circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000186260:+:16:14140543:14140760
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000186260:+:16:14140543:14140760 | ENSG00000186260 | ENST00000571589 | + | 16 | 14140544 | 14140760 | 217 | UGUCUUCAAUAGGCCGUGUUUAAGAGGCGUCUUACACUCCCUGUUGCCAGUGGCUGGAACACAAUGGAUCACACAGGGGCGAUAGACACCGAGGAUGAAGUGGGACCUUUAGCCCAUCUUGCUCCAAGUCCUCAGAGUGAAGCUGUGGCUCAUGAAUUCCAGGAACUCUCCUUGCAGUCCAGUCAAAACUUACCCCCUCUGAACGAAAGGAAAAAUGUGUCUUCAAUAGGCCGUGUUUAAGAGGCGUCUUACACUCCCUGUUGCCAG | circ |
| ENSG00000186260:+:16:14140543:14140760 | ENSG00000186260 | ENST00000571589 | + | 16 | 14140544 | 14140760 | 22 | AAGGAAAAAUGUGUCUUCAAUA | bsj |
| ENSG00000186260:+:16:14140543:14140760 | ENSG00000186260 | ENST00000571589 | + | 16 | 14140344 | 14140553 | 210 | CCUUAUAAUUGCUUGUUUUUAGGUGGGAAGUAUGUUAUUCCUGUUAGAUAAAGAACCUGAGGUCCAGAGAGGGUAAAUGGAUUAGUAUGACCAUACAGCAGUAAAACUGGACUAGAACCCAGGAUUCCCGUUUUAAUACUUUUGUGCAAAUUGGAGAAACAUAACUGCACCAUCUCUUUACACAUUCUUUAUUUUGGCAGUGUCUUCAAU | ie_up |
| ENSG00000186260:+:16:14140543:14140760 | ENSG00000186260 | ENST00000571589 | + | 16 | 14140751 | 14140960 | 210 | AGGAAAAAUGGUGAGUUCAGCAUGUGCAAUGGGAUCUUUGAUUUAAAGAUUUCCCCUCUUUGGGAGCCCAGUUUAUUCCUGUUUGGUAGUAAUAUUUUGGUCAGUUCAGCAGUCUGUCAUGGAGGAGAACCCUGUAGAGGUACUGGAUGGUUUUUAAUAACAGGCAGAAUUUCACUUUCCAUUCAGUUCUUUCAUUAUUUUCCCAGUAAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
