circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000005175:-:12:47679492:47697719
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000005175:-:12:47679492:47697719 | ENSG00000005175 | ENST00000005386 | - | 12 | 47679493 | 47697719 | 993 | GACCGUAUCCUUGAUGAGCUUGACAAAGACGAUAGUACCCAUGAGUCUCUGUCUCAAGAAUCAGAGUCGGAAGAAGAUGGGAUUCAUGUAGAUUCACAAAAGGCUCUUGUUUUAAAAGAAAAGGGCAAUAAAUACUUCAAACAAGGAAAAUAUGAUGAAGCAAUUGACUGCUACACAAAAGGCAUGGAUGCCGAUCCAUAUAAUCCCGUGUUGCCAACGAACAGAGCGUCAGCAUAUUUUAGACUGAAAAAAUUUGCUGUUGCUGAGUCUGAUUGUAAUUUAGCAGUUGCCUUGAAUAGAAGUUAUACAAAGGCUUAUUCCAGACGAGGUGCUGCUCGAUUUGCUUUGCAAAAAUUAGAAGAGGCCAAAAAAGAUUAUGAAAGAGUAUUAGAACUAGAACCAAAUAACUUUGAAGCAACAAAUGAACUCAGGAAAAUCAGUCAGGCUUUAGCAUCCAAAGAAAACUCAUAUCCAAAGGAAGCUGACAUAGUGAUUAAGUCAACAGAAGGAGAGCGAAAGCAAAUUGAAGCACAACAGAAUAAGCAGCAGGCCAUUUCAGAGAAAGAUCGGGGGAAUGGAUUUUUCAAAGAGGGGAAAUAUGAAAGAGCAAUUGAAUGCUAUACUCGAGGGAUAGCAGCAGAUGGUGCUAAUGCCCUUCUUCCAGCUAACAGAGCUAUGGCCUAUCUGAAGAUUCAGAAAUAUGAAGAAGCUGAAAAAGACUGCACACAAGCCAUUUUAUUAGAUGGCUCAUAUUCUAAAGCUUUUGCCAGAAGAGGAACUGCAAGAACAUUUUUGGGAAAGCUAAAUGAGGCAAAACAAGAUUUUGAAACUGUUUUACUUCUGGAACCUGGAAAUAAGCAAGCAGUAACUGAACUCUCCAAAAUUAAAAAGGAAUUAAUUGAGAAAGGACACUGGGAUGAUGUCUUUCUUGAUUCCACACAAAGACAAAAUGUGGUAAAACCCAUUGAUAAUCCACCGCAUCCUGGAUCAACUGACCGUAUCCUUGAUGAGCUUGACAAAGACGAUAGUACCCAUGAGUCUCU | circ |
| ENSG00000005175:-:12:47679492:47697719 | ENSG00000005175 | ENST00000005386 | - | 12 | 47679493 | 47697719 | 22 | CUGGAUCAACUGACCGUAUCCU | bsj |
| ENSG00000005175:-:12:47679492:47697719 | ENSG00000005175 | ENST00000005386 | - | 12 | 47697710 | 47697919 | 210 | GAUCUUGGUUUUCUAAGCUAAGGCAUCUUUUUCUCUGGAUAUAAUCUUAACAAUGAGAGAUUUGGGUUGCUUUGUAAGGCUGUAGGUCCUUAGAUUGUAACAAUUUACAUUCAUUUGAGUGUAUUUUUUUAGUAUGUCUUUUUUUCUUUUUUUUCCUAAUCAUAUAAUUAUGACCCCUGUUUUCCGUCUUUUGAUUUUAGGACCGUAUCC | ie_up |
| ENSG00000005175:-:12:47679492:47697719 | ENSG00000005175 | ENST00000005386 | - | 12 | 47679293 | 47679502 | 210 | UGGAUCAACUGUAAGUAAAGCACUUUCAUUUUUUCCUUGCCAUUUAAAUAUGUUGUAAUAGGAGAAUUGGAAACCAACUAUUGUAUUAAUAAACUUUUUUUUUUUAAUUAUACUUUAAGUUCUAGGGUACAUGUGCACAACGUGCAGAUCUGAUACAUAGGUAUACAUGUGCCAUGUUGGUUUGCUGCACCCAUCAACUCAUCAUUUACA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
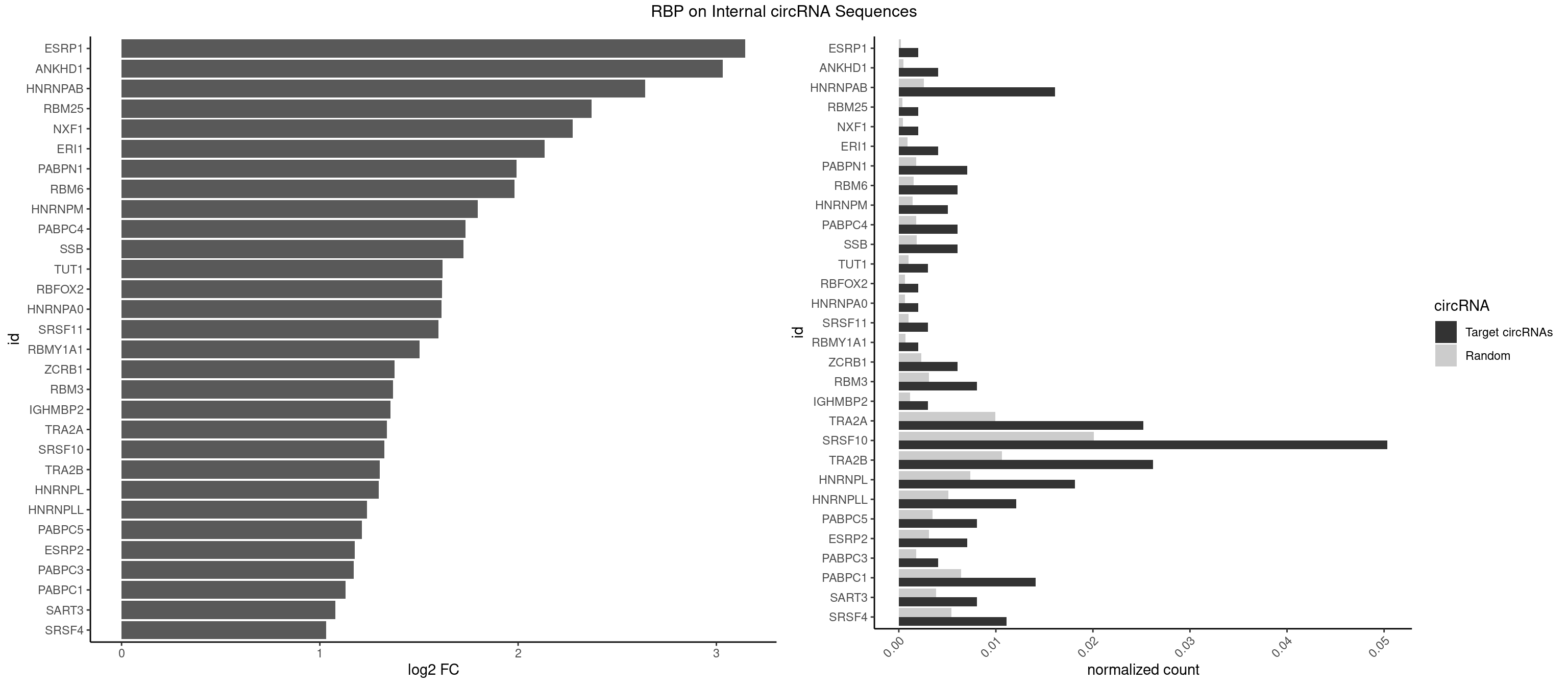
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.002014099 | 0.0002275250 | 3.146037 | AGGGAU | AGGGAU |
| ANKHD1 | 3 | 339 | 0.004028197 | 0.0004927293 | 3.031267 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPAB | 15 | 1782 | 0.016112790 | 0.0025839306 | 2.640567 | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM25 | 1 | 268 | 0.002014099 | 0.0003898359 | 2.369196 | AUCGGG | AUCGGG,CGGGCA,UCGGGC |
| NXF1 | 1 | 286 | 0.002014099 | 0.0004159215 | 2.275751 | AACCUG | AACCUG |
| ERI1 | 3 | 632 | 0.004028197 | 0.0009173461 | 2.134596 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| PABPN1 | 6 | 1222 | 0.007049345 | 0.0017723764 | 1.991804 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM6 | 5 | 1054 | 0.006042296 | 0.0015289102 | 1.982593 | AAUCCA,AUCCAA,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPM | 4 | 999 | 0.005035247 | 0.0014492040 | 1.796802 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| PABPC4 | 5 | 1251 | 0.006042296 | 0.0018144033 | 1.735602 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| SSB | 5 | 1260 | 0.006042296 | 0.0018274462 | 1.725268 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| TUT1 | 2 | 678 | 0.003021148 | 0.0009840095 | 1.618353 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBFOX2 | 1 | 452 | 0.002014099 | 0.0006564894 | 1.617291 | UGACUG | UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.002014099 | 0.0006579386 | 1.614110 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SRSF11 | 2 | 688 | 0.003021148 | 0.0009985015 | 1.597260 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 489 | 0.002014099 | 0.0007101099 | 1.504020 | ACAAGA | ACAAGA,CAAGAC |
| ZCRB1 | 5 | 1605 | 0.006042296 | 0.0023274215 | 1.376364 | AAUUAA,GAAUUA,GAUUAA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM3 | 7 | 2152 | 0.008056395 | 0.0031201361 | 1.368525 | AAAACU,AAGACG,AAGACU,AAUACU,AGACGA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| IGHMBP2 | 2 | 813 | 0.003021148 | 0.0011796520 | 1.356736 | AAAAAA | AAAAAA |
| TRA2A | 24 | 6871 | 0.025176234 | 0.0099589296 | 1.338000 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF10 | 49 | 13860 | 0.050352467 | 0.0200874160 | 1.325770 | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGG,AAGGAA,AAGGAG,ACAAAG,AGACAA,AGAGAA,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAGC,GAGGAA,GAGGGA,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| TRA2B | 25 | 7329 | 0.026183283 | 0.0106226650 | 1.301500 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GAAUUA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPL | 17 | 5085 | 0.018126888 | 0.0073706513 | 1.298267 | AAACAA,AAAUAA,AAAUAC,AACAAA,AAUAAA,ACACAA,CACAAA,CACAAC,CACAAG,CACACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| HNRNPLL | 11 | 3534 | 0.012084592 | 0.0051229360 | 1.238126 | ACCGCA,ACUGCA,AGACGA,CAAACA,CACACA,CACCGC,CAGACG,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC5 | 7 | 2400 | 0.008056395 | 0.0034795387 | 1.211238 | AGAAAA,AGAAAG,AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ESRP2 | 6 | 2150 | 0.007049345 | 0.0031172377 | 1.177221 | GGGAAA,GGGAAU,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC3 | 3 | 1234 | 0.004028197 | 0.0017897669 | 1.170363 | AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC1 | 13 | 4443 | 0.014098691 | 0.0064402624 | 1.130370 | AAAAAA,ACAAAU,ACGAAC,AGAAAA,CAAACA,CAAAUA,CGAACA,CUAACA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SART3 | 7 | 2634 | 0.008056395 | 0.0038186524 | 1.077071 | AAAAAA,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SRSF4 | 10 | 3740 | 0.011077543 | 0.0054214720 | 1.030881 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
RBP on BSJ (Exon Only)
Plot
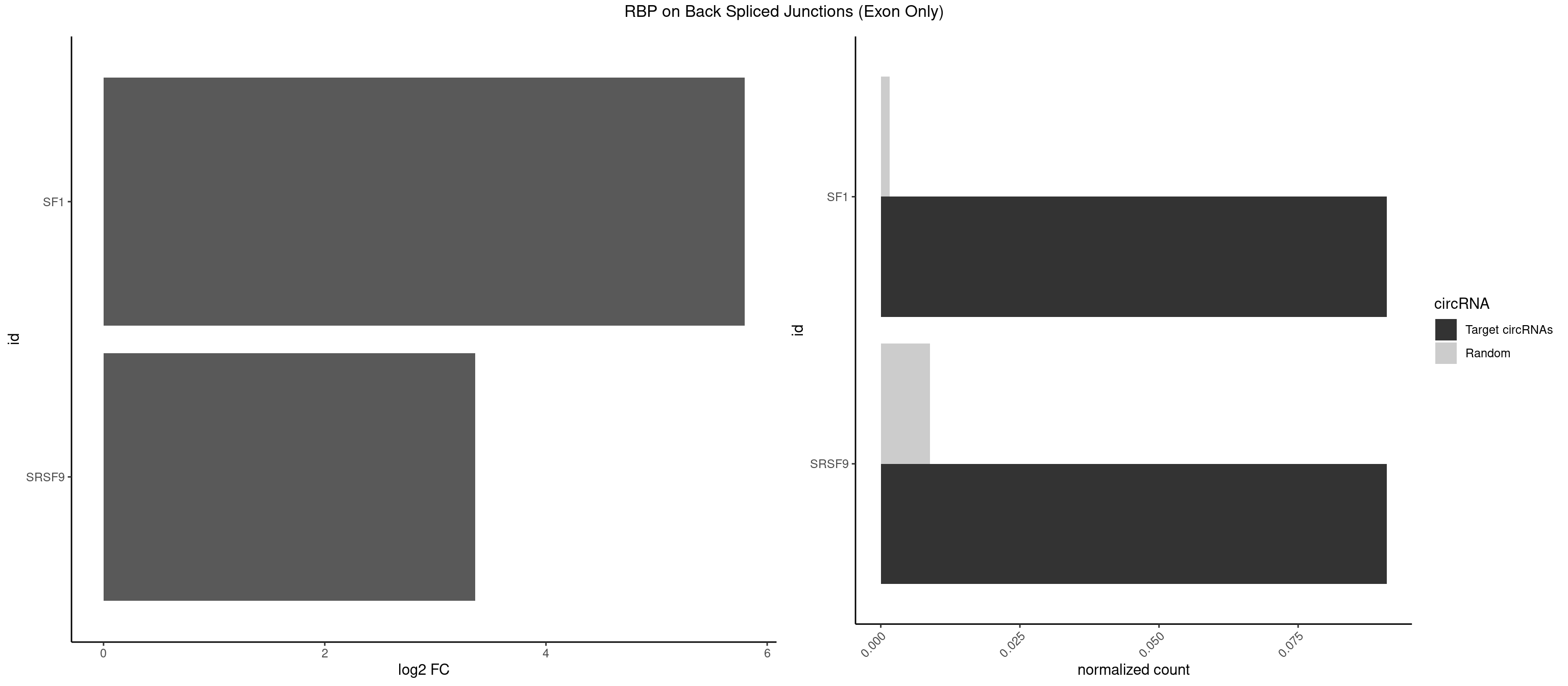
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SF1 | 1 | 24 | 0.09090909 | 0.001637412 | 5.794936 | ACUGAC | ACAGAC,ACAGUC,AGUAAG,AUACUA,CACAGA,CACUGA,CAGUCA,GCUGAC,GCUGCC,UACUGA,UGCUAA,UGCUGA,UGCUGC |
| SRSF9 | 1 | 134 | 0.09090909 | 0.008842023 | 3.361976 | UGACCG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
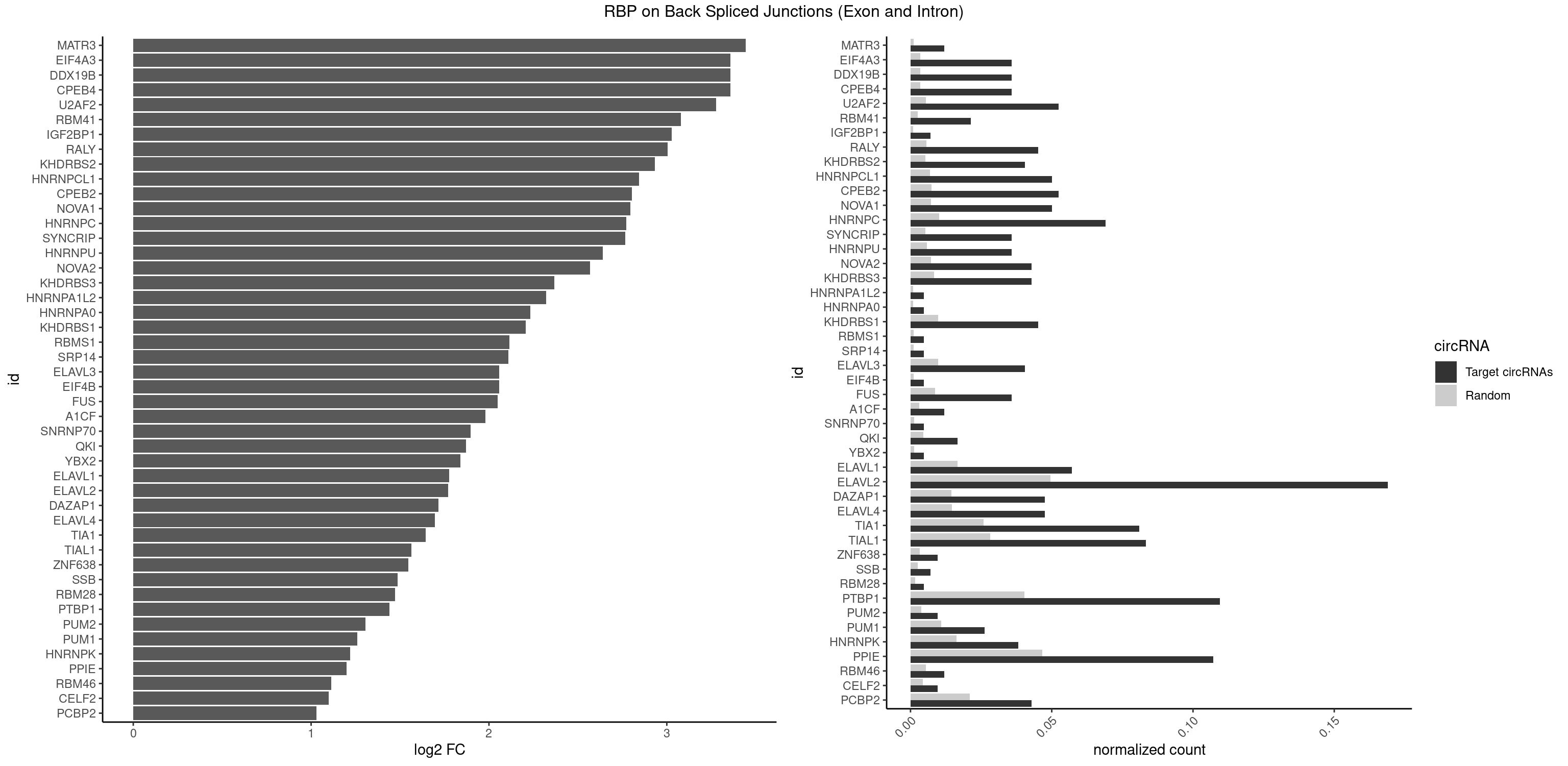
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 4 | 216 | 0.011904762 | 0.0010933091 | 3.444765 | AAUCUU,AUCUUA,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CPEB4 | 14 | 691 | 0.035714286 | 0.0034864974 | 3.356651 | UUUUUU | UUUUUU |
| DDX19B | 14 | 691 | 0.035714286 | 0.0034864974 | 3.356651 | UUUUUU | UUUUUU |
| EIF4A3 | 14 | 691 | 0.035714286 | 0.0034864974 | 3.356651 | UUUUUU | UUUUUU |
| U2AF2 | 21 | 1071 | 0.052380952 | 0.0054010480 | 3.277731 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM41 | 8 | 502 | 0.021428571 | 0.0025342604 | 3.079899 | AUACAU,AUACUU,UACAUG,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IGF2BP1 | 2 | 173 | 0.007142857 | 0.0008766626 | 3.026408 | AAGCAC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RALY | 18 | 1117 | 0.045238095 | 0.0056328094 | 3.005612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| KHDRBS2 | 16 | 1051 | 0.040476190 | 0.0053002821 | 2.932932 | AAUAAA,AUAAAC,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPCL1 | 20 | 1381 | 0.050000000 | 0.0069629182 | 2.844164 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 21 | 1487 | 0.052380952 | 0.0074969770 | 2.804661 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| NOVA1 | 20 | 1428 | 0.050000000 | 0.0071997179 | 2.795916 | AUCAAC,AUUCAU,CAUUCA,UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPC | 28 | 2006 | 0.069047619 | 0.0101118501 | 2.771545 | AUUUUU,CUUUUU,GGAUAU,GGGUAC,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP | 14 | 1041 | 0.035714286 | 0.0052498992 | 2.766140 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPU | 14 | 1136 | 0.035714286 | 0.0057285369 | 2.640263 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA2 | 17 | 1434 | 0.042857143 | 0.0072299476 | 2.567479 | AGGCAU,AUCAAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS3 | 17 | 1646 | 0.042857143 | 0.0082980653 | 2.368689 | AAUAAA,AUAAAC,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| KHDRBS1 | 18 | 1945 | 0.045238095 | 0.0098045143 | 2.206020 | AUUUAA,AUUUAC,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ELAVL3 | 16 | 1928 | 0.040476190 | 0.0097188634 | 2.058214 | AUUUUA,UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| FUS | 14 | 1711 | 0.035714286 | 0.0086255542 | 2.049812 | UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| QKI | 6 | 904 | 0.016666667 | 0.0045596534 | 1.869970 | AAUCAU,ACUCAU,AUCAUA,CUAAUC,UAAUCA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ELAVL1 | 23 | 3309 | 0.057142857 | 0.0166767432 | 1.776736 | UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUGAUU,UUGGUU,UUUAGU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ELAVL2 | 70 | 9826 | 0.169047619 | 0.0495112858 | 1.771600 | AAUUUA,AUACUU,AUUUAA,AUUUAC,AUUUUA,AUUUUU,CAUUUU,CUUUAA,CUUUUU,GAUUUU,GUUUUC,UAAGUU,UACUUU,UAUACU,UAUGUU,UAUUGU,UAUUUU,UCAUUU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUAUAC,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUAGU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| DAZAP1 | 19 | 2878 | 0.047619048 | 0.0145052398 | 1.714965 | AAUUUA,AGUAAA,AGUAUG,UAGGUA,UAGUAU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL4 | 19 | 2916 | 0.047619048 | 0.0146966949 | 1.696047 | UAUUUU,UUGUAU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TIA1 | 33 | 5140 | 0.080952381 | 0.0259018541 | 1.644018 | AUUUUU,CUUUUG,CUUUUU,GUUUUC,UAUUUU,UUUUCU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 34 | 5597 | 0.083333333 | 0.0282043531 | 1.562976 | AUUUUA,AUUUUU,CUUUUG,CUUUUU,GUUUUC,UAUUUU,UUAAAU,UUUAAA,UUUUAA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGGU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PTBP1 | 45 | 8003 | 0.109523810 | 0.0403264813 | 1.441445 | ACUUUC,ACUUUU,AUUUUU,CAUCUU,CUUUUU,GUCUUU,UACUUU,UAUUUU,UCUUUU,UUCCUU,UUCUCU,UUCUUU,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUACAU,UAAAUA,UACAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AGAAUU,AUUGUA,GAAUUG,GUAAUA,GUACAU,UAAAUA,UACAUA,UGUAAU,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPK | 15 | 3244 | 0.038095238 | 0.0163492543 | 1.220386 | ACCCAU,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| PPIE | 44 | 9262 | 0.107142857 | 0.0466696896 | 1.198978 | AAAUAU,AAUAAA,AAUUAU,AAUUUA,AUAAUU,AUAUAA,AUUAAU,AUUAUA,AUUUAA,AUUUUA,AUUUUU,UAAAUA,UAAUAA,UAAUUA,UAUAAU,UAUUAA,UAUUUU,UUAAAU,UUAAUA,UUAAUU,UUUAAA,UUUAAU,UUUUAA,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUCAU,AUCAUA,AUCAUU,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PCBP2 | 17 | 4164 | 0.042857143 | 0.0209844821 | 1.030213 | AAACCA,AACCAA,UUAGGA,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.