circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000117868:-:7:158788003:158799072
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000117868:-:7:158788003:158799072 | ENSG00000117868 | ENST00000275418 | - | 7 | 158788004 | 158799072 | 417 | GUUCAUUUUCCAGACACUGAAAGAGCAGAAUGGCUAAAUAAGACUGUAAAACACAUGUGGCCUUUCAUUUGCCAAUUUAUAGAGAAGUUGUUUCGAGAAACUAUAGAACCAGCCGUGCGGGGAGCAAACACCCACCUUAGCACCUUUAGUUUCACGAAGGUCGACGUGGGCCAGCAGCCCCUCAGGAUCAAUGGUGUUAAGGUAUACACUGAAAAUGUAGACAAAAGGCAAAUUAUUUUGGACCUUCAGAUUAGUUUUGUAGGAAAUUGUGAGAUUGAUUUGGAGAUCAAACGAUAUUUUUGUAGAGCUGGUGUGAAAAGUAUCCAGAUUCAUGGUACCAUGCGGGUGAUCCUGGAACCGUUGAUUGGAGAUAUGCCCUUAGUUGGAGCUUUGUCUAUCUUCUUCCUUAGGAAACCAGUUCAUUUUCCAGACACUGAAAGAGCAGAAUGGCUAAAUAAGACUGUAAA | circ |
| ENSG00000117868:-:7:158788003:158799072 | ENSG00000117868 | ENST00000275418 | - | 7 | 158788004 | 158799072 | 22 | UUAGGAAACCAGUUCAUUUUCC | bsj |
| ENSG00000117868:-:7:158788003:158799072 | ENSG00000117868 | ENST00000275418 | - | 7 | 158799063 | 158799272 | 210 | UCUCAGAACGUAAAGAGCUGGGGUGCCAUGCAUUUCACUUUCCCUCCUUGGAGACCAGUUUCCCAAGCUUUAGAGCAGAGUGUUUUAAAGAAGCAUCUUGUAAACAUAGGACGUAUGAUGAGAAUAUGAAAUUGCCUGCUUGACAUUUUAUUUACAGUUGUUGGGAGUUAAUAAGUCAUAUGUGUGUAUUUUCCUCCUAGGUUCAUUUUC | ie_up |
| ENSG00000117868:-:7:158788003:158799072 | ENSG00000117868 | ENST00000275418 | - | 7 | 158787804 | 158788013 | 210 | UAGGAAACCAGUAAGUCAAUAUUUAUUUCAUUUUUAUUUUCCCAUUUGGUGGCAAAAAUACAUACCCGUGGAAGAAUAUAUAAAGUGGAGAAACAAAAUUUUAUCAACUCACCGUUGUCCCUUUCUGUUGUGUGAAUCCUGUGGGAGUUUUUUGUAUUUCUUUGUGCCUUAACUGAGAGCUUAUCUCUUGAUAAACAGCUCUCUUUCCUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
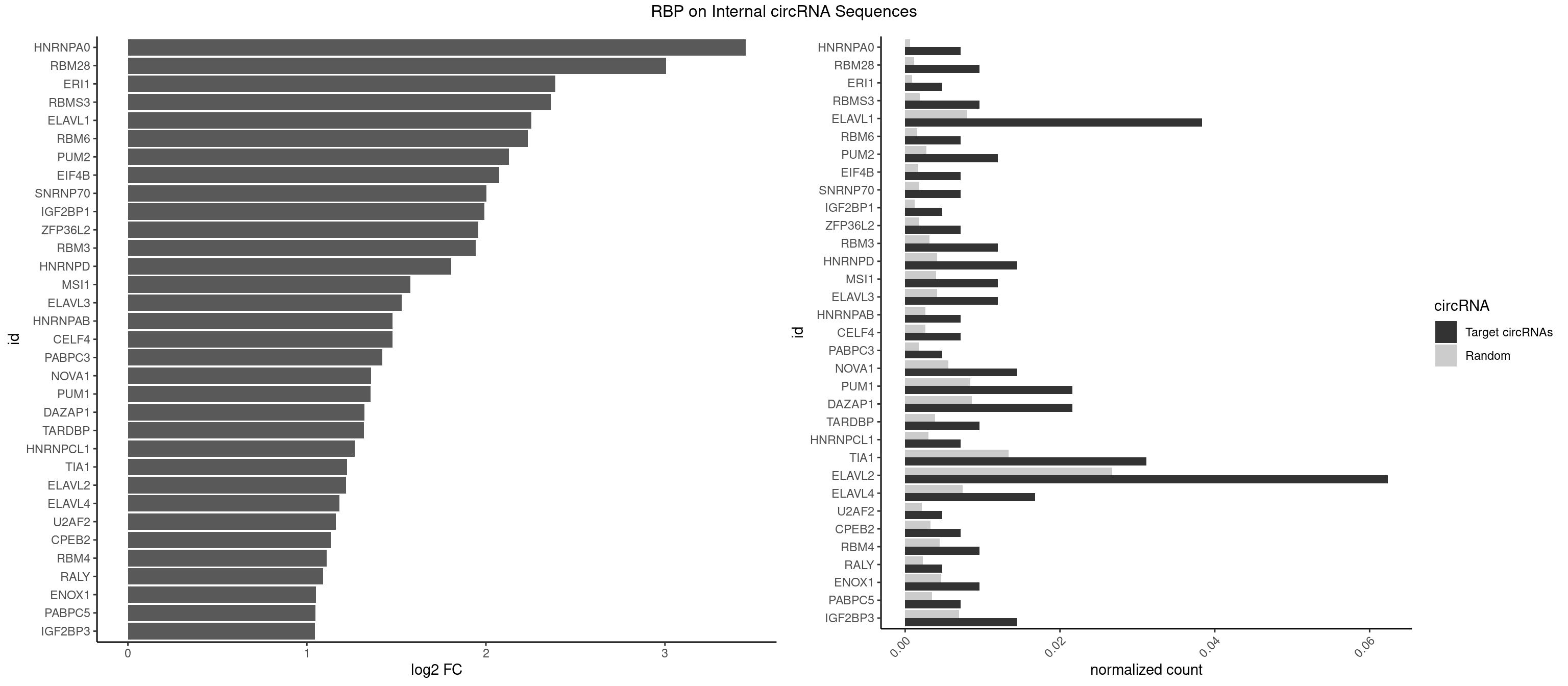
