circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000129515:-:14:34581560:34609742
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000129515:-:14:34581560:34609742 | ENSG00000129515 | ENST00000396526 | - | 14 | 34581561 | 34609742 | 780 | CUUAAAGCAAUAAAUGUAGAUCUUCAAAGUGAUGCUGCUCUGCAGGUGGACAUUUCUGAUGCUCUUAGUGAGCGGGAUAAAGUAAAAUUCACUGUUCACACAAAGAGUUCAUUGCCAAAUUUUAAACAAAACGAGUUUUCAGUUGUUCGGCAACAUGAGGAAUUUAUCUGGCUUCAUGAUUCCUUUGUUGAAAAUGAAGACUAUGCAGGUUAUAUCAUUCCACCAGCACCACCAAGACCUGAUUUUGAUGCUUCAAGGGAAAAACUACAGAAGCUUGGUGAAGGAGAAGGGUCAAUGACGAAGGAAGAAUUCACAAAGAUGAAACAGGAACUGGAAGCUGAAUAUUUGGCAAUAUUCAAGAAGACAGUUGCGAUGCAUGAAGUGUUCCUGUGUCGUGUGGCAGCACAUCCUAUUUUGAGAAGAGAUUUAAAUUUCCAUGUCUUCUUGGAAUAUAAUCAAGAUUUGAGUGUGCGAGGAAAAAAUAAAAAAGAGAAACUUGAAGACUUCUUUAAAAACAUGGUUAAAUCAGCAGAUGGAGUAAUCGUUUCAGGAGUAAAGGAUGUAGAUGAUUUCUUUGAGCACGAACGAACAUUUCUUUUGGAGUAUCAUAACCGAGUUAAGGAUGCAUCUGCUAAAUCUGAUAGAAUGACAAGAUCCCACAAAAGUGCUGCAGAUGAUUACAAUAGAAUUGGUUCUUCAUUAUAUGCUUUAGGAACUCAGGAUUCUACAGAUAUAUGCAAGUUUUUUCUCAAAGUUUCAGAACUGUUCGAUAAAACAAGACUUAAAGCAAUAAAUGUAGAUCUUCAAAGUGAUGCUGCUCUGCAGGUGGA | circ |
| ENSG00000129515:-:14:34581560:34609742 | ENSG00000129515 | ENST00000396526 | - | 14 | 34581561 | 34609742 | 22 | AUAAAACAAGACUUAAAGCAAU | bsj |
| ENSG00000129515:-:14:34581560:34609742 | ENSG00000129515 | ENST00000396526 | - | 14 | 34609733 | 34609942 | 210 | AUACUAUAGAAUUAAGGUAAUAGCGUUGCCCAAAGCAGCUGCUUCAAAAUGUAUAUAGUAUGUAUGUUAAAUGGACAUGAGUACCUCUUUAGACUUAAUAAUCUUUUGUUAUUUUUGUUUUGUUUUUGCAUUGUCUUAAGAGAAAAAAUGAAAUUAUAUAAAACAUGAUUUUCAUGAAGAUACUGGUUUUCUUUUUCUAGCUUAAAGCAA | ie_up |
| ENSG00000129515:-:14:34581560:34609742 | ENSG00000129515 | ENST00000396526 | - | 14 | 34581361 | 34581570 | 210 | UAAAACAAGAGUAAGUACUAAAUUAUAGAGAUAUACUGCAUAAUAUUGUGCCCAGAGAGAGCUCAUGUUAAAGGUUUUUACCACAAUUCAGAAAAUUUUUUUAAAAGAGUGAUCACUAUUGGCCAGGCACGUGGCUCGUGCCUGUAAUCCCAACACUUUGGGAGGCCAAGUGGGGCGGAUCACGAGGUCACGAGAUCAAGACCAUCCUGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
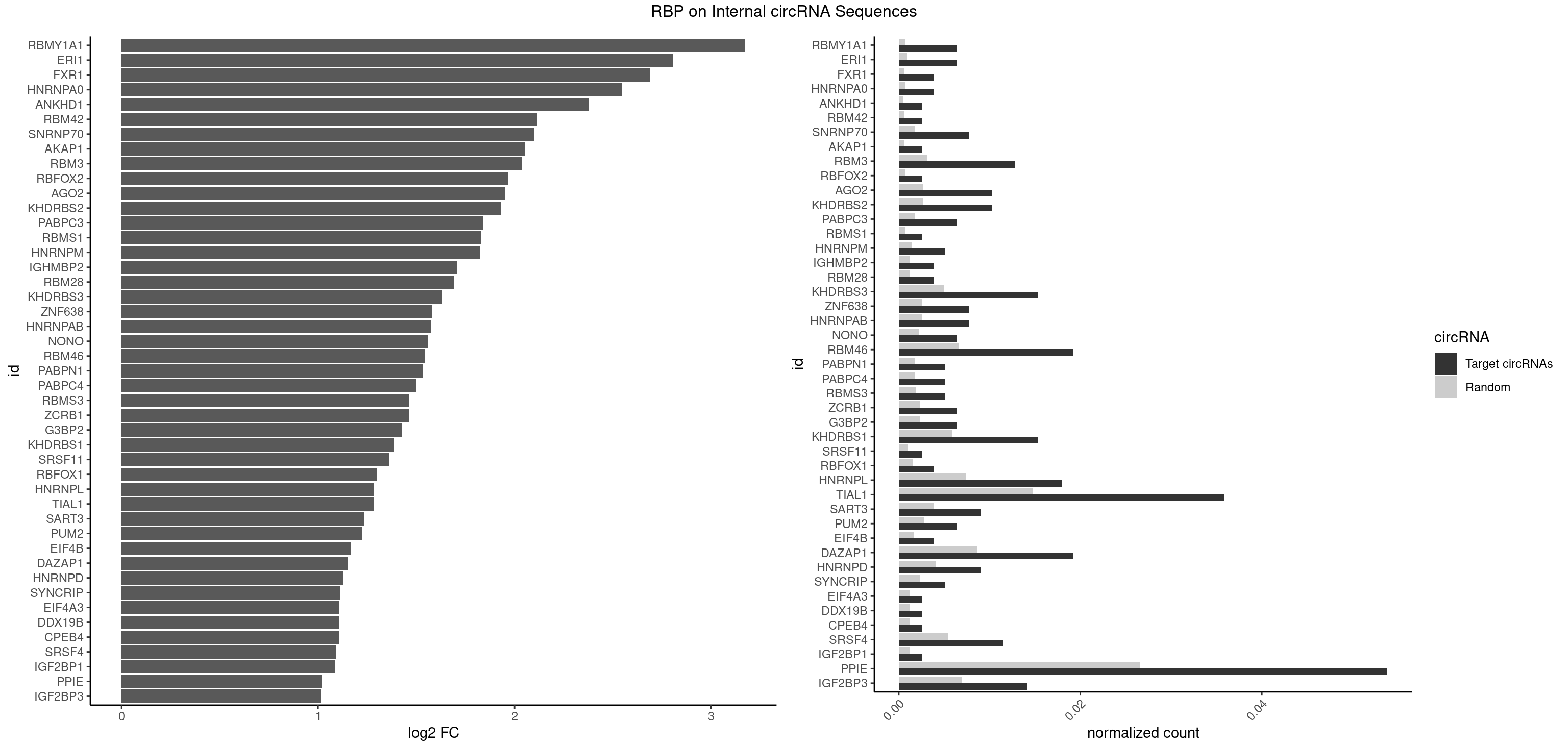
