circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000111880:-:6:88769773:88904955
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000111880:-:6:88769773:88904955 | ENSG00000111880 | ENST00000369485 | - | 6 | 88890495 | 88904955 | 453 | UAUCGAAGCAGCAGUUGCUACUUUUGCCCAAGCCAGACCACCAGGAAUCUACAAGGGUGAUUAUUUGAAGGAACUUUUUCGUCGGUAUGGUGACAUAGAGGAAGCACCACCCCCACCUCUAUUGCCAGAUUGGUGUUUUGAGGAUGAUGAAGACGAAGAUGAGGAUGAGGAUGGAAAGAAGGAAUCAGAACCCGGGUCAAGUGCUUCUUUUGGCAAAAGGAGAAAAGAACGGUUAAAACUGGGCGCUAUUUUCUUGGAAGGUGUUACUGUUAAAGGUGUAACUCAAGUAACAACACAACCAAAGUUAGGAGAGGUACAGCAGAAGUGUCAUCAAUUCUGUGGCUGGGAAGGGUCUGGAUUCCCUGGAGCACAGCCUGUUUCCAUGGACAAGCAAAAUAUUAAACUUUUAGACCUGAAGCCAUACAAAGUAAGCUGGAAAGCAGAUGGUACUCGUAUCGAAGCAGCAGUUGCUACUUUUGCCCAAGCCAGACCACCAGGAAUCU | circ |
| ENSG00000111880:-:6:88769773:88904955 | ENSG00000111880 | ENST00000369485 | - | 6 | 88890495 | 88904955 | 22 | GAUGGUACUCGUAUCGAAGCAG | bsj |
| ENSG00000111880:-:6:88769773:88904955 | ENSG00000111880 | ENST00000369485 | - | 6 | 88904946 | 88905155 | 210 | GAAUACCUUAUAUUAAAACCUUUUCAGAAUGAAUGUGUCCCUUAAUAGAUUGAAUGACUUGAGGGUGAGGAUUGUGAUUUGCCCAUUUUAUAAACUUCUGUUGUACUUUAUAUGGAGCCUUGAAUAUAAUAAGUGAGUUUUUGAUAAGCCUGCAUAAAUAGAGGAUAGCGAGGAAAUUGCAUGGUGUUUGUUUCCUAUAGUAUCGAAGCA | ie_up |
| ENSG00000111880:-:6:88769773:88904955 | ENSG00000111880 | ENST00000369485 | - | 6 | 88890295 | 88890504 | 210 | AUGGUACUCGGUAAGUUAGACUUCUAAAAAAAUUACACAAAUAACCCUAAUGCUUGAAGAAUAUUGUUUUAUUGUUAGAAAAGUUUACACUCUUGAGAGUGUGAAUUUCUAAUGUGAAUAGCAAAAUAAAUGUUUCAAAAAUUAUAUAUUUUUGGUUUUGCUAGUGUUCUAAGCUGUAUAGCACAAGUUAUCUCUCAUUCUGCUAAAUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
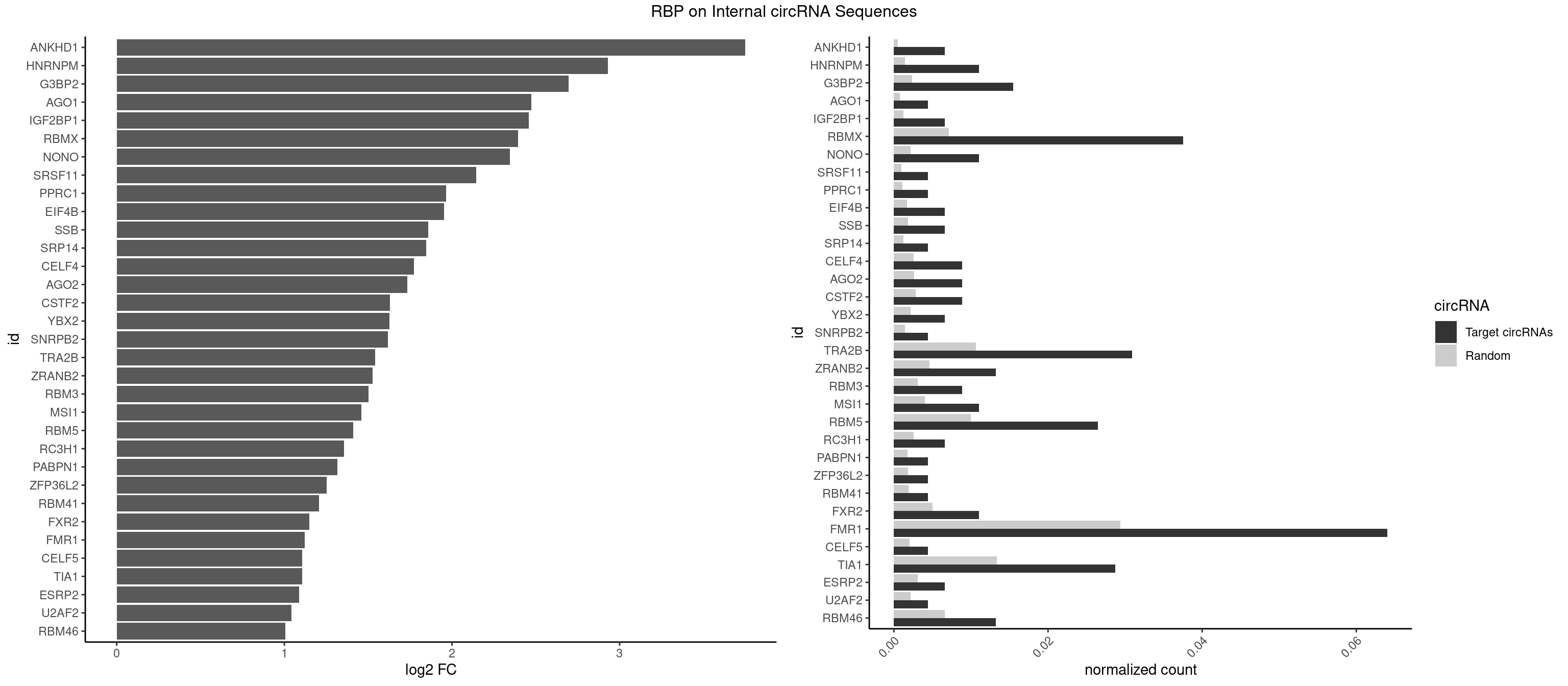
