circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000102606:+:13:111217678:111244294
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000102606:+:13:111217678:111244294 | ENSG00000102606 | ENST00000317133 | + | 13 | 111217679 | 111244294 | 482 | GACAUGACCGAUAAUAGCAACAAUCAACUGGUAGUAAGAGCAAAGUUUAACUUCCAGCAGACCAAUGAGGACGAGCUUUCCUUCUCAAAAGGAGACGUCAUCCAUGUCACCCGUGUGGAAGAGGGAGGCUGGUGGGAGGGCACACUCAACGGCCGGACCGGCUGGUUCCCCAGCAACUACGUGCGCGAGGUCAAGGCCAGCGAGAAGCCUGUGUCUCCCAAAUCAGGAACACUGAAGAGCCCUCCCAAAGGAUUUGAUACGACUGCCAUAAACAAAAGCUAUUACAAUGUGGUGCUACAGAAUAUUUUAGAAACAGAAAAUGAAUAUUCUAAAGAACUUCAGACUGUGCUUUCAACGUACCUACGGCCAUUGCAGACCAGUGAGAAGUUAAGUUCAGCAAACAUUUCAUAUUUAAUGGGAAAUCUAGAAGAAAUAUGUUCUUUCCAGCAAAUGCUCGUACAGUCUUUAGAAGAAUGCACCAAGACAUGACCGAUAAUAGCAACAAUCAACUGGUAGUAAGAGCAAAGUUUAA | circ |
| ENSG00000102606:+:13:111217678:111244294 | ENSG00000102606 | ENST00000317133 | + | 13 | 111217679 | 111244294 | 22 | GAAUGCACCAAGACAUGACCGA | bsj |
| ENSG00000102606:+:13:111217678:111244294 | ENSG00000102606 | ENST00000317133 | + | 13 | 111217479 | 111217688 | 210 | GUGAAGAGGGUGUCCUCUGGGGCUGGAGUGGUGCUGGUGGUGGUCAUGGGGCACUGGAAGGGAGGAAACUGAAAUUUCUCUAAAUGUAUUUCUACUGUUGGGCAAUUAUGCUACCUUUGUACUAGUAAAGUAUAAUCCAUUUUAUAAUGAAGUAGACUGUUCUUGGUAUAAUUUUUCUCCCACUCUUCUUCCCCUUUUAGGACAUGACCG | ie_up |
| ENSG00000102606:+:13:111217678:111244294 | ENSG00000102606 | ENST00000317133 | + | 13 | 111244285 | 111244494 | 210 | AAUGCACCAAGUAAGUAAGAUGCUAAAAAUUUGCAACACUCAGGUUGGUAUAAUUGGUAUUUAAAGGAUUAAGUGUGAAGUAAUUUUAGAAUUCACAUUUCUAAAGAUGCAAACUUAAGUUAUUUUAAAUCUUUUGGGUAAAUCAGCUUUUACAGUAAAAGCUGUAUUUUAGACUGUCUAGUUUGUAUAGCAGGAAUCUUCAGAGAAAGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
