circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000127663:+:19:5039835:5047669
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000127663:+:19:5039835:5047669 | ENSG00000127663 | ENST00000611640 | + | 19 | 5039836 | 5047669 | 485 | AUCAUCCCCCCGAAGGAGUGGAAGCCGCGGCAGACGUAUGAUGACAUCGACGACGUGGUGAUCCCGGCGCCCAUCCAGCAGGUGGUGACGGGCCAGUCGGGCCUCUUCACGCAGUACAAUAUCCAGAAGAAGGCCAUGACAGUGGGCGAGUACCGCCGCCUGGCCAACAGCGAGAAGUACUGUACCCCGCGGCACCAGGACUUUGAUGACCUUGAACGCAAAUACUGGAAGAACCUCACCUUUGUCUCCCCGAUCUACGGGGCUGACAUCAGCGGCUCUUUGUAUGAUGACGACGUGGCCCAGUGGAACAUCGGGAGCCUCCGGACCAUCCUGGACAUGGUGGAGCGCGAGUGCGGCACCAUCAUCGAGGGCGUGAACACGCCCUACCUGUACUUCGGCAUGUGGAAGACCACCUUCGCCUGGCACACCGAGGACAUGGACCUGUACAGCAUCAACUACCUGCACUUUGGGGAGCCUAAGUCCUGAUCAUCCCCCCGAAGGAGUGGAAGCCGCGGCAGACGUAUGAUGACAUCGA | circ |
| ENSG00000127663:+:19:5039835:5047669 | ENSG00000127663 | ENST00000611640 | + | 19 | 5039836 | 5047669 | 22 | CCUAAGUCCUGAUCAUCCCCCC | bsj |
| ENSG00000127663:+:19:5039835:5047669 | ENSG00000127663 | ENST00000611640 | + | 19 | 5039636 | 5039845 | 210 | GCAGGUGAGGCCCUUGUUGGCUGCAGAGUCUUCUGGGGAGUUUGAUAAGGCAAAUGGGGGGGUUCCUCGUGCCCCUGGGGGGUGUCCCGAGGUGCAGAUCUUGGGGGGUGCUGGUCUCUGGGGAGCCGGUGGCACAGUCUGCUCUCUGGCCCUGCCCUGAGGGUCUGAGGGUGCCACUGACUCCCAUCUUGGUCUUGCAGAUCAUCCCCC | ie_up |
| ENSG00000127663:+:19:5039835:5047669 | ENSG00000127663 | ENST00000611640 | + | 19 | 5047660 | 5047869 | 210 | CUAAGUCCUGGUGAGUGUCUGCACUGGCCCUGCCGCCGGCCGGACCGAGAGCCCCUCGGGAGGGAGUCAAUCCCGGGUACACGGCUGGGCGCCGUGGCAGGGGCCCCACCAGGUGAGGCCGCAAAGGUCGGCCUAUGACGGCUGGAGAUCUUCCGGACCGCCUGGGGUCACCCACCAGCUUUGGGGUGGGGGAUGUGCACCCCCAGAGCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
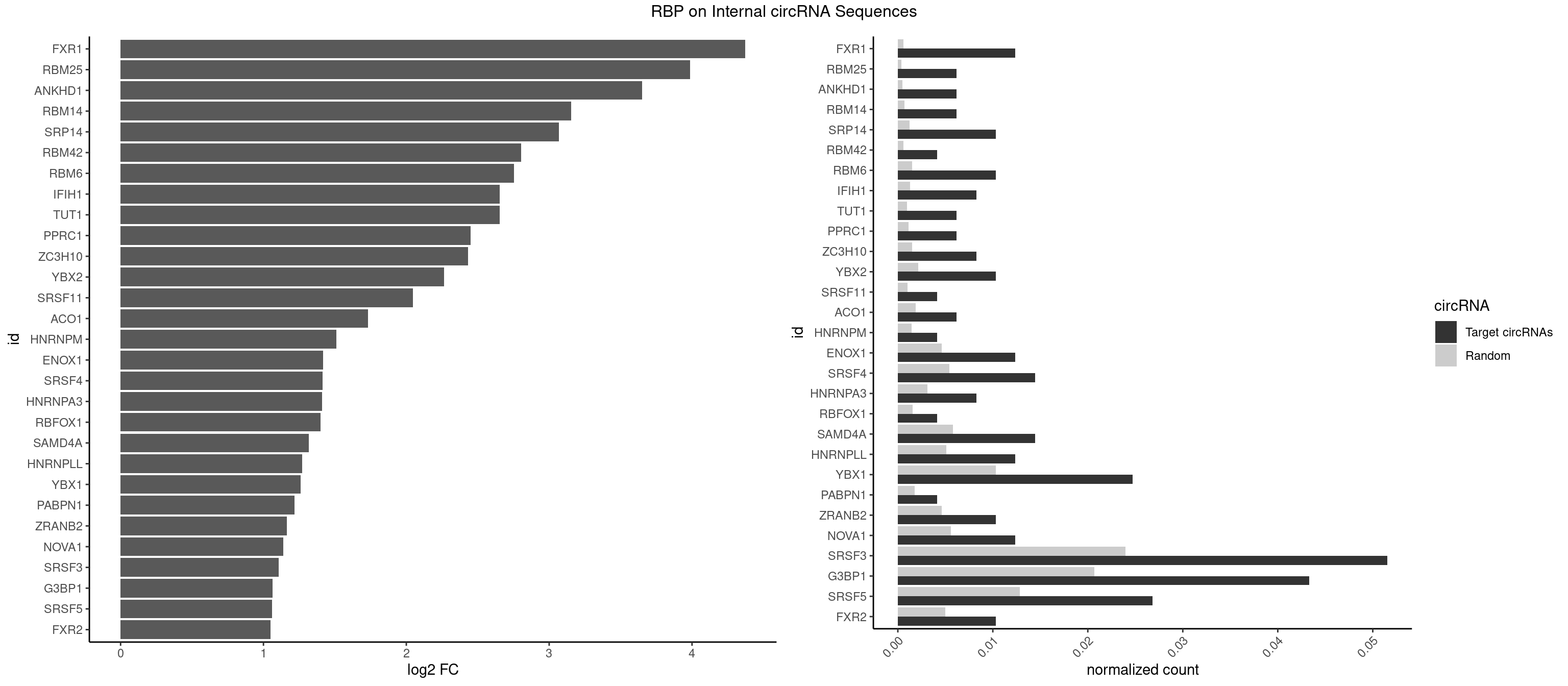
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 5 | 411 | 0.012371134 | 0.0005970720 | 4.372929 | ACGACG,AUGACA,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM25 | 2 | 268 | 0.006185567 | 0.0003898359 | 3.987967 | AUCGGG,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| ANKHD1 | 2 | 339 | 0.006185567 | 0.0004927293 | 3.650039 | AGACGU,GACGUA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM14 | 2 | 478 | 0.006185567 | 0.0006941687 | 3.155548 | CGCGGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| SRP14 | 4 | 847 | 0.010309278 | 0.0012289250 | 3.068475 | CCUGUA,CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM42 | 1 | 407 | 0.004123711 | 0.0005912752 | 2.802042 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM6 | 4 | 1054 | 0.010309278 | 0.0015289102 | 2.753368 | AUCCAG,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| IFIH1 | 3 | 904 | 0.008247423 | 0.0013115296 | 2.652693 | CCGCGG,GCCGCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| TUT1 | 2 | 678 | 0.006185567 | 0.0009840095 | 2.652162 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PPRC1 | 2 | 780 | 0.006185567 | 0.0011318283 | 2.450251 | CGGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| ZC3H10 | 3 | 1053 | 0.008247423 | 0.0015274610 | 2.432808 | CAGCGA,GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| YBX2 | 4 | 1480 | 0.010309278 | 0.0021462711 | 2.264039 | AACAUC,ACAUCA,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SRSF11 | 1 | 688 | 0.004123711 | 0.0009985015 | 2.046107 | AAGAAG | AAGAAG |
