circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000083845:+:19:58392975:58393487
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000083845:+:19:58392975:58393487 | ENSG00000083845 | MSTRG.17847.3 | + | 19 | 58392976 | 58393487 | 339 | GAUUACAUUGCAGUGAAGGAGAAGUAUGCCAAGUACCUGCCUCACAGUGCAGGGCGGUAUGCCGCCAAACGCUUCCGCAAAGCUCAGUGUCCCAUUGUGGAGCGCCUCACUAACUCCAUGAUGAUGCACGGCCGCAACAACGGCAAGAAGCUCAUGACUGUGCGCAUCGUCAAGCAUGCCUUCGAGAUCAUACACCUGCUCACAGGCGAGAACCCUCUGCAGGUCCUGGUGAACGCCAUCAUCAACAGUGGUCCCCGGGAGGACUCCACACGCAUUGGGCGCGCCGGGACUGUGAGACGACAGGCUGUGGAUGUGUCCCCCCUGCGCCGUGUGAACCAGGAUUACAUUGCAGUGAAGGAGAAGUAUGCCAAGUACCUGCCUCACAGUGC | circ |
| ENSG00000083845:+:19:58392975:58393487 | ENSG00000083845 | MSTRG.17847.3 | + | 19 | 58392976 | 58393487 | 22 | GUGUGAACCAGGAUUACAUUGC | bsj |
| ENSG00000083845:+:19:58392975:58393487 | ENSG00000083845 | MSTRG.17847.3 | + | 19 | 58392776 | 58392985 | 210 | CUUGGUAAAGUCACCUGUUCAGGGUUUUUUUCCUCAUGGUGGAUGGGGCUUACAUCCAAGUCCCCUGUUGAGGCAUACCAGGAUCCCAGACAGCUGUUGGACUACCCCGAAUGGGUACUUGAUCUCUCCUCUGGUGUCUGGCCAACGCCCAUGCUGCUCCUGACCAUUUUCCCUUCAUUUCCUGCUCCCUGCUGCCCCAGGAUUACAUUG | ie_up |
| ENSG00000083845:+:19:58392975:58393487 | ENSG00000083845 | MSTRG.17847.3 | + | 19 | 58393478 | 58393687 | 210 | UGUGAACCAGGUGAGCCUGGGGCUUAUGCACGUGGCAGGGUGGACACAGCCACGGGAGUGGGUGGGGUCUUGCCUGGGGCAGGAAGGCUCCUCUCUGUCUGCAUGUUUUACCUGCAGCAUCACUUCCUCUAGGAAGCCUUCCUGGAUUCCCACCCUGCAUAGGGGCUGAUCCAGGCUCUGGGUCCUCUUCGGGAACACAUUUGGCAGAGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
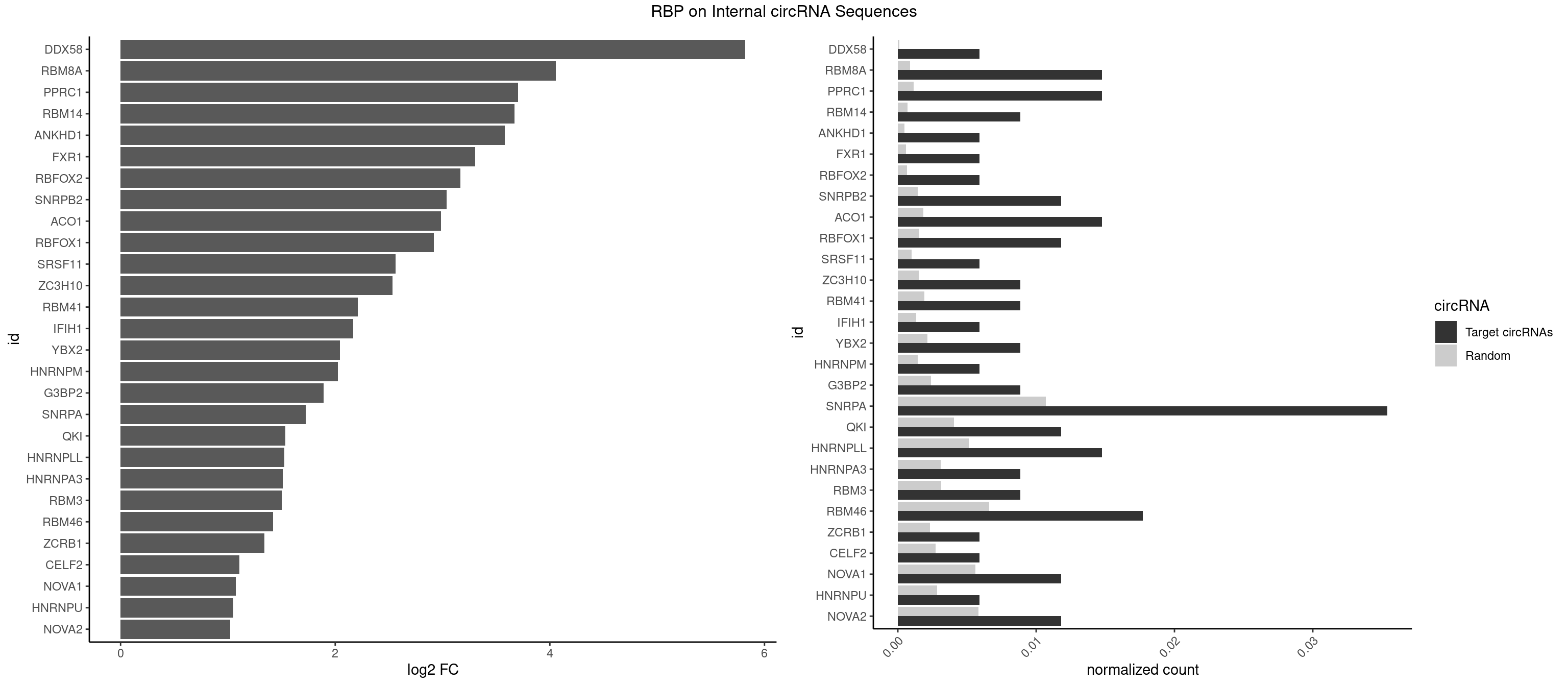
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.005899705 | 0.0001043427 | 5.821241 | GCGCGC | GCGCGC |
| RBM8A | 4 | 611 | 0.014749263 | 0.0008869128 | 4.055707 | CGCGCC,GCGCGC,GUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 4 | 780 | 0.014749263 | 0.0011318283 | 3.703916 | CGCGCC,GCGCGC,GGCGCG,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| RBM14 | 2 | 478 | 0.008849558 | 0.0006941687 | 3.672247 | CGCGCC,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| ANKHD1 | 1 | 339 | 0.005899705 | 0.0004927293 | 3.581776 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| FXR1 | 1 | 411 | 0.005899705 | 0.0005970720 | 3.304666 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBFOX2 | 1 | 452 | 0.005899705 | 0.0006564894 | 3.167799 | UGACUG | UGACUG,UGCAUG |
| SNRPB2 | 3 | 991 | 0.011799410 | 0.0014376103 | 3.036970 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ACO1 | 4 | 1283 | 0.014749263 | 0.0018607779 | 2.986665 | CAGUGA,CAGUGC,CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBFOX1 | 3 | 1077 | 0.011799410 | 0.0015622419 | 2.917025 | AGCAUG,GCAUGC,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRSF11 | 1 | 688 | 0.005899705 | 0.0009985015 | 2.562806 | AAGAAG | AAGAAG |
| ZC3H10 | 2 | 1053 | 0.008849558 | 0.0015274610 | 2.534470 | GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM41 | 2 | 1318 | 0.008849558 | 0.0019115000 | 2.210900 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IFIH1 | 1 | 904 | 0.005899705 | 0.0013115296 | 2.169392 | GGCCGC | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| YBX2 | 2 | 1480 | 0.008849558 | 0.0021462711 | 2.043773 | AACAAC,ACAACG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPM | 1 | 999 | 0.005899705 | 0.0014492040 | 2.025382 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 2 | 1644 | 0.008849558 | 0.0023839405 | 1.892257 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRPA | 11 | 7380 | 0.035398230 | 0.0106965744 | 1.726528 | ACCUGC,AGGAGA,AUGCAC,AUUGCA,CCUGCU,GGUAUG,GUAUGC,UACCUG,UGCACG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| QKI | 3 | 2805 | 0.011799410 | 0.0040664663 | 1.536867 | ACUAAC,AUCAUA,CACUAA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPLL | 4 | 3534 | 0.014749263 | 0.0051229360 | 1.525600 | ACGACA,AGACGA,CAUACA,GACGAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPA3 | 2 | 2140 | 0.008849558 | 0.0031027457 | 1.512060 | AAGGAG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 2 | 2152 | 0.008849558 | 0.0031201361 | 1.503996 | AGACGA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM46 | 5 | 4554 | 0.017699115 | 0.0066011240 | 1.422894 | AUCAUA,AUGAUG,GAUCAU,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 1 | 1605 | 0.005899705 | 0.0023274215 | 1.341910 | GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF2 | 1 | 1886 | 0.005899705 | 0.0027346479 | 1.109288 | AUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| NOVA1 | 3 | 3872 | 0.011799410 | 0.0056127669 | 1.071931 | AUCAAC,AUCAUC,CCCCCC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPU | 1 | 1965 | 0.005899705 | 0.0028491350 | 1.050119 | CCCCCC | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA2 | 3 | 4013 | 0.011799410 | 0.0058171047 | 1.020342 | AGAUCA,AUCAAC,AUCAUC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
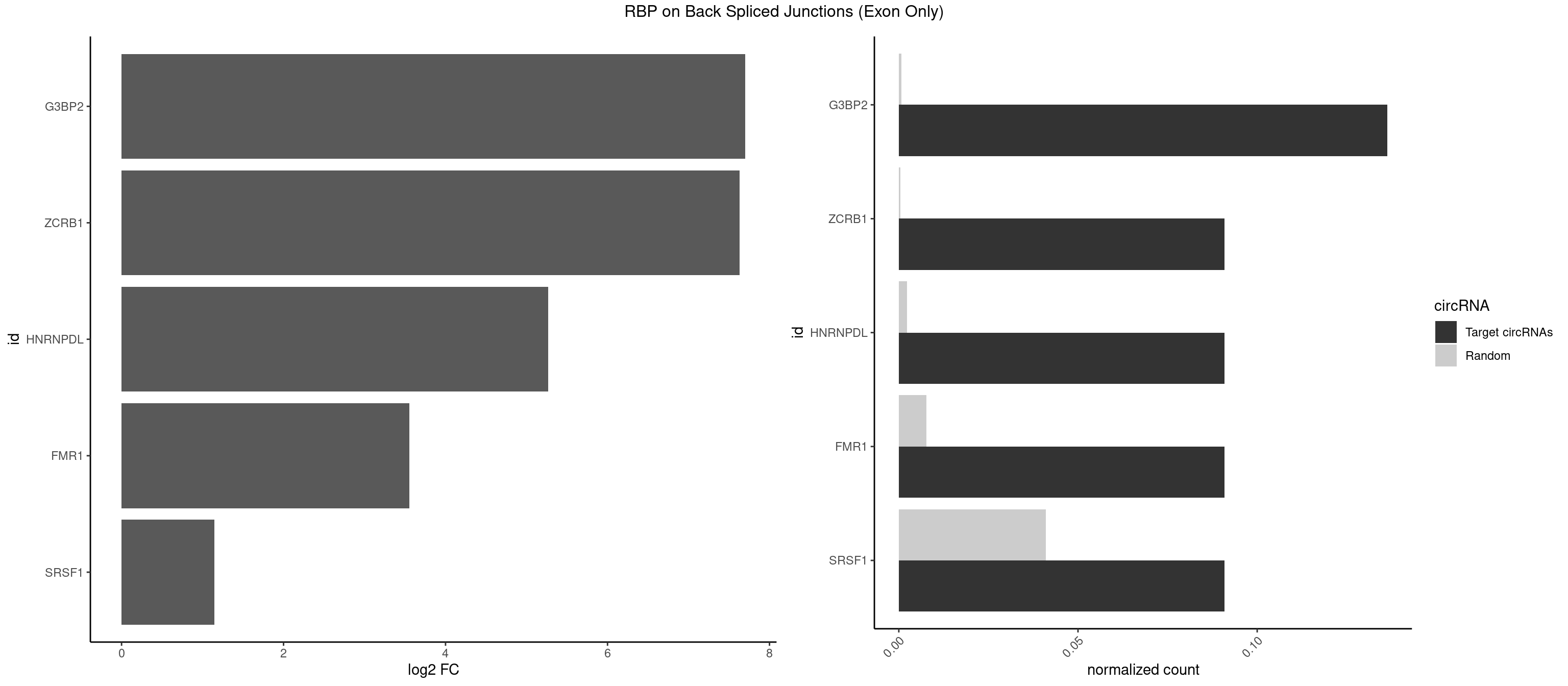
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| G3BP2 | 2 | 9 | 0.13636364 | 0.0006549646 | 7.701826 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUGA,GGAUGG,GGAUUG |
| ZCRB1 | 1 | 6 | 0.09090909 | 0.0004584752 | 7.631437 | GGAUUA | AAUUAA,AUUUAA,GACUUA,GGCUUA |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | GGAUUA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | AGGAUU | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF1 | 1 | 625 | 0.09090909 | 0.0410007860 | 1.148773 | CCAGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
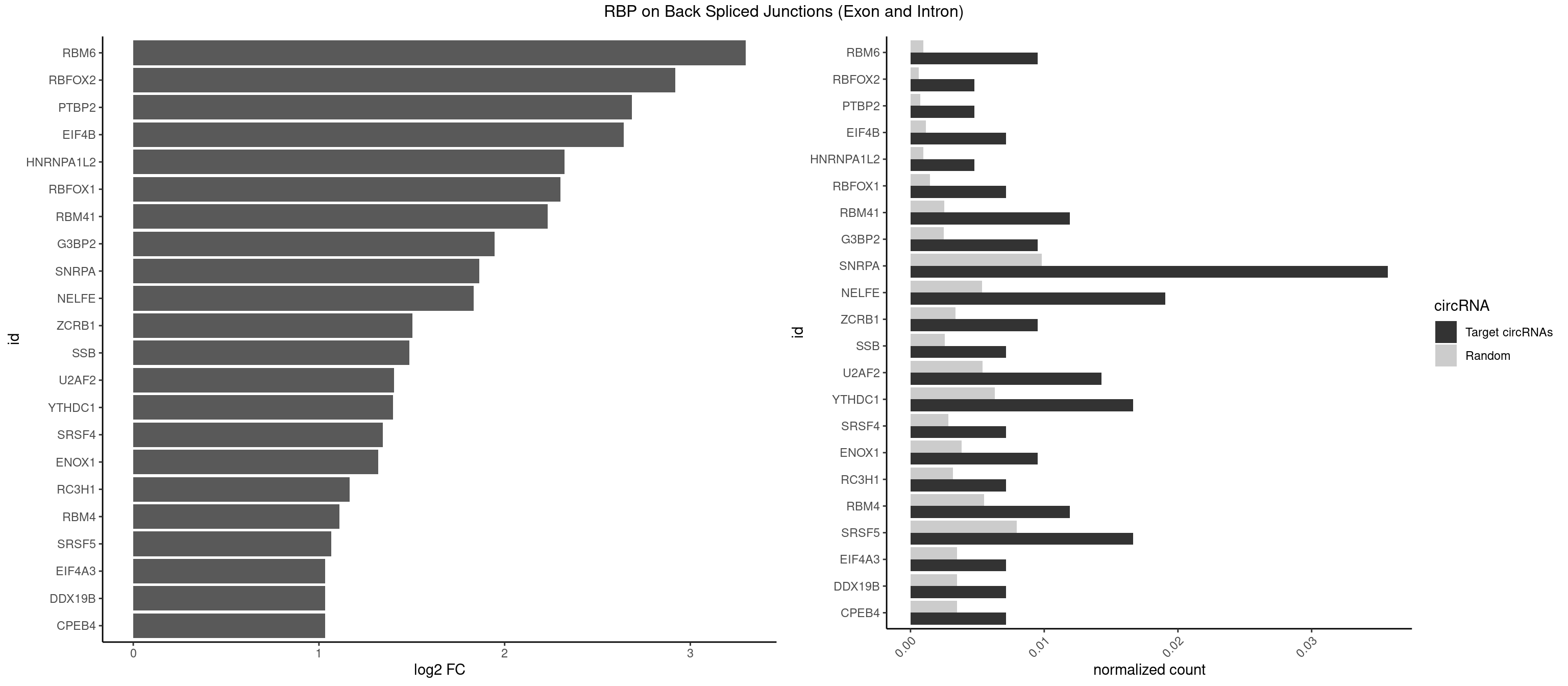
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM6 | 3 | 191 | 0.009523810 | 0.0009673519 | 3.299426 | AUCCAA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | UACAUU,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUGG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRPA | 14 | 1947 | 0.035714286 | 0.0098145909 | 1.863501 | ACCUGC,AUGCAC,AUGCUG,CCUGCU,GAUUCC,UACCUG,UCCUGC,UGCACG,UUACCU,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| NELFE | 7 | 1059 | 0.019047619 | 0.0053405885 | 1.834540 | CUCUCU,CUCUGG,UCUCUC,UCUCUG,UCUGGC,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | GGAUUA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.0054010480 | 1.403262 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.0063230552 | 1.398272 | GACUAC,GCAUAC,GGGUAC,UCCUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGGAAG | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGACAG,CAGACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCCUUC,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CCUUCC,CUUCCU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AGGAAG,CACAGC,UACAUC,UGCAGC,UGCAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.