circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000198162:+:1:117405548:117466427
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198162:+:1:117405548:117466427 | ENSG00000198162 | ENST00000356554 | + | 1 | 117405549 | 117466427 | 610 | AUGAUGAAACAUGCUUGGGAUAACUAUAGGACAUAUGGGUGGGGACAUAAUGAACUCAGACCUAUUGCAAGGAAAGGACACUCCCCUAACAUAUUUGGAAGUUCACAAAUGGGUGCUACCAUAGUAGAUGCUUUGGAUACCCUUUAUAUCAUGGGACUUCAUGAUGAAUUCCUAGAUGGGCAAAGAUGGAUUGAAGACAACCUUGAUUUCAGUGUGAAUUCAGAGGUGUCUGUGUUUGAAGUCAACAUUCGAUUUAUUGGAGGCCUACUUGCAGCAUAUUACCUAUCAGGAGAGGAGAUAUUCAAGAUUAAAGCAGUGCAAUUGGCUGAGAAACUCCUUCCUGCCUUUAACACACCUACUGGGAUUCCUUGGGCAAUGGUGAAUUUGAAAAGUGGAGUAGGGCGAAACUGGGGCUGGGCAUCUGCAGGUAGCAGCAUUCUGGCUGAAUUUGGUACACUACAUAUGGAGUUCAUCCACCUCAGCUACUUGACAGGGGACCUGACUUACUACAAAAAGGUUAUGCACAUUCGGAAACUACUUCAGAAAAUGGAUCGUCCAAAUGGUCUUUAUCCAAAUUAUUUGAACCCCAGAACAGGGCGCUGGGGUCAGUAUGAUGAAACAUGCUUGGGAUAACUAUAGGACAUAUGGGUGGGGACAUAA | circ |
| ENSG00000198162:+:1:117405548:117466427 | ENSG00000198162 | ENST00000356554 | + | 1 | 117405549 | 117466427 | 22 | CUGGGGUCAGUAUGAUGAAACA | bsj |
| ENSG00000198162:+:1:117405548:117466427 | ENSG00000198162 | ENST00000356554 | + | 1 | 117405349 | 117405558 | 210 | CCAUUUUUUUCCUGUCAUAUAGAUGAGAUUAAACAUAGUUAUGUAUGUUUUCAUACUGAUACAAAUGCUAAUUGAAUAAUGUACUAAUAUAGAUCUGGAAGAAUGGGCAAGGAUAUAGUAAUAGAACCCAAAAUUGUAUGAAAUAAAAAAACAGUUUGUGAAAAAUUUAAAUUGAUGGAUUAUAAUUUUUCUCUUUUCAGAUGAUGAAAC | ie_up |
| ENSG00000198162:+:1:117405548:117466427 | ENSG00000198162 | ENST00000356554 | + | 1 | 117466418 | 117466627 | 210 | UGGGGUCAGUGUAAGUAUUCUAGUAUACCUGAGGCUUUCUUAAAAAAUUAUUUGUCUGAAAACUCUUUGCUUGAUGUUGUAAAAAGUACACUACGUGGAGUUAGAAACCCUCAGUGUCUAACCUUGUGGUCUGCUGGUGCUGUUCUUAGGCAUGUUUCUGAUCUCUCUUAGCUUCAGUUUUCCCACUUAUAAAAUGGAAAUGUUAAUAAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
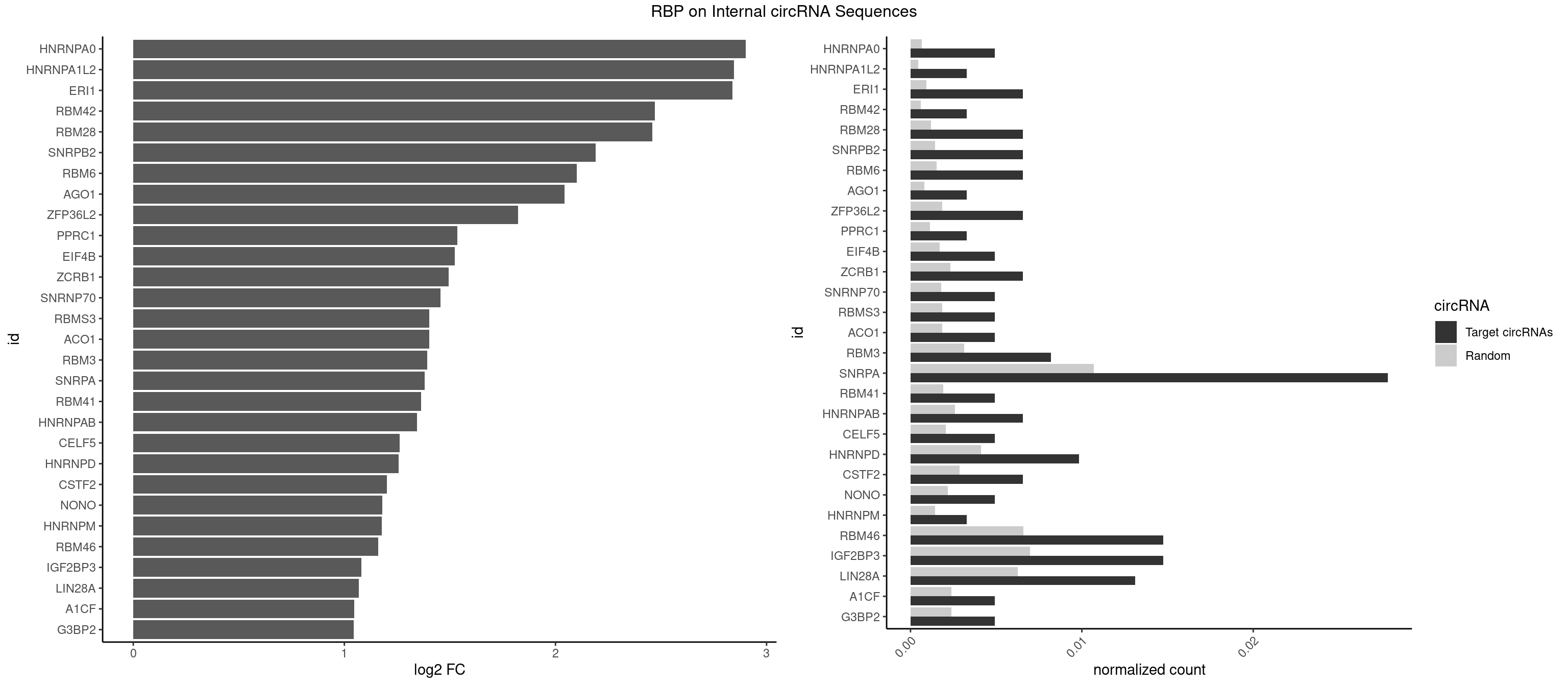
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 2 | 453 | 0.004918033 | 0.0006579386 | 2.902057 | AGAUAU,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| HNRNPA1L2 | 1 | 314 | 0.003278689 | 0.0004564992 | 2.844434 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ERI1 | 3 | 632 | 0.006557377 | 0.0009173461 | 2.837581 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBM42 | 1 | 407 | 0.003278689 | 0.0005912752 | 2.471217 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM28 | 3 | 822 | 0.006557377 | 0.0011926949 | 2.458894 | AGUAGA,AGUAGG,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SNRPB2 | 3 | 991 | 0.006557377 | 0.0014376103 | 2.189446 | AUUGCA,UAUUGC,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM6 | 3 | 1054 | 0.006557377 | 0.0015289102 | 2.100615 | AUCCAA,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| AGO1 | 1 | 548 | 0.003278689 | 0.0007956130 | 2.042980 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ZFP36L2 | 3 | 1277 | 0.006557377 | 0.0018520827 | 1.823970 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PPRC1 | 1 | 780 | 0.003278689 | 0.0011318283 | 1.534464 | GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| EIF4B | 2 | 1179 | 0.004918033 | 0.0017100607 | 1.524034 | UCGGAA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZCRB1 | 3 | 1605 | 0.006557377 | 0.0023274215 | 1.494386 | GACUUA,GAUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SNRNP70 | 2 | 1237 | 0.004918033 | 0.0017941145 | 1.454809 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ACO1 | 2 | 1283 | 0.004918033 | 0.0018607779 | 1.402176 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 2 | 1283 | 0.004918033 | 0.0018607779 | 1.402176 | CUAUAG,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM3 | 4 | 2152 | 0.008196721 | 0.0031201361 | 1.393438 | AAACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SNRPA | 16 | 7380 | 0.027868852 | 0.0106965744 | 1.381505 | AGCAGU,AGGAGA,AGUAGG,AUGCAC,AUUCCU,AUUGCA,GAUACC,GAUUCC,GGAGAU,GUAGGG,UCCUGC,UGCACA,UUACCU,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RBM41 | 2 | 1318 | 0.004918033 | 0.0019115000 | 1.363376 | UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPAB | 3 | 1782 | 0.006557377 | 0.0025839306 | 1.343552 | AAGACA,AGACAA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF5 | 2 | 1415 | 0.004918033 | 0.0020520728 | 1.260999 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPD | 5 | 2837 | 0.009836066 | 0.0041128408 | 1.257946 | AGAUAU,AGUAGG,AUUUAU,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| CSTF2 | 3 | 1967 | 0.006557377 | 0.0028520334 | 1.201128 | GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| NONO | 2 | 1498 | 0.004918033 | 0.0021723567 | 1.178820 | AGAGGA,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPM | 1 | 999 | 0.003278689 | 0.0014492040 | 1.177858 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBM46 | 8 | 4554 | 0.014754098 | 0.0066011240 | 1.160332 | AAUGAA,AUCAUG,AUGAAA,AUGAAU,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| IGF2BP3 | 8 | 4815 | 0.014754098 | 0.0069793662 | 1.079948 | AAACUC,AACACA,AACUCA,AAUUCA,ACACAC,ACACUC,ACAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| LIN28A | 7 | 4315 | 0.013114754 | 0.0062547643 | 1.068163 | AGGAGA,GGAGAU,GGAGUA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| A1CF | 2 | 1642 | 0.004918033 | 0.0023810421 | 1.046488 | CAGUAU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP2 | 2 | 1644 | 0.004918033 | 0.0023839405 | 1.044733 | GGAUAA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
RBP on BSJ (Exon Only)
Plot
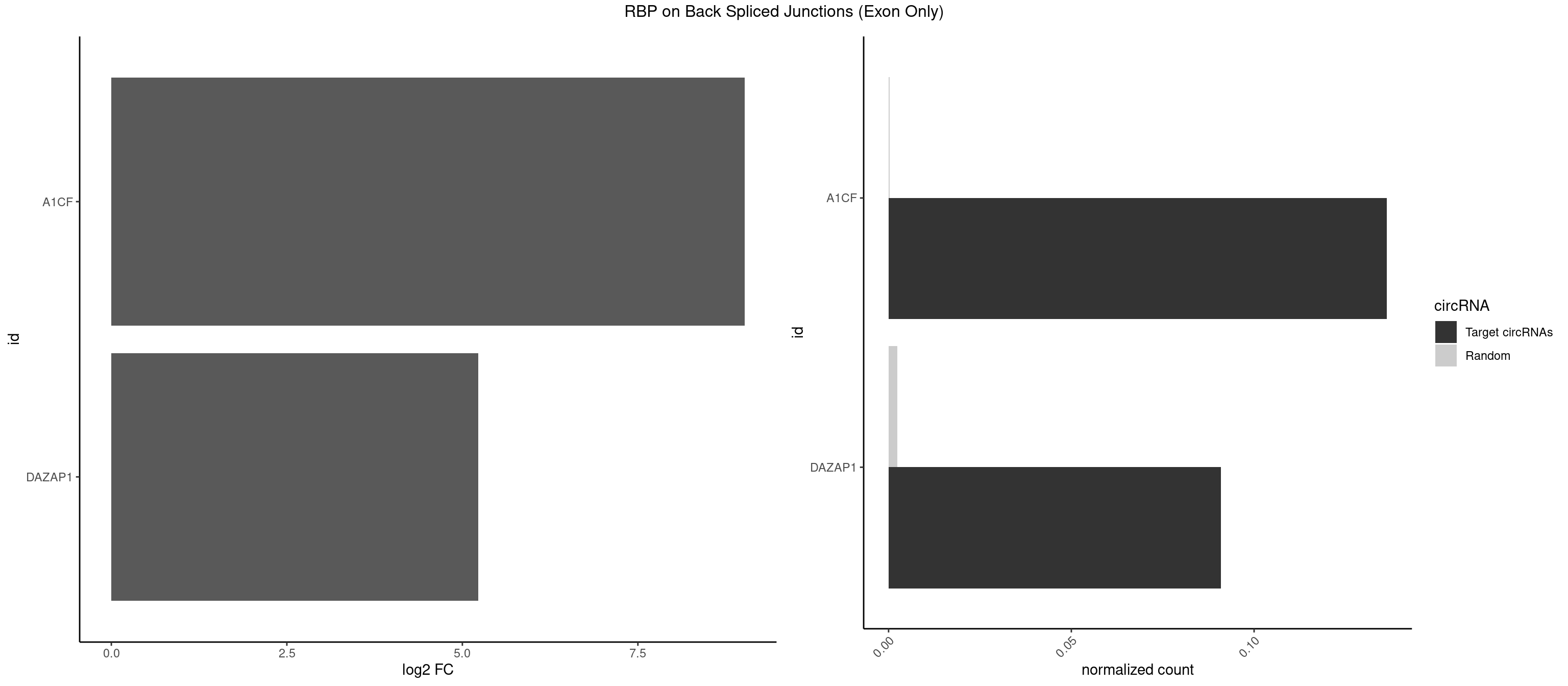
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| A1CF | 2 | 3 | 0.13636364 | 0.0002619859 | 9.023754 | CAGUAU,UCAGUA | AUCAGU,CAGUAU,UAAUUG |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGUAUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
RBP on BSJ (Exon and Intron)
Plot
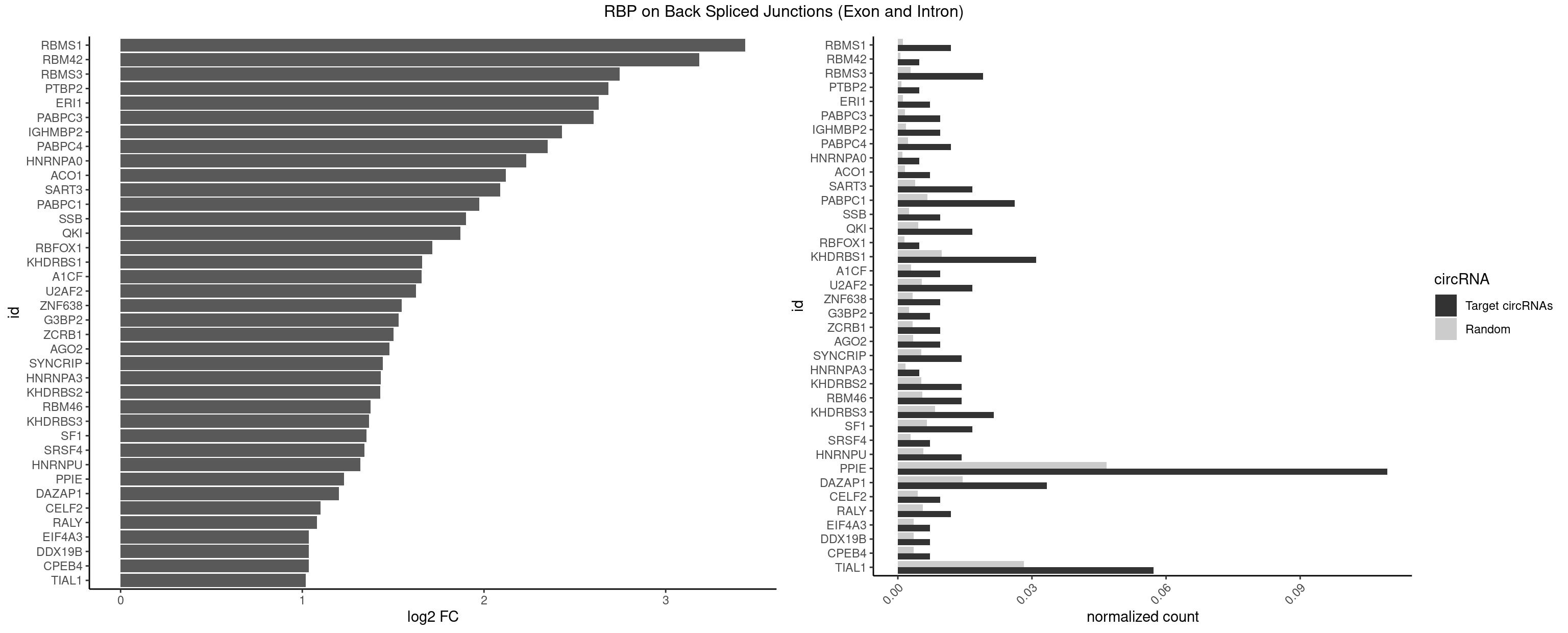
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMS1 | 4 | 217 | 0.011904762 | 0.0010983474 | 3.438132 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBMS3 | 7 | 562 | 0.019047619 | 0.0028365578 | 2.747397 | AAUAUA,AUAUAG,CAUAUA,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGHMBP2 | 3 | 350 | 0.009523810 | 0.0017684401 | 2.429061 | AAAAAA | AAAAAA |
| PABPC4 | 4 | 462 | 0.011904762 | 0.0023327287 | 2.351448 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SART3 | 6 | 777 | 0.016666667 | 0.0039197904 | 2.088117 | AAAAAA,AAAAAC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC1 | 10 | 1321 | 0.026190476 | 0.0066606207 | 1.975314 | AAAAAA,AAAAAC,ACAAAU,ACUAAU,CUAACC,CUAAUA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| QKI | 6 | 904 | 0.016666667 | 0.0045596534 | 1.869970 | ACACUA,ACUAAU,ACUUAU,CUAACC,UAACCU,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| KHDRBS1 | 12 | 1945 | 0.030952381 | 0.0098045143 | 1.658532 | AUAAAA,AUUUAA,CAAAAU,GAAAAC,UAAAAA,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AGUAUA,AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUCUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AUUUAA,GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 | 3 | 677 | 0.009523810 | 0.0034159613 | 1.479247 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| KHDRBS2 | 5 | 1051 | 0.014285714 | 0.0053002821 | 1.430432 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AUGAAA,AUGAUG,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AAAUAA,AAUAAA,AUAAAA,AUUAAA,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACUAAU,ACUUAU,UACUAA,UACUGA,UAGUAA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PPIE | 45 | 9262 | 0.109523810 | 0.0466696896 | 1.230687 | AAAAAA,AAAAAU,AAAAUU,AAAUAA,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAUU,AUUAAA,AUUAUA,AUUAUU,AUUUAA,AUUUUU,UAAAAA,UAAAAU,UAAAUU,UAAUAA,UAAUAU,UAAUUU,UAUAAA,UAUAAU,UUAAAA,UUAAAU,UUAAUA,UUAUAA,UUAUUU,UUUAAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| DAZAP1 | 13 | 2878 | 0.033333333 | 0.0145052398 | 1.200391 | AAAAAA,AAUUUA,AGGAUA,AGUAUA,AGUUAG,AGUUUG,UAGUAA,UAGUAU,UAGUUA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| TIAL1 | 23 | 5597 | 0.057142857 | 0.0282043531 | 1.018655 | AAAUUU,AAUUUU,AGUUUU,AUUUUU,CAGUUU,CUUUUC,GUUUUC,UAAAUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUCAG,UUUUCA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.