circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000115464:-:2:61278164:61288877
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000115464:-:2:61278164:61288877 | ENSG00000115464 | ENST00000398571 | - | 2 | 61278165 | 61288877 | 885 | UGGCAGCUAGACUGUCUUGCUUGCUUGCUGAAGUUAAUAUGCCAGUUUGCAGUAGAUCCAUCCGAUUUGGAUUUAGCUUAUCAUGAUGUCUUUGCCUGGUCUGGUAUAGCGGAAAGCCAUAGGAAAAGAACCUGGCCUGGCAAAUCAAGGAAGGCUGCUGGUGAUCAUGCUAAGGGUCUUCAUAUACCACGAUUAACAGAGGUAUUUCUUGUUCUUGUCCAAGGAACCAGUUUGAUUCAGCGACUUAUGUCUGUUGCUUAUACGUAUGAUAAUCUGGCUCCUAGAGUUUUAAAAGCUCAGUCUGAUCACAGGUCUAGACAUGAAGUUUCACAUUAUUCAAUGUGGCUCUUGGUGAGUUGGGCUCAUUGCUGUUCUUUAGUGAAAUCUAGCCUUGCUGAUAGCGAUCAUUUACAAGAUUGGCUAAAGAAAUUGACUCUCCUUAUUCCUGAGACUGCAGUUCGUCAUGAAUCAUGCAGUGGUCUCUAUAAGUUAUCCCUGUCAGGGCUGGAUGGAGGAGACUCAAUCAAUCGUUCUUUUCUGCUAUUGGCUGCCUCAACAUUAUUGAAAUUUCUUCCUGAUGCUCAAGCACUCAAACCUAUUAGGAUAGAUGAUUAUGAGGAAGAACCAAUAUUAAAACCAGGAUGUAAAGAGUAUUUUUGGUUGUUAUGCAAAUUAGUUGACAACAUACAUAUAAAGGACGCUAGUCAGACAACGCUCCUCGACUUAGAUGCCUUGGCAAGACAUUUGGCUGACUGUAUUCGAAGUAGGGAGAUCCUUGAUCAUCAGGAUGGUAAUGUAGAAGAUGAUGGGCUUACAGGACUCCUAAGGCUUGCAACAAGUGUUGUUAAACACAAACCACCCUUUAAAUUUUCAAGGGAAGGACAGUGGCAGCUAGACUGUCUUGCUUGCUUGCUGAAGUUAAUAUGCCAGUUUGC | circ |
| ENSG00000115464:-:2:61278164:61288877 | ENSG00000115464 | ENST00000398571 | - | 2 | 61278165 | 61288877 | 22 | GGGAAGGACAGUGGCAGCUAGA | bsj |
| ENSG00000115464:-:2:61278164:61288877 | ENSG00000115464 | ENST00000398571 | - | 2 | 61288868 | 61289077 | 210 | UUAUUUCUAGAUACUUUUUCUUCAUUUUUAUAAGAUUGAAUUGGUACUGACCAUAAUAUUUGAGCAGCAGAAUUUUUAUAGUACAUGAUUAAGUACUAGCUAUAAUUAUUUUUAUUUUCUGGUCUUUUAAUUUCAAAAUUGUAUUGUGGCCAUUCUAAAUUAAUGAAAAUAGGGAAUUGACUAUUUCUUCUCAUUUACAGUGGCAGCUAG | ie_up |
| ENSG00000115464:-:2:61278164:61288877 | ENSG00000115464 | ENST00000398571 | - | 2 | 61277965 | 61278174 | 210 | GGAAGGACAGGUAAGUGUUAAGAUAAUCAAAUGUUUCUAUGAAAUGUGAUUGAAAUUUUUUGUUGUUACAUUGUGUAUUAGUCCGUUUUUACACUGCUCAUAAAGGCAUACCCGAGACUGGGCAGUUUACAAAAGAAAAGGGUUUAAUUCGACUCAGAGUUCCAUGUGGCUGGGGAGGCCUCACAAUCGUGCCGGAAGGCAAGUCCGGGC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
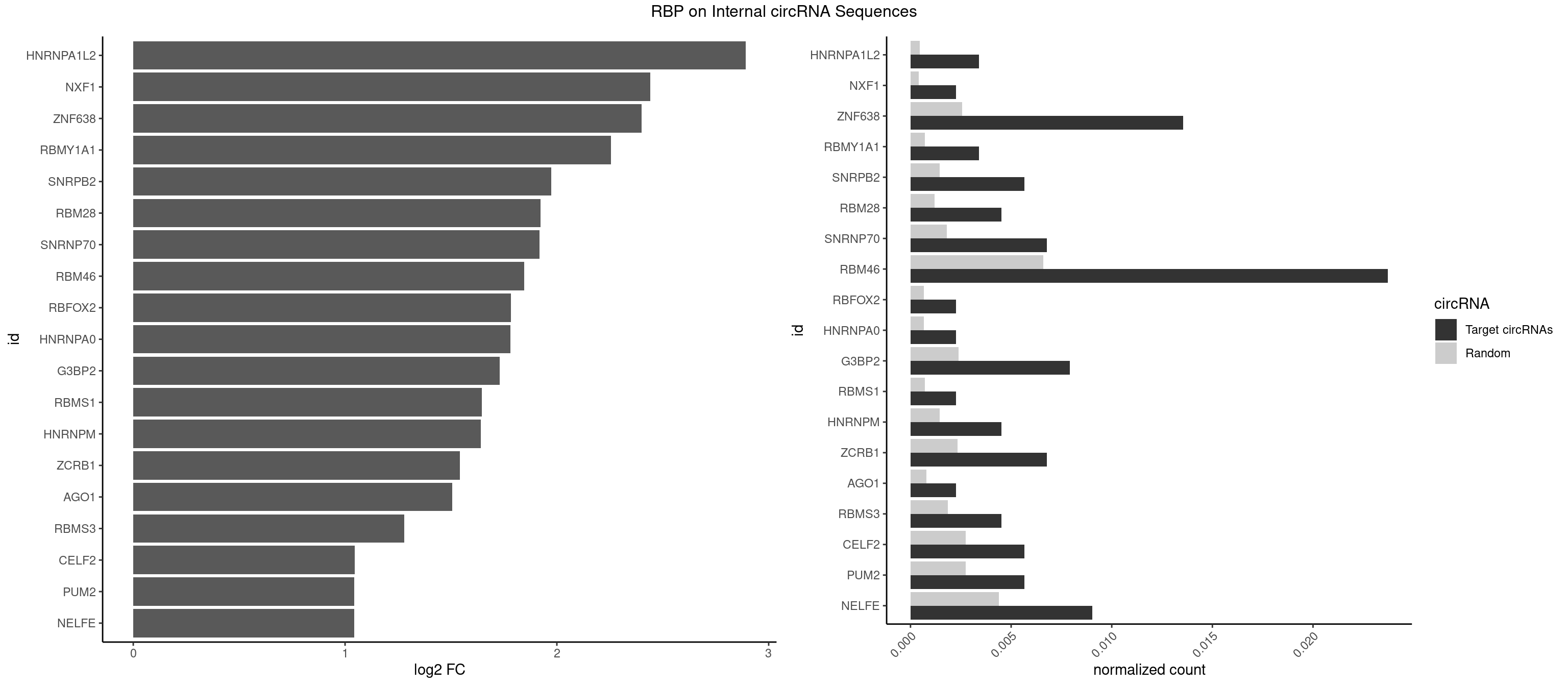
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 314 | 0.003389831 | 0.0004564992 | 2.892529 | GUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| NXF1 | 1 | 286 | 0.002259887 | 0.0004159215 | 2.441867 | AACCUG | AACCUG |
| ZNF638 | 11 | 1773 | 0.013559322 | 0.0025708878 | 2.398946 | CGUUCU,GGUUGU,GUUCGU,GUUCUU,GUUGUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMY1A1 | 2 | 489 | 0.003389831 | 0.0007101099 | 2.255099 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| SNRPB2 | 4 | 991 | 0.005649718 | 0.0014376103 | 1.974506 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM28 | 3 | 822 | 0.004519774 | 0.0011926949 | 1.922026 | AGUAGA,AGUAGG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SNRNP70 | 5 | 1237 | 0.006779661 | 0.0017941145 | 1.917941 | AAUCAA,AUCAAG,AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM46 | 20 | 4554 | 0.023728814 | 0.0066011240 | 1.845856 | AAUCAA,AAUCAU,AUCAAG,AUCAAU,AUCAUG,AUCAUU,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAU,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBFOX2 | 1 | 452 | 0.002259887 | 0.0006564894 | 1.783407 | UGACUG | UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.002259887 | 0.0006579386 | 1.780226 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| G3BP2 | 6 | 1644 | 0.007909605 | 0.0023839405 | 1.730257 | AGGAUA,AGGAUG,GGAUAG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBMS1 | 1 | 497 | 0.002259887 | 0.0007217036 | 1.646772 | AUAUAC | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPM | 3 | 999 | 0.004519774 | 0.0014492040 | 1.640990 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ZCRB1 | 5 | 1605 | 0.006779661 | 0.0023274215 | 1.542481 | GACUUA,GAUUAA,GAUUUA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO1 | 1 | 548 | 0.002259887 | 0.0007956130 | 1.506112 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBMS3 | 3 | 1283 | 0.004519774 | 0.0018607779 | 1.280345 | CAUAUA,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CELF2 | 4 | 1886 | 0.005649718 | 0.0027346479 | 1.046824 | GUCUGU,GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM2 | 4 | 1890 | 0.005649718 | 0.0027404447 | 1.043769 | GUAGAU,UACAUA,UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| NELFE | 7 | 3028 | 0.009039548 | 0.0043896388 | 1.042148 | CUGGCU,GGCUAA,GGUCUC,GUCUCU,UCUGGC,UCUGGU,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
RBP on BSJ (Exon Only)
Plot
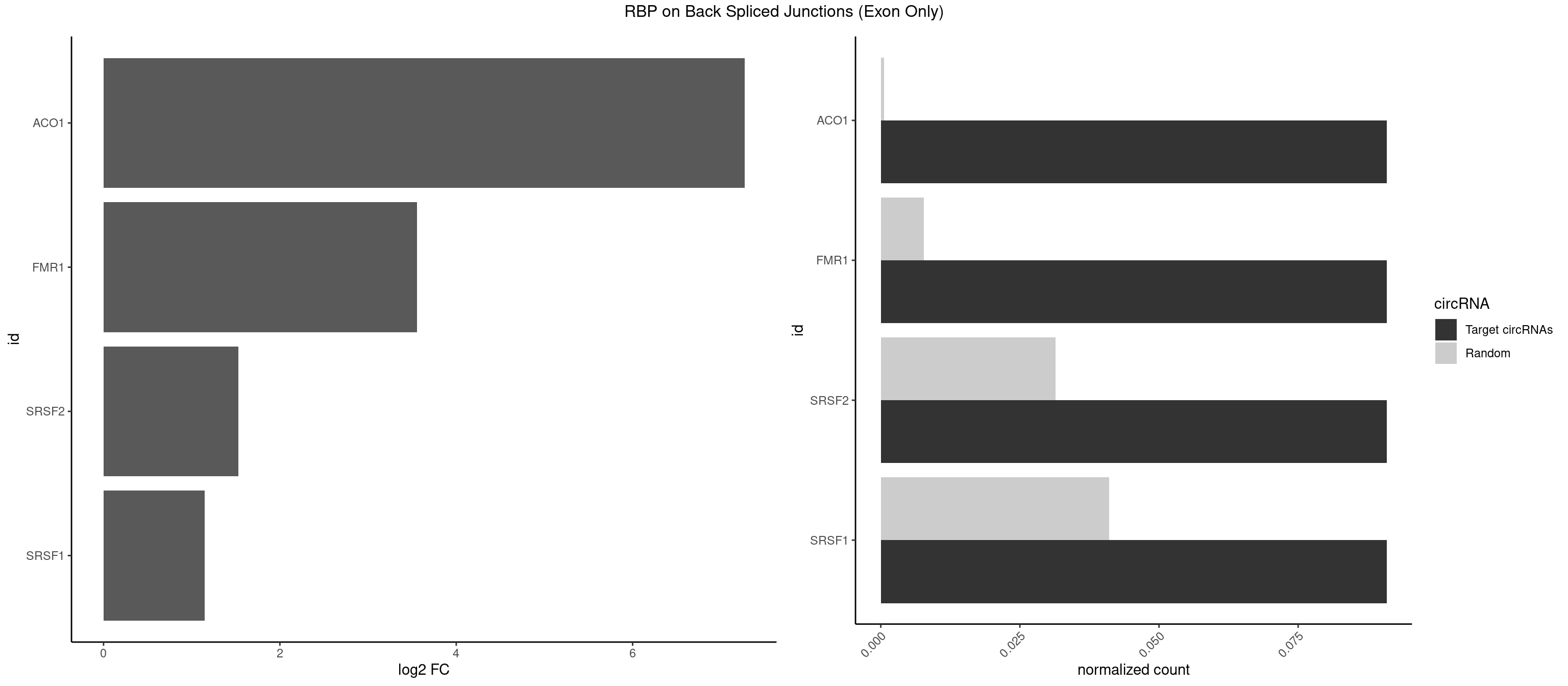
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ACO1 | 1 | 8 | 0.09090909 | 0.0005894682 | 7.268867 | CAGUGG | CAGUGA,CAGUGC,CAGUGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | AGUGGC | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | CAGUGG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
| SRSF1 | 1 | 625 | 0.09090909 | 0.0410007860 | 1.148773 | ACAGUG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
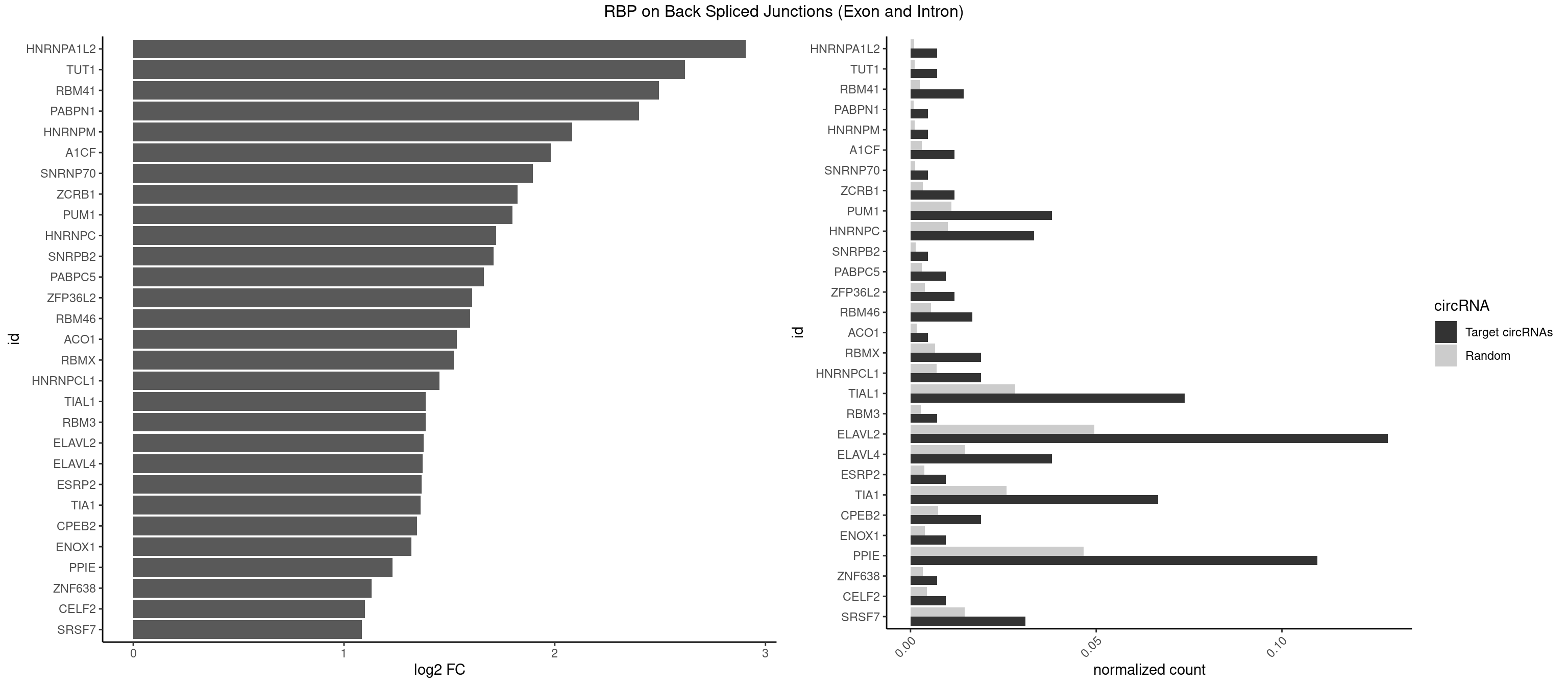
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | AUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | AUACUU,UACAUG,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | AAUUAA,GAUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PUM1 | 15 | 2172 | 0.038095238 | 0.0109482064 | 1.798916 | AAUAUU,AAUGUU,AAUUGU,AGAAUU,AGAUAA,AUUGUA,CAGAAU,GAAUUG,GUACAU,UAAUAU,UAGAUA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPC | 13 | 2006 | 0.033333333 | 0.0101118501 | 1.720919 | AUUUUU,CUUUUU,GCAUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUCAA,AAUGAA,AUCAAA,AUGAAA,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMX | 7 | 1316 | 0.019047619 | 0.0066354293 | 1.521349 | AAGUGU,AGUGUU,AUCAAA,GAAGGA,GGAAGG,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| TIAL1 | 30 | 5597 | 0.073809524 | 0.0282043531 | 1.387889 | AAAUUU,AAUUUU,AUUUUC,AUUUUU,CAGUUU,CUUUUA,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UUAUUU,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | GAGACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ELAVL2 | 53 | 9826 | 0.128571429 | 0.0495112858 | 1.376741 | AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAC,AUUUUC,AUUUUU,CAUUUU,CUUUUA,CUUUUU,GUAUUG,GUUUUU,UAAUUU,UACUUU,UAUUGU,UAUUUG,UAUUUU,UCAUUU,UCUUUU,UGUAUU,UUAAGU,UUAAUU,UUAUUU,UUCAUU,UUGUAU,UUUAAU,UUUAUA,UUUAUU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 15 | 2916 | 0.038095238 | 0.0146966949 | 1.374119 | UAUUUU,UUAUUU,UUGUAU,UUUAUU,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAU,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| TIA1 | 27 | 5140 | 0.066666667 | 0.0259018541 | 1.363910 | AUUUUC,AUUUUU,CUUUUA,CUUUUU,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGGACA,AGUACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PPIE | 45 | 9262 | 0.109523810 | 0.0466696896 | 1.230687 | AAAAUA,AAAAUU,AAAUUA,AAAUUU,AAUAUU,AAUUAA,AAUUAU,AAUUUU,AUAAUA,AUAAUU,AUAUUU,AUUAAU,AUUAUU,AUUUUU,UAAAUU,UAAUAU,UAAUUA,UAAUUU,UAUAAU,UAUUUU,UUAAUU,UUAUAA,UUAUUU,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUUGUU,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAGGAC,AGGACA,AUUGAC,CCGAGA,CGAGAC,GAAGGC,GAGACU,GAUUGA,GGAAGG,UCUUCA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.