circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000152127:+:2:134253094:134254644
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000152127:+:2:134253094:134254644 | ENSG00000152127 | MSTRG.19256.9 | + | 2 | 134253095 | 134254644 | 504 | GCAAUGGGAAAACAGUAAGAGGCAGACCAACAGUGAUCCAGGGCUCUGAAAGCUAAUUGCUUCAAGAUCCUGCUACCAUUUUCUUUUGGGCCGCUUGCAAAGAAGAAUCCUUUGACUGAAGCAUAAUGGAAGUGAGGAAAGGCAACCAGCUGACACAGGAGCCAGAGUGAGACCAGCAGACUCUCACACUCAACCUACACCAUGAAUUUGUGUCUAUCUUCUACGCGUUAAGAGCCAAGGACAGGUGAAGUUGCCAGAGAGCAAUGGCUCUCUUCACUCCGUGGAAGUUGUCCUCUCAGAAGCUGGGCUUUUUCCUGGUGACUUUUGGCUUCAUUUGGGGUAUGAUGCUUCUGCACUUUACCAUCCAGCAGCGAACUCAGCCUGAAAGCAGCUCCAUGCUGCGCGAGCAGAUCCUGGACCUCAGCAAAAGGUACAUCAAGGCACUGGCAGAAGAAAACAGGAAUGUGGUGGAUGGGCCAUACGCUGGAGUCAUGACAGCUUAUGGCAAUGGGAAAACAGUAAGAGGCAGACCAACAGUGAUCCAGGGCUCUGAA | circ |
| ENSG00000152127:+:2:134253094:134254644 | ENSG00000152127 | MSTRG.19256.9 | + | 2 | 134253095 | 134254644 | 22 | GACAGCUUAUGGCAAUGGGAAA | bsj |
| ENSG00000152127:+:2:134253094:134254644 | ENSG00000152127 | MSTRG.19256.9 | + | 2 | 134252895 | 134253104 | 210 | ACAUAGCUGGCUGACCUUUGCCAAGUUAAUCACCUUUUACCUUUAUUUUCUCAUGUUUCUAAUAAAACAGAGACGAUAAUAUUCAUACUUCUUACCAUAUAGAACUUCUGAGGAUUCAGUGAGCAAAGCCACAAAAGAUGGUAUGUCACAAUAUCUGGGAUAUAGCUAGAAUUUAUAAUUUAUUUUUACUCUGUUGAUAGGCAAUGGGAA | ie_up |
| ENSG00000152127:+:2:134253094:134254644 | ENSG00000152127 | MSTRG.19256.9 | + | 2 | 134254635 | 134254844 | 210 | ACAGCUUAUGGUAAGCACUGUUUCUGGGACCUCUCCAUUAAAGUGUGCCUUGGCCUUGAAAUGGCCCUAGAAGCUCCCAGAUAUGGUCUUGUCAUGGACUGAAUGUCCUUUGCCACUGCCUUCAUAAGCAGUGAAUUGCACACAGAGAGGCAUCUUUAGGAAAUUAAAGCAAUGCCAUUUUGGGGGUUCUGAGCUGGUGUAUGCAGACAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
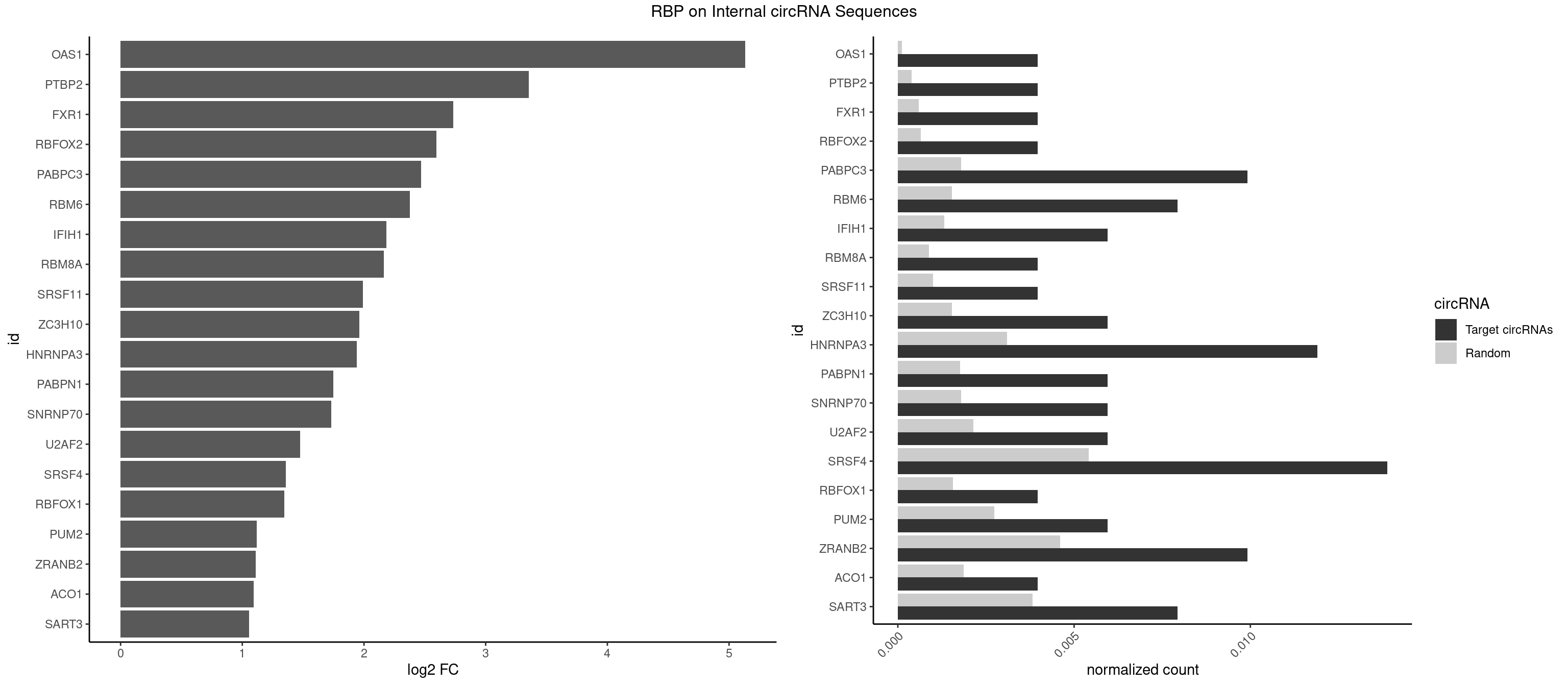
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.003968254 | 0.0001130379 | 5.133626 | GCAUAA | GCAUAA |
| PTBP2 | 1 | 267 | 0.003968254 | 0.0003883867 | 3.352939 | CUCUCU | CUCUCU |
| FXR1 | 1 | 411 | 0.003968254 | 0.0005970720 | 2.732527 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBFOX2 | 1 | 452 | 0.003968254 | 0.0006564894 | 2.595661 | UGACUG | UGACUG,UGCAUG |
| PABPC3 | 4 | 1234 | 0.009920635 | 0.0017897669 | 2.470661 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBM6 | 3 | 1054 | 0.007936508 | 0.0015289102 | 2.376001 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| IFIH1 | 2 | 904 | 0.005952381 | 0.0013115296 | 2.182217 | GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 1 | 611 | 0.003968254 | 0.0008869128 | 2.161640 | UGCGCG | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SRSF11 | 1 | 688 | 0.003968254 | 0.0009985015 | 1.990668 | AAGAAG | AAGAAG |
| ZC3H10 | 2 | 1053 | 0.005952381 | 0.0015274610 | 1.962331 | CAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| HNRNPA3 | 5 | 2140 | 0.011904762 | 0.0031027457 | 1.939921 | AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PABPN1 | 2 | 1222 | 0.005952381 | 0.0017723764 | 1.747782 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 2 | 1237 | 0.005952381 | 0.0017941145 | 1.730195 | AUCAAG,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| U2AF2 | 2 | 1477 | 0.005952381 | 0.0021419234 | 1.474560 | UUUUCC,UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| SRSF4 | 6 | 3740 | 0.013888889 | 0.0054214720 | 1.357175 | AAGAAG,AGAAGA,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBFOX1 | 1 | 1077 | 0.003968254 | 0.0015622419 | 1.344887 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PUM2 | 2 | 1890 | 0.005952381 | 0.0027404447 | 1.119057 | GUACAU,UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZRANB2 | 4 | 3173 | 0.009920635 | 0.0045997733 | 1.108870 | AAAGGU,AGGUAC,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ACO1 | 1 | 1283 | 0.003968254 | 0.0018607779 | 1.092599 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SART3 | 3 | 2634 | 0.007936508 | 0.0038186524 | 1.055441 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
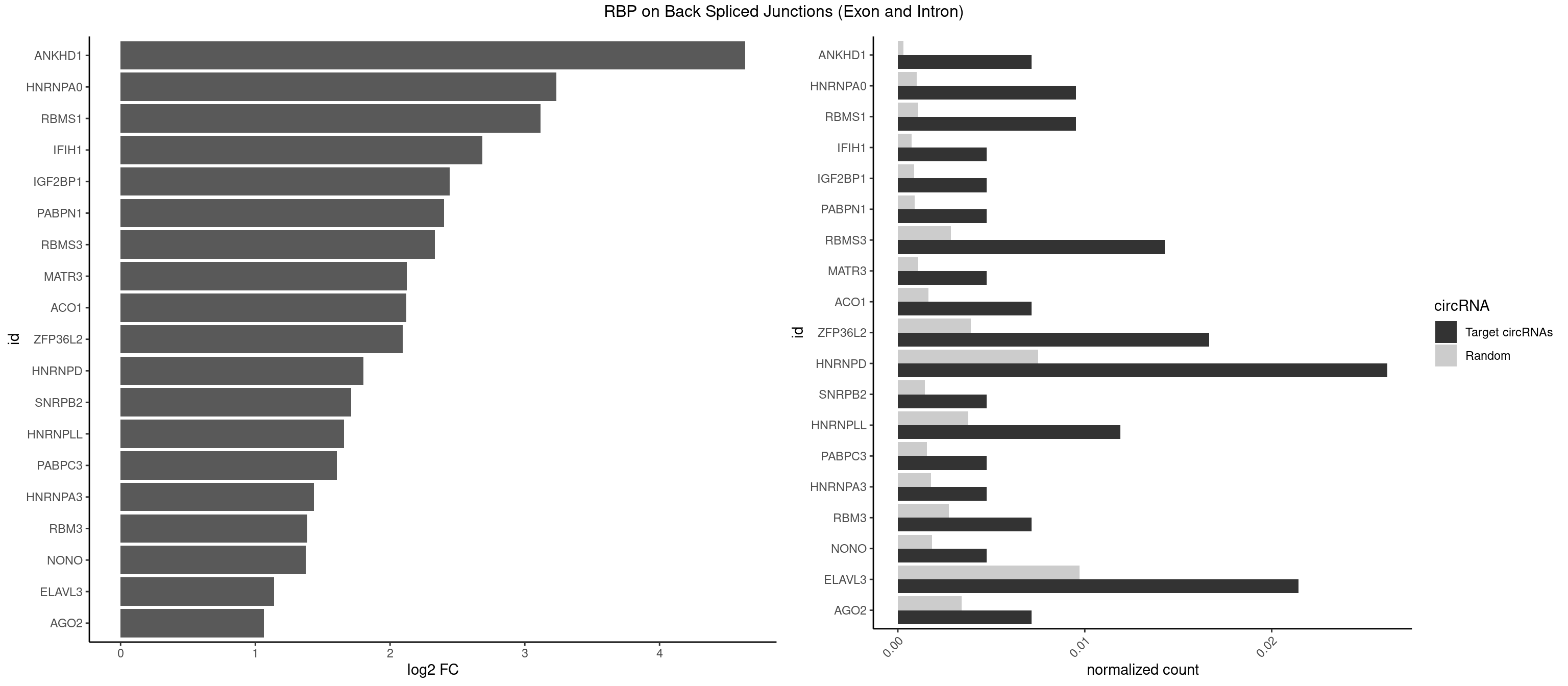
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 56 | 0.007142857 | 0.0002871826 | 4.636461 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBMS3 | 5 | 562 | 0.014285714 | 0.0028365578 | 2.332360 | AUAUAG,CAUAUA,UAUAGA,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZFP36L2 | 6 | 774 | 0.016666667 | 0.0039046755 | 2.093691 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPD | 10 | 1488 | 0.026190476 | 0.0075020153 | 1.803692 | AAUUUA,AGAUAU,AUUUAU,UUAGGA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | AGACGA,CACACA,CACUGC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AGACGA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUAU,UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.