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 4 | 489 | 0.006410256 | 0.0007101099 | 3.174268 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| ERI1 | 4 | 632 | 0.006410256 | 0.0009173461 | 2.804844 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| FXR1 | 2 | 411 | 0.003846154 | 0.0005970720 | 2.687440 | AUGACA,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPA0 | 2 | 453 | 0.003846154 | 0.0006579386 | 2.547392 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| ANKHD1 | 1 | 339 | 0.002564103 | 0.0004927293 | 2.379587 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM42 | 1 | 407 | 0.002564103 | 0.0005912752 | 2.116552 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| SNRNP70 | 5 | 1237 | 0.007692308 | 0.0017941145 | 2.100145 | AAUCAA,AUCAAG,AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| AKAP1 | 1 | 426 | 0.002564103 | 0.0006188101 | 2.050885 | AUAUAU | AUAUAU,UAUAUA |
| RBM3 | 9 | 2152 | 0.012820513 | 0.0031201361 | 2.038773 | AAAACG,AAAACU,AAACGA,AAACUA,AAGACU,AGACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBFOX2 | 1 | 452 | 0.002564103 | 0.0006564894 | 1.965610 | UGCAUG | UGACUG,UGCAUG |
| AGO2 | 7 | 1830 | 0.010256410 | 0.0026534924 | 1.950562 | AAAAAA,AAAGUG,AAGUGC,AGUGCU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| KHDRBS2 | 7 | 1858 | 0.010256410 | 0.0026940701 | 1.928667 | AAUAAA,AUAAAA,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PABPC3 | 4 | 1234 | 0.006410256 | 0.0017897669 | 1.840610 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBMS1 | 1 | 497 | 0.002564103 | 0.0007217036 | 1.828976 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPM | 3 | 999 | 0.005128205 | 0.0014492040 | 1.823193 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| IGHMBP2 | 2 | 813 | 0.003846154 | 0.0011796520 | 1.705055 | AAAAAA | AAAAAA |
| RBM28 | 2 | 822 | 0.003846154 | 0.0011926949 | 1.689191 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| KHDRBS3 | 11 | 3429 | 0.015384615 | 0.0049707696 | 1.629947 | AAAUAA,AAUAAA,AUAAAA,AUAAAG,GAUAAA,UAAAAC,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ZNF638 | 5 | 1773 | 0.007692308 | 0.0025708878 | 1.581150 | GGUUCU,GUUCUU,GUUGUU,UGUUCG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 5 | 1782 | 0.007692308 | 0.0025839306 | 1.573849 | AAGACA,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NONO | 4 | 1498 | 0.006410256 | 0.0021723567 | 1.561121 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM46 | 14 | 4554 | 0.019230769 | 0.0066011240 | 1.542633 | AAUCAA,AAUGAA,AUCAAG,AUCAUA,AUCAUU,AUGAAA,AUGAAG,AUGAUU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| PABPN1 | 3 | 1222 | 0.005128205 | 0.0017723764 | 1.532769 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| PABPC4 | 3 | 1251 | 0.005128205 | 0.0018144033 | 1.498959 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBMS3 | 3 | 1283 | 0.005128205 | 0.0018607779 | 1.462548 | AAUAUA,AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZCRB1 | 4 | 1605 | 0.006410256 | 0.0023274215 | 1.461650 | ACUUAA,AUUUAA,GACUUA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 4 | 1644 | 0.006410256 | 0.0023839405 | 1.427034 | AGGAUG,AGGAUU,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| KHDRBS1 | 11 | 4064 | 0.015384615 | 0.0058910141 | 1.384900 | AUAAAA,AUUUAA,CUAAAU,UAAAAA,UAAAAC,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SRSF11 | 1 | 688 | 0.002564103 | 0.0009985015 | 1.360617 | AAGAAG | AAGAAG |
| RBFOX1 | 2 | 1077 | 0.003846154 | 0.0015622419 | 1.299799 | GCAUGA,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPL | 13 | 5085 | 0.017948718 | 0.0073706513 | 1.284017 | AAACAA,AAAUAA,AACAAA,AAUAAA,ACACAA,CACAAA,CACACA,CACCAC,CACGAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| TIAL1 | 27 | 10182 | 0.035897436 | 0.0147572438 | 1.282457 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUG,CUUUUG,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUAAA,UUUCAG,UUUUAA,UUUUCA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SART3 | 6 | 2634 | 0.008974359 | 0.0038186524 | 1.232745 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PUM2 | 4 | 1890 | 0.006410256 | 0.0027404447 | 1.225972 | GUAGAU,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| EIF4B | 2 | 1179 | 0.003846154 | 0.0017100607 | 1.169369 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| DAZAP1 | 14 | 5964 | 0.019230769 | 0.0086445016 | 1.153562 | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUG,AGGUUA,AGUAAA,AGUUAA,UAGGAA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPD | 6 | 2837 | 0.008974359 | 0.0041128408 | 1.125674 | AAAAAA,AAUUUA,AGAUAU,AUUUAU,UUAGGA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SYNCRIP | 3 | 1634 | 0.005128205 | 0.0023694485 | 1.113903 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| CPEB4 | 1 | 821 | 0.002564103 | 0.0011912456 | 1.105983 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.002564103 | 0.0011912456 | 1.105983 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.002564103 | 0.0011912456 | 1.105983 | UUUUUU | UUUUUU |
| SRSF4 | 8 | 3740 | 0.011538462 | 0.0054214720 | 1.089694 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| IGF2BP1 | 1 | 831 | 0.002564103 | 0.0012057377 | 1.088538 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PPIE | 41 | 18315 | 0.053846154 | 0.0265436196 | 1.020478 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUUU,AAUAAA,AAUAUA,AAUAUU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAUAA,AUAUAU,AUAUUU,AUUAUA,AUUUAA,AUUUAU,AUUUUA,UAAAAA,UAAAAU,UAAAUU,UAUAAU,UAUUUU,UUAAAA,UUAAAU,UUAUAU,UUUAAA,UUUUAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| IGF2BP3 | 10 | 4815 | 0.014102564 | 0.0069793662 | 1.014790 | AAAAAC,AAAACA,AAAUCA,AAAUUC,AACUCA,AAUUCA,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
RBP on BSJ (Exon Only)
Plot
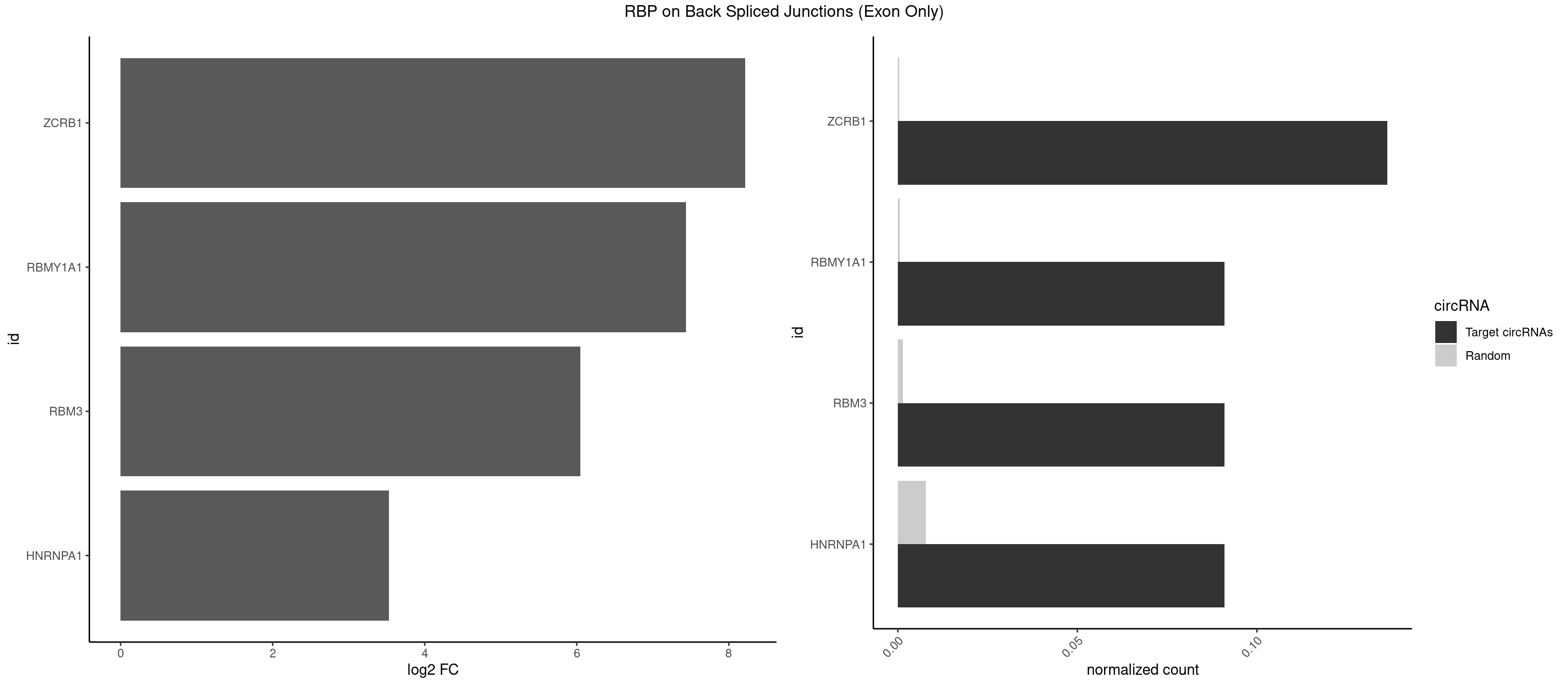
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZCRB1 | 2 | 6 | 0.13636364 | 0.0004584752 | 8.216399 | ACUUAA,GACUUA | AAUUAA,AUUUAA,GACUUA,GGCUUA |
| RBMY1A1 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | CAAGAC | ACAAGA,CAAGAC |
