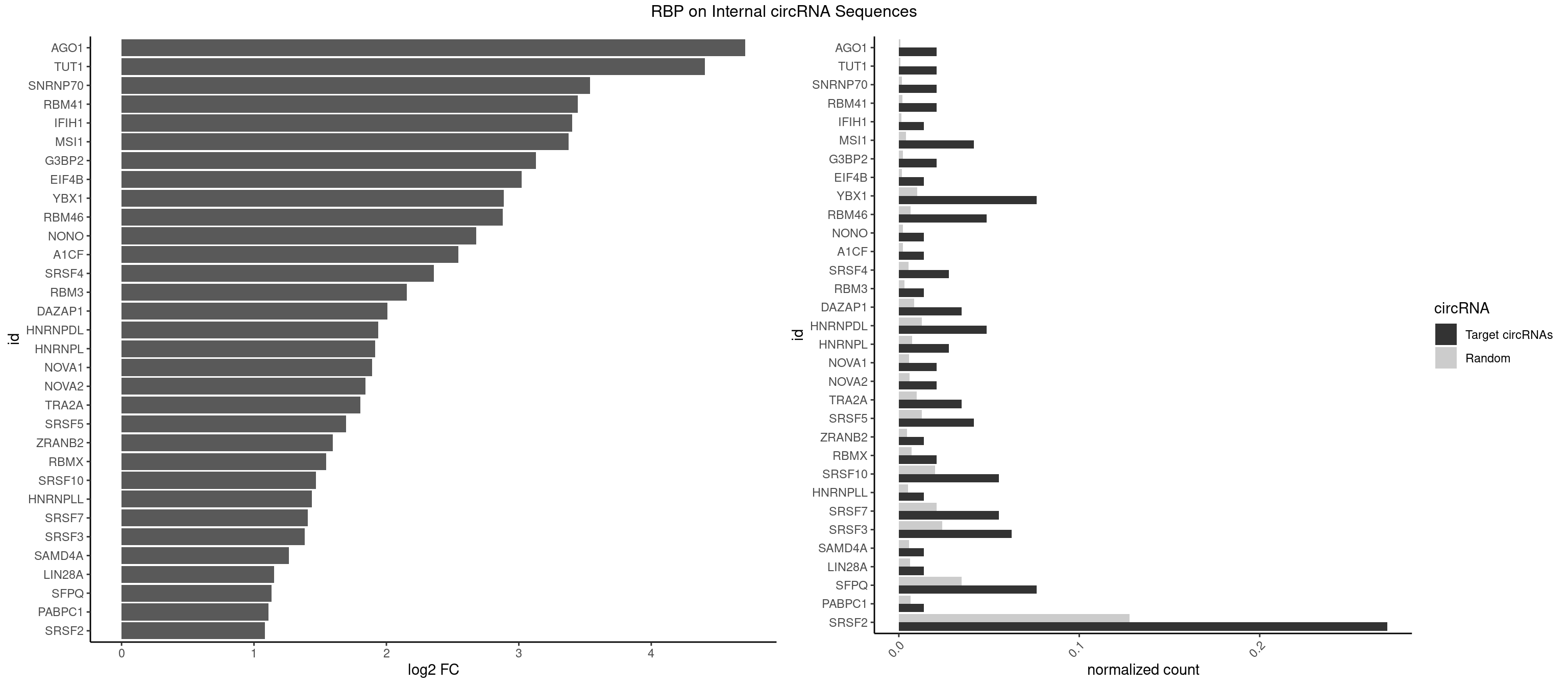
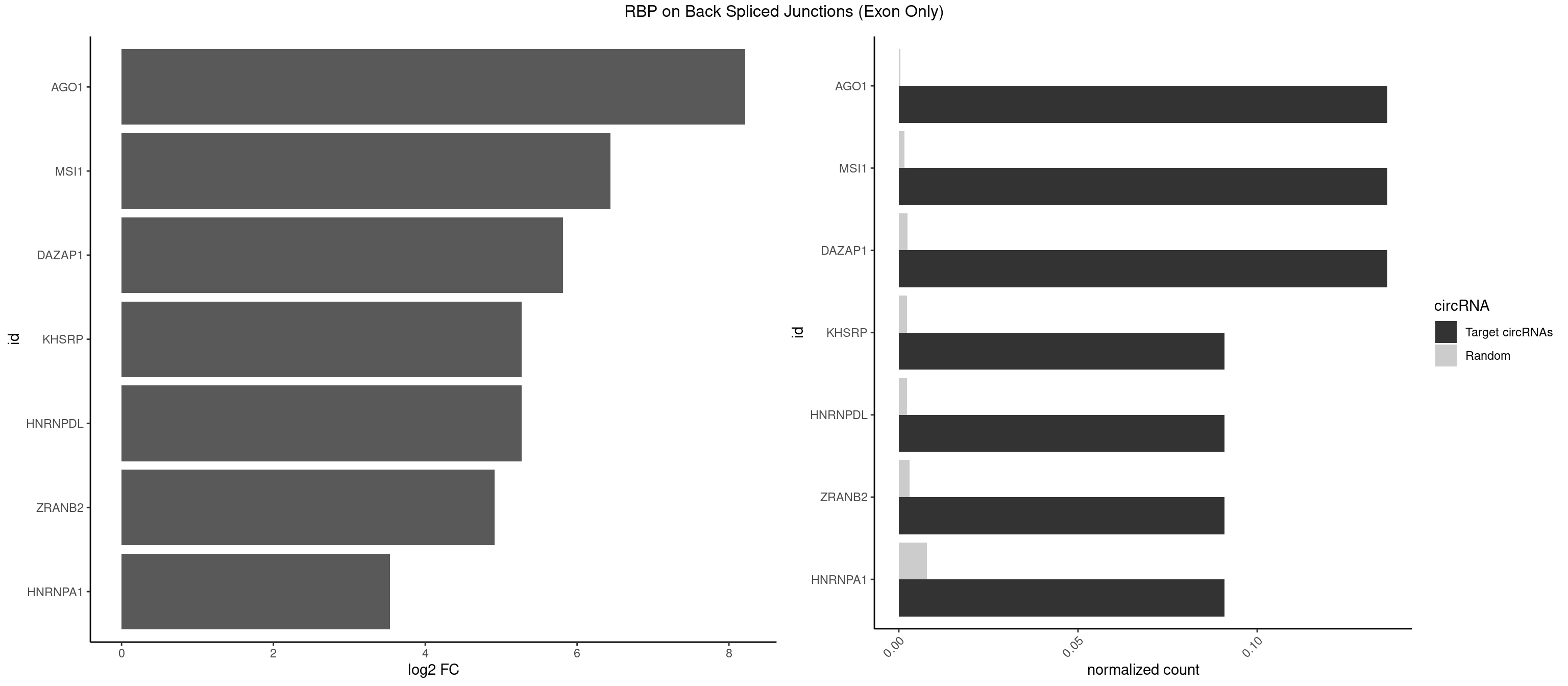
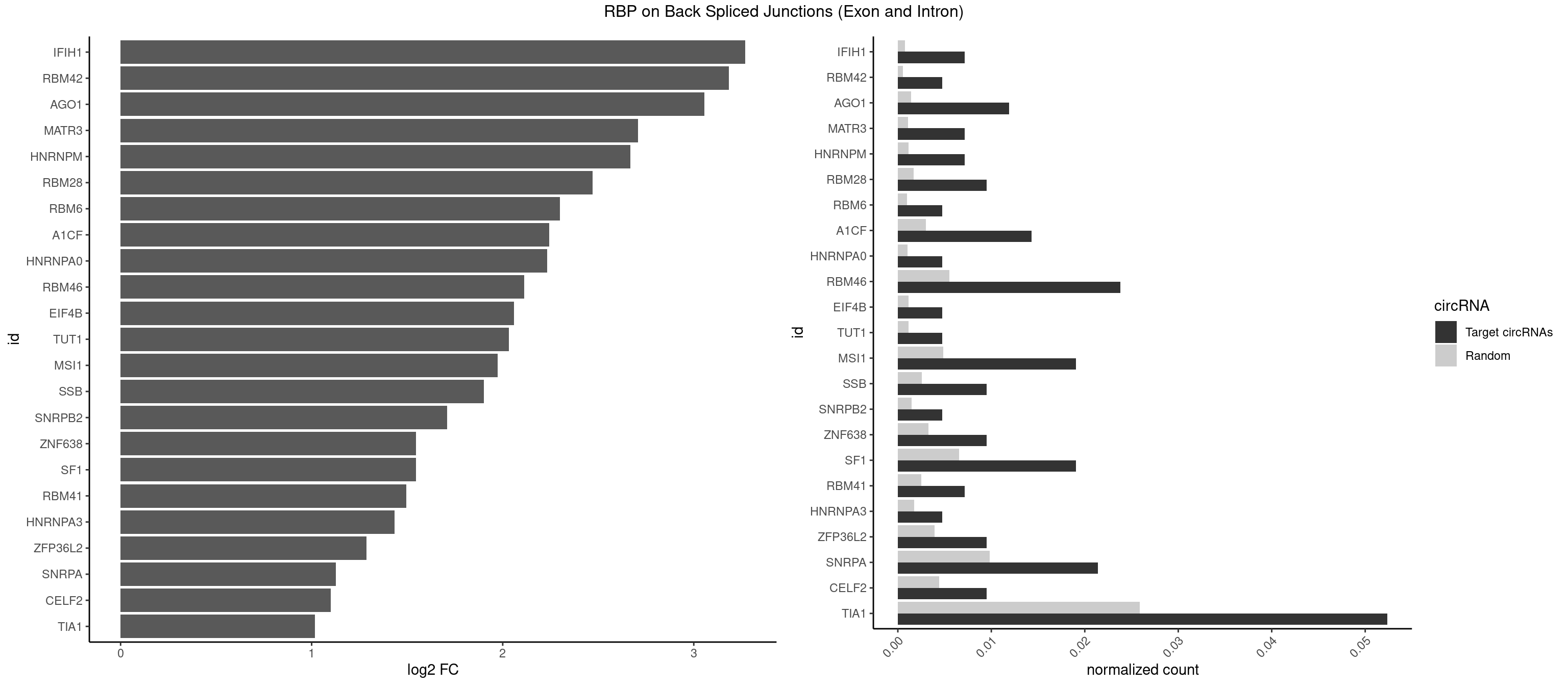
| AGO1 |
2 |
548 |
0.02083333 |
0.0007956130 |
4.710683 |
AGGUAG,GGUAGU |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| TUT1 |
2 |
678 |
0.02083333 |
0.0009840095 |
4.404078 |
AAAUAC,AAUACU |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 |
2 |
1237 |
0.02083333 |
0.0017941145 |
3.537550 |
AUCAAG,GAUCAA |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM41 |
2 |
1318 |
0.02083333 |
0.0019115000 |
3.446117 |
AUACUU,UACUUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IFIH1 |
1 |
904 |
0.01388889 |
0.0013115296 |
3.404609 |
GGCCGC |
CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| MSI1 |
5 |
2770 |
0.04166667 |
0.0040157442 |
3.375154 |
AGGAAG,AGGAGG,AGGUAG,UAGGAG,UAGGUA |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| G3BP2 |
2 |
1644 |
0.02083333 |
0.0023839405 |
3.127474 |
GGAUGA,GGAUUG |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| EIF4B |
1 |
1179 |
0.01388889 |
0.0017100607 |
3.021812 |
UUGGAC |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| YBX1 |
10 |
7119 |
0.07638889 |
0.0103183321 |
2.888153 |
AACCAC,ACCACC,CACCAC,CAGCAA,CCACCA,CCAGCA,UCCAGC |
AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| RBM46 |
6 |
4554 |
0.04861111 |
0.0066011240 |
2.880503 |
AUCAAG,AUGAUG,GAUCAA,GAUGAU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| NONO |
1 |
1498 |
0.01388889 |
0.0021723567 |
2.676598 |
GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| A1CF |
