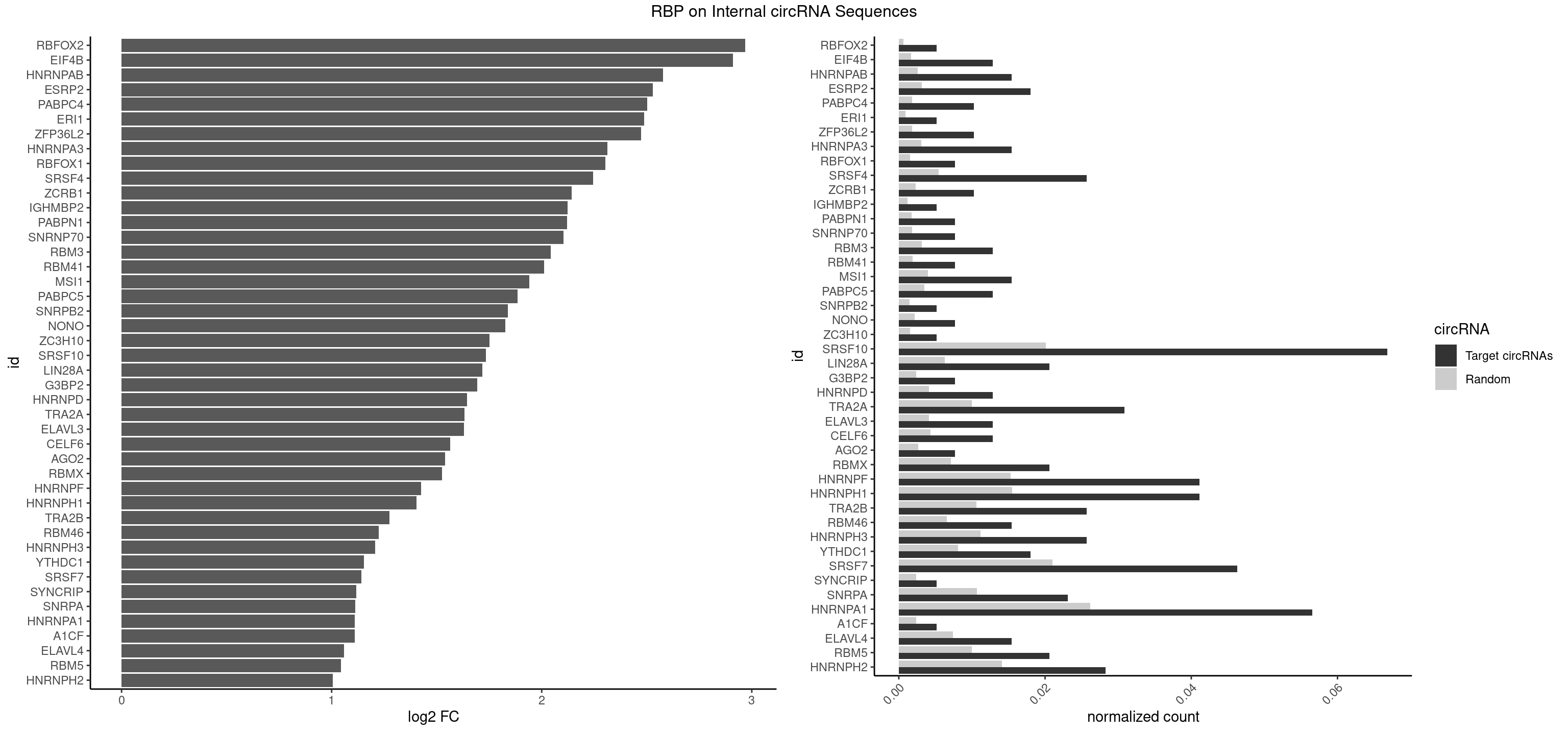
| RBFOX2 |
1 |
452 |
0.005141388 |
0.0006564894 |
2.969314 |
UGCAUG |
UGACUG,UGCAUG |
| EIF4B |
4 |
1179 |
0.012853470 |
0.0017100607 |
2.910039 |
CUUGGA,UUGGAC |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPAB |
5 |
1782 |
0.015424165 |
0.0025839306 |
2.577553 |
AAAGAC,ACAAAG,AGACAA,CAAAGA,GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ESRP2 |
6 |
2150 |
0.017994859 |
0.0031172377 |
2.529245 |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,UGGGAA,UGGGGA |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC4 |
3 |
1251 |
0.010282776 |
0.0018144033 |
2.502663 |
AAAAAA,AAAAAG |
AAAAAA,AAAAAG |
| ERI1 |
1 |
632 |
0.005141388 |
0.0009173461 |
2.486620 |
UUCAGA |
UUCAGA,UUUCAG |
| ZFP36L2 |
3 |
1277 |
0.010282776 |
0.0018520827 |
2.473009 |
AUUUAU,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPA3 |
5 |
2140 |
0.015424165 |
0.0031027457 |
2.313575 |
AAGGAG,CAAGGA,CCAAGG,GCCAAG |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBFOX1 |
2 |
1077 |
0.007712082 |
0.0015622419 |
2.303503 |
GCAUGU,UGCAUG |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRSF4 |
9 |
3740 |
0.025706941 |
0.0054214720 |
2.245401 |
AGAAGA,AGGAAG,GAGGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ZCRB1 |
3 |
1605 |
0.010282776 |
0.0023274215 |
2.143425 |
GAAUUA,GAUUUA,GGCUUA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGHMBP2 |
1 |
813 |
0.005141388 |
0.0011796520 |
2.123797 |
AAAAAA |
AAAAAA |
| PABPN1 |
2 |
1222 |
0.007712082 |
0.0017723764 |
2.121435 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| SNRNP70 |
2 |
1237 |
0.007712082 |
0.0017941145 |
2.103848 |
AAUCAA,GUUCAA |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM3 |
4 |
2152 |
0.012853470 |
0.0031201361 |
2.042477 |
AAGACU,GAAACU,GAGACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM41 |
2 |
1318 |
0.007712082 |
0.0019115000 |
2.012415 |
UACUUG,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MSI1 |
5 |
2770 |
0.015424165 |
0.0040157442 |
1.941453 |
AGGAAG,AGGAGG,AGGUGG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| PABPC5 |
4 |
2400 |
0.012853470 |
0.0034795387 |
1.885190 |
AGAAAG,AGAAAU,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SNRPB2 |
1 |
991 |
0.005141388 |
0.0014376103 |
1.838485 |
GUAUUG |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO |
2 |
1498 |
0.007712082 |
0.0021723567 |
1.827859 |
GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ZC3H10 |
1 |
1053 |
0.005141388 |
0.0015274610 |
1.751022 |
GAGCGA |
CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SRSF10 |
25 |
13860 |
0.066838046 |
0.0200874160 |
1.734378 |
AAAAGA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAGA,AAGGAG,AAGGGA,ACAAAG,AGACAA,AGAGAC,CAAAGA,GAAAGA,GACAAA,GAGAAG,GAGACA,GAGACC,GAGAGA,GAGGAA,GAGGAG |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| LIN28A |
7 |
4315 |
0.020565553 |
0.0062547643 |
1.717202 |
AGGAGA,GGAGAA,GGAGGA,UGGAGA,UGGAGG |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| G3BP2 |
2 |
1644 |
0.007712082 |
0.0023839405 |
1.693772 |
AGGAUG,GGAUGA |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPD |
4 |
2837 |
0.012853470 |
0.0041128408 |
1.643951 |
AAAAAA,AUUUAU,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| TRA2A |
11 |
6871 |
0.030848329 |
0.0099589296 |
1.631130 |
AAGAAA,AGAAAG,AGAAGA,AGGAAG,GAAAGA,GAAGAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ELAVL3 |
4 |
2867 |
0.012853470 |
0.0041563169 |
1.628780 |
AUUUAU,UAUUUU,UUAUUU,UUUAUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF6 |
4 |
2997 |
0.012853470 |
0.0043447134 |
1.564825 |
GUGAGG,GUGGGG,UGUGAG,UGUGGG |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| AGO2 |
2 |
1830 |
0.007712082 |
0.0026534924 |
1.539228 |
AAAAAA,AGUGCU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| RBMX |
7 |
4925 |
0.020565553 |
0.0071387787 |
1.526481 |
AGGAAG,AUCAAA,AUCCCA,UCAAAA |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPF |
15 |
10561 |
0.041131105 |
0.0153064921 |
1.426086 |
AAGGGA,AAGGUG,AGGAAG,AGGGAA,GAGGAA,GGAGGA,GGGAAG,GGGAAU,GGGGAA,UGGGAA,UGUGGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 |
15 |
10717 |
0.041131105 |
0.0155325680 |
1.404933 |
AAGGGA,AAGGUG,AGGAAG,AGGGAA,AUUGGG,GAGGAA,GGAGGA,GGGAAG,GGGAAU,GGGGAA,UGUGGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| TRA2B |
9 |
7329 |
0.025706941 |
0.0106226650 |
1.275012 |
AAGAAC,AGAAGA,AGGAAG,AUAAGA,GAAAGA,GAAUUA,UAAGAA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM46 |
5 |
4554 |
0.015424165 |
0.0066011240 |
1.224409 |
AAUCAA,AUCAAA,AUGAAA,AUGAUG,GAUGAU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPH3 |
9 |
7688 |
0.025706941 |
0.0111429292 |
1.206029 |
AAGGGA,AAGGUG,AGGGAA,AUUGGG,GGAGGA,GGGAAG,GGGAAU,UGUGGG |
AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| YTHDC1 |
6 |
5576 |
0.017994859 |
0.0080822104 |
1.154763 |
GAAUAC,GACUGC,GAGUGC,UAGUAC,UGAUAC,UGGUGC |
GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SRSF7 |
17 |
14481 |
0.046272494 |
0.0209873716 |
1.140633 |
AAAGGA,AGAAGA,AGAGAC,AGAUCU,AGGAAG,GACAAA,GAGACU,GAGAGA,GAGGAA,UCUUCA,UGAGAG |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SYNCRIP |
1 |
1634 |
0.005141388 |
0.0023694485 |
1.117607 |
AAAAAA |
AAAAAA,UUUUUU |
| SNRPA |
8 |
7380 |
0.023136247 |
0.0106965744 |
1.113006 |
AGCAGU,AGGAGA,AUACCU,GAUACC,GCAGUA,UACCUG,UAUGCU |
ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| HNRNPA1 |
21 |
18070 |
0.056555270 |
0.0261885646 |
1.110724 |
AAAAAA,AAAGAG,AAGGAG,AAGGUG,AGGAAG,AUUUAU,CAAAGA,CAAGGA,CCAAGG,GAGGAA,GAGGAG,GCCAAG,GGAGGA,UGGUGC,UUAUUU,UUUAUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| A1CF |
1 |
1642 |
0.005141388 |
0.0023810421 |
1.110565 |
CAGUAU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ELAVL4 |
5 |
5105 |
0.015424165 |
0.0073996354 |
1.059666 |
AAAAAA,AUUUAU,UAUUUU,UUAUUU,UUUAUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| RBM5 |
7 |
6879 |
0.020565553 |
0.0099705232 |
1.044489 |
AAAAAA,AAGGAG,AAGGUG,AGGGAA,CAAGGA,CAAGGG |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPH2 |
10 |
9711 |
0.028277635 |
0.0140746688 |
1.006560 |
AAGGGA,AAGGUG,AGGGAA,AUUGGG,GGAGGA,GGGAAG,GGGAAU,GGGGAA,UGUGGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |