circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000153786:-:16:84990303:84996007
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000153786:-:16:84990303:84996007 | ENSG00000153786 | ENST00000313732 | - | 16 | 84990304 | 84996007 | 418 | AUCCGAUCACAUAGGACAGUAUGCACCUUAAGAUCCUGAAGAAACGGCACAAAAUGUUCAAGUGAUGUUUAGAAAUAACUUGUGAGGGUGCGUCAGGGAAAUCAUGCAGCCAUCAGGACACAGGCUCCGGGACGUCGAGCAUCAUCCUCUCCUGGCUGAAAAUGACAACUAUGACUCUUCAUCGUCCUCCUCCUCCGAGGCUGACGUGGCUGACCGGGUCUGGUUCAUCCGUGACGGCUGCGGCAUGAUCUGUGCUGUCAUGACGUGGCUUCUGGUCGCCUAUGCAGACUUCGUGGUGACUUUCGUCAUGCUGCUGCCUUCCAAAGACUUCUGGUACUCUGUGGUCAACGGGGUCAUCUUUAACUGCUUGGCCGUGCUUGCCCUGUCAUCCCACCUGAGAACCAUGCUCACCGACCCUAUCCGAUCACAUAGGACAGUAUGCACCUUAAGAUCCUGAAGAAACGGCAC | circ |
| ENSG00000153786:-:16:84990303:84996007 | ENSG00000153786 | ENST00000313732 | - | 16 | 84990304 | 84996007 | 22 | UCACCGACCCUAUCCGAUCACA | bsj |
| ENSG00000153786:-:16:84990303:84996007 | ENSG00000153786 | ENST00000313732 | - | 16 | 84995998 | 84996207 | 210 | UACUUGAUUUCCAGGAUUGUGAAUUUACCUAGUAAUCAAAACUUAGAAGAGCAAUGAUUUCAUACUUAAGGAUAGUUCUGAGUGACAGAACUUGAUGCGUCAAAUGUUACGCUCCAAAAAUUAAUCAUACCCUAAGAAAUAUGACACACGUCUGUCAUAAAAUAAAUCACUUUUUCUAAUCAGAUUUUAUUACCUUUCAGAUCCGAUCAC | ie_up |
| ENSG00000153786:-:16:84990303:84996007 | ENSG00000153786 | ENST00000313732 | - | 16 | 84990104 | 84990313 | 210 | CACCGACCCUGUGAGUACUGGCUGCGUCUCUGGGUGGCUCUCUUGUGUCUGGUCUAGUGUCCUCGGGUGUGGCUGACCCUUUGGAGGAAGCAGUGCACAGGGACAUAUUGACAUGGGUGUGGAACAAGAAAGGUGGAUUGAGCUCAUUUUGGGAGCAGAUUCCCUUUAGGCUGAGAAUAAACAUAGGCCAGACCUAGCGCAGUGAAAUAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
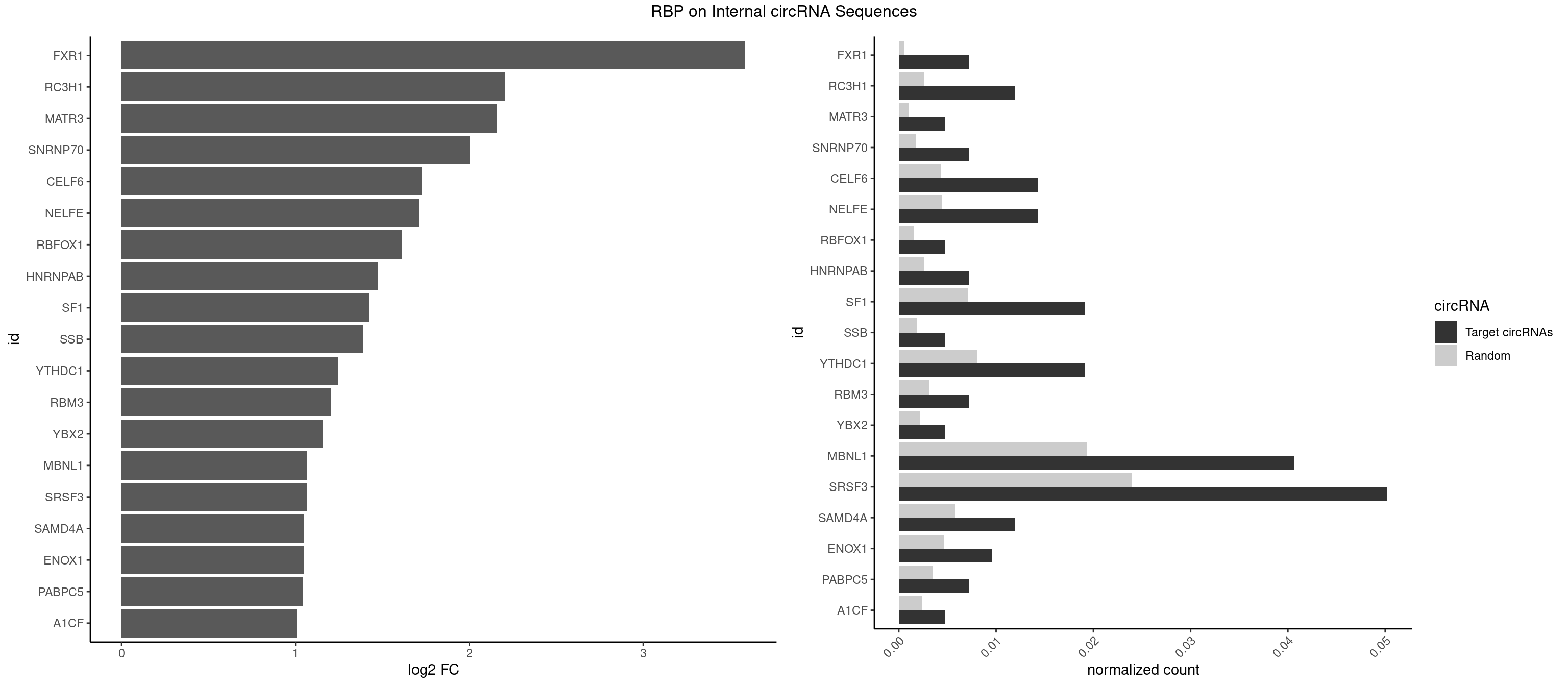
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 2 | 411 | 0.007177033 | 0.000597072 | 3.587411 | AUGACA,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| RC3H1 | 4 | 1786 | 0.011961722 | 0.002589727 | 2.207553 | CUUCUG,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| MATR3 | 1 | 739 | 0.004784689 | 0.001072411 | 2.157567 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SNRNP70 | 2 | 1237 | 0.007177033 | 0.001794114 | 2.000116 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF6 | 5 | 2997 | 0.014354067 | 0.004344713 | 1.724127 | GUGAGG,GUGAUG,GUGGUG,UGUGAG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| NELFE | 5 | 3028 | 0.014354067 | 0.004389639 | 1.709285 | CUGGCU,CUGGUU,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBFOX1 | 1 | 1077 | 0.004784689 | 0.001562242 | 1.614807 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPAB | 2 | 1782 | 0.007177033 | 0.002583931 | 1.473820 | AAAGAC,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SF1 | 7 | 4931 | 0.019138756 | 0.007147474 | 1.420992 | ACCGAC,CACCGA,GCUGAC,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SSB | 1 | 1260 | 0.004784689 | 0.001827446 | 1.388596 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| YTHDC1 | 7 | 5576 | 0.019138756 | 0.008082210 | 1.243675 | GGCUGC,GGGUGC,UCAUGC,UGCUGC,UGGUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBM3 | 2 | 2152 | 0.007177033 | 0.003120136 | 1.201779 | AAGACU,GAAACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| YBX2 | 1 | 1480 | 0.004784689 | 0.002146271 | 1.156593 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| MBNL1 | 16 | 13372 | 0.040669856 | 0.019380204 | 1.069376 | AUGCUC,CUGCCU,CUGCGG,CUGCUG,CUGCUU,CUUGUG,GCUGCG,GCUGCU,GCUUGC,GUGCUG,GUGCUU,UGCGGC,UGCUGC,UGCUGU,UUGCCC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| SRSF3 | 20 | 16536 | 0.050239234 | 0.023965486 | 1.067856 | ACUUCG,ACUUCU,AUCAUC,CAUCAU,CAUCGU,CUCUUC,CUUCAU,GACUUC,GUCAAC,UCAACG,UCAUCC,UCAUCG,UCAUCU,UCGUCC,UCUUCA,UUCAUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| SAMD4A | 4 | 3992 | 0.011961722 | 0.005786671 | 1.047620 | CGGGAC,CGGGUC,CUGGUA,CUGGUC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| ENOX1 | 3 | 3195 | 0.009569378 | 0.004631656 | 1.046897 | AGGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC5 | 2 | 2400 | 0.007177033 | 0.003479539 | 1.044492 | AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 1 | 1642 | 0.004784689 | 0.002381042 | 1.006832 | CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
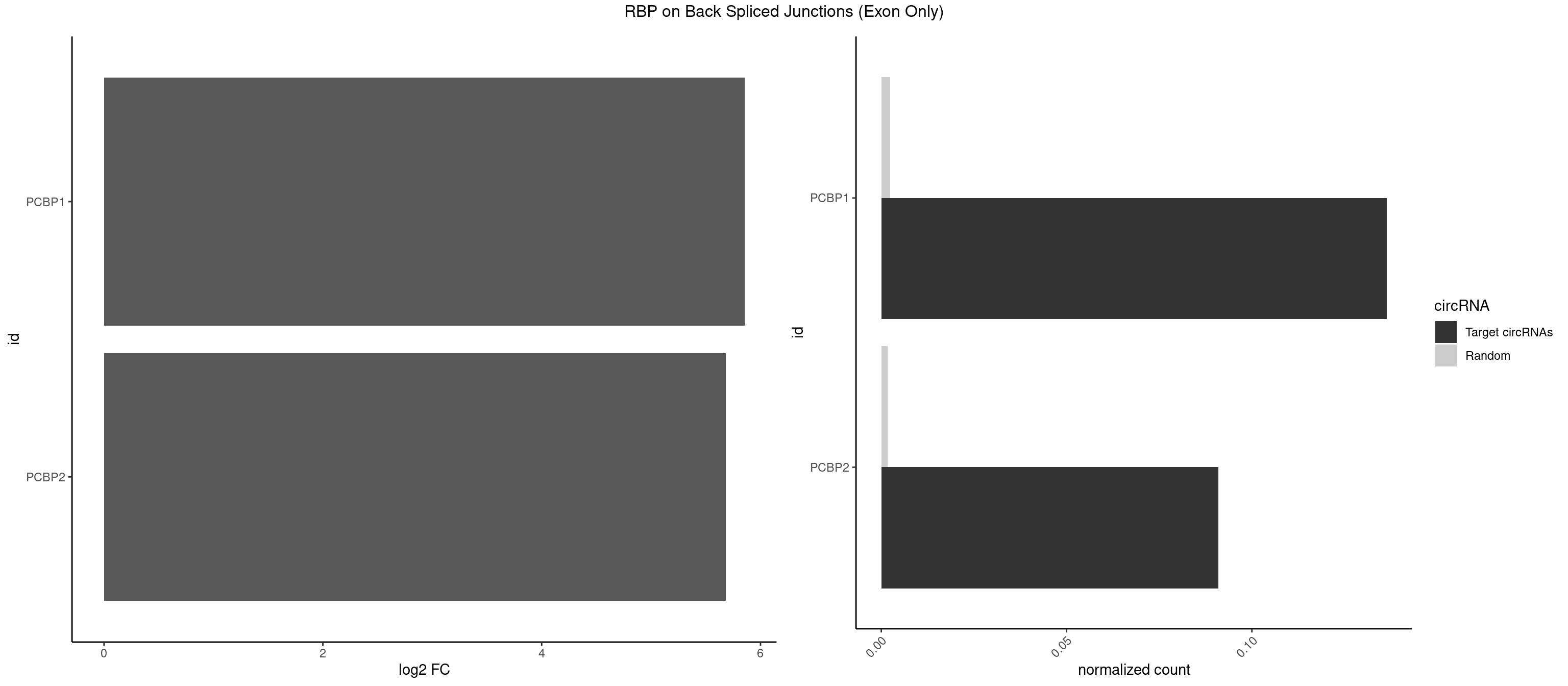
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PCBP1 | 2 | 35 | 0.13636364 | 0.002357873 | 5.853829 | CCUAUC,CUAUCC | AAAAAA,AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,ACCCUC,AUUAAA,CAAACC,CAUACC,CCAACC,CCACCC,CCAUAC,CCAUUC,CCCACC,CCCCAC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,UUAGAG,UUAGGA,UUAGGG |
| PCBP2 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | ACCCUA | AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,AUUAAA,CCAUUC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CUCCCU,CUUCCA,UAACCC,UUAGAG,UUAGGA,UUAGGG |
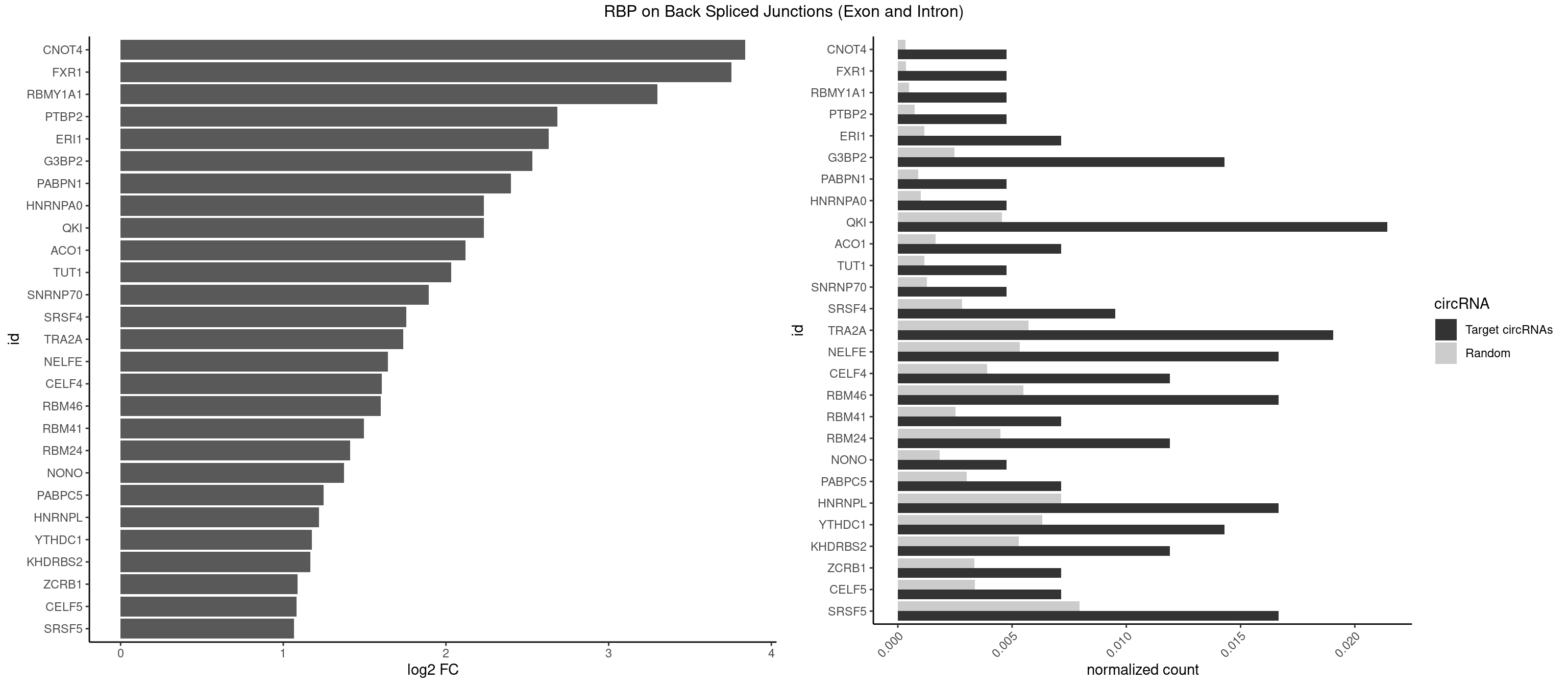
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| G3BP2 | 5 | 490 | 0.014285714 | 0.0024738009 | 2.529772 | AGGAUA,AGGAUU,GGAUAG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| QKI | 8 | 904 | 0.021428571 | 0.0045596534 | 2.232540 | AAUCAU,ACACAC,AUCAUA,CUAAUC,UAAUCA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SRSF4 | 3 | 558 | 0.009523810 | 0.0028164047 | 1.757684 | AGAAGA,AGGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 7 | 1133 | 0.019047619 | 0.0057134220 | 1.737184 | AAGAAA,AGAAAG,AGAAGA,AGGAAG,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUCU,CUCUGG,CUGGCU,GUCUCU,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GGUGUG,GUGUGG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUCAA,AAUCAU,AAUGAU,AUCAAA,AUCAUA,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | GAGUGA,GUGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAAUAA,AAAUAC,AAUAAA,ACACAC,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAGUAC,GGCUGC,UCAUAC,UGAUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AAUAAA,AUAAAA,AUAAAC | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,ACUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AGAAGA,AGGAAG,CACGUC,GAGGAA,UGCGUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.