circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000133059:-:1:205187417:205187806
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000133059:-:1:205187417:205187806 | ENSG00000133059 | ENST00000367162 | - | 1 | 205187418 | 205187806 | 389 | GCCAACUGAGCUGCAUUUCCUUCCCACCUAAGGAAGAGAAGUACCUCCAGCAGAUUGUGGACUGCCUCCCUUGCAUACUGAUCCUCGGCCAGGAUUGUAACGUCAAGUGCCAGCUGUUGAAUCUGCUGUUGGGGGUGCAGGUGCUUCCCACCACCAAGCUGGGCAGUGAGGAGAGCUGUAAGCUUCGGCGCCUCCGCUUCACCUAUGGGACUCAGACUCGGGUCAGCCUGGCGCUCCCUGGACAGUAUGAACUAGUGCACACGCUGGUUGCUCAUCAGGGCAACUGGGAGACCAUCCCUGAGGAGGAUCUGGAGGUCCAAGAGAACAAUGAGGAUGCUGCUCAUGUUUUAGCGGAACUGGAGGUAACGAUGCACCAUGCUCUCUUACAGGCCAACUGAGCUGCAUUUCCUUCCCACCUAAGGAAGAGAAGUACCUCCAG | circ |
| ENSG00000133059:-:1:205187417:205187806 | ENSG00000133059 | ENST00000367162 | - | 1 | 205187418 | 205187806 | 22 | CUCUCUUACAGGCCAACUGAGC | bsj |
| ENSG00000133059:-:1:205187417:205187806 | ENSG00000133059 | ENST00000367162 | - | 1 | 205187797 | 205188006 | 210 | GGAAUUCUGUUUGAUAUGUGAAGAGCUUUUUCUCACUGGGGUUUUUUCCCUGAUAUGUCCUUCAACCAAGGUUCCUAGAAAUUUCUCCUCUAGGACCUUGUCUUCCUCAUGGGAUUUCUGCGUGAGUCUCUCUCUCUCCCCUUCCUCACUCCUCUCUCUCUACUCCUUUCUCUUCCUCCACUCUCCCCUUCCCCAACCAGGCCAACUGAG | ie_up |
| ENSG00000133059:-:1:205187417:205187806 | ENSG00000133059 | ENST00000367162 | - | 1 | 205187218 | 205187427 | 210 | UCUCUUACAGGUACCAAUCUCAUACUUCUAUACCAAUUGUAUACAUAUAUACCAGCUUUCCUUUUAUUUUGACUUUCUAAAAAAGUAAUUCGAUGAUAUAUAGUCUGAACUCUGAGCAGCAGUGGCUAGUGGAGUCAGAGGAACAUAAUCUGGGAAGUUGAAAAACAAACAAGGCUGUAGCAAUACCCAUUCCUGCCUCUCCUUACCAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
