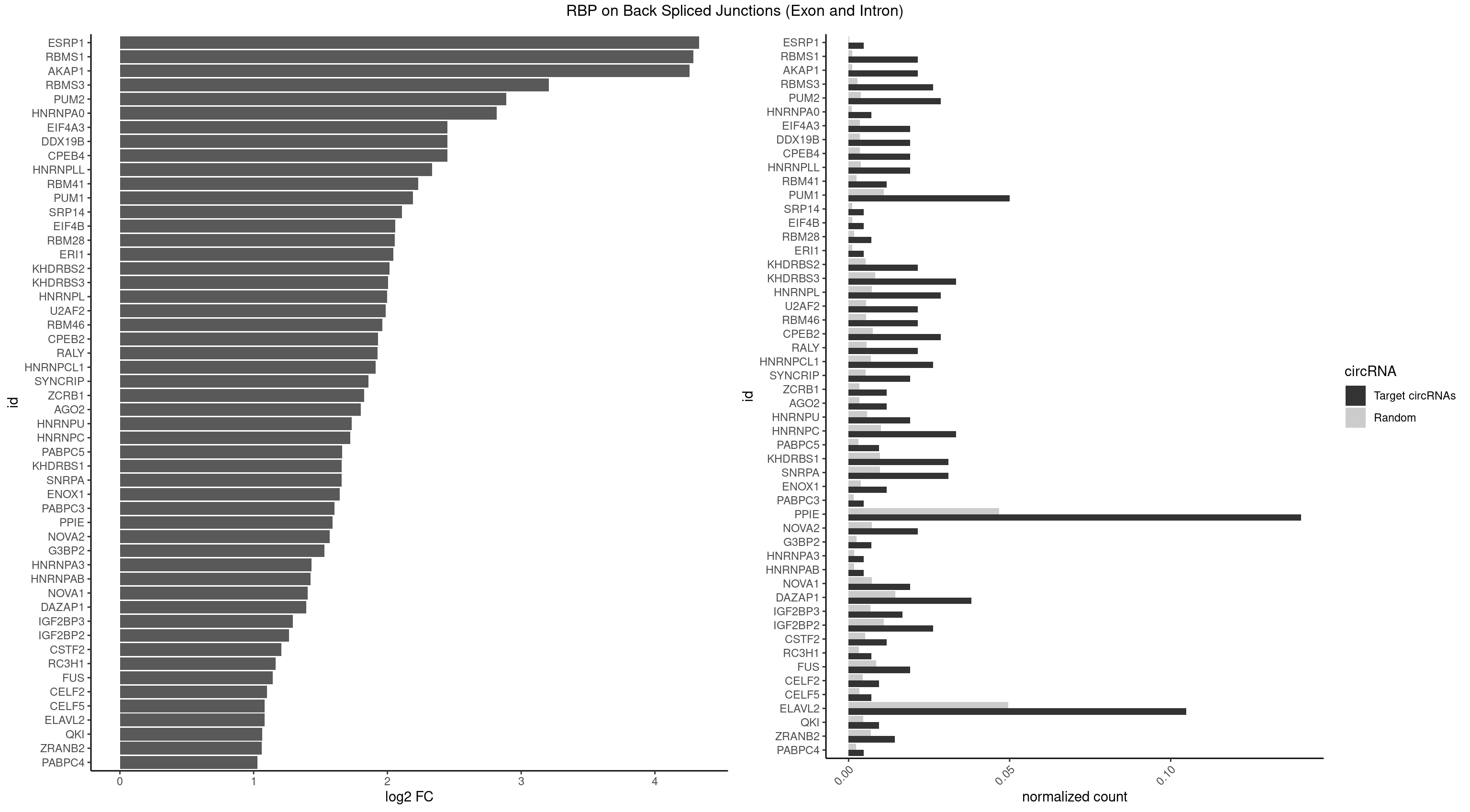
| ESRP1 |
1 |
46 |
0.004761905 |
0.0002367997 |
4.329800 |
AGGGAU |
AGGGAU |
| RBMS1 |
8 |
217 |
0.021428571 |
0.0010983474 |
4.286129 |
AUAUAC,UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 |
8 |
221 |
0.021428571 |
0.0011185006 |
4.259898 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| RBMS3 |
10 |
562 |
0.026190476 |
0.0028365578 |
3.206829 |
AAUAUA,AUAUAU,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PUM2 |
11 |
764 |
0.028571429 |
0.0038542926 |
2.890035 |
GUAUAU,UAAAUA,UAUAUA,UGUAGA,UGUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPA0 |
2 |
200 |
0.007142857 |
0.0010126965 |
2.818299 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| CPEB4 |
7 |
691 |
0.019047619 |
0.0034864974 |
2.449760 |
UUUUUU |
UUUUUU |
| DDX19B |
7 |
691 |
0.019047619 |
0.0034864974 |
2.449760 |
UUUUUU |
UUUUUU |
| EIF4A3 |
7 |
691 |
0.019047619 |
0.0034864974 |
2.449760 |
UUUUUU |
UUUUUU |
| HNRNPLL |
7 |
748 |
0.019047619 |
0.0037736800 |
2.335567 |
ACACAC,ACACCA,CAAACA,CACACA,GCAAAC |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM41 |
4 |
502 |
0.011904762 |
0.0025342604 |
2.231902 |
AUACAU,UACAUG,UACAUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PUM1 |
20 |
2172 |
0.050000000 |
0.0109482064 |
2.191234 |
AAUAUU,AAUGUU,AGAAUU,AGAUAA,GUAUAU,UAAAUA,UAAUAU,UAAUGU,UAUAUA,UGUAGA,UGUAUA,UUAAUG,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| SRP14 |
1 |
218 |
0.004761905 |
0.0011033857 |
2.109602 |
CCUGUA |
CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B |
1 |
226 |
0.004761905 |
0.0011436921 |
2.057840 |
UUGGAA |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 |
2 |
340 |
0.007142857 |
0.0017180572 |
2.055723 |
AGUAGU,UGUAGA |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 |
1 |
228 |
0.004761905 |
0.0011537686 |
2.045185 |
UUUCAG |
UUCAGA,UUUCAG |
| KHDRBS2 |
8 |
1051 |
0.021428571 |
0.0053002821 |
2.015395 |
AAUAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| KHDRBS3 |
13 |
1646 |
0.033333333 |
0.0082980653 |
2.006119 |
AAUAAA,AGAUAA,AUAAAG,AUUAAA,UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPL |
11 |
1419 |
0.028571429 |
0.0071543732 |
1.997676 |
AAACAA,AACAAA,AAUAAA,ACACAA,ACACAC,ACACCA,CACAAA,CACACA,CAUAAA |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| U2AF2 |