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 2 | 453 | 0.007194245 | 0.0006579386 | 3.450818 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM28 | 3 | 822 | 0.009592326 | 0.0011926949 | 3.007656 | UGUAGA,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 632 | 0.004796163 | 0.0009173461 | 2.386343 | UUCAGA | UUCAGA,UUUCAG |
| RBMS3 | 3 | 1283 | 0.009592326 | 0.0018607779 | 2.365975 | CUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ELAVL1 | 15 | 5554 | 0.038369305 | 0.0080503280 | 2.252833 | AUUUAU,UAGUUU,UAUUUU,UGAUUU,UUAGUU,UUAUUU,UUGAUU,UUGUUU,UUUAGU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBM6 | 2 | 1054 | 0.007194245 | 0.0015289102 | 2.234340 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PUM2 | 4 | 1890 | 0.011990408 | 0.0027404447 | 2.129399 | UAAAUA,UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| EIF4B | 2 | 1179 | 0.007194245 | 0.0017100607 | 2.072796 | GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SNRNP70 | 2 | 1237 | 0.007194245 | 0.0017941145 | 2.003571 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGF2BP1 | 1 | 831 | 0.004796163 | 0.0012057377 | 1.991965 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ZFP36L2 | 2 | 1277 | 0.007194245 | 0.0018520827 | 1.957695 | AUUUAU,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM3 | 4 | 2152 | 0.011990408 | 0.0031201361 | 1.942200 | AAACGA,AAACUA,AAGACU,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPD | 5 | 2837 | 0.014388489 | 0.0041128408 | 1.806708 | AAUUUA,AGAUAU,AUUUAU,UUAGGA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| MSI1 | 4 | 2770 | 0.011990408 | 0.0040157442 | 1.578141 | AGUUGG,UAGGAA,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ELAVL3 | 4 | 2867 | 0.011990408 | 0.0041563169 | 1.528503 | AUUUAU,UAUUUU,UUAUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF4 | 2 | 1782 | 0.007194245 | 0.0025839306 | 1.477276 | GGUGUG,GGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 2 | 1782 | 0.007194245 | 0.0025839306 | 1.477276 | AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC3 | 1 | 1234 | 0.004796163 | 0.0017897669 | 1.422109 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| NOVA1 | 5 | 3872 | 0.014388489 | 0.0056127669 | 1.358131 | AACACC,AGCACC,AUUCAU,UUCAUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| PUM1 | 8 | 5823 | 0.021582734 | 0.0084401638 | 1.354535 | AAUUGU,CAGAAU,UAAAUA,UGUAAA,UGUAGA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| DAZAP1 | 8 | 5964 | 0.021582734 | 0.0086445016 | 1.320023 | AAUUUA,AGAUAU,AGGAAA,UAGGAA,UAGUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| TARDBP | 3 | 2654 | 0.009592326 | 0.0038476365 | 1.317908 | GAAUGG,GUUGUU,GUUUUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPCL1 | 2 | 2062 | 0.007194245 | 0.0029897078 | 1.266839 | AUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| TIA1 | 12 | 9206 | 0.031175060 | 0.0133428208 | 1.224329 | AUUUUC,AUUUUG,AUUUUU,CCUUUC,GUUUUG,UAUUUU,UUAUUU,UUUUGG,UUUUGU,UUUUUG | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 25 | 18468 | 0.062350120 | 0.0267653478 | 1.220026 | AAUUUA,AUAUUU,AUUAUU,AUUUAU,AUUUUC,AUUUUG,AUUUUU,CAUUUU,UAUUUU,UCAUUU,UGAUUU,UUAGUU,UUAUUU,UUCAUU,UUGAUU,UUUAGU,UUUAUA,UUUCAU,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 6 | 5105 | 0.016786571 | 0.0073996354 | 1.181781 | AUUUAU,UAUUUU,UUAUUU,UUUGUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| U2AF2 | 1 | 1477 | 0.004796163 | 0.0021419234 | 1.162974 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 2 | 2261 | 0.007194245 | 0.0032780993 | 1.133984 | AUUUUU,CAUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM4 | 3 | 3065 | 0.009592326 | 0.0044432593 | 1.110262 | CUUCCU,CUUCUU,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| RALY | 1 | 1553 | 0.004796163 | 0.0022520629 | 1.090634 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| ENOX1 | 3 | 3195 | 0.009592326 | 0.0046316558 | 1.050353 | CAGACA,UAGACA,UAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC5 | 2 | 2400 | 0.007194245 | 0.0034795387 | 1.047947 | GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 5 | 4815 | 0.014388489 | 0.0069793662 | 1.043747 | AAAACA,AAACAC,AACACA,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
RBP on BSJ (Exon Only)
Plot
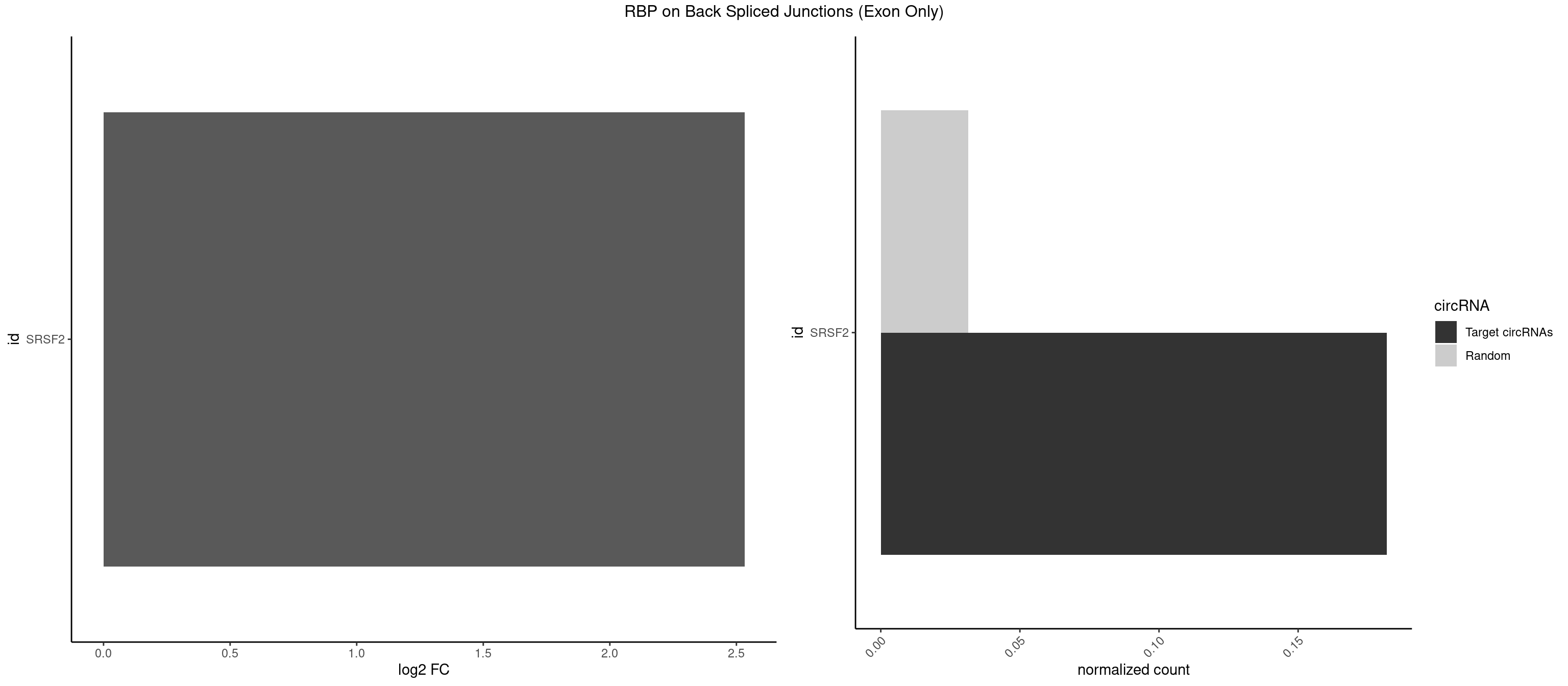
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF2 | 3 | 479 | 0.1818182 | 0.0314383 | 2.531901 | AACCAG,ACCAGU,CCAGUU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
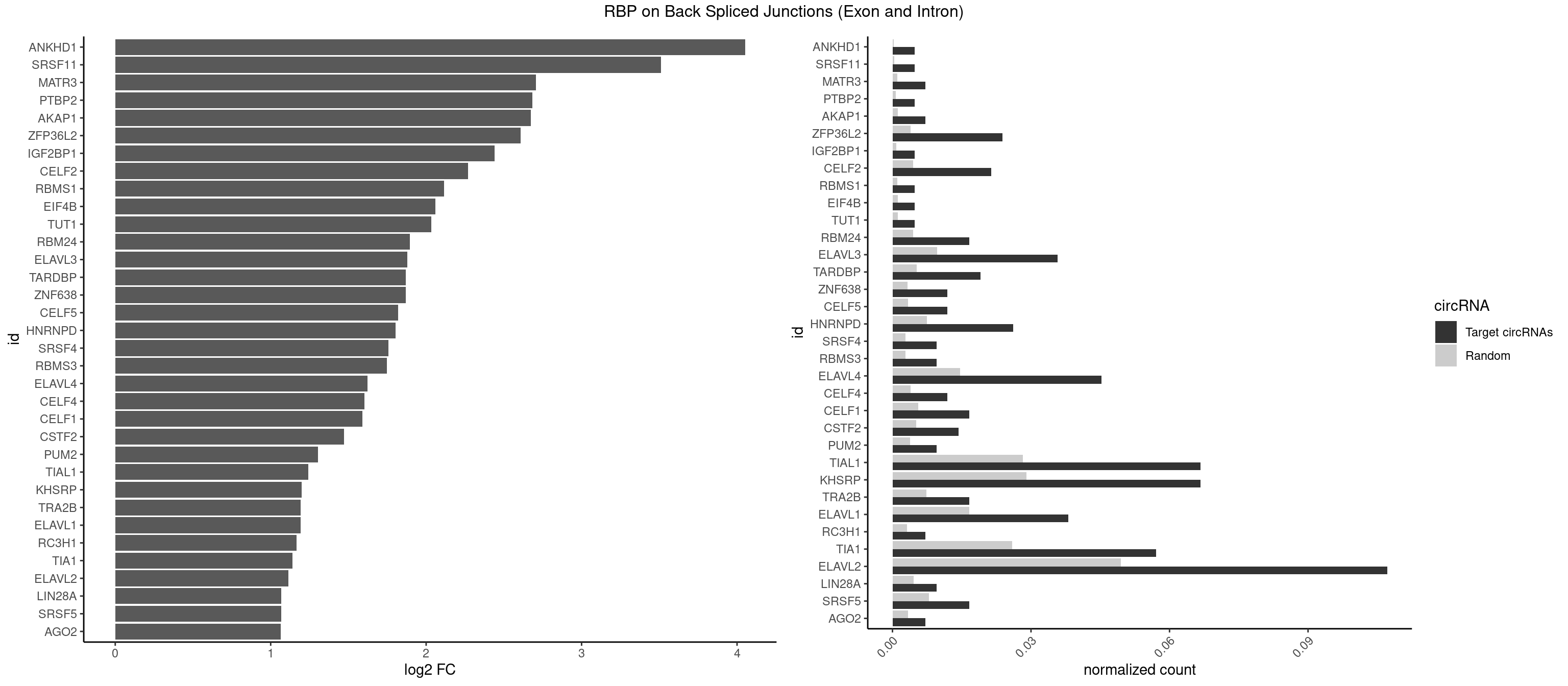
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | GACGUA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| ZFP36L2 | 9 | 774 | 0.023809524 | 0.0039046755 | 2.608264 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | ACCCGU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| CELF2 | 8 | 881 | 0.021428571 | 0.0044437727 | 2.269679 | AUGUGU,GUGUGU,GUUGUU,UAUGUG,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGAGUG,GAGUGU,GUGUGA,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ELAVL3 | 14 | 1928 | 0.035714286 | 0.0097188634 | 1.877642 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| TARDBP | 7 | 1034 | 0.019047619 | 0.0052146312 | 1.868974 | GUGAAU,GUGUGU,GUUGUG,GUUGUU,UGUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | CGUUGU,GUUGUU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPD | 10 | 1488 | 0.026190476 | 0.0075020153 | 1.803692 | AUUUAU,UAUUUA,UUAGAG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SRSF4 | 3 | 558 | 0.009523810 | 0.0028164047 | 1.757684 | AAGAAG,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AAUAUA,AUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ELAVL4 | 18 | 2916 | 0.045238095 | 0.0146966949 | 1.622046 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | GUGUGU,GUUGUG,UGUGUG,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUU,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UACAUA,UAUAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TIAL1 | 27 | 5597 | 0.066666667 | 0.0282043531 | 1.241048 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUU,CAGUUU,GUUUUU,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHSRP | 27 | 5764 | 0.066666667 | 0.0290457477 | 1.198639 | AUAUUU,AUGUGU,AUUUAU,AUUUUA,CCCUCC,CUCCUU,CUGCUU,GUGUAU,GUGUGU,UAGGUU,UAUUUA,UAUUUU,UCCCUC,UGUGUA,UGUGUG,UUAUUU,UUGUAU,UUGUGU,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAGAAG,AAGAAU,AAUAAG,AGGAAA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | AUUUAU,UAUUUA,UAUUUU,UGUUUU,UUAUUU,UUUAUU,UUUGGU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUC,AUUUUU,CCUUUC,CUCCUU,GUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUGU,UUUUUA,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 44 | 9826 | 0.107142857 | 0.0495112858 | 1.113706 | AAUUUU,AUAUUU,AUUUAC,AUUUAU,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUUCU,GUUUUA,GUUUUU,UAUAUA,UAUUUA,UAUUUU,UCAUUU,UGUAUU,UUAUUU,UUCAUU,UUCUUU,UUGUAU,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGAA,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AACAGC,AAGAAG,AUAAAG,GAAGAA,GGAAGA,UACAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.