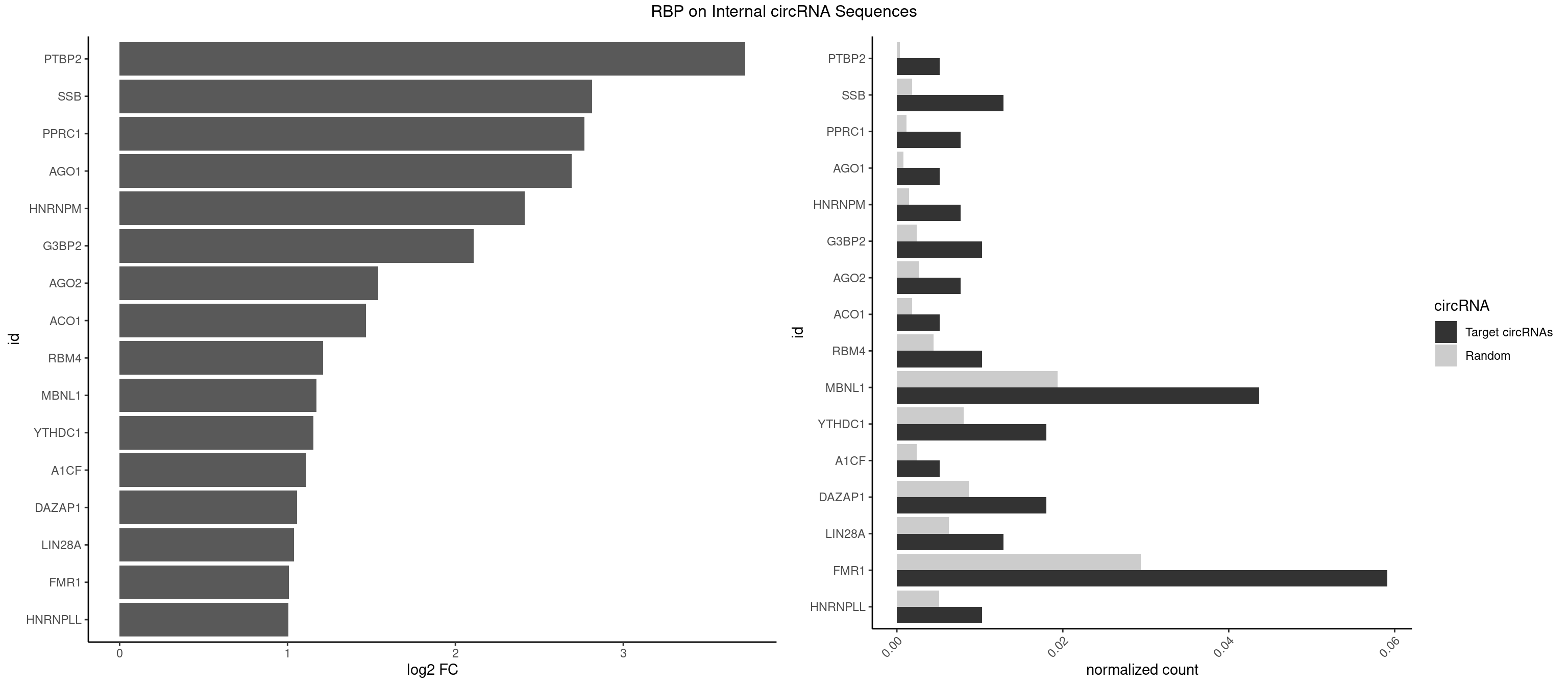
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.005141388 | 0.0003883867 | 3.726592 | CUCUCU | CUCUCU |
| SSB | 4 | 1260 | 0.012853470 | 0.0018274462 | 2.814257 | GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| PPRC1 | 2 | 780 | 0.007712082 | 0.0011318283 | 2.768465 | CGGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| AGO1 | 1 | 548 | 0.005141388 | 0.0007956130 | 2.692019 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPM | 2 | 999 | 0.007712082 | 0.0014492040 | 2.411860 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 3 | 1644 | 0.010282776 | 0.0023839405 | 2.108810 | AGGAUG,AGGAUU,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| AGO2 | 2 | 1830 | 0.007712082 | 0.0026534924 | 1.539228 | AAGUGC,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ACO1 | 1 | 1283 | 0.005141388 | 0.0018607779 | 1.466252 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM4 | 3 | 3065 | 0.010282776 | 0.0044432593 | 1.210540 | CCUUCC,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| MBNL1 | 16 | 13372 | 0.043701799 | 0.0193802045 | 1.173109 | ACGCUG,AUGCUC,CCGCUU,CGCUUC,CUGCCU,CUGCUC,CUGCUG,GCGCUC,GCUGCU,GUGCUU,UCCGCU,UCUGCU,UGCUGC,UGCUGU,UGCUUC,UUGCUC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| YTHDC1 | 6 | 5576 | 0.017994859 | 0.0080822104 | 1.154763 | GACUGC,GCAUAC,GGAUGC,GGGUGC,UAGUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| A1CF | 1 | 1642 | 0.005141388 | 0.0023810421 | 1.110565 | CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| DAZAP1 | 6 | 5964 | 0.017994859 | 0.0086445016 | 1.057730 | AGGAAG,AGGAUG,AGGUAA,AGUAUG,GUAACG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| LIN28A | 4 | 4315 | 0.012853470 | 0.0062547643 | 1.039131 | AGGAGA,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| FMR1 | 22 | 20291 | 0.059125964 | 0.0294072466 | 1.007620 | AAGGAA,ACUGGG,AGCUGG,AGGAAG,AGGAGG,AGGAUG,AGGAUU,CAGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,GAGGAU,GCUGGG,GGACAG,UAAGGA,UAGCGG,UGAGGA | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| HNRNPLL | 3 | 3534 | 0.010282776 | 0.0051229360 | 1.005187 | CACCAC,GCACAC,GCAUAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
RBP on BSJ (Exon Only)
Plot
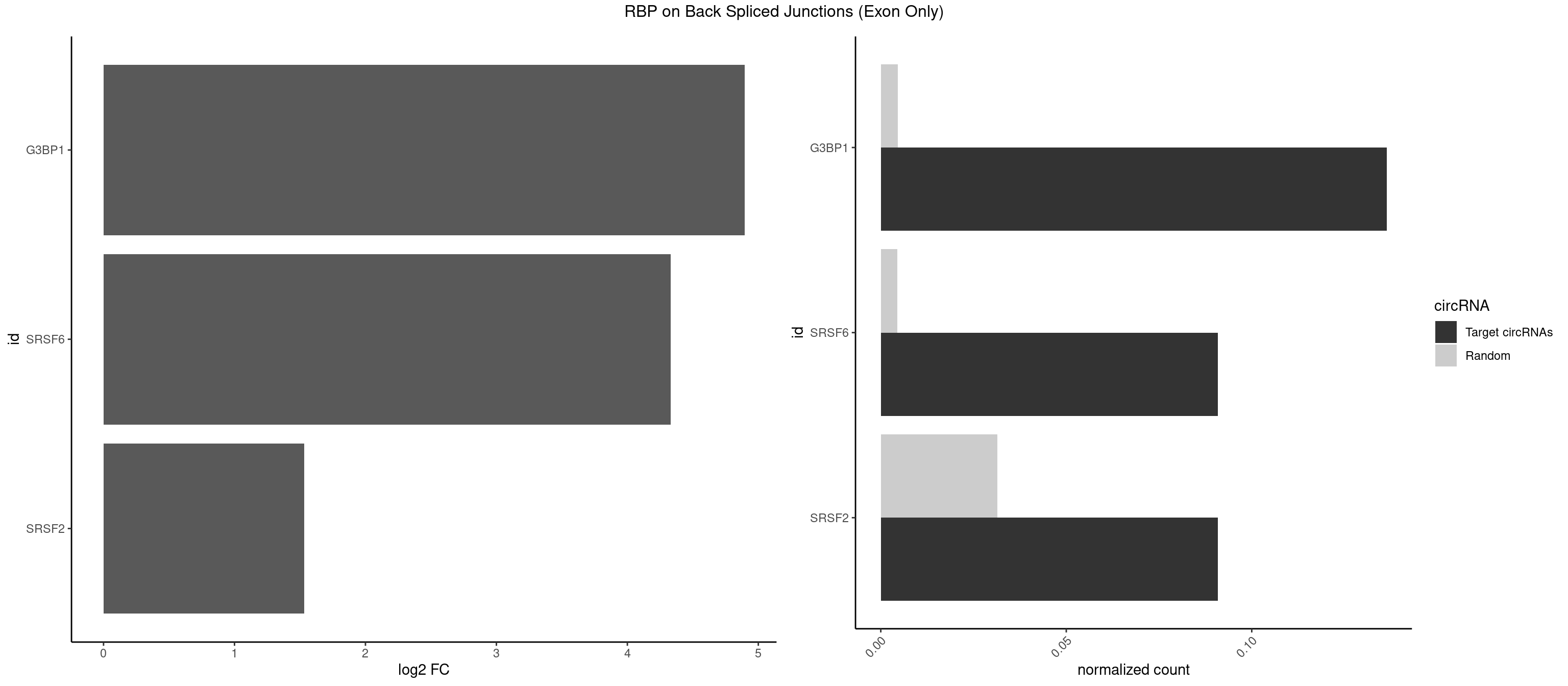
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| G3BP1 | 2 | 69 | 0.13636364 | 0.004584752 | 4.894471 | ACAGGC,CAGGCC | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | UACAGG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | GGCCAA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
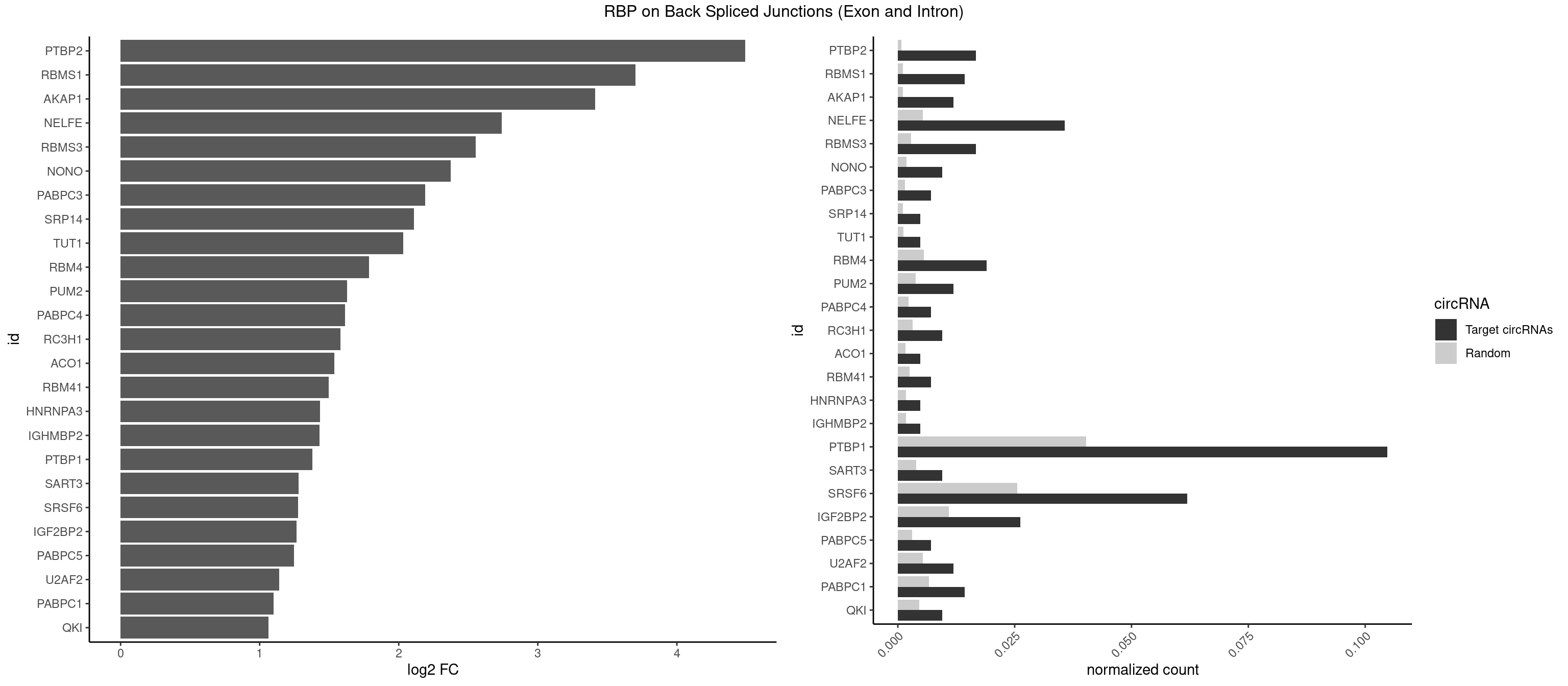
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 6 | 146 | 0.016666667 | 0.0007406288 | 4.492071 | CUCUCU | CUCUCU |
| RBMS1 | 5 | 217 | 0.014285714 | 0.0010983474 | 3.701167 | AUAUAC,AUAUAG,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 4 | 221 | 0.011904762 | 0.0011185006 | 3.411901 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| NELFE | 14 | 1059 | 0.035714286 | 0.0053405885 | 2.741431 | CUCUCU,GUCUCU,UCUCUC,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBMS3 | 6 | 562 | 0.016666667 | 0.0028365578 | 2.554752 | AUAUAG,AUAUAU,CAUAUA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM4 | 7 | 1094 | 0.019047619 | 0.0055169287 | 1.787673 | CCUUCC,CUUCCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | UACAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCCUUC,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACAU,AUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| PTBP1 | 43 | 8003 | 0.104761905 | 0.0403264813 | 1.377315 | ACUUUC,CCUUCC,CCUUUC,CCUUUU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUUCCU,CUUUCC,CUUUCU,CUUUUU,UAUUUU,UCCUCU,UCUCUC,UCUCUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUUCCC,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SRSF6 | 25 | 5069 | 0.061904762 | 0.0255441354 | 1.277058 | CACUGG,CCACUC,CCUCAC,CCUCUC,CUACUC,GAGGAA,GCAGCA,UACAGG,UCAACC,UCACUC,UCACUG,UCUCAC,UCUCUC,UGCGUG,UUACAG,UUUCUC,UUUCUG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| IGF2BP2 | 10 | 2163 | 0.026190476 | 0.0109028617 | 1.264335 | AAAAAC,AAAACA,ACAAAC,CAAACA,CAAUAC,CCAAUC,CCACUC,CCAUUC,GAACUC,GAAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA,AAAAAC,ACAAAC,CAAACA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | CUACUC,CUCAUA,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.