circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000031823:-:19:5941620:5957973
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000031823:-:19:5941620:5957973 | ENSG00000031823 | MSTRG.16270.6 | - | 19 | 5941621 | 5957973 | 180 | AAAAGCCUGCCAUUGCUCCGCCCGUCUUUGUGUUUCAGAAGGAUAAAGGACAAAAGAGGUCAGCUGGCGGCUCCAGUCCUGAAGGCGGAGAAGAUUCUGACAGAGAAGAUGGAAAUUACUGCCCUCCUGUCAAGCGAGAAAGAACAUCCUCUUUAACCCAGUUCCCACCCUCACAGUCAGAAAAGCCUGCCAUUGCUCCGCCCGUCUUUGUGUUUCAGAAGGAUAAAGGA | circ |
| ENSG00000031823:-:19:5941620:5957973 | ENSG00000031823 | MSTRG.16270.6 | - | 19 | 5941621 | 5957973 | 22 | CUCACAGUCAGAAAAGCCUGCC | bsj |
| ENSG00000031823:-:19:5941620:5957973 | ENSG00000031823 | MSTRG.16270.6 | - | 19 | 5957964 | 5958173 | 210 | UCAUUCUGGGUGCAUUUCACAAACCCAGCACACCUGUGCCUCCAGGCAUCCGAGUCAAGACGCUGUUCUAAGUGAUUUGAUGUUCUCUGGAGUUACUAGUUUCAUUUAUAUAUAUGUUUAACAUAUUACAAACAAGUGUAGCUUUGUUUACUGUAUGCAUAUAGUGUUGGGGGGAAAAAAACUAAUUUUUCUUUUUGCAGAAAAGCCUGC | ie_up |
| ENSG00000031823:-:19:5941620:5957973 | ENSG00000031823 | MSTRG.16270.6 | - | 19 | 5941421 | 5941630 | 210 | UCACAGUCAGGUAAAAUAUGCAAACUUACGGAAUGGUGAAAGCGUUUAUGCUUACAAACCCGCCAUUCCCCUCAGUGCCGCGCAUCCGUCUGCUGUGUCCUUCCUGCCCAGCUCCGGUUAGAGUCGAUCUGAGGCUCGGGCUCUGUCAGGAGUGAGGUGAGAGCUUGUCCAGAGUGCAGCAUCAGGGGUCCUCGUCUCGAAUCAGUGACA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
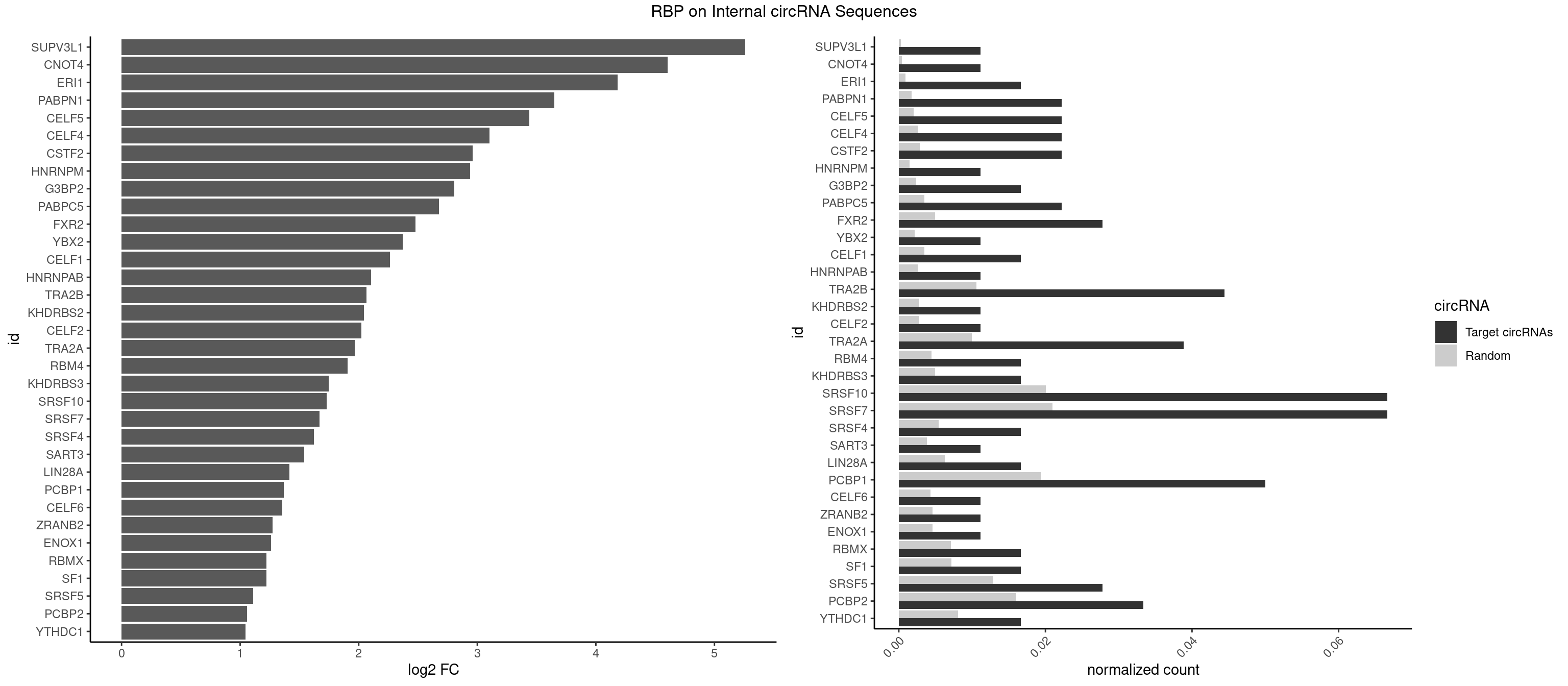
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 199 | 0.01111111 | 0.0002898408 | 5.260599 | CCGCCC | CCGCCC |
| CNOT4 | 1 | 314 | 0.01111111 | 0.0004564992 | 4.605247 | GACAGA | GACAGA |
| ERI1 | 2 | 632 | 0.01666667 | 0.0009173461 | 4.183356 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| PABPN1 | 3 | 1222 | 0.02222222 | 0.0017723764 | 3.648246 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| CELF5 | 3 | 1415 | 0.02222222 | 0.0020520728 | 3.436849 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 3 | 1782 | 0.02222222 | 0.0025839306 | 3.104364 | GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 3 | 1967 | 0.02222222 | 0.0028520334 | 2.961940 | GUGUUU,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPM | 1 | 999 | 0.01111111 | 0.0014492040 | 2.938671 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 2 | 1644 | 0.01666667 | 0.0023839405 | 2.805545 | AGGAUA,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PABPC5 | 3 | 2400 | 0.02222222 | 0.0034795387 | 2.675035 | AGAAAA,AGAAAG,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| FXR2 | 4 | 3434 | 0.02777778 | 0.0049780156 | 2.480289 | GACAAA,GACAGA,GGACAA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| YBX2 | 1 | 1480 | 0.01111111 | 0.0021462711 | 2.372099 | AACAUC | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF1 | 2 | 2391 | 0.01666667 | 0.0034664959 | 2.265416 | UGUGUU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPAB | 1 | 1782 | 0.01111111 | 0.0025839306 | 2.104364 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| TRA2B | 7 | 7329 | 0.04444444 | 0.0106226650 | 2.064857 | AAAGAA,AAGAAC,AGAAGA,AGAAGG,GAAAGA,GAAGGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| KHDRBS2 | 1 | 1858 | 0.01111111 | 0.0026940701 | 2.044144 | GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CELF2 | 1 | 1886 | 0.01111111 | 0.0027346479 | 2.022576 | UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TRA2A | 6 | 6871 | 0.03888889 | 0.0099589296 | 1.965295 | AAAGAA,AAGAGG,AGAAAG,AGAAGA,GAAAGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM4 | 2 | 3065 | 0.01666667 | 0.0044432593 | 1.907275 | CCUCUU,CUCUUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| KHDRBS3 | 2 | 3429 | 0.01666667 | 0.0049707696 | 1.745424 | AUAAAG,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SRSF10 | 11 | 13860 | 0.06666667 | 0.0200874160 | 1.730674 | AAAAGA,AAAGAA,AAAGAG,AAAGGA,AAGAGG,AGAGAA,GAAAGA,GACAAA,GAGAAA,GAGAAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF7 | 11 | 14481 | 0.06666667 | 0.0209873716 | 1.667444 | AAAGGA,AAGGAC,AAGGCG,AGAAGA,AGAGAA,AGGACA,CAGAGA,GAAGGC,GACAAA,GGACAA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SRSF4 | 2 | 3740 | 0.01666667 | 0.0054214720 | 1.620209 | AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SART3 | 1 | 2634 | 0.01111111 | 0.0038186524 | 1.540868 | AGAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| LIN28A | 2 | 4315 | 0.01666667 | 0.0062547643 | 1.413938 | CGGAGA,GGAGAA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PCBP1 | 8 | 13384 | 0.05000000 | 0.0193975949 | 1.366050 | AAAUUA,AAUUAC,ACCCUC,CACCCU,CCACCC,CCCACC,CCCUCC,CCUCUU | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| CELF6 | 1 | 2997 | 0.01111111 | 0.0043447134 | 1.354670 | UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| ZRANB2 | 1 | 3173 | 0.01111111 | 0.0045997733 | 1.272368 | UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ENOX1 | 1 | 3195 | 0.01111111 | 0.0046316558 | 1.262403 | AGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBMX | 2 | 4925 | 0.01666667 | 0.0071387787 | 1.223216 | AGAAGG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SF1 | 2 | 4931 | 0.01666667 | 0.0071474739 | 1.221460 | ACAGUC,CAGUCA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF5 | 4 | 8869 | 0.02777778 | 0.0128544391 | 1.111665 | AGAAGA,AUAAAG,UAAAGG | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| PCBP2 | 5 | 11045 | 0.03333333 | 0.0160079069 | 1.058181 | AAAUUA,AAUUAC,CCCUCC,CCUCCU,UAACCC | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| YTHDC1 | 2 | 5576 | 0.01666667 | 0.0080822104 | 1.044144 | GCCUGC,UACUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
RBP on BSJ (Exon Only)
Plot
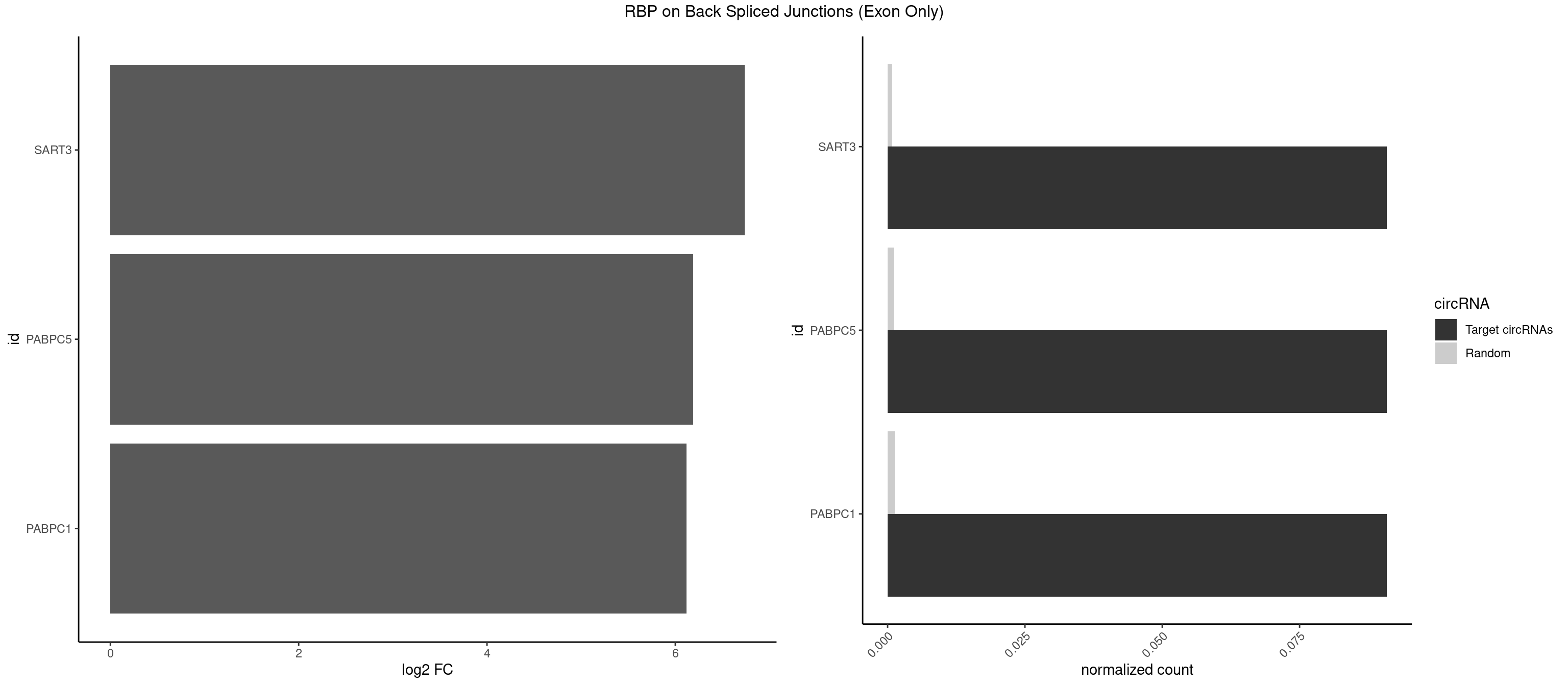
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SART3 | 1 | 12 | 0.09090909 | 0.000851454 | 6.738352 | AGAAAA | AAAAAA,AGAAAA,GAAAAA,GAAAAC |
| PABPC5 | 1 | 18 | 0.09090909 | 0.001244433 | 6.190864 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPC1 | 1 | 19 | 0.09090909 | 0.001309929 | 6.116864 | AGAAAA | AAAAAA,ACAAAC,ACGAAU,AGAAAA,CAAACC,CAAAUA,CGAACA,CUAAUA,GAAAAA,GAAAAC |
RBP on BSJ (Exon and Intron)
Plot
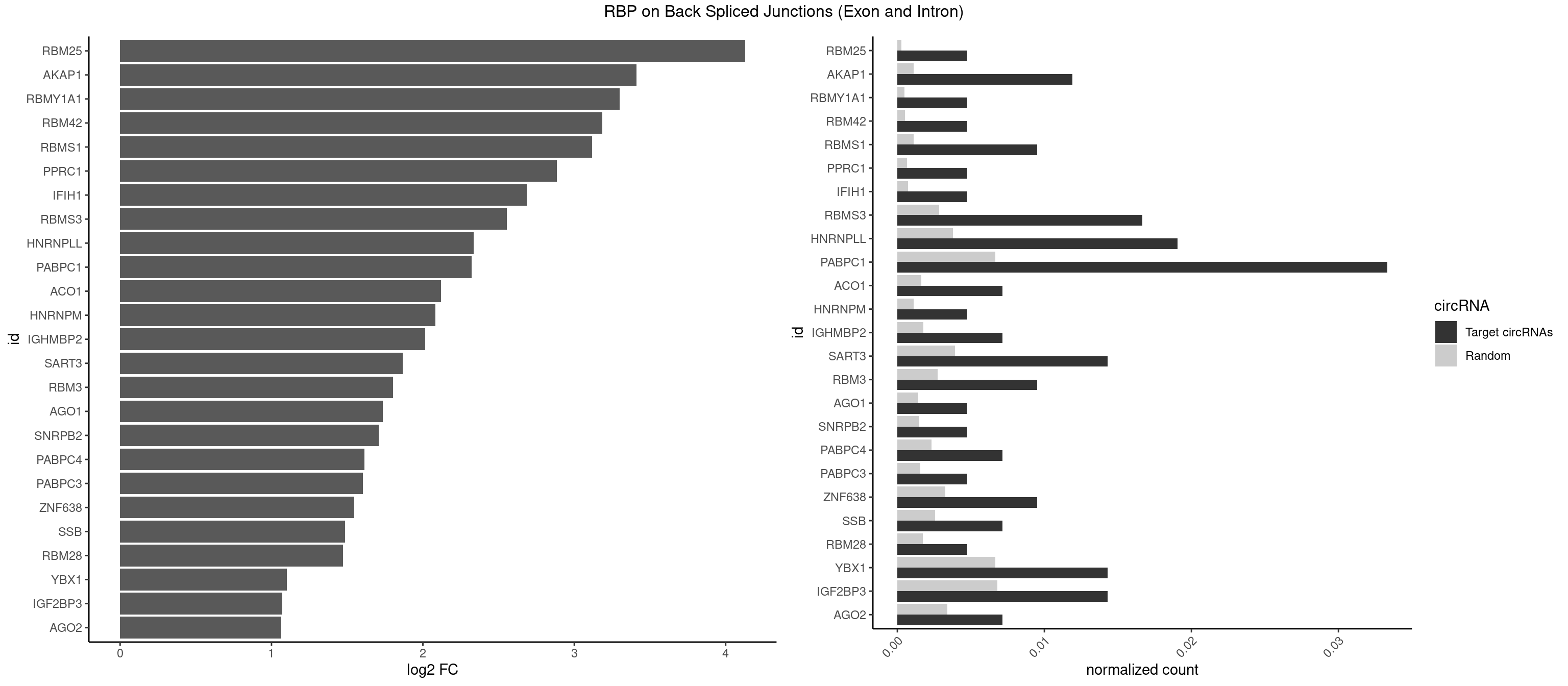
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| AKAP1 | 4 | 221 | 0.011904762 | 0.0011185006 | 3.411901 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PPRC1 | 1 | 127 | 0.004761905 | 0.0006449012 | 2.884389 | CCGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GCCGCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBMS3 | 6 | 562 | 0.016666667 | 0.0028365578 | 2.554752 | AUAUAG,AUAUAU,CAUAUA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPLL | 7 | 748 | 0.019047619 | 0.0037736800 | 2.335567 | ACAAAC,CAAACA,CACACC,GCAAAC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC1 | 13 | 1321 | 0.033333333 | 0.0066606207 | 2.323237 | AAAAAA,AAAAAC,ACAAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CGAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| IGHMBP2 | 2 | 350 | 0.007142857 | 0.0017684401 | 2.014024 | AAAAAA | AAAAAA |
| SART3 | 5 | 777 | 0.014285714 | 0.0039197904 | 1.865725 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA,AAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | GUGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | CACACC,CCAGCA,GAUCUG,GCCUGC,GUCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,ACAAAC,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.