| RBM3 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | AAGACU | AAAACG,AAAACU,AAGACU,AAUACG,AAUACU,AUACUA,GAAACU,GAGACG,GAGACU,GAUACU |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.0078595756 | 3.531901 | GACUUA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
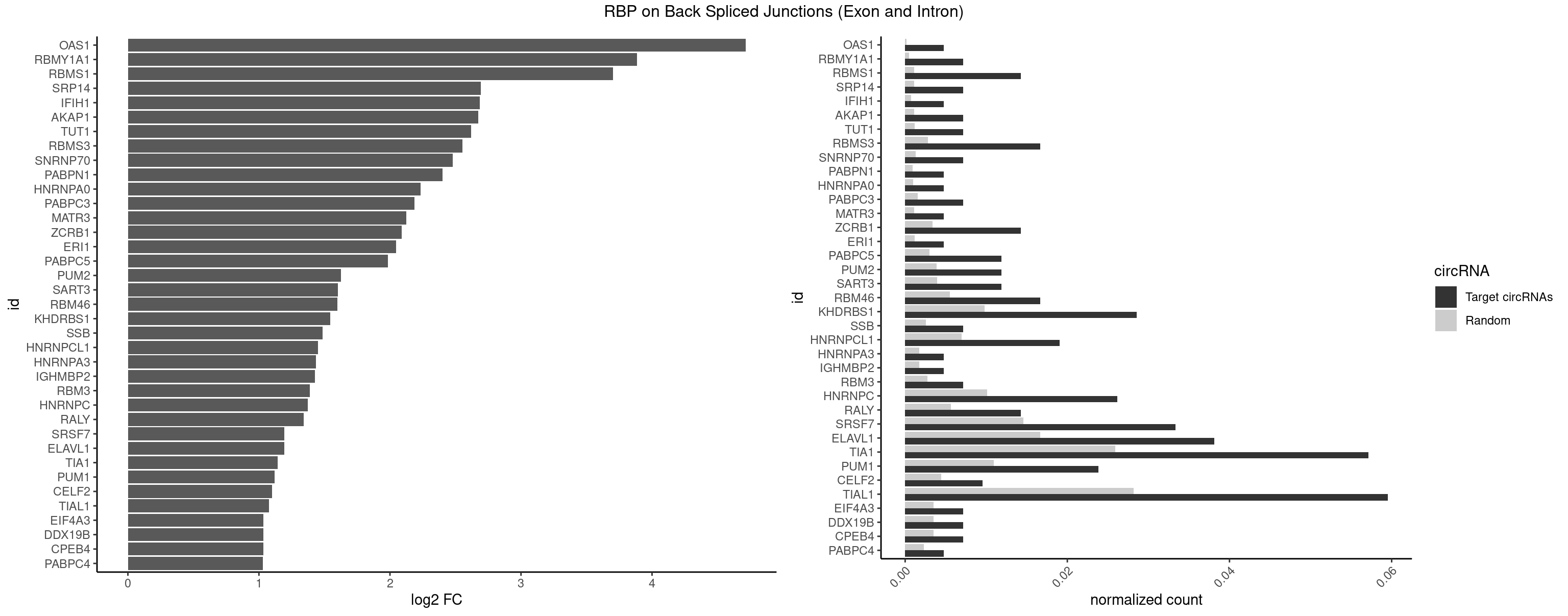
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| RBMY1A1 | 2 | 95 | 0.007142857 | 0.0004836759 | 3.884389 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| RBMS1 | 5 | 217 | 0.014285714 | 0.0010983474 | 3.701167 | AUAUAC,AUAUAG,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | UAUAUA | AUAUAU,UAUAUA |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBMS3 | 6 | 562 | 0.016666667 | 0.0028365578 | 2.554752 | AUAUAG,CUAUAG,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZCRB1 | 5 | 666 | 0.014285714 | 0.0033605401 | 2.087808 | AAUUAA,ACUUAA,GAAUUA,GACUUA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAA,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAUAU,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUGAA,AUCAAG,AUGAAA,AUGAAG,AUGAUU,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| KHDRBS1 | 11 | 1945 | 0.028571429 | 0.0098045143 | 1.543055 | AUAAAA,CAAAAU,CUAAAU,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AUACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RALY | 5 | 1117 | 0.014285714 | 0.0056328094 | 1.342647 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SRSF7 | 13 | 2893 | 0.033333333 | 0.0145808142 | 1.192894 | ACGAGA,AGAGAA,AGAGAG,AGAGAU,AGAUCA,AUAGAA,CAGAGA,CGAGAU,GAGAGA,GAGAUC,UAGAGA,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAUUU,UUGUUU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUC,AUUUUU,CUUUUG,CUUUUU,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UUAUUU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM1 | 9 | 2172 | 0.023809524 | 0.0109482064 | 1.120844 | AAUAUU,AGAAUU,GUAAUA,GUAUAU,UAAUAU,UAUAUA,UGUAAU,UGUAUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AAAUUU,AAUUUU,AUUUUC,AUUUUU,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUUAA,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.