circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000112701:+:6:75621531:75647801
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000112701:+:6:75621531:75647801 | ENSG00000112701 | ENST00000370010 | + | 6 | 75621532 | 75647801 | 477 | CUUUGGCUAGAUCAGAGUCUAAGAGAGAUGGAGGUUUUAAAAAUAAUUGGAGCUUUGAUCAUGAAGAAGAAAGUGAAGGAGAUACAGAUAAAGAUGGGACAAAUCUGCUCAGUGUGGAUGAAGAUGAGGAUUCUGAAACCUCAAAAGGAAAAAAGUUAAAUCGUCGAUCUGAAAUUGUUGCUAAUAGCUCUGGUGAAUUCAUCUUGAAGACAUAUGUAAGACGAAACAAGUCUGAAAGUUUUAAAACUUUGAAAGGCAACCCAAUUGGACUUAACAUGUUGAGCAACAAUAAGAAAUUGAGUGAAAAUACGCAAAAUACGUCAUUAUGUUCUGGAACUGUAGUUCAUGGUAGACGUUUUCAUCAUGCUCAUGCACAGAUACCAGUAGUAAAAACAGCAGCCCAAAGGAAAGAAUACCCACCUCAUGUCCAAAAAGUUGAAAUUAAUCCUGUAAGGUUAAGUCGGCUCCAAGGUGUUGCUUUGGCUAGAUCAGAGUCUAAGAGAGAUGGAGGUUUUAAAAAUAAUUGG | circ |
| ENSG00000112701:+:6:75621531:75647801 | ENSG00000112701 | ENST00000370010 | + | 6 | 75621532 | 75647801 | 22 | CCAAGGUGUUGCUUUGGCUAGA | bsj |
| ENSG00000112701:+:6:75621531:75647801 | ENSG00000112701 | ENST00000370010 | + | 6 | 75621332 | 75621541 | 210 | GAUGAUUAUGAAUUAGUAAAAACAGAUUUUAAUGUCUUCUUUUUCUCUUUGGAGGAAAUCAAAUGUUUACAAAAAAUAGACAAAGGCAACCAAAUUAAAACAAAAUCUAGAAUAAUCAAAGAGCUUGGGAAAUGCACAUAUUUGUUUUAUAUUGAAUAGCUCUAUUAAUGAAUACUUACUGCAGUUUGUUUUUAUUUCAGCUUUGGCUAG | ie_up |
| ENSG00000112701:+:6:75621531:75647801 | ENSG00000112701 | ENST00000370010 | + | 6 | 75647792 | 75648001 | 210 | CAAGGUGUUGGUAAGUGUGCAGUUUUGUUACACCUGUGAAGGAUUUCAAAUUGCUGUAUGAAAAGUACAAUGGAGAUACGAUGCUGGCUUUAGAAUAUUCCCAGGUAUAAUAGAUAUAUAUGUAGAAAACCAUUUUUUUGUAACUGAUUUGAGAAUUUCUUCAAUGGUAACCUGAUACCUGGAACAGUUGCACAGUUAGGUGAUUACAGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
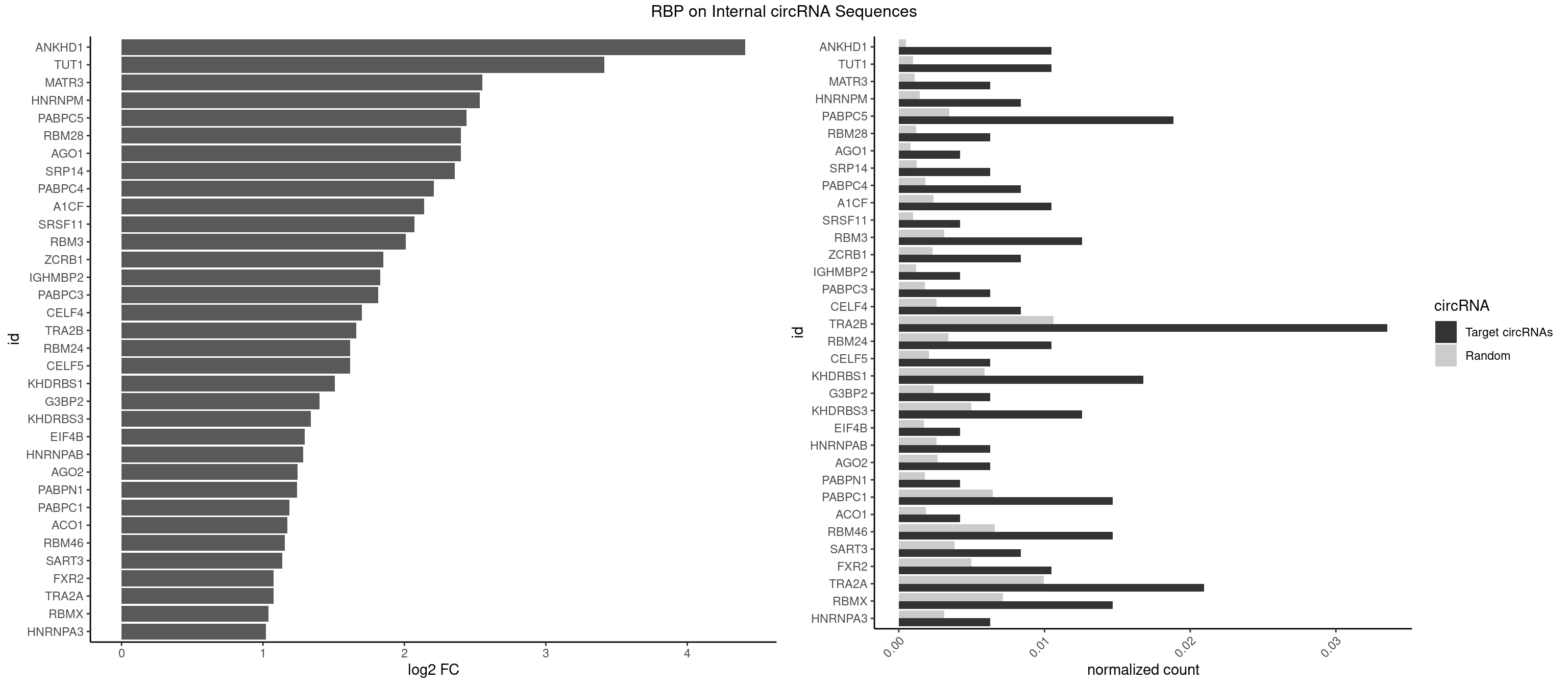
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 4 | 339 | 0.010482180 | 0.0004927293 | 4.411000 | AGACGA,AGACGU,GACGAA,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| TUT1 | 4 | 678 | 0.010482180 | 0.0009840095 | 3.413123 | AAAUAC,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| MATR3 | 2 | 739 | 0.006289308 | 0.0010724109 | 2.552044 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPM | 3 | 999 | 0.008385744 | 0.0014492040 | 2.532678 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| PABPC5 | 8 | 2400 | 0.018867925 | 0.0034795387 | 2.438968 | AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM28 | 2 | 822 | 0.006289308 | 0.0011926949 | 2.398676 | AGUAGU,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| AGO1 | 1 | 548 | 0.004192872 | 0.0007956130 | 2.397800 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 847 | 0.006289308 | 0.0012289250 | 2.355505 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PABPC4 | 3 | 1251 | 0.008385744 | 0.0018144033 | 2.208444 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| A1CF | 4 | 1642 | 0.010482180 | 0.0023810421 | 2.138274 | AUAAUU,GAUCAG,UAAUUG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SRSF11 | 1 | 688 | 0.004192872 | 0.0009985015 | 2.070102 | AAGAAG | AAGAAG |
| RBM3 | 5 | 2152 | 0.012578616 | 0.0031201361 | 2.011292 | AAAACU,AAGACG,AAUACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ZCRB1 | 3 | 1605 | 0.008385744 | 0.0023274215 | 1.849206 | AAUUAA,ACUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGHMBP2 | 1 | 813 | 0.004192872 | 0.0011796520 | 1.829577 | AAAAAA | AAAAAA |
| PABPC3 | 2 | 1234 | 0.006289308 | 0.0017897669 | 1.813130 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| CELF4 | 3 | 1782 | 0.008385744 | 0.0025839306 | 1.698371 | GGUGUU,GUGUGG,GUGUUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TRA2B | 15 | 7329 | 0.033542977 | 0.0106226650 | 1.658865 | AAAGAA,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGGAAA,AUAAGA,GAAAGA,GAAGAA,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM24 | 4 | 2357 | 0.010482180 | 0.0034172229 | 1.617043 | AGUGUG,GAGUGA,GUGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF5 | 2 | 1415 | 0.006289308 | 0.0020520728 | 1.615819 | GUGUGG,GUGUUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| KHDRBS1 | 7 | 4064 | 0.016771488 | 0.0058910141 | 1.509423 | CAAAAU,UAAAAA,UAAAAC,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| G3BP2 | 2 | 1644 | 0.006289308 | 0.0023839405 | 1.399553 | AGGAUU,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| KHDRBS3 | 5 | 3429 | 0.012578616 | 0.0049707696 | 1.339432 | AAAUAA,AGAUAA,AUAAAG,GAUAAA,UAAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| EIF4B | 1 | 1179 | 0.004192872 | 0.0017100607 | 1.293891 | UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPAB | 2 | 1782 | 0.006289308 | 0.0025839306 | 1.283334 | AAGACA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| AGO2 | 2 | 1830 | 0.006289308 | 0.0026534924 | 1.245009 | AAAAAA,AAAGUG | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPN1 | 1 | 1222 | 0.004192872 | 0.0017723764 | 1.242254 | AGAAGA | AAAAGA,AGAAGA |
| PABPC1 | 6 | 4443 | 0.014675052 | 0.0064402624 | 1.188174 | AAAAAA,AAAAAC,ACAAAU,CAAAUC,CUAAUA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ACO1 | 1 | 1283 | 0.004192872 | 0.0018607779 | 1.172033 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM46 | 6 | 4554 | 0.014675052 | 0.0066011240 | 1.152582 | AUCAUG,AUGAAG,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SART3 | 3 | 2634 | 0.008385744 | 0.0038186524 | 1.134875 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| FXR2 | 4 | 3434 | 0.010482180 | 0.0049780156 | 1.074296 | AGACGA,GACAAA,GACGAA,GGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| TRA2A | 9 | 6871 | 0.020964361 | 0.0099589296 | 1.073876 | AAAGAA,AAGAAA,AAGAAG,AGAAAG,AGAAGA,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBMX | 6 | 4925 | 0.014675052 | 0.0071387787 | 1.039616 | AAGAAG,AAGGAA,GAAGGA,UAAGAC,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPA3 | 2 | 2140 | 0.006289308 | 0.0031027457 | 1.019356 | AAGGAG,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
RBP on BSJ (Exon Only)
Plot
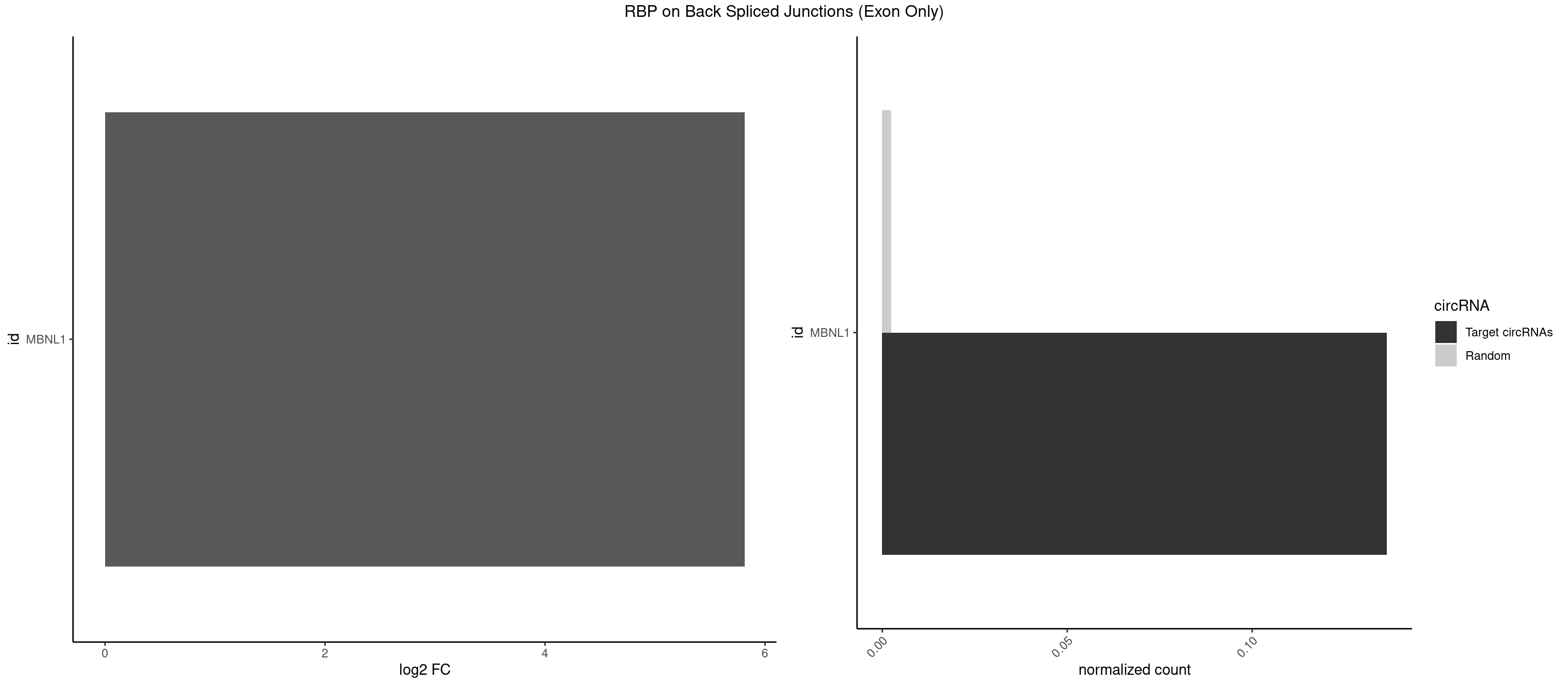
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MBNL1 | 2 | 36 | 0.1363636 | 0.002423369 | 5.814301 | UGCUUU,UUGCUU | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
RBP on BSJ (Exon and Intron)
Plot
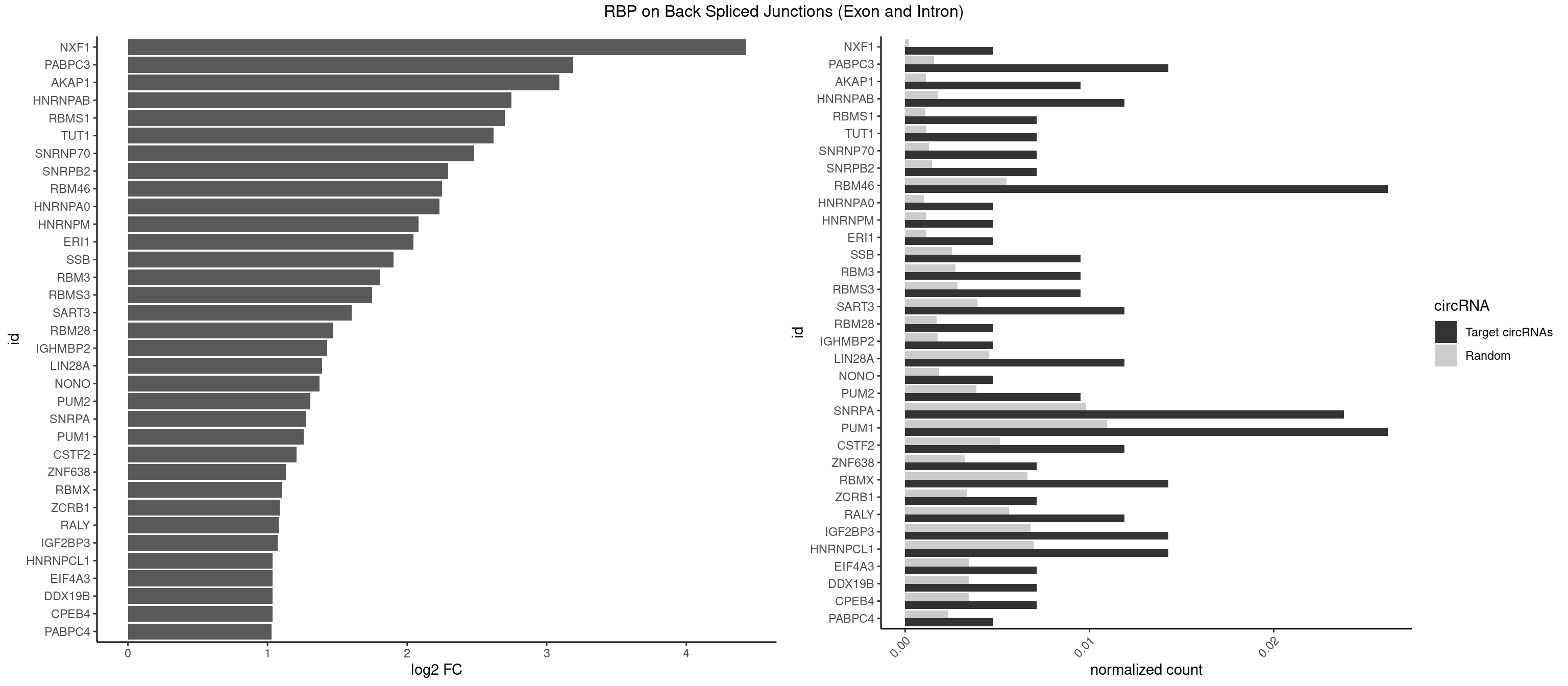
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| PABPC3 | 5 | 310 | 0.014285714 | 0.0015669085 | 3.188580 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| AKAP1 | 3 | 221 | 0.009523810 | 0.0011185006 | 3.089973 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| HNRNPAB | 4 | 351 | 0.011904762 | 0.0017734784 | 2.746885 | ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM46 | 10 | 1091 | 0.026190476 | 0.0055018138 | 2.251063 | AAUCAA,AAUGAA,AUCAAA,AUGAAA,AUGAAU,AUGAUU,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAUACU,AUACGA,GAUACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AAAAAC,AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGAU,GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAGAUA,UAUAUA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SNRPA | 9 | 1947 | 0.023809524 | 0.0098145909 | 1.278539 | AUACCU,AUGCAC,AUGCUG,GAUACC,GGAGAU,UACCUG,UGCACA,UUGCAC | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AAUAUU,AAUGUU,AGAAUU,UAAUGU,UAGAUA,UAUAUA,UGUAGA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUG,GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AAGUGU,ACCAAA,AUCAAA,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,AAAACA,AAAAUC,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.