circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000055917:-:2:20318536:20327378
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000055917:-:2:20318536:20327378 | ENSG00000055917 | MSTRG.18016.1 | - | 2 | 20318537 | 20327378 | 178 | GUUUGAUCUGUGGAUGAAAUGAAUCAUGAUUUUCAAGCUCUUGCAUUAGAAUCUCGGGGAAUGGGAGAGCUUUUGCCUACCAAAAAGUUUUGGGAACCUGAUGAUUCAACAAAAGAUGGACAAAAAGGCAUAUUUCUUGGGGAUGAUGAAUGGAGAGAGACUGCAUGGGGAGCUUCUCGUUUGAUCUGUGGAUGAAAUGAAUCAUGAUUUUCAAGCUCUUGCAUUAGA | circ |
| ENSG00000055917:-:2:20318536:20327378 | ENSG00000055917 | MSTRG.18016.1 | - | 2 | 20318537 | 20327378 | 22 | GGGAGCUUCUCGUUUGAUCUGU | bsj |
| ENSG00000055917:-:2:20318536:20327378 | ENSG00000055917 | MSTRG.18016.1 | - | 2 | 20327369 | 20327578 | 210 | CCUUGACCUAAUGUUUUUCUACAGGUUUUAAGGUAGAUUGCUGUCAUAAACUGGGAAAACCUGUCUUCCCUACAGAAAUCAGUGCUUCCCUUUCUCAGAUCAUGCUAAUAUUAGUCAUAGCAGCAAUAUAAUAUAAAAAUAGUUGAAGAAGUCUGUUUAAACAUUUCUUUGUACUAAUUUUUGUUUUUGUUAUUCAACAGGUUUGAUCUG | ie_up |
| ENSG00000055917:-:2:20318536:20327378 | ENSG00000055917 | MSTRG.18016.1 | - | 2 | 20318337 | 20318546 | 210 | GGAGCUUCUCGUAAGUUACUGGUAUAUGUGUGAGAAUUGUGAAUAUAUUUAGCAUUUUAUAAGUCAUAAUGCACCUUUUUUUUUAGAGUGAGUCUCCAAAUUUUUGCAUUUUUUGUAUAGUUUUGCGCUAUUGCCCAGGCUGGUCUCAAAUAACUGAGCUCAUGCAAUCCUCCUGCCUUGGCCUCCCAAAGUGCUGGGAUGGUAGGCGUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
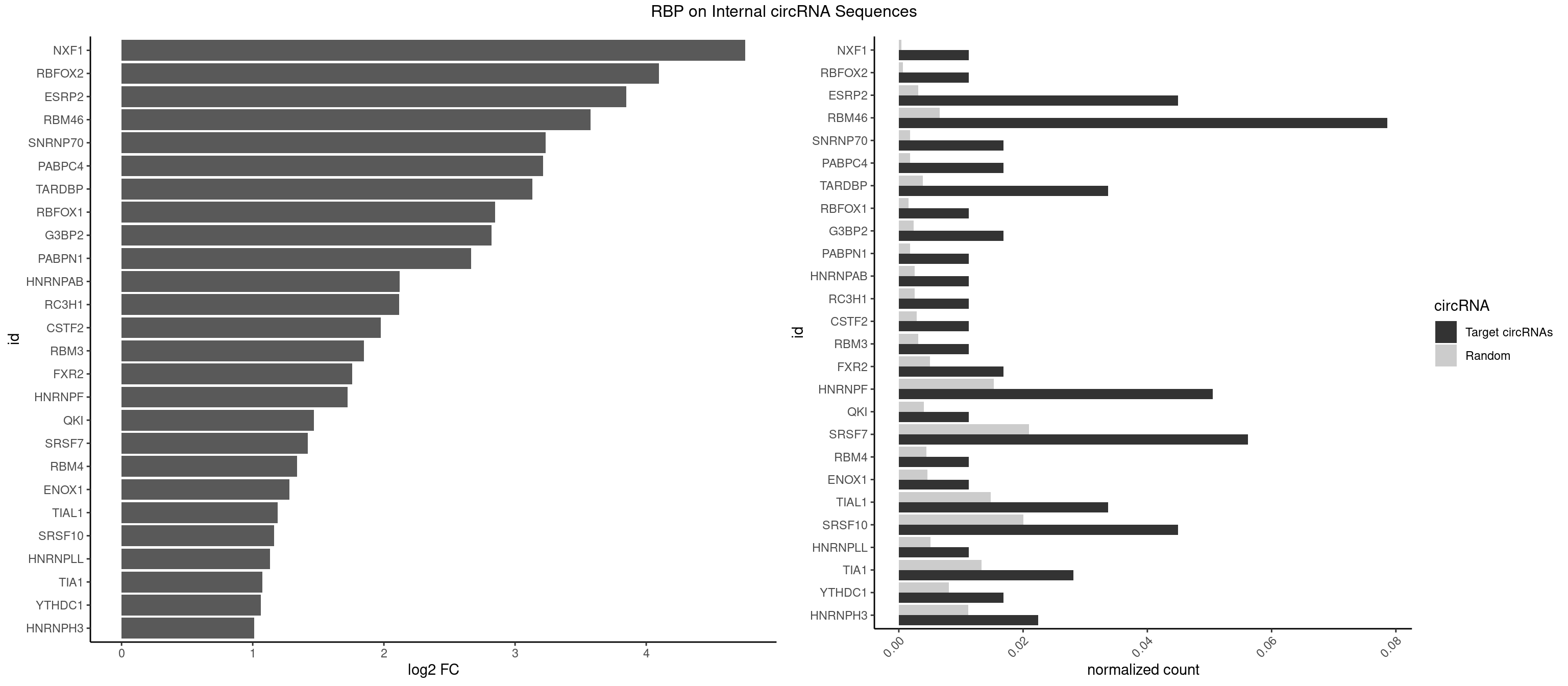
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 286 | 0.01123596 | 0.0004159215 | 4.755668 | AACCUG | AACCUG |
| RBFOX2 | 1 | 452 | 0.01123596 | 0.0006564894 | 4.097207 | UGCAUG | UGACUG,UGCAUG |
| ESRP2 | 7 | 2150 | 0.04494382 | 0.0031172377 | 3.849783 | GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM46 | 13 | 4554 | 0.07865169 | 0.0066011240 | 3.574694 | AAUCAU,AAUGAA,AUCAUG,AUGAAA,AUGAAU,AUGAUG,AUGAUU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SNRNP70 | 2 | 1237 | 0.01685393 | 0.0017941145 | 3.231741 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC4 | 2 | 1251 | 0.01685393 | 0.0018144033 | 3.215518 | AAAAAG | AAAAAA,AAAAAG |
