circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000135250:-:7:105132790:105203785
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000135250:-:7:105132790:105203785 | ENSG00000135250 | ENST00000357311 | - | 7 | 105132791 | 105203785 | 1681 | GCCGGAGCCUCAACAGAAAGCUCCUUUAGUUCCUCCUCCUCCACCGCCACCACCACCACCACCGCCACCUUUGCCAGACCCCACACCCCCGGAGCCAGAGGAGGAGAUCCUGGGAUCAGAUGAUGAGGAGCAAGAGGACCCUGCGGACUACUGCAAAGGUGGAUAUCAUCCAGUGAAAAUUGGAGACCUCUUCAAUGGCCGGUAUCAUGUUAUUAGAAAGCUUGGAUGGGGGCACUUCUCUACUGUCUGGCUGUGCUGGGAUAUGCAGGGGAAAAGAUUUGUUGCAAUGAAAGUUGUAAAAAGUGCCCAGCAUUAUACGGAGACAGCCUUGGAUGAAAUAAAAUUGCUCAAAUGUGUUCGAGAAAGUGAUCCCAGUGACCCAAACAAAGACAUGGUGGUCCAGCUCAUUGACGACUUCAAGAUUUCAGGCAUGAAUGGGAUACAUGUCUGCAUGGUCUUCGAAGUACUUGGCCACCAUCUCCUCAAGUGGAUCAUCAAAUCCAACUAUCAAGGCCUCCCAGUACGUUGUGUGAAGAGUAUCAUUCGACAGGUCCUUCAAGGGUUAGAUUACUUACACAGUAAGUGCAAGAUCAUUCAUACUGACAUAAAGCCGGAAAAUAUCUUGAUGUGUGUGGAUGAUGCAUAUGUGAGAAGAAUGGCAGCUGAGGCCACUGAGUGGCAGAAAGCAGGUGCUCCUCCUCCUUCAGGGUCUGCAGUGAGUACGGCUCCACAGCAGAAACCUAUAGGAAAAAUAUCUAAAAACAAAAAGAAAAAACUGAAAAAGAAACAGAAGAGGCAGGCUGAGUUAUUGGAGAAGCGCCUGCAGGAGAUAGAAGAAUUGGAGCGAGAAGCUGAAAGGAAAAUAAUAGAAGAAAACAUCACCUCAGCUGCACCUUCCAAUGACCAGGAUGGCGAAUACUGCCCAGAGGUGAAACUAAAAACAACAGGAUUAGAGGAGGCGGCUGAGGCAGAGACUGCAAAGGACAAUGGUGAAGCUGAGGACCAGGAAGAGAAAGAAGAUGCUGAGAAAGAAAACAUUGAAAAAGAUGAAGAUGAUGUAGAUCAGGAACUUGCGAACAUAGACCCUACGUGGAUAGAAUCACCUAAAACCAAUGGCCAUAUUGAGAAUGGCCCAUUCUCACUGGAGCAGCAACUGGACGAUGAAGAUGAUGAUGAAGAAGACUGCCCAAAUCCUGAGGAAUAUAAUCUUGAUGAGCCAAAUGCAGAAAGUGAUUACACAUAUAGCAGCUCCUAUGAACAAUUCAAUGGUGAAUUGCCAAAUGGACGACAUAAAAUUCCCGAGUCACAGUUCCCAGAGUUUUCCACCUCGUUGUUCUCUGGAUCCUUAGAACCUGUGGCCUGCGGCUCUGUGCUUUCUGAGGGAUCACCACUUACUGAGCAAGAGGAGAGCAGUCCAUCCCAUGACAGAAGCAGAACGGUUUCAGCCUCCAGUACUGGGGAUUUGCCAAAAGCAAAAACCCGGGCAGCUGACUUGUUGGUGAAUCCCCUGGAUCCGCGGAAUGCAGAUAAAAUUAGAGUAAAAAUUGCUGACCUGGGAAAUGCUUGUUGGGUGCAUAAACACUUCACGGAAGACAUCCAGACGCGUCAGUACCGCUCCAUAGAGGUUUUAAUAGGAGCGGGGUACAGCACCCCUGCGGACAUCUGGAGCACGGCGUGUAUGGCCGGAGCCUCAACAGAAAGCUCCUUUAGUUCCUCCUCCUCCACCGCCAC | circ |
| ENSG00000135250:-:7:105132790:105203785 | ENSG00000135250 | ENST00000357311 | - | 7 | 105132791 | 105203785 | 22 | CGGCGUGUAUGGCCGGAGCCUC | bsj |
| ENSG00000135250:-:7:105132790:105203785 | ENSG00000135250 | ENST00000357311 | - | 7 | 105203776 | 105203985 | 210 | GAUUUUUUUUUUUUUUGGCCUGACUUAUAUGUUGAAACACUACUUGAAUUCAACUAAAAUGGGUGAAGUGACAUUAAAUGACAUUUCUUCUUAGUAUGUGACAAGUUUUAUUUUUUCCCCCAUAUUAAGAAGUGCUCAAAUGCAUCCAUAAUGCAAGAUGUACUUCUAAGUAAAUAGCAAUUUUCUCUCUGCUCUUUCAGGCCGGAGCCU | ie_up |
| ENSG00000135250:-:7:105132790:105203785 | ENSG00000135250 | ENST00000357311 | - | 7 | 105132591 | 105132800 | 210 | GGCGUGUAUGGUAAGGACGGCUGUGCCCUUUGCUGCCAUGGGAAUUGGCUCGUUCCUUUCACACUCUGGAUGGGGCUGAGUCUCUCUGAGGCAUGCGACCUCAGUUUUUCUGACUGUAAGGGUCAUCCACCGUGGGCUGGGUGAGGGGAAGGUUGCUGCCGCAGGCAUCUUAAGAAGUGGAAGGAUCCUCCUCAGGCGGGCCCUGGGUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
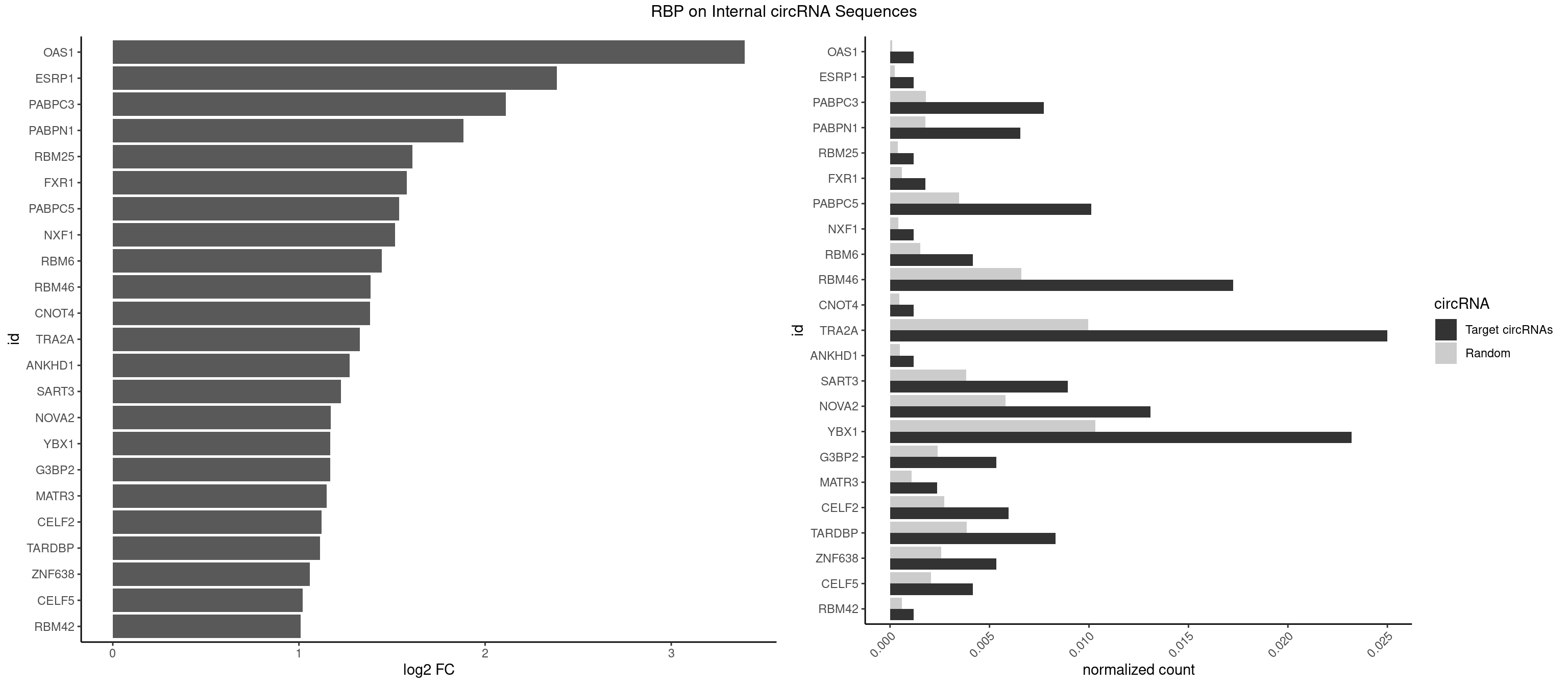
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.001189768 | 0.0001130379 | 3.395802 | GCAUAA | GCAUAA |
| ESRP1 | 1 | 156 | 0.001189768 | 0.0002275250 | 2.386583 | AGGGAU | AGGGAU |
| PABPC3 | 12 | 1234 | 0.007733492 | 0.0017897669 | 2.111348 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPN1 | 10 | 1222 | 0.006543724 | 0.0017723764 | 1.884427 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM25 | 1 | 268 | 0.001189768 | 0.0003898359 | 1.609742 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| FXR1 | 2 | 411 | 0.001784652 | 0.0005970720 | 1.579666 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| PABPC5 | 16 | 2400 | 0.010113028 | 0.0034795387 | 1.539247 | AGAAAA,AGAAAG,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| NXF1 | 1 | 286 | 0.001189768 | 0.0004159215 | 1.516297 | AACCUG | AACCUG |
| RBM6 | 6 | 1054 | 0.004164188 | 0.0015289102 | 1.445532 | AAUCCA,AUCCAA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM46 | 28 | 4554 | 0.017251636 | 0.0066011240 | 1.385950 | AAUGAA,AUCAAA,AUCAAG,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUG,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CNOT4 | 1 | 314 | 0.001189768 | 0.0004564992 | 1.381996 | GACAGA | GACAGA |
| TRA2A | 41 | 6871 | 0.024985128 | 0.0099589296 | 1.327007 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ANKHD1 | 1 | 339 | 0.001189768 | 0.0004927293 | 1.271813 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SART3 | 14 | 2634 | 0.008923260 | 0.0038186524 | 1.224507 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| NOVA2 | 21 | 4013 | 0.013087448 | 0.0058171047 | 1.169811 | ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AUCACC,AUCAUC,GAGACA,GAGGCA,GAGUCA,GAUCAC,UAGAUC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| YBX1 | 38 | 7119 | 0.023200476 | 0.0103183321 | 1.168945 | AACAUC,ACAUCA,ACAUCU,ACCACC,AUCAUC,CACACC,CACCAC,CAGCAA,CAUCUG,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GCCUGC,GGUCUG,GUCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| G3BP2 | 8 | 1644 | 0.005353956 | 0.0023839405 | 1.167257 | AGGAUG,AGGAUU,GGAUAG,GGAUGA,GGAUGG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| MATR3 | 3 | 739 | 0.002379536 | 0.0010724109 | 1.149822 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CELF2 | 9 | 1886 | 0.005948840 | 0.0027346479 | 1.121253 | AUGUGU,GUGUGU,GUUGUU,UAUGUG,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TARDBP | 13 | 2654 | 0.008328376 | 0.0038476365 | 1.114063 | GAAUGG,GUGAAU,GUGUGU,GUUGUG,GUUGUU,UGAAUG,UGUGUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 8 | 