circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000137145:+:9:19352072:19361963
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000137145:+:9:19352072:19361963 | ENSG00000137145 | ENST00000602925 | + | 9 | 19352073 | 19361963 | 1029 | GUGAAGUUCCAUUUCCAAGAGGCAUGAAAGGGCAAGACUUUGAAAAAUCAGAUCAUGGUUCUUCUCAAAAUACCAGCAUGUCUAGCAUCUAUCAGAAUUGUGCAAUGGAGGUUUUGAUGUCCAGUUGUUCACAGUGUAGAGCUUGUGGAGCUUUAGUUUAUGAUGAAGAAAUUAUGGCUGGAUGGACAGCAGAUGACUCAAAUUUGAAUACAGCUUGUCCAUUCUGUAAAAGCAACUUCUUGCCUCUUCUCAAUAUAGAAUUCAAAGAUUUGAGAGGUUCUGCAAGCUUUUUCCUGAAACCAAGUACCUCUGGUGACAGUUUACAAAGUGGAAGCAUUCCAUUGGCAAAUGAAUCCUUGGAGCACAAACCUGUAUCCAGUUUAGCAGAACCUGACUUGAUCAACUUUAUGGACUUCCCAAAACAUAACCAGAUCAUAACUGAAGAAACAGGCUCUGCAGUUGAACCAAGUGAUGAAAUAAAGAGAGCCAGUGGAGAUGUCCAAACUAUGAAAAUUUCAUCUGUGCCUAAUAGUUUAUCAAAGCGAAAUGUGUCUUUGACUCGAAGUCACAGUGUUGGAGGCCCAUUGCAGAAUAUUGACUUUACCCAGCGACCGUUUCAUGGCAUCUCAACAGUUAGUCUUCCAAAUAGUCUGCAGGAAGUUGUGGAUCCUUUAGGAAAAAGACCCAAUCCUCCCCCUGUUUCUGUGCCCUACUUGAGUCCUCUAGUACUCCGUAAAGAACUUGAAUCUUUGCUAGAAAAUGAAGGUGAUCAGGUGAUUCAUACAUCUUCUUUCAUCAAUCAACAUCCAAUCAUUUUCUGGAACCUCGUUUGGUAUUUCAGACGUUUGGACCUUCCUAGUAACUUGCCAGGACUUAUCCUCACAUCUGAACAUUGUAAUGAAGGUGUACAGCUUCCUCUGUCAUCUCUGUCCCAGGAUAGCAAACUUGUGUAUAUUCAGCUGUUAUGGGAUAAUAUCAACCUUCAUCAGGAACCAAGAGAACCUCUGUAUGUCUCAUGGAGGAAUUUUAGUGAAGUUCCAUUUCCAAGAGGCAUGAAAGGGCAAGACUUUGAAAAAUCA | circ |
| ENSG00000137145:+:9:19352072:19361963 | ENSG00000137145 | ENST00000602925 | + | 9 | 19352073 | 19361963 | 22 | GAGGAAUUUUAGUGAAGUUCCA | bsj |
| ENSG00000137145:+:9:19352072:19361963 | ENSG00000137145 | ENST00000602925 | + | 9 | 19351873 | 19352082 | 210 | ACAAUCAUGUAUUUUCCACCUAGAUUUAAUUAACUAUUGUUAACUUUAUCUCUUUCUAUGUAUGCAUACAUUUUUAUACUGAGCCACUUGAAAAACAGUUGCAGACUUUAUUCUGUGUCCCCCCUAAAUACUUUGGCAUGCAUUUUCAAAAAACAAGGACAUUCCUUACUUAAGGUAUUACGAUGUAUAUUCCUUUGUAGGUGAAGUUCC | ie_up |
| ENSG00000137145:+:9:19352072:19361963 | ENSG00000137145 | ENST00000602925 | + | 9 | 19361954 | 19362163 | 210 | AGGAAUUUUAGUAAGUAAAAUAGAAUUAAAGACAUAACAGCCAGGUGCAGUGGCUCGCACCUGUAAUGCCAGCACUUUGGGAGGCCUAGGCUGGCGGACCACUUGAGGUCAGGAGUUGGAGACCAGGCUGGCCAACAUGGUGAAAUCCUGUCUCUAUUAAAAAUACAAAACUUAGCCAGGUGUGGUAGCAGGUGCCUGUAGUCCCAGCUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
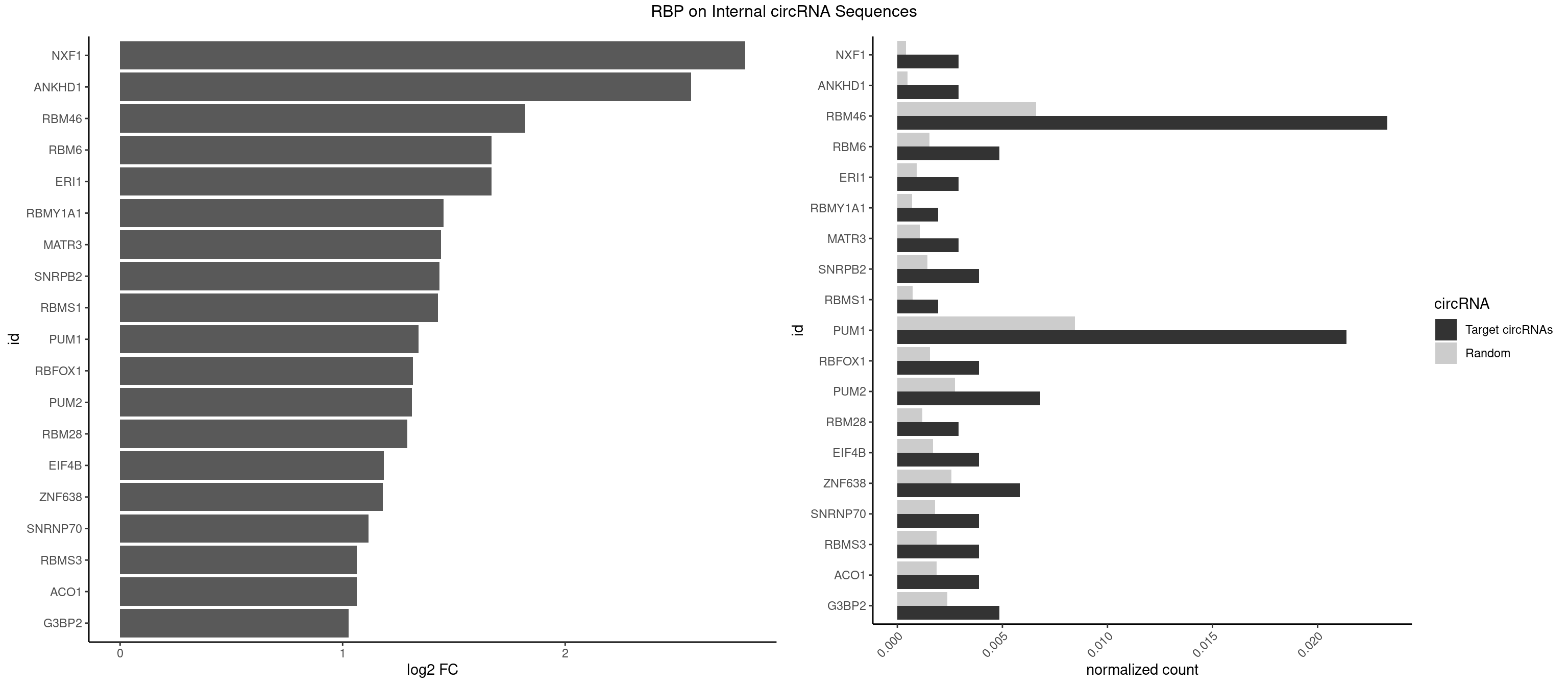
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 2 | 286 | 0.002915452 | 0.0004159215 | 2.809336 | AACCUG | AACCUG |
| ANKHD1 | 2 | 339 | 0.002915452 | 0.0004927293 | 2.564852 | AGACGU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM46 | 23 | 4554 | 0.023323615 | 0.0066011240 | 1.821008 | AAUCAA,AAUCAU,AAUGAA,AUCAAA,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUG,GAUCAA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ERI1 | 2 | 632 | 0.002915452 | 0.0009173461 | 1.668181 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBM6 | 4 | 1054 | 0.004859086 | 0.0015289102 | 1.668181 | AUCCAA,AUCCAG,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBMY1A1 | 1 | 489 | 0.001943635 | 0.0007101099 | 1.452643 | CAAGAC | ACAAGA,CAAGAC |
| MATR3 | 2 | 739 | 0.002915452 | 0.0010724109 | 1.442862 | AAUCUU,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SNRPB2 | 3 | 991 | 0.003887269 | 0.0014376103 | 1.435084 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 1 | 497 | 0.001943635 | 0.0007217036 | 1.429279 