8 |
1071 |
0.021428571 |
0.0054010480 |
1.988224 |
UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| RBM46 |
8 |
1091 |
0.021428571 |
0.0055018138 |
1.961556 |
AAUGAA,AAUGAU,AUCAAU,AUCAUU,AUGAAA,AUGAUU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CPEB2 |
11 |
1487 |
0.028571429 |
0.0074969770 |
1.930192 |
AUUUUU,CAUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY |
8 |
1117 |
0.021428571 |
0.0056328094 |
1.927609 |
UUUUUC,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 |
10 |
1381 |
0.026190476 |
0.0069629182 |
1.911278 |
AUUUUU,CUUUUU,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP |
7 |
1041 |
0.019047619 |
0.0052498992 |
1.859249 |
UUUUUU |
AAAAAA,UUUUUU |
| ZCRB1 |
4 |
666 |
0.011904762 |
0.0033605401 |
1.824774 |
AAUUAA,AUUUAA,GAUUAA,GGAUUA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 |
4 |
677 |
0.011904762 |
0.0034159613 |
1.801175 |
AAAGUG,AAGUGC,AGUGCU,GUGCUU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPU |
7 |
1136 |
0.019047619 |
0.0057285369 |
1.733372 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPC |
13 |
2006 |
0.033333333 |
0.0101118501 |
1.720919 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PABPC5 |
3 |
596 |
0.009523810 |
0.0030078597 |
1.662801 |
AGAAAG,GAAAGU,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| KHDRBS1 |
12 |
1945 |
0.030952381 |
0.0098045143 |
1.658532 |
AUUUAA,CUAAAU,UAAAAA,UUAAAA,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SNRPA |
12 |
1947 |
0.030952381 |
0.0098145909 |
1.657050 |
AGGAGA,AGUAGU,CAGUAG,GGAGAU,GUAGUC,GUAUGC,GUUACC,UACCUG,UUACCU |
ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| ENOX1 |
4 |
756 |
0.011904762 |
0.0038139863 |
1.642167 |
AUACAG,UAUACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC3 |
1 |
310 |
0.004761905 |
0.0015669085 |
1.603618 |
AAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| PPIE |
58 |
9262 |
0.140476190 |
0.0466696896 |
1.589768 |
AAAUAU,AAAUUA,AAUAAA,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUUAA,AUUUUU,UAAAAA,UAAAUA,UAAAUU,UAAUAU,UAAUUU,UAUAAA,UAUAUA,UUAAAA,UUAAUA,UUAAUU,UUAUAU,UUAUUA,UUUAAU,UUUUAA,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| NOVA2 |
8 |
1434 |
0.021428571 |
0.0072299476 |
1.567479 |
AGUCAU,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| G3BP2 |
2 |
490 |
0.007142857 |
0.0024738009 |
1.529772 |
AGGAUU,GGAUUA |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPA3 |
1 |
349 |
0.004761905 |
0.0017634019 |
1.433177 |
AAGGAG |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB |
1 |
351 |
0.004761905 |
0.0017734784 |
1.424957 |
ACAAAG |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NOVA1 |
7 |
1428 |
0.019047619 |
0.0071997179 |
1.403598 |
UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| DAZAP1 |
15 |
2878 |
0.038095238 |
0.0145052398 |
1.393037 |
AAUUUA,AGGAAA,AGGUAA,AGUAAA,AGUAAG,UAGGAA,UAGGAU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| IGF2BP3 |
6 |
1348 |
0.016666667 |
0.0067966546 |
1.294069 |
AAAAAC,ACACAC,CAAACA,CACACA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 |
10 |
2163 |
0.026190476 |
0.0109028617 |
1.264335 |
AAAAAC,ACACAC,CAAAAC,CAAACA,CAACUC,CACACA,GAAUUC,GCAAAC |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| CSTF2 |
4 |
1022 |
0.011904762 |
0.0051541717 |
1.207726 |
GUGUUU,GUUUUG,UGUGUU,UGUUUU |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RC3H1 |
2 |
631 |
0.007142857 |
0.0031841999 |
1.165570 |
CCUUCU,UCUGUG |
CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| FUS |
7 |
1711 |
0.019047619 |
0.0086255542 |
1.142922 |
UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| CELF2 |
3 |
881 |
0.009523810 |
0.0044437727 |
1.099754 |
AUGUGU,GUCUGU,UAUGUG |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 |
2 |
669 |
0.007142857 |
0.0033756550 |
1.081334 |
GUGUUU,UGUGUU |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL2 |
43 |
9826 |
0.104761905 |
0.0495112858 |
1.081285 |
AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUAG,AUUUUU,CAUUUU,CUUUUU,GUUUUA,UAAUUU,UAUAUA,UCAUUU,UGUAUU,UUAAGU,UUAAUU,UUAUAU,UUAUGU,UUCUUU,UUUAAG,UUUAAU,UUUAGU,UUUAUG,UUUUAA,UUUUAG,UUUUUA,UUUUUC,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| QKI |
3 |
904 |
0.009523810 |
0.0045596534 |
1.062615 |
ACACAC,UCUACU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZRANB2 |
5 |
1360 |
0.014285714 |
0.0068571141 |
1.058900 |
AAAGGU,AGGUAA,GGUAAA,GUAAAG,UAAAGG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| PABPC4 |
1 |
462 |
0.004761905 |
0.0023327287 |
1.029520 |
AAAAAG |
AAAAAA,AAAAAG |