circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000141556:+:17:82756164:82800996
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141556:+:17:82756164:82800996 | ENSG00000141556 | ENST00000681983 | + | 17 | 82756165 | 82800996 | 766 | UAAUAAUGGACAAAUACCAGGAGCAGCCUCAUCUGUUGGACCCGCACCUUGAAUGGAUGAUGAACUUGUUGUUGGACAUAGUGCAAGAUCAGACAUCUCCAGCUUCCCUUGUACAUCUGGCUUUUAAAUUUCUUUACAUCAUCACCAAGGUUCGAGGCUAUAAAACAUUUCUUCGUUUAUUUCCUCAUGAAGUUGCCGAUGUAGAGCCUGUUUUAGAUUUGGUCACAAUUCAGAAUCCCAAGGACCAUGAAGCUUGGGAAACCCGCUACAUGCUUUUGCUCUGGCUCUCCGUGACCUGCCUGAUCCCUUUUGAUUUUUCUCGCCUUGACGGGAACCUCCUCACCCAGCCUGGGCAAGCACGAAUGUCCAUAAUGGACCGUAUUCUCCAAAUAGCAGAGUCCUACUUGAUUGUCAGUGACAAGGCCCGAGAUGCAGCUGCUGUCCUUGUGUCCAGAUUUAUCACACGUCCUGAUGUCAAGCAAAGCAAGAUGGCUGAGUUCCUGGACUGGAGCCUGUGCAAUCUGGCCCGUUCCUCCUUCCAGACCAUGCAGGGGGUCAUCACCAUGGAUGGGACGCUGCAGGCCCUGGCACAAAUAUUUAAACAUGGAAAACGUGAAGACUGUUUGCCCUAUGCUGCCACUGUCCUCAGGUGCCUCGAUGGCUGCAGACUCCCUGAGAGCAACCAGACCCUGCUGCGGAAGCUGGGGGUGAAGCUUGUGCAGCGACUGGGGCUGACAUUCCUGAAGCCGAAGGUGGCAGCAUGGAGUAAUAAUGGACAAAUACCAGGAGCAGCCUCAUCUGUUGGACCCGCACCUU | circ |
| ENSG00000141556:+:17:82756164:82800996 | ENSG00000141556 | ENST00000681983 | + | 17 | 82756165 | 82800996 | 22 | GCAGCAUGGAGUAAUAAUGGAC | bsj |
| ENSG00000141556:+:17:82756164:82800996 | ENSG00000141556 | ENST00000681983 | + | 17 | 82755965 | 82756174 | 210 | UCAAAAGGCUAGAGGGAGGUGGGGCAUUCCCUCCAGGUGGCCUGUGUGGCCUCCUUUAGUGAGAGGGGAGGUGUGCUCUUGAGAGCUGGGGAAGAGGUGGAGCUGCCCUGUGGAAGCUAGCAAGGGAAAAUGGGGUUUCUGAGGCUCAUGGGUAUGUUUGGCCUAGUGAUAAUUAAUUUUCAUGUUAUAUUUGUUUUUAGUAAUAAUGGA | ie_up |
| ENSG00000141556:+:17:82756164:82800996 | ENSG00000141556 | ENST00000681983 | + | 17 | 82800987 | 82801196 | 210 | CAGCAUGGAGGUAGGCACCAUGAGGGCGGUGCCUGGGGAGGGCCACGGGGUGGGGAGGGGUUGCUGUGGGGGGCAGGCGCCAUUGAUGAGGGCGGGGCAGGUGCUGUUGGGGCAGGUGCUGUGGGGAGGCAGUCGCCCUGUUGCGGGGUGGGGGAGGCAGGCGCCAUGGGGUGGGGAGGCAGGCGCCUUGGUUCUGGACACAGUUGGGGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
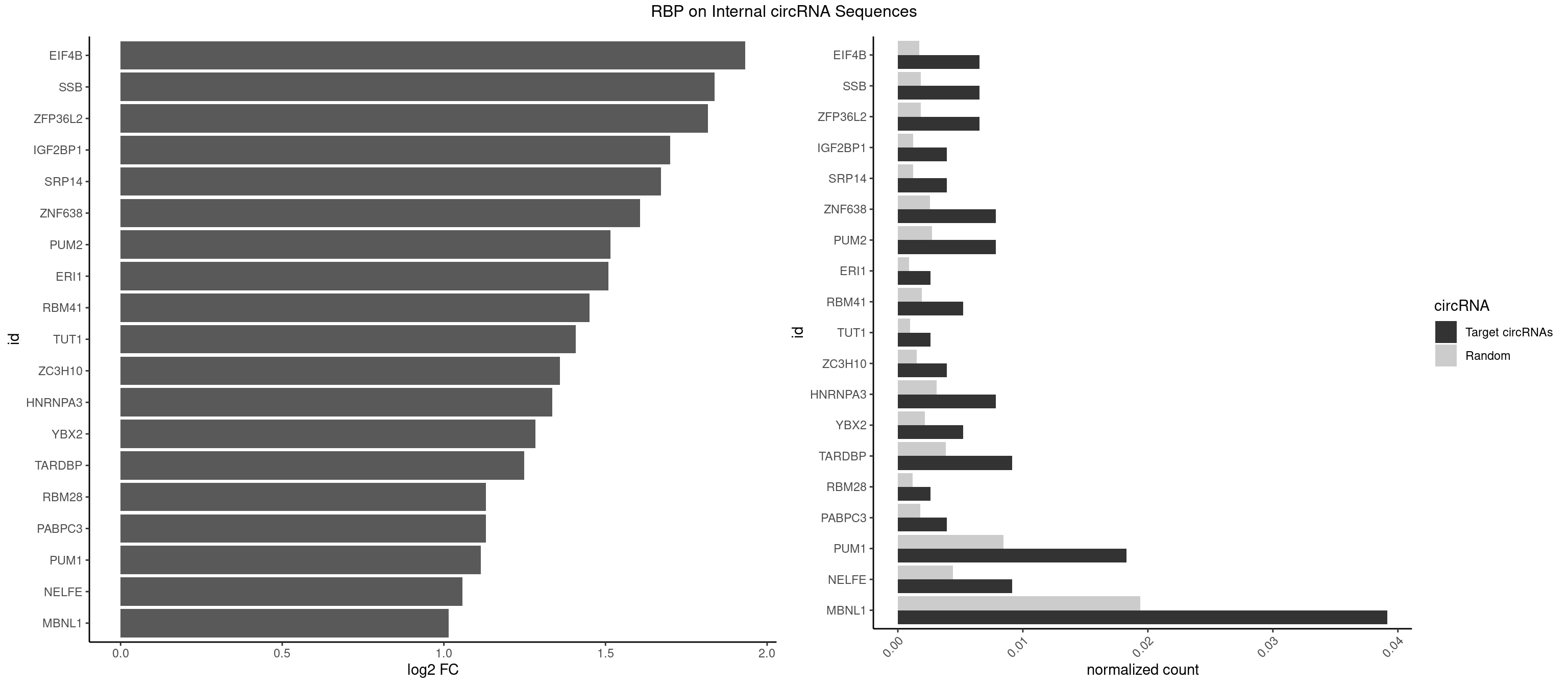
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| EIF4B | 4 | 1179 | 0.006527415 | 0.0017100607 | 1.932464 | GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SSB | 4 | 1260 | 0.006527415 | 0.0018274462 | 1.836683 | CUGUUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ZFP36L2 | 4 | 1277 | 0.006527415 | 0.0018520827 | 1.817363 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| IGF2BP1 | 2 | 831 | 0.003916449 | 0.0012057377 | 1.699630 | AAGCAC,CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRP14 | 2 | 847 | 0.003916449 | 0.0012289250 | 1.672149 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ZNF638 | 5 | 1773 | 0.007832898 | 0.0025708878 | 1.607280 | GGUUCG,GUUGUU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PUM2 | 5 | 1890 | 0.007832898 | 0.0027404447 | 1.515136 | GUACAU,UACAUC,UGUACA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ERI1 | 1 | 632 | 0.002610966 | 0.0009173461 | 1.509046 | UUCAGA | UUCAGA,UUUCAG |
| RBM41 | 3 | 1318 | 0.005221932 | 0.0019115000 | 1.449878 | UACAUG,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TUT1 | 1 | 678 | 0.002610966 | 0.0009840095 | 1.407840 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ZC3H10 | 2 | 1053 | 0.003916449 | 0.0015274610 | 1.358411 | CAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| HNRNPA3 | 5 | 2140 | 0.007832898 | 0.0031027457 | 1.336001 | AGGAGC,CAAGGA,CCAAGG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| YBX2 | 3 | 1480 | 0.005221932 | 0.0021462711 | 1.282751 | ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| TARDBP | 6 | 2654 | 0.009138381 | 0.0038476365 | 1.247966 | GAAUGG,GAAUGU,GUUGUU,UGAAUG,UUGUGC,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM28 | 1 | 822 | 0.002610966 | 0.0011926949 | 1.130359 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPC3 | 2 | 1234 | 0.003916449 | 0.0017897669 | 1.129775 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PUM1 | 13 | 5823 | 0.018276762 | 0.0084401638 | 1.114668 | AAUAUU,CAGAAU,CUUGUA,GUAAUA,GUACAU,GUCCAG,UACAUC,UGUACA,UGUAGA,UGUCCA,UUGUAC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| NELFE | 6 | 3028 | 0.009138381 | 0.0043896388 | 1.057836 | CUCUGG,CUGGCU,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| MBNL1 | 29 | 13372 | 0.039164491 | 0.0193802045 | 1.014962 | ACGCUG,AUGCUU,CCCGCU,CCGCUA,CCUGCU,CGCUGC,CUGCCA,CUGCCU,CUGCGG,CUGCUG,CUUGUG,GCUGCG,GCUGCU,GCUUGU,GCUUUU,GGCUUU,UCUCGC,UGCUGC,UGCUGU,UGCUUU,UUGCCC,UUGCCG,UUGCUC,UUGUGC,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
RBP on BSJ (Exon Only)
Plot
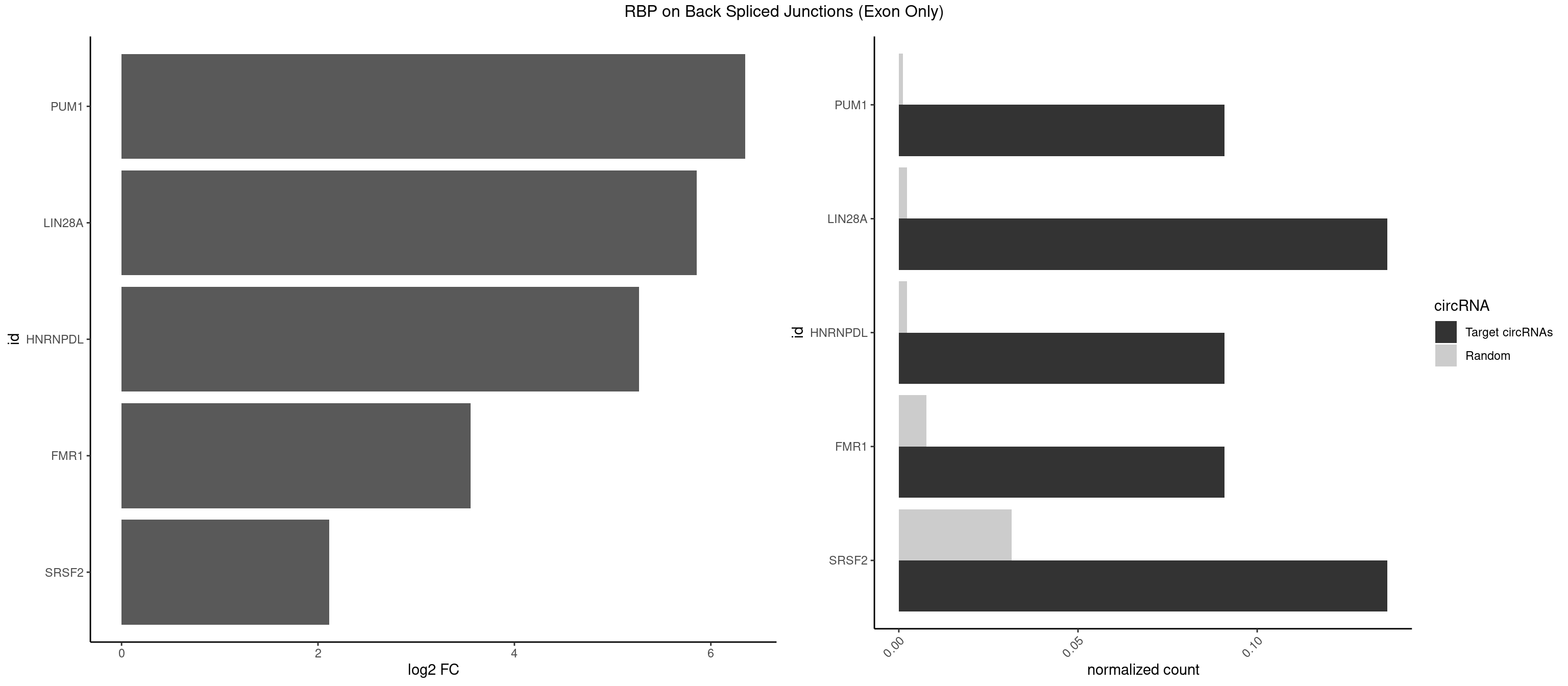
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM1 | 1 | 16 | 0.09090909 | 0.001113440 | 6.351329 | GUAAUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| LIN28A | 2 | 35 | 0.13636364 | 0.002357873 | 5.853829 | GGAGUA,UGGAGU | AGGAGA,AGGAGU,CGGAGG,GGAGAA,GGAGAU,GGAGGA,GGAGGG,UGGAGA,UGGAGG,UGGAGU |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | GGAGUA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | UGGAGU | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF2 | 2 | 479 | 0.13636364 | 0.031438302 | 2.116864 | GGAGUA,UGGAGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
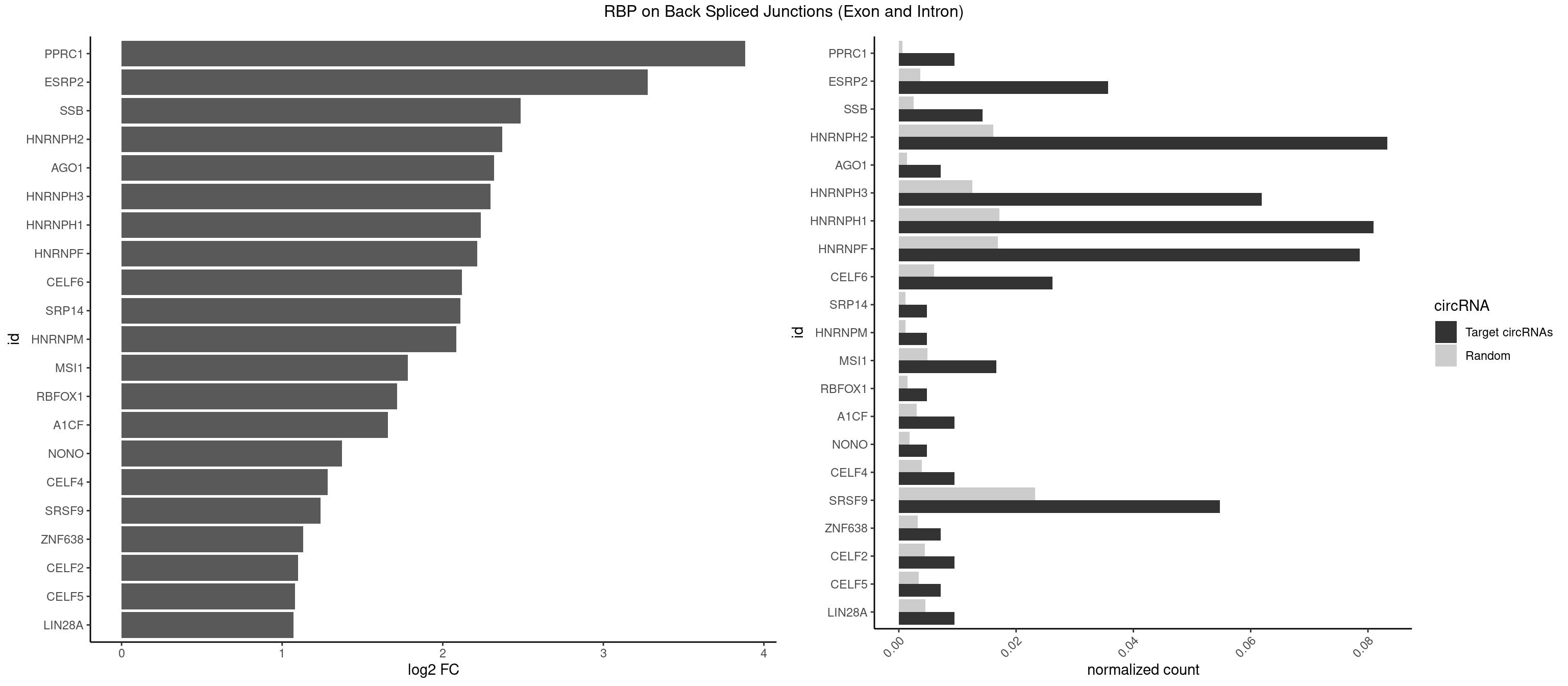
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PPRC1 | 3 | 127 | 0.009523810 | 0.0006449012 | 3.884389 | GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| ESRP2 | 14 | 731 | 0.035714286 | 0.0036880290 | 3.275579 | GGGAAA,GGGAAG,GGGGAA,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SSB | 5 | 505 | 0.014285714 | 0.0025493753 | 2.486358 | GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPH2 | 34 | 3198 | 0.083333333 | 0.0161174929 | 2.370266 | AAGGGA,AGGGAA,AGGGGA,CUGGGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGGAA,GGGGAG,GGGGGA,GGGGGC,GGGGGG,UGGGGG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPH3 | 25 | 2496 | 0.061904762 | 0.0125806127 | 2.298848 | AAGGGA,AGGGAA,AGGGGA,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGGAG,GGGGGC,GGGGGG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 33 | 3406 | 0.080952381 | 0.0171654575 | 2.237565 | AAGGGA,AGGGAA,AGGGGA,CUGGGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGGAA,GGGGAG,GGGGGC,GGGGGG,UGGGGG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPF | 32 | 3360 | 0.078571429 | 0.0169336961 | 2.214108 | AAGGGA,AGGGAA,AGGGGA,AUGGGG,CUGGGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGGAA,GGGGAG,GGGGGC,GGGGGG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| CELF6 | 10 | 1196 | 0.026190476 | 0.0060308343 | 2.118613 | GUGGGG,GUGUGG,UGUGGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| MSI1 | 6 | 961 | 0.016666667 | 0.0048468360 | 1.781850 | AGGUAG,AGGUGG,AGUUGG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GGUGUG,GUGUGG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SRSF9 | 22 | 4608 | 0.054761905 | 0.0232214833 | 1.237713 | GGAAAA,GGAAGC,GGAGGC,GGGAGG,GGGUGG,GGUGCC,GGUGGA,GGUGGC,UGAGAG,UGGAGC,UGGAGG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGGG,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.