circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000156232:+:15:82813102:82826846
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000156232:+:15:82813102:82826846 | ENSG00000156232 | MSTRG.11397.3 | + | 15 | 82813103 | 82826846 | 1161 | GUUAUUCAAGGACACGGAAAAGCCAACACCAUGGUAGCAUUAAUGAACGUUUACCAAGAGGAAGAUGAAGCAUACCAGGAAUUGGUUACCGUGGCAACCAUGUUCUUCCAGUACUUAUUGCAGCCAUUUAGGGCUAUGCGAGAAGUUGCAACUUUAUGUAAGCUUGAUAUUUUGAAGUCUUUGGAUGAGGAUGACCUAGGUCCUAGAAGGGUAGUUGCCCUGGAGAAAGAAGCUGAAGAAUGGACCAGACGGGCUGAAGAAGCUGUCGUCUCUAUUCAGGAUAUCACAGUGAAUUAUUUUAAGGAGACAGUAAAAGCAUUAGCAGGAAUGCAGAAAGAAAUGGAACAGGAUGCGAAGAGAUUUGGUCAGGCUGCCUGGGCCACAGCAAUUCCCAGGUUGGAAAAACUUCAGCUAAUGCUAGCUCGAGAGACUCUGCAACUCAUGAGAGCGAAAGAGUUGUGUUUAAAUCACAAAAGAGCUGAAAUUCAGGGAAAGAUGGAAGAUCUUCCAGAACAAGAAAAAAAUACAAAUGUUGUAGAUGAAUUAGAAAUACAAUUUUAUGAAAUUCAAUUAGAACUAUAUGAAGUUAAAUUUGAGAUAUUAAAAAACGAAGAAAUACUGCUUACUACACAGUUGGACUCUCUUAAAAGACUUAUAAAAGAUGAAAUUUAGAAUCAAGUUCAAAACAAAAACUCGUUGGAAUUUUGAUUGUGAUUUUGCUUAAAAUUAGAGAUUACUUUGGGGAGAAUAGUGGUCUUUGCAAUUUUGAAUCUUCCUACCCAAGAACAUGAAAAACAAGAUGAAGUUGUCUAUUACGAUCCAUGUGAAAAUCCAGAGGAACUUAAAGUCAUUGACUGUGUGGUGGGGCUGCAGGAUGAUAAGAAUUUGGAAGUGAAAGAACUCAGAAGGCAGUGCCAGCAGCUGGAGUCUAAACGGGGCAGGAUCUGUGCCAAAAGAGCCUCUCUCCGGAGUAGAAAGGAUCAGUGCAAAGAAAAUCAUCGGUUCAGAUUGCAACAGGCUGAAGAAAGCAUAAGAUACUCUCGUCAGCAUCACAGUAUUCAGAUGAAAAGAGACAAGAUAAAAGAAGAGGAGCAAAAGAAAAAAGAAUGGAUCAACCAAGAACGUCAAAAAACACUCCAACGAUUGAGAUCAUUUAAAGAUGUUAUUCAAGGACACGGAAAAGCCAACACCAUGGUAGCAUUAAUGAACGU | circ |
| ENSG00000156232:+:15:82813102:82826846 | ENSG00000156232 | MSTRG.11397.3 | + | 15 | 82813103 | 82826846 | 22 | CAUUUAAAGAUGUUAUUCAAGG | bsj |
| ENSG00000156232:+:15:82813102:82826846 | ENSG00000156232 | MSTRG.11397.3 | + | 15 | 82812903 | 82813112 | 210 | AUAUAUCAUACUUUUCUUAACCAUUCUUGUUUUAUUGGGCAUUUUUGAUGGAGGAUGAUAACAUUUUCAUAUUUAAUCAGUAUUUAAAAUUGAUGUAUUAAAAGUUGAGAACAUGAAGGUUUCUUUUGUUUAGUUUUGUUUGUUGGGUAUGUAUUAAAUAUACUGUCCUGACUUGAGCUUUAUUCACAUUUGCUUUCUAGGUUAUUCAAG | ie_up |
| ENSG00000156232:+:15:82813102:82826846 | ENSG00000156232 | MSTRG.11397.3 | + | 15 | 82826837 | 82827046 | 210 | AUUUAAAGAUGUAAGUUCUAUAAACAAUCACCUCAUCUACACUUCUGGGGAAAUAACUGAAGACCACCCUAAAAAAGAUGAAAGUAUGAAGUAUUAAGAACAGGUCAUUCAGAACGUGAAACAUUUAAAAAAUACCAUUAUUUCUGGUAUAUCAAUAAAAAACAACAAAAAUAGAAUAUCAGUGAUUCAAAACCCAUGAUACAAAAUAAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
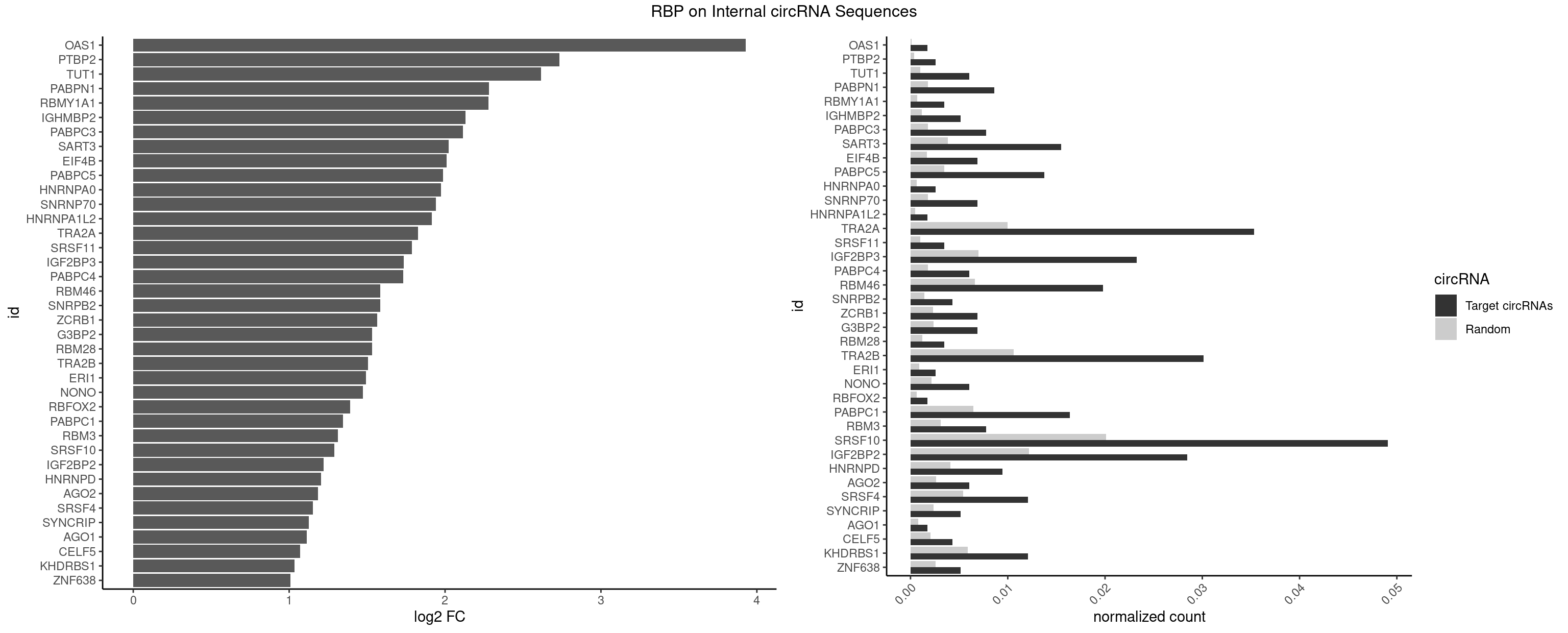
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.001722653 | 0.0001130379 | 3.929753 | GCAUAA | GCAUAA |
| PTBP2 | 2 | 267 | 0.002583979 | 0.0003883867 | 2.734029 | CUCUCU | CUCUCU |
| TUT1 | 6 | 678 | 0.006029285 | 0.0009840095 | 2.615243 | AAAUAC,AAUACU,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPN1 | 9 | 1222 | 0.008613264 | 0.0017723764 | 2.280875 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBMY1A1 | 3 | 489 | 0.003445306 | 0.0007101099 | 2.278518 | ACAAGA | ACAAGA,CAAGAC |
| IGHMBP2 | 5 | 813 | 0.005167959 | 0.0011796520 | 2.131233 | AAAAAA | AAAAAA |
| PABPC3 | 8 | 1234 | 0.007751938 | 0.0017897669 | 2.114785 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SART3 | 17 | 2634 | 0.015503876 | 0.0038186524 | 2.021493 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| EIF4B | 7 | 1179 | 0.006890612 | 0.0017100607 | 2.010585 | GUUGGA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC5 | 15 | 2400 | 0.013781223 | 0.0034795387 | 1.985736 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPA0 | 2 | 453 | 0.002583979 | 0.0006579386 | 1.973570 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| SNRNP70 | 7 | 1237 | 0.006890612 | 0.0017941145 | 1.941360 | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPA1L2 | 1 | 314 | 0.001722653 | 0.0004564992 | 1.915948 | UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| TRA2A | 40 | 6871 | 0.035314384 | 0.0099589296 | 1.826193 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF11 | 3 | 688 | 0.003445306 | 0.0009985015 | 1.786795 | AAGAAG | AAGAAG |
| IGF2BP3 | 26 | 4815 | 0.023255814 | 0.0069793662 | 1.736423 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,AAUACA,AAUUCA,ACACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| PABPC4 | 6 | 1251 | 0.006029285 | 0.0018144033 | 1.732492 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBM46 | 22 | 4554 | 0.019810508 | 0.0066011240 | 1.585482 | AAUCAA,AAUCAU,AAUGAA,AUCAAG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,GAUCAA,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SNRPB2 | 4 | 991 | 0.004306632 | 0.0014376103 | 1.582887 | AUUGCA,UAUUGC,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZCRB1 | 7 | 1605 | 0.006890612 | 0.0023274215 | 1.565899 | ACUUAA,AUUUAA,GAAUUA,GACUUA,GCUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 7 | 1644 | 0.006890612 | 0.0023839405 | 1.531284 | AGGAUA,AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM28 | 3 | 822 | 0.003445306 | 0.0011926949 | 1.530407 | AGUAGA,GAGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TRA2B | 34 | 7329 | 0.030146425 | 0.0106226650 | 1.504841 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AGAAGA,AGAAGG,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAUUA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ERI1 | 2 | 632 | 0.002583979 | 0.0009173461 | 1.494056 | UUCAGA | UUCAGA,UUUCAG |
| NONO | 6 | 1498 | 0.006029285 | 0.0021723567 | 1.472726 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX2 | 1 | 452 | 0.001722653 | 0.0006564894 | 1.391788 | UGACUG | UGACUG,UGCAUG |
| PABPC1 | 18 | 4443 | 0.016365202 | 0.0064402624 | 1.345440 | AAAAAA,AAAAAC,ACAAAU,AGAAAA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM3 | 8 | 2152 | 0.007751938 | 0.0031201361 | 1.312948 | AAAACG,AAAACU,AAACGA,AAGACU,AAUACU,GAGACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF10 | 56 | 13860 | 0.049095607 | 0.0200874160 | 1.289302 | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGAGA,AAGAGG,AAGGAG,AGACAA,AGAGAC,AGAGGA,CAAAGA,GAAAGA,GAGAAA,GAGAAG,GAGACA,GAGAGA,GAGAGC,GAGGAA,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| IGF2BP2 | 32 | 8408 | 0.028423773 | 0.0121863560 | 1.221831 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,AAUACA,AAUUCA,ACACUC,CAAAAC,CAACAC,CAACUC,CAAUUC,GAACUC,GCAUAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| HNRNPD | 10 | 2837 | 0.009474591 | 0.0041128408 | 1.203928 | AAAAAA,AAUUUA,AGAUAU,UUAGAG,UUAGGG,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| AGO2 | 6 | 1830 | 0.006029285 | 0.0026534924 | 1.184095 | AAAAAA,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF4 | 13 | 3740 | 0.012058570 | 0.0054214720 | 1.153302 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SYNCRIP | 5 | 1634 | 0.005167959 | 0.0023694485 | 1.125043 | AAAAAA | AAAAAA,UUUUUU |
| AGO1 | 1 | 548 | 0.001722653 | 0.0007956130 | 1.114493 | GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CELF5 | 4 | 1415 | 0.004306632 | 0.0020520728 | 1.069478 | GUGUGG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| KHDRBS1 | 13 | 4064 | 0.012058570 | 0.0058910141 | 1.033471 | AUAAAA,AUUUAA,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ZNF638 | 5 | 1773 | 0.005167959 | 0.0025708878 | 1.007328 | CGUUGG,GGUUGG,GUUCUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
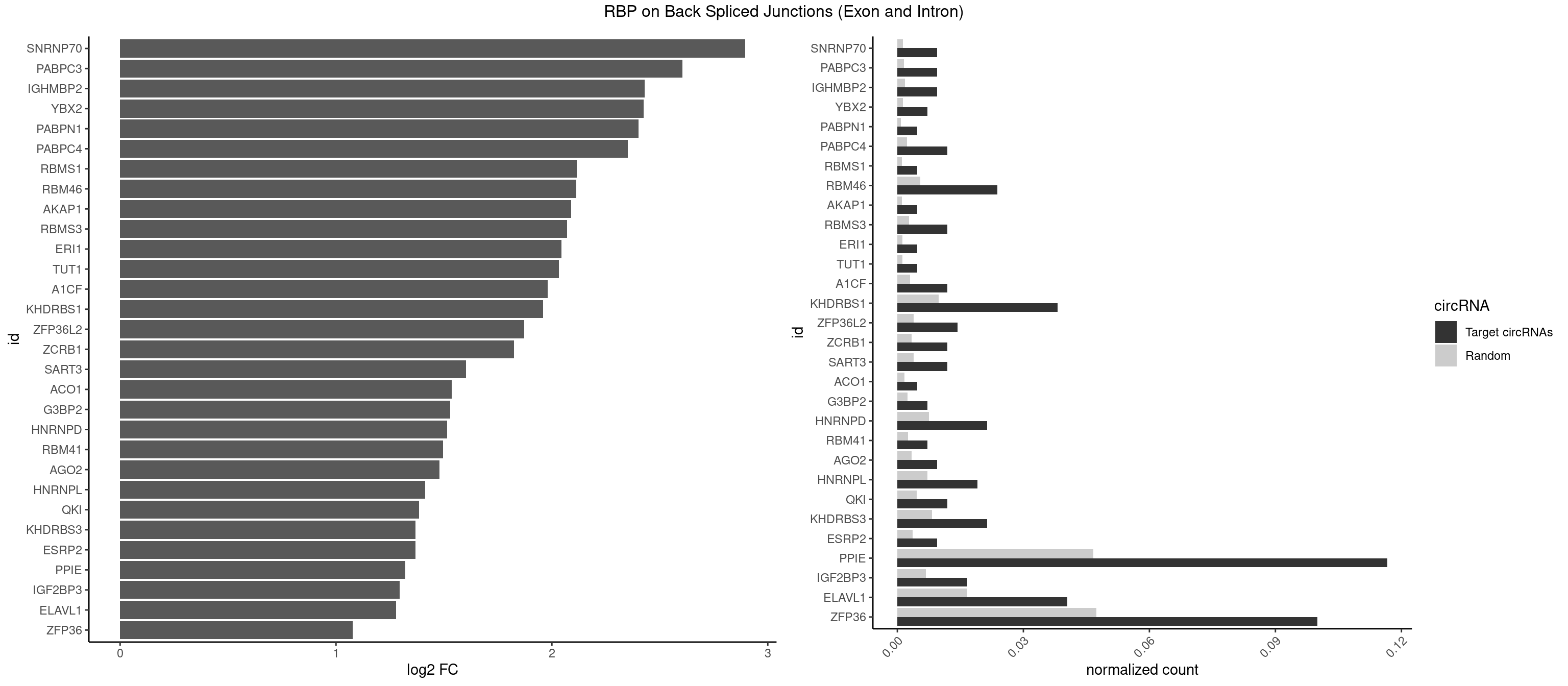
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRNP70 | 3 | 253 | 0.009523810 | 0.0012797259 | 2.895704 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGHMBP2 | 3 | 350 | 0.009523810 | 0.0017684401 | 2.429061 | AAAAAA | AAAAAA |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAAC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| PABPC4 | 4 | 462 | 0.011904762 | 0.0023327287 | 2.351448 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAC | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBM46 | 9 | 1091 | 0.023809524 | 0.0055018138 | 2.113560 | AUCAAU,AUCAUA,AUGAAA,AUGAAG,AUGAUA,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUCAGU,CAGUAU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| KHDRBS1 | 15 | 1945 | 0.038095238 | 0.0098045143 | 1.958093 | AUAAAA,AUUUAA,CAAAAU,CUAAAA,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPD | 8 | 1488 | 0.021428571 | 0.0075020153 | 1.514186 | AAAAAA,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO2 | 3 | 677 | 0.009523810 | 0.0034159613 | 1.479247 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.0071543732 | 1.412713 | AAACAA,AAAUAA,AAAUAC,AACAAA,AAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | AUCAUA,AUCUAC,UAAUCA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AAAUAA,AAUAAA,AUAAAA,AUAAAC,AUUAAA,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PPIE | 48 | 9262 | 0.116666667 | 0.0466696896 | 1.321835 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAUAAA,AAUAUA,AUAAAA,AUAUAU,AUAUUU,AUUAAA,AUUAUU,AUUUAA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAUAAA,UAUUAA,UAUUUA,UUAAAA,UUAAAU,UUAUUU,UUUAAA,UUUAAU,UUUAUU,UUUUAU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| IGF2BP3 | 6 | 1348 | 0.016666667 | 0.0067966546 | 1.294069 | AAAAAC,AAAACA,AAAUAC,ACAAUC,CAAUCA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | UAGUUU,UAUUUA,UGUUUU,UUAGUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUGUU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ZFP36 | 41 | 9408 | 0.100000000 | 0.0474052801 | 1.076880 | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGU,AAAAUA,AAACAA,AAAGUA,AAAUAA,AACAAA,AAUAAA,ACUUCU,ACUUUU,AUAAAC,AUUUAA,UAAACA,UAAAGA,UAAAUA,UAUUUA,UCUUGU,UCUUUU,UUAUUU,UUUAAA,UUUAAU,UUUAUU | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.