1773 | 0.005353956 | 0.0025708878 | 1.058339 | CGUUGU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF5 | 6 | 1415 | 0.004164188 | 0.0020520728 | 1.020953 | GUGUGG,GUGUGU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM42 | 1 | 407 | 0.001189768 | 0.0005912752 | 1.008779 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
RBP on BSJ (Exon Only)
Plot
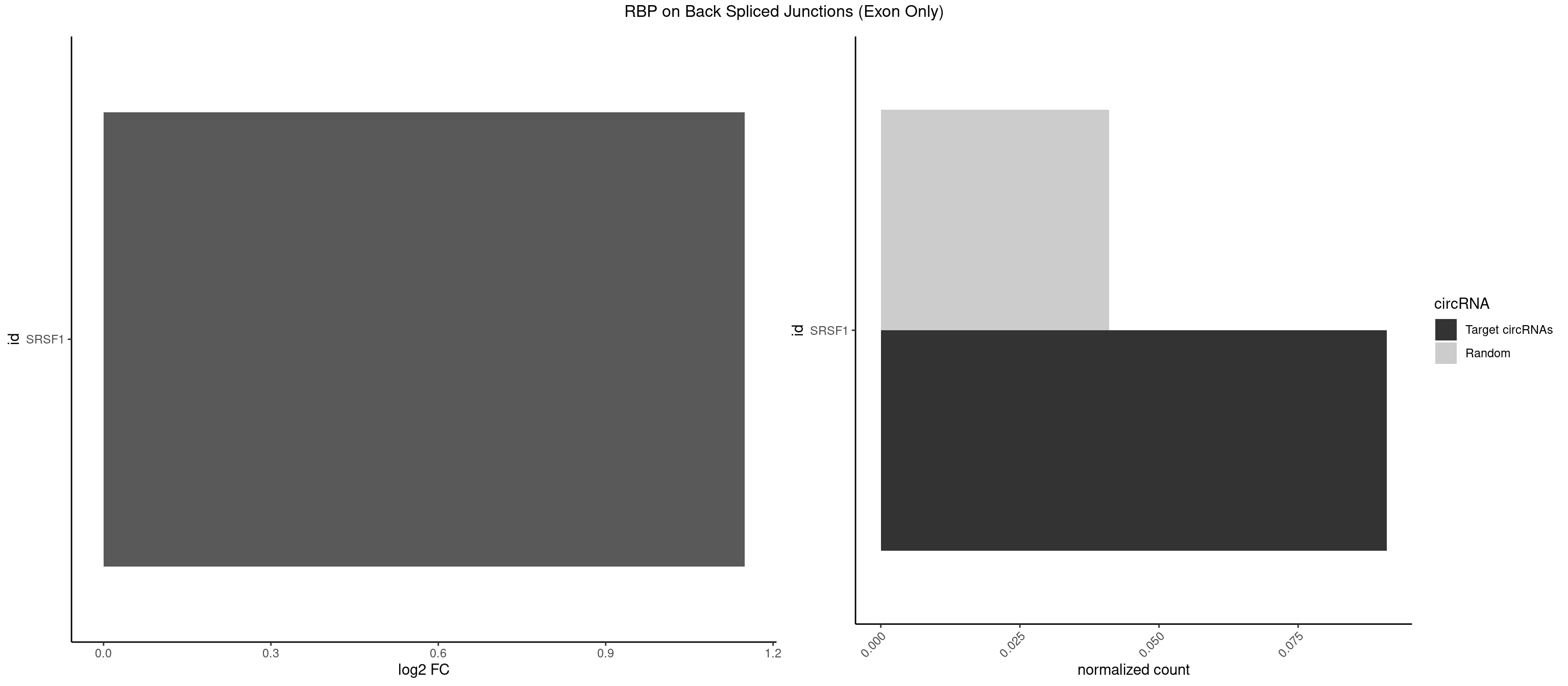
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF1 | 1 | 625 | 0.09090909 | 0.04100079 | 1.148773 | GGCCGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
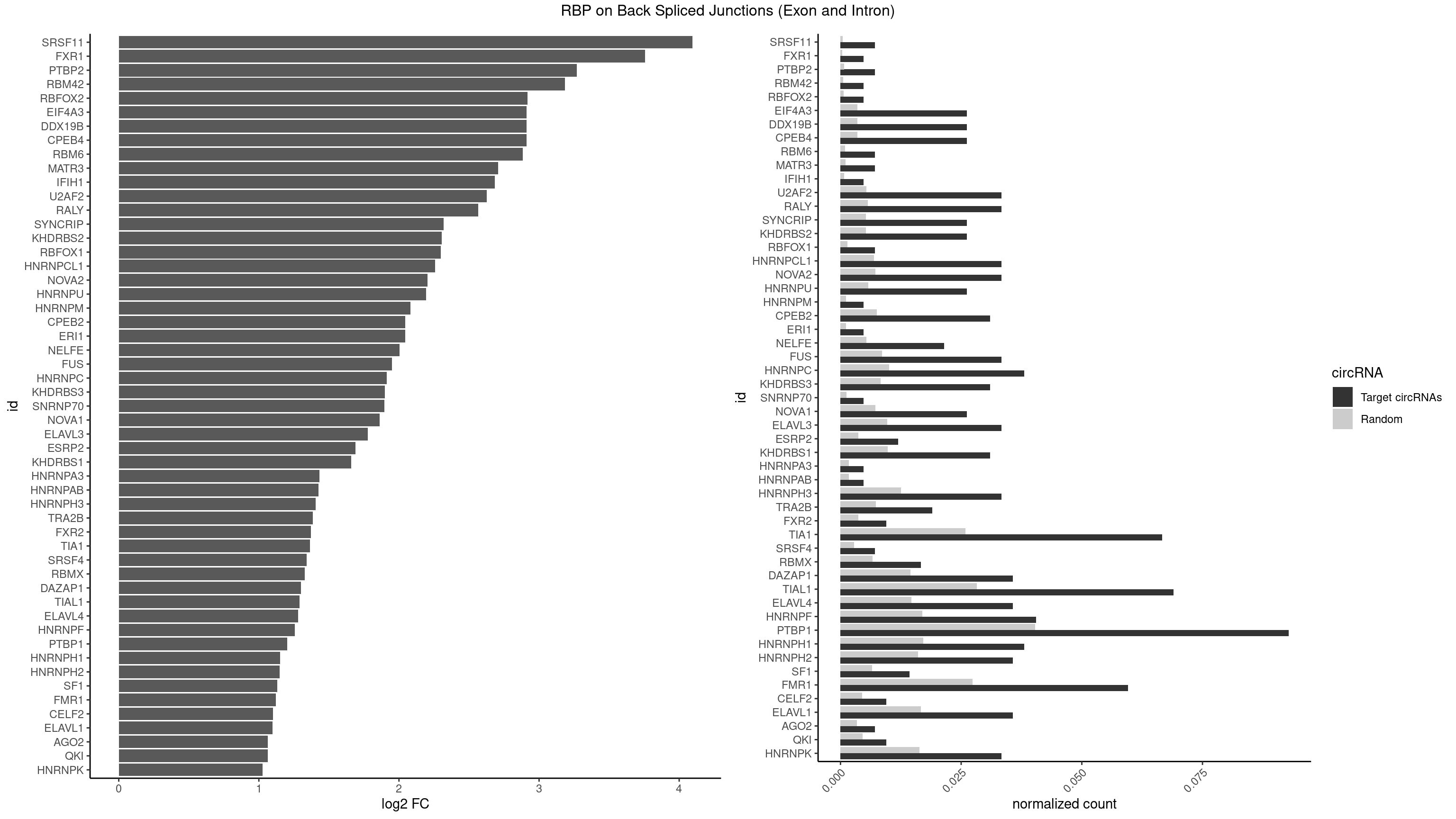
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 2 | 82 | 0.007142857 | 0.0004181782 | 4.094312 | AAGAAG | AAGAAG |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| CPEB4 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| DDX19B | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| EIF4A3 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| U2AF2 | 13 | 1071 | 0.033333333 | 0.0054010480 | 2.625654 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 13 | 1117 | 0.033333333 | 0.0056328094 | 2.565039 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP | 10 | 1041 | 0.026190476 | 0.0052498992 | 2.318681 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 10 | 1051 | 0.026190476 | 0.0053002821 | 2.304901 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGC,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPCL1 | 13 | 1381 | 0.033333333 | 0.0069629182 | 2.259202 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 13 | 1434 | 0.033333333 | 0.0072299476 | 2.204908 | AGGCAU,GAGGCA,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPU | 10 | 1136 | 0.026190476 | 0.0057285369 | 2.192804 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| CPEB2 | 12 | 1487 | 0.030952381 | 