circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000137815:+:15:41464770:41466252
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000137815:+:15:41464770:41466252 | ENSG00000137815 | ENST00000389629 | + | 15 | 41464771 | 41466252 | 227 | AUUUGAAAUCAAGAAAAAACUAAAAACAGCCAAAAAGAAAGAAAAGAAAGAAAAGAAGAAAAAGCAAGAAGAGGAGCAAGAAAAGAAAAAACUGACACAGAUUCAAGAAUCUCAGGUAACAUCCCACAACAAGGAACGGCGUUCCAAGCGGGAUGAGAAACUAGACAAGAAAUCUCAAGCCAUGGAGGAGCUAAAAGCAGAGCGAGAAAAACGAAAGAACAGAACAGAUUUGAAAUCAAGAAAAAACUAAAAACAGCCAAAAAGAAAGAAAAGAAAG | circ |
| ENSG00000137815:+:15:41464770:41466252 | ENSG00000137815 | ENST00000389629 | + | 15 | 41464771 | 41466252 | 22 | GAACAGAACAGAUUUGAAAUCA | bsj |
| ENSG00000137815:+:15:41464770:41466252 | ENSG00000137815 | ENST00000389629 | + | 15 | 41464571 | 41464780 | 210 | AAUGCUAUCAAAUAUUUUAUAUUUUGUUGUAAUUAACAUAUUAAAUAUUUUUAAUGUACUUAAAAGGUACACCAGAGUUUCUUGCAGGAAAGCCAAAAUAUCUAUUAGCUCCUUUCCCUUAGUUCCUUUCUCCUAUCUAAUAGAAAUCAGUUUUUAUAUACAUUUCAUUGCUACUUAAAAACCAAAUAUCUUUUAACCAGAUUUGAAAUC | ie_up |
| ENSG00000137815:+:15:41464770:41466252 | ENSG00000137815 | ENST00000389629 | + | 15 | 41466243 | 41466452 | 210 | AACAGAACAGGUAAGCGAGGGAAAUAUAAUGGCUACUUGUGUCUUUAUAUGAUUUGACUUCCUGGUGUCCAUUUUCAUUUUGCCCUCUUGUUAUAUCUUUCUCAUAUAAAAUUCCAGCACAUACCCAGAAUAGUAUAAACAUCCACAAACCCAUCAUCCAGCCACAGCAGUCAGCUCUUGGCCAGUCUUGUUAAAUCUAUAAUCCUUUCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
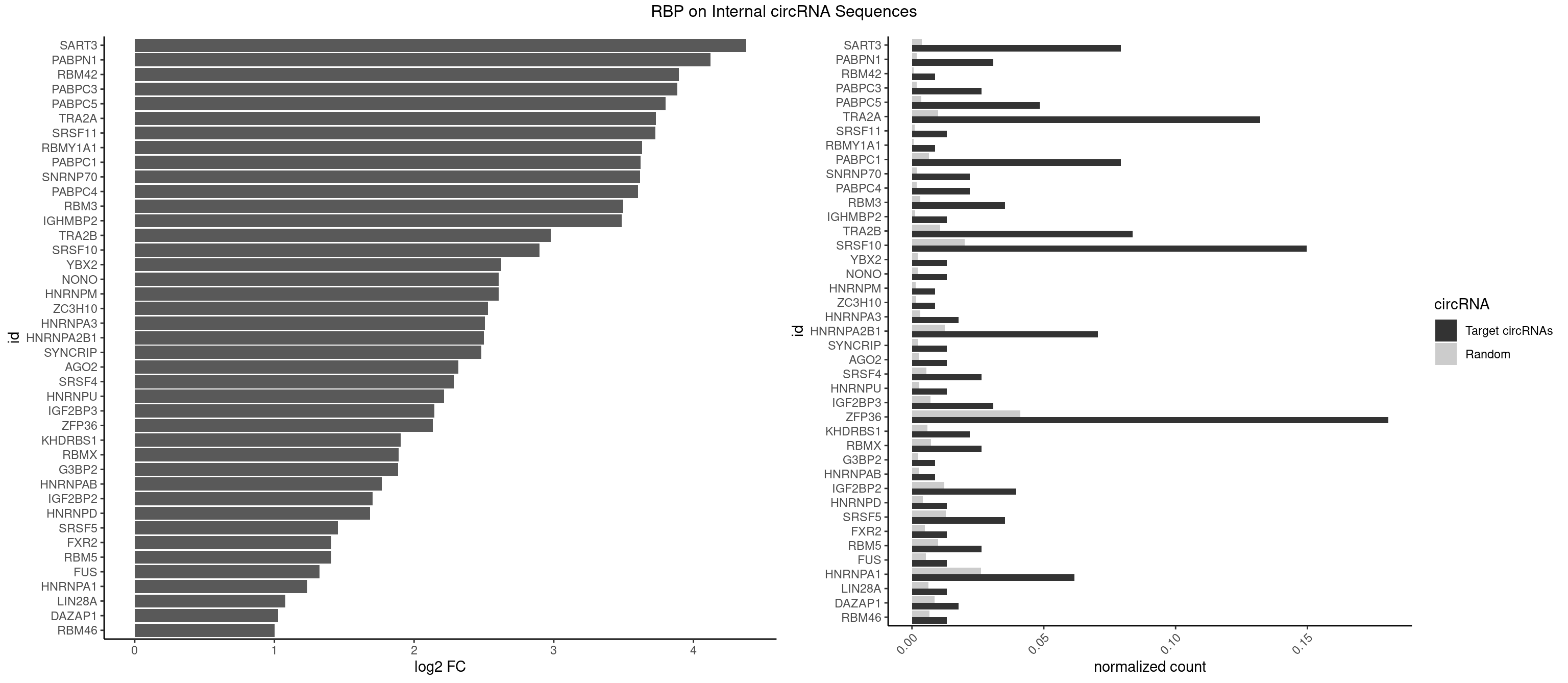
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SART3 | 17 | 2634 | 0.079295154 | 0.0038186524 | 4.376097 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPN1 | 6 | 1222 | 0.030837004 | 0.0017723764 | 4.120906 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM42 | 1 | 407 | 0.008810573 | 0.0005912752 | 3.897334 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPC3 | 5 | 1234 | 0.026431718 | 0.0017897669 | 3.884427 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC5 | 10 | 2400 | 0.048458150 | 0.0034795387 | 3.799771 | AGAAAA,AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2A | 29 | 6871 | 0.132158590 | 0.0099589296 | 3.730136 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,GAAAGA,GAAGAA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF11 | 2 | 688 | 0.013215859 | 0.0009985015 | 3.726362 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 489 | 0.008810573 | 0.0007101099 | 3.633121 | ACAAGA | ACAAGA,CAAGAC |
