circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000083642:+:13:32648753:32652007
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000083642:+:13:32648753:32652007 | ENSG00000083642 | ENST00000315596 | + | 13 | 32648754 | 32652007 | 331 | GGGUAGAAAUAUUUCUGUCAUGGCUCAUUCAAAGACUAGGACCAAUGAUGGAAAAAUUACAUAUCCGCCUGGGGUCAAGGAAAUAUCAGAUAAAAUAUCUAAAGAGGAGAUGGUGAGACGAUUAAAGAUGGUUGUGAAAACUUUUAUGGAUAUGGACCAGGACUCUGAAGAAGAAAAGGAGCUUUAUUUAAACCUAGCUUUACAUCUUGCUUCAGAUUUUUUUCUCAAGCAUCCUGAUAAAGAUGUUCGCUUACUGGUAGCCUGCUGCCUUGCUGAUAUUUUCAGGAUUUAUGCUCCUGAAGCUCCUUACACAUCCCCUGAUAAACUAAAGGGGUAGAAAUAUUUCUGUCAUGGCUCAUUCAAAGACUAGGACCAAUGAUG | circ |
| ENSG00000083642:+:13:32648753:32652007 | ENSG00000083642 | ENST00000315596 | + | 13 | 32648754 | 32652007 | 22 | AUAAACUAAAGGGGUAGAAAUA | bsj |
| ENSG00000083642:+:13:32648753:32652007 | ENSG00000083642 | ENST00000315596 | + | 13 | 32648554 | 32648763 | 210 | AAAGUAAAAAUAAGUUUUGUUUUUCUAAAAAUAAUUUGGGGAAAUGUUCAAUAGAGUGAUUUUCAUUAAAUGGUAAAGUAAAUUACUAUAAAUUAGGAGGUAGUUACAAAUUUUGGUUGGUGGGGAAGGUUACCAUUUAUGUUUGUUAAGAAUGUUUAUAUCUAUUUUUUGGUUUGCAUCUAAUAGUUUUCUUGUUUCAGGGGUAGAAAU | ie_up |
| ENSG00000083642:+:13:32648753:32652007 | ENSG00000083642 | ENST00000315596 | + | 13 | 32651998 | 32652207 | 210 | UAAACUAAAGGCAAGUACUGAUUUAAAUAACUCCAAGAUUGACCGAUACUUUGAUUUAUCUUUCUAACUAAAAUGGGAGUUAAGGUAUUUAGAUGAUUUCCUUGGAUUCUUUGACAUUAGCUACAGUAAUCUUUUGCUAAACUUAAUGUUUUUUUUUUUUUUUAGAACUUAUUUUUGAAAAACUUUCUGGUCUUGAGGUUAAGAAGUAGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
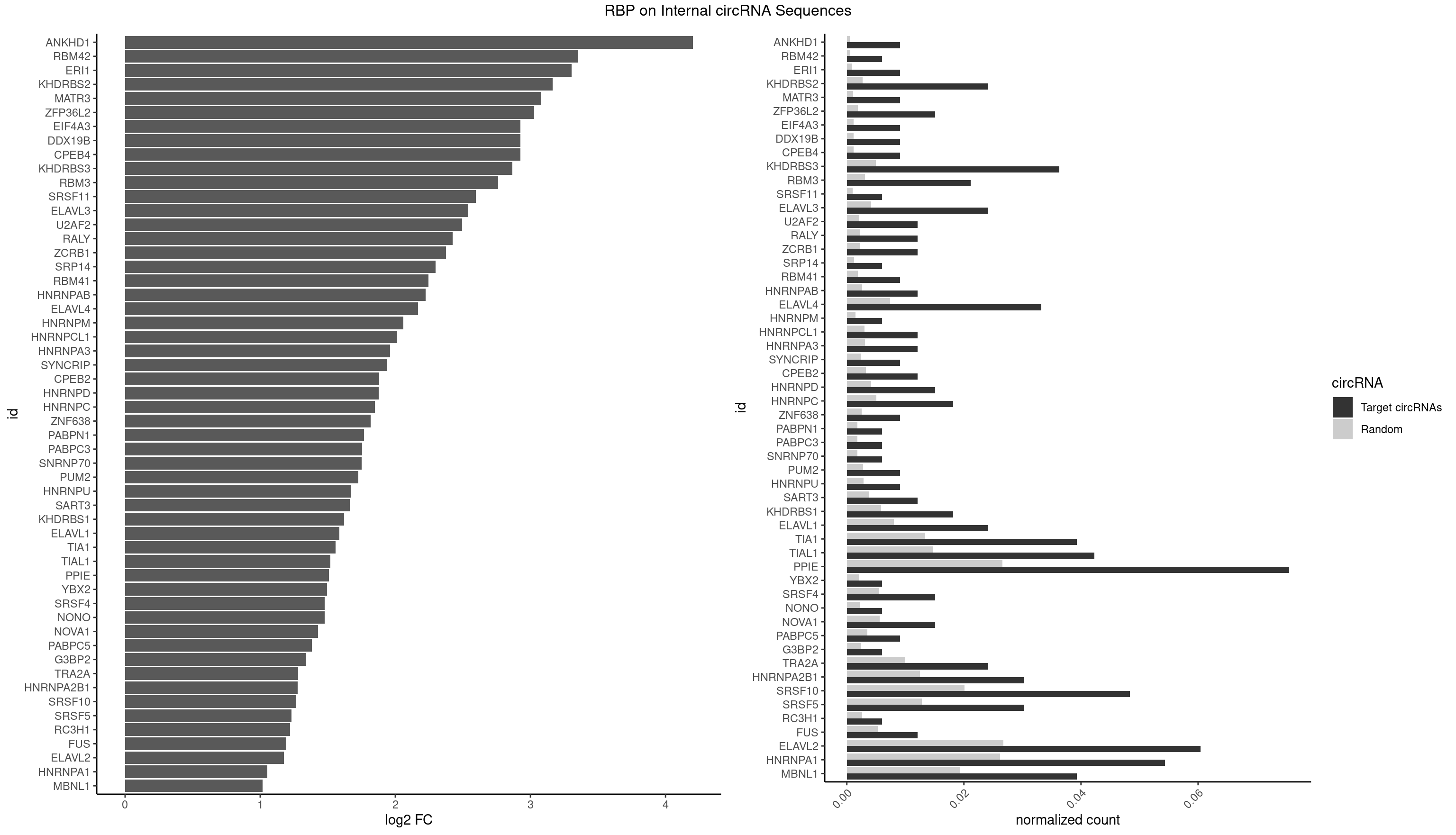
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.009063444 | 0.0004927293 | 4.201192 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM42 | 1 | 407 | 0.006042296 | 0.0005912752 | 3.353195 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| ERI1 | 2 | 632 | 0.009063444 | 0.0009173461 | 3.304521 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| KHDRBS2 | 7 | 1858 | 0.024169184 | 0.0026940701 | 3.165309 | AUAAAA,AUAAAC,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| MATR3 | 2 | 739 | 0.009063444 | 0.0010724109 | 3.079202 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZFP36L2 | 4 | 1277 | 0.015105740 | 0.0018520827 | 3.027876 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CPEB4 | 2 | 821 | 0.009063444 | 0.0011912456 | 2.927588 | UUUUUU | UUUUUU |
| DDX19B | 2 | 821 | 0.009063444 | 0.0011912456 | 2.927588 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 821 | 0.009063444 | 0.0011912456 | 2.927588 | UUUUUU | UUUUUU |
| KHDRBS3 | 11 | 3429 | 0.036253776 | 0.0049707696 | 2.866590 | AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBM3 | 6 | 2152 | 0.021148036 | 0.0031201361 | 2.760843 | AAAACU,AAACUA,AAGACU,AGACGA,AGACUA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF11 | 1 | 688 | 0.006042296 | 0.0009985015 | 2.597260 | AAGAAG | AAGAAG |
| ELAVL3 | 7 | 2867 | 0.024169184 | 0.0041563169 | 2.539791 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| U2AF2 | 3 | 1477 | 0.012084592 | 0.0021419234 | 2.496190 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 3 | 1553 | 0.012084592 | 0.0022520629 | 2.423850 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ZCRB1 | 3 | 1605 | 0.012084592 | 0.0023274215 | 2.376364 | AUUUAA,GAUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRP14 | 1 | 847 | 0.006042296 | 0.0012289250 | 2.297700 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM41 | 2 | 1318 | 0.009063444 | 0.0019115000 | 2.245354 | UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPAB | 3 | 1782 | 0.012084592 | 0.0025839306 | 2.225530 | AAAGAC,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ELAVL4 | 10 | 5105 | 0.033232628 | 0.0073996354 | 2.167074 | AUCUAA,AUUUAU,UAUCUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPM | 1 | 999 | 0.006042296 | 0.0014492040 | 2.059836 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPCL1 | 3 | 2062 | 0.012084592 | 0.0029897078 | 2.015092 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPA3 | 3 | 2140 | 0.012084592 | 0.0031027457 | 1.961551 | AAGGAG,AGGAGC,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SYNCRIP | 2 | 1634 | 0.009063444 | 0.0023694485 | 1.935508 | UUUUUU | AAAAAA,UUUUUU |
| CPEB2 | 3 | 2261 | 0.012084592 | 0.0032780993 | 1.882237 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPD | 4 | 2837 | 0.015105740 | 0.0041128408 | 1.876890 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPC | 5 | 3471 | 0.018126888 | 0.0050316361 | 1.849032 | AUUUUU,GGAUAU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZNF638 | 2 | 1773 | 0.009063444 | 0.0025708878 | 1.817793 | GGUUGU,UGUUCG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPN1 | 1 | 1222 | 0.006042296 | 0.0017723764 | 1.769412 | AGAAGA | AAAAGA,AGAAGA |
| PABPC3 | 1 | 1234 | 0.006042296 | 0.0017897669 | 1.755325 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 1 | 1237 | 0.006042296 | 0.0017941145 | 1.751825 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 2 | 1890 | 0.009063444 | 0.0027404447 | 1.725649 | UACAUA,UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPU | 2 | 1965 | 0.009063444 | 0.0028491350 | 1.669535 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SART3 | 3 | 2634 | 0.012084592 | 0.0038186524 | 1.662033 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| KHDRBS1 | 5 | 4064 | 0.018126888 | 0.0058910141 | 1.621543 | AUAAAA,AUUUAA,GAAAAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ELAVL1 | 7 | 5554 | 0.024169184 | 0.0080503280 | 1.586049 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| TIA1 | 12 | 9206 | 0.039274924 | 0.0133428208 | 1.557545 | AUUUUC,AUUUUU,CUCCUU,CUUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 13 | 10182 | 0.042296073 | 0.0147572438 | 1.519100 | AUUUUC,AUUUUU,CUUUUA,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUCAG,UUUUAU,UUUUCA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PPIE | 24 | 18315 | 0.075528701 | 0.0265436196 | 1.508660 | AAAAAU,AAAAUA,AAAAUU,AAAUAU,AAAUUA,AAUAUU,AUAAAA,AUAUUU,AUUAAA,AUUUAA,AUUUAU,AUUUUU,UAAAAU,UAUUUA,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAU,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| YBX2 | 1 | 1480 | 0.006042296 | 0.0021462711 | 1.493265 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SRSF4 | 4 | 3740 | 0.015105740 | 0.0054214720 | 1.478340 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO | 1 | 1498 | 0.006042296 | 0.0021723567 | 1.475836 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| NOVA1 | 4 | 3872 | 0.015105740 | 0.0056127669 | 1.428313 | CAUUCA,UCAUUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| PABPC5 | 2 | 2400 | 0.009063444 | 0.0034795387 | 1.381163 | AGAAAA,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| G3BP2 | 1 | 1644 | 0.006042296 | 0.0023839405 | 1.341749 | AGGAUU | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TRA2A | 7 | 6871 | 0.024169184 | 0.0099589296 | 1.279106 | AAGAAA,AAGAAG,AAGAGG,AGAAGA,AGAGGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPA2B1 | 9 | 8607 | 0.030211480 | 0.0124747476 | 1.276086 | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AGACUA,AGGAGC,AUUUAA,CAAGGA,GACUAG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SRSF10 | 15 | 13860 | 0.048338369 | 0.0200874160 | 1.266877 | AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGAGG,AAGGAA,AAGGAG,AAGGGG,AGAGGA,CAAAGA,GAGACG,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF5 | 9 | 8869 | 0.030211480 | 0.0128544391 | 1.232830 | AAGAAG,AGAAGA,AUAAAG,CACAUC,GAAGAA,UAAAGG,UACAUA,UACAUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RC3H1 | 1 | 1786 | 0.006042296 | 0.0025897275 | 1.222297 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| FUS | 3 | 3648 | 0.012084592 | 0.0052881452 | 1.192335 | UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL2 | 19 | 18468 | 0.060422961 | 0.0267653478 | 1.174730 | AUAUUU,AUUUAA,AUUUAU,AUUUUC,AUUUUU,CUUUUA,GAUUUA,GAUUUU,UAUUUA,UAUUUU,UUAUUU,UUUAUG,UUUAUU,UUUUAU,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1 | 17 | 18070 | 0.054380665 | 0.0261885646 | 1.054157 | AAAGAG,AAGAGG,AAGGAG,AGACUA,AGAGGA,AGGAGC,AUUUAA,AUUUAU,CAAAGA,CAAGGA,GACUAG,GAGGAG,UUAUUU,UUUAUU,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| MBNL1 | 12 | 13372 | 0.039274924 | 0.0193802045 | 1.019025 | AUGCUC,CCUGCU,CUGCCU,CUGCUG,CUUGCU,UCGCUU,UGCUGC,UGCUUC,UUCGCU,UUGCUG,UUGCUU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
RBP on BSJ (Exon Only)
Plot
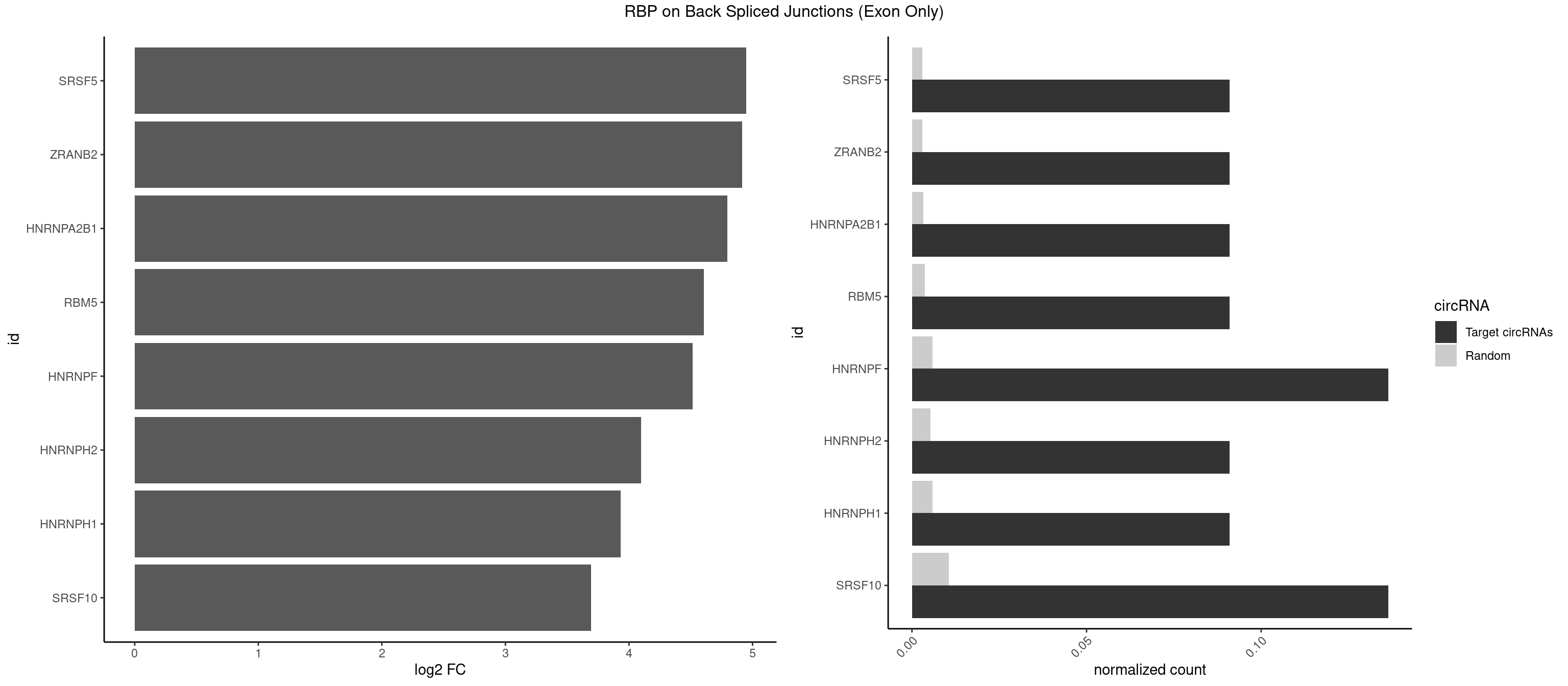
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | UAAAGG | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.003274823 | 4.794936 | AAGGGG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | AAGGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| HNRNPF | 2 | 90 | 0.13636364 | 0.005960178 | 4.515960 | AAGGGG,GGGGUA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.005305214 | 4.098942 | AAGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | AAGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| SRSF10 | 2 | 160 | 0.13636364 | 0.010544931 | 3.692837 | AAAGGG,AAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
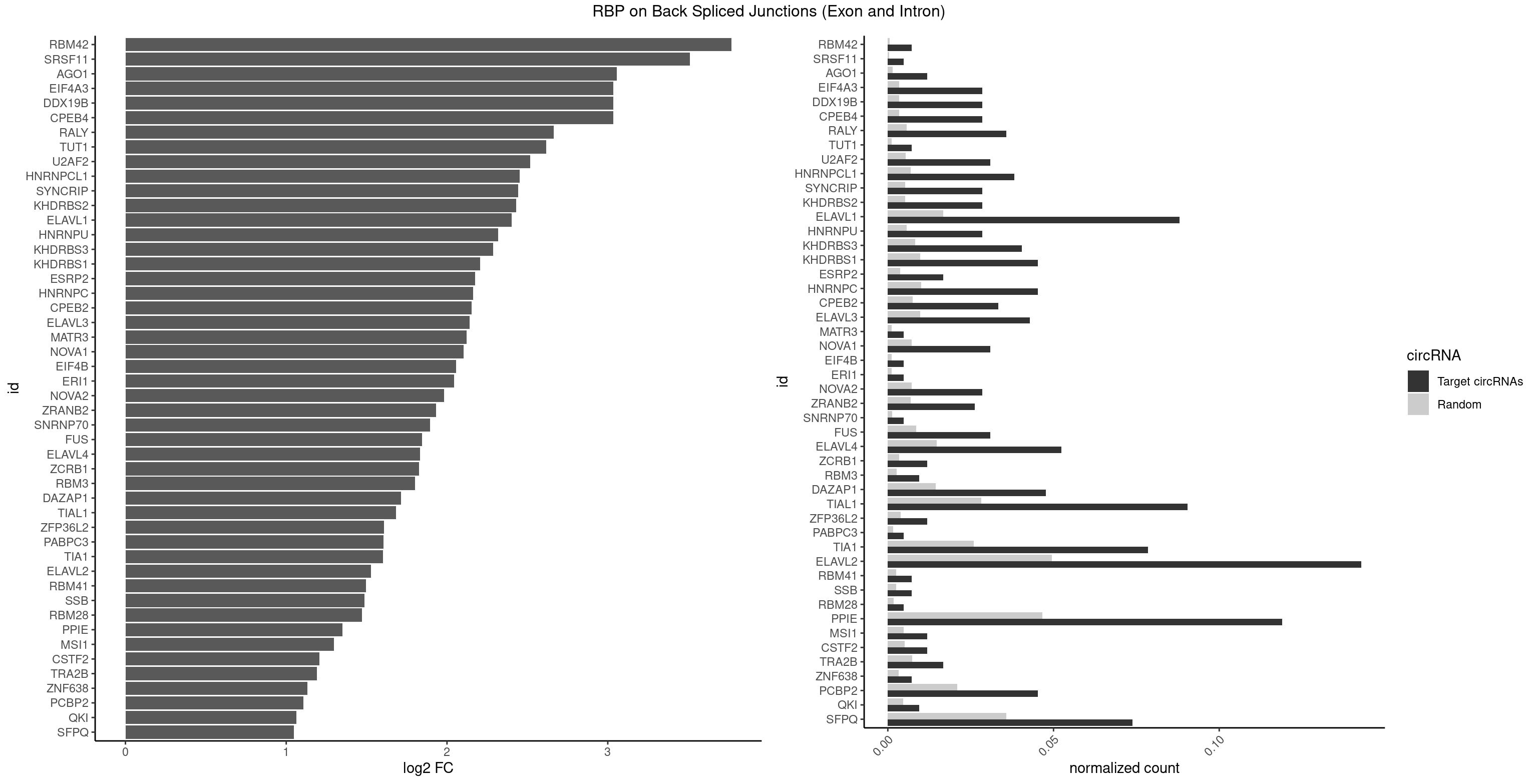
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| AGO1 | 4 | 283 | 0.011904762 | 0.0014308746 | 3.056570 | AGGUAG,GAGGUA,GGUAGU,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CPEB4 | 11 | 691 | 0.028571429 | 0.0034864974 | 3.034723 | UUUUUU | UUUUUU |
| DDX19B | 11 | 691 | 0.028571429 | 0.0034864974 | 3.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 11 | 691 | 0.028571429 | 0.0034864974 | 3.034723 | UUUUUU | UUUUUU |
| RALY | 14 | 1117 | 0.035714286 | 0.0056328094 | 2.664575 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | CGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| U2AF2 | 12 | 1071 | 0.030952381 | 0.0054010480 | 2.518739 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 | 15 | 1381 | 0.038095238 | 0.0069629182 | 2.451847 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP | 11 | 1041 | 0.028571429 | 0.0052498992 | 2.444212 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 11 | 1051 | 0.028571429 | 0.0053002821 | 2.430432 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ELAVL1 | 36 | 3309 | 0.088095238 | 0.0166767432 | 2.401226 | AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPU | 11 | 1136 | 0.028571429 | 0.0057285369 | 2.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| KHDRBS3 | 16 | 1646 | 0.040476190 | 0.0082980653 | 2.286227 | AAAUAA,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| KHDRBS1 | 18 | 1945 | 0.045238095 | 0.0098045143 | 2.206020 | AUUUAA,CUAAAA,UAAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ESRP2 | 6 | 731 | 0.016666667 | 0.0036880290 | 2.176044 | GGGAAA,GGGAAG,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPC | 18 | 2006 | 0.045238095 | 0.0101118501 | 2.161491 | AUUUUU,GGAUUC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 13 | 1487 | 0.033333333 | 0.0074969770 | 2.152585 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ELAVL3 | 17 | 1928 | 0.042857143 | 0.0097188634 | 2.140676 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| NOVA1 | 12 | 1428 | 0.030952381 | 0.0071997179 | 2.104038 | UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| NOVA2 | 11 | 1434 | 0.028571429 | 0.0072299476 | 1.982516 | UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ZRANB2 | 10 | 1360 | 0.026190476 | 0.0068571141 | 1.933369 | AGGUAG,AGGUAU,AGGUUA,GAGGUU,GGUAAA,GUAAAG,UAAAGG,UGGUAA,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| FUS | 12 | 1711 | 0.030952381 | 0.0086255542 | 1.843361 | UGGUGG,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL4 | 21 | 2916 | 0.052380952 | 0.0146966949 | 1.833551 | AUCUAA,AUUUAU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UUAUUU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | ACUUAA,AUUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| DAZAP1 | 19 | 2878 | 0.047619048 | 0.0145052398 | 1.714965 | AGGUAG,AGGUUA,AGUAAA,AGUUAA,UAGUUA,UAGUUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| TIAL1 | 37 | 5597 | 0.090476190 | 0.0282043531 | 1.681620 | AAAUUU,AAUUUU,AGUUUU,AUUUUC,AUUUUG,AUUUUU,CUUUUG,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUCAG,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| TIA1 | 32 | 5140 | 0.078571429 | 0.0259018541 | 1.600949 | AUUUUC,AUUUUG,AUUUUU,CUUUUG,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UUAUUU,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 59 | 9826 | 0.142857143 | 0.0495112858 | 1.528744 | AAUUUG,AAUUUU,AUACUU,AUUUAA,AUUUAG,AUUUAU,AUUUUC,AUUUUG,AUUUUU,CUUUCU,GAUUUA,GAUUUU,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUGUU,UAUUUA,UAUUUU,UCUUUU,UGAUUU,UUAUAU,UUAUGU,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUUAUA,UUUAUG,UUUCAU,UUUCUU,UUUGAU,UUUUAG,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PPIE | 49 | 9262 | 0.119047619 | 0.0466696896 | 1.350981 | AAAAAU,AAAAUA,AAAUAA,AAAUUA,AAAUUU,AAUAAU,AAUUUU,AUAAAU,AUAAUU,AUUAAA,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUUU,UAUAAA,UAUUUA,UAUUUU,UUAAAU,UUAUAU,UUAUUU,UUUAAA,UUUAUA,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAGG,AGGUAG,UAGGAG,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAGAAG,AAGAAU,AAUAAG,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGG,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PCBP2 | 18 | 4164 | 0.045238095 | 0.0209844821 | 1.108215 | AAAUUA,AAUUAC,ACAUUA,AUUAAA,CAUUAA,UUAGGA,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUUAU,AUCUAA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SFPQ | 30 | 7087 | 0.073809524 | 0.0357114067 | 1.047422 | GAGGUA,GUAGUU,GUGAUU,UAAUGU,UAAUUU,UAGUUU,UGAUUU,UGGUCU,UGGUGG,UGGUUG,UGGUUU,UUAAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.