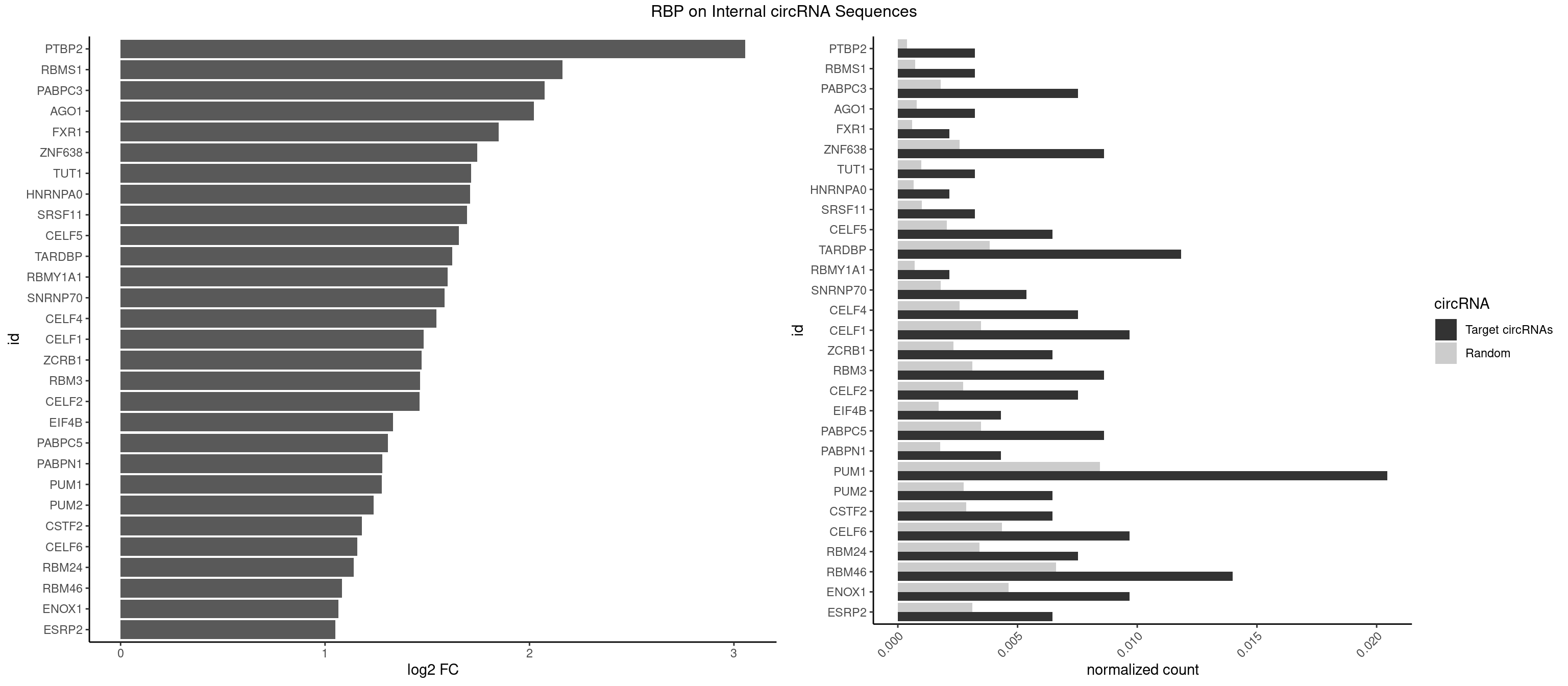
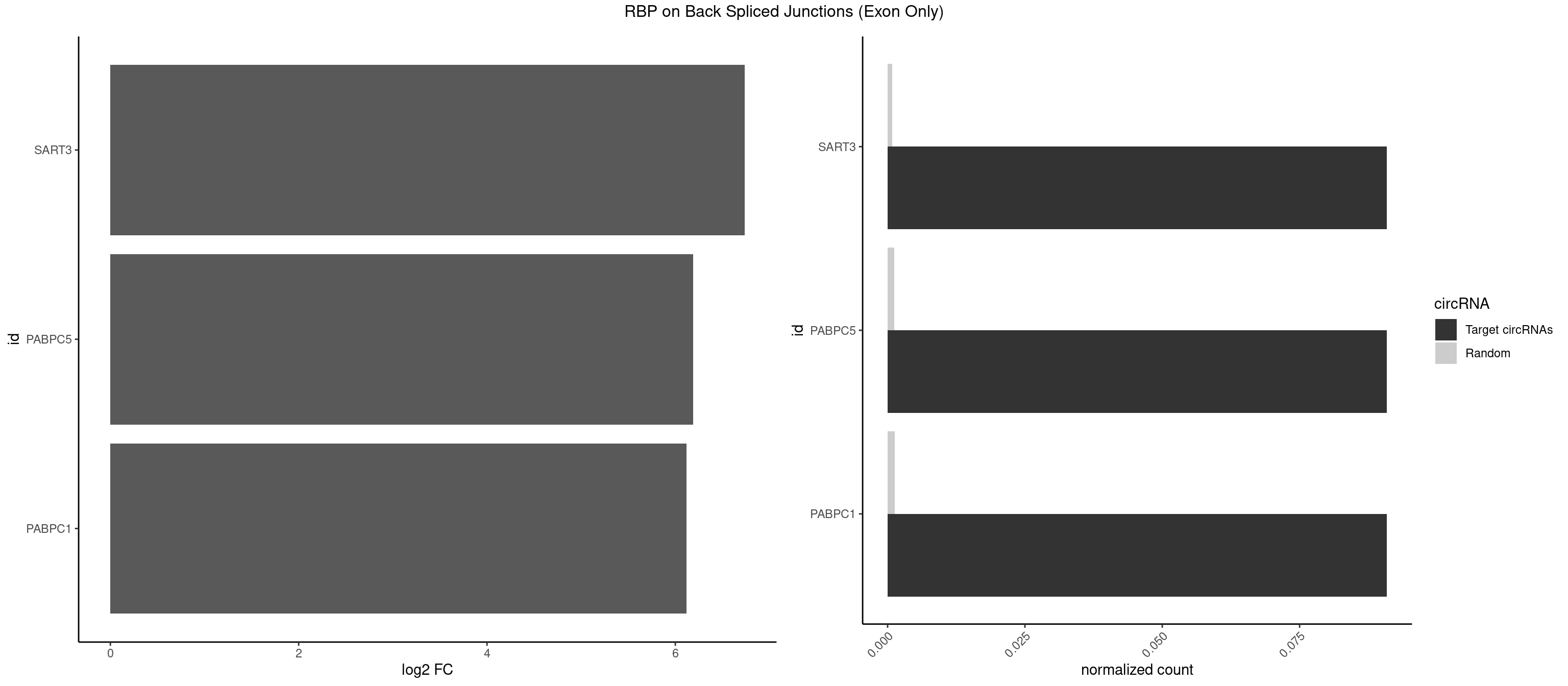
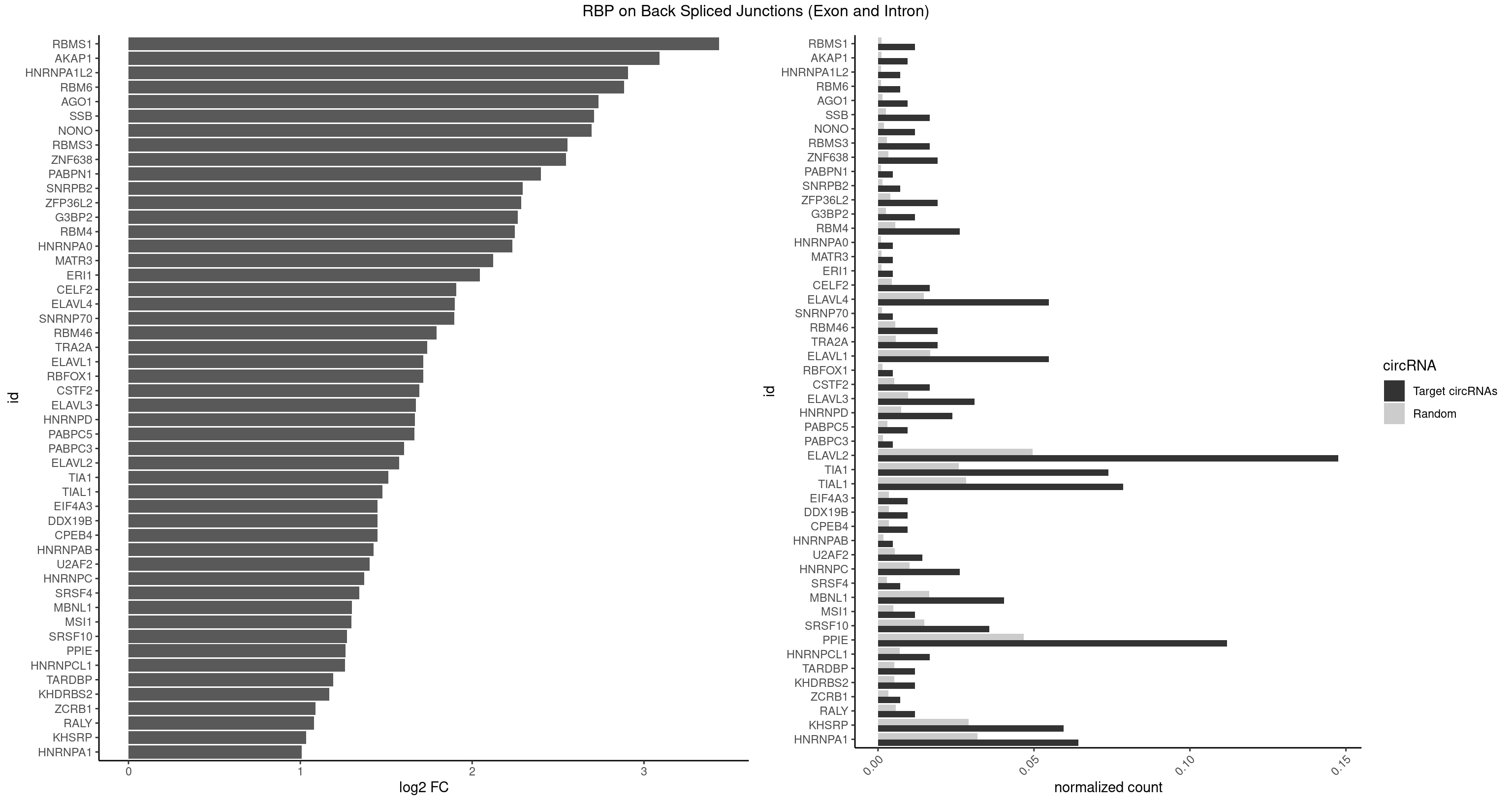
| RBMS1 |
4 |
217 |
0.011904762 |
0.0010983474 |
3.438132 |
AUAUAG,GAUAUA,UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 |
3 |
221 |
0.009523810 |
0.0011185006 |
3.089973 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| HNRNPA1L2 |
2 |
188 |
0.007142857 |
0.0009522370 |
2.907109 |
GUAGGG,UAGGGU |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM6 |
2 |
191 |
0.007142857 |
0.0009673519 |
2.884389 |
AUCCAG,UAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| AGO1 |
3 |
283 |
0.009523810 |
0.0014308746 |
2.734641 |
AGGUAG,GAGGUA,UGAGGU |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SSB |
6 |
505 |
0.016666667 |
0.0025493753 |
2.708750 |
GCUGUU,UGCUGU,UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| NONO |
4 |
364 |
0.011904762 |
0.0018389762 |
2.694564 |
AGAGGA,GAGAGG,GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMS3 |
6 |
562 |
0.016666667 |
0.0028365578 |
2.554752 |
AUAUAG,AUAUAU,UAUAGA,UAUAUA,UAUAUC |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZNF638 |
7 |
646 |
0.019047619 |
0.0032597743 |
2.546767 |
GGUUCU,GUUCUU,GUUGUU,UGUUCU,UGUUGU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPN1 |
1 |
178 |
0.004761905 |
0.0009018541 |
2.400573 |
AAAAGA |
AAAAGA,AGAAGA |
| SNRPB2 |
2 |
288 |
0.007142857 |
0.0014560661 |
2.294425 |
AUUGCA,UAUUGC |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZFP36L2 |
7 |
774 |
0.019047619 |
0.0039046755 |
2.286336 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| G3BP2 |
4 |
490 |
0.011904762 |
0.0024738009 |
2.266737 |
AGGAUG,AGGAUU,GGAUGA,GGAUUG |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM4 |
10 |
1094 |
0.026190476 |
0.0055169287 |
2.247105 |
CCUUCU,CUUCUU,UCCUUC,UUCCUU,UUCUUG |
CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| MATR3 |
1 |
216 |
0.004761905 |
0.0010933091 |
2.122837 |
AUCUUA |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ERI1 |
1 |
228 |
0.004761905 |
0.0011537686 |
2.045185 |
UUCAGA |
UUCAGA,UUUCAG |
| CELF2 |
6 |
881 |
0.016666667 |
0.0044437727 |
1.907109 |
AUGUGU,GUAUGU,GUUGUU,UAUGUU,UGUUGU |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ELAVL4 |
22 |
2916 |
0.054761905 |
0.0146966949 |
1.897681 |
AUUUAU,GUAUCU,UAUUUA,UAUUUU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| SNRNP70 |
1 |
253 |
0.004761905 |
0.0012797259 |
1.895704 |
UUCAAG |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM46 |
7 |
1091 |
0.019047619 |
0.0055018138 |
1.791631 |
AAUCAU,AAUGAU,AUCAUU,AUGAUA,AUGAUG,AUGAUU,GAUGAU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| TRA2A |
7 |
1133 |
0.019047619 |
0.0057134220 |
1.737184 |
AAGAAA,AAGAGG,AGAGGA,AGGAAG,GAAGAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ELAVL1 |
22 |
3309 |
0.054761905 |
0.0166767432 |
1.715335 |
AUUUAU,UAUUUA,UAUUUU,UGUUUU,UUAUUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBFOX1 |
1 |
287 |
0.004761905 |
0.0014510278 |
1.714464 |
GCAUGU |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| CSTF2 |
6 |
1022 |
0.016666667 |
0.0051541717 |
1.693153 |
GUUUUG,UGUGUU,UGUUUU |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ELAVL3 |
12 |
1928 |
0.030952381 |
0.0097188634 |
1.671191 |
AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| HNRNPD |
9 |
1488 |
0.023809524 |
0.0075020153 |
1.666189 |
AAUUUA,AUUUAU,UAUUUA,UUAGAG,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| PABPC5 |
3 |
596 |
0.009523810 |
0.0030078597 |
1.662801 |
AGAAAA,AGAAAU,GAAAAU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPC3 |
1 |
310 |
0.004761905 |
0.0015669085 |
1.603618 |
AAAACC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| ELAVL2 |
61 |
9826 |
0.147619048 |
0.0495112858 |
1.576050 |
AAUUUA,AAUUUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAU,AUUUUC,AUUUUU,CUUUUU,GUUUUU,UAUAUA,UAUAUU,UAUGUU,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUUU,UUAAGU,UUACUU,UUAUAU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGUAU,UUUAAG,UUUACU,UUUAUA,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 |
30 |
5140 |
0.073809524 |
0.0259018541 |
1.510752 |
AUUUUC,AUUUUU,CUCCUU,CUUUUC,CUUUUG,CUUUUU,GUUUUG,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUGU,UUUUUA,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 |
32 |
5597 |
0.078571429 |
0.0282043531 |
1.478087 |
AAAUUU,AAUUUU,AUUUUC,AUUUUU,CUUUUC,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB4 |
3 |
691 |
0.009523810 |
0.0034864974 |
1.449760 |
UUUUUU |
UUUUUU |
| DDX19B |
3 |
691 |
0.009523810 |
0.0034864974 |
1.449760 |
UUUUUU |
UUUUUU |
| EIF4A3 |
3 |
691 |
0.009523810 |
0.0034864974 |
1.449760 |
UUUUUU |
UUUUUU |
| HNRNPAB |
1 |
351 |
0.004761905 |
0.0017734784 |
1.424957 |
CAAAGA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| U2AF2 |
5 |
1071 |
0.014285714 |
0.0054010480 |
1.403262 |
UUUUCC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| HNRNPC |
10 |
2006 |
0.026190476 |
0.0101118501 |
1.372995 |
AUUUUU,CUUUUU,UUUUUA,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF4 |
2 |
558 |
0.007142857 |
0.0028164047 |
1.342647 |
AGGAAG,GAGGAA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| MBNL1 |
16 |
3258 |
0.040476190 |
0.0164197904 |
1.301638 |
CGCUUG,CUUGCU,GCUUGU,GCUUUU,GUGCUU,UCGCUU,UGCUGU,UGCUUC,UGCUUU,UUGCUG,UUGCUU,UUGUGC |
ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| MSI1 |
4 |
961 |
0.011904762 |
0.0048468360 |
1.296424 |
AGGAAG,AGGUAG,AGUUAG,UAGGUG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SRSF10 |
14 |
2937 |
0.035714286 |
0.0148024990 |
1.270661 |
AAAAGA,AAAGAG,AAGAAA,AAGAGA,AAGAGG,AGAGAA,AGAGAG,AGAGGA,CAAAGA,GAGAAA,GAGAGG,GAGGAA |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| PPIE |
46 |
9262 |
0.111904762 |
0.0466696896 |
1.261714 |
AAAAAU,AAAAUU,AAAUUU,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAUAU,AUAUUU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAUUA,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUUA,UAUUUU,UUAUAA,UUAUAU,UUAUUU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPCL1 |
6 |
1381 |
0.016666667 |
0.0069629182 |
1.259202 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| TARDBP |
4 |
1034 |
0.011904762 |
0.0052146312 |
1.190902 |
GUUGUU,GUUUUG,UUGUGC |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| KHDRBS2 |
4 |
1051 |
0.011904762 |
0.0053002821 |
1.167398 |
AUAAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ZCRB1 |
2 |
666 |
0.007142857 |
0.0033605401 |
1.087808 |
ACUUAA,AUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY |
4 |
1117 |
0.011904762 |
0.0056328094 |
1.079612 |
UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| KHSRP |
24 |
5764 |
0.059523810 |
0.0290457477 |
1.035140 |
AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,CUCCUU,GUAUGU,UAUAUU,UAUUUA,UAUUUU,UCCUUC,UGCUUC,UGUAUG,UUAUUU,UUGUAU,UUUAUA,UUUAUU |
ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| HNRNPA1 |
26 |
6340 |
0.064285714 |
0.0319478033 |
1.008781 |
AAAGAG,AAGAGG,AAUUUA,AGAGGA,AGGAAG,AGUUAG,AUAGAA,AUUUAA,AUUUAU,CAAAGA,GAGGAA,GUAGGG,UAGAGA,UAGGGU,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |