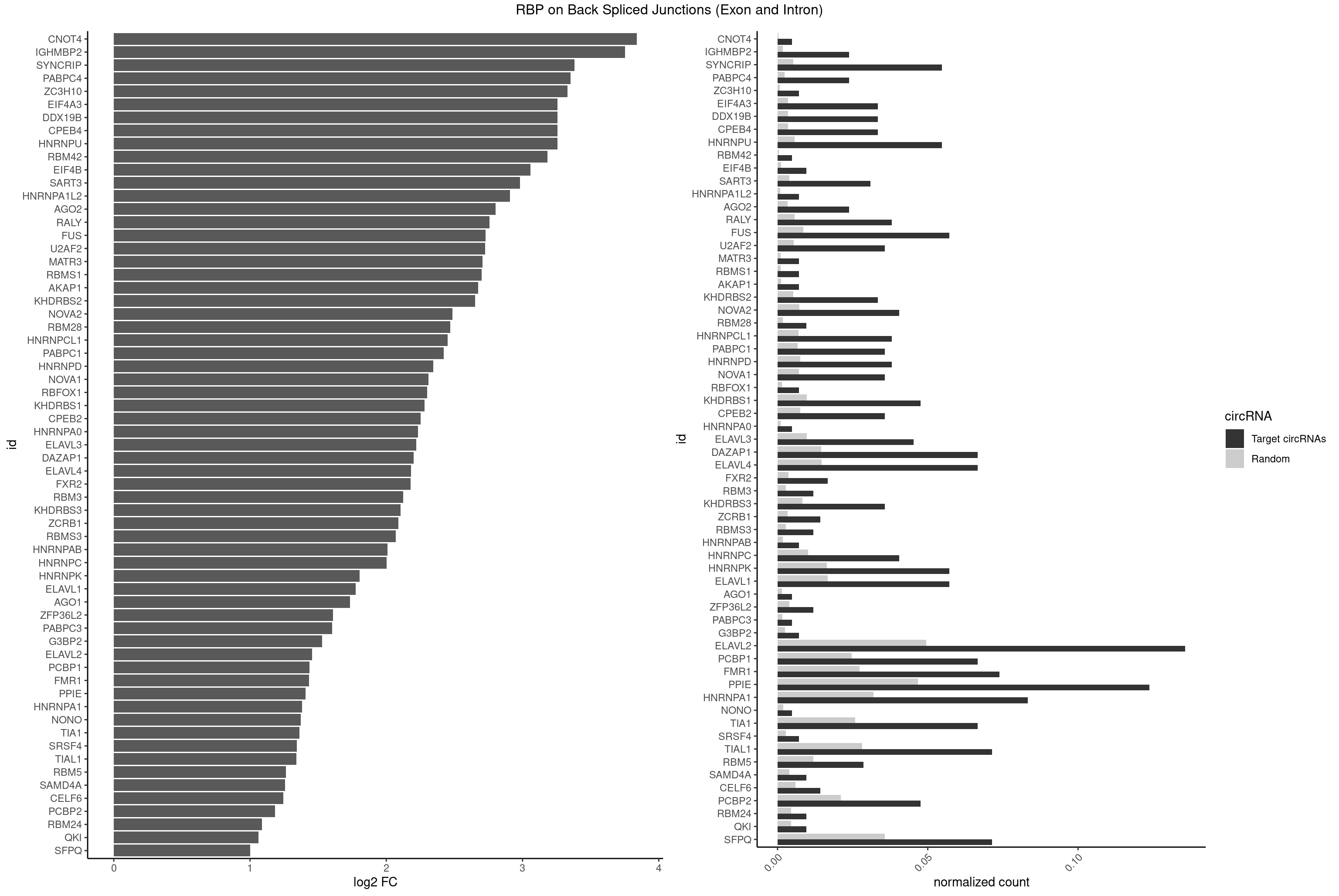
| CNOT4 |
1 |
65 |
0.004761905 |
0.0003325272 |
3.839994 |
GACAGA |
GACAGA |
| IGHMBP2 |
9 |
350 |
0.023809524 |
0.0017684401 |
3.750989 |
AAAAAA |
AAAAAA |
| SYNCRIP |
22 |
1041 |
0.054761905 |
0.0052498992 |
3.382811 |
AAAAAA,UUUUUU |
AAAAAA,UUUUUU |
| PABPC4 |
9 |
462 |
0.023809524 |
0.0023327287 |
3.351448 |
AAAAAA |
AAAAAA,AAAAAG |
| ZC3H10 |
2 |
140 |
0.007142857 |
0.0007103990 |
3.329800 |
CGAGCG,GAGCGA |
CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| CPEB4 |
13 |
691 |
0.033333333 |
0.0034864974 |
3.257115 |
UUUUUU |
UUUUUU |
| DDX19B |
13 |
691 |
0.033333333 |
0.0034864974 |
3.257115 |
UUUUUU |
UUUUUU |
| EIF4A3 |
13 |
691 |
0.033333333 |
0.0034864974 |
3.257115 |
UUUUUU |
UUUUUU |
| HNRNPU |
22 |
1136 |
0.054761905 |
0.0057285369 |
3.256934 |
AAAAAA,UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| RBM42 |
1 |
103 |
0.004761905 |
0.0005239823 |
3.183949 |
AACUAA |
AACUAA,AACUAC,ACUAAG,ACUACG |
| EIF4B |
3 |
226 |
0.009523810 |
0.0011436921 |
3.057840 |
CUUGGA,GUUGGA,UUGGAA |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SART3 |
12 |
777 |
0.030952381 |
0.0039197904 |
2.981202 |
AAAAAA,AAAAAC,AGAAAA,GAAAAA |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPA1L2 |
2 |
188 |
0.007142857 |
0.0009522370 |
2.907109 |
AUAGGG,UAGGGU |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| AGO2 |
9 |
677 |
0.023809524 |
0.0034159613 |
2.801175 |
AAAAAA |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| RALY |
15 |
1117 |
0.038095238 |
0.0056328094 |
2.757684 |
UUUUUC,UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| FUS |
23 |
1711 |
0.057142857 |
0.0086255542 |
2.727884 |
AAAAAA,GGGUGA,UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| U2AF2 |
14 |
1071 |
0.035714286 |
0.0054010480 |
2.725190 |
UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| MATR3 |
2 |
216 |
0.007142857 |
0.0010933091 |
2.707800 |
AAUCUU,AUCUUA |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 |
2 |
217 |
0.007142857 |
0.0010983474 |
2.701167 |
AUAUAC,UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 |
2 |
221 |
0.007142857 |
0.0011185006 |
2.674935 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| KHDRBS2 |
13 |
1051 |
0.033333333 |
0.0053002821 |
2.652825 |
UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| NOVA2 |
16 |
1434 |
0.040476190 |
0.0072299476 |
2.485016 |
ACCACC,GAGACA,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RBM28 |
3 |
340 |
0.009523810 |
0.0017180572 |
2.470761 |
AGUAGU,GUGUAG,UGUAGU |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPCL1 |
15 |
1381 |
0.038095238 |
0.0069629182 |
2.451847 |
CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PABPC1 |
14 |
1321 |
0.035714286 |
0.0066606207 |
2.422773 |
AAAAAA,AAAAAC,ACAAAC,ACUAAU,AGAAAA,GAAAAA |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPD |
15 |
1488 |
0.038095238 |
0.0075020153 |
2.344261 |
AAAAAA,AAUUUA,AUUUAU,UAUUUA,UUAGAG,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| NOVA1 |
14 |
1428 |
0.035714286 |
0.0071997179 |
2.310489 |
ACCACC,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RBFOX1 |
2 |
287 |
0.007142857 |
0.0014510278 |
2.299426 |
AGCAUG,GCAUGC |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| KHDRBS1 |
19 |
1945 |
0.047619048 |
0.0098045143 |
2.280021 |
AUUUAA,CAAAAU,UAAAAA,UAAAAG,UUAAAA,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| CPEB2 |
14 |
1487 |
0.035714286 |
0.0074969770 |
2.252120 |
CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| ELAVL3 |
18 |
1928 |
0.045238095 |
0.0097188634 |
2.218679 |
AUUUAU,AUUUUA,UAUUUA,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| DAZAP1 |
27 |
2878 |
0.066666667 |
0.0145052398 |
2.200391 |
AAAAAA,AAUUUA,AGGUUG,AGUUUA,UAGGAA,UAGUUU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL4 |
27 |
2916 |
0.066666667 |
0.0146966949 |
2.181474 |
AAAAAA,AUUUAU,UAUUUA,UUAUUU,UUUAUU,UUUUAU,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| FXR2 |
6 |
730 |
0.016666667 |
0.0036829907 |
2.178016 |
AGACAA,AGACAG,GACAAA,GACAGA,GACGAG,UGACGA |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RBM3 |
4 |
541 |
0.011904762 |
0.0027307537 |
2.124168 |
AAAACG,AAAACU,AAACUA,GAAACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| KHDRBS3 |
14 |
1646 |
0.035714286 |
0.0082980653 |
2.105654 |
UUAAAC,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ZCRB1 |
5 |
666 |
0.014285714 |
0.0033605401 |
2.087808 |
ACUUAA,AUUUAA,GCUUAA,GGUUUA,GUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBMS3 |
4 |
562 |
0.011904762 |
0.0028365578 |
2.069326 |
AAUAUA,AUAUAU,UAUAUA,UAUAUC |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPAB |
2 |
351 |
0.007142857 |
0.0017734784 |
2.009919 |
AGACAA,GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPC |
16 |
2006 |
0.040476190 |
0.0101118501 |
2.001027 |
CUUUUU,UUUUUC,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPK |
23 |
3244 |
0.057142857 |
0.0163492543 |
1.805348 |
AAAAAA,GCCCAG,UUUUUU |
AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| ELAVL1 |
23 |
3309 |
0.057142857 |
0.0166767432 |
1.776736 |
AUUUAU,UAGUUU,UAUUUA,UGAUUU,UUAGUU,UUAUUU,UUGAUU,UUUAUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| AGO1 |
1 |
283 |
0.004761905 |
0.0014308746 |
1.734641 |
GUAGUA |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ZFP36L2 |
4 |
774 |
0.011904762 |
0.0039046755 |
1.608264 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PABPC3 |
1 |
310 |
0.004761905 |
0.0015669085 |
1.603618 |
AAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| G3BP2 |
2 |
490 |
0.007142857 |
0.0024738009 |
1.529772 |
AGGAUU,GGAUUG |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ELAVL2 |
56 |
9826 |
0.135714286 |
0.0495112858 |
1.454743 |
AAUUUA,AAUUUG,AAUUUU,AUUUAA,AUUUAU,AUUUUA,AUUUUC,CUUUAA,CUUUUA,CUUUUU,GAUUUU,GUUUUU,UAAUUU,UAUAUA,UAUGUU,UAUUUA,UCUUAU,UCUUUU,UGAUUU,UUAAGU,UUAGUU,UUAUGU,UUAUUU,UUCUUU,UUGAUU,UUUAAG,UUUAUA,UUUAUG,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUC,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PCBP1 |
27 |
4891 |
0.066666667 |
0.0246473196 |
1.435535 |
AAAAAA,AAAUUA,CCUCCC,CUUAAA,UUAGAG,UUUUUU |
AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| FMR1 |
30 |
5430 |
0.073809524 |
0.0273629585 |
1.431583 |
AAAAAA,AGCGAA,AGCUGG,AGGAAU,AGGAUU,CAUAGG,GAGCGA,GAGGAU,UGGAGU,UUUUUU |
AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| PPIE |
51 |
9262 |
0.123809524 |
0.0466696896 |
1.407565 |
AAAAAA,AAAAAU,AAAAUU,AAAUUA,AAAUUU,AAUAUA,AAUUUA,AAUUUU,AUAUAU,AUUUAA,AUUUAU,AUUUUA,UAAAAA,UAAUUU,UAUAUA,UAUUUA,UUAAAA,UUAUAA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPA1 |
34 |
6340 |
0.083333333 |
0.0319478033 |
1.383177 |
AAAAAA,AAUUUA,AGAGGA,AGGGUU,AUAGGG,AUUUAA,AUUUAU,UAGGAA,UAGGGU,UUAUUU,UUUAUA,UUUAUU,UUUUUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| NONO |
1 |
364 |
0.004761905 |
0.0018389762 |
1.372636 |
AGAGGA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TIA1 |
27 |
5140 |
0.066666667 |
0.0259018541 |
1.363910 |
AUUUUC,CUUUUA,CUUUUU,GUUUUG,GUUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUGU,UUUUUC,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF4 |
2 |
558 |
0.007142857 |
0.0028164047 |
1.342647 |
GAAGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TIAL1 |
29 |
5597 |
0.071428571 |
0.0282043531 |
1.340583 |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,CUUUUA,CUUUUU,GUUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUC,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM5 |
11 |
2359 |
0.028571429 |
0.0118903668 |
1.264780 |
AAAAAA,AGGGUA,GAGGGU |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| SAMD4A |
3 |
790 |
0.009523810 |
0.0039852882 |
1.256855 |
CUGGAA,CUGGUA,GCUGGU |
CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CELF6 |
5 |
1196 |
0.014285714 |
0.0060308343 |
1.244144 |
GUGAGG,UGUGAG,UGUGAU |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| PCBP2 |
19 |
4164 |
0.047619048 |
0.0209844821 |
1.182216 |
AAAUUA,CCUCCC,CUCCCA,CUUAAA,UUAGAG,UUUUUU |
AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| RBM24 |
3 |
888 |
0.009523810 |
0.0044790407 |
1.088349 |
AGUGUG,GAGUGU,GUGUGA |
AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| QKI |
3 |
904 |
0.009523810 |
0.0045596534 |
1.062615 |
ACUAAU,CUACUC,UCUACU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SFPQ |
29 |
7087 |
0.071428571 |
0.0357114067 |
1.000116 |
AGAGGA,GAAGAA,GGAAGA,GGUAAG,GUAAUU,GUAGUG,GUGAUU,UAAUUU,UAGUGU,UAGUUU,UGAUUU,UGGAAG,UGGAGA,UUAGUU,UUGAUU,UUUUUU |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |