circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000005810:-:13:77055557:77062697
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000005810:-:13:77055557:77062697 | ENSG00000005810 | ENST00000682321 | - | 13 | 77055558 | 77062697 | 975 | CUCUGGCUUGCUCUCGCAUCCCUAUGUGUUCUUGAUCAGGACCACGUAGAUCGUCUCUCCUCGGGGAGAUGGAUGGGAAAGGAUGGACAACAAAAACAAAUGCCUAUGUGUGAUAACCAUGAUGAUGGUGAAACUGCAGCAAUCAUUUUAUGCAAUGUCUGUGGAAAUUUAUGUACAGACUGUGACAGAUUCCUUCACCUUCAUCGAAGAACCAAAACUCAUCAAAGACAGGUCUUCAAAGAAGAAGAAGAAGCUAUAAAGGUUGACCUUCAUGAAGGUUGUGGUAGAACCAAAUUGUUCUGGUUGAUGGCACUGGCAGAUUCUAAAACAAUGAAGGCAAUGGUGGAAUUCCGAGAACACACAGGCAAACCCACCACGAGUAGCUCAGAAGCAUGUCGCUUCUGUGGUUCCAGGAGUGGAACAGAGUUAUCUGCUGUUGGCAGUGUUUGUUCUGAUGCAGAUUGCCAGGAAUACGCUAAGAUAGCCUGUAGUAAGACGCAUCCUUGUGGCCAUCCAUGCGGGGGUGUUAAAAACGAAGAGCACUGUCUGCCCUGUCUACACGGCUGUGACAAAAGUGCCACAAGCCUGAAGCAAGACGCCGAUGACAUGUGCAUGAUAUGUUUCACCGAAGCGCUCUCGGCAGCACCAGCCAUUCAGCUGGAUUGUAGUCACAUAUUCCACUUACAGUGCUGUCGGCGAGUAUUAGAAAAUCGAUGGCUUGGCCCAAGGAUAACAUUUGGAUUUAUAUCUUGUCCCAUUUGCAAGAACAAAAUUAAUCACAUAGUACUAAAAGACCUACUUGAUCCAAUAAAAGAACUCUAUGAGGAUGUCAGAAGAAAAGCCUUAAUGAGAUUGGAAUAUGAAGGUCUGCAUAAGAGUGAAGCUAUCACAACUCCUGGUGUGAGGUUUUAUAAUGACCCAGCUGGCUAUGCAAUGAAUAGAUAUGCAUAUUAUGUGUGCUACAAAUGCAGAAAGCUCUGGCUUGCUCUCGCAUCCCUAUGUGUUCUUGAUCAGGACCACGUAGA | circ |
| ENSG00000005810:-:13:77055557:77062697 | ENSG00000005810 | ENST00000682321 | - | 13 | 77055558 | 77062697 | 22 | AAUGCAGAAAGCUCUGGCUUGC | bsj |
| ENSG00000005810:-:13:77055557:77062697 | ENSG00000005810 | ENST00000682321 | - | 13 | 77062688 | 77062897 | 210 | UGAGUGCCUCCUGUGUGCAUGGCACAUAGGACAGUGAUCGUGUAUGCCUGCUACCACAUCAGGAAGACUUGAGUGCCACUGAGUAUCUAUGUAUUUUCAAAAUCAGCCAGCAUUUAUUGAGCUGUUGUGAUGUGUCACAGUCAGCAUUUCAUAAGUCUUUCCUAAAAAUUCAGUGGUGUAACAGCCUUCCCUUUCAACAGCUCUGGCUUG | ie_up |
| ENSG00000005810:-:13:77055557:77062697 | ENSG00000005810 | ENST00000682321 | - | 13 | 77055358 | 77055567 | 210 | AUGCAGAAAGGUAUGCUAUAAAUUAUACUGAGAAGUUUUAAAAACUAGAGCUUACCUAUAUGAUUAAGAAUUCAAAUUGUACAGUGAUAUCUAAUUAUUCCAUCUUAAGCCUGAAGUUAAAAAUAAGAUAGCUUGGUACAAUGUUUCCCAAUGUUUCCAGUAAAACUUGUUUACUUAACUUGUUUGGGGAUUCUUUAUUUAACCUGAACA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
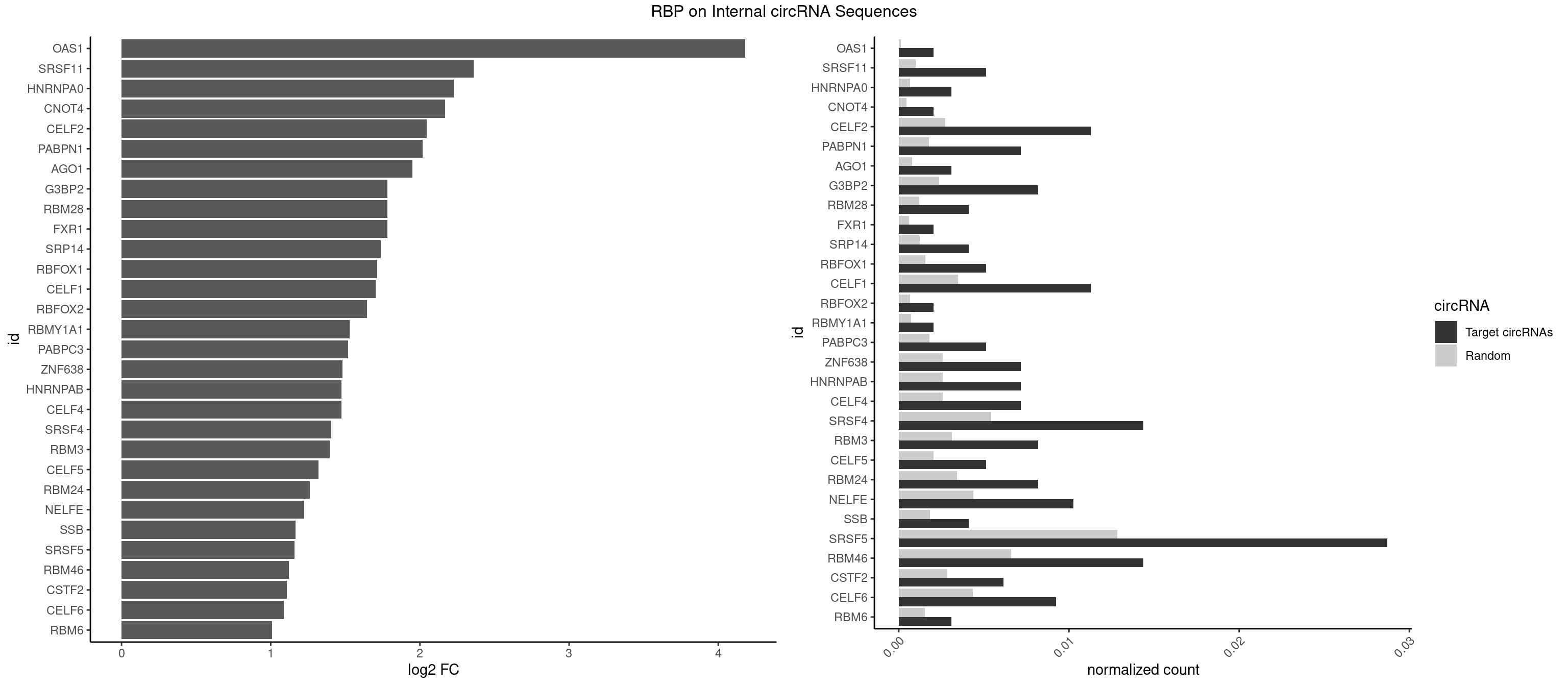
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.002051282 | 0.0001130379 | 4.181647 | GCAUAA | GCAUAA |
| SRSF11 | 4 | 688 | 0.005128205 | 0.0009985015 | 2.360617 | AAGAAG | AAGAAG |
| HNRNPA0 | 2 | 453 | 0.003076923 | 0.0006579386 | 2.225464 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| CNOT4 | 1 | 314 | 0.002051282 | 0.0004564992 | 2.167841 | GACAGA | GACAGA |
| CELF2 | 10 | 1886 | 0.011282051 | 0.0027346479 | 2.044602 | AUGUGU,GUCUGU,UAUGUG,UAUGUU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PABPN1 | 6 | 1222 | 0.007179487 | 0.0017723764 | 2.018196 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| AGO1 | 2 | 548 | 0.003076923 | 0.0007956130 | 1.951350 | GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| G3BP2 | 7 | 1644 | 0.008205128 | 0.0023839405 | 1.783178 | AGGAUA,AGGAUG,GGAUAA,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM28 | 3 | 822 | 0.004102564 | 0.0011926949 | 1.782301 | GAGUAG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FXR1 | 1 | 411 | 0.002051282 | 0.0005970720 | 1.780549 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRP14 | 3 | 847 | 0.004102564 | 0.0012289250 | 1.739129 | CCUGUA,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBFOX1 | 4 | 1077 | 0.005128205 | 0.0015622419 | 1.714836 | AGCAUG,GCAUGA,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| CELF1 | 10 | 2391 | 0.011282051 | 0.0034664959 | 1.702479 | CUGUCU,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBFOX2 | 1 | 452 | 0.002051282 | 0.0006564894 | 1.643682 | UGCAUG | UGACUG,UGCAUG |
| RBMY1A1 | 1 | 489 | 0.002051282 | 0.0007101099 | 1.530412 | CAAGAC | ACAAGA,CAAGAC |
| PABPC3 | 4 | 1234 | 0.005128205 | 0.0017897669 | 1.518682 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 6 | 1773 | 0.007179487 | 0.0025708878 | 1.481614 | GGUUGU,GUUCUU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF4 | 6 | 1782 | 0.007179487 | 0.0025839306 | 1.474313 | GGUGUG,GGUGUU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 6 | 1782 | 0.007179487 | 0.0025839306 | 1.474313 | AAAGAC,AAGACA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SRSF4 | 13 | 3740 | 0.014358974 | 0.0054214720 | 1.405196 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBM3 | 7 | 2152 | 0.008205128 | 0.0031201361 | 1.394917 | AAAACG,AAAACU,AAACGA,AAGACG,AAUACG,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF5 | 4 | 1415 | 0.005128205 | 0.0020520728 | 1.321372 | GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM24 | 7 | 2357 | 0.008205128 | 0.0034172229 | 1.263702 | AGAGUG,GAGUGA,GAGUGG,GUGUGA,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| NELFE | 9 | 3028 | 0.010256410 | 0.0043896388 | 1.224352 | CUCUGG,CUGGCU,CUGGUU,GUCUCU,UCUCUC,UCUGGC,UCUGGU,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SSB | 3 | 1260 | 0.004102564 | 0.0018274462 | 1.166697 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRSF5 | 27 | 8869 | 0.028717949 | 0.0128544391 | 1.159686 | AAGAAG,AGAAGA,AUAAAG,CACAUA,CACGGC,CACGUA,CGCAUC,GAAGAA,UAAAGG,UACAGA,UGCAGA,UGCAGC,UGCAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBM46 | 13 | 4554 | 0.014358974 | 0.0066011240 | 1.121169 | AAUCAU,AAUGAA,AUCAAA,AUCAUU,AUGAAG,AUGAAU,AUGAUA,AUGAUG,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CSTF2 | 5 | 1967 | 0.006153846 | 0.0028520334 | 1.109498 | GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF6 | 8 | 2997 | 0.009230769 | 0.0043447134 | 1.087190 | GUGAGG,UGUGAG,UGUGAU,UGUGGU,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM6 | 2 | 1054 | 0.003076923 | 0.0015289102 | 1.008985 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
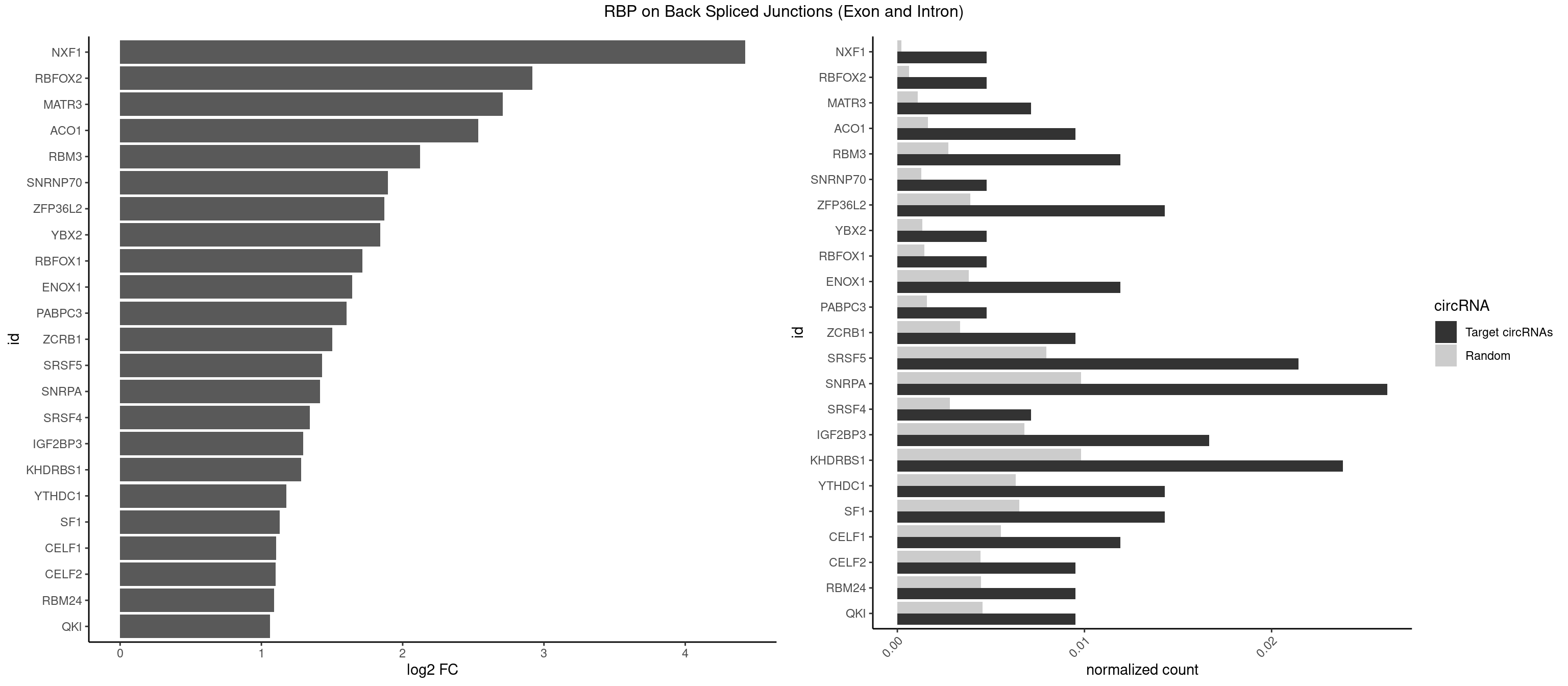
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ACO1 | 3 | 325 | 0.009523810 | 0.0016424829 | 2.535660 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM3 | 4 | 541 | 0.011904762 | 0.0027307537 | 2.124168 | AAAACU,AAACUA,AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AGGACA,GGACAG,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | ACUUAA,AUUUAA,GAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRSF5 | 8 | 1578 | 0.021428571 | 0.0079554615 | 1.429518 | AACAGC,AGGAAG,CAACAG,CACAUA,CACAUC,GGAAGA,UGCAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| SNRPA | 10 | 1947 | 0.026190476 | 0.0098145909 | 1.416042 | CCUGCU,CUGCUA,GGUAUG,GUAUGC,GUUUCC,UAUGCU,UUACCU,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| IGF2BP3 | 6 | 1348 | 0.016666667 | 0.0067966546 | 1.294069 | AAAAAC,AAAAUC,AAAUCA,AAAUUC,AAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUUUAA,CAAAAU,CUAAAA,UAAAAA,UAAAAC,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAGUGC,GCCUGC,UGCUAC,UGGUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACAGUC,CACUGA,CAGUCA,UACUGA,UAUACU | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUUGUG,UGUGUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | AUCUAA,UAACCU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.