circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000145982:+:6:5404541:5431172
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000145982:+:6:5404541:5431172 | ENSG00000145982 | ENST00000648580 | + | 6 | 5404542 | 5431172 | 292 | UUAUUUGCUGGUAUAAAGGAUGGAGAAAGCCUGCAGCUCUUUGAACAAAGUUCUCGCUCUGCGCAUAAACAAGAGACACACACCAUGGAGGCCGUGAAGCUUGUAGAGUUUGAUCUUAAGCAAACGCUUACCAGGCUCAUGGCACAUCUUUUUGGAGAUGAGCUGGAGAUAAGAUGGGUAGACUGCUACUUCCCUUUUACACAUCCUUCCUUUGAGAUGGAGAUCAACUUUCAUGGAGAAUGGCUGGAAGUUCUUGGCUGCGGGGUGAUGGAACAACAACUGGUCAAUUCAGUUAUUUGCUGGUAUAAAGGAUGGAGAAAGCCUGCAGCUCUUUGAACAAAG | circ |
| ENSG00000145982:+:6:5404541:5431172 | ENSG00000145982 | ENST00000648580 | + | 6 | 5404542 | 5431172 | 22 | GGUCAAUUCAGUUAUUUGCUGG | bsj |
| ENSG00000145982:+:6:5404541:5431172 | ENSG00000145982 | ENST00000648580 | + | 6 | 5404342 | 5404551 | 210 | CAUACCAUGUAGCGUAGUGUUUUCUUCAGAGUUGUACUAAGUGAAUGUUAAUUUCAUUACGUUUAUUGACAGGACAUAAUGAGUCUUAUUCCCACAUUAUAACUUUUUAGAGUCAAAAGAUUCUUAGCAGAAUUUUAAAGUUAUCUGAUAUAGAUGGGCCAGGAUAUUUUCUUUAUUUAUACUUUCCUGUUGGUUUACAGUUAUUUGCUG | ie_up |
| ENSG00000145982:+:6:5404541:5431172 | ENSG00000145982 | ENST00000648580 | + | 6 | 5431163 | 5431372 | 210 | GUCAAUUCAGGUAAAAAAGAAUCCCACAUUUUAUUUACACGUGCUCUAAAGGAACCCUCCCUUCUCAGGCAGCCCCGUUGCACACUUGUAGAUAUUUACAUACUUCUAUGCGGGCAGCGGCAUCACUGGUCUCUCUUCAGAUUCCCCUCUAGAUUUGUCUUUGAUACUAAGCCCUGGAAAAACUUAUUUCUCACAUCCGUAUUUGCAAAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
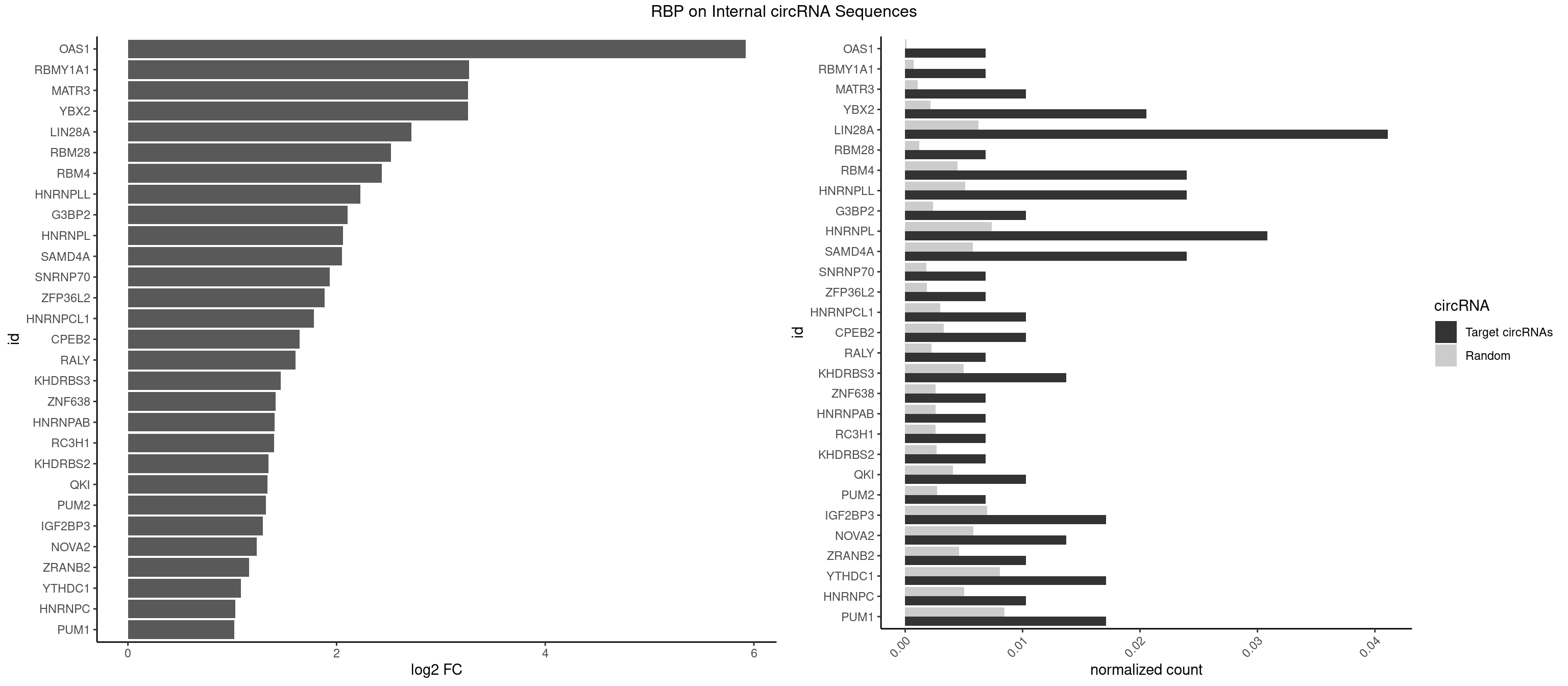
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.006849315 | 0.0001130379 | 5.921081 | GCAUAA | GCAUAA |
| RBMY1A1 | 1 | 489 | 0.006849315 | 0.0007101099 | 3.269845 | ACAAGA | ACAAGA,CAAGAC |
| MATR3 | 2 | 739 | 0.010273973 | 0.0010724109 | 3.260064 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| YBX2 | 5 | 1480 | 0.020547945 | 0.0021462711 | 3.259090 | AACAAC,ACAACA,ACAACU,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| LIN28A | 11 | 4315 | 0.041095890 | 0.0062547643 | 2.715967 | GGAGAA,GGAGAU,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBM28 | 1 | 822 | 0.006849315 | 0.0011926949 | 2.521735 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM4 | 6 | 3065 | 0.023972603 | 0.0044432593 | 2.431696 | CCUUCC,CUCUUU,CUUCCU,UCCUUC,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPLL | 6 | 3534 | 0.023972603 | 0.0051229360 | 2.226344 | ACACAC,ACACCA,CACACA,CACACC,GCAAAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| G3BP2 | 2 | 1644 | 0.010273973 | 0.0023839405 | 2.107574 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPL | 8 | 5085 | 0.030821918 | 0.0073706513 | 2.064093 | AAACAA,AACAAA,ACACAC,ACACCA,CACACA,CACACC,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| SAMD4A | 6 | 3992 | 0.023972603 | 0.0057866714 | 2.050581 | CUGGAA,CUGGUA,CUGGUC,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SNRNP70 | 1 | 1237 | 0.006849315 | 0.0017941145 | 1.932688 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZFP36L2 | 1 | 1277 | 0.006849315 | 0.0018520827 | 1.886811 | UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPCL1 | 2 | 2062 | 0.010273973 | 0.0029897078 | 1.780918 | CUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 2 | 2261 | 0.010273973 | 0.0032780993 | 1.648063 | CCUUUU,CUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY | 1 | 1553 | 0.006849315 | 0.0022520629 | 1.604713 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| KHDRBS3 | 3 | 3429 | 0.013698630 | 0.0049707696 | 1.462491 | AGAUAA,AUAAAC,AUAAAG | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ZNF638 | 1 | 1773 | 0.006849315 | 0.0025708878 | 1.413693 | GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 1 | 1782 | 0.006849315 | 0.0025839306 | 1.406392 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RC3H1 | 1 | 1786 | 0.006849315 | 0.0025897275 | 1.403159 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| KHDRBS2 | 1 | 1858 | 0.006849315 | 0.0026940701 | 1.346172 | AUAAAC | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| QKI | 2 | 2805 | 0.010273973 | 0.0040664663 | 1.337147 | ACACAC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PUM2 | 1 | 1890 | 0.006849315 | 0.0027404447 | 1.321550 | UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| IGF2BP3 | 4 | 4815 | 0.017123288 | 0.0069793662 | 1.294792 | AAUUCA,ACACAC,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| NOVA2 | 3 | 4013 | 0.013698630 | 0.0058171047 | 1.235658 | AGAUCA,AUCAAC,GAGACA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ZRANB2 | 2 | 3173 | 0.010273973 | 0.0045997733 | 1.159359 | GGGUGA,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| YTHDC1 | 4 | 5576 | 0.017123288 | 0.0080822104 | 1.083138 | GACUGC,GCCUGC,GGCUGC,UGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| HNRNPC | 2 | 3471 | 0.010273973 | 0.0050316361 | 1.029895 | CUUUUU,UUUUUG | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM1 | 4 | 5823 | 0.017123288 | 0.0084401638 | 1.020617 | AGAUAA,CUUGUA,UGUAGA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
RBP on BSJ (Exon Only)
Plot
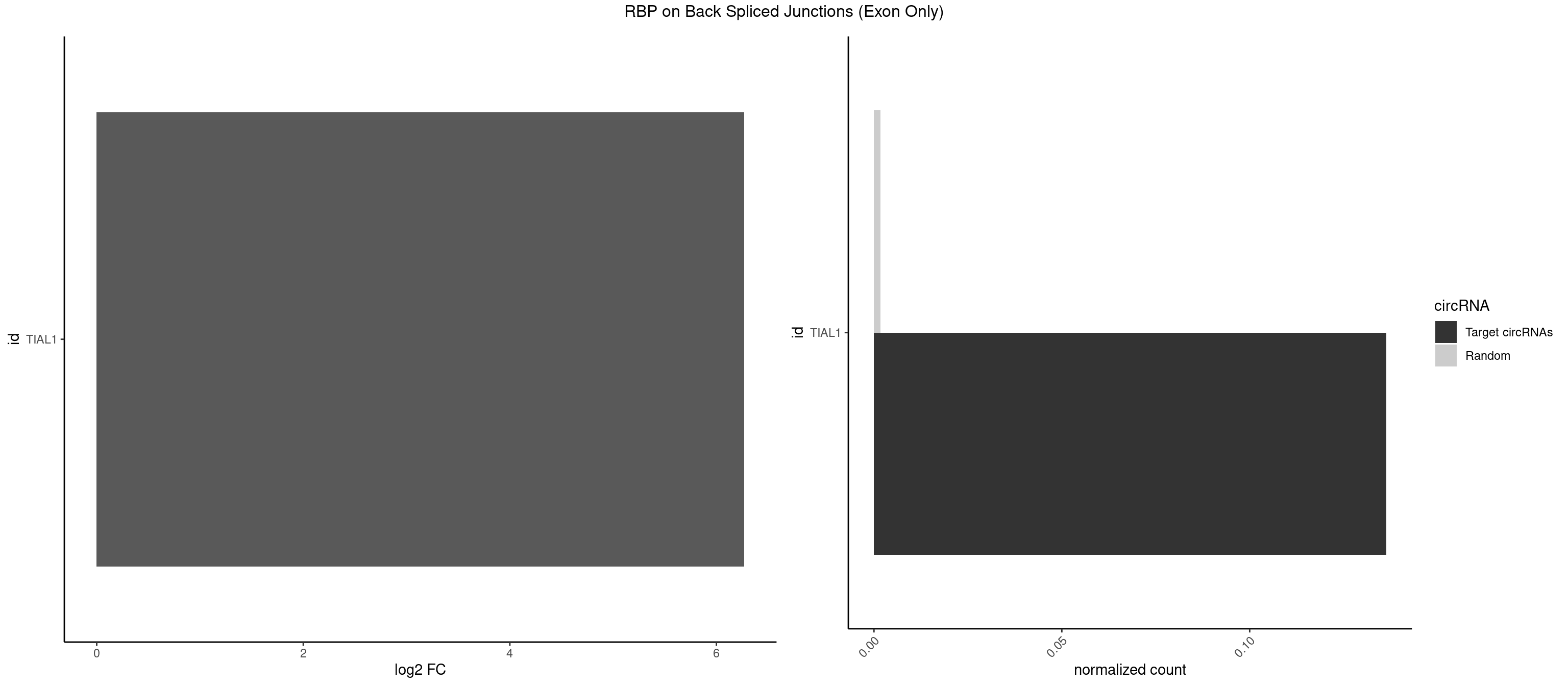
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TIAL1 | 2 | 26 | 0.1363636 | 0.001768405 | 6.268867 | UCAGUU,UUCAGU | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
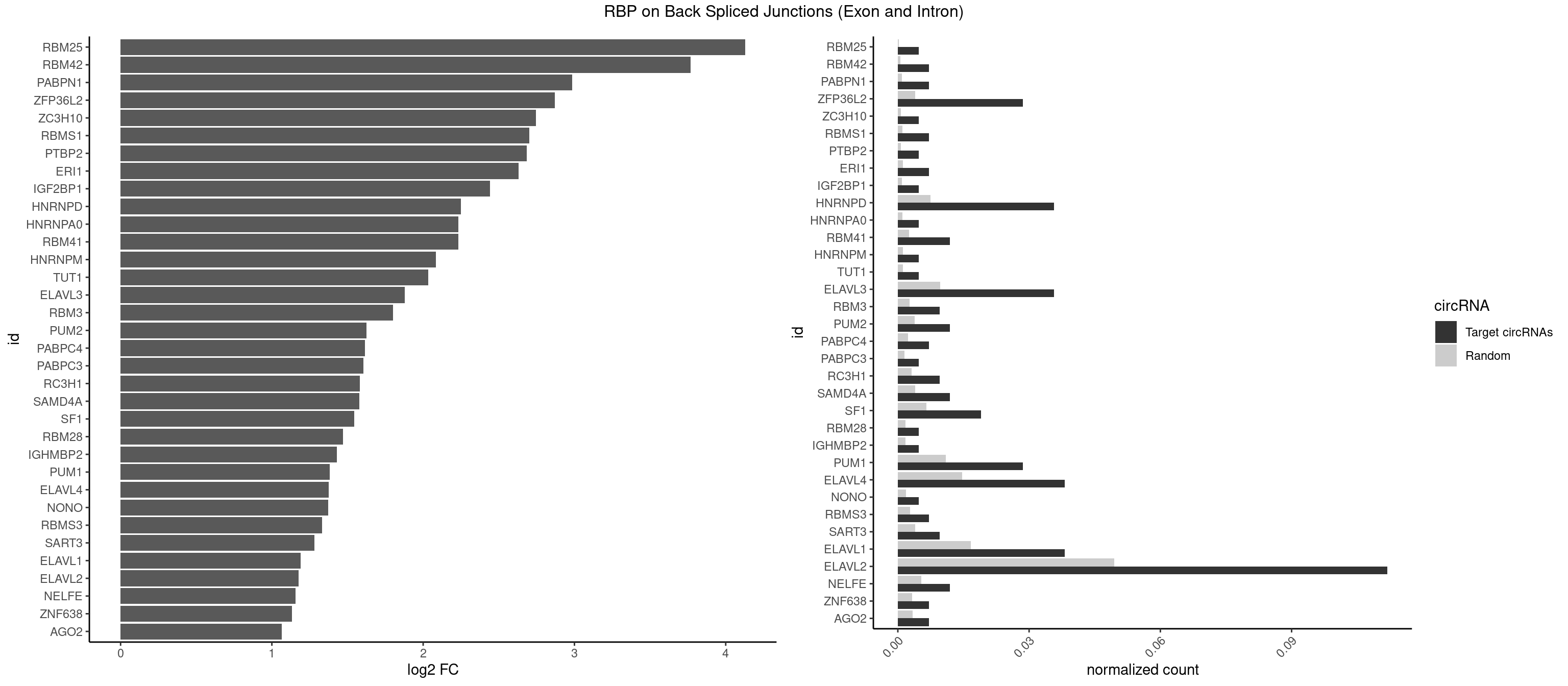
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA | AAAAGA,AGAAGA |
| ZFP36L2 | 11 | 774 | 0.028571429 | 0.0039046755 | 2.871299 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA | UUCAGA,UUUCAG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPD | 14 | 1488 | 0.035714286 | 0.0075020153 | 2.251151 | AAAAAA,AGAUAU,AUUUAU,UAUUUA,UUAGAG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ELAVL3 | 14 | 1928 | 0.035714286 | 0.0097188634 | 1.877642 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AUACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAGAU,UACAUA,UAGAUA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCCUUC,CCUUCU,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CGGGCA,CUGGAA,CUGGUC,GCGGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SF1 | 7 | 1294 | 0.019047619 | 0.0065245869 | 1.545652 | ACUAAG,ACUUAU,AUACUA,UACUAA,UAUACU | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUGUU,ACAUAA,AGAAUU,CAGAAU,CUUGUA,GUAGAU,UACAUA,UAGAUA,UGUAGA,UUGUAC,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ELAVL4 | 15 | 2916 | 0.038095238 | 0.0146966949 | 1.374119 | AAAAAA,AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | AUUUAU,UAUUUA,UAUUUU,UGGUUU,UGUUUU,UUAUUU,UUGGUU,UUUAUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ELAVL2 | 46 | 9826 | 0.111904762 | 0.0495112858 | 1.176442 | AAUUUU,AUACUU,AUAUUU,AUUUAC,AUUUAU,AUUUUA,AUUUUC,CAUUUU,CUUUUU,GUUUUC,UAAUUU,UACUUU,UAUACU,UAUUGA,UAUUUA,UAUUUG,UAUUUU,UCUUAU,UUAAUU,UUAUAC,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUUAUA,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUCU,GGUCUC,GUCUCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.