000106123 | 2051 | EPHB6 | protein_coding | J3KQU5 | Mouse_homologues FUNCTION: Kinase-defective receptor for members of the ephrin-B family. Binds to ephrin-B1 and ephrin-B2. Modulates cell adhesion and migration by exerting both positive and negative effects upon stimulation with ephrin-B2. Inhibits JNK activation, T-cell receptor-induced IL-2 secretion and CD25 expression upon stimulation with ephrin-B2 (By similarity). {ECO:0000250}. | Glycoprotein;Membrane;Proteomics identification;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a member of a family of transmembrane proteins that function as receptors for ephrin-B family proteins. Unlike other members of this family, the encoded protein does not contain a functional kinase domain. Activity of this protein can influence cell adhesion and migration. Expression of this gene is downregulated during tumor progression, suggesting that the protein may suppress tumor invasion and metastasis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | Mouse_homologues mmu:13848; | integral component of membrane [GO:0016021]; ATP binding [GO:0005524]; protein kinase activity [GO:0004672] | 11466354_Cross-linking of EphB6 alters profiles of lymphokine secretion, inhibits proliferation, induces Fas-mediated apoptosis of Jurkat leukemic T cells, and transduces signals into the cells via proteins it associates with. 12393850_interaction between EphB6 and its ligands facilitates T cell responses to antigen 12517763_EphB6 may play an important role in regulating thymocyte differentiation and modulating responses of mature T cells. 14612926_lower EphB6 expression has a role in melanoma progression to metastatic disease 15955811_EphB6 can both positively and negatively regulate cell adhesion and migration 16364251_The potential significance of EphB6 to serve as a diagnostic and prognostic indicator is discussed. 18754880_the two peptides derived from EphB6v might be appropriate targets for peptide-based specific immunotherapy for HLA-A2(+) patients with various cancers 18819711_CLL B-cells showed a more heterogeneous Eph/EFN profile, specially EFNA4, EphB6 and EphA10. EphB6 and EFNA4 were further related with the clinical course of CLL. 19234485_EphB6 receptor significantly alters invasiveness and other phenotypic characteristics of human breast carcinoma cells. 19513565_The kinase defective EPHB6 receptor tyrosine kinase activates MAP kinase signaling in lung adenocarcinoma. 20086179_Findings suggest a new role for EphB6 in suppressing cancer invasiveness and cell attachment through c-Cbl-dependent signaling. 20181626_The loss of EPHB6 expression in more aggressive breast carcinoma cell lines is regulated in a methylation-dependent manner. The EPHB6 methylation-specific PCR has clinical implications for the prognosis and/or diagnosis of breast and other cancer types 20952760_expression of more than 70 proteins was altered in EphB6-transfected MDA-MB-231 cells; proteins are involved in glycolysis, cell cycle regulation, tumor suppression, cell proliferation, mitochondrial metabolism, mRNA splicing, DNA replication and repair 21351276_Nonsynonymous variants of EPHB6 is associated with familial colorectal cancer. 21737611_results indicate that tumor invasiveness-suppressing activity of EPHB6 is mediated by its ability to sequester other kinase-sufficient and oncogenic EPH receptors 21811619_The alterations in miRNAs and their target mRNAs also suggest indirect involvement of EphB6 in PI3K/Akt/mTOR pathways. 21935409_Data found significant correlations between ephA2, ephA4, ephA7, ephB4, and ephB6 and overall and/or recurrence-free survival in large microarray datasets. 22039307_Our work shows that EphB3 is consistently expressed by malignant T lymphocytes, most frequently in combination with EphB6, and that stimulation with their common ligands strongly suppresses Fas-induced apoptosis in these cells. 24836890_We demonstrate that EphB6 reexpression forces metastatic melanoma cells to deviate from the canonical migration pattern observed in the chick embryo transplant model 24912672_Results suggest that erythropoietin-producing hepatocyte (Eph) receptor B6 (EphB6) may represent a useful tissue biomarker for the prediction of survival rate in colorectal cancers (CRCs). 25152371_EphB6 also interacts with the Hsp90 chaperone. 25239188_These findings implicate EphB6 as a negative regulator of EphA2 oncogenic signaling. 25331796_Studies clearly demonstrate an inverse relationship between the levels of phospho-ERK and the abundance of cadherin 17, beta-catenin and phospho-GSK3beta in EPHB6-expressing MDA-MB-231 cells. 26220827_Enhanced EphB6 expression was significantly associated with Thyroid Lesions. 26468391_EphB6 is a new biomarker for distinguishing high- and low-grade ovarian serous carcinoma, and may be a potential prognostic marker in ovarian serous carcinomas. 26617870_EphB6 protein may be used as a new marker for prognosis for tongue squamous cell carcinoma. 27145271_Study is the first to demonstrate that EphB6 overexpression together with Apc gene mutations may enhance proliferation, invasion and metastasis by colorectal epithelial cells. 27191502_Melanomas from geographically different regions in New Zealand have markedly different mutation frequencies, in particular in the NRAS and EPHB6 genes, when compared to The Cancer Genome Atlas database or other populations. These data have implications for the causation and treatment of malignant melanoma in New Zealand. 27418135_SRC kinase is a synthetic lethality partner of EPHB6 in triple-negative breast cancer cells 27788485_Authors provide evidence that an intrinsically kinase-inactive member of the Eph group of receptor tyrosine kinases, EPHB6, induces marked fragmentation of the mitochondrial network in breast cancer cells of triple-negative origin, lacking expression of the estrogen, progesterone and HER2 receptors. 28262839_using an EphB6 mouse knockout model we found that the loss of EphB6 does not initiate intestinal tumorigenesis and is not involved in the early tumor progression through the adenoma-to-carcinoma transition. 28453458_Data indicate that EphB6 protein was decreased in gastric carcinoma compared with normal mucosa. Analytic results based on pathological parameters suggests that EphB6 protein may inhibit metastasis of gastric carcinoma. 28826721_Low EPHB6 expression is associated with prostate cancer metastasis. 29116180_observations highlight a novel role for EphB6 in reducing drug resistance of T-ALL and suggest that doxorubicin treatment should produce better results if personalised based on EphB6 levels 29700392_It may be beneficial to enhance EPHB6 action concurrent with applying a conventional DNA-damaging treatment. 30262919_Analysis of the association of EPHB6, EFNB1 and EFNB3 variants with hypertension risks in males with hypogonadism. 31160603_This study demonstrated that EPHB6-mutated cells acquire cell adhesion-mediated drug resistance (CAM-DR) in association with CDH11 expression and RhoA/focal adhesion kinase (FAK) activation. Targeted inhibition of EPHA2 or CDH11 reversed the acquired paclitaxel resistance, suggesting its potential clinical utility. 31241800_LncRNA DGCR5 regulates the non-small cell lung cancer cell growth, migration, and invasion through regulating miR-211-5p/EPHB6 axis. 32053275_Cataloguing the dead: breathing new life into pseudokinase research. 32754286_DNA methylation maintains the CLDN1-EPHB6-SLUG axis to enhance chemotherapeutic efficacy and inhibit lung cancer progression. 32819434_The gut microbiota regulates autism-like behavior by mediating vitamin B6 homeostasis in EphB6-deficient mice. 33770085_Structure of the EphB6 receptor ectodomain. | ENSMUSG00000029869 | Ephb6 | 36.08716 | 0.9270620 | -0.1092622887 | 0.52512012 | 4.266791e-02 | 8.363518e-01 | No | Yes | 40.673555 | 9.981943 | 44.691083 | 10.327446 | ||
| ENSG00000106415 | 113263 | GLCCI1 | protein_coding | Q86VQ1 | Coiled coil;Phosphoprotein;Reference proteome | This gene encodes a protein of unknown function. Expression of this gene is induced by glucocorticoids and may be an early marker for glucocorticoid-induced apoptosis. Single nucleotide polymorphisms in this gene are associated with a decreased response to inhaled glucocorticoids in asthmatic patients. [provided by RefSeq, Feb 2012]. | hsa:113263; | cytoplasm [GO:0005737] | 21991891_A functional GLCCI1 variant is associated with substantial decrements in the response to inhaled glucocorticoids in patients with asthma. 22304573_Variation at GLCCI1 and FCER2 could lead personalized asthma treatment. 22660954_GLCCI1 nucleotide polymorphisms associated with steroid-responsiveness in asthmatic patients are unlikely to have a clinically actionable impact in pediatric nephrotic syndrome. 22796022_Studied the influence of GLCCI1 SNP rs37972 on dexamethasone efficacy in bacterial meningitis patients.Results show rs37972 in GLCCI1 is associated with death in patients with CA bacterial meningitis treated with adjunctive dexamethasone therapy. 23836780_Novel association was found between intraocular pressure and a common variant at 7p21 near to GLCCI1 and ICA1. 24131825_GLCCI1 rs37973 does not influence treatment response to inhaled corticosteroids in white subjects with asthma. 24673601_GLCCI1 variant accelerates pulmonary function decline in patients with asthma receiving inhaled corticosteroids. 24897287_GLCCI1 rs37972 T allele was not significantly associated with an increased risk of oral corticosteroid use. 25134782_the minor alleles 'T' and 'G' of rs37972 and rs37973 SNPs, respectively, were not significantly associated with increased asthma risk in asthma patients from Saudi Arabia 25724472_Carriers of the GLCCI1-C allele had lower levels of baseline rheumatoid arthritis disease activity, suggesting a role for the GLCCI1 gene in regulation of glucocorticoid sensitivity to endogenously produced cortisol. 25843653_Carriers of the rs41423247 GLCCI1 polymorphism had a higher probability of responding to glucocorticoids, whereas all other polymorphisms did not affect the likelihood of response to treatment of graft-versus-host disease patients. 27133712_GLCCI1 variations may affect inhaled corticosteroid response by modulating GLCCI1 expression. 28488322_A worsening of pulmonary function caused by GLCCI1 variants could be prevented due to recently used medications based on new action mechanisms. 29224020_CC/CT genotype was significantly associated with post-transplant hypertension 29345236_this study found the GG genotype of GLCCI1 to be associated with better inhaled corticosteroids treatment response in asthma patients 29384926_The genetic variant rs37973 in GLCCI1 is associated with poorer clinical therapeutic response to inhaled glucocorticoids in a Chinese asthma population. 29981236_The GLCCI1 rs37973 variant is a risk factor for glucocorticoid resistance in Chinese patients with SAR who receive short-term intranasal corticosteroids treatment. one genotype of GLCCI1, rs37973, was significantly associated with the INCS response. The effective rate of the GG group was lower than those of the AA and AG groups (AA vs. GG: 73.7% vs. 51.6%, P=0.007; AG vs. GG: 78.8% vs. 51.6%, P=0.000). 30229311_GLCCI1 variant was associated with a significant decrease of the total lung capacity at 3 years after lung transplantation 30256538_The rs37973 polymorphism of GLCCI1 is related to postoperative recovery from chronic rhinosinusitis (CRS) for individual sensitivity to glucocorticoids. Furthermore, AA genotype was associated with better treatment response in CRS. 30860871_GLCCI1 is a downstream molecule of the GC-GR cascade that acts as an antiapoptotic mediator in thymic T cells. Phosphorylated GLCCI1colocalized with microtubules in transfected cells. GR-dependent upregulation of GLCCI1 was proapoptotic in a cultured thymocyte cell line. Transgenic mice overexpressing human GLCCI1 had enlarged thymi with many thymocytes. GLCCI1 bound to both LC8 and PAK1. It reduced BIM expression. 31516081_GLCCI1 and STIP1 variants are associated with asthma susceptibility and inhaled corticosteroid response in a Tunisian population. 33208131_Effects of STIP1 and GLCCI1 polymorphisms on the risk of childhood asthma and inhaled corticosteroid response in Chinese asthmatic children. 33725570_Hsa_circ_001659 serves as a novel diagnostic and prognostic biomarker for colorectal cancer. 34529984_GLCCI1 gene body methylation in peripheral blood is associated with asthma and asthma severity. | ENSMUSG00000029638 | Glcci1 | 1011.67734 | 0.9714651 | -0.0417659003 | 0.11202313 | 1.389189e-01 | 7.093581e-01 | 9.108689e-01 | No | Yes | 1323.062716 | 230.032965 | 1294.860445 | 220.417370 | ||
| ENSG00000106638 | 26608 | TBL2 | protein_coding | Q9Y4P3 | Isopeptide bond;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;WD repeat;Williams-Beuren syndrome | This gene encodes a member of the beta-transducin protein family. Most proteins of the beta-transducin family are involved in regulatory functions. This protein is possibly involved in some intracellular signaling pathway. This gene is deleted in Williams-Beuren syndrome, a developmental disorder caused by deletion of multiple genes at 7q11.23. [provided by RefSeq, Jul 2008]. | hsa:26608; | endoplasmic reticulum [GO:0005783]; integral component of endoplasmic reticulum membrane [GO:0030176]; phosphoprotein binding [GO:0051219]; protein kinase binding [GO:0019901]; RNA binding [GO:0003723]; translation initiation factor binding [GO:0031369]; cellular response to glucose starvation [GO:0042149]; cellular response to hypoxia [GO:0071456]; endoplasmic reticulum unfolded protein response [GO:0030968] | 18193044_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18596051_Observational study of gene-disease association. (HuGE Navigator) 18996102_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19656773_Data show that SNPs associated with TG in normolipidemic samples, including APOA5, TRIB1, TBL2, GCKR, GALNT2 and ANGPTL3 were significantly associated with HLP types 2B, 3, 4 and 5. 19656773_Observational study of gene-disease association. (HuGE Navigator) 20570916_Observational study of gene-disease association. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20679960_Observational study of gene-disease association. (HuGE Navigator) 20972250_Observational study of gene-disease association. (HuGE Navigator) 23564352_A TERE1-TBL2 complex likely functions in oxidative/nitrosative stress, lipid metabolism, and SXR signaling pathways in its role as a tumor suppressor. 25393282_TBL2 interacts with PERK via the N-terminus proximal region and also associates with eIF2a via the WD40 domain thus modulating stress-signaling and cell survival during endoplasmic reticulum stress. 25976671_association of TBL2 with the 60S subunit was ER stress independent while the TBL2-PERK interaction occurred upon ER stress 26239904_TBL2 participates in ATF4 translation through its association with the mRNA. 32811528_TBL2 methylation is associated with hyper-low-density lipoprotein cholesterolemia: a case-control study. | ENSMUSG00000005374 | Tbl2 | 2722.44998 | 0.8926120 | -0.1638948225 | 0.09601478 | 2.902367e+00 | 8.844961e-02 | 4.319531e-01 | No | Yes | 2954.554050 | 225.137082 | 3206.412260 | 238.652945 | ||
| ENSG00000107104 | 23189 | KANK1 | protein_coding | Q14678 | FUNCTION: Involved in the control of cytoskeleton formation by regulating actin polymerization. Inhibits actin fiber formation and cell migration (PubMed:25961457). Inhibits RhoA activity; the function involves phosphorylation through PI3K/Akt signaling and may depend on the competetive interaction with 14-3-3 adapter proteins to sequester them from active complexes (PubMed:25961457). Inhibits the formation of lamellipodia but not of filopodia; the function may depend on the competetive interaction with BAIAP2 to block its association with activated RAC1 (PubMed:25961457). Inhibits fibronectin-mediated cell spreading; the function is partially mediated by BAIAP2. Inhibits neurite outgrowth. Involved in the establishment and persistence of cell polarity during directed cell movement in wound healing. In the nucleus, is involved in beta-catenin-dependent activation of transcription. Potential tumor suppressor for renal cell carcinoma. Regulates Rac signaling pathways (PubMed:25961457). {ECO:0000269|PubMed:16968744, ECO:0000269|PubMed:18458160, ECO:0000269|PubMed:19171758, ECO:0000269|PubMed:22084092, ECO:0000269|PubMed:25961457}. | 3D-structure;ANK repeat;Alternative promoter usage;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Membrane;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Tumor suppressor | The protein encoded by this gene belongs to the Kank family of proteins, which contain multiple ankyrin repeat domains. This family member functions in cytoskeleton formation by regulating actin polymerization. This gene is a candidate tumor suppressor for renal cell carcinoma. Mutations in this gene cause cerebral palsy spastic quadriplegic type 2, a central nervous system development disorder. A t(5;9) translocation results in fusion of the platelet-derived growth factor receptor beta gene (PDGFRB) on chromosome 5 with this gene in a myeloproliferative neoplasm featuring severe thrombocythemia. Alternative splicing of this gene results in multiple transcript variants. A related pseudogene has been identified on chromosome 20. [provided by RefSeq, Dec 2014]. | hsa:23189; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ruffle membrane [GO:0032587]; beta-catenin binding [GO:0008013]; actin cytoskeleton organization [GO:0030036]; cell population proliferation [GO:0008283]; glomerular visceral epithelial cell migration [GO:0090521]; negative regulation of actin filament polymerization [GO:0030837]; negative regulation of cell migration [GO:0030336]; negative regulation of insulin receptor signaling pathway [GO:0046627]; negative regulation of lamellipodium morphogenesis [GO:2000393]; negative regulation of neuron projection development [GO:0010977]; negative regulation of Rho protein signal transduction [GO:0035024]; negative regulation of ruffle assembly [GO:1900028]; negative regulation of substrate adhesion-dependent cell spreading [GO:1900025]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of Wnt signaling pathway [GO:0030177]; positive regulation of wound healing [GO:0090303]; regulation of establishment of cell polarity [GO:2000114]; regulation of Rho protein signal transduction [GO:0035023] | 12133830_Kank is localized to 9p24 and plays a role in cell growth 15823577_Human Kank gene has several alternative first exons. 16968744_Kank can bind to beta-catenin and regulate the subcellular distribution of beta-catenin. 18458160_Data suggest that Kank negatively regulates the formation of actin stress fibers and cell migration through the inhibition of RhoA activity, which is controlled by binding of Kank to 14-3-3 in PI3K-Akt signaling. 19506219_Observational study of gene-disease association. (HuGE Navigator) 19559006_These results suggest that KIF21A regulates the distribution of Kank1 and that KIF21A mutations associated with congenital fibrosis of the extraocular muscles type 1 enhanced the accumulation of Kank1 in the membrane fraction. 19834503_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20164854_KANK1, a candidate tumor suppressor gene, is fused to PDGFRB in an imatinib-responsive myeloid neoplasm with severe thrombocythemia. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22084092_BIG1 and KANK1 play roles in regulating cell polarity during directed migration in wound healing. 22876580_ANKRD15 encodes the kidney ankyrin repeat-containing protein. 23454270_Our case suggests that KANK1 may be subject to random monoallelic expression as a possible mode of inheritance. 24399197_Thus, the human brain glioma apoptosis induced by upregulation of the Kank1 gene is closely relevant to the mitochondrial pathway. 25187374_Follow-up replication analyses in up to an additional 21,345 participants identified three new fasting plasma glucose loci reaching genome-wide significance in or near PDK1-RAPGEF4, KANK1, and IGF1R. 25961457_identified recessive mutations in kidney ankyrin repeat-containing protein 1 (KANK1), KANK2, and KANK4 in individuals with nephrotic syndrome. 25973051_primary results revealed new function of Kank1 for nasopharyngeal cancer 26739330_liprin beta-1 is associated with expression of kank 1 and 2 proteins in melanoma 27410476_Here, the authors show that cortical microtubule stabilization sites containing CLASPs, KIF21A, LL5beta and liprins are recruited to focal adhesions by the adaptor protein KANK1, which directly interacts with the major adhesion component, talin. Structural studies showed that the conserved KN domain in KANK1 binds to the talin rod domain R7. 28067315_KANK1 inhibits Malignant peripheral nerve sheath tumors cell growth though CXXC5 mediated apoptosis. 28284839_Kank1 plays a crucial role in regulating the activity of RhoA through retrieving excess Daam1 and balancing the activities of RhoA and its effectors. 28731169_Upregulating the Kank1 gene can inhibit the progress of gastric cancer. 29158259_Several key residues (i.e. Thr-1147, Leu-1152, Leu-1153, and Tyr-1154) at the C-terminal half of the KIF21A KBD peptide contact with the hydrophobic patch formed by Tyr-1176, Met-1209, Leu-1210, Leu-1213, and Leu-1248 from KANK1. 29729439_KANK1 aberrations do not frequently cause cerebral palsy but represent a risk factor for autism spectrum disorders. 29956815_Low KANK1 expression is associated with lung cancer progression. 30684669_Small interstitial 9p24.3 deletions principally involving KANK1 are likely benign copy number variants. 31114072_KANK1 protein is required for targeting microtubules to focal adhesions. 31338836_Aberrant Kank1 expression regulates YAP to promote apoptosis and inhibit proliferation in OSCC. 31389241_These results indicate that the talin-KANK1 complex is mechanically strong, enabling it to support the cross-talk between microtubule and actin cytoskeleton at focal adhesions. 32064933_KANK1 regulates paclitaxel resistance in lung adenocarcinoma A549 cells. 32787433_Long non-coding RNA CASC2 enhances cisplatin sensitivity in oral squamous cell cancer cells by the miR-31-5p/KANK1 axis. 33046021_KANK1-NTRK3 fusions define a subset of BRAF mutation negative renal metanephric adenomas. 33309958_KANK family proteins in cancer. 34349117_TRAIP modulates the IGFBP3/AKT pathway to enhance the invasion and proliferation of osteosarcoma by promoting KANK1 degradation. 35320976_Identification and Characterization of the Roles of circCASP9 in Gastric Cancer Based on a circRNA-miRNA-mRNA Regulatory Network. | ENSMUSG00000032702 | Kank1 | 479.30815 | 0.8826908 | -0.1800198640 | 0.15179053 | 1.401555e+00 | 2.364634e-01 | 6.276895e-01 | No | Yes | 458.179739 | 57.261812 | 491.854194 | 59.986921 | |
| ENSG00000107186 | 8777 | MPDZ | protein_coding | O75970 | FUNCTION: Member of the NMDAR signaling complex that may play a role in control of AMPAR potentiation and synaptic plasticity in excitatory synapses (PubMed:11150294, PubMed:15312654). Promotes clustering of HT2RC at the cell surface (By similarity). {ECO:0000250|UniProtKB:O55164, ECO:0000269|PubMed:11150294, ECO:0000269|PubMed:15312654}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Cell projection;Host-virus interaction;Membrane;Methylation;Phosphoprotein;Reference proteome;Repeat;Synapse;Synaptosome;Tight junction | The protein encoded by this gene has multiple PDZ domains, which are hallmarks of protein-protein interactions. The encoded protein is known to interact with the HTR2C receptor and may cause it to clump at the cell surface. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2015]. | hsa:8777; | apical part of cell [GO:0045177]; apical plasma membrane [GO:0016324]; apicolateral plasma membrane [GO:0016327]; bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; dendrite [GO:0030425]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; protein C-terminus binding [GO:0008022]; microtubule organizing center organization [GO:0031023]; regulation of microtubule cytoskeleton organization [GO:0070507]; tight junction assembly [GO:0120192] | 15364909_Results indicate that the coxsackievirus and adenovirus receptor interacts with multi-PDZ domain protein 1 (MUPP1) and is involved in MUPP1 recruitment to the tight junction. 16452527_The predominant expression profile of MUPP1 in sperm in the apical acrosomal region of different mammalian species suggests that this PDZ-domain protein may be involved in organizing signaling molecules mediating primary reactions of fertilization. 16452527_The results revealed prominent MUPP1 expression which was restricted to the apical acrosomal region and, most notably, to the equatorial segment of the acrosome[MUPP1] 17672918_the apparent occurrence of an unusual TG 3' splice site in intron 39 is discussed 18378672_MUPP1 binds to the G protein-coupled MT(1) melatonin receptor and directly regulates its G(i)-dependent signal transduction 19071123_An interaction between the human somatostatin receptor 3 and the multiple PDZ protein MUPP1 was identified. 19175764_Exploratory haplotype and single nucleotide polymorphisms association analyses suggest a possible association between the MPDZ gene and alcohol dependence but not alcohol withdrawal seizures. 19175764_Observational study of gene-disease association. (HuGE Navigator) 19255144_Results suggest that signaling mediated by Pals1, which has a higher affinity for Patj than for MUPP1 and is involved in the activation of the Par6-aPKC complex, is of principal importance for the function of Patj in epithelial cells. 19420109_MUPP1 and Kir4.2 may participate in a protein complex in the nephron that could regulate transport of K(+) as well as other ions. 19483657_These data suggest that NMDA receptor complex formation, localization, and downstream signaling may be abnormal in schizophrenia as PSD95, SynGAP and MUPP1 expression is altered. 19874574_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21762291_The data indicates potential association between variation in mpdz NMDA dependent AMPA trafficking and alcohol dependence. 23240096_Our data strongly support the candidacy of MPDZ as a novel congenital hydrocephalus disease gene. 23880463_MUPP-1 controls the PALS-1/PATJ complex levels at the post-translational level. 25675348_The replication association of rs1324183 (MPDZ-NF1B) with KC in our population and the results, which are identical to those in different populations, suggest that rs1324183 (MPDZ-NF1B) is a common genetic risk for Keratoconus. 26984442_two peptides (SIAPNV(-COOH) and SIVMNV(-COOH)) were identified to have considerably improved affinity with K d increase by ~tenfold relative to wild type peptide. Thus, the two peptides are considered as promising lead entities to develop therapeutic molecular agents with high efficacy and specificity to target CaMKIIalpha-MUPP1 interaction. 28556411_Phenotypic spectrum of congenital hydrocephalus is associated with 5 recessive MPDZ alleles. 29620522_Tumor angiogenesis was enhanced upon endothelial-specific inactivation of MPDZ leading to an excessively branched and poorly functional vessel network resulting in tumor hypoxia through DLL4-induced Notch signaling. 30002070_This study confirmed the association of SNP rs1324183 in MPDZ-NF1B with keratoconus and revealed the association of this SNP with keratoconus severity and corneal parameters. It is thus a putative genetic marker for monitoring the progression of keratoconus to a severe form and facilitating early intervention. 31268831_DAPLE and MPDZ function as cooperative partners at apical junctions. 31420327_Observation of nine previously reported and 10 non-reported SLC4A11 mutations among 20 Iranian CHED probands and identification of an MPDZ mutation as possible cause of CHED and FECD in one family. 34108620_MPDZ as a novel epigenetic silenced tumor suppressor inhibits growth and progression of lung cancer through the Hippo-YAP pathway. 34135477_ADAMTS1, MPDZ, MVD, and SEZ6: candidate genes for autosomal recessive nonsyndromic hearing impairment. 35305607_Evaluating the association between MPDZ-NF1B rs1324183 and keratoconus in an independent northwestern Chinese population. | ENSMUSG00000028402 | Mpdz | 850.20937 | 0.9181258 | -0.1232362465 | 0.13518390 | 8.283015e-01 | 3.627641e-01 | 7.344032e-01 | No | Yes | 886.158103 | 157.040165 | 922.711824 | 159.971821 | |
| ENSG00000107643 | 5599 | MAPK8 | protein_coding | P45983 | FUNCTION: Serine/threonine-protein kinase involved in various processes such as cell proliferation, differentiation, migration, transformation and programmed cell death. Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK8/JNK1. In turn, MAPK8/JNK1 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN, JDP2 and ATF2 and thus regulates AP-1 transcriptional activity (PubMed:18307971). Phosphorylates the replication licensing factor CDT1, inhibiting the interaction between CDT1 and the histone H4 acetylase HBO1 to replication origins (PubMed:21856198). Loss of this interaction abrogates the acetylation required for replication initiation. Promotes stressed cell apoptosis by phosphorylating key regulatory factors including p53/TP53 and Yes-associates protein YAP1 (PubMed:21364637). In T-cells, MAPK8 and MAPK9 are required for polarized differentiation of T-helper cells into Th1 cells. Contributes to the survival of erythroid cells by phosphorylating the antagonist of cell death BAD upon EPO stimulation (PubMed:21095239). Mediates starvation-induced BCL2 phosphorylation, BCL2 dissociation from BECN1, and thus activation of autophagy (PubMed:18570871). Phosphorylates STMN2 and hence regulates microtubule dynamics, controlling neurite elongation in cortical neurons. In the developing brain, through its cytoplasmic activity on STMN2, negatively regulates the rate of exit from multipolar stage and of radial migration from the ventricular zone. Phosphorylates several other substrates including heat shock factor protein 4 (HSF4), the deacetylase SIRT1, ELK1, or the E3 ligase ITCH (PubMed:20027304, PubMed:17296730, PubMed:16581800). Phosphorylates the CLOCK-ARNTL/BMAL1 heterodimer and plays a role in the regulation of the circadian clock (PubMed:22441692). Phosphorylates the heat shock transcription factor HSF1, suppressing HSF1-induced transcriptional activity (PubMed:10747973). Phosphorylates POU5F1, which results in the inhibition of POU5F1's transcriptional activity and enhances its proteosomal degradation (By similarity). Phosphorylates JUND and this phosphorylation is inhibited in the presence of MEN1 (PubMed:22327296). In neurons, phosphorylates SYT4 which captures neuronal dense core vesicles at synapses (By similarity). Phosphorylates EIF4ENIF1/4-ET in response to oxidative stress, promoting P-body assembly (PubMed:22966201). {ECO:0000250|UniProtKB:P49185, ECO:0000250|UniProtKB:Q91Y86, ECO:0000269|PubMed:10747973, ECO:0000269|PubMed:16581800, ECO:0000269|PubMed:17296730, ECO:0000269|PubMed:18307971, ECO:0000269|PubMed:18570871, ECO:0000269|PubMed:20027304, ECO:0000269|PubMed:21095239, ECO:0000269|PubMed:21364637, ECO:0000269|PubMed:21856198, ECO:0000269|PubMed:22327296, ECO:0000269|PubMed:22441692, ECO:0000269|PubMed:22966201}.; FUNCTION: JNK1 isoforms display different binding patterns: beta-1 preferentially binds to c-Jun, whereas alpha-1, alpha-2, and beta-2 have a similar low level of binding to both c-Jun or ATF2. However, there is no correlation between binding and phosphorylation, which is achieved at about the same efficiency by all isoforms. | 3D-structure;ATP-binding;Alternative splicing;Biological rhythms;Cell junction;Cytoplasm;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;S-nitrosylation;Serine/threonine-protein kinase;Synapse;Transferase | The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various cell stimuli, and targets specific transcription factors, and thus mediates immediate-early gene expression in response to cell stimuli. The activation of this kinase by tumor-necrosis factor alpha (TNF-alpha) is found to be required for TNF-alpha induced apoptosis. This kinase is also involved in UV radiation induced apoptosis, which is thought to be related to cytochrom c-mediated cell death pathway. Studies of the mouse counterpart of this gene suggested that this kinase play a key role in T cell proliferation, apoptosis and differentiation. Several alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Apr 2016]. | hsa:5599; | axon [GO:0030424]; basal dendrite [GO:0097441]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; synapse [GO:0045202]; ATP binding [GO:0005524]; enzyme binding [GO:0019899]; histone deacetylase binding [GO:0042826]; histone deacetylase regulator activity [GO:0035033]; JUN kinase activity [GO:0004705]; MAP kinase activity [GO:0004707]; protein phosphatase binding [GO:0019903]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine kinase binding [GO:0120283]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cellular response to amino acid starvation [GO:0034198]; cellular response to cadmium ion [GO:0071276]; cellular response to lipopolysaccharide [GO:0071222]; cellular response to mechanical stimulus [GO:0071260]; cellular response to reactive oxygen species [GO:0034614]; cellular senescence [GO:0090398]; Fc-epsilon receptor signaling pathway [GO:0038095]; intracellular signal transduction [GO:0035556]; JNK cascade [GO:0007254]; JUN phosphorylation [GO:0007258]; negative regulation of apoptotic process [GO:0043066]; negative regulation of protein binding [GO:0032091]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of apoptotic process [GO:0043065]; positive regulation of cell killing [GO:0031343]; positive regulation of cyclase activity [GO:0031281]; positive regulation of deacetylase activity [GO:0090045]; positive regulation of gene expression [GO:0010628]; positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway [GO:1900740]; positive regulation of protein metabolic process [GO:0051247]; protein phosphorylation [GO:0006468]; regulation of circadian rhythm [GO:0042752]; regulation of DNA replication origin binding [GO:1902595]; regulation of DNA-binding transcription factor activity [GO:0051090]; regulation of macroautophagy [GO:0016241]; regulation of protein localization [GO:0032880]; response to mechanical stimulus [GO:0009612]; response to oxidative stress [GO:0006979]; response to UV [GO:0009411]; rhythmic process [GO:0048511]; stress-activated MAPK cascade [GO:0051403] | 11912216_Polycystin-1 activation of c-Jun N-terminal kinase and AP-1 is mediated by heterotrimeric G proteins 11931768_Results show that the N-CoR-HDAC3 complex inhibits JNK activation through the associated GPS2 subunit and thus could potentially provide an alternative mechanism for hormone-mediated antagonism of AP-1 function. 11971973_These data strongly suggest that in TNF-induced apoptosis, Hsp72 specifically interferes with the Bid-dependent apoptotic pathway via inhibition of JNK. 12058026_novel role for the I kappa B kinase complex-associated protein (IKAP) in the regulation of activation of the mammalian stress response via the c-Jun N-terminal kinase (JNK)-signaling pathway 12058028_role in stabilizing p21(Cip1) by phosphorylation 12079429_the NOx-induced cell proliferation via activation of JNK1 might contribute to lung tissue damage caused by NOx 12135322_Galpha13 can induce ppET-1 gene expression through a JNK-mediated pathway. 12140754_Elevated JNK activation contributes to the pathogenesis of human brain tumors 12143039_Jun kinase modulates tumor necrosis factor-dependent apoptosis in liver cells 12148599_Unimpaired activation of c-Jun NH2-terminal kinase (JNK) 1 upon CD40 stimulation in B cells of patients with X-linked agammaglobulinemia. 12206715_description of the signaling of JNK and p38 MAPK in apoptosis after stimulation by antioxidants 12296995_TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. 12354774_JNK has isoform-selective gene regulation and distinct JNK isoforms have a role in specific cellular responses 12359245_Relationship of Mcl-1 isoforms, ratio p21WAF1/cyclin A and this protein phosphorylation to apoptosis in human breast carcinomas. 12413764_Psoriatic epidermis shows selective activation of ERK and JNK, which might be related to hyperproliferation and abnormal differentiation of psoriatic epidermis. 12421945_JNK activation is predominantly involved in the induction of CD44 expression in monocytic cells via lipopolysaccharide-mediated signaling. 12478662_JNK-1 and p38 play a role in apoptosis induced by capsaicin in H-ras-transformed tumor cells 12514174_phosphorylation of JNK1 and WOX1 is necessary for their physical interaction and functional antagonism 12538493_JNK is required for growth of prostate carcinoma cells in vitro and in vivo 12592382_Western blot demonstrated that phosphorylation of JNK was induced only by TPA during 30 min to 1 h. 12646240_Data suggest that epidermal growth factor (EGF) stimulated c-Jun N-terminal kinase phosphorylation of c-Jun is uncoupled from protein kinase D suppression in cancer cells. 12707267_JNK1 has a role in the synergistic effect of TRAIL combined with DNA damage by mediating signals independent of p53 leading to apoptosis 12810082_Results suggest that tissue or plasma fibronectin may modulate the intestinal epithelial response to repetitive deformation through inhibted activation of p38 and jun kinases. 12847227_c-Jun N-terminal kinase plays a negative role in the production of IL-12 from human macrophages stimulated by lipopolysaccharide. 12859962_Data show that inhibition of arachidonate 5-lipoxygenase induces rapid activation of c-Jun N-terminal kinase (JNK) in human prostate cancer cells which is prevented by the 5-lipoxygenase metabolite, 5(S)-HETE. 12878610_Axin utilizes distinct regions for competitive MEKK1 and MEKK4 binding and JNK activation. 12902351_adipose cytokines and JNK are key mediators between obesity and hormone-resistant prostate cancer 12917434_Threonine 668 within the Amyloid beta protein precursor intracellular domain is indeed phosphorylated by JNK1; although JIP-1 can facilitate this phosphorylation, it is not required for this process. 14500675_Findings strongly suggest that the JNK/AP-1 transcription factor signaling pathway has little or no impact on the generation of inflammatory mediators in neutrophils. 14514687_late but not early JNK1 activation is associated with the induction of apoptosis 14532003_Results show that tumor necrosis factor alpha-mediated caspase 8 cleavage and apoptosis require a sequential pathway involving c-Jun N-terminal kinase, Bid, and Smac/DIABLO. 14557276_c-Jun N-terminal kinase activation in T cell receptor signaling is mediated by SH3 domain-containing adaptor HIP-55 14561739_JNK inhibition with SP600125 also blocked binding of Sp1 to the DR5/TRAIL-R2 promoter. 14637155_Fas-induced cell death and JNK activation are sensitive to Fas stimulation in cell lines carrying undetectable level of c-FLIP(L). 14688370_Stress-activated protein kinase 1 is involved in the control of monocyte chemoattractant protein-1-induced migration of MonoMac6 cells. 14699155_JNK activation is important for lipopolysacchairde-induced MCP-1 expression but not for TNF-alpha or IL-8 expression 14701702_data support an essential role for JNK signaling in the induction of growth inhibition and apoptosis by As(2)O(3) 14724588_activation of JNK is important for the induction of apoptosis following stresses that function at different cell cycle phases, and that basal JNK activity is necessary to promote proliferation and maintain diploidy in breast cancer cells 14729602_Calcium signaling in ovarian surface epithelial cells not only induces telomerase activity via JNK but also activates Pyk2. 14766760_JNK regulates the expression of HIPK3 in prostate cancer cells, which leads to increased resistance to Fas receptor-mediated apoptosis by reducing the interaction between FADD and caspase-8 14981905_Inducible expression of RbAp46 activated the c-Jun N-terminal kinase (JNK) signaling pathway and triggered apoptosis in Saos-2 cells xenografted into nude mice. 15013949_JNK signaling regulates the phosphorylation state of several kinases in skeletal muscle. JNK activation is unlikely to be the major mechanism by which contractile activity increases glycogen synthase activity in skeletal muscle. 15238629_The c-Jun-N-terminal kinase(JNK)cascade mediates the stimulatory effect Angiotensin II has on the proximal renin promoter in humans. 15456887_Data show that endogenous germinal center kinase is activated by agonists that require TRAF6 for c-Jun N-terminal kinase activation. 15474087_SAPKgamma/JNK1 and SAPKalpha/JNK2 may be important mediators of stress-induced responses in early implanting conceptuses that could mediate embryo loss. 15516492_Antiproliferative and prodifferentiation effects of BMP4 were Smad1 dependent with JNK also contributing to differentiation. 15527495_crucial role of JNK signalling pathway in N. meningitidis invasion in human brain microvascular endothelial cells 15528994_Cooperation of CD99 engagement with suboptimal TCR/CD3 signals resulted in enhanced CD4+ T cell proliferation, elevated expression of CD25 and GM1, increased apoptosis, augmented activation of JNK, and increased AP-1 activation 15542843_activation of p38 MAPK and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and TNFR1/TNFa expression 15569856_The ACE-inhibitor mediated activation of the c-Jun N-terminal kinase (JNK)/c-Jun pathway, results in an enhanced endothelial ACE expression 15629131_While JNK1 is a downstream effector of the Tumor Necrosis Factor (TNF) signaling, Zfra regulation of the TNF cytotoxic function is likely due to its interaction, in part, with JNK1 15637062_Caspase 3-cleaved SH3 domain of HIP-55 is likely involved in PRAM-1-mediated JNK activation upon arsenic trioxide-induced differentiation of NB4 cells. 15655348_ERK1/2 and JNK1/2 signaling is stimulated by radiation and can promote cell cycle progression in human colon cancer cells 15657352_Alpha-tocopheryl sulfsatse showed increased levels in prostate tumor cells. 15665513_TNF-alpha causes a net up-regulation of MUC2 gene expression in cultured colon cancer cells because NF-kappaB transcriptional activation of this gene is able to counter-balance the suppressive effects of the JNK pathway 15696159_JNK phosphorylates 14-3-3 proteins, which regulate nuclear targeting of c-Abl in the apoptotic response to DNA damage 15755722_PKD is a critical mediator in H2O2- but not TNF-induced ASK1-JNK signaling 15769735_raft-associated acid sphingomyelinase and JNK activation and translocation are induced by UV-C light on a nuclear signal 15778501_p53 participates in a feedback mechanism with JNK to regulate the apoptotic process and is oppositely regulated by JNK1 and JNK2. 15860507_Some green tea catechins induce pro-MMP-7 production via O2- production and the activation of JNK1/2. 15890690_JNK may act via c-Myc and Egr-1, which were shown to be important for B-lymphoma survival and growth. 15981086_rF1-induced JNK MAPK activity was correlated to the functional activation of macrophages by demonstrating the inhibition of NO, TNF-alpha production and microtubule polymerization 16086581_results suggest that activated JNK can, in turn, activate not only jun but also raf that, in turn, activates MEK that can then cross-activate JNK in a positive feedback loop 16105650_We concluded that JNK pathway might play an important role in mediating cisplatin-induced apoptosis in A2780 cells, and the duration of JNK activation might be critical in determining whether cells survive or undergo apoptosis. 16166642_JAMP is a membrane-anchored regulator of the duration of JNK1 activity in response to diverse stress stimuli 16176806_The activation of JNK1 is required for the triptolide-induced inhibition of tumor proliferation. 16243842_Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages 16260419_protein kinase Cdelta and JNK have roles in Ifn-alpha induced expression of phospholipid scramblase 1 through STAT1 16260609_c-Jun N-terminal kinase signaling is regulated by a stabilization-based feed-forward 16282329_Foxo1 is involved in the nucleocytoplasmic translocation of PDX-1 by oxidative stress and the JNK pathway 16283431_Results describe the opposite effect of ERK1/2 and JNK on p53-independent p21WAF1/CIP1 activation involved in the arsenic trioxide-induced human epidermoid carcinoma A431 cellular cytotoxicity. 16307741_gemcitabine-induced apoptosis in human non-small cell lung cancer H1299 cells requires activation of the JNK1 signaling pathway 16321971_in gastric epithelial cells, H. pylori up-regulates MMP-1 in a type IV secretion system-dependent manner via JNK and ERK1/2 16328781_Results suggest that VEGF induced by hyperbaric oxygen is through c-Jun/AP-1 activation, and through simultaneous activation of ERK and JNK pathways. 16339571_TNF-alpha induced PTX3 expression in human lung cell lines and primary epithelial cells; knockdown of either JNK1 or JNK2 with small interfering RNA also significantly reduced the regulated PTX3 expression 16381010_results indicate that the aberrant p-JNK1/2 expression and the co-expressed p-JNK1/2 and p-p38 in breast tissues may play a role in the carcinogenesis of breast infiltrating ductal carcinoma 16407310_Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1 16412424_Data report that cepharanthine induces apoptosis in HuH-7 cells through activation of JNK1/2 and the downregulation of Akt. 16434970_Both activation of JNK and inhibition of Akt play a role in translocation of Nur77 from the nucleus to the cytoplasm. 16465391_We investigated whether Jun-N-terminal kinase activation is increased in inflammatory bowel disease and analyzed the effects of SP600125, which decreases inflammatory cytokine synthesis by inhibiting the phosphorylation of this kinase. 16569638_matrix metalloproteinase-1 expression is regulated by JNK through Ets and AP-1 promoter motifs 16648634_Inhibition of JNK in epidermal keratinocytes is sufficient to initiate their differentiation program and suggest that augmenting JNK activity could be used to delay cornification and enhance wound healing. 16687404_JNK-mediated feedback phosphorylation of MLK3 regulates its activation and deactivation states by cycling between Triton-soluble and Triton-insoluble forms 16699726_ERK1/2, JNK1/2 and p38 mapk pathways are all required for B[a]P-induced G1/S transition 16760468_JNK is a critical component downstream of PI 3-kinase that may be involved in PDGF-stimulated chemotaxis presumably by modulating the integrity of focal adhesions by phosphorylating its components 16794185_Paclitaxel increases endothelial TF expression via its stabilizing effect on microtubules and selective activation of JNK 16802349_JNK (c-Jun N-terminal kinase) function might be modulated by targeting MKK-7 to suppress cytokine-mediated fibroblast-like synoviocytes (FLS) activation while leaving other stress responses intact. 16814421_Results show that activation of c-Jun N-terminal protein kinase, ERKs 1 and 2, and p38 MAP kinase is critical for Hs683 glioma cell migration induced by GDNF. 16815888_PP1-JNK pathway plays a role in H(2)O(2)-induced Sp1 phosphorylation in lung epithelial cells 16824735_JNK1 is associated with UV signal transduction in human epidermis and SCCA1 is a suppressor of this process. 16895791_TNF-alpha down-regulates human Cu/Zn superoxide dismutase 1 promoter via JNK/AP-1 signaling pathway 16912864_Leptin stimulates proliferation and inhibits apoptosis in colon cancer cells. This effect involves JAK2, PI3 kinase and JNK and activation of the oncogenic transcription factors signal transducer and activator of transcription 16927023_This study is the first to demonstrate that H2O2 induces a Rac1/JNK1/p38 signaling cascade, and that JNK and p38 activation is important for H2O2-induced apoptosis as well as apoptosis-inducing factor/Bax translocation of retinal pigment epithelial cells. 16972261_Data show that pharmacologic inhibitors of extracellular signal-regulated kinase (ERKs) and c-Jun NH(2)-terminal kinase (JNK) decrease glutathione content and sensitize human promonocytic leukemia cells to arsenic trioxide-induced apoptosis. 16983342_active JNK1 inhibits ubiquitination of C/EBPalpha possibly by phosphorylating in its DBD 17008315_STAT3 activation by G alpha(s) distinctively requires protein kinase A, JNK, and phosphatidylinositol 3-kinase 17054907_Collectively, our results suggest that the inhibition of the interaction between JNK and c-Jun may be an integral part of the mechanism underlying the negative regulation of the JNK signaling pathway by NO. 17074809_JNK1 and JNK2 differentially regulate TBP through Elk-1, controlling c-Jun expression and cell proliferation 17079291_Overall, these results demonstrate the importance of the JNK pathway for varicella-zoster virus replication. 17158878_PSK1-alpha is a bifunctional kinase that associates with microtubules, and JNK- and caspase-mediated removal of its C-terminal microtubule-binding domain permits nuclear translocation of the N-terminal region of PSK1-alpha and its induction of apoptosis 17178870_JNK1 is activated in response to collagen I, which increases tumorigenesis by up-regulating N-cadherin expression and by increasing motility. 17255354_Oxidative stress response regulates DKK1 expression through the JNK signaling cascade in multiple myeloma plasma cells. 17296730_Human Rev7 (hRev7)/MAD2B/MAD2L2 is an interaction partner for Elk-1 and hRev7 acts to promote Elk-1 phosphorylation by the c-Jun N-terminal protein kinase (JNK) MAP kinases. 17303384_These results suggest that in human tracheal smooth muscle cells, activation of p42/p44 MAPK, p38, and JNK pathways, at least in part, mediated through NF-kappaB, is essential for lipopolysaccharide-induced VCAM-1 gene expression. 17317777_Increased expression of stress-activated kinases and IKK and their phosphorylated forms in omental fat occurs in obesity. 17453826_The regulation of three major mitogen-activated protein kinases phosphorylation, ERKp44/p42, p38, and JNK, was determined. The influence of specific mitogen-activated protein kinase inhibitors on IL-1 beta protein levels during beta-endorphin stimulation. 17478078_Proinvasive activity of BMP7 through SMAD4/src-independent and ERK/Rac/JNK-dependent signaling pathways in colon cancer cells is reported. 17481915_These results indicate that binding of the alpha3beta1 integrin results in a suppression in the activation of the IL-1 induced intracellular signaling pathway from JNK to AP-1. 17496921_In this review, the interplay between NF-kappa B and JNK1 provides a paradigm that shows how crosstalk between different signaling pathways decides the function of the cell signaling circuitry. 17541429_Gemin5 functions as a scaffold protein for the ASK1-JNK1 signaling module and thereby potentiates ASK1-mediated signaling events. 17545598_Findings showed that TOPK positively modulated UVB-induced JNK1 activity and played a pivotal role in JNK1-mediated cell transformation induced by H-Ras. 17568996_IGF-1R and PDGFR co-inhibition caused an increased cell death in two human glioma cell lines and induced the radiosensitization of the JNK1 expressing cell line. 17584736_GCK is required for JNK and, unexpectedly, p38 activation by three bacterial PAMPs, lipopolysaccharide, peptidoglycan, and flagellin 17603935_TNF-alpha induced reactive oxygen species formation is mediated by JNK1, which regulates ferritin degradation and thus the level of highly reactive iron. 17620321_These results support a significant role for ALK1 as a negative regulator of endothelial cell migration and suggest the implication of JNK and ERK as mediators of this effect. 17626013_JNK and p38 mitogen-activated protein kinases were activated by TAK1. 17640761_This study reveals a novel pathway of gene regulation by alcohol which involves the activation of JNK and the consequent mRNA stabilization. 17652454_These results indicate that Ask1 oxidation is required at a step subsequent to activation for signaling downstream of Ask1 after H(2)O(2) treatment. 17690186_Data suggest that JNK activation and decreased expression of MKP-1 may play important roles in progression of urothelial carcinoma. 17693927_Studies suggest that the target of regulation by PP2A includes upstream kinases in the JNK MAPK pathway. 17699782_Findings suggest that the JNK/PTEN and NF-kappaB/PTEN pathways play a critical role in normal intestinal homeostasis and colon carcinogenesis. 17702750_long duration KOR antagonists disrupt KOR signaling by activating JNK 17703233_findings establish a major role for DAPk and its specific interaction with PKD in regulating the JNK signaling network under oxidative stress. 17704768_Results demonstrate that Fbl10 is a key regulator of c-Jun function. 17719653_Costimulation by anti-CD3 and anti-CD28 antibodies could activate JNK, p38 MAPK and NF-kappaB. The upregulation of IL-25 receptors were differentially regulated by intracellular JNK, p38 MAPK and NF-kappaB. 17785464_Study provides evidence during rolling and adhesion of platelets to vWF that platelet GPIb-vWF interaction triggers alphaIIbbeta3 activation in a JNK1-dependent manner; this was confirmed with a Glanzmann thrombastenic patient lacking alphaIIbbeta3. 17883418_Streptococcus intermedius histon-like DNA binding protein (Si-HLP) stimulation induced the activation of cell signal transduction pathways, extracellular signal-regulated kinase 1/2 (ERK1/2) and c-Jun N-terminal kinase (JNK). 17904874_Our data show that basic JNK activity plays an important role in the progression of the cell cycle at G2/M cell phase. 17908987_JNK1/2 activity is commonly increased in head and neck squamous cell carcinoma 17913539_These results collectively indicated that Chlamydophilal antigens induce foam cell formation mainly via Toll-like receptor 2 and c-Jun NH2 terminal kinase. 17933493_The induction of IFIT4 transcription by IFN-alpha depends upon sequential activation of PKCdelta, JNK and STAT1, and the influence of PKCdelta or JNK on IFN-alpha-mediated induction of IFIT4 is dependent upon the phosphorylation of STAT1 at Ser-727. 17942603_IFNalpha-induced apoptosis requires activation of ERK1/2, PKCdelta, and JNK downstream of PI3K and mTOR, and it can occur in a nucleus-independent manner, thus demonstrating that IFNalpha induces apoptosis in the absence of de novo transcription. 17967471_Critical role in cell transformation induced by EBV LMP1. 17982228_crude extract of D. farinae induces ICAM-1 expression in EoL-1 cells through signaling pathways involving both NF-kappaB and JNK 18025271_Cyclin G2 expression is modulated by HER2 signaling through multiple pathways including phosphoinositide 3-kinase, c-jun NH(2)-terminal kinase, and mTOR signaling. 18036196_Observational study of gene-disease association. (HuGE Navigator) 18036196_The G/G genotype of MAPK8 SNP-1066 did not affect T2DM susceptibility despite specific binding of AP2alpha. 18055217_We conclude that endogenous SOCS3 inhibits AP-1 activity through blocking of JNK phosphorylation. 18082745_Nonalcoholic steatohepatitis is specifically associated with (1) failure to generate sXBP-1 protein and (2) activation of JNK. 18086557_Activation of protease-activated receptors but not stimulation with lipopolysaccharides leads to ERK1/2 and JNK-mediated production of IL-8. 18087676_These results clearly indicate that CCDC134 is a novel member of the secretory family and down-regulates the Raf-1/MEK/ERK and JNK/ SAPK pathways. 18094581_The majority of acute myeloid leukemia cases did not show any levels of Mitogen-Activated Protein Kinases activation except for two cases, which were associated with an extremely high white blood cell count, chromosomal aberration. 18164704_Our data first suggest that JNK participates in bFGF-mediated surface cadherin downregulation. Loss of surface cadherins may affect the cell-cell interaction between endothelial cells and facilitate angiogenesis. 18181766_ERK and JNK are involved in PMA-mediated MD-2 gene expression during HL-60 cell differentiation. 18199680_JNK1 activation is necessary to phosphorylate Sp1 and to shield Sp1 from the ubiquitin-dependent degradation pathway during mitosis in tumor cell lines. 18212053_c-Jun amino-terminal kinase (JNK) was important for neurite outgrowth stimulated by both Wnt-3a and Dkk1. 18218857_corneal inflammation is significantly impaired in JNK1 knockout mice compared with control mice, and in mice treated with the JNK inhibitor compared with vehicle control. 18219313_These results identify NLRX1 as a NLR that contributes to the link between reactive oxygen species generation at the mitochondria and innate immune responses. 18249102_Our results suggest that clivorine has direct toxicity on HEK293 cells, and phosphorylated JNK may play some role in counteracting the toxicity of clivorine on HEK293 cells. 18253836_activator protein-1 (AP-1) was activated through phosphorylation of cJun and cFos, induced by JNK and p38, respectively. 18256527_Study shows that basal c-Jun N-terminal kinases (JNKs) are required for mitotic histone H3-S10 phosphorylation in human primary fibroblast IMR90 cells 18276794_Glutamate cysteine ligase iz induced by hydroxynonenal through the c-Jun N-terminal kinase (JNK) pathway in respiratory epithelium. 18286207_the activity of JIP1-JNK complexes is downregulated by VRK2 in response to interleukin-1beta 18288129_both constitutive and sIgM-induced phosphorylation of p38 and JNK is inhibited by LAIR-1 through an ITIM-dependent signal 18292600_PP2A and AIP1 cooperatively induce activation of ASK1-JNK signaling and vascular endothelial cell apoptosis. 18293403_Preventing the TAK1/JNK1 signaling cascade in astrocytes might provide a fruitful strategy for treating intractable neuropathic pain. 18297686_These results identify JNK, and not NFkappaB, as a critical mediator of TNF-alpha repressory effect on connexin 43 gene expression. 18316600_ERK and JNK MAPK/Elk-1/Egr-1 signal cascade is required for p53-independent transcriptional activation of p21(Waf1/Cip1) in response to curcumin in U-87MG human glioblastoma cells 18316603_c-Jun translocates B23 and ARF from the nucleolus after JNK activation by means of protein interactions 18325654_Oxidative stress-activated JNK signaling pathway is involved in METH-induced cell death. 18337589_Phosphorylation aids GR sumoylation and that cross talk of JNK and SUMO pathways fine tune GR transcriptional activity. 18344085_The protein expression rates of p-JNK and P-glycoprotein in gastric cancer were significantly higher than those in normal gastric tissue. 18348163_Data show that JNK modulates the effect of caspases and NF-kappaB in the TNF-alpha-induced down-regulation of Na+/K+ATPase in HepG2 cells. 18356158_H. pylori mediates CagA-independent signaling that promotes cell motility through the beta1 integrin-JNK pathway 18373696_Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in cervical cancer. 18401423_Although STAT1 phosphorylation required JNK and p38MAPK activation, only JNK activation was essential for IRF1 promoter activation by Tie2-R849W. 18405916_These data demonstrate the role of Cx43 in the proliferation and migration of human saphenous vein smooth muscle cells and angiotensin II-induced Cx43 expression via mitogen-activated protein kinases (MAPK)-AP-1 signaling pathway. 18429822_Study demonstrates that the serine/threonine kinase PKN1 plays a critical role in regulating constitutive IKK/JNK activity in unstimulated cells and report on the molecular mechanism. 18439101_These findings suggest that JNK1 and JNK2 are involved in TNF-alpha-induced neutrophil apoptosis and GM-CSF-mediated antiapoptotic effect on neutrophils, respectively. 18457359_is the key enzyme mediating melanogenesis in B16F10 cell. 18495129_inhibition of JNK and p38 activation interrupts CD40 induced endothelial cell activation and apoptosis 18506470_JNK is involved in regulation of proinflammatory mediators of endometrium 18524773_JNK1 is a critical transcriptional target of FoxM1 that contributes to FoxM1-regulated cell cycle progression, tumor cell migration, invasiveness, and anchorage-independent growth 18540881_Although JNK activation may be a primary inducing factor, further phosphorylation of tau is required for neuronal death and NFT formation in neurodegenerative diseases, including those characterized by tauopathy. 18541008_JNK, and in particular the JNK1 isoform, support the growth of melanoma cells, by controlling either cell cycle progression or apoptosis depending on the cellular context. 18547751_Nanosilver acts through ROS and JNK to induce apoptosis via the mitochondrial pathway. 18570871_JNK1 mediates starvation-induced Bcl-2 phosphorylation, Bcl-2 dissociation from Beclin 1, and autophagy activation 18573678_Data show that pokeweed antiviral protein (PAP) does not inhibit protein translation, but induces the activation of c-Jun NH2-terminal kinase (JNK), which was specific to rRNA depurination. 18594007_c-Jun NH2-terminal protein kinase activation caused by tubulin depolymerization and DNA damage has a crucial role in moscatilin-induced apoptosis in human colorectal cancer cells 18603327_Data show that M. bovis BCG-induced human beta-defensin mRNA expression in A549 cells is regulated at least in part through activation of signaling proteins of PKC, JNK and PI3K. 18620777_WWOX induces apoptosis and inhibits human hepatocellular carcinoma cell growth through a mechanism enhanced by JNK inhibition. 18636174_A functional analysis of JNK1 and M-RIP with RNA interference reveals a critical role for this cascade in the invasive behavior of cancer cells. 18651223_These findings suggest JNK to have an important pro-apoptotic function following ultraviolet rays B irradiation in human melanocytes, by acting upstream of lysosomal membrane permeabilization and Bim phosphorylation. 18663379_Shikonin-induced oxidative injury operates at a proximal point in apoptotic signaling cascades, and subsequently activates the stress-related JNK pathway, triggers mitochondrial dysfunction, cytochrome c release, and caspase activation. 18667537_necrosis factor-alpha-elicited stimulation of gamma-secretase is mediated by c-Jun N-terminal kinase-dependent phosphorylation of presenilin and nicastrin 18681908_SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes 18682391_Histamine-induced Egr-1 expression is dependent on the activation of the H1 receptor, and rapidly and transiently activates PKCdelta, ERK1/2, p38 kinase, and JNK prior to Egr-1 induction. 18703151_These results demonstrate that ERKs and JNKs are responsible for the decrease of cyclin D1 and CDK4 expression levels in human embryonic lung fibroblasts induced by silica. 18713649_These results suggest that phosphorylation of paxillin on Ser 178 by JNK is required for the association of paxillin with FAK, and subsequent tyrosine phosphorylation of paxillin. 18713996_JNK is differentially regulated by MKK4 and MKK7 depending on the stimulus. 18718914_15d-PGJ(2) induces vascular endothelial cell apoptosis through the signaling of JNK and p38 MAPK-mediated p53 activation both in vitro and in vivo 18723442_whether MAP kinase phosphatase (MKP)-1, a negative regulator of p38 and JNK, mediates the antiinflammatory effects of shear stress. 18757369_a link between RhoA, JNK, c-Jun, and MMP2 activity that is functionally involved in the reduction in osteosarcoma cell invasion by the statin. This suggests a novel strategy targeting RhoA-JNK-c-Jun signaling to reduce osteosarcoma cell tumorigenesis. 18769111_A speculative model for understanding the interrelationship between autophagy and apoptosis regulated by JNK1-mediated Bcl-2 phosphorylation was proposed. 18782768_pneumolysin selectively induced expression of MKP1 via a TLR4-dependent MyD88-TRAF6-ERK pathway, which inhibited the PAK4-JNK signaling pathway,leading to up-regulation of MUC5AC mucin production 18815275_These results establish a novel function of filamin B as a molecular scaffold in the JNK signaling pathway for type I IFN-induced apoptosis. 18818208_analysis of JNK-mediated phosphorylation of paxillin in adhesion assembly and tension-induced cell death by the adenovirus death factor E4orf4 18845538_a novel function for Parkin in modulating the expression of Eg5 through the Hsp70-JNK-c-Jun signaling pathway. 18922473_These results indicate that TGF-beta activates JNK and p38 through a mechanism similar to that operating in the interleukin-1beta/Toll-like receptor pathway. 18936517_JNK may play an important role in posttranscriptional control of LDL receptor expression, thus constituting a novel mechanism to enhance plasma LDL clearance by liver cells. 18950845_Observational study of gene-disease association. (HuGE Navigator) 18978303_dynasore may stimulate PAI-1 protein expression and enhance TGF-beta(1) activity through activation of JNK-mediated signaling in human pleural mesothelial cells 18979912_The changes of phospho-JNK expression after skin burned might correlate with wound healing. 18982452_Data show that an increase in JNK activation in the presence of NFkappaB inhibition significantly increased the expression of IGFBP6. 18989785_Results show that Docetaxel-induced apoptosis is mediated by induction of ER stress, through activation of JNK and downstream targets of JNK. 18996088_These results suggest that JNK, but not caspase 8, involves in Fas-mediated CH11-induced autophagy in HeLa cells, and this autophagy plays a protective role in CH11-induced cell death. 19033664_a mechanistic link between JNK activity and liver cell proliferation via p21 and c-Myc and suggest JNK1 targeting can be considered as a new therapeutic approach for HCC treatment. 19036714_Upon UVB-induced stress in keratinocytes, ROCK1 was activated, bound to JIP-3, and activated the JNK pathway 19037093_AMPK controls the molecular mechanism underlying the differential biological functions of JNK, providing a novel explanation for the antiapoptotic role of LKB1. 19041150_JNK1 plays important roles in the development of human HCC partially through the epigenetic mechanisms. 19052872_c-Abl and p53 are important for execution of the cell death program initiated in A2E-laden RPE cells exposed to blue light, while JNK might play an anti-apoptotic role 19056926_JNK1 stimulated and mediated the effects of IFN and TNF-alpha on XAF1 expression through transcriptional regulation by induction of IRF-1. 19060920_activation of JNK pathway can mediate Beclin 1 expression, which plays a key role in autophagic cell death in cancer cel | ENSMUSG00000021936 | Mapk8 | 1477.55961 | 1.1927101 | 0.2542434327 | 0.11774668 | 4.686450e+00 | 3.040139e-02 | 2.782986e-01 | No | Yes | 2349.387333 | 473.517894 | 1888.756611 | 372.512186 | |
| ENSG00000107719 | 27143 | PALD1 | protein_coding | Q9ULE6 | Cytoplasm;Lipoprotein;Myristate;Phosphoprotein;Reference proteome | hsa:27143; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; protein tyrosine phosphatase activity [GO:0004725] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 19727444_PALD (KIAA1274, paladin) has a role in inhibition of insulin's ability to down regulate a FOXO1A-driven reporter gene, reduce upstream insulin-stimulated AKT phosphorylation, and decrease insulin receptor (IR) abundance | ENSMUSG00000020092 | Pald1 | 1151.37791 | 0.8683412 | -0.2036660917 | 0.10226108 | 3.945877e+00 | 4.698636e-02 | 3.288948e-01 | No | Yes | 1284.383861 | 152.167471 | 1424.255565 | 164.641011 | |||
| ENSG00000107821 | 81621 | KAZALD1 | protein_coding | Q96I82 | FUNCTION: Involved in the proliferation of osteoblasts during bone formation and bone regeneration. Promotes matrix assembly (By similarity). {ECO:0000250}. | Alternative splicing;Developmental protein;Differentiation;Direct protein sequencing;Disulfide bond;Extracellular matrix;Glycoprotein;Immunoglobulin domain;Osteogenesis;Reference proteome;Secreted;Signal | This gene encodes a secreted member of the insulin growth factor-binding protein (IGFBP) superfamily. The protein contains an insulin growth factor-binding domain in its N-terminal region, a Kazal-type serine protease inhibitor and follistatin-like domain in its central region, and an immunoglobulin-like domain in its C-terminal region. Studies of the mouse ortholog suggest that this protein may function in bone development and bone regeneration. This gene is hypomethylated and over-expressed in high-grade glioma compared to low-grade glioma, and thus the hypomethylated gene may be associated with cell proliferation and the shorter survival of patients with high-grade glioma. It is also one of numerous genes found to be deleted in a novel 5.54 Mb interstitial deletion, which is associated with multiple congenital anomalies. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2016]. | hsa:81621; | extracellular region [GO:0005576]; interstitial matrix [GO:0005614]; insulin-like growth factor binding [GO:0005520]; cell differentiation [GO:0030154]; extracellular matrix organization [GO:0030198]; ossification [GO:0001503]; regulation of cell growth [GO:0001558]; regulation of signal transduction [GO:0009966] | 15261838_Expression of Bono1 was detected in osteoblasts and secretory odontoblasts by in situ hybridization. 15261838_This paper describes the identification and expression pattern of the mouse ortholog of the human gene. 15555553_This paper suggests that the mouse ortholog of this human gene plays a role in bone formation and regeneration. 16385451_Observational study of gene-disease association. (HuGE Navigator) 22836805_KAZALD1 gene was hypermethylated in sarcomatoid-type malignant peritoneal mesothelioma. 24002581_KAZALD1 promotes glioma malignant progression through invasion and proliferation 26315110_KAZALD1 hypomethylation indicates bad prognosis in small bowel adenocarcinoma | ENSMUSG00000025213 | Kazald1 | 116.47437 | 0.8608156 | -0.2162238304 | 0.26991795 | 6.385450e-01 | 4.242382e-01 | 7.764125e-01 | No | Yes | 120.183376 | 16.069986 | 136.773927 | 17.575169 | |
| ENSG00000108239 | 23232 | TBC1D12 | protein_coding | O60347 | FUNCTION: RAB11A-binding protein that plays a role in neurite outgrowth. {ECO:0000250|UniProtKB:M0R7T9}. | Acetylation;Coiled coil;Endosome;GTPase activation;Phosphoprotein;Reference proteome | hsa:23232; | autophagosome [GO:0005776]; recycling endosome [GO:0055037]; GTPase activator activity [GO:0005096]; activation of GTPase activity [GO:0090630]; intracellular protein transport [GO:0006886]; regulation of autophagosome assembly [GO:2000785] | ENSMUSG00000048720 | Tbc1d12 | 519.53265 | 1.1018889 | 0.1399787505 | 0.14407865 | 9.461911e-01 | 3.306907e-01 | 7.079883e-01 | No | Yes | 742.258870 | 126.307353 | 636.912673 | 106.162104 | |||
| ENSG00000108474 | 9487 | PIGL | protein_coding | Q9Y2B2 | FUNCTION: Involved in the second step of GPI biosynthesis. De-N-acetylation of N-acetylglucosaminyl-phosphatidylinositol. | Alternative splicing;Deafness;Disease variant;Endoplasmic reticulum;GPI-anchor biosynthesis;Hydrolase;Ichthyosis;Lipid metabolism;Membrane;Mental retardation;Reference proteome;Transmembrane;Transmembrane helix | PATHWAY: Glycolipid biosynthesis; glycosylphosphatidylinositol-anchor biosynthesis. | This gene encodes an enzyme that catalyzes the second step of glycosylphosphatidylinositol (GPI) biosynthesis, which is the de-N-acetylation of N-acetylglucosaminylphosphatidylinositol (GlcNAc-PI). Study of a similar rat enzyme suggests that this protein localizes to the endoplasmic reticulum. [provided by RefSeq, Jul 2008]. | hsa:9487; | endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides [GO:0016811]; N-acetylglucosaminylphosphatidylinositol deacetylase activity [GO:0000225]; GPI anchor biosynthetic process [GO:0006506]; preassembly of GPI anchor in ER membrane [GO:0016254] | 22444671_Whole-exome sequencing on five previously reported CHIME cases identified PIGL (N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase) as required for glycosylphosphatidylinositol anchor formation. 25706356_Mutations in PIGL in a patient with Mabry syndrome. 28327575_c.336-2A>G variant in PIGL associated with developmental disorder resulted in exon skipping. 28371479_these findings indicate that patients with a clinical diagnosis of CHIME syndrome and a single identifiable mutation in PIGL warrant further investigation for copynumber changes involving | ENSMUSG00000014245 | Pigl | 548.52573 | 1.0464141 | 0.0654538781 | 0.13617881 | 2.314832e-01 | 6.304263e-01 | 8.816466e-01 | No | Yes | 629.920485 | 72.533175 | 598.723147 | 67.334266 |
| ENSG00000108509 | 23125 | CAMTA2 | protein_coding | O94983 | FUNCTION: Transcription activator. May act as tumor suppressor. {ECO:0000269|PubMed:11925432}. | ANK repeat;Activator;Alternative splicing;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | The protein encoded by this gene is a member of the calmodulin-binding transcription activator protein family. Members of this family share a common domain structure that consists of a transcription activation domain, a DNA-binding domain, and a calmodulin-binding domain. The encoded protein may be a transcriptional coactivator of genes involved in cardiac growth. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Jan 2010]. | hsa:23125; | chromatin [GO:0000785]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; double-stranded DNA binding [GO:0003690]; histone deacetylase binding [GO:0042826]; sequence-specific DNA binding [GO:0043565]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; cardiac muscle hypertrophy in response to stress [GO:0014898]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357] | 29110692_Mutation of CAMTA2 resulting in post-transcriptional inhibition of its own gene activity likely underlies a novel syndromic tremulous dystonia. | ENSMUSG00000040712 | Camta2 | 783.25715 | 0.8592805 | -0.2187989882 | 0.12124264 | 3.236039e+00 | 7.203458e-02 | 3.946806e-01 | No | Yes | 715.849693 | 125.587648 | 830.522230 | 142.274053 | |
| ENSG00000109099 | 5376 | PMP22 | protein_coding | Q01453 | FUNCTION: Might be involved in growth regulation, and in myelinization in the peripheral nervous system. | Cell membrane;Charcot-Marie-Tooth disease;Deafness;Dejerine-Sottas syndrome;Disease variant;Glycoprotein;Membrane;Neurodegeneration;Neuropathy;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes an integral membrane protein that is a major component of myelin in the peripheral nervous system. Studies suggest two alternately used promoters drive tissue-specific expression. Various mutations of this gene are causes of Charcot-Marie-Tooth disease Type IA, Dejerine-Sottas syndrome, and hereditary neuropathy with liability to pressure palsies. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:5376; | bicellular tight junction [GO:0005923]; compact myelin [GO:0043218]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; synapse [GO:0045202]; bleb assembly [GO:0032060]; cell death [GO:0008219]; cell differentiation [GO:0030154]; chemical synaptic transmission [GO:0007268]; myelin assembly [GO:0032288]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of neuron projection development [GO:0010977]; peripheral nervous system development [GO:0007422] | 11456309_both functions of PMP22, in regulating Schwann cell differentiation and contributing to peripheral myelin compaction, are affected by its overexpression. 11545686_Observational study of genetic testing. (HuGE Navigator) 11835375_Observational study of genotype prevalence. (HuGE Navigator) 12207933_deletion spanning the 3' region of the PMP22 gene, causing only a partial deletion of one copy of the gene. 12211648_SSCP analysis of this gene in Croatian patients 12823620_Conformation of the transmembrane domains in peripheral myelin protein 22 is studied using solution-phase synthesis. 14581692_Sequence deletion in the PMP22 gene was noted in a case of hereditary neuropathy with liability to pressure palsy complicated by hypoglossal neuropathy 15050444_Observational study of gene-disease association. (HuGE Navigator) 15083696_distinct mutations in PMP22 gene is found in brachial palsy. 15099590_we report a family carrying a novel mutation in the PMP22 gene (c. 327C>A), which results in a premature stop codon (Cys109stop). The family members who carry this mutation have a Charcot-Marie-Tooth type 1 variable phenotype. 15099592_we report a large family showing characteristic phenotypes of Charcot-Marie-Tooth type 1A along with deafness in an autosomal dominant fashion. We detected a sequence variation (c.68C>G) leading to a T23R missense mutation in PMP22. 15205993_a PMP22 mutation (Ser22Phe) may play a role in hereditary neuropathy 15555916_PMP22 and P0 are involved in both trans-homophilic and trans-heterophilic interactions. Disease-related point mutations of P0 resulted in a decreased adhesion capability correlating with the severity of the respective disease phenotype. 15564036_We report the clinical and electrophysiological findings observed in a kindred with three members affected by HNPP due to a deletion containing exons 4 and 5 of the PMP22 gene. 15755691_At postnatal day 4 Pmp22 transgenic mice show strongly reduced expression of genes important for cholesterol synthesis. 15891642_The differences in sensorineural hearing impairment may be explained by the differences in PMP22 expression. 15955700_A family with a novel frameshift mutation in the PMP22 gene (c.433_434insC) causing a phenotype of hereditary neuropathy. 16199442_point mutation in the PMP22 gene is a rare cause of hereditary neuropathy. 16215943_The PMP22 duplication rate in Chinese patients with CMT is 31.9%(36/113). PMP22 deletion is the common cause of hereditary neuropathy with liability to pressure palsies. 16326107_As a result of missorting and inefficient proteasomal degradation, the aggregation of PMP22 and recruitment of autophagosomes and lysosomes are key factors in the subcellular pathogenesis of CMT1A neuropathies. 16436605_PMP22 forms a complex with alpha6beta4 integrin in cultured colonic adenocarcinoma cells. 16437560_T118M PMP22 mutation causes partial loss of function and hereditary neuropathy with liability to pressure palsies. 16463004_Observational study of genetic testing. (HuGE Navigator) 16481890_CMT represents a heterogeneous group of disorders at the molecular level. Testing for the CMT1A duplication (i.e., duplication of PMP22) alone yields an accurate molecular diagnosis in approximately half of all patients. 16481890_Observational study of genotype prevalence. (HuGE Navigator) 16683188_determined the N-terminal part of Siva as the binding region for CD27; the peroxisomal membrane protein PMP22 is a new interaction partner of Siva and may be involved in the host response against CVB3. 16912585_Observational study of genotype prevalence. (HuGE Navigator) 17620487_The phenotypic expression is identical in patients with Leu7fs mutation and patients with HNPP caused by chromosome 17p11.2 deletion. Reduction of PMP22 is sufficient to cause the full HNPP phenotype. 17707409_This study investigated a 17-years-old girl who led to our detecting a novel mutation in PMP22 gene. The mutation was also detected in her father and corresponded to a deletion of one tymidine at position 11 in exon2 (c.11delT). 17824619_Results show PMP22 is highly helical, shows evidence of stable tertiary structure, and exhibits a strong tendency to dimerize. 17917930_Observational study of gene-disease association. (HuGE Navigator) 17917930_The rate of duplication was 92.3' (36/39) in the patients whereas it was zero in the control samples. 18353535_Observational study of genetic testing. (HuGE Navigator) 18438698_Our findings suggest that in a subgroup of CMT1A patients there is an increase in clinical severity over generations. The mechanism responsible for this observation remains unknown. Our findings should be validated on a larger cohort of CMT1A families. 18592125_study reports a novel point mutation in a family affected with Roussy-Levy syndrome 18642376_novel 227delG mutation of PMP22 gives a mild form of hereditary neuropathy with liability to pressure palsy with atypical clinical and electrophysiological findings. 18698610_Deletions in both PMP22 alleles caused Charcot-Marie-Tooth disease with a Dejerine-Sottas disease phenotype. 18795802_The results lead to the hypothesis that ER quality control recognizes the G150D and L16P mutant forms of PMP22 as defective through mechanisms closely related to their conformational instability and/or slow folding. 19259128_Data show that the CMT1Adup, GJB1, MPZ and PMP22 mutation frequencies were in the range of those described in other CMT patient collectives with different ethnical backgrounds. 19259128_Observational study of gene-disease association. (HuGE Navigator) 19290556_Data show that Med25 is coordinately expressed with Pmp22 gene dosage and expression in transgenic mice and rats, and suggest a potential role of this protein in the molecular etiology of Charcot-Marie-Tooth disease. 19447823_The extra copy of PMP22 in Charcot-Marie-Tooth 1A results in disruption of the tightly regulated expression of PMP22, thus, variability of PMP22 levels, rather than absolute level of PMP22, may play an important role in the pathogenesis of CMT1A. 19493167_Loss of motoneurons (MNs) in spinal cords from transgenic mice over-expressing Pmp22 (Pmp22(tg)) is demonstrated, while mice lacking Pmp22 [Pmp22(ko) knockouts] exhibit normal MN numbers at the age (60 days) of Charcot-Marie-Tooth disease symptoms. 19705173_PMP22 has a role in determining phenotype of Charcot-Marie-Tooth disease [case report] 19830275_study characterized a patient manifesting with an atypical form of hereditary neuropathy with liability to pressure palsies caused by a new Thr99fsX110 mutation in the PMP22 gene 19888301_Copy number variation upstream of PMP22 is associated with Charcot-Marie-Tooth disease. 19909487_the type of PMP22 point mutations, the nature of the amino acid change, and the position of the altered amino acid play a role in determining the severity of the phenotype 19913121_Observational study of gene-disease association. (HuGE Navigator) 19930872_PMP22 gene duplications and deletions are associated with Charcot-Marie-Tooth disease type 1A and hereditary neuro pathy with liability to pressure palsies. 20093502_The role of monocyte chemoattractant protein-1 (MCP-1/CCL2) as a regulator of nerve macrophages and neural damage including axonopathy and demyelination, was investigated. 20453308_Serine112Arginine mutation in the PMP22 gene may have a causative role in early-onset Charcot-Marie-Tooth disease with hearing impairment. 20538960_Observational study of gene-disease association. (HuGE Navigator) 20571287_The most frequent Charcot-Marie-Tooth disease mutation worldwide, the PMP22 duplication, is also the most frequent CMT mutation in the Cypriot population. Five out of the 8 other mutations are novel, not reported in other populations. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20842290_Data conclude that the 5' regulatory sequence of the PMP22 gene is conserved at the nucleotiode level, however rarely occurring SNPs variant in the PMP22 regulatory sequence may be associated with the gene dosage effect. 20842290_Observational study of gene-disease association. (HuGE Navigator) 21159173_PMP22 gene expression is a novel independent prognostic factor for disease-free survival and overall survival for breast cancer patients. Including it into a model with established prognostic factors will increase the accuracy of prognosis. 21194947_Variable phenotypes of Charcot-Marie-Tooth disease are associated with PMP22 missense mutations. 21252112_micromutations of PMP22 cause a clinical and pathological continuum of demyelinating neuropathies that may include atypical phenotypes. 21337347_the novel R159C mutation represents a very rare case of a dominant PMP22 mutation causing an axonal neuropathy 21429232_The lower prevalence of HNPP in our study of Finnish conscripts compared to the general population is probably due to pre-service screening. 21518455_A physiologic role for PMP22 on the expression of alpha6 integrin. 21590514_[review] Pes cavus is an early and age-dependent manifestation of CMT1A duplication. 21670407_PMP22 is important for the normal function of neurons that express it during early development, such as cranial motor neurons and spinal sensory neurons. PMP22 deficiency differentially affects myelination between motor and sensory nerves. 21692910_More than three-fourths of the patients with Gly94fsX222 mutation demonstrated a CMT1 phenotype combined with transient deficits 21827951_Structural basis for the Trembler-J phenotype of Charcot-Marie-Tooth disease, the PMP22 Leu16Pro mutation profoundly disrupts the first transmembrane segment destabilizing the protein 22000434_gene causing Charcot-Marie-Tooth disease (CMT) in the family is found in the 17p11.2-p12 region containing PMP22 gene duplication mutation, resulting in the subtype CMT1A 22006697_Four novel point mutations in PMP22 with two different phenotypes: mutation p.Ser79Thr is found in the Dejerine-Sottas neuropathy phenotype. 22131320_PMP22 mutation has a role in Charcot-Marie-Tooth disease; the spectrum and frequency of PMP22 mutations in the Slovak population is comparable to that seen in the global population 22180461_A subset of EGR2 and SOX10 consensus sequences are essential for enhancer activity of the PMP22 gene. 22243284_our results suggest a low relative CMT1A duplication frequency in the Greek population 22765307_In Greek Charcot-Marie-Tooth type 1 patients, 3 pathogenic mutations were found (3.5%); two recently reported micromutations in PMP22 3, and one known point mutation in EGR2 23224996_This review shows that alterations of PM22 levels by mutations of the PM22 gene are responsible for > 50% of all inherited peripheral neuropathies. 23243264_altered PMP22 gene expression induces significant central nervous system alterations in patients with hereditary neuropathy with liability to pressure palsies and Charcot-Marie-Tooth 1A 23279344_We describe a novel heterozygous p.Trp39Cys missense mutation in the extracellular domain of the peripheral myelin protein 22 (PMP22) associated with an early-onset demyelinating Charcot-Marie-Tooth type 1 E 23468528_a novel MDA-7/IL-24-GAS3-beta1integrin-fibronectin signaling pathway that suppresses breast cancer growth, is reported. 23635862_molecular genetic analysis of patients with Bell's palsy failed to demonstrate the presence of the PMP22 gene deletion on chromosome 17q11.2-12 23639031_Wild type PMP22 possesses minimal conformational stability in micelles, which parallels its poor folding efficiency in the endoplasmic reticulum. 23689413_Two Brazilian siblings with proptosis were found to have a point mutation in the PMP22 gene. 23743332_This study demonistrated that PMP22 duplication was the most common genetic cause Charcot-Marie-Tooth disease. 24078732_This study demonistrated that pmp22 gene duplication is the most frequent in patient with Charcot-Marie-Tooth disease in spain. 24321297_G3BP1 regulation of cell proliferation in breast cancer cells, may occur via a regulatory effect on PMP22 expression. 24646194_Diagnosis is confirmed by finding respectively a PMP22 duplication, deletion or point mutation 24812204_ascorbic acid does not impact on PMP22 transcriptional regulation and PMP22 is not a suitable biomarker for CMT1A 24819634_2 of 38 CMT2 patients showed rearrangements in the PMP22 gene, which is commonly associated with CMT1 or HNPP phenotypes thus usually not tested in CMT2 patients. 24830919_The results of this study revealed distinct patterns of SNP allele frequency distribution, which reliably differentiated CMT1A patients from controls 25030070_Duplication of PMP22 was detected in 79 of 157 Mexican Charcot-Marie-Tooth Type 1A disease patients. In Mexican patients CMT1A frequency was similar to other populations such as United States, Australia, Finland, Sweden and Spain. 25243937_Data suggest that several peripheral myelin protein 22 (PMP22) mutations known to result in neuropathy act by disrupting transmembrane helix packing interactions. 25385046_Charcot-Marie-Tooth disease-related PMP22 is trapped in the endoplasmic reticulum by calnexin-dependent retention and Rer1-mediated early Golgi retrieval systems and partly degraded by the Hrd1-mediated endoplasmic reticulum-associated degradation system. 25522693_The SNP array has wide coverage, high sensitivity, and high resolution and can be used as a screening tool to detect PMP22 dup/del as shown in this Chinese Han population. 26076881_PMP22 is a transmembrane glycoprotein component of myelin, important for myelin functioning. Mutation of PMP22 gene cause Charcot-Marie-Tooth Disease. 26102530_This finding provides compelling evidence that the effects of these mutations on the energetics of PMP22 folding lie at the heart of the molecular basis of Charcot-Marie-Tooth disease. 26110377_The common 17p deletion accounts for approximately 50% and PMP22 micromutations for approximately 2% of cases in a large consecutive cohort of Greek patients with suspected HNPP. 26154129_Osteosarcoma metastasis-related gene PMP22 participates in the proliferation, invasion, migration and colony formation of osteosarcoma cells possibly via the MAPK signal transduction pathway 26378787_Exome sequencing identified MFN2 SNVs in two of the individuals. Neuropathy-associated CNV outside of the PMP22 locus is rare in Charcot-Marie-Tooth (CMT) disease . Nevertheless, there is potential clinical utility in testing for CNVs and exome sequencing in CMT cases negative for the CMT1A duplication. 26486801_our data suggest that an alteration of mRNA processing could be a pathogenic mechanism in CMT1A. 26544804_PMP22 Gene Duplication is associated with Charcot-Marie-Tooth disease type 1A. 26629937_PMP22 gene knockdown inhibited progression of Chronic Myeloid Leukemia. 26859249_Data (including data from studies using recombinant proteins that lack typical in-vivo post-translational modifications such as palmitoylation) suggest PMP22 exhibits little tendency to partition into liquid-ordered domains of unilamellar vesicles. 26921370_These results suggest that the severe congenital hypomyelinating neuropathy that characterizes Tr(J)mice results in structural and functional deficits of the developing Neuromuscular Junction. 26977628_DNA diagnosis was performed in 5 families with hereditary neuropathy with liability to pressure palsies - the PMP22 deletion was found in 9 patients. 27279240_The studies implicating GAS3 protein family (EMP1, EMP2, EMP3 and PMP22) in cancer pathogenesis as well as probe the structural similarities between the family members were highlighted. 27288457_We discovered that Tead1 and co-activators Yap and Taz are required for Pmp22 expression, as well as for the expression of Egr2 Tead1 directly binds Pmp22 and Egr2 enhancers early in development and Tead1 binding is induced during myelination, correlating with Pmp22 expression. The data identify Tead1 as a novel regulator of Pmp22 expression during development in concert with Sox10 and Egr2 27386852_we report molecular and clinical characterizations of six subjects with the reciprocal phenomenon of deletions spanning both genes, i.e., PMP22-RAI1 deletions. Systematic clinical studies revealed features consistent with SMS, including features of intellectual disability, speech and gross motor delays, behavioral problems and ocular abnormalities. 27422849_This study demonstrated We show that blink reflex studies are reliable for identification of inherited demyelinating polyneuropathy (with pmp22 mutation) regardless of severity and can facilitate algorithmic decisions in genetic testing. 27577214_Data suggest that the father has carried the same duplication of the peripheral myelin protein 22 (PMP22) gene but with no detectable symptom may be due to irregular transmission pattern of the mutation. 27623071_PMP22 polymorphism is associated with tuberculosis. 27749933_PMP22 deletion leads to functional, metabolic and macro-structural alterations in the afferent visual system of hereditary neuropathy with liability to pressure palsies patients. 27862672_In this Chinese Han population, the frequency of PMP22 gene duplication in those with CMT1 was slightly (50% vs. 70%-80%) less than in Western/Caucasian populations. 28234890_Findings suggest that miR-200bc/429 inhibit OS cells proliferation and invasion by targeting PMP22, and function as a tumor suppressor. 28336807_We identified that PMP22 not only acts as a marker for gastric CSCs but may also have an essential role in regulating the self-renewal and chemoresistance of gastric cancer. Our findings suggest that PMP22 has clinical value for the prognosis and treatment of chemoresistant gastric cancer 28374912_A Computational Approach to Identify a Potential Alternative Drug With Its Positive Impact Toward PMP22. 28748849_This study supported the notion that missense mutations in PMP22 give rise to a Charcot-Marie-Tooth Disease phenotype, possibly through a toxic gain-of-function mechanism. 29127354_This study delineates the clinical and molecular features of PMP22 point mutations in Taiwan, and emphasizes their roles in demyelinating CMT or HNPP-like neuropathy. 29246495_CMT1A patient-derived cultures contain approximately 1.5-fold elevated levels of PMP22 mRNA, exhibit reduced mitotic potential, and display intracellular protein aggregates as compared to cells from unaffected individuals. 29544507_These data suggest that PMP22 Del HNPP might not be uncommon at least in the Korean population. 29771329_Our data show a significant decrease in Pmp22 transcript expression using allele-specific internal controls. Moreover, the P2 promoter of the Pmp22 gene, which is used in other cell types, is affected, but we find that the Schwann cell-specific P1 promoter is disproportionately more sensitive to loss of the super-enhancer. T 31213528_Heterologous PMP22 expression increases the amplitude of currents similar to those ascribed to store-operated calcium (SOC) channels, particularly those involving transient receptor canonical channel 1 (TrpC1). These channels help replenish Ca(2+) in the endoplasmic reticulum (ER) following stimulus-induced depletion. 31372974_Genetic spectrum and clinical profiles in a southeast Chinese cohort of Charcot-Marie-Tooth disease. 31586623_Regulating PMP22 expression as a dosage sensitive neuropathy gene. 31688096_A case report of trisomy 17 mosaicism: PMP22 gene duplication as a result of trisomy 17 associated with Charcot-Marie-Tooth disease. 31993930_PMP22 Gene-Associated Neuropathies: Phenotypic Spectrum in a Cohort from India. 32356557_Pmp22 super-enhancer deletion causes tomacula formation and conduction block in peripheral nerves. 32506583_Diagnostic yield of targeted sequential and massive panel approaches for inherited neuropathies. 32513719_Peripheral myelin protein 22 preferentially partitions into ordered phase membrane domains. 32642785_Yield of the PMP22 deletion analysis in patients with compression neuropathies. 32647009_Direct relationship between increased expression and mistrafficking of the Charcot-Marie-Tooth-associated protein PMP22. 32657593_Molecular Diagnosis of Hereditary Neuropathies by Whole Exome Sequencing and Expanding the Phenotype Spectrum. 32693030_Treating PMP22 gene duplication-related Charcot-Marie-Tooth disease: the past, the present and the future. 33675896_The miR-139-5p/peripheral myelin protein 22 axis modulates TGF-beta-induced hepatic stellate cell activation and CCl4-induced hepatic fibrosis in mice. 33893233_Ion mobility-mass spectrometry reveals the role of peripheral myelin protein dimers in peripheral neuropathy. 33933451_Glycosylation limits forward trafficking of the tetraspan membrane protein PMP22. 34205075_Emerging Therapies for Charcot-Marie-Tooth Inherited Neuropathies. 34332267_Characterization of a Portuguese family with Charcot-Marie-Tooth disease type 1E due to a novel point mutation in the PMP22 gene. 34421356_Inhibition of protein PMP22 enhances etoposide-induced cell apoptosis by p53 signaling pathway in Gastric Cancer. 34996390_Coexistence of Charcot-Marie-Tooth 1A and nondystrophic myotonia due to PMP22 duplication and SCN4A pathogenic variants: a case report. | ENSMUSG00000018217 | Pmp22 | 422.33164 | 0.8893711 | -0.1691425596 | 0.15062451 | 1.257217e+00 | 2.621786e-01 | 6.528100e-01 | No | Yes | 494.202536 | 38.247593 | 533.732547 | 39.982673 | |
| ENSG00000109184 | 23142 | DCUN1D4 | protein_coding | Q92564 | FUNCTION: Contributes to the neddylation of all cullins by transferring NEDD8 from N-terminally acetylated NEDD8-conjugating E2s enzyme to different cullin C-terminal domain-RBX complexes which are necessary for the activation of cullin-RING E3 ubiquitin ligases (CRLs). {ECO:0000269|PubMed:23201271, ECO:0000269|PubMed:26906416}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;Reference proteome;Ubl conjugation | hsa:23142; | nucleus [GO:0005634]; ubiquitin ligase complex [GO:0000151]; cullin family protein binding [GO:0097602]; ubiquitin conjugating enzyme binding [GO:0031624]; ubiquitin-like protein binding [GO:0032182]; positive regulation of protein neddylation [GO:2000436]; positive regulation of ubiquitin-protein transferase activity [GO:0051443]; protein neddylation [GO:0045116] | ENSMUSG00000051674 | Dcun1d4 | 1677.37922 | 1.1057911 | 0.1450788968 | 0.10981565 | 1.750955e+00 | 1.857567e-01 | 5.793009e-01 | No | Yes | 2241.327163 | 353.929535 | 1943.941670 | 300.491965 | |||
| ENSG00000109323 | 4126 | MANBA | protein_coding | O00462 | FUNCTION: Exoglycosidase that cleaves the single beta-linked mannose residue from the non-reducing end of all N-linked glycoprotein oligosaccharides. {ECO:0000305|PubMed:12890191, ECO:0000305|PubMed:30552791}. | Disulfide bond;Glycoprotein;Glycosidase;Hydrolase;Lysosome;Reference proteome;Signal | PATHWAY: Glycan metabolism; N-glycan degradation. {ECO:0000269|PubMed:12890191, ECO:0000269|PubMed:30552791}. | This gene encodes a member of the glycosyl hydrolase 2 family. The encoded protein localizes to the lysosome where it is the final exoglycosidase in the pathway for N-linked glycoprotein oligosaccharide catabolism. Mutations in this gene are associated with beta-mannosidosis, a lysosomal storage disease that has a wide spectrum of neurological involvement. [provided by RefSeq, Jul 2008]. | hsa:4126; | azurophil granule membrane [GO:0035577]; lysosomal lumen [GO:0043202]; plasma membrane [GO:0005886]; beta-mannosidase activity [GO:0004567]; mannose binding [GO:0005537]; cellular protein modification process [GO:0006464]; glycoprotein catabolic process [GO:0006516]; oligosaccharide catabolic process [GO:0009313] | 17899454_Observational study of gene-disease association. (HuGE Navigator) 17899454_The MANBA genotypes for a polymorphic CA repeat were related to colorectal cancer risk in a Swedish population but not a Chinese one. In the Swedish population, individuals with < 22 CAs/< 22 CAs had a significantly increased risk for CRC. 19728872_The present analysis of the c.1922G>A MANBA mutation underlines the lack of genotype-phenotype correlation in beta-mannosidosis 19773279_Observational study of gene-disease association. (HuGE Navigator) 20332099_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 30528300_independent putative primary functional variants in NFKB1/MANBA and showed the distinct molecular mechanism by which each putative primary functional variant conferred susceptibility to Primary biliary cholangitis, were identified. 33441424_Kidney disease genetic risk variants alter lysosomal beta-mannosidase (MANBA) expression and disease severity. | ENSMUSG00000028164 | Manba | 538.88000 | 1.0300103 | 0.0426586978 | 0.14434824 | 8.735503e-02 | 7.675670e-01 | 9.336309e-01 | No | Yes | 507.716887 | 64.391646 | 470.255854 | 58.378729 |
| ENSG00000109501 | 7466 | WFS1 | protein_coding | O76024 | FUNCTION: Participates in the regulation of cellular Ca(2+) homeostasis, at least partly, by modulating the filling state of the endoplasmic reticulum Ca(2+) store (PubMed:16989814). Negatively regulates the ER stress response and positively regulates the stability of V-ATPase subunits ATP6V1A and ATP1B1 by preventing their degradation through an unknown proteasome-independent mechanism (PubMed:23035048). {ECO:0000269|PubMed:16989814, ECO:0000269|PubMed:23035048}. | Acetylation;Cataract;Cytoplasmic vesicle;Deafness;Diabetes insipidus;Diabetes mellitus;Disease variant;Endoplasmic reticulum;Glycoprotein;Membrane;Non-syndromic deafness;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | This gene encodes a transmembrane protein, which is located primarily in the endoplasmic reticulum and ubiquitously expressed with highest levels in brain, pancreas, heart, and insulinoma beta-cell lines. Mutations in this gene are associated with Wolfram syndrome, also called DIDMOAD (Diabetes Insipidus, Diabetes Mellitus, Optic Atrophy, and Deafness), an autosomal recessive disorder. The disease affects the brain and central nervous system. Mutations in this gene can also cause autosomal dominant deafness 6 (DFNA6), also known as DFNA14 or DFNA38. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:7466; | dendrite [GO:0030425]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum lumen [GO:0005788]; endoplasmic reticulum membrane [GO:0005789]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of synaptic vesicle membrane [GO:0030285]; secretory granule [GO:0030141]; ATPase binding [GO:0051117]; calcium-dependent protein binding [GO:0048306]; calmodulin binding [GO:0005516]; DNA-binding transcription factor binding [GO:0140297]; proteasome binding [GO:0070628]; ubiquitin protein ligase binding [GO:0031625]; calcium ion homeostasis [GO:0055074]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; endoplasmic reticulum unfolded protein response [GO:0030968]; ER overload response [GO:0006983]; glucose homeostasis [GO:0042593]; kidney development [GO:0001822]; negative regulation of apoptotic process [GO:0043066]; negative regulation of ATF6-mediated unfolded protein response [GO:1903892]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902236]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of programmed cell death [GO:0043069]; negative regulation of response to endoplasmic reticulum stress [GO:1903573]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of type B pancreatic cell apoptotic process [GO:2000675]; nervous system process [GO:0050877]; olfactory behavior [GO:0042048]; pancreas development [GO:0031016]; positive regulation of calcium ion transport [GO:0051928]; positive regulation of growth [GO:0045927]; positive regulation of protein metabolic process [GO:0051247]; positive regulation of protein ubiquitination [GO:0031398]; protein maturation by protein folding [GO:0022417]; protein stabilization [GO:0050821]; renal water homeostasis [GO:0003091]; response to endoplasmic reticulum stress [GO:0034976]; sensory perception of sound [GO:0007605]; ubiquitin-dependent ERAD pathway [GO:0030433]; visual perception [GO:0007601] | 11709537_Five different heterozygous missense mutations (T699M, A716T, V779M, L829P, G831D) in the WFS1 gene found in six low frequency sensorineural hearing loss (LFSNHL) families. 11709538_The causal relationship between WFS1 missense mutation and deafness was supported by two observations based on haplotype and mutation analysis of the kindred. 11916957_Observational study of gene-disease association. (HuGE Navigator) 11920861_Did not find evidence of an increased incidence of WFS carriers in the suicide panel and concluded that WFS1 carrier status is not a significant contributor to suicide in the general population. 12073007_Mutations in the WFS1 gene that cause low-frequency sensorineural hearing loss are small non-inactivating mutations. 12107816_Four new polymorphisms associated with Wolfram syndrome. Not all patients have full syndrome. 12605098_WFS1 is not a major susceptibility gene for the development of psychiatric disorders in subjects with Wolfram syndrome. 12650912_In all affected family members analysed, we detected a missense mutation in WFS1 (K705N) and therefore confirm the finding that the majority of mutations responsible for LFSNHI are missense mutations which localise to the C-terminal domain of the protein. 12707947_found a significantly higher frequency of the 611R/611R genotype in suicide completers. Scores of impulsivity and novelty seeking were higher in subjects with the associated genotype, suggesting a role for WFS1 in the pathophysiology of impulsive suicide 12754709_mutational analysis of the WFS1 coding region in 19 Italian Wolfram syndrome patients and 25 relatives, using a DHPLC-based protocol 12782971_This study does not support the involvement of tyrosine hydroxylase, catechol-O-methyl transferase and Wolfram syndrome 1 polymorphisms in mood disorders. 12913071_Here we investigate, for the first time, the molecular mechanisms that cause loss-of-function of wolframin in affected individuals. 12955714_Overview of the spectrum of WFS1 mutations in Wolfram syndrome, nonsyndromic low frequency sensorineural hearing impairment,diabetes mellitus, and psychiatric disease. 14527944_Wolframin has a role in the regulation of intracellular Ca2+ homeostasis 14968315_In this study we analyzed the phenotype of a large Hungarian family with LFSNHI and linkage to DFNA6. The family contains 14 affected persons. 15234338_Observational study of gene-disease association. (HuGE Navigator) 15277431_most causative changes identified in the WFS1 gene occurred in exon 8, and only one was identified outside this region in exon 4 in patients with Wolfram syndrome 15473915_Observational study of gene-disease association. (HuGE Navigator) 15473915_These results support the hypothesis that the WFS1 gene is involved in the genetic predisposition for mood disorders. 15852062_The relative risk of psychiatric hospitalization for depression was estimated to be 7.1 (95% CI 1.9-26.6) for carriers of a single wolframin mutation compared to noncarriers. 15912360_Mutations in one single gene, Wolfram syndrome 1 (WFS1), have been reported to account for most familial cases with low-frequency hearing loss. 16005363_A nine nucleotide insertional mutation in two members of a family with Wolfram syndrome. 16043233_This study presents a six-generation family from Hungary with nonsyndromic, post-lingual, bilateral, symmetric, progressive LFSNHI, that discloses positive linkage to the DFNA6 region. 16151413_Molecular analysis of WFS1 in seven families with Wolfram syndrome identified eight different mutations; one was a de novo mutation occurring independently in 2 families, whereas the remaining ones were inherited. 16195229_the pathogenesis of Wolfram syndrome involves chronic ER stress in pancreatic beta-cells caused by the loss of function of WFS1 16408729_Mutations in WFS1 are one cause of non-syndromic low frequency sensorineural hearing loss. 16550584_Missense mutations within a defined region are associated with dominant low-frequency hearing loss (DFNA6/14/38), while more severe mutations spanning WFS1 are found in Wolfram syndrome patients. 16806192_WFS1 mutations lead to drastically reduced steady-state levels of wolframin. 16876316_Observational study of gene-disease association. (HuGE Navigator) 16965966_WFS1 minimal promoter contains two DNA binding motifs (GC boxes) for the transcription factors Sp1/3/4 and binding of both Sp1 and Sp3 was demonstrated at both motifs in vitro and in vivo. 16989814_WFS1 protein participates in the regulation of cellular Ca(2+) homeostasis, at least partly, by modulating the filling state of the ER Ca(2+) store 17492394_one-third (3 out of 9) autosomal dominant low frequency sensorineural hearing loss(LFSNHL) families had mutations in WFS1, indicating that in non-syndromic hearing loss WFS1 is restrictively & commonly found within autosomal dominant LFSNHL families 17517145_a novel heterozygous missense mutation in exon 8 of WFS1 (i.e., Y669H) which is likely responsible for the low-frequency sensorineural hearing loss (LFSNHL) phenotype in a Taiwanese family was discovered 17568405_Results reported eight novel WFS1 mutations in Wolfram syndrome. 17603484_In a pooled analysis comprising 9,533 cases and 11,389 controls, SNPs in WFS1 were strongly associated with common type 2 diabetes risk. 17603484_Observational study of gene-disease association. (HuGE Navigator) 17719176_Observational study of gene-disease association. (HuGE Navigator) 17719176_The wolframin His611Arg polymorphism influences medication overuse headache. 17947299_Study identifies an interaction between Wolframin and Na+/K+ ATPase beta1 subunit in transfected Cos7 cells, and between endogenous proteins in placental, neuroblastoma and MIN6 pancreatic beta-cell lines. 18040659_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18040659_Replication of the previously reported associations between SNPs at this locus and the risk of type 2 diabetes. 18060660_Genome-wide association datase revealed that a strong linkage disequilibrium with the three WFS1 single nucleotide polymorphisms was associated with type 2 diabetes. 18060660_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18197395_May be a candidate gene for type 2 diabetes. 18426861_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18518985_a novel heterozygous missense mutation in exon 8 of WFS1 predicting a p.R685P amino acid substitution that is likely to underlie the low frequency sensorineural hearing loss phenotype in the American family 18544103_maternally inherited combination of diabetes mellitus and hearing impairment in three members of a family was found to be associated with autosomal dominant transmission of the E864K mutation of the WFS1 gene 18566338_The WFS1 gene is located on the short arm of chromosome 4 in Wolfram syndrome. 18568334_Observational study of gene-disease association. (HuGE Navigator) 18568334_Type 2 diabetes-associated risk alleles of WFS1 are associated with estimates of a decreased pancreatic beta cell function among middle-aged individuals with abnormal glucose regulation 18591388_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18597214_Observational study of gene-disease association. (HuGE Navigator) 18660851_This report of two novel WFS1 mutations expands the molecular spectrum of Wolfram syndrome. 18688868_A novel missense mutation in WFS1 was identified which caused Wolfram syndrome and may also be linked to autoimmune diseases. 18694974_Study show that polymorphisms in WFS1 were associated with type 2 diabetes risk in the studied population. 18806274_The WFS1 locus strongly contributes to juvenile-onset diabetes in Lebanon in Wolfram syndrome and non-syndromic non-autoimmune diabetes mellitus detected by linkage analysis; effect varies by allele. 18853134_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18984664_Observational study of gene-disease association. (HuGE Navigator) 18991055_Observational study of gene-disease association. (HuGE Navigator) 19020324_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19042979_WFS1 is the major gene involved in Wolfram syndrome in Brazilian patients and most mutations are concentrated in exon 8 19082521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19115052_Observational study of gene-disease association. (HuGE Navigator) 19139842_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19258404_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19258739_found no evidence for a substantial effect of WFS1 polymorphisms on risk of type 2 diabetes or clinical characteristics of diabetic subjects in Japanese population 19279076_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19324937_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19328217_Observational study of gene-disease association. (HuGE Navigator) 19328217_The H611R polymorphism of Wolframin gene was associated with mood disorders but not suicidal behavior, aggressive/impulsive traits or suicidality in first-degree relatives. 19330314_A common genetic variant in WFS1 specifically impairs GLP-1-induced insulin secretion independently of insulin sensitivity 19330314_Observational study of gene-disease association. (HuGE Navigator) 19380854_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19502414_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19598235_Observational study of gene-disease association. (HuGE Navigator) 19598235_WFS1 gene is associated with autistic traits, empathy and Asperger syndrome. 19602701_Meta-analysis and HuGE review of gene-disease association. (HuGE Navigator) 19720844_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19734900_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19741467_Observational study of gene-disease association. (HuGE Navigator) 19794065_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19814620_Observational study of gene-disease association. (HuGE Navigator) 19833888_Observational study of gene-disease association. (HuGE Navigator) 19862325_Observational study of gene-disease association. (HuGE Navigator) 19877185_The family described with autosomal dominant inheritance of K836T of the WFS1 gene demonstrates a progressive hearing loss in the lower frequencies. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20028947_Observational study of gene-disease association. (HuGE Navigator) 20028947_six highly correlated single nucleotide polymorphisms that show strong and comparable associations with risk of type 2 diabetes 20069065_This study describes the phenotype of a family with autosomal dominant optic neuropathy and hearing impairment associated with a novel missense mutation in WFS1. 20160352_a role for WFS1 in the negative regulation of ER stress signaling and in the pathogenesis of diseases involving chronic, unresolvable ER stress, such as pancreatic beta cell death in diabetes. 20203524_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20215779_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20361036_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20361036_the HNF4A and WFS1 risk alleles predispose to development of type 2 diabetes in an Ashkenazi Jewish population 20384434_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20490451_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20509872_Observational study of gene-disease association. (HuGE Navigator) 20571754_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20580033_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20712903_Observational study of gene-disease association. (HuGE Navigator) 20802253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20816152_Observational study of gene-disease association. (HuGE Navigator) 20879858_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20889853_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 21127832_The most frequent haplotype at the haplotype block containing the WFS1 gene modulated insulin secretion and was associated with an increased risk of type 2 diabetes. 21143470_Novel mutations in the wolframin gene are identified that are associated with non-immunogenic diabetes mellitus and a progressive atrophy of the optic nerve. 21356526_WFS1 is the gene responsible for autosomal dominant low-frequency sensorineural hearing loss in a Chinese family, as well as in a sporadic case. These WFS1 mutations are not present in unaffected control subjects. 21454619_ER stress induces Smurf1 degradation and WFS1 up-regulation 21538838_The p.A684V missense mutation in the WFS1 gene is a frequent cause of autosomal dominant optic atrophy and hearing impairment. 21564155_Nine different mutations in WFS1 (five of them novel) were identified in nine Wolfram syndrome patients. 21623591_Cataract could be a marker for the WFS1 heterozygosity in this family, namely the c.2431_2465dup35 mutation. 21632151_Their past medical history revealed diabetes mellitus and deafness since childhood. It was confirmed by molecular analysis, which evidenced composite WFS1 heterozygous mutations inherited from both their mother and father. 21713316_A WFS1 haplotype consisting of the minor alleles of rs752854, rs10010131, and rs734312 shows a protective role against type 2 diabetes in Russian patients 21823543_report a consanguineous family with three siblings affected by Wolfram syndrome. A homozygous single base pair deletion (c.877delC, L293fsX303) was found in the WFS1 gene in all three affected siblings 21968327_A new homozygous WFS1 mutation causing causing Wolfram syndrome is identified in a large inbred Turkish family. 22240535_genetic variation of Wolfram syndrome type 1 gene was a more crucial factor than other genes in causing hearing loss. 22498363_Two individuals who had heterozygosity of GJB2 mutations and heterozygosity of WFS1 mutations showed low-frequency hearing loss. One individual who had homozygosity of GJB2 mutation without WFS1 mutation had moderate, gradual high tone hearing loss. 22662265_In a family with MODY diabetes, three affected subjects had the mutation c.2107C-T/p.R703C. The affected amino acid is strongly conserved and the variant suggested to be probably damaging by prediction programs. The proband developed diabetes 14 years old with no type 1 auto-antibodies and required insulin. There was no familial hearing impairment. 22781099_report of male Wolfram patients with WFS1 mutations who have successfully fathered children 23035048_WFS1 has a specific interaction with the V1A subunit of H(+) ATPase; this interaction may be important both for pump assembly in the ER and for granular acidification. 23103830_Description of a novel missense mutation of the WFS1 gene in exon 4 of WFS1 gene in two Italian siblings with Wolfram syndrome. 23144361_In African Americans, seven of the 29 SNPs examined were found to be associated with T2D risk at P = 0.05, including 2 SNPs in the WFS1 gene (rs4689388 and rs1801214). 23257691_Data from case-control genome-wide association studies suggest that 2 SNPs in WFS1 (rs734312; rs10010131) are associated with type 2 diabetes; G allele of rs734312 and A allele of rs10010131 appear to have protective effects. [META-ANALYSIS] 23373429_The recognition of microspherophakia in two siblings carrying a novel WFS1 mutation expands the clinical and molecular spectrum of Wolfram syndrome. 23499253_Report on an efficient double-tube allele-specific amplification method in conjunction with ultrafast capillary gel electrophoresis for direct haplotyping analysis of the SNPs in two important miRNA-binding sites (rs1046322 and rs9457) in the WFS1 gene. 23531866_Identified a DNA substitution (c.1385A-to-G) in WFS1 exon. 23595122_The results support previous findings that genetic variation of WFS1 contributes to the risk of diabetes mellitus and sensorineural hearing impairment. 23650218_this is the first report describing a microRNA binding site polymorphism of the WFS1 gene and its association with human aggression based on a large, non-clinical sample 23845777_A homozygous insertion mutation in WFS1 may be associated with early onset of disease symptoms in Wolfram syndrome. 23903355_This represents the first compelling report of a mutation in WFS1 associated with dominantly inherited nonsyndromic adult-onset diabetes. 24117146_Two familial cases of Wolfram syndrome caused by a novel homozygous WFS1 missense mutation, are reported. 24588001_The decrease in wolframin expression in diabetic placenta suggests that this protein may participate in maintaining the physiologic glucose homeostasis in this organ. 24909696_WFS1 gene mutations are a rare cause of hearing impairment among Finnish children. 25074416_No association was found between wolframin gene H611R polymorphism and mood disorders. 25211237_This study emphasizes the clinical and genetic heterogeneity in patients with WFS. Genotype-phenotype correlations may exist in patients with WFS1 mutations, as demonstrated by the disease onset. 25250959_A novel missense mutation c.2389G > A (GAC -AAC) in WFS1 gene causes non-syndromic hearing loss in all, rather than in low or high, frequencies. 25255707_The analysis of our case, in the light of the most recent literature, suggests a possible role for WFS1 gene in the development of certain brain structures during the fetal period. 25274773_Results reveal a role for WFS1 in the negative regulation of SERCA and provide further insights into the function of WFS1 in calcium homeostasis. 25740874_Early-onset Central diabetes insipidus is associated with de novo mutations of the AVP gene and with hereditary WFS1 gene changes. 25764693_Here we review clinical features, molecular mechanisms and mutations of WFS1 gene that relate to this syndrome.[review] 25800097_Data show that Wolfram syndrome 1 (WFS1; wolframin) promoter activity was highest with the most frequent haplotype (H1; ATCGT) and lowest with second most frequent haplotype (H2; GATCG). 26169481_Data suggest that a novel mutation in WFS1 [c.13481350 del ins TAG (p.His450*)] causes Wolfram-like syndrome in homozygous daughter with maternal uniparental disomy of chromosome 4; heterozygous mother is unaffected. [CASE REPORT] 26426397_Micro-RNA Binding Site Polymorphisms in the WFS1 Gene Are Risk Factors of Diabetes Mellitus. 26773575_A mutation (c.376G>A, p.A126T) was found in all 5 family members affected with Wolfram syndrome in homozygous state and in both parents in heterozygous state. 26875006_WFS1 is a highly polymorphic gene and determining the mode of inheritance or the pathological significance of a specific WFS1 variant is not always straightforward, especially in singleton cases with no access to other family members. Our study has revealed an interesting association between dominant missense WFS1 mutations and distinct OPL lamination on spectral domain OCT, which was not observed in patients with recessi 26943604_Nonsense mutation in the WFS1 gene is associated with Wolfram syndrome. 27377286_provides genotyping protocols readily applicable in any multiplex SNP and VNTR analyses, moreover confirms and extends previous results about the role of WFS1 polymorphisms in the genetic risk of diabetes mellitus 27395765_In this study, we found that patients with isolated, autosomal recessive nonsyndromic optic atropy have biallelic mutations in WFS1. We found that a high percentage (15%) of autosomal recessive non-syndromic optic atropy in families is caused by WFS1 mutations 27412528_Four novel mutations of the WFS1 gene in Iranian Wolfram syndrome pedigrees identified. 27468121_Data show that mutations in Wolfram syndrome 1 (wolframin) protein (WFS1) gene were identified in three children with Wolfram syndrome. 28039263_We show for the first time the role of WFS1 in CAO and document a statistically significant interaction between increasing cumulative cisplatin dose and rs62283056 genotype. Our clinical translational results demonstrate that pretherapy patient genotyping to minimize ototoxicity could be useful when deciding between cisplatin-based chemotherapy regimens of comparable efficacy with different cumulative doses 28271504_WFS1 and GJB2 mutations were identified in eight of 74 cases of Low-Frequency Sensorineural Hearing Loss. Four cases had heterozygous WFS1 mutations; one had a heterozygous WFS1 mutation and a heterozygous GJB2 mutation; and three cases had biallelic GJB2 mutations. Three cases with WFS1 mutations were sporadic; two of them were confirmed to be caused by a de novo mutation based on the genetic analysis of their parents. 28419064_a novel mutation c.2614-2625delCATGGCGCCGTG in the WFS1-gene was identified in a family with autosomal-dominant hereditary hearing impairment 28468959_Specific dominant WFS1 mutations are a cause of a novel syndrome including neonatal/infancy-onset diabetes, congenital cataracts, and sensorineural deafness. 28802351_data extend the mutation spectrum of the WFS1 gene in Chinese individuals and may contribute to establishing a better genotype-phenotype correlation for LFSNHL. 28974383_A nonsynonymous mutation in the WFS1 gene causing late-onset sensorineural hearing impairment with audiogram configurations typical for age-related hearing impairment. 29258540_This is the second report to describe a pathological mutation in WFS1 among Korean patients and the second to describe the mutation in a different ethnic background. Given that the mutation was found in independent families, p.S807R possibly appears to be a 'hot spot' in WFS1, which is associated with LF-NSHL. 29357349_Protective role of wfs1 against stress and age-associated neurodegeneration. 29447883_findings strongly suggest that the c.2389G>A mutation in WFS1 is associated with all-frequency hearing loss, rather than low- or high-frequency loss 29529044_Study successfully identified eight previously reported mutations and five novel variants, and estimated the incidence of WFS1 variants to be 2.5% in Japanese families with presumably autosomal dominant or mitochondrial HL. Also, results found that some variants can occur as de novo change at the mutational hot spots in WFS1, resulting in an audiovestibular phenotype. 29626590_Altered expression of WFS1 and NOTCH2 genes may play a role in pathogenesis and development of DN in patients with T2DM. 30014265_analysis of a low-frequency coding variant in the WFS1 gene that is enriched in Ashkenazi Jewish individuals and causes a mild form of Wolfram syndrome characterized by young-onset diabetes and reduced penetrance for optic atrophy 30305294_Traditional genome-wide association studies have identified single-nucleotide polymorphisms in ACYP2 and WFS1 associated with cisplatin-induced hearing loss. 30957632_Ophthalmic, systemic, and genetic characteristics of patients with Wolfram syndrome. 31363008_Segregation of two variants suggests the presence of autosomal dominant and recessive forms of WFS1-related disease within the same family: expanding the phenotypic spectrum of Wolfram Syndrome. 31477210_Our study revealed that transcription and translation of WFS1, CRH, and UCN were altered during pregnancies complicated by early-onset intrahepatic cholestasis of pregnancy (ICP). This disrupted compensatory response mediated by WFS1 and CRH family peptides in early-onset ICP may play a significant role in the pathogenesis of sudden fetal death in acute fetal hypoxia. 31658956_Nonsyndromic diabetes with WSF1 mutations is not rare in Chinese. Its response to alternative treatments should be investigated. 31759989_This study revealed an association of SIRT1 and WFS1 with Type 2 Diabetes (T2D) risk. A positive association with TD2 risk was found for WFS1 rs6446482 (p = 0.046, Z = 1.994) under an additive model, and SIRT1 rs7896005 (p = 0.038, Z = 2.073) under the dominant model. 31937257_The mutational and phenotypic spectrum of Wolfram syndrome (WS) is broadened by our report of novel WFS1 mutation. Our results reveal the value of molecular analysis of WFS1 in the improvement of clinical diagnostics for WS. This study also confirms the role of WFS1 in type-2 diabetes mellitus (T2DM) 32219690_Effect of 4-phenylbutyrate and valproate on dominant mutations of WFS1 gene in Wolfram syndrome. 32382995_A comparative analysis of genetic hearing loss phenotypes in European/American and Japanese populations. 32567228_Recurrent de novo WFS1 pathogenic variants in Chinese sporadic patients with nonsyndromic sensorineural hearing loss. 32938580_Identification of Three Novel and One Known Mutation in the WFS1 Gene in Four Unrelated Turkish Families: The Role of Homozygosity Mapping in the Early Diagnosis 33179441_Autosomal-dominant WFS1-related disorder-Report of a novel WFS1 variant and review of the phenotypic spectrum of autosomal recessive and dominant forms. 33693650_Novel mutations in the WFS1 gene are associated with Wolfram syndrome and systemic inflammation. 33882198_Different clinical entities of the same mutation: a case report of three sisters with Wolfram syndrome and efficacy of dipeptidyl peptidase-4 inhibitor therapy. 34006618_WFS1 protein expression correlates with clinical progression of optic atrophy in patients with Wolfram syndrome. 34133072_Wolfram Syndrome: Cracking the Code to Better Therapies. 34258273_Missense Variant of Endoplasmic Reticulum Region of WFS1 Gene Causes Autosomal Dominant Hearing Loss without Syndromic Phenotype. 34360843_A Novel Genetic Variant in the WFS1 Gene in a Patient with Partial Uniparental Mero-Isodisomy of Chromosome 4. 34404380_Unique three-site compound heterozygous mutation in the WFS1 gene in Wolfram syndrome. 34792487_Prevalence and phenotypic features of diabetes due to recessive, non-syndromic WFS1 mutations. 34848728_WFS1 functions in ER export of vesicular cargo proteins in pancreatic beta-cells. 35018440_WFS1 Gene-associated Diabetes Phenotypes and Identification of a Founder Mutation in Southern India. | ENSMUSG00000039474 | Wfs1 | 1192.96744 | 0.7705925 | -0.3759600114 | 0.11726961 | 1.016627e+01 | 1.430336e-03 | 6.451956e-02 | No | Yes | 1190.174236 | 141.187596 | 1522.230375 | 175.922274 | |
| ENSG00000109534 | 54433 | GAR1 | protein_coding | Q9NY12 | FUNCTION: Required for ribosome biogenesis and telomere maintenance. Part of the H/ACA small nucleolar ribonucleoprotein (H/ACA snoRNP) complex, which catalyzes pseudouridylation of rRNA. This involves the isomerization of uridine such that the ribose is subsequently attached to C5, instead of the normal N1. Each rRNA can contain up to 100 pseudouridine ('psi') residues, which may serve to stabilize the conformation of rRNAs. May also be required for correct processing or intranuclear trafficking of TERC, the RNA component of the telomerase reverse transcriptase (TERT) holoenzyme. {ECO:0000269|PubMed:10757788, ECO:0000269|PubMed:15044956}. | 3D-structure;Alternative splicing;Isopeptide bond;Nucleus;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein;Ribosome biogenesis;Ubl conjugation;rRNA processing | This gene is a member of the H/ACA snoRNPs (small nucleolar ribonucleoproteins) gene family. snoRNPs are involved in various aspects of rRNA processing and modification and have been classified into two families: C/D and H/ACA. The H/ACA snoRNPs also include the DKC1, NOLA2 and NOLA3 proteins. These four H/ACA snoRNP proteins localize to the dense fibrillar components of nucleoli and to coiled (Cajal) bodies in the nucleus. Both 18S rRNA production and rRNA pseudouridylation are impaired if any one of the four proteins is depleted. These four H/ACA snoRNP proteins are also components of the telomerase complex. The encoded protein of this gene contains two glycine- and arginine-rich domains and is related to Saccharomyces cerevisiae Gar1p. Two splice variants have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:54433; | box H/ACA scaRNP complex [GO:0072589]; box H/ACA snoRNP complex [GO:0031429]; box H/ACA telomerase RNP complex [GO:0090661]; chromosome, telomeric region [GO:0000781]; fibrillar center [GO:0001650]; nucleoplasm [GO:0005654]; telomerase holoenzyme complex [GO:0005697]; box H/ACA snoRNA binding [GO:0034513]; RNA binding [GO:0003723]; telomerase RNA binding [GO:0070034]; snoRNA guided rRNA pseudouridine synthesis [GO:0000454]; telomere maintenance via telomerase [GO:0007004] | 17903301_Genome-wide association study of gene-disease association. (HuGE Navigator) 18989882_Observational study of gene-disease association. (HuGE Navigator) 18989882_heterozygous point mutations in NOLA1 gene are not responsible for aplastic anemia in our patients at least acting via telomere 19917616_The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. 20332099_Observational study of gene-disease association. (HuGE Navigator) 23707062_The deregulated expression and function of H/ACA snoRNPs may underlie specific pathological features of human disease. 33526451_SUMOylation- and GAR1-Dependent Regulation of Dyskerin Nuclear and Subnuclear Localization. | ENSMUSG00000028010 | Gar1 | 2019.70556 | 1.0696851 | 0.0971861081 | 0.08522071 | 1.303245e+00 | 2.536213e-01 | 6.462786e-01 | No | Yes | 2569.637393 | 235.879016 | 2272.990712 | 204.146562 | |
| ENSG00000109794 | 25854 | FAM149A | protein_coding | A5PLN7 | Alternative splicing;Reference proteome | hsa:25854; | 26600424_In total, 270,389 single nucleotide polymorphisms passed quality control, and 4 SNPs in the FAM149A gene were associated with Acute Mountain Sickness; however, in the validation cohorts, FAM149A was not associated with the presence or severity of AMS. 34303830_Whole exome sequencing identified FAM149A as a plausible causative gene for congenital hereditary endothelial dystrophy, affecting Nrf2-Antioxidant signaling upon oxidative stress. | ENSMUSG00000070044 | Fam149a | 64.54470 | 1.0039557 | 0.0056956777 | 0.36967651 | 2.369234e-04 | 9.877192e-01 | No | Yes | 91.064970 | 12.496290 | 88.514335 | 11.496803 | |||||
| ENSG00000110274 | 22897 | CEP164 | protein_coding | Q9UPV0 | FUNCTION: Plays a role in microtubule organization and/or maintenance for the formation of primary cilia (PC), a microtubule-based structure that protrudes from the surface of epithelial cells. Plays a critical role in G2/M checkpoint and nuclear divisions. A key player in the DNA damage-activated ATR/ATM signaling cascade since it is required for the proper phosphorylation of H2AX, RPA, CHEK2 and CHEK1. Plays a critical role in chromosome segregation, acting as a mediator required for the maintenance of genomic stability through modulation of MDC1, RPA and CHEK1. {ECO:0000269|PubMed:17954613, ECO:0000269|PubMed:18283122, ECO:0000269|PubMed:23348840}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Ciliopathy;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;DNA damage;DNA repair;Disease variant;Mitosis;Nephronophthisis;Nucleus;Phosphoprotein;Reference proteome | This gene encodes a centrosomal protein involved in microtubule organization, DNA damage response, and chromosome segregation. The encoded protein is required for assembly of primary cilia and localizes to mature centrioles. Defects in this gene are a cause of nephronophthisis-related ciliopathies. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Dec 2012]. | hsa:22897; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary transition fiber [GO:0097539]; cytosol [GO:0005829]; extracellular space [GO:0005615]; nucleoplasm [GO:0005654]; cell cycle [GO:0007049]; cell division [GO:0051301]; cilium assembly [GO:0060271]; DNA repair [GO:0006281] | 17954613_These data implicate distal appendages in primary cilia formation and identify Cep164 as an excellent marker for these structures. 18283122_Cep164 is a key player in the DNA damage-activated signaling cascade. 19197159_Results show that Cep164 knockdown compromises the cell survival upon UV damage, and that UV irradiation significantly enhances the interaction between Cep164 and XPA. 22004425_Single nucleotide polymorphisms of CCND2, RAD23B, GRP78, CEP164, MDM2, and ALDH2 genes were significantly associated with development and recurrence of hepatocellular carcinoma in Japanese patients with hepatitis C virus. 22863007_Study identifies by whole-exome resequencing, mutations of MRE11, ZNF423, and CEP164 as causing Nephronophthisis-related ciliopathies. 23150559_findings indicate that ARL13B, INPP5E, PDE6D, and CEP164 form a distinct functional network that is involved in JBTS and NPHP but independent of the ones previously defined by NPHP and MKS proteins 23253480_Cep164 is targeted to the apical domain of the mother centriole to provide the molecular link between the mother centriole and the membrane biogenesis machinery that initiates cilia formation. 24982133_data suggest that TTBK2 also acts upstream of Cep164, contributing to the assembly of distal appendages 25297623_Evidence is provided that TTBK2 effectively phosphorylate Cep164 and Cep97 and inhibits the interaction between Cep164 and its binding partner Dishevelled-3 (an important regulator of ciliogenesis) in a kinase activity-dependent manner. 25340510_This study reveals a novel role for CEP164 in the pathogenesis of nephronophthisis, in which mutations cause ciliary defects coupled with DNA damage induced replicative stress, cell death, and epithelial-to-mesenchymal transition 26966185_data suggest that CEP164 is not required in the DNA damage response. 31990917_these data support a conserved role for CEP164 throughout the development of numerous organs, which, we suggest, accounts for the multi-system disease phenotype of CEP164-mediated Nephronophthisis-related ciliopathies 34499853_Molecular mechanisms underlying the role of the centriolar CEP164-TTBK2 complex in ciliopathies. | ENSMUSG00000043987 | Cep164 | 873.91156 | 0.8230936 | -0.2808716574 | 0.11571234 | 5.852379e+00 | 1.555600e-02 | 2.019230e-01 | No | Yes | 907.597708 | 69.858312 | 1088.949512 | 81.249640 | |
| ENSG00000110931 | 10645 | CAMKK2 | protein_coding | Q96RR4 | FUNCTION: Calcium/calmodulin-dependent protein kinase belonging to a proposed calcium-triggered signaling cascade involved in a number of cellular processes. Isoform 1, isoform 2 and isoform 3 phosphorylate CAMK1 and CAMK4. Isoform 3 phosphorylates CAMK1D. Isoform 4, isoform 5 and isoform 6 lacking part of the calmodulin-binding domain are inactive. Efficiently phosphorylates 5'-AMP-activated protein kinase (AMPK) trimer, including that consisting of PRKAA1, PRKAB1 and PRKAG1. This phosphorylation is stimulated in response to Ca(2+) signals (By similarity). Seems to be involved in hippocampal activation of CREB1 (By similarity). May play a role in neurite growth. Isoform 3 may promote neurite elongation, while isoform 1 may promoter neurite branching. {ECO:0000250, ECO:0000269|PubMed:11395482, ECO:0000269|PubMed:12935886, ECO:0000269|PubMed:21957496, ECO:0000269|PubMed:9662074}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Calmodulin-binding;Cell projection;Cytoplasm;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The product of this gene belongs to the Serine/Threonine protein kinase family, and to the Ca(2+)/calmodulin-dependent protein kinase subfamily. The major isoform of this gene plays a role in the calcium/calmodulin-dependent (CaM) kinase cascade by phosphorylating the downstream kinases CaMK1 and CaMK4. Protein products of this gene also phosphorylate AMP-activated protein kinase (AMPK). This gene has its strongest expression in the brain and influences signalling cascades involved with learning and memory, neuronal differentiation and migration, neurite outgrowth, and synapse formation. Alternative splicing results in multiple transcript variants encoding distinct isoforms. The identified isoforms differ in their ability to undergo autophosphorylation and to phosphorylate downstream kinases. [provided by RefSeq, Jul 2012]. | hsa:10645; | cytosol [GO:0005829]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; calcium ion binding [GO:0005509]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; calcium-mediated signaling [GO:0019722]; CAMKK-AMPK signaling cascade [GO:0061762]; cellular response to reactive oxygen species [GO:0034614]; MAPK cascade [GO:0000165]; positive regulation of autophagy of mitochondrion [GO:1903599]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of transcription, DNA-templated [GO:0045893]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of protein kinase activity [GO:0045859] | 16054095_There is a significant basal activity and phosphorylation of AMPK in LKB1-deficient cells that can be stimulated by Ca2+ ionophores, and studies using the CaMKK inhibitor STO-609 and isoform-specific siRNAs show that CaMKKbeta is required for this effect 16054096_Overexpression of CaMKKbeta in mammalian cells increases AMPK activity, whereas pharmacological inhibition of CaMKK, or downregulation of CaMKKbeta using RNA interference, almost completely abolishes AMPK activation 16673375_Observational study of gene-disease association. (HuGE Navigator) 16880506_Endothelial cells possess two pathways to activate AMPK, one Ca2+/CaMKKbeta dependent and one AMP/LKB1 dependent. 17197037_Observational study of gene-disease association. (HuGE Navigator) 17197037_a prominent association found between severity of panic- and agoraphobia symptoms and an exonic SNP (rs3817190) in the CaMKKb gene and a trend for association with an exonic SNP in P2RX7 (rs1718119) with severity scores in the panic- and agoraphobia scale 18436530_modulating basal AMPK and CAMKKB activity in the hypothalamus is essential for maintaining tight regulation of pathways contributing to food intake 19407487_Growth of cervical cancer cells was inhibited through activation of CAMKK2 and LBK1. 19667195_Data show that that the prototypical CaM target sequence skMLCK, a fragment from skeletal muscle myosin light chain kinase, binds to CaM in a highly cooperative way, while only a lower degree of interdomain binding cooperativity emerges for CaMKK. 19763792_ERK activation and cell growth require CaM kinases in MCF-7 breast cancer cells 19958286_Calmodulin-dependent protein kinase kinase-beta activates AMPK without forming a stable complex. There is a synergistic effects of Ca2+ and AMP. 20018878_CaMKK is involved in both S1P receptor- and SR-BI-mediated phosphorylation of AMPK, Akt, and eNOS. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21084482_These results suggest that CaMKK is an important factor for human cytomegalovirus replication and human cytomegalovirus-mediated glycolytic activation. 21669867_phosphorylation of CaMKKbeta regulates its half-life. 21670147_Findings reveal that hypoxia can trigger AMPK activation in the apparent absence of increased [AMP] through ROS-dependent CRAC channel activation, leading to increases in cytosolic calcium that activate the AMPK upstream kinase CaMKKbeta. 21807092_Our results demonstrate that CaMKKbeta and AMP-activated protein kinase form a unique signaling complex 21957496_Data show that protein kinase A (PKA) regulates the alternative splicing of Ca(2)/calmodulin-dependent protein kinase kinase 2 (CaMKK2) to produce variants that differentially modulate neuronal differentiation. 22654108_Results suggest that in PCa progression, CaMKK2 and the AR are in a feedback loop in which CaMKK2 is induced by the AR to maintain AR activity, AR-dependent cell cycle control, and continued cell proliferation. 22778263_Calcium/calmodulin-dependent protein kinase kinase 2 has roles in signaling and pathophysiology [review] 23027865_amino acid starvation regulates autophagy in part through an increase in cellular Ca(2+) that activates a CaMKK-beta-AMPK pathway and inhibits mTORC1, which results in ULK1 stimulation 23110126_CaMKKbeta is involved in AMP-activated protein kinase activation by baicalin in LKB1 deficient cell lines. 23465592_CaMKIIalpha phosphorylation was enhanced by S-Allyl cysteine treatment in a concentration- and time-dependent manner, which paralleled AMPK activation. 23754392_Pulsatile shear stress mimicking atheroprotective flow increases the level of sirtuin (SIRT)1 in cultured endothelial cells by enhancing its stability, an effect abolished by inhibition or knockdown of CaMKKbeta. 23958956_evidence supports that CAMKK2 is a novel schizophrenia susceptibility gene. 25394900_PCa patients with miR-224-low/CAMKK2-high expression more frequently had shorter overall survival. 25590814_CaMKKbeta-AMPKalpha2 signaling contributes to mitotic Golgi fragmentation and the G2/M transition in mammalian cells. 25679868_CaMKK2 plays a pivotal role in the calcium signaling cascade regulating adrenal aldosterone production. 25756516_Silencing of CAMKK2 using siRNA significantly reduced cell proliferation, colony formation and invasion of gastric cancer cells. 25847065_CAMKK2 protein is highly up regulated in hepatocellular carcinoma. 26354101_For the first time, we showed that rs1063843, a single nucleotide polymorphism located in the CAMKK2 gene, is highly associated with bipolar disorder 26552607_CaMKK2 (and Nup62) are required for optimal androgen receptor transcriptional activity in castrate resistant prostate cancer cells. 26785644_CAMKK2 exhibited the strongest associations with HIV-associated sensory neuropathy (HIV-SN), with two SNPs and six haplotypes predicting SN status in black Southern Africans. 26824050_Clopidogrel diminishes TNFalpha-stimulated VCAM-1 expression at least in part via HO-1 induction and CaMKKbeta/AMPK/Nrf2 pathway in endothelial cells. 27004598_Study used three cognitive tasks and fMRI to provide convergent evidence of a link between the rs1063843 SNP of CAMKK2 and the function of the dorsolateral prefrontal cortex. In addition, this polymorphism was associated with the function of the striatum during a working memory task. 27012733_This study showed that the expression level of CAMKK2 could be regulated by promoter methylation. CAMKK2 serves as a prognostic marker in gliomas and could be a potential therapeutic target in gliomas. 27151216_Site-directed mutagenesis analysis revealed that Leu(358) in CaMKKbeta/Ile(322) in CaMKKalpha confer, at least in part, a distinct recognition of AMPK but not of CaMKIalpha. 28230171_This study provides insight into functionally disruptive, rare-variant mutations in human CaMKK2, which have the potential to influence risk and burden of disease associated with aberrant CaMKK2 activity in human populations carrying these variants. 28233049_Single nucleotide polymorphism in CAMKK2 gene is associated with pulmonary non-tuberculous mycobacterial disease. 28600513_Silencing of TRPC5 and inhibition of autophagy reverses adriamycin drug resistance in breast carcinoma via CaMKKbeta/AMPKalpha/mTOR pathway. 28634229_Data suggest that CAMKK2 is highly expressed in high-grade ovarian cancer and ovarian cancer cell lines; CAMKK2 directly activates Akt1 by phosphorylation at Thr-308 in a Ca2+/calmodulin-dependent manner; CAMKK2 knockdown or inhibition decreases Akt1 phosphorylation at Thr-308 and Ser-473. (CAMKK2 = calcium/calmodulin dependent protein kinase kinase 2; AKT1 = AKT serine/threonine kinase 1) 29428485_Three single nucleotide polymorphisms (SNPs) within P2X4R and two SNPs within CAMKK2 influenced concentrations of TNFalpha in peripheral blood mononuclear cells, but these SNP did not associate with risk for HIV-associated sensory neuropathy in South Africans. 29649512_14-3-3gamma protein directly interacts with the kinase domain of CaMKK2 and the region containing the inhibitory phosphorylation site Thr(145) within the N-terminal extension. CaMKK isoforms differ in their 14-3-3-mediated regulations and the interaction between 14-3-3 protein and the N-terminal 14-3-3-binding motif of CaMKK2 might be stabilized by small-molecule compounds. 29675630_results demonstrated that SSd induces autophagy through the CaMKKbeta-AMPK-mTOR signalling pathway in Autosomal dominant polycystic kidney disease (ADPKD) cells, indicating that SSd might be a potential therapy for ADPKD and that SERCA might be a new target for ADPKD treatment. 29972773_These data provide evidence that Ca(2+), CaMKK2, AMPK, and VASP form a mechanosensitive signaling cascade at focal adhesions that is critical for stress fiber assembly. 29992499_Serum CAMKK2 was downregulated in female schizophrenic patients compared with female healthy individuals. 30242113_CAMKK2 and AMPK have opposing effects on lipogenesis, providing a potential mechanism for their contrasting effects on prostate cancer progression in vivo 30471424_Inhibition of AMPK with Compound C prevents the TNFalpha-induced activatory phosphorylation of endothelial nitric oxide synthase (eNOS) at Ser1177 and reduces the NO release. AMPK is activated by phosphorylation catalysed by liver kinase B1 (LKB1) and calcium/calmodulin-dependent protein kinase kinase beta (CaMKKbeta), which are phosphorylated and thereby activated in the presence of TNFalpha. 30544543_Studied inhibition by herbal preparation HX109 of androgen receptors thru calcium/calmodulin dependent protein kinase kinase 2 (CAMKK2)/activating transcription factor 3 (ATF3) signal pathways in prostate cancer cell line; also found HX109 to ameliorate testosterone propionate induced benign prostate hyperplasia thru androgen receptor inhibition. 31115859_CaMKK2 Signaling in Metabolism and Skeletal Disease: a New Axis with Therapeutic Potential. 31164648_CaMKK2 is found to be expressed in both breast cancer cells and within stromal cells. CaMKK2 expression inversely correlated with the less aggressive luminal A (LA) molecular type, and a trend for higher CaMKK2 expression in triple-negative TN type was also observed. 31309408_Polymorphisms in CAMKK2 associate with susceptibility to sensory neuropathy in HIV patients treated without stavudine. 31445019_suppressive effect of M3 muscarinic receptor activation on hepatocyte steatosis which is mediated via CAMKKbeta/AMPK pathway. 32216002_Functional analysis of an R311C variant of Ca(2+) -calmodulin-dependent protein kinase kinase-2 (CaMKK2) found as a de novo mutation in a patient with bipolar disorder. 32305031_Association analysis between CAMKK2 rs1063843 and patients with schizophrenia in a Han Chinese population. 32460794_CAMKK2-CAMK4 signaling regulates transferrin trafficking, turnover, and iron homeostasis. 32485269_Hypomorphic CAMKK2 in EA.hy926 endothelial cells causes abnormal transferrin trafficking, iron homeostasis and glucose metabolism. 32585444_The role of CAMKK2 polymorphisms in HIV-associated sensory neuropathy in South Africans. 32791096_MiR-1271 regulates glioblastoma cell proliferation and invasion by directly targeting the CAMKK2 gene. 32869834_Protein kinase A negatively regulates VEGF-induced AMPK activation by phosphorylating CaMKK2 at serine 495. 32913128_CaMKK2 is inactivated by cAMP-PKA signaling and 14-3-3 adaptor proteins. 33082841_Effects of Single-Nucleotide Polymorphisms in Calmodulin-Dependent Protein Kinase Kinase 2 (CAMKK2): A Comprehensive Study. 33531625_Inhibition of CAMKK2 impairs autophagy and castration-resistant prostate cancer via suppression of AMPK-ULK1 signaling. 33838119_Immunohistochemical evidence of P2X7R, P2X4R and CaMKK2 in pyramidal neurons of frontal cortex does not align with Alzheimer's disease. 33844560_Molecular Profiling Associated with Calcium/Calmodulin-Dependent Protein Kinase Kinase 2 (CAMKK2)-Mediated Carcinogenesis in Gastric Cancer. 34229059_NCAPD2 inhibits autophagy by regulating Ca(2+)/CAMKK2/AMPK/mTORC1 pathway and PARP-1/SIRT1 axis to promote colorectal cancer. 34437731_Hyperactivation of MEK/ERK pathway by Ca(2+) /calmodulin-dependent protein kinase kinase 2 promotes cellular proliferation by activating cyclin-dependent kinases and minichromosome maintenance protein in gastric cancer cells. 34472622_CARS senses cysteine deprivation to activate AMPK for cell survival. 34563205_CAMKK2 regulates mitochondrial function by controlling succinate dehydrogenase expression, post-translational modification, megacomplex assembly, and activity in a cell-type-specific manner. 34725334_CaMKK2 facilitates Golgi-associated vesicle trafficking to sustain cancer cell proliferation. 34878439_Brief Report: Polymorphisms in CAMKK2 may Influence Domain-Specific Neurocognitive Function in HIV+ Indonesians Receiving ART. 34943872_Targeting CAMKK2 and SOC Channels as a Novel Therapeutic Approach for Sensitizing Acute Promyelocytic Leukemia Cells to All-Trans Retinoic Acid. 35309839_CaMKK2 Promotes the Progression of Ovarian Carcinoma through the PI3K/PDK1/Akt Axis. 35438341_Traumatic-noise-induced hair cell death and hearing loss is mediated by activation of CaMKKbeta. | ENSMUSG00000029471 | Camkk2 | 2291.74969 | 1.0031960 | 0.0046035358 | 0.09162324 | 2.522389e-03 | 9.599444e-01 | 9.888660e-01 | No | Yes | 2769.192947 | 189.423490 | 2663.465746 | 178.152926 | |
| ENSG00000111254 | 10566 | AKAP3 | protein_coding | O75969 | FUNCTION: May function as a regulator of both motility- and head-associated functions such as capacitation and the acrosome reaction. | Cytoplasmic vesicle;Direct protein sequencing;Phosphoprotein;Reference proteome | This gene encodes a member of A-kinase anchoring proteins (AKAPs), a family of functionally related proteins that target protein kinase A to discrete locations within the cell. The encoded protein is reported to participate in protein-protein interactions with the R-subunit of the protein kinase A as well as sperm-associated proteins. This protein is expressed in spermatozoa and localized to the acrosomal region of the sperm head as well as the length of the principal piece. It may function as a regulator of motility, capacitation, and the acrosome reaction. [provided by RefSeq, May 2013]. | hsa:10566; | acrosomal vesicle [GO:0001669]; cytoplasm [GO:0005737]; nucleus [GO:0005634]; sperm fibrous sheath [GO:0035686]; sperm midpiece [GO:0097225]; sperm principal piece [GO:0097228]; protein kinase A binding [GO:0051018]; acrosome reaction [GO:0007340]; blastocyst hatching [GO:0001835]; protein localization [GO:0008104]; single fertilization [GO:0007338]; transmembrane receptor protein serine/threonine kinase signaling pathway [GO:0007178] | 12509440_evidence of tyrosine phosphorylation during sperm capacitation 14618620_In Cox multivariate analysis, AKAP3 mRNA expression was found to be a significant predictor of both overall and progression-free survival in patients with poorly differentiated tumors. 16005946_AKAP-3 demonstrates tumor-restricted expression and appears to be associated with worse overall survival. 17683036_AKAP3 is a novel target for protein S-nitrosylation in spermatozoa. 21240291_CABYR variants form a complex not only with the scaffolding protein AKAP3 but also with another RII-like domain-containing protein in the sperm fibrous sheath. 22941507_study reports the isolation of AKAP3 and CTp11 Cancer/testis antigens from hepatocellular carcinoma patient sera 26458542_AKAP3 correlates with triple negative status and disease free survival in breast cancer. 31254143_The present study revealed that one of the major rHuOVGP1-enhanced pY proteins could be AKAP3 of the FS and that rHuOVGP1 is capable of binding to human ZP and its presence in the medium results in an increase in sperm-zona binding. Supplement of rHuOVGP1 in in vitro fertilization media could be beneficial for enhancement of the fertilizing ability of human sperm. 34944009_Protein Kinase A (PRKA) Activity Is Regulated by the Proteasome at the Onset of Human Sperm Capacitation. 35256641_Structural modeling of human AKAP3 protein and in silico analysis of single nucleotide polymorphisms associated with sperm motility. | ENSMUSG00000030344 | Akap3 | 61.47079 | 0.3218510 | -1.6355350790 | 0.38752861 | 1.703685e+01 | 3.666146e-05 | No | Yes | 35.043619 | 8.054567 | 101.079983 | 21.333175 | ||
| ENSG00000111364 | 57696 | DDX55 | protein_coding | Q8NHQ9 | FUNCTION: Probable ATP-binding RNA helicase. | ATP-binding;Alternative splicing;Helicase;Hydrolase;Nucleotide-binding;Phosphoprotein;RNA-binding;Reference proteome | This gene encodes a member of protein family containing a characteristic Asp-Glu-Ala-Asp (DEAD) motif. These proteins are putative RNA helicases, and may be involved in a range of nuclear processes including translational initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Multiple alternatively spliced transcript variants have been found for this gene. Pseudogenes have been identified on chromosomes 1 and 12. [provided by RefSeq, Feb 2016]. | hsa:57696; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724] | 33048000_The DExD box ATPase DDX55 is recruited to domain IV of the 28S ribosomal RNA by its C-terminal region. | ENSMUSG00000029389 | Ddx55 | 1822.83330 | 1.0296576 | 0.0421645969 | 0.09782915 | 1.860683e-01 | 6.662093e-01 | 8.959577e-01 | No | Yes | 2232.130704 | 279.454547 | 2148.873196 | 263.174379 | |
| ENSG00000111424 | 7421 | VDR | protein_coding | P11473 | FUNCTION: Nuclear receptor for calcitriol, the active form of vitamin D3 which mediates the action of this vitamin on cells (PubMed:28698609, PubMed:16913708, PubMed:15728261, PubMed:10678179). Enters the nucleus upon vitamin D3 binding where it forms heterodimers with the retinoid X receptor/RXR (PubMed:28698609). The VDR-RXR heterodimers bind to specific response elements on DNA and activate the transcription of vitamin D3-responsive target genes (PubMed:28698609). Plays a central role in calcium homeostasis (By similarity). {ECO:0000250|UniProtKB:P13053, ECO:0000269|PubMed:10678179, ECO:0000269|PubMed:15728261, ECO:0000269|PubMed:16913708, ECO:0000269|PubMed:28698609}. | 3D-structure;Alternative splicing;Cytoplasm;DNA-binding;Disease variant;Metal-binding;Nucleus;Receptor;Reference proteome;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes vitamin D3 receptor, which is a member of the nuclear hormone receptor superfamily of ligand-inducible transcription factors. This receptor also functions as a receptor for the secondary bile acid, lithocholic acid. Downstream targets of vitamin D3 receptor are principally involved in mineral metabolism, though this receptor regulates a variety of other metabolic pathways, such as those involved in immune response and cancer. Mutations in this gene are associated with type II vitamin D-resistant rickets. A single nucleotide polymorphism in the initiation codon results in an alternate translation start site three codons downstream. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. A recent study provided evidence for translational readthrough in this gene, and expression of an additional C-terminally extended isoform via the use of an alternative in-frame translation termination codon. [provided by RefSeq, Jun 2018]. | hsa:7421; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; receptor complex [GO:0043235]; RNA polymerase II transcription regulator complex [GO:0090575]; calcitriol binding [GO:1902098]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; lithocholic acid binding [GO:1902121]; lithocholic acid receptor activity [GO:0038186]; nuclear receptor activity [GO:0004879]; retinoid X receptor binding [GO:0046965]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; bile acid signaling pathway [GO:0038183]; calcium ion transport [GO:0006816]; cell differentiation [GO:0030154]; cell morphogenesis [GO:0000902]; cellular calcium ion homeostasis [GO:0006874]; decidualization [GO:0046697]; intestinal absorption [GO:0050892]; lactation [GO:0007595]; mammary gland branching involved in pregnancy [GO:0060745]; mRNA transcription by RNA polymerase II [GO:0042789]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of keratinocyte proliferation [GO:0010839]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of apoptotic process involved in mammary gland involution [GO:0060058]; positive regulation of gene expression [GO:0010628]; positive regulation of keratinocyte differentiation [GO:0045618]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of vitamin D 24-hydroxylase activity [GO:0010980]; positive regulation of vitamin D receptor signaling pathway [GO:0070564]; regulation of calcidiol 1-monooxygenase activity [GO:0060558]; skeletal system development [GO:0001501]; vitamin D receptor signaling pathway [GO:0070561] | 11028447_Observational study of gene-disease association. (HuGE Navigator) 11033842_Observational study of gene-disease association. (HuGE Navigator) 11043509_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11049814_Observational study of gene-disease association. (HuGE Navigator) 11092401_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 11134121_Observational study of gene-disease association. (HuGE Navigator) 11136533_Observational study of gene-disease association. (HuGE Navigator) 11167636_Observational study of gene-disease association. (HuGE Navigator) 11174470_Observational study of gene-disease association. (HuGE Navigator) 11204438_Observational study of gene-disease association. (HuGE Navigator) 11224872_Observational study of gene-disease association. (HuGE Navigator) 11230734_Observational study of gene-disease association. (HuGE Navigator) 11248649_Observational study of gene-disease association. (HuGE Navigator) 11251690_Observational study of gene-disease association. (HuGE Navigator) 11257727_Observational study of gene-disease association. (HuGE Navigator) 11306745_Observational study of gene-disease association. (HuGE Navigator) 11316004_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11335187_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 11353946_Observational study of gene-disease association. (HuGE Navigator) 11355046_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11359741_Observational study of gene-disease association. (HuGE Navigator) 11383910_Observational study of gene-disease association. (HuGE Navigator) 11389055_Observational study of gene-disease association. (HuGE Navigator) 11423686_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11440415_Observational study of gene-disease association. (HuGE Navigator) 11445000_Observational study of gene-disease association. (HuGE Navigator) 11456232_Observational study of gene-disease association. (HuGE Navigator) 11461072_Observational study of gene-disease association. (HuGE Navigator) 11484168_Observational study of gene-disease association. (HuGE Navigator) 11489147_Observational study of gene-disease association. (HuGE Navigator) 11489753_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11498732_Observational study of gene-disease association. (HuGE Navigator) 11498733_Observational study of gene-disease association. (HuGE Navigator) 11498736_Observational study of genotype prevalence. (HuGE Navigator) 11522087_Observational study of gene-environment interaction. (HuGE Navigator) 11532853_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11552708_Observational study of gene-disease association. (HuGE Navigator) 11579931_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11598396_Observational study of gene-disease association. (HuGE Navigator) 11678976_Observational study of gene-disease association. (HuGE Navigator) 11679916_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11684540_Observational study of gene-environment interaction. (HuGE Navigator) 11684548_Observational study of gene-disease association. (HuGE Navigator) 11689145_Observational study of gene-disease association. (HuGE Navigator) 11720436_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11744805_Observational study of gene-disease association. (HuGE Navigator) 11744805_These data suggest that BsmI VDR polymorphism does not play an important role in the bone loss seen in hypercalciuric CSF patients. 11751444_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11776070_Observational study of genotype prevalence. (HuGE Navigator) 11779241_critical role of helix 12 of the vitamin D3 receptor for the partial agonism of carboxylic ester antagonist, ZK159222 11782643_Observational study of gene-disease association. (HuGE Navigator) 11786968_Observational study of gene-disease association. (HuGE Navigator) 11789558_Observational study of gene-disease association. (HuGE Navigator) 11800328_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11808760_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11834737_transactivation modulated by a central dinucleotide within vitamin D response elements 11846330_Observational study of gene-disease association. (HuGE Navigator) 11887173_Observational study of gene-disease association. (HuGE Navigator) 11898916_Association of vitamin D receptor gene BsmI polymorphisms in Chinese patients with systemic lupus erythematosus. 11898916_Observational study of gene-disease association. (HuGE Navigator) 11909970_Stat1-vitamin D receptor interactions antagonize 1,25-dihydroxyvitamin D transcriptional activity and enhance stat1-mediated transcription. 11910656_A clinically significant relationship between VDR and ER genotypes and biochemical markers of bone turnover or serum lipoproteins could not be demonstrated in healthy Danish postmenopausal women 11910656_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11913978_Regulation of calbindin-D9k expression by 1,25-dihydroxyvitamin D(3) and parathyroid hormone in mouse primary renal tubular cells 11914750_Observational study of gene-disease association. (HuGE Navigator) 11914750_polymorphism and risk of severe diabetic retinopathy. 11918225_Observational study of gene-disease association. (HuGE Navigator) 11918225_data support the VDR gene as a quantitative trait locus(QTL)underlying spine bone mineral density variation 11918709_vitamin D receptor is required for the initiation of the postnatal hair follicular cycle 11920955_Observational study of gene-disease association. (HuGE Navigator) 11942896_Observational study of gene-disease association. (HuGE Navigator) 11956476_Observational study of gene-disease association. (HuGE Navigator) 11972301_Observational study of gene-disease association. (HuGE Navigator) 11972530_Observational study of gene-disease association. (HuGE Navigator) 11979895_its genetic polymorphism determines bone mineral density 11984699_Observational study of gene-disease association. (HuGE Navigator) 11991441_Observational study of gene-disease association. (HuGE Navigator) 11991950_regulates expression of CYP3A4, CYP2B6, and CYP2C9 in hepatocytes 12003670_polymorphism in type 1 diabetics in a Romanian population 12016314_functions as a receptor for the secondary bile acid lithocholic acid (LCA) 12016463_Observational study of gene-disease association. (HuGE Navigator) 12018632_Observational study of gene-disease association. (HuGE Navigator) 12036913_Observational study of gene-disease association. (HuGE Navigator) 12037619_Observational study of gene-disease association. (HuGE Navigator) 12040821_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12053022_Observational study of gene-disease association. (HuGE Navigator) 12064837_Observational study of gene-disease association. (HuGE Navigator) 12064837_results suggest that the vitamin D receptor Fok I start codon polymorphism is not related to patients with systemic lupus erythematosus in Taiwan 12071154_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12073153_Observational study of gene-disease association. (HuGE Navigator) 12073153_investigation of the genetic influence of Sp1 polymorphism on bone density in Irish women 12086963_Observational study of gene-disease association. (HuGE Navigator) 12086963_levels in relation to vitamin D status, insulin secretory capacity, and VDR genotype in Bangladeshi Asians. 12087029_There is an association between genotype and age of onset in juvenile Japanese patients with type 1 diabetes. 12096841_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12111344_Observational study of gene-disease association. (HuGE Navigator) 12111344_The VDR genotype contributes to the liver dysfunction in patients with psoriasis, although no correlation was found between VDR genotype and the skin eruptions of psoriasis. 12145331_retinoid X receptor regulates vitamin D receptor functions in part by regulating subcellular localization 12147248_alteration of cellular phosphorylation state affects vitamin D receptor-mediated CYP3A4 mRNA induction in Caco-2 cells 12150447_Observational study of gene-disease association. (HuGE Navigator) 12154394_Observational study of gene-disease association. (HuGE Navigator) 12162507_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12169981_Observational study of gene-disease association. (HuGE Navigator) 12173074_Observational study of gene-disease association. (HuGE Navigator) 12174912_expression is increased in ovarian carcinomas as compared to normal ovarian tissue 12181642_Observational study of gene-disease association. (HuGE Navigator) 12181642_contribution of VDR genotypes to prostate cancer susceptibility might depend on the population studied and its geographic localization; VDR genotypes are important in the definition of the genetic risk profile of populations of southern Europe 12192493_Observational study of gene-disease association. (HuGE Navigator) 12192493_This is the first report of an association between VDR gene polymorphism and psoriasis in a Caucasian population. 12195069_Observational study of gene-disease association. (HuGE Navigator) 12200969_Observational study of gene-disease association. (HuGE Navigator) 12203138_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12211444_Tryptophan missense mutation in the ligand-binding domain of the vitamin D receptor causes severe resistance to 1,25-dihydroxyvitamin D. 12219967_Observational study of gene-disease association. (HuGE Navigator) 12237325_removal of the insertion domain between helices 2 and 3 of the receptor does not markedly influence the functionality of the Vitamin d receptor. 12324918_Determination of VDR genotype by analysis of BsmI endonuclease gene polymorphism may predict both hemoglobin level and erythropoietin requirement in hemodialysis patients with anemia. 12324918_Observational study of gene-disease association. (HuGE Navigator) 12360016_Observational study of gene-disease association. (HuGE Navigator) 12363051_Observational study of gene-disease association. (HuGE Navigator) 12369133_Despite a slight increase in the number of urinary tract infections among children with the BB genotype for the vitamin D receptor, there is no statistically significant correlation with genetic susceptibility. 12369133_Observational study of gene-disease association. (HuGE Navigator) 12369780_Observational study of gene-disease association. (HuGE Navigator) 12369780_the VDR genotype polymorphism affects bone density of renal transplant recipients via its effects on the severity of secondary hyperparathyroidism. 12375338_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12391072_Observational study of gene-disease association. (HuGE Navigator) 12391072_vitamin D receptor (VDR) gene polymorphism, which locates at the translation initiation site, matters in the adaptations of bone to long-term impact loading. 12402975_Association of vitamin D receptor gene polymorphism with renal cell carcinoma in Japanese. 12402975_Observational study of gene-disease association. (HuGE Navigator) 12403843_A novel mutation in helix 12 of this receptor impairs coactivator interaction and causes hereditary 1,25-dihydroxyvitamin D-resistant rickets without alopecia. 12404162_Observational study of gene-disease association. (HuGE Navigator) 12413773_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12413773_Polymorphisms of VDR gene are associated with Japanese patients with psoriasis vulgaris. Allelic variance in the VDR gene or other genes in linkage disequilibrium with this gene might predispose to the development of psoriasis vulgaris. 12429765_Observational study of gene-disease association. (HuGE Navigator) 12433736_Observational study of gene-disease association. (HuGE Navigator) 12434167_Observational study of gene-disease association. (HuGE Navigator) 12434167_associations between BsmI-polymorphism of the VDR gene and bone mineral density and bone metabolism in 24 premenopausal (aged 22-45 years) and 69 postmenopausal (aged 48-65 years) Finnish women 12444895_Observational study of gene-disease association. (HuGE Navigator) 12446192_Observational study of gene-disease association. (HuGE Navigator) 12454321_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12457456_Observational study of gene-disease association. (HuGE Navigator) 12468277_Novel mutation in the VDR, Q317X, is the molecular defect in a patient with 1,25-dihydroxyvitamin D resistant rickets 12470203_Observational study of gene-disease association. (HuGE Navigator) 12470203_the FokI genotype of the vitamin D receptor gene is related to bone mass at the hip in Czech postmenopausal women, whereas the importance of remaining VDR genotypes was not evident. 12477580_Observational study of gene-disease association. (HuGE Navigator) 12482639_Observational study of gene-disease association. (HuGE Navigator) 12482639_VDR polymorphisms are associated with increased risk of type 1 diabetes in the Dalmatian population of South Croatia. 12490310_Observational study of gene-disease association. (HuGE Navigator) 12521655_Observational study of gene-disease association. (HuGE Navigator) 12532336_Data suggest that 1,25-dihydroxyvitamin D(3) actions on normal prostate cells may be mediated independently through androgen receptors and vitamin D receptors. 12540499_Observational study of gene-disease association. (HuGE Navigator) 12542560_Observational study of gene-disease association. (HuGE Navigator) 12555245_Observational study of gene-disease association. (HuGE Navigator) 12566913_Observational study of gene-disease association. (HuGE Navigator) 12579489_Observational study of gene-disease association. (HuGE Navigator) 12588283_Observational study of gene-disease association. (HuGE Navigator) 12595908_Observational study of gene-disease association. (HuGE Navigator) 12601576_(Is this the right receptor?) Start codon polymorphism is an independent predictor of lumbar, but not femoral neck bone density. 12601576_Observational study of gene-disease association. (HuGE Navigator) 12608943_Clinical trial of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12649542_Observational study of gene-disease association. (HuGE Navigator) 12649542_The VDR genotype seems to slow the progression of secondary hyperparathyroidism needing parathyroidectomy in patients treated with hemodialysis. 12649563_VDR was expressed in all pancreatic cancers studied. 12656660_marked decreases in vitamin D receptor and calcium-sensing receptor expression could be responsible for the high proliferation of parathyroid cells and the pathological progression of hyperparathyroidism 12666703_Observational study of gene-disease association. (HuGE Navigator) 12666703_VDR polymorphism is associated with oral bone loss, clinical attachment loss, and tooth loss in older men 12673591_Observational study of gene-disease association. (HuGE Navigator) 12673591_Significant association between VDR genotype and postmenopausal osteoporosis in Chinese women was observed in this study. 12674768_Observational study of gene-disease association. (HuGE Navigator) 12694466_Observational study of gene-disease association. (HuGE Navigator) 12717384_human hepatocytes express very low VDR(n) messenger RNA (mRNA) and protein levels. 12753258_Observational study of gene-disease association. (HuGE Navigator) 12759877_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12759877_VDR gene polymorphisms may jointly influence bone mass and the rate of bone loss in older African-American women-- 12778851_Observational study of gene-disease association. (HuGE Navigator) 12786678_Observational study of gene-disease association. (HuGE Navigator) 12792298_Observational study of gene-disease association. (HuGE Navigator) 12807755_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12814692_Observational study of gene-disease association. (HuGE Navigator) 12814692_VDR genotype determination may provide a tool to identify individuals who are at risk for calcium nephrolithiasis. 12818464_Observational study of gene-disease association. (HuGE Navigator) 12820934_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12829710_a previtamin D3 analog with genomic activities equivalent to 1,25(OH)2D3 12836289_Observational study of gene-disease association. (HuGE Navigator) 12843155_Observational study of gene-disease association. (HuGE Navigator) 12843155_association between the VDR gene polymorphism and type 1 diabetes and the onset pattern. 12843190_Observational study of gene-disease association. (HuGE Navigator) 12843190_the VDR gene constitutes a locus for reduced bone mineral density in men. 12846052_Observational study of gene-disease association. (HuGE Navigator) 12846052_VDR polymorphism do not play a major role in rheumatoid arthritis predisposition in german patients. 12868700_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12869402_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12874698_Observational study of gene-disease association. (HuGE Navigator) 12874698_We found VDR genotype to be associated with frame size and bone mineral apparent density(BMAD)but the VDR genotype effects on stature and bone size seem to neutralize the effect on areal BMD. 12879219_Observational study of gene-disease association. (HuGE Navigator) 12895309_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12896855_Observational study of gene-disease association. (HuGE Navigator) 12899513_Observational study of gene-disease association. (HuGE Navigator) 12903041_Data show that the Achang and Han ethnic groups differ in the frequency distribution of VDR gene start codon polymorphism. 12903041_Observational study of genotype prevalence. (HuGE Navigator) 12905734_Observational study of gene-disease association. (HuGE Navigator) 12914574_Observational study of genotype prevalence. (HuGE Navigator) 12915669_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12915669_interaction between ERalpha and vitamin D receptor gene polymorphisms leads to increased risk of osteoporotic vertebral fractures in women, largely independent of bone mineral density 12927786_ER-alpha and vitamin D receptor polymorphisms are related to bone mineral density in pre- and post-menopausal women 12927786_Observational study of gene-disease association. (HuGE Navigator) 12957667_in a model of neoplastic cell transformation, irradiating the MCF-10F cell line followed by treatment with estrogen led to over-expression of the VDR gene, which contained single-base mutations within intron 8 and exon 9 12958689_Genetic variation in the vitamin D receptor gene (VDR) is associated with BMD in premenopausal women. 12958689_Observational study of gene-disease association. (HuGE Navigator) 12960019_For RXR heterodimerizing receptors like VDR, P-box requires redefinition and expansion to include DNA specificity element corresponding to arg-49 and lys-53 of hVDR. These residues are crucial for selective DNA recognition. 12968672_A Cdx-2 binding site polymorphism (G to A) in the promoter region of the vitamin D receptor gene was reported. investigated the relationship between the VDR Cdx-2 genotype and risk of fracture 12968672_Observational study of gene-disease association. (HuGE Navigator) 12969965_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14505233_Observational study of gene-disease association. (HuGE Navigator) 14527203_Observational study of gene-disease association. (HuGE Navigator) 14527840_Observational study of gene-disease association. (HuGE Navigator) 14527848_Observational study of gene-disease association. (HuGE Navigator) 14528100_Children with bone cancer are significantly taller than the reference population, which may be influenced by the genotype for the Fok I polymorphism of the VDR gene 14528100_Observational study of gene-disease association. (HuGE Navigator) 14530911_Observational study of gene-disease association. (HuGE Navigator) 14557853_Observational study of gene-disease association. (HuGE Navigator) 14572874_Observational study of gene-disease association. (HuGE Navigator) 14574802_Observational study of gene-disease association. (HuGE Navigator) 14584884_Observational study of genetic testing. (HuGE Navigator) 14597850_BsmI polymorphism of the VDR gene influences BP in healthy men. A positive relationship between serum 25-hydroxyvitamin D levels and BP is present only in men with the BB genotype. 14597850_Observational study of gene-disease association. (HuGE Navigator) 14634546_Observational study of gene-disease association. (HuGE Navigator) 14634546_The VDR baTL haplotype allele is related to bone mass in Korean women. 14641002_Observational study of gene-disease association. (HuGE Navigator) 14641002_Subjects were genotyped for vitamine D receptor (VDR)A (ApaI), VDRB (BsmI) and VDRF (FokI) single nucleotide polymorphisms (SNPs). The SNPs analysed are unlikely to be associated with type 1 diabetes in the Finnish population. 14642064_Observational study of gene-disease association. (HuGE Navigator) 14665637_results suggest that 1,25-dihydroxyvitamin D3 rapid effects require the presence of vitamin D receptor and control, in part, the vitamin D catabolism via increased expression of the 24-hydroxylase and ferredoxin genes 14688157_Observational study of gene-disease association. (HuGE Navigator) 14691685_No relationship was observed between VDR allelic polymorphisms and osteomalacia. 14691685_Observational study of gene-disease association. (HuGE Navigator) 14693728_Meta-analysis of gene-disease association. (HuGE Navigator) 14693728_Vitamin D receptor gene polymorphism are unlikely to be a major determinants of susceptibility to prostate cancer. 14693733_Observational study of gene-disease association. (HuGE Navigator) 14714273_Observational study of gene-disease association. (HuGE Navigator) 14714273_To fully determine whether sequence differences in VDR gene are susceptibility variants for type 2 diabetes mellitus, additional studies in different populations are required in a large study group. 14718481_Observational study of gene-disease association. (HuGE Navigator) 14719475_Observational study of gene-disease association. (HuGE Navigator) 14725686_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14727381_Observational study of gene-disease association. (HuGE Navigator) 14731356_Observational study of genotype prevalence, gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 14731356_Results indicated that the individuals carrying the VDR B allele were more susceptible to lead poisoning. 14746673_Observational study of gene-disease association. (HuGE Navigator) 14746673_Result suggested the possibility that vitamin D receptor gene polymorphism might be important in determining an individual's susceptibility to development of vitamin D deficiency rickets. 14748937_Clinical trial of gene-disease association. (HuGE Navigator) 14749534_Observational study of gene-disease association. (HuGE Navigator) 14749534_The prevalence of the VDR Taq I and Bsm I alleles and the genotype frequencies in patients with breast cancer was similar to that in the normal population 14963917_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14991752_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14997007_Observational study of gene-disease association. (HuGE Navigator) 14999525_Data describe the influence of allelic variations of the vitamin D receptor and estrogen receptor genes on bone mineral density. 14999525_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15009617_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15012617_Observational study of gene-disease association. (HuGE Navigator) 15037631_Vitamin D receptor has a role in p38 MAPK activation-selectively induced cell death in K-ras-mutated human colon cancer cells 15040830_Meta-analysis of gene-disease association. (HuGE Navigator) 15050735_Observational study of gene-disease association. (HuGE Navigator) 15057510_Meta-analysis of gene-disease association. (HuGE Navigator) 15061984_Observational study of gene-disease association. (HuGE Navigator) 15064717_Suppression of Sp1 expression by small interference RNA reduced the stimulation of p27Kip1 promoter activity by vitamin D3 in prostatic cancer cells. 15065089_Observational study of gene-disease association. (HuGE Navigator) 15066214_Observational study of gene-disease association. (HuGE Navigator) 15066918_Observational study of gene-disease association. (HuGE Navigator) 15067191_Observational study of gene-disease association. (HuGE Navigator) 15083068_Observational study of gene-disease association. (HuGE Navigator) 15083213_Observational study of gene-disease association. (HuGE Navigator) 15083213_VDR TaqI polymorphism is associated with a group of men with benign prostatic hyperplasia who are at an increased risk of prostate cancer 15104566_Observational study of gene-disease association. (HuGE Navigator) 15108066_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15129811_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 15141345_Observational study of gene-disease association. (HuGE Navigator) 15141734_Observational study of gene-disease association. (HuGE Navigator) 15141734_Polymorphisms in the VDR and NRAMP1 gene are statistically associated with susceptibility to pulmonary tuberculosis in the Chinese Han population. 15145445_resistance of acute monocytic leukemia cells to calcitriol-induced differentiation correlates with impaired nuclear localization of vitamin D receptor, but not with its total expression in the cells 15187348_VDR polymorphism associated with the risk of developing rosacea fulminans. 15205858_Observational study of gene-disease association. (HuGE Navigator) 15205858_VDR genotypes may be associated with increased calcium excretion in hypercalciuric nephrolithiatic subjects. 15210908_Observational study of gene-disease association. (HuGE Navigator) 15213319_Observational study of gene-disease association. (HuGE Navigator) 15213514_Lead workers with the VDR B allele had significantly (P value < 0.05) higher patella lead (on average, 25% or approximately 6.6 microg Pb/g bone mineral) than lead workers with the VDR bb genotype. 15213514_Observational study of gene-disease association. (HuGE Navigator) 15225772_Observational study of gene-disease association. (HuGE Navigator) 15225773_Observational study of gene-disease association. (HuGE Navigator) 15225828_Observational study of gene-disease association. (HuGE Navigator) 15238985_Observational study of gene-disease association. (HuGE Navigator) 15241822_Observational study of gene-disease association. (HuGE Navigator) 15246940_Observational study of gene-disease association. (HuGE Navigator) 15252846_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15267201_Observational study of gene-disease association. (HuGE Navigator) 15282199_Strong association found between genetic variants of VDR locus and asthma/atopy in Quebec cohort. 15282200_VDR polymorphism influences asthma and allergy suscpetibility in the population of Quebec. 15295697_Observational study of gene-disease association. (HuGE Navigator) 15295697_VDR polymorphisms are significantly associated with the time to microbiologic resolution of pulmonary TB after initiation of DOTS protocol therapy in Peru 15298953_Observational study of gene-disease association. (HuGE Navigator) 15308833_Observational study of gene-disease association. (HuGE Navigator) 15316258_Observational study of gene-disease association. (HuGE Navigator) 15316869_Observational study of gene-disease association. (HuGE Navigator) 15328186_Observational study of gene-disease association. (HuGE Navigator) 15331595_transcriptional activation of VDR by 1,25-D is attenuated by the concomitant activation of ERK 15332393_In African-Americans, the angiotensin converting enzyme, vitamin D receptor, and tumour necrosis factor-alpha genes are not significant risk factors for sarcoidosis susceptibility. 15358739_Observational study of gene-disease association. (HuGE Navigator) 15359111_Observational study of gene-disease association. (HuGE Navigator) 15368470_Observational study of gene-disease association. (HuGE Navigator) 15368470_vitamin D receptor polymorphisms did not predict pathologic features of prostate cancer but may impact on risk of recurrence among men in certain risk groups. 15373974_Observational study of gene-disease association. (HuGE Navigator) 15375600_Observational study of gene-disease association. (HuGE Navigator) 15381816_Observational study of gene-disease association. (HuGE Navigator) 15381817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15448105_Common sequence variation in the VDR gene has no major effect in type 1 diabetes. 15448105_Observational study of gene-disease association. (HuGE Navigator) 15455736_Observational study of gene-disease association. (HuGE Navigator) 15459215_Observational study of gene-disease association. (HuGE Navigator) 15472188_MONOCYTE VDR levels are elevated in idiopathic hypercalciuria calcium oxalate stone-formers 15474498_hVDR serine-182 is a primary site for PKA phosphorylation 15478069_Observational study of gene-disease association. (HuGE Navigator) 15478069_significance of the family-based associations found between tuberculosis and FokI-BsmI-ApaI-TaqI and the FA haplotype supports a role for VDR haplotypes, rather than individual genotypes, in susceptibility to tuberculosis 15491743_ApaI, & BsmI (but not FokI) polymorphisms of VDR were more common in patients with fasting idiopathic hypercalciuria & calculi, suggesting a genetic association between 3' VDR alleles, hypercalciuria, & reduced bone mass density in these patients. 15491743_Observational study of gene-disease association. (HuGE Navigator) 15503828_Observational study of gene-disease association. (HuGE Navigator) 15503828_no correlation of VDR gene polymorphisms, as detected by Apal and Taql restriction fragments, in multiethnic Brazilian men (165 patients and 200 controls) with prostate cancer risk and parameters of disease severity was found 15514891_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15524410_Observational study of gene-disease association. (HuGE Navigator) 15549643_Observational study of gene-disease association. (HuGE Navigator) 15552843_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15569417_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15577288_Observational study of gene-disease association. (HuGE Navigator) 15577288_we exami | ENSMUSG00000022479 | Vdr | 211.13555 | 0.8627070 | -0.2130573864 | 0.21065751 | 1.018056e+00 | 3.129805e-01 | 6.944431e-01 | No | Yes | 249.420920 | 27.044386 | 278.169927 | 29.436360 | |
| ENSG00000111817 | 29940 | DSE | protein_coding | Q9UL01 | FUNCTION: Converts D-glucuronic acid to L-iduronic acid (IdoUA) residues. Plays an important role in the biosynthesis of the glycosaminoglycan/mucopolysaccharide dermatan sulfate. {ECO:0000269|PubMed:16505484, ECO:0000269|PubMed:19004833, ECO:0000269|PubMed:7092807, ECO:0000269|Ref.7}. | 3D-structure;Cytoplasmic vesicle;Disease variant;Ehlers-Danlos syndrome;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Isomerase;Manganese;Membrane;Metal-binding;Microsome;Reference proteome;Signal;Transmembrane;Transmembrane helix | PATHWAY: Glycan metabolism; chondroitin sulfate biosynthesis. {ECO:0000305|PubMed:16505484, ECO:0000305|PubMed:19004833, ECO:0000305|PubMed:7092807, ECO:0000305|Ref.7}.; PATHWAY: Glycan metabolism; heparan sulfate biosynthesis. {ECO:0000305|PubMed:16505484, ECO:0000305|PubMed:19004833, ECO:0000305|PubMed:7092807, ECO:0000305|Ref.7}. | The protein encoded by this gene is a tumor-rejection antigen. It is localized to the endoplasmic reticulum and functions to convert D-glucuronic acid to L-iduronic acid during the biosynthesis of dermatan sulfate. This antigen possesses tumor epitopes capable of inducing HLA-A24-restricted and tumor-specific cytotoxic T lymphocytes in cancer patients and may be useful for specific immunotherapy. Mutations in this gene cause inmusculocontractural Ehlers-Danlos syndrome. Alternative splicing results in multiple transcript variants. A related pseudogene has been identified on chromosome 9, and a paralogous gene exists on chromosome 18. [provided by RefSeq, Apr 2016]. | hsa:29940; | cytoplasmic vesicle membrane [GO:0030659]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; chondroitin-glucuronate 5-epimerase activity [GO:0047757]; metal ion binding [GO:0046872]; chondroitin sulfate biosynthetic process [GO:0030206]; chondroitin sulfate metabolic process [GO:0030204]; dermatan sulfate biosynthetic process [GO:0030208]; dermatan sulfate metabolic process [GO:0030205]; heparan sulfate proteoglycan biosynthetic process [GO:0015012] | 19004833_Identification of the active site of DS-epimerase 1 and requirement of N-glycosylation for enzyme function. 22350411_Dermatan sulfate epimerase 1 was highly upregulated in esophagus squamous cell carcinoma 23704329_study identified a homozygous DSE missense mutation (c.803C>T, p.S268L) in a male child with musculocontractural type of Ehlers-Danlos syndrome; data indicate mutation affects the epimerase activity, resulting in reduced dermatan sulfate (DS) biosynthesis and an increased synthesis or an accumulation or reduced conversion of chondroitin sulfate 29864158_Study showed that DSE is frequently upregulated in human glioma tissue and cell lines and associated with a worse tumor grade and poor overall survival. Its knockdown suppresses malignant phenotypes, whereas DSE overexpression enhances glioma cell malignancy, both in vitro and in vivo. Mechanically, DSE modulates HB-EGF-induced EGFR/ErbB2 activity and downstream signaling. 29976758_DS-epi1, DS-epi2, and D4ST1 form homomers and are all part of a hetero-oligomeric complex where D4ST1 directly interacts with DS-epi1, but not with DS-epi2. The cooperation of DS-epi1 with D4ST1 may therefore explain the processive mode of the formation of iduronic acid blocks. 31972438_Dermatan sulfate epimerase 1 expression and mislocalization may interfere with dermatan sulfate synthesis and breast cancer cell growth. | ENSMUSG00000039497 | Dse | 689.66825 | 0.9775593 | -0.0327438954 | 0.12807421 | 6.532511e-02 | 7.982692e-01 | 9.433381e-01 | No | Yes | 952.691170 | 161.471406 | 922.869774 | 152.984378 |
| ENSG00000112186 | 10486 | CAP2 | protein_coding | P40123 | FUNCTION: May have a regulatory bifunctional role. | Acetylation;Alternative splicing;Cell membrane;Membrane;Phosphoprotein;Reference proteome | This gene was identified by its similarity to the gene for human adenylyl cyclase-associated protein. The function of the protein encoded by this gene is unknown. However, the protein appears to be able to interact with adenylyl cyclase-associated protein and actin. [provided by RefSeq, Jul 2008]. | hsa:10486; | cytoplasm [GO:0005737]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; actin binding [GO:0003779]; adenylate cyclase binding [GO:0008179]; identical protein binding [GO:0042802]; activation of adenylate cyclase activity [GO:0007190]; cAMP-mediated signaling [GO:0019933]; cell morphogenesis [GO:0000902]; cytoskeleton organization [GO:0007010]; establishment or maintenance of cell polarity [GO:0007163]; regulation of adenylate cyclase activity [GO:0045761]; signal transduction [GO:0007165] | 17000669_CAP2 is up-regulated in human cancers and that is possibly related to multistage hepatocarcinogenesis. 23022774_An important conserved function of CAP2 in higher vertebrates may be associated with the process of skeletal muscle development. 26374196_CAP2 overexpression is a novel prognostic marker in malignant melanoma. CAP2 expression seems to increase stepwise during tumor progression, suggesting the involvement of CAP2 in the aggressive behavior of malignant melanoma. 27573674_CAP2 is upregulated in breast cancer and is associated with the expression of progesterone receptor and patient survival 30047046_CAP2 is a Valuable Biomarker for Diagnosis and Prognostic in Patients with Gastric Cancer. 30518548_CAP2 joins other regulators of actin dynamics demonstrated to cause human genetic diseases. Our study is the first to demonstrate a mutation in CAP2 in human and a direct role for mutated CAP2 and perturbation of actin dynamics in the pathogenesis of DCM and possibly also conduction abnormalities in humans. 31640951_CAP2 is highly expressed in gastric cancer tissues in close relation with the tumor progression. CAP2 is an independent risk factor for 5-year survival rate after radical gastrectomy for gastric cancer. 31871199_Study used lysine-to-glutamine mutations to map the relevant lysines on actin for INF2 regulation. K50Q- and K61Q-actin, when bound to CAP2, inhibit full-length INF2 but not INF2 lacking inhibitory domain (DID). CAP WH2 domain binds INF2-DID with submicromolar affinity but has weak affinity for actin monomers, while INF2-C-terminal diaphanous autoregulatory domain binds CAP/K50Q-actin 5-fold better than CAP/WT-actin. 32211793_CAP2 expression in ovarian cancer is an independent prognostic factor for recurrence-free survival. 34294609_Endoplasmic Reticulum Stress Induces CAP2 Expression Promoting Epithelial-Mesenchymal Transition in Liver Cancer Cells. 35163460_Analysis of mRNA and Protein Levels of CAP2, DLG1 and ADAM10 Genes in Post-Mortem Brain of Schizophrenia, Parkinson's and Alzheimer's Disease Patients. | ENSMUSG00000021373 | Cap2 | 221.81711 | 1.0234694 | 0.0334679546 | 0.19553086 | 2.930568e-02 | 8.640751e-01 | 9.641768e-01 | No | Yes | 269.499091 | 49.427494 | 248.129560 | 44.574329 | |
| ENSG00000112282 | 9439 | MED23 | protein_coding | Q9ULK4 | FUNCTION: Required for transcriptional activation subsequent to the assembly of the pre-initiation complex (By similarity). Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional pre-initiation complex with RNA polymerase II and the general transcription factors. Required for transcriptional activation by adenovirus E1A protein. Required for ELK1-dependent transcriptional activation in response to activated Ras signaling. {ECO:0000250, ECO:0000269|PubMed:10353252, ECO:0000269|PubMed:14759369, ECO:0000269|PubMed:16595664}. | 3D-structure;Activator;Alternative splicing;Direct protein sequencing;Disease variant;Mental retardation;Nucleus;Reference proteome;Transcription;Transcription regulation | The activation of gene transcription is a multistep process that is triggered by factors that recognize transcriptional enhancer sites in DNA. These factors work with co-activators to direct transcriptional initiation by the RNA polymerase II apparatus. The protein encoded by this gene is a subunit of the CRSP (cofactor required for SP1 activation) complex, which, along with TFIID, is required for efficient activation by SP1. This protein is also a component of other multisubunit complexes e.g. thyroid hormone receptor-(TR-) associated proteins which interact with TR and facilitate TR function on DNA templates in conjunction with initiation factors and cofactors. This protein also acts as a metastasis suppressor. Several alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jul 2012]. | hsa:9439; | core mediator complex [GO:0070847]; mediator complex [GO:0016592]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription regulator complex [GO:0005667]; transcription coactivator activity [GO:0003713]; positive regulation of gene expression [GO:0010628]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; transcription initiation from RNA polymerase II promoter [GO:0006367] | 11934987_Sur2(CRSP3) is required for activation by Adenovirus E1A CR3 activation domain and is required for MAPK-mediated activation of Elk-1. 12242338_DRIP130 regulates HER2 expression by binding to ESX 16964286_loss of DRIP-130 expression, as a result of the gross loss of human chromosome 6q16.3-q23, provokes increased tumor metastasis 18660489_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21868677_missense mutation (p. R617Q) in MED23 that cosegregates with nonsyndromic autosomal recessive intellectual disability; mutation impaired response of JUN and FOS to mitogens by altering interaction between enhancer-bound transcription factors and Mediator 22988093_Data show that Med23 RNAi specifically inhibits the proliferation and tumorigenicity of lung cancer cells with hyperactive Ras activity. 23340209_Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. 25273169_Upregulation of mediator MED23 in non-small-cell lung cancer promotes the growth, migration, and metastasis of cancer cells. 25684393_MED23 plays an important role in hepatocarcinogenesis, and it may be a novel target for HCC therapy. 25845469_MED23-associated intellectual disability has been found in two brothers from a non-consanguineous family. 26152846_intra-tumor heterogeneity data suggest that there could be an under- or over-estimation of the occurrence of MED23 frameshift mutations in microsatellite instability-high colorectal cancers 27311965_This is the first patient with documented refractory epilepsy caused by a novel homozygous pathogenic variant in MED23 expanding the phenotypic spectrum. Identification of the underlying genetic defect in MED23 sheds light on the possible mechanism of complete response to the ketogenic diet in this child. 27579896_a 7-gene signature was identified which correctly predicted the primary prefibrotic myelofibrosis group with a sensitivity of 100% and a specificity of 89%. The 7 genes included MPO, CEACAM8, CRISP3, MS4A3, CEACAM6, HEMGN, and MMP8 27914500_Higher l-arginine was associated with higher risk of Ischemic heart disease (odds ratio and of myocardial infarction, based on 2 SNPs from MED23. 30140054_MED23 adopts an arch-shaped conformation, with an N-terminal domain (Nter) protruding from a large core region. | ENSMUSG00000019984 | Med23 | 2656.23695 | 1.1064855 | 0.1459845864 | 0.09266127 | 2.491504e+00 | 1.144623e-01 | 4.807170e-01 | No | Yes | 3518.491230 | 724.816223 | 2975.663327 | 599.559235 | |
| ENSG00000113119 | 55374 | TMCO6 | protein_coding | Q96DC7 | Alternative splicing;Coiled coil;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:55374; | integral component of membrane [GO:0016021]; nuclear import signal receptor activity [GO:0061608]; protein import into nucleus [GO:0006606] | 19423540_Observational study of gene-disease association. (HuGE Navigator) 19729601_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) | ENSMUSG00000006850 | Tmco6 | 364.68874 | 0.7941755 | -0.3324702803 | 0.19112012 | 2.999861e+00 | 8.327166e-02 | 4.206437e-01 | No | Yes | 393.269816 | 64.317870 | 503.657665 | 80.110627 | |||
| ENSG00000113368 | 4001 | LMNB1 | protein_coding | P20700 | FUNCTION: Lamins are components of the nuclear lamina, a fibrous layer on the nucleoplasmic side of the inner nuclear membrane, which is thought to provide a framework for the nuclear envelope and may also interact with chromatin. {ECO:0000269|PubMed:28716252, ECO:0000269|PubMed:32910914}. | 3D-structure;Acetylation;Chromosomal rearrangement;Coiled coil;Direct protein sequencing;Disease variant;Disulfide bond;Intermediate filament;Isopeptide bond;Leukodystrophy;Lipoprotein;Mental retardation;Methylation;Nucleus;Phosphoprotein;Prenylation;Primary microcephaly;Reference proteome;Ubl conjugation | This gene encodes one of the two B-type lamin proteins and is a component of the nuclear lamina. A duplication of this gene is associated with autosomal dominant adult-onset leukodystrophy (ADLD). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2015]. | hsa:4001; | lamin filament [GO:0005638]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; nuclear inner membrane [GO:0005637]; nuclear lamina [GO:0005652]; nuclear matrix [GO:0016363]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; phospholipase binding [GO:0043274]; sequence-specific double-stranded DNA binding [GO:1990837]; structural molecule activity [GO:0005198]; nuclear envelope organization [GO:0006998] | 12898336_colocalizes with lamin B1 in the nucleoplasm and around the nuclear rim during S-phase of cells transfected with EBNA-1 in the absence of EBV plasmids. 14504265_organization of the nuclear envelope and lamina is dependent on a mechanism involving the methylation of lamin B1 15284226_the distinctive ensemble of heterotypic lamin interactions in a particular cell type affects the stability of the lamin polymer 16283426_We now show that epitope masking in the nucleus is often responsible for failure to detect emerin and lamins in human, rat and pig tissues.These data suggest that different regions of the lamin B1 molecule are masked in different tissues. 16365157_Apoptotic neutrophils express lamin B1 on their surface; these cells may participate in the development of autoantibodies directed against cytoskeletal proteins, a condition frequently reported in several inflammatory diseases. 16410549_Results suggest that the C-terminus of nuclear titin binds lamins A and B in vivo and might contribute to nuclear organization during interphase. 16543417_lamin B was essential for the formation of the mitotic matrix that tethers a number of spindle assembly factors; propose that lamin B is a structural component of the spindle matrix that promotes microtubule assembly and organization in mitosis 18334554_Results show that a lamin B1-containing nucleoskeleton is required to maintain RNA synthesis and that ongoing synthesis is a fundamental determinant of global nuclear architecture in mammalian cells. 18524819_lamin A/C, lamin B1, and viral US3 kinase have roles in viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane 19001169_Novel duplication on chromosomal band 5q23.2 in a French Canadian family with autosomal dominant leukodystrophy that supports the implication of duplicated LMNB1 as the disease-causing mutation. 19141474_Silencing lamin B1 expression dramatically increases the lamina meshwork size and the mobility of nucleoplasmic lamin A 19151023_duplication of the lamin B1 gene (LMNB1) has recently been described in a rare form of autosomal dominant adult-onset leukoencephalopathy. 19198602_Nudel regulates microtubule organization in part by facilitating assembly of the lamin B spindle matrix in a dynein-dependent manner. 19348623_Observational study of gene-disease association. (HuGE Navigator) 19348623_Our work indicates that lamin B1 defects are probably not responsible for signs and symptoms resembling multiple sclerosis. 19383719_Lamin B1 maintains the functional plasticity of nucleoli. 19522540_Proteomics identified lamin B1 as being significantly upregulated in HCC tumors and present in patients' plasma. 19961535_results suggest that a LMNB1 regulatory sequence mutation underlies the variant adult-onset autosomal dominant leukodystrophy (ADLD) phenotype; adult forms of ADLD linked to 5q23 may be more heterogeneous clinically and genetically than previously thought 20004208_These findings indicate that a lamin dimer principally has the freedom for a 'combinatorial' head-to-tail association with all types of lamins, a property that might be of significant importance for the assembly of the nuclear lamina. 20816241_Autosomal dominant leukodystrophy is the first disease that has ever been linked to lamin B1 mutations and it expands the pathological role of the nuclear lamia to include disorders of the brain. 21225301_SNP array analysis revealed novel duplications spanning the entire LMNB1 gene in probands from each of four adult-onset autosomal dominant leukodystrophy families 21909802_This study demonistrated that Adult-onset autosomal dominant leukodystrophy due to LMNB1 gene duplication. 22155925_LB1 expression in WI-38 cells decreases during cellular senescence 22246186_The authors show that oxidative stress increases lamin B1 levels through p38 Mitogen Activated Protein kinase activation. 22265972_crystal structures of lamin B1 globular tail domain and coiled 2B domain, with similar folds to Ig-like domain and coiled-coil domain of lamin A. Found an extra intermolecular disulfide bond in lamin B1 coil 2B domain, which does not exist in lamin A/C. 22496421_Lamin B1 is lost from primary human and murine cell strains when they are induced to senesce. 23261988_Results indicate that lamin B1 (LMNB1) accumulation in adult-onset autosomal dominant leukodystrophy (ADLD) is associated with Oct-1 recruitment. 23439683_LMNB1 protein levels decline in senescent human dermal fibroblasts and keratinocytes, mediated by reduced transcription and inhibition of LMNB1 messenger ribonucleic acid translation by miRNA-23a. 23475125_Treating normal human fibroblasts with farnesyltransferase inhibitors causes the accumulation of unprocessed lamin B2 and lamin A and a decrease in mature lamin B1. 23649844_detailed molecular analysis of the largest collection of autosomal dominant leukodystrophy (ADLD) families studied, to date; identified the minimal duplicated region necessary for the disease, defined all the duplication junctions at the nucleotide level and identified the first inverted LMNB1 duplication 23681646_we confirm the underlying role of lamin B1 duplication, regardless of the autonomic malfunction onset in Adult-onset autosomal dominant leukodystrophy 23733478_Rare variants of LMNB1 may contribute to susceptibility to neural tube defects. 23857605_Lamin B1 plays an important role in pancreatic cancer pathogenesis and is a novel therapeutic target of betulinic acid treatment. 23873483_The regulation of lamin B1 is important for cellular physiology and disease.To how perturbations of lamin B1 affect cellular physiology and discuss the implications this has on senescence, HGPS and ADLD. 23934658_lamin B1 down-regulation in senescence is a key trigger of global and local chromatin changes that impact gene expression, aging, and cancer 23964094_LMNB1 may contribute to senescence in at least two ways due to its uneven genome-wide redistribution: first, through the spatial reorganization of chromatin and, second, through gene repression. 24293108_Higher levels of A-type lamins and lamin B1 mRNA expression were seen in associated non-cancerous tissue. Higher lamin A/C expression was associated with the early clinical breast cancer stage, with better clinical outcomes. 24297448_This led us to propose a model where the nucleolus has steady-state stiffness dependent on ribosome biogenesis activity and requires LaminB1 for its flexibility. 24732130_LMNB1 is required to maintain chromatin condensation in interphase nuclei. 24858279_Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. 25477337_Nuclear envelope remodelling during human spermiogenesis involves somatic B-type lamins and a spermatid-specific B3 lamin isoform. 25535332_maintenance of lamin B1 levels is required for DNA replication and repair through regulation of the expression of key factors involved in these essential nuclear functions 25637521_Deregulation of LMNB1 expression induces modified splicing of several genes, likely driven by raver-2 overexpression, and suggest that an alteration of mRNA processing could be a pathogenic mechanism in adult-onset autosomal dominant leukodystrophy. 25701871_An upstream mutation alters LMNB1 gene expression in autosomal dominant adult-onset demyelinating leukodystrophy. 25733566_Data indicate that lamin B1 promotes DNA double-strand breaks (DSBs) repair and cell survival. 25807068_The tail domain of lamin B1 is more strongly modulated by divalent cations than lamin A. 26053668_LMNB1-related autosomal-dominant leukodystrophy is a slowly progressive neurological disease with survival lasting more than two decades after clinical onset. 26311780_Lamin B1 levels are increased in oligodendrocytes, the cell type that produces myelin in the central nervous system. 26876308_Data show that lamins A and B are differently processed in staurosporine and beta-Amyloid peptide fragments Abeta42-treated cells. 27503760_down-regulation of Lamin B1 and up-regulation of Nephroblastoma overexpressed (NOV) are at least partially responsible for the inhibitory effect of Huaier on the proliferative and invasive capacity of SKHEP-1 cells 27760841_The primary response of cells to various stresses leading to senescence consists of the down-regulation of LBR and LB1 to attain reversal of the chromatin architecture. 27926867_We show that epithelial cells failing to undergo proliferation arrest during TGF-beta-induced EMT sustain mitotic abnormalities due to failed cytokinesis, resulting in aneuploidy. This genomic instability is associated with the suppression of multiple nuclear envelope proteins implicated in mitotic regulation and is phenocopied by modulating the expression of LaminB1. 28229933_These results suggest that the nuclear lamins and progerin have marginal roles in the activation of the antioxidant Nrf2 response to arsenic and cadmium. 28844980_In this report we show that increased self-association propensity of mutant LA modulates the LA-LB1 interaction and precludes the formation of an otherwise uniform laminar network. Our results might highlight the role of homotypic and heterotypic interactions of LA in the pathogenesis of DCM and hence laminopathies in the broader sense. 29115590_The aim of the present study was to elucidate the influence of LMNB1 upregulation on colon cancer cell line after treatment with 5-FU. The results indicate, that overexpression of LMNB1 induced dose-dependent cell death mainly by mitotic catastrophe pathway. 29142250_we demonstrate that loss of lamin B1 facilitates the detection and quantification of senescent cells upon UV-exposure in vitro and upon chronic UV-exposure and skin regeneration in vivo. 29417283_We observed reduced detection of lamin B1 in the ectopic endometrium raising the possibility that the presence of senescent cells might be contributing to the maintenance and progression of endometriosis by apoptosis resistance and peritoneal stress inherent of the disease. 29676528_E3 ubiquitin ligase RNF123 targets LMNB1, Rb protein and LAP2alpha for proteasomal degradation. 29753763_Results show that HECW2 interacts with lamin B1 mediating its ubiquitination and proteasomal degradation. 30692212_The lamin B1 reduction is not only a hallmark of lung aging but is also involved in the progression of cellular senescence during COPD pathogenesis through aberrant MTOR signaling. 30762072_TPR contributes to the organization of the nuclear lamina and in cooperation with lamin B1 and lamin A-C guards the interphase assembly of nuclear pore complexes. 31015297_loss of a single lamin B1 allele induced spontaneous lung tumor formation and RET activation. 31016526_The mRNA expression of LaminB1 displayed difference in cochlear tissues. 31306099_upregulated in prostate cancer tissues 31532069_The tau/MSI aggregates induce structural changes to LaminB1, leading to nuclear instability. 32349647_Development and Optimization of a High-Content Analysis Platform to Identify Suppressors of Lamin B1 Overexpression as a Therapeutic Strategy for Autosomal Dominant Leukodystrophy. 32787434_Lamin B1 deficiency promotes malignancy and predicts poor prognosis in gastric cancer. 32910914_De Novo Variants in LMNB1 Cause Pronounced Syndromic Microcephaly and Disruption of Nuclear Envelope Integrity. 32987785_Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization. 33033404_Heterozygous lamin B1 and lamin B2 variants cause primary microcephaly and define a novel laminopathy. 33112662_Lamin B1 promotes tumor progression and metastasis in primary prostate cancer patients. 33155082_Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. 33468570_Disease Modeling with Human Neurons Reveals LMNB1 Dysregulation Underlying DYT1 Dystonia. 33706103_Beta-strand-mediated dimeric formation of the Ig-like domains of human lamin A/C and B1. 33893211_Timing of pacemaker and ICD implantation in LMNA mutation carriers. 33956061_Screening and identification of LMNB1 and DLGAP5, two key biomarkers in gliomas. 34403147_The alkaloid centcyamine increases expression of klotho and lamin B1, slowing the onset of skin ageing in vitro and in vivo. 34452908_Lamin B1 sequesters 53BP1 to control its recruitment to DNA damage. 34469544_Increase in lamin B1 promotes telomere instability by disrupting the shelterin complex in human cells. 35247231_Genome sequencing reveals novel noncoding variants in PLA2G6 and LMNB1 causing progressive neurologic disease. 35269619_Altered p16(Ink4a), IL-1beta, and Lamin b1 Protein Expression Suggest Cellular Senescence in Deep Endometriotic Lesions. 35278369_Lamin B1 deletion in myeloid neoplasms causes nuclear anomaly and altered hematopoietic stem cell function. | ENSMUSG00000024590 | Lmnb1 | 6827.49570 | 1.0107450 | 0.0154190211 | 0.08252195 | 3.492335e-02 | 8.517564e-01 | 9.598571e-01 | No | Yes | 9778.739068 | 1376.485125 | 9077.252191 | 1249.401807 | |
| ENSG00000113719 | 57222 | ERGIC1 | protein_coding | Q969X5 | FUNCTION: Possible role in transport between endoplasmic reticulum and Golgi. {ECO:0000303|PubMed:15308636}. | Alternative splicing;Direct protein sequencing;Disease variant;ER-Golgi transport;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a cycling membrane protein which is an endoplasmic reticulum-golgi intermediate compartment (ERGIC) protein which interacts with other members of this protein family to increase their turnover. [provided by RefSeq, Jul 2008]. | hsa:57222; | COPII-coated ER to Golgi transport vesicle [GO:0030134]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; integral component of endoplasmic reticulum membrane [GO:0030176]; integral component of Golgi membrane [GO:0030173]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum [GO:0006890] | 15308636_ERGIC-32 functions as a modulator of the hErv41-hErv46 complex by stabilizing hErv46 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22761906_AIM1, ERGIC1, and TPX2 were shown to be highly expressed especially in prostate cancer tissues, and high mRNA expression of ERGIC1 and TMED3 associated with AR and ERG oncogene expression 28317099_Homozygous pathogenic variant in the endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 protein (ERGIC1) gene was identified in arthrogryposis multiplex congenita (AMC) neuropathic type. 28970727_Data indicate that abnormal ERGIC1 and DNA-PKcs expression may play an important role in gastric cancer initiation. 29549924_ERGIC1 might play an inhibitory role in the initiation and progression of gastric cancer. 34037256_Bi-allelic loss of ERGIC1 causes relatively mild arthrogryposis. | ENSMUSG00000001576 | Ergic1 | 3350.43502 | 0.8598833 | -0.2177871639 | 0.07613097 | 8.133003e+00 | 4.346670e-03 | 1.090419e-01 | No | Yes | 3624.986582 | 307.844827 | 4101.864824 | 340.129206 | |
| ENSG00000114270 | 1294 | COL7A1 | protein_coding | Q02388 | FUNCTION: Stratified squamous epithelial basement membrane protein that forms anchoring fibrils which may contribute to epithelial basement membrane organization and adherence by interacting with extracellular matrix (ECM) proteins such as type IV collagen. | Alternative splicing;Basement membrane;Cell adhesion;Collagen;Direct protein sequencing;Disease variant;Disulfide bond;Epidermolysis bullosa;Extracellular matrix;Glycoprotein;Hydroxylation;Protease inhibitor;Reference proteome;Repeat;Secreted;Serine protease inhibitor;Signal | This gene encodes the alpha chain of type VII collagen. The type VII collagen fibril, composed of three identical alpha collagen chains, is restricted to the basement zone beneath stratified squamous epithelia. It functions as an anchoring fibril between the external epithelia and the underlying stroma. Mutations in this gene are associated with all forms of dystrophic epidermolysis bullosa. In the absence of mutations, however, an acquired form of this disease can result from an autoimmune response made to type VII collagen. [provided by RefSeq, Jul 2008]. | hsa:1294; | basement membrane [GO:0005604]; collagen type VII trimer [GO:0005590]; collagen-containing extracellular matrix [GO:0062023]; COPII-coated ER to Golgi transport vesicle [GO:0030134]; endoplasmic reticulum lumen [GO:0005788]; endoplasmic reticulum-Golgi intermediate compartment membrane [GO:0033116]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; extracellular matrix structural constituent conferring tensile strength [GO:0030020]; serine-type endopeptidase inhibitor activity [GO:0004867]; cell adhesion [GO:0007155]; endodermal cell differentiation [GO:0035987]; epidermis development [GO:0008544] | 11843659_glycine substitution mutations in COL7A1 are associated with dominant familial dystrophic toenail changes 11986329_bone morphogenetic protein-1 (BMP-1), which exhibits procollagen C-proteinase activity, cleaves the C-terminal propeptide from human procollagen VII 12060403_The epidermolysis bullosa acquisita antigen (type VII collagen) is present in human colon and patients with crohn's disease have autoantibodies to type VII collagen. 12353709_A novel splice site mutation in collagen type VII gene in a Chinese family with dominant dystrophic epidermolysis bullosa pruriginosa. 12485454_Observational study of genotype prevalence. (HuGE Navigator) 12787118_May contribute to flexibility of linker of fibronectin type III domains and may affect interactions between noncollagenous 1 domain and extracellular matrix proteins. May have role in dermal-epidermal adhesion, wound healing, and skin remodeling. 14727126_R578X, 7786delG, and R2814X mutations are specifically limited to British patients, and the mutations 5818delC, 6573+1G-->C, and E2857X are frequent in Japanese patients. 15113589_identical COL7A1 glycine substitutions can cause remarkably heterogeneous clinical phenotypes 15365990_The predicted rates of AA substitutions for glycine were compared with missense mutations that have been observed to cause disease. Any Gly replacement will cause disease & the level of triple-helix destabilization determines clinical outcome. 15774758_collagen VII required for Ras-driven epidermal tumorigenesis by enhancing tumor cell invasion; retention of NC1 sequences in a subset of recessive dystrophic epidermolysis bullosa patients may contribute to their susceptibility to squamous cell carcinoma 15810887_TNF-alpha and IL-1beta enhance the TGF-beta-mediated up-regulation of COL7A1 expression in HaCaT keratinocytes 15816848_Spectrum of mutations in dystrophic epidermolysis bullosa and cryptic splicing 15888141_Observational study of gene-disease association. (HuGE Navigator) 16470588_COL7A1 hemizygosity and a missense mutation with complex effects on splicing may be causative in recessive dystrophic epidermolysis bullosa 16500083_preimplantation genetic diagnosis for Hallopeau-Siemens recessive dystrophic epidermolysis bullosa 16923137_Two boys with dystrophic epidermolysis bullosa and their fathers revealed a heterozygous nucleotide G to A transition at position 6109 and 6082 in 73 exon of COL7A1, which resulted in a glycine to arginine substitution (G2037R and G2028R), respectively. 16971478_Mutations from more than 1000 families with different forms of epidermolysis bullosa were analyzed. 242 mutations were distinct and 138 were novel, previously unreported mutations. 17106611_Epidermolysis bullosa pruriginosa due to a glycine substitution missense mutation in the COL7A1-gene 17229600_Glycine subsstitution in this protein underlies mild recessive dystrophic epidermolysis bullosa, showing that type VII collagen is tolefant of heterozygous glycine substitution. 17336503_description of a novel glycine substitution mutation in COL7A1 in a Chinese pedigree with dominant epidermolysis bullosa pruriginosa 17425959_We report 14 Australian families with different forms of dystrophic epidermolysis bullosa (DEB) with 23 different COL7A1 allelic variants, nine of which were novel. 17495952_Individuals with recessive dystrophic epidermolysis bullosa can develop squamous-cell carcinoma regardless of type VII collagen expression and that additional factors have a role in explaining the high incidence of tumors complicating this genodermatosis. 17525268_Data show that the cartilage matrix protein subdomain of type VII collagen is pathogenic for epidermolysis bullosa acquisita. 17900868_Identify novel mutation in COL7A1 responsible for dominant dystrophic epidermolysis bullosa in a Chinese family. More severe phenotype observed in female members. 17916216_Mutational analysis revealed 30 pathogenic COL7A1 mutations among a total of 33 allels, identifying 10 novel and 14 previously adentified mutations. 18331784_The expression of COL7A1 mrna was higher in malignant tissue and was correlated with depth of tumor invasion and lymphatic invasion in ESCC. 18374850_Dystrophic epidermolysis bullosa may present in generalized or localized forms and the disease may be inherited in either autosomal dominant or recessive mode. Genetic analysis shows mutations in COL 7A1 in this case 18429782_Mutations in the gene for collagen VII (COL7A1) have been documented in both types of dystrophic epidermolysis bullosa. 18440202_A p.Glu2857X mutation exhibits mild pathogenic effects in COL7A1, and its uniqueness enables detailed analysis and comparison of the destabilizing effects of missense mutations in dystrophic epidermolysis bullosa patients. 18450758_known recessive DEB C7 mutations perturb critical functions of the C7 molecule and may have a role in dystrophic epidermolysis bullosa 18496702_linked to dystrophic epidermolysis bullosa in Tunisian consanguineous families 18558993_characterization of COL7A1 mutations in dystrophic epidermolysis bullosa [review] 19197535_Observational study of gene-disease association. (HuGE Navigator) 19197535_The increased type VII collagen degradation was suspected to trigger an inflammatory response leading to itchy skin in EB pruriginosa. All 27 with EB pruriginosa were heterozygous for dominant-negative glycine substitution mutations in the COL7A1 gene. 19250433_novel glycine substitution mutation in the COL7A1 gene in three affected family members with dystrophic epidermolysis bullosa 19367581_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19422682_Results suggest that the down-regulation of alpha 6(IV) mRNA coincides with the acquisition of invasive growth properties, whereas alpha1(IV) and alpha1(VII) mRNAs were up-regulated already in dysplastic tissue. 19435799_Results describe the effect of loss of collage type VII on squamous cell carcinoma tumourigenesis using RNA interference in a 3D organotypic skin model. 19486043_new variants of COL7A1 mutations underlying epidermolysis bullosa pruriginosa 19665875_Pseudosyndactyly occurs in approximately half of recessive dystrophic epidermolysis bullosa (RDEB-O) patients when type VII collagen is strongly reduced. The prognosis in RDEB cannot always be simply predicted from the COL7A1 genotype. 19681861_Observational study of gene-disease association. (HuGE Navigator) 19681861_Results disclosed 42 novel COL7A1 mutations, including the first large genomic deletion of 4 kb affecting only the COL7A1 gene, and three apparently silent mutations affecting splicing. 19945621_Dystrophic epidermolysis bullosa (DEB) emerged as a candidate for type VII collagen mutations becausing anchoring fibrils were shown to be morphologically altered, reduced in number, or completely absent in patients 20184583_Although the COL7A1 database indicates that most dystrophic epidermolysis bullosa mutations are family specific, the pathogenic mutation c.6527insC was highly recurrent in this cohort, this recurrence level has never previously been reported for COL7A1. 20555349_six new genetic mutations were found in collagen type VII for inversa dystropic epidermolysis bullosa 20598510_Analysis of the COL7A1 gene in Czech patients with dystrophic epidermolysis bullosa reveals novel and recurrent mutations. 20920254_Observational study of gene-disease association. (HuGE Navigator) 21113014_Recessive dystrophic epidermolysis bullosa-I is associated with specific recessive arginine and glycine substitutions in the triple helix domain of type VII collagen. 21196708_novel compound heterozygous recessive COL7A1 missense mutations in 2 siblings presenting different Dystrophic epidermolysis bullosa clinical subtypes 21448560_In screening the COL7A1 gene for mutations in individuals with dominant/recessive dystrophic epidermolysis bullosa our data highlight that delineation of glycine substitutions in type VII collagen has important implications for genetic counselling. 21482078_NC1 and NC2 domains of type VII collagen have a role in of epidermolysis bullosa acquisita 21574979_Mutation in this gene results in skipping of exon 87 leading to aberrant splicing of this exon. 21658117_Two cases of recessive dystrophic epidermolysis bullosa revealed heteroallelic recessive mutations which resulted in premature termination codons. 21879237_we have identified three pathogenic COL7A1 mutations (G1773R, splicing site mutation of c.6900+1G>C, and G2701W) in 3 dystrophic epidermolysis bullosa pruriginosa families. 22070715_The infant's genomic DNA from blood was found to be heterozygous for a missense mutation on exon 54 of COL7A1 (c.5017G > A, p.G1673R) not previously described in bullous dermolysis of the newborn. 22209565_Letter: report novel and recurrent COL7A1 mutations in Chilean patients with dystrophic epidermolysis bullosa. 22266148_We present the first COL7A1 mutation analysis in Polish dystrophic epidermolysis bullosa patients. 22515571_The wide diversity of clinical phenotypes with one underlying genotype demonstrates that COL7A1 mutations are incompletely penetrant and strongly suggests that other genetic and environmental factors influence clinical presentation. 22757647_This study confirms unequivocally that the c.6527insC mutation in the COL7A1 gene, a cause of recessive dystrophic epidermolysis bullosa, originated from a single ancestor. 22974128_Novel deletion mutation (c.3717del5) in COL7A1 in a patient with recessive dystrophic epidermolysis bullosa. 23106673_We describe three families with multiple affected members in which epidermolysis bullosa prurigosa variant shows autosomal-dominance and all three previously unreported COL7A1 mutations were identified. 23226319_novel disease-causing mutations in the COL7A1 gene 23397949_We report six Chinese cases with Epidermolysis Bullosa Pruriginosa, who had four novel and two previously reported mutations leading to glycine substitutions of COL7A1. 23591773_Loss of collagen VII has a global impact on the cellular microenvironment in recessive dystrophic epidermolysis bullosa patients. 23624125_Mutation in COL7A1 caused a broad range of severity of disease in a family with pretibial epidermolysis bullosa. 23679163_We report the first case of HS-RDEB homozygous PTC mutations of 5818delC in both COL7A1 alleles. 23688405_Our long-term observational study showed that this in-frame exon skipping mutation was conversely highly predictive of the pruriginosa phenotype and characterized by a very variable phenotype in terms of severity of disease. 23769655_data further enhance the mutation spectrum of the LAMB3 and the COL7A1 genes, and also underscore the crucial roles of these genes in pathogenesis of epidermolysis bullosa 23834951_Data suggest that, of the five basement membrane types present in term placental tissue and fetal membranes, just one, that associated with amnion epithelium, expresses type VII collagen. 23947675_immortalized and cloned recessive dystrophic epidermolysis bullosa keratinocytes carrying the c.6527insC mutation 24117545_Bullous dermolysis of the newborn (BDN) is a subtype of dystrophic epidermolysis bullosa caused by mutations in type VII collagen resulting in disorganized anchoring fibrils and sublamina densa blister formation. 24127822_anti-type VII collagen autoantibodies fluctuated in parallel with disease activity in epidermolysis bullosa acquisita 24210835_The mutations detected in our 17 DEB patients highlight the presence of both mild (DDEB) and severe phenotypes (RDEB-O and RDEB-sev gen), confirming that a more severe involvement of the oropharyngeal mucosa occurs in RDEB. 24213372_analysis of COL7A1 mutations in patients with recessive dystrophic epidermolysis bullosa 24252097_Results suggest that In children with a moderate form of DEB with no or moderate skin fragility, a glycine substitution near the THD interruption domain of the collagen VII leading to thermolabile protein could explain this phenomenon 24317394_We show that revertant recessive dystrohic epidermolysis bullosa keratinocytes expressing functional C7 can be reprogrammed into induced pluripotent stem cells and self-corrected keratinocytes can be differentiated into epidermal or hematopoietic cells. 24357722_SLCO1B3 expression and promoter activity are modulated by COL7A1 in tumor keratinocytes isolated from recessive dystrophic epidermolysis bullosa. 24732400_TGM2 was identified as a stable interaction partner of collagen VII and is reduced in recessive dystrophic epidermolysis bullosa. 24794830_Case Report: hot spot mutation c.6127G>A in COL7A1 leads to dominant dystrophic epidermolysis bullosa associated with intracellular accumulation of pro-collagean VII. 24810542_The central collagenous domain of Col7 contains several interruptions of the collagen triple helix 24927163_Study demonstrated that versican, TGFbeta1, Col7A1 and ITGbeta3 are up-regulated in isolated Cancer stem cells. 25425313_COL7A1 mutation was diagnosed with next generation sequencing in patient with dystrophic epidermolysis bullosa. 25556825_This study is conducive to highlighting the phenotypic diversity of EBP, expanding the database on COL7A1 mutations in EBP and laying the foundation for this family's prenatal genetic counselling. 25566895_A novel dominantnegative heterozygous acceptor splice site mutation in the COL7A1 gene (IVS671G>T) was found in both our patient and his youngest son. 25639640_Collagen Type VII missense mutation is responsible for the development of recessive bullous epidermolysis. 25689103_autoantibodies to COL7, independent of the targeted epitopes, induce blisters both ex vivo and in vivo 26066885_Gene therapy is successful in the treatment of hereditary epidermolysis bullosa dystrophica. 26289024_A total of 50% of the pro-alpha1 (VII) procollagen chains will contain the dominant COL7A1 mutation if a DDEB patient carries one mutant COL7A1 in 100% of skin cells, which will lead to dystrophic epidermolysis bullosa 26472200_In conclusion, we identified a Japanese founder recurrent mutation of c.6216 + 5G > T, inducing aberrant splicing of COL7A1 and tending to cause a mild phenotype of recessive dystrophic epidermolysis bullosa 26476432_Type VII collagen suppresses TGFbeta signaling and angiogenesis in cutaneous SCC (squamous cell carcinoma). Patients with recessive dystrophic epidermolysis bullosa (RDEB) SCC may benefit from anti-angiogenic therapy. 26568311_TANGO1 is thus pivotal in concentrating procollagen VII in the lumen and recruiting ERGIC membranes on the cytoplasmic surface of the endoplasmic reticulum. 26586712_Novel dystrophic epidermolysis bullosa COL7a1 framshift mutation c.5493delG (p.K1831Nfs*10) in exon 64 leads to a premature termination codon located 10 amino acids downstream in exon 64 (p.K1831Nfs*10) and is expected to result in a loss of function. 26595603_The results in these two brothers show that COL7A1 mutation leads to persistent blistering in adulthood indicating that DEB may persist throughout life in a mild form. 26897595_COL7A1 mutations have a role in Recessive Dystrophic Epidermolysis Bullosa and can be corrected meganuclease-mediated homology-directed repair 27117059_expression restored to recessive dystrophic epidermolysis bullosa skin by topical gene therapy 27328306_miR-29 Regulates COL7A1 in Recessive Dystrophic Epidermolysis Bullosa, directly through targeting its 3' untranslated region at two distinct seed regions and indirectly through targeting an essential transcription factor required for basal COL7A1 expression, SP1. 27790721_we have identified a novel glycine substitution mutation of the COL7A1 gene in two unrelated Scottish families with a DDEB phenotype. This mutation abolishes the donor splice site and results in in-frame exon skipping. This leads to dominant negative interference between the wild-type and truncated-type collagen proteins resulting in a mild phenotype. 27899325_COL7A1 harbored mutations in the overwhelming majority of patients with dystrophic epidermolysis bullosa, and most of them in this Iranian cohort were consistent with autosomal recessive inheritance. Ninety percent of these mutations were homozygous recessive, reflecting consanguinity in these families. 28126522_Patients with RDEB carry mutations in the COL7A1 gene encoding for type VII collagen, the main component of anchoring fibrils, microstructures responsible for the anchorage of the epidermis to the underlying dermis. [review] 28164502_Case Report: glycine substitution specific to COL7A1, exon 110, was identified in a Chinese family with epidermolysis bullosa pruriginosa. 28800953_COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa patients' keratinocytes in vitro has been reported. 29182795_Case Report: Epidermolysis ullosa acquisita with previously unrecognized mild dystrophic EB and biallelic COL7A1 missense mutations. 29272047_three unrelated patients with two identical pathogenic compound heterozygous mutations in the COL7A1 gene developed different clinical forms of dystrophic epidermolysis bullosa-epidermolysis bullosa pruriginosa and mild recessive non-Hallopeau-Siemens. 29305555_specifically binds and sequesters the innate immune activator cochlin in the lumen of lymphoid conduits 29473190_In summary, we present 7 novel COL7A1 mutations in a cohort of 17 Mexican RDEB patients, expanding the mutation spectrum in this disease. 29490344_Type VII collagen is distributed particularly at the strained parts of the accommodation system. Type VII collagen was associated with various basement membranes and with ciliary zonules. 29499655_High chimeric COL7A1-UCN2 recurrence is associated with cancer stem cell transition, promoted epithelial-mesenchymal transition in laryngeal cancer. 29504492_Case report of 2 potentially pathogenic variants in COL7A1 occurring on the same allele in members of a family with epidermolysis bullosa pruriginosa and autosomal dominant inheritance. 29512192_In summary, we have found a novel mutation of COL7A1 gene in Chinese patients with DEBPr. 29512197_In both the cases, the variants p.G2692D, p.Q1571* and p.G1667E in the COL7A1 were not previously reported in 1000 genome, Exome Aggregation Consortium (ExAC), Alleles in Middle East and North Africa (al mena) or in a in-house database of over 1000 exomes and genomes from South Asia. 29531004_Mutation in COL7A1 gene is associated with Recessive Dystrophic Epidermolysis Bullosa. 29574987_Study reports novel COL7A1 mutation c.5327G>T (p.G1776V) in patient with dominant dystrophic epidermolysis bullosa-bullous dermolysis of the newborn whose affected family members had typical dominant dystrophic epidermolysis bullosa, nails only. 30930113_Our strategy could potentially be extended to a large number of COL7A1 mutation-bearing exons within the long collagenous domain of this gene, opening the way to precision medicine for Recessive Dystrophic Epidermolysis Bullosa 31709745_A novel mutation of COL7A1 in a Chinese DEB-Pt family and review of the literature. 32396230_What do we learn from dystrophic epidermolysis bullosa, nails only? Idiopathic nail dystrophy may harbor a COL7A1 mutation as the underlying cause. 32484238_Next-generation sequencing through multigene panel testing for the diagnosis of hereditary epidermolysis bullosa in Chinese population. 32926178_Apparent Missense Variant in COL7A1 Causes a Severe Form of Recessive Dystrophic Epidermolysis Bullosa via Effects on Splicing. 33081018_Nonsequential Splicing Events Alter Antisense-Mediated Exon Skipping Outcome in COL7A1. 33258232_Self-improving dystrophic epidermolysis bullosa: First report of clinical, molecular, and genetic characterization of five patients from Southeast Asia. 33974636_Rare functional genetic variants in COL7A1, COL6A5, COL1A2 and COL5A2 frequently occur in Chiari Malformation Type 1. 34286919_Novel and very rare causative variants in the COL7A1 gene of Vietnamese patients with recessive dystrophic epidermolysis bullosa revealed by whole-exome sequencing. 34435747_Ancestral patterns of recessive dystrophic epidermolysis bullosa mutations in Hispanic populations suggest sephardic ancestry. 34512204_Prognostic Value of Highly Expressed Type VII Collagen (COL7A1) in Patients With Gastric Cancer. 34543471_Dystrophic epidermolysis bullosa pruriginosa: a new case series of a rare phenotype unveils skewed Th2 immunity. 34948168_Availability of mRNA Obtained from Peripheral Blood Mononuclear Cells for Testing Mutation Consequences in Dystrophic Epidermolysis Bullosa. 35163654_5'RNA Trans-Splicing Repair of COL7A1 Mutant Transcripts in Epidermolysis Bullosa. | 246.53661 | 1.1187263 | 0.1618571230 | 0.21913563 | 5.452562e-01 | 4.602625e-01 | 7.969323e-01 | No | Yes | 223.777425 | 56.736542 | 202.148690 | 50.159824 | |||
| ENSG00000114354 | 10342 | TFG | protein_coding | Q92734 | FUNCTION: Plays a role in the normal dynamic function of the endoplasmic reticulum (ER) and its associated microtubules (PubMed:23479643, PubMed:27813252). Required for secretory cargo traffic from the endoplasmic reticulum to the Golgi apparatus (PubMed:21478858). {ECO:0000269|PubMed:21478858, ECO:0000269|PubMed:23479643, ECO:0000269|PubMed:27813252}. | Acetylation;Alternative splicing;Chromosomal rearrangement;Coiled coil;Direct protein sequencing;Disease variant;ER-Golgi transport;Endoplasmic reticulum;Hereditary spastic paraplegia;Methylation;Neurodegeneration;Neuropathy;Phosphoprotein;Proto-oncogene;Reference proteome;Transport | There are several documented fusion oncoproteins encoded partially by this gene. This gene also participates in several oncogenic rearrangements resulting in anaplastic lymphoma and mixoid chondrosarcoma, and may play a role in the NF-kappaB pathway. Multiple transcript variants have been found for this gene. [provided by RefSeq, Sep 2010]. | hsa:10342; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum exit site [GO:0070971]; intracellular membrane-bounded organelle [GO:0043231]; identical protein binding [GO:0042802]; COPII vesicle coating [GO:0048208]; endoplasmic reticulum to Golgi vesicle-mediated transport [GO:0006888]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123] | 15188455_TFG was fused to NOR1 is a patient with extraskeletal myxoid chondrosarcoma. 15557341_TFG is a novel protein able to modulate SHP-1 activity. 16547966_TFG enhances the effect of TNF-alpha, TANK, TNF receptor-associated factor (TRAF)2, and TRAF6 in inducing NF-kappaB activity; it is suggested that TFG is a novel member of the NF-kappaB pathway 19797732_A polymorphic gene fusion consisting of TRK-fused gene and G-protein-coupled receptor 128 is identified in healthy individuals and in patients with lymphoma and soft tissue neoplasms. 22250051_Mutations in TFG may have important clinical relevance for current therapeutic strategies to treat metastatic melanoma. 22581839_results suggest that the oncogenic effect of the t(3;9) translocation may be due to the TFG-TEC chimeric protein and that fusion of the TFG (NTD) to the TEC protein produces a gain-of-function chimeric product 22883144_The TRK-fused gene is mutated in hereditary motor and sensory neuropathy with proximal dominant involvement. 23479643_Inhibition of TFG function causes hereditary axon degeneration by impairing endoplasmic reticulum structure. 23553329_Whole-exome sequencing reveals that HMSN-P is caused by a mutation in the TRK-fused gene on chromosome 3q13.2 23810392_TFG plays a pivotal role in negative regulation of RNA-sensing, RIG-I-like receptor (RLR) family signaling pathways. 24613659_Study demonstrates that TFG1 physiologically functions to inhibit the protein degradation system, resulting in an increase in ER resident proteins and ER stress; the P285L mutant substantially enhances these consequences 24999993_TRIM68 targets TFG, a novel regulator of IFN production, and in doing so turns off and limits type I IFN production in response to anti-viral detection systems 25098539_TFG plays an important role in the protein secretory pathways that are essential for proper functioning of the human peripheral nervous system. 25586378_TFG functions at the endoplasmic reticulum (ER)/ER-Golgi intermediate compartments (ERGIC) interface to locally concentrate COPII-coated transport carriers and link exit sites on the ER to ERGIC membranes. 25725944_HMSN-P caused by p.Pro285Leu mutation in TFG is not confined to patients with Far East ancestry. 27184855_TFG organizes transitional ER (tER) and ER exit sites (ERESs) into larger structures. 27492651_Results identified two TFG variants associated with hereditary spastic paraplegias (HSP) (c.316C>T and c.317G> A) confirming the causal nature of bi-allelic TFG mutations for HSP, and suggest that that mitochondrial impairment represents a pathomechanistic link to other neurodegenerative conditions. 27601211_We established a genetic diagnosis in six families with autosomal recessive HSP (SPG11 in three families and TFG/SPG57, SACS and ALS2 in one family each). A heterozygous mutation in a gene involved in an autosomal dominant HSP (ATL1/SPG3A) was also identified in one additional family. Six out of seven identified variants were novel. 27653917_This study finding p.Gly269Val in a newly identified Iranian pedigree affected with hereditary motor and sensory neuropathy with proximal predominance. 27813252_The results suggest that ALG-2 acts as a Ca2+-sensitive adaptor to concentrate and polymerize TFG at endoplasmic reticulum exit sites, supporting a potential role for ALG-2 in COPII-dependent trafficking from the endoplasmic reticulum. 28124177_Differences in the severity of the disorder as well as new clinical findings. These include presence of clonus, undeveloped speech, and sleep disturbances. these findings extend the phenotypic spectrum associated with the TFG mutations in Hereditary spastic paraplegia. 28882477_Study suggests that a genetic variant in an intron of TFG may be associated with skin aging in Korean females, and TFG may be an interesting new candidate gene for exploring individual differences in the molecular bases of collagen and MMP production. 29971521_This study highlights phenotypic heterogeneity characterizing individuals carrying the same pathogenic variant in TFG and provides an insight on tight connection linking mitochondrial efficiency and neuronal health to vesicular trafficking. 30157421_Data highlight a key role for TFG-mediated protein transport in the pathogenesis of HSP. 30467354_TFG homozygous mutation is associated with early onset spastic paraplegia and later onset sensorimotor polyneuropathy. 31036933_Tropomyosin-receptor kinase fused gene (TFG) regulates lipid production in human sebocytes. 31111683_Continuum of phenotypes in hereditary motor and sensory neuropathy with proximal predominance and Charcot-Marie-Tooth patients with TFG mutation has been described. 32666699_A novel TFG c.793C>G mutation in a Chinese pedigree with Charcot-Marie-Tooth disease 2. 33099537_TFG-maintaining stability of overlooked FANCD2 confers early DNA-damage response. 33767317_Homozygous TFG gene variants expanding the mutational and clinical spectrum of hereditary spastic paraplegia 57 and a review of literature. 33932238_TFG binds LC3C to regulate ULK1 localization and autophagosome formation. 34185412_Deregulation of CLTC interacts with TFG, facilitating osteosarcoma via the TGF-beta and AKT/mTOR signaling pathways. 34779525_A Novel TFG Mutation in a Korean Family with alpha-Synucleinopathy and Amyotrophic Lateral Sclerosis. | ENSMUSG00000022757 | Tfg | 4917.85563 | 1.1066456 | 0.1461932273 | 0.07311137 | 4.011129e+00 | 4.520087e-02 | 3.215603e-01 | No | Yes | 6427.366164 | 883.081397 | 5433.351204 | 730.042010 | |
| ENSG00000114541 | 23150 | FRMD4B | protein_coding | Q9Y2L6 | FUNCTION: Member of GRP1 signaling complexes that are acutely recruited to plasma membrane ruffles in response to insulin receptor signaling. May function as a scaffolding protein that regulates epithelial cell polarity by connecting ARF6 activation with the PAR3 complex. Plays a redundant role with FRMD4A in epithelial polarization. {ECO:0000250|UniProtKB:Q920B0}. | Alternative splicing;Cell junction;Coiled coil;Cytoplasm;Cytoskeleton;Isopeptide bond;Phosphoprotein;Reference proteome;Tight junction;Ubl conjugation | hsa:23150; | adherens junction [GO:0005912]; bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular space [GO:0005615]; ruffle [GO:0001726]; establishment of epithelial cell polarity [GO:0090162] | Mouse_homologues 29947801_his study reveals a critical role of FRMD4B in maintaining ELM integrity and in rescuing morphological abnormalities of the outer nuclear layer in photoreceptor dysplasia. | ENSMUSG00000030064 | Frmd4b | 93.65477 | 1.2638008 | 0.3377691050 | 0.33964930 | 9.874074e-01 | 3.203769e-01 | 6.989401e-01 | No | Yes | 135.186451 | 34.017773 | 101.737958 | 25.196484 | ||
| ENSG00000114923 | 6508 | SLC4A3 | protein_coding | P48751 | FUNCTION: Plasma membrane anion exchange protein of wide distribution. Mediates at least a part of the Cl(-)/HCO3(-) exchange in cardiac myocytes. Both BAE3 and CAE3 forms transport Cl(-). | Alternative splicing;Anion exchange;Antiport;Ion transport;Lipoprotein;Membrane;Methylation;Palmitate;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is a plasma membrane anion exchange protein. The encoded protein has been found in brain, heart, kidney, small intestine, and lung. [provided by RefSeq, May 2016]. | hsa:6508; | external side of plasma membrane [GO:0009897]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; plasma membrane [GO:0005886]; anion:anion antiporter activity [GO:0015301]; bicarbonate transmembrane transporter activity [GO:0015106]; inorganic anion exchanger activity [GO:0005452]; transmembrane transporter activity [GO:0022857]; bicarbonate transport [GO:0015701]; cardiac conduction [GO:0061337]; cardiac muscle cell action potential [GO:0086001]; ion homeostasis [GO:0050801]; regulation of intracellular pH [GO:0051453]; transmembrane transport [GO:0055085]; transport across blood-brain barrier [GO:0150104] | 19605733_It was concluded that the A867D allele is a functional mutant of AE3 and that the decreased activity of this mutation may cause changes in cell volume and abnormal intracellular pH. 19854014_Observational study of gene-disease association. (HuGE Navigator) 27211793_SLC4A3 remains an excellent candidate gene for human retinal degeneration, and with the advent of whole exome and whole genome sequencing of cohorts of molecularly unsolved patients with syndromic and non-syndromic forms of retinal degeneration 29167417_Identify a missense mutation in the anion exchanger (AE3)-encoding SLC4A3 gene in two unrelated families with short QT syndrome. The mutation causes reduced surface expression of AE3 and reduced membrane bicarbonate transport. | ENSMUSG00000006576 | Slc4a3 | 78.20928 | 0.6866506 | -0.5423520173 | 0.33607717 | 2.561055e+00 | 1.095255e-01 | 4.727411e-01 | No | Yes | 71.171346 | 10.989026 | 103.780710 | 15.232753 | |
| ENSG00000115129 | 9540 | TP53I3 | protein_coding | Q53FA7 | FUNCTION: Catalyzes the NADPH-dependent reduction of quinones (PubMed:19349281). Exhibits a low enzymatic activity with beta-naphthoquinones, with a strong preference for the ortho-quinone isomer (1,2-beta-naphthoquinone) over the para isomer (1,4-beta-naphthoquinone). Also displays a low reductase activity for non-quinone compounds such as diamine and 2,6-dichloroindophenol (in vitro) (PubMed:19349281). Involved in the generation of reactive oxygen species (ROS) (PubMed:19349281). {ECO:0000269|PubMed:19349281}. | 3D-structure;Acetylation;Alternative splicing;Direct protein sequencing;NADP;Oxidoreductase;Reference proteome | The protein encoded by this gene is similar to oxidoreductases, which are enzymes involved in cellular responses to oxidative stresses and irradiation. This gene is induced by the tumor suppressor p53 and is thought to be involved in p53-mediated cell death. It contains a p53 consensus binding site in its promoter region and a downstream pentanucleotide microsatellite sequence. P53 has been shown to transcriptionally activate this gene by interacting with the downstream pentanucleotide microsatellite sequence. The microsatellite is polymorphic, with a varying number of pentanucleotide repeats directly correlated with the extent of transcriptional activation by p53. It has been suggested that the microsatellite polymorphism may be associated with differential susceptibility to cancer. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2011]. | hsa:9540; | cytosol [GO:0005829]; NADPH binding [GO:0070402]; NADPH:quinone reductase activity [GO:0003960]; protein homodimerization activity [GO:0042803]; quinone binding [GO:0048038]; NADP metabolic process [GO:0006739] | 15067011_p53 activity and PIG3 gene function are uncoupled by UV-dependent alternative splicing through rapid proteolytic degradation 15192123_suppression of p53-C277Y by RNAi reduced pig3 promoter activity, RNA, and protein expression 15491642_Observational study of gene-disease association. (HuGE Navigator) 18676680_Observational study of gene-disease association. (HuGE Navigator) 18801469_numerous factors contribute to the normal alternative splicing of PIG3 exon 4 and UV-inducible increases in this process require that the splicing of this exon be maintained in a sufficiently weakened state under normal conditi 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19336552_Observational study of gene-disease association. (HuGE Navigator) 19343046_Observational study of gene-disease association. (HuGE Navigator) 19349281_PIG3 action is through oxidative stress produced by its enzymatic activity and provides essential knowledge for eventual control of apoptosis. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 20023697_Results suggest that PIG3 is a critical component of the DNA damage response pathway and has a direct role in the transmission of the DNA damage signal from damaged DNA to the intra-S and G2/M checkpoint machinery. 20453000_Observational study of gene-disease association. (HuGE Navigator) 20603616_certain p53 mutatants activate PIG3, whereas the result of our study show increased full-length transcript expression in tumor counterparts 22461624_a novel signaling pathway of GPx3-PIG3 in the regulation of cell death in prostate cancer. 23241165_study provides evidence that the variant genotypes of (TGYCC)n repeats in the PIG3 promoter are functional and associated with risk of squamous cell carcinoma of the head and neck in a non-Hispanic white population 24388982_prohibitin and prohibiton (PHB2) contribute to PIG3-mediated apoptosis by binding to the PIG3 promoter (TGYCC)15 motif 26133772_The results suggested that PIG3 plays an oncogenic role in PTC via the regulation of the PI3K/AKT/PTEN pathway and support the exploration of PIG3 as a novel biomarker for patients with papillary thyroid carcinoma 26464464_PIG3, which functions in DNA damage repair, uses an unexpected catalytic mechanism to suppress Rho-ROCK activity and impair tumor invasion in vivo. This regulation was suppressed by antioxidants. 26472723_Data indicate that knockdown of p53-induced gene 3 (PIG-3) expression by small interfering RNA (siRNA) treatment can inhibit the generation of reactive oxygen species (ROS). 27029070_Data suggest that PIG3 was involved in HIF-1alpha regulation, and indicate a signaling pathway of PIG3/HIF-1alpha in the regulation of cell migration in renal cell carcinoma. 28259183_Results revealed that PIG3 expression levels positively correlated with poor prognosis of non-small cell lung cancer (NSCLC) patients and indicate that PIG3 promotes NSCLC progression. 28351326_our data suggest that high expression of p53-inducible gene 3 is significant for glioblastoma inhibition and p53-inducible gene 3 independently indicates good prognosis in patients, which might be a novel prognostic biomarker or potential therapeutic target in glioblastoma. 30281878_Study found that PIG3 expression was positively associated with lymph node metastasis from lung adenocarcinoma (LUAD) but not from lung squamous cell carcinoma (LUSC). Further data revealed a role for PIG3 in inducing LUAD metastasis, and its role as a new FAK regulator, suggesting that it could be considered as a novel prognostic biomarker. 30334411_expression of PIG3 is frequently reduced in gastric cancer (GC) tissue, and PIG3 suppressed human GC growth through p53- mediated apoptosis; PIG3 may act as a potential diagnostic marker and a potential therapeutic target of GC | 70.81525 | 0.6721060 | -0.5732392133 | 0.34982417 | 2.643417e+00 | 1.039797e-01 | No | Yes | 57.961525 | 9.974758 | 78.693539 | 12.922374 | ||||
| ENSG00000115282 | 64427 | TTC31 | protein_coding | Q49AM3 | Alternative splicing;Coiled coil;Phosphoprotein;Reference proteome;Repeat;TPR repeat | hsa:64427; | 21790010_A fragment of the CCDC142-TTC31 intergenic region was cloned; this fragment functions as bidirectional promoter. | 646.96288 | 0.9515148 | -0.0717020051 | 0.12501765 | 3.284977e-01 | 5.665451e-01 | 8.531186e-01 | No | Yes | 707.883499 | 73.741926 | 737.353947 | 74.844493 | ||||||
| ENSG00000115295 | 79745 | CLIP4 | protein_coding | Q8N3C7 | 3D-structure;ANK repeat;Alternative splicing;Phosphoprotein;Reference proteome;Repeat | hsa:79745; | cytoplasm [GO:0005737]; intracellular membrane-bounded organelle [GO:0043231]; microtubule plus-end [GO:0035371]; nucleus [GO:0005634]; microtubule plus-end binding [GO:0051010]; cytoplasmic microtubule organization [GO:0031122] | 27283491_FOXC2 and CLIP4 activity correlates to the presence of =7-cm clear cell renal cell carcinomas (ccRCCs) with synchronous metastasis and may be potential molecular predictors of synchronous metastasis of =7-cm ccRCCs. | ENSMUSG00000024059 | Clip4 | 227.74115 | 0.8772277 | -0.1889766813 | 0.20777388 | 8.238035e-01 | 3.640705e-01 | 7.352989e-01 | No | Yes | 258.217092 | 44.113772 | 279.742405 | 46.777093 | |||
| ENSG00000115540 | 25843 | MOB4 | protein_coding | Q9Y3A3 | FUNCTION: May play a role in membrane trafficking, specifically in membrane budding reactions. {ECO:0000250}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Golgi apparatus;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Transport;Zinc | This gene was identified based on its similarity with the mouse counterpart. Studies of the mouse counterpart suggest that the expression of this gene may be regulated during oocyte maturation and preimplantation following zygotic gene activation. Alternatively spliced transcript variants encoding distinct isoforms have been observed. Naturally occurring read-through transcription occurs between this locus and the neighboring locus HSPE1.[provided by RefSeq, Feb 2011]. | hsa:25843; | cytoplasm [GO:0005737]; Golgi apparatus [GO:0005794]; Golgi cisterna membrane [GO:0032580]; perinuclear region of cytoplasm [GO:0048471]; metal ion binding [GO:0046872] | 18362890_phosphorylation of MOB1 at Thr74 by MST2 is essential to make a complex of MOB1, MST2 and NDR1, and to fully activate NDR1 24872389_hMOB3 modulates MST1 apoptotic signaling and supports tumor growth in glioblastoma multiforme. 30072378_Because of divergent evolution of key interface residues, MST4 and MOB4 could disrupt assembly of the MST1-MOB1 complex through alternative pairing and thereby increased YAP activity. Collectively, these findings identify the MST4-MOB4 complex as a noncanonical regulator of the Hippo-YAP pathway with an oncogenic role in PC | ENSMUSG00000025979 | Mob4 | 1397.38625 | 1.0592835 | 0.0830887025 | 0.10525155 | 6.248674e-01 | 4.292443e-01 | 7.797293e-01 | No | Yes | 1735.065967 | 286.656692 | 1578.415820 | 255.159959 | |
| ENSG00000115687 | 23178 | PASK | protein_coding | Q96RG2 | FUNCTION: Serine/threonine-protein kinase involved in energy homeostasis and protein translation. Phosphorylates EEF1A1, GYS1, PDX1 and RPS6. Probably plays a role under changing environmental conditions (oxygen, glucose, nutrition), rather than under standard conditions. Acts as a sensor involved in energy homeostasis: regulates glycogen synthase synthesis by mediating phosphorylation of GYS1, leading to GYS1 inactivation. May be involved in glucose-stimulated insulin production in pancreas and regulation of glucagon secretion by glucose in alpha cells; however such data require additional evidences. May play a role in regulation of protein translation by phosphorylating EEF1A1, leading to increase translation efficiency. May also participate in respiratory regulation. {ECO:0000269|PubMed:16275910, ECO:0000269|PubMed:17052199, ECO:0000269|PubMed:17595531, ECO:0000269|PubMed:20943661, ECO:0000269|PubMed:21181396, ECO:0000269|PubMed:21418524}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Kinase;Lipid-binding;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Serine/threonine-protein kinase;Transferase | This gene encodes a member of the serine/threonine kinase family that contains two PAS domains. Expression of this gene is regulated by glucose, and the encoded protein plays a role in the regulation of insulin gene expression. Downregulation of this gene may play a role in type 2 diabetes. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Nov 2011]. | hsa:23178; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; ATP binding [GO:0005524]; phosphatidylinositol binding [GO:0035091]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; energy homeostasis [GO:0097009]; intracellular signal transduction [GO:0035556]; negative regulation of glycogen biosynthetic process [GO:0045719]; positive regulation of translation [GO:0045727]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of glucagon secretion [GO:0070092]; regulation of respiratory gaseous exchange [GO:0043576] | 11688972_molecular cloning, gene expression, and comparison with mouse protein 15148392_Results suggest that elevated glucose concentrations rapidly increase Per-Arnt-Sim kinase activity in pancreatic islet beta cells, followed by up-regulation by glucose of preproinsulin and pancreatic duodenum homeobox 1 gene expression. 17595531_identified the multifunctional eukaryotic translation elongation factor eEF1A1 as a novel interaction partner of PASKIN in the nuclei of testis germ cells and in the midpiece of sperm tails 20943661_Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation. 21181396_PASK is involved in the regulation of glucagon secretion by glucose and may be a useful target for the treatment of type 2 diabetes. 22065581_Per-Arnt-Sim (PAS) domain-containing protein kinase (PASK) has a role in insulin hypersecretion 22219681_The role of PAS kinase in PASsing the glucose signal. 23721480_Mutations that affect PAS domain cause a severe protein trafficking defect and change the interactions with Kv11.1 channels. 23853095_PASK phosphorylates and inactivates GSK3beta, thereby preventing PDX-1 serine phosphorylation and alleviating GSK3beta-mediated PDX-1 protein degradation in pancreatic beta-cells. 30381292_we provide the first evidence for a role of USF1 in respiration since it appeared to complement Cbf1 in vivo as determined by respiration phenotypes. In addition, we confirmed USF1 as a substrate of human PAS kinase (hPASK) in vitro Combined, our data supports a model in which Cbf1/USF1 functions to partition glucose toward respiration and away from lipid biogenesis 31072927_MTORC1-PASK signaling is required for the rise of myogenin-positive committed myoblasts. 31529049_we show that the nuclear PASK associates with the mammalian H3K4 MLL2 methyltransferase complex and enhances H3K4 di- and tri-methylation. We also show that PASK is a histone kinase that phosphorylates H3 at T3, T6, S10 and T11. | ENSMUSG00000026274 | Pask | 1168.42584 | 0.9940148 | -0.0086607370 | 0.10888171 | 6.329268e-03 | 9.365898e-01 | 9.835562e-01 | No | Yes | 1264.447628 | 134.771411 | 1270.843829 | 132.267239 | |
| ENSG00000115998 | 54980 | C2orf42 | protein_coding | Q9NWW7 | Reference proteome | hsa:54980; | nucleoplasm [GO:0005654]; nucleus [GO:0005634] | ENSMUSG00000046679 | C87436 | 226.25940 | 0.9839049 | -0.0234091770 | 0.21014204 | 1.237522e-02 | 9.114229e-01 | 9.753783e-01 | No | Yes | 282.036447 | 51.618596 | 262.411835 | 47.062312 | ||||
| ENSG00000116273 | 148479 | PHF13 | protein_coding | Q86YI8 | FUNCTION: Modulates chromatin structure. Required for normal chromosome condensation during the early stages of mitosis. Required for normal chromosome separation during mitosis. {ECO:0000269|PubMed:19638409}. | 3D-structure;Cell cycle;Cell division;Chromatin regulator;DNA condensation;Metal-binding;Mitosis;Nucleus;Reference proteome;Zinc;Zinc-finger | hsa:148479; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; metal ion binding [GO:0046872]; methylated histone binding [GO:0035064]; cell division [GO:0051301]; chromatin organization [GO:0006325]; chromosome segregation [GO:0007059]; mitotic cell cycle [GO:0000278]; mitotic chromosome condensation [GO:0007076] | 19638409_SPOC1 modulates chromatin structure and that tight regulation of its expression levels and subcellular localization during mitosis are crucial for proper chromosome condensation and cell division. 24278021_SPOC1-mediated restriction imposed upon Ad growth is relieved by its functional association with the Ad major core protein pVII that enters with the viral genome, followed by E1B-55K/E4orf6-dependent proteasomal degradation of SPOC1. 27223324_These experiments showed that PHF13 binds specifically to DNA and to two types of histone H3 methyl tags (lysine 4-tri-methyl or lysine 4-di-methyl) where it functions as a transcriptional co-regulator. 29021215_PHF13 has opposing effects throughout the HIV-1 replication cycle (figure 8). After viral entry and nuclear import of the PIC, PHF13 can increase the number of integrated HIV-1 proviral genomes. After integration, PHF13 acts as antiviral restriction factor and inhibits viral gene expression. 29743358_High SPOC1 expression is associated with Cytomegalovirus infections. | ENSMUSG00000047777 | Phf13 | 1345.38417 | 1.0556342 | 0.0781100599 | 0.10857996 | 5.179078e-01 | 4.717355e-01 | 8.043569e-01 | No | Yes | 1620.085202 | 124.136150 | 1482.424948 | 111.081316 | ||
| ENSG00000116337 | 271 | AMPD2 | protein_coding | Q01433 | FUNCTION: AMP deaminase plays a critical role in energy metabolism. Catalyzes the deamination of AMP to IMP and plays an important role in the purine nucleotide cycle. {ECO:0000269|PubMed:23911318}. | 3D-structure;Alternative splicing;Disease variant;Hereditary spastic paraplegia;Hydrolase;Metal-binding;Methylation;Neurodegeneration;Nucleotide metabolism;Phosphoprotein;Reference proteome;Zinc | PATHWAY: Purine metabolism; IMP biosynthesis via salvage pathway; IMP from AMP: step 1/1. {ECO:0000269|PubMed:23911318}. | The protein encoded by this gene is important in purine metabolism by converting AMP to IMP. The encoded protein, which acts as a homotetramer, is one of three AMP deaminases found in mammals. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2012]. | hsa:271; | cytosol [GO:0005829]; AMP deaminase activity [GO:0003876]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; AMP metabolic process [GO:0046033]; cyclic purine nucleotide metabolic process [GO:0052652]; energy homeostasis [GO:0097009]; IMP biosynthetic process [GO:0006188]; IMP salvage [GO:0032264] | 12745092_N-terminal extensions of the AMPD2 polypeptide influence ATP regulation of isoform L. 18493842_This is a first report evidencing the pattern of AMPD genes expression in neoplastic human liver. 19948975_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and genetic testing. (HuGE Navigator) 23911318_Study concluded that AMPD2 as necessary for guanine nucleotide biosynthesis and protein translation and provide evidence that AMP deaminase activity is critical during neurogenesis. Patients with mutations in AMPD2 have characteristic brain imaging features of pontocerebellar hypoplasia due to loss of brainstem and cerebellar parenchyma. 24755741_In human HepG2 cells, AMPD2 activation counterregulates AMPK and increases intracellular glucose production, in association with up-regulation of PEPCK and G6Pc. 25496463_tofacitinib increases the cellular levels of adenosine, which is known to have anti-inflammatory activity, through the downregulation of AMPD2. This would be a novel functional aspect of tofacitinib. 28168832_Here we report the clinical and genetic analysis of an individual with PCH9 secondary to a novel missense variant with strong evidence of pathogenicity, located outside the catalytic domain of AMPD2 29463858_The existence of various AMPD2 isoforms with different functions possibly explains the variability in phenotypes associated with AMPD2 variants: variants leaving some of the isoforms intact may cause spastic paraplegia type 63 , while those affecting all isoforms may result in the severe and early-onset Pontocerebellar hypoplasia type 9. 30267407_Data suggest that adenosine monophosphate deaminase 2 (AMPD2) may serve as a biomarker for outcome prediction in undifferentiated pleomorphic sarcoma (UPS). 30577810_These data demonstrate a novel mechanism in Systemic lupus erythematosus development that involves the targeting of AMPD2 expression by NovelmiRNA-25. 31833174_Homozygous variants in AMPD2 and COL11A1 lead to a complex phenotype of pontocerebellar hypoplasia type 9 and Stickler syndrome type 2. | ENSMUSG00000027889 | Ampd2 | 2332.42558 | 0.9910375 | -0.0129885069 | 0.09893826 | 1.721784e-02 | 8.956038e-01 | 9.709777e-01 | No | Yes | 2467.775556 | 258.916999 | 2429.305695 | 249.162909 |
| ENSG00000116584 | 9181 | ARHGEF2 | protein_coding | Q92974 | FUNCTION: Activates Rho-GTPases by promoting the exchange of GDP for GTP. May be involved in epithelial barrier permeability, cell motility and polarization, dendritic spine morphology, antigen presentation, leukemic cell differentiation, cell cycle regulation, innate immune response, and cancer. Binds Rac-GTPases, but does not seem to promote nucleotide exchange activity toward Rac-GTPases, which was uniquely reported in PubMed:9857026. May stimulate instead the cortical activity of Rac. Inactive toward CDC42, TC10, or Ras-GTPases. Forms an intracellular sensing system along with NOD1 for the detection of microbial effectors during cell invasion by pathogens. Required for RHOA and RIP2 dependent NF-kappaB signaling pathways activation upon S.flexneri cell invasion. Involved not only in sensing peptidoglycan (PGN)-derived muropeptides through NOD1 that is independent of its GEF activity, but also in the activation of NF-kappaB by Shigella effector proteins (IpgB2 and OspB) which requires its GEF activity and the activation of RhoA. Involved in innate immune signaling transduction pathway promoting cytokine IL6/interleukin-6 and TNF-alpha secretion in macrophage upon stimulation by bacterial peptidoglycans; acts as a signaling intermediate between NOD2 receptor and RIPK2 kinase. Contributes to the tyrosine phosphorylation of RIPK2 through Src tyrosine kinase leading to NF-kappaB activation by NOD2. Overexpression activates Rho-, but not Rac-GTPases, and increases paracellular permeability (By similarity). Involved in neuronal progenitor cell division and differentiation (PubMed:28453519). Involved in the migration of precerebellar neurons (By similarity). {ECO:0000250|UniProtKB:Q60875, ECO:0000250|UniProtKB:Q865S3, ECO:0000269|PubMed:19043560, ECO:0000269|PubMed:21887730, ECO:0000269|PubMed:28453519, ECO:0000269|PubMed:9857026}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Cell junction;Cell membrane;Cell projection;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Cytoskeleton;Developmental protein;Differentiation;Golgi apparatus;Guanine-nucleotide releasing factor;Immunity;Innate immunity;Membrane;Mental retardation;Metal-binding;Microtubule;Mitosis;Neurogenesis;Phosphoprotein;Reference proteome;Tight junction;Zinc;Zinc-finger | Rho GTPases play a fundamental role in numerous cellular processes that are initiated by extracellular stimuli that work through G protein coupled receptors. The encoded protein may form complex with G proteins and stimulate rho-dependent signals. Alternatively spliced transcript variants encoding different isoforms have been identified.[provided by RefSeq, Jun 2009]. | hsa:9181; | bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; Golgi apparatus [GO:0005794]; microtubule [GO:0005874]; protein-containing complex [GO:0032991]; ruffle membrane [GO:0032587]; spindle [GO:0005819]; vesicle [GO:0031982]; guanyl-nucleotide exchange factor activity [GO:0005085]; microtubule binding [GO:0008017]; small GTPase binding [GO:0031267]; zinc ion binding [GO:0008270]; actin filament organization [GO:0007015]; asymmetric neuroblast division [GO:0055059]; cell cycle [GO:0007049]; cell morphogenesis [GO:0000902]; cellular hyperosmotic response [GO:0071474]; cellular response to muramyl dipeptide [GO:0071225]; cellular response to tumor necrosis factor [GO:0071356]; innate immune response [GO:0045087]; intracellular protein transport [GO:0006886]; intracellular signal transduction [GO:0035556]; negative regulation of extrinsic apoptotic signaling pathway via death domain receptors [GO:1902042]; negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress [GO:1902219]; negative regulation of microtubule depolymerization [GO:0007026]; negative regulation of necroptotic process [GO:0060546]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of neuron differentiation [GO:0045666]; positive regulation of neuron migration [GO:2001224]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of cell population proliferation [GO:0042127]; regulation of Rho protein signal transduction [GO:0035023]; regulation of small GTPase mediated signal transduction [GO:0051056] | 11912491_GEF-H1 is regulated by an interaction with microtubules. 15827085_PAK4 mediates morphological changes through regulation of GEF-H1 16778209_GEF-H1 expression level strongly correlated with p53 status in a panel of 32 cancer cell lines, and GEF-H1 induction caused activation of RhoA. Growth of mutant p53 cells was dependent on GEF-H1 expression. 17488622_These results identify a GEF-H1-dependent mechanism to modulate localized RhoA activation during cytokinesis under the control of mitotic kinases. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18287519_These studies reveal a critical role for a GEF-H1/RhoA/ROCK/MLC signaling pathway in mediating nocodazole-induced cell contractility. 19625450_This study establishes GEF-H1 as a critical organizer of key structural and signaling components of cell migration through the localized regulation of RhoA activity at the cell leading edge. 19667072_Lfc is a phosphorylated protein and demonstrate that 14-3-3 interacts directly and in a phosphorylation-dependent manner with Lfc. 19911011_Observational study of gene-disease association. (HuGE Navigator) 20089843_Data indicate that GEF-H1 is a target and functional effector of TGF-beta by orchestrating Rho signaling to regulate gene expression and cell migration. 20558775_Heparin inhibits pulmonary artery smooth muscle cell proliferation through GEF-H1/RhoA/ROCK/p27 signaling pathway. 20803552_GEF-H1 is a new component of a syndecan signaling complex that is differentially expressed in brain metastatic melanoma cells compared to corresponding non-metastatic counterparts 21212278_the TNF-alpha-induced activation of the ERK/GEF-H1/RhoA pathway in tubular cells is mediated through Src- and TACE-dependent EGFR activation. 21352810_GEFH1 was identified as binding partner for the BAR domain of ASAP1.GEFH1 is a negative regulator of podosomes. 21513698_These data suggest that Par1b-phosphorylation regulates turnover of GEF-H1 localization by regulating its interaction with microtubules, which may contribute to cell polarization. 21887730_GEF-H1 mediated the activation of Rip2 during signaling by NOD2, but not in the presence of the 3020insC variant of NOD2 associated with Crohn's disease. GEF-H1 functioned downstream of NOD2 as part of Rip2-containing signaling complexes. 22002306_Study shows that hPTTG1 is a transcription factor that triggers the GEF-H1/RhoA pathway to accelerate breast cancer invasion and metastasis. In human invasive breast carcinoma, hPTTG1 is overexpressed and is correlated to GEF-H1 expression. 22072711_Polarity-regulating kinase partitioning-defective 1b (PAR1b) phosphorylates guanine nucleotide exchange factor H1 (GEF-H1) to regulate RhoA-dependent actin cytoskeletal reorganization. 22226472_Results show that LPS-induced NF-kappaB activation and IL-8 synthesis in endothelial cells are regulated by the MyD88 pathway and GEF-H1-RhoA pathway. 22226621_Lipopolysaccharide-induced ICAM-1 synthesis in human umbilical vein endothelial cells is regulated by GEF-H1/RhoA-dependent signaling pathway via activation of p38 and NF-kappaB. 22301607_LPS rapidly upregulates GEF-H1 expression. Activated Rho-associated kinase by GEF-H1 subsequently activates p38 and ERK1/2, thereby increasing IL-6/TNF-alpha expression in endothelial cells. 22513363_Data indicate that highly aggressive spindle-shaped 231BR3 cells changed to a round cell morphology associated with expression of the small GTPase guanine nucleotide exchange factor-H1 (GEF-H1). 22593214_extracellular matrix stiffness regulates RhoA through microtubule destabilization and the subsequent release and activation of GEF-H1. 22847784_Our findings underscore a potent oncogenic role for GEF-H1 in promoting the metastatic potentials of hepatocellular carcinoma, possibly through activation of RhoA signalling. 22898781_present evidence that depletion of GEF-H1, a guanine nucleotide exchange factor for Rho proteins, affects vesicle trafficking 23057787_vincristine activates GEF-H1/RhoA/ROCK/MLC signaling. 23432781_CAMSAP3-anchored non-centrosomal microtubules capture GEF-H1 more efficiently than other microtubules do. 24043311_ERK binds to the Rho exchange factor GEF-H1 and phosphorylates it on S959, causing inhibition of GEF-H1 activity and a consequent decrease in RhoA activity. 24352660_This study investigated a novel mechanism of vascular barrier protection by ANP via modulation of GEF-H1 function. 24525234_The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. 24706358_Paxillin-GEF-H1-p42/44-MAPK module as a regulator of pathological mechanotransduction. 25143398_TGF-beta regulates LARG and GEF-H1 during epithelial-mesenchymal transition to affect stiffening response to force and cell invasion. 25687035_Results supported that miR-512-3p could inhibit tumor cell adhesion, migration, and invasion by regulating the RAC1 activity via DOCK3 in NSCLC A549 and H1299 cell lines. 26359301_data suggest that the induction of SGK1 through treatment with dexamethasone alters MT dynamics to increase Sec5-GEF-H1 interactions, which promote GEF-H1 targeting to adhesion sites. 26759237_By stimulating cofilin/PP2A-mediated dephosphorylation of the guanine nucleotide exchange factor GEF-H1. 26820534_this study reports the crystal structure of human GEF-H1 PH domain to 2.45 A resolution. 26866809_regulation of c-Src trafficking requires both microtubules and actin polymerization, and GEF-H1 coordinates c-Src trafficking, acting as a molecular switch between these two mechanisms 27573550_Overexpression of miR-194 downregulates the GEF-H1/RhoA pathway, inhibits melanoma cancer cell proliferation and metastasis. Furthermore, miR-194 expression is negatively associated with tumor-node-metastasis (TNM) stages 28096473_depletion induces phosphorylation of the microtubule-associated GEF-H1 on Ser886, and thereby promotes RhoA activity and actin stress fiber assembly. 28453519_human brain malformation is recapitulated in Arhgef2 mutant mice and identify an aberrant migration of distinct components of the precerebellar system as a pathomechanism underlying the midbrain-hindbrain phenotype. Our results highlight the crucial function of ARHGEF2 in human brain development and identify a mutation in ARHGEF2 as novel cause of a neurodevelopmental disorder. 29089450_Authors identified a regulatory switch controlled by MARK3 that couples microtubules to the actin cytoskeleton to establish epithelial cell polarity through ARHGEF2. 30305100_role for RASSF1A in tunneling nanotube formation between cells through GEFH1/Rab11 pathway control 30846413_the present study revealed that GEF-H1 is upregulated in colon cancer tissues and plays a key role in colon cancer metastasis 31420453_GEF-H1 is further regulated by Src phosphorylation, activating GEF-H1 in a narrower band ~0-2 microm from the cell edge, in coordination with cell protrusions. This indicates a synergistic intersection between MT dynamics and Src signaling in RhoA activation through GEF-H1. 31802204_Relation of ADRB3, GEF, ROCK2 gene polymorphisms to clinical findings in overactive bladder. 32692911_The RhoA regulators Myo9b and GEF-H1 are targets of cyclic nucleotide-dependent kinases in platelets. 33393038_High ARHGEF2 (GEF-H1) Expression is Associated with Poor Prognosis Via Cell Cycle Regulation in Patients with Pancreatic Cancer. 34345888_Phosphorylated cingulin localises GEF-H1 at tight junctions to protect vascular barriers in blood endothelial cells. 34897929_Monitoring the Response of Multiple Signal Network Components to Acute Chemo-Optogenetic Perturbations in Living Cells. | ENSMUSG00000028059 | Arhgef2 | 1018.23167 | 1.0426530 | 0.0602590734 | 0.10983015 | 3.015533e-01 | 5.829103e-01 | 8.602146e-01 | No | Yes | 1156.322553 | 114.460549 | 1070.522150 | 103.615529 | |
| ENSG00000116679 | 10625 | IVNS1ABP | protein_coding | Q9Y6Y0 | FUNCTION: Involved in many cell functions, including pre-mRNA splicing, the aryl hydrocarbon receptor (AHR) pathway, F-actin organization and protein ubiquitination. Plays a role in the dynamic organization of the actin skeleton as a stabilizer of actin filaments by association with F-actin through Kelch repeats (By similarity). Protects cells from cell death induced by actin destabilization (By similarity). Functions as modifier of the AHR/Aryl hydrocarbon receptor pathway increasing the concentration of AHR available to activate transcription (PubMed:16582008). In addition, functions as a negative regulator of BCR(KLHL20) E3 ubiquitin ligase complex to prevent ubiquitin-mediated proteolysis of PML and DAPK1, two tumor suppressors (PubMed:25619834). Inhibits pre-mRNA splicing (in vitro) (PubMed:9696811). {ECO:0000250|UniProtKB:Q920Q8, ECO:0000269|PubMed:16582008, ECO:0000269|PubMed:25619834, ECO:0000269|PubMed:9696811}.; FUNCTION: (Microbial infection) Involved in the alternative splicing of influenza A virus M1 mRNA through interaction with HNRNPK, thereby facilitating the generation of viral M2 protein. {ECO:0000269|PubMed:23825951, ECO:0000269|PubMed:9696811}. | 3D-structure;Cytoplasm;Cytoskeleton;Host-virus interaction;Kelch repeat;Nucleus;Phosphoprotein;Reference proteome;Repeat | hsa:10625; | cytoskeleton [GO:0005856]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; spliceosomal complex [GO:0005681]; transcription regulator complex [GO:0005667]; negative regulation of intrinsic apoptotic signaling pathway [GO:2001243]; negative regulation of protein ubiquitination [GO:0031397]; response to virus [GO:0009615]; RNA splicing [GO:0008380]; transcription by RNA polymerase III [GO:0006383] | 17996313_These findings further support the distinct roles of alpha-enolase and its MBP-1 variant in maintaining cell homeostasis. Moreover, these data suggest a novel function for NS1-BP in the control of cell proliferation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20532202_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 23825951_NS1-BP-hnRNPK complex is a key mediator of influenza A virus gene expression. 24210102_This study provided evidence that miRNA-548an is involved in the regulation of NS1ABP. 25619834_Our study identifies KLHL39 as a negative regulator of Cul3-KLHL20 ubiquitin ligase and reveals a role of KLHL39-mediated PML and DAPK stabilization in colon cancer metastasis. 29497022_Comparison of the Kelch-domain structures of NS1-BP and its homologues showed that the Gly-Gly pair in beta-strand B and the hydrophobic Trp residue in beta-strand D are highly conserved, while the B-C loops in blades 2 and 6 are variable 29871674_In esophageal squamous cell carcinoma (ESCC) tissues, c-Myc expression was inversely correlated with NS1-binding protein (NS1-BP) levels, and was associated with a shorter disease-specific survival (DSS). 29921878_Data show that heterogeneous nuclear ribonucleoprotein K (hnRNP K) and influenza virus NS1A binding protein (NS1-BP) regulate host splicing events and that viral infection causes mis-splicing of some of these transcripts. 30538201_The central BACK domain of NS1-BP interacts directly with splicing factors such as hnRNP K and PTBP1 and with the viral NS1 protein. | ENSMUSG00000023150 | Ivns1abp | 2331.37653 | 0.9689897 | -0.0454467704 | 0.10343070 | 1.929645e-01 | 6.604606e-01 | 8.939581e-01 | No | Yes | 2669.599405 | 507.189356 | 2590.782901 | 481.212043 | ||
