circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000283341:+:11:43750910:43754121
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000283341:+:11:43750910:43754121 | ENSG00000283341 | ENST00000532864 | + | 11 | 43750911 | 43754121 | 123 | UUGUCACAGGUAGUACUGAUGGAAUUGGAAAAUCAUAUGCAGAAGAGUUAGCAAAGCAUGGAAUGAAGGUUGUCCUUAUCAGCAGAUCAAAGGAUAAACUUGACCAGGUUUCCAGUGAAAUAAUUGUCACAGGUAGUACUGAUGGAAUUGGAAAAUCAUAUGCAGAAGAGUUA | circ |
| ENSG00000283341:+:11:43750910:43754121 | ENSG00000283341 | ENST00000532864 | + | 11 | 43750911 | 43754121 | 22 | CAGUGAAAUAAUUGUCACAGGU | bsj |
| ENSG00000283341:+:11:43750910:43754121 | ENSG00000283341 | ENST00000532864 | + | 11 | 43750711 | 43750920 | 210 | UAACACUUGGUUUUGAAGUCUUUUUAAAAUUUUAGCUAUUGCGGUGUGUAUUCAACCUUAGAUUUGAACCGCUGGUAUUGUUCUGUCUUUUACAUCAAAGUGUCACACUGGUCCUUUUGUAUGUAAUUGGUGGGUCCUAACUAUGACCAAUUCCUCAAUGUAAUUCUUAGACUAAACUAUUGCUUUUUUCCCUUUCCCAGUUGUCACAGG | ie_up |
| ENSG00000283341:+:11:43750910:43754121 | ENSG00000283341 | ENST00000532864 | + | 11 | 43754112 | 43754321 | 210 | AGUGAAAUAAGUAAGUUCUCACAUCAAACAAAAGGAAUACAUAGCUAAUUAAUGUUUUCAAGCAGUUAUCCGAUACUGACAUAGUUAUUUCUAGUUCAGGAAACUUCUUUUUCAAGGAGAGAUAUCAUUAAAGAGCAUGAAAAAGCACAGGAUGCUUUAAAAAUGGAUCAUCAAUUUUAGGCUGGGUGCGGUGGCUCACGCCUGUAAUCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
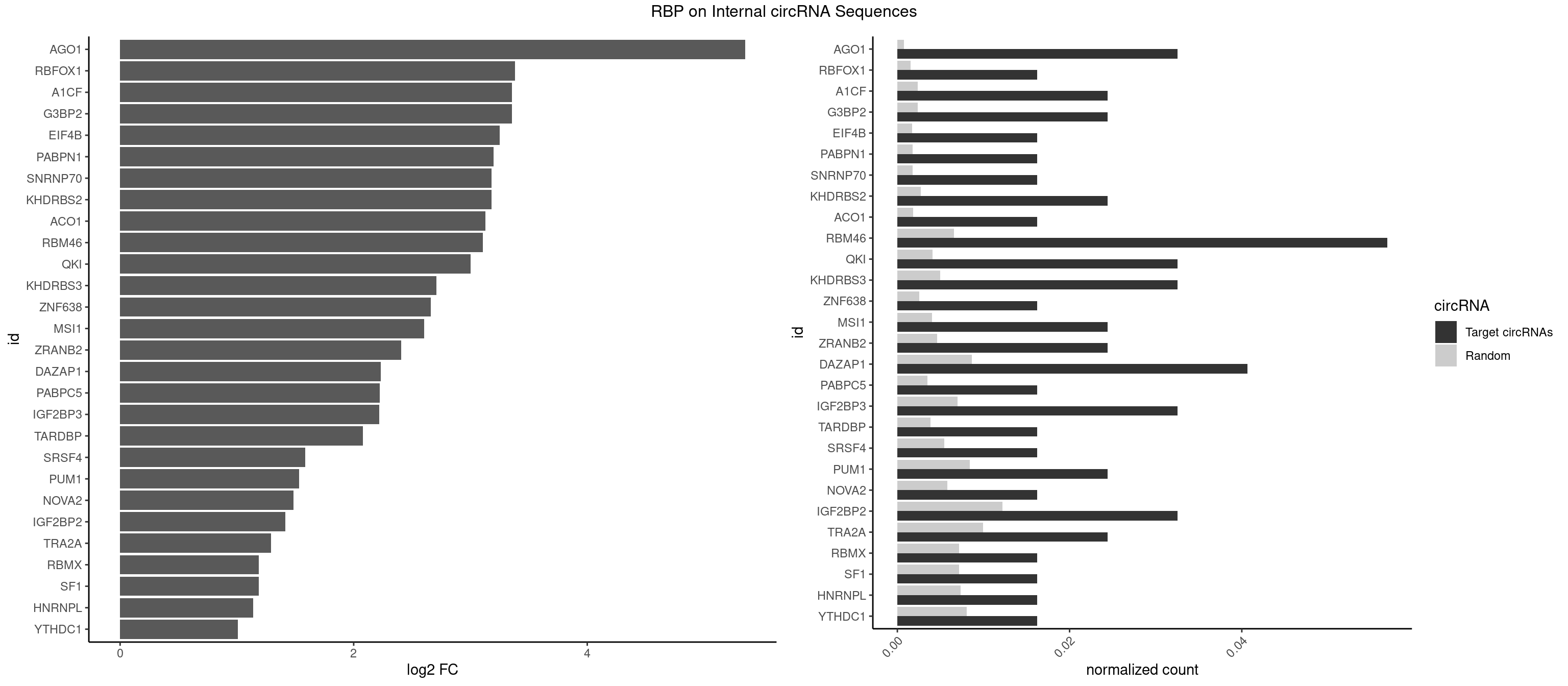
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AGO1 | 3 | 548 | 0.03252033 | 0.000795613 | 5.353131 | AGGUAG,GGUAGU,GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 1 | 1077 | 0.01626016 | 0.001562242 | 3.379652 