circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000149311:+:11:108315822:108335961
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000149311:+:11:108315822:108335961 | ENSG00000149311 | ENST00000683174 | + | 11 | 108315823 | 108335961 | 2262 | GAUCUUCUCUUAGAAAUCUACAGAAGUAUAGGGGAGCCAGAUAGUUUGUAUGGCUGUGGUGGAGGGAAGAUGUUACAACCCAUUACUAGACUACGAACAUAUGAACACGAAGCAAUGUGGGGCAAAGCCCUAGUAACAUAUGACCUCGAAACAGCAAUCCCCUCAUCAACACGCCAGGCAGGAAUCAUUCAGGCCUUGCAGAAUUUGGGACUCUGCCAUAUUCUUUCCGUCUAUUUAAAAGGAUUGGAUUAUGAAAAUAAAGACUGGUGUCCUGAACUAGAAGAACUUCAUUACCAAGCAGCAUGGAGGAAUAUGCAGUGGGACCAUUGCACUUCCGUCAGCAAAGAAGUAGAAGGAACCAGUUACCAUGAAUCAUUGUACAAUGCUCUACAAUCUCUAAGAGACAGAGAAUUCUCUACAUUUUAUGAAAGUCUCAAAUAUGCCAGAGUAAAAGAAGUGGAAGAGAUGUGUAAGCGCAGCCUUGAGUCUGUGUAUUCGCUCUAUCCCACACUUAGCAGGUUGCAGGCCAUUGGAGAGCUGGAAAGCAUUGGGGAGCUUUUCUCAAGAUCAGUCACACAUAGACAACUCUCUGAAGUAUAUAUUAAGUGGCAGAAACACUCCCAGCUUCUCAAGGACAGUGAUUUUAGUUUUCAGGAGCCUAUCAUGGCUCUACGCACAGUCAUUUUGGAGAUCCUGAUGGAAAAGGAAAUGGACAACUCACAAAGAGAAUGUAUUAAGGACAUUCUCACCAAACACCUUGUAGAACUCUCUAUACUGGCCAGAACUUUCAAGAACACUCAGCUCCCUGAAAGGGCAAUAUUUCAAAUUAAACAGUACAAUUCAGUUAGCUGUGGAGUCUCUGAGUGGCAGCUGGAAGAAGCACAAGUAUUCUGGGCAAAAAAGGAGCAGAGUCUUGCCCUGAGUAUUCUCAAGCAAAUGAUCAAGAAGUUGGAUGCCAGCUGUGCAGCGAACAAUCCCAGCCUAAAACUUACAUACACAGAAUGUCUGAGGGUUUGUGGCAACUGGUUAGCAGAAACGUGCUUAGAAAAUCCUGCGGUCAUCAUGCAGACCUAUCUAGAAAAGGCAGUAGAAGUUGCUGGAAAUUAUGAUGGAGAAAGUAGUGAUGAGCUAAGAAAUGGAAAAAUGAAGGCAUUUCUCUCAUUAGCCCGGUUUUCAGAUACUCAAUACCAAAGAAUUGAAAACUACAUGAAAUCAUCGGAAUUUGAAAACAAGCAAGCUCUCCUGAAAAGAGCCAAAGAGGAAGUAGGUCUCCUUAGGGAACAUAAAAUUCAGACAAACAGAUACACAGUAAAGGUUCAGCGAGAGCUGGAGUUGGAUGAAUUAGCCCUGCGUGCACUGAAAGAGGAUCGUAAACGCUUCUUAUGUAAAGCAGUUGAAAAUUAUAUCAACUGCUUAUUAAGUGGAGAAGAACAUGAUAUGUGGGUAUUCCGACUUUGUUCCCUCUGGCUUGAAAAUUCUGGAGUUUCUGAAGUCAAUGGCAUGAUGAAGAGAGACGGAAUGAAGAUUCCAACAUAUAAAUUUUUGCCUCUUAUGUACCAAUUGGCUGCUAGAAUGGGGACCAAGAUGAUGGGAGGCCUAGGAUUUCAUGAAGUCCUCAAUAAUCUAAUCUCUAGAAUUUCAAUGGAUCACCCCCAUCACACUUUGUUUAUUAUACUGGCCUUAGCAAAUGCAAACAGAGAUGAAUUUCUGACUAAACCAGAGGUAGCCAGAAGAAGCAGAAUAACUAAAAAUGUGCCUAAACAAAGCUCUCAGCUUGAUGAGGAUCGAACAGAGGCUGCAAAUAGAAUAAUAUGUACUAUCAGAAGUAGGAGACCUCAGAUGGUCAGAAGUGUUGAGGCACUUUGUGAUGCUUAUAUUAUAUUAGCAAACUUAGAUGCCACUCAGUGGAAGACUCAGAGAAAAGGCAUAAAUAUUCCAGCAGACCAGCCAAUUACUAAACUUAAGAAUUUAGAAGAUGUUGUUGUCCCUACUAUGGAAAUUAAGGUGGACCACACAGGAGAAUAUGGAAAUCUGGUGACUAUACAGUCAUUUAAAGCAGAAUUUCGCUUAGCAGGAGGUGUAAAUUUACCAAAAAUAAUAGAUUGUGUAGGUUCCGAUGGCAAGGAGAGGAGACAGCUUGUUAAGGGCCGUGAUGACCUGAGACAAGAUGCUGUCAUGCAACAGGUCUUCCAGAUGUGUAAUACAUUACUGCAGAGAAACACGGAAACUAGGAAGAGGAAAUUAACUAUCUGUACUUAUAAGGAUCUUCUCUUAGAAAUCUACAGAAGUAUAGGGGAGCCAGAUAGUUUGUA | circ |
| ENSG00000149311:+:11:108315822:108335961 | ENSG00000149311 | ENST00000683174 | + | 11 | 108315823 | 108335961 | 22 | GUACUUAUAAGGAUCUUCUCUU | bsj |
| ENSG00000149311:+:11:108315822:108335961 | ENSG00000149311 | ENST00000683174 | + | 11 | 108315623 | 108315832 | 210 | AUAAGUUGUCCUGCACAGUUCAAACUCGUGUUGUUUGAACUGUAUUUCAGAACUGUAUUUCAGAAUCAUUACAUUUUAUUUCUAUAACAUAACAUUUAGAGUUGGGAGUUACAUAUUGGUAAUGAUACAAUUUAAAAUUUGCUAAAUUUAUAGACCGAUUUUUUUUCCUUCUUCAAUUUUUGUUGUUUCCAUGUUUUCAGGAUCUUCUCU | ie_up |
| ENSG00000149311:+:11:108315822:108335961 | ENSG00000149311 | ENST00000683174 | + | 11 | 108335952 | 108336161 | 210 | UACUUAUAAGGUAACUAUUUGUACUUCUGUUAGUUCACCAAAAACAUAUAAAAGAUGCCAUUUGGUUGGGUGAAGUGGCUCAUGCCCAUAUUCAUAAUGCUUUGGGAGGCCAAGGUGGGAGGAUUGCUUGAGGCCAGGAGUUCGAGACCAGCCUCAGCAACAUAGUGAGACCCCAUCUUGACAAAAAGUUAAAAAAAAAAAAAAAGCCAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
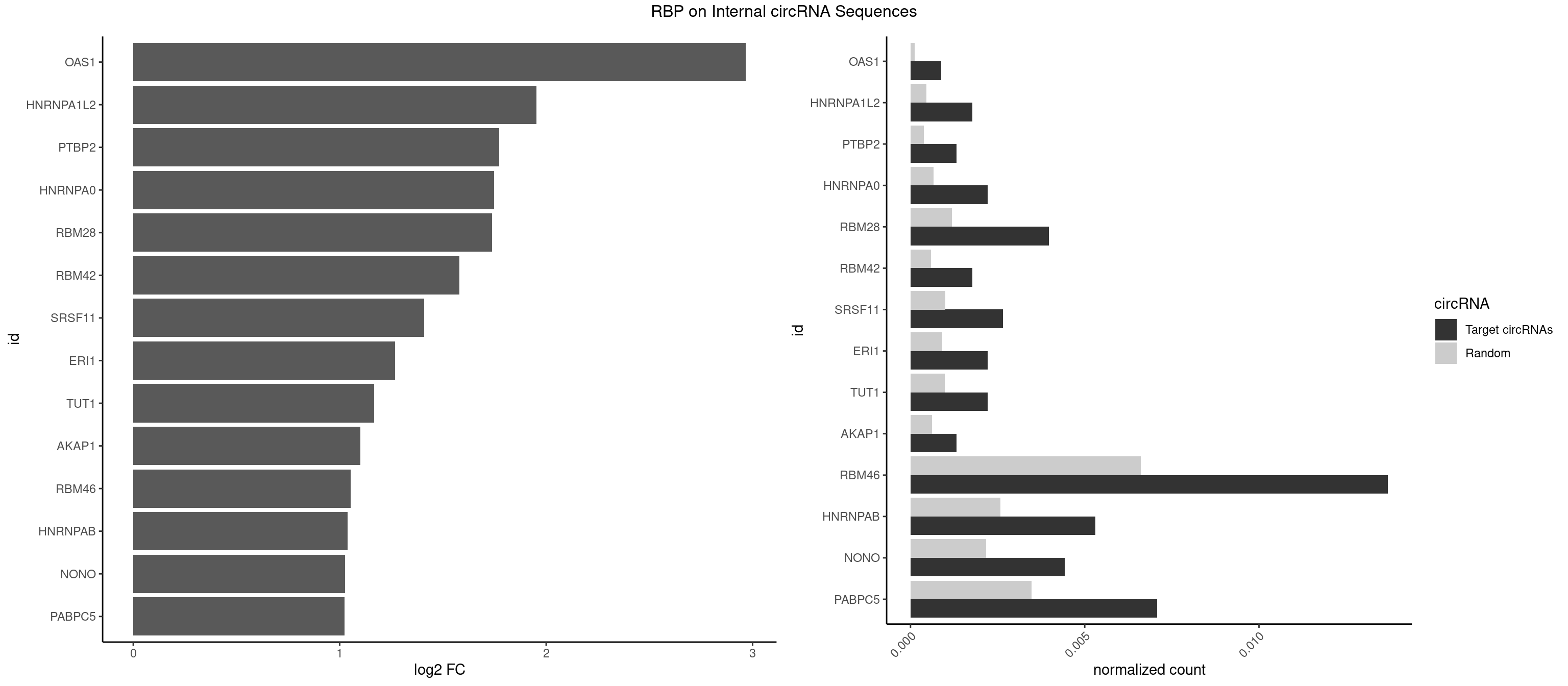
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.0008841733 | 0.0001130379 | 2.967522 | GCAUAA | GCAUAA |
| HNRNPA1L2 | 3 | 314 | 0.0017683466 | 0.0004564992 | 1.953717 | AUAGGG,UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| PTBP2 | 2 | 267 | 0.0013262599 | 0.0003883867 | 1.771798 | CUCUCU | CUCUCU |
| HNRNPA0 | 4 | 453 | 0.0022104332 | 0.0006579386 | 1.748304 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBM28 | 8 | 822 | 0.0039787798 | 0.0011926949 | 1.738101 | AGUAGA,AGUAGG,AGUAGU,GUGUAG,UGUAGA,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM42 | 3 | 407 | 0.0017683466 | 0.0005912752 | 1.580499 | AACUAA,AACUAC,ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRSF11 | 5 | 688 | 0.0026525199 | 0.0009985015 | 1.409527 | AAGAAG | AAGAAG |
| ERI1 | 4 | 632 | 0.0022104332 | 0.0009173461 | 1.268791 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 4 | 678 | 0.0022104332 | 0.0009840095 | 1.167585 | AGAUAC,CAAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| AKAP1 | 2 | 426 | 0.0013262599 | 0.0006188101 | 1.099795 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBM46 | 30 | 4554 | 0.0137046861 | 0.0066011240 | 1.053886 | AAUCAU,AAUGAA,AAUGAU,AUCAAG,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,GAUCAA,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPAB | 11 | 1782 | 0.0053050398 | 0.0025839306 | 1.037796 | AAAGAC,ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NONO | 9 | 1498 | 0.0044208665 | 0.0021723567 | 1.025068 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 | 15 | 2400 | 0.0070733864 | 0.0034795387 | 1.023505 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
RBP on BSJ (Exon Only)
Plot
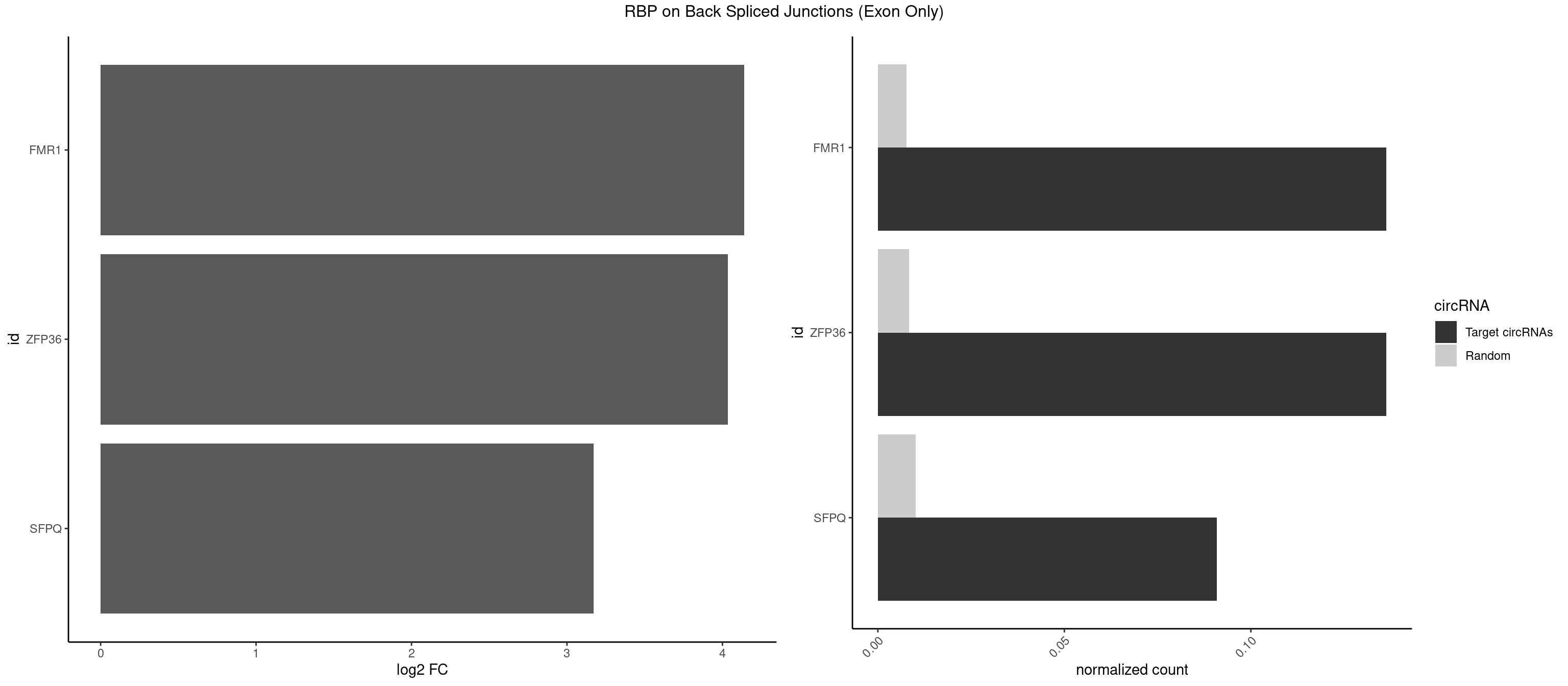
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FMR1 | 2 | 117 | 0.13636364 | 0.007728583 | 4.141111 | AAGGAU,UAAGGA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| ZFP36 | 2 | 126 | 0.13636364 | 0.008318051 | 4.035070 | AUAAGG,UAAGGA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | UAAGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
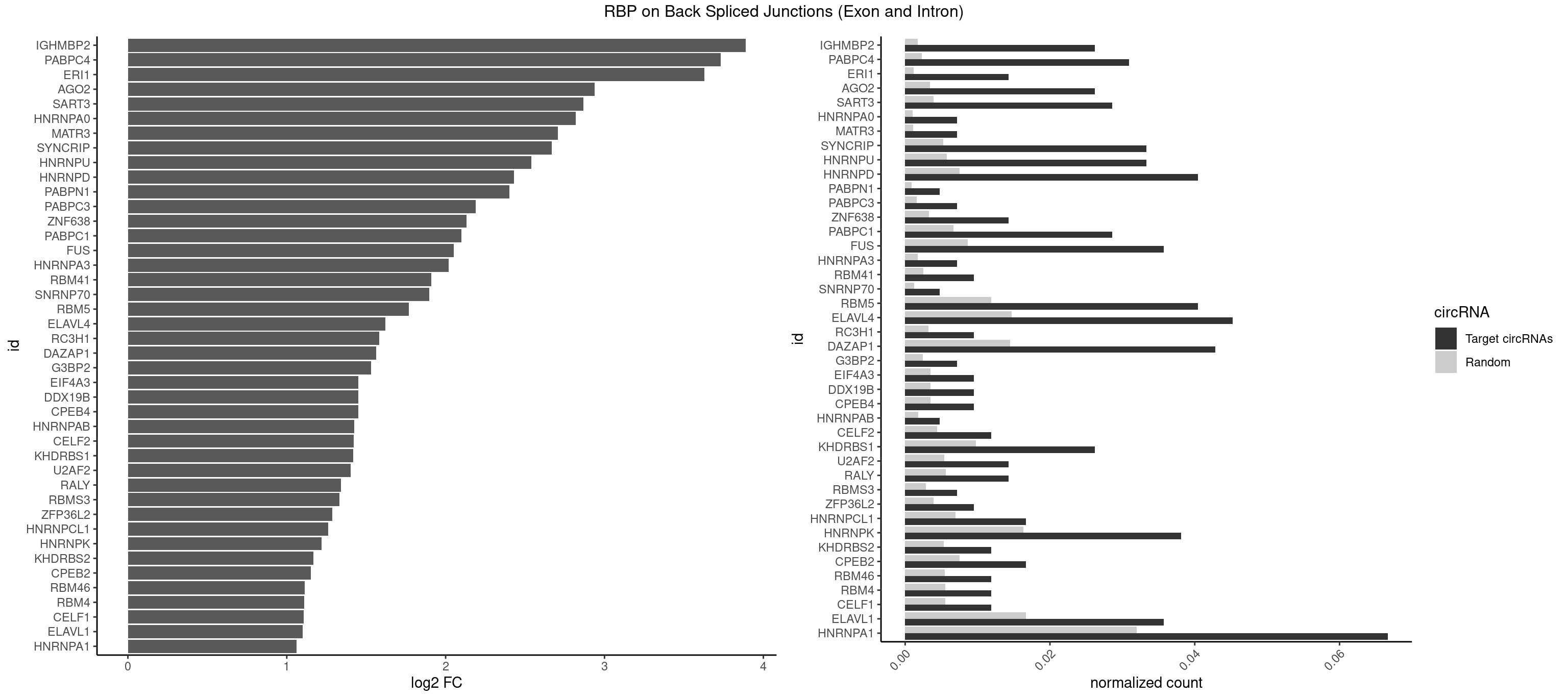
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 10 | 350 | 0.026190476 | 0.0017684401 | 3.888493 | AAAAAA | AAAAAA |
| PABPC4 | 12 | 462 | 0.030952381 | 0.0023327287 | 3.729960 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| ERI1 | 5 | 228 | 0.014285714 | 0.0011537686 | 3.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| AGO2 | 10 | 677 | 0.026190476 | 0.0034159613 | 2.938679 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SART3 | 11 | 777 | 0.028571429 | 0.0039197904 | 2.865725 | AAAAAA,AAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SYNCRIP | 13 | 1041 | 0.033333333 | 0.0052498992 | 2.666604 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| HNRNPU | 13 | 1136 | 0.033333333 | 0.0057285369 | 2.540727 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPD | 16 | 1488 | 0.040476190 | 0.0075020153 | 2.431723 | AAAAAA,AAUUUA,AUUUAU,UUAGAG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 5 | 646 | 0.014285714 | 0.0032597743 | 2.131729 | GGUUGG,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPC1 | 11 | 1321 | 0.028571429 | 0.0066606207 | 2.100845 | AAAAAA,AAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| FUS | 14 | 1711 | 0.035714286 | 0.0086255542 | 2.049812 | AAAAAA,GGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM5 | 16 | 2359 | 0.040476190 | 0.0118903668 | 1.767280 | AAAAAA,AAGGUA,AAGGUG,AGGUAA,CUUCUC,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| ELAVL4 | 18 | 2916 | 0.045238095 | 0.0146966949 | 1.622046 | AAAAAA,AUUUAU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCUUCU,CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| DAZAP1 | 17 | 2878 | 0.042857143 | 0.0145052398 | 1.562962 | AAAAAA,AAUUUA,AGGUAA,AGUUAA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUU,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| KHDRBS1 | 10 | 1945 | 0.026190476 | 0.0098045143 | 1.417524 | AUAAAA,AUUUAA,CUAAAU,UAAAAA,UAAAAG,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.0054010480 | 1.403262 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 5 | 1117 | 0.014285714 | 0.0056328094 | 1.342647 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | CAUAUA,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPK | 15 | 3244 | 0.038095238 | 0.0163492543 | 1.220386 | AAAAAA,ACCCCA,CCCCAU,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | AUUUUU,CAUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUCAU,AAUGAU,AUCAUU,AUGAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUUCU,CUUCUU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUGUUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | AUUUAU,UGUUUU,UUAGUU,UUAUUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPA1 | 27 | 6340 | 0.066666667 | 0.0319478033 | 1.061249 | AAAAAA,AAGGUG,AAUUUA,AUUUAA,AUUUAU,CCAAGG,GCCAAG,GGAGGA,UAGAGU,UAGUGA,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.