circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000092020:-:14:35107303:35110629
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000092020:-:14:35107303:35110629 | ENSG00000092020 | MSTRG.9376.1 | - | 14 | 35107304 | 35110629 | 387 | CUGCCUGCUGAAGAUGAAGUCUUACUACAGAAAUUAAGAGAGGAAUCAAGAGCUGUCUUUCUACAAAGAAAAAGCAGAGAACUGUUAGAUAAUGAAGAAUUACAGAACUUAUGGUUUUUGCUGGACAAACACCAGACACCACCUAUGAUUGGAGAGGAAGCGAUGAUCAAUUACGAAAACUUUUUGAAGGUUGGUGAAAAGGCUGGAGCAAAGUGCAAGCAAUUUUUCACAGCAAAAGUCUUUGCUAAACUCCUUCAUACAGAUUCAUAUGGAAGAAUUUCCAUCAUGCAGUUCUUUAAUUAUGUCAUGAGAAAAGUUUGGCUUCAUCAAACAAGAAUAGGACUCAGUUUAUAUGAUGUCGCUGGGCAGGGGUACCUUCGGGAAUCUCUGCCUGCUGAAGAUGAAGUCUUACUACAGAAAUUAAGAGAGGAAUCAAG | circ |
| ENSG00000092020:-:14:35107303:35110629 | ENSG00000092020 | MSTRG.9376.1 | - | 14 | 35107304 | 35110629 | 22 | UUCGGGAAUCUCUGCCUGCUGA | bsj |
| ENSG00000092020:-:14:35107303:35110629 | ENSG00000092020 | MSTRG.9376.1 | - | 14 | 35110620 | 35110829 | 210 | UUUUCAGGGCAGAGAUCCAGCCUUUCUGAUCACCGAUUCUCUGUGGAUCAGUUUUUGUUAAACAAAUGUUAAGCACAAUUAAGGUGAGAUACAAGAAAUAAUUCGAGGCGGUGGCAUACAGUUAUUUUAUGAGGCCACAUACAUUUUAUAGGAUCCAGUAGUCUAAAAUUUACAUGUAGAAAUUUACCUUGUAUUUUCAGCUGCCUGCUG | ie_up |
| ENSG00000092020:-:14:35107303:35110629 | ENSG00000092020 | MSTRG.9376.1 | - | 14 | 35107104 | 35107313 | 210 | UCGGGAAUCUGUAAGUGUUAACCUUAUAGAUAUUAUAUUAACUCUUAUAGACACUAAAAUGUAAACUGUAGUUAUCACUGGGUGUUGGGAUUACAGGCUGUUCUCAUAAUACUUUGUUUUAUUUCUGUCUGUGUGUGUUUGUGUGUACAUAUUUAGUUUUACUGUAACAAAGCAACUUGUACACUUUCAACUUUUAAAAUUGAACAUCAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
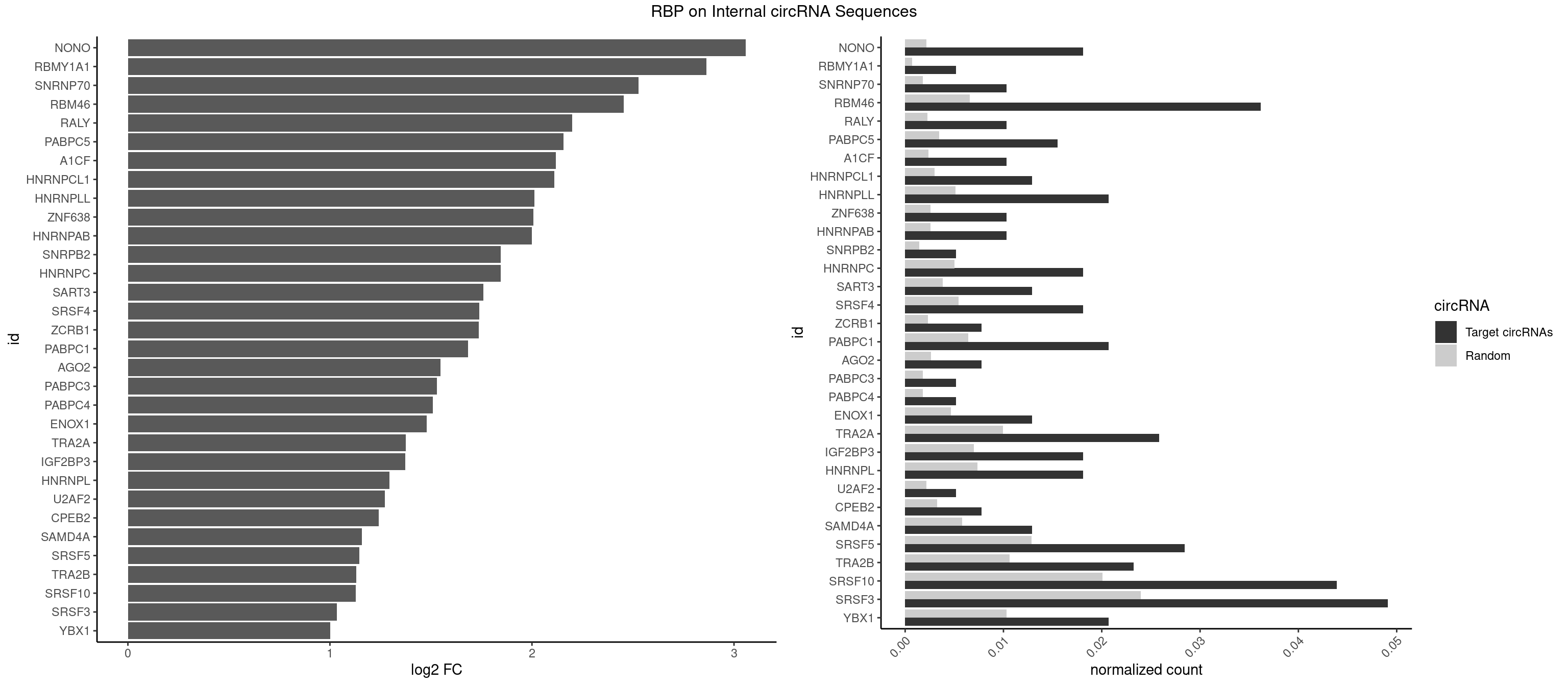
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NONO | 6 | 1498 | 0.018087855 | 0.0021723567 | 3.057688 | AGAGGA,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMY1A1 | 1 | 489 | 0.005167959 | 0.0007101099 | 2.863480 | ACAAGA | ACAAGA,CAAGAC |
