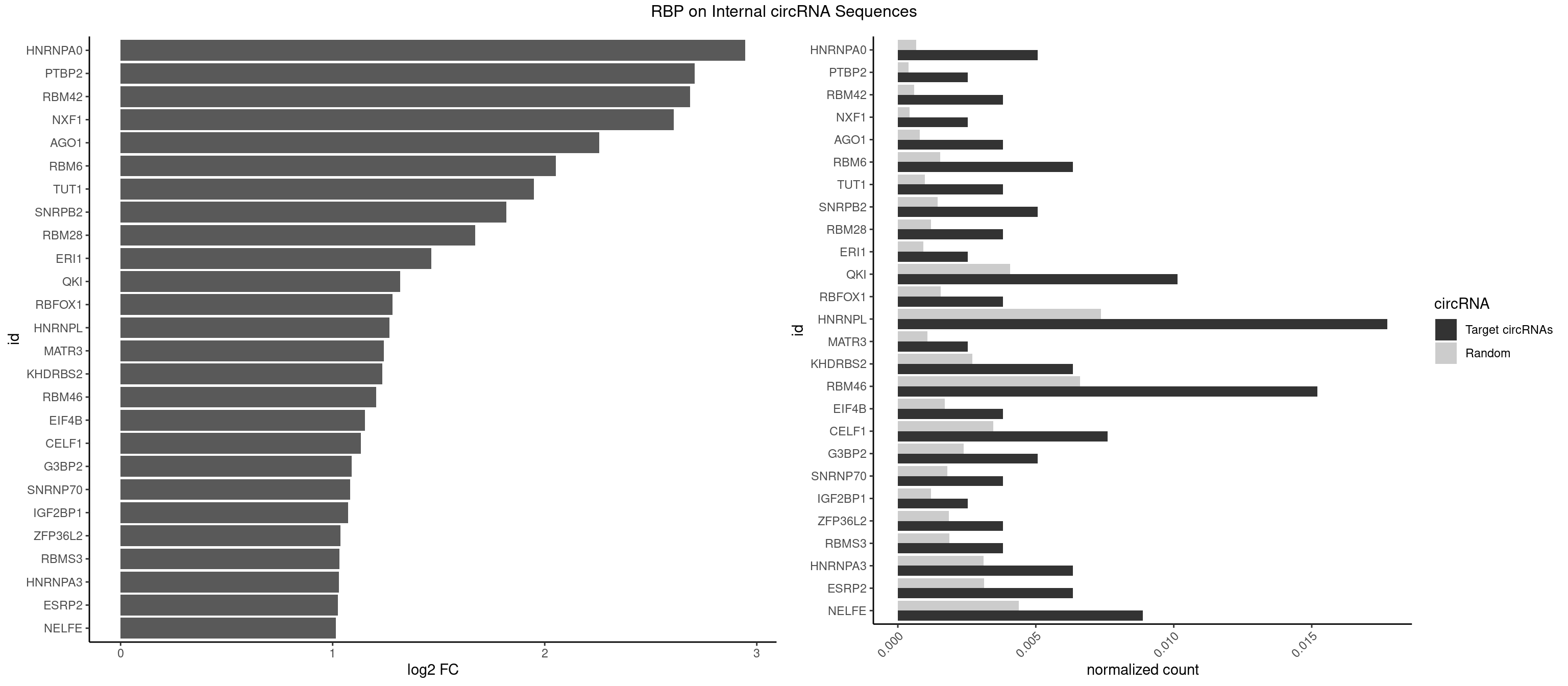
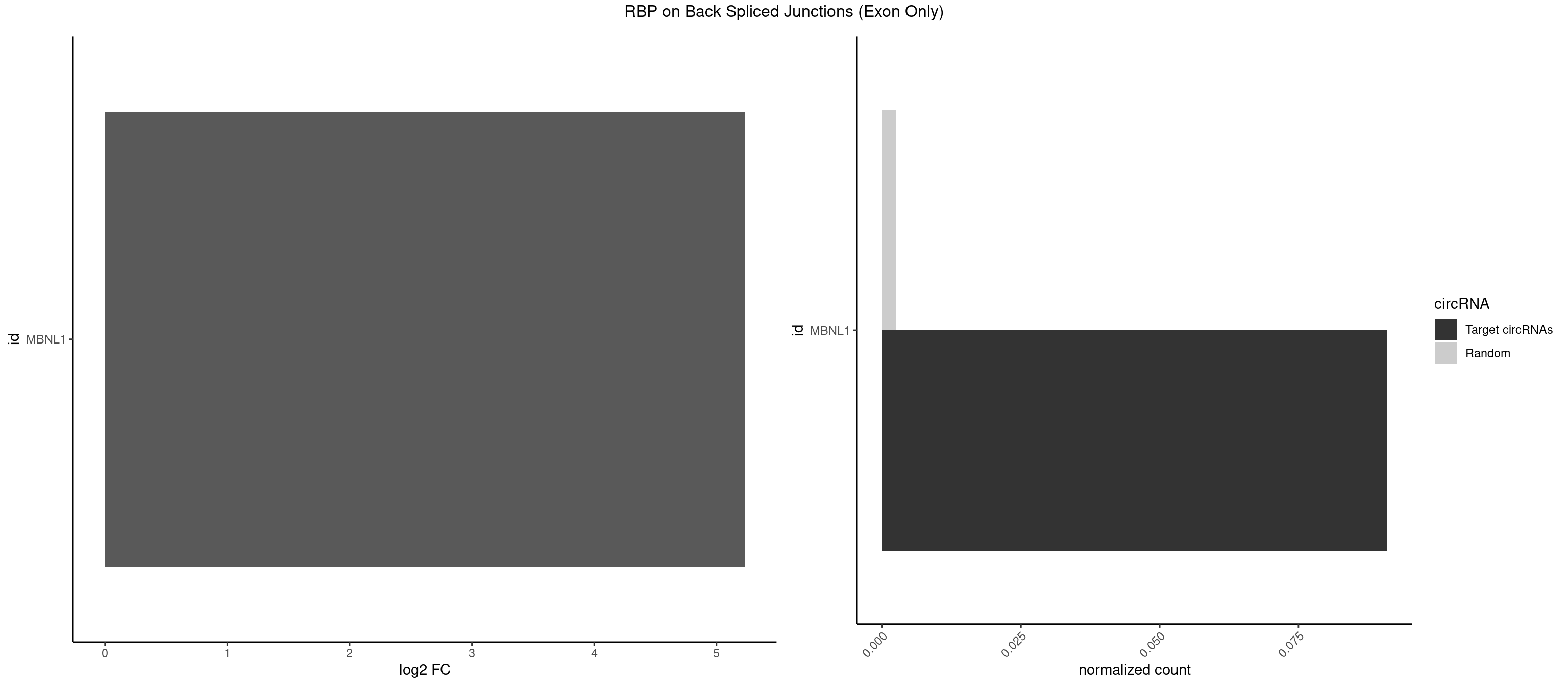
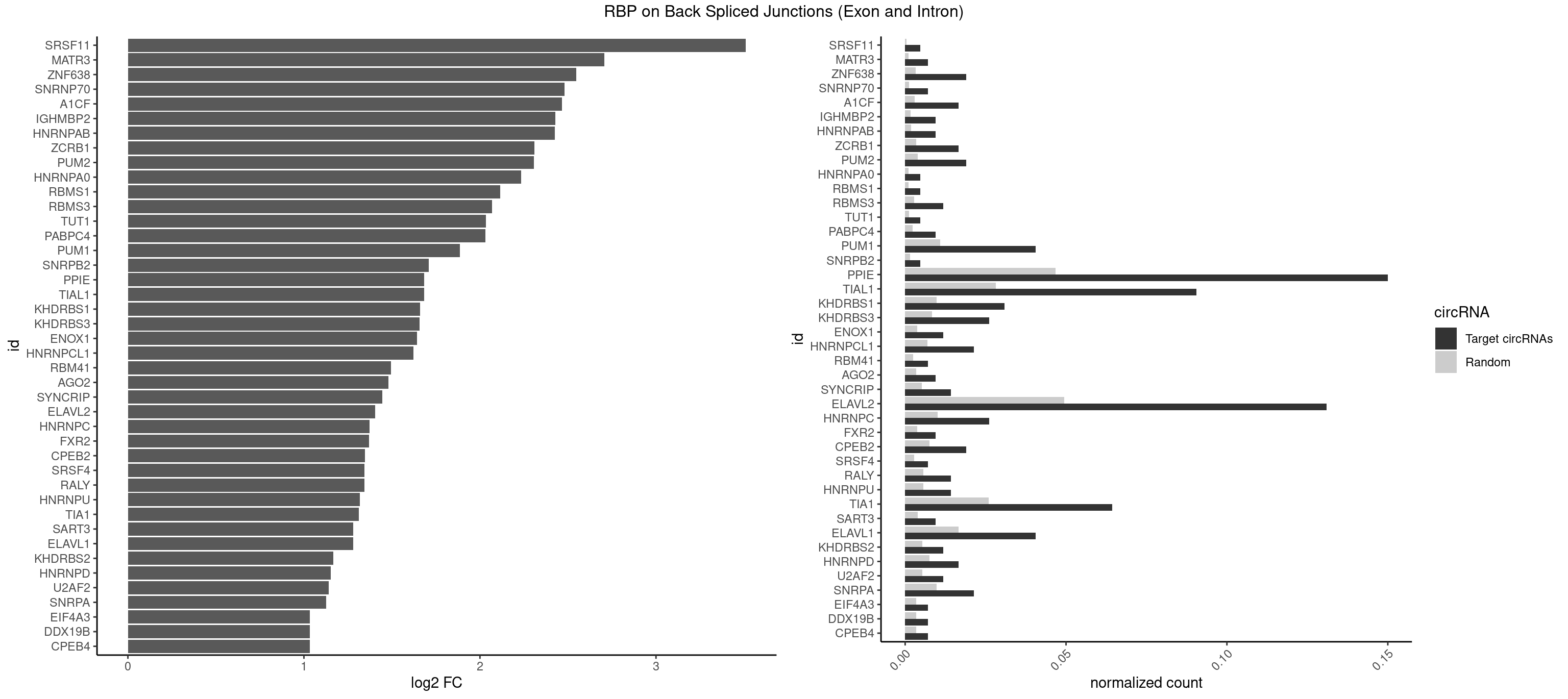
| SRSF11 |
1 |
82 |
0.004761905 |
0.0004181782 |
3.509349 |
AAGAAG |
AAGAAG |
| MATR3 |
2 |
216 |
0.007142857 |
0.0010933091 |
2.707800 |
AUCUUA,CAUCUU |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZNF638 |
7 |
646 |
0.019047619 |
0.0032597743 |
2.546767 |
GGUUCU,GUUCUU,GUUGGU,UGUUCU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SNRNP70 |
2 |
253 |
0.007142857 |
0.0012797259 |
2.480666 |
AUUCAA,GUUCAA |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| A1CF |
6 |
598 |
0.016666667 |
0.0030179363 |
2.465331 |
AUAAUU,CAGUAU,UAAUUA,UAAUUG,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| IGHMBP2 |
3 |
350 |
0.009523810 |
0.0017684401 |
2.429061 |
AAAAAA |
AAAAAA |
| HNRNPAB |
3 |
351 |
0.009523810 |
0.0017734784 |
2.424957 |
AGACAA,CAAAGA,GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZCRB1 |
6 |
666 |
0.016666667 |
0.0033605401 |
2.310201 |
AAUUAA,AUUUAA,GAAUUA,GUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PUM2 |
7 |
764 |
0.019047619 |
0.0038542926 |
2.305073 |
GUAAAU,UAAAUA,UAGAUA,UGUAAA,UGUACA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| RBMS1 |
1 |
217 |
0.004761905 |
0.0010983474 |
2.116204 |
AUAUAG |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 |
4 |
562 |
0.011904762 |
0.0028365578 |
2.069326 |
AAUAUA,AUAUAG,UAUAGA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TUT1 |
1 |
230 |
0.004761905 |
0.0011638452 |
2.032640 |
AAAUAC |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC4 |
3 |
462 |
0.009523810 |
0.0023327287 |
2.029520 |
AAAAAA |
AAAAAA,AAAAAG |
| PUM1 |
16 |
2172 |
0.040476190 |
0.0109482064 |
1.886379 |
AAUUGU,AGAAUU,AGAUAA,AUUGUA,GUAAAU,UAAAUA,UAAUGU,UAGAUA,UGUAAA,UGUAAU,UGUACA,UUAAUG,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| SNRPB2 |
1 |
288 |
0.004761905 |
0.0014560661 |
1.709463 |
AUUGCA |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PPIE |
62 |
9262 |
0.150000000 |
0.0466696896 |
1.684405 |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAUU,AUUAAA,AUUAAU,AUUAUA,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUUA,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUU,UUUAAA,UUUAAU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| TIAL1 |
37 |
5597 |
0.090476190 |
0.0282043531 |
1.681620 |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUU,GUUUUC,UAAAUU,UAUUUU,UUAAAU,UUUAAA,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 |
12 |
1945 |
0.030952381 |
0.0098045143 |
1.658532 |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAU,UAAAAA,UUAAAA,UUAAAU,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| KHDRBS3 |
10 |
1646 |
0.026190476 |
0.0082980653 |
1.658195 |
AAAUAA,AGAUAA,AUAAAA,AUUAAA,GAUAAA,UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ENOX1 |
4 |
756 |
0.011904762 |
