circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000198887:+:9:70297965:70298221
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198887:+:9:70297965:70298221 | ENSG00000198887 | ENST00000361138 | + | 9 | 70297966 | 70298221 | 256 | AUUGAGGAACUUCAGCAGGCUUUAAUAGUAAAGCAAAAUGAAGAGCUUGACCGACAGAGGAGAAUAGGUAAUACCCGCAAAAUGAUAGAGGAUUUGCAAAAUGAACUAAAGACCACGGAAAACUGCGAGAAUCUUCAGCCCCAGAUUGAUGCCAUUACAAAUGAUCUGAGACGGAUUCAGGAUGAAAAGGCAUUAUGUGAAGGCGAAAUAAUUGAUAAGCGAAGAGAGAGGGAAACUCUAGAGAAGGAGAAAAAGAAUUGAGGAACUUCAGCAGGCUUUAAUAGUAAAGCAAAAUGAAGAGCUUGA | circ |
| ENSG00000198887:+:9:70297965:70298221 | ENSG00000198887 | ENST00000361138 | + | 9 | 70297966 | 70298221 | 22 | GGAGAAAAAGAAUUGAGGAACU | bsj |
| ENSG00000198887:+:9:70297965:70298221 | ENSG00000198887 | ENST00000361138 | + | 9 | 70297766 | 70297975 | 210 | UAUAUUAGUGAAUGUGUGAAACAAAGAUACAGGUAGUCUGGUAGAACUGGUAAUGUGCUAGUCUAAUUCUCACAGGACACCUUUUAGUGAGGAAAACACCUAUGUUAGAUUGUUUGCAUGUUAAAAACUAUAUUACUACAGAAGUCUUAUAAACCAAGAGGAAAACAGUCUCAAGUACUAACCUACUUUUGUUUUAUUAGAUUGAGGAAC | ie_up |
| ENSG00000198887:+:9:70297965:70298221 | ENSG00000198887 | ENST00000361138 | + | 9 | 70298212 | 70298421 | 210 | GAGAAAAAGAGUAAGUUUCAUAAAGUUAGAAUGAAAUGUUUAUAAAAUGUAGAUACAUCUCUGUUGAUGAACUUUCUUCUGCUUCUAUAAUUAUUUCUAUAAAAUGUUCAAGUUGCCUUCUUGAGGAAGCUCAACUCAGUUCUUCCAACUUCAUACCUUAAUUAUCUAUAUCUGUCGUGUACUGUAAUUUGCCAUUAUAGCUGACCACUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
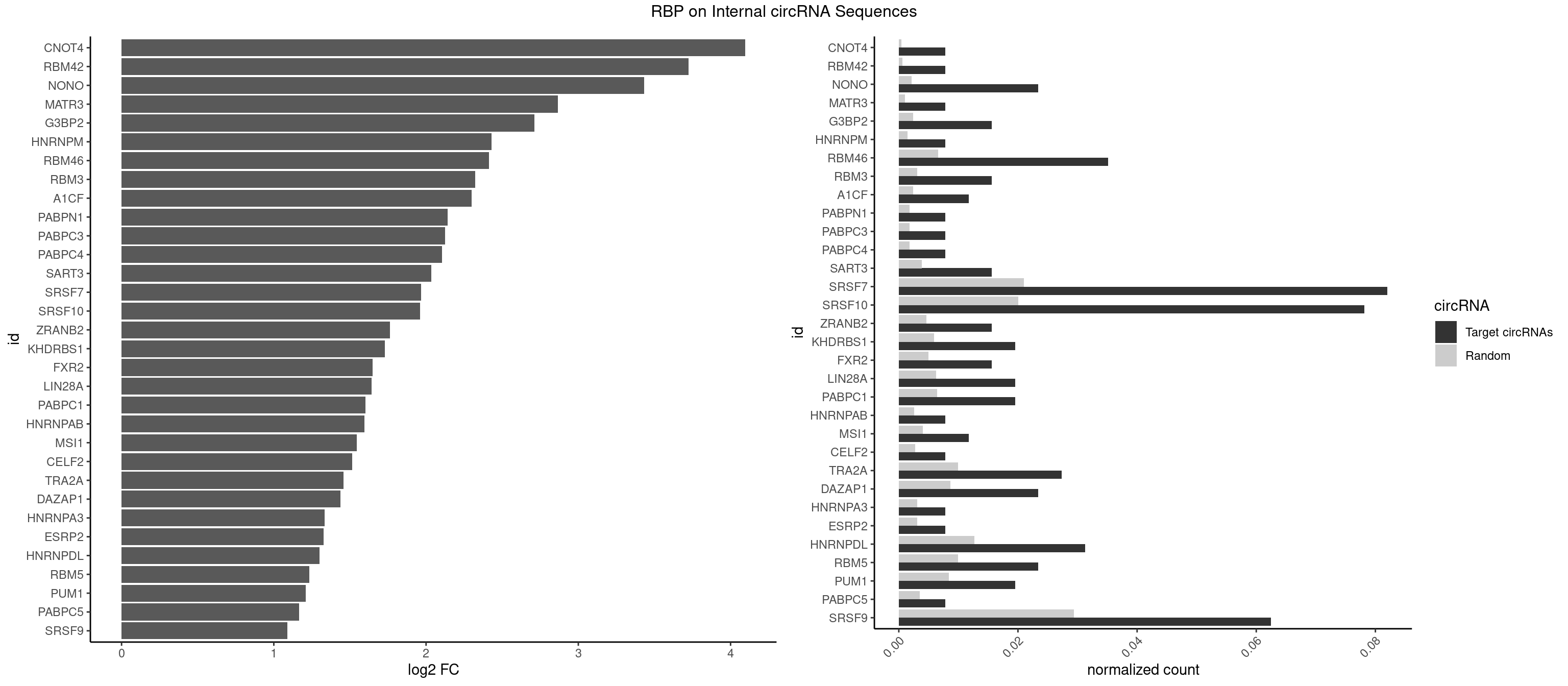
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.00781250 | 0.0004564992 | 4.097100 | GACAGA | GACAGA |
| RBM42 | 1 | 407 | 0.00781250 | 0.0005912752 | 3.723883 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| NONO | 5 | 1498 | 0.02343750 | 0.0021723567 | 3.431486 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| MATR3 | 1 | 739 | 0.00781250 | 0.0010724109 | 2.864926 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| G3BP2 | 3 | 1644 | 0.01562500 | 0.0023839405 | 2.712436 | AGGAUG,AGGAUU,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPM | 1 | 999 | 0.00781250 | 0.0014492040 | 2.430524 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM46 | 8 | 4554 | 0.03515625 | 0.0066011240 | 2.412998 | AAUGAA,AAUGAU,AUGAAA,AUGAAG,AUGAUA,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM3 | 3 | 2152 | 0.01562500 | 0.0031201361 | 2.324175 | AAAACU,GAAACU,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| A1CF | 2 | 1642 | 0.01171875 | 0.0023810421 | 2.299154 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPN1 | 1 | 1222 | 0.00781250 | 0.0017723764 | 2.140099 | AAAAGA | AAAAGA,AGAAGA |
| PABPC3 | 1 | 1234 | 0.00781250 | 0.0017897669 | 2.126013 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC4 | 1 | 1251 | 0.00781250 | 0.0018144033 | 2.106289 | AAAAAG | AAAAAA,AAAAAG |
| SART3 | 3 | 2634 | 0.01562500 | 0.0038186524 | 2.032721 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SRSF7 | 20 | 14481 | 0.08203125 | 0.0209873716 | 1.966652 | AAGGCG,AAUGAU,AGAGAA,AGAGAG,AGAGGA,AGGCGA,AUUGAU,CUAGAG,CUGAGA,GAAGGC,GAGAGA,GAGGAA,GAUAGA,GAUUGA,UAGAGA,UCUUCA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SRSF10 | 19 | 13860 | 0.07812500 | 0.0200874160 | 1.959492 | AAAAGA,AAAGAA,AAAGAC,AAGAGA,AAGGAG,AGAGAA,AGAGAG,AGAGGA,AGAGGG,GAGAAA,GAGAAG,GAGACG,GAGAGA,GAGAGG,GAGGAA,GAGGAG,GAGGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| ZRANB2 | 3 | 3173 | 0.01562500 | 0.0045997733 | 1.764222 | AGGGAA,AGGUAA,GUAAAG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS1 | 4 | 4064 | 0.01953125 | 0.0058910141 | 1.729196 | CAAAAU,GAAAAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| FXR2 | 3 | 3434 | 0.01562500 | 0.0049780156 | 1.650214 | AGACGG,GACAGA,GACGGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| LIN28A | 4 | 4315 | 0.01953125 | 0.0062547643 | 1.642757 | AGGAGA,GGAGAA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PABPC1 | 4 | 4443 | 0.01953125 | 0.0064402624 | 1.600593 | ACAAAU,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPAB | 1 | 1782 | 0.00781250 | 0.0025839306 | 1.596217 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MSI1 | 2 | 2770 | 0.01171875 | 0.0040157442 | 1.545079 | UAGGUA,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF2 | 1 | 1886 | 0.00781250 | 0.0027346479 | 1.514429 | UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TRA2A | 6 | 6871 | 0.02734375 | 0.0099589296 | 1.457149 | AAAGAA,AGAGGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| DAZAP1 | 5 | 5964 | 0.02343750 | 0.0086445016 | 1.438964 | AGGAUG,AGGUAA,AGUAAA,UAGGUA,UAGUAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPA3 | 1 | 2140 | 0.00781250 | 0.0031027457 | 1.332239 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 1 | 2150 | 0.00781250 | 0.0031172377 | 1.325516 | GGGAAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPDL | 7 | 8759 | 0.03125000 | 0.0126950266 | 1.299593 | AACUAA,AAUACC,ACCACG,AUCUGA,CUAGAG,CUUUAA,GAACUA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| RBM5 | 5 | 6879 | 0.02343750 | 0.0099705232 | 1.233078 | AAGGAG,AGGGAA,AGGUAA,GAAGGA,GAGGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| PUM1 | 4 | 5823 | 0.01953125 | 0.0084401638 | 1.210441 | AGAAUU,GAAUUG,GUAAUA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| PABPC5 | 1 | 2400 | 0.00781250 | 0.0034795387 | 1.166888 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SRSF9 | 15 | 20254 | 0.06250000 | 0.0293536261 | 1.090317 | AGGAAC,AGGAGA,AUGAAA,AUGAAC,GAAGGA,GAAGGC,GAUGCC,GGAAAA,GGAGAA,UGAAAA,UGAAGG,UGACCG,UGAUGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
RBP on BSJ (Exon Only)
Plot
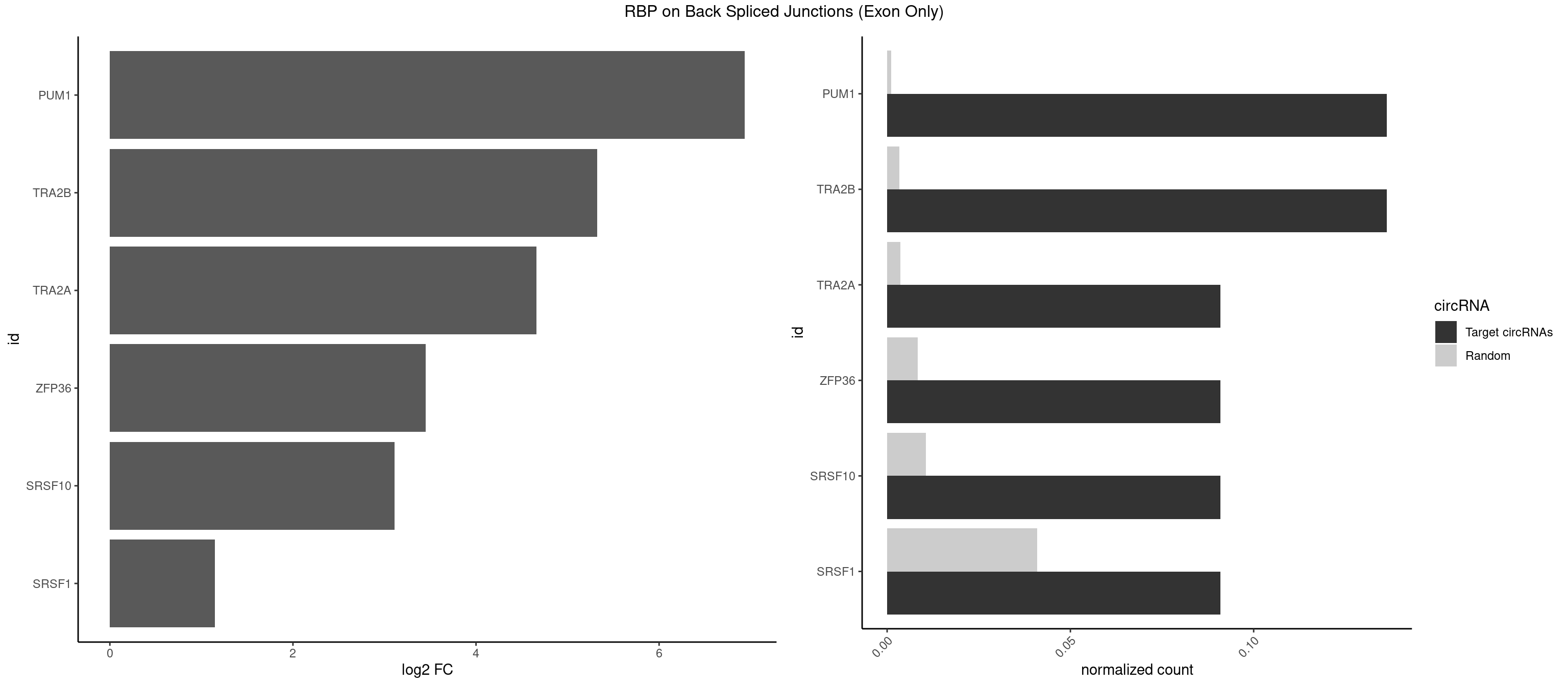
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM1 | 2 | 16 | 0.13636364 | 0.001113440 | 6.936292 | AGAAUU,GAAUUG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| TRA2B | 2 | 51 | 0.13636364 | 0.003405816 | 5.323315 | AAAGAA,AAGAAU | AAAGAA,AAGAAC,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GGAAGG,UAAGAA |
| TRA2A | 1 | 54 | 0.09090909 | 0.003602305 | 4.657432 | AAAGAA | AAAGAA,AAGAAA,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAGAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.010544931 | 3.107875 | AAAGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF1 | 1 | 625 | 0.09090909 | 0.041000786 | 1.148773 | AAAGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
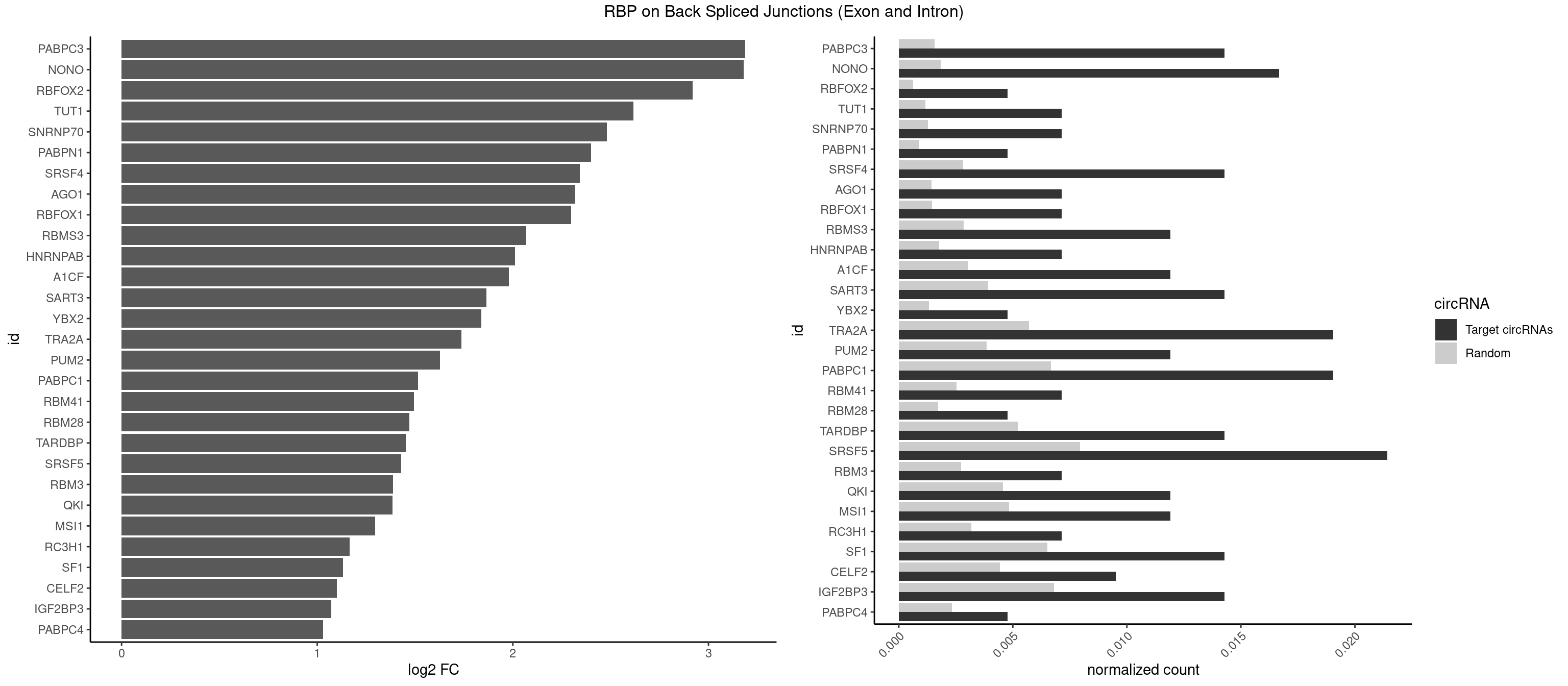
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPC3 | 5 | 310 | 0.014285714 | 0.0015669085 | 3.188580 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| NONO | 6 | 364 | 0.016666667 | 0.0018389762 | 3.179991 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| SRSF4 | 5 | 558 | 0.014285714 | 0.0028164047 | 2.342647 | AGGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | CUAUAU,UAUAGC,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SART3 | 5 | 777 | 0.014285714 | 0.0039197904 | 1.865725 | AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| TRA2A | 7 | 1133 | 0.019047619 | 0.0057134220 | 1.737184 | AAGAGG,AGAGGA,AGGAAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAGAU,UACAUC,UAGAUA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | AAAAAC,ACUAAC,AGAAAA,CUAACC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACAU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGA,GAAUGU,GUGAAU,UGAAUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF5 | 8 | 1578 | 0.021428571 | 0.0079554615 | 1.429518 | AGGAAG,AUAAAG,GAGGAA,UACAGA,UACAUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUAAC,CUAACC,UAACCU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAAG,AGGUAG,AGUAAG,AGUUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCUUCU,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACAGUC,ACUAAC,AGUAAG,GCUGAC,UACUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,UAUGUU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,AAAACA,AAACAC,AACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.