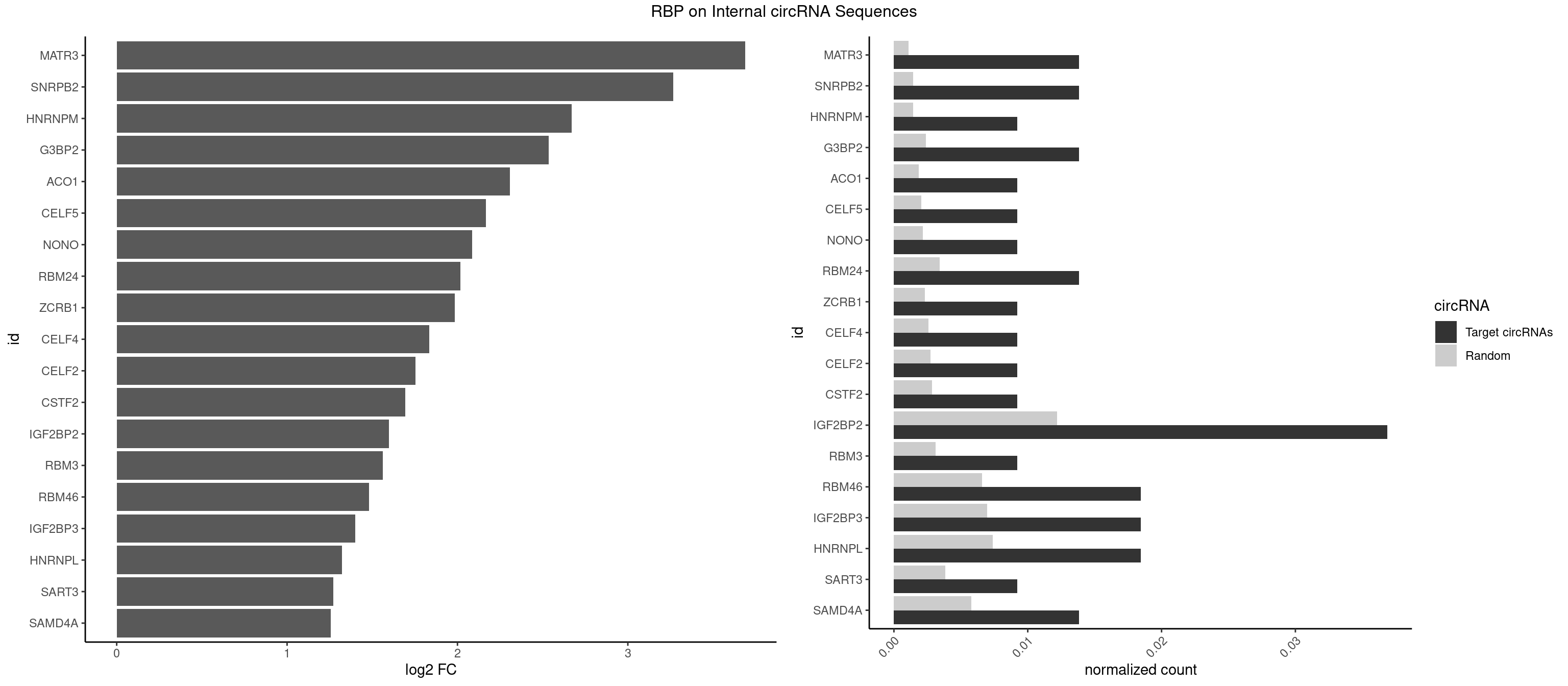
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 2 | 739 | 0.01382488 | 0.001072411 | 3.688338 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SNRPB2 | 2 | 991 | 0.01382488 | 0.001437610 | 3.265523 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPM | 1 | 999 | 0.00921659 | 0.001449204 | 2.668972 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 2 | 1644 | 0.01382488 | 0.002383941 | 2.535847 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ACO1 | 1 | 1283 | 0.00921659 | 0.001860778 | 2.308327 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF5 | 1 | 1415 | 0.00921659 | 0.002052073 | 2.167151 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| NONO | 1 | 1498 | 0.00921659 | 0.002172357 | 2.084972 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM24 | 2 | 2357 | 0.01382488 | 0.003417223 | 2.016371 | AGAGUG,GAGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZCRB1 | 1 | 1605 | 0.00921659 | 0.002327422 | 1.985501 | GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF4 | 1 | 1782 | 0.00921659 | 0.002583931 | 1.834666 | GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF2 | 1 | 1886 | 0.00921659 | 0.002734648 | 1.752878 | AUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CSTF2 | 1 | 1967 | 0.00921659 | 0.002852033 | 1.692242 | GUGUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| IGF2BP2 | 7 | 8408 | 0.03686636 | 0.012186356 | 1.597038 | AACACA,ACACUC,CAAAAC,CACACA,GAACAC,GAACUC,GAAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM3 | 1 | 2152 | 0.00921659 | 0.003120136 | 1.562624 | AAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM46 | 3 | 4554 | 0.01843318 | 0.006601124 | 1.481521 | AUGAAG,AUGAAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| IGF2BP3 | 3 | 4815 | 0.01843318 | 0.006979366 | 1.401137 | AACACA,ACACUC,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPL | 3 | 5085 | 0.01843318 | 0.007370651 | 1.322441 | AACACA,ACACAA,CACACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| SART3 | 1 | 2634 | 0.00921659 | 0.003818652 | 1.271169 | GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SAMD4A | 2 | 3992 | 0.01382488 | 0.005786671 | 1.256462 | CUGGAA,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
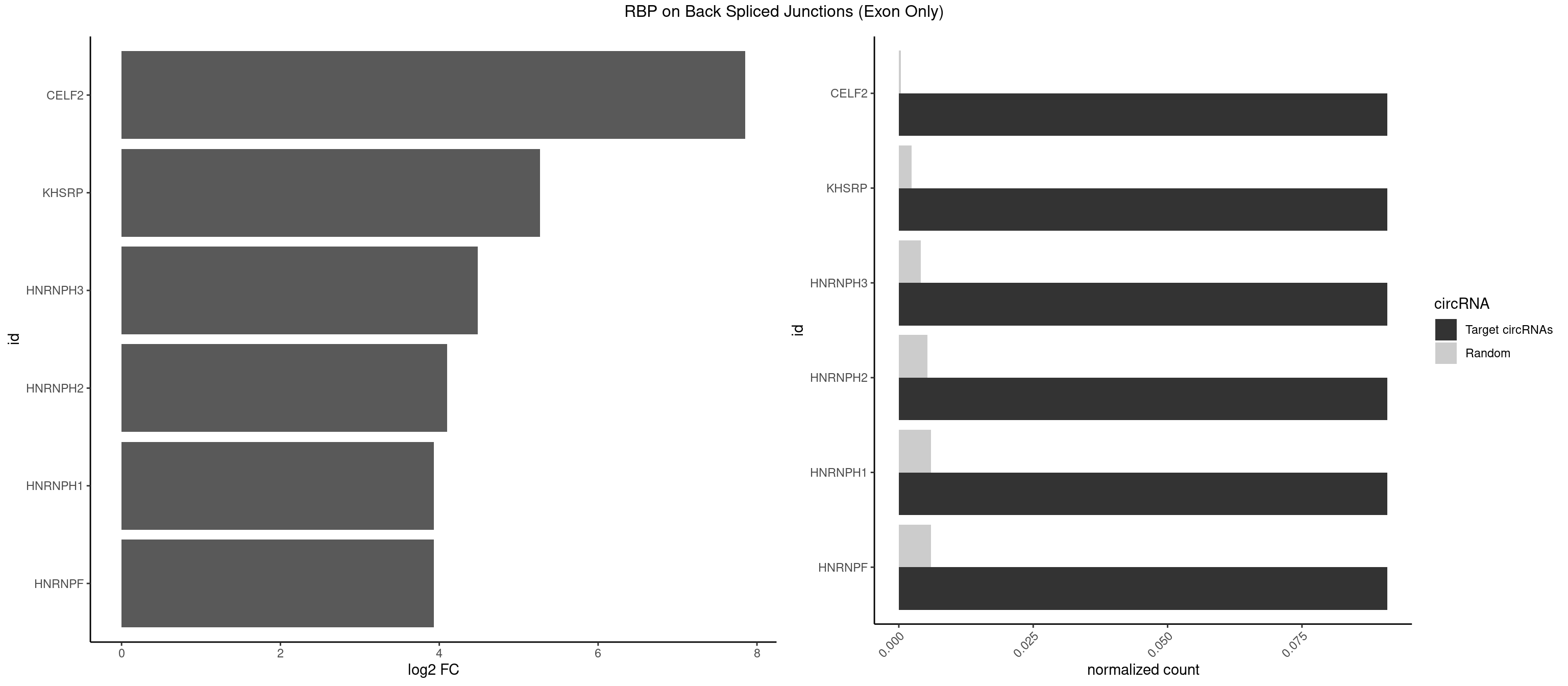
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF2 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | AUGUGU | AUGUGU,GUCUGU,UAUGUU,UGUGUG,UGUUGU |
| KHSRP | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | AUGUGU | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | AAUGUG | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.0053052135 | 4.098942 | AAUGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | AAUGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | AAUGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
RBP on BSJ (Exon and Intron)
Plot
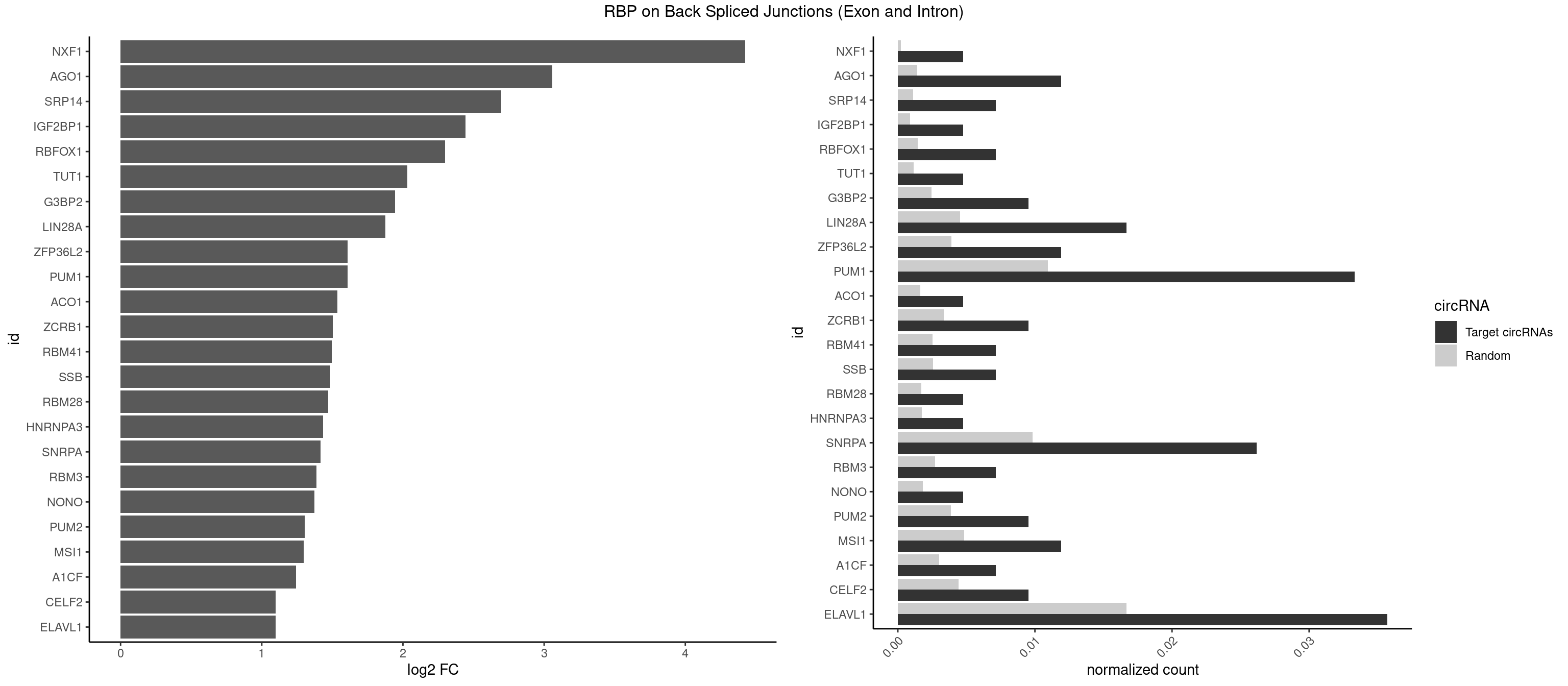
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| AGO1 | 4 | 283 | 0.011904762 | 0.0014308746 | 3.056570 | GAGGUA,GGUAGU,GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUGG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | AGGAGA,GGAGAA,GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PUM1 | 13 | 2172 | 0.033333333 | 0.0109482064 | 1.606271 | AAUAUU,ACAUAA,AGAAUU,AGAUAA,CAGAAU,GUAAAU,GUAAUA,GUCCAG,UAAUAU,UAGAUA,UGUAGA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AUUUAA,GAUUUA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SNRPA | 10 | 1947 | 0.026190476 | 0.0098145909 | 1.416042 | AGCAGU,AGGAGA,AUUCCU,GAUUCC,GCAGUA,UGCACC,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAAAU,UAGAUA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUGG,AGUAAG,UAGGUG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,GUCUGU,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAUUU,UUGAUU,UUGUUU,UUUAUU,UUUGGU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.