circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000070961:-:12:89655678:89656107
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000070961:-:12:89655678:89656107 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89655679 | 89656107 | 429 | AUGUGUAUAUCUCAUGAUUGAUAUGGAGAAACUAGUCAUGGGCCAAAGGUCAAGAUACUUCUCUGGGAAAUGUUGCUGCUGAUGCUGCUUUACAAAGUCAUACAAUGAGUGUUUGGUUUAAGAAAGAUUUUCAUACUUAAAAGAUUUUCAUCUUGGAAAUACAUCAAGUGAAAAUUAAAUUCUUUUGGGAAACAUUUUCCUUCUGAUAUAUUAUACUUGUAAUGGGCGACAUGGCAAACAACUCAGUUGCUUACAGUGGUGUGAAAAACUCUUUGAAGGAAGCUAAUCAUGAUGGAGACUUUGGAAUUACGCUCGCAGAGCUGCGGGCUCUCAUGGAGCUCAGGUCCACAGAUGCAUUACGAAAAAUACAGGAAAGCUAUGGAGAUGUCUAUGGAAUUUGCACCAAAUUGAAAACAUCUCCCAAUGAAGAUGUGUAUAUCUCAUGAUUGAUAUGGAGAAACUAGUCAUGGGCCAAAGGU | circ |
| ENSG00000070961:-:12:89655678:89656107 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89655679 | 89656107 | 22 | UCCCAAUGAAGAUGUGUAUAUC | bsj |
| ENSG00000070961:-:12:89655678:89656107 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89656098 | 89656307 | 210 | CUGAUGUACAGUUUUAUUUAAAAGUUGUACUUUGAUCUUUUAUCAUGCAAAGCACUUUAAAAGCACAAGACUGGAGUUCAAAAAUAUUUAUAUUAUUAAAUUAUCUACUUUAGCUACCUGGUGACAUUUAGAAAAUAUGCUUACAUGCAUAAUACUCUUAAGAUAGAUUUUAUUAACUUUUAAAUAUUUUCUUCUUACAGAUGUGUAUAU | ie_up |
| ENSG00000070961:-:12:89655678:89656107 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89655479 | 89655688 | 210 | CCCAAUGAAGGUGAGUUUUAUUAUGCUUGUUUUUGGUUCUUUGUGCAAAGAGUUCUAUAUAUUUGGCUUAUAAAUGAGCAUGCUUAUAUUAUCUUGGCGAUUGUAUGAUUAUGACAUAAUUAUAGUGACAUUAAAAAUGGGUUAGUUAUCAGAUUAAUACAGUUAAAGUAUUUUUAUAUUAACACAUAAAUGUUGCAAAUUAAUUUAUAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
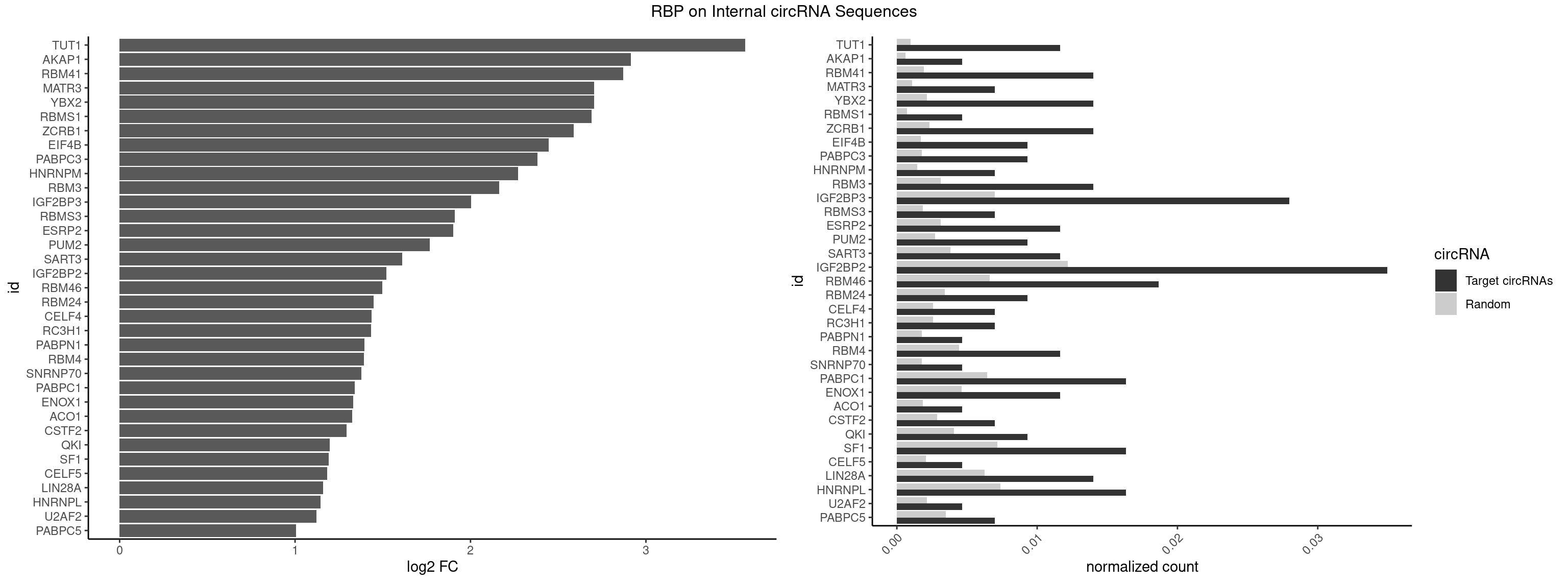
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TUT1 | 4 | 678 | 0.011655012 | 0.0009840095 | 3.566134 | AAAUAC,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| AKAP1 | 1 | 426 | 0.004662005 | 0.0006188101 | 2.913382 | AUAUAU | AUAUAU,UAUAUA |
| RBM41 | 5 | 1318 | 0.013986014 | 0.0019115000 | 2.871208 | AUACAU,AUACUU,UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MATR3 | 2 | 739 | 0.006993007 | 0.0010724109 | 2.705055 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| YBX2 | 5 | 1480 | 0.013986014 | 0.0021462711 | 2.704081 | AACAAC,AACAUC,ACAACU,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMS1 | 1 | 497 | 0.004662005 | 0.0007217036 | 2.691472 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ZCRB1 | 5 | 1605 | 0.013986014 | 0.0023274215 | 2.587180 | AAUUAA,ACUUAA,GAAUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| EIF4B | 3 | 1179 | 0.009324009 | 0.0017100607 | 2.446903 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC3 | 3 | 1234 | 0.009324009 | 0.0017897669 | 2.381179 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPM | 2 | 999 | 0.006993007 | 0.0014492040 | 2.270652 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM3 | 5 | 2152 | 0.013986014 | 0.0031201361 | 2.164304 | AAAACU,AAACUA,GAAACU,GAGACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| IGF2BP3 | 11 | 4815 | 0.027972028 | 0.0069793662 | 2.002817 | AAAAAC,AAAACA,AAACUC,AAAUAC,AAAUUC,AACUCA,AAUACA,CAAACA,CAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RBMS3 | 2 | 1283 | 0.006993007 | 0.0018607779 | 1.910007 | AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ESRP2 | 4 | 2150 | 0.011655012 | 0.0031172377 | 1.902610 | GGGAAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PUM2 | 3 | 1890 | 0.009324009 | 0.0027404447 | 1.766540 | GUAUAU,UACAUC,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SART3 | 4 | 2634 | 0.011655012 | 0.0038186524 | 1.609815 | AAAAAC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| IGF2BP2 | 14 | 8408 | 0.034965035 | 0.0121863560 | 1.520646 | AAAAAC,AAAACA,AAACUC,AAAUAC,AAAUUC,AACUCA,AAUACA,CAAACA,CAACUC,CAUACA,GAAAAC,GCAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM46 | 7 | 4554 | 0.018648019 | 0.0066011240 | 1.498239 | AAUCAU,AAUGAA,AUCAAG,AUCAUG,AUGAAG,AUGAUG,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM24 | 3 | 2357 | 0.009324009 | 0.0034172229 | 1.448126 | GAGUGU,GUGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF4 | 2 | 1782 | 0.006993007 | 0.0025839306 | 1.436346 | GGUGUG,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RC3H1 | 2 | 1786 | 0.006993007 | 0.0025897275 | 1.433113 | CCUUCU,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPN1 | 1 | 1222 | 0.004662005 | 0.0017723764 | 1.395265 | AAAAGA | AAAAGA,AGAAGA |
| RBM4 | 4 | 3065 | 0.011655012 | 0.0044432593 | 1.391260 | CCUUCU,CUCUUU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SNRNP70 | 1 | 1237 | 0.004662005 | 0.0017941145 | 1.377678 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC1 | 6 | 4443 | 0.016317016 | 0.0064402624 | 1.341186 | AAAAAC,CAAACA,CUAAUC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ENOX1 | 4 | 3195 | 0.011655012 | 0.0046316558 | 1.331350 | AAUACA,AUACAG,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ACO1 | 1 | 1283 | 0.004662005 | 0.0018607779 | 1.325045 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CSTF2 | 2 | 1967 | 0.006993007 | 0.0028520334 | 1.293922 | GUGUUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| QKI | 3 | 2805 | 0.009324009 | 0.0040664663 | 1.197175 | AAUCAU,CUAAUC,UAAUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SF1 | 6 | 4931 | 0.016317016 | 0.0071474739 | 1.190872 | CACAGA,UACGAA,UAUACU,UGCUGA,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF5 | 1 | 1415 | 0.004662005 | 0.0020520728 | 1.183869 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| LIN28A | 5 | 4315 | 0.013986014 | 0.0062547643 | 1.160957 | GGAGAA,GGAGAU,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPL | 6 | 5085 | 0.016317016 | 0.0073706513 | 1.146513 | AAACAA,AAAUAC,AAUACA,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| U2AF2 | 1 | 1477 | 0.004662005 | 0.0021419234 | 1.122044 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| PABPC5 | 2 | 2400 | 0.006993007 | 0.0034795387 | 1.007017 | AGAAAG,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
RBP on BSJ (Exon Only)
Plot
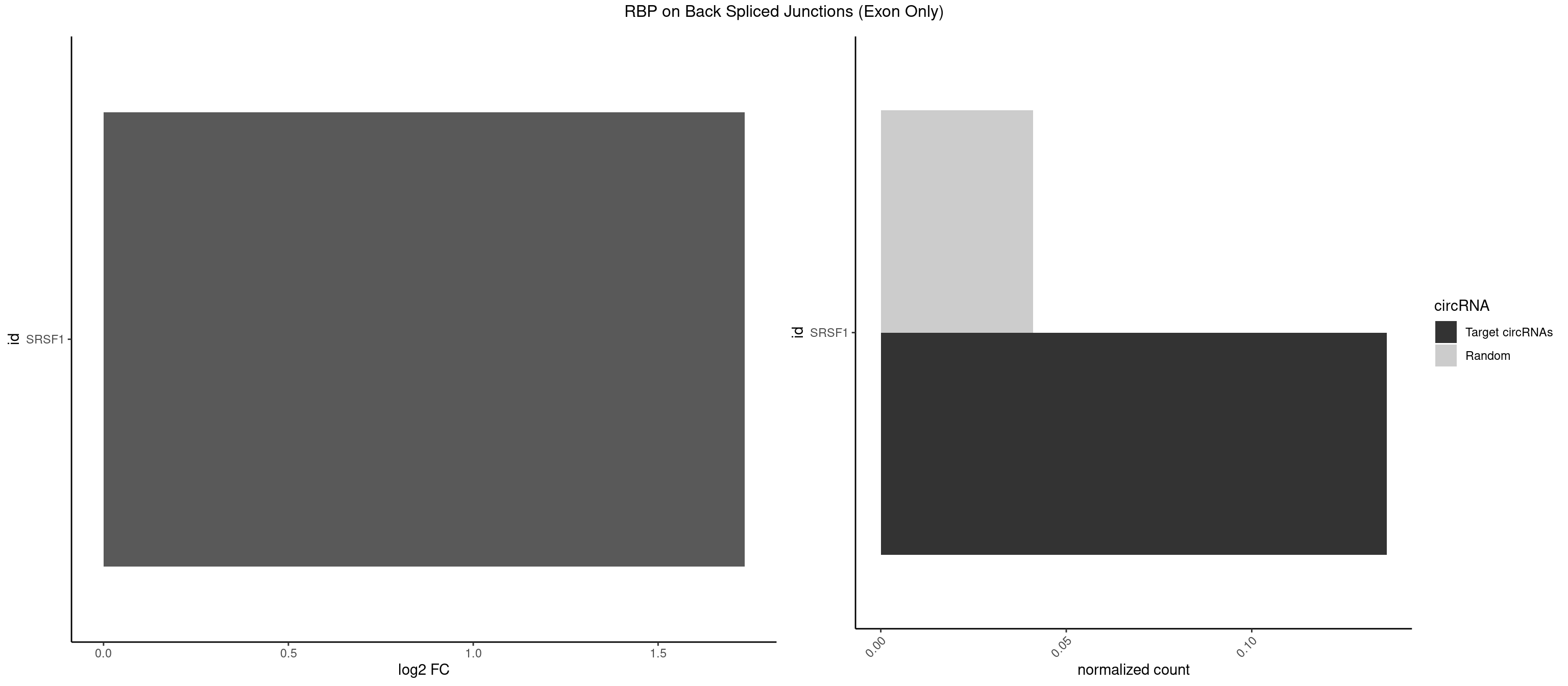
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF1 | 2 | 625 | 0.1363636 | 0.04100079 | 1.733736 | GAAGAU,UGAAGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
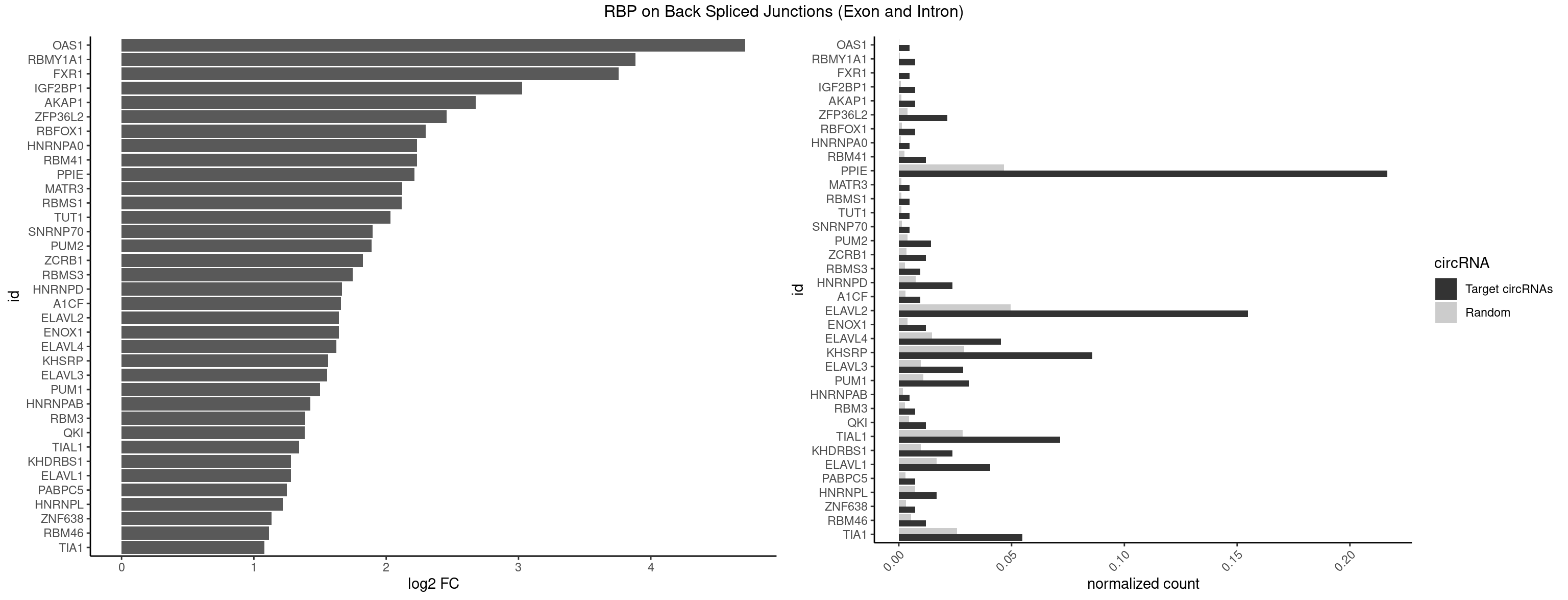
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| RBMY1A1 | 2 | 95 | 0.007142857 | 0.0004836759 | 3.884389 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| IGF2BP1 | 2 | 173 | 0.007142857 | 0.0008766626 | 3.026408 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| ZFP36L2 | 8 | 774 | 0.021428571 | 0.0039046755 | 2.456261 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGC | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | UACAUG,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PPIE | 90 | 9262 | 0.216666667 | 0.0466696896 | 2.214919 | AAAAAU,AAAAUA,AAAUAU,AAAUUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AUAAAU,AUAAUA,AUAAUU,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAUA,UAAAUU,UAAUUA,UAAUUU,UAUAAA,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | GUAUAU,UAAAUA,UAUAUA,UGUACA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | AAUUAA,AUUUAA,GAUUAA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPD | 9 | 1488 | 0.023809524 | 0.0075020153 | 1.666189 | AAUUUA,AUUUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ELAVL2 | 64 | 9826 | 0.154761905 | 0.0495112858 | 1.644221 | AAUUUA,AUAUUU,AUUAUU,AUUUAA,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUU,CUUAUA,CUUUAA,CUUUUA,GAUUUU,GUUUUA,GUUUUU,UAAUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUUAU,UAUUUA,UAUUUG,UAUUUU,UCUUUU,UUAAUU,UUAGUU,UUAUAU,UUAUUU,UUCUUU,UUGUAU,UUUAUA,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAUACA,AUACAG,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ELAVL4 | 18 | 2916 | 0.045238095 | 0.0146966949 | 1.622046 | AUUUAU,UAUCUA,UAUUUA,UAUUUU,UUAUUU,UUGUAU,UUUAUU,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHSRP | 35 | 5764 | 0.085714286 | 0.0290457477 | 1.561209 | AUAUUU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,GUAUAU,GUGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UGUAUA,UGUAUG,UGUGUA,UUAUUA,UUAUUU,UUGUAU,UUUAUA,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| ELAVL3 | 11 | 1928 | 0.028571429 | 0.0097188634 | 1.555714 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PUM1 | 12 | 2172 | 0.030952381 | 0.0109482064 | 1.499356 | AAUAUU,AAUGUU,ACAUAA,AUUGUA,GUAUAU,UAAAUA,UAUAUA,UGUACA,UGUAUA,UUGUAC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | AUCUAC,AUUAAC,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| TIAL1 | 29 | 5597 | 0.071428571 | 0.0282043531 | 1.340583 | AGUUUU,AUUUUA,AUUUUC,AUUUUU,CAGUUU,CUUUUA,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUUUAA,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | AUUUAU,UAUUUA,UAUUUU,UGUUUU,UUAGUU,UUAUUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AACACA,AAUACA,ACAUAA,CACAAG,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUGAA,AUCAUG,AUGAAG,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| TIA1 | 22 | 5140 | 0.054761905 | 0.0259018541 | 1.080117 | AUUUUC,AUUUUU,CUUUUA,GUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUUA,UUUUUG | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.