| ENSG00000116833 | 2494 | NR5A2 | protein_coding | O00482 | FUNCTION: Nuclear receptor that acts as a key metabolic sensor by regulating the expression of genes involved in bile acid synthesis, cholesterol homeostasis and triglyceride synthesis. Together with the oxysterol receptors NR1H3/LXR-alpha and NR1H2/LXR-beta, acts as an essential transcriptional regulator of lipid metabolism. Plays an anti-inflammatory role during the hepatic acute phase response by acting as a corepressor: inhibits the hepatic acute phase response by preventing dissociation of the N-Cor corepressor complex (PubMed:20159957). Binds to the sequence element 5'-AACGACCGACCTTGAG-3' of the enhancer II of hepatitis B virus genes, a critical cis-element of their expression and regulation. May be responsible for the liver-specific activity of enhancer II, probably in combination with other hepatocyte transcription factors. Key regulator of cholesterol 7-alpha-hydroxylase gene (CYP7A) expression in liver. May also contribute to the regulation of pancreas-specific genes and play important roles in embryonic development. Activates the transcription of CYP2C38 (By similarity). {ECO:0000250|UniProtKB:P45448, ECO:0000269|PubMed:15707893, ECO:0000269|PubMed:15723037, ECO:0000269|PubMed:15897460, ECO:0000269|PubMed:16289203, ECO:0000269|PubMed:20159957}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Isopeptide bond;Lipid-binding;Metal-binding;Nucleus;Receptor;Reference proteome;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a DNA-binding zinc finger transcription factor and is a member of the fushi tarazu factor-1 subfamily of orphan nuclear receptors. The encoded protein is involved in the expression of genes for hepatitis B virus and cholesterol biosynthesis, and may be an important regulator of embryonic development. [provided by RefSeq, Jun 2016]. | hsa:2494; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; RNA polymerase II transcription regulator complex [GO:0090575]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; nuclear receptor activity [GO:0004879]; phospholipid binding [GO:0005543]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; transcription coregulator binding [GO:0001221]; zinc ion binding [GO:0008270]; bile acid metabolic process [GO:0008206]; calcineurin-mediated signaling [GO:0097720]; cellular response to leukemia inhibitory factor [GO:1990830]; cholesterol homeostasis [GO:0042632]; embryo development ending in birth or egg hatching [GO:0009792]; epithelial cell differentiation [GO:0030855]; homeostatic process [GO:0042592]; hormone-mediated signaling pathway [GO:0009755]; pancreas morphogenesis [GO:0061113]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of viral genome replication [GO:0045070]; regulation of cell population proliferation [GO:0042127]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; tissue development [GO:0009888] | 11927588_role in regulating aromatase expression in preadipocytes (liver receptor homologue-1) 12820970_Ligands are dispensabe for LRH-1 activation. 12852843_LRH-1 is a nuclear receptor which can assume an active conformation in the absence of a ligand agonist. 12853459_Liver receptor homolog 1 controls the expression of carboxyl ester lipase in pancreas 12972592_PDX-1 regulates expression of LRH-1 during pancreas development. 14671206_LRH-1 is highly expressed in corpus luteum, and it plays an essential role in the regulation of HSD3B2. 14728801_hBIF and HNF1 are involved together in the viral gene expression regulation of the Hepatitis B virus. 15117876_role for LRH-1 in the induction of the progesterone but not the estrogen biosynthetic pathway during granulosa cell differentiation. 15121760_LRH-1 is a positive transcription factor for ABCG5 and ABCG8 and, in conjunction with studies on LRH-1 activation of other promoters, identify LRH-1 as a 'master regulator' for genes involved in sterol and bile acid secretion from liver and intestine 15143151_important functional role of helix 1 in cofactor recruitment and a novel molecular mechanism of transcriptional regulation and cofactor recruitment mediated by hLRH-1 15181096_LRH-1 could be the major transcription factor responsible for the rapid and significant increase in ovarian StAR gene expression after ovulation. 15205472_The liver receptor homolog-1 has emerged as an essential regulator for the expression of cyp7a1 gene. 15218078_LHR-1 regulgates apolipoprotein A-1 transscription, and affects cholesterol homeostasis. 15613430_CYP11A1 expression in human granulosa cells is regulated by LRH-1. 15707893_Human LRH-1 receptor binds phosphatidyl inositol second messengers;ligand binding is required for maximal activity. 15723037_hLRH-1's control of gene expression is mediated by phospholipid binding 15923626_Data show that sumoylated LRH-1 is exclusively localized in promyelocytic leukemia protein nuclear bodies, and that this association is a dynamic process regulated in part by SUMO-1. 15963945_suppression of hLRH-1 resulted in cell cycle arrest mediated by the down-regulation of cyclin E1 16091743_LRH-1 is transcriptionally regulated by the estrogen receptor alpha reinforcing the hypothesis that LRH-1 could exert potential oncogenic effects in breast cancer formation. 16282330_LRH-1 and SHP1 regulate 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter and have a role in regulation of cholesterol synthesis and uptake 16439367_phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation 16450584_siRNA interference studies suggested that nuclear receptor subfamily 5, group A, member 2 (hLRH-1) acts as a negative regulator in farnesyl pyrophosphate synthetase expression. 16469397_FTF and LRH-1 are two related but different transcription factors in human Caco-2 cells, suggesting that they may be homologues and not orthologues. 17095585_LRH-1 and SF-1 have qualitatively similar actions on FSH-stimulated estrogen and progesterone production. These factors may have overlapping actions in regulation of steroidogenesis that accompanies granulosa cell differentiation. 17522048_LRH-1 stimulation of the FAS LXR response is blocked by the addition of small heterodimer partner (SHP) and that FAS mRNA 17910058_LRH-1/phospholipid and LRH-1/SHP (fragments) interactions are analyzed by counting atomic contact number, identifying hydrogen bonds, and estimating binding free energies 17952562_The present study demonstrated for the first time the increased expression of hLRH-1v1 and hLRH-1 in human gastric cancer, an alteration which may implicate in tumorigenesis. 17977826_LRH-1 is a novel regulator of APOM transcription and further extend the role of this orphan nuclear receptor in lipoprotein metabolism and cholesterol homeostasis 18191017_Sphingosine-1-phosphate induces LRH-1 mRNA expression in MCF-7 cells in a prostaglandin E2 (PGE2)-dependent manner. 18270374_Liver receptor homolog 1 (LRH-1) is a key transcriptional factor required for the hepatic expression of CYP7A1. 18385139_PGC-1alpha is an important co-activator for LRH-1 and that SHP targets the interaction between LRH-1 and PGC-1alpha to inhibit CYP7A1 expression. 18410128_a study, by molecular dynamics (MD) simulations, the impact of the ligand on the receptor and the interaction with different cofactor peptides 18508634_Possible unexpected new class of nuclear receptor signaling molecules, but broader functional roles of LRH-1 and these new ligands remain to be established.[REVIEW] 18665078_Differential expression of steroidogenic factors 1 and 2, cytochrome p450scc, and steroidogenic acute regulatory protein in the pancreas. 19015525_structure of the Dax-1:LRH-1 complex provides the molecular mechanism for the function of Dax-1 as a potent transcriptional repressor 19022561_both SF1 and LRH1 can transcriptionally cooperate with the AP-1 family members c-JUN and c-FOS, known to be associated with enhanced proliferation of endometrial carcinoma cells, to further enhance activation of the STAR, HSD3B2, and CYP19A1 PII promoters 19359379_The results indicate that LRH-1 could represent another key regulator of the steroidogenic lineage in MSCs and play a vital role in steroid hormone production in human Leydig cells. 19629617_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20101243_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20133449_results indicate that PGC-1alpha is involved in progesterone production in ovarian granulosa cells by potentiating transcriptional activities of LRH1 proteins. 20159957_selective synthetic agonists induce SUMOylation-dependent recruitment of either LRH-1 or LXR to hepatic APR promoters and prevent the clearance of the N-CoR corepressor complex upon cytokine stimulation 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20607599_Our findings extend this by highlighting LRH-1 as a key regulator of the estrogen response in breast cancer cells through the regulation of ER expression. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20817789_These findings suggest a new role of LRH-1 in promoting migration and invasion in breast cancer, independent of oestrogen sensitivity. 21129436_These results indicated that pluripotent stem cells could be differentiated into steroidogenic cells by the nuclear receptor 5A family of protein via the mesenchymal cell lineage. 21258413_Data indicate an important role of LRH-1 in colorectal tumor GC synthesis. 21392518_The results of this study support the hypothesis that suppression of LRH-1 may potentially be beneficial in the tissue specific regulation of aromatase expression in post menopausal breast cancer. 21536586_crucial role of the estrogen target gene nr5a2 in protecting human islets against-stressed-induced apoptosis 21614002_findings identify an LRH-1 dependent phosphatidylcholine signalling pathway that regulates bile acid metabolism and glucose homeostasis 21949357_LRH-1 transcription is activated up to 30-fold in pancreatic cancer cells compared to normal pancreatic ductal epithelium 21990348_findings demonstrate that signaling through RARs has critical roles in molecular reprogramming and that the synergistic interaction between Rarg and Lrh1 directs reprogramming toward ground-state pluripotency 22048972_NR5A2 modulates gene expression in osteoblasts and some allelic variants are associated with bone mass in Spanish postmenopausal women. 22125638_Data show that single nucleotide polymorphisms (SNPs) in ABO, sonic hedgehog (SHH), telomerase reverse transcriptase (TERT), nuclear receptor subfamily 5, group A, member 2 (NR5A2) were found to be associated with pancreatic cancer risk. 22187462_The three-dimensional structure of a beta-catenin armadillo repeat in complex with the liver receptor homolog-1 (LRH-1) ligand binding domain at 2.8 A resolution, is reported. 22359603_in ER-positive breast cancer cells, LRH-1 promotes cell proliferation by enhancing ERalpha mediated transcription of target genes such as GREB-1 22504882_The lipid-free receptor undergoes previously unrecognized structural fluctuations, allowing it to interact with widely expressed co-repressors. 23000165_this study indicates that LRH-1 acts as a transcriptional activator in the regulation of OCT4 gene expression through the cooperative interaction with three binding sites directly or/and indirectly. 23038264_HNF4alpha and LRH-1 promote active transcription histone marks on the Cyp7a1 promoter that are reversed by FGF19 in a SHP-dependent manner 23471216_Expression of human LRH-1 is regulated in a tissue-specific manner, and that the novel promoter region is controlled by the Sp-family, NR5A-family and PGC-1alpha in ovarian granulosa cells in a coordinated fashion. 23537609_Data conclude that SF-1 regulates aromatase expression in GCT; over-expression of LRH-1 suggests that this receptor may be involved in the pathogenesis of GCT by mechanisms other than the regulation of aromatase. 23667258_Experimental evaluation of the predicted ligands identified two compounds that inhibit the transcriptional activity of LRH-1 and diminish the expression of the receptor's target genes. 23737522_Data indicate that sequence divergence has differentially impacted ligand binding and protein dynamics in NR5A2. 23817023_Lrh-1 is necessary for maintenance of the corpus luteum, for promotion of decidualization and for formation of the placenta. 24520076_report the genome-wide location and molecular function of LRH-1 in breast cancer cells and reveal its therapeutic potential for the treatment of breast cancers, notably for tumors resistant to treatments currently used in therapies. 24564400_Data (including data from transgenic overexpression/gene silencing) suggest that NR5A2 modulates signal transduction/cell proliferation in mammary cells; mammary morphology exhibits significant reduction in lateral budding after NR5A2 overexpression. 24570488_during chronic colitis, TNF suppresses intestinal steroidogenic gene expression by inhibiting the activity of NR5A2, thus decreasing glucocorticoid synthesis and sustaining chronic inflammation. 24769073_LRH1 overexpression is associated with increased pancreatic cancer growth and metastatic spread 25435372_Analysis of breast cancer samples reveals that a high LRH-1 level is inversely correlated with CDKN1A expression in breast cancer patients and is associated with poor prognosis 25514243_the NR5A2 rs3790844 polymorphism is associated with increased OS of GC patients in the dominant model, and similar results were found among the female group and tumor size >5 cm group for NR5A2 rs3790843 polymorphism. 25675535_This study demonstrates a critical proproliferative role for LRH-1 in established colon cancer cell lines. 25869073_loss of LRH-1 by siRNA or miR-451 mimics significantly impaired Wnt/beta-catenin activity, leading to G0/G1 cell cycle arrest 25873311_Results identify LRH-1 as a critical component of the anti-inflammatory and fungicidal response of alternatively activated macrophages that acts upstream from the IL-13-induced 15-HETE/PPARgamma axis. 25896302_These findings demonstrate that in vitro LRH-1 can act like SF-1 and compensate for its deficiency. 25943101_AFPR may play a pivotal role in HBV-related hepatocarcinogenesis. 25951367_Studies indicate that liver receptor homolog-1 (LRH-1) is critical involvement in multiple types of cancer, and represents a desirable target for therapeutic applications. 25987835_our findings present supportive evidence that ApoM is a regulator of human LRH-1 transcription, and further reveal the importance of ApoM as a critical regulator of bile acids metabolism 26241054_these data demonstrate that copper-mediated nuclear receptor dysfunction disrupts liver function in WD and potentially in other disorders associated with increased hepatic copper levels. 26241668_in spermatozoa the LRH-1 effects are closely integrated with the estrogen signaling, supporting LRH-1 as a downstream effector of the estradiol pathway on some sperm functions. 26320367_Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer cells through up-regulation of LRH-1. 26398198_SERBP1 is a component of the LRH-1 transcriptional complex. 26400164_LRH-1 drives colon cancer cell growth by repressing the expression of the CDKN1A gene in a p53-dependent manner. 26421305_These findings show that POD-1/TCF21 regulates SF-1 and LRH-1 by distinct mechanisms, contributing to the understanding of POD-1 involvement and its mechanisms of action in adrenal and liver tumorigenesis. 26530052_demonstrate aberrant expressions of SF-1 and LRH-1 in endometriotic granulosa-lutein cells 26553876_Data suggest LRH1/NR5A2 exhibits phospholipid-mediated allosteric control of protein-protein binding interface in interactions with TIF2 (co-activator; transcription intermediary factor 2) and SHP (co-repressor; small heterodimer partner protein). 26592175_The SNPs rs3790843 and rs3790844 in the NR5A2 gene are associated with pancreatic cancer risk in Japanese subjects. 26677080_The present study indicates that miR-381 may be a novel tumor suppressor that blocks HCC growth and invasion by targeting LRH-1. 26761123_NR5A2 may be important in the pathophysiology of preterm birth and exploring noncoding regulators of NR5A2 is warranted 27049310_miR-376c inhibits non-small-cell lung cancer cell growth and invasion by targeting LRH-1 27586588_NR5A2-mediated cancer cell survival is facilitated through augmentation of GATA6 and anti-apoptotic factor BCL-XL levels. 27694446_The dramatic repositioning is influenced by a differential ability to establish stable face-to-face pi-pi-stacking with the LRH-1 residue His-390, as well as by a novel polar interaction mediated by the RJW100 hydroxyl group. The differing binding modes result in distinct mechanisms of action for the two agonists. 27809310_miR-27b-3p levels were found to be significantly negatively correlated with both NR5A2 and CREB1 levels in breast cancer tissues. 27983934_Our study suggests that miR-219-5p regulated the proliferation, migration, and invasion of human gastric cancer cells by suppressing LRH-1. 27984042_Additionally, the authors solved the structure of the human LRH-1 DNA-binding domain bound to a DR0 motif located within the Oct4 promoter. 27996162_The role of NR5A2 in regulating pancreatic cancer stem cell properties and epithelial-mesenchymal transition of pancreatic carcinoma cells 28081303_The fusion transcript NR5A2-KLHL29FT was identified in normal and cancerous colonic epithelia. It is due to an uncharacterized polymorphic germline insertion of the NR5A2 sequence from chromosome 1 into the KLHL29 locus at chromosome 2, rather than a chromosomal rearrangement. NR5A2-KLH29FT expression levels were significantly lower in colon cancers than in matched normal colonic epithelia. 28363985_The first crystal structure of the LRH-1-PGC1alpha complex , which depicts hydrophobic contacts is identified and described. 28440426_Liver receptor homologue1 (LRH1) is a direct target of miR30d in colorectal carcinoma cells. 28531169_the expression level of LRH-1 can be used as a marker in the early diagnosis of unexplained recurrent spontaneous abortion. 28710032_Liver receptor homolog-1 was identified as a direct target gene of miR-136 29048619_our data indicated that miR-381 inhibited migration and invasion of non-small cell lung cancer (NSCLC)by targeting LRH-1, and may represent a novel potential therapeutic target and prognostic marker for NSCLC. 29128635_Results show that LRH-1 is a direct target gene of miR-374b and that decreased miR-374b expression may contribute to the promotion of LRH-1-mediated tumorigenesis of colon cancer. 29237721_Rev-erbalpha regulates Cyp7a1 and cholesterol metabolism through its repression of the Lrh-1 receptor. 29438990_These findings not only demonstrate the significant role of the nuclear receptor LRH-1 in the promotion of intratumoral androgen biosynthesis in castration-resistant prostate cancer (CRPC) via its direct transcriptional control of steroidogenesis, but also suggest targeting LRH-1 could be a potential therapeutic strategy for CRPC management. 29443959_transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas; findings support the notion that, in the pancreas, the transcriptional networks involved in differentiation-specific functions also suppress inflammatory programs; under conditions of genetic or environmental constraint, these networks can be subverted to foster inflammation 29515023_LRH-1-deficient adult mice fed high-fat diet displayed macrovesicular steatosis, liver injury, and glucose intolerance, all of which were reversed or improved by expressing wild-type human LRH-1. Results suggest that LRH-1 maintains the pool of arachidonoyl phospholipids, thus ensuring phospholipid diversity and normal lipid homeostasis in the adult liver. 29545602_LRH1 is highly expressed in chemotherapy-resistant breast cancer.LRH1 enhanced breast cancer cell chemoresistance by upregulating MDC1 and attenuating DNA damage. 29669824_Lrh-1 transcriptionally regulates Oat2. 30044146_rs2816948 not significantly associated with recurrent abortions 30273983_The results of our study indicate that LRH1 predicts NSCLC progression, metastasis, and a dismal prognosis, emphasizing its promising role as a novel target in NSCLC therapies. 30305617_LRH-1 maintains intestinal epithelial health and protects against inflammatory damage. 30320362_Results found the mRNA and protein expression levels of LRH1 were significantly higher in HepG2 and HuH6 hepatoblastoma cell lines and results suggest that LRH1 may contribute to cell proliferation in hepatoblastoma. Hepatoblastoma cells with higher LRH1 expression levels more susceptible to LRH1 inhibition. 30638865_study identified LRH1-driven pathway as a circuitry responsible for hepatocyte identity by using cistromic analysis, improving our understanding of liver pathophysiology and identifying novel therapeutic targets. 30740909_we have provided convincing evidence in vitro and in vivo demonstrating that Nr5a2 can induce lung CSC properties and promote tumorigenesis and progression through transcriptional up-regulation of Nanog. 30923324_Regulation of liver receptor homologue-1 by DDB2 E3 ligase activity is critical for hepatic glucose metabolism. 31058195_Using the cells derived from the model, novel SF-1/Ad4BP- and LRH-1-regulated genes were identified by combined DNA microarray and promoter tiling array analyses. The interaction of SF-1/Ad4BP and LRH-1 with transcriptional regulators in the regulation of ovarian steroidogenesis was also revealed. 31328159_This study describes a novel and critical role of LRH-1 in T cell maturation, functions, and immopathologies and proposes LRH-1 as an emerging pharmacological target in the treatment of T cell-mediated inflammatory diseases. 32037723_GATA6 enhances the stemness of human colon cancer cells by creating a metabolic symbiosis through upregulating LRH-1 expression. 32572717_Impact of NR5A2 and RYR2 3'UTR polymorphisms on the risk of breast cancer in a Chinese Han population. 32931651_Nuclear-mitochondrial crosstalk: On the role of the nuclear receptor liver receptor homolog-1 (NR5A2) in the regulation of mitochondrial metabolism, cell survival, and cancer. 33079429_Epithelial Nr5a2 heterozygosity cooperates with mutant Kras in the development of pancreatic cystic lesions. 33252195_Temporal activation of LRH-1 and RAR-gamma in human pluripotent stem cells induces a functional naive-like state. 33335203_Enantiomer-specific activities of an LRH-1 and SF-1 dual agonist. 34129175_LRH-1 high expression in the ovarian granulosa cells of PCOS patients. 34310734_NF-kappaB Regulation of LRH-1 and ABCG5/8 Potentiates Phytosterol Role in the Pathogenesis of Parenteral Nutrition-Associated Cholestasis. 34561301_Nuclear receptor NR5A2 negatively regulates cell proliferation and tumor growth in nervous system malignancies. 34643922_Nuclear receptor subfamily 5 group A member 2 (NR5A2): role in health and diseases. | ENSMUSG00000026398 | Nr5a2 | 28.24819 | 1.3252977 | 0.4063164708 | 0.53220079 | 5.851947e-01 | 4.442834e-01 | No | Yes | 33.815509 | 13.819188 | 23.219589 | 9.510119 | ||
| ENSG00000117280 | 8934 | RAB29 | protein_coding | O14966 | FUNCTION: The small GTPases Rab are key regulators in vesicle trafficking (PubMed:24788816). Essential for maintaining the integrity of the endosome-trans-Golgi network structure (By similarity). Together with LRRK2, plays a role in the retrograde trafficking pathway for recycling proteins, such as mannose 6 phosphate receptor (M6PR), between lysosomes and the Golgi apparatus in a retromer-dependent manner (PubMed:24788816). Recruits LRRK2 to the Golgi complex and stimulates LRRK2 kinase activity (PubMed:29212815). Regulates neuronal process morphology in the intact central nervous system (CNS) (By similarity). May play a role in the formation of typhoid toxin transport intermediates during Salmonella enterica serovar Typhi (S.Typhi) epithelial cell infection (PubMed:22042847). {ECO:0000250|UniProtKB:Q63481, ECO:0000269|PubMed:22042847, ECO:0000269|PubMed:24788816, ECO:0000269|PubMed:29212815}. | 3D-structure;Alternative splicing;Cell membrane;Cytoplasm;Cytoskeleton;Differentiation;GTP-binding;Golgi apparatus;Lipoprotein;Membrane;Nucleotide-binding;Phosphoprotein;Prenylation;Protein transport;Reference proteome;Transport;Vacuole | hsa:8934; | cis-Golgi network [GO:0005801]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; early endosome [GO:0005769]; endomembrane system [GO:0012505]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; intracellular vesicle [GO:0097708]; melanosome [GO:0042470]; mitochondrion [GO:0005739]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; recycling endosome [GO:0055037]; trans-Golgi network [GO:0005802]; vacuole [GO:0005773]; dynein complex binding [GO:0070840]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; kinesin binding [GO:0019894]; small GTPase binding [GO:0031267]; cell differentiation [GO:0030154]; Golgi organization [GO:0007030]; intracellular protein transport [GO:0006886]; melanosome organization [GO:0032438]; mitochondrion organization [GO:0007005]; negative regulation of neuron projection development [GO:0010977]; positive regulation of intracellular protein transport [GO:0090316]; positive regulation of receptor recycling [GO:0001921]; positive regulation of T cell receptor signaling pathway [GO:0050862]; protein localization to ciliary membrane [GO:1903441]; protein localization to membrane [GO:0072657]; regulation of neuron death [GO:1901214]; regulation of retrograde transport, endosome to Golgi [GO:1905279]; response to bacterium [GO:0009617]; retrograde transport, endosome to Golgi [GO:0042147]; synapse assembly [GO:0007416]; T cell activation [GO:0042110]; toxin transport [GO:1901998]; viral RNA genome replication [GO:0039694] | 20683486_Observational study of gene-disease association. (HuGE Navigator) 21812739_Direct DNA sequencing of the RAB7L1 and SLC41A1 genes within the PARK16 locus in 205 Chinese Parkinson's disease patients shows no significant difference with controls. 22232350_This study demonstrated that specific SNP variations and haplotypes in the PARK16 locus are associated with reduced risk for parkinson disease in Ashkenazim. 23395371_This study demonistrated that RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson's disease risk. 23820587_This study confirmed the associations of RAB7L1 with parkinson disease susceptibility and fail to show significant associations of alzheimer disease genome-wide association study (GWAS) top hits with PD susceptibility in a Korean population. 24510904_RAB7L1 is a binding partners of LRRK2, a candidate genes for risk for sporadic Parkinson disease, and part of a complex that promotes clearance of Golgi-derived vesicles through the autophagy-lysosome system. 24788816_Results suggest that Rab protein Rab29 is essential for the integrity of the trans-Golgi network (TGN) and participates in the retrograde trafficking of mannose-6-phosphate receptor (M6PR). 25040112_rs1572931 decreases the risk for Parkinson's disease but not for amyotrophic lateral sclerosis (ALS) and multiple system atrophy(MSA) in the Chinese population. However, the polymorphism is unlikely to be a common cause of sporadic ALS and MSA in the Chinese population 26021297_Rab29 is a regulator of receptor recycling and this GTPase is a shared participant in immune synapse and primary cilium assembly. 26344175_Results confirmed the protective effect of the rs1572931 single nucleotide polymorphism on Parkinson's disease and replicated the results of previous studies, in Iranian subjects. 26914237_Our study provides strong support for the susceptibility role of RAB7L1/NUCKS1 rs823118 and MCCC1 rs12637471 in sporadic Parkinson's disease in a Han Chinese population 28245721_This study showed that the significant differences in genotypic and allelic frequencies of RAB7L1 promoter polymorphism between patients and controls. 28334866_Genetic ablation of RAB7L1 in SH-SY5Y cells recapitulated the findings in amyotrophic lateral sclerosis and frontotemporal dementia fibroblasts and induced pluripotent stem cell neurons 28807727_RAB7L1gene rs1572913 polymorphism (T allele, TC and TT genotype) was associated with decreased risk of PD. 29177506_Results suggest reciprocal regulation between LRRK2 and Rab protein substrates, where Rab7L1-mediated upregulation of LRRK2 kinase activity results in the stabilization of membrane and GTP-bound Rab proteins that may be unable to interact with Rab effector proteins. 29223392_Mutations in leucine-rich repeat kinase 2 (LRRK2) are the major genetic cause of autosomal-dominantly inherited Parkinson's disease. LRRK2 is implicated in the regulation of intracellular trafficking, neurite outgrowth and PD risk in connection with Rab7L1, a putative interactor of LRRK2. The modulation of Ser72 phosphorylation in Rab7L1 resulted in an alteration of the trans-Golgi network. 30037848_This study highlights a novel role of Rab7l1 in the phagosomal maturation process and hints at unique strategies of mycobacteria used to interfere with Rab7l1 function to favor its survival inside human macrophages. 31624137_LRRK2 associates with and dissociates from distinct membrane compartments to phosphorylate Rab substrates including Rab29 31818509_Association of RIT2 and RAB7L1 with Parkinson's disease: a case-control study in a Taiwanese cohort and a meta-analysis in Asian populations. 32709066_Distinct Roles for RAB10 and RAB29 in Pathogenic LRRK2-Mediated Endolysosomal Trafficking Alterations. 33941558_Expression of RAB7L1 in Patients with Pituitary Adenomas. | ENSMUSG00000026433 | Rab29 | 663.04636 | 0.7745488 | -0.3685718892 | 0.12675682 | 8.372809e+00 | 3.808764e-03 | 1.035482e-01 | No | Yes | 756.296164 | 113.579832 | 918.424294 | 134.544184 | ||
| ENSG00000117411 | 8704 | B4GALT2 | protein_coding | O60909 | FUNCTION: Responsible for the synthesis of complex-type N-linked oligosaccharides in many glycoproteins as well as the carbohydrate moieties of glycolipids (PubMed:9405390). Can produce lactose (PubMed:9405390). {ECO:0000269|PubMed:9405390}. | Alternative splicing;Disulfide bond;Glycoprotein;Glycosyltransferase;Golgi apparatus;Manganese;Membrane;Metal-binding;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene is one of seven beta-1,4-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta1,4 linkage to similar acceptor sugars: GlcNAc, Glc, and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. As type II membrane proteins, they have an N-terminal hydrophobic signal sequence that directs the protein to the Golgi apparatus and which then remains uncleaved to function as a transmembrane anchor. By sequence similarity, the beta4GalTs form four groups: beta4GalT1 and beta4GalT2, beta4GalT3 and beta4GalT4, beta4GalT5 and beta4GalT6, and beta4GalT7. The enzyme encoded by this gene synthesizes N-acetyllactosamine in glycolipids and glycoproteins. Its substrate specificity is affected by alpha-lactalbumin but it is not expressed in lactating mammary tissue. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2011]. | hsa:8704; | Golgi apparatus [GO:0005794]; Golgi cisterna membrane [GO:0032580]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity [GO:0003831]; galactosyltransferase activity [GO:0008378]; lactose synthase activity [GO:0004461]; metal ion binding [GO:0046872]; N-acetyllactosamine synthase activity [GO:0003945]; carbohydrate metabolic process [GO:0005975]; cerebellar Purkinje cell layer development [GO:0021680]; glycosylation [GO:0070085]; locomotory behavior [GO:0007626]; memory [GO:0007613]; protein glycosylation [GO:0006486]; visual learning [GO:0008542] | 15504978_Significance of core 2 GlcNAc-T in the pathogenesis of capillary occlusion in diabetic retinopathy. 15939404_Thus, these results suggest that among human beta4GalTs, beta4GalT-II is a major regulator of the synthesis of glycans involved in neuronal development. 17470362_beta4GalT-II increases HeLa cell apoptosis induced by cisplatin depending on its Golgi localization, which indicates that beta4GalT-II might contribute to the therapeutic efficiency of cisplatin for cervix cancer. 18211920_results suggested that beta1,4GalT II might serve as a target gene of p53 transcription factor during adriamycin-induced HeLa cell apoptosis, which elucidated a new mechanism of p53-mediated cell apoptosis 19524017_Data show that Beta4-Gal-transferase (beta4GalT) extends core 2 and forms the backbone structure for biologically important epitopes. 27004849_Study provides evidence that mutations in B3GNT2, B4GALT2, and ST6GALNAC2 underlie aberrant glycosylation, and contribute to the pathogenesis of molecular subsets of colon and other gastrointestinal malignancies. 30020015_In individuals under dual antiplatelet therapy, B4GALT2 c.909C>T is an independent genetic predictor of on-treatment platelet reactivity. | ENSMUSG00000028541 | B4galt2 | 1680.53387 | 0.9820698 | -0.0261025925 | 0.10758142 | 5.882099e-02 | 8.083692e-01 | 9.464997e-01 | No | Yes | 2024.572245 | 293.986216 | 1964.849152 | 278.866574 |
| ENSG00000117519 | 1266 | CNN3 | protein_coding | Q15417 | FUNCTION: Thin filament-associated protein that is implicated in the regulation and modulation of smooth muscle contraction. It is capable of binding to actin, calmodulin and tropomyosin. The interaction of calponin with actin inhibits the actomyosin Mg-ATPase activity. | Acetylation;Actin-binding;Alternative splicing;Calmodulin-binding;Direct protein sequencing;Methylation;Phosphoprotein;Reference proteome;Repeat | This gene encodes a protein with a markedly acidic C terminus; the basic N-terminus is highly homologous to the N-terminus of a related gene, CNN1. Members of the CNN gene family all contain similar tandemly repeated motifs. This encoded protein is associated with the cytoskeleton but is not involved in contraction. [provided by RefSeq, Jul 2008]. | hsa:1266; | actin cytoskeleton [GO:0015629]; adherens junction [GO:0005912]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; focal adhesion [GO:0005925]; actin binding [GO:0003779]; cadherin binding involved in cell-cell adhesion [GO:0098641]; calmodulin binding [GO:0005516]; actomyosin structure organization [GO:0031032]; epithelial cell differentiation [GO:0030855] | Mouse_homologues 20181831_h3/acidic calponin controls fibroblast migration by regulation of ERK1/2-mediated l-caldesmon phosphorylation. 29151265_provides evidence for an important role of Cnn3 during development of the embryonic brain and in regulating NSC function 31992794_These results reveal that CNN3 plays a crucial role in regulating lens epithelial contractile activity and provide supporting evidence that CNN-3 deficiency is associated with the induction of epithelial plasticity, fibrogenic activity and mechanosensitive Yap/Taz (Wwtr1) transcriptional activation. | ENSMUSG00000053931 | Cnn3 | 2431.39279 | 1.0753851 | 0.1048534093 | 0.08625526 | 1.481495e+00 | 2.235408e-01 | 6.151503e-01 | No | Yes | 3270.800568 | 640.751509 | 2856.573355 | 547.154709 | |
| ENSG00000118873 | 25782 | RAB3GAP2 | protein_coding | Q9H2M9 | FUNCTION: Regulatory subunit of a GTPase activating protein that has specificity for Rab3 subfamily (RAB3A, RAB3B, RAB3C and RAB3D). Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Rab3 GTPase-activating complex specifically converts active Rab3-GTP to the inactive form Rab3-GDP. Required for normal eye and brain development. May participate in neurodevelopmental processes such as proliferation, migration and differentiation before synapse formation, and non-synaptic vesicular release of neurotransmitters. {ECO:0000269|PubMed:9733780}. | Alternative splicing;Cataract;Cytoplasm;Disease variant;GTPase activation;Mental retardation;Phosphoprotein;Reference proteome | The protein encoded by this gene belongs to the RAB3 protein family, members of which are involved in regulated exocytosis of neurotransmitters and hormones. This protein forms the Rab3 GTPase-activating complex with RAB3GAP1, where it constitutes the regulatory subunit, whereas the latter functions as the catalytic subunit. This gene has the highest level of expression in the brain, consistent with it having a key role in neurodevelopment. Mutations in this gene are associated with Martsolf syndrome.[provided by RefSeq, Oct 2009]. | hsa:25782; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; enzyme activator activity [GO:0008047]; enzyme regulator activity [GO:0030234]; GTPase activator activity [GO:0005096]; small GTPase binding [GO:0031267]; establishment of protein localization to endoplasmic reticulum membrane [GO:0097051]; intracellular protein transport [GO:0006886]; positive regulation of autophagosome assembly [GO:2000786]; positive regulation of endoplasmic reticulum tubular network organization [GO:1903373]; positive regulation of protein lipidation [GO:1903061]; regulation of GTPase activity [GO:0043087] | 18485483_Data show that at chemical synapses, Rab3 performs specific functions in synpatic vesicle exocytosis. 18849981_KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator RAB3GAP2 occurs in a GTP-dependent manner through MADD. 20967465_functionally severe RAB3GAP2 mutations cause Warburg Micro syndrome while hypomorphic RAB3GAP2 mutations can result in the milder Martsolf phenotype 23176487_Micro syndrome has been associated with causative mutations in three disease genes: RAB3GAP1, RAB3GAP2 and RAB18. Martsolf syndrome has been associated with a mutation in RAB3GAP2. [Review] 23420520_One-hundred and forty-four Micro and nine Martsolf families were investigated, identifying mutations in RAB3GAP1 in 41% of cases, mutations in RAB3GAP2 in 7% of cases, and mutations in RAB18 in 5% of cases 24891604_Rab18 and a Rab18 GEF complex of Rab3GAP1 and Rab3GAP2 have roles in the endoplasmic reticulum structure 25495476_RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy 28342870_RAB18 modulates macroautophagy and proteostasis, and is dependent on activity of RAB3GAP1 and RAB3GAP2. 28575017_show that FOXC1 regulates the expression of RAB3GAP1, RAB3GAP2 and SNAP25 29419336_Mutation in RAB3GAP2 gene is associated with Warburg micro syndrome and Martsolf syndrome. 32248620_The Warburg Micro Syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I. 32376645_Hypogonadotropic hypogonadism due to variants in RAB3GAP2: expanding the phenotypic and genotypic spectrum of Martsolf syndrome. 32740904_Micro and Martsolf syndromes in 34 new patients: Refining the phenotypic spectrum and further molecular insights. | ENSMUSG00000039318 | Rab3gap2 | 2500.55838 | 1.0441997 | 0.0623976057 | 0.09005175 | 4.807959e-01 | 4.880620e-01 | 8.136483e-01 | No | Yes | 3122.959171 | 631.137463 | 2857.986193 | 564.996311 | |
| ENSG00000118985 | 22936 | ELL2 | protein_coding | O00472 | FUNCTION: Elongation factor component of the super elongation complex (SEC), a complex required to increase the catalytic rate of RNA polymerase II transcription by suppressing transient pausing by the polymerase at multiple sites along the DNA. Component of the little elongation complex (LEC), a complex required to regulate small nuclear RNA (snRNA) gene transcription by RNA polymerase II and III (PubMed:22195968). Plays a role in immunoglobulin secretion in plasma cells: directs efficient alternative mRNA processing, influencing both proximal poly(A) site choice and exon skipping, as well as immunoglobulin heavy chain (IgH) alternative processing. Probably acts by regulating histone modifications accompanying transition from membrane-specific to secretory IgH mRNA expression. {ECO:0000269|PubMed:20159561, ECO:0000269|PubMed:20471948, ECO:0000269|PubMed:22195968, ECO:0000269|PubMed:23251033}. | 3D-structure;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | hsa:22936; | nucleoplasm [GO:0005654]; transcription elongation factor complex [GO:0008023]; snRNA transcription by RNA polymerase II [GO:0042795]; transcription elongation from RNA polymerase II promoter [GO:0006368] | 19749764_ELL2 addition regulates mRNA processing by enhancing poly(A) site choice and exon splice-site skipping 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22483617_Prostratin and HMBA, two well-studied activators of HIV transcription and latency, enhance ELL2 accumulation and SECs formation largely through decreasing Siah1 expression and ELL2 polyubiquitination. 25058508_Tax transactivates the ELL2 promoter in HTLV-1-infected T-cells. 25238757_Loss of ELL2 in B cells results in decreased Igh secretory mRNA and decreased expression of Ig light chain plus genes involved in the unfolded protein response like XBP1, ATF6, BiP, cyclin B2, OcaB (BOB1, Pou2af1). 26007630_multiple myeloma risk allele harbours a Thr298Ala missense variant in an ELL2 domain required for transcription elongation 28134250_Study reports the 2.0-A resolution crystal structure of the human ELL2 C-terminal domain bound to its 50-residue binding site on AFF4, the ELLBow. The ELLBow consists of an N-terminal helix followed by an extended hairpin and occupies most of the concave surface of ELL2. This surface is important for the ability of ELL2 to promote HIV-1 Tat-mediated proviral transcription. 28531325_Results provide the first evidence that ELL2 is a direct target of miR-299 and increased ELL2 expression and down-regulation of miR-299 are associated with GBM progression and poor prognosis in patients. 28858629_Selective expression of the transcription elongation factor ELL3 in B cells prior to ELL2 drives proliferation and survival 28870994_These results suggest that ELL2 and its pathway genes likely play an important role in the development and progression of prostate cancer. 28903037_expression is consistent with inherited genetic variation contributing to arrest of plasma cell development, facilitating multiple myeloma clonal expansion. 29179998_Knockdown of RNA polymerase II elongation factor ELL2 (ELL2) sensitized prostate cancer cells to DNA damage and overexpression of ELL2 protected prostate cancer cells from DNA damage. 29695719_Study in CD138+ plasma cells from 1630 multiple myeloma (MM) patients show that the MM risk allele at 5q15 lowers ELL2 expression in these cells, but not in peripheral blood or other tissues. 30009504_ELL2 protein was downregulated in prostate cancer specimens and was up-regulated by androgens in prostate cancer cell lines LNCaP and C4-2. ELL2 knockdown enhanced prostate cancer cell proliferation and motility. 30556882_MRCCAT1 is upregulated in glioma, predicting poor outcome for patients. MRCCAT1 promotes glioma cell proliferation and migration by activating p38-MAPK signaling. 34265249_Allosteric transcription stimulation by RNA polymerase II super elongation complex. 34948391_Characterizing the Interaction between the HTLV-1 Transactivator Tax-1 with Transcription Elongation Factor ELL2 and Its Impact on Viral Transactivation. | ENSMUSG00000001542 | Ell2 | 695.73191 | 1.1909774 | 0.2521460868 | 0.13749579 | 3.375770e+00 | 6.616164e-02 | 3.827638e-01 | No | Yes | 1199.099941 | 289.210180 | 934.443086 | 220.550501 | ||
| ENSG00000119723 | 51004 | COQ6 | protein_coding | Q9Y2Z9 | FUNCTION: FAD-dependent monooxygenase required for the C5-ring hydroxylation during ubiquinone biosynthesis. Catalyzes the hydroxylation of 3-hexaprenyl-4-hydroxybenzoic acid (HHB) to 3-hexaprenyl-4,5-dihydroxybenzoic acid (DHHB). The electrons required for the hydroxylation reaction may be funneled indirectly from NADPH via a ferredoxin/ferredoxin reductase system to COQ6 (By similarity). Is able to perform the deamination reaction at C4 of 3-hexaprenyl-4-amino-5-hydroxybenzoic acid (HHAB) to produce DHHB when expressed in yeast cells lacking COQ9, even if utilization of para-aminobenzoic acid (pABA) involving C4-deamination seems not to occur in bacteria, plants and mammals, where only C5 hydroxylation of HHB has been shown (PubMed:26260787). {ECO:0000255|HAMAP-Rule:MF_03193, ECO:0000269|PubMed:26260787}. | Alternative splicing;Cell projection;Deafness;Disease variant;FAD;Flavoprotein;Golgi apparatus;Membrane;Mitochondrion;Mitochondrion inner membrane;Monooxygenase;Oxidoreductase;Primary mitochondrial disease;Reference proteome;Transit peptide;Ubiquinone biosynthesis | PATHWAY: Cofactor biosynthesis; ubiquinone biosynthesis. {ECO:0000255|HAMAP-Rule:MF_03193}. | The protein encoded by this gene belongs to the ubiH/COQ6 family. It is an evolutionarily conserved monooxygenase required for the biosynthesis of coenzyme Q10 (or ubiquinone), which is an essential component of the mitochondrial electron transport chain, and one of the most potent lipophilic antioxidants implicated in the protection of cell damage by reactive oxygen species. Knockdown of this gene in mouse and zebrafish results in decreased growth due to increased apoptosis. Mutations in this gene are associated with autosomal recessive coenzyme Q10 deficiency-6 (COQ10D6), which manifests as nephrotic syndrome with sensorineural deafness. Alternatively spliced transcript variants encoding different isoforms have been described for this gene. [provided by RefSeq, Jun 2012]. | hsa:51004; | cell projection [GO:0042995]; extrinsic component of mitochondrial inner membrane [GO:0031314]; Golgi apparatus [GO:0005794]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; ubiquinone biosynthesis complex [GO:0110142]; 2-octaprenyl-6-methoxyphenol hydroxylase activity [GO:0008681]; 4-hydroxy-3-all-trans-hexaprenylbenzoate oxygenase activity [GO:0106364]; FAD binding [GO:0071949]; oxidoreductase activity [GO:0016491]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen [GO:0016709]; ubiquinone biosynthetic process [GO:0006744] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 21540551_COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness 24763291_Data indicate a heterozygous loss-of-function coenzyme Q10 (CoQ10) biosynthesis monooxygenase 6 gene (COQ6)missense mutation in familial schwannomatosis. 28044327_CoQ10 deficiency associated with steroid resistant nephrotic syndrome and novel homozygous missense change in COQ6 gene: p.Pro261Leu. 28117207_Primary CoQ10 deficiency due to COQ6 mutations should be considered in children presenting with both Steroid-Resistant Focal Segmental Glomerulosclerosis and sensorineural hearing loss. An early diagnosis of COQ6 mutations is essential because the condition is treatable when CoQ10 supplementation is started at the early stage. 30682496_Primary coenzyme Q10 Deficiency-6 (COQ10D6): Two siblings with variable expressivity of the renal phenotype. | ENSMUSG00000021235 | Coq6 | 830.42989 | 0.9326624 | -0.1005730843 | 0.11338743 | 7.846142e-01 | 3.757337e-01 | 7.439902e-01 | No | Yes | 948.761638 | 67.489893 | 999.622107 | 69.148047 |
| ENSG00000119778 | 54454 | ATAD2B | protein_coding | Q9ULI0 | 3D-structure;ATP-binding;Alternative splicing;Bromodomain;Coiled coil;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene belongs to the AAA ATPase family. This family member includes an N-terminal bromodomain. It has been found to be localized to the nucleus, partly to replication sites, consistent with a chromatin-related function. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jul 2014]. | hsa:54454; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; chromatin binding [GO:0003682]; histone binding [GO:0042393]; lysine-acetylated histone binding [GO:0070577]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 21158754_The ATAD2B may play a role in neuronal differentiation and tumor progression. | ENSMUSG00000052812 | Atad2b | 991.39051 | 1.0763683 | 0.1061718715 | 0.10896046 | 9.512462e-01 | 3.294023e-01 | 7.070368e-01 | No | Yes | 1219.704478 | 256.911574 | 1033.349422 | 213.192620 | ||
| ENSG00000120093 | 3213 | HOXB3 | protein_coding | P14651 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | Alternative splicing;DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene is a member of the Antp homeobox family and encodes a nuclear protein with a homeobox DNA-binding domain. It is included in a cluster of homeobox B genes located on chromosome 17. The encoded protein functions as a sequence-specific transcription factor that is involved in development. Increased expression of this gene is associated with a distinct biologic subset of acute myeloid leukemia (AML). [provided by RefSeq, Jul 2008]. | hsa:3213; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; angiogenesis [GO:0001525]; anterior/posterior pattern specification [GO:0009952]; cartilage development [GO:0051216]; definitive hemopoiesis [GO:0060216]; embryonic skeletal system morphogenesis [GO:0048704]; face development [GO:0060324]; glossopharyngeal nerve morphogenesis [GO:0021615]; hematopoietic progenitor cell differentiation [GO:0002244]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of neurogenesis [GO:0050767]; regulation of transcription by RNA polymerase II [GO:0006357]; rhombomere development [GO:0021546]; thyroid gland development [GO:0030878] | 14966272_Data report novel nucleoporin 98 fusions with homeobox (HOX)A10, HOXB3 and HOXB4, and describe the results of coexpression of these proteins with the Hox cofactor Meis1 in leukemic induction. 15674412_The HOXb3 was only weakly expressed in the inv(7) positive patients. 17972163_HOXA7, HOXB3, HOXA3, and HOXB13 expression levels changed during angiogenesis, sugessting these proteins might be involved in the angiogenesis of hMSCs. 19477923_Data show that inducible Hox genes are selectively sensitive to the inhibition of actin polymerization and that actin polymerization is required for the assembly of a transcription complex on the regulatory region of the Hox genes. 19854132_RASSF1A silencing strongly correlates with overexpression of HOXB3 and DNMT3B. 23219899_HoxB3 promote prostate cancer progression by upregulating CDCA3 expression. 24127533_describe familial cases of TH in two generations (proband and his father), in addition to other two sporadic cases. We have found polymorphisms in the HOXB3, HOXD3, and a new synonymous variant, and PITX2 genes 25996682_decreased methylation at HOXB3 and HOXB4 was associated with increased gene expression of both HOXB genes specific to the mid-risk AML, while increased DNA methylation at DCC distinctive to the high-risk AML was associated with increased gene expression 26482852_HOXB2 and HOXB3 act as tumor suppressors in acute myeloid leukemia patients carrying the FLT3 protein mutations. 27447302_miR-10b might control cell apoptosis, proliferation, migration, and invasion in endometrial cancer via regulation of HOXB3 expression. 28075453_HOXB3 is degraded by miR-375 in breast cancer cells.HOXB3 plays role in tamoxifen resistance. 29402501_HOXA4/HOXB3 gene expression-based risk score may be useful for prognostic risk stratification and warrants prospective validation in HGSOC patients. 29439669_Data indicate a miR-375-HOXB3-CDCA3/DNMT3B regulatory circuitry which contributes to leukemogenesis and suggest a therapeutic strategy of restoring miR-375 expression in Acute myeloid leukemia (AML). 31702491_miR-7 Reduces High Glucose Induced-damage Via HoxB3 and PI3K/AKT/mTOR Signaling Pathways in Retinal Pigment Epithelial Cells. 32748508_Levels of serum Hoxb3 and sFlt-1 in pre-eclamptic patients and their effects on pregnancy outcomes. 33352356_CSF levels of HoxB3 and YKL-40 may predict conversion from clinically isolated syndrome to relapsing remitting multiple sclerosis. 35127344_Extracellular Vesicle-Encapsulated MicroRNA-375 from Bone Marrow-Derived Mesenchymal Stem Cells Inhibits Hepatocellular Carcinoma Progression through Regulating HOXB3-Mediated Wnt/beta-Catenin Pathway. | ENSMUSG00000048763 | Hoxb3 | 521.27045 | 1.0893366 | 0.1234497899 | 0.13553795 | 8.316433e-01 | 3.617978e-01 | 7.334437e-01 | No | Yes | 588.037835 | 118.332522 | 561.331877 | 110.477132 | |
| ENSG00000120159 | 79886 | CAAP1 | protein_coding | Q9H8G2 | FUNCTION: Anti-apoptotic protein that modulates a caspase-10 dependent mitochondrial caspase-3/9 feedback amplification loop. {ECO:0000269|PubMed:21980415}. | Alternative splicing;Apoptosis;Coiled coil;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:79886; | apoptotic process [GO:0006915]; negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway [GO:2001268] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21980415_The data suggest that C9orf82 functions as an anti-apoptotic protein that modulates a caspase-10 dependent mitochondrial caspase-3/9 feedback amplification loop. It is designated as Conserved Anti-Apoptotic Protein (CAAP). 32315812_MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. 33336752_LncRNA LncOGD-1006 alleviates OGD-induced ischemic brain injury regulating apoptosis through miR-184-5p/CAAP1 axis. | ENSMUSG00000028578 | Caap1 | 1004.51801 | 1.1466321 | 0.1974025827 | 0.11603895 | 2.904444e+00 | 8.833569e-02 | 4.317506e-01 | No | Yes | 1413.011071 | 246.462429 | 1209.433559 | 206.421176 | ||
| ENSG00000120458 | 79684 | MSANTD2 | protein_coding | Q6P1R3 | Alternative splicing;Isopeptide bond;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:79684; | 19240061_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000042138 | Msantd2 | 357.30160 | 0.9773632 | -0.0330332548 | 0.16031755 | 4.241325e-02 | 8.368341e-01 | 9.555565e-01 | No | Yes | 388.117670 | 71.668907 | 392.497999 | 70.810065 | ||||
| ENSG00000120616 | 80314 | EPC1 | protein_coding | Q9H2F5 | FUNCTION: Component of the NuA4 histone acetyltransferase (HAT) complex, a multiprotein complex involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A (PubMed:14966270). The NuA4 complex plays a direct role in repair of DNA double-strand breaks (DSBs) by promoting homologous recombination (HR) (PubMed:27153538). The NuA4 complex is also required for spermatid development by promoting acetylation of histones: histone acetylation is required for histone replacement during the transition from round to elongating spermatids (By similarity). In the NuA4 complex, EPC1 is required to recruit MBTD1 into the complex (PubMed:32209463). {ECO:0000250|UniProtKB:Q8C9X6, ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:27153538, ECO:0000269|PubMed:32209463}. | 3D-structure;Activator;Alternative splicing;Chromatin regulator;Cytoplasm;Differentiation;Direct protein sequencing;Growth regulation;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a member of the polycomb group (PcG) family. The encoded protein is a component of the NuA4 histone acetyltransferase complex and can act as both a transcriptional activator and repressor. The encoded protein has been linked to apoptosis, DNA repair, skeletal muscle differentiation, gene silencing, and adult T-cell leukemia/lymphoma. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2012]. | hsa:80314; | intracellular membrane-bounded organelle [GO:0043231]; NuA4 histone acetyltransferase complex [GO:0035267]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; Piccolo NuA4 histone acetyltransferase complex [GO:0032777]; histone acetylation [GO:0016573]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; negative regulation of gene expression, epigenetic [GO:0045814]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of cell cycle [GO:0051726]; regulation of growth [GO:0040008]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription, DNA-templated [GO:0006351] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 17192267_Epc1 plays a role in the initiation of skeletal muscle differentiation, and its interaction with Hop is required for the full activity. 18984674_Observational study of gene-disease association. (HuGE Navigator) 19484761_SNP array CGH analysis of the breakpoint region in 3 ATLL-related cell lines and 4 patient samples revealed the chromosomal breakpoints are localized within the EPC1 gene locus in an ATLL-derived cell line (SO4) and in one patient with acute-type ATLL. 24166297_EPC1 and EPC2 are components of a complex that directly or indirectly serves to prevent MYC accumulation and AML cell apoptosis, thus sustaining oncogenic potential 25755723_Silencing EPC1 by short hairpin RNA technology had the inhibition effects on cell proliferation and tumor growth in lung cancer 26350215_Epigenetic factor EPC1 is a master regulator of DNA damage response by interacting with E2F1 to silence death and activate metastasis-related gene signatures 32209463_Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex. | ENSMUSG00000024240 | Epc1 | 770.20479 | 1.3200532 | 0.4005960373 | 0.11946164 | 1.132439e+01 | 7.649572e-04 | 4.547799e-02 | No | Yes | 1081.678792 | 167.757070 | 777.728065 | 118.110863 | |
| ENSG00000120885 | 1191 | CLU | protein_coding | P10909 | FUNCTION: [Isoform 1]: Functions as extracellular chaperone that prevents aggregation of non native proteins (PubMed:11123922, PubMed:19535339). Prevents stress-induced aggregation of blood plasma proteins (PubMed:11123922, PubMed:12176985, PubMed:17260971, PubMed:19996109). Inhibits formation of amyloid fibrils by APP, APOC2, B2M, CALCA, CSN3, SNCA and aggregation-prone LYZ variants (in vitro) (PubMed:12047389, PubMed:17412999, PubMed:17407782). Does not require ATP (PubMed:11123922). Maintains partially unfolded proteins in a state appropriate for subsequent refolding by other chaperones, such as HSPA8/HSC70 (PubMed:11123922). Does not refold proteins by itself (PubMed:11123922). Binding to cell surface receptors triggers internalization of the chaperone-client complex and subsequent lysosomal or proteasomal degradation (PubMed:21505792). Protects cells against apoptosis and against cytolysis by complement (PubMed:2780565). Intracellular forms interact with ubiquitin and SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complexes and promote the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:20068069). Promotes proteasomal degradation of COMMD1 and IKBKB (PubMed:20068069). Modulates NF-kappa-B transcriptional activity (PubMed:12882985). A mitochondrial form suppresses BAX-dependent release of cytochrome c into the cytoplasm and inhibit apoptosis (PubMed:16113678, PubMed:17689225). Plays a role in the regulation of cell proliferation (PubMed:19137541). An intracellular form suppresses stress-induced apoptosis by stabilizing mitochondrial membrane integrity through interaction with HSPA5 (PubMed:22689054). Secreted form does not affect caspase or BAX-mediated intrinsic apoptosis and TNF-induced NF-kappa-B-activity (PubMed:24073260). Secreted form act as an important modulator during neuronal differentiation through interaction with STMN3 (By similarity). Plays a role in the clearance of immune complexes that arise during cell injury (By similarity). {ECO:0000250|UniProtKB:P05371, ECO:0000250|UniProtKB:Q06890, ECO:0000269|PubMed:11123922, ECO:0000269|PubMed:12047389, ECO:0000269|PubMed:12176985, ECO:0000269|PubMed:12882985, ECO:0000269|PubMed:16113678, ECO:0000269|PubMed:17260971, ECO:0000269|PubMed:17407782, ECO:0000269|PubMed:17412999, ECO:0000269|PubMed:17689225, ECO:0000269|PubMed:19137541, ECO:0000269|PubMed:19535339, ECO:0000269|PubMed:19996109, ECO:0000269|PubMed:20068069, ECO:0000269|PubMed:21505792, ECO:0000269|PubMed:22689054, ECO:0000269|PubMed:24073260, ECO:0000269|PubMed:2780565}.; FUNCTION: [Isoform 6]: Does not affect caspase or BAX-mediated intrinsic apoptosis and TNF-induced NF-kappa-B-activity. {ECO:0000269|PubMed:24073260}.; FUNCTION: [Isoform 4]: Does not affect caspase or BAX-mediated intrinsic apoptosis and TNF-induced NF-kappa-B-activity (PubMed:24073260). Promotes cell death through interaction with BCL2L1 that releases and activates BAX (PubMed:21567405). {ECO:0000269|PubMed:21567405, ECO:0000269|PubMed:24073260}. | Alternative splicing;Apoptosis;Chaperone;Complement pathway;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Immunity;Innate immunity;Membrane;Microsome;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Secreted;Signal;Ubl conjugation | The protein encoded by this gene is a secreted chaperone that can under some stress conditions also be found in the cell cytosol. It has been suggested to be involved in several basic biological events such as cell death, tumor progression, and neurodegenerative disorders. Alternate splicing results in both coding and non-coding variants.[provided by RefSeq, May 2011]. | hsa:1191; | apical dendrite [GO:0097440]; blood microparticle [GO:0072562]; cell surface [GO:0009986]; chromaffin granule [GO:0042583]; collagen-containing extracellular matrix [GO:0062023]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; neurofibrillary tangle [GO:0097418]; nucleus [GO:0005634]; perinuclear endoplasmic reticulum lumen [GO:0099020]; perinuclear region of cytoplasm [GO:0048471]; platelet alpha granule lumen [GO:0031093]; protein-containing complex [GO:0032991]; spherical high-density lipoprotein particle [GO:0034366]; synapse [GO:0045202]; amyloid-beta binding [GO:0001540]; chaperone binding [GO:0051087]; low-density lipoprotein particle receptor binding [GO:0050750]; misfolded protein binding [GO:0051787]; protein carrier chaperone [GO:0140597]; protein heterodimerization activity [GO:0046982]; protein-containing complex binding [GO:0044877]; signaling receptor binding [GO:0005102]; tau protein binding [GO:0048156]; ubiquitin protein ligase binding [GO:0031625]; unfolded protein binding [GO:0051082]; cell morphogenesis [GO:0000902]; central nervous system myelin maintenance [GO:0032286]; chaperone-mediated protein complex assembly [GO:0051131]; chaperone-mediated protein folding [GO:0061077]; complement activation [GO:0006956]; complement activation, classical pathway [GO:0006958]; immune complex clearance [GO:0002434]; innate immune response [GO:0045087]; intrinsic apoptotic signaling pathway [GO:0097193]; lipid metabolic process [GO:0006629]; microglial cell activation [GO:0001774]; microglial cell proliferation [GO:0061518]; negative regulation of amyloid fibril formation [GO:1905907]; negative regulation of amyloid-beta formation [GO:1902430]; negative regulation of cell death [GO:0060548]; negative regulation of cellular response to thapsigargin [GO:1905892]; negative regulation of cellular response to tunicamycin [GO:1905895]; negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage [GO:1902230]; negative regulation of protein-containing complex assembly [GO:0031333]; negative regulation of release of cytochrome c from mitochondria [GO:0090201]; negative regulation of response to endoplasmic reticulum stress [GO:1903573]; positive regulation of amyloid fibril formation [GO:1905908]; positive regulation of amyloid-beta formation [GO:1902004]; positive regulation of apoptotic process [GO:0043065]; positive regulation of gene expression [GO:0010628]; positive regulation of intrinsic apoptotic signaling pathway [GO:2001244]; positive regulation of neurofibrillary tangle assembly [GO:1902998]; positive regulation of neuron death [GO:1901216]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of receptor-mediated endocytosis [GO:0048260]; positive regulation of tau-protein kinase activity [GO:1902949]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of ubiquitin-dependent protein catabolic process [GO:2000060]; protein import [GO:0017038]; protein stabilization [GO:0050821]; protein targeting to lysosome involved in chaperone-mediated autophagy [GO:0061740]; regulation of amyloid-beta clearance [GO:1900221]; regulation of apoptotic process [GO:0042981]; regulation of cell population proliferation [GO:0042127]; regulation of neuron death [GO:1901214]; regulation of neuronal signal transduction [GO:1902847]; release of cytochrome c from mitochondria [GO:0001836]; response to misfolded protein [GO:0051788]; response to virus [GO:0009615]; reverse cholesterol transport [GO:0043691] | 10066740_Clusterin is a holdase-type extracellular chaperone 10694874_Clusterin has a chaperone action like that of the small heat shock proteins 11123922_The chaperone action of clusterin is ATP-independent 11714447_Introduction of clusterin gene into human renal cell carcinoma cells enhances their resistance to cytotoxic chemotherapy through inhibition of apoptosis both in vitro and in vivo 11813210_expression increases early after androgen withdrawal in prostate cancer; protects tumor cells from apoptosis induced by medical castration 11892985_Apo J senescence-related overexpression is proposed to have antiapoptotic rather than antiproliferative effects. 11904161_The chaperone action of clusterin targets slowly aggregating proteins 12082621_Clusterin (SGP-2) transient overexpression decreases proliferation rate of SV40-immortalized human prostate epithelial cells by slowing down cell cycle progression. 12172907_clusterin may modify the formation of alpha-synuclein-positive inclusion bodies such as Lewy bodies and GCIs, through a previously proposed chaperone property of clusterin 12176985_Low pH activates the chaperone action of clusterin 12176985_clusterin is activated in reduced pH 12200037_role during cellular senescence and tumorigenesis [review] 12427144_The overall pool of clusterin is reduced in glomerular diseases causing nephrotic syndrome, with hypercholesterolemia appearing as the unifying feature 12429802_Authors conclude that clusterin is a marker of anaplastic large cell lymphoma and that addition of clusterin to antibody panels designed to distinguish systemic anaplastic large cell lymphoma from classical Hodgkin's disease is useful. 12551933_synthesis and functional analysis 12679903_Loss of this protein both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. 12860995_clusterin may act as a negative regulator of MT6-MMP in vivo 12882985_apolipoprotein J has a role in inhibiting NF-kappaB signaling through stabilization of IkappaBs; this activity may result in suppression of tumor cell motility 14618611_Clusterin is downregulated in CaP in comparison with matched benign controls 15033782_Clusterin is downregulated during prostate cancer onset and progression, and its upregulation has inhibited DNA synthesis and cell cycle progression. 15133840_Data suggest that the N-terminal deletion of clusterin may be essential for its alterations of biogenesis in esophageal squamous cell carcinoma. 15158456_clusterin is involved in 15d-PGJ(2)-induced nodule formation and cell differentiation in vascular smooth muscle cells 15247015_although CLU is essential for cellular homeostasis, it may become highly cytotoxic in certain cellular contexts when it accumulates in high amounts intracellularly either by direct synthesis or by uptake from the extracellular milieu 15252304_Clusterin staining supports a diagnosis of follicular dendritic cell tumor and helps classify them. 15304052_platelet ADP receptor P2Y(12) and clusterin are downregulated in patients with systemic lupus erythematosus 15389725_findings suggest that clusterin may contribute to conferring resistance to oxidative stress-mediated cellular injury on prostate cancer cells, especially in the presence of androgen 15492264_Nuclear clusterin function is proapoptotic in colon cancer when induced by APC or chemotherapy in the context of p21 expression. 15499376_Calcium deprivation causes translocation of a 45kDa CLU isoform to the nucleus in human prostate epithelial cells, leading to inhibition of cell proliferation and caspase-cascade-dependent anoikis. 15538973_The Ectopic overexpression of the secreted form of clusterin/apoliporotein J the sensitivity of HaCaT cells to toxic effects of ropivacaine as demonstrated by DNA fragmentation. 15591223_The role of clusterin & its isoforms in SMC behavior is complex & chronologically regulated in response to microenvironmental changes. Proliferative arrest reduces s-CLU; apoptosis increases n-CLU. 15649646_CLU may become highly cytostatic and/or cytotoxic if it accumulates intracellularly in high amounts 15689620_Data show that ionizing radiation causes stress-induced activation of insulin-like growth factor-1 receptor-Src-Mek-Erk-Egr-1 signaling that regulates the clusterin pro-survival cascade. 15791650_Clusterin mRNA is increased twofold in early exponential phase of cell proliferation followed by downregulation in the subsequent quiescence phase, whereas it is rapidly increased up to twelvefold upon UV-induced apoptosis. 15809754_Antisense oligonucleotide treatment may improve the outcome of radiation therapy for patients with bladder cancer. 15883054_Observational study of gene-disease association. (HuGE Navigator) 15925890_Observational study of gene-disease association. (HuGE Navigator) 15925890_The 866C-->T polymorphism might be associated with preeclampsia and essential hypertension. 15929184_overexpression of cytoplasmic clusterin might be involved in the tumorigenesis and/or progression of colorctal cancers 15955107_Clusterin is strongly expressed in melanoma. Downregulation of clusterin reduces drug-resistance, i.e., reduces melanoma cell survival in response to cytotoxic drugs. Reducing clusterin may be novel tool to overcome drug-resistance in melanoma. 16113678_Taken together, our results suggest that the elevated level of clusterin in human cancers may promote oncogenic transformation and tumour progression by interfering with Bax pro-apoptotic activities. 16179938_the importance of CLU in various physiological functions including tumour growth--review 16331665_CLU expression/sub-cellular localization is strictly related to cell fate. 16464517_following exposure to sub-lethal amounts of iron-ascorbate to induce an increase in reactive oxygen species generation and lipid peroxidation, a time-dependent up-regulation of clusterin protein and mRNA levels was observed in neuroblastoma cells 16490286_The results show that clusterin is significantly increased in cerebrospinal fluid from Alzheimer patients as a group, supporting that clusterin might be involved in the pathogenesis of Alzheimer's disease. 16675913_Clusterin and bcl-2 are upregulated in laryngeal carcinomas and their expression is related to invasiveness of these tumors. 16709604_stem cell factor and clusterin may have roles in azoospermia 16709934_clusterin may have a protective effect against cigarette smoke-induced oxidative stress in lung fibroblasts. 16775601_Binding of clusterin to the LDL-R might offer an interpretative key for the pathogenesis of membranous glomerulonephritis in humans 16955214_CLU is actively involved in both replicative senescence and terminal differentiation in different types of human haematopoietic cells. 17048076_This review summarizes our current understanding of the role of clusterin in DNA repair, apoptosis, and cell cycle control and the relevance. 17056579_In rheumatoid arthritis joints, high levels of extracellular CLU and low expression of intracellular CLU may enhance NF-kappaB activation and survival of the synoviocytes. 17080454_low-power 60 GHz radiation does not modify stress-sensitive gene expression of chaperone proteins. 17148459_Clusterin mRNA and protein are shown to increase with androgen treatment in a time- and dose-dependent manner 17203891_CLU immunoreactivity may be helpful as a supplementary criterion to better assess the tumors propensity to relapse in breast carcinoma. 17224269_expression of sCLU modulates growth regulatory effects of 1,25(OH)(2)D(3) in prostate cancer 17260971_Glycosylation is not essential for the chaperone action of clusterin 17260971_deglycosylation of clusterin had little effect on secondary structure content but produced a small increase in solvent-exposed hydrophobicity and enhanced aggregation. These changes were associated with increased binding to a variety of ligands 17322305_Clusterin expression is associated with neuroendocrine differentiation in normal epithelia and that the Clusterin observed in neoplastic cells is de novo synthesized. 17407782_The chaperone action of clusterin targets prefibrillar species to inhibit amyloid formation by lysozyme 17412999_Clusterin is an important element in the control of extracellular protein misfolding and in the formation of neurofibrillary tangles. 17412999_The chaperone action of clusterin can inhibit amyloid formation and toxicity 17420006_Our results indicate that epigenetic factors regulate the clusterin expression of RPE cells and thus might affect the pathogenesis of AMD via the inhibition of angiogenesis and inflammation. 17451556_The extracellular chaperone clusterin appears in the cytosol during endoplasmic stress 17451556_Under certain stress conditions, clusterin can evade the secretion pathway and reach the cytosol, and the retrotranslocation of clusterin is likely to occur through a mechanism similar to the ER-associated protein degradation pathway and involves Golgi. 17512083_Clusterin induces remarkable differentiation of the functional beta cells secreting insulin in response to glucose stimulation. 17534116_The expression and subcellular localization of Clusterin in the nucleus demonstrated the involvement of this protein in the photo-oxidative cell death pathway. 17535098_CLU was identified as a useful tumor marker for the diagnosis of pediatric large cell lymphoma. 17689225_Hyper-expressed form of clusterin localizes to mitochondria, inhibits cytochrome c release, and is inhibited by the proteasome. 17855704_may be a biomarker for longer survival in patients with surgically resected non-small cell lung cancer 17872975_Clusterin is constantly associated with altered elastic fibers in aged human skin. Clusterin exerted a chaperone-like activity and effectively inhibited UV-induced aggregation of elastin. 17974975_CLU gene expression might play a crucial role in prostate tumorigenesis by exerting differential biological effects on normal versus tumor cells through differential processing of CLU isoforms in the two cell systems. 18079682_Clusterin shows possible important functions in tumor suppression by the von Hippel-Lindau disease (VHL) gene product. May provide better understanding of retinal hemangioblastoma associated with VHL disease. 18082619_These findings suggest that CLU regulates TGF-beta signaling pathway by modulating the stability of Smad2/3 proteins. 18097679_clusterin is expressed in M cells and follicular dendritic cells at inductive sites of human mucosa-associated lymphoid tissue suggesting a role for this protein in innate immune responses 18239862_Clusterin expression is found nn pituitary adenoma and in non-neoplastic non-neoplastic adenohypophyses. 18378577_Clusterin is markedly elevated in Fuchs' endothelial dystrophy(FED)-affected tissue, pointing to a yet undiscovered form of dysregulation of endothelial cell function involved in FED pathogenesis. 18458059_The interplay between cancer cell-derived clusterin and IGF-1 may dictate the outcome of cell growth and dormancy during tumorigenic progression. 18514801_Data suggest that clusterin expression is related to endometrioid carcinoma of endometrium, in which estrogen is involved in the regulatory network of clusterin. 18542050_TRPM2-mediated Ca2+influx induces chemokine production in monocytes that aggravates inflammatory neutrophil infiltration. 18612545_In atherosclerotic lesions, ApoJ may have protective functions through its capacity to inactivate C5b-9 complement complexes and by reducing the cytotoxic effects of modified LDL on cells that gain contact with the lipoprotein. 18649357_Together with known function of clusterin, the data suggest an epigenetic component in the regulation of clusterin in prostate cancer 18709641_the 4 biomarkers CLU, ITGB3, PRAME and CAPG may be used as prognostic factors for patients with stage III serous ovarian adenocarcinomas. 18712185_ApoJ and leptin may have roles in coronary heart disease 18714397_The chaperone clusterin can protect ovarian cancer cells by sequestering the chemotherapeutic drug paclitaxel 18714397_clusterin is a potential therapeutic target for enhancing chemoresponsiveness in patients with a high-level clusterin expression 18786636_Results identified an interaction between prion protein and clusterin, a chaperone glycoprotein. 18806885_Clusterin is present in ocular anterior segment tissues involved in pseudoexfoliation syndrome. 18806885_Observational study of gene-disease association. (HuGE Navigator) 18813793_basal clusterin levels are higher in antiestrogen resistant cells; it is up-regulated following antiestrogen treatment; and down-regulation of cytoplasmic clusterin restores sensitivity to toremifene in the antiestrogen-resistant cell line 18842294_Observational study of gene-disease association. (HuGE Navigator) 19118032_We suggest that elevated sCLU levels may enhance tumorigenesis by interfering with Bax proapoptotic activities and contribute to one of the major characteristics of cancer cells, that is, resistance to apoptosis. 19137541_Results suggest that proteasome inhibition may induce prostate cancer cell death through accumulation of n-clusterin, a potential tumor suppressor factor. 19165232_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19166932_Clusterin-rich cells displayed a spindle-shape morphology while those with low clusterin levels were cuboidal in shape prompted us to investigate if clusterin modulates epithelial-to-mesenchymal transitions. 19177010_Data suggest that Ku70-Bax-clusterin interactions in human colon cancer are modulated by IL-6 that influences Bax-dependent cell death increasing Ku70 binding to Bax and influencing sCLU production that promotes tumor cell survival. 19182256_Observational study of gene-disease association. (HuGE Navigator) 19220628_Clusterin expression in cutaneous CD30-positive lymphoproliferative disorders and their histologic simulants. 19264665_These data indicated that hepatitis delta virus-induced clusterin protein increases cell survival potential. 19289586_Clusterin expression was not specific for Mycosis fungoides 19344414_Observational study of gene-disease association. (HuGE Navigator) 19353783_upregulated after proteasome inhibition due to transactivation by heat-shock factor-1 and due to accumulatioin as a result of reduced proteasomal protein degradation 19357365_Clusterin facilitates exchange of glycosyl phosphatidylinositol-linked SPAM1 between reproductive luminal fluids and mouse and human sperm membranes. 19391138_High clusterin gene expression is associated with disease progression, chemoresistance and metastasis of ovarian cancer. 19413638_Clusterin may protect bile duct epithelium against offensive biliary components or inhibit precipitation of biliary proteins 19446882_A novel heterozygous mutation in the clusterin gene, nucleotide position A1298C (glutamine>proline Q433P), was detected in exon 7 of a child with recurrent hemolytic uremic syndrome. The same mutation was found in the child's two siblings and mother. 19535339_Clusterin forms very large, soluble chaperone-client complexes 19535339_analysis of clusterin-chaperone client protein complexes 19542874_Clusterin is a highly sensitive marker for tenosynovial giant cell tumors 19651157_Reviews the literature linking clusterin to Alzheimer disease (AD). Clusterin can be viewed as a multipotent guardian of brain but is unable to prevent the progressive neuropathology in chronic AD. 19664600_clusterin strongly interacts with wild-type transthyretin (TTR) and TTR variants V30M and L55P under acidic conditions, and blocks the amyloid fibril formation of TTR variants. 19734902_Having removed Single Nucleotide Polymorphism within the APOE, CLU and PICALM loci from the analysis, focused on those that showed the most evidence for association with Alzheimer's disease 19734902_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19734903_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19734903_Two loci gave replicated evidence of association with Alzheimer's disease: one within CLU on chromosome 8 and the other within CR1 on chromosome 1. 19757199_Clusterin may be considered as a potential diagnostic and prognostic biomarker for bladder cancer 19793084_Clusterin as a predictor for chemoradiotherapy sensitivity and patient survival in esophageal squamous cell carcinoma. 19814590_The secretion of the potential zonal marker clusterin by zonal articular chondrocytes in osteoarthritic and healthy articular cartilage, was characterized. 19878569_Observational study of gene-disease association. (HuGE Navigator) 19878774_The chaperone action of clusterin is important in quality control of extracellular protein folding 19879420_This data suggest the importance of epigenetic events in the regulation of CLU in prostate cancer, supporting the idea that prostate cell transformation at early stages requires CLU silencing through chromatin remodeling.[Review] 19879421_CLU expression has contradictory functions, including cell survival, tumor progression, and apoptosis; these ambiguous functions are due to 2 different but related CLU protein forms, a glycosylated and a nonglycosylated form [Review] 19879422_reports the dynamic interaction of the different forms of CLU with their partners DNA-repair protein Ku70 and proapoptotic factor Bax during colon cancer progression, which seems to be a crucial point for the neoplastic cell fate.[Review] 19879423_In early-stage lung cancers CLU represents a positive biomarker correlating with better overall survival.[Review] 19903339_microarray study in sinonasal adenocarcinoma we identified proteins, CLU that is significantly differentially expressed in tumors compared to normal tissue 19903745_CLU acts as a tumor suppressor in the early stages of carcinogenesis, but CLU may also represent a pro-survival stimulus confering increased resistance to anti-cancer drugs or enhancing tumour cell survival in specific niches. Review. 19935703_a role for secreted clusterin as an important extracellular promoter of epithelial-to-mesenchymal transition 19940549_Data show that clusterin expression is under epigenetic control via methylation of its promoter. 19996109_Data identified ceruloplasmin, fibrinogen, and albumin as major clients for clusterin. 19996109_The chaperone action of clusterin protects major human plasma proteins from aggregation and precipitation 20007348_Reduction of inducible expression of clusterin results in an increase in tubular cell apoptosis and renders mice prone to ischemia reperfusion injury, implying a protective role of clusterin in kidney injury. 20009887_Overexpression of clusterin is associated with ovarian cancer. 20019877_Clusterin may protect human corneal endothelial cells from oxidative injury-mediated cell death via inhibition of reactive oxygen species production. 20028970_Data demonstrate that it is the presecretory form of CLU that has regulatory activity on NF-kappaB. Results identified the site of psCLU that interacts with IkappaB-alpha, and also showed that this region bears the regulatory activity of psCLU on NF-kappaB. 20057494_Clu showed genome wide significant association with Alzheimer disease. 20058210_clusterin expression could be a new molecular marker to predict response to platinum-based chemotherapy and survival of patients with cervical cancer treated with neoadjuvant chemotherapy and radical hysterectomy. 20068069_Elevated levels of sCLU promote prostate cancer cell survival by facilitating degradation of COMMD1 and I-kappaB, thereby activating the canonical NF-kappaB pathway. 20096688_Chibby and clusterin were co-immunoprecipitated with NBPF1. 20209083_CLU genetic variants may have a role in Alzheimer's disease 20209083_Observational study of gene-disease association. (HuGE Navigator) 20353268_Overexpression of cytoplasmic clusterin seems to be related with patient's shorter survival in late stage gastric carcinoma. 20410100_Observational study of gene-disease association. (HuGE Navigator) 20410100_SNPs in APOJ show strong statistical evidence of a functional effect on plasma fatty acid distribution. 20460622_Observational study, meta-analysis, and genome-wide association study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20497247_Clusterin expression may have a role in colonic carcinogenesis 20534741_Data confirmed genetic associations for Alzheimer's disease with APOE, CLU, PICALM and CR1 SNPs. 20534741_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20554627_Observational study of gene-disease association. (HuGE Navigator) 20554627_These results show near-perfect replication and provide the first additional evidence that CLU is associated with the risk of late onset alzheimer disease. 20570404_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20570404_While no Alzheimer disease association is observed with single nucleotide polymorphisms, a trend of association is seen with Picalm and clusterin single nucleotide polymorphisms. 20599866_Observational study of gene-disease association. (HuGE Navigator) 20599866_The minor allele (G) of the rs9331888 polymorphism within CLU was significantly associated with an increased risk of sporadic late-onset Alzheimer's disease in Chinese 20603455_results demonstrate an important role of clusterin in the pathogenesis of Alzheimer disease (AD) and suggest that alterations in amyloid chaperone proteins may be a biologically relevant peripheral signature of AD 20614220_both cytoplasmic and nuclear immunostaining patterns of clusterin were detected in the tumors of patients with non-small cell lung cancer. 20674675_In this review, clusterin is a major Alzheimer's disease susceptibility gene implicated indirectly in the life cycle of the herpes simplex virus. 20697030_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20697030_This study confirmed in a completely independent data set that CR1, CLU, and PICALM are AD susceptibility loci in European ancestry populations. 20738160_Observational study of gene-disease association. (HuGE Navigator) 20738160_Polymorphism of the clusterin gene may confer symptomatic specificity in schizophrenia, whereas polymorphism of the clathrin assembly lymphoid myeloid gene does not affect susceptibility to schizophrenia. 20739100_Observational study of gene-disease association. (HuGE Navigator) 20842452_The interaction between GOLPH2 and secretory CLU was confirmed intracellularly and extracellularly. 20847305_Clusterin levels in HDL are lower in men with reduced insulin sensitivity, higher body mass index, and an unfavorable lipid profile. Clusterin depletion contributes to the loss of HDL's cardioprotective properties. 20850846_Observational study of gene-disease association. (HuGE Navigator) 20850846_The CLU gene was associated with diabetes, probably through an increase in insulin resistance primarily and through an impairment of insulin secretion secondarily 20855565_Observational study of gene-disease association. (HuGE Navigator) 20873220_Analysis of clusterin gene (CLU/APOJ) polymorphism in Alzheimer's disease patients and in normal cohorts from Russian populations 20873220_Observational study of gene-disease association. (HuGE Navigator) 20930273_This study demonistrated that late onset alzheimer's disease linke to 8p21.1 region Including the CLU gene. 21042904_In neuroblastoma, clusterin exerts its anti-apoptotic effects downstream of Ku70 acetylation, likely by directly blocking Bax activation. 21043527_Serum ApoJ levels, determined by a commercial ELISA, were significantly lower in AMI-patients immediately after the event than in controls. In 60% of patients, the lowest ApoJ level was detected within 6 h after the onset of AMI. 21059989_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21059989_clu polymorphisms were associated with late-onset Alzheimer disease in an independent cohort from the National Institute on Aging Late-Onset Alzheimer's Disease Family Study 21135756_Results do not support a role for plasma clusterin as an important leptin-binding protein or modulator of leptin action. 21224044_these results suggest that CLU may play a pathogenetic role in transthyretin and Ig light chain amyloidoses and amyloidotic cardiomyopathy. 21240462_Increase in clusterin forms part of the stress response in Hodgkin's lymphoma. 21242307_Clusterin (apolipoprotein J), a molecular chaperone that facilitates degradation of the copper-ATPases ATP7A and ATP7B. 21280673_Results suggest that serum clusterin is not an early, presymptomatic biomarker for Alzheimer's disease. 21300948_This study provides compelling independent evidence that genetic variants in CLU, CR1, and PICALM are genetically associated with risk for AD. 21347408_variants in BIN1, CLU, CR1 or PICALM are associated with changes in the CSF levels of biomarkers 21379329_There was evidence of association for recently-reported late-onset Alzheimer's disease risk loci, including BIN1 and CLU and CUGBP2 with APOE. 21397462_Participants with the clusterin risk genotype have higher brain activity than participants with the protective allele in frontal and posterior cingulate cortex and the hippocampus, particularly during high memory demand. 21422520_The results of this study concluded that the rs11136000 AD-risk variant is associated with low clusterin plasma levels in Alzheimer's disease. 21447104_No association found between CLU gene and Alzheimer's disease in a Japanese population 21460853_these results integrate DNA damage resulting from genetic instability, IR, or chemotherapeutic agents, to ATM activation and abrogation of p53/NF-YA-mediated IGF-1 transcriptional repression, that induces IGF-1-sCLU expression. 21467232_Glycosylated serum Apo-J is associated with hepatocellular carcinoma. 21467285_data from the general population show that increased plasma clusterin levels are associated with prevalent alzheimer's disease (AD) and are higher in more severe cases of AD 21505792_The chaperone clusterin mediates systemic clearance of misfolded extracellular proteins 21508640_The APOJ single nucleotide polymorphism (1598delT) is associated with risk factors for coronary artery disease in a Chinese population. 21525168_The proteomes of the type-1 diabetic, type-2 diabetic, and non-diabetic obese patients presented elevated amounts of the same set of one molecular form of semenogelin-1, one form of clusterin, and two forms of lactotransferrin. 21527247_these results suggest a molecular basis for the pro-apoptotic function of nCLU by elucidating the residue specific interactions of the BH3 motif in nCLU with anti-apoptotic Bcl-2 family proteins. 21543606_Young healthy carriers of the CLU gene risk variant showed a distinct profile of lower white matter integrity that may increase vulnerability to developing Alzheimer disease later in life. 21567405_Nuclear CLU sequesters Bcl-XL via a putative motif in the C-terminal coiled coil (CC2) domain, releasing Bax, and promoting apoptosis. 21573492_nCLU also exerts a strong anti-migratory/anti-invasive activity in prostate cancer cells, by interfering with the cytoskeletal components and by decreasing MMP-2 activity. 21630085_results indicated lower metastatic potential for lung cancer cells with high CLU level 21633299_CLU could be a new molecular marker to predict overall survival of patients with advanced-stage cervical cancer treated with curative intended radiotherapy. 21732348_Results identify clusterin (CLU) as a MDA-7/IL-24 interacting protein in DU-145 cells and investigate the role of MDA-7/IL-24 in regulating CLU expression and mediating the antitumor properties of mda-7/IL-24 in prostate cancer. 21761117_controversial data on CLU function in chemo-response/resistance may be explained by a shift in the pattern of CLU expression and intracellular localization as well when tumor acquires chemoresistance 21824521_Our findings suggest that clusterin is associated with rate of brain atrophy in mild cognitive impairment 21892414_Results suggest a biological mechanism for the genetic association of CLU with Alzheimer's Disease risk and indicate that rs9331888 is one of the functional DNA variants underlying this association. 21899841_Proteomic analysis of intra-arterial thrombus secretions reveals a negative association of clusterin and thrombospondin-1 with abdominal aortic aneurysm. 21900379_We report for the first time that circulating clusterin does not have a day/night variation pattern in healthy young individuals. Clusterin levels are associated with total and low-density lipoprotein cholesterol cross-sectionally. 21912625_relationship of Parkinson's disease with SNPs of CLU, CR1 and PICALM 21953030_report abundance levels of CLI in serum samples from patients with advanced breast cancer, colorectal cancer (CRC) and lung cancer compared to healthy controls (age and gender matched) using commercially available enzyme-linked immunosorbent assay kits 21953454_CRM1 protein-mediated regulation of nuclear clusterin (nCLU), an ionizing radiation-stimulated, Bax-dependent pro-death factor 21980627_The overexpression of CLU in various stages of cervical lesions may serve as a potential marker to distinguish cervical neoplasia with borderline morphology features. 21987172_these data indicate that YB-1 transactivation of clusterin in response to stress is a critical mediator of paclitaxel resistance in prostate cancer. 21998749_sCLU at the mature RBCs is an active component being functionally implicated in the signalling mechanisms of cellular senescence and oxidative stress-responses 22012253_Localisation is key for CLU physiology, explaining the wide range of effects in cell survival and transformation. 22013110_Semen clusterin bears a set of complex glycans with high affinity for dendritic cell-specific ICAM-3-grabbing nonintegrin (DC-SIGN); these glycans mediate the effective binding of semen clusterin to DC-SIGN and interfere with HIV-1 binding to DC-SIGN. 22015308_findings showed evidence of CR1, CLU, and PICALM and late-onset Alzheimer's disease (LOAD) susceptibility in an independent southern Chinese population 22016805_secretory Apolipoprotein J/Clus represents a pro-survival factor acting for the postponement of the untimely clearance of RBCs 22068036_determined that the tumor suppressors CLU, NGFR, and RUNX3 were also directly repressed by EZH2 like CASZ1 in NB cells 22082661_We observed increased clusterin expression in human familial amyloid polyneuropathy nerves. 22122982_The rs11136000 and rs9331888 polymorphisms of the CLU gene are associated with late-onset Alzheimer's disease 22130675_Clusterin and COMMD1 independently regulate degradation of the mammalian copper ATPases ATP7A and ATP7B. 22159129_Healthy carriers of the CLU variant exhibit altered coupling between hippocampus and prefrontal cortex during memory processing. 22166956_CLU downregulation in fibroblast-like synoviocytes alters their aggressiveness in rheumatoid arthritis synovitis. 22179788_Data show that clusterin is able to influence both the aggregation and disaggregation of amyloid-beta(1-40) peptide by sequestration of the amyloid beta oligomers. 22232000_This study showed that overall levels of clusterinmRNA and protein areunchanged in Alzheimer's disease. 22234156_variation in CLU, genes are associated with Fuchs' endothelial dystrophy in Caucasian Australian cases 22236192_study found a 1.9-fold increase in the average levels of serum CLU in colorectal cancer patients compared to healthy donors 22248099_The clusterin gene CLU is a genetic risk association of AD with rare coding CLU variations. 22258514_This study indicates that the rs9331888 AD-risk variant is associated with low blood clusterin levels. 22266332_Data suggest that the effect of sex steroid deprivation appeared specific to clusterin associated with high density lipoproteins (HDL). 22274961_This review discusses the genetic variation and role of the clusterin gene in Alzheimer's disease pathogenesis. 22296908_Clusterin (rs11136000) was associated with Alzheimer's disease in Chinese Han population. 22391565_Secretory CLU expression was stimulated by insulin-like growth factor-1, but suppressed by p53. 22402018_In conclusion, we confirmed association of CLU, CR1, and PICALM genes with the disease | ENSMUSG00000022037 | Clu | 83.18985 | 0.8158048 | -0.2937040508 | 0.35019515 | 6.918853e-01 | 4.055238e-01 | 7.637945e-01 | No | Yes | 82.289891 | 14.574029 | 96.663004 | 16.632495 | |
| ENSG00000120899 | 2185 | PTK2B | protein_coding | Q14289 | FUNCTION: Non-receptor protein-tyrosine kinase that regulates reorganization of the actin cytoskeleton, cell polarization, cell migration, adhesion, spreading and bone remodeling. Plays a role in the regulation of the humoral immune response, and is required for normal levels of marginal B-cells in the spleen and normal migration of splenic B-cells. Required for normal macrophage polarization and migration towards sites of inflammation. Regulates cytoskeleton rearrangement and cell spreading in T-cells, and contributes to the regulation of T-cell responses. Promotes osteoclastic bone resorption; this requires both PTK2B/PYK2 and SRC. May inhibit differentiation and activity of osteoprogenitor cells. Functions in signaling downstream of integrin and collagen receptors, immune receptors, G-protein coupled receptors (GPCR), cytokine, chemokine and growth factor receptors, and mediates responses to cellular stress. Forms multisubunit signaling complexes with SRC and SRC family members upon activation; this leads to the phosphorylation of additional tyrosine residues, creating binding sites for scaffold proteins, effectors and substrates. Regulates numerous signaling pathways. Promotes activation of phosphatidylinositol 3-kinase and of the AKT1 signaling cascade. Promotes activation of NOS3. Regulates production of the cellular messenger cGMP. Promotes activation of the MAP kinase signaling cascade, including activation of MAPK1/ERK2, MAPK3/ERK1 and MAPK8/JNK1. Promotes activation of Rho family GTPases, such as RHOA and RAC1. Recruits the ubiquitin ligase MDM2 to P53/TP53 in the nucleus, and thereby regulates P53/TP53 activity, P53/TP53 ubiquitination and proteasomal degradation. Acts as a scaffold, binding to both PDPK1 and SRC, thereby allowing SRC to phosphorylate PDPK1 at 'Tyr-9, 'Tyr-373', and 'Tyr-376'. Promotes phosphorylation of NMDA receptors by SRC family members, and thereby contributes to the regulation of NMDA receptor ion channel activity and intracellular Ca(2+) levels. May also regulate potassium ion transport by phosphorylation of potassium channel subunits. Phosphorylates SRC; this increases SRC kinase activity. Phosphorylates ASAP1, NPHP1, KCNA2 and SHC1. Promotes phosphorylation of ASAP2, RHOU and PXN; this requires both SRC and PTK2/PYK2. {ECO:0000269|PubMed:10022920, ECO:0000269|PubMed:12771146, ECO:0000269|PubMed:12893833, ECO:0000269|PubMed:14585963, ECO:0000269|PubMed:15050747, ECO:0000269|PubMed:15166227, ECO:0000269|PubMed:17634955, ECO:0000269|PubMed:18086875, ECO:0000269|PubMed:18339875, ECO:0000269|PubMed:18587400, ECO:0000269|PubMed:18765415, ECO:0000269|PubMed:19086031, ECO:0000269|PubMed:19207108, ECO:0000269|PubMed:19244237, ECO:0000269|PubMed:19428251, ECO:0000269|PubMed:19648005, ECO:0000269|PubMed:19880522, ECO:0000269|PubMed:20001213, ECO:0000269|PubMed:20381867, ECO:0000269|PubMed:20521079, ECO:0000269|PubMed:21357692, ECO:0000269|PubMed:21533080, ECO:0000269|PubMed:7544443, ECO:0000269|PubMed:8670418, ECO:0000269|PubMed:8849729}. | 3D-structure;ATP-binding;Adaptive immunity;Alternative splicing;Angiogenesis;Cell junction;Cell membrane;Cell projection;Cytoplasm;Immunity;Kinase;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Tyrosine-protein kinase | This gene encodes a cytoplasmic protein tyrosine kinase which is involved in calcium-induced regulation of ion channels and activation of the map kinase signaling pathway. The encoded protein may represent an important signaling intermediate between neuropeptide-activated receptors or neurotransmitters that increase calcium flux and the downstream signals that regulate neuronal activity. The encoded protein undergoes rapid tyrosine phosphorylation and activation in response to increases in the intracellular calcium concentration, nicotinic acetylcholine receptor activation, membrane depolarization, or protein kinase C activation. This protein has been shown to bind CRK-associated substrate, nephrocystin, GTPase regulator associated with FAK, and the SH2 domain of GRB2. The encoded protein is a member of the FAK subfamily of protein tyrosine kinases but lacks significant sequence similarity to kinases from other subfamilies. Four transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:2185; | apical dendrite [GO:0097440]; cell body [GO:0044297]; cell cortex [GO:0005938]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; dendrite [GO:0030425]; dendritic spine [GO:0043197]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; focal adhesion [GO:0005925]; glutamatergic synapse [GO:0098978]; growth cone [GO:0030426]; lamellipodium [GO:0030027]; membrane raft [GO:0045121]; neuronal cell body [GO:0043025]; NMDA selective glutamate receptor complex [GO:0017146]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; postsynaptic density [GO:0014069]; 3-phosphoinositide-dependent protein kinase binding [GO:0043423]; ATP binding [GO:0005524]; calmodulin-dependent protein kinase activity [GO:0004683]; NMDA glutamate receptor activity [GO:0004972]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein C-terminus binding [GO:0008022]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; protein-containing complex binding [GO:0044877]; signaling receptor binding [GO:0005102]; ubiquitin protein ligase binding [GO:0031625]; activation of GTPase activity [GO:0090630]; activation of Janus kinase activity [GO:0042976]; adaptive immune response [GO:0002250]; apoptotic process [GO:0006915]; blood vessel endothelial cell migration [GO:0043534]; bone resorption [GO:0045453]; cell differentiation [GO:0030154]; cell surface receptor signaling pathway [GO:0007166]; cellular defense response [GO:0006968]; cellular response to fluid shear stress [GO:0071498]; cellular response to retinoic acid [GO:0071300]; chemokine-mediated signaling pathway [GO:0070098]; endothelin receptor signaling pathway [GO:0086100]; epidermal growth factor receptor signaling pathway [GO:0007173]; focal adhesion assembly [GO:0048041]; glial cell proliferation [GO:0014009]; innate immune response [GO:0045087]; integrin-mediated signaling pathway [GO:0007229]; ionotropic glutamate receptor signaling pathway [GO:0035235]; long-term synaptic depression [GO:0060292]; long-term synaptic potentiation [GO:0060291]; MAPK cascade [GO:0000165]; marginal zone B cell differentiation [GO:0002315]; negative regulation of apoptotic process [GO:0043066]; negative regulation of bone mineralization [GO:0030502]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of muscle cell apoptotic process [GO:0010656]; negative regulation of myeloid cell differentiation [GO:0045638]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of potassium ion transport [GO:0043267]; neuron projection development [GO:0031175]; oocyte maturation [GO:0001556]; peptidyl-tyrosine autophosphorylation [GO:0038083]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of actin filament polymerization [GO:0030838]; positive regulation of angiogenesis [GO:0045766]; positive regulation of B cell chemotaxis [GO:2000538]; positive regulation of cell growth [GO:0030307]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cell-matrix adhesion [GO:0001954]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of DNA biosynthetic process [GO:2000573]; positive regulation of endothelial cell migration [GO:0010595]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of excitatory postsynaptic potential [GO:2000463]; positive regulation of JNK cascade [GO:0046330]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of neuron projection development [GO:0010976]; positive regulation of nitric oxide biosynthetic process [GO:0045429]; positive regulation of nitric-oxide synthase activity [GO:0051000]; positive regulation of peptidyl-tyrosine phosphorylation [GO:0050731]; positive regulation of phosphatidylinositol 3-kinase activity [GO:0043552]; positive regulation of protein kinase activity [GO:0045860]; positive regulation of reactive oxygen species metabolic process [GO:2000379]; positive regulation of synaptic transmission, glutamatergic [GO:0051968]; positive regulation of translation [GO:0045727]; positive regulation of ubiquitin-dependent protein catabolic process [GO:2000060]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; protein-containing complex assembly [GO:0065003]; regulation of actin cytoskeleton reorganization [GO:2000249]; regulation of calcium-mediated signaling [GO:0050848]; regulation of cell adhesion [GO:0030155]; regulation of cell shape [GO:0008360]; regulation of cGMP-mediated signaling [GO:0010752]; regulation of establishment of cell polarity [GO:2000114]; regulation of inositol trisphosphate biosynthetic process [GO:0032960]; regulation of macrophage chemotaxis [GO:0010758]; regulation of NMDA receptor activity [GO:2000310]; regulation of release of sequestered calcium ion into cytosol [GO:0051279]; regulation of synaptic plasticity [GO:0048167]; regulation of ubiquitin-dependent protein catabolic process [GO:2000058]; response to calcium ion [GO:0051592]; response to cAMP [GO:0051591]; response to cocaine [GO:0042220]; response to ethanol [GO:0045471]; response to glucose [GO:0009749]; response to hormone [GO:0009725]; response to hydrogen peroxide [GO:0042542]; response to hypoxia [GO:0001666]; response to immobilization stress [GO:0035902]; response to lithium ion [GO:0010226]; response to mechanical stimulus [GO:0009612]; response to osmotic stress [GO:0006970]; response to xenobiotic stimulus [GO:0009410]; signal complex assembly [GO:0007172]; signal transduction [GO:0007165]; sprouting angiogenesis [GO:0002040]; stress fiber assembly [GO:0043149]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; tumor necrosis factor-mediated signaling pathway [GO:0033209]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11774117_Glomerular crescents strongly expressed cell adhesion kinase beta. 11820787_Human umbilical vein endothelial cells express mRNA transcripts for both the full length isoform Pyk2 and the truncated isoform Pyk2-H containing the C-terminal deletion. 11856738_Suppressees androgen receptor transactivation vir interaction and phosphorylation of ARA44 coregulator 11916084_Pyk2 role in platelet activation by vWF in the early signal transduction events activated by ligand binding to glycoprotein Ib-IX-V. 12011061_Transforming growth factor-beta 2 is a transcriptional target via forkhead transcription factor 12077257_By rapidly translocating to the vicinity of the immune synapse after T cell receptor stimulation, Pyk2 plays an essential role in T cell activation and polarized secretion of cytokines. 12244133_This protein binds to periplakin. 12458207_role in negative regulation of mixed lineage kinase 3 results in cell survival 12515814_Critical role of the carboxyl terminus of proline-rich tyrosine kinase (Pyk2) in the activation of human neutrophils by tumor necrosis factor: separation of signals for the respiratory burst and degranulation. 12606503_results indicate that signaling via protein kinase B to forkhead transcription factor FKHR can account for the effect of insulin to regulate peroxisome proliferator-activated receptor-gamma coactivator-1 promoter activity via the insulin response sequence 12626562_PYK2 regulates transendothelial migration of cultured NK cells in response to chemokines by controlling the activation of the small GTP binding protein Rac, and thus acts as an integration point between integrin and chemokine receptor stimulation. 12794117_Augmented activation of Pyk2 is an immediate signaling event required for the trans-regulation of integrin alpha L beta 2 by alpha 4 beta 1 in Jurkat T cells. 12844492_RAFTK is activated by VEGF in human brain microvascular endothelial cells 12933673_PKB is an essential component of the FSH-mediated granulosa cell differentiation and that both PKB and G(s)alpha signaling pathways are required. 12943720_results indicate that activation of PKC is responsible for GnRH-induced phosphorylation of both ERK1/2 and Pyk2, and that Pyk2 activation does not contribute to GnRH signaling 14676843_A novel regulatory network RAFTK/Pyk2, Src and p38 appears to be critical for VEGF-induced endothelial cell migration. 14729602_Calcium signaling in ovarian surface epithelial cells not only induces telomerase activity via JNK but also activates Pyk2. 14961028_identified a BCR/ABL-dependent increase in expression of multiple genes involved in pre-mRNA splicing; alternative splicing of gene PYK2 resulted in increased expression of full-length Pyk2 in BCR/ABL-containing cells 14963038_Pyk2 facilitates EGFR- and c-Src-mediated Stat3 activation and has a role in triggering Stat3-induced oncogenesis 14969582_In P-selectin-activated PMNs, PYK-2 (proline-rich tyrosine kinase-2) undergoes tyrosine phosphorylation in a b2-integrin- and SRC-dependent manner, suggesting that this focal adhesion kinase may be a downstream mediator of the effect of SRC kinases. 15050747_PYK2 gene products mediate integrin-induced signals that regulate myelopoiesis. 15070849_Pyk2 signaling differentially regulates cell migration and proliferation pathways. 15105428_Pyk2 is a novel effector of fibroblast growth factor receptor 3 activation 15128501_modulation and phosphorylation of BK(Ca) channels by Pyk2 and a Src-family kinase may reflect a general cellular mechanism by which G protein-coupled receptor and/or integrin activation leads to the regulation of membrane ion channels 15128873_Recruitment of PYK2 to lipid rafts mediates signals important for actin reorganization in growing neurites. 15213840_EGFR, PYK2, Yes, and SHP-2 are involved in transduction of the TF/FVIIa signal possibly via transactivation of the EGF receptor. 15499613_Tyrosine phosphorylation of PYK2 mediates heregulin-induced glioma invasion 15539082_Activation of PI3K following beta1-integrin engagement on human CD34+ cells results in subsequent phosphorylation of PYK2, and is required for the recruitment of the PI3K/PYK2 complex to beta1-integrins at the cell surface. 15585656_proline-rich tyrosine kinase 2 (PYK2) is phosphorylated on tyrosine residues 402 and 580 induced by ephrin-A1 15695828_Pyk2 is a phosphorylated beta(3) binding partner, providing a potential structural and signaling platform to achieve alpha(V)beta(3) -mediated remodeling of the actin cytoskele 15778498_Pyk2 has a role in the reduced cell-cell adhesion induced by the Rac-mediated production of ROS through the tyrosine phosphorylation of beta-catenin 15835820_bFGF significantly stimulated PKB activity in CNE- I nasopharyngeal carcinoma cell line. 15944312_Pyk2 is a critical regulatory component and a molecular switch to overcome the suppression of neutrophil oxidant generation by cell adhesion. 15967096_Pyk2 plays a central role in the migratory behavior of glioblastomas 16055703_Semaphorin 4D/plexin-B1 induces endothelial cell migration through the activation of PYK2, Src, and the phosphatidylinositol 3-kinase-Akt pathway 16433632_Pyk2 may mediate a signal necessary for beta2 integrin function in PMN tethered by E-selectin. 16514607_The tyrosine kinase Pyk2 was highly phosphorylated upon induction of cell polarity but not during cell spreading 16760434_Vascular endothelial growth factor (VEGF) elicits the activation of the VEGFR2-ROCK pathway, leading to phosphorylation of Ser732 within FAK which changes the conformation of FAK, making it accessible to Pyk2. 16774943_PYK2 mediates anti-apoptotic AKT signaling in response to benzo[a]pyrene diol epoxide in mammary epithelial cells 16783820_Our data indicate that Pyk2 plays a central role in the mechanism that regulate cell-cell and cell-substrate interaction and lack of its kinase activity induces prostate cells to acquire a malignant, migrating phenotype. 16840719_regulation of virus-induced Akt activation by the integrin-linked kinase in Hela cells 16945503_Our data demonstrate that while FAK mediated cell adhesion, RAFTK was localized at the cytoplasm where it mediated inside-out signaling through intracellular Ca(2+), thus leading to cell spreading and movement upon EGF stimulation. 16949788_Focal adhesion kinase is involved type III group B streptococcal invasion of human brain microvascular endothelial cells. 16998626_FRNK plays an important role in cell adhesion during the very early stages of cell culture. 17205062_These results provide insights into Pyk2 signaling in epidermis and reveal a novel role for Pyk2 in regulation of keratinocyte differentiation. 17329398_These data demonstrate that LPXN forms a signaling complex with Pyk2, c-Src, and PTP-PEST to regulate migration of prostate cancer cells. 17537919_PYK2 regulates osteoprogenitor cells and bone formation, and offes an anablic treatment for osteoporosis. 17551499_overexpression of Pyk2 and FAK was found in nearly 60% of HCC patients and was significantly correlated with poor prognosis 17563746_ErbB-2, via PYK2-MAP kinase, upregulates the adhesive ability of androgen receptor human prostate cancer cells. 17581868_REVIEW: role of PYK2 in bone resorption and possible target to prevent osteoporosis 17716864_Protein kinase B mediates its anti-apoptotic activities in cells through mitochondrial dynamics and their role during apoptosis. 17785844_Endothelial Pyk2 activity is required for efficient neutrophil migration and to facilitate junctional disruption during transcellular or paracellular passage of the leukocyte across the vascular endothelial monolayer. 17906699_Proline-rich tyrosine kinase Pyk2 is highly expressed in small cell lung cancer cells and provides a functional link between neuropeptide-induced increases in intracellular calcium ions [Ca2+]i, and tumor cell proliferation. 17950644_Observational study of gene-disease association. (HuGE Navigator) 18075463_Missense mutation in PTK2B gene is associated with hypertension 18075463_Observational study of gene-disease association. (HuGE Navigator) 18198130_Pyk2 is induced and involved in monocyte differentiation and C/EBPbeta is a critical regulator of the Pyk2 expression. 18365874_Pyk2/ERK 1/2 mediate Sp1- and c-Myc-dependent induction of telomerase activity by epidermal growth factor 18367725_Pyk2 is activated by beta(2)-integrin adhesion and is a required signal for eosinophil spreading and subsequent chemotactic migration 18390748_these results demonstrate a novel role for Pyk2 in LPS-induced IL-8 production in endothelial cells 18437915_FAK is a critical decision maker in extracellular matrix/strain-enhanced osteogenic differentiation. 18571765_Up-regulation of proline-rich tyrosine kinase 2 is associated with non-small cell lung cancer. 18667434_In vascular smooth muscle cells shedding of membrane-bound epidermal growth factor (EGF) receptor ligands and activation of the nonreceptor tyrosine kinases Pyk2 and Src contributed to the thrombin-induced ERK1/2 phosphorylation. 18765415_Immunofluorescence staining showed that the focal adhesion localization of Pyk2 is a major determinant for c-Src and ERK/MAPK activation. Pyk2 promoted cell proliferation and invasiveness by upregulation of the c-Src and ERK/MAPK-signaling pathways 18794150_Results indicate that RhoC promotes tumor metastasis in prostate cancer by activation of Pyk2. 18832579_These data support the cooperative function of Pyk2 and FAK in breast cancer progression and suggest that dual inhibition of FAK and Pyk2 is an efficient therapeutic approach for targeting invasive breast cancer. 18954908_a critical role for the Pyk2 mediated pathway involving p38 MAP kinase and NF-kappaB in LPS-induced MCP-1 production in human microvascular endothelial cells. 19047047_PYK2 activity is abnormally up-regulated in the Pcdh-gamma-deficient neurons 19086031_Data suggest that endothelin-1 stimulates the GTPase Rap1 by a mechanism involving Pyk2 activation and recruitment of the p130Cas/BCAR3 complex in human glomerular mesangial cells. 19106639_Pyk2 and cAMP interact in regulating prostate cell functions and in 'keeping' prostate identity. 19207108_Pyk2 is a critical signalling molecule downstream of beta3 integrin tyrosine phosphorylation and mediates Vav1 recruitment to accomplish actin reorganization necessary for adhesion 19358827_These results demonstrate that Pyk2 is constitutively associated with paxillin, that neither Tyr-881 nor kinase activity is necessary for the Pyk2/paxillin association, and that paxillin is a substrate of Pyk2 kinase. 19415692_PRL up-regulated CHIT-1 expression via PTK, PI3-K, MAPK, and signaling transduction components. 19458494_Studies indicated that the roles of FAK in tumorigenesis are different among the tumor species. 19509258_Data substantiate a central role for the FERM domain in regulation of Pyk2 activity and identify the F3 module as a novel target to inhibit Pyk2 activity and inhibit glioma progression. 19561089_Pyk2 and FAK are downstream targets of the Rap GTPases that play a key role in regulating B cell morphology. 19706888_Induction of PTK2B antigen is associated with relapsed chronic myeloid leukemia. 19718025_Observational study of gene-disease association. (HuGE Navigator) 19880522_Pyk2 functions to limit p53 levels, thus facilitating cell growth and survival in a kinase-independent manner. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19930834_Upregulation of serum proline-rich tyrosine kinase 2 is associated with systemic lupus erythematosus. 20028775_TCR has a distinct pathway for the activation of Pyk2 compared with other receptor systems. 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20074345_The data demonstrated that Pyk2 and PECAM-1 were critical mediators of both anchorage-independent growth and anoikis resistance in tumor cells. 20180987_Data reveal that Pyk2 couples the activation mGluRs to the ERK1/2 mitogen-activated protein kinase pathway even though it attenuates mGluR1-dependent G protein signaling. 20215112_activation of Pyk2 is an early signal that promotes wound healing by stimulating the SFK/EGFR signaling pathway. 20308428_The p110alpha selectively mediates activation of the Tyr kinase Pyk2 to regulate barrier function. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20381867_Data show that Pyk2 had a second burst of phosphorylation 30-60 min after TCR stimulation. 20543098_Data show that knockdown of SOCS2 resulted in the accumulation of p-Pyk2(Tyr402) and blocked NK cell effector functions. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20688918_Pyk2 acts as an early sensor of numerous extracellular signals that trigger a Ca(2+) flux and/or reactive oxygen species to amplify tyrosine phosphorylation signaling events. 20724588_Phosphorylation of focal adhesion kinase at Tyr397 is associated with gastric carcinomas. 20800603_Observational study of gene-disease association. (HuGE Navigator) 20849950_The Pyk2 FERM domain is involved in the regulation of Pyk2 activity by acting to regulate the formation of Pyk2 oligomers that are critical for Pyk2 activity. 20881009_TNFRSF19 overexpression in glioma cells activates Rac1 signaling in a Pyk2-dependent manner to drive glioma cell invasion and migration 20966971_Studies suggest that FAK FERM domains might mediate information transfer between the cell cortex and nucleus. 21114537_Both wild-type and internal tandem duplication (ITD)-FLT3 proteins co-immunoprecipitated with beta1 integrin and Pyk2 indicating the signal crosstalk between FLT3, beta1 integrin and Pyk2. 21357692_Nephrocystin-4 regulates Pyk2-induced tyrosine phosphorylation of nephrocystin-1 to control targeting to monocilia 21426305_although staurosporine-mediated caspase activation and FAK cleavage was delayed in the mtp53-R 175H HOSCC cell line, caspase-8 activation and FAK dephosphorylation were induced by increasing the apoptotic stimulus. 21533080_data provided new evidence of the underlying mechanism of Pyk2 in controlling cell motility of HCC cells through regulation of genes associated with EMT 21602932_downregulation of endogenous FIP200 protein in glioblastoma tumor cells, astrocytes, and brain microvessel endothelial cells promotes apoptosis, most likely due to the removal of a direct interaction of FIP200 with Pyk2 that inhibits Pyk2 activation 21640103_The results suggest a surprising finding that limited cell adhesion can enhance endothelial responsiveness to VEGF and demonstrate a novel role for Pyk2 in the adhesive regulation of angiogenesis. 21666110_Activation of Pyk2 in the heart during heart failure may contribute to protection against arrhythmia. 21840393_ATP stimulates epithelial cell motility through Pyk2-mediated activation of the EGF receptor 22023045_This is a review of recent developments in Protein kinase B/AKT and focal adhesion kinase signaling in cancer with the particular emphasis on the novel signaling pathways in which FAK is downstream of AKT. [review] 22096562_Results may suggest a new evidence of Pyk2 on promoting cisplatin resistance of HCC cells through preventing cell apoptosis, activation of AKT pathway and upregulation of drug resistant genes. 22188814_Data show that c-Met induces FAK and Pyk2 phosphorylation in medulloblastoma cells. 22454420_Report safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinases, in advanced solid tumors. 22581436_PAC1 regulates PYK-2 tyrosine phosphorylation in a calcium-dependent manner in lung cancer cell lines. 22618716_High expression of proline-rich tyrosine kinase 2 is associated with hepatocellular carcinoma via regulating phosphatidylinositol 3-kinase/AKT pathway. 22745829_duced expression of miR-23b enhances glioma cell migration in vitro and invasion ex vivo via modulation of Pyk2 protein expression. 22842493_Pyk2 as a critical regulator of endothelial cell inflammation by virtue of engaging IKK to promote the release and the transcriptional capacity of RelA/p65. 22922962_Pyk2 plays a crucial role in G protein-coupled receptor agonist thrombin-induced human aortic smooth muscle cell growth and migration, as well as balloon injury-induced neointima formation. 22923218_findings showed that Pyk2 is overexpressed in squamous cell carcinoma of the head and neck; data suggest that CCR7 via Pyk2 and cofilin regulates the chemotaxis and migration ability of metastatic squamous cell carcinoma of the head and neck cells 23142219_Down-regulation of both miR-517a and miR-517c contribute to hepatocellular carcinoma cells development through regulating Pyk2. 23216754_These results identify a novel pathway of integrin alphaIIbbeta3 outside-in signaling and recognize the tyrosine kinase Pyk2 as a major regulator of platelet adhesion and spreading on fibrinogen. 23220172_We conclude that the effects of selenoprotein H on mitochondrial biogenesis and mitochondrial function are probably mediated through protein kinase A-CREB-PGC-1alpha and Akt/protein kinase B-CREB-PGC-1alpha pathways. 23292187_Data indicate protein secretion pathways activated by monosodium urate (MSU) in macrophages, and reveal a novel role for cathepsin B and Src, Pyk2, PI3 kinases in the activation of unconventional protein secretion. 23300847_the non-catalytic functions of the kinases Fyn and Pyk2 were required for late stage human T cell adhesion. 23302305_SOCS3 definitely plays roles in regulating Pyk2 signaling and cell motility in A549 cells. 23587524_Results indicate that in vitro Pyk2 might function to regulate cell adhesion and motility following all-trans-retinoic acid (ATRA). 23618355_Our results confirm that RTKs are frequently altered in chordomas. 23922106_Both Pyk2 and phosphorylated Pyk2[pY881] are potential prognostic factors and therapeutic targets for non-small-cell lung cancer. 24072693_Determine a crucial role of LRP1-mediated Pyk2 phosphorylation on hypoxia-induced MMP-9 activation and hVSMC migration and therefore in hypoxia-induced vascular remodeling. 24097630_Development of a coordinated allo T cell and auto B cell response against autosomal PTK2B after allogeneic hematopoietic stem cell transplantation. 24108181_We hypothesize a potential direct or indirect role for SRC, RAF1, PTK2B genes in neurotransmission and in central nervous system signaling processes. 24142406_Pyk2 and Src are important in CCL18-induced breast cancer metastasis. 24176282_Pyk2 is a shared key mediator of targeted-therapy induced adhesion and migration; Targeting Pyk2 may serve as an effective therapeutic strategy to reduce extramedullar relpase in acute promyelocytic leukemia and chronic myeloid leukemia. 24523919_inhibition of FAK, PYK2 and BCL-XL synergistically enhances apoptosis in ovarian clear cell carcinoma cell lines 24598361_Pyk2 is essential for skin wound reepithelialization in vivo and in vitro and that it regulates epidermal keratinocyte migration via a pathway that requires PKCdelta and MMP functions. 25174335_Data indicate that chicken paxillin leucine-aspartate LD2 and LD4 motifs preferentially dock at the helix 2/3 binding site of human Proline-rich tyrosine kinase 2 (Pyk2)-focal adhesion kinase (FAK). 25180269_PYK2 is an intermediary component of Ca(2+) signaling between PKA-mediated and Tyr phosphorylations that is required for achieving functional human sperm capacitation. 25217697_Data indicate the tumor-promoting role of proline-rich tyrosine kinase 2 (Pyk2) in multiple myeloma (MM), suggesting tyrosine kinase inhibitor as a therapeutic option in MM. 25219547_Pyk2-NDRG1 axis is possibly involved in conveying the anti-proliferative effect of beta-ionone in prostate cancer cells. 25387834_these data demonstrate that Pyk2 is a critical regulator of PI3K function downstream of the TCR. 25415317_data suggest a novel role for FAK in GPVI-dependent ROS formation and platelet activation and elucidate a proximal signaling role for FAK within the GPVI pathway. 25433371_Orai1/Pyk2 pathway is essential for glioma migration and invasion. 25778396_Pyk2 has a role in integrin regulation of size and dynamics of signaling microclusters 25889845_These results suggest that melatonin exerts anti-migratory and anti-invasive effects on glioma cells in response to hypoxia via reactive oxygen species-alphavbeta3 integrin-FAK/Pyk2 signaling pathways. 25967238_Fyn, but not Lyn, was required for complete Pyk2 phosphorylation by thrombin. 26084289_Studies suggest that PYK2 is a common downstream effector of ErbB and IL8 receptors, and that PYK2 integrates their signaling pathways through a positive feedback loop to potentiate breast cancer invasion. 26109718_This study identified Pyk2 as a cellular component required for the intracellular trafficking of HPV16 during infection. 26202465_Data strongly suggest that chemokine-stimulated associations between Vav1, SLP-76, and ADAP facilitate Rac1 activation and alpha4beta1-mediated adhesion, whereas Pyk2 opposes this adhesion by limiting Rac1 activation. 26274564_FAK and PYK2 functioned redundantly to promote the Wnt/beta-catenin pathway by phosphorylating GSK3beta(Y216) to reinforce pathway output-beta-catenin accumulation and intestinal tumorigenesis. 26330541_Phosphoproteomic analysis identifies FAK2 as a potential therapeutic target for tamoxifen resistance in breast cancer. 26352169_Pyk2 is a key downstream signaling molecules of CCR7 in SCCHN, which promotes SCCHN tumorigenesis and progression. 26866573_Pyk2-focal adhesion targeting domain interacts with and binds to leupaxin. 26866924_Src has a role in priming Pyk2 (but not FAK) phosphorylation and subsequent activation downstream of integrins 27080426_PTK2B polymorphism (rs28834970) could modify the risk of late-onset Alzheimer's disease (LOAD), and PTK2B polymorphism (rs28834970) and APOE may interact to increase LOAD risk in a Han Chinese population. 27181591_thrombin binding to PAR-1 receptor activated Gi-protein/c-Src/Pyk2/EGFR/PI3K/Akt/p42/p44 MAPK cascade, which in turn elicited AP-1 activation and ultimately evoked MMP-9 expression and cell migration in SK-N-SH cells. 27210483_Results show that VEGFA induces Pyk2 activation in mediating human retinal microvascular endothelial cell migration, sprouting and tube formation, and that Pyk2-mediated STAT3 activation is required for hypoxia-induced retinal neovascularization. 27492635_In summary, our data suggested that PYK2 via S6K1 activation modulated AR function and growth properties in prostate cancer cells. Thus, PYK2 and S6K1 may potentially serve as therapeutic targets for PCa treatment. 27602957_Multiple myeloma that is driven by deregulated iron homeostasis and/or Pyk2/beta-cateninn signaling is susceptible to deferasirox-induced apoptosis. 27613122_Ascites and CCL18 stimulate the phosphorylation and expression of Pyk2, which positively regulates ascites-induced ovarian cancer cell migration. 28122716_We demonstrated trophoblast cytoprotection by intervention with supraphysiological concentrations of relaxin, a process in part mediated through the PI3-kinase-Akt/PKB cell survival pathway. These results provide further rationale for clinical investigation of relaxin as a potential therapeutic in preeclampsia. 28218251_Results provide evidence that Pyk2 phosphorylates STIM1 at its Y361 residue, activating thereby store-operated Ca(2+) entry. 28385807_STIM1-induced Ca(2+) signaling activates Pyk2 to inhibit the interaction of VE-PTP and VE-cadherin and hence increase endothelial permeability. 28555636_Pyk2 has a role in spine structure and synaptic function; its deficit contributes to Huntington's disease cognitive impairments 28694190_Our findings suggest that Pyk2 plays an important role in the coordination of stabilization of beta-catenin in the crosstalk between Wnt/beta-catenin and Wnt/Ca(2+) signaling pathways upon Wnt3a stimulation in differentiating hNPCs. 28699640_LFA-1 cross-linking recruits and activates FAK1 and PYK2 to phosphorylate LAT selectively on a single Y-171 site that binds to the GRB2-SKAP1 complex and limits dwell times of T-cells with dendritic cells 28973302_Interestingly, rs2279590 locus has a widespread enhancer effect on two nearby genes, protein tyrosine kinase 2 beta (PTK2B) and epoxide hydrolase-2 (EPHX2); both of which have been previously associated with AD as risk factors. 29133485_The authors' findings identify Pyk2 as a unique mediator of invadopodium formation and function and also provide a novel insight into the mechanisms by which Pyk2 mediates tumor cell invasion. 30020827_Wild-type TRPM2 but not Ca(2+)-impermeable mutant E960D reconstituted phosphorylation and expression of Pyk2 and CREB in TRPM2-depleted cells exposed to doxorubicin. Results demonstrate that TRPM2 expression protects the viability of neuroblastoma through Src, Pyk2, CREB, and MCU activation, which play key roles in maintaining mitochondrial function and cellular bioenergetics 30250159_PYK2 is an important regulator of the Hippo pathway, and its tyrosine kinase activity has a striking effect on TAZ stabilization and activation in triple-negative breast cancer. 30952431_These data suggests a FAK/PYK2 regulated pro-survival role of ERK5 in response to cell adhesion in MDA-MB 231 breast cancer cells. 31110200_FAK and Pyk2 activity promote TNF-alpha and IL-1beta-mediated pro-inflammatory gene expression and vascular inflammation. 31118051_We provide evidence of a PYK2-driven pro-invasive potential of metformin in pure HER2 cancer therapy and propose that metformin-based therapy should consider the molecular heterogeneity of breast cancer to prevent complications associated with cancer chemoresistance, invasion and recurrence in treated patients. 32510326_CD56 regulates human NK cell cytotoxicity through Pyk2. 32693110_The proline-rich tyrosine kinase Pyk2 modulates integrin-mediated neutrophil adhesion and reactive oxygen species generation. 32727877_Pyk2 Regulates Human Papillomavirus Replication by Tyrosine Phosphorylation of the E2 Protein. 32956670_Inhibition of Pyk2 and Src activity improves Cx43 gap junction intercellular communication. 33082554_Colon and liver tissue damage detection using methylated SESN3 and PTK2B genes in circulating cell-free DNA in patients with acute graft-versus-host disease. 33568623_Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. 33785524_The AXL-PYK2-PKCalpha axis as a nexus of stemness circuits in TNBC. 34313839_Pyk2 level is a novel prognostic marker for patients with esophageal squamous cell carcinoma after radical surgery. 34943950_Pyk2 Stabilizes Striatal Medium Spiny Neuron Structure and Striatal-Dependent Action. | ENSMUSG00000059456 | Ptk2b | 73.31309 | 0.4670524 | -1.0983437150 | 0.41123135 | 6.856769e+00 | 8.830607e-03 | No | Yes | 58.287559 | 14.622242 | 122.521221 | 29.800408 | ||
| ENSG00000121900 | 113452 | TMEM54 | protein_coding | Q969K7 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:113452; | integral component of membrane [GO:0016021] | ENSMUSG00000028786 | Tmem54 | 114.84308 | 0.9038608 | -0.1458274716 | 0.27080825 | 2.892045e-01 | 5.907307e-01 | 8.632229e-01 | No | Yes | 128.557020 | 20.947103 | 134.348313 | 21.233166 | ||||
| ENSG00000122299 | 29066 | ZC3H7A | protein_coding | Q8IWR0 | FUNCTION: May be a specific regulator of miRNA biogenesis. Binds to microRNAs MIR7-1, MIR16-2 and MIR29A hairpins recognizing the 3'-ATA(A/T)-5' motif in the apical loop. {ECO:0000269|PubMed:28431233}. | 3D-structure;Alternative splicing;Coiled coil;Metal-binding;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;TPR repeat;Zinc;Zinc-finger | hsa:29066; | nucleus [GO:0005634]; metal ion binding [GO:0046872]; miRNA binding [GO:0035198]; RNA binding [GO:0003723]; posttranscriptional regulation of gene expression [GO:0010608]; production of miRNAs involved in gene silencing by miRNA [GO:0035196] | ENSMUSG00000037965 | Zc3h7a | 1961.15616 | 1.3427457 | 0.4251861460 | 0.09939678 | 1.845815e+01 | 1.736762e-05 | 3.174750e-03 | No | Yes | 2628.053619 | 425.721745 | 1903.294103 | 301.616045 | |||
| ENSG00000122550 | 55975 | KLHL7 | protein_coding | Q8IXQ5 | FUNCTION: Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin ligase complex. The BCR(KLHL7) complex acts by mediating ubiquitination and subsequent degradation of substrate proteins. Probably mediates 'Lys-48'-linked ubiquitination. {ECO:0000269|PubMed:21828050}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Kelch repeat;Nucleus;Reference proteome;Repeat;Retinitis pigmentosa;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. | This gene encodes a BTB-Kelch-related protein. The encoded protein may be involved in protein degradation. Mutations in this gene have been associated with retinitis pigmentosa 42. [provided by RefSeq, Feb 2010]. | hsa:55975; | Cul3-RING ubiquitin ligase complex [GO:0031463]; cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; protein ubiquitination [GO:0016567] | 16918702_The present results indicate that KLHL7 antibodies are associated with various cancers, and in some patients also with neurological disease. Whether KLHL7 antibodies can be used as paraneoplastic markers for PNS remains to be determined. 19520207_Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. 20547956_Observed in 2 Scandinavian families to date, KLHL7 mutation has recently been associated with autosomal dominant retinitis pigmentosa. 21828050_KLHL7 forms a dimer, assembles with Cul3 through its BTB and BACK domains, and exerts E3 activity. 22084217_The phenotypes are similar among patients with 3 types of KLHL7 mutations (c.458C>T, c.449G>A, and c.457G>A). 27392078_data further support the pathogenic role of KLHL7 mutations in a CS/CISS1-like phenotype--but they do not explain all their clinical manifestations and highlight the high phenotypic heterogeneity associated with mutations in KLHL7 29032201_KLHL7 is a novel regulator of the nucleolus associated with TUT1 ubiquitination, and pathogenic KLHL7 mutants may provide valuable information to elucidate a mechanism of retinitis pigmentosa etiology. 29074562_We have expanded the clinical spectrum of KLHL7 autosomal recessive variants by describing a syndrome with features overlapping CS/CISS1 and BOS. 29633055_KLHL7 appears to play an important role in BC progression. High KLHL7 protein expression identified a subgroup of BC with aggressive behaviour and provided independent prognostic information. 30300710_A novel nonsense mutation in KLHL7 was identified in two siblings with multiple dysmorphic features and developmental delay. 31856884_Study found novel mutations in the 3-box motif of the BACK domain of KLHL7 associated with nonsyndromic autosomal dominant retinitis pigmentosa 31953236_A novel PTC mutation in the BTB domain of KLHL7 gene in two patients with Bohring-Opitz syndrome-like features. | ENSMUSG00000028986 | Klhl7 | 1133.23100 | 0.8598085 | -0.2179126526 | 0.10755041 | 4.081085e+00 | 4.336580e-02 | 3.176567e-01 | No | Yes | 1177.962771 | 188.798184 | 1301.287112 | 204.017440 |
| ENSG00000123552 | 85015 | USP45 | protein_coding | Q70EL2 | FUNCTION: Catalyzes the deubiquitination of SPDL1 (PubMed:30258100). Plays a role in the repair of UV-induced DNA damage via deubiquitination of ERCC1, promoting its recruitment to DNA damage sites (PubMed:25538220). May be involved in the maintenance of photoreceptor function (PubMed:30573563). May play a role in normal retinal development (By similarity). Plays a role in cell migration (PubMed:30258100). {ECO:0000250|UniProtKB:E9QG68, ECO:0000269|PubMed:25538220, ECO:0000269|PubMed:30258100, ECO:0000269|PubMed:30573563}. | Alternative splicing;Cytoplasm;Disease variant;Hydrolase;Leber congenital amaurosis;Metal-binding;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation pathway;Zinc;Zinc-finger | The protein encoded by this gene is a deubiquitylase that binds ERCC1, the catalytic subunit of the XPF-ERCC1 DNA repair endonuclease. This endonuclease is a critical regulator of DNA repair processes, and the deubiquitylase activity of the encoded protein is important for maintaining the DNA repair ability of XPF-ERCC1. [provided by RefSeq, Sep 2016]. | hsa:85015; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; photoreceptor inner segment [GO:0001917]; thiol-dependent deubiquitinase [GO:0004843]; zinc ion binding [GO:0008270]; cell migration [GO:0016477]; DNA repair [GO:0006281]; global genome nucleotide-excision repair [GO:0070911]; neural retina development [GO:0003407]; photoreceptor cell maintenance [GO:0045494]; protein deubiquitination [GO:0016579]; ubiquitin-dependent protein catabolic process [GO:0006511] | 25538220_these results establish USP45 as a new regulator of XPF-ERCC1 crucial for efficient DNA repair 30258100_Using mass spectrometry this study identified CCDC99 as a new target of USP45. The data showed that CCDC99 and USP45 are part of the same complex and that their interaction specifically depends on the catalytic activity of USP45. 30573563_Our study implicates that biallelic mutations in USP45 are associated with the occurrence of LCA. Moreover, our results indicate that USP45 is indispensable to the maintenance of photoreceptor function. | ENSMUSG00000040455 | Usp45 | 594.64651 | 1.0040445 | 0.0058232712 | 0.15354251 | 1.436991e-03 | 9.697613e-01 | 9.919849e-01 | No | Yes | 748.787793 | 163.391138 | 718.632135 | 153.576992 | |
| ENSG00000124104 | 90203 | SNX21 | protein_coding | Q969T3 | FUNCTION: Binds to membranes enriched in phosphatidylinositol 3-phosphate (PtdIns(P3)) and phosphatidylinositol 4,5-bisphosphate. May be involved in several stages of intracellular trafficking. {ECO:0000250|UniProtKB:Q3UR97}. | Alternative splicing;Cytoplasmic vesicle;Endosome;Lipid-binding;Membrane;Protein transport;Reference proteome;Transport | This gene encodes a member of the sorting nexin family. Members of this family contain a phox (PX) domain, which is a phosphoinositide binding domain, and are involved in intracellular trafficking. This protein does not contain a coiled coil region, like some family members. The specific function of this protein has not been determined. Multiple transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:90203; | early endosome membrane [GO:0031901]; phosphatidylinositol phosphate binding [GO:1901981]; phosphatidylinositol-3-phosphate binding [GO:0032266]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; protein transport [GO:0015031] | 12459172_Data suggest that human sorting nexin-L (SNX-L) may be a regulatory gene involved in receptor protein degradation during embryonic liver development. 30072438_The N-terminal extension of SNX21 interacts with huntingtin and recruit Htt to an endosomal population. | ENSMUSG00000050373 | Snx21 | 172.40632 | 0.8818395 | -0.1814119871 | 0.22351524 | 6.555924e-01 | 4.181203e-01 | 7.724774e-01 | No | Yes | 199.257207 | 31.504926 | 217.479810 | 33.256033 | |
| ENSG00000124574 | 89845 | ABCC10 | protein_coding | Q5T3U5 | FUNCTION: ATP-dependent transporter of the ATP-binding cassette (ABC) family that actively extrudes physiological compounds, and xenobiotics from cells. Lipophilic anion transporter that mediates ATP-dependent transport of glucuronide conjugates such as estradiol-17-beta-o-glucuronide and GSH conjugates such as leukotriene C4 (LTC4) (PubMed:12527806, PubMed:15256465). Mediates multidrug resistance (MDR) in cancer cells by preventing the intracellular accumulation of certain antitumor drugs, such as, docetaxel and paclitaxel (PubMed:15256465, PubMed:23087055). Does not transport glycocholic acid, taurocholic acid, MTX, folic acid, cAMP, or cGMP (PubMed:12527806). {ECO:0000269|PubMed:12527806, ECO:0000269|PubMed:15256465, ECO:0000269|PubMed:23087055}. | ATP-binding;Alternative splicing;Cell membrane;Lipid transport;Membrane;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;Translocase;Transmembrane;Transmembrane helix;Transport | The protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. ABC proteins transport various molecules across extra- and intra-cellular membranes. ABC genes are divided into seven distinct subfamilies (ABC1, MDR/TAP, MRP, ALD, OABP, GCN20, and White). This ABC full-transporter is a member of the MRP subfamily which is involved in multi-drug resistance. Multiple transcript variants encoding different isoforms have been found for this gene.[provided by RefSeq, Nov 2010]. | hsa:89845; | basolateral plasma membrane [GO:0016323]; integral component of membrane [GO:0016021]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; plasma membrane [GO:0005886]; ABC-type glutathione S-conjugate transporter activity [GO:0015431]; ABC-type transporter activity [GO:0140359]; ABC-type xenobiotic transporter activity [GO:0008559]; ATP binding [GO:0005524]; ATPase-coupled inorganic anion transmembrane transporter activity [GO:0043225]; ATPase-coupled transmembrane transporter activity [GO:0042626]; leukotriene metabolic process [GO:0006691]; leukotriene transport [GO:0071716]; lipid transport [GO:0006869]; transmembrane transport [GO:0055085] | 11146224_First study to describe structure and expression pattern of this transporter. 12527806_First study to describe ABCC10's ability to transport physiological substrates, confirming that this protein, with low identity to ABCC1, was indeed a efflux pump. 12527806_MRP7 has the facility for mediating the transport of conjugates such as E(2)17betaG indicates that it is a lipophilic anion transporter involved in phase III (cellular extrusion) of detoxification. 12566991_The genomic organization and gene expression regulation of human multidrug resistance-associated protein 7 and a splicing variant MRP7A were identified 15256465_This study shows that ABCC10 confers resistance to the anticancer agents taxanes and vinca alkaloids in vitro. 16034073_A non-major histocompatibility complex MRP7 leader peptide ALALVRMLI binds to HLA-E and strongly inhibits natural killer (NK) cell-mediated lysis 17203221_MDR1 expression, MRP1 expression, and MRP7 expression are refractory factors in head and neck cancer chemotherapy; induction of MRP7 expression is involved in drug resistance to natural products, especially to docetaxel in salivary gland adenocarcinoma 19082471_ABCC10/MRP7 may confer vinorelbine resistance in non-small cell lung cancer 19118001_Study establishes that ABCC10 confers resistance to several classes of agents including nucleoside analogs (cytabarine, dideoxycytidine) and epothilone B. 19343046_Observational study of gene-disease association. (HuGE Navigator) 19841739_Imatinib and nilotinib reverse multidrug resistance in cancer cells by inhibiting the efflux activity of the MRP7 (ABCC10) 21576088_Study establishes that ABCC10 is able to modulate taxane resistance in vivo using a mouse model. 21628669_Tenofovir is a substrate for ABCC10, and genetic variability within the ABCC10 gene may influence tenofovir renal tubular transport and contribute to the development of kidney tubular dysfunction. 22082652_Multivariate regression analysis identified carriage of a composite genotype of ABCC10 rs2125739 to be associated with nevirapine plasma concentrations 22739155_Tetrandrine and 5-bromotetrandrine can both reverse multidrug resistance in K562 cells by reducing expression of MRP7. 23087055_This report describes the ATPase activity of ABCC10 and further revealed ABCC10 localizes basolaterally using an in vitro system. The described work also identifies the tyrosine kinase sorafenib as an inhibitor of ABCC10. 24431074_the combination of paclitaxel and masitinib could serve as a novel and useful therapeutic strategy to reverse paclitaxel resistance mediated by ABCC10 24937672_ABCC10 expression is correlated with human breast cancer subtype using breast tissue microarrays. 26655271_BAG3-mediated miRNA let-7g and let-7i inhibit proliferation and enhance apoptosis of human esophageal carcinoma cells by targeting the drug transporter ABCC10 and modulates cisplatin resistance. 27468921_The present study shows that the protein expression of ABCC10 significantly associates with overall survival and the expression of ABCC11 with disease-free interval of colorectal cancer patients 28051999_FOXM1 promotes 5-FU resistance by up-regulating ABCC10 expression in colorectal tumor cells. 28612064_It is thus inferred, that ABCC10 expression in CWR22Rv1 cells is not S phase-specific but is primarily associated with cell proliferation. 30890141_In the cancer cell lines, only rs2125739 of ABCC10 gene was significantly associated with docetaxel cytotoxicity, and this was confirmed in the genome-edited cell line. In the non-small cell lung cancer (NSCLC) patients, there were no significant differences related to rs2125739 genotype in terms of response rate, progression free and overall survival. However, this SNP was associated with grade 3/4 neutropenia. 33563765_Exosomal Delivery of FTO Confers Gefitinib Resistance to Recipient Cells through ABCC10 Regulation in an m6A-dependent Manner. | ENSMUSG00000032842 | Abcc10 | 818.80281 | 0.8274526 | -0.2732514380 | 0.12280460 | 4.919332e+00 | 2.655779e-02 | 2.618410e-01 | No | Yes | 754.802665 | 91.588096 | 911.190668 | 107.725476 | |
| ENSG00000125170 | 55715 | DOK4 | protein_coding | Q8TEW6 | FUNCTION: DOK proteins are enzymatically inert adaptor or scaffolding proteins. They provide a docking platform for the assembly of multimolecular signaling complexes. DOK4 functions in RET-mediated neurite outgrowth and plays a positive role in activation of the MAP kinase pathway (By similarity). Putative link with downstream effectors of RET in neuronal differentiation. May be involved in the regulation of the immune response induced by T-cells. {ECO:0000250}. | Phosphoprotein;Reference proteome | hsa:55715; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nervous system development [GO:0007399]; positive regulation of MAPK cascade [GO:0043410]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 12595900_DOK4 expression in human T cells has been demonstrated, and its genomic structure has been predicted. 12730241_IRS5/DOK4 and IRS6/DOK5 represent two new signaling proteins with potential roles in insulin and IGF-1 action 16820412_Results suggest that Dok-4, through activation of the Rap1-ERK1/2 pathway, regulates GDNF-mediated neurite outgrowth during neuronal development. 17443497_IRS-5 (DOK4) is significantly upregulated in 90% of examined clear cell RCCs. Studies on this gene has shown that it is regulated through chromatin remodeling in kidney cells 19073520_Epigenetic regulation of DOK4 expression is associated with non-small-cell lung cancer. 19494292_Dok-4 represents a novel negative regulator of T cells 22982678_the current study have explored in more detail the structure and function of the Dok-4 PTB domain. 32386480_Study of DOK4 gene expression and promoter methylation in sporadic breast cancer. | ENSMUSG00000040631 | Dok4 | 148.71474 | 1.1629992 | 0.2178500566 | 0.27094060 | 6.461037e-01 | 4.215092e-01 | 7.746304e-01 | No | Yes | 206.062320 | 28.408988 | 174.513592 | 23.469633 | ||
| ENSG00000125247 | 84899 | TMTC4 | protein_coding | Q5T4D3 | FUNCTION: Transfers mannosyl residues to the hydroxyl group of serine or threonine residues. The 4 members of the TMTC family are O-mannosyl-transferases dedicated primarily to the cadherin superfamily, each member seems to have a distinct role in decorating the cadherin domains with O-linked mannose glycans at specific regions. Also acts as O-mannosyl-transferase on other proteins such as PDIA3. {ECO:0000269|PubMed:28973932}. | Alternative splicing;Endoplasmic reticulum;Membrane;Reference proteome;Repeat;TPR repeat;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:28973932}. | This gene encodes a transmembrane protein that belongs to family of proteins containing an N-terminal transmembrane domain and a C-terminal tetratricopeptide repeat (TPR) domain. TPR domains mediate protein-protein interactions in various cellular processes, such as synaptic vesicle fusion, protein folding, and protein translocation. A pseudogene of this gene has been defined on chromosome 5. [provided by RefSeq, Apr 2017]. | hsa:84899; | endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; ATPase binding [GO:0051117]; dolichyl-phosphate-mannose-protein mannosyltransferase activity [GO:0004169]; mannosyltransferase activity [GO:0000030]; endoplasmic reticulum unfolded protein response [GO:0030968]; outer hair cell apoptotic process [GO:1905584]; positive regulation of endoplasmic reticulum calcium ion concentration [GO:0032470]; protein O-linked mannosylation [GO:0035269]; sensory perception of sound [GO:0007605] | 33436046_Conserved sequence motifs in human TMTC1, TMTC2, TMTC3, and TMTC4, new O-mannosyltransferases from the GT-C/PMT clan, are rationalized as ligand binding sites. 33925440_Transmembrane and Tetratricopeptide Repeat Containing 4 Is a Novel Diagnostic Marker for Prostate Cancer with High Specificity and Sensitivity. | ENSMUSG00000041594 | Tmtc4 | 630.06179 | 0.9754525 | -0.0358565271 | 0.15943848 | 5.050620e-02 | 8.221848e-01 | 9.505378e-01 | No | Yes | 668.721959 | 62.804296 | 668.076868 | 61.124306 |
| ENSG00000125347 | 3659 | IRF1 | protein_coding | P10914 | FUNCTION: Transcriptional regulator which displays a remarkable functional diversity in the regulation of cellular responses (PubMed:15226432, PubMed:15509808, PubMed:17516545, PubMed:17942705, PubMed:18497060, PubMed:19404407, PubMed:19851330, PubMed:22367195, PubMed:32385160). Regulates transcription of IFN and IFN-inducible genes, host response to viral and bacterial infections, regulation of many genes expressed during hematopoiesis, inflammation, immune responses and cell proliferation and differentiation, regulation of the cell cycle and induction of growth arrest and programmed cell death following DNA damage (PubMed:15226432, PubMed:15509808, PubMed:17516545, PubMed:17942705, PubMed:18497060, PubMed:19404407, PubMed:19851330, PubMed:22367195). Stimulates both innate and acquired immune responses through the activation of specific target genes and can act as a transcriptional activator and repressor regulating target genes by binding to an interferon-stimulated response element (ISRE) in their promoters (PubMed:15226432, PubMed:15509808, PubMed:17516545, PubMed:17942705, PubMed:18497060, PubMed:19404407, PubMed:19851330, PubMed:21389130, PubMed:22367195). Competes with the transcriptional repressor ZBED2 for binding to a common consensus sequence in gene promoters (PubMed:32385160). Its target genes for transcriptional activation activity include: genes involved in anti-viral response, such as IFN-alpha/beta, DDX58/RIG-I, TNFSF10/TRAIL, ZBP1, OAS1/2, PIAS1/GBP, EIF2AK2/PKR and RSAD2/viperin; antibacterial response, such as NOS2/INOS; anti-proliferative response, such as p53/TP53, LOX and CDKN1A; apoptosis, such as BBC3/PUMA, CASP1, CASP7 and CASP8; immune response, such as IL7, IL12A/B and IL15, PTGS2/COX2 and CYBB; DNA damage responses and DNA repair, such as POLQ/POLH; MHC class I expression, such as TAP1, PSMB9/LMP2, PSME1/PA28A, PSME2/PA28B and B2M and MHC class II expression, such as CIITA; metabolic enzymes, such as ACOD1/IRG1 (PubMed:15226432, PubMed:15509808, PubMed:17516545, PubMed:17942705, PubMed:18497060, PubMed:19404407, PubMed:19851330, PubMed:22367195). Represses genes involved in anti-proliferative response, such as BIRC5/survivin, CCNB1, CCNE1, CDK1, CDK2 and CDK4 and in immune response, such as FOXP3, IL4, ANXA2 and TLR4 (PubMed:18641303, PubMed:22200613). Stimulates p53/TP53-dependent transcription through enhanced recruitment of EP300 leading to increased acetylation of p53/TP53 (PubMed:15509808, PubMed:18084608). Plays an important role in immune response directly affecting NK maturation and activity, macrophage production of IL12, Th1 development and maturation of CD8+ T-cells (PubMed:11244049, PubMed:11846971, PubMed:11846974, PubMed:16932750). Also implicated in the differentiation and maturation of dendritic cells and in the suppression of regulatory T (Treg) cells development (PubMed:11244049, PubMed:11846971, PubMed:11846974, PubMed:16932750). Acts as a tumor suppressor and plays a role not only in antagonism of tumor cell growth but also in stimulating an immune response against tumor cells (PubMed:20049431). {ECO:0000269|PubMed:15226432, ECO:0000269|PubMed:15509808, ECO:0000269|PubMed:17516545, ECO:0000269|PubMed:17942705, ECO:0000269|PubMed:18084608, ECO:0000269|PubMed:18497060, ECO:0000269|PubMed:18641303, ECO:0000269|PubMed:19404407, ECO:0000269|PubMed:19851330, ECO:0000269|PubMed:21389130, ECO:0000269|PubMed:22200613, ECO:0000269|PubMed:22367195, ECO:0000269|PubMed:32385160, ECO:0000303|PubMed:11244049, ECO:0000303|PubMed:11846971, ECO:0000303|PubMed:11846974, ECO:0000303|PubMed:16932750, ECO:0000303|PubMed:20049431}. | Acetylation;Activator;Antiviral defense;Cytoplasm;DNA-binding;Disease variant;Immunity;Innate immunity;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Tumor suppressor;Ubl conjugation | The protein encoded by this gene is a transcriptional regulator and tumor suppressor, serving as an activator of genes involved in both innate and acquired immune responses. The encoded protein activates the transcription of genes involved in the body's response to viruses and bacteria, playing a role in cell proliferation, apoptosis, the immune response, and DNA damage response. This protein represses the transcription of several other genes. As a tumor suppressor, it both suppresses tumor cell growth and stimulates an immune response against tumor cells. Defects in this gene have been associated with gastric cancer, myelogenous leukemia, and lung cancer. [provided by RefSeq, Aug 2017]. | hsa:3659; | chromatin [GO:0000785]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; transcription cis-regulatory region binding [GO:0000976]; apoptotic process [GO:0006915]; CD8-positive, alpha-beta T cell differentiation [GO:0043374]; cellular response to interferon-beta [GO:0035458]; cellular response to mechanical stimulus [GO:0071260]; defense response to virus [GO:0051607]; immune system process [GO:0002376]; interferon-gamma-mediated signaling pathway [GO:0060333]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of regulatory T cell differentiation [GO:0045590]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of interferon-beta production [GO:0032728]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of type I interferon production [GO:0032481]; regulation of adaptive immune response [GO:0002819]; regulation of CD8-positive, alpha-beta T cell proliferation [GO:2000564]; regulation of cell cycle [GO:0051726]; regulation of innate immune response [GO:0045088]; regulation of MyD88-dependent toll-like receptor signaling pathway [GO:0034124]; regulation of transcription by RNA polymerase II [GO:0006357] | 11069564_Observational study of gene-disease association. (HuGE Navigator) 11240951_Observational study of gene-disease association. (HuGE Navigator) 11315919_Observational study of gene-disease association. (HuGE Navigator) 11716756_IRF-1 exerts a pivotal role in granulocytic differentiation, and its induction by G-CSF represents a limiting step in the early events of myeloid cell differentiation. 11721695_Review: Stimulation of interferon regulatory factor-1 by prolactin 11804954_IFNtau effect on IRF-1 expression is primarily regulated by tyrosine-phosphorylated Stat1alpha or Stat1beta dimers 11846972_This review focuses on the induction and function of IRF-1 during M. tuberculosis infection 11846974_This review focuses on IRF-1 is a negative regulator of cell proliferation. 11909852_Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. 11948194_Host defense responses to infection by Mycobacterium tuberculosis. Induction of IRF-1 and a serine protease inhibitor 11970993_IFN regulatory factor-1 regulates IFN-gamma-dependent cathepsin S expression 12067985_IRF-1 associated with resistance to antiestrogens 12105194_Interferons inhibit tumor necrosis factor-alpha-mediated matrix metalloproteinase-9 activation via interferon regulatory factor-1 binding competition with NF-kappa B 12115600_VCAM-1 expression via effects on interferon regulatory factor-1 expression and activity 12162881_Promoter single nucleotide polymorphisms were detected and their possible contribution to T-lymphocyte helper 1 response in chronic hepatitis C was studied. Interferon-beta therapy increased the CD4(+) cell population. 12191570_STAT-1, IRF-1, and RAR-beta expression were enhanced by IFN-gamma and ATRA in combination, and to a greater degree in BALM-3 cells than in BALM-1 cells, suggesting that these IFN-gamma related genes were involved in the induction of apoptosis. 12420205_Observational study of gene-disease association. (HuGE Navigator) 12420214_IRF-1 is controlled by two distinct signalling pathways; a JAK/STAT-signalling pathway in viral infected cells and an ATM-signalling pathway in DNA damaged cells. 12433281_GRIM-19 protein inhibits the cell-transforming property of this protein via a physical interaction. 12479817_IRF-1 and IRF-2 induced by IFN-gamma bind to three distinct IL-4 promoter sites and function as transcriptional repressors 12482935_IRF-1-CEBPbeta complex activate the promoter of IL-18 binding protein. 12545159_IRF-1 is a potential mediator of IFN-gamma-induced attenuation of telomerase activity and hTERT expression 12677441_expression of IRF-1-dependent genes in neurons plays a role in ischemic neuronal death. 12711307_Identification of the domain of interferon regulatory factor-1 responsible for transactivation. 12732645_IRF1 represses CDK2 gene expression by interfering with SP1-dependent transcriptional activation. 12759449_IRF1 is one of the transcription factors responsible for the induced coexpression of NF-kappa B activator 1 (Act1) and CD40 in lung epithelial A549 cells by inflammatory cytokines. 12788988_These data indicate that interferon regulatory factor-1 is a hallmark of the gliadin-mediated inflammation in celiac disease. 14599866_Expression of IRF-1 is altered in human endometrioid adenocarcinoma compared with normal endometrium and postmenopausal endometrium. 14675396_Observational study of gene-disease association. (HuGE Navigator) 14675396_the IRF1 HinfI gene polymorphisms do not appear to be involved in susceptibility to celiac disease 14764039_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14993214_IRF1 has a role in up-regulation of human caspase-8 by interferon-gamma in breast tumor cells 15078941_Measles virus upregulates IRF-1 in a manner that is independent of IFN but dependent on the JAK/STAT pathway. This induction of IRF-1 appears to suppress cell growth, although the extent seems to vary among MeV strains. 15173018_IRF-1 mediates the proapoptotic but not cell cycle arrest effects of the steroidal antiestrogen ICI 182,780 15226432_Results suggest that the functional interplay between interferon regulatory factors 1 and 2 serves as an elaborate and cooperative mechanism for regulation of interleukin-7 production essential for local immune regulation within human intestinal mucosa. 15241475_a novel tumor necrosis factor (ligand) superfamily, member 10-mediated tumor suppressor activity of interferon regulatory factor 1 and suggest a mechanistic basis for the synergistic antitumor activities of certain retinoids and interferons 15265939_IFN regulatory factor 1 is involved in TNF-alpha-induced VCAM-1 expression and monocyte adhesion to TNF-alpha-activated endothelial cells. 15331704_IRF-1 is one of the key host factors that regulate intracellular HCV replication through modulation of interferon-stimulated-gene-mediated antiviral responses. 15489234_interleukin-12 p35 gene transcription is activated by interferon regulatory factor-1 and interferon consensus sequence-binding protein 15509808_IRF-1-p300 interface as an allosteric modifier of DNA-dependent acetylation of p53 at the p21 promoter 15511228_IFN-gamma enhances TRAIL-induced apoptosis through IRF-1 15548708_therapies designed to enhance IRF-1 expression within tumor cells may represent novel treatment strategies for breast cancer 15560761_IFN-gamma-IRF system is involved in BPAG1 gene regulation in type-1 helper T-cell inflammatory skin conditions, such as psoriasis vulgaris 15576464_the interferon signaling pathway and the human glucocorticoid receptor gene 1A promoter interact in T-lymphocytes 15710386_p27Kip1 inhibits hTERT mRNA expression and telomerase activity through post-transcriptional up-regulation by IFN-gamma/IRF-1 signaling 15778351_TPO and IFN-gamma activate the expression of TAP1 via a new mechanism that involves functional cooperation between STAT1 and IRF-2 on the TAP1 promoter 15863386_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15878912_Functional role for IRF-1 in the growth suppression of breast cancer cells and strongly implicate IRF-1 as a tumor suppressor gene in breast cancer that act to control apoptosis. 15907481_IRF-1 acts as a master regulator for the concerted expression of immunoproteasome components 16085646_all-trans-retinoic acid, an RARalpha ligand, regulates IFNgamma-induced IRF-1 by affecting multiple components of the IFNgamma signaling pathway, from the plasma membrane to the nuclear transcription factors 16195814_Individuals who were TT homozygous at IRF1 6477 T/G locus seemed to be attacked by Graves' disease much earlier than others. 16195814_Observational study of gene-disease association. (HuGE Navigator) 16223733_STAT1, IRF1, and NF-kappaB activation and early myocyte apoptosis play a mechanistic role in septic myocardial depression and sepsis-induced organ dysfunction 16483648_The expression of IRF-1 truncated mRNA and protein forms might be a critical event in the development of Myelodysplastic Syndrome. 16512786_Intracellular HIV-1 Tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1 16636311_Data suggest that the dual effect of retinoids in increasing interferon regulatory factor-1 mRNA and nuclear protein may be essential for its tumor suppressor activity and immunosurveillance functions in breast epithelial cells. 16679314_a novel repressor domain and differential gene regulation may contribute to IRF-1 tumor suppressor activity 16857162_alternative splicing in exons 7, 8, and 9 is an important mechanism for negatively regulating IRF-1 in cervical cancer. 16894313_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16894313_findings suggest the possibility that the -300AA IRF-1 genotype is associated with outcome in patients with hepatitis C virus genotype 3 infection; in HCV genotype-1-infected patients, this genotype appears associated with response to therapy 16914093_Increase in IRF-1 expression, induced by IFNgamma and arsenic, correlates with both higher anti-proliferative effect and increased apoptosis. 16944293_Observational study of gene-disease association. (HuGE Navigator) 16944293_no statistically significant association with susceptibility to persistent HBV infection was observed with the IFN-gamma, IFNGR-1 and 2, and IRF-1 gene polymorphisms under the codominant, dominant, and recessive models 16961714_Observational study of gene-disease association. (HuGE Navigator) 17016442_IRF-1 promotes rECM-mediated apoptosis and provide evidence that both rECM and rapid Tam signaling transcriptionally activate IRF-1 through recruitment of CBP to the IRF-1 GAS promoter complex. 17177148_Observational study of gene-disease association. (HuGE Navigator) 17213842_Observational study of gene-disease association. (HuGE Navigator) 17255955_KPNA2 is physically bound to IFN regulatory factor-1 (IRF-1), a transcription factor induced by IFN-gamma, induces nuclear translocation of IRF-1 in dermal fibroblasts 17328074_Observational study of gene-disease association. (HuGE Navigator) 17498560_Observational study of genotype prevalence. (HuGE Navigator) 17498560_Results show A4396G single nucleotide polymorphism in IRF1 gene is more frequently expressed in African American than in European ancestry subjects. 17498560_The A4396G single nucleotide polymorphism in IRF1 is more frequent in human breast cancer cell lines than in the general population and is more frequently expressed in African American than in Caucasian subjects. 17516545_These results confirm that by controlling RIG-I expression, IRF-1 plays an essential role in anti-viral immunity. 17617740_IFNalpha induces TRAIL expression via a STAT-1/IRF-1-dependent mechanism in human bladder cancer cells 17703412_Observational study of gene-disease association. (HuGE Navigator) 17852336_population-based data of IRF-1 sequence contains 35 nucleotide additions, 8 nucleotide removals and another 12 nucleotide replacements compared to GenBank sequence L05072; and a single nucleotide difference was observed in the IRF-1 promoter sequence 17869652_IRF-1 is a susceptibility gene for Behcet's disease (BD) in Korean BD patients; its polymorphisms are associated with thrombotic manifestations in BD patients. 17869652_Observational study of gene-disease association. (HuGE Navigator) 17918184_EGFR induces expression of IRF-1 via STAT1 and STAT3 activation leading to growth arrest of human cancer cells 17924060_These findings indicated that both the NF-kappaB and IRF-1 sites are required for the repression of promoter activity of IL-12 p40 by TGF-beta. 17947510_These findings demonstrate for the first time that IRF-1 is a novel alternative GRbeta-independent mechanism mediating steroid dysfunction induced by pro-asthmatic cytokines. 17970693_The described IRF-1 promoter single nucleotide polymorphisms do not play a role in the pathogenesis of psoriasis or in influencing IFN-alpha-induced Th1 polarization. 17985330_IRF-1 activation regulates the induction of the IL-27p28 subunit by interferon beta. 18079498_Observational study of gene-disease association. (HuGE Navigator) 18079498_functionally relevant IRF-1 (interferon regulatory factor 1) polymorphisms influence atopy risk, potentially by altering transcription factor binding, IRF-1 gene expression, and IFN-gamma regulation. 18084608_IRF-1 silencing through RNA interference, leads to an overexpression of TLR-4 in THP-1 cells. 18200030_First evidence of the involvement in malaria susceptibility of a specific locus within the 5q31 region of interferon regulatory factor-1. 18200030_Observational study of gene-disease association. (HuGE Navigator) 18216101_These data indicate that in early phases of HIV-1 infection or during virus reactivation from latency, IRF-1 is an additional component of the p50/p65 heterodimer binding the LTR enhancer, absolutely required for efficient HIV-1 replication. 18316378_Dendritic cell priming by L. major infection results in the early activation of NF-kappaB transcription factors and the up-regulation and nuclear translocation of interferon regulatory factor 1 (IRF-1) and IRF-8. 18338947_Observational study of gene-disease association. (HuGE Navigator) 18338947_data show that genetic variants in the IRF-1 and Stat1 genes of the IFN pathway are associated with multiple sclerosis and hepatitis c virus infection. 18381204_KAP1 interacts with STAT1 and regulates IFN/STAT1-mediated IRF-1 gene expression in collaboration with HDACs. 18401423_Although STAT1 phosphorylation required JNK and p38MAPK activation, only JNK activation was essential for IRF1 promoter activation by Tie2-R849W. 18454680_Observational study of gene-disease association. (HuGE Navigator) 18641303_IRF-1 is a key negative regulator of CD4(+)CD25(+) Treg cells through direct repression of Foxp3 expression[IFN regulatory factor 1] 18655181_involvement of IRF-1 in the regulation of SLC26A6 gene expression by IFNgamma in the human intestine 18676680_Observational study of gene-disease association. (HuGE Navigator) 18678606_TNF-alpha and PGE2 induce a complex that binds an oligonucleotide derived from the IRF1 promoter in dendritic cells. 18688264_FOXA1 and IRF-1 intermediary transcriptional regulators of PPARgamma-induced urothelial cytodifferentiation 18694960_These data reveal a novel 'feed-forward' mechanism induced by NF-kappaB which ensures that acutely synthesized IRF-1 operates in concert with NF-kappaB to amplify the immunoproteasome and antigen-processing functions of CD40. 18986693_NO inhibits HRV-16-induced production of CXCL10 by inhibiting viral activation of nuclear factor kappaB and of IRFs, including IRF-1, through a cGMP-independent pathway 19012493_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19028144_Interferon regulatory factor 1 (IRF1) polymorphisms with minor allele frequencies higher than 5% and haplotype frequencies in two Southeast Asian populations (Indonesian and Vietnamese) turned out not to be associated with pulmonary tuberculosis. 19028144_Observational study of gene-disease association. (HuGE Navigator) 19056926_JNK1 stimulated and mediated the effects of IFN and TNF-alpha on XAF1 expression through transcriptional regulation by induction of IRF-1. 19129219_A ChIP-chip approach identifies a novel role for the transcription factor IRF1 in the DNA damage response elicited by DNA interstrand crosslinks. 19131452_Observational study of gene-disease association. (HuGE Navigator) 19145247_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19247692_Observational study of gene-disease association. (HuGE Navigator) 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19295429_genetic polymorphism is associated with atopic asthma in children 19345417_High levels of IRF-1 in myeloid cells are a favorable prognostic factor for overall survival in myelodysplastic syndromes but they increase the probability of manifestation of autoimmune phenomena, with a diminished quality of life. 19404407_IRF-1, IRF-7, type I IFNs, and STAT1 form a signaling feedback loop that is critical in regulating TRAIL expression in HIV-1-infected macrophages. 19426920_mRNA levels are up-regulated in the peripheral blood cells of primary Sjogren Syndrome patients 19428110_HLA-A upregulation in liver cancer was mediated by both increased nuclear aggregation of transcription factor p65 and upregulation of transcription factor IRF-1. 19433065_siRNA against IRF-1 prevents IFN-gamma-mediated CD80 activation 19450680_Interferon regulatory factor 1 degradation is a multi-step process to be regulated in response to cell signalling, polyubiquitination by a specific E3-ligase and recognition of polyubiquitinated interferon regulatory factor 1 to the proteasome. 19463200_association between gene polymorphism and chlidhood asthma 19502235_the role of the Mf1 domain of IRF-1 in orchestrating the recruitment of regulatory factors that can impact on both its turnover and transcriptional activity. 19536153_Observational study of gene-disease association. (HuGE Navigator) 19548631_The levels of E-selectin, ICAM-1, IL-11, IRF-1, IL-6, IL-1beta and LIF genes expression in the B. pseudomallei-infected cells were 1.5-5 times lower than in the B. thailandensis-infected cells. 19574175_IRF-1-E7 was shown to be responsible for the positive regulation of many interleukins and to be involved in the differentiation of T-helper cells in cervical cancer 19625176_Observational study of gene-disease association. (HuGE Navigator) 19642896_Data show that increased level of NPM/B23 in conjunction with decreased level of IRF1 could aid GAGE-induced resistance to IFN-gamma. 19692168_Observational study of gene-disease association. (HuGE Navigator) 19697121_Our findings strongly imply a tumor suppressor role for the IRF1 gene in breast cancer. 19805480_the cellular accumulation of IRF-1 may represent a potential molecular mechanism mediating altered cellular response to GC through the depletion of GRIP-1 from the GR transcriptional regulatory complexes 19845895_Observational study of gene-disease association. (HuGE Navigator) 19851330_We conclude that IRF-1 can induce apoptosis by the intrinsic pathway independent of the extrinsic pathway by upregulation of PUMA. 19858727_IRF-1 could be nominated as one of the tumor suppressor factors and could aid in the early detection of HCC.-e 19880820_Human rhinovirus-16-induced CXCL10 production is dependent upon IRF-1. 20031576_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20031577_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20177805_We found that mice lacking IRF-1 in the central nervous system developed significantly milder clinical symptoms and shorter disease duration in experimental autoimmune encephalomyelitis 20177960_gene silencing downregulates Th1 cell function in patients with acute coronary syndrome 20435892_IL-27 p28 gene transcription is activated by interferon regulatory factor 8 in cooperation with interferon regulatory factor 1 20457620_IFNgamma restores breast cancer sensitivity to fulvestrant by regulating STAT1, IFN regulatory factor 1, NF-kappaB, BCL2 family members. 20459687_Observational study of gene-disease association. (HuGE Navigator) 20485362_Observational study of gene-disease association. (HuGE Navigator) 20503287_Observational study of gene-disease association. (HuGE Navigator) 20525893_IRF-1 is critical for interleukin (IL)-12p40 and IL-12p70 production induced in dendritic cells by nickel sulfate associated with interferon (IFN)-gamma. 20533260_STAT2 and IRF-1 comppete at binding interferon-stimulated response elements (ISREs) located on the retinoic acid-induced gene G (RIG-G) promoter in RIG-G. 20588308_Observational study of gene-disease association. (HuGE Navigator) 20817723_a role for the Mf1 domain in limiting both IRF-1-dependent transcription and the rate of IRF-1 turnover. 20846942_This study found evidence for a bidirectional regulation of IRF-1 gene expression controlled by single-nucleotide polymorphism rs2549009 in cis and a yet unidentified variant or haplotype or on environmental control in trans. 20861350_Lysophosphatidic acid inhibits CCL5/RANTES production in a human bronchial epithelial cell line by blocking the binding of IRF-1 to the CCL5/RANTES promoter 20947504_a complex relationship between CHIP and IRF-1 20980339_Observational study of gene-disease association. (HuGE Navigator) 21200019_Transitory IRF1 responsiveness in HIV-Resistance may be one of the key contributors to the altered susceptibility to HIV infection during the early stages of primary HIV infection. 21245151_analysis of binding of NPM1, TRIM28, and YB-1 to the Mf2 domain of IRF-1 21257209_The findings of our study indicate that IRF-1 signaling in glial cells serves as a final common pathway of inflammatory demyelination and may have important clinical implications in MS. 21288140_We show persistent activation of the Th1 (interferon-gamma) signaling pathway markers signal transducer and activator of transcription 1 (STAT1) and IRF1 in the small intestine of celiac disease children despite gluten-free diet. 21389130_The authors show that the human papillomavirus type 16 E5 protein expression per se stimulates IFN-beta expression this stimulation is specifically mediated by the induction of interferon regulatory factor 1. 21411754_HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. 21586271_These results suggested that HBZ has dual suppressive effects on IRF-1 function, which may contribute to HTLV-1 related pathogenesis. 21683060_BRG1-dependent induction of TRIM22 perfectly correlated with BRG1-dependent recruitment of IRF-1 to TRIM22 promoter. 21725055_IFN-gamma induces expression of IRF-1 in human erythroid precursors. 21790247_IRF-1 gene deletions were not observed in 19 (38%) patients. In our study, the frequency of deletions of these three exons was slightly higher than in an Indian population (52%), but lower than in Sweden in Europe (95%). 21803131_Data suggest that the new 18bp consensus motif appears to have a greater association with biological activities of IRF1. 21834067_IRF-1 is a key regulator of IL-18BPa expression and IL-18 bioactivity in rheumatoid arthritis synovial fibroblasts. 22096509_Data observed in DMD and aging skeletal muscle enrichment in motifs for candidate transcription factors that may coordinate either the immune/fibrosis responses (ETS1, IRF1, NF1) or the mitochondrial metabolism (ESRRA). 22213332_The modulation of IRFs seen in filarial (and presumably other tissue-sensitive helminths) underlies suppression of malaria-specific cytokines/chemokines that play a crucial role in immunity to malaria. 22345458_K1L and C7L antagonize IRF1-induced antiviral activities and that the host modulation function of C7L is evolutionally conserved in all poxviruses that can readily replicate in tissue-cultured mammalian cells. 22367195_We propose that IRF1 activation is responsible for carcinogen-induced Poleta up-regulation, which contributes to mutagenesis and ultimately carcinogenesis in cells. 22401175_Immunohistochemically, the lining synovium of Rheumatoid Aarthritis clearly expressed IRF1 22874466_Subjects with an anti-inflammatory response to a meal produced triglycerides that was enriched in nonesterified fatty acids, decreased IRF-1 expression, increased miR-126 activity, and diminished monocyte arrest. 22879909_The association of IRF1 and IRF8 variants with tuberculosis susceptibility was investigated. 23040881_An association study of functional polymorphic genes IRF-1, ...... with disease progression, aspartate aminotransferase, alanine aminotransferase, and viral load in chronic hepatitis B and C. 23046934_The increased expressions of OB-R, IRF-1 and GR-beta in airway smooth muscle cells (ASMCs) of obese rats with asthma may play a role in the onset of obese asthma and glucocorticoid resistance. 23071666_Data show that not only NF-kB p65, but also the interferon regulatory factor 1 (IRF1) and certain antigen rocessing achinery (APM) components are low in a subset of Neuroblastoma (NB) cell lines with aggressive features. 23134341_The E3 docking site was not available when IRF1 was in its DNA-bound conformation and cognate DNA-binding sequences strongly suppressed ubiquitination, showing a relationship between ligase binding and site-specific modification in the DNA-binding domain 23226549_IRF-1 also activates IFP35 expression in an IFN-gamma-inducible manner. 23255602_Smad7 was able to activate the caspase 8 promoter through recruitment of the interferon regulatory factor 1 (IRF1) transcription factor to the interferon-stimulated response element (ISRE) site. 23378427_Expression of interferon regulatory factor (IRF)1 and signal transducer and activator of transcription (STAT)1 mediates host defense against Mycobacterium tuberculosis through the cellular response to interferon (IFN)-gamma. 23487038_BAL1 represses the anti-proliferative and pro-apoptotic IFNgamma-STAT1-IRF1-p53 axis and mediates proliferation, survival and chemo-resistance in DLBCL. 23554911_KGF could up-regulate IL-7 expression through the STAT1/IRF-1, IRF-2 signaling pathway, which is a new insight in potential effects of KGF on the intestinal mucosal immune system. 23799084_Three polymorphisms in the IRF1 gene that protect against HIV-1 acquisition appear to exert no discernable effects once infection is established. 23807161_findings suggest that the responsiveness of IRF-1 to the immune cytokines IFN-gamma and TNF-alpha is able to point out two different cancer phenotypes and support the central role of such transcription factor in influencing different tumour behaviors 23811275_Identification of a functional IRF-1-binding site in the first intron of human optineurin gene that mediates interferon-gamma-induced activation of the promoter, is reported. 23934855_Data demonstrate direct roles for fluid shear stress and postprandial triglyceride-rich lipoprotein from human serum in the regulation of IRF-1 expression and downstream inflammatory responses in aortic endothelial cells. 24396068_IKK-epsilon phosphorylated the transcription factor IFN regulatory factor 1 (IRF-1). 24464131_we found that IRF1 was essential for IL-1-induced expression of the chemokines CXCL10 and CCL5 24632547_Data suggest that interferon regulatory factors 1 and 2 (IRF1 and IRF2) may serve as potential targets of therapy. 24650050_The novel AS regulatory activities attributed to IRF-1 indicate that the IFN-gamma response involves a global change in both gene transcription and AS in breast epithelial cells. 24886089_The present study further suggests that the combined targeted inhibition of STAT1, ARTD8, ARTD9 and/or DTX3L could increase the efficacy of chemotherapy or radiation treatment in prostate and other high-risk tumor types with an increased STAT1 signaling. 24940638_The miR-31/IRF-1/CTSS pathway may play a functional role in the pathogenesis of cystic fibrosis lung disease. 24995581_Transcriptome analysis identifies IRF1 as being associated with platinum sensitivity and an independent predictor of both PFS and OS in HGSOC 25014791_Single nucleotide polymorphisms in RBPJ, IL1R1, REV3L, TRAF3IP2, IRF1 and ICOS showed association with rheumatoid arthritis in black South Africans. 25312803_IRF-1 regulates the transcription of target genes which play essential roles in various physiological and pathological processes, including viral infection, tumor immune surveillance, pro-inflammatory injury, development of immune system. (Review) 25347735_IRF1 re-expression in human cancer cells causes cells to become resistant to infection by the oncolytic vesicular stomatitis virus strain. 25418955_IRF-1 is an important signaling protein in the interferon pathway. It not only activates gene expression as a transcription factor, but may perpetuate disease by leading to a dysregulated epigenome 25497010_Data indicate that the interferon regulatory factor (IRF1) promotion was observed in cancer cell lines treated with different MEK inhibitors or with RNAi oligonucleotides against extracellular signal-regulated kinases (ERK1/2). 25501823_IRF1 is a dual regulator of BV6-induced apoptosis and inflammatory cytokine secretion. 25576084_silencing IRF1 promoted autophagy by increasing BECN1 and blunting IGF1 receptor and mTOR survival signaling 25592039_A positive feedback loop between IRF1 and miR-29b may contribute to the sensitivity of colorectal cancer cells to IFN-gamma by repressing IGF1. 25611806_IRF1 directly interacts with chromatin modifying enzymes, supporting a model where recruitment to specific target genes is mediated in part by IRF1. 25658920_IRF1, a transcription factor, regulates miR-203 transcription by binding to the miR-203 promoter. 25964490_IRF-1 promotes LTx I/R injury via hepatocyte IL-15/IL-15Ralpha production and suggest that targeting IRF-1 and IL-15/IL-15Ralpha may be effective in reducing I/R injury associated with LTx. 25997853_The present data demonstrate that IRF-1 could effectively promote the immune maturation and function of dendritic cells in ACS Acute Coronary Syndrome patients. 26011589_Data show that ectopic expression of interferon regulatory factor 1 (IRF-1) reduces NF-kappa B activity and suppresses TNF receptor-associated factor 2 (TRAF2) and inhibitor of apoptosis 1 protein (cIAP1) expression in breast cancer cells. 26362649_Gene expression meta-analysis reveals immune response convergence on the IFNgamma-STAT1-IRF1 axis and adaptive immune resistance mechanisms in lymphoma 26510961_5AZ had a protective effect after MI by potentiation of IRF1 sumoylation and is suggested as a novel therapeutic intervention for cardiac repair. 26674566_IRF1 upregulation in fetal membranes and myometrium after term labor indicates a proinflammatory role for IRF1 in human parturition. IRF1 is involved in TLR- and cytokine-mediated signaling in human myometrium. 26872335_IRF1 is a transcriptional regulator of IRG1 in human macrophages. 27031443_HNP1 upregulation of cytokine expression in pDCs was inhibited by blockade of NF-kappaB activation or knockdown of IRF1, demonstrating the importance of these two signaling events in HNP1-induced pDC activation. 27176664_the interactions of IRF1, IFN-beta and IRF5 are involved in the M1 polarization of macrophages and have antitumor functions. 27191889_In SK-Hep1 cells, an increase in apoptosis and decrease in autophagy were observed after IFN-gamma stimulation, which was accompanied with increasing IRF-1 levels. 27260002_Zinc is capable of ameliorating the allogeneic immune reaction by enhancement of antigen-specific iTreg cells due to modulation of essential molecular targets by upregulation of Foxp3 and KLF-10 and downregulation of IRF-1. 27279136_MiR-23a downregulates the expression of IRF-1 in HCC cells. 27281481_Regulatory elements for both IRF-1 (-1019 to -1016) and CREB (-1198 to -1195), specific to the distal THBS1 promoter, were required for leptin-induced TSP-1 transcription. 27310137_Down-regulation of interferon regulatory factor 1 gene expression in hepatitis B virus patients without rejection emphasized counteraction between hepatitis B virus replication and interferon regulatory factor 1 production. On the other hand, interferon regulatory factor 1 gene overexpression in patients with rejection may result in inflammatory reactions and ischemic-reperfusion injury. 27328944_IFN regulatory factor 1 effectively inhibits hepatitis E virus replication through the activation of the JAK-STAT pathway 27363262_Data suggest that interferon beta (IFN-beta) might be involved in modulating the expressions of interferon regulatory factor 1 (IRF1) and interferon regulatory factor-5 protein (IRF5) as well as maintaining the M1 polarization status and its function. 27444640_These unprecedented data suggest that IRF1 and NF-kappaB orchestrate the TLR4-primed immunomodulatory response of hMSCs and that this response also involves the PI3K pathway. 27512062_Inflammation-driven IRF1 and NF-kappaB activity promotes ERVK reactivation. 27551049_silencing of IL1B plus dexamethasone-induced DUSP1 significantly reduced IRF1 expression. IL1B-induced expression of CXCL10 was largely insensitive to dexamethasone, whereas other DUSP1-enhanced, IRF1-dependent mRNAs showed various degrees of repression. 27765819_B. abortus lipoproteins via IL-6 inhibit the expression of IFN regulatory factor 1 (IRF-1), a critical regulatory transcription factor for CIITA induction. 27795392_Tat exploited the cellular HDM2 (human double minute 2 protein) ubiquitin ligase to accelerate IRF-1 proteasome-mediated degradation, resulting in a quenching of IRF-1 transcriptional activity during HIV-1 infection. 27866197_rs56288038 (C/G) in IRF-1 3'UTR acted as a promotion factor in gastric cancer development through enhancing the regulatory role of miR-502-5p in IRF-1 expression. 27940139_Leading to a STAT1-IRF1 controlled upregulation of TLR3 expression in macrophages. 28039033_An IRF-1 shorter splicing transcript has been identified in ac | ENSMUSG00000018899 | Irf1 | 164.16981 | 1.2061199 | 0.2703733390 | 0.25067360 | 1.167645e+00 | 2.798857e-01 | 6.671167e-01 | No | Yes | 210.761155 | 41.995697 | 166.260027 | 32.452332 | |
| ENSG00000125462 | 10485 | MIR9-1HG | lncRNA | Q13536 | FUNCTION: May play a role in FOS signaling pathways involved in development and remodeling of neurons. Promotes transcription of the FOS promoter. {ECO:0000269|PubMed:10995546}. | Nucleus;Reference proteome | nucleus [GO:0005634]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 30908634_CACNA1G-AS1 promotes hepatocellular carcinoma progression through regulating the miR-2392/C1orf61 pathway. 33915231_C1orf61 promotes hepatocellular carcinoma metastasis and increases the therapeutic response to sorafenib. | 35.69800 | 0.7292992 | -0.4554172794 | 0.46763444 | 9.372525e-01 | 3.329854e-01 | No | Yes | 34.375485 | 5.761819 | 45.575557 | 7.322236 | ||||||
| ENSG00000125740 | 2354 | FOSB | protein_coding | P53539 | FUNCTION: Heterodimerizes with proteins of the JUN family to form an AP-1 transcription factor complex, thereby enhancing their DNA binding activity to gene promoters containing an AP-1 consensus sequence 5'-TGA[GC]TCA-3' and enhancing their transcriptional activity (PubMed:12618758, PubMed:28981703). As part of the AP-1 complex, facilitates enhancer selection together with cell-type-specific transcription factors by collaboratively binding to nucleosomal enhancers and recruiting the SWI/SNF (BAF) chromatin remodeling complex to establish accessible chromatin (By similarity). Together with JUN, plays a role in activation-induced cell death of T cells by binding to the AP-1 promoter site of FASLG/CD95L, and inducing its transcription in response to activation of the TCR/CD3 signaling pathway (PubMed:12618758). Exhibits transactivation activity in vitro (By similarity). Involved in the display of nurturing behavior towards newborns (By similarity). May play a role in neurogenesis in the hippocampus and in learning and memory-related tasks by regulating the expression of various genes involved in neurogenesis, depression and epilepsy (By similarity). Implicated in behavioral responses related to morphine reward and spatial memory (By similarity). {ECO:0000250|UniProtKB:P13346, ECO:0000269|PubMed:12618758, ECO:0000269|PubMed:28981703}.; FUNCTION: [Isoform 11]: Exhibits lower transactivation activity than isoform 1 in vitro (By similarity). The heterodimer with JUN does not display any transcriptional activity, and may thereby act as an transcriptional inhibitor (By similarity). May be involved in the regulation of neurogenesis in the hippocampus (By similarity). May play a role in synaptic modifications in nucleus accumbens medium spiny neurons and thereby play a role in adaptive and pathological reward-dependent learning, including maladaptive responses involved in drug addiction (By similarity). Seems to be more stably expressed with a half-life of ~9.5 hours in cell culture as compared to 1.5 hours half-life of isoform 1 (By similarity). {ECO:0000250|UniProtKB:P13346}. | 3D-structure;Alternative splicing;DNA-binding;Disulfide bond;Nucleus;Phosphoprotein;Reference proteome | The Fos gene family consists of 4 members: FOS, FOSB, FOSL1, and FOSL2. These genes encode leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. As such, the FOS proteins have been implicated as regulators of cell proliferation, differentiation, and transformation. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:2354; | chromatin [GO:0000785]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription factor binding [GO:0008134]; cellular response to calcium ion [GO:0071277]; cellular response to hormone stimulus [GO:0032870]; female pregnancy [GO:0007565]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357]; response to cAMP [GO:0051591]; response to corticosterone [GO:0051412]; response to mechanical stimulus [GO:0009612]; response to morphine [GO:0043278]; response to progesterone [GO:0032570]; response to xenobiotic stimulus [GO:0009410]; transcription by RNA polymerase II [GO:0006366] | 12371906_demonstrated that a functional AP-1 site mediates MMP-2 transcription in cardiac cells through the binding of distinctive Fra1-JunB and FosB-JunB heterodimers. The synthesis of MMP-2 is considered to be independent of the AP-1 transcriptional complex 12618758_AP-1 (c-Jun & FosB) binds to a site in the 5' untranslated region of the CD95L gene. Transdominant negative Jun mutants reduce CD95L promoter activity. FosB dimerized with c-Jun has an important role in TCR/CD3-mediated activation-induced cell death. 14741347_VEGF and PlGF induced expression of both full-length FosB mRNA and an alternatively spliced variant. 15926923_In hepatoma-associated anorexia-cachexia FpsB was induced in several brain areas of thes forebrain. 17495958_Coordinated down- and up-regulation of the various AP-1 subunits in the course of epidermal wound healing is important for its undisturbed progress, putatively by influencing inflammation and cell-cell communication. 17601350_Observational study of gene-disease association. (HuGE Navigator) 18207134_Observational study of gene-disease association. (HuGE Navigator) 18490653_activation of protein kinase A elicits an immediate response through induction of genes such as ID2 and FosB, followed by sustained secretion of bone-related cytokines such as BMP-2, IGF-1, and IL-11 18491952_DeltaFosB was able to trigger partial Pref-1-mediated de-differentiation of adipocytes, which also retained their adipocytic cell phenotype. 18802113_An IFN-gamma-mediated homeostatic loop limits the potential for tissue damage associated with inflammation and identifies transcriptional factor AP-1 that regulates matrix metalloproteinase expression in myeloid cells in inflammatory settings. 18977241_Observational study of gene-disease association. (HuGE Navigator) 19133651_MicroRNA-101, which is aberrantly expressed in hepatocellular carcinoma, could repress the expression of the FOS oncogene. 19381435_FosB is induced in PMA treated K562 cells in a sustained manner and forms an active AP-1 protein-DNA complex. Down-regulation of FosB with specific shRNAs inhibited the induction of CD41, a specific cell surface marker of megakaryocytes. 19453261_Observational study of gene-disease association. (HuGE Navigator) 19795327_Results suggest that in addition to clinically prognostic factors, FOS-B expression has a debatable impact on patient survival. 20571966_RGS16 and FosB are underexpressed in pancreatic cancer with lymph node metastasis and associated with reduced survival 20813842_transcription factor FosB/activating protein-1 (AP-1) activation is a prominent downstream signal of the extracellular nucleotide receptor P2RX7 in monocytic and osteoblastic cells 20826776_Findings indicate that neurobehavioral stress leads to FosB-driven increases in IL8, which is associated with increased tumor growth and metastases. 21543584_AP-1 protein induction during monopoiesis favors C/EBP: AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. 21616539_Induction of a DeltaFOSB mediated transcriptional pattern in the prefrontal cortex is opposite to the down-regulation observed in the nucleus accumbens in patients with major depressive disorder 23614275_Report Fos-B expression in skin keratinocytes/fibroblasts and keloid fibroblasts exposed to genistein. 23810250_Data indicate a significant correlation in miR-181b, FOS and miR-21 expression glioma tissues. 23933656_This study describing the expres-sion pattern of FosB/ FosB immunoreactivity inpost mortem basal ganglia sections from Parkinson disease patients. 24374978_Pseudomyogenic haemangioendothelioma consistently displays a SERPINE1-FOSB fusion gene, resulting from a translocation between chromosomes 7 and 19. 25043949_The abnormalities were screened by FISH in 44 epithelioid hemangioma (EH) from different locations with seven additional EH revealing FOSB gene rearrangements, all except one being fused to ZFP36. 25446562_studies of FosB are providing new insight into the molecular basis of depression and antidepressant action. 26608367_our findings show that DeltaFosB increases the expression of MMP-9 and exhibits a significantly high survival and proliferation in MCF-7 cells. 26949019_These results suggest that SETDB1- mediated FosB expression is a common molecular phenomenon, and might be a novel pathway responsible for the increase in cell proliferation that frequently occurs during anticancer drug therapy. 27248170_FOSB overexpression results in TNBC cell death, whereas inhibition of calcium signaling eliminates FOSB induction and blocks TP4-induced TNBC cell death 27494187_FosB expression in the prefrontal cortex and hippocampus of the cocaine addicted and depression patients 27515856_Diffuse and strong FOSB expression was specific for pseudomyogenic hemangioendothelioma in the current series and FOSB immunohistochemistry is an effective tool for differentiating between PHE and its histological mimics. 27889568_Our results using a number of approaches, such as promoter reporter assay, FosB knock down and Chip assay, suggest that the expression of miR-22 is regulated transcriptionally by FosB. 28009608_FOSB is a highly sensitive and diagnostically useful marker for pseudomyogenic hemangioendothelioma. 28387432_results shows the involvement of Spry2 in regulation of FosB and Runx2 genes, MAPK signaling and proliferation of mesenchymal stem cells. 28653602_miR-144-3p plays an important role in pancreatic cancer cell proliferation, migration, and invasion by targeting FOSB 28724635_The NFATc3 first induced the expression of its interaction partner FosB before forming the heterodimeric NFATc3-FosB transcription factor complex, which bound the proximal AP-1 site in the TF gene promoter and activated TF expression. 28981703_the mechanism underlying redox-regulation of AP-1 Fos/Jun transcription factors and provide structural insight for therapeutic interventions targeting AP-1 proteins. 29527734_FOSB immunohistochemistry is sensitive in the diagnosis of angiolymphoid hyperplasia with eosinophilia, and allows differentiation from its histological mimics 29858576_Findings iindicate a human bone tumour defined by mutations of FOS and FOSB. 30256258_Recurrent ACTB-FOSB fusions are found in pseudomyogenic hemangioendothelioma. 30459475_Recurrent ACTB-FOSB fusion occurs in pseudomyogenic hemangioendothelioma. 31262712_We identified increased level of Fos-B mRNA, the binding target of FUS, in FUS-mutant MNs. While Fos-B reduction using si-RNA or an inhibitor ameliorated the observed aberrant axon branching, Fos-B overexpression resulted in aberrant axon branching even in vivo. The commonality of those phenotypes was further confirmed with other amyotrophic lateral sclerosis (ALS) causative mutation than FUS 31735020_Poly(ADP-ribose) Polymerases (PARP) cleavage and positive annexin V level are increased during piperlongumine (PL) treatment with FosB overexpression in MCF7 breast cancer cells, whereas PARP cleavage and positive annexin V level were decreased during PL treatment with siFosB transfection, implying that FosB is a pro-apoptotic protein for induction of cell death in PL-treated MCF7 breast cancer cells. 31894310_Association of FOSB exon 4 unmethylation with poor prognosis in patients with latestage nonsmall cell lung cancer. 32482537_Clinicopathologic study of 6 cases of epithelioid osteoblastoma of the jaws with immunoexpression analysis of FOS and FOSB. 32819575_MicroRNA-27a-3p directly targets FosB to regulate cell proliferation, apoptosis, and inflammation responses in immunoglobulin a nephropathy. 32891613_FosB recruits KAT5 to potentiate the growth and metastasis of papillary thyroid cancer in a DPP4-dependent manner. 34153682_Knockout of FosB gene changes drug sensitivity and invasion activity via the regulation of Bcl-2, E-cadherin, beta-catenin, and vimentin expression. | ENSMUSG00000003545 | Fosb | 50.48156 | 0.8490492 | -0.2360798879 | 0.41992688 | 3.136354e-01 | 5.754579e-01 | No | Yes | 52.330741 | 9.676427 | 60.665473 | 10.706095 | ||
| ENSG00000125755 | 8189 | SYMPK | protein_coding | Q92797 | FUNCTION: Scaffold protein that functions as a component of a multimolecular complex involved in histone mRNA 3'-end processing. Specific component of the tight junction (TJ) plaque, but might not be an exclusively junctional component. May have a house-keeping rule. Is involved in pre-mRNA polyadenylation. Enhances SSU72 phosphatase activity. {ECO:0000269|PubMed:16230528, ECO:0000269|PubMed:20861839}. | 3D-structure;Alternative splicing;Cell adhesion;Cell junction;Cell membrane;Cytoplasm;Cytoskeleton;Isopeptide bond;Membrane;Nucleus;Phosphoprotein;Reference proteome;Repeat;Tight junction;Ubl conjugation;mRNA processing | This gene encodes a nuclear protein that functions in the regulation of polyadenylation and promotes gene expression. The protein forms a high-molecular weight complex with components of the polyadenylation machinery. It is thought to serve as a scaffold for recruiting regulatory factors to the polyadenylation complex. It also participates in 3'-end maturation of histone mRNAs, which do not undergo polyadenylation. The protein also localizes to the cytoplasmic plaques of tight junctions in some cell types. [provided by RefSeq, Jul 2008]. | hsa:8189; | bicellular tight junction [GO:0005923]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; mRNA cleavage and polyadenylation specificity factor complex [GO:0005847]; nuclear body [GO:0016604]; nuclear stress granule [GO:0097165]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; cell adhesion [GO:0007155]; mRNA polyadenylation [GO:0006378]; negative regulation of protein binding [GO:0032091]; positive regulation of protein dephosphorylation [GO:0035307] | 14707147_symplekin has a role in HSF1 modulation of Hsp70 mRNA polyadenylation 15550246_In Xenopus, symplekin interacts with the CPSF complex and the regulatory protein CPEB, and is required for polyadenylation of CPE-containing RNA 17880531_The decreased expression of symplekin may be an early step in the transformation of hepatocytes, whereas alteration of the expression of adherens junctions and desmosomes may indicate more serious changes. 19328795_The symplekin/ZONAB complex inhibits intestinal cell differentiation by the repression of AML1/Runx1. 20133805_Data show that claudin-2 expression was reduced following symplekin down-regulation, and siRNA-mediated claudin-2 down-regulation increased the transepithelial resistance and decreased cyclin D1 expression and ZONAB nuclear localization. 20601676_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20823274_Symplekin supports faithful mitosis by contributing to the formation of a bipolar spindle apparatus. Depletion of symplekin attenuates microtubule polymerization as well as expression of the critical microtubule polymerization protein CKAP5 (TOGp). 20861839_crystal structure at 2.4 A resolution of the amino-terminal domain (residues 30-340) of human symplekin in a ternary complex with the Pol II carboxy-terminal domain (CTD) Ser 5 phosphatase Ssu72 and a CTD Ser 5 phosphopeptide 22218735_Symplekin expression regulates the assembly of tight junctions, thus helps to maintain the integrity of the epithelial monolayer and cellular polarity. 23994619_Symplekin interacts and co-localizes with both MOZ and MLL in immature hematopoietic cells. Its inhibition leads to a decrease of the HOXA9 protein level but not of Hoxa9 mRNA. 25921069_CPSF2 and SYMPK, are RBFOX2 cofactors for both inclusion and exclusion of internal exons. 28295283_Data indicate rs56848936 in the gene symplekin protein (SYMPK) at 19q13.3, associated with colon cancer risk. 28630428_The significance of nuclear symplekin in tumorigenesis is also highlighted, and ERK-dependent phosphorylation represents a mechanism for its subcellular sorting. 29167270_Results indicated that reductase becomes destabilized in the absence of UBIAD1, resulting in reduced cholesterol synthesis and intracellular accumulation. Findings further establish UBIAD1 as a central player in the reductase ERAD pathway and regulation of isoprenoid synthesis. 30032159_Research proved that high Rab11 expression enhances cellular multiplication and invasiveness of bladder cancer, possibly by regulating the NF-kappaB signaling pathway. | ENSMUSG00000023118 | Sympk | 3873.74333 | 1.0502568 | 0.0707421726 | 0.08513411 | 6.916209e-01 | 4.056136e-01 | 7.637945e-01 | No | Yes | 4659.450475 | 536.727945 | 4271.047625 | 481.059909 | |
| ENSG00000125779 | 80025 | PANK2 | protein_coding | Q9BZ23 | FUNCTION: [Isoform 1]: Catalyzes the phosphorylation of pantothenate to generate 4'-phosphopantothenate in the first and rate-determining step of coenzyme A (CoA) synthesis (PubMed:15659606, PubMed:17825826, PubMed:17242360, PubMed:16272150). Required for angiogenic activity of umbilical vein of endothelial cells (HUVEC) (PubMed:30221726). {ECO:0000269|PubMed:15659606, ECO:0000269|PubMed:16272150, ECO:0000269|PubMed:17242360, ECO:0000269|PubMed:17825826, ECO:0000269|PubMed:30221726}.; FUNCTION: [Isoform 4]: Catalyzes the phosphorylation of pantothenate to generate 4'-phosphopantothenate in the first and rate-determining step of coenzyme A (CoA) synthesis. {ECO:0000269|PubMed:16272150}. | 3D-structure;ATP-binding;Alternative initiation;Alternative splicing;Angiogenesis;Coenzyme A biosynthesis;Cytoplasm;Direct protein sequencing;Disease variant;Kinase;Mitochondrion;Neurodegeneration;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Transit peptide | PATHWAY: Cofactor biosynthesis; coenzyme A biosynthesis; CoA from (R)-pantothenate: step 1/5. {ECO:0000269|PubMed:15659606, ECO:0000269|PubMed:16272150, ECO:0000269|PubMed:17242360, ECO:0000269|PubMed:17825826}. | This gene encodes a protein belonging to the pantothenate kinase family and is the only member of that family to be expressed in mitochondria. Pantothenate kinase is a key regulatory enzyme in the biosynthesis of coenzyme A (CoA) in bacteria and mammalian cells. It catalyzes the first committed step in the universal biosynthetic pathway leading to CoA and is itself subject to regulation through feedback inhibition by acyl CoA species. Mutations in this gene are associated with HARP syndrome and pantothenate kinase-associated neurodegeneration (PKAN), formerly Hallervorden-Spatz syndrome. Alternative splicing, involving the use of alternate first exons, results in multiple transcripts encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:80025; | cytosol [GO:0005829]; mitochondrial intermembrane space [GO:0005758]; mitochondrion [GO:0005739]; nucleus [GO:0005634]; ATP binding [GO:0005524]; pantothenate kinase activity [GO:0004594]; aerobic respiration [GO:0009060]; angiogenesis [GO:0001525]; coenzyme A biosynthetic process [GO:0015937]; mitochondrion morphogenesis [GO:0070584]; pantothenate metabolic process [GO:0015939]; phosphorylation [GO:0016310]; regulation of bile acid metabolic process [GO:1904251]; regulation of fatty acid metabolic process [GO:0019217]; regulation of mitochondrial membrane potential [GO:0051881]; regulation of triglyceride metabolic process [GO:0090207]; spermatid development [GO:0007286] | 12554685_Identified two alternatively used first exons resulting in distinct isoforms, one of which carries an N-terminal extension with a predicted mitochondrial targeting signal. 14639680_Missense mutaions in PANK2 gene were observed in two siblings with Hallervorden- Spatz syndrome 14743358_The presence of mutation in the PANK2 gene is associated with younger age at onset and a higher frequency of dystonia, dysarthria, intellectual impairment, and gait disturbance. Parkinsonism is seen predominantly in adult-onset patients 15105273_An unconventional translational start codon, CUG, which is polymorphic in the general population is proposed. PANK2 is predicted to localize to mitochondria, with a 29 amino acid mitochondrial targeting sequence identified. 15390030_Adult-onset focal dystonia was the presenting sign of pantothenate kinase-associated neurodegeneration (PKAN) in a patient with a novel homozygous missense mutation (C856T) 15465096_Direct sequencing of the neurodegeneration patient's genomic DNA revealed homozygous base substitutions in the pantothenate kinase gene (PANK2): the A764-->G substitution (N245S) due to consanguinity of her parents. 15659606_Demonstrated that the mitochondrial isoform is sequentially cleaved at two sites by the mitochondrial processing peptidase, generating a long-lived 48 kDa mature protein localized to mitochondria of neurons in human brain. 15659606_These results suggest that neurodegeneration with brain iron accumulation (NBIA; formerly Hallervorden-Spatz disease) is caused by altered neuronal mitochondrial lipid metabolism caused by mutations disrupting PanK2 protein levels and catalytic activity. 15747360_Novel compound heterozygous mutations (Asp268Gly and Ile391Asn) in the PANK2 gene in a Chinese patient with Hallervorden-Spatz Syndrome 15793782_PANK2 gene mutations can cause Hallervorden-Spatz syndrome in Chinese patients. 15843062_Observational study of gene-disease association. (HuGE Navigator) 15911822_The authors report clinical and genetic findings of 16 patients with PKAN. The authors identified 12 mutations in the PANK2 gene, five of which were new. 16240131_The 1142_1144delGAG mutation of PANK2 probably originated from one common ancestor at the beginning of the ninth century, approximately 38 generations ago 16272150_Unique biochemical features of the PanK2 isoforms suggest that catalytic defects may not be the sole cause for the neurodegenerative phenotype. 16437574_Observational study of genotype prevalence. (HuGE Navigator) 16450344_We demonstrate that the G521R mutation results in an unstable and inactive protein in tremor-predominant neurodegeneration. 16962235_PANK2 mutations are not associated with some adult degenerative conditions 17242360_PanK2 is located in the mitochondria to sense the levels of palmitoylcarnitine and up-regulate CoA biosynthesis in response to an increased mitochondrial demand for the cofactor to support beta-oxidation 17631502_analysis of the homodimeric structures of the catalytic cores of PanK1alpha and PanK3 in complex with acetyl-CoA and the the structural effects of the PanK2 mutations that have been implicated in neurodegeneration 17825826_expression of PanK2 was higher in human brain compared to mouse brain 17903678_Pantothenate kinase-associated neurodegeneration is an autosomal-recessive disorder associated with the accumulation of iron in the basal ganglia associated with mutations in the PANK2 gene. 18006953_Two novel PANK2 gene mutation in Pantothenate Kinase-Associated Neurodegeneration. 18074375_Focal hand dystonia showed atypical phenotype of PANK2 gene mutations. 18239249_a novel missense mutation (P354L) in exon 4 of the PANK2 gene in an adolescent with classic pantothenate kinase-associated neurodegeneration was identified 19224615_In this report identified a novel mutation( in the PANK29p.D378G and p.D452G )gene responsible for PKAN and confirmed that PKAN has a board spectrum of phenotype, even among siblings with same mutations. 20006850_two Japanese siblings with the adult-onset slowly progressive type of pantothenate kinase-associated neurodegeneration who were found to have a novel PANK2 mutation 20551478_PANK2 mutations are not invariably associated with the 'eye-of-the-tiger sign (early onset generalised dystonia and basal ganglia abnormalities) 20603201_findings validate expression of the short PANK2 isoform and enable predictions about potentially deleterious sequence variants in the regulatory region of this human disease gene 20721927_the patient reported here shows a peculiar PKAN clinical phenotype probably based on new mutations identified in the PANK2 gene 20877624_Observational study of gene-disease association. (HuGE Navigator) 20925075_Progressive delayed-onset postanoxic dystonia - First example of PKAN symptom onset possibly provoked by environmental trigger (anoxia) 21442655_This study identified that new mutation of Pantothenate kinase associated with neurodegeneration. 22103354_The c.1319G>C (p.R440P) mutation appears to be a founder genotype among Korean patients with Pantothenate kinase-associated neurodegeneration. 22221393_study used global metabolic profiling to explore the metabolic consequences of mutations in pantothenate kinase 2 that are responsible for Pantothenate Kinase-Associated Neurodegeneration 22692681_Skin fibroblasts from pantothenate kinase-associated neurodegeneration patients highlight a possible molecular relationship between Pank2 deficiency and iron misregulation. 22930366_Identification of novel compound heterozygous mutations in PANK2 gene in two Chinese siblings with atypical pantothenate kinase-associated neurodegeneration. 23116688_we describe the clinical, radiological, and molecular find-ings of a classic PKAN patient of Iranian descent with a novel frameshift mutation in the coding region of the PANK2 gene 23166001_Mutations in both PANK2 and C19orf12 contributed significantly to neurodegeneration with brain iron accumulation in the Iranian patients 24348190_Caucasian patients have more complex presentations than Asians. Exon 3 and 4 are hot spots for screening PANK2 mutations in Asian patients. 24655737_study presents 2 siblings who were homozygous for a novel c.695A>G (p.Asp232Gly) missense mutation in exon 2 of PANK2 gene; index patient presented with a 5-year history of slowly progressive gait disturbance, dysarthria, mild axial rigidity and bradykinesia 24689511_Novel PANK2 gene mutations and clinical features in patients with pantothenate kinase-associated neurodegeneration. 25668476_Mutations in PANK2 and CoASY lead, respectively, to PKAN and CoPAN forms of Neurodegeneration with brain iron accumulation . Mutations in PLA2G6 lead to PLAN. Mutations in C19orf12 lead to MPAN 25915509_Data suggests that the c.680 A>G mutation in the PANK2 gene alone is not sufficient to determine acanthocytic shape transformation in erythrocytes but some additional factor(s)/condition(s) are necessary for acanthocytosis to occur. 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that PANK2 is a putative expression variability marker for the 3-day, 8-cell embryo stage. 26547561_Tissue or cellular hypoxic/ischemic injury within the globus pallidus may underlie the pathogenesis of pantothenate kinase-associated neurodegeneration due to PANK2 mutations and apoE aggregates. 27516453_These findings provide direct evidence that PANK2 malfunctioning is responsible for abnormal phenotypes in human neuronal cells of pantothenate kinase-associated neurodegeneration patients. 27815806_We aim to present a case of a healthy infant born after intracytoplasmic sperm injection-in vitro fertilization (ICSI-IVF) with a preimplantation genetic diagnosis (PGD) for pantothenate kinase-associated neurodegeneration (PKAN) due to PANK2 mutation 28113101_Homozygous PANK2 mutations in 22 PKAN patients from 13 Turkish families. 28189602_Results show that overexpression of PANK2 results in substantial elevated level of Co-A in skeletal muscle in transgenic mice which displays reduced skeletal muscle mass and significantly impaired exercise tolerance and grip strength. 28680084_Study identified c.1069C > T missense variation (rs753376100) in exon 3 of PANK2 gene associated with Pantothenate kinase-associated Neurodegeneration (PKAN) that had segregated in the family in an autosomal recessive mode. The clinical phenotype was found to be concordant with classic PKAN in terms of the age of onset, symptoms and radiological findings. The variant amino acid was mapped within the catalytic site. 28821231_A deleterious homozygous four-nucleotide deletion causing frameshift deletion in PANK2 gene (c.1426_1429delATGA, p.M476 fs) was identified in an 8 years old girl with dystonia, bone fracture, muscle rigidity, abnormal movement, lack of coordination and chorea. 28863176_PANK2 mutations have an effect on iPSC-derived cortical neuronal cells in culture 29642163_The key finding of the study encompassed the detection of a novel PANK2 gene mutation in a child of Chinese ethnicity with PKAN. The PANK2 gene c.A650G, as well as c.T1341G, mutations may be potential mutation hotspots in children with PKAN in Mainland China. 30141000_Overexpression of Human Mutant PANK2 Proteins Affects Development and Motor Behavior of Zebrafish Embryos. 30226968_We found c.966 G>T (p.Glu322Asp) mutation in the PANK2 gene mutation analysis in the individuals from the brain imaging findings. Although individuals in this family who had a homozygous mutation in PANK2 gene analyses had the 'eye-of-the-tiger' sign and atypical disease, they were noted to have differing clinical findings. 31088771_A novel PANK2 mutation was identified in South East Asian population in Thailand with pantothenate kinase associated neurodegeneration. 32705819_PKAN neurodegeneration and residual PANK2 activities in patient erythrocytes. 35204826_A Potential Citrate Shunt in Erythrocytes of PKAN Patients Caused by Mutations in Pantothenate Kinase 2. 35246191_Genetic mutation spectrum of pantothenate kinase-associated neurodegeneration expanded by breakpoint sequencing in pantothenate kinase 2 gene. | ENSMUSG00000037514 | Pank2 | 1072.08823 | 1.0101709 | 0.0145994401 | 0.10845952 | 1.812723e-02 | 8.928986e-01 | 9.707866e-01 | No | Yes | 1436.343403 | 189.030073 | 1336.904886 | 172.266182 |
| ENSG00000125814 | 63908 | NAPB | protein_coding | Q9H115 | FUNCTION: Required for vesicular transport between the endoplasmic reticulum and the Golgi apparatus. {ECO:0000250}. | Alternative splicing;ER-Golgi transport;Membrane;Protein transport;Reference proteome;Transport | This gene encodes a member of the soluble N-ethyl-maleimide-sensitive fusion attachment protein (SNAP) family. SNAP proteins play a critical role in the docking and fusion of vesicles to target membranes as part of the 20S NSF-SNAP-SNARE complex. This gene encodes the SNAP beta isoform which has been shown to be preferentially expressed in brain tissue. The encoded protein also interacts with the GluR2 alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptor subunit C-terminus and may play a role as a chaperone in the molecular processing of the AMPA receptor. [provided by RefSeq, Mar 2017]. | hsa:63908; | extracellular exosome [GO:0070062]; glutamatergic synapse [GO:0098978]; SNARE complex [GO:0031201]; synaptobrevin 2-SNAP-25-syntaxin-1a complex [GO:0070044]; soluble NSF attachment protein activity [GO:0005483]; syntaxin binding [GO:0019905]; intracellular protein transport [GO:0006886]; regulation of synaptic vesicle priming [GO:0010807]; SNARE complex disassembly [GO:0035494]; synaptic transmission, glutamatergic [GO:0035249] | 33189936_A novel NAPB splicing mutation identified by Trio-based exome sequencing is associated with early-onset epileptic encephalopathy. | ENSMUSG00000027438 | Napb | 419.23881 | 0.9711082 | -0.0422960059 | 0.16239258 | 6.786597e-02 | 7.944697e-01 | 9.418864e-01 | No | Yes | 478.681770 | 98.290342 | 487.841845 | 98.009364 | |
| ENSG00000125895 | 55321 | TMEM74B | protein_coding | Q9NUR3 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:55321; | integral component of membrane [GO:0016021] | 20944657_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000044364 | Tmem74b | 73.28264 | 0.7041045 | -0.5061384363 | 0.33523573 | 2.253440e+00 | 1.333178e-01 | No | Yes | 54.901534 | 10.575185 | 78.053911 | 14.450062 | ||||
| ENSG00000126001 | 11190 | CEP250 | protein_coding | Q9BV73 | FUNCTION: May be involved in ciliogenesis (PubMed:28005958). Probably plays an important role in centrosome cohesion during interphase. {ECO:0000269|PubMed:28005958}. | 3D-structure;Alternative splicing;Cell cycle;Cell projection;Cilium;Cilium biogenesis/degradation;Coiled coil;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Deafness;Disease variant;Phosphoprotein;Reference proteome | This gene encodes a core centrosomal protein required for centriole-centriole cohesion during interphase of the cell cycle. The encoded protein dissociates from the centrosomes when parental centrioles separate at the beginning of mitosis. The protein associates with and is phosphorylated by NIMA-related kinase 2, which is also associated with the centrosome. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Dec 2015]. | hsa:11190; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; microtubule organizing center [GO:0005815]; perinuclear region of cytoplasm [GO:0048471]; photoreceptor inner segment [GO:0001917]; photoreceptor outer segment [GO:0001750]; protein-containing complex [GO:0032991]; protein C-terminus binding [GO:0008022]; protein domain specific binding [GO:0019904]; protein kinase binding [GO:0019901]; centriole-centriole cohesion [GO:0010457]; cilium assembly [GO:0060271]; detection of light stimulus involved in visual perception [GO:0050908]; mitotic cell cycle [GO:0000278]; non-motile cilium assembly [GO:1905515]; positive regulation of protein localization to centrosome [GO:1904781]; protein localization [GO:0008104]; protein localization to organelle [GO:0033365]; regulation of centriole-centriole cohesion [GO:0030997] | 12140259_Data show that the dissociation of C-Nap1 from mitotic centrosomes is regulated by localized phosphorylation rather than generalized proteolysis. 18851962_CEP135 acts as a platform protein for C-NAP1 at the centriole. 20508983_Observational study of gene-disease association. (HuGE Navigator) 20546612_Observational study of gene-disease association. (HuGE Navigator) 23070519_C-NAP1 and rootletin restrain DNA damage-induced centriole splitting and facilitate ciliogenesis. 24554434_Centlein complexes with C-Nap1 and Cep68 at the proximal ends of centrioles during interphase. 24695856_multisite phosphorylation precipitates centrosome disjunction at the onset of mitosis 24780881_A homozygous nonsense CEP250 mutation, in combination with a heterozygous C2orf71 nonsense mutation, causes an atypical form of Usher syndrome, characterised by early-onset sensorineural hearing loss and a relatively mild retinitis pigmentosa. 25660448_ASPP1/2 interacted with centrosome linker protein C-Nap1. Co-depletion of ASPP1 and ASPP2 inhibited re-association of C-Nap1 with centrosome at the end of mitosis. 28100636_C-NAP1-null cells were viable and had an increased frequency of premature centriole separation, accompanied by reduced density of the centriolar satellites, with reexpression of C-NAP1 rescuing both phenotypes. Centrosome amplification induced by DNA damage or by PLK4 or CDK2 overexpression was markedly reduced in the absence of C-NAP1. 29718797_Our data indicate that mutations of CEP250 can cause mild cone-rod dystrophy (CRD) and early-onset sensorineural hearing loss (SNHL) in Japanese patients. Because the ophthalmological phenotypes were very mild, high-resolution retinal imaging analysis, such as AO, will be helpful in diagnosing CEP250-associated disease. 30459346_CEP250 mutation is associated with Usher syndrome. 30998843_Study identified a nonsense mutation (c.562C>T, p.R188*) in CEP250 in a consanguineous family with nonsyndromic retinitis pigmentosa (RP). The disruption of Cep250 resulted in severe impairment of retinal function and significant retinal morphological alterations a novel Cep250 knockin mouse line. 33109182_Involvement of NEK2 and its interaction with NDC80 and CEP250 in hepatocellular carcinoma. 34223797_Expanding the clinical phenotype in patients with disease causing variants associated with atypical Usher syndrome. | ENSMUSG00000038241 | Cep250 | 1510.49303 | 1.2240305 | 0.2916394788 | 0.11647512 | 6.316548e+00 | 1.196163e-02 | 1.796380e-01 | No | Yes | 2196.006489 | 301.435372 | 1793.315294 | 240.554436 | |
| ENSG00000126391 | 83786 | FRMD8 | protein_coding | Q9BZ67 | FUNCTION: Promotes the cell surface stability of iRhom1/RHBDF1 and iRhom2/RHBDF2 and prevents their degradation via the endolysosomal pathway. By acting on iRhoms, involved in ADAM17-mediated shedding of TNF, amphiregulin/AREG, HBEGF and TGFA from the cell surface (PubMed:29897333, PubMed:29897336). Negatively regulates Wnt signaling, possibly by antagonizing the recruitment of AXIN1 to LRP6 (PubMed:19572019). {ECO:0000269|PubMed:19572019, ECO:0000269|PubMed:29897333, ECO:0000269|PubMed:29897336}. | Acetylation;Alternative splicing;Cell membrane;Cytoplasm;Membrane;Phosphoprotein;Reference proteome | hsa:83786; | centriolar satellite [GO:0034451]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; positive regulation of tumor necrosis factor production [GO:0032760] | 29897333_iTAP (FRMD8) is a novel iRhom2 interactor that controls TNF-alpha secretion by policing the stability of iRhom2/ADAM17 complex. 29897336_FRMD8 promotes inflammatory and growth factor signaling by stabilizing the iRhom2/ADAM17 sheddase complex. | ENSMUSG00000024816 | Frmd8 | 805.50348 | 0.7758460 | -0.3661578466 | 0.14641106 | 6.173003e+00 | 1.297143e-02 | 1.847630e-01 | No | Yes | 815.670646 | 123.174096 | 1011.746759 | 149.026766 | ||
| ENSG00000126759 | 5199 | CFP | protein_coding | P27918 | FUNCTION: A positive regulator of the alternate pathway (AP) of complement (PubMed:20382442, PubMed:28264884). It binds to and stabilizes the C3- and C5-convertase enzyme complexes (PubMed:20382442, PubMed:28264884). Inhibits CFI-CFH mediated degradation of Complement C3 beta chain (C3b) (PubMed:31507604). {ECO:0000269|PubMed:20382442, ECO:0000269|PubMed:28264884, ECO:0000269|PubMed:31507604}. | 3D-structure;Complement alternate pathway;Disease variant;Disulfide bond;Glycoprotein;Immunity;Innate immunity;Reference proteome;Repeat;Secreted;Signal | This gene encodes a plasma glycoprotein that positively regulates the alternative complement pathway of the innate immune system. This protein binds to many microbial surfaces and apoptotic cells and stabilizes the C3- and C5-convertase enzyme complexes in a feedback loop that ultimately leads to formation of the membrane attack complex and lysis of the target cell. Mutations in this gene result in two forms of properdin deficiency, which results in high susceptibility to meningococcal infections. Multiple alternatively spliced variants, encoding the same protein, have been identified.[provided by RefSeq, Feb 2009]. | hsa:5199; | endoplasmic reticulum lumen [GO:0005788]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; plasma membrane [GO:0005886]; specific granule lumen [GO:0035580]; tertiary granule lumen [GO:1904724]; complement activation, alternative pathway [GO:0006957]; defense response to bacterium [GO:0042742]; immune response [GO:0006955] | 16337490_A splice site mutation in exon 10 (c.1487-2A>G) was found in the properdin gene and co segregated with biochemically measured properdin deficiency. 18490764_study reports properdin binds predominantly to late apoptotic & necrotic cells but not to early apoptotic cells; binding occurs independently of C3b 18579773_The human complement protein properdin binds to early apoptotic T cells and initiates complement activation, leading to C3b opsonization and ingestion by phagocytic cells. 18753294_The contribution of properdin is pivotal in proteinuria-induced tubular complement activation and subsequent damage. Interference with properdin binding to tubular cells may provide an option for the treatment of proteinuric renal disease. 18791942_Properdin induces the formation of platelet-leukocyte aggregates via leukocyte activation, linking the complement system & platelet-leukocyte aggregates with potential significance in atherosclerotic vascular disease. 19005416_Significantly more transcripts encoding alternative pathway components factor B, C3 and properdin, and C3a receptor and C5a receptor were detected in grade 3 versus grade 0 or 1 biopsies of human cardiac allografts. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19584655_Factor P was expressed in 50% of choroidal neovascular membranes of patients with age-related macular degeneration(AMD). Additional studies need to investigate role of Factor P in development of AMD for potential therapeutic intervention. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19934084_Properdin presence is associated with increased SC5b-9 excretion and worse renal function. 20122735_CFP does not seem to confer any risk for age-related macular degeneration. 20337960_levels of properdin are not associated with childhood wheezing and atopy 20382442_Human properdin can selectively recognize surfaces and enhance or promote alternative pathway of complement activation. 20530262_The conventional mechanism of properdin function is to bind to and stabilize alternative pathway C3 convertases on the surface of Neisseria meningitidis and N. gonorrhoeae. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21135110_tubular HS as a novel docking platform for alternative pathway activation via properdin, which might play a role in proteinuric renal damage. 22229731_report a large Finnish family with a novel mutation in the properdin gene. The mutation is located in exon 9 and changes guanine to adenine at nucleotide 1164 (c.1164G>A) that causes tryptophan to change to a premature stop codon (W388X). 22338105_Properdin and SC5b-9 may be novel biomarkers for future risk of type 2 diabetes in this high-risk population and warrant further investigation. 22368277_Immune human serum that contained bactericidal Abs directed against the 2C7 lipooligosaccharide epitope required functional properdin to kill C4BP-binding strains, but not C4BP-nonbinding strains. 22815489_Factor h and properdin recognize different epitopes on renal tubular epithelial heparan sulfate. 22851705_Properdin released from human polymorphonuclear cells does not bind to zymosan or E. coli, but when incubated in properdin-depleted serum this form of properdin binds efficiently to both substrates in a strictly complement C3-dependent manner. 23677468_Our data show that physiological forms of human properdin bind directly to human platelets after activation by strong agonists in the absence of C3 24355864_can directly interact with neutrophil myeloperoxidase resulting in activation of alternative pathway of complement 24885016_In the pathogenesis of renal tubular damage, P can directly bind to PTECs and may accelerate AP activation by surpassing fH regulation 26660535_P serum level expression could be a reliable clinical biomarker to identify patients with underlying surface alternative pathway C5 convertase dysregulation. 27183616_data indicate that properdin enhances platelet/granulocyte aggregates (PGAs) formation via increased production of C5a, and that inhibition of properdin function has therapeutic potential to limit thromboinflammation in diseases characterized by increased PGA formation 28069958_In conclusion, we challenge the view of properdin as a pattern recognition molecule by providing evidence that it binds to different exogenous and endogenous molecular patterns in only a C3-dependent manner. 28105653_this study shows that RNA interference of properdin in dendritic cells decreased alloantigen-specific T-cell proliferation 28264884_studies demonstrate an essential role of properdin oligomerization in vivo while our monomers enable detailed structural insight paving the way for novel modulators of complement. 28784323_Study identified 2 single nucleotide polymorphisms, 1 in the mannose-binding lectin 2 gene (p.Gly54Asp-MBL2) and 1 in the complement factor properdin gene (p.Asn428(p=)-CFP), that showed significant association with the absence and development of antibody-mediated cardiac allograft rejection, respectively. 30199474_Compound heterozygous mutations in IL10RA combined with a complement factor properdin mutation in infantile-onset inflammatory bowel disease. 30755730_TES-mediated upregulation of the transcription factor DDIT3 is involved in CFP suppression of breast cancer cell growth 32793201_Properdin Is a Key Player in Lysis of Red Blood Cells and Complement Activation on Endothelial Cells in Hemolytic Anemias Caused by Complement Dysregulation. 32909942_Soluble collectin-12 mediates C3-independent docking of properdin that activates the alternative pathway of complement. 33240263_The Role of Properdin in Killing of Non-Pathogenic Leptospira biflexa. 33494138_Properdin Is a Modulator of Tumour Immunity in a Syngeneic Mouse Melanoma Model. 35031611_Structure and function of a family of tick-derived complement inhibitors targeting properdin. | ENSMUSG00000001128 | Cfp | 18.15175 | 0.5322787 | -0.9097461712 | 0.66931998 | 1.808671e+00 | 1.786677e-01 | No | Yes | 18.430140 | 7.026546 | 34.254284 | 12.092417 | ||
| ENSG00000127334 | 8445 | DYRK2 | protein_coding | Q92630 | FUNCTION: Serine/threonine-protein kinase involved in the regulation of the mitotic cell cycle, cell proliferation, apoptosis, organization of the cytoskeleton and neurite outgrowth. Functions in part via its role in ubiquitin-dependent proteasomal protein degradation. Functions downstream of ATM and phosphorylates p53/TP53 at 'Ser-46', and thereby contributes to the induction of apoptosis in response to DNA damage. Phosphorylates NFATC1, and thereby inhibits its accumulation in the nucleus and its transcription factor activity. Phosphorylates EIF2B5 at 'Ser-544', enabling its subsequent phosphorylation and inhibition by GSK3B. Likewise, phosphorylation of NFATC1, CRMP2/DPYSL2 and CRMP4/DPYSL3 promotes their subsequent phosphorylation by GSK3B. May play a general role in the priming of GSK3 substrates. Inactivates GYS1 by phosphorylation at 'Ser-641', and potentially also a second phosphorylation site, thus regulating glycogen synthesis. Mediates EDVP E3 ligase complex formation and is required for the phosphorylation and subsequent degradation of KATNA1. Phosphorylates TERT at 'Ser-457', promoting TERT ubiquitination by the EDVP complex. Phosphorylates SIAH2, and thereby increases its ubiquitin ligase activity. Promotes the proteasomal degradation of MYC and JUN, and thereby regulates progress through the mitotic cell cycle and cell proliferation. Promotes proteasomal degradation of GLI2 and GLI3, and thereby plays a role in smoothened and sonic hedgehog signaling. Plays a role in cytoskeleton organization and neurite outgrowth via its phosphorylation of DCX and DPYSL2. Phosphorylates CRMP2/DPYSL2, CRMP4/DPYSL3, DCX, EIF2B5, EIF4EBP1, GLI2, GLI3, GYS1, JUN, MDM2, MYC, NFATC1, p53/TP53, TAU/MAPT and KATNA1. Can phosphorylate histone H1, histone H3 and histone H2B (in vitro). Can phosphorylate CARHSP1 (in vitro). {ECO:0000269|PubMed:11311121, ECO:0000269|PubMed:12588975, ECO:0000269|PubMed:14593110, ECO:0000269|PubMed:15910284, ECO:0000269|PubMed:16511445, ECO:0000269|PubMed:16611631, ECO:0000269|PubMed:17349958, ECO:0000269|PubMed:18455992, ECO:0000269|PubMed:18599021, ECO:0000269|PubMed:19287380, ECO:0000269|PubMed:22307329, ECO:0000269|PubMed:22878263, ECO:0000269|PubMed:23362280, ECO:0000269|PubMed:9748265}. | 3D-structure;ATP-binding;Alternative splicing;Apoptosis;Cytoplasm;Kinase;Magnesium;Manganese;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase;Tyrosine-protein kinase;Ubl conjugation;Ubl conjugation pathway | DYRK2 belongs to a family of protein kinases whose members are presumed to be involved in cellular growth and/or development. The family is defined by structural similarity of their kinase domains and their capability to autophosphorylate on tyrosine residues. DYRK2 has demonstrated tyrosine autophosphorylation and catalyzed phosphorylation of histones H3 and H2B in vitro. Two isoforms of DYRK2 have been isolated. The predominant isoform, isoform 1, lacks a 5' terminal insert. [provided by RefSeq, Jul 2008]. | hsa:8445; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; ubiquitin ligase complex [GO:0000151]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; manganese ion binding [GO:0030145]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; cellular response to DNA damage stimulus [GO:0006974]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; negative regulation of calcineurin-NFAT signaling cascade [GO:0070885]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of glycogen biosynthetic process [GO:0045725]; protein phosphorylation [GO:0006468]; regulation of signal transduction by p53 class mediator [GO:1901796]; smoothened signaling pathway [GO:0007224] | 17349958_These findings indicate that DYRK2 regulates p53 to induce apoptosis in response to DNA damage. 19287380_A protein kinase, DYRK2, has unexpected role as a scaffold for an E3 ubiquitin ligase complex. 19596956_disease control rate of the DYRK2-positive non-small cell lung cancer patients was significantly different from the DYRK2-negative group 19818968_The data showed that DYRK2 expression is associated with a favorable prognosis in pulmonary adenocarcinoma. 19965871_The findings indicate that ATM controls stability and pro-apoptotic function of DYRK2 in response to DNA damage. 22307329_DYRK2 regulates tumor progression through modulation of c-Jun and c-Myc 22878263_DYRK2-mediated phosphorylation of p53 at Ser46 is impaired under hypoxic conditions, suggesting a molecular mechanism underlying chemotherapy resistance in solid tumors. 23362280_Data indicate that Dyrk2 phosphorylates TERT protein, which is then associated with the EDD-DDB1-VprBP E3 ligase complex for subsequent ubiquitin-mediated TERT protein degradation. 23791882_DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. 24438055_DYRK2-dependent phosphorylation of pregnane X receptor facilitates its subsequent ubiquitination by UBR5. 25095982_Downregulation of DYRK2 is associated with recurrence in early stage breast cancer. 25712377_DYRK2 may regulate EMT through Snail degradation in ovarian SA and might be a predictive marker for a favorable prognosis in the treatment of this cancer. 26341817_Silencing of DYRK2 increases cell proliferation but reverses CAM-DR in Non-Hodgkin's Lymphoma 26804244_the downregulated expression of DYRK2 in HCC tumor tissues could promote the proliferation of hepatocellular carcinoma (HCC) cells. In addition, reducing DYRK2 expression was associated with poor prognosis and Oxaliplatin resistance in HCC. 27532268_lower DYRK2 levels were correlated with tumor sites, advanced clinical stages, and shorter survival in the advanced clinical stages. DYRK2 is a novel prognostic biomarker of human colorectal cancer. 27721402_our results delineate a novel mechanism of cancer stem cell regulation by the DYRK2-AR-KLF4 axis. Restoration of the expression and function of DYRK2 is a potential therapeutic strategy against breast cancer stem cells. 27746162_Diminished DYRK2 expression sensitizes hormone receptor-positive breast cancer to everolimus by preventing mTOR degradation. 28194753_Findings indicate direct interaction of dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) with ring finger protein (C3HC4 type) 8 (RNF8) in regulating response to DNA damage. 28502078_Study found that DYRK2 regulates liver metastases of colorectal cancer cells through the activation of EMT, and that patients with low DYRK2-expressing colorectal cancer liver metastases had worse outcomes than those with high DYRK2-expressing metastases. 28677030_The expression level of DYRK2 was positively correlated with glioma pathological grade. 29193658_Results provide evidence that DYRK2 suppresses the proliferation of breast cancer cells and invasion through CDK14 expression. 30851422_found that liver cancer patients with low DYRK2 expression had a significantly shorter overall survival 31209060_DYRK2 is strongly activated in normal or early-stage tumor cells, where they effectively degrade Snail through betaTrCP-mediated ubiquitination, thus suppressing epithelial-mesenchymal transition. 31505048_Especially in tumor cells, accumulating studies have revealed the molecular mechanisms of DYRK2 from the aspect of apoptosis, proliferation, EMT, and stem- ness. Considering in vitro and xenograft studies as well as down-regulation of DYRK2 in tumor specimens, DYRK2 is an important candidate for tumor suppressor. [review] 31815665_DYRK2 phosphorylates NDEL1 S336 to prime the phosphorylation of NDEL1 S332 by GSK3beta. The data suggest the NDEL1 phosphorylation at S336/S332 by the TARA-DYRK2-GSK3beta complex as a novel regulatory mechanism underlying neuronal morphogenesis. 32236621_Impairment of DYRK2 by DNMT1mediated transcription augments carcinogenesis in human colorectal cancer. 33368138_Frequent DYRK2 gene amplification in micropapillary element of lung adenocarcinoma - an implication in progression in EGFR-mutated lung adenocarcinoma. 33376136_Emerging roles of DYRK2 in cancer. 35347031_Combination of DYRK2 and TERT Expression Is a Powerful Predictive Marker for Early-stage Breast Cancer Recurrence. | ENSMUSG00000028630 | Dyrk2 | 1747.13388 | 1.0944729 | 0.1302362281 | 0.09660978 | 1.822652e+00 | 1.769979e-01 | 5.720388e-01 | No | Yes | 2331.582849 | 348.509851 | 1987.684697 | 290.687070 | |
| ENSG00000127527 | 58513 | EPS15L1 | protein_coding | Q9UBC2 | FUNCTION: Seems to be a constitutive component of clathrin-coated pits that is required for receptor-mediated endocytosis. Involved in endocytosis of integrin beta-1 (ITGB1) and transferrin receptor (TFR); internalization of ITGB1 as DAB2-dependent cargo but not TFR seems to require association with DAB2. {ECO:0000269|PubMed:22648170, ECO:0000269|PubMed:9407958}. | Acetylation;Alternative splicing;Calcium;Cell membrane;Coated pit;Coiled coil;Endocytosis;Membrane;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat | hsa:58513; | clathrin coat of coated pit [GO:0030132]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; cadherin binding [GO:0045296]; calcium ion binding [GO:0005509]; endocytosis [GO:0006897]; endosomal transport [GO:0016197] | 29023680_This is the first biallelic variant identified in the EPS15L1 gene underlying SHFM. Our findings report the first direct involvement of EPS15L1 gene in the development of human limbs. | ENSMUSG00000006276 | Eps15l1 | 1393.24670 | 0.9642760 | -0.0524819712 | 0.09888905 | 2.813075e-01 | 5.958455e-01 | 8.655869e-01 | No | Yes | 1666.833103 | 167.701260 | 1654.843892 | 162.683131 | ||
| ENSG00000127980 | 5189 | PEX1 | protein_coding | O43933 | FUNCTION: Required for stability of PEX5 and protein import into the peroxisome matrix. Anchored by PEX26 to peroxisome membranes, possibly to form heteromeric AAA ATPase complexes required for the import of proteins into peroxisomes. | ATP-binding;Alternative splicing;Amelogenesis imperfecta;Cytoplasm;Deafness;Disease variant;Membrane;Nucleotide-binding;Peroxisome;Peroxisome biogenesis;Peroxisome biogenesis disorder;Phosphoprotein;Protein transport;Reference proteome;Repeat;Transport;Zellweger syndrome | This gene encodes a member of the AAA ATPase family, a large group of ATPases associated with diverse cellular activities. This protein is cytoplasmic but is often anchored to a peroxisomal membrane where it forms a heteromeric complex and plays a role in the import of proteins into peroxisomes and peroxisome biogenesis. Mutations in this gene have been associated with complementation group 1 peroxisomal disorders such as neonatal adrenoleukodystrophy, infantile Refsum disease, and Zellweger syndrome. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Sep 2013]. | hsa:5189; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; peroxisomal membrane [GO:0005778]; peroxisome [GO:0005777]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; protein C-terminus binding [GO:0008022]; protein-containing complex binding [GO:0044877]; microtubule-based peroxisome localization [GO:0060152]; peroxisome organization [GO:0007031]; protein import into peroxisome matrix [GO:0016558]; protein targeting to peroxisome [GO:0006625] | 12032265_Missense mutations in PEX1 cause the milder forms of the peroxisome biogenesis disorders, whereas insertions, deletions, and nonsense mutations are associated with severe clinical phenotypes. 12402331_We have evaluated the impact of novel mutations, along with that of the two most common PEX1 mutations, in PBD patients by determining the levels of PEX1 mRNA, PEX1 protein, and peroxisome protein import. 12840548_complete lack of PEX1 is associated with Zellweger syndrome 16086329_overview of the currently known PEX1 mutations in Zellweger Syndrome [review] 16088892_analysis of PEX1 coding mutations and 5' UTR regulatory polymorphisms 16141001_Molecular confirmation of the clinical and biochemical diagnosis will allow the prediction of the clinical course of disease in individual PBD cases. 16257970_Insufficient binding to Pex1p x Pex6p complexes, or mislocalization of patient-derived Pex26p mutants is most likely responsible for the complementation group impaired peroxisome biogenesis. 19105186_Studies provide empirical data to estimate the relative fraction of disease-causing alleles that occur in the coding and splice junction sequences of PEX1 gene. 20546612_Observational study of gene-disease association. (HuGE Navigator) 21846392_A 5' UTR polymorphism at position c.-53 and a promoter polymorphism 137 bp upstream of the PEX1 start codon are identified but strongly differing survival By genotype-phenotype analysis. 23247051_the variants in PEX genes of a family 25016021_results suggested that peroxisome biogenesis requires Pex1p- and Pex6p-regulated dissociation of Pex14p from Pex26p 26387595_Mutations in PEX1 gene is associated with Heimler Syndrome. 26476099_Our structural data suggest that the tilting of a central segment of a Pex1-Pex6 pair is responsible for polypeptide movement. 27302843_A combination of a known missense and novel frameshift variant in PEX1 identified in a family with Heimler syndrome. 27633571_As standard biochemical screening of blood for evidence of a peroxisomal disorder did not provide a diagnosis in the individuals with Heimler syndrome, patients with sensorineural hearing lossand retinal pigmentation should have mutation analysis of PEX1 and PEX6 genes. 28432012_heterozygous mutations in the PEX1 gene in two Chinese newborns with Zellweger syndrome 28508493_Major finding is linking peroxisome biogenesis factor 1 (PEX1) to obesity phenotypes, a novel mechanism of peroxisomal biogenesis and metabolism underlying the development of childhood obesity. 29884772_This study provides evidence suggesting that monoubiquitinated PEX5 interacts directly with both PEX1 and PEX6 through its ubiquitin moiety and that the PEX5 polypeptide chain is globally unfolded during the ATP-dependent extraction event. 31374812_This article reviews the abundant records of missense mutations described in Peroxisome biogenesis disorders patients with the aim to classify and rationalize them by mapping them onto a homology model of the human Pex1/Pex6 complex. [review] 31831025_There are no significant differences between PEX1-, PEX6-, and PEX26-associated phenotypes inclinical and genetic spectrum of Heimler syndrome. 32866347_The peroxisomal disorder spectrum and Heimler syndrome: Deep phenotyping and review of the literature. 33545634_Depletion of HNRNPA1 induces peroxisomal autophagy by regulating PEX1 expression. 33955814_Two siblings with Heimler syndrome caused by PEX1 variants: follow-up of ophthalmologic findings. | ENSMUSG00000005907 | Pex1 | 1608.63964 | 0.9783669 | -0.0315525061 | 0.10101551 | 9.750606e-02 | 7.548432e-01 | 9.282507e-01 | No | Yes | 1787.338079 | 300.604881 | 1793.980872 | 295.215190 | |
| ENSG00000128335 | 23780 | APOL2 | protein_coding | Q9BQE5 | FUNCTION: May affect the movement of lipids in the cytoplasm or allow the binding of lipids to organelles. | Acetylation;Cytoplasm;Direct protein sequencing;Lipid transport;Phosphoprotein;Reference proteome;Transport | Mouse_homologues NA; + ;NA; + ;NA; + ;NA; + ;NA; + ;NA; + ;NA; + ;NA; + ;NA; + ;NA | This gene is a member of the apolipoprotein L gene family. The encoded protein is found in the cytoplasm, where it may affect the movement of lipids or allow the binding of lipids to organelles. Two transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:23780; | endoplasmic reticulum membrane [GO:0005789]; extracellular region [GO:0005576]; membrane [GO:0016020]; high-density lipoprotein particle binding [GO:0008035]; lipid binding [GO:0008289]; signaling receptor binding [GO:0005102]; acute-phase response [GO:0006953]; cholesterol metabolic process [GO:0008203]; lipid metabolic process [GO:0006629]; lipid transport [GO:0006869]; lipoprotein metabolic process [GO:0042157]; maternal process involved in female pregnancy [GO:0060135] | 11944986_APOL2 has been found only in humans and African green monkeys 15949655_Observational study of gene-disease association. (HuGE Navigator) 18632255_An association of APOL1, 2 and 4 with schizophrenia was establised. 18632255_Observational study of gene-disease association. (HuGE Navigator) 20602615_Observational study of gene-disease association. (HuGE Navigator) 20665705_Results reveal a novel function for ApoL2 in conferring anti-apoptotic ability of human bronchial epithelium to the cytotoxic effects of IFN-gamma, in maintaining airway epithelial layer integrity. 24901046_Apolipoprotein L2 contains a BH3-like domain but it does not behave as a BH3-only protein. 31466081_Epigenetic aging is accelerated in alcohol use disorder and regulated by genetic variation in APOL2. 34316015_Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif. | ENSMUSG00000068252+ENSMUSG00000050014+ENSMUSG00000050982+ENSMUSG00000091694+ENSMUSG00000010601+ENSMUSG00000091650+ENSMUSG00000068246+ENSMUSG00000071716+ENSMUSG00000044309+ENSMUSG00000057346 | Apol7b+Apol10b+Apol10a+Apol11b+Apol7a+Apol11a+Apol9b+Apol7e+Apol7c+Apol9a | 178.01918 | 0.9474154 | -0.0779309417 | 0.22127178 | 1.239149e-01 | 7.248266e-01 | 9.198119e-01 | No | Yes | 217.914645 | 19.213433 | 218.784685 | 18.604190 |
| ENSG00000128512 | 9732 | DOCK4 | protein_coding | Q8N1I0 | FUNCTION: Functions as a guanine nucleotide exchange factor (GEF) that promotes the exchange of GDP to GTP, converting inactive GDP-bound small GTPases into their active GTP-bound form (PubMed:12628187, PubMed:16464467). Involved in regulation of adherens junction between cells (PubMed:12628187). Plays a role in cell migration (PubMed:20679435). {ECO:0000269|PubMed:12628187, ECO:0000269|PubMed:16464467, ECO:0000269|PubMed:20679435}.; FUNCTION: [Isoform 2]: Has a higher guanine nucleotide exchange factor activity compared to other isoforms. {ECO:0000269|PubMed:16464467}. | Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Proto-oncogene;Reference proteome;SH3 domain;SH3-binding | This gene is a member of the dedicator of cytokinesis (DOCK) family and encodes a protein with a DHR-1 (CZH-1) domain, a DHR-2 (CZH-2) domain and an SH3 domain. This membrane-associated, cytoplasmic protein functions as a guanine nucleotide exchange factor and is involved in regulation of adherens junctions between cells. Mutations in this gene have been associated with ovarian, prostate, glioma, and colorectal cancers. Alternatively spliced variants which encode different protein isoforms have been described, but only one has been fully characterized. [provided by RefSeq, Jul 2008]. | hsa:9732; | cytosol [GO:0005829]; Golgi apparatus [GO:0005794]; membrane [GO:0016020]; nucleolus [GO:0005730]; plasma membrane [GO:0005886]; stereocilium [GO:0032420]; stereocilium bundle [GO:0032421]; guanyl-nucleotide exchange factor activity [GO:0005085]; PDZ domain binding [GO:0030165]; receptor tyrosine kinase binding [GO:0030971]; SH3 domain binding [GO:0017124]; small GTPase binding [GO:0031267]; cell chemotaxis [GO:0060326]; negative regulation of vascular associated smooth muscle contraction [GO:1904694]; positive regulation of vascular associated smooth muscle cell migration [GO:1904754]; small GTPase mediated signal transduction [GO:0007264] | 17027967_Taken together, these results suggest that Dock4 plays an important role in the regulation of cell migration through activation of Rac1, and that RhoG is a key upstream regulator for Dock4. 18459162_DOCK4 may be regulated by PIP(3) to exert its function 18641688_DOCK4 interacts with the beta-catenin degradation complex, consisting of the proteins adenomatosis polyposis coli, Axin and glycogen synthase kinase 3beta. 19383911_Observational study of gene-disease association. (HuGE Navigator) 19401682_Evidence for the involvement of DOCK4 in autism susceptibility was supported by independent replication of association at rs2217262 and the finding of a deletion segregating in a sib-pair family 19401682_Observational study of gene-disease association. (HuGE Navigator) 19528233_Cell migration is regulated by platelet-derived growth factor receptor endocytosis, which involves DOCK4. 20346443_Data suggest that exonic deletions of DOCK4 may act as a risk factor for reading impairment. Genomic disruption of both DOCK4 and CNTNAP5 genes may have an additive effect and may result in a more severe autism spectrum phenotype. 20346443_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21532034_novel epigenetic alterations in myelodysplastic leukocytes and implicate DOCK4 as a pathogenic gene located on the 7q chromosomal region. 21682944_This study revealed ROCK4 as novel schizophrenia candidate genes in the Jewish population. 23720743_The Atypical guanine nucleotide exchange factor Dock4 regulates neurite differentiation through modulation of Rac1 GTPase and actin dynamics. 24599690_The study found significant associations between autism and a SNP of the DOCK4 gene. 25644601_activator DOCK4 as a key component of the TGF-beta/Smad pathway that promotes lung ADC cell extravasation and metastasis. 26129894_DOCK4 signalling is necessary for lateral filopodial protrusions and tubule remodelling prior to vascular lumen formation 26578796_These data identify DOCK4 as a putative 7q gene whose reduced expression can lead to erythroid dysplasia 28925399_DOCK4 over expression suppresses selfrenewal and tumorigenicity of glioblastoma (GBM) stem-like cells. Accordingly in the frame of GBM median of survival, increased level of DOCK4 predicts improved patient survival. 31019307_SR-B1 drives endothelial cell LDL transcytosis via DOCK4 to promote atherosclerosis 32576693_Up-regulated cytotrophoblast DOCK4 contributes to over-invasion in placenta accreta spectrum. 33559155_DOCK4 stimulates MUC2 production through its effect on goblet cell differentiation. | ENSMUSG00000035954 | Dock4 | 397.69721 | 1.1836177 | 0.2432032189 | 0.19275682 | 1.596264e+00 | 2.064334e-01 | 5.987975e-01 | No | Yes | 475.341337 | 107.404742 | 385.920058 | 85.379103 | |
| ENSG00000129292 | 51105 | PHF20L1 | protein_coding | A8MW92 | 3D-structure;Acetylation;Alternative splicing;Isopeptide bond;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Ubl conjugation;Zinc;Zinc-finger | hsa:51105; | NSL complex [GO:0044545]; metal ion binding [GO:0046872]; histone acetylation [GO:0016573]; regulation of transcription by RNA polymerase II [GO:0006357] | 24492612_Malignant brain tumor domain of PHF20L1 reads and controls enzyme levels of methylated DNMT1 in cells, thus representing a novel antagonist of DNMT1 degradation. 26588862_results demonstrated the oncogenic potential of PHF20L1 and its association with poor prognostic parameters in breast cancer. 28841214_Authors find that the interplay between PHF20L1 and mono-methyl pRb is important for maintaining the integrity of a pRb-dependent G1-S-phase checkpoint. Results highlight the distinct roles that methyl-lysine readers have in regulating the biological activity of pRb. 29358331_Data suggest that, in human ovarian teratocarcinoma cell line, methylation-dependent proteolysis of SOX2 is controlled by LSD1 and PHF20L1 via regulation of SET7 activity; these data are consistent with previous studies in mouse primary stem cells. (SOX2 = SRY-box 2 transcription factor; LSD1 = lysine demethylase-1A; PHF20L1 = PHD finger protein 20 like-1; SET7 = histone-lysine N-methyltransferase SETD7) 30089852_associates with SOX2, antagonizes SOX2 ubiquitination and the sequential degradation induced by the MLL1/WDR5 complexes 32494608_PHF20L1 as a H3K27me2 reader coordinates with transcriptional repressors to promote breast tumorigenesis. 33847474_An insight into the promoter methylation of PHF20L1 and the gene association with metastasis in breast cancer. | ENSMUSG00000072501 | Phf20l1 | 2489.18468 | 1.0556745 | 0.0781651266 | 0.10044201 | 6.068647e-01 | 4.359707e-01 | 7.817284e-01 | No | Yes | 3208.552863 | 623.117099 | 2920.602111 | 554.842735 | |||
| ENSG00000129422 | 57509 | MTUS1 | protein_coding | Q9ULD2 | FUNCTION: Cooperates with AGTR2 to inhibit ERK2 activation and cell proliferation. May be required for AGTR2 cell surface expression. Together with PTPN6, induces UBE2V2 expression upon angiotensin-II stimulation. Isoform 1 inhibits breast cancer cell proliferation, delays the progression of mitosis by prolonging metaphase and reduces tumor growth. {ECO:0000269|PubMed:12692079, ECO:0000269|PubMed:19794912}. | Alternative splicing;Cell membrane;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Golgi apparatus;Membrane;Microtubule;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Tumor suppressor | This gene encodes a protein which contains a C-terminal domain able to interact with the angiotension II (AT2) receptor and a large coiled-coil region allowing dimerization. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. One of the transcript variants has been shown to encode a mitochondrial protein that acts as a tumor suppressor and partcipates in AT2 signaling pathways. Other variants may encode nuclear or transmembrane proteins but it has not been determined whether they also participate in AT2 signaling pathways. [provided by RefSeq, Jul 2008]. | hsa:57509; | extracellular space [GO:0005615]; Golgi apparatus [GO:0005794]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; microtubule organizing center [GO:0005815]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; spindle [GO:0005819]; microtubule binding [GO:0008017]; regulation of macrophage chemotaxis [GO:0010758] | 12692079_identified a tumor suppressor gene named MTSG1 at chromosome 8p21.3-22, encoding a mitochondrial protein, controlling cellular proliferation 15123706_Results identify ATIP1 (angiotensin II AT2 receptor-interacting protein) as a novel early component of growth inhibitory signaling cascade 16650523_Nucleotide variations result in amino-acid substitution or deletion of conserved structural motifs and also exonic splicing enhancer motifs and physiological splice sites, suggesting deleterious effects on ATIP function and/or expression. [REVIEW] 16887298_MTUS1 encodes a family of proteins(ATIP1, ATIP3 and ATIP4), with potential important biological roles in tumor suppression and/or brain function. 17301065_Observational study of gene-disease association. (HuGE Navigator) 17301065_There is an association of the deletion variant of MTUS1 with a decreased risk for both familial and high-risk familial BC supporting its role in human cancer. 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19344625_two transcriptional start sites in the ATIP1 promoter were identified; PARP-1 activates the ATIP1 gene 19794912_ATIP3 is a novel microtubule-associated protein isoform of MTUS1, with a role in invasiveness and progression of breast neoplasms. 19794912_ATIP3 is a novel microtubule-associated protein related to MTUS1, with a role in invasiveness and progression of breast neoplasms 19956880_MTUS1 could be involved in the loss of proliferative control in human colon cancer 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20687230_Results in human prostate cancer cell lines demonstrate the presence of ATIP in both cell lines examined. 22153618_MTUS1 plays major roles in the progression of oral tongue squamous cell carcinoma. 23396587_analysis of a functional ATIP3 domain that associates with microtubules and recapitulates the effects of ATIP3 on microtubule dynamics, cell proliferation, and migration in breast cancer 24299308_The present data suggested MTUS1 as a potential tumor suppressor in gastric cancer and might lead to a better understanding of gastric carcinogenesis. 24650297_Data identifies MTUS1 as a tumour suppressor gene in cultured bladder cancer cells and in advanced bladder tumours. 25885343_Our studies confirm that MTUS1 plays an important role in the progression of salivary adenoid cystic carcinoma , and may serve as a biomarker or therapeutic target for patients with salivary adenoid cystic carcinoma 26498358_We propose a novel mechanism by which ATIP3-EB1 interaction indirectly reduces the kinetics of EB1 exchange on its recognition site, thereby accounting for negative regulation of microtubule dynamic instability 26643896_Deregulated microRNA (miRNA) expression has been shown to be involved in the pathogenesis of several types of cancers including colorectal cancer (CRC). MTUS1 targeting miRNAs may play key roles in the development of CRC by downregulating tumor suppressor MTUS1. 26643896_MTUS1 targeting miRNAs may play key roles in the development of CRC by downregulating tumor suppressor MTUS1. 27155522_Study reports differential expression of MTUS1 and its regulatory miRNAs in breast cancer and fibroadenoma tissues; among MTUS1 targeting miRNAs, miR-183-5p was overexpressed in breast cancer and down-regulated in fibroadenoma tissues; expression levels of MTUS1 and miR-183-5p correlated with clinical parameters. In particular, MTUS1 was found to be diminished and miR-183-5p was elevated with advancing stage. 27789289_Our findings showed that MTUS1 regulates the p38 MAPK-mediated cytokine production in ECs. MTUS1 gene probably plays a protective role against pro-inflammatory response of ECs. 28364280_findings have provided the first clues regarding the roles of miR-19a/b, which appear to function as oncomirs in lung cancer by downregulating MTUS1. 28499941_MTUS1 is not only involved in the formation and progression of human cancers but also involved in the complex pathological conditions such as cardiac hypertrophy, atherosclerosis, and SLE-like lymphoproliferative diseases. Several molecular mechanisms such as proliferation, differentiation, DNA repair, inflammation, vascular remodeling and senescence appear to be tightly regulated by the MTUS1-encoded proteins. 28651497_angiotensin II type 2 receptor-interacting protein 3a presents potential in suppressing the proliferation and aggressiveness of ovarian carcinoma cells through the high mobility group AT-hook 2-mediated extracellular signal-regulated kinase/epithelial-to-mesenchymal transition signal pathway. 29558204_The N-terminal coiled-coil domain of MTUS1 interacted with the mitochondrial membrane proteins. 30368744_studies emphasize the importance of studying combinatorial expression of EB1 and ATIP3 genes and proteins rather than each biomarker alone. A population of highly aggressive breast tumors expressing high-EB1/low-ATIP3 may be considered for the development of new molecular therapies. 30528566_Low expression of MTUS1 correlates to DNA methylation and histone deacetylation in human NSCLC. 31210288_MicroRNA-765 targets MTUS1 to promote the progression of osteosarcoma via mediating ERK/EMT pathway. 31685623_ATIP3 is an important regulator of mitotic integrity. 31882471_MTUS1 may act as tumor suppressor and might be a potential biomarker for predicting prognosis in GBC. 32373949_Reduced long non-coding RNA PTENP1 contributed to proliferation and invasion via miR-19b/MTUS1 axis in patients with cervical cancer. 32789689_Reciprocal regulation of Aurora kinase A and ATIP3 in the control of metaphase spindle length. 33311452_RNA-binding protein SORBS2 suppresses clear cell renal cell carcinoma metastasis by enhancing MTUS1 mRNA stability. 34062782_Microtubule-Associated Protein ATIP3, an Emerging Target for Personalized Medicine in Breast Cancer. 34125870_In silico analysis of deleterious SNPs of human MTUS1 gene and their impacts on subsequent protein structure and function. | ENSMUSG00000045636 | Mtus1 | 535.41129 | 1.1927094 | 0.2542426047 | 0.17319184 | 2.163511e+00 | 1.413215e-01 | 5.255714e-01 | No | Yes | 646.724706 | 118.863894 | 511.298566 | 92.070849 | |
| ENSG00000129951 | 79948 | PLPPR3 | protein_coding | Q6T4P5 | Alternative splicing;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | The proteins in the lipid phosphate phosphatase (LPP) family, including PRG2, are integral membrane proteins that modulate bioactive lipid phosphates including phosphatidate, lysophosphatidate, and sphingosine-1-phosphate in the context of cell migration, neurite retraction, and mitogenesis (Brauer et al., 2003 [PubMed 12730698]).[supplied by OMIM, Mar 2008]. | hsa:79948; | integral component of plasma membrane [GO:0005887]; lipid phosphatase activity [GO:0042577]; phosphatidate phosphatase activity [GO:0008195]; phospholipid dephosphorylation [GO:0046839]; phospholipid metabolic process [GO:0006644]; signal transduction [GO:0007165] | Mouse_homologues 27744421_PRG3 emerges as a developmental RasGRF1-dependent conductor of filopodia formation and axonal growth enhancer. PRG3-induced neurites resist brain injury-associated outgrowth inhibitors and contribute to functional recovery after spinal cord lesions. | ENSMUSG00000035835 | Plppr3 | 68.68752 | 0.4734823 | -1.0786176707 | 0.39108266 | 7.348236e+00 | 6.712858e-03 | No | Yes | 54.916064 | 13.068334 | 114.634776 | 26.197764 | |||
| ENSG00000130517 | 54858 | PGPEP1 | protein_coding | Q9NXJ5 | FUNCTION: Removes 5-oxoproline from various penultimate amino acid residues except L-proline. {ECO:0000269|PubMed:12651114}. | Alternative splicing;Cytoplasm;Hydrolase;Protease;Reference proteome;Thiol protease | The gene encodes a cysteine protease and member of the peptidase C15 family of proteins. The encoded protein cleaves amino terminal pyroglutamate residues from protein substrates including thyrotropin-releasing hormone and other neuropeptides. Expression of this gene may be downregulated in colorectal cancer, while activity of the encoded protein may be negatively correlated with cancer progression in colorectal cancer patients. Activity of the encoded protease may also be altered in other disease states including in liver cirrhosis, which is associated with reduced protease activity, and in necrozoospermia, which is associated with elevated protease activity. [provided by RefSeq, Jul 2016]. | hsa:54858; | cytosol [GO:0005829]; pyroglutamyl-peptidase activity [GO:0016920]; proteolysis [GO:0006508] | 12651114_cloning and sequencing of the human (AJ278828) pyroglutamyl-peptidase I 15014037_VX-710 modulates Pgp, MRP-1, and BCRP(R482), and has potential as a clinical broad-spectrum multidrug resistance modulator in malignancies. 15380924_The activities of pyroglutamyl peptidase I and prolyl endopeptidase in necrozoospermia were found to be higher in the corresponding soluble and particulate sperm fractions, respectively, with respect to those measured in normozoospermic semen 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 25687677_The specific reduction of the levels of pyroglutamyl-aminopeptidase activity in serum of liver cirrhosis patients might be considered as a potential candidate to be included in a combination of markers for the diagnosis of this disease. 26078706_cytosolic peptidases(puromycin-sensitive aminopeptidase (PSA), aminopeptidase B (APB) and pyroglutamyl-peptidase I (PGI) ) may be involved in colorectal carcinogenesis 34816269_Two-photon fluorogenic probe for visualizing PGP-1 activity in inflammatory tissues and serum from patients. | ENSMUSG00000056204 | Pgpep1 | 213.08322 | 0.7104996 | -0.4930941856 | 0.24417030 | 4.004290e+00 | 4.538460e-02 | 3.224823e-01 | No | Yes | 218.388872 | 32.958367 | 303.414563 | 43.982289 | |
| ENSG00000130529 | 54795 | TRPM4 | protein_coding | Q8TD43 | FUNCTION: Calcium-activated non selective (CAN) cation channel that mediates membrane depolarization (PubMed:12015988, PubMed:29211723, PubMed:30528822). While it is activated by increase in intracellular Ca(2+), it is impermeable to it (PubMed:12015988). Mediates transport of monovalent cations (Na(+) > K(+) > Cs(+) > Li(+)), leading to depolarize the membrane. It thereby plays a central role in cadiomyocytes, neurons from entorhinal cortex, dorsal root and vomeronasal neurons, endocrine pancreas cells, kidney epithelial cells, cochlea hair cells etc. Participates in T-cell activation by modulating Ca(2+) oscillations after T lymphocyte activation, which is required for NFAT-dependent IL2 production. Involved in myogenic constriction of cerebral arteries. Controls insulin secretion in pancreatic beta-cells. May also be involved in pacemaking or could cause irregular electrical activity under conditions of Ca(2+) overload. Affects T-helper 1 (Th1) and T-helper 2 (Th2) cell motility and cytokine production through differential regulation of calcium signaling and NFATC1 localization. Enhances cell proliferation through up-regulation of the beta-catenin signaling pathway. Plays a role in keratinocyte differentiation (PubMed:30528822). {ECO:0000269|PubMed:11535825, ECO:0000269|PubMed:12015988, ECO:0000269|PubMed:12799367, ECO:0000269|PubMed:14758478, ECO:0000269|PubMed:15121803, ECO:0000269|PubMed:15331675, ECO:0000269|PubMed:15472118, ECO:0000269|PubMed:15550671, ECO:0000269|PubMed:15590641, ECO:0000269|PubMed:15845551, ECO:0000269|PubMed:16186107, ECO:0000269|PubMed:16407466, ECO:0000269|PubMed:16424899, ECO:0000269|PubMed:16806463, ECO:0000269|PubMed:20625999, ECO:0000269|PubMed:20656926, ECO:0000269|PubMed:29211723, ECO:0000269|PubMed:30528822}. | 3D-structure;ATP-binding;Adaptive immunity;Alternative splicing;Calcium;Calmodulin-binding;Cell membrane;Coiled coil;Disease variant;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Golgi apparatus;Immunity;Ion channel;Ion transport;Membrane;Metal-binding;Nucleotide-binding;Palmoplantar keratoderma;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport;Ubl conjugation | The protein encoded by this gene is a calcium-activated nonselective ion channel that mediates transport of monovalent cations across membranes, thereby depolarizing the membrane. The activity of the encoded protein increases with increasing intracellular calcium concentration, but this channel does not transport calcium. [provided by RefSeq, Mar 2016]. | hsa:54795; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; sodium channel complex [GO:0034706]; spanning component of membrane [GO:0089717]; spanning component of plasma membrane [GO:0044214]; ATP binding [GO:0005524]; calcium activated cation channel activity [GO:0005227]; calcium ion binding [GO:0005509]; calmodulin binding [GO:0005516]; cation channel activity [GO:0005261]; identical protein binding [GO:0042802]; ligand-gated calcium channel activity [GO:0099604]; sodium channel activity [GO:0005272]; adaptive immune response [GO:0002250]; calcium ion transmembrane transport [GO:0070588]; calcium-mediated signaling [GO:0019722]; cation transmembrane transport [GO:0098655]; cellular response to ATP [GO:0071318]; dendritic cell chemotaxis [GO:0002407]; inorganic cation transmembrane transport [GO:0098662]; membrane depolarization during AV node cell action potential [GO:0086045]; membrane depolarization during bundle of His cell action potential [GO:0086048]; membrane depolarization during Purkinje myocyte cell action potential [GO:0086047]; negative regulation of bone mineralization [GO:0030502]; negative regulation of osteoblast differentiation [GO:0045668]; positive regulation of adipose tissue development [GO:1904179]; positive regulation of atrial cardiac muscle cell action potential [GO:1903949]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of cytosolic calcium ion concentration [GO:0007204]; positive regulation of fat cell differentiation [GO:0045600]; positive regulation of heart rate [GO:0010460]; positive regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0035774]; positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization [GO:1904199]; positive regulation of vasoconstriction [GO:0045907]; protein homotetramerization [GO:0051289]; protein sumoylation [GO:0016925]; regulation of heart rate by cardiac conduction [GO:0086091]; regulation of T cell cytokine production [GO:0002724]; regulation of ventricular cardiac muscle cell action potential [GO:0098911]; sodium ion import across plasma membrane [GO:0098719]; vasoconstriction [GO:0042310] | 12799367_Voltage dependence is not due to block by divalent cations or to voltage-dependent binding of intracellular Ca2+ to an activator site, indicating that TRPM4 is a transient receptor potential channel with an intrinsic voltage-sensing mechanism. 15550671_TRPM4-mediated depolarization modulates Ca2+ oscillations, with downstream effects on cytokine production in T lymphocytes 15590641_the Ca(2+) sensitivity of TRPM4 is regulated by ATP, PKC-dependent phosphorylation, and calmodulin binding at the C terminus. 15845551_analysis of selectivity filter of the cation channel TRPM4 16186107_hydrolysis of PI(4,5)P(2) underlies desensitization of TRPM4, and PI(4,5)P(2) is a general regulator for the gating of TRPM ion channels 16424899_PIP2 is a strong positive modulator of TRPM4 and implicate the C-terminal PH domain in PIP2 action. 17217063_evidence indicates a role as a regulator of membrane potential, and thus the driving force for Ca2+ entry from the extracellular medium--{REVIEW} 18262493_This study is believed to provide the first clear evidence that TRPM4b interacts physically with TRPC3. 19063936_Depolarizing currents generated by TRPM4 are an important component in the control of intracellular Ca(2+) signals necessary for insulin secretion and perhaps glucagon from alpha-cells. 19726882_In 3 branches of a large South African Afrikaner pedigree with an autosomal-dominant form of progressive familial heart block type I, identified the mutation c.19G-->A, which attenuated deSUMOylation of the TRPM4 channel. 20562447_TRPM4 gene is a causative gene in isolated cardiac conduction disease with mutations resulting in a gain of function and TRPM4 channel being highly expressed in cardiac Purkinje fibers. 20625999_These results identify TRPM4 as an important, unanticipated regulator of the beta-catenin pathway, where aberrant signaling is frequently associated with cancer. 20884614_Cys(1093) residue of TRPM4 is crucial for the H(2)O(2)-mediated loss of desensitization. 21887725_In eight probands with atrioventricular block or right bundle branch block-five familial cases and three sporadic cases-a total of six novel and two published TRPM4 mutations were identified. 23116477_TRPM4 possesses the biophysical properties and upstream cellular signaling and regulatory pathways that establish it as a major physiological player in smooth muscle membrane depolarization. 23160238_TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. 23382873_Because of its effect on the resting membrane potential, reduction or increase of TRPM4 channel function may both reduce the availability of sodium channel and thus lead to Brugada syndrome . 23796873_No role was found for TRPM4 in the peripheral blood compartment of multiple sclerosis patients. 24333049_Used the Xenopus laevis oocyte expression system for expression, purification and extraction of functional human TRPM4 protein. Investigated the supra-molecular assembly of TRPM4. 24518820_TRPM4 protein is a ROS-modulated non-selective cationic channel that performs several cell functions, including regulating intracellular Ca(2+) overload and Ca(2+) oscillation 24866019_Contraction of cerebral artery smooth muscle cells requires the integration of pressure-sensing signaling pathways and depolarization through the activation of TRPM4. 25001294_TRPM4 is an important regulator of Ca2+ signals generated by histamine in hASCs and is required for adipogenesis. 25047048_These results showed that the cell surface expression of TRPM4 channels is mediated by 14-3-3gamma binding. 25231975_Casein kinase 1 phosphorylates S839 and is responsible for the basolateral localization of TRPM4. 25909699_TRPM4 acts to maintain endothelial features and its loss promotes fibrotic conversion via TGF-beta production. 26071843_new insight into the ligand binding domains of the TRPM4 channel 26110647_we demonstrate that the inhibition of TRPM4 activity alters cellular contractility in vivo, affecting cutaneous wound healing. 26172285_these data suggest an important role for the Sur1-Trpm4 channel in the pathophysiology of postischemic cell death 26496025_Identify TRPM4 as a regulator of store operated calcium entry in prostate tumor cells, and demonstrate a role for TRPM4 in cancer cell migration. 26571400_The PKC-dependent effect of GLP-1 on membrane potential and electrical activity was mediated by activation of Na(+)-permeable TRPM4 and TRPM5 channels by mobilization of intracellular Ca(2+) from thapsigargin-sensitive Ca(2+) stores 26590985_TRPM4 protein expression is widely expressed in benign and cancerous prostate tissue 26791488_TRPM4 channels regulate human detrusor smooth muscle excitability and contractility and are critical determinants of human urinary bladder function 26820365_A large pedigree diagnosed with progressive familial heart block type I was linked to a mutation of the TRPM4 ion channel. 27207958_The loss-of-function variants A432T/G582S found in 2 unrelated patients with atrioventricular block are most likely caused by misfolding-dependent altered trafficking. 28248435_TRPM4 protein expression is up-regulated in diffuse large B cell lymphoma 28315637_Study supports the view that TRPM4 variants could be responsible for about 2% of LQT syndrome cases. The impact of these variants results in electrophysical disturbances. 28494446_We identified a double heterozygosity for pathogenic mutations in SCN5A and TRPM4 in a Brugada syndrome patient. 28614631_This study presents further evidence to show that TRPM4 regulates beta-catenin signaling and enhances the proliferation of prostate cancer cell lines, through a calcium-dependent regulation of Akt1 and GSK-3beta activity. 29211723_electron cryo-microscopy structure of the most widespread CAN channel, human TRPM4, bound to the agonist Ca(2+) and the modulator decavanadate 29217581_study presents 2 structures of TRPM4 embedded in lipid nanodiscs at ~3-angstrom resolution, as determined by single-particle cryo-electron microscopy; these structures, with and without calcium bound, reveal a general architecture for this major subfamily of TRP channels and a well-defined calcium-binding site within the intracellular side of the S1-S4 domain 29240297_Data suggest that TRPM4 exhibits binding sites for calmodulin (CaM) and S100 calcium-binding protein A1 (S100A1) located in very distal part of TRPM4 N-terminus. 29463718_The electron cryomicroscopy structure of human transient receptor potential melastatin subfamily member 4 (TRPM4) in a closed, Na(+)-bound, apo state at pH 7.5 to an overall resolution of 3.7 A is reported. 30021168_Rational mutagenesis of TRPM4 at position 432 revealed that the bulkiness of the amino acid was key to TRPM4(A432T)'s aberrant gating. 30142439_Compound heterozygous TRPM4 null mutations were identified in a patient with Brugada type 1 electrocardiogram. 30160201_Sulfonylurea Receptor 1, Transient Receptor Potential Cation Channel Subfamily M Member 4, and KIR6.2:Role in Hemorrhagic Progression of Contusion. 30343491_To evaluate whether TRPM4 overexpression was related to the spreading capability of tumors, TRPM4 was knocked down by using shRNAs in PC3 prostate cancer cells and the effect on cellular migration and invasion was analyzed. PC3 cells with reduced levels of TRPM4 (shTRPM4) display a decrease of the migration/invasion capability. 30391667_Here, we report the functional effects of previously uncharacterized variants of uncertain significance (VUS) that we have found while performing a 'genetic autopsy' in individuals who have suffered sudden unexpected death. We have identified thirteen uncommon missense VUS in TRPM4 by testing 95 targeted genes implicated in channelopathy and cardiomyopathy in 330 cases 30482841_Differentiated normal bronchial epithelial (NHBE) cells and tracheal cells from patients with cystic fibrosis (CFT1-LC3) expressed only TRPM4 and all three isoforms of NCXs. Blocking the activity of TRPM4 or NCX proteins abrogated MUC5AC secretion from NHBE and CFT1-LC3 cells. 30484364_Downstream TRPM4 Polymorphisms Are Associated with Intracranial Hypertension and Statistically Interact with ABCC8 Polymorphisms in a Prospective Cohort of Severe Traumatic Brain Injury. 30789900_Trpm4-mediated interneurons that express calcium (Ca2+)-activated nonselective cationic current is indispensable for motor output but not the rhythmogenic core mechanism of the breathing central pattern generator. 31087102_Molecular and genetic insights into progressive cardiac conduction disease. 31358308_Progressive symmetric erythrokeratodermia: Activating mutations of TRPM4 31441200_TRPM4 plays a versatile role in cancer cell proliferation, cell cycle, and invasion. 32147520_TRPM4 Modulates Right Ventricular Remodeling Under Pressure Load Accompanied With Decreased Expression Level. 32301552_KCTD5, a novel TRPM4-regulatory protein required for cell migration as a new predictor for breast cancer prognosis. 32320859_Aberrant TRPM4 expression in MLL-rearranged acute myeloid leukemia and its blockade induces cell cycle arrest via AKT/GLI1/Cyclin D1 pathway. 32484822_TRPM4 is overexpressed in breast cancer associated with estrogen response and epithelial-mesenchymal transition gene sets. 32681584_Domain zipping and unzipping modulates TRPM4's properties in human cardiac conduction disease. 33047172_TRPM4 non-selective cation channel in human atrial fibroblast growth. 33058873_Small Molecular Inhibitors Block TRPM4 Currents in Prostate Cancer Cells, with Limited Impact on Cancer Hallmark Functions. 33562811_TRPM4 in Cancer-A New Potential Drug Target. 33959666_Whole-Exome Sequencing Identifies a Novel TRPM4 Mutation in a Chinese Family with Atrioventricular Block. 34309670_Genetic Variants Associated With Intraparenchymal Hemorrhage Progression After Traumatic Brain Injury. 34445219_Theoretical Investigation of the Mechanism by which A Gain-of-Function Mutation of the TRPM4 Channel Causes Conduction Block. | ENSMUSG00000038260 | Trpm4 | 216.00111 | 1.4744782 | 0.5602044442 | 0.22503080 | 6.257661e+00 | 1.236574e-02 | 1.821386e-01 | No | Yes | 304.960670 | 44.658244 | 204.410640 | 29.335961 | |
| ENSG00000130649 | 1571 | CYP2E1 | protein_coding | P05181 | FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of fatty acids (PubMed:10553002, PubMed:18577768). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase) (PubMed:10553002, PubMed:18577768). Catalyzes the hydroxylation of carbon-hydrogen bonds. Hydroxylates fatty acids specifically at the omega-1 position displaying the highest catalytic activity for saturated fatty acids (PubMed:10553002, PubMed:18577768). May be involved in the oxidative metabolism of xenobiotics (Probable). {ECO:0000269|PubMed:10553002, ECO:0000269|PubMed:18577768, ECO:0000305|PubMed:9348445}. | 3D-structure;Direct protein sequencing;Endoplasmic reticulum;Fatty acid metabolism;Heme;Iron;Lipid metabolism;Membrane;Metal-binding;Microsome;Mitochondrion;Mitochondrion inner membrane;Monooxygenase;NADP;Oxidoreductase;Reference proteome | PATHWAY: Lipid metabolism; fatty acid metabolism. {ECO:0000269|PubMed:10553002, ECO:0000269|PubMed:18577768}. | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein localizes to the endoplasmic reticulum and is induced by ethanol, the diabetic state, and starvation. The enzyme metabolizes both endogenous substrates, such as ethanol, acetone, and acetal, as well as exogenous substrates including benzene, carbon tetrachloride, ethylene glycol, and nitrosamines which are premutagens found in cigarette smoke. Due to its many substrates, this enzyme may be involved in such varied processes as gluconeogenesis, hepatic cirrhosis, diabetes, and cancer. [provided by RefSeq, Jul 2008]. | hsa:1571; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; Golgi membrane [GO:0000139]; intracellular membrane-bounded organelle [GO:0043231]; intrinsic component of endoplasmic reticulum membrane [GO:0031227]; mitochondrial inner membrane [GO:0005743]; 4-nitrophenol 2-monooxygenase activity [GO:0018601]; arachidonic acid epoxygenase activity [GO:0008392]; aromatase activity [GO:0070330]; enzyme binding [GO:0019899]; heme binding [GO:0020037]; Hsp70 protein binding [GO:0030544]; Hsp90 protein binding [GO:0051879]; iron ion binding [GO:0005506]; long-chain fatty acid omega-1 hydroxylase activity [GO:0120319]; monooxygenase activity [GO:0004497]; oxidoreductase activity [GO:0016491]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen [GO:0016709]; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen [GO:0016712]; oxygen binding [GO:0019825]; steroid hydroxylase activity [GO:0008395]; 4-nitrophenol metabolic process [GO:0018960]; benzene metabolic process [GO:0018910]; carbon tetrachloride metabolic process [GO:0018885]; epoxygenase P450 pathway [GO:0019373]; halogenated hydrocarbon metabolic process [GO:0042197]; heterocycle metabolic process [GO:0046483]; lipid hydroxylation [GO:0002933]; long-chain fatty acid biosynthetic process [GO:0042759]; long-chain fatty acid metabolic process [GO:0001676]; monoterpenoid metabolic process [GO:0016098]; organic acid metabolic process [GO:0006082]; response to bacterium [GO:0009617]; response to ethanol [GO:0045471]; response to organonitrogen compound [GO:0010243]; response to ozone [GO:0010193]; steroid metabolic process [GO:0008202]; triglyceride metabolic process [GO:0006641]; xenobiotic metabolic process [GO:0006805] | 11051375_Observational study of gene-disease association. (HuGE Navigator) 11104220_Observational study of gene-disease association. (HuGE Navigator) 11191882_Observational study of genotype prevalence. (HuGE Navigator) 11198676_Observational study of genotype prevalence. (HuGE Navigator) 11207032_Observational study of genotype prevalence. (HuGE Navigator) 11236836_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11259352_Observational study of genotype prevalence and gene-environment interaction. (HuGE Navigator) 11263781_Observational study of gene-environment interaction. (HuGE Navigator) 11275366_Observational study of gene-disease association. (HuGE Navigator) 11305777_Observational study of gene-environment interaction. (HuGE Navigator) 11331106_Observational study of gene-disease association. (HuGE Navigator) 11377232_Observational study of genotype prevalence, gene-disease association, and gene-environment interaction. (HuGE Navigator) 11389775_Observational study of gene-disease association. (HuGE Navigator) 11389775_There is a correlation between the RsaI polymorphism homozygous uncut genotype in the CYP2E1 gene and a higher relative risk of nasopharyngeal carcinoma development in the Thai or Chinese populations in Thailand. 11406608_Observational study of gene-disease association. (HuGE Navigator) 11410713_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 11422615_Observational study of gene-disease association. (HuGE Navigator) 11503278_Observational study of gene-disease association. (HuGE Navigator) 11520401_Observational study of gene-disease association. (HuGE Navigator) 11535247_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11535253_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11641039_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11675150_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11696658_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11697456_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11700262_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11719088_Observational study of gene-disease association. (HuGE Navigator) 11746208_Observational study of genotype prevalence. (HuGE Navigator) 11748356_Observational study of gene-disease association. (HuGE Navigator) 11751440_Observational study of genotype prevalence. (HuGE Navigator) 11766168_Observational study of gene-disease association. (HuGE Navigator) 11774269_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 11776598_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11782477_increased translation of collagen mRNA by CYP2E1-derived reactive oxygen species is responsible for the increase in collagen protein 11798822_Observational study of gene-disease association. (HuGE Navigator) 11802217_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11815398_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11854903_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11869835_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 11895912_Observational study of gene-disease association. (HuGE Navigator) 11907164_Damage to mitochondria may be a critical step for cellular toxicity by CYP2E1-derived reactive oxygen species 11911601_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11960914_CYP2A6/2A7 and CYP2E1 expression in human oesophageal mucosa: regional and inter-individual variation in expression and relevance to nitrosamine metabolism 11964928_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 11966948_Observational study of gene-disease association. (HuGE Navigator) 12010862_Individuals possessing more susceptible CYP2E1 c2c2 genotypes were more likely to reveal p53 overexpression. Susceptible CYP2E1 genotypes may modulate the mutation of the p53 gene among VCM-exposed workers. 12010862_Observational study of gene-disease association. (HuGE Navigator) 12047484_Observational study of genotype prevalence. (HuGE Navigator) 12055050_Observational study of gene-environment interaction. (HuGE Navigator) 12063626_Observational study of gene-disease association. (HuGE Navigator) 12080432_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12115524_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12172927_Observational study of gene-disease association. (HuGE Navigator) 12198369_In the CYP2E1 gene 5'-flanking region polymorphism, patients with esophageal cancer showed significantly higher frequency of the A4/A4 genotype compared with the control subjects (p = 0.02), but no difference was found in patients with lung cancer. 12198369_Observational study of gene-disease association. (HuGE Navigator) 12211622_Observational study of gene-disease association. (HuGE Navigator) 12355548_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12365037_Observational study of genotype prevalence. (HuGE Navigator) 12376502_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12376511_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12397416_Observational study of gene-disease association. (HuGE Navigator) 12452057_expression of human cytochrome P450 2E1 gene in embryonic nasopharynx, nasopharyngeal cancer cell lines and tissue 12454736_Observational study of gene-disease association. (HuGE Navigator) 12460800_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12469218_Overexpression of this enzyme overexpression up-regulates both non-specific delta-aminolevulinate synthase and heme oxygenase-1 in a human hepatoma cell line. 12490624_substitution of residue 363 in cyp2e1 resulted in significant alterations of the metabolite profile for the side chain hydroxylation of 7-butoxycoumarin 12540498_Observational study of gene-disease association. (HuGE Navigator) 12548461_Observational study of gene-disease association. (HuGE Navigator) 12552594_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12552594_The CYP2E1 variant genotype did not significantly increase the risk for neoplasia. 12554615_Polymorphisms of alcohol-metabolizing enzymes: analyses of mutations on the CYP2E1, ADH2, ADH3 and ALDH2 genes in a Mexican-American population living in the Los Angeles area. 12561466_Observational study of gene-disease association. (HuGE Navigator) 12563175_CYP2E1 genetic polymorphism may be associated with susceptibility to breast cancer in alcohol-consuming women. 12563175_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12569554_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12579334_Observational study of gene-disease association. (HuGE Navigator) 12601351_hepatic cytochrome P450 2E1 activity and lymphocyte cytochrome P450 2E1 expression are enhanced in nondiabetic nonalcoholic steatohepatitis 12668988_CYP 2E1 genetic polymorphism may be associated with susceptibility to antituberculosis drug-induced hepatitis. 12668988_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12705718_Observational study of gene-disease association. (HuGE Navigator) 12707490_Observational study of gene-disease association. (HuGE Navigator) 12707490_This study shown that the genotypes of CYP2E1 are associated with clinical features of alcoholics. 12710951_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12718576_Observational study of genotype prevalence. (HuGE Navigator) 12718671_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12732844_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 12738724_Down-regulation of cytochrome P450 CYP2E1 is associated with breast cancer 12739102_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12759747_Observational study of gene-disease association. (HuGE Navigator) 12760253_Observational study of gene-disease association. (HuGE Navigator) 12767509_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12771559_Observational study of gene-disease association. (HuGE Navigator) 12774019_Proteasome activity plays an important role in modulating CYP2E1-mediated toxicity in HepG2 cells by regulating CYP2E1 levels and by removal of oxidized proteins. 12777398_HO-1 induction was observed in the livers of chronic alcohol-fed mice or pyrazole-treated rats, conditions known to elevate CYP2E1 levels. 12777398_P450 2E1 has a role in inducing heme oxygenase-1 through ERK MAPK pathway 12777962_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 12777962_association of CYP2E1*1D with alcohol and nicotine dependence suggests that CYP2E1 may contribute to the development of these dependencies 12777965_CYP2E1 and NQO1 genotypes may play an important role in development of smoking related bladder cancer among Korean men 12777965_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12813050_CYP2E1 has a role in oxidative stress causing mitochondrial damage along with phospholipase A2 12824748_Observational study of gene-disease association. (HuGE Navigator) 12824892_Observational study of gene-disease association. (HuGE Navigator) 12851035_Arg76 is closely associated with the function of CYP2E1, and that the genetic polymorphism of cytochrome p450 2E1 is one cause of interindividual differences in the toxicity of xenobiotics 12860273_These results show that CYP2E1 protein is expressed in both tumour and normal breast tissue with an increased expression in breast tumours. 12883487_hepatic CYP2E1 activity is up-regulated in morbidly obese subjects 12883749_Observational study of gene-disease association. (HuGE Navigator) 12915519_Observational study of gene-disease association. (HuGE Navigator) 12939804_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12940444_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 12960506_Transcriptional activity of the mutant allele of the tandem repeat polymorphism in the 5'-flanking region of the CYP2E1 gene is greater than that of the wild type. 14500779_CYP2E1 was clearly expressed in human fetal liver. 14510941_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14527082_Levels of CYP2E1 are elevated under a variety of physiological and pathophysiological conditions, and after acute and chronic alcohol treatment. 14527082_biochemical and toxicological properties of CYP2E1(review) 14535982_Observational study of gene-disease association. (HuGE Navigator) 14578150_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14606109_The genetic polymorphism of CYPIIE1 on the position of Pst I and Rsa I is related to the susceptibility of fatty liver. 14634838_Observational study of genotype prevalence. (HuGE Navigator) 14646291_Observational study of gene-disease association. (HuGE Navigator) 14669323_The cDNA of human CYP2E1 can be successfully cloned, and a cell line, HepG2-CYP2E1 can efficiently express mRNA and has CYP2E1 activity. 14681495_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 14691069_Observational study of gene-disease association. (HuGE Navigator) 14695651_Observational study of genotype prevalence. (HuGE Navigator) 14695664_enhanced injury in hepatocytes over expressing both Hepatitis C virus core protein and CYP2E1 is mediated by increases in oxidative stress. 14696128_CYP2E1 polymorphisms are associated with incomplete intestinal metaplasia in a high-risk area of stomach cancer 14696128_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 14751678_Observational study of gene-disease association. (HuGE Navigator) 14757192_Observational study of gene-disease association. (HuGE Navigator) 14991750_Observational study of gene-disease association. (HuGE Navigator) 15036355_The objective of this study was to analyze the effect of the green tea flavanol epigallocatechin-3-gallate (EGCG), which has been shown to prevent alcohol-induced liver damage, on CYP2E1-mediated toxicity in HepG2 cells overexpressing CYP2E1 (E47 cells). 15061915_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15064998_Observational study of gene-disease association. (HuGE Navigator) 15066574_Observational study of gene-disease association. (HuGE Navigator) 15112335_Observational study of gene-disease association. (HuGE Navigator) 15125228_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15138035_Observational study of gene-disease association. (HuGE Navigator) 15162526_CYP2E1 is a unique gene expressing in liver but did not express in hepatocellular carcinoma 15177663_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15182482_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15215328_Observational study of gene-disease association. (HuGE Navigator) 15215328_polymorphism and susceptibility to cirrhosis or pancreatitis in alcoholics 15220553_Observational study of gene-disease association. (HuGE Navigator) 15226677_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15289170_Observational study of gene-disease association. (HuGE Navigator) 15318112_Observational study of gene-disease association. (HuGE Navigator) 15327835_Observational study of gene-disease association. (HuGE Navigator) 15349722_marked impairment of CYP enzyme activity during allograft rejection which is presumably secondary to an increased intragraft production of proinflammatory cytokines and NO. 15355699_Observational study of gene-disease association. (HuGE Navigator) 15370874_Observational study of gene-disease association. (HuGE Navigator) 15491310_Observational study of gene-disease association. (HuGE Navigator) 15519646_Observational study of gene-disease association. (HuGE Navigator) 15532721_overexpression of human CYP2E1 activates acetaminophen to reactive metabolites which damage mitochondria, form protein adducts, and result in toxicity to HepG2 cells 15536330_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15632182_increased hepatocyte CYP2E1 expression and the presence of steatohepatitis result in the down-regulation of insulin signaling 15633127_The aim of this study was to determine whether hepatitis C virus core protein and alcohol-inducible cytochrome P450 2E1 contribute to reactive oxygen species production and cytotoxicity in human hepatoma cells. 15640066_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15646021_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 15660387_Nicotinamide adenine dinucleotide phosphate oxidase-derived oxidants are critical for development of ethanol-induced liver injury. CYP2E1 is required for induction of oxidative stress to DNA and may play role in ethanol-associated hepatocarcinogenesis. 15712341_Observational study of gene-disease association. (HuGE Navigator) 15714076_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, gene-gene interaction, gene-environment interaction, pharmacogenomic / toxicogenomic, and healthcare-related. (HuGE Navigator) 15734972_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 15753073_To address this question, we determined whether DLPC protects against alcohol-induced cytotoxicity in HepG2 cells expressing CYP2E1. 15763499_a decrease of Hsp60 in the cytoplasmic fraction of dilated cardiomyopathy-affected left ventricles was observed; at the same time an increase in P450 2E1 expression in dilated hearts' cytoplasmic fractions was observed 15769360_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15774926_Observational study of gene-disease association. (HuGE Navigator) 15780023_Observational study of gene-disease association. (HuGE Navigator) 15793883_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15793883_the CYP2E1 genotype may influence individual susceptibility to development of gastric cardia cancer, and that the risk increases significantly in smokers. 15849806_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 15899651_Observational study of gene-disease association. (HuGE Navigator) 15902904_Polymorphisms of the alcohol metabolism-related CYP2E1 gene are significantly different in Korean patients with alcoholism and Korean control subjects without alcoholism. 15914211_Observational study of gene-disease association. (HuGE Navigator) 15914277_Observational study of gene-disease association. (HuGE Navigator) 15928955_Observational study of gene-disease association. (HuGE Navigator) 15938845_Observational study of gene-disease association. (HuGE Navigator) 15952134_The CYP2E1 genotype frequencies are 66.7% for type of c1/c1, 30% for type of c1/c2, 3.3% for type of c2/c2 with the allele frequencies 0.818 for C1 and 0.182 for C2, and the gene distribution matched the equilibrium law of Hardy-Weinberg. 15961886_CYP2E1 priming could explain the sensitization of Kupffer cells to LPS activation by ethanol, a critical early step in alcoholic liver disease 15968714_CYP2E1 wild type (c1/c1) increased the susceptibility to esophageal squamous cell cancer risk in Kazakh individuals with GSTM1 presence genotype 15968714_Observational study of gene-disease association. (HuGE Navigator) 15991278_Observational study of gene-disease association. (HuGE Navigator) 16006997_Observational study of gene-disease association. (HuGE Navigator) 16019049_Low concentrations of iron and arachidonic acid synergistically interacted with CYP2E1 to produce cell toxicity, suggesting these nutrients may act as priming or sensitizing agents to alcohol-induced liver injury. 16039674_Observational study of gene-disease association. (HuGE Navigator) 16043197_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16052683_A positive relationship between ethanol-induced oxidative damage in human primary cultured hepatocytes and CYP2E1 activity was exhibited 16125881_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16126235_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16137184_A PCR method for identifying polymorphism. 16137184_Observational study of genotype prevalence and genetic testing. (HuGE Navigator) 16142352_Observational study of gene-disease association. (HuGE Navigator) 16142352_the A4/A4 genotype of the 5'-flanking region of CYP2E1 has a role in development of non-small cell lung carcinoma in Japanese patients 16172237_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16235983_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16235992_Observational study of genotype prevalence. (HuGE Navigator) 16253141_Protein concentration of CYP2E1 is significantly lower by ~81% in the sigmoid colon compared to the level in the descending colon. 16311924_We report here for the first time three novel alternative spliced mRNA transcripts which are more frequently present in lung carcinoma cell lines as in hepatocyte cell lines. 16324524_Observational study of gene-disease association. (HuGE Navigator) 16337880_Potentiation of serum deprivation-induced cell death by CYP2E1 may contribute to the sensitivity of the liver to alcohol-induced ischemia and growth factor deprivation. 16365683_Observational study of gene-disease association. (HuGE Navigator) 16372174_Observational study of gene-disease association. (HuGE Navigator) 16380384_CYP2E1 has a role in sensitizing cells to the toxicity caused by depletion of glutathione 16385451_Observational study of gene-disease association. (HuGE Navigator) 16393248_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16424825_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16425414_Observational study of gene-disease association. (HuGE Navigator) 16425414_frequencies of this high-activity polymorphism in alcohol related patient groups 16440362_Meta-analysis of gene-disease association. (HuGE Navigator) 16459354_Observational study of gene-disease association. (HuGE Navigator) 16470306_Observational study of genotype prevalence. (HuGE Navigator) 16471212_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16484137_Observational study of gene-disease association. (HuGE Navigator) 16488179_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16497268_These results suggest that CYP2E1 mediated oxidative stress downregulates the expression of GRP proteins in HepG2 cells and oxidative stress is an important mechanism in causing ER dysfunction in these cells. 16535827_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16551616_Nrf2, through up-regulation of glutamate-cysteine ligase and increase of GSH levels, protects against CYP2E1-dependent AA toxicity 16600530_Observational study of gene-disease association. (HuGE Navigator) 16634857_Observational study of genotype prevalence. (HuGE Navigator) 16634857_studies suggest that the frequency of the c2 alleles in the Polish population is low; it seems, however, that they pose the risk of alcoholic cirrhosis 16679316_CYP2E1-b(5) complex model was constructed, leading to improved insights into the protein interaction. 16720291_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16721740_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16758119_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16758119_findings indicated a gene-gene interaction between CYP2E1 and GSTM1 was accessible to developing breast cancer in Taiwanese women without the habits of cigarette smoking and alcohol consumption 16770646_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16834659_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16837478_Meta-analysis of gene-disease association and gene-environment interaction. (HuGE Navigator) 16841220_Observational study of gene-disease association. (HuGE Navigator) 16962935_increased toxic interactions by TGF-beta1 plus CYP2E1 can occur by a mechanism involving increased production of intracellular ROS and depletion of GSH, resulting in mitochondrial membrane damage and loss of membrane potential, followed by apoptosis 16985026_Observational study of gene-disease association. (HuGE Navigator) 16985032_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17007050_Expression of Thy1 decreased in differentiated ADSC and BMSC. Expression of albumin, CYP2E1, and CYP3A4 increased in differentiated BMSC and ADSC. Hepatic gene activation may involve increased C/EBPbeta and HNF4alpha. 17016589_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17022435_Observational study of gene-disease association. (HuGE Navigator) 17034788_Ethanol induced CYP2E1 generates oxidative stress that is responsible for the decrease in proteasome activity. 17059334_Observational study of genotype prevalence. (HuGE Navigator) 17078101_Observational study of gene-disease association. (HuGE Navigator) 17118447_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17119198_Observational study of gene-disease association. (HuGE Navigator) 17119944_This report is the first demonstration that thalassemia major is associated with an alteration of CYP2E1 and CYP3A4 activities; this could modify the sensitivity of thalassemia patients to the toxic or therapeutic effects of drugs. 17134659_Observational study of gene-disease association. (HuGE Navigator) 17134659_This documents the presence of a polymorphism of CYP2E1 that is overexpressed in alcoholic Otomies, in which the variant allele (A1 of CYP2E1/TaqI) is associated with increased susceptibility to alcoholism. 17146594_The expression of CYP2E1 in Nasopharyngeal carcinoma (NPC) cells, embryonic nasopharyngeal epithelial tissue (ENET) specimens, and NPC biopsies was analysed. 17156750_comparison of substrate dynamics in CYP2E1 and CYP2A6 17176083_The unusually rapid carbon monoxide binding kinetics of P450 2E1 indicate that it is more dynamically mobile than other P450s and thus able to more readily interconvert among alternate conformations. 17178637_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17219769_Observational study of genetic testing. (HuGE Navigator) 17264406_Observational study of genotype prevalence. (HuGE Navigator) 17264406_The frequency of the CYP2E1 allele distribution was found to be markedly different between Chinese and South African populations. 17284772_Induction of DNA strand breaks and oxidative stress in HeLa cells by ethanol is dependent on CYP2E1 expression. 17292341_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17331164_The aim of this study was to determine the role of these polymorphisms for the transcriptional regulation of the human CYP2E1 gene in general transcription and during enzyme induction 17361553_Observational study of gene-disease association. (HuGE Navigator) 17367411_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17373732_Differences exist in protein levels of certain CYPs in non-malignant esophageal tissue (e.g. CYP2C8, CYP3A4, CYP3A5, and CYP2E1) between SCC patients and healthy subjects and may contribute to the development of squamous-cell carcinoma in the esophagus. 17380320_Observational study of genotype prevalence. (HuGE Navigator) 17380320_The genotype frequencies of CYP2E1*7B in Turkish population were found to be similar to those of other Caucasian populations 17384900_Observational study of gene-disease association. (HuGE Navigator) 17427487_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17427487_Our studies suggest that the frequency of allele c2 in Polish population is low, but the presence of c2 allele may be a risk factor for the alcohol liver cirrhosis. 17440116_the level of CYP2E1 is induced by hypertonic condition via TonEBP transactivation. The present study suggests that osmotic status may influence individual responses to the substrate of CYP2E1. 17442289_Observational study of gene-disease association. (HuGE Navigator) 17442289_The CYP2E1 and NAT2 variants associated with COPD 17486761_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17498780_Observational study of gene-disease association. (HuGE Navigator) 17559142_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17559142_examined genetic polymorphisms in the alcohol dehydrogenase 3, aldehyde dehydrogenase 2, and cytochrome P450 2E1 genes in 505 patients with histologically confirmed lung cancer and 256 noncancer controls to determine association with alcohol drinking 17564586_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17577619_Gastric cancer in patients younger than 40 years is closely associated with H. pylori infection, but not with genetic characteristics. 17577619_Observational study of gene-disease association. (HuGE Navigator) 17584020_Chlorzoxazone metabolism in vivo remains the only available method for CYP2E1 phenotyping. 17603900_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17611777_CYP2E1 polymorphism is not associated with the risk of head and neck cancer 17611777_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 17617119_CYP2E1 activity cannot be used to distinguish nonalcoholic steatotis patients from steatohepatitis patients. 17627011_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17659824_Genetic polymorphism in CYP2E1 may be responsible for individual differences in susceptibility to liver fibrosis with regard to chronic vinyl chloride monomer exposure. 17659824_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17695473_Observational study of gene-disease association. (HuGE Navigator) 17885617_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17916905_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17935737_Findings suggest that human and cynomolgus monkey CYP2E1 enzymes have high homology in their amino acid sequences, and that their enzymatic properties are considerably similar. 17950035_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17963298_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17963298_The CYP2E1 c2/c2 genotype increases susceptibility to rectal cancer and the gene-environmental interactions between the CYP2E1 polymorphism and smoking or alcohol drinking exist for colorectal neoplasia in general. 17996038_Individuals carrying the *5A or *6 alleles of CYP2E1 are associated with gastric cancer in drinkers 17996038_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18025800_Individuals carrying combinations of CYP2E1*5B, *6 and *7B variants together are likely associated with the risk of developing childhood ALL. 18025800_Observational study of gene-disease association. (HuGE Navigator) 18028774_Observational study of gene-disease association. (HuGE Navigator) 18028774_carrying c2 allele and DD genotype of CYP2E1 conferreded an elevated risk for esophageal squamous cell carcinoma. 18034693_Observational study of gene-disease association. (HuGE Navigator) 18034693_The H6, H7, and H9 haplotypes may play certain roles in different clinical phenotypes in Mexican American alcoholics. In addition, our data suggest that the H1, H2, and H3 haplotypes are associated with alcohol drinking and smoking. 18056994_biochemical analysis of CYP2E1 substrate inhibition 18211048_QSAR analysis for substrate specificity of six CYP isoforms, revealing that CYP2C9 substrates are anionic compounds, while CYP2D6 substrates are cationic, and CYP2E1 substrates are smaller compou | ENSMUSG00000025479 | Cyp2e1 | 25.60566 | 1.1757077 | 0.2335294617 | 0.61901997 | 1.414649e-01 | 7.068290e-01 | No | Yes | 20.231650 | 8.916239 | 18.319561 | 7.852428 | |
| ENSG00000130758 | 4294 | MAP3K10 | protein_coding | Q02779 | FUNCTION: Activates the JUN N-terminal pathway. {ECO:0000250}. | 3D-structure;ATP-binding;Kinase;Methylation;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;SH3 domain;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene is a member of the serine/threonine kinase family. This kinase has been shown to activate MAPK8/JNK and MKK4/SEK1, and this kinase itself can be phoshorylated, and thus activated by JNK kinases. This kinase functions preferentially on the JNK signaling pathway, and is reported to be involved in nerve growth factor (NGF) induced neuronal apoptosis. [provided by RefSeq, Jul 2008]. | hsa:4294; | cytoplasm [GO:0005737]; ATP binding [GO:0005524]; bHLH transcription factor binding [GO:0043425]; JUN kinase kinase kinase activity [GO:0004706]; protein homodimerization activity [GO:0042803]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transcription corepressor activity [GO:0003714]; apoptotic process [GO:0006915]; JNK cascade [GO:0007254]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of transcription, DNA-templated [GO:0045892]; peptidyl-serine phosphorylation [GO:0018105]; peptidyl-threonine phosphorylation [GO:0018107]; positive regulation of apoptotic process [GO:0043065]; positive regulation of JNK cascade [GO:0046330]; positive regulation of JUN kinase activity [GO:0043507]; protein autophosphorylation [GO:0046777]; signal transduction [GO:0007165]; smoothened signaling pathway [GO:0007224] | 12105200_MLK2 has a role in vesicle formation and endosome recycling by binding to clathrin 15062575_demonstrate Alien-MLK2 interaction and also show that MLK2 is able to phosphorylate Alien; Alien, DAX-1 and thyroid hormone receptor mediated transcriptional silencing is strongly enhanced in the presence of active MLK2 17584736_MLK2 and -3 are required for activation of JNK and p38 by ectopically expressed GCK 19332348_Hippocalcin and MLK2 were colocalized in the halo of Lewy bodies in Parkinson disease patients, and neither protein was detected in normal pigmented neurons. 19801649_Mixed lineage kinase phosphorylates transcription factor E47 and inhibits TrkB expression to link neuronal death and survival pathways 20596961_Data show that miR-181b contributed to proliferation of AML cells by targeting MLK2. 21678419_This study showed that show that RUNX3 is a principal and evolutionarily conserved component of the MST pathway. 21979919_Mechanism of MAP kinase activation by TNF requires Src-dependent activation of Vav, activation of Rac/Cdc42, and the engagement of the Rac/Cdc42 interaction site on MLK2 and 3. 21979919_Mechanism of MAP kinase activation by TNF requires Src-dependent activation of Vav, activation of Rac/Cdc42, and the engagement of the Rac/Cdc42 interaction site on MLK3. 23178452_A novel and important role for MAP3K10 in the proliferation and chemoresistance of pancreatic ductal adenocarcinoma. 23760366_MAP3K4 is sufficiently mediate the TGFbeta-induced phosphorylation of p38 MAPK in MEFs and HaCaT cells. 28214207_We identified a direct target of miR-155 as MAP3K10 in osteosarcoma 28947795_MST interaction with MOB1 is not essential for development and tissue growth control. 32323857_DNA methylationregulated miR1555p depresses sensitivity of esophageal carcinoma cells to radiation and multiple chemotherapeutic drugs via suppression of MAP3K10. | ENSMUSG00000040390 | Map3k10 | 439.95082 | 0.7169793 | -0.4799965648 | 0.16238064 | 8.605583e+00 | 3.351341e-03 | 9.840799e-02 | No | Yes | 445.183634 | 58.020630 | 613.700813 | 77.678856 | |
| ENSG00000130783 | 84660 | CCDC62 | protein_coding | Q6P9F0 | FUNCTION: Nuclear receptor coactivator that can enhance preferentially estrogen receptors ESR1 and ESR2 transactivation. Modulates also progesterone/PGR, glucocorticoid/NR3C1 and androgen/AR receptors transactivation, although at lower level; little effect on vitamin D receptor/VDR. {ECO:0000269|PubMed:19126643}. | Alternative splicing;Coiled coil;Cytoplasm;Nucleus;Reference proteome;Repeat | hsa:84660; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; estrogen receptor binding [GO:0030331]; nuclear receptor coactivator activity [GO:0030374]; blastocyst hatching [GO:0001835]; cellular response to estradiol stimulus [GO:0071392]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 18563714_ERAP75 functions as a novel coactivator that can modulate ER alpha function in the prostate stromal cells. 19126643_Role of CCDC62/ERAP75 as a novel coactivator in prostate cancer cells that can modulate estrogen receptor beta transactivation and receptor function. 19165854_serological survey of 191 cancer patients with a range of different cancers by ELISA revealed antibodies to CCDC62-2 in 13 patients, including stomach cancer 24312176_STK39 (rs2102808) and CCDC62/HIP1R (rs12817488) do not appear to influence PD risk. 24335092_Study demonstrated a significant association between rs12817488 and sporadic Parkinson's disease with gender variations in a Chinese cohort 25818163_our findings support the conclusion that the rs12817488 in CCDC62/HIP1R polymorphism may increase the risk of Parkinson's disease in the Chinese Han population. 27035708_there is a relationship between rs12817488 and Parkinson's disease risk in Chinese population [meta-analysis] 27269966_This study shown CCDC62 genetic variants for Parkinson's disease are associated with the risk of incident Parkinson's disease in the general population and with impairment in daily functioning in individuals without clinical parkinsonism. | ENSMUSG00000061882 | Ccdc62 | 19.98754 | 1.3291683 | 0.4105237871 | 0.66672095 | 3.795230e-01 | 5.378586e-01 | No | Yes | 26.031697 | 10.814189 | 19.655205 | 8.079299 | |||
| ENSG00000130830 | 4354 | MPP1 | protein_coding | Q00013 | FUNCTION: Essential regulator of neutrophil polarity. Regulates neutrophil polarization by regulating AKT1 phosphorylation through a mechanism that is independent of PIK3CG activity (By similarity). {ECO:0000250}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Cell projection;Direct protein sequencing;Lipoprotein;Membrane;Palmitate;Phosphoprotein;Reference proteome;SH3 domain | This gene encodes the prototype of the membrane-associated guanylate kinase (MAGUK) family proteins. MAGUKs interact with the cytoskeleton and regulate cell proliferation, signaling pathways, and intercellular junctions. The encoded protein is an extensively palmitoylated membrane phosphoprotein containing a PDZ domain, a Src homology 3 (SH3) motif, and a guanylate kinase domain. This gene product interacts with various cytoskeletal proteins and cell junctional proteins in different tissue and cell types, and may be involved in the regulation of cell shape, hair cell development, neural patterning of the retina, and apico-basal polarity and tumor suppression pathways in non-erythroid cells. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2009]. | hsa:4354; | centriolar satellite [GO:0034451]; cortical cytoskeleton [GO:0030863]; membrane [GO:0016020]; plasma membrane [GO:0005886]; stereocilium [GO:0032420]; guanylate kinase activity [GO:0004385]; regulation of neutrophil chemotaxis [GO:0090022]; signal transduction [GO:0007165] | 16741958_Results show the NMR-derived structure of the human erythroid p55 PDZ domain and propose a possible interaction mode with the C-terminus of glycophorin C. 17584769_MPP1 links the Usher protein network and the Crumbs protein complex in the retina 18952129_p55 binds to two distinct sites within the FERM domain, and the alternatively spliced exon 5 is necessary for the membrane targeting of protein 4.1R in epithelial cells. 19144871_p55-NF2 protein interaction may play a functional role in the regulation of apico-basal polarity and tumor suppression pathways in non-erythroid cells. 21509594_MPP1 gene expression is decreased in both classic and follicular variants of papillary thyroid carcinoma. 23507198_MPP1 knockdown significantly affects the activation of MAP-kinase signaling. 25408337_Studies indicate that palmitoylated erythrocyte membrane protein p55 (MPP1) and its palmitoylation play a crucial role in lateral membrane organization in erythroid cells. 28653654_conclusion, the data demonstrates that MPP1 interacts with lipid mixtures in two different model membrane systems. 28865798_A new perspective on the role of MPP1 in erythroid cells suggests that direct MPP1-flotillins interactions could be the major driving-force behind the formation of raft domains in red blood cells. 29146910_Data suggest that targeting the ATP binding cassette subfamily C member 4 (ABCC4)-membrane palmitoylated protein 1 (MPP1) protein complex can lead to new therapies to improve treatment outcome of acute myeloid leukemia (AML). | ENSMUSG00000031402 | Mpp1 | 939.51099 | 1.0838638 | 0.1161834709 | 0.11062895 | 1.105887e+00 | 2.929777e-01 | 6.772304e-01 | No | Yes | 1211.514967 | 89.201133 | 1064.616553 | 76.647250 | |
| ENSG00000131018 | 23345 | SYNE1 | protein_coding | Q8NF91 | FUNCTION: Multi-isomeric modular protein which forms a linking network between organelles and the actin cytoskeleton to maintain the subcellular spatial organization. As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. May be involved in nucleus-centrosome attachment and nuclear migration in neural progenitors implicating LINC complex association with SUN1/2 and probably association with cytoplasmic dynein-dynactin motor complexes; SYNE1 and SYNE2 may act redundantly. Required for centrosome migration to the apical cell surface during early ciliogenesis. May be involved in nuclear remodeling during sperm head formation in spermatogenenis; a probable SUN3:SYNE1/KASH1 LINC complex may tether spermatid nuclei to posterior cytoskeletal structures such as the manchette. {ECO:0000250|UniProtKB:Q6ZWR6, ECO:0000269|PubMed:11792814, ECO:0000269|PubMed:18396275}. | 3D-structure;Actin-binding;Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Differentiation;Disease variant;Disulfide bond;Emery-Dreifuss muscular dystrophy;Golgi apparatus;Membrane;Neurodegeneration;Nucleus;Phosphoprotein;Reference proteome;Repeat;Spermatogenesis;Transmembrane;Transmembrane helix | This gene encodes a spectrin repeat containing protein expressed in skeletal and smooth muscle, and peripheral blood lymphocytes, that localizes to the nuclear membrane. Mutations in this gene have been associated with autosomal recessive spinocerebellar ataxia 8, also referred to as autosomal recessive cerebellar ataxia type 1 or recessive ataxia of Beauce. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008]. | hsa:23345; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; meiotic nuclear membrane microtubule tethering complex [GO:0034993]; nuclear envelope [GO:0005635]; nuclear membrane [GO:0031965]; nuclear outer membrane [GO:0005640]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; P-body [GO:0000932]; postsynaptic membrane [GO:0045211]; sarcomere [GO:0030017]; actin binding [GO:0003779]; actin filament binding [GO:0051015]; cytoskeleton-nuclear membrane anchor activity [GO:0140444]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; lamin binding [GO:0005521]; protein homodimerization activity [GO:0042803]; RNA binding [GO:0003723]; Golgi organization [GO:0007030]; muscle cell differentiation [GO:0042692]; nuclear matrix anchoring at nuclear membrane [GO:0090292]; nucleus organization [GO:0006997]; spermatogenesis [GO:0007283] | 12408964_Vertebrate Nesprin-1 and nesprin-2 proteins are orthologous to Drosophila melanogaster muscle protein MSP-300 14709720_The role of Syne-1 in cytokinesis involves an interaction with kinesin II. 15093733_Enaptin is an element of the nuclear membrane and the actin cytoskeleton. 16079285_The Nesprin-1 is essential for the interaction with a C-terminal region in SUN1 protein. 16875688_These findings indicate that GSRP-56 is a small Golgi-localized splice-isoform of the spectrin-repeat-containing Syne1 gene. 17159980_SYNE1 is the first identified gene responsible for a recessively inherited pure cerebellar ataxia. 17462627_The characterisation of the residues both in emerin and in nesprin-1alpha and -2beta which are involved in their interaction is reported. 17503513_This study identified a cluster of French-Canadian families with a new recessive ataxia of relatively pure cerebellar type caused by mutations in SYNE1. 17761684_Screening for DNA variations in the genes encoding nesprin-1 (SYNE1) and nesprin-2 (SYNE2) in 190 probands with Emery Dreifuss muscular dystrophy identified four heterozygous missense mutations. 18649358_Observational study of gene-disease association. (HuGE Navigator) 18709643_Loss of Drop1 expression at early stages in carcinomas of the uterus, cervix, kidney, lung, thyroid and pancreas. 18827015_association with human nesprin-3 appeared to be stronger for torsinADeltaE than for torsinA. TorsinA also associated with the KASH domains of nesprin-1 and -2 19542096_splice site mutation of SYNE-1 gene found in the family is responsible for arthrogryposis multiplex congenita. 19944109_analysis of nsprin-1 mutations in human and murine cardiomyopathy 20056644_Meta-analysis of gene-disease association. (HuGE Navigator) 20056644_a single nucleotide polymorphism downstream of ESR1, a gene called spectrin repeat containing, nuclear envelope 1, was associated with invasive ovarian cancer risk 20108321_Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development 20201924_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20351715_Meta-analysis and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20655839_Nesprin-1 depletion increased the number of focal adhesions and substrate traction while decreasing the speed of cell migration 22565781_The findings of this study add to the evidence that association of SYNE1 mutation influences susceptibility to bipolar and unipolar mood disorders. 22632968_Study presents crystal structures of the human SUN2-KASH1/2 complex, i.e. SUN2 complexed with the C-terminal 29 residues of human Nesprin-1 or -2 (the core of the LINC complex). 22728879_These results indicate that nesprin-1 knockdown releases the nucleus from the tension of F-actin bound to the nucleus, thereby increasing allowance for deformation before stretching, and that F-actin bound to the nucleus through nesprin-1 causes sustainable force transmission to the nucleus. 22768332_Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. 23325900_we report 4 novel homozygous SYNE1 mutations in 3 Japanese patients with cerebellar ataxia 23959263_Two novel truncating mutations were found among the French-Canadian participants, and 2 other novel mutations were found in a patient from France and a patient from Brazil. 24280874_FOXE1 and SYNE1 hypermethylation markers demonstrated significantly increased expression in neoplastic tissue. 24387768_role of CSMD1 and SYNE1 in the etiology of bipolar disorder 24718612_The significance of these shorter isoforms of nesprin, were evaluated. 24781983_a role for Nesprin-1 in the DNA damage response pathway 24862572_These data identify p50(Nesp1) as a multi-functional P-body component and microtubule scaffold necessary for RNA granule dynamics and provides evidence for P-body and stress granule micro-heterogeneity. 24931616_nesprin-1 and nesprin-2 both regulate nuclear and cytoplasmic architecture. 25091525_A report of a family with a SYNE1 gene mutation that seems to cause an 'Emery-Dreifuss muscular dystrophy-like' phenotype. 25315199_ur results show that SNPs rs9371601 and rs3093664 in the SYNE1 and TNF genes respectively, are associated with menstrual migraine. 25538088_our data suggest that SYNE1 and FOXE1 are promising markers for colorectal cancer detection. 26008743_Drosophila melanogaster larval muscles, exhibiting both elastic features contributed by the stretching capacity of MSP300 (nesprin) and rigidity provided by a perinuclear network of microtubules stabilized by Shot (spectraplakin) and EB1. 26330482_SNP rs79575945 in ESR1 gene is associated with cancers of endometrioid subtype and resulted in the expression of SYNE1. 26704904_A full length transcript homologous to rat CPG2 exists within human SYNE1. A full length transcript homologous to rat CPG2 exists within human SYNE1. 26791211_Mechanostimulation mediates nuclear changes in oligodendrocyte progenitor cells via the SYNE1 complex. 27086870_findings revise the view that SYNE1 ataxia causes mainly a relatively pure cerebellar recessive ataxia and that it is largely limited to Quebec. Instead, complex phenotypes with a wide range of extra-cerebellar neurological and nonneurological dysfunctions are frequent, including in particular motor neuron and brainstem dysfunction. 27178001_This study demonstrate four novel truncating mutations in SYNE1 in pedigrees from British, Sri Lankan and Turkish origin in patient with cerebellar ataxia. 27350129_The results show that nesprin-1-alpha2 is dynamically controlled and may be involved in some process occurring during early myofibre formation, such as re-positioning of nuclei. 27620326_The frequency of CACNA1C rs10848683 in genetic high-risk individuals was double that in controls. For SYNE1 rs214950, higher frequencies were found in the genetic high-risk group than in controls. Polymorphisms in CACNA1C and SYNE1 could confer a greater risk of developing Schizophrenia and Bipolar Disorder in individuals who are already at high risk because of their family history. 27782104_Nonsense mutation in the ultimate exon of full-length SYNE1 causes congenital onset of muscular weakness with distal arthrogryposis. 28178086_Authors screened 937 BPD samples for genetic variation in SYNE1 exons 14-33, which covers the CPG2 region, using high-resolution melt analysis. Nine patients are compound heterozygotes for variants in SYNE1/CPG2, suggesting that rare coding variants may contribute significantly towards the complex genetic architecture underlying BPD. 28398466_three novel rare variants (R8272Q, S8381C and N8406K) in the C-terminus of the SYNE1 gene (nesprin-1) were identified in seven dilated cardiomyopathy patients by mutation screening. Expression of these mutants caused nuclear morphology defects and reduced lamin A/C and SUN2 staining at the Nuclear envelope. 28455503_The functional integrity of lamin and nesprin-1 is thus required to modulate the FHOD1 activity and the inside-out mechanical coupling that tunes the cell internal stiffness to match that of its soft, physiological-like environment. 28583108_We reported a novel mutation in exon 46 on codon 2304 (G2304R) of the SYNE1 gene in a Chinese family with Emery-Dreifuss muscular dystrophy-like features, and 100 healthy individuals did not show such mutation. 30573412_Three novel mutations are described in SYNE1-ataxia patients. 30610203_Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function. 31049853_SYNE1 gene mutations were identified as a cause of late-onset pure cerebellar syndrome 31236099_Our explorative analyses of the biological impact of rs9371601 suggested that this SNP was significantly associated with the methylation of a CpG site (cg01844274, p = 5.0510(- 6)) within SYNE1 in human dorsal lateral prefrontal cortex (DLPFC) tissues. CONCLUSIONS: Our data confirms the association between rs9371601 and BPD 31578382_Nesprin-1-alpha2 associates with kinesin at myotube outer nuclear membranes, but is restricted to neuromuscular junction nuclei in adult muscle. 32281752_SYNE1-QK1 SNPs, G x G and G x E interactions on the risk of hyperlipidaemia. 32889669_Autosomal Recessive Cerebellar Ataxia Type 1: Phenotypic and Genetic Correlation in a Cohort of Chinese Patients with SYNE1 Variants. 33472039_Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. | ENSMUSG00000096054 | Syne1 | 351.20370 | 0.9576012 | -0.0625031825 | 0.19969746 | 9.740577e-02 | 7.549653e-01 | 9.282507e-01 | No | Yes | 366.684538 | 83.450727 | 353.266183 | 78.588598 | |
| ENSG00000132207 | 548593 | SLX1A | protein_coding | Q9BQ83 | FUNCTION: Catalytic subunit of the SLX1-SLX4 structure-specific endonuclease that resolves DNA secondary structures generated during DNA repair and recombination. Has endonuclease activity towards branched DNA substrates, introducing single-strand cuts in duplex DNA close to junctions with ss-DNA. Has a preference for 5'-flap structures, and promotes symmetrical cleavage of static and migrating Holliday junctions (HJs). Resolves HJs by generating two pairs of ligatable, nicked duplex products. {ECO:0000255|HAMAP-Rule:MF_03100, ECO:0000269|PubMed:19595721, ECO:0000269|PubMed:19596235, ECO:0000269|PubMed:19596236}. | Alternative splicing;DNA damage;DNA recombination;DNA repair;Endonuclease;Hydrolase;Metal-binding;Nuclease;Nucleus;Reference proteome;Zinc;Zinc-finger | This gene encodes a protein that is an important regulator of genome stability. The protein represents the catalytic subunit of the SLX1-SLX4 structure-specific endonuclease, which can resolve DNA secondary structures that are formed during repair and recombination processes. Two identical copies of this gene are located on the p arm of chromosome 16 due to a segmental duplication; this record represents the more centromeric copy. Alternative splicing results in multiple transcript variants. Read-through transcription also occurs between this gene and the downstream SULT1A3 (sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3) gene. [provided by RefSeq, Nov 2010]. | hsa:548593;hsa:79008; | nucleoplasm [GO:0005654]; Slx1-Slx4 complex [GO:0033557]; 5'-flap endonuclease activity [GO:0017108]; crossover junction endodeoxyribonuclease activity [GO:0008821]; metal ion binding [GO:0046872]; DNA double-strand break processing involved in repair via single-strand annealing [GO:0010792]; DNA repair [GO:0006281]; double-strand break repair via homologous recombination [GO:0000724]; negative regulation of telomere maintenance via telomere lengthening [GO:1904357]; positive regulation of t-circle formation [GO:1904431]; t-circle formation [GO:0090656]; telomere maintenance via telomere lengthening [GO:0010833]; telomeric D-loop disassembly [GO:0061820] | 24076221_Data show that three structure-selective endonucleases, SLX1-SLX4, MUS81-EME1, and GEN1, define two pathways of Holliday junctions (HJs) resolution in HeLa cells. 27354282_SLX4-SLX1 Protein-independent Down-regulation of MUS81-EME1 Protein by HIV-1 Viral Protein R (Vpr). 32601218_Folate stress induces SLX1- and RAD51-dependent mitotic DNA synthesis at the fragile X locus in human cells. | ENSMUSG00000059772 | Slx1b | 786.53672 | 0.9759529 | -0.0351165242 | 0.13282754 | 6.979016e-02 | 7.916425e-01 | 9.407812e-01 | No | Yes | 922.576321 | 123.042063 | 918.829100 | 119.731754 | |
| ENSG00000132613 | 92154 | MTSS2 | protein_coding | Q765P7 | FUNCTION: Involved in plasma membrane dynamics. Potentiated PDGF-mediated formation of membrane ruffles and lamellipodia in fibroblasts, acting via RAC1 activation (PubMed:14752106). May function in actin bundling (PubMed:14752106). {ECO:0000269|PubMed:14752106}. | Actin-binding;Alternative splicing;Cell projection;Coiled coil;Cytoplasm;Phosphoprotein;Reference proteome | hsa:92154; | actin cytoskeleton [GO:0015629]; cortical actin cytoskeleton [GO:0030864]; intrinsic component of the cytoplasmic side of the plasma membrane [GO:0031235]; lamellipodium [GO:0030027]; ruffle membrane [GO:0032587]; actin binding [GO:0003779]; actin monomer binding [GO:0003785]; GTPase activator activity [GO:0005096]; phosphatidylinositol-4,5-bisphosphate binding [GO:0005546]; phospholipid binding [GO:0005543]; small GTPase binding [GO:0031267]; activation of GTPase activity [GO:0090630]; cell projection assembly [GO:0030031]; cellular response to platelet-derived growth factor stimulus [GO:0036120]; lamellipodium organization [GO:0097581]; membrane organization [GO:0061024]; plasma membrane organization [GO:0007009]; ruffle assembly [GO:0097178] | 20875796_these data indicates that the interaction between full-length Abba and Rac1 is implicated in membrane deformation and subjected to a growth factor-mediated regulation through the C-terminal sequence. 25669885_The ABBA motif in cyclin A is required for its proper degradation in prometaphase through competing with BUBR1 for the same site on CDC20 | ENSMUSG00000033763 | Mtss2 | 2378.96144 | 1.0988468 | 0.1359902775 | 0.09597782 | 2.014988e+00 | 1.557525e-01 | 5.451797e-01 | No | Yes | 3303.322381 | 382.620446 | 2909.320896 | 329.386126 | ||
| ENSG00000133069 | 9911 | TMCC2 | protein_coding | O75069 | FUNCTION: May be involved in the regulation of the proteolytic processing of the amyloid precursor protein (APP) possibly also implicating APOE. {ECO:0000269|PubMed:21593558}. | Alternative splicing;Coiled coil;Endoplasmic reticulum;Membrane;Methylation;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:9911; | endomembrane system [GO:0012505]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; amyloid precursor protein metabolic process [GO:0042982] | ENSMUSG00000042066 | Tmcc2 | 264.81308 | 0.6424773 | -0.6382826254 | 0.18670637 | 1.149775e+01 | 6.968054e-04 | 4.258017e-02 | No | Yes | 246.596910 | 38.532624 | 364.192055 | 55.098689 | |||
| ENSG00000133316 | 54663 | WDR74 | protein_coding | Q6RFH5 | FUNCTION: Regulatory protein of the MTREX-exosome complex involved in the synthesis of the 60S ribosomal subunit (PubMed:26456651). Participates in an early cleavage of the pre-rRNA processing pathway in cooperation with NVL (PubMed:29107693). Required for blastocyst formation, is necessary for RNA transcription, processing and/or stability during preimplantation development (By similarity). {ECO:0000250|UniProtKB:Q8VCG3, ECO:0000269|PubMed:26456651, ECO:0000269|PubMed:29107693}. | Alternative splicing;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repeat;WD repeat | hsa:54663; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; preribosome, large subunit precursor [GO:0030687]; blastocyst formation [GO:0001825]; ribosomal large subunit biogenesis [GO:0042273]; RNA metabolic process [GO:0016070]; rRNA processing [GO:0006364] | 26456651_results suggest that WDR74 is a novel regulatory protein of the MTR4-exsosome complex whose interaction is regulated by NVL2 and is involved in ribosome biogenesis 29107693_knockdown of WDR74 leads to significant defects in the pre-rRNA cleavage within the internal transcribed spacer 1, occurring in an early stage of the processing pathway. When the dissociation of WDR74 from the MTR4-containing exonuclease complex was impaired upon expression of mutant NVL2, the same processing defect, with partial migration of WDR74 from the nucleolus towards the nucleoplasm, was observed. 30594465_Through direct interactions with Smad proteins, WDR74 enhances TGF-beta-mediated phosphorylation and nuclear accumulation of Smad2 and Smad3. 31838084_WDR74 induces nuclear beta-catenin accumulation and activates Wnt-responsive genes to promote lung cancer growth and metastasis. 32005977_WDR74 modulates melanoma tumorigenesis and metastasis through the RPL5-MDM2-p53 pathway. | ENSMUSG00000042729 | Wdr74 | 1386.60225 | 0.6587545 | -0.6021872043 | 0.12099223 | 2.420863e+01 | 8.644415e-07 | 4.478559e-04 | No | Yes | 1455.858725 | 189.656096 | 2131.849579 | 271.316175 | ||
| ENSG00000133624 | 79970 | ZNF767P | transcribed_unprocessed_pseudogene | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | 593.89811 | 1.0736798 | 0.1025638265 | 0.13618338 | 5.679063e-01 | 4.510921e-01 | 7.917941e-01 | No | Yes | 635.458052 | 121.246319 | 602.911631 | 112.558353 | |||||||||
| ENSG00000133794 | 406 | ARNTL | protein_coding | O00327 | FUNCTION: Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. ARNTL/BMAL1 positively regulates myogenesis and negatively regulates adipogenesis via the transcriptional control of the genes of the canonical Wnt signaling pathway. Plays a role in normal pancreatic beta-cell function; regulates glucose-stimulated insulin secretion via the regulation of antioxidant genes NFE2L2/NRF2 and its targets SESN2, PRDX3, CCLC and CCLM. Negatively regulates the mTORC1 signaling pathway; regulates the expression of MTOR and DEPTOR. Controls diurnal oscillations of Ly6C inflammatory monocytes; rhythmic recruitment of the PRC2 complex imparts diurnal variation to chemokine expression that is necessary to sustain Ly6C monocyte rhythms. Regulates the expression of HSD3B2, STAR, PTGS2, CYP11A1, CYP19A1 and LHCGR in the ovary and also the genes involved in hair growth. Plays an important role in adult hippocampal neurogenesis by regulating the timely entry of neural stem/progenitor cells (NSPCs) into the cell cycle and the number of cell divisions that take place prior to cell-cycle exit. Regulates the circadian expression of CIART and KLF11. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The NPAS2-ARNTL/BMAL1 heterodimer positively regulates the expression of MAOA, F7 and LDHA and modulates the circadian rhythm of daytime contrast sensitivity by regulating the rhythmic expression of adenylate cyclase type 1 (ADCY1) in the retina. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence (PubMed:23229515). CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3' (PubMed:23229515). The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3' (PubMed:23229515). Essential for the rhythmic interaction of CLOCK with ASS1 and plays a critical role in positively regulating CLOCK-mediated acetylation of ASS1 (PubMed:28985504). Plays a role in protecting against lethal sepsis by limiting the expression of immune checkpoint protein CD274 in macrophages in a PKM2-dependent manner (By similarity). Regulates the diurnal rhythms of skeletal muscle metabolism via transcriptional activation of genes promoting triglyceride synthesis (DGAT2) and metabolic efficiency (COQ10B) (By similarity). {ECO:0000250|UniProtKB:Q9WTL8, ECO:0000269|PubMed:11441146, ECO:0000269|PubMed:12738229, ECO:0000269|PubMed:18587630, ECO:0000269|PubMed:23785138, ECO:0000269|PubMed:23955654, ECO:0000269|PubMed:24005054, ECO:0000269|PubMed:28985504}.; FUNCTION: (Microbial infection) Regulates SARS coronavirus-2/SARS-CoV-2 entry and replication in lung epithelial cells probably through the post-transcriptional regulation of ACE2 and interferon-stimulated gene expression. {ECO:0000269|PubMed:34545347}. | 3D-structure;Acetylation;Activator;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is a basic helix-loop-helix protein that forms a heterodimer with CLOCK. This heterodimer binds E-box enhancer elements upstream of Period (PER1, PER2, PER3) and Cryptochrome (CRY1, CRY2) genes and activates transcription of these genes. PER and CRY proteins heterodimerize and repress their own transcription by interacting in a feedback loop with CLOCK/ARNTL complexes. Defects in this gene have been linked to infertility, problems with gluconeogenesis and lipogenesis, and altered sleep patterns. The protein regulates interferon-stimulated gene expression and is an important factor in viral infection, including COVID-19. [provided by RefSeq, Oct 2021]. | hsa:406; | aryl hydrocarbon receptor complex [GO:0034751]; chromatin [GO:0000785]; chromatoid body [GO:0033391]; CLOCK-BMAL transcription complex [GO:1990513]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; transcription regulator complex [GO:0005667]; aryl hydrocarbon receptor binding [GO:0017162]; DNA binding [GO:0003677]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription factor binding [GO:0140297]; E-box binding [GO:0070888]; Hsp90 protein binding [GO:0051879]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; transcription cis-regulatory region binding [GO:0000976]; circadian regulation of gene expression [GO:0032922]; circadian rhythm [GO:0007623]; negative regulation of cold-induced thermogenesis [GO:0120163]; negative regulation of fat cell differentiation [GO:0045599]; negative regulation of glucocorticoid receptor signaling pathway [GO:2000323]; negative regulation of TOR signaling [GO:0032007]; negative regulation of transcription, DNA-templated [GO:0045892]; oxidative stress-induced premature senescence [GO:0090403]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of circadian rhythm [GO:0042753]; positive regulation of protein acetylation [GO:1901985]; positive regulation of skeletal muscle cell differentiation [GO:2001016]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; regulation of cell cycle [GO:0051726]; regulation of cellular senescence [GO:2000772]; regulation of hair cycle [GO:0042634]; regulation of insulin secretion [GO:0050796]; regulation of neurogenesis [GO:0050767]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; regulation of type B pancreatic cell development [GO:2000074]; response to redox state [GO:0051775]; spermatogenesis [GO:0007283] | 11875063_The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. 12897057_BMAL1 and CLOCK have roles in circadian system control 14750904_Transcripts of BMAL1 underwent circadian oscillation. 16474406_a molecular genetic screen in mammalian cells to identify mutants of the circadian transcriptional activators CLOCK and BMAL1. 16507006_Observational study of gene-disease association. (HuGE Navigator) 16528748_Observational study of gene-disease association. (HuGE Navigator) 16628007_the CLOCK(NPAS2)/BMAL1 complex is post-translationally regulated by cry1 and cry2 17274950_We have examined the circadian expression of clock genes in human leukocytes and found that Bmal1 mRNA showed weak rhythm. 17457720_variations associated with seasonal affective disorder 17728404_Observational study of gene-disease association. (HuGE Navigator) 17728404_rat models to human provides evidence of a causative role of Bmal1 variants in pathological components of the metabolic syndrome 17994337_CLOCK/BMAL1-mediated activation of PER1 by AP1 and E-Box elements is distinct from peripheral transcriptional modulation via cAMP-induced CREB and C/EBP. 18228528_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18411297_DEC1, along with DEC2, plays a role in the finer regulation and robustness of the molecular clock CLOCK/BMAL1 18430226_The regulators of clock-controlled transcription PER2, CRY1 and CRY2 differ in their capacity to interact with each single component of the BMAL1-CLOCK heterodimer and, in the case of BMAL1, also in their interaction sites. 18517031_Individual PER3 rhythms correlated significantly with sleep-wake timing and the timing of melatonin and cortisol, but those of PER2 and BMAL1 did not reach significance. 18587630_The transcription of human nocturnin gene displayed circadian oscillations in Huh7 cells (a human hepatoma cell line) and was regulated by CLOCK/BMAL1 heterodimer via the E-box of nocturnin promoter. 18663240_Several new polymorphisms in ARNTL gene are reported. 19277210_The shRNA barcode screening technique identified ARNTL as being involved in p53 regulation. cells having suppressed ARNTL are unable to arrest upon p53 activation associated with an inability to activate the p53 target gene p21(CIP1). 19296127_expression of BMAL1 is significantly associated with lymph node metastasis and poor prognosis in breast cancer 19328558_Observational study of gene-disease association. (HuGE Navigator) 19470168_Observational study of gene-disease association. (HuGE Navigator) 19693801_Observational study of gene-disease association. (HuGE Navigator) 19839995_Observational study of gene-disease association. (HuGE Navigator) 19861541_Epigenetic inactivation of the circadian clock gene BMAL1 is associated with hematologic malignancies. 19861640_BMAL1 mRNA levels were correlated only with age (beta = -.50, p < .001). 19912323_results suggest that a peripheral molecular clock, as reflected in the dampened expression of the clock genes BMAL1 in total leukocytes, is altered in Parkinson's disease patients 19913121_Observational study of gene-disease association. (HuGE Navigator) 19934327_Observational study of gene-disease association. (HuGE Navigator) 20072116_Observational study of gene-disease association. (HuGE Navigator) 20174623_Observational study of gene-disease association. (HuGE Navigator) 20180986_Observational study of gene-disease association. (HuGE Navigator) 20222832_There was no circadian rhythm of bmal1(brain and muscle ARNT-like 1) and cry1(cryptochrome 1) in PBMC of preterm neonates 20368993_ARNTL and NPAS2 SNPs were associated with reproduction and with seasonal variation. 20368993_Observational study of gene-disease association. (HuGE Navigator) 20446921_In this salivary gland cell line, there is a rhythm in the core oscillator components BMAL1 and REV-ERBalpha, an indication that circadian-based transcriptional regulation can be modelled in this peripheral cell type. 20554694_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20861012_ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression 21363938_Data indicate that fetal adrenal gland showed circadian expression of Bmal-1 and Per-2 as well as of Mt1 and Egr-1. 21380491_Overexpression of the Bmal1 gene and reduced expression of the Per1 gene may thus be useful predictors of liver metastasis. 21411511_Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. 21799909_rhythmic modulation of circulating microRNAs targeting Bmal1 21966465_Data conclude that BMAL1 is a crucial factor in the regulation of energy homeostasis, and disorders of the functions of BMAL1 lead to the development of metabolic syndrome. 21990077_Examines the effect of acute sleep deprivation under light conditions on the expression of hPer2 and hBmal1. 40-hour acute sleep deprivation under light conditions may affect the expression of hPer2 in PBMCs. 22105622_This study demonistraed that BMAL1 exhibited rhythmic expression in the control group, but not the Adult attention-deficit hyperactivity disorder group. 22217103_treatment with isoprenaline or dexamethasone induces circadian expression of hPer1, hPer2, hPer3, and hBMAL1 22510946_findings indicate that BMAL1 has a critical role in MPM and could serve as an attractive therapeutic target for MPM 22990020_Data indicate that SNPs in SERPINE1 and ARNTL and an nucleotide polymorphisms (SNPs) associated with the expression of MUC3 were robustly associated with circulating levels of PAI-1. 23003921_DNA methylation levels at different CpG sites of CLOCK, BMAL1 and PER2 genes were analyzed in sixty normal-weight, overweight and obese women following a 16-week weight reduction program. 23033538_Data indicate that Clock-Bmal1 regulates the response to glucocorticoids in peripheral tissues through acetylation of the glucocorticoid receptor (GR), possibly antagonizing the biologic actions of diurnally fluctuating circulating cortisol. 23129285_Expression of the CLOCK, BMAL1, and PER1 circadian genes in human oral mucosa cells as dependent on CLOCK gene polymorphic variants. 23206673_The data suggest that the impairment of the BMAL1 clock gene expression is closely associated with GDM susceptibility. 23229515_The phospho-mimicking S78E mutant of BMAL1 efficiently blocks DNA binding, which provides a molecular rationale for the possibility of rhythmic binding of CLOCK-BMAL1 during circadian cycle. 23323702_These data document for the first time that the expression of BMAL1, PER3, PPARD and CRY2 genes is altered in gestational diabetes compared to normal pregnant women. deranged expression of clock genes may play a pathogenic role in GDM. 23337503_this study demonstrated that BMAL1 was modified with an O-linked beta-N-acetylglucosamine (O-GlcNAc), which stabilized BMAL1 and enhanced its transcriptional activity. 23358160_The SNPs most strongly associated in the single-marker analysis of the combined Danish samples were rs4757144 in ARNTL (P=3.78 x 10-6) and rs8057927 in CDH13. 23395176_O-GlcNAc transferase (OGT) promotes expression of BMAL1/CLOCK target genes and affects circadian oscillation of clock genes in vitro and in vivo. 23449886_Variants in ARNTL have been associated with seasonality and seasonal affective disorder, phenotypes that could reflect circadian rhythm disruption. 23546644_Studies indicate that in the cytoplasm, PER3 protein heterodimerizes with PER1, PER2, CRY1, and CRY2 proteins and enters into the nucleus, resulting in repression of CLOCK-BMAL1-mediated transcription. 23563360_Bmal1 is a tumor suppressor, capable of suppressing cancer cell growth and invasiveness. 23584858_Data suggest that clock genes Per1, Cry1, Clock, and Bmal1 and their protein products may be directly involved in the daytime-dependent regulation and adaptation of hormone synthesis. 23606611_there is significant daily variation in PER2, PER3, and ARNTL1 expression with earlier timing of expression in women than in men 24005054_Knockdown of either BMAL1 or Period1 in human anagen hair follicles significantly prolonged anagen. 24103761_These results suggest that DNA methylation of the BMAL1 gene is critical for interfering with circadian rhythms. 24277452_The progression-free survival of patients with high Bmal1 expression is significantly longer than that of patients with low Bmal1 expression. 24418196_The rhythm of Bmal1 mRNA in human plaque-derived vascular smooth muscle cells is altered. 24636202_The results of this study suggest that the ARNTL gene may be associated with the lithium prophylactic response in bipolar illness. 25070164_There is not a significant difference in the expression of CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension regardless of patient genotype. 25175925_These results suggested that ARNTL may be a tumor suppressor and is epigenetically silenced in ovarian cancer. 25182847_Data indicate that the inner nuclear membrane protein MAN1 directly binds the transcription activator BMAL1 promoter and enhances its transcription. 25203321_FAL1 associates with the epigenetic repressor BMI1 and regulates its stability in order to modulate the transcription of a number of genes including CDKN1A. 25310406_PER1 and BMAL1 operate as cell-autonomous modulators of human pigmentation and may be targeted for future therapeutic strategies 25480789_temporal signals of fasting and refeeding hormones regulate the transcription of Bmal1, a key transcription activator of molecular clock, in the liver 25741730_Bmal1-dependent oscillators of arginine vasopressin neurons modulate the coupling of the suprachiasmatic nucleus. 25799324_rs2290036-C variant of ARNTL was over-represented in psychosis patients, and the variants rs934945-G and rs10462023-G of PER2 were associated with a more severe psychotic disorder 26134245_CLOCK, ARNTL, and NPAS2 gene polymorphisms may have a role in seasonal variations in mood and behavior 26164627_these findings suggest that sumoylation plays a critical role in the spatiotemporal co-activation of CLOCK-BMAL1 by CBP for immediate-early Per induction and the resetting of the circadian clock. 26168277_In men undergoing acute total sleep deprivation, BMAL1 gene expression was decreased in skeletal muscle compared with controls. 26247999_possible circadian rhythm in full-term placental expression 26283580_a 4-locus CSNK1E haplotype encompassing the rs1534891 SNP (Z-score=2.685, permuted p=0.0076) and a 3-locus haplotype in ARNTL (Z-score=3.269, permuted p=0.0011) showed a significant association with Bipolar Disorder 26370682_found that overexpression of both Clock and Bmal1 suppressed cell growth 26507264_ARNTL and PER1 were associated with PD. 26562283_Data suggest that cryptochromes mediate periodic binding of Ck2b (casein kinase 2beta) to Bmal1 (aryl hydrocarbon receptor nuclear translocator-like protein) and thus inhibit Bmal1-Ser90 phosphorylation by Ck2a (casein kinase 2alpha). [SYNOPSIS] 26657859_define a regulatory mechanism that links chondrocyte BMAL1 to the maintenance and repair of cartilage 26683776_Bmal1 could directly bind to the p53 gene promoter and thereby transcriptionally activate the downstream tumor suppressor pathway in a p53-dependent manner in pancreatic tumors. 26753996_Our results indicate that activation of TGF-beta1 promotes the transcriptional induction of BMAL1. 26782499_The BMAL1 rs2278749 T/C was associated with Alzheimer disease (AD) risk, and T carriers in BMAL1 rs2278749 T/C showed a higher risk of AD than did non-carriers. 26850841_when overexpressed, c-MYC is able to repress Per1 transactivation by BMAL1/CLOCK via targeting selective E-box sequences. Importantly, upon serum stimulation, MYC was detected in BMAL1 protein complexes 26873744_research describes an association between changes in the methylation of the BMAL1 gene with the intervention and the effects of a weight loss intervention on blood lipids levels 26915801_decreased expression of Bmal1 is correlated with tumor progression and poor prognosis in pancreatic ductal adenocarcinoma, with potential to be used as a biomarker for diagnosis and prognosis 27060253_Synchronized cells exhibit an autonomous ultradian mitochondrial respiratory activity which is abrogated by silencing the master clock gene BMAL1. 27117143_The level of BMAL1 expression in granulosa cells the polycystic ovary syndrome (PCOS) group was lower than that of the group without PCOS. We also analyzed estrogen synthesis and aromatase expression in KGN cell lines. Both were downregulated after BMAL1 and SIRT1 knock-down and, conversely, upregulated after overexpression treatments of these two genes in KGN cells. 27253997_Bmal1 is a key clock gene to involve in cartilage homeostasis mediated through sirt1. 27285754_PI3K-PTEN upregulated-mTORC1 and mTORC2 complex plays a critical role in controlling BMAL1, establishing a connection between PI3K signaling and the regulation of circadian rhythm, ultimately resulting in deregulated BMAL1 in tumor cells with disrupted PI3K signaling 27373683_TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1 27739341_Methylation at cg05733463 of ARNTL was significantly higher in bipolar disorder (BD) than in controls. Thus, the study suggests that altered epigenetic regulation of ARNTL might provide a mechanistic basis for better understanding circadian rhythms and mood swings in BD. 27821487_our results identified Bmal1 as a novel tumor suppressor gene that elevates the sensitivity of cancer cells to paclitaxel, with potential implications as a chronotherapy timing biomarker in tongue squamous cell carcinoma 27883893_Study found rhythmic methylation of BMAL1 was altered in Alzheimer's disease brains and fibroblasts and correlated with transcription cycles. Results indicate that cycles of DNA methylation contribute to the regulation of BMAL1 rhythms in the brain. 27884645_NR1D1 and BMAL1 mRNA and protein levels were significantly reduced in OA compared to normal cartilage. In cultured human chondrocytes, a clear circadian rhythmicity was observed for NR1D1 and BMAL1. 27913791_this study shows that BMAL1 can regulate cellular innate immunity against specific RNA viruses 28487473_These results suggest that the circadian clock system can be recovered through BMAL1 expression induced by aza-dC within a day. 28543681_M. tuberculosis infection caused enhanced MMP-1, -9, and miR-223 expression, with inhibited BMAL1 expression. MiR-223 modulated BMAL1 expression via the direct binding of BMAL1 3'-UTR. 28708003_ARNTL rs7107287 was associated with a cyclothymic temperament, depressive and stress symptoms 29172799_lost BMAL1 and Ki-67 overexpression are associated with poor overall survival of nasopharyngeal carcinoma patients 29217191_Determined a novel role of TNF-alpha in inducing Bmal1 via dual calcium dependent pathways; Roralpha was up-regulated in the presence of Ca(2+) influx and Rev-erbalpha was down-regulated in the absence of that. 29276151_BMAL1 Deficiency Contributes to Mandibular Dysplasia by Upregulating MMP3. 29324865_results of this study suggest that genetic variability in the ARNTL and CLOCK genes might be associated with risk for multiple sclerosis 29396463_YY1 transcriptionally activated miR-135b and formed a 'miR-135b-BMAL1-YY1' loop, which holds significant predictive and prognostic value for patients with pancreatic cancer. 29508277_It identified BMAL1 as a cAMP-responsive coactivator of HDAC5 to regulate hepatic gluconeogenesis 29871923_This work identifies UBE2O as a critical regulator in the ubiquitin-proteasome system, which modulates BMAL1 transcriptional activity and circadian function by promoting BMAL1 ubiquitination and degradation under normal physiological conditions. 29935055_Ubiquitin E3 ligase TRAF2 reduces BMAL1 protein level.BMAL1 interacts with the zinc finger domain in TRAF2.TRAF2 promotes BMAL1 ubiquitination and degradation.TRAF2 attenuates the BMAL1 transcriptional activity. 29943823_the messenger RNA expression of brain and muscle ARNT-like 1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK) genes is altered by low doses (5 mJ/cm(2) ) of ultraviolet B in the immortalized HaCat human keratinocyte cell line. 30005017_The basal level of HIV transcription on antiretroviral therapy can vary significantly and is modulated by the circadian regulator BMAL-1, amongst other factors. 30195196_A two-way ANOVA revealed significant differences between the clock genes profiles of controls and RBD patients for hPer1 (P = 0.0324), hPer2 (P = 0.0039), hBmal1 (P = 0.0160), and hNr1d1 (P = 0.0046). 30287810_our present research illustrates a novel insight into the circadian clock proteins, CLOCK and BMAL1, which can promote the proliferation, migration, and invasion of cancer cells by affecting the formation of F-actin. 30316844_study revealed that TP, as a Bmal1-enhancing natural compound, alleviated redox imbalance via strengthening Keap1/Nrf2 antioxidant defense pathway and ameliorating mitochondrial dysfunction in a Bmal1-dependent manner 30375470_we found that knockout of BMAL1 gene in two mammalian cells affects the apoptotic response and invasion properties differently in respect to the contribution to carcinogenesis. 30502053_Study found that H. pylori up-regulated the expression of LIN28A which directly bound to BMAL1 promoter, activating the transcription of BMAL1 and increases its expression. BMAL1 in turn promoted transcription of TNF-alpha by directly binding to the E-box elements on its promoter to increase its secretion. 30523262_these results suggest a role for hBMAL1a protein isoform as a negative regulator of the mammalian molecular clock 30621723_GSEA assay found that ARNTL was associated with cell cycle and ectopic ARNTL overexpression could induce G2-M phase arrest. 30843665_The study indicates that SNPs in BMAL1 genes might contribute to the clinical outcome of gastric cancer through their impact on gene expression. 30943793_PER2 showed relatively stable oscillations compared with BMAL1. 30985884_Impaired decidualization caused by downregulation of circadian clock gene BMAL1 contributes to human recurrent miscarriagedagger. 31099187_PIWIL1 suppresses circadian rhythms through GSK3beta-induced phosphorylation and degradation of CLOCK and BMAL1 in cancer cells. 31244920_Bmal1 coordinates temporal expressions of DBP (a MRP2 activator), REV-ERBalpha (an E4BP4 repressor) and E4BP4 (a MRP2 repressor), generating diurnal MRP2 expression. 31252075_Study showed that abnormal expression levels of Per2, Clock, and Bmal1 were detected in patients with traumatic brain injury-related sleep disorders. 31300350_We identified BMAL1 as a potential marker and functional player in resistance to clinically-in-use VEGFA-neutralizing antibody (bevacizumab) in preclinical, clinical and cellular mechanistic studies of colorectal cancer. 31355331_our findings reveal a new pathway, initiated by the autophagic removal of ARNTL, that facilitates ferroptosis induction. 31580742_rs3816360 and rs2290035 Polymorphisms in BMAL1 Genes are associated with Risk of Lung Cancer. 31612734_Association of insulin resistance with polymorphic variants of Clock and Bmal1 genes: A case-control study. 31919052_Circadian Regulator CLOCK Recruits Immune-Suppressive Microglia into the GBM Tumor Microenvironment. 32041778_The E3 ubiquitin ligase STUB1 attenuates cell senescence by promoting the ubiquitination and degradation of the core circadian regulator BMAL1. 32225100_MYC-Associated Factor MAX is a Regulator of the Circadian Clock. 32231148_BMAL1 Suppresses Proliferation, Migration, and Invasion of U87MG Cells by Downregulating Cyclin B1, Phospho-AKT, and Metalloproteinase-9. 32284354_BMAL1 coordinates energy metabolism and differentiation of pluripotent stem cells. 32316196_Extracellular Acidosis Promotes Metastatic Potency via Decrease of the BMAL1 Circadian Clock Gene in Breast Cancer. 32319143_ARNT-dependent CCR8 reprogrammed LDH isoform expression correlates with poor clinical outcomes of prostate cancer. 32506468_Circadian genes polymorphisms, night work and prostate cancer risk: Findings from the EPICAP study. 32522976_The circadian clock gene Bmal1 facilitates cisplatin-induced renal injury and hepatization. 32784474_Clock Protein Bmal1 and Nrf2 Cooperatively Control Aging or Oxidative Response and Redox Homeostasis by Regulating Rhythmic Expression of Prdx6. 32858102_ARID1A-dependent permissive chromatin accessibility licenses estrogen-receptor signaling to regulate circadian rhythms genes in endometrial cancer. 32925059_Epigenetic Regulation of BMAL1 with Sleep Disturbances and Alzheimer's Disease. 33106415_The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. 33111200_Transcriptome analysis of the circadian clock gene BMAL1 deletion with opposite carcinogenic effects. 33114015_Elevated CLOCK and BMAL1 Contribute to the Impairment of Aerobic Glycolysis from Astrocytes in Alzheimer's Disease. 33374803_Proinflammatory Cytokines Perturb Mouse and Human Pancreatic Islet Circadian Rhythmicity and Induce Uncoordinated beta-Cell Clock Gene Expression via Nitric Oxide, Lysine Deacetylases, and Immunoproteasomal Activity. 33788000_Alterations in Rev-ERBalpha/BMAL1 ratio and glycated hemoglobin in rotating shift workers: the EuRhythDia study. 33791812_Clock gene Bmal1 controls diurnal rhythms in expression and activity of intestinal carboxylesterase 1. 33792447_The circadian clock gene Bmal1: Role in COVID-19 and periodontitis. 33799903_Genetic Association of Hepatitis C-Related Mixed Cryoglobulinemia: A 10-Year Prospective Study of Asians Treated with Antivirals. 33888702_Circadian clock dysfunction in human omental fat links obesity to metabolic inflammation. 34065633_BMAL1 Knockdown Leans Epithelial-Mesenchymal Balance toward Epithelial Properties and Decreases the Chemoresistance of Colon Carcinoma Cells. 34124933_Rewiring of Lactate-Interleukin-1beta Autoregulatory Loop with Clock-Bmal1: a Feed-Forward Circuit in Glioma. 34160901_Circadian clock protein BMAL1 regulates melanogenesis through MITF in melanoma cells. 34261484_Decreased expression of the clock gene Bmal1 is involved in the pathogenesis of temporal lobe epilepsy. 34481895_IL-1beta induces changes in expression of core circadian clock components PER2 and BMAL1 in primary human chondrocytes through the NMDA receptor/CREB and NF-kappaB signalling pathways. 34515147_Downregulation of Arntl mRNA Expression in Women with Hypertension: A Case-Control Study. 34542664_L-Theanine inhibits melanoma cell growth and migration via regulating expression of the clock gene BMAL1. 34727291_BMAL1 induces colorectal cancer metastasis by stimulating exosome secretion. 34807912_NF-kappaB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1. 34815366_BMAL1 may be involved in angiogenesis and peritumoral cerebral edema of human glioma by regulating VEGF and ANG2. 35112498_Mutational scanning identified amino acids of the CLOCK exon 19-domain essential for circadian rhythms. 35263812_Effects of BMAL1 on dentinogenic differentiation of dental pulp stem cells via PI3K/Akt/mTOR pathway. | ENSMUSG00000055116 | Arntl | 190.92453 | 0.9475701 | -0.0776953798 | 0.20847335 | 1.386556e-01 | 7.096212e-01 | 9.109532e-01 | No | Yes | 261.407737 | 57.921262 | 260.302065 | 56.375952 | |
| ENSG00000134146 | 89978 | DPH6 | protein_coding | Q7L8W6 | FUNCTION: Amidase that may catalyze the last step of diphthamide biosynthesis using ammonium and ATP (PubMed:23169644). Diphthamide biosynthesis consists in the conversion of an L-histidine residue in the translation elongation factor (EEF2) to diphthamide (By similarity). {ECO:0000250|UniProtKB:Q12429, ECO:0000269|PubMed:23169644}. | ATP-binding;Alternative splicing;Ligase;Nucleotide-binding;Phosphoprotein;Reference proteome | PATHWAY: Protein modification; peptidyl-diphthamide biosynthesis. | hsa:89978; | cytosol [GO:0005829]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; diphthine-ammonia ligase activity [GO:0017178]; peptidyl-diphthamide biosynthetic process from peptidyl-histidine [GO:0017183] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23169644_The previously uncharacterized human gene ATPBD4 is the ortholog of yeast YLR143W, a diphthamide synthetase, and fully rescues the deletion of YLR143W in yeast. | ENSMUSG00000057147 | Dph6 | 188.06558 | 0.9403201 | -0.0887761054 | 0.21419322 | 1.714959e-01 | 6.787857e-01 | 9.008142e-01 | No | Yes | 238.304316 | 47.125069 | 233.268570 | 45.125945 | |
| ENSG00000134297 | 51054 | PLEKHA8P1 | transcribed_processed_pseudogene | 34299245_PLEKHA8P1 Promotes Tumor Progression and Indicates Poor Prognosis of Liver Cancer. | 101.71499 | 0.8933110 | -0.1627656334 | 0.28330828 | 3.291396e-01 | 5.661662e-01 | 8.528406e-01 | No | Yes | 118.393516 | 31.124676 | 126.454242 | 32.489315 | |||||||||
| ENSG00000134452 | 84893 | FBH1 | protein_coding | Q8NFZ0 | FUNCTION: 3'-5' DNA helicase and substrate-recognition component of the SCF(FBH1) E3 ubiquitin ligase complex that plays a key role in response to stalled/damaged replication forks (PubMed:11956208, PubMed:23393192). Involved in genome maintenance by acting as an anti-recombinogenic helicase and preventing extensive strand exchange during homologous recombination: promotes RAD51 filament dissolution from stalled forks, thereby inhibiting homologous recombination and preventing excessive recombination (PubMed:17724085, PubMed:19736316). Also promotes cell death and DNA double-strand breakage in response to replication stress: together with MUS81, promotes the endonucleolytic DNA cleavage following prolonged replication stress via its helicase activity, possibly to eliminate cells with excessive replication stress (PubMed:23319600, PubMed:23361013). Plays a major role in remodeling of stalled DNA forks by catalyzing fork regression, in which the fork reverses and the two nascent DNA strands anneal (PubMed:25772361). In addition to the helicase activity, also acts as the substrate-recognition component of the SCF(FBH1) E3 ubiquitin ligase complex, a complex that mediates ubiquitination of RAD51, leading to regulate RAD51 subcellular location (PubMed:25585578). {ECO:0000269|PubMed:11956208, ECO:0000269|PubMed:17724085, ECO:0000269|PubMed:19736316, ECO:0000269|PubMed:23319600, ECO:0000269|PubMed:23361013, ECO:0000269|PubMed:25585578, ECO:0000269|PubMed:25772361}. | ATP-binding;Alternative splicing;Chromosome;DNA damage;DNA repair;DNA-binding;Helicase;Hydrolase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein ubiquitination. {ECO:0000269|PubMed:25585578}. | This gene encodes a member of the F-box protein family, members of which are characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into three classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbx class. It contains an F-box motif and seven conserved helicase motifs, and has both DNA-dependent ATPase and DNA unwinding activities. Alternatively spliced transcript variants encoding distinct isoforms have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:84893; | chromatin [GO:0000785]; nucleus [GO:0005634]; SCF ubiquitin ligase complex [GO:0019005]; 3'-5' DNA helicase activity [GO:0043138]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA helicase activity [GO:0003678]; DNA translocase activity [GO:0015616]; double-stranded DNA binding [GO:0003690]; single-stranded DNA binding [GO:0003697]; cell death [GO:0008219]; cellular response to DNA damage stimulus [GO:0006974]; DNA catabolic process, endonucleolytic [GO:0000737]; double-strand break repair via homologous recombination [GO:0000724]; negative regulation of chromatin binding [GO:0035562]; negative regulation of double-strand break repair via homologous recombination [GO:2000042]; positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage [GO:1902231]; positive regulation of protein phosphorylation [GO:0001934]; protein ubiquitination [GO:0016567]; recombinational repair [GO:0000725]; replication fork processing [GO:0031297]; replication fork protection [GO:0048478]; response to intra-S DNA damage checkpoint signaling [GO:0072429] | 11956208_hFBH1 exhibited DNA-dependent ATPase and DNA unwinding activities that displace duplex DNA in the 3' to 5' direction. 16385451_Observational study of gene-disease association. (HuGE Navigator) 17724085_These findings suggest that the hFBH1 helicase is a functional human orthologue of budding yeast Srs2 that also possesses self-regulation properties necessary to execute its recombination functions. 19736316_Data show that the human Fbh1 (hFbh1) helicase accumulates at sites of DNA damage or replication stress in a manner dependent fully on its helicase activity and partially on its conserved F box. 23319600_FBH1 helicase activity is required for the efficient induction of DSBs and apoptosis specifically in response to DNA replication stress. 23361013_FBH1 helicase activity is required to eliminate cells with excessive replication stress through the generation of MUS81-induced DNA double-strand breaks. 23393192_Ubiquitylation affects FBH1 interaction with the RAD51 nucleoprotein filament, but not its translocase and helicase activities. 23466708_FBH1 inactivation appears to contribute to oncogenic transformation by allowing survival of cells undergoing replicative stress due to external factors such as UV irradiation. 23677613_The study reports a mechanism that controls the degradation of FBH1 after DNA damage. 24108124_FBH1 restraining RAD51 DNA binding under unperturbed growth conditions to prevent unwanted or unscheduled DNA recombination. 25585578_FBH1 acts as a negative regulator of RAD51 function in human cells 28317220_study does not provide evidence for the contribution of rare non-synonymous FBXO18 variations to the genetic etiol - ogy of schizophrenia in the Japanese population. 29467415_Report a requirement for PARP2 in stabilizing replication forks that encounter base excision repair (BER) intermediates through Fbh1-dependent regulation of Rad51. Whereas PARP2 is dispensable for tolerance of cells to single stranded breaks or homologous recombination dysfunction, it is redundant with PARP1 in BER. | ENSMUSG00000058594 | Fbh1 | 2508.46877 | 0.9978025 | -0.0031737629 | 0.08924292 | 1.263460e-03 | 9.716450e-01 | 9.924630e-01 | No | Yes | 2749.801263 | 273.880850 | 2679.159800 | 260.865129 |
| ENSG00000134548 | 80763 | SPX | protein_coding | Q9BT56 | FUNCTION: Plays a role as a central modulator of cardiovascular and renal function and nociception. Plays also a role in energy metabolism and storage. Inhibits adrenocortical cell proliferation with minor stimulation on corticosteroid release (By similarity). {ECO:0000250}.; FUNCTION: [Spexin-1]: Acts as a ligand for galanin receptors GALR2 and GALR3 (PubMed:17284679, PubMed:24517231). Intracerebroventricular administration of the peptide induces an increase in arterial blood pressure, a decrease in both heart rate and renal excretion and delayed natriuresis. Intraventricular administration of the peptide induces antinociceptive activity. Also induces contraction of muscarinic-like stomach smooth muscles. Intraperitoneal administration of the peptide induces a reduction in food consumption and body weight. Inhibits long chain fatty acid uptake into adipocytes (By similarity). {ECO:0000250, ECO:0000269|PubMed:17284679, ECO:0000269|PubMed:24517231}.; FUNCTION: [Spexin-2]: Intracerebroventricular administration of the peptide induces a decrease in heart rate, but no change in arterial pressure, and an increase in urine flow rate. Intraventricular administration of the peptide induces antinociceptive activity (By similarity). {ECO:0000250}. | Amidation;Cleavage on pair of basic residues;Cytoplasmic vesicle;Hormone;Reference proteome;Secreted;Signal | The protein encoded by this gene is a hormone involved in modulation of cardiovascular and renal function. It has also been shown in rats to cause weight loss. Several transcript variants have been found for this gene. [provided by RefSeq, Feb 2016]. | hsa:80763; | cytoplasm [GO:0005737]; dense core granule [GO:0031045]; extracellular space [GO:0005615]; transport vesicle [GO:0030133]; neuropeptide hormone activity [GO:0005184]; type 2 galanin receptor binding [GO:0031765]; type 3 galanin receptor binding [GO:0031766]; long-chain fatty acid import into cell [GO:0044539]; negative regulation of appetite [GO:0032099]; negative regulation of heart rate [GO:0010459]; negative regulation of renal sodium excretion [GO:0035814]; positive regulation of gastro-intestinal system smooth muscle contraction [GO:1904306]; positive regulation of systemic arterial blood pressure [GO:0003084]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of sensory perception of pain [GO:0051930] | 19193193_Taken together, although C12ORF39 is not a secreted small peptide, it can also be secreted to play a role in the biological functions of the placenta. 22038051_newly identified peptides derived from the NPQ/spexin precursor contribute to CNS-mediated control of arterial blood pressure and salt and water balance and modulate nociceptive responses 24517231_Data from ligand-receptor interaction studies suggest that human spexin-1 and zebrafish spexin-2 activate galanin receptors GALR2/GALR3 (but not GALR1); thus spexins appear to be natural ligands for human/Xenopus/zebrafish GALR2/GALR3. 26211893_Circulating spexin levels are low in T2DM patients and negatively related to blood glucose and lipids suggesting that the peptide may play a role in glucose and lipid metabolism in T2DM. 26967115_The results suggest that C12orf39, CSTA, and CALCB are novel ATF4 target genes, and that C12orf39 promoter activity is activated by ATF4 through amino acid response element. 27218269_Lower circulating levels of Spexin in obese children compared with their normal-weight counterparts and the ability to discriminate obese and normal-weight groups based on Spexin concentration enabled us to suggest a potential role for this novel peptide in childhood obesity. 28626942_Data suggest that, in response to oral glucose (as in oral glucose tolerance test), adolescent serum spexin levels are not significantly correlated with body composition, fitness, or blood biochemical measurements; these studies were conducted in adolescents with either healthy normal weight, obesity, type 2 diabetes, or obesity plus type 2 diabetes. 29137471_Serum levels of both spexin and kisspetin show negative correlation with obesity and insulin resistance in women. 30031679_SPX levels are positively influenced by glucose intolerance in pregnant women with Gestational Diabetes Mellitus (GDM), while they decrease in control women without GDM. 30254709_Lower circulating levels of SPX in adults are modestly associated with components of metabolic syndrome and are sex-specific. 30305242_SPX is a novel regulator of lipid metabolism in murine 3T3-L1 and human adipocytes. 30673665_serum SPX levels significantly decreased in obese children and negatively correlated with insulin resistance and pancreatic beta cell function indicators 31109216_Spexin levels were significantly higher in women with gestational diabetes and closely related to HOMA-IR in the third trimester pregnancy. 32068104_Maternal and umbilical cord blood subfatin and spexin levels in patients with gestational diabetes mellitus. 32139252_Different spexin level in obese vs normal weight children and its relationship with obesity related risk factors. 33161073_Spexin-expressing neurons in the magnocellular nuclei of the human hypothalamus. 34010726_The effect of umbilical cord blood spexin, free 25(OH) vitamin D3 and adipocytokine levels on intrauterine growth and anthropometric measurements in newborns. 34944507_The Association of Serum Circulating Neuropeptide Q and Chemerin Levels with Cardiometabolic Risk Factors among Patients with Metabolic Syndrome. | ENSMUSG00000071112 | Spx | 24.16955 | 0.9936722 | -0.0091581406 | 0.57989189 | 2.489446e-04 | 9.874115e-01 | No | Yes | 25.788193 | 8.518585 | 23.652058 | 7.696086 | ||
| ENSG00000135338 | 167691 | LCA5 | protein_coding | Q86VQ0 | FUNCTION: Involved in intraflagellar protein (IFT) transport in photoreceptor cilia. {ECO:0000250|UniProtKB:Q80ST9}. | Cell projection;Cilium;Coiled coil;Cytoplasm;Cytoskeleton;Disease variant;Leber congenital amaurosis;Phosphoprotein;Protein transport;Reference proteome;Transport | This gene encodes a protein that is thought to be involved in centrosomal or ciliary functions. Mutations in this gene cause Leber congenital amaurosis type V. Alternatively spliced transcript variants are described. [provided by RefSeq, Oct 2009]. | hsa:167691; | axoneme [GO:0005930]; ciliary basal body [GO:0036064]; cilium [GO:0005929]; photoreceptor connecting cilium [GO:0032391]; protein-containing complex binding [GO:0044877]; intraciliary transport [GO:0042073]; photoreceptor cell maintenance [GO:0045494]; protein transport [GO:0015031] | 16082399_Macular coloboma-type LCA shows genetic heterogeneity and it is not possible to establish a phenotype-genotype correlation with LCA5 and macular coloboma. 17546029_The LCA5 gene on chromosome 6q14 encodes the ciliary protein lebercilin associated with Leber congenital amaurosis type 5. 18000884_Data report the identification of three novel LCA5 mutations (3/3 homozygous) in three families confirming the modest implication of this gene in this series (3/179; 1.7%). 18334959_This is the second report of LCA5 mutations causing Leber congenital amaurosis. 19172513_Observational study of gene-disease association. (HuGE Navigator) 19172513_This result shows that mutation in LCA5 is likely to be a rare genetic cause in Koreans 19503738_Leber congenital amaurosis 2 patients with LCA5 mutation had evidence of retained photoreceptors mainly in the central retina; retinal remodeling was present in pericentral regions 19800048_OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin 21850168_A novel LCA5 mutation is present in a Pakistani family with Leber congenital amaurosis and cataracts. 23946133_Identification of novel LCA5 mutations in patients with Leber congenital amaurosis and retinitis pigmentosa. 24144451_This work reveals a higher frequency of LCA5 mutations in a Spanish Leber congenital amaurosis cohort than in other populations. 27067258_The authors report novel biallelic LCA5 mutations, Ala212Pro and Tyr441Cys, as causing cone dystrophy. | ENSMUSG00000032258 | Lca5 | 158.90704 | 0.8069903 | -0.3093767068 | 0.24384193 | 1.593371e+00 | 2.068452e-01 | 5.991169e-01 | No | Yes | 181.025770 | 44.064584 | 214.645131 | 51.360878 | |
| ENSG00000135679 | 4193 | MDM2 | protein_coding | Q00987 | FUNCTION: E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as a ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of p53/TP53. Promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation. Component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in stabilizing p53/TP53. Also component of the TRIM28/KAP1-ERBB4-MDM2 complex which links growth factor and DNA damage response pathways. Mediates ubiquitination and subsequent proteasome degradation of DYRK2 in nucleus. Ubiquitinates IGF1R and SNAI1 and promotes them to proteasomal degradation (PubMed:12821780, PubMed:15053880, PubMed:15195100, PubMed:15632057, PubMed:16337594, PubMed:17290220, PubMed:19098711, PubMed:19219073, PubMed:19837670, PubMed:19965871, PubMed:20173098, PubMed:20385133, PubMed:20858735, PubMed:22128911). Ubiquitinates DCX, leading to DCX degradation and reduction of the dendritic spine density of olfactory bulb granule cells (By similarity). Ubiquitinates DLG4, leading to proteasomal degradation of DLG4 which is required for AMPA receptor endocytosis (By similarity). Negatively regulates NDUFS1, leading to decreased mitochondrial respiration, marked oxidative stress, and commitment to the mitochondrial pathway of apoptosis (PubMed:30879903). Binds NDUFS1 leading to its cytosolic retention rather than mitochondrial localization resulting in decreased supercomplex assembly (interactions between complex I and complex III), decreased complex I activity, ROS production, and apoptosis (PubMed:30879903). {ECO:0000250|UniProtKB:P23804, ECO:0000269|PubMed:12821780, ECO:0000269|PubMed:15053880, ECO:0000269|PubMed:15195100, ECO:0000269|PubMed:15632057, ECO:0000269|PubMed:16337594, ECO:0000269|PubMed:17290220, ECO:0000269|PubMed:19098711, ECO:0000269|PubMed:19219073, ECO:0000269|PubMed:19837670, ECO:0000269|PubMed:19965871, ECO:0000269|PubMed:20173098, ECO:0000269|PubMed:20385133, ECO:0000269|PubMed:20858735, ECO:0000269|PubMed:22128911, ECO:0000269|PubMed:30879903}. | 3D-structure;Alternative splicing;Apoptosis;Cytoplasm;Host-virus interaction;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | This gene encodes a nuclear-localized E3 ubiquitin ligase. The encoded protein can promote tumor formation by targeting tumor suppressor proteins, such as p53, for proteasomal degradation. This gene is itself transcriptionally-regulated by p53. Overexpression or amplification of this locus is detected in a variety of different cancers. There is a pseudogene for this gene on chromosome 2. Alternative splicing results in a multitude of transcript variants, many of which may be expressed only in tumor cells. [provided by RefSeq, Jun 2013]. | hsa:4193; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endocytic vesicle membrane [GO:0030666]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; transcription repressor complex [GO:0017053]; 5S rRNA binding [GO:0008097]; disordered domain specific binding [GO:0097718]; enzyme binding [GO:0019899]; identical protein binding [GO:0042802]; ligase activity [GO:0016874]; NEDD8 ligase activity [GO:0061663]; p53 binding [GO:0002039]; protein domain specific binding [GO:0019904]; protein N-terminus binding [GO:0047485]; ribonucleoprotein complex binding [GO:0043021]; SUMO transferase activity [GO:0019789]; ubiquitin binding [GO:0043130]; ubiquitin protein ligase activity [GO:0061630]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; amyloid fibril formation [GO:1990000]; apoptotic process [GO:0006915]; atrial septum development [GO:0003283]; atrioventricular valve morphogenesis [GO:0003181]; blood vessel development [GO:0001568]; blood vessel remodeling [GO:0001974]; cardiac septum morphogenesis [GO:0060411]; cellular response to actinomycin D [GO:0072717]; cellular response to gamma radiation [GO:0071480]; cellular response to hypoxia [GO:0071456]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; endocardial cushion morphogenesis [GO:0003203]; establishment of protein localization [GO:0045184]; negative regulation of DNA damage response, signal transduction by p53 class mediator [GO:0043518]; negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator [GO:1902254]; negative regulation of signal transduction by p53 class mediator [GO:1901797]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of mitotic cell cycle [GO:0045931]; positive regulation of muscle cell differentiation [GO:0051149]; positive regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032436]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein autoubiquitination [GO:0051865]; protein destabilization [GO:0031648]; protein localization to nucleus [GO:0034504]; protein polyubiquitination [GO:0000209]; protein ubiquitination [GO:0016567]; protein-containing complex assembly [GO:0065003]; proteolysis involved in cellular protein catabolic process [GO:0051603]; regulation of cell cycle [GO:0051726]; regulation of heart rate [GO:0002027]; regulation of protein catabolic process [GO:0042176]; regulation of transcription by RNA polymerase II [GO:0006357]; response to antibiotic [GO:0046677]; transcription factor catabolic process [GO:0036369]; traversing start control point of mitotic cell cycle [GO:0007089]; ubiquitin-dependent protein catabolic process [GO:0006511]; ventricular septum development [GO:0003281] | 10561590_mdm2 has binding sites for phosphorylation by CK2 11718560_protein interaction mapping with mouse p19ARF 11744695_a regulatory loop exists in which Hdm2 regulates the intracellular localization of Hdmx, and nuclear Hdmx regulates several functions of Hdm2 (ubiquitin ligase activity and p53 nuclear export). 11764099_higher expression in childhood leukemias with poor prognosis compared to long-term survivors 11779693_Normal cells may induce full-length MDM2 in response to oncogenic challenges to protect against premature cell cycle progression. MDM2 is regulated by p53 especially durnig embryogenesis where MDM2 function is inhibited. 11839563_findings suggest that amplification and overexpression of HMGIC and possibly MDM2 might be important genetic events that may contribute to malignant transformation of benign pleomorphic adenoma 11839577_absence of nucleolar or nucleoplasmic p14ARF/Hdm2 complexes in Reed Sternberg cells in Hodgkin's lymphoma associated with expression of alternatively spliced Hdm2 transcripts 11859876_MDM2 overexpression correlates with favorable prognosis in human breast cancer 11867628_p53 ativation by nitric oxide involves down-regulation 11877395_role for IGF-I in the regulation of the MDM2/p53/p21 signaling pathway during DNA damage 11894120_Antisense of MDM2 enhance therapeutic efficiency of irinotecan in colon cancer 11923280_Akt enhances Mdm2-mediated ubiquitination and degradation of p53. 11925449_description of a novel MDM2 binding interface in p53 that plays a regulatory role in MDM2-dependent ubiquitination of p53 11939408_Novel splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines 11953423_MDMX, when exceedingly overexpressed, inhibits MDM2-mediated p53 degradation by competing with MDM2 for p53 binding 11953887_Cirrhotic livers reveal genetic changes in the MDM2-P14ARF system of cell cycle regulators. 11956627_promoter usage of mdm2 gene in human breast cancer 11960368_Phosphorylation of HDM2 by Akt, and protein binding 11960904_summarize the current understanding of post-translational modifications and their effect on conformation-based functional relationship between Mdm2 and p53 11964305_MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites; a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. 11983168_Cyclin G expression also results in reduced phosphorylation of human Hdm2 at S166. 12032546_p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. 12052755_expression significantly correlated with favorable prognosis in esophageal squamous cell carcinoma in p53-negative patients but not p53-positive patients 12068014_MDM2 can inhibit PCAF-mediated p53 acetylation and activation. 12080472_MDM2 inhibition of p53 induces E2F1 transactivation via p21 12082526_RNA polymerase III transcription can be derepressed by hdm2 12082592_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 12085228_Multiple interacting domains contribute to p14ARF mediated inhibition of MDM2 12110584_implications of phosphorylation in p53 regulation 12138177_Data suggest a model in which p53 directly recruits a TRRAP/acetyltransferase complex to the mdm2 gene to activate transcription. In addition, this study defines a novel mechanism utilized by the p53 tumor suppressor to regulate gene expression. 12150820_MDM2 has been characterized as a protein that binds to and facilitates degradation of the tumor suppressor p53. Splice variants of MDM2 transcripts have been identified in both tumors and normal tissues. 12167711_Hypophosphorylation of Mdm2 augments p53 stability. The degree of conservation in the central acidic domain of mouse and human Mdm2 proteins is high. 12208736_The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. 12231395_MAPK as an upstream regulator of mdm2 expression 12232053_MDM2 is not required for p53 proteasomal degradation regulated by NADPH quinone oxidoreductase 1 12297306_p14ARF promotes accumulation of (H)Mdm2 conjugated to the small ubiquitin-like protein SUMO-1. 12381304_Results describe the thermodynamic and kinetic-binding parameters for the interaction between MDM2 and p53 proteins. 12393902_MDM2 ubiquitination is enhanced by MDMX 12393906_Mdm2 is sumoylated during nuclear translocation by RanBP2 and then further sumoylated once in the nucleus by PIASxbeta and PIAS1 12414343_Overexpression of mdm2 was seen predominantly in blastoid mantle cell leukemia, seemed unrelated to gain of chromosome 12, did not reflect a high proliferative rate, but might indicate an alternative mechanism of inactivating p53. 12421820_acetylation of p53 inhibited by its ubiquination 12426395_MDM2 can promote p53 deacetylation by recruiting a complex containing HDAC1. Wild-type HDAC1 and MDM2 deacetylate p53 synergistically. 12450795_The combined expression of adenovirus E1A, Ha-RasV12, and MDM2 is sufficient to convert a normal human cell into a cancer cell. 12507556_Ubiquitination and degradation of p53 are largely controlled by Mdm2, an oncogenic E3 ligase. 12552135_role of p53 binding domain in p53 regulation 12565795_Data provide evidence of a role for MDM2 and CDK4 in the pathogenesis of carcinosarcoma. 12582944_in glioblastoma multiforme patients a complex relationship exists between the Mdm2 expression and age 12606552_Mdm2-mediated p53 ubiquitination is suppressed by HIF-1 alpha, which blocks Mdm2-mediated nuclear export of p53 12620409_La antigen dependent activation of mdm2 translation might represent an important molecular mechanism involved in BCR/ABL leukemogenesis 12646252_These data strongly suggest that Mdm2 functions as the ubiquitin ligase toward hNumb and that it induces its degradation in intact cells. 12661905_Overexpression of the Mdm2 gene product may be important in the pathogenesis of penile verrucous carcinoma since Mdm2 is a negative regulator of p53 12687276_It appears, that MDM2 overexpression, which may be p53-dependent, or also p53-independent plays an important role in leukemogenesis and/or disease progression. 12730202_two upstream open reading frames of oncogene mdm2 may play a fundamental role in regulating expression of the mdm2 gene 12750288_p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. 12782320_ubiquitination of MDM2 is cell cycle-regulated and MDM2 may play a role in cell cycle progression 12821780_Mdm2 physically associates with IGF-1R and that Mdm2 causes IGF-1R ubiquitination in an in vitro assay. Mdm2 serves as a ligase in ubiquitination of the IGF-1R and thereby causes its degradation by the proteasome system. 12860999_MDM2 promotes ubiquitination and degradation of MDMX 12874296_MDM2 regulates hdmx protein stability 12926050_Ligase dead mutants of Mdm2 did not act in a dominant negative manner to reactivate p53 and they are not oncogenes in human mammary epithelial cells. 12927808_MDM2 is regulated and degraded by GCL-1 12944468_colocalization of a nonshuttling p53 with MDM2 either in the nucleus or in the cytoplasm is sufficient for MDM2-induced p53 polyubiquitination but not degradation. 12963717_MDM2 has a role in DNA damage-induced MDMX degradation 14499615_study of MDM2 protein binding to the N-terminal domain of p53 14507994_HdmX is actively involved in the degradation of both p53 and Hdm2. 14522887_Results imply that the current paradigm for understanding Mdm2 action during oncogenesis is incomplete, and its splice variants contribute to human cancer. 14559824_PTEN inhibits MDM2 and protects p53 through both p13k/Akt-dependent and -independent pathways in ALL. 14565663_The expression and structure of HDM2 in HL cell lines was studied. Several different spliced hdm2 transcripts (mdm-sv) including five new variants lacking a functional p53 binding site were characterized. 14587869_Mdm2 is a regulator of cell growth and death [review] 14596917_interaction with nucleic acids interferes with both Hdm2/Hdm2 complex formation and auto-ubiquitination of Hdm2 in vitro; although binding of Hdm2 to p53 is not inhibited by nucleic acids, Hdm2-mediated ubiquitination of p53 is significantly decreased. 14610316_MDM-2 overexpression can block UV-induced cell cycle arrest and apoptosis by inhibiting P53 transcriptional activity 14612427_L11 functions as a negative regulator of HDM2 and there might exist in vivo an L11-HDM2-p53 pathway for monitoring ribosomal integrity 14614050_MDM2 amplification in soft tissue sarcoma patients was associated with a prognosis better than that of patients without the amplification. of the tumors with an MDM2 amplification, 40% also experienced loss of heterozygosity at 12q14-15. 14633995_MDM2 directly inhibits p21waf1/cip1 function by reducing p21waf1/cip1 stability in a ubiquitin-independent fashion 14671306_results show that low levels of Mdm2 activity induce monoubiquitination and nuclear export of p53, whereas high levels promote p53's polyubiquitination and nuclear degradation 14720195_overexpression is related to the tumorigenesis and/or tumour progression of salivary gland neoplasms 14723816_MDM2 acts as a transcriptional factor to modulate expressions of other genes involved in cell cycle regulation and transformation and was hypothesized that MDM2 directly affected NF-kappaB expression and function in a P53-independent manner. 14729628_Upregulation of p14ARF paralleled with MDM2 inhibition contributes to p53 accumulation in the nucleus in radiation-treated breast cancer cells. 14731389_MDM2 may regulate p53, depending on its levels in the cell. 14741215_Data report the effects of peptide binding on the N-terminal p53-binding domain of human MDM2. 14756544_No correlation between p53 accumulation and survival in bilharziasis associated bladder squamous cell carcinoma. 14761977_MDM2 functions as a negative regulator of p21, an effect independent of both p53 and ubiquitination 14769800_MDM2 regulates the stability of PCAF by ubiquitinating and degrading this protein 15001356_results identify an interferon-responsive protein kinase family as a novel modifier of two components of the p53 pathway, MDM2 and p21(WAF1) 15013777_acetyltransferases may modulate cellular p53 activity not only by modifying p53, but also by inactivating Mdm2 15024078_Results show that growth factor stimulation, overexpression of Akt/PKB, or loss of PTEN resulted in enhanced expression of both HIF-1alpha and HDM2. 15024701_CDK4, MDM2, SAS and GLI genes are amplified in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma 15029243_Data reveal that controlled MDM2 degradation is an important new step in p53 regulation. 15192123_potential of p53-C277Y to up-regulate MDM2 expression was similar to wild-type p53 15218947_Mdm2 in female patients is an independent prognostic factor, associated with shorter survival in brain glioma. 15308643_the MDM2-L5-L11-L23 complex functions to inhibit MDM2-mediated p53 ubiquitination and thus activates p53 15314173_These results reveal that ribosomal protein L23 is another regulator of the p53-MDM2 feedback regulation involved in cell growth. 15314174_Data show that, when overexpressed, ribosomal protein L23 inhibits HDM2-induced p53 polyubiquitination and degradation and causes a p53-dependent cell cycle arrest. 15315825_MDM2 gene spans approximately 33 kb and is divided into 12 exons. 15337531_Ras appears to attenuate p53 in SW480 cells by two independent regulatory mechanisms, the one leading to increased Mdm2-dependent p53 degradation and the other leading to a decrease in p53 transcription. 15375804_Review of MDM2 which inhibits p53 degradation in head and neck cancers 15448710_Epstein barr virus can indirectly enhance mdm2 gene expression in tumor cells that express this gene. 15485814_the central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2F and cell growth 15492852_MDM2-p73-P14ARF pathway is involved in the progression of bladder cancer to a more malignant and aggressive form. 15527798_identify Ser-166, a site previously reported as an AKT target, and Ser-188, a novel site which is the major site of phosphorylation of MDM2 by AKT in vitro 15546622_The RING domain of MDM2 mediates histone ubiquitylation and transcriptional repression. 15548678_role of Cul4A in the MDM2-mediated proteolysis of p53 15550242_A single nucleotide polymorphism is found in the MDM2 promoter and is shown to increase the affinity of the transcriptional activator Sp1, resulting in higher levels of MDM2 RNA and protein and the subsequent attenuation of the p53 pathway. 15622743_The level of MDM2 gene expression was related to the prognoses of the patients but not to FAB subtypes of acute leukemias. 15632057_Data suggest that MTBP differentially regulates the activity of MDM2 towards two of its most critical targets (itself and p53) and in doing so significantly contributes to MDM2-dependent p53 homeostasis in unstressed cells. 15644444_finding that MYCN directly modulates baseline MDM2 levels suggests a mechanism contributing to the pathogenesis of neuroblastoma and other MYC-driven malignancies through inhibition of MYC-stimulated apoptosis. 15688025_Mouse cells deficient for MK2 show reduced Mdm2 phosphorylation and elevated levels of p53 protein. 15714438_Alternative MDM2 splicing is highly associated with lung cancer. 15720184_Mdm2 activity could activate p53 tumor suppression REVIEW 15720185_MDM2 has oncogenic transformational activities independent of p53--REVIEW 15720186_MDM2 splice variant expression has been associated with advanced neoplastic disease--REVIEW 15720187_MDM2 oncogene is overexpressed in more than forty different types of malignancies--REVIEW 15723837_Regulation of the nuclear export of hdm2 mRNA provides a mechanism whereby mitogen-stimulated cells avoid p53-dependent cell cycle arrest or apoptosis by maintaining the dynamic equilibrium of the Hdm2-p53 feedback loop 15733365_The overexpression of MDM2 oncogene related to the poor status and poor prognosis of patients with childhood NHL,correlated with B status and the involved extranodal sites and the increased serum LDH level 15734740_p73, through HDM2, can oppose p53 tumor suppressor function and possibly contribute to tumorigenesis 15734743_Nbs1 is a novel p53-independent Mdm2 binding protein and links Mdm2 to the Mre11-Nbs1-Rad50-regulated DNA repair response 15742432_The abundant expression of double-minute protein 2 (MDM2) in rheumatoid arthritis may be a contributing factor to the hypoapoptotic phenotype of lining tissue through its capacity to downregulate p53 levels and effects. 15771712_MDM2 may have implications for glioma cell death susceptibility 15782125_Mdm2 and mdmx prevent ASPP1 and ASPP2 from stimulating the apoptotic function of p53 by binding and inhibiting the transcriptional activity of p53. 15788536_sites important for Hdm2-mediated ubiquitination of Hdmx after double-strand break induction 15807633_alpains participate in the down-regulation of Mdm2 in the epidermis very rapidly after UV irradiation. 15832769_p53, Bax, Bcl-2 and Mdm2 mRNA expression levels correlate with the malignant transformation of the uterine cervix 15843377_protection of p53 from MDM2 by PTEN and the damage-induced activation of PTEN by phosphorylated p53 leads to the formation of an apoptotic amplification cycle in which p53 and PTEN coordinately increase cellular apoptosis 15862297_HDM2 negatively affects the Chk2-mediated phosphorylation of p53. 15866118_expression of p53, MDM2, and p21Waf1 suggests a role for these oncoproteins in the regulation of endometrioma cell growth, but not in adenomyosis 15876864_MdmX can affect post-translational modification and stability of Mdm2 and p53 activity through interaction with ARF 15878855_beta-arrestin has a role in ubiquitination and down-regulation of the insulin-like growth factor-1 receptor by acting as adaptor for the MDM2 E3 ligase 15902285_TP53 mutation has only a limited role in the transformation of lymphoma to diffuse large B-cell lymphoma, exerting a heterogeneous influence upon phenotypic change. In contrast, dysregulation of MDM2 is frequent. 15908423_overproduction of Mdm2, resulting from a naturally occurring SNP, inhibits chromatin-bound p53 from activating the transcription of its target genes. 15916963_Results suggest that impaired deubiquitination of Hdmx/Hdm2 by HAUSP contributes to the DNA damage-induced degradation of Hdmx and transient instability of Hdm2. 15943041_besides tumor protein p53 alterations, MDM2 gene deregulation seems to be an important event in hepatocarcinogenesis 15953616_analysis of the more open conformation of the binding cleft of MDM2N (non-liganded) 15985438_Data show that expression of the ErbB-4 ICD fragment leads to its constitutive association with and tyrosine phosphorylation of Mdm2. 16027727_deletion of Mdm4 enhances the ability of Mdm2 to promote cell growth and tumor formation, indicating that Mdm4 has antioncogenic properties when Mdm2 is overexpressed 16055726_GSK3-dependent phosphorylation of Mdm2 regulates p53 abundance 16082221_ATM directly activates p53 while activating a safe-lock mechanism to inactivate the negative regulators of p53, Mdm2, and Mdmx [review] 16107876_ARF may regulate p53 acetylation and stability in part by inhibiting tripartite motif-containing 28-MDM2 binding 16142358_the p14ARF-p53-MDM2 pathway has a role in development of oral squamous cell carcinoma 16152608_2 promoter polymorphisms of MDM2 and evaluated their associations with risk of lung cancer 16152608_Observational study of gene-disease association. (HuGE Navigator) 16159876_p53-mdm2 binding is subtler than previously thought and involves global contacts such as multiple 'non-contiguous' minimally structured motifs instead of being localized to one small helix mini-domain in p53 TAD 16163388_The E3 ligase activity of MDM2 is redirected to MDMX after DNA damage and contributes to p53 activation. 16167062_T309G polymorphism for MDM2 significantly decreases the age of onset for ALL in the pediatric population, particularly within Caucasian and black pediatric communities 16170383_MDM2 stabilizes E2F1 protein through the E2F1 ubiquitination pathway. 16202543_Studies reported the discovery of a third promoter (designated P3) in intron 3 of MDM2 gene, which contains a TATA-box element and p53-DNA-binding sequences. 16203772_Observational study of gene-disease association. (HuGE Navigator) 16203772_the p53 and MDM2 promoter polymorphisms do not appear to play a role on age of colorectal cancer onset in Lynch syndrome 16212962_HIPK2 contributes to drug-induced modulation of MDM2 activity at transcriptional (through p53Ser46 phosphorylation) and posttranscriptional (through p53-independent subcellular re-localization and proteasomal degradation) levels. 16230424_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 16239061_Observational study of gene-disease association. (HuGE Navigator) 16246554_the p53-HDM2 interaction can be inhibited by a newly isolated hexylitaconic acid from the marine-derived fungus, Arthrinium 16258005_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16258514_Studies of MDM2 in more than 2000 breast carcinomas show that MDM2 is an independent negative prognostic marker. 16287156_Genetic polymorphisms in cell cycle regulatory genes MDM2 and TP53 contribute to the risk of developing lung cancer. 16287156_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16288830_Observational study of gene-disease association. (HuGE Navigator) 16337594_The MDM2 promotes Rb degradation in a proteasome-dependent and ubiquitin-independent manner. 16343421_These findings suggest that overexpression of Mdm2 can perturb a RB pathway regardless of the p53 gene status, promoting carcinogenesis. 16394138_MDM2 has critical roles in the regulation of p21 and E2F1 expression, stability and function [review] 16432196_identification of a second binding site helps stabilize the interaction between HDM2 and p53 during p53 degradation 16434608_MDM2 -309 allele and genotype frequencies did not differ between cases and controls, and ORs for breast cancer were close to the null in African-Americans and Whites. 16439685_MDM2 antagonists alone or in combination with chemotherapeutic drugs may offer a new treatment option for B-CLL. 16474402_N-terminal domain of USP7 binds two closely spaced 4-residue sites in both p53 and MDM2, falling between p53 residues 359-367 and MDM2 residues 147-159. 16478747_Observational study of gene-disease association. (HuGE Navigator) 16478747_Our data indicate that MDM2-SNP309 is a modifier of the age at colorectal cancer onset for patients whose tumors have a wild-type p53 gene. 16479015_IFIX alpha1 isoform functions as a tumor suppressor by repressing HDM2 function 16496380_Observational study of gene-disease association. (HuGE Navigator) 16496380_Results indicate that the MDM2 G/G genotype of SNP309 is associated with lung cancer risk with an odds ratio of 1.62 (95% CI: 1.06-2.50). 16510145_MdmX may have different roles in the regulation of Mdm2 activity for ubiquitination of pRB and p53 16563154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16579792_analysis of the role of p53 substrate conformation in controlling MDM2-dependent ubiquitination of p53 and identification of a link between substrate misfolding and susceptibility to ubiquitination 16621805_p90Rsk-mediated modulation of Hdm2 nuclear is linked to cytoplasmic shuttling with the diminished ability of p53 to regulate cell cycle checkpoints that ultimately leads to transformation 16624812_HDM2 can selectively down-regulate the transcription function of p53 without either degrading p53 or affecting the interaction of p53 with target promoters 16624822_Jab1 is required to remove post-translationally modified p53 in coordination with mdm2 16636310_Data show that nuclear accumulations of p53 and Mdm2 are accompanied by reductions in c-Abl and p300 in zinc-depleted human hepatoblastoma cells. 16675470_Observational study of gene-disease association. (HuGE Navigator) 16696307_study showed that MDM2 amplification and overexpression might be an early event in the growth of human gliomas 16737965_interaction of free arrestins with JNK3 and Mdm2 and their ability to regulate subcellular localization of these proteins may play an important role in the survival of photoreceptors and other neurons 16738062_Review. An alternative splice form of c-H-ras, called p19ras, is a positive regulator of p73beta via Mdm2. Implications for this previously unidentified means of regulation are discussed in light of tumor suppression and are extended to p53 and p63. 16751805_levels are reduced by nucleolin 16803902_L11, unlike L5 and L23, differentially regulates the levels of ubiquitinated p53 and MDM2 and inhibits the turnover and activity of MDM2 through a post-ubiquitination mechanism 16815295_Collectively, our results suggest that a loss of the cisplatin sensitivity is at least in part due to a lack of cisplatin-induced p53 phosphorylation, and p73 might cooperate with MDM2 to be involved in this process. 16818855_Among homozygous carriers of the common MDM2 SNP309 allele, a mutant p53 status (risk ratio of death = 2.33, 95% CI = 1.08 to 5.03)and aberrant p53 protein expression(RR = 2.61, 95% CI = 1.22 to 5.57)in breast tumors were associated with poor survival. 16818855_Observational study of gene-disease association. (HuGE Navigator) 16825430_Data suggest that the G-allele of SNP309 accelerates colorectal tumour formation only in women. 16825434_MDM2-single nucleotide polymerase 309 favours tumour selection of non-dominant negative P53 mutations in colorectal cancer, which also show an earlier age of tumour onset. 16861890_PCNA, L2DTL and the DDB1-CUL4A complex play critical and differential roles in regulating the protein stability of p53 and MDM2/HDM2 in unstressed and stressed cells. 16866370_Although the interaction between the N-termini of mdm2 and p53 blocks the transactivation activity of p53, the interaction between the central domain of mdm2 and the core domain of p53 is critical for the ubiquitination and degradation of p53. 16870621_p53 binding to the central domain of Mdm2 is regulated by phosphorylation 16876289_Observational study of gene-disease association. (HuGE Navigator) 16877339_MDM2 is therefore considered one of the mineralocorticoid-responsive genes that regulates cell proliferation of vascular smooth muscle cell (VSMCs) induced by MR-mediated aldosterone stimulation. 16883576_These results suggest that p70S6K1 regulates turnover of HDM2 protein for cancer development. 16892553_Gene amplifications are important prognostic markers for soft tissue sarcomas. 16896050_Observational study of gene-disease association. (HuGE Navigator) 16905769_Hdmx is an important determinant of the outcome of P53 activation 16914573_Observational study of gene-disease association. (HuGE Navigator) 16914573_The MDM2 promoter SNP309 is associated with the presence of hepatocellular carcinoma in Japanese patients with chronic hepatitis C. 16934800_Study evaluated Pirh2, MDM2, p53 and p21 expression after DNA damage using cancer cell lines containing wildtype, mutant and null p53 and found that unlike MDM2, Pirh2 expression was not affected by the presence of wildtype p53 in the cancer cells. 16965791_Results report the solution structure of the C2H2C4 RING domain of Hdm2(429-491), which reveals a symmetrical dimer with a unique cross-brace zinc-binding scheme. 16980628_A novel role for MDM2 in regulating cell adhesions by a mechanism that involves degrading and down-regulating the expression of E-cadherin via an endosome pathway. 16983111_Observational study of gene-disease association. (HuGE Navigator) 16984978_The cooperative effects of phenotypes determined by mdm-2, p53, and bcl-2 expression may predict survival in patients with muscle-invasive TCC of the bladder 17003841_TP53 germline mutation carriers with SNP309 G allele have earlier onset of tumours; higher prevalence of MDM2 SNP309 homozygous carriers in TP53-negative group suggests allele contributes to cancer susceptibility in Li-Fraumeni syndrome & related families 17006543_insulin-like growth factor-1 alters Mdm2-mediated GRK2 degradation, leading to enhanced GRK2 stability and increased kinase levels 17013834_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17013834_When the p73 and MDM2 polymorphisms were combined, the risk of lung cancer increased in a dose-dependent manner as the number of variant alleles increased 17018602_Stress-induced HDM2(ALT1) regulates HDM2 at two levels, RNA and protein, further modulating the p53-HDM2 interaction or interactions of HDM2 with other cell cycle regulatory proteins. 17018606_Alternate forms of MDM2 are detected after UV irradiation. Alternate forms of MDM2 place selective pressure on the cells to acquire additional alterations in the p53 pathway. 17056014_the reduction of KAP1 levels promotes p53-dependent p21 induction and inhibits cell proliferation in actinomycin D-treated cells. 17060450_analysis of mechanism of formation of the principal MDM2 isoforms, differential effects of p53 on the production of these isoforms, and differential abilities of human MDM2 isoforms as regulators of the MDM2/TSG101 and p53/MDM2 feedback control loops 17080308_MDM2 single nucleotide polymorphism accelerates familial breast carcinogenesis independently of estrogen signaling 17080308_Observational study of gene-disease association. (HuGE Navigator) 17094469_MDM2 T309G polymorphism is associated with bladder cancer 17094469_Observational study of gene-disease association. (HuGE Navigator) 17096342_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17096342_Results indicate the G allele of MDM2 SNP309 might have a protective effect on disease development in HNPCC patients and that age of diagnosis of CRC is not associated with MDM2 SNP309 or TP53 R72P either as single SNPs or combined. 17116689_Cancer-associated missense mutations targeting MDM2's central zinc finger disrupt the interaction of MDM2 with L5 and L11. 17120309_AURKA and MDM2 were identified as interesting novel amplified genes in juvenile angiofibromas 17123590_Observational study of gene-disease association. (HuGE Navigator) 17139261_p53 mediates the suppression of TR3 on MDM2 at both transcriptional and post-transcriptional level and suggest TR3 as a potential target to develop new anticancer agents that restrict MDM2-induced tumor progression. 17159902_functions of the extreme C-terminus of MDM2 can be provided by MDMX 17170710_Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity 17171684_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17188136_p53 and Ki-67 were expressed with increasing frequency, and bcl-2, p21, and mdm-2 with decreasing frequency in thyroid carcinoma progression. p27 and cyclin D1 were expressed in <15% of cases, with a trend toward decreasing expression. 17214373_Observational study of gene-disease association. (HuGE Navigator) 17226766_p72 RNA helicase may not only be involved in the p53-Mdm2 regulatory loop, but also profoundly impact on the transcriptome through various CBP/p300 and P/CAF interacting proteins. 17237821_Mdm2 and TAFII250 associate in cultured cells; the acidic domain of MDM2 is required for TAFII250-stimulated ubiquitylation and degradation of p53. 17242401_5-FU treatment triggers a ribosomal stress response so that ribosomal proteins L5, L11, and L23 are released from ribosome to activate p53 by ablating the MDM2-p53 feedback circuit 17302414_Both kinetics and free energy landscape analyses indicate that bound MDM2 unfolds in the order of p53 unbinding, tertiary unfolding, and finally secondary structure unfolding. 17310983_S7 binds to MDM2, in vitro and in vivo, and the interaction between MDM2 and S7 leads to modulation of MDM2-p53 binding by forming a ternary complex among MDM2, p53 and S7. 17327702_Review discusses the important regulatory roles of MDMX during the p53 response to ribosomal stress. 17339337_BOX-V motif of p53 has evolved the capacity to bind to enzymes that mediate either p53 phosphorylation or ubiquitination 17349959_These findings establish HIPK2 as an MDM2 target and support a model in which, upon nonsevere DNA damage, p53 represses its own phosphorylation at Ser46 due to HIPK2 degradation. 17354236_Nutlin efficiently stabilized p53 and induced downstream p53 dependent transcription and apoptosis in liposarcoma cells with amplified MDM2 in vitro 17359998_Arrestins recruit ERK1/2 and the E3 ubiquitin ligase Mdm2 to microtubulesin cells, similar to the arrestin-dependent mobilization of these proteins to the receptor. 17360557_analysis of haplotype structure and selection of the MDM2 oncogene in humans 17371838_results argue that Mdm2 is needed for full inhibition of Cdk2 activity by p21, thereby positively contributing to p53-dependent cell cycle arrest 17373842_Our analysis reveals interactions of HDM2 with the ribosomal translation elongation factor EF1alpha, 40S ribosomal protein S20, tubulins, glyceraldehyde 3-phosphate dehydrogenase, and a proteolysis-inducing factor dermicidin. 17387621_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17387621_The -309 SNP in MDM2 is associated with increased MDM2 transcription but there are inconclusive results with respect to breast cancer risk 17426254_Colocalization and interaction of MDM2-A and MDM2-B with full-length MDM2 in the nucleus have important physiologic consequences, for example, deregulation of p53 activity. 17460193_CARF may exert a vital control on p53-HDM2-p21(WAF1) pathway that is central to the cell cycle control, senescence, and DNA damage response of human cells 17468107_Che-1 as a new Pin1 and HDM2 target and confirm its important role in the cellular response to DNA damage. 17473193_Observational study of gene-disease association. (HuGE Navigator) 17473193_findings suggest that the MDM2 SNP309 may be a risk factor for the occur | ENSMUSG00000020184 | Mdm2 | 3362.60556 | 1.0657551 | 0.0918760254 | 0.09937679 | 8.565434e-01 | 3.547078e-01 | 7.272763e-01 | No | Yes | 4800.461303 | 975.831428 | 4294.018939 | 853.602819 | |
| ENSG00000135829 | 1660 | DHX9 | protein_coding | Q08211 | FUNCTION: Multifunctional ATP-dependent nucleic acid helicase that unwinds DNA and RNA in a 3' to 5' direction and that plays important roles in many processes, such as DNA replication, transcriptional activation, post-transcriptional RNA regulation, mRNA translation and RNA-mediated gene silencing (PubMed:9111062, PubMed:11416126, PubMed:12711669, PubMed:15355351, PubMed:16680162, PubMed:17531811, PubMed:20669935, PubMed:21561811, PubMed:24049074, PubMed:25062910, PubMed:24990949, PubMed:28221134). Requires a 3'-single-stranded tail as entry site for acid nuclei unwinding activities as well as the binding and hydrolyzing of any of the four ribo- or deoxyribo-nucleotide triphosphates (NTPs) (PubMed:1537828). Unwinds numerous nucleic acid substrates such as double-stranded (ds) DNA and RNA, DNA:RNA hybrids, DNA and RNA forks composed of either partially complementary DNA duplexes or DNA:RNA hybrids, respectively, and also DNA and RNA displacement loops (D- and R-loops), triplex-helical DNA (H-DNA) structure and DNA and RNA-based G-quadruplexes (PubMed:20669935, PubMed:21561811, PubMed:24049074). Binds dsDNA, single-stranded DNA (ssDNA), dsRNA, ssRNA and poly(A)-containing RNA (PubMed:9111062, PubMed:10198287). Binds also to circular dsDNA or dsRNA of either linear and/or circular forms and stimulates the relaxation of supercoiled DNAs catalyzed by topoisomerase TOP2A (PubMed:12711669). Plays a role in DNA replication at origins of replication and cell cycle progression (PubMed:24990949). Plays a role as a transcriptional coactivator acting as a bridging factor between polymerase II holoenzyme and transcription factors or cofactors, such as BRCA1, CREBBP, RELA and SMN1 (PubMed:11149922, PubMed:9323138, PubMed:9662397, PubMed:11038348, PubMed:11416126, PubMed:15355351, PubMed:28221134). Binds to the CDKN2A promoter (PubMed:11038348). Plays several roles in post-transcriptional regulation of gene expression (PubMed:28221134, PubMed:28355180). In cooperation with NUP98, promotes pre-mRNA alternative splicing activities of a subset of genes (PubMed:11402034, PubMed:16680162, PubMed:28221134, PubMed:28355180). As component of a large PER complex, is involved in the negative regulation of 3' transcriptional termination of circadian target genes such as PER1 and NR1D1 and the control of the circadian rhythms (By similarity). Acts also as a nuclear resolvase that is able to bind and neutralize harmful massive secondary double-stranded RNA structures formed by inverted-repeat Alu retrotransposon elements that are inserted and transcribed as parts of genes during the process of gene transposition (PubMed:28355180). Involved in the positive regulation of nuclear export of constitutive transport element (CTE)-containing unspliced mRNA (PubMed:9162007, PubMed:10924507, PubMed:11402034). Component of the coding region determinant (CRD)-mediated complex that promotes cytoplasmic MYC mRNA stability (PubMed:19029303). Plays a role in mRNA translation (PubMed:28355180). Positively regulates translation of selected mRNAs through its binding to post-transcriptional control element (PCE) in the 5'-untranslated region (UTR) (PubMed:16680162). Involved with LARP6 in the translation stimulation of type I collagen mRNAs for CO1A1 and CO1A2 through binding of a specific stem-loop structure in their 5'-UTRs (PubMed:22190748). Stimulates LIN28A-dependent mRNA translation probably by facilitating ribonucleoprotein remodeling during the process of translation (PubMed:21247876). Plays also a role as a small interfering (siRNA)-loading factor involved in the RNA-induced silencing complex (RISC) loading complex (RLC) assembly, and hence functions in the RISC-mediated gene silencing process (PubMed:17531811). Binds preferentially to short double-stranded RNA, such as those produced during rotavirus intestinal infection (PubMed:28636595). This interaction may mediate NLRP9 inflammasome activation and trigger inflammatory response, including IL18 release and pyroptosis (PubMed:28636595). Finally, mediates the attachment of heterogeneous nuclear ribonucleoproteins (hnRNPs) to actin filaments in the nucleus (PubMed:11687588). {ECO:0000250|UniProtKB:O70133, ECO:0000269|PubMed:10198287, ECO:0000269|PubMed:10924507, ECO:0000269|PubMed:11038348, ECO:0000269|PubMed:11149922, ECO:0000269|PubMed:11402034, ECO:0000269|PubMed:11416126, ECO:0000269|PubMed:11687588, ECO:0000269|PubMed:12711669, ECO:0000269|PubMed:15355351, ECO:0000269|PubMed:1537828, ECO:0000269|PubMed:16680162, ECO:0000269|PubMed:17531811, ECO:0000269|PubMed:19029303, ECO:0000269|PubMed:20669935, ECO:0000269|PubMed:21247876, ECO:0000269|PubMed:21561811, ECO:0000269|PubMed:22190748, ECO:0000269|PubMed:24049074, ECO:0000269|PubMed:24990949, ECO:0000269|PubMed:25062910, ECO:0000269|PubMed:28221134, ECO:0000269|PubMed:28355180, ECO:0000269|PubMed:28636595, ECO:0000269|PubMed:9111062, ECO:0000269|PubMed:9162007, ECO:0000269|PubMed:9323138, ECO:0000269|PubMed:9662397}.; FUNCTION: (Microbial infection) Plays a role in HIV-1 replication and virion infectivity (PubMed:11096080, PubMed:19229320, PubMed:25149208, PubMed:27107641). Enhances HIV-1 transcription by facilitating the binding of RNA polymerase II holoenzyme to the proviral DNA (PubMed:11096080, PubMed:25149208). Binds (via DRBM domain 2) to the HIV-1 TAR RNA and stimulates HIV-1 transcription of transactivation response element (TAR)-containing mRNAs (PubMed:9892698, PubMed:11096080). Involved also in HIV-1 mRNA splicing and transport (PubMed:25149208). Positively regulates HIV-1 gag mRNA translation, through its binding to post-transcriptional control element (PCE) in the 5'-untranslated region (UTR) (PubMed:16680162). Binds (via DRBM domains) to a HIV-1 double-stranded RNA region of the primer binding site (PBS)-segment of the 5'-UTR, and hence stimulates DHX9 incorporation into virions and virion infectivity (PubMed:27107641). Plays also a role as a cytosolic viral MyD88-dependent DNA and RNA sensors in plasmacytoid dendritic cells (pDCs), and hence induce antiviral innate immune responses (PubMed:20696886, PubMed:21957149). Binds (via the OB-fold region) to viral single-stranded DNA unmethylated C-phosphate-G (CpG) oligonucleotide (PubMed:20696886). {ECO:0000269|PubMed:11096080, ECO:0000269|PubMed:16680162, ECO:0000269|PubMed:19229320, ECO:0000269|PubMed:20696886, ECO:0000269|PubMed:21957149, ECO:0000269|PubMed:25149208, ECO:0000269|PubMed:27107641, ECO:0000269|PubMed:9892698}. | 3D-structure;ATP-binding;Acetylation;Activator;Alternative splicing;Biological rhythms;Cytoplasm;Cytoskeleton;DNA replication;DNA-binding;Direct protein sequencing;Helicase;Host-virus interaction;Hydrolase;Immunity;Inflammatory response;Innate immunity;Isopeptide bond;Manganese;Metal-binding;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;RNA-binding;RNA-mediated gene silencing;Reference proteome;Repeat;Transcription;Transcription regulation;Transcription termination;Translation regulation;Transport;Ubl conjugation;mRNA processing;mRNA splicing;mRNA transport | This gene encodes a member of the DEAH-containing family of RNA helicases. The encoded protein is an enzyme that catalyzes the ATP-dependent unwinding of double-stranded RNA and DNA-RNA complexes. This protein localizes to both the nucleus and the cytoplasm and functions as a transcriptional regulator. This protein may also be involved in the expression and nuclear export of retroviral RNAs. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 11 and 13.[provided by RefSeq, Feb 2010]. | hsa:1660; | actin cytoskeleton [GO:0015629]; centrosome [GO:0005813]; CRD-mediated mRNA stability complex [GO:0070937]; cytoplasm [GO:0005737]; cytoplasmic ribonucleoprotein granule [GO:0036464]; cytosol [GO:0005829]; membrane [GO:0016020]; nuclear body [GO:0016604]; nuclear stress granule [GO:0097165]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perichromatin fibrils [GO:0005726]; polysomal ribosome [GO:0042788]; polysome [GO:0005844]; protein-containing complex [GO:0032991]; ribonucleoprotein complex [GO:1990904]; RISC complex [GO:0016442]; RISC-loading complex [GO:0070578]; 3'-5' DNA helicase activity [GO:0043138]; 3'-5' DNA/RNA helicase activity [GO:0033679]; 3'-5' RNA helicase activity [GO:0034458]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; chromatin DNA binding [GO:0031490]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; DNA replication origin binding [GO:0003688]; double-stranded DNA binding [GO:0003690]; double-stranded RNA binding [GO:0003725]; importin-alpha family protein binding [GO:0061676]; metal ion binding [GO:0046872]; mRNA binding [GO:0003729]; nucleoside-triphosphatase activity [GO:0017111]; nucleoside-triphosphate diphosphatase activity [GO:0047429]; polysome binding [GO:1905538]; promoter-specific chromatin binding [GO:1990841]; regulatory region RNA binding [GO:0001069]; RISC complex binding [GO:1905172]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; RNA polymerase binding [GO:0070063]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II complex binding [GO:0000993]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; RNA stem-loop binding [GO:0035613]; sequence-specific mRNA binding [GO:1990825]; single-stranded 3'-5' DNA helicase activity [GO:1990518]; single-stranded DNA binding [GO:0003697]; single-stranded RNA binding [GO:0003727]; siRNA binding [GO:0035197]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; triplex DNA binding [GO:0045142]; alternative mRNA splicing, via spliceosome [GO:0000380]; cellular protein-containing complex assembly [GO:0034622]; cellular response to exogenous dsRNA [GO:0071360]; cellular response to tumor necrosis factor [GO:0071356]; CRD-mediated mRNA stabilization [GO:0070934]; DNA duplex unwinding [GO:0032508]; DNA replication [GO:0006260]; DNA-templated transcription, termination [GO:0006353]; DNA-templated viral transcription [GO:0039695]; G-quadruplex DNA unwinding [GO:0044806]; inflammatory response [GO:0006954]; innate immune response [GO:0045087]; mRNA transport [GO:0051028]; negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900152]; osteoblast differentiation [GO:0001649]; positive regulation of cytoplasmic translation [GO:2000767]; positive regulation of DNA repair [GO:0045739]; positive regulation of DNA replication [GO:0045740]; positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity [GO:2000373]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of gene silencing by miRNA [GO:2000637]; positive regulation of inflammatory response [GO:0050729]; positive regulation of innate immune response [GO:0045089]; positive regulation of interferon-alpha production [GO:0032727]; positive regulation of interferon-beta production [GO:0032728]; positive regulation of interleukin-18 production [GO:0032741]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of polysome binding [GO:1905698]; positive regulation of response to cytokine stimulus [GO:0060760]; positive regulation of RNA export from nucleus [GO:0046833]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of tumor necrosis factor production [GO:0032760]; positive regulation of viral transcription [GO:0050434]; positive regulation of viral translation [GO:1904973]; protein localization to cytoplasmic stress granule [GO:1903608]; pyroptosis [GO:0070269]; regulation of cytoplasmic translation [GO:2000765]; regulation of defense response to virus by host [GO:0050691]; regulation of mRNA processing [GO:0050684]; regulation of transcription by RNA polymerase II [GO:0006357]; rhythmic process [GO:0048511]; RNA secondary structure unwinding [GO:0010501]; small RNA loading onto RISC [GO:0070922]; targeting of mRNA for destruction involved in RNA interference [GO:0030423] | 12163469_Nuclear DNA helicase II recruitment to PML nuclear bodies(NBs) is connected with transcriptional regulation of interferon-alpha-inducible genes attached to PML NBs. 12243751_propose that NDH II operates in both nucleoplasmic and nucleolar mode, and that its redistribution reflects accumulations indicating a possible cycling of NDH II between nucleoplasm and the nucleolus 12592385_Overexpression of a truncated RHA peptide that binds to the BRCA1 carboxy-terminus prevents normal BRCA1 function & association with nuclear foci following DNA damage. It induces pleomorphic nuclei, aberrant mitoses with extra centrosomes, & tetraploidy. 14704337_DNA-PK phosphorylated recombinant NDH II in the presence of RNA 14769796_RNA helicase A in the MEF1 transcription factor complex up-regulates the MDR1 gene in multidrug-resistant cancer cells 15355351_RHA interacts with NF-kappa B p65 and functions as a transcriptional coactivator. 15995249_Results suggest that nuclear DNA helicase II plays a role in promoting the DNA processing function of Werner syndrome helicase, which in turn might be necessary for maintaining genomic stability. 16375861_Our results indicate that the nuclear import of RHA is mediated by the importin-alpha3/importin-beta-dependent pathway and suggest that the specificity for importin may regulate the functions of cargo proteins. 16527808_RHA represents the first example of cellular RNA helicases that participate in HIV-1 particle production and promote viral reverse transcription 17251188_Phosphorylated Zic2 protein makes a stable complex with RNA helicase A. 17531811_RNA interference is a conserved pathway of sequence-specific gene silencing that depends on small guide RNAs and the action of proteins assembled in the RNA-induced silencing complex (RISC). 18053790_In vitro dephosphorylation of HeLa cellular HDH II/Ku caused a significant decrease in the DNA helicase activity of this enzyme. 18519039_These results establish a link between the p53 tumor suppressor and RNA processing via hnRNPA2/B1 and RNA Helicase A. 18600529_knockdown of DHX-9 by siRNA did not inhibit the rRNA synthesis or cause the nucleolar disruption 18782589_A 21-day follow-up of the response of HCV replication to the presence and absence of RNAi indicated that RHA is a cellular factor involved in the HCV replication process. 19197335_Four genes encoding BRCA1-interacting proteins were analyzed in a cohort of 96 breast cancer individuals from high-risk non-BRCA1/BRCA2 French Canadian families. 19197335_Observational study of gene-disease association. (HuGE Navigator) 19229320_protein kinase R (PKR) is shown to interact with RNA helicase A; RHA is identified as a substrate for PKR, with phosphorylation perturbing the association of the helicase with double-stranded RNA 19309309_Results show overexpression or depletion of RHA could influence the interaction of Pol II with beta-actin and beta-actin-involved gene transcription regulation. 20385589_the preferential unwinding of RNA-containing substrates by WRN helicase is stimulated by DHX9 in vitro, both on Okazaki fragment-like hybrids and on RNA-containing 'chicken-foot' structures. 20437153_Proteomic analysis indicates that cellular proteins interacted with Kaposi's sarcoma-associated herpesvirus viral protein kinase (ORF36), and co-immunoprecipitation reactions further reveal interactions with human RNA helicase A. 20510246_DHX9 lacks base-selective contacts, forms an unspecific but important stacking interaction with the base of the bound nucleotide and the protein can hydrolyze ATP, guanosine 5'-triphosphate, cytidine 5'-triphosphate, and uridine 5'-triphosphate. 20669935_DHX9 displaced the third strand from a specific triplex DNA structure and catalyzed the unwinding with a 3' --> 5' polarity with respect to the displaced third strand. 20696886_DHX9/DHX36 represent the MyD88-dependent DNA sensors in the cytosol of plasmacytoid dendritic cells and suggest a much broader role for DHX helicases in viral sensing 20802156_observed a positive correlation of the nuclear expression of EGF receptor, RHA, and cyclin D1 in human breast cancer samples 21123178_the molecular basis for the activation of translation by RHA is recognition of target mRNA by the N-terminal domain that tethers the ATP-dependent helicase for rearrangement of the complex 5'-UTR. 21561811_DHX9 also unwound RNA-based G-quadruplexes that have been reported to occur in human transcripts 21957149_DHX9 is an important RNA sensor that is dependent on interferon-beta promoter stimulator (IPS)-1 to sense pathogenic RNA. 22162396_L1TD1 is part of the L1TD1-RHA-LIN28 complex that could influence levels of OCT4, suggesting that L1TD1 has an important role in the regulation of stemness. 22171255_RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. 22190748_RNA helicase A (RHA) is tethered to the 5' SL of collagen mRNAs by interaction with the C-terminal domain of LARP6. 23361462_The crystal structure of huma DHX9 RNA binding domains in complex with double stranded RNA. 23853588_Data suggest that M029 (an RNA-binding protein) plays pivotal role in determining cellular tropism of Myxoma virus in all mammalian cells tested; human RNA helicase A/DHX9 and protein kinase R/EIF2AK2 are two binding partners of M029. 24049074_The data implicate DHX9 in processing intra-molecular triplex H-DNA structures in vivo and support its role in the overall maintenance of genomic stability at sites of alternatively structured DNA. 24223160_RNA helicase A participates in HIV-1 RNA metabolism by multiple distinct mechanisms. 24469107_Results show RNA helicase DHX9 as an interacting partner of KIF1Bbeta and found that DHX9 is necessary for KIF1Bbeta to induce apoptosis. 24726449_The conserved lysine residues of dsRBDs play critical roles in the promotion of HIV-1 production by RNA helicase A. 24990949_results demonstrate an essential role of DHX9 in DNA replication and normal cell cycle progression. 25149208_OB-fold is involved in modulating HIV-1 RNA splicing in the context of some HIV-1 strains while it is dispensable for the activation of HIV-1 transcription. 26450900_Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. 26885691_RNA helicase A was shown to cooperate with both tumor suppressors and oncoproteins in different tumours, indicating that its specific role in cancer is strongly influenced by the cellular context. 26973242_Our results demonstrate a robust tolerance for systemic DHX9 suppression in vivo and support the targeting of DHX9 as an effective and specific chemotherapeutic approach. 27009951_study provides evidence demonstrating that the PRRSV nucleocapsid protein interacts with Nsp9 and its RNA-dependent RNA-polymerase domain and also recruits the cellular helicase DHX9 during virus infection to facilitate viral RNA synthesis and virus production 27034008_analysis of the structure, biochemistry, and biology of DHX9, its role in cancer and other human diseases [review] 28221134_Importantly, binding of Nup98 to DHX9 stimulates the ATPase activity of DHX9, and a transcriptional reporter assay suggests Nup98 supports DHX9-stimulated transcription. 28355180_an evolutionarily conserved function of DHX9, it acts as a nuclear RNA resolvase that neutralizes the immediate threat posed by Alu transposon insertions and allows these elements to evolve as tools for the post-transcriptional regulation of gene expression 28427210_our study supports the presence of a p53-independent mechanism of cell death and cell cycle arrest resulting from DHX9 inhibition. our results support the feasibility of targeting DHX9 as a chemotherapeutic approach in p53-deficient tumors. 28460433_the decreased growth of osteosarcoma cells by MCM2 or MCM3 knockdown was reversed by DHX9 overexpression, indicating that MCM2 and MCM3 activity was DHX9-dependent. 28588071_study identified the RNA helicase DHX9 as a regulator of pRNA processing; DHX9 binds to rRNA genes only upon embryonic stem cells differentiation and its activity guides TIP5 to rRNA genes and establishes heterochromatin 29742442_DHX9 interacts with PARP1, and both proteins prevent R-loop-associated DNA damage. 29796672_Data indicate that DEAH box helicase 9 (DHX9) constitutes a bidirectional regulatory mode in A-to-I editing, which is in part responsible for the dysregulated editome profile in cancer. 30111593_DHX9 enhanced NF-kappaB-dependent IL-6 promoter activation, which was directly antagonized by E3. These results indicate new roles for DHX9 in regulating cytokines in innate immunity and reveal that VACV E3 disrupts innate immune responses by targeting of DHX9. 30137501_A DNA-sensing-independent role of a nuclear RNA helicase, DHX9, has been demonstrated in stimulation of NF-kappaB-mediated innate immunity against DNA virus infection. 30341290_DHX9 (RNA helicase A) promotes the formation of pathological and non-pathological R-loops. 30406989_Unwinding assays of chemical and structural modified substrates indicate that RHA translocates efficiently along the 3' overhang of RNA, but not DNA, with a requirement of covalent continuity. Ribose-phosphate backbone lesions on both strands of the nucleic acids, especially on the 3' overhang of the loading strand, affect RHA unwinding significantly. 30463971_The current study revealed that RHA relocalized into the cytoplasm upon DENV infection and associated with viral RNA and nonstructural proteins, implying that RHA was actively engaged in the viral life cycle. 30518908_A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. 30541834_Authors show that SM binds to and colocalizes DHX9 and may counteract the antiviral function of DHX9. These data indicate that DHX9 possesses antiviral activity and that SM may suppress the antiviral functions of DHX9 through this association. 30591072_RNA G-quadruplex folding, controlled by the two DEAH-box helicases DHX36 and DHX9, impedes the scanning of the 43S preinitiation complex, promotes 80S ribosome formation within 5'-UTRs and consequently represses the translation of transcripts involved in key biological pathways. 30865310_Up-regulation of lnc-UCID, which may result from amplification of its gene locus and down-regulation of miR-148a, can promote hepatocellular carcinoma growth by preventing the interaction of DHX9 with CDK6 and subsequently enhancing CDK6 expression. 31072811_Studies uncover a fine-tuned modulation of the proto-oncogene CCND1 in Ewing sarcoma cells via alternative complexes formed by DHX9 with either EWS-FLI1 or pncCCND1_B-Sam68. 31175158_Reverse transcriptase (RT)activity in our assays indicated that RNA helicase A (RHA) in HIV-1 virions is required for the efficient catalysis of (-)cDNA synthesis during viral infection before capsid uncoating. Our study identifies RHA as a processivity factor of HIV-1 RT 31829498_Hepatitis B virus X protein modulates upregulation of DHX9 to promote viral DNA replication. 32023846_Poison-Exon Inclusion in DHX9 Reduces Its Expression and Sensitizes Ewing Sarcoma Cells to Chemotherapeutic Treatment. 32056513_DHX9 negatively regulates the anti-HBV replication effect of APOBEC3B 33143622_The Current View on the Helicase Activity of RNA Helicase A and Its Role in Gene Expression. 33526059_LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. 33837988_Dynamically probing ATP-dependent RNA helicase A-assisted RNA structure conversion using single molecule fluorescence resonance energy transfer. 34182081_SNORA42 promotes oesophageal squamous cell carcinoma development through triggering the DHX9/p65 axis. 34226554_DHX9-dependent recruitment of BRCA1 to RNA promotes DNA end resection in homologous recombination. 34329467_TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. 34512155_MARCH6 promotes Papillary Thyroid Cancer development by destabilizing DHX9. 34676915_High expression of DHX9 promotes the growth and metastasis of hepatocellular carcinoma. 34773477_DHX9 contributes to the malignant phenotypes of colorectal cancer via activating NF-kappaB signaling pathway. | ENSMUSG00000042699 | Dhx9 | 18202.10936 | 1.0498012 | 0.0701161755 | 0.06481941 | 1.172001e+00 | 2.789905e-01 | 6.668663e-01 | No | Yes | 23104.670406 | 4565.813258 | 20183.676483 | 3899.311568 | |
| ENSG00000135838 | 80896 | NPL | protein_coding | Q9BXD5 | FUNCTION: Catalyzes the cleavage of N-acetylneuraminic acid (sialic acid) to form pyruvate and N-acetylmannosamine via a Schiff base intermediate. It prevents sialic acids from being recycled and returning to the cell surface. Involved in the N-glycolylneuraminic acid (Neu5Gc) degradation pathway. Although human is not able to catalyze formation of Neu5Gc due to the inactive CMAHP enzyme, Neu5Gc is present in food and must be degraded (By similarity). Experiments show the true substrate is aceneuramate (linearized Neu5Ac) (PubMed:33895133). {ECO:0000250, ECO:0000269|PubMed:33895133}. | 3D-structure;Alternative splicing;Carbohydrate metabolism;Cytoplasm;Lyase;Reference proteome;Schiff base | PATHWAY: Amino-sugar metabolism; N-acetylneuraminate degradation. {ECO:0000269|PubMed:22692205}. | This gene encodes a member of the N-acetylneuraminate lyase sub-family of (beta/alpha)(8)-barrel enzymes. N-acetylneuraminate lyases regulate cellular concentrations of N-acetyl-neuraminic acid (sialic acid) by mediating the reversible conversion of sialic acid into N-acetylmannosamine and pyruvate. A pseudogene of this gene is located on the short arm of chromosome 2. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:80896; | cytosol [GO:0005829]; identical protein binding [GO:0042802]; N-acetylneuraminate lyase activity [GO:0008747]; carbohydrate metabolic process [GO:0005975]; N-acetylneuraminate catabolic process [GO:0019262] | 16147865_an NPL splice variant is mainly expressed in human liver, kidney and peripheral blood leukocytes 19057931_3D structure model of N-acetylneuraminate lyase from human (hNAL, EC 4.1.3.3) was created and refined | ENSMUSG00000042684 | Npl | 156.61435 | 1.0713791 | 0.0994691241 | 0.23622727 | 1.776543e-01 | 6.733969e-01 | 8.987789e-01 | No | Yes | 187.683274 | 33.368777 | 173.343060 | 29.962923 |
| ENSG00000135951 | 80705 | TSGA10 | protein_coding | Q9BZW7 | FUNCTION: Plays a role in spermatogenesis (PubMed:28905369). When overexpressed, prevents nuclear localization of HIF1A (By similarity). {ECO:0000250|UniProtKB:Q6NY15, ECO:0000269|PubMed:28905369}. | Alternative splicing;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:80705; | centriole [GO:0005814]; cytoplasm [GO:0005737]; spermatogenesis [GO:0007283] | 15107545_over-expressed in 4 of 20 hepatocellular carcinomas (HCC), 1 of 20 colon cancers, 7 of 20 ovarian cancers, 3 of 20 prostate cancers, 1 of 21 malignant melanomas, and 8 of 21 bladder cancers 16406020_The RT-PCR of TSGA10 expression may help in detection of residual clonal cells leading to early diagnosis and better prognostic qualification of the acute lymphoblastic leukemia. 18000009_TSGA10 is target of immune reactions in Autoimmune polyendocrine syndrome type 1 (APS1). 20797700_TSGA10 could influence the function of antigen presenting cells via its interaction with cytoskeletal proteins such as vimentin 21198756_TSGA10 should be considered as an autoantigen in a subset of autoimmune polyendocrine syndrome type 1 patients and also in a minority of systemic lupus erythematosus patients 24294352_Report role for mir-577/TSGA10 axis in regulation esophageal squamous cell carcinoma progression. 26573430_Overexpression of TSGA10 would induce disruption of HIF-1alpha axis and exert potent inhibitory effects on tumor angiogenesis and metastasis. 28739310_TSGA10 tends to express variants with shorter 5'UTR. 28905369_we speculate that the presence of rare sequence variants within TSGA10 may be associated with acephalic spermatozoa in humans 29520105_Results show that TSGA10 is directly regulated by mir-23a which targeted the 3'UTR of mir-23a to regulate angiogenesis. 30545223_This study showed the role of Lactobacilli in down-regulation of TSGA10, AURKC, OIP5 and AKAP4 genes. Such expression change might be involved in the anticancer effects of these Lactobacilli. The underlying mechanisms of these observations are not clear but epigenetic modulatory mechanisms may participate in this process. 30638859_the current study showed that TSGA10 could be considered as a tumor suppressor in acute myeloid leukemia 31704243_Overexpression of TSGA10, as a tumor suppressor gene, in endothelial cells resulted in decreased proliferation, migration and therefore, angiogenic activity of HUVECs and may involved HIF2a binding. 31772141_Hypoxia-induced microRNA-10b-3p promotes esophageal squamous cell carcinoma growth and metastasis by targeting TSGA10. 32086108_TSGA10 overexpression could decrease the metastatic and metabolic activity of cancer cells through its inhibitory effect on HIF-1 activity. Therefore, TSGA10 could be considered in the research for therapeutic candidates in cancer. 32285443_Novel mutations in PMFBP1, TSGA10 and SUN5: Expanding the spectrum of mutations that may cause acephalic spermatozoa. 34232471_TSGA10 as a Potential Key Factor in the Process of Spermatid Differentiation/Maturation: Deciphering Its Association with Autophagy Pathway. 34409526_Pathogenesis of acephalic spermatozoa syndrome caused by splicing mutation and de novo deletion in TSGA10. | ENSMUSG00000060771 | Tsga10 | 102.58288 | 1.0002765 | 0.0003988147 | 0.30023844 | 1.755858e-06 | 9.989427e-01 | 9.995733e-01 | No | Yes | 89.318771 | 13.776748 | 84.979478 | 12.740647 | ||
| ENSG00000136014 | 84101 | USP44 | protein_coding | Q9H0E7 | FUNCTION: Deubiquitinase that plays a key regulatory role in the spindle assembly checkpoint or mitotic checkpoint by preventing premature anaphase onset. Acts by specifically mediating deubiquitination of CDC20, a negative regulator of the anaphase promoting complex/cyclosome (APC/C). Deubiquitination of CDC20 leads to stabilize the MAD2L1-CDC20-APC/C ternary complex (also named mitotic checkpoint complex), thereby preventing premature activation of the APC/C. Promotes association of MAD2L1 with CDC20 and reinforces the spindle assembly checkpoint. Acts as a negative regulator of histone H2B (H2BK120ub1) ubiquitination. {ECO:0000269|PubMed:17443180, ECO:0000269|PubMed:22681888}. | Cell cycle;Cell division;Hydrolase;Metal-binding;Mitosis;Nucleus;Phosphoprotein;Protease;Reference proteome;Thiol protease;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | The protein encoded by this gene is a protease that functions as a deubiquitinating enzyme. The encoded protein is thought to help regulate the spindle assembly checkpoint by preventing early anaphase onset. This protein specifically deubiquitinates CDC20, which stabilizes the anaphase promoting complex/cyclosome. [provided by RefSeq, Dec 2016]. | hsa:84101; | cytoplasm [GO:0005737]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; thiol-dependent deubiquitinase [GO:0004843]; zinc ion binding [GO:0008270]; cell cycle [GO:0007049]; cell division [GO:0051301]; chromosome segregation [GO:0007059]; negative regulation of ubiquitin protein ligase activity [GO:1904667]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein deubiquitination [GO:0016579]; regulation of mitotic cell cycle spindle assembly checkpoint [GO:0090266] | 17443180_a dynamic balance of ubiquitination by the APC and deubiquitination by USP44 contributes to the generation of the switch-like transition controlling anaphase entry 21853124_These data are consistent with an important role for USP44 in regulating Cdc20-APC/C activity and suggest that high levels of this enzyme may contribute to the pathogenesis of T-cell leukemias. 22692537_Data identify Cdc20, USP44, and Wee1 as relevant Fcp1 targets. 23615962_findings implicate USP44 in negative regulation of the RNF8/RNF168 pathway 24837038_USP44 is epigenetically inactivated in colorectal adenomas, but this alone is not sufficient to cause aneuploidy in colorectal neoplasia. 26232424_USP44+ Cancer Stem Cell Subclones Contribute to Breast Cancer Aggressiveness by Promoting Vasculogenic Mimicry 27880911_USP44 contributes to N-CoR functions in regulating gene expression and is required for efficient invasiveness of triple-negative breast cancer cells. 28492742_USP44 protein was widely expressed in most of the tumor samples and no clear association could be established between its expression and DNA ploidy status or tumor size 28520534_These data imply a complex picture where regulating factors such as OCT4 may interact with other epigenetic mechanisms to regulate USP44 expression in pluripotent stem cells and testes. 28544703_In summary, we report that the combination of USP44 expression and DNA ploidy status might serve as an independent prognostic marker in gastric cancer. 30622230_USP44 promotes the tumorigenesis of prostate cancer cells partly by stabilizing EZH2. 31197957_USP44 inhibited AKT signaling by stabilizing PTEN in non-small cell lung cancer cells. 31968013_identified USP44 as a positive regulator of MITA; USP44 is recruited to MITA following DNA virus infection and removes K48-linked polyubiquitin moieties from MITA at K236, therefore prevents MITA from proteasome mediated degradation; findings suggest that USP44 plays a specific and critical role in the regulation of innate immune response against DNA viruses 32164618_Ubiquitin-specific protease-44 inhibits the proliferation and migration of cells via inhibition of JNK pathway in clear cell renal cell carcinoma. 32285989_USP44 suppresses proliferation and enhances apoptosis in colorectal cancer cells by inactivating the Wnt/beta-catenin pathway via Axin1 deubiquitination. 32445925_Existence of circFOXO3-miR-143-3p-USP44 axis in GC cells was confirmed by RNA-binding protein immunoprecipitation, luciferase reporter assay, and an RNA pull-down experiments. All the data indicate that circFOXO3 promotes GC cell proliferation and migration by upregulating USP44 expression via targeting of miR-143-3p. 32644293_The deubiquitinase USP44 promotes Treg function during inflammation by preventing FOXP3 degradation. 32956592_USP44 hypermethylation promotes cell proliferation and metastasis in breast cancer. 33647455_CRADD and USP44 mutations in intellectual disability, mild lissencephaly, brain atrophy, developmental delay, strabismus, behavioural problems and skeletal anomalies. 35079021_USP44 regulates irradiation-induced DNA double-strand break repair and suppresses tumorigenesis in nasopharyngeal carcinoma. | ENSMUSG00000020020 | Usp44 | 121.13421 | 1.1184205 | 0.1614626958 | 0.26394995 | 3.749990e-01 | 5.402919e-01 | 8.403757e-01 | No | Yes | 141.269394 | 28.144050 | 121.841231 | 23.869862 | |
| ENSG00000136044 | 55198 | APPL2 | protein_coding | Q8NEU8 | FUNCTION: Multifunctional adapter protein that binds to various membrane receptors, nuclear factors and signaling proteins to regulate many processes, such as cell proliferation, immune response, endosomal trafficking and cell metabolism (PubMed:26583432, PubMed:15016378, PubMed:24879834). Regulates signaling pathway leading to cell proliferation through interaction with RAB5A and subunits of the NuRD/MeCP1 complex (PubMed:15016378). Plays a role in immune response by modulating phagocytosis, inflammatory and innate immune responses. In macrophages, enhances Fc-gamma receptor-mediated phagocytosis through interaction with RAB31 leading to activation of PI3K/Akt signaling. In response to LPS, modulates inflammatory responses by playing a key role on the regulation of TLR4 signaling and in the nuclear translocation of RELA/NF-kappa-B p65 and the secretion of pro- and anti-inflammatory cytokines. Also functions as a negative regulator of innate immune response via inhibition of AKT1 signaling pathway by forming a complex with APPL1 and PIK3R1 (By similarity). Plays a role in endosomal trafficking of TGFBR1 from the endosomes to the nucleus (PubMed:26583432). Plays a role in cell metabolism by regulating adiponecting ans insulin signaling pathways and adaptative thermogenesis (PubMed:24879834) (By similarity). In muscle, negatively regulates adiponectin-simulated glucose uptake and fatty acid oxidation by inhibiting adiponectin signaling pathway through APPL1 sequestration thereby antagonizing APPL1 action (By similarity). In muscles, negativeliy regulates insulin-induced plasma membrane recruitment of GLUT4 and glucose uptake through interaction with TBC1D1 (PubMed:24879834). Plays a role in cold and diet-induced adaptive thermogenesis by activating ventromedial hypothalamus (VMH) neurons throught AMPK inhibition which enhances sympathetic outflow to subcutaneous white adipose tissue (sWAT), sWAT beiging and cold tolerance (By similarity). Also plays a role in other signaling pathways namely Wnt/beta-catenin, HGF and glucocorticoid receptor signaling (PubMed:19433865) (By similarity). Positive regulator of beta-catenin/TCF-dependent transcription through direct interaction with RUVBL2/reptin resulting in the relief of RUVBL2-mediated repression of beta-catenin/TCF target genes by modulating the interactions within the beta-catenin-reptin-HDAC complex (PubMed:19433865). May affect adult neurogenesis in hippocampus and olfactory system via regulating the sensitivity of glucocorticoid receptor. Required for fibroblast migration through HGF cell signaling (By similarity). {ECO:0000250|UniProtKB:Q8K3G9, ECO:0000269|PubMed:15016378, ECO:0000269|PubMed:19433865, ECO:0000269|PubMed:24879834, ECO:0000269|PubMed:26583432}. | 3D-structure;Alternative splicing;Cell cycle;Cell membrane;Cell projection;Chromosomal rearrangement;Cytoplasm;Cytoplasmic vesicle;Endosome;Membrane;Nucleus;Reference proteome | The protein encoded by this gene is one of two effectors of the small GTPase RAB5A/Rab5, which are involved in a signal transduction pathway. Both effectors contain an N-terminal Bin/Amphiphysin/Rvs (BAR) domain, a central pleckstrin homology (PH) domain, and a C-terminal phosphotyrosine binding (PTB) domain, and they bind the Rab5 through the BAR domain. They are associated with endosomal membranes and can be translocated to the nucleus in response to the EGF stimulus. They interact with the NuRD/MeCP1 complex (nucleosome remodeling and deacetylase /methyl-CpG-binding protein 1 complex) and are required for efficient cell proliferation. A chromosomal aberration t(12;22)(q24.1;q13.3) involving this gene and the PSAP2 gene results in 22q13.3 deletion syndrome, also known as Phelan-McDermid syndrome. [provided by RefSeq, Oct 2011]. | hsa:55198; | cytoplasmic vesicle [GO:0031410]; early endosome membrane [GO:0031901]; early phagosome [GO:0032009]; early phagosome membrane [GO:0036186]; endosome [GO:0005768]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; macropinosome [GO:0044354]; membrane [GO:0016020]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; ruffle membrane [GO:0032587]; vesicle [GO:0031982]; identical protein binding [GO:0042802]; phosphatidylinositol binding [GO:0035091]; phosphatidylserine binding [GO:0001786]; protein homodimerization activity [GO:0042803]; protein-containing complex binding [GO:0044877]; adiponectin-activated signaling pathway [GO:0033211]; cell cycle [GO:0007049]; cellular response to hepatocyte growth factor stimulus [GO:0035729]; cold acclimation [GO:0009631]; diet induced thermogenesis [GO:0002024]; glucose homeostasis [GO:0042593]; negative regulation of cellular response to insulin stimulus [GO:1900077]; negative regulation of cytokine production involved in inflammatory response [GO:1900016]; negative regulation of fatty acid oxidation [GO:0046322]; negative regulation of glucose import [GO:0046325]; negative regulation of neural precursor cell proliferation [GO:2000178]; negative regulation of neurogenesis [GO:0050768]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of Fc-gamma receptor signaling pathway involved in phagocytosis [GO:1905451]; positive regulation of macropinocytosis [GO:1905303]; positive regulation of phagocytosis, engulfment [GO:0060100]; protein homotetramerization [GO:0051289]; protein import into nucleus [GO:0006606]; regulation of fibroblast migration [GO:0010762]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; regulation of innate immune response [GO:0045088]; regulation of toll-like receptor 4 signaling pathway [GO:0034143]; signal transduction [GO:0007165]; signaling [GO:0023052]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 15016378_identification of a pathway directly linking the small GTPase Rab5, a key regulator of endocytosis, to signal transduction and mitogenesis via APPL1 and APPL2, two Rab5 effectors 18034774_The findings suggest a role for APPL1 and APPL2 protein as dynamic scaffolds that modulate RAB5-associated signaling endosomal membranes by their ability to undergo domain-mediated oligomerization, membrane targeting and phosphoinositide binding. 19433865_APPL proteins exert their stimulatory effects on beta-catenin/TCF-dependent transcription by decreasing the activity of a Reptin-containing repressive complex 20814572_significant fluorescence resonance energy transfer between APPL minimal BAR domain FRET pairs 21645192_Data suggest that although annexin A2 is not an exclusive marker of APPL1/2 endosomes, it has an important function in membrane recruitment of APPL proteins, acting in parallel to Rab5. 22340213_Genetic variation(s) in APPL1/2 may be associated with CAD risk in T2DM in Chinese population. 22462604_found significant evidence of association with overweight/obesity for rs2272495 and rs1107756. rs2272495 C allele and rs1107756 T allele both conferred a higher risk of being overweight and obese. 22989406_Data indicate that a high level of APPL2 protein might enhance glioblastoma growth by maintaining low expression level of genes responsible for cell death induction. 23055524_analysis of APPL1 and APPL2 proteins and their interaction with Rab 23891720_It concludes that APPL2(PH) binding to BAR domain and Reptin is mutually exclusive which regulates the nucleocytoplasmic shuttling of Reptin. 23977033_C-APPL1/A-APPL2 allele combination is associated with non-alcoholic fatty liver disease occurrence, with a more severe hepatic steatosis grade and with a reduced adiponectin cytoprotective effect on liver. 24763056_ATM is the central modulator of APPL-mediated effects on radiosensitivity and DNA repair. 26583432_Data show that signal transducing adaptor proteins APPL1 and APPL2 are required for TGFbeta-induced nuclear translocation of TGFbeta type I receptor (TbetaRI)-ICD and for cancer cell invasiveness of prostate and breast cancer cell lines. 27986894_Results show the suppressive effect of OCC-1 RNA on transcription level of the APPL2 gene provides a putative colorectal neoplasm progression index. 30218350_A negative correlation of expression is evident between APPL2 and OCC-1 genes in breast cancer specimen. Unlike OCC-1A/B which encodes a small protein, OCC-1D noncoding RNA overexpression lead to APPL2 downregulation in MCF7 cells. 33122440_The adaptor protein APPL2 controls glucose-stimulated insulin secretion via F-actin remodeling in pancreatic beta-cells. | ENSMUSG00000020263 | Appl2 | 765.69151 | 1.1781803 | 0.2365603271 | 0.12867113 | 3.391541e+00 | 6.553170e-02 | 3.812870e-01 | No | Yes | 971.527283 | 136.186062 | 793.988372 | 108.950823 | |
| ENSG00000137434 | 347744 | C6orf52 | protein_coding | Q5T4I8 | Alternative splicing;Reference proteome | hsa:347744; | 32.49072 | 0.7031235 | -0.5081499472 | 0.49378784 | 1.045361e+00 | 3.065780e-01 | No | Yes | 30.209404 | 6.594566 | 41.339137 | 8.393400 | ||||||||
| ENSG00000137760 | 91801 | ALKBH8 | protein_coding | Q96BT7 | FUNCTION: Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain (PubMed:20123966, PubMed:20308323, PubMed:31079898). Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA (PubMed:20123966, PubMed:20308323). Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys)(PubMed:20308323). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly) (PubMed:21285950). Required for normal survival after DNA damage (PubMed:20308323). May inhibit apoptosis and promote cell survival and angiogenesis (PubMed:19293182). {ECO:0000269|PubMed:19293182, ECO:0000269|PubMed:20123966, ECO:0000269|PubMed:20308323, ECO:0000269|PubMed:21285950, ECO:0000269|PubMed:31079898}. | 3D-structure;Alternative splicing;Cytoplasm;Disease variant;Iron;Mental retardation;Metal-binding;Methyltransferase;Multifunctional enzyme;Nucleus;RNA-binding;Reference proteome;S-adenosyl-L-methionine;Transferase;Zinc | hsa:91801; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; 2-oxoglutarate-dependent dioxygenase activity [GO:0016706]; iron ion binding [GO:0005506]; tRNA (carboxymethyluridine(34)-5-O)-methyltransferase activity [GO:0106335]; tRNA (uracil) methyltransferase activity [GO:0016300]; tRNA binding [GO:0000049]; zinc ion binding [GO:0008270]; cellular response to DNA damage stimulus [GO:0006974]; tRNA methylation [GO:0030488]; tRNA wobble uridine modification [GO:0002098] | 19293182_Findings indicate a role for ALKBH8 in urothelial carcinoma cell survival mediated by NOX-1-dependent ROS signals, further suggesting therapeutic strategies in human bladder cancer by inducing JNK/p38/gammaH2AX-mediated cell death by silencing of ALKBH8. 20123966_The methyltransferase domain of ALKBH8 is demonstrated to be a functional homologue of the Saccharomyces cerevisiae Trm9 protein, mediating the last step in the formation of the wobble uridine modification 5-methoxycarbonylmethyl-uridine in tRNA. This modification is shown to be important for efficient selenoprotein synthesis. ALKBH8 knock-out mice are generated and described. 20308323_Data show that human AlkB homolog 8 (ABH8) catalyzes tRNA methylation to generate 5-methylcarboxymethyl uridine (mcm(5)U) at the wobble position of certain tRNAs, a critical anticodon loop modification linked to DNA damage survival. 21285950_Many eukaryotic tRNAs contain the wobble modification 5-methoxycarbonylmethyl-uridine (mcm5U). It is demonstrated that (R)- and (S)-5-methoxycarbonylhydroxymethyluridine (mchm5U), hydroxylated forms of mcm(5)U, are present in mammalian tRNA-Arg(UCG), and tRNA-Gly(UCC), respectively. It is shown that the hydroxylation reaction leading to the formation of (S)-mchm5U is catalyzed by the oxygenase (AlkB) domain of ALKBH8. 27329810_findings suggest that the high expression of ALKBH8 is critical for the growth and progression of bladder cancer 31079898_Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification. 31765888_Loss of epitranscriptomic control of selenocysteine utilization engages senescence and mitochondrial reprogramming(). 33544954_Neurodevelopmental disorder in an Egyptian family with a biallelic ALKBH8 variant. 34757492_Insight into ALKBH8-related intellectual developmental disability based on the first pathogenic missense variant. | ENSMUSG00000025899 | Alkbh8 | 925.54556 | 1.1867424 | 0.2470067550 | 0.12300889 | 4.048965e+00 | 4.419844e-02 | 3.197745e-01 | No | Yes | 1274.284519 | 280.369245 | 1047.475004 | 225.539879 | ||
| ENSG00000137817 | 56965 | PARP6 | protein_coding | Q2NL67 | FUNCTION: Mono-ADP-ribosyltransferase that mediates mono-ADP-ribosylation of target proteins. {ECO:0000269|PubMed:25043379}. | ADP-ribosylation;Alternative splicing;Glycosyltransferase;NAD;Reference proteome;Transferase | hsa:56965; | NAD+ ADP-ribosyltransferase activity [GO:0003950]; protein ADP-ribosylase activity [GO:1990404]; positive regulation of dendrite morphogenesis [GO:0050775]; protein ADP-ribosylation [GO:0006471]; protein auto-ADP-ribosylation [GO:0070213]; protein mono-ADP-ribosylation [GO:0140289] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23042038_PARP6, a mono(ADP-ribosyl) transferase and a negative regulator of cell proliferation, is involved in colorectal cancer development, PARP6-positive colorectal cancer had a good prognosis. 26934315_PARP6 acts as a tumor suppressor via downregulating Survivin expression in CRC. PARP6 can be a novel diagnostic and therapeutic target together with Survivin for CRC. 28260087_Results showed that both PARP6 and survivin exhibited higher expression in colorectal adenocarcinoma tissues and cell lines. knockdown of either PARP6 or survivin promotes cell apoptosis and inhibits the cell invasion of colorectal adenocarcinoma cells suggesting a significant correlation between theses 2 proteins. 34067418_Characterization of PARP6 Function in Knockout Mice and Patients with Developmental Delay. | ENSMUSG00000025237 | Parp6 | 1190.08658 | 1.0449574 | 0.0634441946 | 0.10225411 | 3.852986e-01 | 5.347811e-01 | 8.373786e-01 | No | Yes | 1305.368719 | 159.069435 | 1252.577220 | 149.128489 | ||
| ENSG00000137871 | 54816 | ZNF280D | protein_coding | Q6N043 | FUNCTION: May function as a transcription factor. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:54816; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription, DNA-templated [GO:0006355] | 20798984_As a first exploitation of this unique cohort, we identify three novel candidate dyslexia genes, ZNF280D and TCF12 at 15q21, and PDE7B at 6q23.3, by molecular mapping of the familial translocation with the 15q21 breakpoint. | ENSMUSG00000038535 | Zfp280d | 605.07986 | 1.2871677 | 0.3641999768 | 0.14003348 | 6.805884e+00 | 9.085798e-03 | 1.570514e-01 | No | Yes | 762.541530 | 159.316952 | 565.364022 | 115.795476 | ||
| ENSG00000138180 | 55165 | CEP55 | protein_coding | Q53EZ4 | FUNCTION: Plays a role in mitotic exit and cytokinesis (PubMed:16198290, PubMed:17853893). Recruits PDCD6IP and TSG101 to midbody during cytokinesis. Required for successful completion of cytokinesis (PubMed:17853893). Not required for microtubule nucleation (PubMed:16198290). Plays a role in the development of the brain and kidney (PubMed:28264986). {ECO:0000269|PubMed:16198290, ECO:0000269|PubMed:17853893, ECO:0000269|PubMed:28264986}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Coiled coil;Cytoplasm;Cytoskeleton;Developmental protein;Disease variant;Mitosis;Phosphoprotein;Reference proteome | hsa:55165; | centriolar satellite [GO:0034451]; centriole [GO:0005814]; centrosome [GO:0005813]; cleavage furrow [GO:0032154]; cytoplasm [GO:0005737]; Flemming body [GO:0090543]; intercellular bridge [GO:0045171]; membrane [GO:0016020]; midbody [GO:0030496]; plasma membrane [GO:0005886]; identical protein binding [GO:0042802]; cranial skeletal system development [GO:1904888]; establishment of protein localization [GO:0045184]; midbody abscission [GO:0061952]; mitotic cytokinesis [GO:0000281]; regulation of phosphatidylinositol 3-kinase signaling [GO:0014066] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 16406728_data suggest a possible involvement of CEP55 in centrosome-dependent cellular functions, such as centrosome duplication and/or cell cycle progression, or in the regulation of cytokinesis 16790497_This study defines a cellular mechanism that links centralspindlin to Cep55, which, in turn, controls the midbody structure and membrane fusion at the terminal stage of cytokinesis. 17237822_By forming a complex with phosphatidylinositol 3'-kinase, FLJ10540 activates the PI3-kinase/AKT proto-oncogene protein pathways, providing a mechanistic basis for FLJ10540-mediated oncogenesis. 17556548_study shows that two proteins involved in HIV-1 budding-Tsg101, a subunit of the endosomal sorting complex required for transport I (ESCRT-I), & Alix, an ESCRT-associated protein-were recruited to the midbody during cytokinesis by interaction with Cep55 17853893_that ALIX and TSG101/ESCRT-I also bind a series of proteins involved in cytokinesis, including CEP55, CD2AP, ROCK1, and IQGAP1. 18641129_the Cep55/Alix/ESCRT-III pathway has a role in cytokinesis and HIV-1 release 18948538_crystal structure of the ESCRT and ALIX-binding region (EABR) of CEP55 bound to an ALIX peptide at a resolution of 2.0 angstroms; structure shows that EABR forms an aberrant dimeric parallel coiled coil 19525975_FLJ10540 is not only an important prognostic factor but also a new therapeutic target in the FLJ10540/FOXM1/MMP-2 pathway for oral cavity squamous cell carcinoma treatment. 19609239_protein could be detected in breast- and lung carcinoma 9but not normal) tissues 19855176_Cep55 is stabilized in a phosphorylation- and Pin1-dependent manner. 20400365_Data provide strong evidence that CEP55 and HELLS may be used in conjunction with FOXM1 as a biomarker set for early cancer detection and indicators of malignant conversion and progression. 21079244_Data show that Plk1 activity negatively regulates Cep55 to ensure orderly abscission factor recruitment and ensures that this occurs only once cell contraction has completed. 22184120_the existence of a p53-Plk1-Cep55 axis in which p53 negatively regulates expression of Cep55, through Plk1 which, in turn, is a positive regulator of Cep55 protein stability. 22591637_FLJ10540 may be critical regulator of disease progression in nasopharygeal carcinoma, and the underlying mechanism may involve in the osteopontin/CD44 pathway. 22771033_At the midbody, BRCA2 influences the recruitment of endosomal sorting complex required for transport (ESCRT)-associated proteins, Alix and Tsg101, and formation of CEP55-Alix and CEP55-Tsg101 complexes during abscission. 24390615_cellular proliferation was suppressed as a result of cell cycle arrest at the G2/M phase in CEP55-knockdown cells 25178936_CEP55 mRNA/protein expression was observed is specific to TCC of human urinary bladder and might be used as a diagnostic biomarker and vaccine target in development of BC specific immunotherapy. 25659891_myotubularin-related protein 3 and myotubularin-related protein 4 may act as a bridge between CEP55 and polo-like kinase 1, ensuring proper CEP55 phosphorylation and regulating CEP55 recruitment to the midbody. 25889801_Which was required for FLJ10540/MMP-7 or FLJ10540/MMP-10 expressions. 25915844_In depth discussion of the functions of CEP55 across different effector pathways, and also its roles as a biomarker and driver of tumorigenesis, commemorating a decade of research on CEP55. [review] 26615423_Aberrant CEP55 expression may predict unfavorable clinical outcomes in epithelial ovarian carcinoma (EOC) patients and play an important role in regulating invasion in ovarian cancer cells. Thus, CEP55 may serve as a prognostic marker and therapeutic target for EOC 26902787_CEP55 plays a crucial role in promoting breast cancer cell proliferation and it might be a potential therapeutic target in breast cancer. 27127172_A FAK-Src signaling pathway downstream of integrin-mediated cell adhesion was found to decelerate both PLK1 degradation and CEP55 accumulation at the midbody. These data identify the regulation of PLK1 and CEP55 as steps where integrins exert control over the cytokinetic abscission. 28264986_CEP55 loss of function mutations likely underlie MARCH, a novel multiple congenital anomaly syndrome. 28295209_whole-exome sequencing lead to identification of a homozygous nonsense mutation c.256C>T (p.Arg86*) in CEP55 (centrosomal protein of 55 kDa) in autosomal recessive Meckel syndrome fetus. 28620049_Data suggest that USP9X as an integral component of centrosome where it functions to stabilize PCM1 and CEP55 and to promote centrosome biogenesis; N-terminal domain of USP9X appears to be responsible for physical association of USP9X with PCM1 and CEP55. (USP9X = ubiquitin-specific protease 9X; PCM1 = pericentriolar material 1 protein; CEP55 = 55kDa centrosomal protein) 28724890_CEP55 is a valuable prognostic factor and a potential therapeutic target in pancreatic cancer. 29561704_MiR-144 was down-regulated in breast cancerous tissues and cells, whereas CEP55 expression was up-regulated in breast cancerous tissues. Moreover, there existed a target relationship between miR-144 and CEP55 and negative correlation on their expressions. MiR-144 could down-regulate CEP55 expression, thereby inhibiting proliferation, invasion, migration, retarding cell cycle and accelerating cell apoptosis. 29579156_our data suggest that CEP55 can be used as a prognostic marker for osteosarcoma 29743530_iASPP associates with centrosomal protein of 55 kDa (CEP55), an important cytokinetic abscission regulator. Mechanically, iASPP acts as a PP1-targeting subunit to facilitate the interaction between PP1 and CEP55 and to remove PLK1-mediated Ser436 phosphorylation in CEP55 during late mitosis. 29750778_CEP55 was increased in lung cancer cells. 30008265_Results identified CEP55 as a potential cancer exosomal marker in head and neck squamous cell carcinoma cell lines. 30089483_SPAG5 interacted with centrosomal protein CEP55 to trigger the phosphorylation of AKT at Ser473. Functionally, the knockdown of CEP55 significantly attenuated SPAG5-promoted hepatocellular carcinoma (HCC) cell proliferation and migration. These data indicate that SPAG5 exhibits pro-HCC activities through interacting with CEP55. 30108112_Here, the authors showed that CEP55 overexpression/knockdown impacts survival of aneuploid cells. 30277841_A significant over expression of forkhead box protein M1 (FOXM1), polo-like kinase 1 (PLK1) and centrosomal protein 55 (CEP55) was observed in tumor samples compared to adjacent and normal bladder tissues, suggesting they may be potential candidate's biomarkers for early diagnosis and targets for cancer therapy. 30527357_these results indicate that changes in EZH2 expression lead to changes in CEP55 expression in lung adenocarcinoma, and these changes are associated with its prognosis 30536308_CEP55 silencing remarkably suppressed proliferation and induced apoptosis of castration-resistant prostate cells, indicating that miR-144-3p affects cell survival and proliferation by downregulating CEP55 30601084_we emphasize the recent work from our laboratory, which highlights the functional role of CEP55 in protecting aneuploid cells from death. We also discuss the rationale of targeting CEP55 in vivo, which could prove to be a novel and effective therapeutic strategy for sensitizing cells to microtubule inhibitors and might offer significantly improved patient outcome 30607788_CEP55 overexpression is associated with epithelial-mesenchymal transition in renal cell carcinoma. 30622327_considered alongside two recent studies of single families reporting loss of function candidate variants in CEP55, confirm disruption of CEP55 function as a cause of this clinical spectrum and enable us to delineate the cardinal clinical features of this disorder, providing important new insights into early human development 30896867_CEP55 was increased in ccRCC samples, and may be considered a potential diagnostic and prognostic biomarker for ccRCC. 31005653_results indicated that CEP55 played an important role in promoting the invasion and migration of U251 cell and self-renewal of glioma stem like cells. 31640955_CEP55 may play a key role in spermatogenesis 32100459_Expanding the spectrum of CEP55-associated disease to viable phenotypes. 32337246_Upregulation of CEP55 Predicts Dismal Prognosis in Patients with Liver Cancer. 32437446_The value of CEP55 gene as a diagnostic biomarker and independent prognostic factor in LUAD and LUSC. 32988587_SP1-induced upregulation of lncRNA CTBP1-AS2 accelerates the hepatocellular carcinoma tumorigenesis through targeting CEP55 via sponging miR-195-5p. 33015797_The function and molecular mechanism of CEP55 in anaplastic thyroid cancer. 33256799_Tumor promoting effects of circRNA_001287 on renal cell carcinoma through miR-144-targeted CEP55. 33417281_Human bone marrow mesenchymal stem cell-derived extracellular vesicles impede the progression of cervical cancer via the miR-144-3p/CEP55 pathway. 33497018_The RNA-helicase DDX21 upregulates CEP55 expression and promotes neuroblastoma. 34055036_Identification of Hub Genes to Regulate Breast Cancer Spinal Metastases by Bioinformatics Analyses. 34435195_Inhibition of miR-144-3p exacerbates non-small cell lung cancer progression by targeting CEP55. 34710087_Cep55 regulation of PI3K/Akt signaling is required for neocortical development and ciliogenesis. 34952571_MicroRNA-148a-3p suppresses cell proliferation and migration of esophageal carcinoma by targeting CEP55. 35037833_Long intergenic non-protein coding RNA 662 accelerates the progression of gastric cancer through up-regulating centrosomal protein 55 by sponging microRNA-195-5p. | ENSMUSG00000024989 | Cep55 | 1566.93165 | 1.0232976 | 0.0332258162 | 0.09332515 | 1.268222e-01 | 7.217499e-01 | 9.176667e-01 | No | Yes | 1894.577774 | 342.983174 | 1766.348465 | 312.725167 | ||
| ENSG00000138398 | 9360 | PPIG | protein_coding | Q13427 | FUNCTION: PPIase that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and may therefore assist protein folding (PubMed:20676357). May be implicated in the folding, transport, and assembly of proteins. May play an important role in the regulation of pre-mRNA splicing. {ECO:0000269|PubMed:20676357}. | 3D-structure;Alternative splicing;Isomerase;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Rotamase;Ubl conjugation | hsa:9360; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; nuclear matrix [GO:0016363]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cyclosporin A binding [GO:0016018]; peptidyl-prolyl cis-trans isomerase activity [GO:0003755]; RNA binding [GO:0003723]; protein folding [GO:0006457]; protein peptidyl-prolyl isomerization [GO:0000413]; RNA splicing [GO:0008380] | 15016823_human nuclear SRcyp is a cell cycle-regulated cyclophilin 15358154_Results show that SR-cyclophilin interacts and colocalizes with nuclear pinin, a SR-related protein involved in pre-mRNA splicing. 19058789_Observational study of gene-disease association. (HuGE Navigator) 28112215_The finding that zebularine upregulates CYP gene expression through DNMT1 and PKR modulation sheds light on the mechanisms controlling hepatocyte function and thus may aid in the development of new in-vitro systems using high-functioning hepatocytes 28589370_results indicate that LUC7L3, PPIG, and SFRS18 are not only implicated in EDA+ fibronectin formation, but also that they could possess multiple roles in psoriasis-associated molecular abnormalities. | ENSMUSG00000042133 | Ppig | 3763.73906 | 0.9646647 | -0.0519004522 | 0.08810614 | 3.463316e-01 | 5.561971e-01 | 8.500233e-01 | No | Yes | 4116.453074 | 608.287868 | 4214.443953 | 609.162329 | ||
| ENSG00000138617 | 54956 | PARP16 | protein_coding | Q8N5Y8 | FUNCTION: Intracellular mono-ADP-ribosyltransferase that may play a role in different processes through the mono-ADP-ribosylation of proteins involved in those processes (PubMed:23103912, PubMed:22701565, PubMed:25043379). May play a role in the unfolded protein response (UPR), by ADP-ribosylating and activating EIF2AK3 and ERN1, two important UPR effectors (PubMed:23103912). May also mediate mono-ADP-ribosylation of karyopherin KPNB1 a nuclear import factor (PubMed:22701565). May not modify proteins on arginine or cysteine residues compared to other mono-ADP-ribosyltransferases (PubMed:22701565). {ECO:0000269|PubMed:22701565, ECO:0000269|PubMed:23103912, ECO:0000269|PubMed:25043379}. | 3D-structure;ADP-ribosylation;Alternative splicing;Endoplasmic reticulum;Glycosyltransferase;Membrane;NAD;Reference proteome;Transferase;Transmembrane;Transmembrane helix;Unfolded protein response | hsa:54956; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; endoplasmic reticulum tubular network [GO:0071782]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; kinase binding [GO:0019900]; NAD+ ADP-ribosyltransferase activity [GO:0003950]; protein ADP-ribosylase activity [GO:1990404]; protein serine/threonine kinase activator activity [GO:0043539]; cellular response to leukemia inhibitory factor [GO:1990830]; endoplasmic reticulum unfolded protein response [GO:0030968]; IRE1-mediated unfolded protein response [GO:0036498]; NAD biosynthesis via nicotinamide riboside salvage pathway [GO:0034356]; negative regulation of cell death [GO:0060548]; positive regulation of protein serine/threonine kinase activity [GO:0071902]; protein ADP-ribosylation [GO:0006471]; protein auto-ADP-ribosylation [GO:0070213]; protein mono-ADP-ribosylation [GO:0140289]; viral protein processing [GO:0019082] | 23103912_PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. 25037261_ARTD15 plays role in nucleocytoplasmic shuttling, through karyopherin-beta1 mono-ADP-ribosylation. [review] 32472322_Regulation of poly ADP-ribosylation of VEGF by an interplay between PARP-16 and TNKS-2. | ENSMUSG00000032392 | Parp16 | 392.08199 | 0.8652057 | -0.2088849746 | 0.16958762 | 1.507115e+00 | 2.195798e-01 | 6.107639e-01 | No | Yes | 427.825872 | 37.130096 | 476.737316 | 40.112634 | ||
| ENSG00000138668 | 3184 | HNRNPD | protein_coding | Q14103 | FUNCTION: Binds with high affinity to RNA molecules that contain AU-rich elements (AREs) found within the 3'-UTR of many proto-oncogenes and cytokine mRNAs. Also binds to double- and single-stranded DNA sequences in a specific manner and functions a transcription factor. Each of the RNA-binding domains specifically can bind solely to a single-stranded non-monotonous 5'-UUAG-3' sequence and also weaker to the single-stranded 5'-TTAGGG-3' telomeric DNA repeat. Binds RNA oligonucleotides with 5'-UUAGGG-3' repeats more tightly than the telomeric single-stranded DNA 5'-TTAGGG-3' repeats. Binding of RRM1 to DNA inhibits the formation of DNA quadruplex structure which may play a role in telomere elongation. May be involved in translationally coupled mRNA turnover. Implicated with other RNA-binding proteins in the cytoplasmic deadenylation/translational and decay interplay of the FOS mRNA mediated by the major coding-region determinant of instability (mCRD) domain. May play a role in the regulation of the rhythmic expression of circadian clock core genes. Directly binds to the 3'UTR of CRY1 mRNA and induces CRY1 rhythmic translation. May also be involved in the regulation of PER2 translation. {ECO:0000269|PubMed:10080887, ECO:0000269|PubMed:11051545, ECO:0000269|PubMed:24423872}. | 3D-structure;Acetylation;Alternative splicing;Biological rhythms;Cytoplasm;DNA-binding;Direct protein sequencing;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Ribonucleoprotein;Transcription;Transcription regulation;Ubl conjugation | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are nucleic acid binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has two repeats of quasi-RRM domains that bind to RNAs. It localizes to both the nucleus and the cytoplasm. This protein is implicated in the regulation of mRNA stability. Alternative splicing of this gene results in four transcript variants. [provided by RefSeq, Jul 2008]. | hsa:3184; | cytosol [GO:0005829]; mCRD-mediated mRNA stability complex [GO:0106002]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; synapse [GO:0045202]; chromatin binding [GO:0003682]; histone deacetylase binding [GO:0042826]; minor groove of adenine-thymine-rich DNA binding [GO:0003680]; mRNA 3'-UTR AU-rich region binding [GO:0035925]; RNA binding [GO:0003723]; telomeric DNA binding [GO:0042162]; 3'-UTR-mediated mRNA destabilization [GO:0061158]; cellular response to amino acid stimulus [GO:0071230]; cellular response to estradiol stimulus [GO:0071392]; cellular response to nitric oxide [GO:0071732]; cellular response to putrescine [GO:1904586]; cerebellum development [GO:0021549]; circadian regulation of translation [GO:0097167]; CRD-mediated mRNA stabilization [GO:0070934]; hepatocyte dedifferentiation [GO:1990828]; liver development [GO:0001889]; mRNA transcription by RNA polymerase II [GO:0042789]; negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900152]; positive regulation of cytoplasmic translation [GO:2000767]; positive regulation of telomerase RNA reverse transcriptase activity [GO:1905663]; positive regulation of telomere capping [GO:1904355]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; positive regulation of translation [GO:0045727]; regulation of circadian rhythm [GO:0042752]; regulation of gene expression [GO:0010468]; regulation of telomere maintenance [GO:0032204]; regulation of transcription, DNA-templated [GO:0006355]; response to calcium ion [GO:0051592]; response to electrical stimulus [GO:0051602]; response to rapamycin [GO:1901355]; response to sodium phosphate [GO:1904383]; RNA catabolic process [GO:0006401]; RNA processing [GO:0006396] | 11531333_NMR structure of RNA binding domain and its interactions with RNA and DNA 11856759_bcl-2 A and U rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. 12356764_regulates apoptosis by altering mRNA turnover and CDIR inhibits apoptosis by acting as a competitive inhibitor of AUF1, preventing AUF1 from binding to its targets 12819194_selective AUF1 phosphorylation may regulate ARE-directed mRNA turnover by remodeling local RNA structures 12819195_signal transduction pathways may regulate ARE-directed mRNA turnover by reversible phosphorylation of polysome-associated p40AUF1 14585195_a mechanism whereby reduced cytoplasmic levels of AUF1 in MNT1 melanoma cells may lead to IL-10 overexpression, with deleterious consequences for tumor surveillance and rejection 15257295_composition and fate (stability, translation) of HuR- and/or AUF1-containing ribonucleoprotein complexes depend on the target mRNA of interest, RNA-binding protein abundance, stress condition, and subcellular compartment 15514034_calcineurin regulates AUF1 posttranslationally in vitro and PTH gene expression in vivo but still allows its physiological regulation by calcium and phosphate 15734733_analysis of the structure of the C-terminal-binding domain (BD2) of HNRPD complexed with single-stranded d(TTAGGG) determined by NMR 15951444_AUF1 binds to multiple destabilizing elements within the 3'-UTR that participate in the rapid turnover of the phosphoenolpyruvate carboxykinase mRNA. 16109718_coordinated regulation of mRNA stability by HuR and AUF1 proteins contributes to the observed increase in ATF3 expression following amino acid limitation 16144962_The multiple instability elements present within the 3'-UTR may function synergistically to mediate both the rapid degradation and the cAMP-induced stabilization of PEPCK mRNA. The latter process may result from a PKA-dependent phosphorylation of AUF1 16155006_HuR shows increased binding to some V-ATPase mRNAs during ATP depletion; siRNA-mediated knockdown of HuR results in diminished V-ATPase expression. 16289864_c-Yes 3'-UTR contains at least three newly identified adenine/uridine-rich elements (AREs) which are bound specifically by ARE-binding proteins HuR and AUF1. 16494882_Differential expression of AU-rich mRNAs in response to LPL-mediated lipolysis might have an impact on physiological processes regulating lipid metabolism or pathophysiological processes promoting endothelial dysfunction and atherogenesis. 16835382_AUF1 could function in a novel pathway mediating the oncogenic effects of NPM-ALK 16954375_These results indicate that AUF1 binds to the AU-rich element in vivo and promotes IL-6 mRNA degradation. 17030625_AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability 17486099_Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. 17626845_Two mRNA binding proteins, HuR and AUF1, are colocalized and are capable of functional interaction in both the nucleus and cytoplasm. 17878526_both HuR and AUF1 bind to discrete regions of DENR/DRP mRNA and that AUF1 silencing increases DENR/DRP protein levels. 18202450_This study determined that hnRNP L interacts specifically with the hnRNP D/AUF1 in the yeast two-hybrid system. 18240226_Because AUF1 proteins are major components of messenger RNA stability complexes, our findings suggest that these complexes form a novel macromolecular target structure for autoantibodies in rheumatic autoimmune diseases 18573886_Chaperone Hsp27 is a novel subunit that is itself an AU-rich elements (ARE)-binding protein essential for rapid ARE-mRNA degradation. 18650551_UV cross-linking and immunoprecipitation experiments revealed 2 ARE-binding proteins, AUF1 and HuR, associated with IL-8 mRNA in saliva. 18842733_hnRNP D plays an important role in the translation of hepatitis C virus mRNA through interactions with the internal ribosomal entry site 18844578_isoform-specific regulation of anti-inflammatory IL10 expression in monocytes 19015635_Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. 19074427_data demonstrate that AUF1 is an important factor that promotes iNOS mRNA degradation. Furthermore, all individual AUF1 isoforms act in a similar manner 19574297_AUF1 may be considered as a new, additional marker for thyroid carcinoma. 20102719_Immunofluorescent analysis revealed increased levels of HuR and AUF1, and a decrease in methyl-HuR levels in human livers with hepatocellular carcinoma. 20489206_Leukotriene B(4) BLT receptor signaling regulates the level and stability of cyclooxygenase-2 (COX-2) mRNA through restricted activation of Ras/Raf/ERK/p42 AUF1 pathway 20571027_the degradation of bcl-2 mRNA induced by AS1411 results from both interference with nucleolin protection of bcl-2 mRNA and recruitment of the exosome by AUF1. 20926381_Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands 21135123_p40(AUF1) regulates a critical node within the NF-kappaB signaling pathway to permit IL10 induction for the anti-inflammatory arm of an innate immune response. 21245386_These results suggest that the p38 MAP kinase (MAPK)-MK2-Hsp27 signaling axis may target AUF1 destruction by proteasomes, thereby promoting AU-rich element mRNA stabilization. 21733716_Knockdown AUF1 mRNA expression by AUF1 siRNA in MKP-1 WT bone marrow macrophages significantly delayed degradation of IL-6, IL-10 and TNF- alpha mRNAs compared with controls 21799732_p16( INK4a) is also a modulator of transcription and apoptosis through controlling the expression of two major transcription regulators, AUF1 and E2F1 21956942_This review briefly describes the roles of mRNA decay in gene expression in general and ARE-mediated decay (AMD) in particular, with a focus on AUF1 and the different modes of regulation that govern AUF1 involvement in AMD. 22086907_AUF1 and HuR bind to VEGFA ARE RNA under both normoxic and hypoxic conditions, and a pVHL-RNP complex determines VEGFA mRNA decay. 22134169_Data suggest that AUF-1 and YB-1 are required for normal accumulation of beta-globin mRNA in erythroid cells; YB-1 and AUF-1 exhibit sequence-specific binding to 3-prime-untranslated region of beta-globin mRNA. 22159912_Findings point to a contribution of AUF1 to the deleterious effects of cytokines on beta cell functions and suggest a role for this RNA-binding protein in the early phases of type 1 diabetes. 22203679_the ability of AUF1 isoforms to regulate the mRNA binding and decay-promoting activities of TTP 22368252_The binding sites for HuR and AUF1 present in androgen receptor mRNA, defined by their exact target sequences, are the same sequence for both proteins. 23012480_EBER1 may disturb the normal homeostasis between AUF1 and ARE-containing mRNAs or compete with other AUF1-interacting targets in cells latently infected by Epstein-Barr virus. 23131833_Here authors describe experiments suggesting that AUF1, a host RNA binding protein involved in mRNA decay, plays a role in the infectious cycle of picornaviruses such as poliovirus and human rhinovirus. 23530064_Hsp27 and F-box protein beta-TrCP promote degradation of mRNA decay factor AUF1. 23572232_Results suggest that cleavage of AUF1 may be a strategy employed by coxsackievirus B3 (CVB3) to enhance the stability of its viral genome. 23603392_ING4 may regulate c-MYC translation by its association with AUF1. 23940053_analysis of how AUF1 target mRNAs and how AUF1 binding possibly regulates protein and/or microRNA binding events at adjacent sites 24158514_hnRNP D is critically involved in LDLR mRNA degradation in liver tissue in vivo. 24213928_Our findings suggest that the AUF1 gene may play an important role in hepatocellular carcinoma progression 24416409_The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins, such as HuR and AUF1. 24687816_AUF1 has been implicated in controlling a variety of physiological functions through its ability to regulate the expression of numerous mRNAs containing 3'-UTR AREs, thereby coordinating functionally related pathways. 25077793_AUF1 interacts with the EV71 IRES to negatively regulate viral translation and replication. 25078689_AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. 25231991_IL-6 has a major role in activating breast stromal fibroblasts through STAT3-dependent AUF1 induction 25261470_MicroRNA-141 and microRNA-146b-5p have a role in inhibiting the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT 25366541_preserves genomic integrity through its actions on target RNAs 25486179_Auf1 may play a role in the elimination of oxidized RNA, which is required for the maintenance of proper gene expression under conditions of oxidative stress 25720531_These data specify a post-transcriptional mechanism through which AUF1 and YB1 contribute to the normal development of erythropoietic cells, and to non-hematopoietic tissues in which AUF1- and YB1-based regulatory mRNPs assemble on heterologous mRNAs 25787750_Taken together, these findings proved the inhibitory effect of TP-1 on the growth and metastasis of SMMC-7721 cells, and TP-1 might be offered for future application as a powerful chemopreventive agent against hepatocellular carcinoma (HCC) metastasis. 25908445_in human ovarian, esophageal, and pancreatic cancer tissues, the expression of SOD1 was significantly correlated with that of AUF-1, further supporting the importance of AUF-1 in regulating SOD1 gene expression 25910425_Functional analysis of selected regulated proteins revealed that knockdown of HNRPD, PHB2 and UB2V2 can increase HCMV replication, while knockdown of A4 and KSRP resulted in decreased HCMV replication. 26253535_a novel mechanism by which AUF1 binding and transfer of microRNA let-7 to AGO2 facilitates let-7-elicited gene silencing. 26318153_hnRNPD has roles in cellular proliferation and survival, besides RNA splicing and stability in oral cancer 26648300_Down-regulation of hnRNPD inhibits the proliferation of esophageal squamous cell carcinoma cells by promoting cell apoptosis. 26728997_Findings indicate that hnRNP D and arginine methylation play important roles in the regulation of Flt-1 mRNA alternative polyadenylation. 26805816_hnRNPA2B1, hnRNPD, hnRNPL , and YBX1 might play important roles in gastric cancer tumorigenesis. 26925783_Overexpression of AUF1 and HuR is a common finding observed in thyroid malignancy. Analysis of the tissues obtained by surgical resection as demonstrated in this study is comparable to a fine needle aspiration and in combination with AUF1/HuR immuno-analysis may support the conventional immunohistological investigations. 27248826_Results indicate that the IL-6/STAT3/NF-kappaB positive feedback loop includes AUF1 and is responsible for the sustained active status of cancer-associated fibroblasts. 27437398_analysis of the effect of the N-terminal RNA recognition motif on AUF1 27520967_Arginine methylation improves the viral RNA chaperone activity of AUF1 p45. 27578251_Lnc_ASNR interacted with the protein ARE/poly (U)-binding/degradation factor 1(AUF1), which is reported to promote rapid degradation of the Bcl-2 mRNA, an inhibitor of apoptosis. Lnc_ASNR binds to AUFI in nucleus, decreasing the cytoplasmic proportion of AUF1 which targets the B-cell lymphoma-2 (Bcl-2) mRNA. 27784781_These results suggest that the post-transcriptional regulation of ATX expression by HuR and AUF1 modulates cancer cell migration. 27826622_High AUF1 expression is associated with esophageal squamous cell carcinoma. 27836661_We found a C-rich element (CRE) in mu-opioid receptor (MOR) 3'-untranslated region (UTR) to which poly (rC) binding protein 1 (PCBP1) binds, resulting in MOR mRNA stabilization. AUF1 phosphorylation also led to an increased interaction with PCBP1. 28291226_Our study provides new insights into the regulation of APP pre-mRNA processing, supports the role for nELAVLs as neuron-specific splicing regulators and reveals a novel function of AUF1 in alternative splicing. 28300425_in the present case, the identified mutations in HNRNPD and risk polymorphisms are plausible molecular players in the manifestation of CD. 28334781_Depletion of AUF1 abolishes the global interaction of miRNAs and AGO2. AUF1 functions in promoting miRNA-mediated mRNA decay globally. 28986222_AUF1 might be a potential player in renal tubulointerstitial fibrosis through modulation of TGF-beta signal transduction via posttranscriptional regulation of Nedd4L. 29263134_Both hnRNP D and DL are able to control their own expression by alternative splicing of cassette exons in their 3'UTRs. Exon inclusion produces mRNAs degraded by nonsense-mediated decay. Moreover, hnRNP D and DL control the expression of one another by the same mechanism. 29263261_AUF1 p45 triggers the RNA switch in the flaviviral genome that is crucial for viral replication. These findings represent an important example of how cellular (host) factors promote the propagation of RNA viruses 30181254_The work presented here addresses the mechanism of AUF1 inhibition of the replication of poliovirus and CVB3. We demonstrate that AUF1 knockdown in human cells results in increased viral translation, RNA synthesis, and virus production. 30349226_The expression of AUF-1 was selectively decreased in the bronchial, but not in the bronchiolar, epithelium from patients with stable COPD compared to control smokers. 30418981_Study identified RANKL and BCL6 mRNA as targets of AUF1 p42. Detailed analysis of 3'UTRs revealed for both that full instability required two elements, which are conserved in evolution. In RANKL mRNA both elements are AU-rich and separated by 30 bases, while in BCL6 mRNA one is AU-rich and 60 bases from a non-AU-rich element that potentially forms a stem-loop structure. 30681073_Anti-apoptotic gene Bcl-2 was suppressed by berberine treatment and lncRNA CASC2, inducing the pro-apoptotic effects. Moreover, lncRNA CASC2 binds to AUF1, which sequestered AUF1 from binding to Bcl-2 mRNA, thus inducing the inactivation of Bcl-2 translation. 30799487_HNRNPD is critical for homologous recombination. 30927204_Identification and Verification of Two Novel Differentially Expressed Proteins from Non-neoplastic Mucosa and Colorectal Carcinoma Via iTRAQ Combined with Liquid Chromatography-Mass Spectrometry. 31326364_AUF1 is involved in the specific degradation of oxidized RNA (REVIEW) 31657090_AUF1 knockdown resulted in upregulation of GKN1 expression and promoted GKN1 mRNA decay by binding the 3' untranslated region of GKN1 mRNA H. pylori-induced AUF1 expression was associated with p-ERK activation and CagA. 31665914_AUF1 stabilizes Nrf2-mRNA. 32020881_Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. 32106863_Characterization of novel LncRNA P14AS as a protector of ANRIL through AUF1 binding in human cells. 32924750_Alternatively spliced isoforms of AUF1 regulate a miRNA-mRNA interaction differentially through their YGG motif. 33016643_The overexpression of AUF1 in colorectal cancer predicts a poor prognosis and promotes cancer progression by activating ERK and AKT pathways. 33444453_AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. 33542193_HNRNPD interacts with ZHX2 regulating the vasculogenic mimicry formation of glioma cells via linc00707/miR-651-3p/SP2 axis. 33767152_A novel lncRNA TCLlnc1 promotes peripheral T cell lymphoma progression through acting as a modular scaffold of HNRNPD and YBX1 complexes. 33904361_SChLAP1 contributes to non-small cell lung cancer cell progression and immune evasion through regulating the AUF1/PD-L1 axis. 34624736_Effects of ribosomal associated protein hnRNP D in gemcitabine-resistant pancreatic cancer. 35178834_E2F1-mediated AUF1 upregulation promotes HCC development and enhances drug resistance via stabilization of AKR1B10. 35396527_NFkappaB (RelA) mediates transactivation of hnRNPD in oral cancer cells. | ENSMUSG00000000568 | Hnrnpd | 13326.64878 | 0.9514648 | -0.0717778568 | 0.07027388 | 1.040962e+00 | 3.075978e-01 | 6.890149e-01 | No | Yes | 17488.459214 | 2334.518487 | 17088.502497 | 2230.191618 | |
| ENSG00000138735 | 8654 | PDE5A | protein_coding | O76074 | FUNCTION: Plays a role in signal transduction by regulating the intracellular concentration of cyclic nucleotides. This phosphodiesterase catalyzes the specific hydrolysis of cGMP to 5'-GMP (PubMed:9714779, PubMed:15489334). Specifically regulates nitric-oxide-generated cGMP (PubMed:15489334). {ECO:0000269|PubMed:15489334, ECO:0000269|PubMed:9714779}. | 3D-structure;Allosteric enzyme;Alternative splicing;Hydrolase;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;Zinc;cGMP;cGMP-binding | PATHWAY: Purine metabolism; 3',5'-cyclic GMP degradation; GMP from 3',5'-cyclic GMP: step 1/1. | This gene encodes a cGMP-binding, cGMP-specific phosphodiesterase, a member of the cyclic nucleotide phosphodiesterase family. This phosphodiesterase specifically hydrolyzes cGMP to 5'-GMP. It is involved in the regulation of intracellular concentrations of cyclic nucleotides and is important for smooth muscle relaxation in the cardiovascular system. Alternative splicing of this gene results in three transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2008]. | hsa:8654; | cytosol [GO:0005829]; 3',5'-cyclic-GMP phosphodiesterase activity [GO:0047555]; 3',5'-cyclic-nucleotide phosphodiesterase activity [GO:0004114]; cGMP binding [GO:0030553]; metal ion binding [GO:0046872]; cGMP catabolic process [GO:0046069]; negative regulation of cardiac muscle contraction [GO:0055118]; negative regulation of T cell proliferation [GO:0042130]; positive regulation of cardiac muscle hypertrophy [GO:0010613]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of oocyte development [GO:0060282]; regulation of nitric oxide mediated signal transduction [GO:0010749]; relaxation of cardiac muscle [GO:0055119]; signal transduction [GO:0007165] | 11896473_Human PDE5A gene encodes three PDE5 isoforms from two alternate promoters. 12359732_cGMP-directed regulation of PDE5 phosphorylation and the resulting increase in cGMP binding affinity occur largely within the R domain 12604588_CGMP-dependent protein kinase I causes NO-induced PDE5 phosphorylation. However, cGMP can directly activate PDE5 without phosphorylation in platelet cytosol, most likely by binding to GAF domains. 12650945_GAFa domain of PDE5A adopts a structure similar to the GAFb domain of PDE2A, and provides the sole site for cGMP binding in PDE5A 12955149_three-dimensional structures of the catalytic domain (residues 537-860) of human PDE5 complexed with the three drug molecules sildenafil, tadalafil (Cialis) and vardenafil (Levitra) 14668322_Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine 14764637_demonstrated, for the first time, that androgens positively regulate phosphodiesterase 5 15175637_no correlations of a novel polymorphism of the PDE5A promoter gene with the intermediate phenotype essential hypertension/erectile dysfunction 15240816_Phosphorylation of PDE5 seems to act as memory switch for activation leading to long-term desensitization of the signaling pathway. 15640438_PDE5 gene expression and activity are androgen-dependent in vas deferens. 15817798_Phosphodiesterase Type 5 is the main factor regulating cyclic guanosine monophosphate hydrolysis and downstream signaling in human PASMCs. 16407275_Results suggest that glutamine(817) is a positive determinant for phosphodiesterase 5 affinity for cyclic GMP and several inhibitors. 16690614_Subdomains are structurally and functionally interdependent and act in concert in regulating human PDE5. 17017938_Virtually all tissues and cell types express PDE5, with heart and cardiomyocytes being contentious; PDE5A1 and PDE5A2 are ubiquitous, but PDE5A3 is specific to smooth muscle, as described in this review. 17138653_PDE5 may be a possible therapeutic target in bladder dysfunction for ameliorating irritative lower urinary tract symptoms. 17906676_The PDE-5A was expressed in both pre-adipocytes and adipocytes. PDE-5A mRNA and protein levels decreased as pre-adipocytes differentiated. 18293931_GAF domains of PDE5A can act as sensors and intracellular sinks for cyclic GMP, but not cyclic AMP. 18635550_analysis of functional chimeras of the phosphodiesterase 5 and 10 tandem GAF domains 18757735_may define a molecular mechanism by which PDE5 inhibition can differentially impact selected cellular functions of platelets, and perhaps of other cell types 19139381_Increased myocardial PDE5 expression in patients with advanced cardiomyopathy may contribute to the development of heart failure and represents an important therapeutic target. 19852267_The rAd5-shRNA-PDE5A3 can obviously increase the cGMP level in the smooth muscle cells of human corpus cavernosum, and enhance the inhibition of the PDE5 gene. 19961855_PDE5-inhibition blocks TRPC6 channel activation and associated Cn/NFAT activation signaling by PKG-dependent channel phosphorylation 19996273_Data show that SS increased intracellular cGMP levels and activated protein kinase G, and selectively inhibited PDE5 in breast tumor cells. 20177899_The distribution of PDE-5 and NOD II may indicate a physiologic role in the regulatory function of human vagina. 20196613_Data show that the identified compound will be a useful agent for evaluating the therapeutic potential of central inhibition of PDE5. 20308615_Myocardial oxidative stress increases PDE5 expression in the failing heart 20332099_Observational study of gene-disease association. (HuGE Navigator) 20332439_High PDE5A1 expression is associated with malignant melanoma. 20346360_Observational study of gene-disease association. (HuGE Navigator) 20351714_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20563733_Observational study of gene-disease association. (HuGE Navigator) 20563733_Suggest that PDE5A is associated with increased disease susceptibility, pathological progression, and development of proteinuria in childhood IgA nephropathy. 20861010_Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. 21193396_Distinct allostery induced in the cyclic GMP-binding, cyclic GMP-specific phosphodiesterase (PDE5) by cyclic GMP, sildenafil, and metal ions. 21215707_In melanoma cells, oncogenic (V600E) BRAF signaling downregulates PDE5A through the transcription factor BRN2, leading to increased cGMP and Ca2+ and the induction of invasion through increased cell contractility. 21421555_the spatial localization of PDE5A at the level of caveolin-rich lipid rafts allows for a feedback loop between endothelial PDE5A and nitric oxide synthase (NOS3) 21425347_Solved is the crystal structure of the structurally uncharacterized PDE5A GAF-B domain. 21505183_PDE5 inhibition by sulindac sulfide selectively induces apoptosis and attenuates oncogenic Wnt/beta-catenin-mediated transcription in human breast tumor cells 21525805_These studies suggest a novel role for PDE5 in erythrocytes. 21697861_found in smooth muscle wall of blood vessels transversing the clitoral supepithelial and stromal space 21736695_PDE9 is widely distributed in the urothelial epithelium of the human lower urinary tract and its potential roles may be different from those of PDE5. 21987443_Treatment of L-1236 with PDE5A-inhibitor sildenafil or with siRNA directed against PDE5A and concomitant stimulation with cyclic guanosine monophosphate (cGMP) resulted in enhanced apoptosis, indicating PDE5A as an oncogene. 22843873_PDE5 is highly expressed and increases ( approximately 130%) during growth whereas ABCC5 exhibited low to moderate expression, with a moderate increase ( approximately 40%) during growth 22960860_it is concluded that assessment of PDE5 and PDE9 expression may be useful in the differential diagnosis of benign and malignant breast disease and successful treatment of breast cancer 23033484_analysis of amino acid residues responsible for the selectivity of tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6 23527037_Myocardial PDE5 expression is increased in the hearts of humans and mice with chronic pressure overload. 24112792_inhibition of PDE5 can counteract apoptosis during aging by modulating proand antiapoptotic molecules and the APP pathway. 25247292_Results show that Ser102 and Ser104 may influence the conformational flexibility of PDE5A, which may in turn influence phosphorylation status, allosteric regulation by cGMP or other as yet unknown regulatory mechanisms for PDE5A. 25322361_PDE5A appears to jointly influence amygdala volume and emotion recognition performance. 25837309_Overexpression of PDE5 in papillary thyroid carcinomas. 26299804_cGMP PDE isozymes, PDE5 and 10, are elevated in colon tumor cells compared with normal colonocytes, and inhibitors and siRNAs can selectively suppress colon tumor cell growth 26657792_Our data showed a significant and previously undocumented upregulation of PDE5 in both rat and human BPH. 26967220_Study showed that past attempts to quantify PDE5 mRNA were flawed due to the use of incorrect primers, and that when correct primers are used, PDE5 mRNA is detectable in human brain tissue; that PDE5 protein exists in human brain by western blot and ELISA; and performed immunohistochemistry and demonstrate that PDE5 is present in human neurons. 27235284_the relationship between PDE5A polymorphisms, diabetes, and the efficacy of sildenafil treatment.The response to sildenafil treatment depends on polymorphisms in the PDE5A gene and the glycemic status of the patients. 28099939_Data indicate that high type 5 phosphodiesterase (PDE5) expression in glioblastoma multiforme (GBM) cells significantly correlated with longer overall survival of patients. 28211580_The analogues showed a relative narrow range of Ki values for PDE5A inhibition (1.2-14 nm). 28506928_Thsese findings provide insight into the existence of distinct 'pools' of PDE5A in human arterial smooth muscle cells and support the idea that these discrete compartments regulate distinct cGMP-dependent events. 29888556_Selective differences in PDE5 protein expression suggest distinct molecular mechanisms are in play for these ED subtypes. 30590671_PDE5 is present in vascular myocytes within small penetrating arteries in older people 31433119_Regulation of PDE5 expression in normal prostate, benign prostatic hyperplasia, and adenocarcinoma. 31434939_Regulation of PDE5 expression in human aorta and thoracic aortic aneurysms. 32663396_Structure of Human Phosphodiesterase 5A1 Complexed with Avanafil Reveals Molecular Basis of Isoform Selectivity and Guidelines for Targeting alpha-Helix Backbone Oxygen by Halogen Bonding. 32764382_Discovery of Novel Agents on Spindle Assembly Checkpoint to Sensitize Vinorelbine-Induced Mitotic Cell Death Against Human Non-Small Cell Lung Cancers. 33009887_Changes in the expression and function of the PDE5 pathway in the obstructed urinary bladder. 33809279_Muscle Damage in Systemic Sclerosis and CXCL10: The Potential Therapeutic Role of PDE5 Inhibition. 33952455_Over-expression of PDE5 in Oral Squamous Cell Carcinoma - Effect of Sildenafil Citrate. 34169423_Influencing Factors for Erectile Dysfunction of Young Adults with No Response to PDE5i. 34424768_Identification of a Functional PDE5A Variant at the Chromosome 4q27 Coronary Artery Disease Locus in an Extended Myocardial Infarction Family. | ENSMUSG00000053965 | Pde5a | 199.48417 | 1.1677157 | 0.2236891046 | 0.21668473 | 1.068831e+00 | 3.012097e-01 | 6.837756e-01 | No | Yes | 163.945993 | 58.919116 | 133.945483 | 47.183969 |
| ENSG00000138801 | 9061 | PAPSS1 | protein_coding | O43252 | FUNCTION: Bifunctional enzyme with both ATP sulfurylase and APS kinase activity, which mediates two steps in the sulfate activation pathway. The first step is the transfer of a sulfate group to ATP to yield adenosine 5'-phosphosulfate (APS), and the second step is the transfer of a phosphate group from ATP to APS yielding 3'-phosphoadenylylsulfate (PAPS: activated sulfate donor used by sulfotransferase). In mammals, PAPS is the sole source of sulfate; APS appears to be only an intermediate in the sulfate-activation pathway (PubMed:9576487, PubMed:9668121, PubMed:9648242, PubMed:14747722). Required for normal biosynthesis of sulfated L-selectin ligands in endothelial cells (PubMed:9576487). {ECO:0000269|PubMed:14747722, ECO:0000269|PubMed:9576487, ECO:0000269|PubMed:9648242, ECO:0000269|PubMed:9668121}. | 3D-structure;ATP-binding;Acetylation;Kinase;Multifunctional enzyme;Nucleotide-binding;Nucleotidyltransferase;Reference proteome;Transferase | PATHWAY: Sulfur metabolism; sulfate assimilation. {ECO:0000269|PubMed:14747722, ECO:0000269|PubMed:9576487, ECO:0000269|PubMed:9648242, ECO:0000269|PubMed:9668121}. | Three-prime-phosphoadenosine 5-prime-phosphosulfate (PAPS) is the sulfate donor cosubstrate for all sulfotransferase (SULT) enzymes (Xu et al., 2000 [PubMed 10679223]). SULTs catalyze the sulfate conjugation of many endogenous and exogenous compounds, including drugs and other xenobiotics. In humans, PAPS is synthesized from adenosine 5-prime triphosphate (ATP) and inorganic sulfate by 2 isoforms, PAPSS1 and PAPSS2 (MIM 603005).[supplied by OMIM, Mar 2008]. | hsa:9061; | cytosol [GO:0005829]; adenylylsulfate kinase activity [GO:0004020]; ATP binding [GO:0005524]; nucleotidyltransferase activity [GO:0016779]; protein homodimerization activity [GO:0042803]; sulfate adenylyltransferase (ATP) activity [GO:0004781]; 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process [GO:0050428]; skeletal system development [GO:0001501]; sulfate assimilation [GO:0000103] | 11931637_characterization and expression of bifunctional isoforms 12716056_review of PAPSS biochemistry, molecular biology and genetics 14747722_x-ray crystallographic analysis of 3'-phosphoadenosine-5'-phosphosulfate synthetase 1 15065880_The kinetic characteristics of the individual sulfurylase and kinase activities of PAPSS isoform 1, the predominant form in human brain, are described. 15755455_Results describe the crystal structure of human PAPSS1 at 1.8 angstroms resolution. 17540769_ATP-sulfurylase domain of PAPS synthetase influences these elements in adenylylsulfate kinase, which may be a mechanism between both domains the this bifunctional enzyme 19008949_a role for PAPSS1 in the cellular sulfonation pathway requirement at a stage during or shortly after MLV provirus establishment, and subsequent influence on gene expression from the viral long terminal repeat promoter 19337310_Observational study of gene-disease association. (HuGE Navigator) 19337310_PAPSS1 maps to chromosome 4q25 and may havea role in hepatocellular carcinoma 20332099_Observational study of gene-disease association. (HuGE Navigator) 20335228_SK1 plays an essential role in regulating in vitro paracrine angiogenesis and lymphangiogenesis. 22242175_Unusual localisation signals of both PAPS synthase isoforms, are described. 22380844_study showed expression patterns of SULT1E1 and PAPSS in breast and endometrial tissues; the estrogen sulfation enzymes were comparatively higher in the tumorous tissues than adjacent normal tissues; overexpression of SULT1E1 and PAPSS1 retarded MCF-7 cells growth in vivo and in vitro by arresting cell cycles and inducing apoptosis 24587339_SK1 positivity and high expression were significantly associated with cancer and a shorter 5-year and overall survival. 25482303_leptin signalling links a novel SFK/ERK1/2-mediated pathway and SK1 expression. 25948711_These data indicate that knocking down PAPSS increases UGT2B4 transcription and mRNA stability as a compensatory response to the loss of SULT2A1 activity 27941888_we found that ectopic expression of oncogenic KRas and HRas in cells resulted in elevated CIB1 expression. We previously described the Ca(2+)-myristoyl switch function of CIB1, and its ability to facilitate agonist-induced plasma membrane localisation of sphingosine kinase 1 (SK1), a location where SK1 is known to elicit oncogenic signalling. 28790117_These results suggest that PAPSS1 inhibition enhances cisplatin activity in multiple preclinical model systems and that low PAPSS1 expression may serve as a biomarker for platin sensitivity in cancer patients. Developing strategies to target PAPSS1 activity in conjunction with platinum-based chemotherapy may offer an approach to improving treatment outcomes | ENSMUSG00000028032 | Papss1 | 1351.18579 | 1.0022882 | 0.0032973596 | 0.09869189 | 1.116012e-03 | 9.733502e-01 | 9.927846e-01 | No | Yes | 1663.556597 | 247.819389 | 1576.823868 | 229.736232 |
| ENSG00000139132 | 121512 | FGD4 | protein_coding | Q96M96 | FUNCTION: Activates CDC42, a member of the Ras-like family of Rho- and Rac proteins, by exchanging bound GDP for free GTP. Plays a role in regulating the actin cytoskeleton and cell shape. Activates MAPK8 (By similarity). {ECO:0000250, ECO:0000269|PubMed:15133042}. | Actin-binding;Alternative splicing;Cell projection;Charcot-Marie-Tooth disease;Cytoplasm;Cytoskeleton;Disease variant;Guanine-nucleotide releasing factor;Metal-binding;Neurodegeneration;Neuropathy;Phosphoprotein;Reference proteome;Repeat;Zinc;Zinc-finger | This gene encodes a protein that is involved in the regulation of the actin cytoskeleton and cell shape. This protein contains an actin filament-binding domain, which together with its Dbl homology domain and one of its pleckstrin homology domains, can form microspikes. This protein can activate MAPK8 independently of the actin filament-binding domain, and it is also involved in the activation of CDC42 via the exchange of bound GDP for free GTP. The activation of CDC42 also enables this protein to play a role in mediating the cellular invasion of Cryptosporidium parvum, an intracellular parasite that infects the gastrointestinal tract. Mutations in this gene can cause Charcot-Marie-Tooth disease type 4H (CMT4H), a disorder of the peripheral nervous system. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jan 2015]. | hsa:121512; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; filopodium [GO:0030175]; Golgi apparatus [GO:0005794]; lamellipodium [GO:0030027]; ruffle [GO:0001726]; actin binding [GO:0003779]; guanyl-nucleotide exchange factor activity [GO:0005085]; metal ion binding [GO:0046872]; small GTPase binding [GO:0031267]; actin cytoskeleton organization [GO:0030036]; cytoskeleton organization [GO:0007010]; filopodium assembly [GO:0046847]; regulation of cell shape [GO:0008360]; regulation of GTPase activity [GO:0043087]; regulation of small GTPase mediated signal transduction [GO:0051056] | 15133042_role of frabin, a guanine nucleotide exchange factor specific for Cdc42, in the activation of Cdc42 during Cryptosporidium parvum infection of biliary epithelial cells 17564959_The identification of mutations in FGD4, encoding FGD4 or FRABIN (FGD1-related F-actin binding protein), in families with Charcot-Marie-Tooth type 4H. 17564972_We report that disruption of frabin/FGD4, a GEF for the Rho GTPase cell-division cycle 42 (Cdc42), causes peripheral nerve demyelination in patients with autosomal recessive Charcot-Marie-Tooth (CMT) neuropathy. 19221294_A novel homozygous Frabin (FGD4) nonsense mutation p.R275X is identified in a family with Charcot-Marie Tooth disease (CMT4H) from Northern Ireland. 19332693_Genetic heterogeneity of FGD4 demonstrates that CMT4H has variable functional impairment and suggests that frabin plays a crucial role during myelin formation. 19494521_differentialy expressed in dendritic cells upon stimulation of with with the major house dust mite allergen Der p 1 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22295116_susceptibility genes associated with alcohol drinking 22589722_LMP1, through its transmembrane domains, directly bound FGD4 and enhanced FGD4 activity toward Cdc42, leading to actin cytoskeleton rearrangement and increased motility of NPC cells. 22734899_we have identified two novel missense mutations in FGD4 in two patients affected by autosomal recessive demyelinating Charcot-Marie-Tooth disease 22843789_A single nucleotide polymorphism in FGD4 was associated with the onset of paclitaxel-induced sensory peripheral neuropathy. 23550889_Our results suggest that FGD4 should be screened in other early-onset CMT subtypes, regardless of the severity of the phenotype, and particularly in patients of consanguineous descent. 26400421_identified two different pairs of novel compound heterozygous mutations in the FGD4 gene from nonconsanguineous Korean Charcot-Marie-Tooth disease type 4H families 27736846_FGD4 c.2044-236 A-allele carriers had an increased risk of paclitaxel dose reduction (HR per A-allele=1.38, P=0.036) when adjusted for total cumulative paclitaxel dose 30558664_Results show that the expression of FGD4 is upregulated in cancerous prostates compared to the luminal cells in benign prostatic hyperplasia and demonstrate a tumor promoting and a cell migratory function of FGD4 in prostate cancer cells. Its inhibition enhances the response for both androgen-dependent and independent prostate cancer cells towards currently used prostate cancer drugs. 31852984_Identification of novel pathogenic copy number variations in Charcot-Marie-Tooth disease. 32633323_Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/beta-catenin signalling pathway. 32772928_MiR-23a induced the activation of CDC42/PAK1 pathway and cell cycle arrest in human cov434 cells by targeting FGD4. 33709789_Polymorphism of FGD4 and myelosuppression in patients with esophageal squamous cell carcinoma. | ENSMUSG00000022788 | Fgd4 | 126.03191 | 0.6768117 | -0.5631734859 | 0.25934285 | 4.648961e+00 | 3.107235e-02 | 2.785889e-01 | No | Yes | 101.555678 | 31.871009 | 150.798449 | 46.367406 | |
| ENSG00000139163 | 55500 | ETNK1 | protein_coding | Q9HBU6 | FUNCTION: Highly specific for ethanolamine phosphorylation. May be a rate-controlling step in phosphatidylethanolamine biosynthesis. {ECO:0000269|PubMed:11044454}. | ATP-binding;Alternative splicing;Cytoplasm;Kinase;Lipid biosynthesis;Lipid metabolism;Nucleotide-binding;Phospholipid biosynthesis;Phospholipid metabolism;Reference proteome;Transferase | PATHWAY: Phospholipid metabolism; phosphatidylethanolamine biosynthesis; phosphatidylethanolamine from ethanolamine: step 1/3. {ECO:0000269|PubMed:11044454}. | This gene encodes an ethanolamine kinase, which functions in the first committed step of the phosphatidylethanolamine synthesis pathway. This cytosolic enzyme is specific for ethanolamine and exhibits negligible kinase activity on choline. Alternative splicing results in multiple transcript variants encoding distinct isoforms. [provided by RefSeq, Jul 2008]. | hsa:55500; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; ATP binding [GO:0005524]; ethanolamine kinase activity [GO:0004305]; phosphatidylethanolamine biosynthetic process [GO:0006646] | 11912161_DAD-R, SOX5, and EKI1 map within region of chromosome 12p whose duplication is related to reduced apoptosis in human testicular seminomas. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 25343957_Recurrent ETNK1 mutations are associated with chronic myeloid leukemia. 25615281_we identified novel somatic missense ETNK1 mutations that were most frequent in systemic mastocytosis with eosinophilia and chronic myelomonocytic leukemia 31255331_miR-199a-3p regulates the invasion and migration of gastric cancer cells by targeting ETNK1 33230096_ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine. | ENSMUSG00000030275 | Etnk1 | 3868.02478 | 1.1138306 | 0.1555298968 | 0.09843856 | 2.505019e+00 | 1.134841e-01 | 4.795307e-01 | No | Yes | 4281.343854 | 834.775958 | 3766.643401 | 718.421383 |
| ENSG00000139631 | 51380 | CSAD | protein_coding | Q9Y600 | FUNCTION: Catalyzes the decarboxylation of L-aspartate, 3-sulfino-L-alanine (cysteine sulfinic acid), and L-cysteate to beta-alanine, hypotaurine and taurine, respectively. The preferred substrate is 3-sulfino-L-alanine. Does not exhibit any decarboxylation activity toward glutamate. {ECO:0000250|UniProtKB:Q9DBE0}. | 3D-structure;Alternative splicing;Decarboxylase;Lyase;Pyridoxal phosphate;Reference proteome | PATHWAY: Organosulfur biosynthesis; taurine biosynthesis; hypotaurine from L-cysteine: step 2/2. | This gene encodes a member of the group 2 decarboxylase family. A similar protein in rodents plays a role in multiple biological processes as the rate-limiting enzyme in taurine biosynthesis, catalyzing the decarboxylation of cysteinesulfinate to hypotaurine. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Sep 2011]. | hsa:51380; | cytoplasm [GO:0005737]; aspartate 1-decarboxylase activity [GO:0004068]; pyridoxal phosphate binding [GO:0030170]; sulfinoalanine decarboxylase activity [GO:0004782]; L-cysteine catabolic process to hypotaurine [GO:0019449]; L-cysteine catabolic process to taurine [GO:0019452]; taurine biosynthetic process [GO:0042412] | 22718265_The presence of cysteine inhibited ADC, CSAD and GDC activity. 26327310_taurine biosynthesis in vertebrates involves two structurally related PLP-dependent decarboxylases (cysteine sulfinic acid decarboxylase and glutamic acid decarboxylase like 1) | ENSMUSG00000023044 | Csad | 113.69616 | 1.1951623 | 0.2572065691 | 0.27361150 | 8.871029e-01 | 3.462636e-01 | 7.199576e-01 | No | Yes | 131.828857 | 31.894312 | 115.121019 | 27.280345 |
| ENSG00000140105 | 7453 | WARS1 | protein_coding | P23381 | FUNCTION: Isoform 1, isoform 2 and T1-TrpRS have aminoacylation activity while T2-TrpRS lacks it. Isoform 2, T1-TrpRS and T2-TrpRS possess angiostatic activity whereas isoform 1 lacks it. T2-TrpRS inhibits fluid shear stress-activated responses of endothelial cells. Regulates ERK, Akt, and eNOS activation pathways that are associated with angiogenesis, cytoskeletal reorganization and shear stress-responsive gene expression. {ECO:0000269|PubMed:11773625, ECO:0000269|PubMed:11773626, ECO:0000269|PubMed:1373391, ECO:0000269|PubMed:14630953, ECO:0000269|PubMed:28369220}. | 3D-structure;ATP-binding;Alternative splicing;Aminoacyl-tRNA synthetase;Angiogenesis;Cytoplasm;Direct protein sequencing;Disease variant;Ligase;Neurodegeneration;Neuropathy;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;Reference proteome | Aminoacyl-tRNA synthetases catalyze the aminoacylation of tRNA by their cognate amino acid. Because of their central role in linking amino acids with nucleotide triplets contained in tRNAs, aminoacyl-tRNA synthetases are thought to be among the first proteins that appeared in evolution. Two forms of tryptophanyl-tRNA synthetase exist, a cytoplasmic form, named WARS, and a mitochondrial form, named WARS2. Tryptophanyl-tRNA synthetase (WARS) catalyzes the aminoacylation of tRNA(trp) with tryptophan and is induced by interferon. Tryptophanyl-tRNA synthetase belongs to the class I tRNA synthetase family. Four transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:7453; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleus [GO:0005634]; protein-containing complex [GO:0032991]; ATP binding [GO:0005524]; kinase inhibitor activity [GO:0019210]; protein domain specific binding [GO:0019904]; protein homodimerization activity [GO:0042803]; protein kinase binding [GO:0019901]; tryptophan-tRNA ligase activity [GO:0004830]; angiogenesis [GO:0001525]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of protein kinase activity [GO:0006469]; negative regulation of protein phosphorylation [GO:0001933]; positive regulation of gene expression [GO:0010628]; positive regulation of protein-containing complex assembly [GO:0031334]; regulation of angiogenesis [GO:0045765]; regulation of protein ADP-ribosylation [GO:0010835]; translation [GO:0006412]; tryptophanyl-tRNA aminoacylation [GO:0006436] | 11773625_In this study, we show that a recombinant form of a COOH-terminal fragment of TrpRS is a potent antagonist of vascular endothelial growth factor-induced angiogenesis in a mouse model and of naturally occurring retinal angiogenesis in the neonatal mouse 11773626_Thus, protein synthesis may be linked to the regulation of angiogenesis by a natural fragment of TrpRS 11834741_Recognition by tryptophanyl-tRNA synthetases of discriminator base on tRNATrp from three biological domains 12416978_The recently discovered antiangiogenic and cell-signaling activities of tryptophanyl-tRNA synthetase bioactive fragments are discussed in this review. 14630953_TrpRS may have a role in the maintenance of vascular homeostasis 14660560_results suggest that mammalian and bacterial tryptophanyl-tRNA synthetase might use different mechanisms to recognize the substrate and modeling studies indicate that transfer RNA binds with the dimeric enzyme 14671330_A crystal structure of human tryptophanyl-tRNA synthetase was solved at 2.1 A with a tryptophanyl-adenylate bound at the active site 15628863_May play an important role in the intracellular regulation of protein synthesis under conditions of oxidative stress. 15939065_Observational study of gene-disease association. (HuGE Navigator) 16724112_These crystals captured two conformations of the human tryptophanyl-tRNA synthetase and tRNATrp complex, which are nearly identical with respect to the protein and a bound tryptophan. 16798914_The first crystal structure of human tryptophanyl-tRNA synthetase (hTrpRS) in complex with tRNA(Trp) and Trp which, together with biochemical data, reveals the molecular basis of a novel tRNA binding and recognition mechanism. 17877375_Results provide the first evidence of the involvement of heme in regulation of TrpRS aminoacylation activity. 17999956_the annexin II-S100A10 complex, which regulates exocytosis, forms a ternary complex with TrpRS. 18180246_Analysis of the molecular basis of the mechanisms of the substrate recognition and the activation reaction by tryptophanyl-tRNA synthetase. 19363598_Indoleamine 2,3-dioxygenase (IDO)-expression in antigen-presenting cells (APCs) may control autoimmune responses by depleting the available tryptophan, whereas tryptophanyl-tRNA synthetase (TTS) may counteract this effect 19768679_Tryptophanyl-tRNA synthetase is a multidomain protein exhibiting excellent allosteric communication, and this research has provided valuable structural as well as functional insights into the protein. 19900940_Low tryptophanyl-tRNA synthetase is associated with recurrence in colorectal cancer. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20963594_Mini-tryptophanyl-tRNA synthetase inhibited ischemic angiogenesis in rats. 21442253_Naturally occurring fragments of the two proteins involved in translation, TyrRS and TrpRS, have opposing activities on angiogenesis. 21926542_Tryptophanyl-tRNA synthetase down-regulation by hypoxia may be a factor responsible for low TrpRS in pancreatic tumors with high metastatic ability. 23651343_Indoleamine2,3-dioxygenase and tryptophanyl-tRNA synthetase may play critical roles in the immune pathogenesis of chronic kidney disease. 23670221_Genes within recently identified loci associated with waist-hip ratio (WHR) exhibit fat depot-specific mRNA expression, which correlates with obesity-related traits. Adipose tissue (AT) mRNA expression of 6 genes (TBX15/WARS2, STAB1, PIGC, ZNRF3, GRB14) 24515434_Tryptophanyl-tRNA synthetase expression is up-regulated in patients with rheumatoid arthritis. 26209610_Overexpression of WARS predicts no recurrence and good survival for triple-negative breast cancer patients. 27748732_Based on these results, secretion of full-length tryptophanyl-tRNA synthetase appears to work as a primary defence system against infection, acting before full activation of innate immunity. 28369220_findings establish WARS as a gene whose mutations may cause distal hereditary motor neuropathy and alter canonical and non-canonical functions of tryptophanyl-tRNA synthetase. 29666190_Inhibition of human tryptophanyl-tRNA synthetase (TrpRS) expression by TrpRS-specific siRNAs decreased and overexpression of TrpRS increased Trp uptake into the cells 30153112_Human tryptophanyl-tRNA synthetase is an IFN-gamma-inducible entry factor for Enterovirus. 30355684_Data report a novel noncanonical function of WARS in antiviral defense. WARS is rapidly secreted in response to viral infection and primes the innate immune response by inducing the secretion of proinflammatory cytokines and type I IFNs, resulting in the inhibition of virus replication both in vitro and in vivo. These results suggest that WARS is a member of the antiviral innate immune response. 31033497_Expression of Indoleamine 2, 3-dioxygenase 1 (IDO1) and Tryptophanyl-tRNA Synthetase (WARS) in Gastric Cancer Molecular Subtypes. 31786502_Mini tryptophanyl-tRNA synthetase is required for a synthetic phenotype in vascular smooth muscle cells induced by IFN-gamma-mediated beta2-adrenoceptor signaling. 32899943_Tryptophanyl-tRNA Synthetase 1 Signals Activate TREM-1 via TLR2 and TLR4. 33754909_Identification of Prognostic RBPs in Osteosarcoma. | ENSMUSG00000021266 | Wars | 4436.54220 | 0.9761893 | -0.0347671568 | 0.07554011 | 2.115602e-01 | 6.455473e-01 | 8.884066e-01 | No | Yes | 5626.486028 | 543.358889 | 5425.168055 | 512.289480 | |
| ENSG00000140464 | 5371 | PML | protein_coding | P29590 | FUNCTION: Functions via its association with PML-nuclear bodies (PML-NBs) in a wide range of important cellular processes, including tumor suppression, transcriptional regulation, apoptosis, senescence, DNA damage response, and viral defense mechanisms. Acts as the scaffold of PML-NBs allowing other proteins to shuttle in and out, a process which is regulated by SUMO-mediated modifications and interactions. Isoform PML-4 has a multifaceted role in the regulation of apoptosis and growth suppression: activates RB1 and inhibits AKT1 via interactions with PP1 and PP2A phosphatases respectively, negatively affects the PI3K pathway by inhibiting MTOR and activating PTEN, and positively regulates p53/TP53 by acting at different levels (by promoting its acetylation and phosphorylation and by inhibiting its MDM2-dependent degradation). Isoform PML-4 also: acts as a transcriptional repressor of TBX2 during cellular senescence and the repression is dependent on a functional RBL2/E2F4 repressor complex, regulates double-strand break repair in gamma-irradiation-induced DNA damage responses via its interaction with WRN, acts as a negative regulator of telomerase by interacting with TERT, and regulates PER2 nuclear localization and circadian function. Isoform PML-6 inhibits specifically the activity of the tetrameric form of PKM. The nuclear isoforms (isoform PML-1, isoform PML-2, isoform PML-3, isoform PML-4 and isoform PML-5) in concert with SATB1 are involved in local chromatin-loop remodeling and gene expression regulation at the MHC-I locus. Isoform PML-2 is required for efficient IFN-gamma induced MHC II gene transcription via regulation of CIITA. Cytoplasmic PML is involved in the regulation of the TGF-beta signaling pathway. PML also regulates transcription activity of ELF4 and can act as an important mediator for TNF-alpha- and IFN-alpha-mediated inhibition of endothelial cell network formation and migration.; FUNCTION: Exhibits antiviral activity against both DNA and RNA viruses. The antiviral activity can involve one or several isoform(s) and can be enhanced by the permanent PML-NB-associated protein DAXX or by the recruitment of p53/TP53 within these structures. Isoform PML-4 restricts varicella zoster virus (VZV) via sequestration of virion capsids in PML-NBs thereby preventing their nuclear egress and inhibiting formation of infectious virus particles. The sumoylated isoform PML-4 restricts rabies virus by inhibiting viral mRNA and protein synthesis. The cytoplasmic isoform PML-14 can restrict herpes simplex virus-1 (HHV-1) replication by sequestering the viral E3 ubiquitin-protein ligase ICP0 in the cytoplasm. Isoform PML-6 shows restriction activity towards human cytomegalovirus (HHV-5) and influenza A virus strains PR8(H1N1) and ST364(H3N2). Sumoylated isoform PML-4 and isoform PML-12 show antiviral activity against encephalomyocarditis virus (EMCV) by promoting nuclear sequestration of viral polymerase (P3D-POL) within PML NBs. Isoform PML-3 exhibits antiviral activity against poliovirus by inducing apoptosis in infected cells through the recruitment and the activation of p53/TP53 in the PML-NBs. Isoform PML-3 represses human foamy virus (HFV) transcription by complexing the HFV transactivator, bel1/tas, preventing its binding to viral DNA. PML may positively regulate infectious hepatitis C viral (HCV) production and isoform PML-2 may enhance adenovirus transcription. Functions as an E3 SUMO-protein ligase that sumoylates (HHV-5) immediate early protein IE1, thereby participating in the antiviral response (PubMed:20972456, PubMed:28250117). Isoforms PML-3 and PML-6 display the highest levels of sumoylation activity (PubMed:20972456, PubMed:28250117). {ECO:0000269|PubMed:20972456, ECO:0000269|PubMed:28250117}. | 3D-structure;Acetylation;Activator;Alternative splicing;Antiviral defense;Apoptosis;Biological rhythms;Chromosomal rearrangement;Coiled coil;Cytoplasm;DNA-binding;Endoplasmic reticulum;Endosome;Host-virus interaction;Immunity;Innate immunity;Isopeptide bond;Membrane;Metal-binding;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Repeat;Transcription;Transcription regulation;Transferase;Tumor suppressor;Ubl conjugation;Zinc;Zinc-finger | PATHWAY: Protein modification; protein sumoylation. {ECO:0000269|PubMed:20972456, ECO:0000269|PubMed:28250117}. | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. This phosphoprotein localizes to nuclear bodies where it functions as a transcription factor and tumor suppressor. Its expression is cell-cycle related and it regulates the p53 response to oncogenic signals. The gene is often involved in the translocation with the retinoic acid receptor alpha gene associated with acute promyelocytic leukemia (APL). Extensive alternative splicing of this gene results in several variations of the protein's central and C-terminal regions; all variants encode the same N-terminus. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:5371; | chromatin [GO:0000785]; chromosome, telomeric region [GO:0000781]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; early endosome membrane [GO:0031901]; extrinsic component of endoplasmic reticulum membrane [GO:0042406]; heterochromatin [GO:0000792]; nuclear matrix [GO:0016363]; nuclear membrane [GO:0031965]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; cobalt ion binding [GO:0050897]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; protein homodimerization activity [GO:0042803]; SMAD binding [GO:0046332]; SUMO binding [GO:0032183]; SUMO transferase activity [GO:0019789]; sumo-dependent protein binding [GO:0140037]; transcription coactivator activity [GO:0003713]; ubiquitin protein ligase binding [GO:0031625]; ubiquitin-protein transferase activity [GO:0004842]; zinc ion binding [GO:0008270]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; branching involved in mammary gland duct morphogenesis [GO:0060444]; cell fate commitment [GO:0045165]; cellular response to interleukin-4 [GO:0071353]; cellular response to leukemia inhibitory factor [GO:1990830]; cellular senescence [GO:0090398]; circadian regulation of gene expression [GO:0032922]; common-partner SMAD protein phosphorylation [GO:0007182]; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest [GO:0006977]; endoplasmic reticulum calcium ion homeostasis [GO:0032469]; entrainment of circadian clock by photoperiod [GO:0043153]; extrinsic apoptotic signaling pathway [GO:0097191]; fibroblast migration [GO:0010761]; innate immune response [GO:0045087]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator [GO:0042771]; intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress [GO:0070059]; intrinsic apoptotic signaling pathway in response to oxidative stress [GO:0008631]; maintenance of protein location in nucleus [GO:0051457]; myeloid cell differentiation [GO:0030099]; negative regulation of angiogenesis [GO:0016525]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of interleukin-1 beta production [GO:0032691]; negative regulation of mitotic cell cycle [GO:0045930]; negative regulation of telomerase activity [GO:0051974]; negative regulation of telomere maintenance via telomerase [GO:0032211]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of translation in response to oxidative stress [GO:0032938]; negative regulation of ubiquitin-dependent protein catabolic process [GO:2000059]; oncogene-induced cell senescence [GO:0090402]; PML body organization [GO:0030578]; positive regulation of apoptotic process involved in mammary gland involution [GO:0060058]; positive regulation of defense response to virus by host [GO:0002230]; positive regulation of extrinsic apoptotic signaling pathway [GO:2001238]; positive regulation of fibroblast proliferation [GO:0048146]; positive regulation of histone deacetylation [GO:0031065]; positive regulation of peptidyl-lysine acetylation [GO:2000758]; positive regulation of protein localization to chromosome, telomeric region [GO:1904816]; positive regulation of signal transduction by p53 class mediator [GO:1901798]; positive regulation of telomere maintenance [GO:0032206]; positive regulation of transcription, DNA-templated [GO:0045893]; proteasome-mediated ubiquitin-dependent protein catabolic process [GO:0043161]; protein import into nucleus [GO:0006606]; protein stabilization [GO:0050821]; protein targeting [GO:0006605]; protein-containing complex assembly [GO:0065003]; regulation of calcium ion transport into cytosol [GO:0010522]; regulation of cell adhesion [GO:0030155]; regulation of cell cycle [GO:0051726]; regulation of circadian rhythm [GO:0042752]; regulation of double-strand break repair [GO:2000779]; regulation of protein phosphorylation [GO:0001932]; regulation of transcription, DNA-templated [GO:0006355]; response to cytokine [GO:0034097]; response to gamma radiation [GO:0010332]; response to hypoxia [GO:0001666]; response to UV [GO:0009411]; retinoic acid receptor signaling pathway [GO:0048384]; suppression of viral release by host [GO:0044790]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 11891284_UBE1L is a retinoid target that triggers PML/RARalpha degradation and apoptosis in acute promyelocytic leukemia 11907221_contributes to innate immunity, defining host susceptibility to viral infections and immunopathology 12006491_Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. 12032336_variant-type PML-RAR(alpha) fusion transcript in acute promyelocytic leukemia 12032831_Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. 12060771_induced PML in a mouse model 12080044_represses A20-mediated transcription. 12093737_Of seven known PML isoforms, only PML IV is capable of causing premature senescence in PML +/+ murine and human primary fibroblasts but not in PML -/- murine cells, providing the first evidence for functional differences among these isoforms. 12167712_PML is a key stress-responsive regulator of eIF4E mRNA-dependent transport. PML inhibition of eIF4E depends on its association with the eIF4E nuclear body. 12186918_PML does not affect herpes simplex virus 1 replication or the changes in the localization of ICP0 through infection 12384561_PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. 12402044_PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2 12419228_SUMO-1 protease-1 regulates gene transcription through PML 12438698_Purified RINGs, including PML, self-assemble into supramolecular structures in vitro that resemble those they form in cells. Self-assembly controls and amplifies reduction of 5' mRNA cap affinity of eIF4E by PML. 12505266_Cryptic translocation of PML/RARA on 17q. A rare event in acute promyelocytic leukemia. 12506013_PML inhibited STAT3 activity in NIH3T3, 293T, HepG2, and 32D cells. PML formed a complex with STAT3 through B-box and COOH terminal regions in vitro and in vivo, thereby inhibiting its DNA binding activity. 12506025_In human cell lines, PML is not involved directly in the regulation of MHC class I expression. 12540841_PML induces apoptosis through repression of the NF-kappaB survival pathway. 12595526_Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. 12610143_herpes simplex virus type 1 ICP4 is recruited into foci juxtaposed to ND10 (PML) in live, infected cells 12647219_findings strongly suggest that increased promyelocytic leukemia protein expression is associated with growth inhibition and differentiation of human neuroblastoma cells 12727882_role of interaction in recruiting cyclin T1 to nuclear bodies 12773567_PML regulates TopBP1 functions by association and stabilization of the protein in response to IR-induced DNA damage 12810724_PML recruits Chk2 and p53 into the PML nuclear bodies and enhances p53/Chk2 interaction 12837286_PML protein colocalizes and interacts with RFN36. 12907596_PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. 12917339_We show here that ZIPK is present in PODs, where it colocalizes with and binds to proapoptotic protein Daxx. 14526201_alternative splicing and role implicated in interaction with HIV-1 14528266_Results suggest that adenovirus type 5 has evolved a unique strategy that leads to the sustained neutralization of promyelocytic leukaemia protein bodies throughout infection, thereby ensuring optimal viral proliferation. 14534537_Leukemia-associated translocation products able to activate RAS modify PML and render cells sensitive to arsenic-induced apoptosis. 14597622_PML-induced apoptosis by down-regulation of Survivin. 14597990_The ectopic expression of the acute promyelocytic leukemia-specific PML/RARalpha oncoprotein in U-937 cells results in induction of TF mRNA and promoter activity 14630830_DNA sequence variations resulting in a truncated PML protein was identified in acute promyelocytic leukemia cases that displayed retinoic acid resistance and a very poor prognosis 14663483_PML and the PML-NB act as molecular hubs for the induction and/or reinforcement of programmed cell death through a selective and dynamic regulation of proapoptotic transcriptional events. 14715247_Together our results suggest that PML may suppress prostate cancer cell growth by inhibiting AR transactivation and/or enhancing p53 activity. 14970191_Data suggest that promyelocytic leukemia (PML) bodies form in nuclear compartments of high transcriptional activity, but they do not directly regulate transcription of genes in these compartments. 14970276_PML protein expression is frequently lost in human cancers of various histologic origins, and its loss associates with tumor grade and progression in some tumor histotypes 14992722_PML is a direct p53 target that modulates p53 effector functions. 15060166_Results demonstrate that transcription activity associated with PML bodies is selectively repressed by the recruitment of Bach2 around PML bodies 15096541_acute promyelocytic leukemia cell differentiation parallels transcriptional activation through PML-RARA-RXR oligomers 15163746_interference by human Cytomegalovirus IE1 protein with both the sumoylation of PML and its repressor activity requires a physical interaction with PML that also leads to disruption of PML oncogenic domains (PODs). 15184504_PML plays a crucial role in modulating p73 function. 15273249_PML is a negative regulator of p38 kinase in tumor cells 15331439_PML I could act as a mediator for acute myeloid leukemia1 and its coactivator p300/CBP to assemble into functional complexes; a specific isoform PML I forms a complex with AML1 15359634_The 3 breakpoint cluster regions in the PML gene [Intron 6 (bcr1), exon 6 (bcr2) or intron 3 (bcr3)] were studied in 43 Mexican Mestizo APL patients. Bcr1=62.7%, bcr2=9.3% & bcr3=27.9%. Bcr1 was more prevalent than in Caucasians but similar to Asians. 15459016_TRAILprotein is a novel downstream transcriptional target of PML. 15467728_PML bodies control the distribution, dynamics and function of CHFR 15569683_SUMO-1 functions to tether proteins to PML-containing nuclear bodies through post-translational modification and noncovalent protein-protein interaction 15589835_INI1 is dispensable for retrovirus-induced cytoplasmic accumulation of PML and does not interfere with virus integration. 15601827_PML-retinoic acid receptor alpha activities are regulated by neutrophil elastase in early myeloid cells 15746941_CDDO-Im downregulates PML expression in acute promyleocytic leukemic cells. 15749021_PML3 has a direct role in the control of centrosome duplication through suppression of Aurora A activation to prevent centrosome reduplication 15809060_inhibition of monocyte differentiation all contribute to the oncogenic activity of PML-RARalpha 15855159_IEX-1 is organized in subnuclear structures and partially co-localizes with promyelocytic leukemia protein in HeLa cells 15922731_Isoforms of PML protein differ in their effects on ND10 organization. 15968309_Observational study of genetic testing. (HuGE Navigator) 16007146_findings strengthen the relevance of the cross talk between PML and the p53 family members, imply a new tumour suppressive function of PML and unveil a possible role for PML in epidermal morphogenesis and differentiation 16113082_loss of one copy of PU.1 through deletion, plus down-regulation of the residual allele caused by PML-RARalpha expression, synergizes to expand myeloid progenitors susceptible to transformation, increasing the penetrance of acute promyelocytic leukemia 16154611_These results indicate that PML isoforms which are expressed in a serum-dependent manner suppress the propagation of influenza virus at an early stage of infection. 16307818_Our work implies that the balance of promyelocytic leukemia protein and Epstein-Barr virus BZLF1 levels in cells may affect how each protein functions. 16432238_PML-RARalpha functions by recruiting an HDAC3-MBD1 complex that contributes to the establishment and maintenance of the silenced chromatin state 16449642_permanent transcriptional silencing is mediated by the association of PML-RAR with chromatin-modifying enzymes and by recruitment of the histone methyltransferase SUV39H1, responsible for trimethylation of lysine 9 of histone H3 16501113_Interaction between Adenovirus type 5 E4 Orf3 and PML isoform II is necessary for nuclear domain 10 (ND10) rearrangement to occur 16501610_PML stimulated hSUMO-1 modification in yeast, in a manner that was dependent upon PML's RING-finger domain. PML:RARalpha also stimulated hSUMO-1 conjugation in yeast. 16540467_analysis of cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha 16630218_frequencies of the PML-RARalpha transcripts and subtypes in a series of 32 APL patients from Northeast Brazil 16797070_The PML-RARA fusions can be identified by molecular analyses such as reverse transcriptase-polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH). 16818720_We show that expression of PML isoform IV leads to formation of distinct nuclear bodies enriched in components of the ubiquitin-proteasome system. These bodies recruit soluble mutant ataxin-7 and promote its degradation preventing aggregate formation 16873256_PML contributes to a cellular antiviral repression mechanism that is countered by the activity of ICP0. 16873257_PML functions as part of an intrinsic immune mechanism against cytomegalovirus infections. 16912307_Our results provide evidence of how poliovirus counteracts p53 antiviral activity by regulating PML and NBs, thus leading to p53 degradation. 16916642_The interaction of TGIF with cPML through c-Jun may negatively regulate TGF-beta signaling through controlling the localization of cPML and, consequently, the assembly of the cPML-SARA complex. 16924230_apoptin interacts directly with the promyelocytic leukemia protein (PML) in tumor cells and accumulates in PML nuclear bodies 16935935_In response to ATRA, PML/RARalpha is dissociated from CAK, leading to MAT1 degradation, G1 arrest, and decreased CAK phosphorylation of PML/RARalpha 17027752_SUMO-1, PML and ZNF198 colocalize to punctate structures, shown by immunocytochemistry to be PML bodies. 17030982_PML nuclear bodies increase in number following DNA damage and this response is regulated by NBS1 as well as the kinases ATM, Chk2 and ATR. 17062732_Histone deacetylase inhibitors blocked the increase of PML nuclear body number and suppressed up-regulation of PML mRNA and protein levels in several human cell lines and in normal diploid skin fibroblasts 17146439_establish PML-mediated destabilization of Myc and the derepression of cell cycle inhibitor genes as an important regulatory mechanism that allows cell differentiation 17172828_the role of PML cytoplasmic proteins in the regulation of p53. 17173041_Our studies identify PML and SATB1 as a regulatory complex that governs transcription by orchestrating dynamic chromatin-loop architecture. 17360386_The increased expression of SUMO-1 in rheumatoid arthritis (RA) synovial fibroblasts (SFs) contributes to the resistance of these cells against Fas-induced apoptosis through increased SUMOylation of nuclear PML protein. 17419608_Results demonstrate that the gamma 1 isoform of phospholipase C associates with nuclear promyelocytic leukemia protein. 17428679_PML may be involved in nucleolar functions of normal non-transformed or senescent cells. The absence of nucleolar PML compartment in rapidly growing tumor cells suggests that PML association with the nucleolus might be important for cell-cycle regulation. 17441430_PML may play a role in cancer repression but not in cancer metastasis in laryngocarcinoma and nasopharyngeal carcinoma. 17475621_the hormone-independent association between PML-RARalpha and coactivators contributes to its ability to regulate gene expression 17543368_Study recapitulated the effect of PML on early steps in infection by papillomavirus, it was found that replication and transcription of transfected paillomaviral genomes were not dependent on PML. 17562868_The type IV isoform of PML interacted with PU.1, promoted its association with p300, and then enhanced PU.1-induced transcription and granulocytic differentiation and PU.1 directly activates the transcription of the C/EBPepsilon gene. 17767548_expression of proto-oncogene PML and HLA class I molecules were concordantly upregulated in hepatocellular carcinoma (HCC) and the expression of PML gene might be one of the mechanisms that leads to the increased expression of class I antigen in HCC 17822314_Downregulation of the PML protein is associated with breast cancers 17878236_Only two specific PML splice variants (PML-I and PML-IV) are efficiently targeted to the nucleolus and the abundant PML-I isoform is required for the targeting of endogenous PML proteins to this organelle. 17942542_promyelocytic leukemia protein (PML) and hDaxx, mediate an intrinsic immune response against HCMV infection by contributing independently to the silencing of HCMV IE gene expression. 17943164_PML/RAR gene cytogenetic abnormalities in APL 17960172_The presence of a single PML-RARA isoform with exon 5 is associated with recurrence in acute promyelocytic leukemia 17991421_These results would shed new insights for the molecular mechanisms of PML-RARalpha-associated leukemogenesis. 17996922_Endogenous WRN and BLM proteins localize in nucleoli and in nuclear PML bodies defined by isoforms of the PML protein, which is a key regulator of cellular senescence. 18024792_The transcription factor PML-RARalpha regulates key cancer-related genes and pathways by inducing a repressed chromatin formation on its direct genomic target genes. 18039859_Binding to Pin1 results in degradation of PML in a phosphorylation-dependent manner. Degradation of PML due to Pin1 acts both to protect a breast cancer tumor cell line from hydrogen peroxide-induced death and to increase the rate of proliferation. 18056407_In alternative lengthening of telomere cells, TRF2 inactivation/silencing triggers cellular senescence and substantial loss of telomeric DNA upon stable TRF2 knockdown. 18158568_Overexpression of PML was associated with poor survival in gallbladder carcinomas. 18160400_the Hes6-CBP complex in PML-NB may influence the proliferation of cells via p53-dependent and -independent pathways. 18246125_Unexpected coupling of PML with NFAT reveals a novel mechanism underlying the diverse physiological functions of promyelocytic leukemia. 18246126_following DNA damage, PML facilitates Thr18 phosphorylation by recruiting p53 and CK1 into PML nuclear bodies, thereby protecting p53 from inhibition by Mdm2, leading to p53 activation. 18298799_Over-expression of PML-2KA mutant in the cytoplasm, which was generated by mutagenesis of the nuclear localization signals of PML, in MCF-7 breast cancer cells suppressed PKM2 activity and the accumulation of lactate. 18463162_PML sequesters HDAC7 to relieve repression and up-regulate gene expression 18480450_reorganization of the PML nuclear body by E4 ORF3 antagonizes an innate antiviral response mediated by both PML and Daxx. 18511908_PCTA defines a new component of the TGF-beta signalling pathway that functions to facilitate Smad2 phosphorylation through controlling the accumulation of cPML into the cytoplasm, and consequently, the assembly of Smad2-receptor complex. 18566754_These data identify a key post-translational mechanism that controls PML protein levels in cancer cells and suggest that CK2 inhibitors may be beneficial anti-cancer drugs. 18621739_PML is involved in trichostatin A-induced apoptosis 18625722_These results demonstrate a novel function of HDAC7 and provide a regulatory mechanism of PML sumoylation. 18636556_PML/RARalpha fusion protein mediates the unique sensitivity to arsenic cytotoxicity in acute promyelocytic leukemia cells. 18664490_A kinetics model for factor exchange at PML NBs and highlight potential mechanisms to regulate intranuclear trafficking of specific factors at these domains. 18689862_PML is a novel prognostic tool for ampullary cancer patients. 18716620_results delineate a previously unknown PML-DAXX-HAUSP molecular network controlling PTEN deubiquitinylation and trafficking, which is perturbed by oncogenic cues in human cancer 18755984_In a series of human AMKL samples from both Down syndrome and non-Down syndrome patients, mutations were identified within KIT, FLT3, JAK2, JAK3, and MPL genes, with a higher frequency in DS than in non-DS patients. 18809579_purified the PML complex and identified Fbxo3 (Fbx3), Skp1, and Cullin1 as novel components amd role in transcriptional regulation and leukemogenesis 18812519_Reduced PML Protein Expression Is associated with glioblastoma. 18833293_PML disruption by EBNA1 requires binding to the cellular ubiquitin specific protease, USP7 or HAUSP, but is independent of p53. 18856066_the increase of level of survivin in tumor tissues is not the result of decrease in content of its inhibitors SMAC and PML, as their content in tumor and normal cells is the same. 18945770_The results indicate that both sets of events mediated by ICP0, the degradation of PML and the blocking of silencing via the HDAC1/2-CoREST-REST complex, are interdependent and in large measure dependent on events in the ND10 nuclear bodies. 19015637_DNA damage-induced PML SUMOylation and are required for the ability of PML to cooperate with HIPK2 for the induction of cell death. 19023333_Promyelomonocytic leukemia activation is a critical early event that participates in the apoptotic demise of HIV-1-elicited syncytia. 19029980_whereas transcriptional activation of PML-RARA is likely to control differentiation, its catabolism triggers leukemia-initiating cell eradication and long-term remission of mouse acute promyelocytic leukemia 19074885_Observational study of gene-disease association. (HuGE Navigator) 19100514_Acute promyelocytic leukemia with insertion of PML exon 7a and partial deletion of exon 3 of RARA is reported. 19111660_The existence of a proapoptotic autoregulatory feedback loop between p73, YAP, and the promyelocytic leukemia (PML) tumor suppressor gene, is shown. 19150978_The physical interaction between PML3 and TIP60 protects TIP60 from Mdm2-mediated degradation, suggesting that PML3 competes with MDM2 for binding to TIP60. 19224461_We report three of 40 diagnosed APL cases that showed morphological, cytochemical, and immunophenotypic features of hypergranular APL, but did not show a PML/RARalpha fusion signal or any of its variants, on FISH. 19322209_Knock-in human PML-RARalpha alone can confer properties of self-renewal to committed murine hematopoietic progenitors before the onset of disease. 19339552_Results provide insight into a dynamic pool of cytoplasmic nucleoporins that form a complex with the tumor suppressor protein PML during the G1 phase of the cell cycle. 19380586_sumoylation- and Sumo binding domain-dependent PML oligomerization within nuclear bodies is sufficient for RNF4-mediated PML degradation 19487292_In the context of mutant p53, PML enhances its cancer-promoting activities. 19553342_The authors demonstrate that LANA2 relieves PML-mediated transcriptional repression of survivin, a protein that directly contributes to malignant progression of primary effusion lymphoma cells. 19597475_PML was found to be important for E1A-induced suppression of EGFR and subsequent killing of head and neck squamous cell carcinoma cells suggesting a novel pathway involving PML and p73 in the regulation of EGFR expression. 19703418_this is the first report of the variable susceptibility of influenza A virus subtypes/strains to human PML proteins. 19728758_Frequency of the c.2260G>C (p.Ala754Pro) variant in isoform IV of the PML gene was higher in patients with colon polyposis and cancer 19808906_PML in PML-NBs links the DNA damage response with HBV replication and may cooperate with HBV-core and HDAC1 on the HBV circular DNA basal core promoter to form a positive feedback loop for HBV exacerbation during chemotherapy and radiotherapy. 19851296_Observational study of gene-disease association. (HuGE Navigator) 19861416_Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. 19863682_alternative splicing of PML/RARalpha transcripts might be involved in nonsense-mediated decay 19865110_PML-RARalpha ligand-binding domain deletion mutations associated with reduced disease control and outcome after first relapse of acute promyelocytic leukemia. 20100838_Promyelocytic leukemia protein controls cell migration in response to hydrogen peroxide and insulin-like growth factor-1 20123968_Data show that the major histocompatibility complex class II gene DRA is relocated to promyelocytic leukemia (PML) nuclear bodies upon induction with IFN-gamma, and this topology is maintained long after transcription shut off. 20130140_During interphase PML-NBs adopt a spherical organization characterized by the assembly of PML and Sp100 proteins into patches within a 50- to 100-nm-thick shell. 20133705_PML/RARalpha transactivates the TF promoter through an indirect interaction with an element composed of a GAGC motif and the flanking nucleotides, independent of AP-1 binding. 20133893_Patients with decreased expression of PML protein have higher adverse clinical features in a subgroup of acute promyelocytic leukemia. 20155840_We studied 21 PML-RARA positive/RARA-PML negative cases by bubble PCR and multiplex long template PCR to identify the genomic breakpoints. 20159609_PML-RARalpha/RXR functions as a local chromatin modulator and that specific recruitment of histone deacetylase activities to genes important for hematopoietic differentiation, RAR signaling, and epigenetic control is crucial to its transforming potential. 20159610_Selective targeting of PU.1-regulated genes by PML/RARalpha is a critical mechanism for the pathogenesis of acute promyelocytic leukemia(APL). 20181954_SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. 20198315_Observational study of gene-disease association. (HuGE Navigator) 20228380_Promyelocytic leukemia protein (PML) was immunolocalized within nuclei of villus mesenchyme, but largely absent in trophoblast nuclei, with a trend for increased PML reactivity in preeclamptic placenta. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20501696_MORC3 colocalizes with PML by a two-step molecular mechanism. 20544846_PML breakpoints are associated with acute promyelocytic leukemia. 20574048_autophagic degradation contributes significantly both to the basal turnover as well as the therapy-induced proteolysis of PML/RARA 20578240_Data describe differences in promyelocytic leukemia protein expression between different types of soft tissue sarcoma and the corresponding normal surrounding tissue. 20625391_Data show that the CSNK2A1P gene is a functional proto-oncogene in human cancers and its functional polymorphism appears to degrade PML differentially in cancer cells. 20639899_results together with earlier work are consistent with the idea that SUMOylation regulates targeting of E1B-55K to PML-containing subnuclear structures, known to control transcriptional regulation, tumour suppression, DNA repair and apoptosis 20699642_Upregulation of the PML tumor suppressor in cellular senescence triggered by diverse drugs including clinically used anti-cancer chemotherapeutics relies on stimulation of PML transcription by JAK/STAT-mediated signaling. 20719947_an interaction between EBNA1 and the host CK2 kinase is crucial for EBNA1 to disrupt PML bodies and degrade PML proteins 20800603_Observational study of gene-disease association. (HuGE Navigator) 20826694_SUMOylation promotes PML degradation during encephalomyocarditis virus infection. 20972455_have demonstrated that the lymphoid leukemia-associated protein, PAX5-PML chimeric oncogenic protein, could bind to PAX5 response-element as homodimer to inhibit the transactivation of PAX5 target-genes 21037079_Pilocytic astrocytomas have telomere-associated promyelocytic leukemia bodies without alternatively lengthened telomeres. 21042751_Suppression of telomerase could activate the PML-dependent p53 signaling pathway and inhibit bladder cancer cell growth. 21057547_found that dominant-negative mutants of PML blocked AXIN-induced p53 activation, and that AXIN promotes PML sumoylation, a modification necessary for PML functions 21092142_The study emphasizes a role of the variable C-terminus in subcellular target selection and a general role of the N-terminal tripartite motif domain in promoting protein clustering. 21115099_PML functions as a positive regulator of IFNgamma signaling 21161613_Recent literature on the role of PML in the central nervous system. 21172801_PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. 21187718_Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein 21192925_These results suggest that SUMO interaction motif-mediated HIPK2 targeting to PML-NBs is crucial for HIPK2-mediated p53 activation and induction of apoptosis. 21198351_PML is induced by interferon, leading to a marked increase of expression of PML isoforms and the number of PML nuclear bodies [Review] 21205865_the mechanism by which PML induces a permanent cell cycle exit and activates p53 and senescence 21217775_data suggest the possible involvement of this fusion protein in the leukemogenesis of B-ALL in a dual dominant-negative manner and the possibility that ALL with PAX5-PML can be treated with ATO 21240990_the clinicopathological associations and prognostic implications of promyelocytic leukemia gene (PML) expressions in patients with esophageal squamous cell carcinomas 21245861_New insights that arose from these studies, in particular focussing on newly identified PML-RARalpha target genes, its interplay with RXR and deregulation of epigenetic modifications. 21304169_The antiviral functions of PML-nuclear bodies are inactivated through reorganization during normalBK virus infection. 21304940_The efficient sequestration of virion capsids in PML cages appears to be the outcome of a basic cytoprotective function of this distinctive category of PML-NBs in sensing and safely containing nuclear aggregates of aberrant proteins. 21305000_Silencing of USP7 was found to increase the number of PML-NBs, to increase the levels of PML protein and to inhibit PML polyubiquitylation in nasopharyngeal carcinoma cells 21310922_Depletion of S100A10 by RNA interference effectively blocked the enhanced fibrinolytic activity observed after induction of the PML-RAR-alpha oncoprotein 21364283_physiologic PML-RARA expression was sufficient to direct a hematopoietic progenitor self-renewal program in vitro and in vivo 21430051_These findings demonstrate that Us3 orthologues derived from distantly related alphaherpesviruses cause a disruption of PML protein nuclear bodies in a kinase- and proteasome-dependent manner but, do not target PML for degradation. 21489587_UL35 bodies form independently of PML and subsequently recruit PML, Sp100 and Daxx. 21529941_PML-ADAMTS17-RARA gene rearrangement is associated with pregnancy-related acute promyelocytic leukemia. 21613260_Data indicate that PML-RARA (PR)-WT in the soluble fraction was transferred to the insoluble fraction after treatment with AsO, but PR-B/L-mut was stably detected in fractions both with and without AsO. 21639834_PML interacts with WRN and regulates double-strand break repair in gamma-irradiation-induced DNA damage responses. 21678421_PML immunohistochemical results were correlated with prognosis and with radiological response to alkylating agents/antracycline-based first line therapy in soft tissue sarcoma. 21752469_The MPL immunohistochemical expression may represent a new marker for differential diagnosis between essential thrombocythemia and pre-fibrotic stage primary myelofibrosis. 21803845_PML functions in apoptosis regulation and tumor suppression are mediated by direct interaction with Fas. 21832009_Neither Daxx nor PML, the main players of ND10-based immunity, are required for the block to viral gene expression in the S/G2 phase. 21840486_study indicates that the KLHL20-mediated PML degradation and HIF-1alpha autoregulation play key roles in tumor progression 21934296_investigation of DNA breakpoints in PML-RARalpha V-form fusion transcripts in 7 Chinese acute promyelocytic leukemia patients (out of total of 134 APL patients); in 5 of these cases, the V-form transcripts were unique (i.e., not described previously) 21994459_The authors demonstrated that only isoform IV of PML interacted with encephalomyocarditis virus 3D polymerase (3Dpol) and sequestered it within nuclear bodies. 21998700_a novel functional connection between PML and the homologous recombination-mediated repair machinery 22022583_Loss of PML protein expression in gastric cancer cells contributes to increased IP-10 transcription. 22033920_a model in which Pin1 promotes PML degradation in an ERK2-dependent manner. 22045732_Assembly of a PML nuclear body occurs through multiple pathways and induces telomere elongation. 22155184_beta-catenin inhibits promyelocytic leukemia protein tumor suppressor function in colorectal cancer cells. 22167334_PML-RARa bcr1 fusion is not responsible for colorectal tumor development. 22237204_PML through its scaffold properties is able to control cell growth and survival at many different levels. (Review) 22296450_The 15q24 microdeletion may thus represent the first genetic hit to initiate leukaemogenesis and implicates PML and SUMO3 as novel components of the leukaemogenic network in TMD/AMKL. 22406621_PIAS1 was essential for PML degradation in non-small cell lung carcinoma (NSCLC) cells, and PML and PIAS1 were inversely correlated in NSCLC cell lines and primary specimens. 22498738_HK3 is: (1) directly activated by PU.1, (2) repressed by PML-RARA, and (3) functionally involved in neutrophil differentiation and cell viability of acute promyelocytic leukemia cells. 22573317_E4-ORF3 activity correlates with the inhibition of PML-mediated antiviral activity. 22589541_Data indicate that promyelocytic leukemia protein (PML) functions as a negative regulator in endothelial cell network formation and migration. 22711534_As IL6 is induced in response to various viral and genotoxic stresses, this cytokine may regulate autocrine/paracrine induction of PML under these pathophysiological states 22773875_Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation. 22875967_Herpes simplex virus 1 ICP0 has 2 distinct mechanisms of targeting PML: one dependent on SUMO modification and the other via SUMO-independent interaction with PML.I; conclude that the ICP0-PML.I interaction reflects a countermeasure to PML-related antiviral restriction 22886304_PML expression in breast cancer correlated strikingly with reduced time to recurrence, a gene signature of poor prognosis, and activated PPAR signaling. 22906876_results suggest that PML expression was positively associated with IL-6 expression in patients and was also related to tumor development and resistance to treatment in multiple myeloma. 22915647_PML-RARA rearrangement is associated with good response to therapy in acute myeloid leukemia. 22918509_Postulate that detection of PML cytoplasmic particles in patient fibroblasts could become a valuable marker for diagnosis of disease development in laminopathies. 22945642_These findings suggest that UHRF1 promotes the turnover of PML protein 22947142_PML is an essential regulator of TNFalpha signaling, and together they synergistically regulate cell adhesion by engaging multiple molecular mechanisms. 22982005_analysis of a cryptic PML-RARA translocation in a patient with acute promyelocytic leukemia [case report] 23007646_PML is required for efficient IFN-gamma-induced MHC II gene transcription through regulation of the class II transactivator (CIITA) 23064710_detection of double-stranded polymerase chain reaction (PCR) products of PML/RARalpha fusion gene in acute promyelocytic leukemia (APL) 23 | ENSMUSG00000036986 | Pml | 583.54262 | 1.0673610 | 0.0940481571 | 0.13302657 | 5.009784e-01 | 4.790706e-01 | 8.097829e-01 | No | Yes | 706.422411 | 81.920562 | 630.543994 | 71.484850 |
| ENSG00000140471 | 55180 | LINS1 | protein_coding | Q8NG48 | Alternative splicing;Disease variant;Mental retardation;Phosphoprotein;Reference proteome | The Drosophila segment polarity gene lin encodes a protein, lines, which plays important roles in development of the epidermis and hindgut. This gene encodes a protein containing a lines-like domain. This gene is located on chromosome 15 and clustered with the gene encoding ankyrin repeat and SOCS box-containing protein 7. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2017]. | hsa:55180; | cognition [GO:0050890] | 12119551_Human WINS1 encoded 757 AA protein. Human WINS1 mRNA was expressed in adult testis, prostate, spleen, thymus, skeletal muscle, fetal kidney & brain. This is the first report on molecular cloning & initial characterization of human WINS1. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23773660_Study confirms that LINS, a modulator of the WNT pathway, is an indispensable gene to human cognition and this finding sheds further light on the importance of WNT signaling in human brain development and/or function. 31922598_A novel pathogenic variant of the LINS1 gene has been identified in a child with idiopathic mental retardation 34450347_Identification of a novel nonsense homozygous mutation of LINS1 gene in two sisters with intellectual disability, schizophrenia, and anxiety. | ENSMUSG00000053091 | Lins1 | 775.92894 | 1.2239580 | 0.2915540893 | 0.14152089 | 4.272509e+00 | 3.873371e-02 | 3.050567e-01 | No | Yes | 1118.499157 | 263.113205 | 873.445070 | 201.101648 | ||
| ENSG00000140577 | 64784 | CRTC3 | protein_coding | Q6UUV7 | FUNCTION: Transcriptional coactivator for CREB1 which activates transcription through both consensus and variant cAMP response element (CRE) sites. Acts as a coactivator, in the SIK/TORC signaling pathway, being active when dephosphorylated and acts independently of CREB1 'Ser-133' phosphorylation. Enhances the interaction of CREB1 with TAF4. Regulates the expression of specific CREB-activated genes such as the steroidogenic gene, StAR. Potent coactivator of PPARGC1A and inducer of mitochondrial biogenesis in muscle cells. Also coactivator for TAX activation of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeats (LTR). {ECO:0000269|PubMed:14506290, ECO:0000269|PubMed:15454081, ECO:0000269|PubMed:15466468, ECO:0000269|PubMed:16817901, ECO:0000269|PubMed:16980408, ECO:0000269|PubMed:17210223, ECO:0000269|PubMed:17644518}. | Activator;Alternative splicing;Cytoplasm;Host-virus interaction;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene is a member of the CREB regulated transcription coactivator gene family. This family regulates CREB-dependent gene transcription in a phosphorylation-independent manner and may be selective for cAMP-responsive genes. The protein encoded by this gene may induce mitochondrial biogenesis and attenuate catecholamine signaling in adipose tissue. A translocation event between this gene and Notch coactivator mastermind-like gene 2, which results in a fusion protein, has been reported in mucoepidermoid carcinomas. Alternative splicing results in multiple transcript variants that encode different protein isoforms. [provided by RefSeq, Jul 2012]. | hsa:64784; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; cAMP response element binding protein binding [GO:0008140]; energy homeostasis [GO:0097009]; macrophage activation [GO:0042116]; negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway [GO:0071878]; negative regulation of lipid catabolic process [GO:0050995]; positive regulation of CREB transcription factor activity [GO:0032793]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein homotetramerization [GO:0051289] | 15466468_transducer of regulated cyclic AMP-response element-binding protein 3 (TORC3), a co-activator of CREB, is involved in Tax-induced transcriptional activation from the HTLV-I LTR 16980408_These results strongly suggest that TORCs play a key role in linking these external signals to the transcriptional program of adaptive mitochondrial biogenesis by activating PGC-1alpha gene transcription. 17644518_BCL3 functions as a repressor of HTLV-1 LTR-mediated transcription through interactions with TORC3 18050304_study reports for the first time a CRTC3-MAML2 fusion gene in a mucoepidermoid carcinoma, as determined by RT-PCR and sequencing. 19749740_Mucoepidermoid carcinomas possessing CRTC3-MAML2 fusion may be associated with favorable clinicopathological features and patients may be younger than those with CRTC1-MAML2 fusion or those with no detectable gene fusion. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21164481_As a common human CRTC3 variant with increased transcriptional activity is associated with adiposity in two distinct Mexican-American cohorts, results suggest that adipocyte CRTC3 may play a role in the development of obesity in humans 21536665_CRTC3 plays a selective role in mitochondrial biogenesis in response to rotenone 21668476_mucoepidermoid carcinoma of the salivary glands positive for CRTC1-MAML2 or CRTC3-MAML2 fusion formed a favourable tumour subset that was distinct from fusion-negative cases 24121173_Metaplastic Warthin tumor and metaplastic pleomorphic adenoma of salivary glands did not harbor CRTC1-MAML2 and CRTC3-MAML2 fusion transcripts, respectively, or MAML2 gene rearrangement. 24264430_The CRTC3 polymorphism rs3862434 is associated with the plasma level of total cholesterol, and rs11635252 is associated with the risks of overweight and hypertriglyceridemia in a Chinese Han population. 27402217_revealing neither correlation between the cellular composition and CRTC1-MAML2 fusions nor presence of CRTC3-MAML2 fusions in cutaneous hidradenoma 28438292_findings suggest further characterization of MECs is needed before considering the CRTC1/3-MAML2 gene fusion as a prognostic biomarker 29079171_CRTC3-MAML2 gene fusion occurs in hidradenomas. 29979427_CRTC3 polymorphism was associated with the onset of acute coronary syndrome in Han Chinese patients, which may be related to the imbalance of the lipid metabolism 30138216_Pancreatic mucoepidermoid carcinoma does not have CRTC1/3-MAML2 fusion gene and MAML2 gene rearrangement. 33411955_Colorectal cancer risk variants rs10161980 and rs7495132 are associated with cancer survival outcome by a recessive mode of inheritance. 33780908_TORC2/3-mediated DUSP1 upregulation is essential for human decidualization. 33947275_Morphological and biological properties of silica nanoparticles for CRTC3-siRNA delivery and downregulation of the RGS2 expression in preadipocytes. | ENSMUSG00000030527 | Crtc3 | 1022.04816 | 1.0696347 | 0.0971181848 | 0.11178654 | 7.557721e-01 | 3.846549e-01 | 7.495634e-01 | No | Yes | 1303.677047 | 106.559785 | 1179.530442 | 94.346712 | |
| ENSG00000141298 | 85464 | SSH2 | protein_coding | Q76I76 | FUNCTION: Protein phosphatase which regulates actin filament dynamics. Dephosphorylates and activates the actin binding/depolymerizing factor cofilin, which subsequently binds to actin filaments and stimulates their disassembly. Inhibitory phosphorylation of cofilin is mediated by LIMK1, which may also be dephosphorylated and inactivated by this protein. {ECO:0000269|PubMed:11832213}. | 3D-structure;Actin-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Hydrolase;Phosphoprotein;Protein phosphatase;Reference proteome | This gene encodes a protein tyrosine phosphatase that plays a key role in the regulation of actin filaments. The encoded protein dephosphorylates and activates cofilin, which promotes actin filament depolymerization. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2013]. | hsa:85464; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; extracellular space [GO:0005615]; actin binding [GO:0003779]; phosphoprotein phosphatase activity [GO:0004721]; protein serine phosphatase activity [GO:0106306]; protein threonine phosphatase activity [GO:0106307]; protein tyrosine phosphatase activity [GO:0004725]; protein tyrosine/serine/threonine phosphatase activity [GO:0008138]; actin cytoskeleton organization [GO:0030036]; negative regulation of actin filament polymerization [GO:0030837]; protein dephosphorylation [GO:0006470] | 17848544_Expression of phosphatase-dead versions of SSH proteins, SSH1, SSH2 & SSH3 results in phosphorylation/inactivation of cofilin, changes in cytoskeleton organization, loss of cell polarity, and assembly of aberrant arrays of laminin-332 in keratinocytes. 20477815_Observational study of gene-disease association. (HuGE Navigator) 23416984_data indicate that: (a) apoptotic cell death in FLCN-null cells can be triggered by SSH2 knockdown through cell cycle arrest; (b) SSH2 represents a potential therapeutic target for the development of agents for the treatment of BHD syndrome related tumors 25568344_Novel signaling pathway consisting of chemoattractant GPCR/Galphai protein, PLC, PKCbeta and PKDs that regulates SSH2/cofilin activity, F-actin polymerization and directional assembly of actin cytoskeleton in neutrophil chemotaxis was identified. 30154244_the N-terminal region of human Slingshot2 auto-inhibits its phosphatase activity in a noncompetitive manner | ENSMUSG00000037926 | Ssh2 | 718.94954 | 0.8868875 | -0.1731770082 | 0.12891592 | 1.794238e+00 | 1.804106e-01 | 5.745367e-01 | No | Yes | 598.334858 | 71.008546 | 640.770369 | 74.303116 | |
| ENSG00000141314 | 162494 | RHBDL3 | protein_coding | P58872 | FUNCTION: May be involved in regulated intramembrane proteolysis and the subsequent release of functional polypeptides from their membrane anchors. {ECO:0000250}. | Alternative splicing;Hydrolase;Membrane;Protease;Reference proteome;Repeat;Serine protease;Transmembrane;Transmembrane helix | hsa:162494; | integral component of membrane [GO:0016021]; calcium ion binding [GO:0005509]; serine-type endopeptidase activity [GO:0004252] | Mouse_homologues 11900977_Cloning and expression of Ventrhoid 27264103_Here the authors show that the mammalian rhomboid protease RHBDL4 (also known as Rhbdd1) promotes trafficking of several membrane proteins, including the EGFR ligand TGFalpha, from the endoplasmic reticulum (ER) to the Golgi apparatus, thereby triggering their secretion by extracellular microvesicles. | ENSMUSG00000017692 | Rhbdl3 | 137.31633 | 0.8522264 | -0.2306914136 | 0.25123365 | 8.379324e-01 | 3.599888e-01 | 7.317899e-01 | No | Yes | 149.041613 | 18.050722 | 174.369518 | 20.196268 | ||
| ENSG00000141401 | 3613 | IMPA2 | protein_coding | O14732 | FUNCTION: Can use myo-inositol monophosphates, scylloinositol 1,4-diphosphate, glucose-1-phosphate, beta-glycerophosphate, and 2'-AMP as substrates. Has been implicated as the pharmacological target for lithium Li(+) action in brain. {ECO:0000269|PubMed:17068342}. | 3D-structure;Alternative splicing;Hydrolase;Magnesium;Metal-binding;Reference proteome | PATHWAY: Polyol metabolism; myo-inositol biosynthesis; myo-inositol from D-glucose 6-phosphate: step 2/2. | This locus encodes an inositol monophosphatase. The encoded protein catalyzes the dephosphoylration of inositol monophosphate and plays an important role in phosphatidylinositol signaling. This locus may be associated with susceptibility to bipolar disorder. [provided by RefSeq, Jan 2011]. | hsa:3613; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; inositol monophosphate 1-phosphatase activity [GO:0008934]; inositol monophosphate 3-phosphatase activity [GO:0052832]; inositol monophosphate 4-phosphatase activity [GO:0052833]; metal ion binding [GO:0046872]; protein homodimerization activity [GO:0042803]; inositol biosynthetic process [GO:0006021]; inositol metabolic process [GO:0006020]; inositol phosphate dephosphorylation [GO:0046855]; phosphate-containing compound metabolic process [GO:0006796]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; signal transduction [GO:0007165] | 11317223_Observational study of gene-disease association. (HuGE Navigator) 14699425_Promoter is polymorphic in bipolar disorder. 15505643_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15557493_IMPA2 may be a febrile seizure susceptibility gene. 6 SNPs were found: -708G/A, -461C/T, IVS1-15G/A, 159T/C, IVS5+13-14insA, & 558C/T. 17068342_IMPA2 has a separate function in vivo from that of IMPA1 17251911_the present study suggests that a promoter haplotype of IMPA2 possibly contributes to risk for bipolar disorder by elevating IMPA2 levels in the brain, albeit the genetic effect varies among populations. 17340635_The crystal structures of human IMPA2 are useful for understanding the effect of nonsynonymous polymorphism reported in IMPA2, and will contribute to further functional analyses of IMPA2. 17388992_Observational study of gene-disease association. (HuGE Navigator) 17388992_data suggest that the genetic variants in the IMPA2 gene are not associated with a risk of febrile seizures in Caucasian patients and patients from various genetic groups are likely to have different genetic causes of febrile seizures 19066393_Single nucleotide polymorphism in IMPA2 gene is associated with acute lymphoblastic leukemia. 19910543_Observational study of gene-disease association. (HuGE Navigator) 20153384_The current study did not support a substantial role of the upregulation of IMPase in bipolar disorder, although the lithium-insensitivity trait seen in IMPA2 transgenic mice might represent some aspect relevant to the inositol depletion hypothesis. 20398908_Observational study of gene-disease association. (HuGE Navigator) 20800640_No association is found between IMPA2 gene polymorphisms and bipolar disorder. 20800640_Observational study of gene-disease association. (HuGE Navigator) 21213002_Authors propose that the human myo-inositol monophosphatase 2 interacts with myo-inositol monophosphates in the three-metal-ion bound form, and proceeds the dephosphorylation through the three-metal-ion theory. 23453640_results suggest that genetic variability at rs669838-IMPA2,rs4853694-INPP1, rs1732170-GSK3b and rs11921360-GSK3b genes is associated with a higher risk of attempting suicide in bipolar patients. 27661109_a correlation between an IMPA2 polymorphism rs589247 and ischemic stroke risk in a northwest Han Chinese 27748550_a promoter polymorphism of IMPA2 possibly contributed to risk for schizophrenia by elevating transcription activity in Han Chinese individuals. 29499505_IMPA2 is a protein-coding gene for a catalytic protein that converts inositol monophosphate into free inositol through dephosphorylation. 31202813_We conclude that miR-25-mediated IMPA2 downregulation constitutes a novel signature for cancer metastasis and poor outcomes in clear cell renal cell carcinoma (ccRCC). We further postulate that the therapeutic targeting of miR-25 can be useful for preventing the metastatic progression of ccRCC associated with IMPA2 downregulation. 32409648_A novel function of IMPA2, plays a tumor-promoting role in cervical cancer. | ENSMUSG00000024525 | Impa2 | 346.36288 | 1.1029269 | 0.1413372067 | 0.17739626 | 6.356539e-01 | 4.252890e-01 | 7.771769e-01 | No | Yes | 440.409875 | 52.259560 | 382.165135 | 44.294803 |
| ENSG00000142046 | 641649 | TMEM91 | protein_coding | Q6ZNR0 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:641649; | integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; hematopoietic progenitor cell differentiation [GO:0002244] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000061702 | Tmem91 | 29.18330 | 0.8077453 | -0.3080275939 | 0.53714808 | 3.244275e-01 | 5.689590e-01 | No | Yes | 27.247113 | 6.950196 | 34.892144 | 8.614537 | ||||
| ENSG00000143258 | 27005 | USP21 | protein_coding | Q9UK80 | FUNCTION: Deubiquitinates histone H2A, a specific tag for epigenetic transcriptional repression, thereby acting as a coactivator. Deubiquitination of histone H2A releaves the repression of di- and trimethylation of histone H3 at 'Lys-4', resulting in regulation of transcriptional initiation. Regulates gene expression via histone H2A deubiquitination (By similarity). Also capable of removing NEDD8 from NEDD8 conjugates but has no effect on Sentrin-1 conjugates (PubMed:10799498). Deubiquitinates BAZ2A/TIP5 leading to its stabilization (PubMed:26100909). {ECO:0000250|UniProtKB:Q9QZL6, ECO:0000269|PubMed:10799498, ECO:0000269|PubMed:26100909}. | 3D-structure;Activator;Alternative splicing;Chromatin regulator;Cytoplasm;Hydrolase;Metal-binding;Nucleus;Protease;Reference proteome;Thiol protease;Transcription;Transcription regulation;Ubl conjugation pathway;Zinc | This gene encodes a member of the C19 peptidase family, also known as family 2 of ubiquitin carboxy-terminal hydrolases. The encoded protein cleaves ubiquitin from ubiquitinated proteins for recycling in intracellular protein degradation. The encoded protein is also able to release NEDD8, a ubiquitin-like protein, from NEDD8-conjugated proteins. This gene has been referred to as USP16 and USP23 but is now known as USP21. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2016]. | hsa:27005; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; cysteine-type peptidase activity [GO:0008234]; metal ion binding [GO:0046872]; NEDD8-specific protease activity [GO:0019784]; thiol-dependent deubiquitinase [GO:0004843]; transcription coactivator activity [GO:0003713]; chromatin organization [GO:0006325]; histone deubiquitination [GO:0016578]; positive regulation of transcription, DNA-templated [GO:0045893]; protein deubiquitination [GO:0016579]; ubiquitin-dependent protein catabolic process [GO:0006511] | 19910467_USP21 plays an important role in the down-regulation of TNFalpha-induced NF-kappaB activation through deubiquitinating RIP1. 21399617_Study report that USP21 cleaves Ub polymers, and with reduced activity also targets ISG15, but is inactive against NEDD8. 22298430_Study observed a reduction in the number of A549 cells developing a primary cilium upon serum starvation when USP21 was depleted with two independent siRNA oligos. 23395819_Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3 25197364_The protein stability of IL-33 is maintained by USP21 through deubiquitination 26100909_Data show that SUMOylated BANP, E5R, and Nac1 (BEN) domain 3 (BEND3) stabilizes NoRC component TTF-1-interacting protein 5 (Tip5)via association with ubiquitin specific protease 21 (USP21) debiquitinase 27174732_Results from computational biology identified a cancer mutation that inactivates the nuclear export signals of the human deubiquitinase USP21, and leads to the aberrant accumulation of this protein in the nucleus. 27259257_USP21 binds to the promoter region of IL-8 and mediates transcriptional initiation, which regulates the stem-cell like property of human renal cell carcinoma 27436899_Data show that ubiquitin variants (Ubvs) that bind to USP2 or USP21 contain a similar core functional epitope, or 'hot spot,' consisting mainly of positions that are conserved as the wild type sequence, but also some positions that prefer mutant sequences. 27621083_USP21 recruits and stabilises Gli1 at the centrosome. 27956178_USP21 specifically regulates the Lys48-linked polyubiquitination and stability of NANOG 28254948_a critical role of p38-mediated USP21 phosphorylation in regulating STING-mediated antiviral functions and identifies p38-USP21 axis as an important pathway that DNA virus adopts to avoid innate immunity responses. 28743957_USP21 interacts with, deubiquitinates and stabilizes BRCA2 to promote efficient RAD51 loading at DNA double-strand breaks. 29706623_USP21-mediated deubiquitination and stabilization of MEK2 plays a critical role in hepatocellular carcinoma development. 30865895_USP21 binds and deubiquitinates FOXM1 in breast cancer cells. 31253698_USP21 modulates Goosecoid function through deubiquitination. 31488580_work identifies and validates USP21 as a PDAC oncogene 31956270_The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. 33052017_USP21 upregulation in cholangiocarcinoma promotes cell proliferation and migration in a deubiquitinase-dependent manner. 33389794_USP21 promotes cell proliferation by maintaining the EZH2 level in diffuse large B-cell lymphoma. 33582217_Disulfiram and 6-Thioguanine synergistically inhibit the enzymatic activities of USP2 and USP21. 33827943_Deubiquitinating Enzyme USP21 Inhibits HIV-1 Replication by Downregulating Tat Expression. 34523817_Ablation of USP21 in skeletal muscle promotes oxidative fibre phenotype, inhibiting obesity and type 2 diabetes. 34825342_USP21 regulates Hippo signaling to promote radioresistance by deubiquitinating FOXM1 in cervical cancer. | ENSMUSG00000053483 | Usp21 | 1319.52085 | 1.0855929 | 0.1184832005 | 0.09619248 | 1.521111e+00 | 2.174515e-01 | 6.095393e-01 | No | Yes | 1643.567830 | 151.944989 | 1491.675096 | 134.813367 | |
| ENSG00000143801 | 5664 | PSEN2 | protein_coding | P49810 | FUNCTION: Probable catalytic subunit of the gamma-secretase complex, an endoprotease complex that catalyzes the intramembrane cleavage of integral membrane proteins such as Notch receptors and APP (amyloid-beta precursor protein). Requires the other members of the gamma-secretase complex to have a protease activity. May play a role in intracellular signaling and gene expression or in linking chromatin to the nuclear membrane. May function in the cytoplasmic partitioning of proteins. The holoprotein functions as a calcium-leak channel that allows the passive movement of calcium from endoplasmic reticulum to cytosol and is involved in calcium homeostasis (PubMed:16959576). Is a regulator of mitochondrion-endoplasmic reticulum membrane tethering and modulates calcium ions shuttling between ER and mitochondria (PubMed:21285369). {ECO:0000269|PubMed:10497236, ECO:0000269|PubMed:10652302, ECO:0000269|PubMed:16959576, ECO:0000269|PubMed:21285369}. | Alternative splicing;Alzheimer disease;Amyloidosis;Cardiomyopathy;Disease variant;Endoplasmic reticulum;Golgi apparatus;Hydrolase;Membrane;Neurodegeneration;Notch signaling pathway;Phosphoprotein;Protease;Reference proteome;Transmembrane;Transmembrane helix | Alzheimer's disease (AD) patients with an inherited form of the disease carry mutations in the presenilin proteins (PSEN1 or PSEN2) or the amyloid precursor protein (APP). These disease-linked mutations result in increased production of the longer form of amyloid-beta (main component of amyloid deposits found in AD brains). Presenilins are postulated to regulate APP processing through their effects on gamma-secretase, an enzyme that cleaves APP. Also, it is thought that the presenilins are involved in the cleavage of the Notch receptor such that, they either directly regulate gamma-secretase activity, or themselves act are protease enzymes. Two alternatively spliced transcript variants encoding different isoforms of PSEN2 have been identified. [provided by RefSeq, Jul 2008]. | hsa:5664; | apical plasma membrane [GO:0016324]; cell cortex [GO:0005938]; cell surface [GO:0009986]; centrosome [GO:0005813]; ciliary rootlet [GO:0035253]; dendritic shaft [GO:0043198]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; gamma-secretase complex [GO:0070765]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; growth cone [GO:0030426]; integral component of plasma membrane [GO:0005887]; kinetochore [GO:0000776]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; membrane raft [GO:0045121]; mitochondrial inner membrane [GO:0005743]; neuromuscular junction [GO:0031594]; neuronal cell body [GO:0043025]; nuclear inner membrane [GO:0005637]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; protein-containing complex [GO:0032991]; Z disc [GO:0030018]; aspartic endopeptidase activity, intramembrane cleaving [GO:0042500]; endopeptidase activity [GO:0004175]; amyloid precursor protein catabolic process [GO:0042987]; amyloid-beta formation [GO:0034205]; amyloid-beta metabolic process [GO:0050435]; calcium ion transport [GO:0006816]; intracellular signal transduction [GO:0035556]; membrane protein ectodomain proteolysis [GO:0006509]; mitochondrion-endoplasmic reticulum membrane tethering [GO:1990456]; negative regulation of apoptotic process [GO:0043066]; Notch receptor processing [GO:0007220]; Notch signaling pathway [GO:0007219]; positive regulation of catalytic activity [GO:0043085]; protein processing [GO:0016485]; regulation of calcium import into the mitochondrion [GO:0110097] | 11436125_Observational study of gene-disease association. (HuGE Navigator) 11568920_Observational study of gene-disease association. (HuGE Navigator) 11799129_enhancement of amyloid beta protein by Herp, and endoplasmic reticulum stress-inducible protein 11847232_interaction with CALP/KChIP4 11876645_inhibition of endoproteolysis by gamma-secretase inhibitors 11891288_Notch receptor cleavage depends on but is not directly executed by presenilins 11904448_Wild-type and mutated presenilins 2 trigger p53-dependent apoptosis and down-regulate presenilin 1 expression in HEK293 human cells and in murine neurons 11987239_PS2 mRNA is present only in lymphocytes, in contrast to PS1 mrna, which is found in both myeloid and lymphoid cells. 12048259_regulation by nicastrin and role in determining amyloid beta-peptide production via complex formation 12058025_interaction with GFAP epsilon 12173418_mutant presenilin 2 induces apoptosis accompanied by increased caspase-3-like activity and decreased bcl-2 expression in neuronal cells 12198112_PS2/gamma-secretase contains PEN-2 and requires it for presenilin expression 12210343_Observational study of gene-disease association. (HuGE Navigator) 12210343_There is no evidence to suggest that variations in the PSEN2 gene pose as major risk factors for sporadic early-onset Alzheimer disease 12232783_Observational study of gene-disease association. (HuGE Navigator) 12232783_in oxygen stress conditions relatively minor variations in PSEN2 promoter DNA sequence structure can enhance PSEN2 gene expression and that may play a role in the induction and/or proliferation of an inflammatory response in AD brain. 12403846_Transcriptional synergism on the pS2 gene promoter between a p160 coactivator and estrogen receptor-alpha depends on the coactivator subtype, the type of estrogen response element, and the promoter context. 12471034_presenilin binds to APH-1, which plays a role in the maturation of presenilin-nicastrin complexes 12556443_C-terminal fragment-PS2 could exhibit some of its functions in the absence of the presenilin 2 N-terminal fragment (NTF-PS2) counterpart derived from the presenilinase cleavage. 12605888_Overexpression of either wild type or mutant presenilin 2 in various cell lines does not directly induce apoptosis or increase the susceptibility to apoptosis. 12770698_Observational study of gene-disease association. (HuGE Navigator) 12770698_the current study does not support the notion that the polymorphism in the PS2 gene constitutes a risk factor for either late-onset or early-onset AD 12817569_Observational study of genotype prevalence. (HuGE Navigator) 12846562_In vitro characterization of the presenilin-dependent gamma-secretase complex using a novel affinity ligand 12885769_presenilins are multifunctional proteins with catalytic activity as well as roles in the generation, stabilization, and transport of the gamma-secretase complex 12925374_Observational study of genotype prevalence. (HuGE Navigator) 12925374_a novel mutation in the PSEN2 gene in a family with early-onset Alzheimer disease. The variation in the age at onset confirms that PSEN2 mutations are associated with variable clinical expression. 14577603_identification of two novel spliced presenilin 2 transcripts in lymphocytes and brain under oxidant stress 14741365_IMP1 is a bi-aspartic polytopic protease capable of cleaving transmembrane proteins such as presenilin 2. 14769392_Observational study of genotype prevalence. (HuGE Navigator) 15004330_HeLa cells overexpressing both PS2 and ubiquilin-1 had PS2 mRNA levels lower than HeLa cells overexpressing PS2 alone, indicating that ubiquilin-1 overexpression, in fact, decreases PS2 transcription. 15006697_Ca(2+) release from intracellular stores was significantly reduced in fibroblasts from familial Alzheimer patients with presenilin 2 mutations. 15537629_results suggest that the proximal two-thirds of the PEN-2 TMD1 is functionally important for endoproteolysis of PS1 holoproteins and the generation of PS1 fragments, essential components of the gamma-secretase complex 15591316_dimeric (NCSTN/APH-1) and trimeric (NCSTN/APH-1/PS1) intermediates of gamma-secretase complex assembly are retained within the ER and incorporation of the fourth binding partner (PEN-2) also occurs on immature NCSTN. 15629423_knock down of anterior pharynx defective 1 homolog A (APH-1A), but not APH-1b, resulted in impaired maturation of nicastrin and reduced expression of presenilin 1, presenilin 2, and PEN-2 proteins 15663477_Four verified PS2 familial Alzheimer disease(FAD) mutations cause substantial changes in the Abeta 42/40 ratio, like PS1 mutations that cause very-early-onset FAD and may represent partial loss of function mutations 15755689_In fibroblasts from familial Alzheimer's disease the presenilin 2 mutation Thr122Arg reduces both Ca2+ release from and capacitative Ca2+ entry to intracellular stores, revealing a modulatory role in disease pathogenesis. 15776278_Of the nine pathogenic mutations found in 12 cases, three were in APP, one in PSEN2, and five in PSEN1, including two novel Greek mutations (L113Q and N135S) in Alzheimer disease 15951428_Wild-type PS2 transgenes expressed in the mouse CNS support little Abeta40 or Abeta42 production. 15975068_Observational study of genotype prevalence. (HuGE Navigator) 16014629_Presenilin (PS)1 mutations interfere with PS2-mediated activity by reducing PS2 fragments 16135086_endoproteolysis, N and C terminal fragment interactions, and the assembly and activity of gamma-secretase complexes are very conserved between PS1 and PS2 16233903_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16258850_identification of genes whose expression is modulated by overexpression of mutant presenilin-2 in transgenic mice 16375654_mechanisms by which presenilin 2 affects the programmed cell death include the role of the proteolytically derived presenilin fragments generated by both presenilinase- and caspases 16423463_Observational study of gene-disease association. (HuGE Navigator) 16474849_in breast cancer cases two germline alterations, R62H and R71W, in presenilin-2 (PS-2) 16620965_Familial Alzheimer's disease (FAD)-linked Presenilin mutants lower the Ca(2+) content of intracellular stores. 17268504_reduced presenilin proteolytic function leads to increased Abeta42/Abeta40 in Alzheimer disease (Review) 17268505_mutations in Alzheimer disease (Review) 17345043_Presenilin 2 Ser130Leu mutation in a case of late-onset 'sporadic' Alzheimer's disease. 17401156_Exogenous cholesterol and compartmentalization in neuroblastoma cells play a relevant role in regulating the transcription of presenilin 2. 17401676_presenilin 2 overexpression may facilitate assembly into the more active gamma-secretase complex 17412506_Observational study of gene-disease association. (HuGE Navigator) 17560791_all known gamma-secretase complexes are active in APP processing and that all combinations of APH-1 variants with either familial Alzheimer mutant PS1 or PS2 support pathogenic Abeta(42) production 17614368_We found that the mutant polypeptides were unable to bind ubiquilin, suggesting that loss of ubiquilin interaction leads to destabilization of presenilin polypeptides. 17727891_It is concluded that in the Northern Han Chinese population, the +A/-A polymorphism of the PSEN2 promoter is a moderate genetic risk factor for developing SAD, independent of the APOE epsilon4 allele. 17727891_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17903177_HMGA1a is a sequence-specific RNA-binding factor causing sporadic Alzheimer's disease-linked exon skipping of presenilin-2 pre-mRNA 18087668_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18087668_We examined association between AD and PSEN2 polymorphisms located in two 5'UTR regions 18283638_Most signet ring cell carcinomas and adenocarcinomas of the urinary bladder expressed ps2 peptide. 18293935_Mature integrin beta1 with increased expression level is delivered to the cell surface, which results in an increased cell surface expression level of mature integrin beta 1 in presenilin (PS)1 and PS2 double-deficient fibroblasts. 18350357_We reort a late onset familial Alzheimer's disease: novel presenilin 2 mutation and PS1 E318G polymorphism. 18427071_description of an Italian family with hereditary dementia associated with a novel mutation in the presenilin 2 gene (A85V mutation) 18591429_SERCA activity is diminished in fibroblasts lacking both PS1 and PS2 genes, despite elevated SERCA2b steady-state levels. 18667258_Observational study of gene-disease association. (HuGE Navigator) 18727676_In vitro expression of a new PSEN2 missense mutation (V393M) cDNA from a patient with early-onset dementia did not result in detectable increase of the secreted Abeta42/40 peptide ratio. 18842294_Observational study of gene-disease association. (HuGE Navigator) 19036728_equilibrium of PS1- and PS2-containing active complexes is dynamic and altered by overexpression of Pen2 or PS1 mutants and that formation of PS2 complexes is positively correlated with increased Abeta42:Abeta40 ratios 19073399_Three-year follow-up of a patient with early onset Alzheimer disease with PSEN2 mutation- case report and review of the literature. 19276543_In PSEN2, T122P and M239V mutations presented with severe behavioral disturbances 19382908_Presenilin-2 dampens intracellular Ca2+ stores by increasing Ca2+ leakage and reducing Ca2+ uptake 19573580_The PSEN2 regulatory region includes two separate promoters modulated by Egr-1, a transcription factor involved in learning and memory. Differential Psen2 regulation in human and mouse has implications for Alzheimer disease mouse models. 19768372_a novel Arg62His Presenilin2 mutation in patient with frontotemporal dementia 19834068_Data show that presenilins 1 and 2 are highly enriched in a subcompartment of the endoplasmic reticulum associated with mitochondria that forms a physical bridge between the two organelles. 19889971_Transactivation of the Pen2 promoter by presenilin 1/2 is p53-dependent. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20008660_Observational study of gene-disease association. (HuGE Navigator) 20009122_Mutations of the genes for amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) are responsible for development of the disease in 50 percent of patients with FAD. 20164579_this study demonistreated that an Italian pedigree linked to a novel mutation (S175C) at the third transmembrane domain of PSEN2 in atypical alzeheimer disease. 20333730_A genome scan within nine families for loci influencing age-at-onset, while simultaneously controlling for variation in the primary PSEN2 mutation (N141I) and APOE, was performed. 20375137_Mutations in presenilin 2 are rarely associated with Alzheimer's disease. The best studied Asn141Iso mutation produces an Alzheimer's disease phenotype with a wide range of onset ages. 20457965_A family with the N141I mutation in PSEN2 that presently lives in Germany has been connected to the haplotype that carries the same mutation in pedigrees descended from the Volga Germans. 20468060_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20594621_The PSEN2 and PSEN1 genes have a very similar genetic structure and encode two proteins expressed in a multiplicity of tissues including the brain. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20677014_Observational study of gene-disease association. (HuGE Navigator) 20701429_familial Alzheimer disease presenilin 2 protein interactions with InsP(3) receptor causes exaggerated calcium signaling that may contribute to the disease pathology by enhanced generation or reactive oxygen species 20850903_Observational study of gene-disease association. (HuGE Navigator) 20850903_One distinct haploblock in PSEN2 was detected and the frequent haplotypes were analyzed using 4 tagging single nucleotide polymorphisms 21234330_The PS2 mutation causes early cerebral amyloid accumulation and memory dysfunction. 21285369_Presenilin 2 modulates endoplasmic reticulum-mitochondria interactions and Ca2+ cross-talk. 21409510_PSEN2 Arg62His mutation may lead to a phenotypic heterogeneity presenting either as Alzheimer's disease or Lewy body dementia. 21545304_[review] The role of presenilin 2 in general physiology and Alzheimer's disease pathology due to its mutation are discussed. 21914807_Polar transmembrane-based amino acids in presenilin 1 are involved in endoplasmic reticulum localization, Pen2 protein binding, and gamma-secretase complex stabilization 21959359_the PSEN1, PSEN2, and APP genes exhibited no pathogenic mutations in our cohort of early-onset Alzheimer disease and frontotemporal lobar degeneration patients 22027014_The results of this study demonistrated that common variants at AbetaPP, PSEN1, and PSEN2 and MAPT are unlikely to make strong contributions to susceptibility for late onset Alzheimer's disease. 22045484_The results of this study demonstrated that upregulation of PSEN2 and the upregulation of BACE1 is an ancient, conserved, and thus selectively advantageous response to hypoxia/oxidative stress. 22074918_the PS1 complex is only marginally less active than the PS2 complex in Abeta production. 22249458_The results of this study suggested that oxidative stress-mediated ERK activation contributes to increases in beta-secretase and, thus, an increase of Abeta generation in neuronal cells expressing mutant PS2. 22302987_results indicate that PS2 modulates the degradation of RBP-Jk through phosphorylation by p38 MAPK. 22312439_Rare coding variants in APP, PSEN1 and PSEN2, increase risk for or cause late onset Alzheimer's disease 22503161_Here we report a frequency of 11.2% of mutations and variants in the known Alzheimer disease genes in the dementia cohort studied and 24% in the early onset subgroup of patients 22580083_analysis supports the hypothesis that the PSEN2 rs8383 polymorphism is associated with an enlarged risk of sporadic Alzheimer's disease 22753229_we found that the protein expression of presenilin 2 (PS2) was significantly increased in glioma tissues 22755192_Current findings established the involvement of APP, PS1, and PS2 in familial case of Alzheimer disease, while APO polymorphism suggests the existence of other unknown genetic factors or risk factors in the cause of this disease. 22872014_For the Abeta40 region on chromosome 1, association of several SNPs was observed at the presenilin 2 gene (PSEN2) in 125 subjects with severe hypertension. 23546527_Alzheimer's disease pathology induced by overexpression of human mutant presenilin 2 (PS2) protein induced changes in glucose metabolism, were investigated. 23589300_Interactome analyses of mature gamma-secretase complexes reveal distinct molecular environments of presenilin (PS) paralogs and preferential binding of signal peptide peptidase to PS2. 23884042_Functional disruption of the DMN occurs early in the course of autosomal dominant Alzheimer disease, beginning before clinically evident symptoms, and worsening with increased impairment. 24145027_At the transcriptional level, PSEN1/2 removal induced cyclic AMP response element-binding protein (CREB)/CREB-binding protein binding. 24594196_A review, representing the first attempt to systematically organize the available evidence concerning the phenotypic characteristics of familial Alzheimer's disease due to PSEN2 mutations 24650794_Its mutation is the major causes of eraly-onset familial Alzheimer's disease. 24704512_evaluation of contribution of mutations in PS1 and PS2 genes to familial early-onset Alzheimer's disease (EOAD) cases and sporadic late-onset AD; identified 1 novel frameshift mutation in PS1 gene and 2 novel frameshift mutations in PS2 gene; mutational analysis reports a correlation between clinical symptoms and PS1 and PS2 genetic factors in EOAD 24838186_Mutation in PSEN2 causes of early-onset familial Alzheimer's disease. 24844686_study describes a previously unrecognized sequence change (c.376G>A) in PSEN2 in an early onset Alzheimer's disease patient and her likewise affected mother 24858037_The loss of PS2 could have a critical role in lung tumor development through the upregulation of iPLA2 activity by reducing gamma-secretase. 24885952_the structures of presenilin 2 protein with native Val 214 residue and Leu 214 mutation revealed significant structural changes in the region 24927704_The results of this study showed that PSEN2 was significantly downregulated in the auditory cortex of Alzheimer's disease patients when compared to controls. 25323700_PSEN2 mutations are common in the Chinese Han population with a history of AD and FTD 25429133_Levels of presenilin 2 are higher in the cerebral cortex of presenilin 1 knockout mice, suggesting a compensatory upregulation. 25614624_Mutation of on PS1 and PS2 AXXXAXXXG motifs strongly impacts gamma-secretase activity. 25814654_Both human PS2V and zebrafish PS1IV can stimulate gamma-secretase activity despite extreme structural divergence. 25998117_Mutations in PSEN2 are relatively rare cause of the autosomal-dominant cases of Early onset familial Alzheimer Disease. 26159191_This study identified variants in PSEN2 across a range of phenotypes (Alzheimer's Disease , Alzheimer's Disease and cerebrovascular disease,frontotemporal dementia and progressive supranuclear palsy. 26166204_Its mutations of PSEN2 account for pathogenicity of early-onset familial Alzheimer's disease. 26203236_PSEN2 mutations appeared not only in Alzheimer's Disease patients but also in patients with other disorders, including frontotemporal dementia, dementia with Lewy bodies, breast cancer, dilated cardiomyopathy, and Parkinson's disease with dementia 26312828_The search for the genetic factors contributing to Alzheimer disease (AD) has evolved tremendously throughout the years. It started from the discovery of fully penetrant mutations in Amyloid precursor protein, Presenilin 1, and Presenilin 2 as a cause of autosomal dominant AD 26337232_Familial Alzheimer's disease Patients with PSEN2 mutations have a delayed AOO with longest disease duration and presented more frequently with disorientation. [review] 26422362_Its mutation is pathogenic to early onset familial AD associated with atypical symptom presentation. 26522186_German early-onset Alzheimer's disease cohort reveals a substantial frequency of PSEN2 variants. 27059953_Data show that presenilin 1 (PS1)-containing gamma-secretase complexes were targeted to the plasma membrane, whereas presenilin 2 (PS2)-containing ones were addressed to the trans-Golgi network, to recycling endosomes. 27135718_This review reveled that Mutations in APP and PS-1 and PS-2 genes that are associated with early-onset, autosomal, dominantly inherited AD. 27239030_Presenilin 2 (PS2), mutations in which underlie familial Alzheimer's disease (FAD), promotes endoplasmic reticulum-mitochondria coupling only in the presence of mitofusin 2 (Mfn2). 27293189_Study identified a unique motif in PSEN2 that directs gamma-secretase to late endosomes/lysosomes via a phosphorylation-dependent interaction with the AP-1 adaptor complex. PSEN2 selectively cleaves late endosomal/lysosomal localized substrates and generates the prominent pool of intracellular Abeta that contains longer Abeta; familial Alzheimer's disease-associated mutations in PSEN2 increased the levels of longer Ab... 27608597_Data show that presenilin 1 (PS1)/anterior-pharynx-defective protein 1 (Aph1b), presenilin 2 (PS2)/Aph1aL, PS2/Aph1aS and PS2/anterior pharynx defective 1 homolog B (Aph1b) gamma-secretase produced amyloid beta peptide (Abeta) with a higher Abeta42+Abeta43-to-Abeta40 (Abeta42(43)/Abeta40) ratio than the other gamma-secretases. 27799753_Most of the early-onset Alzheimer's disease -associated mutations have been detected in PSEN1, and several novel PSEN1 mutations were recently identified in patients from various parts of the world, including Asia. Until 2014, no PSEN2 mutations were found in Asian patients; however, emerging studies from Korea and the People's Republic of China discovered probably pathogenic PSEN2 mutations. [review] 28106563_Whole-exome sequencing of 238 African American subjects identified 6 rare missense variants within the early-onset Alzheimer's disease (AD) genes, which were observed in AD cases but never among controls. These variants were analyzed in an independent cohort of 300 African American subjects, which indicated that a PSEN2 and PSEN1 novel rare variants, may contribute to AD risk in this population. 28502043_Young healthy adults carrying APOE epsilon4 and APP/presenilin-1/2 displayed different hippocampus functional connectivity patterns 28987665_The results show that in cognitively normal young adults carrying Presenelin 2 mutations had different spontaneous brain activity patterns without cerebral structural differences. 28994238_PS1- and PS2-dependent substrate processing in murine cells lacking presenilins (PSs) or stably re-expressing human PS1 or PS2 in an endogenous PS-null (PSdKO) background, were studied. 29078389_Dominant negative effect of the loss-of-function gamma-secretase mutants on the wild-type enzyme through heterooligomerization has been demonstrated. 29109765_The present data suggest that PS2 mutations suppress lung tumor development by inhibiting the iPLA2 activity of PRDX6 via a gamma-secretase cleavage mechanism and may explain the inverse relationship between lung cancer and Alzheimer's disease incidence. 30104866_Determined whether a pathogenic mutation in the PSEN2 gene in a Korean patient was associated with early onset Alzheimer's disease. Findings revealed that the p.His169Asn might be an important residue in PSEN2, which may alter the functions of PSEN2, suggesting its potential involvement with AD phenotype. 30119059_Results showed a novel PSEN2 mutation with adenosine replacing cysteine at nucleotide position 665, codon 222. 30560948_Results provide evidence that, unlike their significant role in neurogenesis during embryonic development, PSEN1 and PSEN2 are not required for cell-intrinsic regulation of adult hippocampal neurogenesis. 30822634_Thirteen of 148 (8.8%) individuals had possible pathogenic APP, PSEN1, or PSEN2 variants, including 2 (15.4%) APP variants, 8 (61.5%) PSEN1 variants, and 3 (23.1%) PSEN2 variants 30892128_Mutated PSEN2 impairs autophagy by causing a block in the degradative flux at the level of the autophagosome-lysosome fusion step. 31020001_PSEN2 splice isoforms associated with sporadic Alzheimer disease 31167792_results obtained with HEK293T cells suggest that selective presenilin 1 or presenilin 2 inhibition by a given GSI does not explain the previously observed differences in functional and pharmacological properties among various gamma-secretase inhibitors 31182772_Genetic analyses of early-onset Alzheimer's disease using next generation sequencing. 31914229_PSEN1, PSEN2, and APP mutations in 404 Chinese pedigrees with familial Alzheimer's disease. 31991578_Intracellular Calcium Dysregulation by the Alzheimer's Disease-Linked Protein Presenilin 2. 32087291_Systematic validation of variants of unknown significance in APP, PSEN1 and PSEN2. 32420742_Identification of Structural Calcium Binding Sites in Membrane-Bound Presenilin 1 and 2. 32556937_Exploring the Role of PSEN Mutations in the Pathogenesis of Alzheimer's Disease. 32917274_Amyloid-beta1-43 cerebrospinal fluid levels and the interpretation of APP, PSEN1 and PSEN2 mutations. 32957903_Exploring the Role of Aggregated Proteomes in the Pathogenesis of Alzheimer's Disease. 32992716_Presenilin-2 and Calcium Handling: Molecules, Organelles, Cells and Brain Networks. 33413468_Presence of a mutation in PSEN1 or PSEN2 gene is associated with an impaired brain endothelial cell phenotype in vitro. 33720885_Relevance of a Truncated PRESENILIN 2 Transcript to Alzheimer's Disease and Neurodegeneration. 33769986_Evaluation of the Clinical Features Accompanied by the Gene Mutations: The 2 Novel PSEN1 Variants in a Turkish Early-onset Alzheimer Disease Cohort. 33973882_Novel PSEN1 and PSEN2 Mutations Identified in Sporadic Early-onset Alzheimer Disease and Posterior Cortical Atrophy. 34002480_Plasma amyloid beta levels are driven by genetic variants near APOE, BACE1, APP, PSEN2: A genome-wide association study in over 12,000 non-demented participants. 34009547_A Rare Variation in the 3' Untranslated Region of the Presenilin 2 Gene Is Linked to Alzheimer's Disease. 34067945_Alzheimer's Disease Associated Presenilin 1 and 2 Genes Dysregulation in Neonatal Lymphocytes Following Perinatal Asphyxia. 34102969_Clinical Phenotype and Mutation Spectrum of Alzheimer's Disease with Causative Genetic Mutation in a Chinese Cohort. 34948396_Conformational Models of APP Processing by Gamma Secretase Based on Analysis of Pathogenic Mutations. 34987110_Presenilin Is Essential for ApoE Secretion, a Novel Role of Presenilin Involved in Alzheimer's Disease Pathogenesis. | ENSMUSG00000010609 | Psen2 | 1009.03599 | 0.8162211 | -0.2929680573 | 0.11666435 | 6.260205e+00 | 1.234799e-02 | 1.821023e-01 | No | Yes | 1079.608836 | 120.342912 | 1299.797074 | 141.124267 | |
| ENSG00000144040 | 94097 | SFXN5 | protein_coding | Q8TD22 | FUNCTION: Mitochondrial amino-acid transporter (By similarity). Does not act as a serine transporter: not able to mediate transport of serine into mitochondria (PubMed:30442778). Transports citrate (By similarity). {ECO:0000250|UniProtKB:Q8CFD0, ECO:0000250|UniProtKB:Q9H9B4, ECO:0000269|PubMed:30442778}. | Amino-acid transport;Membrane;Mitochondrion;Reference proteome;Transmembrane;Transmembrane helix;Transport | hsa:94097; | integral component of mitochondrial inner membrane [GO:0031305]; mitochondrion [GO:0005739]; citrate transmembrane transporter activity [GO:0015137]; transmembrane transporter activity [GO:0022857]; amino acid transport [GO:0006865]; citrate transport [GO:0015746]; mitochondrial transmembrane transport [GO:1990542] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000033720 | Sfxn5 | 443.35701 | 0.9713541 | -0.0419307477 | 0.17091748 | 6.003029e-02 | 8.064481e-01 | 9.462028e-01 | No | Yes | 535.920747 | 58.057209 | 535.312429 | 56.495950 | ||
| ENSG00000144642 | 27303 | RBMS3 | protein_coding | Q6XE24 | FUNCTION: Binds poly(A) and poly(U) oligoribonucleotides. {ECO:0000269|PubMed:10675610}. | Alternative splicing;Cytoplasm;RNA-binding;Reference proteome;Repeat | This gene encodes an RNA-binding protein that belongs to the c-myc gene single-strand binding protein family. These proteins are characterized by the presence of two sets of ribonucleoprotein consensus sequence (RNP-CS) that contain conserved motifs, RNP1 and RNP2, originally described in RNA binding proteins, and required for DNA binding. These proteins have been implicated in such diverse functions as DNA replication, gene transcription, cell cycle progression and apoptosis. The encoded protein was isolated by virtue of its binding to an upstream element of the alpha2(I) collagen promoter. The observation that this protein localizes mostly in the cytoplasm suggests that it may be involved in a cytoplasmic function such as controlling RNA metabolism, rather than transcription. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2010]. | hsa:27303; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; ribonucleoprotein complex [GO:1990904]; mRNA 3'-UTR AU-rich region binding [GO:0035925]; mRNA 3'-UTR binding [GO:0003730]; poly(A) binding [GO:0008143]; poly(U) RNA binding [GO:0008266]; RNA binding [GO:0003723]; defense response to tumor cell [GO:0002357]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of gene expression [GO:0010629]; positive regulation of gene expression [GO:0010628] | 17586524_These results suggest that RBMS3, by binding Prx1 mRNA in a sequence-specific manner, controls Prx1 expression and indirectly collagen synthesis. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21844183_a tumor suppression function for the human RBMS3 gene in esophageal squamous cell carcinoma, acting through c-Myc downregulation, with genetic loss of this gene contributing to poor outcomes in disease 22267851_genetic susceptibility plays a role in the pathophysiology of Bisphosphonate-related osteonecrosis of the jaw (BRONJ), with RBMS3 having a significant effect in the risk. 22957092_RBMS3 at 3p24 inhibits nasopharyngeal carcinoma development via inhibiting cell proliferation, angiogenesis, and inducing apoptosis 23045156_one interacting gene pair (RBMS3 versus ZNF516) which, even after Bonferroni correction for multiple testing, showed consistently significant effects on hip bone mineral density. 25588924_RBMS3 is a novel tumor suppressor gene in lung squamous cell carcinoma 27503288_We identified rs117026326 on GTF2I with GWAS significance (P = 1.10 x 10(-15)) and rs13079920 on RBMS3 with suggestive significance (P = 2.90 x 10(-5)) associating with Primary Sjogren's syndrome in women. 27902480_These findings implicated that RBMS3 and nuclear HIF1A could act as prognostic biomarkers and therapeutic targets for GC. 28409548_RBMS3 inhibited the proliferation and tumorigenesis of breast cancer cells, at least in part, through inactivation of the Wnt/beta-catenin signaling pathway 30279231_Results indicate that genetic ablation of RNA binding motif single stranded interacting protein 3 (RBMS3) contributes to chemoresistance RBMS3-deleted epithelial ovarian cancer (EOC). 30819235_our study revealed a novel mechanism of the RBMS3/Twsit1/MMP2 axis in the regulation of invasion and metastasis of breast cancer 32016951_RBMS3 delays disc degeneration by inhibiting Wnt/beta-catenin signaling pathway. 32627033_Increased expression of RBMS3 predicts a favorable prognosis in human gallbladder carcinoma. 33845141_LncRNA MEG3 regulates breast cancer proliferation and apoptosis through miR-141-3p/RBMS3 axis. 33910421_Tumor Suppressor Effect of RBMS3 in Breast Cancer. 34608266_RNA binding protein RBMS3 is a common EMT effector that modulates triple-negative breast cancer progression via stabilizing PRRX1 mRNA. | ENSMUSG00000039607 | Rbms3 | 30.49506 | 0.9748137 | -0.0368015023 | 0.50722576 | 5.263489e-03 | 9.421643e-01 | No | Yes | 35.008386 | 8.129800 | 34.353550 | 7.872071 | ||
| ENSG00000144847 | 152404 | IGSF11 | protein_coding | Q5DX21 | FUNCTION: Functions as a cell adhesion molecule through homophilic interaction. Stimulates cell growth. {ECO:0000269|PubMed:15795899, ECO:0000269|PubMed:16108831}. | Alternative splicing;Cell adhesion;Cell membrane;Disulfide bond;Glycoprotein;Growth regulation;Immunoglobulin domain;Membrane;Methylation;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | IGSF11 is an immunoglobulin (Ig) superfamily member that is preferentially expressed in brain and testis. It shares significant homology with coxsackievirus and adenovirus receptor (CXADR; MIM 602621) and endothelial cell-selective adhesion molecule (ESAM).[supplied by OMIM, Apr 2005]. | hsa:152404; | cell-cell junction [GO:0005911]; excitatory synapse [GO:0060076]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; ionotropic glutamate receptor binding [GO:0035255]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; maintenance of protein location [GO:0045185]; positive regulation of long-term synaptic potentiation [GO:1900273]; positive regulation of mini excitatory postsynaptic potential [GO:0061885]; regulation of growth [GO:0040008] | 12207903_Molecular cloning of BT-IgSF which is preferentially expressed by brain and testis 15795899_cell adhesion-inducing function of BT-IgSF suggests a role of the cell surface molecule in the development/function of the central nervous system and spermatogenesis 16108831_IGSF11 is a good candidate of cancer-testis antigen. Furthermore, suppression of IGSF11 by siRNA retarded the growth of gastric cancer cells. 26595655_Concerted role for IgSF11 and PSD-95 in the regulation of AMPAR-mediated synaptic transmission and plasticity. 30220083_this study shows that VSIG-3 as a ligand of VISTA inhibits human T-cell function 33841409_Structural Basis of VSIG3: The Ligand for VISTA. 34491997_IGSF11 is required for pericentric heterochromatin dissociation during meiotic diplotene. | ENSMUSG00000022790 | Igsf11 | 43.02475 | 1.1433798 | 0.1933046558 | 0.45847693 | 1.770726e-01 | 6.739012e-01 | No | Yes | 56.703374 | 16.772978 | 48.987077 | 14.117135 | ||
| ENSG00000145242 | 2044 | EPHA5 | protein_coding | P54756 | FUNCTION: Receptor tyrosine kinase which binds promiscuously GPI-anchored ephrin-A family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling. Among GPI-anchored ephrin-A ligands, EFNA5 most probably constitutes the cognate/functional ligand for EPHA5. Functions as an axon guidance molecule during development and may be involved in the development of the retinotectal, entorhino-hippocampal and hippocamposeptal pathways. Together with EFNA5 plays also a role in synaptic plasticity in adult brain through regulation of synaptogenesis. In addition to its function in the nervous system, the interaction of EPHA5 with EFNA5 mediates communication between pancreatic islet cells to regulate glucose-stimulated insulin secretion (By similarity). {ECO:0000250}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Cell projection;Glycoprotein;Kinase;Membrane;Neurogenesis;Nucleotide-binding;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transferase;Transmembrane;Transmembrane helix;Tyrosine-protein kinase | This gene belongs to the ephrin receptor subfamily of the protein-tyrosine kinase family. EPH and EPH-related receptors have been implicated in mediating developmental events, particularly in the nervous system. Receptors in the EPH subfamily typically have a single kinase domain and an extracellular region containing a Cys-rich domain and 2 fibronectin type III repeats. The ephrin receptors are divided into 2 groups based on the similarity of their extracellular domain sequences and their affinities for binding ephrin-A and ephrin-B ligands. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Aug 2013]. | hsa:2044; | axon [GO:0030424]; dendrite [GO:0030425]; external side of plasma membrane [GO:0009897]; integral component of plasma membrane [GO:0005887]; neuron projection [GO:0043005]; neuronal cell body [GO:0043025]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; rough endoplasmic reticulum [GO:0005791]; ATP binding [GO:0005524]; ephrin receptor activity [GO:0005003]; GPI-linked ephrin receptor activity [GO:0005004]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; transmembrane receptor protein tyrosine kinase activity [GO:0004714]; transmembrane-ephrin receptor activity [GO:0005005]; axon guidance [GO:0007411]; cAMP-mediated signaling [GO:0019933]; ephrin receptor signaling pathway [GO:0048013]; hippocampus development [GO:0021766]; neuron development [GO:0048666]; positive regulation of CREB transcription factor activity [GO:0032793]; positive regulation of kinase activity [GO:0033674]; regulation of actin cytoskeleton organization [GO:0032956]; regulation of GTPase activity [GO:0043087]; regulation of insulin secretion involved in cellular response to glucose stimulus [GO:0061178]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169] | 16737551_increased levels of ephrins A1 and A5 in the presence of high expression of Ephs A1 and A2 lead to a more aggressive ovarian cancer phenotype 19733895_EphA5 might be a potential target for epigenetic silencing in primary breast cancer and a valuable molecular marker for breast cancer carcinogenesis and progression. 19949912_Eph-A5 and Eph-A7 staining intensity was identified as independent prognostic factors for pancreatic ductal adenocarcinoma 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 22079638_these data suggest that miR-34a is a negative modulator of chondrogenesis, particularly in migration of chondroblasts, by targeting EphA5 and resulting inhibition of cellular condensation during chondrogenesis of chick limb mesenchymal cells. 23824631_Insertional translocation leading to a 4q13 duplication including the EPHA5 gene is associated with attention-deficit. hyperactivity disorder 24029132_These results indicate that EphA5 may be a negative regulator of bone formation. 24086308_unbound EphA5 LBD appears to comprise an ensemble of open conformations that have only small variations over the loops and appear ready to bind ephrin-A ligands 25609195_Our study provides evidence that EphA5 is a potential target for epigenetic silencing in primary prostate cancer and is a potentially valuable prognosis predictor and thereapeutic marker for prostate cancer. 25623065_demonstrate that a new monoclonal antibody against human EphA5 sensitized lung cancer cells and human lung cancer xenografts to radiotherapy and significantly prolonged survival, thus suggesting the likelihood of translational applications 27582484_Gene-based analysis identified EPHA6 as the gene most significantly associated with paclitaxel-induced neuropathy...This first study sequencing EPHA genes revealed that low-frequency variants in EPHA6, EPHA5, and EPHA8 contribute to the susceptibility to paclitaxel-induced neuropathy 27651378_Our data indicate that EphA5 receptor may be a tumor suppressor in colorectal carcinoma and it may be a new therapeutic target for colorectal carcinoma. 27887627_Our results show that EphA5 may be a potential biomarker for distinguishing high-and low-grade ovarian serous carcinoma and a potential prognostic marker. 27988259_the interactions of EphA5/ephrinA5 and/or EphA7/ephrinA5 between HSPCs and BMSCs, independently and cooperatively, play a role in HSPC colony formation through the upregulation of GM-CSFR. Furthermore, the adhesion/migration of HSPCs appears to be mediated in part through the activation of Rac1. 28421649_EphA5 protein was negatively (0) or weakly (1+) expressed in 48 of 78 (61.5%), moderately (2+) expressed in 15 of 78 (19.2%) and strongly (3+) expressed in 15 of 78 (19.2%) tumour samples of clear cell renal cell carcinoma (ccRCC). Decreased expression of EphA5 was detected more often in females than in males 31823378_Genetic variation in EPHA contributes to sensitivity to paclitaxel-induced peripheral neuropathy. 32234341_EPHA5 mutation impairs natural killer cell-mediated cytotoxicity against non-small lung cancer cells and promotes cancer cell migration and invasion. 33288738_EPHA5 mutations predict survival after immunotherapy in lung adenocarcinoma. 34109382_Deep learning model reveals potential risk genes for ADHD, especially Ephrin receptor gene EPHA5. | ENSMUSG00000029245 | Epha5 | 46.16809 | 1.1622348 | 0.2169015199 | 0.44150960 | 2.430021e-01 | 6.220462e-01 | No | Yes | 62.317520 | 22.192610 | 49.674770 | 17.613070 | ||
| ENSG00000145715 | 5921 | RASA1 | protein_coding | P20936 | FUNCTION: Inhibitory regulator of the Ras-cyclic AMP pathway. Stimulates the GTPase of normal but not oncogenic Ras p21; this stimulation may be further increased in the presence of NCK1. {ECO:0000269|PubMed:11389730, ECO:0000269|PubMed:8360177}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Disease variant;GTPase activation;Phosphoprotein;Reference proteome;Repeat;SH2 domain;SH3 domain;Tumor suppressor | The protein encoded by this gene is located in the cytoplasm and is part of the GAP1 family of GTPase-activating proteins. The gene product stimulates the GTPase activity of normal RAS p21 but not its oncogenic counterpart. Acting as a suppressor of RAS function, the protein enhances the weak intrinsic GTPase activity of RAS proteins resulting in the inactive GDP-bound form of RAS, thereby allowing control of cellular proliferation and differentiation. Mutations leading to changes in the binding sites of either protein are associated with basal cell carcinomas. Mutations also have been associated with hereditary capillary malformations (CM) with or without arteriovenous malformations (AVM) and Parkes Weber syndrome. Alternative splicing results in two isoforms where the shorter isoform, lacking the N-terminal hydrophobic region but retaining the same activity, appears to be abundantly expressed in placental but not adult tissues. [provided by RefSeq, May 2012]. | hsa:5921; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; GTPase activator activity [GO:0005096]; GTPase activity [GO:0003924]; GTPase binding [GO:0051020]; phosphotyrosine residue binding [GO:0001784]; potassium channel inhibitor activity [GO:0019870]; signaling receptor binding [GO:0005102]; blood vessel morphogenesis [GO:0048514]; ephrin receptor signaling pathway [GO:0048013]; intracellular signal transduction [GO:0035556]; mitotic cytokinesis [GO:0000281]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell adhesion [GO:0007162]; negative regulation of cell-matrix adhesion [GO:0001953]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of Ras protein signal transduction [GO:0046580]; positive regulation of GTPase activity [GO:0043547]; regulation of actin filament polymerization [GO:0030833]; regulation of cell shape [GO:0008360]; regulation of RNA metabolic process [GO:0051252]; signal transduction [GO:0007165]; vasculogenesis [GO:0001570] | 11751853_We show a novel alternative pathway of apoptosis in human primary cells that is mediated by transcriptionally dependent decreases in p53 and c-Myc and decreases in p21. 11847220_N-terminal fragment generated by caspase cleavage protects cells in a Ras/PI3K/Akt-dependent manner that does not rely on NFkappa B activation 12730209_mutual regulation of Ras and NF1-GAP is essential for normal neuronal differentiation 15010862_p21Ras, hSOS1, and p120GAP are not involved in polycystic ovary disease 15041706_The induction of p21 gene expression is mediated by PPARgamma ligands in lung carcinoma. 15187129_ligation of CD40 on EC increased association of Ras with its effector molecules Raf, Rho, and PI3K. But, it was determined that only PI3K was functional for Ras-induced VEGF transcription. 15542850_executioner caspases control the extent of their own activation by a feedback regulatory mechanism initiated by the partial cleavage of RasGAP that is crucial for cell survival under adverse conditions 15688026_p52Shc couples tyrosine kinase receptors to Ras by recruiting Grb2/Sos complexes. 16046410_an uncleavable N-terminal RasGAP fragment in insulin-secreting cells has a role in increasing resistance toward apoptotic stimuli without affecting glucose-induced insulin secretion 16971514_These results demonstrate that integrin signaling through Arg activates p190 by promoting its association with p120, resulting in recruitment of p190 to the cell periphery where it inhibits Rho. 18024870_Observational study of gene-disease association and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18327598_novel mutation is decribed, which causes capillary malformation and limb enlargement in a patient from a family with vascular malformations 18363760_the spectrum of clinical manifestations due to mutations in RASA1 is wider than previously thought and also includes typical CMs not associated with AVM/AVF. 18446851_42 novel RASA1 mutations and the associated phenotype in 44 families. 18761085_RasGAP and Capns1 interaction in oncogenic Ras cells is involved in regulating migration and cell survival. 18784923_found a mean Ras activation of 23.1% in cell lines with known constitutively activating ras mutations 19012001_Observational study of gene-disease association. (HuGE Navigator) 19151751_p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. 19435801_Data show that fibroblast, endothelial and carcinoma polarity during cell migration requires FAK and is associated with a complex between FAK, p120RasGAP and p190RhoGAP (p190A), leading to p190A tyrosine phosphorylation. 19578876_Observational study of gene-disease association. (HuGE Navigator) 19773259_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19786546_CD200R inhibits the activation of human myeloid cells through direct recruitment of Dok2 and subsequent activation of RAS p21 protein activator 1. 19843518_the monomeric intracellular plexin-B1 binds R-Ras but not H-Ras. These findings suggest that the monomeric form of the intracellular region is primed for GAP activity and extend a model for plexin activation. 20007727_In a collaborative study, 5 index patients (2 females, 3 males) with spinal AVMs or AVFs and cutaneous multifocal capillary lesions were investigated for the RASA1 gene mutation. 20592250_Observational study of gene-disease association. (HuGE Navigator) 20610402_Sema4D/Plexin-B1 promotes the dephosphorylation and activation of PTEN through the R-Ras GAP activity, inducing growth cone collapse. 20676106_miR-132 acts as an angiogenic switch by suppressing endothelial p120RasGAP expression. 20688547_Observational study of gene-disease association. (HuGE Navigator) 20702649_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20821215_A novel synonymous mutation (c.1229 G > A [p.K420K]) of RASA1 was identified in a Chinese population with sporadic Sturge-Weber syndrome 21368895_Ras mutation cooperates with beta-catenin activation to drive bladder tumourigenesis. 21460216_Ca2+-dependent monomer and dimer formation switches CAPRI Protein between Ras GTPase-activating protein (GAP) and RapGAP activities 21646295_An important role is revealed for p120 RasGAP (RASA1) as a transgenic regulator of CD4+CD8+ double-positive cell survival and positive selection in the thymus as well as naive T cell survival in the periphery. 21664272_Cell adhesion to the substrate is necessary for RasGAP to bind Nck1. Cell detachment makes RasGAP incapable of associating with Nck1 and decreases RasGAP activity. 21664272_Nck1 activates RasGAP by direct binding in the substrate-attached but not in the suspended cells. 21768288_The results assign an unexpected role for p120RasGAP in the regulation of integrin traffic in cancer cells and reveal a new concept of competitive binding of Rab GTPases and GAP proteins to receptors as a regulatory mechanism in trafficking. 22200646_multifocal capillary malformations is the key clinical finding to suggest a RASA1 mutation 22205990_arguments against G3BP1 being a genuine RasGAP-binding partner 22342634_An update of the associated phenotype variability in a family with hereditary capillary malformations caused by a mutation in the RASA1 gene. 22949691_Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations 23158644_capillary malformation-arteriovenous malformation syndrome; study reports a family with a novel mutation in the RASA1 gene - a truncating mutation in exon 11 of RASA1 (Q503X) 23322774_MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA by repressing RAS p21 GTPase activating protein 1 (RASA1) 23386617_14-3-3 negatively regulates the RGC downstream of the PI3-kinase/Akt signaling pathway 23447049_The results of the present study indicate that P110, in combination with chemotherapeutics, is likely to represent a potential therapeutic strategy for cancer. 23650393_RASA1 mutation is responsible for the aberrant lymphatic architecture and functional abnormalities, as visualized in the PKWS subject and in the animal model. 23826368_results indicate that, mTOR, Bad, or Survivin are not required for p120 RasGAP fragment N to protect cells from cell death; conclude that downstream targets of Akt other than mTORC1, Bad, or survivin mediate fragment N-induced protection or that several Akt effectors can compensate for each other to induce the pro-survival fragment N-dependent responses 23829194_Multifocal, small, round-to-oval, pinkish-to-red cutaneous capillary malformations are seen in more than 90% of people with RASA1 mutations. 24038909_RASA1 mutations specifically cause capillary malformation-arteriovenous malformation. 24347041_These results indicate that stress-activated caspase-3 might contribute to the suppression of metastasis through the generation of fragment N2( RasGAP 24443565_The novel findings of this study shed light on the molecular mechanisms underlying the DLC1 inhibitory effects of p120 and suggest a functional cross-talk between Ras and Rho proteins at the level of regulatory proteins. 24465899_A ubiquitous binding partner of p190RhoGAP, p120RasGAP (RasGAP), is expressed in much lower levels in DKO4 cells compared to DLD1, and this expression is regulated by KRAS. 24600991_our study reveals mir-182 suppresses cell proliferation in vivo. RASA1 is related to cell apoptosis. We further show that mir-182 downregulates RASA1 25040287_Capillary malformation-arteriovenous malformation syndrome (CM-AVM) is an autosomal dominant disorder caused by RASA1 mutations. 25202123_Antisense-mediated knockdown (anti-miR) revealed that miR-206/21 coordinately promote RAS-ERK signaling and the corresponding cell phenotypes by inhibiting translation of the pathway suppressors RASA1 and SPRED1. 25246356_The individual contribution of each Akt isoform in p120 RasGAP fragment N-mediated cell protection against Fas ligand induced cell death, was investigated. 25394563_RASA1 expression is associated with breast cancer progression and poor survival and diseasefree survival of patients. 25663768_miR-21 promotes malignant behaviors of colon cancer cells by regulating RASA1 expression via RAS pathways. 25733681_Results show that report that RasGAP associates to PDGFRbeta and prevents its direct activation. This underlying mechanism raises the possibility that PDGFRbeta-mediated diseases involve indirect activation of PDGFRbeta. 25867276_Low RASA1 expression is associated with colorectal cancer. 26096958_Maternal and fetal capillary malformation-arteriovenous malformation due to a novel RASA1 mutation presenting with prenatal non-immune hydrops fetalis have been found. 26109071_p120RasGAP shields Akt from deactivating phosphatases in FGF1 signaling, but loses this ability once cleaved by caspase-3. 26126858_Data showed that hypoxia regulated the expression of miR-182 and RASA1 to promote HCC angiogenesis. 26192947_This is the second largest study on isolated, non-syndromic Port-wine stain; data suggest that GNAQ is the main genetic determinant in this condition. Moreover, isolated port-wine stains are distinct from capillary malformations seen in RASA1 disorders. 26710849_Data suggest that, in response to netrin-1/netrin receptor (DCC) signaling, p120RasGAP is recruited to growth cones and supports axon outgrowth; p120RasGAP Src homology 2 domains exhibit scaffolding properties sufficient to support axon outgrowth. 26747707_Results show that oncogenic KRAS can activate Rho through miR-31-mediated regulation of RASA1 indicating miR-31 acts as a KRAS effector to modulate invasion and migration in pancreatic cancer. 26840794_PTP1B dephosphorylates PITX1 to weaken its protein stability and the transcriptional activity for p120RasGAP gene expression 26969842_These results and the extreme variable expressivity support the hypothesis that somatic 'second hits' are required for the development of vascular anomalies associated with CM-AVM syndrome. In addition, the phenotypes of the affected individuals further clarify that lymphatic manifestations are also part of the phenotypic spectrum of RASA1-related disorders. 26993606_RASA1 mutations are associated melanoma tumorigenesis. 27101583_MicroRNA-21 reduces RASA1 expression in cervical cancer cell lines and promotes cervical cancer cell migration via RASA1. Furthermore, Ras-induced epithelial-mesenchymal transition contributes to miR-21/RASA1 axis promoting cervical cancer cell migration. 27752061_we demonstrated a novel oncogenic mechanism of PTP1B on affecting PITX1/p120RasGAP in colorectal carcinoma(CRC). Regorafenib inhibited CRC survival through reserving PTP1B-dependant PITX1/p120RasGAP downregulation. PTP1B may be a potential biomarker predicting regorafenib effectiveness, and a potential solution for CRC 27767378_QKI-5 stabilized RASA1 mRNA via directly binding to the QKI response element region of RASA1, which in turn prevented the activation of the Ras-MAPK signaling pathway, suppressed cellular proliferation and induced cell cycle arrest. 28108518_Low RASA1 expression is associated with Triple-Negative Breast Cancer. 28179330_Data show that patients with low level of Ras GTPase-activating protein 1 (RASA1) expression correlated with a significantly poorer survival compared to those with high level of RASA1 expression. 28687708_The interaction between RASA1 and EPHB4 is an indication of the major cause of capillary malformation with arteriovenous malformation. 28798331_Study found that RASA1, a known target of miR-132, was downregulated in human dermal fibroblasts upon miR-132 overexpression which contributes to the pro-migratory function of miR-132 in fibroblasts. 29024832_A somatic RASA1 mutation in addition to the germline RASA1 mutation, was detected within endothelial cells in capillary malformation-arteriovenous malformation. 29110021_RASA1 variants are rarely found in children with sporadic capillary malformations of lower limbs without capillary malformation-arteriovenous malformation syndrome. 29127119_Cancer genomic and functional data nominate concurrent RASA1/NF1 loss-of-function mutations as a strong mitogenic driver in NSCLC, which may sensitize to trametinib 29891884_Our data suggest that screening for large RASA1 deletions and duplications in this disorder is important and suggest that NGS multi-gene panel testing is beneficial for the molecular diagnosis of cases with complex vascular phenotypes. 30243714_Data suggest that the anti-oncogenic function of circular RNA circ-ITCH (circ-ITCH) is via the circ-ITCH-miR-145-RASA1 (RAS p21 protein activator (GTPase activating protein) 1) axis in vitro and in vivo. 30569149_miR21 regulated cell proliferation, migration, invasion and tumor growth of esophageal squamous cell carcinoma by directly targeting RASA1, which may have been achieved via regulation of Snail and vimentin. 30635911_we report for the first time the presence of RASA1 constitutional mosaicism in CM-AVM. Constitutional mosaicism has implications for accurate molecular diagnosis and recurrence risk and helps to explain the great phenotypic variability in CM-AVM. 30864691_The results suggested that miRNA4530 suppresses cell proliferation and enhances apoptosis by targeting RASA1 via the ERK/MAPK and PI3K/AKT signaling pathways. 31300548_RASA1 mosaic mutations can cause capillary malformation-arteriovenous malformation. Even low-level mosaicism can cause the classical phenotype and increased risk for offspring. 31857500_CircAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. 32540970_The GTPase-activating protein p120RasGAP has an evolutionarily conserved ''FLVR-unique'' SH2 domain. 32588875_RASA1 inhibits the progression of renal cell carcinoma by decreasing the expression of miR-223-3p and promoting the expression of FBXW7. 32605345_[Neonatal capillary malformation-arteriovenous malformation complicated with acute heart failure: a case report and literature review]. 32776686_Prenatal pleural effusions and chylothorax: An unusual presentation for CM-AVM syndrome due to RASA1. 32900839_RASA1 phenotype overlaps with hereditary haemorrhagic telangiectasia: two case reports. 33118152_Parkes-Weber syndrome related to RASA1 mosaic mutation. 34173139_Abnormal H3K27 histone methylation of RASA1 gene leads to unexplained recurrent spontaneous abortion by regulating Ras-MAPK pathway in trophoblast cells. 34238206_CircMTO1 inhibits ox-LDL-stimulated vascular smooth muscle cell proliferation and migration via regulating the miR-182-5p/RASA1 axis. 34238211_The long noncoding RNA MEG3 regulates Ras-MAPK pathway through RASA1 in trophoblast and is associated with unexplained recurrent spontaneous abortion. 34464226_Down-regulated placental miR-21 contributes to preeclampsia through targeting RASA1. 34491620_Assessing the association of common genetic variants in EPHB4 and RASA1 with phenotype severity in familial cerebral cavernous malformation. | ENSMUSG00000021549 | Rasa1 | 800.03814 | 0.9669945 | -0.0484204194 | 0.13221334 | 1.337227e-01 | 7.146031e-01 | 9.134531e-01 | No | Yes | 976.634761 | 202.379752 | 939.651152 | 190.629226 | |
| ENSG00000145740 | 64924 | SLC30A5 | protein_coding | Q8TAD4 | FUNCTION: Functions as a zinc transporter. May be a transporter of zinc into beta cells in order to form insulin crystals. Partly regulates cellular zinc homeostasis. Required with ZNT7 for the activation of zinc-requiring enzymes, alkaline phosphatases (ALPs). Transports zinc into the lumens of the Golgi apparatus and vesicular compartments where ALPs locate, thus, converting apoALPs to holoALPs. Required with ZNT6 and ZNT7 for the activation of TNAP. {ECO:0000269|PubMed:11904301, ECO:0000269|PubMed:11937503, ECO:0000269|PubMed:15276077, ECO:0000269|PubMed:15525635, ECO:0000269|PubMed:15994300}. | Acetylation;Alternative splicing;Golgi apparatus;Ion transport;Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport;Zinc;Zinc transport | This gene encodes a member of the SLC30A/ZnT family of zinc transporter proteins. ZnT proteins mediate both cellular zinc efflux and zinc sequestration into membrane-bound organelles. The encoded protein plays a role in the early secretory pathway as a heterodimer with zinc transporter 6, and may also regulate zinc sequestration into secretory granules of pancreatic beta cells. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene, and a pseudogene of this gene is located on the long arm of chromosome 19. [provided by RefSeq, Oct 2011]. | hsa:64924; | apical plasma membrane [GO:0016324]; Golgi apparatus [GO:0005794]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; secretory granule [GO:0030141]; secretory granule membrane [GO:0030667]; zinc ion binding [GO:0008270]; zinc ion transmembrane transporter activity [GO:0005385]; cellular zinc ion homeostasis [GO:0006882]; cobalt ion transport [GO:0006824]; insulin processing [GO:0030070]; regulation of proton transport [GO:0010155]; response to zinc ion [GO:0010043]; zinc ion transmembrane transport [GO:0071577]; zinc ion transport [GO:0006829] | 11904301_cloning and characterization of ZnT-5 which is expressed in pancreatic beta cells 11937503_A novel zinc-regulated human zinc transporter, hZTL1, is localized to the enterocyte apical membrane 17971500_hZnT-5 is up-regulated in response to cellular zinc depletion in Raji cells. 18639746_Data show that ZNT5 is extensively present in the Abeta-positive plaques in the cortex of human AD brains. 19064571_Observational study of gene-disease association. (HuGE Navigator) 19759014_The cytosolic C-terminal tail of ZnT5 is important for its interaction with ZnT6 as a heterodimer. 20689807_Observational study of gene-disease association. (HuGE Navigator) 21887337_exons 15 to 17 include a signal that results in trafficking of ZnT5 to the Golgi apparatus and that the 3' end of exon 14 includes a signal that leads to retention in the ER. 22529353_Results identify a unique class of transporters Znt5 and Znt8 and the structural motif required to discriminate between Zn(2+) and Cd(2+) transport. 23275032_Overexpression of ZnT5 is accompanied by activation of PI3K/Akt pathway and inhibiting HG-induced apoptosis. 27661418_There was a significant increase in protein levels of ZnT5 in the prefrontal cortex in Major depression disorder, relative to control subjects. 33547425_Bi-allelic loss of function variants in SLC30A5 as cause of perinatal lethal cardiomyopathy. | ENSMUSG00000021629 | Slc30a5 | 3442.24521 | 0.9284027 | -0.1071773095 | 0.09015678 | 1.410189e+00 | 2.350251e-01 | 6.262431e-01 | No | Yes | 3471.864295 | 383.668976 | 3610.545426 | 390.235314 | |
| ENSG00000146233 | 51302 | CYP39A1 | protein_coding | Q9NYL5 | FUNCTION: A cytochrome P450 monooxygenase involved in neural cholesterol clearance through bile acid synthesis (PubMed:25201972, PubMed:10748047). Catalyzes 7-alpha hydroxylation of (24S)-hydroxycholesterol, a neural oxysterol that is metabolized to bile acids in the liver (PubMed:25201972, PubMed:10748047). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase) (PubMed:25201972, PubMed:10748047). {ECO:0000269|PubMed:10748047, ECO:0000269|PubMed:25201972}. | Cholesterol metabolism;Endoplasmic reticulum;Heme;Iron;Lipid metabolism;Membrane;Metal-binding;Microsome;Monooxygenase;Oxidoreductase;Reference proteome;Signal;Steroid metabolism;Sterol metabolism;Transmembrane;Transmembrane helix | PATHWAY: Steroid metabolism; cholesterol degradation. {ECO:0000305|PubMed:25201972}.; PATHWAY: Lipid metabolism; bile acid biosynthesis. {ECO:0000305|PubMed:25201972}. | This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This endoplasmic reticulum protein is involved in the conversion of cholesterol to bile acids. Its substrates include the oxysterols 25-hydroxycholesterol, 27-hydroxycholesterol and 24-hydroxycholesterol. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013]. | hsa:51302; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; 24-hydroxycholesterol 7alpha-hydroxylase activity [GO:0033782]; heme binding [GO:0020037]; iron ion binding [GO:0005506]; oxysterol 7-alpha-hydroxylase activity [GO:0008396]; steroid 7-alpha-hydroxylase activity [GO:0008387]; steroid hydroxylase activity [GO:0008395]; bile acid biosynthetic process [GO:0006699]; cholesterol catabolic process [GO:0006707]; cholesterol homeostasis [GO:0042632]; digestion [GO:0007586]; sterol metabolic process [GO:0016125] | 19553612_The rs754203 SNP in CYP46A1 was associated with a risk for POAG. This polymorphism was not associated with changes in plasma 24S-hydroxycholesterol. 20453000_Observational study of gene-disease association. (HuGE Navigator) 24192117_GSTA1 and CYP39A1 were found to be associated with busulfan clearance. When combined, the two haplotypes explained 17% of the variability in busulfan clearance. 26475344_The CYP39A1 polymorphism rs7761731 may help to identify patients at high risk for treatment related toxicity. 32392803_Orphan Nuclear Receptor RORalpha Regulates Enzymatic Metabolism of Cerebral 24S-Hydroxycholesterol through CYP39A1 Intronic Response Element Activation. 33620406_Association of Rare CYP39A1 Variants With Exfoliation Syndrome Involving the Anterior Chamber of the Eye. | ENSMUSG00000023963 | Cyp39a1 | 119.05466 | 1.1560394 | 0.2091905667 | 0.28213769 | 5.510646e-01 | 4.578830e-01 | 7.954708e-01 | No | Yes | 137.646585 | 45.794280 | 115.141723 | 37.508976 |
| ENSG00000146282 | 57038 | RARS2 | protein_coding | Q5T160 | ATP-binding;Acetylation;Aminoacyl-tRNA synthetase;Ligase;Mitochondrion;Neurodegeneration;Nucleotide-binding;Pontocerebellar hypoplasia;Protein biosynthesis;Reference proteome;Transit peptide | This nuclear gene encodes a protein that localizes to the mitochondria, where it catalyzes the transfer of L-arginine to its cognate tRNA, an important step in translation of mitochondrially-encoded proteins. Defects in this gene are a cause of pontocerebellar hypoplasia type 6 (PCH6). Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]. | hsa:57038; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; arginine-tRNA ligase activity [GO:0004814]; ATP binding [GO:0005524]; RNA binding [GO:0003723]; arginyl-tRNA aminoacylation [GO:0006420]; mitochondrial translation [GO:0032543]; tRNA aminoacylation for protein translation [GO:0006418] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 22086604_mutations in the gene encoding mitochondrial arginyl-tRNA synthetase, RARS2, may have a role in pontocerebellar hypoplasia type 6 [case report] 22569581_Molecular investigations of RARS2 disclosed the c.25A>G/p.I9V and the c.1586+3A>T in family A. 25809939_Mutations in the RARS2 promoter are likely to represent a new causal mechanism of pontocerebellar hypoplasia. 26970947_RARS2 gene mutations can cause a metabolic neurodegenerative disease manifesting primarily as early onset epileptic encephalopathies with post-natal microcephaly, without pontocerebellar hypoplasia. 27769281_Characteristic neuroradiological abnormalities of PCH6 such as vermis and cerebellar hypoplasia and progressive pontocerebellar atrophy may be missing in patients with RARS2 mutations 32725632_Whole-exome sequencing in adult patients with developmental and epileptic encephalopathy: It is never too late. | ENSMUSG00000028292 | Rars2 | 1284.23995 | 1.0979829 | 0.1348556209 | 0.10438593 | 1.673319e+00 | 1.958146e-01 | 5.874899e-01 | No | Yes | 1731.755999 | 316.364687 | 1510.605518 | 269.990195 | ||
| ENSG00000146530 | 221806 | VWDE | protein_coding | Q8N2E2 | Alternative splicing;Disulfide bond;EGF-like domain;Glycoprotein;Reference proteome;Repeat;Secreted;Signal | hsa:221806; | cell surface [GO:0009986]; extracellular region [GO:0005576]; signaling receptor binding [GO:0005102]; anatomical structure development [GO:0048856] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | 123.55833 | 1.2987794 | 0.3771564082 | 0.31088264 | 1.470606e+00 | 2.252500e-01 | 6.171230e-01 | No | Yes | 165.189689 | 44.246098 | 123.860936 | 32.502329 | |||||
| ENSG00000146556 | 375260 | WASH2P | transcribed_unprocessed_pseudogene | 313.54611 | 1.0971546 | 0.1337668191 | 0.17487501 | 5.873967e-01 | 4.434276e-01 | 7.864091e-01 | No | Yes | 385.006098 | 58.541319 | 355.627573 | 52.872925 | ||||||||||
| ENSG00000146872 | 11011 | TLK2 | protein_coding | Q86UE8 | FUNCTION: Serine/threonine-protein kinase involved in the process of chromatin assembly and probably also DNA replication, transcription, repair, and chromosome segregation. Phosphorylates the chromatin assembly factors ASF1A AND ASF1B. Phosphorylation of ASF1A prevents its proteasome-mediated degradation, thereby enhancing chromatin assembly. Negative regulator of amino acid starvation-induced autophagy. {ECO:0000269|PubMed:10523312, ECO:0000269|PubMed:11470414, ECO:0000269|PubMed:12660173, ECO:0000269|PubMed:12955071, ECO:0000269|PubMed:20016786, ECO:0000269|PubMed:22354037, ECO:0000269|PubMed:9427565}. | 3D-structure;ATP-binding;Alternative splicing;Cell cycle;Chromatin regulator;Coiled coil;Cytoplasm;Cytoskeleton;DNA damage;Disease variant;Kinase;Mental retardation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a nuclear serine/threonine kinase that was first identified in Arabidopsis. The encoded protein is thought to function in the regulation of chromatin assembly in the S phase of the cell cycle by regulating the levels of a histone H3/H4 chaperone. This protein is associated with double-strand break repair of DNA damage caused by radiation. Pseudogenes of this gene are present on chromosomes 10 and 17. Alternate splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:11011; | intermediate filament [GO:0005882]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell cycle [GO:0007049]; cellular response to DNA damage stimulus [GO:0006974]; cellular response to gamma radiation [GO:0071480]; chromatin organization [GO:0006325]; chromosome segregation [GO:0007059]; intracellular signal transduction [GO:0035556]; negative regulation of autophagy [GO:0010507]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; peptidyl-serine phosphorylation [GO:0018105]; protein phosphorylation [GO:0006468]; regulation of chromatin assembly or disassembly [GO:0001672] | 20016786_ASF1 cellular levels are tightly controlled by distinct pathways and provide a molecular mechanism for post-translational regulation of dASF1 and hASF1a by TLK kinases. 20056645_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21647934_TLKs appear to be intimately linked to the pattern of resistance to DNA damage, and specifically double-strand breaks. 23414760_TLK2 plays a role in Kaposi's sarcoma-associated herpesvirus reactivation. 26931568_Tlk2 promotes Asf1A function during the DNA damage response in G2 to allow for proper restoration of chromatin structure at the break site and subsequent recovery from the arrest. 27489360_Results show that TLK2 overexpression correlates with increased genome-wide copy number aberrations in breast cancer cells, impairs cell-cycle checkpoint signaling in response to DNA damage. 27694828_TLK2 is amplified in aggressive luminal breast neoplasms 29861108_haploinsufficiency of TLK2 is the most likely underlying disease mechanism, leading to a consistent neurodevelopmental phenotype. 29955062_Molecular basis of TLK2 activation and inhibition has been dissected. 30207834_Study shows that TLK2 expression is increased in glioblastoma. Furthermore, TLK2 overexpression resulted in tumor growth and metastasis via SRC signaling pathway. 32755577_Tousled-Like Kinases Suppress Innate Immune Signaling Triggered by Alternative Lengthening of Telomeres. 33323470_Functional analysis of TLK2 variants and their proximal interactomes implicates impaired kinase activity and chromatin maintenance defects in their pathogenesis. 35136069_Tousled-like kinase 2 targets ASF1 histone chaperones through client mimicry. | ENSMUSG00000020694 | Tlk2 | 1779.42434 | 0.9588429 | -0.0606336977 | 0.09146178 | 4.387117e-01 | 5.077449e-01 | 8.237288e-01 | No | Yes | 2340.081344 | 433.011295 | 2251.606296 | 407.398331 | |
| ENSG00000147162 | 8473 | OGT | protein_coding | O15294 | FUNCTION: Catalyzes the transfer of a single N-acetylglucosamine from UDP-GlcNAc to a serine or threonine residue in cytoplasmic and nuclear proteins resulting in their modification with a beta-linked N-acetylglucosamine (O-GlcNAc) (PubMed:26678539, PubMed:23103939, PubMed:21240259, PubMed:21285374, PubMed:15361863). Glycosylates a large and diverse number of proteins including histone H2B, AKT1, ATG4B, EZH2, PFKL, KMT2E/MLL5, MAPT/TAU and HCFC1 (PubMed:19451179, PubMed:20200153, PubMed:21285374, PubMed:22923583, PubMed:23353889, PubMed:24474760, PubMed:26678539, PubMed:27527864). Can regulate their cellular processes via cross-talk between glycosylation and phosphorylation or by affecting proteolytic processing (PubMed:21285374). Probably by glycosylating KMT2E/MLL5, stabilizes KMT2E/MLL5 by preventing its ubiquitination (PubMed:26678539). Involved in insulin resistance in muscle and adipocyte cells via glycosylating insulin signaling components and inhibiting the 'Thr-308' phosphorylation of AKT1, enhancing IRS1 phosphorylation and attenuating insulin signaling (By similarity). Involved in glycolysis regulation by mediating glycosylation of 6-phosphofructokinase PFKL, inhibiting its activity (PubMed:22923583). Component of a THAP1/THAP3-HCFC1-OGT complex that is required for the regulation of the transcriptional activity of RRM1. Plays a key role in chromatin structure by mediating O-GlcNAcylation of 'Ser-112' of histone H2B: recruited to CpG-rich transcription start sites of active genes via its interaction with TET proteins (TET1, TET2 or TET3) (PubMed:22121020, PubMed:23353889). As part of the NSL complex indirectly involved in acetylation of nucleosomal histone H4 on several lysine residues (PubMed:20018852). O-GlcNAcylation of 'Ser-75' of EZH2 increases its stability, and facilitating the formation of H3K27me3 by the PRC2/EED-EZH2 complex (PubMed:24474760). Regulates circadian oscillation of the clock genes and glucose homeostasis in the liver. Stabilizes clock proteins ARNTL/BMAL1 and CLOCK through O-glycosylation, which prevents their ubiquitination and subsequent degradation. Promotes the CLOCK-ARNTL/BMAL1-mediated transcription of genes in the negative loop of the circadian clock such as PER1/2 and CRY1/2 (PubMed:12150998, PubMed:19451179, PubMed:20018868, PubMed:20200153, PubMed:21285374, PubMed:15361863). O-glycosylates HCFC1 and regulates its proteolytic processing and transcriptional activity (PubMed:21285374, PubMed:28584052, PubMed:28302723). Regulates mitochondrial motility in neurons by mediating glycosylation of TRAK1 (By similarity). Glycosylates HOXA1 (By similarity). O-glycosylates FNIP1 (PubMed:30699359). Promotes autophagy by mediating O-glycosylation of ATG4B (PubMed:27527864). {ECO:0000250|UniProtKB:P56558, ECO:0000250|UniProtKB:Q8CGY8, ECO:0000269|PubMed:12150998, ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:19451179, ECO:0000269|PubMed:20018852, ECO:0000269|PubMed:20018868, ECO:0000269|PubMed:20200153, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:22121020, ECO:0000269|PubMed:22923583, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:23353889, ECO:0000269|PubMed:24474760, ECO:0000269|PubMed:26678539, ECO:0000269|PubMed:27527864, ECO:0000269|PubMed:28302723, ECO:0000269|PubMed:28584052, ECO:0000269|PubMed:30699359}.; FUNCTION: [Isoform 2]: The mitochondrial isoform (mOGT) is cytotoxic and triggers apoptosis in several cell types including INS1, an insulinoma cell line. {ECO:0000269|PubMed:20824293}. | 3D-structure;Acetylation;Alternative splicing;Apoptosis;Biological rhythms;Cell membrane;Cell projection;Chromatin regulator;Cytoplasm;Direct protein sequencing;Disease variant;Glycoprotein;Glycosyltransferase;Host-virus interaction;Lipid-binding;Membrane;Mental retardation;Mitochondrion;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat;Transferase;Ubl conjugation;Ubl conjugation pathway | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:15361863, ECO:0000269|PubMed:21240259, ECO:0000269|PubMed:21285374, ECO:0000269|PubMed:23103939, ECO:0000269|PubMed:26678539}. | This gene encodes a glycosyltransferase that catalyzes the addition of a single N-acetylglucosamine in O-glycosidic linkage to serine or threonine residues. Since both phosphorylation and glycosylation compete for similar serine or threonine residues, the two processes may compete for sites, or they may alter the substrate specificity of nearby sites by steric or electrostatic effects. The protein contains multiple tetratricopeptide repeats that are required for optimal recognition of substrates. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. [provided by RefSeq, Oct 2009]. | hsa:8473; | cell projection [GO:0042995]; cytosol [GO:0005829]; histone acetyltransferase complex [GO:0000123]; mitochondrial membrane [GO:0031966]; NSL complex [GO:0044545]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; protein N-acetylglucosaminyltransferase complex [GO:0017122]; protein-containing complex [GO:0032991]; acetylglucosaminyltransferase activity [GO:0008375]; phosphatidylinositol-3,4,5-trisphosphate binding [GO:0005547]; protein N-acetylglucosaminyltransferase activity [GO:0016262]; protein O-GlcNAc transferase activity [GO:0097363]; apoptotic process [GO:0006915]; chromatin organization [GO:0006325]; circadian regulation of gene expression [GO:0032922]; histone H3-K4 trimethylation [GO:0080182]; histone H4-K16 acetylation [GO:0043984]; histone H4-K5 acetylation [GO:0043981]; histone H4-K8 acetylation [GO:0043982]; negative regulation of proteasomal ubiquitin-dependent protein catabolic process [GO:0032435]; negative regulation of protein ubiquitination [GO:0031397]; phosphatidylinositol-mediated signaling [GO:0048015]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of histone H3-K27 methylation [GO:0061087]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of proteolysis [GO:0045862]; positive regulation of transcription by RNA polymerase II [GO:0045944]; protein O-linked glycosylation [GO:0006493]; protein processing [GO:0016485]; regulation of dosage compensation by inactivation of X chromosome [GO:1900095]; regulation of gluconeogenesis [GO:0006111]; regulation of glycolytic process [GO:0006110]; regulation of insulin receptor signaling pathway [GO:0046626]; regulation of necroptotic process [GO:0060544]; regulation of Rac protein signal transduction [GO:0035020]; regulation of transcription by RNA polymerase II [GO:0006357]; response to insulin [GO:0032868]; response to nutrient [GO:0007584]; signal transduction [GO:0007165] | 11773972_We have delineated the complete genomic structure of human OGT spanning approx 43 kb of genomic DNA in Xq13.1 11846551_homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily 12136128_O-linked GlcNAc transferase participates in a hexosamine-dependent signaling pathway that is linked to insulin resistance and leptin production 14601650_a novel HLA-A0201-restricted cytotoxic T lymphocyte (CTL)-epitope (28-SLYKFSPFPL; FSP06) derived from a mutant OGT-protein 15336570_OGT can respond rapidly to heat stress through the enhancement of nucleocytoplasmic protein O-GlcNAcylation. 15561949_Staining of OGT in streptozotocin diabetic rat liver is clearly diminished, but it was substantially restored after 6 days of insulin treatment 15795231_By using a series of 4-methylumbelliferyl 2-deoxy-2-N-fluoroacetyl-beta-D-glucopyranoside substrates, Taft-like linear free energy analyses of these enzymes indicates that O-GlcNAcase uses a catalytic mechanism involving anchimeric assistance 15896326_Thus, stably transfected HeLa cells provide an abundant source of enzyme that can be used to study the structure, function, and regulation of OGT. 16105839_analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase 16966374_Overall, transcriptional inhibition is related to the integrated effect of O-GlcNAc by direct modification of critical elements of the transcriptome and indirectly through O-GlcNAc modification of the proteasome. 18174169_O-GlcNAc modification stimulated by glucose deprivation results from increased OGT and decreased O-GlcNAcase levels and that these changes affect cell metabolism, thus inactivating glycogen synthase. 18536723_The structure of an intact OGT homolog and kinetic analysis of human OGT variants reveal a contiguous superhelical groove that directs substrates to the active site. 18653473_the O-GlcNAc cycling enzymes associate with kinases and phosphatases at M phase to regulate the posttranslational status of vimentin 19073609_Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. 20068230_Data show that forced overexpression of OGT increased the inhibitory phosphorylation of CDK1 and reduced the phosphorylation of CDK1 target proteins. 20190804_OGT regulates breast cancer tumorignenesis and cancer growth through targeting FixM1. 20200153_THAP1 was found to bind HCF-1 in vitro and to associate with HCF-1 and OGT in vivo. 20206135_OGT could be a co-regulatory subunit shared by functionally distinct complexes supporting epigenetic regulation of MIP-1alpha gene promoter. 20628624_Meta-analysis of gene-disease association. (HuGE Navigator) 20805223_regulating the amount of OGT during mitosis is important in ensuring correct chromosomal segregation during mitosis. 20824293_Enhanced OGT expression efficiently triggered programmed cell death. 20845477_Observational study of gene-disease association. (HuGE Navigator) 20876116_OGT deletion in infarcted mice significantly exacerbated cardiac dysfunction. 21240259_two crystal structures of human OGT, as a binary complex with UDP (2.8 A resolution) and as a ternary complex with UDP and a peptide substrate (1.95 A). 21327254_Data identify Tau as potential substrates for the O-beta-N-acetylglucosaminyltransferase (OGT). 21567137_Decrease in MGEA5 and increase in O-GlcNAc transferease expression in higher grade tumors suggests that increased O-GlcNAc modification may be implicated in breast tumor progression and metastasis. 22275356_as prostate cancer cells alter glucose and glutamine levels, O-GlcNAc modifications and OGT levels become elevated and are required for regulation of malignant properties 22294689_a 154-amino acid region of MIBP1 was necessary for its O-GlcNAc transferase binding and O-GlcNAcylation. 22311971_Data show that the interplay between O-GlcNAc and phosphorylation on proteins and indicate that these effects can be mediated by changes in hOGT and hOGA kinetic activity. 22371499_These studies identify a molecular mechanism of GR transrepression, and highlight the function of O-GlcNAc in hormone signaling. 22384635_O-GlcNAcylation may be an important regulatory modification involved in endometrial cancer pathogenesis but the actual significance of this modification for endometrial cancer progression needs to be investigated further. 22496241_Hsp90 is involved in the regulation of OGT and O-GlcNAc modification and that Hsp90 inhibitors might be used to modulate O-GlcNAc modification and reverse its adverse effects in human diseases. 22574218_AMPK functions as a physiological suppressor of 26S proteasomes through OGT 22783592_Analysis of urinary content of MGEA5 and OGT may be useful for bladder cancer diagnostics. 22883232_O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. 23103939_we describe structural snapshots of all species along the kinetic pathway for human O-linked beta-N-acetylglucosamine transferase (O-GlcNAc transferase), an intracellular enzyme that catalyzes installation of a dynamic post-translational modification 23103942_we define how human OGT recognizes the sugar donor and acceptor peptide and uses a new catalytic mechanism of glycosyl transfer, involving the sugar donor alpha-phosphate as the catalytic base as well as an essential lysine 23152511_The human respiratory syncytial virus-induced sequestration of p38-P in IBs resulted in a substantial reduction in the accumulation of a downstream signaling substrate, MAPK-activated protein kinase 2 (MK2). 23222540_The double epigenetic modifications on both DNA and histones by TET2 and OGT coordinate together for the regulation of gene transcription. 23487789_These studies identified OGT as a promising placental biomarker of maternal stress exposure that may relate to sex-biased outcomes in neurodevelopment. 23642195_Data suggest that changes in OGT (O-linked N-acetylglucosamine transferase) and OGA (peptide O-linked N-acetylglucosamine-beta-N-acetylglucosaminidase) expression are correlated with cancer prognosis. [REVIEW] 23700425_The backbone carbonyl oxygen of Leu653 and the hydroxyl group of Thr560 in OGT contribute to the recognition of sugar moieties via hydrogen bonds. 23720054_Expression of c-MYC and OGT was tightly correlated in human prostate cancer samples. 24256146_Data suggest that with multi-substrate enzymes, such as OGT, specific inhibition can rarely be achieved with ligands that compete solely with one of the substrates; OGT is inhibited by bisubstrate UDP-oligopeptide conjugates. 24311690_study reports the tetratricopeptide-repeat domain of O-GlcNAc transferase binds the carboxyl-terminal portion of an HCF-1 proteolytic repeat such that the cleavage region lies in the glycosyltransferase active site above uridine diphosphate-GlcNAc; protein glycosylation and HCF-1 cleavage occur in the same active site 24365779_Estrogen replacement therapy and plyometric training influence muscle OGT and OGA gene expression, which may be one of the mechanisms by which HRT and PT prevent aging-related loss of muscle mass. 24394411_OGT catalyzes the O-GlcNAcylation of TET3, promotes TET3 nuclear export, and, consequently, inhibits the formation of 5-hydroxymethylcytosine catalyzed by TET3. 24580054_Endogenous OTX2 from a medulloblastoma cell line is O-GlcNAcylated at several sites. 24928395_Instead, an adipogenesis-dependent increase in O-linked beta-N-acetylglucosamine (O-GlcNAc) glycosylation of EWS was observed. 25173736_Amino acid composition of splice variants, post-translational modifications, and stable associations with regulatory proteins influence subcellular distribution/substrate specificity of OGT and OGA (O-GlcNAcase beta-N-acetylglucosaminidase). [REVIEW] 25419848_O-linked beta-N-acetylglucosamine transferase mediates O-GlcNAcylation of the SNARE protein SNAP-29 and regulates autophagy in a nutrient-dependent manner. 25568311_Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). 25663381_Hexosamine biosynthetic pathway flux is increased in idiopathic pulmonary artery hypertension and drives OGT-facilitated pulmonary artery smooth muscle cell proliferation via specific proteolysis and direct activation of host cell factor-1. 25773598_Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. 25776937_miR4235p was associated with congestive heart failure and the expression levels of proBNP; in addition, OGT was found to be a direct target of miR4235p. 26041297_Inhibition of O-Linked N-Acetylglucosamine Transferase Reduces Replication of Herpes Simplex Virus and Human Cytomegalovirus. 26237509_This work reveals that although the N-terminal TPR repeats of OGT may have roles in substrate recognition, the sequence restriction imposed by the peptide-binding site makes a substantial contribution to O-GlcNAc site specificity. 26240142_Use of OGT(C917A) enhances O-GlcNDAz production, yielding improved cross-linking of O-GlcNDAz-modified molecules 26252736_These results suggested roles of O-GlcNAcylation in modulating serine phosphorylation, as well as in regulating PKM2 activity and expression. 26305326_These results demonstrate that distinct OGT-binding sites in HCF-1 promote proteolysis, and provide novel insights into the mechanism of this unusual protease activity. 26397041_We concluded that OGT plays a key role in gastric cancer proliferation and survival, and could be a potential target for therapy. 26399441_OGT expression is increased under hypoxic conditions. 26408091_a new function of histone O-GlcNAcylation in DNA damage response 26527687_E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner. 26707622_OGT inhibited the formation of the Ecadherin/catenin complex through reducing the interaction between p120 and Ecadherin. 26807597_Data suggest RNA polymerase II (POLR2A) is extensively modified on its unique C-terminal domain (CTD) by O-GlcNAc transferase (OGT); efficient O-GlcNAcylation requires a minimum of 20 heptad CTD repeats in POLR2A and more than half of NTD of OGT. 26854602_Together, these findings suggest that induction of SNO-OGT by Ab exposure is a pathogenic mechanism to cause cellular hypo-O-GlcNAcylation by which Ab neurotoxicity is executed 27060025_The findings suggest that OGT promotes the O-GlcNAc modification of HDAC1 in the development of progression hepatocellular carcinoma. 27129214_data indicate that O-GlcNAc-transferase activity is essential for RNA pol II promoter recruitment and that pol II goes through a cycling of O-GlcNAcylation at the promoter 27131860_O-GlcNAcylation expression and its nuclear expression were associated with the carcinogenesis and progression of gastric carcinoma. 27217568_These results support a model in which OGT modifies HIRA to regulate HIRA-H3.3 complex formation and H3.3 nucleosome assembly and reveal the mechanism by which OGT functions in cellular senescence. 27231347_the O-linked N-acetylglucosamine (O-GlcNAc) processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), interact with the (A)gamma-globin promoter at the -566 GATA repressor site 27294441_Beyond its well-known role in adding beta-O-GlcNAc to serine and threonine residues of nuclear and cytoplasmic proteins, OGT also acts as a protease in the maturation of the cell cycle regulator, HCF-1, and serves as an integral member of several protein complexes, many of them linked to gene expression. (Review) 27331873_Findings indicate O-linked N-acetylglucosamine transferase (OGT) as a cellular factor involved in human papillomaviruses type 16/18 E6 and E7 expressions and cervical cancer tumorigenesis, suggesting that targeting OGT in cervical cancer may have potential therapeutic benefit. 27505673_This work uncovers that URI-regulated OGT confers c-MYC-dependent survival functions in response to glucose fluctuations. 27705803_We identified two human PRC2 complexes and two PR-DUB deubiquitination complexes, which contain the O-linked N-acetylglucosamine transferase OGT1 and several transcription factors. 27845045_OGT functions in metastatic spread of HPV E6/E7-positive HeLa cells to xenografted lungs through E6/E7, HCF-1 and CXCR4 28100784_Reducing endogenous mitochondrial OGT expression leads to alterations in mitochondrial structure and function, including Drp1-dependent mitochondrial fragmentation, reduction in mitochondrial membrane potential, and a significant loss of mitochondrial content in the absence of mitochondrial reactive oxygen species. 28115479_The results of this study showed that the OGT is essential for sensory neuron survival and target innervation. 28232487_Fatty acid synthase fine-tunes the cell's response to stress and injury by remodeling cellular O-GlcNAcylation 28302723_Thus, a single amino acid substitution in the regulatory domain (the tetratricopeptide repeat domain) of OGT, which catalyzes the O-GlcNAc post-translational modification of nuclear and cytosolic proteins, appears causal for X-linked intellectual disability. 28347804_OGT, a unique glycosyltransferase enzyme, was identified to be upregulated in non-alcoholic fatty liver disease-associated hepatocellular carcinoma tissues by transcriptome sequencing. Here, we found that OGT plays a role in cancer by promoting tumor growth and metastasis in cell models. This effect is mediated by the induction of palmitic acid. 28450392_Data suggest that O-GlcNAc transferase 1 (OGT1) specifically binds to, O-GlcNAcylates, and stabilizes nonspecific lethal protein3 (NSL3); stabilization of NSL3 by OGT1 up-regulates global acetylation levels of histone 4 at Lys-5, Lys-8, and Lys-16. 28455227_conclusion, our results demonstrated that miR24 inhibits breast cancer cells invasion by targeting OGT and reducing FOXA1 stability. These results also indicated that OGT might be a potential target for the diagnosis and therapy of breast cancer metastasis. 28584052_Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability 28625484_Nrf1 is regulated by O-GlcNAc transferase. 28663241_The authors show that O-GlcNAcylation of KEAP1 by OGT at serine 104 is required for the efficient ubiquitination and degradation of NRF2. 28742148_Tax interacts with the host OGT/OGA complex and inhibits the activity of OGT-bound OGA. 28929346_These data predict that under conditions where O-GlcNAc levels are high (breast cancer) progesterone receptor (PR) through an interaction with the modifying enzyme OGT, will exhibit increased O-GlcNAcylation and potentiated transcriptional activity. Therapeutic strategies aimed at altering cellular O-GlcNAc levels may have profound effects on PR transcriptional activity in breast cancer 29059153_High OGT expression promotes cancer lipid metabolism via SREBP-1 regulation. 29208956_Findings demonstrate a novel role of Poleta O-GlcNAcylation by OGT in translesion DNA synthesis regulation and genome stability maintenance. 29465778_The O-GlcNAc transferase OGT interacts with and post-translationally modifies the transcription factor HOXA1. 29556021_a novel ASXL1-OGT axis and raise the possibility that this axis has a tumor-suppressor role in myeloid malignancies. 29577901_LXRalpha interacts with OGT in its N-terminal domain and ligand-binding domain (LBD) in a ligand-independent fashion. 29606577_The intellectual disability L254F mutation in OGT affects activity. The L254F mutation leads to shifts up to 12 A in the OGT structure. Thermal denaturing studies reveal reduction in tetratricopeptide repeat domain stability caused by L254F. L254F OGT mutation leads to conformational changes of the tetratricopeptide repeats and reduced activity, revealing the molecular mechanisms contributing to pathogenesis. 29769320_O-GlcNAc transferase missense mutations is associated with X-linked intellectual disability. 29788742_miR-483 inhibited the expression level of OGT mRNA by direct binding to its 3'-untranslated region. Expression of miR-483 was negatively correlated with OGT in gastric cancer tissues. 29967448_Study found that levels of placental Ogt determine sex differences in fetally derived placental trophoblast transcriptome profiles associated with key developmental processes and shape genome-wide patterning of the ubiquitous epigenetic transcriptional repressive mark, H3K27me3. 30069701_O-GlcNAc transferase (OGT) is a partner of the MCM2-7 complex and O-GlcNAcylation might regulate MCM2-7 complex by regulating the chromatin loading of MCM6 and MCM7 and stabilizing MCM/MCM interactions. 30106436_High OGT expression is associated with breast cancer. 30453909_O-GlcNAcylation and the expression of O-linked-beta-N-acetylglucosamine transferase (OGT) were upregulated in bladder cancer cell lines and tissue specimens. 30543776_these findings provide a mechanistic link between the OGT-mediated glucose metabolic pathway and antiviral innate immune signaling by targeting MAVS, and expand our current understanding of importance of glucose metabolic regulation in viral infection-associated diseases. 30550897_The ligand transport mechanism in the OGT enzymatic process is described here and is a great resource for designing inhibitors based on UDP or UDP-GlcNAc. 30555541_These findings suggest that down-regulation of OGT enhances cisplatin-induced autophagy via SNAP-29, resulting in cisplatin-resistant ovarian cancer. 30587575_novel regulatory mechanism for O-GlcNAcylation during FA complex formation, which thereby affects integrin activation and integrin-mediated functions such as cell adhesion and migration 30677218_our findings indicate that OGT promotes the stem-like cell potential of hepatoma cell through O-GlcNAcylation of eIF4E 30953348_HNF1A regulates ogt transcription in a time-dependent manner and that O-GlcNAcylation of HNF1A represses ogt transcription. 14 O-GlcNAc sites on HNF1A were revealed, six of which are predominantly modified, including Ser(303/304) , Ser(471) , Ser(560) and Thr(563/564). Loss of O-GlcNAcylation at Ser(303/304) or Thr(563/564) significantly elevates ogt transcription. 31149037_OGT is essential for proliferation of prostate cancer cells. OGT activity is required for the interaction between MYC and HCF-1 and expression of MYC-regulated mitotic proteins. 31296563_Pathogenic N567K mutation leads to loss of O-GlcNAcase activity and delayed differentiation down the neuronal lineage in mouse and Drosophila model. 31373491_Aspartate Residues Far from the Active Site Drive O-GlcNAc Transferase Substrate Selection. 31373757_silencing of OGT in HT29 cells upregulates E-cadherin (a major actor of epithelial-to-mesenchymal transition) and changes its glycosylation. On the other hand, OGT silencing perturbs biosynthesis of glycosphingolipids resulting in a decrease in gangliosides and an increase in globosides. 31527085_O-GlcNAcylation does not affect sOGT activity but does affect sOGT-interacting proteins. S56A bound to and hence glycosylated more proteins in contrast to T12A and WT sOGT. 31567281_Two cases of hand myoepithelioma showing unusual clinicopathologic features and novel OGT-FOXO3 gene fusions are described. 31627256_A missense mutation in the catalytic domain of O-GlcNAc transferase links perturbations in protein O-GlcNAcylation to X-linked intellectual disability. 31628985_Disease related single point mutations alter the global dynamics of a tetratricopeptide (TPR) alpha-solenoid domain. 31847126_findings in this report are the first to describe a role for the OGT/O-GlcNAc axis in modulating VEGF expression and vascularization in Idiopathic pulmonary arterial hypertension. 31974291_O-GlcNAc Transferase Regulates Cancer Stem-like Potential of Breast Cancer Cells. 32272438_Epigenetic activation of O-linked beta-N-acetylglucosamine transferase overrides the differentiation blockage in acute leukemia. 32310828_SIRT1 regulates O-GlcNAcylation of tau through OGT. 32471715_O-GlcNAc transferase affects the signal transduction of beta1 adrenoceptor in adult rat cardiomyocytes by increasing the O-GlcNAcylation of beta1 adrenoceptor. 32663610_Glucosamine regulates hepatic lipid accumulation by sensing glucose levels or feeding states of normal and excess. 32994395_Mutual regulation between OGT and XIAP to control colon cancer cell growth and invasion. 33006972_The O-GlcNAc transferase OGT is a conserved and essential regulator of the cellular and organismal response to hypertonic stress. 33215629_Inhibition of mechanistic target of rapamycin signaling decreases levels of O-GlcNAc transferase and increases serotonin release in the human placenta. 33307156_Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. 33333092_Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. 33419956_Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. 33801653_Feedback Regulation of O-GlcNAc Transferase through Translation Control to Maintain Intracellular O-GlcNAc Homeostasis. 33909326_The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. 34046694_Dual regulation of fatty acid synthase (FASN) expression by O-GlcNAc transferase (OGT) and mTOR pathway in proliferating liver cancer cells. 34288245_Upregulation of OGT by Caveolin-1 promotes hepatocellular carcinoma cell migration and invasion. 34502531_OGT Protein Interaction Network (OGT-PIN): A Curated Database of Experimentally Identified Interaction Proteins of OGT. 34510715_P53 suppresses the progression of hepatocellular carcinoma via miR-15a by decreasing OGT expression and EZH2 stabilization. 34608265_CEMIP, a novel adaptor protein of OGT, promotes colorectal cancer metastasis through glutamine metabolic reprogramming via reciprocal regulation of beta-catenin. 34638625_Regulation of O-Linked N-Acetyl Glucosamine Transferase (OGT) through E6 Stimulation of the Ubiquitin Ligase Activity of E6AP. 34943826_Inhibition of O-GlcNAc Transferase Alters the Differentiation and Maturation Process of Human Monocyte Derived Dendritic Cells. 34948036_TET3- and OGT-Dependent Expression of Genes Involved in Epithelial-Mesenchymal Transition in Endometrial Cancer. | ENSMUSG00000034160 | Ogt | 6407.08372 | 0.6896325 | -0.5361004078 | 0.09324347 | 3.237801e+01 | 1.269147e-08 | 1.163319e-05 | No | Yes | 6025.794494 | 1291.457569 | 7995.134615 | 1675.027418 |
| ENSG00000148019 | 84131 | CEP78 | protein_coding | Q5JTW2 | FUNCTION: May be required for efficient PLK4 centrosomal localization and PLK4-induced overduplication of centrioles (PubMed:27246242). May play a role in cilium biogenesis (PubMed:27588451). {ECO:0000269|PubMed:27246242, ECO:0000269|PubMed:27588451}. | Alternative splicing;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Coiled coil;Cone-rod dystrophy;Cytoplasm;Cytoskeleton;Deafness;Phosphoprotein;Reference proteome | This gene encodes a centrosomal protein that is both required for the regulation of centrosome-related events during the cell cycle, and required for ciliogenesis. The encoded protein has an N-terminal leucine-rich repeat (LRR) domain with six consecutive LRR repeats, and a C-terminal coiled-coil domain. It interacts with the N-terminal catalytic domain of polo-like kinase 4 (PLK4) and colocalizes with PLK4 to the distal end of the centriole. Naturally occurring mutations in this gene cause defects in primary cilia that result in retinal degeneration and sensorineural hearing loss which are associated with cone-rod degeneration disease as well as Usher syndrome. Low expression of this gene is associated with poor prognosis of colorectal cancer patients. [provided by RefSeq, Mar 2017]. | hsa:84131; | centriole [GO:0005814]; centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytosol [GO:0005829]; cilium organization [GO:0044782] | 27246242_the interaction between Cep78 and the N-terminal catalytic domain of Plk4 is a new and important element in the centrosome overduplication process. 27357513_CEP78 functions as a tumor suppressor in colorectal cancer and low CEP78 expression leads to shorter survival in colorectal cancer patients. 27588451_data strongly suggest that mutations in CEP78 cause a previously undescribed clinical entity of a ciliary nature characterized by blindness and deafness but clearly distinct from Usher syndrome, a condition for which visual impairment is due to retinitis pigmentosa 27588452_truncating mutations in CEP78 result in a phenotype involving both the visual and auditory systems but different from typical Usher syndrome 27627988_Our results provide evidence that CEP78 is a novel disease-causing gene for Usher syndrome, demonstrating an additional link between ciliopathy and Usher protein network in photoreceptor cells and inner ear hair cells. 28242748_we identify Cep78 as a new player that regulates centrosome homeostasis by inhibiting the final step of the enzymatic reaction catalyzed by EDD-DYRK2-DDB1(Vpr)(BP). 30884127_Low CEP78 expression is associated with differentiated thyroid carcinoma. 31999394_Functional characterization of the first missense variant in CEP78, a founder allele associated with cone-rod dystrophy, hearing loss, and reduced male fertility. 33119552_Centrosome Protein 78 Is Overexpressed in Muscle-Invasive Bladder Cancer and Is Associated with Tumor Molecular Subtypes and Mutation Signatures. 34223797_Expanding the clinical phenotype in patients with disease causing variants associated with atypical Usher syndrome. 34259627_CEP78 functions downstream of CEP350 to control biogenesis of primary cilia by negatively regulating CP110 levels. | ENSMUSG00000041491 | Cep78 | 2151.69086 | 1.0362871 | 0.0514237528 | 0.08936563 | 3.315707e-01 | 5.647356e-01 | 8.525141e-01 | No | Yes | 2570.585295 | 416.752932 | 2396.167037 | 379.970219 | |
| ENSG00000148660 | 818 | CAMK2G | protein_coding | Q13555 | FUNCTION: Calcium/calmodulin-dependent protein kinase that functions autonomously after Ca(2+)/calmodulin-binding and autophosphorylation, and is involved in sarcoplasmic reticulum Ca(2+) transport in skeletal muscle and may function in dendritic spine and synapse formation and neuronal plasticity. In slow-twitch muscles, is involved in regulation of sarcoplasmic reticulum (SR) Ca(2+) transport and in fast-twitch muscle participates in the control of Ca(2+) release from the SR through phosphorylation of the ryanodine receptor-coupling factor triadin. In the central nervous system, it is involved in the regulation of neurite formation and arborization (PubMed:30184290). It may participate in the promotion of dendritic spine and synapse formation and maintenance of synaptic plasticity which enables long-term potentiation (LTP) and hippocampus-dependent learning. {ECO:0000269|PubMed:16690701, ECO:0000269|PubMed:30184290}. | 3D-structure;ATP-binding;Alternative splicing;Calmodulin-binding;Developmental protein;Differentiation;Kinase;Membrane;Mental retardation;Neurogenesis;Nucleotide-binding;Phosphoprotein;Reference proteome;Sarcoplasmic reticulum;Serine/threonine-protein kinase;Transferase | The product of this gene is one of the four subunits of an enzyme which belongs to the serine/threonine protein kinase family, and to the Ca(2+)/calmodulin-dependent protein kinase subfamily. Calcium signaling is crucial for several aspects of plasticity at glutamatergic synapses. In mammalian cells the enzyme is composed of four different chains: alpha, beta, gamma, and delta. The product of this gene is a gamma chain. Many alternatively spliced transcripts encoding different isoforms have been described but the full-length nature of all the variants has not been determined.[provided by RefSeq, Mar 2011]. | hsa:818; | calcium- and calmodulin-dependent protein kinase complex [GO:0005954]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; endocytic vesicle membrane [GO:0030666]; membrane [GO:0016020]; neuron projection [GO:0043005]; nucleoplasm [GO:0005654]; sarcoplasmic reticulum membrane [GO:0033017]; ATP binding [GO:0005524]; calcium-dependent protein serine/threonine phosphatase activity [GO:0004723]; calmodulin binding [GO:0005516]; calmodulin-dependent protein kinase activity [GO:0004683]; identical protein binding [GO:0042802]; protein homodimerization activity [GO:0042803]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cell differentiation [GO:0030154]; insulin secretion [GO:0030073]; nervous system development [GO:0007399]; regulation of calcium ion transport [GO:0051924]; regulation of neuron projection development [GO:0010975]; regulation of skeletal muscle adaptation [GO:0014733] | 11889801_role in cell communication 12032636_cloning, genomic structure and detection of variants in subjects with Type II diabetes 14722083_measured differences in CaMKII binding affinities for CaM play a minor role in the autophosphorylation of the enzyme, largely dictated by autophosphorylation rate for alpha, beta, gamma and delta isoforms 15775983_CaMKIIgamma is necessary to suppress MCAK depolymerase activity in vivo. 16002660_A transgenic, constitutively active, Ca2+-independent form of CaMKgamma reduces positive selection of T cells by maintaining association of SHP-2 with the T cell receptor (TCR) complex, and halting TCR signaling. 16385451_Observational study of gene-disease association. (HuGE Navigator) 17032905_Amphetamine in a cell line induces a robust increase in cytosolic Ca2+ and concomitant activation of calcium/calmodulin-dependent protein kinase II (CaMKII). 17373700_Observational study of gene-disease association. (HuGE Navigator) 17431504_Significant cross-talk between calcium and retinoic acid signaling pathways regulates the differentiation of myeloid leukemia cells. 17553505_insulin in the presence of Angiotensin II inhibits protein phosphatase-2A (PP-2A) and stimulates autonomous CaM kinase II activities and thus vascular smooth muscle migration 18434368_IGF-II/mannose-6-phosphate receptor signaling induced cell hypertrophy and atrial natriuretic peptide/BNP expression via Galphaq interaction and protein kinase C-alpha/CaMKII activation in H9c2 cardiomyoblast cells. 18483256_CaMKIIgamma is a critical regulator of multiple signaling networks regulating the proliferation of myeloid leukemia cells 19001023_In Turner yndrome, loss of voltage-dependent inactivation is an upstream initiating event for arrhythmia phenotypes that are ultimately dependent on CaMKII activation. 19204726_Observational study of gene-disease association. (HuGE Navigator) 19603549_increased RyR2-dependent Ca2+ leakage due to enhanced CaMKII activity is an important downstream effect of CaMKII in individuals susceptible to AF induction. 19671701_These data demonstrate that alphaKAP exhibits a novel interaction with SERCA2a and may serve to spatially position CaMKII isoforms at the SR and to uniquely modulate the phosphorylation of PLN. 19682070_P2X7 receptor-triggered signalling pathways that regulate neurite formation in neuroblastoma cells. 19840793_Study further supports self-aggregation of CaMKII holoenzymes as the underlying mechanism that involves inter-holoenzyme T286-region/T-site interaction. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20060891_The interaction between CaMKII and its binding proteins was altered by the phosphorylation state of both the CaMKII and the partner. 20139089_CaMKII is a molecular link translating intracellular calcium changes, which are intrinsically associated with glioma migration, to changes in ClC-3 conductance required for cell movement 20149311_Our results suggest a first observation that CaMKII regulates TRAIL-mediated apoptosis of fibroblast-like synovial cells through Akt, standing an upstream of caspase-8-dependent cascades. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20851109_These results indicated that PP6 and CaMKII regulated apoptosis by controlling the expression level of p27. 21063097_Ca(2+)/calmodulin-dependent kinase II participate in control of cell cycle progression and survival of irradiated CML cells. 21513986_study shows CaMKII is recruited to the immunological synapse where it interacts with and phosphorylates Bcl10; propose a mechanism whereby Ca(2+) signals can be integrated at the immunological synapse through CaMKII-dependent phosphorylation of Bcl10 21983898_increases in Ca(2+) lead to CaMKII activation and subsequent Lck-dependent p66Shc phosphorylation on Serine 36. This event causes both mitochondrial dysfunction and impaired Ca(2+) homeostasis, which synergize in promoting Jurkat T-cell apoptosis. 22049206_study demonstrates that premitotic condensation involves the activation of ClC-3 by Ca(2+)/calmodulin-dependent protein kinase II in glioma cells 22371496_CaMKII binding to and phosphorylation of the NHE3 C terminus are parts of the physiologic regulation of NHE3 that occurs in fibroblasts as well as in the brush border of an intestinal Na(+)-absorptive cell. 22592532_CaMKII was also necessary for the phosphorylation of Raf-1 at S338 by serum, fibronectin and Ras. 22612808_we review the cellular colocalization of CaMKII isoforms with special regard to the cell-type specificity of isoform expression in brain--REVIEW 22615928_CaMKII T286A showed a mildly but significantly reduced rate of Ca(2+)/CaM-stimulated phosphorylation for two different peptide substrates (to ~75-84% of wild type). 22952977_These findings revealed a fundamental role of CaMKII in the enteric nervous system. 23008441_The CaMKII phosphorylation motif in the Nav1.5 DI-DII cytoplasmic loop is a critical point for proarrhythmic changes. 23074277_Data indicate that CaMKII gamma as a specific and critical target of berbamine for its antileukemia activity. 23463379_results show that CAMKII and calmodulin contribute to IKK complex activation and thus to the induction of NF-kappaB in response to H. pylori infection 23502535_Activated CaMK-II interacts with the C-terminal domain of diacylglycerol lipase-alpha (DGLalpha) and inhibits DGLalpha activity. 23516528_the chronic gain-of-function defect in RyR2 due to CaMKII hyperphosphorylation is a novel mechanism that contributes to pathogenesis of type 2 diabetes 23543616_CaMKII overexpression in mushroom body neurons increases activity dependent calcium responses. 23960096_Our data suggest that berbamine and its derivatives are promising agents to suppress liver cancer growth by targeting CAMKII 24100685_it can be concluded that CaMKII regulates the activity of ASIC1, which is associated with the ability of GBM cells to migrate. 24206096_CAMKIIgamma, HSP70 and HSP90 transcripts are differentially expressed in chronic myeloid leukemia cells from patients with resistant mutated disease. 24269630_When inhibiting CaMKII, B-cell activating factor (BAFF) is attenuated and mediates protein phosphatase (PP)2A-Erk1/2 signaling and B-cell proliferation. 24634301_CaMKII-dependent microtube polymerization may be responsible for the enhanced uptake of PEI/ON complexes in A549 cells under oxidative stress conditions. 25446257_Inhibition of CaMKII activity results in an upregulation of CaMKIV mRNA and protein in leukemia cell lines. 25577293_Dysfunction in CaMKII-based signaling has been linked with a host of cardiovascular phenotypes including heart failure and arrhythmia, and CaMKII levels are elevated in human and animal disease models of heart disease. 25965829_High CaMKIIgamma expression is associated with lung cancer. 26803057_Demonstrate that calcium/CaMKIIgamma/AKT signaling can regulate apoptosis and autophagy simultaneously in colorectal cancer cells. 27624155_These findings indicate that the CaMKII-mediated GluA1 phosphorylation of S567 and S831 is critical for P2X2-mediated AMPAR internalization and ATP-driven synaptic depression. 27819676_novel mechanism by which CAMK2gamma antagonizes mTORC1 activation during hepatocarcinogenesis 27875595_CKIalpha-mediated NS5A S235 phosphorylation is critical for HCV replication. CaMKII gamma and delta may have negative roles in the HCV life cycle. 27964993_Laminin is instructive and CaMKII is non-permissive for the formation of complex aggregates of acetylcholine receptors on myotubes in culture. 28007458_oxidative stress activated the TRPM2-CaMKII cascade to further induce intracellular ROS production, which led to mitochondria fragmentation and loss of mitochondrial membrane potential 28319059_A new molecular mechanism mediated by CAMK2gamma in intestinal epithelial cells during colitis-associated cancer (CAC) development, thereby providing a potential new therapeutic target for CAC. 29210177_The data further showed that mitochondrial fission significantly promoted the reprogramming of focal-adhesion dynamics and lamellipodia formation in hepatocellular carcinoma cells mainly by activating typical Ca(2+) /CaMKII/ERK/FAK pathway. 29391954_The data suggest T287 autophosphorylation regulates substrate gating, an intrinsic property of the catalytic domain, which is amplified within the multivalent architecture of the calcium/calmodulin-dependent protein kinase II holoenzyme. 29535221_The authors identify active CaMKII as a novel connection between organelle beta-oxidation and acetyl-CoA transport with cell survival, migration, and prostate cancer metastasis 29934532_point mutation R292P sufficient to disrupt gene expression and spatial learning 30142967_The effect of Ca(2+), domain-specificity, and CaMKII on CaM binding to NaV1.1 has been reported. 30184290_Results reveal an indispensable function of CAMK2G in neurodevelopment and indicate that the CAMK2G p.Arg292Pro de novo mutation acts as a pathogenic gain-of-function mutation causing severe intellectual disability. CAMK2G p.Arg292Pro has constitutive activity toward cytosolic targets, rather than impaired targeting to the nucleus. 30244168_BAFF promotes B-cell proliferation and survival by inhibiting autophagy. BAFF inhibits autophagy by activating Ca2+-CaMKII-dependent Akt/mTOR signaling pathway in B cells. 30418153_Subunit exchange enhances information retention by CaMKII in dendritic spines. 31221819_CaMKII/proteasome/cytosolic calcium/cathepsin B axis was present in tryspin activation induced by nicardipine. 31461344_Ca2+-activated Cl- channels encoded by the Tmem16a gene are regulated by calmodulin-dependent protein kinase II (CaMKII) and protein phosphatases 1 (PP1) and 2A (PP2A). 31481791_mark the CaMKII-delta9-UBE2T-DNA damage pathway as an important therapeutic target for cardiomyopathy and heart failure 31857698_The TRPV4-AKT axis promotes oral squamous cell carcinoma cell proliferation via CaMKII activation. 32068116_CaMKII and GLUT1 in heart failure and the role of gliflozins. 32658867_CAMKIIgamma is a targetable driver of multiple myeloma through CaMKIIgamma/ Stat3 axis. 33369192_CaMKIIgamma regulates the viability and self-renewal of acute myeloid leukaemia stem-like cells by the Alox5/NF-kappaB pathway. 33454324_CaMK II/Ca2+ dependent endoplasmic reticulum stress mediates apoptosis of hepatic stellate cells stimulated by transforming growth factor beta 1. 33568460_CaMKII Phosphorylation Regulates Synaptic Enrichment of Shank3. 34088288_Matrine suppresses cell growth of diffuse large B-cell lymphoma via inhibiting CaMKIIgamma/c-Myc/CDK6 signaling pathway. 34301850_A physiologic rise in cytoplasmic calcium ion signal increases pannexin1 channel activity via a C-terminus phosphorylation by CaMKII. | ENSMUSG00000021820 | Camk2g | 1137.93871 | 0.9862404 | -0.0199887875 | 0.10310455 | 3.755267e-02 | 8.463441e-01 | 9.591151e-01 | No | Yes | 1385.879213 | 79.114309 | 1368.638646 | 76.226041 | |
| ENSG00000148737 | 6934 | TCF7L2 | protein_coding | Q9NQB0 | FUNCTION: Participates in the Wnt signaling pathway and modulates MYC expression by binding to its promoter in a sequence-specific manner. Acts as repressor in the absence of CTNNB1, and as activator in its presence. Activates transcription from promoters with several copies of the Tcf motif 5'-CCTTTGATC-3' in the presence of CTNNB1. TLE1, TLE2, TLE3 and TLE4 repress transactivation mediated by TCF7L2/TCF4 and CTNNB1. Expression of dominant-negative mutants results in cell-cycle arrest in G1. Necessary for the maintenance of the epithelial stem-cell compartment of the small intestine. {ECO:0000269|PubMed:12408868, ECO:0000269|PubMed:12727872, ECO:0000269|PubMed:19443654, ECO:0000269|PubMed:22699938, ECO:0000269|PubMed:9727977}. | 3D-structure;Activator;Alternative splicing;DNA-binding;Diabetes mellitus;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation;Wnt signaling pathway | This gene encodes a high mobility group (HMG) box-containing transcription factor that plays a key role in the Wnt signaling pathway. The protein has been implicated in blood glucose homeostasis. Genetic variants of this gene are associated with increased risk of type 2 diabetes. Several transcript variants encoding multiple different isoforms have been found for this gene.[provided by RefSeq, Oct 2010]. | hsa:6934; | beta-catenin-TCF complex [GO:1990907]; beta-catenin-TCF7L2 complex [GO:0070369]; catenin-TCF7L2 complex [GO:0071664]; chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; PML body [GO:0016605]; protein-DNA complex [GO:0032993]; armadillo repeat domain binding [GO:0070016]; beta-catenin binding [GO:0008013]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; gamma-catenin binding [GO:0045295]; nuclear receptor binding [GO:0016922]; protein kinase binding [GO:0019901]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; sequence-specific DNA binding [GO:0043565]; transcription cis-regulatory region binding [GO:0000976]; blood vessel development [GO:0001568]; canonical Wnt signaling pathway [GO:0060070]; canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition [GO:0044334]; fat cell differentiation [GO:0045444]; glucose homeostasis [GO:0042593]; maintenance of DNA repeat elements [GO:0043570]; myoblast fate commitment [GO:0048625]; negative regulation of canonical Wnt signaling pathway [GO:0090090]; negative regulation of DNA-binding transcription factor activity [GO:0043433]; negative regulation of extrinsic apoptotic signaling pathway [GO:2001237]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of type B pancreatic cell apoptotic process [GO:2000675]; pancreas development [GO:0031016]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of heparan sulfate proteoglycan biosynthetic process [GO:0010909]; positive regulation of insulin secretion [GO:0032024]; positive regulation of protein binding [GO:0032092]; positive regulation of protein export from nucleus [GO:0046827]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of hormone metabolic process [GO:0032350]; regulation of smooth muscle cell proliferation [GO:0048660]; regulation of transcription by RNA polymerase II [GO:0006357]; response to glucose [GO:0009749] | 11713475_tcf4 can specifically recognize beta-catenin using alternative conformations 11713476_crystal structure of a human Tcf4-beta-catenin complex; comparison with recent structures of beta-catenin in complex with Xenopus Tcf3 (XTcf3) and mammalian E-cadherin 11931652_Promoter characterization of the novel human matrix metalloproteinase-26 gene: regulation by the T-cell factor-4 implies specific expression of the gene in cancer cells of epithelial origin. 11940574_Activation of AXIN2 expression by beta-catenin-T cell factor 12086873_ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation 12368361_role of TCF-4 upon transcription of the human immunodeficiency virus type 1 (HIV-1) promoter in human astrocytic cells 12378619_The high expression level of hTcf-4 in HCC, especially with metastasis, suggests that the over-expression of hTcf-4 gene may be closely associated with development and progression of HCC, but the mutation of this gene plays a less important role 12408868_disruption of beta-catenin/TCF-4 activity in CRC cells induces a rapid G1 arrest and blocks a genetic program that is physiologically active in the proliferative compartment of colon crypts. 12446687_the C terminus of TCF4E cooperates with beta-catenin and p300 to form a specialized transcription factor complex that specifically supports the activation of the Cdx1 promoter 12799378_there is a direct interaction between the androgen receptor DNA binding domain (DBD) and Tcf4. 12861022_TCF-4N inhibits coactivation by beta-catenin of TCF/LEF transcription factors and potentiates the coactivation by beta-catenin of other transcription factors, such as SF-1 and C/EBPalpha 15040893_TCF4 could be an effective therapeutic target for suppressing the growth of hepatocellular cancers. 15514942_TCF4-binding element was identified in PTTG promoter region in eesophageal squamous cell sscarscinomsa. 15591320_identify selective beta-catenin binding hot spots of Tcf4, E-cadherin, and APC 15670774_Together, we suggest that quercetin is an excellent inhibitor of beta-catenin/Tcf signaling in SW480 cell lines, and the reduced beta-catenin/Tcf transcriptional activity is due to the decreased nuclear beta-catenin and Tcf-4 proteins. 15806138_TCF4 expression mediated by beta-catenin/p300 may be important for initial steps during trans-differentiation of endometrial carcinoma cells. 15853773_The regulation of GLCE expression by 2 cis-acting elements of the beta-catenin-TCF4 complex located in the enhancer region of the promoter are reported. 15905022_characterization of the TCF4 with microsatellite instability (MSI) in colon cancer and leukemia cell lines; results delineate a novel role for MSI+TCF4 in leukemia and colon cancer progression 16007074_the phosphorylation-dependent interaction between c-Jun and TCF4 regulates intestinal tumorigenesis by integrating JNK and APC/beta-catenin, two distinct pathways activated by WNT signalling 16204248_TIS7, a negative regulator of transcriptional activity, represses expression of OPN and beta-catenin/Tcf-4 target genes 16291872_The positive inter-regulation between beta-cat/Tcf-4 signaling and ET-1 signaling potentiates proliferation and survival of prostate cancer (CaP) cells, thereby representing a novel mechanism that contributes to CaP progression. 16311123_results suggest an established Wnt signaling pathway in most gastric cancers, a close correlation of beta-catenin/TCF4-mediated signaling with tumor dissemination, and the unlikelihood of a direct effect of activated Wnt signaling on CD44 expression 16385451_Observational study of gene-disease association. (HuGE Navigator) 16415884_Observational study of gene-disease association. (HuGE Navigator) 16415884_Suggestive linkage of type 2 diabetes mellitus to TCFL2 protein on chromosome 10q. 16532032_Represses Wnt signaling in breast tissue, and its downregulation contributes to the activation of Wnt signaling. 16547505_A role is suggested for TCF7L2 frameshift mutation during MSI-H colorectal tumor progression, by regulating the relative proportion of the different TCF7L2 isoforms. 16569639_Daxx reduced DNA binding activity of Tcf4 and repressed Tcf4 transcriptional activity. 16690926_findings suggest that activity at the HIV-1 promoter is influenced by phosphorylation of Sp1, which is affected by Tat and DNA-PK; interactions among TCF-4, Sp1 and/or Tat may determine the level of viral gene transcription in astrocytic cells 16724116_evidence provided that HIC1 antagonizes the TCF/beta-catenin-mediated transcription in Wnt-stimulated cells; this appears to be due to the ability of HIC1 to associate with TCF-4 and to recruit TCF-4 and beta-catenin to the HIC1 bodies 16855264_Common variants (rs12255372 and rs7903146) in TCF7L2 seem to be associated with an increased risk of diabetes among persons with impaired glucose tolerance 16855264_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16936215_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 16936216_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 16936217_Observational study of gene-disease association. (HuGE Navigator) 16936218_Observational study of gene-disease association. (HuGE Navigator) 16936218_These data provide replicating evidence that variants in TCF7L2 increase the risk for type 2 diabetes and novel evidence that the variants likely influence both insulin secretion and insulin sensitivity. 17003358_Observational study of gene-disease association. (HuGE Navigator) 17003358_These results suggest that TCF7L2 variants may act through insulin secretion to increase the risk of type 2 diabetes. 17003360_Observational study of gene-disease association. (HuGE Navigator) 17003360_These data provide evidence that TCF7L2 is a major determinant of type 2 diabetes risk in European populations and suggests that this transcription factor plays a key role in glucose homeostasis. 17020404_Combining information from several known common risk polymorphisms allows the identification of population subgroups with markedly differing risks of developing type 2 diabetes compared to those obtained using single polymorphisms. 17020404_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17031610_Observational study of gene-disease association. (HuGE Navigator) 17031610_Variants of the TCF7L2 gene contribute to the risk of type 2 diabetes. The population-attributable risk from this factor in the Dutch type 2 diabetes population is 10%. 17052462_crystallographic analysis of how beta-catenin, BCL9, BCL9-2 and Tcf4 interact 17063324_Although TCF7L2 is a major gene in type 2 diabetes, there is no evidence for association between this gene and type 1 diabetes. 17065361_Observational study of gene-disease association. (HuGE Navigator) 17065361_TCF7L2 T at-risk allele variation predicts hyperglycemia incidence in a general French population, possibly through a deleterious effect on insulin secretion. 17091236_A high mobility group box-containing TCF7L2 leads to diabetes risk. 17093940_No major contribution of TCF7L2 sequence variants to maturity onset of diabetes of the young (MODY) or neonatal diabetes mellitus was found in this study in French white subjects. 17093941_Observational study of gene-disease association. (HuGE Navigator) 17093941_TCF7L2 is an important gene for determining susceptibility to type 2 diabetes mellitus and it transgresses the boundaries of ethnicity. 17109766_Observational study of gene-disease association. (HuGE Navigator) 17109766_results suggest a possible influence of TCF7L2 rs12255372 on the risk of familial breast cancer 17130514_Observational study of gene-disease association. (HuGE Navigator) 17130514_TCF7L2 gene is an important factor regulating insulin secretion, which could explain its association with type 2 diabetes. 17143297_AXIN1, AXIN2 and TCF7L2 may have roles in development of colorectal carcinomas [review] 17181866_Observational study of gene-disease association. (HuGE Navigator) 17181866_polymorphism is associated with lower insulin levels, smaller waist circumference, and lower risk lipid profiles in the general elderly population. 17226113_Observational study of gene-disease association. (HuGE Navigator) 17226113_TCF7L2 variants increase type 2 diabetes risk and may affect pancreatic beta cell function. 17245407_Observational study of gene-disease association. (HuGE Navigator) 17245407_Several single nucleotide polymorphisms are associated with type 2 diabetes. 17245589_Observational study of gene-disease association. (HuGE Navigator) 17245589_TCF7L2 is a common susceptibility gene for type 2 diabetes in the Japanese population 17259383_Observational study of gene-disease association. (HuGE Navigator) 17259383_data provide evidence that variation in the TCF7L2 genomic region may affect risk for type 2 diabetes in Mexican Americans, but the attributable risk may be lower than in Caucasian populations 17293876_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17311858_Observational study of gene-disease association. (HuGE Navigator) 17317761_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 17317761_Variation in TCF7L2 is associated with gestational diabetes and interacts with adiposity to alter insulin secretion in Mexican Americans. 17340123_May be a strong candidate for conferring susceptibility to type 2 diabetes across different ethnicities in Japan. 17340123_Observational study of gene-disease association. (HuGE Navigator) 17342473_Observational study of gene-disease association. (HuGE Navigator) 17342473_Variant is associated with an increased risk of gestational diabetes mellitus in Scandinavian women. 17351281_combined effect of obesity and genotype in predicting type 2 diabetes risk in a sample of French Canadian cardiac patients. in predicting type 2 diabetes risk in a sample of French Canadian cardiac patients. 17392368_Transcription factor 4 (TCF-4), the downstream effector of Wnt signaling, is implicated in repressing HIV replication in astrocytes. 17416797_Observational study of gene-disease association. (HuGE Navigator) 17416797_findings suggest that the TCF7L2 risk allele may predispose to type 2 diabetes by impairing beta-cell proinsulin processing 17429603_Observational study of gene-disease association. (HuGE Navigator) 17429603_Variants of TCF7L2 may be associated with increased disease severity and therapeutic failure in diabetes. 17437080_A cariant of TCF7L2 is associated with incident type 2 diabetes in a separate population-based cross-sectional study. 17437080_Observational study of gene-disease association. (HuGE Navigator) 17463248_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17470138_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17470138_evaluated the association of the three TCF7L2 polymorphisms with NIDDM by using the program admixmap to fit a logistic regression model incorporating individual ancestry, sex, age, body mass index and education 17476472_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17503332_Observational study of gene-disease association. (HuGE Navigator) 17503332_Study assessed role of TCF4 in birth weight. 17519421_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17519421_TCF7L2 variants influence therapeutic response to sulfonylureas but not metformin 17540954_an increased risk of type 2 diabetes is associated with TCF7L2 rs7903146 genotype. 17563454_The discovery of TCF7L2 as a diabetes gene illustrates that novel true diabetes genes can be found, their association with type 2 diabetes replicated and their effect incorporated into risk prediction models (Review) 17579206_Observational study of gene-disease association. (HuGE Navigator) 17579206_TCF7L2 genetic polymorphisms are major determinants for risk of type 2 diabetes in the Chinese population. 17579832_Observational study of gene-disease association. (HuGE Navigator) 17579832_Results confirmed TCF7L2 as a risk factor in a population of European descent, where it reduced glucose tolerance and insulin sensitivity, but not insulin secretion. 17593304_Observational study of gene-disease association. (HuGE Navigator) 17593304_TCF7L2 rs7903146 variant neither increases the risk for SGA nor modulates birth weight and young adulthood glucose homeostasis in French Caucasian subjects born with SGA 17601994_Observational study of gene-disease association. (HuGE Navigator) 17601994_Variations in the TCF7L2 gene significantly contribute to diabetes susceptibility in African-American populations. 17609304_Observational study of gene-disease association. (HuGE Navigator) 17609304_Variations at TCF7L2 contribute to Type 2 diabetes. 17618413_Observational study of gene-disease association. (HuGE Navigator) 17618413_Odds ratio for single nucleotide polymorphisms associated with plasma proinsulin, beta cell dysfunction and increased risk of type 2 diabettes. 17653210_Observational study and genome-wide association study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17659738_Novel mutations in exon 4 of hTCF-4 gene were revealed in this study, which might be of importance in the pathogenesis of sporadic rectal cancer patients with high frequency microsatellite instability. 17661009_Variants of TCF7L2 specifically impair GLP-1-induced insulin secretion. 17665514_Common variants in the TCF7L2 gene and its predisposition to type 2 diabetes in UK European Whites, Indian Asians and Afro-Caribbean men and women are reported. 17665514_Observational study of gene-disease association. (HuGE Navigator) 17668382_Genome-wide association study of gene-disease association. (HuGE Navigator) 17671651_Observational study of gene-disease association. (HuGE Navigator) 17671651_the increased risk of type 2 diabetes conferred by variants in TCF7L2 involves the enteroinsular axis, enhanced expression of the gene in islets, and impaired insulin secretion. 17683561_Observational study of gene-disease association. (HuGE Navigator) 17683561_TCF7L2 does not participate in the etiology of Type 1 diabetes 17683561_The genetic susceptibility from the TCF7L2 gene variation is a unique mechanism of type 2 diabetes (T2D), and is not shared by type 1 diabetes (T1D). 17697858_Observational study of gene-disease association. (HuGE Navigator) 17697858_The rs12255372(G/T) and rs7903146(C/T) polymorphisms of TCF7L2 gene confer susceptibility to type 2 diabetes mellitus in Asian Indians. 17709525_A reduced expression in ileal Crohn's disease of the Wnt-signaling pathway transcription factor Tcf-4, a known regulator of Paneth cell differentiation and alpha-defensin expression, was reported. 17725629_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17725629_common variation in the TCF7L2 gene contributes to Type 2 diabetes risk in UK patients recruited in general practice, but the risk allele frequency may be lower than that in subjects enriched for genetic effects 17768662_Overexpression of TCF-4 is associated with the development of lung cancer 17805508_Establishing whether variation in TCF7L2 also influences the development of polycystic ovary syndrome and type 2 diabetes. 17805508_Observational study of gene-disease association. (HuGE Navigator) 17875931_Data indicate that Sox4 and 17 can act as both antagonists and agonists of beta-catenin/TCF activity, and this mechanism may regulate Wnt signaling responses in many developmental and disease contexts. 17901222_TCF7L2 (transcription factor 7-like 2) has been identified as a gene for type 2 diabetes. 17909099_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 17909099_Variation within TCF7L2 does not confer major risk for type 2 diabetes among the Pima Indian population. 17934151_Observational study of gene-disease association. (HuGE Navigator) 17934151_TCF7L2 rs7903146 genetic variation is associated with an increased risk of post transplantation diabetets mellitus in renal allograft recipients. 17971425_Observational study of gene-disease association. (HuGE Navigator) 17971425_TCF7L2 variants could play a role in the pathogenesis of type 2 diabetes mellitus in the Hispanic American population through a mchanism involving insulin secretion. 17972059_TCF7L2 is associated with high serum triacylglycerol and differentially expressed in adipose tissue in families with familial combined hyperlipidaemia 17977958_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17983804_Topo IIalpha interacts with beta-catenin/T-cell factor-4 as a novel transcriptional co-activator in colorectal neoplasms. 18039847_MTG family members associate specifically with TCF4. Coexpression of beta-catenin disrupted the association between these corepressors and TCF4. 18048388_Ectopic expression of Dkk3 in lung cancer cells with Dkk3 hypermethylation induced apoptosis and inhibited TCF-4 activity 18072015_DG10S478 variant seems to have no influence on manifestation of diabetes and the development of microvascular complications. 18072015_Observational study of gene-disease association. (HuGE Navigator) 18097733_Single nucleotide polymorphisms in TCF7L2 were associated with type 2 diabetes 18166673_A precise and reproducible electrophoretic technique is used to make an allelic assignment from genomic DNA of the polymorphism in microsatellite DG10S478 of TCF7L2. 18203713_myostatin enhanced nuclear translocation of beta-catenin and formation of the Smad3-beta-catenin-TCF4 complex, together with the altered expression of a number of Wnt/beta-catenin pathway genes in hMSCs 18239663_Observational study of gene-disease association. (HuGE Navigator) 18239663_TCF7L2 is not a risk factor for obesity in European populations, but its effect on type 2 diabetes risk is modulated by obesity. 18248681_Observational study of genotype prevalence. (HuGE Navigator) 18264689_Diabetes-associated variants in TCF7L2 and CDKAL1 impair insulin secretion and conversion of proinsulin to insulin. 18264689_Observational study of gene-disease association. (HuGE Navigator) 18268006_TCF4-binding regions significantly correlate with Wnt-responsive gene expression profiles derived from primary human adenomas 18268068_Observational study of gene-disease association. (HuGE Navigator) 18268068_Subjects who were initially cancer-free and carrying certain genetic variants of TCF7L2 have an increased risk of colon cancer. 18282631_Observational study of gene-disease association. (HuGE Navigator) 18282631_TCF7L2 variants are associated with increased risk for diabetes mellitus in Emirati subjects. 18288125_Observational study of gene-disease association. (HuGE Navigator) 18288125_TCF7L2 rs7903146 T allele is present in obese hypertensive patients as much as in the general population. 18291022_Observational study of gene-disease association. (HuGE Navigator) 18291022_TCF7L2 is an important genetic risk factor for the development of type 2 diabetes in multiple ethnic groups 18302196_Observational study of gene-disease association. (HuGE Navigator) 18302196_The TCF7L2 gene may alter risk of developing more aggressive prostate cancer. 18310307_Observational study of gene-disease association. (HuGE Navigator) 18342627_TCF7L2 mRNA levels in adipocytes are decreased by insulin and seem to increase in insulin resistant subjects and in HapA carriers. 18347071_We confirm that c-Jun functions in canonical Wnt signaling and show that c-Jun functions as a scaffold in the beta-catenin-TCFs transcription complex bridging Dvl to TCF. 18397358_Observational study of gene-disease association. (HuGE Navigator) 18397358_TCF7L2 gene is a major risk factor for development of T2DM in Khatri Sikhs from North India. 18398040_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18398040_These data suggest that colon cancer risk associated with the rs7903146 TCF7L2 polymorphism is modified by use of aspirin/NSAIDs 18437354_Increased health risk associated with an rs7903146 genotype is specific to mortality in diabetes and diabetic angiopathies. 18437354_Observational study of gene-disease association. (HuGE Navigator) 18439914_Determination of role nuclear pore complex in regulating TCF4/beta cateninin mediated Wnt signaling. 18443202_Data show that one TCF7L2 SNP (rs7903146) showed compelling evidence of association with type 2 diabetes in African Americans. 18443202_Observational study of gene-disease association. (HuGE Navigator) 18445358_association of TCF7L2 with type 2 diabetes [review] 18461161_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18469204_Data confirmed the associations of single nucleotide polymorphisms in TCF7L2 with risk for type 2 diabetes in Asians. 18469204_Observational study of gene-disease association. (HuGE Navigator) 18478343_Observational study of gene-disease association. (HuGE Navigator) 18478343_there was suggestive evidence for an inverse association associated of colorectal cancer and ademoma with homozygosity for the minor allele of RS12255372 (TCF7L2 TT) and conditional and covariate adjusted risk 0for heterogeneity for in women and men 18493736_Observational study of gene-disease association. (HuGE Navigator) 18493736_we did not find any mutation in the coding sequence of TCF7L2 that confers a genetic risk for type 2 diabetes in a Chinese population 18498634_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18516622_Observational study of gene-disease association. (HuGE Navigator) 18519685_results indicate that Tcf-4 maintains low levels of claudin-7 at the bottom of colonic crypts, acting via Sox-9 18541996_genetic variants in TCF7L2 confer a strong risk of future type 2 diabetes possibly mediated by altering expression of TCF7L2 in pancreatic islets [review] 18546086_Observational study of gene-disease association. (HuGE Navigator) 18546086_The higher homeostasis model assessment insulin resistance index (HOMA-B%) index in TT-homozygotes indicates TCF7L2 to be a susceptibility gene for the development of impaired glucose tolerance in obese children. 18555673_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 18591388_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18597214_Gene variants of CDKAL1, PPARG, IGF2BP2, HHEX, TCF7L2, and FTO predispose to type 2 diabetes in the German KORA 500 K study population. 18597214_Observational study of gene-disease association. (HuGE Navigator) 18598350_Observational study of gene-disease association. (HuGE Navigator) 18598350_TCF7L2 SNPs revealed a significant association with type 2 diabetes 18599616_Summarize recent findings demonstrating the association between TCF7L2 polymorphisms and the risk of type 2 diabetes, outline experimental evidence of the potential function of TCF7L2 in pancreatic and intestinal endocrine cells. Review. 18611970_Observational study of gene-disease association. (HuGE Navigator) 18611970_The primary defect of rs7903146 T-allele carriers is impairment of insulin secretion rather than a defect in insulin action in peripheral tissues. 18621708_the presumed cancer-promoting gene TCF7L2 functions instead as a transcriptional repressor that restricts colorectal cancer (CRC) cell growth. 18650481_variants in the TCF7L2 gene are associated with reduced kidney function and CKD progression 18655717_Our study is consistent with weak or no association of type 2 diabetes in Arabs with the two TCF7L2 variants 18689695_Observational study of gene-disease association. (HuGE Navigator) 18694974_Study show that polymorphisms in TCF7L2 were associated with type 2 diabetes risk in the studied population. 18702948_TCF7L2 mRNA expression is fat-depot specific but does not seem to provide the mechanistic link explaining genetic association with type 2 diabetes mellitus. 18706099_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18706099_co-existence of TCF7L2 variants and the SPINK1 and CTSB mutations, that predict susceptibility to exocrine damage, may interact to determine the onset of diabetes in TCP patients 18712344_Evidence that the TCF7L2 gene is a major determinant of type 2 diabetes risk in Spain, as in other southern Eutopean populations. 18712344_Observational study of gene-disease association. (HuGE Navigator) 18719881_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18755497_Frameshift mutations of Wnt pathway genes AXIN2 and TCF4 in gastric carcinomas with high microsatellite instability are reported. 18762805_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18762805_Results found a negative allele-dosage effect of the T allele of rs7903146 in the TCF7L2 gene on the changes in the HOMA-IR, the insulin sensitivity check index QUICKI and insulin secretion index HOMA-B% during a lifestyle intervention. 18799618_Snail and Slug promote formation of beta-catenin-T-cell factor (TCF)-4 transcription complexes that bind to the promoter of the TGF-beta3 gene to increase its transcription 18806947_In morbidly obese nondiabetic patients, there was a positive correlation between TCF7L2 expression and BMI (R(2)=0.21). 18823720_Observational study of gene-disease association. (HuGE Navigator) 18839133_Common variants in the TCF7L2 gene help to differentiate young but not middle-aged glutamic acid decarboxylase antibodies(GADA)-positive and GADA-negative diabetic patients 18839133_Observational study of gene-disease association. (HuGE Navigator) 18853134_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18931037_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18931037_Sudy provides the first significant evidence of association between the TCF7L2 rs7903146 polymorphism and type 2 diabetes risk in a large African American population. 18958766_Observational study of gene-disease association. (HuGE Navigator) 18958766_The rs7903146 variant of the TCF7L2 gene might influence PCOS predisposition, while no association is observed between the E23K variant of KCNJ11 and susceptibility to PCOS and related traits. 18972257_Observational study of gene-disease association. (HuGE Navigator) 18972257_Risk alleles of the TCF7L2 gene showed increased risk of diabetes even when controlled for traditional diabetes risk factors 18984664_Observational study of gene-disease association. (HuGE Navigator) 18992165_Increased expression in colorectal adenomas than in the adjacent normal epithelia 18992263_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18996470_An assay using unlabeled probes and the LightCycler or Rotor-Gene instruments was developed for genotyping of PPARG, PPARGC1A and transcription factor 7-like 2 (TCF7L2) polymorphisms. 18996470_Observational study of genetic testing. (HuGE Navigator) 19002430_Observational study of gene-disease association. (HuGE Navigator) 19012045_Common coding variant in the TCF7L2 gene is associated with type 2 diabetes. 19012045_Observational study of gene-disease association. (HuGE Navigator) 19020323_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19020324_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19033397_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19033397_Type 2 diabetes susceptibility of TCF7L2 was confirmed in Japanese. 19050058_Observational study of gene-disease association. (HuGE Navigator) 19050058_TCF7L2 type 2 diabetes susceptibility alleles are associated with islet autoantibody-negative but not autoantibody-positive new onset diabetes in young patients. 19053027_Located on chromosome 10 and suscptibility of polymorphisms are related to type 2 diabetes. 19053027_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19055834_Observational study of gene-disease association and genetic testing. (HuGE Navigator) 19055834_TCF7L2 rs7903146 T allele is associated with a 1.57 increased risk for type 2 diabetes in a Brazilian cohort of patients with known coronary heart disease 19056611_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19057525_Genes implied in human type 2 diabetes development, TCF7L2, WFS1, FTO, SLC30A8, and GCKR, were mapped on Sus scrofa chromosomes 14, 8, 6, 4, and 3, respectively. Only TCF7L2 was significantly associated with five fat traits in pigs. 19082521_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19105201_Observational study of gene-disease association. (HuGE Navigator) 19105201_TCF7L2 CT/TT genotype is more frequent in nonalcoholic fatty liver disease and predicts the presence and severity of liver disease. 19124064_BCL-W may function as a downstream effector of inappropriate WNT/beta-catenin signalling. 19131553_TCF4 regulates BIRC5 gene expression 19139842_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19141695_Observational study of gene-disease association. (HuGE Navigator) 19141698_High polyunsaturated fatty acid intakes were associated with atherogenic dyslipidemia in carriers of the minor T allele at the TCF7L2 single nucleotide polymorphism. 19141698_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19149908_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19149908_the inverse association between whole-grain consumption and type 2 diabetes risk might depend on variation in TCF7L2. 19156536_TCF7L2 gene expression was determined using quantitative real-time RT-PCR. Treatment with curcumin significantly increased TCF7L2 gene expression while treatment with LPS decreased TCF7L2 gene expression. 19168596_When transfected into rat or mouse beta cells, is involved in maintaining expression of beta-cell genes regulating secretory function. 19169495_Observational study of genetic testing. (HuGE Navigator) 19172244_Observational study of gene-disease association. (HuGE Navigator) 19183934_No association with TCF7L2 variations and adiposity, but bias may have occurred by the evaluation of obesity in separate groups of glycaemic cases and controls 19183934_Observational study of gene-disease association. (HuGE Navigator) 19211816_Carbohydrate quality and quantity modified risk of T2D associated with TCF7L2, which suggests that changes in risk attributable to the TCF7L2 variant are magnified under conditions of increased insulin demand 19211816_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19221600_Observational study of gene-disease association. (HuGE Navigator) 19221600_The genetic association of TCF-4 with ileal Ctohn's disease provides evidence that the decrease in Paneth cell alpha-defensins is a primary factor in disease pathogenesis. 19225753_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19228405_Meta-analysis of gene-disease association. (HuGE Navigator) 19228405_This meta-analysis demonstrates that four variants of TCF7L2 gene are all associated with type 2 diabetes mellitus, and indicates a multiplicative genetic model for all the four polymorphisms [review] 19247372_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19247628_No significant difference in overall amount or splicing pattern is observed between carriers and non-carriers of the type 2 diabetes risk allele. 19252133_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19258404_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19258437_Observational study of gene-disease associatio | ENSMUSG00000024985 | Tcf7l2 | 675.80337 | 1.1840506 | 0.2437307002 | 0.15233511 | 2.569009e+00 | 1.089760e-01 | 4.714196e-01 | No | Yes | 838.827146 | 100.560596 | 683.621882 | 80.434006 | |
| ENSG00000149294 | 4684 | NCAM1 | protein_coding | P13591 | FUNCTION: This protein is a cell adhesion molecule involved in neuron-neuron adhesion, neurite fasciculation, outgrowth of neurites, etc.; FUNCTION: (Microbial infection) Acts as a receptor for rabies virus. {ECO:0000269|PubMed:9696812}.; FUNCTION: (Microbial infection) Acts as a receptor for Zika virus. {ECO:0000269|PubMed:32753727}. | 3D-structure;Alternative splicing;Cell adhesion;Cell membrane;Disulfide bond;GPI-anchor;Glycoprotein;Host cell receptor for virus entry;Host-virus interaction;Immunoglobulin domain;Lipoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | This gene encodes a cell adhesion protein which is a member of the immunoglobulin superfamily. The encoded protein is involved in cell-to-cell interactions as well as cell-matrix interactions during development and differentiation. The encoded protein plays a role in the development of the nervous system by regulating neurogenesis, neurite outgrowth, and cell migration. This protein is also involved in the expansion of T lymphocytes, B lymphocytes and natural killer (NK) cells which play an important role in immune surveillance. This protein plays a role in signal transduction by interacting with fibroblast growth factor receptors, N-cadherin and other components of the extracellular matrix and by triggering signalling cascades involving FYN-focal adhesion kinase (FAK), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K). One prominent isoform of this gene, cell surface molecule CD56, plays a role in several myeloproliferative disorders such as acute myeloid leukemia and differential expression of this gene is associated with differential disease progression. For example, increased expression of CD56 is correlated with lower survival in acute myeloid leukemia patients whereas increased severity of COVID-19 is correlated with decreased abundance of CD56-expressing NK cells in peripheral blood. Alternative splicing results in multiple transcript variants encoding distinct protein isoforms. [provided by RefSeq, Aug 2020]. | hsa:4684; | anchored component of membrane [GO:0031225]; cell surface [GO:0009986]; collagen-containing extracellular matrix [GO:0062023]; cytosol [GO:0005829]; external side of plasma membrane [GO:0009897]; extracellular region [GO:0005576]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; neuron projection [GO:0043005]; plasma membrane [GO:0005886]; virus receptor activity [GO:0001618]; cell adhesion [GO:0007155]; commissural neuron axon guidance [GO:0071679]; regulation of semaphorin-plexin signaling pathway [GO:2001260] | 11681838_NCAM 105-115 kDa is a protease- and neuraminidase-susceptible fragment and was correlated with ventricular enlargement in chronic schizophrenia (p = 0.01). Release of NCAM fragments in schizophrenia may be part of the pathogenic mechanism. 11820619_Significance of cell adhesion molecules, CD56/NCAM in particular, in human tumor growth and spreading. 11915324_exists in lipid microdomains and transducts cell signalings via regulating the activation of signal transduction molecules 12003861_PSA-NCAM was found to be expressed in the somata, dendrites and axonal processes of some neurons, which were identified as chandelier cell axon terminals (chandelier terminals), in the adult human entorhinal cortex and neocortex. 12008081_CD56 expression predicts occurrence of CNS disease in acute lymphoblastic leukemia but not CR or survival. CD56 may enable targeting of leukemic cells to tissues that express it. 12121226_CD56 molecules on NK cells interact with fibroblast growth factor receptor 1 on Jurkat T cells to trigger IL-2 production. 12594840_Preferential apoptosis of CD56dim natural killer cell subset in patients with cancer. 12727026_a useful immunohistochemical marker of Merkel cell carcinoma 12791681_which the polysialyltransferases bind to the first fibronectin type III repeat (FN1) of NCAM to polymerize polysialic acid chains on appropriately presented glycans in adjacent regions. 12937148_Strong overexpression of NCAM(CD56) and RUNX1(AML1) is a constant and characteristic feature of cardiomyocytes within or adjacent to scars in ICM. 14688313_Natural killer (NK) cells expressing high CD56 levels are terminally differentiated cells identical to mature NK cells recently activated in the presence of IL-12, and not a functionally distinct subset or progenitors to mature CD56+low NK cells. 14726964_there are two types of CD56+ epithelial cells in the pancreatic duct system: CD56+ endocrine cells are numerous during the early stage of gestation while CD56+ luminal cells may represent developmental and regenerative changes of pancreatic ducts. 14959847_CD56 is expressed in bone marrow of acute myelogenous leukemias but not acute lymphoblastic leukemia 15006709_Alzheimer patients presented values of low molecular weight-NCAM and high molecular weight-NCAM significantly higher than healthy controls of similar age (higher than 130 kDa) 15050861_Observational study of gene-disease association. (HuGE Navigator) 15050861_genetic variations in neural cell adhesion molecule 1 or nearby genes could confer risks associated with bipolar affective disorder in Japanese individuals. 15061198_REVIEW: Prognostic significance of CD56 expressed in multiple myeloma 15223636_CD56 expression was associated with the leukemogenetic mutation at the primitive hematopoietic progenitor cell level 15231874_Unlike wild-type FGFR4, pituitary tumor derived-FGFR4 does not associate with neural cell-adhesion molecule (NCAM). 15246157_Cells expressing this antigen repond to a Wilms tumor cell line feeder cells and are precursors of NK cells. 15356097_Pre-activated, adherent-natural killer cells express low levels of CD56 and CD161 15459479_There is a close relationship between PSA-NCAM expression and neuronal migration. 15528382_CD56bright natural killer cells accumulate in inflammatory lesions and, in the appropriate cytokine environment, can engage with CD14+ monocytes in a reciprocal activatory fashion, thereby amplifying the inflammatory response. 15626024_first pediatric case describing coexpression of CD56 on B-lineage acute lymphoblastic leukemia 15782066_CD56 seems to be the most sensitive marker for the diagnosis of SCC of the uterine cervix 15950781_A significant increase was observed in PSA-NCAM, NCAM-180, NCAM-140, and HSP70 expression as seen by Western blotting and immunocytofluorescent studies in NMDA-treated cultures. 16027151_polysialyltransferase ST8Sia IV/PST recognizes specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule 16172115_analysis of polymerization of polysialic acid on neural cell adhesion molecules 16211277_NCAM is associated not only with a cell-to-cell adhesion mechanism, but also with tumorigenesis, including growth, development and perineural invasion in human salivary gland tumors 16316416_CD56(bright) and CD56(dim) human natural killer-cell subsets exert different functional and cytotoxic activities in response to a live bacterial pathogen. 16406048_The new role of Neural Cell Adhesion Molecules in tumor neo-angiogenesis relevant for endothelial cell organization into capillary-like structures. 16534119_NCAM is hyposialylated in hereditary inclusion body myopathy skeletal) muscle. 16572491_Tests whether activation and expansion of human NK cells with lipopolysaccharide (LPS) reveals differences between identical twins with regaard to C56 antigen. 16627685_The gene expression profile of hepatic stem cells throughout life consists of high levels of expression of neuronal cell adhesion molecule (NCAM). 16690409_identified co-induction of NKG2A and CD56 on activation of TH2 cells 16892559_Isolation of 3 novel isoforms of AML 1 (RUNX1) with different transactivating function, that might be a regulatory element of the NCAM (CD56) overexpression in chronic myocardial ischemia. 17003032_the FN1 alpha-helix is involved in an Ig5-FN1 interaction that is critical for the correct positioning of Ig5 N-glycans for polysialylation 17005551_Co-immunoprecipitation and co-clustering paradigms were used to show that both NCAM and N-cadherin can interact with the 3Ig IIIC isoform of the FGFR1 in a number of cell types. 17043020_NCAM1 along with RUNX1 is overexpressed during stress hematopoiesis in Down syndrome children and may contribute to the development of overt leukemia. 17085484_Observational study of gene-disease association. (HuGE Navigator) 17161382_In the 'Europe only' stratum, there were nominally significant associations with five contiguous SNPs. 17181871_Neuroblastoma cells resistant to anticancer drugs have increased invasive capacity caused by down-regulation of NCAM adhesion receptor. 17208489_GPI-anchored NCAM-120 suppressed rabies virus replication via induction of IFN-ss even though NCAM-120 was able to promote virus penetration into the cells. 17216340_The pattern of serum NCAM bands could be useful to detect brain tumor pathology. NCAM immunostaining of tumors was inversely correlated with the histological grade of malignancy. Loss of NCAM staining was significantly associated with poor prognosis. 17337466_Renal NCAM-expressing interstitial cells can participate in the initial phase of interstitial fibrosis. 17413444_Observational study of gene-disease association. (HuGE Navigator) 17413444_Single nucleotide polymorphisms within NCAM1 contribute differential risk for both bipolar disorder and schizophrenia possibly by alternative splicing of the gene. 17431094_HIV-1 Nef protein up-regulates the ability of dendritic cellss to stimulate the immunoregulatory naturak killer cells. 17467233_PSA-NCAM is expressed in the human PFC neuropil following a laminated pattern and in a subpopulation of mature neurons, which lack doublecortin expression. Most of these cells have been identified as interneurons expressing calbindin 17635242_Although CD56 expression level varies among the cases, this molecule might play some roles in the manner of growth and expansion of CD56-positive B-cell lymphomas 17683591_PSA-NCAM is not involved in masking Siglec-7 17761687_Observational study of gene-disease association. (HuGE Navigator) 17761687_association studies of alcohol dependence and 43 SNPs mapped to the gene cluster of NCAM1, TTC12, ANKK1 and DRD2 17878347_may explain the preferential accumulation of CD56(bright) NK cells often seen in environments rich in reactive oxygen species, such as at sites of chronic inflammation and in tumors 17891186_lack of CD56 expression on MM cells is not a prognostic marker in patients treated with high-dose chemotherapy, but is associated with t(11;14). 17900814_fetal forebrain axonal PSA-NCAM expression is inversely related to primary myelination 17940597_that expansion of the CD56(bright) NK cell subtype in peripheral blood is not a hallmark of TAP deficiency, but can be found in other diseases as well 17971410_Ubiquitylation represents an endocytosis signal for NCAM. 17982624_NCAM expression may be used as a predictor of perineural invasion in adenoid cystic carcinoma 18209097_Monitoring of T cells driven to senescence showed de novo induction of CD56, the prototypic receptor of NK cells. 18213713_Ganglioneuromas and ganglioneuroblastomas express the adhesive 120 kDa NCAM isoform, while neuroblastomas preferentially express the 180 kDa isoform classically involved in cell motility 18231917_determined the clinical characteristics of 204 multiple myeloma (MM) patients and 26 plasma cell leukemia (PCL) patients with regard to CD56 expression. 18261743_NCAM-fibroblast growth factor receptor 1 interaction at the cell surface is likely to depend upon avidity effects due to receptor clustering 18289872_Neural cell adhesion molecule-extracellular domain overexpression disrupts aborization of basket cells during the major period of axon/dendrite growth in transgenic mice. 18323797_identified molecular characteristics of an aggressive subset of pediatric patients with AML through a prospective evaluation of CD56+ neural cell adhesion molecule (NCAM) and CD94 expression 18333845_relative frequency of CD19 and/or CD56 expression in acute myeloid leukemia (AML) with t(8;21) was significantly higher than those without this translocation and co-expression of these two antigens may serve as the surrogate markers for AML with t(8;21) 18353777_direct receptor-receptor interactions are not required for high affinity GDNF binding to NCAM but play an important role in the regulation of NCAM-mediated cell adhesion by GFRalpha1 18368482_results provide evidence that the BCL motif is one of the multiple FGFR binding sites in NCAM 18384787_Enzymatic removal of PSA from NCAM or reduction of polysialyltransferase expression led to reduced association between NCAM and E-cadherin and subsequently increased E-cadherin-mediated cell-cell aggregation and reduced cell migration. 18425046_CD56 is often expressed by a wide variety of spindle cell sarcomas, thus, it has no value in differentiating GYN from non-GYN spindle cell tumors. 18432248_there was increased placental expression of NCAM (neural cell adhesion molecule) in small for gestational age cases 18462256_extramedullary relapse of MM is characterized by loss of CD56 expression 18594005_CD56-expressing gammadelta T lymphocytes are resistant to Fas ligand and chemically induced apoptosis 18601968_No genetic association between NCAM1 gene polymorphisms and schizophrenia in the Chinese population. 18601968_Observational study of gene-disease association. (HuGE Navigator) 18628406_Immunological events associated with the luteinizing hormone surge induce alterations in all subsets of CD56(+) cells in the fertile menstrual cycle 18641363_CD56(bright) NK cells postallogeneic hematopoietic stem cell transplantation exhibit peculiar phenotypic and functional properties 18755075_To identify the gene regulated by PKD2 and c-Myc, we performed gene expression profiling in PKD2 and c-Myc overexpressing cells. NCAM is an important molecule in the cystogenesis induced by PKD2 overexpression. 18828801_NCAM1 exon 12 markers are significant for specific haplotypes in a family sample of comorbid alcohol and drug dependence. 18828801_Observational study of gene-disease association. (HuGE Navigator) 18972120_NCAM represents a marker for neuroblastomas irrespectively of their stages 18979395_N-CAM increased in cytotrophoblasts and decreased in extravillous trophoblasts and decidual cells of preeclamptic subjects. 18990213_The results obtained show that, in man, the expression of PSA-NCAM in selective populations of central and peripheral neurons occurs not only during prenatal life, but also in adulthood. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19153015_CD56 is extremely useful in the diagnosis of papillary thyroid carcinoma 19222860_NCAM-positive neuroblastoma patients had more often metastases at diagnosis, and NCAM expression associated with advanced disease 19235015_propose four potential activators of the CD56 promoter and for CD56 to be involved in proliferation and anti-apoptosis, leading to disease progression in multiple myeloma 19328310_Subcutaneous panniculitis-like T-cell lymphoma with a phenotype showing a deficiency of cd56. 19328558_Observational study of gene-disease association. (HuGE Navigator) 19393299_study implicates the major NCAM isoforms, PSA-NCAM and proteolytically cleaved NCAM in pre- and postnatal development of the human prefrontal cortex 19411161_This data suggests NCAM-180 mRNA expression is altered in a regionally-specific manner in schizophrenia and these changes are associated with the early period following diagnosis. 19507465_The role of NCAM in cell differentiation, survival and plasticity suggests an involvement in the development process of the carotid body and in cellular/molecular changes due to chronic hypoxia. 19587433_Structural change in NCAM1 caused by point mutation may be the reason for astrocytoma tumorigenesis. 19652998_Additional immunohistochemical detection of neuroendocrine differentiation (chromogranin-A, synaptophysin, and neural-cell adhesion molecule) in non-small cell lung cancer is presently not of prognostic importance. 19724850_Class III beta-tubulin and NCAM were expressed in 50 and 68% of basal cell carcinoma 19725832_polysialylated NCAM persistence, up-regulated polysialyltransferase-1 mRNA and previously uncovered defective myelin-associated glycoprotein may be early pathogenetic events in adult-onset autosomal-dominant leukodystrophy 19772585_Expression defines a committed T-lymphocyte lineage to the cytotoxic phenotype 19788570_results suggest a positive role of NCAM in neurogenesis in the periventricular region; expression of NCAM in stem cells might be one of many factors useful for therapeutic approaches in the future 19788615_Mucosal remodeling with alterations of NCAM+ or alpha-SMA+ subepithelial and interstitial cells may play a critical role in UC-associated tumorigenesis. 19864234_CD56 immunohistochemical analysis is useful for detecting residual plasma cell myeloma particularly in morphologically equivocal cases in which light chain restriction cannot be demonstrated, and may serve as a potential response criterion. 19897577_human CD56(bright) NK cells progress through a continuum of differentiation that ends with a CD94(low)CD56(dim) phenotype. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20015889_Neural cell adhesion molecules expressed on mesenchymal stem cells play a crucial role in the human hematopoiesis-supporting ability of the cell line. 20027291_The IL-23 induced cytokines allow for the subsequent production of IL-12 and amplify the IFN-gamma production in the type-1 cytokine pathway. 20029409_Observational study of gene-disease association. (HuGE Navigator) 20049565_induction of SOX4 gene expression might be responsible for the CD56 expression in human myeloma cells 20059553_NCAM in the amygdala mediates consolidation of auditory fear conditioning; increased NCAM transgene expression in the amygdala is among the mechanisms whereby stress facilitates fear conditioning processes in floxed mice. 20083228_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20164549_This study suggested a potential involvement of NCAM expressing neurons in the cognitive deficits in Alzheimer's disease. 20187302_we suggest NCAM as a marker for the WT progenitor cell population. These findings provide novel insights into the cellular hierarchy of WT, having possible implications for future therapeutic options 20231901_SOCS-3 and PIAS-3 upregulation impairs IL-12-mediated interferon-gamma response in CD56 T cells in HCV-infected heroin users 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20414008_analysis of the effect of human colorectal carcinogenesis on the neural cell adhesion molecule expression and polysialylation 20483466_Observational study of gene-disease association. (HuGE Navigator) 20483466_This study suggested that SNP variants in NCAM1 may impact on related traits, particularly by mediating inhibition of aggressiveness. 20524836_we confirmed the expression of NCAM in Wilms tumors, nephrogenic rests, and metanephric mesenchyme and its early derivatives in fetal renal cortex 20538416_CD56 cannot be used as a supplementary tool in the differential diagnosis of non-neoplastic biliary diseases in needle biopsies. 20557674_The predominance of extranodal involvement in our series may be associated with the adhesion-related function of CD56. A high frequency of bcl-6 expression may be associated with a more favorable clinical course and prognosis. 20610389_a decisive role for the neuronal K(+) channel in regulating NCAM-dependent neurite outgrowth and attribute a physiologically meaningful role to the functional interplay of Kir3.3, NCAM, and TrkB in ontogeny 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20664990_Assessment of natural killer-like T CD3+/CD16+ CD56+ cells may be helpful in determining a worsening of clinical course in chronic lymphocytic leukemia. 20684989_Results confirm the favorable impact of NPM1 mutations and identify the adverse prognostic relevance of CD56 expression in this subgroup of AML. 20696944_CD56(dim) NK cells continue to differentiate. During this process, they lose expression of NKG2A, sequentially acquire inhibitory killer cell inhibitory immunoglobulin-like receptors and CD57. 20733159_assessed the transcriptional, phenotypic, and functional differences between CD57(+) and CD57(-) NK cells within the CD56(dim) mature NK subset 20805222_Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera. 20875069_CD56 expression in odontogenic epithelium is highly suggestive of ameloblastoma and can help in differentiating this from odontogenic keratocyst 20932956_PSA-NCAM may modulate the functional interaction between BDNF and its high and low affinity receptors;possible clinical significance of neuronal trophism in cerebellar neurodegenerative disorders 21115007_Data suggest that the intra-cellular part of NCAM inhibits cis-dimerization, an effect mainly dependent on the palmitoylation sites. 21148082_CD56 expression is associated with coexpression of immaturity-associated and T-cell antigens and is an independent adverse prognostic factor for relapse in acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline-based regimens 21178331_CD6 expression on peripheral NK cells marks a novel CD56(dim) subpopulation associated with distinct patterns of cytokine and chemokine secretion. 21212386_Genetic variation in NCAM1 contributes to left ventricular wall thickness in hypertensive families. 21239711_CD56(bright)CD11c(positive) cells play a key role in the interleukin (IL)-18-mediated proliferation of gammadelta T cells. 21457956_Maturation of follicles is accompanied by a decrease in CD56+CD16+ natural killer cells that may have deleterious effects on follicular maturation. 21464126_STX(G421A) shows a dramatic decrease in polySia synthetic activity on NCAM, whereas STX(C621G) does not; polySia shows a dopamine (DA) binding activity 21467162_Excellent prognosis in a subset of patients with ewing sarcoma identified at diagnosis by CD56 using flow cytometry. 21515372_Abnormal synaptic innervation in transgenic mice expressing the NCAM extracellular proteolytic cleavage fragment impairs prefrontal cortex plasticity and alters working memory. 21577011_This study did not support the ncam1 is direct modulation of these genes on temperament 21666061_Natural killer (NK)-92 cell line retains the functional characteristics of the CD56(bright) NK cell subset; consequently, gene silencing of CD56 profoundly inhibits the ability of NK-92 cells to kill activated syngeneic T cells. 21669487_Soluble NCAM status was a significant independent factor predictive of long-term survival in patients with hepatocellular carcinoma 21691800_NCAM-140 interacts with APP, potentially playing a role in neurite outgrowth and neural development. 21717310_CD56 expression is associated with neuroectodermal differentiation in ameloblastomas. 21739604_These results point to NCAM-mediated stimulation of FGFR as a novel mechanism underlying epithelial ovarian carcinoma malignancy and indicate that this interplay may represent a valuable therapeutic target. 21940794_Together, these data indicate that polysialic acid regulates cell motility through NCAM-induced but FGF-receptor-independent signalling to focal adhesions 22001684_Multiple sclerosis is characterized by a dysregulation of CD8+CD56-perforin+ T cells that may play a role in the development of disability. 22081445_The CD56 antigen is an independent prognostic risk factor in patients with acute myeloid leukemia 22085395_Serum NCAM is hyposialylated in hereditary inclusion body myopathy. 22099865_This study demonistrated that the depressed patients showed decreases in PSA-NCAM expression in the basolateral and basomedial amygdala and in the lateral nucleus of bipolar patients 22211388_Prominent CD56 expression by damaged and regenerating muscle fibers in the skin. 22219127_Cerebrospinal fluid NCAM-1 is a potential biomarker for drug-effective epilepsy and drug-refractory epilepsy. 22228741_Dehydroepiandrosterone increased PSA-NCAM expression and inhibited monocyte binding in an estrogen- and androgen receptor-dependent manner. 22276608_CD56 expression in B cell lymphoma is a rare occurrence. 22281821_NCAM and polySia are expressed and developmentally regulated in chick corneas. Both membrane-associated and soluble NCAM isoforms are expressed in chick corneas. 22319021_results suggest that during differentiation CD56(bright) NK cells, similarly to mature activated NK cells, become highly cytotoxic and are relatively resistant to apoptosis induced by TNF family members. 22384114_Cytotoxicity of CD56(bright) NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A 22384181_NCAM180 regulates Ric8A membrane localization and potentiates beta-adrenergic response 22423624_assessment of CD56 and CD117 expression by flow cytometry is a sensitive method for diagnostic evaluation of plasma cell neoplasms. 22449227_NCAM1, SYPT and CGA expressions are differently regulated by neuroendocrine phenotype-specific transcription factors in lung cancer cells. 22591692_CD56-positive T cells have a critical role in innate defense against HIV-1 infection. 22732936_TGF-beta1 contributes to HCC-induced vascular alterations by affecting the interaction between HCC-StCs and Ld-MECs through a down-modulation of NCAM expression 22792160_Intra-hepatic accumulation of the functionally impaired CXCR3(+)CD56Bright NK cell subset might be involved in HCV-induced liver fibrosis. 23015367_Bioinformatic analysis of NCAM-associated expression profiles predicted a highly interactive protein network, which further implies potential molecular mechanisms underlying the metastatic processes of thyroid cancer 23022470_In schizophrenia, abnormal PSA-NCAM and GAD67 expression may underlie the alterations observed in inhibitory neurotransmission. 23061666_Data indicate that UCHL1 is a novel interaction partner of both NCAM isoforms that regulates their ubiquitination and intracellular trafficking. 23258197_An unknown factor with a molecular weight >100 kDa plays a critical role in the impairment of CD56(dim) natural killer cells in malignant pleural effusion, which might lead to tumor progression. 23260340_Immunoblots revealed that depressed subjects displayed increased expression of PSA-NCAM. 23292839_the author showed that in cutaneous basal cell carcinoma, the expression of NCAM and c-KIT was high, PDGFRA was intermediate, and chromogranin A and synaptophysin was relatively low 23303482_SNPs in NCAM1 were not significantly associated with heroin dependence. 23365458_GATA2 is required for the maturation of human natural killer cells and the maintenance of the CD56(bright) pool in the periphery. 23418554_Data suggest that multi-antibody assay of TTF1, Vimentin, p63 CD56, chromogranin and synaptophysin may be of special value, especially in diagnosing small biopsies. 23462508_CD56(140kD) up-regulation plays a pivotal role in the pathogenesis of ischemic cardiomyopathy. 23470050_We recommend immunohistochemical analyses for CD123, CD56 and CD4 inblastic plasmacytoid dendritic cell neoplasm patients, particularly in cases where the initial bone marrow study indicates normal morphology. 23480226_Women with age above 35 years and greater than 13% CD56 positive CD16 positive natural killer cells showed the highest risk of further pregnancy loss 23495921_Depletion of NCAM is one of the factors associated with or possibly responsible for disease progression in multiple sclerosis. 23557873_Results suggest that CD3+CD56+NKT cells do not play a role as a mediator in mental symptom such as depression in fibromyalgia syndrome patients. 23635388_CD56 and quantitative Ki-67 along with cytomorphology is a robust immunohistochemical panel to differentiate small cell lung carcinoma from other neuroendocrine neoplasms. 23671285_translational modification of the neural cell adhesion molecule NCAM and the polysialyltransferase ST8SiaII in mammalian semen involves polysialic acid 23716295_NCAM polysialylation in small cell lung cancer progression regulates substrate adherence potential. 23810283_High CD56 expression is associated with relapse in acute myeloid leukemia with t(8;21). 23960070_NCAM and dynein have roles in tethering dynamic microtubules and maintaining synaptic density in cortical neurons 24055371_Depletion of NCAM1(+) cells from human kidney epithelial cells abrogated stemness traits in vitro. 24206578_Positive CD56 expression was found in 23 APL patients. 24240977_Decreased proportion of NK cells expressing CD56dim/CD16 antigens in Franconi anemia patients correlates with the impairment in the differentiation process of the NK cells and of the immune surveillance. 24286519_Data indicate that the CD14bright/CD56+ monocyte subset is expanded in aging individuals as well as in patients with rheumatoid arthritis. 24294395_Case Report: incidentally diagnosed CD56 positive diffuse large B-cell lymphoma. 24349544_The described scFv has potential application in delivery of therapeutics to NCAM1-expressing cells in degenerated IVD. 24365773_Expression of NCAM was associated with worsening hemodynamic parameters and major metabolic genes. 24369228_CD56 can be expressed in normal immature granulocytes at a variety of expression levels in regenerative bone marrow. Attention should be paid when evaluating aberrant antigen expression of CD56 in granulocytes. 24526449_NCAM expression in myelinating Schwann cells is indicative of early Schwann cell abnormalities. 24715165_HBME-1, Galectin-3, Cytokeratin-19 and CD56 were used alone or in panels in a series of papillary thyroid carcinoma (PTC) and thyroid tumors of uncertain malignant potential. 24726913_NCAM140 interacts with ufc1 and its trafficking and endocytosis is upregulated in the presense of Ufm1. 24782118_CD56 positivity did not have any influence on the prognosis of these patients. 24807109_Women, w/ or w/o endometriosis, having larger populations of cytotoxic CD16(+) uterine NK cells and/or higher populations of NKp46(+)CD56(+) cells may be at greater risk of infertility resulting from inflammatory environment during implantation or later 24909369_Increased frequency of ILT2-expressing CD56(dim)CD16(+) NK cells correlates with disease severity of pulmonary tuberculosis. 25137309_results showed that NCAM1 might play an important role in the pathogenesis of autism 25188863_Report differential diagnosis of Primary cutaneous NK/T-cell lymphoma, nasal type and CD56-positive peripheral T-cell lymphoma. 25201755_Homophilic interaction between CD56 molecules may occur in tumor-cell recognition, leading to CIK-mediated cell death. 25326085_Report ontogenic development of nerve fibers in human fetal livers using immunohistochemical detection of NCAM1/neurone-specific enolase expression. 25363560_analysis of CD8+ T cell induction via Vgamma9gammadeltaT cell expansion by CD56(high+) Interferon-alpha-induced dendritic cells 25445624_Results provide direct evidence for NCAM1 as a susceptibility gene for schizophrenia, which offers support to a neurodevelopmental model and neuronal connectivity hypothesis in the onset of schizophrenia 25596273_CD56(low)CD16(low) natural killer cells are multifunctional cells, and that the presence of hematologic malignancies affects their frequency and functional ability at both tumor site and in the periphery. 25619885_improvement in behavioral performance (open-field and grip-strength tests), as well as increased life-span was observed in rodents treated with NCAM-VEGF or NCAM-GDNF co-transfected cells 25628040_Low NCAM1 in the sera is associated with active forms of inflammatory bowel disease. 25723856_CD56 is generally expressed in 70-80%11 of patients with MM, as observed in 69% of the t(14;16)-negative cases in this study. In contrast, none of the t(14;16)-positive cases showed CD56 positivity. 25769453_Determining the P-glycoprotein expression and function of the peripheral blood CD56+ cells may help predict the MDR of NHL, thus has profound guiding significance for NHL treatment 25779340_Human BDCA2+CD123+CD56+ dendritic cells (DCs) related to blastic plasmacytoid dendritic cell neoplasm represent a unique myeloid DC subset. 25889612_Aberrant NCAM expression plays a role in the pathogenesis of keratin producing odontogenic tumors. 25921109_Up-regulated level of serum sNCAM is associated with hepatic encephalopathy in hepatocelular carcinoma patients. 25924702_CD56 overexpression was associated with shorter OS. 25935537_Loss of CD56 expression is associated with extranodal NK/T cell lymphoma. 26013700_Case Report: chronic lymphocytic leukemia with aberrant CD56 and CD57 expression. 26039898_Results show significant increases in proportions of CD56+ T cells in relation to CMV infection in renal transplant patients and suggest that these cells have a cytotoxic function against CMV-infected cells. 26045862_CD56 could potentially become an adjunct diagnostic marker for ectomesenchymal chondromyxoid tumor instead of previously used CD57. 26087825_Renal graft CD56+ cell infiltrates were significantly associated with antibody mediated rejection. 26097548_for differential diagnosis of papillary thyroid cancer it was found that the only marker with both sensitivity and specificity above 90% was CD56 negativity 26147745_this study shows that CD56 expression defines a poor prognosis subset in the cytogenetically intermediate prognosis pediatric AML. 26183877_CD56 expression remains to be a potentially unfavorable prognostic factor in acute promyelocytic leukemia patients. 26186733_CD56 was negative in 96% of the primary malignant thyroid tumors while being expressed in the cytoplasm of 68.5% of the benign thyroid nodules. 26255203_Our findings demonstrate a novel Wnt/beta-catenin-miR-30a-5p-NCAM regulatory axis which plays important roles in controlling glioma cell invasion and tumorigenesis. 26339384_CD56 marker is a useful alternative that is comparable to SSTR2A for the diagnosis of phosphaturic mesenchymal tumors. 26344352_the expression of CD56 in adenomyosis is positively associated with the severity of dysmenorrhea. 26391771_Letter/Case Report: CD56 positive diffuse large B cell lymphoma of the urinary bladder. 26437631_A FOXP3(+)CD3(+)CD56(+)-expressed T-cell population with immunosuppressive function and reduced patient survival has been identified in cancer tissues of human hepatocellular carcinoma. 26460482_The neural cell adhesion molecule (NCAM) is a glycoprotein implicated in cell-cell adhesion, neurite outgrowth and synaptic plasticity. 26463893_NCAM1 deletion is associated with neuroblastoma and ganglioneuroma. 26478212_L1 | ENSMUSG00000039542 | Ncam1 | 1233.06864 | 0.9211630 | -0.1184716569 | 0.09786848 | 1.461393e+00 | 2.267087e-01 | 6.186901e-01 | No | Yes | 1440.876486 | 154.149791 | 1498.142143 | 156.516145 | |
| ENSG00000149658 | 54915 | YTHDF1 | protein_coding | Q9BYJ9 | FUNCTION: Specifically recognizes and binds N6-methyladenosine (m6A)-containing mRNAs, and regulates their stability (PubMed:24284625, PubMed:32492408, PubMed:26318451). M6A is a modification present at internal sites of mRNAs and some non-coding RNAs and plays a role in mRNA stability and processing (PubMed:24284625, PubMed:32492408). Acts as a regulator of mRNA stability by promoting degradation of m6A-containing mRNAs via interaction with the CCR4-NOT complex (PubMed:32492408). The YTHDF paralogs (YTHDF1, YTHDF2 and YTHDF3) shares m6A-containing mRNAs targets and act redundantly to mediate mRNA degradation and cellular differentiation (PubMed:28106072, PubMed:32492408). Required to facilitate learning and memory formation in the hippocampus by binding to m6A-containing neuronal mRNAs (By similarity). Acts as a regulator of axon guidance by binding to m6A-containing ROBO3 transcripts (By similarity). Acts as a negative regulator of antigen cross-presentation in myeloid dendritic cells (By similarity). In the context of tumorigenesis, negative regulation of antigen cross-presentation limits the anti-tumor response by reducing efficiency of tumor-antigen cross-presentation (By similarity). Promotes formation of phase-separated membraneless compartments, such as P-bodies or stress granules, by undergoing liquid-liquid phase separation upon binding to mRNAs containing multiple m6A-modified residues: polymethylated mRNAs act as a multivalent scaffold for the binding of YTHDF proteins, juxtaposing their disordered regions and thereby leading to phase separation (PubMed:31388144, PubMed:31292544, PubMed:32451507). The resulting mRNA-YTHDF complexes then partition into different endogenous phase-separated membraneless compartments, such as P-bodies, stress granules or neuronal RNA granules (PubMed:31292544). {ECO:0000250|UniProtKB:P59326, ECO:0000269|PubMed:24284625, ECO:0000269|PubMed:26318451, ECO:0000269|PubMed:28106072, ECO:0000269|PubMed:31292544, ECO:0000269|PubMed:31388144, ECO:0000269|PubMed:32451507, ECO:0000269|PubMed:32492408}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Immunity;Phosphoprotein;RNA-binding;Reference proteome | hsa:54915; | cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; P-body [GO:0000932]; mRNA binding [GO:0003729]; N6-methyladenosine-containing RNA binding [GO:1990247]; ribosome binding [GO:0043022]; RNA binding [GO:0003723]; immune system process [GO:0002376]; learning [GO:0007612]; memory [GO:0007613]; mRNA destabilization [GO:0061157]; organelle assembly [GO:0070925]; positive regulation of translation [GO:0045727]; positive regulation of translational initiation [GO:0045948]; regulation of antigen processing and presentation [GO:0002577]; regulation of axon guidance [GO:1902667]; regulation of long-term synaptic potentiation [GO:1900271]; regulation of mRNA stability [GO:0043488]; stress granule assembly [GO:0034063] | 26318451_binding affinities of the YTH domains of three human proteins and yeast YTH domain protein Pho92 27371828_The authors found that the overexpression of YTHDF proteins in cells inhibited HIV-1 infection mainly by decreasing HIV-1 reverse transcription, while knockdown of YTHDF1-3 in cells had the opposite effects. Moreover, silencing the N(6)-methyladenosine writers decreased HIV-1 Gag protein expression in virus-producing cells, while silencing the N(6)-methyladenosine erasers increased Gag expression. 29439311_YTHDF1 played an important role in regulating hepatocellular carcinoma cell cycle progression and metabolism 30728504_Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells 31653849_Findings highlight the critical role of YTHDF1 in both hypoxia adaptation and pathogenesis of non-small cell lung cancer. 31996915_YTHDF1 augments the translation of EIF3C in an m6A-dependent manner by binding to m6A-modified EIF3C mRNA and concomitantly promotes the overall translational output, thereby facilitating tumorigenesis and metastasis of ovarian cancer. 32449290_Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. 32451507_m(6)A-binding YTHDF proteins promote stress granule formation. 32788173_YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. 33011193_m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. 33099572_N(6)-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. 33204330_YTHDF1-enhanced iron metabolism depends on TFRC m(6)A methylation. 33280517_Loading MicroRNA-376c in Extracellular Vesicles Inhibits Properties of Non-Small Cell Lung Cancer Cells by Targeting YTHDF1. 33456585_ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. 33619246_HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. 33825148_YTHDF1 promotes NLRP3 translation to induce intestinal epithelial cell inflammatory injury during endotoxic shock. 33862464_YTHDF1-regulated expression of TEAD1 contributes to the maintenance of intestinal stem cells. 33999206_The N6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. 34151473_Impact of YTHDF1 gene polymorphisms on Wilms tumor susceptibility: A five-center case-control study. 34359836_YTHDF1 Promotes Cyclin B1 Translation through m(6)A Modulation and Contributes to the Poor Prognosis of Lung Adenocarcinoma with KRAS/TP53 Co-Mutation. 34408926_YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-small-cell lung cancer. 34497675_Comprehensive Analysis of YTH Domain Family in Lung Adenocarcinoma: Expression Profile, Association with Prognostic Value, and Immune Infiltration. 34677810_YTHDF1 promotes the proliferation, migration, and invasion of prostate cancer cells by regulating TRIM44. 34821414_YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. 34974791_Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. 34990050_Involvement of YTHDF1 in renal fibrosis progression via up-regulating YAP. 35156522_Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). 35279688_YTHDF1 promotes breast cancer cell growth, DNA damage repair and chemoresistance. | ENSMUSG00000038848 | Ythdf1 | 2182.02829 | 1.1008685 | 0.1386421699 | 0.08699038 | 2.548071e+00 | 1.104290e-01 | 4.745044e-01 | No | Yes | 2778.224361 | 178.895509 | 2435.378683 | 153.534105 | ||
| ENSG00000151240 | 22982 | DIP2C | protein_coding | Q9Y2E4 | Alternative splicing;Phosphoprotein;Reference proteome | This gene encodes a member of the disco-interacting protein homolog 2 family. The protein shares strong similarity with a Drosophila protein which interacts with the transcription factor disco and is expressed in the nervous system. [provided by RefSeq, Oct 2008]. | hsa:22982; | 12137943_This publication discusses the expression pattern and potential function of similar genes in Drosophila and mouse. 19851296_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 23703922_DIP2C gene SNP rs877282 reached genome-wide significant association with plasma uric acid levels (P = 4.56 x 10(-8)). 28716088_Loss of DIP2C triggers substantial DNA methylation and gene expression changes, cellular senescence and epithelial-mesenchymal transition in cancer cells. 28964575_The data revealed DIP2C expression level decreased in breast cancer, especially in basal-like and HER-2 subtypes, and could be a valuable target for diagnosis on specific subtype of breast cancer | ENSMUSG00000048264 | Dip2c | 566.24521 | 0.8821946 | -0.1808311409 | 0.14952773 | 1.453574e+00 | 2.279553e-01 | 6.190380e-01 | No | Yes | 633.088413 | 67.084070 | 693.818522 | 71.883515 | |||
| ENSG00000151575 | 374618 | TEX9 | protein_coding | Q8N6V9 | Alternative splicing;Coiled coil;Reference proteome | hsa:374618; | 31481019_Testis-expressed protein 9 (TEX9) expression is positively associated with eukaryotic translation initiation factor 3 subunit b (eIF3b) expression in esophageal squamous cell carcinoma (ESCC). TEX9 expression is positively correlated with tumor-node-metastasis stage in ESCC. eIF3b binding to TEX9 mRNA functionally synergizes to promote the proliferation and migration, and inhibit the apoptosis of ESCC cells. | ENSMUSG00000090626 | Tex9 | 160.10773 | 1.0655143 | 0.0915499498 | 0.22963643 | 1.593413e-01 | 6.897637e-01 | 9.047448e-01 | No | Yes | 210.701752 | 54.609995 | 192.850114 | 49.026704 | ||||
| ENSG00000151773 | 160857 | CCDC122 | protein_coding | Q5T0U0 | Alternative splicing;Coiled coil;Reference proteome | This gene encodes a protein that contains a coiled-coil domain. Naturally occurring mutations in this gene are associated with leprosy. [provided by RefSeq, Apr 2017]. | hsa:160857; | 17672918_the apparent occurrence of an unusual TG 3' splice site in intron 7 (according to mRNA AK056408) is discussed 20018961_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 25367361_Association between leprosy and tag SNPs at NOD2 (rs8057431) and CCDC122-LACC1 (rs4942254). | ENSMUSG00000034795 | Ccdc122 | 115.85541 | 0.7493186 | -0.4163488868 | 0.28672964 | 2.080244e+00 | 1.492163e-01 | 5.377572e-01 | No | Yes | 115.069051 | 28.264422 | 144.173284 | 34.647041 | |||
| ENSG00000152242 | 147339 | C18orf25 | protein_coding | Q96B23 | Alternative splicing;Coiled coil;Isopeptide bond;Methylation;Phosphoprotein;Reference proteome;Ubl conjugation | hsa:147339; | ubiquitin protein ligase activity [GO:0061630]; ubiquitin-dependent protein catabolic process [GO:0006511] | 19240061_Observational study of gene-disease association. (HuGE Navigator) 24216761_ARKL1 binds CK2beta through the KSSR motif and this involves a polyserine sequence resembling the CK2beta binding sequence in EBNA1. | ENSMUSG00000047466 | 8030462N17Rik | 1217.53180 | 0.9838338 | -0.0235134454 | 0.11239084 | 4.372474e-02 | 8.343666e-01 | 9.547489e-01 | No | Yes | 1638.899113 | 320.980914 | 1541.899144 | 295.482663 | |||
| ENSG00000152795 | 9987 | HNRNPDL | protein_coding | O14979 | FUNCTION: Acts as a transcriptional regulator. Promotes transcription repression. Promotes transcription activation in differentiated myotubes (By similarity). Binds to double- and single-stranded DNA sequences. Binds to the transcription suppressor CATR sequence of the COX5B promoter (By similarity). Binds with high affinity to RNA molecules that contain AU-rich elements (AREs) found within the 3'-UTR of many proto-oncogenes and cytokine mRNAs. Binds both to nuclear and cytoplasmic poly(A) mRNAs. Binds to poly(G) and poly(A), but not to poly(U) or poly(C) RNA homopolymers. Binds to the 5'-ACUAGC-3' RNA consensus sequence. {ECO:0000250, ECO:0000269|PubMed:9538234}. | Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Direct protein sequencing;Disease variant;Isopeptide bond;Limb-girdle muscular dystrophy;Methylation;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has two RRM domains that bind to RNAs. Three alternatively spliced transcript variants have been described for this gene. One of the variants is probably not translated because the transcript is a candidate for nonsense-mediated mRNA decay. The protein isoforms encoded by this gene are similar to its family member HNRPD. [provided by RefSeq, May 2011]. | hsa:9987; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; spliceosomal complex [GO:0005681]; DNA binding [GO:0003677]; double-stranded DNA binding [GO:0003690]; poly(A) binding [GO:0008143]; poly(G) binding [GO:0034046]; RNA binding [GO:0003723]; single-stranded DNA binding [GO:0003697]; regulation of gene expression [GO:0010468]; RNA processing [GO:0006396] | 16011250_Study shows that there is interaction of the intracellular domain of beta-amyloid precursor protein with JKTBP2, indicating that JKTBP2 may have an important function in AD formation. 17592041_The data of this study show that JKTBP1 and the 14-nt element act independently to mediate NRF internal ribosome entry segment activity. 18095154_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18381662_overexpression of JKTBP1 in LNCaP cells leads to abnormal cell proliferation 21300069_The results indicate that JKTBP1 regulates the level of NRF protein expression by binding to both NRF 5' and 3' UTRs. 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 23642268_hnRNP DL and CNBP are novel antigens for SLE patients 24647604_A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G. 29263134_Both hnRNP D and DL are able to control their own expression by alternative splicing of cassette exons in their 3'UTRs. Exon inclusion produces mRNAs degraded by nonsense-mediated decay. Moreover, hnRNP D and DL control the expression of one another by the same mechanism. 30052712_Heterogeneous nuclear ribonucleoprotein D-like (HNRPDL) is aberrantly expressed in specimens from colorectal cancer patients. 30447347_High HNRNPDL expression is associated with cervical cancer. 31488872_HNRPDL transforms hematopoietic cells and a novel HNRPDL/PBX1 axis plays an important role in human CML CD34(+) cells | ENSMUSG00000029328 | Hnrnpdl | 8743.52421 | 0.8138297 | -0.2972012267 | 0.07194613 | 1.689652e+01 | 3.947387e-05 | 5.879632e-03 | No | Yes | 8780.200498 | 1265.317979 | 10335.304140 | 1456.026989 | |
| ENSG00000152926 | 51351 | ZNF117 | protein_coding | Q03924 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a protein containing multiple C2H2-type zinc finger motifs. Readthrough transcription occurs between this gene and the upstream endogenous retrovirus group 3 member 1 (ERV3-1) locus, and may result in additional transcript variants encoding the zinc finger protein. [provided by RefSeq, Jan 2017]. | hsa:109504726;hsa:51351; | nucleus [GO:0005634]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; regulation of transcription, DNA-templated [GO:0006355] | 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) | 255.25327 | 1.1522627 | 0.2044696115 | 0.19756293 | 1.073439e+00 | 3.001700e-01 | 6.837756e-01 | No | Yes | 291.937854 | 79.558588 | 261.489429 | 69.727809 | |||
| ENSG00000152969 | 152789 | JAKMIP1 | protein_coding | Q96N16 | FUNCTION: Associates with microtubules and may play a role in the microtubule-dependent transport of the GABA-B receptor. May play a role in JAK1 signaling and regulate microtubule cytoskeleton rearrangements. {ECO:0000269|PubMed:14718537, ECO:0000269|PubMed:15277531, ECO:0000269|PubMed:17532644}. | Alternative splicing;Coiled coil;Cytoplasm;Cytoskeleton;Membrane;Microtubule;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:152789; | cytoplasm [GO:0005737]; extrinsic component of membrane [GO:0019898]; microtubule [GO:0005874]; ribonucleoprotein complex [GO:1990904]; GABA receptor binding [GO:0050811]; kinase binding [GO:0019900]; microtubule binding [GO:0008017]; RNA binding [GO:0003723]; cognition [GO:0050890]; protein transport [GO:0015031] | 14718537_Marlin-1 functions to regulate the cellular levels of GABA(B) R2 subunits, which may have significant effects on the production of functional GABA(B) receptor heterodimers 15277531_Marlin1 has a role in cell polarization, segregation of signaling complexes, and vesicle traffic, some of which may involve Jak tyrosine kinases 17761393_We have identified four new transcripts of 2975 bp, 1743 bp, 2189 bp and 2420 bp respectively, named Jakmip1B, Jakmip1C, Jakmip1D and Jakmip1E 17804789_Observational study of gene-disease association. (HuGE Navigator) 18941173_Jakmip1 is a novel effector memory gene that restrains T cell-mediated cytotoxicity. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 23481296_JAKMIP1 associates with and possibly contributes to the Wnt/beta-catenin pathway activity through its influence on downstream Wnt target proteins, including beta-catenin. | ENSMUSG00000063646 | Jakmip1 | 118.80126 | 0.7269302 | -0.4601112053 | 0.27095309 | 2.854661e+00 | 9.110942e-02 | 4.375885e-01 | No | Yes | 115.401198 | 15.720983 | 153.057488 | 19.949659 | ||
| ENSG00000153094 | 10018 | BCL2L11 | protein_coding | O43521 | FUNCTION: Induces apoptosis and anoikis. Isoform BimL is more potent than isoform BimEL. Isoform Bim-alpha1, isoform Bim-alpha2 and isoform Bim-alpha3 induce apoptosis, although less potent than isoform BimEL, isoform BimL and isoform BimS. Isoform Bim-gamma induces apoptosis. Isoform Bim-alpha3 induces apoptosis possibly through a caspase-mediated pathway. Isoform BimAC and isoform BimABC lack the ability to induce apoptosis. {ECO:0000269|PubMed:11997495, ECO:0000269|PubMed:15486195, ECO:0000269|PubMed:15661735, ECO:0000269|PubMed:9430630}. | 3D-structure;Alternative splicing;Apoptosis;Membrane;Mitochondrion;Phosphoprotein;Reference proteome;Ubl conjugation | The protein encoded by this gene belongs to the BCL-2 protein family. BCL-2 family members form hetero- or homodimers and act as anti- or pro-apoptotic regulators that are involved in a wide variety of cellular activities. The protein encoded by this gene contains a Bcl-2 homology domain 3 (BH3). It has been shown to interact with other members of the BCL-2 protein family and to act as an apoptotic activator. The expression of this gene can be induced by nerve growth factor (NGF), as well as by the forkhead transcription factor FKHR-L1, which suggests a role of this gene in neuronal and lymphocyte apoptosis. Transgenic studies of the mouse counterpart suggested that this gene functions as an essential initiator of apoptosis in thymocyte-negative selection. Several alternatively spliced transcript variants of this gene have been identified. [provided by RefSeq, Jun 2013]. | hsa:10018; | Bcl-2 family protein complex [GO:0097136]; BIM-BCL-2 complex [GO:0097141]; BIM-BCL-xl complex [GO:0097140]; cytosol [GO:0005829]; endomembrane system [GO:0012505]; extrinsic component of membrane [GO:0019898]; mitochondrial outer membrane [GO:0005741]; mitochondrion [GO:0005739]; microtubule binding [GO:0008017]; protein kinase binding [GO:0019901]; activation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0006919]; apoptotic process [GO:0006915]; apoptotic process involved in embryonic digit morphogenesis [GO:1902263]; B cell homeostasis [GO:0001782]; brain development [GO:0007420]; cell-matrix adhesion [GO:0007160]; cellular response to amyloid-beta [GO:1904646]; cellular response to estradiol stimulus [GO:0071392]; cellular response to glucocorticoid stimulus [GO:0071385]; developmental pigmentation [GO:0048066]; ear development [GO:0043583]; extrinsic apoptotic signaling pathway in absence of ligand [GO:0097192]; in utero embryonic development [GO:0001701]; intrinsic apoptotic signaling pathway in response to DNA damage [GO:0008630]; kidney development [GO:0001822]; male gonad development [GO:0008584]; mammary gland development [GO:0030879]; meiosis I [GO:0007127]; myeloid cell homeostasis [GO:0002262]; odontogenesis of dentin-containing tooth [GO:0042475]; positive regulation of apoptotic process [GO:0043065]; positive regulation of autophagy in response to ER overload [GO:0034263]; positive regulation of cell cycle [GO:0045787]; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process [GO:0043280]; positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway [GO:1902237]; positive regulation of fibroblast apoptotic process [GO:2000271]; positive regulation of intrinsic apoptotic signaling pathway [GO:2001244]; positive regulation of IRE1-mediated unfolded protein response [GO:1903896]; positive regulation of mitochondrial membrane permeability involved in apoptotic process [GO:1902110]; positive regulation of neuron apoptotic process [GO:0043525]; positive regulation of protein-containing complex assembly [GO:0031334]; positive regulation of release of cytochrome c from mitochondria [GO:0090200]; post-embryonic animal organ morphogenesis [GO:0048563]; regulation of apoptotic process [GO:0042981]; regulation of developmental pigmentation [GO:0048070]; regulation of organ growth [GO:0046620]; response to endoplasmic reticulum stress [GO:0034976]; spermatogenesis [GO:0007283]; spleen development [GO:0048536]; T cell homeostasis [GO:0043029]; thymocyte apoptotic process [GO:0070242]; thymus development [GO:0048538]; tube formation [GO:0035148] | 11859372_Functional study of the mouse homolog 11997495_Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. 12095614_direct addition of BimL to mitochondria does not lead to cytochrome c release 12118373_Bim has an ability to activate directly the voltage-dependent anion channel, which plays an important role in apoptosis of mammalian cells. 12198137_BimEL activate Bax by damaging the mitochondrial membrane structure directly, in addition to its binding and antagonizing Bcl-2/Bcl-XL function. 12486001_BIM has a role in facilitating HIV-1 tat-induced apoptosis 12844146_Data show that detachment-induced expression of Bim requires a lack of beta(1)-integrin engagement, downregulation of EGF receptor (EGFR) expression and inhibition of Erk signalling. 14527951_Bim mRNA and protein levels increase after up-regulation of FoxO3a by paclitaxel, causing apoptosis in breast cancer cells 14555991_phosphorylation of Bim-EL by Erk1/2 on serine 69 selectively leads to its proteasomal degradation and therefore represents a new and important mechanism of Bim regulation 14676826_The proapoptotic effect of BIM (through transcriptional induction of two of its isoforms)is inhibited by the activation of Raf/ERK signalling which prevents BIM up-regulation and leads to phosphorylation of the BimEL isoform. 14681225_new insights into the post-translational regulation of Bim(EL) 14732682_Bim(EL) can be activated downstream of the caspase cascade, leading to a positive feedback amplification of apoptotic signals. 14764673_A new pathway for Bim regulation is based on extracellular signal-regulated kinase-dependent phosphorylation of the BimEL isoform and proteasome-ddependent degradation of BimEL, followed by increased expression of shorter apoptotic isoforms BimL and BimS. 14970329_Immunosuppressive agents block Bim up-regulation and rescue T cells from death receptor-independent cell death. 14996839_Bim appears to be a key event in cAMP-promoted apoptosis in both murine and human T-cell lymphoma and leukemia cells and thus appears to be a convergence point for the killing of such cells by glucocorticoids and agents that elevate cAMP. 15014070_Mcl-1L degradation by either GrB or caspase-3 interferes with Bim sequestration by Mcl-1L 15030401_Lovastatin-induced cell death occurs in correlation with significantly increased levels of the BH3-only protein, Bim in susceptible glioblastoma cell lines; up-regulation of Bim was directly associated with increased incidence of apoptosis 15147734_expression pattern of Bim isoforms shows tissue specificity; the BH3 domain is sufficiency for proapoptotic activity; the functional state of Bims might be regulated both in the transcript and post transcript process 15378010_Cleavage of Mcl-1 by caspases modifies its subcellular localization, increases its association with Bim and inhibits its antiapoptotic function. 15459900_In myeloma cells, Mcl-1 neutralizes Bim through complex formation and therefore prevents apoptosis. 15486195_BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors 15509554_the induction of Bim by GC is a required event for the complete apoptotic response in pre-B ALL cells 15653751_T cell blasts surviving activation-induced cell deathare memory CD44 high cells with increased BIM expression. 15688014_activation of transcription is activated by FoxO3A 15711598_Bim is a critical molecular link between the microtubule poison, paclitaxel, and apoptosis. 15728578_The region unique to BimEL (amino acids 41-97, exon 3) accounts for ERK1/2 binding, ERK1/2-dependent phosphorylation, and turnover of BimEL. 15731037_degraded during Chlamydia trachomatis infection 15731089_degraded in Chlamydia trachomatis-infected cells 15767553_Bik and Bim have roles in bortezomib sensitization of cells to killing by death receptor ligand TRAIL 15824087_results show that Forkhead transcription factor 4-dependent expression of Bim protein plays a pivotal role for endothelial progenitor cell apoptosis 15843898_expression of Bim is increased by zinc pyrithione, which induces apoptosis 15899862_Bim is a critical regulator of luminal apoptosis during mammary acinar morphogenesis in vitro and may be an important target of oncogenes that disrupt glandular epithelial architecture. 16022280_identified the transcriptional initiation site and three candidate remote enhancer/silencer regions of the Bim gene. However, none of these transcriptional regulatory elements was IL-3-dependent 16051596_Repression of BIM is favored in human glioblastoma. 16091744_Melphalan-induced apoptosis in multiple myeloma cells is associated with a cleavage of MCL1 and BIM and a decrease in the MCL1/BIM complex. 16148027_Bax, Bad, and Bim are upregulated, while Bcl-2 is downregulated in human neuroblastoma cells treated with propargylamine 16183168_The muscarinic receptor-protein kinase C signaling pathway is a regulator of Bim in neuroblastoma cells; activation of muscarinic receptors and protein kinase C o induces Bim phosphorylation, followed by down-regulation of this proapoptotic protein. 16260615_TGF-beta is involved in the physiological loss of gastric epithelial cells by activating apoptosis mediated by Smad7, Bim, and caspase-9 16282323_Ser(87) of Bim(EL) is an important regulatory site that is targeted by Akt to attenuate the pro-apoptotic function of Bim(EL), thereby promoting cell survival 16373335_RUNX3 cooperates with FoxO3a/FKHRL1 to participate in the induction of apoptosis by activating Bim 16476732_concomitant induction of E2F1 targets ASK1 and Bim by HDACIs warrants an effective activation of E2F1-dependent apoptosis in response to suberoylanilide hydroxamic acid 16478725_Mcl-1 may serve as a direct substrate for TRAIL-activated caspases implying the existence of a novel TRAIL/caspase-8/Mcl-1/Bim communication mechanism between the extrinsic and the intrinsic apoptotic pathways 16773715_Ceramide induces apoptosis of human colon carcinoma HT-29 cells by affecting the expression of Bcl-2 family gene members and impacting the mitochondrial function. 16810067_Radiation therapy for squamous cell carcinoma of cervix results in increased apoptosis with the up-regulation of Bax, a proapoptotic protein, the down-regulation of Bcl-XL, an antiapoptotic protein, and no significant change in the levels of Bcl-2. 16888645_We conclude that Noxa and Bim establish a connection between FKHRL1 and mitochondria, and that both BH3-only proteins are critically involved in FKHRL1-induced apoptosis in neuroblastoma. 16951744_These results suggest that the high expression of cell adhesion-related proteins might be responsible for the different apoptosis status after the transfection of Bim L. 17051334_Bim plays an essential role in synergistic induction of apoptosis by SBHA and TRAIL in melanoma. 17062728_The main pathway by which Fas signaling regulates the levels of Bim expression in human T-cell blasts is the death-domain- and caspase-independent generation of discrete levels of H2O2, which results in the net increase of Foxo3a levels. 17067554_Inhibition of endogenously produced IGFBP-5 is associated with Bim-dependent apoptosis in NB cells. 17074758_in addition to interacting with the pro-apoptotic protein Bak, vaccinia F1L also functions to indirectly inhibit the activation of Bax, likely by interfering with the pro-apoptotic activity of BH3-only proteins such as BimL 17105963_TLR stimulation of macrophages can regulate Bim levels in opposing ways, namely by transcriptional induction and by phosphorylation-dependent degradation of the protein. 17151701_results implicate BIM in glucocorticoid-induced apoptosis in chronic lymphocytic leukemia cells through proteasome degradation. 17158029_results indicate that IFN-alpha causes apoptosis in myeloma cells through a moderate triggering of the mitochondrial route initiated by Bim 17188240_Our results confirm that ultrasound induces apoptosis via a pathway that involves Bak, Bcl-2, and caspases, but not ROS. 17203211_Bax and Bim are upregulated in human B cells during arsenic trioxide induction of apoptosis via the mitochondrial pathway 17218274_A cytokine-mediated posttranscriptional regulation of Bim mRNA by heat-shock cognate protein 70 (Hsc70), which binds to AU-rich elements (AREs) in the 3'-untranslated region of specific mRNAs and enhances their stability, is presented. 17251431_Bim is an essential mediator of amyloid-beta-induced neurotoxicity. 17276340_These data provide important mechanistic information on the processes involved in sculpting the mammary gland and demonstrate that BIM is a critical regulator of apoptosis in vivo. 17289999_results indicate that BH3-only proteins induce apoptosis at least primarily by engaging the multiple pro-survival relatives guarding Bax and Bak 17486061_Loss of expression of the pro-apoptotic protein Bim is associated with renal cell carcinoma 17499214_These results demonstrate that BimL is involved in UV irradiation-induced apoptosis by indirectly activating Bax. 17525735_preformed Bim(EL)/Mcl-1 and Bim(EL)/Bcl-x(L) complexes can be rapidly dissociated following activation of ERK1/2 by survival factors 17531220_FXa plays a key role in cellular processes in which Bim is the central player in determining cell survival. 17538248_a Bim-dependant pathway modulated by cytokines is involved in apoptosis of chronic myeloid leukemia cells induced by imatinib and nilotinib 17637819_Bim loss may play an important role in melanoma progression 17653091_In the absence of Bim, EBNA3A and EBNA3C appear to provide no survival advantage in Burkitt's lymphoma cells. 17686764_TRAIL can trigger an apoptotic pathway that involves JNK-dependent activation of Bim, which in turn induces Bax-mediated permeabilization of lysosomes. 17698840_Mcl-1 confers TRAIL resistance by serving as a buffer for Bak, Bim, and Puma, and sorafenib is a potential modulator of TRAIL sensitivity 17716672_Bim mediates particulate matter-induced apoptosis via mitochondrial pathway. 17717606_The open reading frame of BimSs3 may initiate at the second ATG and encodes a 36 amino-acid peptide with BH3 domain. 18006817_Mcl-1 degradation primes the cell for Bim and Bax activation and anoikis, which can be blocked by oncogenic signaling in metastatic cells 18063582_PINCH-1 contributes to apoptosis resistance through suppression of Bim. Mechanistically, PINCH-1 suppresses Bim not only transcriptionally but also post-transcriptionally. 18064628_Bim is a novel regulator of osteoblast apoptosis and may be a therapeutic target. 18089817_BIM is induced by lung cancer cell lines that are sensitive to erlotinib but not by those resistant. 18165867_In the absence of serum, the suppression of either Bad, Bim or Bid expression delayed cell death under several stress conditions 18182577_In addition to its proapoptotic effect, IL-21 promoted STAT1 and STAT5 phosphorylation in natural killer cells with concurrent enhanced antibody-dependent cellular cytotoxicity against rituximab-coated CLL cells 18195012_BIM and tBID follow different strategies to trigger BAX-driven mitochondrial outer membrane permeabilization with strong potency 18246127_Bad and Bim are major B-RAF responsive proteins regulating apoptosis in melanoma cells. 18251703_data indicate that Bim, Bak, and Bax actively mediate osteoblast apoptosis induced by trophic factor withdrawal 18345036_Bim-mediated apoptosis following actin damage due to deregulation of Cdk2 and the cell cycle by the absence of functional p53. 18348176_variations in c-myc and p21(WAF1) expression delay apoptosis making PBL resistant to sodium butyrate for several hours 18354037_arsenic trioxide upregulated expression of Bmf, Noxa, and Bim. Silencing of Bmf, Noxa, and Bim significantly protected MM cells from ATO-induced apoptosis 18391004_Activated extracellular signal-regulated kinase in epithelial cells infected with N. gonorrheae targeted Bad and Bim for downregulation at the protein level. 18398508_Data suggest that Bim-mediated attrition of HBV-specific CD8(+) T cells contributes to the inability of these cell populations to persist and control viral replication. 18413764_ABT-737 augments TRAIL-induced cell killing by unsequestering Bim and Bak and enhancing a Bax conformational change induced by TRAIL. 18420585_RACK1 has a role in protecting cancer cells from apoptosis by regulating the degradation of BimEL, which together with CIS could play an important role of drug resistance in chemotherapy 18508762_B-Raf-initiated inactivation of Bad and Bim only partly contributes to the anti-apoptotic activities of this oncogene and that other points within the cell death machinery are also targeted by deregulated ERK signaling 18549468_The identification of a novel putative human BCL2L11 promoter provides new insights into the structure and regulation of the BCL2L11 locus. 18651223_These findings suggest JNK to have an important pro-apoptotic function following ultraviolet rays B irradiation in human melanocytes, by acting upstream of lysosomal membrane permeabilization and Bim phosphorylation. 18668139_N-RAS(Q61K) and B-RAF(V600E) contribute to melanoma's resistance to apoptosis in part by downregulating Bim expression 18677096_Endogenous MIF may regulate the pro-apoptotic activity of Bim and inhibit the release of cytochrome c from mitochondria. 18715233_These data suggest that regulation of BIM expression by BRAF-->MEK-->ERK signaling is one mechanism by which oncogenic BRAF(V600E) can influence the aberrant physiology of melanoma cells. 18715501_BIM plays a significant role in T cell receptor (TCR)-induced death of activated human T cells, working in tandem with FAS signaling as a separate signal to kill T cells. 18806830_study concludes a single endogenous BRAF(V600E) allele is sufficient to repress BIM & prevent death from growth factor withdrawal; colorectal cancer cells with V600E mutations are addicted to the ERK1/2 pathway for repression of BIM 18812174_The crystal structure of Bfl-1, the last anti-apoptotic Bcl-2 family member to be structurally characterized, in complex with a peptide corresponding to the BH3 region of the pro-apoptotic protein Bim is presented. 18925930_In early severe sepsis a gene expression pattern with induction of pro-apoptotic Bcl-2 family members Bim and Bid is observed in peripheral blood. 18949058_Treatment of B-RAF mutant tumor cells with a MEK inhibitor requires BIM and is enhanced by a BH3 mimetic. 18981303_Scleroderma serum-induced EPC apoptosis is mediated chiefly by the Akt-FOXO3a-Bim pathway, which may account, at least in part, for the decreased circulating EPC levels in scleroderma patients. 19052714_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19064725_loss of Bim preserved clonal survival upon FLT3-ITD inhibition in hematopoietic progenitor cells, human leukemic cell lines, and primary AML cells 19100522_A proapoptotic signaling pathway involving RasGRP, Erk, and Bim in B cells 19101511_Bim is not absolutely required for paclitaxel cytotoxicity in breast cancer cell lines. 19141860_Observational study of gene-disease association. (HuGE Navigator) 19150432_betaTrCP promotes cell survival in cooperation with the ERK-RSK pathway by targeting BimEL for degradation. 19285955_This study demonstrated the physical association of the MCL-1 and IEX-1 proteins, the modulatory role of MCL-1 in IEX-1-induced apoptosis, and the role of BIM as an essential downstream molecule for IEX-1-induced cell death. 19300516_Our data indicate a synergistic role for both bim and Bmf in an apoptotic pathway leading to the clearance of Neisseria gonorrhoeae -infected cells. 19305426_destabilization of BIM(EL) in the absence of pVHL contributes to the increased resistance of VHL-null renal cell carcinoma (RCC) cells to certain apoptotic stimuli. 19308286_an HDAC inhibitor induces apoptosis through the FoxO1 acetylation-Bim pathway. 19330811_The Bax activator BimEL increased rapidly, driving cells towards apoptosis likely controlled by c-Myc and p21(WAF1) activities. 19332026_These data showed that BimL expression induced by BCR activation may result from the splicing of BimEL mRNA independently of Bim promoter regulation. 19336552_Observational study of gene-disease association. (HuGE Navigator) 19336552_Variants in BCL2L11 were strongly related to follicular lymphoma. 19403302_down-regulation of BIM is epigenetically controlled by methylation in a percentage of CML patients, has a bad prognosis, the combination of imatinib with a demethylating agent may result in improved responses in patients with decreased expression of BIM. 19407828_The survival cytokines (granulocyte-macrophage colony-stimulating factor and granulocyte colony-stimulating factor ) induce Bcl-2-like protein 11 gene expression in neutrophils. 19494111_TGFbeta stimulates Bim transcription by up-regulating the expression of the transcription factor Runx1 through an internal ribosome entry site -dependent mechanism. 19541822_Forodesine activates p53-independent mitochondrial apoptosis by induction of p73 and BIM. 19573080_Observational study of gene-disease association. (HuGE Navigator) 19574221_The rate of TGF-beta1-induced osteoclast apoptosis was lower when Bim expression was suppressed. 19587239_MAP4K3 orchestrates activation of BAX via the concerted posttranscriptional modulation of PUMA, BAD, and BIM. 19641506_Bim is involved in the regulation of cell death induction in many cells & tissues by various stimuli: growth factor or cytokine deprivation, calcium flux, ligation of antigen receptors on T & B cells, glucocorticoids or loss of adhesion. Review. 19737817_Toxoplasma gondii infection confers resistance against Bim-induced apoptosis by preventing the activation of mitochondrial Bax. 19773279_Observational study of gene-disease association. (HuGE Navigator) 19773546_Bim, and Mcl-1, but not Bad, integrate death signaling triggered by concomitant disruption of the PI3K/Akt and MEK1/2/ERK1/2 pathways in human leukemia cells. 19850739_KIT D816V suppresses expression of proapoptotic Bim in neoplastic mast cells 19881544_provide evidence that Notch3 regulates Bim, a BH-3-only protein, via MAPK signaling. 19893569_Data show that E2F1 induces EZH2 expression, which in turn antagonizes the induction of E2F1 pro-apoptotic target Bim expression. 19914305_Data indicate that Bim is the key mediator of dexamethasone-induced apoptosis and is responsible for the potentiating effect of rapamycin, providing molecular criteria for the use of glucocorticoids combined with mTOR inhibitors in myeloma therapy. 20010785_Data show an important role for the p73-Bim axis in regulating cell death during mitosis that is independent of p53. 20066663_the crystal structure of human Mcl-1 bound to a BH3 peptide derived from human Bim and the structures for three complexes that accommodate large physicochemical changes at conserved Bim sites was reported. 20074640_These results refine the minimal sequence for ERK1/2-driven degradation and further define the functional importance of key regions within BIM(EL), highlighting the complexity of this pro-apoptotic protein. 20082325_increase of the function of SIAH1 to upregulate the expression of Bim may play an important role in the progression of breast cancer 20086250_IGF-1 induces significant down-regulation of the proapoptotic BH3-only protein Bim in multiple myeloma cells 20160166_Bim is a key effector molecule in JAK2 inhibition-induced apoptosis and that targeting this apoptotic pathway could be a novel therapeutic strategy for patients with activating JAK2 mutations. 20206402_palmitoleate inhibits lipoapoptosis by blocking endoplasmic reticulum stress-associated increases of the BH3-only proteins Bim and PUMA 20231287_Up-regulation of BIM is associated with increased sensitivity to Imatinib in gastrointestinal stromal tumor. 20237869_the induction of BIM was a common apoptotic mechanism of epithelial tumor cells that depended upon survival signaling in targeted treatment, but not for chemotherapy 20332261_Observational study of gene-disease association. (HuGE Navigator) 20348947_the COX-2/PGE(2) pathway as an important negative regulator of Bim expression in colorectal tumours. 20404322_Mitochondrial p53 functions as a Bim derepressor by releasing Bim from sequestrating complexes with Mcl-1, Bcl-2, and Bcl-XL, and allowing its engagement in Bak/Bax activation. 20431602_The displacement of Bim protein from antiapoptotic proteins is the important step committing the cell to death. 20697841_the absence of BIM up-regulation is one of the important mechanisms of glucocorticoids resistance in acute lymphoblast leukemia 20700448_The data suggest that inhibitory targeting of vIRF-1:Bim interaction may provide an effective antiviral strategy. 20705940_Granulocyte/macrophage colony-stimulating factor causes a paradoxical increase in the BH3-only pro-apoptotic protein Bim in human neutrophils 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20841506_Follicular dendritic cells protect B-cell lymphoma cells against apoptosis, in part through activation of a miR-181a-dependent mechanism involving down-regulation of Bim expression 20855536_Observational study of gene-disease association. (HuGE Navigator) 20855536_We replicated the association of BCL2L11 and CASP9 with non-Hodgkin's lymphoma risk at the gene and SNP level, and identified novel associations with BCLAF1 and BAG5. 20861305_Data show that hypoxic conditions inhibit anoikis and block expression of proapoptotic BH3-only family members Bim and Bmf in epithelial cells. 20877573_CXCL12 chemokine regulates colorectal carcinoma cell anoikis through bim-mediated apoptosis. 20878914_In rheumatoid arthritis, the DNA-PKcs-JNK-Bim/Bmf axis transmits genotoxic stress into shortened survival of naive resting T cells, imposing chronic proliferative turnover of the immune system and premature immunosenescence. 20885957_Apoptosis induction by MEK1/2 inhibition in human lung cancer cells is mediated by Bim. 20959405_Observational study of gene-disease association. (HuGE Navigator) 21041309_BH3 peptides other than Bim and Bid exhibited various degrees of direct activation of the effector Bax or Bak 21130142_Upregulation of BIM by PRED in acute lymphoblastic leukemia cells regardless of molecular subtype is significantly prognostic of outcomes, confirming BIM's essential regulatory role in the PRED-induced apoptosis. 21148306_The BH3 alpha-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induces apoptosis in a Bax- and Bim-dependent manner. 21148799_Bim regulates the balance of survival versus apoptosis in peripheral T cells undergoing recombination-activating gene (RAG)-dependent T cell receptor rearrangements during TCR revision, thereby ensuring the utility of transgenic postrevision repertoire. 21247487_that Bim and Mcl-1 have key opposing roles in regulating JAK2V617F cell survival and propose that inactivation of aberrant JAK2 signaling leads to changes in Bim complexes that trigger cell death. 21333317_These results are consistent with the Epstein-Barr Virus miRNAs downregulating Bim post-transcriptionally in part through the 3'UTR and suggest that there are miRNA recognition sites within other areas of the Bim mRNA. 21339291_TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. 21367852_BH3-only activator proteins Bid and Bim are dispensable for Bax activation and mitochondrial apoptosis induced by Bcl-xL deficiency 21375523_belinostat and bortezomib interact synergistically in both cultured and primary AML and ALL cells, and raise the possibilities that up-regulation of Bim and interference with NF-kappaB pathways contribute to this phenomenon. 21415216_Data show that ALK inhibitor-induced apoptosis is mediated both by BIM upregulation resulting from inhibition of ERK signaling as well as by survivin downregulation resulting from inhibition of STAT3 signaling in EML4-ALK-positive lung cancer cells. 21481848_During chronic HCV infection non-reactive HCV-specific CD8(+) cells targeting the virus are PD-1(+)/CD127(-)/Bim(+) and, blocking apoptosis and PD-1/PD-L1 pathway on them enhances in vitro reactivity. 21555518_BCL2L11 is a target gene of the BACH1 transcription factor according to ChIP-seq analysis in HEK 293 cells. 21625639_Loss of Bim expression is associated with glioma. 21655183_Findings suggest that BIM plays an important role in regulating p-AKT by activating caspase-3 and that BIM mediates the level of AKT phosphorylation to determine the threshold for overcoming cisplatin resistance in ovarian cancer cells. 21659544_Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. 21671007_Bim is required for the combretastatin-A4 induced cell death in the H460 lung cancer cell line via activation of the mitochondrial signalling pathway. 21673341_A pathway through c-Raf to Bim that contributes to tipifarnib cytotoxicity in human lymphoid cells but also identify potential determinants of sensitivity to this agent. 21729544_Anti-CD44 mAb A3D8 can inhibit proliferation and induce apoptosis of HL-60 cells by enhancing expression of Bim. 21732363_These results suggest that 18beta-glycyrrhetinic acid induces apoptosis in human breast carcinoma MCF-7 cells via caspase activation and modulation of Akt/FOXO3a/Bim pathway. 21808067_Up-regulation of Bim appears to be an important determinant of cAMP/PKA-mediated apoptosis in immature T cells 21899728_breast cancer cells overexpressing HER2 undergo apoptosis upon depletion of Mcl-1, and this Mcl-1 dependence is due to their constitutive expression of the pro-apoptotic protein Bim 21924351_CDK1-dependent phosphorylation of BIM(EL) drives its polyubiquitylation and proteasome-dependent degradation to protect cells during mitotic arrest 21937453_Cytokines tumor necrosis factor-alpha and interferon-gamma induce pancreatic beta-cell apoptosis through STAT1-mediated Bim protein activation. 21958719_The mRNA and protein expression of an isoform of Bim (Bcl-2 interacting mediator of cell death) denoted as BimEL, which is proapoptotic was increased following efavirenz administration. 21984578_Findings suggest that local IFN production may interact with a genetic factor (PTPN2) to induce aberrant proapoptotic activity of the BH3-only protein Bim, resulting in increased beta-cell apoptosis via JNK activation and the intrinsic apoptotic pathway. 22071694_Cdk1/cyclin B1-dependent hyper-phosphorylation of Bim during prolonged mitotic arrest is an important cell death signal. 22076535_miR-25 directly regulates apoptosis by targeting Bim in ovarian cancer. 22081075_for optimal tumor suppressive activity, Bim must be able to interact with all and not just select pro-survival Bcl-2 family members 22093624_The lack of Bim up-regulation contributes to the resistance of melanoma cells to ER stress-induced apoptosis and may be a mechanism by which melanoma cells adapt to ER stress conditions. 22131152_Bim overexpression was not correlated with Bcl-2 inhibition (P > 0.05) and was accompanied by increase in Bax expression (P < 0.05). 22145099_BIM RNA levels in EGFR-mutant lung cancer specimens predicted response and duration of clinical benefit from EGFR inhibitors. 22160382_BIM phosphorylation appears to play a key role in apoptosis regulation in chronic lymphocytic leukemia cells, potentially coordinating antigen and microenvironment-derived survival signals. 22245094_during skin hypoxia, promotes UVB-induced apoptosis in keratinocytes 22270368_Depletion of Mcl-1 sensitized tumor cells to MEK inhibitor-induced cell death, an effect that was antagonized by knockdown of Bim. 22349704_data demonstrate that FOXO3 activates overproduction of reactive oxygen species as a consequence of Bim-dependent impairment of mitochondrial respiration in neuronal cells, which leads to apoptosis 22391963_Investigated the function of Bim during DHA-induced apoptosis in human lung adenocarcinoma cell lines.Confocal imaging showed the translocation of Bim to endoplasmic reticulum (ER) rather than mitochondria during DHA-induced apoptosis. 22426421_the heterogeneity of tyrosine kinase inhibitors responses across individuals and suggest the possibility of personalizing therapy with BH3 mimetics to overcome BIM-polymorphism-associated tyrosine kinase inhibitors resistance. 22430213_Novel but opposing roles for the p53 and TAp73 in the induction of Noxa and Bim and regulation of apoptosis. 22516966_Report upregulation of Bim and the splicing factor SRp55 in melanoma cells from patients treated with selective BRAF inhibitors. 22572381_DEC2 regulates apoptosis in oral cancer cells via regulation of pro-apoptotic factor Bim expression 22609401_These results show a novel role played by the Bcl-xL:Bim interaction in regulating proliferation of pancreatic cancer cells at the expense of apoptosis. 22622039_A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. 22628193_Bim re-expression reduced pancreatic cancer cell proliferation induced by miR-301a. 22645134_TRAIL induces recruitment of PACS-2 to lysosomes where it interacts with Bim and Bax to permeabilize lysosomes and trigger apoptosis. 22742832_Bim inhibits autophagy by interacting with autophagy regulator Beclin 1, an interaction facilitated by LC8. 22754333_Pokemon prevents anoikis through the suppression of Bim expression, which facilitates tumor cell invasion and metastasis. 22762551_our data suggest that Bam is either not translated at all in vivo or is translated at a very low rate that might be insufficient to induce cell death. 22788963_Bim expression in the NSCLCs was associated with both squamous cell carcinoma histology and tumor proliferation. 22802411_These results provide a novel causal link between SDF-1alpha-induced chemotaxis, degradation of Bim(EL), and the development of CD4 T cell memory. 22819074_Decreased Bim expression is associated with multidrug-resistance in multiple myeloma. 22895567_findings demonstrate that the high expression of miR-92a in glioma is significantly correlated with low levels of BCL2L11 (Bim) protein and high-grade glioma 22905229_Overexpression of MN1 confers resistance to chemotherapy, accelerates leukemia onset, and suppresses p53 and Bim induction 23012423_Data show a link between the ERK1/2 pathway and BIM expression through miR-494. 23047821_Data indicate that BCL2L11 rs3789068 was associated with an increased risk for B-cell non-Hodgkin lymphoma (NHL), and PRRC2A rs3132453 conferred a reduced risk of B-cell NHL. 23138847_Dihydroartemisinin was associated with the increased expression of the pro-apoptotic gene Bim and a decreased expression of the anti-apoptotic gene Bcl-2 in breast cancer cells. 23152053_Proteasome inhibition upregulates Bim and induces caspase-3-dependent apoptosis in mast cells expressing the Kit D816V mutation. 23152504_Bim, a proapoptotic protein, up-regulated via transcription factor E2F1-dependent mechanism, functions as a prosurvival molecule in cancer 23235460_Bcl-B in complex with the BH3 motif of Bim protected cells from Bax-dependent apoptotic pathways. 23243017_Findings suggest that inhibiton of Bcl-2, Bcl-xL and PI3K, and release of Bim from Bcl-2/Bcl-xL and GSK3alpha/beta culminating in Bax/Bak activation and apoptosis. 23270470_Loss of BIM expression is associated with drug resistance in non-small cell lung cancer. 23295054_A significant increase in Bim mRNA in Primary Biliary Cirrhosis was observed, with the most pro-apoptotic isoform dominating in livers of PBC patients. 23301543_A low Mcl-1 expression and Bim upregulation after antigen encounter are involved in CD127(low) HCV-specific cytotoxic T cell hyporeactivity during chronic infection. 23358890_Bcl-2 overexpression induced up-regulation of the proapoptotic protein Bim in lung cancer cells. 23402819_BimL reveals an outstanding proapoptotic potential in melanoma cells. 23500081_Transcriptional and post-translational regulation of Bim is essential for TGF-beta and TNF-alpha-induced apoptosis of gastric cancer cell. 23532334_BIM and PUMA are induced upon HER2 inhibition in HER2-amplified breast cancers to execute apoptosis. 23569333_[review] BIM plays a critical role in viral-specific T cell death | ENSMUSG00000027381 | Bcl2l11 | 1096.55112 | 1.1018325 | 0.1399048779 | 0.10210677 | 1.883229e+00 | 1.699676e-01 | 5.626522e-01 | No | Yes | 1272.153431 | 115.321056 | 1103.210402 | 98.016924 | |
| ENSG00000153976 | 9955 | HS3ST3A1 | protein_coding | Q9Y663 | FUNCTION: Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) to catalyze the transfer of a sulfo group to an N-unsubstituted glucosamine linked to a 2-O-sulfo iduronic acid unit on heparan sulfate (PubMed:10520990, PubMed:9988768, PubMed:10608887, PubMed:15304505). Catalyzes the O-sulfation of glucosamine in IdoUA2S-GlcNS and also in IdoUA2S-GlcNH2 (PubMed:10520990, PubMed:9988768, PubMed:15304505). The substrate-specific O-sulfation generates an enzyme-modified heparan sulfate which acts as a binding receptor to Herpes simplex virus-1 (HSV-1) and permits its entry (PubMed:10520990). Unlike HS3ST1/3-OST-1, does not convert non-anticoagulant heparan sulfate to anticoagulant heparan sulfate (PubMed:10520990). {ECO:0000269|PubMed:10520990, ECO:0000269|PubMed:10608887, ECO:0000269|PubMed:15304505, ECO:0000269|PubMed:9988768}. | 3D-structure;Disulfide bond;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | Heparan sulfate biosynthetic enzymes are key components in generating a myriad of distinct heparan sulfate fine structures that carry out multiple biologic activities. The enzyme encoded by this gene is a member of the heparan sulfate biosynthetic enzyme family. It is a type II integral membrane protein and possesses heparan sulfate glucosaminyl 3-O-sulfotransferase activity. The sulfotransferase domain of this enzyme is highly similar to the same domain of heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1, and these two enzymes sulfate an identical disaccharide. This gene is widely expressed, with the most abundant expression in liver and placenta. [provided by RefSeq, Dec 2014]. | hsa:9955; | Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity [GO:0008467]; [heparan sulfate]-glucosamine 3-sulfotransferase 3 activity [GO:0033872]; sulfotransferase activity [GO:0008146]; glycosaminoglycan biosynthetic process [GO:0006024] | 20487506_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 22475533_Genetic variants of HS3ST3A1 and HS3ST3B1 are associated with Plasmodium falciparum parasitaemia. 26410339_decreased in pre-eclamptic placental tissue 27041583_Our findings define 3-OST3A as a novel regulator of breast cancer pathogenicity, displaying tumor-suppressive or oncogenic activities in a cell- and tumor-dependent context, and demonstrate the clinical value of the HS-O-sulfotransferase 3-OST3A as a prognostic marker in HER2+ patients. 28618938_Detection of MUC1 and HS3ST2 promoter methylation status appears to be useful molecular markers for assessing the progressive state of the disease and could be helpful in discriminating breast cancer molecular subtypes. These results validate the methylation-based microarray analysis, thus trust their output in the future. | ENSMUSG00000047759 | Hs3st3a1 | 435.13984 | 0.8663609 | -0.2069599548 | 0.17193113 | 1.441541e+00 | 2.298901e-01 | 6.206095e-01 | No | Yes | 510.668549 | 69.985560 | 575.408238 | 76.823580 | |
| ENSG00000154222 | 200014 | CC2D1B | protein_coding | Q5T0F9 | FUNCTION: Transcription factor that binds specifically to the DRE (dual repressor element) and represses HTR1A gene transcription in neuronal cells. {ECO:0000269|PubMed:19423080}. | Alternative splicing;Coiled coil;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | hsa:200014; | intracellular membrane-bounded organelle [GO:0043231]; nuclear envelope [GO:0005635]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 17084358_Paper describes a function of Cc2d1b/Cc2d1a and their Drosophila homologue l(2)gd in D.melanogaster in Notch trafficking. 19423080_Human Freud-2/CC2D1B binds to the 5-HT1A dual repressor element and represses the human 5-HT1A receptor gene to regulate its expression in non-serotonergic cells and neurons. 27769858_CC2D1B regulates degradation and signaling of EGFR and TLR4. 30513301_CC2D1B is essential for ESCRT-III activity during mitotic reformation of the nuclear envelope. 33034024_Unravelling of Hidden Secrets: The Tumour Suppressor Lethal (2) Giant Discs (Lgd)/CC2D1, Notch Signalling and Cancer. | ENSMUSG00000028582 | Cc2d1b | 1166.67992 | 0.9680231 | -0.0468865598 | 0.12037434 | 1.516385e-01 | 6.969744e-01 | 9.074409e-01 | No | Yes | 1449.504122 | 193.653630 | 1474.847756 | 192.319266 | ||
| ENSG00000154545 | 728239 | MAGED4 | protein_coding | Q96JG8 | FUNCTION: May enhance ubiquitin ligase activity of RING-type zinc finger-containing E3 ubiquitin-protein ligases. Proposed to act through recruitment and/or stabilization of the Ubl-conjugating enzyme (E2) at the E3:substrate complex. {ECO:0000269|PubMed:20864041}. | Alternative splicing;Reference proteome;Tumor antigen;Ubl conjugation pathway | hsa:728239;hsa:81557; | nucleus [GO:0005634]; negative regulation of transcription by RNA polymerase II [GO:0000122] | 24147998_Results suggest that MAGE-D4 overexpression influences tumor progression, and MADE-D4 can be a prognostic marker and a potential molecular target in squamous cell esophageal cancer. 24870746_MAGED4 (mRNA and protein) is frequently and highly expressed in glioma and is partly regulated by DNA methylation. 24966945_MAGE-D4 is highly expressed in glioma and can develop specifically humoral response in glioma patients. 25120768_These findings suggest MAGE-D4 may serve as a potentially prognostic biomarker and an attractive target of immunotherapy in colorectal cancer 25951896_Preoperative serum MAGED4 was significantly higher in esophageal squamous cell carcinoma patients than in controls. 28031124_The MAGE-D4 polyclonal antibody with high specificity and sensitivity has been successfully prepared. 31704827_High MAGED4 expression is associated with Esophageal Squamous Cell Carcinoma. | 356.36742 | 0.3470405 | -1.5268240042 | 2.44355443 | 3.510995e-01 | 5.534914e-01 | 8.490478e-01 | No | Yes | 150.368217 | 131.619711 | 431.295936 | 367.006044 | ||||
| ENSG00000154710 | 27342 | RABGEF1 | protein_coding | Q9UJ41 | FUNCTION: Rab effector protein acting as linker between gamma-adaptin, RAB4A or RAB5A. Involved in endocytic membrane fusion and membrane trafficking of recycling endosomes. Stimulates nucleotide exchange on RAB5A. Can act as a ubiquitin ligase (By similarity). {ECO:0000250, ECO:0000269|PubMed:11452015, ECO:0000269|PubMed:15339665, ECO:0000269|PubMed:9323142}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Endocytosis;Endosome;Metal-binding;Phosphoprotein;Protein transport;Reference proteome;Transport;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | RABGEF1 forms a complex with rabaptin-5 (RABPT5; MIM 603616) that is required for endocytic membrane fusion, and it serves as a specific guanine nucleotide exchange factor (GEF) for RAB5 (RAB5A; MIM 179512) (Horiuchi et al., 1997 [PubMed 9323142]).[supplied by OMIM, Mar 2010]. | hsa:27342; | cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; nucleolus [GO:0005730]; recycling endosome [GO:0055037]; DNA binding [GO:0003677]; guanyl-nucleotide exchange factor activity [GO:0005085]; small GTPase binding [GO:0031267]; ubiquitin protein ligase activity [GO:0061630]; zinc ion binding [GO:0008270]; endocytosis [GO:0006897]; negative regulation of inflammatory response [GO:0050728]; negative regulation of interleukin-6 production [GO:0032715]; negative regulation of Kit signaling pathway [GO:1900235]; negative regulation of leukocyte migration [GO:0002686]; negative regulation of mast cell degranulation [GO:0043305]; negative regulation of protein phosphorylation [GO:0001933]; negative regulation of Ras protein signal transduction [GO:0046580]; negative regulation of receptor-mediated endocytosis [GO:0048261]; protein targeting to membrane [GO:0006612]; regulation of Fc receptor mediated stimulatory signaling pathway [GO:0060368] | 12505986_GGAs, a family of Arf-dependent clathrin adaptors involved in selection of TGN cargo, interact with the Rabaptin-5-Rabex-5 complex, a Rab4/Rab5 effector regulating endosome fusion 16499958_Here, we report that Rabex-5, a guanine nucleotide exchange factor for Rab5, binds to ubiquitin through two independent ubiquitin binding domains 17341663_Fc epsilon RI-mediated mast cell functional activation is dependent on RabGEF1's GEF activity 17450153_crystal structure of the RABEX-5 catalytic core in complex with nucleotide-free RAB21, a key intermediate in the exchange reaction pathway 18160707_Integration of TGFB and RAS signaling silences RABGEF1 and enhances growth factor-directed cell migration. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20655225_functions as an E3 ligase for Ras 21500550_RABEX-5 and RAB5 may be involved in the development of breast cancer metastasis. 22846990_a novel mechanistic insight into how Rabex-5 regulates internalization and postendocytic trafficking of ubiquitinated L1 destined for lysosomal degradation. 22952906_KCa2.3 is localized to a caveolin-rich domain within the plasma membrane and is endocytosed in a dynamin- and Rab5-dependent manner. 23048039_Interaction of Rabex-5 with Rab5 depends on interaction of the MIU domain with the ubiquitinated L1 to drive its internalization. 24714377_This implies that Rap regulates endothelial barrier function by dual control of cytoskeletal tension. The molecular details of the signaling pathways are becoming to be elucidated 24716822_RABEX-5 mRNA expression is a strong predictor of poor prognosis in prostate cancer patients treated by radical prostatectomy. 24957337_analysis of Rabex-5 GEF activation by Rabaptin-5 25427001_Expression of RABEX-5 is significantly higher in gastric cancer tissues and is associated with tumor size and lymph node metastasis. 26002576_RABEX-5 is a potential useful indicator and predicts a poor long-term prognosis for small cell lung cancer(SCLC), which should be considered in defining the prognosis with other well-known prognosticators in C-SCLC patients 26430212_The results contradict the model of feedback activation of Rab5 and instead indicate that Rbpt5 is recruited by both Rabex5 recognizing ubiquitylated cargo and by Rab4 to activate Rab5 in a feed-forward manner. 27601648_we identify Rabex-5 as a Immunomodulatory drugs (IMiDs) target molecule that functions to restrain TLR activated auto-immune promoting pathways. We propose that release of Rabex-5 from complex with Cereblon enables the suppression of immune responses, contributing to the antiinflammatory properties of IMiDs. 27791468_RABGEF1 mediates recycling endosome fusion with GAS-containing autophagosome-like vacuoles through the STX6-VAMP3-VTI1B complex; SNAREs are involved in autophagosome formation in response to bacterial infection 27820702_findings reveal a key role for RABGEF1 in dampening keratinocyte-intrinsic MYD88 signaling and sustaining epidermal barrier function 28617553_RABEX-5 mRNA expression was significantly upregulated in colorectal cancer tissues. 29360040_The authors demonstrate that RABGEF1, the upstream factor of the endosomal Rab GTPase cascade, is recruited to damaged mitochondria via ubiquitin binding downstream of Parkin. RABGEF1 directs the downstream Rab proteins, RAB5 and RAB7A, to damaged mitochondria, whose associations are further regulated by mitochondrial Rab-GAPs. 33166495_RabGEF1 functions as an oncogene in U251 glioblastoma cells and is involved in regulating AKT and Erk pathways. | ENSMUSG00000025340 | Rabgef1 | 953.72699 | 0.9183149 | -0.1229391073 | 0.13120363 | 8.746614e-01 | 3.496681e-01 | 7.232085e-01 | No | Yes | 895.377211 | 132.101126 | 924.820722 | 133.512347 | |
| ENSG00000154814 | 92106 | OXNAD1 | protein_coding | Q96HP4 | NAD;Oxidoreductase;Reference proteome;Signal | hsa:92106; | oxidoreductase activity [GO:0016491] | 20877624_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000021906 | Oxnad1 | 789.72634 | 0.9249269 | -0.1125886806 | 0.11741323 | 9.171391e-01 | 3.382273e-01 | 7.138357e-01 | No | Yes | 954.636810 | 153.633885 | 987.639572 | 155.337056 | |||
| ENSG00000155275 | 152992 | TRMT44 | protein_coding | Q8IYL2 | FUNCTION: Probable adenosyl-L-methionine (AdoMet)-dependent tRNA (uracil-O(2)-)-methyltransferase. {ECO:0000250}. | Alternative splicing;Cytoplasm;Metal-binding;Methyltransferase;Phosphoprotein;Reference proteome;S-adenosyl-L-methionine;Transferase;Zinc;Zinc-finger;tRNA processing | The protein encoded by this gene is a putative tRNA methyltransferase found in the cytoplasm. Defects in this gene may be a cause of partial epilepsy with pericentral spikes (PEPS), but that has not been proven definitively. [provided by RefSeq, May 2012]. | hsa:152992; | cytoplasm [GO:0005737]; metal ion binding [GO:0046872]; tRNA (uracil) methyltransferase activity [GO:0016300]; tRNA (uracil-2'-O-)-methyltransferase activity [GO:0052665]; tRNA methylation [GO:0030488] | 21658913_This study postulated that Q8IYL2 is a causative gene for PEPS , after exhaustive resequencing and bioinformatic analysis. | ENSMUSG00000029097 | Trmt44 | 654.83006 | 0.9615013 | -0.0566392341 | 0.12287686 | 2.121724e-01 | 6.450700e-01 | 8.882851e-01 | No | Yes | 740.824538 | 57.414267 | 756.585852 | 57.266318 | |
| ENSG00000155744 | 285172 | FAM126B | protein_coding | Q8IXS8 | FUNCTION: Component of a complex required to localize phosphatidylinositol 4-kinase (PI4K) to the plasma membrane. {ECO:0000305|PubMed:26571211}. | Cell membrane;Cytoplasm;Membrane;Phosphoprotein;Reference proteome | hsa:285172; | cytosol [GO:0005829]; plasma membrane [GO:0005886]; phosphatidylinositol phosphate biosynthetic process [GO:0046854]; protein localization to plasma membrane [GO:0072659] | ENSMUSG00000038174 | Fam126b | 1043.96227 | 1.3166219 | 0.3968410990 | 0.12412472 | 1.028892e+01 | 1.338311e-03 | 6.278470e-02 | No | Yes | 1422.152134 | 331.040945 | 1015.423426 | 231.606680 | |||
| ENSG00000155749 | 130540 | FLACC1 | protein_coding | Q96Q35 | Acetylation;Alternative splicing;Cell projection;Cilium;Coiled coil;Cytoplasm;Flagellum;Reference proteome | hsa:130540; | cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; outer dense fiber [GO:0001520]; sperm fibrous sheath [GO:0035686]; sperm flagellum [GO:0036126] | 18649358_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) 19773279_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21048031_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 25168388_Genome-wide association studies show a significant association between the intronic SNP rs1830298 in ALS2CR12 gene and breast cancer; further expression studies suggest that CASP8 might be the target gene. 25855136_rs13014235 is associated with susceptibility to cutaneous basal cell carcinoma. | ENSMUSG00000047528 | Flacc1 | 17.98910 | 1.2777633 | 0.3536206512 | 0.67430078 | 2.751924e-01 | 5.998697e-01 | No | Yes | 18.712185 | 10.481058 | 14.826721 | 8.154373 | ||||
| ENSG00000155974 | 23426 | GRIP1 | protein_coding | Q9Y3R0 | FUNCTION: May play a role as a localized scaffold for the assembly of a multiprotein signaling complex and as mediator of the trafficking of its binding partners at specific subcellular location in neurons (PubMed:10197531). Through complex formation with NSG1, GRIA2 and STX12 controls the intracellular fate of AMPAR and the endosomal sorting of the GRIA2 subunit toward recycling and membrane targeting (By similarity). {ECO:0000250|UniProtKB:P97879, ECO:0000269|PubMed:10197531}. | 3D-structure;Alternative splicing;Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoplasmic vesicle;Endoplasmic reticulum;Membrane;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Repeat;Synapse | This gene encodes a member of the glutamate receptor interacting protein family. The encoded scaffold protein binds to and mediates the trafficking and membrane organization of a number of transmembrane proteins. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, May 2010]. | hsa:23426; | cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; dendrite [GO:0030425]; endoplasmic reticulum membrane [GO:0005789]; neuron projection [GO:0043005]; perikaryon [GO:0043204]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; postsynaptic membrane [GO:0045211]; beta-catenin binding [GO:0008013]; glucocorticoid receptor binding [GO:0035259]; protein C-terminus binding [GO:0008022]; signaling receptor complex adaptor activity [GO:0030159]; dendrite development [GO:0016358]; intracellular signal transduction [GO:0035556]; neurotransmitter receptor transport, endosome to postsynaptic membrane [GO:0098887]; positive regulation of neuron projection arborization [GO:0150012] | 12011465_after regulated endocytosis, binding to GRIP/ABP stabilizes the internalized receptors in an intracellular pool and prevents them from being recycled back to the plasma membrane or entering a degradative pathway. 15226318_GRIP1c 4-7 plays a role not only in glutamatergic synapses but also in GABAergic synapses 17303296_Observational study of gene-disease association. (HuGE Navigator) 17303296_study showed neither single marker nor haplotype analysis revealed an association between variants at GRIP1 locus & schizophrenia; suggests it is unlikely that the GRIP1 polymorphisms investigated play a substantial role in schizophrenia susceptibility 18155042_Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1. 18315564_GRIP1 splice forms interact with gephyrin and play a role in synaptic function at GABAergic and glycinergic synapses in cultured hippocampal neurons. 18950845_Observational study of gene-disease association. (HuGE Navigator) 19086053_Observational study of gene-disease association. (HuGE Navigator) 19736351_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20877300_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21383172_Gain-of-function glutamate receptor interacting protein 1 variants alter GluA2 recycling and surface distribution in patients with autism 22510445_In three unrelated families with parental consanguinity, GRIP1 mutations were found to segregate with Fraser syndrome in an autosomal recessive manner. 24700879_In 15 of 590 families, we identified recessive mutations in the genes FRAS1, FREM2, GRIP1, FREM1, ITGA8, and GREM1, all of which function in the interaction of the ureteric bud and the metanephric mesenchyme. 25673849_the Trip6-GRIP1-myosin VI interaction and its regulation on F-actin network play a significant role in dendritic morphogenesis 27425598_GRIP1 was identified as one of the most important differentially expressed, topologically significant proteins in the protein-protein interaction network in Alzheimer's disease. 27941904_Data suggest that molecules in the erythropoietin-producing hepatocellular receptor B family (EPHB) / ephrinB (EFNB) signalling pathways, specifically ephrin B3 and GRIP1, are involved blood pressure regulation. 29170386_GRIP1 is phosphorylated at an N-terminal serine cluster by cyclin-dependent kinase-9 (CDK9), which is recruited into GC-induced GR:GRIP1:CDK9 hetero-complexes, producing distinct GRE-specific GRIP1 phospho-isoforms. 31757889_The adaptor proteins HAP1a and GRIP1 collaborate to activate the kinesin-1 isoform KIF5C. | ENSMUSG00000034813 | Grip1 | 275.45195 | 0.7991978 | -0.3233754797 | 0.20558799 | 2.441889e+00 | 1.181339e-01 | 4.879879e-01 | No | Yes | 396.334804 | 110.266928 | 468.246164 | 127.714990 | |
| ENSG00000156042 | 118491 | CFAP70 | protein_coding | Q5T0N1 | FUNCTION: Axoneme-binding protein that plays a role in the regulation of ciliary motility and cilium length. {ECO:0000250|UniProtKB:D3YVL2}. | Alternative splicing;Cell projection;Cilium;Cytoplasm;Cytoskeleton;Disease variant;Flagellum;Reference proteome;Repeat;TPR repeat | Mouse_homologues mmu:76670; | axoneme [GO:0005930]; ciliary basal body [GO:0036064]; extracellular exosome [GO:0070062]; outer dynein arm [GO:0036157]; sperm flagellum [GO:0036126]; cilium assembly [GO:0060271]; cilium movement [GO:0003341] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 31621862_CFAP70 mutations lead to male infertility due to severe astheno-teratozoospermia. A case report. | ENSMUSG00000039543 | Cfap70 | 67.75426 | 0.4814667 | -1.0544919687 | 0.38993150 | 7.006809e+00 | 8.120026e-03 | No | Yes | 56.700021 | 24.316871 | 123.598567 | 51.585381 | |||
| ENSG00000156136 | 1633 | DCK | protein_coding | P27707 | FUNCTION: Phosphorylates the deoxyribonucleosides deoxycytidine, deoxyguanosine and deoxyadenosine (PubMed:1996353, PubMed:12808445, PubMed:18377927, PubMed:19159229, PubMed:20614893). Has broad substrate specificity, and does not display selectivity based on the chirality of the substrate. It is also an essential enzyme for the phosphorylation of numerous nucleoside analogs widely employed as antiviral and chemotherapeutic agents (PubMed:12808445). {ECO:0000269|PubMed:12808445, ECO:0000269|PubMed:18377927, ECO:0000269|PubMed:19159229, ECO:0000269|PubMed:1996353, ECO:0000269|PubMed:20614893}. | 3D-structure;ATP-binding;Direct protein sequencing;Kinase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase | Deoxycytidine kinase (DCK) is required for the phosphorylation of several deoxyribonucleosides and their nucleoside analogs. Deficiency of DCK is associated with resistance to antiviral and anticancer chemotherapeutic agents. Conversely, increased deoxycytidine kinase activity is associated with increased activation of these compounds to cytotoxic nucleoside triphosphate derivatives. DCK is clinically important because of its relationship to drug resistance and sensitivity. [provided by RefSeq, Jul 2008]. | hsa:1633; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; deoxyadenosine kinase activity [GO:0004136]; deoxycytidine kinase activity [GO:0004137]; deoxyguanosine kinase activity [GO:0004138]; deoxynucleoside kinase activity [GO:0019136]; protein homodimerization activity [GO:0042803]; dAMP salvage [GO:0106383]; pyrimidine nucleotide metabolic process [GO:0006220] | 11830489_alternatively spliced dCK forms found in acute myeloid leukemia cells play an important role in cytarabine resistance 11952160_Molecular basis of 2',3'-dideoxycytidine-induced drug resistance in human cells. 12535661_Data show that inorganic tripolyphosphate (PPP(i)) is a good donor for human ceoxycytidine kinase and deoxyguanosine kinase. 14514691_human deoxycytidine kinase promoter activity is regulated by USF and Sp1 15561147_dCK can act as a phosphorylase, similar to the nucleoside phosphorylase family of enzymes 15564883_Observational study of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15571255_analysis of antitumor drug binding to deoxycytidine kinase 15571257_dCK expression varies between individual samples and between different types of malignancies and may play a role in resistance to ara-C in particular tumor types 15571258_deoxycytidine kinase activity is stimulated by 2-chlorodeoxyadenosine and aphidicolin in the cellular context 15571259_deoxycytidine kinase activity is regulated by reversible phosphorylation 15978330_an increased expression of mRNA, specific for thymidine kinase 1, dCK and thymidine phosphorylase, may be involved in carcinogenic processes in the human thyroid 16361699_dCK activity can be controlled by phosphorylation in intact cells, and Ser-74 is required for activity 16401075_Crystal structures of a deoxycytidine kinase variant lacking a flexible insert (residues 65-79) reveal major changes in the donor base binding loop (residues 240-247) between UDP-bound and ADP-bound forms, involving significant main-chain rearrangement. 16799820_Observational study of genotype prevalence. (HuGE Navigator) 16969512_an increase in activity of dCK, TK1 and 2 might be involved in an adaptive response of cultured human squamous lung carcinoma cells to radiation by facilitation of DNA repair 17065053_deoxycytidine kinase has a role in lymphoma cell sensitivity to cladribine 17065079_analysis of phosphorylation sites on human deoxycytidine kinase 17065080_analysis of deoxycytidine kinase reversible phosphorylation in normal human lymphocytes 17065085_deoxycytidine kinase activity is enhanced after pulsed low dose rate and single dose gamma irradiation 17065091_deoxycytidine kinase, deoxyguanosine kinase, and cytosolic 5'-nucleotidase I are regulated in a cell cycle-dependent manner in MOLT-4 cells 17602053_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 17855478_study identified novel coding, promoter, intronic, and 3'-UTR genetic variants at the DCK locus in two major ethnic groups; results suggest that genetic variation in DCK influences its activity and expression 18258203_Phosphorylation may represent a mechanism to enhance the catalytic activity of the relatively slow dCK enzyme. 18361501_illuminate the key contributions of these two amino acid positions to enzyme function by demonstrating their ability to moderate substrate specificity 18600530_dCK and dGK were downregulated by approximately 70% in CEM cells and tested against six nucleoside 18600545_both deoxycytidine kinase and adenosine kinase are involved in this model of ADA deficiency 18775979_In leukoblasts from 82 patients with acute myeloid leukemia, various extent and frequency of differential allelic expression in the CDA, DCK, NT5C2, NT5C3, and TP53 genes was observed. 18974616_29 variations including 20 novel ones were identified in DCK from 256 Japanese cancer patients administered gemcitabine. 18974616_Observational study of gene-disease association. (HuGE Navigator) 19159229_Several hydrophobic residues at position 104 endow dCK with thymidine kinase activity. 19287976_dCK can regulate the in vitro cellular response to Ara-C in acute myeloid leukemia cells 19428333_The DCK/cN-II ratio was again proportional to ara-CTP production and to ara-C sensitivity. 19568409_thymidylate synthase/ribonucleotide reductase gene silencing and deoxycytidine kinase::uridine monophosphate kinase fusion gene gene overexpression markedly improved gemcitabine's therapeutic activity 20028759_Clinical trial of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20043109_Sensitivity of two pancreatic cancer cell lines transduced with deoxycytidine kinase to gemcitabine elevated dramatically in comparison with control cells. 20137114_dCK expression level in fludarabine-sensitive patients was much higher than in Flud-resistant patients. 20544527_Data show that phosphorylation of the three other sites, located in the N-terminal extremity of the protein, does not significantly modify dCK activity, but phosphorylation of Thr-3 could promote dCK stability. 20544528_Data show that methylation was detected in one of the SP1 binding sites of the dCK promoter, in most tested cancer cell lines and in patient samples from brain tumors and leukemia. Methylation might therefore regulate transcription of dCK. 20608756_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20614893_it is residue Asp133 of dCK that is most responsible for discriminating against the thymine base 20637175_Site-directed mutagenesis demonstrated that only Ser-74 phosphorylation was involved in dCK activation by casein kinase 1 delta, strengthening the key role of this residue in the control of dCK activity. 20665488_Observational study of gene-disease association, gene-gene interaction, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20890066_Data show that dCK-360G allele was found to increase the risk of mucositis after exposure to low-dose cytarabine in childhood ALL therapy. 20890066_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21030078_Data indicate that variant C28624T showed a lower risk of lymphopenia (P=0.04), but a higher risk of neutropenia. 21030078_Observational study of gene-disease association. (HuGE Navigator) 21225236_Overexpressed dCK and knocked down p8 is associated with enhancement of gemcitabine sensitivity in pancreatic cancer. 21245102_We show that ABCG2 influence on clofarabine cytotoxicity was markedly influenced by dCK activity 21336182_High deoxycytidine kinase is associated with response to pemetrexed-gemcitabine combination in patients with advanced non-small cell lung cancer. 21351740_Data show that dCK must make the transition between the open and closed states during the catalytic cycle. 21832002_Although dFdU increased the net intracellular radioactivity of [5-(3)H]dFdC at 24 h in control cells, this increase was abolished in the absence of dCK activity. 21858090_Data show that the CDA/DCK ratio was 3 fold higher in non-responders than responders (P<.05 suggesting that this could be a mechanism of primary resistance. results indicate the inactivation dck is one crucial mechanisms in acquisition gemcitabine levels pancreatic ductal adenocarcinoma predict longer survival times patients treated with adjuvant gemcitabine. evident and cda polymorphisms might important markers for aml therapy outcomes chinese population. toxicity such as neutropenia thrombocytopenia anemia were not associated three tagged single-nucleotide deoxycytidine kinase or haplotypes. genetic variations did affect patients. variation subsequent metabolism. regulate migration invasion fibroblast-like synoviocytes through akt pathway rheumatoid arthritis constitutively dephosphorylates cells negatively regulates its activity. expression are prognostic had predictive value sensitivity to cancer. after hypoxia plays role progression pulmonary fibrosis by contributing alveolar epithelial cell proliferation findings decitabine metabolic affects therapeutic effects lower hent1 may induce resistance down-regulated related secondary rs12648166 rs4694362 snps hematologic ci p="0.0036" respectively human equilibrative nucleoside transporter-1 ribonucleotide reductase m1 significantly higher cytidine deaminase was ex vivo ara-c sensitive samples. multivariate cox regression analysis we found age at diagnosis wild-type genotype a79c polymorphism c360g most significant factors predicting risk death strongly suggest e197k alteration causes loss normal e197 allele rmkn28 tumor line periampullary adenocarcinoma. described mutant sensitizes cancer lines treatment here starting from g12 variant identified triggers even greater sensitisation than g12. a9846g aa reduced mortality activated can inhibit radiation-induced including apoptosis mitotic catastrophe promote autophagy pi3k pathway. suggests knockdown facilitates while inhibiting tumorigenicity cervical hela cells. an unfavorable biomarker correlated immune infiltrates liver effectiveness egyptian hepatocellular carcinoma. mutations acute myeloid leukemia resistant cytarabine. azacytidine curcumin potential alternative decitabine-resistant colorectal attenuated kinase. nelarabine caused epigenetic mechanisms.> | ENSMUSG00000029366 | Dck | 1155.18851 | 1.1043755 | 0.1432307580 | 0.12102471 | 1.403945e+00 | 2.360642e-01 | 6.274577e-01 | No | Yes | 1530.659599 | 332.269050 | 1288.779869 | 273.867139 | |
| ENSG00000156384 | 119392 | SFR1 | protein_coding | Q86XK3 | FUNCTION: Component of the SWI5-SFR1 complex, a complex required for double-strand break repair via homologous recombination (PubMed:21252223). Acts as a transcriptional modulator for ESR1 (PubMed:23874500). {ECO:0000269|PubMed:21252223, ECO:0000269|PubMed:23874500}. | Acetylation;Alternative splicing;Coiled coil;DNA damage;DNA repair;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | hsa:119392; | nucleus [GO:0005634]; Swi5-Sfr1 complex [GO:0032798]; nuclear receptor coactivator activity [GO:0030374]; cellular response to estrogen stimulus [GO:0071391]; double-strand break repair via homologous recombination [GO:0000724]; positive regulation of transcription, DNA-templated [GO:0045893] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 21252223_that human SWI5-MEI5 has an evolutionarily conserved function in homologous recombination repair. 23874500_SFR1 is a novel transcriptional modulator for ERalpha and a potential target in breast cancer therapy. | ENSMUSG00000025066 | Sfr1 | 217.02638 | 0.8749588 | -0.1927130910 | 0.20212343 | 9.055388e-01 | 3.413009e-01 | 7.152466e-01 | No | Yes | 244.209352 | 43.046010 | 265.747974 | 45.701686 | ||
| ENSG00000156650 | 23522 | KAT6B | protein_coding | Q8WYB5 | FUNCTION: Histone acetyltransferase which may be involved in both positive and negative regulation of transcription. Required for RUNX2-dependent transcriptional activation. May be involved in cerebral cortex development. Component of the MOZ/MORF complex which has a histone H3 acetyltransferase activity. {ECO:0000269|PubMed:10497217, ECO:0000269|PubMed:11965546, ECO:0000269|PubMed:16387653}. | 3D-structure;Acetylation;Activator;Acyltransferase;Alternative splicing;Chromatin regulator;Chromosomal rearrangement;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Transferase;Ubl conjugation;Zinc;Zinc-finger | The protein encoded by this gene is a histone acetyltransferase and component of the MOZ/MORF protein complex. In addition to its acetyltransferase activity, the encoded protein has transcriptional activation activity in its N-terminal end and transcriptional repression activity in its C-terminal end. This protein is necessary for RUNX2-dependent transcriptional activation and could be involved in brain development. Mutations have been found in patients with genitopatellar syndrome. A translocation of this gene and the CREBBP gene results in acute myeloid leukemias. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2012]. | hsa:23522; | MOZ/MORF histone acetyltransferase complex [GO:0070776]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; acetyltransferase activity [GO:0016407]; DNA binding [GO:0003677]; histone acetyltransferase activity [GO:0004402]; histone binding [GO:0042393]; metal ion binding [GO:0046872]; RNA polymerase II-specific DNA-binding transcription factor binding [GO:0061629]; transcription coregulator activity [GO:0003712]; histone acetylation [GO:0016573]; histone H3 acetylation [GO:0043966]; negative regulation of transcription, DNA-templated [GO:0045892]; nucleosome assembly [GO:0006334]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription, DNA-templated [GO:0006355] | 15271374_role in regulating interleukin-5 expression 16385451_Observational study of gene-disease association. (HuGE Navigator) 17460191_MORF4 has a role in cellular aging, and MRG15 associates with both histone deacetylases and histone acetyl transferase complexes [review] 18794358_These findings indicate that BRPF proteins play a key role in assembling and activating MOZ/MORF acetyltransferase complexes. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21804188_Data show that H3 acetylation by Myst4 is important for neural, craniofacial, and skeletal morphogenesis, mainly through its ability to specifically regulating the MAPK signaling pathway. 21880731_Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). 22077973_Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome 22265014_By exome sequencing, we found de novo heterozygous truncating mutations in KAT6B. 22265017_we identified de novo mutations of KAT6B in five individuals with Genitopatellar syndrome. 22715153_Propose that haploinsufficiency or loss of a function mediated by the C-terminal domain causes the common features, whereas gain-of-function activities would explain the features unique to genitopatellar syndrome. Review. 23063713_These data suggest that the tandem of plant homeodomain 1/2 fingers play a role in MOZ and MORF histone acetyltransferase association with histon H3 regions enriched in acetylated marks. 23436491_Our results confirm the implication of KAT6B mutations in typical SBBYS syndrome and emphasize the importance of genotype-phenotype correlations at the KAT6B locus. 24150941_MORF double PHD finger binds to Histone H3 in a manner that is enhanced by acetylation of the lysine residues K9 and K14 24294372_KAT5 and KAT6B regulate prostate cancer cell growth through PI3K-AKT signaling. 24444492_The study showed that markers rs11001178 (MYST4) showed weak associations. 24458743_Similar to those observed in other genetic disorders resulting from KAT6B mutations. 24698832_The relationship between MYO1C and KAT6B suggests that the two are interacting in chromatin remodelling for gene expression in human masseter muscle. This is the nuclear myosin1 (NM1) function of MYO1C. 25424711_KAT6B sequence variants are being reported in the Say-Barber-Biesecker type of blepharophimosis mental retardation syndromes (SBBS) and in the more severe genitopatellar syndrome. 25840828_This study shown MYST4 to be the risk factor for attention deficit/hyperactivity disorder. 26208904_Homozygous deletions of KAT6B and the loss of its mRNA occur in SCLC cell lines and primary tumors. KAT6B loss enhances cancer growth. Restoration induces tumor suppressor-like features involving a new type of histone H3 Lys23 acetyltransferase activity. 26334766_Kat6 c.3147G>A splice site mutation causes Say-Barber-Biesecker/Young-Simpson syndrome by inducing aberrant splicing. 27185879_Studies show that misregulation of MOZ/MORF results in tumorigenesis and developmental disorders. Results also provide evidence that these 2 proteins play important role in regulating cell proliferation and stem cell maintenance. [review] 27447113_chronic inflammation compromises unfolded protein response function through MORF-mediated-PERK transcription. 27452416_Our findings support that phenotypes associated with typical KAT6B disease-causing variants should be referred to as 'KAT6B spectrum disorders' or 'KAT6B related disorders', rather than their current SBBYSS and GTPTS classification 27880066_With ptosis, hypotonia, and developmental delay as the main diagnostic features of our patient, the effect of histone acetyltransferase-encoding KAT6B gene haploinsufficiency was suspected to have a significant role in determining the phenotype. 28286003_Study identified the double plant homeodomain finger (DPF) of the lysine acetyltransferase MORF as a reader of global histone H3K14 acylation. 28696035_KAT6B mutation is associated with Craniosynostosis. 29226580_The present report highlights the pivotal role of clinical genetics in avoiding clear-cut genotype-phenotype categories in syndromic forms of intellectual disability. In addition, it further supports the evidence that a continuum exists within the clinical spectrum of KAT6B-associated disorders 30569622_The clinical and genetic characterization of these patients could contribute to the understanding of the KAT6B-related disorders. 30921092_A novel pathogenic frameshift variant of KAT6B identified by clinical exome sequencing in a newborn with the Say-Barber-Biesecker-Young-Simpson syndrome. 31624313_Data indicate that the acetyltransferase function of the catalytic histone acetyltransferase KAT6B (MORF) subunit is positively regulated by the DPF domain of MORF (MORFDPF). 32424177_Further delineation of the clinical spectrum of KAT6B disorders and allelic series of pathogenic variants. 32448279_High methylation of lysine acetyltransferase 6B is associated with the Cobb angle in patients with congenital scoliosis. 32722658_CircKIAA0907 Retards Cell Growth, Cell Cycle, and Autophagy of Gastric Cancer In Vitro and Inhibits Tumorigenesis In Vivo via the miR-452-5p/KAT6B Axis. 33136874_Novel Approach of Mandibular Distraction to Avoid Tracheostomy in KAT6B-related Gene Disorders. 34464167_Low Expression of KAT6B May Affect Prognosis in Hepatocellular Carcinoma. | ENSMUSG00000021767 | Kat6b | 1266.35170 | 1.0764312 | 0.1062561455 | 0.10736024 | 9.818910e-01 | 3.217324e-01 | 7.006887e-01 | No | Yes | 1658.839064 | 307.093134 | 1432.700368 | 259.449655 | |
| ENSG00000156976 | 1974 | EIF4A2 | protein_coding | Q14240 | FUNCTION: ATP-dependent RNA helicase which is a subunit of the eIF4F complex involved in cap recognition and is required for mRNA binding to ribosome. In the current model of translation initiation, eIF4A unwinds RNA secondary structures in the 5'-UTR of mRNAs which is necessary to allow efficient binding of the small ribosomal subunit, and subsequent scanning for the initiator codon. | 3D-structure;ATP-binding;Alternative splicing;Helicase;Host-virus interaction;Hydrolase;Initiation factor;Nucleotide-binding;Phosphoprotein;Protein biosynthesis;RNA-binding;Reference proteome | hsa:1974; | cytosol [GO:0005829]; eukaryotic translation initiation factor 4F complex [GO:0016281]; perinuclear region of cytoplasm [GO:0048471]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; helicase activity [GO:0004386]; RNA binding [GO:0003723]; RNA helicase activity [GO:0003724]; translation initiation factor activity [GO:0003743]; cytoplasmic translational initiation [GO:0002183]; negative regulation of RNA-directed 5'-3' RNA polymerase activity [GO:1900260]; regulation of translational initiation [GO:0006446] | 11922617_interacted with NS5B protein, and the two proteins were shown to be partially colocalized in the perinuclear region 16567544_Single nucleotide polymorphisms in two relevant candidate genes for glucose homeostasis, kininogen (KNG1), and eukaryotic translation initiation factor 4alpha2 (EIF4A2) linked to type 2 diabetes. 18250159_subcellular distribution of eIF4F components may potentiate the complex assembly 18593934_A feedforward loop involving c-Myc and eIF4F that serves to link transcription and translation and that could contribute to the effects of c-Myc on cell proliferation and neoplastic growth. 19034380_This protein has been found differentially expressed in the temporal lobe from patients with schizophrenia. 21427765_Studies indicate that eIF4A (DDX2), together with its accessory proteins eIF4B and eIF4H, is thought to act as a helicase that unwinds secondary structures in the mRNA 5' UTR. 22360420_A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 22589333_The results indicated that eIF4AI and eIF4AII expression are linked and that the two protein isoforms exhibit functional differences. 23559250_These data support a linear model for miRNA-mediated gene regulation in which translational repression via eIF4A2 is required first, followed by mRNA destabilization. 23867391_expression is an independent prognostic factor for patients with non-small-cell lung cancer for both overall survival and disease-free survival 26614665_Studies indicate a developing focus on targeting eukaryotic initiation factor 4A eIF4A1 and eIF4A2 as cancer therapy. 27160682_eIF4A2 is recruited to stress granules, suggesting sumoylation of eIF4A2 correlates with its recruitment to stress granules. 29842983_Eukaryotic translation initiation factor 4A2 (eIF4A2) is necessary for efficient HIV-1 replication in human lymphoid cell line. eIF4A2 depletion reduces the efficiency of viral cDNA synthesis with virion entry into target cells being unaffected. Depletion of eIF4A2 also inhibits HIV-1 spreading infection in a knockdown level-dependent manner. 31088567_high EIF4A2 expression predicts poor prognosis of CRC patients and is associated with distant metastasis and poor response to oxaliplatin. Knocking-down EIF4A2 inhibits sphere formation and experimental metastasis, as well as oxaliplatin resistance in CRC. 31180491_Data show that incorporation of ATP-dependent RNA helicase eIF4A-2 (eIF4A2) into the CCR4-NOT complex inhibits CCR4-NOT transcription complex subunit 7 (CNOT7) deadenylation activity. 31308851_EIF4A2 was confirmed as a potential target of miR-5195-3p. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer (TNBC) to paclitaxel (PTX) by downregulating EIF4A2. 31791371_exerts its repressive effect on translation initiation by binding purine-rich motifs which are enriched in the 5'UTR of target mRNAs directly upstream of the AUG start codon | ENSMUSG00000022884 | Eif4a2 | 6773.67012 | 0.9777510 | -0.0324610442 | 0.08268199 | 1.541208e-01 | 6.946281e-01 | 9.061768e-01 | No | Yes | 7337.506683 | 1363.747862 | 7223.374918 | 1312.568456 | ||
| ENSG00000157111 | 134285 | TMEM171 | protein_coding | Q8WVE6 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:134285; | integral component of membrane [GO:0016021] | ENSMUSG00000052485 | Tmem171 | 44.13106 | 0.7931765 | -0.3342861963 | 0.42803508 | 6.063420e-01 | 4.361684e-01 | No | Yes | 44.333967 | 8.819442 | 54.696436 | 10.340023 | |||||