1 |
1642 |
0.01388889 |
0.0023810421 |
2.544266 |
UGAUCA |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SRSF4 |
3 |
3740 |
0.02777778 |
0.0054214720 |
2.357175 |
AGGAAG,GAGGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBM3 |
1 |
2152 |
0.01388889 |
0.0031201361 |
2.154250 |
AAUACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| DAZAP1 |
4 |
5964 |
0.03472222 |
0.0086445016 |
2.006005 |
AGGAAA,AGGAAG,AGGUAG,UAGGUA |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPDL |
6 |
8759 |
0.04861111 |
0.0126950266 |
1.937023 |
ACCACG,ACCAGA,ACUUUA,CACCAG,CUUUAG,UUUAGG |
AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| HNRNPL |
3 |
5085 |
0.02777778 |
0.0073706513 |
1.914067 |
AAAUAC,CACCAC,CACGAC |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| NOVA1 |
2 |
3872 |
0.02083333 |
0.0056127669 |
1.892110 |
ACCACC |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 |
2 |
4013 |
0.02083333 |
0.0058171047 |
1.840521 |
ACCACC |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| TRA2A |
4 |
6871 |
0.03472222 |
0.0099589296 |
1.801797 |
AAGAGG,AGGAAG,GAAGAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF5 |
5 |
8869 |
0.04166667 |
0.0128544391 |
1.696627 |
AGGAAG,CCGCAG,CGCAGG,GAGGAA,GGAAGA |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| ZRANB2 |
1 |
3173 |
0.01388889 |
0.0045997733 |
1.594297 |
AGGUAG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBMX |
2 |
4925 |
0.02083333 |
0.0071387787 |
1.545145 |
AGGAAG,AGUAAC |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SRSF10 |
