circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000145495:+:5:10415487:10417404
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000145495:+:5:10415487:10417404 | ENSG00000145495 | ENST00000274140 | + | 5 | 10415488 | 10417404 | 317 | UAUUUGCUGGCCGUUGGUUAAUGUCGUUUUGGACGGGGACUGCCAAAAUCCAUGAGCUCUACACAGCUGCUUGUGGUCUCUAUGUUUGCUGGCUAACCAUAAGGGCUGUGACGGUGAUGGUGGCAUGGAUGCCUCAGGGACGCAGAGUGAUCUUCCAGAAGGUUAAAGAGUGGUCUCUCAUGAUCAUGAAGACUUUGAUAGUUGCGGUGCUGUUGGCUGGAGUUGUCCCUCUCCUUCUGGGGCUCCUGUUUGAGCUGGUCAUUGUGGCUCCCCUGAGGGUUCCCUUGGAUCAGACUCCUCUUUUUUAUCCAUGGCAGUAUUUGCUGGCCGUUGGUUAAUGUCGUUUUGGACGGGGACUGCCAAAAUC | circ |
| ENSG00000145495:+:5:10415487:10417404 | ENSG00000145495 | ENST00000274140 | + | 5 | 10415488 | 10417404 | 22 | AUCCAUGGCAGUAUUUGCUGGC | bsj |
| ENSG00000145495:+:5:10415487:10417404 | ENSG00000145495 | ENST00000274140 | + | 5 | 10415288 | 10415497 | 210 | UACCUUGUUUUGGUUGUGAUUUCAUCAAAUUGUGUACUCUUUAGACAUUGUGAUUUUUGUCUUUUGUUUUUUUGAUGAAGACAUUGAUAUAUCGAAUGUGAUAUCUUUUUCCUAAUGUGAAAAUACUAACAUAACAACAUGAAGAUGACAAUUCUCUUUACCUAGCCACAUGUCUCAUUUAUCAGCUUUUCUAUUUUCAGUAUUUGCUGG | ie_up |
| ENSG00000145495:+:5:10415487:10417404 | ENSG00000145495 | ENST00000274140 | + | 5 | 10417395 | 10417604 | 210 | UCCAUGGCAGGUAAAUGUAUGUCUUUUGCUCAUGUUAUUUCAUUAAGGAUUUGAGAUCAGAAAAUAAUCCUAGCCAGGCAUGGGGCUUACGCCUAUAAUCUAGCACUUUGGGAGGCUGAGGUGGGAGGACUGCUUGCACCCAGGAGUUCGAGACCAGCCUGGGCAGCAUGGCAAGACCUAGUCUCUAUAAAAAAUUGGCUCAUGCGUCUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
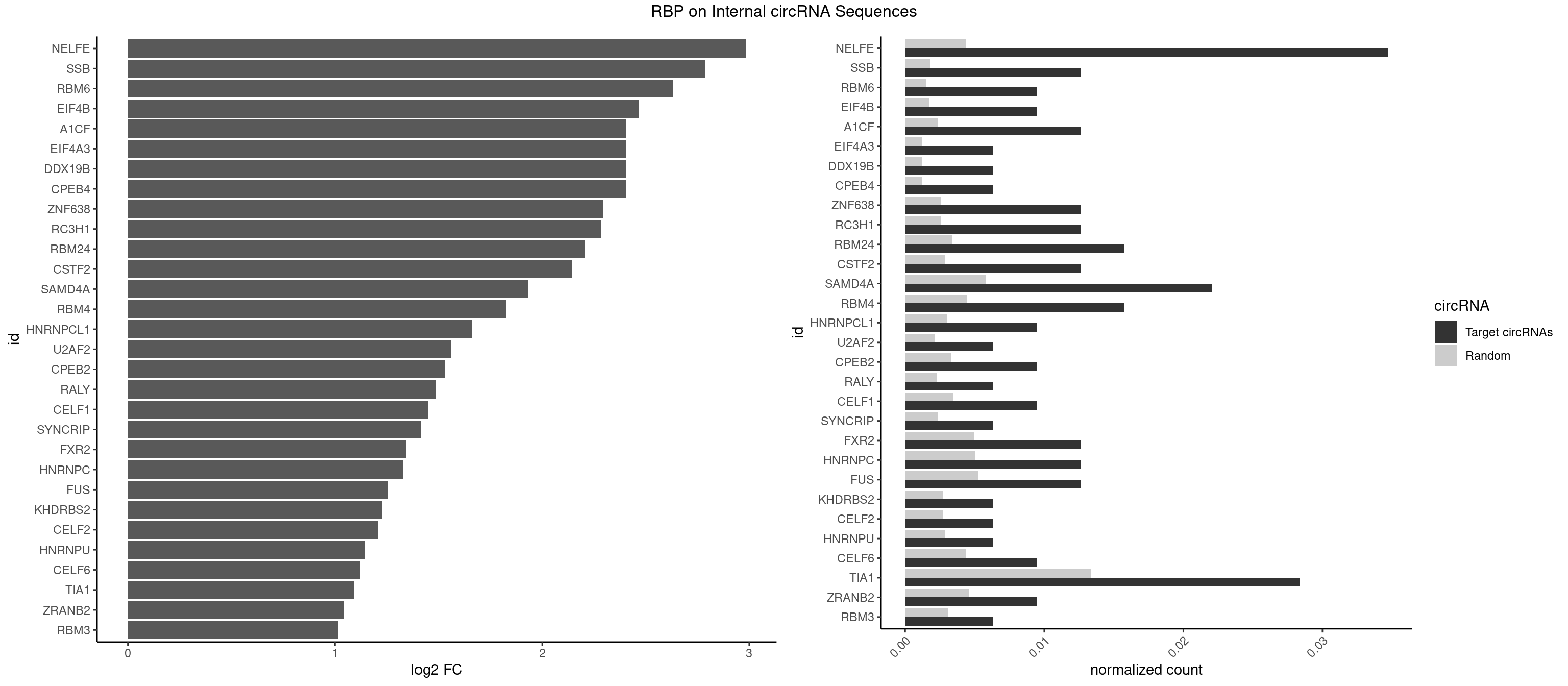
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NELFE | 10 | 3028 | 0.034700315 | 0.004389639 | 2.982775 | CUGGCU,GCUAAC,GGCUAA,GGUCUC,GUCUCU,UCUCUC,UGGCUA,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SSB | 3 | 1260 | 0.012618297 | 0.001827446 | 2.787616 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM6 | 2 | 1054 | 0.009463722 | 0.001528910 | 2.629904 | AAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| EIF4B | 2 | 1179 | 0.009463722 | 0.001710061 | 2.468360 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| A1CF | 3 | 1642 | 0.012618297 | 0.002381042 | 2.405852 | CAGUAU,GAUCAG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CPEB4 | 1 | 821 | 0.006309148 | 0.001191246 | 2.404974 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.006309148 | 0.001191246 | 2.404974 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.006309148 | 0.001191246 | 2.404974 | UUUUUU | UUUUUU |
| ZNF638 | 3 | 1773 | 0.012618297 | 0.002570888 | 2.295179 | CGUUGG,GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 3 | 1786 | 0.012618297 | 0.002589727 | 2.284645 | CCUUCU,CUUCUG,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM24 | 4 | 2357 | 0.015772871 | 0.003417223 | 2.206549 | AGAGUG,GAGUGA,GAGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CSTF2 | 3 | 1967 | 0.012618297 | 0.002852033 | 2.145454 | GUUUUG,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SAMD4A | 6 | 3992 | 0.022082019 | 0.005786671 | 1.932066 | CUGGCC,CUGGUC,GCUGGA,GCUGGC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM4 | 4 | 3065 | 0.015772871 | 0.004443259 | 1.827755 | CCUCUU,CCUUCU,CUCUUU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPCL1 | 2 | 2062 | 0.009463722 | 0.002989708 | 1.662403 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| U2AF2 | 1 | 1477 | 0.006309148 | 0.002141923 | 1.558538 | UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 2 | 2261 | 0.009463722 | 0.003278099 | 1.529548 | CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY | 1 | 1553 | 0.006309148 | 0.002252063 | 1.486198 | UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CELF1 | 2 | 2391 | 0.009463722 | 0.003466496 | 1.448930 | UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SYNCRIP | 1 | 1634 | 0.006309148 | 0.002369448 | 1.412894 | UUUUUU | AAAAAA,UUUUUU |
| FXR2 | 3 | 3434 | 0.012618297 | 0.004978016 | 1.341875 | GACGGG,GGACGG,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPC | 3 | 3471 | 0.012618297 | 0.005031636 | 1.326418 | CUUUUU,UUUUUA,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FUS | 3 | 3648 | 0.012618297 | 0.005288145 | 1.254683 | CGGUGA,UGGUGG,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| KHDRBS2 | 1 | 1858 | 0.006309148 | 0.002694070 | 1.227658 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CELF2 | 1 | 1886 | 0.006309148 | 0.002734648 | 1.206090 | UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| HNRNPU | 1 | 1965 | 0.006309148 | 0.002849135 | 1.146921 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| CELF6 | 2 | 2997 | 0.009463722 | 0.004344713 | 1.123147 | GUGAUG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| TIA1 | 8 | 9206 | 0.028391167 | 0.013342821 | 1.089378 | CUCCUU,CUUUUU,GUUUUG,UCUCCU,UUUUAU,UUUUGG,UUUUUA,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZRANB2 | 2 | 3173 | 0.009463722 | 0.004599773 | 1.040845 | AGGUUA,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBM3 | 1 | 2152 | 0.006309148 | 0.003120136 | 1.015836 | AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
RBP on BSJ (Exon Only)
Plot
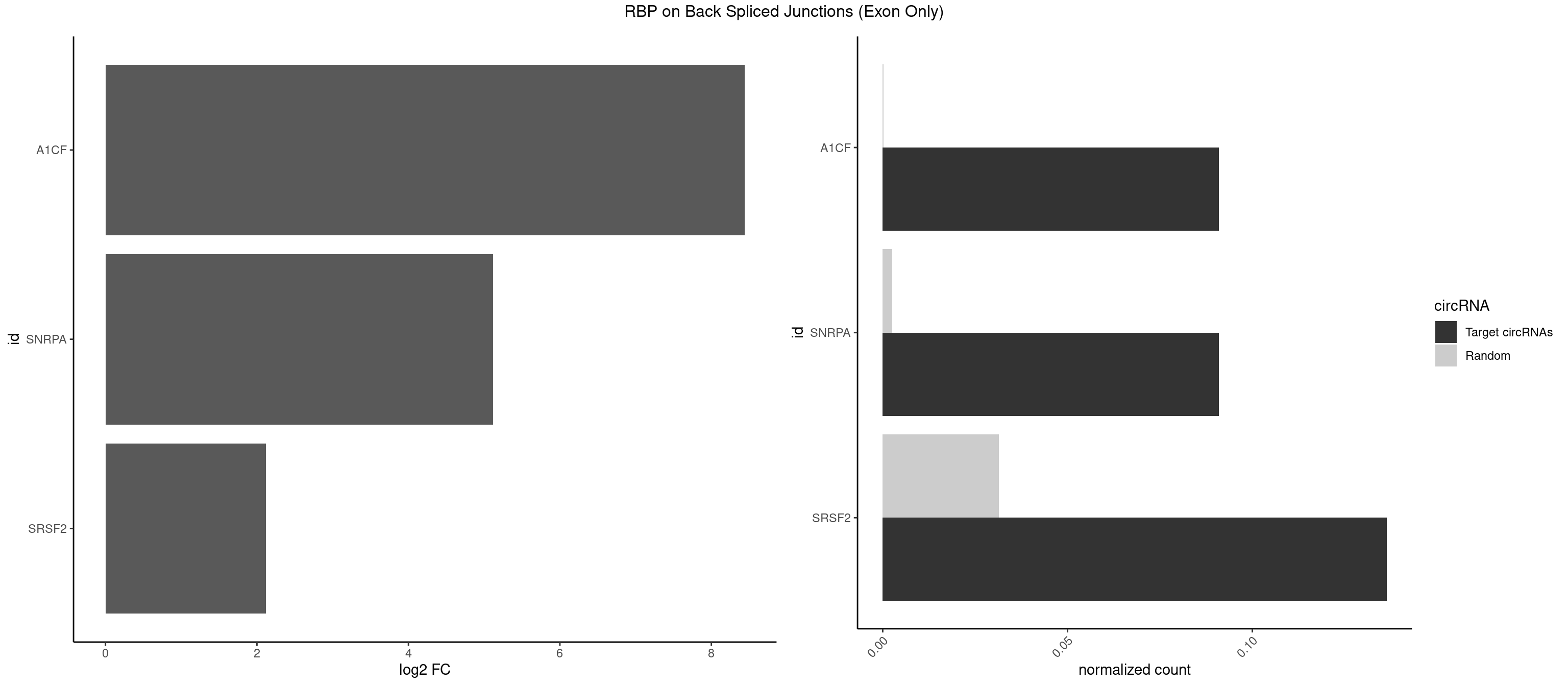
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| A1CF | 1 | 3 | 0.09090909 | 0.0002619859 | 8.438792 | CAGUAU | AUCAGU,CAGUAU,UAAUUG |
| SNRPA | 1 | 39 | 0.09090909 | 0.0026198585 | 5.116864 | GCAGUA | AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,GAGCAG,GAUACC,GCAGUA,GGAGAU,GGGUAU,GUAGGC,GUAGUG,UAUGCU,UCCUGC,UGCACG,UUCCUG,UUGCAC |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | GCAGUA,GGCAGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
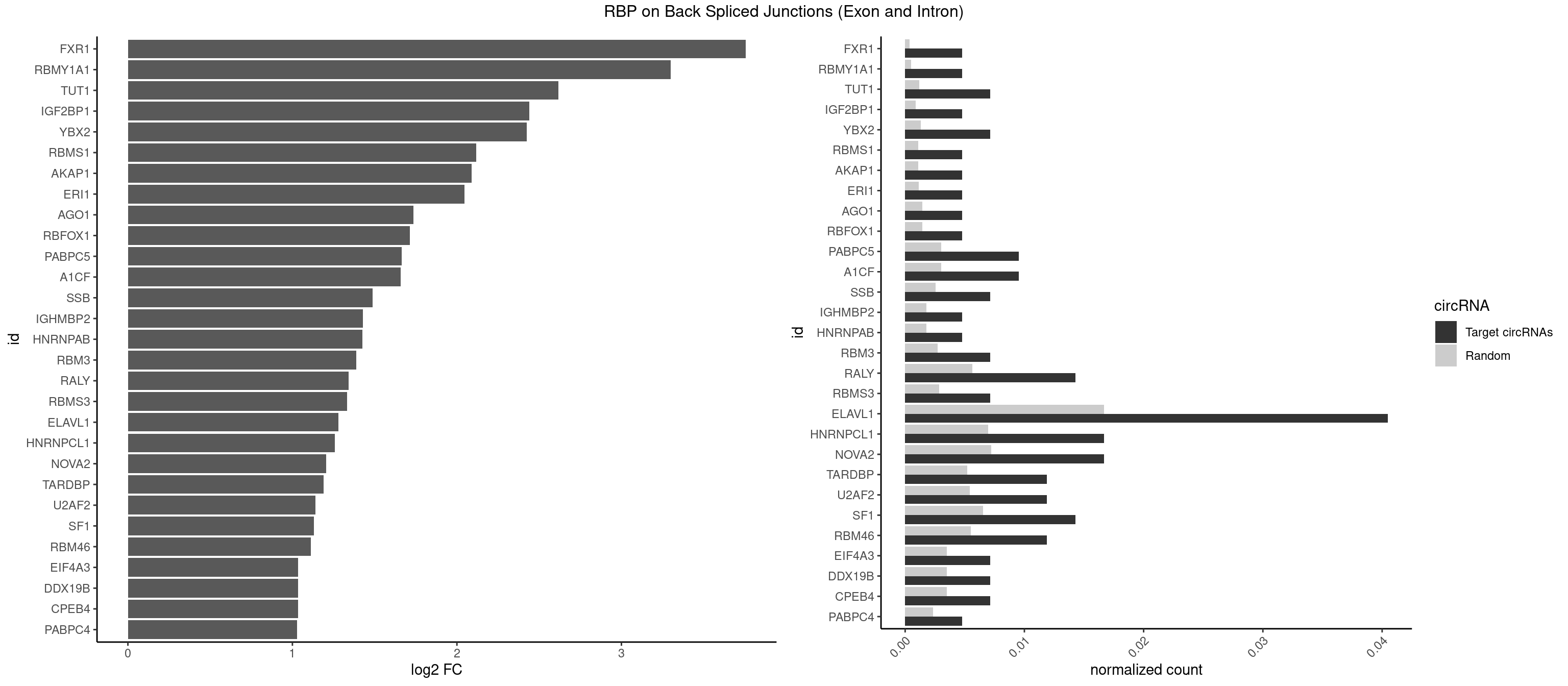
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAAC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | CAGUAU,GAUCAG,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAUACU,AUACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RALY | 5 | 1117 | 0.014285714 | 0.0056328094 | 1.342647 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | AUUUAU,UAUUUU,UGAUUU,UGUUUU,UUAUUU,UUGGUU,UUGUUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AGACAU,AGAUCA,AGGCAU,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGU,GUUGUG,GUUUUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUAAC,AUACUA,CUAACA,UAACAA,UACUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AUCAAA,AUGAAG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.