circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000229807:-:X:73829067:73831274
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000229807:-:X:73829067:73831274 | ENSG00000229807 | ENST00000429829 | - | X | 73829068 | 73831274 | 373 | AUCUUCCUCAGAAGAAUAGGCUUGUUGUUUUACAGUGUUAGUGAUCCAUUCCCUUUGACGAUCCCUAGGUGGAGAUGGGGCAUGAGGAUCCUCCAGGGGAAAAGCUCACUACCACUGGGCAACAACCCUAGGUCAGGAGGUUCUGUCAAGAUACUUUCCUGGUCCCAGAUAGGAAGAUAAAGUCUCAAAAACAACCACCACACGUCAAGCUCUUCAUUGUUCCUAUCUGCCAAAUCAUUAUACUUCCUACAAGCAGUGCAGAGAGCUGAGUCUUCAGCAGGUCCAAGAAAUUUGAACACACUGAAGGAAGUCAGCCUUCCCACCUGAAGAUCAACAUGCCUGGCACUCUAGCACUUGAGGAUAGCUGAAUGAAAUCUUCCUCAGAAGAAUAGGCUUGUUGUUUUACAGUGUUAGUGAUCCAUU | circ |
| ENSG00000229807:-:X:73829067:73831274 | ENSG00000229807 | ENST00000429829 | - | X | 73829068 | 73831274 | 22 | AGCUGAAUGAAAUCUUCCUCAG | bsj |
| ENSG00000229807:-:X:73829067:73831274 | ENSG00000229807 | ENST00000429829 | - | X | 73831265 | 73831474 | 210 | GGAAUGAAUAGGAUAAUUUUUUUUUUUUUUGCUAAGUUGGUAGCCAGAAUAUAACAGCUCCGCACAACUGUAAAUGUCCACUCUUCAAUCCACAUGAAGAAAAGGGUAAAAAUAUGGUUGAACUCAACCACUAGUUGCCCAUUAGAACAGACUUUCCCAGUGUACUGCAUUUCAAUACUUUUUCUUUUAUCUCUUUUCAGAUCUUCCUCA | ie_up |
| ENSG00000229807:-:X:73829067:73831274 | ENSG00000229807 | ENST00000429829 | - | X | 73828868 | 73829077 | 210 | GCUGAAUGAAGUAAGUUGUUGAUGUUGCAGUCCUGUGAGGAUCACUUCAGAACUGUUAUAACAGCUGUUUUUUGGGAGCUGGUGUUGGAUGGGGUGUGUUGGUCUAAUGUGAAGUGGGGCUAAAUGUGAGAUGGAAAGAUGACCAGUCUUCCAUAUUACUGACUGGGUUCACUGAAGCAACUCAAAGACAUUAUGGUCUUCUUACCAGUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
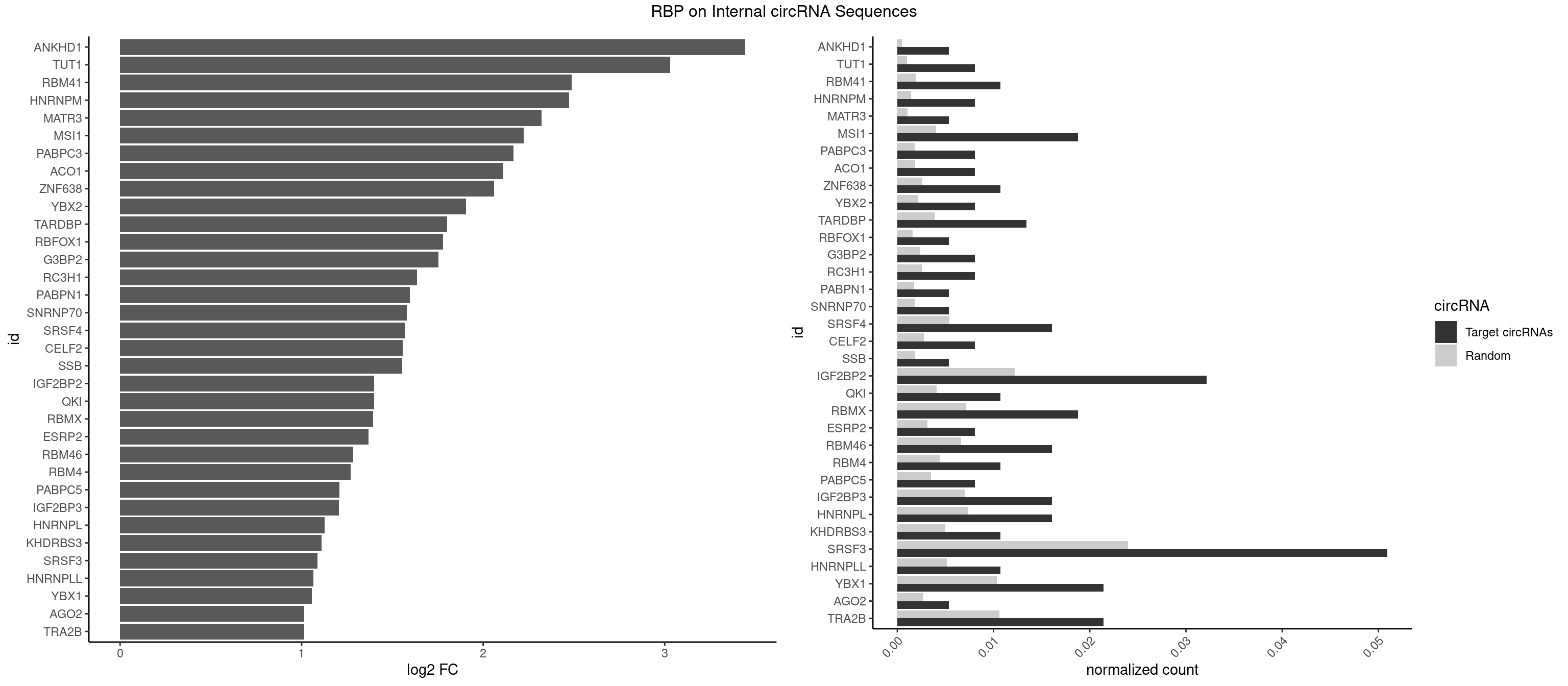
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 339 | 0.005361930 | 0.0004927293 | 3.443885 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| TUT1 | 2 | 678 | 0.008042895 | 0.0009840095 | 3.030971 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM41 | 3 | 1318 | 0.010723861 | 0.0019115000 | 2.488047 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 2 | 999 | 0.008042895 | 0.0014492040 | 2.472454 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| MATR3 | 1 | 739 | 0.005361930 | 0.0010724109 | 2.321895 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| MSI1 | 6 | 2770 | 0.018766756 | 0.0040157442 | 2.224440 | AGGAAG,AGGAGG,AGGUGG,UAGGAA,UAGGUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| PABPC3 | 2 | 1234 | 0.008042895 | 0.0017897669 | 2.167943 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 2 | 1283 | 0.008042895 | 0.0018607779 | 2.111809 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZNF638 | 3 | 1773 | 0.010723861 | 0.0025708878 | 2.060486 | GGUUCU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX2 | 2 | 1480 | 0.008042895 | 0.0021462711 | 1.905883 | AACAAC | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| TARDBP | 4 | 2654 | 0.013404826 | 0.0038476365 | 1.800708 | GAAUGA,GUUGUU,UGAAUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBFOX1 | 1 | 1077 | 0.005361930 | 0.0015622419 | 1.779135 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 2 | 1644 | 0.008042895 | 0.0023839405 | 1.754367 | AGGAUA,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RC3H1 | 2 | 1786 | 0.008042895 | 0.0025897275 | 1.634915 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPN1 | 1 | 1222 | 0.005361930 | 0.0017723764 | 1.597067 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 1 | 1237 | 0.005361930 | 0.0017941145 | 1.579481 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SRSF4 | 5 | 3740 | 0.016085791 | 0.0054214720 | 1.569030 | AGAAGA,AGGAAG,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| CELF2 | 2 | 1886 | 0.008042895 | 0.0027346479 | 1.556360 | GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SSB | 1 | 1260 | 0.005361930 | 0.0018274462 | 1.552924 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| IGF2BP2 | 11 | 8408 | 0.032171582 | 0.0121863560 | 1.400520 | AAAAAC,AAAACA,AAAUCA,AACACA,ACACAC,CAAAUC,CCACAC,CCAUUC,GAAAUC,GAACAC,GCACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| QKI | 3 | 2805 | 0.010723861 | 0.0040664663 | 1.398977 | AAUCAU,ACACAC,CACACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBMX | 6 | 4925 | 0.018766756 | 0.0071387787 | 1.394430 | AAGGAA,AGGAAG,AGUGUU,GAAGGA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ESRP2 | 2 | 2150 | 0.008042895 | 0.0031172377 | 1.367447 | GGGAAA,GGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM46 | 5 | 4554 | 0.016085791 | 0.0066011240 | 1.285003 | AAUCAU,AAUGAA,AUCAUU,AUGAAA,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM4 | 3 | 3065 | 0.010723861 | 0.0044432593 | 1.271134 | CCUUCC,CUUCCU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PABPC5 | 2 | 2400 | 0.008042895 | 0.0034795387 | 1.208819 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 5 | 4815 | 0.016085791 | 0.0069793662 | 1.204619 | AAAAAC,AAAACA,AAAUCA,AACACA,ACACAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPL | 5 | 5085 | 0.016085791 | 0.0073706513 | 1.125923 | AAACAA,AACACA,ACACAC,ACCACA,CACCAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| KHDRBS3 | 3 | 3429 | 0.010723861 | 0.0049707696 | 1.109283 | AGAUAA,AUAAAG,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SRSF3 | 18 | 16536 | 0.050938338 | 0.0239654858 | 1.087794 | ACCACC,ACGAUC,AUCAAC,AUCUUC,CACCAC,CACUAC,CAGAGA,CCACCA,CUACAA,CUCUUC,CUUCAG,CUUCAU,GAUCAA,UACUUC,UCUUCA,UCUUCC,UGUCAA | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| HNRNPLL | 3 | 3534 | 0.010723861 | 0.0051229360 | 1.065782 | ACACAC,ACCACA,CACCAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| YBX1 | 7 | 7119 | 0.021447721 | 0.0103183321 | 1.055615 | AACCAC,ACCACA,ACCACC,CAACCA,CACCAC,CCACAC,CCACCA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| AGO2 | 1 | 1830 | 0.005361930 | 0.0026534924 | 1.014860 | UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| TRA2B | 7 | 7329 | 0.021447721 | 0.0106226650 | 1.013679 | AAGAAU,AAGGAA,AGAAGA,AGGAAG,GAAGAA,GAAGGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
