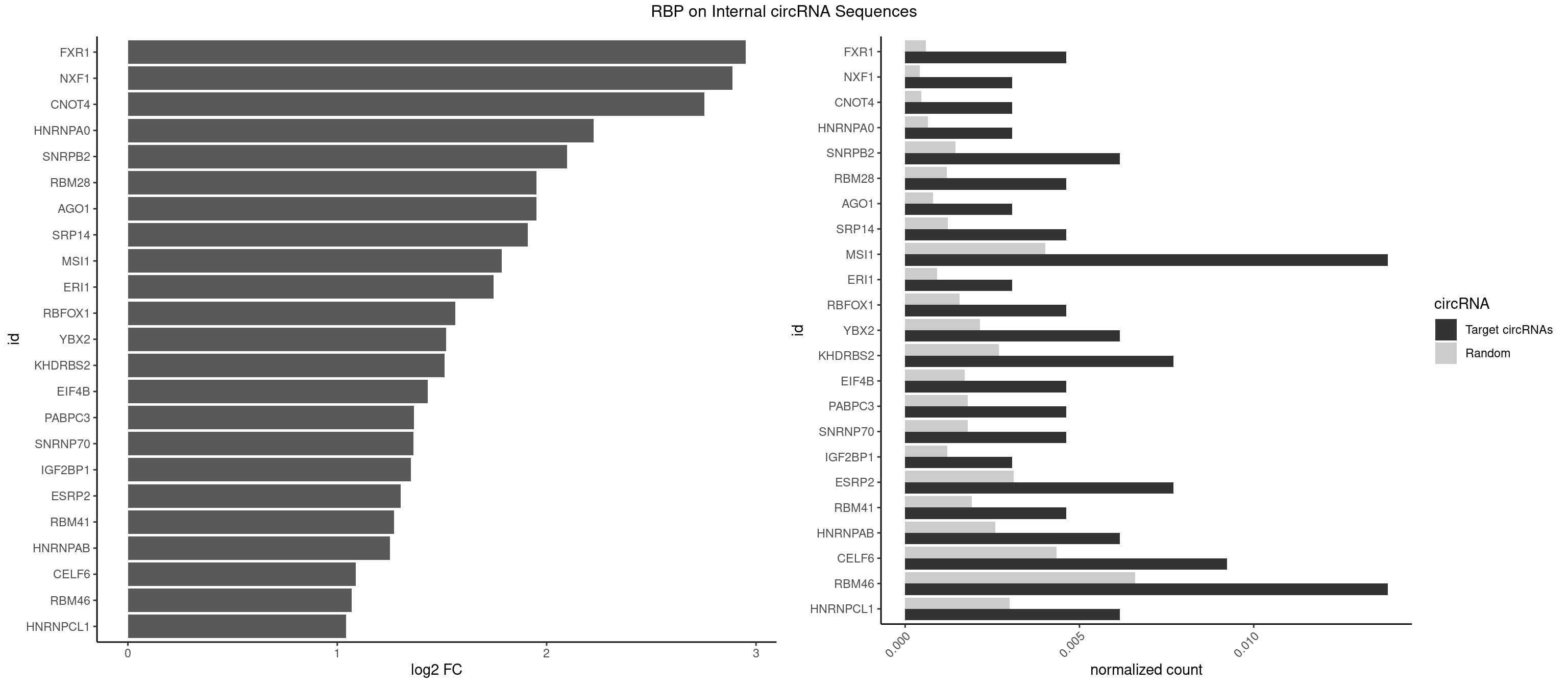
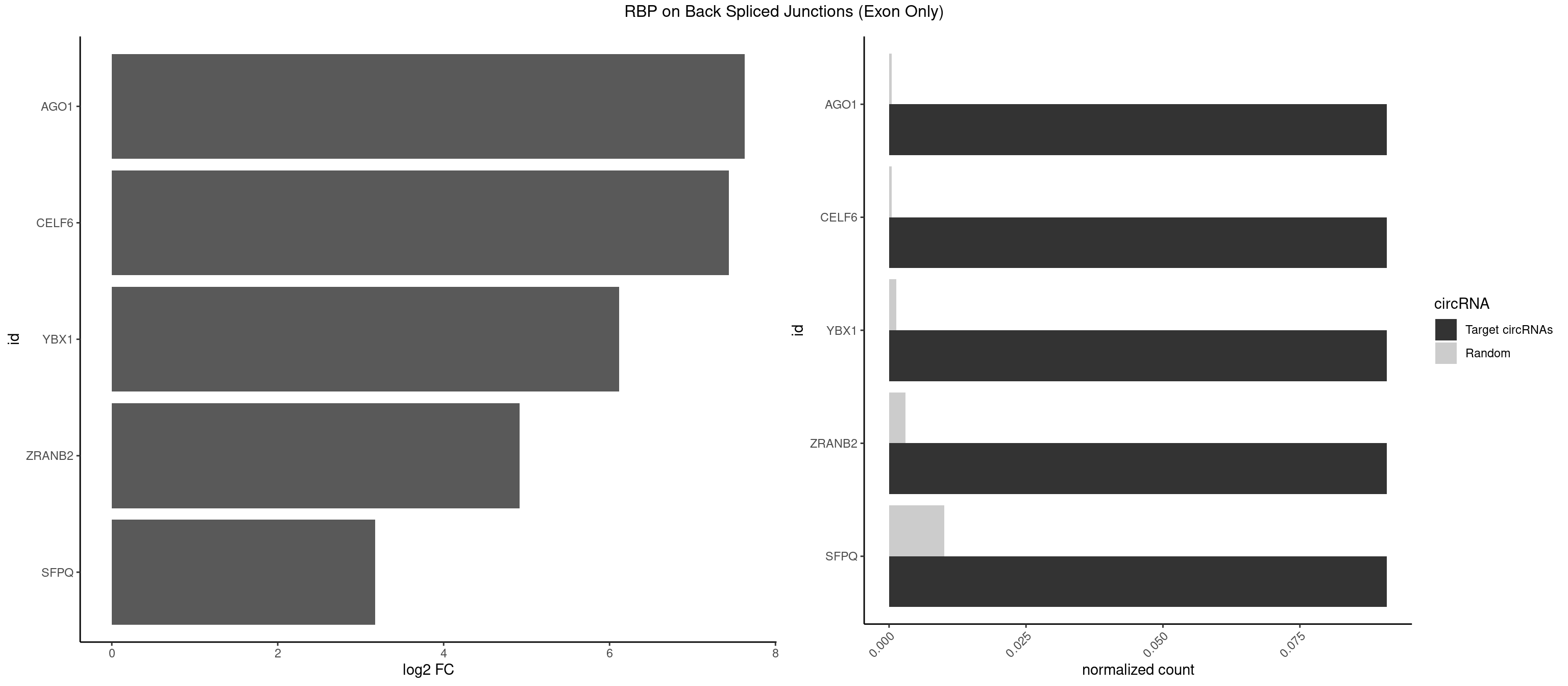
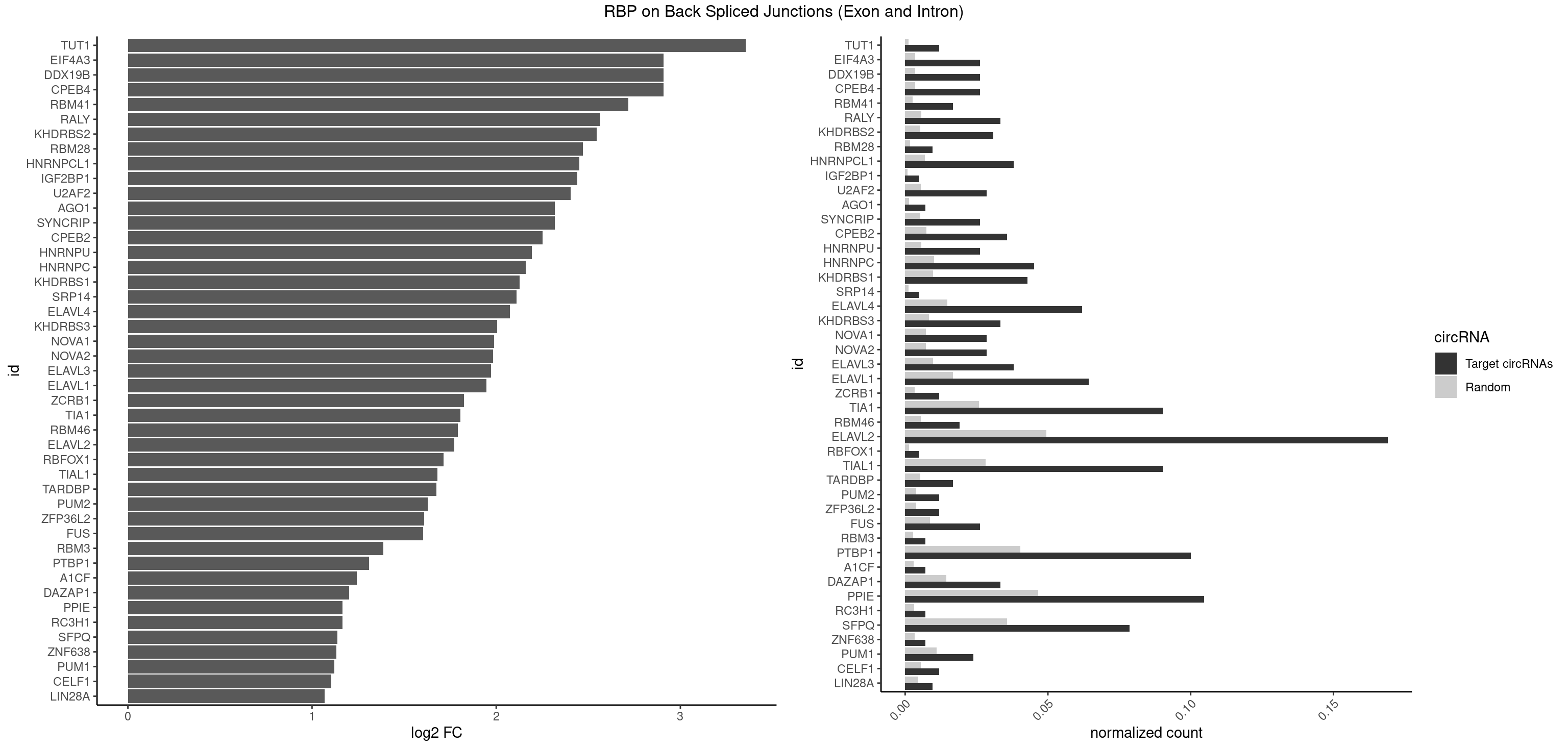
| TUT1 |
4 |
230 |
0.011904762 |
0.0011638452 |
3.354568 |
AAAUAC,AAUACU |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| CPEB4 |
10 |
691 |
0.026190476 |
0.0034864974 |
2.909192 |
UUUUUU |
UUUUUU |
| DDX19B |
10 |
691 |
0.026190476 |
0.0034864974 |
2.909192 |
UUUUUU |
UUUUUU |
| EIF4A3 |
10 |
691 |
0.026190476 |
0.0034864974 |
2.909192 |
UUUUUU |
UUUUUU |
| RBM41 |
6 |
502 |
0.016666667 |
0.0025342604 |
2.717329 |
AUACUU,UACUUG,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RALY |
13 |
1117 |
0.033333333 |
0.0056328094 |
2.565039 |
UUUUUC,UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| KHDRBS2 |
12 |
1051 |
0.030952381 |
0.0053002821 |
2.545909 |
AAUAAA,AUAAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM28 |
3 |
340 |
0.009523810 |
0.0017180572 |
2.470761 |
AGUAGA,GAGUAG,UGUAGG |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPCL1 |
15 |
1381 |
0.038095238 |
0.0069629182 |
2.451847 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| IGF2BP1 |
1 |
173 |
0.004761905 |
0.0008766626 |
2.441445 |
GCACCC |
AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| U2AF2 |
11 |
1071 |
0.028571429 |
0.0054010480 |
2.403262 |
UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| AGO1 |
2 |
283 |
0.007142857 |
0.0014308746 |
2.319604 |
GAGGUA,UGAGGU |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SYNCRIP |
10 |
1041 |
0.026190476 |
0.0052498992 |
2.318681 |
UUUUUU |
AAAAAA,UUUUUU |
| CPEB2 |
14 |
1487 |
0.035714286 |
0.0074969770 |
2.252120 |
AUUUUU,CCUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPU |
10 |
1136 |
0.026190476 |
0.0057285369 |
2.192804 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPC |
18 |
2006 |
0.045238095 |
0.0101118501 |
2.161491 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 |
17 |
1945 |
0.042857143 |
0.0098045143 |
2.128018 |
AUAAAA,UAAAAA,UUAAAA,UUUUAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SRP14 |
1 |
218 |
0.004761905 |
0.0011033857 |
2.109602 |
GCCUGU |
CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ELAVL4 |
25 |
2916 |
0.061904762 |
0.0146966949 |
2.074559 |
AUUUAU,UAUUUU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS3 |
13 |
1646 |
0.033333333 |
0.0082980653 |
2.006119 |
AAUAAA,AUAAAA,UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| NOVA1 |
11 |
1428 |
0.028571429 |
0.0071997179 |
1.988561 |
UCAGUC,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 |
11 |
1434 |
0.028571429 |
0.0072299476 |
1.982516 |
AGGCAU,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ELAVL3 |
15 |
1928 |
0.038095238 |
0.0097188634 |
1.970751 |
AUUUAU,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ELAVL1 |
26 |
3309 |
0.064285714 |
0.0166767432 |
1.946661 |
AUUUAU,UAGUUU,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAUUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ZCRB1 |
4 |
666 |
0.011904762 |
0.0033605401 |
1.824774 |
ACUUAA,GGAUUA,GGUUUA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| TIA1 |
37 |
5140 |
0.090476190 |
0.0259018541 |
1.804483 |
AUUUUC,AUUUUU,CUUUUA,CUUUUG,CUUUUU,GUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM46 |
7 |
1091 |
0.019047619 |
0.0055018138 |
1.791631 |
AAUGAA,AUCAAU,AUGAAU,AUGAUU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ELAVL2 |
70 |
9826 |
0.169047619 |
0.0495112858 |
1.771600 |
AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUUAU,AUUUUC,AUUUUU,CUUAUA,CUUUCU,CUUUUA,CUUUUU,GUUUUA,GUUUUU,UACUUU,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UCUUAU,UCUUUU,UGAUUU,UUAAUU,UUACUU,UUAUAU,UUAUUG,UUAUUU,UUCUUU,UUGUAU,UUUACU,UUUAUA,UUUAUG,UUUAUU,UUUCUU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBFOX1 |
1 |
287 |
0.004761905 |
0.0014510278 |
1.714464 |
GCAUGA |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TIAL1 |
37 |
5597 |
0.090476190 |
0.0282043531 |
1.681620 |
AAUUUU,AGUUUU,AUUUUC,AUUUUU,CUUUUA,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TARDBP |
6 |
1034 |
0.016666667 |
0.0052146312 |
1.676328 |
GAAUGA,GAAUGU,UGAAUG |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PUM2 |
4 |
764 |
0.011904762 |
0.0038542926 |
1.627001 |
UAAAUA,UGUAAA,UGUACA,UGUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZFP36L2 |
4 |
774 |
0.011904762 |
0.0039046755 |
1.608264 |
AUUUAU,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| FUS |
10 |
1711 |
0.026190476 |
0.0086255542 |
1.602353 |
UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| RBM3 |
2 |
541 |
0.007142857 |
0.0027307537 |
1.387202 |
AAUACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PTBP1 |
41 |
8003 |
0.100000000 |
0.0403264813 |
1.310201 |
ACUUUU,AUUUUC,AUUUUU,CCUUUU,CUCUUG,CUUUCC,CUUUCU,CUUUUU,GUCUUA,GUCUUU,UACUUU,UAUUUU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCUU,UUCUUU,UUUCUU,UUUUCU,UUUUUC,UUUUUU |
ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| A1CF |
2 |
598 |
0.007142857 |
0.0030179363 |
1.242939 |
UAAUUG,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| DAZAP1 |
13 |
2878 |
0.033333333 |
0.0145052398 |
1.200391 |
AGGUAA,UAGGAA,UAGUUU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| PPIE |
43 |
9262 |
0.104761905 |
0.0466696896 |
1.166556 |
AAAAAU,AAAAUA,AAAUAU,AAUAAA,AAUUUU,AUAAAA,AUAAAU,AUAUUU,AUUUAU,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAUAAA,UAUAUU,UAUUUU,UUAAAA,UUAAUU,UUAUAU,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| RC3H1 |
2 |
631 |
0.007142857 |
0.0031841999 |
1.165570 |
CCUUCU,CUUCUG |
CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SFPQ |
32 |
7087 |
0.078571429 |
0.0357114067 |
1.137620 |
AGAGAG,AGAGGU,CAGGCA,CUGGGA,GAAGCA,GAGGUA,GGAGAG,GGAGGA,GGUCUG,GUCUGG,UAAUUG,UAGUUU,UCUGGA,UGAUUG,UGAUUU,UGGAAG,UGGAGG,UGGUCU,UGGUUU,UUAAUU,UUGGUU,UUUUUU |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| ZNF638 |
2 |
646 |
0.007142857 |
0.0032597743 |
1.131729 |
GUUCUU,UGUUCU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PUM1 |
9 |
2172 |
0.023809524 |
0.0109482064 |
1.120844 |
AAUGUU,CUUGUA,UAAAUA,UGUAAA,UGUACA,UGUAUA,UUGUAC,UUGUAG |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CELF1 |
4 |
1097 |
0.011904762 |
0.0055320435 |
1.105654 |
GUUUGU,UGUGUU,UGUUUG,UUGUGU |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| LIN28A |
3 |
900 |
0.009523810 |
0.0045395002 |
1.069005 |
AGGAGA,GGAGGA,UGGAGG |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |