circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000106683:+:7:74096621:74099238
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000106683:+:7:74096621:74099238 | ENSG00000106683 | ENST00000418310 | + | 7 | 74096622 | 74099238 | 456 | GUGUUGUGACUGCAGUGCCUCCCUGUCGCACCAGUACUAUGAGAAGGAUGGGCAGCUCUUCUGCAAGAAGGACUACUGGGCCCGCUAUGGCGAGUCCUGCCAUGGGUGCUCUGAGCAAAUCACCAAGGGACUGGUUAUGGUGGCUGGGGAGCUGAAGUACCACCCCGAGUGUUUCAUCUGCCUCACGUGUGGGACCUUUAUCGGUGACGGGGACACCUACACGCUGGUGGAGCACUCCAAGCUGUACUGCGGGCACUGCUACUACCAGACUGUGGUGACCCCCGUCAUCGAGCAGAUCCUGCCUGACUCCCCUGGCUCCCACCUGCCCCACACCGUCACCCUGGUGUCCAUCCCAGCCUCAUCUCAUGGCAAGCGUGGACUUUCAGUCUCCAUUGACCCCCCGCACGGCCCACCGGGCUGUGGCACCGAGCACUCACACACCGUCCGCGUCCAGGGGUGUUGUGACUGCAGUGCCUCCCUGUCGCACCAGUACUAUGAGAAGGAUG | circ |
| ENSG00000106683:+:7:74096621:74099238 | ENSG00000106683 | ENST00000418310 | + | 7 | 74096622 | 74099238 | 22 | CGCGUCCAGGGGUGUUGUGACU | bsj |
| ENSG00000106683:+:7:74096621:74099238 | ENSG00000106683 | ENST00000418310 | + | 7 | 74096422 | 74096631 | 210 | ACCUGGAAGGCAGAGGUUGCAGUGAGCCGAGAUCGAGCCACUGCACUGCAGCCUGGCGACAGAGCAAGACUCCGUCUCAAAAAACAAACAAAAAGAAAACUUGUUCUAAUUCUUACAAAGGUGCCUGUAGCCGAGGCAGGGGCCCAGGUGAGGUGGAGGAGGGCGGGAGUGGACGUCUCAGCCCGGCCCCUCUCCUGCAGGUGUUGUGAC | ie_up |
| ENSG00000106683:+:7:74096621:74099238 | ENSG00000106683 | ENST00000418310 | + | 7 | 74099229 | 74099438 | 210 | GCGUCCAGGGGUGAGUGGCCGGCCUGCCGAGGCUGCCGUCGGUGUGGCUAUGGCUGUUGAUGUGGGUGGCAGAGUCUGGCACUGGGGGCCCUGAAAAUGAAUGGGCGAGUGUUUGGGUACAGAUGGGGCCCAGUUCUGACAACCUGGUUUGCCAGAUUUCUGGCCCAGUCAUUCCUCUGAAUACCAUUACAAAUGCCAGAUACAAUAAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
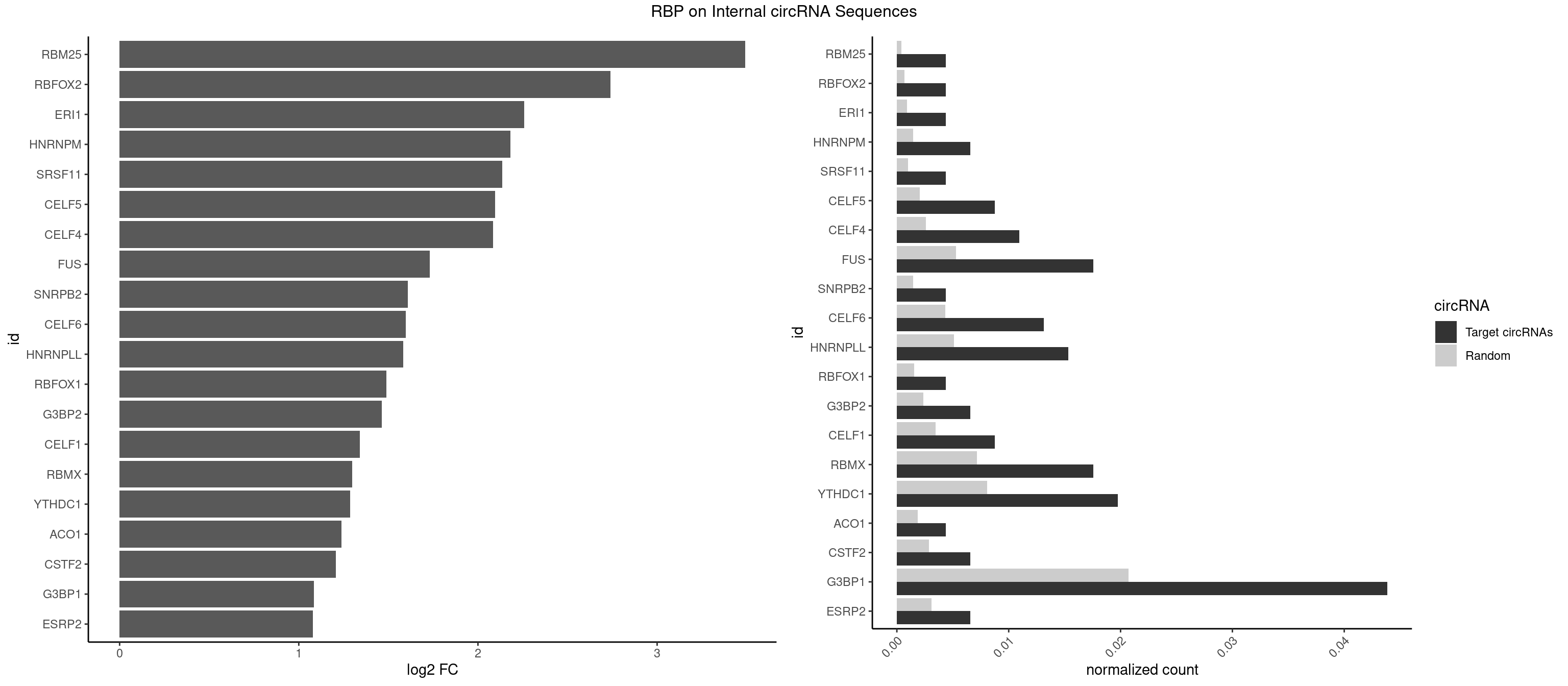
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 268 | 0.004385965 | 0.0003898359 | 3.491956 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBFOX2 | 1 | 452 | 0.004385965 | 0.0006564894 | 2.740051 | UGACUG | UGACUG,UGCAUG |
| ERI1 | 1 | 632 | 0.004385965 | 0.0009173461 | 2.257356 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPM | 2 | 999 | 0.006578947 | 0.0014492040 | 2.182596 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| SRSF11 | 1 | 688 | 0.004385965 | 0.0009985015 | 2.135058 | AAGAAG | AAGAAG |
| CELF5 | 3 | 1415 | 0.008771930 | 0.0020520728 | 2.095812 | GUGUGG,GUGUUG,GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 1782 | 0.010964912 | 0.0025839306 | 2.085255 | GGUGUU,GUGUGG,GUGUUG,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| FUS | 7 | 3648 | 0.017543860 | 0.0052881452 | 1.730132 | CGGUGA,GGGUGC,GGGUGU,UGGUGA,UGGUGG,UGGUGU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| SNRPB2 | 1 | 991 | 0.004385965 | 0.0014376103 | 1.609222 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF6 | 5 | 2997 | 0.013157895 | 0.0043447134 | 1.598596 | GUGGUG,GUGUGG,GUGUUG,UGUGGG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPLL | 6 | 3534 | 0.015350877 | 0.0051229360 | 1.583278 | ACACAC,ACUGCA,CACACA,CACACC,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBFOX1 | 1 | 1077 | 0.004385965 | 0.0015622419 | 1.489276 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 2 | 1644 | 0.006578947 | 0.0023839405 | 1.464509 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CELF1 | 3 | 2391 | 0.008771930 | 0.0034664959 | 1.339416 | GUGUUG,GUUGUG,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBMX | 7 | 4925 | 0.017543860 | 0.0071387787 | 1.297217 | AAGAAG,AGAAGG,AGUGUU,AUCCCA,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| YTHDC1 | 8 | 5576 | 0.019736842 | 0.0080822104 | 1.288069 | GACUAC,GACUGC,GGGUGC,UACUAC,UACUGC,UCCUGC,UGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| ACO1 | 1 | 1283 | 0.004385965 | 0.0018607779 | 1.236988 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CSTF2 | 2 | 1967 | 0.006578947 | 0.0028520334 | 1.205866 | GUGUUG,GUGUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| G3BP1 | 19 | 14282 | 0.043859649 | 0.0206989801 | 1.083335 | ACACGC,ACCCCC,CACCGG,CCACAC,CCACCC,CCACCG,CCAUCC,CCCACA,CCCACC,CCCCAC,CCCCCC,CCCCCG,CCCCGC,CCCGCA,CCGCAC,CGGCCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| ESRP2 | 2 | 2150 | 0.006578947 | 0.0031172377 | 1.077589 | GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
RBP on BSJ (Exon Only)
Plot
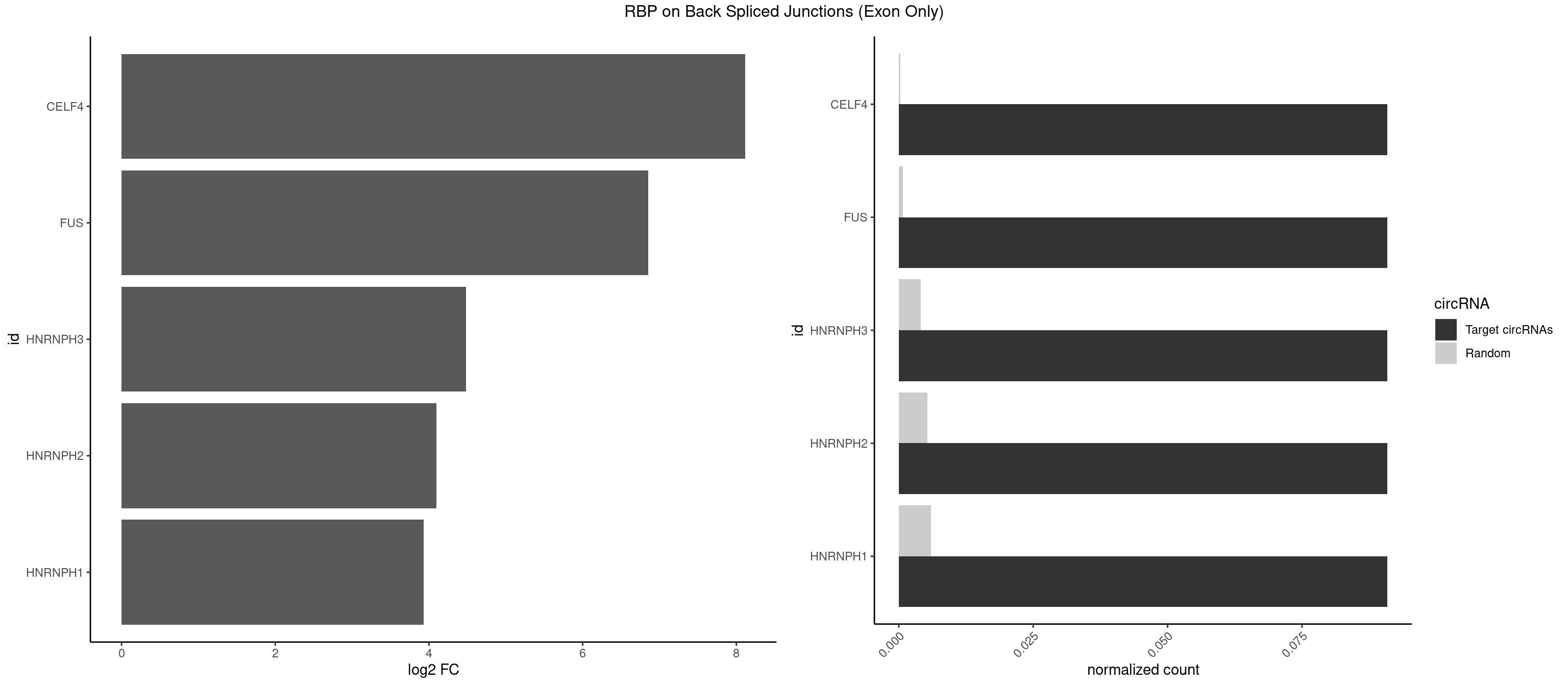
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF4 | 1 | 4 | 0.09090909 | 0.0003274823 | 8.116864 | GGUGUU | GGUGUG,GUGUUG,UGUGUG |
| FUS | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | GGGUGU | AAAAAA,CGGUGG,GGGUGA,GGGUGG,GGGUGU |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | GGGUGU | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.0053052135 | 4.098942 | GGGUGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | GGGUGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
RBP on BSJ (Exon and Intron)
Plot
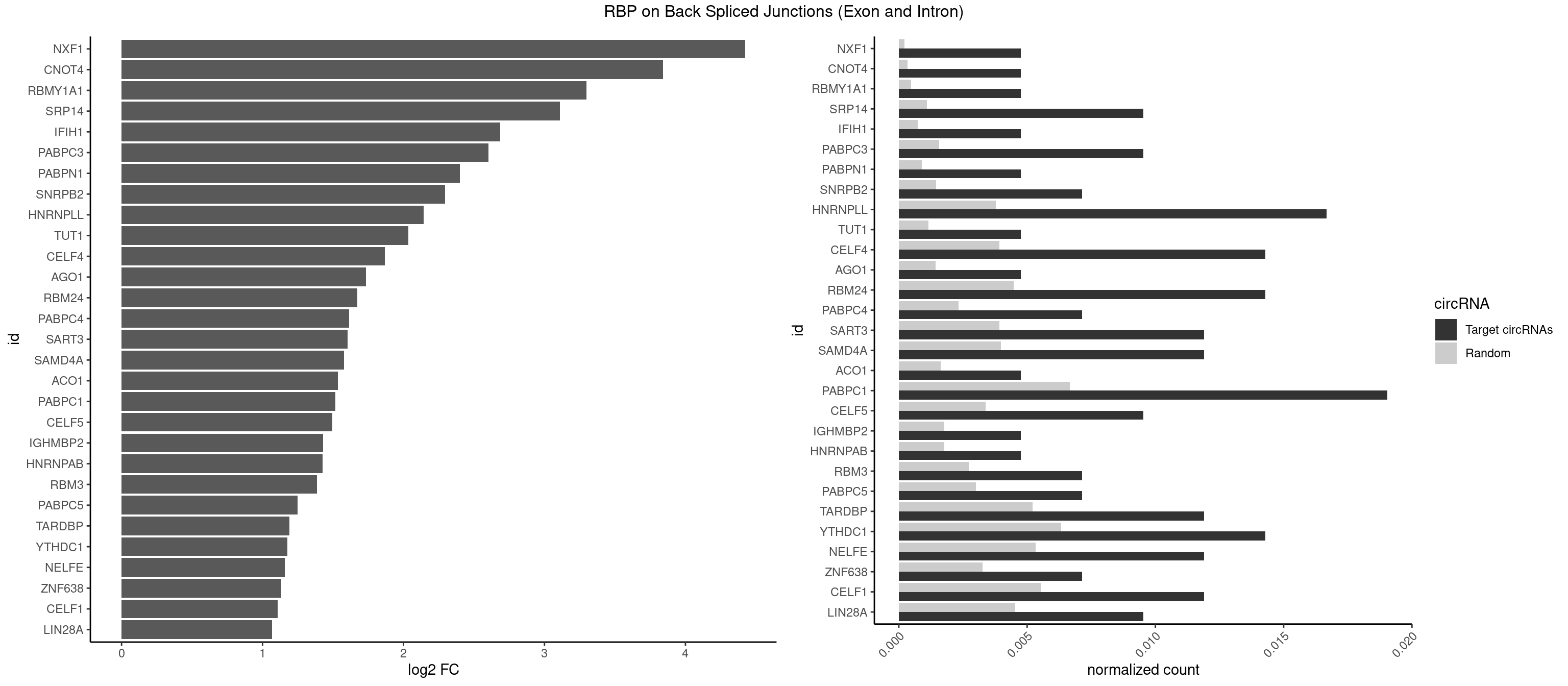
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| SRP14 | 3 | 218 | 0.009523810 | 0.0011033857 | 3.109602 | CCUGUA,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPLL | 6 | 748 | 0.016666667 | 0.0037736800 | 2.142922 | ACAAAC,ACUGCA,CAAACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GGUGUG,GGUGUU,GUGUGG,GUGUUG,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBM24 | 5 | 888 | 0.014285714 | 0.0044790407 | 1.673311 | GAGUGG,GAGUGU,GUGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AAAAAC,AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CUGGAA,CUGGCA,CUGGCC,GCGGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PABPC1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | AAAAAA,AAAAAC,ACAAAC,ACAAAU,AGAAAA,CAAACA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGG,GUGUUG,GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGG,GUUGUG,UGAAUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAAUAC,GCCUGC,GGCUGC,GGGUAC,UCCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUGGUU,UCUGGC,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUGUUG,GUUGUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGGA,GGAGGG,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.