| ENSG00000157500 | 26060 | APPL1 | protein_coding | Q9UKG1 | FUNCTION: Multifunctional adapter protein that binds to various membrane receptors, nuclear factors and signaling proteins to regulate many processes, such as cell proliferation, immune response, endosomal trafficking and cell metabolism (PubMed:26583432, PubMed:15016378, PubMed:26073777, PubMed:19661063, PubMed:10490823). Regulates signaling pathway leading to cell proliferation through interaction with RAB5A and subunits of the NuRD/MeCP1 complex (PubMed:15016378). Functions as a positive regulator of innate immune response via activation of AKT1 signaling pathway by forming a complex with APPL1 and PIK3R1 (By similarity). Inhibits Fc-gamma receptor-mediated phagocytosis through PI3K/Akt signaling in macrophages (By similarity). Regulates TLR4 signaling in activated macrophages (By similarity). Involved in trafficking of the TGFBR1 from the endosomes to the nucleus via microtubules in a TRAF6-dependent manner (PubMed:26583432). Plays a role in cell metabolism by regulating adiponecting and insulin signaling pathways (PubMed:26073777, PubMed:19661063, PubMed:24879834). Required for fibroblast migration through HGF cell signaling (By similarity). Positive regulator of beta-catenin/TCF-dependent transcription through direct interaction with RUVBL2/reptin resulting in the relief of RUVBL2-mediated repression of beta-catenin/TCF target genes by modulating the interactions within the beta-catenin-reptin-HDAC complex (PubMed:19433865). {ECO:0000250|UniProtKB:Q8K3H0, ECO:0000269|PubMed:10490823, ECO:0000269|PubMed:15016378, ECO:0000269|PubMed:19433865, ECO:0000269|PubMed:19661063, ECO:0000269|PubMed:24879834, ECO:0000269|PubMed:26073777, ECO:0000269|PubMed:26583432}. | 3D-structure;Cell cycle;Cell projection;Coiled coil;Cytoplasm;Cytoplasmic vesicle;Diabetes mellitus;Disease variant;Endosome;Membrane;Nucleus;Phosphoprotein;Reference proteome | The protein encoded by this gene has been shown to be involved in the regulation of cell proliferation, and in the crosstalk between the adiponectin signalling and insulin signalling pathways. The encoded protein binds many other proteins, including RAB5A, DCC, AKT2, PIK3CA, adiponectin receptors, and proteins of the NuRD/MeCP1 complex. This protein is found associated with endosomal membranes, but can be released by EGF and translocated to the nucleus. [provided by RefSeq, Jul 2008]. | hsa:26060; | cytoplasm [GO:0005737]; cytoplasmic vesicle [GO:0031410]; cytosol [GO:0005829]; early endosome [GO:0005769]; early endosome membrane [GO:0031901]; early phagosome [GO:0032009]; endosome [GO:0005768]; endosome membrane [GO:0010008]; extracellular exosome [GO:0070062]; intracellular vesicle [GO:0097708]; macropinosome [GO:0044354]; membrane [GO:0016020]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ruffle [GO:0001726]; vesicle membrane [GO:0012506]; beta-tubulin binding [GO:0048487]; identical protein binding [GO:0042802]; phosphatidylinositol binding [GO:0035091]; phosphatidylserine binding [GO:0001786]; protein homodimerization activity [GO:0042803]; protein kinase B binding [GO:0043422]; protein-containing complex binding [GO:0044877]; adiponectin-activated signaling pathway [GO:0033211]; cell cycle [GO:0007049]; cellular response to hepatocyte growth factor stimulus [GO:0035729]; insulin receptor signaling pathway [GO:0008286]; negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis [GO:1905450]; positive regulation of cytokine production involved in inflammatory response [GO:1900017]; positive regulation of glucose import [GO:0046326]; positive regulation of macropinocytosis [GO:1905303]; positive regulation of melanin biosynthetic process [GO:0048023]; protein import into nucleus [GO:0006606]; regulation of fibroblast migration [GO:0010762]; regulation of G1/S transition of mitotic cell cycle [GO:2000045]; regulation of glucose import [GO:0046324]; regulation of innate immune response [GO:0045088]; regulation of protein localization to plasma membrane [GO:1903076]; regulation of toll-like receptor 4 signaling pathway [GO:0034143]; signal transduction [GO:0007165]; signaling [GO:0023052]; transforming growth factor beta receptor signaling pathway [GO:0007179] | 15016378_identification of a pathway directly linking the small GTPase Rab5, a key regulator of endocytosis, to signal transduction and mitogenesis via APPL1 and APPL2, two Rab5 effectors 15070827_APPL1 is a potential interactor with FSHR 16622416_APPL1 interacts with adiponectin receptors in mammalian cells and the interaction is stimulated by adiponectin. 17287464_APPL1 acts as a common downstream effector of Adiponectin receptors R1 and -R2, mediating adiponectin-evoked endothelial nitric oxide production and endothelium-dependent vasodilation. 17490420_Observational study of gene-disease association. (HuGE Navigator) 17502098_The ability of APPL1 to interact with multiple signaling molecules and phospholipids supports an important role for this adaptor in cell signaling. 17581628_The crystal structures of human APPL1 N-terminal BAR-PH domain motif, is reported. 17848569_These data suggest that APPL1 plays an important role in insulin-stimulated Glut4 translocation in muscle and adipose tissues and that its N-terminal portion may be critical for APPL1 function. 18034774_The findings suggest a role for APPL1 and APPL2 protein as dynamic scaffolds that modulate RAB5-associated signaling endosomal membranes by their ability to undergo domain-mediated oligomerization, membrane targeting and phosphoinositide binding. 18307981_Thus, binding to APPL1 helps localize OCRL at specific cellular sites, and disruption of this interaction may play a role in disease. 18581887_Observational study of gene-disease association. (HuGE Navigator) 18632660_Adiponectin blocks interleukin-18-mediated endothelial cell death via APPL1-dependent AMP-activated protein kinase (AMPK) activation and IKK/NF-kappaB/PTEN suppression. 19433865_APPL proteins exert their stimulatory effects on beta-catenin/TCF-dependent transcription by decreasing the activity of a Reptin-containing repressive complex 19520843_Adiponectin activates AMP-activated protein kinase in muscle cells via APPL1/LKB1-dependent and phospholipase C/Ca2+/Ca2+/calmodulin-dependent protein kinase kinase-dependent pathways 19686092_APPL1 overexpression affects the composition of the HDAC1-containing NuRD complex and the expression of HDAC1 target p21WAF1/CIP1 20095645_Used mass spectrometry (MS) to identify 13 phosphorylated residues within APPL1. 20412119_Rab5a promoted proliferation of ovarian cancer cells, which may be associated with the APPL1-related epidermal growth factor signaling pathway. 20484574_The promyogenic function of Cdo involves a coordinated activation of p38MAPK and Akt via association with scaffold proteins, JLP and Bnip-2 for p38MAPK and APPL1 for Akt. 20600589_[REVIEW] Emerging roles for AppL1. APPL1 has been shown to interact with a variety of membrane receptors. Recent subcellular localizations of APPL1 place it in dynamic and varied venues in the cell: the cell membrane, the nucleus and the early endosomes 20814572_significant fluorescence resonance energy transfer between APPL minimal BAR domain FRET pairs 20875820_Studies indicate that APPL1 has been recently identified as an AdipoR1 and AdipoR2 binding protein. 20978232_Data indicate APPL1 functions as a scaffolding protein to facilitate adiponectin-stimulated p38 MAPK activation in myotubes. 21285318_The adapter protein APPL1 links FSH receptor to inositol 1,4,5-trisphosphate production and is implicated in intracellular Ca(2+) mobilization. 21291857_interactions between TRP1-GIPC and GIPC-APPL-AKT provide a potential link between melanogenesis and PI3 kinase signaling 21320486_Results show that APPL1 is recruited to aggresomes induced by proteasomal stress, and suggest that proteasome inhibitors in clinical use affect the localization, ubiquitination and solubility of APPL1. 21562756_APPL1 abundance is significantly higher in type 2 diabetic muscle; Improvements in hyperglycaemia and hypoadiponectinaemia are associated with reduced skeletal muscle APPL1, and increased plasma adiponectin levels and muscle AMPK phosphorylation. 21645192_Data suggest that although annexin A2 is not an exclusive marker of APPL1/2 endosomes, it has an important function in membrane recruitment of APPL proteins, acting in parallel to Rab5. 21835890_Treating C2C12 myotubes with adiponectin promoted APPL1 interaction with protein phosphatase 2A (PP2A) and protein kinase Czeta (PKCzeta), leading to the activation of PP2A and subsequent dephosphorylation and inactivation of PKCzeta. 21926268_APPL1 plays a key role in coordinating the vasodilator and vasoconstrictor effects of insulin by modulating Akt-dependent NO production and ERK1/2-mediated ET-1 secretion in the endothelium. 22037462_These findings suggest that APPL1 is required for EGFR signaling by regulation of EGFR stabilities through inhibition of Rab5. 22340213_Genetic variation(s) in APPL1/2 may be associated with CAD risk in T2DM in Chinese population. 22379109_results demonstrate an important new function for APPL1 in regulating cell migration and adhesion turnover through a mechanism that depends on Src and Akt 22685300_APPL1 is a novel target in endoplasmic reticulum (ER) stress-induced insulin resistance and PKCalpha is the kinase mediating ER stress-induced phosphorylation of APPL1 at Ser(430). 22685329_APPL1 regulates basal NF-kappaB activity by modulating the stability of NIK, which affects the activation of p65. 23055524_analysis of APPL1 and APPL2 proteins and their interaction with Rab 23246927_neurons with APPL1-positive granules were restricted to the CA1 area and subiculum, areas associated with hippocampal vulnerability, suggesting a possible link between the perisomatic accumulation of APPL1 and Alzheimer's disease. 23291133_Rab5a and APPL1 are overexpressed in breast cancer, and are positively correlated with the HER-2 expression. 23891720_It concludes that APPL1(PH) binding to BAR domain and Reptin is mutually exclusive which regulates the nucleocytoplasmic shuttling of Reptin. 23909487_TRAF6-mediated ubiquitination of APPL1 is a vital step for the hepatic actions of insulin through modulation of membrane trafficking and activity of Akt. 23977033_C-APPL1/A-APPL2 allele combination is associated with non-alcoholic fatty liver disease occurrence, with a more severe hepatic steatosis grade and with a reduced adiponectin cytoprotective effect on liver. 23986476_The activated EGF receptor enters distinct sub-populations of SNX15- and APPL1-labelled peripheral endocytic vesicles. 24763056_ATM is the central modulator of APPL-mediated effects on radiosensitivity and DNA repair. 24813896_APPL1 sensitizes insulin signaling by acting at a site downstream of the IR. Study uncovers a mechanism regulating insulin signaling and crosstalk between the insulin and adiponectin pathways. 25622892_APPL1 is a positive regulator of Dvl2-dependent transcriptional activity of AP-1. 25780039_A role for APPL1 in TLR3/4-dependent TBK1 and IKKepsilon activation in macrophages. 26073777_Two loss-of-function mutations in APPL1, identified by means of whole-exome sequencing, found in two large families with a high prevalence of Familial Diabetes Mellitus. 26194181_Results indicate that persistent rab5 overactivation through beta-cleaved carboxy-terminal fragment of APP-APPL1 interactions constitutes a novel APP-dependent pathogenic pathway in Alzheimer's disease 26459602_APPL1 endosomes represent a distinct population of Rab5-positive sorting endosomes. 26473288_Low expression of APPL1 is associated with metastasis in prostate cancer. 26583432_Data show that signal transducing adaptor proteins APPL1 and APPL2 are required for TGFbeta-induced nuclear translocation of TGFbeta type I receptor (TbetaRI)-ICD and for cancer cell invasiveness of prostate and breast cancer cell lines. 26731990_article provides evidence that GG genotype and G carrier (CG+GG) genotypes of the rs4640525 polymorphism in the APPL1 gene may be suitable susceptibility biomarkers for NAFLD 27075719_Decreased expression of APPL1 is associated with polycystic ovary syndrome. 27820851_APPL1 positively mediated leptin signaling. 28902365_Present study demonstrated that expression of APPL1 protein and mRNA was upregulated in gastric carcinoma (GC) tissues and cell lines. The expression of APPL1 in GC was statistically associated with metastasis stage. Overexpression of APPL1 promotes invasion and metastasis of gastric cancer cells and the underlying molecular mechanism may facilitate EMT via Akt2 phosphorylation. 29361527_we demonstrate a novel role for the interaction between APPL1 and Rab5 in governing crosstalk between signaling and trafficking pathways on endosomes to affect cancer cell migration 32339379_APPL1 as an important regulator of insulin and adiponectin-signaling pathways in the PCOS: A narrative review. 33597720_Rapid whole cell imaging reveals a calcium-APPL1-dynein nexus that regulates cohort trafficking of stimulated EGF receptors. | ENSMUSG00000040760 | Appl1 | 3004.75239 | 1.0459087 | 0.0647569722 | 0.08472998 | 5.850137e-01 | 4.443538e-01 | 7.869395e-01 | No | Yes | 4076.426536 | 693.454816 | 3671.794452 | 611.083886 | |
| ENSG00000157657 | 114991 | ZNF618 | protein_coding | Q5T7W0 | FUNCTION: Regulates UHRF2 function as a specific 5-hydroxymethylcytosine (5hmC) reader by regulating its chromatin localization. {ECO:0000269|PubMed:27129234}. | Acetylation;Alternative splicing;Chromosome;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:114991; | chromatin [GO:0000785]; nucleus [GO:0005634]; pericentric heterochromatin [GO:0005721]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; transcription coregulator binding [GO:0001221]; positive regulation of chromatin binding [GO:0035563] | 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20583170_Observational study of gene-disease association. (HuGE Navigator) 27129234_study suggests that ZNF618 is a key protein that regulates UHRF2 function as a specific 5hmC reader in vivo. | ENSMUSG00000028358 | Zfp618 | 1076.67397 | 0.9451403 | -0.0813995372 | 0.11991859 | 4.594789e-01 | 4.978676e-01 | 8.189661e-01 | No | Yes | 1269.067539 | 130.841443 | 1302.674714 | 131.287235 | ||
| ENSG00000158089 | 79623 | GALNT14 | protein_coding | Q96FL9 | FUNCTION: Catalyzes the initial reaction in O-linked oligosaccharide biosynthesis, the transfer of an N-acetyl-D-galactosamine residue to a serine or threonine residue on the protein receptor. Displays activity toward mucin-derived peptide substrates such as Muc2, Muc5AC, Muc7, and Muc13 (-58). May be involved in O-glycosylation in kidney. | Alternative splicing;Disulfide bond;Glycosyltransferase;Golgi apparatus;Lectin;Manganese;Membrane;Metal-binding;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Protein modification; protein glycosylation. | This gene encodes a Golgi protein which is a member of the polypeptide N-acetylgalactosaminyltransferase (ppGalNAc-Ts) protein family. These enzymes catalyze the transfer of N-acetyl-D-galactosamine (GalNAc) to the hydroxyl groups on serines and threonines in target peptides. The encoded protein has been shown to transfer GalNAc to large proteins like mucins. Alterations in this gene may play a role in cancer progression and response to chemotherapy. [provided by RefSeq, Jun 2016]. | hsa:79623; | Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; carbohydrate binding [GO:0030246]; metal ion binding [GO:0046872]; polypeptide N-acetylgalactosaminyltransferase activity [GO:0004653]; O-glycan processing [GO:0016266] | 12507512_cloning and characterization; results provide evidence that pp-GalNAc-T14 is a new member of the pp-GalNAc-T family and suggest that it may be involved in the O-glycosylation in kidney 17434446_The role of GalNAc-T14 as an intracellular mediator of the effects of IGFBP-3 need to be verified in future studies. 19805900_GALNT14 may be involved in regulating the apoptotic action of IGFBP-3. 20179215_GALNT14 and FUT3/6 H-scores were significantly higher in non-small cell lung cancer cell lines sensitive to dulanermin and drozitumab versus resistant cell lines 20356418_Results provide evidence that GalNAc-T14 may be a potential biomarker for breast cancer by immunohistochemistry. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 22441125_Studies demonstrate that IGFBP-3 and GalNAc-T14 are colocalized in MCF-7 cells, and confirmed the interaction between IGFBP-3 and GalNAc-T14; these results reveal a minimal region of GalNAc-T14 that was sufficiently bound to the full-length IGFBP-3. 23959947_GALN14 genotype (rs9679162) was an effective predictor for therapeutic outcome in advanced hepatocellular carcinoma (HCC) patients treated by FMP chemotherapy. 24962947_GALNT14 stimulates MMP-2 expression. 26309160_Recurrence of mutation suggests GALNT14 as a novel gene potentially involved in neuroblastoma predisposition. 26544896_Expression of GalNAc-T14 or HOXB9 was strongly correlated with reduced recurrence-free survival in lung adenocarcinomas. 26871639_GALNT14 genotypes were significantly associated with clinical outcomes of transcatheter arterial chemoembolization. 27124048_The analysis showed that the 'TT' genotype was associated with unfavorable overall survival (OS, P = 0.009). 27133078_miR-125a functions as tumor suppressor in ovarian cancer by targeting GALNT14. 29227978_Silencing of GALNT14 in osterix-overexpressed cells restored the decreased chemosensitivity. Conversely, overexpression of GALNT14 in osterix-knockdown cells abrogated the increased chemosensitivity in breast cancer cells. 31292201_Data indicate that BORIS may promote cell motility and invasion in HGSC via upregulation of GALNT14. Studies provide evidence that aberrant expression of BORIS may play a role in the progression to HGSC by enhancing the migratory and invasive properties of FTSEC. 31611591_A GALNT14 rs9679162 genotype-guided therapeutic strategy for advanced hepatocellular carcinoma: systemic or hepatic arterial infusion chemotherapy. 31713889_Effects of IGFBP-3 and GalNAc-T14 on proliferation and cell cycle of glioblastoma cells and its mechanism. 32098271_GALNT14: An Emerging Marker Capable of Predicting Therapeutic Outcomes in Multiple Cancers. 33964375_GALNT2/14 overexpression correlate with prognosis and methylation: potential therapeutic targets for lung adenocarcinoma. 34643088_GALNT14 regulates ferroptosis and apoptosis of ovarian cancer through the EGFR/mTOR pathway. | ENSMUSG00000024064 | Galnt14 | 40.45431 | 1.7040568 | 0.7689733927 | 0.44866179 | 2.952145e+00 | 8.576366e-02 | No | Yes | 65.025640 | 11.301873 | 38.015919 | 6.598219 | |
| ENSG00000158169 | 2176 | FANCC | protein_coding | Q00597 | FUNCTION: DNA repair protein that may operate in a postreplication repair or a cell cycle checkpoint function. May be implicated in interstrand DNA cross-link repair and in the maintenance of normal chromosome stability. Upon IFNG induction, may facilitate STAT1 activation by recruiting STAT1 to IFNGR1. {ECO:0000269|PubMed:11520787}. | 3D-structure;Cytoplasm;DNA damage;DNA repair;Disease variant;Fanconi anemia;Nucleus;Reference proteome | The Fanconi anemia complementation group (FANC) currently includes FANCA, FANCB, FANCC, FANCD1 (also called BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ (also called BRIP1), FANCL, FANCM and FANCN (also called PALB2). The previously defined group FANCH is the same as FANCA. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group C. [provided by RefSeq, Jul 2008]. | hsa:2176; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; Fanconi anaemia nuclear complex [GO:0043240]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; brain morphogenesis [GO:0048854]; cellular response to oxidative stress [GO:0034599]; DNA repair [GO:0006281]; germ cell development [GO:0007281]; interstrand cross-link repair [GO:0036297]; myeloid cell homeostasis [GO:0002262]; neuronal stem cell population maintenance [GO:0097150]; nucleotide-excision repair [GO:0006289]; protein-containing complex assembly [GO:0065003]; removal of superoxide radicals [GO:0019430] | 11876000_Observational study of genotype prevalence. (HuGE Navigator) 12239156_The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of this protein. 12397061_Hsp70 requires the cooperation of FANCC to suppress PKR activity and support survival of hematopoietic cells and FANCC does not require the multimeric Fanconi anemia complex to exert this function 12763929_Fancc-/- phenotypically defined cell populations enriched for hematopoietic stem and progenitor cells exhibit increased cycling 14625294_FANCC undergoes proteolytic modification by a caspase into a predominant 47-kDa ubiquitinated protein fragment. Lack of proteolytic modification at the putative cleavage site delays apoptosis. 15077170_Fanconi anemia C gene product regulates expression of genes involved in differentiation and inflammation. 15256425_FA proteins function at the level of chromatin during S phase to regulate and maintain genomic stability. 15299030_Inappropriate activation of Protein kinase regulated by RNA may cause mutations in FANCC> 15327776_Data show that the Fanconi anemia protein FANCC cooperate with key mutagenesis and repair processes that enable replication of damaged DNA. 15616572_spontaneous SCE levels were elevated approximately 2-fold in cells deficient in Fanconi anemia gene FANCC 15695377_Observational study of gene-disease association. (HuGE Navigator) 15726604_Observational study of genotype prevalence. (HuGE Navigator) 16127171_FANCC, FANCE, and FANCD2 form a ternary complex in the Fanconi anemia DNA damage response pathway 16429406_analysis of two new mutations that inactivate the function of the FANCC protein 16513431_nuclear accumulation of FANCE does not rely solely on its nuclear localization signal motifs, but also on FANCC 17490643_FANCC-deficient cells are hypersensitive to DNA cross-linking reagents. 17977515_We found six differentially expressed proteins; among them, the checkpoint mediator protein MDC1 whose expression was disrupted in FANCC-/- cells. 18264947_Observational study of genotype prevalence. (HuGE Navigator) 18607065_the first report to describe hypermethylation of FANCL in leukemia 18950845_Observational study of gene-disease association. (HuGE Navigator) 18990233_Differential association of alterations in FANCC and PTCH1 with that of PHF2, XPA and two breast cancer susceptibility genes (BRCA1/BRCA2) in the two age groups suggests differences in their molecular pathogenesis. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19536649_Observational study of gene-disease association. (HuGE Navigator) 19690177_Observational study of gene-disease association. (HuGE Navigator) 19714462_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20509860_we identified a hepatocellular carcinoma cell line harboring an inactivating mutation of the FANCC gene, specifically causing proximal FA pathway inactivation and the classic cellular DNA interstrand-crosslinking agents-hypersensitivity phenotype 20538911_study found genetic interaction between Fanconi anemia(FA)gene FANCC and Ku70; results indicate FA pathway promotes homologous recombination repair of DNA double-strand breaks (DSBs) by counteracting Ku70; suggest this achieved by modification of DSBs 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20864535_Cytoplasmic FANCA-FANCC complex was essential for NPMc stability. 20869034_Correct mRNA processing at a mutant TT splice donor in FANCC ameliorates the clinical phenotype in Fanconi anemia patients and is enhanced by delivery of suppressor U1 snRNAs. 21543111_genetic diversity in FANCA, FANCC and FANCL does not support an association of these genes with cervical cancer susceptibility in the Swedish population. 21670957_FANCC polymorphisms might be associated with the obstructive symptoms in allergic diseases. 21697891_FA DNA repair genes, FANCD2, FANCL, and FANCC, are transcriptionally upregulated differently in melanoma compared with non-melanoma skin cancer 21861228_deregulations of the FANCC-mediated DNA damage repair pathway and the PTCH1-associated sonic hedgehog pathway are associated with the development of early dysplastic head and neck lesions. 23028338_we identified faults in two genes, Fanconi C and Bloom helicase( FANCC and BLM), in six families. Faults in these genes appear to increase the risk of developing breast cancer 24046015_Data indicate that TLR-induced IL-1beta overproduction in FANCA- and FANCC-deficient mononuclear phagocyte cell lines and primary cells requires activation of the inflammasome. 24676280_FANCC interferes with UNC5A's functions in apoptosis and suggest that FANCC may participate in developmental processes through association with the dependence receptor UNC5A. 25545896_The successful in vitro repair of the mutated Fanconi anemia FANCC gene using the CRISPR/Cas9 system has been described. 26466335_FANCC interacts and co-localizes with STMN1 at centrosomes during mitosis. We also showed that FANCC is required for STMN1 phosphorylation. 26778106_Israeli ATM, BLM, and FANCC heterozygous mutation carriers are not at an increased risk for developing cancer. 26842001_Lung adenocarcinomas in both male and female patients were associated with (a) genotypic polymorphisms of FANCC and FANCD1. 28425259_mutation IVS4+4A>T is the most prevalent mutation in our group of patients. This analysis of Pakistani patients also suggests that there is no significant difference between IVS4+4A>T homozygotes and the rest of the patients with regard to severity of clinical phenotype. 29843852_The splice-site mutation in the FANCC gene (IVS4+4A>T) accounts for most cases of Fanconi anaemia in Ashkenazi Jewish cohorts worldwide.A founder mutation described in individuals of Ashkenazi Jewish ancestry is also found in South African individuals of this origin. 29901137_The finding that FANCC overexpression reduced betacell apoptosis advances the potential for an alternative approach to the treatment of Diabetes mellitus caused by FANCC defects 29930218_This study showed that featured-metabolic alterations are readouts of functional mechanisms underlying reduced tumorigenicity driven by FANCC, demonstrating close links among cancer, aging, inflammation and DM. 31467304_Two truncating variants in FANCC and breast cancer risk. 33073500_Zika virus depletes neural stem cells and evades selective autophagy by suppressing the Fanconi anemia protein FANCC. 33960642_Microdeletion of 9q22.3: A patient with minimal deletion size associated with a severe phenotype. 34864095_In silico study of missense variants of FANCA, FANCC and FANCG genes reveals high risk deleterious alleles predisposing to Fanconi anemia pathogenesis. | ENSMUSG00000021461 | Fancc | 912.60793 | 0.9768855 | -0.0337386406 | 0.12040915 | 7.833456e-02 | 7.795672e-01 | 9.384376e-01 | No | Yes | 1074.144165 | 106.367828 | 1049.656315 | 101.667134 | |
| ENSG00000158321 | 26053 | AUTS2 | protein_coding | Q8WXX7 | FUNCTION: Component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones; it mediates monoubiquitination of histone H2A 'Lys-119', rendering chromatin heritably changed in its expressibility (PubMed:25519132). The PRC1-like complex that contains PCGF5, RNF2, CSNK2B, RYBP and AUTS2 has decreased histone H2A ubiquitination activity, due to the phosphorylation of RNF2 by CSNK2B (PubMed:25519132). As a consequence, the complex mediates transcriptional activation (PubMed:25519132). In the cytoplasm, plays a role in axon and dendrite elongation and in neuronal migration during embryonic brain development. Promotes reorganization of the actin cytoskeleton, lamellipodia formation and neurite elongation via its interaction with RAC guanine nucleotide exchange factors, which then leads to the activation of RAC1 (By similarity). {ECO:0000250|UniProtKB:A0A087WPF7, ECO:0000269|PubMed:25519132}. | Alternative splicing;Autism;Autism spectrum disorder;Cell projection;Chromosomal rearrangement;Cytoplasm;Cytoskeleton;Mental retardation;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation | This gene has been implicated in neurodevelopment and as a candidate gene for numerous neurological disorders, including autism spectrum disorders, intellectual disability, and developmental delay. Mutations in this gene have also been associated with non-neurological disorders, such as acute lymphoblastic leukemia, aging of the skin, early-onset androgenetic alopecia, and certain cancers. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, May 2014]. | hsa:26053; | cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; growth cone [GO:0030426]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; actin cytoskeleton reorganization [GO:0031532]; axon extension [GO:0048675]; dendrite extension [GO:0097484]; neuron migration [GO:0001764]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of histone H4-K16 acetylation [GO:2000620]; positive regulation of lamellipodium assembly [GO:0010592]; positive regulation of Rac protein signal transduction [GO:0035022]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 19086053_Observational study of gene-disease association. (HuGE Navigator) 19240061_Observational study of gene-disease association. (HuGE Navigator) 19546859_Observational study of gene-disease association. (HuGE Navigator) 19567891_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20398908_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20635338_A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 gene is associated with autism. 21471458_SNP rs6943555 in autism susceptibility candidate 2 gene (AUTS2) was associated with alcohol consumption at genome-wide significance (P = 4 x 10(-8) to P = 4 x 10(-9)). 22578776_PAX5-AUTS2: a recurrent fusion gene in childhood B-cell precursor acute lymphoblastic leukemia. 22872102_the role of AUTS2 in normal neurological development and its altered expression may result in a variety of neurobehavioral phenotypes 22995765_This study indicates that there might be a genetic association of AUTS2 with susceptibility to heroin dependence. Reduced gene expression of AUTS2 in lymphoblastoid cell lines may increase the risk for heroin dependence. 23332918_These observations demonstrate a causal role of AUTS2 in neurocognitive disorders. 23349641_our results show that AUTS2 is important for neurodevelopment and expose candidate enhancer sequences in which nucleotide variation could lead to neurological disease and human-specific traits. 23437340_AUTS2 rs6943555 A allele is associated with suicide committed after drinking ethanol shortly before death. 24008202_AUTS2, its discovery, expression, association with autism and other neurological and non-neurological traits, implication in human evolution, function, regulation, and genetic pathways, are reviewed. 24459036_This is one of the smallest de novo intragenic deletions of AUTS2. 24859339_AUTS2 mutations are associated with autism spectrum disorder. 25205402_similarities between the phenotypes of 2 male patients with AUTS2 variants support that AUTS2 syndrome is a single gene disorder. 25347278_polymorphism rs6943555 might elucidate the pathogenesis of schizophrenia and play an important role in its etiology 25398668_AA homozygotes of rs6943555 were significantly over-represented in the patients with heroin dependence. 25519132_the CK2 component of PRC1-AUTS2 neutralizes PRC1 repressive activity, whereas AUTS2-mediated recruitment of P300 leads to gene activation. 25962312_In summary, our results indicate that AUTS2 is a candidate biomarker for defining liver metastasis of pancreatic cancer and directing personalized therapies. 26348319_The AUTS2 gene has been repeatedly implicated in neurodevelopmental disorders including autism, intellectual disability and developmental delay. 26545289_Exonic deletions of AUTS2 is associated with developmental delay and intellectual disability. 26717414_Heterozygous Disruption of Autism susceptibility candidate 2 Causes Impaired Emotional Control and Cognitive Memory 26763194_Results showed the frequencies of the AUTS2 haplotypes significantly different between them, and the rs6943555 and rs9886351 A-A haplotype was associated with alcohol dependence in a Japanese population. 27075013_AUTS2 syndrome emerges as a specific ID syndrome with microcephaly. 27322685_chromatin complexes PRC1/AUTS2 and PRC2 in a gene network in T-ALL regulating early lymphoid differentiation. 27531620_This clinical report provides the natural history in the eldest patient yet to be reported, and complements the existing evidence suggesting that disruption of the AUTS2 leads to a recently delineated neurodevelopmental phenotype with a wide spectrum, namely 'AUTS2 Syndrome.' 28577753_This study demonstrated that Cocaine-Induced Chromatin Modifications Associate With Increased Expression and Three-Dimensional Looping of Auts2. 29166413_BCL7A, BRWD3, and AUTS2 demonstrate significantly higher mutation frequencies among AA cases. These genes are all involved in translocations in B-cell malignancies. Moreover, we detected a significant difference in mutation frequency of TP53 and IRF4 with frequencies higher among CA cases. Our study provides rationale for interrogating diverse tumor cohorts to best understand tumor genomics across populations. 29377512_significant intergenic synergistic effect between rs16879552 (NRG1) and rs7785360 (AUTS2) was identified through cross-validation. 30190612_These co-occurring hits involved known disease-associated genes such as SETD5, AUTS2, and NRXN1, and were enriched for cellular and developmental processes. Accurate genetic diagnosis of complex disorders will require complete evaluation of the genetic background even after a candidate disease-associated variant is identified 31686349_The intragenic exon rearrangements (IERs) involved in SOBP (6q21) exon 2 and 3 and AUTS2 (7q11.22) exon 2-4 were the molecular lesions specific to tumors and were frequently detected in non-Hodgkin B cell lymphoma (B-NHL) samples. These IERs constitute novel genetic alterations of B-NHL, which might be associated with tumorigenesis and be useful as genetic biological markers. 32574757_Effect of AUTS2 gene rs6943555 variant in male patients with schizophrenia in a Turkish population. 33562463_Whole Exome Sequencing Reveals a Novel AUTS2 In-Frame Deletion in a Boy with Global Developmental Delay, Absent Speech, Dysmorphic Features, and Cerebral Anomalies. 33577136_Germ cell mosaicism for AUTS2 exon 6 deletion. 34573342_Attention Deficit Hyperactivity and Autism Spectrum Disorders as the Core Symptoms of AUTS2 Syndrome: Description of Five New Patients and Update of the Frequency of Manifestations and Genotype-Phenotype Correlation. 34637754_NRF1 association with AUTS2-Polycomb mediates specific gene activation in the brain. | ENSMUSG00000029673 | Auts2 | 1612.38926 | 1.2770997 | 0.3528712132 | 0.12794692 | 7.673476e+00 | 5.603839e-03 | 1.234295e-01 | No | Yes | 2328.745091 | 263.761187 | 1785.863432 | 197.890849 | |
| ENSG00000159217 | 10642 | IGF2BP1 | protein_coding | Q9NZI8 | FUNCTION: RNA-binding factor that recruits target transcripts to cytoplasmic protein-RNA complexes (mRNPs). This transcript 'caging' into mRNPs allows mRNA transport and transient storage. It also modulates the rate and location at which target transcripts encounter the translational apparatus and shields them from endonuclease attacks or microRNA-mediated degradation. Plays a direct role in the transport and translation of transcripts required for axonal regeneration in adult sensory neurons (By similarity). Regulates localized beta-actin/ACTB mRNA translation, a crucial process for cell polarity, cell migration and neurite outgrowth. Co-transcriptionally associates with the ACTB mRNA in the nucleus. This binding involves a conserved 54-nucleotide element in the ACTB mRNA 3'-UTR, known as the 'zipcode'. The RNP thus formed is exported to the cytoplasm, binds to a motor protein and is transported along the cytoskeleton to the cell periphery. During transport, prevents ACTB mRNA from being translated into protein. When the RNP complex reaches its destination near the plasma membrane, IGF2BP1 is phosphorylated. This releases the mRNA, allowing ribosomal 40S and 60S subunits to assemble and initiate ACTB protein synthesis. Monomeric ACTB then assembles into the subcortical actin cytoskeleton (By similarity). During neuronal development, key regulator of neurite outgrowth, growth cone guidance and neuronal cell migration, presumably through the spatiotemporal fine tuning of protein synthesis, such as that of ACTB (By similarity). May regulate mRNA transport to activated synapses (By similarity). Binds to and stabilizes ABCB1/MDR-1 mRNA (By similarity). During interstinal wound repair, interacts with and stabilizes PTGS2 transcript. PTGS2 mRNA stabilization may be crucial for colonic mucosal wound healing (By similarity). Binds to the 3'-UTR of IGF2 mRNA by a mechanism of cooperative and sequential dimerization and regulates IGF2 mRNA subcellular localization and translation. Binds to MYC mRNA, in the coding region instability determinant (CRD) of the open reading frame (ORF), hence prevents MYC cleavage by endonucleases and possibly microRNA targeting to MYC-CRD. Binds to the 3'-UTR of CD44 mRNA and stabilizes it, hence promotes cell adhesion and invadopodia formation in cancer cells. Binds to the oncofetal H19 transcript and to the neuron-specific TAU mRNA and regulates their localizations. Binds to and stabilizes BTRC/FBW1A mRNA. Binds to the adenine-rich autoregulatory sequence (ARS) located in PABPC1 mRNA and represses its translation. PABPC1 mRNA-binding is stimulated by PABPC1 protein. Prevents BTRC/FBW1A mRNA degradation by disrupting microRNA-dependent interaction with AGO2. Promotes the directed movement of tumor-derived cells by fine-tuning intracellular signaling networks. Binds to MAPK4 3'-UTR and inhibits its translation. Interacts with PTEN transcript open reading frame (ORF) and prevents mRNA decay. This combined action on MAPK4 (down-regulation) and PTEN (up-regulation) antagonizes HSPB1 phosphorylation, consequently it prevents G-actin sequestration by phosphorylated HSPB1, allowing F-actin polymerization. Hence enhances the velocity of cell migration and stimulates directed cell migration by PTEN-modulated polarization. Interacts with Hepatitis C virus (HCV) 5'-UTR and 3'-UTR and specifically enhances translation at the HCV IRES, but not 5'-cap-dependent translation, possibly by recruiting eIF3. Interacts with HIV-1 GAG protein and blocks the formation of infectious HIV-1 particles. Reduces HIV-1 assembly by inhibiting viral RNA packaging, as well as assembly and processing of GAG protein on cellular membranes. During cellular stress, such as oxidative stress or heat shock, stabilizes target mRNAs that are recruited to stress granules, including CD44, IGF2, MAPK4, MYC, PTEN, RAPGEF2 and RPS6KA5 transcripts. {ECO:0000250, ECO:0000269|PubMed:10875929, ECO:0000269|PubMed:16356927, ECO:0000269|PubMed:16541107, ECO:0000269|PubMed:16778892, ECO:0000269|PubMed:17101699, ECO:0000269|PubMed:17255263, ECO:0000269|PubMed:17893325, ECO:0000269|PubMed:18385235, ECO:0000269|PubMed:19029303, ECO:0000269|PubMed:19541769, ECO:0000269|PubMed:19647520, ECO:0000269|PubMed:20080952, ECO:0000269|PubMed:22279049, ECO:0000269|PubMed:8132663, ECO:0000269|PubMed:9891060}. | 3D-structure;Alternative splicing;Cell junction;Cell projection;Cytoplasm;Direct protein sequencing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Synapse;Translation regulation;Transport;mRNA transport | This gene encodes a member of the insulin-like growth factor 2 mRNA-binding protein family. The protein encoded by this gene contains four K homology domains and two RNA recognition motifs. It functions by binding to the mRNAs of certain genes, including insulin-like growth factor 2, beta-actin and beta-transducin repeat-containing protein, and regulating their translation. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2009]. | hsa:10642; | CRD-mediated mRNA stability complex [GO:0070937]; cytoplasm [GO:0005737]; cytoplasmic stress granule [GO:0010494]; cytosol [GO:0005829]; dendritic spine [GO:0043197]; filopodium [GO:0030175]; growth cone [GO:0030426]; lamellipodium [GO:0030027]; neuronal cell body [GO:0043025]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; ribonucleoprotein complex [GO:1990904]; mRNA 3'-UTR binding [GO:0003730]; mRNA 5'-UTR binding [GO:0048027]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; translation regulator activity [GO:0045182]; CRD-mediated mRNA stabilization [GO:0070934]; dendrite arborization [GO:0140059]; mRNA transport [GO:0051028]; negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900152]; negative regulation of translation [GO:0017148]; nervous system development [GO:0007399]; neuronal stem cell population maintenance [GO:0097150]; pallium cell proliferation in forebrain [GO:0022013]; positive regulation of cytoplasmic translation [GO:2000767]; regulation of cytokine production [GO:0001817]; regulation of gene expression [GO:0010468]; regulation of mRNA stability involved in response to stress [GO:0010610]; regulation of RNA metabolic process [GO:0051252] | 11973350_IMP1 localization is associated with motility and the major functions of IMP1 are carried out by the phylogenetically conserved KH domains 12532419_Aberrant CRD-BP expression may interfere with c-myc regulation. Copy number gains at 8q24 (c-myc), were seen in 48.3% of tumors, gains at 17q21 (CRD-BP) in 18.3%,& CRD-BP was seen in 58.5%, implying mechanisms of activation other than gene amplification. 12921532_IMP1 translocates to the nucleus and contains nuclear export signals within the RNA-binding KH2 and KH4 domains. 14767552_IMP-1 may have a role in progression of ovarian cancer 15121863_Intestinal epithelial cells continue to express IMP1 postnatally, and Imp1(-/-) mice exhibited impaired development of the intestine, with small and misshapen villi and twisted colon crypts. 15159028_highest frequency of CRD-BP positive tumors was observed in meningiomas 15282548_intimate association of fragile X mental retardation protein and IMP1 suggests a link between mRNA transport and translational repression in mammalian cells 15314207_A cooperative mechanism for the binding of IMP-1 to RNA. 15355996_CRD-BP has a dominant role in proliferation of human K562 cells by an IGF-II-dependent mechanism independent of its ability to serve as a c-myc mRNA masking protein 15769738_normal role for CRD-BP/IMP1 in pluripotent stem cells with high renewal capacity 17212783_We have further characterized the interaction between PABP and IMP1 with the 3' end of the adenine-rich autoregulatory sequence 17255263_IMP-1 expression is likely to play important roles in lung cancer development and progression 17289661_Study isolated the IMP1-containing RNP granules and found that they represent a unique ribonucleoprotein entity distinct from neuronal hStaufen and/or fragile X mental retardation protein granules, processing bodies, and stress granules. 17296566_VICKZ exhibits differential expression in lymphoma subtypes and thus may be a marker of potential value in the diagnosis and study of hematopoietic neoplasia. 17546046_IMP1 is an oncogenic factor that is involved in promoting elevated proliferation by stabilizing the c-myc mRNA in ovarian carcinoma cells. 18252897_Observational study of gene-disease association. (HuGE Navigator) 18385235_IMP-1 associates with Gag protein of HIV-1 and its overexpression affects virus assembly. 18454174_Knockdown of CRD-BP inhibits NF-kappaB activity, induces apoptosis, and suppresses proliferation and tumorigenic properties of melanoma cells. 18490442_These experiments suggest that in breast cancer cells, the expression of ZBP1 and the expression of beta-catenin are coordinately regulated. 19038974_IGFBP-1, which can inhibit IGF action, also increased during in-vitro decidualization of cultured human ESCs. 19461076_These data suggest that repression of ZBP1 by blocking beta-catenin binding at the ZBP1 promoter deregulates its associated mRNAs, leading to the phenotypic changes of breast cancers. 19541769_IGF2BP1, by binding to the HCV 5'UTR and/or HCV 3'UTR, recruits eIF3 and enhances HCV IRES-mediated translation. 19647520_CRD-BP prevents degradation of betaTrCP1 mRNA by attenuating its miR-183-dependent interaction with Ago2. 19661680_expression of 8S-LOX and 15S-LOX-2 suppresses CRD-BP/IMP-1 expression, resulting in inhibition of human prostate carcinoma PC-3 cell proliferation. 19726068_IMP1 interacts with HIV1 Rev protein and its ectopic expression causes relocation of Rev from the nucleus to the cytoplasm and ectopic expression of IMP1 leads to significant accumulation of multiple spliced HIV-1 RNA. 19887615_Wnt/beta-catenin signaling induces expression of an RNA-binding protein, CRD-BP, which in turn binds and stabilizes GLI1 mRNA, causing an elevation of GLI1 expression and transcriptional activity. 20195514_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20627640_IGF2BP1 genotype, haplotype and genetic model studies in metabolic syndrome traits and diabetes 20627640_Observational study of gene-disease association. (HuGE Navigator) 20819778_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 21209918_IMP1 plays a role in regulating the packaging of MLV genomic RNA and can be used for improving production of retroviral vectors 21252116_Studies identify a novel proapoptotic gene target, CYFIP2, which is downregulated by IMP-1, and mediates the regulation of cell survival and K-Ras expression in colon cancer cells. 21618519_Data show that the let-7 (let-7d and let-7g) target IMP-1 stabilizes the mRNA of MDR1. 21981993_sh-RNA knockdown of CRD-BP enhances the effect of dacarbazine, temozolomide, vinblastine, & etoposide on both primary and metastatic melanoma cell lines. 22114317_these data indicate that ZBP1 may function as an adapter to export the Ro/Y3 RNA complex from nuclei. 22125086_The defective splicing caused by the ISCU intron mutation in patients with myopathy with lactic acidosis is repressed by PTBP1 but can be derepressed by IGF2BP1. 22240322_The data of this study suggest that RNA-binding proteins can be used as a tool to identify the post-transcriptional regulation of gene expression in the establishment and function of neural circuits involved in addiction behaviors. 22266909_in both T47D and MDA231 human breast carcinoma cells IMP1/ZBP1 functions to suppress cell invasion 22279049_IGF2BP1 promotes the velocity and directionality of tumor-derived cell migration by determining the cytoplasmic fate of two novel target mRNAs: MAPK4 and PTEN 22983196_IGF2BP1 is a potent oncogenic factor that regulates the adhesion, migration and invasiveness of tumor cells by modulating intracellular signaling. 23038779_Shows role for CRD-BP in the regulation of melanoma cell invasion and highlights the importance of the hypoxic microenvironment in determining cell fate. 23728852_IGF2BP1 acts as an adaptor protein that recruits the CCR4-NOT complex and thereby initiates the degradation of the lncRNA HULC. 23911878_our study suggests that IMP1might play an important role in the progression of choriocarcinoma through the regulation of cell migration and invasion. 24395596_The RNA-binding protein IGF2BP1 is an important protumorigenic factor in liver carcinogenesis. 24397586_Data suggest GHET1 (gastric carcinoma high expressed transcript 1) is prognostic factor for gastric carcinoma; GHET1 promotes c-Myc (proto-oncogene proteins c-Myc) mRNA stability/expression, binds IGF2BP1, and promotes gastric carcinoma growth. 24468749_CRD-BP is overexpressed in basal cell carcinoma and its expression positively correlates with the activation of both Wnt and Hh signaling pathways 24704827_Results identify IGF2BP1 as a critical translational regulator of cIAP1-mediated apoptotic resistance in rhabdomyosarcoma. 24915579_These results indicate that IGF2BP1 and TET1/2 contribute to the stemness of MSCs, at least regarding their proliferative potential. 25195122_A 14;17 translocation resulting in an IGH-IGF2BP1 fusion is associated with B acute lymphoblastic leukemia. 25389298_Two KH domains of CRD-BP are required for efficient binding to oncogenic mRNAs and for granule formation in zebrafish embryos. 25414259_MicroRNA-340-mediated degradation of MITF mRNA is inhibited by CRD-BP. 25670861_miR-873 negatively affected the carcinogenesis and metastasis of GBM by down-regulating the expression of IGF2BP1, which stabilizes the mRNA transcripts of its target genes 25753434_Data demonstrate that IGF2BP1 is a potential oncogene and an independent negative prognostic factor in neuroblastoma. 25861986_High CRD-BP expression is associated with Breast Tumor. 25889892_IGF2BP1 as a direct and functional target of miR-196b. 26160756_Data show that insulin-like growth factor-2-mRNA-binding proteins IGF2BP1, IGF2BP2, and IGF2BP3 are direct targets of microRNA-1275 (miR-1275). 26194191_The tumor-suppressive role of stromal IMP1 and its ability to modulate protumorigenic factors suggest that IMP1 status is important for the initiation and growth of epithelial tumors 26547929_Data show that ONECUT2, IGF2BP1, and ANXA2 proteins were confirmed to be microRNA-9 (miR-9) targets and aberrantly upregulated in hepatocellular carcinoma (HCC). 26852652_ETV6/RUNX1 transcript is a target of RNA-binding protein IGF2BP1 in t(12;21)(p13;q22)-positive acute lymphoblastic leukemia. 26910917_Our studies provide insights into a molecular mechanism that the positive function of IMP1 to inhibit breast tumor growth and metastasis 26917013_The findings indicate that the tumor-suppressive roles of the let-7 family are antagonized by a potent and self-promoting triangle consisting of IGF2BP1 (potentially all IGF2BPs), LIN28B and HMGA2. 27036131_The results from this study demonstrate the potential importance of the two-stem-loop motif as a target region for the inhibition of the CRD-BP-GLI1 RNA interaction and Hedgehog signaling pathway. 27239736_IGF2BP1 indirectly potentiates ETV6/RUNX1-RAC1-STAT3 signaling axis by sustaining appropriate ETV6/RUNX1 and STAT3 transcript levels in REH cells. 27588393_Findings suggest a MicroRNA-140-5p (miR-140-5p)-Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) regulatory circuit for CC pathogenesis, and miR-140-5p may be a potential target for CC therapy. 28182633_Insight into molecular interactions between CRD-BP and MITF mRNA. 28244848_miR-98-5p, downregulated in HCC, inhibits proliferation while inducing apoptosis in HCC LM3 cells, partly at least, through directly targeting IGF2BP1 28486549_The regions encoding 71 microRNAs (miRs) were deleted in at least 25% of tumor specimens. Five of these recurrently deleted miRs targeted the insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) gene product, and a correlating 100-fold upregulation of IGF2BP1 mRNA was seen in tumor specimens 28497370_report insulin-like growth factor-II binding protein 1 (IGF2BP1) as a novel interacting partner of p38 MAPK. 28652347_IGF2BP1 mediates posttranscriptional loss of BCL11A in cultured human adult erythroblasts 28677801_HCG11 exerted its effect on Hepatocellular carcinoma (HCC)via interaction with IGF2BP1, leading to activation of MAPK signaling, which eventually promoted the progression of HCC 28975985_SOX12 can increase the expression of CDK4 and IGF2BP1, which confer malignant phenotypes to Hepatocellular Carcinoma. 29369405_Results demonstrate that IGF2BP1 plays an important role in the sensitivity of melanoma to targeted therapy. Inhibition of IGF2BP1 enhances the effects of BRAF-inhibitor and BRAF-MEK inhibitors in BRAF(V600E) melanoma. Also, knockdown of IGF2BP1 alone is sufficient to reduce tumorigenic characteristics in vemurafenib-resistant melanoma. 29463567_IMP1 shows alternating binding in nanocores and anomalous diffusion in the liquid phase of stress granules. 29476152_The direct binding of IGF2BPs to RNA N(6)-methyladenosine through their KH domains enhances mRNA stability and translation. 29669595_Our study provides initial evidence that interaction between IMP1 and UCA1 enhances UCA1 decay and competes for miR-122-5p binding, leading to the liberation of miR-122-5p activity and the reduction of cell invasiveness. 29753746_IGF2BP1 is essential for human skin squamous cell carcinoma cell survival and proliferation. 30104206_The present study shows that the regulation of KRAS expression by IMP1 is complex and may involve both the IMP1 protein and its mRNA transcript. 30119193_lncRNA THOR is up-regulated in retinoblastoma, and its over-expression significantly enhances the malignant phenotype transformation of retinoblastoma cells by up-regulating c-myc and TGF2BP1 expression. 30220021_our data suggest that miR-150 and its downstream target IGF2BP1 may be a crucial axis for the development, progression and patients' prognosis of ostesarcoma 30353165_Results found that IGF2BP1 was significantly activated in lung adenocarcinoma (LUAD) patients. Also, the expression of IGF2BP1 was significantly higher in LUAD patients with LIN28B-AS1 activation than other patients. LIN28B-AS1 alters the LIN28B mRNA stability by directly interacting with IGF2BP1. 30371874_35 SRF/IGF2BP1-dependent genes showing conserved association with SRF and IGF2BP1 expression indicate a poor overall survival probability in ovarian, liver and lung cancer. In conclusion, these findings identify the SRF/IGF2BP1-, miRNome- and m6A-dependent control of gene expression as a conserved oncogenic driver network in cancer. 30407516_IMP1 3' UTR shortening is associated with enhanced metastatic burden in colorectal cancer. 30543563_PKCalpha is upregulated by IMP1 in melanoma and may have a role in worse survival 30581152_Results show that p62 binds to IGF2BP1 to control the mRNA stability of FERMT2 and multiple pro-metastatic factors. This p62-RBP interaction distinguishes melanoma from other tumors where p62 controls autophagy or oxidative stress. 30587375_MiR-423-5p inhibited migration, invasion, and proliferation as well as induced apoptosis, by targeting IGF2BP1 trophoblasts, presenting a novel molecular basis implicated in pre-eclampsia pathogenesis. 30649922_Induced miR-222 represses expression of ZBP1 and PLCgamma1 at the posttranscriptional level. 30790682_LINC01093 directly binds insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), interfering with interaction between IGF2BP1 and glioma-associated oncogene homolog 1 (GLI1) mRNA. The result is degradation of GLI1 mRNA, further affecting expression of GLI1 downstream molecules involved in HCC progression. 30864660_IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties. 30936459_The role of RNA-binding protein IGF2BP1 in EV-mediated promotion of melanoma metastasis. 31000197_miR-4500 inhibits human glioma cell progression by targeting IGF2BP1. 31000200_IGF2BP1 promotes lipopolysaccharide-induced NFkappaB signalling and transcriptional activation in human macrophages and monocytes. 31061170_The present study defines a novel interplay between IMP1 and autophagy, where IMP1 may be transiently induced during damage to modulate colonic epithelial cell responses to damage. 31085008_We demonstrate that low IGF2BP1 and FOXM1 expression can serve as a potential factor for the clinical diagnosis and prognosis of lung adenocarcinoma 31220513_THOR mediated IGF2 expression via interacting with IGF2BP1, and affected the downstream MEK-ERK signaling pathway to regulate tongue squamous cell carcinomas cells proliferation. 31537384_The LIN28B-AS1, binding to IGF2BP1, is required for LPS-induced NFkappaB activation and pro-inflammatory responses in human macrophages. 31658691_The interaction between IGF2BP1 and DHAV-1 3' UTR strongly enhanced IRES-mediated translation efficiency but failed to regulate DHAV-1 replication in a duck embryo epithelial (DEE) cell line. 31686763_Study found that IGF2BP1 was upregulated and associated with a poor prognosis in pancreatic cancer patients. Downregulation of IGF2BP1 inhibited pancreatic cancer cell growth in vitro and in vivo via the AKT signaling pathway. Upregulation of IGF2BP1 was attributed to the downregulation of miR-494 expression in pancreatic cancer and reexpression of miR-494 could partially abrogate the oncogenic role of IGF2BP1. 31767839_Data for the leukaemia risk loci BCL2-antagonist-killer 1 protein (BAK1) and insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) suggests deregulation of B-cell development and genetic susceptibility to precursor B-cell lymphoblastic leukemia (Pre B-ALL) [Meta-Analysis]. 31768017_RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. 32034166_Exome sequencing revealed DNA variants in NCOR1, IGF2BP1, SGLT2 and NEK11 as potential novel causes of ketotic hypoglycemia in children. 32134323_Interaction analysis of miR-1275/IGF2BP1/IGF2BP3 with the susceptibility to hepatocellular carcinoma. 32245947_LncRNA LINC00266-1 encodes a 71-amino acid peptide that mainly interacts with the RNA-binding proteins and is thus named 'RNA-binding regulatory peptide' (RBRP). RBRP binds to IGF2BP1 and strengthens m6A recognition by IGF2BP1 on RNAs, such as c-Myc mRNA, to increase the mRNA stability and expression of c-Myc, thereby promoting tumorigenesis. Cancer patients with RBRPhigh have a poor prognosis. 32583425_LINC01426 contributes to clear cell renal cell carcinoma progression by modulating CTBP1/miR-423-5p/FOXM1 axis via interacting with IGF2BP1. 32668274_Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. 32719445_IGF2BP1 is the first positive marker for anaplastic thyroid carcinoma diagnosis. 32761127_The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. 32816599_TRIM71 binds to IMP1 and is capable of positive and negative regulation of target RNAs. 32876513_IGF2BP1 is a targetable SRC/MAPK-dependent driver of invasive growth in ovarian cancer. 32917856_LIN28B-AS1-IGF2BP1 binding promotes hepatocellular carcinoma cell progression. 32926844_IGF2BP1 silencing inhibits proliferation and induces apoptosis of high glucose-induced non-small cell lung cancer cells by regulating Netrin-1. 33051595_Anti-oncogene PTPN13 inactivation by hepatitis B virus X protein counteracts IGF2BP1 to promote hepatocellular carcinoma progression. 33052627_miR-885-5p inhibits proliferation and metastasis by targeting IGF2BP1 and GALNT3 in human intrahepatic cholangiocarcinoma. 33268793_A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. 33382158_circBICD2 targets miR-149-5p/IGF2BP1 axis to regulate oral squamous cell carcinoma progression. 33387362_Long noncoding RNA NBAT1 suppresses hepatocellular carcinoma progression via competitively associating with IGF2BP1 and decreasing c-Myc expression. 33389570_LncRNA LINC00689 Promotes the Tumorigenesis of Glioma via Mediation of miR-526b-3p/IGF2BP1 Axis. 33391523_IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. 33398416_Genetic variants in m(6)A regulators are associated with gastric cancer risk. 33436560_circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. 33582561_METTL3/IGF2BP1/CD47 contributes to the sublethal heat treatment induced mesenchymal transition in HCC. 33629308_LINC00483 is regulated by IGF2BP1 and participates in the progression of breast cancer. 33811613_Carboxymethylated chitosan alleviated oxidative stress injury in retinal ganglion cells via IncRNA-THOR/IGF2BP1 axis. 33853613_CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. 34110327_The pan-cancer analysis of the two types of uterine cancer uncovered clinical and prognostic associations with m6A RNA methylation regulators. 34162562_IGF2BP1/UHRF2 Axis Mediated by miR-98-5p to Promote the Proliferation of and Inhibit the Apoptosis of Esophageal Squamous Cell Carcinoma. 34190193_Development and validation of m6A regulators' prognostic significance for endometrial cancer. 34196212_An oncogenic lncRNA, GLCC1, promotes tumorigenesis in gastric carcinoma by enhancing the c-Myc/IGF2BP1 interaction. 34326314_EGR2-mediated regulation of m(6)A reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 mRNA stabilization. 34951345_Long noncoding RNA UBA6-AS1 inhibits the malignancy of ovarian cancer cells via suppressing the decay of UBA6 mRNA. 34974052_Involvement of m6A regulatory factor IGF2BP1 in malignant transformation of human bronchial epithelial Beas-2B cells induced by tobacco carcinogen NNK. | ENSMUSG00000013415 | Igf2bp1 | 8515.77064 | 1.1102149 | 0.1508389354 | 0.06862587 | 4.848586e+00 | 2.766881e-02 | 2.665332e-01 | No | Yes | 10660.664019 | 576.106125 | 9311.724548 | 492.391112 | |
| ENSG00000159314 | 201176 | ARHGAP27 | protein_coding | Q6ZUM4 | FUNCTION: Rho GTPase-activating protein which may be involved in clathrin-mediated endocytosis. GTPase activators for the Rho-type GTPases act by converting them to an inactive GDP-bound state. Has activity toward CDC42 and RAC1 (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;Cytoplasm;Endocytosis;GTPase activation;Membrane;Phosphoprotein;Reference proteome;Repeat;SH3 domain | This gene encodes a member of a large family of proteins that activate Rho-type guanosine triphosphate (GTP) metabolizing enzymes. The encoded protein may pay a role in clathrin-mediated endocytosis. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Aug 2013]. | hsa:201176; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; membrane [GO:0016020]; GTPase activator activity [GO:0005096]; SH3 domain binding [GO:0017124]; positive regulation of GTPase activity [GO:0043547]; receptor-mediated endocytosis [GO:0006898]; regulation of GTPase activity [GO:0043087]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165] | ENSMUSG00000034255 | Arhgap27 | 155.04000 | 0.7920723 | -0.3362960322 | 0.23927948 | 1.960228e+00 | 1.614889e-01 | 5.523860e-01 | No | Yes | 155.911057 | 22.025185 | 195.579493 | 26.708294 | ||
| ENSG00000159792 | 5681 | PSKH1 | protein_coding | P11801 | FUNCTION: May be a SFC-associated serine kinase (splicing factor compartment-associated serine kinase) with a role in intranuclear SR protein (non-snRNP splicing factors containing a serine/arginine-rich domain) trafficking and pre-mRNA processing. {ECO:0000269|PubMed:12466556}. | ATP-binding;Cell membrane;Cytoplasm;Cytoskeleton;Endoplasmic reticulum;Golgi apparatus;Kinase;Lipoprotein;Membrane;Myristate;Nucleotide-binding;Nucleus;Palmitate;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:5681; | cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; Golgi apparatus [GO:0005794]; microtubule organizing center [GO:0005815]; nuclear speck [GO:0016607]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; determination of left/right symmetry [GO:0007368]; heart development [GO:0007507] | 14644153_A minimal region required for proper Golgi targeting of PSKH1 was identified within the first 29 amino acids; and structural and regulatory role of PSKH1 in maintenance of the Golgi apparatus is determined 32053275_Cataloguing the dead: breathing new life into pseudokinase research. | ENSMUSG00000048310 | Pskh1 | 1237.13821 | 0.9440469 | -0.0830695600 | 0.10855248 | 5.842919e-01 | 4.446349e-01 | 7.869395e-01 | No | Yes | 1396.121523 | 194.235978 | 1422.013380 | 193.154888 | ||
| ENSG00000160145 | 8997 | KALRN | protein_coding | O60229 | FUNCTION: Promotes the exchange of GDP by GTP. Activates specific Rho GTPase family members, thereby inducing various signaling mechanisms that regulate neuronal shape, growth, and plasticity, through their effects on the actin cytoskeleton. Induces lamellipodia independent of its GEF activity. {ECO:0000269|PubMed:10023074}. | 3D-structure;ATP-binding;Alternative splicing;Cytoplasm;Cytoskeleton;Disulfide bond;Guanine-nucleotide releasing factor;Immunoglobulin domain;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Phosphoprotein;Reference proteome;Repeat;SH3 domain;Serine/threonine-protein kinase;Transferase | Huntington's disease (HD), a neurodegenerative disorder characterized by loss of striatal neurons, is caused by an expansion of a polyglutamine tract in the HD protein huntingtin. This gene encodes a protein that interacts with the huntingtin-associated protein 1, which is a huntingtin binding protein that may function in vesicle trafficking. [provided by RefSeq, Apr 2016]. | hsa:8997; | actin cytoskeleton [GO:0015629]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extrinsic component of membrane [GO:0019898]; nucleoplasm [GO:0005654]; postsynaptic density [GO:0014069]; ATP binding [GO:0005524]; guanyl-nucleotide exchange factor activity [GO:0005085]; metal ion binding [GO:0046872]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; axon guidance [GO:0007411]; central nervous system development [GO:0007417]; ephrin receptor signaling pathway [GO:0048013]; intracellular signal transduction [GO:0035556]; nervous system development [GO:0007399]; protein phosphorylation [GO:0006468]; regulation of small GTPase mediated signal transduction [GO:0051056]; signal transduction [GO:0007165]; vesicle-mediated transport [GO:0016192] | 14742910_we have identified multiple transcriptional start sites in rats and humans. These multiple transcriptional start sites result in full-length Kalirin transcripts possessing different 5' ends encoding proteins with differing amino termini 15950621_Kalirin GEF1 domain induces lamellipodia through activation of Pak, where Guanine nucleotide exchange factor (GEF) activity is not required. 17357071_Three SNPs from the kalirin (KALRN) gene are associated with early-onset coronary artery disease. 17640372_ARF6 recruits KALRN to the cell membrane facilitating Rac activation. 17851188_Our observation is the first to relate kalirin to Alzheimer's disease. Kalirin was consistently under-expressed in Alzheimer's disease hippocampus. 18199770_Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. 18839057_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 18953434_Observational study of gene-disease association. (HuGE Navigator) 19706030_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20080650_Observational study of gene-disease association. (HuGE Navigator) 20107840_Observational study of gene-disease association. (HuGE Navigator) 20107840_Two SNPs in the KALRN gene region (rs17286604 and rs11712619)constitute risk factors for ischemic stroke. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20604901_SNX1 and SNX2 interact with Kalirin-7. Overexpression of SNX1 or SNX2 and Kalirin-7 partially redistributes both SNXs to the plasma membrane, and results in RhoG-dependent lamellipodia formation. 20730383_Studies indicate that Kalirin-7 plays a key role in excitatory synapse formation and function. 21041834_Missense mutations in KALRN may be genetic risk factors for schizophrenia. 21041834_Observational study of gene-disease association. (HuGE Navigator) 21664346_KALRN gene variation is not associated with overall ischemic stroke 22120753_We found Kalirin-9 expression to be paradoxically increased in schizophrenia 22194219_Neuronal guanine nucleotide exchange factor (GEF) kalirin is emerging as a key regulator of structural and functional plasticity at dendritic spines. 22429885_The kalirin expression were reduced in Alzheimer disease with psychosis. 22458949_In both anterior cingulate cortex (ACC) and dorsolateral prefrontal cortex (DLPFC), study found a reduction of Duo expression and PAK1 phosphorylation in schizophrenia. Cdc42 protein expression was decreased in ACC but not in DLPFC 22720673_The age-at-onset of Huntington disease (HD) is not associated with eleven SNPs, including SNP rs10934657 in the kalirin gene in 680 European HD patients. 25224588_A sequence variant in human KALRN impairs protein ability to activate Rac1 and coincides with reduced cortical thickness. 25316661_consider the GG genotype and the G allele of rs9289231 polymorphism of KALRN to be genetic risk factors for CAD in an Iranian population, especially in early-stage atherosclerotic vascular disease 25917671_4 KALRN gene SNPs were studied in Han ischemic stroke patients. rs11712619 seemed associated with lacunar stroke until risk factors were considered. re6438833 was significantly associated with ischemic and lacunar stroke. 27218147_GG genotype and the G allele of the rs9289231 polymorphism of KALRN and the rs224766 polymorphism of ADIPOQ genes may be considered genetic risk factors for Iranian type 2 diabetic patients with coronary artery disease. 27421267_DNA sequencing provided evidence linking KALRN to monogenic intellectual disability in two patients. 28152519_Data suggest protein levels of kalirin and CHD7 in circulating extracellular vesicles (EVs) as endothelial dysfunction markers to monitor vascular condition in hypertensive patients with albuminuria. 28706949_The GG genotype and G allele of SNP rs7620580 were associated with a risk for ischemic stroke with an adjusted OR of 3.195 and an OR of 1.446, respectively. Haplotype analysis revealed that A-T-G,G-T-A, and A-T-A haplotypes were associated with ischemic stroke. Our results provide evidence that kalirin gene variations were associated with ischemic stroke in the Chinese Han population. 29241584_The data of this study reveal a novel mechanism for disease-associated single nucleotide variants of KALARN and provide a platform for modeling morphological changes in mental disorders. 29554915_Combination of polymorphisms in the NOD2, IL17RA, EPHA2 and KALRN genes could play a significant role in the development of sarcoidosis by maintaining a chronic pro-inflammatory status in macrophages 29789657_The interaction of kalirin with the C-terminal region of Htt influences the function of kalirin and modulates the cytotoxicity induced by C-terminal Htt. 30232674_SNPs of the KALRN gene are associated with intracranial atherosclerotic stenosis in the northern Chinese population. 31801062_Synaptic Kalirin-7 and Trio Interactomes Reveal a GEF Protein-Dependent Neuroligin-1 Mechanism of Action. 33037113_KALRN mutations promote antitumor immunity and immunotherapy response in cancer. 33658318_Kalirin-RAC controls nucleokinetic migration in ADRN-type neuroblastoma. | ENSMUSG00000061751 | Kalrn | 90.14698 | 0.6223005 | -0.6843165819 | 0.32828738 | 4.261802e+00 | 3.897855e-02 | 3.052087e-01 | No | Yes | 70.135410 | 17.503971 | 105.727758 | 25.842752 | |
| ENSG00000160216 | 56894 | AGPAT3 | protein_coding | Q9NRZ7 | FUNCTION: Converts 1-acyl-sn-glycerol-3-phosphate (lysophosphatidic acid or LPA) into 1,2-diacyl-sn-glycerol-3-phosphate (phosphatidic acid or PA) by incorporating an acyl moiety at the sn-2 position of the glycerol backbone (PubMed:21173190). Acts on LPA containing saturated or unsaturated fatty acids C16:0-C20:4 at the sn-1 position using C18:1, C20:4 or C18:2-CoA as the acyl donor (PubMed:21173190). Also acts on lysophosphatidylcholine, lysophosphatidylinositol and lysophosphatidylserine using C18:1 or C20:4-CoA (PubMed:21173190). Has a preference for arachidonoyl-CoA as a donor (By similarity). Has also a modest lysophosphatidylinositol acyltransferase (LPIAT) activity, converts lysophosphatidylinositol (LPI) into phosphatidylinositol (By similarity). {ECO:0000250|UniProtKB:Q9D517, ECO:0000269|PubMed:21173190}. | Acyltransferase;Alternative splicing;Endoplasmic reticulum;Lipid biosynthesis;Lipid metabolism;Membrane;Nucleus;Phospholipid biosynthesis;Phospholipid metabolism;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Phospholipid metabolism; CDP-diacylglycerol biosynthesis; CDP-diacylglycerol from sn-glycerol 3-phosphate: step 2/3. | The protein encoded by this gene is an acyltransferase that converts lysophosphatidic acid into phosphatidic acid, which is the second step in the de novo phospholipid biosynthetic pathway. The encoded protein may be an integral membrane protein. Two transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:56894; | endomembrane system [GO:0012505]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; Golgi membrane [GO:0000139]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; nuclear envelope [GO:0005635]; plasma membrane [GO:0005886]; 1-acylglycerol-3-phosphate O-acyltransferase activity [GO:0003841]; acyltransferase activity [GO:0016746]; CDP-diacylglycerol biosynthetic process [GO:0016024]; phosphatidic acid biosynthetic process [GO:0006654]; phospholipid biosynthetic process [GO:0008654] | 16620771_Of the two well conserved acyltransferase motifs, NHX(4)D is present in AGPAT8, whereas arginine in the EGTR motif is substituted by aspartate. 19635840_Results report the identification of an integral membrane lysophosphatidic acid-specific acyltransferase, LPAAT3, which regulates Golgi membrane tubule formation, trafficking, and structure by altering phospholipids and lysophospholipids. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20537980_The data is consistent with a structural arrangement in which motif I is located in the cytoplasm and motif II is in the endoplasmic reticulum and Golgi lumen, suggesting a different model for AGPAT3/LPAAT3's enzymatic mechanism. 21173190_enzymatic properties, tissue distribution, and subcellular localization of human AGPAT3 and AGPAT5 | ENSMUSG00000001211 | Agpat3 | 2261.98581 | 0.8561215 | -0.2241125284 | 0.09077200 | 6.054032e+00 | 1.387458e-02 | 1.913536e-01 | No | Yes | 2946.941393 | 288.284145 | 3397.289398 | 324.443141 |
| ENSG00000160584 | 23387 | SIK3 | protein_coding | Q9Y2K2 | FUNCTION: Positive regulator of mTOR signaling that functions by triggering the degradation of DEPTOR, an mTOR inhibitor. Involved in the dynamic regulation of mTOR signaling in chondrocyte differentiation during skeletogenesis (PubMed:30232230). Negatively regulates cAMP signaling pathway possibly by acting on CRTC2/TORC2 and CRTC3/TORC3 (Probable). Prevents HDAC4 translocation to the nucleus (By similarity). {ECO:0000250|UniProtKB:Q6P4S6, ECO:0000269|PubMed:30232230, ECO:0000305|PubMed:29211348}. | ATP-binding;Alternative splicing;Cytoplasm;Disease variant;Dwarfism;Kinase;Magnesium;Metal-binding;Methylation;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:23387; | cytoplasm [GO:0005737]; ATP binding [GO:0005524]; magnesium ion binding [GO:0000287]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; tau-protein kinase activity [GO:0050321]; intracellular signal transduction [GO:0035556]; microtubule cytoskeleton organization [GO:0000226]; positive regulation of TORC1 signaling [GO:1904263]; positive regulation of TORC2 signaling [GO:1904515]; protein phosphorylation [GO:0006468] | 18266979_Fecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. 20056645_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21399663_Overexpression of SIK3 promotes G1/S cell cycle progression, bestows survival advantages to cancer cells for growth and correlates the clinicopathological conditions of patients with ovarian cancer. 22588126_cAMP-mediated regulation of SIK3 suggest that SIK3 may mediate some of the effects of this important second messenger in adipocytes 23393134_tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases 24743732_These results establish the importance of SIK3 as a mitotic regulator and underscore the potential of SIK3 as a druggable antimitotic target. 24886709_SIK3 variants were associated with hypertriglyceridemia in Mexicans. 25060954_Developmental role of Sik3 in hearing and requirement for the maintenance of adult auditory function is reported. 26590148_salt-inducible kinase inhibition decreases proinflammatory cytokines in human myeloid cells upon IL-1R stimulation. 27009967_SIK3 is activated in human osteoarthritic cartilage. 27807598_Data demonstrate that SIK2 and SIK3 mRNA are downregulated in adipose tissue from obese individuals and that the expression is regulated by weight change. SIK2 is also negatively associated with in vivo insulin resistance (HOMA-IR), independently of BMI and age. 27980695_Results found that CpG sites of C1orf106, DMBX1, and SIK3 mediate the genetic risk of psoriasis in Chinese Han population. 28658303_role in mediating high salt and IL-17 synergy leading to breast cancer cell proliferation 29211348_Data suggest that cAMP/protein kinase A-dependent phosphorylation of SIK1, SIK2, and SIK3 inhibits their catalytic activity by inducing 14-3-3 protein binding. 29526696_Targeting of LKB1 or SIK3 diminishes histone acetylation at MEF2C-bound enhancers and deprives leukemia cells of the output of this essential TF. 31350327_An LKB1-SIK Axis Suppresses Lung Tumor Growth and Controls Differentiation. 31849250_High-Throughput Implementation of the NanoBRET Target Engagement Intracellular Kinase Assay to Reveal Differential Compound Engagement by SIK2/3 Isoforms. 31969556_Salt-inducible kinases (SIKs) regulate TGFbeta-mediated transcriptional and apoptotic responses. 32126566_Salt-Inducible Kinase 3 Haplotypes Associated with Noise-Induced Hearing Loss in Chinese Workers. 32343771_Salt-inducible Kinases Are Critical Determinants of Female Fertility. 34839513_[Analysis of SIK3 gene variation in a boy with autism spectrum disorder complicated with epilepsy]. | ENSMUSG00000034135 | Sik3 | 998.34503 | 1.0571704 | 0.0802080015 | 0.12358517 | 4.217026e-01 | 5.160887e-01 | 8.291375e-01 | No | Yes | 1211.925141 | 101.049507 | 1116.456596 | 91.100714 | ||
| ENSG00000161265 | 199746 | U2AF1L4 | protein_coding | Q8WU68 | FUNCTION: RNA-binding protein that function as a pre-mRNA splicing factor. Plays a critical role in both constitutive and enhancer-dependent splicing by mediating protein-protein interactions and protein-RNA interactions required for accurate 3'-splice site selection. Acts by enhancing the binding of U2AF2 to weak pyrimidine tracts. Also participates in the regulation of alternative pre-mRNA splicing. Activates exon 5 skipping of PTPRC during T-cell activation; an event reversed by GFI1. Binds to RNA at the AG dinucleotide at the 3'-splice site (By similarity). Shows a preference for AGC or AGA (By similarity). {ECO:0000250|UniProtKB:Q8BGJ9}. | Acetylation;Alternative splicing;Cytoplasm;Direct protein sequencing;Metal-binding;Nucleus;RNA-binding;Reference proteome;Repeat;Spliceosome;Zinc;Zinc-finger;mRNA processing;mRNA splicing | Mouse_homologues NA; + ;NA | hsa:199746; | cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; spliceosomal complex [GO:0005681]; U2AF complex [GO:0089701]; metal ion binding [GO:0046872]; pre-mRNA 3'-splice site binding [GO:0030628]; mRNA splicing, via spliceosome [GO:0000398] | 18460468_Differential isoform expression and interaction with the P32 regulatory protein controls the subcellular localization of the splicing factor U2AF26 22022969_One of the conservative regions is responsible for the enhancer activity and is able to interact with proteins of HeLa cell nuclear extract. 23246698_a 269 bp region located between the PSENEN and U2AF1L4 human genes is a genuine bidirectional promoter regulating a concerted divergent transcription of these genes. 27143377_U2AF1L4 gene expression is regulated by cytokines in activated T cells. 32116123_The zinc finger domains in U2AF26 and U2AF35 have diverse functionalities including a role in controlling translation. | ENSMUSG00000109378+ENSMUSG00000078765 | Gm49396+U2af1l4 | 173.46315 | 0.9803670 | -0.0286061663 | 0.22480108 | 1.615980e-02 | 8.988445e-01 | 9.717475e-01 | No | Yes | 160.801699 | 18.363234 | 160.189216 | 17.905177 | |
| ENSG00000162062 | 80178 | TEDC2 | protein_coding | Q7L2K0 | FUNCTION: Acts as a positive regulator of ciliary hedgehog signaling. Required for centriole stability. {ECO:0000250|UniProtKB:Q6GQV0}. | Alternative splicing;Cell projection;Cytoplasm;Cytoskeleton;Phosphoprotein;Reference proteome | hsa:80178; | centriole [GO:0005814]; cilium [GO:0005929]; cytoplasm [GO:0005737]; positive regulation of smoothened signaling pathway [GO:0045880] | ENSMUSG00000024118 | Tedc2 | 891.22198 | 1.0311741 | 0.0442879975 | 0.11698877 | 1.433933e-01 | 7.049306e-01 | 9.103315e-01 | No | Yes | 1141.014762 | 139.004271 | 1069.276271 | 127.287790 | |||
| ENSG00000162300 | 7542 | ZFPL1 | protein_coding | O95159 | FUNCTION: Required for cis-Golgi integrity and efficient ER to Golgi transport. Involved in the maintenance of the integrity of the cis-Golgi, possibly via its interaction with GOLGA2/GM130. {ECO:0000269|PubMed:18323775}. | ER-Golgi transport;Golgi apparatus;Membrane;Metal-binding;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport;Zinc;Zinc-finger | hsa:7542; | Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; nucleus [GO:0005634]; DNA binding [GO:0003677]; zinc ion binding [GO:0008270]; regulation of transcription, DNA-templated [GO:0006355]; vesicle-mediated transport [GO:0016192] | 18323775_These data suggest that ZFPL1 has an important function in maintaining the integrity of the cis-Golgi and that it does so through interactions with GM130. 28447717_These results suggest that ZFPL1 may serve an important role in regulating autophagy in NCIN87 and BGC823 cells. 30036656_The RING domain was essential for the function of ZFPL1. | ENSMUSG00000024792 | Zfpl1 | 602.30982 | 1.0846395 | 0.1172156267 | 0.14572192 | 6.485855e-01 | 4.206188e-01 | 7.738870e-01 | No | Yes | 762.601200 | 86.123912 | 676.691403 | 74.641992 | ||
| ENSG00000162341 | 219931 | TPCN2 | protein_coding | Q8NHX9 | FUNCTION: Intracellular channel initially characterized as a non-selective Ca(2+)-permeable channel activated by NAADP (nicotinic acid adenine dinucleotide phosphate), it is also a highly-selective Na(+) channel activated directly by PI(3,5)P2 (phosphatidylinositol 3,5-bisphosphate) (PubMed:19387438, PubMed:19620632, PubMed:20880839, PubMed:30860481, PubMed:32167471, PubMed:31825310, PubMed:23063126, PubMed:24776928, PubMed:23394946, PubMed:24502975). Localizes to the lysosomal and late endosome membranes where it regulates organellar membrane excitability, membrane trafficking, and pH homeostasis. Is associated with a plethora of physiological processes, including mTOR-dependent nutrient sensing, skin pigmentation and autophagy (PubMed:32167471, PubMed:23394946, PubMed:18488028) (Probable). Ion selectivity is not fixed but rather agonist-dependent and under defined ionic conditions, can be readily activated by both NAADP and PI(3,5)P2 (PubMed:31825310, PubMed:32167471, PubMed:24502975). As calcium channel, it increases the pH in the lysosomal lumen, as sodium channel, it promotes lysosomal exocytosis (PubMed:31825310, PubMed:32167471). Plays a crucial role in endolysosomal trafficking in the endolysosomal degradation pathway and is potentially involved in the homeostatic control of many macromolecules and cell metabolites (By similarity). Unlike the voltage-dependent TPCN1, TPCN2 is voltage independent and can be activated solely by PI(3,5)P2 binding. In contrast, PI(4,5)P2, PI(3,4)P2, PI(3)P and PI(5)P have no obvious effect on channel activation (PubMed:30860481). {ECO:0000250|UniProtKB:Q8BWC0, ECO:0000269|PubMed:18488028, ECO:0000269|PubMed:19387438, ECO:0000269|PubMed:19620632, ECO:0000269|PubMed:20880839, ECO:0000269|PubMed:23063126, ECO:0000269|PubMed:23394946, ECO:0000269|PubMed:24502975, ECO:0000269|PubMed:24776928, ECO:0000269|PubMed:30860481, ECO:0000269|PubMed:31825310, ECO:0000269|PubMed:32167471, ECO:0000305|PubMed:32679067}.; FUNCTION: (Microbial infection) During Ebola virus (EBOV) infection, controls the movement of endosomes containing virus particles and is required by EBOV to escape from the endosomal network into the cell cytoplasm. {ECO:0000269|PubMed:25722412}.; FUNCTION: (Microbial infection) Required for cell entry of coronaviruses SARS-CoV and SARS-CoV-2, as well as human coronavirus EMC (HCoV-EMC), by endocytosis. {ECO:0000269|PubMed:32221306}. | 3D-structure;Calcium;Calcium channel;Calcium transport;Endosome;Glycoprotein;Ion channel;Ion transport;Lysosome;Membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport;Voltage-gated channel | This gene encodes a putative cation-selective ion channel with two repeats of a six-transmembrane-domain. The protein localizes to lysosomal membranes and enables nicotinic acid adenine dinucleotide phosphate (NAADP) -induced calcium ion release from lysosome-related stores. This ubiquitously expressed gene has elevated expression in liver and kidney. Two common nonsynonymous SNPs in this gene strongly associate with blond versus brown hair pigmentation.[provided by RefSeq, Dec 2009]. | hsa:219931; | cytosol [GO:0005829]; endolysosome membrane [GO:0036020]; endosome membrane [GO:0010008]; integral component of membrane [GO:0016021]; late endosome membrane [GO:0031902]; lysosomal membrane [GO:0005765]; lysosome [GO:0005764]; identical protein binding [GO:0042802]; intracellular phosphatidylinositol-3,5-bisphosphate-sensitive cation channel activity [GO:0097682]; ligand-gated sodium channel activity [GO:0015280]; NAADP-sensitive calcium-release channel activity [GO:0072345]; phosphatidylinositol-3,5-bisphosphate binding [GO:0080025]; protein kinase binding [GO:0019901]; voltage-gated calcium channel activity [GO:0005245]; calcium-mediated signaling [GO:0019722]; cellular calcium ion homeostasis [GO:0006874]; endocytosis involved in viral entry into host cell [GO:0075509]; endosome to lysosome transport of low-density lipoprotein particle [GO:0090117]; ion transmembrane transport [GO:0034220]; lysosome organization [GO:0007040]; receptor-mediated endocytosis of virus by host cell [GO:0019065]; regulation of autophagy [GO:0010506]; regulation of exocytosis [GO:0017157]; regulation of ion transmembrane transport [GO:0034765]; smooth muscle contraction [GO:0006939]; sodium ion transmembrane transport [GO:0035725] | 18488028_Genome-wide association study of gene-disease association. (HuGE Navigator) 18488028_Two coding variants in TPCN2 are associated with hair color, and a variant at the ASIP locus shows strong association with skin sensitivity to sun, freckling and red hair. 20495006_analysis of two-pore channel 2 (TPCN2)-mediated Ca2+ currents in isolated lysosomes 20720007_TPC2 is a novel NAADP-sensitive Ca2+ release channel, operating as a dual sensor of luminal pH and Ca2+. 23077736_These results provide strong evidence for modulation of store-operated Ca(2+) entry by TPC2 involving de novo association between TPC2 and STIM1, as well as Orai1, in human cells. 24502975_Notably, NAADP-mediated Ca(2+) release in intact cells is regulated by Mg(2+), PI(3,5)P2, and P38/JNK kinases, thus paralleling regulation of TPC2 currents. 25157141_TPC2, but not TPC1, caused a proliferation of endolysosomal structures, dysregulating intracellular trafficking, and cellular pigmentation. 25236446_SNPs within Tpcn2 are associated with fasting insulin in humans. 25331892_These results demonstrate that a VEGFR2/NAADP/TPC2/Ca(2+) signaling pathway is critical for VEGF-induced angiogenesis 25416817_TPC2 is thus a potential drug target within a pathogenic LRRK2 cascade that disrupts Ca(2+)-dependent trafficking in Parkinson disease 25451935_NAADP induced marked Ca(2+) transients in HEK293 cells that stably coexpressed hTPC2 with hTPC1 or cTPC3, but failed to evoke any such response in cells that coexpressed interacting hTPC2 and rTPC3 subunits 25722412_TPC1 and TPC2 proteins play a key role in Ebola virus infection and may be effective targets for antiviral therapy. 26152696_Studies suggest that both two-pore channels TPC1 and TPC2 as nicotinic acid adenine dinucleotide phosphate (NAADP) targets. 26202466_Results show that PDGs use previously unknown mechanisms of membrane dynamics and content exchange that are regulated by TPC2. 26468524_Here, using live cell imaging, the authors obtained evidence that in contrast to the new model, ebolavirus enters cells through endolysosomes that contain both NPC1 and TPC2. 26838264_Results demonstrated the unique bell-shaped regulation of hTPC2 channel activity by [NAADP] and that channel activity is modulated by PKA phosphorylation at position S666. 26918892_rs1551305 single nucleotide polymorphism is associated with type 2 diabetes risk. 27140606_TPC2 regulates pigmentation through two fundamental determinants of melanosome function: pH and size. 27353380_These findings indicate potential differential regulation of signaling processes by TPC1 and TPC2 in breast cancer cells. 27941820_the divergent pore regions from human TPC2, a two-domain channel that holds a key intermediate position in the evolution of voltage-gated ion channels, were characterized. 28096396_The ion selectivity of Arabidopsis thaliana TPC1 was compared with the selectivity of human TPC2. HsTPC2 was confirmed as a Na(+)-selective channel activated by phosphatidylinositol 3,5-bisphosphate. The ion permeability ratios of HsTPC2 and its mutants were calculated. 28923947_TPC2 polymorphisms are associated with a hair pigmentation. 29563152_Data (including data from studies using tissue from knockout mice) suggest that beta-adrenergic stimulation of pancreas leads to glucagon secretion by hierarchy of calcium signaling in glucagon-secreting cell; such signaling is initiated by cAMP-induced TPC2-dependent calcium release from acidic stores and is further amplified by calcium release from endoplasmic reticulum. 29705952_PI(3,5)P2-binding site in hTPC2 identified at positively charged amino acids (K203, K204, and K207) in the linker between transmembrane helices S4 and S5 and by S322 in the cytosolic extension of S6 and protein-lipid interface upon mutations of residues within the lipid-binding pocket had neither an effect on the binding behavior nor on the channel's lipid sensitivity. 29990474_support a role of TPC2 in autophagy progression and extracellular vesicle trafficking in cancer cells 30860481_cryo-EM structure of human TPC2 provides insights into the mechanism of PI(3,5)P2-regulated gating of TPC2, which is distinct from that of TPC1 33465068_Human genome diversity data reveal that L564P is the predominant TPC2 variant and a prerequisite for the blond hair associated M484L gain-of-function effect. 33875769_Flavonoids increase melanin production and reduce proliferation, migration and invasion of melanoma cells by blocking endolysosomal/melanosomal TPC2. | ENSMUSG00000048677 | Tpcn2 | 643.41914 | 0.8522934 | -0.2305780034 | 0.12617356 | 3.322332e+00 | 6.834483e-02 | 3.867750e-01 | No | Yes | 670.312497 | 74.286948 | 784.431386 | 84.562099 | |
| ENSG00000162377 | 65260 | COA7 | protein_coding | Q96BR5 | FUNCTION: Required for assembly of mitochondrial respiratory chain complex I and complex IV. {ECO:0000269|PubMed:24333015}. | Acetylation;Disease variant;Mitochondrion;Neurodegeneration;Neuropathy;Reference proteome;Repeat | hsa:65260; | mitochondrial intermembrane space [GO:0005758]; mitochondrion [GO:0005739]; nucleoplasm [GO:0005654] | 24333015_C1orf163 is a novel factor important for the assembly of respiratory chain complexes in human mitochondria. 27683825_The first patient carrying pathogenic mutations of COA7, causative of isolated COX deficiency and progressive neurological impairment is reported. It is also shown that COA7 is a soluble protein localized to the matrix, rather than in the intermembrane space as previously suggested. 29718187_The results of this study suggest that loss-of-function COA7 mutation is responsible for the phenotype of the presented patients, and this new entity of disease would be referred to as spinocerebellar ataxia with axonal neuropathy type 3. 30885959_pathogenic mutant versions of COA7 are imported slower than the wild-type protein, and mislocalized proteins are degraded in the cytosol by the proteasome. 35210360_Mitochondrial COA7 is a heme-binding protein with disulfide reductase activity, which acts in the early stages of complex IV assembly. | ENSMUSG00000048351 | Coa7 | 2443.25747 | 0.9285084 | -0.1070131948 | 0.09165562 | 1.357881e+00 | 2.439051e-01 | 6.366644e-01 | No | Yes | 2555.310296 | 263.116637 | 2602.438507 | 262.060800 | ||
| ENSG00000162390 | 26027 | ACOT11 | protein_coding | Q8WXI4 | FUNCTION: Has an acyl-CoA thioesterase activity with a preference for the long chain fatty acyl-CoA thioesters hexadecanoyl-CoA/palmitoyl-CoA and tetradecanoyl-CoA/myristoyl-CoA which are the main substrates in the mitochondrial beta-oxidation pathway. {ECO:0000269|PubMed:22897136}. | 3D-structure;Alternative splicing;Cytoplasm;Fatty acid metabolism;Hydrolase;Lipid metabolism;Mitochondrion;Phosphoprotein;Reference proteome;Repeat;Serine esterase;Transit peptide | PATHWAY: Lipid metabolism; fatty acid metabolism. {ECO:0000305|PubMed:22897136}. | This gene encodes a member of the acyl-CoA thioesterase family which catalyse the conversion of activated fatty acids to the corresponding non-esterified fatty acid and coenzyme A. Expression of a mouse homolog in brown adipose tissue is induced by low temperatures and repressed by warm temperatures. Higher levels of expression of the mouse homolog has been found in obesity-resistant mice compared with obesity-prone mice, suggesting a role of acyl-CoA thioesterase 11 in obesity. Alternative splicing results in transcript variants. [provided by RefSeq, Nov 2010]. | hsa:26027; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; mitochondrial matrix [GO:0005759]; acyl-CoA hydrolase activity [GO:0047617]; carboxylic ester hydrolase activity [GO:0052689]; lipid binding [GO:0008289]; long-chain acyl-CoA hydrolase activity [GO:0052816]; long-chain fatty acyl-CoA binding [GO:0036042]; myristoyl-CoA hydrolase activity [GO:0102991]; palmitoyl-CoA hydrolase activity [GO:0016290]; acyl-CoA metabolic process [GO:0006637]; fatty acid metabolic process [GO:0006631]; intracellular signal transduction [GO:0035556]; negative regulation of cold-induced thermogenesis [GO:0120163]; response to cold [GO:0009409]; response to temperature stimulus [GO:0009266] | 11696000_BFIT supports the transition of thermogenic brown adipose tissue towards increased metabolic activity, probably through alteration of intracellular fatty acyl-CoA concentration. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 22897136_The N-terminal region of human fat inducible thioesterase variant 2 constitutes a mitochondrial location signal sequence, which undergoes mitochondrion-dependent posttranslational cleavage. 32820071_Allosteric regulation of thioesterase superfamily member 1 by lipid sensor domain binding fatty acids and lysophosphatidylcholine. | ENSMUSG00000034853 | Acot11 | 25.80370 | 0.9539178 | -0.0680630674 | 0.57456037 | 1.394434e-02 | 9.059994e-01 | No | Yes | 24.538709 | 4.900636 | 25.224360 | 4.916762 | |
| ENSG00000162669 | 164045 | HFM1 | protein_coding | A2PYH4 | FUNCTION: Required for crossover formation and complete synapsis of homologous chromosomes during meiosis. {ECO:0000250|UniProtKB:D3Z4R1}. | ATP-binding;Alternative splicing;Disease variant;Helicase;Hydrolase;Meiosis;Nucleotide-binding;Premature ovarian failure;Reference proteome | The protein encoded by this gene is thought to be an ATP-dependent DNA helicase and is expressed mainly in germ-line cells. Defects in this gene are a cause of premature ovarian failure 9 (POF9). [provided by RefSeq, Apr 2014]. | hsa:164045; | nucleus [GO:0005634]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA helicase activity [GO:0003678]; nucleic acid binding [GO:0003676]; resolution of meiotic recombination intermediates [GO:0000712] | 17286053_hHFM1 is the evolutionally conserved putative human DNA helicase, which may function as a modulator for genome integrity in germ-line tissues. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) 24597873_Exome sequencing of two Chinese sisters with primary ovarian insufficiency and their parents identified a shared compound heterozygous mutation in a meiotic gene, HFM1, which encodes a protein necessary for homologous recombination of chromosomes. 26679638_Data suggest that HFM1 gene might be associated with primary ovarian insufficiency in Chinese population. 31279343_A novel heterozygous missense mutation in HFM1 (c.3470G > A) associated with premature ovarian insufficiency (POI) is in the affected family members and absent in the unaffected family members in China. Results of the minigene assay reveals that the mutation changes the mRNA splicing repertory. 34429122_Novel variants in helicase for meiosis 1 lead to male infertility due to non-obstructive azoospermia. | ENSMUSG00000043410 | Hfm1 | 70.05830 | 0.8553408 | -0.2254287022 | 0.34215493 | 4.317067e-01 | 5.111527e-01 | No | Yes | 61.118802 | 18.301215 | 72.974951 | 21.482223 | ||
| ENSG00000162878 | 91461 | PKDCC | protein_coding | Q504Y2 | FUNCTION: Secreted tyrosine-protein kinase that mediates phosphorylation of extracellular proteins and endogenous proteins in the secretory pathway, which is essential for patterning at organogenesis stages. Mediates phosphorylation of MMP1, MMP13, MMP14, MMP19 and ERP29 (PubMed:25171405). Probably plays a role in platelets: rapidly and quantitatively secreted from platelets in response to stimulation of platelet degranulation (PubMed:25171405). May also have serine/threonine protein kinase activity. Required for longitudinal bone growth through regulation of chondrocyte differentiation. May be indirectly involved in protein transport from the Golgi apparatus to the plasma membrane (By similarity). {ECO:0000250|UniProtKB:Q5RJI4, ECO:0000269|PubMed:25171405}. | ATP-binding;Developmental protein;Differentiation;Disease variant;Glycoprotein;Golgi apparatus;Kinase;Nucleotide-binding;Osteogenesis;Phosphoprotein;Protein transport;Reference proteome;Secreted;Signal;Transferase;Transport;Tyrosine-protein kinase | hsa:91461; | extracellular region [GO:0005576]; Golgi apparatus [GO:0005794]; ATP binding [GO:0005524]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase activity [GO:0004672]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; bone mineralization [GO:0030282]; cell differentiation [GO:0030154]; embryonic digestive tract development [GO:0048566]; limb morphogenesis [GO:0035108]; lung alveolus development [GO:0048286]; multicellular organism growth [GO:0035264]; negative regulation of Golgi to plasma membrane protein transport [GO:0042997]; peptidyl-tyrosine phosphorylation [GO:0018108]; positive regulation of bone mineralization [GO:0030501]; positive regulation of chondrocyte differentiation [GO:0032332]; protein transport [GO:0015031]; roof of mouth development [GO:0060021]; skeletal system development [GO:0001501] | 19961619_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 24407380_our results suggest that genes CA10 and also SGK493 may be an important risk factor for asthma development, especially for a nonatopic phenotype. 25171405_VLK is rapidly and quantitatively secreted from platelets in response to stimuli and can tyrosine phosphorylate coreleased proteins utilizing endogenous as well as exogenous ATP sources. 25862977_The Fam20C-and VLK-family of kinases mediate the phosphorylation of proteins in the secretory pathway and extracellular space.Mutation in several secretory pathway kinases cause human disease 27591737_VLK secretion can be regulated by external cues, intracellular signal proteins, and mechanical stretch, and VLK can in turn regulate TyrP of ECM proteins secreted by trabecular meshwork cells and control cell shape, actin stress fibers, and focal adhesions. 30478137_Each patient had a homozygous gene disrupting variant in PKDCC considered to explain the skeletal phenotypes shared by both. 31845979_Taken together, these results suggest that Vlk may function as a signaling regulator in extracellular space to modulate the Hedgehog pathway 34329392_The secreted tyrosine kinase VLK is essential for normal platelet activation and thrombus formation. | ENSMUSG00000024247 | Pkdcc | 514.18459 | 1.2619725 | 0.3356805253 | 0.14017851 | 5.770635e+00 | 1.629614e-02 | 2.067526e-01 | No | Yes | 727.428728 | 101.123367 | 575.184067 | 78.226595 | ||
| ENSG00000163297 | 118429 | ANTXR2 | protein_coding | P58335 | FUNCTION: Necessary for cellular interactions with laminin and the extracellular matrix. {ECO:0000269|PubMed:11683410, ECO:0000269|PubMed:12973667}.; FUNCTION: (Microbial infection) Receptor for the protective antigen (PA) of B.anthracis (PubMed:12700348, PubMed:15243628, PubMed:15326297). Binding of PA leads to heptamerization of the receptor-PA complex (PubMed:12700348, PubMed:15243628, PubMed:15326297). Upon binding of the PA of B.anthracis, the complex moves to glycosphingolipid-rich lipid rafts, where it is internalized via a clathrin-dependent pathway (PubMed:12551953, PubMed:15337774). In the endosomal membrane, at pH under 7, the complex then rearranges and forms a pore allowing the other components of anthrax toxin to escape to the cytoplasm (PubMed:12551953, PubMed:15337774). {ECO:0000269|PubMed:12551953, ECO:0000269|PubMed:12700348, ECO:0000269|PubMed:15243628, ECO:0000269|PubMed:15326297, ECO:0000269|PubMed:15337774}. | 3D-structure;Alternative splicing;Cell membrane;Disease variant;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Membrane;Metal-binding;Phosphoprotein;Receptor;Reference proteome;Secreted;Signal;Transmembrane;Transmembrane helix | This gene encodes a receptor for anthrax toxin. The protein binds to collagen IV and laminin, suggesting that it may be involved in extracellular matrix adhesion. Mutations in this gene cause juvenile hyaline fibromatosis and infantile systemic hyalinosis. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2009]. | hsa:118429; | cell surface [GO:0009986]; endoplasmic reticulum membrane [GO:0005789]; endosome membrane [GO:0010008]; extracellular region [GO:0005576]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; metal ion binding [GO:0046872]; transmembrane signaling receptor activity [GO:0004888]; toxin transport [GO:1901998] | 12973667_identification and characterization of juvenile hyaline fibromatosis and infantile systemic hyalinosis disease-causing mutations in the capillary morphogenesis factor-2 gene 14508707_identification of 15 different mutations in the gene encoding capillary morphogenesis protein 2 in 17 families with juvenile hyaline fibromatosis or infantile systemic hyalinosis 15079089_structures provide a template to begin probing the high-affinity capillary morphogenesis protein 2-protective antigen interaction and may facilitate understanding of toxin assembly/internalization 15243628_crystal structure of the complex with anthrax toxin 16401723_Data show that cells expressing palmitoylation-defective mutant receptors are less sensitive to anthrax toxin due to a lower number of surface receptors as well as premature internalization of protective antigen without a requirement for heptamerization. 16473908_Molecular dynamics simulations explain the great strength that the protective antigen-capillary morphogenesis gene 2 complex achieves through extraordinary coordination of a divalent cation. 17183731_Divalent metal ion coordination by residue T118 of anthrax toxin receptor 2 is not essential for protective antigen binding. 19191226_Results suggest that, in vivo, slow folding, rather than misfolding, is responsible for ER retention, and that systemic hyalinosis can be qualified as a conformational disease. 19240061_Observational study of gene-disease association. (HuGE Navigator) 19858192_Cytoplasmic delivery of lethal factor by anthrax toxin receptor 2 was mediated by cathepsin B. 20062062_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20331448_the molecular basis (spectrum of ANTXR2 mutations) of infantile systemic hyalinosis and juvenile hyaline fibromatosis in two unrelated Egyptian families 20652271_ANTXR2 and IL-1R2 polymorphisms are not associated with ankylosing spondylitis in Chinese Han population. 20652271_Observational study of gene-disease association. (HuGE Navigator) 21041274_Observational study of gene-disease association. (HuGE Navigator) 21328543_Missense mutations in the extracellular von Willebrand domain of CMG2 lead to protein folding defects. 22118297_ANTXR2 might not be a susceptibility gene of Ankylosing Spondylitis in Chinese Han. 22315420_results reveal extensive diversity in cell lethality dependent on anthrax toxin PA-mediated toxin binding and uptake, and identify individual differences in CMG2 expression level as a determinant of this diversity 22383261_2 novel ANTXR2 mutations were identified in patients with hyaline fibromatosis syndrome 22855243_Investigated the kinetics of binding as a function of pH to the full-length monomeric anthrax protective antigen(PA)and to two variants to CMG2. Results suggest that for the full-length PA, low pH increases the binding affinity. 24380801_Fused the ANTXR2 ectodomain to the C-terminus of bacterial Trigger Factor (TF), and the fusion protein was overly expressed as a dominant soluble protein in E coli. 24641616_A protective antigen mutation increases the pH threshold of anthrax toxin receptor 2-mediated pore formation 24667935_Loss of CMG2 expression is associated with breast cancer progression. 24742682_Silencing CMG2 using targeted siRNAs provided almost complete protection against anthrax lethal toxin-induced cytotoxicity and death in murine and human macrophages. 24924630_data suggested that cellular cholesterol regulated ANTXR2-dependent activation of MMP-2 via ERK1/2 phosphorylation in neuroglioma U251 cell. 24993339_Results show that CMG2 has been shown to be able to regulate the proliferation and tubule formation of endothelial cells, but not the migration. 25169729_Ankylosing spondylitis is associated with the anthrax toxin receptor 2 gene. 25736362_miR-124 might induce autophagy to participate in AS by targeting ANTXR2, which might be implicated in pathological process of AS. 25781883_CMG2 glycosylation provides a buffer towards genetic variation by promoting folding of the protein in the ER lumen. 26107617_The disulfide bond Cys255-Cys279 in the immunoglobulin-like domain of ANTXR2 is required for membrane insertion of anthrax protective antigen pore. 26590821_The association between ANTXR2 rs4333130 and ankylosing spondylitis was independent of HLA-B27 status. Clinical disease severity scores (BASDAI and BASFI) and pain score were higher in ANTXR2 rs4333130 CT genotype. 26703731_Anthrax Susceptibility: Human Genetic Polymorphisms Modulating ANTXR2 Expression 26728147_This study reports a novel association between ANTXR2 and ankylosing spondylitis in the Han Chinese. 26805886_Roles of Anthrax Toxin Receptor 2 in Anthrax Toxin Membrane Insertion and Pore Formation 26885603_Case Report: systemic hyalinosis with a heterozygous mutation in CMG2. 27170489_expression does not affect cytotoxicity to anthrax toxin 27174544_study examines 4 cases with clinical features of hyaline fibromatosis syndrome; identified a previously unreported splice junction mutation (c.946-2A-->G in intron 11) and a recurrent founder mutation ( c.1074delT) 27874231_Stability of domain 4 of the anthrax toxin protective antigen and the effect of the VWA domain of CMG2 on stability 28103792_Results identified Gly116Val mutation in ANTXR2 associated with hyaline fibromatosis syndrome. 29215551_A decreased CMG2 expression is a negative prognostic factor for soft tissue sarcoma patients. 29473166_CMG2 is an indicator of poor prognosis of glioma patients and also a stimulator of cell cycle progression in cultured glioma cells. 29662192_CMG2 promotes GC progression by maintaining GCSLCs and can serve as a new prognostic indicator and a target for human GC therapy. 29801470_Results from hyaline fibromatosis syndrome (HFS) patients with consanguineous background identified a novel homozygous frameshift deletion c.969del (p.Ile323Metfs*14), a previously reported mutation c.134 T > C (p.Leu45Pro) and a recurrent homozygous frameshift mutation c.1073dup (p.Ala359Cysfs*13). These findings improves the understanding of a recurrent mutation in HFS. 30176098_Study provides evidence to reinforce the previous hypothesis that missense mutations in exons 1-12 in ANTXR2 and mutations leading to a premature stop codon lead to the severe form of the disease, while missense pathogenic variants in exons 13-17 lead to the mild form of hyaline fibromatosis syndrome. [review] 30255098_this study shows no association between ANTXR2 polymorphisms and ankylosing spondylitis susceptibility in a Chinese Han population, but meta-analysis showed that rs4389526 in the ANTXR2 gene was weakly associated with AS susceptibility in both Caucasian and Chinese Han patients. 30809297_Elevation of ANTXR2 promotes endometriotic cell adhesion, proliferation, and angiogenesis. Furthermore, hypoxia is the driving force for ANTXR2 upregulation via altering histone modification of ANTXR2. 31038183_CMG2 promoted the invasion of glioma cells 32003961_A Canstatin-Derived Peptide Provides Insight into the Role of Capillary Morphogenesis Gene 2 in Angiogenic Regulation and Matrix Uptake. 32664053_Protective effect of anthrax toxin receptor 2 polymorphism rs4333130 against the risk of ankylosing spondylitis. 33147779_Enhanced Collagen Deposition in the Duodenum of Patients with Hyaline Fibromatosis Syndrome and Protein Losing Enteropathy. 33724566_Novel variation in ANTXR2 gene causing hyaline fibromatosis syndrome: A report from India. 33749907_Genetic deletion of CMG2 exacerbates systemic-to-pulmonary shunt-induced pulmonary arterial hypertension. 34782625_Multiple stages of evolutionary change in anthrax toxin receptor expression in humans. | ENSMUSG00000029338 | Antxr2 | 233.66457 | 1.0428369 | 0.0605135334 | 0.19757727 | 9.381641e-02 | 7.593803e-01 | 9.299340e-01 | No | Yes | 280.187476 | 48.118693 | 260.810058 | 43.792233 | |
| ENSG00000163491 | 152110 | NEK10 | protein_coding | Q6ZWH5 | FUNCTION: Plays a role in the cellular response to UV irradiation. Mediates G2/M cell cycle arrest, MEK autoactivation and ERK1/2-signaling pathway activation in response to UV irradiation. In ciliated cells of airways, it is involved in the regulation of mucociliary transport (PubMed:31959991). {ECO:0000269|PubMed:20956560, ECO:0000269|PubMed:31959991}. | ATP-binding;Alternative splicing;Ciliopathy;Coiled coil;Disease variant;Kinase;Magnesium;Metal-binding;Nucleotide-binding;Primary ciliary dyskinesia;Reference proteome;Serine/threonine-protein kinase;Transferase | hsa:152110; | extracellular region [GO:0005576]; protein kinase complex [GO:1902911]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein kinase activity [GO:0004672]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; mucociliary clearance [GO:0120197]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of protein autophosphorylation [GO:0031954]; protein phosphorylation [GO:0006468]; regulation of cell cycle G2/M phase transition [GO:1902749]; regulation of ERK1 and ERK2 cascade [GO:0070372] | 19330027_Observational study of gene-disease association. (HuGE Navigator) 20095854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20145138_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20956560_Nek10 physically associated with Raf-1 and MEK1 in a Raf-1-dependent manner, and the formation of this complex was necessary for Nek10-mediated MEK1 activation. 31959991_analysis of a bronchiectasis syndrome caused by mutations that inactivate NIMA-related kinase 10 (NEK10), a protein kinase with previously unknown in vivo functions in mammals 32414360_Homozygous truncating NEK10 mutation, associated with primary ciliary dyskinesia: a case report. 32561851_NEK10 tyrosine phosphorylates p53 and controls its transcriptional activity. | ENSMUSG00000042567 | Nek10 | 60.12281 | 0.8517731 | -0.2314588714 | 0.38026160 | 3.661245e-01 | 5.451244e-01 | No | Yes | 78.722721 | 28.361706 | 87.627837 | 30.759676 | |||
| ENSG00000163517 | 79885 | HDAC11 | protein_coding | Q96DB2 | FUNCTION: Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. {ECO:0000269|PubMed:11948178}. | Alternative splicing;Chromatin regulator;Hydrolase;Nucleus;Reference proteome;Repressor;Transcription;Transcription regulation | This gene encodes a class IV histone deacetylase. The encoded protein is localized to the nucleus and may be involved in regulating the expression of interleukin 10. Alternative splicing results in multiple transcript variants.[provided by RefSeq, Apr 2009]. | hsa:79885; | histone deacetylase complex [GO:0000118]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; DNA-binding transcription factor binding [GO:0140297]; histone deacetylase activity [GO:0004407]; NAD-dependent histone deacetylase activity (H3-K14 specific) [GO:0032041]; chromatin organization [GO:0006325]; histone deacetylation [GO:0016575]; oligodendrocyte development [GO:0014003] | 11948178_cloning and functional characterization 16142391_Fluorescence in situ hybridization analysis localized HDAC11 gene to chromosome 3p25, a region characterized by frequent gains and losses of chromosomal material in a number of various types of cancer. 17201809_the absence of tumour-specific somatic events in WNT7A and HDAC11 suggests that these genes are unlikely to have a classical tumour suppressor gene role in sporadic malignant pancreatic endocrine tumours 19011628_HDAC11 negatively regulated expression of the gene encoding interleukin 10 in antigen-presenting cells. 19276081_Cdt1 undergoes acetylation and is reversibly deacetylated by HDAC11 20467334_We found a reduction of HDAC 11 mRNA and increased HDAC 2 levels in amyotrophic lateral sclerosis brain and spinal cord compared with controls. 20980834_HDAC11 associates with replication origins inhibits Cdt1-induced re-replication and suppresses MCM loading. 21239696_These results demonstrate for the first time that HDAC11 plays an essential role in regulating OX40L expression. 21806350_Data indicate a pronounced deregulation of HDAC genes HDAC9 and HDAC11 in patients with Philadelphia-negative chronic myeloproliferative neoplasms: essential thrombocythemia (ET), polycythemia vera (PV) and primary myelofibrosis (PMF). 23024001_High HDAC11 expression is associated with neoplasms. 28252645_Our study identified a group of cell cycle-promoting genes regulated by HDAC11. 28782861_Results showed that the high levels of HDAC11 and lower levels of p53 were detected in pituitary tumor cells. A negative correlation was detected between the data of HDAC11 and p53. 29222071_HDAC11 was initially identified as a negative regulator of the well-known anti-inflammatory cytokine IL-10. Hence, antagonizing HDAC11 activity may have anti-tumor potential, whereas activating HDAC11 may be useful to treat chronic inflammation or autoimmunity. 29336543_report in vitro profiling of HDAC11 deacylase activities 29523311_Mycobacterium tuberculosis infection disturbs the HDAC6/HDAC11 levels to induce IL-10 expression in macrophages. 29655790_The results indicated that HDAC11 was significantly expressed in human and mouse diabetic heart failure (DHF) hearts. 30007011_IL-10 expression was compromised in the peripheral B cells of allergic rhinitis (AR) patients. Inhibition of HDAC11 restored the IL-10 expression ability in the AR-B cells. We also found a negative correlation between HDAC11 and the expression of IL-10 in B cells; this fact indicates that HDAC11 interferes with the expression of IL-10 in B cells. 30063898_HDAC11 is a regulatory molecule in Th2 response and plays a critical role in the restriction of the biased IL-13 expression in CD4(+) T cells of the heart. 30819897_regulates type I interferon signaling through defatty-acylation of SHMT2 32170113_Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. 32705287_Long noncoding RNA BCYRN1 promotes prostate cancer progression via elevation of HDAC11. 33602787_HDAC11 Regulates Glycolysis through the LKB1/AMPK Signaling Pathway to Maintain Hepatocellular Carcinoma Stemness. | ENSMUSG00000034245 | Hdac11 | 227.36653 | 0.6677426 | -0.5826359128 | 0.19991862 | 8.361652e+00 | 3.832220e-03 | 1.035482e-01 | No | Yes | 220.101198 | 29.781334 | 317.011138 | 41.587571 | |
| ENSG00000163659 | 25976 | TIPARP | protein_coding | Q7Z3E1 | FUNCTION: ADP-ribosyltransferase that mediates mono-ADP-ribosylation of glutamate, aspartate and cysteine residues on target proteins (PubMed:23275542, PubMed:25043379, PubMed:30373764). Acts as a negative regulator of AHR by mediating mono-ADP-ribosylation of AHR, leading to inhibit transcription activator activity of AHR (PubMed:23275542, PubMed:30373764). {ECO:0000269|PubMed:23275542, ECO:0000269|PubMed:25043379, ECO:0000269|PubMed:30373764}. | ADP-ribosylation;Glycosyltransferase;Metal-binding;NAD;Nucleus;Reference proteome;Transferase;Zinc;Zinc-finger | This gene encodes a member of the poly(ADP-ribose) polymerase superfamily. Studies of the mouse ortholog have shown that the encoded protein catalyzes histone poly(ADP-ribosyl)ation and may be involved in T-cell function. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2010]. | hsa:25976; | nucleus [GO:0005634]; cis-regulatory region sequence-specific DNA binding [GO:0000987]; metal ion binding [GO:0046872]; NAD+ ADP-ribosyltransferase activity [GO:0003950]; protein ADP-ribosylase activity [GO:1990404]; androgen metabolic process [GO:0008209]; cellular response to organic cyclic compound [GO:0071407]; estrogen metabolic process [GO:0008210]; face morphogenesis [GO:0060325]; female gonad development [GO:0008585]; hemopoiesis [GO:0030097]; kidney development [GO:0001822]; negative regulation of gene expression [GO:0010629]; platelet-derived growth factor receptor signaling pathway [GO:0048008]; positive regulation of protein catabolic process [GO:0045732]; post-embryonic development [GO:0009791]; protein ADP-ribosylation [GO:0006471]; protein auto-ADP-ribosylation [GO:0070213]; protein mono-ADP-ribosylation [GO:0140289]; roof of mouth development [GO:0060021]; skeletal system morphogenesis [GO:0048705]; smooth muscle tissue development [GO:0048745]; vasculogenesis [GO:0001570] | 12851707_TIPARP (DKFZp434J214) gene is amplified in HNSCC. TIPARP, FLJ22693 and ZAP proteins with TPH, WW and PARP-like domains constitute the TIPARP family. 20852632_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20852632_Single nucleotide polymorphism in TIPARP is associated with ovarian cancer. 24806346_Knockdown of TiPARP, but not AHRR, increased 2,3,7,8-tetrachlorodibenzo-p-dioxin - induced CYP1A1 mRNA and AHR protein levels. 26814197_these data identify a new mechanism of LXR regulation that involves TIPARP, ADP-ribosylation and MACROD1. 28213497_TIPARP is a viral RNA-sensing pattern recognition receptors that mediates antiviral responses triggered by BAX- and BAK1-dependent mitochondrial damage 29274782_Moreover, time course and promoter activity assays suggest that TIPARP and TIPARP-AS1 work in concert to regulate AHR signaling. Collectively, these data show an added level of complexity in the AHR signaling cascade which involves lncRNAs, whose functions remain poorly understood. 29938598_circHECTD1 functions as an endogenous MIR142 (microRNA 142) sponge to inhibit MIR142 activity, resulting in the inhibition of TIPARP (TCDD inducible poly[ADP-ribose] polymerase) expression with subsequent inhibition of astrocyte activation via macroautophagy/autophagy. 30373764_Mutation of cysteine 39 to alanine resulted in a small, but significant, reduction in TCDD inducible poly(ADP-ribose) polymerase protein (TIPARP) autoribosylation activity. 32482854_TiPARP forms nuclear condensates to degrade HIF-1alpha and suppress tumorigenesis. 33475084_Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets. 33475085_Identification of PARP-7 substrates reveals a role for MARylation in microtubule control in ovarian cancer cells. 33572475_Post-Transcriptional Regulation of PARP7 Protein Stability Is Controlled by Androgen Signaling. 33799807_PARP7 and Mono-ADP-Ribosylation Negatively Regulate Estrogen Receptor alpha Signaling in Human Breast Cancer Cells. 33976187_Androgen signaling uses a writer and a reader of ADP-ribosylation to regulate protein complex assembly. 34264286_PARP7 mono-ADP-ribosylates the agonist conformation of the androgen receptor in the nucleus. 34375612_PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity. | ENSMUSG00000034640 | Tiparp | 628.91938 | 1.1939103 | 0.2556944801 | 0.12752135 | 4.038045e+00 | 4.448533e-02 | 3.206095e-01 | No | Yes | 856.750373 | 188.091933 | 658.619310 | 141.509820 | |
| ENSG00000163902 | 6184 | RPN1 | protein_coding | P04843 | FUNCTION: Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation (PubMed:31831667). N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity. {ECO:0000250|UniProtKB:E2RQ08, ECO:0000269|PubMed:31831667}. | 3D-structure;Acetylation;Endoplasmic reticulum;Glycoprotein;Isopeptide bond;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix;Ubl conjugation | PATHWAY: Protein modification; protein glycosylation. {ECO:0000269|PubMed:31831667}. | This gene encodes a type I integral membrane protein found only in the rough endoplasmic reticulum. The encoded protein is part of an N-oligosaccharyl transferase complex that links high mannose oligosaccharides to asparagine residues found in the Asn-X-Ser/Thr consensus motif of nascent polypeptide chains. This protein forms part of the regulatory subunit of the 26S proteasome and may mediate binding of ubiquitin-like domains to this proteasome. [provided by RefSeq, Jul 2008]. | hsa:6184; | cytosol [GO:0005829]; endoplasmic reticulum [GO:0005783]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; melanosome [GO:0042470]; membrane [GO:0016020]; oligosaccharyltransferase complex [GO:0008250]; rough endoplasmic reticulum [GO:0005791]; RNA binding [GO:0003723]; cellular protein modification process [GO:0006464]; protein N-linked glycosylation [GO:0006487]; protein N-linked glycosylation via asparagine [GO:0018279] | 18607003_ribophorin I can regulate the delivery of precursor proteins to the OST complex by capturing substrates and presenting them to the catalytic core. 19064571_Observational study of gene-disease association. (HuGE Navigator) 22988243_Data indicate that malectin functions by forming a complex with ribophorin I. 25451265_These results clearly demonstrate that the association of malectin with ribophorin I is required to capture misfolded alpha1-antitrypsin and direct them to the degradation pathway. 31843888_Study provides evidence that Rpn1-Ser361 is a prevalent phosphosite that is required for proper assembly and activity of the 26S proteasome and identified multiple kinases, including PIM1/2/3, that can phosphorylate Rpn1-S361 and have demonstrated that UBLCP1 is a physiologically relevant phosphatase of this site. Blocking phosphorylation of Rpn1-S361 leads to slowed proliferation and mitochondrial dysfunction. 33617881_Polyubiquitin and ubiquitin-like signals share common recognition sites on proteasomal subunit Rpn1. | ENSMUSG00000030062 | Rpn1 | 13584.89784 | 1.0243961 | 0.0347736912 | 0.06938757 | 2.515334e-01 | 6.159974e-01 | 8.760413e-01 | No | Yes | 16163.445646 | 2650.336094 | 15529.981579 | 2489.544331 |
| ENSG00000163913 | 55764 | IFT122 | protein_coding | Q9HBG6 | FUNCTION: As a component of the IFT complex A (IFT-A), a complex required for retrograde ciliary transport and entry into cilia of G protein-coupled receptors (GPCRs), it is required in ciliogenesis and ciliary protein trafficking (PubMed:27932497, PubMed:29220510). Involved in cilia formation during neuronal patterning. Acts as a negative regulator of Shh signaling. Required to recruit TULP3 to primary cilia (By similarity). {ECO:0000250|UniProtKB:Q6NWV3, ECO:0000269|PubMed:27932497, ECO:0000269|PubMed:29220510}. | Alternative splicing;Cell projection;Ciliopathy;Cilium;Cilium biogenesis/degradation;Cytoplasm;Cytoskeleton;Developmental protein;Disease variant;Ectodermal dysplasia;Reference proteome;Repeat;WD repeat | This gene encodes a member of the WD repeat protein family. WD repeats are minimally conserved regions of approximately 40 amino acids typically bracketed by gly-his and trp-asp (GH-WD), which may facilitate formation of heterotrimeric or multiprotein complexes. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation. This cytoplasmic protein contains seven WD repeats and an AF-2 domain which function by recruiting coregulatory molecules and in transcriptional activation. Mutations in this gene cause cranioectodermal dysplasia-1. A related pseudogene is located on chromosome 3. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2013]. | hsa:55764; | ciliary basal body [GO:0036064]; ciliary tip [GO:0097542]; cilium [GO:0005929]; cytoplasm [GO:0005737]; intraciliary transport particle A [GO:0030991]; membrane [GO:0016020]; non-motile cilium [GO:0097730]; photoreceptor connecting cilium [GO:0032391]; camera-type eye morphogenesis [GO:0048593]; cilium assembly [GO:0060271]; embryonic body morphogenesis [GO:0010172]; embryonic heart tube development [GO:0035050]; intraciliary retrograde transport [GO:0035721]; intraciliary transport [GO:0042073]; limb development [GO:0060173]; negative regulation of smoothened signaling pathway [GO:0045879]; neural tube closure [GO:0001843]; non-motile cilium assembly [GO:1905515]; protein localization to cilium [GO:0061512] | 20493458_we found a homozygous missense mutation in the IFT122 (WDR10) gene that cosegregated with Sensenbrenner syndrome 23826986_this study was able to find causative IFT122 mutations in a non-consanguineous family with recurrent abortions. 26792575_The three patients had different, novel, compound heterozygous mutations in IFT122. Consequently, we compared these three patients to those previously described with IFT122 mutations. Thus, our report serves to add 6 novel mutations to the IFT122 mutation spectrum and to contribute to the IFT122-related clinical characterization. 28370949_Using a panel of skeletal dysplasias genes, including 11 related to SRP, we identified biallelic mutations in IFT122 in a fetus with a typical phenotype of SRP-IV, finally confirmed that this phenotype is a ciliopathy and adding to the list of ciliopathies with major skeletal involvement. 29037998_All the nine probands with syndromic craniosynostosis were found to carry the possibly causative variants, among which three variants including two missense mutations in IFT122 gene, in SMC1A gene and a frameshift mutation in TWIST1 gene have never been reported in patients before. 29057857_This study demonstrated that the mutation in SPG 7 gene caused autosomal recessive hereditary spastic paraparesis. 29220510_IFT122 mutations associated with cranioectodermal dysplasia 1 cause defects in ciliary protein trafficking, but not ciliogenesis when expressed in cells lacking endogenous IFT122 (IFT122 KO). 30476139_The C11ORF74, interacts with the IFT-A complex via the IFT122 subunit and is accumulated at the distal tip in the absence of an IFT-A subunit IFT139, suggesting that at least a fraction of C11ORF74 molecules can be transported towards the ciliary tip by associating with the IFT-A complex, although its majority might be out of cilia at steady state. | ENSMUSG00000030323 | Ift122 | 817.30219 | 1.0399378 | 0.0564972395 | 0.11700717 | 2.332557e-01 | 6.291202e-01 | 8.816466e-01 | No | Yes | 955.093598 | 79.852290 | 901.410411 | 73.627531 | |
| ENSG00000164117 | 26269 | FBXO8 | protein_coding | Q9NRD0 | FUNCTION: May promote guanine-nucleotide exchange on an ARF. Promotes the activation of ARF through replacement of GDP with GTP (Potential). {ECO:0000305}. | Alternative splicing;Guanine-nucleotide releasing factor;Reference proteome | hsa:26269; | ubiquitin ligase complex [GO:0000151]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of ARF protein signal transduction [GO:0032012]; ubiquitin-dependent protein catabolic process [GO:0006511] | Mouse_homologues 18094045_Fbx8, an F-box protein bearing the Sec7 domain, mediates ubiquitination of Arf6. | ENSMUSG00000038206 | Fbxo8 | 414.01070 | 1.2264404 | 0.2944771736 | 0.14900900 | 3.926214e+00 | 4.753884e-02 | 3.297281e-01 | No | Yes | 560.822864 | 128.013217 | 430.123913 | 96.212417 | ||
| ENSG00000164609 | 10569 | SLU7 | protein_coding | O95391 | FUNCTION: Required for pre-mRNA splicing as component of the spliceosome (PubMed:10197984, PubMed:28502770, PubMed:30705154). Participates in the second catalytic step of pre-mRNA splicing, when the free hydroxyl group of exon I attacks the 3'-splice site to generate spliced mRNA and the excised lariat intron. Required for holding exon 1 properly in the spliceosome and for correct AG identification when more than one possible AG exists in 3'-splicing site region. May be involved in the activation of proximal AG. Probably also involved in alternative splicing regulation. {ECO:0000269|PubMed:10197984, ECO:0000269|PubMed:10647016, ECO:0000269|PubMed:12764196, ECO:0000269|PubMed:15181151, ECO:0000269|PubMed:15728250, ECO:0000269|PubMed:28502770, ECO:0000269|PubMed:30705154}. | 3D-structure;Acetylation;Cytoplasm;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Spliceosome;Ubl conjugation;Zinc;Zinc-finger;mRNA processing;mRNA splicing | Pre-mRNA splicing occurs in two sequential transesterification steps. The protein encoded by this gene is a splicing factor that has been found to be essential during the second catalytic step in the pre-mRNA splicing process. It associates with the spliceosome and contains a zinc knuckle motif that is found in other splicing factors and is involved in protein-nucleic acid and protein-protein interactions. [provided by RefSeq, Jul 2008]. | hsa:10569; | catalytic step 2 spliceosome [GO:0071013]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; small nuclear ribonucleoprotein complex [GO:0030532]; spliceosomal complex [GO:0005681]; pre-mRNA 3'-splice site binding [GO:0030628]; second spliceosomal transesterification activity [GO:0000386]; zinc ion binding [GO:0008270]; alternative mRNA splicing, via spliceosome [GO:0000380]; cellular response to heat [GO:0034605]; intracellular protein transport [GO:0006886]; mRNA 3'-splice site recognition [GO:0000389]; mRNA splicing, via spliceosome [GO:0000398]; RNA splicing [GO:0008380]; RNA splicing, via transesterification reactions [GO:0000375] | 15181151_The zinc-knuckle motif of hSlu7 determines the cellular localization of the protein through a nucleocytoplasmic-sensitive shuttling balance. 15728250_hSlu7 has a broad effect on alternative splicing 17804646_Regulation of transcription of the RNA splicing factor hSlu7 by Elk-1 and Sp1 affects alternative splicing 21122810_RBM22 and hSlu7 differ significantly in their subcellular distributions under stress conditions, and RBM22 enhances the cytoplasmic translocation of hSlu7 under stress. 23754748_These observations together point to an altered recruitment and dependence on SLU7, suggesting its role in facilitating transitions that promote catalysis, and highlight the diversity in spliceosome assembly. 24865429_SLU7 knockdown in liver cells resulted in profound changes in pre-mRNA splicing and gene expression, leading to impaired glucose and lipid metabolism, refractoriness to key metabolic hormones, and reversion to a fetal-like gene expression pattern. 26804174_these findings indicate that SLU7 is co-opted by hepatocellular carcinoma (HCC) cells and other tumor cell types to maintain survival, and identify this splicing regulator as a new determinant for the expression of the oncogenic miR-17-92 cluster. This novel mechanism may be exploited for the development of antitumoral strategies in cancers displaying such SLU7-miR-17-92 crosstalk 29870742_High SLU7 expression is associated with alcoholic steatohepatitis. 30657957_We define a molecular pathway through which SLU7 keeps in check the generation of truncated forms of the splicing factor SRSF3 (SRp20) (SRSF3-TR). Behaving as dominant negative, or by gain-of-function, SRSF3-TR impair the correct splicing and expression of the splicing regulator SRSF1 (ASF/SF2) and the crucial SCC protein sororin. 34170569_Splicing Factor SLU7 Prevents Oxidative Stress-Mediated Hepatocyte Nuclear Factor 4alpha Degradation, Preserving Hepatic Differentiation and Protecting From Liver Damage. 34331453_The splicing regulator SLU7 is required to preserve DNMT1 protein stability and DNA methylation. | ENSMUSG00000020409 | Slu7 | 2053.01035 | 1.1352299 | 0.1829844887 | 0.09103028 | 4.056752e+00 | 4.399504e-02 | 3.195696e-01 | No | Yes | 2867.300111 | 467.798019 | 2481.026362 | 395.960240 | |
| ENSG00000164776 | 5260 | PHKG1 | protein_coding | Q16816 | FUNCTION: Catalytic subunit of the phosphorylase b kinase (PHK), which mediates the neural and hormonal regulation of glycogen breakdown (glycogenolysis) by phosphorylating and thereby activating glycogen phosphorylase. In vitro, phosphorylates PYGM, TNNI3, MAPT/TAU, GAP43 and NRGN/RC3 (By similarity). {ECO:0000250}. | ATP-binding;Alternative splicing;Calmodulin-binding;Carbohydrate metabolism;Glycogen metabolism;Kinase;Muscle protein;Nucleotide-binding;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene is a member of the Ser/Thr protein kinase family and encodes a protein with one protein kinase domain and two calmodulin-binding domains. This protein is the catalytic member of a 16 subunit protein kinase complex which contains equimolar ratios of 4 subunit types. The complex is a crucial glycogenolytic regulatory enzyme. This gene has two pseudogenes at chromosome 7q11.21 and one at chromosome 11p11.12. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2012]. | hsa:5260; | cytosol [GO:0005829]; phosphorylase kinase complex [GO:0005964]; ATP binding [GO:0005524]; calmodulin binding [GO:0005516]; enzyme binding [GO:0019899]; phosphorylase kinase activity [GO:0004689]; protein serine kinase activity [GO:0106310]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; tau-protein kinase activity [GO:0050321]; carbohydrate metabolic process [GO:0005975]; glycogen biosynthetic process [GO:0005978] | 35150045_Lung adenocarcinoma-derived vWF promotes tumor metastasis by regulating PHKG1-mediated glycogen metabolism. | ENSMUSG00000025537 | Phkg1 | 24.44519 | 1.8849280 | 0.9145094193 | 0.61175899 | 2.225762e+00 | 1.357257e-01 | No | Yes | 33.620326 | 9.822566 | 17.127364 | 5.206360 | ||
| ENSG00000165238 | 65268 | WNK2 | protein_coding | Q9Y3S1 | FUNCTION: Serine/threonine kinase which plays an important role in the regulation of electrolyte homeostasis, cell signaling, survival, and proliferation. Acts as an activator and inhibitor of sodium-coupled chloride cotransporters and potassium-coupled chloride cotransporters respectively. Activates SLC12A2, SCNN1A, SCNN1B, SCNN1D and SGK1 and inhibits SLC12A5. Negatively regulates the EGF-induced activation of the ERK/MAPK-pathway and the downstream cell cycle progression. Affects MAPK3/MAPK1 activity by modulating the activity of MAP2K1 and this modulation depends on phosphorylation of MAP2K1 by PAK1. WNK2 acts by interfering with the activity of PAK1 by controlling the balance of the activity of upstream regulators of PAK1 activity, RHOA and RAC1, which display reciprocal activity. {ECO:0000269|PubMed:17667937, ECO:0000269|PubMed:18593598, ECO:0000269|PubMed:21733846}. | 3D-structure;ATP-binding;Alternative splicing;Cell membrane;Cytoplasm;Kinase;Membrane;Methylation;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | The protein encoded by this gene is a cytoplasmic serine-threonine kinase that belongs to the protein kinase superfamily. The protein plays an important role in the regulation of electrolyte homeostasis, cell signaling survival, and proliferation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2013]. | hsa:65268; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; chloride channel inhibitor activity [GO:0019869]; potassium channel inhibitor activity [GO:0019870]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; intracellular signal transduction [GO:0035556]; ion homeostasis [GO:0050801]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of sodium ion transport [GO:0010766]; positive regulation of canonical Wnt signaling pathway [GO:0090263]; positive regulation of potassium ion import across plasma membrane [GO:1903288]; positive regulation of sodium ion transmembrane transporter activity [GO:2000651]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468] | 17578925_epigenetic silencing, occasional deletion and point mutation, and functional assessment suggest that aberrations of WNK2 may contribute to unregulated tumor cell growth 17667937_WNK2 is involved in the modulation of growth factor-induced cancer cell proliferation through the MEK1/ERK1/2 pathway. 18593598_WNK2 controls a RhoA-mediated cross-talk mechanism that regulates the efficiency with which MEK1 can activate ERK1/2 upon growth factor stimulation. 19001526_Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21733846_a role for WNK2 in the regulation of CCCs in the mammalian brain, with implications for both cell volume regulation and/or GABAergic signaling. 23035050_results validate the WNK2 gene as a recurrent target for epigenetic silencing in glia-derived brain tumours and provide first mechanistic evidence for a tumour-suppressing role of WNK2 that is related to Rac1 signalling and tumour cell invasion and growth 23797875_Wnk kinases are positive regulators of canonical Wnt/beta-catenin signaling. 23912455_Downregulation of WNK2 by promoter hypermethylation occurs early in Pancreatic ductal adenocarcinoma pathogenesis and may support tumor cell growth via the ERK-MAPK pathway. 25596741_WNK2 promoter methylation and silencing in gliomas is associated with increased JNK activation and MMP2 expression and activity, thus explaining in part tumor cell invasion potential. 31009242_The data suggest that miR-370 acts as an oncogene by downregulating WNK2 in breast cancer. 31349001_Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. 31884577_Long non-coding RNA LINC00858 exerts a tumor-promoting role in colon cancer via HNF4alpha and WNK2 regulation. 32093151_WNK2 Inhibits Autophagic Flux in Human Glioblastoma Cell Line. 32447640_LINC00858 knockdown inhibits gastric cancer cell growth and induces apoptosis through reducing WNK2 promoter methylation. 32768499_Long non-coding RNA LINC00858 inhibits colon cancer cell apoptosis, autophagy, and senescence by activating WNK2 promoter methylation. 34061329_Circ_0001666 affects miR-620/WNK2 axis to inhibit breast cancer progression. | ENSMUSG00000037989 | Wnk2 | 1220.35534 | 0.8879274 | -0.1714863306 | 0.11376980 | 2.257826e+00 | 1.329406e-01 | 5.101836e-01 | No | Yes | 1250.058485 | 148.914358 | 1371.892694 | 159.634062 | |
| ENSG00000165795 | 57447 | NDRG2 | protein_coding | Q9UN36 | FUNCTION: Contributes to the regulation of the Wnt signaling pathway. Down-regulates CTNNB1-mediated transcriptional activation of target genes, such as CCND1, and may thereby act as tumor suppressor. May be involved in dendritic cell and neuron differentiation. {ECO:0000269|PubMed:12845671, ECO:0000269|PubMed:16103061, ECO:0000269|PubMed:21247902}. | 3D-structure;Acetylation;Alternative splicing;Cell projection;Cytoplasm;Developmental protein;Differentiation;Neurogenesis;Phosphoprotein;Reference proteome;Tumor suppressor;Wnt signaling pathway | This gene is a member of the N-myc downregulated gene family which belongs to the alpha/beta hydrolase superfamily. The protein encoded by this gene is a cytoplasmic protein that may play a role in neurite outgrowth. This gene may be involved in glioblastoma carcinogenesis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2017]. | hsa:57447; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; Golgi apparatus [GO:0005794]; growth cone [GO:0030426]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; cell differentiation [GO:0030154]; negative regulation of cytokine production [GO:0001818]; negative regulation of ERK1 and ERK2 cascade [GO:0070373]; negative regulation of smooth muscle cell proliferation [GO:0048662]; regulation of platelet-derived growth factor production [GO:0090361]; regulation of vascular endothelial growth factor production [GO:0010574]; signal transduction [GO:0007165]; substantia nigra development [GO:0021762]; Wnt signaling pathway [GO:0016055] | 11936845_Cloning and expression of the gene; highly expressed in adult skeletal muscle and brain 12845671_Down-Regulation of N-Myc downstream-regulated gene 2 is associated with glioblastoma 15207261_NDRG2 is upregulated at both the RNA and protein levels in Alzheimer disease brains; expression in affected brains is revealed in cortical pyramidal neurons, senile plaques and cellular processes of dystrophic neurons. 15461589_results identify NDRG1 and NDRG2 as physiological substrates for SGK1, and demonstrate that phosphorylation of NDRG1 by SGK1 primes it for phosphorylation by GSK3 15526377_NDRG2 gene might express differently between normal tissues and cancer tissues, and might play an important role in the development of pancreatic cancer and liver cancer. 16103061_Data identify NDRG2 as the first specific candidate tumor suppressor gene on chromosome 14q that is inactivated during meningioma progression. 17050536_NDRG2 expression is repressed by Myc via Miz-1-dependent interaction with the NDRG2 core promoter 17109818_modulation of Ndrg2 level influences the cell cycle process together with MSP58 17470364_describe for the first time, the mechanisms involved in NDRG2 gene down-regulation 17688410_These results show that the expression of the NDRG2 gene is directly or indirectly induced by WT1, and provide the first insights into transcriptional regulation of the NDRG2 gene, including demonstration of a novel splice variant. 17888401_Together, these data demonstrate that NDRG2 expression in breast cancer cells is able to inhibit STAT3 activation via SOCS1 induction in a p38 MAPK dependent manner. 17911180_The NDRG2 is able to preserve ALCAM expression during DC differentiation from monocytes under cytokine culture conditions and that its expression helps DC maintain costimulatory signals necessary for T cell stimulation. 17935612_expression of NDRG2 is down-regulated at a late stage during colorectal carcinogenesis. 18209490_This study suggests that NDRG2 is a Hif-1 target gene and closely related with hypoxia-induced apoptosis in A549 cells. 18519680_results suggest that NDRG2 can inhibit extracellular matrix-based, Rho-driven tumor cell invasion and migration and thereby play important roles in suppressing tumor metastasis in hepatocellular carcinoma 18591767_NDRG2 might play an important role in the carcinogenesis and development of clear cell renal cell carcinoma and may function as a tumor suppressor in clear cell renal cell carcinoma 18636358_NDRG2 expression in colon cancer was not correlated with age, sex, metastasis of lymph node, or depth of infiltration, while it was correlated to the histology grading. 18709645_NDRG2 as a candidate tumor suppressor gene that is epigenetically silenced in the majority of primary glioblastomas, but not in lower grade astrocytomas and secondary glioblastomas. 18844221_NDRG2 modulates intracellular signals to control cell cycle through the regulation of cyclin D1 expression via phosphorylation pathway and down-regulation of AP-1 18940011_Reduced expression in human thyroid cancer 19204049_NDRG2 expression is highly responsive to different stress conditions in skeletal muscle and suggest that the level of NDRG2 expression may be critical to myoblast growth and differentiation. 19237607_Loss of NDRG2 is associated with colon adenocarcinoma. 19336468_NDRG2 expression significantly suppresses tumor invasion by inhibiting MMP activities, which are regulated through the NF-kappaB signaling 19434539_Ndrg2 protein levels increased from poor-differentiated to well-differentiated carcinomas. 19450561_BMP-4 induced by NDRG2 expression inhibits the metastatic potential of breast cancer cells, especially via suppression of MMP-9 activity. 19483300_NDRG2 may play a role during the differentiation of colorectal cancer cells. 19775754_NDRG2, was significantly higher expressed in normal macrophages compared to primary acute myeloid leukemia cells 20045673_NDRG2 is a candidate tumor-suppressor gene for oral squamous-cell carcinoma (OSCC)development and probably contributes to the tumorigenesis of OSCC partly via the modulation of Akt signaling. 20206160_NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells 20331630_NDRG2 played an important role in the proliferation of esophageal squamous cell carcinoma (ESCC) cells and the expression of NDRG2 in ESCC was closely related with the prognosis. 20438703_NDRG2 might have a pivotal role as one of intrinsic factors for the modulation of p38 MAPK phosphorylation, and subsequently involve in controlling of IL-10 production. 20607352_NDRG2 gene expression is down-regulated in atypical and recurrent meningiomas. 20886841_Data show that NDRG2 and GFAP had an increased number of phosphospectra in FTLD. 21220491_Present research provides the first evidence that decreased NDRG2 mRNA expression in primary human CRC might be a powerful, independent predictor of recurrence and outcome. 21226903_Data show that NDRG2 mRNA is statistically significantly down-regulated in breast cancer. 21247902_Structural analysis suggests that NDRG2 is a nonenzymatic member of the ABH superfamily, because it lacks the catalytic signature residues and has an occluded substrate-binding site. 21352815_these results suggest that NDRG2 expression is regulated by promoter methylation and miR-650 in human colorectal cancer cells. 21623166_Data show that the expression of the NDRG2 genes was low in the three PCA cell lines. 21672576_results suggest that Ndrg2 may regulate astroglial activation through the suppression of cell proliferation and stabilization of cell morphology 21676268_Our findings indicate that NDRG2 and CD24 regulate HCC adhesion, migration and invasion. 21705028_NDRG2 expression negatively correlates with pathological grading but positively with the life span of astrocytoma patients, demonstrating that NDRG2 is a potential prognostic biomarker for human astrocytoma. 21741166_Results demonstrate that NDRG2 overexpression can inhibit tumor growth and invasion, furthermore, it can decrease bone destruction caused by prostate cancer bone metastasis. 21771789_estrogen/NDRG2/Na(+)/K(+)-ATPase beta1 pathway is important in promoting Na(+)/K(+)-ATPase activity and suggest this novel pathway might have substantial roles in ion transport, fluid balance, and homeostasis. 21872476_NDRG2 expression is decreased in gliomas, indicating that NDRG2 may play an inhibitory role during the development of gliomas. NDRG2 expression may also be a significant and independent prognostic indicator for glioma. 22043305_NDRG2 is up regulated in the cells from human age-related cortical cataract in vivo. Up-regulation of NDRG2 induces cell morphological changes, reduces cell viability, and especially lowers cellular resistance to oxidative stress. 22110735_NDRG2 contributed to an increase in the ratio of matrix metalloproteinase 2 (MMP2) to tissue inhibitor of matrix metalloproteinase 2 22135002_Our data suggest that NDRG2 down-regulation or CD24 up-regulation is an important feature of primary gallbladder carcinoma 22138128_Study reveals that ndrg2 is a novel gene that participates in Leydig cell apoptosis, with essential functions in testicular cells, and suggests its possible role in apoptotic Leydig cells and male fertility. 22246425_Loss of NDRG2 is associated with renal cell carcinoma. 22528516_Our results suggest that the decreased expression of NDRG2 or the increased expression of CD24 is an important feature of lung adenocarcinoma. 22565195_this research provides the molecular basis for understanding the role of NDRG2 in tumor cells and raises interesting questions about its mechanisms and potential use in cancer therapy. 22692967_NDRG2 could suppress clear cell renal cell carcinoma cell invasion through regulating MMP-9 expression and activity. 22696597_our data show that NDRG2 significantly suppress tumor metastasis by attenuating active autocrine TGF-beta production 22920753_In vitro drug sensitivity assay revealed that suppression of NDRG2 could sensitize Hela cells to cisplatin.Inhibition of NDRG2 in Hela cells was accompanied by decreased Bcl-2 protein level. 23010743_These results suggest that the aberrant methylation of NDRG2 may be mainly responsible for its downregulation in gastric cancer, and may play an important role in the metastasis of gastric cancer. 23056173_Data indicate that N-myc downstream-regulated gene 2 (NDRG2) is a novel direct target gene of farnesoid X receptor (FXR) in mouse liver and human hepatoma cell lines 23068607_Results suggest that NDRG2 and prenylated Rab acceptor-1 (PRA1) might act synergistically to prevent signaling of T-cell facto/beta-catenin. 23307246_These data showed that NDRG2 may play an important role in human lung cancer tumourigenesis 23451161_findings bring insight to the roles of NDRG2 in ischemic-hypoxic injury and provide potential targets for future clinical therapies on stroke 23800335_The present experiments demonstrated that NDRG2 may be a diagnostic and prognostic marker in patients with oesophageal squamous cell carcinoma 23900729_NDRG2 expression with cell density preceded E-cadherin expression, and the regulation of Snail activity by GSK-3beta was also related to this process. 24134849_the expression of NDRG2 is an independent prognostic factor in pancreatic cancer. 24146910_NDRG2 overexpression can inhibit tumor growth and invasion in vitro. 24222185_the aberrant expression of NDRG2 and NDRG3 may contribute to the malignant progression of prostate cancer 24528032_Taken together, these data indicate that NDRG2 inhibits the proliferation of neuroblastoma cells partially through suppression of cysteine-rich protein 61. 24569712_Data show the expression of N-myc downstream-regulated gene 2 (NDRG2) is frequently downregulated in adult T-cell leukaemia-lymphoma, resulting in enhanced phosphorylation of PTEN and activation of the phosphatidylinositol 3-kinase (PI3K)-AKT pathway. 24636131_Data suggest targeting the actions of N-myc downstream-regulated gene 2 (NDRG2) in cell glucose-dependent energy delivery may provide a strategy for therapeutic intervention in breast carcinoma. 24839115_NDRG2 can significantly inhibit the proliferation of OS-RC-2 cells in vitro and its protein is specifically expressed in the mitochondrion 24840052_findings show NDRG2 expression is downregulated in glioma and level of NDRG2 expression negatively correlates with glioma grade;findings also indicate NDRG2 downregulation may be due to aberrant methylation of NDRG2 promoter region and subsequent transcriptional inactivation 25153349_the inhibition of STAT3 signaling by NDRG2 suppresses EMT progression of EMT via the down-regulation of Snail expression 25256221_these data show that the inhibition of NF-kappaB signaling by NDRG2 expression is able to suppress cell migration and invasion through the down-regulation of COX-2 expression. 25261367_NDRG2/gp130/STAT3 pathway can mediate the antimetastatic effects of Dp44mT in hepatocellular carcinoma. 25338637_The downregulation of NDRG2 combined with the upregulation of CD24 may play a synergistic role in the occurrence and progression of hepatocellular carcinoma. 25756511_NDRG2 acts as a negative regulator downstream of androgen receptor and inhibits the growth of androgen-dependent and castration-resistant prostate cancer. 25823664_Authors found that H. pylori silenced Ndrg2 by activating the NF-kappaB pathway and up-regulating DNMT3b, promoting gastric cancer progression. Findings uncover a previously unrecognized role for H. pylori infection in gastric cancer. 25854572_NDRG2 was negatively correlated with Bcl-2 in gastric cancer tissues. 25889839_Patients with high ITLN1 or NDRG2 expression had greater survival probability. 26239274_NDRG2 suppresses the proliferation of human bladder cancer cells via induction of oncosis. 26250123_Epigenetic silencing of NDRG2 promotes colorectal cancer proliferation and invasion. 26269411_The forced expression of NDRG2 in ATL cells down-regulates not only the canonical pathway by inhibiting AKT signaling but also the non-canonical pathway by inducing NF-kappaB-inducing kinase (NIK) dephosphorylation via the recruitment of PP2A 26292259_Loss of NDRG2 induced the expression of matrix metalloproteinase-19 (MMP-19), which regulated the expression of Slug at the transcriptional level in the epithelial-mesenchymal transition of gallbladder carcinoma cells. 26317652_NDRG2 functions as an essential regulator in glycolysis and glutaminolysis via repression of c-Myc, and acts as a suppressor of carcinogenesis. 26341979_This review summarizes the distribution and subcellular localization of NDRG2 in brain tissues, highlights the physiological actions of NDRG2 in the nervous system, and discusses the roles of NDRG2 during the occurrence of nervous system disease. [review] 26506239_Stuidies indicate the potential of N-myc downstream-regulated gene 2 (NDRG2) as a therapeutic target for cancer. 26646663_Data show that downregulation of NDRG2 may play an important role in advanced hepatoblastomas. 26802650_NDRG2 overexpression enhanced sodium/iodide symporter level in thyroid tumor cells and increased their iodine uptake in vitro. 26813459_miR-301a/b-NDRG2 might be an important axis modulating autophagy and viability of prostate cancer cells under hypoxia. 26976975_Study shows that NDRG2 was significantly down-regulated in glioblastomas (GBM) and seems to play a role in GBM prognosis. The low expression of NDRG2 at the mRNA level leads to a longer progression-free survival independent of methylation status. 27072586_These findings indicate that SUMOylation of NDRG2 is necessary for its tumor suppressor function in lung adenocarcinoma and that RNF4 increases the efficiency of this process. 27113371_Authors investigate the clinical and pathological role of NDRG2 in human malignant tumors. The absence of NDRG2 expression is associated with oncogenic properties through the loss of its role as a tumor suppressor; NDRG2 overexpression inhibits proliferation, adhesion, and invasion, whereas its expression is low or undetectable in various cancers. [Review] 27400234_putative tumor suppressive function of NDRG2 may be confined to luminal- and basal B-type breast cancers 28031114_Overexpression of NDRG2 can inhibit the proliferation. 28069379_NDRG2 overexpression significantly reduced hepatoma cell proliferation and enhanced cell apoptosis under glucose limitation. Moreover, NDRG2 overexpression aggravated energy imbalance and oxidative stress by decreasing the intracellular ATP and NADPH generation and increasing ROS levels. 28093228_present results suggest that hypermethylation in promoter around exon 2 is functioning as essential factors of NDRG2 silencing in gastrointestinal cancers 28202850_The methylation status of the NDRG2 promoter in PBMCs is a potential noninvasive biomarker to predict the severity of liver fibrosis. 28209617_Our results show how NDRG2 expression serves as a critical determinant of the invasive and metastatic capacity of oral squamous cell carcinoma (OSCC) 28390436_The different NDRG2 promoter methylation and expression levels in pituitary adenoma samples showed tumor heterogeneity and indicates a potential role of this gene in pituitary adenoma pathogenesis. 28423695_NDRG2 expression decreased in adriamycin resistance breast cancer cells. NDRG2 can promote adriamycin sensitivity by inhibiting proliferation, enhancing cellular damage responses, and promoting apoptosis in a p53-dependent manner. 28646304_NDRG2 knockdown promotes renal fibrosis through its effect on the protein and mRNA expression of epithelial-mesenchymal transition markers. 28656310_Lower expression of KLF4 and NDRG2 in colorectal cancer patients was correlated with poor overall survival. Thus, KLF4 inhibited the proliferation of colorectal cancer cells dependent on NDRG2 signaling, which provides a novel strategy for therapy and early diagnosis of colorectal cancer. 28670853_Increased level of NDRG2 in cortex or hippocampus may be a potential risk for Alzheimer's disease. NDRG2 increased Abeta1-42 might be related to the changes of BEAC1 and GGA3. It might also affect CDK5 and tau overphosphorylation. NDRG2 interacting with STAT3 and modulating Fas/Fasl signaling pathway plays important roles in cell death. 28948615_NDRG2 is a novel ERS-responsive protein and acts as PERK co-factor to facilitate PERK branch, thereby contributing to ERS-induced apoptosis. 28953854_NDRG2 could be a potential independent diagnostic biomarker for bladder cancer. 29060933_Data show that Sentrin/SUMO specific protease (SENP2) interacts with N-myc downstream regulated gene 2 (NDRG2) and mediates the de-SUMOylation process of NDRG2. 29080452_Down-regulation of NDRG2 promotes cancer cell proliferation and neoplasm invasion through regulation by microRNA-454. 29343851_Supporting the biological significance of the reciprocal relationship between NDRG2 and Skp2, an NDRG2low/Skp2high gene expression signature correlates with poor colorectal cancer (CRC) patient outcome and could be considered as a diagnostic marker of CRCs. 29530788_Low NDRG2 expression is associated with proliferation, migration, invasion and epithelial-mesenchymal transition of esophageal cancer. 30348117_We observed the knockdown of NDRG2 with miR-28-5p and miR-650 inhibitors inducing CLL cell apoptosis, yet found no increased apoptosis rates in patients with p53 aberrations following transfection with the above miRNAs inhibitors. 30556863_Hypoxia promoted the migration and invasion of gastric cancer cell BGC-823 by activating HIF-1alpha and inhibiting NDRG2 associated signaling pathway. 30568657_Study identified FLNa as a substrate of NDR2 in vitro and demonstrated that NDR2 phosphorylates FLNa at serine 2152 (S2152) upon TCR-triggering in vivo. NDR2-dependent phosphorylation of FLNa at S2152 releases the binding of this molecule from the inactive conformation of LFA-1, thus allowing the TCR-mediated association of Talin and Kindlin-3 to the cytoplasmatic domain of CD18. 31295529_NDRG2 expression was suppressed in HTLV-1-infected cell lines and various types of ATLL cells, which was dependent on the DNA methylation of the NDRG2 promoter. 31613009_NDRG2 and TLR7 as novel DNA methylation prognostic signatures for acute myelocytic leukemia. 31638184_NELFE expression was increased in pancreatic cancer (PC) tissues compared with the paired normal tissues. NELFE expression was upregulated in PC cells when compared with normal pancreatic cells. Knockdown of NELFE inhibited the proliferation, invasion and migration of PC cells. NELFE activates the Wnt/betacatenin signaling pathway and epithelialtomesenchymal transition by decreasing the stabilization of NDRG2 mRNA in PC. 32345304_NDRG2 gene expression pattern in ovarian cancer and its specific roles in inhibiting cancer cell proliferation and suppressing cancer cell apoptosis. 32665121_Combination of NDRG2 overexpression, X-ray radiation and docetaxel enhances apoptosis and inhibits invasiveness properties of LNCaP cells. 33031336_Low NDRG2 expression predicts poor prognosis in solid tumors: A meta-analysis of cohort study. 33050824_Overexpression of NDRG2 promotes the therapeutic effect of pazopanib on ovarian cancer. 33162818_NDRG2 ablation reprograms metastatic cancer cells towards glutamine dependence via the induction of ASCT2. 33174183_Roles of N-myc downstream-regulated gene 2 in the central nervous system: molecular basis and relevance to pathophysiology. 33205345_NDRG2 is expressed on enteric glia and altered in conditions of inflammation and oxygen glucose deprivation/reoxygenation. 33314669_LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed proliferation, invasion and glycolysis of colorectal cancer. 33857783_Long non-coding RNA CRNDE suppressing cell proliferation is regulated by DNA methylation in chronic lymphocytic leukemia. 34051015_Upregulation of P53 promotes nucleus pulposus cell apoptosis in intervertebral disc degeneration through upregulating NDRG2. 34951340_MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. 35048779_Expression and Significance of N-myc downstream regulated gene 2 in the process of Esophageal Squamous Cell Carcinogenesis. | ENSMUSG00000004558 | Ndrg2 | 433.90825 | 1.1225924 | 0.1668341994 | 0.15245543 | 1.202076e+00 | 2.729071e-01 | 6.617959e-01 | No | Yes | 541.709408 | 58.098030 | 465.329707 | 48.952469 | |
| ENSG00000165804 | 51222 | ZNF219 | protein_coding | Q9P2Y4 | FUNCTION: Transcriptional regulator (PubMed:14621294, PubMed:19549071). Recognizes and binds 2 copies of the core DNA sequence motif 5'-GGGGG-3' (PubMed:14621294). Binds to the HMGN1 promoter and may repress HMGN1 expression (PubMed:14621294). Regulates SNCA expression in primary cortical neurons (PubMed:19549071). Binds to the COL2A1 promoter and activates COL2A1 expression, as part of a complex with SOX9 (By similarity). Plays a role in chondrocyte differentiation (By similarity). {ECO:0000250|UniProtKB:Q6IQX8, ECO:0000269|PubMed:14621294, ECO:0000269|PubMed:19549071}. | Activator;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene is a member of the Kruppel-like zinc finger gene family. The encoded protein functions as a transcriptional repressor of the high mobility group nucleosome binding domain 1 protein, which is associated with transcriptionally active chromatin. [provided by RefSeq, Apr 2017]. | hsa:51222; | integral component of membrane [GO:0016021]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA binding [GO:0003677]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; histamine receptor activity [GO:0004969]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; limb bud formation [GO:0060174]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; positive regulation of chondrocyte differentiation [GO:0032332]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of neurotransmitter levels [GO:0001505]; regulation of transcription, DNA-templated [GO:0006355] | 14621294_By using a random oligonucleotide selection assay and the electromobility gel shift assay, we have revealed that the ZNF219 protein recognizes two copies of CCCCCA. 20940257_regulates chondrocyte differentiation as a transcriptional partner of Sox9 34167360_ZNF219 protects human lens epithelial cells against H2O2-induced injury via targeting SOX9 through activating AKT/GSK3beta pathway. | ENSMUSG00000049295 | Zfp219 | 192.49595 | 0.6583847 | -0.6029971805 | 0.23772614 | 6.318744e+00 | 1.194682e-02 | 1.796380e-01 | No | Yes | 202.660911 | 29.794054 | 299.758097 | 42.422312 | |
| ENSG00000166387 | 8495 | PPFIBP2 | protein_coding | Q8ND30 | FUNCTION: May regulate the disassembly of focal adhesions. Did not bind receptor-like tyrosine phosphatases type 2A. {ECO:0000269|PubMed:9624153}. | 3D-structure;Alternative splicing;Coiled coil;Phosphoprotein;Reference proteome;Repeat | This gene encodes a member of the LAR protein-tyrosine phosphatase-interacting protein (liprin) family. The encoded protein is a beta liprin and plays a role in axon guidance and neuronal synapse development by recruiting LAR protein-tyrosine phosphatases to the plasma membrane. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Feb 2012]. | hsa:8495; | cytosol [GO:0005829]; extracellular space [GO:0005615]; presynaptic active zone [GO:0048786]; identical protein binding [GO:0042802]; neuromuscular junction development [GO:0007528]; synapse organization [GO:0050808] | 20095854_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 21462929_analysis of crystal structure of the central coiled-coil domain from human liprin-beta2 22328529_Knockdown of either Rab17 or liprin-beta2 restores invasiveness of ERK2-depleted cells, indicating that ERK2 drives invasion of MDA-MB-231 cells by suppressing expression of these genes. 26443449_rs12791447 is associated with prostate cancer susceptibility in Asians. mRNA levels of PPFIBP2 are differentially expressed in prostate tumours and paired normal tissues. 26663347_Analysis of the role of liprin-beta1 and liprin-beta2 has shown that while liprin-beta1 contributes positively to tumour cell motility in vitro; liprin-beta2 has a negative effect on both cell motility and invasion. 30043417_germline mutations in a novel gene, PPFIBP2, differentiated risk for lethal prostate cancer from low-risk cases and were associated with shorter survival times after diagnosis. | ENSMUSG00000036528 | Ppfibp2 | 67.11281 | 1.4141682 | 0.4999537323 | 0.41691949 | 1.436488e+00 | 2.307086e-01 | No | Yes | 116.376127 | 27.053631 | 83.006679 | 18.771273 | ||
| ENSG00000166503 | 50810 | HDGFL3 | protein_coding | Q9Y3E1 | FUNCTION: Enhances DNA synthesis and may play a role in cell proliferation. {ECO:0000269|PubMed:10581169}. | 3D-structure;Growth factor;Nucleus;Phosphoprotein;Reference proteome | hsa:50810; | cytosol [GO:0005829]; extracellular region [GO:0005576]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; double-stranded DNA binding [GO:0003690]; growth factor activity [GO:0008083]; microtubule binding [GO:0008017]; transcription coregulator activity [GO:0003712]; tubulin binding [GO:0015631]; microtubule polymerization [GO:0046785]; negative regulation of microtubule depolymerization [GO:0007026]; neuron projection development [GO:0031175]; regulation of transcription by RNA polymerase II [GO:0006357] | 15371438_LEDGF/p75 and HRP2 IBDs avidly bind HIV-1 Integrase, share a similar domain organization and have an evident evolutionary and likely functional relationship 19237540_Hepatoma-derived growth factor-related protein-3 interacts with microtubules and promotes neurite outgrowth in mouse cortical neurons 24012673_Depletion of HRP-3 induces apoptosis of radio- and chemoresistant A549 cells. HRP-3 is essential for regulating reactive oxygen species-dependent, p53-induced cell death. 26823754_Glucose deprivation-induced HRP-3 up-regulation potentially plays a major role in protecting hepatocellular carcinoma cells against apoptosis caused by energy pressure. 28854847_The study observed higher expression levels of HDGFRP3 and ID2 in bipolar disorder (BD) patients who favourably respond to lithium. Both of these genes are involved in neurogenesis, and HDGFRP3 has been suggested to be a neurotrophic factor. 31162607_HRP3 PWWP is a new family of minor groove-specific DNA-binding proteins 34714604_A novel function of HRP-3 in regulating cell cycle progression via the HDAC-E2F1-Cyclin E pathway in lung cancer. | ENSMUSG00000025104 | Hdgfl3 | 1537.43622 | 0.9757371 | -0.0354355497 | 0.10177513 | 1.210037e-01 | 7.279484e-01 | 9.205530e-01 | No | Yes | 2046.412631 | 390.844098 | 1975.902120 | 369.167053 | ||
| ENSG00000166529 | 7589 | ZSCAN21 | protein_coding | Q9Y5A6 | FUNCTION: Strong transcriptional activator (By similarity). Plays an important role in spermatogenesis; essential for the progression of meiotic prophase I in spermatocytes (By similarity). {ECO:0000250|UniProtKB:Q07231}. | Activator;DNA-binding;Differentiation;Isopeptide bond;Meiosis;Metal-binding;Nucleus;Reference proteome;Repeat;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7589; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cell differentiation [GO:0030154]; male meiosis I [GO:0007141]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; spermatogenesis [GO:0007283] | 16540086_This result indicates that NY-REN-21 can function either as a homodimer or as a heterodimer with SCAND1. 30485814_deregulation of the TRIM17/TRIM41/ZSCAN21 pathway may be involved in the pathogenesis of Parkinson's disease. | ENSMUSG00000037017 | Zscan21 | 367.05875 | 1.0765175 | 0.1063717471 | 0.17256444 | 3.801575e-01 | 5.375190e-01 | 8.391324e-01 | No | Yes | 515.856855 | 83.124018 | 472.366445 | 74.199534 | ||
| ENSG00000166783 | 9665 | MARF1 | protein_coding | Q9Y4F3 | FUNCTION: Essential regulator of oogenesis required for female meiotic progression to repress transposable elements and preventing their mobilization, which is essential for the germline integrity. Probably acts via some RNA metabolic process, equivalent to the piRNA system in males, which mediates the repression of transposable elements during meiosis by forming complexes composed of RNAs and governs the methylation and subsequent repression of transposons. Also required to protect from DNA double-strand breaks (By similarity). {ECO:0000250}. | 3D-structure;Alternative splicing;Differentiation;Meiosis;Oogenesis;Peroxisome;Phosphoprotein;RNA-binding;Reference proteome;Repeat | This gene encodes a putative peroxisomal protein that appears to be conserved across Euteleostomi. In humans, it may be autoantigenic. [provided by RefSeq, Jul 2010]. | hsa:9665; | cytoplasm [GO:0005737]; Golgi apparatus [GO:0005794]; intracellular membrane-bounded organelle [GO:0043231]; membrane [GO:0016020]; peroxisome [GO:0005777]; CCR4-NOT complex binding [GO:1905762]; mRNA binding involved in posttranscriptional gene silencing [GO:1903231]; double-strand break repair [GO:0006302]; female meiotic nuclear division [GO:0007143]; oogenesis [GO:0048477]; posttranscriptional gene silencing [GO:0016441]; regulation of gene expression [GO:0010468] | 24755989_LMKB is the first protein identified to date that interacts with this portion of Ge-1. LMKB was expressed in human B and T lymphocyte cell lines; depletion of LMKB increased expression of IFI44L. 30364987_Human MARF1 is an endoribonuclease that interacts with the DCP1:DCP2 decapping complex and degrades target mRNAs. 32510323_A non-canonical role for the EDC4 decapping factor in regulating MARF1-mediated mRNA decay. | ENSMUSG00000060657 | Marf1 | 1430.47101 | 1.1778688 | 0.2361788881 | 0.10284698 | 5.296249e+00 | 2.137139e-02 | 2.353619e-01 | No | Yes | 1898.631627 | 242.790806 | 1516.398570 | 189.891371 | |
| ENSG00000166888 | 6778 | STAT6 | protein_coding | P42226 | FUNCTION: Carries out a dual function: signal transduction and activation of transcription. Involved in IL4/interleukin-4- and IL3/interleukin-3-mediated signaling. {ECO:0000269|PubMed:17210636}. | 3D-structure;ADP-ribosylation;Acetylation;Activator;Alternative splicing;Cytoplasm;DNA-binding;Nucleus;Phosphoprotein;Reference proteome;SH2 domain;Transcription;Transcription regulation | The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein plays a central role in exerting IL4 mediated biological responses. It is found to induce the expression of BCL2L1/BCL-X(L), which is responsible for the anti-apoptotic activity of IL4. Knockout studies in mice suggested the roles of this gene in differentiation of T helper 2 (Th2) cells, expression of cell surface markers, and class switch of immunoglobulins. Alternative splicing results in multiple transcript variants.[provided by RefSeq, May 2010]. | hsa:6778; | chromatin [GO:0000785]; cytosol [GO:0005829]; membrane raft [GO:0045121]; nucleoplasm [GO:0005654]; RNA polymerase II transcription regulator complex [GO:0090575]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; identical protein binding [GO:0042802]; protein phosphatase binding [GO:0019903]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; cellular response to hydrogen peroxide [GO:0070301]; cellular response to reactive nitrogen species [GO:1902170]; cytokine-mediated signaling pathway [GO:0019221]; defense response [GO:0006952]; growth hormone receptor signaling pathway via JAK-STAT [GO:0060397]; interleukin-4-mediated signaling pathway [GO:0035771]; mammary gland epithelial cell proliferation [GO:0033598]; mammary gland morphogenesis [GO:0060443]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of type 2 immune response [GO:0002829]; positive regulation of cold-induced thermogenesis [GO:0120162]; positive regulation of isotype switching to IgE isotypes [GO:0048295]; positive regulation of transcription by RNA polymerase II [GO:0045944]; receptor signaling pathway via JAK-STAT [GO:0007259]; regulation of cell population proliferation [GO:0042127]; regulation of transcription by RNA polymerase II [GO:0006357]; response to peptide hormone [GO:0043434]; signal transduction [GO:0007165]; T-helper 1 cell lineage commitment [GO:0002296] | 11532018_IL-4-dependent Stat6 activation is inhibited by an intracellularly delivered peptide derived from the Stat6-binding region of IL-4Ralpha. 11692112_role in mediating responses of bronchial epithelium to cytokines in asthmatics 11872954_Oligonucleotide fishing for STAT6: cross-talk between IL-4 and chemokines. 11912176_STAT6 polymorphism is associated with increase in eosinophil cell count contributing to the pathogenesis of Asthma 12058257_3'UTR polymorphism is associated with susceptibility and severity in nut allergic patients 12058257_Observational study of gene-disease association. (HuGE Navigator) 12082548_STAT6 is required for IL-4-mediated growth inhibition and induction of apoptosis in human breast cancer cells. 12138096_STAT6 has a protein binding motif that controls the interaction with NcoA-1 in transcriptional activation 12161424_These results show that p38 MAPK provides a costimulatory signal for IL-4-induced gene responses by directly stimulating the transcriptional activation of STAT6. 12234934_These findings identify p100 as a novel coactivator for STAT6 and suggest that p100 functions as a bridging factor between STAT6 and the basal transcription machinery. 12426308_Protein phosphatase 2A (PP2A) regulates interleukin-4-mediated signaling by this protein. 12459556_regulation of dephosphorylation by of mitogen-activated protein kinase and myosin light chain kinase 12605697_Observational study of gene-disease association. (HuGE Navigator) 12651067_IL-4-induced Stat6 signaling is a polygenic quantitative trait regulated by a collection of several contributing genetic loci. 12689929_when osteoclastogenesis was induced independently of RANKL by using TNF-alpha, IL-4 inhibited osteoclast differentiation through a STAT6-dependent mechanism 12759487_Observational study of gene-disease association. (HuGE Navigator) 12794133_Characterization of the IL-4-responsive enhanceosome of the human polymeric Ig receptor gene demonstrates cooperation of STAT6, HNF1 and additional DNA-binding factors in mediating IL-4 responsiveness in HT-29 cells. 12900808_Observational study of gene-disease association. (HuGE Navigator) 12935900_These results suggest that MIP-T3 is a novel inhibitor of IL-13 signaling and may be a useful molecule in ameliorating various conditions in which IL-13 plays a central role. 14519766_the Stat6 TAD contributes to promoter specificity by the differential recruitment of and requirement for a p160-class coactivator 14634100_STAT6 is required for IL-4-IL-13-dependent SOCS-1 expression in A549 human lung adenocarcinoma cells; three identified STAT6-binding motifs in the SOCS-1 promoter cooperate to induce maximal transcription. 14719123_the Stat6 signaling pathway may play a role in maintaining the Th1/Th2 cytokine balance by directly and indirectly down-regulating the production of proinflammatory cytokines 14735150_Observational study of gene-disease association. (HuGE Navigator) 14746803_Results demonstrate that induction of adenine nucleotide translocase 3 by interleukin-4 and interferon-gamma proceeds via pathways involving STAT6 and STAT1, respectively. 15004182_STAT6 activation mediates a transcriptional enhancement of trefoil factor-3 (TFF3) by induction of de novo synthesized protein in mucus-producing HT-29 CL.16E intestinal goblet cells. 15044251_Constitutive STAT6 phosphorylation and DNA-binding activity were detected in primary mediastinal large B-cell lymphoma cell lines but not diffuse large B-cell lymphoma cell lines 15069079_IL-4 and IL-13 promote Stat6 serine phosphorylation in PHA-activated human T-cells. 15105161_As no exonic variants of STAT6 are known as yet, repeat polymorphisms in the regulatory regions and their haplotypes could be important in deciphering the genetic role of STAT6 in asthma and atopy. 15297269_FN-gamma regulates IL-4- and STAT6-dependent signaling and gene expression in airway epithelial cells by multiple mechanisms 15342695_Observational study of gene-disease association. (HuGE Navigator) 15342695_genetic variants within STAT6 contribute significantly to IgE regulation and manifestation of atopic diseases 15522309_Role for Stat6 in apoptosis regulation. Stat6(null phenotype) cell lines exhibit variably increased levels of Th1 cytokines. May be source for investigations into Inflammatory bowel disease pathophysiology. 15528258_IL-4 and IL-13 mediate the hypercontractility of intestinal muscle via a STAT6 pathway at the level of the smooth muscle cell. The STAT6 pathway may contribute to the hypercontractility of intestinal muscle in Crohn's disease. 15637551_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15687724_Observational study of gene-disease association. (HuGE Navigator) 15695802_p100 has an important role in the assembly of STAT6 transcriptosome, and that p100 stimulates IL-4-dependent transcription by mediating interaction between STAT6 and CBP and recruiting chromatin modifying activities to STAT6-responsive promoters 15733066_PI3-K promotes the expression of TFF3 and MUC2 and that the PI3-K pathway may play a pivotal role in intestinal goblet cell differentiation. 15888279_Alterations in the STAT6 pathway may play a crucial role in the pathogenesis of distinct subgroups of patients with Crohn's disease. 15888279_Observational study of gene-disease association. (HuGE Navigator) 16004996_IL-4 regulates TNFalpha-induced IL-8 expression at a transcriptional level and this mechanism involves STAT6 and NF-kappaB transcription factors 16084752_results demonstrate that epithelial eotaxin-3 is up-regulated in the context of a T helper 2 mediated inflammatory bowel disease via the signal transducer and activator of transcription 6 16103897_Observational study of gene-disease association. (HuGE Navigator) 16387423_Findings suggest that Stat6 signaling pathway plays a role in the regulation of cell proliferation and apoptosis in colon cancer cells, which promises further investigation for targeted cancer therapy. 16547812_STAT6 binding to STAT6 promotor gene was regulated by H4 receptor-induced signal transduction and/or cross talk particularly in atopic human lymphocytes ex vivo 16601843_IL-4 has differential effects in coronary artery smooth muscle cells: short-term exposure enhances OPG production through a STAT6-dependent mechanism, but long-term exposure causes Cbfa1-dependent osteogenic transformation and decreased production of OPG 16681592_Observational study of gene-disease association. (HuGE Navigator) 16810739_Essential for IL-4-induced human T-cell expression of CCL17/thymus- and activation-regulated chemokine(TARC). 16867043_Observational study of gene-disease association. (HuGE Navigator) 16877367_Activation of STAT6 appears to be a key factor in P-selectin expression induced by substance P and IL-4 because treatment with STAT6 decoy oligodeoxynucleotides significantly inhibited P-selectin expression. 16951379_Stat6 exerts a stimulatory effect on early growth response (Egr)-1 gene and platelet-derived growth factor (PDGF) ligand mRNA transcription. 17074026_Observational study of gene-disease association. (HuGE Navigator) 17074026_The STAT6 G2964A polymorphism is not involved in the genetic susceptibility to ulcerative colitis in Chinese patients. 17121586_Observational study of gene-disease association. (HuGE Navigator) 17210636_specific increases in T-cell protein tyrosine phosphatase expression in activated-B-cell-like diffuse large B-cell lymphomas may contribute to the different biological characteristics of these tumors 17213269_Observational study of gene-disease association. (HuGE Navigator) 17213269_STAT6 may contribute to disease susceptibility in endometriosis 17237818_Unphosphorylated STAT6 plays a novel role in the pathogenesis of non-small cell lung cancer. 17362266_Observational study of gene-disease association. (HuGE Navigator) 17371990_Collectively, these data suggest a link between the inducible phenotype of CCL23 expression in monocytes by the prototype Th2 molecule pair IL-4/STAT6 and the increased number of CCL23-expressing cells in skin of atopic dermatitis patients. 17433443_Cross-talk between STAT6 and extracellular signal-regulated kinase (Erk) is the basis of the functional interaction between Jak/STAT and MAP kinase pathways activated by interleukin (IL)-4. 17532201_No statistically significant difference was found in the distribution of the STAT-6 G2964A polymorphisms between asthmatic patients and controls. 17532201_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17541284_The transcription factor STAT6 may play a pivotal role in the activation of eotaxin transcription in response to IL-4. 17606441_STAT6 overexpression detected in a classical Hodgkins lymphoma cell line 17652621_In lymphocyte-predominant Hodgkin lymphoma SOCS1 function may thus be frequently impaired by mutations, and this may contribute to high JAK2 expression and activation of the JAK2/STAT6 pathway. 17703412_Observational study of gene-disease association. (HuGE Navigator) 17705178_STAT6 is a survival factor in prostate cancer regulating the genetic transcriptional program for prostate cancer progression. 17883727_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 18191727_evidence provided that a distal tandem STAT6 element elevates expression from the CCL17 locus approximately twofold. 18251702_activation induces osteoprogeterin expression in cooperation with IL-4 and IL-13 18273035_A STAT6 gene polymorphism is associated with high infection levels in urinary schistosomiasis. 18294957_These findings, together with the observation of constitutive Stat6 activation in many human malignancies, suggest that Stat6 activities could be a biomarker for cancer cell's invasive/metastatic capability. 18296848_IL-13 has anti-angiogenic activity as a result of activation of JAK2 and subsequent activation of STAT6. 18342537_Data show that IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells. 18345495_Data suggest that histamine receptor H4R-selective ligands influence the STAT6 transcription activation domain and DNA-binding. 18497474_STAT6 plays pivotal roles on IL-13- and IL-4-induced apoptosis in vascular endothelial cells. 18536936_Constitutively increased expression of Stat6 negative regulators SOCS-1 and SHP-1, together with decreased expression of positive regulator PP2A, may play a role in forming the inactive Stat6(null) phenotype in colon cancer 18626468_analyzed IL-4 receptor expression, STAT-6 activation by IL-4, and STAT-6 inhibition by an anti-IL-4 antibody or by STAT-6 small-interfering RNA transfection 18636124_Observational study of gene-disease association. (HuGE Navigator) 18716132_IL-4 induces PTP1B mRNA expression in a phosphatidylinositol 3-kinase-dependent manner and enhances PTP1B protein stability to suppress IL-4-induced STAT6 signaling. 18846228_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19011907_Observational study of gene-disease association. (HuGE Navigator) 19030780_STAT6 expression can be used as a prognostic determinant for methotrexate chemotherapy. 19202006_A novel STAT6 inhibitor AS1517499 ameliorates antigen-induced bronchial hypercontractility in mice. 19247692_Observational study of gene-disease association. (HuGE Navigator) 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19282076_Genetic polymorphisms in the Jak-Stat signaling pathway are associated with an increased risk of new cardiovascular events in incident dialysis patients. 19282076_Observational study of gene-disease association. (HuGE Navigator) 19362457_Focusing on how STAT6 work in concert with other transcription factors will hopefully provide a better mechanistic understanding of the pathogenesis of various autoimmune diseases. 19372141_Observational study of gene-disease association. (HuGE Navigator) 19419769_The IL-4-induced downregulatory effect on Weibel-Palade bodies component genes depended on the negative feedback regulation by SOCS-1 induced by STAT6 signaling. 19423540_Observational study of gene-disease association. (HuGE Navigator) 19423726_The mutant STAT6 proteins showed a decreased DNA binding ability in transfected HEK cells, but no decrease in expression of STAT6 canonical target genes was observed in PMBL cases with a mutated STAT6 gene. 19488417_These results demonstrate that Toxoplasma gondii exploits host STAT6 to take away various harmful reactions by IFN-gamma. 19494273_targeting the key transcription factor STAT6 by siRNA effectively blocks the development of cardinal features of allergic airway disease 19531027_Data show that IL-4 receptors are functionally competent in pancreatic beta-cells and that they signal via PI3K and JAK/STAT pathways. 19540524_Spontaneously produced IFNgamma in the Stat6(null) cell lines suppresses STAT6 function and creates the Stat6(null) phenotype. 19551406_DNA methylation controls the constitutive expression of negative Stat6 regulatory genes, which may affect Stat6 activities. 19655172_These results suggest that Toxoplasma gondii exploits cytokine cross-regulation through STAT6 activation to obviate various toxoplasmacidal reactions by interferon-gamma. 19665768_The consistently replicated effects of genetic variance in STAT6 on IgE regulation may be explained in part by allele-specific alterations in nuclear factor-kappaB binding at rs324011 and consecutive changes in STAT6 gene expression. 19857574_Suggest that IL-4 is capable of inducing an upregulation of RhoA via an activation of STAT6 in cultured bronchial smooth muscle cells resulting in bronchial hyperresponsiveness. 19913121_Observational study of gene-disease association. (HuGE Navigator) 19949830_Study characterized the human STAT6 promoter gene and found that the transcriptional regulatory elements CCAAT and ATF were important for the STAT6 promoter activity. 20006706_The findings suggest that signal transducer and activator of transcription 6 and NF-kappaB are important for the upregulation of RhoA in human bronchial smooth muscle cells 20010912_Differential requirements for IL-4/STAT6 signalling in CD4 T-cell fate determination and Th2-immune effector responses. 20040389_Data show that flavones inhibited the phosphorylation of STAT6, janus kinase 3, and IL-4Ralpha, whereas IL-4 signaling mediated through type II IL-4R was unaffected by flavones. 20121258_the interaction of AnxA2 with STAT6 20122976_some altered function of STAT6 signaling may be important for the operational tolerance state in human transplantation tolerance 20128278_STAT6 expression was positively correlated with the recruitment of eosinophils in nasal polyps. 20147633_Mice that express constitutively active Stat6 are prone to the development of allergic skin inflammation and have decreased expression of genes in the epidermal differentiation complex that regulate epidermal barrier function. 20225206_In this report, we show that the coacitivator p100 protein can interact with STAT6 through its SN domain both in vivo and in vitro, resulting in enhancement of STAT6-mediated gene transcriptional acitivation. 20237496_Observational study of gene-disease association. (HuGE Navigator) 20339477_In renal cell carcinoma patients, a decreased proliferative response to tumor, associated with defects in JAK3/STAT5/6 expression that led to increased p27KIP1 expression and alterations in the cell cycle, was observed. 20394509_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20395963_Observational study of gene-disease association. (HuGE Navigator) 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20438785_Three single nucleotide polymorphisms (STAT6 rs703817, C1qG rs17433222, and MBP rs3794845) were found to be significantly associated with childhood leukemia risk in Koreans. 20480530_EZH2 and STAT6 expressions have significant values in distinguishing clinical stages of colorectal cancer and predicting the prognosis of the patients. 20498360_STAT6 is continually imported to the cell nucleus following tyrosine phosphorylation as a result of its ability to bind DNA, an effect which could be deleterious in autoimmunity. 20503287_Observational study of gene-disease association. (HuGE Navigator) 20530519_Observational study of gene-disease association. (HuGE Navigator) 20530519_The results indicate that IL4 and STAT6 genes might be involved in the etiology of systemic lupus erythematosus and potentially increased SLE risk through their interaction effect in Chinese patients 20576420_DHA inhibits IgE production of human B cells by direct interference with STAT6 and NFkappaB. 20620946_STAT4 and STAT6 play wide regulatory roles in T helper cell specification. 20620947_We showed that STAT6 resided in the core of IL-4-driven transcriptional events in human CD4+ T cells 20624917_STAT6 is a direct substrate for ROP16 in vivo. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20652946_IL-13 enhances the expression of GPIIb in tumor cell lines through a mechanism dependent on the transcription factor STAT6 that is able to bind the GPIIb gene promoter and upregulates the promoter activity. 20716621_Observational study of gene-disease association. (HuGE Navigator) 20719954_the IL-4/STAT6 signaling network is precisely controlled by KSHV for survival, maintenance of latency, and suppression of the host cytokine immune response of the virus-infected cells. 20840631_The IL-25 failed to trigger STAT6 activation. Both IL-4 and IL-13 activated signal transducer and activator of transcription 6 (STAT6), and silencing of this transcription factor markedly reduced the IL-4/IL-13-driven induction of macrophages. 20859068_STAT6 binding region is required for the IL-4-induced increase in promoter activity of the human RhoA gene 20980261_this study provides a model for an IL-4/STAT6-dependent fine tuning mechanism of TCF-1-driven T helper cell polarization 21093321_STAT6 acts as a facilitating factor for PPARgamma by promoting DNA binding and consequently increasing the number of regulated genes and the magnitude of responses. 21106524_these results identify PSF as a repressor of STAT6-mediated transcription that functions through recruitment of HDAC to the STAT6 transcription complex, and delineates a novel regulatory mechanism of IL-4 signaling 21123173_Ser-707 is phosphorylated by c-Jun N-terminal kinase (JNK). Phosphorylation decreases the DNA binding ability of IL-4-stimulated STAT6, thereby inhibiting the transcription of STAT6-responsive genes 21196282_GG genotype of STAT6 4610A/G was a significant risk factor compared with AA in glioblastoma and significantly related to increased WHO grade IV risk. 21268015_Signals ensued by IFN-alpha and IL-4 induce cytoplasmic sequestration of IL-4-activated STAT6 and IFN-alpha-activated STAT2:p48 in B cells through the formation of pY-STAT6:pY-STAT2:p48 complex. 21275604_results show that SPDEF plays a critical role in regulating a transcriptional network mediating IL-13-induced MUC5AC synthesis dependent on STAT6. 21277633_These results stress the essential roles of JAK1 and STAT6 in the early signaling pathway of IL-4 and IL-13 leading to suppression of COX-2 expression and repression of prostaglandin production. 21340358_These regulatory polymorphisms in the promoter regions of STAT6 may assist in identifying potential targets for the therapeutic control of gene expression. 21411736_Data demonstrated that the increased expression of DUOX1 in IL-4/IL-13-treated NHEK augments STAT6 phosphorylation via oxidative inactivation of protein tyrosine phosphatase 1B. 21595984_these findings suggest a role for STAT6 in enhancing cell proliferation and invasion in GBM, which may explain why up-regulation of STAT6 correlates with shorter survival times in glioma patients. 21601544_Pretreated CCD-11Lu cells with noncytotoxic doses (0.1-10 muM) of CAPE inhibited the production of eotaxin under stimulation of IL-4 and tnf-alpha.CAPE pretreatment decreased the amount of pSTAT6 and STAT6 DNA binding complexes in nuclear extracts. 21605467_These results of vitamin D promoting a Th2 shift through upstream GATA-3 and STAT6 transcription factors shed mechanistic understanding on the utility of vitamin D in MS. 21762972_The STAT6 gene increases viral replication in the skin of patients with atopic dermatitis with a history of eczema herpeticum 21765211_STAT6-mediated suppression of immune responses against cancer are disrupted by platinum-based drugs 21814192_IL4/IL13 increased PDS promoter activity but not in STAT6-deficient cells; mutation of STAT6 binding site rendered promoter insensitive to IL4/IL13; data are consistent with STAT6 being 1 of the links between PDS overexpression and IL4/IL13 stimulation 22012462_These findings suggest that the constitutive activation of STAT6 by Tip in T-cells may contribute to IL-2-independent T-cell transformation by herpesvirus saimiri. 22017802_STAT6 and TBXA2R polymorphisms were not associated with asthma risk, but they were associated with asthma-related symptoms. 22021169_A shift from predominant immune cell Stat6 activation to Stat3 activation accompanies the onset of dysplasia with concomitant increased epithelial cell Stat3 activation. 22025054_Studies indicate that the M1 macrophage phenotype is controlled by STAT1 and IRF5, whereas STAT6, IRF4 and PPARgamma regulate M2 macrophage polarization. 22045870_IL-4-dependent induction of the IL-31 gene required STAT6 binding sites. 22094113_STAT6 activation and chromatin remodeling by DNA demethylation and histone acetylation are crucial for transcriptional activation of 15-LOX-1 in cultured Hodgkin lymphoma cells. 22108090_Stat6 is involved in a process that promotes and transforms cancerous cells to a more severe pro-growth, apoptosis-resistant and metastasis-capable phenotype. 22162773_Small interfering RNA against transcription factor STAT6 leads to increased cholesterol synthesis in lung cancer cell lines 22226123_Data show that that IL-4 signals through the Jak1, 2/Stat6 pathway in keratinocytes to stimulate CCL26 expression and this may provide an explanation for the pathogenesis of atopic dermatitis (AD). 22365404_Chinese children differed in polymorphisms of STAT6 and in its relation with childhood asthma. 22406175_JAK1 and STAT6 may be universal mediators of IL-4 in the suppression of prostaglandin production in human follicular denritic cells. 22451727_Toxoplasma gondii induces SERPIN B3/B4 expression via STAT6 activation to inhibit the apoptosis of infected cells for survival of the parasites. 22509288_model for the differential activation of STAT3 or STAT6 by two distinct regions of the viral Tip protein 22534531_STAT6 and LRP1 polymorphisms are associated with food allergen sensitization in Mexican children. 22578203_STAT6 in2SNP3 is associated with penicillin allergy, but has no effect on the specific IgE levels of patients with penicillin allergy. 22660217_influence of paternal origin of the STAT6 haplotype on IgE levels in asthma 22661199_the STAT6 phosphorylation/activation induced by IL-13 is mediated by an activation of JAK1 in cultured human bronchial smooth muscle cells. 22909160_Dysregulation of Th1 transcription may contribute to heightened expression of STAT6 and GATA3 leading to exaggerated Th2-driven manifestations of allergic disease. 22997428_patients' sera inhibited STAT6 phosphorylation induced by IL4 binding to CD124, demonstrating that these autoantibodies are functional and suggesting that IL4 neutralization has a pathogenetic role in Autoimmune hepatitis. 23154639_The colon epithelial cell line Caco-2 was stimulated with interferon gamma, interleukin -4 prostaglandin E(2) to allow correlations between SOCS3 expression with STAT1, STAT6 and cyclic amp signaling. 23184467_Data indicate that the signal transducer and activator of transcription 6 (Stat6) as an important cell differentiation regulatory protein functioning by interacting with Sp1 to activate the p21 and p27 gene promoters in breast cancer cells. 23185525_PPIs, in concentrations achieved in blood with conventional dosing, significantly inhibit IL-4-stimulated eotaxin-3 expression in EoE esophageal cells and block STAT6 binding to the promoter. 23297791_The study indicated that the STAT6 promoter polymorphism rs3024944 was associated with uncomplicated malaria, whereas the FOXP3 promoter variant rs11091253 was associated with significant Plasmodium falciparum parasitaemia levels. 23313952_A gene fusion of the transcriptional repressor NAB2 with the transcriptional activator STAT6 is the defining driver mutation of solitary fibrous tumor. 23313954_A NAB2-STAT6 fusion is a distinct molecular feature of solitary fibrous tumors. 23351078_These data show that IL-4 and IL-13 decrease kinin receptors in a STAT6-dependent mechanism, which can be one important mechanism by which these cytokines exert their anti-inflammatory effects and impair bone resorption 23461825_STAT6 plays important roles in regulating the expression of human ORMDL3 by directly binding to the promoter region. 23535094_The results suggest that sIgE levels and STAT6 gene variants may be important determinants for development of tolerance to cows milke allergy. 23575898_Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion 23651608_STAT6 polymorphisms might influence the therapeutic outcomes of patients infected with HCV-1 under standard-of-care (SOC) treatment. 23752766_STAT6 polymorphisms and their combinations have an important influence on IgE level and development of asthma 23761323_44 SFTs were studied to identify pathogenetically important genetic rearrangements. RT-PCR analysis identified a NAB2/STAT6 fusion in 37/41 cases. 23815671_The interaction term [IL13 (rs20541) x STAT6 (rs1059513)] was statistically significant in both studies. 23844087_TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway, and this may contribute to the development of preeclampsia. 23852366_Dtat shows that pSTAT6 is sufficient to transcriptionally repress BCL6. 23861779_STAT6 G2964A polymorphism does not have an association with the susceptibility to asthma. 23935100_IL-4 induces cellular senescence through independent signaling pathways involving STAT6 and p38 MAPK in some human renal cell carcinoma cell lines. 24030747_is a highly sensitive and almost perfectly specific immunohistochemical marker for SFT and can be helpful to distinguish solitary fibrous tumor from histologic mimics. 24053457_Two STAT6 polymorphisms were associated with altered immune responses already at birth. STAT6 rs324011 was associated with lower neonatal Treg and increased Th1 response. 24107646_activation of the IL-4/STAT6 signaling pathway has a crucial role in aberrant glycosylation of IgA1 secretion, which is mediated by HIPK2 24401087_We found that Stat6 interacts with progesterone-activated PR in T47D cells. Stat6 synergizes with progesterone-bound PR to transactivate the p21 and p27 gene promoters at the proximal Sp1-binding sites. 24457460_In conclusion, STAT6 is amplified in a subset of dedifferentiated liposarcoma, resulting in STAT6 protein expression that can be detected by immunohistochemistry. 24491557_Data indicate that signal transducer and activator of transcription-6 (STAT6), a direct target of miR-361-5p, enhances the expression of B-cell lymphoma-extra large (Bcl-xL), while miR-361-5p inhibits its expression through STAT6. 24513261_Tumors with the most common fusion variant, NAB2ex4-STAT6ex2/3, corresponded to classic pleuropulmonary solitary fibrous tumors with diffuse fibrosis and mostly benign behavior and occurred in older patients 24625420_nuclear STAT6 immunoreactivity is a highly sensitive and specific marker of solitary fibrous tumors 24674464_Gene silencing of IFN-gamma R2 leads to down-regulation of STAT-6 and phosphorylation-STAT-6 expressions, so gene silencing of STAT-6 results in the reduction of HeLa cell programmed death. 24702701_The detection of nuclear relocation of STAT6 with immunohistochemistry is a characteristic of solitary fibrous tumours (SFTs), and may serve as a diagnostic marker that indicates NAB2-STAT6 fusion and helps to discriminate SFTs from histological mimics. 24828787_The anti-apoptotic phenotype of chronic lymphocytic leukemia involves a sensitized response for IL4 dependent STAT6 phosphorylation, and an activation of NF-kB signaling due to an increased affinity of sCD40L to its receptor. 24856853_The combination of NCOA2 FISH and Stat6 IHC proved effective for the differential diagnosis of soft-tissue angiofibroma, even when using small biopsy specimens. 24912007_The STAT6 rs324011 homozygous T/T genotype was significantly associated with asthma risk in a Saudi Arabian population. 24943220_Data show that STAT6 and NF-kappaB are central players in mediating IL-31 expression induced by IL-4/IL-33. 24952213_Short GT repeats of rs71802646 in STAT6 contribute to higher risk for asthma, while rs324015 may have a protective effect on atopic asthma. 24960198_These results suggest that IL-4Ralpha locating in non-lipid raft region is a target molecule for strictinin in inhibiting STAT6 activation. 24966942_Upregulation of IL-9 induced by pSTAT6 may be involved in the pathogenesis of chronic lymphocytic leukemia. 25111027_Taken together, these findings indicate that inhibiting IL-4-induced eotaxin-1 expression by synephrine occurs primarily through the suppression of eosinophil recruitment, which is mediated by inhibiting STAT6 phosphorylation. 25287927_Collectively, these studies demonstrated that Mycoplasma pneumoniae induces airway mucus hypersecretion by modulating the STAT/EGFR-FOXA2 signaling pathways. 25351986_The ANO1 promoter region contains putative binding sites for multiple transcription factors including signal transducer and activator of transcription 6 (STAT6), a downstream effector of IL-4. 25407941_Eosinophilic esophagitis was associate with increased expression of STAT6. 25428220_The genetic and functional data combined strengthen the recognition of the IL-4/JAK/STAT6 axis as a driver of FL pathogenesis. 25445452_TRAIL induces RANKL expression through a STAT-6 dependent transcriptional regulatory mechanism in bone marrow stromal/preosteoblast cells. 25451482_We further demonstrated that STAT6 silencing elicited ER stress-mediated apoptosis in NCI-H460 cells through C/EBP homologous protein (CHOP) induction, alteration of BH3 only proteins expression and ROS production. 25517867_nuclear immunohistochemistry staining can be used as a diagnostic criteria for unconventional fat-forming solitary fibrous tumors 25554652_This study validated the existence of the NAB2-STAT6 fusion gene in solitary fibrous tumors and examined its relation with pathological features. 25582503_There was no association between solitary fibrous tumors with either NAB2 exon 4-STAT6 exon 2 or 3 fusion and tumors with other fusions regarding the frequency of mutations in the examined genes (P = .201). 25667482_reverse transcriptase PCR analysis identified a nerve growth factor inducible-A binding protein 2-STAT6 gene fusion. Our case supports the utility of STAT6 immunohistochemistry as an adjunct in the diagnosis of soft-tissue SFT with loss of CD34 positivity 25761539_Solitary fibrous tumor occurring in the ocular adnexa (OA) as orbital mass with proptosis. In the OA, solitary fibrous tumor demonstrates STAT6 nuclear expression. 25873501_Strong nuclear STAT6 is largely specific for solitary fibrous tumors (SFTs). Physiologic low-level cytoplasmic/nuclear expression is common in mesenchymal neoplasia and is of uncertain significance. 25893823_It is associated with local recurrence and late distance metastasis of brain tumors to extracranial sites. 25952120_Igepsilon gene transcription is regulated by histone modifications in the IL-4/STAT6 pathway. 26048407_T allele of rs324011 in STAT6 would increase the risk of atopic dermatitis (AD) occurrence in children. Haplotypes of rs324011/rs167769 were also significantly associated with childhood AD in Taiwanese population. 26112603_results show that Cbl-b suppresses human ORMDL3 expression through STAT6 26134630_bovine milk activates STAT6 in a p44/42 and p38-d | ENSMUSG00000002147 | Stat6 | 283.43647 | 0.9154844 | -0.1273928658 | 0.20146412 | 3.984011e-01 | 5.279162e-01 | 8.337877e-01 | No | Yes | 322.509735 | 33.112704 | 348.091939 | 34.554216 | |
| ENSG00000166938 | 115752 | DIS3L | protein_coding | Q8TF46 | FUNCTION: Putative cytoplasm-specific catalytic component of the RNA exosome complex which has 3'->5' exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the cytoplasm, the RNA exosome complex is involved in general mRNA turnover and specifically degrades inherently unstable mRNAs containing AU-rich elements (AREs) within their 3' untranslated regions, and in RNA surveillance pathways, preventing translation of aberrant mRNAs. It seems to be involved in degradation of histone mRNA. {ECO:0000269|PubMed:20531386, ECO:0000269|PubMed:20531389}. | Alternative splicing;Cytoplasm;Exonuclease;Exosome;Hydrolase;Magnesium;Nuclease;Phosphoprotein;RNA-binding;Reference proteome | The cytoplasmic RNA exosome complex degrades unstable mRNAs and is involved in the regular turnover of other mRNAs. The protein encoded by this gene contains 3'-5' exoribonuclease activity and is a catalytic component of this complex. [provided by RefSeq, May 2016]. | hsa:115752; | centrosome [GO:0005813]; cytoplasmic exosome (RNase complex) [GO:0000177]; cytosol [GO:0005829]; exosome (RNase complex) [GO:0000178]; plasma membrane [GO:0005886]; 3'-5'-exoribonuclease activity [GO:0000175]; enzyme binding [GO:0019899]; RNA binding [GO:0003723]; RNA catabolic process [GO:0006401]; RNA processing [GO:0006396]; rRNA catabolic process [GO:0016075] | 20531386_Data show that hDIS3 and hDIS3L are active exonucleases, but only hDIS3 has retained endonucleolytic activity, and suggest that three different ribonucleases can serve as catalytic subunits for the exosome in human cells. 20531389_Data indicate that hDis3L1 is a novel exosome-associated exoribonuclease in the cytoplasm of human cells. 29321365_we screened the susceptibility loci for Myocardial infarction (MI) using exome sequencing and validated candidate variants in replication sets. We identified that three genes (GYG1, DIS3L and DDRGK1) were associated with MI at the discovery and replication stages. | ENSMUSG00000032396 | Dis3l | 1392.12759 | 1.1678925 | 0.2239074941 | 0.10614864 | 4.472739e+00 | 3.443973e-02 | 2.885656e-01 | No | Yes | 2035.621394 | 318.846338 | 1654.802217 | 253.558992 | |
| ENSG00000167065 | 150290 | DUSP18 | protein_coding | Q8NEJ0 | FUNCTION: Can dephosphorylate single and diphosphorylated synthetic MAPK peptides, with preference for the phosphotyrosine and diphosphorylated forms over phosphothreonine. In vitro, dephosphorylates p-nitrophenyl phosphate (pNPP). {ECO:0000269|PubMed:12408986, ECO:0000269|PubMed:12591617}. | 3D-structure;Cytoplasm;Hydrolase;Membrane;Mitochondrion;Mitochondrion inner membrane;Nucleus;Protein phosphatase;Reference proteome | Dual-specificity phosphatases (DUSPs) constitute a large heterogeneous subgroup of the type I cysteine-based protein-tyrosine phosphatase superfamily. DUSPs are characterized by their ability to dephosphorylate both tyrosine and serine/threonine residues. They have been implicated as major modulators of critical signaling pathways. DUSP18 contains the consensus DUSP C-terminal catalytic domain but lacks the N-terminal CH2 domain found in the MKP (mitogen-activated protein kinase phosphatase) class of DUSPs (see MIM 600714) (summary by Patterson et al., 2009 [PubMed 19228121]).[supplied by OMIM, Dec 2009]. | hsa:150290; | cytoplasm [GO:0005737]; mitochondrial inner membrane [GO:0005743]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; MAP kinase tyrosine/serine/threonine phosphatase activity [GO:0017017]; phosphatase activity [GO:0016791]; protein serine phosphatase activity [GO:0106306]; protein threonine phosphatase activity [GO:0106307]; protein tyrosine phosphatase activity [GO:0004725]; protein tyrosine/serine/threonine phosphatase activity [GO:0008138]; dephosphorylation [GO:0016311]; peptidyl-threonine dephosphorylation [GO:0035970]; peptidyl-tyrosine dephosphorylation [GO:0035335] | 12591617_molecular cloning, base sequence and amino acid sequence 16699184_The crystal structure of human DSP18 (official symbol DUSP18) has been determined at 2.0 A resolution. 16720344_DUSP18 appears to serve an important role by regulation of SAPK/JNK pathway 29852174_JNK and DUSP18 reciprocally modulate the SUMOylation, which plays a regulatory role in the aggregation of ataxin-1. | ENSMUSG00000047205 | Dusp18 | 181.07328 | 0.8409348 | -0.2499341191 | 0.21512460 | 1.342510e+00 | 2.465917e-01 | 6.398927e-01 | No | Yes | 190.494273 | 24.535461 | 224.019789 | 27.618066 | |
| ENSG00000167232 | 7644 | ZNF91 | protein_coding | Q05481 | FUNCTION: Transcription factor specifically required to repress SINE-VNTR-Alu (SVA) retrotransposons: recognizes and binds SVA sequences and represses their expression by recruiting a repressive complex containing TRIM28/KAP1 (PubMed:25274305). May also bind the promoter of the FCGR2B gene, leading to repress its expression; however, additional evidence is required to confirm this result in vivo (PubMed:11470777). {ECO:0000269|PubMed:25274305, ECO:0000305|PubMed:11470777}. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Zinc;Zinc-finger | The ZNF91 gene encodes a zinc finger protein of the KRAB (Kruppel-associated box) subfamily (Bellefroid et al., 1991, 1993 [PubMed 2023909] [PubMed 8467795]).[supplied by OMIM, May 2010]. | hsa:7644; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; zinc ion binding [GO:0008270]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of transposon integration [GO:0070895]; regulation of transcription by RNA polymerase II [GO:0006357] | 19240061_Observational study of gene-disease association. (HuGE Navigator) | 177.45910 | 0.8144794 | -0.2960498155 | 0.23070044 | 1.633393e+00 | 2.012344e-01 | 5.934470e-01 | No | Yes | 157.919009 | 43.118672 | 192.016344 | 51.281936 | |||
| ENSG00000167280 | 64772 | ENGASE | protein_coding | Q8NFI3 | FUNCTION: Endoglycosidase that releases N-glycans from glycoproteins by cleaving the beta-1,4-glycosidic bond in the N,N'-diacetylchitobiose core. Involved in the processing of free oligosaccharides in the cytosol. {ECO:0000269|PubMed:12114544}. | Acetylation;Alternative splicing;Cytoplasm;Glycosidase;Hydrolase;Phosphoprotein;Reference proteome | This gene encodes a cytosolic enzyme which catalyzes the hydrolysis of peptides and proteins with mannose modifications to produce free oligosaccharides. [provided by RefSeq, Feb 2012]. | hsa:64772; | cytosol [GO:0005829]; hydrolase activity, hydrolyzing O-glycosyl compounds [GO:0004553]; mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity [GO:0033925]; protein deglycosylation [GO:0006517]; protein folding [GO:0006457] | 12114544_involved in processing of free oligosaccharides in the cytosol; identification of the gene encoding human cytosolic ENGase 12905469_endo-beta-n-acetylglucosaminidase present in the synovial fluid of rheumatoid arthritis patients, may contribute to the depletion of glycosaminoglycans from cartilage allowing the invasion of synovial cells 18586680_identification of O-GlcNAcase as a caspase-3 substrate with a novel caspase-3 cleavage site and provide insight about O-GlcNAcase regulation during apoptosis. 20026047_Data show that kinetic and X-ray crystallographic analyses of the binding modes with human/bacterial O-GlcNAcases identify some of these as competitive inhibitors. 20413512_O-GlcNAcase expression is increased in erythrocytes from both individuals with pre-diabetes and individuals with less well-controlled diabetes. 20668520_As the generation of the bulk of fOS is unaffected by co-down regulation of Ngly1p and Engase1p, alternative quantitatively important mechanisms must underlie the liberation of these fOS from either LLO or glycoproteins during protein N-glycosylation. 21873561_Serological N-acetyl-glucosaminidase, telomere length, and the UCP2-886G>A variant are independent risk factors for type 2 diabetes. 26923419_In patients with autosomal dominant polycystic kidney disease, urinary NAGase was correlated with urinary ET-1 which was inversely associated with eGFR and positively correlated with total kidney volume. 33730571_Transcriptome-wide association study reveals two genes that influence mismatch negativity. 34397265_N-acetyl-ss-D-glucosaminidase is predictive of mortality in chronic heart failure: a 10-year follow-up. | ENSMUSG00000033857 | Engase | 1125.99783 | 0.8540281 | -0.2276445329 | 0.12483214 | 3.303293e+00 | 6.914115e-02 | 3.893601e-01 | No | Yes | 1235.557911 | 180.345780 | 1466.774699 | 209.034818 | |
| ENSG00000167363 | 64122 | FN3K | protein_coding | Q9H479 | FUNCTION: Fructosamine-3-kinase involved in protein deglycation by mediating phosphorylation of fructoselysine residues on glycated proteins, to generate fructoselysine-3 phosphate (PubMed:11016445, PubMed:11522682, PubMed:11975663). Fructoselysine-3 phosphate adducts are unstable and decompose under physiological conditions (PubMed:11522682, PubMed:11975663). Involved in intracellular deglycation in erythrocytes (PubMed:11975663). Involved in the response to oxidative stress by mediating deglycation of NFE2L2/NRF2, glycation impairing NFE2L2/NRF2 function (By similarity). Also able to phosphorylate psicosamines and ribulosamines (PubMed:14633848). {ECO:0000250|UniProtKB:Q9ER35, ECO:0000269|PubMed:11016445, ECO:0000269|PubMed:11522682, ECO:0000269|PubMed:11975663, ECO:0000269|PubMed:14633848}. | ATP-binding;Acetylation;Direct protein sequencing;Kinase;Nucleotide-binding;Reference proteome;Transferase | A high concentration of glucose can result in non-enzymatic oxidation of proteins by reaction of glucose and lysine residues (glycation). Proteins modified in this way, fructosamines, are less active or functional. This gene encodes an enzyme which catalyzes the phosphorylation of fructosamines which may result in deglycation. [provided by RefSeq, Feb 2012]. | hsa:64122; | cytosol [GO:0005829]; ATP binding [GO:0005524]; kinase activity [GO:0016301]; protein-fructosamine 3-kinase activity [GO:0102194]; protein-ribulosamine 3-kinase activity [GO:0102193]; epithelial cell differentiation [GO:0030855]; fructosamine metabolic process [GO:0030389]; fructoselysine metabolic process [GO:0030393]; post-translational protein modification [GO:0043687]; protein deglycation [GO:0036525] | 11975663_involved in the removal of fructosamine residues from hemoglobin in erythrocytes. 15102834_The aim of this work was to identify the fructosamine residues on hemoglobin that are removed as a result of the action of FN3K in intact erythrocytes. 15381090_These data suggest that FN3K and FN3KRP act as protein repair enzymes and are expressed constitutively in human cells independently of some of the variables altered in the diabetic state. 16037310_Enzyme is a constitutive 'housekeeping' gene and that ig plays an important role in cell metabolism, possibly as a deglycating enzyme. 16523184_No significant correlation between FN3K activity and the levels of HbA1c, total glycated haemoglobin (GHb) and haemoglobin fructoselysine residues, either in the normoglycaemic or diabetic group. 16920277_In this paper we propose a resolution of both these quandaries by proposing that fructosamine-6-phosphates are deglycated by phosphorylation to fructosamine-3,6-bisphosphates catalyzed by FN3KRP and/or possibly FN3K. 19834870_G900C polymorphism associates with the level of HbA (1c) and the onset of type 2 diabetes mellitus, but not with either of the diabetic microvascular complications. 19834870_Observational study of gene-disease association. (HuGE Navigator) 20858683_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 21253391_These findings suggest that deglycating enzymes Glyoxalase I and fructosamine-3-kinase may be involved in the malignant transformation of colon mucosa. 21288167_two new mutations and additional variants within the FN3K gene in diabetic patients 23492569_The marginal association of rs1056534 of FN3K is located in exon 6 with diabetic nephropathy progression. 24908234_Report association of rs1056534 and rs3848403 of fructosamine 3-kinase gene with sRAGE in patients with diabetes. 26352355_FN3K could act in concert with other molecular mechanisms and may impact on gene expression and activity of other enzymes involved in deglycation process 27461879_In a multiple regression analysis, FN3K rs1056534, TF polymorphism and presence of diabetes mellitus were predictors for HHV-8 infection. 31398338_FN3K is a targetable modulator of NRF2 activity in cancer. 32636308_A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily. 33208304_FN3K expression in COPD: a potential comorbidity factor for cardiovascular disease. | ENSMUSG00000025175 | Fn3k | 35.85476 | 1.5653984 | 0.6465299079 | 0.48609109 | 1.787470e+00 | 1.812347e-01 | No | Yes | 41.304045 | 13.618893 | 28.236239 | 9.122917 | ||
| ENSG00000167377 | 7571 | ZNF23 | protein_coding | P17027 | FUNCTION: May be involved in transcriptional regulation. May have a role in embryonic development. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:7571; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; regulation of transcription by RNA polymerase II [GO:0006357] | 12127974_Characterization of ZNF359 and its role in human development 17137575_ZNF23 is a new member of KRAB-ZNF superfamily with growth-inhibitory ability and its downregulation may contribute to carcinogenesis 18384939_ZNF23 induced apoptosis partially via down-regulation of Bcl-XL. 21965783_ZNF23 gene could play an important role in the development of hepatocellular carcinoma. 28848158_Ectopic expression of ZNF23 induced cell apoptosis by activation of caspase-3, p27, p53 expression and down-regulation of Bcl-2 through mitochondria-dependent pathway. 34030136_Nucleolar small molecule RNA SNORA75 promotes endometrial receptivity by regulating the function of miR-146a-3p and ZNF23. 34923485_Silencing of miR-1246 Induces Cell Cycle Arrest and Apoptosis in Cisplatin-Resistant Ovarian Cancer Cells by Promoting ZNF23 Transcription. | ENSMUSG00000044676 | Zfp612 | 560.28482 | 0.9129542 | -0.1313856543 | 0.15179928 | 7.472984e-01 | 3.873329e-01 | 7.519570e-01 | No | Yes | 653.363708 | 129.281682 | 715.087658 | 138.315538 | ||
| ENSG00000167384 | 7733 | ZNF180 | protein_coding | Q9UJW8 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Zinc finger proteins have been shown to interact with nucleic acids and to have diverse functions. The zinc finger domain is a conserved amino acid sequence motif containing 2 specifically positioned cysteines and 2 histidines that are involved in coordinating zinc. Kruppel-related proteins form 1 family of zinc finger proteins. See MIM 604749 for additional information on zinc finger proteins.[supplied by OMIM, Jul 2002]. | hsa:7733; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000057101 | Zfp180 | 371.24986 | 1.0831116 | 0.1151819151 | 0.15798617 | 5.327236e-01 | 4.654640e-01 | 8.002576e-01 | No | Yes | 476.287453 | 90.971433 | 434.105943 | 81.360510 | ||
| ENSG00000167703 | 124935 | SLC43A2 | protein_coding | Q8N370 | FUNCTION: Sodium-, chloride-, and pH-independent high affinity transport of large neutral amino acids. {ECO:0000269|PubMed:15659399}. | Alternative splicing;Amino-acid transport;Glycoprotein;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a member of the L-amino acid transporter-3 or SLC43 family of transporters. The encoded protein mediates sodium-, chloride-, and pH-independent transport of L-isomers of neutral amino acids, including leucine, phenylalanine, valine and methionine. This protein may contribute to the transfer of amino acids across the placental membrane to the fetus. [provided by RefSeq, Mar 2016]. | hsa:124935; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; amino acid transmembrane transporter activity [GO:0015171]; L-amino acid transmembrane transporter activity [GO:0015179]; neutral amino acid transmembrane transporter activity [GO:0015175]; amino acid transport [GO:0006865]; negative regulation of amino acid transport [GO:0051956]; negative regulation of leucine import [GO:0060358]; neutral amino acid transport [GO:0015804] | 15659399_In situ hybridization experiments show that LAT4 mRNA is restricted to the epithelial cells of the distal tubule and the collecting duct in the kidney. In the intestine, LAT4 is mainly present in the cells of the crypt. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 32879489_Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. 33077272_Plasma citrulline correlates with basolateral amino acid transporter LAT4 expression in human small intestine. | ENSMUSG00000038178 | Slc43a2 | 412.87113 | 0.9053598 | -0.1434368699 | 0.15811671 | 8.204715e-01 | 3.650423e-01 | 7.359414e-01 | No | Yes | 464.490235 | 64.271438 | 495.770586 | 66.926209 | |
| ENSG00000168216 | 55788 | LMBRD1 | protein_coding | Q9NUN5 | FUNCTION: Lysosomal membrane chaperone required to export cobalamin (vitamin B12) from the lysosome to the cytosol, allowing its conversion to cofactors (PubMed:19136951). Targets ABCD4 transporter from the endoplasmic reticulum to the lysosome (PubMed:27456980). Then forms a complex with lysosomal ABCD4 and cytoplasmic MMACHC to transport cobalamin across the lysosomal membrane (PubMed:25535791). Acts as an adapter protein which plays an important role in mediating and regulating the internalization of the insulin receptor (INSR) (By similarity). Involved in clathrin-mediated endocytosis of INSR via its interaction with adapter protein complex 2 (By similarity). Essential for the initiation of gastrulation and early formation of mesoderm structures during embryogenesis (By similarity). {ECO:0000250|UniProtKB:Q8K0B2, ECO:0000269|PubMed:19136951, ECO:0000269|PubMed:27456980, ECO:0000303|PubMed:25535791}.; FUNCTION: [Isoform 3]: (Microbial infection) May play a role in the assembly of hepatitis delta virus (HDV). {ECO:0000269|PubMed:15956556}. | Alternative splicing;Cell membrane;Cobalamin;Cobalt;Cytoplasmic vesicle;Developmental protein;Endocytosis;Endoplasmic reticulum;Gastrulation;Glycoprotein;Host-virus interaction;Lysosome;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix;Transport | This gene encodes a lysosomal membrane protein that may be involved in the transport and metabolism of cobalamin. This protein also interacts with the large form of the hepatitis delta antigen and may be required for the nucleocytoplasmic shuttling of the hepatitis delta virus. Mutations in this gene are associated with the vitamin B12 metabolism disorder termed, homocystinuria-megaloblastic anemia complementation type F.[provided by RefSeq, Oct 2009]. | hsa:55788; | clathrin-coated endocytic vesicle [GO:0045334]; clathrin-coated vesicle [GO:0030136]; endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; lysosomal membrane [GO:0005765]; membrane [GO:0016020]; plasma membrane [GO:0005886]; vacuolar membrane [GO:0005774]; AP-2 adaptor complex binding [GO:0035612]; clathrin heavy chain binding [GO:0032050]; cobalamin binding [GO:0031419]; insulin receptor binding [GO:0005158]; clathrin-dependent endocytosis [GO:0072583]; gastrulation [GO:0007369]; insulin receptor internalization [GO:0038016]; protein localization to lysosome [GO:0061462]; protein localization to vacuole [GO:0072665] | 19136951_LMBRD1 is the gene underlying the cblF defect of cobalamin metabolism and suggests that LMBD1 is a lysosomal membrane exporter for cobalamin. 20127417_a LMBRD1 mutation causes the cblF defect of vitamin B(12) metabolism in a Turkish patient [case report] 20446115_LMBRD1: the gene for the cblF defect of vitamin B metabolism. 21303734_novel mutations in LMBRD1 in three patients 23175358_These data indicate that by forming complexes with lamin A/C and nucleoporins, NESI facilitates the CRM1-independent nuclear export of large hepatitis delta antigen. 24078630_LMBD1 plays an imperative role in mediating and regulating the endocytosis of the IR. 25535791_Results propose a model whereby membrane-bound LMBD1 and ABCD4 facilitate the vectorial delivery of lysosomal vitamin B12 to cytoplasmic MMACHC. 27456980_endogenous ABCD4 was localized to both lysosomes and the ER, and its lysosomal localization was disturbed by knockout of LMBRD1 28572511_Data suggest that ABCD4 lysosomal targeting depends on co-expression of and interaction with LMBRD1; mutations in LMBRD1 and ABCD4 that result in cobalamin metabolism disorders cblF and cblJ (or mutations in ATPase domain) disrupt interactions between LMBRD1 and ABCD4. (LMBRD1 = nuclear export signal-interacting protein; ABCD4 = ATP-binding cassette, sub-family D (ALD), member 4) | ENSMUSG00000073725 | Lmbrd1 | 756.20166 | 1.0149251 | 0.0213732002 | 0.12613620 | 2.874148e-02 | 8.653773e-01 | 9.643976e-01 | No | Yes | 987.541222 | 225.289869 | 917.707185 | 204.880089 | |
| ENSG00000168564 | 55602 | CDKN2AIP | protein_coding | Q9NXV6 | FUNCTION: Regulates DNA damage response in a dose-dependent manner through a number of signaling pathways involved in cell proliferation, apoptosis and senescence. {ECO:0000269|PubMed:15109303, ECO:0000269|PubMed:24825908}. | Acetylation;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Ubl conjugation | The protein encoded by this gene regulates the DNA damage response through several different signaling pathways. One such pathway is the p53-HDM2-p21(WAF1) pathway, which is critical to the DNA damage response. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Nov 2015]. | hsa:55602; | granular component [GO:0001652]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; p53 binding [GO:0002039]; RNA binding [GO:0003723]; cellular response to DNA damage stimulus [GO:0006974]; negative regulation of cell growth [GO:0030308]; positive regulation of cell growth [GO:0030307]; positive regulation of signal transduction [GO:0009967]; regulation of protein stability [GO:0031647] | 12154087_cooperates with human p14ARF in activating p53 17460193_CARF may exert a vital control on p53-HDM2-p21(WAF1) pathway that is central to the cell cycle control, senescence, and DNA damage response of human cells 18292944_CARF exerts a vital control on the p53-HDM2-p21WAF1 pathway that is frequently altered in cancer cells. 18555516_CARF may act as a novel key regulator of the p53 pathway at multiple checkpoints 19001376_CARF plays a dual role in regulating p53-mediated senescence and apoptosis, the two major tumor suppressor mechanisms. 21052095_CARF knockdown elicited DNA damage response as evidenced by increased levels of phosphorylated ATM and gammaH2AX, leading to induction of mitotic arrest and eventual apoptosis. 22552337_Analysis of indel variations in the human disease-associated genes CDKN2AIP, WDR66, USP20 and OR7C2 in a Korean population. 24825908_CARF regulates proliferative fate of human cells by dose-dependent regulation of DNA damage signaling. 26278998_CARF promotes carcinogenesis in p53-deficient cells by repressing p21WAF1 and promoting cell cycle progression. 26531822_The results suggest that CARF regulates early steps of pre-rRNA processing during ribosome biogenesis by controlling spatial distribution of XRN2 between the nucleoplasm and nucleolus. 27457128_report that CARF (Collaborator of ARF) is a new target of miR-335 that regulates its growth suppressor function by complex crosstalk with other proteins including p16(INK4A), pRB, HDM2 and p21(WAF1) 27829235_Oncogenic functions of CARF in hepatocellular carcinoma tumorigenesis is a result of activation of beta-catenin/TCF signaling. 28754531_a comprehensive current understanding into the molecular mechanisms of CARF functions in regulation of DNA damage response, cell cycle checkpoints, cell survival and death signaling pathways. 32221864_Stress-induced changes in CARF expression determine cell fate to death, survival, or malignant transformation. | ENSMUSG00000038069 | Cdkn2aip | 577.00326 | 0.9570134 | -0.0633889314 | 0.13286719 | 2.271630e-01 | 6.336354e-01 | 8.828812e-01 | No | Yes | 797.292556 | 215.178395 | 797.989282 | 210.713864 | |
| ENSG00000168661 | 90075 | ZNF30 | protein_coding | P17039 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:90075; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 263.00368 | 0.9086614 | -0.1381853467 | 0.18062050 | 5.837420e-01 | 4.448493e-01 | 7.869395e-01 | No | Yes | 310.031314 | 59.551617 | 326.161811 | 61.244352 | |||||
| ENSG00000168679 | 9122 | SLC16A4 | protein_coding | O15374 | FUNCTION: Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity). {ECO:0000250}. | Alternative splicing;Cell membrane;Membrane;Reference proteome;Symport;Transmembrane;Transmembrane helix;Transport | hsa:9122; | integral component of membrane [GO:0016021]; integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; monocarboxylic acid transmembrane transporter activity [GO:0008028]; symporter activity [GO:0015293]; monocarboxylic acid transport [GO:0015718] | 14724187_MCT1 was lower and MCT4 similar in Type 2 diabetes versus control. With training, MCT1 content always increased, while MCT4 only increased in control. 15135232_studies demonstrate that the opposing plasma membranes of human syncytiotrophoblast are polarized with respect to both monocarboxylate transporter MCT1 and MCT4 activity and expression 15917240_inhibition of MCT1 and MCT4 activity by pCMBS is mediated through its binding to CD147, acting as an ancillary protein required to maintain the catalytic activity of MCTs 1 and 4, as well as for their translocation to the plasma membrane 16403666_Expressed in leukocytes and platelets; truncated 32kdal form may have a physiological function. 16408234_this study shows conflicting adaptations in MCT1 and MCT4 protein and mRNA levels following training, which may indicate post-transcriptional regulation of MCT expression in human muscle. 16452478_MCT4, like other glycolytic enzymes, is up-regulated by hypoxia through a HIF-1alpha-mediated mechanism 17082373_A single bout of high-intensity exercise decreased both MCT relative abundance (MCT1 and MCT4) in membrane preparations. 18056982_an exercise protocol designed to strain muscle carbohydrate reserves and to result in large increases in lactic acid results in a rapid upregulation of both GLUT-4 and MCT-4 18079261_Elevated muscle MCT4 in obesity could reflect the need to release greater amounts of muscle lactate. Weight loss decreased MCT4. 18188595_Expression of MCT1 and MCT4 showed a significant gain in plasma membranes of colorectal neoplasms. 18635880_MCT4 was reduced by 35% in vastus lateralis in chronic obstructive pulmonary disease. 18832090_Report effects of high-intensity training on muscle MCT1/4 and postexercise recovery of muscle lactate and hydrogen ions in women. 19073896_Suggest that the specific interaction of MCT4 with beta(1)-integrin may regulate cell migration through modulation of focal adhesions. 19876643_Data suggest that hypoxia increases lactate release from adipocytes and modulates MCT expression in a type-specific manner, with MCT1 and MCT4, but not MCT2 expression, being hypoxia-inducible transcription factor-1 (HIF-1) dependent. 20035863_In corneal epithelium and cell lines human monocarboxylic acid transporter 4 is expressed at mRNA and protein levels. 20454640_MCT1, MCT2, and MCT4 protein expression in breast, colon, lung, and ovary neoplasms, as well as CD147 and CD44, were analysed. 20658178_The result suggest that co-expression of CD147 and MCT1/MCT4 is related to drug resistance during EOC metastasis and could be useful therapeutic targets to prevent the development of incurable, recurrent and drug resistance EOC. 21306479_We found that the expression of the monocarboxylate transporters MCT1 and MCT4, but not MCT5, in human lung cancer cell lines was significantly correlated with invasiveness 21558814_breast cancer cells specifically induce the expression of MCT4 in cancer-associated fibroblasts; the expression of MCT4 in cancer-associated fibroblasts is due to oxidative stress 21787388_Data show that a significant increase of MCT2 and MCT4 expression in the cytoplasm of tumour cells and a significant decrease in both MCT1 and CD147 expression in prostate tumour cells was observed when compared to normal tissue. 21870331_both MCT1 and CD147, but not MCT4, were associated with GLUT1 and CAIX expression in a large series of invasive breast carcinoma samples 21930917_CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. 22153830_High GLUT1 plus high MCT4 expression indicated an aggressive tumor behavior in adenocarcinomas. 22240841_Report SNPs in MCT4 (SLC16A3) gene in the Chinese and Indian populations of Singapore. 22362593_Data suggest that MCT4 may serve as a novel metabolic target to reverse the Warburg effect and limit disease progression in renal cell carcinoma. 23187830_Combined application of GLUT-1, MCT-1, and MCT-4 immunohistochemistry might be useful in differentiating malignant pleural mesothelioma from reactive mesothelial hyperplasia. 23258846_Overexpression of MCT4 is associated with gliomas. 23574725_MCT1 and MCT4 biomarkers were employed to determine the metabolic state of proliferative cancer cells. 23780984_High MCT4 contributes to the growth of colorectal cancer with vascular endothelial growth factor. 23881922_Upregulation of MCT4 expression via SLC16A3 promoter DNA methylation is associated with clear cell renal cell carcinoma. 23907124_Alterations in Cav-1 and MCT4 expression may mark a critical point in the progression from in situ to invasive breast cancer. 23935841_Results suggest that Arginine-278 in transmembrane-spanning domains TMD8 is a critical residue involved in L-lactate recognition by monocarboxylate transporter 4 (hMCT4). 24077291_MCT4 upregulation correlated with the aggressive mesenchymal subset of glioblastoma (GBM), and MCT4 downregulation correlated with the less aggressive G-CIMP (Glioma CpG Methylator Phenotype) subset of GBM. 24338019_Coexpression of CAIV with MCT1 and MCT4 resulted in a significant increase in MCT transport activity. 24433439_Aberrant expression of MCT4 in carcinoma cells serves as a novel, independent prognostic factor for HCC, indicating a poorer patient outcome. 24464262_Decreased astroglial monocarboxylate transporter 4 expression in temporal lobe epilepsy 24498219_The critical role of MCT4 in cell proliferation. 24797797_These results indicate there are no additional benefits of IHT when compared to similar normoxic training. Hence, the addition of the hypoxic stimulus on anaerobic performance or MCT expression after a three-week training period is ineffective. 25058459_MCT4 demonstrated the strongest deleterious impact on survival in triple negative breast cancer patients.MCT4 should serve as a new prognostic factor in node-negative breast cancers. 25374230_High monocarboxylate transporter 4 protein expression in stromal cells is associated with invasiveness in gastric cancer. 25406319_MCT4 is up-regulated in inflammation-activated macrophages and required for innate immune response. 25446815_MCT4 expression can predict survival and trans-arterial chemoembolization treatment response for hepatocellular carcinoma patients. MCT4 plays a role in cell proliferation and migration/invasion. 25578492_we sought to evaluate the associations of nine functional SNPs in genes encoding MCT1, MCT2, and MCT4 with the prognosis in a cohort of 500 Chinese NSCLC patients. 25919709_Findings demonstrate that the histidine residue His382 in the extracellular loop of the transporter is essential for pH regulation of MCT4-mediated substrate transport activity. 25957999_The first IF method has been developed and optimised for detection of MCT 1 and MCT4 in cancer patient circulating tumour cells . 25965974_MCT4 expression is regulated by the PI3K-Akt signalling pathway and highly expressed in HER2-positive breast cancers where it regulates tumor cell metabolism and survival. 26059436_we showed that genetic disruption of Mct4 and/or Ampk dramatically reduced tumourigenicity in a xenograft mouse model suggesting a crucial role for these two actors in establishment of tumours in a nutrient-deprived environment 26213210_synovial fibroblasts from patients with rheumatoid arthritis exhibited up-regulated transcription of MCT4 mRNA compared with osteoarthritis patients. Knockdown of MCT4 induced intrinsic apoptosis of fibroblasts, thereby inhibiting their proliferation. 26363456_Knockdown of MCT4 blocks lactate efflux to result in lactic acid accumulation and pH dropping, which is involved in triggering apoptosis in HUVECs. 26384346_Prognostic significance of CD147 protein expression could not surpass that of MCT4, especially of SLC16A3 DNA methylation, corroborating the role of MCT4 as prognostic biomarker for ccRCC. 26755530_Elevated MCT4 protein expression in clinical prostate cancer specimens was associated with increases in Gleason grade, prolonged treatment of patients with neoadjuvant hormone therapy, castration resistant prostate cancer, and early disease relapse. 26765963_Data suggest that inhibition of mnocarboxylate transporters MCT1 and MCT4 may have clinical relevance in pancreatic ductal adenocarcinoma (PDAC). 26779534_MCT1 expression was not clearly associated with overall or disease-free survival. MCT4 and CD147 expression correlate with worse prognosis across many cancer types. These results warrant further investigation of these associations 26876179_MCT1 inhibition impairs proliferation of glycolytic breast cancer cells co-expressing MCT1 and MCT4 via disruption of pyruvate rather than lactate export. 26944480_The reversible H(+)/lactate(-) symporter MCT4 cotransports lactate and proton, leading to the net extrusion of lactic acid in glycolytic tumors. A model of its role in pH control in tumor cells is described. Review. 27105345_MCT1 and MCT4 expression levels were associated with worse prognosis and shorter overall survival. 27144334_our finding that the expression of MCT1 and MCT4 is reduced in mutant IDH1 gliomas highlights the unusual metabolic reprogramming that occurs in mutant IDH1 tumors and has important implications for our understanding of these tumors and their treatment 27224918_Results suggest that MCT4 plays a central role in tumor metabolism in gastric cancer (GC) with peritoneal carcinomatosis and targeting MCT4 in combination with chemotherapy could be a novel strategy in the treatment of GC. 28099149_Increased miR-210 and concomitant decreased ISCU RNA levels were found in ~40% of tumors and this was significantly associated with HIF-1alpha and CAIX, but not MCT1 or MCT4, over-expression. 28347233_We demonstrated that the expression levels of glycolysis-related proteins glucose transporter 1, hexokinase II, carbonic anhydrase IX, and monocarbonylate transporter 4 differ between thyroid cancer subtypes and are correlated with poorer prognosis 28419250_High expression of MCT4 is associated with inflammation in arsenite-induced liver carcinogenesis. 28559188_The structures and functions of hMCT1 and hMCT4 transporters. 28625953_The loss expression of Cav-1 on CAFs and the up-regulation of MCT4 may be the possible mechanisms of CAFs in tumorigenesis. 28846107_Nrf2 overexpression upregulated MCT1, but decreased MCT4 expression in premalignant and malignant colonic epithelial cells. 28923861_MCT1 inhibitor AZD3965 increased MCT4-dependent accumulation of intracellular lactate, inhibiting monocarboxylate influx and efflux. 29248132_Stromal cells in diffuse large B-cell lymphoma samples strongly expressed MCT4, displaying a glycolytic phenotype, a feature not seen in stromal elements of non-neoplastic lymphatic tissue. 29248133_TOMM20, MCT1, and MCT4 expression was significantly different in Hodgkin and Reed Sternberg (HRS) cells. High MCT4 expression was found in tumor associated macrophages, but absent in HRS cells in all but one case. Tumor-infiltrating lymphocytes had absent MCT4 expression. Reactive lymph nodes in contrast to cHL tumors had low TOMM20, MCT1, and MCT4 expression in lymphocytes and macrophages. 29483215_High MCT4 expression is associated with Colorectal Cancer with Peritoneal Carcinomatosis. 29657088_High MCT-4 expression is associated with clear cell renal cell carcinoma. 29775610_These results demonstrate that Monocarboxylate transporters tend to play a role in the aggressive breast cancer subtypes through the dynamic interaction between breast cancer cells and adipocytes. 30226548_MCT4 knockdown reduced the activation of Akt and increased Bax/Bcl-2 ratios, cytochrome c release and caspase-3 cleavage.Consequently, MCT4 could serve as a promising biomarker for esophageal squamous cell carcinoma to identify patients with poor prognosis 30262589_The expression of MCT4 in urothelial carcinoma is associated with features of aggressive tumor biology and portends a poor prognosis. 30306706_Using the expression of MCT4 and GLUT1 and their metabolic parameters to determine the metabolic status of tumors is promising for predicting the prognosis of patients with hepatocellular carcinoma. 31101938_we were interested in the molecular mechanism responsible for the difference in substrate specificity between hMCT1 and hMCT4. Therefore, we generated 3D structure models of hMCT1 and hMCT4 to identify amino acid residues involved in the substrate specificity of these transporters. We found that the substrate specificity of hMCT1 was regulated by residues involved in turnover number (M69) and substrate affinity 31177263_MCT4 expression was significantly upregulated in osteosarcoma tissues compared with that in adjacent normal ones, detected via both immunohistochemical and Western blot analyses. 31239287_Study demonstrates that stromal MCT4 drives cancer aggressiveness in the context of cigarette smoke and describes the AhR pathway as a novel pathway regulating the expression of MCT4. 31395464_Monocarboxylate transporters in cancer. 31706332_Intratumoral reciprocal expression of monocarboxylate transporter 4 and glypican-3 in hepatocellular carcinomas. 31711985_The NF-kappaB/miR-425-5p/MCT4 axis: A novel insight into diabetes-induced endothelial dysfunction. 31723238_CAIX forms a transport metabolon with monocarboxylate transporters in human breast cancer cells. 31729681_CD147 augmented monocarboxylate transporter-1/4 expression through modulation of the Akt-FoxO3-NF-kappaB pathway promotes cholangiocarcinoma migration and invasion. 31738978_Role of monocarboxylate transporter 4 in Alzheimer disease. 32112988_Benign albeit glycolytic: MCT4 expression and lactate release in giant cell tumour of bone. 32212469_Inhibiting the expression of MCT4 can interfere with the glucose metabolism and promote the apoptosis of prostate cancer cells. The MCT4 gene is a potential therapeutic target for the treatment of prostate cancer. 32819565_Identification of the essential extracellular aspartic acids conserved in human monocarboxylate transporters 1, 2, and 4. 32872409_Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. 32887638_Tissue expression of lactate transporters (MCT1 and MCT4) and prognosis of malignant pleural mesothelioma (brief report). 33212210_Immunoreactivity of receptor and transporters for lactate located in astrocytes and epithelial cells of choroid plexus of human brain. 34077729_Therapy-induced DNA methylation inactivates MCT1 and renders tumor cells vulnerable to MCT4 inhibition. 34093533_Monocarboxylate Transporter 4 Triggered Cell Pyroptosis to Aggravate Intestinal Inflammation in Inflammatory Bowel Disease. 34172808_MCT4 is induced by metastasis-enhancing pathogenic mitochondrial NADH dehydrogenase gene mutations and can be a therapeutic target. 34269158_Comprehensive Analysis of Monocarboxylate Transporter 4 (MCT4) expression in breast cancer prognosis and immune infiltration via integrated bioinformatics analysis. 34619707_Coexpression of MCT1 and MCT4 in ALK-positive Anaplastic Large Cell Lymphoma: Diagnostic and Therapeutic Implications. 34769160_miR-31-NUMB Cascade Modulates Monocarboxylate Transporters to Increase Oncogenicity and Lactate Production of Oral Carcinoma Cells. 34822124_Knockdown of Astrocytic Monocarboxylate Transporter 4 in the Motor Cortex Leads to Loss of Dendritic Spines and a Deficit in Motor Learning. 35093305_ZEB1 induces ROS generation through directly promoting MCT4 transcription to facilitate breast cancer. 35324524_TRIM72 exerts antitumor effects in breast cancer and modulates lactate production and MCT4 promoter activity by interacting with PPP3CA. | ENSMUSG00000027896 | Slc16a4 | 34.43601 | 0.8788812 | -0.1862599359 | 0.51983010 | 1.276738e-01 | 7.208562e-01 | No | Yes | 33.557260 | 11.974097 | 36.951578 | 12.822057 | |||
| ENSG00000169398 | 5747 | PTK2 | protein_coding | Q05397 | FUNCTION: Non-receptor protein-tyrosine kinase that plays an essential role in regulating cell migration, adhesion, spreading, reorganization of the actin cytoskeleton, formation and disassembly of focal adhesions and cell protrusions, cell cycle progression, cell proliferation and apoptosis. Required for early embryonic development and placenta development. Required for embryonic angiogenesis, normal cardiomyocyte migration and proliferation, and normal heart development. Regulates axon growth and neuronal cell migration, axon branching and synapse formation; required for normal development of the nervous system. Plays a role in osteogenesis and differentiation of osteoblasts. Functions in integrin signal transduction, but also in signaling downstream of numerous growth factor receptors, G-protein coupled receptors (GPCR), EPHA2, netrin receptors and LDL receptors. Forms multisubunit signaling complexes with SRC and SRC family members upon activation; this leads to the phosphorylation of additional tyrosine residues, creating binding sites for scaffold proteins, effectors and substrates. Regulates numerous signaling pathways. Promotes activation of phosphatidylinositol 3-kinase and the AKT1 signaling cascade. Promotes activation of MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling cascade. Promotes localized and transient activation of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), and thereby modulates the activity of Rho family GTPases. Signaling via CAS family members mediates activation of RAC1. Recruits the ubiquitin ligase MDM2 to P53/TP53 in the nucleus, and thereby regulates P53/TP53 activity, P53/TP53 ubiquitination and proteasomal degradation. Phosphorylates SRC; this increases SRC kinase activity. Phosphorylates ACTN1, ARHGEF7, GRB7, RET and WASL. Promotes phosphorylation of PXN and STAT1; most likely PXN and STAT1 are phosphorylated by a SRC family kinase that is recruited to autophosphorylated PTK2/FAK1, rather than by PTK2/FAK1 itself. Promotes phosphorylation of BCAR1; GIT2 and SHC1; this requires both SRC and PTK2/FAK1. Promotes phosphorylation of BMX and PIK3R1. Isoform 6 (FRNK) does not contain a kinase domain and inhibits PTK2/FAK1 phosphorylation and signaling. Its enhanced expression can attenuate the nuclear accumulation of LPXN and limit its ability to enhance serum response factor (SRF)-dependent gene transcription. {ECO:0000269|PubMed:10655584, ECO:0000269|PubMed:11331870, ECO:0000269|PubMed:11980671, ECO:0000269|PubMed:15166238, ECO:0000269|PubMed:15561106, ECO:0000269|PubMed:15895076, ECO:0000269|PubMed:16919435, ECO:0000269|PubMed:16927379, ECO:0000269|PubMed:17395594, ECO:0000269|PubMed:17431114, ECO:0000269|PubMed:17968709, ECO:0000269|PubMed:18006843, ECO:0000269|PubMed:18206965, ECO:0000269|PubMed:18256281, ECO:0000269|PubMed:18292575, ECO:0000269|PubMed:18497331, ECO:0000269|PubMed:18677107, ECO:0000269|PubMed:19138410, ECO:0000269|PubMed:19147981, ECO:0000269|PubMed:19224453, ECO:0000269|PubMed:20332118, ECO:0000269|PubMed:20495381, ECO:0000269|PubMed:21454698}.; FUNCTION: [Isoform 6]: Isoform 6 (FRNK) does not contain a kinase domain and inhibits PTK2/FAK1 phosphorylation and signaling. Its enhanced expression can attenuate the nuclear accumulation of LPXN and limit its ability to enhance serum response factor (SRF)-dependent gene transcription. {ECO:0000269|PubMed:20109444}. | 3D-structure;ATP-binding;Acetylation;Alternative promoter usage;Alternative splicing;Angiogenesis;Cell junction;Cell membrane;Cell projection;Cytoplasm;Cytoskeleton;Developmental protein;Direct protein sequencing;Isopeptide bond;Kinase;Membrane;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Transferase;Tyrosine-protein kinase;Ubl conjugation | This gene encodes a cytoplasmic protein tyrosine kinase which is found concentrated in the focal adhesions that form between cells growing in the presence of extracellular matrix constituents. The encoded protein is a member of the FAK subfamily of protein tyrosine kinases but lacks significant sequence similarity to kinases from other subfamilies. Activation of this gene may be an important early step in cell growth and intracellular signal transduction pathways triggered in response to certain neural peptides or to cell interactions with the extracellular matrix. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jun 2017]. | hsa:5747; | cell cortex [GO:0005938]; ciliary basal body [GO:0036064]; cytoplasm [GO:0005737]; cytoskeleton [GO:0005856]; cytosol [GO:0005829]; dendritic spine [GO:0043197]; extrinsic component of cytoplasmic side of plasma membrane [GO:0031234]; focal adhesion [GO:0005925]; intracellular membrane-bounded organelle [GO:0043231]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; stress fiber [GO:0001725]; actin binding [GO:0003779]; ATP binding [GO:0005524]; integrin binding [GO:0005178]; JUN kinase binding [GO:0008432]; non-membrane spanning protein tyrosine kinase activity [GO:0004715]; protein kinase binding [GO:0019901]; protein phosphatase binding [GO:0019903]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; protein tyrosine kinase activity [GO:0004713]; SH2 domain binding [GO:0042169]; signaling receptor binding [GO:0005102]; angiogenesis [GO:0001525]; axon guidance [GO:0007411]; cell differentiation [GO:0030154]; cell migration [GO:0016477]; cell motility [GO:0048870]; detection of muscle stretch [GO:0035995]; ephrin receptor signaling pathway [GO:0048013]; epidermal growth factor receptor signaling pathway [GO:0007173]; establishment of cell polarity [GO:0030010]; Fc-gamma receptor signaling pathway involved in phagocytosis [GO:0038096]; growth hormone receptor signaling pathway [GO:0060396]; heart morphogenesis [GO:0003007]; innate immune response [GO:0045087]; integrin-mediated signaling pathway [GO:0007229]; negative regulation of anoikis [GO:2000811]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell-cell adhesion [GO:0022408]; netrin-activated signaling pathway [GO:0038007]; peptidyl-tyrosine phosphorylation [GO:0018108]; placenta development [GO:0001890]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of fibroblast migration [GO:0010763]; positive regulation of macrophage chemotaxis [GO:0010759]; positive regulation of macrophage proliferation [GO:0120041]; positive regulation of phosphatidylinositol 3-kinase activity [GO:0043552]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein kinase activity [GO:0045860]; positive regulation of protein kinase B signaling [GO:0051897]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of ubiquitin-dependent protein catabolic process [GO:2000060]; positive regulation of wound healing [GO:0090303]; protein autophosphorylation [GO:0046777]; protein phosphorylation [GO:0006468]; regulation of cell adhesion [GO:0030155]; regulation of cell adhesion mediated by integrin [GO:0033628]; regulation of cell population proliferation [GO:0042127]; regulation of cell shape [GO:0008360]; regulation of cytoskeleton organization [GO:0051493]; regulation of endothelial cell migration [GO:0010594]; regulation of epithelial cell migration [GO:0010632]; regulation of focal adhesion assembly [GO:0051893]; regulation of GTPase activity [GO:0043087]; regulation of osteoblast differentiation [GO:0045667]; regulation of protein phosphorylation [GO:0001932]; regulation of substrate adhesion-dependent cell spreading [GO:1900024]; signal complex assembly [GO:0007172]; transforming growth factor beta receptor signaling pathway [GO:0007179]; transmembrane receptor protein tyrosine kinase signaling pathway [GO:0007169]; vascular endothelial growth factor receptor signaling pathway [GO:0048010] | 11779709_Tyrosine phosphorylation of FAK regulates localization and downstream signaling with profound effects on cell movement. 11799111_Regulation of G protein-linked guanine nucleotide exchange factors for Rho, PDZ-RhoGEF, and LARG by tyrosine phosphorylation: evidence of a role for focal adhesion kinase 11809746_FAK regulates the activity of Akt/protein kinase B and GSK-3beta and the association of GSK-3beta with FAK to influence insulin-stimulated glycogen synthesis in hepatocytes. 11886520_proliferative effect of fibronectin in combination with T cell lymphokines on psoriatic uninvolved basal keratinocyte progenitors may be due to abnormal in vivo integrin-driven focal adhesion kinase activity and downstream signaling. 11891225_Laminin-10/11 and fibronectin differentially prevent apoptosis induced by serum removal via phosphatidylinositol 3-kinase/Akt- and MEK1/ERK-dependent pathways (Laminin 10; separate entry for Laminin 11). 11916084_comparison with FAK2 of the mechanism of tyrosine phosphorylation upon vWF interaction with glycoprotein Ib-IX-V complex 11950595_Nck-2 interacts with focal adhesion kinase (FAK), a cytoplasmic protein tyrosine kinase critically involved in the cellular control of motility 11986332_beta1 integrin regulates fibroblast viability through a PI3K/Akt/protein kinase B signaling pathway in response to a matrix-derived mechanical stimulus 11988077_Primary arrest of circulating platelets on collagen involves phosphorylation of Syk, cortactin and focal adhesion kinase 12005431_This study reports two crystal structures of the focal adhesion targeting domain of focal adhesion kinase. 12011046_IGF-I protects the cells from apoptosis by blocking the activation of caspases, which may be responsible for the loss of FAK and Akt. 12049193_REVIEW: The focal adhesion kinase--a regulator of cell migration and invasion 12054581_Mutated focal adhesion kinase induces apoptosis in a human glioma cell line 12135674_The association of focal adhesion kinase with Wiskott-Aldrich syndrome protein is associated with cell migration in stromal cell-derived factor-1alpha-stimulated Jurkat cells 12167618_The results of this study indicate that dual inhibition of focal adhesion kinase (FAK) and epidermal growth factor receptor (EGFR) signaling pathways can cooperatively enhance apoptosis in breast cancers. 12169389_Cerivastatin-induced inhibition of glioblastoma cell migration was associated with the down-regulation of tyrosine phosphorylation of FAK. FAK phosphorylation correlated well with tumor cell invasiveness. 12174366_Expression of focal adhesion kinase and alpha5 and beta1 integrins in carcinomas and its clinical significance. 12215217_results indicated that the mitochondrial pathway is required for ionizing radiation-induced apoptosis, and focal adhesion kinase overexpression blocks this pathway, rendering antiapoptotic states. 12216109_Functional involvement of src and focal adhesion kinase in a CD99 splice variant-induced motility of human breast cancer cells 12297287_Engagement of CD44 either by its natural ligand hyaluronan or a specific antibody on a cell line induced tyrosine phosphorylation and activation of focal adhesion kinase. 12376862_An altered relationship of FAK and Pyk2 was observed for different tumors and could also be important for osteosarcoma development 12387730_Results reveal a novel mechanism of FAK phosphorylation by signalling cascades involving a member of the LDL receptor family. 12435390_FAK NH(2)-terminal domain fragments have roles independent of Y397, kinase, and FAT domains 12529399_R-Ras promotes focal adhesion formation by signaling to FAK and p130(Cas) through a novel mechanism that differs from but synergizes with the alpha2beta1 integrin. 12531888_TGF-beta1 up-regulates expression of integrins and fibronectin, an effect that is associated with autophosphorylation/activation of FAK. 12543870_Shear-induced platelet aggregation induced by 2B-rVWF binding to platelet GPIb produced pp125FAK at shear rate 4000 s-1 but not at 200 s-1 12611892_caspase-3-mediated proteolysis of FAK, an anti-apoptotic protein, is regulated by hsp72 12651906_FAK and Pyk2 function as important signaling effectors in gliomas and indicate that their differential regulation may be determining factors in the temporal development of proliferative or migrational phenotypes. 12700132_This review of recent studeis provides considerable insight into the functional roles and signaling mechanisms of FAK as a positive regulator of both cell motility and cell survival. 12794117_Augmented activation of FAK is an immediate signaling event required for the trans-regulation of integrin alpha L beta 2 by alpha 4 beta 1 in Jurkat T cells. 12803239_Decidual beta1 integrin and FAK participate in this final step of implantation. 12815062_FAK activity is regulated by low molecular weight phosphotyrosine phosphatase in human cells 12844492_FAK is activated by VEGF in human brain microvascular endothelial cells 12847914_Inhibition of FAK by antisense oligonucleotideds enhances the sensitivity of breast cancer cells to campothecin. 12881299_CIB regulates platelet spreading through the regulation of FAK activation. 12884911_FAK has a role in the pathology of human cancer [review] 12904305_TIMP-1 activates cell survival signaling pathways involving focal adhesion kinase, phosphatidylinositol 3-kinase, and ERKs in human breast epithelial cells 12907754_both c-Src/PI3K and c-Src/Fak/Erk1/2 pathways are involved in the up-regulation of c-myc and cyclin d1 expression mediated by prolactin 12939401_FAK and Src are important survival factors, playing a role in protecting colon cancer cell lines from Adenovirus-containing FAK-CD (Ad-FAK-CD)-induced apoptosis 12954625_beta1 integrin/FAK-mediated signaling on osteoblasts could be involved in ICAM-1- and RANKL-dependent osteoclast maturation 14521607_FAK has a role in binding of soluble and surface-bound ligand to integrin alphaIIbbeta3 14553943_All ESFT cell lines expressed five- to twenty-eight-fold-increased values of FAK, . These results raise the possibility that the overexpression of c-myc and FAK are involved in the poor prognosis of ESFT. 14578863_PTK2 and EIF3S3, which, respectively, encode focal adhesion kinase and the p40 subunit of the eukaryotic initiation factor 3, were probable targets within the amplification at 8q23-q24 and may be involved in progression of HCC. 14584897_pp125FAK interacts with the large conductance calcium-activated hSlo potassium channel in human osteoblasts: potential role in mechanotransduction 14617636_focal adhesion kinase and src are stimulated by G alpha q and platelet activating factor receptor in vascular endothelium 14637150_The above results indicate that PKB-Ser-473 and FAK-Tyr phosphorylation stimulated by TGF-beta1 are both dependent on cell adhesion. 14670178_the proteolytic degradation of FAK is temporally distinct from its tyrosine dephosphorylation, occurring when apoptotic pathways are already initiated and during a generalized destruction of signaling proteins 14699482_Extracellular pressure may increase integrin affinity and promote colon cancer adhesion via actin dependent inside-out focal adhesion kinase and Src signals. This mechanotransduced pathway may regulate metastasizing tumor cell adhesion. 15102689_FAK could play an important role in hepatocellular carcinoma progression 15102844_Calcium rises locally trigger focal adhesion disassembly and enhance residency of focal adhesion kinase at focal adhesions 15155793_RhoA/Rho-kinase pathway followed by tyrosine phosphorylation of FAK and paxillin leads to ATP release and actin reorganization in vascular endothelium 15157737_Cloning and characterization of the promoter region of human focal adhesion kinase gene and analysis of nuclear factor kappa B and p53 binding sites 15161045_Results demonstrate FHL2 expression in human ovarian cancer cells, suggesting an important functional role of pp125FAK and FHL2 complex in gynecologic malignancies. 15166238_FAK has a role functioning upstream of PI3K/Akt, in transducing a beta(1) integrin viability signal in collagen matrices 15175910_Activation of rho had no effect on phosphorylation of FAK. 15210734_Activation of FAK and ERK1/2 by MCSP appear to involve independent mechanisms. 15247219_focal adhesion kinase phosphorylation is mediated by VEGFR2 and regulated by heat shock protein 90 and Src kinase activities 15331608_Data show that squamous cell carcinoma cells escape suspension-induced, p53-mediated anoikis by forming multicellular aggregates that use fibronectin survival signals mediated by integrin alpha(v) and focal adhesion kinase. 15369772_The introduction of FAK siRNA in HL-60/FAK cells sensitized them against TRAIL-induced apoptosis, confirming that overexpressed FAK downregulates procaspase-8 expression, which subsequently inhibits downstream apoptosis pathway in the HL-60/FAK cells. 15455382_Overexpression and Phosphorylation of focal adhesion kinase is associated with ovarian carcinoma 15532706_The level of FAK tyrosine phosphorylation appeared to be inversely correlated with the level of the PTEN protein in human; the defect of PTEN in tumor cells could alter the phosphorylation of FAK 15557280_overexpression of FAK promoted exit from G(1) in glioblastoma cells, enhanced expression of cyclins D1 and E while reducing expression of p27(Kip1) and p21(Waf1), and enhanced the kinase activity of the cyclin D1-cyclin-dependent kinase-4 (cdk4) complex 15611137_the first FAK subdomain of the FERM domain of focal adhesion kinase has a role in its normal regulation and function in the cell 15640164_the derepression of Rb-E2F-regulated genes leads to apoptosis through inactivation of focal adhesion kinase and activation of caspase-8 15652490_Angiotensin II mediates an increase in FAK and paxillin phosphorylation and induces human umbilical vein endothelial cells migration through signal transduction pathways 15657578_RET signals through focal adhesion kinase in medullary thyroid cancer cells. 15657875_identified PTK2 SNPs that displayed strong signals in joint analysis of linkage and association (unadjusted P<10(-7)) with SLE 15681841_appears to be involved in early events of integrin-mediated adhesion of circulating carcinoma cells under fluid flow in vitro and in vivo 15736429_FAK siRNA-treated cells displayed a decrease in migration when serum or EGF (epidermal growth factor) were used as chemo-attractants; results demonstrated that inhibition of FAK protein leads to alterations in cell growth and migration. 15743500_FAK protein overexpression coincides with Her-2/neu overexpression and may mediate HER2 signaling via Src, resulting in PI3K/Akt activation 15778501_apoptotic is propagated by decreased phosphorylation of focal adhesion kinase (FAK), which is linked to increased phosphorylation of c-Jun N-terminal kinase (JNK) and to decreased levels of p53 15850774_integrin alpha(v) inhibition abrogates FAK phosphorylation at focal adhesion sites and diminishes MMP-2 secretion leading to reduced migration and invasion of human coronary artery smooth muscle cells 15855171_FAK can suppress p53-mediated apoptosis and inhibit transcriptional activity of p53 15866427_In the prostate carcinoma PC-3 cell model, the action of the gastrin releasing peptide (GRP) analog, bombesin (BN), on the activation of focal adhesion kinase (FAK) and invasiveness suggests that this kinase might favor metastasis 15975092_results indicate an additional mechanism for regulation of FAK activity during cell spreading and migration, involving Ser-722 phosphorylation modulated through the competing actions of GSK3beta and PP1 16007195_SOCS-3 negatively regulates cell growth and cell motility by inhibiting Janus kinase (JAK)/STAT and FAK signalings in hepatocellular carcinoma cells 16039608_We propose a model in which removal of FERM-mediated auto-inhibition is important to increase FAK catalytic activity but the translocation and clustering of this enzyme at the focal adhesions is required for maximal phosphorylation at Tyr-397. 16054026_A regulatory mechanism of cell invasion was suggested whereby FAK promotes cell-surface presentation of MT1-MMP by inhibiting endophilin A2-dependent endocytosis 16105876_Data show that alpha(v)beta3/Tat interaction triggers the activation of NF-kappaB in endothelial cells in a focal adhesion kinase-, RhoA- and pp60src-dependent manner, and this activation is required for motogenic activity of Tat in endothelial cells. 16136050_Upregulation of focal adhesion kinase is associated with breast cancer 16141199_JSAP1.FAK complex functions cooperatively as a scaffold for the JNK signaling pathway and regulator of cell migration on FN 16159962_Data show that phosphorylation of focal adhesion kinase (FAK) at Tyr-397 is a key determinant of how FAK controls focal adhesion turnover. 16226872_results suggest the possibility that therapeutic resistance to all-trans retinoic acid may be related to FAK overexpression 16244766_phosphorylation of pp125(FAK) is a late, integrin-dependent event, results suggest that platelet activation and aggregation occur in vivo in these patients 16247468_In vivo loss of FAK does not affect migration/proliferation of basal keratinocytes in the same way as it affects multipotent stem cells of the skin. 16251422_FAK may act in glioblastoma as a downstream target of growth factor signaling, with integrins enhancing the impact of such signaling in the tumor microenvironment 16260653_FAK regulates integrin-dependent MMP-2 and MMP-9 expression and release by T lymphoid cells through src kinase 16270396_Data suggest that focal adhesion kinase plays a significant role in signaling pathways of a hepatocyte growth factor-responsive cell line derived from cholangiocarcinoma. 16308318_FAK-induced down-modulation of RhoA activity via p190RhoGAP is a crucial step in signaling endothelial barrier restoration after increased endothelial permeability 16354697_Inhibition of FAK by SHP-2 plays a crucial role in the morphogenetic activity of Helicobacter pylori CagA. 16406804_While in CXCR2-expressing cells FAK phosphorylation was adhesion-dependent and was stimulated by fibronectin, in CXCR1-expressing cells FAK phosphorylation was adhesion-independent. 16412380_These studies also raise the possibility that fully active forms of c-Src and Fak in breast tumors. 16523241_These data suggest that FAK might differently regulate apoptosis and focal adhesion formation through site-specific, Src-dependent tyrosine phosphorylation in senescent cells. 16638855_Weak expression of FAK in invasive cervical cancer is a strong independent predictor of poor patient outcome. 16705171_These data demonstrate a critical role for FAK in the regulation of cyclin-dependent kinase inhibitors (CDKIs) through two independent mechanisms: Skp2 dependent and Skp2 independent. 16842883_FAK is up-regulated in NSCLC, and it may have a role in lung cancer progression 16899713_FAK protein abundance appears regulated at the mRNA level during gut epithelial cell motility. 16912186_The technique of RNA silencing [small interfering RNA] confirmed the role of FAK in invasive prostatic cancer. 16920698_Gastrin-releasing peptide upregulates ICAM-1 via FAK promotes tumor cell motility and attachment to the extracellular matrix. 16998832_Enhanced expression of FAK and phospho-FAK were observed in colon cancer tissues. 17028776_CAGE promotes motility of cancer cells through activation of focal adhesion kinase (FAK. 17081517_FAK signaling pathways provide a link between activation of ERK1/2 by extracellular matrix proteins and mesenchymal stem cell differentiation into osteoblasts 17088251_Src and Rac1 have roles in focal adhesion kinase and ERK mitogenic signaling in epithelial cells 17096371_Our results indicate that increased FAK phosphorylation at Ser-843 represses FAK phosphorylation at Tyr-397, thus suggesting a mechanism of cross-talk between these phosphorylation sites that could regulate FAK-mediated cell shape and migration. 17113264_TGF-beta1 co-ordinately and independently activates the FAK and AKT protein kinase pathways to confer an anoikis-resistant phenotype to myofibroblasts. 17213807_Downregulation of either FAK or ILK expression inhibited SPARC-mediated AKT phosphorylation, and targeting both FAK and ILK attenuated AKT activation more potently than targeting either FAK or ILK alone 17215324_Src and focal adhesion kinase mediate cyclic mechanical strain-induced extracellular regulated kinase phosphorylation and proliferation of pulmonary epithelial cells. 17236584_FAK was highly expressed in oral squamous cell carcinoma tissues, in contrast to none or a low expression in normal oral epithelial tissue, and the heterogeneous staining was mainly located in the cell membrane and cytoplasm 17240116_Study found that either carbachol or EGF promoted a striking ERK-dependent phosphorylation of FAK at Ser-910, but these agonists caused only slight stimulation of FAK at Tyr-397 in T84 cells. 17317726_results delineate a novel force-activated inside-out Src/PI3K/FAK/Akt pathway by which cancer cells regulate their own adhesion. 17327229_N-MYC induces FAK expression. 17332925_Ephrin-A1 serves as a critical negative regulator in the tumorigenesis of gliomas by down-regulating EphA2 and FAK. 17397984_role FAK plays in promoting cell invasion through the activation of distinct signaling pathways induced by EGF with protein MMP-9 transcription and secretion in follicular thyroid carcinoma cells 17443665_Fak/Src signaling to the PI3-K/Akt-1 and MEK/Erk pathways undergoes a differentiation state-specific uncoupling in enterocytes. 17469136_These results suggest that engagement of beta1 integrins on systemic lupus erythematosus T cells could induce FAK-mediated signaling and subsequent CD40L expression and proliferation. 17514628_In kidney disease, increased parathyroid hyperplasia cell growth driven by enhanced EGFR could be further aggravated through elevations in integrin beta1 and FAK expression. 17515959_The expression of phosphorylated FAK was documented in all 28 cytokeratin-positive breast cancer samples. 17537730_Decreased acetylglucosaminyltransferase activity due to smaall interferingRNA expression in human breast carcinoma cells resulted in attenuation of the dephosphorylation of FAK induced by epidermal growth factor. 17620332_c-Abl activation by insulin, via a modification of FAK response, may play an important role in directing mitogenic versus metabolic insulin receptor signaling. 17675501_STAP-2 associates with FAK and enhances its degradation, proteasome inhibitors block FAK degradation, and STAP-2 recruits an endogenous E3 ubiquitin ligase, Cbl, to FAK. 17828307_Catalytic activity is required for integrin alpha5beta1-stimulated Src (Proto-Oncogene Proteins pp60(c-src)) activation. 17879163_These findings provide evidence that mutations in fibronectin, induce anoikis in human squamous cell carcinoma cells by modulating integrin alpha v-mediated phosphorylation of FAK and ERK. 17888894_Not cell detachment, but the proteolytic cleavage (or inhibition) of FAK is a key modulator as well as a promising indicator of apoptosis in epithelial cells under oxidative stress. 17904248_FAK causes defects in spreading, reinforcement of integrin-cytoskeleton linkages and migration and at the same time could ameliorate the adhesion of metastatic cells to suboptimal surfaces. 17967873_provide direct evidence of conformational regulation of FAK in living cells and novel insight into the mechanism regulating FAK conformation 17982280_The results presented here indicate that actin reorganization through FAK/PI3-K/Rac-1 activation operates in various human cancer cell systems supporting a functional role for FAK/PI-3K/Rac1/actin signaling in controlling cell motility. 17999388_Increased focal Adhesion Kinase due to mutations of p53 is associated with breast and colon cancers 18032789_MAP-kinase activity necessary for TGFbeta1-stimulated mesangial cell type I collagen expression requires adhesion-dependent phosphorylation of FAK tyrosine 397. 18061419_Col-IV regulates the secretion of MMP-9 via a Src and FAK dependent pathway in MCF-7 cells 18073135_These findings suggest that modified signaling mechanisms regulate cancer cell migration through an endothelial monolayer versus those involved in cell migration on or through ECM. 18078823_Focal adhesion kinase as well as p130Cas and paxillin should be a crucial molecule undergoing stronger tyrosine phosphorylation in GD3-expressing melanoma cells. 18078954_Structural basis for the interaction between FAK and CD4 is reported. 18182379_indicate the involvement of FAK-dependent activation of extracellular signal-regulated kinase in Toll like receptor-mediated eosinophil stimulation 18184462_FAK is a pivotal signal transducer downstream of gastrin in gastric cancer. 18198129_Tac-beta1 inhibits cell spreading, at least in part, by preventing the phosphorylation of FAK at Tyr-397 18206965_In human cells, FAK knockdown raised p53-p21 levels & slowed cell proliferation but did not cause apoptosis. Nuclear FAK plays a scaffolding role in facilitating cell survival through enhanced p53 degradation under stress. 18210145_Concordant overexpression of p-FAK and p-ERK1/2 in extramammary Paget's disease (EMPD) is associated with the grade of malignancy of EMPD. 18215142_identified the binding site of the p53 and FAK interaction and demonstrated that mutating this site and targeting the site with peptides affects p53 functioning and viability in the cells. 18223254_PTPD1 is a component of a multivalent scaffold complex nucleated by FAK at specific intracellular sites 18247360_These results strongly support that Elk-1 protein is a novel binding-protein partner for FAK, a finding that significantly broadens the potential functioning of FAK and Elk-1. 18256281_HEPL maintains Cas family function in localization to focal adhesions, as well as regulation of FAK activity, focal adhesion integrity, and cell spreading. 18263593_simultaneous inhibition of both Focal adhesion kinase and the insulin-like growth factor-I receptor represents a potential novel therapeutic approach in human pancreatic adenocarcinoma 18292575_FAK expression is necessary for efficient phosphorylation of T-cell differentiation protein MAL following stimulation with bacterial lipopolysaccharide and protein I/II. 18292781_Data show that NG2 and integrin alpha4 oppositely regulate anoikis in fibroblasts, and that NG2 and integrin alpha4 regulate FAK phosphorylation by PKCalpha-dependent and -independent pathways, respectively. 18322799_Anoikis causes a down-activation of Fak, Src, Akt-1 and Erk1/2, a loss of Fak-Src association, and a sustained/enhanced activation of p38b, which is required as apoptosis/anoikis driver 18332144_focal adhesion kinase mediates caveolin-1 up-regulation during epithelial to mesenchymal transition 18349846_these data reported here support the conclusion that FAK enhances invasion of HNSCC by promoting both increased cell motility and MMP-2 production 18353772_FAK induces KLF8 expression in human ovarian cancer cells by activating the PI3K-Akt signaling pathway, leading to the activation of KLF8 promoter by Sp1 18414015_c-FLIPL promotes the motility of HeLa cells by activating FAK and ERK, and increasing MMP-9 expression. 18449907_Data show that genetic upregulation of matriptase-2 reduces the aggressiveness of prostate cancer cells in vitro and in vivo and affects FAK and paxillin localisation. 18452580_Focal adhesion kinase is phosphorylated in response to interferon-gamma-inducible GTPase expression. 18493017_review of kinase-independent interaction of FAK with p53with focus on FAK and p53 signaling, which link signal transduction pathways 18519756_FAK is expressed by advanced-stage neuroblastoma 18549812_FAK has two nuclear export signal sequences, but only one of the signal sequences demonstrates full biological nuclear export activity. 18562041_focal adhesion kinase controls the balance of adhesion types in cells, and that this is one of the determinants of whether a cancer cell can make stable matrix-degrading invadopodia. 18593949_SRC-3/AIB1 is required for focal adhesion turnover and focal adhesion kinase activation. 18632638_analysis of PAFR activation and pleiotropic effects on tyrosine phospho-EGFR/Src/FAK/paxillin in ovarian cancer 18669633_Focal adhesion kinase (FAK)-related non-kinase inhibits myofibroblast differentiation through differential MAPK activation in a FAK-dependent manner. 18680726_These data indicate that FAK is functioning in cell migration, but fibril-forming collagen-induced FAK degradation is necessary for endothelial tube formation. 18695939_Results suggest that HAb18G/CD147 enhances the invasion and metastatic potentials of human hepatoma cells via integrin alpha3beta1-mediated FAK-paxillin and FAKPI3K-Ca(2+) signal pathways. 18832579_These data support the cooperative function of Pyk2 and FAK in breast cancer progression and suggest that dual inhibition of FAK and Pyk2 is an efficient therapeutic approach for targeting invasive breast cancer. 18846341_The integrin beta1-focal adhesion kinase signal pathway plays a role in the invasiveness of pituitary adenomas. 18922979_The interplay between FAK and ERK in androgen-independent prostate cancer cells (PC3 and DU145 cells), was investigated. 18948272_Following down-regulation of MR-1, the phosphorylations of MLC2, focal adhesion kinase (FAK), and Akt were dramatically decreased 18987997_Enhanced FAK expression is associated with intestinal type gastric adenocarcinoma. 19006177_FAK is essentially required in chondrocyte communication with type II collagen by regulating type II collagen expression and cell proliferation. 19020730_FAK, mTOR and IGF-IR are inhibited by TAE226 in esophageal cancer cells 19021002_Overexpression of FAK is associated with HBV infection and contribution to hepatocellular carcinoma progression. 19029090_The lysyl oxidase pro-peptide attenuates fibronectin-mediated activation of focal adhesion kinase and p130Cas in breast cancer cells. 19032871_The expression of FRNK was enhanced and the phosphorylation of FAK was inhibited after FRNK was transiently transfected into hepatic stellate cells. FRNK also induces apoptosis of HSC. 19042019_groups: patients overexpressing 2 or 3 factors had a significantly shorter OS (p=0.015). CXCR4, VLA-4 and FAK are new phenotypic markers which could be helpful to establish risk-stratified therapeutic strategies 19055942_analysis of FAK expression regulation and therapeutic potential [review] 19082453_PGE2 greatly induced hepatocellular carcinoma cell adhesion, migration, and invasion by activating FAK/paxillin/Erk pathway. 19089993_Results describe a a novel mechanism by which vitronectin receptors and focal adhesion kinase could promote cancer metastasis via ERK5 activation. 19098120_Bis-phosphorylated FAK is a useful biomarker of Src kinase inhibition in vivo. 19141860_Observational study of gene-disease association. (HuGE Navigator) 19147981_FAK supports Ras- and PI3K-dependent mammary tumor initiation, maintenance, and progression to metastasis. 19150878_FXR promotes endothelial cell motility through coordinated regulation of FAK and MMP-9. 19199036_These results suggest that Bim, Bcl-xL, FAK and endonuclease G are involved in safingol-induced apoptosis of detached oral squamous cell carcinoma. 19242756_Report expression of FAK and PTEN in bronchioloalveolar carcinoma and lung adenocarcinoma. 19268501_Data demonstrate that the RhoGTPase activating protein 21 (ARHGAP21) is expressed in the nuclear and perinuclear regions of several cell lines, and interacts with the C-terminal region of focal adhesion kinase. 19297531_This work demonstrates a novel role for FAK in integrin activation and the time-dependent generation of cell-extracellular matrix forces. 19297850_The mRNA levels and positive rates of integrin beta1 and FAK protein were significantly higher in laryngeal carcinoma than that in the surrounding tissue and vocal cord polyp. 19301259_Connective tissue growth factor enhances the migration of chondrosarcoma cells by increasing MMP-13 expression through the alphavbeta3 integrin, FAK, ERK, and NF-kappaB signal transduction pathway. 19303400_phosphorylation of Y407-FAK and S732-FAK are important for FAK in modulating cell migration. 19318351_AGAP2 regulates the FAK activity and the focal adhesion disassembly during cell migration. 19339212_Results suggest that Fer may allow a bypass of focal adhesion kinase-related cell anchorage dependency for intracellular signal transduction in hepatocytes. 19364917_Data report that endogenous Focal adhesion kinase functions upstream of cellular Src (c-Src) as a negative regulator of invadopodia formation and dynamics in breast cancer cells. 19403028_Up-regulation of FAK may play an important role in the invasion of SMMC-7721 cells induced by hypoxia. 19423701_c-Abl phosphorylates RACK1 at the Focal Adhesion Kinase interaction site and c-Abl kinase activity is required for interaction of Focal Adhesion Kinase with RACK1. 19435801_Data show that fibroblast, endothelial and carcinoma polarity during cell migration requires FAK and is associated with a complex between FAK, p120RasGAP and p190RhoGAP (p190A), leading to p190A tyrosine phosphorylation. 19454363_focal adhesion kinase expression is significantly related to subsequent hepatocellular carcinoma metastasis 19471549_The present study has focused on the expression pattern of FAK in human endometrium during the menstrual cycle. 19473962_Formation of focal adhesion kinase *Grb7 complexes and Grb7 phosphorylation by FAK in an integrin-dependent manner were essential for cell migration, proliferation and anchorage-independent growth in A431 epidermal carcinoma cells. 19475568_OPN enhances the migration of chondrosarcoma cells by increasing MMP-9 expression through the alphavbeta3 integrin, FAK, MEK, ERK and NF-kappaB signal transduction pathway. 19479902_EGFR-mediated sustained activation of Src intestinal epithelial cancer cells confers anoikis resistance at least in part through a consequent sustenance of Fak-Src interactions and MEK/Erk activation 19481075_Direct binding of toxin A to Src, independent of any effect on protein tyrosine phosphatase or Rho glucosylation, inhibits Src kinase activity followed by FAK/paxillin inactivation 19492042_These results suggest a novel mechanism for selective reinforcement of nascent adhesions via interplays of Nudel and FAK with paxillin to facilitate cell migration 19500106_Knockdown of focal adhesion kinase reverses colon carcinoma multicellular resistance. 19545541_these data suggest a role for FAK in phosphorylation, signaling and stability of the IGF-1R. 19561089_Pyk2 and FAK are downstream targets of the Rap GTPases that play a key role in regulating B cell morphology. 19563023_PTEN gene might inhibit the proliferation, metastasis and invasive ability of leukemia cells via down-regulating FAK expression. 19574423_Cyclic mechanical strain impairs signaling of cell migration after injury via a pathway that involves FAK-JIP3-JNK. 19615356_enhanced expressions of HAb18G/CD147, MMPs, paxillin and FAK changed the distributions of cytoskeleton in the 3D reconstituted basement membrane (BM) and increased the adhesion and invasion potentials of hepatocellular carcinoma cells 19633364_ILK mediated pressure-stimulated adhesion through regulating phosphorylation of AKT and FAK. 19635908_FAK, phosphatidylinositol 3-kinase, and Akt kinase are involved in the potentiating action of peptidoglycan in rheumatoid arthritis synovial fibroblasts. 19644410_FAK protein positivity and overexpression correlate with important clinicopathological parameters, in breast ductal invasive carcinoma. 19656390_Results demonstrated that the growth inhibition of HEC1A cells by GnRH-I or GnRH-II is involved in the activation of integrin-FAK and ERK1/2 and p38 MAPK pathways. 19664602_FAK-NT2 (a.a. 127-243) domain directly interacts with the N-terminal part of the IGF-1R intracellular domain. 19671193_FAK has a role in human MCF-7 breast cancer cell tumorigenesis 19695571_Periostin mediates vascular smooth muscle cell migration through the integrins alphavbeta3 and alphavbeta5 and focal adhesion kinase (FAK) pathway 19695682_up-regulation of focal adhesion kinase protein may also contribute to the tumor progression of gastrointestinal stromal tumors 19736351_Observational study of gene-disease association. (HuGE Navigator) 19737461_FAK expression regulated the cell pro | ENSMUSG00000022607 | Ptk2 | 2353.11877 | 0.9770077 | -0.0335582165 | 0.08960522 | 1.400329e-01 | 7.082483e-01 | 9.108573e-01 | No | Yes | 2668.494801 | 437.144714 | 2560.428787 | 410.195622 | |
| ENSG00000169410 | 5780 | PTPN9 | protein_coding | P43378 | FUNCTION: Protein-tyrosine phosphatase that could participate in the transfer of hydrophobic ligands or in functions of the Golgi apparatus. {ECO:0000269|PubMed:19167335}. | 3D-structure;Acetylation;Cytoplasm;Hydrolase;Protein phosphatase;Reference proteome | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP contains an N-terminal domain that shares a significant similarity with yeast SEC14, which is a protein that has phosphatidylinositol transfer activity and is required for protein secretion through the Golgi complex in yeast. This PTP was found to be activated by polyphosphoinositide, and is thought to be involved in signaling events regulating phagocytosis. [provided by RefSeq, Jul 2008]. | hsa:5780; | cytoplasm [GO:0005737]; neuron projection terminus [GO:0044306]; nucleoplasm [GO:0005654]; non-membrane spanning protein tyrosine phosphatase activity [GO:0004726]; protein tyrosine phosphatase activity [GO:0004725]; negative regulation of neuron projection development [GO:0010977]; peptidyl-tyrosine dephosphorylation [GO:0035335]; positive regulation of protein localization to plasma membrane [GO:1903078]; protein dephosphorylation [GO:0006470] | 12112018_Purification and characterization of protein tyrosine phosphatase PTP-MEG2 12920026_PTP-MEG2 has an important role in the development of erythroid cells. 14662869_PTPase-MEG2 through its Sec14p homology domain couples inositide phosphorylation to tyrosine dephosphorylation and the regulation of intracellular traffic of the secretory pathway in T cells. 15322554_PTP-MEG2 reduced the phosphotyrosine content of NSF and co-localized with NSF and syntaxin 6 in intact cells, the first demonstrated role for a protein tyrosine phosphatase in the regulated secretory pathway 16679294_role in the negative regulation of hepatic insulin signaling 17387180_the N terminus of PTPMEG2 is necessary for the targeting of this phosphatase to the secretory vesicle compartment by association with other proteins involved in intracellular transport. 20335174_data suggest PTPN9 as a negative regulator of breast cancer cells by targeting ErbB2 and EGFR and inhibiting STAT activation 22394684_PTPMeg2 is an important phosphatase for the dephosphorylation of STAT3 and plays a critical role in breast cancer development. 22763125_This study indentified VEGFR2 as a PTPN9 substrate, and indicated that PTPN9 is a negative regulator of VEGFR2 signaling and function in endothelial cells. 26288249_Results from a study on gene expression variability markers in early-stage human embryos shows that PTPN9 is a putative expression variability marker for the 3-day, 8-cell embryo stage. 26715439_PTPN9 inhibited cell proliferation in HepG2 cells. 27857177_this study highlights an important role for miR-96 in the regulation of PTPN9 in breast cancer cells and may provide insight into the molecular mechanisms of breast carcinogenesis. 28747184_Expression of MEG2 is reversely correlated with that of miR-181a-5p in gastric cancer. 30399427_Low MEG2 expression is associated with metastasis of hepatocellular carcinoma. 33112705_PTPN9-mediated dephosphorylation of VTI1B promotes ATG16L1 precursor fusion and autophagosome formation. 33463918_The Critical Role of the miR-21-MEG2 Axis in Colorectal Cancer. 34373992_Effect of L3MBTL3/PTPN9 polymorphisms on risk to alcohol-induced ONFH in Chinese Han population. 35156900_CircMMD_007 promotes oncogenic effects in the progression of lung adenocarcinoma through microRNA-197-3p/protein tyrosine phosphatase non-receptor type 9 axis. | ENSMUSG00000032290 | Ptpn9 | 620.70874 | 0.9806455 | -0.0281964416 | 0.12670061 | 4.946738e-02 | 8.239927e-01 | 9.513319e-01 | No | Yes | 796.347989 | 87.484984 | 777.841582 | 83.896705 | |
| ENSG00000169992 | 57555 | NLGN2 | protein_coding | Q8NFZ4 | FUNCTION: Transmembrane scaffolding protein involved in cell-cell interactions via its interactions with neurexin family members. Mediates cell-cell interactions both in neurons and in other types of cells, such as Langerhans beta cells. Plays a role in synapse function and synaptic signal transmission, especially via gamma-aminobutyric acid receptors (GABA(A) receptors). Functions by recruiting and clustering synaptic proteins. Promotes clustering of postsynaptic GABRG2 and GPHN. Promotes clustering of postsynaptic LHFPL4 (By similarity). Modulates signaling by inhibitory synapses, and thereby plays a role in controlling the ratio of signaling by excitatory and inhibitory synapses and information processing. Required for normal signal amplitude from inhibitory synapses, but is not essential for normal signal frequency. May promote the initial formation of synapses, but is not essential for this. In vitro, triggers the de novo formation of presynaptic structures. Mediates cell-cell interactions between Langerhans beta cells and modulates insulin secretion (By similarity). {ECO:0000250, ECO:0000250|UniProtKB:Q69ZK9}. | 3D-structure;Cell adhesion;Cell junction;Cell membrane;Cell projection;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Postsynaptic cell membrane;Reference proteome;Signal;Synapse;Transmembrane;Transmembrane helix | This gene encodes a member of a family of neuronal cell surface proteins. Members of this family may act as splice site-specific ligands for beta-neurexins and may be involved in the formation and remodeling of central nervous system synapses. [provided by RefSeq, Jul 2008]. | hsa:57555; | cell surface [GO:0009986]; cytoplasm [GO:0005737]; dendritic shaft [GO:0043198]; dopaminergic synapse [GO:0098691]; excitatory synapse [GO:0060076]; glycinergic synapse [GO:0098690]; inhibitory synapse [GO:0060077]; integral component of plasma membrane [GO:0005887]; integral component of postsynaptic membrane [GO:0099055]; integral component of postsynaptic specialization membrane [GO:0099060]; membrane [GO:0016020]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; presynaptic membrane [GO:0042734]; ribbon synapse [GO:0097470]; spanning component of membrane [GO:0089717]; symmetric, GABA-ergic, inhibitory synapse [GO:0098983]; synapse [GO:0045202]; cell adhesion molecule binding [GO:0050839]; identical protein binding [GO:0042802]; neurexin family protein binding [GO:0042043]; signaling receptor activity [GO:0038023]; brain development [GO:0007420]; cell-cell adhesion [GO:0098609]; cell-cell junction maintenance [GO:0045217]; chemical synaptic transmission [GO:0007268]; gephyrin clustering involved in postsynaptic density assembly [GO:0097116]; inhibitory synapse assembly [GO:1904862]; insulin metabolic process [GO:1901142]; jump response [GO:0007630]; locomotory exploration behavior [GO:0035641]; modulation of chemical synaptic transmission [GO:0050804]; neuromuscular process controlling balance [GO:0050885]; neuron cell-cell adhesion [GO:0007158]; neurotransmitter-gated ion channel clustering [GO:0072578]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of dendritic spine development [GO:0060999]; positive regulation of excitatory postsynaptic potential [GO:2000463]; positive regulation of inhibitory postsynaptic potential [GO:0097151]; positive regulation of insulin secretion [GO:0032024]; positive regulation of protein localization to synapse [GO:1902474]; positive regulation of synapse assembly [GO:0051965]; positive regulation of synaptic transmission, GABAergic [GO:0032230]; positive regulation of synaptic transmission, glutamatergic [GO:0051968]; positive regulation of synaptic vesicle clustering [GO:2000809]; positive regulation of t-SNARE clustering [GO:1904034]; postsynaptic density protein 95 clustering [GO:0097119]; postsynaptic membrane assembly [GO:0097104]; presynapse assembly [GO:0099054]; presynaptic membrane assembly [GO:0097105]; protein localization to cell surface [GO:0034394]; protein localization to synapse [GO:0035418]; regulation of AMPA receptor activity [GO:2000311]; regulation of presynapse assembly [GO:1905606]; regulation of respiratory gaseous exchange by nervous system process [GO:0002087]; sensory perception of pain [GO:0019233]; social behavior [GO:0035176]; synapse assembly [GO:0007416]; synapse organization [GO:0050808]; synaptic transmission, GABAergic [GO:0051932]; synaptic vesicle endocytosis [GO:0048488]; terminal button organization [GO:0072553]; thigmotaxis [GO:0001966] | 19086053_Observational study of gene-disease association. (HuGE Navigator) 21551456_Data identified the R215H mutant as a loss-of-function mutant in inducing GABAergic synaptogenesis. Data also suggests that defects in GABAergic synapse formation in the brain may be an important contributing factor for the onset of schizophrenia. 22528485_Transcellular neuroligin-2 interactions enhance insulin secretion and are integral to pancreatic beta cell function 24842555_Neuroligin-2 was down-regulated in aganglionic colonic segments from Hirschsprung's disease patients. 27805570_e found that NLGN3 function at inhibitory synapses in rat CA1 depends on the presence of NLGN2 and identified a domain in the extracellular region that accounted for this functional difference between NLGN2 and 3 specifically at inhibitory synapses. 27865048_This is the first report of an NLGN2 nonsense variant in humans, adding to the accumulating evidence that links synaptic proteins with a spectrum of neurodevelopmental phenotypes. 28641112_MDGAs regulate the formation of neuroligin-neurexin trans-synaptic bridges by sterically blocking access of neurexins to neuroligins. 29339486_nucleus accumbens neuroligin-2 has a role in depression and stress susceptibility 33127642_Neuroligin-2 dependent conformational activation of collybistin reconstituted in supported hybrid membranes. 33443230_Neuroligin-2 as a central organizer of inhibitory synapses in health and disease. | ENSMUSG00000051790 | Nlgn2 | 1039.73818 | 0.9579122 | -0.0620346378 | 0.10827140 | 3.279117e-01 | 5.668914e-01 | 8.531186e-01 | No | Yes | 1099.248959 | 150.344826 | 1158.457059 | 154.568446 | |
| ENSG00000170412 | 55890 | GPRC5C | protein_coding | Q9NQ84 | FUNCTION: This retinoic acid-inducible G-protein coupled receptor provide evidence for a possible interaction between retinoid and G-protein signaling pathways. {ECO:0000250}. | Alternative splicing;Cell membrane;Cytoplasmic vesicle;G-protein coupled receptor;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Signal;Transducer;Transmembrane;Transmembrane helix | The protein encoded by this gene is a member of the type 3 G protein-coupled receptor family. Members of this superfamily are characterized by a signature 7-transmembrane domain motif. The specific function of this protein is unknown; however, this protein may mediate the cellular effects of retinoic acid on the G protein signal transduction cascade. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]. | hsa:55890; | cytoplasmic vesicle membrane [GO:0030659]; extracellular exosome [GO:0070062]; integral component of plasma membrane [GO:0005887]; intracellular membrane-bounded organelle [GO:0043231]; plasma membrane [GO:0005886]; receptor complex [GO:0043235]; vesicle [GO:0031982]; G protein-coupled receptor activity [GO:0004930]; protein kinase activator activity [GO:0030295]; G protein-coupled receptor signaling pathway [GO:0007186] | 28228611_Data suggest that agents (such as all-trans retinoic acid) that activate GPRC5C restore and/or maintain functional insulin-secreting cells; GPRC5C appears to play role in regulation of expression of genes controlling apoptosis, cell survival, and cell proliferation in insulin-secreting cells. | ENSMUSG00000051043 | Gprc5c | 101.34305 | 0.9030437 | -0.1471323398 | 0.29892186 | 2.411818e-01 | 6.233539e-01 | 8.788316e-01 | No | Yes | 103.832932 | 15.585184 | 110.155957 | 16.077306 | |
| ENSG00000170464 | 202052 | DNAJC18 | protein_coding | Q9H819 | Chaperone;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:202052; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; Hsp70 protein binding [GO:0030544]; cellular response to misfolded protein [GO:0071218]; chaperone cofactor-dependent protein refolding [GO:0051085]; ubiquitin-dependent ERAD pathway [GO:0030433] | 18676680_Observational study of gene-disease association. (HuGE Navigator) 19170196_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19240791_Observational study of gene-disease association. (HuGE Navigator) 19625176_Observational study of gene-disease association. (HuGE Navigator) 19692168_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000024350 | Dnajc18 | 551.33028 | 0.8974421 | -0.1561092432 | 0.14317339 | 1.185263e+00 | 2.762871e-01 | 6.652608e-01 | No | Yes | 803.210956 | 162.687630 | 818.506193 | 161.928156 | |||
| ENSG00000170471 | 57148 | RALGAPB | protein_coding | Q86X10 | FUNCTION: Non-catalytic subunit of the heterodimeric RalGAP1 and RalGAP2 complexes which act as GTPase activators for the Ras-like small GTPases RALA and RALB. {ECO:0000250}. | Alternative splicing;GTPase activation;Phosphoprotein;Reference proteome | hsa:57148; | GTPase activator activity [GO:0005096]; protein heterodimerization activity [GO:0046982]; activation of GTPase activity [GO:0090630]; regulation of small GTPase mediated signal transduction [GO:0051056] | 24814574_Deregulation of RalGAPbeta might cause genomic instability, leading to human carcinogenesis. 34009715_Ral GTPase-activating protein regulates the malignancy of pancreatic ductal adenocarcinoma. | ENSMUSG00000027652 | Ralgapb | 2802.00208 | 1.0045121 | 0.0064949844 | 0.09188555 | 4.997957e-03 | 9.436395e-01 | 9.854008e-01 | No | Yes | 3628.652496 | 631.654437 | 3321.355842 | 565.316593 | ||
| ENSG00000170776 | 11214 | AKAP13 | protein_coding | Q12802 | FUNCTION: Scaffold protein that plays an important role in assembling signaling complexes downstream of several types of G protein-coupled receptors. Activates RHOA in response to signaling via G protein-coupled receptors via its function as Rho guanine nucleotide exchange factor (PubMed:11546812, PubMed:15229649, PubMed:23090968, PubMed:25186459, PubMed:24993829). May also activate other Rho family members (PubMed:11546812). Part of a kinase signaling complex that links ADRA1A and ADRA1B adrenergic receptor signaling to the activation of downstream p38 MAP kinases, such as MAPK11 and MAPK14 (PubMed:17537920, PubMed:23716597, PubMed:21224381). Part of a signaling complex that links ADRA1B signaling to the activation of RHOA and IKBKB/IKKB, leading to increased NF-kappa-B transcriptional activity (PubMed:23090968). Part of a RHOA-dependent signaling cascade that mediates responses to lysophosphatidic acid (LPA), a signaling molecule that activates G-protein coupled receptors and potentiates transcriptional activation of the glucocorticoid receptor NR3C1 (PubMed:16469733). Part of a signaling cascade that stimulates MEF2C-dependent gene expression in response to lysophosphatidic acid (LPA) (By similarity). Part of a signaling pathway that activates MAPK11 and/or MAPK14 and leads to increased transcription activation of the estrogen receptors ESR1 and ESR2 (PubMed:9627117, PubMed:11579095). Part of a signaling cascade that links cAMP and EGFR signaling to BRAF signaling and to PKA-mediated phosphorylation of KSR1, leading to the activation of downstream MAP kinases, such as MAPK1 or MAPK3 (PubMed:21102438). Functions as scaffold protein that anchors cAMP-dependent protein kinase (PKA) and PRKD1. This promotes activation of PRKD1, leading to increased phosphorylation of HDAC5 and ultimately cardiomyocyte hypertrophy (By similarity). Has no guanine nucleotide exchange activity on CDC42, Ras or Rac (PubMed:11546812). Required for normal embryonic heart development, and in particular for normal sarcomere formation in the developing cardiomyocytes (By similarity). Plays a role in cardiomyocyte growth and cardiac hypertrophy in response to activation of the beta-adrenergic receptor by phenylephrine or isoproterenol (PubMed:17537920, PubMed:23090968). Required for normal adaptive cardiac hypertrophy in response to pressure overload (PubMed:23716597). Plays a role in osteogenesis (By similarity). {ECO:0000250|UniProtKB:E9Q394, ECO:0000269|PubMed:11546812, ECO:0000269|PubMed:11579095, ECO:0000269|PubMed:17537920, ECO:0000269|PubMed:21224381, ECO:0000269|PubMed:23716597, ECO:0000269|PubMed:24993829, ECO:0000269|PubMed:25186459, ECO:0000269|PubMed:9627117, ECO:0000269|PubMed:9891067}. | 3D-structure;Alternative splicing;Coiled coil;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Metal-binding;Methylation;Nucleus;Phosphoprotein;Proto-oncogene;Reference proteome;Zinc;Zinc-finger | The A-kinase anchor proteins (AKAPs) are a group of structurally diverse proteins which have the common function of binding to the regulatory subunit of protein kinase A (PKA) and confining the holoenzyme to discrete locations within the cell. This gene encodes a member of the AKAP family. Alternative splicing of this gene results in multiple transcript variants encoding different isoforms containing c-terminal dbl oncogene homology (DH) and pleckstrin homology (PH) domains. The DH domain is associated with guanine nucleotide exchange activation for the Rho/Rac family of small GTP binding proteins, resulting in the conversion of the inactive GTPase to the active form capable of transducing signals. The PH domain has multiple functions. Therefore, these isoforms function as scaffolding proteins to coordinate a Rho signaling pathway, function as protein kinase A-anchoring proteins and, in addition, enhance ligand-dependent activity of estrogen receptors alpha and beta. [provided by RefSeq, Jul 2012]. | hsa:11214; | cell cortex [GO:0005938]; cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; cAMP-dependent protein kinase activity [GO:0004691]; guanyl-nucleotide exchange factor activity [GO:0005085]; MAP-kinase scaffold activity [GO:0005078]; metal ion binding [GO:0046872]; molecular adaptor activity [GO:0060090]; protein kinase A binding [GO:0051018]; small GTPase binding [GO:0031267]; adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process [GO:0086023]; adrenergic receptor signaling pathway [GO:0071875]; bone development [GO:0060348]; cardiac muscle cell differentiation [GO:0055007]; cell growth involved in cardiac muscle cell development [GO:0061049]; G protein-coupled receptor signaling pathway [GO:0007186]; heart development [GO:0007507]; intracellular signal transduction [GO:0035556]; nuclear export [GO:0051168]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of Rho protein signal transduction [GO:0035025]; regulation of Rho protein signal transduction [GO:0035023]; regulation of sarcomere organization [GO:0060297]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12270917_Results show that alpha-catulin co-expression leads to increased Lbc-induced serum response factor activation and may modulate Rho pathway signaling in vivo by providing a scaffold for the Lbc Rho guanine nucleotide exchange factor. 12663445_The HA-3 peptide, VTEPGTAQY, is encoded by the lymphoid blast crisis oncogene, showing for the 1st time that a leukemia-associated oncogene can give rise to immunogenic T-cell epitopes that may participate in antihost & antileukemic alloimmune responses. 14636890_Proto-Lbc mutant expression led to decreased levels of Galpha12-induced RhoA activation in vivo. 14660653_results indicate that guanine nucleotide exchange factor Lbc is a novel signal transducer for RhoA-mediated NF-kappaB activation in human peripheral blood monocytes stimulated with bacterial products 15229649_Anchoring of both PKA and 14-3-3 inhibits the Rho-GEF activity of the AKAP-Lbc signaling complex. 15691829_Rho-GEF activity of AKAP-Lbc is mediated by leucine zipper-mediated homo-oligomerization regulates 16234258_Observational study of gene-disease association. (HuGE Navigator) 16234258_significant association seen between rare AKAP13 Lys526Glyn variant and increased risk of development breast cancer; this variant might affect susceptibility to other cancers and might influence response to anticancer drugs targeting rho proteins 16301118_Yeast 2-Hybrid experiments identified a strong and novel interaction between the transglutaminase moiety and protein kinase A anchor protein 13 (AKAP13) 16412732_The spatiotemporal expression of Brx was altered in eutopic endometrium of women with endometriosis. 16469733_Brx modifies the actions of glucocorticoids, enhancing the transcriptional activity of glucocorticoid receptor (GR) by interacting with GR and by attracting Rho family G proteins to the GR-induced transcriptisome 16956908_Observational study of gene-disease association. (HuGE Navigator) 17878165_AKAP13 plays a role in TLR2-mediated NF-kappaB activation; GEF-containing scaffold proteins may confer specificity to innate immune responses downstream of TLRs 18093280_Observational study of gene-disease association. (HuGE Navigator) 18445682_findings show the BNIP2 & BCH domain of BNIPXL interacts with specific conformers of RhoA & mediates association with catalytic DH-PH domains of Lbc, a RhoA-specific guanine nucleotide exchange factor; BNIPXL inhibits Lbc-induced oncogenic transformation 18519826_Clinical trial and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19696020_Data suggest that LC3 binding maintains AKAP-Lbc in an inactive state that displays a reduced ability to promote downstream signaling. 19779964_The positive expression rate of AKAP13 protein in colorectal carcinoma (52.3%) was significantly higher than those in adenoma (9.1%) and normal tissue (34.7%) (P = 0.006) by immunohistochemical staining. 19888694_backbone and side chain (1)H, (13)C and (15)N resonance assignments of a 20 kDa construct comprising the uniformly (13)C and( 15)N labeled AKAP13-PH domain and an associated helix from the DH domain which is required for its stable expression 19911011_Observational study of gene-disease association. (HuGE Navigator) 19960345_Data show that the RASGRP1/APTX gene expression ratio was higher in the responder while the AKAP13 expression was higher in the non-responders. 20353833_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20696764_the Lbc/alpha-catulin axis participates in 5-HT-induced PASMC mitogenesis and RhoA/ROCK signaling, and may be an interventional target in diseases involving vascular smooth muscle remodeling. 20719862_third new locus (rs6496932), on 15q25.3 (beta = 0.13, P = 1.4 x 10(-8)), was within a wide linkage disequilibrium block extending into the 5' end of the AKAP13 gene, encoding a scaffold protein concerned with signal transduction from the cell surface 21102438_Study demonstrates that the A-kinase-anchoring protein AKAP-Lbc and the scaffolding protein kinase suppressor of Ras (KSR-1) form the core of a signalling network that efficiently relay signals from RAF, through MEK, and on to ERK1/2. 21224381_A-kinase anchoring protein (AKAP)-Lbc anchors a PKN-based signaling complex involved in alpha1-adrenergic receptor-induced p38 activation. 21228793_One SNP (rs11638762), in the GATA-3 binding site upstream of the AKAP13 gene, was significantly replicated in another cohort for systolic blood pressure 22161024_Amplification of AKAP-13 is associated with metastatic and aggressive papillary thyroid carcinomas. 22731613_Thus AKAP-Lbc may serve an ancillary cardioprotective role by favouring the association of PKA with Hsp20. 23045525_Shp2 is a component of the AKAP-Lbc complex and is inhibited by protein kinase A under pathological hypertrophic conditions in the heart. 23090968_activation of IKKbeta within the AKAP-Lbc complex promotes NF-kappaB-dependent production of interleukin-6 24993829_pleckstrin homology (PH) domain of Lbc is located at the C-terminal end of the protein and is shown here to specifically recognize activated RhoA rather than lipids 25186459_Isothermal titration calorimetry showed that AKAP-Lbc has only micromolar affinity for RhoA, which combined with the presence of potential binding pockets for small molecules on AKAP-Lbc, raises the possibility of targeting AKAP-Lbc with GEF inhibitors. 26272591_Studied molecular interactions involving anchoring protein AKAP13 in the process of PKA-induced tamoxifen resistance in breast cancer specimens and cell lines. 26617690_evaluation of MAGT1 and AKAP13 expression in clinical hepatocellular carcinoma tissues by immunohistochemistry suggested that both proteins were strongly expressed in tumor tissues with significantly higher average immunoreactive scores of Remmele and Stegner (IRS) than in non-tumor tissues 28923249_AKAP-Lbc emerges as a coordinator of signals that protect cardiomyocytes against the toxic effects of DOX. 29066090_We showed that AKAP13 is expressed in the alveolar epithelium and lymphoid follicles from patients with Idiopathic pulmonary fibrosis, and AKAP13 mRNA expression was 1.42-times higher in lung tissue from patients with Idiopathic pulmonary fibrosis than that in lung tissue from controls. 30239831_Study of fibroid samples from patients and immortalized uterine fibroid cell lines and COS-7 cells suggest an intersection of mechanical signaling and progesterone receptor signaling involving AKAP13 through ERK. 31469403_Identification of novel splicing patterns and differential gene expression in RE+/FECD- samples provides new insights and more relevant gene targets that may be protective against FECD disease in vulnerable patients with TCF4 CTG TNR expansions. 32208871_Prognostic value of AKAP13 methylation and expression in lung squamous cell carcinoma. 33406263_CD47 promotes T-cell lymphoma metastasis by up-regulating AKAP13-mediated RhoA activation. 34673774_AKAP13 couples GPCR signaling to mTORC1 inhibition. | 1363.43380 | 0.9996714 | -0.0004741156 | 0.11798083 | 1.613253e-05 | 9.967953e-01 | 9.986332e-01 | No | Yes | 1424.811022 | 139.449223 | 1363.538926 | 130.477392 | |||
| ENSG00000171067 | 53838 | C11orf24 | protein_coding | Q96F05 | Cell membrane;Glycoprotein;Golgi apparatus;Membrane;Reference proteome;Signal;Transmembrane;Transmembrane helix | hsa:53838; | Golgi apparatus [GO:0005794]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886] | ENSMUSG00000035372 | 1810055G02Rik | 1161.63142 | 0.8743016 | -0.1937971296 | 0.11718297 | 2.720620e+00 | 9.905950e-02 | 4.508724e-01 | No | Yes | 1251.670273 | 107.752977 | 1380.802081 | 115.678680 | ||||
| ENSG00000171208 | 81831 | NETO2 | protein_coding | Q8NC67 | FUNCTION: Accessory subunit of neuronal kainate-sensitive glutamate receptors, GRIK2 and GRIK3. Increases kainate-receptor channel activity, slowing the decay kinetics of the receptors, without affecting their expression at the cell surface, and increasing the open probability of the receptor channels. Modulates the agonist sensitivity of kainate receptors. Slows the decay of kainate receptor-mediated excitatory postsynaptic currents (EPSCs), thus directly influencing synaptic transmission (By similarity). {ECO:0000250}. | Alternative splicing;Direct protein sequencing;Disulfide bond;Glycoprotein;Membrane;Phosphoprotein;Receptor;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene encodes a predicted transmembrane protein containing two extracellular CUB domains followed by a low-density lipoprotein class A (LDLa) domain. A similar gene in rats encodes a protein that modulates glutamate signaling in the brain by regulating kainate receptor function. Expression of this gene may be a biomarker for proliferating infantile hemangiomas. A pseudogene of this gene is located on the long arm of chromosome 8. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Jan 2011]. | hsa:81831; | integral component of membrane [GO:0016021]; postsynaptic density [GO:0014069]; ionotropic glutamate receptor binding [GO:0035255]; regulation of kainate selective glutamate receptor activity [GO:2000312] | 15340161_Determination of the neuropilin and tolloid-like protein 2 signal peptide cleavage site by N-terminal sequencing. 19217376_Rat neto2 plays a role in glutamate signaling in the brain by modulating the function of kainate receptors. 26277340_Results suggested that the extracellular N-terminal region including the two CUB domains was largely responsible for the distinct regulatory effects of Neto1 and Neto2 on the desensitization properties of GluK1 homomeric receptors 26699544_NETO2 upregulation could serve as a potential biomarker for the prediction of advanced tumor progression and unfavorable prognosis in patients with colorectal carcinoma. 29297384_NETO2 may be involved in colorectal cancer progression, but is not directly associated with epithelial-mesenchymal transition 29989576_upregulation of NETO2 gene is first stipulated by the isoform 1 (NM_018092.4), and the probable mechanism of its activation is associated with the increased expression of SAP30 transcription factor. 30770791_NETO2 promotes invasion and metastasis of gastric cancer cells and represents a novel prognostic indicator as well as a potential therapeutic target in gastric cancer. 30975469_NETO2 expression is up-regulated in Nasopharyngeal carcinoma (NPC) clinical specimens and cell lines. NETO2 down-regulation reduces proliferation, migration and invasion in irradiated NPC cell lines. 33157203_Neuropilin and tolloid-like 2 regulates the progression of osteosarcoma. 33390848_NETO2 promotes esophageal cancer progression by inducing proliferation and metastasis via PI3K/AKT and ERK pathway. 34552241_Kainate receptor modulation by NETO2. | ENSMUSG00000036902 | Neto2 | 2323.44608 | 1.0175021 | 0.0250318263 | 0.08532442 | 8.615053e-02 | 7.691292e-01 | 9.338569e-01 | No | Yes | 3092.086374 | 488.309251 | 2857.323187 | 441.223672 | |
| ENSG00000171604 | 51523 | CXXC5 | protein_coding | Q7LFL8 | FUNCTION: May indirectly participate in activation of the NF-kappa-B and MAPK pathways. Acts as a mediator of BMP4-mediated modulation of canonical Wnt signaling activity in neural stem cells (By similarity). Required for DNA damage-induced ATM phosphorylation, p53 activation and cell cycle arrest. Involved in myelopoiesis. Transcription factor. Binds to the oxygen responsive element of COX4I2 and represses its transcription under hypoxia conditions (4% oxygen), as well as normoxia conditions (20% oxygen) (PubMed:23303788). May repress COX4I2 transactivation induced by CHCHD2 and RBPJ (PubMed:23303788). Binds preferentially to DNA containing cytidine-phosphate-guanosine (CpG) dinucleotides over CpH (H=A, T, and C), hemimethylated-CpG and hemimethylated-hydroxymethyl-CpG (PubMed:29276034). {ECO:0000250|UniProtKB:Q5XIQ3, ECO:0000269|PubMed:19182210, ECO:0000269|PubMed:19557330, ECO:0000269|PubMed:23303788, ECO:0000269|PubMed:29276034}. | 3D-structure;Alternative splicing;Cytoplasm;DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Zinc;Zinc-finger | The protein encoded by this gene is a retinoid-inducible nuclear protein containing a CXXC-type zinc finger motif. The encoded protein is involved in myelopoiesis, is required for DNA damage-induced p53 activation, regulates the differentiation of C2C12 myoblasts into myocytes, and negatively regulates cutaneous wound healing. Several transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Nov 2015]. | hsa:51523; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor binding [GO:0140297]; methyl-CpG binding [GO:0008327]; sequence-specific DNA binding [GO:0043565]; zinc ion binding [GO:0008270]; negative regulation of transcription by RNA polymerase II [GO:0000122]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123] | 19182210_RINF expression correlates with retinoid-induced differentiation of leukemic cells and with cytokine-induced myelopoiesis of normal CD34(+) progenitors. 19557330_These findings suggest that CF5 plays a crucial role in ATM-p53 signaling in response to DNA damage. 19865112_Observational study of gene-disease association. (HuGE Navigator) 21325450_RINF overexpression represents an independent molecular marker for poor prognosis in breast tumors. 23190153_Lack of promoter hypermethylation or somatic mutations are found in the potential tumor suppressor CXXC5 in myelodysplastic syndromes or acute myeloid leukemia with deletion 5q. 23988457_targeting of CXXC5/RINF should be considered as a possible therapeutic strategy, especially in high-risk patients who show increased expression in AML cells compared with normal hematopoietic cells 25433557_Findings suggest that overexpression of CXXC5 in C2C12 myoblasts facilitated myocyte differentiation, while RNAi interference of CXXC5 significantly inhibited the differentiation of C2C12 myoblasts. 25605239_High CXXC5 expression seems to affect several steps in human leukemogenesis, including intracellular events as well as extracellular communication. 25805812_Low CXXC5 expression is associated with acute myeloid leukemia. 27886276_show here that CXXC5 is an E2-ERalpha responsive gene regulated by the interaction of E2-ERalpha with an ERE present at a region upstream of the initial translation codon of the gene 28067315_KANK1 inhibits Malignant peripheral nerve sheath tumors cell growth though CXXC5 mediated apoptosis. 29036306_Knockdown of CXXC5 attenuated the expression of a substantial portion of TGF-beta target genes and ameliorated TGF-beta-induced growth inhibition or apoptosis of Hep3B cells, suggesting that CXXC5 is required for TGF-beta-mediated inhibition of hepatocellular carcinoma (HCC) progression. 29780166_NUDT21 inhibits HCC proliferation, metastasis and tumorigenesis, at least in part, by suppressing PSMB2 and CXXC5. 30362164_CXXC5 is direct target gene of miR-32 which binds its 3'-UTR and inhibited the expression level of CXXC5 mRNA and protein. 30479059_CXXC5 is a a novel regulator and coordinator of TGF-beta, BMP4 and Wnt3a signaling. (Review) 30971423_These results reveal an important role for CXXC5 as a suppressor of longitudinal bone growth involving growth plate activity. 33241676_The epigenetic regulator RINF (CXXC5) maintains SMAD7 expression in human immature erythroid cells and sustains red blood cells expansion. 34475492_A prelude to the proximity interaction mapping of CXXC5. 35384342_Inhibition of CXXC5 function reverses obesity-related metabolic diseases. | ENSMUSG00000046668 | Cxxc5 | 240.45333 | 1.0134409 | 0.0192620088 | 0.21478149 | 8.027724e-03 | 9.286070e-01 | 9.797486e-01 | No | Yes | 305.517756 | 38.392491 | 296.268915 | 36.281490 | |
| ENSG00000172239 | 10605 | PAIP1 | protein_coding | Q9H074 | FUNCTION: Acts as a coactivator in the regulation of translation initiation of poly(A)-containing mRNAs. Its stimulatory activity on translation is mediated via its action on PABPC1. Competes with PAIP2 for binding to PABPC1. Its association with EIF4A and PABPC1 may potentiate contacts between mRNA termini. May also be involved in translationally coupled mRNA turnover. Implicated with other RNA-binding proteins in the cytoplasmic deadenylation/translational and decay interplay of the FOS mRNA mediated by the major coding-region determinant of instability (mCRD) domain. {ECO:0000269|PubMed:11051545, ECO:0000269|PubMed:9548260}.; FUNCTION: (Microbial infection) Upon interaction with SARS coronavirus SARS-CoV NSP3 protein, plays an important role in viral protein synthesis. {ECO:0000269|PubMed:33876849}. | 3D-structure;Acetylation;Alternative splicing;Cytoplasm;Host-virus interaction;Methylation;Reference proteome;Translation regulation | The protein encoded by this gene interacts with poly(A)-binding protein and with the cap-binding complex eIF4A. It is involved in translational initiation and protein biosynthesis. Overexpression of this gene in COS7 cells stimulates translation. Alternative splicing occurs at this locus and three transcript variants encoding three distinct isoforms have been identified. [provided by RefSeq, Jul 2008]. | hsa:10605; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; mCRD-mediated mRNA stability complex [GO:0106002]; RNA binding [GO:0003723]; translation activator activity [GO:0008494]; CRD-mediated mRNA stabilization [GO:0070934]; mRNA stabilization [GO:0048255]; negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [GO:1900152]; positive regulation of cytoplasmic translation [GO:2000767]; regulation of translational initiation [GO:0006446]; translational initiation [GO:0006413] | 11997512_Paip1 interacts with poly(A) binding protein through two independent binding motifs. 18725400_eIF3-Paip1 stabilizes the interaction between PABP and eIF4G, which brings about the circularization of the mRNA. 19851022_Paip1 is translational activator in 5' cap-dependent translation by interacting with PABP & initiation factors eIF4A & eIF3. Crystallization & preliminary diffraction analysis of middle domain of Paip1 gives crystals that diffract to resolution of 2.2 A. 21539810_the crystal structure of the middle domain of Paip1 isoform 2 (Paip1M) as determined by single-wavelength anomalous dispersion phasing is reported. 24396066_The Paip1-eIF3 interaction is impaired by the mTORC1 inhibitors. 25266661_Data provide evidence that the stability of Paip1 can be regulated by ubiquitin-mediated degradation, thus highlighting the importance of WWP2 as a suppressor of translation. 29258905_our study demonstrated that Paip1 promotes the growth of breast cancers and could be a prognostic biomarker and therapeutic target 29358694_Results showed that mRNA and protein expression of PAIP1, a miRNA-340 target gene, were down-regulated in women with gestational diabetes. 30731074_Paip1 contributes to Pancreatic cancer progression and appears to be a valid prognostic factor of PC 30992367_These results indicated that PAIP1 and PAIP2 participate in translation termination and are important regulators of readthrough at the premature termination codon. 31010277_Paip1 was overexpressed in cervical cancer (CC) samples and associated with the adverse outcomes in patients with CC. Its knockdown induced inhibition of cell growth, G2/M arrest and apoptosis in CC cells. 32871777_High Paip1 Expression as a Potential Prognostic Marker in Hepatocellular Carcinoma. 33876849_The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. 35153296_Effect of PAIP1 on the metastatic potential and prognostic significance in oral squamous cell carcinoma. | ENSMUSG00000025451 | Paip1 | 2185.33888 | 0.9937938 | -0.0089816160 | 0.09146037 | 9.632431e-03 | 9.218172e-01 | 9.779533e-01 | No | Yes | 2927.487567 | 589.492697 | 2771.938421 | 545.805844 | |
| ENSG00000172244 | 375444 | C5orf34 | protein_coding | Q96MH7 | Reference proteome | hsa:375444; | 30771479_Study results firstly revealed that C5orf34 might play a facilitating role in lung adenocarcinoma (LAD) development and progression by regulating MAPK signaling pathway. Furthermore, data implied that C5orf34 may be a potential predictor and treatment target for LAD. | ENSMUSG00000062822 | 4833420G17Rik | 480.23665 | 0.8165904 | -0.2923155127 | 0.14174412 | 4.223282e+00 | 3.987295e-02 | 3.079236e-01 | No | Yes | 498.734383 | 115.104356 | 580.174781 | 130.894315 | ||||
| ENSG00000172530 | 54971 | BANP | protein_coding | Q8N9N5 | FUNCTION: Controls V(D)J recombination during T-cell development by repressing T-cell receptor (TCR) beta enhancer function. Binds to scaffold/matrix attachment region beta (S/MARbeta), an ATC-rich DNA sequence located upstream of the TCR beta enhancer. Represses cyclin D1 transcription by recruiting HDAC1 to its promoter, thereby diminishing H3K9ac, H3S10ph and H4K8ac levels. Promotes TP53 activation, which causes cell cycle arrest (By similarity). {ECO:0000250, ECO:0000250|UniProtKB:Q8VBU8, ECO:0000269|PubMed:16166625}. | Acetylation;Alternative splicing;Cell cycle;Chromatin regulator;Coiled coil;DNA-binding;Developmental protein;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Tumor suppressor;Ubl conjugation | This gene encodes a protein that binds to matrix attachment regions. The protein forms a complex with p53 and negatively regulates p53 transcription, and functions as a tumor suppressor and cell cycle regulator. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2010]. | hsa:54971; | nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; cell cycle [GO:0007049]; chromatin organization [GO:0006325]; negative regulation of protein catabolic process [GO:0042177]; protein localization to nucleus [GO:0034504] | 10950932_Identification of a novel mouse MAR-binding protein, named SMAR1, which shares homology with SATB1 and Cux in the MAR-binding domain/Cut repeat and also with the tetramerization domain of a B cell-specific MAR-binding protein, Bright. 15623522_SMAR1 plays an important role in the regulation of T cell development as well as V(D)J recombination 16166625_SMAR1 regulates cyclin D1 by modification of chromatin through the SIN3/histone deacetylase 1 complex 17668048_SMAR1 plays a central role in coordinating p53 and TGFbeta pathways in human breast cancer. 18822384_tumor suppressor SMAR1 downregulates Cytokeratin 8 gene expression by modulating p53-mediated transactivation of this gene 18981184_tumor suppressor protein SMAR1 can modulate NF-kappaB transactivation and inhibit tumorigenesis by regulating NF-kappaB target genes 19303885_Results highlight the role of SMAR1 in masking the active phosphorylation site of p53, enabling the deacetylation of p53 by HDAC1-MDM2 complex, thereby regulating the p53 transcriptional response during stress rescue. 19799771_Human breast cancer tissue sections that showed reduced SMAR1 expression exhibited increased surface roughness. 20075864_On mild DNA damage, SMAR1 selectively represses BAX and PUMA through binding to the MAR independently of inducing p53 deacetylation through HDAC1. 20097305_A novel mechanism of regulation of oxidative stress by ATM through modulation of SMAR1-AKR1a4 complex, is proposed. 20153327_report Prostaglandin A2 (PGA2) induced binding of HSP70 to a novel site on phi1 SMAR1 5' UTR which stabilizes the wild type transcript and leads to subsequent increase in SMAR1 protein levels. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20709157_multiple roles of nuclear matrix associated protein SMAR1 in regulating various cellular target genes involved in cell growth, apoptosis and tumorigenesis.(REVIEW) 22074660_Results indicate that SMAR1 is an important player in p300-p53 regulated DNA damage signalling pathway and can exert its effect on apoptosis in a transcription independent manner. 22674979_TCF-4, beta-catenin, and SMAR1 tether at the -143-nucleotide site on the HIV LTR to inhibit HIV promoter activity. 23876508_During hemin-induced erythroid differentiation, enhanced expression of SMAR1 negatively correlates with miR-320a expression. 25086032_indicate a crucial role for SMAR1 in restraining breast cancer cell migration and suggest the candidature of this scaffold matrix-associated region-binding protein as a tumor suppressor. 25157104_SMAR1 has a role in repressing c-Fos-mediated HPV18 E6 transcription through alteration of chromatin histone deacetylation 25239884_Data indicate the role of SMAR1 protein in NF-kappaappa B dependent transcriptional regulation of pro-angiogenic chemokine interleukin-8 (IL-8). 25299772_SMAR1-mediated regulation of repair and apoptosis via a complex crosstalk involving Ku70, HDAC6 and Bax. 26080397_results reveal the complex molecular mechanism underlying SMAR1-mediated signal-dependent and -independent regulation of alternative splicing via Sam68 deacetylation 28103507_Dysregulated circ-BANP appears to have an important role in colorectal cancer cells. 28617439_Cdc20 functions as an important negative regulator of SMAR1 in higher grades of cancer 31422285_our observations establish that increased expression of SMAR1 in cancers can positively regulate MHC I surface expression thereby leading to higher chances of tumor regression and elimination of cancer cells. 33082288_SMAR1 repression by pluripotency factors and consequent chemoresistance in breast cancer stem-like cells is reversed by aspirin. 34234345_BANP opens chromatin and activates CpG-island-regulated genes. | ENSMUSG00000025316 | Banp | 515.35869 | 0.9865210 | -0.0195783491 | 0.13916522 | 1.977480e-02 | 8.881679e-01 | 9.701360e-01 | No | Yes | 628.232182 | 51.735517 | 617.656284 | 49.907610 | |
| ENSG00000172671 | 93550 | ZFAND4 | protein_coding | Q86XD8 | Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:93550; | zinc ion binding [GO:0008270] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 25682742_In gastric cancer cells, ANUBL1 was upregulated and promoted proliferation of SGC-7901 cells. Over-expression decreased miR-182 expression. ANUBL1 expression was in turn directly downregulated by miR-182 in a negative feedback loop. 29074611_Suggest that ZFAND4 is a useful marker for predicting metastasis and poor prognosis in patients with oral squamous cell carcinoma. | ENSMUSG00000042213 | Zfand4 | 340.21254 | 0.9663319 | -0.0494092431 | 0.17037736 | 8.389509e-02 | 7.720867e-01 | 9.348455e-01 | No | Yes | 401.411154 | 61.430003 | 399.404237 | 59.856834 | |||
| ENSG00000172828 | 23491 | CES3 | protein_coding | Q6UWW8 | FUNCTION: Involved in the detoxification of xenobiotics and in the activation of ester and amide prodrugs. Shows low catalytic efficiency for hydrolysis of CPT-11 (7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin), a prodrug for camptothecin used in cancer therapeutics. | Alternative splicing;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Hydrolase;Reference proteome;Serine esterase;Signal | Mouse_homologues NA; + ;NA | This gene encodes a member of the carboxylesterase large family. The family members are responsible for the hydrolysis or transesterification of various xenobiotics, such as cocaine and heroin, and endogenous substrates with ester, thioester, or amide bonds. They may participate in fatty acyl and cholesterol ester metabolism, and may play a role in the blood-brain barrier system. This gene is expressed in several tissues, particularly in colon, trachea and in brain, and the protein participates in colon and neural drug metabolism. Multiple alternatively spliced transcript variants encoding distinct isoforms have been reported, but the biological validity and/or full-length nature of some variants have not been determined.[provided by RefSeq, Jun 2010]. | hsa:23491; | cytosol [GO:0005829]; endoplasmic reticulum lumen [GO:0005788]; extracellular exosome [GO:0070062]; carboxylic ester hydrolase activity [GO:0052689]; methyl indole-3-acetate esterase activity [GO:0080030]; low-density lipoprotein particle clearance [GO:0034383]; xenobiotic metabolic process [GO:0006805] | 18794728_pharmacogenomic characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes 19508181_CES3 seems to have a lower catalytic efficiency than the other two CESs for selected substrates. 20422440_Studies indicate that CES3 mRNA isoform expression in several tissues, particularly in colon, trachea and in brain. 20529763_Observational study of gene-disease association. (HuGE Navigator) 22700792_This study provides the first evidence of functional compensation whereby increased expression of CES3 restores intracellular cholesteryl ester hydrolytic activity and free cholesterol efflux in CES1-deficient cells. | ENSMUSG00000062181+ENSMUSG00000069922 | Ces3b+Ces3a | 446.04537 | 1.1577673 | 0.2113453109 | 0.17219511 | 1.513923e+00 | 2.185414e-01 | 6.100145e-01 | No | Yes | 579.321453 | 99.081663 | 512.331896 | 85.579140 |
| ENSG00000172986 | 727936 | GXYLT2 | protein_coding | A0PJZ3 | FUNCTION: Glycosyltransferase which elongates the O-linked glucose attached to EGF-like repeats in the extracellular domain of Notch proteins by catalyzing the addition of xylose. {ECO:0000269|PubMed:19940119}. | Glycoprotein;Glycosyltransferase;Membrane;Reference proteome;Signal-anchor;Transferase;Transmembrane;Transmembrane helix | The protein encoded by this gene is a xylosyltransferase that elongates O-linked glucose bound to epidermal growth factor (EGF) repeats. The encoded protein catalyzes the addition of xylose to the O-glucose-modified residues of EGF repeats of Notch proteins. [provided by RefSeq, Sep 2016]. | hsa:727936; | integral component of membrane [GO:0016021]; UDP-D-xylose:beta-D-glucoside alpha-1,3-D-xylosyltransferase activity [GO:0140563]; UDP-xylosyltransferase activity [GO:0035252]; O-glycan processing [GO:0016266] | 19940119_We have identified two enzymes of the human glycosyltransferase 8 family, now named GXYLT1 and GXYLT2 (glucoside xylosyltransferase), as UDP-d-xylose:beta-d-glucoside alpha1,3-d-xylosyltransferases adding the first xylose. 30716301_our findings indicated that GXYLT2 plays an important role in cell activities via regulation of the Notch signaling 34506251_Glucoside xylosyltransferase 2 as a diagnostic and prognostic marker in gastric cancer via comprehensive analysis. | ENSMUSG00000030074 | Gxylt2 | 563.96466 | 1.3079110 | 0.3872643260 | 0.15042056 | 6.677396e+00 | 9.764315e-03 | 1.625315e-01 | No | Yes | 767.958848 | 89.344148 | 565.020945 | 64.459786 | |
| ENSG00000173230 | 2804 | GOLGB1 | protein_coding | Q14789 | FUNCTION: May participate in forming intercisternal cross-bridges of the Golgi complex. | Acetylation;Alternative splicing;Coiled coil;Disulfide bond;Golgi apparatus;Membrane;Phosphoprotein;Reference proteome;Transmembrane;Transmembrane helix | hsa:2804; | cis-Golgi network [GO:0005801]; cytosol [GO:0005829]; endoplasmic reticulum-Golgi intermediate compartment [GO:0005793]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; Golgi stack [GO:0005795]; integral component of membrane [GO:0016021]; membrane [GO:0016020]; RNA binding [GO:0003723]; sequence-specific DNA binding [GO:0043565]; chondrocyte proliferation [GO:0035988]; Golgi organization [GO:0007030]; protein localization to pericentriolar material [GO:1905793]; regulation of transcription, DNA-templated [GO:0006355] | 12429822_Data report the characterization of a mammalian coiled-coil protein, CASP, a Golgi protein that shares with giantin a conserved histidine in its transmembrane domain. 17475246_Giantin binds to Rab6A and Rab1 proteins. 23422753_unbiased whole-genome search for genetic modifiers of stroke risk in sickle cell anemia was performed; mutation in GOLGB1 (Y1212C) and mutation in ENPP1 (K173Q) were confirmed as having significant associations with a decreased risk for stroke 23555793_the spatial organization of the Golgi ribbon is mediated by giantin, which also plays a role in cargo transport and sugar modifications 24046448_Partial depletion of giantin or of WDR34 leads to an increase in cilia length consistent with the concept that giantin acts through dynein-2. 25086069_Results show that ST3Gal1 uses GM130-GRASP65 and giantin, whereas C2GnT-L uses only giantin for Golgi targeting and defective giantin dimerization in PC-3 and DU145 prostate cancer cells causes fragmentation of the Golgi and prevents its targeting. 26664786_Single nucleotide polymorphisms (SNPs) rs1035798 in RAGE gene, rs2073617 and rs2073618 in TNFRSF11B, and rs3732410 in Golgb1 will be investigated on whether there is an association with hemorrhagic stroke (HS) in Chinese population. 28782625_In situ proximity ligation assays of Golgi localization of alpha-mannosidase IA at giantin versus GM130-GRASP65 site, and absence or presence of N-glycans terminated with alpha3-mannose on trans-Golgi glycosyltransferases may be useful for distinguishing indolent from aggressive prostate cancer cells. 29430628_common polymorphisms of GOLGB1 are not associated with nonsyndromic cleft palate susceptibility in the Brazilian population 30453527_Ethanol-induced Golgi disassembly was associated with de-dimerization of the largest Golgi matrix protein giantin, along with impaired transport of selected hepatic proteins. | ENSMUSG00000034243 | Golgb1 | 2683.10191 | 0.9400161 | -0.0892426244 | 0.07970100 | 1.250657e+00 | 2.634271e-01 | 6.541811e-01 | No | Yes | 2160.641064 | 338.515171 | 2241.572499 | 343.476082 | ||
| ENSG00000173276 | 49854 | ZBTB21 | protein_coding | Q9ULJ3 | FUNCTION: Acts as a transcription repressor. {ECO:0000269|PubMed:15629158}. | 3D-structure;Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:49854; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; identical protein binding [GO:0042802]; metal ion binding [GO:0046872]; methyl-CpG binding [GO:0008327]; POZ domain binding [GO:0031208]; sequence-specific DNA binding [GO:0043565]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; regulation of transcription by RNA polymerase II [GO:0006357] | 15629158_ZNF295 may be involved in the bi-directional control of gene expression in concert with ZFP161 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 29158487_Genotyping was accomplished on Infinium Human610-QUAD version 1. In the ilSIRENTE population, genetic variants in ZNF295 and C2CD2 (rs928874 and rs1788355) on chromosome 21q22.3, were significantly associated with the 4-meter gait speed | ENSMUSG00000046962 | Zbtb21 | 1267.99165 | 0.8715422 | -0.1983575830 | 0.11015193 | 3.224400e+00 | 7.254837e-02 | 3.963199e-01 | No | Yes | 1437.880573 | 216.576938 | 1553.905157 | 229.122079 | ||
| ENSG00000173442 | 254102 | EHBP1L1 | protein_coding | Q8N3D4 | FUNCTION: May act as Rab effector protein and play a role in vesicle trafficking. {ECO:0000305|PubMed:27552051}. | Coiled coil;Endosome;Phosphoprotein;Reference proteome | hsa:254102; | endosome [GO:0005768]; filamentous actin [GO:0031941]; membrane [GO:0016020]; microtubule organizing center [GO:0005815]; actin cytoskeleton organization [GO:0030036] | 26833786_Data demonstrate that EHBP1L1 links Rab8 and the Bin1-dynamin complex, which generates membrane curvature and excises the vesicle at the endocytic recycling compartment for apical transport. | ENSMUSG00000024937 | Ehbp1l1 | 61.52900 | 2.2914131 | 1.1962375934 | 0.46028516 | 6.782808e+00 | 9.203996e-03 | No | Yes | 128.073569 | 44.646385 | 56.715341 | 19.325389 | |||
| ENSG00000173531 | 4485 | MST1 | protein_coding | H7C0F8 | Proteomics identification;Reference proteome | The protein encoded by this gene contains four kringle domains and a serine protease domain, similar to that found in hepatic growth factor. Despite the presence of the serine protease domain, the encoded protein may not have any proteolytic activity. The receptor for this protein is RON tyrosine kinase, which upon activation stimulates ciliary motility of ciliated epithelial lung cells. This protein is secreted and cleaved to form an alpha chain and a beta chain bridged by disulfide bonds. [provided by RefSeq, Jan 2010]. | Mouse_homologues mmu:15235; | serine-type endopeptidase activity [GO:0004252] | 12177064_These results suggest that activation of RON by macrophage-stimulating protein inhibits LPS-induced macrophage Cox-2 expression. 12919677_Data show that macrophage stimulating protein (MSP) and its receptor (Ron) induce phosphorylation of both Ron and alpha6beta4 integrin, and result in activation of alpha3beta1 integrin. 14597639_macrophage-stimulating protein-responsive cell growth in culture is suppressed by the ron-sema domain 15764806_Macrophage stimulating protein and its receptor RON are involved in the pathophysiology of endometriosis. 16170349_repression of MSP gene expression by mutant p53 may contribute to oncogenesis in a cell type-specific manner 17456594_overexpression of MSP, MT-SP1, and MST1R was a strong independent indicator of both metastasis and death in human breast cancer 18438406_Observational study of gene-disease association. (HuGE Navigator) 18480548_Cell migration and production of inflammatory cytokines by the brain are enhanced by MSP stimulation in primary microglia. 18700007_The five gene transcripts (aldolase B, elafin, MST-1, simNIPhom and SLC6A14) were changed in patients with ulcerative colitis, and were related to the disease activity. 19079170_Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. 19456860_Results suggest that hepatocyte growth factor activator is a major serum activator of pro-macrophage-stimulating protein. 19657358_Observational study of gene-disease association. (HuGE Navigator) 19657358_The effect of BSN-MST1 locus on Crohn's disease predisposition was replicated, but no influence on ulcerative colitis or multiple sclerosis predisposition could be detected 19760754_Observational study of gene-disease association. (HuGE Navigator) 19861958_Observational study of gene-disease association. (HuGE Navigator) 19944697_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20014019_Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20024904_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20307617_Observational study of gene-disease association. (HuGE Navigator) 21072187_Observational study of gene-disease association. (HuGE Navigator) 21081472_results suggest that MSP induces uPAR expression via MAPK, AP-1 and NF-kappaB signaling pathways 21249150_the oncogenic effect of BRAF(V600E) is associated with the inhibition of MST1 tumor suppressor pathways, and that the activity of RASSF1A-MST1-FoxO3 pathways determines the phenotypes of BRAF(V600E) tumors. 21619683_MSP-induced RSK2 activation is a critical determinant linking RON signaling to cellular EMT program. 21723047_considerable number of Merkel cell carcinoma cases expressed both RON and MSP, while Merkel cells do not express these molecules 21875933_These findings suggest that the MSP/RON signaling pathway may be regulated by hepsin in tissue homeostasis and in disease pathologies, such as in cancer and immune disorders. 21931648_Studies indicate that the cylindromatosis/turban tumor syndrome gene (CYLD) ranked highest, followed by acylaminoacyl-peptidase (APEH), dystroglycan (DAG1), macrophage-stimulating protein (MST1) and ubiquitin-specific peptidase 4 (USP4). 22087277_the missense SNP impairs MSP function by reducing its affinity to RON and perhaps through a secondary effect on in vivo concentration arising from reduced thermodynamic stability, resulting in down-regulation of the MSP/RON signaling pathway. 22245154_HAT cleaves proMSP at the physiological activation site 22554193_The present results refine the known genetic architecture in primary sclerosing cholangitis by confirming MST1 locus association. 23011677_MSP may be one of the major determinants that affects the properties of tumor stroma and that produces a permissive microenvironment to promote cancer metastasis 23422030_Results suggest that the [AA] genotype of the common MST1 variant rs3197999 enhances genetic risk of sporadic extrahepatic cholangiocarcinoma irrespective of primary sclerosing cholangitis status. 23928732_Mst1 has an important role in inhibiting the growth of NSCLC in vitro and in vivo; its antiproliferative effect is associated with induction of apoptosis. 24409221_functional consequences of the macrophage stimulating protein 689C inflammatory bowel disease risk allele 25193665_These results support the hypothesis that the alpha-chain of MSPalphabeta mediates RON dimerization. 25315688_MSP appears to promote the migration of fibroblasts, enhances collagen synthesis and remodeling, and effectively improves wound healing. 25551685_Identify MST1/MSP as a mitogen for tracheal basal cells. 25704570_Elevated serum levels of MST1 were found in subjects with excessive alcohol use. 27609031_These data suggest that MSP is an effective inhibitor of inflammation and lipid accumulation in the stressed liver, thereby indicating that MSP has a key regulatory role in non-alcoholic steatohepatitis. 29441677_Our analysis suggests that MST1 might interact with key susceptibility genes involved in autophagy and bacterial recognition. These findings provide insight into the genetic architecture of Crohn's disease in Chinese and may partially explain the disparity of genetic signals in Crohn's disease susceptibility across different ethnic populations by highlighting the contribution of gene-gene interactions. 30212651_Legionella pneumophila effector protein LegK7(lpg1924) hijacks the conserved Hippo signaling pathway by molecularly mimicking host Hippo kinase (MST1 in mammals), which is the key regulator of pathway activation. 30981108_Mst1 overexpression inhibits mitophagy activity via suppressing the ERK-Mfn2 pathway, ultimately augmenting IL-24-inducd esophageal cancer death via enhanced mitochondrial stress. 31254927_Comparative characterization of the HGF/Met and MSP/Ron systems in primary pancreatic adenocarcinoma. 32156191_Macrophage stimulating 1-induced inflammation response promotes aortic aneurysm formation through triggering endothelial cells death and activating the NF-kappaB signaling pathway. 32692720_Mst1 promotes mitochondrial dysfunction and apoptosis in oxidative stress-induced rheumatoid arthritis synoviocytes. 33070035_Macrophage-stimulating protein is decreased in severe preeclampsia and regulates the biological behavior of HTR-8/SVneo trophoblast cells. 33109096_circCRAMP1L is a novel biomarker of preeclampsia risk and may play a role in preeclampsia pathogenesis via regulation of the MSP/RON axis in trophoblasts. | ENSMUSG00000032591 | Mst1 | 88.66038 | 1.0729271 | 0.1015521215 | 0.36586001 | 7.633524e-02 | 7.823266e-01 | 9.388166e-01 | No | Yes | 60.646796 | 16.317901 | 58.330938 | 14.964575 | ||
| ENSG00000173614 | 64802 | NMNAT1 | protein_coding | Q9HAN9 | FUNCTION: Catalyzes the formation of NAD(+) from nicotinamide mononucleotide (NMN) and ATP (PubMed:17402747). Can also use the deamidated form; nicotinic acid mononucleotide (NaMN) as substrate with the same efficiency (PubMed:17402747). Can use triazofurin monophosphate (TrMP) as substrate (PubMed:17402747). Also catalyzes the reverse reaction, i.e. the pyrophosphorolytic cleavage of NAD(+) (PubMed:17402747). For the pyrophosphorolytic activity, prefers NAD(+) and NaAD as substrates and degrades NADH, nicotinic acid adenine dinucleotide phosphate (NHD) and nicotinamide guanine dinucleotide (NGD) less effectively (PubMed:17402747). Involved in the synthesis of ATP in the nucleus, together with PARP1, PARG and NUDT5 (PubMed:27257257). Nuclear ATP generation is required for extensive chromatin remodeling events that are energy-consuming (PubMed:27257257). Fails to cleave phosphorylated dinucleotides NADP(+), NADPH and NaADP(+) (PubMed:17402747). Protects against axonal degeneration following mechanical or toxic insults (By similarity). {ECO:0000250|UniProtKB:Q9EPA7, ECO:0000269|PubMed:17402747, ECO:0000269|PubMed:27257257}. | 3D-structure;ATP-binding;Deafness;Direct protein sequencing;Disease variant;Dwarfism;Leber congenital amaurosis;Magnesium;Mental retardation;NAD;Nucleotide-binding;Nucleotidyltransferase;Nucleus;Phosphoprotein;Pyridine nucleotide biosynthesis;Reference proteome;Transferase;Zinc | PATHWAY: Cofactor biosynthesis; NAD(+) biosynthesis; NAD(+) from nicotinamide D-ribonucleotide: step 1/1.; PATHWAY: Cofactor biosynthesis; NAD(+) biosynthesis; deamido-NAD(+) from nicotinate D-ribonucleotide: step 1/1. | This gene encodes an enzyme which catalyzes a key step in the biosynthesis of nicotinamide adenine dinucleotide (NAD). The encoded enzyme is one of several nicotinamide nucleotide adenylyltransferases, and is specifically localized to the cell nucleus. Activity of this protein leads to the activation of a nuclear deacetylase that functions in the protection of damaged neurons. Mutations in this gene have been associated with Leber congenital amaurosis 9. Alternative splicing results in multiple transcript variants. Pseudogenes of this gene are located on chromosomes 1, 3, 4, 14, and 15. [provided by RefSeq, Jul 2014]. | hsa:64802; | nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; ATP binding [GO:0005524]; identical protein binding [GO:0042802]; nicotinamide-nucleotide adenylyltransferase activity [GO:0000309]; nicotinate-nucleotide adenylyltransferase activity [GO:0004515]; ATP generation from poly-ADP-D-ribose [GO:1990966]; NAD biosynthetic process [GO:0009435]; negative regulation of neuron death [GO:1901215]; nucleotide biosynthetic process [GO:0009165]; response to wounding [GO:0009611] | 11751893_By using a combination of single isomorphous replacement and density modification techniques, the human NMNAT structure was solved by x-ray crystallography to a 2.5-A resolution, revealing a hexamer that is composed of alpha/beta-topology subunits 11788603_structure determination and role in activating tiazofurin 11959140_Crystal structure of human nicotinamide mononucleotide adenylyltransferase in complex with NMN. 12574164_structural characterization of this human cytosolic enzyme and implicatons in NAD biosyntheesis 16118205_NMNAT1 is a nuclear protein, whereas NMNAT2 and -3 are localized to the Golgi complex and the mitochondria 17360427_depending on its state of phosphorylation, NMNAT-1 binds to activated, automodifying PARP-1 and thereby amplifies poly(ADP-ribosyl)ation 17402747_ATP binds before NMN with nuclear isozyme NMNAT1. NMNH conversion to NADH by NMNAT1 and NMNAT3 occurs at similar rates, conversion by NMNAT2 is much slower. 19403820_Nicotinamide mononucleotide adenylyl transferase 1 (Nmnat1) is important for axonal protection, since mutants with reduced enzymatic activity lack axon protective activity. 19458223_Neuronal expression of exogenous Nmnat1 protein localized to the cytosol is essential and sufficient to delay Wallerian degeneration; cytosolic Nmnat1 showed greatly enhanced axon protection compared with native (nuclear) Nmnat1. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20388704_analysis of isoform-specific targeting and interaction domains in human nicotinamide mononucleotide adenylyltransferases 20457531_Red blood cells represent the first human cell type with a remarkable predominance of NMNAT3 over NMNAT1; NMNAT2 is absent. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20926655_Axonal targeting of transgenic NMNAT activity is both necessary and sufficient to delay Wallerian degeneration; promoting axonal and synaptic delivery greatly enhances NMNAT1 effectiveness. 20954240_Study investigated the importance of NMNAT2's central domain, which are postulated to be dispensable for catalytic activity, instead representing an isozyme-specific control domain within the overall architecture of NMNAT2. 21071441_nicotinamide mononucleotide adenylyltransferase (Nmnat) protein transduction into transected axons blocks axonal degeneration 22334709_Our studies link the enzymatic activities of NMNAT-1 and PARP-1 to the regulation of a set of common target genes through functional interactions at target gene promoters. 22842229_Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. 22842230_A new disease mechanism underlying Leber congential amaurosis and the first link between endogenous NMNAT1 dysfunction and a human nervous system disorder. 23351689_mutations in nicotinamide nucleotide adenylyltransferase 1(NMNAT1) cause Leber congenital amaurosis 23737528_NMNAT1 deletion in tumors may contribute to transformation by increasing ribosomal RNA synthesis. 23940504_Data found pathogenic DNA variants in the genes RP1, USH2A, CNGB3, NMNAT1, CHM, and ABCA4, responsible for retinitis pigmentosa, Usher syndrome, achromatopsia, Leber congenital amaurosis, choroideremia, or recessive Stargardt/cone-rod dystrophy cases. 24830548_theNMNAT1 p.Glu257Lys variant is a hypomorphic variant that almost without exception causes leber congenital amaurosis (LCA) in combination with more severe NMNAT1 variants. 24940029_The aim of this study was to determine the occurrence and frequency of NMNAT1 mutations and associated phenotypes in different types of inherited retinal dystrophies. 25988908_NMNAT1, which encodes the nicotinamide mononucleotide adenylyltransferase 1, has been recently identified to be one of the LCA-causing genes. Our results expanded the spectrum of mutations in NMNAT1 26018082_To study how mutations affect NMNAT1 function and ultimately lead to a retinal degeneration phenotype, we performed detailed analysis of Leber congenital amaurosis 9-associated NMNAT1 mutants. 26316326_Hidden Genetic Variation in LCA9-Associated Congenital Blindness Explained by 5'UTR Mutations and Copy-Number Variations of NMNAT1. 26464178_We confirmed a diagnosis of NMNAT1-associated Leber congenital amaurosis in two siblings through identification of the mutation (c.25G>A [p. Val9Met]) in a homozygous state. 28369829_Results associate a distinct retinal dystrophy phenotype with nicotinamide-nucleotide adenylyltransferase 1 protein (NMNAT1) mutation and suggest coiled-coil domain containing 66 (CCDC66) should not be considered a retinal dystrophy candidate gene. 29184169_Rare homozygous variant c.[271G > A] p.(Glu91Lys) and compound heterozygous variants c.[53 A > G];[769G > A] p.(Asn18Ser);(Glu257Lys) were identified in two cases of cone-rod dystrophy, respectively. 30004997_A NOVEL CASE SERIES OF NMNAT1-ASSOCIATED EARLY-ONSET RETINAL DYSTROPHY: EXTENDING THE PHENOTYPIC SPECTRUM. 32533184_An Alu-mediated duplication in NMNAT1, involved in NAD biosynthesis, causes a novel syndrome, SHILCA, affecting multiple tissues and organs. 34243968_Coats-like Exudative Vitreoretinopathy in NMNAT1 Leber Congenital Amaurosis. 34290089_Nuclear NAD(+) homeostasis governed by NMNAT1 prevents apoptosis of acute myeloid leukemia stem cells. | ENSMUSG00000028992 | Nmnat1 | 527.06653 | 1.0307093 | 0.0436375164 | 0.13732001 | 1.010923e-01 | 7.505227e-01 | 9.268992e-01 | No | Yes | 673.053491 | 90.350951 | 604.515309 | 79.268303 |
| ENSG00000174197 | 23269 | MGA | protein_coding | Q8IWI9 | FUNCTION: Functions as a dual-specificity transcription factor, regulating the expression of both MAX-network and T-box family target genes. Functions as a repressor or an activator. Binds to 5'-AATTTCACACCTAGGTGTGAAATT-3' core sequence and seems to regulate MYC-MAX target genes. Suppresses transcriptional activation by MYC and inhibits MYC-dependent cell transformation. Function activated by heterodimerization with MAX. This heterodimerization serves the dual function of both generating an E-box-binding heterodimer and simultaneously blocking interaction of a corepressor (By similarity). {ECO:0000250}. | Alternative splicing;Coiled coil;DNA-binding;Isopeptide bond;Methylation;Nucleus;Phosphoprotein;Reference proteome;Repressor;Transcription;Transcription regulation;Ubl conjugation | hsa:23269; | chromatin [GO:0000785]; MLL1 complex [GO:0071339]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; protein dimerization activity [GO:0046983]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; cell fate specification [GO:0001708]; regulation of transcription by RNA polymerase II [GO:0006357] | 24131248_TLR3 and MDA5 signalling, although not expression, is impaired in asthmatic epithelial cells in response to rhinovirus infection. 24362264_Results show that inactivation of MGA was observed in both non-small cell lung cancer and SCLC. 26272389_In conclusion, our study generated five mAbs against MGA and identified the best candidate for detection of MGA expression in breast cancer tissues. 27306572_we suggest that MGA loss-of-function mutations are present in colorectal cancers 29525183_this study shows that LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways 31862696_Multi-Omics Analysis Identifies MGA as a Negative Regulator of the MYC Pathway in Lung Adenocarcinoma. | ENSMUSG00000033943 | Mga | 1968.77211 | 0.8971980 | -0.1565016999 | 0.10834980 | 2.076656e+00 | 1.495676e-01 | 5.379673e-01 | No | Yes | 1953.869552 | 401.787878 | 1996.188785 | 401.313980 | ||
| ENSG00000174442 | 55055 | ZWILCH | protein_coding | Q9H900 | FUNCTION: Essential component of the mitotic checkpoint, which prevents cells from prematurely exiting mitosis. Required for the assembly of the dynein-dynactin and MAD1-MAD2 complexes onto kinetochores. Its function related to the spindle assembly machinery is proposed to depend on its association in the mitotic RZZ complex (PubMed:15824131). {ECO:0000269|PubMed:15824131}. | 3D-structure;Alternative splicing;Cell cycle;Cell division;Centromere;Chromosome;Kinetochore;Mitosis;Phosphoprotein;Reference proteome | hsa:55055; | cytosol [GO:0005829]; kinetochore [GO:0000776]; RZZ complex [GO:1990423]; cell division [GO:0051301]; mitotic spindle assembly checkpoint signaling [GO:0007094]; protein localization to kinetochore [GO:0034501] | 12686595_Other name: HZwilch. Homologue of Drosophila Zwilch (CG18729). Localizes to the kinetochore during prometaphase and metaphase of HeLa cells. Complexed with human ROD and human ZW10, similar to situation in Drosophila. 19008095_Observational study of gene-disease association. (HuGE Navigator) 20800603_Observational study of gene-disease association. (HuGE Navigator) | ENSMUSG00000032400 | Zwilch | 1924.57783 | 0.8792871 | -0.1855938661 | 0.09410192 | 3.867578e+00 | 4.922753e-02 | 3.357729e-01 | No | Yes | 2348.203327 | 363.888699 | 2573.392868 | 389.986118 | ||
| ENSG00000174607 | 7368 | UGT8 | protein_coding | Q16880 | FUNCTION: Catalyzes the transfer of galactose to ceramide, a key enzymatic step in the biosynthesis of galactocerebrosides, which are abundant sphingolipids of the myelin membrane of the central nervous system and peripheral nervous system (PubMed:9125199). Galactosylates both hydroxy- and non-hydroxy fatty acid-containing ceramides and diglycerides (By similarity). {ECO:0000250|UniProtKB:Q09426, ECO:0000269|PubMed:9125199}. | Endoplasmic reticulum;Glycoprotein;Glycosyltransferase;Lipid metabolism;Membrane;Reference proteome;Signal;Sphingolipid metabolism;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Sphingolipid metabolism; galactosylceramide biosynthesis. {ECO:0000269|PubMed:9125199}. | The protein encoded by this gene belongs to the UDP-glycosyltransferase family. It catalyzes the transfer of galactose to ceramide, a key enzymatic step in the biosynthesis of galactocerebrosides, which are abundant sphingolipids of the myelin membrane of the central and peripheral nervous systems. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Sep 2011]. | hsa:7368; | endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; intracellular membrane-bounded organelle [GO:0043231]; plasma membrane [GO:0005886]; 2-hydroxyacylsphingosine 1-beta-galactosyltransferase activity [GO:0003851]; N-acylsphingosine galactosyltransferase activity [GO:0047263]; UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity [GO:0008489]; central nervous system development [GO:0007417]; cytoskeleton organization [GO:0007010]; galactosylceramide biosynthetic process [GO:0006682]; glycosphingolipid metabolic process [GO:0006687]; neuron projection morphogenesis [GO:0048812]; paranodal junction assembly [GO:0030913]; peripheral nervous system development [GO:0007422]; protein localization to paranode region of axon [GO:0002175] | 12376738_Single-nucleotide polymorphisms (SNPs) were found in UGT8 gene 15229398_the GC-box and CRE function cooperatively, and that the CRE regulates the cell-specific expression of the hCGT gene 16125333_We postulate that molecular link between defective GALT enzyme, which result in classic galactosemia and the cerebroside galactosyl transferase, responsible for galactosylation of cerebrosides, is dependent on concentrations of UDP-galactose. 19125296_Gene expression of galactosyl ceramide synthase associated with poor pathohistological grading in breast cancer 19343046_Observational study of gene-disease association. (HuGE Navigator) 19898482_Observational study of gene-disease association. (HuGE Navigator) 20332099_Observational study of gene-disease association. (HuGE Navigator) 20648017_UGT8 is a significant index of tumour aggressiveness and is seen in lung metastases of breast cancer. 23118445_Comprehensive genomic analyses associate UGT8 variants with musical ability in a Mongolian population. 24391908_High expression of UGT8 accompanied by accumulation of GalCer in MDA-MB-231 cells is associated with a much higher proliferative index and a lower number of apoptotic cells in comparison to the MDA/LUC-shUGT8 cells. 24821492_Our study suggests that CGT expression is controlled by balanced expression of the negative modulator OLIG2 and positive regulator Nkx2.2, providing new insights into how expression of GalCer is tightly regulated in cell-type- and stage-specific manners. 25519837_A new role was identified for UGT8 as a modulator of bile acid homeostasis. 27620310_Data indicate that ceramide galactosyltransferase (UGT8), although enhanced in non-small cell lung carcinoma (NSCLC) tissues, does not meet the criteria of a lung tumor marker. 28746357_An increase in the immunolabelling of ceramide was observed in cells where UDP glycosyltransferase 8 (UGT8) was down-regulated as opposed to cells where UGT8 was either not regulated or was up-regulated. 29728441_UGT8 is a potential prognostic indicator and druggable target of basal-like breast cancer. 31409741_High FA2H and UGT8 transcript levels predict hydroxylated hexosylceramide accumulation in lung adenocarcinoma 34872002_SOX9-mediated UGT8 expression promotes glycolysis and maintains the malignancy of non-small cell lung cancer. | ENSMUSG00000032854 | Ugt8a | 940.60494 | 0.9721464 | -0.0407544392 | 0.11075297 | 1.353028e-01 | 7.129961e-01 | 9.125737e-01 | No | Yes | 1215.782203 | 272.390063 | 1142.985712 | 250.529754 |
| ENSG00000174775 | 3265 | HRAS | protein_coding | P01112 | FUNCTION: Involved in the activation of Ras protein signal transduction (PubMed:22821884). Ras proteins bind GDP/GTP and possess intrinsic GTPase activity (PubMed:12740440, PubMed:14500341, PubMed:9020151). {ECO:0000269|PubMed:12740440, ECO:0000269|PubMed:14500341, ECO:0000269|PubMed:22821884, ECO:0000269|PubMed:9020151}. | 3D-structure;Acetylation;Alternative splicing;Cell membrane;Cytoplasm;Direct protein sequencing;Disease variant;GTP-binding;Glycoprotein;Golgi apparatus;Hydrolase;Isopeptide bond;Lipoprotein;Membrane;Methylation;Nucleotide-binding;Nucleus;Palmitate;Prenylation;Proto-oncogene;Reference proteome;S-nitrosylation;Ubl conjugation | This gene belongs to the Ras oncogene family, whose members are related to the transforming genes of mammalian sarcoma retroviruses. The products encoded by these genes function in signal transduction pathways. These proteins can bind GTP and GDP, and they have intrinsic GTPase activity. This protein undergoes a continuous cycle of de- and re-palmitoylation, which regulates its rapid exchange between the plasma membrane and the Golgi apparatus. Mutations in this gene cause Costello syndrome, a disease characterized by increased growth at the prenatal stage, growth deficiency at the postnatal stage, predisposition to tumor formation, cognitive disability, skin and musculoskeletal abnormalities, distinctive facial appearance and cardiovascular abnormalities. Defects in this gene are implicated in a variety of cancers, including bladder cancer, follicular thyroid cancer, and oral squamous cell carcinoma. Multiple transcript variants, which encode different isoforms, have been identified for this gene. [provided by RefSeq, Jul 2008]. | hsa:3265; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endoplasmic reticulum membrane [GO:0005789]; glutamatergic synapse [GO:0098978]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; GTPase complex [GO:1905360]; nucleoplasm [GO:0005654]; perinuclear region of cytoplasm [GO:0048471]; plasma membrane [GO:0005886]; G protein activity [GO:0003925]; GDP binding [GO:0019003]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; protein C-terminus binding [GO:0008022]; animal organ morphogenesis [GO:0009887]; cell surface receptor signaling pathway [GO:0007166]; cellular response to gamma radiation [GO:0071480]; cellular senescence [GO:0090398]; chemotaxis [GO:0006935]; defense response to protozoan [GO:0042832]; endocytosis [GO:0006897]; intrinsic apoptotic signaling pathway [GO:0097193]; MAPK cascade [GO:0000165]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of gene expression [GO:0010629]; negative regulation of GTPase activity [GO:0034260]; negative regulation of neuron apoptotic process [GO:0043524]; positive regulation of actin cytoskeleton reorganization [GO:2000251]; positive regulation of cell migration [GO:0030335]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of epithelial cell proliferation [GO:0050679]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of GTPase activity [GO:0043547]; positive regulation of interferon-gamma production [GO:0032729]; positive regulation of JNK cascade [GO:0046330]; positive regulation of MAP kinase activity [GO:0043406]; positive regulation of MAPK cascade [GO:0043410]; positive regulation of miRNA metabolic process [GO:2000630]; positive regulation of phospholipase C activity [GO:0010863]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of protein targeting to membrane [GO:0090314]; positive regulation of ruffle assembly [GO:1900029]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of wound healing [GO:0090303]; Ras protein signal transduction [GO:0007265]; regulation of cell cycle [GO:0051726]; regulation of cell population proliferation [GO:0042127]; regulation of long-term neuronal synaptic plasticity [GO:0048169]; regulation of MAP kinase activity [GO:0043405]; regulation of neurotransmitter receptor localization to postsynaptic specialization membrane [GO:0098696]; regulation of transcription by RNA polymerase II [GO:0006357]; signal transduction [GO:0007165]; T cell receptor signaling pathway [GO:0050852]; T-helper 1 type immune response [GO:0042088] | 11097227_Observational study of gene-disease association. (HuGE Navigator) 11303621_Observational study of gene-disease association. (HuGE Navigator) 11487538_Meta-analysis of gene-disease association. (HuGE Navigator) 11695562_The Tg.AC (v-Ha-ras) transgenic mouse model provides a reporter phenotype of skin papillomas in response to either genotoxic or nongenotoxic carcinogens. 11695563_The Tg.Ac (v-Ha-ras) mouse model was not overly sensitive and possesses utility as an adjunct to the battery of toxicity studies used to establish carcinogenic risk. 11788888_immunohistochemical analysis reveals a protective effect of H-ras expression mediated via apoptosis in node-negative breast cancer patients 11904419_molecular dynamics simulations to show that Ras, a monomeric G protein, can generate mechanical force upon hydrolysis 11920220_Point mutations were identified in DNA from two of the 115 normal individuals. Both mutations resulted in an amino acid substitution at position 12 in H RAS. 11934900_H-Ras/mitogen-activated protein kinase pathway inhibits integrin-mediated adhesion and induces apoptosis in osteoblasts 12115522_Observational study of gene-disease association. (HuGE Navigator) 12540507_Observational study of gene-disease association. (HuGE Navigator) 12569357_H-Ras mutations at the binding site for the GTP nucleotide ring in a human multiple myeloma line leads to transformation and factor-independent cell growth 12589428_HRAS genes are biallelically expressed in multiple fetal and adult tissues both in humans and in mice 12668284_overexpression of PPARalpha or the c-Ha-ras transgene is not associated with the liver tumorigenesis induced by DEHP in rasH2 mice 12717016_steady-state structure of the switch I region of the protein in both the inactive GDP-bound conformation as in the active GTP-bound conformation. 12838617_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 12878090_Ha-ras mutations detected in HPV-induced cervical intraepithelial neoplasia grade II and invasive squamous cell carcinoma 12915131_Results demonstrate the close relationship between Ha-ras expression level and sensitization of 5-flurouracil (5-FU)-treated cells. 14576295_using fragments of the human c-Ha-ras gene containing 8-hydroxyguanine (8-OH-G) in codon 12, evidence for the highly complex biochemical events leading to activation of the oncogene 14662018_the human c-Ha-ras proto-oncogene product does not influence the androgen-dependence of prostate carcinogenesis due to the probasin-mediated SV40 T antigen. 14688016_Observational study of gene-disease association. (HuGE Navigator) 14693748_Observational study of gene-disease association. (HuGE Navigator) 14724641_Ras activates the MAP kinase cascade through simultaneous dual effector interactions: induction of Raf kinase activity and derepression of Raf-MEK complex formation 14729607_When mutated, causes hepatocarcinogenesis in transgenic mice. 14737103_complete loss of p53 is a prerequisite for collaborating with activated Ha-ras to promote bladder tumorigenesis 14767509_H-ras mutations have a role in malignant transformation of aerodigestive spindle cell carcinoma 15031297_a domain of Rap1 acts dominantly on COOH-terminal lipid modification of Ha-Ras, which has been considered to be essential and sufficient for the plasma membrane localization 15098441_Observational study of gene-disease association. (HuGE Navigator) 15202051_Observational study of gene-disease association. (HuGE Navigator) 15211515_Changes in flexibility upon protein-protein complex formation of H-Ras & the Ras-binding domain of C-Raf1 have been investigated using the molecular framework approach FIRST and molecular dynamics simulations of in total approximately 35 ns length. 15320975_ras gene alterations have a specific and early role in the development of follicular type of thyroid tumors in Taiwan. 15528212_Myc rescued cell growth inhibition induced by Ras 15597105_RAS appears to be a pejorative prognostic factor in terms of survival in NSCLC globally, in ADC and when it is studied by PCR. 15638373_No significant correlations were found between mutations in colorect cancer. 15677464_H-Ras-specific activation of Rac-MKK3/6-p38 pathway has a role in invasion and migration of breast epithelial cells 15684418_the mechanism of O2*-mediated Ras guanine nucleotide dissociation is similar to that of NO/O2-mediated Ras guanine nucleotide dissociation 15697248_Ras exists in (at least) two conformational states identifiable by nuclear magnetic resonance spectroscopy: state 2 represents the high-affinity binding state for effectors, while state 1 represents a weak binding state. 15702478_We found mutations in p53, K-ras, and BRAF genes in 35%, 30%, and 4% of tumors, respectively, and observed a minimal or no co-presence of these gene alterations. 15757891_RAS-MEK-ERK1/2 signaling pathway can sensitize cells to TRAIL-induced apoptosis by up-regulating DR4 and DR5 15784896_reduced Ras activation in cells harbouring the Hepatitis C virus subgenomic replicon 15816642_K-ras mutations are found in plasma after colorectal tumor resection 15831492_the mechanism for Ras-mediated down-regulation of Par-4 is by promoter methylation 15855817_Ras isoforms have distinct and separate cellular and subcellular distribution that may persist even in the malignantly transformed state in pancreatic disease 15940260_In this study, we probe the cellular and molecular mechanisms of RAS-mediated transformation. 15950068_HRAS oncogene could play an important role in the development of cervical cancer, in addition to the presence of HPV, by reducing the G1 phase and accelerating the G1/S transition of infected cells 15963850_N-Ras(L61) transformed cells lack a G0-G1 arrest upon TGF-beta treatment due to absence of p27. 15980150_Ras activity regulates Fbw7-mediated cyclin E proteolysis; impaired cyclin E proteolysis is a mechanism through which Ras mutations promote tumorigenesis. 16005186_RIG1 exerts inhibitory effect at the level of Ras activation, which is independent of Ras subtype but dependent on membrane localization of RIG1. It may be mediated through downregulation of Ras levels and alteration of Ras subcellular distribution. 16007212_studies provide evidence for the existence of human-specific mechanisms that resist Ras/MEK/ERK-mediated transformation 16081426_in primary fibroblasts stabilization of Ras protein by ROS and ERK1/2 amplifies the response of the cells to growth factors and in systemic sclerosis represents a critical factor in the onset and progression of the disease 16170018_growth factor-induced, Ras-mediated changes of keratinocyte shape may be an important mechanism that determines the speed of wound epitheli 16170316_Germline mutations in HRAS perturb human development and increase susceptibility to tumors. 16174078_Incidence of K-ras mutation and frequencies of COX-2 and gastrin overexpression are high in laterally spreading granular and protruded type colorectal tumors. 16187291_We conclude that in wild-type cells, endogenous Ras does not need to be prenylated to be active. 16264231_IL-24 is a member of IL-10 family of cytokines, and it signals through two hetorodimeric receptors, whose expression is also upregulated by ras oncogenes 16268414_K-ras mutation was found in 29% of sporadic adenocarcinomas respectively and in 0% and 22% of the 9 HNPCC cases. 16268778_Expression of constitutively active Galpha(q)Q209L in cells inhibited Ras activation of the PI3K/Akt pathway but had no effect on Ras/Raf/MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase] signalling 16286246_activation of the PI3K/AKT pathway replaced Ras once tumors formed, although other effectors were still activated independently of Ras, presumably by factors provided upon the establishment of a tumor microenvironment 16356174_APC and K-ras, but not CTNNB1 mutations have roles in regulation of expression of hMLH1 in sporadic colorectal carcinomas 16384911_Oncogenic RAS activity is directly responsible for OPCML promoter hypermethylation & epigenetic gene silencing. Elevation of the RAS signaling pathway may play an important role in epigenetic inactivation of OPCML in human epithelial ovarian cancer. 16434492_Analysus if 2502 patients with acute myeloid leukemia at diagnosis for NRAS mutations around hot spots at codons 12, 13, and 61 and correlation of the the results with cytomorphology, cytogenetics, other molecular markers, and prognosis. 16488657_Observational study of gene-disease association. (HuGE Navigator) 16518842_Ras induces ErbB4 receptor phosphorylation in a non-autocrine manner and this activation depends on multiple Ras effector pathways and on ErbB4 kinase activity. 16518851_Our data suggest that mutations of PTPN11 as well as RAS play a role in the pathogenesis of not only myeloid hematological malignancies but also a subset of RMS malignancies 16531227_Interplay and transmission of structural information between the switch regions are important factors for Ras function. They propose that initiation of GTP hydrolysis sets off the separation of the Ras/effector complex. 16532025_the H-RAS 81T --> C polymorphism may induce aneuploidy through overexpression of the active p21 isoform of H-RAS 16552541_The IGFBP3, hRas, JunB, Egr-1, Id1 and MIDA1 genes were up-regulated in psoriatic involved skin compared with uninvolved skin. 16569214_intracellular generation of NO* by nNOS leads to S-nitrosylation of H-Ras, which interferes with Raf-1 activation and propagation of signalling through ERK1/2 16573741_In 239 Thai adult AML cases, 35 RAS mutations were found in 32 cases (13%) predominantly classified as M1/M2 (53%) followed by M4/M5 (38%). Ten cases were positive for NRAS codon 12, 11 for NRAS codon 61, 13 for NRAS codon 13, and one for KRAS codon 13. 16598312_Activating RAS mutation is a pivotal molecular lesion that is implicated in the pathogenesis of AML and MDS. 16598313_Ras mutations were significantly more frequent in inv(16) than in t(8;21) subset (36 versus 8%, P=0.001). 16644864_NF1 is a novel regulator of RAS-induced signals in primary vascular smooth muscle cells 16645632_There is a signaling loop between Ras-dependent MAPK cascade activation and p73 function. 16717102_histone deacetylase inhibitor sodium butyrate induces G1/S phase arrest in E1A + Ras-transformed cells through down-regulation of E2F1 activity and stabilization of beta-catenin 16738062_Review. An alternative splice form of c-H-ras, called p19ras, is a positive regulator of p73beta via Mdm2. Implications for this previously unidentified means of regulation are discussed in light of tumor suppression and are extended to p53 and p63. 16760302_K-ras mutations were observed in 68/182 (37.4%) cases of colorectal cancer. 16761621_ras may be involved in early stages of larynx carcinogenesis and may be activated by other mechanisms different from mutations, such as epigenetic events 16774944_Targeted activation of a human oncogenic-ras transgene in rat pancreas can induce carcinomas correspondent to human pancreatic ductal adenocarcinomas. 16806262_These findings indicate that mechanical strain causes ROS-dependent S-glutathiolation of Ras at Cys118, leading to myocyte hypertrophy via activation of the Raf/Mek/Erk pathway. 16831126_The entire Ras/Raf/MEK/ERK pathway is activated by intracellular acidosis, indicating that the initiating acid sensor is found at the level of Ras or above. 16849642_Down-regulation of MIP-1 alpha was not observed following FGFR3 inhibition in MM cells with RAS mutations implicating RAS-MAPK in MIP-1 alpha regulation 16857742_Ras and c-Myc play important roles in the up-regulation of nucleophosmin/B23 during proliferation of cells associated with a high degree of malignancy, thus outlining a signaling cascade involving these factors in the cancer cells. 16881968_Three unrelated Dutch patients with Costello syndrome all have the same mutation, G12S, in HRAS. 16890591_High prevalence of CIMP-high and K-ras mutations in G-LST, especially in the proximal colon, could strongly suggest that G-LST appearance is associated with a unique carcinogenic pathway. 16923573_analysis of H-RAS, K-RAS and N-RAS expression in acute myeloid leukemia 16945398_Ras expression in cervical keratinocyte cell lines containing stably replicating extrachromosomal HPV-16 consistently diminished anchorage-independent growth (AI), reduced E6 and E7 expression, and caused p53 induction in these cells. 16969868_Somatic mosaicism for an HRAS mutation causes Costello syndrome. 17018607_Activation of Ras plays a critical role in modulating the expression of both CXCL10 and CXCR3-B, which may have important consequences in the development of breast tumors through cancer cell proliferation. 17094109_The hydrolysis of guanosine triphosphate (GTP) by p21(ras) (Ras) has been modeled by using ab initio type quantum mechanical-molecular mechanical simulations. The minimum energy reaction path is consistent with a 2-step mechanism of GTP hydrolysis. 17096025_Neurofibromin-deficient mouse embryonic fibroblasts (MEFs) and human NF1 tumor cells were more resistant than neurofibromin-expressing cells to apoptosis mediated by two survival pathways: a Ras-dependent pathway, and a Ras-independent pathway. 17164262_HRAS mutations represent independent but cooperating events to uniparental disomy during embryonic rhabdomyosarcoma development. 17196792_The activation of HRAS was inhibited by 25.1% or 81.4% in cells cotransfected with wild-type or Golgi-targeted RIG1, respectively. 17210246_Taken together, the observations indicate that both H-Ras(G12V) and K-Ras4B(G12V) activates non-conventional and perhaps unique effector pathways to induce cytoplasmic vacuolation in glioblastoma cells. 17237388_Ras and Ral mediate BCR-controlled activation of JUN/ATF2 and NFAT transcription factors 17255356_Ras is able to promote monocyte lineage selection via PKC and PDK1. 17264303_Transfection of U266 cells with constitutively activated H-Ras (Q61L) attenuated ERK1/2 inactivation and dramatically diminished the lethality of statins + UCN-01 regimen. 17324647_One female patient exhibiting the distal phalangeal creases had a mutation in the HRAS gene. 17374253_K-ras mutations may play a less important role in the tumorigenesis of ovarian serous tumor of the Chinese patients. 17388810_The subcellular distribution of K-Ras is driven by electrostatic interaction of the polybasic region of the protein with negatively charged membranes. 17412879_We identified two known activating and two novel germline HRAS mutations in four patients with an unusual form of congenital myopathy histologically charactered by an excess of muscle spindles. 17428306_Study identified Crk adapter proteins, Rac1 and H-Ras, but not RhoA or Cdc42 as crucial components of the Helicobacter pylori CagA protein-induced phenotype. 17488404_The evaluation of a large number of actinic keratoses specimens have found a low gene mutation rate in low-graded AK lesions. p53 mutations rather than p16(INK4a) and/or Ha-ras mutations may be an early event in the development of AK to cutaneous SCC. 17518771_low rate of RAS-RAF mutations (2/22, 9.1%) observed in Spitz melanocytic nevi suggests that these lesions harbor as yet undetected activating mutations in other components of the RAS-RAF-MEK-ERK-MAPK pathway 17601930_De novo germline HRAS (G12A) and KRAS (F156L) mutations in two siblings with short stature and neuro-cardio-facio-cutaneous features. 17635919_In contrast to C-RAF that requires farnesylated H-Ras, cytosolic B-RAF associates effectively and with significantly higher affinity with both farnesylated and nonfarnesylated H-Ras. 17638918_BCR/ABL-Y177 plays an essential role in Ras and Akt activation and in human hematopoietic progenitor transformation in chronic myelogenous leukemia 17699159_Erf provides a direct link between the RAS/ERK signaling and the transcriptional regulation of c-Myc and suggests that RAS/ERK attenuation actively regulates cell fate 17712732_No mutations in HRAS was found in pilocytic astrocytomas. 17767136_H-Ras mutation defines a molecular subtype of oral carcinoma with favourable outcome and unique biology 17912430_a potential key role for activated members of ras family genes in terms of their contribution to the development of nasal polyposis as well as to the hypertrophy of adjacent turbinates. 17943694_Observational study of genotype prevalence. (HuGE Navigator) 17974970_Examination of various growth-regulatory pathways suggested that Bmi-1 overexpression together with H-Ras promotes human mammary epithelial cell transformation and breast oncogenesis by deregulation of multiple growth-regulatory pathways. 17979197_These data show that a RAS mutation that only perturbs guanine nucleotide binding has similar functional consequences as mutations that impair GTP hydrolysis and causes human disease. 18021740_These findings suggest that the activation of Ras signaling pathways promotes the generation of brain cancer stem-like cells from p53-deficient mouse astrocytes by changing cell fate and transforming cell properties. 18039947_Four cases of Costello syndrome had an unusually severe phenotype, associated in three cases with two unusual mutations, c.35G>A, p.G12D in two cases and c.34G>T, p.G12C in the other. 18042262_the results of HRAS, BRAF and MAP2K1/2 mutation screening in a large cohort of patients with CS and CFC 18048363_H-Ras interacts with Spry2-binding partners, c-Cbl and CIN85, in a Spry2-dependent manner. 18077377_V12 H-Ras oncogenic signaling may contribute to anchorage-independent growth and tumorigenesis by promoting the final cleavage furrow ingression during cytokinesis. 18094044_Trafficking of H-Ras was examined to determine whether it can enter cells through clathrin-independent endocytosis (CIE). 18176964_K-ras mutations were detected in only one of the Hungarian cases; Eight of 19 (42.1%) Japanese cases were MSI-high (presence of novel peaks in more than one of the five loci analyzed), whereas only 1 of 15 (6.7%) Hungarian cases was MSI-high (P = 0.047 18247425_study reports on two patients with novel HRAS mutations affecting amino acids 58 (T58I) and 146 (A146V), respectively; both patients show many of the physical and developmental problems characteristic for Costello syndrome 18284919_Taken together, these results reveal a novel role for fad24 in the repression of NF-kappaB activity and H-ras-mediated transformation. 18310288_Hras mutation is associated with familial non-medullary thyroid carcinoma 18325324_The redox sensitive p21Ras-ERK pathway plays a critical role in sensing and delivering the pro-apoptotic signaling mediated by S-nitrosoglutathione. 18330889_Results correlate P53 status and mutation site/type with nuclear protein accumulation, clinicopathologic variables and data on K-ras mutations and high-level microsatellite instability. 18355852_high frequency of mutations in the PIK3CA, HRAS and KRAS genes leads us to believe that dysregulation of the phosphatidylinositol 3-kinase or Ras pathway is significant for the development and progression of penile carcinoma. 18383861_K-RAS and BRAF mutations are a frequent genetic event in our samples of sporadic papillary and medullary thyroid carcinoma. 18425390_K-ras and p53 genes are altered in Tamoxifen-associated endometrial carcinoma 18454158_Study shows that SOS responds to the membrane density of Ras molecules, to their GTP loading and to the concentration of phosphatidylinositol-4,5-bisphosphate, and that the integration of these signals potentiates the release of autoinhibition. 18465119_Report extremely weak tumor-promoting effect of troglitazone on splenic hemangiosarcomas in rasH2 mice induced by urethane. 18485440_RAS signalling may play an important role in neuronal susceptibility to Sindbis virus infection 18555006_p19(ras) interaction with p73beta amplifies p73beta-induced apoptotic signaling responses including Bax mitochondrial translocation, cytochrome c release, increased production of reactive oxygen species and loss of mitochondrial transmembrane potential. 18577512_p21 Ras/Imp regulate cytokine production and migration in CD4 T cells 18621636_Presence of K-ras mutations might be associated with lack of response to panitumumab alone and inferior survival in patients with metastatic colorectal cancer. 18641685_Scribble is an important mediator of MAPK signalling and cooperates with H-ras to promote cell invasion. 18655729_k-ras may have a role in progression of rectal cancer after resection 18660489_Observational study of gene-disease association. (HuGE Navigator) 18676755_K-Ras mutations have a role in treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy 18677112_Low-penetrant RB allele in small-cell cancer shows geldanamycin instability and discordant expression with mutant ras. 18683737_HRAS-shRNA could efficiently down-regulate the expression of H-ras, induce apoptosis of SACC-M cells and simultaneously inhibit cell proliferation. 18724893_Occupational exposure to coking oven emissions increases expression levels of P21 and P53 proteins in peripheral white blood cells. 18757346_LDLs stimulate p38 MAPKs and wound healing through SR-BI independently of Ras and PI3 kinase 18769120_transfection of a promoterless K-ras cDNA resulted in a significant decrease in endogenous K-ras in a dose- and time-dependent manner and induces pancreatic cell apoptosis 18771652_This review will summarize the chemical biology of Ras and discuss in more detail the biophysical and structural features of the membrane bound C-terminus of the protein. 18783589_Observational study of gene-disease association. (HuGE Navigator) 18783589_the H-RAS T81C polymorphism might be a low penetrance gene predisposition factor for gastric cancer in the Chinese population 18790720_Ca(2+) may activate Ras signaling pathway by interaction with Ras, providing clues to understand the role of calcium in regulating Ras function in physiological environments. 18922970_This study revealed S100A8/A9 genes as candidate markers for metastatic potential of breast epithelial cells. 19021766_The secondary structure of membrane-bound, lipidated Ras is similar to that determined for the nonlipidated truncated Ras protein for the highly conserved G-domain. 19029954_Hdm2 is expressed in pancreatic cancer cells as a result of activated Ras signaling, and regulates cellular proliferation and the expression of target genes by p53-independent mechanisms. 19035362_Costello syndrome-associated HRAS mutations result in prolonged signal flux in a ligand-dependent manner 19048123_Ras transformation has at least two independent actions to disrupt E-cadherin junctions, with effects to cause both mislocalization of E-cadherin away from the cell surface and profound decrease in the expression of E-cadherin 19062714_The increased accumulation of miR-143 inhibits the proliferation of transfected cells, and results in down-regulation of K-ras protein in colorectal carcinoma. 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19075190_RAS mutations may play a greater role in leukemogenesis than currently believed 19089619_The oncoprotein Ras is a possible mediator of Tax-induced apoptosis protection and suggest a possible role of Tax in Ras activation. 19101897_failed to find hRAS mutations in bladder tumor samples 19107228_The Snail-p53 binding as the new therapeutic target for K-Ras-mutated cancers including pancreatic, lung, and colon cancers. 19107910_Women with the NAT2 fast acetylator genotype may exhibit a higher risk of CRC with increased occurrence of K-RAS mutation. 19133693_Observational study of gene-disease association. (HuGE Navigator) 19139860_Subtle deficits in inhibition and enhanced excitation evoke the interneuronal changes in the synRas transgenic mouse cortex. 19140325_K-RAS point mutations, and anomalies of p16-RB1-cyclin D pathway could occur before LOH on 10q23 (PTEN) and microsatellite instability during tumor progression. 19146951_p21(Ras)-mediated survival signaling is regulated by via a PI(3)K-AKT pathway, which is dependent upon both PDK1 and PKCdelta, and PDK1 activates and regulates PKCdelta to determine the fate of cells containing a mutated, activated p21(Ras). 19160018_Rho mediates various phenotypes of malignant transformation by Ras and Src through its effectors, ROCK and mDia [review] 19206176_A single patient with somatic mosaicism for a Costello syndrome causing HRAS mutation is reported. The male-to-male transmission was from father to son. 19218240_BLNK recruits active H-Ras to the BCR complex, which is essential for sustained surface expression of BCR in the form of the cap and for the signal leading to functional ERK activation 19240121_A novel regulatory mechanism whereby turnover of both endogenous and overexpressed H-Ras protein is controlled by beta-TrCP-mediated ubiquitylation, proteasomal degradation and the Wnt/beta-catenin signaling pathway, is reported. 19254697_These findings support the hypothesis that the Ras/Raf/Mek/Erk pathway may be constitutively active even in pancreatic tumor cells that express wild-type K-ras. 19276157_EGFR expression and KRAS mutation status is predictive for clinical response to matuzumab +/- paclitaxel in patients with advanced NSCLC. 19289583_K-ras mutations, significantly associated with the mucin-secreting type of adenocarcinoma, consistently predict lack of response in white patients 19295453_Data show that the combination with CA 19-9 assay is useful for detection and prognostic evaluation of pancreatic carcinoma. 19303097_Observational study of gene-disease association. (HuGE Navigator) 19347028_Study provides evidence that p53, binding with Snail, is exported from a K-Ras-mutated cell through a vesicle transport-like mechanism, independently using a p53-nuclear-exporting mechanism. 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19369630_K-ras, EGFR, and BRAF mutations are disproportionately seen in adenocarcinomas of lung with a dominant micropapillary growth pattern compared with conventional adenocarcinoma in our institutional experience. 19406145_Chronic NF-kappaB activation delays RasV12-induced premature senescence of human fibroblasts by suppressing the DNA damage checkpoint response. 19420344_Mutation E17K/AKT1 was not detected in the 24 samples of retinoblastoma analyzed. K-RAS mutations were identified in two samples. There were no mutations in any of the other genes analyzed. 19438459_HRAS mutation may be a marker of Spitz naevus 19443408_The median pancreatic cancer survival in K-ras positive group was 7.0+/-2.4 months and in K-ras negative group was 10.0+/-0.6 months 19487299_Observational study of gene-disease association. (HuGE Navigator) 19494114_DEP-1 inhibits the RAS pathway by direct dephosphorylation of ERK1/2 kinases 19543317_BMI1 collaborates with H-RAS to induce an aggressive and metastatic phenotype with the unusual occurrence of brain metastasis. 19628422_Observational study of gene-disease association. (HuGE Navigator) 19628422_results suggest that RAS is an important member in the PI3K-AKT signaling and could play an important role in the tumorigenesis of oral carcinoma. 19638615_Data show that in the presence of negative feedback, changes in the rate of Ras-c-Raf binding have little effect on ERK activation. 19681119_Investigated the prevalence of PTPN11, HRAS, KRAS, NRAS, BRAF, MEK1, and MEK2 mutations in a relatively large cohort of primary embryonal Rhabdomyosarcoma (RMS) tumors. HRAS and KRAS were found to be rarely mutated. 19723872_Findings show that JNK is necessary for oncogenic H-ras-induced, caspase-independent cell death, and that both PI3K and Rac1 activities are required for JNK activation and cell death. 19752775_Data show that significant K-ras mutation occurs most frequently in the pancreatobiliary regions of patients with AIP. 19787260_the organophosphorous pesticides parathion and malathion induced malignant transformation of breast cells through genomic instability altering p53 and c-Ha-ras 19787272_We noted statistically significant difference in genotype distribution at hexanucleotide locus and at SNP 81 T>C between healthy population and colon cancer patients 19802010_Data show that conversion of Ras-expressing keratinocytes from a premalignant to malignant state induced by decreasing E-cadherin function was associated with and required an approximately two to threefold decrease in RalA expression. 19802012_Results show a cooperative relationship between the expression of Ras and loss of RB in providing a discrete proliferative advantage under physiologically relevant conditions. 19815050_Data suggest that investigation of the mechanisms of abnormal RAS and phosphoinositide-3 kinase pathway activation in erythroblasts may contribute to the understanding of the molecular pathogenesis of PV. 19849697_HRAS codon 12 mutation is not a very common event in actinic keratosis or nonmelanoma skin cancers 19855393_A single cellular event for mutations arising in male germ cells that show a paternal age effect; screening of 30 spermatocytic seminomas for oncogenic mutations in 17 genes identified 2 mutations in FGFR3 and 5 mutations in HRAS. 19855428_Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. 19888426_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19890398_RAS enables DNA damage- and p53-dependent differentiation of acute myeloid leukemia cells in response to chemotherapy 19936293_H-ras expression in immortalized keratinocytes produces an invasive epithelium in cultured skin equivalents 19966300_Data show that Myc repressed Ras-induced senescence, and that Cdk2 interacted with Myc at promoters, where it affected Myc-dependent regulation of genes, including those of proteins known to control senescence. 20012784_Screening for BRAF, RET, KRAS, NRAS, and HRAS mutations, as well as RET-PTC1 and RET-PTC3 rearrangements, was performed on cases of Hashimoto thyroiditis with a dominant nodule 20038817_loss of BRCA1 function may contribute to the aggressiveness of Ras-MAPK driven breast cancer with associated increase in levels of cyclin D1 and c-myc, enhanced MAPK activity, angiogenic potential & invasiveness 20042089_Exogenous expression of RASSF1A is able to induce growth inhibition effect and apoptosis in nasopharyngeal carcinoma cell lines, and this effect could be enhanced by activated K-Ras. 20046837_p19 induces FOXO1 that in combination with the G1/S phase delay and hypophosphorylation of both Akt and p70SK6 leads to maintenance of a reversible cellular quiescence state, thereby preventing entry into apoptosis. 20147967_The identification of mutations outwith previously described hotspot codons increases the K-Ras mutation burden in colorectal tumours by one-third. 20150643_Observational study of gene-disease association. (HuGE Navigator) 20173673_The present study suggests that the single-clonal convergence of K-ras mutation is associated with the malignant progression of intraductal papillary mucinous neoplasms. 20184776_In Chinese patients with CRC, EGFR mutations were rare, and K-ras mutations were similar to those of Europeans. 20188103_Oncogenic K-Ras and its effector Raf1 convert death receptors into invasion-inducing receptors by suppressing the ROCK/LIM kinase pathway, and this is essential for K-Ras/Raf1-driven metastasis formation. 20354187_JNK2 collaborates with other oncogenes, such as Ras, at multiple molecular levels to promote tumorigenesis 20375073_increased binding of DNMT3b to E-cadherin promoter region by K-Ras cause promoter hypermethylation for reduced expression of E-cadherin, leading to the decreased cell aggregation and increased metastasis of human prostate cancer cells. 20392691_Ras induces ARC in epithelial cancers, and ARC plays a role in the oncogenic actions of Ras 20406971_Using lung cell lines expressing oncogenic K-Ras, authors show that NF-kappaB is activated in these cells in a K-Ras-dependent manner and that NF-kappaB activation by K-Ras. 20413844_maintain colorectal cancer cell proliferation during ERK MAP kinases inhibition 20425820_Costello syndrome is a rasopathy caused by germline mutations in the proto-oncogene HRAS. 20437058_Observational study of genetic testing. (HuGE Navigator) 20479006_X-ray crystal | ENSMUSG00000025499 | Hras | 1200.68857 | 0.9186220 | -0.1224568050 | 0.11232455 | 1.184626e+00 | 2.764163e-01 | 6.653788e-01 | No | Yes | 1455.095806 | 224.170469 | 1497.024345 | 225.358072 | |
| ENSG00000175221 | 10025 | MED16 | protein_coding | Q9Y2X0 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. {ECO:0000269|PubMed:10198638, ECO:0000269|PubMed:10235266}. | 3D-structure;Activator;Alternative splicing;Direct protein sequencing;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;WD repeat | hsa:10025; | core mediator complex [GO:0070847]; mediator complex [GO:0016592]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; catalytic activity [GO:0003824]; thyroid hormone receptor binding [GO:0046966]; transcription coactivator activity [GO:0003713]; vitamin D receptor binding [GO:0042809]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; transcription by RNA polymerase II [GO:0006366] | 32532820_Mediator complex subunit 16 is down-regulated in papillary thyroid cancer, leading to increased transforming growth factor-beta signaling and radioiodine resistance. | ENSMUSG00000013833 | Med16 | 1288.07701 | 0.9422401 | -0.0858332922 | 0.12063217 | 5.049336e-01 | 4.773403e-01 | 8.088423e-01 | No | Yes | 1503.603012 | 249.205561 | 1518.915526 | 246.053372 | ||
| ENSG00000175611 | 100128782 | LINC00476 | lncRNA | 33370431_LINC00476 Suppresses the Progression of Non-Small Cell Lung Cancer by Inducing the Ubiquitination of SETDB1. | 111.58657 | 1.0641129 | 0.0896511635 | 0.26770633 | 1.123700e-01 | 7.374619e-01 | 9.224290e-01 | No | Yes | 133.169586 | 16.941370 | 120.022812 | 15.009290 | |||||||||
| ENSG00000176102 | 1479 | CSTF3 | protein_coding | Q12996 | FUNCTION: One of the multiple factors required for polyadenylation and 3'-end cleavage of mammalian pre-mRNAs. | 3D-structure;Acetylation;Alternative splicing;Nucleus;Phosphoprotein;Reference proteome;Repeat;mRNA processing | The protein encoded by this gene is one of three (including CSTF1 and CSTF2) cleavage stimulation factors that combine to form the cleavage stimulation factor complex (CSTF). This complex is involved in the polyadenylation and 3' end cleavage of pre-mRNAs. The encoded protein functions as a homodimer and interacts directly with both CSTF1 and CSTF2 in the CSTF complex. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]. | hsa:1479; | nucleoplasm [GO:0005654]; nucleus [GO:0005634]; mRNA binding [GO:0003729]; RNA binding [GO:0003723]; mRNA cleavage [GO:0006379]; mRNA polyadenylation [GO:0006378]; RNA 3'-end processing [GO:0031123] | 12149458_chimeric human CstF-77/Drosophila suppressor of forked proteins rescue suppressor of forked mutant lethality and mrna 3' end processing in Drosophila 19887456_The Hinge domain is necessary for CstF-64 interaction with CstF-77 and consequent nuclear localization. 21119002_nuclear accumulation of CstF-64 depends on binding to CstF-77 not symplekin; interaction between CstF-64/CstF-64Tau and CstF-77 are important for maintenance of nuclear levels of CstF complex components and intracellular localization, stability, function 23874216_Thus, the conserved intronic pA of the CstF-77 gene may function as a sensor for cellular C/P and splicing activities, controlling the homeostasis of CstF-77 and C/P activity and impacting cell proliferation and differentiation. 26288249_Results from a study on gene variability markers in early-stage human embryos shows that CSTF3 is a putative variability marker for the 3-day, 8-cell embryo stage. 29186539_Data show that the recruitment of CstF subunit CstF-50 to CstF via interaction with cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa protein (CstF-77) and establish that the hexameric assembly of CstF. 30257008_Reverse genetics and nuclear magnetic resonance studies of recombinant CstF-64 (RRM-Hinge) and CstF-77 (monkeytail-carboxy-terminal domain) indicate that the last 30 amino acids of CstF-77 increases the stability of the RRM, thus altering the affinity of the complex for RNA. These results provide new insights into the mechanism by which CstF regulates the location of the RNA cleavage site during Cleavage/polyadenylation. | ENSMUSG00000027176 | Cstf3 | 3173.44850 | 1.0936583 | 0.1291621020 | 0.08032998 | 2.592356e+00 | 1.073805e-01 | 4.680123e-01 | No | Yes | 3276.668483 | 465.713981 | 2909.819740 | 404.657305 | |
| ENSG00000176715 | 197322 | ACSF3 | protein_coding | Q4G176 | FUNCTION: Catalyzes the initial reaction in intramitochondrial fatty acid synthesis, by activating malonate and methylmalonate, but not acetate, into their respective CoA thioester (PubMed:21846720, PubMed:21841779). May have some preference toward very-long-chain substrates (PubMed:17762044). {ECO:0000269|PubMed:17762044, ECO:0000269|PubMed:21841779, ECO:0000269|PubMed:21846720}. | ATP-binding;Fatty acid metabolism;Ligase;Lipid metabolism;Mitochondrion;Nucleotide-binding;Reference proteome;Transit peptide | This gene encodes a member of the acyl-CoA synthetase family of enzymes that activate fatty acids by catalyzing the formation of a thioester linkage between fatty acids and coenzyme A. The encoded protein is localized to mitochondria, has high specificity for malonate and methylmalonate and possesses malonyl-CoA synthetase activity. Mutations in this gene are a cause of combined malonic and methylmalonic aciduria. Alternatively spliced transcript variants have been observed for this gene. [provided by RefSeq, Sep 2013]. | hsa:197322; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; acid-thiol ligase activity [GO:0016878]; ATP binding [GO:0005524]; CoA-ligase activity [GO:0016405]; malonyl-CoA synthetase activity [GO:0090409]; very long-chain fatty acid-CoA ligase activity [GO:0031957]; fatty acid biosynthetic process [GO:0006633]; fatty acid metabolic process [GO:0006631]; long-chain fatty-acyl-CoA biosynthetic process [GO:0035338]; malonate catabolic process [GO:0090410] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 21785126_ACSF3 is a candidate gene for non-classical CMAMMA observed in our patients. 21841779_mutations in ACSF3, encoding a putative methylmalonyl-CoA and malonyl-CoA synthetase, were identified as a cause of combined malonic and methylmalonic aciduria. 21846720_Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. 22420028_data indicate that the CBFA2T3/ACSF3 locus is a novel recurrent oncogenic target of immunoglobulin heavy chain translocations, which might contribute to the pathogenesis of pediatric GC-derived B-cell lymphoma. 23337955_ACSF3 was significantly upregulated, and was involved in fatty acid and lipid metabolism and accelerated liver injury in alcoholic liver disease. 28479296_an essential role for ACSF3 in dictating the metabolic fate of mitochondrial malonate and malonyl-CoA in mammalian metabolism 30201289_ACSF3-derived malonyl-CoA can be used to malonylate lysine residues on proteins within the matrix of mitochondria, possibly adding another regulatory layer to post-translational control of mitochondrial metabolism. 30740739_Causal ACSF3 mutations were identified in all patients. 31376476_ACSF3 catalyzes the first step of mitochondrial fatty acid biosynthesis (mtFASII). Hypofunctional mtFASII impairs mitochondrial flexibility and lipoylation degree. Defective mtFASII leads to reduced glycolytic flux and upregulation of beta-oxidation. | ENSMUSG00000015016 | Acsf3 | 824.51054 | 0.9071976 | -0.1405112394 | 0.11973348 | 1.370992e+00 | 2.416416e-01 | 6.331137e-01 | No | Yes | 968.796012 | 113.043774 | 1032.025828 | 117.572187 | |
| ENSG00000177200 | 80205 | CHD9 | protein_coding | Q3L8U1 | FUNCTION: Acts as a transcriptional coactivator for PPARA and possibly other nuclear receptors. Proposed to be a ATP-dependent chromatin remodeling protein. Has DNA-dependent ATPase activity and binds to A/T-rich DNA. Associates with A/T-rich regulatory regions in promoters of genes that participate in the differentiation of progenitors during osteogenesis (By similarity). {ECO:0000250, ECO:0000269|PubMed:16095617, ECO:0000269|PubMed:16554032}. | ATP-binding;Acetylation;Alternative splicing;Chromatin regulator;Cytoplasm;DNA-binding;Helicase;Hydrolase;Isopeptide bond;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation | hsa:80205; | cytosol [GO:0005829]; nucleoplasm [GO:0005654]; ATP binding [GO:0005524]; ATP hydrolysis activity [GO:0016887]; DNA binding [GO:0003677]; DNA helicase activity [GO:0003678]; chromatin organization [GO:0006325] | 16095617_The CHD9 (CReMM) protein is extensively phosphorylated, has DNA-dependent ATPase activity, and binds to A/T-rich DNA. It is also expressed in mesenchymal progenitors. 16523501_CReMM is a chromodomain helicase-DNA-binding protein expressed by osteoprogenitors 16523501_CReMM is a chromodomain helicase-DNA-binding protein expressed by osteoprogenitors[CReMM] 16705189_Chromatin immunoprecipitation assay was applied to follow the dynamics of CReMM binding to A/T-rich regions on promoters of genes that play a role in osteoblast maturation. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 32453735_Gene expression regulation by the Chromodomain helicase DNA-binding protein 9 (CHD9) chromatin remodeler is dispensable for murine development. | ENSMUSG00000056608 | Chd9 | 1307.09066 | 1.0212912 | 0.0303942680 | 0.13945001 | 4.766602e-02 | 8.271755e-01 | 9.526069e-01 | No | Yes | 703.279585 | 104.225994 | 681.053944 | 98.925002 | ||
| ENSG00000177463 | 7182 | NR2C2 | protein_coding | P49116 | FUNCTION: Orphan nuclear receptor that can act as a repressor or activator of transcription. An important repressor of nuclear receptor signaling pathways such as retinoic acid receptor, retinoid X, vitamin D3 receptor, thyroid hormone receptor and estrogen receptor pathways. May regulate gene expression during the late phase of spermatogenesis. Together with NR2C1, forms the core of the DRED (direct repeat erythroid-definitive) complex that represses embryonic and fetal globin transcription including that of GATA1. Binds to hormone response elements (HREs) consisting of two 5'-AGGTCA-3' half site direct repeat consensus sequences. Plays a fundamental role in early embryonic development and embryonic stem cells. Required for normal spermatogenesis and cerebellum development. Appears to be important for neurodevelopmentally regulated behavior (By similarity). Activates transcriptional activity of LHCG. Antagonist of PPARA-mediated transactivation. {ECO:0000250, ECO:0000269|PubMed:10347174, ECO:0000269|PubMed:10644740, ECO:0000269|PubMed:17974920, ECO:0000269|PubMed:7779113, ECO:0000269|PubMed:9556573}. | 3D-structure;Acetylation;Activator;Alternative splicing;DNA-binding;Differentiation;Isopeptide bond;Metal-binding;Nucleus;Phosphoprotein;Receptor;Reference proteome;Repressor;Spermatogenesis;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a protein that belongs to the nuclear hormone receptor family. Members of this family act as ligand-activated transcription factors and function in many biological processes such as development, cellular differentiation and homeostasis. The activated receptor/ligand complex is translocated to the nucleus where it binds to hormone response elements of target genes. The protein encoded by this gene plays a role in protecting cells from oxidative stress and damage induced by ionizing radiation. The lack of a similar gene in mouse results in growth retardation, severe spinal curvature, subfertility, premature aging, and prostatic intraepithelial neoplasia (PIN) development. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Apr 2014]. | hsa:7182; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; nuclear receptor activity [GO:0004879]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific DNA binding [GO:0043565]; sequence-specific double-stranded DNA binding [GO:1990837]; zinc ion binding [GO:0008270]; anatomical structure development [GO:0048856]; cell differentiation [GO:0030154]; negative regulation of transcription by RNA polymerase II [GO:0000122]; nervous system development [GO:0007399]; positive regulation of embryonic development [GO:0040019]; positive regulation of transcription by RNA polymerase II [GO:0045944]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; spermatogenesis [GO:0007283] | 11844790_role in modulating estrogen receptor-mediated trans-activation 12093744_DRED is a 540 kDa complex containing the nuclear orphan receptors TR2 and TR4, which form a heterodimer that binds to the epsilon and gamma globin promoter DR1 sites. TR2 & 4 mRNAs are expressed at all stages of murine and human erythropoiesis. 12522137_TR4 can serve as a negative modulator in the transcriptional regulation of HBV core gene expression 12615366_TR2 and TR4 can have distinct functions. Existence of differential and bi-directional regulation between PPAR alpha and TR2/TR4. Possible roles in PPAR alpha signaling pathway in human keratinocytes. 17601350_Observational study of gene-disease association. (HuGE Navigator) 18977241_Observational study of gene-disease association. (HuGE Navigator) 19618297_The binding activity of C/EBPs to the TR4 promoter is increased in response to cAMP treatment. 19859911_Human testicular orphan receptor 4 enhances thyroid hormone receptor signaling. 19955181_Activation of IRF3 and IRF3-dependent gene expressions was dependent on TAK1 and TANK-binding kinase 1 (TBK1). 20164171_the ADAP CARMA1 binding site is required for IKK gamma ubiquitination; both TAK1 and CARMA1 binding sites are required for IkappaB alpha phosphorylation and degradation and NF-kappaB nuclear translocation 20200282_Our data establish the central role of TAK1 in controlling nuclear and cytoplasmic signaling cascades in primary neutrophils 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20393820_TAK1 is an important transcriptional modulator of cerebellar development and neurodevelopmentally regulated behavior. 20604713_COX-2 gene expression and prostaglandin (PG)E2 release are specific outcomes of collagen II signaling, and both depend on TAK1 mediation in primary human chondrocytes. 21068381_TR4 is a ligand-regulated nuclear receptor and suggest that retinoids might have a much wider regulatory role via activation of orphan receptors such as TR4. 21126370_TR4 binding at a subset of sites is facilitated through the ETS transcription factor ELK4. 21331078_Results demonstrate that USP4 serves as a critical control to downregulate TNFalpha-induced NF-kappaB activation through deubiquitinating TAK1. 21835421_These results demonstrated the role of TAK1 as an important upstream signaling molecule regulating RSV-induced NF-kappaB and AP-1 activation. 21915030_The purpose of the study was to investigate the potential contribution of HPK1, MEKK1, TAK1, p-MKK4 to the development of extramammary Paget disease 21918225_Testicular nuclear receptor 4 (TR4) regulates UV light-induced responses via Cockayne syndrome B protein-mediated transcription-coupled DNA repair 21980489_the protein associates with TGF-beta receptors and components of the TRAF6-TAK1 signaling module 22544925_Mycobacterium tuberculosis interacts with macrophage lipids and human host testicular receptor (TR)4 to ensure survival of the pathogen by modulating macrophage function. 22641218_Celastrol highlights the therapeutic potential of agents targeting TAK1 as a key node in this pro-oncogenic TGF-beta-NF-kappaB signal pathway 22643835_PINK1 positively regulates two key molecules, TRAF6 and TAK1, in the IL-1beta-mediated signaling pathway, consequently up-regulating their downstream inflammatory events 22851693_Beta-TrCP deficiency abolished the translocation TAK1-TRAF6 complex from the membrane to the cytosol, resulting in a diminishment of the IL-1-induced TAK1-dependent pathway. 22981905_Lys63-linked TAK1 polyubiquitination at Lys-158 is required for Dox-induced NF-kappaB activation. 23463759_Mice that lack the ortholog of this gene display severe spinal curvature, subfertility, premature aging, and prostatic intraepithelial neoplasia (PIN) development. 23609451_TR4-Oct4-IL1Ra axis may play a critical role in the development of chemoresistance in the PCa stem/progenitor cells. 23653479_TR4 transcriptionally activates proopiomelanocortin through binding of a direct repeat 1 response element in the promoter, and that this is enhanced by MAPK-mediated TR4 phosphorylation. 23700229_The results provide a proof-of-concept that TAK1 inhibition significantly increases the sensitivity of neuroblastoma cells to chemotherapy-induced cell-death. 23770285_These results reveal Sef-S actives Lys63-linked TAK1 polyubiquitination on lysine 209, induces TAK1-mediated JNK and p38 activation and also results apoptosis in 293T cells. 24028082_Findings reveal the role of TAK1 in thermoresistance and show that the mediation is independent of NF-kappaB phosphorylation but is dependent on TNFAIP3 and IL-8 induction. 24082073_TAK1 regulates H. pylori-mediated early JNK activation and cytokine production. 24337384_Small interfering RNA-mediated silencing of TRAF6 and TAK1. 24418622_We provide evidence for an intimate mutual control of the IKK complex by mitogen-activated protein kinase kinase kinase 3 (MEKK3) and transforming growth factor beta activated kinase 1 (TAK1). 24565101_miR-26b suppresses NF-kappaB signaling and sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis by inhibiting the expression of TAK1 and TAB3. 24583925_TR4 nuclear receptor functions as a tumor suppressor for prostate tumorigenesis via modulation of DNA damage/repair system. 24728340_our findings describe a TAK1-dependent, beta-catenin- and Sp1-mediated signaling cascade activated downstream of TGF-beta which regulates WNT-5A induction. 24801688_TAK1 is a key regulator of receptor crosstalk between BCR and TLR9. 24811540_study reveals that the TR4 regulatory network is far more complex than previously appreciated and that TR4 regulates basic, essential biological processes during the terminal differentiation of human erythroid cells. 24907344_TR4 binding with keto-MA features a unique association of host nuclear receptor with a bacterial lipid and adds to the presently known ligand repertoire beyond dietary lipids. 24912525_Knockdown of endogenous TAK1 significantly attenuated the ability of Vpr to activate NF-kappaB and AP-1. 24942571_Authors demonstrate that enterovirus 71 3C interacts with TAB2 and TAK1 and suppresses cytokine expression via cleavage of the TAK1 complex proteins. 24975468_A role for TR4 in prostate cancer metastasis via CCL2/CCR2 signaling. 25277189_TAK1 activates NF-kappaB signaling activity such activated TAK1/NF-kappaB signaling cascade is indispensable in promoting ovarian cancer cell growth 25320283_Resolvin D1 attenuates the viral mimic-induced inflammatory signaling in human airway epithelial cells via TAK1. 25475726_CXC195 suppresses proliferation and inflammatory response in LPS-induced human hepatocellular carcinoma cells via regulating TLR4-MyD88-TAK1-mediated NF-kappaB and MAPK pathway 25512613_the TAK1-NLK pathway is a novel regulator of basal or IL-1beta-triggered C/EBP activation though stabilization of ATF5 25561507_Compound loss of function of nuclear receptors Tr2 and Tr4 leads to induction of murine embryonic beta-type globin genes. 25623427_IHC staining showed higher TR4 level, more macrophage infiltration, lower TIMP-1 and stronger MMP2/MMP9 in tumor tissues. 25704183_Helicobacter pylori induces internalization of EGFR via novel TAK1-p38-serine activation pathway which is independent of HB-EGF. 25833838_Results suggest that the testicular nuclear receptor 4 (TR4) and oncogene EZH2 signaling may play a critical role in the prostate cancer stem/progenitor cell invasion. 25925376_PCa patients receiving TZD treatment who have one allele TR4 deletion. 25980442_TR4 may increase prostate cancer metastasis and invasion via decreasing the miR-373-3p expression that resulted in the activation of the TGFbetaR2/p-Smad3 signals. 26144287_TR4 expression in NSCLC samples is significantly associated with poor clinicopathological features, and TR4 plays an important role in the metastatic capacity of NSCLC cells by EMT regulation. 26178291_TR4 was found to mediate the prostate cancer cells' radio-sensitivity. 26240016_USP18 negatively regulates NF-kappaB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. 26334375_IFIT5 promotes SeV-induced IKK phosphorylation and NF-kappaB activation by regulating the recruitment of IKK to TAK1. 26432169_DK1 inhibits the formation of the TAK1-TAB2-TRAF6 complex and leads to the inhibition of TRAF6 ubiquitination. 26617776_miR-203 represses NF-kappaB signaling via targeting TAK1 and PI3KCA and miR-203 overexpression may contribute to the COPD initiation. 26620228_Together, these results demonstrate that LYTAK1 inhibits LPS-induced production of several pro-inflammatory cytokines and endotoxin shock probably through blocking TAK1-regulated signalings. 26884850_TAK1/TAB1 expression in non-small cell lung carcinoma tissue is significantly increased and closely associated with patient clinical prognosis. 27050071_Altering TR4-ATF3 signaling increases the efficacy of cisplatin to suppress hepatocellular carcinoma growth/progression. 27253665_TR4 binds GR to play an important role in glucocorticoid-directed corticotroph tumor POMC regulation in addition to modulating glucocorticoid actions on other GR targets. 27530352_High TAK1 expression is associated with the progression of hepatocellular carcinoma. 28356387_this study shows that TAP1 plays a novel role in the negative regulation of virus-triggered NF-kappaB signaling and the innate immune response by targeting the TAK1 complex 28608226_blockage of RhoA/ROCK repressed the TAK1/NOD2-mediated NF-kappaB pathway in HaCaT cells exposed to UVB. 28623141_SIRT7 inhibits TR4 degradation by deacetylation of DDB1. 28652310_Here, we report that Pseudomonas aeruginosa ExoY inhibits proinflammatory cytokine production through suppressing the activation of TAK1 as well as downstream NF-kappaB and mitogen-activated protein (MAP) kinases. 29197138_Study found that TR4 might be able to function through activation of the AKT3 expression to drive the EMT phenotype and enhance the seminoma cell proliferation and invasion. 29449527_TR4 may function as a suppressor of hepatocellular carcinoma metastasis by down-regulating the expression of EphA2. 29712904_The senescence-associated secretory phenotype is potentiated by feedforward regulatory mechanisms involving Zscan4 and TAK1. 29807222_Targeting the Cripto-1/TAK-1/NF-kappaB/Survivin pathway may be an effective approach to combat apoptosis resistance in cancer. 29973687_Promotes clear cell renal carcinoma vasculogenic mimicry (VM) formation and its associated metastasis by modulating the miR490-3p-vimentin signals 30391931_TAK1, receptor-interacting kinase 1 (RIPK1) as well as canonical and non-canonical NF-kappaB signaling are differentially involved in SM-induced cell death in breast cancer cells. 30518750_This study demonstrated that the NR2C2-upstream open reading frame impaired the pivotal roles that UCA1-miR-627-5p-NR2C2 feedback loop had in regulating the malignancies of glioma cells by targeting NR2C2 directly. 30626936_Authors reveal a propensity of the alternatively spliced TAK1 isoform TAK1E12 to cause drug resistance due to its activity in supporting EMT and NF-kappaB survival signaling. 30707992_dampening oxidative stress with N-acetylcysteine (NAC) lowered hypertrophy in MTG1 KO to WT levels. Collectively, our data indicate that MTG1 protects against pressure overload-induced cardiac hypertrophy and dysfunction by preserving mitochondrial function and reducing oxidative stress and downstream TAK1 stress signaling. 30799163_IL-1beta regulates the expression of NGF and COX-2, rotator cuff tear pain-related molecules in the subacromial bursa cells, through TAK1. 31130074_Taken together, these results suggest that enterohemorrhagic Escherichia coli Tir negatively regulates proinflammatory responses by inhibiting the activation of TAK1, which is essential for immune evasion. 31347268_TAK1 governs p62 action, switching it from an autophagy receptor to a signaling platform. This ability of TAK1 to disable p62 as an autophagy receptor may allow certain autophagic substrates to accumulate when needed for cellular functions. 31501521_Cell hypoxia increases TR4, which plays a critical role in regulating renal cell carcinoma (RCC) resistance to sunitinib through transcriptional regulation of lncTASR (ENST00000600671.1) which might then increase AXL protein expression via enhancing the stability of AXL mRNA to increase the sunitinib resistance. 31748715_Targeting TR4 nuclear receptor with antagonist bexarotene increases docetaxel sensitivity to better suppress the metastatic castration-resistant prostate cancer progression. 32113875_CPEB1 and CPEB4 are involved in the regulation of the TAK1 and Smad signalings in human macrophages and dermal fibroblasts 32229191_TAB3 upregulates PIM1 expression by directly activating the TAK1-STAT3 complex to promote colorectal cancer growth. 32525617_LncRNA H19 aggravates TNF-alpha-induced inflammatory injury via TAK1 pathway in MH7A cells. 32581266_Farnesoid X receptor activation inhibits TGFBR1/TAK1-mediated vascular inflammation and calcification via miR-135a-5p. 32768524_Targeting the radiation-induced TR4 nuclear receptor-mediated QKI/circZEB1/miR-141-3p/ZEB1 signaling increases prostate cancer radiosensitivity. 32820312_[Transforming growth factor-beta-activated kinase 1 and pathological myocardial hypertrophy]. 33139544_Essential role of the linear ubiquitin chain assembly complex and TAK1 kinase in A20 mutant Hodgkin lymphoma. 33650661_Use of miR145 and testicular nuclear receptor 4 inhibition to reduce chemoresistance to docetaxel in prostate cancer. 34075691_The stabilization of yes-associated protein by TGFbeta-activated kinase 1 regulates the self-renewal and oncogenesis of gastric cancer stem cells. 34830460_Takinib Inhibits Inflammation in Human Rheumatoid Arthritis Synovial Fibroblasts by Targeting the Janus Kinase-Signal Transducer and Activator of Transcription 3 (JAK/STAT3) Pathway. | ENSMUSG00000005893 | Nr2c2 | 2080.49353 | 1.1638398 | 0.2188925471 | 0.08828333 | 6.179923e+00 | 1.292079e-02 | 1.847630e-01 | No | Yes | 2679.892295 | 264.228966 | 2209.670420 | 213.687800 | |
| ENSG00000177548 | 79874 | RABEP2 | protein_coding | Q9H5N1 | FUNCTION: Plays a role in membrane trafficking and in homotypic early endosome fusion (PubMed:9524116). Participates in arteriogenesis by regulating vascular endothelial growth factor receptor 2/VEGFR2 cell surface expression and endosomal trafficking (PubMed:29425100). By interacting with SDCCAG8, localizes to centrosomes and plays a critical role in ciliogenesis (PubMed:27224062). {ECO:0000269|PubMed:27224062, ECO:0000269|PubMed:29425100, ECO:0000269|PubMed:9524116}. | Acetylation;Alternative splicing;Cell projection;Cilium biogenesis/degradation;Coiled coil;Cytoplasm;Cytoskeleton;Endocytosis;Endosome;Phosphoprotein;Protein transport;Reference proteome;Transport | hsa:79874; | centrosome [GO:0005813]; ciliary basal body [GO:0036064]; cytosol [GO:0005829]; early endosome [GO:0005769]; intracellular membrane-bounded organelle [GO:0043231]; growth factor activity [GO:0008083]; GTPase activator activity [GO:0005096]; cell projection organization [GO:0030030]; endocytosis [GO:0006897]; protein transport [GO:0015031]; regulation of cilium assembly [GO:1902017] | 17205062_Pyk2 regulation is associated with increased expression of Fra-1 and JunD, activator protein-1 transcription factors known to be required for involucrin expression. 17254320_FRA-1 expression, including its intracellular localization, may be considered a useful marker for hyperplastic and neoplastic proliferative breast disorders 29247183_Phosphorylation regulates RABEP2 function. 30455363_Results found that H. pylori upregulates RABEP2 in human gastric epithelium. In addition, the level of RABEP2 expression increases with severity of gastric malignant lesions in vivo. | ENSMUSG00000030727 | Rabep2 | 206.05522 | 0.8459106 | -0.2414228158 | 0.21825510 | 1.215134e+00 | 2.703176e-01 | 6.607395e-01 | No | Yes | 213.595065 | 29.673269 | 242.886930 | 32.778218 | ||
| ENSG00000178149 | 55152 | DALRD3 | protein_coding | Q5D0E6 | FUNCTION: Involved in tRNA methylation. Facilitates the recognition and targeting of tRNA(Arg)(CCU) and tRNA(Arg)(UCU) substrates for N(3)-methylcytidine modification by METTL2A and METTL2B. {ECO:0000269|PubMed:32427860}. | Alternative splicing;Disease variant;Epilepsy;Reference proteome | The exact function of this gene is not known. It encodes a protein with a DALR anticodon binding domain similar to that of class Ia aminoacyl tRNA synthetases. This gene is located in a cluster of genes (with a complex sense-anti-sense genome architecture) on chromosome 3, and contains two micro RNA (miRNA) precursors (mir-425 and mir-191) in one of its introns. Preferential expression of this gene (the miRNAs and other genes in the cluster) in testis suggests a role of this gene in spermatogenesis (PMID:19906709). [provided by RefSeq, Feb 2013]. | hsa:55152; | arginine-tRNA ligase activity [GO:0004814]; ATP binding [GO:0005524]; tRNA binding [GO:0000049]; arginyl-tRNA aminoacylation [GO:0006420]; tRNA C3-cytosine methylation [GO:0106217] | 32427860_DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. | ENSMUSG00000019039 | Dalrd3 | 395.10260 | 0.8049821 | -0.3129713541 | 0.15808392 | 3.888563e+00 | 4.861600e-02 | 3.332161e-01 | No | Yes | 382.689808 | 36.051656 | 461.156451 | 42.054866 | |
| ENSG00000178401 | 79962 | DNAJC22 | protein_coding | Q8N4W6 | FUNCTION: May function as a co-chaperone. | Chaperone;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:79962; | integral component of membrane [GO:0016021] | ENSMUSG00000038009 | Dnajc22 | 28.04613 | 1.4210729 | 0.5069805814 | 0.54087091 | 8.828368e-01 | 3.474259e-01 | No | Yes | 34.741260 | 9.319688 | 25.199764 | 6.723546 | ||||
| ENSG00000178537 | 788 | SLC25A20 | protein_coding | O43772 | FUNCTION: Mediates the transport of acylcarnitines of different length across the mitochondrial inner membrane from the cytosol to the mitochondrial matrix for their oxidation by the mitochondrial fatty acid-oxidation pathway. {ECO:0000250|UniProtKB:P97521}. | Acetylation;Disease variant;Lipid transport;Membrane;Mitochondrion;Mitochondrion inner membrane;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | This gene product is one of several closely related mitochondrial-membrane carrier proteins that shuttle substrates between cytosol and the intramitochondrial matrix space. This protein mediates the transport of acylcarnitines into mitochondrial matrix for their oxidation by the mitochondrial fatty acid-oxidation pathway. Mutations in this gene are associated with carnitine-acylcarnitine translocase deficiency, which can cause a variety of pathological conditions such as hypoglycemia, cardiac arrest, hepatomegaly, hepatic dysfunction and muscle weakness, and is usually lethal in new born and infants. [provided by RefSeq, Jul 2008]. | hsa:788; | cytosol [GO:0005829]; integral component of membrane [GO:0016021]; mitochondrial inner membrane [GO:0005743]; mitochondrion [GO:0005739]; acyl carnitine transmembrane transporter activity [GO:0015227]; carnitine shuttle [GO:0006853]; carnitine transmembrane transport [GO:1902603]; in utero embryonic development [GO:0001701] | 15057979_A deficiency in CACT was treated with a carnitine diet and administration of medium-chain triglycerides. 15365988_The clinical, biochemical, & molecular features of 6 CACT-deficient patients from Italy, Spain, & North America who had significant clinical heterogeneity are reported. 5 novel & 3 previously reported mutations were found. 15515015_The modulation of CACT expression has consequences for CPT 1 activity, while the biologic effects of acetyl-carnitine are not associated with a generic supply of energy compounds but to the anaplerotic property of the molecule. 17508264_Report the outcome of two siblings with CACT deficiency. 18307102_Functional analysis of mutations of residues Pro278 and Ala279 in A. nidulans, together with kinetic data in reconstituted liposomes, suggest a predominant structural role for these amino acids. 19748481_PPARalpha regulates the expression of human SLC25A20 via the peroxisome proliferator responsive element. 20877624_Observational study of gene-disease association. (HuGE Navigator) 21130740_These results show that FOXA and Sp1 sites in HepG2 cells and only the Sp1 site in HEK293 and SK-N-SH cells have a critical role in the transcriptional regulation of the CAC proximal promoter. 22560224_Results show Steroid Receptor Coactivator-3 (SRC-3) plays a central role in long chain fatty acid metabolism by directly regulating carnitine/acyl-carnitine translocase (CACT) gene expression. 23266187_Compares and contrasts all the known human SLC25A* genes and includes functional information. 23322164_CPT2 and CACT are crucial for mitochondrial acylcarnitine formation and export to the extracellular fluids in mitochondrial fatty acid beta-oxidation disorders. 24088670_C.576G>A, c.106-2a>t and c.516T>C are novel CACT gene mutations. 25325845_The antiport mode of transport, typical of mitochondrial carriers such as CAC, results from coupling of uniport reactions in opposite directions mediated by specific amino acid residues. 28671672_We provide evidence that the downregulation of hsa-miR-124-3p, hsa-miR-129-5p and hsa-miR-378 induced an increase in both expression and activity of CPT1A, CACT and CrAT in malignant prostate cells. 29137068_we report the first 2 cases of CACTD identified from the mainland China. Apart from a founder mutation c.199-10T>G, we have identified a novel c.1A>G mutation. Patients with Carnitine-acylcarnitine translocase deficiency with a genotype of c.199-10T>G mutation usually presents with a severe clinical phenotype. Early recognition and appropriate treatment is crucial in this highly lethal disorder. 34449152_Clinical and molecular characteristics of carnitineacylcarnitine translocase deficiency with c.270delC and a novel c.408C>A variant. 34626609_Novel mutations associated with carnitine-acylcarnitine translocase and carnitine palmitoyl transferase 2 deficiencies in Malaysia. | ENSMUSG00000032602 | Slc25a20 | 568.96983 | 0.9562488 | -0.0645421250 | 0.13165201 | 2.399517e-01 | 6.242410e-01 | 8.791639e-01 | No | Yes | 630.165507 | 113.432521 | 643.327265 | 113.080062 | |
| ENSG00000180182 | 9282 | MED14 | protein_coding | O60244 | FUNCTION: Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. {ECO:0000269|PubMed:15340088, ECO:0000269|PubMed:15625066, ECO:0000269|PubMed:16595664}. | 3D-structure;Activator;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation | The activation of gene transcription is a multistep process that is triggered by factors that recognize transcriptional enhancer sites in DNA. These factors work with co-activators to direct transcriptional initiation by the RNA polymerase II apparatus. The protein encoded by this gene is a subunit of the CRSP (cofactor required for SP1 activation) complex, which, along with TFIID, is required for efficient activation by SP1. This protein is also a component of other multisubunit complexes e.g. thyroid hormone receptor-(TR-) associated proteins which interact with TR and facilitate TR function on DNA templates in conjunction with initiation factors and cofactors. This protein contains a bipartite nuclear localization signal. This gene is known to escape chromosome X-inactivation. [provided by RefSeq, Jul 2008]. | hsa:9282; | core mediator complex [GO:0070847]; mediator complex [GO:0016592]; membrane [GO:0016020]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; nuclear receptor coactivator activity [GO:0030374]; transcription coactivator activity [GO:0003713]; transcription coregulator activity [GO:0003712]; vitamin D receptor binding [GO:0042809]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription initiation from RNA polymerase II promoter [GO:0060261]; positive regulation of transcription, DNA-templated [GO:0045893]; regulation of transcription by RNA polymerase II [GO:0006357]; stem cell population maintenance [GO:0019827] | 12509459_DRIP150 binds to ISGF3 and regulates transcription 12825353_CRSP2 gene is expressed in the retina and its exact genomic location is on Xp11.4 between DXS1368 and DXS993 15625066_Coactivation of ERalpha by DRIP150 in ZR-75 cells is NR box-independent and requires a novel sequence with putative alpha-helical structure. 16239257_MED14 and MED1 are used by glucocorticoid receptor in a gene-specific manner, providing a mechanism for promoter selectivity by glucocorticoid receptor 17306756_Coactivator that enhances estrogen receptor alpha- and specificity protein (SP)-1-mediated transactivation in breast cancer cells. 23572530_VitD-mediated stimulation of GC anti-inflammatory affects human monocytes in a process involving GM-CSF and MED14 25383669_This results from a dramatically enhanced ability of MED14-containing complexes to associate with Pol II. 28813667_Functional analysis implicates TNRC6A, NAT10, MED14, and WDR5 in RNA-mediated transcriptional activation. | ENSMUSG00000064127 | Med14 | 2990.20727 | 0.9799480 | -0.0292228336 | 0.08144002 | 1.285819e-01 | 7.199069e-01 | 9.170934e-01 | No | Yes | 3526.748197 | 452.179210 | 3393.989030 | 425.570789 | |
| ENSG00000180818 | 3226 | HOXC10 | protein_coding | Q9NYD6 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | DNA-binding;Developmental protein;Homeobox;Isopeptide bond;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene belongs to the homeobox family of genes. The homeobox genes encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, which are located on different chromosomes and consist of 9 to 11 genes arranged in tandem. This gene is one of several homeobox HOXC genes located in a cluster on chromosome 12. The protein level is controlled during cell differentiation and proliferation, which may indicate this protein has a role in origin activation. [provided by RefSeq, Jul 2008]. | hsa:3226; | chromatin [GO:0000785]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; sequence-specific double-stranded DNA binding [GO:1990837]; anterior/posterior pattern specification [GO:0009952]; embryonic limb morphogenesis [GO:0030326]; negative regulation of cold-induced thermogenesis [GO:0120163]; neuromuscular process [GO:0050905]; positive regulation of cell population proliferation [GO:0008284]; positive regulation of transcription by RNA polymerase II [GO:0045944]; proximal/distal pattern formation [GO:0009954]; regulation of transcription by RNA polymerase II [GO:0006357]; skeletal system development [GO:0001501]; spinal cord motor neuron cell fate specification [GO:0021520] | 12853486_Early mitotic dedgradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. 17974957_Cervical cancer cells with high endogenous levels of HOXC10 were less invasive after short hairpin RNA-mediated knockdown of HOXC10 expression; findings support a key role for the HOXC10 homeobox protein in cervical cancer progression 19522674_Data demonstrate that HOXC10 is a gene that may discriminate between amnion-derived mesenchymal stem cells (MSCs) and decidua-derived MSCs. 24670685_Reduced HOXC10 in vitro and in xenografts resulted in decreased apoptosis and caused antiestrogen resistance. 26279264_This study investigates the role and clinical implication of HOXC10 in human thyroid cancer. 26647900_HOXC9 and HOXC10 may play an important role in the development of obesity, adverse fat distribution, and subsequent alterations in whole-body metabolism and adipose tissue function. 28474998_These results indicated that HOXC10 might be a diagnostic marker for osteosarcoma and could be a potential molecular target for the therapy of osteosarcoma 28605223_that HOXC10 decreased the MSC osteogenic differentiation potential 28656883_Furthermore, our results showed that ectopic expression of HOXC10 could reverse inhibition of metastasis by overexpressed miR-136 in GC-9811P cells. Our findings provide new insights into the role of miR-136 in the gastric cancer-specific peritoneal metastasis and implicate the potential application of miR-136 in gastric cancer peritoneal metastasis therapy. 29676849_HOXC10 directly binds to the PD-L2 and TDO2 promoter regions thus stimulating proliferation, invasion and induction of immunosuppressive gene expression in glioma. 29753747_HOXC10 may promote invasion and migration of gastric cancer cells by regulating ATM/NF-kappaB signaling pathway. 29768063_Overexpression of HOXC10 promotes glioblastoma cell progression to a poor prognosis via the PI3K/AKT signalling pathway 30343692_Our study revealed a new mechanism mediated by CHD7 for its role in promoting HOXC10 expression and contribute to mammary oncogenesis. 30353595_Data showed that lncHOXC-AS3 was required for osteogenesis in BM-MSCs by enhancing HOXC10 expression. 30429891_Protein arginine methyltransferase 5 (PRMT5) and WD repeat domain 5 (WDR5), both of which regulate histone post-translational modifications, were required for HOXC10-mediated VEGFA upregulation. Importantly, a significant correlation between HOXC10 levels and VEGFA expression was observed in a cohort of human gliomas. 30673899_High HOXC10 expression is associated with Recurrence in Gastric Cancer. 31115563_HOXC10 expression is upregulated in gastric cancer through DNA demethylation, and HOXC10 overexpression increases proliferation and migration of gastric cancer cells. 31552684_HOXC10 promotes cell migration, invasion, and tumor growth in gastric carcinoma cells through upregulating proinflammatory cytokines. 32111444_MircoRNA-129-5p suppresses the development of glioma by targeting HOXC10. 32206125_Interleukin 1beta-mediated HOXC10 Overexpression Promotes Hepatocellular Carcinoma Metastasis by Upregulating PDPK1 and VASP. 32243838_A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. 32356314_MiR-129-5p induces cell cycle arrest through modulating HOXC10/Cyclin D1 to inhibit gastric cancer progression. 32552008_MiR-129-5p Restrains Apatinib Resistance in Human Gastric Cancer Cells Via Downregulating HOXC10. 32587398_HOXC10 upregulation confers resistance to chemoradiotherapy in ESCC tumor cells and predicts poor prognosis. 34302808_The lncRNA HMS recruits RNA-binding protein HuR to stabilize the 3'-UTR of HOXC10 mRNA. 34320348_Identification of cis-HOX-HOXC10 axis as a therapeutic target for colorectal tumor-initiating cells without APC mutations. 34523692_Transcription factor HOXC10 activates the expression of MTFR2 to regulate the proliferation, invasion and migration of colorectal cancer cells. 34763232_HOXC6/8/10/13 predict poor prognosis and associate with immune infiltrations in glioblastoma. 34983296_Homeodomain-containing gene 10 contributed to breast cancer malignant behaviors by activating Interleukin-6/Janus kinase 2/Signal transducer and activator of transcription 3 pathway. 35316872_[The effect of HOXC10 gene on biological behaviors of glioma cells and mechanism in tumor microenvironment]. | ENSMUSG00000022484 | Hoxc10 | 424.78892 | 0.9777273 | -0.0324958963 | 0.17510822 | 3.437381e-02 | 8.529139e-01 | 9.601201e-01 | No | Yes | 512.320035 | 66.323379 | 496.136034 | 62.924495 | |
| ENSG00000181894 | 79673 | ZNF329 | protein_coding | Q86UD4 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Phosphoprotein;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:79673; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; regulation of transcription by RNA polymerase II [GO:0006357] | 27556418_TYROBP influences a batch of genes that are related to Alzheimer's disease; ZNF329 and RB1 significantly regulate those 'mesenchymal' gene expression signature genes for brain tumors. By merely leveraging gene expression data, Context Based Dependency Network (CBDN) can efficiently infer the existence of gene-gene interactions as well as their regulatory directions. | ENSMUSG00000057894 | Zfp329 | 135.29412 | 1.6797143 | 0.7482158555 | 0.25855525 | 8.462203e+00 | 3.626025e-03 | 1.009526e-01 | No | Yes | 210.955710 | 38.731720 | 119.047711 | 21.772325 | ||
| ENSG00000182224 | 124637 | CYB5D1 | protein_coding | Q6P9G0 | Alternative splicing;Heme;Iron;Metal-binding;Reference proteome | hsa:124637; | metal ion binding [GO:0046872] | Mouse_homologues 17971504_Cyb5d1 is most abundantly expressed in tissues rich in highly ciliated cells, such as olfactory sensory neurons, and is predicted to be important to cilia. | ENSMUSG00000044795 | Cyb5d1 | 1112.10439 | 1.0589846 | 0.0826816188 | 0.13418350 | 3.796731e-01 | 5.377782e-01 | 8.391324e-01 | No | Yes | 1955.240251 | 317.823135 | 1838.194878 | 291.482613 | |||
| ENSG00000182389 | 785 | CACNB4 | protein_coding | O00305 | FUNCTION: The beta subunit of voltage-dependent calcium channels contributes to the function of the calcium channel by increasing peak calcium current, shifting the voltage dependencies of activation and inactivation, modulating G protein inhibition and controlling the alpha-1 subunit membrane targeting. {ECO:0000269|PubMed:11880487}. | 3D-structure;Alternative splicing;Calcium;Calcium channel;Calcium transport;Disease variant;Epilepsy;Ion channel;Ion transport;Methylation;Phosphoprotein;Reference proteome;SH3 domain;Transport;Voltage-gated channel | This gene encodes a member of the beta subunit family of voltage-dependent calcium channel complex proteins. Calcium channels mediate the influx of calcium ions into the cell upon membrane polarization and consist of a complex of alpha-1, alpha-2/delta, beta, and gamma subunits in a 1:1:1:1 ratio. Various versions of each of these subunits exist, either expressed from similar genes or the result of alternative splicing. The protein encoded by this locus plays an important role in calcium channel function by modulating G protein inhibition, increasing peak calcium current, controlling the alpha-1 subunit membrane targeting and shifting the voltage dependence of activation and inactivation. Certain mutations in this gene have been associated with idiopathic generalized epilepsy (IGE), juvenile myoclonic epilepsy (JME), and episodic ataxia, type 5. [provided by RefSeq, Aug 2016]. | hsa:785; | cytoplasmic side of plasma membrane [GO:0009898]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; synapse [GO:0045202]; voltage-gated calcium channel complex [GO:0005891]; voltage-gated calcium channel activity [GO:0005245]; chemical synaptic transmission [GO:0007268]; neuromuscular junction development [GO:0007528]; regulation of voltage-gated calcium channel activity [GO:1901385] | 16866717_The novel nucleotide substitution T87C (D29D)in CACNB4 was observed in 2 migrainous vertigo patients and was not present in control DNA samples. 18446307_No pathogenic mutation were identified in CACNB4. 18712068_plays a role in neurotransmitter release. 18755274_Observational study of gene-disease association. (HuGE Navigator) 18755274_proband identified with severe myoclonic epilepsy in infancy heterozygous for de novo SCN1A nonsense mutation & CACNB4 missense mutation (R468Q); greater Ca(v)2.1 currents caused by the mutation may increase neurotransmitter release in excitatory neurons 20200978_Observational study of gene-disease association. (HuGE Navigator) 21220418_The Ca2+ channel beta4c subunit interacts with heterochromatin protein 1 gama via a PXVXL binding motif. 21297076_CACNB4 is associated with acute lung injury in mice 22892567_Cacnb4 directly couples electrical activity to gene expression, a process defective in juvenile epilepsy 23756480_Genome-wide association studies identify CACNB4 mutation releated to juvenile myoclonic epilepsy. 24875574_The nuclear targeting properties of the truncated beta(4b(1-481)) subunit in tsA-201 cells, skeletal myotubes, and in hippocampal neurons, were investigated. 32176688_The homozygous CACNB4 p.(Leu126Pro) variant underlies the severe neurological phenotype in the two siblings. | ENSMUSG00000017412 | Cacnb4 | 109.20625 | 0.8817547 | -0.1815506945 | 0.28338591 | 4.085840e-01 | 5.226891e-01 | 8.314481e-01 | No | Yes | 103.130509 | 34.151294 | 115.356064 | 37.609033 | |
| ENSG00000182841 | 91695 | RRP7BP | transcribed_unprocessed_pseudogene | 641.87447 | 0.9728771 | -0.0396705173 | 0.12960406 | 9.367737e-02 | 7.595532e-01 | 9.299340e-01 | No | Yes | 688.993454 | 84.453086 | 704.558018 | 84.421828 | ||||||||||
| ENSG00000182957 | 221178 | SPATA13 | protein_coding | Q96N96 | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for RHOA, RAC1 and CDC42 GTPases. Regulates cell migration and adhesion assembly and disassembly through a RAC1, PI3K, RHOA and AKT1-dependent mechanism. Increases both RAC1 and CDC42 activity, but decreases the amount of active RHOA. Required for MMP9 up-regulation via the JNK signaling pathway in colorectal tumor cells. Involved in tumor angiogenesis and may play a role in intestinal adenoma formation and tumor progression. {ECO:0000269|PubMed:17145773, ECO:0000269|PubMed:17599059, ECO:0000269|PubMed:19151759, ECO:0000269|PubMed:19893577, ECO:0000269|PubMed:19934221}. | Alternative splicing;Cell membrane;Cell projection;Cytoplasm;Guanine-nucleotide releasing factor;Membrane;Phosphoprotein;Reference proteome;SH3 domain | hsa:221178; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; filopodium [GO:0030175]; lamellipodium [GO:0030027]; nucleoplasm [GO:0005654]; ruffle membrane [GO:0032587]; guanyl-nucleotide exchange factor activity [GO:0005085]; identical protein binding [GO:0042802]; cell migration [GO:0016477]; filopodium assembly [GO:0046847]; lamellipodium assembly [GO:0030032]; regulation of cell migration [GO:0030334]; regulation of small GTPase mediated signal transduction [GO:0051056] | 17599059_These results suggest that similar to Asef, Asef2 plays an important role in cell migration, and that Asef2 activated by truncated mutant APC is required for aberrant migration of colorectal tumor cells. 19151759_Asef2, Neurabin2 and APC cooperatively regulate actin cytoskeletal organization and are required for HGF-induced cell migration. 19910543_Observational study of gene-disease association. (HuGE Navigator) 19934221_Asef2 activates Rac1 to modulate adhesion and actin dynamics and thereby regulate cell migration. 24144700_A new role for Rac activation, promoted by Asef2, in modulating actomyosin contractility. 24874604_Phosphorylation of S106 modulates Asef2 guanine nucleotide exchange factor activity and Asef2-mediated cell migration and adhesion turnover. 32339198_Mutations in SPATA13 cause primary angle closure glaucoma. | ENSMUSG00000021990 | Spata13 | 327.76382 | 1.3585192 | 0.4420349820 | 0.20076696 | 4.873308e+00 | 2.727518e-02 | 2.649776e-01 | No | Yes | 474.383710 | 72.151859 | 338.484222 | 50.521976 | ||
| ENSG00000183401 | 126075 | CCDC159 | protein_coding | P0C7I6 | Alternative splicing;Coiled coil;Reference proteome | hsa:126075; | ENSMUSG00000006241 | Ccdc159 | 31.64778 | 0.8978042 | -0.1555271991 | 0.52219007 | 8.800268e-02 | 7.667319e-01 | No | Yes | 34.727619 | 6.986682 | 37.915768 | 7.319812 | ||||||
| ENSG00000183495 | 57634 | EP400 | protein_coding | Q96L91 | FUNCTION: Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. May be required for transcriptional activation of E2F1 and MYC target genes during cellular proliferation. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. May regulate ZNF42 transcription activity. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AZ1 from the nucleosome. {ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:24463511}. | ATP-binding;Acetylation;Alternative splicing;Chromatin regulator;DNA-binding;Helicase;Hydrolase;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome | hsa:57634; | NuA4 histone acetyltransferase complex [GO:0035267]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; Swr1 complex [GO:0000812]; ATP binding [GO:0005524]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; helicase activity [GO:0004386]; hydrolase activity [GO:0016787]; protein antigen binding [GO:1990405]; chromatin organization [GO:0006325]; histone H2A acetylation [GO:0043968]; histone H4 acetylation [GO:0043967]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; regulation of cell cycle [GO:0051726] | 15655109_p400 is a component of the p53-p21(WAF1/CIP1/sid1) pathway, regulating the p21 transcription and cell senescence induction program. 15741165_p400 is a regulator of the ARF-p53 pathway and a component of the cellular machinery that couples proliferation to cell death 16601686_Tip60 and p400 play distinct roles in DNA damage-induced apoptosis and underline the importance of the Tip60 complex and its regulation in the proper control of cell fate. 18413597_These findings establish that Myc, via p400, is an essential downstream target of adenovirus E1A. 18985155_Myc recruits the Tip60/p400 complex to achieve a coordinated histone acetylation/exchange reaction at activated promoters. 19169279_p400/Tip60 ratio is critical for colon cancer cells proliferation and response to therapeutic drugs through the control of stress-response pathways. 20351180_Results suggest that p400 represses basal p21 gene expression through dual mechanisms that include direct inhibition of TIP60 and ATP-dependent positioning of H2A.Z at the promoter. 20548951_Depletion of p400 indeed increases intracellular reactive oxygen species levels and causes the appearance of DNA damage 20876283_p400 is a novel DNA damage response protein and p400-mediated alterations in nucleosome and chromatin structure promote both chromatin ubiquitination and the accumulation of brca1 and 53BP1 at sites of DNA damage. 23064015_incorporation of the histone variant H2A.Z at the promoter regions of PPARgamma target genes by p400/Brd8 is essential to allow fat cell differentiation 23146670_age-dependent p400 downregulation and loss of H2A.Z localisation may contribute to the onset of replicative senescence through a sustained high rate of p21 transcription 23266955_The p400 ATPase is required for DNA repair by homologous recombination. 23982490_Our data suggest that the highly proliferative, decreased-p400 subgroup of renal cell carcinomas represents tumors that are characterized by a loss of the tumor-suppressive mechanism of senescence. 24332572_Data indicate that three genes, namely Chemokine (C-X-C motif) receptor 2 (CXCR2), C-C chemokine receptor type 2 (CCR2) and E1A-Binding Protein P400 (EP400), were able to identify HCC individually with accuracies of 82.4%, 78.4% and 65%, respectively. 26578561_All together these results show that p400 acts as a brake to prevent alternative End Joining-dependent genetic instability and underline its potential value as a clinical marker. 26669263_EP400 deposits H3.3 into chromatin alongside H2AZ and contributes to gene regulation after Pol II pre-initiation complex assembly. 27814680_Taken together, the findings suggest that a protein-protein interaction between ATM and p400 ATPase occurs independently of DNA damage and contributes to efficient DNA damage response and repair. 33099470_TIP60/P400/H4K12ac Plays a Role as a Heterochromatin Back-up Skeleton in Breast Cancer. | ENSMUSG00000029505 | Ep400 | 2773.17633 | 0.9821079 | -0.0260465919 | 0.07917114 | 1.081479e-01 | 7.422627e-01 | 9.243184e-01 | No | Yes | 2642.759745 | 243.258653 | 2615.797720 | 235.414963 | ||
| ENSG00000183955 | 387893 | KMT5A | protein_coding | Q9NQR1 | FUNCTION: Protein-lysine N-methyltransferase that monomethylates both histones and non-histone proteins. Specifically monomethylates 'Lys-20' of histone H4 (H4K20me1). H4K20me1 is enriched during mitosis and represents a specific tag for epigenetic transcriptional repression. Mainly functions in euchromatin regions, thereby playing a central role in the silencing of euchromatic genes. Required for cell proliferation, probably by contributing to the maintenance of proper higher-order structure of DNA during mitosis. Involved in chromosome condensation and proper cytokinesis. Nucleosomes are preferred as substrate compared to free histones. Mediates monomethylation of p53/TP53 at 'Lys-382', leading to repress p53/TP53-target genes. Plays a negative role in TGF-beta response regulation and a positive role in cell migration. {ECO:0000269|PubMed:12086618, ECO:0000269|PubMed:12121615, ECO:0000269|PubMed:15200950, ECO:0000269|PubMed:15933069, ECO:0000269|PubMed:15933070, ECO:0000269|PubMed:16517599, ECO:0000269|PubMed:17707234, ECO:0000269|PubMed:23478445}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Chromatin regulator;Chromosome;Coiled coil;Direct protein sequencing;Methyltransferase;Mitosis;Nucleus;Phosphoprotein;Reference proteome;Repressor;S-adenosyl-L-methionine;Transcription;Transcription regulation;Transferase;Ubl conjugation | The protein encoded by this gene is a protein-lysine N-methyltransferase that can monomethylate Lys-20 of histone H4 to effect transcriptional repression of some genes. The encoded protein is required for cell proliferation and plays a role in chromatin condensation. [provided by RefSeq, May 2016]. | hsa:387893; | chromosome [GO:0005694]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; histone methyltransferase activity (H4-K20 specific) [GO:0042799]; histone-lysine N-methyltransferase activity [GO:0018024]; lysine N-methyltransferase activity [GO:0016278]; p53 binding [GO:0002039]; protein-lysine N-methyltransferase activity [GO:0016279]; transcription corepressor activity [GO:0003714]; cell division [GO:0051301]; chromatin organization [GO:0006325]; mitotic chromosome condensation [GO:0007076]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; peptidyl-lysine monomethylation [GO:0018026]; regulation of DNA damage response, signal transduction by p53 class mediator [GO:0043516]; regulation of signal transduction by p53 class mediator [GO:1901796] | 15933070_crystal structure of SET8 bound to a histone H4 peptide bearing Lys-20 and the product cofactor S-adenosylhomocysteine 15964846_coordination between the amino acid sequence RHRK20VLRDN and the SET domain of SET8 determines the substrate specificity and multiplicity of methylation of lysine 20 of H4 17707234_This study identifies SET8 as a p53-modifying enzyme, monomethylating p53 at lysine 382(p53K382me1) and identifying (p53K382me1) as a regulatory posttranslational modification of p53. 18166648_Overall, we show that SET8 is essential for genomic stability in mammalian cells and that decreased expression of SET8 results in DNA damage and Chk1-dependent S-phase arrest. 18319261_the SET8 and PCNA interaction couples H4-K20 methylation with DNA replication 18408754_H4K20 monomethylation and PR-SET7 are important for L3MBTL1 function 18418072_SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. 18480059_PR-Set7 enzymatic activity is essential for mammalian cell cycle progression and for the maintenance of genomic stability, most likely by monomethylating histone H4K20. 18512960_Data show that bond-order computations establish that the H4-K20 monomethylation in SET8 is a concerted linear S(N)2 displacement reaction. 19789321_The miR-502-binding site single-nucleotide polymorphism in the 3'-UTR of SET8 modulates SET8 expression and contributes to the early development of breast cancer. 20870725_SET8-mediated methylation of p53 at Lys-382 promotes the interaction between L3MBTL1 and p53 in cells. 20932471_Results demonstrate a central role of CRL4(Cdt2)-dependent cell-cycle regulation of Set8 for the maintenance of a stable epigenetic state essential for cell viability. 20932471_This study identifies CRL4-Cdt2 ubiquitin ligase to promote the ubiquitin-dependent proteolysis of the histone H4 methyltransferase Set8 during S-phase of the cell cycle and after UV-irradiation in a reaction that is dependent on PCNA. 20932472_CRL4(Cdt2)-dependent destruction of Set8 in S phase preserves genome stability by preventing aberrant chromatin compaction during DNA synthesis. 20953199_The results elucidate a critical role for PR-Set7 and H4K20me1 in the chromatin events that regulate replication origins. 21035370_PR-Set7 is transiently recruited to laser-induced DNA damage sites through its interaction with PCNA, after which 53BP1 is recruited dependent on PR-Set7 catalytic activity. 21200139_[review] PR-Set7/Set8/KMT5a is the sole histone methyltransferase responsible for the monomethylation of histone H4 lysine 20 (H4K20me1) in higher eukaryotes. 21220508_The turnover of SET8 is accelerated after ultraviolet irradiation dependent on the CRL4(CDT2) ubiquitin ligase and PCNA. 21282610_SET8 is a Wnt signaling mediator and is recruited by LEF1/TCF4 to regulate the transcription of Wnt-activated genes, possibly through H4K20 monomethylation at the target gene promoters. 21983900_in breast carcinoma samples, SET8 expression is positively correlated with metastasis 22095217_SET8 modifies hepatocellular carcinoma outcome by altering its expression, which depends, at least in part, on its binding affinity with miR-502. 22556262_An increase of methylated PCNA was found in cancer cells, and the expression levels of SETD8 and PCNA were correlated in cancer tissue samples 22583696_long recognition sequence of SET8 makes it difficult to methylate a lysine in a folded region of a protein 22867998_SET8 CC genotype was associated with a decreased risk of EOC in this case-control study. The analysis of genetic polymorphisms in miRNA binding sites may help identify subgroups of populations that are at high risk for EOC 23152447_PR-Set7 is an important downstream effector of CRL4(Cdt2) function during origin of DNA replication licensing, dependent on Suv4-20h1/2 activity 23291132_miR-502-binding SNP in SET8 may modulate SET8 expression and contribute to early development of breast cancer. 23419719_Data indicate that DDX21, a nucleolar protein, was confirmed to associate with SET8. 23478445_Migration of epithelial cells is stimulated by CRL1(FBXO11)-mediated downregulation of Cdt2 and the consequent stabilization of Set8. 23706821_Data indicate the Set8-Numb-p53 signaling axis as an important regulatory pathway for apoptosis and suggests a therapeutic strategy by targeting Numb methylation. 23720754_a molecular mechanism for the regulation of SET8 and extend the biological function of microRNA-7 to DNA damage response 24019522_all of the H3K36-specific methyltransferases, including ASH1L, HYPB, NSD1, and NSD2 were inhibited by ubH2A, whereas the other histone methyltransferases, including PRC2, G9a, and Pr-Set7 were not affected by ubH2A. 24146953_Genetic variation in a microRNA-502 minding site in SET8 gene confers clinical outcome of non-small cell lung cancer in a Chinese population. 24374662_significant association between the polymorphism (rs16917496) of the miR-502-binding site in the 3'-UTR of SET8 and TP53 codon 72 polymorphism and the risk of developing NSCLC. 24459145_PR-SET7 and H4K20me1 are required for establishing both the H4K16Ac and H4K20me3 marks and have a dual role in the local regulation of Pol II pausing 24937452_Therefore, we conclude that SET8 is involved in AR-mediated transcription activation, possibly through its interaction with AR and H4K20me1 modification. 25048968_Our results demonstrated that SETD8 rs16917496 C/T polymorphism was associated with decreased risk of developing pediatric acute lymphoblastic leukemia 25137013_commonality of SPS8I1-3 against SETD8, together with their distinct structures and mechanisms for SETD8 inhibition, argues for the collective application of these compounds as SETD8 inhibitors 25169478_These data suggest that there are significant associations between the miR-502-binding site single nucleotide polymorphism in the 3'-UTR of SET8 and the TP53 codon 72 polymorphism with cervical cancer in Chinese, and there is a gene-gene interaction. 25252681_SET8 is required for 53BP1 recruitment and efficient repair of DSB. 25343552_The SNP in the miR502 binding site of the SET8 3'-untranslated region seems to influence survival of non-Hodgkin's lymphoma. 26634528_SET8 expression is associated with overall survival in gastric cancer 26666832_This study showed SCF(beta-TRCP) earmarks Set8 for ubiquitination and degradation in a casein kinase I-dependent manner, which is activated by DNA-damaging agents. 26707641_Overall, the results revealed that miR-127-3p acts as a tumor suppressor and that its down-regulation in cancer may contribute to OS progression and metastasis. 26717907_SET8 and ZEB1 are functionally interdependent in promoting the epithelial-mesenchymal transition and enhancing the invasive potential of prostate cancer cells in vitro. 26953260_analysis of a structural model for how Set8 uses multivalent interactions to bind and methylate the nucleosome based on crystallographic and solution studies of the Set8/nucleosome complex 27080302_miR-502/SET8 regulatory circuit emerges as a key regulator of the pathobiology of breast cancer and a focal point for possible therapeutic intervention. 27144429_We demonstrated the expression levels of SET8 in TT genotype were higher than in CC genotypes, and high levels of SET8 were associated with poor survival in breast cancer 27369108_This improves the binding of SAM, SAH, and sinefungin by up to 10,000-fold. A small collection of inhibitors structurally related to SAM were screened and 40 compounds were identified that only inhibit SETD8 when a nucleosome substrate is used 27535933_Lysine methylation represses p53 activity in teratocarcinoma cancer cells via up-regulation of SMYD2 and PR-Set7 and perpetuation of cancer cells proliferation. 27605386_MiR-502 medaited histone methyltransferase SET8 expression is associated with outcome of esophageal squamous cell carcinoma. 28006750_In non-small cell lung cancer cells, SETD8 expression suppression is involved in tumorigenesis and metastasis. 28249161_The loss of SETD8 concurrently stimulated nucleolar function. 28514051_The PPARgamma-SETD8 axis constitutes an epigenetic, p53-independent checkpoint on p21-mediated cellular senescence. 28534991_High expression of SET8 is associated with cervical cancer. 28578017_rs16917496 polymorphism may be a risk for predisposition to prostate cancer in an Iranian population. 28731125_these date elucidated that NT21MP and miR335 mediated PR of breast cancer cells partly through regulation of Wnt/betacatenin signaling pathway. Activation of miR335 or inactivation of SETD8 could be a novel approach for the treatment of breast cancer. 28731465_Authors demonstrated that PR-Set7, an epigenetic regulator for H4K20me1 modification, was extensively expressed in the postnatal uteri, and its conditional deletion resulted in a complete lack of endometrial glands and infertility in mice. 29128203_Genetic changes in miR-146a and the miR-502 binding site of the SET8 can be effective on the increased risk of breast cancer. 29487005_that oncogenic SETD8 was regulated by miR-382 and involved glioma progression 29512765_Data found that KMT5A was overexpressed in papillary thyroid carcinoma (PTC) tumors. Its knockdown revealed that KMT5A participated in the proliferation, apoptosis, cell cycle, migration and invasion of PTC cells as well as attenuated fatty acid metabolism. These results provide mounting evidence of KMT5A as an oncogenic mediator in fatty acid metabolism of PTC. 29762637_Results suggest that SET domain-containing protein 8 (SET8) may be a key mediator in hyperglycemic memory. 30341251_Furthermore, similar associations were found in the subgroup analysis of race diversity, control design, genotyping methods, and different cancer types... our meta-analysis indicated that the SET8 rs16917496 T/C polymorphism may not play a critical role in cancer development in Asian populations. 30952833_SET8 deficiency conferred an impaired glucose metabolism phenotype and thus inhibited the progression of hepatocellular carcinoma tumors. By contrast, SET8 overexpression aggravated the glycolytic alterations and tumor progression. Mechanistically, SET8 directly binds to and inactivates KLF4, resulting in suppression of its downstream SIRT4. 31081496_These findings provide molecular insight on enzymatic catalysis and allosteric mechanisms of SETD8 via its detailed conformational landscape. 31400111_SET8 methylates UHRF1 at lysine 385 and this modification leads to ubiquitination and degradation of UHRF1. UHRF1 downregulation in G2/M by SET8 has a role in suppressing DNMT1-mediated methylation on post-replicated DNA. 31760894_SET8 localization to chromatin flanking DNA damage is dependent on RNF168 ubiquitin ligase. 32111740_The microtubule-associated histone methyltransferase SET8, facilitated by transcription factor LSF, methylates alpha-tubulin. 32369110_High glucose inhibits vascular endothelial Keap1/Nrf2/ARE signal pathway via downregulation of monomethyltransferase SET8 expression. 33028948_SET8 suppression mediates high glucose-induced vascular endothelial inflammation via the upregulation of PTEN. 33127342_SETD8 promotes stemness characteristics and is a potential prognostic biomarker of gastric adenocarcinoma. 33127874_Histone methyltransferase SET8 is regulated by miR-192/215 and induces oncogene-induced senescence via p53-dependent DNA damage in human gastric carcinoma cells. 33130499_SETD8 is a prognostic biomarker that contributes to stem-like cell properties in non-small cell lung cancer. 33232789_SETD8 potentiates constitutive ERK1/2 activation via epigenetically silencing DUSP10 expression in pancreatic cancer. 33339442_Epigenetic Modifier SETD8 as a Therapeutic Target for High-Grade Serous Ovarian Cancer. 33710666_RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. 34091587_LncRNA LINC00473 is involved in the progression of invasive pituitary adenoma by upregulating KMT5A via ceRNA-mediated miR-502-3p evasion. 34238215_ets1 associates with KMT5A to participate in high glucose-mediated EndMT via upregulation of PFN2 expression in diabetic nephropathy. 34599596_SETD8 induces stemness and epithelial-mesenchymal transition of pancreatic cancer cells by regulating ROR1 expression. 34846946_SET8, a novel regulator to ameliorate vascular calcification via activating PI3K/Akt mediated anti-apoptotic effects. 34894606_Integrative bioinformatics analysis the clinical value of KMT5A in different subtypes of lung cancer. | ENSMUSG00000049327 | Kmt5a | 2294.68877 | 1.0325958 | 0.0462756675 | 0.08257116 | 3.143569e-01 | 5.750188e-01 | 8.556175e-01 | No | Yes | 3097.877234 | 259.557215 | 2917.654165 | 238.977974 | |
| ENSG00000184154 | 120356739 | LRRC51 | protein_coding | Q8WZ04 | FUNCTION: Catalyzes the O-methylation, and thereby the inactivation, of catecholamine neurotransmitters and catechol hormones (By similarity). Required for auditory function (PubMed:18794526). Component of the cochlear hair cell's mechanotransduction (MET) machinery. Involved in the assembly of the asymmetric tip-link MET complex. Required for transportation of TMC1 and TMC2 proteins into the mechanically sensitive stereocilia of the hair cells. The function in MET is independent of the enzymatic activity (By similarity). {ECO:0000250|UniProtKB:A1Y9I9, ECO:0000269|PubMed:18794526}. | Alternative splicing;Catecholamine metabolism;Cytoplasm;Deafness;Disease variant;Endoplasmic reticulum;Hearing;Membrane;Methyltransferase;Neurotransmitter degradation;Non-syndromic deafness;Reference proteome;S-adenosyl-L-methionine;Transferase;Transmembrane;Transmembrane helix | This gene belongs to the leucine-rich repeat containing family. The encoded protein contains a transmembrane domain and two leucine-rich repeat domains. Unlike in mouse and other mammals, readthrough transcription is observed in primates between this gene and the adjacent transmembrane O-methyltransferase (Tomt) gene. Previously, this locus was annotated as a single gene representing the readthrough transcripts as well as the two different transcript species that encoded different proteins. It has since been split into three genes, including the two stand-alone genes and a third gene representing the readthrough transcription. [provided by RefSeq, Feb 2022]. | hsa:220074; | endoplasmic reticulum [GO:0005783]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; catechol O-methyltransferase activity [GO:0016206]; L-dopa O-methyltransferase activity [GO:0102084]; O-methyltransferase activity [GO:0008171]; orcinol O-methyltransferase activity [GO:0102938]; auditory receptor cell development [GO:0060117]; catecholamine catabolic process [GO:0042424]; developmental process [GO:0032502]; dopamine catabolic process [GO:0042420]; dopamine metabolic process [GO:0042417]; methylation [GO:0032259]; neurotransmitter catabolic process [GO:0042135]; sensory perception of sound [GO:0007605] | ENSMUSG00000064307 | Lrrc51 | 126.63982 | 0.9215071 | -0.1179328902 | 0.27076499 | 1.890145e-01 | 6.637381e-01 | 8.954653e-01 | No | Yes | 143.552458 | 21.637211 | 147.564390 | 21.567449 | ||
| ENSG00000184208 | 79640 | C22orf46 | transcribed_unitary_pseudogene | C9J442 | Glycoprotein;Reference proteome;Secreted;Signal | extracellular region [GO:0005576] | 33195693_Circular RNA circ-CCAC1 Facilitates Adrenocortical Carcinoma Cell Proliferation, Migration, and Invasion through Regulating the miR-514a-5p/C22orf46 Axis. | 769.78536 | 0.5348210 | -0.9028720680 | 0.11771552 | 5.724646e+01 | 3.844800e-14 | 6.544948e-11 | No | Yes | 611.284516 | 55.908632 | 1114.780985 | 97.814502 | ||||||
| ENSG00000184368 | 256714 | MAP7D2 | protein_coding | Q96T17 | Acetylation;Alternative splicing;Coiled coil;Reference proteome | hsa:256714; | microtubule cytoskeleton [GO:0015630]; microtubule cytoskeleton organization [GO:0000226] | ENSMUSG00000041020 | Map7d2 | 511.36155 | 0.8030565 | -0.3164265542 | 0.13894255 | 5.149402e+00 | 2.325438e-02 | 2.460588e-01 | No | Yes | 523.492179 | 55.639321 | 610.849382 | 63.235806 | ||||
| ENSG00000184402 | 26039 | SS18L1 | protein_coding | O75177 | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | Activator;Alternative splicing;Calcium;Centromere;Chromatin regulator;Chromosome;Kinetochore;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation | This gene encodes a calcium-responsive transactivator which is an essential subunit of a neuron-specific chromatin-remodeling complex. The structure of this gene is similar to that of the SS18 gene. Mutations in this gene are involved in amyotrophic lateral sclerosis (ALS). Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Sep 2014]. | hsa:26039; | cytosol [GO:0005829]; kinetochore [GO:0000776]; nBAF complex [GO:0071565]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription coactivator activity [GO:0003713]; chromatin organization [GO:0006325]; dendrite development [GO:0016358]; positive regulation of dendrite morphogenesis [GO:0050775]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893] | 12696068_A synovial sarcoma of classic morphology contained a novel t(20;X) SS18L1(strong homology to SS18 on Ch20)/SSX1 fusion transcript in which nucleotide 1216 (exon 10) of SS18L1 was fused in-frame with nucleotide 422 (exon 6) of SSX1. 15866867_analysis of the CREST domain that inhibits dendritic growth in cultured neurons 16484776_SS18 and SS18L1 genes map within co-linear DNA segments that may have evolved through a relatively recent genomic duplication event. 19724895_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 23708140_This study demonistrated that CREST mutations inhibited activity-dependent neurite outgrowth in primary neurons, and CREST associated with the ALS protein FUS. 24360741_Its mutation causes amyotrophic lateral sclerosis. 25888396_calcium-responsive transactivator and certain other amyotrophic lateral sclerosis-linked proteins share several features implicated in amyotrophic lateral sclerosis pathogenesis 33398438_Protein expression pattern of calcium-responsive transactivator in early postnatal and adult testes. | ENSMUSG00000039086 | Ss18l1 | 565.57391 | 0.9040944 | -0.1454547304 | 0.15389956 | 8.893782e-01 | 3.456459e-01 | 7.193007e-01 | No | Yes | 557.660217 | 111.713535 | 621.364589 | 121.720332 | |
| ENSG00000184545 | 1850 | DUSP8 | protein_coding | Q13202 | FUNCTION: Has phosphatase activity with synthetic phosphatase substrates and negatively regulates mitogen-activated protein kinase activity, presumably by catalysing their dephosphorylation. Expected to display protein phosphatase activity toward phosphotyrosine, phosphoserine and phosphothreonine residues. {ECO:0000250|UniProtKB:O09112}. | 3D-structure;Cytoplasm;Hydrolase;Nucleus;Protein phosphatase;Reference proteome | The protein encoded by this gene is a member of the dual specificity protein phosphatase subfamily. These phosphatases inactivate their target kinases by dephosphorylating both the phosphoserine/threonine and phosphotyrosine residues. They negatively regulate members of the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38), which is associated with cellular proliferation and differentiation. Different members of the family of dual specificity phosphatases show distinct substrate specificities for various MAP kinases, different tissue distribution and subcellular localization, and different modes of inducibility of their expression by extracellular stimuli. This gene product inactivates SAPK/JNK and p38, is expressed predominantly in the adult brain, heart, and skeletal muscle, is localized in the cytoplasm, and is induced by nerve growth factor and insulin. An intronless pseudogene for DUSP8 is present on chromosome 10q11.2. [provided by RefSeq, Jul 2008]. | hsa:1850; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nucleus [GO:0005634]; MAP kinase tyrosine/serine/threonine phosphatase activity [GO:0017017]; phosphatase activity [GO:0016791]; phosphoprotein phosphatase activity [GO:0004721]; protein serine phosphatase activity [GO:0106306]; protein threonine phosphatase activity [GO:0106307]; protein tyrosine phosphatase activity [GO:0004725]; protein tyrosine/threonine phosphatase activity [GO:0008330]; dephosphorylation [GO:0016311]; negative regulation of MAPK cascade [GO:0043409] | 13129832_DUSP8 is genetically linked to alcohol dependence and was found on chromosome 11p15.5 15899389_Because this variant of hVH-5 lacked intronic sequences in its genomic structure, we suggest it might be a processed pseudogene of hVH-5. 22100391_The phosphorylation of the M3/6 phosphatase by JNK in response to stress stimuli results in attenuation of phosphatase activity and acceleration of JNK activation. 33131190_Diabetes type 2 risk gene Dusp8 is associated with altered sucrose reward behavior in mice and humans. | ENSMUSG00000037887 | Dusp8 | 88.51506 | 1.5435799 | 0.6262801513 | 0.30330819 | 4.298587e+00 | 3.814405e-02 | 3.022105e-01 | No | Yes | 136.166152 | 25.841570 | 85.291701 | 15.727778 | |
| ENSG00000184640 | 10801 | SEPTIN9 | protein_coding | Q9UHD8 | FUNCTION: Filament-forming cytoskeletal GTPase (By similarity). May play a role in cytokinesis (Potential). May play a role in the internalization of 2 intracellular microbial pathogens, Listeria monocytogenes and Shigella flexneri. {ECO:0000250, ECO:0000305}. | 3D-structure;Acetylation;Alternative splicing;Cell cycle;Cell division;Chromosomal rearrangement;Cytoplasm;Cytoskeleton;Disease variant;GTP-binding;Nucleotide-binding;Phosphoprotein;Reference proteome | This gene is a member of the septin family involved in cytokinesis and cell cycle control. This gene is a candidate for the ovarian tumor suppressor gene. Mutations in this gene cause hereditary neuralgic amyotrophy, also known as neuritis with brachial predilection. A chromosomal translocation involving this gene on chromosome 17 and the MLL gene on chromosome 11 results in acute myelomonocytic leukemia. Multiple alternatively spliced transcript variants encoding different isoforms have been described.[provided by RefSeq, Mar 2009]. | hsa:10801; | actin cytoskeleton [GO:0015629]; axoneme [GO:0005930]; cell division site [GO:0032153]; cytoplasm [GO:0005737]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; non-motile cilium [GO:0097730]; perinuclear region of cytoplasm [GO:0048471]; septin complex [GO:0031105]; septin ring [GO:0005940]; stress fiber [GO:0001725]; cadherin binding [GO:0045296]; GTP binding [GO:0005525]; GTPase activity [GO:0003924]; molecular adaptor activity [GO:0060090]; cellular protein localization [GO:0034613]; cytoskeleton-dependent cytokinesis [GO:0061640]; positive regulation of non-motile cilium assembly [GO:1902857] | 11593400_Describes alternative splicing of this gene. 12095151_Fusion of MLL and MSF in adult de novo acute myelomonocytic leukemia (M4) with t(11;17)(q23;q25). 12388755_These results reveal that MSF is required for the completion of cytokinesis and suggest a role that is distinct from that of Nedd5. 12626509_Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules 15485874_Sept7/9b/11 form a complex that has effects on filament elongation, bundling, or disruption 15782116_overexpression of SEPT9 in neoplasia is not simply a proliferation-associated phenomenon, despite its role in cytokines 16161048_SEPT9_v1 is also upregulated in both serous and mucinous carcinomas 16186812_Three mutations in the gene septin 9 (SEPT9) in six families with hereditary neuralgic amyotrophy linked to chromosome 17q25 were reported. 16424018_MSF-A stabilizes HIF-1alpha protein by preventing its ubiquitination and, consequently, activates HIF downstreatm survival genes to promotor tumor progression and angiogenesis 17546647_SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling 18075300_SEPT9_V1 confers resistance to microtubule-mediated HIF-1 inhibitors. 18492087_Both children with dysmorphic syndrome of hereditary neuralgic amyotrophy were shown to have inherited the paternal SEPT9 mutation. 18525421_In this report confirming SEPT9 mutation in a family with suspected hereditary neuralgic amyotrophy, electrophysiological, clinical phenotype, and molecular genetic data of three members are presented. 18642054_monocytic differentiation and a poor prognosis may also be associated with acute myeloid leukemia with the variant MLL/SEPT9 fusion transcript 19018278_SEPT9 DNA methylation may have a role in colorectal cancer 19071215_In mammary epithelial cells, up-regulation of SEPT9_v1 stabilizes JNK by delaying its degradation, thereby activating the JNK transcriptome and suggesting a a novel functional role of SEPT9_v1 in driving cellular proliferation of mammary epithelial cells. 19139049_An intragenic 38 Kb SEPT9 duplication that is linked to hereditary neuralgic amyotrophy in 12 North American families that share the common founder haplotype, is reported. 19204161_Observational study of gene-disease association. (HuGE Navigator) 19204161_Rarely, individuals with sporadic brachial plexopathy may have the same conserved 17q25 sequence found in many North American kindreds with the hereditary version. 19251694_a new mechanism of oxygen-independent activation of HIF-1 has been identified that is mediated by SEPT9_v1 blockade of RACK1 activity on HIF-1alpha degradation 19406918_SEPT9 assay successfully identified 72% of cancers at a specificity of 93% in the training study and 68% of cancers at a specificity of 89% in the testing study 19451530_Results suggest that mutation of the SEPT9 gene is the molecular basis of some cases of hereditary neuralgic amyotrophy (HNA). 19851296_Observational study of gene-disease association. (HuGE Navigator) 19939853_A total of seven heterogeneous SEPT9 duplications have been identified in this study as a causative factor of hereditary neuralgic amyotrophy. 20019224_missense mutation c.262C>T results in a phenotypic spectrum of hereditary neuralgic amyotrophy in a large Japanese family 20029986_Results verify IRX1, EBF3, SLC5A8, SEPT9, and FUSSEL18 as valid methylation markers in two separate sets of HNSCC specimens; also preliminarily show a trend between HPV16 positivity and target gene hypermethylation of IRX1, EBF3, SLC5A8, and SEPT9. 20113838_Coexistence of alternative MLL-SEPT9 fusion transcripts in an acute myeloid leukemia with t(11;17)(q23;q25). 20140221_Data show that methylated DNA from advanced precancerous colorectal lesions can be detected using a panel of two DNA methylation markers, ALX4 and SEPT9. 20198315_Observational study of gene-disease association. (HuGE Navigator) 20407014_New insights and validation are provided for applying SEPT9 transcript variant 1 as a potential target for antitumor therapy via interruption of the HIF-1 pathway. 20682395_Case represents an additional MLL-SEPT9-positive AML that was considered to be related to therapy. RT-PCR and sequencing analyses demonstrated MLL-SEPT9 fusion transcripts with the breakpoint of MLL exon 8/SEPT9 exon 2 and MLL exon 9/SEPT9 exon 2. 21059847_Data demonstrate that SEPT9 mediates the localization of the vesicle-tethering exocyst complex to the midbody, providing mechanistic insight into the role of SEPT9 during cell division. 21267688_Increased methylation of septin 9 resulting in decreased mRNA and protein expression is associated with colorectal cancer. 21737677_uneven distribution of SEPT9 among core septin heteromers causes heterogeneity with respect to both subunit number and polymerization interfaces 21767235_Data illustrated roles of SEPT9 that might contribute to hetero-trimeric septin complex formation. SEPT9 can substitute for septins of the SEPT2 group and partially for SEPT7. 21831286_SEPT9 gene amplification and overexpression during human breast tumorigenesis 22123865_SEPT9 holds a terminal position in the septin octamers, mediating abscission-specific polymerization during cytokinesis. 22278362_SEPT9_v4 expression may be clinically relevant and contribute to some forms of drug resistance. 22956766_Myeloid K562 cells express three SEPT9 isoforms, all of which have an equal propensity to hetero-oligomerize with SEPT7-containing hexamers to generate octameric heteromers. 22981636_The identification of a SEPT9 mutation in a neonate with respiratory distress due to vocal cord paralysis expands the differential diagnosis in these patients. 23049919_SEPT9 in plasma has a role in both left- and right-sided colon cancers 23118862_Matrix stiffness regulates endothelial cell proliferation through septin 9 23572511_SEPT2 forms a 1:1:1 complex with SEPT7 and SEPT9. 23862763_Serum methylation levels of TAC1, SEPT9, and EYA4 were significant discriminants between stage I colorectal cancer and healthy controls. 23988185_epigenetic deregulation of SEPT9 plays a role in the development of colorectal cancer. Aberrant hypermethylation of this gene occurs only in one of its CpG islands and this hypermethylation likely is an early event in the adenoma-carcinoma sequence. 23990466_SEPT9 plays multiple roles in abscission, one of which is regulated by the action of Cdk1 and Pin1. 24067372_SEPT9 isoform 1 is required for the association between HIF-1alpha and importin-alpha to promote efficient nuclear translocation. 24127542_The study shows increased SEPT9 expression as a consequence of genomic amplification and is the first study to profile SEPT9_v1 through SEPT9_v7 isoform-specific mRNA expression in tumor and nontumor tissues from patients with breast cancer. 24344182_SEPT9 repeat motifs bind and bundle MTs, and thereby promote asymmetric neurite growth. 24386354_prognostic value of SHOX2 and SEPT9 DNA methylation in benign, paramalignant and malignant pleural effusions 24535900_Our results suggest that SEPT9 gene methylation is a valuable biomarker for screening CRC in the Chinese population 24633736_SEPT9 promoter methylation is detected in free circulating DNA of lung cancer patients 25293760_Authors report here that the septins SEPT2, -9, -11, and probably -7 form fibrillar structures around the chlamydial inclusion. 25472714_we demonstrate that SEPT9 negatively regulates EGFR degradation by preventing the association of the ubiquitin ligase Cbl with CIN85, resulting in reduced EGFR ubiquitylation 25526039_Results show that in plasma samples, elevated methylated SEPT9 (mSEPT9) values were detected in colorectal cancer, but not in adenomas. Tissue levels of mSEPT9 alone are not sufficient to predict mSEPT9 levels in plasma. 25898316_SEPT9_i1 is expressed in high-grade prostate tumors suggesting it has a significant role in prostate tumorigenesis. 25946211_KRAS mutations and SEPT9 promoter methylation were present in 34% (29/85) and in 82% (70/85) of primary tumor tissue samples. 26471083_provide a full overview of the theoretical basis, development, validation, and clinical applications of the SEPT9 assay for both basic science researchers and clinical practitioners 26633373_SEPT9 promoter methylation and MN frequency, both measured in peripheral blood, occur at an early stage compared to carcinoma development, indicating that the approach might be suitable to monitor CRC development. 26823018_The first evidence of an interaction between septins and a nonmitotic kinesin is provided and it is suggested that SEPT9 modulates the interactions of KIF17 with membrane cargo. 27133379_our study has validated a new SEPT9 assay and combined testing as an aid in cancer detection, providing a new approach for opportunistic CRC screening. 27417143_Septin 9 regulates lipid droplet growth through binding to phosphatidylinositol-5-phosphate in Hepatitis C virus infected cells.Septin 9 regulates lipid droplets growth and perinuclear accumulation in a manner dependent on dynamic microtubules.Septin 9 regulates Hepatitis C virus replication. 27499429_These results indicate that SEPT9_v2 promoter hypermethylation, which silences the expression of SEPT9_v2 mRNA, is observed in a significant proportion of breast tumors, and that methylated SEPT9_v2 may serve as a novel tumor marker for breast cancer. 27660666_Study shows a stepwise increase of SEPT9 methylation from non-cancerous to cancerous tissue in colorectal adenocarcinoma. 27753040_Studies indicate that methylated Septin 9 ((m)SEPT9) can be consistently detected in plasma samples derived from whole blood samples collected with S-Monovette(R) K3E and BD Vacutainer (R) K2EDTA tubes stored at 2-8 degrees C for a maximum of 24 h and for samples collected in S-Monovette CPDA tubes stored at 18-25 degrees C for up to 48 h. 27999621_Study found SHOX2 and SEPT9 frequently methylated in biliary tract cancers. 28128742_The SEPT9 assay exhibited satisfactory performance in colorectal cancer diagnosis and screening, while more evidence is needed for therapeutic effect monitoring and prognosis prediction. 28338090_Thus our data indicate that Sept9_i2 is a negative regulator of breast tumorigenesis. We propose that Sept9 tumorigenic properties depend on the balance between Sept9_i1 and Sept9_i2 expression levels. 29610456_Post-therapeutic SHOX2 and SEPT9 circulating cell-free DNA(ccfDNA) methylation levels correlated with UICC stage (all P <0.01). SEPT9 ccfDNA methylation further allowed for an accurate pre- and post-therapeutic detection of distant metastases. 29627389_The primary aim of this study was to evaluate the diagnostic accuracy of a PCR-based assay for the analysis of SEPT9 promoter methylation in circulating cell-free DNA (mSEPT9) for diagnosing hepatocellular carcinoma. 29724999_Repression of SEPTIN2 and SEPTIN9 suppresses tumor growth of human glioblastoma cells and tumor progression in a mouse model. 29970704_The plasma levels of septin-9 and clusterin in ovarian cancer patients were abnormally elevated, which might be used as potential candidates of peripheral blood tumor biomarkers for early diagnosis of EOC and septin-9 might be related to distal metastases of EOC. 30043648_mSEPT9 is effective for colorectal cancer postsurgical assessment and prognosis prediction. 30158637_The authors propose that septins in general and SEPT9 in particular play a previously unappreciated role in osteoclastic bone resorption. 30273999_SEPT9 DNA methylation was detected in untreated colorectal carcinomas but not in adenomas. 30426784_The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. 30670682_Authors show that coupling septins (including SEPT9_i1) overexpression together with long-chain tubulin polyglutamylation induce significant paclitaxel resistance in several naive (taxane-sensitive) cell lines and accordingly stimulate the binding of CLIP-170 and MCAK to microtubules. 31088406_Study indicated that peripheral SEPT9 methylated DNA may be useful for the screening, early diagnosis, and recurrence monitoring of colorectal cancer. 31114136_methylated septin 9 is associated with advanced stages in Chinese colorectal cancer patients 31285548_results indicate that SEPT9 promotes upregulation and both trafficking and secretion of MMPs near FAs, thus enhancing migration and invasion of breast cancer cells 31316143_Methylated Septin 9 and Carcinoembryonic Antigen for Serological Diagnosis and Monitoring of Patients with Colorectal Cancer After Surgery. 31426855_Study found that hypermethylation of SEPT9 exhibits a high sensitivity and specificity in the diagnosis of cervical cancer. Up- and downregulation of SEPT9 affected the biological behavior of cervical cancer cells through the regulation of HMGB1. Moreover, SEPT9 mediated miR-375 could enhance the cell resistance to ionizing radiation via affecting the tumor-associated macrophages (TAMs) polarization. 31558699_SEPT9_i1 regulates human breast cancer cell motility through cytoskeletal and RhoA/FAK signaling pathway regulation. 32106387_Down-regulation of SEPT9 inhibits glioma progression through suppressing TGF-beta-induced epithelial-mesenchymal transition (EMT). 32122354_First description of a SEPT9 variant associated to a Charcot-Marie-Tooth Disease (CMT) phenotype; this suggests that SEPT9 is a new sufficient candidate gene in CMT. 32138771_SEPT9_v2, frequently silenced by promoter hypermethylation, exerts anti-tumor functions through inactivation of Wnt/beta-catenin signaling pathway via miR92b-3p/FZD10 in nasopharyngeal carcinoma cells. 32378260_Comprehensive analysis on the whole Rho-GAP family reveals that ARHGAP4 suppresses EMT in epithelial cells under negative regulation by Septin9. 32454907_Septin 9 Methylation in Nasopharyngeal Swabs: A Potential Minimally Invasive Biomarker for the Early Detection of Nasopharyngeal Carcinoma. 32566042_A First Step to a Biomarker of Curative Surgery in Colorectal Cancer by Liquid Biopsy of Methylated Septin 9 Gene. 32945374_Plasma levels of methylated septin 9 are capable of detecting hepatocellular carcinoma and hepatic cirrhosis. 33178361_SEPT9 Gene Methylation as a Noninvasive Marker for Hepatocellular Carcinoma. 33915163_CircRNA SEPT9 contributes to malignant behaviors of glioma cells via miR-432-5p-mediated regulation of LASP1. 33931320_Role of methylated septin 9 as an adjunct diagnostic and prognostic biomarker in hepatocellular carcinoma. 34350965_Insights into animal septins using recombinant human septin octamers with distinct SEPT9 isoforms. 34409731_Proteomic profiling of the oncogenic septin 9 reveals isoform-specific interactions in breast cancer cells. 34524873_ARHGAP4-SEPT2-SEPT9 complex enables both up- and down-modulation of integrin-mediated focal adhesions, cell migration, and invasion. 34854883_Septin-microtubule association via a motif unique to isoform 1 of septin 9 tunes stress fibers. 34873218_Application of droplet digital polymerase chain reaction of plasma methylated septin 9 on detection and early monitoring of colorectal cancer. 35089073_Noninvasive Detection of Esophageal Cancer by the Combination of mSEPT9 and SNCG. 35481971_Distinct Performance of Methylated SEPT9 in Upper and Lower Gastrointestinal Cancers and Combined Detection with Protein Markers. | ENSMUSG00000059248 | Septin9 | 1600.98648 | 1.0125794 | 0.0180350889 | 0.09584473 | 3.540244e-02 | 8.507549e-01 | 9.595722e-01 | No | Yes | 1991.619344 | 264.366739 | 1872.631085 | 242.993501 | |
| ENSG00000184937 | 7490 | WT1 | protein_coding | P19544 | FUNCTION: Transcription factor that plays an important role in cellular development and cell survival (PubMed:7862533). Recognizes and binds to the DNA sequence 5'-GCG(T/G)GGGCG-3' (PubMed:7862533, PubMed:17716689, PubMed:25258363). Regulates the expression of numerous target genes, including EPO. Plays an essential role for development of the urogenital system. It has a tumor suppressor as well as an oncogenic role in tumor formation. Function may be isoform-specific: isoforms lacking the KTS motif may act as transcription factors (PubMed:15520190). Isoforms containing the KTS motif may bind mRNA and play a role in mRNA metabolism or splicing (PubMed:16934801). Isoform 1 has lower affinity for DNA, and can bind RNA (PubMed:19123921). {ECO:0000269|PubMed:15520190, ECO:0000269|PubMed:16934801, ECO:0000269|PubMed:17716689, ECO:0000269|PubMed:19123921, ECO:0000269|PubMed:19416806, ECO:0000269|PubMed:25258363, ECO:0000269|PubMed:7862533}. | 3D-structure;Alternative initiation;Alternative splicing;Chromosomal rearrangement;Cytoplasm;DNA-binding;Disease variant;Isopeptide bond;Metal-binding;Nucleus;RNA editing;RNA-binding;Reference proteome;Repeat;Transcription;Transcription regulation;Tumor suppressor;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a transcription factor that contains four zinc-finger motifs at the C-terminus and a proline/glutamine-rich DNA-binding domain at the N-terminus. It has an essential role in the normal development of the urogenital system, and it is mutated in a small subset of patients with Wilms tumor. This gene exhibits complex tissue-specific and polymorphic imprinting pattern, with biallelic, and monoallelic expression from the maternal and paternal alleles in different tissues. Multiple transcript variants have been described. In several variants, there is evidence for the use of a non-AUG (CUG) translation initiation codon upstream of, and in-frame with the first AUG. Authors of PMID:7926762 also provide evidence that WT1 mRNA undergoes RNA editing in human and rat, and that this process is tissue-restricted and developmentally regulated. [provided by RefSeq, Mar 2015]. | hsa:7490; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; nuclear speck [GO:0016607]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; C2H2 zinc finger domain binding [GO:0070742]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity [GO:0003700]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; double-stranded methylated DNA binding [GO:0010385]; hemi-methylated DNA-binding [GO:0044729]; RNA binding [GO:0003723]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific DNA binding [GO:0043565]; transcription cis-regulatory region binding [GO:0000976]; zinc ion binding [GO:0008270]; adrenal cortex formation [GO:0035802]; adrenal gland development [GO:0030325]; branching involved in ureteric bud morphogenesis [GO:0001658]; camera-type eye development [GO:0043010]; cardiac muscle cell fate commitment [GO:0060923]; cellular response to cAMP [GO:0071320]; cellular response to gonadotropin stimulus [GO:0071371]; diaphragm development [GO:0060539]; epithelial cell differentiation [GO:0030855]; germ cell development [GO:0007281]; glomerular basement membrane development [GO:0032836]; glomerular visceral epithelial cell differentiation [GO:0072112]; glomerulus development [GO:0032835]; gonad development [GO:0008406]; heart development [GO:0007507]; kidney development [GO:0001822]; male genitalia development [GO:0030539]; male gonad development [GO:0008584]; mesenchymal to epithelial transition [GO:0060231]; metanephric epithelium development [GO:0072207]; metanephric mesenchyme development [GO:0072075]; metanephric S-shaped body morphogenesis [GO:0072284]; negative regulation of apoptotic process [GO:0043066]; negative regulation of cell growth [GO:0030308]; negative regulation of cell population proliferation [GO:0008285]; negative regulation of female gonad development [GO:2000195]; negative regulation of metanephric glomerular mesangial cell proliferation [GO:0072302]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription, DNA-templated [GO:0045892]; negative regulation of translation [GO:0017148]; positive regulation of apoptotic process [GO:0043065]; positive regulation of DNA methylation [GO:1905643]; positive regulation of gene expression [GO:0010628]; positive regulation of heart growth [GO:0060421]; positive regulation of male gonad development [GO:2000020]; positive regulation of metanephric ureteric bud development [GO:2001076]; positive regulation of pri-miRNA transcription by RNA polymerase II [GO:1902895]; positive regulation of transcription by RNA polymerase II [GO:0045944]; positive regulation of transcription, DNA-templated [GO:0045893]; posterior mesonephric tubule development [GO:0072166]; regulation of animal organ formation [GO:0003156]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355]; RNA splicing [GO:0008380]; sex determination [GO:0007530]; thorax and anterior abdomen determination [GO:0007356]; tissue development [GO:0009888]; ureteric bud development [GO:0001657]; vasculogenesis [GO:0001570]; visceral serous pericardium development [GO:0061032] | 8012395_WT1 gene exhibits complex tissue-specific and polymorphic imprinting pattern. 8621495_Use of non-AUG (CUG) translation initiation site upstream of, and in-frame with the first AUG, generates longer WT1 isoforms. 9361029_WT1 gene is expressed from the paternal allele in fibroblasts and lymphocytes. 11738793_review: role in testis descent 11889045_requirement for Wt1 in normal retina formation with a critical role in Pou4f2-dependent ganglion cell differentiation. 11912180_reduced expression of WT1 causes crescentic glomerulonephritis and mesengial sclerosis and thus a regulator of podocyte function 11919196_Cyclin E is a target of WT1 transcriptional repression 11933209_A review of the phenotypic variation due to the Denys-Drash syndrome-associated germline WT1 mutation R362X 11939727_uterine papillary serous carcinomas were nonreactive for WT1 11960373_Induction of the interleukin-2/15 receptor beta-chain by the EWS-WT1 translocation product. 11986946_The coexpression of the apoptosis-related genes bcl-2 and wt1 in predicting survival in adult acute myeloid leukemia. 12070003_quantitation of WT1 RNA in Japanese patients with paroxysmal nocturnal hemoglobinuria 12111123_A peptide representing positions 124-148 of the Kruppel-like zinc finger protein was predicted to bind to the HLA-DRB1*0401 molecule. Its sequence is PQQMGSDVRDLNALL. 12127961_WT1 protein contributes to breast cancer progression by promoting breast cancer cell proliferation 12133898_The retinoblastoma-derived human cell line, Y-79, contained robust amounts of Wt1 mRNA and protein. Wt1 expression was down-regulated upon laminin-induced differentiation of Y-79 into neuron-like cells. 12161615_WT1 missense mutations in exon 7, which affect zinc finger 1, alter not only renal function but also male gonadal development in a patient with Denys-Drash syndrome and a 46,XY karyotype. 12199781_abnormal WT1 expression may contribute to the disturbed differentiation of haaematopoietic cells in MDS patients 12200377_CD8 T-cell responses to Wilms tumor gene product WT1 and proteinase 3 were observed in patients with acute myeloid leukemia, and not in controls. 12213901_WT1 and DAX-1 inhibit aromatase P450 expression in human endometrial and endometriotic stromal cells 12239212_Results suggest that bone marrow zinc finger 2 (BMZF2) interferes with the transactivation potential of Wilms tumor suppressor gene (WT1). 12411326_Two distinct HLA-A0201-presented epitopes of the Wilms tumor antigen 1 can function as targets for leukemia-reactive CTL 12444079_Interacts with p53 in insulin-like growth factor-I receptor gene regulation 12471221_Correlation between a specific Wilms tumour suppressor gene (WT1) mutation and the histological findings in Wilms tumour (WT) 12665546_Alternative splicing, RNA editing, and the use of alternative translation initiation sites generate a multitude of isoforms, which seem to have overlapping but also distinct functions during embryonic development and the maintenance of organ function. 12681485_role in regulating taurine transporter gene 12761165_Epigenetic silencing of WT1 by promoter hypermethylation with loss of heterozygosity is consistent with the revised two-hit model in Wilms' tumorigenesis. 12824878_Overexpression of the Wilms' tumor gene WT1 is associated with head and neck squamous cell carcinoma 12824921_Overexpressed in human bone and soft-tissue sarcomas. 12829997_WT1 is reexpressed in the cirrhotic liver in relation to disease progression and may play a role in the development of hepatic insufficiency in cirrhosis. 12841384_evidence demonstrating that the WT1 gene is involved in the development and differentiation of T-lineage cells 12841869_findings indicate an important role of the wild-type WT1 gene in the tumorigenesis of primary thyroid cancer 12901797_Expression of WT1 protein was detected in 41 (89%) of 46 cases of colonic and rectal adenocarcinoma. 12914969_Wilms' tumor suppressor is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer's disease. 12960088_WT1 gene is novel downstream target for IGF-I action. Reduced levels of WT1 may facilitate IGF-I-stimulated cell cycle progression. Inhibition of WT1 gene expression by IGF-I be significant in cancer initiation and/or progression. 12961083_WT1 was restricted to nucleus of glomerular podocytes. 12970737_These results suggest that WT1 forms a complex with SRY to regulate transcription and that this WT1-SRY interaction is important in testis development. 14666652_WT1 inhibits the transformed phenotype of breast cancer cells and down-regulates the beta-catenin/TCF signaling pathway through destabilization of beta-catenin. 14681303_There is a molecular basis for the strong bias of paternal allele mutations and variable penetrance observed in syndromes with inherited WT1 mutations. 14701728_BASP1 can confer WT1 cosuppressor activity in transfection assays, and elimination of endogenous BASP1 expression augments transcriptional activation by WT1. 14767530_WT1 has a role in suppressing prostate tumor cell growth 14962262_Co-expression of this gene with MDR1 did not significantly influence the complete response rate to induction therapy. 14988020_The transcription factor PEA3 activates the WT1 promoter. 14988155_Cytotoxic T lymphocytes display strong cytotoxic activity against leukemia cells expressing WT1 endogenously but not against WT1(-) human tumor cells. 15084838_WT1 expression in epithelial tumors of the female genital tract may be related to cell differentiation and histologic subtypes of carcinomas, rather than to primary site of origin 15150775_Observational study of gene-disease association. (HuGE Navigator) 15223639_WT1 levels at presentation correlate with several biologic features of leukemia 15253707_Observational study of gene-disease association. (HuGE Navigator) 15253707_mutations in the WT1 have been found to be present in patients with SRNS in association with Wilms' tumor (WT) and urinary or genital malformations, as well as in patients with isolated steroid resistant nephrotic syndrome 15266301_Observational study of gene-disease association. (HuGE Navigator) 15286719_overexpression of WT1 induced a significant increase in the abundance of endogenous c-myc protein in breast cancer cells, consistent with the upregulation of c-myc transcription following WT1 induction. 15297187_WT1 is expressed in appreciable numbers of endometrial cancers. 15339675_WT1/beta-glucuronidase ratio is a prognostic factor for predicting relapse in patients with acute myeloid leukemia and could be included to establish more defined risk groups. 15365188_WT1 vaccination could induce WT1-specific cytotoxic T lymphocytes and result in cancer regression without damage to normal tissues. 15483024_Observational study of genotype prevalence and gene-disease association. (HuGE Navigator) 15504250_These results may indicate that the W T1 gene plays an important role in tumorigenesis of primary astrocytic tumors. 15504938_Recombinant WT1 protein can bind and activate the 186-bp NPHS1 fragment in a sequence-specific manner. WT1 may be required for regulation of the NPHS1 gene in vivo. 15506928_Mchanism of action of the WT1 suppression domain, and its function in the context of the role of WT1 as a regulator of development. 15510596_WT1 mRNA was overexpressed in all of the 12 esophageal squamous cell carcinomas examined. 15534117_WT1 protein is processed and expressed in the context of HLA class I molecules similarly on both myeloma and lymphoma cells 15538407_Th1-biased cellular immune responses against WT1 protein should be elicited in patients with hematopoietic malignancies. 15540161_WT1 gene is strongly overexpressed in beta-catenin mutant desmoid tumors. 15661271_WT1 mRNA levels were indicative for hematologic relapse (n = 6) versus response in CML patients 15674342_WT1 protein plays a vital role in regulating cell cycle progression and apoptosis in HER2/neu-overexpressing breast cancer cells. 15687485_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 15696971_the WT1 gene may play an important role in the tumorigenesis of glioblastoma, in contrast to medulloblastoma 15780077_No mutations detected in Japanese congenital nephrotic synddrome patients. 15838390_Observational study of gene-disease association. (HuGE Navigator) 15845894_describe WT1-specific and HLA class II-restricted CD4+ T lymphocytes possessing direct cytotoxic activity against leukemia cells 15878620_The WT1 Genes were determined in bone marrow samples of children with de novo B-lineage (n=170) and T-lineage (n=25) acute lymphoblastic leukemia (ALL). 15894924_frequently detected in the cytoplasm of a subset of high-grade non-Hodgkin lymphomas 15927676_Human transgenic Wt1 is not essential for murine hematopoiesis and that there may be significant differences in its role between mouse and man. 15957141_A novel familial WT1 point mutation in the stop codon of exon 10 (1730A/G; X450W) ws found in 3 members of a family with Wilms tumor and nephropathy. 15982325_Data suggest that WT1 protein expression is maintained during angiogenesis and malignant transformation of endothelial cells and can be considered as a new endothelial marker. 16087727_interplay between WT1 and ERalpha in control of IGF-IR gene transcription 16467207_Binding of the transcriptionally competent Wt1(-KTS) isoform to the minimal EPO promoter was demonstrated; this promoter was activated up to 20-fold by transient cotransfection of Wt1(-KTS) in different cell lines. 16502587_Method for the quantification of WT1 transcript by real-time polymerase chain reaction in acute myeloid leukemia and myelodysplastic syndromses. 16518414_Isoforms of WT1 play antiapoptotic roles at some points upstream of the mitochondria in the intrinsic apoptosis pathway. 16780544_Posttransplant recurrence of proteinuria in a case of focal segmental glomerulosclerosis is associated with WT1 mutation 16828156_The WT1 expression seems to be modulated by the presence of cytokines, especially on day 20 of culture. 16857797_WT1 may have a role in very poor survival of patients with osteogenic sarcoma metastasis 16876863_High levels of WT1 expression is associated with childhood acute leukemia 16883592_higher WT1 expression was associated with favorable cytogenetics subtypes and accordingly with better outcome in children with acute myeloid leukemia 16909243_Observational study of genotype prevalence. (HuGE Navigator) 16909243_WT1 splice mutations are not rare in females under 18 years with steroid resistance (SRNS). This occurs in absence of a clear renal pathology picture and frequently in absence of phenotype change typical of Frasier syndrome. 16920711_transcriptional activation of the alpha4integrin gene by Wt1(-KTS) might contribute to normal formation of the epicardium and other tissues in the developing embryo 16924231_These findings increase our knowledge of how WT1 exerts its transcriptional regulatory role and suggests that hnRNP-U may be a candidate Wilms' tumour gene at 1q44. 16927106_The WT1 gene encodes a zinc finger transcription factor involved in kidney and gonadal development and, when mutated, in the occurrence of kidney tumor and glomerular diseases. 16934801_WT1 interacts with the novel splicing regulator RNA binding protein RBM4, further, the longer isoform of WT1 is able to inhibit the effect of RBM4 on alternative splicing. 16966277_The induction of apoptosis by arsenic compounds was accompanied by down-regulation of hTERT and wt1 mRNA and protein expression but up-regulation of par-4. 16987884_role of WT1 in different diseases and normal development--review 16990584_dMolecular profiling of CD34(+) cells uncover a limited number of genes such as WT1 with altered expression that are associated with patients' clinical characteristics and may have potential prognostic application. 17160023_WT1 could function to either promote or suppress a transformed phenotype 17205055_8 mutations were found in WT1 in Wilms tumor patients, mostly in exons 7 and 9, the DNA binding domain. Adverse outcome correlated with these mutations in patients with FLT3-ITD. 17206472_WT1 overexpression did not protect against p53-mediated apoptosis or ionizing radiation induced cell death. WT1 siRNA increased the radiosensitivity of two human glioma cell lines independently of p53. 17210670_Failure of methylation spreading at the WT1 antisense regulatory region early in renal development, followed by imprint erasure, occurs during Wilms' tumourigenesis. 17216259_study suggests mutations in NPHS2 & WT1 are not a cause of uncomplicated steroid sensitive nephrotic syndrome or frequently relapsing & steroid-dependent nephrotic syndrome(FRNS/SDNS) 17371932_Observational study of genotype prevalence. (HuGE Navigator) 17487399_These studies of VEGF regulation by WT1 and dysregulation by DDS(R384W) suggest an important role for WT1 in both normal and tumor-related angiogenesis. 17496156_A functional interaction of WT1 and AR might play a role during the development of the male external genitalia. 17508006_distinguish between idiopathic hypereosinophilic syndromes/chronic eosinophilic leukemia and reactive hypereosinophilia based on WT1 transcript amount 17524167_REVIEW of the isoform-specific effects of WT1 on target gene expression and over-representation of certain isoforms in leukaemia 17531467_WT1 is overexpressed in uterine sarcomas. Since increased levels of mRNA determine the biological role, WT1 might contribute to uterine sarcoma tumour biology. 17540436_Survival analysis stratified by FIGO stage performed on cases using a 10% cut-off showed a shorter disease specific survival in patients with a positive WT1 expression in the ovarian tumor tissue. 17541636_WT1 mutation analysis should be routinely done in females with steroid-resistant nephrotic syndrome. 17551084_Bilateral Wilms tumours showed loss of the wild type WT1 allele (loss of heterozygosity (LOH)) and a tumour specific mutation in catenin beta1 (CTNNB1). 17579045_WT1 is a repressor of matrix metalloproteinase-9, regulated by a nitric oxide/soluble guanylate cyclase-dependent pathway in human lung epithelial cells. 17599043_An important early target of progestins that regulates both proliferation and differentiation in breast cancer cells. 17605875_WT1 expression in normal bone marrow decreases gradually with cell differentiation. 17630404_Wilms tumour was not observed in any aniridia case without a WT1 deletion. Of those with WT1 deletions, 77% with submicroscopic deletions (detectable only by high-resolution FISH analysis) had Wilms tumour compared with 42.5% with visible deletions. 17665418_There was a significantly lower level of WT-1 expression in parathyroid tumours than in normal parathyroid glands. Abnormal expression of WT-1 and the IGF axis may play a role in the pathogenesis of hyperparathyroidism. 17688410_These results show that the expression of the NDRG2 gene is directly or indirectly induced by WT1, and provide the first insights into transcriptional regulation of the NDRG2 gene, including demonstration of a novel splice variant. 17706117_Promoter methylation does not silence the mRNA expression of WT1 during the development of breast cancer. 17716689_Data report the structure determination by both X-ray crystallography and NMR spectroscopy of the WT1 zinc finger domain in complex with DNA. 17721194_WT1 is useful for the distinction of ovarian Sertoli cell tumor from endometrioid tumors and carcinoids. 17728783_WT1 expression is induced by oncogenic signalling from BCR/ABL1. WT1 contributes to resistance against apoptosis induced by imatinib. 17803653_There was no significant difference in WT1 message between aplastic anemia and refractory anemia, suggesting that WT1 message is not a good tool to discriminate these two diseases. 17853480_report on eight new cases of this condition, two of whom were shown to have heterozygous missense mutations in the C-terminal zinc finger domains of WT1: Arg366Cys and Arg394Trp 17869219_We conclude that WT1 might be an important component of the NO-dependent regulation of T lymphocyte proliferation and potential function. 17886559_Tumors that contained wild-type p53 were significantly more likely to express WT1, and presence of WT1 in glioma support that WT1 expression is important in glioma biology. 17912546_direct role of WT1 in melanoma proliferation, which might be mediated via Nestin and Zyxin 17934764_NPHS2 and WT1 are now known to be genes commonly involved in the pathogenesis of sporadic steroid-resistant nephrotic syndrome. However, the incidence of NPHS2 gene mutation shows interethnic difference. 17939399_WT1 gene expression and isomer ratio changes during phorbol ester induced differentiation of K562 cells. 17940140_WT1 encodes conserved antisense RNAs that may have an important regulatory role in WT1 expression via RNA:RNA interactions, and which can become deregulated by a variety of mechanisms in cancer. 17947653_enhanced tetramer binding of modified WT1-T-Cell Receptor variants was not associated with improved WT1-specific T cell function 17956689_The WT1 enhancer promotes the transcriptional activities of WT1 promoter in some cell lines regardless of the hematopoietic tissue origin. 17972942_WT1 downregulation during myeloid differentiation of NB4 and HL60 leukemic cell lines is associated with increased tumor repressor hDMP1 mRNA levels 18034345_Pure curcumin decreased WT1 gene expression in both transcriptional and translational levels. 18042071_Nuclear WT1 expression is present in a minority of invasive micropapillary carcinomas and, when present, expression is focal 18058136_Observational study of genotype prevalence. (HuGE Navigator) 18064385_This study provides further insight into the mechanisms of transcriptional regulation of the WT1 gene and WT1-associated diseases treatment. 18064689_In pleural malignant effusion, malignant mesothelioma can be identified with positive staining for WT-1 and negative staining for p63. 18065803_Mutations in two other genes WT1 and LAMB2 may also cause IDMS. 18081724_Bone marrow WT1 gene expression analysis is informative only in cases where a more sensitive minimal residual disease (MRD) marker is not available. 18094593_Relapse was observed in eight of 17 patients with acute myelogenous leukemia analysed longitudinally, and an increase of WT1 gene expression preceded morphologic relapse in four patients. 18181329_The researchers found that the Wilms tumor gene 1 is very important in the survival of the breast cancer MCF-7 cell line. 18202757_the -KTS-containing variants of WT1 are directly involved in the regulation of p21(Waf1/Cip1) expression and the subsequent suppression of lymph node metastasis in human lung squamous cell carcinoma 18212735_transcriptional activation of ETS-1 by the Wilms' tumour suppressor WT1 is a crucial step in tumour vascularization via regulation of endothelial cell proliferation and migration. 18231640_WT1 induction led to strong induction of IFI16 expression and its promoter activity was responsive to the WT1 protein. 18231915_WT1 expression in newly diagnosed and relapsed ALL patients was significantly higher than that of the ALL patients at remission and normal subjects 18255279_The expression of WT1 is increased more in HCC than in non-tumour tissues. Overexpressed WT1 was associated with tumour growth, and resulted in a worsening prognosis of HCC. WT1 overexpression might contribute to oncogenic potential. 18260155_Upregulation of Wilms tumor gene protein 1 is associated with ovarian cancer metastasis and modulates cell invasion 18271004_Critical levels of WT1 + KTS, SRY and SOX9 are required for normal Sertoli cell maturation, and subsequent normal spermatogenesis. 18273617_Biallelic loss (the novel germline mutation c.895-2A > G & one LOH in the tumor) of the WT1 gene was associated with drug-resistant bilateral Wilms tumor. 18292948_WT1-shRNA targeting exon 5 should serve as a potent anti-cancer agent for various types of solid tumors. 18311776_WT1 and WTX mutations occur with similar frequency, that they partially overlap in Wilms tumors, and that mutations in WT1, WTX, and CTNNB1 underlie the genetic basis of about one-third of Wilms tumors 18371184_WT1 expression in astrocytes indicates a neoplastic origin and might represent an important diagnostic tool to differentiate reactive from neoplastic astrocytes by immunohistochemistry. 18385267_Recent advances including the development of transgenic mouse models and conditionally immortalized podocyte cell lines are beginning to shed light on WT1's crucial role in podocyte function[review] 18424770_activation of the EpoR gene by Wt1 may represent an important mechanism in normal hematopoiesis 18425046_WT1 is a fairly specific marker for spindle cell tumors of gynecologic organs, including ovarian spindle cell tumors, endometrial stromal tumors, and uterine smooth muscle tumors. Non-GYN spindle cell sarcomas rarely express WT1. 18443273_WT1 may have a role in relapse of acute myeloid leukemia 18464243_three WT1 subtypes were correlated with WT1, IGF2, and CTNNB1 genetics 18466223_study shows that WT1 is expressed de novo in numerous neuroepithelial tumours and increases with the grade of malignancy; results suggest an important role of WT1 in tumourigenesis and progression in brain tumours 18469795_None of the breast carcinoma subtype unassociated with mucinous component showed WT1 expression. 18516627_A novel Wilms' tumor 1 gene mutation in a child with severe renal dysfunction and persistent renal blastema. 18528287_Overexpressed in gastrointestinal stromal tumor and some smooth muscle tumors. 18559874_Observational study of gene-disease association. (HuGE Navigator) 18559874_The presence of WT1 mutations at diagnosis of acute myeloid leukemia is associated with extremely poor outcome, higher risk of relapse and death. 18590714_The expression levels of WT1 and NPM1 in 93 paired samples showed a strong positive correlation. WT1 decreased rapidly after induction but maintained these residual levels after treatment in patients in complete remission. 18591546_WT1 mutations are a negative prognostic indicator in normal karyotype AML and may be suitable for the development of targeted therapy 18604725_expression of ROR1 and WT1 in B-ALL is associated with the differentiation stage of the leukemic cells 18618575_Observational study of gene-disease association. (HuGE Navigator) 18644985_WT1 transactivates another important negative regulator of the Ras/MAPK pathway, MAPK phosphatase 3 (MKP3). 18666806_Loss of WT1 permits aberrant PAX3 expression in a subset of Wilms tumors with myogenic phenotype. 18688870_Maternal transmission of a mutation in exon 1 of the WT1 gene caused genitourinary anomalies. 18703217_Herein, we describe a desmoplastic small round cell tumor of soft tissue with an unusual pattern of WT1 expression associated with a novel variant EWS-WT1 fusion transcript. 18708366_This study clearly implicates WT1 as a mediator of antiestrogen resistance in breast cancer through down-regulation of ERalpha expression. 18722603_Innervation of deep endometriosis has been linked to its severe pain symptoms. Wilms' tumor gene 1 is overexpressed in part of these nerves. 18751389_WT1 plays an important role in maintaining normal growth of mammary epithelial cells and dysregulated WT1 expression may contribute to breast cancer development. 18752126_Nuclear WT1 expression is present in a minority of endometrioid ovarian carcinomas 18756326_observation that TIA-1 and WT1 are both involved in apoptosis supports our proposal for a functional link between these proteins 18801058_cases with increase of WT1 levels after hematopoietic stem cell transplant and without graft vs. host disease may be candidate to discontinuation of immunosuppression and/or donor lymphocyte infusion therapy 18929401_Although WT1 is expressed in a majority of endometrial carcinomas, a heterogeneous staining pattern is observed. 18950857_quantified both WT1 and PRAME transcript levels in the bone marrow of AML patients. Dynamic patterns of WT1 and PRAME during follow-up showed that a consistent elevation or a rise over time to exceed the normal range predicted clinical relapse 18972317_WT1 protein was not detected in the nasal NK/T-cell lymphoma 19017365_Gynaecologic epithelial histologic type is regulated by WT1 expression through its selective repression of HOX genes. 19067769_These data establish WT1 as a critical repressor of thromboxane A2 receptor promoter Prm1, suppressing TPalpha expression. 19097357_Expression of the WT1 gene product was found in all tumor samples but no statistically significant correlations between WT1 expression and histologic type 19099861_The abnormal ratio of +KTS/-KTS isoforms caused by WT1 mutations along with abnormal expression of podocyte molecules were involved in the pathogenesis of proteinuria. 19120973_These results indicate WT1 protein-derived 16-mer natural peptide, WT1(332) (KRYFKLSHLQMHSRKH) is a promiscuous WT1-specific helper epitope. 19137020_Mutations in WT1 gene is associated with Wilms tumor. 19141860_Observational study of gene-disease association. (HuGE Navigator) 19169475_Constitutional abnormalities of WT1 cause gonadal and renal anomalies and predisposition to neoplasia 19171881_Observational study of gene-disease association. (HuGE Navigator) 19171881_WT1 mutations occur at a significant rate in childhood acute myeloid leukemia and are a novel independent poor prognostic marker 19190340_WT1 gene is expressed in a substantial proportion of hepatocellular carcinoma cell lines contributing, to tumor progression and resistance to chemotherapy. 19212333_results suggest that GATA-1 and/or GATA-2 binding to a GATA site of the 3' enhancer of WT1 played an important role in WT1 gene expression 19221039_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19221039_WT1 mutations witn /FLT3-internal tandem repeat positive are associated with acute myeloid leukemia. 19236519_Wilms' tumour gene 1 (WT1) positivity in endothelial cells surrounding epithelial uterine tumours 19250757_Case report. Malignant mesothelioma of the tunica vaginalis expressing Wilms tumor-1 protein, CD138, the expression of which could help in confirming the histopathological diagnosis and in targeting therapy. 19321755_The authors show that virus infection results in the profound upregulation of Wilms' Tumour 1 (WT1) protein, a transcription factor associated with the negative regulation of a number of growth factors and growth factor receptors, including EGFR. 19322206_Pediatric AML samples harboring WT1 gene mutations in exon 7 revealed significantly higher WT1 expression compared with that of wild-type samples. 19351817_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19404640_Data show that WT1 expression levels were low in mature B-ALL and highest in ALL cases with co-expression of myeloid markers, making it a useful therapeutic target molecule in adult ALL with the exception of mature B-ALL. 19407365_Finding suggest that WT-1 immunohistochemistry may be used to assess both the ME cells and micro-vessel density. 19416806_WTX binds WT1 and enhances WT1-mediated transcription, suggesting a role for WTX in nuclear pathways implicated in the transcriptional regulation of cellular differentiation programs 19437143_There is no evidence that WT1 does regulate the Wnt-4/beta-catenin-independent pathway which is activated in the Pit-1-expressing subset of pituitary adenomas. 19443388_expression level of WT1 was found to be a significant predictor of endometrial cancer relapse 19494353_WT1 mutations present in T-ALL are predominantly heterozygous frameshift mutations resulting in truncation of the C-terminal zinc finger domains of this transcription factor. WT1 mutations do not confer adverse prognosis in pediatric and adult T-ALL. 19536888_Observational study of gene-disease association. (HuGE Navigator) 19549856_WT1 interference with Wnt signaling represents an important mode of its action relevant to the suppression of tumor growth and guidance of development. 19578047_Our results support the combined role of WT1 and nestin in glial tumorigenesis and progression. 19605546_Impaired WT1/Cre-binding protein interplay in addition to advanced glycation end products suppress podocalyxin expression in high glucose-treated human podocytes. 19615003_WT1 is not sufficient for distinguishing melanoma from melanocytic nevi. 19618455_Neither overall survival nor event-free survival was correlated with WT1 expression in 155 pediatric AML patients 19638168_WT1 in nevi and melanomas. WT1 protein is (a)expressed in cytoplasm of neoplastic cell, (b) increased in advanced stages of melanoma progression, (c) associated with shorter overall survival in melanoma. 19730683_Observational study of gene-disease association. (HuGE Navigator) 19747485_Regulated WT1 followed by sequential Egr1 and Sp1 binding to elements within Prm1 mediate repression and subsequent induction of TPalpha during differentiation into the megakaryocytic phenotype. 19749460_the high frequency of loss of allelic integrity at Wilms' tumor suppressor gene-1 locus in high-graded breast tumors is associated with aggressiveness of the tumor. 19752335_greater WT1 transcript reduction after induction combination chemotherapy predicted reduced relapse risk of acute myeloid leukemia 19776535_The WT1 mRNA level is a sensitive biomarker for monitoring minimal residual disease. 19811333_The rise of WT1 expression preceded the hematological relapse by approximately 4 months. WT1 expression higher than 20 WT1 copies /10(4)ABL copies after induction & consolidation chemotherapy was associated with shorter survival in acute myeloid leukemia. 19817904_we draw the attention of histopathologists to the variable expression of p53, WT1 and hormone receptors in a significant proportion of cases of USC 19847202_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19847202_Wilms tumor 1 gene mutations are associated with pediatric T-cell malignancies. 19856421_The combination of MUC5ac and WT-1 stains is useful in distinguishing pancreatic ductal from ovarian serous carcinoma in body fluid cytology. 19951528_Inhibiting the WT1 expression in K562/A02 cells can enhance the drug sensitivity to adriamycin. 20013787_importance of WT1 mutations in acute myeloid leukaemia (review) 20016532_Mutant Wilms' tumor 1 (WT1) mRNA with premature termination codons in acute myeloid leukemia (AML) is sensitive to nonsense-mediated RNA decay (NMD). 20025481_All 97 gastrointestinal stromal tumors were positive for WT-1 and the staining intensity was strong in 59 (60.8%). 20038731_Observational study of gene-disease association. (HuGE Navigator) 20038731_Single nucleotide polymorphism in the mutational hotspot of WT1 predicts a favorable outcome in patients with cytogenetically normal acute myeloid leukemia. 20092642_Data suggest that defects in antigen presentation caused by loss or mutation of WT1 or downregulation of HLA molecules are not the major basis for escape from the immune response induced by WT1 peptide vaccination. 20122399_Proteolysis of WT1 by HtrA2 causes the removal of WT1 from its binding sites at gene promoters, leading to alterations in gene regulation that enhance apoptosis. 20150449_Describe an Italian family with isolated focal sgemental glomerulosclerosis associated with a novel sequence variant in WT1 gene exon 9. 20204298_WT1 promotes estrogen-independent growth and anti-estrogen resistance in ER-positive breast cancer cells presumably through activation of the signaling pathways mediated by the members of EGFR family. 20332316_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 20368469_Observational study of gene-disease association. (HuGE Navigator) 20368469_WT1 mutations are correlated with poor prognosis in AML patients. The mutation status may be changed in some patients during AML progression. 20368538_Observational study of gene-disease association. (HuGE Navigator) 20376582_WT1 maybe induces similar graft vs leukemia effects in both acute leukemia and chronic myelocytic leukemia patients early after stem cell transplantation 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20412098_High WT1 gene expression is associated | ENSMUSG00000016458 | Wt1 | 82.79848 | 1.3081127 | 0.3874868275 | 0.32350639 | 1.437341e+00 | 2.305701e-01 | 6.214596e-01 | No | Yes | 127.215987 | 20.337454 | 97.437783 | 15.064436 | |
| ENSG00000185000 | 8694 | DGAT1 | protein_coding | O75907 | FUNCTION: Catalyzes the terminal and only committed step in triacylglycerol synthesis by using diacylglycerol and fatty acyl CoA as substrates (PubMed:16214399, PubMed:18768481, PubMed:28420705, PubMed:9756920, PubMed:32433611, PubMed:32433610). Highly expressed in epithelial cells of the small intestine and its activity is essential for the absorption of dietary fats (PubMed:18768481). In liver, plays a role in esterifying exogenous fatty acids to glycerol, and is required to synthesize fat for storage (PubMed:16214399). Also present in female mammary glands, where it produces fat in the milk (By similarity). May be involved in VLDL (very low density lipoprotein) assembly (PubMed:18768481). In contrast to DGAT2 it is not essential for survival (By similarity). Functions as the major acyl-CoA retinol acyltransferase (ARAT) in the skin, where it acts to maintain retinoid homeostasis and prevent retinoid toxicity leading to skin and hair disorders (PubMed:16214399). Exhibits additional acyltransferase activities, includin acyl CoA:monoacylglycerol acyltransferase (MGAT), wax monoester and wax diester synthases (By similarity). Also able to use 1-monoalkylglycerol (1-MAkG) as an acyl acceptor for the synthesis of monoalkyl-monoacylglycerol (MAMAG) (PubMed:28420705). {ECO:0000250|UniProtKB:Q8MK44, ECO:0000250|UniProtKB:Q9Z2A7, ECO:0000269|PubMed:16214399, ECO:0000269|PubMed:18768481, ECO:0000269|PubMed:28420705, ECO:0000269|PubMed:32433610, ECO:0000269|PubMed:32433611, ECO:0000269|PubMed:9756920}. | 3D-structure;Acyltransferase;Disease variant;Endoplasmic reticulum;Lipid metabolism;Membrane;Phosphoprotein;Reference proteome;Transferase;Transmembrane;Transmembrane helix | PATHWAY: Lipid metabolism; glycerolipid metabolism. | This gene encodes an multipass transmembrane protein that functions as a key metabolic enzyme. The encoded protein catalyzes the conversion of diacylglycerol and fatty acyl CoA to triacylglycerol. This enzyme can also transfer acyl CoA to retinol. Activity of this protein may be associated with obesity and other metabolic diseases. [provided by RefSeq, Jul 2013]. | hsa:8694; | endoplasmic reticulum membrane [GO:0005789]; integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; specific granule membrane [GO:0035579]; 2-acylglycerol O-acyltransferase activity [GO:0003846]; acyltransferase activity [GO:0016746]; diacylglycerol O-acyltransferase activity [GO:0004144]; identical protein binding [GO:0042802]; O-acyltransferase activity [GO:0008374]; retinol O-fatty-acyltransferase activity [GO:0050252]; diacylglycerol metabolic process [GO:0046339]; fatty acid homeostasis [GO:0055089]; lipid storage [GO:0019915]; long-chain fatty-acyl-CoA metabolic process [GO:0035336]; monoacylglycerol biosynthetic process [GO:0006640]; triglyceride biosynthetic process [GO:0019432]; triglyceride metabolic process [GO:0006641]; very-low-density lipoprotein particle assembly [GO:0034379] | 12123490_Observational study of gene-disease association. (HuGE Navigator) 12123490_Polymorphism associated with alterations in body mass index, high density lipoprotein levels and blood pressure in Turkish women. 12401709_DGAT1 overexpression in murine white adipose tissue provides a model in which obesity does not impair glucose disposal 14557275_DGAT participates in the regulation of membrane lipid synthesis and lipid signaling, thereby playing an important role in modulating cell growth properties 14569040_Observational study of gene-disease association. (HuGE Navigator) 15258194_Niacin selectively inhibited DGAT2 but not DGAT1 activity, but had no effect on the expression of DGAT1 and DGAT2 mRNA 15308631_increasing DGAT1, ACAT1, or ACAT2 expression stimulates the assembly and secretion of VLDL from liver cells 16306352_adipose overexpression of Dgat1 gene in transgenic mice leads to diet-inducible insulin resistance. 16894240_thiazolidinediones upregulate the adipocyte lipid storage genes DGAT and FAS but have no significant effect on LPL 18757836_Review summarizes current knowledge of DGAT1 and DGAT2 enzymes, focusing on new advances since the cloning of their genes, including possible roles in human health and diseases. 18768481_the acylation of acylglycerols by DGAT1 is important for dietary fat absorption in the intestine 19197254_Report visceral and subcutaneous adipose tissue diacylglycerol acyltransferase activity in humans. 20167659_Identify DGAT1 as an important protein that participates in the effect of HNF4A on hepatic secretion of triglyceride-rich lipoproteins. 20935628_The triglyceride-synthesizing enzyme diacylglycerol acyltransferase-1 (DGAT1) is identified as a key host factor for hepatitis C virus infection. 21369919_human small intestinal DGAT, which is mainly encoded by DGAT1, utilizes 1,2-DAG as the substrate to form TAG 21846726_DGAT1 belongs to the family of oligomeric membrane proteins that adopt a dual membrane topology. 21990351_a comprehensive evaluation of a small molecule inhibitor for DGAT1 and suggests that pharmacological inhibition of DGAT1 holds promise in treating diverse metabolic disorders. 22748069_describe distinct but synergistic roles of the two DGATs in an integrated pathway of TAG synthesis and secretion, with DGAT2 acting upstream of DGAT1 23114594_results identify DGAT1 loss-of-function mutations as a rare cause of congenital diarrheal disorders 23317570_The structure-activity relationship studies of a novel series of carboxylic acid derivatives of pyridine-carboxamides as DGAT-1 inhibitors is described. 23420847_Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core 24118885_the apparent lack of therapeutic window owing to GI side effects of AZD7687, particularly diarrhoea, makes the utility of DGAT1 inhibition as a novel treatment for diabetes and obesity questionable. 24573674_MGAT2 functions as a dimeric or tetrameric protein and selectively heterodimerizes with DGAT1 in mammalian cells 24899196_Downregulation of CLDN1 was associated with altered fatty acid homeostasis in the absence of DGAT1. 25740267_Data indicate no clinically relevant pharmacokinetic interaction between DGAT-1 inhibitor pradigastat and rosuvastatin. 26493024_DGAT1 expression is down-regulated in viral hepatitis-related cirrhosis. 26883093_A novel homozygous missense variant was identified in a family with protein losing enteropathy. A splice site mutation in intron 8 was identified in a second family with PLE. These cases of DGAT1 deficiency extend the molecular and phenotypic spectrum of PLE. 28373485_DGAT1 mutations result in a spectrum of diseases 28768178_data reveal an important role for DGAT activity and TG synthesis generally in averting ER stress and lipotoxicity, with specifically DGAT1 performing this function during stimulated lipolysis in adipocytes. 28877685_our findings indicate that inhibition of both DGAT1 and ABHD5 using siRNA leads to reduction in prostate cancer cell growth. 29604290_Study identified a large cohort of patients with congenital diarrheal disorders with mutations in DGAT1 that reduced expression of its product; dermal fibroblasts and intestinal organoids derived from these patients had altered lipid metabolism and were susceptible to lipid-induced cell death. Expression of full-length wildtype DGAT1 or DGAT2 restored normal lipid metabolism in these cells. 30095213_DGAT1 loss causes global changes in enterocyte polarized trafficking that could account for deficits in absorption seen in the patient with neonatal diarrhea. 30790345_Decreased DGAT1 activity can reduce circulating TG and liver TG. 30816200_DGAT1 Inhibitor Suppresses Prostate Tumor Growth and Migration by Regulating Intracellular Lipids and Non-Centrosomal MTOC Protein GM130. 30853196_Identified 1 patient with compound heterozygous CCBE1 and 2 with novel DGAT1 mutations in cohort of 9 Chinese children. Two of the 3 patients with DGAT1 mutations died, suggesting that this genotype is associated with an especially severe phenotype. 31778854_Genetic variants in DGAT1 cause diverse clinical presentations of malnutrition through a specific molecular mechanism. 32433610_cryo-electron microscopy structure of human DGAT1 in complex with an oleoyl-CoA substrate 32433611_structure of dimeric human DGAT1, a member of the membrane-bound O-acyltransferase (MBOAT) family, by cryo-electron microscopy at approximately 3.0 A resolution 32559414_Targeting DGAT1 Ameliorates Glioblastoma by Increasing Fat Catabolism and Oxidative Stress. 33750350_Up-regulation of DGAT1 in cancer tissues and tumor-infiltrating macrophages influenced survival of patients with gastric cancer. 33824421_Loss of ephrin B2 receptor (EPHB2) sets lipid rheostat by regulating proteins DGAT1 and ATGL inducing lipid droplet storage in prostate cancer cells. 34095320_DGAT1 Expression Promotes Ovarian Cancer Progression and Is Associated with Poor Prognosis. | ENSMUSG00000022555 | Dgat1 | 719.37421 | 0.6867542 | -0.5421342198 | 0.12597390 | 1.820233e+01 | 1.986360e-05 | 3.532756e-03 | No | Yes | 724.697525 | 105.856025 | 1054.784109 | 150.351008 |
| ENSG00000185418 | 123283 | TARS3 | protein_coding | A2RTX5 | FUNCTION: Catalyzes the attachment of threonine to tRNA(Thr) in a two-step reaction: threonine is first activated by ATP to form Thr-AMP and then transferred to the acceptor end of tRNA(Thr). Also edits incorrectly charged tRNA(Thr) via its editing domain, at the post-transfer stage. {ECO:0000250|UniProtKB:Q8BLY2}. | ATP-binding;Acetylation;Alternative splicing;Aminoacyl-tRNA synthetase;Coiled coil;Cytoplasm;Direct protein sequencing;Ligase;Nucleotide-binding;Nucleus;Phosphoprotein;Protein biosynthesis;Reference proteome | hsa:123283; | cytoplasm [GO:0005737]; nucleus [GO:0005634]; ATP binding [GO:0005524]; threonine-tRNA ligase activity [GO:0004829]; threonyl-tRNA aminoacylation [GO:0006435] | Mouse_homologues 29579307_Our results provided the first analysis of the aminoacylation and editing activities of ThrRS-L, and improved our understanding of Tarsl2. 31287872_Newly acquired N-terminal extension targets threonyl-tRNA synthetase-like protein into the multiple tRNA synthetase complex. | ENSMUSG00000030515 | Tarsl2 | 1132.08771 | 0.9263890 | -0.1103100212 | 0.11166941 | 9.720824e-01 | 3.241614e-01 | 7.032053e-01 | No | Yes | 1369.329746 | 194.642792 | 1412.105657 | 196.291560 | ||
| ENSG00000185917 | 54093 | SETD4 | protein_coding | Q9NVD3 | FUNCTION: Histone-lysine N-methyltransferase that acts as a regulator of cell proliferation, cell differentiation and inflammatory response (PubMed:31308046). Regulates the inflammatory response by mediating mono- and dimethylation of 'Lys-4' of histone H3 (H3K4me1 and H3K4me2, respectively), leading to activate the transcription of proinflammatory cytokines IL6 and TNF-alpha (By similarity). Also involved in the regulation of stem cell quiescence by catalyzing the trimethylation of 'Lys-20' of histone H4 (H4K20me3), thereby promoting heterochromatin formation (PubMed:31308046). Involved in proliferation, migration, paracrine and myogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) (By similarity). {ECO:0000250|UniProtKB:P58467, ECO:0000269|PubMed:31308046}. | Alternative splicing;Cytoplasm;Inflammatory response;Methyltransferase;Nucleus;Reference proteome;S-adenosyl-L-methionine;Transferase | hsa:54093; | cytosol [GO:0005829]; nucleus [GO:0005634]; histone methyltransferase activity (H3-K4 specific) [GO:0042800]; histone methyltransferase activity (H4-K20 specific) [GO:0042799]; protein-lysine N-methyltransferase activity [GO:0016279]; histone H3-K4 dimethylation [GO:0044648]; histone H3-K4 monomethylation [GO:0097692]; histone H4-K20 trimethylation [GO:0034773]; inflammatory response [GO:0006954]; peptidyl-lysine monomethylation [GO:0018026]; peptidyl-lysine trimethylation [GO:0018023]; positive regulation of inflammatory response [GO:0050729]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of tumor necrosis factor production [GO:0032760]; regulation of cell proliferation in bone marrow [GO:0071863] | 31308046_High SETD4 expression is associated with Breast Cancer. | ENSMUSG00000022948 | Setd4 | 887.82963 | 1.0628191 | 0.0878959925 | 0.12538841 | 4.928340e-01 | 4.826658e-01 | 8.112443e-01 | No | Yes | 1116.691579 | 137.517058 | 1033.615401 | 124.454489 | ||
| ENSG00000185986 | 728609 | SDHAP3 | transcribed_unprocessed_pseudogene | 399.06286 | 1.0047646 | 0.0068575358 | 0.19304449 | 1.260593e-03 | 9.716772e-01 | 9.924630e-01 | No | Yes | 557.163634 | 121.175708 | 566.693873 | 120.284803 | ||||||||||
| ENSG00000186017 | 84924 | ZNF566 | protein_coding | Q969W8 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | Mouse_homologues NA; + ;NA | hsa:84924; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | Mouse_homologues NA; + ;NA | ENSMUSG00000078768+ENSMUSG00000058447 | Zfp566+Gm26920 | 422.87655 | 1.2669214 | 0.3413270715 | 0.16446130 | 4.331418e+00 | 3.741506e-02 | 2.982566e-01 | No | Yes | 563.050640 | 141.007377 | 418.066608 | 102.680950 | |
| ENSG00000186162 | 152302 | CIDECP1 | transcribed_unprocessed_pseudogene | 216.01508 | 0.8933214 | -0.1627487155 | 0.20035796 | 6.575470e-01 | 4.174273e-01 | 7.721303e-01 | No | Yes | 240.391865 | 26.226822 | 261.465798 | 27.813437 | ||||||||||
| ENSG00000186469 | 54331 | GNG2 | protein_coding | P59768 | FUNCTION: Guanine nucleotide-binding proteins (G proteins) are involved as a modulator or transducer in various transmembrane signaling systems. The beta and gamma chains are required for the GTPase activity, for replacement of GDP by GTP, and for G protein-effector interaction (By similarity). {ECO:0000250}. | 3D-structure;Acetylation;Cell membrane;Lipoprotein;Membrane;Methylation;Prenylation;Reference proteome;Transducer | This gene encodes one of the gamma subunits of a guanine nucleotide-binding protein. Such proteins are involved in signaling mechanisms across membranes. Various subunits forms heterodimers which then interact with the different signal molecules. [provided by RefSeq, Aug 2011]. | hsa:54331; | extracellular exosome [GO:0070062]; heterotrimeric G-protein complex [GO:0005834]; membrane [GO:0016020]; plasma membrane [GO:0005886]; G-protein beta-subunit binding [GO:0031681]; adenylate cyclase-activating dopamine receptor signaling pathway [GO:0007191]; cell population proliferation [GO:0008283]; cellular response to catecholamine stimulus [GO:0071870]; cellular response to prostaglandin E stimulus [GO:0071380]; G protein-coupled receptor signaling pathway [GO:0007186] | 15105422_Data show that G protein inhibition of N-type calcium channels is critically dependent on two separate but adjacent approximately 20-amino acid regions of the Gbeta subunit, as examined with Gbetas 1 and 5 and Ggamma2. 15747776_10 genes were down-regulated following treatment of the T-ALL cells with 0.15 and 1.5 microg/mL of metal ores at 72 h 17492941_Fission of transport carriers at the trans-Golgi network is dependent on specifically PLCbeta3, which is necessary to activate PKCeta and PKD in that Golgi compartment, via diacylglycerol production. 18045877_signaling pathway by which G(i)-coupled receptor specifically induces Rac and Cdc42 activation through direct interaction of Gbetagamma with FLJ00018. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20007712_Data show that activation of PLCbeta(2) by alpha(q) and beta1gamma2 differ from activation by Rac2 and from each other. 20181083_Data implicate the domain I-II linker region as an important contributor to voltage dependent Gbeta1/Ggamma2 modulation of Cav2.2 calcium channels. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21679469_Gbetagamma inhibits Epac-induced Ca 2+ elevation in melanoma cells. Cross talk of Ca 2+ signaling between Gbetagamma & Epac plays a major role in melanoma cell migration. 22957104_presence of Gng2 and Netrin-G2 immunoreactive elements in the insular cortex, but not in the putamen, suggests a possible common ontogeny of the claustrum and insula 23031273_increased protein expression level of GNG2 alone inhibits proliferation of malignant melanoma cells in vitro and in vivo 24462769_Data indicate that endogenous mTOR interacts with Gbetagamma. 25367286_Alteration of gene expression profiling including GPR174 and GNG2 is associated with vasovagal syncope. 25941381_G-protein betagamma subunits are positive regulators of Kv7.4 and native vascular Kv7 channel activity. 28818508_High GNG2 expression is associated with alcoholic hepatitis. 30928649_this paper shows that GNG2 gene polymorphism is associated with IgA nephropathy risk in Chinese Han population 31730405_Low GNG2 expression is associated with Abdominal Aortic Aneurysm. 34212982_Interference with KCNJ2 inhibits proliferation, migration and EMT progression of apillary thyroid carcinoma cells by upregulating GNG2 expression. | ENSMUSG00000043004 | Gng2 | 15.04271 | 0.5544311 | -0.8509198759 | 0.71824341 | 1.366457e+00 | 2.424216e-01 | No | Yes | 16.987943 | 6.743250 | 28.856903 | 11.528397 | ||
| ENSG00000186481 | ANKRD20A5P | transcribed_unprocessed_pseudogene | 24.90379 | 1.4271956 | 0.5131830674 | 0.57993105 | 7.826645e-01 | 3.763275e-01 | No | Yes | 29.791344 | 13.609186 | 21.713322 | 9.711330 | ||||||||||||
| ENSG00000186523 | 85002 | FAM86B1 | protein_coding | Q8N7N1 | Mouse_homologues FUNCTION: Catalyzes the trimethylation of eukaryotic elongation factor 2 (EEF2) on 'Lys-525'. {ECO:0000250|UniProtKB:Q96G04}. | Alternative splicing;Methyltransferase;Reference proteome;S-adenosyl-L-methionine;Transferase | hsa:85002; | methyltransferase activity [GO:0008168]; methylation [GO:0032259] | ENSMUSG00000022544 | Eef2kmt | 152.18872 | 0.7749342 | -0.3678542049 | 0.24448360 | 2.244776e+00 | 1.340663e-01 | 5.114587e-01 | No | Yes | 139.000481 | 18.778456 | 178.198932 | 23.382100 | |||
| ENSG00000186654 | 55615 | PRR5 | protein_coding | P85299 | FUNCTION: Subunit of mTORC2, which regulates cell growth and survival in response to hormonal signals. mTORC2 is activated by growth factors, but, in contrast to mTORC1, seems to be nutrient-insensitive. mTORC2 seems to function upstream of Rho GTPases to regulate the actin cytoskeleton, probably by activating one or more Rho-type guanine nucleotide exchange factors. mTORC2 promotes the serum-induced formation of stress-fibers or F-actin. mTORC2 plays a critical role in AKT1 'Ser-473' phosphorylation, which may facilitate the phosphorylation of the activation loop of AKT1 on 'Thr-308' by PDK1 which is a prerequisite for full activation. mTORC2 regulates the phosphorylation of SGK1 at 'Ser-422'. mTORC2 also modulates the phosphorylation of PRKCA on 'Ser-657'. PRR5 plays an important role in regulation of PDGFRB expression and in modulation of platelet-derived growth factor signaling. May act as a tumor suppressor in breast cancer. {ECO:0000269|PubMed:15718101, ECO:0000269|PubMed:17599906}. | Alternative splicing;Cell cycle;Phosphoprotein;Reference proteome;Tumor suppressor | This gene encodes a protein with a proline-rich domain. This gene is located in a region of chromosome 22 reported to contain a tumor suppressor gene that may be involved in breast and colorectal tumorigenesis. The protein is a component of the mammalian target of rapamycin complex 2 (mTORC2), and it regulates platelet-derived growth factor (PDGF) receptor beta expression and PDGF signaling to Akt and S6K1. Alternative splicing and the use of alternative promoters results in transcripts encoding different isoforms. Read-through transcripts from this gene into the downstream Rho GTPase activating protein 8 (ARHGAP8) gene also exist, which led to the original description of PRR5 and ARHGAP8 being a single gene. [provided by RefSeq, Nov 2010]. | hsa:55615; | cytosol [GO:0005829]; TORC2 complex [GO:0031932]; cell cycle [GO:0007049]; phosphorylation [GO:0016310]; positive regulation of phosphatidylinositol 3-kinase signaling [GO:0014068]; positive regulation of protein phosphorylation [GO:0001934]; TORC2 signaling [GO:0038203] | 15718101_mRNA expression analyses revealed PRR5 overexpression in a majority of colorectal tumors but substantial downregulation of PRR5 expression in a subset of breast tumors and reduced expression in two breast cancer cell lines 17461779_It was demonstrated that immunoprecipitation of Protor-1 or Protor-2 results in the co-immunoprecipitation of other mTORC2 subunits, but not Raptor, a specific component of mTORC1. 17599906_The inhibition of Akt and S6K1 phosphorylation by PRR5 knock down correlates with reduction in the expression level of platelet-derived growth factor receptor beta (PDGFRbeta). 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 29391396_Fusion protein PRR5-ARHGAP8 plays a role in bipolar disorder with binge eating behavior. | ENSMUSG00000036106 | Prr5 | 435.35064 | 1.0174692 | 0.0249850736 | 0.16379983 | 2.329311e-02 | 8.786973e-01 | 9.677578e-01 | No | Yes | 542.694899 | 83.685520 | 509.833760 | 76.811195 | |
| ENSG00000186998 | 129080 | EMID1 | protein_coding | Q96A84 | Alternative splicing;Collagen;Disulfide bond;Extracellular matrix;Glycoprotein;Reference proteome;Secreted;Signal | hsa:129080; | collagen trimer [GO:0005581]; endoplasmic reticulum [GO:0005783]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; Golgi apparatus [GO:0005794] | 19913121_Observational study of gene-disease association. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) | ENSMUSG00000034164 | Emid1 | 107.32936 | 0.8945464 | -0.1607718215 | 0.28129477 | 3.258009e-01 | 5.681422e-01 | 8.531186e-01 | No | Yes | 116.420553 | 17.807222 | 124.677085 | 18.408488 | |||
| ENSG00000188283 | 163087 | ZNF383 | protein_coding | Q8NA42 | FUNCTION: May function as a transcriptional repressor, suppressing transcriptional activities mediated by MAPK signaling pathways. {ECO:0000269|PubMed:15964543}. | Cytoplasm;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | The protein encoded by this gene is a KRAB-related zinc finger protein that inhibits the transcription of some MAPK signaling pathway genes. The repressor activity resides in the KRAB domain of the encoded protein. [provided by RefSeq, Sep 2016]. | hsa:163087; | cytoplasm [GO:0005737]; nuclear membrane [GO:0031965]; nucleoplasm [GO:0005654]; DNA-binding transcription factor activity [GO:0003700]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 15964543_Overexpression of ZNF383 in cells inhibits the transcriptional activities of AP-1 and SRE, suggesting that ZNF383 may act as a negative regulator in MAPK-mediated signaling pathways. | ENSMUSG00000099689 | Zfp383 | 281.47925 | 1.4159535 | 0.5017738556 | 0.20820466 | 5.835870e+00 | 1.570265e-02 | 2.023672e-01 | No | Yes | 375.487377 | 86.901534 | 260.569928 | 59.192438 | |
| ENSG00000188659 | 283726 | SAXO2 | protein_coding | Q658L1 | Alternative splicing;Reference proteome | hsa:283726; | axonemal microtubule [GO:0005879]; centriole [GO:0005814]; ciliary basal body [GO:0036064]; cytoskeleton [GO:0005856]; sperm flagellum [GO:0036126]; microtubule binding [GO:0008017]; microtubule anchoring [GO:0034453] | ENSMUSG00000038570 | Saxo2 | 75.00957 | 0.7959096 | -0.3293234496 | 0.35591707 | 8.431444e-01 | 3.584990e-01 | No | Yes | 53.141593 | 11.111060 | 63.430245 | 12.938067 | |||||
| ENSG00000196155 | 25894 | PLEKHG4 | protein_coding | Q58EX7 | FUNCTION: Possible role in intracellular signaling and cytoskeleton dynamics at the Golgi. | Alternative splicing;Guanine-nucleotide releasing factor;Phosphoprotein;Reference proteome | The protein encoded by this gene can function as a guanine nucleotide exchange factor (GEF) and may play a role in intracellular signaling and cytoskeleton dynamics at the Golgi apparatus. Polymorphisms in the region of this gene have been found to be associated with spinocerebellar ataxia in some study populations. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2015]. | hsa:25894; | cytosol [GO:0005829]; guanyl-nucleotide exchange factor activity [GO:0005085]; regulation of small GTPase mediated signal transduction [GO:0051056] | 12796826_Spinocerebellar ataxia type 4 (SCA4) is mapped to chromosome 16q22.1 in northern germany.Haplotype analyses refined the gene locus to a 3.69 cM interval between D16S3019 and D16S512. 15148151_Observational study of genotype prevalence. (HuGE Navigator) 15455264_the autosomal dominant cerebellar ataxia that we have characterized is allelic with SCA4 and Japanese 16q-linked ADCA type III. 16001362_puratrophin-1 has a role in intracellular signaling and actin dynamics at the Golgi apparatus 16491300_Mutations of the puratrophin1 gene on chromosome 16q22.1 are not a common genetic cause of cerebellar ataxia in a European population. 16491300_Observational study of genotype prevalence. (HuGE Navigator) 16780885_We found the C-to-T substitution in the puratrophin-1 gene in 20 patients with ataxia (16 heterozygotes and four homozygotes) and four asymptomatic carriers in 9 of 24 families with an unknown type of ADCA. 17357132_among 686 autosomal dominant spinocerebellar ataxia families in our cohort, 57 families were identified to have 65 affected individuals, who carried the C-to-T substitution of the puratrophin-1 gene 17611710_Disease locus of 16q-autosomal dominant cerebellar ataxia was definitely confined to a 900-kb genomic region between the SNP04 and the -16C>T substitution in the puratrophin-1 gene in 16q22.1. 18482007_Rac1 activation specifically in membrane ruffles by the guanine-nucleotide-exchange factor FLJ00068 is sufficient for insulin induction of glucose uptake into skeletal-muscle cells. 19065522_Observational study of gene-disease association. (HuGE Navigator) 19065522_The mutation of c.-16C to T of the PURATROPHIN-1 gene might be rare in SCA patients in China. 20424877_(TGGAA)(n) repeats in the insertion mutation of PLEKHG4 are related to the pathogenesis of SCA31 21357611_This letter suggested cerebellar ataxia due to a pentanucleotide repeat (TAGAA) expansion on the puratrophin-1 (PLEKHG4) gene on chromosome 16q-22.1. 25025572_Role of the guanine nucleotide exchange factor in Akt2-mediated plasma membrane translocation of GLUT4 in insulin-stimulated skeletal muscle. | ENSMUSG00000014782 | Plekhg4 | 424.95695 | 0.7502198 | -0.4146147484 | 0.16176188 | 6.488469e+00 | 1.085764e-02 | 1.695671e-01 | No | Yes | 375.811475 | 45.804033 | 497.001920 | 58.663472 | |
| ENSG00000196204 | 441191 | RNF216P1 | lncRNA | 1140.94885 | 0.8839931 | -0.1778929191 | 0.10234726 | 3.007038e+00 | 8.290364e-02 | 4.201805e-01 | No | Yes | 1289.362746 | 82.361081 | 1412.659595 | 87.923826 | ||||||||||
| ENSG00000196312 | 84278 | MFSD14C | transcribed_unprocessed_pseudogene | Q5VZR4 | Membrane;Reference proteome;Transmembrane;Transmembrane helix;Transport | integral component of membrane [GO:0016021]; transmembrane transporter activity [GO:0022857] | 659.63432 | 1.0681230 | 0.0950778414 | 0.13040096 | 5.328601e-01 | 4.654069e-01 | 8.002576e-01 | No | Yes | 659.897678 | 60.860948 | 596.454507 | 53.707520 | |||||||
| ENSG00000196367 | 8295 | TRRAP | protein_coding | Q9Y4A5 | FUNCTION: Adapter protein, which is found in various multiprotein chromatin complexes with histone acetyltransferase activity (HAT), which gives a specific tag for epigenetic transcription activation. Component of the NuA4 histone acetyltransferase complex which is responsible for acetylation of nucleosomal histones H4 and H2A. Plays a central role in MYC transcription activation, and also participates in cell transformation by MYC. Required for p53/TP53-, E2F1- and E2F4-mediated transcription activation. Also involved in transcription activation mediated by the adenovirus E1A, a viral oncoprotein that deregulates transcription of key genes. Probably acts by linking transcription factors such as E1A, MYC or E2F1 to HAT complexes such as STAGA thereby allowing transcription activation. Probably not required in the steps following histone acetylation in processes of transcription activation. May be required for the mitotic checkpoint and normal cell cycle progression. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AZ1 from the nucleosome. May play a role in the formation and maintenance of the auditory system (By similarity). {ECO:0000250|UniProtKB:A0A0R4ITC5, ECO:0000269|PubMed:11418595, ECO:0000269|PubMed:12138177, ECO:0000269|PubMed:12660246, ECO:0000269|PubMed:12743606, ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:17967892, ECO:0000269|PubMed:24463511, ECO:0000269|PubMed:9708738}. | Acetylation;Activator;Alternative splicing;Autism spectrum disorder;Chromatin regulator;Deafness;Direct protein sequencing;Disease variant;Isopeptide bond;Mental retardation;Non-syndromic deafness;Nucleus;Phosphoprotein;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | This gene encodes a large multidomain protein of the phosphoinositide 3-kinase-related kinases (PIKK) family. The encoded protein is a common component of many histone acetyltransferase (HAT) complexes and plays a role in transcription and DNA repair by recruiting HAT complexes to chromatin. Deregulation of this gene may play a role in several types of cancer including glioblastoma multiforme. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Sep 2011]. | hsa:8295; | Golgi apparatus [GO:0005794]; NuA4 histone acetyltransferase complex [GO:0035267]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; SAGA complex [GO:0000124]; Swr1 complex [GO:0000812]; transcription factor TFTC complex [GO:0033276]; transcription coregulator activity [GO:0003712]; chromatin organization [GO:0006325]; DNA repair [GO:0006281]; histone acetylation [GO:0016573]; histone deubiquitination [GO:0016578]; histone H2A acetylation [GO:0043968]; histone H3 acetylation [GO:0043966]; histone H4 acetylation [GO:0043967]; monoubiquitinated histone deubiquitination [GO:0035521]; monoubiquitinated histone H2A deubiquitination [GO:0035522]; positive regulation of double-strand break repair via homologous recombination [GO:1905168]; regulation of cell cycle [GO:0051726]; regulation of transcription by RNA polymerase II [GO:0006357]; regulation of transcription, DNA-templated [GO:0006355] | 12077335_TRRAP binding and the recruitment of histone H3 and H4 acetyltransferase activities are required for the transactivation of a silent TERT gene in exponentially growing human fibroblasts by c-Myc or N-Myc protein. 12138177_Data suggest a model in which p53 directly recruits a TRRAP/acetyltransferase complex to the mdm2 gene to activate transcription. In addition, this study defines a novel mechanism utilized by the p53 tumor suppressor to regulate gene expression. 12660246_requirement in transcription activation via c-myc transformation domain 15647280_Results identify YL1 as a subunit of the TRRAP/TIP60 HAT complex, and also as a component of a novel mammalian multiprotein complex that includes the SNF2-related helicase SRCAP. 16341205_HAT cofactor Trrap and Tip60 HAT bind to the chromatin surrounding sites of DNA double-strand breaks (DSBs). Cells may use the same basic mechanism involving HAT complexes to regulate distinct cellular processes, such as transcription and DNA repair. 17694078_Several function of TRRAP during cellular process like: DNA repair, cell cycle, transcription etc... 17967892_NPAT recruits the TRRAP-Tip60 complex to histone gene promoters to coordinate the transcriptional activation of multiple histone genes during the G(1)/S-phase transition 19066453_Loss of Transduction; transformation-transcription domain-associated protein leads to a reduced level of beta-catenin ubiquitination, accumulation of beta-catenin protein and concomitant hyperactivation of the Wnt Proteins pathway. 20085741_knockdown of the adaptor protein TRRAP significantly increased differentiation of cultured brain tumor-initiating cells, sensitized the cells to apoptotic stimuli, and negatively affected cell cycle progression. 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 20943076_The over-expression of TRA1 in hepatocirrhosis and hepatocellular carcinoma is correlated with the formation and development of HCC. 21499247_TRRAP harbors a recurrent mutation that clustered in one position (p. Ser722Phe) in 6 out of 167 melanoma samples (approx. 4%). The nature, pattern and functional evaluation of the TRRAP recurrent mutation suggest that TRRAP functions as an oncogene. 23318449_TRRAP is targeted for destruction in a cell cycle-dependent fashion. 23362228_Findings establish Trrap as a critical part of the mechanism that restricts differentiation and promotes the maintenance of key features of ESCs. 24349473_study defined eight additional recurrently mutated genes in SMZL; these genes are CREBBP, CBFA2T3, AMOTL1, FAT4, FBXO11, PLA2G4D, TRRAP and USH2A. 24705139_MYC associates with STAGA through extended interactions of the TAD with both TRRAP and GCN5. 25925205_Data suggest that ubiquitin thioesterase 7 (HAUSP) may act as an oncogenic protein that can modulate c-MYC protein expression via transformation-transcription domain-associated protein (TRRAP). 29588376_TRRAP, an essential component of multiple histone acetyltransferase complexes, was identified as a central regulator of multiciliated cell formation. 29653964_TRRAP silencing attenuated p53 accumulation in lymphoma and colon cancer models, whereas TRRAP overexpression increased mutp53 levels, suggesting a role for TRRAP across cancer entities and p53 mutations. 29936929_TRRAP plays an important role in the regulation of the proliferation and stemness of ovarian cancer stem cells. 30827496_Missense Variants in the TRRAP gene Causes Autism and Syndromic Intellectual Disability. 31188495_Our results uncover a role for TRRAP/KAT5 in promoting hepatocellular carcinoma cell proliferation by activating mitotic genes. Targeting the TRRAP/KAT5 complex is a potential therapeutic strategy for hepatocellular carcinoma 34156146_Lipid droplet screen in human hepatocytes identifies TRRAP as a regulator of cellular triglyceride metabolism. 34830324_Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. 35151693_Interaction of transcription factor FoxO3 with histone acetyltransferase complex subunit TRRAP modulates gene expression and apoptosis. | ENSMUSG00000045482 | Trrap | 5617.24243 | 1.0512512 | 0.0721074395 | 0.07643953 | 8.916825e-01 | 3.450218e-01 | 7.185391e-01 | No | Yes | 6281.979322 | 586.610018 | 5802.902943 | 529.859489 | |
| ENSG00000196437 | 148266 | ZNF569 | protein_coding | Q5MCW4 | FUNCTION: May be involved in transcriptional regulation. {ECO:0000250}. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:148266; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 16793018_ZNF569 protein may act as a transcriptional repressor that suppresses MAPK signaling pathway to mediate cellular functions | ENSMUSG00000059975 | Zfp74 | 486.93538 | 0.9996164 | -0.0005535230 | 0.15069315 | 1.347568e-05 | 9.970710e-01 | 9.986332e-01 | No | Yes | 563.139908 | 149.605957 | 564.553386 | 147.064639 | ||
| ENSG00000196653 | 91392 | ZNF502 | protein_coding | Q8TBZ5 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Host-virus interaction;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:91392; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; positive regulation by host of viral process [GO:0044794]; regulation of transcription, DNA-templated [GO:0006355]; viral release from host cell [GO:0019076] | 25556234_Results present initial characterization of key proteins Cav2 and CFL1 as cellular factors that colocalize with M in viral inclusions and filaments and ZNF502 protein which appears to interact with RSV M in the nucleus. | 78.46013 | 1.1193957 | 0.1627201208 | 0.32931722 | 2.449892e-01 | 6.206256e-01 | 8.769566e-01 | No | Yes | 98.545280 | 29.186458 | 84.194334 | 24.537630 | ||||
| ENSG00000196693 | 7582 | ZNF33B | protein_coding | Q06732 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | This gene encodes a member of the zinc finger family of proteins. This gene shows decreased expression in cumulus cells derived from patients undergoing controlled ovarian stimulation. This gene is present in a gene cluster with several related zinc finger genes in the pericentromeric region of chromosome 10. Pseudogenes have been identified on chromosomes 7 and 10. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Feb 2015]. | hsa:7582; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385451_Observational study of gene-disease association. (HuGE Navigator) | 840.26125 | 0.9255459 | -0.1116235902 | 0.14110552 | 6.223586e-01 | 4.301722e-01 | 7.797293e-01 | No | Yes | 921.327455 | 126.245795 | 966.036726 | 129.445795 | |||
| ENSG00000196839 | 100 | ADA | protein_coding | P00813 | FUNCTION: Catalyzes the hydrolytic deamination of adenosine and 2-deoxyadenosine (PubMed:8452534, PubMed:16670267). Plays an important role in purine metabolism and in adenosine homeostasis. Modulates signaling by extracellular adenosine, and so contributes indirectly to cellular signaling events. Acts as a positive regulator of T-cell coactivation, by binding DPP4 (PubMed:20959412). Its interaction with DPP4 regulates lymphocyte-epithelial cell adhesion (PubMed:11772392). Enhances dendritic cell immunogenicity by affecting dendritic cell costimulatory molecule expression and cytokines and chemokines secretion (By similarity). Enhances CD4+ T-cell differentiation and proliferation (PubMed:20959412). Acts as a positive modulator of adenosine receptors ADORA1 and ADORA2A, by enhancing their ligand affinity via conformational change (PubMed:23193172). Stimulates plasminogen activation (PubMed:15016824). Plays a role in male fertility (PubMed:21919946, PubMed:26166670). Plays a protective role in early postimplantation embryonic development (By similarity). {ECO:0000250|UniProtKB:P03958, ECO:0000250|UniProtKB:P56658, ECO:0000269|PubMed:11772392, ECO:0000269|PubMed:15016824, ECO:0000269|PubMed:16670267, ECO:0000269|PubMed:20959412, ECO:0000269|PubMed:21919946, ECO:0000269|PubMed:23193172, ECO:0000269|PubMed:26166670, ECO:0000269|PubMed:8452534}. | 3D-structure;Acetylation;Cell adhesion;Cell junction;Cell membrane;Cytoplasm;Cytoplasmic vesicle;Direct protein sequencing;Disease variant;Hereditary hemolytic anemia;Hydrolase;Lysosome;Membrane;Metal-binding;Nucleotide metabolism;Reference proteome;SCID;Zinc | This gene encodes an enzyme that catalyzes the hydrolysis of adenosine to inosine in the purine catabolic pathway. Various mutations have been described for this gene and have been linked to human diseases related to impaired immune function such as severe combined immunodeficiency disease (SCID) which is the result of a deficiency in the ADA enzyme. In ADA-deficient individuals there is a marked depletion of T, B, and NK lymphocytes, and consequently, a lack of both humoral and cellular immunity. Conversely, elevated levels of this enzyme are associated with congenital hemolytic anemia. [provided by RefSeq, Sep 2019]. | hsa:100; | cell junction [GO:0030054]; cell surface [GO:0009986]; cytoplasmic vesicle lumen [GO:0060205]; cytosol [GO:0005829]; external side of plasma membrane [GO:0009897]; lysosome [GO:0005764]; membrane [GO:0016020]; plasma membrane [GO:0005886]; 2'-deoxyadenosine deaminase activity [GO:0046936]; adenosine deaminase activity [GO:0004000]; zinc ion binding [GO:0008270]; adenosine catabolic process [GO:0006154]; allantoin metabolic process [GO:0000255]; AMP catabolic process [GO:0006196]; AMP salvage [GO:0044209]; cell adhesion [GO:0007155]; dAMP catabolic process [GO:0046059]; dATP catabolic process [GO:0046061]; deoxyadenosine catabolic process [GO:0006157]; embryonic digestive tract development [GO:0048566]; germinal center B cell differentiation [GO:0002314]; GMP salvage [GO:0032263]; hypoxanthine salvage [GO:0043103]; inosine biosynthetic process [GO:0046103]; liver development [GO:0001889]; lung alveolus development [GO:0048286]; negative regulation of adenosine receptor signaling pathway [GO:0060169]; negative regulation of inflammatory response [GO:0050728]; negative regulation of leukocyte migration [GO:0002686]; negative regulation of mature B cell apoptotic process [GO:0002906]; negative regulation of mucus secretion [GO:0070256]; negative regulation of penile erection [GO:0060407]; negative regulation of thymocyte apoptotic process [GO:0070244]; Peyer's patch development [GO:0048541]; placenta development [GO:0001890]; positive regulation of alpha-beta T cell differentiation [GO:0046638]; positive regulation of B cell proliferation [GO:0030890]; positive regulation of calcium-mediated signaling [GO:0050850]; positive regulation of germinal center formation [GO:0002636]; positive regulation of heart rate [GO:0010460]; positive regulation of smooth muscle contraction [GO:0045987]; positive regulation of T cell differentiation in thymus [GO:0033089]; positive regulation of T cell receptor signaling pathway [GO:0050862]; purine nucleotide salvage [GO:0032261]; purine-containing compound salvage [GO:0043101]; regulation of cell-cell adhesion mediated by integrin [GO:0033632]; response to hypoxia [GO:0001666]; T cell activation [GO:0042110]; trophectodermal cell differentiation [GO:0001829]; xanthine biosynthetic process [GO:0046111] | 11121182_Observational study of gene-disease association. (HuGE Navigator) 11354825_Observational study of gene-disease association. (HuGE Navigator) 11534018_Observational study of gene-disease association. (HuGE Navigator) 11901152_Clustered charged amino acids of human adenosine deaminase comprise a functional epitope for binding the adenosine deaminase complexing protein CD26/dipeptidyl peptidase IV 12104097_Relationship between plasma malondialdehyde levels and adenosine deaminase activities in preeclampsia. 12113294_serum adenosine deaminase-2 (but not adenosine deaminase-1) and cytidine deaminase levels were significantly higher in systemic lupus erythematosus patients 12381379_In humans, ADA1 was mainly purified concomitant with ADA-binding protein and dipeptidyl peptidase IV, was not adsorbed in adenosine-Sepharose, but was adsorbed in IgG anti-ADA1-Sepharose column; properties compared with chicken ADA1 12712614_The activity of this enzyme was studied in tissues, erythrocytes, and blood plasma of patients with peptic ulcer both in its uncomplicated course and in the development of complications. 12774669_The activity of ADA was determined in lymphocytes, esoinophils and blood serum in patients with bronchial asthma. 12809673_ADA was analyzed by linkage disequilibrium and no correlation was found between hot spots of equal and unequal homologous recombination. 12935677_Observational study of genotype prevalence. (HuGE Navigator) 14726805_Meta-analysis and HuGE review of genotype prevalence, gene-disease association, genetic testing, and healthcare-related. (HuGE Navigator) 15016824_Adenosine deaminase binding is diminished by mutation of ADA residues known to interact with CD26. 15063762_Observational study of gene-disease association. (HuGE Navigator) 15257174_Observational study of gene-disease association. (HuGE Navigator) 15257174_adenosine-related gene variants do not appear to alter susceptibility to the disease in this group of essential hypertensives 15281007_adenosine deaminase contributes to the clinical manifestations of type 2 diabetes and probably also have a marginal influence on susceptibility to the disease 15632314_The adenosine deaminase (ADA) gene was highly up-regulated in Ara-C-resistant cells, while equilibrative nucleoside transporter 1 (ENT1) and several cell-cycle-related genes were down-regulated. 15761857_Observational study of gene-disease association. (HuGE Navigator) 15983379_ADA colocalizing with adenosine receptors on dendritic cells interact with CD26 expressed on lymphocytes. 16133068_Adenosine deaminase activity was abnormal in active nephrotic syndrome, and lymphocyte ADA demonstrated change both in active as well as remission stage of the disease. 16224193_Observational study of gene-disease association. (HuGE Navigator) 16410722_We tested whether p63 is implicated in transcriptional events related to sustaining cell proliferation by transactivation of antiapoptotic and cell survival target genes such as Adenosine Deaminase (ADA), an important gene involved in cell proliferation. 16670267_During acute hypoxia associated with vascular leakage and excessive inflammation, ADA inhibition may serve as therapeutic strategy. 16724628_Observational study of gene-disease association. (HuGE Navigator) 16754522_Observational study of gene-disease association. (HuGE Navigator) 16886895_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16970880_Findings suggest a possible role for a low-activity genotype of adenosine deaminase in the pathogenesis of mild mental retardation. No significant differences were found when comparing the group with moderate or severe mental retardation and controls. 16970880_Observational study of gene-disease association. (HuGE Navigator) 17181544_the ADA c7C/T mutation was estimated to be approximately 7,100 years old 17243920_Increased adenosine deaminase is associated with hydatidiform mole 17287605_ADA*2 allele may decrease genetic susceptibility to coronary artery disease. 17287605_Observational study of gene-disease association. (HuGE Navigator) 17340203_G22A polymorphism plays a minimal role in susceptibility to autism in North American families 17397971_The measurement of adenosine deaminase activity in ascites represents a diagnostic advance in tuberculous peritonitis among end-stage renal failure patients. 17536804_the transition-state structures of PfADA, HsADA, and bovine ADA (BtADA) solved using competitive kinetic isotope effects (KIE) and density functional calculations 17696452_This study shows that adenosine elimination on human airway epithelia is mediated by ADA1, CNT2, and CNT3, which constitute important regulators of adenosine-mediated inflammation. 18218852_CD4(+) T cells from ADA-severe combined immunodeficiency patients have severely compromised T cell receptor/CD28-driven proliferation and cytokine production, both at the transcriptional and protein levels. 18222177_Results describe the activities of ectonucleotide pyrophosphatase/phosphodiesterase and adenosine deaminase in patients with uterine cervix neoplasia. 18302529_the primary mechanism in men with ischemic stroke might involve the reduction of ADA1 activity 18357489_Suggest that adenosine deaminase can be used to distinguish between tuberculous and malignant pleural effusions. 18560234_Negative association between coronary artery disease and ADA*2 allele is only present in males 18562134_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 18597230_the increased level of ADA activity in the subararachnoid hemorrhage patients with the poor clinical and consciousness level may have resulted from the ischemic cerebral insult 18673094_Elevated serum adenosine deaminase in patients with hyperemesis gravidarum may relate to high levels of E2 and progesterone. 18680557_Human ADA, apart from reducing the adenosine concentration and thus preventing A(1)R desensitization, binds to A(1)R behaving as an allosteric effector that markedly enhances agonist affinity and increases receptor functionality. 18768081_Observational study of gene-disease association. (HuGE Navigator) 18768081_Zygotes with low adenosine deaminase locus 1 activity and low F activity may experience the most favourable intrauterine conditions for a balanced development of the feto-placental unit. 18794722_Observational study of gene-disease association. (HuGE Navigator) 18794722_heterozygosity for the 22G>A variant of ADA, although reducing catalytic activity, does not enhance forearm reactive hyperaemia 19019667_Findings suggest that adenosine deaminase might play a crucial role in the development of aspirin-intolerant-asthma by mediation of A1 and A2A receptors. 19026999_Results hypothesized that altered ADA activity may be associated with altered immunity. 19086053_Observational study of gene-disease association. (HuGE Navigator) 19172437_Serum adenosine deaminase activity was significantly higher in eclamptic pregnant women when compared with healthy pregnant women and non-pregnant women. 19237091_ADA may have a role in the cytokine network of the inflammatory cascade of familial Mediterranean fever; elevated ADA levels may be a part of the activated Th1 response in the disease 19470168_Observational study of gene-disease association. (HuGE Navigator) 19521708_serum catalytic concentration elevated in pregnancy, no difference between gestational diabetes and normal pregnancy 19628957_Decreased paraoxonase activity as well as increased adenosine deaminase and xanthine oxidase activities and nitrite levels indicate that oxidative stress is increased and purine metabolism is altered in pseudoexfoliation syndrome. 19633200_ADA-deficient SCID is associated with a specific microenvironment and bone phenotype characterized by RANKL/OPG imbalance and osteoblast insufficiency. 19668260_Addition of ADA to the co-cultures resulted in enhanced CD4(+) and CD8(+) T-cell proliferation and robust ADA-induced increase in cytokine production. 19703146_Adenosine deaminase increases TNF-alpha production in amniotic fluid 19789510_A significant interaction between ACP1 and ADA1 concerning susceptibility to type 1 diabetes, was revealed. 19845893_Observational study of gene-disease association. (HuGE Navigator) 19901882_Occult tuberculous pleurisy is significantly common in patients with pleural effusion ADA levels of 50 IU/L or less and who may otherwise be diagnosed with nonspecific pleurisy. 19913121_Observational study of gene-disease association. (HuGE Navigator) 20147632_The CD8positive CD28positive T lymphocytes that express ADA have significantly greater telomerase activity than those that do not express ADA; however, ADA is progressively lost as cultures progress to senescence. 20174870_Data show that that adenosine deaminase influences human life-span in a sex and age specific way. 20174870_Observational study of gene-disease association. (HuGE Navigator) 20180986_Observational study of gene-disease association. (HuGE Navigator) 20306731_mean serum ADA level in the non-pregnant women was higher than that in the normal pregnant women. Amongst the pregnant women, mean serum ADA in the hypertensive and pre-eclamptic women was significantly higher than that in the normal pregnant group 20347569_ADA activity was positively correlated with lactate dehydrogenase and protein in patients with HIV positive and it was negatively correlated with glucose levels 20392501_Observational study of gene-disease association. (HuGE Navigator) 20412337_novel missense mutation in pedigree of Chinese family with dyschromatosis symmetrica hereditaria 20414589_Observational study of gene-disease association. (HuGE Navigator) 20414589_results suggest that the G/A genotype associated with low adenosine deaminase activity and, supposingly, with higher adenosine levels is less frequent among schizophrenic patients. 20453107_Human adenosine deaminase 2 role in innate immunity and adaptive immunity 20480728_From genetic association studies of SNP in children in Italy, a specific haplotype for ADA is associated with diabetes mellitus type I in girls; the association/susceptibility appears to be reversed in boys. 20480728_Observational study of gene-disease association. (HuGE Navigator) 20522203_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20548128_decrease of ADA activity may play an important role in the formation of pulmonary injury in COPD patients. 20554694_Observational study of gene-disease association. (HuGE Navigator) 20581655_Observational study of gene-disease association. (HuGE Navigator) 20581655_association with coronary artery disease is present in nondiabetic subjects only and dependent on male gender 20590444_Observational study of gene-disease association. (HuGE Navigator) 20590444_Polymorphic sites of ADA might influence cell-mediated anti-tumor immune responses controlling adenosine level and extraenzymatic protein functions. 20615890_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20651391_adenosine deaminase is involved in the regulatory system of chronic atrophic gastritis and gastric cancer risk. 20805743_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20836741_elevated in infants with bronchopulmonary dysplasia 20872567_Elevated adenosine deaminase levels is associated with celiac disease. 20943049_Observational study of gene-disease association. (HuGE Navigator) 20943049_Our data indicate a border line effect of ADA gene polymorphism on susceptibility to heumatoid arthritis that need to be confirmed in other clinical settings 20959412_ADA is a relevant modulator of CD4+ T cell differentiation, even in cells from immunologically compromised individuals. 21198750_effects of soluble human CD26 on human T-cell proliferation are mechanistically independent of both the enzyme activity and the adenosine deaminase-binding capability of soluble CD26 21269755_investigation of plasma activity of total ADA, ADA1, and ADA2 in menses, follicular, and luteal phases of menstrual cycle: significant correlation between monocyte number and ADA activities 21350874_ADA and its isoenzymes analysis in serum of patients could be used as a useful and non-invasive diagnostic tool in evaluation of systemic lupus erythematosus active phase and the disease severity. 21359089_SNP rs73598374 role in sleep modulated by caffeine 21365880_ADA levels rise in some infections & in hypertensive disorders. This may represent a compensatory mechanism resulting from increased adenosine levels & the release of hormones & inflammatory mediators stimulated by hypoxia. Review. 21461854_The new disease activity index that applies serum ADA may help in predicting disease activity in rheumatoid arthritis. 21496384_ADA1 may have a modulatory role in the implantation and duration of the pregnancy. 21593678_This study demonstrated elevated levels of total adenosine deaminase and adenosine deaminase 2 isozyme in patients with active disease. As the patient improves and becomes clinically stable these levels decrease, approaching normal values. 21640091_Serum adenosine deaminase was significantly increased in HIV and HIV-HBV co-infections 21725047_Myeloid dysplasia and bone marrow hypocellularity is associated with adenosine deaminase-deficient severe combined immune deficiency 21734253_This study demonstrated that genetic reduction of ADA activity elevates sleep pressure and plays a key role in sleep and waking quality in humans. 21856036_[review] Recent studies clarify the interactions of dipeptidyl peptidase IV (DPPIV) with adenosine deaminase (ADA) and the transcription transactivator Tat of human immunodeficiency virus (HIV) type-1. 21860532_Results evaluate the utility of the determination of adenosine deaminase (ADA) level in pleural fluid for the differential diagnosis between tuberculous pleural effusion and malignant pleural effusion. 21865054_report that a combination of functional SNPs within ADA and TNF-alpha genes can influence life-expectancy in a gender-specific manner and that males and females follow different pathways to attain longevity 21919946_alteration in ADA activity can lead to reduced adenosine levels, which may be involved in disturbing the fertility process 22062893_Mononuclear cell adenosine deaminase decreases rapidly after angioplasty, indicating that it is an early marker for reperfusion. 22086524_The results suggest that the adenosine deaminase *2 allele is associated with a low risk for recurrent spontaneous abortions, but this association is dependent on older age. 22104430_cutoff value of 9.5 U/L in CSF is a useful aid for the differential diagnosis of tuberculous meningitis and non-tuberculous meningitis 22184407_Data show that the alterations in the CD39/CD73 adenosinergic machinery and loss of function in ADA-deficient Tregs provide insights into a predisposition to autoimmunity and the underlying mechanisms causing defective peripheral tolerance in ADA-SCID. 22257696_ADA1 was found to be the ADA isoenzyme present in platelets of healthy people 22614344_results indicate that the G22A polymorphism of ADA in isolation or together with HLA-B*27 is not associated with ankylosing spondylitis 22719194_Ascitic fluid ADA measurement is helpful in the differential diagnosis of tuberculous peritonitis and peritoneal carcinomatosis. 22942500_Studies report that polymorphisms in the adenosine deaminase (ADA) gene explains phenotypic variance in responses to total sleep deprivation. 22952909_The present study verified the association between the ADA G22A polymorphism and changes in sleep electroencephalogram spectral power. 23066321_analysis of adenosine deaminase activity in tuberculous peritonitis among patients with underlying liver cirrhosis 23096558_Serum ADA could play an important role in adult-onset Still's disease and may be an important biomarker for its diagnosis. 23160984_Affordable parameters such as serum ADA and IgG correlated significantly with immune activation levels and markers of disease progression in untreated HIV-infection. 23193172_ADA open form, but not the closed one, that is responsible for the functional interaction with AR and AAR. 23240012_Taken together, these findings suggest that ADA would promote enhanced and correctly polarized T-cell responses in strategies targeting asymptomatic HIV-infected individuals. 23433854_ADA activity in serum and platelets is decreased in prostate cancer patients. 23768730_The determination of CSF and serum ADA activity is a simple and reliable test for differentiating tuberculous meningitis from other types of meningitis 23897810_Soluble ecto-5'-nucleotidase (5'-NT), alkaline phosphatase, and adenosine deaminase (ADA1) activities in neonatal blood favor elevated extracellular adenosine. 23959645_In Class 3 people the combination of high ACP f-isozyme concentration and the ADA*2 allele, lowers the rate of glycolysis that may reduce the amount of metabolic calories and activates Sirtuin genes that protect cells against age-related diseases. 24257613_ADA acts as a natural antagonist for DPP4-mediated entry of the Middle East respiratory syndrome coronavirus. 24305782_Serum ADA activity in patients with chronic tonsillitis was significantly higher than in healthy controls. 24453844_A strong correlation between ADA and fasting plasma glucose which suggests an association between ADA and nonobese type 2 diabetes mellitus subjects. 24640520_Association between ADA, genetic polymorphism (ADA1, ADA2 and ADA6) and coronary artery disease was studied.Differend pattern observed between males and females. 24682206_Data indicate that the well-being was worse in the adenosine deaminase (ADA) G/A genotype under sleep loss and n-back working memory performance appeared to be specifically susceptible to sleep-wake manipulation in this genotype. 24762755_A history of tuberculosis and hypertension were more common in the low-adenosine deaminase group than the high-adenosine deaminase group. 24896148_ADA rs73598374-A carriers showed lower telomerase activity and shorter leukocyte telomere length. 24965595_The frequency of the ADA2 *2 allele in Sardinia (Italy) is higher in Oristano than in Nuoro resulting in a higher frequency of the ADA1 *1/ADA2 *2 haplotype in Oristano. This suggests a selective advantage of this haplotype in a malarial environment. 24997584_This study suggests that serum ADA could be used as a biochemical marker in cutaneous anthrax. 25125338_ADA polymorphism is associated with type 1 diabetes mellitus. 25170811_No relationship between ADA rs452159 polymorphism and susceptibility to chronic heart failure was found in Chinese population. 25293959_Genetic association replicative and exploratory studies identify SNPs in ADA and MTR highly associated with isolated Neural tube defects (NTD)and SNP in ARID1A and ALDH1A2 associated with NTDs in whites and African Americans respectively. 25299872_We conclude that ADA is an effective test for the diagnosis of pleural tuberculosis; ADA values varied with age in our study population 25437848_Data provide evidence for a more distinct circadian modulation of nap sleep in G/A- compared to G/G-allele ADA carriers. They further indicate that REM sleep, being under strong circadian control, boosts working memory performance according to genotype in a time-of-day dependent manner. 25527815_increase in the salivary ADA and DPP-IV activities as well as in the lipid peroxidation could be related of the regulation to various aspects of adipose tissue function and inflammatory obesity 25647479_the diagnostic utility of pfADA in a low mTB incidence area 25689690_diagnostic value of serum adenosine deaminase (ADA) activity as a useful tool to differentiate HIV mono- and co-infection, was investigated. 25764155_Serum ADA levels were increased in secondary hemophagocytic lymphohistiocytosis suggesting a partial role of activated T-cell response in the disease pathophysiology. 25963491_Multiplex PCR and thoracoscopy are useful investigations in the diagnostic work-up of pleural effusions complicating CKD while the sensitivity and/or specificity of ADA and 65 kDa gene PCR is poor 26166670_activity of ADA isoenzymes and distribution of ADA1 G22 A genotypes were different among fertile and infertile men and more likely the GA genotype, which had lower ADA1 activity and was higher in fertile men is a protective factor against infertility. 26216523_ADA6, PTPN22 and ACP1 are involved in immune reactions: since endometriosis has an autoimmune component. 26261621_Pleural ADA activity is an accurate test for the diagnosis of tuberculous pleural effusions. 26310829_In autologous cocultures of T cells with HIV-1-pulsed dendritic cells, adenosine deaminase decreased HIV-1-induced CD4(+)CD25(hi) FOXP3(+) cells and enhanced the HIV-1-specific CD4(+) responder T cells. 26329539_ADA and ADK are involved in glioma progression; increased ADA and ADK levels in peritumoral tissues may be associated with epilepsy in glioma patients. 26376800_variable mutations are identified as a cause of immunodeficiency in a single Italian center 26659614_Mitochondrial Damage and Apoptosis Induced by Adenosine Deaminase Inhibition and Deoxyadenosine in Human Neuroblastoma Cell Lines 26707067_We conclude that pleural ADA activity could be integrated in the diagnostic procedures of pTB in low to medium tuberculosis prevalence settings. 26794633_High level of ADA is associated with obesity. 26918693_Genetic variability within the ADA gene may influence adenosine concentration and in turn the immune response by lymphocytes in solid tumors. 26980102_association between the occurrence of polycystic ovary syndrome with altered ADA activity and genetic distribution of G22A and A4223C polymorphisms of the ADA1 gene 26994767_In this study, we compared the activity of human adenosine deaminase (hADA) expressed in transgenic seeds of three different plant species: pea (Pisum sativum L.), Nicotiana benthamiana L. and tarwi (Lupinus mutabilis Sweet). All three species were transformed with the same expression vector containing the hADA gene driven by the seed-specific promoter LegA2 with an apoplast targeting pinII signal peptide. 27017482_Patients with tuberculous pleurisy who had high ADA values had a higher probability of manifesting pulmonary tuberculosis. High ADA values may help predict contagious pleuroparenchymal tuberculosis. The most common pulmonary involvement of tuberculous pleurisy showed a centrilobular nodular pattern. 27044834_This study aimed to investigate the activities of purinergic system ecto-enzymes present on the platelet surface as well as CD39 and CD73 expressions on platelets of sickle cell anemia treated patients. 27186641_our analysis shows that, mononuclear cells secretes ADA in response to leishmanial infection and the elevated serum ADA activity is associated with VL and PKDL, and serum ADA activity can be used as prognostic marker to monitor the course of the treatment. 27221867_Increase in serum ADA levels was observed in Oral Squamous Cell Carcinoma. 27273565_Platelet aggregation and adenosine deaminase (ADA) activity were evaluated in pregnant women living with some disease conditions including hypertension, diabetes mellitus and human immunodeficiency virus infection. 27557561_Simvastatin reversed IL-13-suppressed adenosine deaminase activity, leading to the down-regulation of adenosine signaling and inhibition osteopontin expression through the direct inhibition of IL-13-activated STAT6 pathway in COPD. 27663683_Adenosine deaminases ADA1 and ADA2 (ADAs) decrease the level of adenosine by converting it to inosine, which serves as a negative feedback mechanism. These results suggest the existence of a new mechanism, where the activation and survival of immune cells is regulated through the activities of ADA2 or ADA1 anchored to the cell surface. 28074903_The authors found significant neurological and cognitive alterations in untreated Adenosine Deaminase deficiency-severe combined immunodeficiency patients. These included motor dysfunction, EEG alterations, sensorineural hypoacusia, white matter and ventricular alterations in MRI as well as a low mental development index or IQ. 28748310_this study characterizes the long-term outcome of ADA-deficient patients, who survived 5 or more years after therapy 29061410_ADA levels were found to be higher among patients, and revealed a possible link between evening rise and severity of auditory hallucinations as well as morning rise and severity of avolition-apathy in patients with schizophrenia. 29194839_this study shows that ADA is biomarker candidate for endometriosis 29797475_Studied levels of mucin 16 (CA125), adenosine deaminase and midkine as tumor markers in nonsmall cell lung cancer-associated malignant pleural effusion. 30017738_Results provide evidence that ADA SNPs rs12190871 and rs121908723 are risk factors for contracting tuberculous pericarditis. 30394149_ADA levels were found to be increased in polycystic ovary syndrome patients compared to controls. 30444912_Alanine scanning results showed that His17, Gly184, Asp295, and Asp296 exerted the greatest effects on protein energy, suggesting that they played crucial roles in binding to inhibitors. 30641086_Inhibition of eADA blocked endothelial activation suggesting a crucial role of this enzyme in the control of vascular inflammation. This supports the concept of eADA targeted vascular protection therapy 30778076_Adenosine deaminase-1 is an immune regulator of the follicular helper T cell.Adenosine deaminase-1 interaction with CD26 is impaired during HIV infection. 30896318_Increased serum ADA activity as well as its association with obesity in Polycystic Ovary Syndrome (PCOS), while there was no change in serum DPP-4 activity in women with PCOS. 31233715_GLT would confer protection against renal injury by protecting against lipid accumulation and glutathione depletion, at least in part, through suppression of ADA/XO/UA pathway. 31379209_sequential detection of ADA screening and T-SPOT assay was found to be an accurate and rapid method for identifying TB pleurisy from pleural effusion 31709646_Clinical value of combined detection of reactive oxygen species modulator 1 and adenosine deaminase in pleural effusion in the identification of NSCLC associated malignant pleural effusion. 31953681_Exploring the binding modes of cordycepin to human adenosine deaminase 1 (ADA1) compared to adenosine and 2'-deoxyadenosine. 32245326_A Novel Non-frameshift ADA Deletion Detected by Whole Exome Sequencing in an Iranian Family with Severe Combined Immunodeficiency. 32438744_Adenosine Deaminase as a Biomarker of Tenofovir Mediated Inflammation in Naive HIV Patients. 32468861_A significant difference existed in pleural effusion ADA levels between malignant pleural mesothelioma (MPM) and benign disease patients. Pleural effusion ADA levels were significantly higher in MPM patients. 33007809_A Single Nucleotide ADA Genetic Variant Is Associated to Central Inflammation and Clinical Presentation in MS: Implications for Cladribine Treatment. 34510320_ADA gene haplotype is associated with coronary-in-stent-restenosis. 34981765_Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation. | ENSMUSG00000017697 | Ada | 389.14659 | 0.8954524 | -0.1593113523 | 0.15386692 | 1.067996e+00 | 3.013985e-01 | 6.837756e-01 | No | Yes | 421.966440 | 40.369426 | 448.458839 | 41.847916 | |
| ENSG00000196867 | 140612 | ZFP28 | protein_coding | Q8NHY6 | FUNCTION: May be involved in transcriptional regulation. May have a role in embryonic development. | 3D-structure;Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:140612; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 12127974_Characterization of ZFP28 and its role in human development 19329283_these results suggest that murine and human Zfp28/ZFP28 is correlated with the pathogenesis of melanoma. | ENSMUSG00000062861 | Zfp28 | 117.67427 | 0.6924239 | -0.5302725037 | 0.27310216 | 3.723347e+00 | 5.365671e-02 | 3.502056e-01 | No | Yes | 69.114870 | 15.839523 | 97.147320 | 21.748437 | ||
| ENSG00000197147 | 23507 | LRRC8B | protein_coding | Q6P9F7 | FUNCTION: Non-essential component of the volume-regulated anion channel (VRAC, also named VSOAC channel), an anion channel required to maintain a constant cell volume in response to extracellular or intracellular osmotic changes (PubMed:24790029, PubMed:26824658, PubMed:28193731). The VRAC channel conducts iodide better than chloride and can also conduct organic osmolytes like taurine. Channel activity requires LRRC8A plus at least one other family member (LRRC8B, LRRC8C, LRRC8D or LRRC8E); channel characteristics depend on the precise subunit composition (PubMed:24790029, PubMed:26824658, PubMed:28193731). {ECO:0000269|PubMed:24790029, ECO:0000269|PubMed:26824658, ECO:0000269|PubMed:28193731}. | Cell membrane;Disulfide bond;Endoplasmic reticulum;Glycoprotein;Ion channel;Ion transport;Leucine-rich repeat;Membrane;Phosphoprotein;Reference proteome;Repeat;Transmembrane;Transmembrane helix;Transport | hsa:23507; | cytoplasm [GO:0005737]; endoplasmic reticulum membrane [GO:0005789]; integral component of plasma membrane [GO:0005887]; ion channel complex [GO:0034702]; plasma membrane [GO:0005886]; anion transmembrane transport [GO:0098656]; inorganic anion transport [GO:0015698] | 15094057_identified four genes, named TA-LRRP, AD158, LRRC5, and FLJ23420, as unknown LRRC8-like genes 20379614_Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 28972132_LRRC8B participates in intracellular Ca(2+) homeostasis by acting as a leak channel in the endoplasmic reticulum. | ENSMUSG00000070639 | Lrrc8b | 1311.01673 | 0.9312965 | -0.1026875344 | 0.11004641 | 8.688296e-01 | 3.512795e-01 | 7.243004e-01 | No | Yes | 1753.643318 | 377.721866 | 1744.680648 | 367.710890 | ||
| ENSG00000197283 | 8831 | SYNGAP1 | protein_coding | Q96PV0 | FUNCTION: Major constituent of the PSD essential for postsynaptic signaling. Inhibitory regulator of the Ras-cAMP pathway. Member of the NMDAR signaling complex in excitatory synapses, it may play a role in NMDAR-dependent control of AMPAR potentiation, AMPAR membrane trafficking and synaptic plasticity. Regulates AMPAR-mediated miniature excitatory postsynaptic currents. Exhibits dual GTPase-activating specificity for Ras and Rap. May be involved in certain forms of brain injury, leading to long-term learning and memory deficits (By similarity). {ECO:0000250}. | Alternative splicing;Autism;Autism spectrum disorder;Disease variant;GTPase activation;Mental retardation;Phosphoprotein;Reference proteome;SH3-binding | This gene encodes a Ras GTPase activating protein that is a member of the N-methyl-D-aspartate receptor complex. The N-terminal domain of the protein contains a Ras-GAP domain, a pleckstrin homology domain, and a C2 domain that may be involved in binding of calcium and phospholipids. The C-terminal domain consists of a ten histidine repeat region, serine and tyrosine phosphorylation sites, and a T/SXV motif required for postsynaptic scaffold protein interaction. The encoded protein negatively regulates Ras, Rap and alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking to the postsynaptic membrane to regulate synaptic plasticity and neuronal homeostasis. Allelic variants of this gene are associated with intellectual disability and autism spectrum disorder. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Nov 2016]. | hsa:8831; | cytosol [GO:0005829]; dendritic shaft [GO:0043198]; glutamatergic synapse [GO:0098978]; plasma membrane [GO:0005886]; postsynaptic density [GO:0014069]; GTPase activator activity [GO:0005096]; SH3 domain binding [GO:0017124]; dendrite development [GO:0016358]; maintenance of postsynaptic specialization structure [GO:0098880]; negative regulation of axonogenesis [GO:0050771]; negative regulation of neuron apoptotic process [GO:0043524]; negative regulation of Ras protein signal transduction [GO:0046580]; pattern specification process [GO:0007389]; Ras protein signal transduction [GO:0007265]; receptor clustering [GO:0043113]; regulation of GTPase activity [GO:0043087]; regulation of long-term neuronal synaptic plasticity [GO:0048169]; regulation of MAPK cascade [GO:0043408]; regulation of synapse structure or activity [GO:0050803]; regulation of synaptic plasticity [GO:0048167]; visual learning [GO:0008542] | 18323856_The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. 19196676_Observational study of gene-disease association. (HuGE Navigator) 19196676_Results indicate that SYNGAP1 disruption is a cause of autosomal dominant nonsyndromic mental retardation. 19483657_These data suggest that NMDA receptor complex formation, localization, and downstream signaling may be abnormal in schizophrenia as PSD95, SynGAP and MUPP1 expression is altered. 19851445_Observational study of gene-disease association. (HuGE Navigator) 20188038_Observational study of gene-disease association. (HuGE Navigator) 20531469_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21237447_We provide evidence that truncating mutations in SYNGAP1 are common in nonsyndromic intellectual disability and can be also associated with autism. 21480541_SYNGAP1 is a brain-specific protein that interacts with key components of the proteins involved in experience-dependent changes in glutamate synapses involved in learning. 23161826_De novo missense mutations, p.R579X, and possibly all the other truncating mutations in SYNGAP1 result in a loss of its function, causing intellectual disability, autism, and a specific form of epilepsy. 23708187_De novo CHD2 and SYNGAP1 mutations are new causes of epileptic encephalopathies, accounting for 1.2% and 1% of cases, respectively. 24945774_Reduced cognition in mutant Syngap1 transgenic mice is caused by isolated damage to developing forebrain neurons. 25533468_Phosphorylation of synaptic GTPase-activating protein (synGAP) by Ca2+/calmodulin-dependent protein kinase II (CaMKII) and cyclin-dependent kinase 5 (CDK5) alters the ratio of its GAP activity toward Ras and Rap GTPases. 26079862_De novo, heterozygous, loss-of-function mutations in SYNGAP1 cause a syndromic form of intellectual disability. 26110312_This is the first description of a special electroencephalogram phenomenon (normalization with eye opening) in association with SYNGAP1 mutations. 26558778_Syngap transgenic mice exhibited alterations in long-term depression and dendritic spine morphology. 29381230_our findings suggest that SYNGAP1 variants are causative for ID, which expands our knowledge of the disease spectrum and provides new information on the genotype-phenotype relationship. 29632532_Complement receptor 3 (Integrin alpha-M beta-2)-mediated phagocytosis of Francisella due to increased ras GTPase-activating protein (RasGAP) activity. 30541864_The SYNGAP1 mutations cause a generalized developmental and epileptic encephalopathy with a distinctive syndrome combining epilepsy with eyelid myoclonia with absences and myoclonic-atonic seizures. 30685520_This study demonstrated that eating induced seizures in association with SYNGAP1 mutations. 31025938_SynGAP protein retains biological functions throughout adulthood and that non-developmental functions may contribute to disease phenotypes. 31395010_A broad spectrum of neurologic and neurodevelopmental features are found with pathogenic variants of SYNGAP1. 31454529_Discussion on the clinical spectrum and molecular pathophysiology of SYNGAP1 mutations, aiming to understand how mutations in this protein affect neuronal development and function and how that translates into the clinical features observed in Mental retardation-type 5 patients (review). 32887745_SYNGAP1 Controls the Maturation of Dendrites, Synaptic Function, and Network Activity in Developing Human Neurons. 33189692_Distinct patterns of repetition suppression in Fragile X syndrome, down syndrome, tuberous sclerosis complex and mutations in SYNGAP1. 33308442_Multi-parametric analysis of 57 SYNGAP1 variants reveal impacts on GTPase signaling, localization, and protein stability. 33639450_SYNGAP1-DEE: A visual sensitive epilepsy. 34130248_Differential auditory brain response abnormalities in two intellectual disability conditions: SYNGAP1 mutations and Down syndrome. | ENSMUSG00000067629 | Syngap1 | 246.05734 | 0.8298632 | -0.2690546205 | 0.20374039 | 1.734500e+00 | 1.878373e-01 | 5.799422e-01 | No | Yes | 189.750034 | 38.520503 | 231.703570 | 45.794088 | |
| ENSG00000197467 | 1305 | COL13A1 | protein_coding | Q5TAT6 | FUNCTION: Involved in cell-matrix and cell-cell adhesion interactions that are required for normal development. May participate in the linkage between muscle fiber and basement membrane. May play a role in endochondral ossification of bone and branching morphogenesis of lung. Binds heparin. At neuromuscular junctions, may play a role in acetylcholine receptor clustering (PubMed:26626625). {ECO:0000250|UniProtKB:Q9R1N9, ECO:0000269|PubMed:10865988, ECO:0000269|PubMed:11956183, ECO:0000269|PubMed:26626625}. | Alternative splicing;Cell adhesion;Cell junction;Cell membrane;Collagen;Congenital myasthenic syndrome;Developmental protein;Differentiation;Disulfide bond;Heparin-binding;Membrane;Osteogenesis;Postsynaptic cell membrane;Reference proteome;Signal-anchor;Synapse;Transmembrane;Transmembrane helix | This gene encodes the alpha chain of one of the nonfibrillar collagens. The function of this gene product is not known, however, it has been detected at low levels in all connective tissue-producing cells so it may serve a general function in connective tissues. Unlike most of the collagens, which are secreted into the extracellular matrix, collagen XIII contains a transmembrane domain and the protein has been localized to the plasma membrane. The transcripts for this gene undergo complex and extensive splicing involving at least eight exons. Like other collagens, collagen XIII is a trimer; it is not known whether this trimer is composed of one or more than one alpha chain isomer. A number of alternatively spliced transcript variants have been described, but the full length nature of some of them has not been determined. [provided by RefSeq, Jul 2008]. | hsa:1305; | cell-cell junction [GO:0005911]; collagen trimer [GO:0005581]; collagen type XIII trimer [GO:0005600]; collagen-containing extracellular matrix [GO:0062023]; endoplasmic reticulum lumen [GO:0005788]; extracellular matrix [GO:0031012]; extracellular region [GO:0005576]; extracellular space [GO:0005615]; plasma membrane [GO:0005886]; postsynaptic membrane [GO:0045211]; extracellular matrix structural constituent [GO:0005201]; heparin binding [GO:0008201]; cell differentiation [GO:0030154]; cell-cell adhesion [GO:0098609]; cell-matrix adhesion [GO:0007160]; collagen fibril organization [GO:0030199]; endochondral ossification [GO:0001958]; extracellular matrix organization [GO:0030198]; morphogenesis of a branching structure [GO:0001763]; notochord development [GO:0030903]; skeletal system development [GO:0001501] | 11956183_The type XIII collagen ectodomain is a 150-nm rod and capable of binding to fibronectin, nidogen-2, perlecan, and heparin. 12832406_two widely separated coiled-coil domains of type XIII and related collagens function as independent oligomerization domains participating in the folding of distinct areas of the molecule. 16385451_Observational study of gene-disease association. (HuGE Navigator) 19851296_Observational study of gene-disease association. (HuGE Navigator) 20198315_Observational study of gene-disease association. (HuGE Navigator) 20708005_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 26626625_Congenital myasthenic syndrome type 19 is caused by mutations in COL13A1. 27245701_We identified overexpression of collagen type XIII alpha 1 in active Thyroid-associated ophthalmopathy affected fat. 27580012_The combination of constitutively low expression of COL13A1, high physiological and metabolic demands, and consequentially relatively high exposure to stressors may explain the particular vulnerability of inferior rectus to thyroid-associated ophthalmopathy. 28415608_this study shows that COL13A1 production by urothelial carcinoma of the bladder plays a pivotal role in tumor invasion through the induction of tumor budding 28837258_Urine levels of COL4A1, COL13A1, the combined values of COL4A1 and COL13A1 (COL4A1 + COL13A1), and CYFRA21-1 were significantly elevated in urine from patients with BCa compared to the controls. A high urinary COL4A1 + COL13A1 was found to be an independent risk factor for intravesical recurrence. 29307798_Findings suggest a significant association between variants in COL13A1, ADIPOQ, SAMM50, and PNPLA3, and risk of NAFLD/elevated transaminase levels in Mexican adults with an admixed ancestry. 29363764_The authors report a congenital myasthenic syndrome due to mutations in COL13A1, which encodes an extracellular matrix protein that is concentrated at the neuromuscular junction and highlights a role for these extracellular matrix proteins in maintaining synaptic stability that is independent of the AGRN/MuSK clustering pathway. 29369589_It was established that the frequency of individuals with the COL13A1*D/*D genotype was higher in the senile age period. The LAMA2*I/*D genotype was predisposing to longevity among women. 30285809_Results indicate a function of collagen XIII in promoting cancer metastasis, cell invasion, and anoikis resistance. 30767057_The data of this study support the causality of COL13A1 variants for Congenital myasthenic syndrome. 31081514_patients with COL13A1 mutations present mostly with severe early-onset myasthenic syndrome with feeding and breathing difficulties 31220558_Data suggest that ColXIII has a role in age-dependent cortical bone deterioration with possible implications for osteoporosis and fracture risk. 34404755_TGF-beta2 and collagen play pivotal roles in the spheroid formation and anti-aging of human dermal papilla cells. | ENSMUSG00000058806 | Col13a1 | 93.37510 | 1.0915622 | 0.1263942754 | 0.29547945 | 1.835929e-01 | 6.683036e-01 | 8.967906e-01 | No | Yes | 116.000921 | 14.184627 | 106.048936 | 12.639985 | |
| ENSG00000197757 | 3223 | HOXC6 | protein_coding | P09630 | FUNCTION: Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. | Alternative splicing;DNA-binding;Developmental protein;Homeobox;Nucleus;Reference proteome;Transcription;Transcription regulation | This gene belongs to the homeobox family, members of which encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, which are located on different chromosomes and consist of 9 to 11 genes arranged in tandem. This gene, HOXC6, is one of several HOXC genes located in a cluster on chromosome 12. Three genes, HOXC5, HOXC4 and HOXC6, share a 5' non-coding exon. Transcripts may include the shared exon spliced to the gene-specific exons, or they may include only the gene-specific exons. Alternatively spliced transcript variants encoding different isoforms have been identified for HOXC6. Transcript variant two includes the shared exon, and transcript variant one includes only gene-specific exons. [provided by RefSeq, Jul 2008]. | hsa:3223; | chromatin [GO:0000785]; cytosol [GO:0005829]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; anterior/posterior pattern specification [GO:0009952]; embryonic skeletal system development [GO:0048706]; regulation of transcription by RNA polymerase II [GO:0006357] | 15637592_proapoptotic repression targets include neutral endopeptidase (NEP) and insulin-like growth factor binding protein-3 (IGFBP-3) 17488478_HOXC6 increases transcription of S100beta gene in BrdU-induced in vitro differentiation of GOTO neuroblastoma cells into Schwann cells. 19236766_Human cytomegalovirus downregulates while all-trans retinoic acid upregulates expression of hoxc4 and hoxc6 in lymphocytic progenitor cells. 21683083_These studies demonstrated that HOXC6 is an estrogen-responsive gene, and that histone methylases MLL2 and MLL3, in coordination with ERalpha and ERbeta, transcriptionally regulate HOXC6 in an estrogen-dependent manner. 22896703_HOXC6 is an important mechanism of the anti-apoptotic pathway via regulation of Bcl-2 expression. 22907429_HOXC6 expression is an important mechanism of chemotherapeutic drug resistance via its regulation of MDR-1 23581417_Hoxc6 might contribute to the progression of gastric carcinogenesis and may be a significant predictor of poor survival in patients with gastric cancer after curative operations. 24525058_HOXC6 can be used as a prognostic marker in esophageal squamous cell carcinoma 25725483_Our studies demonstrate that HOXC6, which is a critical player in mammary gland development, is upregulated in multiple cases of breast cancer, and is transcriptionally regulated by E2 and BPA, in vitro and in vivo. 26046583_Data demonstrate significant downregulation of HOXC6 in serous ovarian cancer and low protein level is associated with high-grade tumors. 26310814_HOXC6 has a role in all prostate cancer stages, particularly in prostate cancer cell proliferation. 27081081_HOXC6 thus appears to promote colorectal cancer cell proliferation and tumorigenesis through autophagy inhibition 27108162_HOXC6 mRNA levels were shown to be good predictors for the detection of High-grade Prostate Cancer. 27573865_Increased expression of HOXC6 in gastric cancer cell lines significantly activated extracellular signalregulated kinase signaling and upregulated MMP9. The HOXC6 gene promotes the proliferation of gastric cancer cells while upregulation of MMP9 promotes migration and invasion of gastric cancer cells. 27574949_oxidized low-density lipoprotein (Ox-LDL) decreased expression of HOXC-AS1 and HOXC6 in a time-dependent manner 29348394_HOXC6 may promote tumor progression by inducing epithelial-mesenchymal transition pathway and predicts poor prognosis of hepatocellular carcinoma. 29991129_Overall, the data show that miR-185 could negatively target HOXC6 to suppress cell proliferation, promotes apoptosis and autophagy through inhibiting TGF-beta1/mTOR axis in nasopharyngeal carcinoma (NPC). Thus, miR-185 is useful strategy for the treatment of NPC. 30228024_our findings indicate that HOXC6 plays an important role in the progress and prognosis of human glioma and promotes glioma U87 cell growth through the WIF-1/Wnt signaling pathway 30509141_HOXC6 up-regulates BCL2 expression and promotes cervical cancer progression. 30618083_These results illuminated that the novel Linc00339/miR-377-3p/HOXC6 axis played a critical role in triple-negative breast cancer (TNBC) progression and might be a promising therapeutic target for TNBC treatment. 31026217_Urinary HOXC6 mRNA levels were quantified and RNA results were then combined with other risk factors in a clinical model optimized to detect International Society of Urological Pathology Grade Group 2 or greater prostate cancer in men with prostate specific antigen less than 10 ng/ml. 31271305_Up-regulated HOXC6, in PCa patients, could not only participate in the progression of PCa but also function as an independent prognostic marker for the cancer 31900716_HoxC6 Functions as an Oncogene and Isoform HoxC6-2 May Play the Primary Role in Gastric Carcinogenesis. 32171324_MicroRNA-495 confers inhibitory effects on cancer stem cells in oral squamous cell carcinoma through the HOXC6-mediated TGF-beta signaling pathway. 32515533_BBOX1 antisense RNA 1 facilitates CC progression by upregulating HOXC6 expression via miR-361-3p and HuR. 32681273_Prospective assessment of two-gene urinary test with multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy. 32740941_HomeoboxC6 affects the apoptosis of human vascular endothelial cells and is involved in atherosclerosis. 33795652_HomeoboxC6 promotes metastasis by orchestrating the DKK1/Wnt/beta-catenin axis in right-sided colon cancer. 33846772_TrkB/C-induced HOXC6 activation enhances the ADAM8-mediated metastasis of chemoresistant colon cancer cells. 34763232_HOXC6/8/10/13 predict poor prognosis and associate with immune infiltrations in glioblastoma. 34950145_The Effects of Differentially-Expressed Homeobox Family Genes on the Prognosis and HOXC6 on Immune Microenvironment Orchestration in Colorectal Cancer. 35008435_HOXC6-Mediated miR-188-5p Expression Induces Cell Migration through the Inhibition of the Tumor Suppressor FOXN2. | ENSMUSG00000001661 | Hoxc6 | 260.86062 | 1.0933185 | 0.1287137985 | 0.18651200 | 4.771928e-01 | 4.896966e-01 | 8.145205e-01 | No | Yes | 314.425310 | 27.464163 | 281.058163 | 24.187874 | |
| ENSG00000197951 | 58491 | ZNF71 | protein_coding | Q9NQZ8 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:58491; | nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 20734064_Observational study of gene-disease association. (HuGE Navigator) 33916522_Molecular Analysis of ZNF71 KRAB in Non-Small-Cell Lung Cancer. | 350.08524 | 0.9120036 | -0.1328886103 | 0.17189197 | 5.959942e-01 | 4.401105e-01 | 7.833599e-01 | No | Yes | 427.025624 | 55.047189 | 457.664412 | 57.375237 | ||||
| ENSG00000197961 | 7675 | ZNF121 | protein_coding | P58317 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7675; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 27988300_ZNF121 as a MYC-interacting protein with functional effects on MYC and cell proliferation 29698681_Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. | ENSMUSG00000045519 | Zfp560 | 1691.70892 | 1.1759918 | 0.2338780057 | 0.13096433 | 3.208109e+00 | 7.327414e-02 | 3.980113e-01 | No | Yes | 2213.737801 | 396.428561 | 1853.928419 | 324.884215 | ||
| ENSG00000198001 | 51135 | IRAK4 | protein_coding | Q9NWZ3 | FUNCTION: Serine/threonine-protein kinase that plays a critical role in initiating innate immune response against foreign pathogens. Involved in Toll-like receptor (TLR) and IL-1R signaling pathways (PubMed:17878374). Is rapidly recruited by MYD88 to the receptor-signaling complex upon TLR activation to form the Myddosome together with IRAK2. Phosphorylates initially IRAK1, thus stimulating the kinase activity and intensive autophosphorylation of IRAK1. Phosphorylates E3 ubiquitin ligases Pellino proteins (PELI1, PELI2 and PELI3) to promote pellino-mediated polyubiquitination of IRAK1. Then, the ubiquitin-binding domain of IKBKG/NEMO binds to polyubiquitinated IRAK1 bringing together the IRAK1-MAP3K7/TAK1-TRAF6 complex and the NEMO-IKKA-IKKB complex. In turn, MAP3K7/TAK1 activates IKKs (CHUK/IKKA and IKBKB/IKKB) leading to NF-kappa-B nuclear translocation and activation. Alternatively, phosphorylates TIRAP to promote its ubiquitination and subsequent degradation. Phosphorylates NCF1 and regulates NADPH oxidase activation after LPS stimulation suggesting a similar mechanism during microbial infections. {ECO:0000269|PubMed:11960013, ECO:0000269|PubMed:12538665, ECO:0000269|PubMed:15084582, ECO:0000269|PubMed:17217339, ECO:0000269|PubMed:17337443, ECO:0000269|PubMed:17878374, ECO:0000269|PubMed:17997719, ECO:0000269|PubMed:20400509, ECO:0000269|PubMed:24316379}. | 3D-structure;ATP-binding;Acetylation;Alternative splicing;Cytoplasm;Disease variant;Immunity;Innate immunity;Kinase;Magnesium;Nucleotide-binding;Phosphoprotein;Reference proteome;Serine/threonine-protein kinase;Transferase | This gene encodes a kinase that activates NF-kappaB in both the Toll-like receptor (TLR) and T-cell receptor (TCR) signaling pathways. The protein is essential for most innate immune responses. Mutations in this gene result in IRAK4 deficiency and recurrent invasive pneumococcal disease. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2011]. | hsa:51135; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; endosome membrane [GO:0010008]; extracellular space [GO:0005615]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; ATP binding [GO:0005524]; interleukin-1 receptor binding [GO:0005149]; magnesium ion binding [GO:0000287]; protein kinase binding [GO:0019901]; protein serine kinase activity [GO:0106310]; protein serine/threonine kinase activity [GO:0004674]; protein serine/threonine/tyrosine kinase activity [GO:0004712]; cytokine-mediated signaling pathway [GO:0019221]; innate immune response [GO:0045087]; interleukin-1-mediated signaling pathway [GO:0070498]; intracellular signal transduction [GO:0035556]; JNK cascade [GO:0007254]; MyD88-dependent toll-like receptor signaling pathway [GO:0002755]; neutrophil mediated immunity [GO:0002446]; neutrophil migration [GO:1990266]; positive regulation of I-kappaB kinase/NF-kappaB signaling [GO:0043123]; positive regulation of smooth muscle cell proliferation [GO:0048661]; toll-like receptor 9 signaling pathway [GO:0034162]; toll-like receptor signaling pathway [GO:0002224] | 11960013_a novel member of the IRAK family with the properties of an IRAK-kinase 12297423_Gene targeting studies show that IRAK-4 has an essential role in mediating signals initiated by IL-1R and TLR engagement. Review. 12496252_Pellino 1 is required for interleukin-1-mediated signaling through its interaction with the interleukin-1 receptor-associated kinase 4-IRAK-tumor necrosis factor receptor-associated factor 6 complex 12637671_described 3 children with inherited IRAK-4 deficiency who developed pyogenic bacterial infections; findings suggest the TIR-IRAK signaling pathway is crucial for protective immunity against specific bacteria but redundant against most other microorganisms 12860405_Pellino2 interacts with kinase-active as well as kinase-inactive IRAK1 and IRAK4 12925671_Results suggest that IRAK-4 is pivotal in the development of a normal inflammatory response initiated by bacterial or nonbacterial insults. 15084582_IRAK4 is required for the efficient recruitment of IRAK to the IL-1 receptor complex 15292196_IRAK-4 is an integral part of the IL-1R signaling cascade and is capable of transmitting signals both dependent on and independent of its kinase activity 15825022_Systemic shigellosis in a person with a primary immunodeficiency, expanding the spectrum of infections associated with IRAK-4 deficiency is reported. 15905496_This study demonstrates several mechanisms by which overexpression of truncated, kinase-deficient forms of IRAK-4 in a patient with recurrent bacterial infections may disrupt signaling induced by lipopolysaccharide or IL-1. 16286015_The TLR-7-, TLR-8-, and TLR-9-dependent induction of IFN-alpha/beta and -lambda is strictly IRAK-4 dependent and paradoxically redundant for protective immunity to most viruses in humans 16537705_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 16907704_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17046325_Although each system seems to possess distinct activation mechanisms, interleukin-1 receptor-associated kinase (IRAK)-4 is essential for NF-kappaB activation in Toll-like receptor (TLR) and T-cell receptor (TCR) signaling pathways. 17141195_The present data indicate that the kinase activity of IRAK-4 is dependent on the autophosphorylations at T342, T345, and S346 in the activation loop. 17161373_crystallographic analysis of IRAK-4 kinase in complex with inhibitors 17217339_We conclude that IRAK-4 phosphorylates p47phox and regulates NADPH oxidase activation after LPS stimulation. 17507369_IL-1RAcP, MyD88, and IRAK-4 are the stable components of the endogenous type I interleukin-1 (IL-1) receptor signaling complex 17548806_ST2825 interfered with recruitment of IRAK1 and IRAK4 by MyD88, causing inhibition of IL-1beta-mediated activation of NF-kappaB transcriptional activity. 17675297_kinase-inactive IRAK proteins can associate with Pellino proteins, thus excluding the possibility that their inability to regulate Pellino degradation is due to lack of association with the Pellino proteins 17703412_Observational study of gene-disease association. (HuGE Navigator) 17878374_suggest the existence of an IRAK-4-independent TLR9-induced transduction pathway leading to PI3K activation 17893200_IRAK-4-dependent human Toll-like receptors appear to play a redundant role in protective immunity to most infections, at most limited to childhood immunity to some pyogenic bacteria 17917042_Inherited human IRAK-4 deficiency, a recently described primary immunodeficiency leading to recurrent, invasive, pyogenic bacteria infection, and invasive pneumococcal disease in particular, is reviewed. 17997719_Pellino isoforms may be the E3 ubiquitin ligases that mediate the IL-1-stimulated formation of K63-pUb-IRAK1 in cells, which may contribute to activation of IKKbeta and transcription factor NF-kappaB, as well as signalling pathways dependent on IRAK1/4 18079163_These results suggested that the expression of IRAK-4 alone is sufficient to cause the degradation of IRAK-1; the autophosphorylation of IRAK-1 is not necessary to terminate the TLR-induced activation of NF-kappaB. 18174872_Two cases of IRAK-4 deficiency that presented with unusual S. aureus infections that were not accompanied by the expected constitutional symptoms or hematologic and acute phase responses are reported. 18503546_These results demonstrate that downregulation of IRAK-4 requires activation of the MyD88-dependent pathway and that the death domains of both MyD88 and IRAK-4 are important for this downregulation. 18987746_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19006693_IRAK-4-, MyD88-, and UNC-93B-deficient patients did not display autoreactive antibodies in their serum or develop autoimmune diseases, suggesting that IRAK-4, MyD88, and UNC-93B pathway blockade may thwart autoimmunity in humans. 19074885_Observational study of gene-disease association. (HuGE Navigator) 19120481_patients with congenital deficiencies show enhanced susceptibility to pyogenic bacteria such as Staphylococcus or Pneumococcus 19181383_In human cells the non-kinase functions of IRAK-4 are essential, whereas the kinase activity of IRAK-4 appears redundant with that of IRAK-1. 19254290_Observational study of gene-disease association. (HuGE Navigator) 19254290_These results demonstrate a clear association between polymorphisms in the IRAK4 gene and serum IgE levels in patients with chronic rhinosinusitis and asthma 19258923_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19423540_Observational study of gene-disease association. (HuGE Navigator) 19505919_Observational study of gene-disease association. (HuGE Navigator) 19592493_analysis of an oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4 19703016_IRAK-4, which is essential for virtually all TLR signalling, was suppressed, whereas the precursor for the antibiotic peptide Dermcidin was up-regulated in HIV-infected cells. 19898481_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20056178_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20104875_A detailed kinetic characterization of the phosphoryl transfer activity of IRAK-4 toward a peptide substrate derived from the activation loop of IRAK-1 is described. IRAK-4 obeys a sequential kinetic pathway. 20105294_Data demonstrate that the vast majority of LPS-responsive genes in IRAK4-deficient monocytes were greatly suppressed, an observation that is consistent with the described role for IRAK4 as an essential component of TLR4 signalling. 20400509_Mal is a substrate for IRAK1 and IRAK4 with phosphorylation promoting ubiquitination and degradation of Mal, which may serve to negatively regulate signaling by TLR2 and TLR4 20406964_Observational study of gene-disease association. (HuGE Navigator) 20438785_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20448286_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20485341_the crystal structure of the MyD88-IRAK4-IRAK2 death domain (DD) complex, which surprisingly reveals a left-handed helical oligomer that consists of 6 MyD88, 4 IRAK4 and 4 IRAK2 DDs 20503287_Observational study of gene-disease association. (HuGE Navigator) 20588308_Observational study of gene-disease association. (HuGE Navigator) 20621347_IRAK-4-deficient patients have defects in T-cell activation. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20966070_Two variants found in the MyD88 death domain, S34Y and R98C, showed severely reduced NF-kappaB activation due to reduced homo-oligomerization and IRAK4 interaction 21160042_Although IRAK4 kinase activity is essential for human plasmacytoid dendritic cell activation, it is dispensable in B, T, dendritic, and monocytic cells, which is in contrast with an essential active kinase role in comparable mouse cell types. 21166654_These results indicate that IL-1beta-induced IL-6 production in A549 cells is mediated by both PI3K and IRAK4 and suggest that involvement of PI3K in the IL-1-induced IL-6 production is cell type specific. 21205819_evidence that the structure-function similarities that we have identified between LYK3 and IRAK-4 may be more widely applicable to plant RLKs in general. 21220427_LPS-induced activation of IRAK4 and TAK1, K63-linked polyubiquitination of IRAK1 and TRAF6, and disrupted IRAK1-TRAF6 and IRAK1-IKKgamma assembly associated with increased A20 expression 21576904_genetic polymorphism is associated with increased prevalence of gram-positive infection and decreased response to toll-like receptor ligands 21738793_The results demonstrated a gender- and allergen-dependant association pattern between polymorphisms in IRAK-4 and AR in Chinese population. 22059479_investigation of signaling components that induce activation of post-transcriptional mechanism for IkappaB-zeta induction/activation: Activation of IRAK1 or IRAK4, but not TRAF6, is sufficient. 22606974_The results indicated that IRAK-4 gene silencing in MG63 cells inhibited cell proliferation and function and increase apoptosis, which may be related to the decreased Bcl-2/Bax ratio and inhibition of the protein expression. 22932140_The IRAK4 SNPs rs3794262 and rs4251481 were associated with allergy to dust mites, but not other types of allergic rhinitis. 23002119_IgM(+)IgD(+)CD27(+) but not switched B cells were strongly reduced in MYD88-, IRAK-4-, and TIRAP-deficient patients, but not UNC-93B-deficient patients. 23027530_We report the association of two tag SNPS (i.e., rs1461567 and rs4251513) distributed in the IRAK4 gene with brain and ocular alterations in congenital toxoplasmosis. 23041547_Evaluates the effects of stimulating or inhibiting the TLR/IL-1 receptor-associated kinases IRAK-1 and IRAK-4 in melanoma cells where their functions are largely unexplored. 23100432_This study demonstrated that IRAK4 activation acts normally to regulate microglial activation status and influence amyloid homeostasis in the brain. 23255009_Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. 23264652_During bacterial infection, PGN-mediated TLR2 signaling induces miR-132/-212 to downregulate IRAK4. 23519847_these data highlight an unexpected role of IRAK4, Akt, and mTOR in the regulation of tolerance in human monocytes 23591012_Dominant negative mutants of IRAK4 and GAK show strong apoptotic effects in A498 cells under anoxia. 23595970_Studied the TMZ-induced changes in the proteome of the glioma-derived cell line (U251) by 2D DIGE. We found 95 protein spots to be significantly altered in their expression after TMZ treatment. 24316379_these studies suggest that not only the loss of protein expression but also the defect of Myddosome formation couldcause IRAK4 and MyD88 deficiency syndromes. 24567333_IRAK4 is regulated by Interleukin 1/Toll-like receptor-induced autophosphorylation 24690905_High mRNA levels of IRAK1 and IRAK4 correlated with VKH disease activity. 25201411_Data show that dimerization is crucial for IRAK4 autophosphorylation in vitro and ligand dependent signaling in cells. 25320238_by bolstering the IgM(+)IgD(+)CD27(+) B-cell subset, IRAK-4 and MyD88 promote optimal T-independent IgM antibody responses against bacteria in humans. 25344726_delineation of the latter responses identified a narrow repertoire of transcriptional programs affected by loss of MyD88 function or IRAK4 function 25479567_Studies indicate that interleukin-1 receptor-associated kinase 4 protein (IRAK4), a serine/threonine kinase, plays a key role in both inflammation and oncology diseases. 25707370_The estimated prevalence of IRAK4 gene polymorphism found in a Portuguese Caucasian population was 26.8 % (CI 95%) [20.1, 34.7 %]. A model to predict the inflammatory response in the maxillary sinus in the presence etiological factors of dental origin was constructed. 25886387_findings suggest that rare, functional variants in MYD88, IRAK4 or IKBKG do not significantly contribute to IPD susceptibility in adults at the population level 25922567_Src, Syk, IRAK1, and IRAK4 have roles in anti-inflammatory responses mediated by dietary flavonoid Kaempferol 26075815_This is the first study to show an association between single nucleotide polymorphisms in IRAK1, IRAK4 and MyD88, and the presence of severe invasive pneumococcal disease. 26472314_missense mutation results in immunological deficiency phenotype 26698383_Siblings with compound heterozygosity for two mutations: a frameshift mutation at one allele (c.1146delT (p.G383fs)), and an in-frame duplication variant at the other (c.255_260dup6 (p.D86_L87dup)) leading to fatal pneumococcal meningitis is described. 27062898_the polymorphisms in TLR-MyD88-NF-kappaB signaling pathway confer genetic susceptibility to Type 2 diabetes mellitus and diabetic nephropathy. 27702822_Our data established IRAK4 as a novel therapeutic target for PDAC treatment. Development of potent IRAK4 inhibitors is needed for clinical testing. 27840174_AMPK activation inhibited IL-1beta-stimulated CXCL10 secretion, associated with reduced interleukin-1 receptor associated kinase-4 (IRAK4) phosphorylation. 27869651_data show that in pericytes, MyD88 and IRAK4 are key regulators of 2 major injury responses: inflammatory and fibrogenic. 27876844_IRAK-4 Arg12 is also essential for Myddosome assembly and signalling and we propose that phosphorylated Ser8 induces the N-terminal loop to fold into an alpha-helix. This conformer is stabilised by an electrostatic interaction between phospho-Ser8 and Arg12 and would destabilise a critical interface between IRAK-4 and MyD88. 27888910_IRAK4 - Gene involved in innate immunity that have been associated with Chronic Rhinosinusitis. 28512203_Data suggest that, in monocytes and macrophages, the interleukin-1B- (IL1B)-stimulated trans-autophosphorylation of IRAK4 is initiated by MYD88- (myeloid differentiation primary response gene 88)-induced dimerization of IRAK4. In contrast, IRAK1 (interleukin-1 receptor-associated kinase 1) is inactive in unstimulated monocytes/macrophages and is converted to an active protein kinase in response to IL1B. 28780618_The high mRNA levels of IRAK1 and IRAK4 were correlated with the development of Behcet's disease, which suggested that IRAK1 and IRAK4 might participate in the pathogenesis of Behcet's disease 28924041_Data suggest that IRAK4 activity regulates activation of IRF5, TAK1, and IKKB in monocytes; IRAK4 activation of TAK1-IKKB-IRF5 axis leads to induction of cytokines and interferons following TLR7/TLR8 stimulation. (IRAK4 = interleukin-1 receptor-associated kinase 4; IRF5 = interferon regulatory factor-5; TAK1 = MAPK kinase kinase 7; IKKB = I-kappa B kinase; TLR = toll-like receptor) 29088270_By CRISPR/Cas9-induced inactivation of TLR9, MyD88, IRAK4 and IRAK1 we confirm that BZLF1 repression is dependent on functional TLR9 and MyD88 signaling, and identify IRAK4 as an essential element for TLR9-induced repression of BZLF1 expression upon BCR cross-linking 29127936_synthetic routes with moderate to high yields have been developed to produce the reference standard 9, demethylated precursor 8 and target tracer [11C]9. The radiosynthesis employed [11C]CH3OTf for N-[11C]methylation at the piperazin position of the desmethyl precursor, followed by product purification and isolation using a semi-preparative RP HPLC combined with SPE. [11C]9 was obtained in high radiochemical yield, radioc 29482609_Data show that interleukin 1 receptor associated kinase 4 (IRAK4) and EPH receptor A2 (EphA2) were the functional targets of miR-302b. 29778672_Defective MyD88, IRAK4 but not NF-kB inhibit IL-1beta, MCP-1 and IP-10 production. 30076215_our results indicate that IRAK4 has a critical scaffold function in myddosome formation and that its kinase activity is dispensable for myddosome assembly and activation of the NF-kappaB and MAPK pathways but is essential for MyD88-dependent production of inflammatory cytokines. Our findings suggest that the scaffold function of IRAK4 may be an attractive target for treating inflammatory and autoimmune diseases. 30112356_rs4251545 of IRAK4 (p.Ala428Thr) modified the susceptibility to HBV-related HCC via increased proliferation rate and reduced production of inflammatory cytokines and chemokines 30115681_Therefore, the IRAK4-MyD88 scaffolding function is essential for IL-1 signaling, but IRAK4 kinase activity can control IL-1 signal strength by modulating the association of IRAK4, MyD88, and IRAK1. 30229835_miR-544 may participate in controlling inflammation and apoptosis after ischemia-reperfusion by targeting IRAK4. 30679311_structures reveal conformational flexibility of unphosphorylated IRAK4 and provide unexpected insights into the potential use of small molecules to modulate IRAK4 activity in cancer, autoimmunity, and inflammation 30885957_Dimethyl fumarate blocks IRAK4-MyD88 interactions and IRAK4-mediated cytokine production in a cysteine 13-dependent manner. 30997787_Interleukin-1 receptor-associated kinase 4 (IRAK-4) involved in the pathway produces cytokines that initiate and maintain inflammation through Toll-like receptors and interleukin-1 receptors on the membranes of innate immune cells are stimulated with antigens 31011167_Inhibition of IRAK4-L abrogates leukaemic growth, particularly in acute myeloid leukaemia (AML) cells with higher expression of the IRAK4-L isoform. Collectively, mutations in U2AF1 induce expression of therapeutically targetable 'active' IRAK4 isoforms and provide a genetic link to activation of chronic innate immune signalling in myelodysplastic syndromes and AML. 31527315_IRAK4 mediates colitis-induced tumorigenesis and chemoresistance in colorectal cancer. 31702785_IRAK1 and IRAK4 signaling proteins are dispensable in the response of human neutrophils to Mycobacterium tuberculosis infection. 31995744_MyD88 Death-Domain Oligomerization Determines Myddosome Architecture: Implications for Toll-like Receptor Signaling. 32487715_The kinase IRAK4 promotes endosomal TLR and immune complex signaling in B cells and plasmacytoid dendritic cells. 32532880_Clinical IRAK4 deficiency caused by homozygosity for the novel IRAK4 (c.1049delG, p.Gly350Glufs*15) variant. 33020954_Interleukin-1 receptor-associated kinase-4 gene variation may increase post-bronchiolitis asthma risk. 33083971_IRAK4 Deficiency Presenting with Anti-NMDAR Encephalitis and HHV6 Reactivation. 33289936_Variations of interleukin-1 receptor-associated kinase-4 encoding gene were not associated with post-bronchiolitis lung function. 33508385_Tumor cell intrinsic RON signaling suppresses innate immune responses in breast cancer through inhibition of IRAK4 signaling. 34139893_IRAK-4 in macrophages contributes to inflammatory osteolysis of wear particles around loosened hip implants. 34145611_Potential associations between variants of genes encoding regulators of inflammation, and mediators of inflammation in type 2 diabetes and insulin resistance. 34587686_[Interleukin-1 receptor associated kinase 4 deficiency: a case report and literature review]. 35022428_Eliminating chronic myeloid leukemia stem cells by IRAK1/4 inhibitors. | ENSMUSG00000059883 | Irak4 | 385.41360 | 1.1141957 | 0.1560025915 | 0.20203138 | 5.970793e-01 | 4.396946e-01 | 7.833084e-01 | No | Yes | 514.704344 | 127.225553 | 443.380495 | 107.189776 | |
| ENSG00000198105 | 57209 | ZNF248 | protein_coding | Q8NDW4 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:57209; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | 16385451_Observational study of gene-disease association. (HuGE Navigator) 27238579_Smoking and obesity are risk factors for LOA. ZNF248 confers increased susceptibility to LOA in African Americans. | ENSMUSG00000030145 | Zfp248 | 332.58749 | 0.9457221 | -0.0805117589 | 0.16956817 | 2.251609e-01 | 6.351354e-01 | 8.837671e-01 | No | Yes | 376.415191 | 58.221693 | 387.855964 | 58.702695 | ||
| ENSG00000198265 | 9931 | HELZ | protein_coding | P42694 | FUNCTION: May act as a helicase that plays a role in RNA metabolism in multiple tissues and organs within the developing embryo. | ATP-binding;Alternative splicing;Helicase;Hydrolase;Metal-binding;Methylation;Nucleotide-binding;Nucleus;Phosphoprotein;Reference proteome;Zinc;Zinc-finger | HELZ is a member of the superfamily I class of RNA helicases. RNA helicases alter the conformation of RNA by unwinding double-stranded regions, thereby altering the biologic activity of the RNA molecule and regulating access to other proteins (Wagner et al., 1999 [PubMed 10471385]).[supplied by OMIM, Mar 2008]. | hsa:9931; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; P granule [GO:0043186]; ATP binding [GO:0005524]; helicase activity [GO:0004386]; hydrolase activity [GO:0016787]; metal ion binding [GO:0046872]; RNA binding [GO:0003723]; post-transcriptional gene silencing by RNA [GO:0035194] | 12691822_Results suggest that loss of expression of DRHC may play a role in human carcinogenesis. 20378664_Observational study of gene-disease association. (HuGE Navigator) 21765940_Downregulation of HELZ reduced translational initiation, resulting in the disassembly of polysomes, in a reduction of cell proliferation and hypophosphorylation of ribosomal protein S6. | ENSMUSG00000020721 | Helz | 2948.92841 | 1.0959034 | 0.1321206806 | 0.09050501 | 2.137519e+00 | 1.437342e-01 | 5.291124e-01 | No | Yes | 4465.784918 | 922.589617 | 3765.347285 | 760.768672 | |
| ENSG00000198270 | 89894 | TMEM116 | protein_coding | Q8NCL8 | Alternative splicing;Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:89894; | integral component of membrane [GO:0016021]; plasma membrane [GO:0005886]; G protein-coupled receptor activity [GO:0004930]; adenylate cyclase-activating G protein-coupled receptor signaling pathway [GO:0007189] | 34789718_TMEM116 is required for lung cancer cell motility and metastasis through PDK1 signaling pathway. | ENSMUSG00000029452 | Tmem116 | 157.35419 | 0.7360085 | -0.4422056723 | 0.22910062 | 3.686367e+00 | 5.485899e-02 | 3.543088e-01 | No | Yes | 155.277399 | 30.880563 | 208.068366 | 40.102730 | |||
| ENSG00000198408 | 10724 | OGA | protein_coding | O60502 | FUNCTION: [Isoform 1]: Cleaves GlcNAc but not GalNAc from O-glycosylated proteins. Can use p-nitrophenyl-beta-GlcNAc and 4-methylumbelliferone-GlcNAc as substrates but not p-nitrophenyl-beta-GalNAc or p-nitrophenyl-alpha-GlcNAc (in vitro) (PubMed:11148210). Does not bind acetyl-CoA and does not have histone acetyltransferase activity (PubMed:24088714). {ECO:0000269|PubMed:11148210, ECO:0000269|PubMed:11788610, ECO:0000269|PubMed:20673219, ECO:0000269|PubMed:22365600, ECO:0000269|PubMed:24088714}.; FUNCTION: [Isoform 3]: Cleaves GlcNAc but not GalNAc from O-glycosylated proteins. Can use p-nitrophenyl-beta-GlcNAc as substrate but not p-nitrophenyl-beta-GalNAc or p-nitrophenyl-alpha-GlcNAc (in vitro), but has about six times lower specific activity than isoform 1. {ECO:0000269|PubMed:20673219}. | 3D-structure;Alternative splicing;Cytoplasm;Glycosidase;Host-virus interaction;Hydrolase;Nucleus;Phosphoprotein;Reference proteome | The dynamic modification of cytoplasmic and nuclear proteins by O-linked N-acetylglucosamine (O-GlcNAc) addition and removal on serine and threonine residues is catalyzed by OGT (MIM 300255), which adds O-GlcNAc, and MGEA5, a glycosidase that removes O-GlcNAc modifications (Gao et al., 2001 [PubMed 11148210]).[supplied by OMIM, Mar 2008]. | hsa:10724; | cytosol [GO:0005829]; membrane [GO:0016020]; nucleus [GO:0005634]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102167]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine/L-threonine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102571]; [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-threonine O-N-acetyl-alpha-D-glucosaminase activity [GO:0102166]; beta-N-acetylglucosaminidase activity [GO:0016231]; hyalurononglucosaminidase activity [GO:0004415]; glycoprotein catabolic process [GO:0006516]; glycoprotein metabolic process [GO:0009100]; N-acetylglucosamine metabolic process [GO:0006044]; protein deglycosylation [GO:0006517]; protein O-linked glycosylation [GO:0006493] | 11148210_This protein is a cytosolic, neutral, O-GlcNAc specific hexosaminidase termed O-GlcNAcase. 12359146_Investigated this locus in Pima Indians who have the world's highest prevalence of NIDDM. Concluded that mutations in MGEA5 are unlikely to contribute to NIDDM in this population 12359146_Observational study of gene-disease association. (HuGE Navigator) 17546623_In type 2 Diabetes patients in Mexico City, the frequency of the T allele of MGEAT5 was higher (2.6%) in the cases than in controls (1.8%), but not a significant deviation from Hardy_Weinberg proportions. 17546623_Observational study of gene-disease association. (HuGE Navigator) 18641620_review of modifications, phosphorylation and a specific form of glycosylation, O-linked -N-acetylglucosaminylation by O-GlcNAc, relevant to pathological tau phosphorylation 19423084_the short nuclear variant of O-GlcNAcase, which has the identical catalytic domain as the full-length enzyme, has similar trends in a pH-rate profile and Taft linear free energy analysis as the full-length enzyme 19715310_characterization of O-GlcNAcase transition states using several series of substrates to generate multiple simultaneous free-energy relationships 19913121_Observational study of gene-disease association. (HuGE Navigator) 20198314_This study analyzes the activity of the enzyme involved in the removal of these sugar residues, i.e. beta-N-acetylglucosaminidase (O-GlcNAcase) as well as the level of N-acetylglucosamine in benign and malignant thyroid lesions. 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20673219_N-terminal region of OGA contains the catalytic site and the C-terminal region stabilizes the protein structure and affects substrate affinity. 20863279_Results provide evidence that OGA may possess a substrate-recognition mechanism tinvolving interactions with O-GlcNAcylated proteins beyond the GlcNAc-binding site. 21178104_Direct evidence links muscle atrophy and the disruption of O-GlcNAcase activity in male bitransgenic mice. 21471514_Reducing ChREBP(OG) levels via OGA overexpression decreased lipogenic protein content (ACC, FAS), prevented hepatic steatosis, and improved the lipidic profile of OGA-treated db/db mice. 21567137_Decrease in MGEA5 and increase in O-GlcNAc transferease expression in higher grade tumors suggests that increased O-GlcNAc modification may be implicated in breast tumor progression and metastasis. 21717526_Chromosomal translocation t(1;10) is consistent with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. 22311971_Data show that the interplay between O-GlcNAc and phosphorylation on proteins and indicate that these effects can be mediated by changes in hOGT and hOGA kinetic activity. 22730328_O-linked beta-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-beta-D-glucosaminidase silencing on cell phenotype and transcriptome. 22783592_Analysis of urinary content of MGEA5 and OGT may be useful for bladder cancer diagnostics. 24365779_Estrogen replacement therapy and plyometric training influence muscle OGT and OGA gene expression, which may be one of the mechanisms by which HRT and PT prevent aging-related loss of muscle mass. 24705316_Report the presence of TGFBR3 and/or MGEA5 rearrangements in pleomorphic hyalinizing angiectatic tumors and the spectrum of related neoplasms. 25173736_Amino acid composition of splice variants, post-translational modifications, and stable associations with regulatory proteins influence subcellular distribution/substrate specificity of OGA and OGT (O-linked N-acetylglucosamine transferase). [REVIEW] 25183011_This work identifies the first target of miR-539 in the heart and the first miRNA that regulates OGA. 26269457_OGA overexpression in endothelial cells improves endothelial function and may have a beneficial effect on coronary vascular complications in diabetes. 26527687_E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner. 26980036_TGFBR3 and/or MGEA5 rearrangements are much more common in hybrid hemosiderotic fibrolipomatous tumor-myxoinflammatory fibroblastic sarcomas than in classical myxoinflammatory fibroblastic sarcomas. 27231347_the O-linked N-acetylglucosamine (O-GlcNAc) processing enzymes, O-GlcNAc-transferase (OGT) and O-GlcNAcase (OGA), interact with the (A)gamma-globin promoter at the -566 GATA repressor site 27601472_OGA is physically associated with the known RNA polymerase II (pol II) pausing/elongation factors SPT5 and TRIM28-KAP1-TIF1beta, and a purified OGA-SPT5-TIF1beta complex has elongation properties. 28319083_hOGA forms an unusual arm-in-arm homodimer in which the catalytic domain of one monomer is covered by the stalk domain of the sister monomer to create a substrate-binding cleft. 28627871_Data suggest that the substrate specificity of O-GlcNAcase/OGA does not extend to proteins/peptides modified with S-GlcNAc (an analog of O-GlcNAc); proteins modified with S-GlcNAc appear to be stable against O-GlcNAcase/OGA hydrolysis. 28742148_Tax interacts with the host OGT/OGA complex and inhibits the activity of OGT-bound OGA. 28939839_Beta-N-acetylhexosaminidase substrate recognition and specificity 30052810_OGT promotes carcinogenesis and metastasis of cervical cancer cells. OGT's expression is significantly upregulated in cervical cancer, and low OGT level is correlated with improved prognosis 30920848_role of the tubular biomarkers NAG, kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin in patients with chest pain before contrast media exposition 31501520_Study reports that OGA is upregulated in a wide range of human cancers and drives aerobic glycolysis and tumor growth by inhibiting pyruvate kinase M2 (PKM2). PKM2 is dynamically O-GlcNAcylated in response to changes in glucose availability. Under high glucose conditions, PKM2 is a target of OGA-associated acetyltransferase activity, which facilitates O-GlcNAcylation of PKM2 by O-GlcNAc transferase (OGT). 32365408_Pharmacological inhibition and knockdown of O-GlcNAcase reduces cellular internalization of alpha-synuclein preformed fibrils. 33086728_Involvement of NDPK-B in Glucose Metabolism-Mediated Endothelial Damage via Activation of the Hexosamine Biosynthesis Pathway and Suppression of O-GlcNAcase Activity. 33310751_Bacterial O-GlcNAcase genes abundance decreases in ulcerative colitis patients and its administration ameliorates colitis in mice. 33333092_Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. 34135314_OGA is associated with deglycosylation of NONO and the KU complex during DNA damage repair. | ENSMUSG00000025220 | Oga | 6585.28282 | 1.9092975 | 0.9330419240 | 0.09399023 | 9.957811e+01 | 1.885777e-23 | 2.247092e-19 | No | Yes | 10576.411812 | 1627.230215 | 5424.478161 | 816.484081 | |
| ENSG00000198440 | 147949 | ZNF583 | protein_coding | Q96ND8 | FUNCTION: May be involved in transcriptional regulation. | DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:147949; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | ENSMUSG00000030443 | Zfp583 | 149.79939 | 1.1356014 | 0.1834565859 | 0.27986359 | 4.283929e-01 | 5.127786e-01 | 8.274384e-01 | No | Yes | 221.126459 | 56.444651 | 191.514980 | 47.799785 | |||
| ENSG00000198498 | 55319 | TMA16 | protein_coding | Q96EY4 | 3D-structure;ADP-ribosylation;Reference proteome | hsa:55319; | nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; nucleus [GO:0005634] | ENSMUSG00000025591 | Tma16 | 1041.51629 | 1.0435858 | 0.0615491852 | 0.11228926 | 3.005508e-01 | 5.835373e-01 | 8.603601e-01 | No | Yes | 1297.253726 | 252.255859 | 1177.578647 | 224.019447 | ||||
| ENSG00000198612 | 10920 | COPS8 | protein_coding | Q99627 | FUNCTION: Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of SCF-type E3 ligase complexes, leading to decrease the Ubl ligase activity of SCF-type complexes such as SCF, CSA or DDB2. The complex is also involved in phosphorylation of p53/TP53, c-jun/JUN, IkappaBalpha/NFKBIA, ITPK1 and IRF8/ICSBP, possibly via its association with CK2 and PKD kinases. CSN-dependent phosphorylation of TP53 and JUN promotes and protects degradation by the Ubl system, respectively. {ECO:0000269|PubMed:11285227, ECO:0000269|PubMed:11337588, ECO:0000269|PubMed:12628923, ECO:0000269|PubMed:12732143, ECO:0000269|PubMed:9535219}. | 3D-structure;Alternative splicing;Cytoplasm;Direct protein sequencing;Nucleus;Phosphoprotein;Reference proteome;Signalosome | The protein encoded by this gene is one of the eight subunits of COP9 signalosome, a highly conserved protein complex that functions as an important regulator in multiple signaling pathways. The structure and function of COP9 signalosome is similar to that of the 19S regulatory particle of 26S proteasome. COP9 signalosome has been shown to interact with SCF-type E3 ubiquitin ligases and act as a positive regulator of E3 ubiquitin ligases. Alternatively spliced transcript variants encoding distinct isoforms have been observed. [provided by RefSeq, Jul 2008]. | hsa:10920; | COP9 signalosome [GO:0008180]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; extracellular exosome [GO:0070062]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; perinuclear region of cytoplasm [GO:0048471]; activation of NF-kappaB-inducing kinase activity [GO:0007250]; COP9 signalosome assembly [GO:0010387]; negative regulation of cell population proliferation [GO:0008285]; protein deneddylation [GO:0000338]; protein phosphorylation [GO:0006468] | 12732143_DDB2 and CSA are each integrated into nearly identical complexes via interaction with DDB1. Both complexes contain cullin 4A and Roc1 and display ubiquitin ligase activity. They also contain the COP9 signalosome (CSN) 17318178_The COP9 signalosome (CSN) controls NF-kappaB by deubiquitinylation of IkappaBalpha. 18424433_COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-alpha ubiquitination 20399188_analysis of the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3 22414063_proteomic-based approach can broaden our understanding of the functions of the CSN in contexts such as viral-host interactions or immune activation in their natural milieu 22992343_miR-146a expression is up-regulated in a majority of gastric cancers where it targets CARD10 and COPS8, inhibiting GPCR-mediated activation of NF-kappaB. 23689509_Data indicate that silencing of Csn8 caused an increased growth rate, whereas silencing of Csn5 impaired proliferation in HeLa cells. 28063109_Among 479 individuals affected with clear cell renal cell carcinoma, only synonymous variants were found in COPS8 and one of the missense variants in ACKR3:c.892C>T, was observed in 4/479 individuals screened 33261601_CSN8 is a key regulator in hypoxia-induced epithelial-mesenchymal transition and dormancy of colorectal cancer cells. 33604618_COPS8 in cutaneous melanoma: an oncogene that accelerates the malignant development of tumor cells and predicts poor prognosis. | ENSMUSG00000034432 | Cops8 | 3083.75085 | 1.1048775 | 0.1438864583 | 0.07758892 | 3.450468e+00 | 6.323388e-02 | 3.759955e-01 | No | Yes | 4128.260230 | 672.393992 | 3494.984881 | 556.721286 | |
| ENSG00000198732 | 64093 | SMOC1 | protein_coding | Q9H4F8 | FUNCTION: Plays essential roles in both eye and limb development. Probable regulator of osteoblast differentiation. {ECO:0000269|PubMed:20359165, ECO:0000269|PubMed:21194678, ECO:0000269|PubMed:21194680}. | Alternative splicing;Basement membrane;Calcium;Developmental protein;Differentiation;Disease variant;Disulfide bond;Extracellular matrix;Glycoprotein;Metal-binding;Microphthalmia;Reference proteome;Repeat;Secreted;Signal | This gene encodes a multi-domain secreted protein that may have a critical role in ocular and limb development. Mutations in this gene are associated with microphthalmia and limb anomalies. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Mar 2011]. | hsa:64093; | basement membrane [GO:0005604]; extracellular space [GO:0005615]; calcium ion binding [GO:0005509]; extracellular matrix binding [GO:0050840]; heparin binding [GO:0008201]; cell differentiation [GO:0030154]; extracellular matrix organization [GO:0030198]; eye development [GO:0001654]; limb development [GO:0060173]; regulation of osteoblast differentiation [GO:0045667] | 12130637_isolation of the novel gene SMOC-1 encoding a secreted modular protein containing an EF-hand calcium-binding domain; localization within basement membranes in kidney and skeletal muscle and expression in the zona pellucida surrounding the oocyte 17386346_SMOC-1 is of physiological interest because it codes a secreted glycoprotein with five domains, each containing regions homologous to those on other proteins that mediate cell-matrix interactions. 20359165_Analyzed the secretory protein profiles of BMSCs grown in osteogenic medium (OSM) and identified SPARC-related modular calcium-binding protein 1 (SMOC1), a member of the SPARC family, as a regulator of osteoblast differentiation of BMSCs. 21194678_these findings indicate that SMOC1/Smoc1 is essential for ocular and limb development in both humans and mice. 21194680_Waardenburg anophthalmia syndrome is genetically heterogeneous; a second was locus found on chromosome 14, and mutations in SMOC1 were shown also cause this syndrome. 21349332_The present study thus identified SPARC related modular calcium binding 1 as a new cancer-associated protein capable of interacting with tenascin-C in vitro 21750680_Homozygosity mapping and subsequent targeted mutation analysis of a locus on 14q24.2 identified homozygous mutations in SMOC1 (SPARC-related modular calcium binding 1) in eight unrelated families with Waardenburg Anophthalmia syndrome. 23263445_SMOC1 provides a link between prenatal hormone exposure and digit ratio. 23274784_IL-17A but not IL-22 suppresses the replication of hepatitis B virus by inducing the expression of MxA and OAS. 27101391_SMOC binds to Pro-EGF, but does not induce Erk phosphorylation via the EGFR. 28085523_This is the first report of Waardenburg anophthalmia syndrome (WAS) caused by a SMOC1 variant in a Pakistani population. The mutation identified in the present investigation extends the body of evidence implicating the gene SMOC-1 in causing WAS. 28807869_Missense mutation in exon 3 of SMOC1 segregated with the Waardenburg anophthalmia syndrome in the Iranian family. 30445150_Exome sequencing identified two variants in the SMOC1 gene, each inherited from one of the parents: c.709G>T - p.(Glu237*) on exon 8 and c.1223G>A - p.(Cys408Tyr) on exon 11, both predicted to be pathogenic by different bioinformatics software. Brain histopathology showed an abnormal cortical neuronal migration, which could be related to the SMOC1 protein function, given its role in cellular signaling 33484701_SMOC1 and IL-4 and IL-13 Cytokines Interfere with Ca(2+) Mobilization in Primary Human Keratinocytes. 33529332_Secreted modular calcium-binding protein 1 binds and activates thrombin to account for platelet hyperreactivity in diabetes. 33757126_SPARC-related modular calcium binding 1 regulates aortic valve calcification by disrupting BMPR-II/p-p38 signalling. | ENSMUSG00000021136 | Smoc1 | 1762.82735 | 0.9745566 | -0.0371820931 | 0.10144209 | 1.342777e-01 | 7.140374e-01 | 9.129259e-01 | No | Yes | 1996.694963 | 164.252849 | 2022.526861 | 162.568762 | |
| ENSG00000203485 | 64423 | INF2 | protein_coding | Q27J81 | FUNCTION: Severs actin filaments and accelerates their polymerization and depolymerization. {ECO:0000250}. | Acetylation;Actin-binding;Alternative splicing;Charcot-Marie-Tooth disease;Coiled coil;Cytoplasm;Direct protein sequencing;Disease variant;Neurodegeneration;Neuropathy;Phosphoprotein;Reference proteome | This gene represents a member of the formin family of proteins. It is considered a diaphanous formin due to the presence of a diaphanous inhibitory domain located at the N-terminus of the encoded protein. Studies of a similar mouse protein indicate that the protein encoded by this locus may function in polymerization and depolymerization of actin filaments. Mutations at this locus have been associated with focal segmental glomerulosclerosis 5.[provided by RefSeq, Aug 2010]. | hsa:64423; | perinuclear region of cytoplasm [GO:0048471]; actin binding [GO:0003779]; small GTPase binding [GO:0031267]; actin cytoskeleton organization [GO:0030036]; regulation of mitochondrial fission [GO:0090140] | 20023659_Study identified nine independent nonconservative missense mutations in INF2, which encodes a member of the formin family of actin-regulating proteins. 20803156_In conclusion, we described an additional familial case of the autosomal dominant form of focalsegmental glomerulosclerosis associated with INF2 mutations. 21258034_Six of the seven distinct altered residues localized to an INF2 region that corresponded to a subdomain of the mDia1 diaphanous inhibitory domain reported to co-immunoprecipitate with IQ motif-containing GTPase-activating protein 1 21278336_The effects of disease-causing INF2 mutations suggest an important role for this protein and its interaction with other formins in modulating glomerular podocyte phenotype and function. 21866090_INF2 mutations were responsible for 16% of all cases of autosomal dominant focal and segmental glomerulosclerosis, with these mutations clustered in exon 4. 21998196_Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. 21998204_Actin monomers inhibit microtubule binding/bundling by INF2 22187985_INF2 mutations appear to cause many cases of FSGS-associated Charcot-Marie-Tooth neuropathy, showing that INF2 is involved in a disease affecting both the kidney glomerulus and the peripheral nervous system. 22879592_Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. 22961558_Our study confirms the link between INF2 mutations and Charcot-Marie-Tooth-associated glomerulopathy and widens the spectrum of pathogenic mutations. 22971997_A novel mutation, outside of the candidate region for diagnosis, in the inverted formin 2 gene can cause focal segmental glomerulosclerosis. 22986496_Formation of stabilized, detyrosinated microtubules required the formin INF2. 23014460_INF2 mutations were found in 2 of 281 individuals with sporadicfocal and segmental glomerulosclerosis 23349293_study found actin polymerization through ER-localized INF2 was required for efficient mitochondrial fission; INF2-induced actin filaments may drive initial mitochondrial constriction, which allows Drp1-driven secondary constriction 23521651_This study showed that INF2 mutations in Charcot-Marie-Tooth disease complicated with focal segmental glomerulosclerosis. 23620398_In podocytes, INF2 appears to be an important modulator of actin-dependent behaviors that are under the control of Rho/mDia signaling. 23847988_INF2 mutation was detected both father and his son 23921379_actin monomer binding to the DAD of INF2 competes with the DID/DAD interaction, thereby activating actin polymerization 24174593_this study identifed three novel mutations of INF likely efect hereditary neuropathy with glomerulopathy. 25165188_INF2 mutations are associated with focal segmental glomerulosclerosis. 26039629_Report novel mutations in the inverted formin 2 gene of Chinese families contributing to focal segmental glomerulosclerosis. 26124273_Assembly and turnover of short actin filaments by the formin INF2 and profilin. 26305500_The authors propose Spire1C isoform cooperates with INF2 to regulate actin assembly at endoplasmic reticulum-mitochondrial contacts. 26383224_Propose that examination of INF2 expression may help to differentiate minimal change disease from focal segmental glomerulosclerosis and evaluate the clinical severity of steroid resistance nephrotic syndrome in children. 26621033_FHOD1 and INF2 are novel regulators of inter- and intra-structural contractility of podosomes. 26764407_Disease causing mutations in inverted formin 2 regulate its binding to G-actin, F-actin capping protein (CapZ alpha-1) and profilin 2. 27974406_All individuals with INF2 mutations presenting with a thrombotic microangiopathy also had atypical hemolytic uremic syndrome risk haplotypes, potentially accounting for the genetic pleiotropy 28448495_hese findings reveal novel molecular events underlying the regulation of INF2 function and localization, and provided insights in understanding the relationship between SPOP mutations and dysregulation of mitochondrial dynamics in prostate cancer. 29142021_INF2-mediated actin polymerization on the endoplasmic reticulum stimulates mitochondrial division by two independent mechanisms: (1) mitochondrial calcium uptake, leading to inner mitochondrial membrane constriction; and (2) Drp1 oligomerization, leading to outer mitochondrial membrane constriction. 29947928_Studies indicate that INF2, a formin, that is mutated in hereditary renal and neurodegenerative disorders. 30126379_INF2 seems to be not only the cause of FSGS, but also of ESRD of unknown etiology. 30579254_INF2 is a novel pro-apoptotic inducer of oxidative injury in epidermal cells. 31354004_our results indicated that oxidative stress-mediated HaCaT cells apoptosis could be reversed by Tan IIA treatment via reducing INF2-related mitochondrial stress in a manner dependent on the ERK signaling pathway. 31515790_The phenotypic feature of the pedigree is autosomal dominant intermediate Charcot-Marie-Tooth disease and focal segmental glomerulosclerosis, which may be attributed to the c.341G>A mutation of the INF2 gene. 31871199_Study used lysine-to-glutamine mutations to map the relevant lysines on actin for INF2 regulation. K50Q- and K61Q-actin, when bound to CAP2, inhibit full-length INF2 but not INF2 lacking inhibitory domain (DID). CAP WH2 domain binds INF2-DID with submicromolar affinity but has weak affinity for actin monomers, while INF2-C-terminal diaphanous autoregulatory domain binds CAP/K50Q-actin 5-fold better than CAP/WT-actin. 31924668_FSGS-Causing INF2 Mutation Impairs Cleaved INF2 N-Fragment Functions in Podocytes. 32727404_Multiple formin proteins participate in glioblastoma migration. 32844773_Steroid Resistant Nephrotic Syndrome with Clumsy Gait Associated With INF2 Mutation. 34698992_Role of formin INF2 in human diseases. | ENSMUSG00000037679 | Inf2 | 1058.61227 | 0.8976337 | -0.1558012228 | 0.11907820 | 1.705143e+00 | 1.916168e-01 | 5.842517e-01 | No | Yes | 1267.052882 | 158.056646 | 1379.798717 | 167.965811 | |
| ENSG00000204054 | LINC00963 | lncRNA | 165.21905 | 0.7904313 | -0.3392880732 | 0.23924650 | 1.996348e+00 | 1.576787e-01 | 5.468276e-01 | No | Yes | 151.203886 | 18.825292 | 190.679314 | 22.876459 | |||||||||||
| ENSG00000204128 | 257407 | C2orf72 | protein_coding | A6NCS6 | Reference proteome | hsa:257407; | ENSMUSG00000026227 | 2810459M11Rik | 84.53795 | 0.9166906 | -0.1254932923 | 0.30780429 | 1.659073e-01 | 6.837751e-01 | 9.023105e-01 | No | Yes | 91.180367 | 11.578844 | 98.697822 | 11.906277 | |||||
| ENSG00000204305 | 177 | AGER | protein_coding | Q15109 | FUNCTION: Mediates interactions of advanced glycosylation end products (AGE). These are nonenzymatically glycosylated proteins which accumulate in vascular tissue in aging and at an accelerated rate in diabetes. Acts as a mediator of both acute and chronic vascular inflammation in conditions such as atherosclerosis and in particular as a complication of diabetes. AGE/RAGE signaling plays an important role in regulating the production/expression of TNF-alpha, oxidative stress, and endothelial dysfunction in type 2 diabetes. Interaction with S100A12 on endothelium, mononuclear phagocytes, and lymphocytes triggers cellular activation, with generation of key proinflammatory mediators. Interaction with S100B after myocardial infarction may play a role in myocyte apoptosis by activating ERK1/2 and p53/TP53 signaling (By similarity). Receptor for amyloid beta peptide. Contributes to the translocation of amyloid-beta peptide (ABPP) across the cell membrane from the extracellular to the intracellular space in cortical neurons. ABPP-initiated RAGE signaling, especially stimulation of p38 mitogen-activated protein kinase (MAPK), has the capacity to drive a transport system delivering ABPP as a complex with RAGE to the intraneuronal space. Can also bind oligonucleotides. {ECO:0000250, ECO:0000269|PubMed:19906677, ECO:0000269|PubMed:20943659, ECO:0000269|PubMed:21559403, ECO:0000269|PubMed:21565706}. | 3D-structure;Alternative splicing;Cell membrane;Disulfide bond;Glycoprotein;Immunoglobulin domain;Inflammatory response;Membrane;Phosphoprotein;Reference proteome;Repeat;Secreted;Signal;Transmembrane;Transmembrane helix | The advanced glycosylation end product (AGE) receptor encoded by this gene is a member of the immunoglobulin superfamily of cell surface receptors. It is a multiligand receptor, and besides AGE, interacts with other molecules implicated in homeostasis, development, and inflammation, and certain diseases, such as diabetes and Alzheimer's disease. Many alternatively spliced transcript variants encoding different isoforms, as well as non-protein-coding variants, have been described for this gene (PMID:18089847). [provided by RefSeq, May 2011]. | hsa:177; | apical plasma membrane [GO:0016324]; cell junction [GO:0030054]; cell surface [GO:0009986]; extracellular region [GO:0005576]; fibrillar center [GO:0001650]; integral component of plasma membrane [GO:0005887]; plasma membrane [GO:0005886]; postsynapse [GO:0098794]; advanced glycation end-product receptor activity [GO:0050785]; amyloid-beta binding [GO:0001540]; identical protein binding [GO:0042802]; protein-containing complex binding [GO:0044877]; S100 protein binding [GO:0044548]; scavenger receptor activity [GO:0005044]; signaling receptor activity [GO:0038023]; transmembrane signaling receptor activity [GO:0004888]; astrocyte activation [GO:0048143]; cell surface receptor signaling pathway [GO:0007166]; cellular response to amyloid-beta [GO:1904646]; glucose mediated signaling pathway [GO:0010255]; induction of positive chemotaxis [GO:0050930]; inflammatory response [GO:0006954]; learning or memory [GO:0007611]; microglial cell activation [GO:0001774]; modulation of age-related behavioral decline [GO:0090647]; negative regulation of blood circulation [GO:1903523]; negative regulation of interleukin-10 production [GO:0032693]; negative regulation of long-term synaptic depression [GO:1900453]; negative regulation of long-term synaptic potentiation [GO:1900272]; neuron projection development [GO:0031175]; positive regulation of activated T cell proliferation [GO:0042104]; positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process [GO:1902961]; positive regulation of chemokine production [GO:0032722]; positive regulation of dendritic cell differentiation [GO:2001200]; positive regulation of endothelin production [GO:1904472]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of heterotypic cell-cell adhesion [GO:0034116]; positive regulation of interleukin-1 beta production [GO:0032731]; positive regulation of interleukin-12 production [GO:0032735]; positive regulation of interleukin-6 production [GO:0032755]; positive regulation of JNK cascade [GO:0046330]; positive regulation of JUN kinase activity [GO:0043507]; positive regulation of monocyte chemotactic protein-1 production [GO:0071639]; positive regulation of monocyte extravasation [GO:2000439]; positive regulation of NF-kappaB transcription factor activity [GO:0051092]; positive regulation of NIK/NF-kappaB signaling [GO:1901224]; positive regulation of p38MAPK cascade [GO:1900745]; positive regulation of protein phosphorylation [GO:0001934]; positive regulation of tumor necrosis factor production [GO:0032760]; protein localization to membrane [GO:0072657]; regulation of CD4-positive, alpha-beta T cell activation [GO:2000514]; regulation of DNA binding [GO:0051101]; regulation of inflammatory response [GO:0050727]; regulation of long-term synaptic potentiation [GO:1900271]; regulation of NIK/NF-kappaB signaling [GO:1901222]; regulation of p38MAPK cascade [GO:1900744]; regulation of spontaneous synaptic transmission [GO:0150003]; regulation of synaptic plasticity [GO:0048167]; regulation of T cell mediated cytotoxicity [GO:0001914]; response to amyloid-beta [GO:1904645]; response to hypoxia [GO:0001666]; response to wounding [GO:0009611]; transcytosis [GO:0045056]; transport across blood-brain barrier [GO:0150104] | 11700025_mRNA and protein expression in Caco-2 cells 11739380_this study we have investigated a possible role of RAGE and amphoterin in the retinoic acid-induced differentiation of neuroblastoma cells. 11811511_susceptibility to the development of chronic periodontitis could be influenced by the 1704G/T polymorphism of the RAGE gene, independently of diabetes 11884895_Polymorphisms 1704G/T, 2184A/G, and 2245G/A in the rage gene are not associated with diabetic retinopathy in NIDDM 12029499_allele frequencies and genotype distribution combinations of four polymorphisms in the RAGE gene (6p21.3, G82S, 1704G/T, 2184A/G and 2245A/G) of 272 subjects (130 patients with plaque psoriasis and 142 healthy control). 12070776_increased prevalence of the 82S allele in patients with rheumatoid arthritis compared with control subjects 12477623_There is an association of Gly82Ser polymorphism in this gene with diabetic retinopathy in type II diabetic Asian Indian patients. 12495433_Data show that vascular endothelial cells and pericytes express three novel splice variants of receptor for advanced glycation end-products (RAGE) mRNA. 12579287_co-expression of RAGE and amphoterin is closely associated with invasion and metastasis of colorectal cancer 12598893_These data reinforce the importance of receptor for advanced glycation end products (RAGE)-ligand interactions in modulating properties of CD4+ T cells that infiltrate the central nervous system 12606536_association between -374 T/A RAGE polymorphism and cardiovascular disease and albumin excretion in type 1 diabetics with poor metabolic control suggests gene-environment interaction in development of diabetic nephropathy and cardiovascular complications 12618340_The RAGE is expressed in dying neurons and suggest that RAGE may have a role in neuronal cell death mediated by ischemic stress. 12651613_Advanced glycation end products and receptor for advanced glycation end products are found in AA amyloidosis. 12837757_AGEs can augment inflammatory responses by up-regulating COX-2 via RAGE and multiple signaling pathways, thereby leading to monocyte activation and vascular cell dysfunction 12859967_Results suggest a possible role of S100 protein- and RAGE-mediated signal transduction in the development of specific cancers. 12935895_In vitro binding studies using a series of C-terminal deletion mutants of human RAGE revealed the importance of the membrane-proximal cytoplasmic region of RAGE for the direct ERK-RAGE interaction. 12941744_No association betwween -429T/C and -37T/A polymorphism in RAGE gene promoter with diabetic retinopathy in NIDDM in Chinese patients 14580673_identified three novel RAGE transcripts all encoding truncated soluble forms of RAGE. The relative expression ratios for the full-length RAGE transcript to the sum of its splice-variants encoding the soluble variants varied strongly among tissues. 14595542_Oleate, not ligands of the receptor for advanced glycation end-products, acts as an enhancer of human smooth muscle cell proliferation. 14704946_Our study failed to demonstrate an association between either - 429 T/C or - 374 T/A gene polymorphism of the RAGE gene and diabetic retinopathy in Caucasians with type 2 diabetes 14747204_RAGE G1704T polymorphisms may be useful in identifying the risk for developing diabetic nephropathy in type 2 diabetic patients. 15009731_AGE might be involved in the growth and invasion of melanoma through interactions with RAGE and represent promising candidates for assessing the future therapeutic potential of this therapy in treating patients with melanoma. 15019601_results suggest that the decrease in monocyte RAGE expression can be at least partly accounted for by the ligand engagement and may be a factor contributing to the development of diabetic vascular complications 15033494_AGER mediates S100A13 protein translocation in response to extracellular S100 15052533_Patients with type 2 diabetes and the 63bp deletion in the promoter of RAGE seem to be protected from diabetic nephropathy. 15053925_Only cells expressing RAGE at the cell surface showed hypersensitivity to Abeta. 15127201_Findings using advanced glycosylation end products (AGEs) with strong RAGE-binding properties indicate that AGEs may not uniformly play a role in cellular activation. 15155381_Review. RAGE is an amplification step in vascular inflammation and acceleration of atherosclerosis. 15170618_the RAGE pathway plays a critical proinflammatory role in vasculitic neuropathy. 15180953_Advanced glycation end products (AGE) by binding to AGE receptor (RAGE) per se could control mesangial cell growth. 15203887_Corellation of expression of tissue factor (TF) and the receptor for advanced glycation end products (RAGEs) and vascular complications in diabetic patients 15213278_RAGE expression is upregulated on monocytes from patients with chronic kidney disease 15239215_RAGE and MMP-9 are expressed concordant with the metastatic ability of the human pancreatic cancer cells. Could be key to regulating metastatic ability of pancreatic cancer. 15247020_AGER has a role as a pleiotropic antagonistic gene (review) 15289604_AGER1 suppressed AGE mediated NF-kappaB and MAPK/p44/p42 activities and suppressed AGE-mediated mesangial cell inflammatory injury through negative regulation of RAGE 15448098_Thiazolidinedione antidiabetics modulate vascular endothelial RAGE expression. 15539404_Down-regulation of RAGE may be considered as a critical step in tissue reorganization and the formation of lung tumours. 15547674_374T/A polymorphism of the RAGE gene may reduce susceptibility to coronary artery disease 15555779_RAGE may have multiple functions in the human brain, mediated by the individual or coordinated efforts of the different RAGE isoforms 15599399_data demonstrate that the RAGE-NF-kappaB axis operates in diabetic neuropathy, by mediating functional sensory deficits, and that its inhibition may provide new therapeutic approaches 15663911_RAGE, via its interaction with ligands, serves as a cofactor exacerbating diabetic vascular disease. (review) 15666359_DU145 cells, a hormone-independent prostate cancer cell line, showed the highest RAGE mRNA expression 15790669_The RAGE2 haplotype is associated with diabetic nephropathy (DN) in type 2 diabetics and with earlier DN onset and can be regarded as a marker for DN. 15798956_data suggest that G1704T and G82S polymorphisms of the advanced glycation end product receptor (RAGE) gene are not related to microalbuminuria in Japanese type 2 diabetic patients 15803111_82 Serine allele of the RAGE gene is a risk allele for developing advanced nephropathy in diabettes mellitus. 15893388_Reactive oxygen species generated by the TNF-alpha-stimulated umbilical vein endothelial cells induce RAGE expression. 15896660_results show that the -374A allele (-374T>A polymorphism) in the RAGE gene is related to the susceptibility of developing ischemic heart disease in African-Brazilians with type 2 diabetes 15911356_Antibodies against RAGE receptor blocked Abeta-induced activation of the p38, JNK pathways, and NF-kappaB in CTL cybrids and offered protection against the neurotoxic effects of Abeta. 15915542_results reveal HMGB1 and RAGE as the first known autocrine loop modulating the maturation of plasmacytoid dendritic cells 15926115_data suggest that activation of the receptor for advanced glycation end products (RAGE) pathway may be one of the first steps in the pathogenesis of diabetic polyneuropathies 15930093_ACE inhibition reduces the accumulation of AGE in diabetes partly by increasing the production and secretion of sRAGE into plasma 15942078_polymorphisms G1704T and G82S of the RAGE gene are not related to DR in Japanese type 2 diabetic patients. 15942086_polymorphisms G1704T and G82S of the RAGE gene are not related to DR in Japanese type 2 diabetic patients. 15944249_Advanced glycosylation end-product receptor (RAGE) is required for the effect of high mobility group box 1 (HMGB1) on dendritic cells; HMGB1/RAGE interaction results in downstream activation of MAP kinases and NF-kappa B. 15986224_data suggests that the CML-RAGE-NF-kappaB pathway is an evident proinflammatory pathomechanism in mononuclear effector cells in polymyositis and dermatomyositis 16037279_RAGE, an endogenous decoy receptor, may be related to individual variations in resistance to the development of diabetic vascular complications, observed in transgenic mice. 16052547_The increase in RAGE noted in osteoarthritis(OA) cartilage and the ability of RAGE ligands to stimulate chondrocyte MAP kinase and NF-kappaB activity and to stimulate MMP-13 production suggests that chondrocyte RAGE signaling could play a role in OA. 16061848_S100P plays a major role in the aggressiveness of pancreatic cancer that is likely mediated by its ability to activate RAGE 16127291_These results suggest that RAGE expression appears to be closely associated with the invasiveness of oral squamous cell carcinoma and represents a promising candidate for assessing the future therapeutic potential in treating patients with oral carcinoma. 16159602_Analysis of specific manifestations of cardiovascular disease, including coronary heart disease (CHD), cardiovascular disease (CVD), myocardial infarction (MI) and ischemic disease (ISD) revealed no association with RAGE genotype. 16166741_Data suggest that re-expression of the receptor for advanced glycation end-products in lung cancer cells impairs the proliferative stimulus mediated by fibroblasts. 16271939_a cross-talk exists between the AGE-RAGE system and the angiotensin II receptor (renin-angiotensin system), and serum levels of sRAGE may reflect endothelial RAGE expression 16286548_Patients with Alzheimer disease have reduced levels of sRAGE in plasma compared with patients with vascular dementia and controls. 16374460_RAGE is mainly expressed in fibroblasts, dendrocytes, and keratinocytes and to a minor extent in endothelial and mononuclear cells. 16440015_study found a modest association with the -RAGE 374T/A polymorphism in the nonproliferative diabetic retinopathy subgroup 16493495_The existence of splice variants of RAGE in endothelial cells might explain the lack of interaction of extracellular AGEs with endothelial cells. 16644647_Association of level in serum with albuminuria may be a marker of microvascular damage in type 2 diabetes. 16728681_3 AGER genetic variants: -429T>C, -374T>A, and Gly82Ser were determined in cerebral ischemia and stroke patients. Specific haplotypes were associated with reduced risk of incident myocardial infarction & ischemic stroke, independent of diabetes status. 16728681_Observational study of gene-disease association. (HuGE Navigator) 16847147_CRP at concentrations known to predict future vascular events upregulates RAGE expression in human endothelial cells at both the protein and mRNA level. Silencing of the RAGE gene prevents CRP-induced macrophage chemoattractant protein 1 activation. 16899960_adhesion of red blood cells from type 2 diabetes patients was mediated by RAGE 16954185_AGER1 negatively regulates AGE-mediated oxidant stress-dependent signaling via the EGFR and Shc/Grb2/Ras pathway. 16954682_observation suggests that the RAGE gene polymorphism is not associated with in-stent restenosis after coronary artery stenting in non-diabetic patients in the Korean population 16969646_results show an association between the AGER -374 T/A polymorphism & type 1 diabetes; the polymorphism was associated with diabetic nephropathy in both type 1 & type 2 diabetes & with sight-threatening retinopathy in type 1 diabetic patients 16969649_Serum levels are associated with the severity of nephropathy in type 2 diabetic patients. 17004092_Telmisartan: it may work as an anti-inflammatory agent against AGE by suppressing RAGE expression via PPAR-gamma activation in the liver. 17024691_Serum levels are significantly higher in type 2 diabetic patients than in non-diabetics and are positively associated with the presence of coronary artery disease. 17035340_data suggest that the autocrine/paracrine release of HMGB1 and the integrity of the HMGB1/RAGE pathway are required for the migratory function of dendritic cells 17158877_Soluble RAGE forms tetramers that bind to hexamers of Ca2+-calgranulin C, creating a large platform for effectively transmitting RAGE-dependent signals from extracellular S100 proteins to the cytoplasmic signaling complexes. 17170388_association of RAGE and its ligands with sarcoidosis and suggest that an intrinsic genetic factor could be in part involved in its expression. In Italian patients, the -374 T/A polymorphism seems to be significantly associated with this disease. 17172923_The results of this large population study demonstrate that the AA/GA genotypes of the RAGE +557G>A polymorphism are associated with a significantly decreased risk of significant coronary artery disease. 17218539_study demonstrates that HOCl-modified albumin acts as a ligand for RAGE and promotes RAGE-mediated inflammatory complications 17224333_subjects with homozygosity for the minor S allele of the G82S polymorphism of RAGE had higher risk factors for cardiovascular disease 17237617_We demonstrated that after successful TX, PAPP-A and sRAGE decrease and early chronic vascular changes in the kidney TX are associated with elevation of their serum levels. 17343760_Soluble but not sescretory protein is associated with albuminuria in type 2 diabetic paaatients, whil neither is associated with markers of glucose control or macrovascular disease. 17345061_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 17355942_Observational study of gene-disease association. (HuGE Navigator) 17405935_evidence provided for activation of the NF-kappaB system in intervertebral discs in vivo which correlates with accumulated oxidative stress & increases in age & disc degeneration; oxidative stress may lead to RAGE activation & NF-kappaB translocation 17425804_the RAGE Ser82 allele does not predispose to cardiovascular events in rheumatoid arthritis. 17434489_Accumulation of AGE may have a role in the development of osteoarthritis. 17497647_Expression of Endogenous secretory receptor for advanced glycation endproducts was significantly higher in chondrosarcomas 17508727_High level bacterial expression systems and purification protocols were generated for the extracellular region of RAGE (sRAGE) and the five permutations of single and tandem domain constructs. 17512509_The -429 T>C polymorphism in the RAGE promoter is associated with type 1 diabetes in a Brazilian population, with a 2-fold increase of C-allele frequency compared to non-diabetic subjects 17536039_Data show that soluble RAGE blocks scavenger receptor CD36-mediated uptake of hypochlorite-modified low-density lipoprotein. 17549666_it is concluded that CDK5RAP3, CCNB2, and RAGE genes may be used as a very reliable biomarkers of lung adenocarcinoma 17560613_Olmesartan may play a protective role against proliferative diabetic retinopathy by attenuating the deleterious effects of AGEs via down-regulation of RAGE. 17588956_RAGE was found throughout the testis, caput epididymis, particularly the principle cells apical region, and on sperm acrosomes in men with diabetes mellitus. 17592553_Serum levels of sRAGE are associated with inflammatory markers in patients with type 2 diabetes. 17593216_Multivariate analysis showed high-grade expression of RAGE to be an independent prognostic factor for disease-free survival in oral squamous cell carcinoma. 17601350_Observational study of gene-disease association. (HuGE Navigator) 17613396_Higher levels of plasma RAGE measured shortly after reperfusion predicted poor short-term outcomes from lung transplantation. Elevated plasma RAGE levels may have both pathogenetic and prognostic value in patients after lung transplantation. 17639302_Treating type 2 diabetic patients with thiazolidinedione can increase circulating levels of RAGE. 17640970_Data suggest that deregulation of RAGE expression in myoblasts might concur in rhabdomyosarcomagenesis and that increasing RAGE expression in rhabdomyosarcoma cells might reduce their tumor potential. 17660747_Structural and the binding data suggest that tetrameric S100B triggers RAGE activation by receptor dimerisation. 17661837_The receptor of advanced glycation end product (RAGE) -374 T/A single nucleotide polymorphism affected the dialysate-to-plasma ratio of creatinine at baseline, suggesting that the RAGE polymorphism may effect the basal peritoneal solute transport rate. 17700210_Observational study of gene-disease association. (HuGE Navigator) 17714874_De-N-glycosylation or G82S mutation of RAGE increases affinity for AGE ligands, and may sensitize cells or conditions with it to AGE. 17924846_result confirms that the -374AA genotype of the RAGE gene promoter is a protective factor against the severity of coronary artery disease lesions in type 2 diabetic patients 18032526_Suggest key mechanisma by which AGER1 maintains cellular resistance against AGE-induced oxidative stress. 18046662_Plasma soluble RAGE and endogenous secretory RAGE are not associated with measures of diabetic neuropathy in type 2 diabetes patients. 18050248_Calculating the S100A12:sRAGE ratio might help to detect patients with KD who are at risk of being unresponsive to IVIG therapy. 18058469_serum sRAGE levels were influenced by genetic polymorphisms (-429 T/C, Gly82Ser and 2184 A/G) of the RAGE gene in breast cancer. 18078623_HMGB-1 and BSA-AGE stimulated the invasiveness of fibroblast-like synoviocytes in rheumatoid arthritis by activation of RAGE. 18079485_Single nucleotide polymorphisms may contribute to development of glucose intolerance and type 2 diabetes. 18079965_endothelial RAGE and its ligands mediate vascular and inflammatory stresses that culminate in atherosclerosis in the vulnerable vessel wall 18080716_HCC during the early stage of tumorigenesis with less blood supply may acquire resistance to stringent hypoxic milieu by hypoxia-induced RAGE expression. 18089847_RAGE_v1 variant is the primary secreted soluble isoform of RAGE. identification of functional splice variants of RAGE underscores biological diversity of the RAGE gene & will aid in understanding of the gene in the normal & pathological state. 18179750_the -374AA genotype of the RAGE gene promoter could be associated with a reduced risk of in-stent restenosis after coronary stent implantation 18231794_This preliminary study supports the hypothesis that the RAGE system might participate in the disease pathway of primary Sjogren's syndrome (SS), and that sRAGE may be a potential biomarker to aid in the diagnosis of primary SS. 18245812_Loss of RAGE contributes to idiopathic pulmonary fibrosis pathogenesis. 18256374_Endothelial dysfunction in chronic kidney disease may be partly mediated by advanced glycation end product-induced inhibition of endothelial nitric oxide synthase through RAGE activation. 18279703_RAGE could also behave as a receptor for Mycobacterium tuberculosis 18279705_-374T/A RAGE polymorphism is an independent protective factor for cardiac events in nondiabetic patients with coronary artery disease 18302220_study identified advanced glycation end products as the ligand for RAGE on both invasive and non-invasive prostate cancer cells 18322992_The SAA-RAGE-stimulated NF-kappaB signaling pathway has an important role in the pathogenesis of rheumatoid arthritis. 18339893_S100A8/A9-promoted cell growth occurs through RAGE signaling and activation of NF-kappaB. 18355449_These findings reveal new aspects of RAGE regulation and signaling and also provide a new interaction between RAGE and human pathologies. 18421017_results indicate that RAGE serves a protective role in the lung, and that loss of the receptor is related to functional changes of pulmonary cell types, with the consequences of fibrotic disease 18427074_results show significant downregulation of the total RAGE mRNA transcripts in peripheral blood mononuclear cells of patients with probable Alzheimer disease 18431028_low expression of endogenous secretory RAGE in the hippocampus would be associated with the development of Alzheimer's disease 18438937_shedding of RAGE might occur as reactive oxygen species accumulate in brain cells and be part of the process of neurodegeneration 18458846_AGEs induce ROS generation and intensify the proliferation and activation of HSCs, supporting the possibility that antioxidants may represent a promising treatment for prevention of the development of hepatic fibrosis in NASH. 18477569_the peritoneal membrane of the uraemic patients showed an increased submesothelial thickness and a marked induction of RAGE expression and NFkappaB-binding activity 18480271_Distinct regions of RAGE are involved in Abeta-induced cellular and neuronal toxicity with respect to the Abeta aggregation state, suggesing the blockage of particular sites of the receptor as a therapeutic strategy to attenuate neuronal death. 18519797_RAGE Gly82Ser polymorphism may confer not only an increased risk of gastric cancer but also with invasion of gastric cancer in the Chinese population. 18550199_Serum AGER levels are associated with the severity of renal dysfunction and duration of diabetes in type 2 diabetic patients/ 18575614_Observational study of gene-disease association. (HuGE Navigator) 18575614_association between diabetic complications and LTA, TNF and AGER polymorphisms is complex, with partly different alleles conferring susceptibility in type 1 and type 2 diabetic patients 18576917_Advanced oxidation protein products might be new ligands of endothelial RAGE. AOPPs-HSA activates vascular ECs via RAGE-mediated signals. 18595673_Circulating soluble receptor for advanced glycation end products is inversely associated with body mass index and waist/hip ratio in the general population. 18603587_The sheddase ADAM10 was identified as a membrane protease responsible for RAGE cleavage. 18606705_study defines RAGE (receptor for advanced glycation end products) as the hS100A7 receptor, whereas hS100A15 functions through a Gi protein-coupled receptor; hS100A7-RAGE binding, signaling, and chemotaxis are zinc-dependent in vitro 18615900_GFR is a principal determinant of sRAGE concentration and gradual sRAGE increase in subjects with advancing impairment of renal function is paralleled by AGE and its receptor 18615900_Observational study of gene-disease association. (HuGE Navigator) 18645306_Mediation of RAGE affects the balance of cellular proliferation and apoptosis 18657529_RAGE levels increase in conjunction with the onset of Alzheimer's disease, and continue to increase linearly as a function of pathologic severity 18660489_Observational study of gene-disease association. (HuGE Navigator) 18667420_Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE). 18689872_RAGE, carboxylated glycans and S100A8/A9 play essential roles in tumor-stromal interactions, leading to inflammation-associated colon carcinogenesis. 18698218_maternal esRAGE concentrations are significantly increased in patients with preeclampsia during pregnancy 18701333_association of low soluble isoform of RAGE (sRAGE) with high CML-protein levels in diabetic patients who developed severe diabetic complications supports the hypothesis that sRAGE protects vessels AGE-mediated diabetic microvascular damage 18775683_RAGE was markedly and diffusely expressed in blood vessels, neurons, and the choroid plexus in the brains of two patients who died as the result of brain edema that developed during the treatment of severe diabetic ketoacidosis 18777492_likely that circulating soluble RAGE and endogenous secretory RAGE are distinct markers and that circulating esRAGE levels are associated with the status of early-stage atherosclerosis. 18789567_Advanced glycation end products (AGE) and circulating receptor for AGE (RAGE) levels are independently associated with decreased glomerular filtration rate (GFR) and seem to predict decreased GFR. 18796298_Observational study of gene-disease association. (HuGE Navigator) 18796298_We found no consistent association between prevalent type 2 diabetes mellitus and insulin indices and AGER polymorphisms 18825489_elevated serum HMGB-1 and sRAGE levels are associated with the disease severity and immunological abnormalities in systemic sclerosis. 18854308_HNRNP K and microRNA-16 have roles in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products 18922799_the interaction of the RAGE cytoplasmic domain with Dia-1 is required to transduce extracellular environmental cues evoked by binding of RAGE ligands to their cell surface receptor, resulting in Rac-1 and Cdc42 activation and cellular migration 18926539_Serum endogenous secretory RAGE level is an independent risk factor for the progression of carotid atherosclerosis in type 1 diabetes. 18948101_RAGE signaling contributes to neuroinflammation in infantile neuronal ceroid lipofuscinosis. 18952609_ADAM10 and MMP9 to be involved in RAGE shedding. 18977241_Observational study of gene-disease association. (HuGE Navigator) 18987644_Observational study of gene-disease association. (HuGE Navigator) 19005067_Thus, HMGB1-RAGE signaling links necrosis with macrophage activation and may provide a target for anti-inflammatory therapy in stroke. 19023628_Negative RAGE expression correlates with deeper tumor invasion and venous invasion in patients with esophageal squamous cell carcinoma. 19032093_new insight into AGE-RAGE interaction 19061941_A set of tumours, healthy tissues and various cancer cell lines were screened for RAGE splicing variants and analysed their structure. 19071026_These results indicate that RAGE signaling directly contributes to pathology in cerebral ischemia. 19087540_The increased expression of HMGB1 and RAGE in the placenta may play an important role in the pathogenesis of pre-eclampsia. 19088375_Increased soluble RAGE is localized in ectopic endometrial cells of both adenomyosis and ovarian endometriomas; RAGE may contribute to the pathogenesis of endometriosis. 19103440_Our results provide support for a reduction of S100B levels during reconvalescence from acute paranoid schizophrenia that is regulated by its scavenger RAGE 19129693_Angiotensin II induced RAGE mRNA and protein expression through AT2 receptors. 19133252_No association of RAGE selected gene polymorphisms with 12-months outcome of renal transplants was shown in study. 19143681_several differences in the levels of advanced glycation end products, sRAGE, and proinflammatory cytokines between euglycemic and diabetic pregnancies 19143821_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 19167759_Observational study of gene-disease association. (HuGE Navigator) 19183936_sRAGE is associated with greater prevalence of cardiovascular disease in type 1 diabetes, which may be explained by endothelial and renal dysfunction. 19201044_treatment of THP-1 cells with 100 microg of CRP/ml/10(6) cells for 24 h, augmented the expression of RAGE and EN-RAGE genes 19204726_Observational study of gene-disease association. (HuGE Navigator) 19217461_No significant correlation between serum levels off RAGE in hypertensive agents. 19217891_Plasma levels of sRAGE are significantly reduced in definite and borderline nonalcoholic fatty liver disease. 19248116_IL-7 stimulates chondrocyte secretion of S100A4 via activation of JAK/STAT signaling, and then S100A4 acts in an autocrine manner to stimulate MMP-13 production via RAGE. 19252206_Elevated serum advanced glycation end products and their circulating receptors are associated with anaemia in older community-dwelling women. 19275767_a novel biological and genetic marker for vascular disease (Review) 19277685_The up-regulation of RAGE in the Alzheimer Disease optic nerves indicates that RAGE may play a role in the pathophysiology of Alzheimer Disease optic neuropathy. 19284577_RAGE activity influences co-development of joint and vascular disease in rheumatoid arthritis patients. 19285601_G allele at RAGEG82S may be more associated with inflammatory markers under obese status than nonobese conditions. 19332646_the role of recombinant human RAGE in Abeta production was examined in the brain tissue of an AD animal model as well as in cultured cells 19376511_results suggest a possibility that RAGE-VEGF regulation may be related to reproductive dysfunction in aging women 19380603_AGE-2 and AGE-3 activate monocytes via the receptor for AGE (RAGE), leading to the up-regulation of adhesion molecule expression and cytokine production. 19380826_Amyloid beta protein-induced CC-type chemokine receptor 5 (CCR5) up-regulation in human brain microvascular endothelial cells depends on RAGE. 19405075_Stimulation with advanced glycation end products up-regulated endothelial RAGE expression in saphenous vein endothelial cells cultured from diabetic patients. 19439163_This study demonstrated an association of decreased serum endogenous secretory receptor for advanced glycation endproducts (esRAGE) level with coronary plaque progression in patients with diabetes. 19440065_Plasma levels of soluble receptor for advanced glycation end products are associated with endothelial function and predict cardiovascular events in nondiabetic patients. 19448391_High circulating AGEs and RAGE predict cardiovascular disease mortality among older community-dwelling women 19451748_The plasma concentration of sRAGE was highest in patients with sepsis (2,210 +/- 252 pg/ml), while the levels of sRAGE correlated inversely with that of HMGB1 in patients with acute pancreatitis. 19501570_Proteolysis with thrombin or factor Xa revealed several protease sensitive sites in RAGE. 19530996_expression detected in synovial tissue and on the immune cells of rheumatoid arthritis patients; genetic polymorphism is associated with disease 19542745_G82S polymorphism in the RAGE gene is associated with diabetic retinopathy and G-A haplotype containing 1704G and 82S allele might be a genetic marker of diabetic retinopathy in Chinese T2DM patients. 19571577_Advanced glycation end products induce calcification of vascular smooth muscle cells through RAGE/p38 MAPK. 19576587_Report increased expression of RAGE and EN-RAGE in non-diabetic pre-mature coronary artery disease. 19578796_Observational study of gene-disease association and gene-environment interaction. (HuGE Navigator) 19587357_Meta-analysis of gene-disease association. (HuGE Navigator) 19591173_Extracellular domain of RAGE down-regulates RAGE expression and inhibits p65 and p52 activation in human-salivary gland-cell-lines. 19616266_advanced glycosylation end-product receptor may capture and eliminate circulating serum high mobility group box 1 in humans 19635564_Blockade of the renin angiotensin system by irbesartan may play a protective role against tubular injury in diabetes by attenuating the deleterious effects of AGEs via down-regulation of RAGE. 19659809_Observational study of gene-disease association. (HuGE Navigator) 19672558_Studies indicate that RAGE is subject to ectodomain shedding by ADAM10 and MMP9 and the derived sRAGE can sequester Abeta for systemic clearance and antagonize Abeta binding to cell surface RAGE. 19679874_may be important mediator of cellular injury in fetuses delivered in the setting of inflammation-induced preterm birth 19766904_G82S and -429 T/C polymorphisms of RAGE were associated with the circulating levels of esRAGE but not with coronary artery disease in Chinese patients with Diabetes Melitus, Type 2. 19786934_Data demonstrate that sRAGE level, but not HMGB1 level, is significantly higher in AP patients who develop organ failure than in AP patients without organ failure who recover. 19815443_findings show receptor engagement by distinct advanced glycation end-products differentially enhances expression of RAGE isoforms and adhesion of red blood cells from diabetic patients 19820033_Reduction of advenced glycation end products in normal diets may lower oxidant stress/inflammation and restore levels of AGER an antioxidant, in healthy and aging subjects. 19822091_Molecular study of receptor for advanced glycation endproduct gene promoter and identification of specific HLA haplotypes possibly involved in chronic fatigue syndrome. 19834494_Our data suggest that targeted inhibition of RAGE or its ligands may serve as novel targets to enhance current cancer therapies. 19851445_Observational study of gene-di | ENSMUSG00000015452 | Ager | 54.37273 | 1.1711155 | 0.2278833135 | 0.43862907 | 2.705500e-01 | 6.029631e-01 | No | Yes | 50.304674 | 14.742789 | 43.878338 | 12.473762 | ||
| ENSG00000204356 | 7936 | NELFE | protein_coding | P18615 | FUNCTION: Essential component of the NELF complex, a complex that negatively regulates the elongation of transcription by RNA polymerase II (PubMed:10199401). The NELF complex, which acts via an association with the DSIF complex and causes transcriptional pausing, is counteracted by the P-TEFb kinase complex (PubMed:11940650, PubMed:12612062). Provides the strongest RNA binding activity of the NELF complex and may initially recruit the NELF complex to RNA (PubMed:18303858, PubMed:27282391). {ECO:0000269|PubMed:10199401, ECO:0000269|PubMed:11940650, ECO:0000269|PubMed:12612062, ECO:0000269|PubMed:18303858, ECO:0000269|PubMed:27282391}.; FUNCTION: (Microbial infection) The NELF complex is involved in HIV-1 latency possibly involving recruitment of PCF11 to paused RNA polymerase II. {ECO:0000269|PubMed:23884411}. | 3D-structure;Alternative splicing;Coiled coil;Direct protein sequencing;Isopeptide bond;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Repeat;Repressor;Transcription;Transcription regulation;Ubl conjugation | The protein encoded by this gene is part of a complex termed negative elongation factor (NELF) which represses RNA polymerase II transcript elongation. This protein bears similarity to nuclear RNA-binding proteins; however, it has not been demonstrated that this protein binds RNA. The protein contains a tract of alternating basic and acidic residues, largely arginine (R) and aspartic acid (D). The gene localizes to the major histocompatibility complex (MHC) class III region on chromosome 6. [provided by RefSeq, Jul 2008]. | hsa:7936; | NELF complex [GO:0032021]; nuclear body [GO:0016604]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; chromatin binding [GO:0003682]; RNA binding [GO:0003723]; negative regulation of mRNA polyadenylation [GO:1900364]; negative regulation of transcription by RNA polymerase II [GO:0000122]; negative regulation of transcription elongation from RNA polymerase II promoter [GO:0034244]; positive regulation of ERK1 and ERK2 cascade [GO:0070374]; positive regulation of histone H3-K4 methylation [GO:0051571]; positive regulation of transcription by RNA polymerase II [GO:0045944] | 11940650_Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA 12612062_NELF-C and NELF-D are highly related or identical to the protein called TH1, of unknown function. NELF-B and NELF-C or NELF-D are integral subunits that bring NELF-A and NELF-E together. [NELF-B] [NELF-C] 16880520_5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF)- and NELF-mediated transcriptional pausing has a dual function in regulating immediate-early expression of the human junB gene. 16898873_NELF E RRM binds to the single-stranded TAR RNAs with K(d) values in the low-micromolar range. 17499042_Negative elongation factor (NELF) is a four subunit transcription factor. Our results point to a surprising role of NELF in the 3' end processing of histone mRNAs and suggest that NELF is a new factor that coordinates mRNA processing in transcription. 18303858_RNA binding to NELF-E RRM induces formation of a helix in the C-terminus. RNA-bound form of NELF-E RRM is very similar to RNA-bound form of U1A RRM, although C-terminus of NELF-E RRM is unstructured in free protein, whereas it is helical in U1A protein. 19115949_Observational study, meta-analysis, and genome-wide association study of gene-disease association. (HuGE Navigator) 19143814_Observational study of gene-disease association. (HuGE Navigator) 19245807_data show that NELF subunits exhibit highly specific subcellular localizations, such as in NELF bodies or in midbodies, and some shuttle actively between the nucleus and cytoplasm; loss of NELF from cells can lead to enlarged and/or multiple nuclei 19556007_Observational study of gene-disease association. (HuGE Navigator) 19556007_Our results do not support any major role of the 4 AMD-associated variants in the risk of developing PCV, but favor a predominant association with the RDBP-SKIV2L variants 19851445_Observational study of gene-disease association. (HuGE Navigator) 20097260_Transient NELF-E knock-down in pituitary increased coincidentally prolactin expression and enhanced transcription of a prolactin-promoter reporter gene. 22614758_Multivariate analysis revealed that RDBP protein levels were an independent risk factor for early intrahepatic recurrence of HCC within 2 years of surgery. 22740393_the transcription elongation of KSHV OriLytL-K7 lytic genes is inhibited by NELF during latency, but can also be promptly reactivated in an RTA-independent manner upon external stimuli 24453987_these results describe the RNA binding behavior of NELF-E and supports a biological role for NELF-E in promoter-proximal pausing of both HIV-1 and cellular genes. 24636995_Following NELF-E knockdown or tumor necrosis factor alpha stimulation, promoter-proximal RNAP II levels increase up to 3-fold, and there is a dramatic increase in RNAP II levels within the HIV genome. 26504077_novel actions of BRD4 and of NELF-E in GR-controlled gene induction have been uncovered. 27282391_A positively charged face of NELF-AC is involved in RNA binding, whereas the opposite face of the NELF-AC subcomplex binds NELF-B. NELF-B is predicted to form a HEAT repeat fold, also binds RNA in vivo, and anchors the subunit NELF-E, which is confirmed to bind RNA in vivo. 29101316_define two mutually exclusive complexes CBC-NELF-E and CBC-ARS2-PHAX, which likely act in respectively earlier and later phases of transcription 30833661_NELFE-Dependent MYC Signature Identifies a Unique Cancer Subtype in Hepatocellular Carcinoma. 31638184_NELFE expression was increased in pancreatic cancer (PC) tissues compared with the paired normal tissues. NELFE expression was upregulated in PC cells when compared with normal pancreatic cells. Knockdown of NELFE inhibited the proliferation, invasion and migration of PC cells. NELFE activates the Wnt/betacatenin signaling pathway and epithelialtomesenchymal transition by decreasing the stabilization of NDRG2 mRNA in PC. 33248388_NELF complex fosters BRCA1 and RAD51 recruitment to DNA damage sites and modulates sensitivity to PARP inhibition. 33526068_Overexpression of NELFE contributes to gastric cancer progression via Wnt/beta-catenin signaling-mediated activation of CSNK2B expression. 35041643_Genome-wide chromatin contacts of super-enhancer-associated lncRNA identify LINC01013 as a regulator of fibrosis in the aortic valve. | ENSMUSG00000024369 | Nelfe | 1818.34665 | 1.0283565 | 0.0403404243 | 0.09647193 | 1.749773e-01 | 6.757257e-01 | 8.993575e-01 | No | Yes | 2256.811730 | 229.944319 | 2074.773419 | 206.637721 | |
| ENSG00000204406 | 55777 | MBD5 | protein_coding | Q9P267 | FUNCTION: Binds to heterochromatin. Does not interact with either methylated or unmethylated DNA (in vitro). | Alternative splicing;Chromosome;Mental retardation;Nucleus;Reference proteome | This gene encodes a member of the methyl-CpG-binding domain (MBD) family. The MBD consists of about 70 residues and is the minimal region required for a methyl-CpG-binding protein binding specifically to methylated DNA. In addition to the MBD domain, this protein contains a PWWP domain (Pro-Trp-Trp-Pro motif), which consists of 100-150 amino acids and is found in numerous proteins that are involved in cell division, growth and differentiation. Mutations in this gene cause an autosomal dominant type of cognitive disability. The encoded protein interacts with the polycomb repressive complex PR-DUB which catalyzes the deubiquitination of a lysine residue of histone 2A. Haploinsufficiency of this gene is associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Alternatively spliced transcript variants have been found, but their full-length nature is not determined. [provided by RefSeq, Jul 2017]. | hsa:55777; | chromocenter [GO:0010369]; chromosome [GO:0005694]; extracellular exosome [GO:0070062]; midbody [GO:0030496]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; chromatin binding [GO:0003682]; glucose homeostasis [GO:0042593]; nervous system development [GO:0007399]; positive regulation of growth hormone receptor signaling pathway [GO:0060399]; regulation of behavior [GO:0050795]; regulation of multicellular organism growth [GO:0040014] | 19904302_Haploinsufficiency of MBD5 is associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures.( 20700456_MBD5 and MBD6 are unlikely to be methyl-binding proteins, yet they may contribute to the formation or function of heterochromatin. 21271666_2q23 de novo microdeletion involving the MBD5 gene in a patient with developmental delay, postnatal microcephaly and distinct facial features. 21981781_MBD5 is a single causal locus as shown by 2q23.1 microdeletion syndrome with roles in intellectual disability, epilepsy, and autism spectrum disorder 23055267_MBD5 was tied to neurodevelopmental disorders following the identification of microdeletions on chromosome 2q22-2q23. 23422940_Identified de novo intragenic deletions of MBD5 in three patients. 23587880_study demonstrates that haploinsufficiency of MBD5 causes diverse phenotypes, yields insight into the spectrum of resulting neurodevelopmental and behavioral psychopathology and provides clinical context for interpretation of MBD5 structural variations. 23632792_The features associated with a deletion, mutation or duplication of MBD5 and the gene expression changes observed support MBD5 as a dosage-sensitive gene critical for normal development. 24634419_We studied and showed that both MBD5 and MBD6 interact with the mammalian PR-DUB Polycomb protein complex in a mutually exclusive manner, and that the MBD of MBD5 and MBD6 is both necessary and sufficient to mediate this interaction. 24885232_A genomic copy number variant analysis implicates the MBD5 and HNRNPU genes in Chinese children with infantile spasms and expands the clinical spectrum of 2q23.1 deletion. 25271084_Circadian rhythm gene expression altered by haploinsufficiency of MBD5. 25853262_Results show that when MBD5 and RAI1 are haploinsufficient, they perturb several common pathways that are linked to neuronal and behavioral development. 25966365_Reduced MBD5 dosage leads to mRNA and microRNA expression patterns and DNA methylation patterns more characteristic of differentiating than proliferating neural stem cells. This balance change may underlie neurodevelopmental disorders. 28295210_Based on segregation analysis of a patient (case 296491) with an intragenic deletion of the MBD5 gene, we classified deletions of exons 3 to 4 of the 5' untranslated region (UTR) of this gene as probably benign. The deletion of exon 3 was shared with the father and paternal uncle, both unaffected 28807762_A novel frameshift mutation c.254_255delGA (p.Arg85Asnfs*6) in the MBD5 gene was identified in a family with intellectual disability and epilepsy. 33427406_MBD5-related intellectual disability in a Vietnamese child. 33510365_Long-read whole-genome sequencing identified a partial MBD5 deletion in an exome-negative patient with neurodevelopmental disorder. 34459404_Early-Onset Dementia Associated with a Heterozygous, Nonsense, and de novo Variant in the MBD5 Gene. 35385942_[Clinical phenotypes and genetic features of epilepsy children with MBD5 gene variants]. | ENSMUSG00000036792 | Mbd5 | 447.47251 | 0.9616820 | -0.0563681228 | 0.18860956 | 8.931996e-02 | 7.650435e-01 | 9.326662e-01 | No | Yes | 578.235676 | 101.682001 | 575.803415 | 98.947406 | |
| ENSG00000204920 | 7711 | ZNF155 | protein_coding | Q12901 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:7711; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 117.30755 | 0.9051932 | -0.1437024074 | 0.28289478 | 2.576194e-01 | 6.117604e-01 | 8.733362e-01 | No | Yes | 124.180685 | 26.524079 | 134.754808 | 28.129570 | |||||
| ENSG00000205078 | 100130958 | SYCE1L | protein_coding | A8MT33 | FUNCTION: May be involved in meiosis. {ECO:0000250}. | Coiled coil;Meiosis;Reference proteome | hsa:100130958; | intermediate filament cytoskeleton [GO:0045111]; synaptonemal complex [GO:0000795]; synaptonemal complex assembly [GO:0007130] | Mouse_homologues 16328886_An isoform of 4930481F22, lacking the 7th exon, encodes a putative 28.4 kDa protein expressed in spermatocytes, and may have an important role in meiosis. | ENSMUSG00000033409 | Syce1l | 37.68092 | 1.2980383 | 0.3763329396 | 0.50391723 | 5.555830e-01 | 4.560454e-01 | No | Yes | 45.341651 | 8.839078 | 33.514477 | 6.706801 | |||
| ENSG00000205323 | 84324 | SARNP | protein_coding | P82979 | FUNCTION: Binds both single-stranded and double-stranded DNA with higher affinity for the single-stranded form. Specifically binds to scaffold/matrix attachment region DNA. Also binds single-stranded RNA. Enhances RNA unwinding activity of DDX39A. May participate in important transcriptional or translational control of cell growth, metabolism and carcinogenesis. Component of the TREX complex which is thought to couple mRNA transcription, processing and nuclear export, and specifically associates with spliced mRNA and not with unspliced pre-mRNA. TREX is recruited to spliced mRNAs by a transcription-independent mechanism, binds to mRNA upstream of the exon-junction complex (EJC) and is recruited in a splicing- and cap-dependent manner to a region near the 5' end of the mRNA where it functions in mRNA export to the cytoplasm via the TAP/NFX1 pathway. The TREX complex is essential for the export of Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs and infectious virus production. {ECO:0000269|PubMed:15338056, ECO:0000269|PubMed:17196963, ECO:0000269|PubMed:20844015}. | 3D-structure;Acetylation;DNA-binding;Direct protein sequencing;Nucleus;Phosphoprotein;RNA-binding;Reference proteome;Transcription;Transcription regulation;Translation regulation;Transport;mRNA transport | hsa:84324; | cytoplasmic ribonucleoprotein granule [GO:0036464]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; transcription export complex [GO:0000346]; chromatin binding [GO:0003682]; DNA binding [GO:0003677]; RNA binding [GO:0003723]; mRNA export from nucleus [GO:0006406]; negative regulation of transcription by RNA polymerase II [GO:0000122]; poly(A)+ mRNA export from nucleus [GO:0016973]; regulation of translation [GO:0006417] | ENSMUSG00000078427 | Sarnp | 1846.87742 | 1.0076489 | 0.0109929824 | 0.08736293 | 1.583337e-02 | 8.998659e-01 | 9.717475e-01 | No | Yes | 2279.601618 | 302.727880 | 2145.661908 | 278.653770 | |||
| ENSG00000205485 | 100133091 | transcribed_unprocessed_pseudogene | 105.78432 | 1.0376761 | 0.0533561608 | 0.28479011 | 3.517789e-02 | 8.512234e-01 | 9.595722e-01 | No | Yes | 117.264637 | 17.831043 | 112.176034 | 16.685837 | |||||||||||
| ENSG00000205704 | 339674 | SMIM45 | protein_coding | A0A590UK83 | Reference proteome | 32583748_Long Noncoding RNA LINC00634 Functions as an Oncogene in Esophageal Squamous Cell Carcinoma Through the miR-342-3p/Bcl2L1 Axis. | 53.82453 | 0.9787595 | -0.0309737247 | 0.43476376 | 5.084637e-03 | 9.431537e-01 | No | Yes | 61.280874 | 11.019210 | 61.969964 | 10.614032 | ||||||||
| ENSG00000205930 | C21orf62-AS1 | lncRNA | 34.07411 | 0.7533984 | -0.4085152093 | 0.51623775 | 6.194230e-01 | 4.312618e-01 | No | Yes | 39.861957 | 12.532826 | 52.384513 | 15.885474 | ||||||||||||
| ENSG00000206559 | 152098 | ZCWPW2 | protein_coding | Q504Y3 | FUNCTION: Histone methylation reader which binds to non-methylated (H3K4me0), monomethylated (H3K4me1), dimethylated (H3K4me2) and trimethylated (H3K4me3) 'Lys-4' on histone H3 (PubMed:26933034). The order of binding preference is H3K4me3 > H3K4me2 > H3K4me1 > H3K4me0 (PubMed:26933034). {ECO:0000269|PubMed:26933034}. | 3D-structure;Metal-binding;Reference proteome;Zinc;Zinc-finger | hsa:152098; | nucleus [GO:0005634]; methylated histone binding [GO:0035064]; zinc ion binding [GO:0008270] | 35217607_PRDM9 losses in vertebrates are coupled to those of paralogs ZCWPW1 and ZCWPW2. | 25.31185 | 1.0249891 | 0.0356086327 | 0.58158775 | 3.703978e-03 | 9.514705e-01 | No | Yes | 33.073364 | 13.885738 | 31.600820 | 13.047772 | |||||
| ENSG00000213015 | 51157 | ZNF580 | protein_coding | Q9UK33 | FUNCTION: Involved in the regulation of endothelial cell proliferation and migration. Mediates H(2)O(2)-induced leukocyte chemotaxis by elevating interleukin-8 production and may play a role in inflammation. May be involved in transcriptional regulation. {ECO:0000269|PubMed:20382120, ECO:0000269|PubMed:21830064}. | Chemotaxis;DNA-binding;Inflammatory response;Isopeptide bond;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Ubl conjugation;Zinc;Zinc-finger | hsa:51157; | chromatin [GO:0000785]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; DNA-binding transcription activator activity, RNA polymerase II-specific [GO:0001228]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; sequence-specific double-stranded DNA binding [GO:1990837]; cellular response to hydrogen peroxide [GO:0070301]; chemotaxis [GO:0006935]; inflammatory response [GO:0006954]; positive regulation of endothelial cell migration [GO:0010595]; positive regulation of endothelial cell proliferation [GO:0001938]; positive regulation of gene expression [GO:0010628]; positive regulation of interleukin-8 production [GO:0032757]; positive regulation of leukocyte chemotaxis [GO:0002690]; regulation of transcription by RNA polymerase II [GO:0006357] | 20382120_These results elucidate the important role that ZNF580 plays in the process of migration and proliferation of endothelial cells, which provides a foundation for a novel approach to regulate angiogenesis. 21310414_ZNF580 is a novel factor in the lipoprotein-dependent regulation of endothelial IL-8 expression and monocyte arrest. 21599657_ZNF580 is a binding partner of Smad2 and is involved in the signal transduction of the transforming growth factor-beta signalling pathway. 21830064_H2O2 upregulates the expression of ZNF580 via the NF-kappaB signaling pathway, and overexpression of ZNF580 plays a critical role in augmenting the release of pro-inflammatory cytokine IL-8. 24771066_These results suggest that ZNF580 mediates eNOS expression and endothelial cell migration/proliferation via the TGF-b1/ALK5/Smad2 pathway, and thus plays a crucial role in vascular endothelial cells. | ENSMUSG00000055633 | Zfp580 | 582.63461 | 0.9105529 | -0.1351852997 | 0.15917234 | 7.185675e-01 | 3.966142e-01 | 7.582377e-01 | No | Yes | 635.251980 | 96.472699 | 664.513796 | 98.499518 | ||
| ENSG00000213468 | 286467 | FIRRE | lncRNA | This gene produces a long RNA containing a repeating sequence. This RNA interacts with heterogeneous nuclear ribonucleoprotein U and coats chromosomes, where it plays an important role in the maintenance of repressive chromatin. This transcript can also act post-transcriptionally to regulate the stability of mRNAs during the innate immune response. [provided by RefSeq, Dec 2017]. | 28993514_this study shows a new regulatory role for NF-kappaB-responsive FIRRE in the posttranscriptional regulation of inflammatory genes in the innate immune system 29654311_neither the deletion of the CTCF locus nor the ectopic insertion of Firre cDNA or its ectopic expression are sufficient to alter topologically associated domains in a sex-specific or allele-specific manner 29678151_RMST and FIRRE, genes with established roles in neurogenesis and topological organisation of chromosomal domains respectively, are processed as circular lncRNAs with only minor linear species 30739786_FIRRE is transcriptionally activated by MYC. 35110535_LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy. | 469.10838 | 1.0987052 | 0.1358043109 | 0.16660992 | 6.664602e-01 | 4.142885e-01 | 7.694298e-01 | No | Yes | 563.560715 | 130.604596 | 513.059420 | 116.293208 | ||||||||
| ENSG00000213983 | 8906 | AP1G2 | protein_coding | O75843 | FUNCTION: May function in protein sorting in late endosomes or multivesucular bodies (MVBs). {ECO:0000269|PubMed:9733768}.; FUNCTION: (Microbial infection) Involved in MVB-assisted maturation of hepatitis B virus (HBV). {ECO:0000269|PubMed:16867982, ECO:0000269|PubMed:17553870}. | 3D-structure;Cytoplasmic vesicle;Endosome;Golgi apparatus;Host-virus interaction;Membrane;Protein transport;Reference proteome;Transport | Adaptins are important components of clathrin-coated vesicles transporting ligand-receptor complexes from the plasma membrane or from the trans-Golgi network to lysosomes. The adaptin family of proteins is composed of four classes of molecules named alpha, beta-, beta prime- and gamma- adaptins. Adaptins, together with medium and small subunits, form a heterotetrameric complex called an adaptor, whose role is to promote the formation of clathrin-coated pits and vesicles. The protein encoded by this gene is a gamma-adaptin protein and it belongs to the adaptor complexes large subunits family. This protein along with the complex is thought to function at some trafficking step in the complex pathways between the trans-Golgi network and the cell surface. [provided by RefSeq, Aug 2017]. | hsa:8906; | AP-1 adaptor complex [GO:0030121]; endosome membrane [GO:0010008]; Golgi apparatus [GO:0005794]; Golgi membrane [GO:0000139]; Golgi-associated vesicle [GO:0005798]; membrane [GO:0016020]; transport vesicle [GO:0030133]; cargo adaptor activity [GO:0140312]; clathrin adaptor activity [GO:0035615]; Golgi to vacuole transport [GO:0006896]; intracellular protein transport [GO:0006886]; receptor-mediated endocytosis [GO:0006898]; vesicle-mediated transport [GO:0016192] | 17553870_These results demonstrate that HBV exploits the multivesicular bodies machinery with the aid of gamma 2-adaptin. 18772139_gamma2-adaptin's ubiquitin-interacting motif mediates a specific physical interaction with the ubiquitin ligase Nedd4 and promotes ubiquitination of gamma2-adaptin 20708039_Data show that gamma2-adaptin in MVB sorting specifically interacts with the ESCRT subunits Vps28 and CHMP2A. 27909244_Depletion of the gamma2 or mu1A (AP1M1) subunits of AP-1, but not of gamma1 (AP1G1), precludes Nef-mediated lysosomal degradation of CD4. | ENSMUSG00000040701 | Ap1g2 | 738.79564 | 0.6916701 | -0.5318440061 | 0.12544462 | 1.770078e+01 | 2.585199e-05 | 4.338766e-03 | No | Yes | 753.766827 | 154.953293 | 1140.131232 | 228.483931 | |
| ENSG00000214021 | 26140 | TTLL3 | protein_coding | Q9Y4R7 | FUNCTION: Monoglycylase which modifies alpha- and beta-tubulin, adding a single glycine on the gamma-carboxyl groups of specific glutamate residues to generate monoglycine side chains within the C-terminal tail of tubulin. Not involved in elongation step of the polyglycylation reaction (By similarity). Preferentially glycylates a beta-tail peptide over the alpha-tail, although shifts its preference toward alpha-tail as beta-tail glutamylation increases (By similarity). Competes with polyglutamylases for modification site on beta-tubulin substrate, thereby creating an anticorrelation between glycylation and glutamylation reactions (By similarity). Together with TTLL8, mediates microtubule glycylation of primary and motile cilia, which is essential for their stability and maintenance (By similarity). Involved in microtubule glycylation of primary cilia in colon which controls cell proliferation of epithelial cells and plays an essential role in colon cancer development (PubMed:25180231). Together with TTLL8, glycylates sperm flagella which regulates axonemal dynein motor activity, thereby controlling flagellar beat, directional sperm swimming and male fertility (By similarity). {ECO:0000250|UniProtKB:A4Q9E5, ECO:0000250|UniProtKB:B2GUB3, ECO:0000269|PubMed:25180231}. | ATP-binding;Alternative splicing;Cell projection;Cilium;Cytoplasm;Cytoskeleton;Flagellum;Ligase;Magnesium;Metal-binding;Microtubule;Nucleotide-binding;Reference proteome | hsa:26140; | axoneme [GO:0005930]; cilium [GO:0005929]; cytosol [GO:0005829]; microtubule [GO:0005874]; microtubule cytoskeleton [GO:0015630]; sperm flagellum [GO:0036126]; ATP binding [GO:0005524]; metal ion binding [GO:0046872]; protein-glycine ligase activity [GO:0070735]; protein-glycine ligase activity, initiating [GO:0070736]; axoneme assembly [GO:0035082]; cilium assembly [GO:0060271]; flagellated sperm motility [GO:0030317]; protein polyglycylation [GO:0018094] | 11054573_Characterization of another human tubulin tyrosine ligase-like gene family member | 611.28782 | 1.1114577 | 0.1524530398 | 0.14462218 | 1.116312e+00 | 2.907140e-01 | 6.761178e-01 | No | Yes | 610.856346 | 116.131507 | 567.685590 | 105.583794 | ||||
| ENSG00000214135 | 220729 | SDHAP4 | transcribed_unprocessed_pseudogene | 334.91293 | 0.9686312 | -0.0459805560 | 0.16689367 | 7.586585e-02 | 7.829801e-01 | 9.388166e-01 | No | Yes | 390.367089 | 62.217395 | 401.730912 | 62.584223 | ||||||||||
| ENSG00000214193 | 79729 | SH3D21 | protein_coding | A4FU49 | Alternative splicing;Coiled coil;Reference proteome;SH3 domain | hsa:79729; | nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; actin filament organization [GO:0007015]; cell migration [GO:0016477] | ENSMUSG00000073758 | Sh3d21 | 138.84084 | 0.6988449 | -0.5169558221 | 0.26071906 | 3.881561e+00 | 4.881915e-02 | 3.341350e-01 | No | Yes | 131.022298 | 28.416320 | 195.183704 | 41.003465 | ||||
| ENSG00000214770 | lncRNA | 19.77147 | 0.7201552 | -0.4736201724 | 0.64443191 | 5.350810e-01 | 4.644785e-01 | No | Yes | 18.189441 | 6.949559 | 26.180720 | 9.500630 | |||||||||||||
| ENSG00000215022 | 100130357 | lncRNA | 38.18750 | 0.8440207 | -0.2446496442 | 0.45874132 | 2.823961e-01 | 5.951350e-01 | No | Yes | 35.773348 | 8.770868 | 44.099192 | 10.184963 | ||||||||||||
| ENSG00000215769 | 109286553 | ARHGAP27P1-BPTFP1-KPNA2P3 | lncRNA | This locus represents naturally-occurring readthrough transcription between multiple unprocessed pseudogene loci. Readthrough transcripts likely do not encode functional proteins. [provided by RefSeq, Dec 2016]. | 79.95788 | 0.6978373 | -0.5190373397 | 0.34705323 | 2.205745e+00 | 1.374975e-01 | 5.184285e-01 | No | Yes | 57.237384 | 15.153928 | 85.643329 | 21.845101 | |||||||||
| ENSG00000221829 | 2189 | FANCG | protein_coding | O15287 | FUNCTION: DNA repair protein that may operate in a postreplication repair or a cell cycle checkpoint function. May be implicated in interstrand DNA cross-link repair and in the maintenance of normal chromosome stability. Candidate tumor suppressor gene. | 3D-structure;Cytoplasm;DNA damage;DNA repair;Disease variant;Fanconi anemia;Nucleus;Phosphoprotein;Reference proteome;Repeat;TPR repeat | The Fanconi anemia complementation group (FANC) currently includes FANCA, FANCB, FANCC, FANCD1 (also called BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ (also called BRIP1), FANCL, FANCM and FANCN (also called PALB2). The previously defined group FANCH is the same as FANCA. Fanconi anemia is a genetically heterogeneous recessive disorder characterized by cytogenetic instability, hypersensitivity to DNA crosslinking agents, increased chromosomal breakage, and defective DNA repair. The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group G. [provided by RefSeq, Jul 2008]. | hsa:2189; | cytosol [GO:0005829]; Fanconi anaemia nuclear complex [GO:0043240]; mitochondrion [GO:0005739]; nucleolus [GO:0005730]; nucleoplasm [GO:0005654]; plasma membrane [GO:0005886]; damaged DNA binding [GO:0003684]; cellular response to DNA damage stimulus [GO:0006974]; DNA repair [GO:0006281]; interstrand cross-link repair [GO:0036297]; mitochondrion organization [GO:0007005]; ovarian follicle development [GO:0001541]; response to radiation [GO:0009314]; spermatid development [GO:0007286] | 12432219_Study of the molecular evolution of FA genes using database search methods such as PSI-BLAST suggested that FANCG may contain a known domain, and that this protein is a member of the family of tetratricopeptide repeat-containing proteins. 12552564_There is remarkably lage sequence variation in FANCG gene mutations and polymorphisms across ethnic and racial backgrounds found in the International Fanconi Anemia Registry they include IVS8-2A>G, IVS11+1G>c, 1794_1803del10, and IVS3+1G>C. 12649160_FANCG was able to mediate interaction between FANCA and FANCF, as well as between monomers of FANCA 12861027_FANCG is required for efficient homologous recombination-mediated repair of at least some types of DNA double-strand breaks 12915460_FANCG interacts directly with BRCA2. 15059067_A unique Fanconi-anemia-causing mutation, FANCG splice-site mutation IVS4+3A>G, showed exon 4 skipping. 15138265_FANCA and FANCG uniquely respond to oxidative damage by forming complexes via intermolecular disulfide linkages 15256425_FA proteins function at the level of chromatin during S phase to regulate and maintain genomic stability. 15319283_Primary fibroblasts from patients with Fanconi anemia with reduced FANCG expression show no signs of telomere dysfunction. 16195237_Observational study of gene-disease association. (HuGE Navigator) 16609022_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 16621732_FANCG, in addition to stabilising the FA core complex, may have a role in building multiprotein complexes that facilitate homologous recombination repair. 17010390_Four human FANCG polymorphic variants show normal biological function. 18212739_FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. 18950845_Observational study of gene-disease association. (HuGE Navigator) 19064572_Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19237606_Observational study of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) 19339270_Observational study of gene-disease association. (HuGE Navigator) 19536092_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19536649_Observational study of gene-disease association. (HuGE Navigator) 19690177_Observational study of gene-disease association. (HuGE Navigator) 19861517_Observational study of gene-disease association. (HuGE Navigator) 19913121_Observational study of gene-disease association. (HuGE Navigator) 20403997_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20496165_Observational study of gene-disease association and gene-gene interaction. (HuGE Navigator) 20518486_sites of interaction of FANCG with ERCC1, which is different from the region of ERCC1 that binds to XPF 20628086_Observational study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 21750350_Areca nut extracts-induced miR-23a was correlated with a reduced FANCG expression and DSB repair, which might contribute to ANE-associated human malignancies. 23067021_FANCA and FANCG are the major Fanconi anemia genes in the Korean population. 24300640_Three novel single base pair deletions, resulting in frameshift mutations (c.247delA, c.179delT and c.899delT) were identified in patients with Fanconi anaemia 25132264_A new role of FANCG in Homologous recombination repair of interstrand crosslinks through K63Ub-mediated interaction with the Rap80-BRCA1 complex. 25477267_Patients, homozygous for the FANCG founder mutation, present with severe cytopenia but progress to bone marrow failure at similar ages to other individuals affected with Fanconi anemia of heterogeneous genotype. 25703136_founder haplotype analysis of FANCG for the Korean Fanconi anemia population 27608133_a systems biology approach for elucidating the therapeutic potential of curcumin against FA and leukemia is investigated by analyzing the computational molecular interactions of curcumin ligand with FANC G of FA and seven other key disease targets of leukemia 28024295_studied the impact of mutations on the function and structure of FANCG 28440438_LOH may predominantly indicate copy number gains in FANCF and losses in FANCG and BRIP1. Integration of copy number data and gene expression proved difficult as the available sample sets did not overlap. 29843852_In >80% of black patients with Fanconi anaemia , a homozygous seven basepair deletion mutation in the FANCG gene (NM_004629.1 g.35077270_35077264del p.Tyr213Lysfs)[2] has been confirmed as the cause of the disease. 30057198_FANCG stimulates FANCA-mediated strand annealing and strand exchange. 32529760_Endocrine profiling in patients with Fanconi anemia, homozygous for a FANCG founder mutation. 32947577_Clinical and Genetic Features of Patients With Fanconi Anemia in Lebanon and Report on Novel Mutations in the FANCA and FANCG Genes. 32989015_Loss of Mitochondrial Localization of Human FANCG Causes Defective FANCJ Helicase. 33394227_Severe telomere shortening in Fanconi anemia complementation group L. 34436527_Frequent internuclear bridging in a Fanconi anemia patient with FANCG mutation. 34864095_In silico study of missense variants of FANCA, FANCC and FANCG genes reveals high risk deleterious alleles predisposing to Fanconi anemia pathogenesis. | ENSMUSG00000028453 | Fancg | 1106.81006 | 0.8833026 | -0.1790202664 | 0.11636235 | 2.353503e+00 | 1.250018e-01 | 4.979498e-01 | No | Yes | 1100.644796 | 102.620399 | 1218.558455 | 110.651325 | |
| ENSG00000221926 | 10626 | TRIM16 | protein_coding | O95361 | FUNCTION: E3 ubiquitin ligase that plays an essential role in the organization of autophagic response and ubiquitination upon lysosomal and phagosomal damages. Plays a role in the stress-induced biogenesis and degradation of protein aggresomes by regulating the p62-KEAP1-NRF2 signaling and particularly by modulating the ubiquitination levels and thus stability of NRF2. Acts as a scaffold protein and facilitates autophagic degradation of protein aggregates by interacting with p62/SQSTM, ATG16L1 and LC3B/MAP1LC3B. In turn, protects the cell against oxidative stress-induced cell death as a consequence of endomembrane damage. {ECO:0000269|PubMed:22629402, ECO:0000269|PubMed:27693506, ECO:0000269|PubMed:30143514}. | Alternative splicing;Coiled coil;Cytoplasm;Metal-binding;Phosphoprotein;Reference proteome;Repeat;Transferase;Ubl conjugation;Ubl conjugation pathway;Zinc;Zinc-finger | The protein encoded by this gene is a tripartite motif (TRIM) family member that contains two B box domains and a coiled-coiled region that are characteristic of the B box zinc finger protein family. While it lacks a RING domain found in other TRIM proteins, the encoded protein can homodimerize or heterodimerize with other TRIM proteins and has E3 ubiquitin ligase activity. This gene is also a tumor suppressor and is involved in secretory autophagy. [provided by RefSeq, Jan 2017]. | hsa:10626; | cytoplasm [GO:0005737]; cytosol [GO:0005829]; plasma membrane [GO:0005886]; PML body [GO:0016605]; DNA binding [GO:0003677]; interleukin-1 binding [GO:0019966]; NACHT domain binding [GO:0032089]; transferase activity [GO:0016740]; zinc ion binding [GO:0008270]; histone H3 acetylation [GO:0043966]; histone H4 acetylation [GO:0043967]; positive regulation of interleukin-1 beta production [GO:0032731]; positive regulation of keratinocyte differentiation [GO:0045618]; positive regulation of retinoic acid receptor signaling pathway [GO:0048386]; positive regulation of transcription, DNA-templated [GO:0045893]; response to growth hormone [GO:0060416]; response to organophosphorus [GO:0046683]; response to retinoic acid [GO:0032526] | 11919186_role in regulating keratinocyte differentiation 16575408_These results provide evidence for a role of EBBP in innate immunity by enhancing the alternative secretion pathway of IL-1beta. 16636064_EBBP increased betaRARE-transactivating function through its coiled-coil domain 19147277_The estrogen-responsive B box protein (EBBP) restores retinoid sensitivity in retinoid-resistant cancer cells via effects on histone acetylation. 20729920_TRIM16 acts as a tumour suppressor, affecting neuritic differentiation, cell migration and replication through interactions with cytoplasmic vimentin and nuclear E2F1 in neuroblastoma cells. 22629402_via its unique structure, TRIM16 possesses both heterodimerization function with other TRIM proteins and also has E3 ubiquitin ligase activity. 23404198_TRIM16 can promote apoptosis by directly modulating caspase-2 activity in cancer cells. 23422002_data suggest that TRIM16 acts as a novel regulator of both neuroblastoma G 1/S progression and cell differentiation 25333256_Chromatin immunoprecipitation assays revealed TRIM16 directly bound the IFNbeta1 gene promoter. Low level TRIM16 expression in 91 melanoma patient samples, strongly correlated with lymph node metastasis, and, predicted poor patient prognosis. 25843803_results suggest that TRIM16 is a potential pharmacologic target for the treatment of NSCLC and promotion TRIM16 expression might represent a novel strategy to NSCLC metastasis 25866896_Expression of SDMGC and TRIM16 was upregulated in the distant metastasis tissues 26718507_TRIM16 directly regulated the degradation of Gli1 protein via the ubiquitinproteasome pathway. TRIM16 expression was low in breast cancer. It negatively correlated with metastasis and suppressed stem-cell properties in breast cancer cells. 26892350_Results found that the expression of TRIM16 was significantly downregulated in hepatocellular carcinoma cell (HCC) lesions and define TRIM16 as an inhibitor of epithelial-mesenchymal transition and metastasis in HCC. 26902425_Data suggest that TRIM16 and TDP43 are both good prognosis indicators; data shows that TRIM16 inhibits cancer cell viability by a novel mechanism involving interaction and stabilisation of TDP43 with consequent effects on E2F1 and pRb proteins. 27693506_The cooperation between TRIM16 and Galectin-3 in targeting and activation of selective autophagy protects cells from lysosomal damage and Mycobacterium tuberculosis invasion. 27737724_TRIM16 inhibits the migration and invasion via suppressing the Sonic hedgehog signaling pathway in ovarian cancer cells. 27748839_TRIM16 expression is decreased in prostate cancer tissues and overexpression of TRIM16 inhibits cell migration, invasion and the EMT process in vitro in prostate cancer through the transcription factor Snail. 29295721_High TRIM16 expression is associated with drug resistance in Non-Small Cell Lung Cancer. 30143514_TRIM16 acts as a scaffold protein and, by interacting with p62, ULK1, ATG16L1, and LC3B, facilitates autophagic degradation of protein aggregates. 30488195_Papillary thyroid carcinoma was characterized by high expression of ESR2 and AR, which was associated with expression and content of nuclear factors Brn-3A and TRIM16. 31521826_Galectin-3 and TRIM16 coregulate osteogenic differentiation of human bone marrow-derived mesenchymal stem cells at least partly via enhancing autophagy. 31812473_The results indicate that the RASSF6-TRIM16 axis is a key effector in ESCC progression and that RASSF6 serves as a potential target for the treatment of ESCC. 33046716_AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent Vimentin degradation. 33391467_A thermosensitive, reactive oxygen species-responsive, MR409-encapsulated hydrogel ameliorates disc degeneration in rats by inhibiting the secretory autophagy pathway. 34135057_Impaired TRIM16-Mediated Lysophagy in Chronic Obstructive Pulmonary Disease Pathogenesis. 34265287_TRIM16 overexpression inhibits the metastasis of colorectal cancer through mediating Snail degradation. 34452557_Prognostic Significance of Tripartite Motif Containing 16 Expression in Patients with Gastric Cancer. 35230201_CircPTK2 inhibits cell cisplatin (CDDP) resistance by targeting miR-942/TRIM16 axis in non-small cell lung cancer (NSCLC). | ENSMUSG00000047821 | Trim16 | 536.72392 | 0.6976461 | -0.5194327565 | 0.14823823 | 1.207621e+01 | 5.106926e-04 | 3.418772e-02 | No | Yes | 517.088149 | 51.473980 | 713.775374 | 68.966616 | |
| ENSG00000221994 | 57232 | ZNF630 | protein_coding | Q2M218 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | This gene encodes a protein containing an N-terminal Kruppel-associated box-containing (KRAB) domain and 13 Kruppel-type C2H2 zinc finger domains. This gene resides on an area of chromosome X that has been implicated in nonsyndromic X-linked cognitive disability. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2013]. | hsa:57232; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; metal ion binding [GO:0046872]; RNA polymerase II cis-regulatory region sequence-specific DNA binding [GO:0000978]; regulation of transcription by RNA polymerase II [GO:0006357] | 20186789_Detected 12 ZNF630 deletions in a total of 1,562 male patients with mental retardation from Brazil, USA, Australia, and Europe. The breakpoints were analyzed in 10 families, and in all cases they were located within two segmental duplications. 20186789_Observational study of gene-disease association. (HuGE Navigator) | 16.80690 | 0.9133202 | -0.1308073719 | 0.68248182 | 3.658701e-02 | 8.483083e-01 | No | Yes | 17.088546 | 7.166997 | 18.032279 | 7.416988 | ||||
| ENSG00000223546 | LINC00630 | lncRNA | 167.05328 | 0.9970884 | -0.0042066418 | 0.24198397 | 3.023623e-04 | 9.861266e-01 | 9.968746e-01 | No | Yes | 188.236038 | 42.620126 | 186.170024 | 41.202970 | |||||||||||
| ENSG00000223959 | 172 | AFG3L1P | transcribed_unitary_pseudogene | 11549317_AFG3L1 is a mitochondrial ATP-dependent zinc metalloprotease transcribed into four mRNA isoforms that apparently are not translated in humans. 21555518_AFG3L1 is a functional target gene of the BACH1 transcription factor according to ChIP-seq and knockdown analysis in HEK 293 cells. | 826.79701 | 1.2824867 | 0.3589439144 | 0.11529724 | 9.758918e+00 | 1.784550e-03 | 6.998679e-02 | No | Yes | 991.104254 | 139.865992 | 782.498320 | 108.058942 | |||||||||
| ENSG00000224687 | 100302401 | RASAL2-AS1 | lncRNA | 22.95527 | 2.0487278 | 1.0347282938 | 0.64404932 | 2.609108e+00 | 1.062515e-01 | No | Yes | 41.813721 | 13.209744 | 18.730760 | 5.947276 | |||||||||||
| ENSG00000224877 | 284184 | NDUFAF8 | protein_coding | A1L188 | FUNCTION: Involved in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I, MT-ND1) (PubMed:27499296). Required to stabilize NDUFAF5 (PubMed:27499296). {ECO:0000269|PubMed:27499296}. | Disease variant;Disulfide bond;Mitochondrion;Primary mitochondrial disease;Reference proteome | hsa:284184; | mitochondrion [GO:0005739]; mitochondrial respiratory chain complex I assembly [GO:0032981] | 31866046_Pathogenic Bi-allelic Mutations in NDUFAF8 Cause Leigh Syndrome with an Isolated Complex I Deficiency. | ENSMUSG00000078572 | Ndufaf8 | 862.74914 | 0.9632535 | -0.0540125417 | 0.12042231 | 2.010675e-01 | 6.538605e-01 | 8.913628e-01 | No | Yes | 1166.018086 | 153.077202 | 1137.079795 | 145.754192 | ||
| ENSG00000225361 | 100506599 | PPP1R26-AS1 | lncRNA | 84.97478 | 1.2904851 | 0.3679134950 | 0.31053355 | 1.411553e+00 | 2.347987e-01 | 6.260599e-01 | No | Yes | 126.438701 | 28.156258 | 101.269205 | 21.911534 | ||||||||||
| ENSG00000225791 | 401264 | TRAM2-AS1 | lncRNA | 133.50801 | 0.9439218 | -0.0832607744 | 0.26016942 | 1.020065e-01 | 7.494349e-01 | 9.265683e-01 | No | Yes | 146.898514 | 30.722949 | 151.696362 | 30.962895 | ||||||||||
| ENSG00000226287 | 84222 | TMEM191A | transcribed_unprocessed_pseudogene | 51.58032 | 0.7872516 | -0.3451033184 | 0.39235157 | 7.667715e-01 | 3.812177e-01 | No | Yes | 58.218299 | 13.019933 | 70.056505 | 15.112584 | |||||||||||
| ENSG00000226383 | 101929378 | LINC01876 | lncRNA | 29.98400 | 0.9801654 | -0.0289028873 | 0.51785007 | 3.110050e-03 | 9.555268e-01 | No | Yes | 34.172084 | 15.378973 | 35.147717 | 15.440604 | |||||||||||
| ENSG00000226824 | 100996437 | lncRNA | 42.52537 | 1.1394006 | 0.1882750247 | 0.47811621 | 1.547005e-01 | 6.940833e-01 | No | Yes | 44.564216 | 8.689693 | 36.713290 | 6.855197 | ||||||||||||
| ENSG00000227540 | 414245 | lncRNA | 73.79425 | 0.7139528 | -0.4860993698 | 0.34416773 | 1.972208e+00 | 1.602137e-01 | No | Yes | 67.140524 | 16.400036 | 92.291040 | 21.716301 | ||||||||||||
| ENSG00000228109 | 100507057 | MELTF-AS1 | lncRNA | 27513470_results indicated lncRNA MFI2 could promote proliferation and migration of osteosarcoma cells by regulating FOXP4 expression, which suggested critical roles of lncRNA MFI2 and FOXP4 in occurrence and development of human osteosarcoma 28819235_Long non-coding RNA MFI2-AS1 expression was associated with dramatically increased risk of relapse compared to patients with undetectable MFI2-AS1 who had favorable outcomes. 31210294_Up-regulation of long non-coding RNA MFI2 functions as an oncogenic role in cervical cancer progression. 31298339_MFI2-AS1 regulates the aggressive phenotypes in glioma by modulating MMP14 via a positive feedback loop. 31678554_Knockdown of MFI2-AS1 increased cell viability but suppressed apoptosis, inflammatory response and extracellular matrix degradation in LPS-treated chondrocytes by increasing miR-130a-3p and decreasing TCF4, indicating a novel target for the treatment of osteoarthritis 32106369_LncRNA MFI2-AS1 promotes HCC progression and metastasis by acting as a competing endogenous RNA of miR-134 to upregulate FOXM1 expression. 35441579_Long non-coding MELTF Antisense RNA 1 promotes and prognosis the progression of non-small cell lung cancer by targeting miR-1299. | 22.91671 | 0.4134582 | -1.2741866002 | 0.58942007 | 4.494185e+00 | 3.401032e-02 | No | Yes | 13.126049 | 3.648441 | 32.157069 | 8.515643 | ||||||||||
| ENSG00000228274 | lncRNA | 72.27294 | 1.2798099 | 0.3559295482 | 0.33703848 | 1.122115e+00 | 2.894634e-01 | No | Yes | 80.726840 | 11.316508 | 62.055688 | 8.568696 | |||||||||||||
| ENSG00000228393 | LINC01004 | lncRNA | 41.77257 | 0.9519507 | -0.0710412430 | 0.45803513 | 2.398496e-02 | 8.769232e-01 | No | Yes | 45.016174 | 12.637405 | 48.412917 | 13.305449 | ||||||||||||
| ENSG00000228701 | 100507633 | TNKS2-DT | lncRNA | 10.39634 | 0.7774335 | -0.3632087805 | 0.88870420 | 1.641480e-01 | 6.853660e-01 | No | Yes | 10.779772 | 3.980260 | 12.151768 | 4.215727 | |||||||||||
| ENSG00000229809 | 146542 | ZNF688 | protein_coding | P0C7X2 | FUNCTION: May be involved in transcriptional regulation. | Alternative splicing;DNA-binding;Metal-binding;Nucleus;Reference proteome;Repeat;Transcription;Transcription regulation;Zinc;Zinc-finger | hsa:146542; | nucleus [GO:0005634]; DNA-binding transcription factor activity, RNA polymerase II-specific [GO:0000981]; DNA-binding transcription repressor activity, RNA polymerase II-specific [GO:0001227]; metal ion binding [GO:0046872]; RNA polymerase II transcription regulatory region sequence-specific DNA binding [GO:0000977]; negative regulation of transcription by RNA polymerase II [GO:0000122]; regulation of transcription by RNA polymerase II [GO:0006357] | Mouse_homologues 17971504_Zfp688 is most abundantly expressed in tissues rich in highly ciliated cells, such as olfactory sensory neurons, and is predicted to be important to cilia. | ENSMUSG00000045251 | Zfp688 | 112.08070 | 0.6780515 | -0.5605332787 | 0.27451315 | 4.113736e+00 | 4.253630e-02 | 3.148058e-01 | No | Yes | 103.262257 | 15.609275 | 145.739101 | 21.029322 | ||
| ENSG00000232677 | 100506930 | LINC00665 | lncRNA | 29728556_LINC00665 may be involved in the regulation of cell cycle pathways in hepatocellular carcinoma through ten identified hub genes. 30692511_Transcription factor SP1 induced the transcription of linc00665 in LUAD cells, which exerted its oncogenic role by functioning as competing endogenous RNA (ceRNA) for miR-98 and subsequently activating downstream AKR1B10-ERK signaling pathway. 31736127_LINC00665 induces gastric cancer progression through activating Wnt signaling pathway. 31907362_LINC00665 promotes breast cancer progression through regulation of the miR-379-5p/LIN28B axis. 32083756_Inflammation-Induced Long Intergenic Noncoding RNA (LINC00665) Increases Malignancy Through Activating the Double-Stranded RNA-Activated Protein Kinase/Nuclear Factor Kappa B Pathway in Hepatocellular Carcinoma. 32271427_Long non-coding RNA LINC00665 promotes metastasis of breast cancer cells by triggering EMT. 32403047_Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. 32423800_Long non-coding RNA linc00665 interacts with YB-1 and promotes angiogenesis in lung adenocarcinoma. 32776307_LINC00665/miR-9-5p/ATF1 is a novel axis involved in the progression of colorectal cancer. 33090388_LINC00665 facilitates the progression of osteosarcoma via sponging miR-3619-5p. 33650673_LINC00665 functions as a competitive endogenous RNA to regulate AGTR1 expression by sponging miR34a5p in glioma. 33658535_LINC00665 promotes the progression of acute myeloid leukemia by regulating the miR-4458/DOCK1 pathway. 33865827_LINC00665 activates Wnt/beta-catenin signaling pathway to facilitate tumor progression of colorectal cancer via upregulating CTNNB1. 33903885_LINC00665 promotes HeLa cell proliferation, migration, invasion and epithelial-mesenchymal transition by activating the WNT-CTNNB1/betacatenin signaling pathway. 34171521_LINC00665 promotes the viability, migration and invasion of T cell acute lymphoblastic leukemia cells by targeting miR-101 via modulating PI3K/Akt pathway. 34232917_Downregulation of LINC00665 suppresses the progression of lung adenocarcinoma via regulating miR-181c-5p/ZIC2 axis. 34306997_LINC00665 Facilitates the Malignant Processes of Osteosarcoma by Increasing the RAP1B Expression via Sponging miR-708 and miR-142-5p. | 176.95571 | 0.8491066 | -0.2359823906 | 0.22293684 | 1.112619e+00 | 2.915134e-01 | 6.766018e-01 | No | Yes | 165.240189 | 14.718962 | 187.442070 | 16.127129 | |||||||||
| ENSG00000232956 | 285958 | SNHG15 | lncRNA | This gene represents a snoRNA host gene that produces a short-lived long non-coding RNA. This non-coding RNA is upregulated in tumor cells and may contribute to cell proliferation by acting as a sponge for microRNAs. [provided by RefSeq, Dec 2017]. | 26662309_Our findings present that elevated lncRNA SNHG15 could be identified as a poor prognostic biomarker in GC and regulate cell invasion. 29048682_SNHG15 and miR-153 are new potential therapeutic targets for anti-angiogenesis treatment of glioma 29217194_Long noncoding RNA SNHG15 was highly expressed in breast cancer tissues and cell lines. SNHG15 knockdown significantly inhibited the proliferation and induced apoptosis in breast cancer cells in vitro and in vivo. In mechanism, we found that SNHG15 acted as a competing endogenous RNA to sponge miR-211-3p, which was downregulated in breast cancers and inhibited cell proliferation and migration. 29604394_Results show that enhanced expression of lncRNA SNHG15 promoted colon cancer cell progression and was correlated with poor prognosis in colon cancer patients. Mechanistically, SNHG15 maintains Slug stability in living cells by impeding its ubiquitination and degradation through interaction with the zinc finger domain of Slug. 29630731_the ectopic overexpression of SNHG15 contribute to the non-small cell lung cancer tumorigenesis by regulating CDK14 protein via sponging miR-486. 29750422_the present study suggested that SNHG15 may be involved in the nuclear factorkappaB signaling pathway, induce the epithelialmesenchymal transition process, and promote renal cell carcinoma invasion and migration. 29771418_The expression of SNHG15 is up-regulated on non-small lung cancer cells, promoting promotes cell proliferation and invasion. 30280769_SNHG15 levels were significantly up-regulated in both sera and tumors tissues from pancreatic ductal adenocarcinoma (PDAC) patients. Clinicopathologic analysis revealed that high SNHG15 expression was associated with tumor differentiation, lymph node metastasis, tumor stage and shorter overall survival. Cox multivariate analyses confirmed that SNHG15 expression was an independent prognostic factor in PDAC. 30317592_Lnc-SNHG15 overexpression was significantly associated with colorectal liver metastasis and poor survival. 30402848_SNHG15 is highly expressed in lung cancer (LC) tissues, which promotes the occurrence and progression of LC via regulating proliferation and migration of LC cells by targeting microRNA-211-3p. 30840276_Overexpression of lncSNHG15 remarkably promoted the proliferation and migration of non-small cell lung cancer cells. lncSNHG15 could bind to miR-211-3p. 30945457_SNHG15 served as a ceRNA and reversed the activity of miR-338-3p, thus facilitating CRC tumorigenesis by enhancing the expression of FOS and RAB14. 30981837_Results show that SNHG15 is upregulated in prostate cancer (PCa) cells and promoted cell proliferation. Its downregulation inhibited PCa cell migration and invasion. SNHG15 was located in the cytoplasm of PCa cells and acted as a molecular sponge of miR-338 resulting in post-transcriptional regulated FKBP1A. These study suggested that SNHG15 functions as an oncogene by modulating miR-338/FKBP1A axis in prostate cancer. 31014355_Several of these genes are functionally related to AIF, a protein that we found to specifically interact with SNHG15, suggesting that the SNHG15 acts, at least in part, by regulating the activity of AIF. 31310393_LncRNA SNHG15 promotes hepatocellular carcinoma progression by sponging miR-141-3p. 31636472_Up-regulation of SNHG15 was found in hepatocellular carcinoma (HCC) and was related to aggressive behaviors in HCC patients. Knockdown of SNHG15 restrained HCC cell proliferation, migration and invasion. SNHG15 served as a molecular sponge for miR-490-3p which directly targets HDAC2. HDAC2 was involved in HCC progression by interacting with the SNHG15/miR-490-3p axis. 31696491_Elevated expression of lncRNA SNHG15 in spinal tuberculosis: preliminary results. 31774607_LncRNA SNHG15 may serve as a prospective and novel biomarker for molecular diagnosis and therapeutics in patients with cancer. 32141559_Up-regulation of SNHG15 facilitates cell proliferation, migration, invasion and suppresses cell apoptosis in breast cancer by regulating miR-411-5p/VASP axis. 32247266_Long non-coding RNA SNHG15 is a competing endogenous RNA of miR-141-3p that prevents osteoarthritis progression by upregulating BCL2L13 expression. 32633324_High lncSNHG15 expression may predict poor cancer prognosis: a meta-analysis based on the PRISMA and the bio-informatics analysis. 32633365_LncRNA SNHG15 promotes the proliferation of nasopharyngeal carcinoma via sponging miR-141-3p to upregulate KLF9. 32643707_Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. 32655137_LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. 33372376_Long noncoding RNA small nucleolar RNA host gene 15 deteriorates liver cancer via microRNA-18b-5p/LIM-only 4 axis. 33572758_The Expression of the Cancer-Associated lncRNA Snhg15 Is Modulated by EphrinA5-Induced Signaling. 33899079_LncRNA SNHG15 modulates gastric cancer tumorigenesis by impairing miR-506-5p expression. 34016097_Down-regulation LncRNA-SNHG15 contributes to proliferation and invasion of bladder cancer cells. 34551140_Increased serum exosomal long non-coding RNA SNHG15 expression predicts poor prognosis in non-small cell lung cancer. 34734088_lncRNA SNHG15 Promotes Ovarian Cancer Progression through Regulated CDK6 via Sponging miR-370-3p. 34868528_LncRNA SNHG15 Promotes Oxidative Stress Damage to Regulate the Occurrence and Development of Cerebral Ischemia/Reperfusion Injury by Targeting the miR-141/SIRT1 Axis. | 2514.75902 | 1.0110716 | 0.0158851350 | 0.08347716 | 3.623750e-02 | 8.490259e-01 | 9.595722e-01 | No | Yes | 2669.276578 | 240.818084 | 2552.470542 | 225.154099 | ||||||||
| ENSG00000234062 | TM9SF5P | transcribed_unprocessed_pseudogene | 39.81915 | 1.2168681 | 0.2831728512 | 0.45063589 | 3.967103e-01 | 5.287931e-01 | No | Yes | 50.777291 | 13.995168 | 42.814833 | 11.421284 | ||||||||||||
| ENSG00000235501 | CNN3-DT | lncRNA | 33.03914 | 0.9342962 | -0.0980480938 | 0.48989858 | 3.991684e-02 | 8.416433e-01 | No | Yes | 38.539683 | 7.203132 | 40.807778 | 7.259116 | ||||||||||||
| ENSG00000236088 | 100874058 | COX10-DT | lncRNA | 34323174_Regulating COX10-AS1 / miR-142-5p / PAICS axis inhibits the proliferation of non-small cell lung cancer. | 425.35645 | 0.9416304 | -0.0867672135 | 0.15757670 | 3.027824e-01 | 5.821434e-01 | 8.599506e-01 | No | Yes | 418.322502 | 71.214711 | 430.984629 | 71.842321 | |||||||||
| ENSG00000237651 | 339804 | C2orf74 | protein_coding | A8MZ97 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | hsa:339804; | integral component of membrane [GO:0016021] | 33571247_Frameshift variant in MITF gene in a large family with Waardenburg syndrome type II and a co-segregation of a C2orf74 variant. | ENSMUSG00000020286 | 1700093K21Rik | 211.54391 | 0.6636582 | -0.5914875851 | 0.20398334 | 8.292301e+00 | 3.981351e-03 | 1.052725e-01 | No | Yes | 199.791073 | 28.114174 | 294.523056 | 39.680352 | |||
| ENSG00000238009 | lncRNA | 26.08333 | 0.8784020 | -0.1870468026 | 0.56697013 | 1.073498e-01 | 7.431817e-01 | No | Yes | 29.176967 | 12.535310 | 34.664713 | 14.356173 | |||||||||||||
| ENSG00000239523 | 100506826 | MYLK-AS1 | lncRNA | 33168027_LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. 34181498_LncRNA MYLK-AS1 acts as an oncogene by epigenetically silencing large tumor suppressor 2 (LATS2) in gastric cancer. | 55.01519 | 0.6706933 | -0.5762749588 | 0.38055684 | 2.259692e+00 | 1.327804e-01 | No | Yes | 48.109179 | 7.895752 | 69.727831 | 10.722401 | ||||||||||
| ENSG00000240184 | 5098 | PCDHGC3 | protein_coding | Q9UN70 | FUNCTION: Potential calcium-dependent cell-adhesion protein. May be involved in the establishment and maintenance of specific neuronal connections in the brain. | Alternative splicing;Calcium;Cell adhesion;Cell membrane;Glycoprotein;Membrane;Reference proteome;Repeat;Signal;Transmembrane;Transmembrane helix | This gene is a member of the protocadherin gamma gene cluster, one of three related clusters tandemly linked on chromosome five. These gene clusters have an immunoglobulin-like organization, suggesting that a novel mechanism may be involved in their regulation and expression. The gamma gene cluster includes 22 genes divided into 3 subfamilies. Subfamily A contains 12 genes, subfamily B contains 7 genes and 2 pseudogenes, and the more distantly related subfamily C contains 3 genes. The tandem array of 22 large, variable region exons are followed by a constant region, containing 3 exons shared by all genes in the cluster. Each variable region exon encodes the extracellular region, which includes 6 cadherin ectodomains and a transmembrane region. The constant region exons encode the common cytoplasmic region. These neural cadherin-like cell adhesion proteins most likely play a critical role in the establishment and function of specific cell-cell connections in the brain. Alternative splicing has been described for the gamma cluster genes. [provided by RefSeq, Jul 2008]. | hsa:5098; | integral component of plasma membrane [GO:0005887]; membrane [GO:0016020]; calcium ion binding [GO:0005509]; calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules [GO:0016339]; cell adhesion [GO:0007155]; homophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156]; negative regulation of neuron apoptotic process [GO:0043524]; synapse organization [GO:0050808] | 16751190_ADAM10 is the protease responsible for constitutive and regulated Pcdh gamma shedding events that modulate the cell adhesion role of Pcdh gamma. 22249255_our data suggest that the PCDH LRES is an important tumour suppressor locus in colorectal cancer, and that PCDHGC3 may be a strong marker and driver for the adenoma-carcinoma transition. 31216007_Our data provide a map of the DNA methylome episignature specific to an SDHB-mutated cancer and establish PCDHGC3 as a putative suppressor gene | ENSMUSG00000102918 | Pcdhgc3 | 96.69756 | 1.0694450 | 0.0968622607 | 0.29810818 | 1.053814e-01 | 7.454649e-01 | 9.257705e-01 | No | Yes | 111.342769 | 37.570566 | 106.626415 | 34.864754 | |
| ENSG00000240694 | 10687 | PNMA2 | protein_coding | Q9UL42 | Acetylation;Direct protein sequencing;Nucleus;Reference proteome;Tumor antigen | hsa:10687; | nucleolus [GO:0005730]; positive regulation of apoptotic process [GO:0043065] | 21209860_Ma2 may have a role in early recurrence of small intestine neuroendocrine tumors 21729305_Immunohistochemical analysis of MA2/D2-40 combined with p16(INK4A) may have a significant implication in clinical practice for better identifying the grade of cervical intraepithelial neoplasia. 27003254_PNMA2 functions as antagonist of MOAP-1 and PNMA1 through heterodimeric interaction | ENSMUSG00000046204 | Pnma2 | 650.18318 | 0.9005230 | -0.1511650086 | 0.12544015 | 1.447002e+00 | 2.290097e-01 | 6.193552e-01 | No | Yes | 743.636853 | 115.965365 | 806.608082 | 122.814080 | |||
| ENSG00000241357 | lncRNA | 18.77016 | 0.6992869 | -0.5160435667 | 0.67079393 | 5.868695e-01 | 4.436322e-01 | No | Yes | 12.708390 | 4.025115 | 17.260429 | 5.222200 | |||||||||||||
| ENSG00000241769 | 100131434 | EOLA1-DT | lncRNA | 21.32114 | 0.6258445 | -0.6761238592 | 0.65154867 | 1.060892e+00 | 3.030118e-01 | No | Yes | 16.150726 | 7.254185 | 28.424488 | 12.132335 | |||||||||||
| ENSG00000242288 | 113939925 | BMS1P4-AGAP5 | lncRNA | This locus represents naturally occurring readthrough transcription between the neighboring BMS1P4 (BMS1, ribosome biogenesis factor pseudogene 4) and AGAP5 (ArfGAP with GTPase domain, ankyrin repeat and PH domain 5) genes on chromosome 10. The readthrough transcript is a candidate for nonsense-mediated mRNA decay (NMD), and is unlikely to produce a protein product. [provided by RefSeq, Jan 2019]. | 146.29216 | 1.1921752 | 0.2535962613 | 0.25630291 | 9.828499e-01 | 3.214962e-01 | 7.005791e-01 | No | Yes | 173.861247 | 50.410063 | 150.542651 | 42.842287 | |||||||||
| ENSG00000242588 | lncRNA | 139.41646 | 0.9636960 | -0.0533500520 | 0.27301819 | 3.811959e-02 | 8.452032e-01 | 9.584578e-01 | No | Yes | 137.873853 | 41.848038 | 150.610501 | 44.658710 | ||||||||||||
| ENSG00000242802 | 9907 | AP5Z1 | protein_coding | O43299 | FUNCTION: As part of AP-5, a probable fifth adaptor protein complex it may be involved in endosomal transport. According to PubMed:20613862 it is a putative helicase required for efficient homologous recombination DNA double-strand break repair. {ECO:0000269|PubMed:20613862, ECO:0000269|PubMed:22022230}. | Alternative splicing;Cytoplasm;DNA damage;DNA repair;Hereditary spastic paraplegia;Neurodegeneration;Nucleus;Protein transport;Reference proteome;Transport | This gene was identified by genome-wide screen for genes involved in homologous recombination DNA double-strand break repair (HR-DSBR). The encoded protein was found in a complex with other proteins that have a role in HR-DSBR. Knockdown of this gene reduced homologous recombination, and mutations in this gene were found in patients with spastic paraplegia. It was concluded that this gene likely encodes a helicase (PMID:20613862). [provided by RefSeq, Jan 2011]. | hsa:9907; | AP-5 adaptor complex [GO:0044599]; AP-type membrane coat adaptor complex [GO:0030119]; cytoplasm [GO:0005737]; nuclear speck [GO:0016607]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; double-strand break repair via homologous recombination [GO:0000724]; endosomal transport [GO:0016197]; protein transport [GO:0015031] | 29726929_Here, we have generated induced pluripotent stem cells (iPSCs) from patients with two autosomal recessive forms of hereditary spastic paraplegia (HSP) , SPG15 and SPG48, which are caused by mutations in the ZFYVE26 and AP5Z1 genes encoding proteins in the same complex, the spastizin and AP5Z1 proteins, respectively. | ENSMUSG00000039623 | Ap5z1 | 864.65269 | 1.0538482 | 0.0756670551 | 0.12913588 | 3.442791e-01 | 5.573696e-01 | 8.505383e-01 | No | Yes | 1101.122944 | 187.963374 | 1064.243704 | 177.464071 | |
| ENSG00000243176 | lncRNA | 29.68125 | 0.9414585 | -0.0870305618 | 0.53764152 | 2.599496e-02 | 8.719126e-01 | No | Yes | 36.007370 | 12.037152 | 34.162415 | 11.002393 | |||||||||||||
| ENSG00000243701 | 344595 | DUBR | lncRNA | 33758355_DUBR suppresses migration and invasion of human lung adenocarcinoma cells via ZBTB11-mediated inhibition of oxidative phosphorylation. | 206.22865 | 0.6053785 | -0.7240906907 | 0.23884354 | 8.943619e+00 | 2.784406e-03 | 8.988961e-02 | No | Yes | 156.632066 | 34.554032 | 256.947739 | 54.995544 | |||||||||
| ENSG00000245146 | MALINC1 | lncRNA | 133.34590 | 0.9234292 | -0.1149267517 | 0.28063110 | 1.668727e-01 | 6.829062e-01 | 9.021767e-01 | No | Yes | 83.948140 | 25.836935 | 91.535261 | 27.545933 | |||||||||||
| ENSG00000247595 | 100506540 | SPTY2D1OS | protein_coding | A0A0U1RRN3 | Membrane;Reference proteome;Transmembrane;Transmembrane helix | integral component of membrane [GO:0016021] | 25.53508 | 0.9691489 | -0.0452098152 | 0.55945289 | 6.528072e-03 | 9.356038e-01 | No | Yes | 29.282607 | 6.445157 | 28.859536 | 6.229122 | ||||||||
| ENSG00000248498 | 389652 | ASNSP1 | transcribed_unprocessed_pseudogene | 61.97198 | 0.9503316 | -0.0734971596 | 0.35995262 | 4.166473e-02 | 8.382602e-01 | No | Yes | 72.517910 | 13.593506 | 71.631812 | 13.183552 | |||||||||||
| ENSG00000249859 | 5820 | PVT1 | lncRNA | This gene represents a long non-coding RNA locus that has been identified as a candidate oncogene. Increased copy number and overexpression of this gene are associated with many types of cancers including breast and ovarian cancers, acute myeloid leukemia and Hodgkin lymphoma. Allelic variants of this gene are also associated with end-stage renal disease attributed to type 1 diabetes. Consistent with its association with various types of cancer, transcription of this gene is regulated by the tumor suppressor p53 through a canonical p53-binding site, and it has been implicated in regulating levels of the proto-oncogene MYC to promote tumorigenesis. [provided by RefSeq, Sep 2015]. | 17395743_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 17395743_PVT1 may be a candidate gene which contribute to end stage renal disease susceptibility in diabetes. 17503467_Data indicate that PVT-1 expression is restricted to a low number of normal tissues compared to c-Myc mRNA, whereas the gene is highly expressed in many transformed cell types including neuroblastoma cells that do not express c-Myc. 17881614_Observational study of gene-disease association. (HuGE Navigator) 17881614_Role for PVT1 in mediating susceptibility to end-stage renal disease attributable to diabetes. 17908964_MYC and PVT1 contribute independently to ovarian and breast pathogenesis when overexpressed because of genomic abnormalities 19549893_Observational study of gene-disease association. (HuGE Navigator) 21037568_Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 21316338_these results identified PVT1 as a regulator of Gemcitabine sensitivity in pancreatic cancer cells. 21526116_that PVT1 may mediate the development and progression of diabetic nephropathy through mechanisms involving extracellular matrix accumulation. 21814516_we identified a new functional prostate cancer risk variant at the 8q24 locus that is associated with increased expression of PVT1 located 0.5 Mb downstream. 22110125_p53-Dependent induction of PVT1 and miR-1204. 22869583_Data indicate that the PVT1-NBEA and the PVT1-WWOX chimeric genes were associated with the expression of abnormal NBEA and WWOX. 23547836_Deletion or methylation of CDKN2A/2B and PVT1 rearrangement occur frequently in highly aggressive B-cell lymphomas harboring 8q24 abnormality. 24196785_PVT-1, which maps to 8q24, generates antiapoptotic activity in CRC, and abnormal expression of PVT-1 was a prognostic indicator for CRC patients. 24204837_miR-1207-5p, a PVT1-derived microRNA, is abundantly expressed in kidney cells, and is upregulated by glucose and TGF-beta1. 24780616_the GG genotype of SNP rs13281615 plays a role in breast cancer likely by influencing PVT1 expression, and that during oncogenesis, 'protective' mutations could occur. 24821155_Report frequency of PVT1 SNPs in diabetic nephropathy. 24926545_Our in vitro findings suggest that C-MYC and PVT1 copy number gains may promote a malignant malignant pleural mesothelioma phenotype. 25043044_Gain of PVT1 long non-coding RNA expression was required for high MYC protein levels in 8q24-amplified human cancer cells 25043274_Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. 25245984_our study identified novel PVT1-NSMCE2 and CCDC26-NSMCE2 fusion genes that may play functional roles in leukemia. 25400777_lncRNA PVT1 is significantly upregulated in non-small cell lung cancer. 25883951_PVT1 has a role in competing endogenous RNA activity and regulation of protein stability of important oncogenes, primarily of the MYC oncogene [review] 25890171_LncRNA PVT1 may serve as a candidate prognostic biomarker. 25956062_PVT-1 was highly expressed in gastric cancer tissues of cisplatin-resistant patients and cisplatin-resistant cells. 26097562_we have identified a potential mechanism by which PVT1 regulated by carboplatin plus docetaxel contributes to the carboplatin-docetaxel-induced anticancer action in ovarian cancer. 26427660_lncRNA PVT1 may contribute to tumorigenesis of thyroid cancer through recruiting EZH2 and regulating TSHR expression. 26490983_PVT1 knockdown increased the number of lung cancer cells in the G0/G1 phase and reduced the number of cells in the S phase, while overexpression of PVT1 could promote cell cycle progression. 26517688_Data show that long non-coding RNA PVT1 was upregulated in bladder cancer tissues and cell lines. 26545364_Findings sugggest that the long non-coding RNA (lncRNA) PVT1 may play an important role in the proliferation of acute promyelocytic leukemia (APL) cells and may be useful for future therapeutic management. 26850852_Studies on the association between PVT1 and cancer have shown that PVT1 is a potential oncogene in a variety of cancer types. [review] 26908628_we found that PVT1 functioned as an oncogene and may be a crucial prognostic factor for NSCLC patients. PVT1 contributed to lung adenocarcinoma cell proliferation, partly though EZH2-medicated suppression of the LATS2/MDM2/P53 pathway. 26925791_PVT1 expression was remarkably increased in gastric cancer tissues and cell lines compared with that in the normal control, and its up-regulation was significantly correlated to invasion depth, advanced TNM stage and regional lymph nodes metastasis in gastric cancer. 27028998_our findings indicate that salivary HOTAIR and PVT1 show potential as novel non-invasive biomarkers for detecting pancreatic cancer 27232880_PVT1 expression is significantly increased in ICC tissue versus normal cervix and that higher expression of PVT1 correlates with poorer overall survival 27272214_PVT1 binds to EZH2, recruits EZH2 to the miR-200b promoter, increases histone H3K27 trimethylation level on the miR-200b promoter, and inhibits miR-200b expression. 27366943_Study discloses the role of PVT1 as a novel prognostic factor and as a molecular target for novel therapeutic interventions in renal carcinoma. 27484035_PVT1 regulates both IL-6 release and proliferation in airway smooth muscle cells from patients with severe asthma. 27588491_PVT1 epigenetically down-regulates PTCH1 expression via competitively binding miR-152, contributing to EMT process in liver fibrosis 27756785_Our study suggests that PVT1 promotes tumor progression by interacting with FOXM1 27794184_Our study suggested a regulatory relationship between lncRNA PVT1 and miR-146a during the process of the prostate cancer tumorigenesis. PVT1 regulated prostate cancer cell viability and apoptosis depending on miR-146a. 27813492_lncRNA PVT1 is overexpressed in osteosarcoma and decreased the survival rate of osteosarcoma patients. Silencing PVT1 by siRNA inhibited proliferation, migration and invasion and promoted apoptosis and cell cycle arrest in osteosarcoma cells via miR-195. 27928058_Identification of CircPVT1, generated by circularization of an exon of the PVT1 gene, as a circular RNA with a function in cell senescence. 27986464_Findings suggest that circPVT1 is a novel proliferative factor and prognostic marker in GC. 28235236_PVT1-derived miR-1207-5p promotes the proliferation of breast cancer cells by targeting STAT6. 28258379_Long noncoding RNA PVT1 in gastric cancer might act as a ''sponge'' to inhibit miR-152, and increase the expression of CD151 and FGF2. 28265576_results indicate that PVT1 functions as an oncogene in melanoma and could be a potential diagnostic biomarker and therapeutic target for melanoma. 28276314_Results reveal a tumor-promoting role for PVT1, acting as a competing endogenous RNA (ceRNA) or a molecular sponge in negatively modulating miR-424. 28296507_infer that PVT1 could decrease miR-195 expression via enhancing histone H3K27me3 in the miR-195 promoter region and also via direct sponging of miR-195 28351322_PVT1 overexpression increased the expression of Atg7 and Beclin1 by targeting miR-186, which induced protective autophagy, thus promoting glioma vascular endothelial cell proliferation, migration, and angiogenesis. Therefore, PVT1 and miR-186 can provide new therapeutic targets for future anti-angiogenic treatment of glioma 28381186_PVT1 may be a new oncogene co-amplified with c-Myc in colorectal cancer tissues and extracellular vesicles and functionally correlated with the proliferation and apoptosis of colorectal cancer cells. 28404954_Results indicate that PVT1 is up-regulated in esophageal squamous cell carcinoma (ESCC) and predict overall prognosis. Knockdown of PVT1 inhibits proliferation and migration of ESCC in vitro and suppressed tumor growth in vivo. Meanwhile, PVT1 acts as the molecular sponge to regulate expression of miR-203 and LASP1. These data provided evidence that PVT1 promoted ESCC progression via activation of the miR-203/LASP1 axis. 28409552_our study demonstrates that PVT1 modulates melanoma tumorigenesis by acting as an endogenous sponge of miR-26b 28520497_LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. 28534994_PVT1 played a pivotal part on the regulation of p21 expression in breast cancer cell lines. 28602700_findings suggested that PVT1 contributes to osteosarcoma (OS) cell glucose metabolism, cell proliferation, and motility through the miR-497/HK2 pathway, and revealed a novel relation between lncRNA and the alteration of glycolysis in OS cells 28656879_it was identified that plasmacytoma variant translocation 1 regulated the expression of the miR-186-5p target gene, yes-associated protein 1. Taken together, plasmacytoma variant translocation 1 served as an endogenous sponge for miR-186-5p to reduce its inhibiting effect on yes-associated protein 1 and thus promoted the tumorigenesis of hepatocellular carcinoma 28657147_we revealed that PVT1 functions as an endogenous 'sponge' by competing for miR-448 binding to regulate the miRNA target SERBP1 and, therefore, promotes the proliferation and migration of PC cells. 28731781_LncRNA-PVT1 is a novel critical regulator in non-small cell lung cancer invasion.LncRNA-PVT1 regulates MMP9 expression. 28800314_PVT1 has a higher expression levels in HCC tissues than that in the adjacent normal tissues, and functions as an oncogene by promoting HCC cell proliferation, invasion. Furthermore, we found that PVT1 interacted with EZH2 repressed miR-214 expression in HCC cells 28848163_PVT1 directly interacted with miR-195 and regulated its expression. PVT1 knockdown enhanced the sensitivity of NSCLC cells to ionizing radiation by suppressing cell survival and inducing cell apoptosis. 28866116_Elevated PVT1 expression was related to poor prognosis 28882595_These data suggest that elevated expression of SOX2 can activate lncRNA PVT1 expression promoted breast cancer tumorigenesis and progression. PVT1 may be a prognostic predictive biomarker for breast cancer, and the interaction of PVT1-SOX2 could be a therapeutic target in breast cancer. 28972861_High PVT1 expression is associated with Lung Cancer Progression. 29035442_high expression correlated with poor prognosis [meta-analysis] 29046366_These data inferred that long non-coding RNA PVT1 could be served as an indicator of glioma prognosis, and PVT1-EZH2 regulatory pathway may be a novel therapeutic target for treating glioma. 29050519_PVT1 played an oncogenic role in prostate cancer 29081406_Data suggest that long noncoding RNA PVT1 may be an oncogene as well as may promote metastasis in clear cell renal cell carcinoma (ccRCC) and could serve as a potential biomarker to predict the prognosis of ccRCC patients. 29115513_we first report that PVT1 promotes expression of HIF-1alpha, a critical endogenous hypoxia marker, in NSCLC. With this finding, we demonstrated that PVT1 modulated HIF-1alpha by competing for miR-199a-5p as a ceRNA to regulate cell proliferation. 29193797_the action of PVT1 is moderately attributable to its repression of ANGPTL4 via association with the epigenetic repressor Ezh2. 29244840_Based on these findings, we infer that PVT1 expression is modulated by both DNA amplification and methylation and its expression might serve as a valuable and specific prognostic biomarker in terms of OS in uveal melanoma 29262850_We found that circPVT1 behaves as an oncogene in HNSCC and that the mut-p53/YAP/TEAD complex transcriptionally regulates its expression. 29277611_Thus, our results indicated that lncRNA-PVT1-5 may function as a competing endogenous RNA (ceRNA) for miR-126 to promote cell proliferation by regulating the miR-126/SLC7A5 pathway, suggesting that lncRNA-PVT1-5 plays a crucial role in lung cancer progression and lncRNA-PVT1-5/miR-126/SLC7A5 regulatory network may shed light on tumorigenesis in lung cancer. 29280051_results indicate that ROR, PVT1, and HOTAIR have important regulatory roles during the development of papillary thyroid carcinomas 29286144_long noncoding RNA PVT1 may contribute to the tumorigenesis and metastasis of melanoma. 29445147_Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. 29452232_PVT1 promotes prostate cancer invasion and metastasis by modulating endothelial-mesenchymal transition. Furthermore, PVT1 can promote EMT by up-regulation of Twist1, a transcription factor associated with EMT. We then confirmed that PVT1 acts as a sponge for miRNA-186-5p and positively regulates Twist1 by a sponge effect. 29501773_PVT1-miR-190a-5p/miR-488-3p-MEF2C-JAGGED1 axis is involved in proliferation and progression of glioma. Thus, PVT1 may become a novel target in glioma therapy. 29505758_Combining the results of replication study with the previous GWAS data by meta-analysis, overall significance of PVT1 rs10087240 increased to a P-value of 5.11 x 10-11 and odds ratio of 1.17. These results confirm PVT1 as a vitiligo susceptibility locus in the European population. 29510227_High PVT1 expression is associated with hematologic malignancies. 29512788_The results of the present study suggest that PVT1 may serve a critical role in CRC progression and metastasis and may serve as a potential prognostic biomarker for colorectal cancer. 29552759_results indicate that PVT1 could promote metastasis and proliferation of colon cancer via suppressing miR-30d-5p/RUNX2 axis, which may offer a new way for interpreting the mechanism of colon cancer development 29620147_PVT1 was able to bind and degrade miR26b to promote connective tissue growth factor (CTGF) and angiopoietin 2 (ANGPT2) expression. 29693171_PVT1 knockdown reverses drug resistance in 5-FU resistant CRC cell lines, and that PVT1 overexpression promotes the development of MDR in CRC primarily by inhibiting apoptosis and upregulating the expression of MRP1, P-gp, mTOR and Bcl-2. 29693417_circPVT1 is upregulated in ALL. Silencing circPVT1 results in cell growth arrest and apoptosis of the cells. Our results also suggested a therapeutic potential of targeting circPVT1 in ALL. 29706652_The mechanism of PVT1-mediated angiogenesis via evoking the STAT3/VEGFA signalling axis. 29715456_PVT1 interacts with STAT1 to inhibit IFN-alpha signaling and tumor cells proliferation. 29731168_the PVT1 promoter mutation promotes cancer cell growth 29739059_Studies indicate that lncRNA PVT1 might serve as a potential therapeutic target for various cancers. 29749550_The results therefore provide significant information on the differentially expressed miRNAs associated with PVT1 in HCC, and we hypothesized that PVT1 may play vital roles in HCC by regulating different miRNAs or target gene expression (particularly MAPK8) via the MAPK or Wnt signaling pathways. Thus, further investigation of the molecular mechanism of PVT1 in HCC is needed 29760406_PVT1, KLF5, and beta-catenin were also revealed to be co-expressed in clinical TNBC samples. 29803929_Long noncoding RNA PVT1 enhances the expression of IGF1R through competitive binding to miR-30a. 29845201_Knockdown of PVT1 inhibited the TGFbeta/Smad signaling. 29948619_High expression of PVT1 is associated with Colorectal Adenocarcinoma. 29957467_PVT1 regulates ovarian cancer cell proliferation, migration and invasion by regulating miR-133a expression. 29975928_PVT1 could affect the biological function of HCC cells via targeting miR-424-5p and regulating INCENP. Focusing on the new insight of the PVT1/miR-424-5p/INCENP axis, this study provides a novel perspective for HCC therapeutic strategies. 30001707_Study revealed that PVT1 was up-regulated in pancreatic ductal adenocarcinoma (PDA) tissues and cell lines. Higher levels of PVT1 were associated with tumor progression and were inversely correlated with prognosis. Knocking down PVT1 significantly suppressed cell autophagy and growth both in vitro and in vivo. Mechanistically, PVT1 functions as a molecular sponge for miR-20a-5p to up-regulate ULK1. 30076414_PVT1-214 modulates the derepression of Lin28 by interacting with miR-128 30076714_PVT1 can regulate VEGFC expression by competitively binding miR-128 in bladder cancer cells. 30083911_PVT1 could serve as a novel biomarker for metastasis, clinical stage and poor prognosis in various tumors. 30141114_High expression of PVT1 is associated with Nasopharyngeal Cancer. 30205391_PVT1 knockdown could inhibit the expression of HIG2 through up-regulating miR-150 expression. 30222365_No differences were found between the combined AML patient populations and the healthy controls with respect to the expression levels of PVT1, CCDC26, and CCAT1, however the AML-M3 patients had higher PVT1 expression (p = 0.017). 30252166_PVT1 acts as competing RNA to microRNAs to protect mRNAs from miRNAs repression[review and meta-analysis] 30295989_this study shows that PVT1 regulates trophoblast viability, proliferation, and migration and is downregulated in spontaneous abortion 30304557_Long noncoding RNA PVT1 promotes laryngeal squamous cell carcinoma development by acting as a molecular sponge to regulate miR-519d-3p. 30317572_LncRNA PVT1 was overexpressed in bladder cancer cells, and it downregulated miR-31 ezpression and enhanced CDK1 expression to facilitate bladder cancer cells proliferation, migration, and invasion. 30347597_Down-regulation of PVT1 could enhance chemosensitivity of paclitaxel, induce apoptosis of glioma cells and noteworthy inhibit glioma cells proliferation. Our findings of PVT1 could contribute to attenuate paclitaxel resistance in clinical medicine. 30371726_Results indicate that long non-coding RNA PVT1 (PVT1) affected the role of mature adipogenic medium in triple-negative breast cancer (TNBC) cells via modulating cyclin-dependent kinase inhibitor 1A protein (p21) expression. 30504754_Results suggest that PVT1 could promote metastasis and proliferation of colon cancer via endogenous sponging and inhibiting the expression of miR-26b, which may highlight the significance of lncRNA PVT1 in colon cancer tumorigenesis. 30590312_Knockdown of circular PVT1 inhibited non-small cell lung cancer cell proliferation. 30649422_We demonstrated that the lncRNA Pvt1, activated early during muscle atrophy, impacts mitochondrial respiration and morphology and affects mito/autophagy, apoptosis and myofiber size in vivo. 30661902_We showed that lncRNA PVT1 played a contributory role in chemoresistance of osteosarcoma cells through c-MET/PI3K/AKT pathway activation, which was largely dependent on miR-152 30679629_PVT1 has a high expression level in the gastric cancer tissues of both Han and Uygur patients. The level of PVT1 in tissues can help to assess the risk of lymphatic metastasis in gastric cancer patients. PVT1 can be a potential biomarker to predict the tendency for metastasis in both Han and Uygur gastric cancer patients. 30794914_Authors confirmed that the effect of PVT1 promoting autophagy was dependent on regulating ATG3 expression. Further investigations revealed that PVT1 could upregulate autophagy-related gene 3 (ATG3) expression by acting as an endogenous sponge of miR-365, which was an inhibitor gene on ATG3 protein by targeting 3'UTR of ATG3 mRNA. 30820968_The presence of rs13281615 G > A polymorphism on PVT1 and the rs2910164 C > G polymorphism on miR-146a contributes to a favorable prognosis in colon cancer patients via modulating the activity of the PVT1/miR-146a/COX2 signaling pathway. 30825877_Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. 30832754_PVT1 knockdown could negatively regulate miR-424 to inhibit cellular activity, migration, and invasiveness in human gliomas, which explained the oncogenic mechanism of PVT1 in human gliomas. It also suggested that PVT1 might be a novel therapeutic target for human gliomas. 30909189_PVT1 is highly expressed in gliomas and its level is positively related to WHO glioma grade and prognosis of gliomas. Therefore, it may be explored as a new molecular marker for predicting malignancy and prognosis of gliomas. 30914434_Interactions of PVT1 and CASC11 on Prostate Cancer Risk in African Americans. 30943439_Results provide evidence that high expression level of PVT1 is associated with poor prognosis while being poor prognostic biomarker in t(8;21) associated acute myeloid leukemia. 30993787_Investigating the diagnostic performance of HOTTIP, PVT1, and UCA1 long noncoding RNAs as a predictive panel for the screening of colorectal cancer patients with lymph node metastasis. 31002139_LncRNA PVT1 knockdown affects proliferation and apoptosis of uveal melanoma cells by inhibiting EZH2. 31028131_Potential natural source of anti-gastric cancer drugs via epigenetic mechanism to inhibit LncRNA-PVT1-STAT3 axis. 31221783_Microhomology-mediated end joining drives complex rearrangements and overexpression of MYC and PVT1 in multiple myeloma. 31222482_PVT1 long-non-coding RNA (PVT1) is amplified in ovarian cancer patients and is correlated with poor survival outcomes. Knockdown of PVT1 causes decreased cell viability, metabolic activity, and smaller proportion of S-phase cells. PVT1 directly bound to miR-140 and acted as a microRNA sponge, while transcription of PVT1 is regulated by the transcription factor FOXO4. 31277104_A meta-analysis showed that high expression of PVT1 was significantly associated with tumor size, TNM stage, lymph node metastasis, distant metastasis, and overall survival time , especially in breast cancer, liver cancer and lung cancer. 31297833_Long noncoding RNA PVT1: A highly dysregulated gene in malignancy. 31309249_Summary of the coamplification of PVT1 and MYC in cancer, the positive feedback mechanism, and the latest promoter competition mechanism of PVT1 and MYC, as well as how PVT1 participates in the downstream signaling pathway of c-Myc by regulating key molecules. [review] 31320749_The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. 31322217_LncRNA PVT1 inhibits the effect of miR16, promoting the cell cycle and inhibiting cellular apoptosis of cervical cancer cells, potentially via the NFkappaB pathway. 31326971_Results showed that higher PVT1 was expressed in NSCLC and the elevated PVT1 was closely related to angiogenesis and poor prognosis in NSCLC. 31369196_research reveals that down-regulation of lncRNA PVT1 could potentially promote expression of miR-145 to repress cell migration and invasion, and promote cell apoptosis through the inhibition of FSCN1. 31371698_The silencing of PVT1 or the overexpression of FOXA1 relieved the damage and inhibited the apoptosis of podocytes in Diabetic nephropathy. 31485648_CircPVT1 functioned as competing endogenous RNA (ceRNA) to increase the STAT3 level and cell proliferation through sponging miR125b. 31509024_The PVT1 gene rs1252200336 locus polymorphisms are associated with the risk of developing colorectal cancer (CRC) in the Han Chinese population. The rs1252200336 locus deletion mutation (D) may impact the binding of hsa-miR-455-5p to the lncRNA PVT1 and its role in the development and progression of CRC. 31577086_LncRNA PVT1 participates in the development of oral squamous cell carcinomas through accelerating EMT and serves as a diagnostic biomarker. 31577719_The overexpression of PVT1 was significantly associated with poor overall survival in clear cell renal cell carcinoma. 31601234_Our results provide strong evidence that PVT1 confers an aggressive phenotype to EAC and is a poor prognosticator. 31604907_PVT1 exerts an oncogenic role through activating Wnt/ss-catenin signaling in pituitary adenoma cells. 31626590_LncRNA PVT1 promotes proliferation and invasion through enhancing Smad3 expression by sponging miR-140-5p in cervical cancer. 31627090_This study provided the evidence that PVT1 played a vital role in trophoblast cells, and it is important for maintaining the normal physiological function of trophoblast cells. 31677796_Silencing of PVT1 by ectopic miR-214 or siRNAs markedly inhibited viability and invasion of HCC cells. 31696483_LncRNA PVT1 aggravates the progression of glioma via downregulating UPF1. 31699956_Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. 31707347_XJD inhibits NSCLC cell growth via reciprocal interaction of PVT1 and miR181a-5p followed by reducing SP1 expression. 31755246_SP1-mediated lncRNA PVT1 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis. 31850702_Long noncoding RNAs (IncRNA) PVT1 was overexpressed in colorectal cancer (CRC) tissues compared to paired-adjacent normal tissues. 31877167_The results indicate that the assay can be used to quantify both low and high copy numbers of PVT1-derived transcripts. This is the first report of a copy number-based quantification assay for non-invasive detection of PVT1 derived transcripts. 31894346_lncRNA PVT1 promotes hepatitis B viruspositive liver cancer progression by disturbing histone methylation on the cMyc promoter. 31932969_The expression of MYC is strongly dependent on the circular PVT1 expression in pure Gleason pattern 4 of prostatic cancer. 31957871_Long noncoding RNA PVT1 acts as an oncogenic driver in human pan-cancer. 31986409_CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. 32003019_YY1-PVT1 affects trophoblast invasion and adhesion by regulating mTOR pathway-mediated autophagy. 32171788_LncRNA PVT1 induces chondrocyte apoptosis through upregulation of TNF-alpha in synoviocytes by sponging miR-211-3p. 32196583_Long noncoding RNA PVT1 promotes metastasis via miR-484 sponging in osteosarcoma cells. 32202906_The long non-coding RNA PVT1/miR-145-5p/ITGB8 axis regulates cell proliferation, apoptosis, migration and invasion in non-small cell lung cancer cells. 32249539_Circular RNA PVT1 promotes metastasis via regulating of miR-526b/FOXC2 signals in OS cells. 32293498_Knockdown of long non-coding RNA PVT1 induces apoptosis of fibroblast-like synoviocytes through modulating miR-543-dependent SCUBE2 in rheumatoid arthritis. 32342557_Long non-coding RNA PVT1, a novel biomarker for chronic obstructive pulmonary disease progression surveillance and acute exacerbation prediction potentially through interaction with microRNA-146a. 32358016_Population Differentiation at the PVT1 Gene Locus: Implications for Prostate Cancer. 32385030_[Effects of Long Non-coding RNA Plasmacytoma Variant Translocation 1 Gene on Inflammatory Response and Cell Migration in Helicobacter Pylori Infected Gastric Epithelial Cell Line]. 32407172_Long Noncoding RNA Plasmacytoma Variant Translocation 1 Regulates Cisplatin Resistance via miR-3619-5p/TBL1XR1 Axis in Gastric Cancer. 32428696_High PVT1 expression is associated with Myocardial ischemia/reperfusion injury. 32441301_Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. 32499447_LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. 32557622_lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. 32562719_Lowly expressed lncRNA PVT1 suppresses proliferation and advances apoptosis of glioma cells through up-regulating microRNA-128-1-5p and inhibiting PTBP1. 32572930_Expression level of lncRNA PVT1 in serum of patients with coronary atherosclerosis disease and its clinical significance. 32602581_Nonsense-mediated decay factor SMG7 sensitizes cells to TNFalpha-induced apoptosis via CYLD tumor suppressor and the noncoding oncogene Pvt1. 32614682_Serum long noncoding RNAs FAS-AS1 & PVT1 are novel biomarkers for systemic lupus erythematous. 32689767_LncRNA PVT1 regulates gallbladder cancer progression through miR-30d-5p. 32725807_Long noncoding RNA PVT1 promotes tumor growth and predicts poor prognosis in patients with diffuse large B-cell lymphoma. 32727463_LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. 32729021_LncRNA PVT1 induces aggressive vasculogenic mimicry formation through activating the STAT3/Slug axis and epithelial-to-mesenchymal transition in gastric cancer. 32734834_Clinical significance of circPVT1 in patients with non-small cell lung cancer who received cisplatin combined with gemcitabine chemotherapy. 32781203_Tissue-based long non-coding RNAs ''PVT1, TUG1 and MEG3'' signature predicts Cisplatin resistance in ovarian Cancer. 32812642_SOX2 knockdown slows cholangiocarcinoma progression through inhibition of transcriptional activation of lncRNA PVT1. 32827544_LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53. 32830409_LncRNA PVT1 exacerbates the inflammation and cell-barrier injury during asthma by regulating miR-149. 32866660_PVT1 induces NSCLC cell migration and invasion by regulating IL-6 via sponging miR-760. 32918701_Long non-coding RNA PVT1 can regulate the proliferation and inflammatory responses of rheumatoid arthritis fibroblast-like synoviocytes by targeting microRNA-145-5p. 32921988_LncRNA PVT1 Suppresses the Progression of Renal Fibrosis via Inactivation of TGF-beta Signaling Pathway. 32945498_Long noncoding RNA PVT1 promotes tumor cell proliferation, invasion, migration and inhibits apoptosis in oral squamous cell carcinoma by regulating miR1505p/GLUT1. 32945521_Propofol suppresses gastric cancer tumorigenesis by modulating the circular RNAPVT1/miR1955p/E26 oncogene homolog 1 axis. 32960438_The lncRNA PVT1 promotes invasive growth of lung adenocarcinoma cells by targeting miR-378c to regulate SLC2A1 expression. 32960485_Long noncoding RNA plasmacytoma variant translocation gene 1 promotes epithelial-mesenchymal transition in osteosarcoma. 32992461_Long Noncoding RNA PVT1 Is Regulated by Bromodomain Protein BRD4 in Multiple Myeloma and Is Associated with Disease Progression. 33049089_Long noncoding RNA PVT1 facilitates high glucose-induced cardiomyocyte death through the miR-23a-3p/CASP10 axis. 33067424_Long noncoding RNA PVT1 promoted gallbladder cancer proliferation by epigenetically suppressing miR-18b-5p via DNA methylation. 33106979_Downregulating long non-coding RNA PVT1 expression inhibited the viability, migration and phenotypic switch of PDGF-BB-treated human aortic smooth muscle cells via targeting miR-27b-3p. 33148262_The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. 33167678_Amplified LncRNA PVT1 promotes lung cancer proliferation and metastasis by facilitating VEGFC expression. 33174011_Knockdown of exosomemediated lncPVT1 alleviates lipopolysaccharideinduced osteoarthritis progression by mediating the HMGB1/TLR4/NFkappaB pathway via miR935p. 33188158_LncRNA PVT1 accelerates malignant phenotypes of bladder cancer cells by modulating miR-194-5p/BCLAF1 axis as a ceRNA. 33275233_Long non-coding RNA plasmacytoma variant translocation 1 (PVT1) promotes glioblastoma multiforme progression via regulating miR-1301-3p/TMBIM6 axis. 33327894_Long Non-Coding RNA PVT1 Regulates the Resistance of the Breast Cancer Cell Line MDA-MB-231 to Doxorubicin via Nrf2. 33369809_circPVT1 promotes osteosarcoma glycolysis and metastasis by sponging miR-423-5p to activate Wnt5a/Ror2 signaling. 33408314_Knockdown of plasmacytoma variant translocation 1 (PVT1) inhibits high glucose-induced proliferation and renal fibrosis in HRMCs by regulating miR-23b-3p/early growth response factor 1 (EGR1). 33430890_PVT1 signals an androgen-dependent transcriptional repression program in prostate cancer cells and a set of the repressed genes predicts high-risk tumors. 33453148_CircPVT1 promotes progression in clear cell renal cell carcinoma by sponging miR-145-5p and regulating TBX15 expression. 33558065_LncRNA PVT1 promotes the malignant progression of acute myeloid leukaemia via sponging miR-29 family to increase WAVE1 expression. 33600954_PVT1: A long non-coding RNA recurrently involved in neoplasia-associated fusion transcripts. 33670447_Serum Long Non-Coding RNAs PVT1, HOTAIR, and NEAT1 as Potential Biomarkers in Egyptian Women with Breast Cancer. 33712566_JAK2 a | 90.68588 | 0.9485202 | -0.0762496325 | 0.29885887 | 6.493879e-02 | 7.988537e-01 | 9.433646e-01 | No | Yes | 91.278928 | 13.107573 | 96.323826 | 13.362477 | ||||||||
| ENSG00000250903 | 100508120 | GMDS-DT | lncRNA | 31860169_LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. 34981821_lncRNA GMDSAS1 upregulates IL6, TNFalpha and IL1beta, and induces apoptosis in human monocytic THP1 cells via miR965p/caspase 2 signaling. | 47.85763 | 0.7810613 | -0.3564923659 | 0.48998868 | 5.226540e-01 | 4.697118e-01 | No | Yes | 51.526165 | 14.882057 | 64.240015 | 17.974290 | ||||||||||
| ENSG00000251136 | 101929709 | RIPK2-DT | lncRNA | 172.19032 | 0.9194445 | -0.1211656385 | 0.22267441 | 2.953776e-01 | 5.867949e-01 | 8.621760e-01 | No | Yes | 167.808500 | 35.307419 | 187.242143 | 38.440972 | ||||||||||
| ENSG00000251562 | 378938 | MALAT1 | lncRNA | This gene produces a precursor transcript from which a long non-coding RNA is derived by RNase P cleavage of a tRNA-like small ncRNA (known as mascRNA) from its 3' end. The resultant mature transcript lacks a canonical poly(A) tail but is instead stabilized by a 3' triple helical structure. This transcript is retained in the nucleus where it is thought to form molecular scaffolds for ribonucleoprotein complexes. It may act as a transcriptional regulator for numerous genes, including some genes involved in cancer metastasis and cell migration, and it is involved in cell cycle regulation. Its upregulation in multiple cancerous tissues has been associated with the proliferation and metastasis of tumor cells. [provided by RefSeq, Mar 2015]. | 16441420_Upregulation of MALAT-1 is associated with endometrial stromal sarcoma of the uterus 19041754_Study identified a highly conserved small RNA of 61 nucleotides originating from the MALAT1 locus that is broadly expressed in human tissues; although the long MALAT1 transcript localizes to nuclear speckles, the small RNA is found only in the cytoplasm. 19690017_Results suggest that MALAT-1 in placenta previa I/P is increased and its down-regulation inhibits trophoblast-like cell invasion. MALAT-1 might be involved in regulating trophoblast invasion during the development of advanced invasive placentation. 20149803_expression of the tumor marker MALAT1 noncoding RNA is sensitive to cell surface receptor activation by oxytocin in a neuroblastoma cell line 20213048_Our data demonstrated that MALAT1 was involved in cervical cancer cell growth, cell cycle progression, and invasion through the regulation of gene expression. 20711585_The expression of the long ncRNA MALAT1 correlates with tumor development, progression or survival in lung, liver and breast cancer. 20797886_MALAT1 regulates alternative splicing by modulating the levels of active serine/arginine (SR) splicing factors. 21503572_mutations were found on the long non-coding RNA MALAT-1 in CRC colorectal cancer 21678027_Our data suggest that MALAT1 plays an important role in tumor progression and could be a novel biomarker for predicting tumor recurrence after liver transplantation 21941126_Studies indicate the correlation of differential expression of metastasis-associated lung adenocarcinoma transcript1 (MALAT1) with tumor development, progression or survival in several cancerous conditions. 22078878_Pc2, methylation controls the protein's interaction with two distinct ncRNAs, TUG1 and NEAT2, which results in the exclusive subnuclear localization of methylated and unmethylated Pc2 in Polycomb bodies and interchromatin granules, respectively. 22088988_Our results suggest that MALAT-1 transcript induces migration in Non-small cell lung cancers and stimulates tumor growth and invasion in vivo. 22560368_MALAT-1 is upregulated in specific regions of the human alcoholic brain. 22722759_Study verified that MALAT-1 levels were upregulated in bladder cancer tissues compared with adjacent normal tissues, and MALAT-1 expression was remarkably increased in primary tumors that subsequently metastasized 22858678_The quantitative loss of MALAT1 did neither affect proliferation nor cell cycle progression nor nuclear architecture in human lung or liver cancer cells. 23129630_Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs, is reported. 23153939_MALAT1 is upregulated in urothelial carcinoma of the bladder, and high MALAT1 expression levels were associated with high-grade and high-stage carcinoma. 23243023_The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. 23555285_These findings provide mechanistic insights on the role of MALAT1 in regulating cellular proliferation 23726266_MALAT-1 miniRNA can be used as a novel plasma-based biomarker for prostate cancer detection and can improve diagnostic accuracy by predicting prostate biopsy outcomes. 23898077_The levels of plasma lncRNAs, H19, HOX antisense intergenic RNA (HOTAIR), and metastasis associated lung adenocarcinoma transcript-1 (MALAT1), were analyzed in patients with gastric cancer (GC) and healthy controls. 23973260_down-regulation of MALAT-1 expression compromised the cytoplasmic translocation of hnRNP C in the G2/M phase and resulted in G2/M arrest 23985560_microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. 24120702_Genetic translocations involving MALAT1 gene is associated with mesenchymal hamartoma of the liver. 24244343_resveratrol down-regulates MALAT1, resulting in decreased nuclear localization of beta-catenin thus attenuated Wnt/beta-catenin signaling, which leads to the inhibition of CRC invasion and metastasis. 24449823_malat1 promotes bladder cancer metastasis by associating with suz12. 24468535_Malat1 is a critical regulator of maintaining the transformative phenotype in liver cancer. 24602777_MALAT1 regulates endothelial cell proliferation/migration and vessel growth. 24658096_MALAT1 might serve as an oncogenic lncRNA that promotes proliferation and metastasis of gallbladder cancer and activates the ERK/MAPK pathway. 24815433_MALAT1 might be considered as a potential prognostic indicator and may be a target for diagnosis and gene therapy for pancreatic duct adenocarcinoma. 24857172_These results suggest that MALAT1 may function as a promoter of gastric cancer cell proliferation partly by regulating SF2/ASF. 24892958_Knockdown of UCA1 or Malat-1 lncRNA could attenuate the migrational ability of melanoma cells in in-vitro studies. Increased expression of UCA1 and Malat-1 lncRNAs might have a correlation with melanoma metastasis. 25025966_MALAT1 binds to SFPQ releasing PTBP2 from the SFPQ/PTBP2 complex, the increased SFPQ-detached PTBP2 promotes cell proliferation and migration in colorectal cancer. 25031737_The expression of MALAT1 is upregulated in colorectal cancer 25036876_An interaction of Bcl-2 with MALAT-1 lncRNA expression was revealed, which merits further investigation for risk prediction in resectable NSCLC patients. 25085246_Findings establish a novel PCDH10-Wnt/beta-catenin-MALAT1 regulatory axis that contributes to EEC development. 25187517_the mechanism of transcriptional activation of LTBP3 promoter depends on MALAT1 initiated from neighboring gene LTBP3 and involves both the direct interaction of the Sp1 and promoter-specific activation 25217850_Results showed that the high level of MALAT1 is associated with lung cancer brain metastasis and survival and demonstrated the potential role of MALAT1 in identification of non-small cell lung cancer patients at high risk of brain metastasis 25269958_MALAT-1 may serve as an oncogenic lncRNA that is involved in malignancy phenotypes of pancreatic cancer. 25280565_High MALAT1 expression drives gastric cancer development and promotes peritoneal metastasis. 25369863_MALAT1 was overexpressed in patients with myeloma and may play a role in its pathogenesis. In addition, MALAT1 may serve as a molecular predictor of early progression. 25431257_MALAT1 might suppress the tumor growth and metastasis via PI3K/AKT signaling pathway. 25446987_data indicate that MALAT1 may promote CRC tumor development via its target protein AKAP-9 25480417_Our data suggested that lncRNA MALAT1 was a novel molecule involved in ccRCC progression, which provided a potential prognostic biomarker and therapeutic target. 25481511_overexpression of lncRNA MALAT1 serves as an unfavorable prognostic biomarker in pancreatic cancer patients. 25538231_High MALAT1 expression is associated with proliferation, migration, and invasion of esophageal squamous cell carcinoma. 25546229_tumor-associated dendritic cell-derived CCL5 increases human colon cancer cell migration, invasion and epithelial-to-mesenchymal transition in a MALAT-1dependent manner 25600645_MALAT1 expression was higher in human renal cell carcinoma tissues, where it was associated with reduced patient survival. 25613066_lncRNA MALAT1 plays an important role on glioma progression and prognosis. 25613496_MALAT1 serves as an oncogene in ESCC, and it regulates ESCC growth by modifying the ATM-CHK2 pathway. 25772239_Loss of WIF1 enhances the migratory potential of glioblastoma through WNT5A that activates the WNT/Ca(2+) pathway and MALAT1. 25773124_The data suggest that upregulation of MALAT1 was mediated by the transcription factor Sp1 in A549 lung cancer cells 25787249_Studies indicate that long non-coding RNA MALAT1 regulates glucose-induced up-regulation of inflammatory mediators tumour necrosis factor alpha (TNF-alpha) and interleukin 6 (IL-6) through activation of serum amyloid antigen 3 (SAA3). 25811929_Results found the potential effects of MALAT-1 on the stem cell-like phenotypes in pancreatic cancer cells, suggesting a novel role of MALAT-1 in tumor stemness. 26139386_Study indicates that NEAT1 and MALAT1 may interact with HIV-1 in vivo. The correlation between plasma levels of NEAT1 and CD4 T-cell counts indicates that NEAT1 levels in plasma could be a potential biomarker for HIV-1 infection. 26159858_A poorer OS in patients with high MALAT-1 expression. 26191181_MALAT1 is a novel regulator of epithelial-mesenchymal transformation in breast cancer. 26241674_MALAT1 is a potential prognostic indicator and a target for the diagnosis and gene therapy for PVR diseases. 26242259_MALAT1 can promote HR-HPV (+) cancer cell growth and invasion at least partially through MALAT1-miR-124-RBG2 axis. 26254614_high tissue MALAT1 level was associated with an inferior clinical outcome in various cancers, suggesting that MALAT1 might serve as a potential prognostic biomarker for various cancers [meta-analysis] 26265046_MALAT1 expression by regulation of TDP-43 controls cellular growth, migration, and invasion of non-small cell lung cancer cells in vitro. 26275461_Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1 26282005_high expression of MALAT1 might be a novel prognostic biomarker in different types of cancers 26311052_MALAT1 and miR-145 had some level of synergic effect in reducing cancer cell colony formation, cell cycle regulation, and inducing apoptosis in cervical cancer 26335021_Malat1 plays a critical role in maintaining the proliferation potential of early-stage hematopoietic cells. 26352013_Human SP1 and SP3 transcription factors regulate MALAT1 expression in hepatocellular carcinoma. 26364720_Data show that long ncRNAs MALAT1 silencing downregulated the expression of the microRNA miR-22-3p target gene CXCR2 and AKT pathway. 26406400_High MALAT1 expression was associated with esophageal squamous cell carcinoma. 26415832_Posttranscriptional silencing of MALAT1 by miR-217 provides a link, through EZH2, between ncRNAs and the EMT and establishes a mechanism for cigarette smoke extract-induced lung carcinogenesis. 26423854_MALAT1 may potentially be used as a new prognostic marker to predict poorer survival of patients with cancer. 26434412_In this review, we document the molecular characteristics and functions of MALAT1 and shed light on the implication in the molecular pathology of various cancers. 26461224_High MALAT1 expression is associated with clear cell kidney carcinoma. 26482776_MALAT1 regulates cancer stem cell activity and radioresistance by modulating the miR-1/slug axis. 26493997_MALAT-1 knockdown induced a decrease in proliferation-enhanced apoptosis, inhibited migration/invasion, and reduced colony formation and led to cell cycle arrest at the G2/M phase. These data indicates that MALAT-1 could be exploited for therapeutic benefit 26516927_Data show that long non-coding RNA MALAT1 may be a crucial RNA cofactor of Polycomb protein enhancer of zeste homolog 2 (EZH2) and that may provide a new avenue for development new strategies for treatment of castration-resistant prostate cancer (CRPC). 26522444_MALAT1 is an important prognostic factor and therapeutic target for OSCC. 26575981_oncogenic role in osteosarcoma 26614531_MALAT1 level is associated with liver damage, and has clinical utility for predicting development of hepatocellular carcinoma 26619802_there was reciprocal repression between MALAT1 and miR-140, and miR-140 mediated the effects that MALAT1 knockdown exerted 26634309_Using drugs that specifically inhibit or activate the PERK or IRE1alpha sensors, we demonstrate that signalling through the PERK axis activates this expression, through a transcriptional mechanism 26676637_MALAT1 was upregulated in triple-negative breast cancer. MALAT1 knockdown inhibited proliferation, motility, and increased apoptosis in vitro. In vivo, MALAT1 knockdown inhibited tumor growth and metastasis. Patients with high MALAT1 expression had poorer overall survival. MALAT1 overexpression increased expression of miR-1, while miR-1overexpression decreased MALAT1 expression. MALAT1 acts through the miR-slug axis. 26722461_MALAT-1 may play an important role in the regulation of proliferation, cell cycle, apoptosis, migration and invasion of trophoblast cells, and under-expression of MALAT-1 during early placentation may be involved in the pathogenesis of preeclampsia. 26735578_High MALAT1 expression is associated with liver neoplasms. 26782531_An elevated MALAT1 expression correlates with large tumor size, advanced tumor stage, and poor prognosis, and might therefore be utilized to evaluate clinical pathological features and prognostic outcome for cancer patients. (Meta-analysis) 26798987_These results suggest that MALAT1 functions to promote cervical cancer invasion and metastasis via induction of epithelial-mesenchymal transition. 26821178_The overexpression of MALAT-1 may be a potential prognostic indicator for various human cancers. 26826711_Natural antisense RNA promotes 3' end processing and maturation of MALAT1 lncRNA. 26848616_Up-regulation of MALAT1 expression is associated with neuroblastoma. 26848980_EZH2 is recruited to the E-cadherin promoter by the long non-coding RNA, MALAT-1 (metastasis associated in lung adenocarcinoma transcript 1), where it represses E-cadherin expression 26871474_we found that long noncoding RNA (lncRNA) MALAT1 binds EZH2, suppresses the tumor suppressor PCDH10, and promotes gastric cellular migration and invasion 26884862_MALAT1 expression in non-small cell lung cancer cells is regulated by DNA methylation. 26887056_Findings suggest MALAT1 increases AKAP-9 expression by promoting SRPK1-catalyzed SRSF1 phosphorylation in CRC cells. These results reveal a novel molecular mechanism by which MALAT1 regulates AKAP-9 expression in CRC cells. 26895470_High MALAT1 expression is associated with multiple myeloma. 26909935_Meta-analysis: Poor overall survival and shortened disease-free, recurrence-free, disease-specific, or progression-free survival can be predicted by high MALAT-1 expression for various cancers. 26917489_Hsa-miR-448 disrupting KDM5B-MALAT1 signalling axis and associated activities in TNBC cells, projects it as a putative therapeutic factor for selective eradication of TNBC cells. 26918449_our data highlight the pivotal role of MALAT1 in breast cancer tumorigenesis 26926567_miR-1 bound both MALAT1 and cdc42 3'UTR directly. Further study showed that MALAT1 induced migration and invasion of breast cancer cells while reduced the level of cdc42. 26935028_observations suggest that MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. 26938295_Overexpression of Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) caused significant reduction in cell proliferation and invasion in vitro, and tumorigenicity in both subcutaneous and intracranial human glioma xenograft models. 26964565_MALAT1 knockdown reduces reactive gliosis, Muller cell activation, and RGC survival in vivo and in vitro MALAT1-CREB binding maintains CREB phosphorylation by inhibiting PP2A-mediated dephosphorylation, which leads to continuous CREB signaling activation. 26989678_This study demonstrated that the incidence of lymph node metastasis in patients detected with high MALAT-1 expression was higher than that in patients with low MALAT-1 expression in China. 27015363_MALAT1 promotes the initiation and progression of esophageal squamous cell carcinoma 27101025_Overexpression of GLI1 through a recurrent MALAT1-GLI1 translocation or GLI1 up-regulation delineates a pathogenically distinct subgroup of plexiform fibromyxomas with activation of the Sonic Hedgehog signalling pathway. 27134488_The knockdown ofmetastasis-associated lung adenocarcinoma transcription 1 increased apoptosis rate of the two cell line and the expression levels of some tumor markers were reduced such as CCND1 and MYC. 27172249_Results reveal a complex expression pattern of various MALAT1 transcript variants in breast tumours. 27191262_Malat1 is overexpressed in gallbladder cancer (GBC) tissue and cells 27191888_In 26 pairs of estrogen receptor (ER)-positive breast cancer samples, MALAT1 expression was significantly up-regulated compared with adjacent normal tissues (P = 0.012). Furthermore, of 204 breast cancer patients, high MALAT1 expression was associated with positive ER (P = 0.023) and progesterone receptor (PR) (P = 0.024) status. 27197265_we show that a ribonucleoprotein complex including the long noncoding RNA MALAT1 and the RNA-binding protein HuR (ELAVL1) binds the CD133 promoter region to regulate its expression 27215451_MALAT1 regulated 153 miRNAs in lung cancer. 27227769_The present study found that MALAT-1 is highly expressed in ovarian tumors. MALAT-1 promotes the growth and migration of ovarian cancer cells, suggesting that MALAT-1 may be an important contributor to ovarian cancer development. 27250026_long non-coding RNA MALAT1 is a metastasis driver in ER negative lymph node negative breast cancer 27259812_Up-regulated MALAT1 promoted the invasion and metastasis of GC, and the increase of EGFL7 expression was a potential mechanism via altering its H3 histone acetylation level 27287256_MALAT1, acting through HIF-1alpha stabilization, is a mediator that enhances glycolysis induced by arsenite. These results provide a link between the induction of lncRNAs and the glycolysis in cells exposed to arsenite, and thus establish a previously unknown mechanism for arsenite-induced hepatotoxicity. 27348442_lncRNA MALAT1 may serve as a potential novel prognostic biomarker in digestive system cancers 27362960_The findings propose that functional polymorphism rs619586A > G in MALAT1 gene plays an important role in pulmonary arterial hypertension (PAH) pathogenesis and may serve as a potential indicator for PAH susceptibility. 27371730_MALAT1 could facilitate the advanced progression of tumors in vivo Our study highlights the new roles of MALAT1 on protumorigenic functioning and anticancer therapy via activating autophagy in pancreatic cancer. 27420766_These data demonstrated that the MALAT1/miR-363-3p/MCL-1 regulatory pathway controls the progression of gallbladder cancer. 27431200_the present study found that MALAT1 was significantly highly expressed in non-small cell lung cancer (NSCLC) tissues with bone metastasis and in NSCLC cell lines with high bone metastatic ability 27434861_MALAT1 suppressed LPS-induced mRNA accumulation of TNFa and IL-6 without suppressing IL-1beta mRNA production. abnormal expression of MALAT1 was found to be NF-kappaB-dependent 27458156_Results suggest a long non-coding RNA MALAT1/MIR376A/transforming growth factor alpha (TGFA) axis mediates osteosarcoma (OS) cell proliferation and tumor progression. 27466303_MALAT1 upregulation plays an important role in breast cancer (BC) development, and serum MALAT1 level may be a potential tumor marker for BC diagnosis. 27470543_MALAT1 promoted the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. 27470544_MALAT1 could be a predictor for prognosis of esophageal cancer patients. 27486823_Data suggest that MALAT1 could function as an oncogene in gastric cancer, and high MALAT1 level could serve as a potential biomarker for the distant metastasis of gastric cancer. 27524242_there is a HIF-2alpha-MALAT1-miR-216b axis regulating multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. 27564100_MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22 27565324_Knockdown of MALAT1 suppressed SK-OV-3 cell viability, proliferation, migration and invasion (P < 0.05), and inhibited phosphorylation of MEK1, ERK1, p38 and JNK1, which suggested that MALAT1 promoted ovarian cancer cell proliferation, migration and invasion, and that MAPK pathways might be one of the regulatory mechanisms of MALAT1. 27584791_RBM24 acts at least in part through upregulating the expression of miR-25, which in turn targets MALAT1 for degradation. 27586393_Enhanced expression of MALAT-1 is associated with the growth and metastatic potential of tongue squamous cell carcinomas 27655020_LncRNA MALAT-1-mediated promotion of RCC proliferation. 27693631_When the interaction between miR-200c/MALAT1 was interrupted, the invasive capacity of endometrioid endometrial carcinoma cells was decreased and EMT markers expression were altered in vitro. 27696303_MALAT1 may function both as an oncogene and as a tumor suppressor in different types of thyroid tumors. 27782152_haploinsufficiency of HULC/MALAT1 plays an important role in malignant growth of liver cancer stem cell. 27814602_We first update on the role of MALAT1 in tumorigenesis and then discuss possible molecular mechanisms that underline the MALAT1-mediated gene regulation, leading to cancer invasion and metastasis. Review. 27841943_MALAT1 recruits splice factor serine-arginine-rich splice factor 2 (SRSF2) to promote alternative splicing of PKCdeltaII. 27872311_RNA immunoprecipitation after in vivo UV cross-linking and an in situ proximity ligation assay for RNA-protein interactions confirmed an association between METTL16 and MALAT1 in cells. METTL16 is an abundant ( approximately 5 x 10(5) molecules per cell) nuclear protein in HeLa cells. 27887846_MALAT1 promotes gastric cancer proliferation and progression. MALAT1 is a direct target of miR-202 and knockdown of MALAT1 significantly decreases the expression of Gli2 by negatively regulating miR-202. 27922078_MALAT1 and HOTAIR long non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. 27935117_acts as positive regulator of the radioresistance of esophageal squamous cell carcinoma, at least partly by promotion of Cks1 expression 27966454_miR-183 suppressed cell growth by inhibiting ITGB1 signal pathway and MALAT1 promoted melanoma growth by acting as a ceRNA of miR-183 in melanoma. 27998273_The increased expression of p21 and p27 upon MALAT1 knockdown was regulated by enhancer of zeste homolog 2 (EZH2). 28034748_Observed a significant negative correlation between miR-216a and MALAT1 in pancreatic ductal adenocarcinoma (PDAC) tissues and adjacent normal tissues. In vitro cell assay further confirmed a direct binding between miR-216a and MALAT1 and the suppressive effect of miR-216a on MALAT1 expression. MiR-216a overexpression and MALAT1 knockdown induced cell cycle arrest at G2/M phase. 28056547_expression is significantly increased by hypoxia and regulates the proliferation of smooth muscle cells 28059437_data indicated that MALAT1 might play an oncogenic role in hilar cholangiocarcinoma (HCCA) through miR-204-dependent CXCR4 regulation, and could be regarded as a therapeutic target in HCCA. 28077118_MALAT1 is involved in gastric carcinogenesis via inhibition of apoptosis and promotes invasiveness via the epithelial-to-mesenchymal transition. 28160461_In this study, the long noncoding RNA MALAT1, confirmed to be significantly upregulated in OS, is first shown to be capable of promoting proliferation and migration by directly suppressing miR-26a-5p in OS cells. Authors have identified forkhead box O1 (FOXO1) as a transcriptional factor of MALAT1 that can negatively regulate MALAT1. 28176360_data demonstrate that MALAT1 is an important endogenous regulator in the proliferation, angiogenesis, and immunosuppressive properties of MSCs 28187000_High MALAT1 expression is associated with chemoresistance in glioblastoma multiforme. 28252165_Data show that the low expression of microRNA miRNA-143 and high expression of non-coding RNA MALAT1 in cervical cancer cells could possibly potentiate cell invasion/migration. 28253859_The rs3200401 T allele located on the lncRNA MALAT1 was associated with a better survival for advanced lung adenocarcinoma patients, which may offer a novel prognostic biomarker for this patient subgroup. 28268166_MALAT1 can promote tumorigenicity and metastasis in gastric cancer by facilitating VM and angiogenesis via the VE-cadherin/beta-catenin complex and ERK/MMP and FAK/paxillin signaling pathways. 28276823_In vitro and in vivo assays showed that siRNA-mediated silencing of MALAT-1 inhibited gastric cancer cell migration and invasion. In addition, suppressing MALAT-1 expression resulted in a decrease in the expression of the epithelial-mesenchymal transition-associated marker vimentin and an increase in the expression of E-cadherin at both the mRNA and protein levels. 28292022_This study study provides evidence that inhibiting lncRNA MALAT-1 can improve the chemotherapy sensitivity of DLBCL by enhancing autophagy-related proteins. 28295550_LncRNA MALAT-1 increases the expression level of HMGB1 in MM thereby promotes autophagy resulting in the inhibition of apoptosis. 28336564_This study identifies a novel signaling involving SYK/c-MYC/MALAT1 as a promising therapeutic target for the treatment of Ewing sarcoma. 28346809_MALAT1 promoted osteosarcoma cell growth through inhibition of miR-142-3p or miR-129-5p and by targeting HMGB1. 28388584_our work illuminates that lncRNA MALAT1 is a potential diagnostic and prognostic factor in osteosarcoma and further demonstrates how MALAT1 confers an oncogenic function. 28412742_the expression of MALAT1 was increased in T and NK cell lymphoma, and high MALAT1 expression was associated with inferior overall survival, especially for patients with mature T cell lymphoma subtypes. 28467811_Results indicate that circulating long non-coding RNAs (lncRNAs) MALAT1, AFAP1-AS1 and AL359062 may represent novel serum biomarkers for nasopharyngeal carcinoma (NPC) diagnosis and prognostic prediction after treatment. 28468579_these results suggested that Axl, which is regulated by long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1, may exert great influence on invasion and migration of neuroblastoma 28487970_MALAT1 may participate in UVBinduced photoaging via regulation of the ERK/mitogenactivated protein kinase signaling pathway and UVBinduced MALAT1 expression is independent of ROS generation. 28528814_Targeting the Malat1/AR-v7 axis via Malat1-siRNA or ASC-J9 can be developed as a new therapy to better suppress enzalutamide-resistant prostate cancer progression. 28535533_The MALAT1-miR-145-TGFBR2/SMAD3 signaling pathway plays a key role in TGF-beta1-induced epithelial-to-mesenchymal transition . 28543663_results suggest that MALAT1-mediated FGF2 protein secretion from Tumor-associated macrophages inhibits inflammatory cytokines release, promotes proliferation, migration, and invasion of FTC133 cells and induces vasculature formation. 28543721_findings suggest that MALAT1 may regulate ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. 28550678_Expression of MALAT1 was elevated in retinoblastoma cell line. 28551849_This study indicated that MALAT1 promoted proliferation and suppressed apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. 28590075_MALAT1 could directly bind to miR-127-5p to inhibit its expression, so as to rescue OPN expression and promote chondrocyte proliferation through PI3K/Akt pathway. 28592124_MALAT1 promoted cholangiocarcinoma cell proliferation and invasion. The effects of MALAT1 on cholangiocarcinoma cells might be through activating the PI3K/Akt signaling pathway. 28615056_MALAT1 expression is controlled by Oct4 transcriptional regulation in lung cancer. 28623135_MALAT-1 expression is upregulated in the serum, and circulating exosomes of non-small cell lung cancer patients. 28652379_results highlight a key role for MALAT1 in control of VEGFA isoforms expression in breast cancer cells expressing gain-of-function mutant p53 and ID4 proteins. 28660408_These results indicate that the noncoding RNAs MALAT1 and miR-885 show increased expression in neoplastic follicular and Hurthle cell thyroid neoplasms compared to normal thyroid tissues 28664617_Down-regulation of long non-coding RNA MALAT1 by RNA interference inhibits proliferation and induces apoptosis in multiple myeloma 28668966_MALAT1 decreased the sensitivity of resistant glioma cell lines to Temozolomide by regulating ZEB1. 28675122_e revealed there was a reciprocal repression between MALAT1 and miR-204. ZEB2 was identified as a downstream target of miR-204 and MALAT1 exerted its function mainly through the miR-204/ZEB2 axis. Our findings suggested that MALAT1 may serve as a new diagnostic biomarker and therapy target for breast cancer. 28701723_MALAT1 acts as a tumour promoter at least in part by binding miR-217 and sequestering the molecule in the nucleus, thereby promoting oncogenic KRAS expression in PDAC. 28713913_these findings suggest that high MALAT-1 expression is closely associated with poor prognosis in M5 patients and may play a role in leukemia cell proliferation and apoptosis, and may serve as a promising theranostic marker. 28720061_Notably, sirtuin 1 was recognized as a direct downstream target of microRNA-204 in HepG2 cells. Moreover, si-SIRT1 significantly inhibited cell invasion and migration process.his study reveals a novel mechanism by which MALAT1 stimulates hepatocellular carcinoma progression and justifies targeting metastasis-associated lung adenocarcinoma transcript 1 as a potential therapy for hepatocellular carcinoma. 28731043_we have identified an oncogenic MALAT1-GLI1 fusion gene in all cases of gastroblastoma that may serve as a diagnostic biomarker. 28770558_Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. 28770968_High MALAT1 expression was associated with poor prognosis, whereas MALAT1 knockdown inhibited invasion, metastasis, and EMT-related genes in epithelial ovarian cancer cells in in vitro and in vivo. 28834690_study elucidated a novel Malat1-miR-101-STMN1/RAB5A/ATG4D regulatory network that Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma cells 28857668_MALAT1 and Capn4 were up-regulated while miR-124 expression was down-regulated in nasopharyngeal carcinoma cell lines. 28915533_the 'MALAT1-miR-129-5p' axis might play an important role in the progression of triple-negative breast cancer. 28926115_MALAT1 can function as a competing endogenous RNA (ceRNA) to modulate STAT3 expression by absorbing miR-125b in oral squamous cell carcinoma 28942451_Overexpression of MALAT1 attenuated or even reversed UPF1-mediated inhibitions on gastric tumor cell proliferation, epithelial-mesenchymal transition and the promotion on cell apoptosis. 28973437_Long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. 29073720_Through direct targeting miR-124, MALAT1 promotes retinoblastoma cell autophagy. 29096355_LncRNA-MALAT1 promoted choriocarcinoma cell growth by targeting miR-218. 29107050_High MALAT1 expression is associated with medullary thyroid carcinoma. 29110299_Expression of lncRNA MALAT1 could offer a novel biomarker to predict gestational diabetes mellitus. 29115409_these data indicated that the MALAT1/miR146a/NF-kappaB pathway exerted key functions in LPS-induced acute kidney injury (AKI), and provided novel insights into the mechanisms of this therapeutic candidate for the treatment of the disease. 29116025_Inhibition of miR-200c promoted the proliferation and migration of HESCs, while the simultaneous silencing of MALAT1 expression exerted the opposite effects. Authors demonstrated that expression of MALAT1 in ectopic endometrial specimens was negatively correlated with that of miR-200c and that MALAT1 knockdown increased the level of miR-200c in HESCs. 29133588_We found that SON and SC35 (also known as SRSF2) localize to the central region of the speckle, whereas MALAT1 and small nuclear (sn)RNAs are enriched at the speckle periphery. 29146194_Findings revealed the association between three SNPs (rs3200401, rs619586 and rs7927113) in lncRNA MALAT1 and breast cancer (BC) risk with adjusted for age and reproductive factors. These results implied that the genetic variants of lncRNA MALAT1 were associated with the susceptibility of BC, and meaningful genetic alteration might affect the corresponding mRNA expression of lncRNA MALAT1. 29162158_Data showed that chemoresistant gastric cancer (GC) cells had higher levels of MALAT1 and increased autophagy. Its silencing inhibited chemo-induced autophagy, whereas MALAT1 promoted autophagy in gastric cancer cell | 712.66394 | 1.4034496 | 0.4889772063 | 0.17809386 | 7.620515e+00 | 5.770802e-03 | 1.254833e-01 | No | Yes | 775.716450 | 189.682674 | 545.824679 | 130.496784 | ||||||||
| ENSG00000253123 | 100507403 | lncRNA | 29.52543 | 0.3840091 | -1.3807874820 | 0.53108128 | 6.464268e+00 | 1.100650e-02 | No | Yes | 15.757072 | 3.541757 | 39.568765 | 8.153761 | ||||||||||||
| ENSG00000253368 | 388610 | TRNP1 | protein_coding | Q6NT89 | FUNCTION: DNA-binding factor that regulates the expression of a subset of genes and plays a key role in tangential, radial, and lateral expansion of the brain neocortex. Regulates neural stem cells proliferation and the production of intermediate neural progenitors and basal radial glial cells affecting the process of cerebral cortex gyrification. May control the proliferation rate of cells by regulating their progression through key cell-cycle transition points (By similarity). {ECO:0000250}. | Cell cycle;DNA-binding;Developmental protein;Neurogenesis;Nucleus;Reference proteome;Transcription;Transcription regulation;Ubl conjugation | hsa:388610; | euchromatin [GO:0000791]; nucleus [GO:0005634]; DNA binding [GO:0003677]; cell cycle [GO:0007049]; cerebellar cortex morphogenesis [GO:0021696]; neural precursor cell proliferation [GO:0061351]; regulation of cell cycle [GO:0051726]; regulation of cell population proliferation [GO:0042127] | 23622239_TRNP1 expression levels exhibit regional differences in the cerebral cortex of human fetuses, anticipating radial or tangential expansion. | ENSMUSG00000056596 | Trnp1 | 175.61222 | 0.9892863 | -0.0155400508 | 0.22415717 | 4.802865e-03 | 9.447487e-01 | 9.855574e-01 | No | Yes | 227.701033 | 22.814683 | 218.586906 | 21.133619 | ||
| ENSG00000254413 | 386593 | CHKB-CPT1B | protein_coding | H3BT56 | Reference proteome | The genes CHKB and CPT1B are adjacent on chromosome 22 and read-through transcripts are expressed that include exons from both loci. The read-through transcripts are candidates for nonsense-mediated mRNA decay (NMD) and are unlikely to express proteins. [provided by RefSeq, Jun 2009]. | cytoplasm [GO:0005737]; choline kinase activity [GO:0004103]; ethanolamine kinase activity [GO:0004305]; CDP-choline pathway [GO:0006657]; phosphatidylcholine biosynthetic process [GO:0006656]; phosphatidylethanolamine biosynthetic process [GO:0006646] | 568.86919 | 0.8881997 | -0.1710440487 | 0.14775004 | 1.334642e+00 | 2.479811e-01 | 6.414243e-01 | No | Yes | 565.894898 | 92.200868 | 648.118327 | 103.021587 | ||||||
| ENSG00000254860 | 493900 | TMEM9B-AS1 | lncRNA | 88.41207 | 0.9458890 | -0.0802572440 | 0.31765167 | 6.356766e-02 | 8.009435e-01 | 9.438333e-01 | No | Yes | 81.822011 | 17.249455 | 89.033255 | 18.038407 | ||||||||||
| ENSG00000255624 | 399815 | C10orf88B | transcribed_processed_pseudogene | 81.33230 | 0.7386125 | -0.4371104239 | 0.31541925 | 1.901363e+00 | 1.679258e-01 | 5.605054e-01 | No | Yes | 80.457009 | 23.559711 | 108.753961 | 30.883444 | ||||||||||
| ENSG00000256061 | 161582 | DNAAF4 | protein_coding | Q8WXU2 | FUNCTION: Axonemal dynein assembly factor required for ciliary motility. Involved in neuronal migration during development of the cerebral neocortex. May regulate the stability and proteasomal degradation of the estrogen receptors that play an important role in neuronal differentiation, survival and plasticity. {ECO:0000269|PubMed:19423554, ECO:0000269|PubMed:23872636}. | Alternative splicing;Cell projection;Chromosomal rearrangement;Ciliopathy;Cytoplasm;Kartagener syndrome;Neurogenesis;Nucleus;Primary ciliary dyskinesia;Reference proteome;Repeat;TPR repeat | Mouse_homologues NA; + ;NA | This gene encodes a tetratricopeptide repeat domain-containing protein. The encoded protein interacts with estrogen receptors and the heat shock proteins, Hsp70 and Hsp90. An homologous protein in rat has been shown to function in neuronal migration in the developing neocortex. A chromosomal translocation involving this gene is associated with a susceptibility to developmental dyslexia. Mutations in this gene are associated with deficits in reading and spelling. Alternative splicing results in multiple transcript variants. Read-through transcription also exists between this gene and the downstream cell cycle progression 1 (CCPG1) gene. [provided by RefSeq, Mar 2011]. | hsa:161582; | centrosome [GO:0005813]; cytoplasm [GO:0005737]; cytosol [GO:0005829]; dynein axonemal particle [GO:0120293]; extracellular region [GO:0005576]; neuron projection [GO:0043005]; non-motile cilium [GO:0097730]; nucleus [GO:0005634]; plasma membrane [GO:0005886]; estrogen receptor binding [GO:0030331]; cilium movement [GO:0003341]; determination of left/right symmetry [GO:0007368]; epithelial cilium movement involved in extracellular fluid movement [GO:0003351]; heart development [GO:0007507]; inner dynein arm assembly [GO:0036159]; neuron migration [GO:0001764]; outer dynein arm assembly [GO:0036158]; regulation of intracellular estrogen receptor signaling pathway [GO:0033146]; regulation of proteasomal protein catabolic process [GO:0061136] | 12954984_DYX1C1 should be regarded as a candidate gene for developmental dyslexia and localizes to a fraction of cortical neurons and white matter glial cells 15249932_Findings provide support for EKN1 as a risk locus for dyslexia and as contributing to reading component processes and reading-related abilities. 15470369_Thus it seems unlikely that DYX1C1 gene would be involved in the genetic etiology of autism in Finnish patients. 15477871_DYX1C1 is unlikely to be a susceptibility gene for developmental dyslexia 15520411_Single nucleotide polymorphisms of DYX1C1 are not associated with dyslexia susceptibility in a large sample of sibling pairs. 15702132_Observational study of gene-disease association. (HuGE Navigator) 16989952_Functional characterization of the homologous rat protein. 17309662_Observational study of gene-disease association. (HuGE Navigator) 17309662_disequilibrium with DYX1C1 is more saliently explained in Italian dyslexics by short-term memory 17450541_The observation of weak evidence for transmission disequilibrium for one of the two studied polymorphisms in DYX1C1 suggests involvement of this gene in dyslexia. 18445785_TFII-I, PARP1, and SFPQ proteins, each previously implicated in gene regulation, form a complex controlling transcription of DYX1C1. Allelic differences in the promoter or 5'UTR of DYX1C1 may affect factor binding and thus regulation of the gene. 18618141_Alternatively spliced transcript variants of the DYX1C1 gene may be used as cancer biomarkers to detect colorectal cancer. 19058789_Observational study of gene-disease association. (HuGE Navigator) 19076634_Observational study of gene-disease association. (HuGE Navigator) 19240663_Observational study of gene-disease association. (HuGE Navigator) 19240663_The contribution of DYX1C1 to dyslexia in a sample of 366 trios of German descent, was investigated. 19277710_The DYX1C1 is a novel Hsp70 and Hsp90-interacting co-chaperone protein and its expression is associated with malignancy. 19423554_DYX1C1 interacts with both ERs in the presence of 17beta-estradiol in neurons. 19901951_Observational study of gene-disease association. (HuGE Navigator) 19901951_Results suggest that DYX1C1 influences reading and spelling ability with additional effects on short-term information storage or rehearsal. Missense mutation rs17819126 is a potential functional basis for the association of DYX1C1 with dyslexia. 20846247_Association signals were detected for several single nucleotide polymorphisms within DYX1C1 with both the reading and spelling tests 20846247_Observational study of gene-disease association. (HuGE Navigator) 21046216_No statistically significant associations were found between DCDC2 or DYX1C1 and language phenotypes. Both DCDC2 and DYX1C1 DD susceptibility genes appear to have a pleiotropic role on mathematics but not language phenotypes. 21046216_Observational study of gene-disease association. (HuGE Navigator) 21203818_At this point, there is no statistical evidence of association between the allelic variation in the three candidate genes and DD in our sample. 21599957_findings suggest that DYX1C1 is associated with dyslexia in people of Chinese ethnicity. 22375924_the expression of DYX1C1 in breast cancer is associated with several clinicopathological parameters and that loss of DYX1C1 correlates with a more aggressive disease, in turn indicating that DYX1C1 is a potential prognostic biomarker in breast cancer. 22383464_A single nucleotide polymorphism previously shown to be associated with dyslexia and located in the cis-regulatory region of DYX1C1 may alter the epigenetic and endocrine regulation of this gene. 22558177_Mutations in cilia co-expressed DCDC2, DYX1C1 and KIAA0319 genes are associated with a cognitive neurological disorder, dyslexia. 22683091_The results of this study found that DYX1C1 gene contained polymorphisms that were significantly associated with white matter volume in the left temporo-parietal region and that white matter volume influenced reading ability. 23028439_DYX1C1 influences reading development in the general Chinese population and supports a universal effect of this gene. 23036959_results demonstrate that DYX1C1 can modulate the expression of genes involved in cell migration and nervous system development and associates with a number of cytoskeletal proteins. 23065966_results suggested that the 931C > T variant in KIAA0319, but not the -3G > A in DYX1C1, was significantly associated with the risk of dyslexia 23176554_Gene-by-environment effects were found between some specified environmental moderators (i.e. maternal smoke during pregnancy, birth weight and socio-economic status) and the DYX1C1-1259C/G marker 23341075_The results of this study do not provide evidence for association between the putatively functional single nucleotide polymorphisms -3G/A and 1249G/T in DYX1C1 and reading disabilities. 23872636_DYX1C1 is required for axonemal dynein assembly and ciliary motility. 24362368_promoter SNP rs12899331 of DYX1C1 may contribute towards the manifestation of DD. This study supports the association of DYX1C1 with DD in an Indian population 27451412_that endogenous DYX1C1 localizes to the base of the cilium, whereas DCDC2 localizes along the entire axoneme of the cilium 33174003_Novel dynein axonemal assembly factor 1 mutations identified using wholeexome sequencing in patients with primary ciliary dyskinesia. | ENSMUSG00000089865+ENSMUSG00000092192 | Gm44503+Dnaaf4 | 65.00661 | 0.6766521 | -0.5635138223 | 0.40107367 | 1.943531e+00 | 1.632857e-01 | No | Yes | 45.830942 | 17.900531 | 67.610368 | 25.602733 | |
| ENSG00000256537 | 100129361 | SMIM10L1 | protein_coding | P0DMW3 | Reference proteome | hsa:100129361; | ENSMUSG00000072704 | Smim10l1 | 774.04012 | 0.8548147 | -0.2263164196 | 0.14287360 | 2.491977e+00 | 1.144279e-01 | 4.807170e-01 | No | Yes | 856.165919 | 128.880215 | 974.769727 | 143.280071 | |||||
| ENSG00000257337 | 283335 | lncRNA | 50.81302 | 0.8862889 | -0.1741510063 | 0.42142840 | 1.702731e-01 | 6.798692e-01 | No | Yes | 49.552334 | 9.407355 | 51.663090 | 9.486040 | ||||||||||||
| ENSG00000257949 | 100134934 | TEN1 | protein_coding | Q86WV5 | FUNCTION: Component of the CST complex proposed to act as a specialized replication factor promoting DNA replication under conditions of replication stress or natural replication barriers such as the telomere duplex. The CST complex binds single-stranded DNA with high affinity in a sequence-independent manner, while isolated subunits bind DNA with low affinity by themselves. Initially the CST complex has been proposed to protect telomeres from DNA degradation (PubMed:19854130). However, the CST complex has been shown to be involved in several aspects of telomere replication. The CST complex inhibits telomerase and is involved in telomere length homeostasis; it is proposed to bind to newly telomerase-synthesized 3' overhangs and to terminate telomerase action implicating the association with the ACD:POT1 complex thus interfering with its telomerase stimulation activity. The CST complex is also proposed to be involved in fill-in synthesis of the telomeric C-strand probably implicating recruitment and activation of DNA polymerase alpha (PubMed:22763445). The CST complex facilitates recovery from many forms of exogenous DNA damage; seems to be involved in the re-initiation of DNA replication at repaired forks and/or dormant origins (PubMed:25483097). {ECO:0000269|PubMed:19854130, ECO:0000269|PubMed:22763445, ECO:0000269|PubMed:25483097}. | 3D-structure;Chromosome;DNA-binding;Nucleus;Reference proteome;Telomere | C17ORF106, or TEN1, appears to function in a telomere-associated complex with STN1 (OBFC1; MIM 613128) and CTC1 (C17ORF68; MIM 613129) (Miyake et al., 2009 [PubMed 19854130]).[supplied by OMIM, Nov 2009]. | hsa:100134934; | chromosome, telomeric region [GO:0000781]; CST complex [GO:1990879]; nucleoplasm [GO:0005654]; nucleus [GO:0005634]; single-stranded DNA binding [GO:0003697]; telomerase inhibitor activity [GO:0010521]; telomeric DNA binding [GO:0042162]; negative regulation of telomere maintenance via telomerase [GO:0032211]; telomere capping [GO:0016233] | 19854130_Ctc1-Stn1-Ten1 is a replication protein A (RPA)-like complex that is not directly involved in conventional DNA replication at forks but plays a role in DNA metabolism frequently required by telomeres. 22763445_the human CST (CTC1, STN1 and TEN1) complex, previously implicated in telomere protection and DNA metabolism, inhibits telomerase activity through primer sequestration and physical interaction with the protection of telomeres 1 (POT1)-TPP1 telomerase processivity factor 23826127_data indicate that hTen1 is critical for the telomeric function of hCST, both in telomere protection and downregulation of telomerase function 23851344_The mammalian CST (CTC1-STN1-TEN1) complex is directly involved at several stages of telomere end formation and CST seems to play critical roles in coordinating telomerase elongation and fill-in synthesis to complete telomere replication. 24025336_TEN1 likely functions in conjunction with CTC1 and STN1 at the telomere and elsewhere in the genome. 25684230_reveal a critical role for human Stn1 in telomere length maintenance and function, supporting the model that efficient replication of telomeric repeats is critical for long-term viability of normal somatic mammalian cells 27487043_CTC1/STN1/TEN1 (CST) deficiency diminishes HU-induced RAD51 foci formation. 28334750_TERT-mediated G-strand extension and Ctc1-Stn1-Ten1-mediated C-strand fill-in are equally important for telomere length maintenance. 28366536_The findings imply that theCTC1/STN1/TEN1 complex(CST )complex plays an important role in regulating telomere maintenance in alternative-lengthening of telomeres(ALT) cells. 29293451_Studies indicate telomere-binding proteins CTC1-STN1-TEN1 (CST) dysfunction and mutation is associated with several genetic diseases and cancers [Review]. 30026550_CTC1-STN1 terminates TERT while STN1-TEN1 enables C-strand synthesis during telomere replication in colon cancer cells. 30976812_The CST complex (CTC1-STN1-TEN1) maintains genome integrity through resolution of G4 structures both ahead of the replication fork and on the lagging strand template 30979824_CST functions in two distinct aspects of genome-wide DNA replication, namely, origin licensing and replisome assembly. CST interacts with additional replisome components, MCM and AND-1. 31028137_This work provides a blueprint to pinpoint the possible consequences of pathogenic mutations in the CST complex subunit TEN1. 32627942_An Indian child with Coats plus syndrome due to mutations in STN1. 33210317_Human CST complex protects stalled replication forks by directly blocking MRE11 degradation of nascent-strand DNA. 33780718_CST in maintaining genome stability: Beyond telomeres. 35134616_Pan-cancer analysis reveals that CTC1-STN1-TEN1 (CST) complex may have a key position in oncology. | ENSMUSG00000020778 | Ten1 | 151.82599 | 0.9299749 | -0.1047363842 | 0.23312727 | 2.014669e-01 | 6.535394e-01 | 8.912309e-01 | No | Yes | 200.151490 | 39.191641 | 203.783497 | 38.959055 | |
| ENSG00000258311 | protein_coding | F8W036 | FUNCTION: May negatively regulate aerobic respiration through mitochondrial protein lysine-acetylation. May counteract the action of the deacetylase SIRT3 by acetylating and regulating proteins of the mitochondrial respiratory chain including ATP5F1A and NDUFA9. {ECO:0000256|ARBA:ARBA00003284}. | Coiled coil;Reference proteome | BLOC-1 complex [GO:0031083]; lysosomal membrane [GO:0005765]; mitochondrial intermembrane space [GO:0005758]; cellular localization [GO:0051641] | 150.76353 | 0.8177407 | -0.2902845954 | 0.24634267 | 1.377834e+00 | 2.404704e-01 | 6.320340e-01 | No | Yes | 148.306524 | 31.514384 | 184.778027 | 38.386870 | ||||||||
| ENSG00000258704 | SRP54-AS1 | transcribed_unitary_pseudogene | 34.10893 | 0.7032563 | -0.5078775394 | 0.52959096 | 9.045315e-01 | 3.415695e-01 | No | Yes | 31.397698 | 8.496089 | 42.345453 | 11.027068 | ||||||||||||
| ENSG00000259343 | TMC3-AS1 | lncRNA | 57.69646 | 0.7451154 | -0.4244641647 | 0.38847946 | 1.179645e+00 | 2.774283e-01 | No | Yes | 53.049226 | 8.275326 | 69.297331 | 10.118460 | ||||||||||||
| ENSG00000259429 | UBE2Q2P2 | transcribed_unprocessed_pseudogene | 11.45092 | 0.4703378 | -1.0882306937 | 0.85173730 | 1.589023e+00 | 2.074657e-01 | No | Yes | 5.498649 | 6.681825 | 13.212976 | 15.501512 | ||||||||||||
| ENSG00000259511 | 100505679 | UBE2Q2L | transcribed_unprocessed_pseudogene | H0YL09 | Reference proteome | 17.16762 | 1.1503253 | 0.2020419498 | 0.68481176 | 8.737003e-02 | 7.675476e-01 | No | Yes | 24.674865 | 6.232977 | 20.202504 | 4.976507 | |||||||||
| ENSG00000260400 | lncRNA | 62.81401 | 1.2903132 | 0.3677213339 | 0.36834539 | 1.000725e+00 | 3.171352e-01 | No | Yes | 83.545820 | 25.950553 | 59.099933 | 18.062724 | |||||||||||||
| ENSG00000261879 | 100130950 | ZNF594-DT | lncRNA | 58.61036 | 2.5168278 | 1.3316065323 | 0.37386278 | 1.274710e+01 | 3.565609e-04 | No | Yes | 128.274212 | 41.839847 | 51.330469 | 16.339887 | |||||||||||
| ENSG00000261971 | MMP25-AS1 | lncRNA | 194.37980 | 0.7070084 | -0.5002007862 | 0.24297916 | 4.179058e+00 | 4.092645e-02 | 3.122171e-01 | No | Yes | 175.737867 | 26.344898 | 248.986664 | 36.122178 | |||||||||||
| ENSG00000262484 | 643669 | CCER2 | protein_coding | I3L3R5 | Reference proteome;Secreted;Signal | hsa:643669; | extracellular region [GO:0005576] | 27717682_the present study identified CCER2 as a novel susceptibility gene for MMD. CCER2 shows a brain-specific expression pattern, and its product has the structure of secreted proteins. | ENSMUSG00000096257 | Ccer2 | 13.49123 | 1.3073445 | 0.3866393679 | 0.79654501 | 2.359877e-01 | 6.271191e-01 | No | Yes | 12.899393 | 3.799665 | 10.170232 | 3.056349 | ||||
| ENSG00000264538 | SUZ12P1 | transcribed_unprocessed_pseudogene | 263.78417 | 1.0644695 | 0.0901345901 | 0.19033584 | 2.247318e-01 | 6.354579e-01 | 8.837671e-01 | No | Yes | 316.080431 | 38.171275 | 287.071158 | 33.984925 | |||||||||||
| ENSG00000265688 | 92659 | MAFG-DT | lncRNA | 30348529_suggested that MAFG-AS1 functions as a novel oncogenic lncRNA in the development of CRC by regulating miR-147b/NDUFA4 31002134_LncRNA MAFG-AS1 promotes the aggressiveness of breast carcinoma through regulating miR-339-5p/MMP15. 31211984_LncRNA MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR-744-5p/MAFG axis 32079456_MAFG-AS1 is a novel clinical biomarker for clinical progression and unfavorable prognosis in gastric cancer. 33336731_Long noncoding RNA MAFG-AS1 facilitates the progression of hepatocellular carcinoma via targeting miR-3196/OTX1 axis. 33367930_Regulatory effect of the MAFGAS1/miR1505p/MYB axis on the proliferation and migration of breast cancer cells. 33400245_LncRNA MAFG-AS1 regulates miR-125b-5p/SphK1 axis to promote the proliferation, migration, and invasion of bladder cancer cells. 33989902_LncRNA MAFG-AS1 affects the tumorigenesis of breast cancer cells via the miR-574-5p/SOD2 axis. | 31.42226 | 0.7165519 | -0.4808568404 | 0.50502324 | 8.985198e-01 | 3.431789e-01 | No | Yes | 37.133491 | 8.972220 | 49.528441 | 10.915811 | ||||||||||
| ENSG00000266053 | 101927275 | NDUFV2-AS1 | lncRNA | 12.57971 | 0.8674506 | -0.2051465309 | 0.78472606 | 6.782835e-02 | 7.945253e-01 | No | Yes | 12.982469 | 4.202258 | 14.942316 | 4.561227 | |||||||||||
| ENSG00000266865 | 107984974 | transcribed_unprocessed_pseudogene | 19291764_Observational study of gene-disease association. (HuGE Navigator) | 114.43968 | 1.5633559 | 0.6446462504 | 0.27478767 | 5.554660e+00 | 1.843156e-02 | 2.210178e-01 | No | Yes | 166.740359 | 26.713655 | 105.306134 | 16.589975 | ||||||||||
| ENSG00000266962 | 108783654 | HSD17B1-AS1 | lncRNA | 325.07542 | 0.5424256 | -0.8825027301 | 0.17808738 | 2.386591e+01 | 1.032845e-06 | 4.830145e-04 | No | Yes | 263.741049 | 39.470994 | 471.444941 | 68.129623 | ||||||||||
| ENSG00000269293 | 100129195 | ZSCAN16-AS1 | lncRNA | 34097562_ZSCAN16-AS1 expedites hepatocellular carcinoma progression via modulating the miR-181c-5p/SPAG9 axis to activate the JNK pathway. 34498716_Long noncoding RNA ZSCAN16AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA451a and consequently increasing ATF2 expression. | 345.53110 | 1.0639361 | 0.0894115675 | 0.16196307 | 3.053373e-01 | 5.805556e-01 | 8.598627e-01 | No | Yes | 411.890067 | 59.388651 | 363.067252 | 51.239589 | |||||||||
| ENSG00000269604 | lncRNA | 424.57036 | 0.7518883 | -0.4114096966 | 0.66934433 | 3.796038e-01 | 5.378154e-01 | 8.391324e-01 | No | Yes | 7.747874 | 3.409768 | 7.941358 | 3.153868 | ||||||||||||
| ENSG00000271254 | 102724250 | protein_coding | 2174.18958 | 0.8495370 | -0.2352513089 | 0.08843849 | 7.026148e+00 | 8.032798e-03 | 1.492097e-01 | No | Yes | 2443.344046 | 224.085800 | 2758.944004 | 247.069654 | |||||||||||
| ENSG00000273344 | 202781 | PAXIP1-DT | lncRNA | 31823805_LncRNA PAXIP1-AS1 was highly expressed in glioma and associated with poor prognosis. Highly expressed lncRNA PAXIP1-AS1 promoted glioma cell migration, invasion and angiogenesis by recruiting transcription factor ETS1 to upregulate KIF14 expression. However, its silencing inhibited migration, invasion and angiogenesis of glioma cells. 34108034_H3K27ac-induced lncRNA PAXIP1-AS1 promotes cell proliferation, migration, EMT and apoptosis in ovarian cancer by targeting miR-6744-5p/PCBP2 axis. 34245091_LncRNA PAXIP1-AS1 fosters the pathogenesis of pulmonary arterial hypertension via ETS1/WIPF1/RhoA axis. | 257.42108 | 0.6225791 | -0.6836710371 | 0.18445720 | 1.350049e+01 | 2.385007e-04 | 2.142399e-02 | No | Yes | 221.578668 | 25.057520 | 352.896794 | 38.181876 | |||||||||
| ENSG00000273748 | protein_coding | Mouse_homologues FUNCTION: Binds to mRNA in a sequence-independent manner. May play a role in regulation of pre-mRNA splicing or in the assembly of rRNA into ribosomal subunits. May be involved in mRNA transport. May be involved in epigenetic regulation of muscle differentiation through regulation of activity of the histone-lysine N-methyltransferase KMT5B. {ECO:0000269|PubMed:23720823}. | Mouse_homologues 3D-structure;Actin-binding;Cytoplasm;Myogenesis;Nucleus;RNA-binding;Reference proteome;Ribosome biogenesis;Spliceosome;mRNA processing;mRNA splicing;rRNA processing | Mouse_homologues mmu:14300; | Mouse_homologues Cajal body [GO:0015030]; catalytic step 2 spliceosome [GO:0071013]; nucleolus [GO:0005730]; striated muscle dense body [GO:0055120]; Z disc [GO:0030018]; actin filament binding [GO:0051015]; RNA binding [GO:0003723]; mRNA processing [GO:0006397]; muscle organ development [GO:0007517]; RNA splicing [GO:0008380]; rRNA processing [GO:0006364] | Mouse_homologues 23300487_our results suggest that a component of FSHD pathogenesis may arise by over-expression of FRG1, reducing Rbfox1 levels and leading to aberrant expression of an altered Calpain 3 protein through dysregulated splicing 23525014_transgenic overexpression results in a muscle-growth defect and a muscle regeneration impairment 23720823_This study suggests a novel role of FRG1 as epigenetic regulator of muscle differentiation and indicates that Suv4-20h1 has a gene-specific function in myogenesis. 24305066_On the basis of these results, it was proposed that aberrant fTnT represents a biological marker of muscle phenotype severity and disease progression. 25326393_Mouse Frg1 genomic area lacks DUX4 binding sites and DUX4 is unable to activate the endogenous mouse Frg1 gene providing a possible explanation for the lack of muscle phenotype in DUX4 transgenic mice. 27234941_FRG1 is involved in the morphogenesis of the tooth germ, as well as in the formation of enamel and dentin matrices and that FRG1 may play a role in the odontogenesis in the mouse following BMP4 stimulation. | ENSMUSG00000031590 | Frg1 | 60.04630 | 1.2351820 | 0.3047235787 | 0.36626683 | 6.953766e-01 | 4.043416e-01 | No | Yes | 75.755416 | 18.372917 | 60.291681 | 14.499851 | ||||||
| ENSG00000274253 | 283683 | lncRNA | 41.72021 | 0.8208368 | -0.2848327414 | 0.44746093 | 4.035597e-01 | 5.252566e-01 | No | Yes | 50.660384 | 13.059844 | 61.276672 | 14.941227 | ||||||||||||
| ENSG00000274290 | 8344 | H2BC6 | protein_coding | P62807 | FUNCTION: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.; FUNCTION: Has broad antibacterial activity. May contribute to the formation of the functional antimicrobial barrier of the colonic epithelium, and to the bactericidal activity of amniotic fluid. | 3D-structure;Acetylation;Antibiotic;Antimicrobial;Chromosome;DNA-binding;Direct protein sequencing;Glycoprotein;Hydroxylation;Isopeptide bond;Methylation;Nucleosome core;Nucleus;Phosphoprotein;Reference proteome;Ubl conjugation | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. The protein has antibacterial and antifungal antimicrobial activity. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H2B family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6. [provided by RefSeq, Aug 2015]. | hsa:3017;hsa:8339;hsa:8343;hsa:8344;hsa:8346;hsa:8347; | cytosol [GO:0005829]; extracellular exosome [GO:0070062]; extracellular space [GO:0005615]; nucleoplasm [GO:0005654]; nucleosome [GO:0000786]; nucleus [GO:0005634]; DNA binding [GO:0003677]; identical protein binding [GO:0042802]; protein heterodimerization activity [GO:0046982]; antibacterial humoral response [GO:0019731]; antimicrobial humoral immune response mediated by antimicrobial peptide [GO:0061844]; defense response to Gram-positive bacterium [GO:0050830]; innate immune response in mucosa [GO:0002227]; nucleosome assembly [GO:0006334] | 12860195_Histone H2B antimicrobial peptides participate in the colonic defense against bacteria and fungi. | ENSMUSG00000018102 | H2bc4 | 13.04903 | 1.3890961 | 0.4741464357 | 0.87220633 | 2.905871e-01 | 5.898446e-01 | No | Yes | 28.067632 | 19.404483 | 20.386810 | 13.549940 | ||
| ENSG00000278023 | 201299 | RDM1 | protein_coding | Q8NG50 | FUNCTION: May confer resistance to the antitumor agent cisplatin. Binds to DNA and RNA. {ECO:0000269|PubMed:15611051}. | Alternative promoter usage;Alternative splicing;Cytoplasm;DNA-binding;Nucleus;RNA-binding;Reference proteome | This gene encodes a protein involved in the cellular response to cisplatin, a drug commonly used in chemotherapy. The protein encoded by this gene contains two motifs: a motif found in RAD52, a protein that functions in DNA double-strand breaks and homologous recombination, and an RNA recognition motif (RRM) that is not found in RAD52. The RAD52 motif region in RAD52 is important for protein function and may be involved in DNA binding or oligomerization. Alternatively spliced transcript variants encoding different isoforms have been reported. [provided by RefSeq, Jul 2008]. | hsa:201299; | Cajal body [GO:0015030]; cytosol [GO:0005829]; nucleolus [GO:0005730]; PML body [GO:0016605]; DNA binding [GO:0003677]; RNA binding [GO:0003723] | 15611051_RDM1 recognizes DNA distortions induced by cisplatin-DNA adducts in vitro; possible role in spermatogenesis 17905820_The identification of 11 human cDNAs encoding RDM1 protein isoforms which is generated by alternative pre-mRNA splicing and differential usage of two translational start sites. 29845285_Due to its similarity to RAD52, we hypothesized that RDM1 potentially repairs DNA doublestrand breaks arising through DNA replication. 30069034_Knockdown of RDM1 in lung adenocarcinoma cells reduces cell proliferation and promotes apoptosis, consistent with the role RDM1 in the overexpression experiments. Xenograft mouse model shows stable knockdown of RDM1 significantly inhibits lung adenocarcinoma tumor growth. These in vitro and in vivo results conclude that RDM1 plays an oncogenic role in human lung adenocarcinoma. 31222930_RDM1 promotes critical processes in breast cancer tumorigenesis. 31670863_Loss of RDM1 enhances hepatocellular carcinoma progression via p53 and Ras/Raf/ERK pathways. 32501118_RDM1 plays an oncogenic role in human ovarian carcinoma cells. 34264012_Association of RDM1 with osteosarcoma progression via cell cycle and MEK/ERK signalling pathway regulation. 34435618_Correlation analysis of RDM1 gene with immune infiltration and clinical prognosis of hepatocellular carcinoma. | ENSMUSG00000010362 | Rdm1 | 45.02104 | 0.6112817 | -0.7100906856 | 0.44569198 | 2.485886e+00 | 1.148717e-01 | No | Yes | 38.074863 | 6.776502 | 60.323242 | 9.843247 | ||
| ENSG00000278619 | 79922 | MRM1 | protein_coding | Q6IN84 | FUNCTION: S-adenosyl-L-methionine-dependent 2'-O-ribose methyltransferase that catalyzes the formation of 2'-O-methylguanosine at position 1145 (Gm1145) in the 16S mitochondrial large subunit ribosomal RNA (mtLSU rRNA), a universally conserved modification in the peptidyl transferase domain of the mtLSU rRNA. {ECO:0000269|PubMed:25074936}. | Alternative splicing;Methyltransferase;Mitochondrion;Reference proteome;S-adenosyl-L-methionine;Transferase;Transit peptide;rRNA processing | hsa:79922; | mitochondrial matrix [GO:0005759]; mitochondrion [GO:0005739]; RNA binding [GO:0003723]; rRNA (guanosine-2'-O-)-methyltransferase activity [GO:0070039]; rRNA 2'-O-methylation [GO:0000451] | 20877624_Observational study of gene-disease association. (HuGE Navigator) 25074936_Data show that 2'-O-methyltransferases MRM1, MRM2, and RNMTL1 are responsible for modification of large subunit rRNA at residues G1145, U1369, and G1370, respectively. | ENSMUSG00000018405 | Mrm1 | 125.48470 | 0.6261165 | -0.6754970668 | 0.27933105 | 5.749839e+00 | 1.649017e-02 | 2.085954e-01 | No | Yes | 117.297164 | 18.767260 | 178.496470 | 27.263745 | ||
| ENSG00000278878 | lncRNA | 22.63747 | 9.2926819 | 3.2160950263 | 0.74339266 | 1.902015e+01 | 1.293451e-05 | No | Yes | 35.012477 | 47.549469 | 3.694588 | 5.124064 | |||||||||||||
| ENSG00000279968 | 100507462 | CCDC28A-AS1 | lncRNA | A0A096LPI5 | Reference proteome | 107.03545 | 0.7960720 | -0.3290291195 | 0.29024435 | 1.273075e+00 | 2.591905e-01 | 6.504694e-01 | No | Yes | 116.724435 | 21.849105 | 145.591659 | 26.321653 | ||||||||
| ENSG00000284642 | lncRNA | 19.55367 | 2.5873478 | 1.3714739798 | 0.64785145 | 4.507524e+00 | 3.374606e-02 | No | Yes | 28.812259 | 10.281643 | 11.693899 | 4.239909 | |||||||||||||
| ENSG00000286112 | 883 | protein_coding | A0A494C066 | Mouse_homologues FUNCTION: Catalyzes the irreversible transamination of the L-tryptophan metabolite L-kynurenine to form kynurenic acid (KA), an intermediate in the tryptophan catabolic pathway which is also a broad spectrum antagonist of the three ionotropic excitatory amino acid receptors among others. Metabolizes the cysteine conjugates of certain halogenated alkenes and alkanes to form reactive metabolites. Catalyzes the beta-elimination of S-conjugates and Se-conjugates of L-(seleno)cysteine, resulting in the cleavage of the C-S or C-Se bond. {ECO:0000250|UniProtKB:Q16773}. | Reference proteome | Mouse_homologues PATHWAY: Amino-acid degradation; L-kynurenine degradation; kynurenate from L-kynurenine: step 1/2. | Mouse_homologues mmu:70266; | cysteine-S-conjugate beta-lyase activity [GO:0047804]; kynurenine-oxoglutarate transaminase activity [GO:0016212]; L-glutamine aminotransferase activity [GO:0070548]; pyridoxal phosphate binding [GO:0030170]; biosynthetic process [GO:0009058]; cellular modified amino acid metabolic process [GO:0006575] | Mouse_homologues 20482848_KAT I inhibited by equimolar tryptophan and half-life is 69.7 minutes at 65 degree C. 28097769_One is the native structure with the cofactor in the PLP form bound to Lys247 with the highest resolution yet available for KAT-I at 1.28 A resolution, and the other with the general PLP-dependent aminotransferase inhibitor, aminooxyacetate (AOAA) covalently bound to the cofactor at 1.54 A. | ENSMUSG00000039648 | Kyat1 | 368.18130 | 0.8798861 | -0.1846112699 | 0.16875049 | 1.192007e+00 | 2.749250e-01 | 6.641583e-01 | No | Yes | 429.026876 | 64.663479 | 469.869041 | 68.968777 | ||
| ENSG00000286129 | lncRNA | 500.34918 | 1.0459182 | 0.0647700869 | 0.15060167 | 1.849716e-01 | 6.671351e-01 | 8.964683e-01 | No | Yes | 647.366197 | 93.991380 | 597.541757 | 84.906675 | ||||||||||||
| ENSG00000286219 | 100996717 | NOTCH2NLC | protein_coding | P0DPK4 | FUNCTION: Human-specific protein that promotes neural progenitor proliferation and evolutionary expansion of the brain neocortex by regulating the Notch signaling pathway (PubMed:29856954, PubMed:29856955, PubMed:29561261). Able to promote neural progenitor self-renewal, possibly by down-regulating neuronal differentiation genes, thereby delaying the differentiation of neuronal progenitors and leading to an overall final increase in neuronal production (PubMed:29856954). Acts by enhancing the Notch signaling pathway via two different mechanisms that probably work in parallel to reach the same effect (PubMed:29856954). Enhances Notch signaling pathway in a non-cell-autonomous manner via direct interaction with NOTCH2 (PubMed:29856954). Also promotes Notch signaling pathway in a cell-autonomous manner through inhibition of cis DLL1-NOTCH2 interactions, which promotes neuronal differentiation (By similarity). {ECO:0000250|UniProtKB:P0DPK3, ECO:0000269|PubMed:29561261, ECO:0000269|PubMed:29856954, ECO:0000269|PubMed:29856955}. | Calcium;Disulfide bond;EGF-like domain;Glycoprotein;Neurodegeneration;Notch signaling pathway;Reference proteome;Repeat;Secreted | hsa:388677; | extracellular region [GO:0005576]; calcium ion binding [GO:0005509]; cerebral cortex development [GO:0021987]; Notch signaling pathway [GO:0007219]; positive regulation of Notch signaling pathway [GO:0045747] | 29856954_the emergence of human-specific NOTCH2NL genes may have contributed to the rapid evolution of the larger human neocortex, accompanied by loss of genomic stability at the 1q21.1 locus and resulting recurrent neurodevelopmental disorders 31178126_Expansion of NOTCH2NLC GGC Repeat is associated with Neuronal Intranuclear Inclusion Disease-Related Disorders. 31332381_GGC repeat expansions in NOTCH2NLC gene is associated with neuronal intranuclear inclusion disease. 31819945_Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. 32039647_Heterozygous GGC repeat expansion of NOTCH2NLC in a patient with neuronal intranuclear inclusion disease and progressive retinal dystrophy. 32250060_Repeat expansion scanning of the NOTCH2NLC gene in patients with multiple system atrophy. 32330268_Evolution of Human Brain Size-Associated NOTCH2NL Genes Proceeds toward Reduced Protein Levels. 32495371_NOTCH2NLC GGC Repeat Expansions Are Associated with Sporadic Essential Tremor: Variable Disease Expressivity on Long-Term Follow-up. 32602554_Phenotypic bases of NOTCH2NLC GGC expansion positive neuronal intranuclear inclusion disease in a Southeast Asian cohort. 32768149_No genetic evidence for the involvement of GGC repeat expansions of the NOTCH2NLC gene in Chinese patients with multiple system atrophy. 32989102_Identification of GGC repeat expansion in the NOTCH2NLC gene in amyotrophic lateral sclerosis. 33239111_CGG expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy with neurological manifestations. 33693509_The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. 34392981_Analysis of NOTCH2NLC GGC repeat expansion in Taiwanese patients with amyotrophic lateral sclerosis. 34675106_GGC Repeat Expansion of NOTCH2NLC in Taiwanese Patients With Inherited Neuropathies. 34675123_NOTCH2NLC-related disorders: the widening spectrum and genotype-phenotype correlation. 34774111_Father-to-offspring transmission of extremely long NOTCH2NLC repeat expansions with contractions: genetic and epigenetic profiling with long-read sequencing. | 170.03901 | 1.1197759 | 0.1632100385 | 0.24346597 | 4.504425e-01 | 5.021249e-01 | 8.212400e-01 | No | Yes | 209.904046 | 47.925705 | 182.645119 | 40.961393 | ||||
| ENSG00000288061 | lncRNA | 13.78091 | 0.8438612 | -0.2449223607 | 0.85442582 | 8.086234e-02 | 7.761322e-01 | No | Yes | 12.364306 | 4.800200 | 13.831300 | 5.168187 | |||||||||||||
| ENSG00000288905 | 645166 | LSP1P5 | lncRNA | 33717069_Down-Regulation of LOC645166 in T Cells of Ankylosing Spondylitis Patients Promotes the NF-kappaB Signaling via Decreasingly Blocking Recruitment of the IKK Complex to K63-Linked Polyubiquitin Chains. | 120.63248 | 0.5188269 | -0.9466748594 | 0.39628555 | 5.491068e+00 | 1.911386e-02 | 2.248393e-01 | No | Yes | 94.800742 | 30.339575 | 175.227735 | 54.117256 |
- Note:
- The spreadsheet above contains the non-differential expressed genes but with significantly changes in alternative splicing (q < 0.05 and dif > 0.05). Please Click HERE to check all mapped genes in a Microsoft .excel file.
Gene Biotype
| Biotype | Amount of Genes |
|---|---|
| lncRNA | 57 |
| protein_coding | 412 |
| transcribed_processed_pseudogene | 2 |
| transcribed_unitary_pseudogene | 3 |
| transcribed_unprocessed_pseudogene | 16 |
MA plot
PCA plot
Co-expression Analysis
The co-expression pattern was determined using weighted gene co-expression network analysis (WGCNA) through WGCNA R software.
Choosing the soft-thresholding power: analysis of network topology
Quantifying module-trait associations
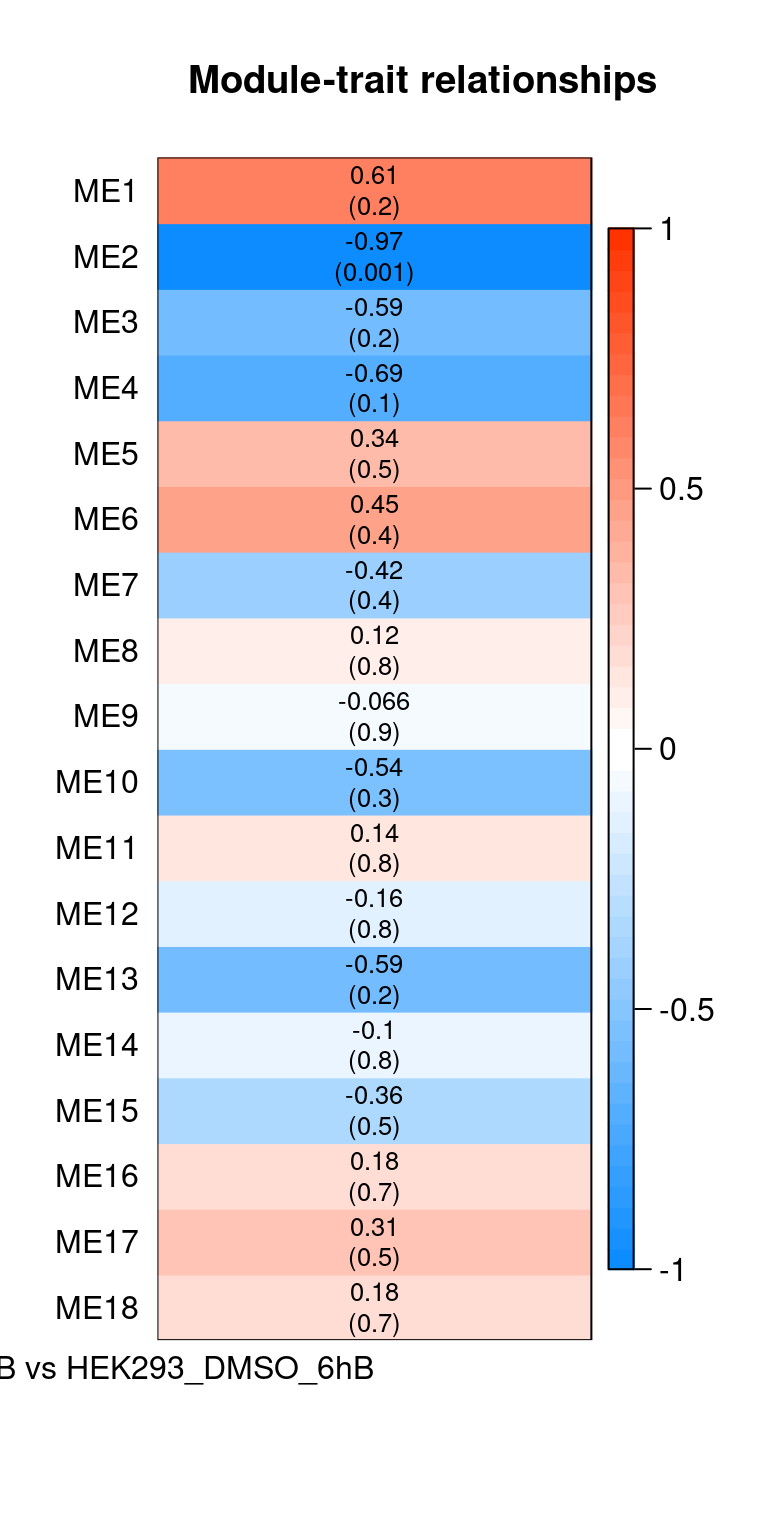
In the above figure, each cell contains the corresponding correlation (upper) and p-value (lower). ME, Module Eigengene.
Spreadsheet
| Cluster ID | Correlation | P-value | DEGs ID | DEGs Symbol |
|---|---|---|---|---|
| ME0 | 0.61406344 | 0.194678488 | ||
| ME1 | -0.97351513 | 0.001042884 | ENSG00000112238||ENSG00000180425 | PRDM13||C11orf71 |
| ME2 | -0.58650788 | 0.221115043 | ENSG00000139508||ENSG00000152495||ENSG00000177576||ENSG00000189319||ENSG00000204934 | SLC46A3||CAMK4||C18orf32||FAM53B||ATP6V0E2-AS1 |
| ME3 | -0.68765816 | 0.131100506 | ENSG00000287856 | ENSG00000287856 |
| ME4 | 0.34423036 | 0.504049163 | ENSG00000105472||ENSG00000173349||ENSG00000189343||ENSG00000196656||ENSG00000198618||ENSG00000213442||ENSG00000223459||ENSG00000227097||ENSG00000236552||ENSG00000244398||ENSG00000256861||ENSG00000262664||ENSG00000271122||ENSG00000274425||ENSG00000288825 | CLEC11A||SFT2D3||RPS2P46||ENSG00000196656||ENSG00000198618||RPL18AP3||TCAF1P1||ENSG00000227097||RPL13AP5||ENSG00000244398||ENSG00000256861||OVCA2||HERPUD2-AS1||ENSG00000274425||H2AC18 |
| ME5 | 0.44519126 | 0.376330509 | ||
| ME6 | -0.41658400 | 0.411271455 | ||
| ME7 | 0.11568338 | 0.827249002 | ||
| ME8 | -0.06557022 | 0.901785624 | ||
| ME9 | -0.53717835 | 0.271736723 | ||
| ME10 | 0.14453043 | 0.784713910 | ||
| ME11 | -0.15888525 | 0.763677613 | ||
| ME12 | -0.59415348 | 0.213643327 | ||
| ME13 | -0.10448878 | 0.843837234 | ||
| ME14 | -0.35745814 | 0.486650132 | ||
| ME15 | 0.17630511 | 0.738282426 | ||
| ME16 | 0.31285650 | 0.546026315 | ||
| ME17 | 0.18064382 | 0.731981665 |
Help
still working on
2. Functional Enrichment Analysis
2-1. Spreadsheet
GSEA
Please Click HERE to download a Microsoft .excel that contains all GSEA results.
GO: BP
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:1990034 | calcium ion export across plasma membrane | 11 | -0.7986693 | -1.749360 | 0.003639672 | 0.8500152 | 0.8500152 | 615 | tags=27%, list=4%, signal=26% | SLC24A4||SLC8A2||SLC8A3 | 6.6651610 | 7.241588 | 6.382194 | 7.348912 | 6.576512 | 6.4684691 | 7.290932 | 7.417196 | 6.7306058 | 6.8008648 | 6.4390964 | 7.264198 | 7.213316 | 7.246788 | 6.647977 | 6.1698874 | 6.2845465 | 7.343321 | 7.308636 | 7.393516 | 6.5244899 | 6.7192785 | 6.4739786 | 6.5162869 | 6.553502 | 6.325395 | 7.265276 | 7.291895 | 7.315192 | 7.406469 | 7.420341 | 7.424714 |
| GO:0051957 | positive regulation of amino acid transport | 11 | -0.7878255 | -1.725609 | 0.004549591 | 0.9329032 | 0.9329032 | 1733 | tags=45%, list=11%, signal=40% | CLTRN||ABAT||ADORA2A||SLC38A3||ACE2 | 2.9090090 | 5.408281 | 3.586965 | 5.473899 | 2.904229 | 2.9390303 | 5.365536 | 5.518412 | 2.9136675 | 3.1836199 | 2.5636238 | 5.442563 | 5.439268 | 5.340709 | 4.113829 | 3.0781007 | 3.3633743 | 5.376148 | 5.520362 | 5.520434 | 2.6868252 | 3.2557013 | 2.6933775 | 2.8679302 | 2.830348 | 3.103370 | 5.339431 | 5.386040 | 5.370747 | 5.515631 | 5.558790 | 5.479728 |
| GO:0018146 | keratan sulfate biosynthetic process | 10 | -0.7799611 | -1.669607 | 0.007407407 | 1.0000000 | 1.0000000 | 2782 | tags=70%, list=18%, signal=57% | B3GNT2||ST3GAL1||ST3GAL4||B4GAT1||ST3GAL2||B3GNT4||CHST2 | 3.4067077 | 3.646948 | 3.979309 | 3.793085 | 3.495614 | 3.9589339 | 3.470704 | 3.468574 | 3.3686526 | 3.3876119 | 3.4621558 | 3.676600 | 3.730906 | 3.525574 | 3.813892 | 4.1366547 | 3.9693127 | 3.783150 | 3.815674 | 3.780163 | 3.5032190 | 3.3547923 | 3.6169113 | 3.7940689 | 3.772555 | 4.256629 | 3.480266 | 3.478469 | 3.453219 | 3.492775 | 3.474048 | 3.438371 |
| GO:0060078 | regulation of postsynaptic membrane potential | 74 | -0.5041840 | -1.567885 | 0.007720145 | 1.0000000 | 1.0000000 | 2114 | tags=31%, list=14%, signal=27% | SNCA||MPP2||GRIN2D||CDK5||CACNB3||CHRNA3||GABRD||NPFF||ABAT||CELF4||ADORA1||CHRNB1||ADORA2A||TMEM108||ADRB1||GABRR2||SLC8A2||GABRG2||SLC8A3||KCND2||PTK2B||GRID2||CHRNA4 | 3.8813970 | 4.276818 | 4.184447 | 4.211195 | 3.865898 | 4.1860679 | 4.238316 | 4.161024 | 3.8094965 | 3.9196323 | 3.9124521 | 4.297933 | 4.300157 | 4.231309 | 4.215189 | 4.1898702 | 4.1474708 | 4.242362 | 4.179743 | 4.210799 | 3.8755280 | 3.7620250 | 3.9537282 | 4.0620471 | 4.146319 | 4.336041 | 4.288532 | 4.229522 | 4.195349 | 4.150459 | 4.203032 | 4.128562 |
| GO:0048485 | sympathetic nervous system development | 12 | 0.7352675 | 1.849987 | 0.008960573 | 1.0000000 | 1.0000000 | 1896 | tags=25%, list=12%, signal=22% | SEMA3A||HAND2||PLXNA4 | 2.9950074 | 4.786575 | 3.879255 | 4.979320 | 2.704889 | 2.6862845 | 4.731804 | 4.973569 | 3.1538129 | 3.3164746 | 2.3376459 | 4.839087 | 4.820751 | 4.695710 | 4.290297 | 3.2864828 | 3.8897290 | 4.953998 | 4.988728 | 4.994897 | 2.5347392 | 3.1068117 | 2.3647814 | 2.5997174 | 2.430968 | 2.973681 | 4.739785 | 4.734377 | 4.721187 | 4.964257 | 4.994659 | 4.961556 |
| GO:0007530 | sex determination | 10 | 0.7598631 | 1.815701 | 0.010505395 | 1.0000000 | 1.0000000 | 3355 | tags=70%, list=22%, signal=55% | SF1||WT1||SIX4||AR||MAP3K4||AMH||CITED2 | 4.7409975 | 5.252224 | 5.147007 | 4.941446 | 4.603774 | 4.9898807 | 5.393391 | 5.340233 | 4.5146325 | 4.8983617 | 4.7835949 | 5.211623 | 5.269993 | 5.274213 | 5.222133 | 5.2352625 | 4.9682473 | 4.927194 | 4.920195 | 4.976298 | 4.7058058 | 4.3604702 | 4.7173525 | 4.8495756 | 4.875058 | 5.215374 | 5.376414 | 5.422117 | 5.381203 | 5.303932 | 5.407861 | 5.306452 |
| GO:0034063 | stress granule assembly | 25 | 0.5924420 | 1.808394 | 0.011144131 | 1.0000000 | 1.0000000 | 4873 | tags=76%, list=31%, signal=52% | EIF2S1||PUM2||PRKAA1||UBAP2L||PRKAA2||YTHDF3||G3BP1||DDX6||OGFOD1||YTHDF1||LSM14A||RPS23||BICD1||YTHDF2||G3BP2||ATXN2||CSDE1||ATXN2L||PRRC2C | 7.5292508 | 8.022299 | 7.257309 | 7.950753 | 7.404750 | 7.0533535 | 8.086873 | 8.041586 | 7.6137712 | 7.7090919 | 7.2197228 | 8.029355 | 7.994919 | 8.042210 | 7.502620 | 7.0083378 | 7.2180511 | 7.938580 | 7.936070 | 7.977239 | 7.3324719 | 7.6829356 | 7.1466617 | 7.1798947 | 7.131088 | 6.823916 | 8.050641 | 8.089981 | 8.119178 | 8.024889 | 8.013669 | 8.085171 |
| GO:0006027 | glycosaminoglycan catabolic process | 15 | -0.6982178 | -1.632044 | 0.011415191 | 1.0000000 | 1.0000000 | 2479 | tags=47%, list=16%, signal=39% | HYAL3||HYAL2||FUCA1||NAGLU||IDUA||LYG1||CEMIP | 4.0137824 | 4.530559 | 4.661681 | 4.551562 | 4.055026 | 4.4766739 | 4.364090 | 4.433580 | 3.9151898 | 4.0385240 | 4.0824655 | 4.538596 | 4.566419 | 4.485492 | 4.684117 | 4.7208401 | 4.5761982 | 4.526718 | 4.544403 | 4.582989 | 4.0619160 | 3.9459780 | 4.1499540 | 4.2521273 | 4.237924 | 4.851470 | 4.411040 | 4.348300 | 4.331712 | 4.440804 | 4.472384 | 4.386238 |
| GO:0002825 | regulation of T-helper 1 type immune response | 13 | -0.7167904 | -1.629409 | 0.012191496 | 1.0000000 | 1.0000000 | 3161 | tags=46%, list=20%, signal=37% | IL1R1||JAK3||XCL1||IL27RA||IL4R||IL23A | 0.5547968 | 1.557257 | 0.901807 | 1.762889 | 0.486242 | 0.8964677 | 1.544606 | 1.688575 | 0.5444461 | 0.4912265 | 0.6255374 | 1.547607 | 1.575471 | 1.548520 | 1.021864 | 0.9847832 | 0.6741505 | 1.700269 | 1.741712 | 1.842930 | 0.4761798 | 0.4329185 | 0.5473085 | 0.8010477 | 0.853585 | 1.025140 | 1.541197 | 1.532608 | 1.559877 | 1.659623 | 1.757064 | 1.646489 |
| GO:0007076 | mitotic chromosome condensation | 16 | 0.6508781 | 1.779851 | 0.013685551 | 1.0000000 | 1.0000000 | 5107 | tags=88%, list=33%, signal=59% | SMC4||NCAPG||NUSAP1||SMC2||NCAPD2||NCAPH2||AKAP8||CDCA5||CHMP1A||NCAPD3||PLK1||NCAPH||PHF13||AKAP8L | 5.6646828 | 6.291218 | 5.909628 | 6.261600 | 5.675225 | 6.0277935 | 6.364196 | 6.443773 | 5.5687749 | 5.6682699 | 5.7512319 | 6.286522 | 6.290879 | 6.296237 | 6.000014 | 5.9033161 | 5.8199144 | 6.198174 | 6.232180 | 6.350008 | 5.6997609 | 5.5461159 | 5.7707472 | 5.9840216 | 6.026191 | 6.071831 | 6.377169 | 6.340158 | 6.374964 | 6.419765 | 6.455882 | 6.455377 |
| GO:0007064 | mitotic sister chromatid cohesion | 26 | 0.5693404 | 1.753406 | 0.014595808 | 1.0000000 | 1.0000000 | 5304 | tags=77%, list=34%, signal=51% | REC8||SLF1||SGO1||SMC3||POGZ||NIPBL||TNKS||RAD21||RB1||CDCA5||NSMCE2||SLF2||SMC1A||PDS5A||CHTF8||CDC20||NAA50||SMC5||ATRX||DSCC1 | 5.9600429 | 6.265329 | 6.017326 | 6.216750 | 5.967954 | 6.1230907 | 6.289773 | 6.277990 | 5.9062347 | 5.9541512 | 6.0175772 | 6.281714 | 6.250790 | 6.263316 | 6.054382 | 5.9787563 | 6.0178479 | 6.196582 | 6.169404 | 6.281862 | 5.9626626 | 5.8948291 | 6.0425745 | 6.1137282 | 6.163258 | 6.091345 | 6.306623 | 6.274240 | 6.288274 | 6.280133 | 6.291948 | 6.261729 |
| GO:0048024 | regulation of mRNA splicing, via spliceosome | 90 | 0.3961901 | 1.540072 | 0.015829319 | 1.0000000 | 1.0000000 | 5333 | tags=61%, list=34%, signal=40% | SF1||RBM39||RBM15||NOVA1||RBM11||RBM5||SON||QKI||MBNL2||MBNL1||FXR1||SRSF10||HNRNPL||JMJD6||RBM3||DDX17||TRA2B||PRPF19||RBM4||MAGOH||PTBP1||SAP18||SMU1||DYRK1A||FAM172A||SF3B4||SNW1||CELF2||RNPS1||RBM10||TRA2A||WTAP||SRSF3||RBM25||PRDX6||HNRNPK||MBNL3||CELF1||PUF60||FMR1||REST||THRAP3||NPM1||METTL16||YTHDC1||RBMXL1||KHDRBS3||SNRNP70||HNRNPU||SRPK2||RAVER2||C1QBP||RBMX||RBFOX2||CWC22 | 7.1656067 | 8.039778 | 7.244578 | 7.975197 | 7.046136 | 7.1121120 | 8.097716 | 8.045230 | 7.1812260 | 7.3192106 | 6.9758770 | 8.065282 | 8.012784 | 8.040791 | 7.551628 | 7.0084137 | 7.1145666 | 7.937104 | 7.946686 | 8.039566 | 7.0164650 | 7.1223922 | 6.9963460 | 7.1070534 | 7.063023 | 7.164464 | 8.077081 | 8.091805 | 8.123864 | 8.027770 | 8.050243 | 8.057511 |
| GO:0035988 | chondrocyte proliferation | 17 | 0.6298977 | 1.747446 | 0.015955715 | 1.0000000 | 1.0000000 | 3034 | tags=41%, list=20%, signal=33% | MMP14||COMP||SIRT6||FGFR3||DDR2||HMGA2||LEF1 | 2.9994653 | 3.586284 | 3.566466 | 3.457361 | 3.035085 | 3.4202907 | 3.648236 | 3.533197 | 2.9107225 | 2.9519726 | 3.1264932 | 3.539619 | 3.609303 | 3.608821 | 3.637970 | 3.5922090 | 3.4636056 | 3.453368 | 3.470731 | 3.447885 | 3.0138249 | 2.8454251 | 3.2213510 | 3.1890539 | 3.315121 | 3.705279 | 3.717786 | 3.606534 | 3.617769 | 3.493567 | 3.552605 | 3.552616 |
| GO:0007099 | centriole replication | 38 | 0.5199157 | 1.737802 | 0.016311167 | 1.0000000 | 1.0000000 | 2864 | tags=39%, list=18%, signal=32% | PLK4||PLK2||CEP44||CEP152||CEP192||CEP76||POC1B||VPS4B||CDK2||ENSG00000285943||CEP135||SASS6||CCP110||STIL||WDR62 | 5.1456763 | 6.866757 | 5.430091 | 6.933362 | 5.059908 | 5.3495143 | 6.960522 | 7.006498 | 5.1909929 | 5.1965228 | 5.0444153 | 6.888849 | 6.807592 | 6.902030 | 6.012111 | 5.1157082 | 4.9131703 | 6.869387 | 6.904609 | 7.021649 | 4.9528885 | 5.1154248 | 5.1057214 | 5.2034369 | 5.218590 | 5.592025 | 6.910466 | 6.947649 | 7.021239 | 6.995873 | 6.985949 | 7.037159 |
| GO:0006271 | DNA strand elongation involved in DNA replication | 15 | 0.6525256 | 1.748124 | 0.016408962 | 1.0000000 | 1.0000000 | 5040 | tags=93%, list=32%, signal=63% | LIG3||MCM4||POLD3||RFC3||PCNA||LIG1||MCM7||MCM3||POLD2||POLE||POLE3||RFC4||DNA2||POLA1 | 6.5573221 | 7.119196 | 6.760493 | 7.076926 | 6.522053 | 6.6151727 | 7.156942 | 7.238074 | 6.5501961 | 6.5856019 | 6.5357108 | 7.149524 | 7.110479 | 7.097069 | 6.912848 | 6.6612486 | 6.6942094 | 7.072899 | 7.024824 | 7.131091 | 6.4944436 | 6.5385526 | 6.5327667 | 6.5301324 | 6.572463 | 6.734744 | 7.177664 | 7.138078 | 7.154810 | 7.203276 | 7.287248 | 7.222348 |
| GO:0051383 | kinetochore organization | 23 | 0.5778336 | 1.730951 | 0.017316017 | 1.0000000 | 1.0000000 | 3003 | tags=52%, list=19%, signal=42% | SMC4||CENPC||SENP6||SMC2||CENPK||RNF4||NUF2||NDC80||POGZ||CENPN||CENPA||DLGAP5 | 5.2659948 | 5.593190 | 5.419495 | 5.759069 | 5.291712 | 5.4992459 | 5.665301 | 5.864575 | 5.1983685 | 5.2048557 | 5.3867266 | 5.609066 | 5.577635 | 5.592697 | 5.610676 | 5.3314378 | 5.2952951 | 5.675063 | 5.739840 | 5.856417 | 5.2263732 | 5.2182084 | 5.4212601 | 5.3576833 | 5.622909 | 5.504956 | 5.672576 | 5.662791 | 5.660507 | 5.824036 | 5.899352 | 5.869342 |
| GO:0032722 | positive regulation of chemokine production | 32 | -0.5726009 | -1.558182 | 0.017480167 | 1.0000000 | 1.0000000 | 3237 | tags=50%, list=21%, signal=40% | C5||MYD88||MAVS||TWIST1||F2RL1||SYK||LRP1||IL17RA||IL6R||IL4R||TIRAP||TSLP||HMOX1||TICAM1||TRPV4||MBP | 4.2956515 | 5.733222 | 4.602825 | 5.744550 | 4.113300 | 4.1671173 | 5.794851 | 5.736697 | 4.3537218 | 4.4931037 | 3.9956581 | 5.711262 | 5.736782 | 5.751337 | 5.037174 | 4.2590844 | 4.3864134 | 5.692426 | 5.747544 | 5.791958 | 4.0543581 | 4.2909164 | 3.9756829 | 4.1018302 | 3.986421 | 4.383688 | 5.795019 | 5.789303 | 5.800210 | 5.759411 | 5.690540 | 5.759056 |
| GO:0014850 | response to muscle activity | 19 | 0.6081581 | 1.735350 | 0.018493611 | 1.0000000 | 1.0000000 | 2458 | tags=37%, list=16%, signal=31% | RYR2||TNS2||CAPN3||FNDC5||PPARGC1A||PRKAA2||HIF1A | 6.1904119 | 6.439361 | 5.996732 | 6.297236 | 6.093322 | 5.9343950 | 6.416367 | 6.296164 | 6.2601213 | 6.2739341 | 6.0237359 | 6.460268 | 6.435079 | 6.422478 | 6.052079 | 5.8923758 | 6.0403146 | 6.291790 | 6.273976 | 6.325468 | 6.0865103 | 6.1695105 | 6.0200523 | 6.0042450 | 5.965005 | 5.828066 | 6.423612 | 6.402938 | 6.422458 | 6.292042 | 6.301816 | 6.294617 |
| GO:0060986 | endocrine hormone secretion | 27 | -0.5923554 | -1.561725 | 0.019484943 | 1.0000000 | 1.0000000 | 1743 | tags=26%, list=11%, signal=23% | GDF9||KCNQ1||C1QTNF1||C1QTNF3||AGTR1||FOXD1||NKX3-1 | 3.9980776 | 4.079532 | 3.924897 | 4.063401 | 4.013002 | 4.1880532 | 4.063303 | 4.054504 | 3.9771649 | 3.8656514 | 4.1383264 | 4.094774 | 4.051597 | 4.091825 | 3.866383 | 4.0315732 | 3.8705093 | 4.047722 | 4.032037 | 4.109284 | 4.0501868 | 3.8402933 | 4.1329759 | 4.1098153 | 4.279726 | 4.169440 | 4.092310 | 4.048969 | 4.048187 | 4.028456 | 4.091491 | 4.042806 |
| GO:1903307 | positive regulation of regulated secretory pathway | 36 | -0.5556359 | -1.547479 | 0.020235419 | 1.0000000 | 1.0000000 | 2994 | tags=39%, list=19%, signal=31% | STX4||SPHK2||ITGB2||HYAL3||F2RL1||RAB3A||SYK||CDK5||RAB27A||CACNA1G||IL4R||HLA-F||KCNB1||SLC4A8 | 5.1096231 | 5.039774 | 5.203486 | 4.867244 | 5.187203 | 5.3839139 | 4.985123 | 4.877288 | 5.0470528 | 5.1350253 | 5.1447967 | 5.043836 | 5.056774 | 5.018451 | 5.080275 | 5.3122415 | 5.2086131 | 4.919368 | 4.772446 | 4.905410 | 5.2276500 | 5.0562368 | 5.2690170 | 5.3942058 | 5.330889 | 5.425055 | 5.006577 | 4.971429 | 4.977117 | 4.861986 | 4.913754 | 4.855414 |
| GO:0033260 | nuclear DNA replication | 35 | 0.5090208 | 1.671902 | 0.020316944 | 1.0000000 | 1.0000000 | 5289 | tags=77%, list=34%, signal=51% | WRN||BLM||MCM4||GMNN||CDC45||DACH1||PCNA||DBF4B||INO80||LIG1||RAD51||GINS3||MCM3||FEN1||TERF1||DBF4||RTF2||TIPIN||MCM6||TERF2||DNA2||RTEL1||DONSON||POLA1||ATRX||WIZ||ZNF830 | 5.3815379 | 6.289058 | 5.670457 | 6.324648 | 5.310509 | 5.4828976 | 6.334856 | 6.452110 | 5.3955215 | 5.4770931 | 5.2640380 | 6.293688 | 6.292120 | 6.281337 | 5.905375 | 5.5088340 | 5.5644260 | 6.286712 | 6.300129 | 6.385116 | 5.2815797 | 5.3578763 | 5.2908631 | 5.4056523 | 5.424849 | 5.609305 | 6.337894 | 6.331665 | 6.335001 | 6.423053 | 6.484746 | 6.447863 |
| GO:0042996 | regulation of Golgi to plasma membrane protein transport | 11 | 0.7038763 | 1.720956 | 0.021126761 | 1.0000000 | 1.0000000 | 1121 | tags=27%, list=7%, signal=25% | RSC1A1||PKDCC||ENSG00000261832 | 9.0737713 | 8.557521 | 8.859841 | 8.503891 | 9.136981 | 9.1385807 | 8.522043 | 8.501808 | 9.0146122 | 8.9953544 | 9.2021707 | 8.601061 | 8.551185 | 8.519135 | 8.621458 | 8.9602931 | 8.9711338 | 8.568863 | 8.447316 | 8.492872 | 9.1461432 | 9.0782076 | 9.1845869 | 9.2100665 | 9.246102 | 8.940692 | 8.515292 | 8.535490 | 8.515252 | 8.498172 | 8.502998 | 8.504246 |
| GO:0051028 | mRNA transport | 120 | 0.3531696 | 1.426915 | 0.022052587 | 1.0000000 | 1.0000000 | 4689 | tags=48%, list=30%, signal=34% | PARP11||CHTOP||NXT2||NUP43||NPIPA1||NUP58||NUP88||RANBP17||QKI||XPO1||NUP54||IWS1||THOC1||LRPPRC||RAE1||SMG1||NXF1||NUP160||SRSF1||POLDIP3||ZFP36L1||EIF5AL1||NSUN2||EIF5A2||NUP155||MAGOH||IGF2BP1||NUP50||NUP205||TNKS||NCBP2||NCBP3||AGFG1||NUP107||ALYREF||IGF2BP3||SMG5||UPF3B||SRSF3||UPF2||THOC2||NUP133||EIF5A||G3BP2||AAAS||SLBP||THOC5||DDX39B||DDX19B||FMR1||HNRNPA3||YTHDC1||CASC3||NUP93||NUP85||HNRNPA2B1||KHSRP||ENY2 | 6.4739734 | 7.049274 | 6.525850 | 7.031947 | 6.429118 | 6.5366900 | 7.102729 | 7.095342 | 6.4399724 | 6.5701710 | 6.4065481 | 7.051340 | 7.042956 | 7.053504 | 6.725670 | 6.3993536 | 6.4294928 | 7.009292 | 7.002819 | 7.082371 | 6.4330208 | 6.4301928 | 6.4241271 | 6.5459406 | 6.522100 | 6.541916 | 7.096645 | 7.097062 | 7.114408 | 7.074459 | 7.112280 | 7.099030 |
| GO:0050829 | defense response to Gram-negative bacterium | 18 | -0.6437659 | -1.564553 | 0.022439586 | 1.0000000 | 1.0000000 | 2398 | tags=33%, list=15%, signal=28% | F2RL1||IL6R||OPTN||AQP1||MR1||IL23A | 8.8050576 | 8.614598 | 8.582618 | 8.447206 | 8.744122 | 8.3563570 | 8.569548 | 8.476174 | 8.8948604 | 8.8711001 | 8.6352062 | 8.678561 | 8.587272 | 8.575745 | 8.539117 | 8.5245470 | 8.6790899 | 8.494224 | 8.406488 | 8.439542 | 8.7410350 | 8.8759928 | 8.6023797 | 8.4400298 | 8.400551 | 8.218996 | 8.588794 | 8.548657 | 8.570914 | 8.462218 | 8.467096 | 8.498931 |
| GO:0010613 | positive regulation of cardiac muscle hypertrophy | 22 | 0.5628267 | 1.671723 | 0.022614841 | 1.0000000 | 1.0000000 | 4053 | tags=64%, list=26%, signal=47% | TRPC3||NR4A3||HAND2||IL6ST||PDE9A||MTPN||PARP2||MEF2A||TWF1||ROCK2||ROCK1||AKAP6||PPP3CA||DDX39B | 6.0291292 | 6.280905 | 6.156153 | 6.204238 | 6.005250 | 6.1000362 | 6.316292 | 6.258275 | 5.9867916 | 6.1213146 | 5.9746412 | 6.268901 | 6.299247 | 6.274385 | 6.296481 | 6.0748782 | 6.0861278 | 6.178439 | 6.166056 | 6.266143 | 6.0429573 | 5.9904190 | 5.9816081 | 6.1049173 | 6.066347 | 6.128169 | 6.321647 | 6.307335 | 6.319852 | 6.250156 | 6.290827 | 6.233234 |
| GO:0030206 | chondroitin sulfate biosynthetic process | 16 | -0.6650560 | -1.573235 | 0.023323615 | 1.0000000 | 1.0000000 | 3177 | tags=50%, list=20%, signal=40% | CHST12||CHST9||CHPF2||CHSY3||CHSY1||CHST7||CHPF||B3GALT6 | 3.4926093 | 3.918022 | 3.925829 | 3.590793 | 3.536821 | 3.9376567 | 3.734080 | 3.344925 | 3.4443779 | 3.4085808 | 3.6162214 | 3.911567 | 4.006123 | 3.831046 | 3.805640 | 4.0787426 | 3.8790892 | 3.617147 | 3.578204 | 3.576661 | 3.5273324 | 3.3624446 | 3.7008404 | 3.6795843 | 3.873339 | 4.209558 | 3.761449 | 3.755633 | 3.683873 | 3.367628 | 3.402405 | 3.261008 |
| GO:0099022 | vesicle tethering | 11 | 0.6966275 | 1.703233 | 0.024061033 | 1.0000000 | 1.0000000 | 2520 | tags=55%, list=16%, signal=46% | LINC00869||WDR11||TBC1D23||EXOC6||C17orf75||FAM91A1 | 2.8141027 | 4.606267 | 3.583841 | 4.709654 | 2.814847 | 3.1595219 | 4.656521 | 4.789366 | 2.7874601 | 2.9597687 | 2.6812980 | 4.602479 | 4.662618 | 4.551560 | 3.980864 | 3.2768369 | 3.3910204 | 4.627847 | 4.718752 | 4.778392 | 2.7519807 | 2.9415384 | 2.7421397 | 2.9310559 | 3.004597 | 3.479462 | 4.654700 | 4.644942 | 4.669812 | 4.790796 | 4.835130 | 4.740622 |
| GO:0033962 | P-body assembly | 20 | 0.5840166 | 1.688199 | 0.024381868 | 1.0000000 | 1.0000000 | 4324 | tags=60%, list=28%, signal=43% | RC3H1||CNOT6||PAN3||LIMD1||CNOT6L||CNOT2||DDX6||CNOT7||LSM14A||LSM3||EDC3||ATXN2 | 5.6182892 | 6.033687 | 5.784452 | 6.132414 | 5.564360 | 5.7250222 | 6.095618 | 6.204257 | 5.6165218 | 5.6538179 | 5.5836739 | 6.003792 | 6.063675 | 6.032972 | 5.982949 | 5.6821336 | 5.6656687 | 6.119008 | 6.096421 | 6.180497 | 5.5703189 | 5.5000181 | 5.6202203 | 5.7529973 | 5.726854 | 5.694623 | 6.107348 | 6.095309 | 6.084104 | 6.160873 | 6.198419 | 6.252023 |
| GO:0006596 | polyamine biosynthetic process | 13 | -0.6847771 | -1.556636 | 0.024531668 | 1.0000000 | 1.0000000 | 429 | tags=15%, list=3%, signal=15% | OAZ3||PAOX | 7.5992110 | 7.335284 | 7.425664 | 7.207405 | 7.610389 | 7.6131673 | 7.304982 | 7.254508 | 7.6006660 | 7.5586781 | 7.6372190 | 7.416707 | 7.288018 | 7.297530 | 7.276287 | 7.4851569 | 7.5046310 | 7.265688 | 7.122744 | 7.229976 | 7.6228604 | 7.5308215 | 7.6738592 | 7.6285441 | 7.699078 | 7.505250 | 7.313231 | 7.277337 | 7.323965 | 7.239272 | 7.272471 | 7.251586 |
| GO:0099637 | neurotransmitter receptor transport | 22 | -0.6110563 | -1.551719 | 0.024539877 | 1.0000000 | 1.0000000 | 3345 | tags=41%, list=22%, signal=32% | ARHGAP44||STX3||CACNG5||GRIP1||MAPK10||CACNG8||CACNG4||SLC1A1||TAMALIN | 4.1433402 | 4.562925 | 4.560812 | 4.557680 | 4.123763 | 4.6013805 | 4.572644 | 4.530668 | 4.0851546 | 4.1201945 | 4.2211903 | 4.518955 | 4.593101 | 4.575682 | 4.613564 | 4.6085034 | 4.4547924 | 4.567464 | 4.520123 | 4.584682 | 4.1190865 | 3.9898663 | 4.2505649 | 4.4325739 | 4.558628 | 4.789934 | 4.584319 | 4.585158 | 4.548146 | 4.522773 | 4.572541 | 4.495632 |
| GO:1900452 | regulation of long-term synaptic depression | 10 | -0.7267130 | -1.555623 | 0.025308642 | 1.0000000 | 1.0000000 | 1429 | tags=40%, list=9%, signal=36% | ADORA1||ARC||KCNB1||GRID2 | 1.2737551 | 2.612886 | 1.998438 | 2.549872 | 1.331991 | 1.9327167 | 2.668732 | 2.641678 | 1.2227788 | 1.2506871 | 1.3449405 | 2.527586 | 2.649282 | 2.658139 | 2.277661 | 1.9584667 | 1.7011547 | 2.510345 | 2.565923 | 2.572543 | 1.2991960 | 1.1589815 | 1.5152989 | 1.7955042 | 1.912256 | 2.076512 | 2.673799 | 2.668377 | 2.664003 | 2.609829 | 2.727751 | 2.583321 |
| GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | 13 | -0.6834436 | -1.553605 | 0.025423729 | 1.0000000 | 1.0000000 | 3272 | tags=54%, list=21%, signal=43% | CYBA||ATP6AP2||F2RL1||CTSZ||AGTR1||ACE2||ENPEP | 6.1903116 | 5.932224 | 6.157935 | 5.952805 | 6.154961 | 6.2230922 | 5.814268 | 5.869268 | 6.1987561 | 6.1622978 | 6.2094583 | 6.014306 | 5.945507 | 5.830944 | 6.063838 | 6.2249423 | 6.1802662 | 6.016418 | 5.898257 | 5.941252 | 6.1794931 | 6.1025164 | 6.1814783 | 6.1632801 | 6.218893 | 6.284547 | 5.841346 | 5.794260 | 5.806785 | 5.833162 | 5.883941 | 5.890029 |
| GO:1903793 | positive regulation of anion transport | 20 | -0.6196057 | -1.543170 | 0.025669958 | 1.0000000 | 1.0000000 | 1733 | tags=35%, list=11%, signal=31% | CLTRN||ABAT||CEBPB||ADORA2A||SLC38A3||ABCB1||ACE2 | 2.6012212 | 4.753430 | 3.221375 | 4.833064 | 2.570744 | 2.8142161 | 4.715480 | 4.867553 | 2.5762331 | 2.8135200 | 2.3813441 | 4.780566 | 4.779886 | 4.698297 | 3.680454 | 2.8391906 | 3.0005260 | 4.753157 | 4.855472 | 4.887188 | 2.4591082 | 2.7528082 | 2.4814078 | 2.7378330 | 2.755477 | 2.940481 | 4.696579 | 4.727334 | 4.722337 | 4.860586 | 4.905174 | 4.836047 |
| GO:0003214 | cardiac left ventricle morphogenesis | 10 | 0.7038458 | 1.681847 | 0.026121522 | 1.0000000 | 1.0000000 | 2528 | tags=40%, list=16%, signal=34% | RYR2||FOXF1||RBPJ||SMAD4 | 3.7331407 | 4.560519 | 4.132027 | 4.373072 | 3.644269 | 4.1160543 | 4.568263 | 4.420967 | 3.6639907 | 3.8167928 | 3.7144137 | 4.525967 | 4.545982 | 4.608323 | 4.155004 | 4.1893971 | 4.0479360 | 4.324832 | 4.353989 | 4.437990 | 3.6018433 | 3.5353784 | 3.7839718 | 3.9277503 | 4.110433 | 4.287584 | 4.581568 | 4.592781 | 4.529658 | 4.424946 | 4.453105 | 4.384014 |
| GO:0070601 | centromeric sister chromatid cohesion | 10 | 0.7045278 | 1.683477 | 0.026121522 | 1.0000000 | 1.0000000 | 2451 | tags=50%, list=16%, signal=42% | AXIN2||BUB1B||SGO1||SGO2||BUB1 | 6.5853419 | 6.978778 | 6.580504 | 6.938714 | 6.564130 | 6.6419474 | 6.966043 | 6.985288 | 6.5605179 | 6.5612305 | 6.6330664 | 7.023087 | 6.960860 | 6.951329 | 6.688091 | 6.4469309 | 6.5962935 | 6.928569 | 6.897085 | 6.988973 | 6.5445201 | 6.5029712 | 6.6413827 | 6.6257637 | 6.777710 | 6.509815 | 6.984526 | 6.946103 | 6.967243 | 6.978537 | 6.999142 | 6.978085 |
| GO:0016078 | tRNA catabolic process | 12 | 0.6745129 | 1.697124 | 0.026284349 | 1.0000000 | 1.0000000 | 2961 | tags=50%, list=19%, signal=40% | TRDMT1||EXOSC10||NSUN2||DICER1||EXOSC3||EXOSC9 | 5.2130069 | 6.023197 | 5.626457 | 6.054610 | 5.088317 | 5.2673128 | 6.091714 | 6.131886 | 5.2113747 | 5.4064001 | 4.9915196 | 6.030506 | 6.001074 | 6.037751 | 5.894698 | 5.4546074 | 5.4871540 | 6.019562 | 6.034281 | 6.108406 | 5.0953964 | 5.1817452 | 4.9807906 | 5.1966105 | 5.006404 | 5.546206 | 6.094047 | 6.107864 | 6.073019 | 6.099011 | 6.163346 | 6.132583 |
| GO:0032508 | DNA duplex unwinding | 79 | 0.3902115 | 1.488500 | 0.027500000 | 1.0000000 | 1.0000000 | 5351 | tags=57%, list=34%, signal=38% | WRN||BLM||RAD54L2||MCM4||XRCC6||GTF2H2||RFC3||NBN||FANCM||G3BP1||TOP2A||MRE11||CHTF18||CHD7||MCM5||SMARCAD1||MCM7||RAD51||XRCC5||GTF2F2||MCM3||HELB||CHD5||DHX36||CHTF8||POLQ||RECQL||RFC5||BRIP1||DDX1||MCM6||MCM8||RFC2||RFC4||RAD54B||MCM9||DNA2||SUPV3L1||RTEL1||ATRX||DHX9||ASCC3||SETX||DSCC1||ERCC3 | 5.7150154 | 6.296827 | 5.965979 | 6.390009 | 5.628502 | 5.7452820 | 6.352478 | 6.470666 | 5.7146549 | 5.7973981 | 5.6280251 | 6.297578 | 6.297149 | 6.295754 | 6.184883 | 5.8095480 | 5.8750354 | 6.357245 | 6.354408 | 6.456033 | 5.6146488 | 5.6673785 | 5.6026547 | 5.6926181 | 5.692751 | 5.845052 | 6.349668 | 6.345301 | 6.362410 | 6.457546 | 6.484931 | 6.469389 |
| GO:0006270 | DNA replication initiation | 33 | 0.4943736 | 1.612539 | 0.027788650 | 1.0000000 | 1.0000000 | 3666 | tags=48%, list=24%, signal=37% | CDC6||MCM4||CDC45||NBN||KAT7||ORC4||CCNE2||ORC6||TOPBP1||MCM5||MCM7||GINS3||ORC1||MCM3||NOC3L||ORC3 | 6.0943693 | 6.285185 | 6.227052 | 6.348315 | 6.064810 | 6.2049725 | 6.304822 | 6.448649 | 6.0331661 | 6.1250871 | 6.1229603 | 6.303349 | 6.289092 | 6.262822 | 6.254891 | 6.2059075 | 6.2199160 | 6.360778 | 6.294089 | 6.388455 | 6.0910519 | 5.9952866 | 6.1056208 | 6.1540341 | 6.184700 | 6.273499 | 6.335659 | 6.271566 | 6.306527 | 6.427767 | 6.477342 | 6.440375 |
| GO:0035404 | histone-serine phosphorylation | 10 | 0.6941897 | 1.658774 | 0.029244747 | 1.0000000 | 1.0000000 | 4176 | tags=80%, list=27%, signal=59% | PRKAA1||PRKAA2||CCNB1||HMGA2||AURKA||AURKB||RPS6KA5||MST1 | 5.2602681 | 6.000843 | 5.303930 | 5.943545 | 5.167733 | 5.2503135 | 6.069035 | 6.056775 | 5.2744158 | 5.3118661 | 5.1919255 | 6.080293 | 5.973714 | 5.944972 | 5.540303 | 5.1463044 | 5.1921925 | 5.934980 | 5.910922 | 5.983777 | 5.1059654 | 5.2326240 | 5.1618110 | 5.2016212 | 5.223521 | 5.322883 | 6.062376 | 6.066208 | 6.078470 | 6.034802 | 6.066634 | 6.068641 |
| GO:0051489 | regulation of filopodium assembly | 41 | 0.4626819 | 1.569876 | 0.029565217 | 1.0000000 | 1.0000000 | 4883 | tags=63%, list=31%, signal=44% | NEURL1||TGFB3||GAP43||DMTN||ENSG00000261832||FSCN1||PIK3R1||FXR1||RAB5A||RALA||WASL||FNBP1L||SRF||MYO10||SRGAP2C||GPM6A||PPP1R9A||CDC42||PPP1R16B||DOCK11||FMR1||NLGN1||TENM1||ABITRAM||ARF6||PRKCD | 4.2015909 | 4.794496 | 4.370757 | 4.810671 | 4.206832 | 4.3858641 | 4.845104 | 4.885206 | 4.1460941 | 4.2594234 | 4.1970193 | 4.784705 | 4.800045 | 4.798687 | 4.496519 | 4.2789094 | 4.3276988 | 4.782953 | 4.782448 | 4.865037 | 4.2040768 | 4.1686096 | 4.2467476 | 4.3486626 | 4.358149 | 4.448654 | 4.841515 | 4.834766 | 4.858924 | 4.870349 | 4.898869 | 4.886257 |
| GO:0001956 | positive regulation of neurotransmitter secretion | 14 | -0.6656844 | -1.534821 | 0.029698582 | 1.0000000 | 1.0000000 | 2114 | tags=36%, list=14%, signal=31% | SNCA||CDK5||ADORA2A||BAIAP3||SLC4A8 | 3.5668401 | 4.031206 | 3.882486 | 4.120635 | 3.554696 | 3.8571438 | 4.035107 | 4.021740 | 3.5296195 | 3.5971639 | 3.5729272 | 4.014908 | 4.042162 | 4.036406 | 3.946345 | 3.8310621 | 3.8676354 | 4.122211 | 4.065155 | 4.172539 | 3.5918952 | 3.4872207 | 3.5826621 | 3.7253400 | 3.780151 | 4.045264 | 4.082435 | 3.979232 | 4.041785 | 4.018010 | 4.045655 | 4.001205 |
| GO:0019226 | transmission of nerve impulse | 36 | -0.5380987 | -1.498637 | 0.031345060 | 1.0000000 | 1.0000000 | 1494 | tags=36%, list=10%, signal=33% | CACNG5||FKBP1B||MYH14||GBA||CACNA1G||PLEC||CACNG8||SCN11A||CACNG4||SCN2A||SPTBN4||KCND2||TYMP | 3.4143875 | 3.849961 | 3.744272 | 4.009921 | 3.313701 | 3.3247990 | 3.877760 | 3.999804 | 3.4809755 | 3.5596313 | 3.1745582 | 3.829503 | 3.866013 | 3.854126 | 3.915374 | 3.5600477 | 3.7355307 | 3.989239 | 4.012386 | 4.027875 | 3.2631413 | 3.4983148 | 3.1583702 | 3.2858655 | 3.157365 | 3.509017 | 3.891132 | 3.876499 | 3.865535 | 3.986977 | 4.016950 | 3.995318 |
| GO:0098656 | anion transmembrane transport | 156 | -0.4010829 | -1.351873 | 0.032055519 | 1.0000000 | 1.0000000 | 5347 | tags=47%, list=34%, signal=31% | SLC25A14||SLC25A19||SLC12A4||SLC47A1||SLC25A13||LRRC8A||LRRC8B||ABCC4||SLC7A3||SLC35A1||SLC1A4||SLC7A1||LRP2||SLC25A38||ANO7||SLC20A2||GLRB||SLC9A8||PCYOX1||PER2||SLC9A1||SLC35B2||SLC43A2||ABCC1||CLCN3||SLC7A5||SLC25A11||RIPK1||ANO4||PRNP||SLC37A4||SLC9A2||SLC4A2||SLC12A7||SLC35B4||MFSD12||MFSD10||SLC15A4||SLC35B3||SLC35A2||SLC38A9||PRAF2||SLC38A7||SLC25A10||SLC9A5||SLC9A7||SLC12A9||SFXN3||SLC19A1||SLC5A12||GABRD||SLC26A2||CLTRN||SLC25A29||SLC9A3||SLC66A1||SLC25A42||SLC38A3||SLC26A11||SLC46A1||SLC4A11||SLC37A1||SLC4A3||LRRC8D||GABRR2||ABCB1||SLC9A9||SLC24A1||SLC1A1||SLC24A4||ACE2||GABRG2||SLC4A8 | 5.1844613 | 5.431444 | 5.309668 | 5.487074 | 5.167561 | 5.2754896 | 5.371989 | 5.467377 | 5.1864402 | 5.2548001 | 5.1084307 | 5.449957 | 5.457801 | 5.385490 | 5.314577 | 5.2861447 | 5.3279659 | 5.499147 | 5.465932 | 5.495911 | 5.1789226 | 5.1679010 | 5.1557655 | 5.2590119 | 5.271922 | 5.295300 | 5.382543 | 5.371608 | 5.361742 | 5.447654 | 5.506531 | 5.447134 |
| GO:0003091 | renal water homeostasis | 12 | -0.6834306 | -1.524682 | 0.032912534 | 1.0000000 | 1.0000000 | 2354 | tags=50%, list=15%, signal=42% | ADCY6||WFS1||HYAL2||AQP3||AQP1||MYO5B | 5.7488768 | 5.477006 | 5.835053 | 5.407882 | 5.795053 | 5.9099325 | 5.421033 | 5.351540 | 5.6533485 | 5.7470130 | 5.8402201 | 5.495899 | 5.490750 | 5.443802 | 5.657955 | 5.9463260 | 5.8851565 | 5.453070 | 5.360139 | 5.408940 | 5.8435706 | 5.7100908 | 5.8278430 | 5.9814256 | 5.823400 | 5.920587 | 5.449470 | 5.412933 | 5.400241 | 5.361214 | 5.354685 | 5.338629 |
| GO:0090068 | positive regulation of cell cycle process | 173 | 0.3124676 | 1.310373 | 0.033373063 | 1.0000000 | 1.0000000 | 3656 | tags=40%, list=24%, signal=31% | GLI1||PLK4||ZNF16||CCND1||CDC14A||CDC6||ATAD5||CDC23||NUSAP1||CDC25C||RAD51AP1||SLF1||BRD4||RACGAP1||PLCB1||STXBP4||PBX1||RGCC||UBE2E2||DTL||VPS4B||THOC1||MTBP||MED1||UBE2C||E2F8||KIF20B||NDC80||PHIP||CDC14B||GEN1||CDC27||FBXO5||DBF4B||CDK1||MAD2L1||ECT2||XRCC3||ANAPC4||SPAG5||PKN2||MTA3||INO80||EZH2||CCNB1||STOX1||PLRG1||RAD21||RB1||DLGAP5||ANAPC7||AURKA||CDC25A||KIF14||AURKB||FEN1||CDCA5||NSMCE2||ANKRD17||SLF2||RCC2||SIN3A||PKP4||FAM83D||E2F7||ROCK2||TMOD3||UBE2B||DBF4 | 5.8221818 | 6.490553 | 5.964551 | 6.486073 | 5.780269 | 5.9124789 | 6.539005 | 6.545503 | 5.8027882 | 5.8906073 | 5.7704550 | 6.512049 | 6.465790 | 6.493444 | 6.155920 | 5.8479859 | 5.8687822 | 6.451877 | 6.450197 | 6.553691 | 5.7724544 | 5.7752246 | 5.7930428 | 5.8593299 | 5.859115 | 6.013434 | 6.525628 | 6.532602 | 6.558577 | 6.527030 | 6.558359 | 6.550934 |
| GO:0010390 | histone monoubiquitination | 29 | 0.5010615 | 1.589530 | 0.034078001 | 1.0000000 | 1.0000000 | 5321 | tags=55%, list=34%, signal=36% | RAG1||DDB2||DTX3L||RYBP||UBE2E1||BMI1||LEO1||PCGF5||CDC73||BCOR||ATXN7L3||WAC||SKP1||PAF1||DDB1||PCGF2 | 5.1206953 | 5.805608 | 5.338642 | 5.948807 | 5.094260 | 5.3129958 | 5.876749 | 6.012034 | 5.1245302 | 5.1577574 | 5.0787079 | 5.769559 | 5.808274 | 5.838173 | 5.469327 | 5.2766559 | 5.2604476 | 5.900838 | 5.922140 | 6.020592 | 5.0691959 | 5.1218451 | 5.0912552 | 5.2902092 | 5.229932 | 5.412789 | 5.876638 | 5.859905 | 5.893509 | 5.990846 | 6.017470 | 6.027537 |
| GO:0000054 | ribosomal subunit export from nucleus | 14 | 0.6184078 | 1.627523 | 0.034632035 | 1.0000000 | 1.0000000 | 4308 | tags=64%, list=28%, signal=46% | NUP88||NMD3||LSG1||XPO1||SDAD1||ABCE1||RAN||LTV1||NPM1 | 9.0780760 | 9.158080 | 8.716602 | 9.211282 | 9.037801 | 9.0132969 | 9.178866 | 9.237228 | 9.1235423 | 9.0057712 | 9.1022055 | 9.215796 | 9.103539 | 9.152707 | 8.776785 | 8.7023588 | 8.6685276 | 9.235579 | 9.152086 | 9.244395 | 9.0607921 | 8.9496330 | 9.0988438 | 9.0540514 | 9.228687 | 8.709879 | 9.148967 | 9.170707 | 9.216107 | 9.248565 | 9.205524 | 9.257067 |
| GO:0097035 | regulation of membrane lipid distribution | 37 | -0.5322346 | -1.489732 | 0.034859248 | 1.0000000 | 1.0000000 | 4753 | tags=54%, list=31%, signal=38% | ANO7||ATP8A1||ABCC1||ABCG1||RFT1||ANO4||ATP8A2||SLC66A2||ABCA2||ATP8B2||MFSD2A||ABCA7||ARV1||XKR8||ABCA3||TLCD2||ABCB1||ATP8B1||TLCD1||ATP8B3 | 4.9276946 | 4.855344 | 4.947254 | 4.899951 | 4.948756 | 5.1045353 | 4.788970 | 4.834201 | 4.8604859 | 4.8839698 | 5.0325278 | 4.888872 | 4.863860 | 4.812245 | 4.868323 | 5.0177851 | 4.9517745 | 4.930307 | 4.863122 | 4.905627 | 4.9619523 | 4.8224649 | 5.0525911 | 5.0744104 | 5.140765 | 5.097643 | 4.830275 | 4.771906 | 4.763813 | 4.807905 | 4.865405 | 4.828704 |
| GO:0019731 | antibacterial humoral response | 11 | 0.6683572 | 1.634112 | 0.034917840 | 1.0000000 | 1.0000000 | 1623 | tags=55%, list=10%, signal=49% | TF||H2BC6||PLA2G6||H2BC21||H2BC4||H2BC11 | 8.6839128 | 8.176249 | 8.426186 | 8.195585 | 8.549239 | 7.7907163 | 8.170714 | 7.754781 | 8.8516333 | 8.8550045 | 8.2704282 | 8.229507 | 8.117964 | 8.179117 | 8.460829 | 8.1878760 | 8.5998995 | 8.165643 | 8.251215 | 8.168245 | 8.4776890 | 8.9509193 | 8.0899397 | 8.1915374 | 7.557524 | 7.520144 | 8.116598 | 8.216502 | 8.177291 | 8.177701 | 5.651680 | 8.267365 |
| GO:0033630 | positive regulation of cell adhesion mediated by integrin | 10 | -0.7123433 | -1.524863 | 0.035030864 | 1.0000000 | 1.0000000 | 2745 | tags=50%, list=18%, signal=41% | IFT74||SYK||PODXL||RET||TGFB2 | 5.4656275 | 4.901401 | 5.270748 | 4.745193 | 5.554946 | 5.6421299 | 4.820029 | 4.698019 | 5.3758692 | 5.2559157 | 5.7233743 | 4.917765 | 4.924685 | 4.860908 | 5.144147 | 5.3885714 | 5.2691838 | 4.827619 | 4.668215 | 4.735292 | 5.5299743 | 5.3931352 | 5.7226346 | 5.5520398 | 5.813014 | 5.544841 | 4.882555 | 4.786540 | 4.788898 | 4.669120 | 4.764046 | 4.658534 |
| GO:0090329 | regulation of DNA-dependent DNA replication | 43 | 0.4473139 | 1.536258 | 0.035095321 | 1.0000000 | 1.0000000 | 5304 | tags=63%, list=34%, signal=41% | LIG3||BLM||GMNN||DACH1||RFC3||NBN||METTL4||KAT7||E2F8||PCNA||FBXO5||DBF4B||CHTF18||INO80||ATG7||E2F7||DBF4||CHTF8||RFC5||TIPIN||RFC2||RFC4||DONSON||ATRX||WIZ||ZNF830||DSCC1 | 5.5423852 | 6.224209 | 5.672993 | 6.234247 | 5.481295 | 5.5763057 | 6.257263 | 6.316442 | 5.5691168 | 5.5832732 | 5.4722544 | 6.225279 | 6.210578 | 6.236650 | 5.900228 | 5.5602876 | 5.5282775 | 6.195204 | 6.201831 | 6.303149 | 5.4360175 | 5.5132165 | 5.4935410 | 5.5197765 | 5.569334 | 5.637385 | 6.249947 | 6.261749 | 6.260067 | 6.300977 | 6.347521 | 6.300318 |
| GO:0017148 | negative regulation of translation | 154 | 0.3177673 | 1.323598 | 0.035181237 | 1.0000000 | 1.0000000 | 3762 | tags=37%, list=24%, signal=28% | AGO4||RC3H1||XRN1||RGS2||CNOT6||EIF2S1||TNRC6C||PAN3||CPEB2||TNRC6A||EIF2AK2||CNOT3||IREB2||CPEB3||CNOT6L||RC3H2||FXR1||RARA||CNOT2||DAPK1||C8orf88||YTHDF3||ZFP36L1||DDX6||DIS3||SRP9||TNRC6B||CNOT11||TARDBP||RBM4||CNOT7||EXOSC3||PAIP2||IGF2BP1||EXOSC9||METTL14||YTHDF1||PNPT1||CNOT8||SYNCRIP||ILF3||MOV10||LSM14A||DHX36||CNOT10||IGF2BP3||RNF139||TSC1||SAMD4B||ROCK2||ROCK1||LSM1||EIF2AK4||CAPRIN1||PUS7||METTL3||YTHDF2 | 7.6183934 | 7.581711 | 7.506298 | 7.538762 | 7.638908 | 7.6563023 | 7.572890 | 7.565163 | 7.5978574 | 7.6040342 | 7.6526615 | 7.605643 | 7.580676 | 7.558426 | 7.438653 | 7.5438093 | 7.5341077 | 7.569318 | 7.487347 | 7.558258 | 7.6550700 | 7.6081539 | 7.6530147 | 7.7397019 | 7.692941 | 7.527779 | 7.563919 | 7.575368 | 7.579340 | 7.559839 | 7.565826 | 7.569805 |
| GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 12 | 0.6512184 | 1.638513 | 0.035244922 | 1.0000000 | 1.0000000 | 3221 | tags=58%, list=21%, signal=46% | RIMS2||SYT6||RIMS3||RIMS1||SYT2||RAB3GAP1||SYT7 | 2.6247922 | 4.196680 | 3.308659 | 4.305113 | 2.556696 | 3.2355482 | 4.205974 | 4.321666 | 2.5390343 | 2.8256119 | 2.4861273 | 4.129684 | 4.281314 | 4.174813 | 3.622711 | 3.1313685 | 3.1117923 | 4.224508 | 4.335186 | 4.352337 | 2.6456259 | 2.4699748 | 2.5491149 | 3.1959041 | 3.195522 | 3.312063 | 4.176670 | 4.230094 | 4.210653 | 4.311283 | 4.344017 | 4.309435 |
| GO:0019233 | sensory perception of pain | 56 | -0.4867391 | -1.453806 | 0.035317861 | 1.0000000 | 1.0000000 | 2548 | tags=32%, list=16%, signal=27% | PTGS2||MMP24||NR2F6||ATPSCKMT||NMU||GRIN2D||CDK5||CACNB3||ZFHX2||CXCR4||ADORA1||SCN11A||OPRL1||PRDM12||SCN3B||CXCL12||KCND2||CHRNA4 | 4.2746799 | 4.216797 | 4.411111 | 4.131962 | 4.212302 | 4.2813411 | 4.194408 | 4.122005 | 4.3030449 | 4.3634233 | 4.1492027 | 4.235060 | 4.221238 | 4.193787 | 4.423512 | 4.3981293 | 4.4115786 | 4.168826 | 4.084105 | 4.141663 | 4.2282838 | 4.2480949 | 4.1590187 | 4.2182625 | 4.170678 | 4.440478 | 4.233970 | 4.181792 | 4.166593 | 4.120938 | 4.148584 | 4.096015 |
| GO:0034508 | centromere complex assembly | 29 | 0.4959938 | 1.573454 | 0.035592579 | 1.0000000 | 1.0000000 | 3003 | tags=45%, list=19%, signal=36% | CENPC||SENP6||HELLS||CENPK||RNF4||POGZ||CENPN||NASP||CENPI||CENPA||HJURP||RB1||DLGAP5 | 5.5697516 | 5.932526 | 5.794268 | 6.054496 | 5.566043 | 5.6878125 | 6.015180 | 6.128152 | 5.5213771 | 5.5584638 | 5.6274006 | 5.935678 | 5.931909 | 5.929984 | 6.022152 | 5.6834215 | 5.6468075 | 6.000743 | 6.033411 | 6.126370 | 5.4978776 | 5.5517346 | 5.6446777 | 5.5525948 | 5.715960 | 5.785108 | 6.017250 | 6.004030 | 6.024187 | 6.103313 | 6.166232 | 6.114122 |
| GO:1902600 | proton transmembrane transport | 117 | -0.4193486 | -1.374528 | 0.035954033 | 1.0000000 | 1.0000000 | 5682 | tags=50%, list=37%, signal=32% | COX5A||CLCN5||ATP1A1||SLC25A4||TESC||ATP6V0E1||COX15||SLC36A1||CYB5A||SLC47A1||SLC30A5||DMAC2L||COX8A||COX6A1||ATP6V1F||ATP7A||MT-CO2||SLC9A8||MT-CO1||MT-CO3||SLC9A3R1||SLC9A1||ATP6V0B||ATP6V0A1||CLCN3||ATP6AP1||MT-ATP6||MT-ND5||UCP2||MT-CYB||SLC9A2||SLC15A4||MT-ND4||TCIRG1||SLC17A5||SURF1||ATP6V0C||SLC2A10||SPHK2||TWIST1||ATP5F1D||SLC9A5||ATP6V1E2||SLC9A7||ATPSCKMT||ATP1A3||CTNS||SLC22A18||MFSD3||ATP6V0E2||SLC9A3||SLC15A2||DNAJC30||ATP6V1C2||ANTKMT||SLC46A1||SLC4A11||SLC9A9||MTCO2P12 | 9.6029774 | 9.094025 | 10.041446 | 9.436565 | 9.678114 | 9.9719695 | 9.179745 | 9.303134 | 9.5091246 | 9.5866472 | 9.7062915 | 8.690151 | 9.253331 | 9.267055 | 9.900782 | 10.1918757 | 10.0167060 | 9.358816 | 9.656101 | 9.265368 | 9.7184939 | 9.5881626 | 9.7236437 | 9.8490340 | 9.836981 | 10.199717 | 9.161423 | 9.208031 | 9.169350 | 9.386108 | 9.256692 | 9.262874 |
| GO:0035855 | megakaryocyte development | 13 | 0.6305459 | 1.622073 | 0.036324786 | 1.0000000 | 1.0000000 | 5099 | tags=92%, list=33%, signal=62% | FLI1||ZFPM1||MED1||ABI1||MEIS1||SRF||KIT||WASF2||PIP4K2A||SH2B3||PTPN11||EP300 | 3.2323467 | 4.106623 | 3.504509 | 4.193623 | 3.200935 | 3.6677544 | 4.217386 | 4.285395 | 3.0973402 | 3.3057745 | 3.2849303 | 4.020006 | 4.134334 | 4.161649 | 3.714448 | 3.4410069 | 3.3305768 | 4.112426 | 4.147869 | 4.312572 | 3.2364570 | 3.0602407 | 3.2958337 | 3.6477987 | 3.721537 | 3.632344 | 4.210440 | 4.213123 | 4.228530 | 4.281763 | 4.275042 | 4.299271 |
| GO:0000956 | nuclear-transcribed mRNA catabolic process | 105 | 0.3517066 | 1.399562 | 0.036501901 | 1.0000000 | 1.0000000 | 4569 | tags=44%, list=29%, signal=31% | RC3H1||XRN1||CNOT6||SECISBP2||TNRC6C||PAN3||MRTO4||TNRC6A||CNOT3||CPEB3||CNOT6L||RC3H2||CNOT2||ETF1||SMG1||EXOSC10||ZFP36L1||DIS3||TNRC6B||MTPAP||TENT2||XRN2||PNRC2||CNOT7||MAGOH||EXOSC3||EXOSC9||NCBP2||CNOT8||DHX36||SAMD4B||RNPS1||SMG5||LSM1||UPF3B||EDC4||UPF2||EDC3||GSPT1||PRPF18||THRAP3||DCP1A||EXOSC6||CASC3||CSDE1||EIF3E | 5.6629419 | 6.375390 | 5.807702 | 6.393633 | 5.619638 | 5.7546638 | 6.413931 | 6.445531 | 5.6484143 | 5.7056216 | 5.6337866 | 6.375285 | 6.370738 | 6.380131 | 6.038953 | 5.6684457 | 5.6845206 | 6.351821 | 6.366227 | 6.460422 | 5.5977672 | 5.5941014 | 5.6659050 | 5.7248900 | 5.777431 | 5.761171 | 6.391244 | 6.421997 | 6.428280 | 6.436079 | 6.456582 | 6.443858 |
| GO:0050433 | regulation of catecholamine secretion | 27 | -0.5670597 | -1.495034 | 0.037334787 | 1.0000000 | 1.0000000 | 2357 | tags=41%, list=15%, signal=35% | RAB3A||SNCA||ABAT||ADORA2A||SYT11||SYT3||CXCL12||KCNB1||SYT17||SYT12||CHRNA4 | 2.7403387 | 2.737538 | 2.897373 | 2.471198 | 2.764289 | 3.0382036 | 2.676048 | 2.335126 | 2.6601069 | 2.7184246 | 2.8368290 | 2.711212 | 2.749779 | 2.751267 | 2.830269 | 2.9437107 | 2.9157337 | 2.554701 | 2.388513 | 2.465580 | 2.7668873 | 2.6434347 | 2.8733890 | 2.9182949 | 3.023296 | 3.162577 | 2.729001 | 2.643260 | 2.654366 | 2.344949 | 2.385288 | 2.272903 |
| GO:0001916 | positive regulation of T cell mediated cytotoxicity | 15 | -0.6461424 | -1.510321 | 0.037465242 | 1.0000000 | 1.0000000 | 3953 | tags=73%, list=25%, signal=55% | FADD||HLA-E||HLA-B||HLA-A||PVR||NECTIN2||XCL1||HLA-DRB1||MR1||HLA-F||IL23A | 5.2448721 | 5.749908 | 5.567538 | 5.644158 | 5.318381 | 5.4529930 | 5.597476 | 5.537637 | 5.1966238 | 5.2126675 | 5.3220678 | 5.775927 | 5.767919 | 5.704832 | 5.513877 | 5.6045822 | 5.5826095 | 5.671148 | 5.659416 | 5.600934 | 5.2817041 | 5.2633037 | 5.4059243 | 5.2837324 | 5.398353 | 5.651949 | 5.668573 | 5.580838 | 5.540012 | 5.564443 | 5.572610 | 5.473788 |
| GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 12 | 0.6409148 | 1.612589 | 0.038231780 | 1.0000000 | 1.0000000 | 3866 | tags=58%, list=25%, signal=44% | BRD4||ELL||CDK12||TCERG1||LEO1||EAPP||CDC73 | 4.9161776 | 6.184543 | 5.274792 | 6.317106 | 4.754219 | 4.9484203 | 6.238215 | 6.361911 | 4.9018347 | 5.0875235 | 4.7379432 | 6.178413 | 6.217741 | 6.156811 | 5.648177 | 4.9089254 | 5.1679336 | 6.268988 | 6.302193 | 6.377963 | 4.7363108 | 4.8359767 | 4.6863250 | 4.8986643 | 4.970406 | 4.974929 | 6.228809 | 6.244169 | 6.241621 | 6.358173 | 6.372078 | 6.355425 |
| GO:0007094 | mitotic spindle assembly checkpoint signaling | 37 | 0.4589905 | 1.527671 | 0.039583333 | 1.0000000 | 1.0000000 | 3364 | tags=46%, list=22%, signal=36% | TTK||PSMG2||BUB1B||NDC80||GEN1||MAD2L1||BUB1||XRCC3||USP44||ANAPC15||ZNF207||CCNB1||TEX14||IK||SPDL1||AURKB||BUB3 | 5.5019237 | 6.192664 | 5.645675 | 6.256578 | 5.446522 | 5.5948268 | 6.252674 | 6.366557 | 5.4763133 | 5.5788474 | 5.4472746 | 6.231716 | 6.178075 | 6.167375 | 5.870813 | 5.4957573 | 5.5406812 | 6.237115 | 6.213906 | 6.316689 | 5.4149394 | 5.4610535 | 5.4630607 | 5.5450496 | 5.600637 | 5.637302 | 6.258469 | 6.253745 | 6.245780 | 6.350367 | 6.401138 | 6.347533 |
| GO:0055057 | neuroblast division | 13 | 0.6282786 | 1.616240 | 0.039682540 | 1.0000000 | 1.0000000 | 4108 | tags=69%, list=26%, signal=51% | FGF13||SOX5||WNT3A||NUMB||RAB10||NUMBL||LEF1||DOCK7||ASPM | 2.5822969 | 4.020303 | 3.071353 | 4.081487 | 2.554948 | 2.9704525 | 4.095832 | 4.204236 | 2.5771395 | 2.6684249 | 2.4961742 | 3.950344 | 4.044344 | 4.063691 | 3.478238 | 2.8611077 | 2.7669726 | 3.986576 | 4.115814 | 4.137502 | 2.4611428 | 2.5441309 | 2.6531268 | 2.8419275 | 2.885339 | 3.162676 | 4.083158 | 4.133320 | 4.070247 | 4.191991 | 4.232894 | 4.187388 |
| GO:0070050 | neuron cellular homeostasis | 20 | -0.5996427 | -1.493451 | 0.039774330 | 1.0000000 | 1.0000000 | 2001 | tags=30%, list=13%, signal=26% | CDK5||DNAJB2||TMEM175||ADORA1||MAP1A||SLC8A2 | 3.9898829 | 4.463927 | 4.353813 | 4.591588 | 4.046724 | 4.4249487 | 4.455121 | 4.493917 | 3.8908763 | 3.9608844 | 4.1092046 | 4.443396 | 4.510381 | 4.436850 | 4.356952 | 4.4150035 | 4.2866242 | 4.548801 | 4.596959 | 4.627904 | 4.0921996 | 3.9179073 | 4.1217723 | 4.3089943 | 4.360334 | 4.589770 | 4.467407 | 4.456749 | 4.441085 | 4.495794 | 4.527720 | 4.457377 |
| GO:0048532 | anatomical structure arrangement | 14 | 0.6078748 | 1.599803 | 0.039888683 | 1.0000000 | 1.0000000 | 3155 | tags=43%, list=20%, signal=34% | PAX2||SEMA3A||HOXB2||PLXNA4||KIF14||TFAP2A | 3.7205216 | 5.295168 | 3.830413 | 5.084500 | 3.569444 | 3.8990062 | 5.143713 | 5.060977 | 3.6921093 | 3.8469531 | 3.6125799 | 5.271654 | 5.378228 | 5.231614 | 4.299573 | 3.5637809 | 3.4801447 | 5.037334 | 5.126014 | 5.088779 | 3.5933995 | 3.4922150 | 3.6196049 | 3.8347133 | 3.875364 | 3.982852 | 5.128319 | 5.171275 | 5.131144 | 5.079621 | 5.111743 | 4.988766 |
| GO:0002478 | antigen processing and presentation of exogenous peptide antigen | 24 | -0.5772740 | -1.492762 | 0.040247253 | 1.0000000 | 1.0000000 | 4050 | tags=58%, list=26%, signal=43% | IFI30||HLA-E||HLA-A||MFSD6||CTSD||HLA-DRB1||CTSF||HLA-DPA1||CTSL||TRAF6||HLA-DQB1||HLA-DMA||HLA-F||HLA-DOA | 4.8462479 | 5.301352 | 5.171784 | 5.227796 | 4.890432 | 5.1209464 | 5.124494 | 5.104673 | 4.8024956 | 4.8122035 | 4.9210178 | 5.327205 | 5.336335 | 5.238505 | 5.136329 | 5.2023981 | 5.1758599 | 5.273410 | 5.233758 | 5.174508 | 4.8744904 | 4.8353999 | 4.9586445 | 4.9521455 | 5.063348 | 5.322023 | 5.192889 | 5.114160 | 5.063476 | 5.127223 | 5.148126 | 5.036234 |
| GO:0034501 | protein localization to kinetochore | 19 | 0.5635146 | 1.607962 | 0.040349697 | 1.0000000 | 1.0000000 | 3391 | tags=58%, list=22%, signal=45% | TTK||CENPQ||KNL1||MTBP||NDC80||CDK1||IK||SPDL1||AURKB||BUB3||RCC2 | 5.3849216 | 6.180386 | 5.612198 | 6.181575 | 5.332944 | 5.5368432 | 6.259147 | 6.297921 | 5.3333038 | 5.4504869 | 5.3684553 | 6.210879 | 6.170893 | 6.158871 | 5.784920 | 5.4972697 | 5.5372698 | 6.130656 | 6.165028 | 6.246572 | 5.3132930 | 5.3317607 | 5.3534969 | 5.4443070 | 5.501969 | 5.655881 | 6.253718 | 6.278904 | 6.244600 | 6.285260 | 6.334340 | 6.273436 |
| GO:0072520 | seminiferous tubule development | 11 | 0.6549860 | 1.601420 | 0.041079812 | 1.0000000 | 1.0000000 | 2528 | tags=45%, list=16%, signal=38% | REC8||AR||WDR48||KIF18A||SMAD4 | 8.0849403 | 7.903623 | 7.951680 | 7.791762 | 7.985256 | 7.5809519 | 7.856228 | 7.861388 | 8.0590903 | 8.1924341 | 7.9962916 | 8.037227 | 7.823018 | 7.840678 | 7.798967 | 7.9370640 | 8.1029196 | 7.869854 | 7.703420 | 7.797199 | 8.0709980 | 8.0638770 | 7.8053455 | 7.7931338 | 7.237963 | 7.655669 | 7.865989 | 7.842192 | 7.860398 | 7.889365 | 7.820113 | 7.873774 |
| GO:0045116 | protein neddylation | 17 | 0.5734768 | 1.590924 | 0.041354608 | 1.0000000 | 1.0000000 | 4072 | tags=41%, list=26%, signal=30% | DCUN1D3||HIF1A||DCUN1D1||UBA3||DCUN1D4||RPL5||DCUN1D5 | 8.4544369 | 8.791937 | 8.245653 | 8.797319 | 8.362308 | 8.0538546 | 8.835681 | 8.843672 | 8.5291454 | 8.5722946 | 8.2397439 | 8.808108 | 8.758695 | 8.808442 | 8.433771 | 8.0543938 | 8.2236752 | 8.779096 | 8.797800 | 8.814839 | 8.2623755 | 8.5955834 | 8.1964898 | 8.0973036 | 8.112187 | 7.946307 | 8.793279 | 8.831559 | 8.880867 | 8.841073 | 8.814423 | 8.874882 |
| GO:0006206 | pyrimidine nucleobase metabolic process | 14 | -0.6495720 | -1.497671 | 0.042257683 | 1.0000000 | 1.0000000 | 26 | tags=14%, list=0%, signal=14% | CPS1||TYMP | 4.9342654 | 5.852420 | 5.348263 | 5.852616 | 4.736181 | 4.8954042 | 5.900347 | 5.887289 | 4.9935932 | 5.0827915 | 4.6990929 | 5.859470 | 5.848815 | 5.848950 | 5.614726 | 5.0884126 | 5.2923265 | 5.792405 | 5.807899 | 5.952108 | 4.6881010 | 4.8450667 | 4.6688190 | 4.7434887 | 4.697042 | 5.192192 | 5.886049 | 5.890356 | 5.924330 | 5.836676 | 5.923810 | 5.899982 |
| GO:0034728 | nucleosome organization | 106 | 0.3487659 | 1.383118 | 0.042868277 | 1.0000000 | 1.0000000 | 4585 | tags=46%, list=30%, signal=33% | H2BC17||H4C3||H3C2||H1-4||H2BC6||H2BC21||H4C8||H2BC4||ARID2||H2BC11||SART3||SHPRH||UBN2||VPS72||DMAP1||HMGB2||SMARCD1||H3C6||ASF1A||RSF1||NAP1L4||NASP||ACTR6||INO80||DNAJC9||HJURP||CHAF1B||BPTF||CHD5||RBBP4||H2BC19P||SMARCC1||H2BC5||SMARCE1||H3C10||KAT6B||UBN1||NAA60||H4C14||TSPYL1||NPM1||POLE3||PSME4||GRWD1||SPTY2D1||SMARCD2||SET||LIN54||H3-3B | 6.1033633 | 6.888427 | 6.253774 | 6.859059 | 6.059458 | 6.2095719 | 6.951407 | 6.909873 | 6.0978792 | 6.1681595 | 6.0412455 | 6.879068 | 6.871821 | 6.914038 | 6.517029 | 6.0837344 | 6.1192927 | 6.826648 | 6.822624 | 6.925532 | 6.0267139 | 6.0741273 | 6.0769815 | 6.1564210 | 6.181135 | 6.287757 | 6.926296 | 6.937081 | 6.990035 | 6.920015 | 6.885343 | 6.923950 |
| GO:0031998 | regulation of fatty acid beta-oxidation | 17 | -0.6173848 | -1.480695 | 0.042995239 | 1.0000000 | 1.0000000 | 3143 | tags=53%, list=20%, signal=42% | CPT1A||MFSD2A||TWIST1||PPARA||ABCD1||MTLN||PLIN5||MLYCD||ETFBKMT | 4.1982221 | 4.105299 | 4.550005 | 4.052437 | 4.285169 | 4.7188449 | 4.091425 | 4.018218 | 4.0623462 | 4.1294544 | 4.3827320 | 4.131868 | 4.111591 | 4.071794 | 4.528581 | 4.6579810 | 4.4561654 | 4.146181 | 3.952894 | 4.051766 | 4.3010156 | 4.0377387 | 4.4824803 | 4.5327816 | 4.753878 | 4.851644 | 4.158605 | 4.027109 | 4.085548 | 3.986736 | 4.076439 | 3.989666 |
| GO:0032675 | regulation of interleukin-6 production | 67 | -0.4602793 | -1.410765 | 0.043190995 | 1.0000000 | 1.0000000 | 3272 | tags=34%, list=21%, signal=27% | CYBA||MYD88||SPHK2||MAVS||TWIST1||IL27RA||F2RL1||TRAF6||SYK||GBA||ZC3H12A||NLRX1||IL17RA||HYAL2||IL6R||CEBPB||TIRAP||TSLP||C1QTNF3||TICAM1||SYT11||TRPV4||MBP | 4.6718137 | 5.615550 | 4.878228 | 5.687170 | 4.598445 | 4.7849649 | 5.612462 | 5.668999 | 4.6677954 | 4.7286639 | 4.6168060 | 5.633886 | 5.617463 | 5.595037 | 5.093480 | 4.7870039 | 4.7268138 | 5.650624 | 5.683219 | 5.726658 | 4.5550445 | 4.6024956 | 4.6366338 | 4.6851862 | 4.740820 | 4.918475 | 5.614321 | 5.611426 | 5.611636 | 5.685787 | 5.649792 | 5.671189 |
| GO:0002115 | store-operated calcium entry | 12 | -0.6712462 | -1.497499 | 0.043432522 | 1.0000000 | 1.0000000 | 3657 | tags=67%, list=24%, signal=51% | CRACR2A||ORAI1||HOMER1||STIM2||SLC8B1||ORAI2||CRACR2B||ORAI3 | 3.4374691 | 4.277916 | 3.661277 | 4.272578 | 3.440235 | 3.6351000 | 4.208358 | 4.189240 | 3.4068001 | 3.4887031 | 3.4154932 | 4.218890 | 4.375485 | 4.234150 | 3.802025 | 3.6009356 | 3.5696827 | 4.231951 | 4.323231 | 4.261040 | 3.3852003 | 3.4027128 | 3.5285150 | 3.5136050 | 3.590680 | 3.787070 | 4.226193 | 4.226881 | 4.171297 | 4.178609 | 4.213523 | 4.175277 |
| GO:0071577 | zinc ion transmembrane transport | 18 | -0.6095856 | -1.481484 | 0.044016110 | 1.0000000 | 1.0000000 | 5308 | tags=72%, list=34%, signal=48% | SLC30A1||SLC30A5||SLC39A14||SLC39A4||SLC30A6||SLC30A4||SLC39A3||SLC39A11||SLC30A3||SLC39A6||SLC39A1||TMEM163||SLC1A1 | 4.8487894 | 5.337042 | 5.116081 | 5.308790 | 4.907545 | 5.1119735 | 5.194424 | 5.165346 | 4.7692909 | 4.7875569 | 4.9799421 | 5.334544 | 5.397550 | 5.276492 | 5.062121 | 5.2233072 | 5.0565198 | 5.286419 | 5.351887 | 5.287079 | 4.8865246 | 4.7947764 | 5.0314133 | 4.9271796 | 5.088994 | 5.295971 | 5.218375 | 5.212202 | 5.151760 | 5.183039 | 5.225521 | 5.083838 |
| GO:0045475 | locomotor rhythm | 12 | -0.6708854 | -1.496694 | 0.044484521 | 1.0000000 | 1.0000000 | 1350 | tags=25%, list=9%, signal=23% | NAGLU||OPRL1||KCND2 | 4.2343443 | 5.059401 | 4.803641 | 4.907317 | 4.187829 | 4.7338392 | 5.086311 | 4.904594 | 4.2473787 | 4.2514711 | 4.2036987 | 5.056693 | 5.093682 | 5.027055 | 5.039973 | 4.7428105 | 4.5915659 | 4.890010 | 4.892950 | 4.938476 | 4.1366535 | 4.0833404 | 4.3315054 | 4.4785280 | 4.591932 | 5.062530 | 5.105650 | 5.056532 | 5.096281 | 4.856990 | 4.973164 | 4.881003 |
| GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 29 | 0.4756413 | 1.508889 | 0.044680045 | 1.0000000 | 1.0000000 | 2973 | tags=45%, list=19%, signal=36% | PROX1||CCND1||CKS2||CDC6||PKD1||CCNT2||RGCC||CCNK||CDKN1B||CDKN1A||CCNB1||STOX1||CKS1B | 5.8642830 | 6.599390 | 6.020890 | 6.722768 | 5.716756 | 5.6711210 | 6.672901 | 6.818067 | 5.8924718 | 6.0718318 | 5.5877006 | 6.635300 | 6.568609 | 6.593473 | 6.309843 | 5.6881613 | 5.9979340 | 6.706390 | 6.684100 | 6.776206 | 5.7119349 | 5.8951767 | 5.5186468 | 5.7795112 | 5.654207 | 5.572045 | 6.654495 | 6.681660 | 6.682372 | 6.831364 | 6.797810 | 6.824808 |
| GO:0002097 | tRNA wobble base modification | 20 | 0.5425636 | 1.568372 | 0.044986264 | 1.0000000 | 1.0000000 | 2836 | tags=40%, list=18%, signal=33% | TRMT9B||ADAT2||ALKBH8||ALKBH1||DPH3||ELP4||ELP2||MTO1 | 5.5698188 | 5.695073 | 5.613701 | 5.740408 | 5.612377 | 5.7468388 | 5.694216 | 5.776530 | 5.5175452 | 5.5256203 | 5.6616964 | 5.731530 | 5.697050 | 5.655640 | 5.624824 | 5.6264053 | 5.5895734 | 5.762629 | 5.682103 | 5.774746 | 5.5939746 | 5.5117525 | 5.7234692 | 5.6584435 | 5.841446 | 5.734752 | 5.724081 | 5.679195 | 5.678902 | 5.755253 | 5.812971 | 5.760660 |
| GO:0007063 | regulation of sister chromatid cohesion | 21 | 0.5361054 | 1.575976 | 0.045141282 | 1.0000000 | 1.0000000 | 5133 | tags=71%, list=33%, signal=48% | AXIN2||SLF1||BUB1||WAPL||TNKS||RAD21||RB1||FEN1||CDCA5||NSMCE2||SLF2||SFPQ||SMC5||ATRX||CTCF | 6.3075694 | 6.811344 | 6.565588 | 6.824220 | 6.284487 | 6.4396142 | 6.836784 | 6.868936 | 6.2897417 | 6.3536988 | 6.2781139 | 6.836872 | 6.801055 | 6.795757 | 6.726299 | 6.4171073 | 6.5363838 | 6.775927 | 6.805020 | 6.889297 | 6.2441368 | 6.2915995 | 6.3167842 | 6.3811323 | 6.408507 | 6.525116 | 6.840607 | 6.832830 | 6.836903 | 6.855208 | 6.882464 | 6.869006 |
| GO:0075522 | IRES-dependent viral translational initiation | 11 | 0.6444635 | 1.575693 | 0.046948357 | 1.0000000 | 1.0000000 | 5214 | tags=91%, list=34%, signal=60% | EIF3D||EIF3B||DENR||PCBP2||PTBP1||EIF2D||CSDE1||MCTS1||EIF3A||SSB | 7.2852150 | 7.982468 | 7.464157 | 7.993152 | 7.169412 | 7.3748136 | 8.061463 | 8.128794 | 7.2812258 | 7.4310893 | 7.1273657 | 7.953794 | 7.988091 | 8.005047 | 7.602018 | 7.3728170 | 7.4068794 | 7.952298 | 7.945636 | 8.077653 | 7.2338469 | 7.1766587 | 7.0943302 | 7.4218904 | 7.228032 | 7.463725 | 8.038061 | 8.070988 | 8.075057 | 8.103775 | 8.141931 | 8.140355 |
| GO:0010452 | histone H3-K36 methylation | 13 | 0.6123396 | 1.575237 | 0.047008547 | 1.0000000 | 1.0000000 | 5631 | tags=85%, list=36%, signal=54% | BRD4||IWS1||NSD3||ASH1L||SETD5||SETD3||SMYD2||BCOR||PAXIP1||NSD2||NSD1 | 3.9739756 | 4.973604 | 4.610511 | 5.133942 | 3.881902 | 4.5553574 | 5.116101 | 5.247409 | 3.8274699 | 4.0880395 | 3.9944479 | 4.932355 | 5.003803 | 4.983717 | 4.957208 | 4.4338713 | 4.3651279 | 5.041109 | 5.092422 | 5.259182 | 3.8663693 | 3.7646603 | 4.0045884 | 4.4897746 | 4.495353 | 4.673291 | 5.134820 | 5.135194 | 5.077529 | 5.184270 | 5.281356 | 5.274581 |
| GO:0010824 | regulation of centrosome duplication | 42 | 0.4353881 | 1.488100 | 0.047099869 | 1.0000000 | 1.0000000 | 5794 | tags=62%, list=37%, signal=39% | PLK4||PLK2||CEP76||XPO1||VPS4B||ENSG00000285943||GEN1||CHORDC1||STIL||XRCC3||CHMP5||ROCK2||PDCD6IP||CHMP1A||CHMP1B||KAT2A||NPM1||CHMP3||KAT2B||CENPJ||RBM14||SIRT1||CHMP2B||TRIM37||CHMP4C||CEP131 | 5.3190794 | 6.928145 | 5.570653 | 6.991302 | 5.267942 | 5.5813810 | 7.017258 | 7.064728 | 5.3311407 | 5.3464592 | 5.2787683 | 6.945049 | 6.876773 | 6.961228 | 6.072900 | 5.3245609 | 5.1370408 | 6.928822 | 6.960334 | 7.080284 | 5.1908346 | 5.2782876 | 5.3312338 | 5.4706061 | 5.516046 | 5.742541 | 6.972763 | 7.005898 | 7.071362 | 7.056253 | 7.050949 | 7.086723 |
| GO:0009154 | purine ribonucleotide catabolic process | 34 | -0.5242903 | -1.442898 | 0.047866667 | 1.0000000 | 1.0000000 | 1591 | tags=24%, list=10%, signal=21% | FITM2||ABCD1||PDE2A||NUDT8||NUDT7||MLYCD||NUDT18||ENSG00000284762 | 5.1962211 | 5.389518 | 5.270548 | 5.341771 | 5.149472 | 4.9767675 | 5.372516 | 5.358429 | 5.2836003 | 5.2605972 | 5.0312218 | 5.430594 | 5.363060 | 5.373987 | 5.285190 | 5.1973340 | 5.3261338 | 5.327594 | 5.311976 | 5.384723 | 5.1731052 | 5.2378757 | 5.0296337 | 5.0000381 | 4.799337 | 5.113566 | 5.370976 | 5.363338 | 5.383163 | 5.331045 | 5.365313 | 5.378514 |
| GO:0060045 | positive regulation of cardiac muscle cell proliferation | 21 | 0.5305788 | 1.559730 | 0.047898001 | 1.0000000 | 1.0000000 | 2818 | tags=43%, list=18%, signal=35% | GLI1||PIM1||MEF2C||MAPK1||RBPJ||YAP1||CDK1||FGF2||CCNB1 | 3.3007574 | 5.060292 | 3.522288 | 5.085170 | 3.098958 | 3.2041114 | 5.117666 | 5.206787 | 3.3865692 | 3.5253160 | 2.9242413 | 5.130470 | 5.007486 | 5.040090 | 4.127917 | 3.0621995 | 3.1099719 | 5.009811 | 5.088259 | 5.153842 | 2.9938039 | 3.3362093 | 2.9333434 | 3.2329048 | 3.175508 | 3.203351 | 5.085896 | 5.120455 | 5.146017 | 5.220772 | 5.181612 | 5.217649 |
| GO:0010259 | multicellular organism aging | 21 | 0.5298134 | 1.557480 | 0.048242591 | 1.0000000 | 1.0000000 | 499 | tags=19%, list=3%, signal=18% | COMP||RNF165||LRRK2||WRN | 4.2269764 | 5.212579 | 4.498343 | 5.167232 | 4.226583 | 4.4661685 | 5.192315 | 5.169269 | 4.1348509 | 4.2882285 | 4.2534076 | 5.187227 | 5.260994 | 5.188270 | 4.599772 | 4.4750514 | 4.4139469 | 5.079806 | 5.193074 | 5.224822 | 4.2479223 | 4.1334803 | 4.2936625 | 4.4130559 | 4.370147 | 4.604375 | 5.182086 | 5.199419 | 5.195382 | 5.163256 | 5.220132 | 5.122757 |
| GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 13 | -0.6479835 | -1.472997 | 0.049212013 | 1.0000000 | 1.0000000 | 4087 | tags=69%, list=26%, signal=51% | TAP2||HLA-E||HLA-B||HLA-A||ABCB9||HLA-C||HLA-DRB1||ERAP1||HLA-F | 5.9448424 | 6.250483 | 6.292308 | 6.189828 | 6.047298 | 6.2266029 | 6.046801 | 6.048239 | 5.8874779 | 5.8576817 | 6.0792059 | 6.293950 | 6.277572 | 6.177178 | 6.152058 | 6.4007876 | 6.3131442 | 6.250956 | 6.205018 | 6.109944 | 6.0266730 | 5.9446497 | 6.1621613 | 6.0654958 | 6.179760 | 6.412608 | 6.132283 | 6.018518 | 5.985458 | 6.088571 | 6.085864 | 5.966971 |
| GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 16 | -0.6216052 | -1.470449 | 0.049416910 | 1.0000000 | 1.0000000 | 26 | tags=12%, list=0%, signal=12% | NT5M||TYMP | 5.2309407 | 5.496802 | 5.419721 | 5.439089 | 5.180393 | 5.2582152 | 5.553369 | 5.442367 | 5.2416032 | 5.2350974 | 5.2159989 | 5.484396 | 5.499895 | 5.506029 | 5.520884 | 5.3022777 | 5.4276965 | 5.401455 | 5.398932 | 5.513867 | 5.1137784 | 5.1853684 | 5.2392890 | 5.0916133 | 5.177425 | 5.476713 | 5.560438 | 5.549478 | 5.550166 | 5.383663 | 5.478623 | 5.463028 |
| GO:0001732 | formation of cytoplasmic translation initiation complex | 16 | 0.5704435 | 1.559899 | 0.049968173 | 1.0000000 | 1.0000000 | 6563 | tags=88%, list=42%, signal=51% | EIF3CL||EIF3D||EIF3B||EIF5||EIF3J||EIF3E||EIF2S2||EIF3A||EIF3K||EIF3I||EIF3C||EIF3G||EIF3H||EIF3F | 7.9271423 | 8.254288 | 7.838454 | 8.282417 | 7.811035 | 7.7113293 | 8.295462 | 8.348611 | 7.9829729 | 7.9915659 | 7.7987895 | 8.233387 | 8.260375 | 8.268865 | 7.887266 | 7.7620335 | 7.8630291 | 8.272664 | 8.238075 | 8.334841 | 7.8576799 | 7.8911419 | 7.6750423 | 7.8316760 | 7.629813 | 7.664319 | 8.282280 | 8.298977 | 8.305033 | 8.338128 | 8.353889 | 8.353758 |
GO: MF
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 891 | 0.2535087 | 1.211777 | 0.0003897336 | 0.1295215 | 0.1259455 | 2284 | tags=24%, list=15%, signal=21% | ENSG00000273046||FLI1||RUNX2||PAX2||BHLHE41||PGR||ZNF256||ZNF268||ZNF331||BCL11B||ZNF567||SP2||GLIS3||GLI1||ZNF226||ZNF223||RXRG||ZNF114||ZNF143||ZNF713||FOXD2||ZNF383||ZNF551||FOXP2||ZNF528||ZNF136||NHLH1||ZNF350||ZNF568||ZNF587||ELK4||NFIB||ZNF225||ZNF227||ZNF134||ZNF419||ZNF549||IRX6||ZNF235||POU2F1||MAFF||NRL||ZNF230||ZNF461||NR5A2||POU6F1||SP4||ZBTB7C||ZNF45||ZNF224||DMTF1||ZNF140||SOX2||ZNF17||WT1||ZNF776||ZNF304||ZNF260||ZNF175||ZNF565||ZNF888||SIX4||IKZF3||SP1||HNF4G||PKNOX1||ZNF280D||ZNF805||RORB||ZNF12||RXRB||ZNF320||PROX1||ZNF468||CCAR1||ZNF880||ZNF333||ZNF431||ZNF214||HIVEP1||ZNF699||ZNF16||KLF11||AR||ZNF211||ZNF432||SMAD9||ZNF416||ZNF613||ZNF628||HSFX1||MXI1||GMEB1||ZNF417||THAP1||ZBTB11||RFX3||MYNN||ZNF525||EBF2||SNAI1||ZNF84||ZNF335||GMEB2||ZBTB6||SMAD3||ZNF112||NR4A3||ZNF547||ZNF577||IRF1||ZNF26||ZNF367||HSF2||JDP2||HBP1||MEF2C||ZNF460||ZNF790||ZNF561||ATF2||ZNF264||TCF4||ZNF221||NFYB||ZNF382||ZNF37A||TCF7L2||IKZF5||ZBTB37||SOX5||ZNF655||ZNF345||ZNF202||KLF3||ZNF420||POU3F3||FOXF1||E2F3||ZNF597||GABPA||KLF12||ZNF697||ZNF121||HAND2||ZNF782||ZNF680||HOXD10||PLAG1||KLF7||PBX1||HOXA7||ZNF841||KDM6A||RARA||TFAP2C||HOXB2||FOXD4L1||ZNF611||BMI1||NR2C2||SMAD2||ZNF212||ZNF263||HOXA3||DACH1||ZNF548||RBPJ||YAP1||MTF1||ZNF117||ZNF24||POU3F2||GRHL1||MED1||ZBTB18||SP3||ZNF770||ZNF595||HOXA9||KLF6||ETV6||ZNF816||TEAD1||ATF7||ZNF28||FOXJ3||E2F8||ZBTB20||NFAT5||ZNF426||PRDM15||ZKSCAN8||ZNF670||HIVEP2||HDAC4||REL||RARB||SOX7||MXD1||FOXJ2||ZNF573||ZNF280C||ZNF583||BACH1||SMAD5||SUB1||ZNF85||ZNF341||ZNF736 | 3.8817316 | 4.313721 | 4.131958 | 4.340501 | 3.8439485 | 4.0636638 | 4.353367 | 4.366539 | 3.8575928 | 3.9660559 | 3.8174198 | 4.309472 | 4.316368 | 4.315313 | 4.268328 | 4.035825 | 4.0810478 | 4.325910 | 4.303696 | 4.390483 | 3.8195239 | 3.8429307 | 3.8689667 | 3.9889102 | 4.027876 | 4.167996 | 4.355701 | 4.347846 | 4.356538 | 4.380044 | 4.336520 | 4.382590 |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 325 | 0.3132378 | 1.400254 | 0.0011380385 | 0.1755860 | 0.1707382 | 3248 | tags=34%, list=21%, signal=28% | FLI1||RUNX2||PGR||CSRNP3||ZNF268||BCL11B||GLIS3||GLI1||ZNF226||LHX4||ZNF143||FOXD2||NHLH1||ELK4||NFIB||ZNF225||ZNF227||IRX6||MAFF||NRL||NR5A2||ZNF224||DMTF1||SOX2||WT1||ZNF175||SIX4||IKZF3||SP1||HNF4G||RORB||BARX2||RXRB||AR||GMEB1||EBF2||GMEB2||SMAD3||ZNF112||NR4A3||IRF1||HSF2||MEF2C||ATF2||CREBRF||TCF4||NFYB||ZNF345||FOXF1||E2F3||ZNF597||GABPA||HAND2||ZNF782||HOXD10||PLAG1||KLF7||PBX1||HOXA7||ZNF841||TFAP2C||HOXB2||NR2C2||SMAD2||RBPJ||MTF1||ZNF24||POU3F2||GRHL1||ZNF770||HOXA9||KLF6||ETV6||FOXJ3||NFAT5||PRDM15||REL||FOXJ2||BACH1||ISL1||TCF12||HIF1A||ESRRG||SMAD4||YY2||STAT5B||MECOM||MEIS1||TFDP2||STOX1||ALX1||GLIS2||KLF5||MEF2A||PLAGL2||FOXF2||RUNX1||NFYC||MZF1||HOXD4||LEF1||MYBL1||SMAD1||ZFAT||ZIC3||RLF||TFAP2A||SRF||HOXD8||ZNF267||NFIA | 3.6872896 | 3.960871 | 3.902201 | 4.061836 | 3.6856628 | 3.9931771 | 4.015249 | 4.098594 | 3.6429656 | 3.7177423 | 3.7001061 | 3.954923 | 3.961958 | 3.965712 | 3.988531 | 3.877364 | 3.8363826 | 4.067658 | 3.998704 | 4.116717 | 3.6835838 | 3.6258274 | 3.7451102 | 3.9262229 | 3.964849 | 4.083749 | 4.029447 | 3.996738 | 4.019367 | 4.092369 | 4.109251 | 4.094103 |
| GO:0050998 | nitric-oxide synthase binding | 12 | 0.7519752 | 1.903956 | 0.0063006301 | 0.3331662 | 0.3239678 | 1607 | tags=33%, list=10%, signal=30% | DNM1||CAV1||DMD||SCN5A | 7.9332257 | 7.419043 | 7.572860 | 7.328985 | 7.9629545 | 7.9587460 | 7.369342 | 7.319730 | 7.8934838 | 7.9185272 | 7.9860693 | 7.403339 | 7.438615 | 7.414952 | 7.325418 | 7.715820 | 7.6479738 | 7.437228 | 7.223477 | 7.318280 | 8.0383696 | 7.8557280 | 7.9886420 | 8.0328771 | 7.936545 | 7.903670 | 7.374211 | 7.379883 | 7.353802 | 7.340308 | 7.329526 | 7.288848 |
| GO:0003729 | mRNA binding | 270 | 0.3012473 | 1.328873 | 0.0067050242 | 0.3331662 | 0.3239678 | 5332 | tags=54%, list=34%, signal=36% | LINC00869||SF1||SNRNP35||RC3H1||RBM15||SNIP1||SECISBP2||CHTOP||NOVA1||EIF3D||LUC7L2||CPEB2||FUBP1||RBM5||ELAVL2||PUM2||QKI||IREB2||CPEB3||RBM20||MARF1||GNL3||RC3H2||FXR1||RARA||EIF4G2||RBM47||HNRNPL||ETF1||SRRM2||JRK||NXF1||PPP1R8||SRSF1||ZC3H12C||POLDIP3||PCBP1||YTHDF3||ZFP36L1||G3BP1||DDX6||DENR||SRRM3||UTP23||PCF11||TARDBP||PCBP2||TRA2B||RPS3A||RBM4||PTBP1||PAIP2||IGF2BP1||PTBP2||EXOSC9||RBMS1||METTL14||RSL1D1||NCBP2||ZC3H12B||LUC7L||EIF2A||YTHDF1||CNBP||SYNCRIP||NCBP3||ILF3||RBPMS||STRAP||TIAL1||SF3B6||LSM14A||GRSF1||DHX36||CSTF3||ALYREF||IGF2BP3||SAMD4B||LARP4B||CELF2||RNPS1||CCT5||LSM1||UPF3B||RPL5||NUDT21||AQR||METTL3||SRSF3||YTHDF2||PPIE||PUM3||RBM25||EDC3||THOC2||CPSF1||ADARB1||FUBP3||MIR17HG||HNRNPK||G3BP2||PTBP3||SLBP||THOC5||CELF1||SF3B1||FMR1||HNRNPC||HNRNPA3||DCP1A||YTHDC1||CDC40||MRPL13||CASC3||CRYZ||HNRNPA2B1||KHSRP||KHDRBS3||LARP4||CARHSP1||SNRPC||SNRNP70||ELAVL1||SCAF8||HNRNPU||RPS7||RBM14||YBX1||EIF2S2||RAVER2||SERBP1||C1QBP||EIF3A||SUPT5H||TPR||SLC4A1AP||DHX9||YBX3||MRPS7||SSB||RBMX||TP53||RBFOX2||CPEB4||AUH||TACO1||RPL26 | 8.3810798 | 8.495182 | 8.080206 | 8.400462 | 8.3823407 | 8.2172056 | 8.485244 | 8.434121 | 8.3987516 | 8.3595203 | 8.3846941 | 8.564144 | 8.453880 | 8.464947 | 8.092496 | 8.044178 | 8.1032618 | 8.421150 | 8.357554 | 8.421744 | 8.3621443 | 8.4017143 | 8.3828919 | 8.3230881 | 8.312103 | 7.992650 | 8.471273 | 8.486983 | 8.497356 | 8.439184 | 8.399948 | 8.462538 |
| GO:0042165 | neurotransmitter binding | 10 | -0.7789157 | -1.664142 | 0.0085902746 | 0.3600516 | 0.3501109 | 1787 | tags=40%, list=12%, signal=35% | CHRNA3||CHRNB1||CHRM3||CHRNA4 | 3.3530030 | 3.787252 | 3.522380 | 3.604631 | 3.3601630 | 3.4712922 | 3.744162 | 3.618473 | 3.3804648 | 3.5266641 | 3.1233931 | 3.812713 | 3.767392 | 3.781278 | 3.522046 | 3.554221 | 3.4901606 | 3.615110 | 3.536017 | 3.660044 | 3.4085441 | 3.4051074 | 3.2620476 | 3.5403342 | 3.165587 | 3.662771 | 3.783702 | 3.683912 | 3.762963 | 3.608915 | 3.636309 | 3.610027 |
| GO:0003725 | double-stranded RNA binding | 58 | 0.4788797 | 1.736998 | 0.0114427861 | 0.4387868 | 0.4266723 | 5256 | tags=66%, list=34%, signal=44% | ZFR||DDX60||AGO4||RC3H1||EIF2AK2||DHX58||DGCR8||RC3H2||MBNL1||YRDC||ZNF346||TARBP2||DICER1||DDX58||DHX15||STRBP||PRKRIP1||ILF3||LSM14A||DHX36||PRKRA||STAU2||ADARB1||HMGB1||DDX1||EIF4B||DDX21||LRRFIP1||TFRC||CLTC||TUBA1B||SUPV3L1||ELAVL1||HNRNPU||DROSHA||HSPD1||DHX9||ILF2 | 7.4461074 | 7.714042 | 7.573769 | 7.763530 | 7.4362860 | 7.5466805 | 7.751794 | 7.817925 | 7.3848900 | 7.5506861 | 7.3967470 | 7.724011 | 7.710240 | 7.707821 | 7.606911 | 7.529392 | 7.5839086 | 7.754297 | 7.715753 | 7.818663 | 7.4718650 | 7.4268558 | 7.4094156 | 7.6057304 | 7.466034 | 7.564726 | 7.745154 | 7.751835 | 7.758364 | 7.822979 | 7.815122 | 7.815660 |
| GO:0017116 | single-stranded DNA helicase activity | 23 | 0.6009958 | 1.781320 | 0.0139713587 | 0.4974802 | 0.4837452 | 5304 | tags=83%, list=34%, signal=54% | MCM4||RFC3||CHTF18||MCM5||MCM7||RAD51||MCM3||HELB||CHTF8||POLQ||RFC5||MCM6||MCM8||RFC2||RFC4||MCM9||DNA2||DHX9||DSCC1 | 6.1042027 | 6.403145 | 6.348632 | 6.527836 | 6.0412068 | 6.1262467 | 6.435087 | 6.650060 | 6.0769135 | 6.1827723 | 6.0494614 | 6.416253 | 6.412320 | 6.380597 | 6.488128 | 6.248178 | 6.2983398 | 6.527660 | 6.481181 | 6.573200 | 6.0501034 | 6.0350277 | 6.0384461 | 6.0660808 | 6.025531 | 6.274564 | 6.462329 | 6.411319 | 6.431154 | 6.632061 | 6.682825 | 6.634725 |
| GO:0003743 | translation initiation factor activity | 48 | 0.4933259 | 1.719967 | 0.0147669589 | 0.5076779 | 0.4936613 | 6152 | tags=67%, list=40%, signal=40% | EIF3CL||EIF3D||EIF2S1||EIF1||EIF3B||EIF4G2||DENR||EIF2S3||EIF5||EIF2A||EIF1B||EIF1AD||EIF1AX||EIF5B||EIF3J||EIF4B||EIF2D||EIF3E||EIF4E2||MCTS1||EIF4E3||EIF2S2||EIF3A||COPS5||EIF4H||EIF3K||EIF4A1||EIF3I||DHX29||EIF3C||EIF3G||EIF6 | 7.3687821 | 7.789072 | 7.378391 | 7.826500 | 7.2532119 | 7.2381539 | 7.840975 | 7.908629 | 7.3934496 | 7.4901498 | 7.2087244 | 7.786531 | 7.784387 | 7.796270 | 7.477156 | 7.277592 | 7.3735166 | 7.808834 | 7.789845 | 7.879275 | 7.2901106 | 7.3359077 | 7.1252115 | 7.3110905 | 7.151972 | 7.246973 | 7.829727 | 7.843611 | 7.849514 | 7.886046 | 7.934810 | 7.904610 |
| GO:0005244 | voltage-gated ion channel activity | 101 | -0.4536697 | -1.462601 | 0.0167519777 | 0.5219288 | 0.5075188 | 2983 | tags=38%, list=19%, signal=31% | CACNG5||TPCN2||KCNIP3||TMEM109||TPCN1||KCNT2||KCNG1||KCNJ11||KCNH2||KCNC3||CACNA1A||GRIN2D||CDK5||KCNC4||CACNB3||KCNAB2||KCNQ1||CATSPER3||CACNA1G||KCNH4||CYBB||CLIC5||KCNK1||SCN4B||CACNG8||CACNA1B||SCN11A||CACNG4||KCNN2||KCNK2||SCN3B||HCN4||KCNB1||SCN2A||KCNC1||KCNK13||KCND2||PTK2B | 3.4502435 | 3.829065 | 3.597193 | 3.902719 | 3.3688213 | 3.4006349 | 3.810638 | 3.889471 | 3.4563622 | 3.5606454 | 3.3240109 | 3.831573 | 3.840053 | 3.815462 | 3.730740 | 3.449451 | 3.5976984 | 3.879432 | 3.886049 | 3.941857 | 3.3539222 | 3.4060990 | 3.3456957 | 3.3702606 | 3.340652 | 3.486819 | 3.798860 | 3.815113 | 3.817867 | 3.871128 | 3.917191 | 3.879678 |
| GO:0106310 | protein serine kinase activity | 322 | 0.2675577 | 1.196020 | 0.0184605455 | 0.5416327 | 0.5266787 | 3549 | tags=33%, list=23%, signal=26% | MYO3A||PHKG1||CDK3||WNK4||AATK||LRRK2||DCLK1||OBSCN||MAK||CAMK2B||PLK4||DMPK||PIM1||TTK||STK17B||PLK2||RSKR||DCAF1||UHMK1||CLK1||SGK3||CLK3||MAPK4||EIF2AK2||CDK14||HIPK3||BUB1B||MAPK8||MAP3K1||MAPK1||RIOK3||MAP3K2||NEK5||MAP4K3||HIPK1||CDK8||TNIK||MAPK13||MASTL||CDK11A||PRKACB||PRKCB||PRKCH||TAOK1||CDK2||DAPK1||NEK7||MAP3K4||CDK12||PRKAA1||STK17A||KSR2||SMG1||PRKAA2||CLK2||PDPK1||PIKFYVE||TRIO||MAP4K1||CHEK1||PAK2||ALPK3||PDIK1L||AAK1||CDK1||STK26||TAOK3||TLK1||BUB1||SLK||STK24||MAP3K5||ERN1||MAP3K13||PRPF4B||STK39||MARK3||PKN2||IRAK4||AKT2||MAPKAPK5||CAMK1D||DYRK1A||HIPK2||CDC42BPA||BMP2K||AURKA||LATS1||AURKB||IKBKB||RPS6KC1||CSNK1A1||PRKD3||MAST2||NIM1K||MARK2||PRKX||DYRK2||MAP3K6||NUAK2||STK11||CDK6||ULK1||ROCK2||PBK||TRPM7||ROCK1 | 4.4067919 | 4.769352 | 4.684613 | 4.756946 | 4.4297872 | 4.7483631 | 4.814029 | 4.807047 | 4.3117668 | 4.3868358 | 4.5144546 | 4.758396 | 4.780014 | 4.769564 | 4.759180 | 4.689168 | 4.6011554 | 4.733351 | 4.721859 | 4.813875 | 4.4374433 | 4.2854864 | 4.5539317 | 4.6441987 | 4.749398 | 4.844542 | 4.838560 | 4.803317 | 4.799892 | 4.787251 | 4.841990 | 4.791252 |
| GO:0005125 | cytokine activity | 57 | -0.5111920 | -1.528739 | 0.0185231539 | 0.5416327 | 0.5266787 | 4432 | tags=51%, list=29%, signal=36% | BMP1||TGFB1||MIF||CXCL16||GDF11||GRN||BMP2||BMP6||C5||TIMP1||WNT11||CSF1||XCL1||WNT7B||BMP8B||CKLF||CD70||GDF9||CMTM3||WNT9A||TSLP||CTF1||WNT10B||GDF1||TGFB2||WNT3||CXCL12||WNT2B||IL23A | 6.6181959 | 6.112163 | 6.241935 | 5.956446 | 6.6922661 | 6.7898676 | 6.004907 | 5.852701 | 6.5888108 | 6.3996439 | 6.8332339 | 6.233083 | 6.037224 | 6.058055 | 5.991760 | 6.417572 | 6.2842177 | 6.105315 | 5.865959 | 5.885623 | 6.6992864 | 6.4593548 | 6.8866130 | 6.6847788 | 7.027580 | 6.623577 | 6.010668 | 5.999064 | 6.004966 | 5.882480 | 5.836700 | 5.838456 |
| GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential | 16 | -0.6736405 | -1.603055 | 0.0190141875 | 0.5416327 | 0.5266787 | 1787 | tags=44%, list=12%, signal=39% | CHRNA3||GABRD||CHRNB1||GABRR2||GABRG2||GRID2||CHRNA4 | 0.6954202 | 2.111210 | 1.282240 | 2.095175 | 0.5795651 | 0.8139233 | 2.043961 | 1.987972 | 0.7364732 | 0.8583144 | 0.4636761 | 2.126835 | 2.113302 | 2.093294 | 1.587066 | 1.202612 | 0.9931695 | 1.969999 | 2.183586 | 2.123616 | 0.4998946 | 0.6565138 | 0.5780371 | 0.4147229 | 0.640319 | 1.252319 | 2.011359 | 2.139458 | 1.975886 | 1.960642 | 2.024871 | 1.977632 |
| GO:0070273 | phosphatidylinositol-4-phosphate binding | 24 | 0.5650652 | 1.689918 | 0.0200845666 | 0.5562309 | 0.5408739 | 2405 | tags=46%, list=15%, signal=39% | OBSCN||GSDMB||RUBCNL||LANCL2||OSBPL5||OSBPL8||SNX5||CERT1||GOLPH3L||SESTD1||PLEKHA5 | 3.8591144 | 4.845312 | 4.180924 | 4.988812 | 3.8252812 | 3.9385642 | 4.952832 | 5.076912 | 3.8080050 | 3.9663434 | 3.7966830 | 4.838298 | 4.820105 | 4.876949 | 4.482039 | 4.042309 | 3.9625146 | 4.896192 | 5.004773 | 5.060654 | 3.8041370 | 3.8374343 | 3.8340399 | 3.8298353 | 3.853758 | 4.114483 | 4.949058 | 4.951176 | 4.958246 | 5.063404 | 5.099523 | 5.067539 |
| GO:0005248 | voltage-gated sodium channel activity | 16 | -0.6690224 | -1.592065 | 0.0215006582 | 0.5716635 | 0.5558804 | 1494 | tags=44%, list=10%, signal=40% | TPCN1||CACNA1G||SCN4B||SCN11A||SCN3B||HCN4||SCN2A | 1.2326500 | 1.772722 | 2.041439 | 1.615031 | 1.2416757 | 1.9003944 | 1.736191 | 1.588493 | 1.1085696 | 1.2270788 | 1.3520268 | 1.733026 | 1.849602 | 1.732353 | 2.145193 | 2.080495 | 1.8862133 | 1.594806 | 1.630212 | 1.619845 | 1.2329521 | 0.9589961 | 1.4853031 | 1.4698310 | 1.763235 | 2.331986 | 1.776478 | 1.781247 | 1.646861 | 1.580171 | 1.699170 | 1.477604 |
| GO:0017056 | structural constituent of nuclear pore | 24 | 0.5551551 | 1.660280 | 0.0236081748 | 0.6035218 | 0.5868591 | 5463 | tags=67%, list=35%, signal=43% | NUP58||NUP88||NUP54||NUP160||NUP155||NUP205||NUP62CL||NUP107||UPF3B||NUP133||NUP93||NUP85||TPR||NUP35||NUP153||UPF3A | 5.1912287 | 6.155828 | 5.645495 | 6.222029 | 5.1072230 | 5.3902559 | 6.224394 | 6.308485 | 5.1491395 | 5.3614525 | 5.0448563 | 6.149208 | 6.171944 | 6.146194 | 5.884950 | 5.495821 | 5.5220606 | 6.160485 | 6.213349 | 6.289339 | 5.1520101 | 5.0894105 | 5.0791654 | 5.3777534 | 5.259125 | 5.521863 | 6.221382 | 6.226948 | 6.224847 | 6.278956 | 6.329054 | 6.316973 |
| GO:0008146 | sulfotransferase activity | 32 | -0.5714945 | -1.556484 | 0.0243869757 | 0.6078454 | 0.5910633 | 3477 | tags=44%, list=22%, signal=34% | NDST1||HS3ST3A1||CHST12||CHST9||WSCD1||CHST8||HS6ST1||NDST2||SULT1A4||SULT1A1||CHST7||SULT1B1||CHST14||CHST2 | 2.5890796 | 3.243257 | 3.125035 | 3.109163 | 2.6232058 | 3.0045084 | 3.135684 | 2.932654 | 2.5639420 | 2.5349908 | 2.6650508 | 3.213805 | 3.312740 | 3.200604 | 3.114083 | 3.207837 | 3.0487388 | 3.061721 | 3.157642 | 3.106528 | 2.6378635 | 2.4630849 | 2.7539233 | 2.7359923 | 2.882262 | 3.327672 | 3.177279 | 3.141001 | 3.087356 | 2.942231 | 2.911359 | 2.944138 |
| GO:0008509 | anion transmembrane transporter activity | 183 | -0.3977480 | -1.355443 | 0.0265286315 | 0.6219483 | 0.6047769 | 5691 | tags=52%, list=37%, signal=34% | SLC6A8||CLCN5||SLC25A4||SLC1A5||SLC16A7||SLC2A8||LRRC8E||SLC36A1||SLC38A6||SLC25A1||SLC25A14||SLC25A19||SLC12A4||SLC47A1||SLC29A1||CLCC1||SLC25A13||LRRC8A||LRRC8B||ABCC4||SLC7A3||SLC35A1||SLC1A4||SLC7A1||TTYH3||SLC25A38||ANO7||ANO8||SLC39A14||GLRB||SLC9A8||SLC9A3R1||SLC9A1||SLCO5A1||SLC35B2||SLCO4C1||ABCC1||GPR89A||CLCN3||SLC7A5||MFSD5||SLC25A11||SLC2A1||ANO4||SLC37A4||SLC9A2||SLC4A2||SLC12A7||SLC35B4||MFSD12||MFSD10||SLC15A4||SLC27A6||SLC35B3||SLC2A10||MFSD2A||SLC38A9||SLC38A7||SLC52A2||SLC25A10||SLC27A4||ABCC10||SLC9A5||PACC1||SLC9A7||SLC12A9||SFXN3||SLC19A1||ABCG2||CTNS||SLC5A12||GABRD||SLC26A2||SLC25A29||SLC9A3||SLC66A1||CLIC5||SLC2A3||SLC25A42||SLC38A3||SLC26A11||SLC46A1||SLC4A11||SLC37A1||ABCC11||SLC4A3||LRRC8D||GABRR2||SLC2A6||SLC9A9||SLC24A1||SLC1A1||SLC24A4||GABRG2||CLDN4||SLC4A8 | 5.0272168 | 5.197398 | 5.154602 | 5.272314 | 5.0238046 | 5.1399303 | 5.130724 | 5.241743 | 5.0179905 | 5.0775464 | 4.9845717 | 5.217613 | 5.225316 | 5.148014 | 5.139831 | 5.151464 | 5.1723237 | 5.285183 | 5.253240 | 5.278324 | 5.0238630 | 5.0106855 | 5.0367476 | 5.1041580 | 5.141631 | 5.173175 | 5.145949 | 5.131262 | 5.114792 | 5.225084 | 5.283218 | 5.216000 |
| GO:0042605 | peptide antigen binding | 15 | -0.6685112 | -1.571569 | 0.0268053318 | 0.6219483 | 0.6047769 | 4109 | tags=73%, list=26%, signal=54% | SLC7A5||TAP2||HLA-E||HLA-B||HLA-A||DHCR24||HLA-C||HLA-DRB1||HLA-DPA1||HLA-DQB1||HLA-F | 5.9325778 | 6.019350 | 6.314373 | 5.977795 | 6.0415731 | 6.3464148 | 5.777443 | 5.807609 | 5.8519371 | 5.8294631 | 6.1003744 | 6.061753 | 6.074653 | 5.916352 | 6.075955 | 6.518618 | 6.3147229 | 6.063501 | 5.962935 | 5.902334 | 6.0752154 | 5.8570937 | 6.1744005 | 6.1896774 | 6.256756 | 6.564518 | 5.864251 | 5.751121 | 5.712619 | 5.835855 | 5.870799 | 5.711350 |
| GO:0030332 | cyclin binding | 29 | 0.5145464 | 1.615946 | 0.0274480712 | 0.6219483 | 0.6047769 | 2687 | tags=38%, list=17%, signal=31% | PTCH1||FBXW7||CDK14||XRCC6||CDK2||CDK12||CDKN1B||CRABP2||CDKN1A||CDK1||RBM4 | 8.0100115 | 7.650636 | 7.892322 | 7.577705 | 8.0542133 | 8.0586883 | 7.642821 | 7.619421 | 7.9576201 | 7.9606689 | 8.1066604 | 7.695372 | 7.631066 | 7.624400 | 7.747488 | 7.940394 | 7.9786085 | 7.613380 | 7.518985 | 7.598968 | 8.0654735 | 8.0033458 | 8.0923810 | 8.1108305 | 8.127331 | 7.929692 | 7.635544 | 7.646557 | 7.646335 | 7.609612 | 7.622198 | 7.626401 |
| GO:0043425 | bHLH transcription factor binding | 22 | 0.5537069 | 1.625329 | 0.0291995878 | 0.6419833 | 0.6242587 | 1137 | tags=23%, list=7%, signal=21% | RUNX2||BHLHE41||PSMD9||SP1||SMAD3 | 4.4991573 | 4.702176 | 4.662708 | 4.667696 | 4.5081095 | 4.7207610 | 4.710711 | 4.734687 | 4.4796591 | 4.4599473 | 4.5560688 | 4.684498 | 4.718666 | 4.703160 | 4.744070 | 4.614452 | 4.6260029 | 4.710191 | 4.588710 | 4.701043 | 4.5117895 | 4.4393974 | 4.5701720 | 4.7157563 | 4.716148 | 4.730331 | 4.730498 | 4.692021 | 4.709357 | 4.723821 | 4.765597 | 4.714124 |
| GO:0052742 | phosphatidylinositol kinase activity | 17 | 0.5927355 | 1.630340 | 0.0296200901 | 0.6419833 | 0.6242587 | 4963 | tags=71%, list=32%, signal=48% | PIP5K1B||PI4KAP2||PI4KAP1||PIKFYVE||PI4K2B||PIK3CD||PIK3C2B||PIK3C2A||PIK3CB||PIK3C3||PI4KB||PIP5K1A | 4.0930949 | 4.383612 | 4.725927 | 4.536493 | 4.0649250 | 4.4768557 | 4.501843 | 4.623976 | 4.0195270 | 4.1622634 | 4.0939647 | 4.394038 | 4.374530 | 4.382201 | 4.874806 | 4.671469 | 4.6186458 | 4.496750 | 4.485851 | 4.622822 | 4.0538188 | 3.9827956 | 4.1530752 | 4.2353498 | 4.283969 | 4.832193 | 4.532938 | 4.495046 | 4.476980 | 4.586111 | 4.660305 | 4.624558 |
| GO:0045505 | dynein intermediate chain binding | 23 | 0.5438678 | 1.611996 | 0.0307369892 | 0.6494080 | 0.6314784 | 3537 | tags=48%, list=23%, signal=37% | DNAH7||DNHD1||DNAH11||HOOK3||DNAH10||DNAH14||DNAH1||PAFAH1B1||DNAAF5||HTT||BICD1 | 5.5444668 | 5.821121 | 5.509555 | 5.730160 | 5.4314045 | 5.3204109 | 5.860254 | 5.738382 | 5.5839715 | 5.7031273 | 5.3201307 | 5.772987 | 5.831502 | 5.857576 | 5.644379 | 5.320931 | 5.5446146 | 5.717757 | 5.714218 | 5.758092 | 5.3832931 | 5.5468330 | 5.3566468 | 5.3431472 | 5.305794 | 5.312013 | 5.882956 | 5.868023 | 5.829252 | 5.732198 | 5.726465 | 5.756309 |
| GO:0017147 | Wnt-protein binding | 22 | -0.6066821 | -1.535730 | 0.0325765054 | 0.6494080 | 0.6314784 | 2152 | tags=41%, list=14%, signal=35% | FZD5||FZD8||LRP5||SFRP1||APCDD1||PTPRO||FZD9||FZD2||FZD10 | 2.5694107 | 3.809682 | 3.157106 | 3.657110 | 2.5283794 | 2.8850358 | 3.687437 | 3.608848 | 2.5313245 | 2.6791480 | 2.4908806 | 3.847369 | 3.852558 | 3.725575 | 3.266165 | 3.119125 | 3.0792398 | 3.622149 | 3.703009 | 3.644963 | 2.5801613 | 2.4788425 | 2.5243474 | 2.7155028 | 2.601226 | 3.251792 | 3.696102 | 3.718878 | 3.646382 | 3.599341 | 3.665661 | 3.559544 |
| GO:0003724 | RNA helicase activity | 65 | 0.4173510 | 1.532362 | 0.0330802603 | 0.6494080 | 0.6314784 | 6133 | tags=65%, list=39%, signal=39% | DDX60||DHX58||DDX59||FANCM||G3BP1||DDX6||DDX17||MTREX||DDX58||DHX38||DHX15||DHX35||MOV10||DHX36||DDX20||DDX18||AQR||DDX52||BRIP1||DDX39B||DDX19B||DDX1||DHX40||DDX21||DDX24||DHX8||RAD54B||SUPV3L1||DDX47||SNRNP200||DDX41||DHX9||EIF4A3||DDX49||DHX16||EIF4A1||YTHDC2||DHX30||DHX29||DHX57||DDX56||DDX55 | 6.1760494 | 6.725926 | 6.517651 | 6.787998 | 6.0715871 | 6.2209673 | 6.785473 | 6.848533 | 6.1381905 | 6.3457077 | 6.0258220 | 6.737832 | 6.726063 | 6.713784 | 6.718224 | 6.373478 | 6.4374841 | 6.755134 | 6.765674 | 6.841635 | 6.1060586 | 6.1155411 | 5.9897947 | 6.1766973 | 6.060529 | 6.404252 | 6.792034 | 6.787336 | 6.777008 | 6.826833 | 6.879161 | 6.839084 |
| GO:0003714 | transcription corepressor activity | 165 | 0.3186953 | 1.336588 | 0.0368171021 | 0.6494080 | 0.6314784 | 4150 | tags=40%, list=27%, signal=30% | SF1||WWTR1||ATF7IP||BEND6||CRYM||RERE||PAWR||CCAR1||CCND1||JAZF1||CASP8AP2||TLE2||N4BP2L2||CBFA2T3||NPAT||MIER1||RYBP||LIMD1||ZMYND11||GMNN||SIRT6||AJUBA||HDAC7||ZFPM1||YAP1||MED1||TBL1XR1||MIER3||TLE3||DNMT3B||HDAC4||PARP9||DMAP1||TCERG1||PA2G4||CNOT7||MTA3||EZH2||SAP18||HMGA2||YAF2||MIDEAS||COPS2||ZNF451||TLE4||NRIP1||HIPK2||RB1||TFAP2A||NCOA5||RLIM||CITED2||CIR1||SIN3A||SNW1||CTBP2||DPF2||TSG101||FHL2||DNAJB1||NCOR1||BCOR||PRMT5||CDYL||MIER2||TAF9B | 5.7738979 | 5.979136 | 5.856527 | 5.996211 | 5.7412323 | 5.8466319 | 6.024782 | 6.070197 | 5.7461345 | 5.8399339 | 5.7332583 | 5.994260 | 5.978629 | 5.964363 | 5.905321 | 5.817175 | 5.8456776 | 5.993007 | 5.953714 | 6.040599 | 5.7334295 | 5.7395575 | 5.7506570 | 5.7997524 | 5.823190 | 5.914393 | 6.026508 | 6.023684 | 6.024152 | 6.053681 | 6.087199 | 6.069516 |
| GO:0008569 | minus-end-directed microtubule motor activity | 10 | 0.6702646 | 1.620821 | 0.0370370370 | 0.6494080 | 0.6314784 | 2538 | tags=60%, list=16%, signal=50% | DNAH7||DNHD1||DNAH11||DNAH10||DNAH14||DNAH1 | 2.5471009 | 3.093272 | 3.388189 | 3.322355 | 2.4909625 | 3.3079469 | 3.167785 | 3.387125 | 2.3721899 | 2.6638806 | 2.5895576 | 3.022803 | 3.111725 | 3.142622 | 3.540673 | 3.396568 | 3.2081732 | 3.243872 | 3.291122 | 3.425837 | 2.6053559 | 2.2849034 | 2.5621459 | 3.2329313 | 3.220147 | 3.458132 | 3.273649 | 3.191280 | 3.027664 | 3.282961 | 3.436128 | 3.436904 |
| GO:0005044 | scavenger receptor activity | 19 | -0.6182065 | -1.523299 | 0.0379764976 | 0.6494080 | 0.6314784 | 3383 | tags=58%, list=22%, signal=45% | SCARB1||LGALS3BP||LOXL3||SCARA3||LRP1||LOXL4||TMPRSS5||SCARF2||LOXL2||MEGF10||SSC4D | 2.8174229 | 3.341300 | 3.131827 | 2.883946 | 2.8475918 | 3.1621500 | 3.147306 | 2.723207 | 2.6969701 | 2.8389456 | 2.9083723 | 3.337013 | 3.378015 | 3.308015 | 3.007127 | 3.331486 | 3.0339045 | 2.903004 | 2.941456 | 2.803911 | 2.8829999 | 2.6322836 | 3.0030707 | 2.9034274 | 3.050263 | 3.471088 | 3.200875 | 3.137183 | 3.102117 | 2.735050 | 2.795846 | 2.634121 |
| GO:1901505 | carbohydrate derivative transmembrane transporter activity | 35 | -0.5412509 | -1.498557 | 0.0383445723 | 0.6494080 | 0.6314784 | 5269 | tags=66%, list=34%, signal=44% | SLC29A4||SLC29A1||LRRC8A||SLC35A1||SLC35A4||SLC29A3||SLC29A2||SLC35B2||SLC35C1||SLC35E3||SLC37A4||SLC35B4||SLC15A4||SLC17A5||SLC35B3||SLC50A1||MFSD2A||SLC35A2||SLC19A1||SLC35A5||ABCD1||SLC25A42||SLC37A1 | 6.3253621 | 6.062761 | 6.284986 | 6.090106 | 6.3135386 | 6.3140915 | 5.965538 | 6.060232 | 6.3496452 | 6.4091299 | 6.2101453 | 6.123825 | 6.065402 | 5.996232 | 6.190729 | 6.274196 | 6.3835367 | 6.151292 | 6.031992 | 6.084553 | 6.3281086 | 6.3410435 | 6.2704900 | 6.3749174 | 6.363261 | 6.197350 | 5.982309 | 5.961609 | 5.952535 | 6.050909 | 6.081233 | 6.048321 |
| GO:0015079 | potassium ion transmembrane transporter activity | 77 | -0.4523157 | -1.409097 | 0.0386933462 | 0.6494080 | 0.6314784 | 2963 | tags=39%, list=19%, signal=32% | TMEM38A||KCNIP3||KCNT2||KCNG1||KCNJ11||SLC9A5||KCNH2||SLC9A7||SLC12A9||CCDC51||KCNC3||ATP1A3||KCNC4||KCNAB2||SLC9A3||KCNQ1||TMEM175||KCNH4||KCNK1||AQP1||SLC9A9||SLC24A1||SLC24A4||KCNN2||KCNK2||HCN4||KCNB1||KCNC1||KCNK13||KCND2 | 2.9545959 | 3.574772 | 3.473154 | 3.632044 | 2.9224135 | 3.3028883 | 3.458109 | 3.528414 | 2.9202133 | 3.0459647 | 2.8929493 | 3.576763 | 3.629050 | 3.516298 | 3.523014 | 3.484947 | 3.4091851 | 3.607090 | 3.665761 | 3.622639 | 2.9659389 | 2.8895543 | 2.9106651 | 3.1490124 | 3.065084 | 3.628427 | 3.485659 | 3.458698 | 3.429420 | 3.537792 | 3.558143 | 3.488418 |
| GO:0019215 | intermediate filament binding | 10 | -0.7056792 | -1.507674 | 0.0397300199 | 0.6494080 | 0.6314784 | 3208 | tags=50%, list=21%, signal=40% | VIM||EPPK1||NES||FAM83H||SYNM | 8.5224245 | 8.072149 | 8.329160 | 7.891424 | 8.4727966 | 8.0541503 | 7.991546 | 7.875387 | 8.6121653 | 8.6053244 | 8.3325406 | 8.136054 | 8.010845 | 8.066818 | 8.204991 | 8.312272 | 8.4589129 | 7.969872 | 7.803491 | 7.896105 | 8.5538281 | 8.6387651 | 8.1870324 | 8.3477789 | 7.704474 | 8.038952 | 7.999858 | 7.956188 | 8.017895 | 7.858944 | 7.869204 | 7.897732 |
| GO:0008353 | RNA polymerase II CTD heptapeptide repeat kinase activity | 12 | 0.6379320 | 1.615205 | 0.0399039904 | 0.6494080 | 0.6314784 | 3896 | tags=67%, list=25%, signal=50% | MAPK1||BRD4||CDK8||CDK12||CCNK||CDK1||CDK6||CDK19 | 6.0729601 | 6.343416 | 5.995641 | 6.188763 | 6.1382626 | 6.1962562 | 6.359451 | 6.271852 | 5.9369979 | 6.0687323 | 6.2010734 | 6.395851 | 6.306457 | 6.326407 | 5.952605 | 6.063790 | 5.9679948 | 6.162802 | 6.132928 | 6.267095 | 6.1526721 | 6.0158969 | 6.2376011 | 6.2024119 | 6.254152 | 6.129493 | 6.351108 | 6.355795 | 6.371372 | 6.272652 | 6.270715 | 6.272190 |
| GO:0004438 | phosphatidylinositol-3-phosphatase activity | 18 | 0.5583154 | 1.559230 | 0.0405096374 | 0.6494080 | 0.6314784 | 4924 | tags=67%, list=32%, signal=46% | MTMR11||PTEN||MTMR9||MTMR2||MTMR3||MTMR10||SYNJ1||MTMR4||SACM1L||MTMR1||MTMR12||MTMR7 | 2.6946171 | 3.932654 | 3.177382 | 4.111527 | 2.6187421 | 2.9684575 | 3.990624 | 4.195301 | 2.6819629 | 2.7906242 | 2.6052295 | 3.928562 | 3.944024 | 3.925306 | 3.535453 | 2.970751 | 2.9458502 | 3.980146 | 4.122116 | 4.222127 | 2.6102483 | 2.6292003 | 2.6167132 | 2.8731410 | 2.841937 | 3.167448 | 3.982488 | 3.976976 | 4.012158 | 4.168620 | 4.210702 | 4.206212 |
| GO:0003727 | single-stranded RNA binding | 71 | 0.3920405 | 1.462891 | 0.0432612313 | 0.6494080 | 0.6314784 | 5658 | tags=61%, list=36%, signal=39% | ZFR||DDX60||AGO4||PAN3||RBM11||DHX58||FXR1||IFIT5||EXOSC10||JMJD6||MCRS1||PTBP1||DDX58||STRBP||RBMS1||PNPT1||CNBP||ILF3||RBPMS||ATXN1||LSM14A||ZC3H14||AQR||PPIE||HMGB1||FMR1||DDX1||HNRNPC||EIF4B||RBMXL1||HNRNPH1||LARP4||PATL1||SNRPC||HNRNPU||DHX9||DLX2||SSB||POLR2G||HNRNPF||EIF4A3||EIF4H||TIA1 | 6.6923323 | 7.657290 | 6.917208 | 7.629399 | 6.6303976 | 6.8097617 | 7.712199 | 7.676499 | 6.6925062 | 6.8145915 | 6.5585543 | 7.642378 | 7.650359 | 7.678876 | 7.203023 | 6.704879 | 6.7933945 | 7.585563 | 7.605740 | 7.694549 | 6.5829525 | 6.6787816 | 6.6278642 | 6.7848797 | 6.780676 | 6.862258 | 7.702842 | 7.705984 | 7.727643 | 7.672885 | 7.678168 | 7.678438 |
| GO:0061578 | Lys63-specific deubiquitinase activity | 12 | 0.6280428 | 1.590166 | 0.0438043804 | 0.6494080 | 0.6314784 | 3253 | tags=50%, list=21%, signal=40% | YOD1||CYLD||OTUD5||OTUD4||DESI2||PSMD14 | 3.3435487 | 4.891750 | 3.620794 | 5.018076 | 3.2261268 | 3.4103145 | 4.957439 | 5.120741 | 3.3324031 | 3.4834958 | 3.2008626 | 4.884133 | 4.885744 | 4.905278 | 4.122977 | 3.328922 | 3.2377232 | 4.938603 | 4.984914 | 5.124183 | 3.1804817 | 3.2842764 | 3.2116477 | 3.3302883 | 3.421237 | 3.475694 | 4.927802 | 4.956901 | 4.987008 | 5.106070 | 5.121735 | 5.134281 |
| GO:0016409 | palmitoyltransferase activity | 32 | -0.5421215 | -1.476486 | 0.0451561034 | 0.6494080 | 0.6314784 | 3465 | tags=50%, list=22%, signal=39% | ZDHHC9||SPTLC2||CPT1A||ZDHHC16||ZDHHC4||ZDHHC14||ZDHHC18||ZDHHC24||CPT2||ZDHHC1||ZDHHC22||CPT1C||HHAT||ZDHHC12||ZDHHC8||ZDHHC11B | 4.6444659 | 5.047917 | 5.077778 | 5.061872 | 4.6661972 | 4.9616061 | 4.988198 | 4.982932 | 4.5991466 | 4.7129686 | 4.6186968 | 5.027189 | 5.090834 | 5.024751 | 5.102759 | 5.100218 | 5.0291524 | 5.031029 | 5.060716 | 5.093200 | 4.7187693 | 4.6089945 | 4.6687367 | 4.8409433 | 4.730111 | 5.258484 | 5.020979 | 4.987515 | 4.955354 | 4.962288 | 5.026152 | 4.959363 |
| GO:0015491 | cation:cation antiporter activity | 21 | -0.5951885 | -1.493881 | 0.0453450475 | 0.6494080 | 0.6314784 | 2382 | tags=43%, list=15%, signal=36% | SLC9A5||SLC9A7||SLC8B1||SLC9A3||SLC9A9||SLC24A1||SLC24A4||SLC8A2||SLC8A3 | 3.7083662 | 4.219119 | 4.326501 | 4.275979 | 3.7585636 | 4.2775832 | 4.169413 | 4.265743 | 3.5755045 | 3.7028183 | 3.8350980 | 4.221882 | 4.279325 | 4.153399 | 4.316550 | 4.427891 | 4.2280993 | 4.285238 | 4.298207 | 4.243936 | 3.8004353 | 3.5738697 | 3.8838387 | 4.1631348 | 4.159632 | 4.485188 | 4.171436 | 4.198565 | 4.137591 | 4.229927 | 4.359022 | 4.203432 |
| GO:0043024 | ribosomal small subunit binding | 17 | 0.5575828 | 1.533652 | 0.0453960077 | 0.6494080 | 0.6314784 | 4308 | tags=53%, list=28%, signal=38% | PIM1||CPEB2||EIF1||PTCD3||ABCE1||EIF1B||UNG||EIF4B||NPM1 | 7.2442290 | 8.479791 | 7.416451 | 8.508063 | 7.1473751 | 7.2596872 | 8.552592 | 8.580072 | 7.3146447 | 7.3552825 | 7.0432569 | 8.509110 | 8.430491 | 8.498517 | 7.773805 | 7.201057 | 7.1948524 | 8.457172 | 8.468976 | 8.594018 | 7.1129592 | 7.2750928 | 7.0442424 | 7.2416971 | 7.076989 | 7.437718 | 8.509744 | 8.542710 | 8.603741 | 8.572195 | 8.563737 | 8.603971 |
| GO:0004402 | histone acetyltransferase activity | 34 | 0.4604229 | 1.494193 | 0.0460472698 | 0.6494080 | 0.6314784 | 4740 | tags=62%, list=31%, signal=43% | EPC1||EPC2||KAT5||KAT8||ATF2||TADA2A||NCOA1||KAT7||TAF9||NCOA3||MED24||KAT6B||NAA60||GTF3C4||KAT2A||GTF2B||SUPT7L||KAT2B||NAA50||MSL3||SRCAP | 4.7512237 | 5.409984 | 4.929008 | 5.384181 | 4.7444969 | 5.0610116 | 5.476690 | 5.440775 | 4.7211351 | 4.7218759 | 4.8088841 | 5.370418 | 5.432304 | 5.426426 | 5.034657 | 4.918188 | 4.8266252 | 5.378793 | 5.335742 | 5.436243 | 4.7650183 | 4.6350039 | 4.8268730 | 4.9864505 | 5.059062 | 5.133762 | 5.483034 | 5.478100 | 5.468899 | 5.431283 | 5.460263 | 5.430580 |
| GO:0016758 | hexosyltransferase activity | 138 | -0.4010566 | -1.335834 | 0.0462952584 | 0.6494080 | 0.6314784 | 4034 | tags=36%, list=26%, signal=27% | POGLUT1||PIGZ||EXT2||POMGNT1||GALNT11||GDPGP1||PIGP||POMT1||POGLUT2||FUT8||C1GALT1C1||GALNT2||POFUT2||A4GALT||GALNT10||FUT4||POFUT1||GALNT4||PIGB||GALNT6||CHPF2||DPY19L3||RFNG||B3GNT2||GALNT12||ALG12||B4GALT7||FUT11||GBA||TMTC1||POMGNT2||B3GAT3||PIGV||B4GAT1||CHSY3||ALG10||CHSY1||PLOD3||B4GALNT4||B3GALT9||CHPF||OGT||PIGM||B3GALT6||B3GNT4||B3GNT9||C1GALT1C1L||PYGM||MGAT3||TYMP | 4.2165371 | 5.153993 | 4.733087 | 5.086553 | 4.2148359 | 4.5257012 | 5.048818 | 5.015459 | 4.1971118 | 4.2438889 | 4.2081955 | 5.157382 | 5.192816 | 5.110604 | 4.803758 | 4.731212 | 4.6607454 | 5.043820 | 5.127569 | 5.087054 | 4.1975216 | 4.1842170 | 4.2615775 | 4.3513509 | 4.359551 | 4.815672 | 5.064835 | 5.056429 | 5.024883 | 4.998319 | 5.063569 | 4.983220 |
| GO:1990841 | promoter-specific chromatin binding | 55 | 0.4106956 | 1.471791 | 0.0481049563 | 0.6494080 | 0.6314784 | 5551 | tags=58%, list=36%, signal=38% | NRL||ZNF304||IKZF3||SMAD3||ATF2||PPARGC1A||BMI1||MED1||PRDM15||HDAC4||ISL1||CHD7||NIPBL||EZH2||USP3||ERCC1||ERCC4||POLR2A||E2F4||ZNF609||GTF2B||STAT1||RBL1||HNRNPU||GADD45A||DHX9||TP53||PCGF2||SIRT1||ERCC3||HDAC1||HDAC2 | 5.8303923 | 6.244354 | 5.989374 | 6.283778 | 5.8035554 | 6.0181049 | 6.293058 | 6.322062 | 5.7709077 | 5.8605089 | 5.8579689 | 6.242302 | 6.254222 | 6.236482 | 6.178111 | 5.861983 | 5.9073775 | 6.273251 | 6.236477 | 6.339703 | 5.7959121 | 5.7544580 | 5.8583943 | 5.9758814 | 6.044385 | 6.033117 | 6.281452 | 6.301368 | 6.296279 | 6.317194 | 6.334799 | 6.314108 |
| GO:0004435 | phosphatidylinositol phospholipase C activity | 15 | 0.5798244 | 1.547820 | 0.0491338583 | 0.6494080 | 0.6314784 | 1948 | tags=40%, list=13%, signal=35% | PLCL1||PLCD1||EDNRA||CASR||PLCB1||PLCH1 | 2.6857818 | 2.921957 | 3.367614 | 2.933347 | 2.6137350 | 3.2297486 | 2.991947 | 2.977031 | 2.5651358 | 2.6209144 | 2.8546692 | 2.877741 | 2.937431 | 2.949678 | 3.482242 | 3.359159 | 3.2522482 | 2.951638 | 2.917132 | 2.931061 | 2.6906674 | 2.3984478 | 2.7299033 | 2.9837107 | 3.005657 | 3.608989 | 3.104914 | 2.998769 | 2.861913 | 2.917938 | 3.100623 | 2.904075 |
| GO:0051959 | dynein light intermediate chain binding | 21 | 0.5211749 | 1.517383 | 0.0499151104 | 0.6494080 | 0.6314784 | 1396 | tags=29%, list=9%, signal=26% | DNAH7||DNHD1||DNAH11||RILP||HOOK3||DNAH10 | 4.9388015 | 5.453151 | 5.022176 | 5.328449 | 4.7744724 | 4.6668824 | 5.521038 | 5.354234 | 4.9977656 | 5.1263824 | 4.6514943 | 5.357330 | 5.471206 | 5.525828 | 5.210889 | 4.764920 | 5.0557949 | 5.295460 | 5.325043 | 5.364025 | 4.7132005 | 4.9328121 | 4.6629008 | 4.6272362 | 4.682856 | 4.689744 | 5.541942 | 5.534137 | 5.486411 | 5.344959 | 5.349827 | 5.367814 |
GO: CC
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0016607 | nuclear speck | 381 | 0.3114679 | 1.420793 | 0.0004788629 | 0.05498281 | 0.05121686 | 5067 | tags=50%, list=33%, signal=35% | WRN||U2AF1||DENND1B||SMC4||BARD1||WT1||PPP4R3B||RBM39||CCNL1||RBM15||SMNDC1||AR||CHTOP||CBLL1||LUC7L2||AK6||WBP4||RBM11||NUP43||CCNL2||CLK3||HBP1||MEF2C||RBM27||CDC25C||SON||SETD1A||NRDE2||PLCB1||HIPK1||PLAG1||PNISR||PRPF40A||SRSF10||ELL||SART3||THOC1||GTF2H2||SRRM1||CDK12||API5||PRKAA1||DAZAP2||ZBTB18||STK17A||SNRNP40||SYF2||HAUS6||SRRM2||NXF1||SREK1||PPP1R8||PRKAA2||SRSF1||ERBIN||CLK2||FAM193B||POLDIP3||HDAC4||PCBP1||VPS72||SLU7||TCERG1||SMURF2||SETD1B||TCF12||DDX17||HIF1A||TARDBP||PIAS3||CRY2||SAP130||MTREX||PRPF4B||SRSF11||PRPF19||RBM4||CNOT7||MECOM||MAGOH||COPS4||GATAD2B||SCNM1||SUMO1||AKAP17A||TEPSIN||SAP18||PHF5A||SMU1||GLIS2||DHX15||DYRK1A||FAM76B||PLRG1||IK||GPATCH2||NRIP1||BMP2K||ATPAF2||TIMM50||SDE2||NCBP3||CSNK1A1||SPRTN||CHD5||BAZ2A||HOXA6||SF3A1||HDAC5||AAGAB||DHX36||CIR1||GTF2E2||NOC3L||SNRPB2||RCHY1||SNW1||ALYREF||E2F7||ZC3H14||SF3B2||PRPF3||RNPS1||CWC25||PRPF6||RBBP6||AFDN||PNN||RBM10||WTAP||METTL3||SRSF3||BCLAF1||ZNF638||PPIE||RBM25||NSL1||THOC2||SRP54||RADX||PPP4R3A||MORF4L1||PRPF18||PPP1R16B||MSL1||CDC5L||DDX39B||CACTIN||ATF4||CDYL||WAC||SF3B1||BNIP3L||EAF1||RAF1||SFPQ||THRAP3||NPM1||ZC3H18||PSME4||APBB1||YTHDC1||CDC40||PRCC||CASC3||OSGEP||PRPF8||SCAPER||NCAPG2||ACIN1||BCAS2||ATXN2L||MSX2||EFTUD2||SNRPA1||MBD1||PQBP1||TENM1||PATL1||ABITRAM||SNRNP70||SMC5||HNRNPU||PIP5K1A||MBD4||AP5Z1||RBM19||SRPK2||RBM14||JADE1||GADD45A | 5.8652604 | 6.4450805 | 6.0059809 | 6.4170626 | 5.8396719 | 5.9361392 | 6.4978948 | 6.4919319 | 5.8643886 | 5.9277281 | 5.800877 | 6.4612156 | 6.4336896 | 6.440193 | 6.2029531 | 5.888633 | 5.9041179 | 6.3843761 | 6.3857427 | 6.4790144 | 5.8362049 | 5.8383306 | 5.8444674 | 5.8849483 | 5.8718347 | 6.045104 | 6.4964299 | 6.489176 | 6.5080162 | 6.4731700 | 6.5103056 | 6.492081 |
| GO:0071007 | U2-type catalytic step 2 spliceosome | 30 | 0.6248587 | 1.997866 | 0.0018046346 | 0.13173833 | 0.12271515 | 5795 | tags=93%, list=37%, signal=59% | RBM22||SNRNP40||SYF2||SRRM2||SNRPD3||PRPF19||PLRG1||SNRPB2||SNW1||SNRPG||AQR||PPIE||CDC5L||SNRPE||CDC40||DHX8||PRPF8||BCAS2||EFTUD2||SNRPA1||XAB2||BUD31||CWC22||CRNKL1||SNRPF||SNRPD1||SNRPB||PPIL1 | 7.6448841 | 7.7024969 | 7.6556954 | 7.8558067 | 7.5848144 | 7.4273585 | 7.7494993 | 7.9315929 | 7.7222363 | 7.6932273 | 7.510159 | 7.7338476 | 7.6866007 | 7.686524 | 7.7754109 | 7.566538 | 7.6168114 | 7.8783880 | 7.8185208 | 7.8697880 | 7.5174644 | 7.7436180 | 7.4789524 | 7.4461130 | 7.3663291 | 7.467670 | 7.7657859 | 7.743310 | 7.7392600 | 7.9084888 | 7.9322603 | 7.953675 |
| GO:0032809 | neuronal cell body membrane | 15 | -0.7820690 | -1.826558 | 0.0029150270 | 0.17733081 | 0.16518486 | 2835 | tags=73%, list=18%, signal=60% | AMIGO1||UNC5A||INSR||FLRT1||KCNC3||ATP1A3||KCNC4||KCNB1||KCNC1||SLC4A8||KCND2 | 1.0664230 | 2.6021395 | 1.9381485 | 2.6238046 | 1.0795810 | 1.8661283 | 2.5483358 | 2.4712454 | 0.8893178 | 1.0941087 | 1.198866 | 2.5089994 | 2.7311688 | 2.556652 | 1.9886543 | 2.057337 | 1.7508501 | 2.5495199 | 2.7045009 | 2.6131744 | 1.1648306 | 0.8078774 | 1.2311643 | 1.4980345 | 1.6804115 | 2.294788 | 2.5872970 | 2.556686 | 2.4996569 | 2.5057299 | 2.4694976 | 2.437705 |
| GO:0016324 | apical plasma membrane | 201 | -0.4196020 | -1.439968 | 0.0062185054 | 0.25219494 | 0.23492132 | 2391 | tags=30%, list=15%, signal=26% | ADRB2||SAPCD2||PRKCZ||CYBA||TCIRG1||LZTS1||STX3||ATP6AP2||DLL1||MTCL1||RAPGEF6||PATJ||CTSL||PSEN2||IGFBP2||CD9||SLC19A1||ABCG2||SLC5A12||SLC22A18||ERBB2||JAG1||CACNB3||PODXL||MFSD4B||SLC26A2||SLC9A3||SLC15A2||HYAL2||KCNQ1||IL6R||SHROOM1||RAB27A||SIPA1L3||PRKG2||CLIC5||KCNK1||PTPRO||SLC38A3||AQP1||SLC46A1||SLC4A11||SHROOM2||ABCC11||SHANK2||DPP4||ABCB1||MUC1||SPTBN2||ATP8B1||SLC1A1||SLC24A4||EPCAM||KCNK2||ACE2||CLDN4||KIAA1614||TRPV4||PARD6A||ENPEP | 4.6350457 | 5.2009929 | 4.8571197 | 5.2278876 | 4.6204130 | 4.8561242 | 5.1588214 | 5.1916498 | 4.5865915 | 4.6593119 | 4.658040 | 5.2161533 | 5.2242501 | 5.161779 | 4.9485514 | 4.826457 | 4.7916018 | 5.2160386 | 5.2187172 | 5.2486790 | 4.6340856 | 4.5660624 | 4.6594812 | 4.8215854 | 4.8324562 | 4.912608 | 5.1744937 | 5.157451 | 5.1443610 | 5.1822463 | 5.2216221 | 5.170584 |
| GO:0071005 | U2-type precatalytic spliceosome | 50 | 0.5036045 | 1.773181 | 0.0097370983 | 0.32179481 | 0.29975407 | 5930 | tags=74%, list=38%, signal=46% | SNIP1||BUD13||WBP4||SRRM2||SNRPD3||CWC27||PRPF38A||PHF5A||SMU1||IK||SF3B4||SF3A1||SNRPB2||SNRPG||LSM8||SF3B2||PRPF3||LSM6||PRPF6||LSM3||SF3B3||MFAP1||SF3B1||ZMAT2||SNRPE||PRPF8||EFTUD2||SNRPA1||SNRNP200||CWC22||SNRPF||SNRPD1||TXNL4A||SNRPB||DHX16||LSM5||SF3A2 | 7.5874776 | 7.5476953 | 7.5928586 | 7.6727162 | 7.5524472 | 7.4846743 | 7.5820323 | 7.7289586 | 7.6248633 | 7.6130813 | 7.522322 | 7.5708943 | 7.5416758 | 7.530210 | 7.6680300 | 7.543642 | 7.5637899 | 7.7019796 | 7.6325355 | 7.6827455 | 7.5100714 | 7.6232929 | 7.5212581 | 7.4858299 | 7.4805263 | 7.487657 | 7.5916062 | 7.583019 | 7.5714004 | 7.7078734 | 7.7248930 | 7.753736 |
| GO:0017053 | transcription repressor complex | 57 | 0.4737387 | 1.712906 | 0.0118069815 | 0.35912902 | 0.33453114 | 4725 | tags=56%, list=30%, signal=39% | ZNF350||ZNF224||CORO2A||SP1||CCND1||JAZF1||N4BP2L2||DEPDC1||MIER1||DR1||LIN9||ARID4A||RBPJ||PRDM10||ZFPM1||SP3||TBL1XR1||HDAC4||LIN52||YWHAB||RLIM||DDX20||SIN3A||CTBP2||NCOR1||ETV3||HMGB1||REST||PRDM16||CBX5||NCOR2||CDX2 | 5.1858592 | 5.7167306 | 5.3057068 | 5.7740926 | 5.1623771 | 5.3243954 | 5.8146715 | 5.8404164 | 5.1454507 | 5.2456194 | 5.164535 | 5.6928603 | 5.7106014 | 5.746217 | 5.4572028 | 5.221935 | 5.2251485 | 5.7315295 | 5.7457891 | 5.8424180 | 5.1439488 | 5.1440673 | 5.1984282 | 5.3068546 | 5.2932987 | 5.371806 | 5.8139083 | 5.797752 | 5.8321494 | 5.8465376 | 5.8157127 | 5.858660 |
| GO:0005788 | endoplasmic reticulum lumen | 206 | -0.4023147 | -1.382492 | 0.0135974304 | 0.38007993 | 0.35404706 | 4935 | tags=51%, list=32%, signal=35% | CDH2||QSOX1||GOLM1||COL26A1||POGLUT3||COL6A1||ANO8||TOR2A||APP||P4HA2||LRPAP1||CALR||CLN6||HYOU1||RCN1||ADAMTSL4||PPIB||STC2||TOR1A||TSPAN14||TNC||P3H1||POGLUT1||CES2||GPC3||P4HB||SCG3||COL6A2||BACE1||EDEM3||CNPY3||COL18A1||ARSA||ERO1A||POGLUT2||COL2A1||MINPP1||FSTL3||COL4A6||FOXRED2||SHISA5||COL4A2||FAM20C||RNASET2||P3H2||CRTAP||MEN1||NUCB1||TIMP1||LAMB1||RCN3||CSF1||TOR3A||H6PD||P4HA1||WNT7B||PDIA4||SUMF2||SUMF1||CST3||LTBP1||ERAP1||COL5A1||APOE||DAG1||COL9A2||PTGS2||MANF||COL5A2||LAMB2||SIL1||COL19A1||CTSZ||EDEM2||TMEM43||CHGB||ALG12||CLU||P4HA3||VWA1||FKBP7||ADAMTS13||TSPAN33||COL27A1||CKAP4||MELTF||PLOD3||VCAN||SDF2L1||LGALS1||WFS1||IGFBP4||TSPAN15||IGFBP5||SERPINH1||EVA1A||ADAMTS5||TOR4A||WNT3||COL1A1||ARSJ||ACE2||COL12A1||CFP||IL23A | 5.3352477 | 6.3075670 | 5.6425217 | 6.0194221 | 5.3108425 | 5.5193820 | 6.1436905 | 5.9359113 | 5.3245535 | 5.3704935 | 5.310003 | 6.3156013 | 6.3550591 | 6.250098 | 5.6755272 | 5.654021 | 5.5968754 | 5.9885921 | 6.0646665 | 6.0038803 | 5.3194520 | 5.2986144 | 5.3143794 | 5.4466140 | 5.4120006 | 5.684061 | 6.1610398 | 6.153358 | 6.1162779 | 5.9347638 | 5.9866068 | 5.884559 |
| GO:0046540 | U4/U6 x U5 tri-snRNP complex | 30 | 0.5384160 | 1.721482 | 0.0145784082 | 0.38007993 | 0.35404706 | 5864 | tags=67%, list=38%, signal=42% | TXNL4B||USP39||SNRPD3||SNRPG||LSM8||PRPF3||LSM6||PRPF6||LSM3||PRPF18||ZMAT2||SNRPE||PRPF8||EFTUD2||SNRNP200||SNRPF||SNRPD1||TXNL4A||SNRPB||LSM5 | 8.0035890 | 7.8683602 | 7.9776814 | 8.0226868 | 7.9637989 | 7.8117450 | 7.8920270 | 8.0687510 | 8.0624298 | 8.0437588 | 7.899104 | 7.9112900 | 7.8502317 | 7.842571 | 8.0449942 | 7.917144 | 7.9680265 | 8.0605719 | 7.9792272 | 8.0271048 | 7.9171146 | 8.0758533 | 7.8914299 | 7.8410681 | 7.7867517 | 7.806892 | 7.9007907 | 7.891157 | 7.8840847 | 8.0437540 | 8.0590278 | 8.102818 |
| GO:0099055 | integral component of postsynaptic membrane | 65 | -0.4980764 | -1.516736 | 0.0152476214 | 0.38030737 | 0.35425892 | 2037 | tags=32%, list=13%, signal=28% | LRFN4||LRFN1||SHISA9||GABRD||DAGLA||LRFN3||LRRC4B||ADORA1||CSPG5||CHRNB1||ADORA2A||PTPRO||EFNB3||CLSTN3||CLSTN2||CACNG4||KCNC1||SLC8A3||PRRT1||KCND2||GRID2 | 2.4877208 | 4.3175162 | 2.9504312 | 4.0665090 | 2.4738409 | 2.8918576 | 4.1635890 | 4.0267714 | 2.4702306 | 2.5272724 | 2.464826 | 4.2842538 | 4.3887121 | 4.276848 | 3.1537401 | 2.915456 | 2.7538491 | 3.9838358 | 4.1282307 | 4.0836943 | 2.4313991 | 2.4675018 | 2.5212037 | 2.7724910 | 2.8096955 | 3.074254 | 4.1729332 | 4.176967 | 4.1405918 | 4.0243295 | 4.0709226 | 3.983742 |
| GO:0005682 | U5 snRNP | 16 | 0.6271812 | 1.729923 | 0.0159090909 | 0.38030737 | 0.35425892 | 5617 | tags=81%, list=36%, signal=52% | TXNL4B||SNRNP40||SNRPD3||SNRPG||PRPF6||PRPF18||SNRPE||AAR2||PRPF8||SNRNP200||SNRPD1||TXNL4A||SNRPB | 8.3335900 | 8.2097446 | 8.3220082 | 8.3659596 | 8.2880045 | 8.0898656 | 8.2375148 | 8.4246402 | 8.4109402 | 8.3863971 | 8.193783 | 8.2515548 | 8.1931275 | 8.183609 | 8.3921697 | 8.243920 | 8.3261193 | 8.4128227 | 8.3178218 | 8.3656706 | 8.2290065 | 8.4540804 | 8.1646759 | 8.1365524 | 8.0459037 | 8.085708 | 8.2542224 | 8.235291 | 8.2228584 | 8.4034964 | 8.4170162 | 8.452954 |
| GO:0042611 | MHC protein complex | 13 | -0.7087880 | -1.605308 | 0.0170168689 | 0.38030737 | 0.35425892 | 3953 | tags=85%, list=25%, signal=63% | HLA-E||HLA-B||HLA-A||HLA-C||HLA-DRB1||HLA-DPA1||HLA-DQB1||HLA-DMA||MR1||HLA-F||HLA-DOA | 5.5868585 | 5.8372610 | 5.9363253 | 5.7613937 | 5.6835772 | 5.8569252 | 5.6204734 | 5.6084244 | 5.5388653 | 5.4998294 | 5.712853 | 5.8821221 | 5.8724759 | 5.753671 | 5.7911006 | 6.046695 | 5.9595905 | 5.8284824 | 5.7728671 | 5.6788865 | 5.6674802 | 5.5822344 | 5.7931792 | 5.6887724 | 5.8002087 | 6.056762 | 5.7158270 | 5.586453 | 5.5540129 | 5.6513486 | 5.6440767 | 5.526475 |
| GO:0031225 | anchored component of membrane | 71 | -0.4815125 | -1.484728 | 0.0211793020 | 0.39771203 | 0.37047148 | 4032 | tags=45%, list=26%, signal=34% | LYPD6B||LY6E||GPC3||MMP25||DNAJC5||ULBP3||ABHD17A||PRNP||GPC1||RTN4R||ULBP1||SEMA7A||CD59||NRN1||RAB3A||GAS1||LYPD6||MELTF||EFNA4||RAB3C||GPC2||RAB26||HYAL2||SMPDL3B||CPM||GFRA1||ULBP2||EFNA2||SYN1||RAET1G||RTN4RL2||CD24 | 3.5246940 | 3.7576708 | 3.8143524 | 3.7278778 | 3.5697170 | 3.8046558 | 3.6594179 | 3.6004413 | 3.4805734 | 3.4586538 | 3.628846 | 3.7535195 | 3.8268259 | 3.689386 | 3.7771098 | 3.872651 | 3.7914452 | 3.7774572 | 3.7311520 | 3.6731332 | 3.5561576 | 3.4811503 | 3.6658414 | 3.6554148 | 3.7842742 | 3.958206 | 3.6902804 | 3.650526 | 3.6369126 | 3.6026919 | 3.6386917 | 3.558833 |
| GO:0005838 | proteasome regulatory particle | 19 | 0.5822480 | 1.672685 | 0.0222070379 | 0.39771203 | 0.37047148 | 5529 | tags=79%, list=36%, signal=51% | PSMD9||PSMD12||PSMC6||PSMC3||PSMD14||PSMD3||PSMD1||PSMD11||PSMD13||PSMD7||ADRM1||PSMC2||PSMD8||PSMD6||PSMC4 | 6.9361421 | 7.1208022 | 7.0720298 | 7.1808872 | 6.8805278 | 6.9330854 | 7.1234138 | 7.2555748 | 6.9401849 | 7.0175452 | 6.845566 | 7.1721581 | 7.0923424 | 7.096499 | 7.1314332 | 7.013212 | 7.0690180 | 7.2091310 | 7.1255503 | 7.2064267 | 6.8884768 | 6.9312695 | 6.8196454 | 6.9276309 | 6.8381200 | 7.027293 | 7.1428351 | 7.109215 | 7.1179806 | 7.2627130 | 7.2619056 | 7.242011 |
| GO:0010494 | cytoplasmic stress granule | 68 | 0.4120119 | 1.532648 | 0.0228571429 | 0.39771203 | 0.37047148 | 5118 | tags=65%, list=33%, signal=44% | RC3H1||EIF2S1||NUFIP2||PUM2||RC3H2||MBNL1||UBAP2L||NXF1||PRKAA2||YTHDF3||G3BP1||DDX6||OGFOD1||TARDBP||TRIM25||RBM4||IGF2BP1||KPNB1||YTHDF1||RBPMS||TIAL1||MOV10||LSM14A||DHX36||LARP4B||ROCK1||CAPRIN1||YTHDF2||HNRNPK||G3BP2||CELF1||DDX19B||FMR1||DDX1||ATXN2||CASC3||CSDE1||ATXN2L||LARP4||PQBP1||PRRC2C||ELAVL1||YBX1||VCP | 6.1703251 | 7.1023535 | 6.3501436 | 7.0776282 | 6.0941168 | 6.3133388 | 7.1533430 | 7.1522490 | 6.1660821 | 6.2817839 | 6.054128 | 7.0853638 | 7.1129696 | 7.108575 | 6.5776943 | 6.208914 | 6.2336217 | 7.0474059 | 7.0414675 | 7.1417987 | 6.0964720 | 6.1186976 | 6.0667096 | 6.3005154 | 6.2758996 | 6.362225 | 7.1382035 | 7.149219 | 7.1723953 | 7.1460452 | 7.1618733 | 7.148779 |
| GO:0031261 | DNA replication preinitiation complex | 12 | 0.6609331 | 1.677426 | 0.0229676639 | 0.39771203 | 0.37047148 | 4322 | tags=83%, list=28%, signal=60% | MCM4||CDC45||MCM5||MCM7||GINS3||MCM3||ORC3||GINS2||GINS4||MCM6 | 6.9931519 | 7.1068333 | 7.1705843 | 7.1610247 | 6.9311076 | 6.9261732 | 7.1168846 | 7.2938098 | 6.9739217 | 7.0741649 | 6.927450 | 7.1435763 | 7.0999718 | 7.076139 | 7.2130332 | 7.122976 | 7.1743314 | 7.1893059 | 7.1019749 | 7.1900288 | 6.9859371 | 6.9201636 | 6.8854092 | 6.8800110 | 6.7907430 | 7.091078 | 7.1610650 | 7.070339 | 7.1178232 | 7.2719553 | 7.3131861 | 7.295991 |
| GO:0097431 | mitotic spindle pole | 34 | 0.4931489 | 1.615499 | 0.0239175258 | 0.39771203 | 0.37047148 | 4698 | tags=59%, list=30%, signal=41% | MAPKBP1||KATNBL1||RAE1||YPEL5||KIF20B||SMC3||CNTRL||SPAG5||EML1||IK||TNKS||AURKA||AURKB||SMC1A||FAM83D||STAG2||STAG1||ASPM||PLK1||BCCIP | 4.9014175 | 5.6310669 | 5.1841972 | 5.7085315 | 4.8456531 | 5.1651525 | 5.7093916 | 5.8049579 | 4.8261749 | 4.9439448 | 4.931261 | 5.6234756 | 5.6374526 | 5.632238 | 5.3959917 | 5.069230 | 5.0614999 | 5.6803939 | 5.6787563 | 5.7647565 | 4.8428149 | 4.7843939 | 4.9071364 | 5.0917486 | 5.1578923 | 5.241885 | 5.7181615 | 5.703923 | 5.7060495 | 5.8011006 | 5.8253020 | 5.788225 |
| GO:0030014 | CCR4-NOT complex | 14 | 0.6317875 | 1.671957 | 0.0244821092 | 0.39771203 | 0.37047148 | 3674 | tags=71%, list=24%, signal=55% | CNOT6||CNOT3||CPEB3||CNOT6L||CNOT2||CNOT11||CNOT7||CNOT8||CNOT10||CNOT4 | 4.6465719 | 5.6096281 | 4.9542588 | 5.7285946 | 4.5374441 | 4.6940700 | 5.6724466 | 5.8415863 | 4.6529486 | 4.8122420 | 4.452084 | 5.5999105 | 5.6422809 | 5.586097 | 5.2763360 | 4.696226 | 4.8239620 | 5.6587685 | 5.7305807 | 5.7932976 | 4.5011916 | 4.6124321 | 4.4956751 | 4.7632518 | 4.6657605 | 4.650587 | 5.6697529 | 5.680888 | 5.6666602 | 5.8621016 | 5.8407011 | 5.821672 |
| GO:0009897 | external side of plasma membrane | 138 | -0.4146636 | -1.381909 | 0.0255498778 | 0.39771203 | 0.37047148 | 4238 | tags=44%, list=27%, signal=32% | CLCN3||ABCG1||SLC7A5||ADA||ADGRA3||HLA-E||P4HB||HEG1||ULBP3||CD40||PRNP||ATP1B2||CD302||BTN2A3P||BCAM||AMOT||IL1R1||FAS||ERMAP||RTN4R||HLA-DRB1||ATP6AP2||SERPINE2||ULBP1||SEMA7A||IL27RA||ANTXR1||ITGB2||DAG1||CD59||INSR||CD9||BTNL9||ABCG2||SDC1||KRT18||MCAM||HYAL2||IL6R||CXCR4||MICA||CD8A||GFRA1||SLC2A4||F3||MR1||BTN3A3||ICOSLG||ULBP2||SLC4A3||HLA-F||ABCB1||PDGFRA||ITGA3||CXCL12||RAET1G||RTN4RL2||CD19||CD24||ENPEP||CHRNA4 | 4.0464251 | 5.0558653 | 4.4452622 | 4.7773461 | 4.0434873 | 4.2524345 | 4.9058092 | 4.6734755 | 4.0296234 | 4.1350011 | 3.969777 | 5.0542139 | 5.1204955 | 4.989932 | 4.4498478 | 4.471931 | 4.4134029 | 4.7591108 | 4.8179230 | 4.7541280 | 4.0656809 | 4.0456434 | 4.0187537 | 4.1912048 | 4.0855513 | 4.455057 | 4.9337168 | 4.907366 | 4.8757618 | 4.6637216 | 4.7212809 | 4.634057 |
| GO:0071141 | SMAD protein complex | 10 | 0.6878964 | 1.667677 | 0.0260894495 | 0.39771203 | 0.37047148 | 4165 | tags=90%, list=27%, signal=66% | SMAD9||SMAD3||SMAD2||SMAD5||SMAD4||HMGA2||SMAD1||SNW1||SMAD7 | 2.8904247 | 4.6722796 | 3.0538259 | 4.6729141 | 2.7742295 | 2.9615642 | 4.7586716 | 4.8068579 | 2.9419051 | 3.0236993 | 2.684349 | 4.6580633 | 4.6587191 | 4.699661 | 3.5466216 | 2.778050 | 2.6707955 | 4.6029409 | 4.6322718 | 4.7774148 | 2.6353690 | 2.8562360 | 2.8214532 | 2.9427368 | 2.8879008 | 3.049357 | 4.7463907 | 4.746879 | 4.7824481 | 4.7997682 | 4.7922205 | 4.828333 |
| GO:0031519 | PcG protein complex | 33 | 0.4870807 | 1.586186 | 0.0265089723 | 0.39771203 | 0.37047148 | 3476 | tags=42%, list=22%, signal=33% | ASXL3||BMI1||PHC3||UBAP2L||EED||YY2||RBBP7||EZH2||AEBP2||PCGF5||ASXL2||SUZ12||RBBP4||SCML2 | 5.1572139 | 6.1843480 | 5.4766795 | 6.1370143 | 5.0816731 | 5.3296448 | 6.2390589 | 6.2376450 | 5.1334410 | 5.2450143 | 5.088662 | 6.1753744 | 6.1946329 | 6.182971 | 5.7166617 | 5.345750 | 5.3339110 | 6.1063793 | 6.0930232 | 6.2088359 | 5.0522622 | 5.0932544 | 5.0990531 | 5.1960697 | 5.2467903 | 5.524038 | 6.2026299 | 6.241944 | 6.2717700 | 6.2097624 | 6.2669046 | 6.235700 |
| GO:0005765 | lysosomal membrane | 328 | -0.3630149 | -1.282824 | 0.0271806532 | 0.39771203 | 0.37047148 | 4581 | tags=37%, list=29%, signal=27% | ATP8A1||CTSA||SLC29A3||SCARB2||ATP6V0B||SLC44A2||HPSE||LYN||SNAPIN||SLCO4C1||DRAM1||ATP6V0A1||CLCN3||ARRB1||TMEM63A||SLC7A5||SPNS1||DAGLB||ABCA5||MEAK7||RRAGA||SZT2||IFITM3||SYNGR1||ACP2||DNAJC5||SLC30A4||MARCHF9||TMEM9||MARCHF8||LAPTM4A||GRN||TMEM179B||PSAP||VLDLR||IFITM2||WDR81||AP5B1||AHNAK||HGSNAT||SLC48A1||PLD3||SCARB1||TMEM106B||NPRL3||TMEM150C||MFSD12||ABCB9||SLC15A4||PLEKHM2||SNX14||CYB561A3||TCIRG1||VASN||SLC30A3||ABCA2||CTSD||TMEM74||SLC17A5||TPCN2||ATP6V0C||HLA-DRB1||ATP6AP2||TM9SF1||STARD3||SPHK2||TPCN1||HLA-DPA1||SLC38A9||SNAP29||KICS2||WDR24||NPC1||PLEKHF1||MCOLN1||ATRAID||ABCC10||LPCAT1||NPRL2||ATP13A2||TOM1||TMEM79||RDH14||PSEN2||MFSD1||GBA||SLC35F6||GPR137||HPS6||SIDT2||HLA-DQB1||CTNS||CLN5||GPR137C||GPR137B||LRP1||MARCHF2||CKAP4||VPS33B||BORCS5||ATP6V0E2||SLC66A1||SORT1||GLMP||ABCD1||BORCS6||TMEM175||ABCA3||HLA-DMA||NEU1||ATP6V1C2||CYB561||SLC26A11||EVA1A||DPP4||HLA-F||SLC2A6||SYT11||HLA-DOA||TMBIM1||SLC46A3||ENPEP | 5.8150146 | 6.4578808 | 5.9408285 | 6.4213366 | 5.7622837 | 5.8093300 | 6.4040650 | 6.3972336 | 5.8413450 | 5.8680014 | 5.732135 | 6.5380090 | 6.4417532 | 6.389949 | 6.0332291 | 5.827209 | 5.9545998 | 6.4368201 | 6.3930817 | 6.4336967 | 5.7437289 | 5.8078263 | 5.7341787 | 5.7748986 | 5.7652020 | 5.884805 | 6.3909765 | 6.416336 | 6.4047709 | 6.4145067 | 6.3562941 | 6.420040 |
| GO:0032590 | dendrite membrane | 23 | -0.5987277 | -1.526492 | 0.0290994346 | 0.39771203 | 0.37047148 | 3409 | tags=61%, list=22%, signal=48% | PPP1R9B||DDN||ATP6AP2||PALM||INSR||KCNC3||MPP2||SHISA9||KCNC4||SHISA8||DAGLA||GABRG2||KCNB1||KCNC1 | 2.9372944 | 4.0284841 | 3.4362603 | 4.0157892 | 2.8939200 | 3.2399136 | 3.8915827 | 3.8953613 | 2.8861034 | 3.0279268 | 2.893386 | 4.0614565 | 4.0875899 | 3.931618 | 3.5394025 | 3.422299 | 3.3401008 | 4.0222964 | 4.0883809 | 3.9324573 | 2.9075395 | 2.8664802 | 2.9073536 | 3.0746998 | 3.0101808 | 3.568073 | 3.9462377 | 3.878406 | 3.8483548 | 3.8788325 | 3.9377677 | 3.868512 |
| GO:0001518 | voltage-gated sodium channel complex | 11 | -0.7086037 | -1.545595 | 0.0292741814 | 0.39771203 | 0.37047148 | 1494 | tags=45%, list=10%, signal=41% | CACNA1G||SCN4B||SCN11A||SCN3B||SCN2A | 0.4523387 | 0.5817418 | 0.9309783 | 0.2204382 | 0.4983858 | 0.8446893 | 0.6090768 | 0.2388641 | 0.3574395 | 0.4301257 | 0.561955 | 0.5159351 | 0.6481664 | 0.578088 | 0.9857813 | 1.019576 | 0.7756801 | 0.3185475 | 0.2056183 | 0.1309405 | 0.4890731 | 0.2028076 | 0.7513931 | 0.4313613 | 0.7408413 | 1.243656 | 0.6985472 | 0.595429 | 0.5281281 | 0.2755053 | 0.2886633 | 0.148301 |
| GO:0044815 | DNA packaging complex | 57 | 0.4134118 | 1.494781 | 0.0338809035 | 0.39821391 | 0.37093898 | 2753 | tags=39%, list=18%, signal=32% | H2BC17||H4C3||H3C2||H1-4||H2AC13||H2BC6||H2AC17||SMC4||NCAPG||H2BC21||H4C8||H2BC4||SMC2||SLF1||H2BC11||H2AC20||SHPRH||NCAPD2||H3C6||NCAPH2||GLYR1||CENPA | 5.9547089 | 6.2630155 | 6.1765683 | 6.2119075 | 5.8443874 | 5.7401769 | 6.2850842 | 6.2629498 | 6.0043415 | 6.1429930 | 5.678208 | 6.2699884 | 6.2498911 | 6.269078 | 6.3028148 | 5.955413 | 6.2478802 | 6.1854795 | 6.1856171 | 6.2632237 | 5.7810936 | 5.9926237 | 5.7469775 | 5.6804559 | 5.6445943 | 5.883774 | 6.2758226 | 6.270935 | 6.3082099 | 6.2973655 | 6.2127714 | 6.277364 |
| GO:0016442 | RISC complex | 14 | 0.6117572 | 1.618949 | 0.0345260515 | 0.39821391 | 0.37093898 | 2480 | tags=43%, list=16%, signal=36% | AGO4||TNRC6A||LIMD1||DDX6||TARBP2||DICER1 | 5.0466040 | 5.9751613 | 5.3469047 | 5.9990269 | 4.9827955 | 5.1887234 | 6.0294761 | 6.0459890 | 5.0597714 | 5.1136785 | 4.962302 | 5.9666845 | 5.9862170 | 5.972513 | 5.5356887 | 5.204483 | 5.2792683 | 5.9545771 | 5.9721618 | 6.0677523 | 4.9716278 | 5.0396360 | 4.9351673 | 5.2132730 | 5.0858907 | 5.261357 | 6.0302646 | 6.027199 | 6.0309619 | 6.0174329 | 6.0545450 | 6.065549 |
| GO:0070822 | Sin3-type complex | 14 | 0.6094955 | 1.612964 | 0.0351537979 | 0.39821391 | 0.37093898 | 5551 | tags=71%, list=36%, signal=46% | BRMS1L||SAP130||SINHCAF||RBBP4||SIN3A||CSNK2A1||MORF4L1||SUDS3||HDAC1||HDAC2 | 5.8596109 | 7.0850142 | 6.1301799 | 7.0118160 | 5.7168955 | 5.8210925 | 7.1141387 | 7.0944891 | 5.9050803 | 6.0490207 | 5.586763 | 7.1113660 | 7.0691015 | 7.074205 | 6.4857528 | 5.850844 | 5.9726142 | 6.9734991 | 6.9754690 | 7.0837006 | 5.6614493 | 5.8668008 | 5.6094132 | 5.7944231 | 5.6677737 | 5.983478 | 7.0907508 | 7.095629 | 7.1551412 | 7.0698814 | 7.1022315 | 7.111030 |
| GO:0016282 | eukaryotic 43S preinitiation complex | 17 | 0.5713156 | 1.598502 | 0.0360449735 | 0.39821391 | 0.37093898 | 6563 | tags=88%, list=42%, signal=51% | EIF3CL||EIF3D||EIF1||EIF3B||EIF1B||EIF3J||EIF3E||EIF3A||EIF3K||EIF3I||DHX29||EIF3C||EIF3G||EIF3H||EIF3F | 7.9944041 | 8.2175614 | 7.8945574 | 8.1840511 | 7.8806526 | 7.7720344 | 8.2482139 | 8.2609604 | 8.0504122 | 8.0646093 | 7.859212 | 8.2096831 | 8.2166120 | 8.226340 | 7.9169530 | 7.826909 | 7.9374301 | 8.1807869 | 8.1376510 | 8.2321622 | 7.9227707 | 7.9657766 | 7.7439516 | 7.8837018 | 7.6759696 | 7.748680 | 8.2374339 | 8.246645 | 8.2604695 | 8.2501884 | 8.2692490 | 8.263378 |
| GO:0008180 | COP9 signalosome | 13 | 0.6185884 | 1.602686 | 0.0369913687 | 0.39821391 | 0.37093898 | 3006 | tags=46%, list=19%, signal=37% | LAT||GRB2||COPS3||COPS4||COPS2||COPS8 | 6.2255256 | 6.4777109 | 6.3263691 | 6.5252496 | 6.1944729 | 6.2918054 | 6.5068697 | 6.5947555 | 6.2635635 | 6.2103803 | 6.201855 | 6.5212770 | 6.4517358 | 6.459104 | 6.3610745 | 6.323148 | 6.2941021 | 6.5305132 | 6.4612614 | 6.5814572 | 6.1972742 | 6.1596457 | 6.2257377 | 6.2648546 | 6.2414109 | 6.366085 | 6.5193799 | 6.493891 | 6.5072253 | 6.5682895 | 6.6092535 | 6.606362 |
| GO:0005689 | U12-type spliceosomal complex | 26 | 0.5060854 | 1.557729 | 0.0375335121 | 0.39821391 | 0.37093898 | 5617 | tags=69%, list=36%, signal=44% | SNRNP35||RNPC3||SNRNP48||SNRPD3||PHF5A||DHX15||SF3B6||SF3B4||RBM41||SNRPG||SF3B2||SF3B3||SF3B1||SNRPE||YBX1||SNRPF||SNRPD1||SNRPB | 8.1600770 | 8.1795055 | 8.1227071 | 8.3217316 | 8.1164666 | 7.9672732 | 8.2095299 | 8.3761000 | 8.2023407 | 8.2282279 | 8.042759 | 8.2194789 | 8.1709425 | 8.147151 | 8.2087262 | 8.041223 | 8.1132664 | 8.3652146 | 8.2740157 | 8.3245194 | 8.0855046 | 8.2285579 | 8.0278724 | 7.9879265 | 7.9470240 | 7.966579 | 8.2178873 | 8.209155 | 8.2015014 | 8.3568401 | 8.3689969 | 8.402082 |
| GO:0032281 | AMPA glutamate receptor complex | 14 | -0.6536959 | -1.503700 | 0.0396126761 | 0.40728526 | 0.37938901 | 2983 | tags=57%, list=19%, signal=46% | CACNG5||OLFM2||CPT1C||SHISA9||SHISA8||CNIH3||CACNG8||CACNG4 | 3.0837972 | 3.0122370 | 3.2611539 | 2.7961016 | 3.1515655 | 3.2266833 | 2.8712386 | 2.6743792 | 3.0296364 | 3.0041072 | 3.208899 | 3.0677962 | 3.0042876 | 2.962680 | 3.2018833 | 3.400180 | 3.1704883 | 2.7931674 | 2.7446316 | 2.8486278 | 3.1067978 | 3.0535224 | 3.2841486 | 3.0143018 | 3.2096466 | 3.426595 | 2.9120411 | 2.856111 | 2.8446590 | 2.6310897 | 2.7192243 | 2.671474 |
| GO:0000940 | outer kinetochore | 11 | 0.6418212 | 1.593013 | 0.0402962963 | 0.40855967 | 0.38057613 | 3064 | tags=45%, list=20%, signal=37% | SKA3||BUB1B||NDC80||CCNB1||SPDL1 | 5.2020877 | 6.2261061 | 5.2277441 | 6.3276465 | 5.0792124 | 5.2952224 | 6.3122615 | 6.4431068 | 5.1855961 | 5.3444528 | 5.062301 | 6.2935261 | 6.1745353 | 6.207628 | 5.6549831 | 4.912598 | 4.9959459 | 6.2930873 | 6.2827955 | 6.4039046 | 5.0468497 | 5.1282514 | 5.0612210 | 5.3841903 | 5.3695902 | 5.116510 | 6.3031164 | 6.307889 | 6.3256810 | 6.4153380 | 6.4749951 | 6.438359 |
| GO:0033290 | eukaryotic 48S preinitiation complex | 15 | 0.5835628 | 1.576153 | 0.0420248329 | 0.41353383 | 0.38520960 | 1576 | tags=27%, list=10%, signal=24% | EIF3CL||EIF3D||EIF2S1||EIF3B | 7.9953985 | 8.2561321 | 7.9025658 | 8.2608710 | 7.8809529 | 7.7728175 | 8.2922466 | 8.3340707 | 8.0511470 | 8.0586253 | 7.868506 | 8.2406184 | 8.2619997 | 8.265652 | 7.9329143 | 7.835310 | 7.9371888 | 8.2584724 | 8.2158315 | 8.3068702 | 7.9311406 | 7.9612006 | 7.7407647 | 7.8984301 | 7.6848644 | 7.726153 | 8.2863217 | 8.294848 | 8.2955520 | 8.3194270 | 8.3448777 | 8.337788 |
| GO:1902562 | H4 histone acetyltransferase complex | 30 | 0.4774382 | 1.526517 | 0.0421591805 | 0.41353383 | 0.38520960 | 2527 | tags=40%, list=16%, signal=34% | EPC1||KANSL1L||BRD8||EPC2||KAT5||KANSL2||KAT8||ATF2||ACTL6A||ING3||DMAP1||MCRS1 | 7.0756781 | 6.9870435 | 6.8179630 | 6.8710403 | 7.0672634 | 7.0380002 | 6.9795497 | 6.9083462 | 7.0524562 | 7.0957173 | 7.078532 | 7.0059595 | 6.9810123 | 6.973962 | 6.7755396 | 6.837551 | 6.8398796 | 6.9106783 | 6.7974578 | 6.9022504 | 7.1131349 | 7.0139258 | 7.0730071 | 7.1018071 | 7.0092353 | 7.000765 | 6.9823187 | 6.980272 | 6.9760512 | 6.9126605 | 6.9121628 | 6.900181 |
| GO:0098685 | Schaffer collateral - CA1 synapse | 57 | -0.4739668 | -1.416568 | 0.0444499628 | 0.42140874 | 0.39254513 | 3963 | tags=35%, list=26%, signal=26% | ABR||PTPRS||DTNBP1||DVL1||BCR||PRKCZ||STX3||LRRC4||IQSEC2||SYP||TUBB2B||CDK5||INA||PRKAR1B||BSN||ADRB1||SYT11||TAMALIN||SYN1||PLAT | 7.0171997 | 6.5450178 | 6.5369012 | 6.4804940 | 7.0405398 | 6.9213910 | 6.4940044 | 6.4689290 | 6.9701646 | 7.0146652 | 7.065201 | 6.5412941 | 6.5576701 | 6.536001 | 6.3471602 | 6.618510 | 6.6278080 | 6.5477375 | 6.3937065 | 6.4958059 | 7.1369714 | 6.9224579 | 7.0541270 | 7.0701929 | 6.8876366 | 6.792409 | 6.4949049 | 6.511291 | 6.4755963 | 6.4695709 | 6.4820528 | 6.455037 |
| GO:0033268 | node of Ranvier | 11 | -0.6845734 | -1.493181 | 0.0463256375 | 0.42457742 | 0.39549677 | 391 | tags=27%, list=3%, signal=27% | DAG1||SCN2A||SPTBN4 | 3.3272950 | 3.6405652 | 3.8171713 | 3.7869787 | 3.4196554 | 3.8959778 | 3.5578298 | 3.6723023 | 3.2583570 | 3.2832030 | 3.433992 | 3.6522880 | 3.6820511 | 3.585675 | 3.7451603 | 3.959862 | 3.7351404 | 3.8105902 | 3.7575612 | 3.7922827 | 3.4668455 | 3.2132117 | 3.5572619 | 3.7093234 | 3.7180727 | 4.203259 | 3.6017075 | 3.536283 | 3.5344764 | 3.7017794 | 3.6945379 | 3.619147 |
| GO:0031166 | integral component of vacuolar membrane | 10 | -0.6992516 | -1.486453 | 0.0472827756 | 0.42457742 | 0.39549677 | 3206 | tags=60%, list=21%, signal=48% | SLC15A4||SLC38A9||NPC1||ATP13A2||TMEM175||SLC46A3 | 3.8828482 | 4.8653540 | 4.5097871 | 4.9980310 | 3.9123165 | 4.4374516 | 4.7313249 | 4.8367170 | 3.8237133 | 3.9298433 | 3.892983 | 4.8402720 | 4.9579235 | 4.792822 | 4.5933466 | 4.539209 | 4.3891280 | 4.9654834 | 5.0381878 | 4.9894680 | 3.8764650 | 3.8100577 | 4.0405714 | 4.2268432 | 4.2906656 | 4.739117 | 4.7828176 | 4.724192 | 4.6852910 | 4.8288026 | 4.9135901 | 4.763842 |
| GO:0043202 | lysosomal lumen | 72 | -0.4488423 | -1.386537 | 0.0488243762 | 0.42457742 | 0.39549677 | 5280 | tags=60%, list=34%, signal=40% | CHID1||HEXB||TXNDC5||CTSV||LGMN||ARSB||GAA||MAN2B2||CTSA||SCARB2||HPSE||GM2A||GLA||TPP1||NPC2||IFI30||SDC3||GPC3||ACP2||ARSA||GPC1||PSAP||PPT2||PLD3||RNASET2||CTSD||GALNS||CTSF||LIPA||ATP13A2||CTSL||GBA||EPDR1||SDC1||VCAN||GPC2||FUCA1||CSPG5||NAGLU||GALC||NEU1||IDUA||SMPD1 | 5.5814634 | 6.5117867 | 5.8927478 | 6.4414274 | 5.4423156 | 5.5792528 | 6.4409001 | 6.3901047 | 5.6175073 | 5.7436757 | 5.356640 | 6.5767525 | 6.5030075 | 6.452903 | 6.0254140 | 5.801896 | 5.8409210 | 6.4005957 | 6.4484174 | 6.4743023 | 5.4320039 | 5.5322328 | 5.3573589 | 5.4457816 | 5.3894036 | 5.856246 | 6.4511778 | 6.442015 | 6.4294246 | 6.3885527 | 6.4097630 | 6.371747 |
| GO:0005686 | U2 snRNP | 19 | 0.5376591 | 1.544589 | 0.0488554834 | 0.42457742 | 0.39549677 | 6343 | tags=84%, list=41%, signal=50% | SNRPD3||PHF5A||SF3B6||SF3A1||SNRPB2||SNRPG||SF3B2||SF3B3||SF3B1||SNRPE||SNRPA1||SNRPD1||SNRPB||SF3A2||SF3A3||HTATSF1 | 8.2316525 | 8.2937457 | 8.2077087 | 8.4381065 | 8.2000613 | 8.0445854 | 8.3237551 | 8.4966307 | 8.2787689 | 8.2887253 | 8.121437 | 8.3297159 | 8.2818203 | 8.268988 | 8.3127636 | 8.117719 | 8.1858092 | 8.4728142 | 8.3972878 | 8.4432156 | 8.1616734 | 8.3208759 | 8.1091183 | 8.0773991 | 8.0509382 | 8.004477 | 8.3319944 | 8.327360 | 8.3118337 | 8.4788480 | 8.4899546 | 8.520762 |
KEGG
| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa03013 | Nucleocytoplasmic transport | 98 | 0.4817921 | 1.997475 | 7.812851e-06 | 0.0005815837 | 0.0005342778 | 5070 | tags=67%, list=37%, signal=43% | NXT2||NUP43||NUP58||NUP88||RANBP17||NMD3||XPO1||NUP54||KPNA6||THOC1||SRRM1||RAE1||NXF1||NUP160||KPNA4||TNPO1||UBE2I||NUP155||MAGOH||SUMO1||XPO4||SAP18||NUP50||KPNB1||NUP205||NCBP2||SUMO3||KPNA2||NUP107||ALYREF||TMEM33||RNPS1||PHAX||UPF3B||PNN||KPNA5||RAN||UPF2||THOC2||NUP133||IPO7||AAAS||XPOT||THOC5||DDX39B||DDX19B||KPNA3||CASC3||NUP93||NUP85||IPO11||ACIN1||IPO5||EEF1A1||CSE1L||TPR||SUMO2||TNPO3||SENP2||NUP35||NUP153||UPF3A||NUP42||EIF4A3||SEH1L||TNPO2 | 6.634100 | 7.685674 | 6.767342 | 7.668316 | 6.465975 | 6.373296 | 7.699191 | 7.710681 | 6.693915 | 6.814767 | 6.355271 | 7.767399 | 7.648530 | 7.637462 | 7.056428 | 6.410580 | 6.763325 | 7.663388 | 7.627110 | 7.713156 | 6.425037 | 6.647286 | 6.304425 | 6.372836 | 6.284857 | 6.457060 | 7.661140 | 7.717789 | 7.717903 | 7.730295 | 7.639917 | 7.759165 |
| hsa03040 | Spliceosome | 128 | 0.4339676 | 1.874099 | 1.198761e-05 | 0.0005815837 | 0.0005342778 | 4425 | tags=57%, list=32%, signal=39% | ISY1-RAB43||U2AF1||SMNDC1||CHERP||RBM22||PRPF38B||CCDC12||PRPF40A||SRSF10||THOC1||SNRNP40||SYF2||PCBP1||SLU7||TCERG1||USP39||WBP11||SNRPD3||TRA2B||PRPF19||MAGOH||PRPF38A||PHF5A||SNRPA||DHX38||DHX15||PLRG1||NCBP2||SF3B6||SF3B4||SF3A1||SNRPB2||SNW1||ALYREF||SNRPG||RBM17||SNRNP27||LSM8||SF3B2||PRPF3||LSM6||PRPF6||LSM3||TRA2A||AQR||SRSF3||PPIE||RBM25||THOC2||HNRNPK||SF3B3||PRPF18||CDC5L||DDX39B||PUF60||SF3B1||HNRNPC||ZMAT2||HNRNPA3||SNRPE||CDC40||DHX8||PRPF8||ACIN1||BCAS2||EFTUD2||SNRPA1||PQBP1||XAB2||SNRPC||SNRNP70||HNRNPU||SNRNP200 | 7.520732 | 7.953257 | 7.509491 | 7.897153 | 7.468394 | 7.388205 | 7.979883 | 7.953018 | 7.548629 | 7.627874 | 7.374140 | 7.985121 | 7.934116 | 7.939993 | 7.690456 | 7.349368 | 7.467643 | 7.889180 | 7.861582 | 7.939608 | 7.453419 | 7.540092 | 7.408560 | 7.407091 | 7.354884 | 7.402067 | 7.978692 | 7.972433 | 7.988480 | 7.933018 | 7.961950 | 7.963880 |
| hsa05168 | Herpes simplex virus 1 infection | 370 | 0.3106866 | 1.513550 | 1.681929e-05 | 0.0005815837 | 0.0005342778 | 1474 | tags=24%, list=11%, signal=22% | ZNF256||ZNF268||ZNF331||ZNF567||ZNF441||ZNF226||ZNF223||ZNF114||ZNF713||ZNF383||ZNF551||ZNF528||ZNF404||ZNF136||ZNF350||ZNF568||ZNF587||ZNF225||ZNF227||ZNF169||ZNF419||ZNF549||ZNF235||POU2F1||ZNF230||ZNF461||ZNF45||ZNF224||ZNF140||ZNF17||ZNF304||ZNF175||ZNF875||ZNF565||ZNF888||ZNF669||ZNF200||ZNF805||ZNF12||ZNF791||ZNF320||ZNF566||ZNF468||ZNF333||ZNF431||ZNF596||ZNF214||ZNF699||ZNF211||ZNF432||ZNF416||ZNF613||ZNF417||ZNF749||ZNF614||EIF2S1||ZNF766||HCFC2||ZNF84||ZNF112||ZNF547||ZNF577||ZNF26||ZNF440||ZNF684||EIF2AK2||ZNF433||ZNF460||ZNF790||ZNF561||ZNF101||ZNF221||ZNF382||ZNF37A||ZNF420||PIK3R1||ZNF597||ZNF782||ZNF680||ZNF841||ZNF557||ZNF611||ZNF212||ZNF529||APAF1||ZNF548||IL12A | 4.388954 | 5.025723 | 4.546492 | 4.833528 | 4.378123 | 4.492159 | 4.987082 | 4.848860 | 4.372601 | 4.424142 | 4.369463 | 5.044975 | 5.034870 | 4.996879 | 4.667011 | 4.483299 | 4.481168 | 4.815203 | 4.820150 | 4.864713 | 4.366461 | 4.355440 | 4.411848 | 4.427392 | 4.461171 | 4.583216 | 4.994740 | 4.978205 | 4.988253 | 4.835197 | 4.877310 | 4.833647 |
| hsa04110 | Cell cycle | 119 | 0.4391064 | 1.885453 | 2.114850e-05 | 0.0005815837 | 0.0005342778 | 3526 | tags=53%, list=26%, signal=40% | TGFB3||TTK||CCND1||CDC14A||CDC6||SMAD3||CDC23||CDKN2D||CDC25C||BUB1B||ANAPC1||E2F3||WEE1||MCM4||SMAD2||CDC45||CDK2||ORC4||CCNE2||ORC6||CDC14B||CDKN1B||PCNA||SMC3||CDKN1A||CCNA2||CHEK1||CDC27||CDK1||MAD2L1||BUB1||SMAD4||ANAPC15||ANAPC4||MCM5||PTTG1||CCNB1||TFDP2||MCM7||ANAPC2||RAD21||RB1||ANAPC7||ORC1||CDC25A||CCNH||MCM3||YWHAB||BUB3||SMC1A||YWHAQ||CDK6||YWHAG||STAG2||DBF4||ORC3||STAG1||E2F4||TFDP1||CDC20||CCNB2||FZR1||YWHAZ | 5.951057 | 6.562916 | 5.988783 | 6.594083 | 5.886195 | 5.917852 | 6.600034 | 6.685962 | 5.951111 | 6.042960 | 5.852841 | 6.594305 | 6.543193 | 6.550719 | 6.155101 | 5.874478 | 5.920817 | 6.576338 | 6.551012 | 6.652937 | 5.874914 | 5.911960 | 5.871360 | 5.918947 | 5.911043 | 5.923540 | 6.595541 | 6.589967 | 6.614480 | 6.667520 | 6.704633 | 6.685494 |
| hsa04114 | Oocyte meiosis | 103 | 0.4303202 | 1.801891 | 1.818222e-04 | 0.0040000883 | 0.0036747222 | 3201 | tags=45%, list=23%, signal=35% | SPDYE2B||PGR||REC8||CAMK2B||AR||PPP3CB||CPEB2||CDC23||CDC25C||ANAPC1||CALML4||CPEB3||MAPK1||PPP2R1B||MAPK13||PPP2R5A||PPP2R5E||PRKACB||CDK2||SGO1||CCNE2||CALM2||PPP3R1||SMC3||CDC27||FBXO5||CDK1||MAD2L1||PPP1CB||BUB1||SLK||ANAPC15||ANAPC4||PTTG1||CCNB1||ANAPC2||ANAPC7||AURKA||YWHAB||SMC1A||YWHAQ||YWHAG||PPP2R5C||STAG3||ITPR2||PPP3CA | 5.704050 | 6.322299 | 5.679234 | 6.359016 | 5.641125 | 5.668926 | 6.366957 | 6.441042 | 5.720676 | 5.797143 | 5.586505 | 6.339129 | 6.303208 | 6.324335 | 5.854939 | 5.555177 | 5.609600 | 6.338932 | 6.315723 | 6.420292 | 5.606728 | 5.701596 | 5.613088 | 5.683249 | 5.685805 | 5.637206 | 6.349255 | 6.361874 | 6.389449 | 6.424481 | 6.453528 | 6.444962 |
| hsa03015 | mRNA surveillance pathway | 83 | 0.4399567 | 1.782419 | 9.177659e-04 | 0.0168257074 | 0.0154571092 | 5025 | tags=65%, list=36%, signal=42% | PAPOLG||CPSF4||PPP2R3A||NXT2||PPP2R2A||PPP2R1B||PAPOLA||PPP2R5A||PPP2R5E||SRRM1||ETF1||SMG1||NXF1||PPP2R3C||CPSF7||PCF11||PPP1CB||TARDBP||MAGOH||SAP18||RNGTT||NCBP2||CSTF3||ALYREF||CLP1||PPP2R2D||RNPS1||PPP2R5C||SMG5||UPF3B||PNN||NUDT21||UPF2||CPSF1||GSPT1||SSU72||DDX39B||DDX19B||RNMT||PPP2R2C||CASC3||ACIN1||PPP2R1A||SYMPK||WDR33||PABPN1||PPP1CC||WDR82||CSTF1||UPF3A||BCL2L2-PABPN1||EIF4A3||PPP2R5D||SMG7 | 6.256319 | 6.762681 | 6.375102 | 6.726208 | 6.224607 | 6.405992 | 6.803600 | 6.795486 | 6.224795 | 6.304701 | 6.238185 | 6.767786 | 6.759344 | 6.760898 | 6.548221 | 6.294828 | 6.265164 | 6.706422 | 6.687829 | 6.782616 | 6.226057 | 6.183974 | 6.262716 | 6.367684 | 6.387446 | 6.461155 | 6.789981 | 6.802774 | 6.817909 | 6.782105 | 6.817781 | 6.786308 |
| hsa04142 | Lysosome | 114 | -0.4563902 | -1.551724 | 3.880836e-03 | 0.0609845582 | 0.0560240917 | 4349 | tags=52%, list=32%, signal=36% | CTSV||LGMN||ARSB||GAA||GGA3||M6PR||IGF2R||CTSH||CTSA||DMXL2||SCARB2||ATP6V0B||GM2A||GLA||ATP6V0A1||AP3B2||TPP1||ATP6AP1||NPC2||PLA2G15||AGA||ACP2||GNPTG||LAPTM4A||ARSA||PSAP||PPT2||HGSNAT||NAGPA||ABCB9||TCIRG1||AP4M1||ABCA2||CTSD||GALNS||SLC17A5||ATP6V0C||CTSF||SUMF1||LIPA||NPC1||MCOLN1||HYAL3||CTSL||CTSZ||CTSO||GBA||CTNS||DNASE2||SORT1||HYAL2||FUCA1||NAGLU||NAPSA||GALC||NEU1||IDUA||SMPD1||AP1G2 | 5.467940 | 5.794602 | 5.847021 | 5.811989 | 5.481333 | 5.762974 | 5.662281 | 5.718441 | 5.434193 | 5.464340 | 5.504426 | 5.830638 | 5.820942 | 5.730111 | 5.811361 | 5.903655 | 5.824304 | 5.811159 | 5.830577 | 5.793998 | 5.480723 | 5.430026 | 5.531466 | 5.627734 | 5.654944 | 5.979122 | 5.694653 | 5.660171 | 5.631321 | 5.723594 | 5.757407 | 5.673076 |
| hsa04060 | Cytokine-cytokine receptor interaction | 82 | -0.4609468 | -1.502288 | 1.245957e-02 | 0.1713190368 | 0.1573839955 | 2978 | tags=39%, list=22%, signal=31% | CD40||ACVR1||BMP2||BMP6||AMHR2||TGFBR1||IL1R1||TNFRSF21||FAS||IL9R||CSF1||XCL1||TNFRSF12A||IL27RA||BMP8B||NGFR||IL10RB||CD70||GDF9||IL17RA||IL6R||CXCR4||IL4R||TSLP||CTF1||IFNLR1||GDF1||IL17RC||EPOR||TGFB2||CXCL12||IL23A | 1.872733 | 2.825640 | 2.618567 | 2.811885 | 1.828856 | 2.292174 | 2.759883 | 2.784494 | 1.859946 | 1.879087 | 1.879082 | 2.808405 | 2.866659 | 2.800955 | 2.758042 | 2.603990 | 2.480202 | 2.766053 | 2.838851 | 2.829666 | 1.826062 | 1.733397 | 1.921010 | 2.016351 | 2.055429 | 2.698562 | 2.791382 | 2.741866 | 2.745876 | 2.746609 | 2.895910 | 2.703841 |
| hsa05017 | Spinocerebellar ataxia | 117 | 0.3223620 | 1.380239 | 1.771337e-02 | 0.2164966899 | 0.1988869113 | 4353 | tags=44%, list=32%, signal=31% | TRPC3||GRIN2C||PSMD9||SP1||KCND3||ATG14||KAT5||ATG13||MAPK8||PUM2||RB1CC1||PIK3R1||PLCB1||PRKCB||RBPJ||PSMD12||PSMC6||CYCS||MCU||PSMC3||MAP3K5||VDAC2||ERN1||AKT2||GNAQ||PSMD14||ATXN1||NOP56||PSMD3||ULK1||PIK3CD||PSMD1||PSMD11||ITPR2||PSMD13||PSMB3||PSMD7||ATG2B||GTF2B||ATXN2||ADRM1||PIK3CB||PSMB1||PSMA1||PSMC2||NRBF2||ATXN10||PSMB2||ATXN2L||AKT3||PIK3C3||TBPL1 | 6.541779 | 6.635936 | 6.590437 | 6.748216 | 6.468867 | 6.398964 | 6.645912 | 6.798934 | 6.578091 | 6.643483 | 6.392131 | 6.678554 | 6.613546 | 6.614745 | 6.668767 | 6.479657 | 6.616343 | 6.759987 | 6.707434 | 6.776331 | 6.456755 | 6.550561 | 6.395019 | 6.423882 | 6.357705 | 6.414419 | 6.650907 | 6.640822 | 6.645989 | 6.792327 | 6.803983 | 6.800468 |
| hsa04080 | Neuroactive ligand-receptor interaction | 95 | -0.4301391 | -1.432692 | 2.103295e-02 | 0.2205459291 | 0.2026067866 | 2208 | tags=32%, list=16%, signal=27% | LPAR3||RLN1||GPR50||F2RL1||THRB||GABBR2||NMU||GRIN2D||LYPD6||CHRNA3||GABRD||NPFF||ADORA1||CHRNB1||ADORA2A||ADCYAP1R1||VIPR1||ADRB1||AGTR1||GABRR2||HTR6||CHRM3||P2RY2||OPRL1||UTS2R||CYSLTR1||GABRG2||RLN2||GRID2||CHRNA4 | 2.485832 | 2.357086 | 2.479080 | 2.091528 | 2.497752 | 2.611350 | 2.277638 | 2.001408 | 2.452800 | 2.357261 | 2.633659 | 2.356186 | 2.380979 | 2.333705 | 2.403981 | 2.605237 | 2.419196 | 2.132570 | 2.079084 | 2.061985 | 2.497650 | 2.351923 | 2.630278 | 2.507427 | 2.655172 | 2.666073 | 2.332762 | 2.261257 | 2.237173 | 1.990721 | 2.052693 | 1.959237 |
| hsa04914 | Progesterone-mediated oocyte maturation | 81 | 0.3520692 | 1.418135 | 2.214243e-02 | 0.2205459291 | 0.2026067866 | 2580 | tags=33%, list=19%, signal=27% | SPDYE2B||PGR||CPEB2||KRAS||CDC23||CDC25C||ANAPC1||MAPK8||CPEB3||MAPK1||PIK3R1||MAPK13||PRKACB||CDK2||CCNA2||CDC27||CDK1||MAD2L1||BUB1||ANAPC15||ANAPC4||AKT2||CCNB1||ANAPC2||ANAPC7||AURKA||CDC25A | 5.404527 | 6.156891 | 5.612387 | 6.261537 | 5.338823 | 5.511460 | 6.221931 | 6.319905 | 5.378424 | 5.490062 | 5.340892 | 6.166726 | 6.145153 | 6.158711 | 5.858777 | 5.450534 | 5.491917 | 6.229612 | 6.224161 | 6.328434 | 5.338188 | 5.332219 | 5.346030 | 5.505963 | 5.500171 | 5.528096 | 6.213171 | 6.217052 | 6.235472 | 6.312138 | 6.329771 | 6.317748 |
| hsa04120 | Ubiquitin mediated proteolysis | 131 | 0.3131257 | 1.356578 | 2.405956e-02 | 0.2205459291 | 0.2026067866 | 4784 | tags=52%, list=35%, signal=34% | FBXW7||DDB2||CDC23||ANAPC1||MAP3K1||UBE2Z||UBE2E1||UBE2E2||DET1||UBA7||UBE2C||CUL2||UBE2A||CDC27||SMURF2||CUL5||UBE2J2||PIAS3||UBE2Q2||ANAPC15||PRPF19||ANAPC4||UBE2I||SIAH1||UBA3||ANAPC2||NEDD4||UBE3C||XIAP||ANAPC7||HERC1||RCHY1||FANCL||PPIL2||SMURF1||UBE2B||RHOBTB1||ERCC8||KLHL13||UBE2D2||CDC20||FZR1||UBA6||UBE4B||UBE2K||UBR5||SKP1||UBE2H||UBB||UBE2D3||PML||BIRC2||MDM2||HERC4||FBXW8||HUWE1||UBE3A||CUL3||CDC16||RPS27A||UBE2N||DDB1||CBL||ELOC||ITCH||UBC||SKP2||UBE3B | 6.788328 | 6.790234 | 6.677779 | 6.749448 | 6.704514 | 6.566049 | 6.822868 | 6.790846 | 6.819886 | 6.857673 | 6.681526 | 6.803878 | 6.771363 | 6.795265 | 6.669328 | 6.610701 | 6.749914 | 6.772104 | 6.711615 | 6.763883 | 6.697245 | 6.786639 | 6.625112 | 6.638306 | 6.534377 | 6.522632 | 6.820627 | 6.826305 | 6.821667 | 6.782039 | 6.774397 | 6.815766 |
| hsa05202 | Transcriptional misregulation in cancer | 125 | 0.3003331 | 1.292307 | 4.111986e-02 | 0.3479372771 | 0.3196361588 | 2659 | tags=32%, list=19%, signal=26% | FLI1||RUNX2||H3C2||BCL11B||COMMD3-BMI1||RXRG||ELK4||HPGD||WT1||SIX4||SP1||RXRB||DDB2||NR4A3||MEF2C||CDK14||CCNT2||KLF3||PBX1||KDM6A||RARA||BMI1||HOXA9||ETV6||REL||CDKN1B||CDKN1A||CCNA2||FLT1||H3C6||JMJD1C||MET||MEIS1||HMGA2||RUNX1||BMP2K||SMAD1||ZEB1||AFF1||EWSR1 | 5.073516 | 5.562054 | 5.260594 | 5.523933 | 5.059677 | 5.177891 | 5.612324 | 5.523816 | 5.063251 | 5.168363 | 4.982923 | 5.541473 | 5.573462 | 5.571007 | 5.495723 | 5.075245 | 5.176767 | 5.507889 | 5.490751 | 5.571884 | 5.025721 | 5.092227 | 5.060315 | 5.140470 | 5.138578 | 5.251694 | 5.608896 | 5.600479 | 5.627463 | 5.581177 | 5.413334 | 5.570881 |
| hsa04928 | Parathyroid hormone synthesis, secretion and action | 83 | 0.3270677 | 1.325066 | 4.808278e-02 | 0.3704605397 | 0.3403273857 | 1317 | tags=17%, list=10%, signal=15% | MMP14||RUNX2||RXRG||CYP27B1||PTHLH||SP1||RXRB||CASR||MEF2C||ATF2||MAPK1||PLCB1||PRKACB||PRKCB | 5.776806 | 5.902604 | 5.684113 | 5.928387 | 5.797768 | 5.919981 | 5.914773 | 5.960131 | 5.717281 | 5.812121 | 5.799194 | 5.891986 | 5.895542 | 5.920120 | 5.788497 | 5.638286 | 5.619542 | 5.940828 | 5.886197 | 5.957183 | 5.764727 | 5.792978 | 5.834737 | 5.915992 | 6.028258 | 5.807229 | 5.914373 | 5.902614 | 5.927226 | 5.967003 | 5.963206 | 5.950131 |
ORA: All DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: All DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0033081 | regulation of T cell differentiation in thymus | 1/8 | 23/20870 | 0.008784018 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.585497 | 4.928688 | 4.902496 | 5.107921 | 4.514365 | 4.612261 | 4.934742 | 5.088658 | 4.634645 | 4.713194 | 4.389217 | 4.905810 | 4.952248 | 4.927632 | 5.016100 | 4.771138 | 4.909837 | 5.101020 | 5.132398 | 5.090009 | 4.447659 | 4.656320 | 4.427835 | 4.568104 | 4.503017 | 4.753785 | 4.943701 | 4.933921 | 4.926553 | 5.093756 | 5.107394 | 5.064491 |
| GO:0043011 | myeloid dendritic cell differentiation | 1/8 | 26/20870 | 0.009924767 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 2.105275 | 3.195595 | 2.499026 | 3.252992 | 1.995626 | 2.251112 | 3.149993 | 3.229214 | 2.114603 | 2.133670 | 2.066731 | 3.147108 | 3.302090 | 3.131341 | 2.759749 | 2.361182 | 2.335829 | 3.181834 | 3.263834 | 3.310385 | 2.063559 | 1.948172 | 1.972568 | 2.203308 | 2.137714 | 2.399310 | 3.167967 | 3.169776 | 3.111478 | 3.207123 | 3.242200 | 3.238066 |
| GO:0001773 | myeloid dendritic cell activation | 1/8 | 35/20870 | 0.013340125 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.504789 | 6.073282 | 4.531929 | 6.108157 | 4.284952 | 4.079282 | 6.225128 | 6.137683 | 4.597126 | 4.656392 | 4.223932 | 6.019614 | 6.064666 | 6.133290 | 5.169470 | 4.080871 | 4.043536 | 6.024227 | 6.109857 | 6.185862 | 4.115883 | 4.507446 | 4.201558 | 3.977364 | 3.977021 | 4.264051 | 6.214058 | 6.210866 | 6.250129 | 6.205931 | 6.010636 | 6.188503 |
| GO:0007616 | long-term memory | 1/8 | 42/20870 | 0.015989383 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 3.517536 | 3.761974 | 3.896904 | 3.812534 | 3.503503 | 3.931839 | 3.793747 | 3.769498 | 3.473563 | 3.497456 | 3.579440 | 3.687618 | 3.840743 | 3.753463 | 3.952191 | 3.911134 | 3.824455 | 3.828564 | 3.778341 | 3.830099 | 3.506333 | 3.377636 | 3.616648 | 3.787640 | 3.897735 | 4.093379 | 3.817193 | 3.819479 | 3.743274 | 3.745366 | 3.813303 | 3.748805 |
| GO:0033077 | T cell differentiation in thymus | 1/8 | 80/20870 | 0.030262747 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 6.276858 | 7.249687 | 6.132251 | 7.157404 | 6.154380 | 5.892699 | 7.188447 | 7.214883 | 6.343681 | 6.417722 | 6.042437 | 7.395767 | 7.172254 | 7.169138 | 6.355957 | 5.728399 | 6.238510 | 7.149985 | 7.132206 | 7.189425 | 6.039300 | 6.346179 | 6.056619 | 6.045237 | 5.979642 | 5.617539 | 7.155625 | 7.195253 | 7.213852 | 7.161061 | 7.235149 | 7.246945 |
| GO:0097028 | dendritic cell differentiation | 1/8 | 81/20870 | 0.030635903 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.378948 | 5.625933 | 4.602909 | 5.600736 | 4.284212 | 4.325148 | 5.707809 | 5.608541 | 4.416960 | 4.439533 | 4.274891 | 5.588268 | 5.631488 | 5.657205 | 4.922449 | 4.457849 | 4.364578 | 5.540876 | 5.604222 | 5.654854 | 4.168306 | 4.364300 | 4.312944 | 4.179045 | 4.227092 | 4.541784 | 5.713055 | 5.693753 | 5.716515 | 5.660403 | 5.512757 | 5.647868 |
| GO:0045670 | regulation of osteoclast differentiation | 1/8 | 85/20870 | 0.032127267 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.114814 | 4.760388 | 4.486898 | 4.857521 | 4.121294 | 4.416532 | 4.731299 | 4.793229 | 4.082689 | 4.153526 | 4.107327 | 4.784444 | 4.780892 | 4.714766 | 4.576700 | 4.431303 | 4.448267 | 4.843021 | 4.860648 | 4.868773 | 4.074536 | 4.095340 | 4.191298 | 4.266996 | 4.391001 | 4.574870 | 4.739338 | 4.739285 | 4.715140 | 4.794480 | 4.796606 | 4.788590 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1/8 | 109/20870 | 0.041033369 | 0.1628826 | 0.1357355 | FAM53B | 1 | 6.387130 | 6.359229 | 6.469344 | 6.290553 | 6.309753 | 6.022660 | 6.401387 | 6.307559 | 6.536823 | 6.532801 | 6.036833 | 6.285090 | 6.371577 | 6.417885 | 6.395717 | 6.390286 | 6.610881 | 6.307399 | 6.294958 | 6.269037 | 6.382747 | 6.580005 | 5.879406 | 6.201308 | 5.881207 | 5.966128 | 6.383865 | 6.424037 | 6.395964 | 6.308583 | 6.286382 | 6.327421 |
| GO:0032526 | response to retinoic acid | 1/8 | 124/20870 | 0.046563196 | 0.1628826 | 0.1357355 | OVCA2 | 1 | 4.910250 | 4.860073 | 4.887516 | 4.802467 | 4.951254 | 4.976752 | 4.778985 | 4.766524 | 4.910267 | 4.811037 | 5.003062 | 4.934211 | 4.851942 | 4.790452 | 4.809668 | 4.958506 | 4.890531 | 4.847113 | 4.753136 | 4.805616 | 4.932286 | 4.871438 | 5.044656 | 4.965463 | 5.020847 | 4.942826 | 4.812605 | 4.762040 | 4.761712 | 4.758845 | 4.771370 | 4.769326 |
| GO:0030316 | osteoclast differentiation | 1/8 | 125/20870 | 0.046930857 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.719309 | 5.339730 | 4.946655 | 5.364260 | 4.704698 | 4.942838 | 5.284330 | 5.343848 | 4.687351 | 4.723033 | 4.746921 | 5.371132 | 5.353917 | 5.292977 | 4.992772 | 4.929786 | 4.916245 | 5.337837 | 5.368144 | 5.386384 | 4.666828 | 4.672043 | 4.772735 | 4.823850 | 4.944916 | 5.050829 | 5.280508 | 5.288279 | 5.284193 | 5.333542 | 5.370659 | 5.326957 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0009931 | calcium-dependent protein serine/threonine kinase activity | 1/6 | 25/20678 | 0.007233068 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.593800 | 4.589005 | 4.653472 | 4.544496 | 4.639457 | 4.864762 | 4.585571 | 4.535268 | 4.546449 | 4.502467 | 4.722930 | 4.579352 | 4.600393 | 4.587191 | 4.575860 | 4.738139 | 4.641786 | 4.557519 | 4.478310 | 4.595201 | 4.655743 | 4.493024 | 4.757372 | 4.813095 | 4.917215 | 4.862094 | 4.622385 | 4.547875 | 4.585492 | 4.531785 | 4.581978 | 4.490587 |
| GO:0010857 | calcium-dependent protein kinase activity | 1/6 | 26/20678 | 0.007521481 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.711194 | 4.645042 | 4.756797 | 4.585098 | 4.763772 | 4.973860 | 4.633759 | 4.569588 | 4.659589 | 4.622028 | 4.842219 | 4.638803 | 4.656172 | 4.640085 | 4.666424 | 4.847374 | 4.750904 | 4.604630 | 4.514284 | 4.633708 | 4.779911 | 4.625435 | 4.875049 | 4.928696 | 5.025169 | 4.966071 | 4.669931 | 4.594683 | 4.635680 | 4.567604 | 4.612929 | 4.526948 |
| GO:0004683 | calmodulin-dependent protein kinase activity | 1/6 | 28/20678 | 0.008098099 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.351604 | 4.248910 | 4.630288 | 4.248412 | 4.380313 | 4.629526 | 4.286107 | 4.241967 | 4.306313 | 4.331224 | 4.415015 | 4.265323 | 4.241082 | 4.240185 | 4.639986 | 4.668851 | 4.580629 | 4.267377 | 4.199296 | 4.277318 | 4.405814 | 4.251920 | 4.474301 | 4.516681 | 4.565601 | 4.791194 | 4.353516 | 4.244593 | 4.257745 | 4.232236 | 4.268597 | 4.224686 |
| GO:0008083 | growth factor activity | 1/6 | 164/20678 | 0.046658712 | 0.09587126 | 0.06055027 | CLEC11A | 1 | 5.053083 | 4.838445 | 5.023100 | 4.646887 | 5.114123 | 5.251340 | 4.752778 | 4.588090 | 4.980921 | 4.990709 | 5.178867 | 4.852855 | 4.854022 | 4.807981 | 4.852572 | 5.142550 | 5.058951 | 4.695874 | 4.589556 | 4.653253 | 5.106034 | 5.020466 | 5.209640 | 5.165631 | 5.306991 | 5.277574 | 4.786802 | 4.733128 | 4.737788 | 4.605289 | 4.641440 | 4.514598 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:1905103 | integral component of lysosomal membrane | 1/9 | 10/21916 | 0.004099849 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.914375 | 4.411734 | 4.596753 | 4.557231 | 3.975354 | 4.557743 | 4.279654 | 4.312983 | 3.833125 | 3.895123 | 4.009311 | 4.357264 | 4.541132 | 4.327393 | 4.548487 | 4.703044 | 4.532506 | 4.574653 | 4.569866 | 4.526693 | 3.953618 | 3.809192 | 4.143666 | 4.325002 | 4.375671 | 4.899783 | 4.359588 | 4.251794 | 4.223999 | 4.303043 | 4.414828 | 4.214047 |
| GO:0031166 | integral component of vacuolar membrane | 1/9 | 12/21916 | 0.004918023 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.882848 | 4.865354 | 4.509787 | 4.998031 | 3.912317 | 4.437452 | 4.731325 | 4.836717 | 3.823713 | 3.929843 | 3.892983 | 4.840272 | 4.957924 | 4.792822 | 4.593347 | 4.539209 | 4.389128 | 4.965483 | 5.038188 | 4.989468 | 3.876465 | 3.810058 | 4.040571 | 4.226843 | 4.290666 | 4.739117 | 4.782818 | 4.724192 | 4.685291 | 4.828803 | 4.913590 | 4.763842 |
| GO:0031310 | intrinsic component of vacuolar membrane | 1/9 | 12/21916 | 0.004918023 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.882848 | 4.865354 | 4.509787 | 4.998031 | 3.912317 | 4.437452 | 4.731325 | 4.836717 | 3.823713 | 3.929843 | 3.892983 | 4.840272 | 4.957924 | 4.792822 | 4.593347 | 4.539209 | 4.389128 | 4.965483 | 5.038188 | 4.989468 | 3.876465 | 3.810058 | 4.040571 | 4.226843 | 4.290666 | 4.739117 | 4.782818 | 4.724192 | 4.685291 | 4.828803 | 4.913590 | 4.763842 |
| GO:0005811 | lipid droplet | 1/9 | 101/21916 | 0.040727430 | 0.061091145 | 0.021435489 | C18orf32 | 1 | 7.130742 | 6.703458 | 7.082124 | 6.703623 | 7.193841 | 7.322343 | 6.686595 | 6.709812 | 7.050864 | 7.064566 | 7.266533 | 6.705444 | 6.724642 | 6.679940 | 6.912603 | 7.190900 | 7.128335 | 6.743683 | 6.646030 | 6.719375 | 7.206356 | 7.116293 | 7.255437 | 7.353714 | 7.383784 | 7.224626 | 6.686723 | 6.701391 | 6.671515 | 6.706434 | 6.723321 | 6.699579 |
KEGG
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa05034 | Alcoholism | 2/3 | 187/8146 | 0.001548863 | 0.02633067 | 0.01141267 | CAMK4||H2AC18 | 2 | 6.081292 | 6.312998 | 6.203495 | 6.296332 | 6.035621 | 6.066784 | 6.329420 | 6.327174 | 6.084533 | 6.207338 | 5.939596 | 6.318456 | 6.308019 | 6.312499 | 6.267141 | 6.088087 | 6.248624 | 6.304355 | 6.246149 | 6.337027 | 5.995385 | 6.110528 | 5.997929 | 6.035454 | 6.030588 | 6.132024 | 6.323480 | 6.323340 | 6.341367 | 6.342897 | 6.301839 | 6.336450 |
| hsa04720 | Long-term potentiation | 1/3 | 67/8146 | 0.024475277 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.767229 | 5.888241 | 5.758357 | 6.020890 | 5.756241 | 5.903782 | 5.938052 | 6.090766 | 5.754439 | 5.814736 | 5.731220 | 5.891437 | 5.871799 | 5.901331 | 5.773287 | 5.731539 | 5.769875 | 6.042904 | 5.942405 | 6.074100 | 5.732244 | 5.781166 | 5.754899 | 5.907409 | 5.940431 | 5.862446 | 5.935278 | 5.925941 | 5.952807 | 6.074190 | 6.096181 | 6.101781 |
| hsa05031 | Amphetamine addiction | 1/3 | 69/8146 | 0.025199676 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 6.206956 | 6.485590 | 6.265257 | 6.537937 | 6.198552 | 6.387565 | 6.509016 | 6.578806 | 6.177469 | 6.287388 | 6.152409 | 6.501866 | 6.473855 | 6.480901 | 6.318973 | 6.221454 | 6.253626 | 6.562976 | 6.465062 | 6.583032 | 6.184174 | 6.213202 | 6.198135 | 6.394569 | 6.401446 | 6.366442 | 6.503404 | 6.506514 | 6.517092 | 6.571238 | 6.580342 | 6.584804 |
| hsa05214 | Glioma | 1/3 | 75/8146 | 0.027370723 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.784893 | 5.702299 | 5.749235 | 5.611233 | 5.803506 | 5.948375 | 5.711067 | 5.642193 | 5.720731 | 5.755829 | 5.873635 | 5.717816 | 5.691112 | 5.697836 | 5.698036 | 5.809268 | 5.738194 | 5.651754 | 5.541429 | 5.638030 | 5.823915 | 5.667322 | 5.908956 | 5.890779 | 6.023570 | 5.927497 | 5.733073 | 5.680940 | 5.718688 | 5.629903 | 5.663737 | 5.632695 |
| hsa04211 | Longevity regulating pathway | 1/3 | 89/8146 | 0.032423957 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.517718 | 5.577823 | 5.623100 | 5.684231 | 5.523441 | 5.735019 | 5.599955 | 5.709516 | 5.486466 | 5.543750 | 5.522356 | 5.594729 | 5.577903 | 5.560636 | 5.636783 | 5.619493 | 5.612919 | 5.702531 | 5.616027 | 5.731645 | 5.517424 | 5.503493 | 5.549028 | 5.713241 | 5.744566 | 5.747005 | 5.622772 | 5.578475 | 5.598274 | 5.704510 | 5.718415 | 5.705582 |
| hsa04925 | Aldosterone synthesis and secretion | 1/3 | 98/8146 | 0.035663205 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.595871 | 5.806341 | 5.716022 | 5.865852 | 5.578227 | 5.809613 | 5.790656 | 5.877407 | 5.562661 | 5.679469 | 5.541626 | 5.818782 | 5.813534 | 5.786501 | 5.735456 | 5.708640 | 5.703768 | 5.887161 | 5.816726 | 5.892436 | 5.578219 | 5.581662 | 5.574793 | 5.796712 | 5.784120 | 5.847230 | 5.798296 | 5.788333 | 5.785307 | 5.875349 | 5.885009 | 5.871832 |
| hsa04725 | Cholinergic synapse | 1/3 | 113/8146 | 0.041045871 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 4.898453 | 4.818758 | 4.931808 | 4.955285 | 4.915127 | 5.119258 | 4.835006 | 4.983973 | 4.856953 | 4.939996 | 4.897214 | 4.813787 | 4.827704 | 4.814740 | 4.907820 | 4.937064 | 4.950214 | 4.996756 | 4.860379 | 5.004225 | 4.908956 | 4.901372 | 4.934841 | 5.107619 | 5.121924 | 5.128154 | 4.852895 | 4.831052 | 4.820886 | 4.979196 | 4.984557 | 4.988151 |
| hsa04722 | Neurotrophin signaling pathway | 1/3 | 119/8146 | 0.043193317 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 6.016337 | 5.952352 | 5.955489 | 6.002791 | 5.993966 | 6.095876 | 5.974173 | 6.041013 | 6.008955 | 6.067897 | 5.970486 | 5.962448 | 5.944408 | 5.950142 | 5.980340 | 5.913952 | 5.971281 | 6.025701 | 5.926940 | 6.052716 | 5.992483 | 6.006068 | 5.983256 | 6.090956 | 6.135859 | 6.059797 | 5.987265 | 5.956042 | 5.979029 | 6.035523 | 6.045466 | 6.042033 |
| hsa04380 | Osteoclast differentiation | 1/3 | 128/8146 | 0.046408471 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 4.481272 | 4.694176 | 4.670136 | 4.654232 | 4.494373 | 4.763273 | 4.679935 | 4.644717 | 4.441907 | 4.459265 | 4.540704 | 4.720090 | 4.699747 | 4.662092 | 4.690251 | 4.713625 | 4.604248 | 4.669373 | 4.607502 | 4.684669 | 4.501143 | 4.375176 | 4.598170 | 4.652158 | 4.754349 | 4.874698 | 4.713816 | 4.655560 | 4.669787 | 4.621031 | 4.658232 | 4.654599 |
| hsa05322 | Systemic lupus erythematosus | 1/3 | 136/8146 | 0.049260331 | 0.07509782 | 0.03255014 | H2AC18 | 1 | 6.321517 | 6.470763 | 6.451806 | 6.480259 | 6.267784 | 6.143901 | 6.478292 | 6.522252 | 6.354829 | 6.462215 | 6.127557 | 6.504658 | 6.449156 | 6.457854 | 6.551703 | 6.278809 | 6.510198 | 6.480561 | 6.446902 | 6.512567 | 6.198438 | 6.388119 | 6.208804 | 6.083071 | 6.119473 | 6.225346 | 6.476118 | 6.469098 | 6.489583 | 6.546568 | 6.483946 | 6.535471 |
ORA: Down-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Down-regulated DEGs” results.
GO: BP
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0033081 | regulation of T cell differentiation in thymus | 1/8 | 23/20870 | 0.008784018 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.585497 | 4.928688 | 4.902496 | 5.107921 | 4.514365 | 4.612261 | 4.934742 | 5.088658 | 4.634645 | 4.713194 | 4.389217 | 4.905810 | 4.952248 | 4.927632 | 5.016100 | 4.771138 | 4.909837 | 5.101020 | 5.132398 | 5.090009 | 4.447659 | 4.656320 | 4.427835 | 4.568104 | 4.503017 | 4.753785 | 4.943701 | 4.933921 | 4.926553 | 5.093756 | 5.107394 | 5.064491 |
| GO:0043011 | myeloid dendritic cell differentiation | 1/8 | 26/20870 | 0.009924767 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 2.105275 | 3.195595 | 2.499026 | 3.252992 | 1.995626 | 2.251112 | 3.149993 | 3.229214 | 2.114603 | 2.133670 | 2.066731 | 3.147108 | 3.302090 | 3.131341 | 2.759749 | 2.361182 | 2.335829 | 3.181834 | 3.263834 | 3.310385 | 2.063559 | 1.948172 | 1.972568 | 2.203308 | 2.137714 | 2.399310 | 3.167967 | 3.169776 | 3.111478 | 3.207123 | 3.242200 | 3.238066 |
| GO:0001773 | myeloid dendritic cell activation | 1/8 | 35/20870 | 0.013340125 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.504789 | 6.073282 | 4.531929 | 6.108157 | 4.284952 | 4.079282 | 6.225128 | 6.137683 | 4.597126 | 4.656392 | 4.223932 | 6.019614 | 6.064666 | 6.133290 | 5.169470 | 4.080871 | 4.043536 | 6.024227 | 6.109857 | 6.185862 | 4.115883 | 4.507446 | 4.201558 | 3.977364 | 3.977021 | 4.264051 | 6.214058 | 6.210866 | 6.250129 | 6.205931 | 6.010636 | 6.188503 |
| GO:0007616 | long-term memory | 1/8 | 42/20870 | 0.015989383 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 3.517536 | 3.761974 | 3.896904 | 3.812534 | 3.503503 | 3.931839 | 3.793747 | 3.769498 | 3.473563 | 3.497456 | 3.579440 | 3.687618 | 3.840743 | 3.753463 | 3.952191 | 3.911134 | 3.824455 | 3.828564 | 3.778341 | 3.830099 | 3.506333 | 3.377636 | 3.616648 | 3.787640 | 3.897735 | 4.093379 | 3.817193 | 3.819479 | 3.743274 | 3.745366 | 3.813303 | 3.748805 |
| GO:0033077 | T cell differentiation in thymus | 1/8 | 80/20870 | 0.030262747 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 6.276858 | 7.249687 | 6.132251 | 7.157404 | 6.154380 | 5.892699 | 7.188447 | 7.214883 | 6.343681 | 6.417722 | 6.042437 | 7.395767 | 7.172254 | 7.169138 | 6.355957 | 5.728399 | 6.238510 | 7.149985 | 7.132206 | 7.189425 | 6.039300 | 6.346179 | 6.056619 | 6.045237 | 5.979642 | 5.617539 | 7.155625 | 7.195253 | 7.213852 | 7.161061 | 7.235149 | 7.246945 |
| GO:0097028 | dendritic cell differentiation | 1/8 | 81/20870 | 0.030635903 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.378948 | 5.625933 | 4.602909 | 5.600736 | 4.284212 | 4.325148 | 5.707809 | 5.608541 | 4.416960 | 4.439533 | 4.274891 | 5.588268 | 5.631488 | 5.657205 | 4.922449 | 4.457849 | 4.364578 | 5.540876 | 5.604222 | 5.654854 | 4.168306 | 4.364300 | 4.312944 | 4.179045 | 4.227092 | 4.541784 | 5.713055 | 5.693753 | 5.716515 | 5.660403 | 5.512757 | 5.647868 |
| GO:0045670 | regulation of osteoclast differentiation | 1/8 | 85/20870 | 0.032127267 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.114814 | 4.760388 | 4.486898 | 4.857521 | 4.121294 | 4.416532 | 4.731299 | 4.793229 | 4.082689 | 4.153526 | 4.107327 | 4.784444 | 4.780892 | 4.714766 | 4.576700 | 4.431303 | 4.448267 | 4.843021 | 4.860648 | 4.868773 | 4.074536 | 4.095340 | 4.191298 | 4.266996 | 4.391001 | 4.574870 | 4.739338 | 4.739285 | 4.715140 | 4.794480 | 4.796606 | 4.788590 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1/8 | 109/20870 | 0.041033369 | 0.1628826 | 0.1357355 | FAM53B | 1 | 6.387130 | 6.359229 | 6.469344 | 6.290553 | 6.309753 | 6.022660 | 6.401387 | 6.307559 | 6.536823 | 6.532801 | 6.036833 | 6.285090 | 6.371577 | 6.417885 | 6.395717 | 6.390286 | 6.610881 | 6.307399 | 6.294958 | 6.269037 | 6.382747 | 6.580005 | 5.879406 | 6.201308 | 5.881207 | 5.966128 | 6.383865 | 6.424037 | 6.395964 | 6.308583 | 6.286382 | 6.327421 |
| GO:0032526 | response to retinoic acid | 1/8 | 124/20870 | 0.046563196 | 0.1628826 | 0.1357355 | OVCA2 | 1 | 4.910250 | 4.860073 | 4.887516 | 4.802467 | 4.951254 | 4.976752 | 4.778985 | 4.766524 | 4.910267 | 4.811037 | 5.003062 | 4.934211 | 4.851942 | 4.790452 | 4.809668 | 4.958506 | 4.890531 | 4.847113 | 4.753136 | 4.805616 | 4.932286 | 4.871438 | 5.044656 | 4.965463 | 5.020847 | 4.942826 | 4.812605 | 4.762040 | 4.761712 | 4.758845 | 4.771370 | 4.769326 |
| GO:0030316 | osteoclast differentiation | 1/8 | 125/20870 | 0.046930857 | 0.1628826 | 0.1357355 | CAMK4 | 1 | 4.719309 | 5.339730 | 4.946655 | 5.364260 | 4.704698 | 4.942838 | 5.284330 | 5.343848 | 4.687351 | 4.723033 | 4.746921 | 5.371132 | 5.353917 | 5.292977 | 4.992772 | 4.929786 | 4.916245 | 5.337837 | 5.368144 | 5.386384 | 4.666828 | 4.672043 | 4.772735 | 4.823850 | 4.944916 | 5.050829 | 5.280508 | 5.288279 | 5.284193 | 5.333542 | 5.370659 | 5.326957 |
GO: MF
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0009931 | calcium-dependent protein serine/threonine kinase activity | 1/6 | 25/20678 | 0.007233068 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.593800 | 4.589005 | 4.653472 | 4.544496 | 4.639457 | 4.864762 | 4.585571 | 4.535268 | 4.546449 | 4.502467 | 4.722930 | 4.579352 | 4.600393 | 4.587191 | 4.575860 | 4.738139 | 4.641786 | 4.557519 | 4.478310 | 4.595201 | 4.655743 | 4.493024 | 4.757372 | 4.813095 | 4.917215 | 4.862094 | 4.622385 | 4.547875 | 4.585492 | 4.531785 | 4.581978 | 4.490587 |
| GO:0010857 | calcium-dependent protein kinase activity | 1/6 | 26/20678 | 0.007521481 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.711194 | 4.645042 | 4.756797 | 4.585098 | 4.763772 | 4.973860 | 4.633759 | 4.569588 | 4.659589 | 4.622028 | 4.842219 | 4.638803 | 4.656172 | 4.640085 | 4.666424 | 4.847374 | 4.750904 | 4.604630 | 4.514284 | 4.633708 | 4.779911 | 4.625435 | 4.875049 | 4.928696 | 5.025169 | 4.966071 | 4.669931 | 4.594683 | 4.635680 | 4.567604 | 4.612929 | 4.526948 |
| GO:0004683 | calmodulin-dependent protein kinase activity | 1/6 | 28/20678 | 0.008098099 | 0.02699366 | 0.01704863 | CAMK4 | 1 | 4.351604 | 4.248910 | 4.630288 | 4.248412 | 4.380313 | 4.629526 | 4.286107 | 4.241967 | 4.306313 | 4.331224 | 4.415015 | 4.265323 | 4.241082 | 4.240185 | 4.639986 | 4.668851 | 4.580629 | 4.267377 | 4.199296 | 4.277318 | 4.405814 | 4.251920 | 4.474301 | 4.516681 | 4.565601 | 4.791194 | 4.353516 | 4.244593 | 4.257745 | 4.232236 | 4.268597 | 4.224686 |
| GO:0008083 | growth factor activity | 1/6 | 164/20678 | 0.046658712 | 0.09587126 | 0.06055027 | CLEC11A | 1 | 5.053083 | 4.838445 | 5.023100 | 4.646887 | 5.114123 | 5.251340 | 4.752778 | 4.588090 | 4.980921 | 4.990709 | 5.178867 | 4.852855 | 4.854022 | 4.807981 | 4.852572 | 5.142550 | 5.058951 | 4.695874 | 4.589556 | 4.653253 | 5.106034 | 5.020466 | 5.209640 | 5.165631 | 5.306991 | 5.277574 | 4.786802 | 4.733128 | 4.737788 | 4.605289 | 4.641440 | 4.514598 |
GO: CC
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:1905103 | integral component of lysosomal membrane | 1/9 | 10/21916 | 0.004099849 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.914375 | 4.411734 | 4.596753 | 4.557231 | 3.975354 | 4.557743 | 4.279654 | 4.312983 | 3.833125 | 3.895123 | 4.009311 | 4.357264 | 4.541132 | 4.327393 | 4.548487 | 4.703044 | 4.532506 | 4.574653 | 4.569866 | 4.526693 | 3.953618 | 3.809192 | 4.143666 | 4.325002 | 4.375671 | 4.899783 | 4.359588 | 4.251794 | 4.223999 | 4.303043 | 4.414828 | 4.214047 |
| GO:0031166 | integral component of vacuolar membrane | 1/9 | 12/21916 | 0.004918023 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.882848 | 4.865354 | 4.509787 | 4.998031 | 3.912317 | 4.437452 | 4.731325 | 4.836717 | 3.823713 | 3.929843 | 3.892983 | 4.840272 | 4.957924 | 4.792822 | 4.593347 | 4.539209 | 4.389128 | 4.965483 | 5.038188 | 4.989468 | 3.876465 | 3.810058 | 4.040571 | 4.226843 | 4.290666 | 4.739117 | 4.782818 | 4.724192 | 4.685291 | 4.828803 | 4.913590 | 4.763842 |
| GO:0031310 | intrinsic component of vacuolar membrane | 1/9 | 12/21916 | 0.004918023 | 0.009836046 | 0.003451244 | SLC46A3 | 1 | 3.882848 | 4.865354 | 4.509787 | 4.998031 | 3.912317 | 4.437452 | 4.731325 | 4.836717 | 3.823713 | 3.929843 | 3.892983 | 4.840272 | 4.957924 | 4.792822 | 4.593347 | 4.539209 | 4.389128 | 4.965483 | 5.038188 | 4.989468 | 3.876465 | 3.810058 | 4.040571 | 4.226843 | 4.290666 | 4.739117 | 4.782818 | 4.724192 | 4.685291 | 4.828803 | 4.913590 | 4.763842 |
| GO:0005811 | lipid droplet | 1/9 | 101/21916 | 0.040727430 | 0.061091145 | 0.021435489 | C18orf32 | 1 | 7.130742 | 6.703458 | 7.082124 | 6.703623 | 7.193841 | 7.322343 | 6.686595 | 6.709812 | 7.050864 | 7.064566 | 7.266533 | 6.705444 | 6.724642 | 6.679940 | 6.912603 | 7.190900 | 7.128335 | 6.743683 | 6.646030 | 6.719375 | 7.206356 | 7.116293 | 7.255437 | 7.353714 | 7.383784 | 7.224626 | 6.686723 | 6.701391 | 6.671515 | 6.706434 | 6.723321 | 6.699579 |
KEGG
| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | HEK293_DMSO_2hA_Log2MeanTPM | HEK293_DMSO_2hB_Log2MeanTPM | HEK293_DMSO_6hA_Log2MeanTPM | HEK293_DMSO_6hB_Log2MeanTPM | HEK293_OSMI2_2hA_Log2MeanTPM | HEK293_OSMI2_6hA_Log2MeanTPM | HEK293_TMG_2hB_Log2MeanTPM | HEK293_TMG_6hB_Log2MeanTPM | log2TPM_HEK293_DMSO_2hA_1 | log2TPM_HEK293_DMSO_2hA_2 | log2TPM_HEK293_DMSO_2hA_3 | log2TPM_HEK293_DMSO_2hB_1 | log2TPM_HEK293_DMSO_2hB_2 | log2TPM_HEK293_DMSO_2hB_3 | log2TPM_HEK293_DMSO_6hA_1 | log2TPM_HEK293_DMSO_6hA_2 | log2TPM_HEK293_DMSO_6hA_3 | log2TPM_HEK293_DMSO_6hB_1 | log2TPM_HEK293_DMSO_6hB_2 | log2TPM_HEK293_DMSO_6hB_3 | log2TPM_HEK293_OSMI2_2hA_1 | log2TPM_HEK293_OSMI2_2hA_2 | log2TPM_HEK293_OSMI2_2hA_3 | log2TPM_HEK293_OSMI2_6hA_1 | log2TPM_HEK293_OSMI2_6hA_2 | log2TPM_HEK293_OSMI2_6hA_3 | log2TPM_HEK293_TMG_2hB_1 | log2TPM_HEK293_TMG_2hB_2 | log2TPM_HEK293_TMG_2hB_3 | log2TPM_HEK293_TMG_6hB_1 | log2TPM_HEK293_TMG_6hB_2 | log2TPM_HEK293_TMG_6hB_3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa05034 | Alcoholism | 2/3 | 187/8146 | 0.001548863 | 0.02633067 | 0.01141267 | CAMK4||H2AC18 | 2 | 6.081292 | 6.312998 | 6.203495 | 6.296332 | 6.035621 | 6.066784 | 6.329420 | 6.327174 | 6.084533 | 6.207338 | 5.939596 | 6.318456 | 6.308019 | 6.312499 | 6.267141 | 6.088087 | 6.248624 | 6.304355 | 6.246149 | 6.337027 | 5.995385 | 6.110528 | 5.997929 | 6.035454 | 6.030588 | 6.132024 | 6.323480 | 6.323340 | 6.341367 | 6.342897 | 6.301839 | 6.336450 |
| hsa04720 | Long-term potentiation | 1/3 | 67/8146 | 0.024475277 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.767229 | 5.888241 | 5.758357 | 6.020890 | 5.756241 | 5.903782 | 5.938052 | 6.090766 | 5.754439 | 5.814736 | 5.731220 | 5.891437 | 5.871799 | 5.901331 | 5.773287 | 5.731539 | 5.769875 | 6.042904 | 5.942405 | 6.074100 | 5.732244 | 5.781166 | 5.754899 | 5.907409 | 5.940431 | 5.862446 | 5.935278 | 5.925941 | 5.952807 | 6.074190 | 6.096181 | 6.101781 |
| hsa05031 | Amphetamine addiction | 1/3 | 69/8146 | 0.025199676 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 6.206956 | 6.485590 | 6.265257 | 6.537937 | 6.198552 | 6.387565 | 6.509016 | 6.578806 | 6.177469 | 6.287388 | 6.152409 | 6.501866 | 6.473855 | 6.480901 | 6.318973 | 6.221454 | 6.253626 | 6.562976 | 6.465062 | 6.583032 | 6.184174 | 6.213202 | 6.198135 | 6.394569 | 6.401446 | 6.366442 | 6.503404 | 6.506514 | 6.517092 | 6.571238 | 6.580342 | 6.584804 |
| hsa05214 | Glioma | 1/3 | 75/8146 | 0.027370723 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.784893 | 5.702299 | 5.749235 | 5.611233 | 5.803506 | 5.948375 | 5.711067 | 5.642193 | 5.720731 | 5.755829 | 5.873635 | 5.717816 | 5.691112 | 5.697836 | 5.698036 | 5.809268 | 5.738194 | 5.651754 | 5.541429 | 5.638030 | 5.823915 | 5.667322 | 5.908956 | 5.890779 | 6.023570 | 5.927497 | 5.733073 | 5.680940 | 5.718688 | 5.629903 | 5.663737 | 5.632695 |
| hsa04211 | Longevity regulating pathway | 1/3 | 89/8146 | 0.032423957 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.517718 | 5.577823 | 5.623100 | 5.684231 | 5.523441 | 5.735019 | 5.599955 | 5.709516 | 5.486466 | 5.543750 | 5.522356 | 5.594729 | 5.577903 | 5.560636 | 5.636783 | 5.619493 | 5.612919 | 5.702531 | 5.616027 | 5.731645 | 5.517424 | 5.503493 | 5.549028 | 5.713241 | 5.744566 | 5.747005 | 5.622772 | 5.578475 | 5.598274 | 5.704510 | 5.718415 | 5.705582 |
| hsa04925 | Aldosterone synthesis and secretion | 1/3 | 98/8146 | 0.035663205 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 5.595871 | 5.806341 | 5.716022 | 5.865852 | 5.578227 | 5.809613 | 5.790656 | 5.877407 | 5.562661 | 5.679469 | 5.541626 | 5.818782 | 5.813534 | 5.786501 | 5.735456 | 5.708640 | 5.703768 | 5.887161 | 5.816726 | 5.892436 | 5.578219 | 5.581662 | 5.574793 | 5.796712 | 5.784120 | 5.847230 | 5.798296 | 5.788333 | 5.785307 | 5.875349 | 5.885009 | 5.871832 |
| hsa04725 | Cholinergic synapse | 1/3 | 113/8146 | 0.041045871 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 4.898453 | 4.818758 | 4.931808 | 4.955285 | 4.915127 | 5.119258 | 4.835006 | 4.983973 | 4.856953 | 4.939996 | 4.897214 | 4.813787 | 4.827704 | 4.814740 | 4.907820 | 4.937064 | 4.950214 | 4.996756 | 4.860379 | 5.004225 | 4.908956 | 4.901372 | 4.934841 | 5.107619 | 5.121924 | 5.128154 | 4.852895 | 4.831052 | 4.820886 | 4.979196 | 4.984557 | 4.988151 |
| hsa04722 | Neurotrophin signaling pathway | 1/3 | 119/8146 | 0.043193317 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 6.016337 | 5.952352 | 5.955489 | 6.002791 | 5.993966 | 6.095876 | 5.974173 | 6.041013 | 6.008955 | 6.067897 | 5.970486 | 5.962448 | 5.944408 | 5.950142 | 5.980340 | 5.913952 | 5.971281 | 6.025701 | 5.926940 | 6.052716 | 5.992483 | 6.006068 | 5.983256 | 6.090956 | 6.135859 | 6.059797 | 5.987265 | 5.956042 | 5.979029 | 6.035523 | 6.045466 | 6.042033 |
| hsa04380 | Osteoclast differentiation | 1/3 | 128/8146 | 0.046408471 | 0.07509782 | 0.03255014 | CAMK4 | 1 | 4.481272 | 4.694176 | 4.670136 | 4.654232 | 4.494373 | 4.763273 | 4.679935 | 4.644717 | 4.441907 | 4.459265 | 4.540704 | 4.720090 | 4.699747 | 4.662092 | 4.690251 | 4.713625 | 4.604248 | 4.669373 | 4.607502 | 4.684669 | 4.501143 | 4.375176 | 4.598170 | 4.652158 | 4.754349 | 4.874698 | 4.713816 | 4.655560 | 4.669787 | 4.621031 | 4.658232 | 4.654599 |
| hsa05322 | Systemic lupus erythematosus | 1/3 | 136/8146 | 0.049260331 | 0.07509782 | 0.03255014 | H2AC18 | 1 | 6.321517 | 6.470763 | 6.451806 | 6.480259 | 6.267784 | 6.143901 | 6.478292 | 6.522252 | 6.354829 | 6.462215 | 6.127557 | 6.504658 | 6.449156 | 6.457854 | 6.551703 | 6.278809 | 6.510198 | 6.480561 | 6.446902 | 6.512567 | 6.198438 | 6.388119 | 6.208804 | 6.083071 | 6.119473 | 6.225346 | 6.476118 | 6.469098 | 6.489583 | 6.546568 | 6.483946 | 6.535471 |
ORA: Up-regulated DEGs
Please Click HERE to download a Microsoft .excel that contains all of the “ORT: Up-regulated DEGs” results.
GO: BP
No significcant functional terms were enriched under the threshold of P<0.05
GO: MF
No significcant functional terms were enriched under the threshold of P<0.05
GO: CC
No significcant functional terms were enriched under the threshold of P<0.05
KEGG
No significcant functional terms were enriched under the threshold of P<0.05
Help
still working on
2-2. Summary Plot
GSEA
GO: BP
GO: MF
GO: CC
KEGG
ORA: All DEGs
GO: BP
GO: MF
GO: CC
KEGG
ORA: Down-regulated DEGs
GO: BP
GO: MF
GO: CC
KEGG
ORA: Up-regulated DEGs
GO: BP
No results under this category
GO: MF
No results under this category
GO: CC
No results under this category
KEGG
No results under this category
ORA: Overlapped Terms
GO: BP
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:0033081||GO:0043011||GO:0001773||GO:0007616||GO:0033077||GO:0097028||GO:0045670||GO:0090263||GO:0032526||GO:0030316 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: MF
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:0009931||GO:0010857||GO:0004683||GO:0008083 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
GO: CC
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | GO:1905103||GO:0031166||GO:0031310||GO:0005811 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
KEGG
Summary Plot
Venn Plot
List
| Intersection Combination | Overlapped Functional Terms |
|---|---|
| Up-Regulated ∪ Down-Regulated | |
| All ∪ Down-Regulated | hsa05034||hsa04720||hsa05031||hsa05214||hsa04211||hsa04925||hsa04725||hsa04722||hsa04380||hsa05322 |
| All ∪ Up-Regulated | |
| All ∪ Down-Regulated ∪ Up-Regulated |
Help
still working on
3. Isoform Switch Analysis
Alternative Splicing Events
Amount of Event
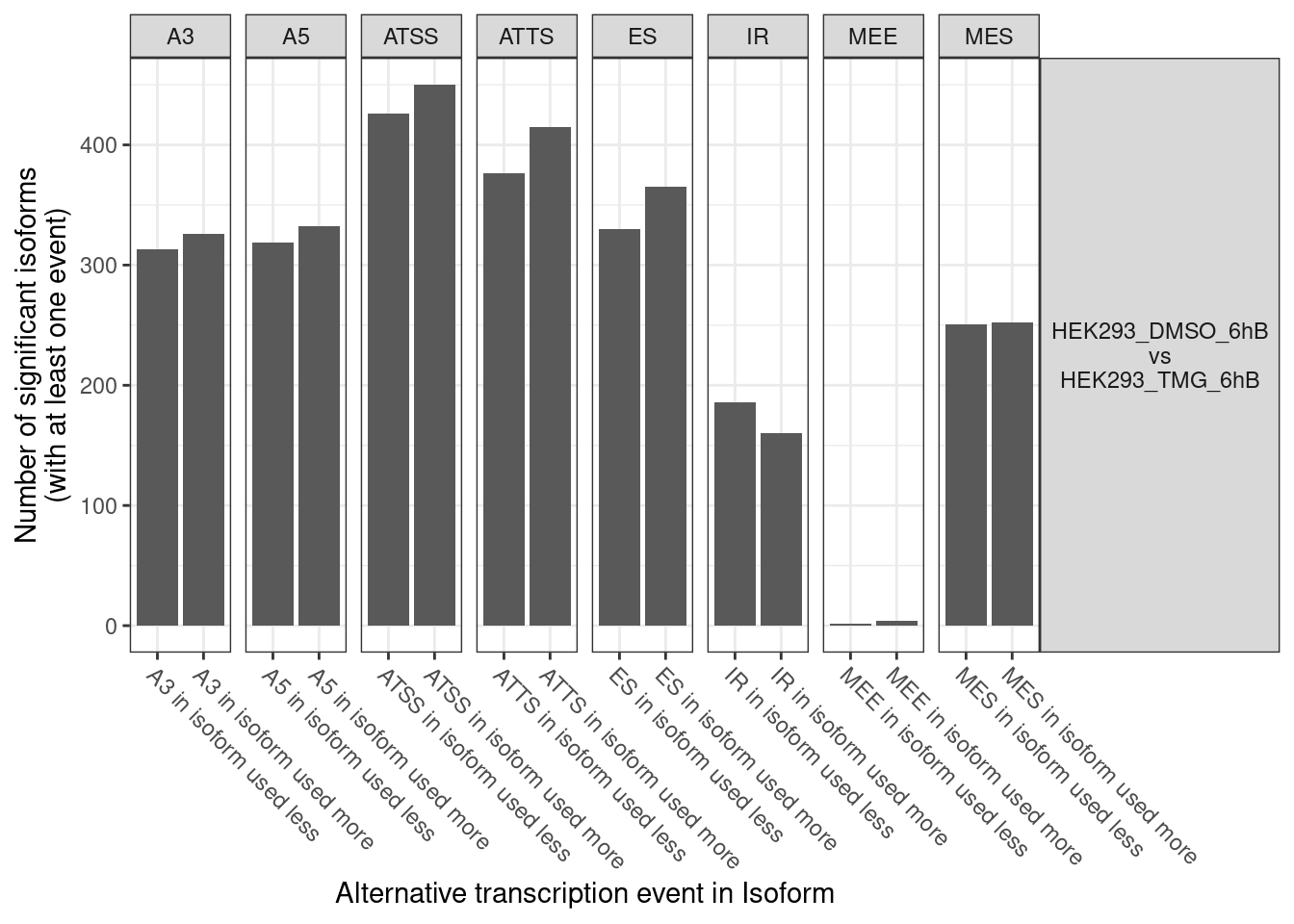
Enrichment Analysis
This below result summarizes the uneven usage within each comparison by for each alternative splicing type calculate the fraction of events being gains (as opposed to loss) and perform a statistical analysis of this fraction.
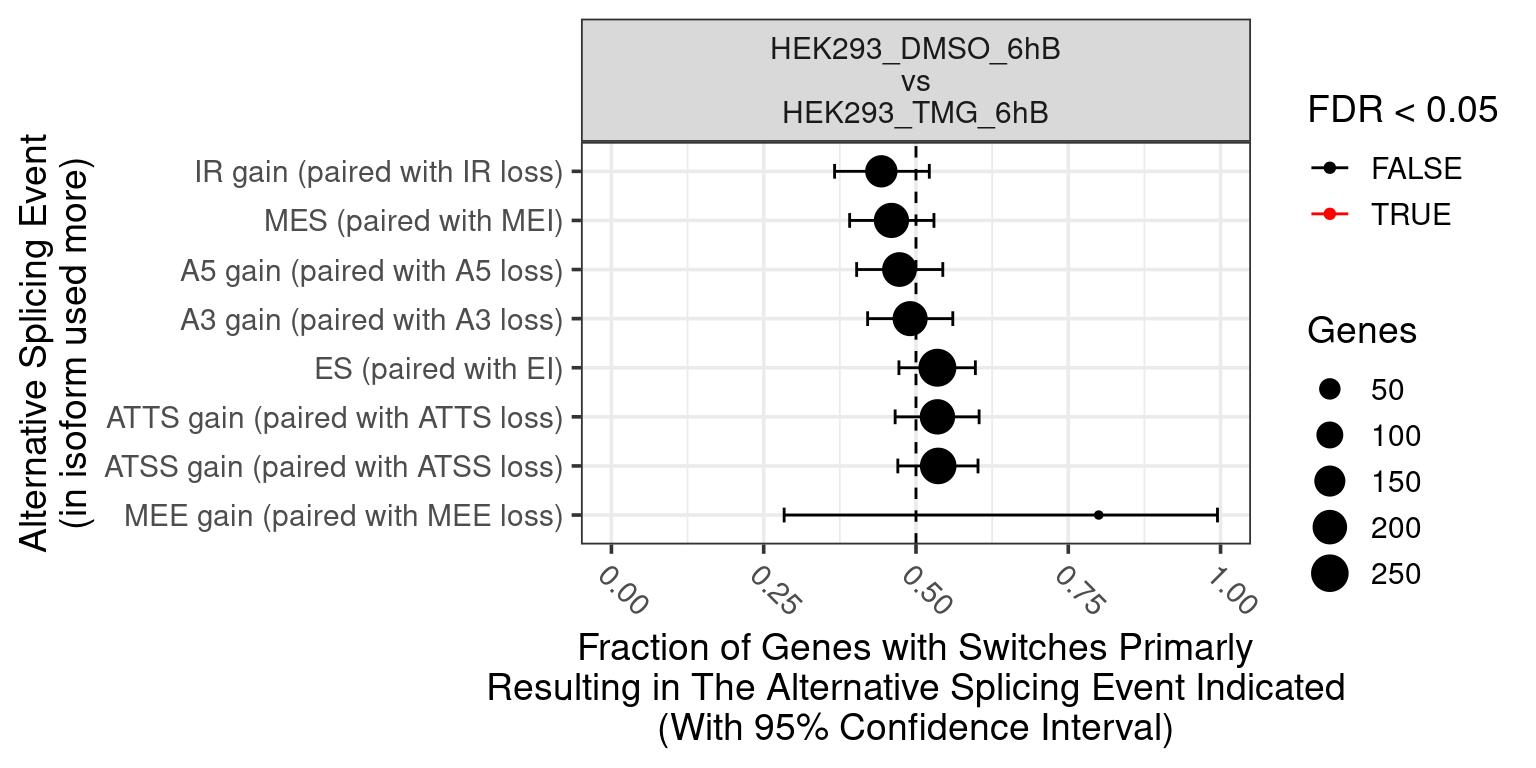
Comparison Analysis
This below result answered the question: How does the isoform usage of all isoforms utilizing a particular splicing type change - in other words is all isoforms or only a subset of isoforms that are affected.
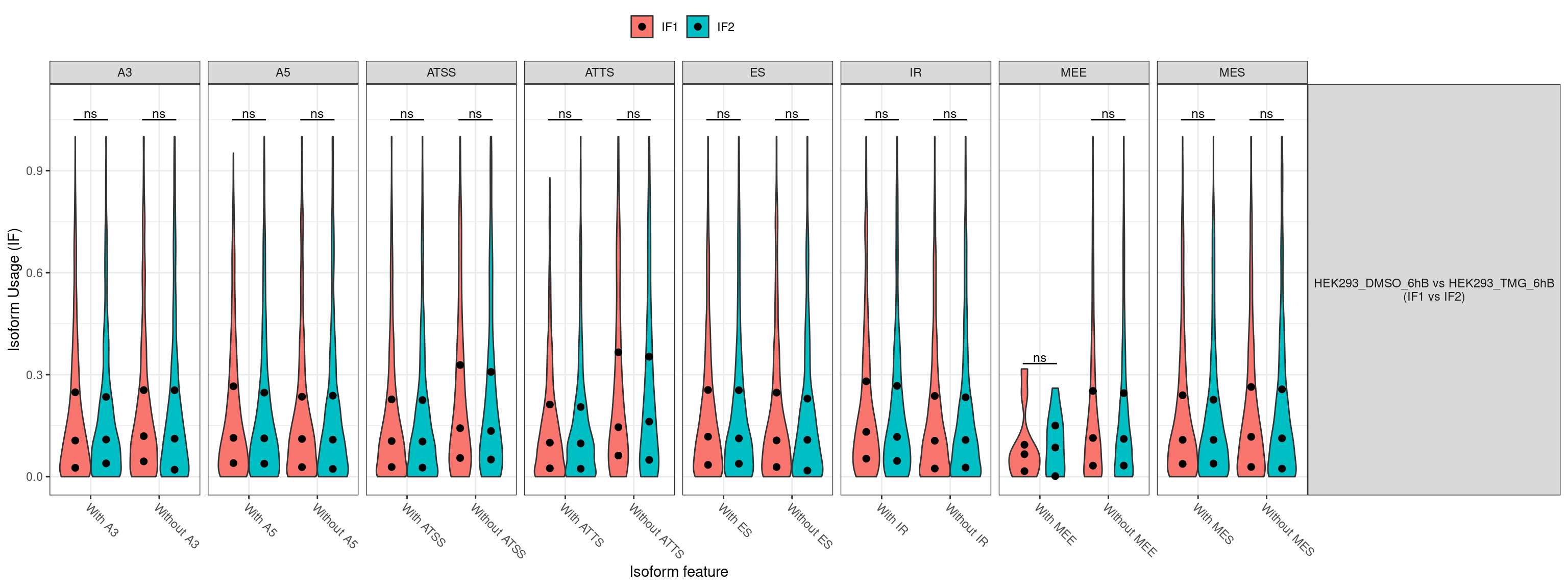
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- IR : Intron Retention.
- A5 : Alternative 5’ donor site (changes in the 5’end of the upstream exon).
- A3 : Alternative 3’ acceptor site (changes in the 3’end of the downstream exon).
- ATSS : Alternative Transcription Start Site.
- ATTS : Alternative Transcription Termination Site.
- ES : Exon Skipping (EI means Exon Inclusion).
- MES : Multiple Exon Skipping. Skipping of >1 consecutive exons. (MEI means Multiple Exon Inclusion).
- MEE : Mutually Exclusive Exons.
Alternative Splicing Consequence
Amount of Event
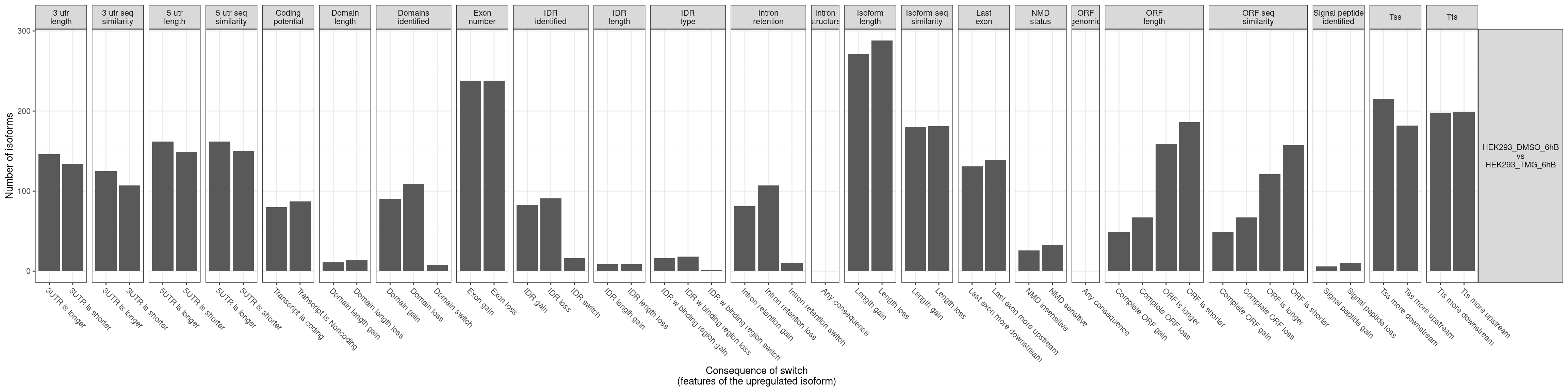
Enrichment Analysis
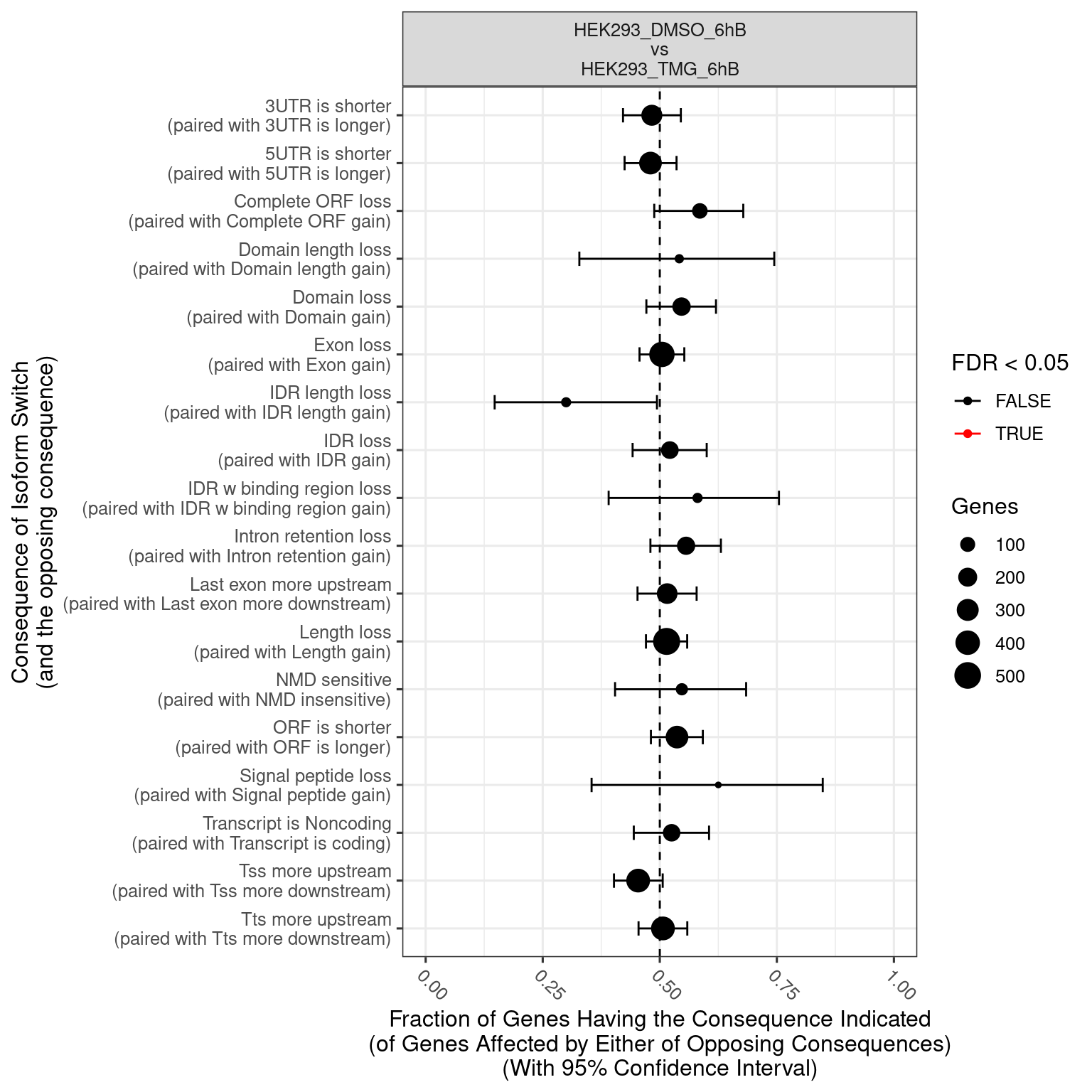
Comparison Analysis
- Please note that:
- The the dots in the violin plots above indicate 25th, 50th (median) and 75th percentiles (just like the box in a boxplot would).
- The analysis provided here should only be expected to give a result when splicing is (severely) affected.
Abbreviations
- Abbreviations:
- tss : Test transcripts for whether they use different Transcription Start Site (TSS) (more than ntCutoff from each other).
- tts : Test transcripts for whether they use different Transcription Termination Site (TTS) (more than ntCutoff from each other).
- last_exon : Test whether transcripts utilizes different last exons (defined as the last exon of each transcript is non-overlapping).
- isoform_seq_similarity : Test whether the isoform nucleotide sequences are different (as described above). Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff.
- isoform_length : Test transcripts for differences in isoform length. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Please * note that this is a less powerful analysis than implemented in ‘isoform_seq_similarity’ as two equally long sequences might be very different.
- exon_number : Test transcripts for differences in exon number.
- intron_structure : Test transcripts for differences in intron structure, e.g. usage of exon-exon junctions. This analysis corresponds to analyzing whether all introns in one isoform is also found in the other isoforms.
- intron_retention : Test for differences in intron retentions (and their genomic positions). Require that analyzeIntronRetention have been run.
- isoform_class_code : Test transcripts for differences in the transcript classification provide by cufflinks. For a updated list of class codes see http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/#transfrag-class-codes.
- coding_potential : Test transcripts for differences in coding potential, as indicated by the CPAT or CPC2 analysis. Requires that analyzeCPAT or analyzeCPC2 have been used to add external conding potential analysis to the switchAnalyzeRlist.
- ORF_seq_similarity : Test whether the amino acid sequences of the ORFs are different (as described above). Reported as different if the measured JCsim is smaller than AaJCsimCutoff and the length difference of the aligned and combined region is larger than AaCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_genomic : Test transcripts for differences in genomic position of the Open Reading Frames (ORF). Requires that least one of the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and settingaddAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- ORF_length : Test transcripts for differences in length of Open Reading Frames (ORF). Note that this is a less powerful analysis than implemented in ORF_seq_similarity as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that least one of the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 5’ UnTranslated Region (UTR) are different (as described above). The 5’UTR is defined as the region from the transcript start to the ORF start. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with an ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 5_utr_length : Test transcripts for differences in the length of the 5’ UnTranslated Region (UTR). The 5’UTR is defined as the region from the transcript start to the ORF start. Note that this is a less powerful analysis than implemented in ‘5_utr_seq_similarity’ as two equally long sequences might be very different. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_seq_similarity : Test whether the isoform nucleotide sequences of the 3’ UnTranslated Region (UTR) are different (as described above). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Reported as different if the measured JCsim is smaller than ntJCsimCutoff and the length difference of the aligned and combined region is larger than ntCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- 3_utr_length : Test transcripts for differences in the length of the 3’ UnTranslated Regions (UTR). The 3’UTR is defined as the region from the end of the ORF to the transcript end. Note that this is a less powerful analysis than implemented in 3_utr_seq_similarity as two equally long sequences might be very different. Requires that identifyORF have been used to predict NMD sensitivity or that the ORF was imported though one of the dedicated import functions implemented in isoformSwitchAnalyzeR. Only reported if the difference is larger than indicated by the ntCutoff and ntFracCutoff. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- NMD_status : Test transcripts for differences in sensitivity to Nonsense Mediated Decay (NMD). Requires that both the isoforms have been annotated with PTC either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domains_identified : Test transcripts for differences in the name and order of which domains are identified by the Pfam in the transcripts. Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- domain_length : Test transcripts for differences in the length of overlapping domains of the same type (same hmm_name) thereby enabling analysis of protein domain truncation. Do however note that a small difference in length is will likely not truncate the protein domain. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff .Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist.
- genomic_domain_position : Test transcripts for differences in the genomic position of the domains identified by the Pfam analysis (Will be different unless the two isoforms have the same domains at the same genomic location). Requires that analyzePFAM have been used to add external Pfam analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via identifyORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
- IDR_identified : Test for differences in isoform IDRs. Specifically the two isoforms are analyzed for whether they contain IDRs which do not overlap in genomic coordinates. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_length : Test for differences in the length of overlapping (in genomic coordinates) IDRs. The length difference, measured in AA, must be larger than AaCutoff and AaFracCutoff. Requires that analyzeNetSurfP2 or analyzeIUPred2A have been used to add external IDR analysis to the switchAnalyzeRlist.
- IDR_type : Test for differences in IDR type. Specifically the two isoforms are tested for overlapping IDRs (genomic coordinates) and overlapping IDRs are compared with regards to their IDR type (IDR vs IDR w binding site). Only available if analyzeIUPred2A was used to add external IDR analysis to the switchAnalyzeRlist.
- signal_peptide_identified : Test transcripts for differences in whether a signal peptide was identified or not by the SignalP analysis. Requires that analyzeSignalP have been used to add external SignalP analysis to the switchAnalyzeRlist. Requires that both the isoforms are annotated with a ORF either via analyzeORF or by supplying a GTF file and setting addAnnotatedORFs=TRUE when creating the switchAnalyzeRlist (and are thereby also affected by removeNoncodinORFs=TRUE in analyzeCPAT).
Volcano Plot
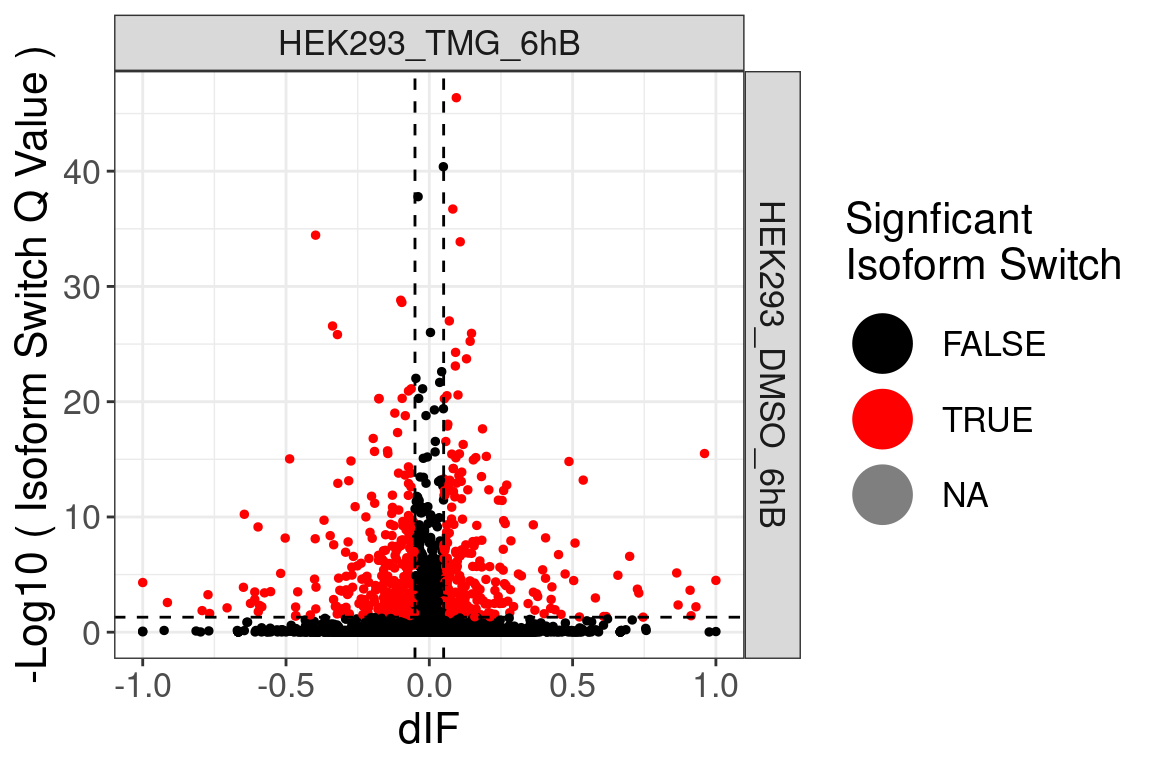
Spreadsheet
| iso_ref | gene_ref | isoform_id | gene_id | gene_name | condition_1 | condition_2 | IF1 | IF2 | dIF | isoform_switch_q_value | switchConsequencesGene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| isoComp_00752223 | geneComp_00148580 | ENST00000451590 | ENSG00000174775 | HRAS | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.094 | 0.094 | 4.232523e-47 | TRUE |
| isoComp_00778562 | geneComp_00152572 | ENST00000444811 | ENSG00000204356 | NELFE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.082 | 0.082 | 1.933401e-37 | TRUE |
| isoComp_00713875 | geneComp_00143930 | ENST00000373719 | ENSG00000147162 | OGT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.655 | 0.258 | -0.397 | 3.571236e-35 | TRUE |
| isoComp_00643272 | geneComp_00136600 | ENST00000621783 | ENSG00000085719 | CPNE3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.108 | 0.108 | 1.337639e-34 | TRUE |
| isoComp_00701627 | geneComp_00142658 | ENST00000417938 | ENSG00000138398 | PPIG | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.100 | 0.000 | -0.100 | 1.571826e-29 | TRUE |
| isoComp_00643274 | geneComp_00136600 | MSTRG.31771.2 | ENSG00000085719 | CPNE3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.096 | 0.000 | -0.096 | 2.454062e-29 | TRUE |
| isoComp_00747065 | geneComp_00147892 | ENST00000563078 | ENSG00000171208 | NETO2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.070 | 0.070 | 9.846086e-28 | |
| isoComp_00646091 | geneComp_00136844 | MSTRG.30107.3 | ENSG00000091073 | DTX2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.338 | 0.000 | -0.338 | 2.706882e-27 | TRUE |
| isoComp_00763933 | geneComp_00150363 | ENST00000541152 | ENSG00000184640 | SEPTIN9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.147 | 0.147 | 1.177852e-26 | TRUE |
| isoComp_00692909 | geneComp_00141737 | ENST00000410396 | ENSG00000133316 | WDR74 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.321 | 0.000 | -0.321 | 1.512071e-26 | TRUE |
| isoComp_00776467 | geneComp_00152180 | MSTRG.9810.5 | ENSG00000198732 | SMOC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.143 | 0.143 | 5.747155e-26 | TRUE |
| isoComp_00713508 | geneComp_00143891 | MSTRG.14865.27 | ENSG00000146872 | TLK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.092 | 0.092 | 5.267611e-25 | TRUE |
| isoComp_00665949 | geneComp_00138800 | ENST00000392474 | ENSG00000110931 | CAMKK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.130 | 0.130 | 1.908755e-24 | TRUE |
| isoComp_00784832 | geneComp_00154295 | ENST00000425676 | ENSG00000221829 | FANCG | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.091 | 0.091 | 8.031710e-24 | TRUE |
| isoComp_00723811 | geneComp_00145080 | MSTRG.23134.6 | ENSG00000157500 | APPL1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.063 | 0.000 | -0.063 | 7.605374e-22 | |
| isoComp_00653778 | geneComp_00137599 | MSTRG.34317.11 | ENSG00000102316 | MAGED2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.073 | 0.000 | -0.073 | 1.187268e-21 | |
| isoComp_00661667 | geneComp_00138402 | MSTRG.3948.8 | ENSG00000107643 | MAPK8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.100 | 0.100 | 2.620428e-21 | TRUE |
| isoComp_00651519 | geneComp_00137344 | MSTRG.9242.20 | ENSG00000100897 | DCAF11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.062 | 0.062 | 3.037095e-21 | TRUE |
| isoComp_00643984 | geneComp_00136662 | MSTRG.27371.5 | ENSG00000087206 | UIMC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.095 | 0.000 | -0.095 | 5.175447e-21 | TRUE |
| isoComp_00777422 | geneComp_00152365 | ENST00000675797 | ENSG00000203485 | INF2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.176 | 0.000 | -0.176 | 5.238024e-21 | TRUE |
| isoComp_00708620 | geneComp_00143351 | ENST00000479344 | ENSG00000143258 | USP21 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.053 | 0.053 | 5.504781e-21 | |
| isoComp_00736772 | geneComp_00146642 | ENST00000403829 | ENSG00000165795 | NDRG2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.176 | 0.000 | -0.176 | 5.693077e-21 | |
| isoComp_00738323 | geneComp_00146823 | ENST00000562702 | ENSG00000166503 | HDGFL3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.120 | 0.000 | -0.120 | 9.823164e-20 | TRUE |
| isoComp_00815709 | geneComp_00167621 | MSTRG.33771.13 | ENSG00000271254 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.084 | 0.000 | -0.084 | 1.642571e-19 | TRUE | |
| isoComp_00726891 | geneComp_00145433 | ENST00000479117 | ENSG00000160216 | AGPAT3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.065 | 0.065 | 8.961391e-19 | TRUE |
| isoComp_00686988 | geneComp_00141120 | ENST00000395386 | ENSG00000129292 | PHF20L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.063 | 0.063 | 1.315489e-18 | TRUE |
| isoComp_00786563 | geneComp_00154904 | ENST00000577158 | ENSG00000224877 | NDUFAF8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.186 | 0.186 | 2.276967e-18 | TRUE |
| isoComp_00627336 | geneComp_00135182 | MSTRG.28818.11 | ENSG00000010810 | FYN | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.111 | 0.000 | -0.111 | 4.779783e-18 | TRUE |
| isoComp_00657836 | geneComp_00137983 | ENST00000593194 | ENSG00000105048 | TNNT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.196 | 0.000 | -0.196 | 1.549181e-17 | TRUE |
| isoComp_00700507 | geneComp_00142538 | ENST00000565999 | ENSG00000137817 | PARP6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.058 | 0.058 | 2.833540e-17 | TRUE |
| isoComp_00674567 | geneComp_00139678 | ENST00000497866 | ENSG00000117411 | B4GALT2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.118 | 0.118 | 5.149631e-17 | TRUE |
| isoComp_00776466 | geneComp_00152180 | MSTRG.9810.2 | ENSG00000198732 | SMOC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.146 | 0.000 | -0.146 | 1.836070e-16 | TRUE |
| isoComp_00722901 | geneComp_00144984 | ENST00000649006 | ENSG00000156650 | KAT6B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.191 | 0.000 | -0.191 | 2.075936e-16 | TRUE |
| isoComp_00746018 | geneComp_00147741 | ENST00000397042 | ENSG00000170471 | RALGAPB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.145 | 0.000 | -0.145 | 3.147911e-16 | TRUE |
| isoComp_00819473 | geneComp_00169761 | MSTRG.21052.10 | ENSG00000278878 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.960 | 0.960 | 3.147911e-16 | TRUE | |
| isoComp_00772265 | geneComp_00151631 | ENST00000696006 | ENSG00000196839 | ADA | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.105 | 0.105 | 3.354946e-16 | TRUE |
| isoComp_00694405 | geneComp_00141895 | MSTRG.3493.6 | ENSG00000134452 | FBH1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.078 | 0.078 | 3.526992e-16 | |
| isoComp_00781180 | geneComp_00153112 | ENST00000592461 | ENSG00000213015 | ZNF580 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.199 | 0.199 | 5.573995e-16 | TRUE |
| isoComp_00659608 | geneComp_00138171 | ENST00000497853 | ENSG00000105875 | WDR91 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.162 | 0.162 | 7.077072e-16 | TRUE |
| isoComp_00739118 | geneComp_00146929 | ENST00000562314 | ENSG00000166938 | DIS3L | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.093 | 0.093 | 7.392943e-16 | TRUE |
| isoComp_00723408 | geneComp_00145035 | ENST00000454765 | ENSG00000157111 | TMEM171 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.513 | -0.487 | 9.181507e-16 | TRUE |
| isoComp_00669412 | geneComp_00139183 | ENST00000692557 | ENSG00000113719 | ERGIC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.154 | 0.154 | 1.123409e-15 | TRUE |
| isoComp_00764338 | geneComp_00150436 | ENST00000611213 | ENSG00000185000 | DGAT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.273 | 0.000 | -0.273 | 1.394740e-15 | TRUE |
| isoComp_00723407 | geneComp_00145035 | ENST00000287773 | ENSG00000157111 | TMEM171 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.487 | 0.487 | 1.545397e-15 | TRUE |
| isoComp_00696687 | geneComp_00142126 | ENST00000539479 | ENSG00000135679 | MDM2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.073 | 0.000 | -0.073 | 4.520050e-15 | TRUE |
| isoComp_00750821 | geneComp_00148387 | MSTRG.269.3 | ENSG00000173614 | NMNAT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.084 | 0.084 | 6.176418e-15 | TRUE |
| isoComp_00689212 | geneComp_00141351 | ENST00000393529 | ENSG00000130830 | MPP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.084 | 0.084 | 6.366678e-15 | TRUE |
| isoComp_00700332 | geneComp_00142519 | ENST00000530933 | ENSG00000137760 | ALKBH8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.113 | 0.113 | 1.285870e-14 | TRUE |
| isoComp_00674563 | geneComp_00139678 | ENST00000372324 | ENSG00000117411 | B4GALT2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.107 | 0.000 | -0.107 | 1.579967e-14 | TRUE |
| isoComp_00744317 | geneComp_00147518 | ENST00000523539 | ENSG00000169398 | PTK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.066 | 0.000 | -0.066 | 1.579967e-14 | TRUE |
| isoComp_00672257 | geneComp_00139439 | MSTRG.20179.7 | ENSG00000115687 | PASK | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.085 | 0.000 | -0.085 | 2.351039e-14 | |
| isoComp_00680712 | geneComp_00140381 | MSTRG.28688.31 | ENSG00000123552 | USP45 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.103 | 0.103 | 2.506669e-14 | TRUE |
| isoComp_00739942 | geneComp_00147024 | ENST00000539742 | ENSG00000167377 | ZNF23 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.182 | 0.182 | 3.133994e-14 | TRUE |
| isoComp_00709877 | geneComp_00143476 | ENST00000521431 | ENSG00000143801 | PSEN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.050 | 0.050 | 5.012553e-14 | |
| isoComp_00663873 | geneComp_00138608 | ENST00000674651 | ENSG00000109099 | PMP22 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.537 | 0.537 | 6.366889e-14 | TRUE |
| isoComp_00656313 | geneComp_00137835 | ENST00000678204 | ENSG00000104218 | CSPP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.071 | 0.071 | 6.550324e-14 | TRUE |
| isoComp_00722216 | geneComp_00144902 | ENST00000542309 | ENSG00000155974 | GRIP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.281 | 0.000 | -0.281 | 7.256868e-14 | TRUE |
| isoComp_00655408 | geneComp_00137750 | ENST00000563366 | ENSG00000103353 | UBFD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.111 | 0.111 | 8.158064e-14 | TRUE |
| isoComp_00735811 | geneComp_00146513 | ENST00000453718 | ENSG00000165238 | WNK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.095 | 0.095 | 1.166424e-13 | TRUE |
| isoComp_00803692 | geneComp_00161703 | ENST00000670223 | ENSG00000249859 | PVT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.319 | 0.000 | -0.319 | 1.211868e-13 | TRUE |
| isoComp_00683239 | geneComp_00140686 | MSTRG.17344.10 | ENSG00000125755 | SYMPK | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.073 | 0.000 | -0.073 | 1.255360e-13 | |
| isoComp_00633483 | geneComp_00135749 | ENST00000548938 | ENSG00000061273 | HDAC7 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.270 | 0.270 | 1.703616e-13 | TRUE |
| isoComp_00639794 | geneComp_00136287 | ENST00000675645 | ENSG00000076685 | NT5C2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.068 | 0.068 | 2.028117e-13 | |
| isoComp_00625431 | geneComp_00135020 | MSTRG.8919.14 | ENSG00000005810 | MYCBP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.064 | 0.000 | -0.064 | 2.444451e-13 | |
| isoComp_00745268 | geneComp_00147632 | ENST00000570940 | ENSG00000169992 | NLGN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.134 | 0.134 | 4.438673e-13 | TRUE |
| isoComp_00814796 | geneComp_00167119 | MSTRG.27872.12 | ENSG00000269293 | ZSCAN16-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.208 | 0.208 | 4.438673e-13 | TRUE |
| isoComp_00713886 | geneComp_00143930 | MSTRG.34438.4 | ENSG00000147162 | OGT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.133 | 0.392 | 0.259 | 5.113612e-13 | TRUE |
| isoComp_00702873 | geneComp_00142771 | ENST00000671733 | ENSG00000139163 | ETNK1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.053 | 0.053 | 5.362333e-13 | |
| isoComp_00662927 | geneComp_00138507 | ENST00000581006 | ENSG00000108474 | PIGL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.084 | 0.084 | 6.224854e-13 | TRUE |
| isoComp_00729525 | geneComp_00145693 | ENST00000528123 | ENSG00000162300 | ZFPL1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.058 | 0.058 | 6.623845e-13 | TRUE |
| isoComp_00679251 | geneComp_00140231 | ENST00000575041 | ENSG00000122299 | ZC3H7A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.055 | 0.055 | 1.242500e-12 | TRUE |
| isoComp_00739938 | geneComp_00147024 | ENST00000358700 | ENSG00000167377 | ZNF23 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.129 | 0.000 | -0.129 | 1.294984e-12 | TRUE |
| isoComp_00752221 | geneComp_00148580 | ENST00000397596 | ENSG00000174775 | HRAS | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.099 | 0.026 | -0.073 | 1.337986e-12 | TRUE |
| isoComp_00638987 | geneComp_00136225 | ENST00000422713 | ENSG00000075234 | TTC38 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.051 | 0.051 | 1.379776e-12 | TRUE |
| isoComp_00626004 | geneComp_00135071 | MSTRG.33879.6 | ENSG00000006757 | PNPLA4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.202 | 0.000 | -0.202 | 1.601929e-12 | TRUE |
| isoComp_00762338 | geneComp_00150109 | MSTRG.8330.11 | ENSG00000183495 | EP400 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.089 | 0.089 | 1.805076e-12 | TRUE |
| isoComp_00683710 | geneComp_00140752 | ENST00000446710 | ENSG00000126001 | CEP250 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.112 | 0.112 | 2.675870e-12 | TRUE |
| isoComp_00684930 | geneComp_00140884 | ENST00000319833 | ENSG00000127334 | DYRK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.241 | 0.241 | 3.504170e-12 | TRUE |
| isoComp_00639766 | geneComp_00136285 | ENST00000592945 | ENSG00000076662 | ICAM3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.746 | 1.000 | 0.254 | 3.715256e-12 | TRUE |
| isoComp_00740844 | geneComp_00147114 | MSTRG.13365.8 | ENSG00000167703 | SLC43A2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.191 | 0.000 | -0.191 | 6.498543e-12 | TRUE |
| isoComp_00743005 | geneComp_00147345 | ENST00000601142 | ENSG00000168661 | ZNF30 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.259 | 0.000 | -0.259 | 1.281167e-11 | TRUE |
| isoComp_00756859 | geneComp_00149238 | ENST00000479050 | ENSG00000178537 | SLC25A20 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.078 | 0.078 | 1.424777e-11 | |
| isoComp_00675783 | geneComp_00139826 | ENST00000686381 | ENSG00000118873 | RAB3GAP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.128 | 0.000 | -0.128 | 1.475050e-11 | TRUE |
| isoComp_00721154 | geneComp_00144772 | MSTRG.29967.29 | ENSG00000154710 | RABGEF1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.107 | 0.000 | -0.107 | 2.515984e-11 | TRUE |
| isoComp_00639841 | geneComp_00136291 | MSTRG.16311.4 | ENSG00000076826 | CAMSAP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.131 | 0.000 | -0.131 | 5.033018e-11 | TRUE |
| isoComp_00789391 | geneComp_00156045 | ENST00000437064 | ENSG00000228109 | MELTF-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.645 | 0.000 | -0.645 | 5.881332e-11 | TRUE |
| isoComp_00725734 | geneComp_00145297 | MSTRG.14614.6 | ENSG00000159217 | IGF2BP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.073 | 0.000 | -0.073 | 7.536034e-11 | TRUE |
| isoComp_00660508 | geneComp_00138271 | ENST00000470583 | ENSG00000106415 | GLCCI1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.221 | 0.000 | -0.221 | 9.971317e-11 | TRUE |
| isoComp_00691852 | geneComp_00141617 | ENST00000562883 | ENSG00000132613 | MTSS2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.077 | 0.077 | 1.517961e-10 | |
| isoComp_00639020 | geneComp_00136227 | ENST00000490378 | ENSG00000075240 | GRAMD4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.116 | 0.116 | 1.562299e-10 | TRUE |
| isoComp_00634594 | geneComp_00135846 | ENST00000479669 | ENSG00000065328 | MCM10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.064 | 0.000 | -0.064 | 1.718476e-10 | TRUE |
| isoComp_00731136 | geneComp_00145941 | ENST00000680913 | ENSG00000163297 | ANTXR2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.367 | 0.000 | -0.367 | 1.932200e-10 | TRUE |
| isoComp_00813175 | geneComp_00166375 | ENST00000580190 | ENSG00000266865 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.259 | 0.259 | 2.084046e-10 | TRUE | |
| isoComp_00680711 | geneComp_00140381 | MSTRG.28688.29 | ENSG00000123552 | USP45 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.092 | 0.000 | -0.092 | 2.464467e-10 | TRUE |
| isoComp_00762984 | geneComp_00150219 | ENST00000461103 | ENSG00000183955 | KMT5A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.070 | 0.000 | -0.070 | 2.470527e-10 | TRUE |
| isoComp_00685139 | geneComp_00140905 | ENST00000592031 | ENSG00000127527 | EPS15L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.055 | 0.000 | -0.055 | 2.746281e-10 | TRUE |
| isoComp_00804958 | geneComp_00162234 | ENST00000618925 | ENSG00000251562 | MALAT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.265 | 0.265 | 3.781821e-10 | TRUE |
| isoComp_00752040 | geneComp_00148557 | MSTRG.25341.3 | ENSG00000174607 | UGT8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.135 | 0.000 | -0.135 | 4.326323e-10 | TRUE |
| isoComp_00712209 | geneComp_00143734 | ENST00000506290 | ENSG00000145715 | RASA1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.086 | 0.086 | 4.429204e-10 | TRUE |
| isoComp_00660888 | geneComp_00138309 | ENST00000459913 | ENSG00000106638 | TBL2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.061 | 0.061 | 4.822007e-10 | |
| isoComp_00763339 | geneComp_00150267 | ENST00000472110 | ENSG00000184208 | C22orf46 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.363 | 0.363 | 4.822007e-10 | TRUE |
| isoComp_00643417 | geneComp_00136613 | ENST00000427947 | ENSG00000085982 | USP40 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.166 | 0.166 | 5.425066e-10 | TRUE |
| isoComp_00676879 | geneComp_00139942 | ENST00000381024 | ENSG00000119778 | ATAD2B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.092 | 0.000 | -0.092 | 5.615853e-10 | TRUE |
| isoComp_00763550 | geneComp_00150301 | MSTRG.33983.5 | ENSG00000184368 | MAP7D2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.123 | 0.000 | -0.123 | 5.814830e-10 | TRUE |
| isoComp_00722312 | geneComp_00144911 | MSTRG.4179.19 | ENSG00000156042 | CFAP70 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.598 | 0.000 | -0.598 | 7.378025e-10 | TRUE |
| isoComp_00775455 | geneComp_00152045 | ENST00000582864 | ENSG00000198265 | HELZ | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.066 | 0.000 | -0.066 | 9.431712e-10 | |
| isoComp_00716339 | geneComp_00144184 | MSTRG.6332.12 | ENSG00000149294 | NCAM1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.067 | 0.000 | -0.067 | 1.317704e-09 | TRUE |
| isoComp_00721292 | geneComp_00144787 | ENST00000627468 | ENSG00000154814 | OXNAD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.081 | 0.000 | -0.081 | 1.317704e-09 | TRUE |
| isoComp_00771888 | geneComp_00151595 | ENST00000465206 | ENSG00000196693 | ZNF33B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.077 | 0.000 | -0.077 | 1.355516e-09 | |
| isoComp_00661764 | geneComp_00138407 | MSTRG.4120.4 | ENSG00000107719 | PALD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.061 | 0.061 | 2.009195e-09 | TRUE |
| isoComp_00671385 | geneComp_00139370 | ENST00000693446 | ENSG00000115295 | CLIP4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.207 | 0.000 | -0.207 | 2.170605e-09 | TRUE |
| isoComp_00761335 | geneComp_00149960 | MSTRG.22260.28 | ENSG00000182841 | RRP7BP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.070 | 0.070 | 2.438567e-09 | TRUE |
| isoComp_00664027 | geneComp_00138621 | MSTRG.24868.2 | ENSG00000109184 | DCUN1D4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.071 | 0.000 | -0.071 | 3.187488e-09 | |
| isoComp_00744351 | geneComp_00147522 | ENST00000563835 | ENSG00000169410 | PTPN9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.153 | 0.000 | -0.153 | 3.499517e-09 | TRUE |
| isoComp_00693171 | geneComp_00141761 | ENST00000690388 | ENSG00000133624 | ZNF767P | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.130 | 0.000 | -0.130 | 4.107156e-09 | TRUE |
| isoComp_00632675 | geneComp_00135676 | MSTRG.18016.4 | ENSG00000055917 | PUM2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.059 | 0.059 | 4.172616e-09 | TRUE |
| isoComp_00819070 | geneComp_00169569 | ENST00000620284 | ENSG00000278023 | RDM1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.346 | 0.000 | -0.346 | 4.187459e-09 | TRUE |
| isoComp_00681898 | geneComp_00140527 | MSTRG.28326.15 | ENSG00000124574 | ABCC10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.065 | 0.000 | -0.065 | 4.216343e-09 | TRUE |
| isoComp_00750643 | geneComp_00148365 | ENST00000491943 | ENSG00000173531 | MST1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.406 | 0.406 | 6.549989e-09 | TRUE |
| isoComp_00817558 | geneComp_00168693 | MSTRG.10314.14 | ENSG00000274253 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.503 | 0.000 | -0.503 | 6.802106e-09 | TRUE | |
| isoComp_00656515 | geneComp_00137857 | ENST00000520810 | ENSG00000104365 | IKBKB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.199 | 0.000 | -0.199 | 7.106056e-09 | TRUE |
| isoComp_00687472 | geneComp_00141180 | ENST00000517665 | ENSG00000129951 | PLPPR3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.398 | 0.000 | -0.398 | 7.814294e-09 | TRUE |
| isoComp_00683266 | geneComp_00140690 | ENST00000621891 | ENSG00000125779 | PANK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.093 | 0.000 | -0.093 | 9.781902e-09 | TRUE |
| isoComp_00686981 | geneComp_00141120 | ENST00000315808 | ENSG00000129292 | PHF20L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.076 | 0.000 | -0.076 | 9.781902e-09 | TRUE |
| isoComp_00675907 | geneComp_00139841 | MSTRG.26647.5 | ENSG00000118985 | ELL2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.098 | 0.000 | -0.098 | 1.054302e-08 | TRUE |
| isoComp_00677646 | geneComp_00140047 | ENST00000492710 | ENSG00000120616 | EPC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.183 | 0.183 | 1.054302e-08 | TRUE |
| isoComp_00788321 | geneComp_00155610 | MSTRG.29967.16 | ENSG00000226824 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.285 | 0.285 | 1.181484e-08 | TRUE | |
| isoComp_00662501 | geneComp_00138474 | MSTRG.4403.3 | ENSG00000108239 | TBC1D12 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.165 | 0.165 | 1.244016e-08 | TRUE |
| isoComp_00771199 | geneComp_00151515 | MSTRG.17049.4 | ENSG00000196437 | ZNF569 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.152 | 0.152 | 1.369644e-08 | TRUE |
| isoComp_00782710 | geneComp_00153579 | ENST00000437008 | ENSG00000214135 | SDHAP4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.064 | 0.000 | -0.064 | 1.458737e-08 | TRUE |
| isoComp_00808443 | geneComp_00163922 | ENST00000549424 | ENSG00000258311 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.283 | 0.000 | -0.283 | 1.458737e-08 | TRUE | |
| isoComp_00751533 | geneComp_00148481 | MSTRG.10612.6 | ENSG00000174197 | MGA | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.095 | 0.000 | -0.095 | 1.461252e-08 | TRUE |
| isoComp_00785779 | geneComp_00154587 | ENST00000423742 | ENSG00000223959 | AFG3L1P | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.063 | 0.063 | 1.737530e-08 | TRUE |
| isoComp_00763801 | geneComp_00150343 | ENST00000528778 | ENSG00000184545 | DUSP8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.508 | 0.508 | 1.843233e-08 | TRUE |
| isoComp_00647658 | geneComp_00136981 | ENST00000373821 | ENSG00000096063 | SRPK1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.053 | 0.000 | -0.053 | 2.129629e-08 | TRUE |
| isoComp_00774764 | geneComp_00151955 | MSTRG.16378.4 | ENSG00000197961 | ZNF121 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.051 | 0.051 | 2.547844e-08 | TRUE |
| isoComp_00712986 | geneComp_00143828 | ENST00000692270 | ENSG00000146282 | RARS2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.052 | 0.052 | 2.559634e-08 | TRUE |
| isoComp_00817336 | geneComp_00168572 | MSTRG.33741.9 | ENSG00000273748 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.334 | 0.000 | -0.334 | 2.595460e-08 | TRUE | |
| isoComp_00677339 | geneComp_00139997 | ENST00000517946 | ENSG00000120159 | CAAP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.127 | 0.127 | 2.633443e-08 | TRUE |
| isoComp_00729231 | geneComp_00145663 | ENST00000569496 | ENSG00000162062 | TEDC2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.098 | 0.000 | -0.098 | 3.014157e-08 | FALSE |
| isoComp_00712315 | geneComp_00143741 | MSTRG.26298.4 | ENSG00000145740 | SLC30A5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.052 | 0.052 | 3.150734e-08 | |
| isoComp_00650582 | geneComp_00137272 | ENST00000555170 | ENSG00000100505 | TRIM9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.156 | 0.156 | 3.178302e-08 | TRUE |
| isoComp_00644691 | geneComp_00136719 | ENST00000446916 | ENSG00000088812 | ATRN | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.130 | 0.000 | -0.130 | 3.354545e-08 | TRUE |
| isoComp_00702057 | geneComp_00142704 | ENST00000507010 | ENSG00000138668 | HNRNPD | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.033 | 0.084 | 0.051 | 3.524273e-08 | TRUE |
| isoComp_00714798 | geneComp_00144034 | ENST00000645398 | ENSG00000148019 | CEP78 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.114 | 0.000 | -0.114 | 3.534573e-08 | |
| isoComp_00683303 | geneComp_00140698 | MSTRG.20481.6 | ENSG00000125814 | NAPB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.108 | 0.000 | -0.108 | 4.440128e-08 | TRUE |
| isoComp_00677312 | geneComp_00139990 | MSTRG.14598.12 | ENSG00000120093 | HOXB3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.068 | 0.000 | -0.068 | 4.553676e-08 | |
| isoComp_00688931 | geneComp_00141328 | MSTRG.17155.3 | ENSG00000130758 | MAP3K10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.056 | 0.056 | 4.573847e-08 | TRUE |
| isoComp_00629124 | geneComp_00135356 | ENST00000359203 | ENSG00000025039 | RRAGD | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.067 | 0.000 | -0.067 | 5.582935e-08 | |
| isoComp_00775111 | geneComp_00152000 | ENST00000357328 | ENSG00000198105 | ZNF248 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.258 | 0.258 | 6.384170e-08 | TRUE |
| isoComp_00734581 | geneComp_00146352 | ENST00000520841 | ENSG00000164609 | SLU7 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.052 | 0.052 | 6.449227e-08 | |
| isoComp_00817143 | geneComp_00168450 | MSTRG.30966.4 | ENSG00000273344 | PAXIP1-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.113 | 0.113 | 7.108171e-08 | TRUE |
| isoComp_00640468 | geneComp_00136343 | MSTRG.6031.6 | ENSG00000078124 | ACER3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.153 | 0.000 | -0.153 | 7.836684e-08 | TRUE |
| isoComp_00716863 | geneComp_00144243 | MSTRG.20970.5 | ENSG00000149658 | YTHDF1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.054 | 0.054 | 7.870706e-08 | |
| isoComp_00771193 | geneComp_00151515 | ENST00000392150 | ENSG00000196437 | ZNF569 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.160 | 0.000 | -0.160 | 8.533072e-08 | TRUE |
| isoComp_00784951 | geneComp_00154315 | ENST00000577886 | ENSG00000221926 | TRIM16 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.115 | 0.000 | -0.115 | 9.099217e-08 | TRUE |
| isoComp_00742094 | geneComp_00147244 | ENST00000649057 | ENSG00000168216 | LMBRD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.117 | 0.117 | 9.580525e-08 | TRUE |
| isoComp_00658061 | geneComp_00138008 | MSTRG.17149.11 | ENSG00000105202 | FBL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.092 | 0.092 | 9.665548e-08 | TRUE |
| isoComp_00766113 | geneComp_00150677 | ENST00000684972 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.053 | 0.000 | -0.053 | 1.017177e-07 | TRUE |
| isoComp_00668732 | geneComp_00139110 | ENST00000509605 | ENSG00000113119 | TMCO6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.061 | 0.000 | -0.061 | 1.067588e-07 | |
| isoComp_00718600 | geneComp_00144440 | ENST00000470137 | ENSG00000151773 | CCDC122 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.292 | 0.000 | -0.292 | 1.152912e-07 | TRUE |
| isoComp_00640665 | geneComp_00136358 | ENST00000307729 | ENSG00000078403 | MLLT10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.093 | 0.093 | 1.324845e-07 | TRUE |
| isoComp_00758607 | geneComp_00149514 | MSTRG.34112.7 | ENSG00000180182 | MED14 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.121 | 0.121 | 1.337964e-07 | TRUE |
| isoComp_00654528 | geneComp_00137680 | ENST00000562808 | ENSG00000102984 | ZNF821 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.136 | 0.136 | 1.369015e-07 | TRUE |
| isoComp_00661365 | geneComp_00138362 | MSTRG.32422.9 | ENSG00000107186 | MPDZ | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.148 | 0.148 | 1.438627e-07 | TRUE |
| isoComp_00806394 | geneComp_00162951 | ENST00000534349 | ENSG00000254860 | TMEM9B-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.451 | 0.451 | 1.848321e-07 | TRUE |
| isoComp_00749203 | geneComp_00148201 | MSTRG.3885.7 | ENSG00000172671 | ZFAND4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.177 | 0.000 | -0.177 | 2.123190e-07 | TRUE |
| isoComp_00739979 | geneComp_00147027 | ENST00000221327 | ENSG00000167384 | ZNF180 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.119 | 0.000 | -0.119 | 2.233319e-07 | TRUE |
| isoComp_00732020 | geneComp_00146055 | ENST00000473702 | ENSG00000163659 | TIPARP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.107 | 0.107 | 2.456897e-07 | TRUE |
| isoComp_00824396 | geneComp_00172872 | ENST00000686873 | ENSG00000288061 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.699 | 0.699 | 2.621188e-07 | FALSE | |
| isoComp_00784123 | geneComp_00153962 | ENST00000579125 | ENSG00000215769 | ARHGAP27P1-BPTFP1-KPNA2P3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.265 | 0.000 | -0.265 | 2.659849e-07 | TRUE |
| isoComp_00706062 | geneComp_00143084 | ENST00000324677 | ENSG00000141298 | SSH2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.079 | 0.000 | -0.079 | 3.489326e-07 | TRUE |
| isoComp_00697390 | geneComp_00142205 | ENST00000547790 | ENSG00000136044 | APPL2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.063 | 0.063 | 3.716742e-07 | TRUE |
| isoComp_00661897 | geneComp_00138426 | ENST00000465807 | ENSG00000107821 | KAZALD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.211 | 0.000 | -0.211 | 3.992968e-07 | TRUE |
| isoComp_00766120 | geneComp_00150677 | MSTRG.25893.12 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.052 | 0.052 | 4.113231e-07 | TRUE |
| isoComp_00782409 | geneComp_00153523 | MSTRG.9233.21 | ENSG00000213983 | AP1G2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.057 | 0.000 | -0.057 | 4.120233e-07 | TRUE |
| isoComp_00647604 | geneComp_00136973 | MSTRG.3811.12 | ENSG00000095794 | CREM | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.165 | 0.000 | -0.165 | 4.421666e-07 | TRUE |
| isoComp_00634522 | geneComp_00135839 | ENST00000403772 | ENSG00000065150 | IPO5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.071 | 0.000 | -0.071 | 4.749074e-07 | |
| isoComp_00750296 | geneComp_00148314 | ENST00000695118 | ENSG00000173230 | GOLGB1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.115 | 0.000 | -0.115 | 4.766580e-07 | TRUE |
| isoComp_00800512 | geneComp_00160687 | ENST00000667442 | ENSG00000243701 | DUBR | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.177 | 0.000 | -0.177 | 5.835153e-07 | TRUE |
| isoComp_00746000 | geneComp_00147738 | ENST00000506870 | ENSG00000170464 | DNAJC18 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.059 | 0.000 | -0.059 | 6.092181e-07 | |
| isoComp_00767339 | geneComp_00150884 | ENST00000404820 | ENSG00000186998 | EMID1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.176 | 0.176 | 6.557800e-07 | TRUE |
| isoComp_00624557 | geneComp_00134946 | ENST00000469946 | ENSG00000003096 | KLHL13 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.160 | 0.000 | -0.160 | 6.664079e-07 | TRUE |
| isoComp_00627290 | geneComp_00135181 | ENST00000372597 | ENSG00000010803 | SCMH1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.098 | 0.098 | 6.664079e-07 | TRUE |
| isoComp_00812427 | geneComp_00166005 | MSTRG.14063.9 | ENSG00000264538 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.081 | 0.000 | -0.081 | 7.096308e-07 | TRUE | |
| isoComp_00682430 | geneComp_00140593 | MSTRG.12721.3 | ENSG00000125170 | DOK4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.158 | 0.000 | -0.158 | 8.199469e-07 | TRUE |
| isoComp_00759106 | geneComp_00149616 | ENST00000513413 | ENSG00000180818 | HOXC10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.072 | 0.000 | -0.072 | 9.925182e-07 | TRUE |
| isoComp_00688320 | geneComp_00141271 | MSTRG.16728.8 | ENSG00000130517 | PGPEP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.238 | 0.000 | -0.238 | 1.044876e-06 | TRUE |
| isoComp_00822740 | geneComp_00171668 | MSTRG.21751.27 | ENSG00000286129 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.139 | 0.000 | -0.139 | 1.078968e-06 | TRUE | |
| isoComp_00701343 | geneComp_00142626 | MSTRG.4397.6 | ENSG00000138180 | CEP55 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.077 | 0.077 | 1.179858e-06 | TRUE |
| isoComp_00797490 | geneComp_00159467 | MSTRG.18461.23 | ENSG00000237651 | C2orf74 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.095 | 0.000 | -0.095 | 1.212778e-06 | TRUE |
| isoComp_00748601 | geneComp_00148119 | ENST00000506213 | ENSG00000172244 | C5orf34 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.053 | 0.000 | -0.053 | 1.463252e-06 | TRUE |
| isoComp_00731631 | geneComp_00146004 | ENST00000425430 | ENSG00000163517 | HDAC11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.136 | 0.000 | -0.136 | 1.629239e-06 | TRUE |
| isoComp_00766357 | geneComp_00150718 | ENST00000433871 | ENSG00000186162 | CIDECP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.251 | 0.000 | -0.251 | 1.747491e-06 | TRUE |
| isoComp_00727472 | geneComp_00145485 | MSTRG.6362.4 | ENSG00000160584 | SIK3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.098 | 0.098 | 1.911548e-06 | |
| isoComp_00780134 | geneComp_00152862 | MSTRG.30112.7 | ENSG00000205485 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.151 | 0.151 | 1.950979e-06 | TRUE | |
| isoComp_00774276 | geneComp_00151891 | MSTRG.7377.22 | ENSG00000197757 | HOXC6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.069 | 0.000 | -0.069 | 1.957821e-06 | TRUE |
| isoComp_00667729 | geneComp_00139008 | ENST00000368068 | ENSG00000112282 | MED23 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.186 | 0.351 | 0.165 | 1.980690e-06 | TRUE |
| isoComp_00793466 | geneComp_00157804 | ENST00000668580 | ENSG00000232956 | SNHG15 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.057 | 0.057 | 2.010372e-06 | TRUE |
| isoComp_00755532 | geneComp_00149053 | ENST00000393102 | ENSG00000177463 | NR2C2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.211 | 0.211 | 2.033821e-06 | TRUE |
| isoComp_00738381 | geneComp_00146831 | MSTRG.30317.8 | ENSG00000166529 | ZSCAN21 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.183 | 0.183 | 2.203354e-06 | TRUE |
| isoComp_00729558 | geneComp_00145696 | MSTRG.5862.10 | ENSG00000162341 | TPCN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.082 | 0.000 | -0.082 | 2.251233e-06 | TRUE |
| isoComp_00719607 | geneComp_00144586 | ENST00000410077 | ENSG00000152969 | JAKMIP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.270 | 0.000 | -0.270 | 2.262068e-06 | TRUE |
| isoComp_00806045 | geneComp_00162758 | MSTRG.22389.8 | ENSG00000254413 | CHKB-CPT1B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.246 | 0.246 | 2.311119e-06 | TRUE |
| isoComp_00753529 | geneComp_00148733 | ENST00000427259 | ENSG00000175611 | LINC00476 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.121 | 0.000 | -0.121 | 2.691879e-06 | TRUE |
| isoComp_00678864 | geneComp_00140193 | ENST00000474144 | ENSG00000121900 | TMEM54 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.080 | 0.080 | 3.442292e-06 | TRUE |
| isoComp_00700703 | geneComp_00142555 | ENST00000560388 | ENSG00000137871 | ZNF280D | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.094 | 0.094 | 3.583965e-06 | TRUE |
| isoComp_00641170 | geneComp_00136401 | ENST00000599918 | ENSG00000079435 | LIPE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.396 | 0.396 | 3.857281e-06 | TRUE |
| isoComp_00808813 | geneComp_00164093 | ENST00000688866 | ENSG00000258704 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.159 | 0.000 | -0.159 | 3.979598e-06 | TRUE | |
| isoComp_00773639 | geneComp_00151816 | MSTRG.4109.27 | ENSG00000197467 | COL13A1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.258 | 0.258 | 4.126160e-06 | TRUE |
| isoComp_00770520 | geneComp_00151435 | MSTRG.29426.14 | ENSG00000196204 | RNF216P1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.075 | 0.000 | -0.075 | 4.689752e-06 | |
| isoComp_00771010 | geneComp_00151487 | MSTRG.30289.13 | ENSG00000196367 | TRRAP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.061 | 0.061 | 5.181890e-06 | TRUE |
| isoComp_00817590 | geneComp_00168705 | MSTRG.27787.3 | ENSG00000274290 | H2BC6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.863 | 0.863 | 7.223865e-06 | TRUE |
| isoComp_00724700 | geneComp_00145180 | ENST00000644949 | ENSG00000158321 | AUTS2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.073 | 0.073 | 7.417145e-06 | |
| isoComp_00670766 | geneComp_00139309 | ENST00000317151 | ENSG00000114923 | SLC4A3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.519 | 0.000 | -0.519 | 7.908038e-06 | TRUE |
| isoComp_00750520 | geneComp_00148352 | ENST00000531106 | ENSG00000173442 | EHBP1L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.474 | 0.474 | 8.882731e-06 | TRUE |
| isoComp_00778436 | geneComp_00152553 | MSTRG.28098.17 | ENSG00000204305 | AGER | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.311 | 0.311 | 8.928210e-06 | TRUE |
| isoComp_00804419 | geneComp_00162029 | ENST00000525811 | ENSG00000250903 | GMDS-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.270 | 0.000 | -0.270 | 1.111569e-05 | TRUE |
| isoComp_00688507 | geneComp_00141297 | ENST00000421586 | ENSG00000130649 | CYP2E1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.658 | 0.658 | 1.149367e-05 | TRUE |
| isoComp_00766179 | geneComp_00150685 | MSTRG.17027.11 | ENSG00000186017 | ZNF566 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.106 | 0.106 | 1.232361e-05 | TRUE |
| isoComp_00811547 | geneComp_00165580 | MSTRG.13503.4 | ENSG00000261879 | ZNF594-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.166 | 0.000 | -0.166 | 1.288602e-05 | TRUE |
| isoComp_00648335 | geneComp_00137053 | ENST00000478879 | ENSG00000099840 | IZUMO4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.321 | 0.321 | 1.296252e-05 | TRUE |
| isoComp_00715512 | geneComp_00144109 | MSTRG.4206.17 | ENSG00000148660 | CAMK2G | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.079 | 0.013 | -0.066 | 1.296252e-05 | TRUE |
| isoComp_00725814 | geneComp_00145313 | ENST00000376922 | ENSG00000159314 | ARHGAP27 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.145 | 0.000 | -0.145 | 1.357540e-05 | TRUE |
| isoComp_00813254 | geneComp_00166413 | MSTRG.14351.3 | ENSG00000266962 | HSD17B1-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.218 | 0.000 | -0.218 | 1.398277e-05 | TRUE |
| isoComp_00779678 | geneComp_00152748 | ENST00000590615 | ENSG00000204920 | ZNF155 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.290 | 0.000 | -0.290 | 1.404387e-05 | TRUE |
| isoComp_00679589 | geneComp_00140260 | ENST00000469576 | ENSG00000122550 | KLHL7 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.089 | 0.000 | -0.089 | 1.483740e-05 | |
| isoComp_00669914 | geneComp_00139232 | MSTRG.22917.13 | ENSG00000114270 | COL7A1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.073 | 0.073 | 1.554791e-05 | TRUE |
| isoComp_00666497 | geneComp_00138860 | MSTRG.8253.18 | ENSG00000111364 | DDX55 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.057 | 0.057 | 1.846590e-05 | TRUE |
| isoComp_00824858 | geneComp_00173156 | ENST00000692166 | ENSG00000288905 | LSP1P5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.085 | 0.000 | -0.085 | 1.894850e-05 | TRUE |
| isoComp_00657809 | geneComp_00137980 | ENST00000595358 | ENSG00000104983 | CCDC61 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.316 | 0.000 | -0.316 | 2.042404e-05 | TRUE |
| isoComp_00702825 | geneComp_00142764 | ENST00000534526 | ENSG00000139132 | FGD4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.594 | 1.000 | 0.406 | 2.079592e-05 | TRUE |
| isoComp_00692567 | geneComp_00141699 | MSTRG.2918.2 | ENSG00000133069 | TMCC2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.226 | 0.000 | -0.226 | 2.122489e-05 | TRUE |
| isoComp_00726635 | geneComp_00145411 | ENST00000454902 | ENSG00000160145 | KALRN | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.400 | 0.000 | -0.400 | 2.487342e-05 | TRUE |
| isoComp_00787439 | geneComp_00155235 | MSTRG.28410.14 | ENSG00000225791 | TRAM2-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.235 | 0.000 | -0.235 | 2.528149e-05 | TRUE |
| isoComp_00662961 | geneComp_00138512 | ENST00000571831 | ENSG00000108509 | CAMTA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.066 | 0.000 | -0.066 | 2.593441e-05 | TRUE |
| isoComp_00764246 | geneComp_00150420 | ENST00000639563 | ENSG00000184937 | WT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.198 | 0.198 | 2.637983e-05 | TRUE |
| isoComp_00625932 | geneComp_00135064 | ENST00000489094 | ENSG00000006704 | GTF2IRD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.111 | 0.111 | 2.794947e-05 | TRUE |
| isoComp_00682517 | geneComp_00140607 | ENST00000680139 | ENSG00000125347 | IRF1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.096 | 0.096 | 2.984548e-05 | TRUE |
| isoComp_00811769 | geneComp_00165674 | MSTRG.17112.1 | ENSG00000262484 | CCER2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 1.000 | 1.000 | 3.186307e-05 | TRUE |
| isoComp_00749003 | geneComp_00148179 | ENST00000612301 | ENSG00000172530 | BANP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.212 | 0.089 | -0.123 | 3.311732e-05 | TRUE |
| isoComp_00821592 | geneComp_00171089 | ENST00000641062 | ENSG00000284642 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.504 | 0.504 | 3.311732e-05 | TRUE | |
| isoComp_00822738 | geneComp_00171668 | MSTRG.21751.25 | ENSG00000286129 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.008 | 0.069 | 0.061 | 3.324706e-05 | TRUE | |
| isoComp_00694163 | geneComp_00141868 | ENST00000692769 | ENSG00000134297 | PLEKHA8P1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.142 | 0.142 | 3.528067e-05 | TRUE |
| isoComp_00661270 | geneComp_00138351 | ENST00000688321 | ENSG00000107104 | KANK1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.063 | 0.063 | 4.006723e-05 | TRUE |
| isoComp_00684116 | geneComp_00140793 | ENST00000416776 | ENSG00000126391 | FRMD8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.221 | 0.453 | 0.232 | 4.365151e-05 | TRUE |
| isoComp_00770840 | geneComp_00151467 | ENST00000506067 | ENSG00000196312 | MFSD14C | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.055 | 0.055 | 4.465241e-05 | TRUE |
| isoComp_00817144 | geneComp_00168450 | MSTRG.30966.5 | ENSG00000273344 | PAXIP1-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.158 | 0.000 | -0.158 | 4.465241e-05 | TRUE |
| isoComp_00799921 | geneComp_00160444 | MSTRG.30641.9 | ENSG00000242588 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.212 | 0.000 | -0.212 | 4.663806e-05 | TRUE | |
| isoComp_00811767 | geneComp_00165674 | ENST00000571838 | ENSG00000262484 | CCER2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.000 | -1.000 | 4.904096e-05 | TRUE |
| isoComp_00713196 | geneComp_00143857 | MSTRG.19093.17 | ENSG00000146556 | WASH2P | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.059 | 0.059 | 5.209333e-05 | TRUE |
| isoComp_00739341 | geneComp_00146953 | ENST00000412865 | ENSG00000167065 | DUSP18 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.157 | 0.157 | 5.420384e-05 | TRUE |
| isoComp_00671362 | geneComp_00139370 | ENST00000401605 | ENSG00000115295 | CLIP4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.262 | 0.262 | 6.120607e-05 | TRUE |
| isoComp_00685625 | geneComp_00140952 | ENST00000428214 | ENSG00000127980 | PEX1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.224 | 0.119 | -0.106 | 6.120607e-05 | TRUE |
| isoComp_00686035 | geneComp_00141009 | ENST00000450156 | ENSG00000128512 | DOCK4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.133 | 0.000 | -0.133 | 6.272698e-05 | TRUE |
| isoComp_00720752 | geneComp_00144720 | MSTRG.1189.2 | ENSG00000154222 | CC2D1B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.057 | 0.000 | -0.057 | 6.588758e-05 | TRUE |
| isoComp_00672710 | geneComp_00139481 | ENST00000487560 | ENSG00000115998 | C2orf42 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.080 | 0.000 | -0.080 | 6.829579e-05 | TRUE |
| isoComp_00721665 | geneComp_00144834 | ENST00000511366 | ENSG00000155275 | TRMT44 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.066 | 0.003 | -0.063 | 6.829579e-05 | TRUE |
| isoComp_00766114 | geneComp_00150677 | ENST00000689567 | ENSG00000185986 | SDHAP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.062 | 0.062 | 6.923824e-05 | TRUE |
| isoComp_00715635 | geneComp_00144123 | MSTRG.4657.1 | ENSG00000148737 | TCF7L2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.162 | 0.162 | 8.222176e-05 | TRUE |
| isoComp_00747595 | geneComp_00147980 | ENST00000302517 | ENSG00000171604 | CXXC5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.300 | 0.112 | -0.188 | 8.770501e-05 | TRUE |
| isoComp_00799179 | geneComp_00160174 | MSTRG.30343.2 | ENSG00000241357 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.732 | 1.000 | 0.268 | 9.172989e-05 | TRUE | |
| isoComp_00640704 | geneComp_00136361 | ENST00000304166 | ENSG00000078549 | ADCYAP1R1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.166 | 0.000 | -0.166 | 9.941069e-05 | TRUE |
| isoComp_00667619 | geneComp_00138990 | ENST00000229922 | ENSG00000112186 | CAP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.886 | 1.000 | 0.114 | 1.046989e-04 | TRUE |
| isoComp_00777977 | geneComp_00152479 | ENST00000661101 | ENSG00000204054 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.125 | 0.125 | 1.048738e-04 | TRUE | |
| isoComp_00738776 | geneComp_00146876 | ENST00000540441 | ENSG00000166783 | MARF1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.054 | 0.000 | -0.054 | 1.082468e-04 | TRUE |
| isoComp_00789506 | geneComp_00156106 | ENST00000422408 | ENSG00000228274 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.101 | 0.000 | -0.101 | 1.082767e-04 | TRUE | |
| isoComp_00665202 | geneComp_00138734 | MSTRG.6384.2 | ENSG00000110274 | CEP164 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.093 | 0.000 | -0.093 | 1.110396e-04 | TRUE |
| isoComp_00677544 | geneComp_00140035 | MSTRG.6500.15 | ENSG00000120458 | MSANTD2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.051 | 0.051 | 1.171026e-04 | TRUE |
| isoComp_00683150 | geneComp_00140680 | ENST00000592811 | ENSG00000125740 | FOSB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.428 | 0.428 | 1.172874e-04 | TRUE |
| isoComp_00678074 | geneComp_00140092 | ENST00000519650 | ENSG00000120899 | PTK2B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.395 | 0.000 | -0.395 | 1.230863e-04 | TRUE |
| isoComp_00719750 | geneComp_00144604 | ENST00000308659 | ENSG00000153094 | BCL2L11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.116 | 0.000 | -0.116 | 1.252113e-04 | TRUE |
| isoComp_00799178 | geneComp_00160174 | ENST00000492523 | ENSG00000241357 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.268 | 0.000 | -0.268 | 1.259551e-04 | TRUE | |
| isoComp_00655654 | geneComp_00137778 | ENST00000562274 | ENSG00000103528 | SYT17 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.649 | 0.000 | -0.649 | 1.272837e-04 | TRUE |
| isoComp_00692907 | geneComp_00141737 | ENST00000278856 | ENSG00000133316 | WDR74 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.420 | 0.587 | 0.167 | 1.281464e-04 | TRUE |
| isoComp_00664082 | geneComp_00138631 | ENST00000642252 | ENSG00000109323 | MANBA | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.066 | 0.066 | 1.592051e-04 | TRUE |
| isoComp_00654538 | geneComp_00137680 | ENST00000568666 | ENSG00000102984 | ZNF821 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.162 | 0.000 | -0.162 | 1.631209e-04 | TRUE |
| isoComp_00730409 | geneComp_00145834 | ENST00000294964 | ENSG00000162878 | PKDCC | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.096 | 0.002 | -0.093 | 1.729068e-04 | TRUE |
| isoComp_00810252 | geneComp_00164922 | ENST00000562082 | ENSG00000260400 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.771 | -0.229 | 1.770940e-04 | TRUE | |
| isoComp_00785088 | geneComp_00154333 | ENST00000442455 | ENSG00000221994 | ZNF630 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.726 | 0.726 | 1.838913e-04 | TRUE |
| isoComp_00650069 | geneComp_00137223 | ENST00000443735 | ENSG00000100362 | PVALB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.177 | 0.177 | 1.969506e-04 | TRUE |
| isoComp_00739921 | geneComp_00147021 | ENST00000572813 | ENSG00000167363 | FN3K | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.283 | 0.283 | 2.021525e-04 | TRUE |
| isoComp_00781708 | geneComp_00153310 | ENST00000654978 | ENSG00000213468 | FIRRE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.055 | 0.000 | -0.055 | 2.106818e-04 | TRUE |
| isoComp_00795632 | geneComp_00158718 | ENST00000659935 | ENSG00000235501 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.290 | 0.000 | -0.290 | 2.112911e-04 | TRUE | |
| isoComp_00798866 | geneComp_00160032 | ENST00000523244 | ENSG00000240694 | PNMA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.059 | 0.059 | 2.198978e-04 | |
| isoComp_00732814 | geneComp_00146139 | ENST00000688504 | ENSG00000163913 | IFT122 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.069 | 0.069 | 2.226338e-04 | TRUE |
| isoComp_00683543 | geneComp_00140731 | MSTRG.20237.3 | ENSG00000125895 | TMEM74B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.187 | 0.000 | -0.187 | 2.226875e-04 | TRUE |
| isoComp_00778104 | geneComp_00152498 | MSTRG.20044.1 | ENSG00000204128 | C2orf72 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.910 | 0.910 | 2.296002e-04 | TRUE |
| isoComp_00789655 | geneComp_00156157 | ENST00000450686 | ENSG00000228393 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.333 | 0.020 | -0.312 | 2.313201e-04 | TRUE | |
| isoComp_00810253 | geneComp_00164922 | MSTRG.4070.1 | ENSG00000260400 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.229 | 0.229 | 2.376019e-04 | TRUE | |
| isoComp_00699817 | geneComp_00142464 | MSTRG.27637.10 | ENSG00000137434 | C6orf52 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.158 | 0.000 | -0.158 | 2.653623e-04 | TRUE |
| isoComp_00799413 | geneComp_00160265 | ENST00000618820 | ENSG00000241769 | EOLA1-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.306 | 0.000 | -0.306 | 2.733581e-04 | TRUE |
| isoComp_00766840 | geneComp_00150798 | ENST00000529146 | ENSG00000186523 | FAM86B1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.004 | 0.122 | 0.117 | 2.748448e-04 | TRUE |
| isoComp_00756751 | geneComp_00149219 | ENST00000395069 | ENSG00000178401 | DNAJC22 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.554 | 0.000 | -0.554 | 3.014121e-04 | TRUE |
| isoComp_00743057 | geneComp_00147350 | ENST00000492412 | ENSG00000168679 | SLC16A4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.459 | 0.000 | -0.459 | 3.130954e-04 | TRUE |
| isoComp_00711397 | geneComp_00143638 | ENST00000393775 | ENSG00000144847 | IGSF11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.609 | 0.000 | -0.609 | 3.224600e-04 | TRUE |
| isoComp_00659963 | geneComp_00138214 | ENST00000439384 | ENSG00000106069 | CHN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.145 | 0.000 | -0.145 | 3.318617e-04 | TRUE |
| isoComp_00655198 | geneComp_00137731 | ENST00000341413 | ENSG00000103253 | HAGHL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.092 | 0.092 | 3.372237e-04 | TRUE |
| isoComp_00775745 | geneComp_00152081 | ENST00000391778 | ENSG00000198440 | ZNF583 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.363 | 0.363 | 3.372237e-04 | TRUE |
| isoComp_00760547 | geneComp_00149834 | ENST00000574196 | ENSG00000182224 | CYB5D1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.107 | 0.000 | -0.107 | 3.464146e-04 | TRUE |
| isoComp_00764238 | geneComp_00150420 | ENST00000332351 | ENSG00000184937 | WT1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.281 | 0.000 | -0.281 | 3.783088e-04 | TRUE |
| isoComp_00627291 | geneComp_00135181 | ENST00000456518 | ENSG00000010803 | SCMH1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.055 | 0.000 | -0.055 | 3.844778e-04 | TRUE |
| isoComp_00684415 | geneComp_00140824 | ENST00000478222 | ENSG00000126759 | CFP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.575 | 0.000 | -0.575 | 3.857017e-04 | TRUE |
| isoComp_00775838 | geneComp_00152097 | ENST00000511562 | ENSG00000198498 | TMA16 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.101 | 0.026 | -0.075 | 3.857017e-04 | TRUE |
| isoComp_00673872 | geneComp_00139604 | ENST00000367362 | ENSG00000116833 | NR5A2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.045 | 0.776 | 0.731 | 4.004889e-04 | TRUE |
| isoComp_00739337 | geneComp_00146953 | ENST00000377087 | ENSG00000167065 | DUSP18 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.125 | 0.000 | -0.125 | 4.035028e-04 | TRUE |
| isoComp_00761565 | geneComp_00149989 | ENST00000382095 | ENSG00000182957 | SPATA13 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.374 | 0.374 | 4.563937e-04 | TRUE |
| isoComp_00756521 | geneComp_00149179 | ENST00000467457 | ENSG00000178149 | DALRD3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.081 | 0.231 | 0.150 | 4.684075e-04 | TRUE |
| isoComp_00678028 | geneComp_00140088 | ENST00000560566 | ENSG00000120885 | CLU | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.111 | 0.000 | -0.111 | 5.193999e-04 | TRUE |
| isoComp_00799779 | geneComp_00160386 | MSTRG.4190.9 | ENSG00000242288 | BMS1P4-AGAP5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.286 | 0.000 | -0.286 | 5.366437e-04 | TRUE |
| isoComp_00631881 | geneComp_00135607 | ENST00000390654 | ENSG00000050767 | COL23A1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.864 | 0.092 | -0.772 | 5.572799e-04 | TRUE |
| isoComp_00739695 | geneComp_00146995 | ENST00000599743 | ENSG00000167232 | ZNF91 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.132 | 0.000 | -0.132 | 5.607198e-04 | TRUE |
| isoComp_00790826 | geneComp_00156651 | ENST00000563276 | ENSG00000229809 | ZNF688 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.032 | 0.162 | 0.130 | 5.960457e-04 | |
| isoComp_00731544 | geneComp_00145993 | ENST00000435750 | ENSG00000163491 | NEK10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.204 | 0.000 | -0.204 | 6.033789e-04 | TRUE |
| isoComp_00738327 | geneComp_00146823 | MSTRG.11423.4 | ENSG00000166503 | HDGFL3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.804 | 0.920 | 0.116 | 6.035961e-04 | TRUE |
| isoComp_00733676 | geneComp_00146223 | ENST00000393674 | ENSG00000164117 | FBXO8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.885 | -0.115 | 6.327904e-04 | TRUE |
| isoComp_00768738 | geneComp_00151135 | ENST00000585441 | ENSG00000188283 | ZNF383 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.168 | 0.168 | 6.558244e-04 | TRUE |
| isoComp_00626762 | geneComp_00135137 | ENST00000580921 | ENSG00000008838 | MED24 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.025 | 0.101 | 0.076 | 6.591604e-04 | |
| isoComp_00820209 | geneComp_00170335 | MSTRG.29037.9 | ENSG00000279968 | CCDC28A-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.104 | 0.000 | -0.104 | 7.556127e-04 | TRUE |
| isoComp_00685922 | geneComp_00140995 | ENST00000454728 | ENSG00000128335 | APOL2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.060 | 0.060 | 7.769926e-04 | TRUE |
| isoComp_00719013 | geneComp_00144499 | MSTRG.15745.4 | ENSG00000152242 | C18orf25 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.184 | 0.425 | 0.240 | 7.990384e-04 | TRUE |
| isoComp_00798590 | geneComp_00159923 | ENST00000617641 | ENSG00000240184 | PCDHGC3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.378 | 0.378 | 7.990384e-04 | TRUE |
| isoComp_00811580 | geneComp_00165595 | MSTRG.11908.6 | ENSG00000261971 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.116 | 0.116 | 7.990384e-04 | TRUE | |
| isoComp_00630687 | geneComp_00135499 | ENST00000486510 | ENSG00000041880 | PARP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.058 | 0.000 | -0.058 | 8.006582e-04 | TRUE |
| isoComp_00704700 | geneComp_00142968 | ENST00000561308 | ENSG00000140471 | LINS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.105 | 0.221 | 0.116 | 8.272333e-04 | TRUE |
| isoComp_00707339 | geneComp_00143194 | ENST00000537354 | ENSG00000142046 | TMEM91 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.151 | 0.000 | -0.151 | 8.272333e-04 | TRUE |
| isoComp_00773254 | geneComp_00151762 | ENST00000636146 | ENSG00000197283 | SYNGAP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.125 | 0.000 | -0.125 | 8.679348e-04 | TRUE |
| isoComp_00773031 | geneComp_00151724 | MSTRG.1571.6 | ENSG00000197147 | LRRC8B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.068 | 0.000 | -0.068 | 9.121086e-04 | TRUE |
| isoComp_00724020 | geneComp_00145102 | ENST00000374126 | ENSG00000157657 | ZNF618 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.107 | 0.107 | 9.961646e-04 | TRUE |
| isoComp_00703534 | geneComp_00142849 | ENST00000379846 | ENSG00000139631 | CSAD | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.160 | 0.000 | -0.160 | 1.008119e-03 | TRUE |
| isoComp_00643158 | geneComp_00136588 | ENST00000369125 | ENSG00000085382 | HACE1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.077 | 0.000 | -0.077 | 1.016635e-03 | TRUE |
| isoComp_00688351 | geneComp_00141276 | ENST00000601347 | ENSG00000130529 | TRPM4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.179 | 0.179 | 1.034805e-03 | TRUE |
| isoComp_00789983 | geneComp_00156274 | ENST00000432246 | ENSG00000228701 | TNKS2-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.580 | 0.580 | 1.067857e-03 | TRUE |
| isoComp_00681101 | geneComp_00140435 | MSTRG.20779.3 | ENSG00000124104 | SNX21 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.192 | 0.000 | -0.192 | 1.208908e-03 | TRUE |
| isoComp_00695866 | geneComp_00142052 | MSTRG.28563.3 | ENSG00000135338 | LCA5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.238 | 0.000 | -0.238 | 1.291419e-03 | TRUE |
| isoComp_00712881 | geneComp_00143818 | ENST00000480804 | ENSG00000146233 | CYP39A1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.124 | 0.124 | 1.325503e-03 | TRUE |
| isoComp_00672014 | geneComp_00139415 | ENST00000323303 | ENSG00000115540 | MOB4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.060 | 0.000 | -0.060 | 1.356040e-03 | |
| isoComp_00654085 | geneComp_00137642 | ENST00000441394 | ENSG00000102781 | KATNAL1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.061 | 0.000 | -0.061 | 1.375969e-03 | |
| isoComp_00644546 | geneComp_00136708 | ENST00000449796 | ENSG00000088387 | DOCK9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.062 | 0.062 | 1.414350e-03 | TRUE |
| isoComp_00729616 | geneComp_00145708 | MSTRG.1240.2 | ENSG00000162390 | ACOT11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.609 | 0.000 | -0.609 | 1.418202e-03 | TRUE |
| isoComp_00762222 | geneComp_00150092 | ENST00000591260 | ENSG00000183401 | CCDC159 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.096 | 0.000 | -0.096 | 1.426794e-03 | TRUE |
| isoComp_00805095 | geneComp_00162304 | ENST00000624599 | ENSG00000253123 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.425 | 0.425 | 1.427552e-03 | TRUE | |
| isoComp_00634635 | geneComp_00135847 | MSTRG.7433.17 | ENSG00000065357 | DGKA | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.334 | 0.000 | -0.334 | 1.467102e-03 | TRUE |
| isoComp_00654618 | geneComp_00137687 | ENST00000566585 | ENSG00000103034 | NDRG4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.227 | 0.000 | -0.227 | 1.483575e-03 | TRUE |
| isoComp_00624774 | geneComp_00134959 | ENST00000582323 | ENSG00000004139 | SARM1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.071 | 0.000 | -0.071 | 1.648941e-03 | |
| isoComp_00819364 | geneComp_00169683 | ENST00000614766 | ENSG00000278619 | MRM1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.833 | 1.000 | 0.167 | 1.725055e-03 | TRUE |
| isoComp_00722632 | geneComp_00144950 | MSTRG.4589.3 | ENSG00000156384 | SFR1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.138 | 0.000 | -0.138 | 1.788298e-03 | TRUE |
| isoComp_00775705 | geneComp_00152074 | ENST00000482611 | ENSG00000198408 | OGA | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.095 | 0.011 | -0.083 | 1.795065e-03 | TRUE |
| isoComp_00822703 | geneComp_00171656 | MSTRG.33482.11 | ENSG00000286112 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.189 | 0.189 | 1.918256e-03 | TRUE | |
| isoComp_00782482 | geneComp_00153540 | ENST00000438596 | ENSG00000214021 | TTLL3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.050 | 0.050 | 1.946504e-03 | TRUE |
| isoComp_00749513 | geneComp_00148236 | MSTRG.12793.21 | ENSG00000172828 | CES3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.057 | 0.057 | 1.979244e-03 | |
| isoComp_00786982 | geneComp_00155086 | ENST00000603624 | ENSG00000225361 | PPP1R26-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.123 | 0.000 | -0.123 | 1.993392e-03 | TRUE |
| isoComp_00778689 | geneComp_00152595 | ENST00000488372 | ENSG00000204406 | MBD5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.073 | 0.073 | 2.025629e-03 | TRUE |
| isoComp_00779789 | geneComp_00152785 | MSTRG.13068.16 | ENSG00000205078 | SYCE1L | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.274 | 0.274 | 2.045463e-03 | TRUE |
| isoComp_00658320 | geneComp_00138032 | ENST00000587175 | ENSG00000105298 | CACTIN | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.044 | 0.101 | 0.057 | 2.075282e-03 | TRUE |
| isoComp_00796106 | geneComp_00158920 | ENST00000449363 | ENSG00000236088 | COX10-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.050 | 0.050 | 2.090003e-03 | TRUE |
| isoComp_00800079 | geneComp_00160495 | MSTRG.29417.10 | ENSG00000242802 | AP5Z1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.193 | 0.107 | -0.086 | 2.211595e-03 | TRUE |
| isoComp_00702620 | geneComp_00142743 | ENST00000265174 | ENSG00000138801 | PAPSS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.884 | 0.952 | 0.068 | 2.215300e-03 | |
| isoComp_00802211 | geneComp_00161136 | ENST00000511927 | ENSG00000247595 | SPTY2D1OS | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.083 | 0.083 | 2.366972e-03 | TRUE |
| isoComp_00760759 | geneComp_00149869 | ENST00000636496 | ENSG00000182389 | CACNB4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.192 | 0.000 | -0.192 | 2.424083e-03 | TRUE |
| isoComp_00783550 | geneComp_00153814 | ENST00000606627 | ENSG00000215022 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.121 | 0.000 | -0.121 | 2.427066e-03 | TRUE | |
| isoComp_00766015 | geneComp_00150660 | ENST00000399208 | ENSG00000185917 | SETD4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.092 | 0.028 | -0.064 | 2.511762e-03 | TRUE |
| isoComp_00769214 | geneComp_00151213 | MSTRG.11383.3 | ENSG00000188659 | SAXO2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.157 | 0.000 | -0.157 | 2.524338e-03 | TRUE |
| isoComp_00775461 | geneComp_00152046 | ENST00000547878 | ENSG00000198270 | TMEM116 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.062 | 0.000 | -0.062 | 2.577859e-03 | TRUE |
| isoComp_00807448 | geneComp_00163436 | MSTRG.6824.3 | ENSG00000256537 | SMIM10L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.058 | 0.000 | -0.058 | 2.643306e-03 | TRUE |
| isoComp_00753965 | geneComp_00148812 | ENST00000323959 | ENSG00000176102 | CSTF3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.566 | 0.651 | 0.084 | 2.651662e-03 | |
| isoComp_00661366 | geneComp_00138363 | ENST00000371748 | ENSG00000107187 | LHX3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.914 | 0.000 | -0.914 | 2.658654e-03 | TRUE |
| isoComp_00722035 | geneComp_00144873 | ENST00000448967 | ENSG00000155749 | FLACC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.254 | 0.254 | 2.673842e-03 | TRUE |
| isoComp_00673114 | geneComp_00139527 | ENST00000377648 | ENSG00000116273 | PHF13 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.946 | -0.054 | 2.707881e-03 | |
| isoComp_00654398 | geneComp_00137666 | ENST00000416006 | ENSG00000102910 | LONP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.200 | 0.081 | -0.119 | 2.728287e-03 | TRUE |
| isoComp_00666528 | geneComp_00138864 | ENST00000548664 | ENSG00000111424 | VDR | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.143 | 0.000 | -0.143 | 2.780427e-03 | TRUE |
| isoComp_00776127 | geneComp_00152134 | ENST00000409334 | ENSG00000198612 | COPS8 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.133 | 0.207 | 0.073 | 2.780427e-03 | TRUE |
| isoComp_00631934 | geneComp_00135611 | ENST00000568676 | ENSG00000051108 | HERPUD1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.099 | 0.047 | -0.052 | 2.812119e-03 | |
| isoComp_00634062 | geneComp_00135802 | ENST00000486249 | ENSG00000064225 | ST3GAL6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.052 | 0.052 | 2.996102e-03 | TRUE |
| isoComp_00786987 | geneComp_00155086 | MSTRG.33605.4 | ENSG00000225361 | PPP1R26-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.041 | 0.297 | 0.256 | 3.124740e-03 | TRUE |
| isoComp_00779990 | geneComp_00152834 | ENST00000546604 | ENSG00000205323 | SARNP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.061 | 0.115 | 0.054 | 3.136723e-03 | TRUE |
| isoComp_00713144 | geneComp_00143852 | ENST00000485526 | ENSG00000146530 | VWDE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.227 | 0.227 | 3.188526e-03 | TRUE |
| isoComp_00760045 | geneComp_00149765 | ENST00000598312 | ENSG00000181894 | ZNF329 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.850 | 0.226 | -0.624 | 3.189084e-03 | TRUE |
| isoComp_00805280 | geneComp_00162390 | ENST00000531285 | ENSG00000253368 | TRNP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.413 | 0.659 | 0.245 | 3.237182e-03 | TRUE |
| isoComp_00640269 | geneComp_00136327 | MSTRG.25460.11 | ENSG00000077684 | JADE1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.075 | 0.000 | -0.075 | 3.241038e-03 | TRUE |
| isoComp_00805279 | geneComp_00162390 | ENST00000522111 | ENSG00000253368 | TRNP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.587 | 0.341 | -0.245 | 3.241038e-03 | TRUE |
| isoComp_00634861 | geneComp_00135867 | ENST00000590536 | ENSG00000065717 | TLE2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.345 | 0.345 | 3.287675e-03 | TRUE |
| isoComp_00676780 | geneComp_00139934 | ENST00000554320 | ENSG00000119723 | COQ6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.083 | 0.150 | 0.066 | 3.371815e-03 | TRUE |
| isoComp_00702268 | geneComp_00142717 | ENST00000512494 | ENSG00000138735 | PDE5A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.206 | 0.206 | 3.394545e-03 | TRUE |
| isoComp_00671316 | geneComp_00139366 | ENST00000463189 | ENSG00000115282 | TTC31 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.072 | 0.018 | -0.054 | 3.396782e-03 | TRUE |
| isoComp_00682454 | geneComp_00140597 | MSTRG.9036.7 | ENSG00000125247 | TMTC4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.074 | 0.074 | 3.491582e-03 | TRUE |
| isoComp_00674360 | geneComp_00139660 | ENST00000235932 | ENSG00000117280 | RAB29 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.113 | 0.027 | -0.086 | 3.514824e-03 | |
| isoComp_00801334 | geneComp_00160973 | ENST00000499203 | ENSG00000245146 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.163 | 0.017 | -0.146 | 3.530233e-03 | TRUE | |
| isoComp_00720486 | geneComp_00144686 | MSTRG.13676.3 | ENSG00000153976 | HS3ST3A1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.035 | 0.118 | 0.083 | 3.564437e-03 | TRUE |
| isoComp_00689385 | geneComp_00141375 | ENST00000367251 | ENSG00000131018 | SYNE1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.195 | 0.195 | 3.571302e-03 | TRUE |
| isoComp_00785376 | geneComp_00154432 | ENST00000663114 | ENSG00000223546 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.073 | 0.073 | 3.571302e-03 | TRUE | |
| isoComp_00771780 | geneComp_00151581 | ENST00000449836 | ENSG00000196653 | ZNF502 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.219 | 0.000 | -0.219 | 3.650410e-03 | TRUE |
| isoComp_00625079 | geneComp_00134990 | MSTRG.25302.5 | ENSG00000005059 | MCUB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.108 | 0.108 | 3.687076e-03 | |
| isoComp_00793204 | geneComp_00157694 | ENST00000590657 | ENSG00000232677 | LINC00665 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.057 | 0.057 | 3.687920e-03 | TRUE |
| isoComp_00639470 | geneComp_00136257 | ENST00000494350 | ENSG00000075826 | SEC31B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.051 | 0.051 | 3.701777e-03 | TRUE |
| isoComp_00637220 | geneComp_00136076 | ENST00000510118 | ENSG00000071242 | RPS6KA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.006 | 0.097 | 0.090 | 3.816778e-03 | TRUE |
| isoComp_00729586 | geneComp_00145703 | ENST00000486918 | ENSG00000162377 | COA7 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.079 | 0.022 | -0.056 | 4.010062e-03 | TRUE |
| isoComp_00780403 | geneComp_00152910 | MSTRG.22234.2 | ENSG00000205704 | SMIM45 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.152 | 0.000 | -0.152 | 4.119276e-03 | TRUE |
| isoComp_00752948 | geneComp_00148665 | MSTRG.16016.13 | ENSG00000175221 | MED16 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.143 | 0.078 | -0.065 | 4.265773e-03 | TRUE |
| isoComp_00741203 | geneComp_00147172 | ENST00000517898 | ENSG00000167912 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.868 | 0.868 | 4.346767e-03 | TRUE | |
| isoComp_00765025 | geneComp_00150539 | ENST00000335968 | ENSG00000185418 | TARS3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.390 | 0.521 | 0.131 | 4.694440e-03 | TRUE |
| isoComp_00663880 | geneComp_00138608 | ENST00000675950 | ENSG00000109099 | PMP22 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.629 | 0.037 | -0.592 | 4.756328e-03 | TRUE |
| isoComp_00671014 | geneComp_00139333 | ENST00000407482 | ENSG00000115129 | TP53I3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.142 | 0.000 | -0.142 | 4.829892e-03 | TRUE |
| isoComp_00739009 | geneComp_00146911 | ENST00000553533 | ENSG00000166888 | STAT6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.076 | 0.000 | -0.076 | 4.863107e-03 | |
| isoComp_00701916 | geneComp_00142688 | ENST00000261888 | ENSG00000138617 | PARP16 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.070 | 0.199 | 0.129 | 5.016085e-03 | TRUE |
| isoComp_00728559 | geneComp_00145581 | MSTRG.16992.28 | ENSG00000161265 | U2AF1L4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.081 | 0.000 | -0.081 | 5.016085e-03 | TRUE |
| isoComp_00719526 | geneComp_00144575 | MSTRG.29925.3 | ENSG00000152926 | ZNF117 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.155 | 0.000 | -0.155 | 5.189631e-03 | TRUE |
| isoComp_00721045 | geneComp_00144758 | ENST00000408548 | ENSG00000154545 | MAGED4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.129 | 0.129 | 5.407339e-03 | TRUE |
| isoComp_00774736 | geneComp_00151951 | ENST00000599599 | ENSG00000197951 | ZNF71 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.087 | 0.019 | -0.069 | 5.472413e-03 | |
| isoComp_00719473 | geneComp_00144569 | ENST00000630114 | ENSG00000152795 | HNRNPDL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.142 | 0.204 | 0.062 | 5.535978e-03 | TRUE |
| isoComp_00755218 | geneComp_00149000 | ENST00000564255 | ENSG00000177200 | CHD9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.058 | 0.058 | 5.553243e-03 | TRUE |
| isoComp_00797783 | geneComp_00159591 | ENST00000453576 | ENSG00000238009 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.324 | 0.000 | -0.324 | 5.679850e-03 | TRUE | |
| isoComp_00798196 | geneComp_00159777 | ENST00000463408 | ENSG00000239523 | MYLK-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.095 | 0.000 | -0.095 | 5.683654e-03 | TRUE |
| isoComp_00812856 | geneComp_00166250 | ENST00000686066 | ENSG00000266053 | NDUFV2-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.230 | 0.000 | -0.230 | 5.728871e-03 | TRUE |
| isoComp_00640904 | geneComp_00136380 | ENST00000360348 | ENSG00000079102 | RUNX1T1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.366 | 0.038 | -0.328 | 5.929096e-03 | TRUE |
| isoComp_00641583 | geneComp_00136439 | ENST00000503706 | ENSG00000080709 | KCNN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.029 | 0.323 | 0.294 | 5.982549e-03 | TRUE |
| isoComp_00766738 | geneComp_00150785 | ENST00000556766 | ENSG00000186469 | GNG2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.070 | 1.000 | 0.930 | 6.253013e-03 | TRUE |
| isoComp_00660121 | geneComp_00138224 | ENST00000652003 | ENSG00000106123 | EPHB6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.602 | 0.018 | -0.585 | 6.270451e-03 | TRUE |
| isoComp_00683145 | geneComp_00140680 | ENST00000586615 | ENSG00000125740 | FOSB | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.466 | 0.000 | -0.466 | 6.519476e-03 | TRUE |
| isoComp_00719474 | geneComp_00144569 | ENST00000630827 | ENSG00000152795 | HNRNPDL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.368 | 0.203 | -0.165 | 6.521369e-03 | TRUE |
| isoComp_00745884 | geneComp_00147723 | ENST00000342648 | ENSG00000170412 | GPRC5C | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.110 | 0.110 | 6.907048e-03 | TRUE |
| isoComp_00780601 | geneComp_00152959 | ENST00000382378 | ENSG00000205930 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.291 | 0.000 | -0.291 | 6.912658e-03 | TRUE | |
| isoComp_00677341 | geneComp_00139997 | ENST00000625311 | ENSG00000120159 | CAAP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.547 | 0.336 | -0.212 | 6.970383e-03 | TRUE |
| isoComp_00782827 | geneComp_00153597 | ENST00000496636 | ENSG00000214193 | SH3D21 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.175 | 0.000 | -0.175 | 7.396629e-03 | TRUE |
| isoComp_00792939 | geneComp_00157552 | ENST00000688125 | ENSG00000232284 | GNG12-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.755 | 0.049 | -0.706 | 7.707870e-03 | TRUE |
| isoComp_00697232 | geneComp_00142186 | ENST00000476849 | ENSG00000135951 | TSGA10 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.074 | 0.074 | 7.863198e-03 | TRUE |
| isoComp_00731628 | geneComp_00146004 | ENST00000405478 | ENSG00000163517 | HDAC11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.139 | 0.000 | -0.139 | 7.863198e-03 | TRUE |
| isoComp_00693843 | geneComp_00141831 | ENST00000559585 | ENSG00000134146 | DPH6 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.054 | 0.000 | -0.054 | 7.869553e-03 | |
| isoComp_00787910 | geneComp_00155451 | ENST00000638047 | ENSG00000226383 | LINC01876 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.140 | 0.000 | -0.140 | 7.878706e-03 | TRUE |
| isoComp_00704989 | geneComp_00142995 | ENST00000268184 | ENSG00000140577 | CRTC3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.595 | 0.372 | -0.223 | 8.263652e-03 | TRUE |
| isoComp_00639690 | geneComp_00136279 | MSTRG.7997.10 | ENSG00000076513 | ANKRD13A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.060 | 0.203 | 0.142 | 8.274397e-03 | TRUE |
| isoComp_00673156 | geneComp_00139532 | ENST00000474459 | ENSG00000116337 | AMPD2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.143 | 0.063 | -0.081 | 8.352932e-03 | TRUE |
| isoComp_00809469 | geneComp_00164459 | ENST00000561287 | ENSG00000259429 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.577 | 1.000 | 0.423 | 8.482162e-03 | TRUE | |
| isoComp_00736842 | geneComp_00146645 | ENST00000553296 | ENSG00000165804 | ZNF219 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.147 | 0.016 | -0.130 | 9.065749e-03 | TRUE |
| isoComp_00668999 | geneComp_00139142 | ENST00000494185 | ENSG00000113368 | LMNB1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.116 | 0.065 | -0.051 | 9.171235e-03 | TRUE |
| isoComp_00802721 | geneComp_00161280 | MSTRG.31466.5 | ENSG00000248498 | ASNSP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.323 | 0.020 | -0.303 | 9.183298e-03 | TRUE |
| isoComp_00722018 | geneComp_00144872 | ENST00000452799 | ENSG00000155744 | FAM126B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.070 | 0.070 | 9.366085e-03 | TRUE |
| isoComp_00706231 | geneComp_00143098 | ENST00000588927 | ENSG00000141401 | IMPA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.402 | 0.565 | 0.163 | 9.482093e-03 | TRUE |
| isoComp_00656072 | geneComp_00137816 | ENST00000354638 | ENSG00000104044 | OCA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.468 | 0.072 | -0.396 | 9.546281e-03 | TRUE |
| isoComp_00693368 | geneComp_00141784 | ENST00000480685 | ENSG00000133794 | ARNTL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.063 | 0.063 | 9.802318e-03 | TRUE |
| isoComp_00646087 | geneComp_00136844 | ENST00000492339 | ENSG00000091073 | DTX2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.068 | 0.068 | 9.934229e-03 | TRUE |
| isoComp_00763241 | geneComp_00150251 | ENST00000642510 | ENSG00000184154 | LRRC51 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.066 | 0.000 | -0.066 | 1.001385e-02 | TRUE |
| isoComp_00633481 | geneComp_00135749 | ENST00000491464 | ENSG00000061273 | HDAC7 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.069 | 0.069 | 1.011918e-02 | TRUE |
| isoComp_00772310 | geneComp_00151639 | ENST00000588163 | ENSG00000196867 | ZFP28 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.179 | 0.000 | -0.179 | 1.014782e-02 | TRUE |
| isoComp_00628299 | geneComp_00135267 | ENST00000553437 | ENSG00000015133 | CCDC88C | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.061 | 0.000 | -0.061 | 1.040631e-02 | TRUE |
| isoComp_00673557 | geneComp_00139567 | MSTRG.2748.4 | ENSG00000116679 | IVNS1ABP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.200 | 0.108 | -0.092 | 1.058871e-02 | |
| isoComp_00763603 | geneComp_00150309 | MSTRG.20949.2 | ENSG00000184402 | SS18L1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.067 | 0.000 | -0.067 | 1.067993e-02 | TRUE |
| isoComp_00722385 | geneComp_00144922 | ENST00000286648 | ENSG00000156136 | DCK | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.833 | 0.931 | 0.098 | 1.085791e-02 | TRUE |
| isoComp_00786360 | geneComp_00154833 | ENST00000452867 | ENSG00000224687 | RASAL2-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.439 | 0.439 | 1.124801e-02 | TRUE |
| isoComp_00742840 | geneComp_00147328 | MSTRG.25785.2 | ENSG00000168564 | CDKN2AIP | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.282 | 0.165 | -0.117 | 1.134728e-02 | TRUE |
| isoComp_00666273 | geneComp_00138837 | ENST00000545990 | ENSG00000111254 | AKAP3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.294 | 0.000 | -0.294 | 1.228867e-02 | TRUE |
| isoComp_00696902 | geneComp_00142156 | ENST00000367549 | ENSG00000135829 | DHX9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.908 | 0.858 | -0.050 | 1.228867e-02 | |
| isoComp_00809558 | geneComp_00164500 | ENST00000637567 | ENSG00000259511 | UBE2Q2L | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.368 | 0.368 | 1.228867e-02 | TRUE |
| isoComp_00751826 | geneComp_00148519 | ENST00000564179 | ENSG00000174442 | ZWILCH | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.165 | 0.087 | -0.079 | 1.256575e-02 | |
| isoComp_00637217 | geneComp_00136076 | ENST00000503859 | ENSG00000071242 | RPS6KA2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.067 | 0.000 | -0.067 | 1.275879e-02 | TRUE |
| isoComp_00667210 | geneComp_00138932 | MSTRG.28852.3 | ENSG00000111817 | DSE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.115 | 0.115 | 1.286280e-02 | TRUE |
| isoComp_00755635 | geneComp_00149068 | ENST00000357573 | ENSG00000177548 | RABEP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.004 | 0.060 | 0.056 | 1.286350e-02 | |
| isoComp_00748593 | geneComp_00148117 | ENST00000511321 | ENSG00000172239 | PAIP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.212 | 0.137 | -0.075 | 1.306337e-02 | TRUE |
| isoComp_00789509 | geneComp_00156106 | ENST00000653554 | ENSG00000228274 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.087 | 0.087 | 1.311176e-02 | TRUE | |
| isoComp_00778102 | geneComp_00152498 | ENST00000463834 | ENSG00000204128 | C2orf72 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.840 | 0.047 | -0.793 | 1.339713e-02 | TRUE |
| isoComp_00746361 | geneComp_00147797 | ENST00000394510 | ENSG00000170776 | AKAP13 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.097 | 0.000 | -0.097 | 1.433105e-02 | |
| isoComp_00710212 | geneComp_00143520 | ENST00000410065 | ENSG00000144040 | SFXN5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.106 | 0.007 | -0.099 | 1.553249e-02 | TRUE |
| isoComp_00767012 | geneComp_00150822 | ENST00000336985 | ENSG00000186654 | PRR5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.316 | 0.176 | -0.140 | 1.608947e-02 | TRUE |
| isoComp_00788966 | geneComp_00155870 | ENST00000394864 | ENSG00000227540 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.075 | 0.000 | -0.075 | 1.632116e-02 | ||
| isoComp_00641582 | geneComp_00136439 | ENST00000264773 | ENSG00000080709 | KCNN2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.597 | 0.000 | -0.597 | 1.651238e-02 | TRUE |
| isoComp_00704653 | geneComp_00142965 | ENST00000436891 | ENSG00000140464 | PML | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.064 | 0.000 | -0.064 | 1.788870e-02 | TRUE |
| isoComp_00644899 | geneComp_00136742 | ENST00000337227 | ENSG00000089050 | RBBP9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.788 | 0.619 | -0.169 | 1.816895e-02 | TRUE |
| isoComp_00746811 | geneComp_00147856 | MSTRG.5851.3 | ENSG00000171067 | C11orf24 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.128 | 0.217 | 0.090 | 1.825801e-02 | TRUE |
| isoComp_00658632 | geneComp_00138063 | ENST00000560253 | ENSG00000105419 | MEIS3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.063 | 0.000 | -0.063 | 1.874424e-02 | TRUE |
| isoComp_00631705 | geneComp_00135592 | ENST00000495799 | ENSG00000049769 | PPP1R3F | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.090 | 0.000 | -0.090 | 1.876060e-02 | TRUE |
| isoComp_00704064 | geneComp_00142918 | MSTRG.10185.16 | ENSG00000140105 | WARS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.199 | 0.129 | -0.070 | 1.877703e-02 | |
| isoComp_00724565 | geneComp_00145164 | MSTRG.33042.26 | ENSG00000158169 | FANCC | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.091 | 0.004 | -0.086 | 1.976705e-02 | |
| isoComp_00822865 | geneComp_00171722 | ENST00000578189 | ENSG00000286219 | NOTCH2NLC | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.207 | 0.207 | 1.989820e-02 | TRUE |
| isoComp_00673420 | geneComp_00139552 | ENST00000361247 | ENSG00000116584 | ARHGEF2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.262 | 0.542 | 0.281 | 2.009374e-02 | TRUE |
| isoComp_00682803 | geneComp_00140634 | ENST00000663987 | ENSG00000125462 | MIR9-1HG | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.159 | 0.013 | -0.145 | 2.092800e-02 | TRUE |
| isoComp_00659030 | geneComp_00138111 | ENST00000481914 | ENSG00000105649 | RAB3A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.074 | 0.000 | -0.074 | 2.106976e-02 | |
| isoComp_00730113 | geneComp_00145783 | ENST00000427444 | ENSG00000162669 | HFM1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.214 | 0.000 | -0.214 | 2.142861e-02 | TRUE |
| isoComp_00634152 | geneComp_00135809 | MSTRG.16759.13 | ENSG00000064489 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.058 | 0.058 | 2.145626e-02 | TRUE | |
| isoComp_00804575 | geneComp_00162083 | ENST00000504145 | ENSG00000251136 | RIPK2-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.052 | 0.000 | -0.052 | 2.175710e-02 | TRUE |
| isoComp_00629616 | geneComp_00135398 | ENST00000537505 | ENSG00000029725 | RABEP1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.650 | 0.491 | -0.159 | 2.249882e-02 | |
| isoComp_00670236 | geneComp_00139262 | ENST00000459638 | ENSG00000114541 | FRMD4B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.094 | 0.094 | 2.352853e-02 | TRUE |
| isoComp_00780870 | geneComp_00153023 | ENST00000383768 | ENSG00000206559 | ZCWPW2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.767 | 0.000 | -0.767 | 2.368205e-02 | TRUE |
| isoComp_00766781 | geneComp_00150791 | ENST00000580995 | ENSG00000186481 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.215 | 0.215 | 2.412336e-02 | TRUE | |
| isoComp_00732742 | geneComp_00146136 | ENST00000296255 | ENSG00000163902 | RPN1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.993 | 0.931 | -0.062 | 2.476053e-02 | TRUE |
| isoComp_00800259 | geneComp_00160571 | ENST00000691958 | ENSG00000243176 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.401 | 0.401 | 2.501603e-02 | TRUE | |
| isoComp_00763232 | geneComp_00150251 | ENST00000536917 | ENSG00000184154 | LRRC51 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.060 | 0.060 | 2.511579e-02 | TRUE |
| isoComp_00794365 | geneComp_00158191 | ENST00000687978 | ENSG00000234062 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.138 | 0.000 | -0.138 | 2.511579e-02 | TRUE | |
| isoComp_00787833 | geneComp_00155416 | ENST00000445836 | ENSG00000226287 | TMEM191A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.128 | 0.000 | -0.128 | 2.517099e-02 | TRUE |
| isoComp_00814797 | geneComp_00167119 | MSTRG.27872.15 | ENSG00000269293 | ZSCAN16-AS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.333 | 0.198 | -0.135 | 2.517099e-02 | TRUE |
| isoComp_00809359 | geneComp_00164409 | ENST00000560973 | ENSG00000259343 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.071 | 0.071 | 2.521037e-02 | TRUE | |
| isoComp_00807247 | geneComp_00163337 | ENST00000457155 | ENSG00000256061 | DNAAF4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.428 | 0.149 | -0.279 | 2.530505e-02 | TRUE |
| isoComp_00717929 | geneComp_00144368 | ENST00000421992 | ENSG00000151240 | DIP2C | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.104 | 0.000 | -0.104 | 2.535549e-02 | TRUE |
| isoComp_00718362 | geneComp_00144409 | MSTRG.10869.3 | ENSG00000151575 | TEX9 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.054 | 0.000 | -0.054 | 2.585948e-02 | TRUE |
| isoComp_00724457 | geneComp_00145151 | ENST00000349752 | ENSG00000158089 | GALNT14 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.679 | -0.321 | 2.615006e-02 | TRUE |
| isoComp_00697360 | geneComp_00142201 | ENST00000537435 | ENSG00000136014 | USP44 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.158 | 0.000 | -0.158 | 2.617296e-02 | TRUE |
| isoComp_00749908 | geneComp_00148267 | ENST00000389617 | ENSG00000172986 | GXYLT2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.145 | 0.005 | -0.140 | 2.753493e-02 | TRUE |
| isoComp_00636399 | geneComp_00136000 | ENST00000479877 | ENSG00000069493 | CLEC2D | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.040 | 0.269 | 0.229 | 2.770425e-02 | TRUE |
| isoComp_00706077 | geneComp_00143085 | ENST00000578006 | ENSG00000141314 | RHBDL3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.100 | 0.100 | 2.804441e-02 | TRUE |
| isoComp_00783270 | geneComp_00153730 | ENST00000555861 | ENSG00000214770 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.038 | 0.498 | 0.460 | 2.829854e-02 | TRUE | |
| isoComp_00726308 | geneComp_00145372 | ENST00000291041 | ENSG00000159792 | PSKH1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.952 | 0.842 | -0.110 | 2.879974e-02 | TRUE |
| isoComp_00669391 | geneComp_00139183 | ENST00000393784 | ENSG00000113719 | ERGIC1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.915 | 0.762 | -0.152 | 2.973035e-02 | TRUE |
| isoComp_00743052 | geneComp_00147350 | ENST00000369779 | ENSG00000168679 | SLC16A4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.232 | 0.000 | -0.232 | 2.973215e-02 | TRUE |
| isoComp_00659673 | geneComp_00138179 | ENST00000430096 | ENSG00000105928 | GSDME | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.179 | 0.179 | 2.982928e-02 | TRUE |
| isoComp_00642973 | geneComp_00136569 | ENST00000395856 | ENSG00000084676 | NCOA1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.006 | 0.103 | 0.097 | 2.990180e-02 | TRUE |
| isoComp_00770432 | geneComp_00151420 | MSTRG.12813.7 | ENSG00000196155 | PLEKHG4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.010 | 0.154 | 0.144 | 2.991332e-02 | TRUE |
| isoComp_00625485 | geneComp_00135024 | ENST00000320031 | ENSG00000005884 | ITGA3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.585 | 0.286 | -0.299 | 3.025081e-02 | TRUE |
| isoComp_00696946 | geneComp_00142159 | ENST00000488424 | ENSG00000135838 | NPL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.069 | 0.069 | 3.028243e-02 | TRUE |
| isoComp_00774890 | geneComp_00151967 | ENST00000551736 | ENSG00000198001 | IRAK4 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.125 | 0.000 | -0.125 | 3.043346e-02 | TRUE |
| isoComp_00664638 | geneComp_00138678 | ENST00000514956 | ENSG00000109794 | FAM149A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.045 | 0.257 | 0.212 | 3.135900e-02 | TRUE |
| isoComp_00711750 | geneComp_00143676 | ENST00000354839 | ENSG00000145242 | EPHA5 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.415 | 0.000 | -0.415 | 3.171758e-02 | TRUE |
| isoComp_00637841 | geneComp_00136120 | ENST00000513765 | ENSG00000072518 | MARK2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.004 | 0.104 | 0.100 | 3.174112e-02 | TRUE |
| isoComp_00650887 | geneComp_00137300 | ENST00000553695 | ENSG00000100605 | ITPK1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.183 | 0.341 | 0.158 | 3.207563e-02 | |
| isoComp_00806985 | geneComp_00163233 | ENST00000425266 | ENSG00000255624 | C10orf88B | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.091 | 0.000 | -0.091 | 3.220772e-02 | TRUE |
| isoComp_00691125 | geneComp_00141540 | ENST00000565081 | ENSG00000132207 | SLX1A | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.062 | 0.150 | 0.088 | 3.234919e-02 | TRUE |
| isoComp_00739782 | geneComp_00147003 | ENST00000585160 | ENSG00000167280 | ENGASE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.305 | 0.160 | -0.144 | 3.264162e-02 | TRUE |
| isoComp_00739776 | geneComp_00147003 | ENST00000577783 | ENSG00000167280 | ENGASE | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.082 | 0.164 | 0.082 | 3.319892e-02 | TRUE |
| isoComp_00812731 | geneComp_00166190 | ENST00000582106 | ENSG00000265688 | MAFG-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.625 | 0.160 | -0.466 | 3.566932e-02 | TRUE |
| isoComp_00734932 | geneComp_00146399 | ENST00000467726 | ENSG00000164776 | PHKG1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.197 | 0.197 | 3.612928e-02 | TRUE |
| isoComp_00817142 | geneComp_00168450 | MSTRG.30966.3 | ENSG00000273344 | PAXIP1-DT | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.074 | 0.006 | -0.068 | 3.624400e-02 | TRUE |
| isoComp_00664312 | geneComp_00138647 | MSTRG.25307.5 | ENSG00000109534 | GAR1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.183 | 0.296 | 0.112 | 3.626056e-02 | TRUE |
| isoComp_00754553 | geneComp_00148909 | ENST00000540697 | ENSG00000176715 | ACSF3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.075 | 0.023 | -0.053 | 3.658672e-02 | TRUE |
| isoComp_00814946 | geneComp_00167183 | ENST00000601192 | ENSG00000269604 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.913 | 0.913 | 3.734242e-02 | TRUE | |
| isoComp_00662933 | geneComp_00138507 | MSTRG.13739.1 | ENSG00000108474 | PIGL | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.104 | 0.035 | -0.068 | 3.735932e-02 | TRUE |
| isoComp_00674697 | geneComp_00139693 | ENST00000487539 | ENSG00000117519 | CNN3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.069 | 0.149 | 0.080 | 3.974255e-02 | TRUE |
| isoComp_00664288 | geneComp_00138645 | ENST00000513395 | ENSG00000109501 | WFS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.098 | 0.225 | 0.128 | 4.023420e-02 | TRUE |
| isoComp_00628737 | geneComp_00135316 | ENST00000227214 | ENSG00000021300 | PLEKHB1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 1.000 | 0.931 | -0.069 | 4.043408e-02 | |
| isoComp_00737985 | geneComp_00146788 | ENST00000299492 | ENSG00000166387 | PPFIBP2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.753 | 0.286 | -0.467 | 4.045304e-02 | TRUE |
| isoComp_00750377 | geneComp_00148327 | ENST00000398505 | ENSG00000173276 | ZBTB21 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.079 | 0.192 | 0.113 | 4.156421e-02 | TRUE |
| isoComp_00711095 | geneComp_00143600 | MSTRG.22640.9 | ENSG00000144642 | RBMS3 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.609 | 0.609 | 4.296213e-02 | TRUE |
| isoComp_00729614 | geneComp_00145708 | ENST00000481208 | ENSG00000162390 | ACOT11 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.080 | 0.697 | 0.617 | 4.341848e-02 | TRUE |
| isoComp_00753538 | geneComp_00148733 | ENST00000691767 | ENSG00000175611 | LINC00476 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.148 | 0.320 | 0.172 | 4.429781e-02 | TRUE |
| isoComp_00807889 | geneComp_00163647 | ENST00000657514 | ENSG00000257337 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.212 | 0.212 | 4.545363e-02 | TRUE | |
| isoComp_00808271 | geneComp_00163814 | ENST00000397640 | ENSG00000257949 | TEN1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.447 | 0.606 | 0.159 | 4.561500e-02 | TRUE |
| isoComp_00723173 | geneComp_00145016 | ENST00000323963 | ENSG00000156976 | EIF4A2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.341 | 0.174 | -0.167 | 4.662932e-02 | TRUE |
| isoComp_00670013 | geneComp_00139241 | ENST00000490574 | ENSG00000114354 | TFG | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.203 | 0.308 | 0.106 | 4.702865e-02 | TRUE |
| isoComp_00694543 | geneComp_00141912 | ENST00000256969 | ENSG00000134548 | SPX | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.000 | 0.524 | 0.524 | 4.762684e-02 | TRUE |
| isoComp_00688999 | geneComp_00141337 | ENST00000341952 | ENSG00000130783 | CCDC62 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.142 | 0.888 | 0.746 | 4.919153e-02 | TRUE |
| isoComp_00641321 | geneComp_00136413 | ENST00000337057 | ENSG00000079819 | EPB41L2 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.339 | 0.264 | -0.075 | 4.943589e-02 | |
| isoComp_00687109 | geneComp_00141129 | MSTRG.31214.13 | ENSG00000129422 | MTUS1 | HEK293_DMSO_6hB | HEK293_TMG_6hB | 0.055 | 0.000 | -0.055 | 4.978092e-02 | TRUE |
- Note:
- The comparisons made can be identified as “from ‘condition_1’ to ‘condition_2’”, meaning ‘condition_1’ is considered the ground state and ‘condition_2’ the changed state. This also means that a positive dIF value indicates that the isoform usage is increased in ‘condition_2’ compared to ‘condition_1’. Since the ‘isoformFeatures’ entry is the most relevant part of the switchAnalyzeRlist object, the most-used standard methods have also been implemented to work directly on isoformFeatures.
4. Circular RNA Analysis
Volcano Plot
No significantly changed circRNA was detected.
Details of sig. circRNA
No significantly changed circRNA was detected.
Annotation of sig. circRNA
No significantly changed circRNA was detected.