circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000135049:-:9:85692688:85692813
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000135049:-:9:85692688:85692813 | ENSG00000135049 | ENST00000357081 | - | 9 | 85692689 | 85692813 | 125 | CCUUACCAAUAAUUCUAGGAUCGUAGGACUCCUGGCUCAACUGGAGAAGAUCAAUGCUGAGCCUUCAGAAUCAGACACUGCCCGAUAUGUUACAUCAAAAAUUCUUCAUCUGGCUCAGAGUCAAGCCUUACCAAUAAUUCUAGGAUCGUAGGACUCCUGGCUCAACUGGAGAAGA | circ |
| ENSG00000135049:-:9:85692688:85692813 | ENSG00000135049 | ENST00000357081 | - | 9 | 85692689 | 85692813 | 22 | UCAGAGUCAAGCCUUACCAAUA | bsj |
| ENSG00000135049:-:9:85692688:85692813 | ENSG00000135049 | ENST00000357081 | - | 9 | 85692804 | 85693013 | 210 | GUUUCUCACUUAAGUCAGUAGAAUUUAAGCUAUAGAACAAUAUUAAGCUAUAUUAAGCUAUAGAACAGAGUAAGGAAGUGCGACGUGUAAAUGCUUUCUGUUUUUAAAAGAGUAGAAUUGUUUUUUAAACAGUGAUCGUUAUAGUAUUGAAUUUACACCAUUGAUUUAGAAUGUGCCCUAACAUUCUACGUUCUUUUUAGCCUUACCAAU | ie_up |
| ENSG00000135049:-:9:85692688:85692813 | ENSG00000135049 | ENST00000357081 | - | 9 | 85692489 | 85692698 | 210 | CAGAGUCAAGGUAAACAUUGAUCUCUUCUUUGUUUUUGAAAUGCUUUUUUAAUUCUUUAAAAGUGGAUGCUAAAAAUGUUCAAACUUCUAAUAAUGGAUGAUUUUCCACAAAAAUUGAACUUUCAGGCCGGGUGUGGUGGCUCACACCUGUAAUCCCAGCACUUUGGGAGGCCAAGGUGGGCAGAUCAUGAAGUCAGUAGUUUUGAGACC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
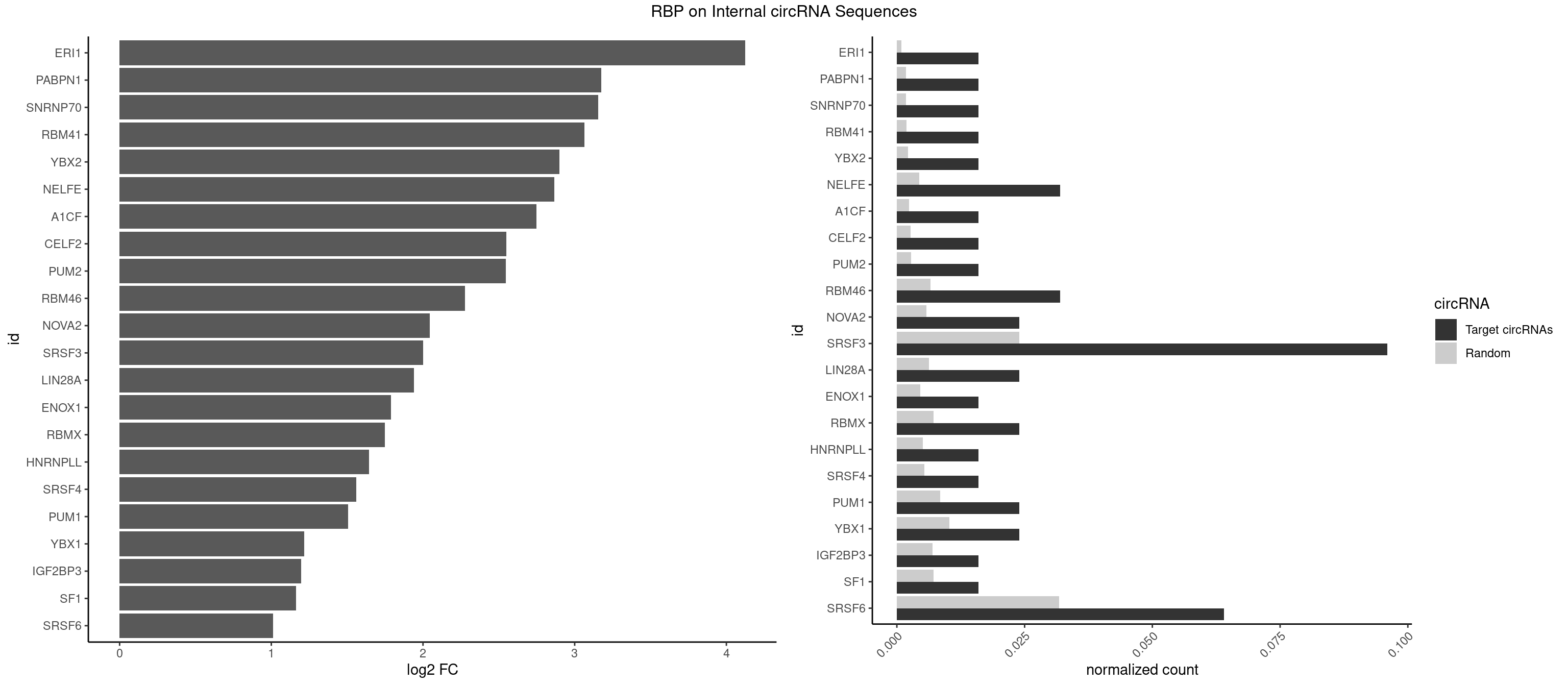
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ERI1 | 1 | 632 | 0.016 | 0.0009173461 | 4.124462 | UUCAGA | UUCAGA,UUUCAG |
| PABPN1 | 1 | 1222 | 0.016 | 0.0017723764 | 3.174315 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 1 | 1237 | 0.016 | 0.0017941145 | 3.156728 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM41 | 1 | 1318 | 0.016 | 0.0019115000 | 3.065295 | UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX2 | 1 | 1480 | 0.016 | 0.0021462711 | 2.898168 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NELFE | 3 | 3028 | 0.032 | 0.0043896388 | 2.865898 | CUGGCU,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| A1CF | 1 | 1642 | 0.016 | 0.0023810421 | 2.748407 | AUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CELF2 | 1 | 1886 | 0.016 | 0.0027346479 | 2.548645 | UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM2 | 1 | 1890 | 0.016 | 0.0027404447 | 2.545590 | UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM46 | 3 | 4554 | 0.032 | 0.0066011240 | 2.277288 | AUCAAA,AUCAAU,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| NOVA2 | 2 | 4013 | 0.024 | 0.0058171047 | 2.044661 | AGAUCA,GAGUCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SRSF3 | 11 | 16536 | 0.096 | 0.0239654858 | 2.002076 | ACAUCA,CAUCAA,CUUCAG,CUUCAU,GAUCAA,UACAUC,UCAGAG,UCAUCU,UCUUCA,UUCAUC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| LIN28A | 2 | 4315 | 0.024 | 0.0062547643 | 1.940007 | GGAGAA,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ENOX1 | 1 | 3195 | 0.016 | 0.0046316558 | 1.788472 | CAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBMX | 2 | 4925 | 0.024 | 0.0071387787 | 1.749285 | AUCAAA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPLL | 1 | 3534 | 0.016 | 0.0051229360 | 1.643029 | CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SRSF4 | 1 | 3740 | 0.016 | 0.0054214720 | 1.561315 | AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PUM1 | 2 | 5823 | 0.024 | 0.0084401638 | 1.507691 | CAGAAU,UACAUC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| YBX1 | 2 | 7119 | 0.024 | 0.0103183321 | 1.217825 | ACAUCA,CAUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| IGF2BP3 | 1 | 4815 | 0.016 | 0.0069793662 | 1.196904 | AAAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SF1 | 1 | 4931 | 0.016 | 0.0071474739 | 1.162567 | UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF6 | 7 | 21880 | 0.064 | 0.0317100317 | 1.013133 | AGAAGA,AUCGUA,CCUGGC,CUUCAG,GAUCAA,UCCUGG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
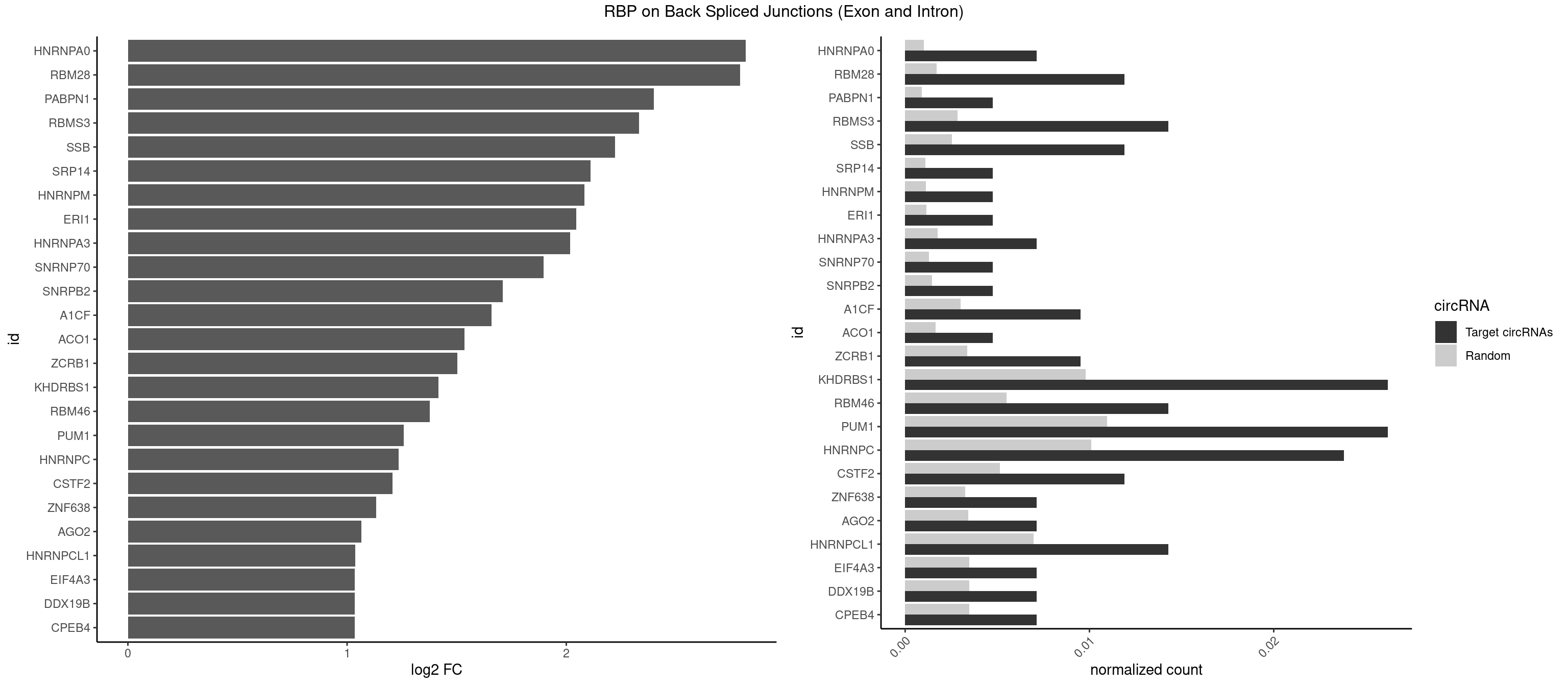
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM28 | 4 | 340 | 0.011904762 | 0.0017180572 | 2.792689 | AGUAGA,AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBMS3 | 5 | 562 | 0.014285714 | 0.0028365578 | 2.332360 | CUAUAG,CUAUAU,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | UCAGUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | ACUUAA,AUUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| KHDRBS1 | 10 | 1945 | 0.026190476 | 0.0098045143 | 1.417524 | AUUUAA,AUUUAC,CUAAAA,UAAAAA,UAAAAG,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AUCAUG,AUGAAG,AUGAUU,GAUCAU,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AAUAUU,AAUGUU,AAUUGU,AGAAUU,GAAUUG,GUAAAU,UGUAAA,UGUAAU,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.0101118501 | 1.235492 | CUUUUU,UUUUUA,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | CGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.