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.006622517 | 0.0004927293 | 3.748512 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPM | 4 | 999 | 0.011037528 | 0.0014492040 | 2.929084 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 6 | 1644 | 0.015452539 | 0.0023839405 | 2.696424 | AGGAUG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| AGO1 | 1 | 548 | 0.004415011 | 0.0007956130 | 2.472278 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| IGF2BP1 | 2 | 831 | 0.006622517 | 0.0012057377 | 2.457463 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RBMX | 16 | 4925 | 0.037527594 | 0.0071387787 | 2.394203 | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,GAAGGA,GGAAGG,GUAACA,UAACAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| NONO | 4 | 1498 | 0.011037528 | 0.0021723567 | 2.345084 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SRSF11 | 1 | 688 | 0.004415011 | 0.0009985015 | 2.144581 | AAGAAG | AAGAAG |
| PPRC1 | 1 | 780 | 0.004415011 | 0.0011318283 | 1.963762 | GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| EIF4B | 2 | 1179 | 0.006622517 | 0.0017100607 | 1.953332 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SSB | 2 | 1260 | 0.006622517 | 0.0018274462 | 1.857551 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRP14 | 1 | 847 | 0.004415011 | 0.0012289250 | 1.845020 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF4 | 3 | 1782 | 0.008830022 | 0.0025839306 | 1.772850 | GGUGUU,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO2 | 3 | 1830 | 0.008830022 | 0.0026534924 | 1.734525 | AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CSTF2 | 3 | 1967 | 0.008830022 | 0.0028520334 | 1.630426 | GUGUUU,GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| YBX2 | 2 | 1480 | 0.006622517 | 0.0021462711 | 1.625547 | AACAAC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SNRPB2 | 1 | 991 | 0.004415011 | 0.0014376103 | 1.618744 | UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TRA2B | 13 | 7329 | 0.030905077 | 0.0106226650 | 1.540698 | AAAGAA,AAGAAC,AAGAAG,AAGGAA,AGAAGG,AGGAAG,GAAAGA,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ZRANB2 | 5 | 3173 | 0.013245033 | 0.0045997733 | 1.525817 | AAAGGU,AGGUAC,CGGUAU,GGGUGA,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBM3 | 3 | 2152 | 0.008830022 | 0.0031201361 | 1.500808 | AAAACU,AAGACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| MSI1 | 4 | 2770 | 0.011037528 | 0.0040157442 | 1.458678 | AGGAAG,AGUAAG,AGUUAG,UAGGAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBM5 | 11 | 6879 | 0.026490066 | 0.0099705232 | 1.409710 | AAGGAA,AAGGAG,AAGGUG,AGGGUG,CAAGGG,GAAGGA,GAAGGG,GAAGGU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| RC3H1 | 2 | 1786 | 0.006622517 | 0.0025897275 | 1.354579 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPN1 | 1 | 1222 | 0.004415011 | 0.0017723764 | 1.316732 | AAAAGA | AAAAGA,AGAAGA |
| ZFP36L2 | 1 | 1277 | 0.004415011 | 0.0018520827 | 1.253269 | UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM41 | 1 | 1318 | 0.004415011 | 0.0019115000 | 1.207712 | UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| FXR2 | 4 | 3434 | 0.011037528 | 0.0049780156 | 1.148774 | AGACGA,GACAAG,GACGAA,GGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| FMR1 | 28 | 20291 | 0.064017660 | 0.0294072466 | 1.122298 | AAGGAA,AAGGAG,ACUGGG,AGCAGC,AGCUGG,AGGAAG,AGGAAU,AGGAUG,CUGGGC,GAAGGA,GAAGGG,GACAAG,GAGGAU,GCUGGG,GGACAA,GGCUGG,GUGGCU,UGAGGA,UGGCUG | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| CELF5 | 1 | 1415 | 0.004415011 | 0.0020520728 | 1.105335 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TIA1 | 12 | 9206 | 0.028697572 | 0.0133428208 | 1.104865 | AUUUUC,CUUUUA,CUUUUG,CUUUUU,GUUUUG,UAUUUU,UUAUUU,UUUUCG,UUUUCU,UUUUGG,UUUUUC | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ESRP2 | 2 | 2150 | 0.006622517 | 0.0031172377 | 1.087111 | GGGAAG,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| U2AF2 | 1 | 1477 | 0.004415011 | 0.0021419234 | 1.043510 | UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| RBM46 | 5 | 4554 | 0.013245033 | 0.0066011240 | 1.004668 | AUCAAU,AUGAAG,AUGAUG,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
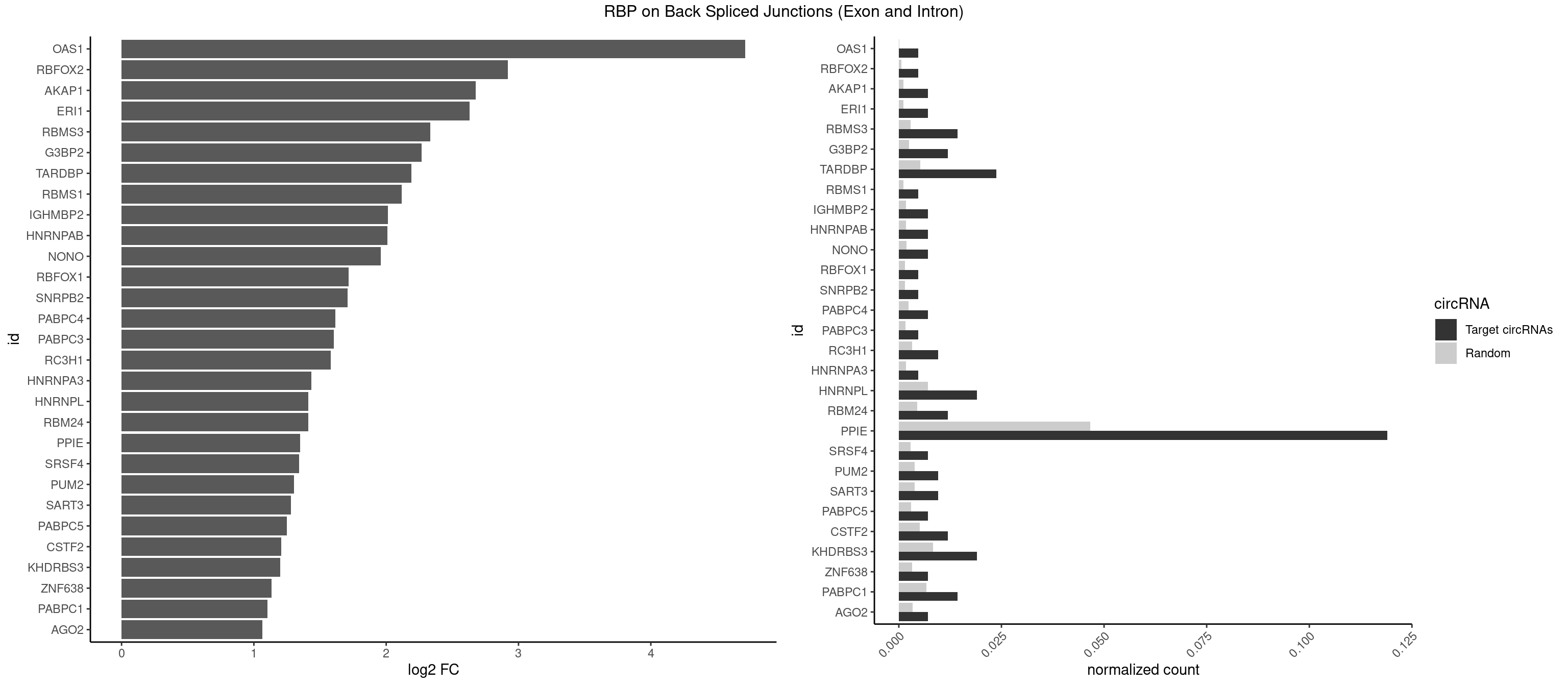
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBMS3 | 5 | 562 | 0.014285714 | 0.0028365578 | 2.332360 | AAUAUA,AUAUAU,CUAUAG,UAUAGC,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUA,AGGAUU,GGAUAG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TARDBP | 9 | 1034 | 0.023809524 | 0.0052146312 | 2.190902 | GAAUGA,GAAUGU,GUGAAU,GUUUUG,UGAAUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| IGHMBP2 | 2 | 350 | 0.007142857 | 0.0017684401 | 2.014024 | AAAAAA | AAAAAA |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CUUCUG,UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.0071543732 | 1.412713 | AAAUAA,AAUAAA,ACACAA,CACAAA,CACAAG,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,AGUGUG,GAGUGU,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PPIE | 49 | 9262 | 0.119047619 | 0.0466696896 | 1.350981 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUUA,AAAUUU,AAUAAA,AAUAUA,AAUAUU,AAUUAU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAUA,AUUUUA,AUUUUU,UAAAAA,UAAAUA,UAAAUU,UAAUAA,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAUA,UUAUAA,UUAUAU,UUUAUA,UUUAUU,UUUUAU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAAAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,AGAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUU,GUUUUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AAAUAA,AAUAAA,AUAAAC,AUUAAA,UAAAAC,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA,ACAAAU,AGAAAA,CAAAUA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.