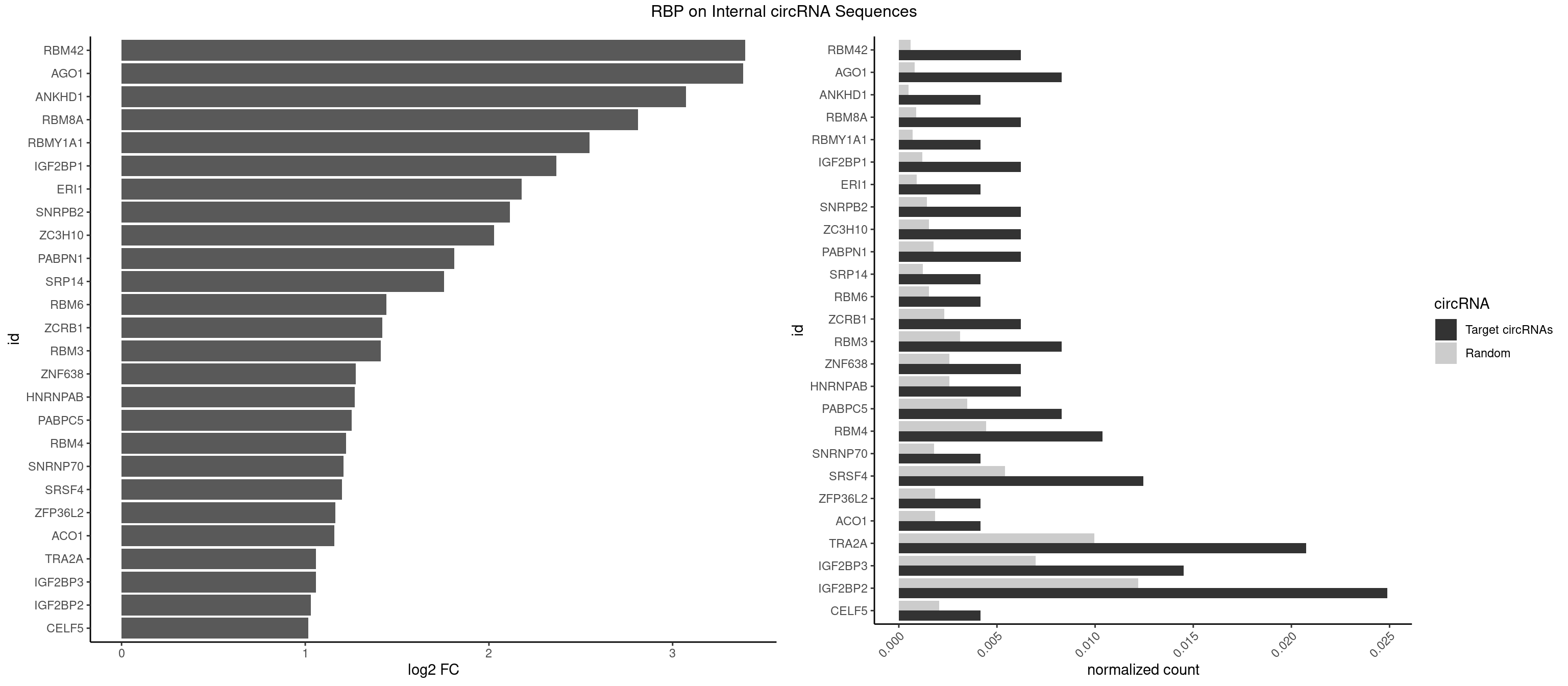
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 2 | 407 | 0.006224066 | 0.0005912752 | 3.395956 | AACUAC,ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| AGO1 | 3 | 548 | 0.008298755 | 0.0007956130 | 3.382756 | GGUAGU,GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ANKHD1 | 1 | 339 | 0.004149378 | 0.0004927293 | 3.074028 | AGACGU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM8A | 2 | 611 | 0.006224066 | 0.0008869128 | 2.810993 | GUGCGC,UGCGCG | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBMY1A1 | 1 | 489 | 0.004149378 | 0.0007101099 | 2.546781 | CAAGAC | ACAAGA,CAAGAC |
| IGF2BP1 | 2 | 831 | 0.006224066 | 0.0012057377 | 2.367941 | ACCCGU,CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ERI1 | 1 | 632 | 0.004149378 | 0.0009173461 | 2.177357 | UUCAGA | UUCAGA,UUUCAG |
| SNRPB2 | 2 | 991 | 0.006224066 | 0.0014376103 | 2.114185 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZC3H10 | 2 | 1053 | 0.006224066 | 0.0015274610 | 2.026722 | CAGCGA,CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 2 | 1222 | 0.006224066 | 0.0017723764 | 1.812172 | AGAAGA | AAAAGA,AGAAGA |
| SRP14 | 1 | 847 | 0.004149378 | 0.0012289250 | 1.755498 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM6 | 1 | 1054 | 0.004149378 | 0.0015289102 | 1.440391 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZCRB1 | 2 | 1605 | 0.006224066 | 0.0023274215 | 1.419125 | AUUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM3 | 3 | 2152 | 0.008298755 | 0.0031201361 | 1.411286 | AUACGA,GAGACG,GAUACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ZNF638 | 2 | 1773 | 0.006224066 | 0.0025708878 | 1.275591 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 2 | 1782 | 0.006224066 | 0.0025839306 | 1.268290 | AAGACA,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 | 3 | 2400 | 0.008298755 | 0.0034795387 | 1.253999 | AGAAAA,AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM4 | 4 | 3065 | 0.010373444 | 0.0044432593 | 1.223205 | CCUUCU,GCGCGA,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SNRNP70 | 1 | 1237 | 0.004149378 | 0.0017941145 | 1.209623 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SRSF4 | 5 | 3740 | 0.012448133 | 0.0054214720 | 1.199173 | AGAAGA,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ZFP36L2 | 1 | 1277 | 0.004149378 | 0.0018520827 | 1.163746 | UAUUUA | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ACO1 | 1 | 1283 | 0.004149378 | 0.0018607779 | 1.156989 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| TRA2A | 9 | 6871 | 0.020746888 | 0.0099589296 | 1.058832 | AAAGAA,AAGAAA,AAGAGG,AGAAGA,GAAGAA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| IGF2BP3 | 6 | 4815 | 0.014522822 | 0.0069793662 | 1.057154 | AAAUCA,ACAAUC,ACACUC,CAAACA,CAAUCA,CACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 11 | 8408 | 0.024896266 | 0.0121863560 | 1.030663 | AAAUCA,ACAAUC,ACACUC,CAAACA,CAAAUC,CAAUCA,CACUCA,GAAAUC,GAACAC,GCAAAC,GCACAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| CELF5 | 1 | 1415 | 0.004149378 | 0.0020520728 | 1.015813 | GUGUGG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
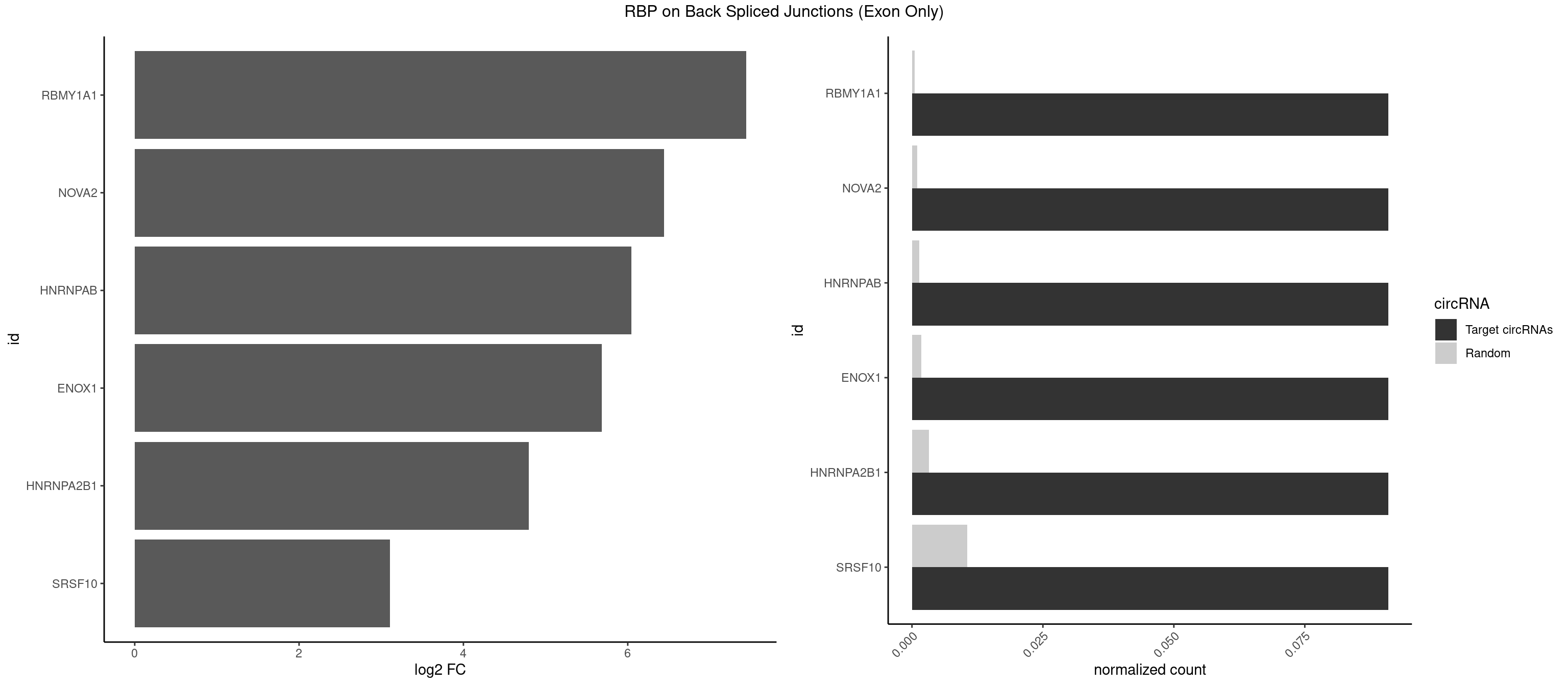