| ACO1 | 2 | 1283 | 0.006185567 | 0.0018607779 | 1.733000 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPM | 1 | 999 | 0.004123711 | 0.0014492040 | 1.508683 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ENOX1 | 5 | 3195 | 0.012371134 | 0.0046316558 | 1.417378 | AGGACA,AGUACA,GUACAG,UGGACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SRSF4 | 6 | 3740 | 0.014432990 | 0.0054214720 | 1.412614 | AAGAAG,AGAAGA,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPA3 | 3 | 2140 | 0.008247423 | 0.0031027457 | 1.410398 | AAGGAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBFOX1 | 1 | 1077 | 0.004123711 | 0.0015622419 | 1.400326 | GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SAMD4A | 6 | 3992 | 0.014432990 | 0.0057866714 | 1.318565 | CGGGCC,CUGGAA,CUGGAC,CUGGCA,CUGGCC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPLL | 5 | 3534 | 0.012371134 | 0.0051229360 | 1.271935 | CACACC,CAGACG,GACGAC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| YBX1 | 11 | 7119 | 0.024742268 | 0.0103183321 | 1.261768 | AACAUC,ACAUCA,ACAUCG,ACCACC,AUCAUC,CACACC,CAUCAU,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| PABPN1 | 1 | 1222 | 0.004123711 | 0.0017723764 | 1.218258 | AGAAGA | AAAAGA,AGAAGA |
| ZRANB2 | 4 | 3173 | 0.010309278 | 0.0045997733 | 1.164309 | GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| NOVA1 | 5 | 3872 | 0.012371134 | 0.0056127669 | 1.140194 | ACCACC,AUCAAC,AUCAUC,CCCCCC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| SRSF3 | 24 | 16536 | 0.051546392 | 0.0239654858 | 1.104913 | ACAUCA,ACAUCG,ACCACC,ACGACG,ACUUCG,AUCAAC,AUCAUC,AUCGAC,CAUCAA,CAUCAU,CAUCGA,CCUCUU,CUCUUC,CUUCAC,UACAAU,UACAGC,UACUUC,UCAUCC,UCAUCG,UCUUCA | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| G3BP1 | 20 | 14282 | 0.043298969 | 0.0206989801 | 1.064773 | ACACGC,ACGCAG,ACGCCC,CACGCA,CACGCC,CAUCGG,CCAUCC,CCCAUC,CCCCCC,CCCCCG,CCCCGC,CCCGGC,CCCUAC,CCGCCG,CCUCCG,CGGCAC,CGGCAG,UCGGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| SRSF5 | 12 | 8869 | 0.026804124 | 0.0128544391 | 1.060188 | AACAGC,AAGAAG,AGAAGA,CAACAG,CGCGGC,GAAGAA,GGAAGA,UACAGC,UGCGGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| FXR2 | 4 | 3434 | 0.010309278 | 0.0049780156 | 1.050301 | GACGGG,UGACAG,UGACGA,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
RBP on BSJ (Exon Only)
Plot
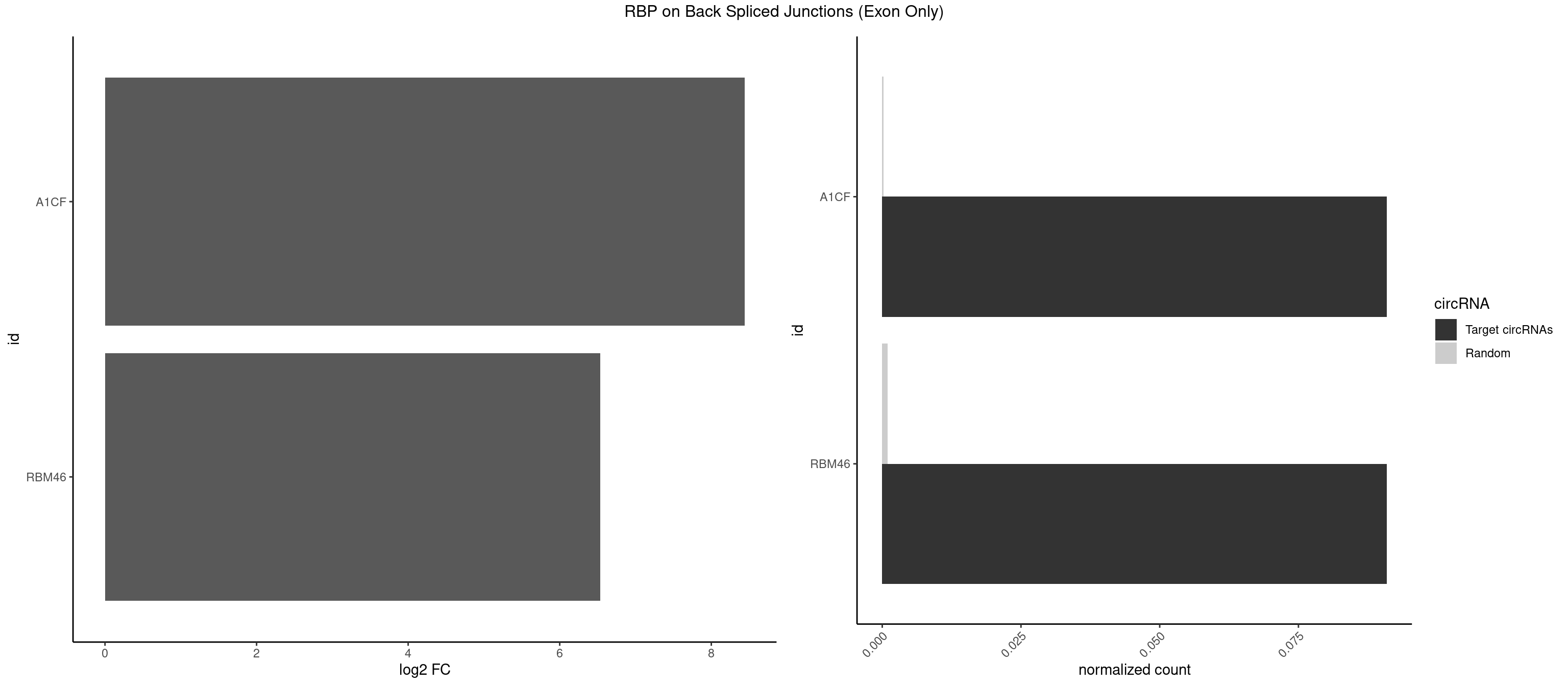
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| A1CF | 1 | 3 | 0.09090909 | 0.0002619859 | 8.438792 | UGAUCA | AUCAGU,CAGUAU,UAAUUG |
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | GAUCAU | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
RBP on BSJ (Exon and Intron)
Plot
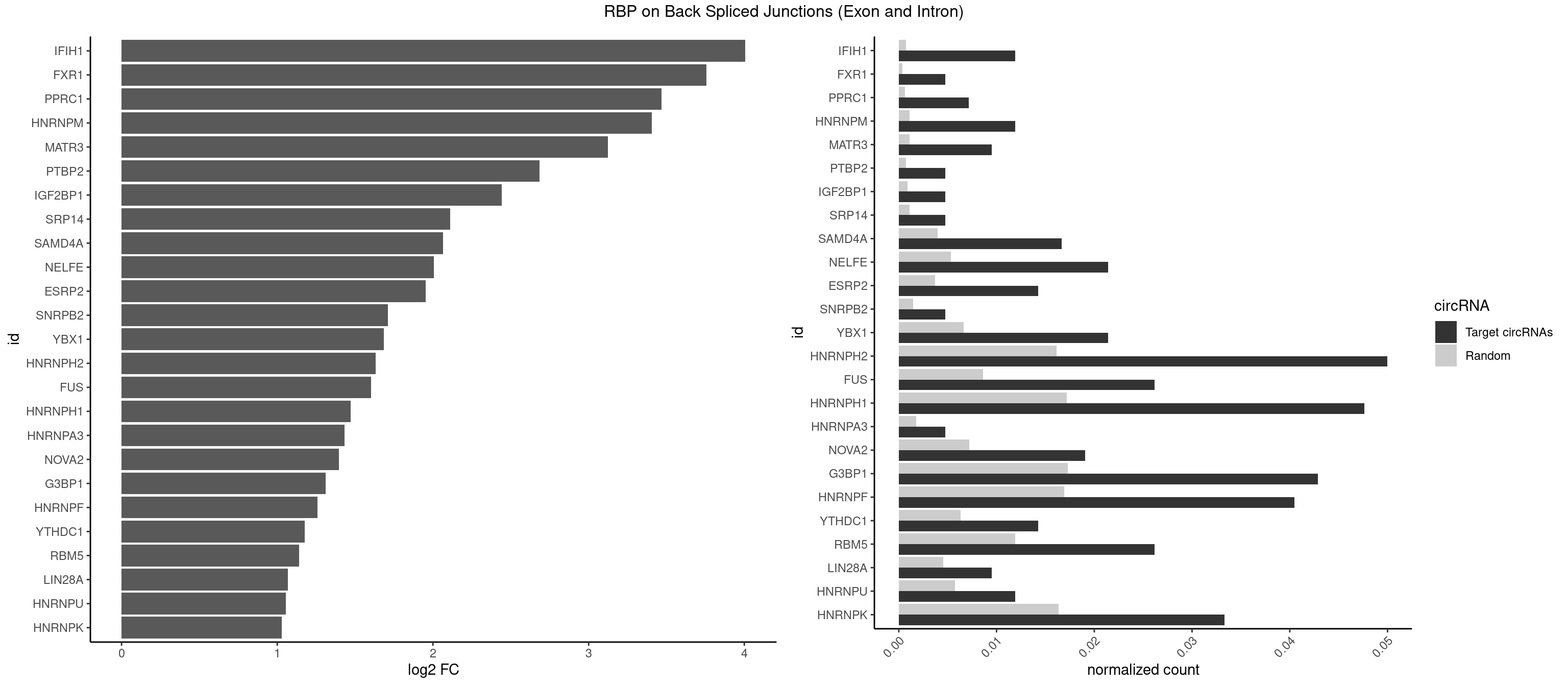
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IFIH1 | 4 | 146 | 0.011904762 | 0.0007406288 | 4.006644 | GGCCCU,GGCCGC | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| HNRNPM | 4 | 222 | 0.011904762 | 0.0011235389 | 3.405417 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| MATR3 | 3 | 216 | 0.009523810 | 0.0010933091 | 3.122837 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SAMD4A | 6 | 790 | 0.016666667 | 0.0039852882 | 2.064210 | CGGGUA,CUGGCC,CUGGUC,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| NELFE | 8 | 1059 | 0.021428571 | 0.0053405885 | 2.004465 | CUCUCU,CUCUGG,GGUCUC,GUCUCU,UCUCUG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ESRP2 | 5 | 731 | 0.014285714 | 0.0036880290 | 1.953651 | GGGGAG,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| YBX1 | 8 | 1321 | 0.021428571 | 0.0066606207 | 1.685807 | AUCAUC,CCACCA,CCCUGC,GGUCUG,GUCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| HNRNPH2 | 20 | 3198 | 0.050000000 | 0.0161174929 | 1.633301 | CUGGGG,GGAGGG,GGGAGG,GGGGAG,GGGGGA,GGGGGG,GGGUGU,UGGGGG,UGGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| FUS | 10 | 1711 | 0.026190476 | 0.0086255542 | 1.602353 | CGGUGG,GGGGGG,GGGUGC,GGGUGG,GGGUGU,UGGUGA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPH1 | 19 | 3406 | 0.047619048 | 0.0171654575 | 1.472030 | CUGGGG,GGAGGG,GGGAGG,GGGGAG,GGGGGG,GGGUGU,UGGGGG,UGGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | AGAUCA,AUCAUC,GAGUCA,GGGGGG | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| G3BP1 | 17 | 3431 | 0.042857143 | 0.0172914148 | 1.309480 | ACCCAC,ACCCCC,AGGCCC,AGGCCG,CCCACC,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCUC,CCCUCG,CCGCCG,CCGGCC,CCUCGG,CGGCCG,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| HNRNPF | 16 | 3360 | 0.040476190 | 0.0169336961 | 1.257177 | AUGGGG,CUGGGG,GGAGGG,GGGAGG,GGGAUG,GGGGAG,GGGGGG,UGGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GGCUGC,GGGUAC,GGGUGC,UCGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBM5 | 10 | 2359 | 0.026190476 | 0.0118903668 | 1.139249 | AGGGAG,AGGGUG,GAGGGA,GAGGGU,GGGGGG,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGAU,GGAGGG,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | GGGGGG | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPK | 13 | 3244 | 0.033333333 | 0.0163492543 | 1.027741 | ACCCCC,CAUCCC,CCCCAC,CCCCCA,GCCCCU,GGGGGG,UCCCAU,UCCCCC,UCCCGA | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.