0.0038139863 |
1.642167 |
AAUACA,AGGACA,UAGACA,UGUACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPCL1 |
8 |
1381 |
0.021428571 |
0.0069629182 |
1.621772 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RBM41 |
2 |
502 |
0.007142857 |
0.0025342604 |
1.494937 |
AUACAU,UACAUG |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO2 |
3 |
677 |
0.009523810 |
0.0034159613 |
1.479247 |
AAAAAA |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SYNCRIP |
5 |
1041 |
0.014285714 |
0.0052498992 |
1.444212 |
AAAAAA,UUUUUU |
AAAAAA,UUUUUU |
| ELAVL2 |
54 |
9826 |
0.130952381 |
0.0495112858 |
1.403213 |
AAUUUA,AAUUUU,AUUUAA,AUUUAC,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUAA,CUUUUA,CUUUUU,GAUUUU,GUUUUA,GUUUUC,UAAGUU,UAUUUA,UAUUUU,UCAUUU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUAGUU,UUAUGU,UUCAUU,UUCUUU,UUGUAU,UUUAAG,UUUAAU,UUUAUG,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPC |
10 |
2006 |
0.026190476 |
0.0101118501 |
1.372995 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FXR2 |
3 |
730 |
0.009523810 |
0.0036829907 |
1.370661 |
AGACAA,GACAAA,GGACAA |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CPEB2 |
7 |
1487 |
0.019047619 |
0.0074969770 |
1.345230 |
AUUUUU,CAUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY |
5 |
1117 |
0.014285714 |
0.0056328094 |
1.342647 |
UUUUUC,UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| SRSF4 |
2 |
558 |
0.007142857 |
0.0028164047 |
