circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000048405:-:7:127373341:127377329
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000048405:-:7:127373341:127377329 | ENSG00000048405 | ENST00000265827 | - | 7 | 127373342 | 127377329 | 1837 | GAACUAAACAACUUAAGCAUAUUUUAUUAAAAGAUGUGGACACUAUUUUUGAAUGUAAGUUAUGCCGCAGUCUCUUCAGAGGAUUACCAAAUUUAAUUACCCAUAAAAAAUUCUACUGCCCACCAAGUCUCCAGAUGGAUGACAACCUUCCUGAUGUAAAUGAUAAACAAAGCCAAGCCAUAAAUGAUCUCCUAGAAGCCAUAUAUCCAAGUGUGGACAAACGAGAAUAUAUUAUUAAGCUAGAACCCAUAGAAACUAAUCAAAAUGCAGUAUUUCAAUAUAUUUCGAGGACUGAUAAUCCUAUUGAAGUCACAGAGUCAAGCAGUACUCCUGAACAAACCGAAGUUCAGAUACAGGAAACUAGCACUGAACAGUCAAAAACAGUACCGGUUACAGAUACAGAGGUGGAAACUGUAGAGCCCCCUCCUGUUGAGAUUGUUACAGAUGAAGUUGCACCUACAUCUGAUGAACAACCUCAGGAGUCGCAGGCUGACUUGGAAACUUCUGACAAUUCUGAUUUUGGUCACCAGUUGAUAUGUUGUCUUUGUAGAAAAGAAUUCAAUUCUAGACGAGGUGUUCGCCGUCACAUUCGAAAAGUACACAAGAAAAAGAUGGAAGAACUAAAAAAGUACAUUGAAACACGAAAGAAUCCAAACCAAUCCUCUAAAGGACGCAGUAAGAAUGUUCUAGUUCCAUUAAGUAGGAGUUGUCCAGUAUGUUGUAAAUCAUUUGCUACAAAAGCGAAUGUAAGGAGGCAUUUUGAUGAAGUUCAUAGAGGACUAAGGAGGGAUUCAAUUACUCCUGAUAUAGCAACAAAGCCUGGGCAACCUUUGUUCCUGGAUUCUAUUUCUCCUAAAAAAUCUUUUAAGACUCGAAAACAAAAGUCUUCUUCAAAGGCUGAAUACAAUUUAACUGCAUGCAAAUGCCUCCUUUGCAAGAGGAAAUAUAGUUCACAAAUAAUGCUUAAAAGACAUAUGCAAAUUGUCCACAAGAUAACUCUUUCUGGAACAAACUCUAAAAGAGAAAAAGGCCCUAAUAAUACUGCCAACAGUUCAGAAAUAAAAGUUAAAGUUGAACCAGCAGAUUCUGUAGAAUCUUCACCCCCUUCCAUUACCCAUUCUCCACAGAAUGAAUUAAAGGGAACAAAUCAUUCAAAUGAAAAAAAGAACACACCGGCAGCACAGAAAAAUAAAGUUAAACAAGACUCUGAAAGCCCUAAAUCAACUAGUCCGUCGGCUGCAGGUGGCCAGCAAAAAACCAGAAAACCAAAACUUUCAGCUGGCUUUGACUUUAAGCAACUUUACUGUAAACUUUGUAAACGUCAGUUUACUUCCAAACAGAACUUGACUAAACACAUCGAGUUGCACACAGAUGGAAAUAACAUUUAUGUUAAAUUCUACAAGUGUCCUCUUUGCACUUAUGAAACUCGUCGGAAACGUGAUGUGAUACGACAUAUAACUGUGGUUCAUAAAAAGUCAUCUCGUUAUCUUGGGAAAAUAACAGCCAGUUUAGAGAUCAGAGCUAUAAAAAAGCCUAUUGAUUUUGUUCUAAAUAAAGUGGCAAAAAGAGGCCCUUCGAGGGAUGAAGCAAAACAUAGUGAUUCAAAACAUGAUGGCACUUCUAACUCUCCUAGUAAAAAGUAUGAAGUAGCUGACGUCGGUAUUGAAGUAAAAGUCACAAAAAACUUUUCUCUUCACAGAUGCAAUAAAUGUGGAAAGGCAUUUGCCAAAAAGACUUACCUUGAACAUCAUAAGAAAACUCAUAAGGCAAAUGCUUCCAAUUCACCUGAAGGAAACAAAACCAAAGGCCGAAGUACAAGAUCUAAGGCUCUUGUCUGGAACUAAACAACUUAAGCAUAUUUUAUUAAAAGAUGUGGACACUAUUUUU | circ |
| ENSG00000048405:-:7:127373341:127377329 | ENSG00000048405 | ENST00000265827 | - | 7 | 127373342 | 127377329 | 22 | GCUCUUGUCUGGAACUAAACAA | bsj |
| ENSG00000048405:-:7:127373341:127377329 | ENSG00000048405 | ENST00000265827 | - | 7 | 127377320 | 127377529 | 210 | UUUCCAAAUGAUAGUAGGGUUUUGCUUAGAAUUUAGUCUAUAACUUAGUUCUAAAUAUACUGUGGUGGAAAUUGGCAAUGCCUAUAUUUUGUUUUCAAAGAGUAACUGCACUUUUGGUCUGUCAACAGUGGUUGUCCAGUUUAAAAGUGCAUGUUAAAGUUACCUUUUGUGUUAAGUUUUUCUUUUCUUUUUUCUCUCAGGAACUAAACA | ie_up |
| ENSG00000048405:-:7:127373341:127377329 | ENSG00000048405 | ENST00000265827 | - | 7 | 127373142 | 127373351 | 210 | CUCUUGUCUGGUGAGGAACAGUUAACAGAGUUUUGCUUUUUUCCCCCCAUCGAACUAAAAAAAAAAUCAUUUGACCAUAAUUUAUAGCUGGUUCCAUUUUAACACGUUUGCUUCCAUAUAUCUCAUGGCAAUGGGAACUGCAAGAGUAAUGUGCAUAUUGCAUUUACCUCUUCAGUGACCUUUAUUCCAGUGGCUUGGGAACAAAAGUUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
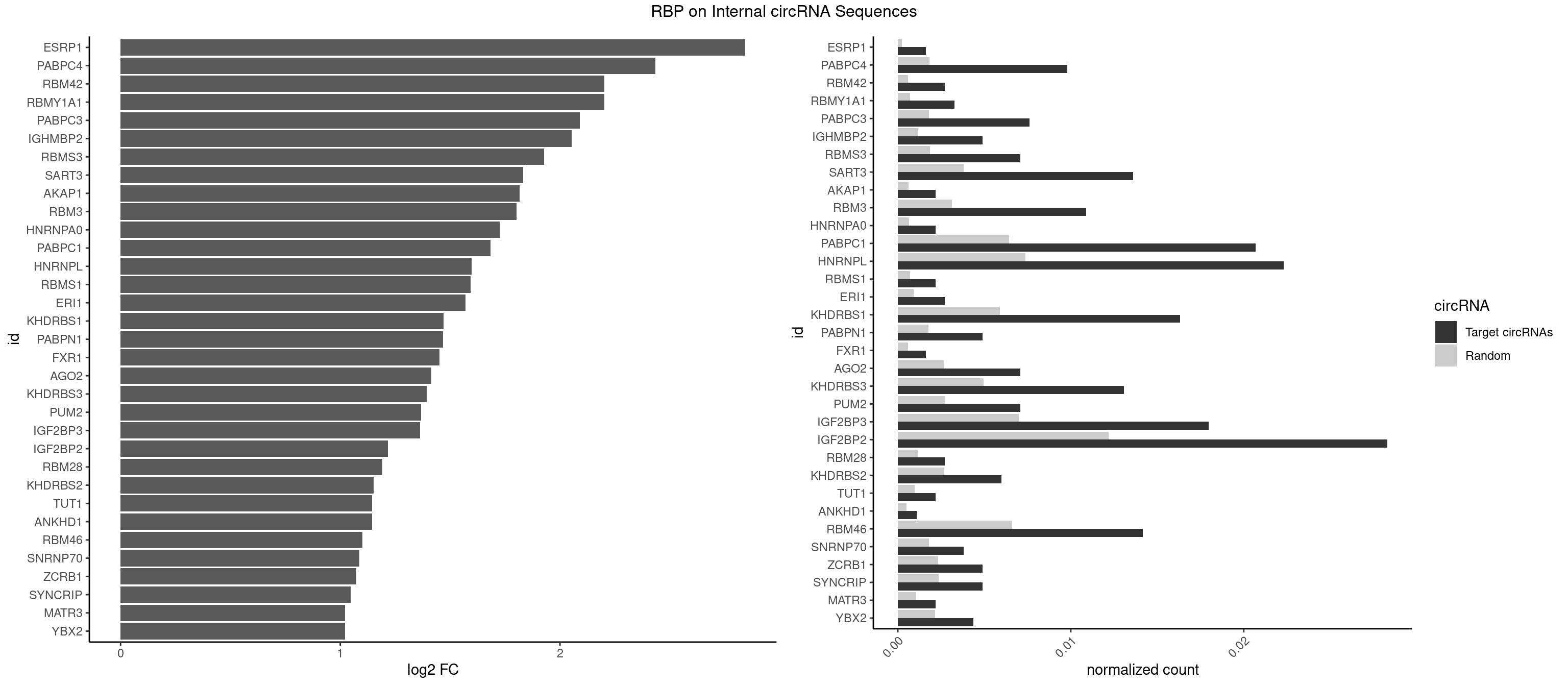
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 2 | 156 | 0.001633097 | 0.0002275250 | 2.843514 | AGGGAU | AGGGAU |
| PABPC4 | 17 | 1251 | 0.009798585 | 0.0018144033 | 2.433078 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBM42 | 4 | 407 | 0.002721829 | 0.0005912752 | 2.202675 | AACUAA,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBMY1A1 | 5 | 489 | 0.003266195 | 0.0007101099 | 2.201497 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| PABPC3 | 13 | 1234 | 0.007621121 | 0.0017897669 | 2.090232 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGHMBP2 | 8 | 813 | 0.004899292 | 0.0011796520 | 2.054212 | AAAAAA | AAAAAA |
| RBMS3 | 12 | 1283 | 0.007076756 | 0.0018607779 | 1.927182 | AAUAUA,AUAUAG,AUAUAU,CAUAUA,UAUAGC,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SART3 | 24 | 2634 | 0.013609145 | 0.0038186524 | 1.833441 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| AKAP1 | 3 | 426 | 0.002177463 | 0.0006188101 | 1.815080 | AUAUAU | AUAUAU,UAUAUA |
| RBM3 | 19 | 2152 | 0.010887316 | 0.0031201361 | 1.802968 | AAAACU,AAACGA,AAACUA,AAGACU,AAUACU,AGACGA,AUACGA,GAAACG,GAAACU,GAUACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPA0 | 3 | 453 | 0.002177463 | 0.0006579386 | 1.726624 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| PABPC1 | 37 | 4443 | 0.020685901 | 0.0064402624 | 1.683456 | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CUAAUA,CUAAUC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPL | 40 | 5085 | 0.022318998 | 0.0073706513 | 1.598408 | AAACAA,AAACAC,AAAUAA,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACGA,CACAAA,CACAAG,CACACA,CACACC,CACGAA,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBMS1 | 3 | 497 | 0.002177463 | 0.0007217036 | 1.593170 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ERI1 | 4 | 632 | 0.002721829 | 0.0009173461 | 1.569038 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| KHDRBS1 | 29 | 4064 | 0.016330974 | 0.0058910141 | 1.471023 | AUAAAA,AUUUAA,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| PABPN1 | 8 | 1222 | 0.004899292 | 0.0017723764 | 1.466888 | AAAAGA | AAAAGA,AGAAGA |
| FXR1 | 2 | 411 | 0.001633097 | 0.0005970720 | 1.451634 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| AGO2 | 12 | 1830 | 0.007076756 | 0.0026534924 | 1.415196 | AAAAAA,AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| KHDRBS3 | 23 | 3429 | 0.013064780 | 0.0049707696 | 1.394142 | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAUA,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PUM2 | 12 | 1890 | 0.007076756 | 0.0027404447 | 1.368678 | GUAAAU,GUACAU,UAAAUA,UACAUC,UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| IGF2BP3 | 32 | 4815 | 0.017964072 | 0.0069793662 | 1.363946 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACACAC,ACAUUC,CAAACA,CACACA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 51 | 8408 | 0.028307022 | 0.0121863560 | 1.215893 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACACAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAAUUC,CACACA,CAUUCA,CCAAAC,CCAAUC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC,GCACAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM28 | 4 | 822 | 0.002721829 | 0.0011926949 | 1.190351 | AGUAGG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| KHDRBS2 | 10 | 1858 | 0.005988024 | 0.0026940701 | 1.152293 | AAUAAA,AUAAAA,AUAAAC,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| TUT1 | 3 | 678 | 0.002177463 | 0.0009840095 | 1.145904 | AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ANKHD1 | 1 | 339 | 0.001088732 | 0.0004927293 | 1.143781 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM46 | 25 | 4554 | 0.014153511 | 0.0066011240 | 1.100376 | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAUA,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SNRNP70 | 6 | 1237 | 0.003810561 | 0.0017941145 | 1.086731 | AAUCAA,AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZCRB1 | 8 | 1605 | 0.004899292 | 0.0023274215 | 1.073841 | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GCUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SYNCRIP | 8 | 1634 | 0.004899292 | 0.0023694485 | 1.048022 | AAAAAA | AAAAAA,UUUUUU |
| MATR3 | 3 | 739 | 0.002177463 | 0.0010724109 | 1.021791 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| YBX2 | 7 | 1480 | 0.004354927 | 0.0021462711 | 1.020816 | AACAAC,AACAUC,ACAACU,ACAUCA,ACAUCG,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
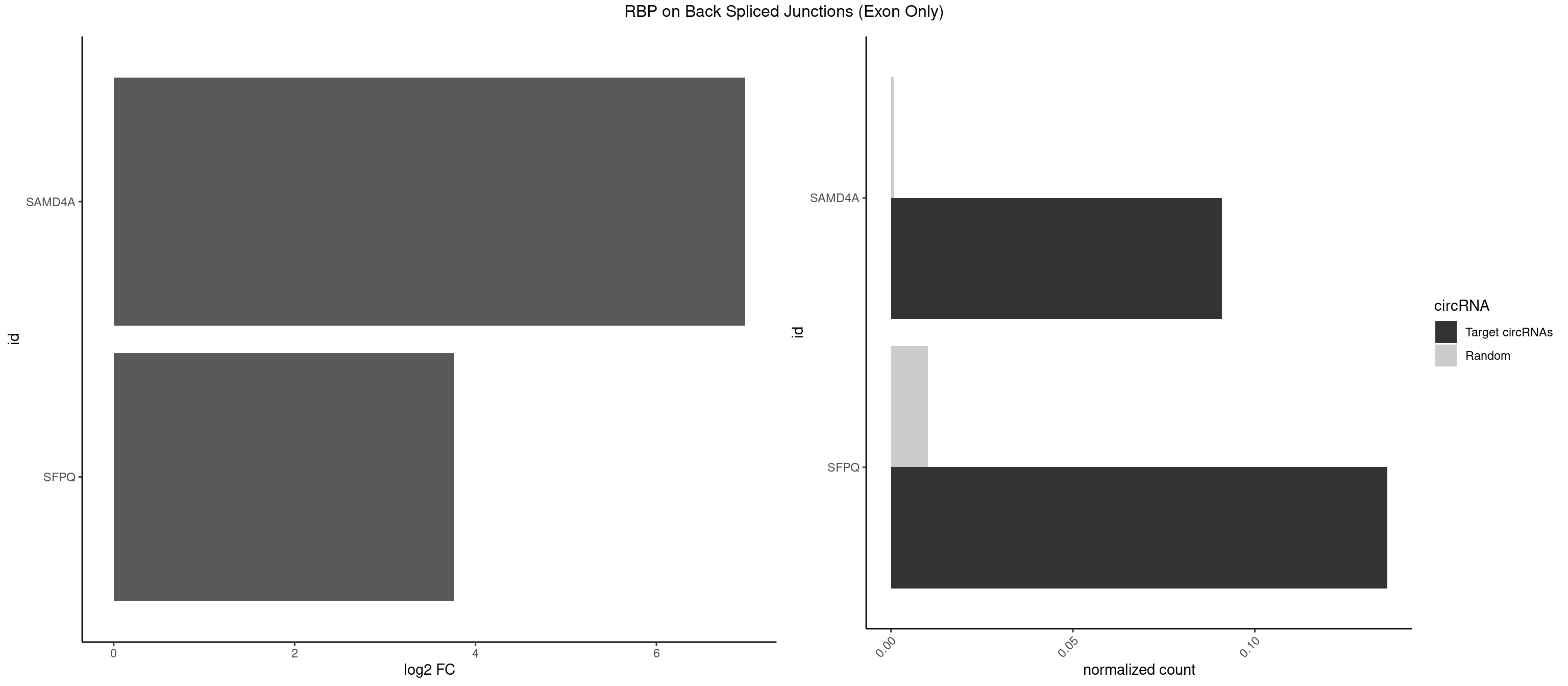
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SAMD4A | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | CUGGAA | CGGGAA,CGGGAC,CGGGCA,CGGGUA,CUGGAA,CUGGCA,CUGGCC,CUGGUA,GCUGGU |
| SFPQ | 2 | 153 | 0.13636364 | 0.0100864553 | 3.756968 | GUCUGG,UCUGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
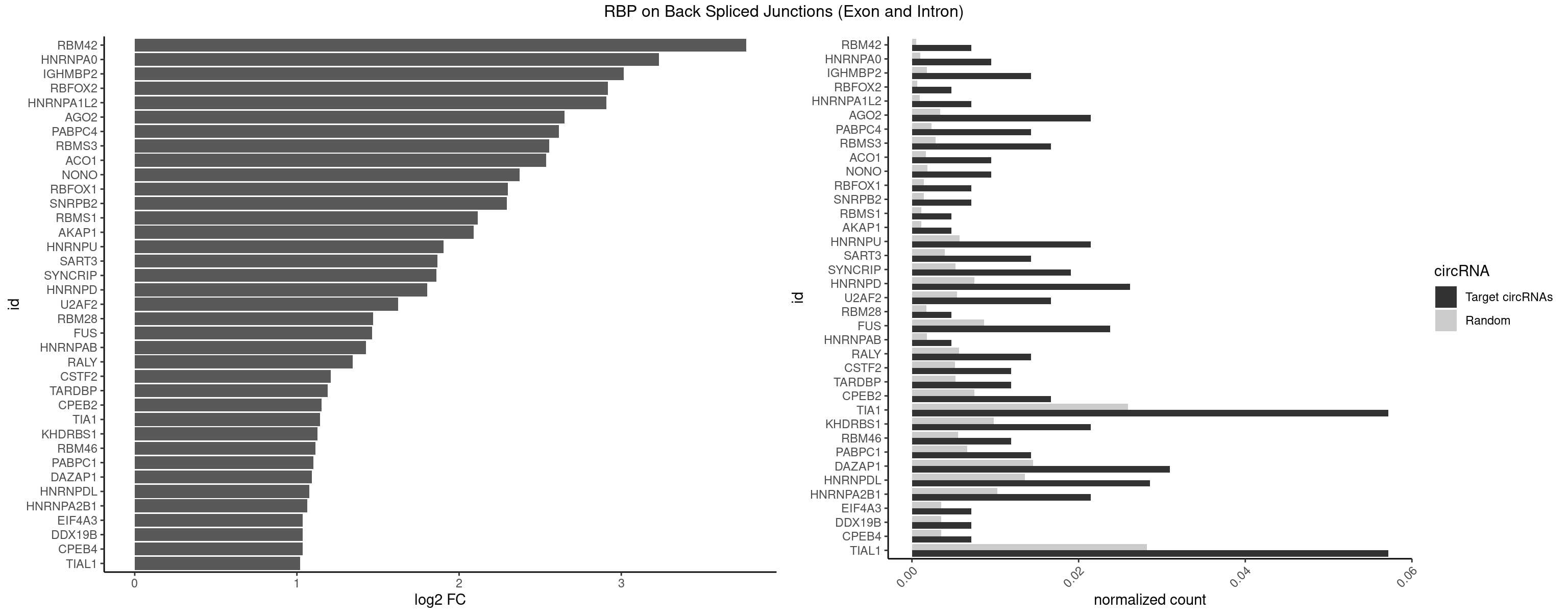
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| IGHMBP2 | 5 | 350 | 0.014285714 | 0.0017684401 | 3.014024 | AAAAAA | AAAAAA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | GUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| AGO2 | 8 | 677 | 0.021428571 | 0.0034159613 | 2.649172 | AAAAAA,AAAGUG,AAGUGC,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPC4 | 5 | 462 | 0.014285714 | 0.0023327287 | 2.614483 | AAAAAA | AAAAAA,AAAAAG |
| RBMS3 | 6 | 562 | 0.016666667 | 0.0028365578 | 2.554752 | AAUAUA,AUAUAU,CAUAUA,CUAUAU,UAUAGC,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ACO1 | 3 | 325 | 0.009523810 | 0.0016424829 | 2.535660 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | AUUGCA,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAC | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| HNRNPU | 8 | 1136 | 0.021428571 | 0.0057285369 | 1.903297 | AAAAAA,CCCCCC,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SART3 | 5 | 777 | 0.014285714 | 0.0039197904 | 1.865725 | AAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SYNCRIP | 7 | 1041 | 0.019047619 | 0.0052498992 | 1.859249 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| HNRNPD | 10 | 1488 | 0.026190476 | 0.0075020153 | 1.803692 | AAAAAA,AAUUUA,AGUAGG,AUUUAU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FUS | 9 | 1711 | 0.023809524 | 0.0086255542 | 1.464850 | AAAAAA,UGGUGA,UGGUGG,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RALY | 5 | 1117 | 0.014285714 | 0.0056328094 | 1.342647 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUUUUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUG,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UUUAUU,UUUUCU,UUUUGG,UUUUGU,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUUUAC,CUAAAA,CUAAAU,UAAAAA,UAAAAG,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUCAU,AAUGAU,AUCAUU,AUGAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| DAZAP1 | 12 | 2878 | 0.030952381 | 0.0145052398 | 1.093476 | AAAAAA,AAUUUA,AGUAGG,AGUUAA,AGUUUA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | AACUAA,AAUUUA,ACUGCA,AGUAGG,CCUUUA,GAACUA,UAACAG | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | AAUUUA,AGGAAC,AGUAGG,GUAGGG,UAGGGU,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| TIAL1 | 23 | 5597 | 0.057142857 | 0.0282043531 | 1.018655 | AGUUUU,AUUUUA,AUUUUG,CAGUUU,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAUUUU,UUCAGU,UUUAAA,UUUAUU,UUUUAA,UUUUCA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.