circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000141127:+:17:18865468:18911251
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141127:+:17:18865468:18911251 | ENSG00000141127 | MSTRG.13816.21 | + | 17 | 18865469 | 18911251 | 861 | GAGGAAUUUUGUUGUCAGAGAAUAAAAGGAGGUUGUCCAUAAUUGACUUUAAGCAGCAAUCAGUAAAACAUUGAGCUCUUCAGCUCCGCCUUUCUUGCUCUGAAAAUUGGAAAACCAAGAAGGUUUUGAUGUUUUGUGUGACGCCACCUGAAUUAGAAACCAAGAUGAACAUAACCAAAGGUGGUCUGGUGUUGUUUUCAGCAAACUCGAAUUCAUCAUGUAUGGAGCUAUCAAAGAAAAUUGCAGAGCGGCUAGGGGUGGAGAUGGGCAAAGUGCAGGUUUACCAGGAACCUAACAGAGAAACAAGAGUACAAAUUCAAGAGUCUGUGAGGGGAAAAGAUGUUUUCAUCAUCCAAACUGUUUCGAAGGACGUGAACACCACCAUCAUGGAGCUCCUGAUCAUGGUGUAUGCAUGUAAGACCUCUUGUGCCAAGAGCAUCAUUGGCGUGAUACCCUACUUUCCUUACAGCAAGCAGUGCAAGAUGAGAAAAAGAGGCUCCAUUGUCUCUAAAUUGCUGGCUUCCAUGAUGUGCAAAGCUGGUCUAACUCAUCUUAUUACUAUGGAUUUACACCAGAAGGAAAUUCAGGGCUUCUUCAAUAUUCCUGUUGACAAUUUAAGAGCAUCUCCCUUCUUAUUACAGUAUAUUCAAGAAGAGAUCCCAGAUUACAGGAAUGCAGUAAUCGUGGCCAAGUCUCCAGCCUCGGCGAAGAGGGCACAGUCUUUUGCUGAGCGCCUGCGCCUGGGAAUUGCAGUGAUUCAUGGAGAGGCGCAGGAUGCCGAGUCGGACUUGGUGGAUGGACGGCAUUCCCCACCCAUGGUCAGAAGUGUGGCUGCCAUCCACCCCAGCCUGGAGAUCCCCAGAGGAAUUUUGUUGUCAGAGAAUAAAAGGAGGUUGUCCAUAAUUGACUUU | circ |
| ENSG00000141127:+:17:18865468:18911251 | ENSG00000141127 | MSTRG.13816.21 | + | 17 | 18865469 | 18911251 | 22 | GGAGAUCCCCAGAGGAAUUUUG | bsj |
| ENSG00000141127:+:17:18865468:18911251 | ENSG00000141127 | MSTRG.13816.21 | + | 17 | 18865269 | 18865478 | 210 | CGUGUUCCACUUGAGGAAAAUGAAGAUGAGAGGUCAGUUUUAGACAUGACACAGGGCAGCUGAAAGGCCGGGAAAGGUUUUAUGGUAAACAAGGAUUACAGGAAAAGCUGGGUGUAGAUGGGUCAUUUAUUCUUUGUCUAAGGAUUAAUCAGUCCUUGUUCUCGCUAAUGGCUCUGCCAUUUUUCUGUGUGUUAAUCCAGGAGGAAUUUU | ie_up |
| ENSG00000141127:+:17:18865468:18911251 | ENSG00000141127 | MSTRG.13816.21 | + | 17 | 18911242 | 18911451 | 210 | GAGAUCCCCAGUAAGUGUGCUGCUGCCUCUUUCCCACUUUCCUCUGAUUUUAAGGCUGACUACACUGUUGUCUGUGUGGUUUUUUUCCUCAUUCUUCGUUUUUUUUUAGUUGGCAAAAACUCAUUAACACCUUUCUUGGCCAGAUAGUAGCGAAUGCAUUUCUGAUUACCUUGUAAAUUGUAAGGCCGUUUAGAAGCAGUCUGUCCUUCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
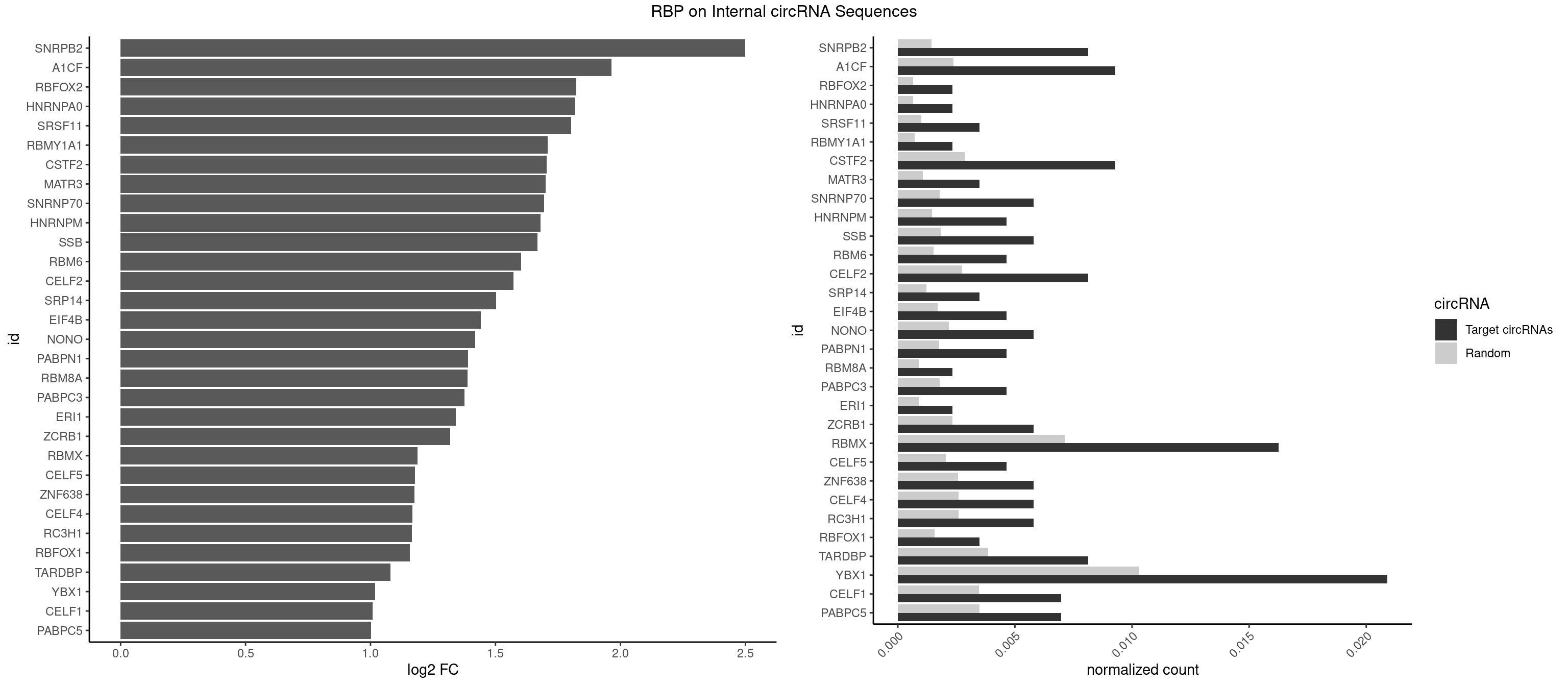
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPB2 | 6 | 991 | 0.008130081 | 0.0014376103 | 2.499597 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| A1CF | 7 | 1642 | 0.009291521 | 0.0023810421 | 1.964322 | AGUAUA,AUAAUU,AUCAGU,CAGUAU,UAAUUG,UCAGUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBFOX2 | 1 | 452 | 0.002322880 | 0.0006564894 | 1.823071 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.002322880 | 0.0006579386 | 1.819890 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SRSF11 | 2 | 688 | 0.003484321 | 0.0009985015 | 1.803041 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 489 | 0.002322880 | 0.0007101099 | 1.709801 | ACAAGA | ACAAGA,CAAGAC |
| CSTF2 | 7 | 1967 | 0.009291521 | 0.0028520334 | 1.703924 | GUGUUG,GUUUUG,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| MATR3 | 2 | 739 | 0.003484321 | 0.0010724109 | 1.700020 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SNRNP70 | 4 | 1237 | 0.005807201 | 0.0017941145 | 1.694571 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPM | 3 | 999 | 0.004645761 | 0.0014492040 | 1.680654 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| SSB | 4 | 1260 | 0.005807201 | 0.0018274462 | 1.668014 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM6 | 3 | 1054 | 0.004645761 | 0.0015289102 | 1.603411 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CELF2 | 6 | 1886 | 0.008130081 | 0.0027346479 | 1.571915 | GUCUGU,GUUGUU,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SRP14 | 2 | 847 | 0.003484321 | 0.0012289250 | 1.503481 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 3 | 1179 | 0.004645761 | 0.0017100607 | 1.441867 | GUCGGA,UCGGAC,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| NONO | 4 | 1498 | 0.005807201 | 0.0021723567 | 1.418582 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPN1 | 3 | 1222 | 0.004645761 | 0.0017723764 | 1.390230 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM8A | 1 | 611 | 0.002322880 | 0.0008869128 | 1.389051 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PABPC3 | 3 | 1234 | 0.004645761 | 0.0017897669 | 1.376143 | AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ERI1 | 1 | 632 | 0.002322880 | 0.0009173461 | 1.340377 | UUUCAG | UUCAGA,UUUCAG |
| ZCRB1 | 4 | 1605 | 0.005807201 | 0.0023274215 | 1.319110 | AUUUAA,GAAUUA,GAUUUA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBMX | 13 | 4925 | 0.016260163 | 0.0071387787 | 1.187593 | AAGAAG,AAGGAA,AAGUGU,ACCAAA,AGAAGG,AUCAAA,AUCCCA,AUCCCC,GAAGGA,UAAGAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF5 | 3 | 1415 | 0.004645761 | 0.0020520728 | 1.178833 | GUGUGG,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ZNF638 | 4 | 1773 | 0.005807201 | 0.0025708878 | 1.175576 | GGUUGU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF4 | 4 | 1782 | 0.005807201 | 0.0025839306 | 1.168276 | GGUGUU,GUGUGG,GUGUUG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RC3H1 | 4 | 1786 | 0.005807201 | 0.0025897275 | 1.165043 | CCCUUC,CCUUCU,UCCCUU,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBFOX1 | 2 | 1077 | 0.003484321 | 0.0015622419 | 1.157260 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TARDBP | 6 | 2654 | 0.008130081 | 0.0038476365 | 1.079297 | GUUGUU,GUUUUG,UGUGUG,UUGUGC,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| YBX1 | 17 | 7119 | 0.020905923 | 0.0103183321 | 1.018702 | ACACCA,ACCACC,AUCAUC,CACCAC,CAGCAA,CAUCAU,CCACCA,CCUGCG,GCCUGC,GGUCUG,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| CELF1 | 5 | 2391 | 0.006968641 | 0.0034664959 | 1.007399 | GUGUUG,UGUGUG,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPC5 | 5 | 2400 | 0.006968641 | 0.0034795387 | 1.001981 | AGAAAA,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
RBP on BSJ (Exon Only)
Plot
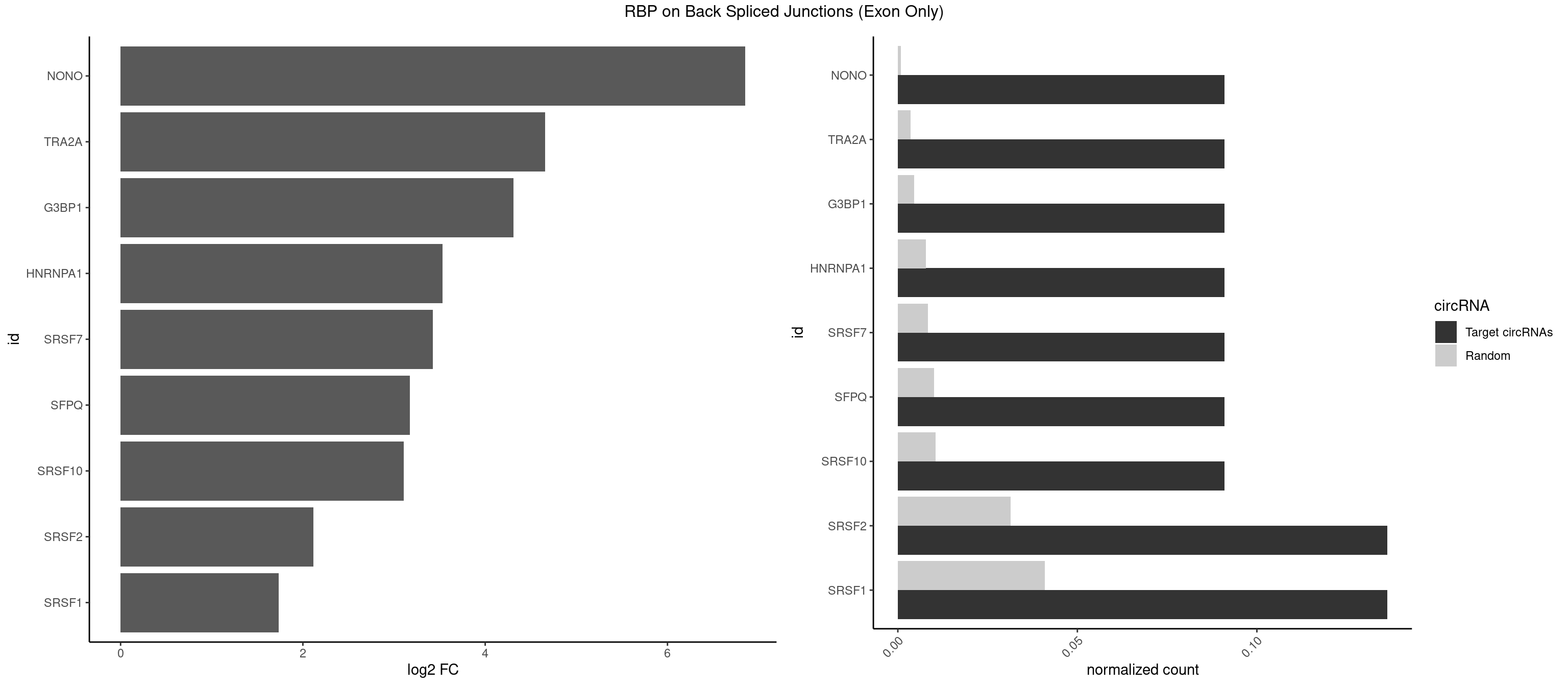
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NONO | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | AGAGGA | AGAGGA,AGGAAC,GAGGAA |
| TRA2A | 1 | 54 | 0.09090909 | 0.0036023055 | 4.657432 | AGAGGA | AAAGAA,AAGAAA,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| G3BP1 | 1 | 69 | 0.09090909 | 0.0045847524 | 4.309509 | CCCCAG | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.0078595756 | 3.531901 | AGAGGA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SRSF7 | 1 | 128 | 0.09090909 | 0.0084490438 | 3.427565 | AGAGGA | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | AGAGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | AGAGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | AGAGGA,CAGAGG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
| SRSF1 | 2 | 625 | 0.13636364 | 0.0410007860 | 1.733736 | AGAGGA,CAGAGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
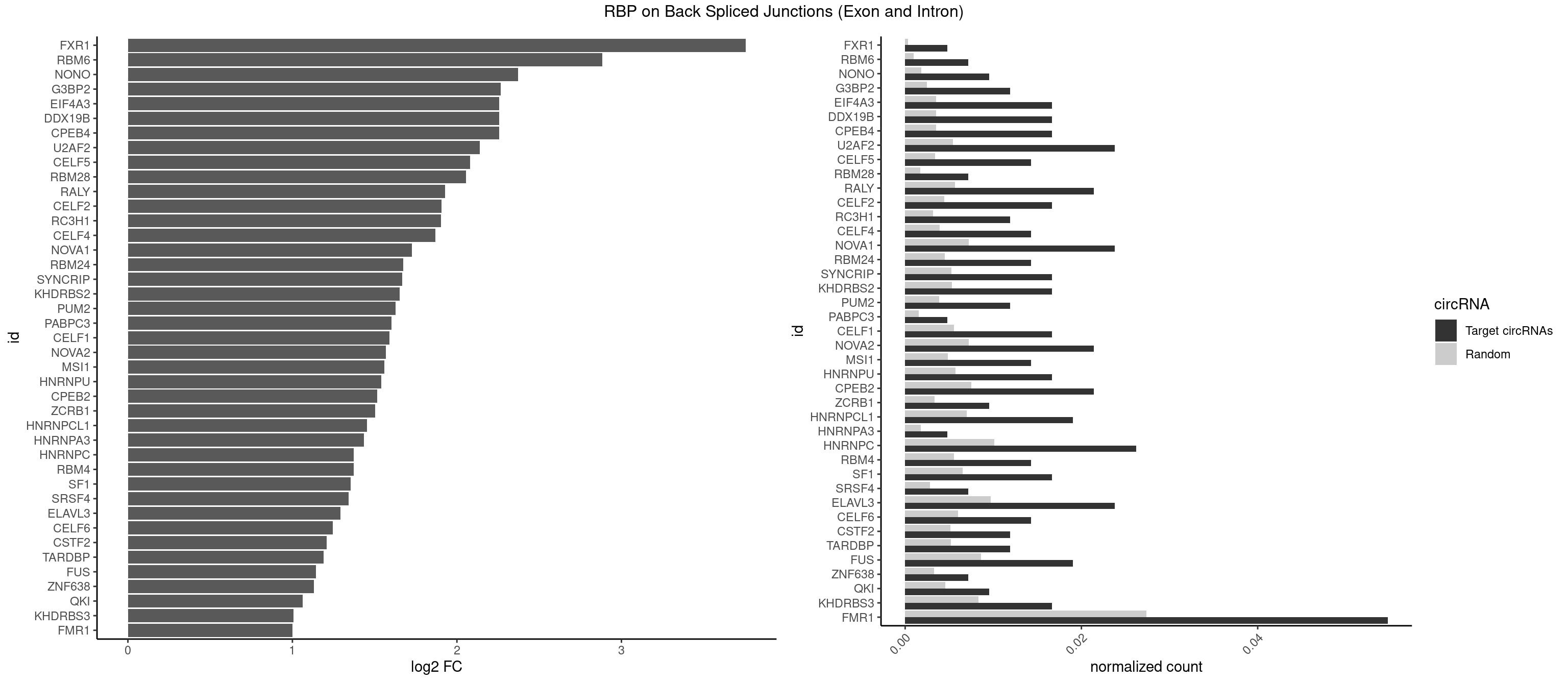
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CPEB4 | 6 | 691 | 0.016666667 | 0.0034864974 | 2.257115 | UUUUUU | UUUUUU |
| DDX19B | 6 | 691 | 0.016666667 | 0.0034864974 | 2.257115 | UUUUUU | UUUUUU |
| EIF4A3 | 6 | 691 | 0.016666667 | 0.0034864974 | 2.257115 | UUUUUU | UUUUUU |
| U2AF2 | 9 | 1071 | 0.023809524 | 0.0054010480 | 2.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUGG,GUGUGU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | GUGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RALY | 8 | 1117 | 0.021428571 | 0.0056328094 | 1.927609 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CELF2 | 6 | 881 | 0.016666667 | 0.0044437727 | 1.907109 | GUCUGU,GUGUGU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CCUUCU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GUGUGG,GUGUGU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| NOVA1 | 9 | 1428 | 0.023809524 | 0.0071997179 | 1.725526 | AACACC,UCAGUC,UCAUUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RBM24 | 5 | 888 | 0.014285714 | 0.0044790407 | 1.673311 | AGUGUG,GUGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SYNCRIP | 6 | 1041 | 0.016666667 | 0.0052498992 | 1.666604 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 6 | 1051 | 0.016666667 | 0.0053002821 | 1.652825 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAAAU,GUAGAU,UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | GUGUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.0072299476 | 1.567479 | AACACC,AGACAU,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGGAGG,AGUAAG,AGUUGG,UAGUAG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPU | 6 | 1136 | 0.016666667 | 0.0057285369 | 1.540727 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CAUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM4 | 5 | 1094 | 0.014285714 | 0.0055169287 | 1.372636 | CCUCUU,CCUUCU,CUCUUU,UCCUUC,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | AGUAAG,AUUAAC,GCUGAC,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ELAVL3 | 9 | 1928 | 0.023809524 | 0.0097188634 | 1.292679 | AUUUAU,AUUUUA,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGUGG,UGUGGU,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,UGUGUG,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGUGU,UGUGUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| FUS | 7 | 1711 | 0.019047619 | 0.0086255542 | 1.142922 | GGGUGU,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUCAU,AUUAAC,UAAUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FMR1 | 22 | 5430 | 0.054761905 | 0.0273629585 | 1.000948 | AAGGAU,AGCGAA,AGCUGG,AGGAAU,AGGAGG,AGGAUU,CAGCUG,CUAAGG,GCAGCU,GCUGGG,GGCUGA,UAAGGA,UAGCGA,UGAGGA,UUUUUU | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.