1.342647 |
AAGAAG,GAAGAA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPU |
5 |
1136 |
0.014285714 |
0.0057285369 |
1.318335 |
AAAAAA,UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| TIA1 |
26 |
5140 |
0.064285714 |
0.0259018541 |
1.311443 |
AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUU,GUUUUA,GUUUUC,GUUUUG,UAUUUU,UCUCCU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SART3 |
3 |
777 |
0.009523810 |
0.0039197904 |
1.280762 |
AAAAAA |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ELAVL1 |
16 |
3309 |
0.040476190 |
0.0166767432 |
1.279236 |
AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGUUUU,UUAGUU,UUGUUU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| KHDRBS2 |
4 |
1051 |
0.011904762 |
0.0053002821 |
1.167398 |
AUAAAA,GAUAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPD |
6 |
1488 |
0.016666667 |
0.0075020153 |
1.151615 |
AAAAAA,AAUUUA,AUUUAU,UAUUUA |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| U2AF2 |
4 |
1071 |
0.011904762 |
0.0054010480 |
1.140228 |
UUUUCC,UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| SNRPA |
8 |
1947 |
0.021428571 |
0.0098145909 |
1.126536 |
AUUGCA,CCUGCU,CUGCUA,GUAUGC,UCCUGC,UUCCUG,UUUCCU |
ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| CPEB4 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| DDX19B |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| EIF4A3 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |