circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000196814:+:9:126340507:126386658
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000196814:+:9:126340507:126386658 | ENSG00000196814 | ENST00000361171 | + | 9 | 126340508 | 126386658 | 328 | GACCAGUCCACCAUGCCUGAAGUCAAAGACCUCUCAGAAGCCUUGCCAGAAACGUCAAUGGAUCCCAUCACGGGAGUCGGGGUGGUGGCUUCUCGGAACCGAGCCCCGACAGGCUAUGACGUAGUUGCACAGACAGCAGAUGGUGUGGAUGCUGACCUCUGGAAAGACGGCUUAUUUAAAUCCAAGGUUACCAGAUACCUGUGUUUCACAAGAUCAUUUUCCAAAGAAAAUAGUCAUCUGGGGAACGUGUUAGUAGAUAUGAAGCUCAUUGACAUCAAGGACACACUGCCUGUGGGCUUCAUCCCAAUUCAGGAGACGGUGGACACACGACCAGUCCACCAUGCCUGAAGUCAAAGACCUCUCAGAAGCCUUGCCAGA | circ |
| ENSG00000196814:+:9:126340507:126386658 | ENSG00000196814 | ENST00000361171 | + | 9 | 126340508 | 126386658 | 22 | GGUGGACACACGACCAGUCCAC | bsj |
| ENSG00000196814:+:9:126340507:126386658 | ENSG00000196814 | ENST00000361171 | + | 9 | 126340308 | 126340517 | 210 | CUGAGAAGAGUGGGAAACAGAGGUGAUUUGAAUUCUUGACCUGGGGAAAGGGUCACAUAGGGUCAGGACUCAUUCAACCAGUACAGGUAUGUGAAACUCAGGGUAUUUGCCCCAAGAUUCUGGUUCUGUUUGAGACUUUAUAUUAUUCACAUAAAUGCUAUGUUUGAAUUUUGUGUUUUCUUUUUCCUUUGCUGAACCAGGACCAGUCCA | ie_up |
| ENSG00000196814:+:9:126340507:126386658 | ENSG00000196814 | ENST00000361171 | + | 9 | 126386649 | 126386858 | 210 | GUGGACACACGUGAGUCAUCCUUUAGUAGCACUCAUCACAAUGAGCGUUUUCUUCCUCCAGGUAUAUUUGUAGGUGUUUUUCUAUGUGCAUUUUUCUUCAGAAACACUUUUGGAGGCCUUACUACAUGCCCAUGAACAAACCCUGCUGCUUUUGAUGUGUGUCUGGCUGGGUUCUCCAAGGAGUUUGCAGAGUAUUCUGGGUAGCAGCAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
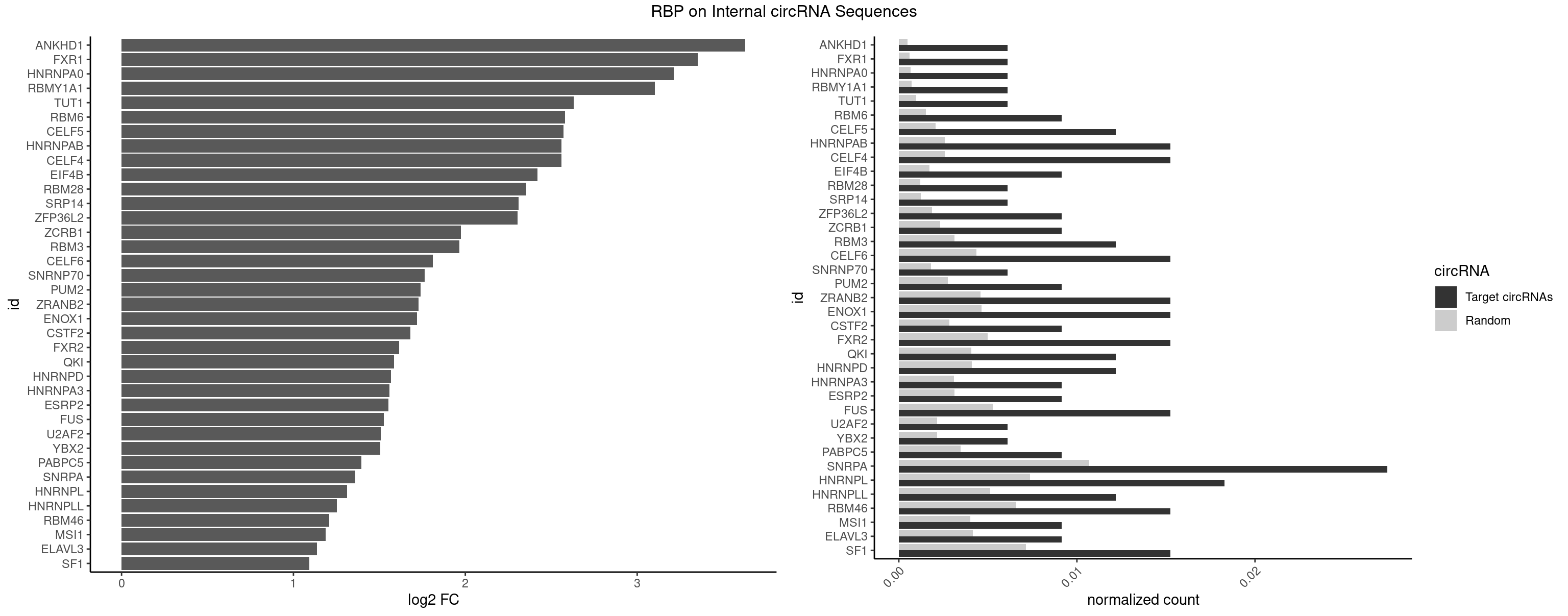
