circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000114541:-:3:69432633:69432761
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000114541:-:3:69432633:69432761 | ENSG00000114541 | ENST00000459638 | - | 3 | 69432634 | 69432761 | 128 | GCUUUGAUGAGAAAUCCAGAAGAAAGAGUGUAUGAAAGAAAAAGAUGACAUCUAUGCAUAGCAAACAAAAUCUGAAGAUUAUUACAGUAAAAACAAAAGAAAGGCAAGAGAAUACUUACUAGUUAAAGGCUUUGAUGAGAAAUCCAGAAGAAAGAGUGUAUGAAAGAAAAAGAUGACA | circ |
| ENSG00000114541:-:3:69432633:69432761 | ENSG00000114541 | ENST00000459638 | - | 3 | 69432634 | 69432761 | 22 | ACUAGUUAAAGGCUUUGAUGAG | bsj |
| ENSG00000114541:-:3:69432633:69432761 | ENSG00000114541 | ENST00000459638 | - | 3 | 69432752 | 69432961 | 210 | GACUAGCCUCAAUUUCAUCUGUAUUUCGAGAGAACUCUAUUCUAAAUCUCAUGGAAUUAGGUGAAUUUCUAAAACUGUGAUUAAACUGUAGUCAGCCAGGGGAAAAUUAGAAAUAAAUUAGAAGUGUCACUGACUUGAGAUUUAUAGUUUCUCCUCCUGUGCCUACAGAAAUUAAGGAGCUUCUCUCUUCUGUAUUACAGGCUUUGAUGA | ie_up |
| ENSG00000114541:-:3:69432633:69432761 | ENSG00000114541 | ENST00000459638 | - | 3 | 69432434 | 69432643 | 210 | CUAGUUAAAGGUAAGUUGACGUUUCUGUAAAUGCUGAACUCUUUGAUAUGUUAGAUAGAAUAGAAAAGUGAGUUUAUGUGUGUUUGCAUGUGAGUACACAUUGACUUGUCAAGCGGGGCUUGGACAGGAUGUUAUAUGAUAUAUGAUAUGGCAUAUUAACAAAAUAUUUACAUUACUAGCAAAUGAUGAAGAAAAUUAAUAAAUUAAGUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
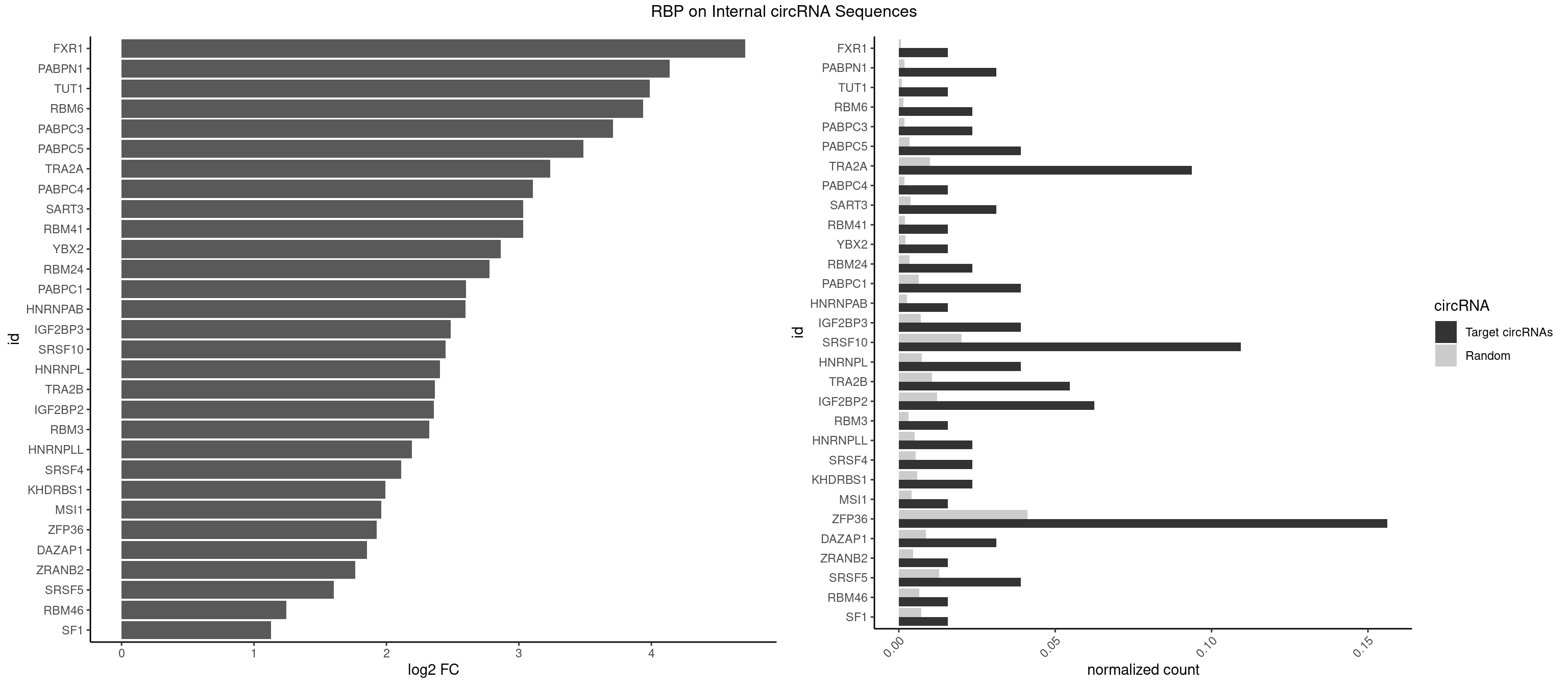
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 411 | 0.0156250 | 0.0005970720 | 4.709807 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| PABPN1 | 3 | 1222 | 0.0312500 | 0.0017723764 | 4.140099 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| TUT1 | 1 | 678 | 0.0156250 | 0.0009840095 | 3.989040 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.0234375 | 0.0015289102 | 3.938243 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PABPC3 | 2 | 1234 | 0.0234375 | 0.0017897669 | 3.710975 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC5 | 4 | 2400 | 0.0390625 | 0.0034795387 | 3.488816 | AGAAAA,AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2A | 11 | 6871 | 0.0937500 | 0.0099589296 | 3.234756 | AAAGAA,AAGAAA,AGAAAG,AGAAGA,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PABPC4 | 1 | 1251 | 0.0156250 | 0.0018144033 | 3.106289 | AAAAAG | AAAAAA,AAAAAG |
| SART3 | 3 | 2634 | 0.0312500 | 0.0038186524 | 3.032721 | AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM41 | 1 | 1318 | 0.0156250 | 0.0019115000 | 3.031079 | AUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX2 | 1 | 1480 | 0.0156250 | 0.0021462711 | 2.863952 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM24 | 2 | 2357 | 0.0234375 | 0.0034172229 | 2.777922 | AGAGUG,GAGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPC1 | 4 | 4443 | 0.0390625 | 0.0064402624 | 2.600593 | AAAAAC,AGAAAA,CAAACA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPAB | 1 | 1782 | 0.0156250 | 0.0025839306 | 2.596217 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| IGF2BP3 | 4 | 4815 | 0.0390625 | 0.0069793662 | 2.484616 | AAAAAC,AAAACA,AAAAUC,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SRSF10 | 13 | 13860 | 0.1093750 | 0.0200874160 | 2.444919 | AAAAGA,AAAGAA,AAAGAG,AAGAAA,AAGAGA,AGAGAA,GAAAGA,GAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPL | 4 | 5085 | 0.0390625 | 0.0073706513 | 2.405920 | AAACAA,AACAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| TRA2B | 6 | 7329 | 0.0546875 | 0.0106226650 | 2.364065 | AAAGAA,AGAAGA,GAAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| IGF2BP2 | 7 | 8408 | 0.0625000 | 0.0121863560 | 2.358589 | AAAAAC,AAAACA,AAAAUC,CAAACA,GAAAUC,GAAUAC,GCAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM3 | 1 | 2152 | 0.0156250 | 0.0031201361 | 2.324175 | AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPLL | 2 | 3534 | 0.0234375 | 0.0051229360 | 2.193776 | CAAACA,GCAAAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SRSF4 | 2 | 3740 | 0.0234375 | 0.0054214720 | 2.112062 | AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| KHDRBS1 | 2 | 4064 | 0.0234375 | 0.0058910141 | 1.992231 | CAAAAU,UAAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| MSI1 | 1 | 2770 | 0.0156250 | 0.0040157442 | 1.960117 | UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZFP36 | 19 | 28400 | 0.1562500 | 0.0411588414 | 1.924582 | AAAAAC,AAAAAG,AAAACA,AAAAGA,AAACAA,AAAGAA,AACAAA,AAGAAA,AGAAAG,AGCAAA,AGUAAA,CAAACA | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| DAZAP1 | 3 | 5964 | 0.0312500 | 0.0086445016 | 1.854002 | AGUAAA,AGUUAA,UAGUUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ZRANB2 | 1 | 3173 | 0.0156250 | 0.0045997733 | 1.764222 | UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| SRSF5 | 4 | 8869 | 0.0390625 | 0.0128544391 | 1.603518 | AGAAGA,GAAGAA,UAAAGG,UGCAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBM46 | 1 | 4554 | 0.0156250 | 0.0066011240 | 1.243073 | AUGAAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SF1 | 1 | 4931 | 0.0156250 | 0.0071474739 | 1.128351 | UACUAG | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
RBP on BSJ (Exon Only)
Plot
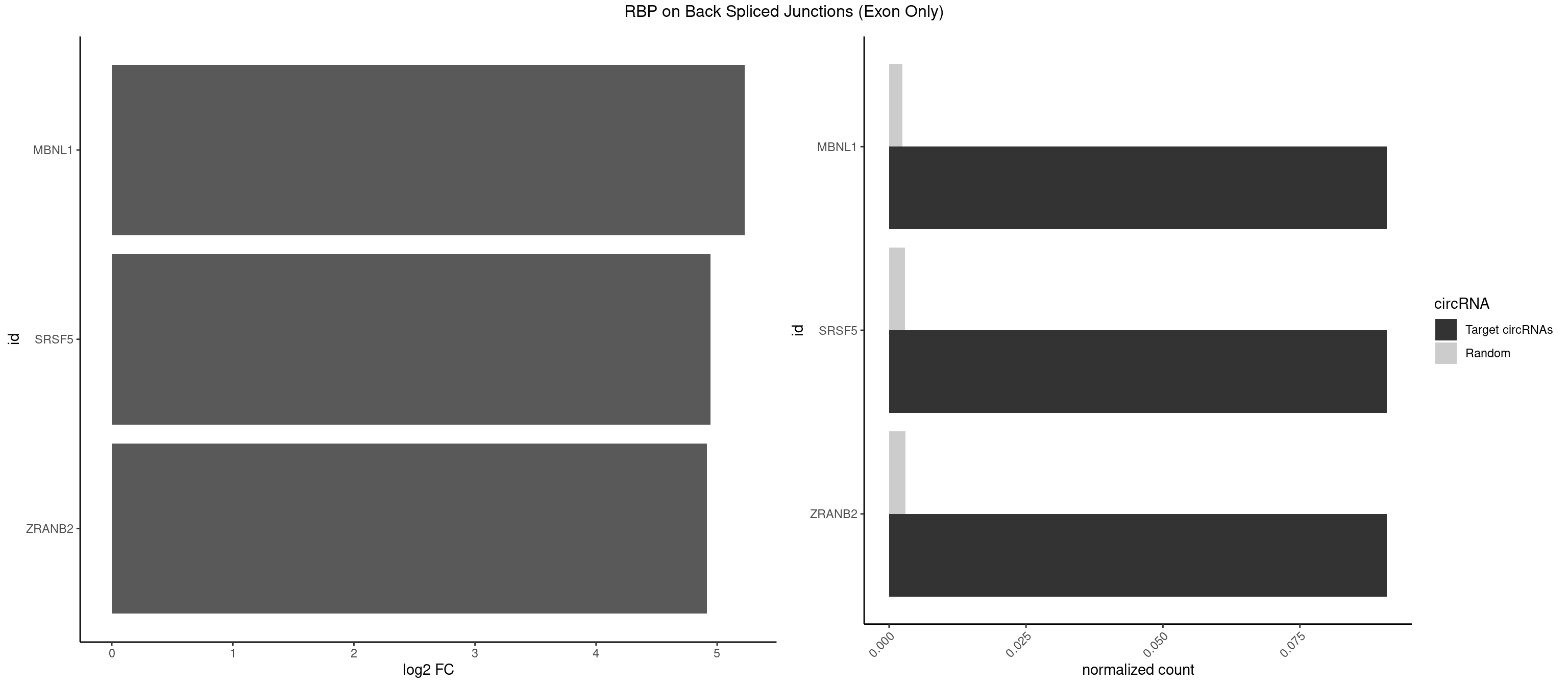
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MBNL1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | GGCUUU | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | UAAAGG | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
RBP on BSJ (Exon and Intron)
Plot
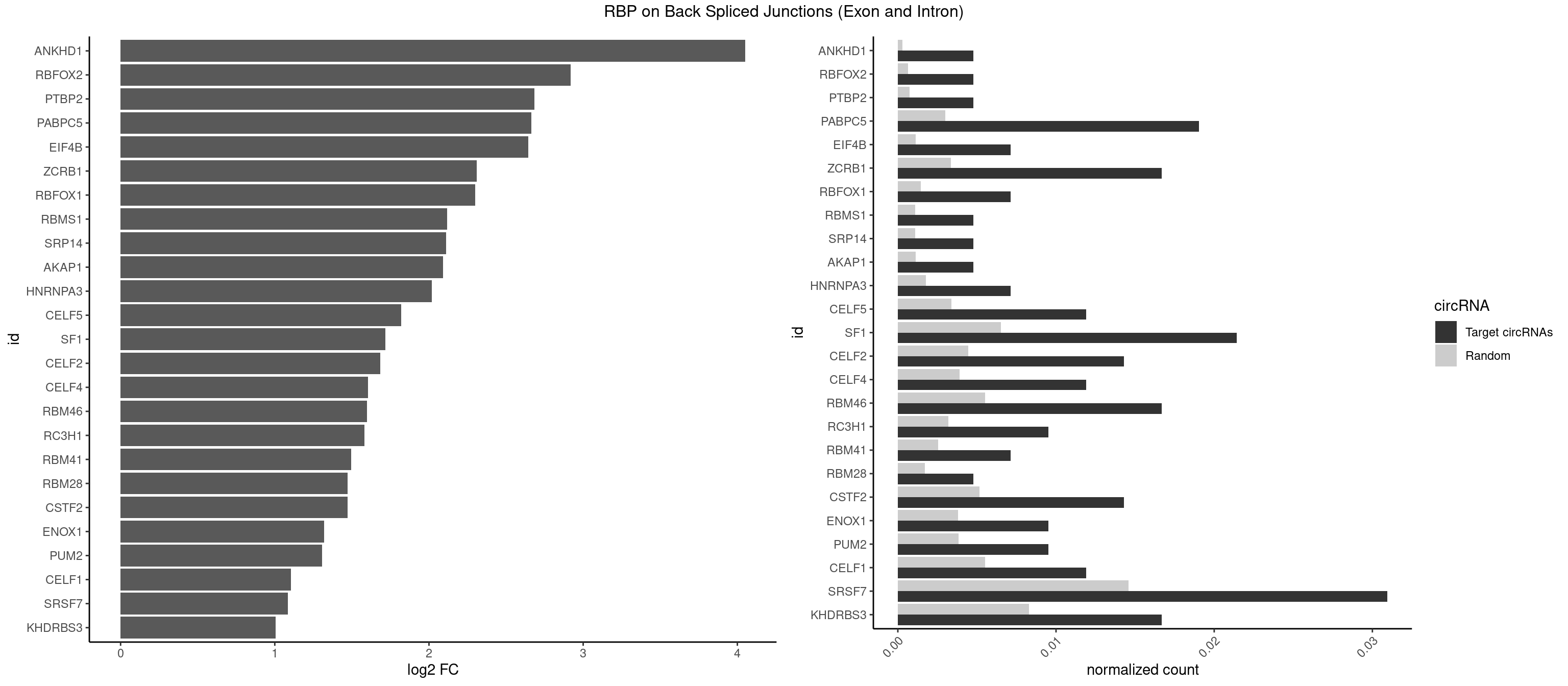
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| PABPC5 | 7 | 596 | 0.019047619 | 0.0030078597 | 2.662801 | AGAAAA,AGAAAU,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZCRB1 | 6 | 666 | 0.016666667 | 0.0033605401 | 2.310201 | AAUUAA,GAAUUA,GAUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,AGGAGC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SF1 | 8 | 1294 | 0.021428571 | 0.0065245869 | 1.715577 | ACUAGC,ACUGAC,AUUAAC,CACUGA,UAACAA,UACUAG,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,GUGUGU,UAUGUG,UAUGUU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUGAU,AUGAAG,AUGAUA,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGUACA,GGACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAAAU,UAGAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUGUGU,UGUGUG,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAUGAU,AGAGAA,AUAGAA,AUUGAC,CGAGAG,CUCUUC,GAAGAA,GAGAGA,GAUAGA,UCGAGA,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAAUAA,AAUAAA,AUUAAA,UAAAAC,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.