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | CAAGAC | ACAAGA,CAAGAC |
| NOVA2 | 1 | 15 | 0.09090909 | 0.0010479434 | 6.438792 | AGACAU | ACCACC,AGACAU,AGAUCA,AGGCAU,AGUCAU,CUAGAU,GAGACA,GAGGCA,GAGUCA,UAGAUC |
| HNRNPAB | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA |
| ENOX1 | 1 | 26 | 0.09090909 | 0.0017684045 | 5.683904 | AAGACA | AAGACA,AGACAG,AGGACA,AGUACA,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,UAGACA,UGGACA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | CCAAGA | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | AAGACA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
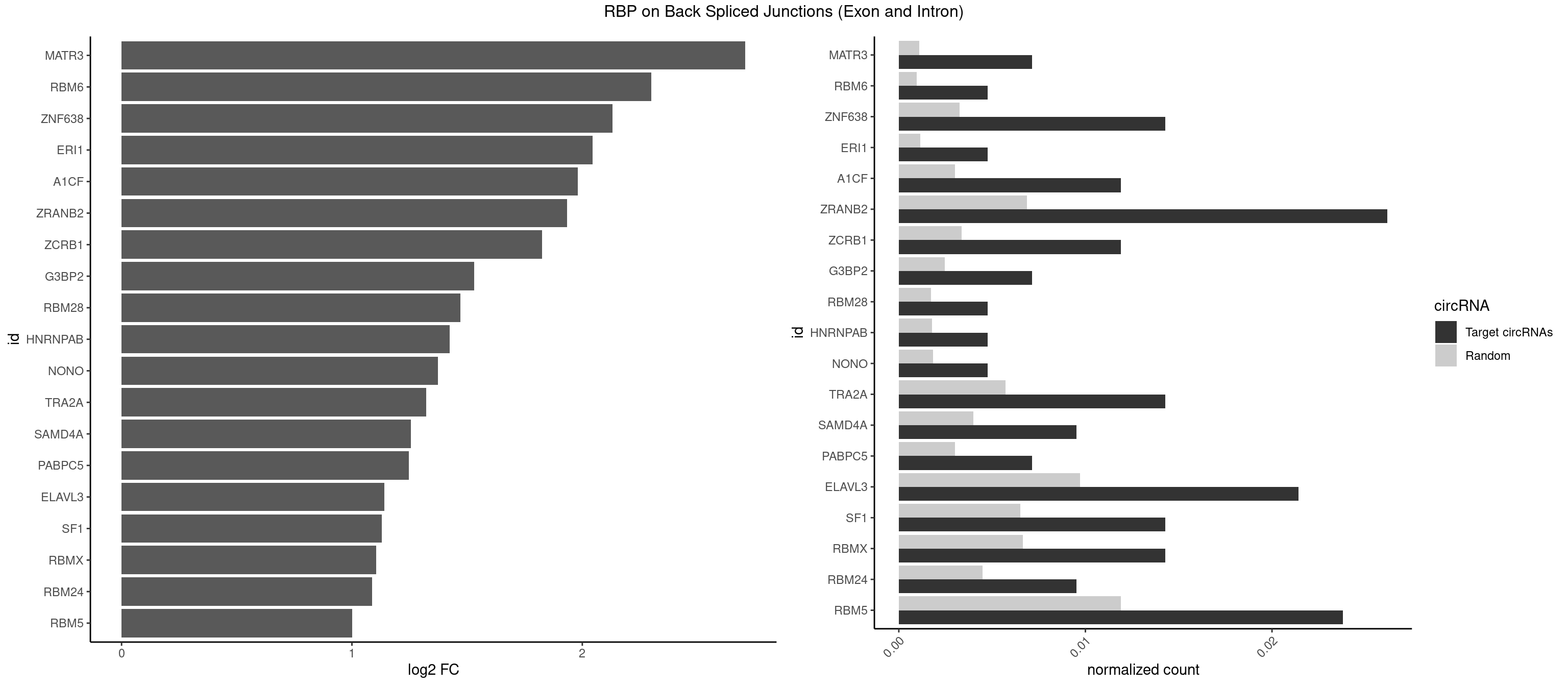
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | AAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZNF638 | 5 | 646 | 0.014285714 | 0.0032597743 | 2.131729 | GGUUGG,GUUCUU,GUUGGU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AGUAUA,AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZRANB2 | 10 | 1360 | 0.026190476 | 0.0068571141 | 1.933369 | GGGUAA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | ACUUAA,AUUUAA,GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AAGAGG,AGAAAG,GAAAGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAA,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUUA,UAUUUA,UAUUUU,UUAUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | AGUAAG,UACUAG,UAGUAA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AAGUAA,AAGUGU,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | AGUGUG,GAGUGG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AGGGAG,AGGGUG,CUCUUC,GAAGGG,GAGGGU,GGUGGU,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.