circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000172915:+:13:35040932:35045401
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000172915:+:13:35040932:35045401 | ENSG00000172915 | ENST00000379939 | + | 13 | 35040933 | 35045401 | 429 | CUGGUUGGUGGAGAAUUUGACUUGGAGAUGAACUUUAUUAUCCAGGAUGCUGAGAGUAUAACAUGUAUGACAGAGCUUUUGGAGCACUGUGAUGUAACAUGUCAAGCAGAAAUAUGGAGCAUGUUUACAGCCAUUCUACGAAAAAGUGUUCGGAAUUUACAGACUAGCACAGAAGUUGGGCUAAUUGAACAAGUAUUGCUGAAAAUGAGUGCUGUAGAUGACAUGAUAGCAGAUCUUCUAGUUGAUAUGUUGGGGGUUCUUGCCAGCUACAGCAUCACUGUCAAGGAGUUGAAGCUUUUGUUCAGCAUGCUUCGAGGAGAAAGUGGAAUCUGGCCAAGACAUGCAGUAAAAUUAUUAUCAGUUCUUAAUCAGAUGCCACAGAGACACGGUCCUGAUACUUUUUUCAAUUUCCCUGGUUGUAGCGCUGCGCUGGUUGGUGGAGAAUUUGACUUGGAGAUGAACUUUAUUAUCCAGGAUGC | circ |
| ENSG00000172915:+:13:35040932:35045401 | ENSG00000172915 | ENST00000379939 | + | 13 | 35040933 | 35045401 | 22 | GUAGCGCUGCGCUGGUUGGUGG | bsj |
| ENSG00000172915:+:13:35040932:35045401 | ENSG00000172915 | ENST00000379939 | + | 13 | 35040733 | 35040942 | 210 | UAGAUUUCUGUUUGCUAGUUUCGAUGCUCACAAUGCCUUUCACUUAUUUGUAGCUCUUAUACUUUUGAUCAAAAUUAAUUUUAUACCAGAAUCCUUGAAAUUUUUUAGAGUAGUAGCUUUGAAUUCUUGUUAUUAUUUCAUUCUACUGUCAUCGAUAACCUCCCAUGUUAACACUAUUGUUUCUUUCUGUUUACUUUCAGCUGGUUGGUG | ie_up |
| ENSG00000172915:+:13:35040932:35045401 | ENSG00000172915 | ENST00000379939 | + | 13 | 35045392 | 35045601 | 210 | UAGCGCUGCGGUAAGUUUUAAAUACAUGUGCUGAUUUUUAUUUAUUUAUUUUUAGGUCACCUUUAGUAAGGUUUAACUUACAUAUAAUAAAAUUUACCAGUUUAGAAAGCACAACUUGAUGAGUUUGGAAGUCAUACAACCACUACUAUAUAAUGAUUUGAACAAUUUCAUCACUCCCCAAAAUUCCCUUGUGCCUGUUGGAAGAUAUUC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
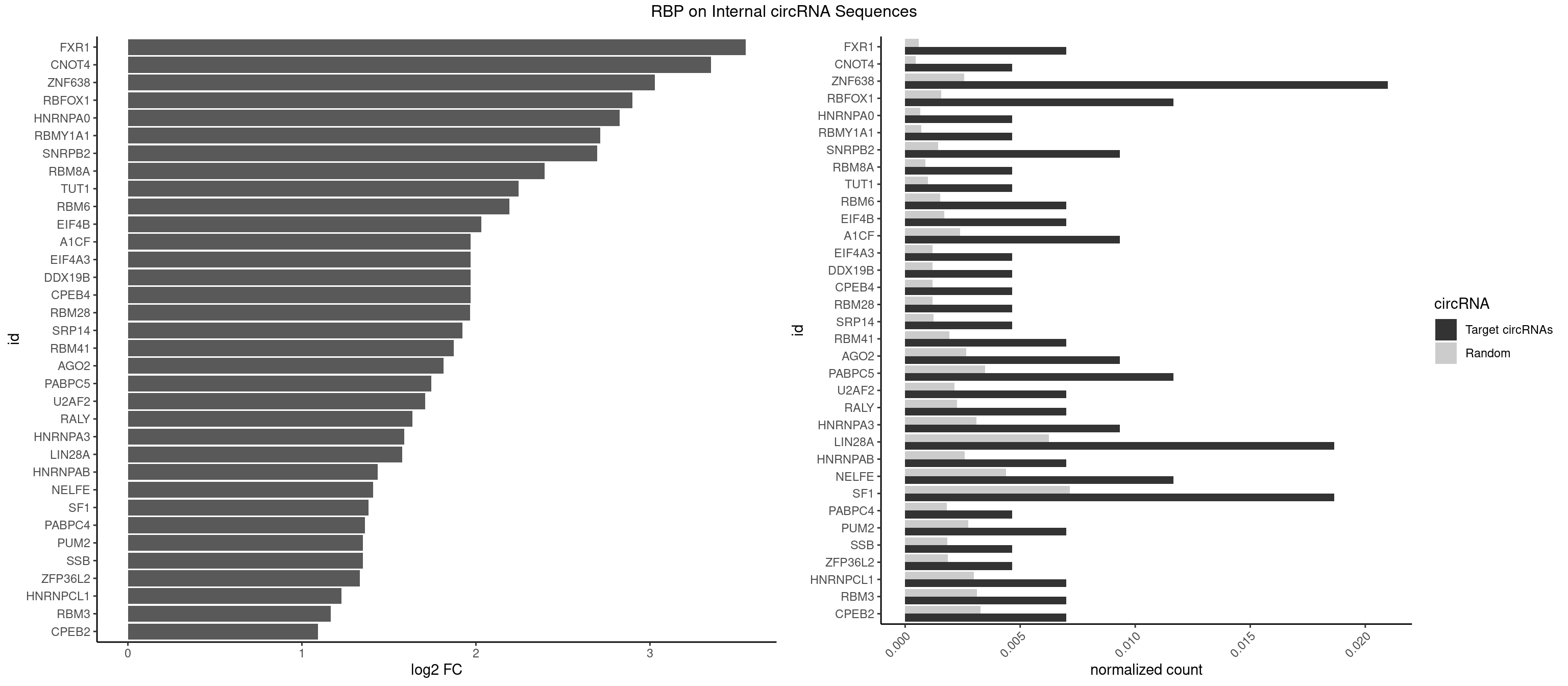
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 2 | 411 | 0.006993007 | 0.0005970720 | 3.549936 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| CNOT4 | 1 | 314 | 0.004662005 | 0.0004564992 | 3.352266 | GACAGA | GACAGA |
| ZNF638 | 8 | 1773 | 0.020979021 | 0.0025708878 | 3.028609 | GGUUCU,GGUUGG,GGUUGU,GUUCUU,GUUGGU,UGUUCG,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBFOX1 | 4 | 1077 | 0.011655012 | 0.0015622419 | 2.899261 | AGCAUG,GCAUGC,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.004662005 | 0.0006579386 | 2.824926 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.004662005 | 0.0007101099 | 2.714836 | CAAGAC | ACAAGA,CAAGAC |
| SNRPB2 | 3 | 991 | 0.009324009 | 0.0014376103 | 2.697278 | GUAUUG,UAUUGC,UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM8A | 1 | 611 | 0.004662005 | 0.0008869128 | 2.394086 | UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| TUT1 | 1 | 678 | 0.004662005 | 0.0009840095 | 2.244206 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.006993007 | 0.0015289102 | 2.193409 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| EIF4B | 2 | 1179 | 0.006993007 | 0.0017100607 | 2.031865 | CUUGGA,UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| A1CF | 3 | 1642 | 0.009324009 | 0.0023810421 | 1.969357 | AGUAUA,AUCAGU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CPEB4 | 1 | 821 | 0.004662005 | 0.0011912456 | 1.968480 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.004662005 | 0.0011912456 | 1.968480 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.004662005 | 0.0011912456 | 1.968480 | UUUUUU | UUUUUU |
| RBM28 | 1 | 822 | 0.004662005 | 0.0011926949 | 1.966725 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRP14 | 1 | 847 | 0.004662005 | 0.0012289250 | 1.923554 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM41 | 2 | 1318 | 0.006993007 | 0.0019115000 | 1.871208 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO2 | 3 | 1830 | 0.009324009 | 0.0026534924 | 1.813058 | AAAGUG,AGUGCU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPC5 | 4 | 2400 | 0.011655012 | 0.0034795387 | 1.743982 | AGAAAG,AGAAAU,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| U2AF2 | 2 | 1477 | 0.006993007 | 0.0021419234 | 1.707006 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 2 | 1553 | 0.006993007 | 0.0022520629 | 1.634666 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPA3 | 3 | 2140 | 0.009324009 | 0.0031027457 | 1.587405 | AAGGAG,CAAGGA,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| LIN28A | 7 | 4315 | 0.018648019 | 0.0062547643 | 1.575995 | AGGAGA,AGGAGU,GGAGAA,GGAGAU,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPAB | 2 | 1782 | 0.006993007 | 0.0025839306 | 1.436346 | AAGACA,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NELFE | 4 | 3028 | 0.011655012 | 0.0043896388 | 1.408776 | CUGGUU,GGCUAA,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SF1 | 7 | 4931 | 0.018648019 | 0.0071474739 | 1.383517 | ACAGAC,ACUAGC,CACAGA,UACGAA,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PABPC4 | 1 | 1251 | 0.004662005 | 0.0018144033 | 1.361455 | AAAAAG | AAAAAA,AAAAAG |
| PUM2 | 2 | 1890 | 0.006993007 | 0.0027404447 | 1.351503 | GUAGAU,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SSB | 1 | 1260 | 0.004662005 | 0.0018274462 | 1.351122 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ZFP36L2 | 1 | 1277 | 0.004662005 | 0.0018520827 | 1.331802 | UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPCL1 | 2 | 2062 | 0.006993007 | 0.0029897078 | 1.225908 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RBM3 | 2 | 2152 | 0.006993007 | 0.0031201361 | 1.164304 | AGACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CPEB2 | 2 | 2261 | 0.006993007 | 0.0032780993 | 1.093053 | CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
RBP on BSJ (Exon Only)
Plot
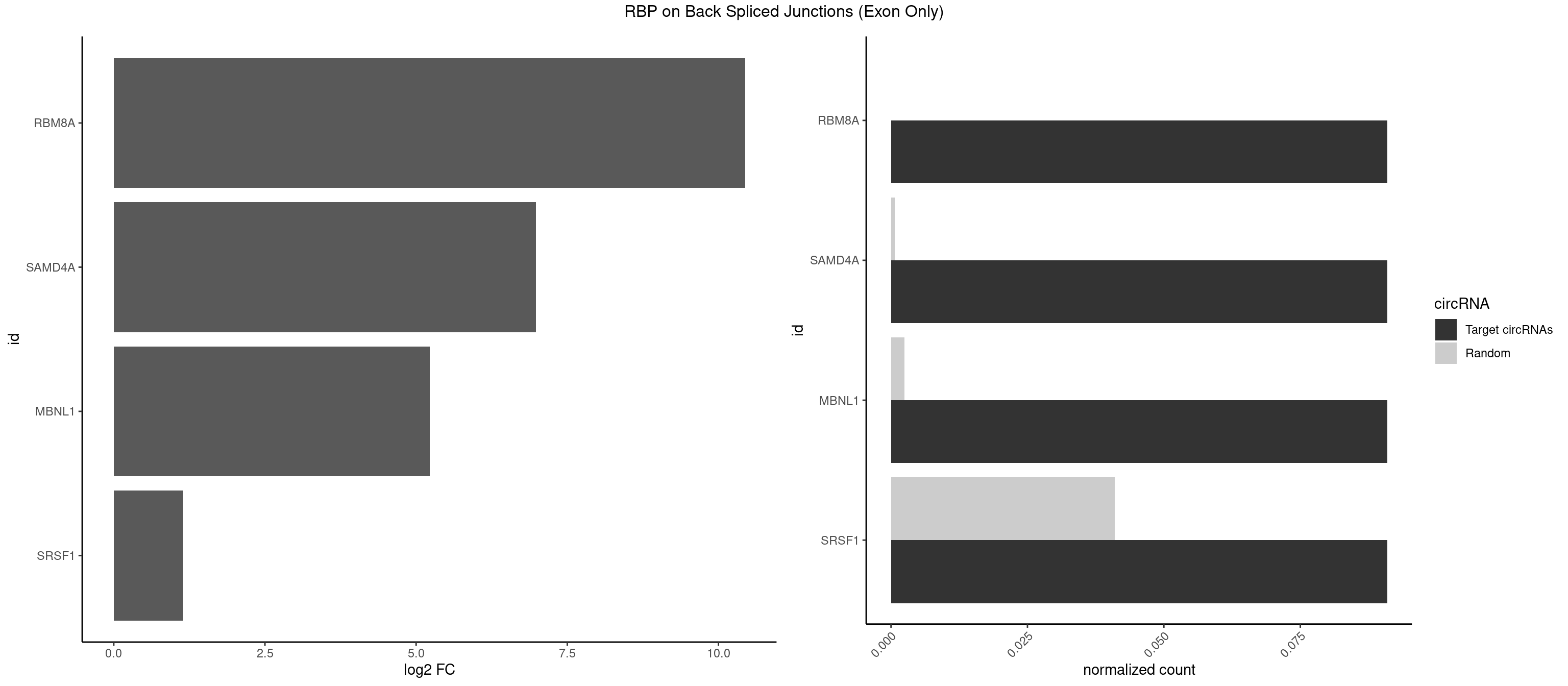
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 1 | 0 | 0.09090909 | 6.549646e-05 | 10.438792 | UGCGCU | |
| SAMD4A | 1 | 10 | 0.09090909 | 7.204611e-04 | 6.979360 | GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGUA,CUGGAA,CUGGCA,CUGGCC,CUGGUA,GCUGGU |
| MBNL1 | 1 | 36 | 0.09090909 | 2.423369e-03 | 5.229338 | GCGCUG | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
| SRSF1 | 1 | 625 | 0.09090909 | 4.100079e-02 | 1.148773 | CGCUGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
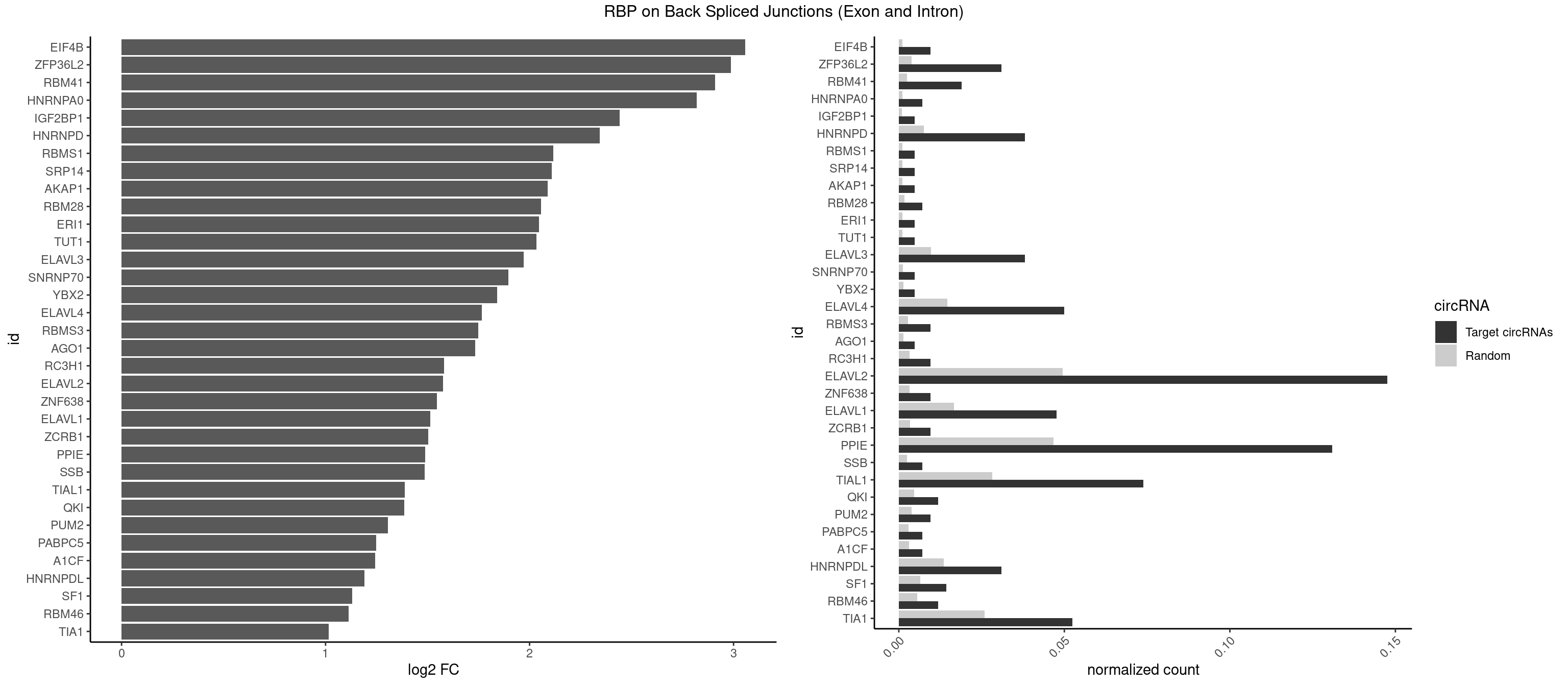
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| EIF4B | 3 | 226 | 0.009523810 | 0.0011436921 | 3.057840 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZFP36L2 | 12 | 774 | 0.030952381 | 0.0039046755 | 2.986776 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM41 | 7 | 502 | 0.019047619 | 0.0025342604 | 2.909974 | AUACAU,AUACUU,UACAUG,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPD | 15 | 1488 | 0.038095238 | 0.0075020153 | 2.344261 | AAUUUA,AGAUAU,AUUUAU,UAUUUA,UUAGAG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | UAUAUA | AUAUAU,UAUAUA |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ELAVL3 | 15 | 1928 | 0.038095238 | 0.0097188634 | 1.970751 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ELAVL4 | 20 | 2916 | 0.050000000 | 0.0146966949 | 1.766436 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | CAUAUA,CUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ELAVL2 | 61 | 9826 | 0.147619048 | 0.0495112858 | 1.576050 | AAUUUA,AAUUUU,AUACUU,AUUAUU,AUUUAC,AUUUAU,AUUUUA,AUUUUU,CUUAUA,CUUUCU,GAUUUU,GUUUUA,UAAGUU,UAAUUU,UACUUU,UAUACU,UAUAUA,UAUUAU,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCUUAU,UGAUUU,UUAAUU,UUACUU,UUAUAC,UUAUUU,UUCAUU,UUCUUU,UUUACU,UUUAGU,UUUAUA,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUGG,GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ELAVL1 | 19 | 3309 | 0.047619048 | 0.0166767432 | 1.513701 | AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PPIE | 54 | 9262 | 0.130952381 | 0.0466696896 | 1.488485 | AAAAUU,AAAUUA,AAAUUU,AAUAAA,AAUUAA,AAUUUA,AAUUUU,AUAAAA,AUAAUA,AUAUAA,AUUAAU,AUUAUU,AUUUAU,AUUUUA,AUUUUU,UAAAAU,UAAAUA,UAAUAA,UAAUUU,UAUAAU,UAUAUA,UAUUAU,UAUUUA,UAUUUU,UUAAAU,UUAAUU,UUAUUA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| TIAL1 | 30 | 5597 | 0.073809524 | 0.0282043531 | 1.387889 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUU,CAGUUU,CUUUUG,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACACUA,ACUUAU,UAACCU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAAAUA,UACAUA,UAUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPDL | 12 | 2689 | 0.030952381 | 0.0135530028 | 1.191438 | AAUUUA,ACCAGA,AGAUAU,AGUAGC,AUACCA,CCUUUA,CUUUAG,GAGUAG,GCUAGU,GUAGCU,UUUAGG | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUUAU,AGUAAG,UAGUAA,UAUACU,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUGAU,AUCAAA,AUGAUU,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUU,CCUUUC,CUUUUG,GUUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.