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 339 | 0.006097561 | 0.0004927293 | 3.629365 | GACGUA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| FXR1 | 1 | 411 | 0.006097561 | 0.0005970720 | 3.352255 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPA0 | 1 | 453 | 0.006097561 | 0.0006579386 | 3.212207 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.006097561 | 0.0007101099 | 3.102118 | ACAAGA | ACAAGA,CAAGAC |
| TUT1 | 1 | 678 | 0.006097561 | 0.0009840095 | 2.631488 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.009146341 | 0.0015289102 | 2.580691 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CELF5 | 3 | 1415 | 0.012195122 | 0.0020520728 | 2.571150 | GUGUGG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 1782 | 0.015243902 | 0.0025839306 | 2.560593 | GGUGUG,GUGUGG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 4 | 1782 | 0.015243902 | 0.0025839306 | 2.560593 | AAAGAC,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| EIF4B | 2 | 1179 | 0.009146341 | 0.0017100607 | 2.419147 | CUCGGA,UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 1 | 822 | 0.006097561 | 0.0011926949 | 2.354007 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRP14 | 1 | 847 | 0.006097561 | 0.0012289250 | 2.310835 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ZFP36L2 | 2 | 1277 | 0.009146341 | 0.0018520827 | 2.304046 | UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ZCRB1 | 2 | 1605 | 0.009146341 | 0.0023274215 | 1.974462 | AUUUAA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM3 | 3 | 2152 | 0.012195122 | 0.0031201361 | 1.966623 | AAGACG,GAAACG,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF6 | 4 | 2997 | 0.015243902 | 0.0043447134 | 1.810899 | GUGGUG,GUGUGG,UGUGGG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| SNRNP70 | 1 | 1237 | 0.006097561 | 0.0017941145 | 1.764960 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 2 | 1890 | 0.009146341 | 0.0027404447 | 1.738785 | GUAGAU,UAGAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZRANB2 | 4 | 3173 | 0.015243902 | 0.0045997733 | 1.728598 | AGGUUA,GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ENOX1 | 4 | 3195 | 0.015243902 | 0.0046316558 | 1.718632 | AGACAG,AGGACA,CAGACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| CSTF2 | 2 | 1967 | 0.009146341 | 0.0028520334 | 1.681204 | GUGUUU,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| FXR2 | 4 | 3434 | 0.015243902 | 0.0049780156 | 1.614590 | AGACAG,AGACGG,GACAGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| QKI | 3 | 2805 | 0.012195122 | 0.0040664663 | 1.584457 | ACACAC,CACACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD | 3 | 2837 | 0.012195122 | 0.0041128408 | 1.568097 | AGAUAU,UAUUUA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPA3 | 2 | 2140 | 0.009146341 | 0.0031027457 | 1.559649 | CAAGGA,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 2 | 2150 | 0.009146341 | 0.0031172377 | 1.552927 | GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| FUS | 4 | 3648 | 0.015243902 | 0.0052881452 | 1.527399 | CGGUGG,GGGUGG,UGGUGG,UGGUGU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| U2AF2 | 1 | 1477 | 0.006097561 | 0.0021419234 | 1.509325 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| YBX2 | 1 | 1480 | 0.006097561 | 0.0021462711 | 1.506400 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PABPC5 | 2 | 2400 | 0.009146341 | 0.0034795387 | 1.394299 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SNRPA | 8 | 7380 | 0.027439024 | 0.0106965744 | 1.359080 | AGGAGA,AUACCU,AUGCUG,GAUACC,GUUACC,UACCUG,UGCACA,UUGCAC | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| HNRNPL | 5 | 5085 | 0.018292683 | 0.0073706513 | 1.311403 | ACACAC,ACACGA,CACAAG,CACGAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| HNRNPLL | 3 | 3534 | 0.012195122 | 0.0051229360 | 1.251261 | ACACAC,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM46 | 4 | 4554 | 0.015243902 | 0.0066011240 | 1.207449 | AUCAAG,AUCAUU,AUGAAG,GAUCAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| MSI1 | 2 | 2770 | 0.009146341 | 0.0040157442 | 1.187527 | UAGUAG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ELAVL3 | 2 | 2867 | 0.009146341 | 0.0041563169 | 1.137889 | UAUUUA,UUAUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SF1 | 4 | 4931 | 0.015243902 | 0.0071474739 | 1.092727 | ACAGAC,CACAGA,GCUGAC,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
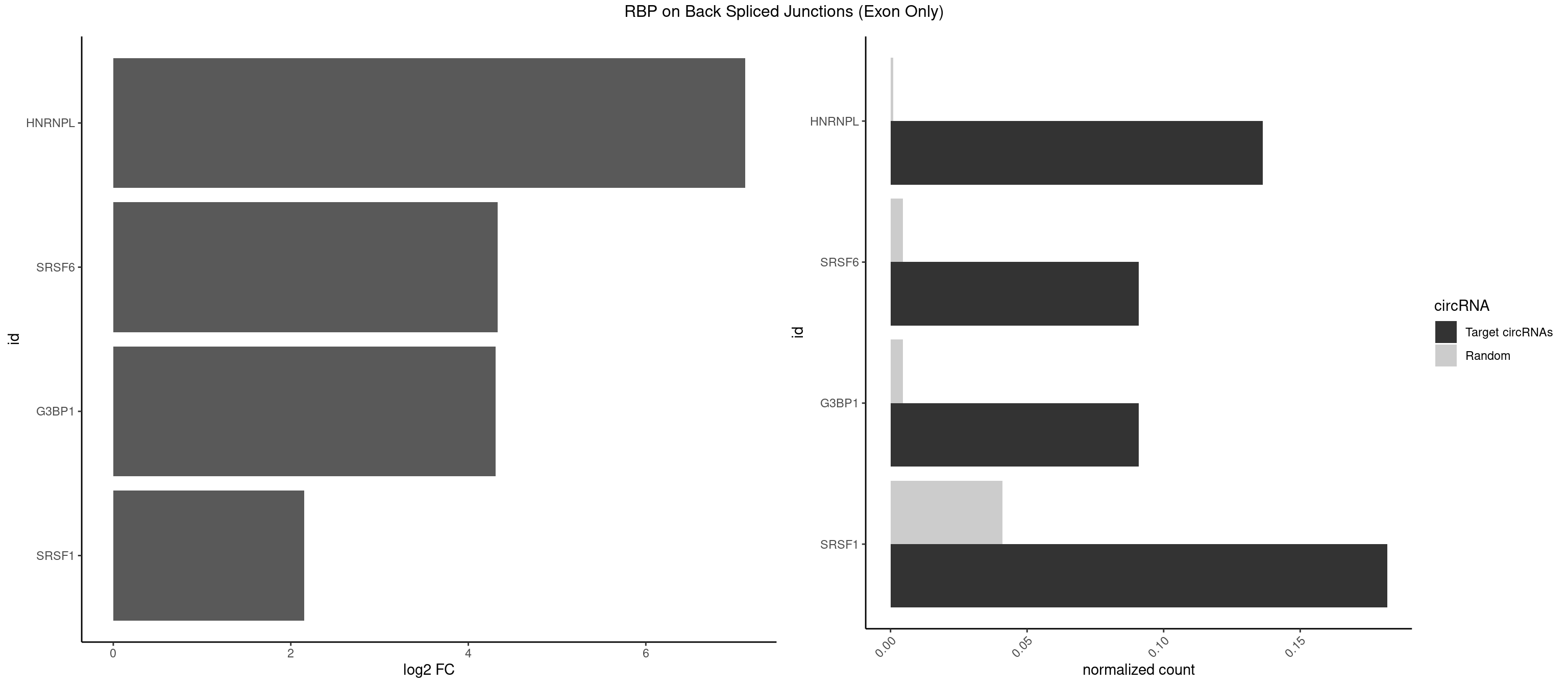
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPL | 2 | 14 | 0.13636364 | 0.0009824469 | 7.116864 | ACACGA,CACGAC | AAAUAA,AAAUAC,AACAAA,ACACCA,ACAUAA,ACAUAC,CACAAC,CACCAC,CAUAAA,CAUACA |
| SRSF6 | 1 | 68 | 0.09090909 | 0.0045192560 | 4.330267 | CACACG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
| G3BP1 | 1 | 69 | 0.09090909 | 0.0045847524 | 4.309509 | CACACG | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| SRSF1 | 3 | 625 | 0.18181818 | 0.0410007860 | 2.148773 | ACACGA,CACACG,CGACCA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
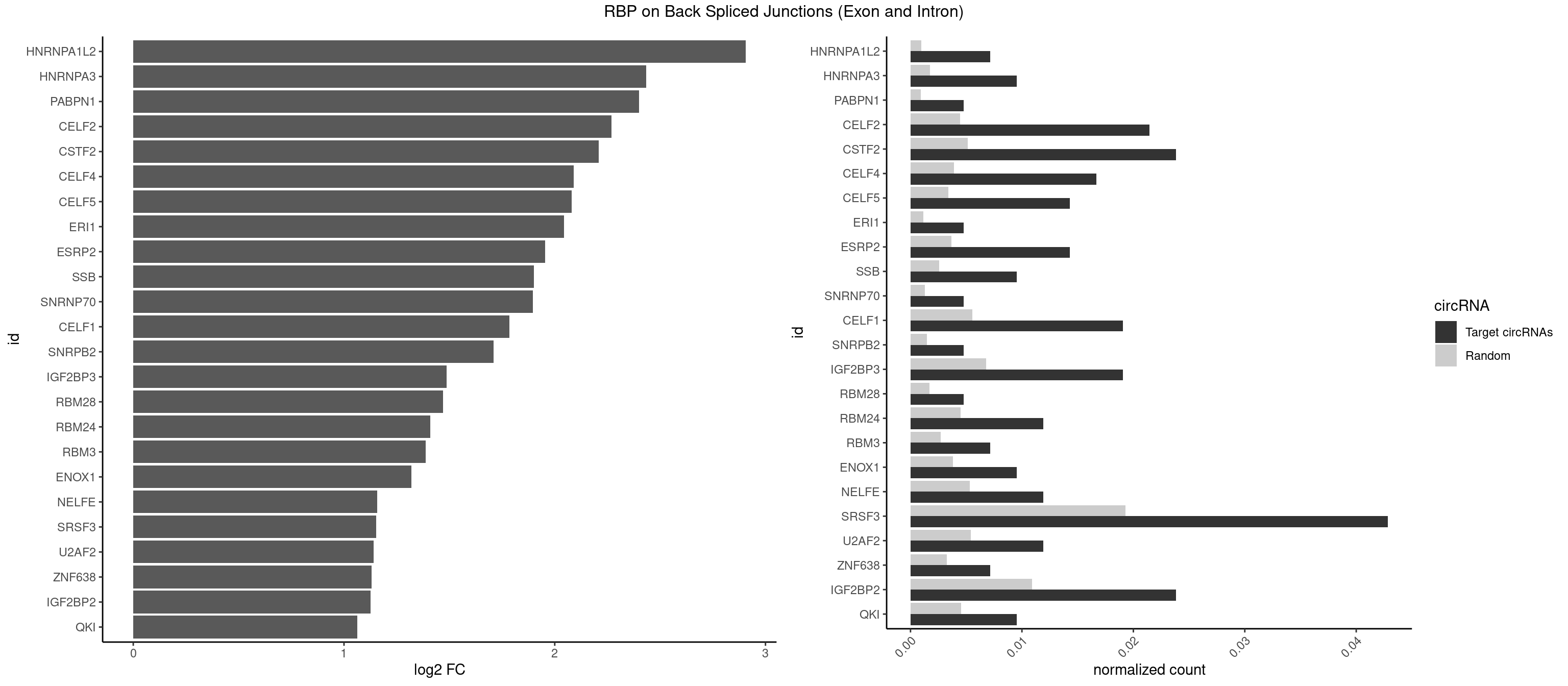
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | AUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA3 | 3 | 349 | 0.009523810 | 0.0017634019 | 2.433177 | AAGGAG,CAAGGA,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| CELF2 | 8 | 881 | 0.021428571 | 0.0044437727 | 2.269679 | AUGUGU,GUAUGU,GUGUGU,UAUGUG,UAUGUU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CSTF2 | 9 | 1022 | 0.023809524 | 0.0051541717 | 2.207726 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF4 | 6 | 776 | 0.016666667 | 0.0039147521 | 2.089973 | GGUGUU,GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| ESRP2 | 5 | 731 | 0.014285714 | 0.0036880290 | 1.953651 | GGGAAA,GGGGAA,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF1 | 7 | 1097 | 0.019047619 | 0.0055320435 | 1.783726 | GUGUGU,UGUCUG,UGUGUG,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| IGF2BP3 | 7 | 1348 | 0.019047619 | 0.0067966546 | 1.486714 | AAACAC,AAACUC,AACUCA,ACAAAC,ACACAC,CACUCA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,GAGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGUACA,GUACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUGGCU,CUGGUU,UCUGGC,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SRSF3 | 17 | 3825 | 0.042857143 | 0.0192765014 | 1.152692 | ACUACA,ACUUUA,CAUCAC,CAUUCA,CUCCAA,CUUCAG,CUUUAU,UCACAA,UCAUCA,UCAUCC,UCUCCA,UCUUCA,UCUUCC,UUCAAC,UUCUCC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| IGF2BP2 | 9 | 2163 | 0.023809524 | 0.0109028617 | 1.126832 | AAACAC,AAACUC,AACUCA,ACAAAC,ACACAC,CACUCA,CAUUCA,GAAUUC,GCACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACACAC,ACUCAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.