circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000144749:-:3:66405197:66462509
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000144749:-:3:66405197:66462509 | ENSG00000144749 | ENST00000273261 | - | 3 | 66405198 | 66462509 | 942 | AAACCUGAGUUACAACAAACUCUCUGAGAUUGACCCUGCUGGUUUUGAGGACUUGCCGAACCUACAGGAAGUGUACCUCAAUAAUAAUGAGUUGACAGCGGUACCAUCCCUGGGCGCUGCUUCAUCACAUGUCGUCUCUCUCUUUCUGCAGCACAACAAGAUUCGCAGCGUGGAGGGGAGCCAGCUGAAGGCCUACCUUUCCUUAGAAGUGUUAGAUCUGAGUUUGAACAACAUCACGGAAGUGCGGAACACCUGCUUUCCACACGGACCGCCUAUAAAGGAGCUCAACCUGGCAGGCAAUCGGAUUGGCACCCUGGAGUUGGGAGCAUUUGAUGGUCUGUCACGGUCGCUGCUAACUCUUCGCCUGAGCAAAAACAGGAUCACCCAGCUUCCUGUAAGAGCAUUCAAGCUACCCAGGCUGACACAACUGGACCUCAAUCGGAACAGGAUUCGGCUGAUAGAGGGCCUCACCUUCCAGGGGCUCAACAGCUUGGAGGUGCUGAAGCUUCAGCGAAACAACAUCAGCAAACUGACAGAUGGGGCCUUCUGGGGACUGUCCAAGAUGCAUGUGCUGCACCUGGAGUACAACAGCCUGGUAGAAGUGAACAGCGGCUCGCUCUACGGCCUCACGGCCCUGCAUCAGCUCCACCUCAGCAACAAUUCCAUCGCUCGCAUUCACCGCAAGGGCUGGAGCUUCUGCCAGAAGCUGCAUGAGUUGGUCCUGUCCUUCAACAACCUGACACGGCUGGACGAGGAGAGCCUGGCCGAGCUGAGCAGCCUGAGUGUCCUGCGUCUCAGCCACAAUUCCAUCAGCCACAUUGCGGAGGGUGCCUUCAAGGGACUCAGGAGCCUGCGAGUCUUGGAUCUGGACCAUAACGAGAUUUCGGGCACAAUAGAGGACACGAGCGGCGCCUUCUCAGGGCUCGACAGCCUCAGCAAGCUAAACCUGAGUUACAACAAACUCUCUGAGAUUGACCCUGCUGGUUUUGAGG | circ |
| ENSG00000144749:-:3:66405197:66462509 | ENSG00000144749 | ENST00000273261 | - | 3 | 66405198 | 66462509 | 22 | CUCAGCAAGCUAAACCUGAGUU | bsj |
| ENSG00000144749:-:3:66405197:66462509 | ENSG00000144749 | ENST00000273261 | - | 3 | 66462500 | 66462709 | 210 | CUUGGAGUGCCUUAUUUAUGGAUGUAGGUGGGGAGGGGGUAUGUCUGUAAAUCUCAGUAUCAAUACGGUUACUUAAUACAUGAUCAUGGGAUGGGGGCUUGGAUUUGAGGGUGGGGGAGCCAGGAGAAGAGAAGAUUUCUGAAUUCUGGGUAUGAUGCCAGGUUGUUGAUUCCGUUUUUUUCUCUCUCUGUCUUUACCAGAAACCUGAGU | ie_up |
| ENSG00000144749:-:3:66405197:66462509 | ENSG00000144749 | ENST00000273261 | - | 3 | 66404998 | 66405207 | 210 | UCAGCAAGCUGUGAGUAUCCGCCGCACGGAGCUCCGCCGGCAAAUGCGCGGGACACACUUGGGUGGGAGAUGUUUCUGUGGCGCCCCCCGCGCCUCUCUGCUCUGGGCCCAGGCGGAAGAGGAGCUGGCUUUUGUUUGGGGCAGAGCUUGUUUUGUUUGUGGAGAAGAGUUUGAAGAAGAGGGUGGGGGACACCCCACUCCUUGAUGGCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
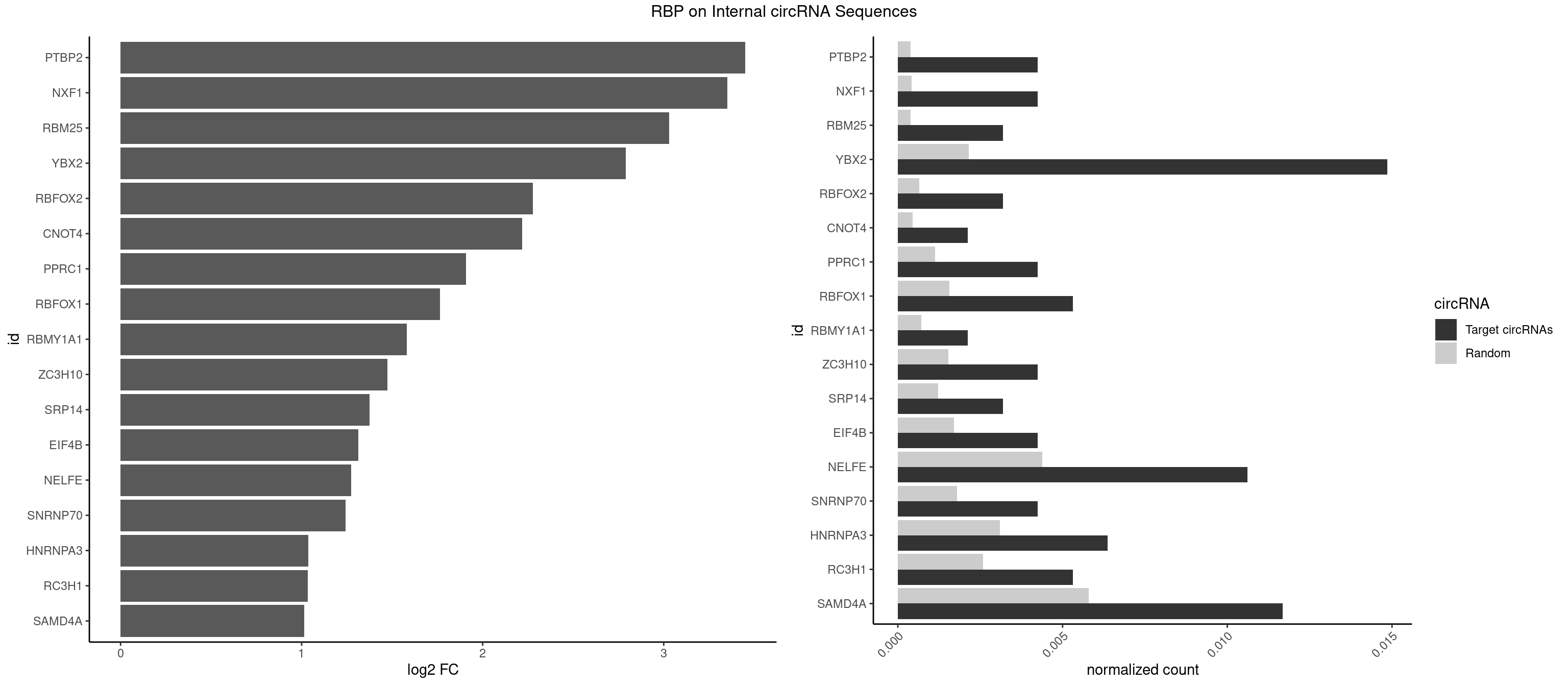
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 3 | 267 | 0.004246285 | 0.0003883867 | 3.450635 | CUCUCU | CUCUCU |
| NXF1 | 3 | 286 | 0.004246285 | 0.0004159215 | 3.351818 | AACCUG | AACCUG |
| RBM25 | 2 | 268 | 0.003184713 | 0.0003898359 | 3.030225 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| YBX2 | 13 | 1480 | 0.014861996 | 0.0021462711 | 2.791724 | AACAAC,AACAUC,ACAACA,ACAACU,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBFOX2 | 2 | 452 | 0.003184713 | 0.0006564894 | 2.278320 | UGCAUG | UGACUG,UGCAUG |
| CNOT4 | 1 | 314 | 0.002123142 | 0.0004564992 | 2.217517 | GACAGA | GACAGA |
| PPRC1 | 3 | 780 | 0.004246285 | 0.0011318283 | 1.907546 | CGGCGC,GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| RBFOX1 | 4 | 1077 | 0.005307856 | 0.0015622419 | 1.764511 | GCAUGA,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBMY1A1 | 1 | 489 | 0.002123142 | 0.0007101099 | 1.580087 | ACAAGA | ACAAGA,CAAGAC |
| ZC3H10 | 3 | 1053 | 0.004246285 | 0.0015274610 | 1.475066 | CAGCGA,CGAGCG,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SRP14 | 2 | 847 | 0.003184713 | 0.0012289250 | 1.373767 | CCUGUA,CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 3 | 1179 | 0.004246285 | 0.0017100607 | 1.312154 | CUUGGA,UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| NELFE | 9 | 3028 | 0.010615711 | 0.0043896388 | 1.274027 | CUCUCU,CUGGUU,GCUAAC,GUCUCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SNRNP70 | 3 | 1237 | 0.004246285 | 0.0017941145 | 1.242929 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPA3 | 5 | 2140 | 0.006369427 | 0.0031027457 | 1.037618 | AAGGAG,AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RC3H1 | 4 | 1786 | 0.005307856 | 0.0025897275 | 1.035329 | CCUUCU,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SAMD4A | 10 | 3992 | 0.011677282 | 0.0057866714 | 1.012899 | CGGGCA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
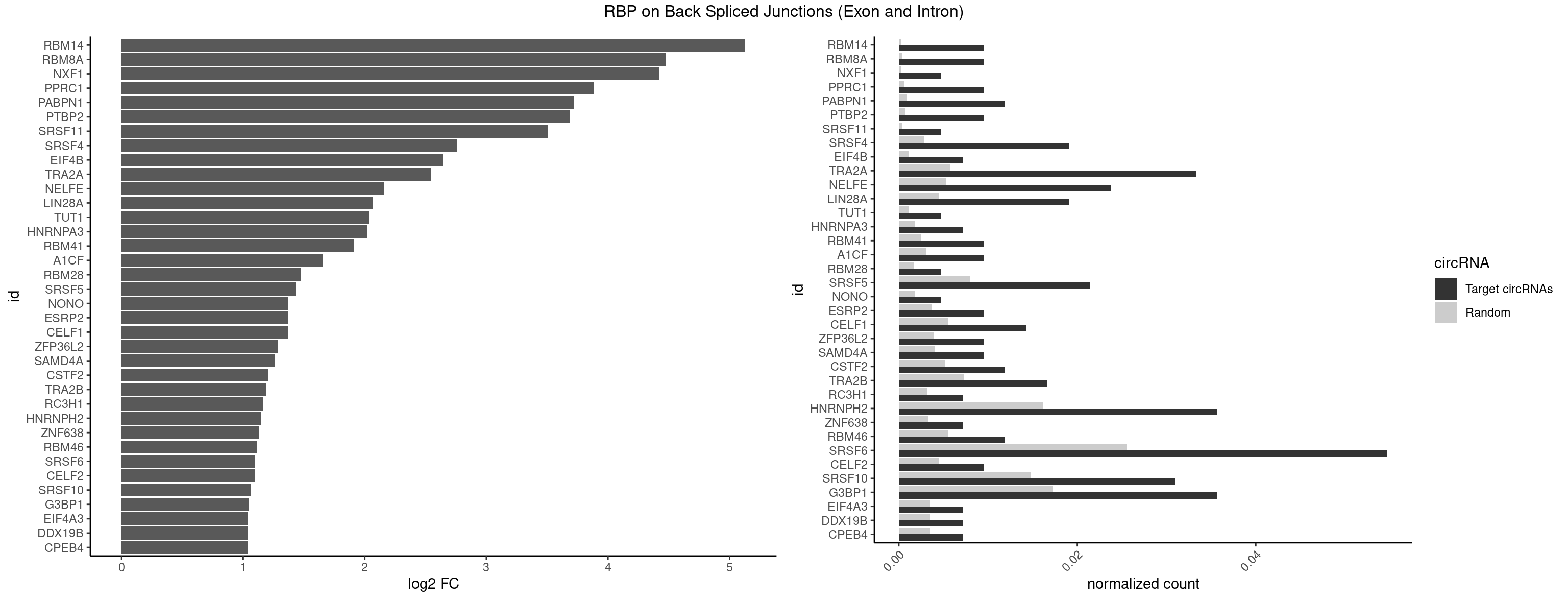
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM14 | 3 | 53 | 0.009523810 | 0.0002720677 | 5.129501 | CGCGCC,CGCGGG,GCGCGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM8A | 3 | 84 | 0.009523810 | 0.0004282547 | 4.474998 | AUGCGC,CGCGCC,UGCGCG | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| PPRC1 | 3 | 127 | 0.009523810 | 0.0006449012 | 3.884389 | CCGCGC,CGCGCC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| PABPN1 | 4 | 178 | 0.011904762 | 0.0009018541 | 3.722501 | AGAAGA | AAAAGA,AGAAGA |
| PTBP2 | 3 | 146 | 0.009523810 | 0.0007406288 | 3.684716 | CUCUCU | CUCUCU |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| SRSF4 | 7 | 558 | 0.019047619 | 0.0028164047 | 2.757684 | AAGAAG,AGAAGA,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TRA2A | 13 | 1133 | 0.033333333 | 0.0057134220 | 2.544539 | AAGAAG,AAGAGG,AGAAGA,AGAGGA,GAAGAA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| NELFE | 9 | 1059 | 0.023809524 | 0.0053405885 | 2.156468 | CUCUCU,CUCUGG,CUGGCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| LIN28A | 7 | 900 | 0.019047619 | 0.0045395002 | 2.069005 | AGGAGA,GGAGAA,GGAGAU,GGAGGG,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACAUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | CAGUAU,UCAGUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRSF5 | 8 | 1578 | 0.021428571 | 0.0079554615 | 1.429518 | AAGAAG,AGAAGA,CACGGA,GAAGAA,GGAAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | CUGUCU,GUUUGU,UGUCUG,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | AUUUAU,UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CGGGAC,GCGGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAGAAG,AGAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPH2 | 14 | 3198 | 0.035714286 | 0.0161174929 | 1.147874 | GAGGGG,GGAGGG,GGGAGG,GGGGAG,GGGGCU,GGGGGA,GGGGGC,UGGGGG,UGGGUG,UUGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGU,GUUGUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AUCAAU,AUCAUG,AUGAUG,GAUCAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SRSF6 | 22 | 5069 | 0.054761905 | 0.0255441354 | 1.100181 | AACCUG,AAGAAG,AGAAGA,CACGGA,CCACUC,CCUCUC,CUCUGG,GAAGAA,GGAAGA,UCUCAG,UCUCUC,UCUCUG,UGUGGA,UUCUGG,UUUCUC,UUUCUG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,GUCUGU,GUUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SRSF10 | 12 | 2937 | 0.030952381 | 0.0148024990 | 1.064210 | AAGAAG,AAGAGA,AAGAGG,AGAGAA,AGAGGA,AGAGGG,GAGAAG,GAGGAG,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| G3BP1 | 14 | 3431 | 0.035714286 | 0.0172914148 | 1.046445 | AUCCGC,CCAGGC,CCCAGG,CCCCAC,CCCCCC,CCCCCG,CCCCGC,CCGCAC,CCGCCG,CCGGCA,CUCCGC,UCCGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.