RBP on BSJ (Exon Only)
Plot
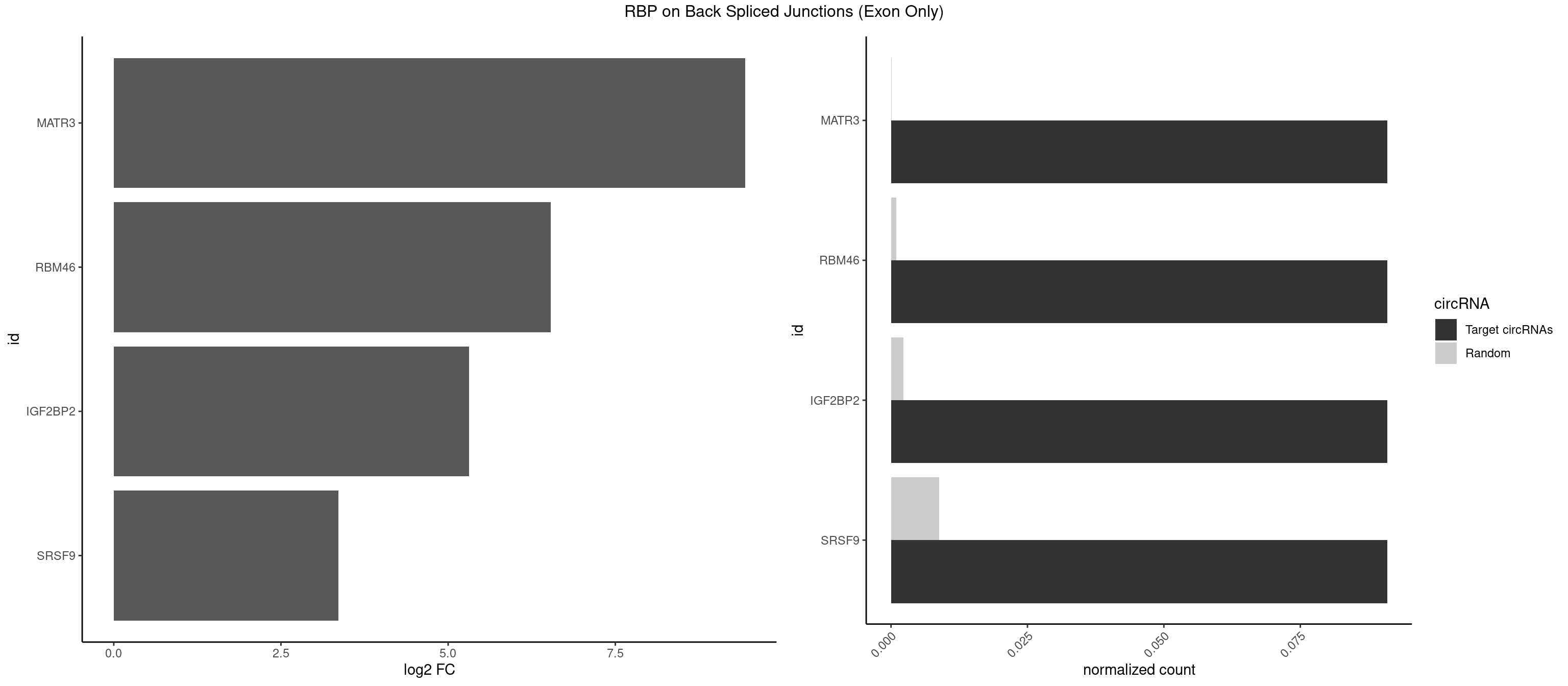
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 1 | 1 | 0.09090909 | 0.0001309929 | 9.438792 | AAUCUU | CAUCUU |
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | AUGAAA | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
| IGF2BP2 | 1 | 34 | 0.09090909 | 0.0022923762 | 5.309509 | GAAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
| SRSF9 | 1 | 134 | 0.09090909 | 0.0088420225 | 3.361976 | AUGAAA | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
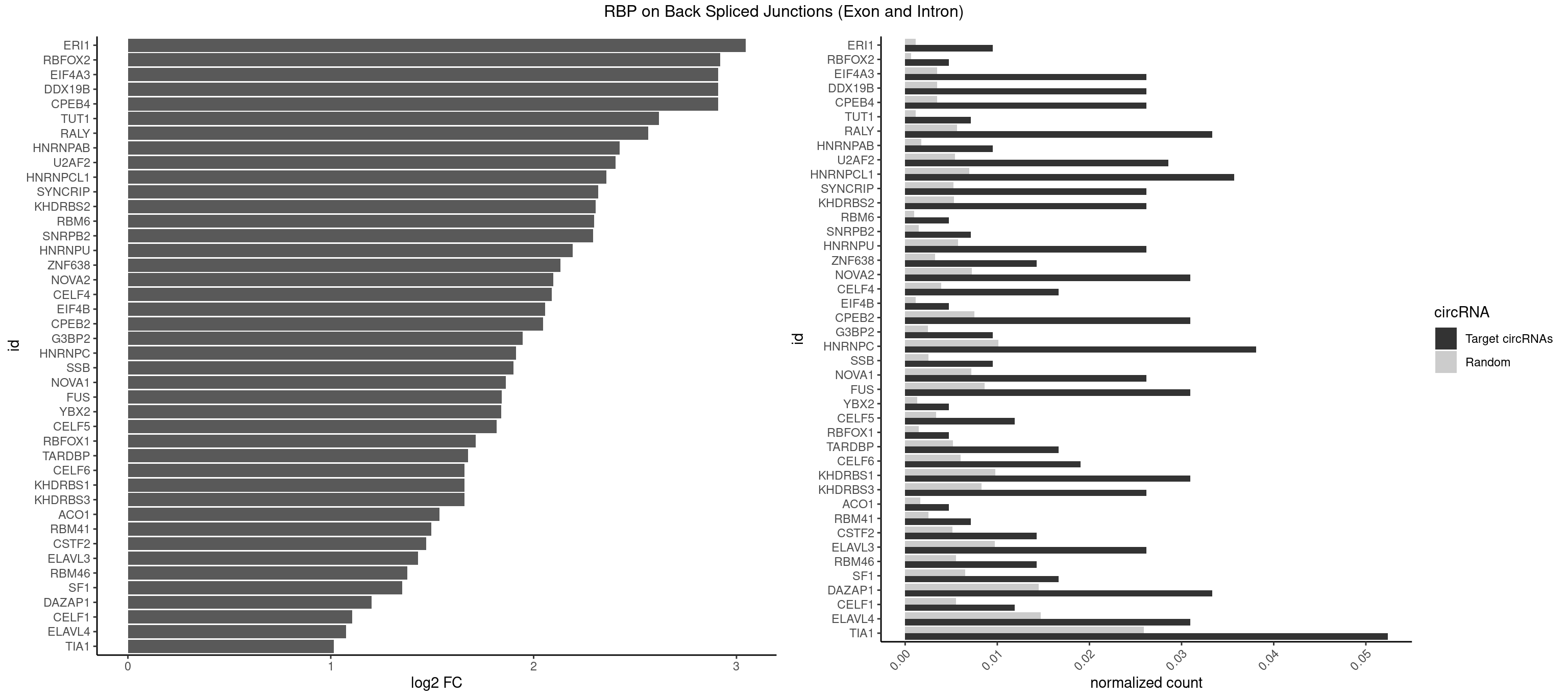
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ERI1 | 3 | 228 | 0.009523810 | 0.0011537686 | 3.045185 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| CPEB4 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| DDX19B | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| EIF4A3 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAUACU,CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RALY | 13 | 1117 | 0.033333333 | 0.0056328094 | 2.565039 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPAB | 3 | 351 | 0.009523810 | 0.0017734784 | 2.424957 | AAAGAC,AAGACA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| U2AF2 | 11 | 1071 | 0.028571429 | 0.0054010480 | 2.403262 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 | 14 | 1381 | 0.035714286 | 0.0069629182 | 2.358737 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP | 10 | 1041 | 0.026190476 | 0.0052498992 | 2.318681 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 10 | 1051 | 0.026190476 | 0.0053002821 | 2.304901 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | AAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPU | 10 | 1136 | 0.026190476 | 0.0057285369 | 2.192804 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ZNF638 | 5 | 646 | 0.014285714 | 0.0032597743 | 2.131729 | GUUGGU,GUUGUU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| NOVA2 | 12 | 1434 | 0.030952381 | 0.0072299476 | 2.097993 | AGACAU,GAUCAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| CELF4 | 6 | 776 | 0.016666667 | 0.0039147521 | 2.089973 | GGUGUG,GGUGUU,GUGUGU,GUGUUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CPEB2 | 12 | 1487 | 0.030952381 | 0.0074969770 | 2.045670 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUA,GGAUAA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPC | 15 | 2006 | 0.038095238 | 0.0101118501 | 1.913564 | AUUUUU,CUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| NOVA1 | 10 | 1428 | 0.026190476 | 0.0071997179 | 1.863030 | UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| FUS | 12 | 1711 | 0.030952381 | 0.0086255542 | 1.843361 | GGGUGU,UGGUGU,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,GUGUUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GAAUGA,GUGUGU,GUUGUU,UGAAUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF6 | 7 | 1196 | 0.019047619 | 0.0060308343 | 1.659181 | GUGAGG,GUGGGG,GUGUUG,UGUGAG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| KHDRBS1 | 12 | 1945 | 0.030952381 | 0.0098045143 | 1.658532 | CUAAAU,UAAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| KHDRBS3 | 10 | 1646 | 0.026190476 | 0.0082980653 | 1.658195 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ELAVL3 | 10 | 1928 | 0.026190476 | 0.0097188634 | 1.430183 | UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AAUGAA,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACAGAC,ACUGAC,AGUAAG,CACUGA,UACUGA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| DAZAP1 | 13 | 2878 | 0.033333333 | 0.0145052398 | 1.200391 | AGGAUA,AGUAAG,UAGGAU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUGUGU,GUGUUG,UGUGUU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ELAVL4 | 12 | 2916 | 0.030952381 | 0.0146966949 | 1.074559 | UCUAAU,UUUUAU,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUU,CUUUUA,CUUUUC,CUUUUU,GUUUUU,UUUUAU,UUUUCU,UUUUGG,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.