| PABPC1 | 17 | 4443 | 0.079295154 | 0.0064402624 | 3.622041 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SNRNP70 | 4 | 1237 | 0.022026432 | 0.0017941145 | 3.617892 | AAUCAA,AUCAAG,AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC4 | 4 | 1251 | 0.022026432 | 0.0018144033 | 3.601669 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBM3 | 7 | 2152 | 0.035242291 | 0.0031201361 | 3.497627 | AAAACG,AAAACU,AAACGA,AAACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| IGHMBP2 | 2 | 813 | 0.013215859 | 0.0011796520 | 3.485837 | AAAAAA | AAAAAA |
| TRA2B | 18 | 7329 | 0.083700441 | 0.0106226650 | 2.978089 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AGAAGA,GAAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF10 | 33 | 13860 | 0.149779736 | 0.0200874160 | 2.898479 | AAAAGA,AAAGAA,AAGAAA,AAGAAG,AAGAGG,AAGGAA,AGACAA,AGAGGA,GAAAGA,GAGAAA,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| YBX2 | 2 | 1480 | 0.013215859 | 0.0021462711 | 2.622366 | AACAUC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 2 | 1498 | 0.013215859 | 0.0021723567 | 2.604937 | AGAGGA,AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPM | 1 | 999 | 0.008810573 | 0.0014492040 | 2.603975 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 1 | 1053 | 0.008810573 | 0.0015274610 | 2.528100 | GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| HNRNPA3 | 3 | 2140 | 0.017621145 | 0.0031027457 | 2.505690 | AGGAGC,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPA2B1 | 15 | 8607 | 0.070484581 | 0.0124747476 | 2.498297 | AAGAAG,AAGGAA,ACUAGA,AGGAAC,AGGAGC,CAAGAA,CAAGGA,CUAGAC,UAGACA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SYNCRIP | 2 | 1634 | 0.013215859 | 0.0023694485 | 2.479647 | AAAAAA | AAAAAA,UUUUUU |
| AGO2 | 2 | 1830 | 0.013215859 | 0.0026534924 | 2.316306 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF4 | 5 | 3740 | 0.026431718 | 0.0054214720 | 2.285514 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPU | 2 | 1965 | 0.013215859 | 0.0028491350 | 2.213674 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| IGF2BP3 | 6 | 4815 | 0.030837004 | 0.0069793662 | 2.143495 | AAAAAC,AAAACA,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ZFP36 | 40 | 28400 | 0.180616740 | 0.0411588414 | 2.133657 | AAAAAA,AAAAAC,AAAAAG,AAAACA,AAAAGA,AAAAGC,AAAGAA,AAAGCA,AACAAG,AAGAAA,AAGCAA,AAGGAA,ACAAGG,AGAAAG,CAAGGA | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| KHDRBS1 | 4 | 4064 | 0.022026432 | 0.0058910141 | 1.902648 | CUAAAA,UAAAAA,UAAAAG | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBMX | 5 | 4925 | 0.026431718 | 0.0071387787 | 1.888521 | AAGAAG,AAGGAA,AUCCCA,GUAACA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| G3BP2 | 1 | 1644 | 0.008810573 | 0.0023839405 | 1.885888 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPAB | 1 | 1782 | 0.008810573 | 0.0025839306 | 1.769668 | AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| IGF2BP2 | 8 | 8408 | 0.039647577 | 0.0121863560 | 1.701966 | AAAAAC,AAAACA,AAAUCA,GAAAUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| HNRNPD | 2 | 2837 | 0.013215859 | 0.0041128408 | 1.684063 | AAAAAA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SRSF5 | 7 | 8869 | 0.035242291 | 0.0128544391 | 1.455041 | AACAGC,AAGAAG,AGAAGA,CACAGA,GAAGAA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| FXR2 | 2 | 3434 | 0.013215859 | 0.0049780156 | 1.408628 | AGACAA,GACAAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RBM5 | 5 | 6879 | 0.026431718 | 0.0099705232 | 1.406529 | AAAAAA,AAGGAA,AGGUAA,CAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| FUS | 2 | 3648 | 0.013215859 | 0.0052881452 | 1.321437 | AAAAAA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPA1 | 13 | 18070 | 0.061674009 | 0.0261885646 | 1.235726 | AAAAAA,AAGAGG,ACUAGA,AGAGGA,AGGAGC,CAAGGA,CUAGAC,GAGGAG,GGAGGA,UAGACA | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| LIN28A | 2 | 4315 | 0.013215859 | 0.0062547643 | 1.079243 | GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| DAZAP1 | 3 | 5964 | 0.017621145 | 0.0086445016 | 1.027453 | AAAAAA,AGGUAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| RBM46 | 2 | 4554 | 0.013215859 | 0.0066011240 | 1.001487 | AAUCAA,AUCAAG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
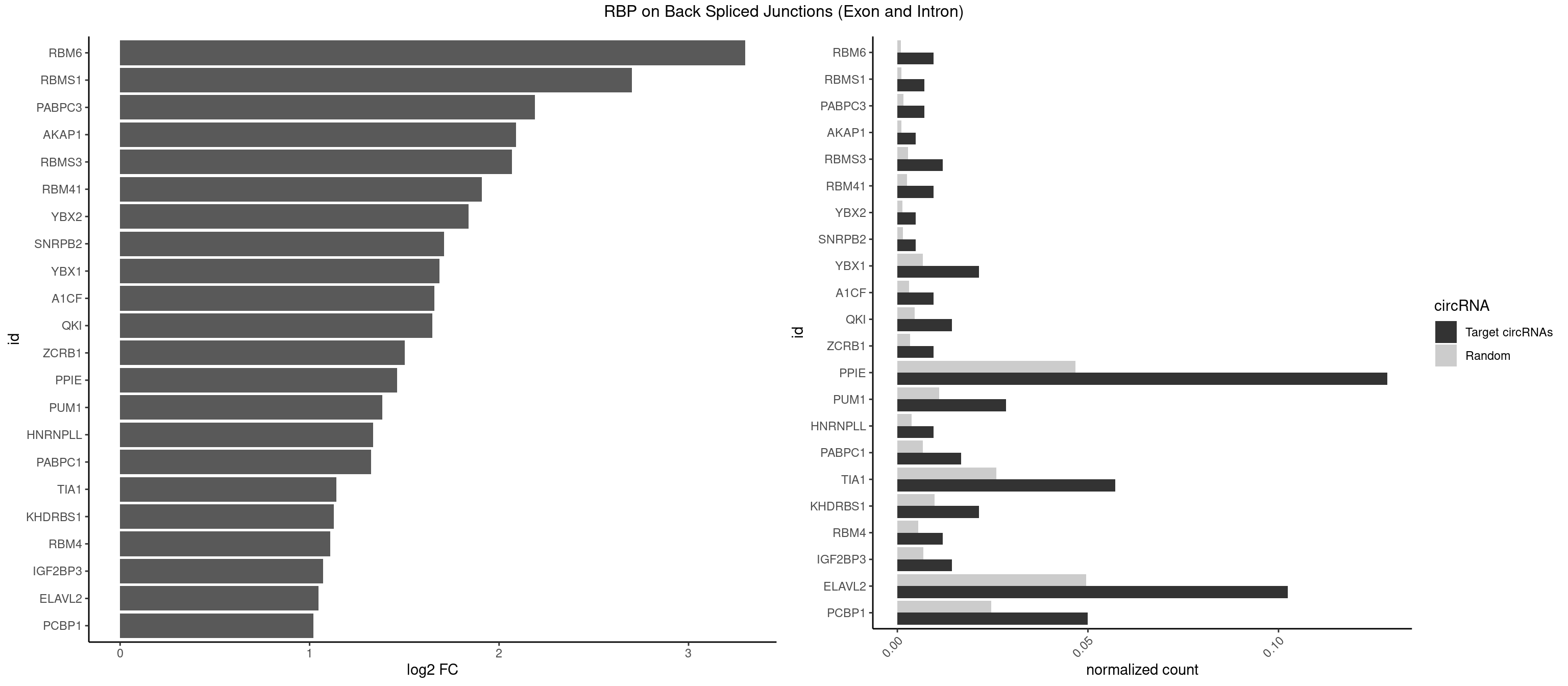
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM6 | 3 | 191 | 0.009523810 | 0.0009673519 | 3.299426 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAC,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAAAC,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | UAUAUA | AUAUAU,UAUAUA |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,CAUAUA,UAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACAUU,UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | AACAUC | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| YBX1 | 8 | 1321 | 0.021428571 | 0.0066606207 | 1.685807 | AACAUC,ACACCA,AUCAUC,CAUCAU,CCACAA,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AGUAUA,AUCAGU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AUCUAA,AUUAAC,CUCAUA,UCAUAU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,ACUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PPIE | 53 | 9262 | 0.128571429 | 0.0466696896 | 1.462012 | AAAAUA,AAAAUU,AAAUAU,AAUAUA,AAUAUU,AAUUAA,AUAAAA,AUAUAA,AUAUUA,AUAUUU,AUUAAA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAUUA,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUAU,UUUAAU,UUUAUA,UUUUAA,UUUUAU,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUAUU,CAGAAU,CCAGAA,UAAAUA,UAAUGU,UAUAUA,UGUAAU,UGUCCA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACAAAC,ACACCA,ACAUAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | AAAAAC,ACAAAC,CAAACC,CAAAUA,CUAAUA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUCUCC,UUUUAU,UUUUGU,UUUUUA | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUAAAA,CAAAAU,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CUUCCU,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,AAAUCA,AAAUUC,ACAAAC,ACAUAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ELAVL2 | 42 | 9826 | 0.102380952 | 0.0495112858 | 1.048118 | AUAUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUCU,CUUUUA,GUUUUU,UAUAUA,UAUAUU,UAUUUU,UCAUUU,UCUUUU,UGAUUU,UUAGUU,UUAUAU,UUCAUU,UUUAAU,UUUAUA,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PCBP1 | 20 | 4891 | 0.050000000 | 0.0246473196 | 1.020497 | AAACCA,AAAUUC,AACCAA,AAUUAA,AAUUCC,ACCAAA,AUUAAA,AUUCCA,CAAACC,CAUACC,CCCUCU,CCCUUA,CCUAUC,CCUCUU,CCUUUC,CUUAAA,CUUUCC | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.