| TARDBP | 5 | 2654 | 0.03370787 | 0.0038476365 | 3.131041 | GAAUGG,GUUUUG,UGAAUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBFOX1 | 1 | 1077 | 0.01123596 | 0.0015622419 | 2.846433 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 2 | 1644 | 0.01685393 | 0.0023839405 | 2.821665 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PABPN1 | 1 | 1222 | 0.01123596 | 0.0017723764 | 2.664366 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPAB | 1 | 1782 | 0.01123596 | 0.0025839306 | 2.120484 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RC3H1 | 1 | 1786 | 0.01123596 | 0.0025897275 | 2.117251 | UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CSTF2 | 1 | 1967 | 0.01123596 | 0.0028520334 | 1.978060 | GUUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM3 | 1 | 2152 | 0.01123596 | 0.0031201361 | 1.848442 | GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| FXR2 | 2 | 3434 | 0.01685393 | 0.0049780156 | 1.759443 | GACAAA,GGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPF | 8 | 10561 | 0.05056180 | 0.0153064921 | 1.723904 | AUGGGA,AUGGGG,GGAAUG,GGGAAU,GGGAUG,GGGGAA,GGGGAG,UGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| QKI | 1 | 2805 | 0.01123596 | 0.0040664663 | 1.466275 | AAUCAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SRSF7 | 9 | 14481 | 0.05617978 | 0.0209873716 | 1.420529 | AGAGAC,AGAGAG,GACAAA,GAGACU,GAGAGA,GGACAA,UCAACA,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| RBM4 | 1 | 3065 | 0.01123596 | 0.0044432593 | 1.338433 | UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ENOX1 | 1 | 3195 | 0.01123596 | 0.0046316558 | 1.278523 | UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| TIAL1 | 5 | 10182 | 0.03370787 | 0.0147572438 | 1.191662 | AGUUUU,AUUUUC,CUUUUG,UUUUCA | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF10 | 7 | 13860 | 0.04494382 | 0.0200874160 | 1.161831 | AAAAGA,AGAGAC,AGAGAG,GACAAA,GAGAGA,GAGAGC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPLL | 1 | 3534 | 0.01123596 | 0.0051229360 | 1.133080 | ACUGCA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| TIA1 | 4 | 9206 | 0.02808989 | 0.0133428208 | 1.073987 | AUUUUC,CUUUUG,GUUUUG,UUUUGG | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| YTHDC1 | 2 | 5576 | 0.01685393 | 0.0080822104 | 1.060263 | GACUGC,GCCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| HNRNPH3 | 3 | 7688 | 0.02247191 | 0.0111429292 | 1.011994 | GGAAUG,GGGAAU,GGGGAG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
RBP on BSJ (Exon Only)
Plot
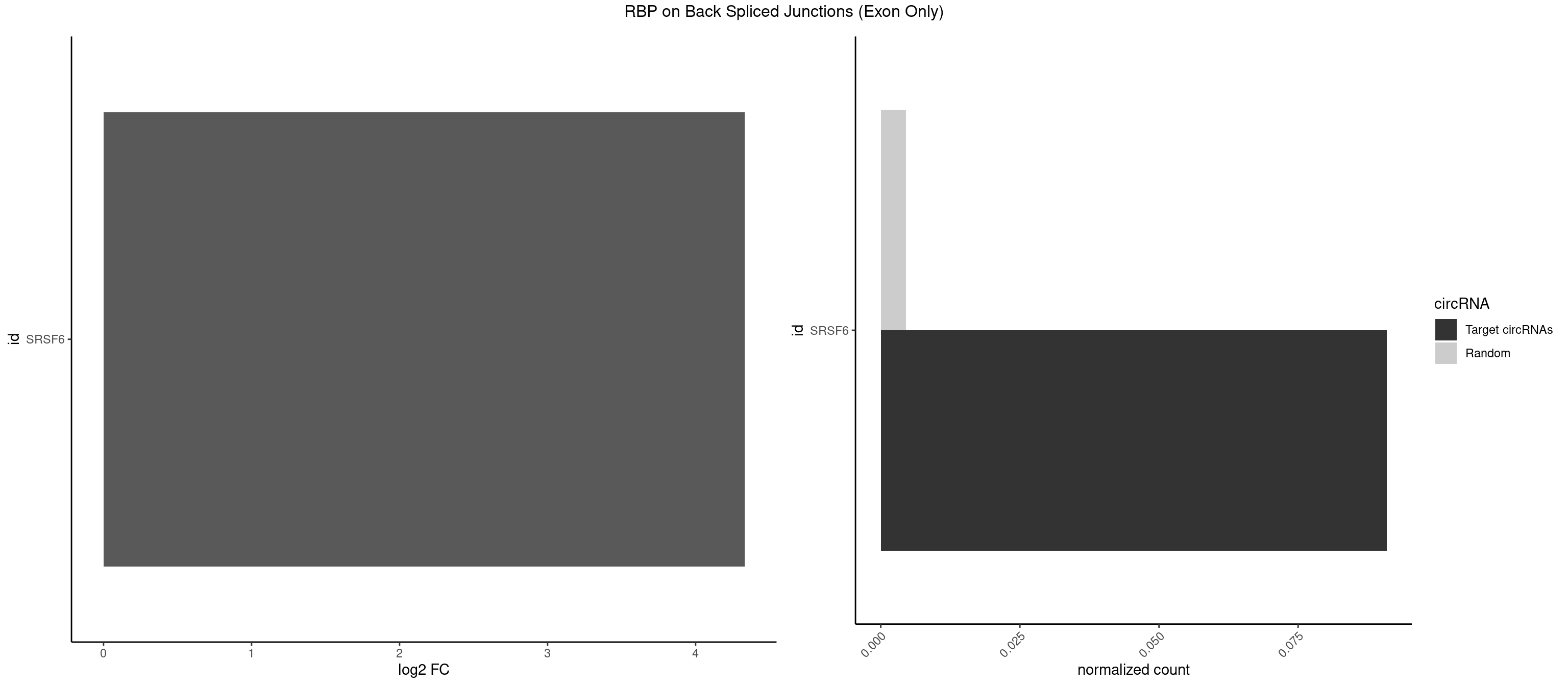
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | UUCUCG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
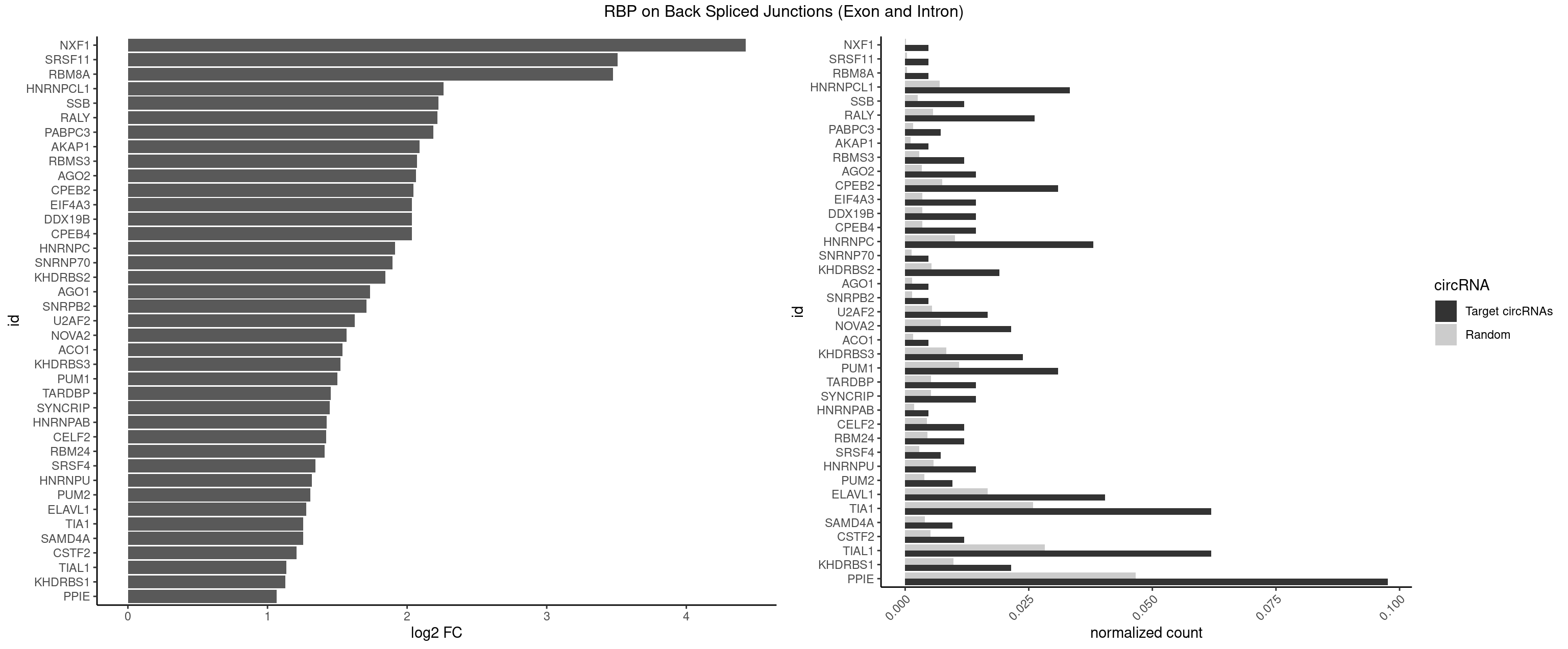
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| HNRNPCL1 | 13 | 1381 | 0.033333333 | 0.0069629182 | 2.259202 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RALY | 10 | 1117 | 0.026190476 | 0.0056328094 | 2.217116 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,AUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAGUG,AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CPEB2 | 12 | 1487 | 0.030952381 | 0.0074969770 | 2.045670 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| CPEB4 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| DDX19B | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| HNRNPC | 15 | 2006 | 0.038095238 | 0.0101118501 | 1.913564 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| KHDRBS2 | 7 | 1051 | 0.019047619 | 0.0053002821 | 1.845470 | AUAAAA,AUAAAC,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.0072299476 | 1.567479 | AGAUCA,AGUCAU,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| KHDRBS3 | 9 | 1646 | 0.023809524 | 0.0082980653 | 1.520692 | AAAUAA,AUAAAA,AUAAAC,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PUM1 | 12 | 2172 | 0.030952381 | 0.0109482064 | 1.499356 | AAUAUU,AAUGUU,AAUUGU,AGAAUU,GAAUUG,GUAGAU,GUAUAU,UAAUAU,UAAUGU,UGUAUA,UUGUAC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GUGAAU,GUUUUG,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | AUGUGU,GUCUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,GAGUGA,GUGUGA,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AAGAAG,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAGAU,GUAUAU,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | UAGUUU,UAUUUA,UGUUUU,UUGUUU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| TIA1 | 25 | 5140 | 0.061904762 | 0.0259018541 | 1.256995 | AUUUUU,CCUUUC,CUUUUU,GUUUUA,GUUUUG,GUUUUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGUA,CUGGUC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TIAL1 | 25 | 5597 | 0.061904762 | 0.0282043531 | 1.134133 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUU,CUUUUU,GUUUUU,UUUAAA,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUAAAA,GAAAAC,UAAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| PPIE | 40 | 9262 | 0.097619048 | 0.0466696896 | 1.064677 | AAAAAU,AAAAUA,AAAUAA,AAAUUU,AAUAUA,AAUAUU,AAUUUU,AUAAAA,AUAAUA,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUUUA,AUUUUU,UAAAAA,UAAUAU,UAAUUU,UAUAAA,UAUAAU,UAUAUU,UAUUUA,UUAUAA,UUUAAA,UUUAUA,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.