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| A1CF | 2 | 1642 | 0.02439024 | 0.002381042 | 3.356639 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP2 | 2 | 1644 | 0.02439024 | 0.002383941 | 3.354884 | AGGAUA,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| EIF4B | 1 | 1179 | 0.01626016 | 0.001710061 | 3.249222 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPN1 | 1 | 1222 | 0.01626016 | 0.001772376 | 3.197585 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 1 | 1237 | 0.01626016 | 0.001794114 | 3.179998 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| KHDRBS2 | 2 | 1858 | 0.02439024 | 0.002694070 | 3.178445 | AUAAAC,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ACO1 | 1 | 1283 | 0.01626016 | 0.001860778 | 3.127364 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM46 | 6 | 4554 | 0.05691057 | 0.006601124 | 3.107913 | AAUCAU,AAUGAA,AUCAAA,AUCAUA,AUGAAG,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| QKI | 3 | 2805 | 0.03252033 | 0.004066466 | 2.999494 | AAUCAU,AUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| KHDRBS3 | 3 | 3429 | 0.03252033 | 0.004970770 | 2.709801 | AAAUAA,AUAAAC,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ZNF638 | 1 | 1773 | 0.01626016 | 0.002570888 | 2.661003 | GGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| MSI1 | 2 | 2770 | 0.02439024 | 0.004015744 | 2.602565 | AGGUAG,AGUUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZRANB2 | 2 | 3173 | 0.02439024 | 0.004599773 | 2.406670 | AGGUAG,AGGUUU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| DAZAP1 | 4 | 5964 | 0.04065041 | 0.008644502 | 2.233415 | AGGAUA,AGGUAG,AGGUUG,AGUUAG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| PABPC5 | 1 | 2400 | 0.01626016 | 0.003479539 | 2.224374 | GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 3 | 4815 | 0.03252033 | 0.006979366 | 2.220174 | AAAAUC,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| TARDBP | 1 | 2654 | 0.01626016 | 0.003847636 | 2.079297 | GAAUGA | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF4 | 1 | 3740 | 0.01626016 | 0.005421472 | 1.584585 | AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PUM1 | 2 | 5823 | 0.02439024 | 0.008440164 | 1.530961 | AAUUGU,GAAUUG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| NOVA2 | 1 | 4013 | 0.01626016 | 0.005817105 | 1.482969 | AGAUCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| IGF2BP2 | 3 | 8408 | 0.03252033 | 0.012186356 | 1.416075 | AAAAUC,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| TRA2A | 2 | 6871 | 0.02439024 | 0.009958930 | 1.292242 | AGAAGA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBMX | 1 | 4925 | 0.01626016 | 0.007138779 | 1.187593 | AUCAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SF1 | 1 | 4931 | 0.01626016 | 0.007147474 | 1.185836 | UACUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| HNRNPL | 1 | 5085 | 0.01626016 | 0.007370651 | 1.141478 | AAAUAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| YTHDC1 | 1 | 5576 | 0.01626016 | 0.008082210 | 1.008520 | UAGUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
RBP on BSJ (Exon Only)
Plot
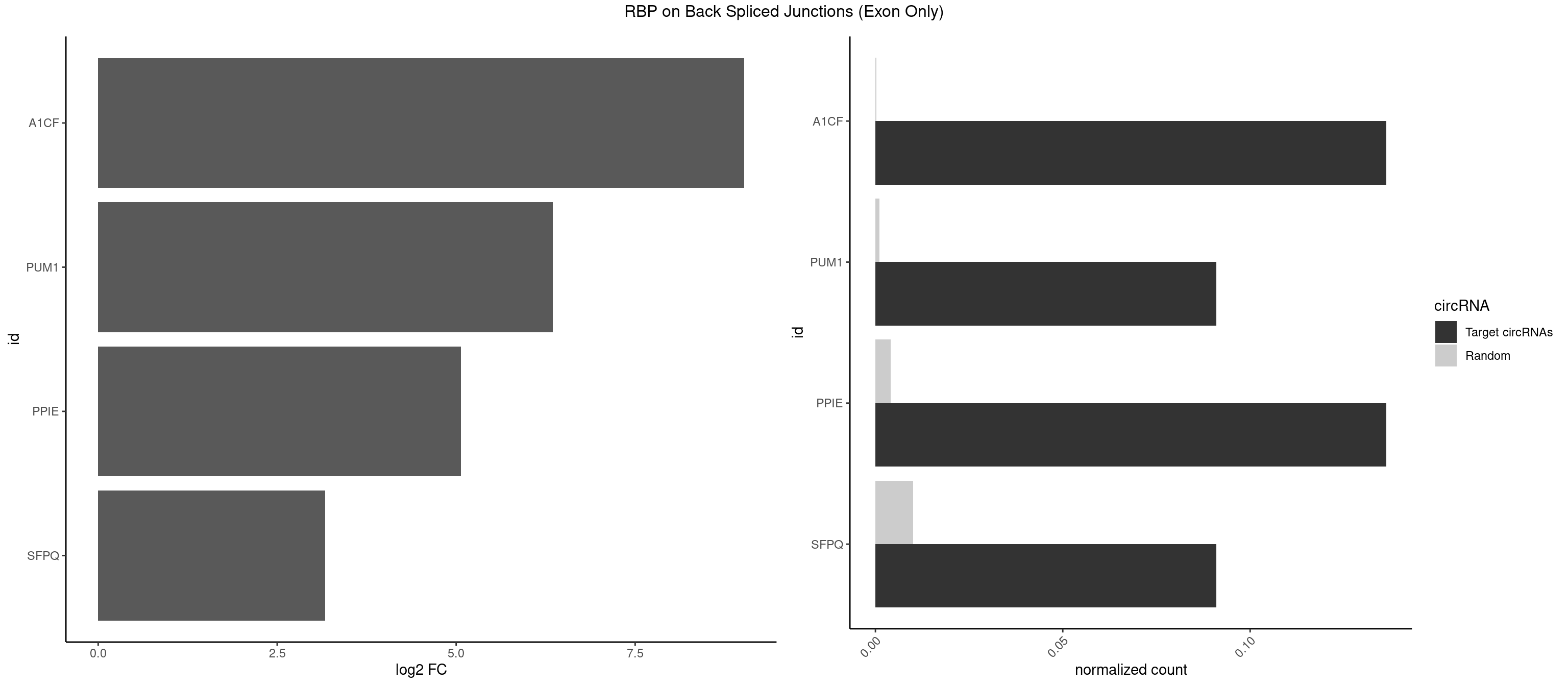