0.0074969770 | 2.045670 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| NELFE | 8 | 1059 | 0.021428571 | 0.0053405885 | 2.004465 | CUCUCU,CUCUGG,GUCUCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| FUS | 13 | 1711 | 0.033333333 | 0.0086255542 | 1.950277 | GGGUGA,GGGUGU,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPC | 15 | 2006 | 0.038095238 | 0.0101118501 | 1.913564 | AUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS3 | 12 | 1646 | 0.030952381 | 0.0082980653 | 1.899203 | AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| NOVA1 | 10 | 1428 | 0.026190476 | 0.0071997179 | 1.863030 | UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| ELAVL3 | 13 | 1928 | 0.033333333 | 0.0097188634 | 1.778106 | UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ESRP2 | 4 | 731 | 0.011904762 | 0.0036880290 | 1.690617 | GGGAAG,GGGAAU,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| KHDRBS1 | 12 | 1945 | 0.030952381 | 0.0098045143 | 1.658532 | CUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPH3 | 13 | 2496 | 0.033333333 | 0.0125806127 | 1.405763 | AGGGGA,GAGGGG,GGAAGG,GGGAAG,GGGAAU,GGGCUG,GGGGCU,GGGUGU,UGGGUG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| TRA2B | 7 | 1447 | 0.019047619 | 0.0072954454 | 1.384543 | AAGAAG,GAAGGA,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | GACAAG,GGACGG,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| TIA1 | 27 | 5140 | 0.066666667 | 0.0259018541 | 1.363910 | AUUUUC,AUUUUU,CCUUUC,GUUUUA,GUUUUU,UAUUUU,UCCUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AAGAAG | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AAGAAG,AAGUAA,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| DAZAP1 | 14 | 2878 | 0.035714286 | 0.0145052398 | 1.299927 | AGGUUG,AGUAAA,AGUAUG,UAGUAU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| TIAL1 | 28 | 5597 | 0.069047619 | 0.0282043531 | 1.291674 | AAUUUU,AGUUUU,AUUUUC,AUUUUU,CAGUUU,GUUUUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUUAUU,UUUCAG,UUUUAU,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 14 | 2916 | 0.035714286 | 0.0146966949 | 1.281010 | UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPF | 16 | 3360 | 0.040476190 | 0.0169336961 | 1.257177 | AGGGGA,AUGGGA,AUGGGG,GAAGGU,GAGGGG,GAUGGG,GGAAGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGCU,UGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| PTBP1 | 38 | 8003 | 0.092857143 | 0.0403264813 | 1.203285 | AUUUUC,AUUUUU,CAUCUU,CCUUUC,CUCUCU,CUCUUU,CUUCUU,GCUGUG,UAUUUU,UCCCCC,UCUCUC,UCUUCU,UCUUUC,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| HNRNPH1 | 15 | 3406 | 0.038095238 | 0.0171654575 | 1.150102 | AGGGGA,GAAGGU,GAGGGG,GGAAGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGCU,GGGUGU,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH2 | 14 | 3198 | 0.035714286 | 0.0161174929 | 1.147874 | AGGGGA,GAGGGG,GGAAGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGCU,GGGUGU,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUUAU,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| FMR1 | 24 | 5430 | 0.059523810 | 0.0273629585 | 1.121243 | AAGGAU,CGGCUG,CUGAGG,GAAGGA,GACAAG,GCUGAG,GCUGGG,GGCUGA,GGCUGG,GGGCUG,UAAGGA,UGACAA,UGCGAC,UUUUUU | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUG,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | UAUUUU,UUAUUU,UUUAUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAGUGC,AGUGCU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACACUA,ACUUAU,CACACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPK | 13 | 3244 | 0.033333333 | 0.0163492543 | 1.027741 | CCCCAU,CCCCCA,UCCCCC,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.