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PUM1 | 21 | 5823 | 0.021379981 | 0.0084401638 | 1.340918 | AAUAUU,AAUUGU,ACAUAA,AGAAUU,AUUGUA,CAGAAU,GAAUUG,GUAUAU,GUCCAG,UAAUAU,UACAUC,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBFOX1 | 3 | 1077 | 0.003887269 | 0.0015622419 | 1.315139 | AGCAUG,GCAUGA,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PUM2 | 6 | 1890 | 0.006802721 | 0.0027404447 | 1.311702 | GUAUAU,UACAUC,UGUAAA,UGUACA,UGUAGA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM28 | 2 | 822 | 0.002915452 | 0.0011926949 | 1.289495 | GUGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| EIF4B | 3 | 1179 | 0.003887269 | 0.0017100607 | 1.184710 | CUUGGA,GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZNF638 | 5 | 1773 | 0.005830904 | 0.0025708878 | 1.181453 | GGUUCU,GUUCUU,GUUGUU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SNRNP70 | 3 | 1237 | 0.003887269 | 0.0017941145 | 1.115485 | AAUCAA,AUUCAA,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ACO1 | 3 | 1283 | 0.003887269 | 0.0018607779 | 1.062851 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 3 | 1283 | 0.003887269 | 0.0018607779 | 1.062851 | AAUAUA,AUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| G3BP2 | 4 | 1644 | 0.004859086 | 0.0023839405 | 1.027337 | AGGAUA,GGAUAA,GGAUAG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
RBP on BSJ (Exon Only)
Plot
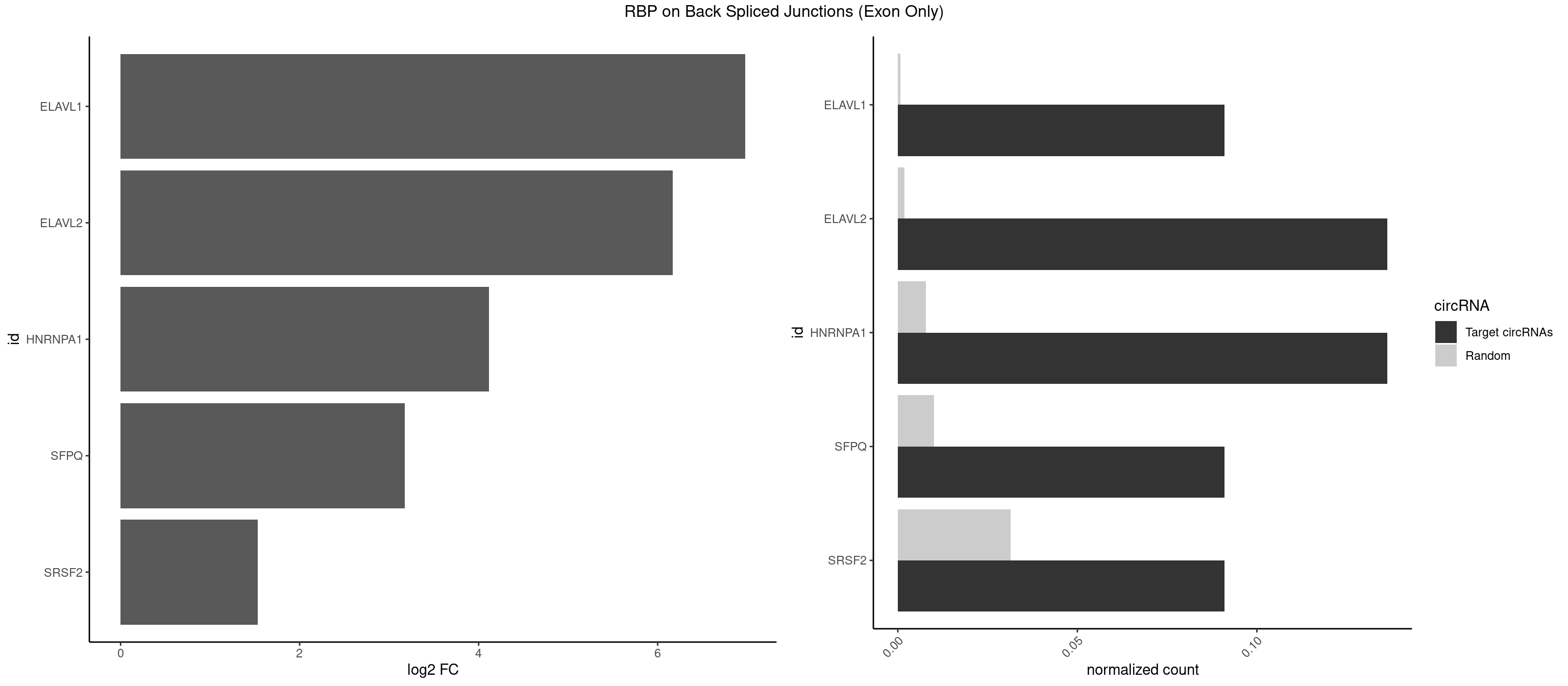
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ELAVL1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UUUAGU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| ELAVL2 | 2 | 28 | 0.13636364 | 0.0018993974 | 6.165773 | UUUAGU,UUUUAG | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