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| A1CF | 2 | 3 | 0.13636364 | 0.0002619859 | 9.023754 | AUAAUU,UAAUUG | AUCAGU,CAGUAU,UAAUUG |
| PUM1 | 1 | 16 | 0.09090909 | 0.0011134399 | 6.351329 | AAUUGU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| PPIE | 2 | 61 | 0.13636364 | 0.0040607807 | 5.069558 | AAUAAU,AUAAUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUUAA,UAAAAA,UAAAUA,UAAAUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUUA |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | UAAUUG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
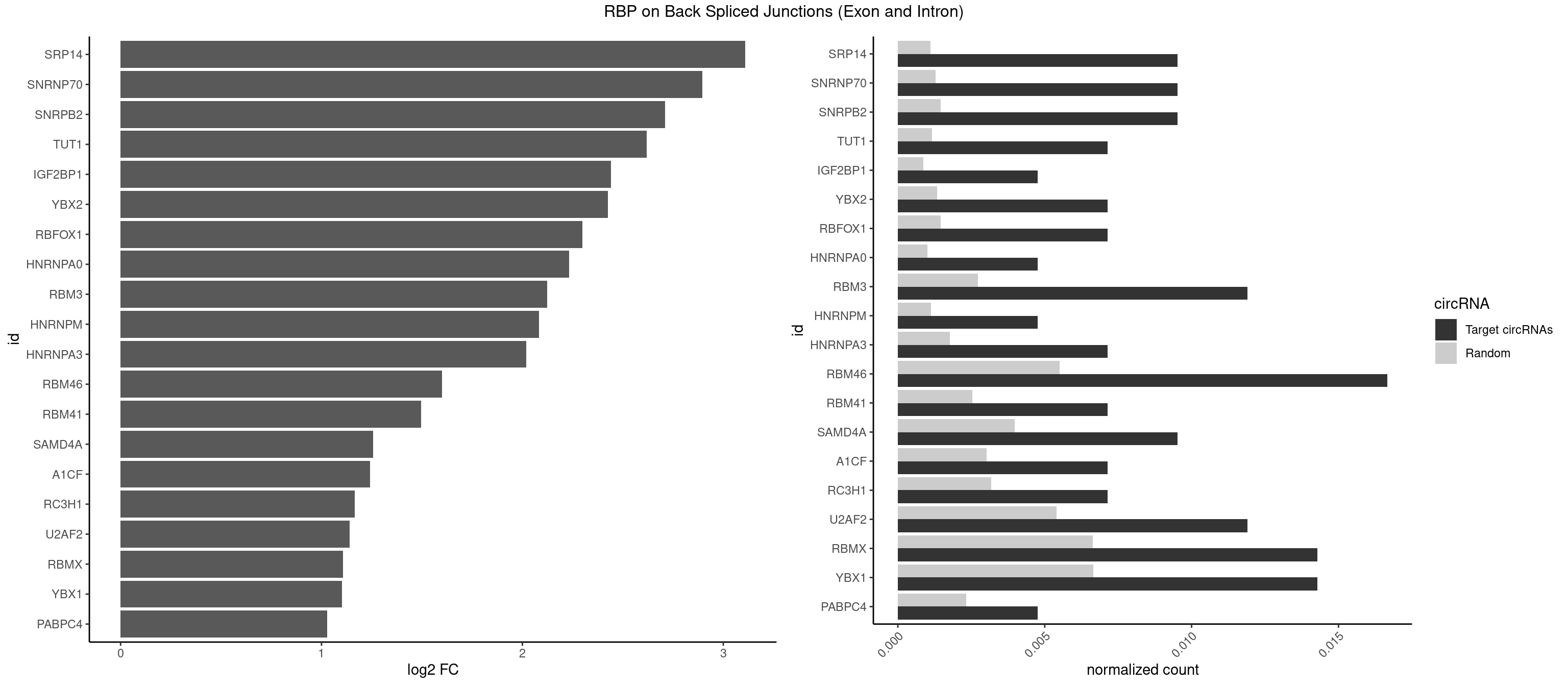
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 3 | 218 | 0.009523810 | 0.0011033857 | 3.109602 | CCUGUA,CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SNRNP70 | 3 | 253 | 0.009523810 | 0.0012797259 | 2.895704 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SNRPB2 | 3 | 288 | 0.009523810 | 0.0014560661 | 2.709463 | GUAUUG,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | CGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM3 | 4 | 541 | 0.011904762 | 0.0027307537 | 2.124168 | AAACUA,AGACUA,GAAACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AUCAAA,AUCAAU,AUCAUU,AUGAAA,GAUCAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACAU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGUA,CUGGUC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUA,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AAGGAA,AAGUAA,AAGUGU,AUCAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACAUCA,AUCAUC,UGCGGU | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.