7 |
13860 |
0.05555556 |
0.0200874160 |
1.467639 |
AAGAGA,AAGAGG,AGAGAG,GAGAGA,GAGGAA |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPLL |
1 |
3534 |
0.01388889 |
0.0051229360 |
1.438888 |
CACCAC |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SRSF7 |
7 |
14481 |
0.05555556 |
0.0209873716 |
1.404410 |
AGAGAG,AGAGAU,AGGAAG,GAGAGA,GAGGAA |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SRSF3 |
8 |
16536 |
0.06250000 |
0.0239654858 |
1.382898 |
ACCACC,ACUUUA,AGAGAU,CACCAC,CCACCA,GAUCAA |
AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| SAMD4A |
1 |
3992 |
0.01388889 |
0.0057866714 |
1.263126 |
CGGGUA |
CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| LIN28A |
1 |
4315 |
0.01388889 |
0.0062547643 |
1.150904 |
GGAGGA |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SFPQ |
10 |
24050 |
0.07638889 |
0.0348548043 |
1.132005 |
AAGAGG,AGAGAG,GAAGAG,GAGGAA,GGAAGA,GGAGGA,GUAGUG,UAGUGG,UGAUGG |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| PABPC1 |
1 |
4443 |
0.01388889 |
0.0064402624 |
1.108740 |
CAAAUA |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SRSF2 |
38 |
88240 |
0.27083333 |
0.1278792060 |
1.082624 |
AAGAGA,ACCACC,AGAGAU,AGCUCC,AGGAAG,AGGAGG,AUGAUG,CCACCA,CCGCAG,CGCAGG,CUGAUG,GAACCA,GAGAGA,GAGAUG,GAGGAA,GAUGAU,GAUGCU,GAUGGA,GCCGCA,GCUCCA,GCUGAU,GGAACC,GGAGGA,GGCCGC,GGCUGA,GGGUAU,GUAACC,UAACCA,UCCAGC,UGAUGC,UGAUGG |
AAAAGA,AAAGAG,AAAGGA,AACCAG,AACGAG,AAGAAG,AAGAGA,AAGCAG,AAGCGG,AAGCUG,AAGGAG,AAGGCG,AAGUAC,AAUACC,AAUCCC,AAUGCC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACCCGC,ACCCGG,ACCGGU,ACCGUA,ACCUCC,ACCUGC,ACGAGA,ACGAGG,ACGCUG,ACGGAG,ACGUGU,ACUACC,ACUACU,ACUAGA,ACUCAA,ACUCCC,ACUCCU,ACUGUA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGACUA,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCCC,AGCCGA,AGCCUC,AGCGGA,AGCGUG,AGCUCC,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGA,AGGCGU,AGGCUG,AGGGUA,AGUACG,AGUAGA,AGUAGG,AGUAGU,AGUAUC,AGUCGA,AGUGAC,AGUGGU,AGUGUU,AGUUCC,AUACCG,AUCACC,AUCACU,AUCCCC,AUCCCG,AUCCCU,AUCCGG,AUCGAG,AUCGCU,AUGAAU,AUGAUG,AUGCCG,AUGCGU,AUGCUG,AUGGAG,AUUACC,AUUACU,AUUAGU,AUUCCA,AUUCCC,AUUCCU,AUUGAU,CACCUC,CAGAGA,CAGAGG,CAGAGU,CAGCUA,CAGUAG,CAGUCC,CAGUGG,CAGUGU,CCACCA,CCACCG,CCACUA,CCACUG,CCAGAA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCACA,CCCCCA,CCCCCG,CCCCUA,CCCCUG,CCCGCA,CCCGCU,CCCGUG,CCCUUG,CCGAGA,CCGCAG,CCGCUA,CCGGAU,CCGGUG,CCUCAG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGAA,CGAGAG,CGAGAU,CGAGCA,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGG,CGCAGU,CGCCCU,CGCUGC,CGGAGA,CGGAGU,CGUAAG,CGUGAU,CGUGCG,CGUGUA,CGUGUU,CUAACG,CUACCA,CUACCG,CUACUA,CUACUG,CUAGAA,CUAGAC,CUCAAG,CUCCAA,CUCCCA,CUCCCG,CUCCUA,CUCCUG,CUCGAG,CUCGUG,CUGAUG,CUGCAA,CUGCUA,CUGUUA,GAAAGG,GAACCA,GAACGA,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GAAUGC,GACCAC,GACCCC,GACCCG,GACCGG,GACCGU,GACCUG,GACGCU,GACGGA,GACUAC,GACUAG,GACUCA,GACUCC,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGG,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCGU,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUAU,GAGUGA,GAUCAC,GAUCCC,GAUCCG,GAUCGA,GAUCGC,GAUGAU,GAUGCG,GAUGCU,GAUGGA,GAUUAC,GAUUAG,GAUUCC,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCAAA,GCCACC,GCCACU,GCCCAC,GCCCCC,GCCCCU,GCCCUU,GCCGAG,GCCGCA,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGAGC,GCGAGU,GCGCAG,GCGGAG,GCGGUC,GCGUGU,GCUACC,GCUACU,GCUCCA,GCUCCC,GCUCCU,GCUCGA,GCUCGU,GCUGAU,GCUGCA,GCUGCU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGACUG,GGAGAA,GGAGAG,GGAGAU,GGAGCC,GGAGCG,GGAGGA,GGAGUA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAA,GGCCAC,GGCCCA,GGCCCC,GGCCGC,GGCCUC,GGCGAG,GGCGUG,GGCUAC,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUAAG,GGUACG,GGUAGG,GGUAUG,GGUCAC,GGUCAG,GGUCCC,GGUCGC,GGUGAG,GGUGAU,GGUUAA,GGUUAC,GGUUCC,GGUUGG,GGUUGU,GUAACC,GUAAGC,GUAAGU,GUACGA,GUACGC,GUAGGC,GUAGGG,GUAGUC,GUAUCC,GUAUGC,GUCACC,GUCACU,GUCAGU,GUCCCC,GUCCCU,GUCCUC,GUCGAU,GUCGCC,GUCUAA,GUGAGC,GUGAUG,GUGCAG,GUGGUC,GUGUAA,GUUAAU,GUUACC,GUUACG,GUUACU,GUUCCA,GUUCCC,GUUCCG,GUUCCU,GUUCGA,GUUCGU,GUUCUG,GUUGGC,GUUGUU,GUUUCG,UAACCA,UAAGCU,UAAGUA,UACCAG,UACGAG,UACGCU,UACGUG,UAGACU,UAGGCU,UAUCCC,UAUGCC,UAUGCU,UCACCA,UCACCG,UCACUA,UCACUG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCCCA,UCCCCG,UCCCCU,UCCCGC,UCCCGU,UCCCUA,UCCCUG,UCCGGA,UCCUCA,UCCUCC,UCCUGC,UCCUGU,UCGAGA,UCGAGU,UCGCCG,UCGCUG,UCGUAA,UCGUGC,UCUAAC,UCUGUA,UGAAUG,UGAGCU,UGAUCG,UGAUGC,UGAUGG,UGCAGA,UGCAGU,UGCCGC,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUAC,UGUUCC,UGUUCG,UUAAUG,UUACCA,UUACCG,UUACGU,UUACUA,UUACUG,UUAGUG,UUCCAG,UUCCCA,UUCCCG,UUCCCU,UUCCGG,UUCCUA,UUCCUG,UUCGAG,UUCGUG,UUCUGU,UUGAUG,UUGGCG,UUGUUG,UUUCGA |