| HNRNPA1 | 2 | 119 | 0.13636364 | 0.0078595756 | 4.116864 | AGUGAA,UAGUGA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | UUAGUG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | UUAGUG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
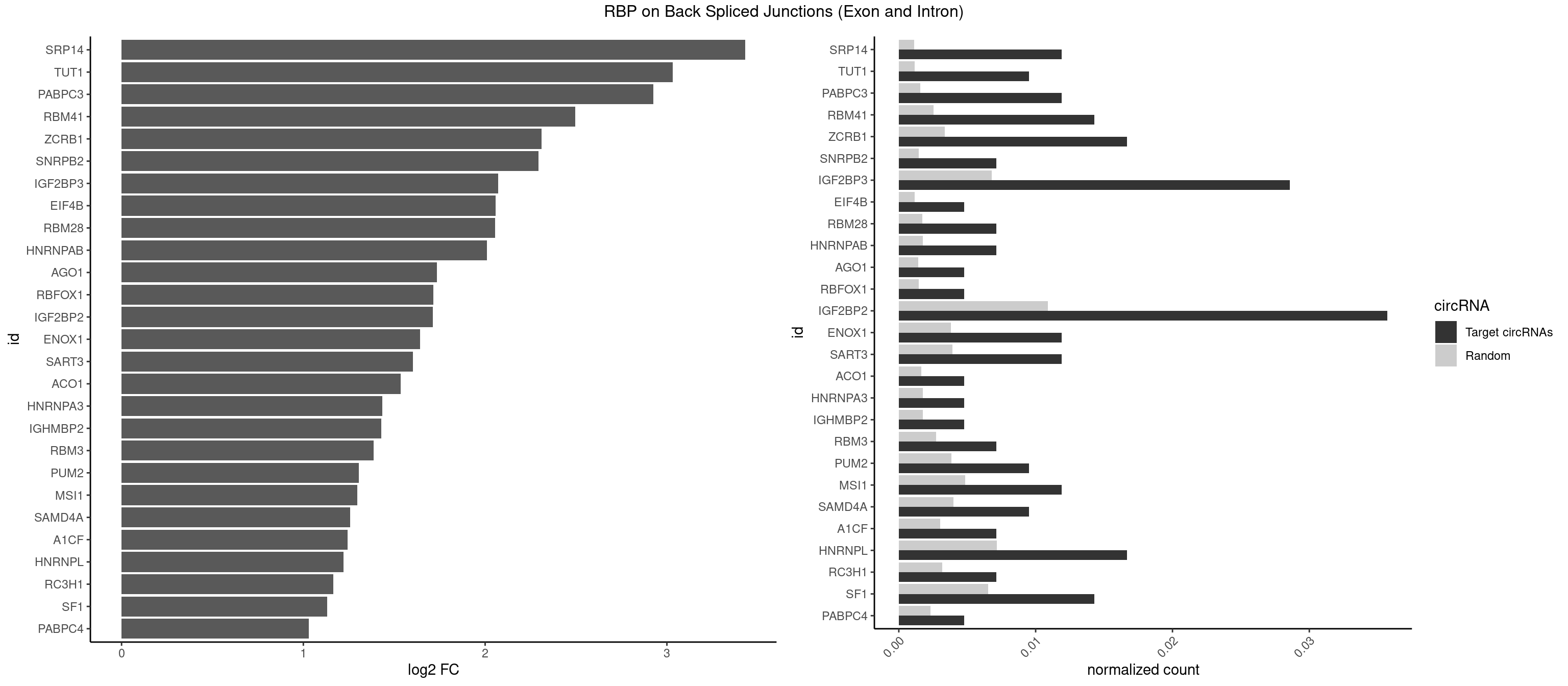
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 4 | 218 | 0.011904762 | 0.001103386 | 3.431530 | CCUGUA,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| TUT1 | 3 | 230 | 0.009523810 | 0.001163845 | 3.032640 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC3 | 4 | 310 | 0.011904762 | 0.001566909 | 2.925546 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBM41 | 5 | 502 | 0.014285714 | 0.002534260 | 2.494937 | AUACAU,AUACUU,UACAUU,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ZCRB1 | 6 | 666 | 0.016666667 | 0.003360540 | 2.310201 | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.001456066 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| IGF2BP3 | 11 | 1348 | 0.028571429 | 0.006796655 | 2.071676 | AAAAAC,AAAACA,AAAUAC,AAUACA,ACAAUC,ACAUUC,CAAUCA,CAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| EIF4B | 1 | 226 | 0.004761905 | 0.001143692 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 2 | 340 | 0.007142857 | 0.001718057 | 2.055723 | UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.001773478 | 2.009919 | AAAGAC,AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| AGO1 | 1 | 283 | 0.004761905 | 0.001430875 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.001451028 | 1.714464 | GCAUGC | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| IGF2BP2 | 14 | 2163 | 0.035714286 | 0.010902862 | 1.711794 | AAAAAC,AAAACA,AAAUAC,AAUACA,ACAAUC,ACAUUC,CAAAAC,CAAUCA,CAUACA,GAAAUC,GCAUAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| ENOX1 | 4 | 756 | 0.011904762 | 0.003813986 | 1.642167 | AAGACA,AAUACA,AGGACA,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SART3 | 4 | 777 | 0.011904762 | 0.003919790 | 1.602690 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.001642483 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.001763402 | 1.433177 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.001768440 | 1.429061 | AAAAAA | AAAAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.002730754 | 1.387202 | AAAACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PUM2 | 3 | 764 | 0.009523810 | 0.003854293 | 1.305073 | GUAUAU,UAAAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| MSI1 | 4 | 961 | 0.011904762 | 0.004846836 | 1.296424 | AGUAAG,AGUUGG,UAGGUG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SAMD4A | 3 | 790 | 0.009523810 | 0.003985288 | 1.256855 | CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| A1CF | 2 | 598 | 0.007142857 | 0.003017936 | 1.242939 | UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.007154373 | 1.220068 | AAACAA,AAAUAC,AAUACA,ACAUAA,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.003184200 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.006524587 | 1.130615 | AGUAAG,AUUAAC,UACUGA,UAGUAA,UAUACU | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PABPC4 | 1 | 462 | 0.004761905 | 0.002332729 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.