| SNRNP70 | 3 | 1237 | 0.010335917 | 0.0017941145 | 2.526323 | AAUCAA,AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM46 | 13 | 4554 | 0.036175711 | 0.0066011240 | 2.454238 | AAUCAA,AAUGAA,AUCAAA,AUCAAG,AUCAAU,AUCAUG,AUGAAG,AUGAUG,AUGAUU,GAUCAA,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RALY | 3 | 1553 | 0.010335917 | 0.0022520629 | 2.198347 | UUUUUC,UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| PABPC5 | 5 | 2400 | 0.015503876 | 0.0034795387 | 2.155661 | AGAAAA,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 3 | 1642 | 0.010335917 | 0.0023810421 | 2.118001 | UAAUUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPCL1 | 4 | 2062 | 0.012919897 | 0.0029897078 | 2.111518 | AUUUUU,CUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPLL | 7 | 3534 | 0.020671835 | 0.0051229360 | 2.012624 | ACAAAC,ACACCA,CAAACA,CACCAC,CAUACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ZNF638 | 3 | 1773 | 0.010335917 | 0.0025708878 | 2.007328 | GGUUGG,GUUCUU,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 3 | 1782 | 0.010335917 | 0.0025839306 | 2.000027 | ACAAAG,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRPB2 | 1 | 991 | 0.005167959 | 0.0014376103 | 1.845922 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPC | 6 | 3471 | 0.018087855 | 0.0050316361 | 1.845922 | AUUUUU,CUUUUU,GGGUAC,UUUUUC,UUUUUG | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SART3 | 4 | 2634 | 0.012919897 | 0.0038186524 | 1.758459 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SRSF4 | 6 | 3740 | 0.018087855 | 0.0054214720 | 1.738265 | AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ZCRB1 | 2 | 1605 | 0.007751938 | 0.0023274215 | 1.735824 | AAUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPC1 | 7 | 4443 | 0.020671835 | 0.0064402624 | 1.682475 | ACAAAC,AGAAAA,CAAACA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| AGO2 | 2 | 1830 | 0.007751938 | 0.0026534924 | 1.546665 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPC3 | 1 | 1234 | 0.005167959 | 0.0017897669 | 1.529823 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC4 | 1 | 1251 | 0.005167959 | 0.0018144033 | 1.510099 | AAAAAG | AAAAAA,AAAAAG |
| ENOX1 | 4 | 3195 | 0.012919897 | 0.0046316558 | 1.479995 | AUACAG,CAGACA,CAUACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| TRA2A | 9 | 6871 | 0.025839793 | 0.0099589296 | 1.375532 | AAAGAA,AAGAAA,AGAGGA,AGGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| IGF2BP3 | 6 | 4815 | 0.018087855 | 0.0069793662 | 1.373853 | AAACAC,AAACUC,ACAAAC,CAAACA,CAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPL | 6 | 5085 | 0.018087855 | 0.0073706513 | 1.295157 | AAACAA,AAACAC,ACACCA,CACCAC,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| U2AF2 | 1 | 1477 | 0.005167959 | 0.0021419234 | 1.270688 | UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 2 | 2261 | 0.007751938 | 0.0032780993 | 1.241697 | AUUUUU,CUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| SAMD4A | 4 | 3992 | 0.012919897 | 0.0057866714 | 1.158789 | CGGGAA,CUGGAC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SRSF5 | 10 | 8869 | 0.028423773 | 0.0128544391 | 1.144831 | AGGAAG,CACAGC,GAAGAA,GAGGAA,GGAAGA,UACAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| TRA2B | 8 | 7329 | 0.023255814 | 0.0106226650 | 1.130446 | AAAGAA,AAGAAU,AGGAAG,GAAGAA,GAAUUA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF10 | 16 | 13860 | 0.043927649 | 0.0200874160 | 1.128837 | AAAGAA,AAGAAA,AAGAGA,ACAAAG,AGAGAA,AGAGAG,AGAGGA,CAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAGG,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF3 | 18 | 16536 | 0.049095607 | 0.0239654858 | 1.034636 | AACUUU,ACCACC,ACUACA,AUUCAU,CACCAC,CAGAGA,CAUCAA,CAUCAU,CUACAA,CUACAG,CUUCAU,GAUCAA,GCUUCA,UACAAA,UCAUCA,UUACGA,UUCAUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| YBX1 | 7 | 7119 | 0.020671835 | 0.0103183321 | 1.002457 | ACACCA,ACCACC,CACCAC,CAGCAA,CAUCAU,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
RBP on BSJ (Exon Only)
Plot
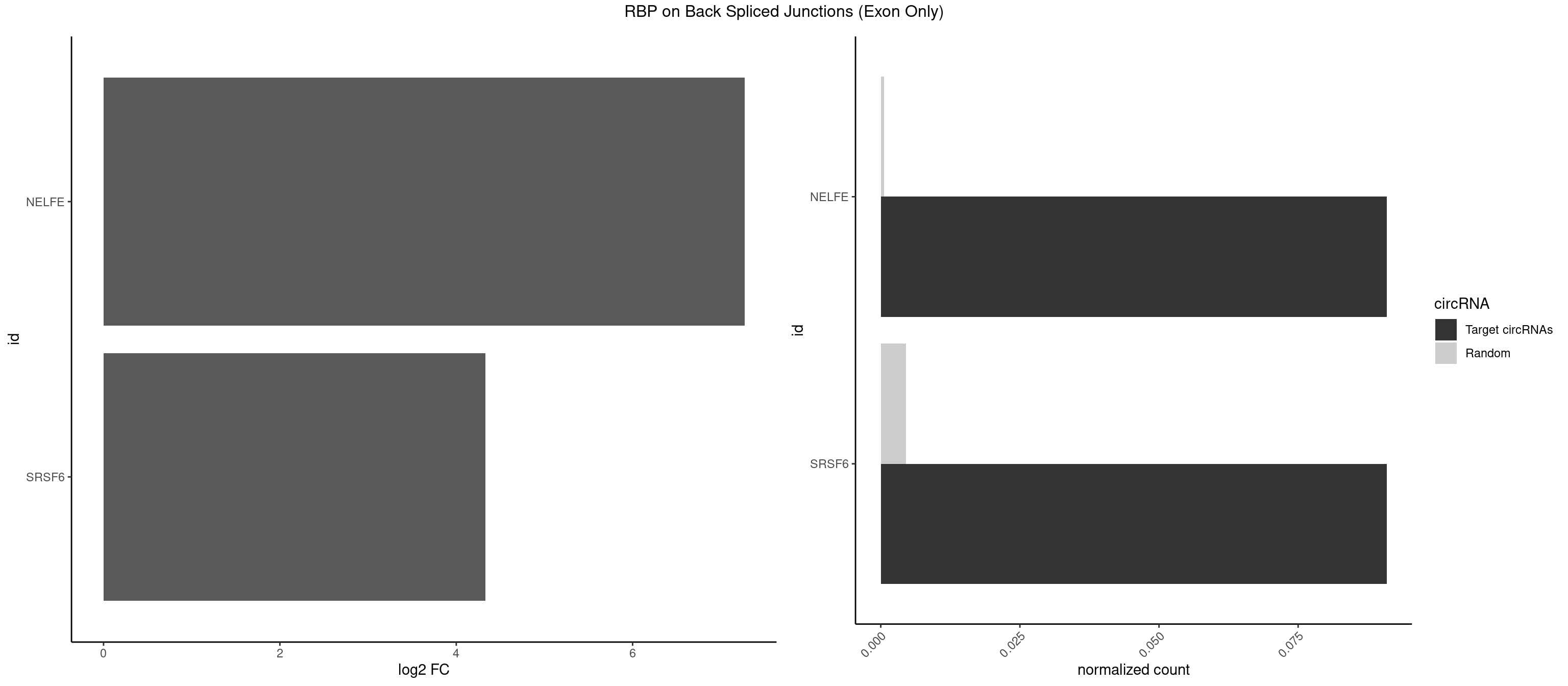
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NELFE | 1 | 8 | 0.09090909 | 0.0005894682 | 7.268867 | UCUCUG | CUCUGG,CUGGCU,CUGGUU,UCUGGC,UCUGGU |
| SRSF6 | 1 | 68 | 0.09090909 | 0.0045192560 | 4.330267 | UCUCUG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
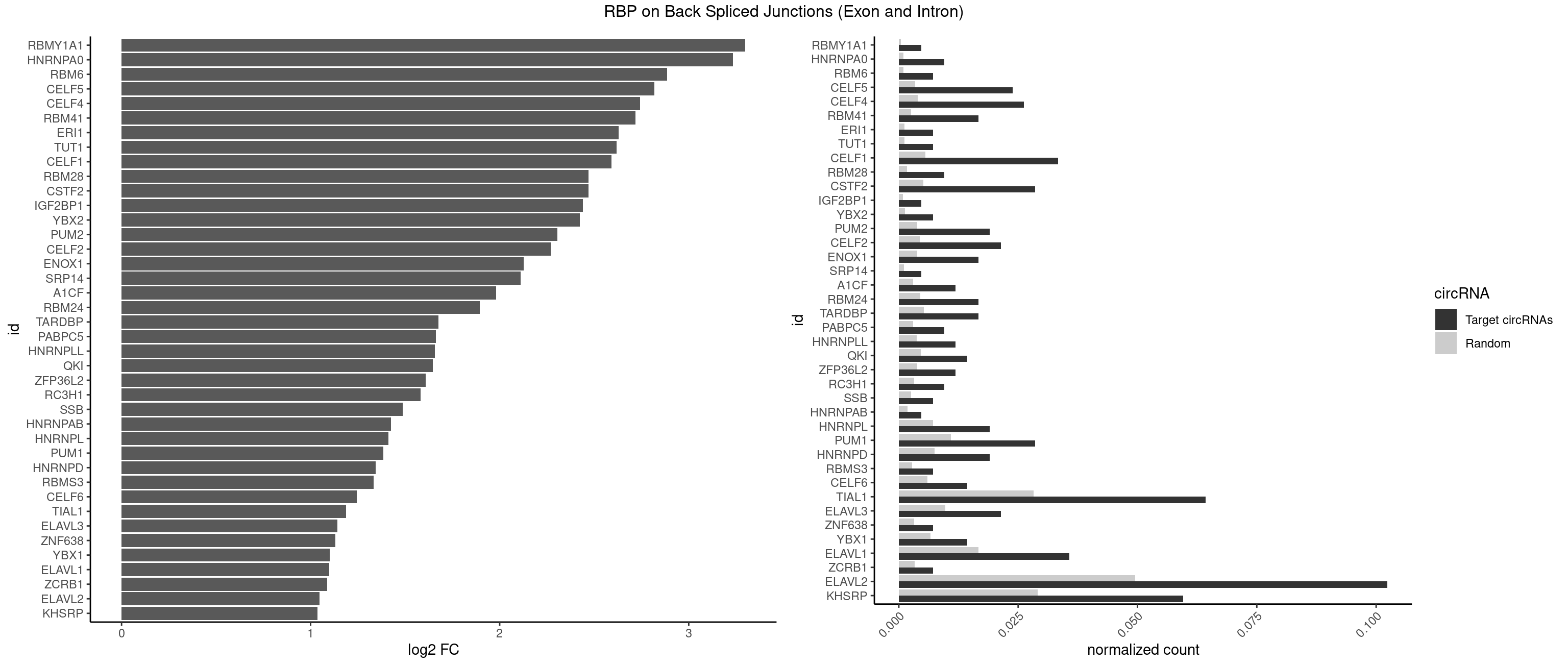
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CELF5 | 9 | 669 | 0.023809524 | 0.0033756550 | 2.818299 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 10 | 776 | 0.026190476 | 0.0039147521 | 2.742049 | GGUGUU,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 6 | 502 | 0.016666667 | 0.0025342604 | 2.717329 | AUACAU,AUACUU,UACAUG,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| CELF1 | 13 | 1097 | 0.033333333 | 0.0055320435 | 2.591081 | CUGUCU,GUGUGU,GUGUUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CSTF2 | 11 | 1022 | 0.028571429 | 0.0051541717 | 2.470761 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM28 | 3 | 340 | 0.009523810 | 0.0017180572 | 2.470761 | AGUAGU,UGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PUM2 | 7 | 764 | 0.019047619 | 0.0038542926 | 2.305073 | GUACAU,UACAUA,UAGAUA,UGUAAA,UGUACA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CELF2 | 8 | 881 | 0.021428571 | 0.0044437727 | 2.269679 | GUCUGU,GUGUGU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ENOX1 | 6 | 756 | 0.016666667 | 0.0038139863 | 2.127594 | AUACAG,CAUACA,UAGACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,AUCAGU,GAUCAG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACAUAC,CAUACA,GCAUAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | ACACUA,AUUAAC,CACUAA,CUCAUA,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.0071543732 | 1.412713 | AAACAA,AAAUAA,AACAAA,ACAUAC,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUGUU,CUUGUA,GUACAU,UACAUA,UAGAUA,UGUAAA,UGUACA,UGUAGA,UUGUAC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPD | 7 | 1488 | 0.019047619 | 0.0075020153 | 1.344261 | AAUUUA,AGAUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGUUG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| TIAL1 | 26 | 5597 | 0.064285714 | 0.0282043531 | 1.188580 | AAAUUU,AGUUUU,AUUUUA,AUUUUC,CAGUUU,CUUUUA,GUUUUU,UAUUUU,UCAGUU,UUAUUU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AACAUC,ACAUCA,CAUCAU,GCCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | UAGUUU,UAUUUA,UAUUUU,UGUUUU,UUAGUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUGUU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| ELAVL2 | 42 | 9826 | 0.102380952 | 0.0495112858 | 1.048118 | AAUUUA,AUACUU,AUAUUU,AUUUAC,AUUUAG,AUUUUA,AUUUUC,CAUUUU,CUUAUA,CUUUCU,CUUUUA,GUUUUA,GUUUUU,UACUUU,UAGUUA,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCUUAU,UGUAUU,UUAGUU,UUAUAU,UUAUUU,UUGUAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUUAA,UUUUAU,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHSRP | 24 | 5764 | 0.059523810 | 0.0290457477 | 1.035140 | AUAUUU,AUUUUA,CAGCCU,CUGCCU,CUGUGU,GUGUGU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UGUGUA,UGUGUG,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.