circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000130695:+:1:26257596:26259802
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000130695:+:1:26257596:26259802 | ENSG00000130695 | MSTRG.607.1 | + | 1 | 26257597 | 26259802 | 438 | CUUCAGAAUGGAGCCAUCUGCCACCAUCCUGCUGCUUUUGGUCCUUCACUGCCCAUCUUAGAGCCAGCACAGUGGAUCAGCAUCUUGAACAGUAAUGAACACCUUCUGAAGGAAAAAGAGCUUCUCAUUGACAAGCAGAGGAAACACAUCUCUCAGCUGGAGCAGAAAGUGCGAGAGAGCGAACUGCAAGUCCACAGUGCCCUCUUGGGCCGCCCUGCCCCCUUUGGUGAUGUCUGCUUGCUGAGGCUACAGGAAUUGCAGCGAGAAAACACUUUCUUACGUGCACAGUUUGCACAGAAGACAGAAGCCUUGAGCAGAGAAAAGAUUGACCUUGAAAAGAAACUCUCUGCUUCUGAAGUUGAAGUCCAGCUCAUCAGAGAGUCGCUCAAAGUGGCGUUGCAGAAGCAUUCUGAGGAAGUGAAGAAACAGGAAGAAAGGCUUCAGAAUGGAGCCAUCUGCCACCAUCCUGCUGCUUUUGGUCCUUCACU | circ |
| ENSG00000130695:+:1:26257596:26259802 | ENSG00000130695 | MSTRG.607.1 | + | 1 | 26257597 | 26259802 | 22 | AGGAAGAAAGGCUUCAGAAUGG | bsj |
| ENSG00000130695:+:1:26257596:26259802 | ENSG00000130695 | MSTRG.607.1 | + | 1 | 26257397 | 26257606 | 210 | AGGGAGUGUUCAGUUGGCAGAAGCCUUCCUGGCAGGUUCUUUCCAGUCAUGCCUCAGAGCUAAAAUUAUUUCAUAAACAUUGGGCCUCAUCAGGCUGAGCCAAGUUGGGCUUUGACUACAGACCCAGCUGAUUACUGCUCAGGUGUUUUCAGGCGUGUAUGGGUCAAGAUCAAGCUCAUCUGGGCCUACUUGCCUUUCAGCUUCAGAAUG | ie_up |
| ENSG00000130695:+:1:26257596:26259802 | ENSG00000130695 | MSTRG.607.1 | + | 1 | 26259793 | 26260002 | 210 | GGAAGAAAGGGUGAGCUGAGUAGCUGAUAGCCCUAGUUCACAAAGGAGUUGGCAGUCAGCUGUCUACUCUAAAGCCUAAGCUGUUUCUGGCCUGGAACUAAGAGGAGACAAUUGGGUGAGAUCCUCCACUGGCUUCUCACACAGAAUCGUCACAGAUCUCCUUUUUGCUAGAAAUGGAACUCUCCUCCCUCCCUCAUUUUUGCUUCAUCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
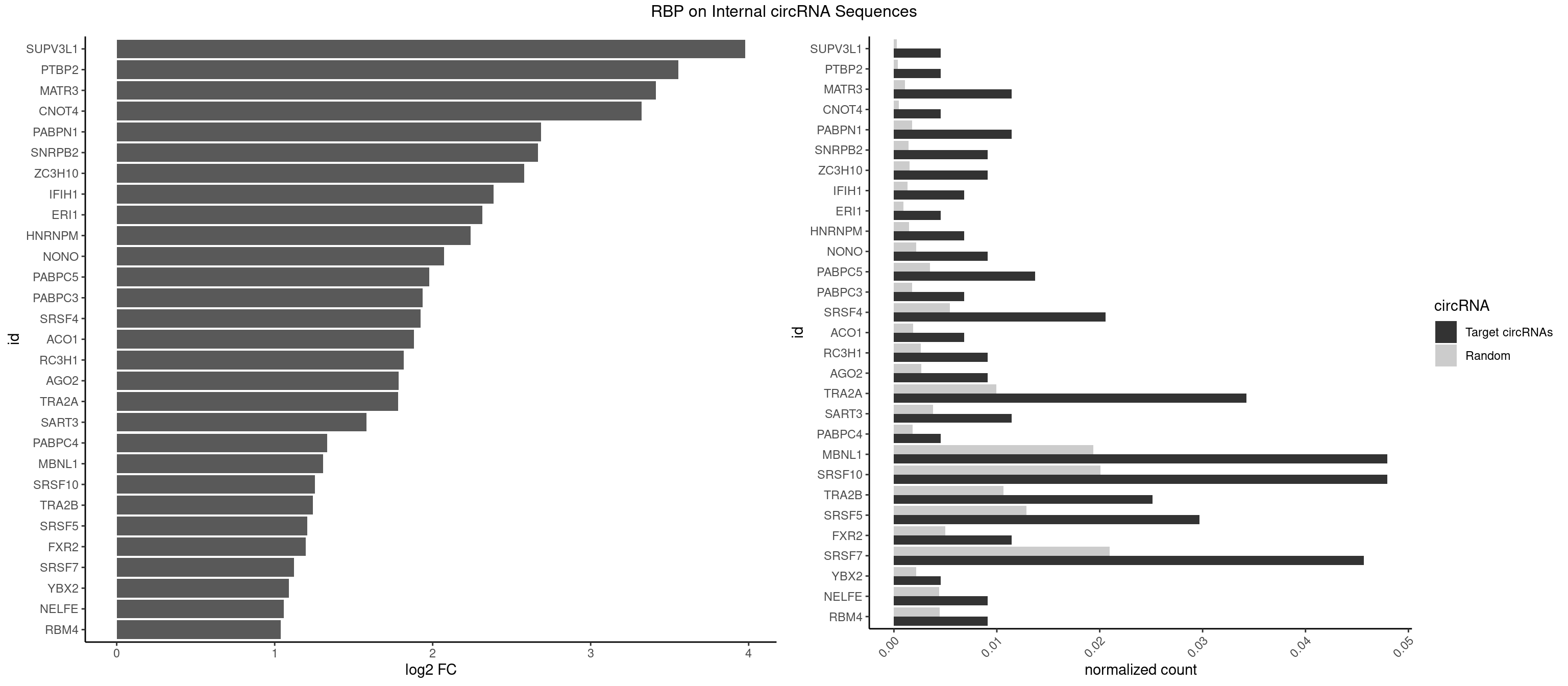
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 199 | 0.004566210 | 0.0002898408 | 3.977665 | CCGCCC | CCGCCC |
| PTBP2 | 1 | 267 | 0.004566210 | 0.0003883867 | 3.555432 | CUCUCU | CUCUCU |
| MATR3 | 4 | 739 | 0.011415525 | 0.0010724109 | 3.412067 | AUCUUA,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CNOT4 | 1 | 314 | 0.004566210 | 0.0004564992 | 3.322313 | GACAGA | GACAGA |
| PABPN1 | 4 | 1222 | 0.011415525 | 0.0017723764 | 2.687240 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| SNRPB2 | 3 | 991 | 0.009132420 | 0.0014376103 | 2.667325 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZC3H10 | 3 | 1053 | 0.009132420 | 0.0015274610 | 2.579862 | CAGCGA,GAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| IFIH1 | 2 | 904 | 0.006849315 | 0.0013115296 | 2.384709 | GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ERI1 | 1 | 632 | 0.004566210 | 0.0009173461 | 2.315459 | UUCAGA | UUCAGA,UUUCAG |
| HNRNPM | 2 | 999 | 0.006849315 | 0.0014492040 | 2.240699 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| NONO | 3 | 1498 | 0.009132420 | 0.0021723567 | 2.071736 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 | 5 | 2400 | 0.013698630 | 0.0034795387 | 1.977064 | AGAAAA,AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPC3 | 2 | 1234 | 0.006849315 | 0.0017897669 | 1.936188 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRSF4 | 8 | 3740 | 0.020547945 | 0.0054214720 | 1.922238 | AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ACO1 | 2 | 1283 | 0.006849315 | 0.0018607779 | 1.880054 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RC3H1 | 3 | 1786 | 0.009132420 | 0.0025897275 | 1.818197 | CCUUCU,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| AGO2 | 3 | 1830 | 0.009132420 | 0.0026534924 | 1.783105 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| TRA2A | 14 | 6871 | 0.034246575 | 0.0099589296 | 1.781897 | AAAGAA,AAGAAA,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SART3 | 4 | 2634 | 0.011415525 | 0.0038186524 | 1.579862 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC4 | 1 | 1251 | 0.004566210 | 0.0018144033 | 1.331502 | AAAAAG | AAAAAA,AAAAAG |
| MBNL1 | 20 | 13372 | 0.047945205 | 0.0193802045 | 1.306803 | CCUGCU,CUGCCA,CUGCCC,CUGCUG,CUGCUU,CUUGCU,GCUGCU,GCUUGC,GCUUUU,UCGCUC,UCUGCU,UGCUGC,UGCUUC,UGCUUU,UUGCUG | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| SRSF10 | 20 | 13860 | 0.047945205 | 0.0200874160 | 1.255095 | AAAAGA,AAAGAA,AAAGAG,AAGAAA,AAGACA,AAGGAA,AGAGAA,AGAGAG,AGAGGA,GAGAAA,GAGAGA,GAGAGC,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| TRA2B | 10 | 7329 | 0.025114155 | 0.0106226650 | 1.241355 | AAAGAA,AAGGAA,AGAAGA,AGGAAA,AGGAAG,GAAGAA,GAAGGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF5 | 12 | 8869 | 0.029680365 | 0.0128544391 | 1.207242 | AGAAGA,AGGAAG,CACAGA,CACAUC,GAAGAA,GAGGAA,GGAAGA,UGCAGA,UGCAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| FXR2 | 4 | 3434 | 0.011415525 | 0.0049780156 | 1.197355 | AGACAG,GACAAG,GACAGA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRSF7 | 19 | 14481 | 0.045662100 | 0.0209873716 | 1.121476 | AGAAGA,AGAGAA,AGAGAG,AGAGGA,AGGAAG,AUUGAC,CAGAGA,CGAGAG,CUUCAC,GAAGAA,GAGAGA,GAGGAA,GAUUGA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| YBX2 | 1 | 1480 | 0.004566210 | 0.0021462711 | 1.089165 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NELFE | 3 | 3028 | 0.009132420 | 0.0043896388 | 1.056895 | CUCUCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 3 | 3065 | 0.009132420 | 0.0044432593 | 1.039379 | CCUCUU,CCUUCU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
RBP on BSJ (Exon Only)
Plot
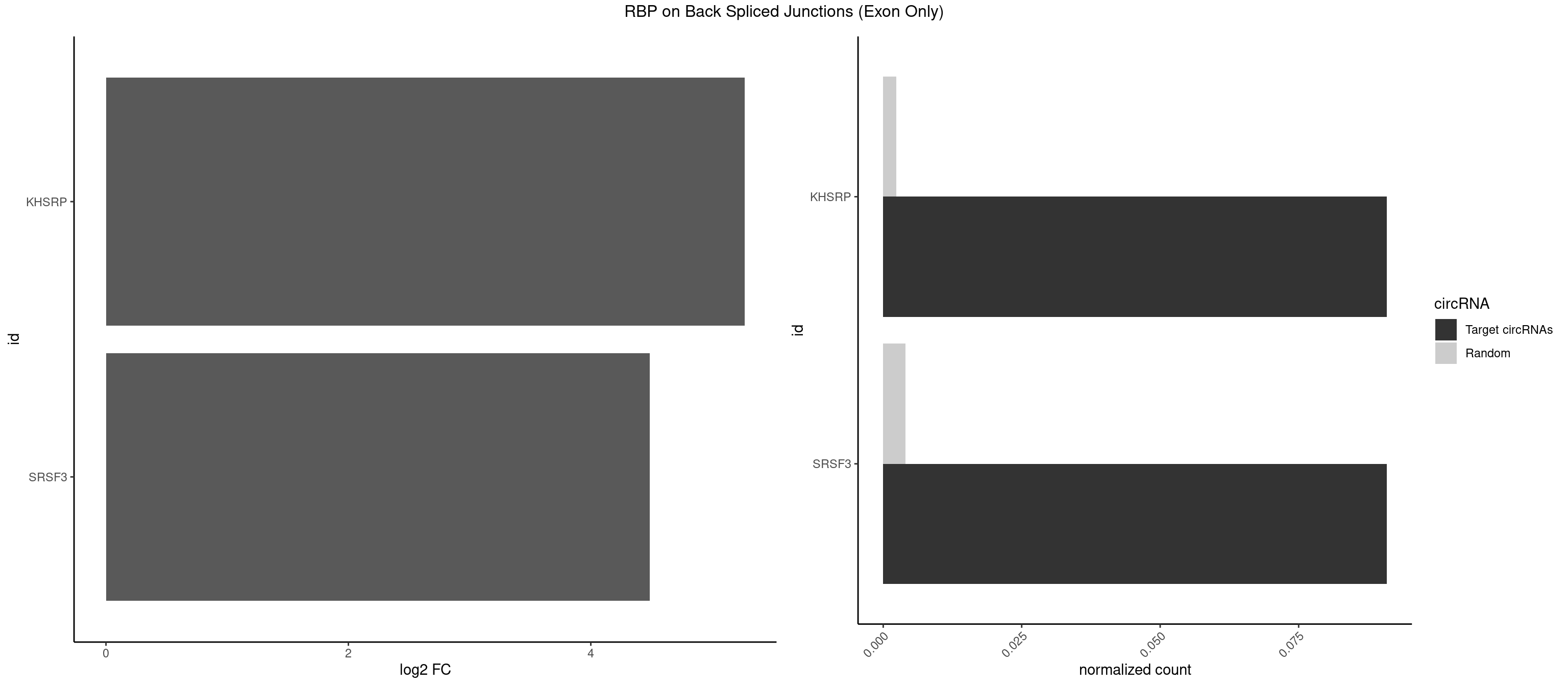
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| KHSRP | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | GGCUUC | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| SRSF3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | GCUUCA | AACGAU,ACCACC,ACUACG,AGAGAU,CACAAC,CACAUC,CACCAC,CAGAGA,CAUCAC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CUACAG,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUUAU,GACUUC,GAGAUU,UACAGC,UCAGAG,UCUCCA,UCUUCA,UCUUCC,UGUCAA,UUCGAC,UUCUCC,UUCUUC |
RBP on BSJ (Exon and Intron)
Plot
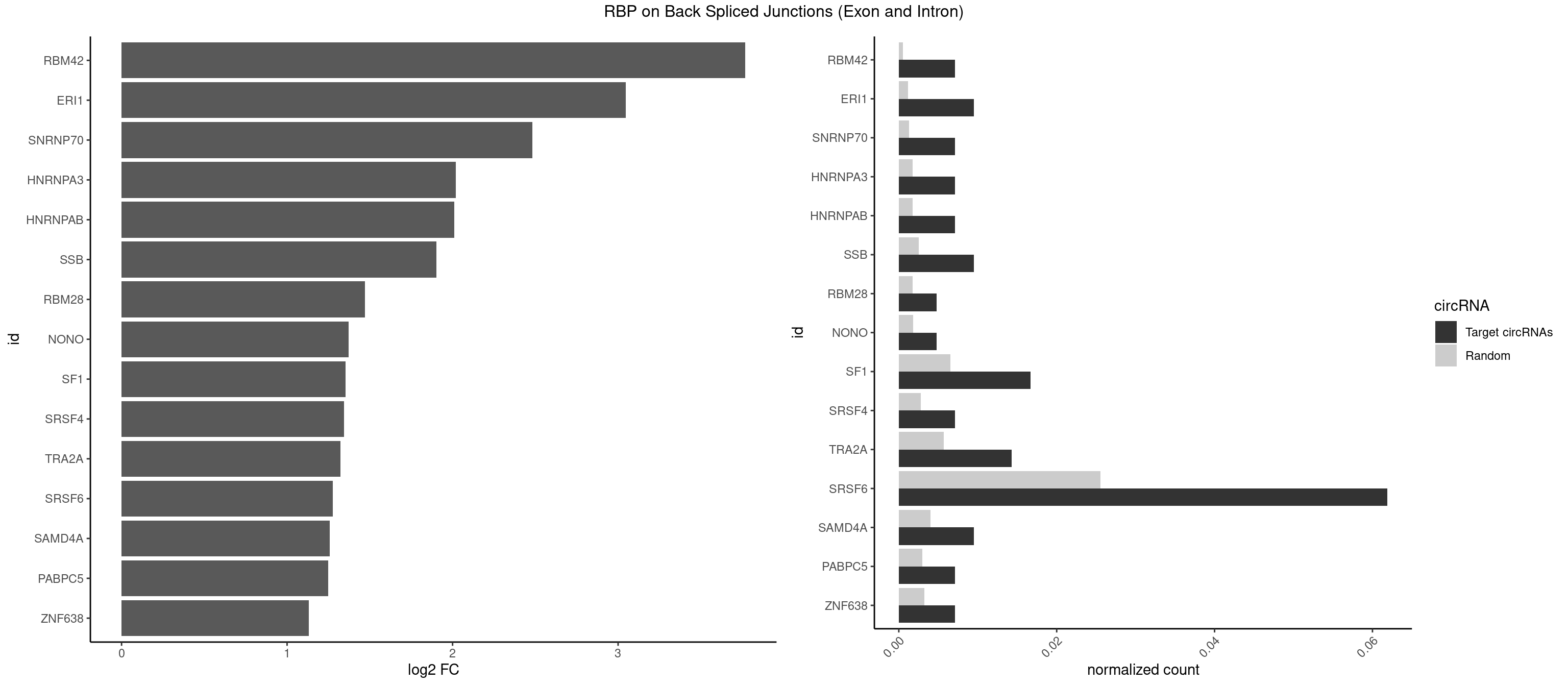
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | AACUAA,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| ERI1 | 3 | 228 | 0.009523810 | 0.0011537686 | 3.045185 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | ACAAAG,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACAGAC,ACUAAG,CACAGA,CAGUCA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AAGAAA,AAGAGG,AGAAAG,AGAGGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF6 | 25 | 5069 | 0.061904762 | 0.0255441354 | 1.277058 | CACUGG,CCACUG,CCUCAG,CCUGGC,CUACAG,CUACUC,CUCAGG,CUCAUC,CUUCAG,CUUCUC,GAAGAA,GAUCAA,GCUCAU,GGAAGA,UCACAC,UCACAG,UCCUGG,UCUCAC,UUACUG,UUCAGG,UUCUGG,UUUCAG,UUUCUG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAA,CUGGCA,CUGGCC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.